Compositions and methods for transient delivery of nucleases
Gao , et al.
U.S. patent number 10,584,321 [Application Number 15/550,452] was granted by the patent office on 2020-03-10 for compositions and methods for transient delivery of nucleases. This patent grant is currently assigned to University of Massachusetts. The grantee listed for this patent is University of Massachusetts. Invention is credited to Guangping Gao, Dan Wang, Phillip D. Zamore.







| United States Patent | 10,584,321 |
| Gao , et al. | March 10, 2020 |
Compositions and methods for transient delivery of nucleases
Abstract
The disclosure in some aspects relates to recombinant adeno-associated viruses having nuclease grafted to one or more capsid proteins. In some aspects, the disclosure relates to isolated AAV capsid proteins having terminally grafted nucleases and isolated nucleic acids encoding the same. Recent approaches to delivering nucleases to cells for gene editing have focused on delivering of expression vectors engineered to express the nucleases in target cells. However, these approaches have proved to be problematic in many instances due to genotoxicity resulting from to prolonged expression of gene editing system in vivo. To prevent such off-target genotoxicity due to prolonged presence of a gene editing system, several studies explored delivery of mRNA or protein instead of delivering the gene coding for the nucleases in cell culture.
| Inventors: | Gao; Guangping (Westborough, MA), Zamore; Phillip D. (Northborough, MA), Wang; Dan (Belchertown, MA) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Applicant: |
|
||||||||||
| Assignee: | University of Massachusetts
(Boston, MA) |
||||||||||
| Family ID: | 56614963 | ||||||||||
| Appl. No.: | 15/550,452 | ||||||||||
| Filed: | February 12, 2016 | ||||||||||
| PCT Filed: | February 12, 2016 | ||||||||||
| PCT No.: | PCT/US2016/017886 | ||||||||||
| 371(c)(1),(2),(4) Date: | August 11, 2017 | ||||||||||
| PCT Pub. No.: | WO2016/131009 | ||||||||||
| PCT Pub. Date: | August 18, 2016 |
Prior Publication Data
| Document Identifier | Publication Date | |
|---|---|---|
| US 20180037877 A1 | Feb 8, 2018 | |
| US 20180179501 A9 | Jun 28, 2018 | |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | Issue Date | ||
|---|---|---|---|---|---|
| 62115928 | Feb 13, 2015 | ||||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | A61K 48/005 (20130101); C12N 7/00 (20130101); C12N 9/22 (20130101); C12N 15/11 (20130101); C07K 14/005 (20130101); C12N 15/907 (20130101); C12N 2750/14142 (20130101); C12N 2310/20 (20170501); C12N 2750/14122 (20130101); C07K 2319/735 (20130101); C12N 2750/14121 (20130101); A61K 38/00 (20130101); C12N 2750/14143 (20130101) |
| Current International Class: | C12N 9/22 (20060101); C07K 14/005 (20060101); C12N 7/00 (20060101); C12N 15/11 (20060101); C12N 15/90 (20060101); A61K 48/00 (20060101); A61K 38/00 (20060101) |
References Cited [Referenced By]
U.S. Patent Documents
| 5478745 | December 1995 | Samulski et al. |
| 5871982 | February 1999 | Wilson et al. |
| 6001650 | December 1999 | Colosi |
| 6156303 | December 2000 | Russell et al. |
| 6177403 | January 2001 | Stedman |
| 6251677 | June 2001 | Wilson et al. |
| 6399575 | June 2002 | Smith et al. |
| 6485966 | November 2002 | Gao et al. |
| 6498244 | December 2002 | Patel et al. |
| 6544786 | April 2003 | Xiao et al. |
| 6953690 | October 2005 | Gao et al. |
| 6962815 | November 2005 | Bartlett |
| 7022519 | April 2006 | Gao et al. |
| 7198951 | April 2007 | Gao et al. |
| 7235393 | June 2007 | Gao et al. |
| 7238526 | July 2007 | Wilson et al. |
| 7282199 | October 2007 | Gao et al. |
| 7427396 | September 2008 | Arbetman et al. |
| 7456015 | November 2008 | Bohn et al. |
| 7906111 | March 2011 | Wilson et al. |
| 8222221 | July 2012 | Corey et al. |
| 8524446 | September 2013 | Gao et al. |
| 8734809 | May 2014 | Gao et al. |
| 9217155 | December 2015 | Gao et al. |
| 9249424 | February 2016 | Wolf et al. |
| 9701984 | July 2017 | Gao et al. |
| 10370432 | August 2019 | Esteves et al. |
| 2001/0016355 | August 2001 | Samulski et al. |
| 2002/0019050 | February 2002 | Gao et al. |
| 2002/0164783 | November 2002 | Feldhaus |
| 2002/0192823 | December 2002 | Bartlett |
| 2003/0040101 | February 2003 | Wilson et al. |
| 2003/0103939 | June 2003 | Engelhardt et al. |
| 2003/0110526 | June 2003 | Brown et al. |
| 2003/0119191 | June 2003 | Gao et al. |
| 2003/0138772 | July 2003 | Gao et al. |
| 2003/0207259 | November 2003 | Gao et al. |
| 2003/0228282 | December 2003 | Gao et al. |
| 2004/0101514 | May 2004 | Liu et al. |
| 2004/0219528 | November 2004 | Morris et al. |
| 2005/0014262 | January 2005 | Gao et al. |
| 2005/0032219 | February 2005 | Aubourg et al. |
| 2005/0037988 | February 2005 | Zamore et al. |
| 2005/0137153 | June 2005 | McSwiggen et al. |
| 2005/0197313 | September 2005 | Roelvink |
| 2005/0255086 | November 2005 | Davidson et al. |
| 2005/0255089 | November 2005 | Chiorini et al. |
| 2005/0287122 | December 2005 | Bartlett et al. |
| 2006/0018841 | January 2006 | Arbetman et al. |
| 2006/0063174 | March 2006 | Turner et al. |
| 2006/0093589 | May 2006 | Warrington et al. |
| 2006/0153826 | July 2006 | Arnould et al. |
| 2006/0189515 | August 2006 | Beliveau et al. |
| 2006/0189564 | August 2006 | Burright et al. |
| 2006/0228800 | October 2006 | Lin et al. |
| 2006/0292117 | December 2006 | Loiler et al. |
| 2007/0004042 | January 2007 | Gao et al. |
| 2007/0036760 | February 2007 | Wilson et al. |
| 2007/0243526 | October 2007 | Kay et al. |
| 2007/0253936 | November 2007 | Kay et al. |
| 2007/0292410 | December 2007 | Cashman et al. |
| 2008/0042783 | February 2008 | Vitek et al. |
| 2008/0075737 | March 2008 | Gao et al. |
| 2008/0075740 | March 2008 | Gao et al. |
| 2008/0292595 | November 2008 | Arbetman et al. |
| 2009/0042828 | February 2009 | Xu et al. |
| 2009/0111766 | April 2009 | Atkinson et al. |
| 2009/0149409 | June 2009 | Bohn et al. |
| 2009/0197338 | August 2009 | Vandenberghe et al. |
| 2009/0215879 | August 2009 | DiPrimio et al. |
| 2009/0239240 | September 2009 | Chu |
| 2010/0104561 | April 2010 | Zhong et al. |
| 2010/0186103 | July 2010 | Gao et al. |
| 2010/0227909 | September 2010 | Cleary et al. |
| 2010/0323001 | December 2010 | Pachuk |
| 2011/0171262 | July 2011 | Bakker et al. |
| 2011/0172293 | July 2011 | Fish et al. |
| 2011/0212520 | September 2011 | Davidson et al. |
| 2011/0258716 | October 2011 | Baltimore et al. |
| 2011/0306550 | December 2011 | Vitek et al. |
| 2012/0041048 | February 2012 | Weinberg et al. |
| 2012/0077870 | March 2012 | Blanks et al. |
| 2012/0137379 | May 2012 | Gao et al. |
| 2012/0270930 | October 2012 | Van Der Maarel et al. |
| 2012/0309050 | December 2012 | Kumon et al. |
| 2013/0030042 | January 2013 | Couto |
| 2013/0101558 | April 2013 | Gao et al. |
| 2013/0109742 | May 2013 | Hewitt et al. |
| 2013/0142861 | June 2013 | Tsou et al. |
| 2013/0195801 | August 2013 | Gao et al. |
| 2013/0281516 | October 2013 | Gao et al. |
| 2013/0310443 | November 2013 | Srivastave et al. |
| 2013/0323226 | December 2013 | Wilson et al. |
| 2014/0010861 | January 2014 | Bancel et al. |
| 2014/0142161 | May 2014 | Flotte et al. |
| 2014/0142288 | May 2014 | Davidson et al. |
| 2014/0147418 | May 2014 | Chiorini et al. |
| 2014/0179770 | June 2014 | Zhang |
| 2014/0201857 | July 2014 | Fahrenkrug et al. |
| 2014/0205599 | July 2014 | Willemsen et al. |
| 2014/0335054 | November 2014 | Gao et al. |
| 2015/0065560 | March 2015 | Bjorklund et al. |
| 2015/0258180 | September 2015 | Mahuran et al. |
| 2016/0000794 | January 2016 | Chiorini et al. |
| 2016/0017005 | January 2016 | Asokan et al. |
| 2016/0060624 | March 2016 | Davidson et al. |
| 2016/0076054 | March 2016 | Auricchio et al. |
| 2016/0153005 | June 2016 | Zhang et al. |
| 2016/0185832 | June 2016 | Drivas et al. |
| 2016/0194374 | July 2016 | Wijnholds et al. |
| 2016/0272976 | September 2016 | Kaspar |
| 2017/0029785 | February 2017 | Zhao et al. |
| 2017/0114340 | April 2017 | Mueller et al. |
| 2017/0165377 | June 2017 | Gao et al. |
| 2018/0265863 | September 2018 | Esteves et al. |
| 2019/0038773 | February 2019 | Esteves et al. |
| 2261242 | Dec 2010 | EP | |||
| 2453923 | May 2012 | EP | |||
| 2468891 | Jun 2012 | EP | |||
| 2008-538286 | Oct 2008 | JP | |||
| WO 2003/042397 | May 2003 | WO | |||
| WO 2003/093460 | Nov 2003 | WO | |||
| WO 2004/108922 | Dec 2004 | WO | |||
| WO 2005/033321 | Apr 2005 | WO | |||
| WO 2006/031267 | Mar 2006 | WO | |||
| WO 2006/066066 | Jun 2006 | WO | |||
| WO 2006/119432 | Nov 2006 | WO | |||
| WO 2007/000668 | Jan 2007 | WO | |||
| WO 2007/027775 | Mar 2007 | WO | |||
| WO 2007/127264 | Nov 2007 | WO | |||
| WO 2008/091703 | Jul 2008 | WO | |||
| WO 2008/125846 | Oct 2008 | WO | |||
| WO 2008/147839 | Dec 2008 | WO | |||
| WO 2008/150897 | Dec 2008 | WO | |||
| WO 2009/043936 | Apr 2009 | WO | |||
| WO 2009/109665 | Sep 2009 | WO | |||
| WO 2009/137006 | Nov 2009 | WO | |||
| WO 2009/146178 | Dec 2009 | WO | |||
| WO 2010/027446 | Mar 2010 | WO | |||
| WO 2010/034314 | Apr 2010 | WO | |||
| WO 2010/071454 | Jun 2010 | WO | |||
| WO 2010/099383 | Sep 2010 | WO | |||
| WO 2010/129021 | Nov 2010 | WO | |||
| WO 2010/138263 | Dec 2010 | WO | |||
| WO 2011/008823 | Jan 2011 | WO | |||
| WO 2011/094198 | Aug 2011 | WO | |||
| WO 2012/123430 | Sep 2012 | WO | |||
| WO 2013/055865 | Apr 2013 | WO | |||
| WO 2013/123503 | Aug 2013 | WO | |||
| WO 2013/170078 | Nov 2013 | WO | |||
| WO 2013/190059 | Dec 2013 | WO | |||
| WO 2014/160092 | Oct 2014 | WO | |||
| WO 2014/186746 | Nov 2014 | WO | |||
| WO 2014/197748 | Nov 2014 | WO | |||
| WO 2014/197748 | Dec 2014 | WO | |||
| WO 2015/121501 | Aug 2015 | WO | |||
| WO 2015/164786 | Oct 2015 | WO | |||
| WO 2015/168666 | Nov 2015 | WO | |||
| WO 2016/054554 | Apr 2016 | WO | |||
| WO 2016/065001 | Apr 2016 | WO | |||
| WO 2017/023724 | Feb 2017 | WO | |||
| WO 2017/136536 | Aug 2017 | WO | |||
| WO 2018/226785 | Dec 2018 | WO | |||
Other References
|
Buning, Gene Therapy, 10: 1142-1151, 2003. cited by examiner . Encyclopaedia Britannica, "nuclease" retrived from https://www.britannica.com/science/nuclease on Jul. 7, 2019. cited by examiner . Schaefer et al., Nat Methods, 14(6): 547-548, May 30, 2017. cited by examiner . Gaj et al., Trends Biotechnol., 31(7): 397-405, 2013. cited by examiner . Adachi et al., Drawing a high-resolution functional map of adeno-associated virus capsid by massively parallel sequencing. Nat Commun. 2014;5:3075. doi: 10.1038/ncomms4075. cited by applicant . Afione et al., In vivo model of adeno-associated virus vector persistence and rescue. J Virol. May 1996;70(5):3235-41. cited by applicant . Ahmed et al., A Single Intravenous rAAV Injection as Late as P20 Achieves Efficacious and Sustained CNS Gene Therapy in Canavan Mice. Mol Ther. Jul. 2, 2013. doi: 10.1038/mt.2013.138. [Epub ahead of print]. cited by applicant . Akache et al., The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J Virol. Oct. 2006;80(19):9831-6. cited by applicant . Arbetman et al., Novel caprine adeno-associated virus (AAV) capsid (AAV-Go.1) is closely related to the primate AAV-5 and has unique tropism and neutralization properties. J Virol. Dec. 2005;79(24):15238-45. cited by applicant . Arbuthnot et al., Hepatic delivery of RNA interference activators for therapeutic application. Curr Gene Ther. Apr. 2009;9(2):91-103. cited by applicant . Asokan et al., The AAV vector toolkit: poised at the clinical crossroads. Mol Ther. Apr. 2012;20(4):699-708. doi: 10.1038/mt.2011.287. Epub Jan. 24, 2012. cited by applicant . Baek et al., AAV-mediated gene delivery in adult GM1-gangliosidosis mice corrects lysosomal storage in CNS and improves survival. PLoS One. Oct. 18, 2010;5(10):e13468. doi: 10.1371/journal.pone.0013468. cited by applicant . Bals et al., Transduction of well-differentiated airway epithelium by recombinant adeno-associated virus is limited by vector entry. J Virol. Jul. 1999;73(7):6085-8. cited by applicant . Berns et al., Biology of adeno-associated virus. Curr Top Microbiol Immunol. 1996;218:1-23. cited by applicant . Berns et al., Detection of adeno-associated virus (AAV)-specific nucleotide sequences in DNA isolated from latently infected Detroit 6 cells. Virology. Dec. 1975;68(2):556-60. cited by applicant . Beutler et al., AAV for pain: steps towards clinical translation. Gene Ther. Apr. 2009;16(4):461-9. Epub Mar. 5, 2009. cited by applicant . Bish et al., Adeno-associated virus (AAV) serotype 9 provides global cardiac gene transfer superior to AAV1, AAV6, AAV7, and AAV8 in the mouse and rat. Hum Gene Ther. Dec. 2008;19(12):1359-68. doi: 10.1089/hum.2008.123. cited by applicant . Borel et al., Recombinant AAV as a platform for translating the therapeutic potential of RNA interference. Mol Ther. Apr. 2014;22(4):692-701. doi:10.1038/mt.2013.285. Epub Dec. 19, 2013. cited by applicant . Bourdenx et al., Systemic gene delivery to the central nervous system using Adeno-associated virus. Front Mol Neurosci. Jun. 2, 2014;7:50. doi: 10.3389/fnmol.2014.00050.eCollection 2014. cited by applicant . Brantly et al., Phase I trial of intramuscular injection of a recombinant adeno-associated virus serotype 2 alphal-antitrypsin (AAT) vector in AAT-deficient adults. Hum Gene Ther. Dec. 2006;17(12):1177-86. cited by applicant . Buning et al., Receptor targeting of adeno-associated virus vectors. Gene Ther. Jul. 2003;10(14):1142-51. cited by applicant . Calcedo et al., Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J Infect Dis. Feb. 1, 2009;199(3):381-90. cited by applicant . Carter et al., Adeno-associated virus gene expression and regulation. CRC Handbook of parvoviruses. 1990:227-54. cited by applicant . Carter, in "Handbook of Parvoviruses", ed., P. Tijsser, CRC Press, pp. 155-168 (1990). cited by applicant . Cearley et al., Expanded repertoire of AAV vector serotypes mediate unique patterns of transduction in mouse brain. Mol Ther. Oct. 2008;16(10):1710-8. doi: 10.1038/mt.2008.166. Epub Aug. 19, 2008. cited by applicant . Cearley et al., Transduction characteristics of adeno-associated virus vectors expressing cap serotypes 7, 8, 9, and Rh10 in the mouse brain. Mol Ther. Mar. 2006;13(3):528-37. Epub Jan. 18, 2006. cited by applicant . Chadderton et al., Improved retinal function in a mouse model of dominant retinitis pigmentosa following AAV-delivered gene therapy. Mol Ther. Apr. 2009;17(4):593-9. Epub Jan. 27, 2009. cited by applicant . Chen et al., Comparative study of anti-hepatitis B virus RNA interference by double-stranded adeno-associated virus serotypes 7, 8, and 9. Mol Ther. Feb. 2009;17(2):352-9. Epub Dec. 9, 2008. cited by applicant . Chen et al., Efficient transduction of vascular endothelial cells with recombinant adeno-associated virus serotype 1 and 5 vectors. Hum Gene Ther. Feb. 2005;16(2):235-47. cited by applicant . Chen et al., Molecular signatures of disease brain endothelia provide new sites for CNS-directed enzyme therapy. Nat Med. Oct. 2009;15(10):1215-8. doi: 10.1038/nm.2025. Epub Sep. 13, 2009. cited by applicant . Cheng et al., Development of optimized AAV3 serotype vectors: mechanism of high-efficiency transduction of human liver cancer cells. Gene Ther. Apr. 2012;19(4):375-84. doi: 10.1038/gt.2011.105. Epub Jul. 21, 2011. cited by applicant . Chiorini et al., Cloning and characterization of adeno-associated virus type 5. J Virol. Feb. 1999;73(2):1309-19. cited by applicant . Chirmule et al., Humoral immunity to adeno-associated virus type 2 vectors following administration to murine and nonhuman primate muscle. J Virol. Mar. 2000;74(5):2420-5. cited by applicant . Choi et al., Effects of adeno-associated virus DNA hairpin structure on recombination. J Virol. Jun. 2005;79(11):6801-7. cited by applicant . Choudhury et al., Identification of Novel vectors capable of CNS transduction in adult mice after single round selection using DNA shuffled AAV capsid library. Mol Ther. May 1, 2013;21(1):S1/. cited by applicant . Cideciyan et al., Human RPE65 gene therapy for Leber congenital amaurosis: persistence of early visual improvements and safety at 1 year. Hum Gene Ther. Sep. 2009;20(9):999-1004. cited by applicant . Conlon et al., Efficient hepatic delivery and expression from a recombinant adeno-associated virus 8 pseudotyped alpha1-antitrypsin vector. Mol Ther. Nov. 2005;12(5):867-75. Epub Aug. 8, 2005. cited by applicant . Conlon et al., Ribozyme Approaches towards Down-Regulation of Pi*Z Mutant Human a-1 Anti-Trypsin. Mol. Therapy. 2004;9:S333. Abstract 875. cited by applicant . Conrad et al., Safety of single-dose administration of an adeno-associated virus (AAV)-CFTR vector in the primate lung. Gene Ther. Aug. 1996;3(8):658-68. cited by applicant . Cruz et al., In vivo post-transcriptional gene silencing of alpha-1 antitrypsin by adeno-associated virus vectors expressing siRNA. Lab Invest. Sep. 2007;87(9):893-902. Epub Jun. 25, 2007. cited by applicant . Cruz et al., The promise of gene therapy for the treatment of alpha-1 antitrypsin deficiency. Pharmacogenomics. Sep. 2007;8(9):1191-8. cited by applicant . Davidoff et al., Sex significantly influences transduction of murine liver by recombinant adeno-associated viral vectors through an androgen-dependent pathway. Blood. Jul. 15, 2003;102(2):480-8. Epub Mar. 13, 2003. cited by applicant . Davidson et al., Recombinant adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous system. Proc Natl Acad Sci U S A. Mar. 28, 2000;97(7):3428-32. cited by applicant . Daya et al., Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev. Oct. 2008;21(4):583-93. doi: 10.1128/CMR.00008-08. cited by applicant . Duque et al., Intravenous administration of self-complementary AAV9 enables transgene delivery to adult motor neurons. Mol Ther. Jul. 2009;17(7):1187-96. doi: 10.1038/mt.2009.71. Epub Apr. 14, 2009. cited by applicant . Ehlert et al., Cellular toxicity following application of adeno-associated viral vector-mediated RNA interference in the nervous system. BMC Neurosci. Feb. 18, 2010;11:20. cited by applicant . Fechner et al., Cardiac-targeted RNA interference mediated by an AAV9 vector improves cardiac function in coxsackievirus B3 cardiomyopathy. J Mol Med (Berl). Sep. 2008;86(9):987-97. doi: 10.1007/s00109-008-0363-x. Epub Jun. 12, 2008. cited by applicant . Feigin et al., Modulation of metabolic brain networks after subthalamic gene therapy for Parkinson's disease. Proc Natl Acad Sci U S A. Dec. 4, 2007;104(49):19559-64. Epub Nov. 27, 2007. cited by applicant . Fischer et al., Successful transgene expression with serial doses of aerosolized rAAV2 vectors in rhesus macaques. Mol Ther. Dec. 2003;8(6):918-26. cited by applicant . Fisher et al., Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J Virol. Jan. 1996;70(1):520-32. cited by applicant . Flotte et al., Gene therapy for alpha-1 antitrypsin deficiency. Hum Mol Genet. Apr. 15, 2011;20(R1):R87-92. doi: 10.1093/hmg/ddr156. Epub Apr. 16, 2011. cited by applicant . Flotte et al., Phase I trial of intramuscular injection of a recombinant adeno-associated virus alpha 1-antitrypsin (rAAV2-CB-hAAT) gene vector to AAT-deficient adults. Hum Gene Ther. Jan. 2004;15(1):93-128. cited by applicant . Flotte, Recombinant adeno-associated virus (AAV) gene therapy vectors for, cystic fibrosis (CF), alpha-1-antitrypsin deficiency (AAT) and fatty oxidation disorders (FAO). Umass Medical School. Interdisciplinary Graduate Program. Last accessed at http://www.umassmed.edu/igp/faculty/flotte.cfm?start=0& on Aug. 27, 2009. cited by applicant . Foust et al., Intravascular AAV9 preferentially targets neonatal-neurons and adult-astrocytes. Nature Biotechnology, 27; 59-65 2009. cited by applicant . Foust et al., Over the barrier and through the blood: to CNS delivery we go. Cell Cycle. Dec. 15, 2009;8(24):4017-8. cited by applicant . Fraldi et al., Functional correction of CNS lesions in an MPS-IIIA mouse model by intracerebral AAV-mediated delivery of sulfamidase and SUMF1 genes. Hum Mol Genet. Nov. 15, 2007;16(22):2693-702. Epub Aug. 27, 2007. cited by applicant . Fu et al., Self-complementary adeno-associated virus serotype 2 vector: global distribution and broad dispersion of AAV-mediated transgene expression in mouse brain. Mol Ther. Dec. 2003;8(6):911-7. cited by applicant . Gadalla et al., Improved survival and reduced phenotypic severity following AAV9/MECP2 gene transfer to neonatal and juvenile male Mecp2 knockout mice. Mol Ther. Jan. 2013;21(1):18-30. doi:10.1038/mt.2012.200. Epub Sep. 25, 2012. cited by applicant . Gao et al., Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc Natl Acad Sci U S A. May 13, 2003;100(10):6081-6. Epub Apr. 25, 2003. cited by applicant . Gao et al., Adeno-associated virus-mediated gene transfer to nonhuman primate liver can elicit destructive transgene-specific T cell responses. Hum Gene Ther. Sep. 2009;20(9):930-42. doi: 10.1089/hum.2009.060. cited by applicant . Gao et al., Biology of AAV serotype vectors in liver-directed gene transfer to nonhuman primates. Mol Ther. Jan. 2006;13(1):77-87. Epub Oct. 10, 2005. cited by applicant . Gao et al., Clades of Adeno-associated viruses are widely disseminated in human tissues. J Virol. Jun. 2004;78(12):6381-8. cited by applicant . Gao et al., Erythropoietin gene therapy leads to autoimmune anemia in macaques. Blood. May 1, 2004;103(9):3300-2. Epub Dec. 24, 2003. cited by applicant . Gao et al., Inadvertent gene transfer of co-packaged rep and cap sequences during the production of AAV vector and its potential impact on vector performance. Molecular Therapy. May 2008;16(Suppl. 1):S105-S106. Abstract 279. cited by applicant . Gao et al., New recombinant serotypes of AAV vectors. Curr Gene Ther. Jun. 2005;5(3):285-97. cited by applicant . Gao et al., Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci U S A. Sep. 3, 2002;99(18):11854-9. Epub Aug. 21, 2002. cited by applicant . Gao et al., Purification of recombinant adeno-associated virus vectors by column chromatography and its performance in vivo. Hum Gene Ther. Oct. 10, 2000;11(15):2079-91. cited by applicant . Gao et al., RAAV-mediated targeting in adult mice and its potential in generating animal models of tissue-specific somatic transgenics or knock down. Molecular Therapy. May 2008;16(1):S118-S119. Abstract 316. cited by applicant . Genbank Submission; Accession No. ADZ26851; Wilson et al.; Jun. 30, 2005. cited by applicant . Genbank Submission; Accession No. AF028705.1; Rutledge et al.; Jan. 12, 1998. cited by applicant . Genbank Submission; NCBI Accession No. AAB95450; Rutledge et al.; Jan. 12, 1998. cited by applicant . Genbank Submission; NCBI, Accession No. AAS99264; Gao et al.; Jun. 24, 2004. cited by applicant . Genbank Submission; NCBI, Accession No. ABA71701; Schmidt et al.; May 10, 2006. cited by applicant . Genbank Submission; NCBI, Accession No. ACB55301; Vandenberghe et al.; Jul. 31, 2008. cited by applicant . Genbank Submission; NCBI, Accession No. ACB55310; Vandenberghe et al.; Jul. 31, 2008. cited by applicant . Genbank Submission; NCBI, Accession No. AY530579.10; 2004. cited by applicant . Genbank Submission; NCBI, Accession No. NP_049542; Xiao et al.; Mar. 11, 2010. cited by applicant . Genbank Submission; NCBI, Accession No. YP_680426; Ruffing et al.; Nov. 19, 2010. cited by applicant . Grimm et al., Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. May 25, 2006;441(7092):537-41. cited by applicant . Grimm, Small silencing RNAs: state-of-the-art. Adv Drug Deliv Rev. Jul. 25, 2009;61(9):672-703. doi: 10.1016/j.addr.2009.05.002. Epub May 7, 2009. cited by applicant . Hauswirth et al., Treatment of leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: short-term results of a phase I trial. Hum Gene Ther. Oct. 2008;19(10):979-90. cited by applicant . Hernandez et al., Latent adeno-associated virus infection elicits humoral but not cell-mediated immune responses in a nonhuman primate model. J Virol. Oct. 1999;73(10):8549-58. cited by applicant . Hildinger et al., Hybrid vectors based on adeno-associated virus serotypes 2 and 5 for muscle-directed gene transfer. J Virol. Jul. 2001;75(13):6199-203. cited by applicant . Iida et al., Systemic Delivery of Tyrosine-Mutant AAV Vectors Results in Robust Transduction of Neurons in Adult Mice. BioMed Res Int. 2013;2013. cited by applicant . Iwamoto et al., Global diffuse distribution in the brain and efficient gene delivery to the dorsal root ganglia by intrathecal injection of adeno-associated viral vector serotype 1. J Gene Med. Jun. 2009;11(6):498-505. doi: 10.1002/jgm.1325. cited by applicant . Jakobsson et al., Lentiviral vectors for use in the central nervous system. Mol Ther. Mar. 2006;13(3):484-93. Epub Jan. 3, 2006. cited by applicant . Janson et al., Clinical protocol. Gene therapy of Canavan disease: AAV-2 vector for neurosurgical delivery of aspartoacylase gene (ASPA) to the human brain. Hum Gene Ther. Jul. 20, 2002;13(11):1391-412. cited by applicant . Koornneef et al., Apolipoprotein B knockdown by AAV-delivered shRNA lowers plasma cholesterol in mice. Mol Ther. Apr. 2011;19(4):731-40. doi:10.1038/mt.2011.6. Epub Feb. 8, 2011. cited by applicant . Kota et al., AAV8-Mediated Delivery of miR-26a inhibits cancer cell proliferation and induces tumor-specific apoptosis in a liver cancer model. Mol. Therapy. May 2009. 17(1):S300. Abstract 783. cited by applicant . Kotin et al., Organization of adeno-associated virus DNA in latently infected Detroit 6 cells. Virology. Jun. 1989;170(2):460-7. cited by applicant . Kotin et al., Site-specific integration by adeno-associated virus. Proc Natl Acad Sci U S A. Mar. 1990;87(6):2211-5. cited by applicant . Lawlor et al., Efficient gene delivery and selective transduction of glial cells in the mammalian brain by AAV serotypes isolated from nonhuman primates. Mol Ther. Oct. 2009;17(10):1692-702. doi: 10.1038/mt.2009.170. cited by applicant . Lebherz et al., Gene therapy with novel adeno-associated virus vectors substantially diminishes atherosclerosis in a murine model of familial hypercholesterolemia. J Gene Med. Jun. 2004;6(6):663-72. cited by applicant . Leone et al., Aspartoacylase gene transfer to the mammalian central nervous system with therapeutic implications for Canavan disease. Ann Neurol. Jul. 2000;48(1):27-38. Erratum in: Ann Neurol Sep. 2000;48(3):398. Bilianuk L [corrected to Bilaniuk L]. cited by applicant . Li et al., Efficient and Targeted Transduction of Nonhuman Primate Liver With Systemically Delivered Optimized AAV3B Vectors. Mol Ther. Dec. 2015;23(12):1867-76. doi: 10.1038/mt.2015.174. Epub Sep. 25, 2015. cited by applicant . Li et al., Ex vivo transduction and transplantation of bone marrow cells for liver gene delivery of alpha1-antitrypsin. Mol Ther. Aug. 2010;18(8):1553-8. Epub Jun. 15, 2010. cited by applicant . Li et al., Protein trans-splicing as a means for viral vector-mediated in vivo gene therapy. Hum Gene Ther. Sep. 2008;19(9):958-64. doi: 10.1089/hum.2008.009. cited by applicant . Lin et al., Impact of preexisting vector immunity on the efficacy of adeno-associated virus-based HIV-1 Gag vaccines. Hum Gene Ther. Jul. 2008;19(7):663-9. cited by applicant . Liu et al., Biological Differences in rAAV Transduction of Airway Epithelia in Humans and in Old World Non-human Primates. Mol Ther. Dec. 2007;15(12):2114-23. Epub Jul. 31, 2007. cited by applicant . Liu et al., Comparative biology of rAAV transduction in ferret, pig and human airway epithelia. Gene Ther. Nov. 2007;14(21):1543-8. Epub Aug. 30, 2007. cited by applicant . Liu et al., Species-specific differences in mouse and human airway epithelial biology of recombinant adeno-associated virus transduction. Am J Respir Cell Mol Biol. Jan. 2006;34(1):56-64. Epub Sep. 29, 2005. cited by applicant . Loiler et al., Targeting recombinant adeno-associated virus vectors to enhance gene transfer to pancreatic islets and liver. Gene Ther. Sep. 2003;10(18):1551-8. cited by applicant . Lowenstein, Crossing the rubicon. Nat Biotechnol. Jan. 2009;27(1):42-4. cited by applicant . Lux et al., Green fluorescent protein-tagged adeno-associated virus particles allow the study of cytosolic and nuclear trafficking. J Virol. Sep. 2005;79(18):11776-87. cited by applicant . Ma et al., Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. Apr. 2010;28(4):341-7. doi: 10.1038/nbt.1618. Epub Mar. 28, 2010. cited by applicant . Maguire et al., Directed evolution of adeno-associated virus for glioma cell transduction. J Neurooncol. Feb. 2010;96(3):337-47. doi:10.1007/s11060-009-9972-7. Epub Jul. 19, 2009. cited by applicant . Maguire et al., Gene therapy for the nervous system: challenges and new strategies. Neurotherapeutics. Oct. 2014;11(4):817-39. doi: 10.1007/s13311-014-0299-5. cited by applicant . Manfredsson et al., AAV9: a potential blood-brain barrier buster. Mol Ther. Mar. 2009;17(3):403-5. cited by applicant . Manno et al., Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med. Mar. 2006;12(3):342-7. Epub Feb. 12, 2006. Erratum in: Nat Med. May 2006;12(5):592. Rasko, John [corrected to Rasko, John Je]; Rustagi, Pradip K [added]. cited by applicant . Matalon et al., Adeno-associated virus-mediated aspartoacylase gene transfer to the brain of knockout mouse for canavan disease. Mol Ther. May 2003;7(5 Pt 1):580-7. cited by applicant . McCarty et al., Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. Dec. 2003;10(26):2112-8. cited by applicant . McCarty et al., Integration of adeno-associated virus (AAV) and recombinant AAV vectors. Annu Rev Genet. 2004;38:819-45. cited by applicant . McCarty et al., Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. Aug. 2001;8(16):1248-54. cited by applicant . McCarty, Self-complementary AAV vectors; advances and applications. Mol Ther. Oct. 2008;16(10):1648-56. Epub Aug. 5, 2008. cited by applicant . McCurdy et al., Sustained normalization of neurological disease after intracranial gene therapy in a feline model. Sci Transl Med. Apr. 9, 2014;6(231):231ra48. doi: 10.1126/scitranslmed.3007733. cited by applicant . Meijer et al., Controlling brain tumor growth by intraventricular administration of an AAV vector encoding IFN-beta. Cancer Gene Ther. Aug. 2009;16(8):664-71. doi: 10.1038/cgt.2009.8. Epub Feb. 6, 2009. cited by applicant . Mietzsch et al., OneBac 2.0: Sf9 Cell Lines for Production of AAV5 Vectors with Enhanced Infectivity and Minimal Encapsidation of Foreign DNA. Hum Gene Ther. Oct. 2015;26(10):688-97. doi:10.1089/hum.2015.050. Epub Aug. 6, 2015. cited by applicant . Mingozzi et al., CD8(+) T-cell responses to adeno-associated virus capsid in humans. Nat Med. Apr. 2007;13(4):419-22. Epub Mar. 18, 2007. cited by applicant . Moss et al., Repeated adeno-associated virus serotype 2 aerosol-mediated cystic fibrosis transmembrane regulator gene transfer to the lungs of patients with cystic fibrosis: a multicenter, double-blind, placebo-controlled trial. Chest. Feb. 2004;125(2):509-21. cited by applicant . Mueller et al., Development of Simultaneous Gene Augmentation and Reduction of Mutant Gene Expression with a Single Recombinant AAV for Alpha-1 Antitrypsin Disease. Molecular Therapy May 2009;17(1):S391-S392. Abstract 1030. cited by applicant . Mueller et al., In Vivo AAV Delivered Allele Specific shRNA for the Knockdown of Alpha-1 Antitrypsin. Molecular Therapy May 2010;18(1):S22. Abstract 53. cited by applicant . Mueller et al., In Vivo Allele Specific Knockdown of Mutant Alpha-1 Antitrypsin Using Recombinant AAV Delivered shRNA. Molecular Therapy May 2009;17(1):S313. Abstract 817. cited by applicant . Mueller et al., Sustained miRNA-mediated knockdown of mutant AAT with simultaneous augmentation of wild-type AAT has minimal effect on global liver miRNA profiles. Mol Ther. Mar. 2012;20(3):590-600. Epub Jan. 17, 2012. cited by applicant . Mueller et al., The pros and cons of immunomodulatory IL-10 gene therapy with recombinant AAV in a Cftr-/- -dependent allergy mouse model. Gene Ther. Feb. 2009;16(2):172-83. Epub Sep. 25, 2008. cited by applicant . Mueller et al., Using rAAV Delivered miRNAs to Knockdown Misfolded Human Alpha 1 Antitrypsin in a Transgenic Mouse Model. Molecular Therapy May 2010;18(1):S21. Abstract 51. cited by applicant . NCBI Blast Protein Sequence. RID-09JSKF33114. Alignment of Seq ID Nos. 87, 179. 2016. cited by applicant . O'Reilly et al., RNA interference-mediated suppression and replacement of human rhodopsin in vivo. Am J Hum Genet. Jul. 2007;81(1):127-35. Epub May 23, 2007. cited by applicant . Passini et al., CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J Clin Invest. Apr. 2010;120(4):1253-64. doi: 10.1172/JCI41615. Epub Mar. 15, 2010. cited by applicant . Pertin et al., Efficacy and specificity of recombinant adeno-associated virus serotype 6 mediated gene transfer to drg neurons through different routes of delivery. Poster sessions. Eur J. Pain. 2009;13:S74. Abstract 229. cited by applicant . Pfeifer et al., Pharmacological potential of RNAi--focus on miRNA. Pharmacol Ther. Jun. 2010;126(3):217-27. doi: 10.1016/j.pharmthera.2010.03.006. Epub Apr. 11, 2010. cited by applicant . Scallan et al., Human immunoglobulin inhibits liver transduction by AAV vectors at low AAV2 neutralizing titers in SCID mice. Blood. Mar. 1, 2006;107(5):1810-7. Epub Oct. 25, 2005. cited by applicant . Schattgen et al., Cutting Edge: DNA in the Lung Microenvironment during Influenza Virus Infection Tempers Inflammation by Engaging the DNA Sensor AIM2. J Immunol. Jan. 1, 2016;196(1):29-33. doi:10.4049/jimmunol.1501048. cited by applicant . Schnepp et al., Characterization of adeno-associated virus genomes isolated from human tissues. J Virol. Dec. 2005;79(23):14793-803. cited by applicant . Seiler et al., Adeno-associated virus types 5 and 6 use distinct receptors for cell entry. Hum Gene Ther. Jan. 2006;17(1):10-9. cited by applicant . Snyder et al., Comparison of adeno-associated viral vector serotypes for spinal cord and motor neuron gene delivery. Hum Gene Ther. Sep. 2011;22(9):1129-35. doi: 10.1089/hum.2011.008. Epub Jul. 25, 2011. cited by applicant . Sondhi et al., Enhanced survival of the LINCL mouse following CLN2 gene transfer using the rh.10 rhesus macaque-derived adeno-associated virus vector. Mol Ther. Mar. 2007;15(3):481-91. Epub Dec. 19, 2006. cited by applicant . Song et al., Intramuscular administration of recombinant adeno-associated virus 2 alpha-1 antitrypsin (rAAV-SERPINA1) vectors in a nonhuman primate model: safety and immunologic aspects. Mol Ther. Sep. 2002;6(3):329-35. cited by applicant . Stoica et al., Targeting Human SOD1 Using AAV mediated RNAi in a mouse model of amyotrophic lateral sclerosis. Mol ther. Jun. 2013;21(1):S149. cited by applicant . Storek et al., Intrathecal long-term gene expression by self-complementary adeno-associated virus type 1 suitable for chronic pain studies in rats. Mol Pain. Jan. 30, 2006;2:4. cited by applicant . Storkebaum et al., Treatment of motoneuron degeneration by intracerebroventricular delivery of VEGF in a rat model of ALS. Nat Neurosci. Jan. 2005;8(1):85-92. Epub Nov. 28, 2004. cited by applicant . Suckau et al., 851. The Effect of Genome Size and Design of scAAV Vectors on Efficiency of shRNA Expression and Gene Knockdown. May 1, 2007;15(1):S325. cited by applicant . Tenenbaum et al., Recombinant AAV-mediated gene delivery to the central nervous system. J Gene Med. Feb. 2004;6 Suppl 1:S212-22. cited by applicant . Tomar et al., Use of adeno-associated viral vector for delivery of small interfering RNA. Oncogene. Aug. 28, 2003;22(36):5712-5. cited by applicant . Towne et al., Systemic AAV6 delivery mediating RNA interference against SOD1: neuromuscular transduction does not alter disease progression in fALS mice. Mol Ther. Jun. 2008;16(6):1018-25. doi:10.1038/mt.2008.73. Epub Apr. 15, 2008. cited by applicant . Truong et al., Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res. Jul. 27, 2015;43(13):6450-8. doi: 10.1093/nar/gkv601. Epub Jun. 16, 2015. cited by applicant . Vandenberghe et al., Heparin binding directs activation of T cells against adeno-associated virus serotype 2 capsid. Nat Med. Aug. 2006;12(8):967-71. Epub Jul. 16, 2006. cited by applicant . Vandenberghe et al., Tailoring the AAV vector capsid for gene therapy. Gene Ther. Mar. 2009;16(3):311-9. Epub Dec. 4, 2008. cited by applicant . Vandendriessche et al., Efficacy and safety of adeno-associated viral vectors based on serotype 8 and 9 vs. lentiviral vectors for hemophilia B gene therapy. J Thromb Haemost. Jan. 2007;5(1):16-24. Epub Sep. 26, 2006. cited by applicant . Virella-Lowell et al., Enhancing rAAV vector expression in the lung. J Gene Med. Jul. 2005;7(7):842-50. cited by applicant . Vulchanova et al., Differential adeno-associated virus mediated gene transfer to sensory neurons following intrathecal delivery by direct lumbar puncture. Mol Pain. May 28, 2010;6:31. doi: 10.1186/1744-8069-6-31. cited by applicant . Wang et al., Neuroprotective effects of glial cell line-derived neurotrophic factor mediated by an adeno-associated virus vector in a transgenic animal model of amyotrophic lateral sclerosis. J Neurosci. Aug. 15, 2002;22(16):6920-8. cited by applicant . Wang et al., Rescue and replication of adeno-associated virus type 2 as well as vector DNA sequences from recombinant plasmids containing deletions in the viral inverted terminal repeats: selective encapsidation of viral genomes in progeny virions. J Virol. Mar. 1996;70(3):1668-77. cited by applicant . Wang et al., Somatically Repairing Compound Heterozygous Recessive Mutations by Chromosomal Cut-and-Paste for in Vivo Gene Therapy. May 2016. 24(1):S289. Abstract 733. cited by applicant . Wang et al., Sustained correction of disease in naive and AAV2-pretreated hemophilia B dogs: AAV2/8-mediated, liver-directed gene therapy. Blood. Apr. 15, 2005;105(8):3079-86. Epub Jan. 6, 2005. cited by applicant . Wang et al., The design of vectors for RNAi delivery system. Curr Pharm Des. 2008;14(13):1327-40. cited by applicant . Wang et al., The potential of adeno-associated viral vectors for gene delivery to muscle tissue. Expert Opin Drug Deliv. Mar. 2014;11(3):345-64. doi: 10.1517/17425247.2014.871258. Epub Jan. 3, 2014. cited by applicant . Wang et al., Widespread spinal cord transduction by intrathecal injection of rAAV delivers efficacious RNAi therapy for amyotrophic lateral sclerosis. Hum Mol Genet. Feb. 1, 2014;23(3):668-81. doi: 10.1093/hmg/ddt454. Epub Sep. 18, 2013. cited by applicant . Watson et al., Intrathecal administration of AAV vectors for the treatment of lysosomal storage in the brains of MPS I mice. Gene Ther. Jun. 2006;13(11):917-25. cited by applicant . Weismann et al., Systemic AAV9 gene transfer in adult GM1 gangliosidosis mice reduces lysosomal storage in CNS and extends lifespan. Hum Mol Genet. Aug. 1, 2015;24(15):4353-64. doi: 10.1093/hmg/ddv168. Epub May 10, 2015. cited by applicant . Weismann, Approaches and Considerations Towards a Safe and Effective Adena-Associated Virus Mediated Therapeutic Intervention for GM 1-Gangliosidosis: A Dissertation. University Massachusetts Medical School. Aug. 5, 2014. cited by applicant . Wu et al., Alpha2,3 and alpha2,6 N-linked sialic acids facilitate efficient binding and transduction by adeno-associated virus types 1 and 6. J Virol. Sep. 2006;80(18):9093-103. cited by applicant . Xie et al., Isolation of transcriptionally active novel AAV capsid sequences from chimpanzee tissues for vector development. Meeting Abstract: 12th Annual Meeting of the American Society of Gene Therapy. May 1, 2009. Abstract 91. cited by applicant . Xie et al., 676. DNA Sequences Encoding shRNAs Can Replace Mutant ITR in scAAV Genome for Efficient Replication and Packaging and Transcribe shRNAs by pol III Promoter Activity of wt ITR for Efficient Gene Silencing Mol Therapy. May 2015;23(1):S269. cited by applicant . Xie et al., Characterization of positioning effect of pol III-shRNA transcription unit in scAAV vector genome on the packaging efficiency and functionality of shRNA silencing. Molecular Therapy. May 2010;18(1): S262. Abstract 671. cited by applicant . Xie et al., MicroRNA regulated tissue specific transduction by rAAV vector. Molecular Therapy. May 2009;17(1): S279. Abstract 732. cited by applicant . Xie et al., MicroRNA-regulated, systemically delivered rAAV9: a step closer to CNS-restricted transgene expression. Mol Ther. Mar. 2011;19(3):526-35. doi: 10.1038/mt.2010.279. Epub Dec. 21, 2010. cited by applicant . Xie et al., rAAV-mediated delivery of micro RNA scavengers leads to efficient and stable knock-down of cognate micro RNAs, upregulation of their natural target genes and phenotypic changes in mice. Molecular Therapy. May 2010;18(1): S140. Abstract 362. cited by applicant . Xie et al., Short DNA Hairpins Compromise Recombinant Adeno-Associated Virus Genome Homogeneity. Mol Ther. Jun. 7, 2017;25(6):1363-1374. doi: 10.1016/j.ymthe.2017.03.028. Epub Apr. 24, 2017. cited by applicant . Xu et al., Delivery of MDR1 small interfering RNA by self-complementary recombinant adeno-associated virus vector. Mol Ther. Apr. 2005;11(4):523-30. cited by applicant . Yan et al., Unique biologic properties of recombinant AAV1 transduction in polarized human airway epithelia. J Biol Chem. Oct. 6, 2006;281(40):29684-92. Epub Aug. 9, 2006. cited by applicant . Zabner et al., Adeno-associated virus type 5 (AAV5) but not AAV2 binds to the apical surfaces of airway epithelia and facilitates gene transfer. J Virol. Apr. 2000;74(8):3852-8. cited by applicant . Zhang et al., Characterization of 12 AAV vectors for intravascular delivery to target CNS and detarget non-CNS tissues by mirna regulation: implications in treatment of canavan disease. Molecular Therapy. May 2010;18(1): S174. Abstract 450. cited by applicant . Zhong et al., Chimpanzee-derived novel natural variants of aav9: vector development and interrogation of correlations between capsid structure and vector biology. Molecular Therapy. May 2010;18(1): S24. Abstract 58. cited by applicant . Zincarelli et al., Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. Jun. 2008;16(6):1073-80. doi: 10.1038/mt.2008.76. Epub Apr. 15, 2008. cited by applicant . Zolotukhin et al., Production and purification of serotype 1, 2, and 5 recombinant adeno-associated viral vectors. Methods. Oct. 2002;28(2):158-67. cited by applicant . Invitation to Pay Additional Fees dated Dec. 17, 2015, for Application No. PCT/US2015/053798. cited by applicant . International Search Report and Written Opinion dated Feb. 12, 2016, for Application No. PCT/US2015/053798. cited by applicant . International Preliminary Report on Patentability dated Apr. 13, 2017, for Application No. PCT/US2015/053798. cited by applicant . Meister et al., Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. Mar. 2004;10(3):544-50. cited by applicant. |
Primary Examiner: Ton; Thaian N.
Attorney, Agent or Firm: Wolf, Greenfield & Sacks, P.C.
Parent Case Text
RELATED APPLICATIONS
This application is a National Stage Application of PCT/US2016/017886, filed Feb. 12, 2016, entitled "COMPOSITIONS AND METHODS FOR TRANSIENT DELIVERY OF NUCLEASES", which claims the benefit under 35 U.S.C. .sctn. 119(e) of U.S. Provisional Application Ser. No. 62/115,928, entitled "COMPOSITIONS AND METHODS FOR TRANSIENT DELIVERY OF NUCLEASES" filed on Feb. 13, 2015, the entire contents of each application which are incorporated herein by reference.
Claims
What is claimed is:
1. An adeno-associated virus (AAV) capsid protein having a terminally grafted nuclease.
2. The AAV capsid protein of claim 1, wherein the capsid protein is a VP2 capsid protein.
3. The AAV capsid protein of claim 2, wherein the terminally grafted nuclease is grafted to the N-terminus of the VP2 capsid protein, or wherein the terminally grafted nuclease is grafted to the C-terminus of the VP2 capsid protein.
4. The AAV capsid protein of claim 1, wherein the nuclease is selected from: Transcription Activator-like Effector Nucleases (TALENs), Zinc Finger Nucleases (ZFNs), engineered meganuclease, re-engineered homing endonucleases and a Cas-family nuclease.
5. The AAV capsid protein of claim 2, further comprising a linker conjugated to the C-terminus of the terminally grafted nuclease and the N-terminus of the VP2 protein, or further comprising a linker conjugated to the N-terminus of the terminally grafted nuclease and the C-terminus of the VP2 protein.
6. The AAV capsid protein of claim 1, wherein the AAV capsid protein having a terminally grafted nuclease is of: (i) an AAV1, AAV2, AAV3, AAV4, AAV5, AAV6, AAV7, AAV8, AAV9, AAV10, AAV11, or AAV12 serotype; or (ii) a serotype derived from a non-human primate.
7. A recombinant adeno-associated virus (rAAV) comprising the capsid protein of claim 1.
8. The rAAV of claim 7, wherein the rAAV comprises a transgene encoding a guide RNA.
9. The rAAV of claim 7, wherein the AAV is an empty viral particle with no transgene.
10. An in vitro method of targeting genome editing in a cell, the method comprising: delivering to the cell a first recombinant adeno associated virus (rAAV) having a terminally-grafted Cas-family endonuclease on at least one capsid protein, wherein the terminally-grafted Cas-family endonuclease is directed to a genomic cleavage site by a guide RNA.
11. The method of claim 10, wherein the first rAAV comprises a transgene encoding the guide RNA.
12. The method of claim 10 further comprising administering a second rAAV having a transgene encoding a guide RNA that directs the Cas-family endonuclease to a cleavage site in a target nucleic acid.
13. A composition comprising: (i) a first recombinant adeno-associated virus (rAAV) having an terminally-grafted nuclease on at least one capsid protein; and (ii) a second rAAV having a transgene encoding a guide RNA that directs the nuclease to a cleavage site in a target nucleic acid.
14. An adeno-associated virus (AAV) capsid protein having a terminally grafted nuclease or fragment thereof, wherein the nuclease or fragment thereof comprises a terminally grafted intein.
15. The AAV capsid protein of claim 14, wherein the capsid protein is a VP2 capsid protein, or wherein the intein is IntN or IntC.
16. The composition of claim 13, wherein the nuclease is selected from: Transcription Activator-like Effector Nucleases (TALENs), Zinc Finger Nucleases (ZFNs), engineered meganuclease, re-engineered homing endonucleases and a Cas-family nuclease.
17. The composition of claim 13, wherein the capsid protein is a VP2 capsid protein.
18. The AAV capsid protein of claim 14, wherein the nuclease is selected from: Transcription Activator-like Effector Nucleases (TALENs), Zinc Finger Nucleases (ZFNs), engineered meganuclease, re-engineered homing endonucleases and a Cas-family nuclease.
Description
FIELD OF THE INVENTION
The disclosure in some aspects relates to isolated nucleic acids, compositions, and kits useful for protein delivery to cells.
BACKGROUND
Recently, gene editing using designer DNA sequence-specific nucleases emerged as a technology for both basic biomedical research and therapeutic development. Platforms based on three distinct types of endonucleases have been developed for gene editing, namely the zinc finger nuclease (ZFN), the transcription activator-like effector nuclease (TALEN), and the clustered regularly interspaced short palindromic repeat (CRISPR) associated endonuclease 9 (cas9). Each nuclease is capable of inducing a DNA double-stranded break (DSB) at specific DNA loci, thus triggering two DNA repair pathways. The non-homologous end joining (NHEJ) pathway generates random insertion/deletion (indel) mutations at the DSB, whereas the homology-directed repair (HDR) pathway repairs the DSB with the genetic information carried on a donor template. Therefore, these gene editing platforms are capable of manipulating genes at specific genomic loci in multiple ways, such as disrupting gene function, repairing a mutant gene to normal, and inserting DNA material.
Transforming the gene editing technology into therapeutic uses encounters several obstacles, including the concern over safety. Certain gene editing platforms have been shown to induce off-target DSBs throughout genomes, which is associated with genotoxicity. Such off-target effects not only stem from the intrinsic ambiguity of DNA sequence recognition by nucleases, but also attribute to the prolonged presence of an active gene editing system in a given cell. As a result, off-target DSBs accumulate over time, and ultimately lead to genotoxicity.
SUMMARY
Recent approaches to delivering nucleases to cells for gene editing have focused on delivering of expression vectors engineered to express the nucleases in target cells. However, these approaches have proved to be problematic in many instances due to genotoxicity resulting from to prolonged expression of gene editing system in vivo. To prevent such off-target genotoxicity due to prolonged presence of a gene editing system, several studies explored delivery of mRNA or protein instead of delivering the gene coding for the nucleases in cell culture. As a result, the gene editing system functions only in a short period of time until the nuclease mRNA or protein is naturally degraded inside cells, which has been shown to reduce off-target effects. However, delivery of mRNA or protein in vivo is a significant task, and the delivery efficiency is very limited with conventional techniques. In contrast, the present disclosure overcomes such genotoxicity and delivery issues by using viruses for transiently delivering nucleases to cells thereby fulfilling the task of inducing permanent gene editing in a transient manner such that the nucleases will degrade naturally. In some embodiments, the disclosure relates to the uses of a viral vector (e.g., an AAV) as a delivery vehicle to carry a nuclease (e.g., a Cas9 protein or other designer nuclease proteins) to cells. In some embodiments, to avoid the potential genotoxicity due to prolonged expression of gene editing system in vivo, methods are provided herein to transiently deliver an endonuclease protein using recombinant adeno-associated viruses. In some embodiments, AAV capsid is used as a delivery vehicle to carry the Cas9 protein or other designer nuclease proteins.
In some aspects, the disclosure relates to an adeno-associated virus (AAV) capsid protein having a terminally grafted nuclease.
In some embodiments, the capsid protein is a VP2 capsid protein. In some embodiments, the terminally grafted nuclease is grafted to the N-terminus of the VP2 capsid protein. In some embodiments, the terminally grafted nuclease is grafted to the C-terminus of the VP2 capsid protein.
In some embodiments, the nuclease is selected from: Transcription Activator-like Effector Nucleases (TALENs), Zinc Finger Nucleases (ZFNs), engineered meganuclease, re-engineered homing endonucleases and a Cas-family nuclease. In some embodiments, the nuclease is a Cas-family nuclease selected from the group consisting of Cas9 and Cas7. In some embodiments, the nuclease is represented by SEQ ID NO: 2. In some embodiments, the nuclease is a polypeptide encoded by the nucleic acid sequence represented by SEQ ID NO: 1.
In some embodiments, the AAV capsid protein further comprises a linker conjugated to the C-terminus of the terminally grafted nuclease and the N-terminus of the VP2 protein. In some embodiments, the AAV capsid protein further comprises a linker conjugated to the N-terminus of the terminally grafted nuclease and the C-terminus of the VP2 protein.
In some embodiments, the AAV capsid protein hays an terminally grafted nuclease is of a serotype derived from a non-human primate. In some embodiments, the AAV capsid protein has an terminally grafted nuclease is selected from: AAV1, AAV2, AAV3, AAV4, AAV5, AAV6, AAV7, AAV8, AAV9, AAV10, AAV11, and AAV12.
In some aspects, the disclosure relates to a recombinant adeno-associated virus (rAAV) comprising an adeno-associated virus (AAV) capsid protein having a terminally grafted nuclease.
In some embodiments, the rAAV comprises a transgene. In some embodiments, the transgene encodes a guide RNA. In some embodiments, the guide RNA directs the nuclease to a cleavage site in a target nucleic acid.
In some embodiments, the AAV is an empty viral particle with no transgene.
In some aspects, the disclosure provides a composition comprising an rAAV as described by this document. In some embodiments, the composition further comprises a pharmaceutically acceptable carrier.
In some aspects, the disclosure relates to a nucleic acid encoding an AAV capsid protein having an terminally grafted nuclease. In some embodiments, a host cell contains the nucleic acid. In some embodiments, the host cell contains a nucleic acid encodes an AAV VP2 capsid protein having an terminally grafted nuclease. In some embodiments, the host cell further comprises one or more nucleic acids encoding VP1 and VP3 capsid proteins.
In some aspects, the disclosure relates to a composition comprising a host cell as described by this document and a sterile cell culture medium. In some aspects, the disclosure relates to a composition comprising a host cell as described by this document and a cryopreservative.
In some aspects, the disclosure relates to an isolated nucleic acid comprising a sequence represented by SEQ ID NO: 3.
In some aspects, the disclosure relates to an isolated nucleic acid encoding an AAV capsid protein having an amino acid sequence selected from the group consisting of: SEQ ID NOs: 2 and 4.
In some aspects, the disclosure relates to an isolated AAV capsid protein comprising an amino acid sequence selected from the group consisting of: SEQ ID NOs: 2 and 4.
In some aspects, the disclosure relates to a composition comprising an isolated AAV capsid protein as described by this document. In some embodiments, the composition further comprises a pharmaceutically acceptable carrier.
In some aspects, the disclosure relates to a kit for producing a rAAV, the kit comprising: a container housing an isolated nucleic acid having a sequence of SEQ ID NO: 1 or 3. In some embodiments, the kit further comprises instructions for producing the rAAV. In some embodiments, the kit further comprises at least one container housing a recombinant AAV vector, wherein the recombinant AAV vector comprises a transgene.
In some aspects, the disclosure relates to a kit comprising: a container housing a recombinant AAV having an isolated AAV capsid protein having an amino acid sequence as set forth in SEQ ID NO: 2 or 4.
In some aspects, the disclosure relates to a method of targeting genome editing in a cell, the method comprising: delivering to the cell a first recombinant adeno associated virus (rAAV) having an terminally-grafted nuclease on at least one capsid protein, wherein when present in the cell, the terminally-grafted nuclease is directed to a genomic cleavage site by a guide RNA.
In some embodiments of the method, the first rAAV comprises a transgene encoding the guide RNA.
In some embodiments, the method further comprises administering a second rAAV having a transgene encoding a guide RNA that directs the nuclease to a cleavage site in a target nucleic acid.
In some embodiments, the cell is present in a subject, and the first rAAV or second rAAV is administered to the subject intravenously, intravascularly, transdermally, intraocularly, intrathecally, orally, intramuscularly, subcutaneously, intranasally, or by inhalation, thereby delivering the first rAAV or second rAAV to the cell. In some embodiments, the subject is selected from a mouse, a rat, a rabbit, a dog, a cat, a sheep, a pig, and a non-human primate. In some embodiments, the subject is a human.
In some aspects, the disclosure relates to a composition comprising: i.) a first recombinant adeno-associated virus (rAAV) having an terminally-grafted nuclease on at least one capsid protein; and ii.) a second rAAV having a transgene encoding a guide RNA that directs the nuclease to a cleavage site in a target nucleic acid.
In some embodiments, the first rAAV is an empty viral particle. In some embodiments, the first rAAV has an terminally-grafted nuclease that is grafted to the C-terminus of a VP2 capsid protein of the rAAV.
In some aspects, the disclosure relates to an adeno-associated virus (AAV) capsid protein having a terminally grafted nuclease or fragment thereof, wherein the nuclease or fragment thereof comprises a terminally grafted intein.
In some embodiments, the capsid protein is a VP2 capsid protein. In some embodiments, the intein is IntN or IntC. In some embodiments, the capsid protein is represented by any one of SEQ ID NO: 7 to 9.
Each of the limitations of the disclosure can encompass various embodiments of the disclosure. It is, therefore, anticipated that each of the limitations of the disclosure involving any one element or combinations of elements can be included in each aspect of the disclosure. This disclosure is not limited in its application to the details of construction and the arrangement of components set forth in the following description or illustrated in the drawings. The disclosure is capable of other embodiments and of being practiced or of being carried out in various ways.
BRIEF DESCRIPTION OF DRAWINGS
The accompanying drawings are not intended to be drawn to scale. In the drawings, each identical or nearly identical component that is illustrated in various figures is represented by a like numeral. For purposes of clarity, not every component may be labeled in every drawing. In the drawings:
FIGS. 1A-1B shows the SpCas9-VP2 fusion protein is produced in HEK293 cells. FIG. 1A shows the HA tagged SpCas9 is fused to the N-terminus of VP2. The expression of this fusion protein is driven by the CMV promoter. BGHpA: bovine growth hormone polyadenylation signal. FIG. 1B depicts western blotting using anti-HA antibody showing the HA-tagged fusion protein (.about.230 kD, arrow) produced from transiently transfected HEK293 cells. HA tagged SpCas9 (.about.162 kD) is marked by the triangle. Star indicates a band of unknown origin, a likely degradation product from the fusion protein.
FIGS. 2A-2B show the SpCas9-VP2 fusion protein mediates gene editing in HEK293 cells. FIG. 2A shows the DNA repair reporter construct. The mutant GFP (GFPmut) carries a disruptive insertion (Ins), followed by out-of-frame (+3 frame) T2A and mCherry. In the presence of a functional gene editing system targeting Ins, +1 insertion by NHEJ shifts the T2A and mCherry to in-frame, resulting mCherry fluorescence. FIG. 2B shows the results of a reporter assay in HEK293 cells by co-transfection of the reporter construct and various plasmid as indicated. Both mCherry fluorescence and bright field images are shown. Scale bar=50 .mu.M.
FIGS. 3A-3B show alternative strategies utilizing intein-mediated protein trans-splicing (PTS). FIG. 3A shows the N-terminus and C-terminus Npu DnaE intein (IntN and IntC,) are fused with SpCas9 and VP2, respectively. The IntC-VP2 is packaged into AAV virion. PTS occurs between the SpCas9-IntN fusion protein and the IntC-AAV chimeric virion to produce the SpCas9-AAV virion. FIG. 3B shows that in the first AAV vector, the AAV genome encodes the N-terminal portion of SpCas9 (SpCas9N) fused with IntN. The second AAV vector carries IntC and the C-terminal portion of SpCas9 fused to VP2. In vivo transduction of the first AAV vector produces the fusion protein SpCas9N-IntN, which is followed by delivery of the second vector. PTS occurs to reconstitute the full-length SpCas9 protein.
FIG. 4 shows an expression construct comprising a nucleic acid sequence encoding SpCas9 nuclease N-terminally fused to VP2 capsid protein.
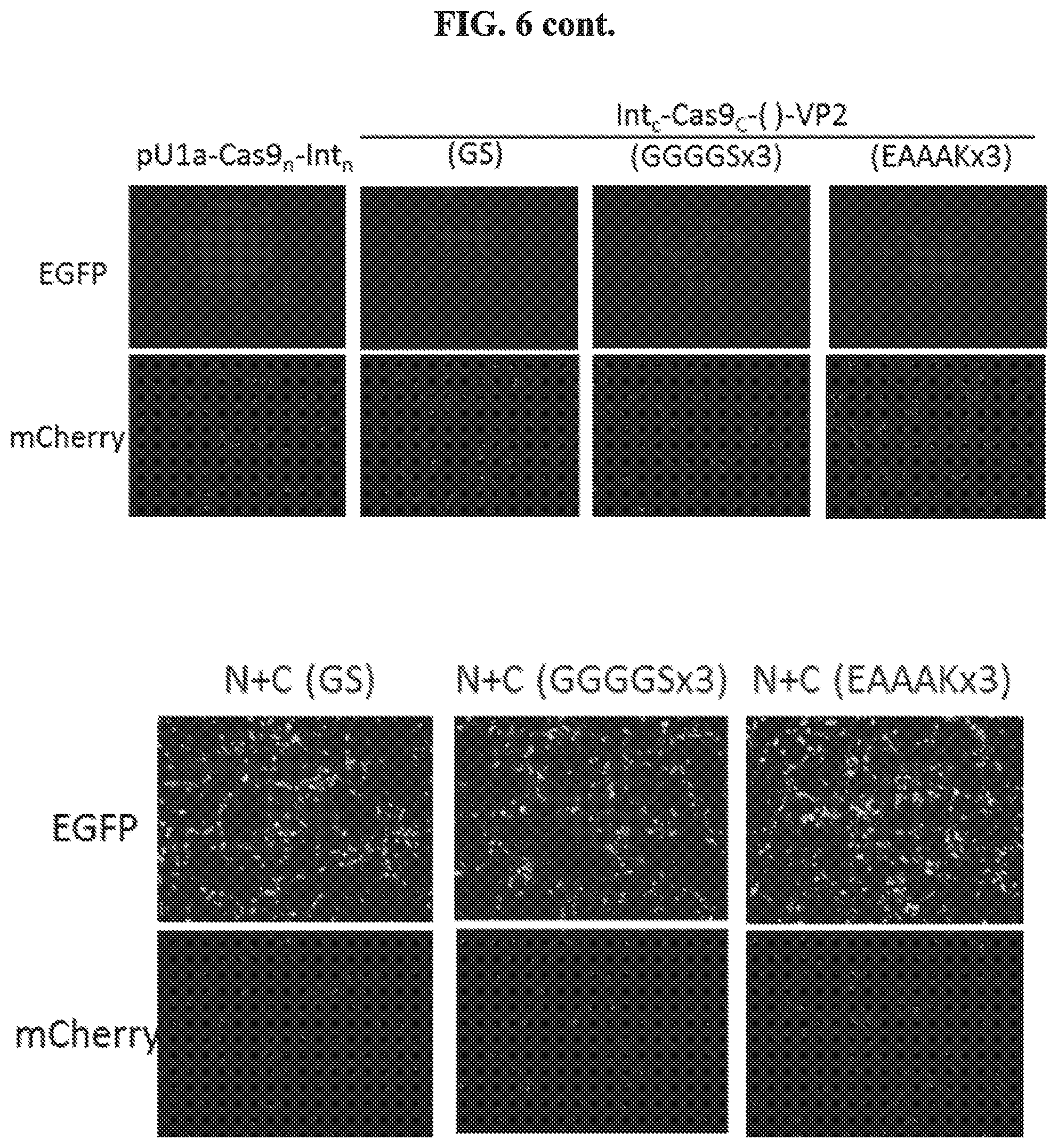
FIG. 5 shows co-transfection of Split Cas9 parts in HEK293 cells reconstituted SpCas9 and VP2 fusion protein, as measured by Western blot. Ctrl: pCMV-SpCas9-(EAAAKx3)-VP2; N: pU1a-Cas9.sub.n-Int.sub.n; C part: Int.sub.cCas9.sub.c-( )-VP2; HA tag is present in SpCas9 N-terminal. The designation "( )" refers to a linker sequence (e.g., GS, GGGGSx3, EAAAKx3).
FIG. 6 shows co-transfection of Split Cas9 parts in HEK293 cells reconstituted gene editing function. Cells were transfected with EGFP-ON reporter, pU1a-Cas9.sub.n-Int.sub.n, and Int.sub.c-Cas9.sub.C-( )-VP2. EGFP reports Cas9 cleavage and NHEJ repair; mCherry is constitutively expressed as control.
FIG. 7 shows incorporation of Int.sub.C-SpCas9.sub.c polypeptide onto rAAV2 capsid. Cells were transfected with plasmid encoding VP1 and VP3 proteins, and a plasmid encoding Int.sub.c-SpCas9.sub.c-( )-VP2. Purified rAAV particles were examined by silver staining.
DETAILED DESCRIPTION
Genome editing is a powerful tool for the interrogation and manipulation of biological functions within cells. For example, genome editing allows for the repair of mutant genes to normal function, disruption of gene function and the insertion of genetic material (e.g. DNA), all at specific genomic loci. However, several challenges associated with the delivery and prolonged expression of nucleases in cells, such as genotoxicity due to off-target cleavage of DNA, has limited the therapeutic effectiveness of gene editing platforms. The instant disclosure overcomes current limitations by providing compositions and methods that improve delivery of genome editing nucleases. Accordingly, in some aspects, the disclosure relates to viral proteins comprising a terminally grafted nucleases.
As used herein, "genome editing" refers to adding, disrupting or changing genomic sequences (e.g., a gene sequence). In some embodiments, genome editing is performed using engineered proteins and related molecules. In some aspects, genome editing comprises the use of engineered nucleases to cleave a target genomic locus. In some embodiments, genome editing further comprises inserting, deleting, mutating or substituting nucleic acid residues at a cleaved locus. In some embodiments, inserting, deleting, mutating or substituting nucleic acid residues at a cleaved locus is accomplished through endogenous cellular mechanisms such as homologous recombination (HR) and non-homologous end joining (NHEJ). Exemplary genome editing technologies include, but are not limited to Transcription Activator-like Effector Nucleases (TALENs), Zinc Finger Nucleases (ZFNs), engineered meganuclease re-engineered homing endonucleases, the CRISPR/Cas system. In some embodiments, the gene editing technologies are proteins or molecules related to TALENs, including but not limited to transcription activator-like effectors (TALEs) and restriction endonucleases (e.g. FokI). In some embodiments, the gene editing technologies are proteins or molecules related to ZFNs, including but not limited to proteins comprising the Cys.sub.2His.sub.2 fold group (for example Zif268 (EGR1)), and restriction endonucleases (e.g. FokI). In some embodiments, the gene editing technologies are proteins or molecules related to the CRISPR/Cas system, including but not limited to Cas9, Cas6, dCas9, CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA).
As used herein, the terms "endonuclease" and "nuclease" refer to an enzyme that cleaves a phosphodiester bond or bonds within a polynucleotide chain. Nucleases may be naturally occurring or genetically engineered. Genetically engineered nucleases are particularly useful for genome editing and are generally classified into four families: zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), engineered meganucleases and CRISPR-associated proteins (Cas nucleases). In some embodiments, the nuclease is a ZFN. In some embodiments, the ZFN comprises a FokI cleavage domain. In some embodiments, the ZFN comprises Cys.sub.2His.sub.2 fold group. In some embodiments, the nuclease is a TALEN. In some embodiments, the TALEN comprises a FokI cleavage domain. In some embodiments, the nuclease is an engineered meganuclease.
The term "CRISPR" refers to "clustered regularly interspaced short palindromic repeats", which are DNA loci containing short repetitions of base sequences. CRISPR loci form a portion of a prokaryotic adaptive immune system that confers resistance to foreign genetic material. Each CRISPR loci is flanked by short segments of "spacer DNA", which are derived from viral genomic material. In the Type II CRISPR system, spacer DNA hybridizes to transactivating RNA (tracrRNA) and is processed into CRISPR-RNA (crRNA) and subsequently associates with CRISPR-associated nucleases (Cas nucleases) to form complexes that recognize and degrade foreign DNA. In certain embodiments, the nuclease is a CRISPR-associated nuclease (Cas nuclease). Examples of CRISPR nucleases include, but are not limited to Cas9, Cas6 and dCas9. dCas9 is an engineered Cas protein that binds to a target locus but does not cleave said locus. In some embodiments, the nuclease is Cas9. In some embodiments, the Cas9 is derived from the bacteria S. pyogenes (SpCas9).
For the purpose of genome editing, the CRISPR system can be modified to combine the tracrRNA and crRNA in to a single guide RNA (sgRNA) or just (gRNA). As used herein, the term "guide RNA" or "gRNA" refers to a polynucleotide sequence that is complementary to a target sequence in a cell and associates with a Cas nuclease, thereby directing the Cas nuclease to the target sequence. In some embodiments, a gRNA ranges between 1 and 30 nucleotides in length. In some embodiments, a gRNA ranges between 5 and 25 nucleotides in length. In some embodiments, a gRNA ranges between 10 and 20 nucleotides in length. In some embodiments, a gRNA ranges between 14 and 18 nucleotides in length.
Aspects of the disclosure relate to SpCas9 grafted to an AAV2 capsid protein, VP2. However, in some embodiments, the same strategy can be applied in other contexts. For example, the SpCas9 can be replaced with any modified SpCas9 such as mutated or truncated forms, Cas9 proteins from other species and nucleases used in other gene editing platforms such as ZFNs and TALENs. In some embodiments, a nuclease terminally grafted to an AAV2 capsid protein may also be fused to another functional domain, for example single guide RNA (sgRNA).
Similarly, the AAV2 capsid protein VP2 may be replaced with VP2 of other AAV serotypes (e.g., AAV3, AAV3b, AAV4, AAV5, AAV6, AAV7, AAV8, AAV9, AAVrh8, AAV10, and variants thereof), or a suitable capsid protein of any viral vector. Thus, in some aspects, the disclosure relates to the viral delivery of a nuclease. Examples of viral vectors include retroviral vectors (e.g. Maloney murine leukemia virus, MML-V), adenoviral vectors (e.g. AD100), lentiviral vectors (HIV and FIV-based vectors), herpesvirus vectors (e.g. HSV-2). In some embodiments, the disclosure relates to adeno-associated viruses (AAVs). In some embodiments, a nuclease is grafted to or replaces all or a portion of a viral glycoprotein.
In some embodiments, SpCas-VP2 is incorporated into AAV2 capsid to form AAV virion. In some embodiments, the start codon of VP2 is mutated in the cap gene from the trans AAV production plasmid. In some embodiments, when Cas9 is fused to the N-terminus of VP2, the resulting Cas9-VP2 fusion protein is functional with respect to both productive AAV assembly and being an active component of the CRISPR/Cas9 gene editing system.
In some embodiments, a catalytically deficient form of the cas9 protein (dCas9) is fused with a C-terminal peptide domain that either activates or represses gene expression. In such embodiments, such a dCas9-effector fusion protein binds DNA in a sgRNA-guided manner.
In some aspects, the disclosure relates to the discovery that inteins can be utilized to rejoin (e.g., reconstitute) fragments or portions of gene editing proteins to generate a functional gene editing protein that is grafted onto an AAV capsid protein. As used herein, "intein" refers to a self-splicing protein intron (e.g., peptide) that ligates flanking N-terminal and C-terminal exteins (e.g., fragments to be joined). The use of certain inteins for joining heterologous protein fragments is described, for example, in Wood et al., J. Biol. Chem. 289(21); 14512-9 (2014). For example, when fused to separate protein fragments, the inteins IntN and IntC recognize each other, splice themselves out and simultaneously ligate the flanking N- and C-terminal exteins of the protein fragments to which they were fused, thereby reconstituting a full length protein from the two protein fragments. Other suitable inteins will be apparent to a person of skill in the art.
A nuclease protein fragment (e.g., Cas9 fragment) can vary in length. In some embodiments, a protein fragment ranges from 2 amino acids to about 1000 amino acids in length. In some embodiments, a protein fragment ranges from about 5 amino acids to about 500 amino acids in length. In some embodiments, a protein fragment ranges from about 20 amino acids to about 200 amino acids in length. In some embodiments, a protein fragment ranges from about 10 amino acids to about 100 amino acids in length. Suitable protein fragments of other lengths will be apparent to a person of skill in the art.
In some embodiments, a portion or fragment of a nuclease (e.g., a fragment of Cas9) is fused to an intein. The nuclease can be fused to the N-terminus or the C-terminus of the intein. In some embodiments, a portion or fragment of a nuclease (e.g., a fragment of Cas9) is fused to an intein and fused to an AAV capsid protein. The intein, nuclease and capsid protein can be fused together in any arrangement (e.g., nuclease-intein-capsid, intein-nuclease-capsid, capsid-intein-nuclease, etc.). In some embodiments, the N-terminus of an intein is fused to the C-terminus of a nuclease (e.g., Cas9) and the C-terminus of the intein is fused to the N-terminus of an AAV capsid protein.
In some embodiments, the IntN/IntC system is used to join fragments of a nuclease. In some embodiments, IntC is fused to the N-terminus of a nuclease (e.g., Cas9) fragment that is grafted to an AAV capsid protein. In some embodiments, IntN is fused to the C-terminus of a nuclease (e.g., Cas9) fragment. In some embodiments, a fragment of a nuclease fused to an intein is represented by SEQ ID NO: 6. In some embodiments, an AAV capsid protein comprising an intein fused to a fragment of a nuclease that has been terminally grafted to the AAV capsid protein is represented by any one of SEQ ID NO: 7 to 9.
Isolated AAV Capsid Proteins and Nucleic Acids Encoding the Same
AAVs disclosed herein are useful for creating vectors that facilitate delivery of nucleases to cells for human gene editing applications. Protein and amino acid sequences as well as other information regarding the AAVs capsid are set forth in the sequence listing.
In some embodiments, an AAV capsid having a terminally graft nuclease is provided that has an amino acid sequence represented by SEQ ID NO: 4. In some embodiments, an AAV capsid having a terminally graft nuclease is provided that is encoded by a nucleic acid sequence represented by SEQ ID NO: 3.
An example of an isolated nucleic acid that encodes an AAV capsid protein having a terminally graft nuclease is a nucleic acid having a sequence of: SEQ ID NO: 3 as well as nucleic acids having substantial homology thereto. In some embodiments, isolated nucleic acids that encode AAV capsids are provided that encode the VP2 protein portion of the amino acid sequence represented by SEQ ID NO: 3.
In some embodiments, nucleic acids are provided that encode an AAV capsid having a nuclease grafted within its capsid sequence (e.g., a AAV9 capsid) and up to 5, up to 10, up to 20, up to 30, up to 40, up to 50, up to 100 other amino acid alternations.
In some embodiments, a fragment (portion) of an isolated nucleic acid encoding a AAV capsid sequence may be useful for constructing a nucleic acid encoding a desired capsid sequence. Fragments may be of any appropriate length (e.g., at least 9, at least 18, at least 36, at least 72, at least 144, at least 288, at least 576, at least 1152 or more nucleotides in length). For example, a fragment of nucleic acid sequence encoding a variant amino acid (compared with a known AAV serotype) may be used to construct, or may be incorporated within, a nucleic acid sequence encoding an AAV capsid sequence to alter the properties of the AAV capsid. For example, a nucleic sequence encoding an AAV variant may comprise n amino acid variants (e.g., in which n=1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) compared with a known AAV serotype (e.g., AAV9). A recombinant cap sequence may be constructed having one or more of the n amino acid variants by incorporating fragments of a nucleic acid sequence comprising a region encoding a variant amino acid into the sequence of a nucleic acid encoding the known AAV serotype. The fragments may be incorporated by any appropriate method, including using site directed mutagenesis. In some embodiments, polypeptide fragments that are not normally present in AAV capsid proteins may be incorporated into a recombinant cap sequence. In some embodiments, the polypeptide fragment is grafted onto the recombinant cap sequence. Thus, new AAV variants may be created having new properties.
As used herein, "grafting" refers to joining or uniting of at least two polymeric molecules. In some embodiments, the term grafting refers joining or uniting of at least two polymeric molecules such that one of the at least two molecules is inserted within another of the at least two molecules. In some embodiments, the term grafting refers to joining or uniting of at least two polymeric molecules such that one of the at least two molecules is appended to another of the at least two molecules. In some embodiments, the term grafting refers joining or uniting of at least two nucleic acid molecules such that one of the at least two nucleic acid molecules is inserted within another of the at least two nucleic acid molecules. In some embodiments, the term grafting refers to joining or uniting of at least two nucleic acid molecules such that one of the at least two molecules is appended to another of the at least two nucleic acid molecules.
In some embodiments, a grafted nucleic acid molecule encodes a chimeric protein. In some embodiments, a grafted nucleic acid molecule encodes a chimeric protein, such that one polypeptide is effectively inserted into another polypeptide (e.g. not directly conjugated before the N-terminus or after the C-terminus), thereby creating a contiguous fusion of two polypeptides. In some embodiments, a grafted nucleic acid molecule encodes a chimeric protein, such that one polypeptide is effectively appended to another polypeptide (e.g. directly conjugated before the N-terminus or after the C-terminus), thereby creating a contiguous fusion of two polypeptides. In some embodiments, the term grafting refers to joining or uniting of at least two polypeptides, or fragments thereof, such that one of the at least two polypeptides or fragments thereof is inserted within another of the at least two polypeptides or fragments thereof. In some embodiments, the term grafting refers to joining or uniting of at least two polypeptides or fragments thereof such that one of the at least two polypeptides or fragments thereof is appended to another of the at least two polypeptides or fragments thereof.
In some embodiments, the instant disclosure relates to an adeno-associated virus (AAV) capsid protein comprising a AAV capsid protein having an N-terminally grafted nuclease.
In some embodiments, the AAV capsid protein further comprises a linker. Non-limiting examples of linkers include flexible linkers (e.g. glycine-rich linkers), rigid linkers (e.g. [EAAK].sub.n, where n>2), and cleavable linkers (e.g. protease-sensitive sequences). Other linkers are disclosed, for example in Chen et al., Fusion protein linkers: Property, design and functionality. Advanced drug delivery reviews, 2013. In some embodiments, the linker is conjugated to the C-terminus of a terminally grafted nuclease (e.g., an N-terminally grafted nuclease). In some embodiments, the linker is conjugated to the N-terminus of the terminally grafted nuclease (e.g., an N-terminally grafted nuclease). In some embodiments, one linker is conjugated to the N-terminus of the terminally grafted nuclease and a second linker is conjugated to the C-terminus of the terminally grafted nuclease.
In some embodiments, the linker is a glycine-rich linker. In some embodiments, the linker comprises at least one polypeptide repeat, each repeat comprising at least 80% glycine residues. In some embodiments, the polypeptide repeat comprises GGGS (SEQ ID NO: 5). In some embodiments, the linker comprises a formula selected from the group consisting of: [G].sub.n, [G].sub.nS, [GS].sub.n, and [GGSG].sub.n, wherein G is glycine and wherein n is an integer greater than one (e.g., 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 or more).
In some cases, fragments of capsid proteins disclosed herein are provided. Such fragments may at least 10, at least 20, at least 50, at least 100, at least 200, at least 300, at least 400, at least 500 or more amino acids in length. In some embodiments, chimeric capsid proteins are provided that comprise one or more fragments of one or more capsid proteins disclosed herein.
"Homology" refers to the percent identity between two polynucleotides or two polypeptide moieties. The term "substantial homology", when referring to a nucleic acid, or fragment thereof, indicates that, when optimally aligned with appropriate nucleotide insertions or deletions with another nucleic acid (or its complementary strand), there is nucleotide sequence identity in about 90 to 100% of the aligned sequences. When referring to a polypeptide, or fragment thereof, the term "substantial homology" indicates that, when optimally aligned with appropriate gaps, insertions or deletions with another polypeptide, there is nucleotide sequence identity in about 90 to 100% of the aligned sequences. The term "highly conserved" means at least 80% identity, preferably at least 90% identity, and more preferably, over 97% identity. In some cases, highly conserved may refer to 100% identity. Identity is readily determined by one of skill in the art by, for example, the use of algorithms and computer programs known by those of skill in the art.
As described herein, alignments between sequences of nucleic acids or polypeptides are performed using any of a variety of publicly or commercially available Multiple Sequence Alignment Programs, such as "Clustal W", accessible through Web Servers on the internet. Alternatively, Vector NTI utilities may also be used. There are also a number of algorithms known in the art which can be used to measure nucleotide sequence identity, including those contained in the programs described above. As another example, polynucleotide sequences can be compared using BLASTN, which provides alignments and percent sequence identity of the regions of the best overlap between the query and search sequences. Similar programs are available for the comparison of amino acid sequences, e.g., the "Clustal X" program, BLASTP. Typically, any of these programs are used at default settings, although one of skill in the art can alter these settings as needed. Alternatively, one of skill in the art can utilize another algorithm or computer program which provides at least the level of identity or alignment as that provided by the referenced algorithms and programs. Alignments may be used to identify corresponding amino acids between two proteins or peptides. A "corresponding amino acid" is an amino acid of a protein or peptide sequence that has been aligned with an amino acid of another protein or peptide sequence. Corresponding amino acids may be identical or non-identical. A corresponding amino acid that is a non-identical amino acid may be referred to as a variant amino acid.
Alternatively for nucleic acids, homology can be determined by hybridization of polynucleotides under conditions which form stable duplexes between homologous regions, followed by digestion with single-stranded-specific nuclease(s), and size determination of the digested fragments. DNA sequences that are substantially homologous can be identified in a Southern hybridization experiment under, for example, stringent conditions, as defined for that particular system. Defining appropriate hybridization conditions is within the skill of the art.
A "nucleic acid" sequence refers to a DNA or RNA sequence. In some embodiments, proteins and nucleic acids of the disclosure are isolated. As used herein, the term "isolated" means artificially produced. As used herein with respect to nucleic acids, the term "isolated" means: (i) amplified in vitro by, for example, polymerase chain reaction (PCR); (ii) recombinantly produced by cloning; (iii) purified, as by cleavage and gel separation; or (iv) synthesized by, for example, chemical synthesis. An isolated nucleic acid is one which is readily manipulable by recombinant DNA techniques well known in the art. Thus, a nucleotide sequence contained in a vector in which 5' and 3' restriction sites are known or for which polymerase chain reaction (PCR) primer sequences have been disclosed is considered isolated but a nucleic acid sequence existing in its native state in its natural host is not. An isolated nucleic acid may be substantially purified, but need not be. For example, a nucleic acid that is isolated within a cloning or expression vector is not pure in that it may comprise only a tiny percentage of the material in the cell in which it resides. Such a nucleic acid is isolated, however, as the term is used herein because it is readily manipulable by standard techniques known to those of ordinary skill in the art. As used herein with respect to proteins or peptides, the term "isolated" refers to a protein or peptide that has been isolated from its natural environment or artificially produced (e.g., by chemical synthesis, by recombinant DNA technology, etc.).
The skilled artisan will also realize that conservative amino acid substitutions may be made to provide functionally equivalent variants, or homologs of the capsid proteins. In some aspects the disclosure embraces sequence alterations that result in conservative amino acid substitutions. As used herein, a conservative amino acid substitution refers to an amino acid substitution that does not alter the relative charge or size characteristics of the protein in which the amino acid substitution is made. Variants can be prepared according to methods for altering polypeptide sequence known to one of ordinary skill in the art such as are found in references that compile such methods, e.g. Molecular Cloning: A Laboratory Manual, J. Sambrook, et al., eds., Second Edition, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., 1989, or Current Protocols in Molecular Biology, F. M. Ausubel, et al., eds., John Wiley & Sons, Inc., New York. Conservative substitutions of amino acids include substitutions made among amino acids within the following groups: (a) M, I, L, V; (b) F, Y, W; (c) K, R, H; (d) A, G; (e) S, T; (f) Q, N; and (g) E, D. Therefore, one can make conservative amino acid substitutions to the amino acid sequence of the proteins and polypeptides disclosed herein.
Recombinant AAVs
In some aspects, the disclosure provides isolated AAVs. As used herein with respect to AAVs, the term "isolated" refers to an AAV that has been artificially produced or obtained. Isolated AAVs may be produced using recombinant methods. Such AAVs are referred to herein as "recombinant AAVs". Recombinant AAVs (rAAVs) preferably have tissue-specific targeting capabilities, such that a nuclease and/or transgene of the rAAV will be delivered specifically to one or more predetermined tissue(s). The AAV capsid is an important element in determining these tissue-specific targeting capabilities. Thus, an rAAV having a capsid appropriate for the tissue being targeted can be selected Methods for obtaining recombinant AAVs having a desired capsid protein are well known in the art. (See, for example, US 2003/0138772), the contents of which are incorporated herein by reference in their entirety). Typically the methods involve culturing a host cell which contains a nucleic acid sequence encoding an AAV capsid protein; a functional rep gene; a recombinant AAV vector composed of, AAV inverted terminal repeats (ITRs) and a transgene; and sufficient helper functions to permit packaging of the recombinant AAV vector into the AAV capsid proteins. In some embodiments, capsid proteins are structural proteins encoded by the cap gene of an AAV. AAVs comprise three capsid proteins, virion proteins 1 to 3 (named VP1, VP2 and VP3), all of which are transcribed from a single cap gene via alternative splicing. In some embodiments, the molecular weights of VP1, VP2 and VP3 are respectively about 87 kDa, about 72 kDa and about 62 kDa. In some embodiments, upon translation, capsid proteins form a spherical 60-mer protein shell around the viral genome. In some embodiments, the functions of the capsid proteins are to protect the viral genome, deliver the genome and interact with the host. In some aspects, capsid proteins deliver the viral genome to a host in a tissue specific manner. In some embodiments, the a terminally grafted nuclease is present on all three capsid proteins (e.g. VP1, VP2, VP3) of a rAAV. In some embodiments, the terminally grafted nuclease is present on two of the capsid proteins (e.g. VP2 and VP3) of a rAAV. In some embodiments, the terminally grafted nuclease is present on a single capsid protein of a rAAV. In some embodiments, the terminally grafted nuclease is present on the VP2 capsid protein of the rAAV.
In some embodiments, the instant disclosure relates to an adeno-associated virus (AAV) capsid protein comprising: an AAV capsid protein having an N-terminally grafted nuclease, wherein the AAV capsid protein is not of an AAV2 serotype. In some embodiments, the AAV capsid protein is of an AAV serotype selected from the group consisting of AAV3, AAV4, AAV5, AAV6, AAV8, AAVrh8 AAV9, and AAV10. In some embodiments, the capsid protein having an N-terminally grafted nuclease is a viral protein 2 (VP2) capsid protein. In some embodiments, the AAV capsid protein having a terminally grafted nuclease is of a serotype derived from a non-human primate. In some embodiments, the AAV capsid protein having a terminally grafted nuclease is of a AAVrh8 serotype. In some embodiments, the AAV capsid protein having an N-terminally grafted nuclease is of an AAV9, optionally AAV9.47, serotype.
In some aspects, the instant disclosure relates to the location within an AAV capsid protein where a nuclease is grafted. In some embodiments, the nuclease is N-terminally grafted to the capsid protein. In some embodiments, the nuclease is C-terminally grafted to a capsid protein. In some embodiments, a nuclease that is C-terminally grafted to a capsid protein (e.g., VP2) resides within the viral particle, and the viral particle does not contain a genome, e.g., a nucleic acid harboring a transgene.
The components to be cultured in the host cell to package a rAAV vector in an AAV capsid may be provided to the host cell in trans. Alternatively, any one or more of the required components (e.g., recombinant AAV vector, rep sequences, cap sequences, and/or helper functions) may be provided by a stable host cell which has been engineered to contain one or more of the required components using methods known to those of skill in the art. Most suitably, such a stable host cell will contain the required component(s) under the control of an inducible promoter. However, the required component(s) may be under the control of a constitutive promoter. Examples of suitable inducible and constitutive promoters are provided herein, in the discussion of regulatory elements suitable for use with the transgene. In still another alternative, a selected stable host cell may contain selected component(s) under the control of a constitutive promoter and other selected component(s) under the control of one or more inducible promoters. For example, a stable host cell may be generated which is derived from 293 cells (which contain E1 helper functions under the control of a constitutive promoter), but which contain the rep and/or cap proteins under the control of inducible promoters. Still other stable host cells may be generated by one of skill in the art.
In some embodiments, the instant disclosure relates to a host cell containing a nucleic acid that comprises a coding sequence encoding a nuclease terminally grafted to a capsid protein that is operably linked to a promoter. In some embodiments, the instant disclosure relates to a composition comprising the host cell described above. In some embodiments, the composition comprising the host cell above further comprises a cryopreservative.
The recombinant AAV vector, rep sequences, cap sequences, and helper functions required for producing the rAAV of the disclosure may be delivered to the packaging host cell using any appropriate genetic element (vector). The selected genetic element may be delivered by any suitable method, including those described herein. The methods used to construct any embodiment of this disclosure are known to those with skill in nucleic acid manipulation and include genetic engineering, recombinant engineering, and synthetic techniques. See, e.g., Sambrook et al, Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Press, Cold Spring Harbor, N.Y. Similarly, methods of generating rAAV virions are well known and the selection of a suitable method is not a limitation on the present disclosure. See, e.g., K. Fisher et al, J. Virol., 70:520-532 (1993) and U.S. Pat. No. 5,478,745.
In some embodiments, recombinant AAVs may be produced using the triple transfection method (described in detail in U.S. Pat. No. 6,001,650). Typically, the recombinant AAVs are produced by transfecting a host cell with an recombinant AAV vector (comprising a transgene) to be packaged into AAV particles, an AAV helper function vector, and an accessory function vector. An AAV helper function vector encodes the "AAV helper function" sequences (i.e., rep and cap), which function in trans for productive AAV replication and encapsidation. Preferably, the AAV helper function vector supports efficient AAV vector production without generating any detectable wild-type AAV virions (i.e., AAV virions containing functional rep and cap genes). Non-limiting examples of vectors suitable for use with the present disclosure include pHLP19, described in U.S. Pat. No. 6,001,650 and pRep6cap6 vector, described in U.S. Pat. No. 6,156,303, the entirety of both incorporated by reference herein. The accessory function vector encodes nucleotide sequences for non-AAV derived viral and/or cellular functions upon which AAV is dependent for replication (i.e., "accessory functions"). The accessory functions include those functions required for AAV replication, including, without limitation, those moieties involved in activation of AAV gene transcription, stage specific AAV mRNA splicing, AAV DNA replication, synthesis of cap expression products, and AAV capsid assembly. Viral-based accessory functions can be derived from any of the known helper viruses such as adenovirus, herpesvirus (other than herpes simplex virus type-1), and vaccinia virus.
In some aspects, the disclosure provides transfected host cells. The term "transfection" is used to refer to the uptake of foreign DNA by a cell, and a cell has been "transfected" when exogenous DNA has been introduced inside the cell membrane. A number of transfection techniques are generally known in the art. See, e.g., Graham et al. (1973) Virology, 52:456, Sambrook et al. (1989) Molecular Cloning, a laboratory manual, Cold Spring Harbor Laboratories, New York, Davis et al. (1986) Basic Methods in Molecular Biology, Elsevier, and Chu et al. (1981) Gene 13:197. Such techniques can be used to introduce one or more exogenous nucleic acids, such as a nucleotide integration vector and other nucleic acid molecules, into suitable host cells.
A "host cell" refers to any cell that harbors, or is capable of harboring, a substance of interest. Often a host cell is a mammalian cell. A host cell may be used as a recipient of an AAV helper construct, an AAV minigene plasmid, an accessory function vector, or other transfer DNA associated with the production of recombinant AAVs. The term includes the progeny of the original cell which has been transfected. Thus, a "host cell" as used herein may refer to a cell which has been transfected with an exogenous DNA sequence. It is understood that the progeny of a single parental cell may not necessarily be completely identical in morphology or in genomic or total DNA complement as the original parent, due to natural, accidental, or deliberate mutation.
As used herein, the term "cell line" refers to a population of cells capable of continuous or prolonged growth and division in vitro. Often, cell lines are clonal populations derived from a single progenitor cell. It is further known in the art that spontaneous or induced changes can occur in karyotype during storage or transfer of such clonal populations. Therefore, cells derived from the cell line referred to may not be precisely identical to the ancestral cells or cultures, and the cell line referred to includes such variants.
As used herein, the terms "recombinant cell" refers to a cell into which an exogenous DNA segment, such as DNA segment that leads to the transcription of a biologically-active polypeptide or production of a biologically active nucleic acid such as an RNA, has been introduced.
As used herein, the term "vector" includes any genetic element, such as a plasmid, phage, transposon, cosmid, chromosome, artificial chromosome, virus, virion, etc., which is capable of replication when associated with the proper control elements and which can transfer gene sequences between cells. Thus, the term includes cloning and expression vehicles, as well as viral vectors. In some embodiments, useful vectors are contemplated to be those vectors in which the nucleic acid segment to be transcribed is positioned under the transcriptional control of a promoter. A "promoter" refers to a DNA sequence recognized by the synthetic machinery of the cell, or introduced synthetic machinery, required to initiate the specific transcription of a gene. The phrases "operatively positioned," "under control" or "under transcriptional control" means that the promoter is in the correct location and orientation in relation to the nucleic acid to control RNA polymerase initiation and expression of the gene. The term "expression vector or construct" means any type of genetic construct containing a nucleic acid in which part or all of the nucleic acid encoding sequence is capable of being transcribed. In some embodiments, expression includes transcription of the nucleic acid, for example, to generate a biologically-active polypeptide product or functional RNA (e.g., guide RNA) from a transcribed gene.
The foregoing methods for packaging recombinant vectors in desired AAV capsids to produce the rAAVs of the disclosure are not meant to be limiting and other suitable methods will be apparent to the skilled artisan.
Recombinant AAV Vectors
"Recombinant AAV (rAAV) vectors" of the disclosure are typically composed of, at a minimum, a transgene and its regulatory sequences, and 5' and 3' AAV inverted terminal repeats (ITRs). It is this recombinant AAV vector which is packaged into a capsid protein and delivered to a selected target cell. In some embodiments, the transgene is a nucleic acid sequence, heterologous to the vector sequences, which encodes a polypeptide, protein, functional RNA molecule (e.g., gRNA) or other gene product, of interest. The nucleic acid coding sequence is operatively linked to regulatory components in a manner which permits transgene transcription, translation, and/or expression in a cell of a target tissue.
In some embodiments, the instant disclosure relates to a recombinant AAV (rAAV) comprising a capsid protein having an N-terminally grafted nuclease, wherein the N-terminally grafted nuclease is present only in the VP2 capsid protein. In some embodiments, the rAAV comprises a capsid protein having an amino acid sequence represented by SEQ ID NO: 4.
The AAV sequences of the vector typically comprise the cis-acting 5' and 3' inverted terminal repeat sequences (See, e.g., B. J. Carter, in "Handbook of Parvoviruses", ed., P. Tijsser, CRC Press, pp. 155 168 (1990)). The ITR sequences are about 145 bp in length. Preferably, substantially the entire sequences encoding the ITRs are used in the molecule, although some degree of minor modification of these sequences is permissible. The ability to modify these ITR sequences is within the skill of the art. (See, e.g., texts such as Sambrook et al, "Molecular Cloning. A Laboratory Manual", 2d ed., Cold Spring Harbor Laboratory, New York (1989); and K. Fisher et al., J Virol., 70:520 532 (1996)). An example of such a molecule employed in the present disclosure is a "cis-acting" plasmid containing the transgene, in which the selected transgene sequence and associated regulatory elements are flanked by the 5' and 3' AAV ITR sequences. The AAV ITR sequences may be obtained from any known AAV, including presently identified mammalian AAV types.
In addition to the major elements identified above for the recombinant AAV vector, the vector also includes conventional control elements necessary which are operably linked to the transgene in a manner which permits its transcription, translation and/or expression in a cell transfected with the plasmid vector or infected with the virus produced by the disclosure. As used herein, "operably linked" sequences include both expression control sequences that are contiguous with the gene of interest and expression control sequences that act in trans or at a distance to control the gene of interest.
Expression control sequences include appropriate transcription initiation, termination, promoter and enhancer sequences; efficient RNA processing signals such as splicing and polyadenylation (polyA) signals; sequences that stabilize cytoplasmic mRNA; sequences that enhance translation efficiency (i.e., Kozak consensus sequence); sequences that enhance protein stability; and when desired, sequences that enhance secretion of the encoded product. A great number of expression control sequences, including promoters which are native, constitutive, inducible and/or tissue-specific, are known in the art and may be utilized.
As used herein, a nucleic acid sequence (e.g., coding sequence) and regulatory sequences are said to be "operably" linked when they are covalently linked in such a way as to place the expression or transcription of the nucleic acid sequence under the influence or control of the regulatory sequences. If it is desired that the nucleic acid sequences be translated into a functional protein, two DNA sequences are said to be operably linked if induction of a promoter in the 5' regulatory sequences results in the transcription of the coding sequence and if the nature of the linkage between the two DNA sequences does not (1) result in the introduction of a frame-shift mutation, (2) interfere with the ability of the promoter region to direct the transcription of the coding sequences, or (3) interfere with the ability of the corresponding RNA transcript to be translated into a protein. Thus, a promoter region would be operably linked to a nucleic acid sequence if the promoter region were capable of effecting transcription of that DNA sequence such that the resulting transcript might be translated into the desired protein or polypeptide. Similarly two or more coding regions are operably linked when they are linked in such a way that their transcription from a common promoter results in the expression of two or more proteins having been translated in frame. In some embodiments, operably linked coding sequences yield a fusion protein. In some embodiments, operably linked coding sequences yield a functional RNA (e.g., gRNA).
For nucleic acids encoding proteins, a polyadenylation sequence generally is inserted following the transgene sequences and before the 3' AAV ITR sequence. A rAAV construct useful in the present disclosure may also contain an intron, desirably located between the promoter/enhancer sequence and the transgene. One possible intron sequence is derived from SV-40, and is referred to as the SV-40 T intron sequence. Another vector element that may be used is an internal ribosome entry site (IRES). An IRES sequence is used to produce more than one polypeptide from a single gene transcript. An IRES sequence would be used to produce a protein that contain more than one polypeptide chains. Selection of these and other common vector elements are conventional and many such sequences are available [see, e.g., Sambrook et al, and references cited therein at, for example, pages 3.18 3.26 and 16.17 16.27 and Ausubel et al., Current Protocols in Molecular Biology, John Wiley & Sons, New York, 1989]. In some embodiments, a Foot and Mouth Disease Virus 2A sequence is included in polyprotein; this is a small peptide (approximately 18 amino acids in length) that has been shown to mediate the cleavage of polyproteins (Ryan, M D et al., EMBO, 1994; 4: 928-933; Mattion, N M et al., J Virology, November 1996; p. 8124-8127; Furler, S et al., Gene Therapy, 2001; 8: 864-873; and Halpin, C et al., The Plant Journal, 1999; 4: 453-459). The cleavage activity of the 2A sequence has previously been demonstrated in artificial systems including plasmids and gene therapy vectors (AAV and retroviruses) (Ryan, M D et al., EMBO, 1994; 4: 928-933; Mattion, N M et al., J Virology, November 1996; p. 8124-8127; Furler, S et al., Gene Therapy, 2001; 8: 864-873; and Halpin, C et al., The Plant Journal, 1999; 4: 453-459; de Felipe, P et al., Gene Therapy, 1999; 6: 198-208; de Felipe, P et al., Human Gene Therapy, 2000; 11: 1921-1931; and Klump, H et al., Gene Therapy, 2001; 8: 811-817).
The precise nature of the regulatory sequences needed for gene expression in host cells may vary between species, tissues or cell types, but shall in general include, as necessary, 5' non-transcribed and 5' non-translated sequences involved with the initiation of transcription and translation respectively, such as a TATA box, capping sequence, CAAT sequence, enhancer elements, and the like. Especially, such 5' non-transcribed regulatory sequences will include a promoter region that includes a promoter sequence for transcriptional control of the operably joined gene. Regulatory sequences may also include enhancer sequences or upstream activator sequences as desired. The vectors of the disclosure may optionally include 5' leader or signal sequences. The choice and design of an appropriate vector is within the ability and discretion of one of ordinary skill in the art.
Examples of constitutive promoters include, without limitation, the retroviral Rous sarcoma virus (RSV) LTR promoter (optionally with the RSV enhancer), the cytomegalovirus (CMV) promoter (optionally with the CMV enhancer) [see, e.g., Boshart et al, Cell, 41:521-530 (1985)], the SV40 promoter, the dihydrofolate reductase promoter, the .beta.-actin promoter, the phosphoglycerol kinase (PGK) promoter, and the EF1.alpha. promoter [Invitrogen].
Inducible promoters allow regulation of gene expression and can be regulated by exogenously supplied compounds, environmental factors such as temperature, or the presence of a specific physiological state, e.g., acute phase, a particular differentiation state of the cell, or in replicating cells only. Inducible promoters and inducible systems are available from a variety of commercial sources, including, without limitation, Invitrogen, Clontech and Ariad. Many other systems have been described and can be readily selected by one of skill in the art. Examples of inducible promoters regulated by exogenously supplied promoters include the zinc-inducible sheep metallothionine (MT) promoter, the dexamethasone (Dex)-inducible mouse mammary tumor virus (MMTV) promoter, the T7 polymerase promoter system (WO 98/10088); the ecdysone insect promoter (No et al, Proc. Natl. Acad. Sci. USA, 93:3346-3351 (1996)), the tetracycline-repressible system (Gossen et al, Proc. Natl. Acad. Sci. USA, 89:5547-5551 (1992)), the tetracycline-inducible system (Gossen et al, Science, 268:1766-1769 (1995), see also Harvey et al, Curr. Opin. Chem. Biol., 2:512-518 (1998)), the RU486-inducible system (Wang et al, Nat. Biotech., 15:239-243 (1997) and Wang et al, Gene Ther., 4:432-441 (1997)) and the rapamycin-inducible system (Magari et al, J. Clin. Invest., 100:2865-2872 (1997)). Still other types of inducible promoters which may be useful in this context are those which are regulated by a specific physiological state, e.g., temperature, acute phase, a particular differentiation state of the cell, or in replicating cells only.
In another embodiment, the native promoter for the transgene will be used. The native promoter may be preferred when it is desired that expression of the transgene should mimic the native expression. The native promoter may be used when expression of the transgene must be regulated temporally or developmentally, or in a tissue-specific manner, or in response to specific transcriptional stimuli. In a further embodiment, other native expression control elements, such as enhancer elements, polyadenylation sites or Kozak consensus sequences may also be used to mimic the native expression.
In some embodiments, the regulatory sequences impart tissue-specific gene expression capabilities. In some cases, the tissue-specific regulatory sequences bind tissue-specific transcription factors that induce transcription in a tissue specific manner. Such tissue-specific regulatory sequences (e.g., promoters, enhancers, etc.) are well known in the art. Exemplary tissue-specific regulatory sequences include, but are not limited to the following tissue specific promoters: a liver-specific thyroxin binding globulin (TBG) promoter, an insulin promoter, a glucagon promoter, a somatostatin promoter, a pancreatic polypeptide (PPY) promoter, a synapsin-1 (Syn) promoter, a creatine kinase (MCK) promoter, a mammalian desmin (DES) promoter, a .alpha.-myosin heavy chain (a-MHC) promoter, or a cardiac Troponin T (cTnT) promoter. Other exemplary promoters include Beta-actin promoter, hepatitis B virus core promoter, Sandig et al., Gene Ther., 3:1002-9 (1996); alpha-fetoprotein (AFP) promoter, Arbuthnot et al., Hum. Gene Ther., 7:1503-14 (1996)), bone osteocalcin promoter (Stein et al., Mol. Biol. Rep., 24:185-96 (1997)); bone sialoprotein promoter (Chen et al., J. Bone Miner. Res., 11:654-64 (1996)), CD2 promoter (Hansal et al., J. Immunol., 161:1063-8 (1998); immunoglobulin heavy chain promoter; T cell receptor .alpha.-chain promoter, neuronal such as neuron-specific enolase (NSE) promoter (Andersen et al., Cell. Mol. Neurobiol., 13:503-15 (1993)), neurofilament light-chain gene promoter (Piccioli et al., Proc. Natl. Acad. Sci. USA, 88:5611-5 (1991)), and the neuron-specific vgf gene promoter (Piccioli et al., Neuron, 15:373-84 (1995)), among others which will be apparent to the skilled artisan.
In some embodiments, one or more bindings sites for one or more of miRNAs are incorporated in a transgene of a rAAV vector, to inhibit the expression of the transgene in one or more tissues of an subject harboring the transgene. The skilled artisan will appreciate that binding sites may be selected to control the expression of a transgene in a tissue specific manner. For example, binding sites for the liver-specific miR-122 may be incorporated into a transgene to inhibit expression of that transgene in the liver. The target sites in the mRNA may be in the 5' UTR, the 3' UTR or in the coding region. Typically, the target site is in the 3' UTR of the mRNA. Furthermore, the transgene may be designed such that multiple miRNAs regulate the mRNA by recognizing the same or multiple sites. The presence of multiple miRNA binding sites may result in the cooperative action of multiple RISCs and provide highly efficient inhibition of expression. The target site sequence may comprise a total of 5-100, 10-60, or more nucleotides. The target site sequence may comprise at least 5 nucleotides of the sequence of a target gene binding site.
Recombinant AAV Vector: Transgene Coding Sequences
The composition of the transgene sequence of the rAAV vector will depend upon the use to which the resulting vector will be put. For example, one type of transgene sequence includes a reporter sequence, which upon expression produces a detectable signal. In another example, the transgene encodes a therapeutic protein or therapeutic functional RNA. In another example, the transgene encodes a protein or functional RNA that is intended to be used for research purposes, e.g., to create a somatic transgenic animal model harboring the transgene, e.g., to study the function of the transgene product. In another example, the transgene encodes a protein or functional RNA that is intended to be used to create an animal model of disease. Appropriate transgene coding sequences will be apparent to the skilled artisan.
Also contemplated herein are methods of delivering a transgene to a subject using the rAAVs described herein. In some embodiments, the instant disclosure relates to a method for delivering a transgene to a subject comprising administering a rAAV to a subject, wherein the rAAV comprises: (i) a capsid protein having a terminally grafted nuclease, e.g., a nuclease having a sequence set forth as SEQ ID NO: 2, and optionally (ii) at least one transgene, e.g., a transgene encoding a gRNA, and wherein the rAAV infects cells of a target tissue of the subject. In some embodiments of the method, at least one transgene encodes a single guide RNA, a CRISPR RNA (crRNA), and/or a trans-activating crRNA (tracrRNA).
In some embodiments, the rAAV vectors may comprise a transgene, wherein the transgene is a gRNA. In some embodiments, the gRNA targets a nucleic acid sequence that causes disease in a subject. For example, expression of the huntingtin (Htt) gene causes Huntington's disease. Without wishing to be bound by any particular theory, a gRNA targeting the Htt gene directs Cas9 cleavage of the gene, thereby preventing its expression. Other similar genes (disease-associated or otherwise) can be targeted.
Recombinant AAV Administration Methods
The rAAVs may be delivered to a subject in compositions according to any appropriate methods known in the art. The rAAV, preferably suspended in a physiologically compatible carrier (i.e., in a composition), may be administered to a subject, i.e. host animal, such as a human, mouse, rat, cat, dog, sheep, rabbit, horse, cow, goat, pig, guinea pig, hamster, chicken, turkey, or a non-human primate (e.g., Macaque). In some embodiments a host animal does not include a human.
Delivery of the rAAVs to a mammalian subject may be by, for example, intramuscular injection or by administration into the bloodstream of the mammalian subject. Administration into the bloodstream may be by injection into a vein, an artery, or any other vascular conduit. In some embodiments, the rAAVs are administered into the bloodstream by way of isolated limb perfusion, a technique well known in the surgical arts, the method essentially enabling the artisan to isolate a limb from the systemic circulation prior to administration of the rAAV virions. A variant of the isolated limb perfusion technique, described in U.S. Pat. No. 6,177,403, can also be employed by the skilled artisan to administer the virions into the vasculature of an isolated limb to potentially enhance transduction into muscle cells or tissue. Moreover, in certain instances, it may be desirable to deliver the virions to the CNS of a subject. By "CNS" is meant all cells and tissue of the brain and spinal cord of a vertebrate. Thus, the term includes, but is not limited to, neuronal cells, glial cells, astrocytes, cereobrospinal fluid (CSF), interstitial spaces, bone, cartilage and the like. Recombinant AAVs may be delivered directly to the CNS or brain by injection into, e.g., the ventricular region, as well as to the striatum (e.g., the caudate nucleus or putamen of the striatum), spinal cord and neuromuscular junction, or cerebellar lobule, with a needle, catheter or related device, using neurosurgical techniques known in the art, such as by stereotactic injection (see, e.g., Stein et al., J Virol 73:3424-3429, 1999; Davidson et al., PNAS 97:3428-3432, 2000; Davidson et al., Nat. Genet. 3:219-223, 1993; and Alisky and Davidson, Hum. Gene Ther. 11:2315-2329, 2000).
Aspects of the instant disclosure relate to compositions comprising a recombinant AAV comprising a capsid protein having a terminally grafted (e.g., N-terminally grafted or C-terminally grafted) nuclease. In some embodiments, the nuclease is terminally grafted onto a capsid protein. In some embodiments, the a terminally grafted nuclease is present on all three capsid proteins (e.g. VP1, VP2, VP3) of the rAAV. In some embodiments, the terminally grafted nuclease is present on two of the capsid proteins (e.g. VP2 and VP3) of the rAAV. In some embodiments, the terminally grafted nuclease is present on a single capsid protein of the rAAV. In some embodiments, the terminally grafted nuclease is present on the VP2 capsid protein of the rAAV. In some embodiments, the composition further comprises a pharmaceutically acceptable carrier.
The compositions of the disclosure may comprise an rAAV alone, or in combination with one or more other viruses (e.g., a second rAAV encoding having one or more different transgenes). In some embodiments, a composition comprises 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more different rAAVs each having one or more different transgenes.
Suitable carriers may be readily selected by one of skill in the art in view of the indication for which the rAAV is directed. For example, one suitable carrier includes saline, which may be formulated with a variety of buffering solutions (e.g., phosphate buffered saline). Other exemplary carriers include sterile saline, lactose, sucrose, calcium phosphate, gelatin, dextran, agar, pectin, peanut oil, sesame oil, and water. The selection of the carrier is not a limitation of the present disclosure.
Optionally, the compositions of the disclosure may contain, in addition to the rAAV and carrier(s), other conventional pharmaceutical ingredients, such as preservatives, or chemical stabilizers. Suitable exemplary preservatives include chlorobutanol, potassium sorbate, sorbic acid, sulfur dioxide, propyl gallate, the parabens, ethyl vanillin, glycerin, phenol, and parachlorophenol. Suitable chemical stabilizers include gelatin and albumin.
The rAAVS are administered in sufficient amounts to transfect the cells of a desired tissue and to provide sufficient levels of gene transfer and expression without undue adverse effects. Conventional and pharmaceutically acceptable routes of administration include, but are not limited to, direct delivery to the selected organ (e.g., intraportal delivery to the liver), oral, inhalation (including intranasal and intratracheal delivery), intraocular, intravenous, intramuscular, subcutaneous, intradermal, intratumoral, and other parental routes of administration. Routes of administration may be combined, if desired.
The dose of rAAV virions required to achieve a particular "therapeutic effect," e.g., the units of dose in genome copies/per kilogram of body weight (GC/kg), will vary based on several factors including, but not limited to: the route of rAAV virion administration, the level of gene or RNA expression required to achieve a therapeutic effect, the specific disease or disorder being treated, and the stability of the gene or RNA product. One of skill in the art can readily determine a rAAV virion dose range to treat a patient having a particular disease or disorder based on the aforementioned factors, as well as other factors that are well known in the art.
An effective amount of an rAAV is an amount sufficient to target infect an animal, target a desired tissue. In some embodiments, an effective amount of an rAAV is an amount sufficient to produce a stable somatic transgenic animal model. The effective amount will depend primarily on factors such as the species, age, weight, health of the subject, and the tissue to be targeted, and may thus vary among animal and tissue. For example, an effective amount of the rAAV is generally in the range of from about 1 ml to about 100 ml of solution containing from about 10.sup.9 to 10.sup.16 genome copies. In some cases, a dosage between about 10.sup.11 to 10.sup.13 rAAV genome copies is appropriate. In certain embodiments, 10.sup.12 or 10.sup.13 rAAV genome copies is effective to target heart, liver, and pancreas tissues. In some cases, stable transgenic animals are produced by multiple doses of an rAAV.
In some embodiments, rAAV compositions are formulated to reduce aggregation of AAV particles in the composition, particularly where high rAAV concentrations are present (e.g., .about.10.sup.13 GC/ml or more). Methods for reducing aggregation of rAAVs are well known in the art and, include, for example, addition of surfactants, pH adjustment, salt concentration adjustment, etc. (See, e.g., Wright F R, et al., Molecular Therapy (2005) 12, 171-178, the contents of which are incorporated herein by reference.)
Formulation of pharmaceutically-acceptable excipients and carrier solutions is well-known to those of skill in the art, as is the development of suitable dosing and treatment regimens for using the particular compositions described herein in a variety of treatment regimens.
Typically, these formulations may contain at least about 0.1% of the active compound or more, although the percentage of the active ingredient(s) may, of course, be varied and may conveniently be between about 1 or 2% and about 70% or 80% or more of the weight or volume of the total formulation. Naturally, the amount of active compound in each therapeutically-useful composition may be prepared is such a way that a suitable dosage will be obtained in any given unit dose of the compound. Factors such as solubility, bioavailability, biological half-life, route of administration, product shelf life, as well as other pharmacological considerations will be contemplated by one skilled in the art of preparing such pharmaceutical formulations, and as such, a variety of dosages and treatment regimens may be desirable.
In certain circumstances it will be desirable to deliver the rAAV-based therapeutic constructs in suitably formulated pharmaceutical compositions disclosed herein either subcutaneously, intraopancreatically, intranasally, parenterally, intravenously, intramuscularly, intrathecally, or orally, intraperitoneally, or by inhalation. In some embodiments, the administration modalities as described in U.S. Pat. Nos. 5,543,158; 5,641,515 and 5,399,363 (each specifically incorporated herein by reference in its entirety) may be used to deliver rAAVs. In some embodiments, a preferred mode of administration is by portal vein injection.
The pharmaceutical forms suitable for injectable use include sterile aqueous solutions or dispersions and sterile powders for the extemporaneous preparation of sterile injectable solutions or dispersions. Dispersions may also be prepared in glycerol, liquid polyethylene glycols, and mixtures thereof and in oils. Under ordinary conditions of storage and use, these preparations contain a preservative to prevent the growth of microorganisms. In many cases the form is sterile and fluid to the extent that easy syringability exists. It must be stable under the conditions of manufacture and storage and must be preserved against the contaminating action of microorganisms, such as bacteria and fungi. The carrier can be a solvent or dispersion medium containing, for example, water, ethanol, polyol (e.g., glycerol, propylene glycol, and liquid polyethylene glycol, and the like), suitable mixtures thereof, and/or vegetable oils. Proper fluidity may be maintained, for example, by the use of a coating, such as lecithin, by the maintenance of the required particle size in the case of dispersion and by the use of surfactants. The prevention of the action of microorganisms can be brought about by various antibacterial and antifungal agents, for example, parabens, chlorobutanol, phenol, sorbic acid, thimerosal, and the like. In many cases, it will be preferable to include isotonic agents, for example, sugars or sodium chloride. Prolonged absorption of the injectable compositions can be brought about by the use in the compositions of agents delaying absorption, for example, aluminum monostearate and gelatin.
For administration of an injectable aqueous solution, for example, the solution may be suitably buffered, if necessary, and the liquid diluent first rendered isotonic with sufficient saline or glucose. These particular aqueous solutions are especially suitable for intravenous, intramuscular, subcutaneous and intraperitoneal administration. In this connection, a sterile aqueous medium that can be employed will be known to those of skill in the art. For example, one dosage may be dissolved in 1 ml of isotonic NaCl solution and either added to 1000 ml of hypodermoclysis fluid or injected at the proposed site of infusion, (see for example, "Remington's Pharmaceutical Sciences" 15th Edition, pages 1035-1038 and 1570-1580). Some variation in dosage will necessarily occur depending on the condition of the host. The person responsible for administration will, in any event, determine the appropriate dose for the individual host.
Sterile injectable solutions are prepared by incorporating the active rAAV in the required amount in the appropriate solvent with various of the other ingredients enumerated herein, as required, followed by filtered sterilization. Generally, dispersions are prepared by incorporating the various sterilized active ingredients into a sterile vehicle which contains the basic dispersion medium and the required other ingredients from those enumerated above. In the case of sterile powders for the preparation of sterile injectable solutions, the preferred methods of preparation are vacuum-drying and freeze-drying techniques which yield a powder of the active ingredient plus any additional desired ingredient from a previously sterile-filtered solution thereof.
The rAAV compositions disclosed herein may also be formulated in a neutral or salt form. Pharmaceutically-acceptable salts, include the acid addition salts (formed with the free amino groups of the protein) and which are formed with inorganic acids such as, for example, hydrochloric or phosphoric acids, or such organic acids as acetic, oxalic, tartaric, mandelic, and the like. Salts formed with the free carboxyl groups can also be derived from inorganic bases such as, for example, sodium, potassium, ammonium, calcium, or ferric hydroxides, and such organic bases as isopropylamine, trimethylamine, histidine, procaine and the like. Upon formulation, solutions will be administered in a manner compatible with the dosage formulation and in such amount as is therapeutically effective. The formulations are easily administered in a variety of dosage forms such as injectable solutions, drug-release capsules, and the like.
As used herein, "carrier" includes any and all solvents, dispersion media, vehicles, coatings, diluents, antibacterial and antifungal agents, isotonic and absorption delaying agents, buffers, carrier solutions, suspensions, colloids, and the like. The use of such media and agents for pharmaceutical active substances is well known in the art. Supplementary active ingredients can also be incorporated into the compositions. The phrase "pharmaceutically-acceptable" refers to molecular entities and compositions that do not produce an allergic or similar untoward reaction when administered to a host.
Delivery vehicles such as liposomes, nanocapsules, microparticles, microspheres, lipid particles, vesicles, and the like, may be used for the introduction of the compositions of the present disclosure into suitable host cells. In particular, the rAAV vector delivered transgenes may be formulated for delivery either encapsulated in a lipid particle, a liposome, a vesicle, a nanosphere, or a nanoparticle or the like.
Such formulations may be preferred for the introduction of pharmaceutically acceptable formulations of the nucleic acids or the rAAV constructs disclosed herein. The formation and use of liposomes is generally known to those of skill in the art. Recently, liposomes were developed with improved serum stability and circulation half-times (U.S. Pat. No. 5,741,516). Further, various methods of liposome and liposome like preparations as potential drug carriers have been described (U.S. Pat. Nos. 5,567,434; 5,552,157; 5,565,213; 5,738,868 and 5,795,587).
Liposomes have been used successfully with a number of cell types that are normally resistant to transfection by other procedures. In addition, liposomes are free of the DNA length constraints that are typical of viral-based delivery systems. Liposomes have been used effectively to introduce genes, drugs, radiotherapeutic agents, viruses, transcription factors and allosteric effectors into a variety of cultured cell lines and animals. In addition, several successful clinical trials examining the effectiveness of liposome-mediated drug delivery have been completed.
Liposomes are formed from phospholipids that are dispersed in an aqueous medium and spontaneously form multilamellar concentric bilayer vesicles (also termed multilamellar vesicles (MLVs). MLVs generally have diameters of from 25 nm to 4 .mu.m. Sonication of MLVs results in the formation of small unilamellar vesicles (SUVs) with diameters in the range of 200 to 500 ANG., containing an aqueous solution in the core.
Alternatively, nanocapsule formulations of the rAAV may be used. Nanocapsules can generally entrap substances in a stable and reproducible way. To avoid side effects due to intracellular polymeric overloading, such ultrafine particles (sized around 0.1 .mu.m) should be designed using polymers able to be degraded in vivo. Biodegradable polyalkyl-cyanoacrylate nanoparticles that meet these requirements are contemplated for use.
In addition to the methods of delivery described above, the following techniques are also contemplated as alternative methods of delivering the rAAV compositions to a host. Sonophoresis (e.g., ultrasound) has been used and described in U.S. Pat. No. 5,656,016 as a device for enhancing the rate and efficacy of drug permeation into and through the circulatory system. Other drug delivery alternatives contemplated are intraosseous injection (U.S. Pat. No. 5,779,708), microchip devices (U.S. Pat. No. 5,797,898), ophthalmic formulations (Bourlais et al., 1998), transdermal matrices (U.S. Pat. Nos. 5,770,219 and 5,783,208) and feedback-controlled delivery (U.S. Pat. No. 5,697,899).
Kits and Related Compositions
The agents described herein may, in some embodiments, be assembled into pharmaceutical or diagnostic or research kits to facilitate their use in therapeutic, diagnostic or research applications. A kit may include one or more containers housing the components of the disclosure and instructions for use. Specifically, such kits may include one or more agents described herein, along with instructions describing the intended application and the proper use of these agents. In certain embodiments agents in a kit may be in a pharmaceutical formulation and dosage suitable for a particular application and for a method of administration of the agents. Kits for research purposes may contain the components in appropriate concentrations or quantities for running various experiments.
In some embodiments, the instant disclosure relates to a kit for producing a rAAV, the kit comprising a container housing an isolated nucleic acid having a sequence of SEQ ID NO: 1 or SEQ ID NO: 3. In some embodiments, the kit further comprises instructions for producing the rAAV. In some embodiments, the kit further comprises at least one container housing a recombinant AAV vector, wherein the recombinant AAV vector comprises a transgene.
In some embodiments, the instant disclosure relates to a kit comprising a container housing a recombinant AAV having an isolated AAV capsid protein having an amino acid sequence as set forth in any of SEQ ID NO: 4.
The kit may be designed to facilitate use of the methods described herein by researchers and can take many forms. Each of the compositions of the kit, where applicable, may be provided in liquid form (e.g., in solution), or in solid form, (e.g., a dry powder). In certain cases, some of the compositions may be constitutable or otherwise processable (e.g., to an active form), for example, by the addition of a suitable solvent or other species (for example, water or a cell culture medium), which may or may not be provided with the kit. As used herein, "instructions" can define a component of instruction and/or promotion, and typically involve written instructions on or associated with packaging of the disclosure. Instructions also can include any oral or electronic instructions provided in any manner such that a user will clearly recognize that the instructions are to be associated with the kit, for example, audiovisual (e.g., videotape, DVD, etc.), Internet, and/or web-based communications, etc. The written instructions may be in a form prescribed by a governmental agency regulating the manufacture, use or sale of pharmaceuticals or biological products, which instructions can also reflects approval by the agency of manufacture, use or sale for animal administration.
The kit may contain any one or more of the components described herein in one or more containers. As an example, in one embodiment, the kit may include instructions for mixing one or more components of the kit and/or isolating and mixing a sample and applying to a subject. The kit may include a container housing agents described herein. The agents may be in the form of a liquid, gel or solid (powder). The agents may be prepared sterilely, packaged in syringe and shipped refrigerated. Alternatively it may be housed in a vial or other container for storage. A second container may have other agents prepared sterilely. Alternatively the kit may include the active agents premixed and shipped in a syringe, vial, tube, or other container. The kit may have one or more or all of the components required to administer the agents to an animal, such as a syringe, topical application devices, or iv needle tubing and bag, particularly in the case of the kits for producing specific somatic animal models.
In some cases, the methods involve transfecting cells with total cellular DNAs isolated from the tissues that potentially harbor proviral AAV genomes at very low abundance and supplementing with helper virus function (e.g., adenovirus) to trigger and/or boost AAV rep and cap gene transcription in the transfected cell. In some cases, RNA from the transfected cells provides a template for RT-PCR amplification of cDNA and the detection of novel AAVs. In cases where cells are transfected with total cellular DNAs isolated from the tissues that potentially harbor proviral AAV genomes, it is often desirable to supplement the cells with factors that promote AAV gene transcription. For example, the cells may also be infected with a helper virus, such as an Adenovirus or a Herpes Virus. In a specific embodiment, the helper functions are provided by an adenovirus. The adenovirus may be a wild-type adenovirus, and may be of human or non-human origin, preferably non-human primate (NHP) origin. Similarly adenoviruses known to infect non-human animals (e.g., chimpanzees, mouse) may also be employed in the methods of the disclosure (See, e.g., U.S. Pat. No. 6,083,716). In addition to wild-type adenoviruses, recombinant viruses or non-viral vectors (e.g., plasmids, episomes, etc.) carrying the necessary helper functions may be utilized. Such recombinant viruses are known in the art and may be prepared according to published techniques. See, e.g., U.S. Pat. Nos. 5,871,982 and 6,251,677, which describe a hybrid Ad/AAV virus. A variety of adenovirus strains are available from the American Type Culture Collection, Manassas, Va., or available by request from a variety of commercial and institutional sources. Further, the sequences of many such strains are available from a variety of databases including, e.g., PubMed and GenBank.
Cells may also be transfected with a vector (e.g., helper vector) which provides helper functions to the AAV. The vector providing helper functions may provide adenovirus functions, including, e.g., E1a, E1b, E2a, E4ORF6. The sequences of adenovirus gene providing these functions may be obtained from any known adenovirus serotype, such as serotypes 2, 3, 4, 7, 12 and 40, and further including any of the presently identified human types known in the art. Thus, in some embodiments, the methods involve transfecting the cell with a vector expressing one or more genes necessary for AAV replication, AAV gene transcription, and/or AAV packaging.
In some cases, a capsid gene can be used to construct and package recombinant AAV vectors, using methods well known in the art, to determine functional characteristics associated with the novel capsid protein encoded by the gene. For example, novel isolated capsid genes can be used to construct and package recombinant AAV (rAAV) vectors comprising a reporter gene (e.g., B-Galactosidase, GFP, Luciferase, etc.). The rAAV vector can then be delivered to an animal (e.g., mouse) and the tissue targeting properties of the novel isolated capsid gene can be determined by examining the expression of the reporter gene in various tissues (e.g., heart, liver, kidneys) of the animal. Other methods for characterizing the novel isolated capsid genes are disclosed herein and still others are well known in the art.
The kit may have a variety of forms, such as a blister pouch, a shrink wrapped pouch, a vacuum sealable pouch, a sealable thermoformed tray, or a similar pouch or tray form, with the accessories loosely packed within the pouch, one or more tubes, containers, a box or a bag. The kit may be sterilized after the accessories are added, thereby allowing the individual accessories in the container to be otherwise unwrapped. The kits can be sterilized using any appropriate sterilization techniques, such as radiation sterilization, heat sterilization, or other sterilization methods known in the art. The kit may also include other components, depending on the specific application, for example, containers, cell media, salts, buffers, reagents, syringes, needles, a fabric, such as gauze, for applying or removing a disinfecting agent, disposable gloves, a support for the agents prior to administration etc.
The instructions included within the kit may involve methods for detecting a latent AAV in a cell. In addition, kits of the disclosure may include, instructions, a negative and/or positive control, containers, diluents and buffers for the sample, sample preparation tubes and a printed or electronic table of reference AAV sequence for sequence comparisons.
EXAMPLES
Overview
To avoid the potential genotoxicity due to prolonged expression gene editing components, an endonuclease protein is transiently delivered and degrades naturally in the cell. Specifically, AAV capsid is used as a delivery vehicle to carry a Cas9 protein or other designer nuclease protein. AAV capsid consists of 60 copies of three capsid proteins, VP1, VP2 and VP3, at a ratio of 1:1:18. Although AAV capsid adopts a tightly packed structure, it has been shown that the VP2 protein with an N-terminal fusion protein can be incorporated into AAV capsid, and such a chimeric AAV is infectious.
Example 1
Results provided herein indicate that when Cas9 is fused to the N-terminus of VP2 (FIG. 1A), the resulting Cas9-VP2 fusion protein is functional.
A plasmid expressing S. pyogenes Cas9 (SpCas9; SEQ ID NOs: 1 and 2) fused with AAV2 VP2 was constructed (FIG. 1A). The resulting construct is represented by SEQ ID NO: 3 and the fusion protein is represented by SEQ ID NO: 4. Transfection of the construct into HEK293 cells yields a fusion protein product of expected size, as demonstrated by western blotting (FIG. 1B). A fluorescence reporter assay as illustrated in FIG. 2A was used to test if SpCas9-VP2 can function in gene editing. In the reporter construct, the GFP is disrupted by an insertion. The downstream out-of-frame T2A, when shifted to in-frame, mediates translation termination and re-initiation to produce mCherry reporter protein. In the presence of the sgRNA targeting the insertion in the GFP sequence and a functional SpCas9, indels by NHEJ shift the downstream T2A and mCherry to in-frame, thus giving mCherry fluorescence signal. Using this reporter system, SpCas9-VP2 induction of NHEJ by co-transfection in HEK293 cells was tested (FIG. 2B). Negative control cells expressing SpCas9 only or sgRNA only did not induce mCherry signal. Positive control cells, co-expressing sgRNA and SpCas9 yielded mCherry signal. When sgRNA and the SpCas9-VP2 fusion were co-expressed, mCherry fluorescence was also observed, demonstrating that the SpCas-VP2 fusion protein behaves similarly as SpCas9 in inducing gene editing and NHEJ (FIG. 2B).
SpCas-VP2 can be also incorporated into AAV2 capsid to form AAV virion. The start codon of VP2 is mutated in the cap gene from the trans AAV production plasmid. The omission of VP2 expression from this plasmid in HEK293 cells is validated by western blotting using an antibody targeting a C-terminal epitope shared by VP1, VP2 and VP3. Small-scale AAV production is performed using the VP2 null-trans plasmid and the SpCas9-VP2 in replacement of the original trans plasmid to examine the presence of Cas9 protein covalently linked to the outer surface of AAV2 virion. ELISA is performed using antibodies recognizing a fully assembled AAV2 virion and the HA-tagged SpCas9. Alternatively, immuno electron microscopy is performed to visualize the presence of HA-tagged SpCas9 immunoreactivity outside of AAV2 virion. Next, a small-scale AAV production-infection assay is performed to validate that SpCas9-AAV delivers the SpCas9 into HEK293 cells and mediates gene editing. The same reporter system as illustrated in FIG. 2B is used for this assay.
Serials of in vivo experiments using SpCas9-AAV2 expressing EGFP and sgRNA targeting the mouse ROSA26 locus (SpCas9-AAV2-EGFP-sgROSA26) obtained from large-scale production are next performed. The tropism of SpCas9-AAV2 is characterized in mice by systemic delivery. Wild-type C57BL/6J mice are injected with SpCas9-AAV2-EGFP-sgROSA26 at postnatal day 1 (P1) via facial vein and at 8 weeks old via tail vein, respectively. The mice are sacrificed 3 weeks after injection and fixed. Tissues including liver, heart, skeletal muscle, pancreas, adrenal gland, kidneys, spleen, brain, and spinal cord are analyzed for EGFP expression by immunofluorescence staining. The best transduced tissue(s) are selected to demonstrate SpCas9-AAV mediated gene editing of ROSA26 locus in vivo in another group of mice treated in the same manner, from which fresh tissues are harvested and genomic DNA extracted. The gene editing events represented by random indels near the sgRNA targeting site in the ROSA26 locus are investigated using Surveyor assay and single DNA molecule sequencing.
To demonstrate the improved safety profile of the SpCas9 transiently delivered using SpCas9-AAV2 and contrast with prolonged expression of SpCas9 from a conventional rAAV2 vector, SpCas9-AAV2 are packaged with transgene cassettes expressing sgRNAs with reported off-target effects in mouse genome, and inject into mice. The gene editing events at both on- and off-target genomic DNA loci are analyzed by Surveyor assay and single DNA molecule sequencing. Transient delivery of SpCas9 significantly reduces the chance of off-target effect.
Example 2
Intein-mediated protein trans-splicing (PTS) is used to fuse SpCas9 protein with VP2 after AAV assembly as illustrated in FIG. 3A. For example, the naturally split intein Npu DnaE, which has the most robust trans-splicing activity identified so far, is used to fuse SpCas9 protein with VP2 by PTS. The SpCas9 carries IntN, and VP2 carries IntC. Since IntC comprises only 36 amino acid residues, an IntC-VP2 fusion is amenable to AAV assembly. First, IntC-AAV2 virion and SpCas9-IntN protein are produced and purified separately, and then incubated to allow for PTS to occur in vitro. Intein-mediated PTS is a spontaneous reaction and does not require other co-factors. The fast kinetic nature of Npu DnaE split intein produces SpCas9-AAV2 fusion protein rapidly. The fusion protein is further purified by dialysis.
Alternatively, IntC is fused with a truncated C-terminal portion of SpCas9 onto AAV2 capsid to allow for in vivo transduction. The rest portion of SpCas9 and IntN are encoded by a transgene expression cassette as rAAV genome (FIG. 3B). Co-delivery of the two portions of SpCas9 reconstitutes the full-length, functional SpCas9. Importantly, as the IntC-SpCas9C protein is degraded naturally, the long-term expression of SpCas9N-IntN only is non-functional, thus mitigating off-target effects.
FIG. 4 shows one example of an expression construct comprising a nucleic acid sequence encoding SpCas9 nuclease N-terminally fused to VP2 capsid protein.
Example 3
Gene editing platforms, such as the Cas9/sgRNA system, have been shown to induce off-target DNA double-stranded breaks (DSBs) throughout genomes, which is associated with toxicity. Such off-target effects not only stem from ambiguity of DNA sequence recognition by nucleases, but also can be attributed to the prolonged presence of an active gene editing system in a given cell. As a result, off-target DSBs accumulate over time, and ultimately lead to genotoxicity. To mitigate the potential toxicity due to prolonged expression of gene editing system in vivo, transient delivery of endonuclease protein, which induces permanent gene editing followed by natural degradation of the endonuclease, was examined. Specifically, the VP2 protein of AAV capsid was used as a protein delivery vehicle to ferry the Cas9 protein in vivo.
A sensitive gene editing reporter plasmid was constructed. Co-transfection of the reporter plasmid and a plasmid expressing the SpCas9-VP2 fusion protein induced gene editing in HEK293 cells. An rAAV packaging system was modified to include a plasmid expressing VP1 and VP3, and another plasmid expressing either the SpCas9-VP2 fusion protein or the EGFP-VP2 fusion protein. EGFP-AAV2 (EGFP protein grafted on the AAV2 capsid) was successfully produced. However, rAAV particles carrying SpCas9 protein were not produced, likely because the large size of SpCas9 protein interfered with the AAV packaging process.
The transgene encoding SpCas9 was split into halves to utilize split intein-mediated protein trans-splicing (PTS) to transiently reconstitute the full-length SpCas9 (FIG. 3A). When the two parts of a split intein (termed Int.sub.N and Int.sub.C, respectively) fuse, the split intein mediates PTS, resulting in the generation of a fusion protein with the intein being spliced out. Plasmids expressing the fusion proteins SpCas9.sub.N-Int.sub.N and Int.sub.C-SpCas9.sub.C-VP2 (pU1a-Cas9.sub.n-Int.sub.n and Int.sub.cCas9.sub.c-( )-VP2, respectively) were generated. The designation "( )" in the Int.sub.c-Cas9.sub.c plasmid represents the presence of a linker sequence (e.g., GS, GGGGSx3 or EAAAKx3). Results show productive intein-mediated reconstitution of SpCas9-VP2 protein in HEK293 cells by co-transfection (FIG. 5). Importantly, co-transfection of plasmids expressing SpCas9.sub.N-Int.sub.N and Int.sub.C-SpCas9.sub.C-VP2 in HEK293 cells led to gene editing based on the EGFP-ON reporter assay, as shown in FIG. 6. EGFP fluorescence reports Cas9 cleavage and NHEJ repair and mCherry is constitutively expressed as a control.
The Int.sub.C-SpCas9.sub.C protein to be grafted on VP2 is equal or smaller than EGFP. Since EGFP-AAV2 successfully produced, rAAV packaging of the Int.sub.C-SpCas9.sub.C-VP2 was investigated. Guided by structural analysis, SpCas9 split sites close to the C-terminus of SpCas9 were strategically screened and identified. Cells were transfected with a plasmid encoding VP1 and VP3 proteins, and a second plasmid encoding Int.sub.c-Cas9.sub.c-( )-VP2. Results indicate successful incorporation of Int.sub.c-SpCas9.sub.c-VP2 polypeptide onto rAAV2 capsid (FIG. 7).
TABLE-US-00001 SEQUENCES >SEQ ID NO: 1 SpCas9 nucleic acid sequence ATGTACCCATACGATGTTCCAGATTACGCTTCGCCGAAGAAAAAGCGCAAGGTC GAAGCGTCCGACAAGAAGTACAGCATCGGCCTGGACATCGGCACCAACTCTGTG GGCTGGGCCGTGATCACCGACGAGTACAAGGTGCCCAGCAAGAAATTCAAGGTG CTGGGCAACACCGACCGGCACAGCATCAAGAAGAACCTGATCGGAGCCCTGCTG TTCGACAGCGGCGAAACAGCCGAGGCCACCCGGCTGAAGAGAACCGCCAGAAG AAGATACACCAGACGGAAGAACCGGATCTGCTATCTGCAAGAGATCTTCAGCAA CGAGATGGCCAAGGTGGACGACAGCTTCTTCCACAGACTGGAAGAGTCCTTCCT GGTGGAAGAGGATAAGAAGCACGAGCGGCACCCCATCTTCGGCAACATCGTGGA CGAGGTGGCCTACCACGAGAAGTACCCCACCATCTACCACCTGAGAAAGAAACT GGTGGACAGCACCGACAAGGCCGACCTGCGGCTGATCTATCTGGCCCTGGCCCA CATGATCAAGTTCCGGGGCCACTTCCTGATCGAGGGCGACCTGAACCCCGACAA CAGCGACGTGGACAAGCTGTTCATCCAGCTGGTGCAGACCTACAACCAGCTGTTC GAGGAAAACCCCATCAACGCCAGCGGCGTGGACGCCAAGGCCATCCTGTCTGCC AGACTGAGCAAGAGCAGACGGCTGGAAAATCTGATCGCCCAGCTGCCCGGCGAG AAGAAGAATGGCCTGTTCGGCAACCTGATTGCCCTGAGCCTGGGCCTGACCCCCA ACTTCAAGAGCAACTTCGACCTGGCCGAGGATGCCAAACTGCAGCTGAGCAAGG ACACCTACGACGACGACCTGGACAACCTGCTGGCCCAGATCGGCGACCAGTACG CCGACCTGTTTCTGGCCGCCAAGAACCTGTCCGACGCCATCCTGCTGAGCGACAT CCTGAGAGTGAACACCGAGATCACCAAGGCCCCCCTGAGCGCCTCTATGATCAA GAGATACGACGAGCACCACCAGGACCTGACCCTGCTGAAAGCTCTCGTGCGGCA GCAGCTGCCTGAGAAGTACAAAGAGATTTTCTTCGACCAGAGCAAGAACGGCTA CGCCGGCTACATTGACGGCGGAGCCAGCCAGGAAGAGTTCTACAAGTTCATCAA GCCCATCCTGGAAAAGATGGACGGCACCGAGGAACTGCTCGTGAAGCTGAACAG AGAGGACCTGCTGCGGAAGCAGCGGACCTTCGACAACGGCAGCATCCCCCACCA GATCCACCTGGGAGAGCTGCACGCCATTCTGCGGCGGCAGGAAGATTTTTACCCA TTCCTGAAGGACAACCGGGAAAAGATCGAGAAGATCCTGACCTTCCGCATCCCC TACTACGTGGGCCCTCTGGCCAGGGGAAACAGCAGATTCGCCTGGATGACCAGA AAGAGCGAGGAAACCATCACCCCCTGGAACTTCGAGGAAGTGGTGGACAAGGGC GCTTCCGCCCAGAGCTTCATCGAGCGGATGACCAACTTCGATAAGAACCTGCCCA ACGAGAAGGTGCTGCCCAAGCACAGCCTGCTGTACGAGTACTTCACCGTGTATA ACGAGCTGACCAAAGTGAAATACGTGACCGAGGGAATGAGAAAGCCCGCCTTCC TGAGCGGCGAGCAGAAAAAGGCCATCGTGGACCTGCTGTTCAAGACCAACCGGA AAGTGACCGTGAAGCAGCTGAAAGAGGACTACTTCAAGAAAATCGAGTGCTTCG ACTCCGTGGAAATCTCCGGCGTGGAAGATCGGTTCAACGCCTCCCTGGGCACATA CCACGATCTGCTGAAAATTATCAAGGACAAGGACTTCCTGGACAATGAGGAAAA CGAGGACATTCTGGAAGATATCGTGCTGACCCTGACACTGTTTGAGGACAGAGA GATGATCGAGGAACGGCTGAAAACCTATGCCCACCTGTTCGACGACAAAGTGAT GAAGCAGCTGAAGCGGCGGAGATACACCGGCTGGGGCAGGCTGAGCCGGAAGC TGATCAACGGCATCCGGGACAAGCAGTCCGGCAAGACAATCCTGGATTTCCTGA AGTCCGACGGCTTCGCCAACAGAAACTTCATGCAGCTGATCCACGACGACAGCC TGACCTTTAAAGAGGACATCCAGAAAGCCCAGGTGTCCGGCCAGGGCGATAGCC TGCACGAGCACATTGCCAATCTGGCCGGCAGCCCCGCCATTAAGAAGGGCATCC TGCAGACAGTGAAGGTGGTGGACGAGCTCGTGAAAGTGATGGGCCGGCACAAGC CCGAGAACATCGTGATCGAAATGGCCAGAGAGAACCAGACCACCCAGAAGGGA CAGAAGAACAGCCGCGAGAGAATGAAGCGGATCGAAGAGGGCATCAAAGAGCT GGGCAGCCAGATCCTGAAAGAACACCCCGTGGAAAACACCCAGCTGCAGAACGA GAAGCTGTACCTGTACTACCTGCAGAATGGGCGGGATATGTACGTGGACCAGGA ACTGGACATCAACCGGCTGTCCGACTACGATGTGGACCATATCGTGCCTCAGAGC TTTCTGAAGGACGACTCCATCGACAACAAGGTGCTGACCAGAAGCGACAAGAAC CGGGGCAAGAGCGACAACGTGCCCTCCGAAGAGGTCGTGAAGAAGATGAAGAA CTACTGGCGGCAGCTGCTGAACGCCAAGCTGATTACCCAGAGAAAGTTCGACAA TCTGACCAAGGCCGAGAGAGGCGGCCTGAGCGAACTGGATAAGGCCGGCTTCAT CAAGAGACAGCTGGTGGAAACCCGGCAGATCACAAAGCACGTGGCACAGATCCT GGACTCCCGGATGAACACTAAGTACGACGAGAATGACAAGCTGATCCGGGAAGT GAAAGTGATCACCCTGAAGTCCAAGCTGGTGTCCGATTTCCGGAAGGATTTCCAG TTTTACAAAGTGCGCGAGATCAACAACTACCACCACGCCCACGACGCCTACCTGA ACGCCGTCGTGGGAACCGCCCTGATCAAAAAGTACCCTAAGCTGGAAAGCGAGT TCGTGTACGGCGACTACAAGGTGTACGACGTGCGGAAGATGATCGCCAAGAGCG AGCAGGAAATCGGCAAGGCTACCGCCAAGTACTTCTTCTACAGCAACATCATGA ACTTTTTCAAGACCGAGATTACCCTGGCCAACGGCGAGATCCGGAAGCGGCCTCT GATCGAGACAAACGGCGAAACCGGGGAGATCGTGTGGGATAAGGGCCGGGATTT TGCCACCGTGCGGAAAGTGCTGAGCATGCCCCAAGTGAATATCGTGAAAAAGAC CGAGGTGCAGACAGGCGGCTTCAGCAAAGAGTCTATCCTGCCCAAGAGGAACAG CGATAAGCTGATCGCCAGAAAGAAGGACTGGGACCCTAAGAAGTACGGCGGCTT CGACAGCCCCACCGTGGCCTATTCTGTGCTGGTGGTGGCCAAAGTGGAAAAGGG CAAGTCCAAGAAACTGAAGAGTGTGAAAGAGCTGCTGGGGATCACCATCATGGA AAGAAGCAGCTTCGAGAAGAATCCCATCGACTTTCTGGAAGCCAAGGGCTACAA AGAAGTGAAAAAGGACCTGATCATCAAGCTGCCTAAGTACTCCCTGTTCGAGCT GGAAAACGGCCGGAAGAGAATGCTGGCCTCTGCCGGCGAACTGCAGAAGGGAA ACGAACTGGCCCTGCCCTCCAAATATGTGAACTTCCTGTACCTGGCCAGCCACTA TGAGAAGCTGAAGGGCTCCCCCGAGGATAATGAGCAGAAACAGCTGTTTGTGGA ACAGCACAAGCACTACCTGGACGAGATCATCGAGCAGATCAGCGAGTTCTCCAA GAGAGTGATCCTGGCCGACGCTAATCTGGACAAAGTGCTGTCCGCCTACAACAA GCACCGGGATAAGCCCATCAGAGAGCAGGCCGAGAATATCATCCACCTGTTTAC CCTGACCAATCTGGGAGCCCCTGCCGCCTTCAAGTACTTTGACACCACCATCGAC CGGAAGAGGTACACCAGCACCAAAGAGGTGCTGGACGCCACCCTGATCCACCAG AGCATCACCGGCCTGTACGAGACACGGATCGACCTGTCTCAGCTGGGAGGCGAC AGCCCCAAGAAGAAGAGAAAGGTGGAGGCCAGC >SEQ ID NO: 2 SpCas9 amino acid sequence MYPYDVPDYASPKKKRKVEASDKKYSIGLDIGTNSVGWAVITDEYKVPSKKFKVLG NTDRHSIKKNLIGALLFDSGETAEATRLKRTARRRYTRRKNRICYLQEIFSNEMAKVD DSFFHRLEESFLVEEDKKHERHPIFGNIVDEVAYHEKYPTIYHLRKKLVDSTDKADLR LIYLALAHMIKFRGHFLIEGDLNPDNSDVDKLFIQLVQTYNQLFEENPINASGVDAKA ILSARLSKSRRLENLIAQLPGEKKNGLFGNLIALSLGLTPNFKSNFDLAEDAKLQLSKD TYDDDLDNLLAQIGDQYADLFLAAKNLSDAILLSDILRVNTEITKAPLSASMIKRYDE HHQDLTLLKALVRQQLPEKYKEIFFDQSKNGYAGYIDGGASQEEFYKFIKPILEKMD GTEELLVKLNREDLLRKQRTFDNGSIPHQIHLGELHAILRRQEDFYPFLKDNREKIEKI LTFRIPYYVGPLARGNSRFAWMTRKSEETITPWNFEEVVDKGASAQSFIERMTNFDK NLPNEKVLPKHSLLYEYFTVYNELTKVKYVTEGMRKPAFLSGEQKKAIVDLLFKTNR KVTVKQLKEDYFKKIECFDSVEISGVEDRFNASLGTYHDLLKIIKDKDFLDNEENEDI LEDIVLTLTLFEDREMIEERLKTYAHLFDDKVMKQLKRRRYTGWGRLSRKUNGIRD KQSGKTILDFLKSDGFANRNFMQLIHDDSLTFKEDIQKAQVSGQGDSLHEHIANLAG SPAIKKGILQTVKVVDELVKVMGRHKPENIVIEMARENQTTQKGQKNSRERMKRIEE GIKELGSQILKEHPVENTQLQNEKLYLYYLQNGRDMYVDQELDINRLSDYDVDHIVP QSFLKDDSIDNKVLTRSDKNRGKSDNVPSEEVVKKMKNYWRQLLNAKLITQRKFDN LTKAERGGLSELDKAGFIKRQLVETRQITKHVAQILDSRMNTKYDENDKLIREVKVIT LKSKLVSDFRKDFQFYKVREINNYHHAHDAYLNAVVGTALIKKYPKLESEFVYGDY KVYDVRKMIAKSEQEIGKATAKYFFYSNIMNFFKTEITLANGEIRKRPLIETNGETGEI VWDKGRDFATVRKVLSMPQVNIVKKTEVQTGGFSKESILPKRNSDKLIARKKDWDP KKYGGFDSPTVAYSVLVVAKVEKGKSKKLKSVKELLGITIMERSSFEKNPIDFLEAK GYKEVKKDLIIKLPKYSLFELENGRKRMLASAGELQKGNELALPSKYVNFLYLASHY EKLKGSPEDNEQKQLFVEQHKHYLDEIIEQISEFSKRVILADANLDKVLSAYNKHRDK PIREQAENIIHLFTLTNLGAPAAFKYFDTTIDRKRYTSTKEVLDATLIHQSITGLYETRI DLSQLGGDSPKKKRKVEAS >SEQ ID NO: 3 SpCas9-VP2 fusion nucleic acid ATGTACCCATACGATGTTCCAGATTACGCTTCGCCGAAGAAAAAGCGCAAGGTC GAAGCGTCCGACAAGAAGTACAGCATCGGCCTGGACATCGGCACCAACTCTGTG GGCTGGGCCGTGATCACCGACGAGTACAAGGTGCCCAGCAAGAAATTCAAGGTG CTGGGCAACACCGACCGGCACAGCATCAAGAAGAACCTGATCGGAGCCCTGCTG TTCGACAGCGGCGAAACAGCCGAGGCCACCCGGCTGAAGAGAACCGCCAGAAG AAGATACACCAGACGGAAGAACCGGATCTGCTATCTGCAAGAGATCTTCAGCAA CGAGATGGCCAAGGTGGACGACAGCTTCTTCCACAGACTGGAAGAGTCCTTCCT GGTGGAAGAGGATAAGAAGCACGAGCGGCACCCCATCTTCGGCAACATCGTGGA CGAGGTGGCCTACCACGAGAAGTACCCCACCATCTACCACCTGAGAAAGAAACT GGTGGACAGCACCGACAAGGCCGACCTGCGGCTGATCTATCTGGCCCTGGCCCA CATGATCAAGTTCCGGGGCCACTTCCTGATCGAGGGCGACCTGAACCCCGACAA CAGCGACGTGGACAAGCTGTTCATCCAGCTGGTGCAGACCTACAACCAGCTGTTC GAGGAAAACCCCATCAACGCCAGCGGCGTGGACGCCAAGGCCATCCTGTCTGCC AGACTGAGCAAGAGCAGACGGCTGGAAAATCTGATCGCCCAGCTGCCCGGCGAG AAGAAGAATGGCCTGTTCGGCAACCTGATTGCCCTGAGCCTGGGCCTGACCCCCA ACTTCAAGAGCAACTTCGACCTGGCCGAGGATGCCAAACTGCAGCTGAGCAAGG ACACCTACGACGACGACCTGGACAACCTGCTGGCCCAGATCGGCGACCAGTACG CCGACCTGTTTCTGGCCGCCAAGAACCTGTCCGACGCCATCCTGCTGAGCGACAT CCTGAGAGTGAACACCGAGATCACCAAGGCCCCCCTGAGCGCCTCTATGATCAA GAGATACGACGAGCACCACCAGGACCTGACCCTGCTGAAAGCTCTCGTGCGGCA GCAGCTGCCTGAGAAGTACAAAGAGATTTTCTTCGACCAGAGCAAGAACGGCTA CGCCGGCTACATTGACGGCGGAGCCAGCCAGGAAGAGTTCTACAAGTTCATCAA GCCCATCCTGGAAAAGATGGACGGCACCGAGGAACTGCTCGTGAAGCTGAACAG AGAGGACCTGCTGCGGAAGCAGCGGACCTTCGACAACGGCAGCATCCCCCACCA GATCCACCTGGGAGAGCTGCACGCCATTCTGCGGCGGCAGGAAGATTTTTACCCA TTCCTGAAGGACAACCGGGAAAAGATCGAGAAGATCCTGACCTTCCGCATCCCC TACTACGTGGGCCCTCTGGCCAGGGGAAACAGCAGATTCGCCTGGATGACCAGA AAGAGCGAGGAAACCATCACCCCCTGGAACTTCGAGGAAGTGGTGGACAAGGGC GCTTCCGCCCAGAGCTTCATCGAGCGGATGACCAACTTCGATAAGAACCTGCCCA ACGAGAAGGTGCTGCCCAAGCACAGCCTGCTGTACGAGTACTTCACCGTGTATA ACGAGCTGACCAAAGTGAAATACGTGACCGAGGGAATGAGAAAGCCCGCCTTCC TGAGCGGCGAGCAGAAAAAGGCCATCGTGGACCTGCTGTTCAAGACCAACCGGA AAGTGACCGTGAAGCAGCTGAAAGAGGACTACTTCAAGAAAATCGAGTGCTTCG ACTCCGTGGAAATCTCCGGCGTGGAAGATCGGTTCAACGCCTCCCTGGGCACATA CCACGATCTGCTGAAAATTATCAAGGACAAGGACTTCCTGGACAATGAGGAAAA CGAGGACATTCTGGAAGATATCGTGCTGACCCTGACACTGTTTGAGGACAGAGA GATGATCGAGGAACGGCTGAAAACCTATGCCCACCTGTTCGACGACAAAGTGAT GAAGCAGCTGAAGCGGCGGAGATACACCGGCTGGGGCAGGCTGAGCCGGAAGC TGATCAACGGCATCCGGGACAAGCAGTCCGGCAAGACAATCCTGGATTTCCTGA AGTCCGACGGCTTCGCCAACAGAAACTTCATGCAGCTGATCCACGACGACAGCC TGACCTTTAAAGAGGACATCCAGAAAGCCCAGGTGTCCGGCCAGGGCGATAGCC TGCACGAGCACATTGCCAATCTGGCCGGCAGCCCCGCCATTAAGAAGGGCATCC TGCAGACAGTGAAGGTGGTGGACGAGCTCGTGAAAGTGATGGGCCGGCACAAGC CCGAGAACATCGTGATCGAAATGGCCAGAGAGAACCAGACCACCCAGAAGGGA CAGAAGAACAGCCGCGAGAGAATGAAGCGGATCGAAGAGGGCATCAAAGAGCT GGGCAGCCAGATCCTGAAAGAACACCCCGTGGAAAACACCCAGCTGCAGAACGA GAAGCTGTACCTGTACTACCTGCAGAATGGGCGGGATATGTACGTGGACCAGGA ACTGGACATCAACCGGCTGTCCGACTACGATGTGGACCATATCGTGCCTCAGAGC TTTCTGAAGGACGACTCCATCGACAACAAGGTGCTGACCAGAAGCGACAAGAAC CGGGGCAAGAGCGACAACGTGCCCTCCGAAGAGGTCGTGAAGAAGATGAAGAA CTACTGGCGGCAGCTGCTGAACGCCAAGCTGATTACCCAGAGAAAGTTCGACAA TCTGACCAAGGCCGAGAGAGGCGGCCTGAGCGAACTGGATAAGGCCGGCTTCAT CAAGAGACAGCTGGTGGAAACCCGGCAGATCACAAAGCACGTGGCACAGATCCT GGACTCCCGGATGAACACTAAGTACGACGAGAATGACAAGCTGATCCGGGAAGT GAAAGTGATCACCCTGAAGTCCAAGCTGGTGTCCGATTTCCGGAAGGATTTCCAG TTTTACAAAGTGCGCGAGATCAACAACTACCACCACGCCCACGACGCCTACCTGA ACGCCGTCGTGGGAACCGCCCTGATCAAAAAGTACCCTAAGCTGGAAAGCGAGT TCGTGTACGGCGACTACAAGGTGTACGACGTGCGGAAGATGATCGCCAAGAGCG AGCAGGAAATCGGCAAGGCTACCGCCAAGTACTTCTTCTACAGCAACATCATGA ACTTTTTCAAGACCGAGATTACCCTGGCCAACGGCGAGATCCGGAAGCGGCCTCT GATCGAGACAAACGGCGAAACCGGGGAGATCGTGTGGGATAAGGGCCGGGATTT TGCCACCGTGCGGAAAGTGCTGAGCATGCCCCAAGTGAATATCGTGAAAAAGAC CGAGGTGCAGACAGGCGGCTTCAGCAAAGAGTCTATCCTGCCCAAGAGGAACAG CGATAAGCTGATCGCCAGAAAGAAGGACTGGGACCCTAAGAAGTACGGCGGCTT CGACAGCCCCACCGTGGCCTATTCTGTGCTGGTGGTGGCCAAAGTGGAAAAGGG CAAGTCCAAGAAACTGAAGAGTGTGAAAGAGCTGCTGGGGATCACCATCATGGA AAGAAGCAGCTTCGAGAAGAATCCCATCGACTTTCTGGAAGCCAAGGGCTACAA AGAAGTGAAAAAGGACCTGATCATCAAGCTGCCTAAGTACTCCCTGTTCGAGCT GGAAAACGGCCGGAAGAGAATGCTGGCCTCTGCCGGCGAACTGCAGAAGGGAA ACGAACTGGCCCTGCCCTCCAAATATGTGAACTTCCTGTACCTGGCCAGCCACTA TGAGAAGCTGAAGGGCTCCCCCGAGGATAATGAGCAGAAACAGCTGTTTGTGGA ACAGCACAAGCACTACCTGGACGAGATCATCGAGCAGATCAGCGAGTTCTCCAA GAGAGTGATCCTGGCCGACGCTAATCTGGACAAAGTGCTGTCCGCCTACAACAA GCACCGGGATAAGCCCATCAGAGAGCAGGCCGAGAATATCATCCACCTGTTTAC CCTGACCAATCTGGGAGCCCCTGCCGCCTTCAAGTACTTTGACACCACCATCGAC CGGAAGAGGTACACCAGCACCAAAGAGGTGCTGGACGCCACCCTGATCCACCAG AGCATCACCGGCCTGTACGAGACACGGATCGACCTGTCTCAGCTGGGAGGCGAC AGCCCCAAGAAGAAGAGAAAGGTGGAGGCCAGCGAATTGGCTCCGGGAAAAAA GAGGCCGGTAGAGCACTCTCCTGTGGAGCCAGACTCCTCCTCGGGAACCGGAAA GGCGGGCCAGCAGCCTGCAAGAAAAAGATTGAATTTTGGTCAGACTGGAGACGC AGACTCAGTACCTGACCCCCAGCCTCTCGGACAGCCACCAGCAGCCCCCTCTGGT CTGGGAACTAATACGATGGCTACAGGCAGTGGCGCACCAATGGCAGACAATAAC GAGGGCGCCGACGGAGTGGGTAATTCCTCGGGAAATTGGCATTGCGATTCCACA TGGATGGGCGACAGAGTCATCACCACCAGCACCCGAACCTGGGCCCTGCCCACC TACAACAACCACCTCTACAAACAAATTTCCAGCCAATCAGGAGCCTCGAACGAC AATCACTACTTTGGCTACAGCACCCCTTGGGGGTATTTTGACTTCAACAGATTCC ACTGCCACTTTTCACCACGTGACTGGCAAAGACTCATCAACAACAACTGGGGATT CCGACCCAAGAGACTCAACTTCAAGCTCTTTAACATTCAAGTCAAAGAGGTCACG CAGAATGACGGTACGACGACGATTGCCAATAACCTTACCAGCACGGTTCAGGTG TTTACTGACTCGGAGTACCAGCTCCCGTACGTCCTCGGCTCGGCGCATCAAGGAT GCCTCCCGCCGTTCCCAGCAGACGTCTTCATGGTGCCACAGTATGGATACCTCAC CCTGAACAACGGGAGTCAGGCAGTAGGACGCTCTTCATTTTACTGCCTGGAGTAC TTTCCTTCTCAGATGCTGCGTACCGGAAACAACTTTACCTTCAGCTACACTTTTGA GGACGTTCCTTTCCACAGCAGCTACGCTCACAGCCAGAGTCTGGACCGTCTCATG AATCCTCTCATCGACCAGTACCTGTATTACTTGAGCAGAACAAACACTCCAAGTG GAACCACCACGCAGTCAAGGCTTCAGTTTTCTCAGGCCGGAGCGAGTGACATTCG GGACCAGTCTAGGAACTGGCTTCCTGGACCCTGTTACCGCCAGCAGCGAGTATCA AAGACATCTGCGGATAACAACAACAGTGAATACTCGTGGACTGGAGCTACCAAG TACCACCTCAATGGCAGAGACTCTCTGGTGAATCCGGGCCCGGCCATGGCAAGC CACAAGGACGATGAAGAAAAGTTTTTTCCTCAGAGCGGGGTTCTCATCTTTGGGA AGCAAGGCTCAGAGAAAACAAATGTGGACATTGAAAAGGTCATGATTACAGACG AAGAGGAAATCAGGACAACCAATCCCGTGGCTACGGAGCAGTATGGTTCTGTAT CTACCAACCTCCAGAGAGGCAACAGACAAGCAGCTACCGCAGATGTCAACACAC AAGGCGTTCTTCCAGGCATGGTCTGGCAGGACAGAGATGTGTACCTTCAGGGGC CCATCTGGGCAAAGATTCCACACACGGACGGACATTTTCACCCCTCTCCCCTCAT GGGTGGATTCGGACTTAAACACCCTCCTCCACAGATTCTCATCAAGAACACCCCG GTACCTGCGAATCCTTCGACCACCTTCAGTGCGGCAAAGTTTGCTTCCTTCATCAC ACAGTACTCCACGGGACAGGTCAGCGTGGAGATCGAGTGGGAGCTGCAGAAGGA AAACAGCAAACGCTGGAATCCCGAAATTCAGTACACTTCCAACTACAACAAGTC TGTTAATGTGGACTTTACTGTGGACACTAATGGCGTGTATTCAGAGCCTCGCCCC ATTGGCACCAGATACCTGACTCGTAATCTGTAA >SEQ ID NO: 4 SpCas9-VP2 fusion protein MYPYDVPDYASPKKKRKVEASDKKYSIGLDIGTNSVGWAVITDEYKVPSKKFKVLG NTDRHSIKKNLIGALLFDSGETAEATRLKRTARRRYTRRKNRICYLQEIFSNEMAKVD DSFFHRLEESFLVEEDKKHERHPIFGNIVDEVAYHEKYPTIYHLRKKLVDSTDKADLR LIYLALAHMIKFRGHFLIEGDLNPDNSDVDKLFIQLVQTYNQLFEENPINTASGVDAKA ILSARLSKSRRLENLIAQLPGEKKNGLFGNLIALSLGLTPNFKSNFDLAEDAKLQLSKD TYDDDLDNLLAQIGDQYADLFLAAKNLSDAILLSDILRVNTEITKAPLSASMIKRYDE HHQDLTLLKALVRQQLPEKYKEIFFDQSKNGYAGYIDGGASQEEFYKFIKPILEKMD GTEELLVKLNREDLLRKQRTFDNGSIPHQIHLGELHAILRRQEDFYPFLKDNREKIEKI LTFRIPYYVGPLARGNSRFAWMTRKSEETITPWNFEEVVDKGASAQSFIERMTNFDK NLPNEKVLPKHSLLYEYFTVYNELTKVKYVTEGMRKPAFLSGEQKKAIVDLLFKTNR KVTVKQLKEDYFKKIECFDSVEISGVEDRFNASLGTYHDLLKIIKDKDFLDNEENEDI LEDIVLTLTLFEDREMIEERLKTYAHLFDDKVMKQLKRRRYTGWGRLSRKLINGIRD KQSGKTILDFLKSDGFANRNFMQLIHDDSLTFKEDIQKAQVSGQGDSLHEHIANLAG SPAIKKGILQTVKVVDELVKVMGRHKPENIVIEMARENQTTQKGQKNSRERMKRIEE GIKELGSQILKEHPVENTQLQNEKLYLYYLQNGRDMYVDQELDINRLSDYDVDHIVP QSFLKDDSIDNKVLTRSDKNRGKSDNVPSEEVVKKMKNYWRQLLNAKLITQRKFDN LTKAERGGLSELDKAGFIKRQLVETRQITKHVAQILDSRMNTKYDENDKLIREVKVIT LKSKLVSDFRKDFQFYKVREINNYHHAHDAYLNAVVGTALIKKYPKLESEFVYGDY KVYDVRKMIAKSEQEIGKATAKYFFYSNIMNFFKTEITLANGEIRKRPLIETNGETGEI VWDKGRDFATVRKVLSMPQVNIVKKTEVQTGGFSKESILPKRNSDKLIARKKDWDP KKYGGFDSPTVAYSVLVVAKVEKGKSKKLKSVKELLGITIMERSSFEKNPIDFLEAK GYKEVKKDLIIKLPKYSLFELENGRKRMLASAGELQKGNELALPSKYVNFLYLASHY EKLKGSPEDNEQKQLFVEQHKHYLDEIIEQISEFSKRVILADANLDKVLSAYNKHRDK PIREQAENIIHLFTLTNLGAPAAFKYFDTTIDRKRYTSTKEVLDATLIHQSITGLYETRI DLSQLGGDSPKKKRKVEASELAPGKKRPVEHSPVEPDSSSGTGKAGQQPARKRLNFG
QTGDADSVPDPQPLGQPPAAPSGLGTNTMATGSGAPMADNNEGADGVGNSSGNWH CDSTWMGDRVITTSTRTWALPTYNNHLYKQISSQSGASNDNHYFGYSTPWGYFDFN RFHCHFSPRDWQRLINNNWGFRPKRLNFKLFNIQVKEVTQNDGTTTIANNLTSTVQV FTDSEYQLPYVLGSAHQGCLPPFPADVFMVPQYGYLTLNNGSQAVGRSSFYCLEYFP SQMLRTGNNFTFSYTFEDVPFHSSYAHSQSLDRLMNPLIDQYLYYLSRTNTPSGTTTQ SRLQFSQAGASDIRDQSRNWLPGPCYRQQRVSKTSADNNNSEYSWTGATKYHLNGR DSLVNPGPAMASHKDDEEKFFPQSGVLIFGKQGSEKTNVDIEKVMITDEEEIRTTNPV ATEQYGSVSTNLQRGNRQAATADVNTQGVLPGMVWQDRDVYLQGPIWAKIPHTDG HFHPSPLMGGFGLKHPPPQILIKNTPVPANPSTTFSAAKFASFITQYSTGQVSVEIEWE LQKENSKRWNPEIQYTSNYNKSVNVDFTVDTNGVYSEPRPIGTRYLTRNL >SEQ ID NO: 5 Linker sequence GGGS >SEQ ID NO: 6 Cas9.sub.n-Int.sub.n MYPYDVPDYASPKKKRKVEASDKKYSIGLDIGTNSVGWAVITDEYKVPSKKFKVLG NTDRHSIKKNLIGALLFDSGETAEATRLKRTARRRYTRRKNRICYLQEIFSNEMAKVD DSFFHRLEESFLVEEDKKHERHPIFGNIVDEVAYHEKYPTIYHLRKKLVDSTDKADLR LIYLALAHMIKFRGHFLIEGDLNPDNSDVDKLFIQLVQTYNQLFEENPINTASGVDAKA ILSARLSKSRRLENLIAQLPGEKKNGLFGNLIALSLGLTPNFKSNFDLAEDAKLQLSKD TYDDDLDNLLAQIGDQYADLFLAAKNLSDAILLSDILRVNTEITKAPLSASMIKRYDE HHQDLTLLKALVRQQLPEKYKEIFFDQSKNGYAGYIDGGASQEEFYKFIKPILEKMD GTEELLVKLNREDLLRKQRTFDNGSIPHQIHLGELHAILRRQEDFYPFLKDNREKIEKI LTFRIPYYVGPLARGNSRFAWMTRKSEETITPWNFEEVVDKGASAQSFIERMTNFDK NLPNEKVLPKHSLLYEYFTVYNELTKVKYVTEGMRKPAFLSGEQKKAIVDLLFKTNR KVTVKQLKEDYFKKIECFDSVEISGVEDRFNASLGTYHDLLKIIKDKDFLDNEENEDI LEDIVLTLTLFEDREMIEERLKTYAHLFDDKVMKQLKRRRYTGWGRLSRKLINGIRD KQSGKTILDFLKSDGFANRNFMQLIHDDSLTFKEDIQKAQVSGQGDSLHEHIANLAG SPAIKKGILQTVKVVDELVKVMGRHKPENIVIEMARENQTTQKGQKNSRERMKRIEE GIKELGSQILKEHPVENTQLQNEKLYLYYLQNGRDMYVDQELDINRLSDYDVDHIVP QSFLKDDSIDNKVLTRSDKNRGKSDNVPSEEVVKKMKNYWRQLLNAKLITQRKFDN LTKAERGGLSELDKAGFIKRQLVETRQITKHVAQILDSRMNTKYDENDKLIREVKVIT LKSKLVSDFRKDFQFYKVREINNYHHAHDAYLNAVVGTALIKKYPKLESEFVYGDY KVYDVRKMIAKSEQEIGKATAKYFFYSNIMNFFKTEITLANGEIRKRPLIETNGETGEI VWDKGRDFATVRKVLSMPQVNIVKKTEVQTGGFSKESILPKRNSDKLIARKKDWDP KKYGGFDSPTVAYSVLVVAKVEKGKSKKLKSVKELLGITIMERSSFEKNPIDFLEAK GYKEVKKDLIIKLPKYSLFELENGRKCLSYETEILTVEYGLLPIGKIVEKRIECTVYSV DNNGNIYTQPVAQWHDRGEQEVFEYCLEDGSLIRATKDHKFMTVDGQMLPIDEIFER ELDLMRVDNLPN >SEQ ID NO: 7 Int.sub.c-Cas9.sub.c-GS-VP2 MIKIATRKYLGKQNVYDIGVERDHNFALKNGFIASNCMLASAGELQKGNELALPSK YVNFLYLASHYEKLKGSPEDNEQKQLFVEQHKHYLDEIIEQISEFSKRVILADANLDK VLSAYNKHRDKPIREQAENIIHLFTLTNLGAPAAFKYFDTTIDRKRYTSTKEVLDATLI HQSITGLYETRIDLSQLGGDSPKKKRKVEASGSAPGKKRPVEHSPVEPDSSSGTGKAG QQPARKRLNFGQTGDADSVPDPQPLGQPPAAPSGLGTNTMATGSGAPMADNNEGA DGVGNSSGNWHCDSTWMGDRVITTSTRTWALPTYNNHLYKQISSQSGASNDNHYF GYSTPWGYFDFNRFHCHFSPRDWQRLINNNWGFRPKRLNFKLFNIQVKEVTQNDGT TTIANNLTSTVQVFTDSEYQLPYVLGSAHQGCLPPFPADVFMVPQYGYLTLNNGSQA VGRSSFYCLEYFPSQMLRTGNNFTFSYTFEDVPFHSSYAHSQSLDRLMNPLIDQYLYY LSRTNTPSGTTTQSRLQFSQAGASDIRDQSRNWLPGPCYRQQRVSKTSADNNNSEYS WTGATKYHLNGRDSLVNPGPAMASHKDDEEKFFPQSGVLIFGKQGSEKTNVDIEKV MITDEEEIRTTNPVATEQYGSVSTNLQRGNRQAATADVNTQGVLPGMVWQDRDVY LQGPIWAKIPHTDGHFHPSPLMGGFGLKHPPPQILIKNTPVPANPSTTFSAAKFASFITQ YSTGQVSVEIEWELQKENSKRWNPEIQYTSNYNKSVNVDFTVDTNGVYSEPRPIGTR YLTRNL >SEQ ID NO: 8 Int.sub.c-Cas9.sub.c-GGGGSGGGGSGGGGS-VP2 MIKIATRKYLGKQNVYDIGVERDHNFALKNGFIASNCMLASAGELQKGNELALPSK YVNFLYLASHYEKLKGSPEDNEQKQLFVEQHKHYLDEIIEQISEFSKRVILADANLDK VLSAYNKHRDKPIREQAENIIHLFTLTNLGAPAAFKYFDTTIDRKRYTSTKEVLDATLI HQSITGLYETRIDLSQLGGDSPKKKRKVEASGGGGSGGGGSGGGGSAPGKKRPVEHS PVEPDSSSGTGKAGQQPARKRLNFGQTGDADSVPDPQPLGQPPAAPSGLGTNTMAT GSGAPMADNNEGADGVGNSSGNWHCDSTWMGDRVITTSTRTWALPTYNNHLYKQI SSQSGASNDNHYFGYSTPWGYFDFNRFHCHFSPRDWQRLINNNWGFRPKRLNFKLF NIQVKEVTQNDGTTTIANNLTSTVQVFTDSEYQLPYVLGSAHQGCLPPFPADVFMVP QYGYLTLNNGSQAVGRSSFYCLEYFPSQMLRTGNNFTFSYTFEDVPFHSSYAHSQSL DRLMNPLIDQYLYYLSRTNTPSGTTTQSRLQFSQAGASDIRDQSRNWLPGPCYRQQR VSKTSADNNNSEYSWTGATKYHLNGRDSLVNPGPAMASHKDDEEKFFPQSGVLIFG KQGSEKTNVDIEKVMITDEEEIRTTNPVATEQYGSVSTNLQRGNRQAATADVNTQG VLPGMVWQDRDVYLQGPIWAKIPHTDGHFHPSPLMGGFGLKHPPPQILIKNTPVPAN PSTTFSAAKFASFITQYSTGQVSVEIEWELQKENSKRWNPEIQYTSNYNKSVNVDFTV DTNGVYSEPRPIGTRYLTRNL >SEQ ID NO: 9 Int.sub.c-Cas9.sub.c-EAAAKEAAAKEAAAK-VP2 MIKIATRKYLGKQNVYDIGVERDHNFALKNGFIASNCMLASAGELQKGNELALPSK YVNFLYLASHYEKLKGSPEDNEQKQLFVEQHKHYLDEIIEQISEFSKRVILADANLDK VLSAYNKHRDKPIREQAENIIHLFTLTNLGAPAAFKYFDTTIDRKRYTSTKEVLDATLI HQSITGLYETRIDLSQLGGDSPKKKRKVEASEAAAKEAAAKEAAAKAPGKKRPVEH SPVEPDSSSGTGKAGQQPARKRLNFGQTGDADSVPDPQPLGQPPAAPSGLGTNTMAT GSGAPMADNNEGADGVGNSSGNWHCDSTWMGDRVITTSTRTWALPTYNNHLYKQI SSQSGASNDNHYFGYSTPWGYFDFNRFHCHFSPRDWQRLINNNWGFRPKRLNFKLF NIQVKEVTQNDGTTTIANNLTSTVQVFTDSEYQLPYVLGSAHQGCLPPFPADVFMVP QYGYLTLNNGSQAVGRSSFYCLEYFPSQMLRTGNNFTFSYTFEDVPFHSSYAHSQSL DRLMNPLIDQYLYYLSRTNTPSGTTTQSRLQFSQAGASDIRDQSRNWLPGPCYRQQR VSKTSADNNNSEYSWTGATKYHLNGRDSLVNPGPAMASHKDDEEKFFPQSGVLIFG KQGSEKTNVDIEKVMITDEEEIRTTNPVATEQYGSVSTNLQRGNRQAATADVNTQG VLPGMVWQDRDVYLQGPIWAKIPHTDGHFHPSPLMGGFGLKHPPPQILIKNTPVPAN PSTTFSAAKFASFITQYSTGQVSVEIEWELQKENSKRWNPEIQYTSNYNKSVNVDFTV DTNGVYSEPRPIGTRYLTRNL >SEQ ID NO: 10 pU1a-Cas9.sub.c-Int.sub.c CCTGCAGGCAGCTGCGCGCTCGCTCGCTCACTGAGGCCGCCCGGGCAAAGCCCG GGCGTCGGGCGACCTTTGGTCGCCCGGCCTCAGTGAGCGAGCGAGCGCGCAGAG AGGGAGTGGCCAACTCCATCACTAGGGGTTCCTGCGGCCTCTAGAATGGAGGCG GTACTATGTAGATGAGAATTCAGGAGCAAACTGGGAAAAGCAACTGCTTCCAAA TATTTGTGATTTTTACAGTGTAGTTTTGGAAAAACTCTTAGCCTACCAATTCTTCT AAGTGTTTTAAAATGTGGGAGCCAGTACACATGAAGTTATAGAGTGTTTTAATGA GGCTTAAATATTTACCGTAACTATGAAATGCTACGCATATCATGCTGTTCAGGCT CCGTGGCCACGCAACTCATACTACCGGTGCCACCATGTACCCATACGATGTTCCA GATTACGCTTCGCCGAAGAAAAAGCGCAAGGTCGAAGCGTCCGACAAGAAGTAC AGCATCGGCCTGGACATCGGCACCAACTCTGTGGGCTGGGCCGTGATCACCGAC GAGTACAAGGTGCCCAGCAAGAAATTCAAGGTGCTGGGCAACACCGACCGGCAC AGCATCAAGAAGAACCTGATCGGAGCCCTGCTGTTCGACAGCGGCGAAACAGCC GAGGCCACCCGGCTGAAGAGAACCGCCAGAAGAAGATACACCAGACGGAAGAA CCGGATCTGCTATCTGCAAGAGATCTTCAGCAACGAGATGGCCAAGGTGGACGA CAGCTTCTTCCACAGACTGGAAGAGTCCTTCCTGGTGGAAGAGGATAAGAAGCA CGAGCGGCACCCCATCTTCGGCAACATCGTGGACGAGGTGGCCTACCACGAGAA GTACCCCACCATCTACCACCTGAGAAAGAAACTGGTGGACAGCACCGACAAGGC CGACCTGCGGCTGATCTATCTGGCCCTGGCCCACATGATCAAGTTCCGGGGCCAC TTCCTGATCGAGGGCGACCTGAACCCCGACAACAGCGACGTGGACAAGCTGTTC ATCCAGCTGGTGCAGACCTACAACCAGCTGTTCGAGGAAAACCCCATCAACGCC AGCGGCGTGGACGCCAAGGCCATCCTGTCTGCCAGACTGAGCAAGAGCAGACGG CTGGAAAATCTGATCGCCCAGCTGCCCGGCGAGAAGAAGAATGGCCTGTTCGGC AACCTGATTGCCCTGAGCCTGGGCCTGACCCCCAACTTCAAGAGCAACTTCGACC TGGCCGAGGATGCCAAACTGCAGCTGAGCAAGGACACCTACGACGACGACCTGG ACAACCTGCTGGCCCAGATCGGCGACCAGTACGCCGACCTGTTTCTGGCCGCCAA GAACCTGTCCGACGCCATCCTGCTGAGCGACATCCTGAGAGTGAACACCGAGAT CACCAAGGCCCCCCTGAGCGCCTCTATGATCAAGAGATACGACGAGCACCACCA GGACCTGACCCTGCTGAAAGCTCTCGTGCGGCAGCAGCTGCCTGAGAAGTACAA AGAGATTTTCTTCGACCAGAGCAAGAACGGCTACGCCGGCTACATTGACGGCGG AGCCAGCCAGGAAGAGTTCTACAAGTTCATCAAGCCCATCCTGGAAAAGATGGA CGGCACCGAGGAACTGCTCGTGAAGCTGAACAGAGAGGACCTGCTGCGGAAGCA GCGGACCTTCGACAACGGCAGCATCCCCCACCAGATCCACCTGGGAGAGCTGCA CGCCATTCTGCGGCGGCAGGAAGATTTTTACCCATTCCTGAAGGACAACCGGGA AAAGATCGAGAAGATCCTGACCTTCCGCATCCCCTACTACGTGGGCCCTCTGGCC AGGGGAAACAGCAGATTCGCCTGGATGACCAGAAAGAGCGAGGAAACCATCAC CCCCTGGAACTTCGAGGAAGTGGTGGACAAGGGCGCTTCCGCCCAGAGCTTCAT CGAGCGGATGACCAACTTCGATAAGAACCTGCCCAACGAGAAGGTGCTGCCCAA GCACAGCCTGCTGTACGAGTACTTCACCGTGTATAACGAGCTGACCAAAGTGAA ATACGTGACCGAGGGAATGAGAAAGCCCGCCTTCCTGAGCGGCGAGCAGAAAAA GGCCATCGTGGACCTGCTGTTCAAGACCAACCGGAAAGTGACCGTGAAGCAGCT GAAAGAGGACTACTTCAAGAAAATCGAGTGCTTCGACTCCGTGGAAATCTCCGG CGTGGAAGATCGGTTCAACGCCTCCCTGGGCACATACCACGATCTGCTGAAAATT ATCAAGGACAAGGACTTCCTGGACAATGAGGAAAACGAGGACATTCTGGAAGAT ATCGTGCTGACCCTGACACTGTTTGAGGACAGAGAGATGATCGAGGAACGGCTG AAAACCTATGCCCACCTGTTCGACGACAAAGTGATGAAGCAGCTGAAGCGGCGG AGATACACCGGCTGGGGCAGGCTGAGCCGGAAGCTGATCAACGGCATCCGGGAC AAGCAGTCCGGCAAGACAATCCTGGATTTCCTGAAGTCCGACGGCTTCGCCAAC AGAAACTTCATGCAGCTGATCCACGACGACAGCCTGACCTTTAAAGAGGACATC CAGAAAGCCCAGGTGTCCGGCCAGGGCGATAGCCTGCACGAGCACATTGCCAAT CTGGCCGGCAGCCCCGCCATTAAGAAGGGCATCCTGCAGACAGTGAAGGTGGTG GACGAGCTCGTGAAAGTGATGGGCCGGCACAAGCCCGAGAACATCGTGATCGAA ATGGCCAGAGAGAACCAGACCACCCAGAAGGGACAGAAGAACAGCCGCGAGAG AATGAAGCGGATCGAAGAGGGCATCAAAGAGCTGGGCAGCCAGATCCTGAAAG AACACCCCGTGGAAAACACCCAGCTGCAGAACGAGAAGCTGTACCTGTACTACC TGCAGAATGGGCGGGATATGTACGTGGACCAGGAACTGGACATCAACCGGCTGT CCGACTACGATGTGGACCATATCGTGCCTCAGAGCTTTCTGAAGGACGACTCCAT CGACAACAAGGTGCTGACCAGAAGCGACAAGAACCGGGGCAAGAGCGACAACG TGCCCTCCGAAGAGGTCGTGAAGAAGATGAAGAACTACTGGCGGCAGCTGCTGA ACGCCAAGCTGATTACCCAGAGAAAGTTCGACAATCTGACCAAGGCCGAGAGAG GCGGCCTGAGCGAACTGGATAAGGCCGGCTTCATCAAGAGACAGCTGGTGGAAA CCCGGCAGATCACAAAGCACGTGGCACAGATCCTGGACTCCCGGATGAACACTA AGTACGACGAGAATGACAAGCTGATCCGGGAAGTGAAAGTGATCACCCTGAAGT CCAAGCTGGTGTCCGATTTCCGGAAGGATTTCCAGTTTTACAAAGTGCGCGAGAT CAACAACTACCACCACGCCCACGACGCCTACCTGAACGCCGTCGTGGGAACCGC CCTGATCAAAAAGTACCCTAAGCTGGAAAGCGAGTTCGTGTACGGCGACTACAA GGTGTACGACGTGCGGAAGATGATCGCCAAGAGCGAGCAGGAAATCGGCAAGG CTACCGCCAAGTACTTCTTCTACAGCAACATCATGAACTTTTTCAAGACCGAGAT TACCCTGGCCAACGGCGAGATCCGGAAGCGGCCTCTGATCGAGACAAACGGCGA AACCGGGGAGATCGTGTGGGATAAGGGCCGGGATTTTGCCACCGTGCGGAAAGT GCTGAGCATGCCCCAAGTGAATATCGTGAAAAAGACCGAGGTGCAGACAGGCGG CTTCAGCAAAGAGTCTATCCTGCCCAAGAGGAACAGCGATAAGCTGATCGCCAG AAAGAAGGACTGGGACCCTAAGAAGTACGGCGGCTTCGACAGCCCCACCGTGGC CTATTCTGTGCTGGTGGTGGCCAAAGTGGAAAAGGGCAAGTCCAAGAAACTGAA GAGTGTGAAAGAGCTGCTGGGGATCACCATCATGGAAAGAAGCAGCTTCGAGAA GAATCCCATCGACTTTCTGGAAGCCAAGGGCTACAAAGAAGTGAAAAAGGACCT GATCATCAAGCTGCCTAAGTACTCCCTGTTCGAGCTGGAAAACGGCCGGAAGTGT CTGTCGTATGAGACCGAGATCCTGACCGTGGAGTATGGACTGCTGCCGATTGGAA AGATTGTGGAGAAGCGCATTGAGTGCACCGTGTACAGCGTGGATAACAATGGCA ACATCTATACACAGCCAGTGGCCCAGTGGCACGACCGCGGAGAGCAGGAGGTCT TCGAGTACTGCCTGGAGGATGGCAGCCTGATTCGCGCCACCAAGGATCATAAGTT CATGACGGTGGACGGACAGATGCTGCCCATCGATGAGATTTTTGAGCGCGAGCT GGATCTGATGCGCGTGGATAACCTGCCGAATTAAGAATTCGATCTTTTTCCCTCT GCCAAAAATTATGGGGACATCATGAAGCCCCTTGAGCATCTGACTTCTGGCTAAT AAAGGAAATTTATTTTCATTGCAATAGTGTGTTGGAATTTTTTGTGTCTCTCACTC GGCGGCCGCAGGAACCCCTAGTGATGGAGTTGGCCACTCCCTCTCTGCGCGCTCG CTCGCTCACTGAGGCCGGGCGACCAAAGGTCGCCCGACGCCCGGGCTTTGCCCG GGCGGCCTCAGTGAGCGAGCGAGCGCGCAGCTGCCTGCAGG >SEQ ID NO: 11 Int.sub.c-Cas9.sub.c-GS-VP2 GTTGACATTGATTATTGACTAGTTATTAATAGTAATCAATTACGGGGTCATTAGTT CATAGCCCATATATGGAGTTCCGCGTTACATAACTTACGGTAAATGGCCCGCCTG GCTGACCGCCCAACGACCCCCGCCCATTGACGTCAATAATGACGTATGTTCCCAT AGTAACGCCAATAGGGACTTTCCATTGACGTCAATGGGTGGACTATTTACGGTAA ACTGCCCACTTGGCAGTACATCAAGTGTATCATATGCCAAGTACGCCCCCTATTG ACGTCAATGACGGTAAATGGCCCGCCTGGCATTATGCCCAGTACATGACCTTATG GGACTTTCCTACTTGGCAGTACATCTACGTATTAGTCATCGCTATTACCATGGTGA TGCGGTTTTGGCAGTACATCAATGGGCGTGGATAGCGGTTTGACTCACGGGGATT TCCAAGTCTCCACCCCATTGACGTCAATGGGAGTTTGTTTTGGCACCAAAATCAA CGGGACTTTCCAAAATGTCGTAACAACTCCGCCCCATTGACGCAAATGGGCGGTA GGCGTGTACGGTGGGAGGTCTATATAAGCAGAGCTCTCTGGCTAACTAGAGAAC CCACTGCTTACTGGCTTATCGAAATTAATACGACTCACTATAGGGAGACCCAAGC TGGCTAGCGCCACCATGATCAAGATTGCCACGCGCAAGTACCTGGGCAAGCAGA ACGTGTACGACATCGGAGTGGAGCGCGATCACAACTTTGCCCTGAAGAATGGCT TTATTGCCTCGAACTGTATGCTGGCCTCTGCCGGCGAACTGCAGAAGGGAAACGA ACTGGCCCTGCCCTCCAAATATGTGAACTTCCTGTACCTGGCCAGCCACTATGAG AAGCTGAAGGGCTCCCCCGAGGATAATGAGCAGAAACAGCTGTTTGTGGAACAG CACAAGCACTACCTGGACGAGATCATCGAGCAGATCAGCGAGTTCTCCAAGAGA GTGATCCTGGCCGACGCTAATCTGGACAAAGTGCTGTCCGCCTACAACAAGCACC GGGATAAGCCCATCAGAGAGCAGGCCGAGAATATCATCCACCTGTTTACCCTGA CCAATCTGGGAGCCCCTGCCGCCTTCAAGTACTTTGACACCACCATCGACCGGAA GAGGTACACCAGCACCAAAGAGGTGCTGGACGCCACCCTGATCCACCAGAGCAT CACCGGCCTGTACGAGACACGGATCGACCTGTCTCAGCTGGGAGGCGACAGCCC CAAGAAGAAGAGAAAGGTGGAGGCCAGCGGATCCGCTCCGGGAAAAAAGAGGC CGGTAGAGCACTCTCCTGTGGAGCCAGACTCCTCCTCGGGAACCGGAAAGGCGG GCCAGCAGCCTGCAAGAAAAAGATTGAATTTTGGTCAGACTGGAGACGCAGACT CAGTACCTGACCCCCAGCCTCTCGGACAGCCACCAGCAGCCCCCTCTGGTCTGGG AACTAATACGATGGCTACAGGCAGTGGCGCACCAATGGCAGACAATAACGAGGG CGCCGACGGAGTGGGTAATTCCTCGGGAAATTGGCATTGCGATTCCACATGGATG GGCGACAGAGTCATCACCACCAGCACCCGAACCTGGGCCCTGCCCACCTACAAC AACCACCTCTACAAACAAATTTCCAGCCAATCAGGAGCCTCGAACGACAATCAC TACTTTGGCTACAGCACCCCTTGGGGGTATTTTGACTTCAACAGATTCCACTGCC ACTTTTCACCACGTGACTGGCAAAGACTCATCAACAACAACTGGGGATTCCGACC CAAGAGACTCAACTTCAAGCTCTTTAACATTCAAGTCAAAGAGGTCACGCAGAA TGACGGTACGACGACGATTGCCAATAACCTTACCAGCACGGTTCAGGTGTTTACT GACTCGGAGTACCAGCTCCCGTACGTCCTCGGCTCGGCGCATCAAGGATGCCTCC CGCCGTTCCCAGCAGACGTCTTCATGGTGCCACAGTATGGATACCTCACCCTGAA CAACGGGAGTCAGGCAGTAGGACGCTCTTCATTTTACTGCCTGGAGTACTTTCCT TCTCAGATGCTGCGTACCGGAAACAACTTTACCTTCAGCTACACTTTTGAGGACG TTCCTTTCCACAGCAGCTACGCTCACAGCCAGAGTCTGGACCGTCTCATGAATCC TCTCATCGACCAGTACCTGTATTACTTGAGCAGAACAAACACTCCAAGTGGAACC ACCACGCAGTCAAGGCTTCAGTTTTCTCAGGCCGGAGCGAGTGACATTCGGGACC AGTCTAGGAACTGGCTTCCTGGACCCTGTTACCGCCAGCAGCGAGTATCAAAGAC ATCTGCGGATAACAACAACAGTGAATACTCGTGGACTGGAGCTACCAAGTACCA CCTCAATGGCAGAGACTCTCTGGTGAATCCGGGCCCGGCCATGGCAAGCCACAA GGACGATGAAGAAAAGTTTTTTCCTCAGAGCGGGGTTCTCATCTTTGGGAAGCAA GGCTCAGAGAAAACAAATGTGGACATTGAAAAGGTCATGATTACAGACGAAGAG GAAATCAGGACAACCAATCCCGTGGCTACGGAGCAGTATGGTTCTGTATCTACCA ACCTCCAGAGAGGCAACAGACAAGCAGCTACCGCAGATGTCAACACACAAGGCG TTCTTCCAGGCATGGTCTGGCAGGACAGAGATGTGTACCTTCAGGGGCCCATCTG GGCAAAGATTCCACACACGGACGGACATTTTCACCCCTCTCCCCTCATGGGTGGA TTCGGACTTAAACACCCTCCTCCACAGATTCTCATCAAGAACACCCCGGTACCTG CGAATCCTTCGACCACCTTCAGTGCGGCAAAGTTTGCTTCCTTCATCACACAGTA CTCCACGGGACAGGTCAGCGTGGAGATCGAGTGGGAGCTGCAGAAGGAAAACA GCAAACGCTGGAATCCCGAAATTCAGTACACTTCCAACTACAACAAGTCTGTTAA TGTGGACTTTACTGTGGACACTAATGGCGTGTATTCAGAGCCTCGCCCCATTGGC ACCAGATACCTGACTCGTAATCTGTAAGAATTAAACCCGCTGATCAGCCTCGACT GTGCCTTCTAGTTGCCAGCCATCTGTTGTTTGCCCCTCCCCCGTGCCTTCCTTGAC CCTGGAAGGTGCCACTCCCACTGTCCTTTCCTAATAAAATGAGGAAATTGCATCG CATTGTCTGAGTAGGTGTCATTCTATTCTGGGGGGTGGGGTGGGGCAGGACAGCA AGGGGGAGGATTGGGAAGACAATAGCAGGCATGCTGGGGATGCGGTGGGCTCTA TGG >SEQ ID NO: 12 Int.sub.c-Cas9.sub.c-GGGGSx3-VP2 GTTGACATTGATTATTGACTAGTTATTAATAGTAATCAATTACGGGGTCATTAGTT CATAGCCCATATATGGAGTTCCGCGTTACATAACTTACGGTAAATGGCCCGCCTG GCTGACCGCCCAACGACCCCCGCCCATTGACGTCAATAATGACGTATGTTCCCAT AGTAACGCCAATAGGGACTTTCCATTGACGTCAATGGGTGGACTATTTACGGTAA ACTGCCCACTTGGCAGTACATCAAGTGTATCATATGCCAAGTACGCCCCCTATTG ACGTCAATGACGGTAAATGGCCCGCCTGGCATTATGCCCAGTACATGACCTTATG
GGACTTTCCTACTTGGCAGTACATCTACGTATTAGTCATCGCTATTACCATGGTGA TGCGGTTTTGGCAGTACATCAATGGGCGTGGATAGCGGTTTGACTCACGGGGATT TCCAAGTCTCCACCCCATTGACGTCAATGGGAGTTTGTTTTGGCACCAAAATCAA CGGGACTTTCCAAAATGTCGTAACAACTCCGCCCCATTGACGCAAATGGGCGGTA GGCGTGTACGGTGGGAGGTCTATATAAGCAGAGCTCTCTGGCTAACTAGAGAAC CCACTGCTTACTGGCTTATCGAAATTAATACGACTCACTATAGGGAGACCCAAGC TGGCTAGCGCCACCATGATCAAGATTGCCACGCGCAAGTACCTGGGCAAGCAGA ACGTGTACGACATCGGAGTGGAGCGCGATCACAACTTTGCCCTGAAGAATGGCT TTATTGCCTCGAACTGTATGCTGGCCTCTGCCGGCGAACTGCAGAAGGGAAACGA ACTGGCCCTGCCCTCCAAATATGTGAACTTCCTGTACCTGGCCAGCCACTATGAG AAGCTGAAGGGCTCCCCCGAGGATAATGAGCAGAAACAGCTGTTTGTGGAACAG CACAAGCACTACCTGGACGAGATCATCGAGCAGATCAGCGAGTTCTCCAAGAGA GTGATCCTGGCCGACGCTAATCTGGACAAAGTGCTGTCCGCCTACAACAAGCACC GGGATAAGCCCATCAGAGAGCAGGCCGAGAATATCATCCACCTGTTTACCCTGA CCAATCTGGGAGCCCCTGCCGCCTTCAAGTACTTTGACACCACCATCGACCGGAA GAGGTACACCAGCACCAAAGAGGTGCTGGACGCCACCCTGATCCACCAGAGCAT CACCGGCCTGTACGAGACACGGATCGACCTGTCTCAGCTGGGAGGCGACAGCCC CAAGAAGAAGAGAAAGGTGGAGGCCAGCGGTGGCGGCGGTTCAGGCGGAGGTG GCTCTGGGGGCGGGGGTTCTGCTCCGGGAAAAAAGAGGCCGGTAGAGCACTCTC CTGTGGAGCCAGACTCCTCCTCGGGAACCGGAAAGGCGGGCCAGCAGCCTGCAA GAAAAAGATTGAATTTTGGTCAGACTGGAGACGCAGACTCAGTACCTGACCCCC AGCCTCTCGGACAGCCACCAGCAGCCCCCTCTGGTCTGGGAACTAATACGATGGC TACAGGCAGTGGCGCACCAATGGCAGACAATAACGAGGGCGCCGACGGAGTGG GTAATTCCTCGGGAAATTGGCATTGCGATTCCACATGGATGGGCGACAGAGTCAT CACCACCAGCACCCGAACCTGGGCCCTGCCCACCTACAACAACCACCTCTACAA ACAAATTTCCAGCCAATCAGGAGCCTCGAACGACAATCACTACTTTGGCTACAGC ACCCCTTGGGGGTATTTTGACTTCAACAGATTCCACTGCCACTTTTCACCACGTGA CTGGCAAAGACTCATCAACAACAACTGGGGATTCCGACCCAAGAGACTCAACTT CAAGCTCTTTAACATTCAAGTCAAAGAGGTCACGCAGAATGACGGTACGACGAC GATTGCCAATAACCTTACCAGCACGGTTCAGGTGTTTACTGACTCGGAGTACCAG CTCCCGTACGTCCTCGGCTCGGCGCATCAAGGATGCCTCCCGCCGTTCCCAGCAG ACGTCTTCATGGTGCCACAGTATGGATACCTCACCCTGAACAACGGGAGTCAGGC AGTAGGACGCTCTTCATTTTACTGCCTGGAGTACTTTCCTTCTCAGATGCTGCGTA CCGGAAACAACTTTACCTTCAGCTACACTTTTGAGGACGTTCCTTTCCACAGCAG CTACGCTCACAGCCAGAGTCTGGACCGTCTCATGAATCCTCTCATCGACCAGTAC CTGTATTACTTGAGCAGAACAAACACTCCAAGTGGAACCACCACGCAGTCAAGG CTTCAGTTTTCTCAGGCCGGAGCGAGTGACATTCGGGACCAGTCTAGGAACTGGC TTCCTGGACCCTGTTACCGCCAGCAGCGAGTATCAAAGACATCTGCGGATAACAA CAACAGTGAATACTCGTGGACTGGAGCTACCAAGTACCACCTCAATGGCAGAGA CTCTCTGGTGAATCCGGGCCCGGCCATGGCAAGCCACAAGGACGATGAAGAAAA GTTTTTTCCTCAGAGCGGGGTTCTCATCTTTGGGAAGCAAGGCTCAGAGAAAACA AATGTGGACATTGAAAAGGTCATGATTACAGACGAAGAGGAAATCAGGACAACC AATCCCGTGGCTACGGAGCAGTATGGTTCTGTATCTACCAACCTCCAGAGAGGCA ACAGACAAGCAGCTACCGCAGATGTCAACACACAAGGCGTTCTTCCAGGCATGG TCTGGCAGGACAGAGATGTGTACCTTCAGGGGCCCATCTGGGCAAAGATTCCAC ACACGGACGGACATTTTCACCCCTCTCCCCTCATGGGTGGATTCGGACTTAAACA CCCTCCTCCACAGATTCTCATCAAGAACACCCCGGTACCTGCGAATCCTTCGACC ACCTTCAGTGCGGCAAAGTTTGCTTCCTTCATCACACAGTACTCCACGGGACAGG TCAGCGTGGAGATCGAGTGGGAGCTGCAGAAGGAAAACAGCAAACGCTGGAAT CCCGAAATTCAGTACACTTCCAACTACAACAAGTCTGTTAATGTGGACTTTACTG TGGACACTAATGGCGTGTATTCAGAGCCTCGCCCCATTGGCACCAGATACCTGAC TCGTAATCTGTAAGAATTAAACCCGCTGATCAGCCTCGACTGTGCCTTCTAGTTG CCAGCCATCTGTTGTTTGCCCCTCCCCCGTGCCTTCCTTGACCCTGGAAGGTGCCA CTCCCACTGTCCTTTCCTAATAAAATGAGGAAATTGCATCGCATTGTCTGAGTAG GTGTCATTCTATTCTGGGGGGTGGGGTGGGGCAGGACAGCAAGGGGGAGGATTG GGAAGACAATAGCAGGCATGCTGGGGATGCGGTGGGCTCTATGG >SEQ ID NO: 13 Int.sub.c-Cas9.sub.c-EAAAKx3-VP2 GTTGACATTGATTATTGACTAGTTATTAATAGTAATCAATTACGGGGTCATTAGTT CATAGCCCATATATGGAGTTCCGCGTTACATAACTTACGGTAAATGGCCCGCCTG GCTGACCGCCCAACGACCCCCGCCCATTGACGTCAATAATGACGTATGTTCCCAT AGTAACGCCAATAGGGACTTTCCATTGACGTCAATGGGTGGACTATTTACGGTAA ACTGCCCACTTGGCAGTACATCAAGTGTATCATATGCCAAGTACGCCCCCTATTG ACGTCAATGACGGTAAATGGCCCGCCTGGCATTATGCCCAGTACATGACCTTATG GGACTTTCCTACTTGGCAGTACATCTACGTATTAGTCATCGCTATTACCATGGTGA TGCGGTTTTGGCAGTACATCAATGGGCGTGGATAGCGGTTTGACTCACGGGGATT TCCAAGTCTCCACCCCATTGACGTCAATGGGAGTTTGTTTTGGCACCAAAATCAA CGGGACTTTCCAAAATGTCGTAACAACTCCGCCCCATTGACGCAAATGGGCGGTA GGCGTGTACGGTGGGAGGTCTATATAAGCAGAGCTCTCTGGCTAACTAGAGAAC CCACTGCTTACTGGCTTATCGAAATTAATACGACTCACTATAGGGAGACCCAAGC TGGCTAGCGCCACCATGATCAAGATTGCCACGCGCAAGTACCTGGGCAAGCAGA ACGTGTACGACATCGGAGTGGAGCGCGATCACAACTTTGCCCTGAAGAATGGCT TTATTGCCTCGAACTGTATGCTGGCCTCTGCCGGCGAACTGCAGAAGGGAAACGA ACTGGCCCTGCCCTCCAAATATGTGAACTTCCTGTACCTGGCCAGCCACTATGAG AAGCTGAAGGGCTCCCCCGAGGATAATGAGCAGAAACAGCTGTTTGTGGAACAG CACAAGCACTACCTGGACGAGATCATCGAGCAGATCAGCGAGTTCTCCAAGAGA GTGATCCTGGCCGACGCTAATCTGGACAAAGTGCTGTCCGCCTACAACAAGCACC GGGATAAGCCCATCAGAGAGCAGGCCGAGAATATCATCCACCTGTTTACCCTGA CCAATCTGGGAGCCCCTGCCGCCTTCAAGTACTTTGACACCACCATCGACCGGAA GAGGTACACCAGCACCAAAGAGGTGCTGGACGCCACCCTGATCCACCAGAGCAT CACCGGCCTGTACGAGACACGGATCGACCTGTCTCAGCTGGGAGGCGACAGCCC CAAGAAGAAGAGAAAGGTGGAGGCCAGCGAGGCAGCAGCCAAAGAGGCCGCTG CCAAGGAGGCAGCGGCTAAAGCTCCGGGAAAAAAGAGGCCGGTAGAGCACTCT CCTGTGGAGCCAGACTCCTCCTCGGGAACCGGAAAGGCGGGCCAGCAGCCTGCA AGAAAAAGATTGAATTTTGGTCAGACTGGAGACGCAGACTCAGTACCTGACCCC CAGCCTCTCGGACAGCCACCAGCAGCCCCCTCTGGTCTGGGAACTAATACGATGG CTACAGGCAGTGGCGCACCAATGGCAGACAATAACGAGGGCGCCGACGGAGTG GGTAATTCCTCGGGAAATTGGCATTGCGATTCCACATGGATGGGCGACAGAGTC ATCACCACCAGCACCCGAACCTGGGCCCTGCCCACCTACAACAACCACCTCTACA AACAAATTTCCAGCCAATCAGGAGCCTCGAACGACAATCACTACTTTGGCTACAG CACCCCTTGGGGGTATTTTGACTTCAACAGATTCCACTGCCACTTTTCACCACGTG ACTGGCAAAGACTCATCAACAACAACTGGGGATTCCGACCCAAGAGACTCAACT TCAAGCTCTTTAACATTCAAGTCAAAGAGGTCACGCAGAATGACGGTACGACGA CGATTGCCAATAACCTTACCAGCACGGTTCAGGTGTTTACTGACTCGGAGTACCA GCTCCCGTACGTCCTCGGCTCGGCGCATCAAGGATGCCTCCCGCCGTTCCCAGCA GACGTCTTCATGGTGCCACAGTATGGATACCTCACCCTGAACAACGGGAGTCAG GCAGTAGGACGCTCTTCATTTTACTGCCTGGAGTACTTTCCTTCTCAGATGCTGCG TACCGGAAACAACTTTACCTTCAGCTACACTTTTGAGGACGTTCCTTTCCACAGC AGCTACGCTCACAGCCAGAGTCTGGACCGTCTCATGAATCCTCTCATCGACCAGT ACCTGTATTACTTGAGCAGAACAAACACTCCAAGTGGAACCACCACGCAGTCAA GGCTTCAGTTTTCTCAGGCCGGAGCGAGTGACATTCGGGACCAGTCTAGGAACTG GCTTCCTGGACCCTGTTACCGCCAGCAGCGAGTATCAAAGACATCTGCGGATAAC AACAACAGTGAATACTCGTGGACTGGAGCTACCAAGTACCACCTCAATGGCAGA GACTCTCTGGTGAATCCGGGCCCGGCCATGGCAAGCCACAAGGACGATGAAGAA AAGTTTTTTCCTCAGAGCGGGGTTCTCATCTTTGGGAAGCAAGGCTCAGAGAAAA CAAATGTGGACATTGAAAAGGTCATGATTACAGACGAAGAGGAAATCAGGACAA CCAATCCCGTGGCTACGGAGCAGTATGGTTCTGTATCTACCAACCTCCAGAGAGG CAACAGACAAGCAGCTACCGCAGATGTCAACACACAAGGCGTTCTTCCAGGCAT GGTCTGGCAGGACAGAGATGTGTACCTTCAGGGGCCCATCTGGGCAAAGATTCC ACACACGGACGGACATTTTCACCCCTCTCCCCTCATGGGTGGATTCGGACTTAAA CACCCTCCTCCACAGATTCTCATCAAGAACACCCCGGTACCTGCGAATCCTTCGA CCACCTTCAGTGCGGCAAAGTTTGCTTCCTTCATCACACAGTACTCCACGGGACA GGTCAGCGTGGAGATCGAGTGGGAGCTGCAGAAGGAAAACAGCAAACGCTGGA ATCCCGAAATTCAGTACACTTCCAACTACAACAAGTCTGTTAATGTGGACTTTAC TGTGGACACTAATGGCGTGTATTCAGAGCCTCGCCCCATTGGCACCAGATACCTG ACTCGTAATCTGTAAGAATTAAACCCGCTGATCAGCCTCGACTGTGCCTTCTAGT TGCCAGCCATCTGTTGTTTGCCCCTCCCCCGTGCCTTCCTTGACCCTGGAAGGTGC CACTCCCACTGTCCTTTCCTAATAAAATGAGGAAATTGCATCGCATTGTCTGAGT AGGTGTCATTCTATTCTGGGGGGTGGGGTGGGGCAGGACAGCAAGGGGGAGGAT TGGGAAGACAATAGCAGGCATGCTGGGGATGCGGTGGGCTCTATGG
This disclosure is not limited in its application to the details of construction and the arrangement of components set forth in this description or illustrated in the drawings. The disclosure is capable of other embodiments and of being practiced or of being carried out in various ways. Also, the phraseology and terminology used herein is for the purpose of description and should not be regarded as limiting. The use of "including," "comprising," or "having," "containing," "involving," and variations thereof herein, is meant to encompass the items listed thereafter and equivalents thereof as well as additional items.
Having thus described several aspects of at least one embodiment of this disclosure, it is to be appreciated various alterations, modifications, and improvements will readily occur to those skilled in the art. Such alterations, modifications, and improvements are intended to be part of this disclosure, and are intended to be within the spirit and scope of the disclosure. Accordingly, the foregoing description and drawings are by way of example only.
SEQUENCE LISTINGS
1
1314197DNAArtificial SequenceSynthetic Polynucleotide 1atgtacccat acgatgttcc agattacgct tcgccgaaga aaaagcgcaa ggtcgaagcg 60tccgacaaga agtacagcat cggcctggac atcggcacca actctgtggg ctgggccgtg 120atcaccgacg agtacaaggt gcccagcaag aaattcaagg tgctgggcaa caccgaccgg 180cacagcatca agaagaacct gatcggagcc ctgctgttcg acagcggcga aacagccgag 240gccacccggc tgaagagaac cgccagaaga agatacacca gacggaagaa ccggatctgc 300tatctgcaag agatcttcag caacgagatg gccaaggtgg acgacagctt cttccacaga 360ctggaagagt ccttcctggt ggaagaggat aagaagcacg agcggcaccc catcttcggc 420aacatcgtgg acgaggtggc ctaccacgag aagtacccca ccatctacca cctgagaaag 480aaactggtgg acagcaccga caaggccgac ctgcggctga tctatctggc cctggcccac 540atgatcaagt tccggggcca cttcctgatc gagggcgacc tgaaccccga caacagcgac 600gtggacaagc tgttcatcca gctggtgcag acctacaacc agctgttcga ggaaaacccc 660atcaacgcca gcggcgtgga cgccaaggcc atcctgtctg ccagactgag caagagcaga 720cggctggaaa atctgatcgc ccagctgccc ggcgagaaga agaatggcct gttcggcaac 780ctgattgccc tgagcctggg cctgaccccc aacttcaaga gcaacttcga cctggccgag 840gatgccaaac tgcagctgag caaggacacc tacgacgacg acctggacaa cctgctggcc 900cagatcggcg accagtacgc cgacctgttt ctggccgcca agaacctgtc cgacgccatc 960ctgctgagcg acatcctgag agtgaacacc gagatcacca aggcccccct gagcgcctct 1020atgatcaaga gatacgacga gcaccaccag gacctgaccc tgctgaaagc tctcgtgcgg 1080cagcagctgc ctgagaagta caaagagatt ttcttcgacc agagcaagaa cggctacgcc 1140ggctacattg acggcggagc cagccaggaa gagttctaca agttcatcaa gcccatcctg 1200gaaaagatgg acggcaccga ggaactgctc gtgaagctga acagagagga cctgctgcgg 1260aagcagcgga ccttcgacaa cggcagcatc ccccaccaga tccacctggg agagctgcac 1320gccattctgc ggcggcagga agatttttac ccattcctga aggacaaccg ggaaaagatc 1380gagaagatcc tgaccttccg catcccctac tacgtgggcc ctctggccag gggaaacagc 1440agattcgcct ggatgaccag aaagagcgag gaaaccatca ccccctggaa cttcgaggaa 1500gtggtggaca agggcgcttc cgcccagagc ttcatcgagc ggatgaccaa cttcgataag 1560aacctgccca acgagaaggt gctgcccaag cacagcctgc tgtacgagta cttcaccgtg 1620tataacgagc tgaccaaagt gaaatacgtg accgagggaa tgagaaagcc cgccttcctg 1680agcggcgagc agaaaaaggc catcgtggac ctgctgttca agaccaaccg gaaagtgacc 1740gtgaagcagc tgaaagagga ctacttcaag aaaatcgagt gcttcgactc cgtggaaatc 1800tccggcgtgg aagatcggtt caacgcctcc ctgggcacat accacgatct gctgaaaatt 1860atcaaggaca aggacttcct ggacaatgag gaaaacgagg acattctgga agatatcgtg 1920ctgaccctga cactgtttga ggacagagag atgatcgagg aacggctgaa aacctatgcc 1980cacctgttcg acgacaaagt gatgaagcag ctgaagcggc ggagatacac cggctggggc 2040aggctgagcc ggaagctgat caacggcatc cgggacaagc agtccggcaa gacaatcctg 2100gatttcctga agtccgacgg cttcgccaac agaaacttca tgcagctgat ccacgacgac 2160agcctgacct ttaaagagga catccagaaa gcccaggtgt ccggccaggg cgatagcctg 2220cacgagcaca ttgccaatct ggccggcagc cccgccatta agaagggcat cctgcagaca 2280gtgaaggtgg tggacgagct cgtgaaagtg atgggccggc acaagcccga gaacatcgtg 2340atcgaaatgg ccagagagaa ccagaccacc cagaagggac agaagaacag ccgcgagaga 2400atgaagcgga tcgaagaggg catcaaagag ctgggcagcc agatcctgaa agaacacccc 2460gtggaaaaca cccagctgca gaacgagaag ctgtacctgt actacctgca gaatgggcgg 2520gatatgtacg tggaccagga actggacatc aaccggctgt ccgactacga tgtggaccat 2580atcgtgcctc agagctttct gaaggacgac tccatcgaca acaaggtgct gaccagaagc 2640gacaagaacc ggggcaagag cgacaacgtg ccctccgaag aggtcgtgaa gaagatgaag 2700aactactggc ggcagctgct gaacgccaag ctgattaccc agagaaagtt cgacaatctg 2760accaaggccg agagaggcgg cctgagcgaa ctggataagg ccggcttcat caagagacag 2820ctggtggaaa cccggcagat cacaaagcac gtggcacaga tcctggactc ccggatgaac 2880actaagtacg acgagaatga caagctgatc cgggaagtga aagtgatcac cctgaagtcc 2940aagctggtgt ccgatttccg gaaggatttc cagttttaca aagtgcgcga gatcaacaac 3000taccaccacg cccacgacgc ctacctgaac gccgtcgtgg gaaccgccct gatcaaaaag 3060taccctaagc tggaaagcga gttcgtgtac ggcgactaca aggtgtacga cgtgcggaag 3120atgatcgcca agagcgagca ggaaatcggc aaggctaccg ccaagtactt cttctacagc 3180aacatcatga actttttcaa gaccgagatt accctggcca acggcgagat ccggaagcgg 3240cctctgatcg agacaaacgg cgaaaccggg gagatcgtgt gggataaggg ccgggatttt 3300gccaccgtgc ggaaagtgct gagcatgccc caagtgaata tcgtgaaaaa gaccgaggtg 3360cagacaggcg gcttcagcaa agagtctatc ctgcccaaga ggaacagcga taagctgatc 3420gccagaaaga aggactggga ccctaagaag tacggcggct tcgacagccc caccgtggcc 3480tattctgtgc tggtggtggc caaagtggaa aagggcaagt ccaagaaact gaagagtgtg 3540aaagagctgc tggggatcac catcatggaa agaagcagct tcgagaagaa tcccatcgac 3600tttctggaag ccaagggcta caaagaagtg aaaaaggacc tgatcatcaa gctgcctaag 3660tactccctgt tcgagctgga aaacggccgg aagagaatgc tggcctctgc cggcgaactg 3720cagaagggaa acgaactggc cctgccctcc aaatatgtga acttcctgta cctggccagc 3780cactatgaga agctgaaggg ctcccccgag gataatgagc agaaacagct gtttgtggaa 3840cagcacaagc actacctgga cgagatcatc gagcagatca gcgagttctc caagagagtg 3900atcctggccg acgctaatct ggacaaagtg ctgtccgcct acaacaagca ccgggataag 3960cccatcagag agcaggccga gaatatcatc cacctgttta ccctgaccaa tctgggagcc 4020cctgccgcct tcaagtactt tgacaccacc atcgaccgga agaggtacac cagcaccaaa 4080gaggtgctgg acgccaccct gatccaccag agcatcaccg gcctgtacga gacacggatc 4140gacctgtctc agctgggagg cgacagcccc aagaagaaga gaaaggtgga ggccagc 419721399PRTArtificial SequenceSynthetic Polypeptide 2Met Tyr Pro Tyr Asp Val Pro Asp Tyr Ala Ser Pro Lys Lys Lys Arg1 5 10 15Lys Val Glu Ala Ser Asp Lys Lys Tyr Ser Ile Gly Leu Asp Ile Gly 20 25 30Thr Asn Ser Val Gly Trp Ala Val Ile Thr Asp Glu Tyr Lys Val Pro 35 40 45Ser Lys Lys Phe Lys Val Leu Gly Asn Thr Asp Arg His Ser Ile Lys 50 55 60Lys Asn Leu Ile Gly Ala Leu Leu Phe Asp Ser Gly Glu Thr Ala Glu65 70 75 80Ala Thr Arg Leu Lys Arg Thr Ala Arg Arg Arg Tyr Thr Arg Arg Lys 85 90 95Asn Arg Ile Cys Tyr Leu Gln Glu Ile Phe Ser Asn Glu Met Ala Lys 100 105 110Val Asp Asp Ser Phe Phe His Arg Leu Glu Glu Ser Phe Leu Val Glu 115 120 125Glu Asp Lys Lys His Glu Arg His Pro Ile Phe Gly Asn Ile Val Asp 130 135 140Glu Val Ala Tyr His Glu Lys Tyr Pro Thr Ile Tyr His Leu Arg Lys145 150 155 160Lys Leu Val Asp Ser Thr Asp Lys Ala Asp Leu Arg Leu Ile Tyr Leu 165 170 175Ala Leu Ala His Met Ile Lys Phe Arg Gly His Phe Leu Ile Glu Gly 180 185 190Asp Leu Asn Pro Asp Asn Ser Asp Val Asp Lys Leu Phe Ile Gln Leu 195 200 205Val Gln Thr Tyr Asn Gln Leu Phe Glu Glu Asn Pro Ile Asn Ala Ser 210 215 220Gly Val Asp Ala Lys Ala Ile Leu Ser Ala Arg Leu Ser Lys Ser Arg225 230 235 240Arg Leu Glu Asn Leu Ile Ala Gln Leu Pro Gly Glu Lys Lys Asn Gly 245 250 255Leu Phe Gly Asn Leu Ile Ala Leu Ser Leu Gly Leu Thr Pro Asn Phe 260 265 270Lys Ser Asn Phe Asp Leu Ala Glu Asp Ala Lys Leu Gln Leu Ser Lys 275 280 285Asp Thr Tyr Asp Asp Asp Leu Asp Asn Leu Leu Ala Gln Ile Gly Asp 290 295 300Gln Tyr Ala Asp Leu Phe Leu Ala Ala Lys Asn Leu Ser Asp Ala Ile305 310 315 320Leu Leu Ser Asp Ile Leu Arg Val Asn Thr Glu Ile Thr Lys Ala Pro 325 330 335Leu Ser Ala Ser Met Ile Lys Arg Tyr Asp Glu His His Gln Asp Leu 340 345 350Thr Leu Leu Lys Ala Leu Val Arg Gln Gln Leu Pro Glu Lys Tyr Lys 355 360 365Glu Ile Phe Phe Asp Gln Ser Lys Asn Gly Tyr Ala Gly Tyr Ile Asp 370 375 380Gly Gly Ala Ser Gln Glu Glu Phe Tyr Lys Phe Ile Lys Pro Ile Leu385 390 395 400Glu Lys Met Asp Gly Thr Glu Glu Leu Leu Val Lys Leu Asn Arg Glu 405 410 415Asp Leu Leu Arg Lys Gln Arg Thr Phe Asp Asn Gly Ser Ile Pro His 420 425 430Gln Ile His Leu Gly Glu Leu His Ala Ile Leu Arg Arg Gln Glu Asp 435 440 445Phe Tyr Pro Phe Leu Lys Asp Asn Arg Glu Lys Ile Glu Lys Ile Leu 450 455 460Thr Phe Arg Ile Pro Tyr Tyr Val Gly Pro Leu Ala Arg Gly Asn Ser465 470 475 480Arg Phe Ala Trp Met Thr Arg Lys Ser Glu Glu Thr Ile Thr Pro Trp 485 490 495Asn Phe Glu Glu Val Val Asp Lys Gly Ala Ser Ala Gln Ser Phe Ile 500 505 510Glu Arg Met Thr Asn Phe Asp Lys Asn Leu Pro Asn Glu Lys Val Leu 515 520 525Pro Lys His Ser Leu Leu Tyr Glu Tyr Phe Thr Val Tyr Asn Glu Leu 530 535 540Thr Lys Val Lys Tyr Val Thr Glu Gly Met Arg Lys Pro Ala Phe Leu545 550 555 560Ser Gly Glu Gln Lys Lys Ala Ile Val Asp Leu Leu Phe Lys Thr Asn 565 570 575Arg Lys Val Thr Val Lys Gln Leu Lys Glu Asp Tyr Phe Lys Lys Ile 580 585 590Glu Cys Phe Asp Ser Val Glu Ile Ser Gly Val Glu Asp Arg Phe Asn 595 600 605Ala Ser Leu Gly Thr Tyr His Asp Leu Leu Lys Ile Ile Lys Asp Lys 610 615 620Asp Phe Leu Asp Asn Glu Glu Asn Glu Asp Ile Leu Glu Asp Ile Val625 630 635 640Leu Thr Leu Thr Leu Phe Glu Asp Arg Glu Met Ile Glu Glu Arg Leu 645 650 655Lys Thr Tyr Ala His Leu Phe Asp Asp Lys Val Met Lys Gln Leu Lys 660 665 670Arg Arg Arg Tyr Thr Gly Trp Gly Arg Leu Ser Arg Lys Leu Ile Asn 675 680 685Gly Ile Arg Asp Lys Gln Ser Gly Lys Thr Ile Leu Asp Phe Leu Lys 690 695 700Ser Asp Gly Phe Ala Asn Arg Asn Phe Met Gln Leu Ile His Asp Asp705 710 715 720Ser Leu Thr Phe Lys Glu Asp Ile Gln Lys Ala Gln Val Ser Gly Gln 725 730 735Gly Asp Ser Leu His Glu His Ile Ala Asn Leu Ala Gly Ser Pro Ala 740 745 750Ile Lys Lys Gly Ile Leu Gln Thr Val Lys Val Val Asp Glu Leu Val 755 760 765Lys Val Met Gly Arg His Lys Pro Glu Asn Ile Val Ile Glu Met Ala 770 775 780Arg Glu Asn Gln Thr Thr Gln Lys Gly Gln Lys Asn Ser Arg Glu Arg785 790 795 800Met Lys Arg Ile Glu Glu Gly Ile Lys Glu Leu Gly Ser Gln Ile Leu 805 810 815Lys Glu His Pro Val Glu Asn Thr Gln Leu Gln Asn Glu Lys Leu Tyr 820 825 830Leu Tyr Tyr Leu Gln Asn Gly Arg Asp Met Tyr Val Asp Gln Glu Leu 835 840 845Asp Ile Asn Arg Leu Ser Asp Tyr Asp Val Asp His Ile Val Pro Gln 850 855 860Ser Phe Leu Lys Asp Asp Ser Ile Asp Asn Lys Val Leu Thr Arg Ser865 870 875 880Asp Lys Asn Arg Gly Lys Ser Asp Asn Val Pro Ser Glu Glu Val Val 885 890 895Lys Lys Met Lys Asn Tyr Trp Arg Gln Leu Leu Asn Ala Lys Leu Ile 900 905 910Thr Gln Arg Lys Phe Asp Asn Leu Thr Lys Ala Glu Arg Gly Gly Leu 915 920 925Ser Glu Leu Asp Lys Ala Gly Phe Ile Lys Arg Gln Leu Val Glu Thr 930 935 940Arg Gln Ile Thr Lys His Val Ala Gln Ile Leu Asp Ser Arg Met Asn945 950 955 960Thr Lys Tyr Asp Glu Asn Asp Lys Leu Ile Arg Glu Val Lys Val Ile 965 970 975Thr Leu Lys Ser Lys Leu Val Ser Asp Phe Arg Lys Asp Phe Gln Phe 980 985 990Tyr Lys Val Arg Glu Ile Asn Asn Tyr His His Ala His Asp Ala Tyr 995 1000 1005Leu Asn Ala Val Val Gly Thr Ala Leu Ile Lys Lys Tyr Pro Lys 1010 1015 1020Leu Glu Ser Glu Phe Val Tyr Gly Asp Tyr Lys Val Tyr Asp Val 1025 1030 1035Arg Lys Met Ile Ala Lys Ser Glu Gln Glu Ile Gly Lys Ala Thr 1040 1045 1050Ala Lys Tyr Phe Phe Tyr Ser Asn Ile Met Asn Phe Phe Lys Thr 1055 1060 1065Glu Ile Thr Leu Ala Asn Gly Glu Ile Arg Lys Arg Pro Leu Ile 1070 1075 1080Glu Thr Asn Gly Glu Thr Gly Glu Ile Val Trp Asp Lys Gly Arg 1085 1090 1095Asp Phe Ala Thr Val Arg Lys Val Leu Ser Met Pro Gln Val Asn 1100 1105 1110Ile Val Lys Lys Thr Glu Val Gln Thr Gly Gly Phe Ser Lys Glu 1115 1120 1125Ser Ile Leu Pro Lys Arg Asn Ser Asp Lys Leu Ile Ala Arg Lys 1130 1135 1140Lys Asp Trp Asp Pro Lys Lys Tyr Gly Gly Phe Asp Ser Pro Thr 1145 1150 1155Val Ala Tyr Ser Val Leu Val Val Ala Lys Val Glu Lys Gly Lys 1160 1165 1170Ser Lys Lys Leu Lys Ser Val Lys Glu Leu Leu Gly Ile Thr Ile 1175 1180 1185Met Glu Arg Ser Ser Phe Glu Lys Asn Pro Ile Asp Phe Leu Glu 1190 1195 1200Ala Lys Gly Tyr Lys Glu Val Lys Lys Asp Leu Ile Ile Lys Leu 1205 1210 1215Pro Lys Tyr Ser Leu Phe Glu Leu Glu Asn Gly Arg Lys Arg Met 1220 1225 1230Leu Ala Ser Ala Gly Glu Leu Gln Lys Gly Asn Glu Leu Ala Leu 1235 1240 1245Pro Ser Lys Tyr Val Asn Phe Leu Tyr Leu Ala Ser His Tyr Glu 1250 1255 1260Lys Leu Lys Gly Ser Pro Glu Asp Asn Glu Gln Lys Gln Leu Phe 1265 1270 1275Val Glu Gln His Lys His Tyr Leu Asp Glu Ile Ile Glu Gln Ile 1280 1285 1290Ser Glu Phe Ser Lys Arg Val Ile Leu Ala Asp Ala Asn Leu Asp 1295 1300 1305Lys Val Leu Ser Ala Tyr Asn Lys His Arg Asp Lys Pro Ile Arg 1310 1315 1320Glu Gln Ala Glu Asn Ile Ile His Leu Phe Thr Leu Thr Asn Leu 1325 1330 1335Gly Ala Pro Ala Ala Phe Lys Tyr Phe Asp Thr Thr Ile Asp Arg 1340 1345 1350Lys Arg Tyr Thr Ser Thr Lys Glu Val Leu Asp Ala Thr Leu Ile 1355 1360 1365His Gln Ser Ile Thr Gly Leu Tyr Glu Thr Arg Ile Asp Leu Ser 1370 1375 1380Gln Leu Gly Gly Asp Ser Pro Lys Lys Lys Arg Lys Val Glu Ala 1385 1390 1395Ser35997DNAArtificial SequenceSynthetic Polynucleotide 3atgtacccat acgatgttcc agattacgct tcgccgaaga aaaagcgcaa ggtcgaagcg 60tccgacaaga agtacagcat cggcctggac atcggcacca actctgtggg ctgggccgtg 120atcaccgacg agtacaaggt gcccagcaag aaattcaagg tgctgggcaa caccgaccgg 180cacagcatca agaagaacct gatcggagcc ctgctgttcg acagcggcga aacagccgag 240gccacccggc tgaagagaac cgccagaaga agatacacca gacggaagaa ccggatctgc 300tatctgcaag agatcttcag caacgagatg gccaaggtgg acgacagctt cttccacaga 360ctggaagagt ccttcctggt ggaagaggat aagaagcacg agcggcaccc catcttcggc 420aacatcgtgg acgaggtggc ctaccacgag aagtacccca ccatctacca cctgagaaag 480aaactggtgg acagcaccga caaggccgac ctgcggctga tctatctggc cctggcccac 540atgatcaagt tccggggcca cttcctgatc gagggcgacc tgaaccccga caacagcgac 600gtggacaagc tgttcatcca gctggtgcag acctacaacc agctgttcga ggaaaacccc 660atcaacgcca gcggcgtgga cgccaaggcc atcctgtctg ccagactgag caagagcaga 720cggctggaaa atctgatcgc ccagctgccc ggcgagaaga agaatggcct gttcggcaac 780ctgattgccc tgagcctggg cctgaccccc aacttcaaga gcaacttcga cctggccgag 840gatgccaaac tgcagctgag caaggacacc tacgacgacg acctggacaa cctgctggcc 900cagatcggcg accagtacgc cgacctgttt ctggccgcca agaacctgtc cgacgccatc 960ctgctgagcg acatcctgag agtgaacacc gagatcacca aggcccccct gagcgcctct 1020atgatcaaga gatacgacga gcaccaccag gacctgaccc tgctgaaagc tctcgtgcgg 1080cagcagctgc ctgagaagta caaagagatt ttcttcgacc agagcaagaa cggctacgcc 1140ggctacattg acggcggagc cagccaggaa gagttctaca agttcatcaa gcccatcctg 1200gaaaagatgg acggcaccga ggaactgctc gtgaagctga acagagagga cctgctgcgg 1260aagcagcgga ccttcgacaa cggcagcatc ccccaccaga tccacctggg agagctgcac 1320gccattctgc ggcggcagga agatttttac ccattcctga aggacaaccg ggaaaagatc 1380gagaagatcc tgaccttccg catcccctac tacgtgggcc ctctggccag gggaaacagc 1440agattcgcct ggatgaccag aaagagcgag gaaaccatca ccccctggaa cttcgaggaa 1500gtggtggaca agggcgcttc cgcccagagc ttcatcgagc ggatgaccaa cttcgataag 1560aacctgccca acgagaaggt gctgcccaag cacagcctgc tgtacgagta cttcaccgtg 1620tataacgagc tgaccaaagt gaaatacgtg accgagggaa tgagaaagcc cgccttcctg 1680agcggcgagc agaaaaaggc catcgtggac ctgctgttca agaccaaccg gaaagtgacc 1740gtgaagcagc tgaaagagga ctacttcaag aaaatcgagt gcttcgactc cgtggaaatc 1800tccggcgtgg aagatcggtt caacgcctcc ctgggcacat accacgatct gctgaaaatt 1860atcaaggaca aggacttcct ggacaatgag gaaaacgagg acattctgga agatatcgtg 1920ctgaccctga cactgtttga ggacagagag atgatcgagg aacggctgaa aacctatgcc 1980cacctgttcg acgacaaagt gatgaagcag ctgaagcggc ggagatacac cggctggggc 2040aggctgagcc ggaagctgat caacggcatc cgggacaagc agtccggcaa gacaatcctg 2100gatttcctga agtccgacgg cttcgccaac agaaacttca tgcagctgat ccacgacgac
2160agcctgacct ttaaagagga catccagaaa gcccaggtgt ccggccaggg cgatagcctg 2220cacgagcaca ttgccaatct ggccggcagc cccgccatta agaagggcat cctgcagaca 2280gtgaaggtgg tggacgagct cgtgaaagtg atgggccggc acaagcccga gaacatcgtg 2340atcgaaatgg ccagagagaa ccagaccacc cagaagggac agaagaacag ccgcgagaga 2400atgaagcgga tcgaagaggg catcaaagag ctgggcagcc agatcctgaa agaacacccc 2460gtggaaaaca cccagctgca gaacgagaag ctgtacctgt actacctgca gaatgggcgg 2520gatatgtacg tggaccagga actggacatc aaccggctgt ccgactacga tgtggaccat 2580atcgtgcctc agagctttct gaaggacgac tccatcgaca acaaggtgct gaccagaagc 2640gacaagaacc ggggcaagag cgacaacgtg ccctccgaag aggtcgtgaa gaagatgaag 2700aactactggc ggcagctgct gaacgccaag ctgattaccc agagaaagtt cgacaatctg 2760accaaggccg agagaggcgg cctgagcgaa ctggataagg ccggcttcat caagagacag 2820ctggtggaaa cccggcagat cacaaagcac gtggcacaga tcctggactc ccggatgaac 2880actaagtacg acgagaatga caagctgatc cgggaagtga aagtgatcac cctgaagtcc 2940aagctggtgt ccgatttccg gaaggatttc cagttttaca aagtgcgcga gatcaacaac 3000taccaccacg cccacgacgc ctacctgaac gccgtcgtgg gaaccgccct gatcaaaaag 3060taccctaagc tggaaagcga gttcgtgtac ggcgactaca aggtgtacga cgtgcggaag 3120atgatcgcca agagcgagca ggaaatcggc aaggctaccg ccaagtactt cttctacagc 3180aacatcatga actttttcaa gaccgagatt accctggcca acggcgagat ccggaagcgg 3240cctctgatcg agacaaacgg cgaaaccggg gagatcgtgt gggataaggg ccgggatttt 3300gccaccgtgc ggaaagtgct gagcatgccc caagtgaata tcgtgaaaaa gaccgaggtg 3360cagacaggcg gcttcagcaa agagtctatc ctgcccaaga ggaacagcga taagctgatc 3420gccagaaaga aggactggga ccctaagaag tacggcggct tcgacagccc caccgtggcc 3480tattctgtgc tggtggtggc caaagtggaa aagggcaagt ccaagaaact gaagagtgtg 3540aaagagctgc tggggatcac catcatggaa agaagcagct tcgagaagaa tcccatcgac 3600tttctggaag ccaagggcta caaagaagtg aaaaaggacc tgatcatcaa gctgcctaag 3660tactccctgt tcgagctgga aaacggccgg aagagaatgc tggcctctgc cggcgaactg 3720cagaagggaa acgaactggc cctgccctcc aaatatgtga acttcctgta cctggccagc 3780cactatgaga agctgaaggg ctcccccgag gataatgagc agaaacagct gtttgtggaa 3840cagcacaagc actacctgga cgagatcatc gagcagatca gcgagttctc caagagagtg 3900atcctggccg acgctaatct ggacaaagtg ctgtccgcct acaacaagca ccgggataag 3960cccatcagag agcaggccga gaatatcatc cacctgttta ccctgaccaa tctgggagcc 4020cctgccgcct tcaagtactt tgacaccacc atcgaccgga agaggtacac cagcaccaaa 4080gaggtgctgg acgccaccct gatccaccag agcatcaccg gcctgtacga gacacggatc 4140gacctgtctc agctgggagg cgacagcccc aagaagaaga gaaaggtgga ggccagcgaa 4200ttggctccgg gaaaaaagag gccggtagag cactctcctg tggagccaga ctcctcctcg 4260ggaaccggaa aggcgggcca gcagcctgca agaaaaagat tgaattttgg tcagactgga 4320gacgcagact cagtacctga cccccagcct ctcggacagc caccagcagc cccctctggt 4380ctgggaacta atacgatggc tacaggcagt ggcgcaccaa tggcagacaa taacgagggc 4440gccgacggag tgggtaattc ctcgggaaat tggcattgcg attccacatg gatgggcgac 4500agagtcatca ccaccagcac ccgaacctgg gccctgccca cctacaacaa ccacctctac 4560aaacaaattt ccagccaatc aggagcctcg aacgacaatc actactttgg ctacagcacc 4620ccttgggggt attttgactt caacagattc cactgccact tttcaccacg tgactggcaa 4680agactcatca acaacaactg gggattccga cccaagagac tcaacttcaa gctctttaac 4740attcaagtca aagaggtcac gcagaatgac ggtacgacga cgattgccaa taaccttacc 4800agcacggttc aggtgtttac tgactcggag taccagctcc cgtacgtcct cggctcggcg 4860catcaaggat gcctcccgcc gttcccagca gacgtcttca tggtgccaca gtatggatac 4920ctcaccctga acaacgggag tcaggcagta ggacgctctt cattttactg cctggagtac 4980tttccttctc agatgctgcg taccggaaac aactttacct tcagctacac ttttgaggac 5040gttcctttcc acagcagcta cgctcacagc cagagtctgg accgtctcat gaatcctctc 5100atcgaccagt acctgtatta cttgagcaga acaaacactc caagtggaac caccacgcag 5160tcaaggcttc agttttctca ggccggagcg agtgacattc gggaccagtc taggaactgg 5220cttcctggac cctgttaccg ccagcagcga gtatcaaaga catctgcgga taacaacaac 5280agtgaatact cgtggactgg agctaccaag taccacctca atggcagaga ctctctggtg 5340aatccgggcc cggccatggc aagccacaag gacgatgaag aaaagttttt tcctcagagc 5400ggggttctca tctttgggaa gcaaggctca gagaaaacaa atgtggacat tgaaaaggtc 5460atgattacag acgaagagga aatcaggaca accaatcccg tggctacgga gcagtatggt 5520tctgtatcta ccaacctcca gagaggcaac agacaagcag ctaccgcaga tgtcaacaca 5580caaggcgttc ttccaggcat ggtctggcag gacagagatg tgtaccttca ggggcccatc 5640tgggcaaaga ttccacacac ggacggacat tttcacccct ctcccctcat gggtggattc 5700ggacttaaac accctcctcc acagattctc atcaagaaca ccccggtacc tgcgaatcct 5760tcgaccacct tcagtgcggc aaagtttgct tccttcatca cacagtactc cacgggacag 5820gtcagcgtgg agatcgagtg ggagctgcag aaggaaaaca gcaaacgctg gaatcccgaa 5880attcagtaca cttccaacta caacaagtct gttaatgtgg actttactgt ggacactaat 5940ggcgtgtatt cagagcctcg ccccattggc accagatacc tgactcgtaa tctgtaa 599741998PRTArtificial SequenceSynthetic Polypeptide 4Met Tyr Pro Tyr Asp Val Pro Asp Tyr Ala Ser Pro Lys Lys Lys Arg1 5 10 15Lys Val Glu Ala Ser Asp Lys Lys Tyr Ser Ile Gly Leu Asp Ile Gly 20 25 30Thr Asn Ser Val Gly Trp Ala Val Ile Thr Asp Glu Tyr Lys Val Pro 35 40 45Ser Lys Lys Phe Lys Val Leu Gly Asn Thr Asp Arg His Ser Ile Lys 50 55 60Lys Asn Leu Ile Gly Ala Leu Leu Phe Asp Ser Gly Glu Thr Ala Glu65 70 75 80Ala Thr Arg Leu Lys Arg Thr Ala Arg Arg Arg Tyr Thr Arg Arg Lys 85 90 95Asn Arg Ile Cys Tyr Leu Gln Glu Ile Phe Ser Asn Glu Met Ala Lys 100 105 110Val Asp Asp Ser Phe Phe His Arg Leu Glu Glu Ser Phe Leu Val Glu 115 120 125Glu Asp Lys Lys His Glu Arg His Pro Ile Phe Gly Asn Ile Val Asp 130 135 140Glu Val Ala Tyr His Glu Lys Tyr Pro Thr Ile Tyr His Leu Arg Lys145 150 155 160Lys Leu Val Asp Ser Thr Asp Lys Ala Asp Leu Arg Leu Ile Tyr Leu 165 170 175Ala Leu Ala His Met Ile Lys Phe Arg Gly His Phe Leu Ile Glu Gly 180 185 190Asp Leu Asn Pro Asp Asn Ser Asp Val Asp Lys Leu Phe Ile Gln Leu 195 200 205Val Gln Thr Tyr Asn Gln Leu Phe Glu Glu Asn Pro Ile Asn Ala Ser 210 215 220Gly Val Asp Ala Lys Ala Ile Leu Ser Ala Arg Leu Ser Lys Ser Arg225 230 235 240Arg Leu Glu Asn Leu Ile Ala Gln Leu Pro Gly Glu Lys Lys Asn Gly 245 250 255Leu Phe Gly Asn Leu Ile Ala Leu Ser Leu Gly Leu Thr Pro Asn Phe 260 265 270Lys Ser Asn Phe Asp Leu Ala Glu Asp Ala Lys Leu Gln Leu Ser Lys 275 280 285Asp Thr Tyr Asp Asp Asp Leu Asp Asn Leu Leu Ala Gln Ile Gly Asp 290 295 300Gln Tyr Ala Asp Leu Phe Leu Ala Ala Lys Asn Leu Ser Asp Ala Ile305 310 315 320Leu Leu Ser Asp Ile Leu Arg Val Asn Thr Glu Ile Thr Lys Ala Pro 325 330 335Leu Ser Ala Ser Met Ile Lys Arg Tyr Asp Glu His His Gln Asp Leu 340 345 350Thr Leu Leu Lys Ala Leu Val Arg Gln Gln Leu Pro Glu Lys Tyr Lys 355 360 365Glu Ile Phe Phe Asp Gln Ser Lys Asn Gly Tyr Ala Gly Tyr Ile Asp 370 375 380Gly Gly Ala Ser Gln Glu Glu Phe Tyr Lys Phe Ile Lys Pro Ile Leu385 390 395 400Glu Lys Met Asp Gly Thr Glu Glu Leu Leu Val Lys Leu Asn Arg Glu 405 410 415Asp Leu Leu Arg Lys Gln Arg Thr Phe Asp Asn Gly Ser Ile Pro His 420 425 430Gln Ile His Leu Gly Glu Leu His Ala Ile Leu Arg Arg Gln Glu Asp 435 440 445Phe Tyr Pro Phe Leu Lys Asp Asn Arg Glu Lys Ile Glu Lys Ile Leu 450 455 460Thr Phe Arg Ile Pro Tyr Tyr Val Gly Pro Leu Ala Arg Gly Asn Ser465 470 475 480Arg Phe Ala Trp Met Thr Arg Lys Ser Glu Glu Thr Ile Thr Pro Trp 485 490 495Asn Phe Glu Glu Val Val Asp Lys Gly Ala Ser Ala Gln Ser Phe Ile 500 505 510Glu Arg Met Thr Asn Phe Asp Lys Asn Leu Pro Asn Glu Lys Val Leu 515 520 525Pro Lys His Ser Leu Leu Tyr Glu Tyr Phe Thr Val Tyr Asn Glu Leu 530 535 540Thr Lys Val Lys Tyr Val Thr Glu Gly Met Arg Lys Pro Ala Phe Leu545 550 555 560Ser Gly Glu Gln Lys Lys Ala Ile Val Asp Leu Leu Phe Lys Thr Asn 565 570 575Arg Lys Val Thr Val Lys Gln Leu Lys Glu Asp Tyr Phe Lys Lys Ile 580 585 590Glu Cys Phe Asp Ser Val Glu Ile Ser Gly Val Glu Asp Arg Phe Asn 595 600 605Ala Ser Leu Gly Thr Tyr His Asp Leu Leu Lys Ile Ile Lys Asp Lys 610 615 620Asp Phe Leu Asp Asn Glu Glu Asn Glu Asp Ile Leu Glu Asp Ile Val625 630 635 640Leu Thr Leu Thr Leu Phe Glu Asp Arg Glu Met Ile Glu Glu Arg Leu 645 650 655Lys Thr Tyr Ala His Leu Phe Asp Asp Lys Val Met Lys Gln Leu Lys 660 665 670Arg Arg Arg Tyr Thr Gly Trp Gly Arg Leu Ser Arg Lys Leu Ile Asn 675 680 685Gly Ile Arg Asp Lys Gln Ser Gly Lys Thr Ile Leu Asp Phe Leu Lys 690 695 700Ser Asp Gly Phe Ala Asn Arg Asn Phe Met Gln Leu Ile His Asp Asp705 710 715 720Ser Leu Thr Phe Lys Glu Asp Ile Gln Lys Ala Gln Val Ser Gly Gln 725 730 735Gly Asp Ser Leu His Glu His Ile Ala Asn Leu Ala Gly Ser Pro Ala 740 745 750Ile Lys Lys Gly Ile Leu Gln Thr Val Lys Val Val Asp Glu Leu Val 755 760 765Lys Val Met Gly Arg His Lys Pro Glu Asn Ile Val Ile Glu Met Ala 770 775 780Arg Glu Asn Gln Thr Thr Gln Lys Gly Gln Lys Asn Ser Arg Glu Arg785 790 795 800Met Lys Arg Ile Glu Glu Gly Ile Lys Glu Leu Gly Ser Gln Ile Leu 805 810 815Lys Glu His Pro Val Glu Asn Thr Gln Leu Gln Asn Glu Lys Leu Tyr 820 825 830Leu Tyr Tyr Leu Gln Asn Gly Arg Asp Met Tyr Val Asp Gln Glu Leu 835 840 845Asp Ile Asn Arg Leu Ser Asp Tyr Asp Val Asp His Ile Val Pro Gln 850 855 860Ser Phe Leu Lys Asp Asp Ser Ile Asp Asn Lys Val Leu Thr Arg Ser865 870 875 880Asp Lys Asn Arg Gly Lys Ser Asp Asn Val Pro Ser Glu Glu Val Val 885 890 895Lys Lys Met Lys Asn Tyr Trp Arg Gln Leu Leu Asn Ala Lys Leu Ile 900 905 910Thr Gln Arg Lys Phe Asp Asn Leu Thr Lys Ala Glu Arg Gly Gly Leu 915 920 925Ser Glu Leu Asp Lys Ala Gly Phe Ile Lys Arg Gln Leu Val Glu Thr 930 935 940Arg Gln Ile Thr Lys His Val Ala Gln Ile Leu Asp Ser Arg Met Asn945 950 955 960Thr Lys Tyr Asp Glu Asn Asp Lys Leu Ile Arg Glu Val Lys Val Ile 965 970 975Thr Leu Lys Ser Lys Leu Val Ser Asp Phe Arg Lys Asp Phe Gln Phe 980 985 990Tyr Lys Val Arg Glu Ile Asn Asn Tyr His His Ala His Asp Ala Tyr 995 1000 1005Leu Asn Ala Val Val Gly Thr Ala Leu Ile Lys Lys Tyr Pro Lys 1010 1015 1020Leu Glu Ser Glu Phe Val Tyr Gly Asp Tyr Lys Val Tyr Asp Val 1025 1030 1035Arg Lys Met Ile Ala Lys Ser Glu Gln Glu Ile Gly Lys Ala Thr 1040 1045 1050Ala Lys Tyr Phe Phe Tyr Ser Asn Ile Met Asn Phe Phe Lys Thr 1055 1060 1065Glu Ile Thr Leu Ala Asn Gly Glu Ile Arg Lys Arg Pro Leu Ile 1070 1075 1080Glu Thr Asn Gly Glu Thr Gly Glu Ile Val Trp Asp Lys Gly Arg 1085 1090 1095Asp Phe Ala Thr Val Arg Lys Val Leu Ser Met Pro Gln Val Asn 1100 1105 1110Ile Val Lys Lys Thr Glu Val Gln Thr Gly Gly Phe Ser Lys Glu 1115 1120 1125Ser Ile Leu Pro Lys Arg Asn Ser Asp Lys Leu Ile Ala Arg Lys 1130 1135 1140Lys Asp Trp Asp Pro Lys Lys Tyr Gly Gly Phe Asp Ser Pro Thr 1145 1150 1155Val Ala Tyr Ser Val Leu Val Val Ala Lys Val Glu Lys Gly Lys 1160 1165 1170Ser Lys Lys Leu Lys Ser Val Lys Glu Leu Leu Gly Ile Thr Ile 1175 1180 1185Met Glu Arg Ser Ser Phe Glu Lys Asn Pro Ile Asp Phe Leu Glu 1190 1195 1200Ala Lys Gly Tyr Lys Glu Val Lys Lys Asp Leu Ile Ile Lys Leu 1205 1210 1215Pro Lys Tyr Ser Leu Phe Glu Leu Glu Asn Gly Arg Lys Arg Met 1220 1225 1230Leu Ala Ser Ala Gly Glu Leu Gln Lys Gly Asn Glu Leu Ala Leu 1235 1240 1245Pro Ser Lys Tyr Val Asn Phe Leu Tyr Leu Ala Ser His Tyr Glu 1250 1255 1260Lys Leu Lys Gly Ser Pro Glu Asp Asn Glu Gln Lys Gln Leu Phe 1265 1270 1275Val Glu Gln His Lys His Tyr Leu Asp Glu Ile Ile Glu Gln Ile 1280 1285 1290Ser Glu Phe Ser Lys Arg Val Ile Leu Ala Asp Ala Asn Leu Asp 1295 1300 1305Lys Val Leu Ser Ala Tyr Asn Lys His Arg Asp Lys Pro Ile Arg 1310 1315 1320Glu Gln Ala Glu Asn Ile Ile His Leu Phe Thr Leu Thr Asn Leu 1325 1330 1335Gly Ala Pro Ala Ala Phe Lys Tyr Phe Asp Thr Thr Ile Asp Arg 1340 1345 1350Lys Arg Tyr Thr Ser Thr Lys Glu Val Leu Asp Ala Thr Leu Ile 1355 1360 1365His Gln Ser Ile Thr Gly Leu Tyr Glu Thr Arg Ile Asp Leu Ser 1370 1375 1380Gln Leu Gly Gly Asp Ser Pro Lys Lys Lys Arg Lys Val Glu Ala 1385 1390 1395Ser Glu Leu Ala Pro Gly Lys Lys Arg Pro Val Glu His Ser Pro 1400 1405 1410Val Glu Pro Asp Ser Ser Ser Gly Thr Gly Lys Ala Gly Gln Gln 1415 1420 1425Pro Ala Arg Lys Arg Leu Asn Phe Gly Gln Thr Gly Asp Ala Asp 1430 1435 1440Ser Val Pro Asp Pro Gln Pro Leu Gly Gln Pro Pro Ala Ala Pro 1445 1450 1455Ser Gly Leu Gly Thr Asn Thr Met Ala Thr Gly Ser Gly Ala Pro 1460 1465 1470Met Ala Asp Asn Asn Glu Gly Ala Asp Gly Val Gly Asn Ser Ser 1475 1480 1485Gly Asn Trp His Cys Asp Ser Thr Trp Met Gly Asp Arg Val Ile 1490 1495 1500Thr Thr Ser Thr Arg Thr Trp Ala Leu Pro Thr Tyr Asn Asn His 1505 1510 1515Leu Tyr Lys Gln Ile Ser Ser Gln Ser Gly Ala Ser Asn Asp Asn 1520 1525 1530His Tyr Phe Gly Tyr Ser Thr Pro Trp Gly Tyr Phe Asp Phe Asn 1535 1540 1545Arg Phe His Cys His Phe Ser Pro Arg Asp Trp Gln Arg Leu Ile 1550 1555 1560Asn Asn Asn Trp Gly Phe Arg Pro Lys Arg Leu Asn Phe Lys Leu 1565 1570 1575Phe Asn Ile Gln Val Lys Glu Val Thr Gln Asn Asp Gly Thr Thr 1580 1585 1590Thr Ile Ala Asn Asn Leu Thr Ser Thr Val Gln Val Phe Thr Asp 1595 1600 1605Ser Glu Tyr Gln Leu Pro Tyr Val Leu Gly Ser Ala His Gln Gly 1610 1615 1620Cys Leu Pro Pro Phe Pro Ala Asp Val Phe Met Val Pro Gln Tyr 1625 1630 1635Gly Tyr Leu Thr Leu Asn Asn Gly Ser Gln Ala Val Gly Arg Ser 1640 1645 1650Ser Phe Tyr Cys Leu Glu Tyr Phe Pro Ser Gln Met Leu Arg Thr 1655 1660 1665Gly Asn Asn Phe Thr Phe Ser Tyr Thr Phe Glu Asp Val Pro Phe 1670 1675 1680His Ser Ser Tyr Ala His Ser Gln Ser Leu Asp Arg Leu Met Asn 1685 1690 1695Pro Leu Ile Asp Gln Tyr Leu Tyr Tyr Leu Ser Arg Thr Asn Thr 1700 1705 1710Pro Ser Gly Thr Thr Thr Gln Ser Arg Leu Gln Phe Ser Gln Ala 1715 1720 1725Gly Ala Ser Asp Ile Arg Asp Gln Ser Arg Asn Trp Leu Pro Gly 1730 1735 1740Pro Cys Tyr Arg Gln Gln Arg Val Ser Lys Thr Ser Ala Asp Asn 1745 1750 1755Asn Asn Ser Glu Tyr Ser Trp Thr Gly Ala Thr Lys Tyr His Leu 1760 1765 1770Asn Gly Arg Asp Ser Leu Val Asn Pro Gly Pro Ala Met Ala Ser 1775 1780 1785His Lys Asp Asp Glu Glu Lys Phe Phe Pro Gln Ser Gly Val Leu 1790 1795 1800Ile Phe Gly Lys Gln Gly Ser Glu Lys Thr Asn Val Asp Ile Glu 1805 1810 1815Lys Val Met Ile Thr Asp Glu Glu Glu
Ile Arg Thr Thr Asn Pro 1820 1825 1830Val Ala Thr Glu Gln Tyr Gly Ser Val Ser Thr Asn Leu Gln Arg 1835 1840 1845Gly Asn Arg Gln Ala Ala Thr Ala Asp Val Asn Thr Gln Gly Val 1850 1855 1860Leu Pro Gly Met Val Trp Gln Asp Arg Asp Val Tyr Leu Gln Gly 1865 1870 1875Pro Ile Trp Ala Lys Ile Pro His Thr Asp Gly His Phe His Pro 1880 1885 1890Ser Pro Leu Met Gly Gly Phe Gly Leu Lys His Pro Pro Pro Gln 1895 1900 1905Ile Leu Ile Lys Asn Thr Pro Val Pro Ala Asn Pro Ser Thr Thr 1910 1915 1920Phe Ser Ala Ala Lys Phe Ala Ser Phe Ile Thr Gln Tyr Ser Thr 1925 1930 1935Gly Gln Val Ser Val Glu Ile Glu Trp Glu Leu Gln Lys Glu Asn 1940 1945 1950Ser Lys Arg Trp Asn Pro Glu Ile Gln Tyr Thr Ser Asn Tyr Asn 1955 1960 1965Lys Ser Val Asn Val Asp Phe Thr Val Asp Thr Asn Gly Val Tyr 1970 1975 1980Ser Glu Pro Arg Pro Ile Gly Thr Arg Tyr Leu Thr Arg Asn Leu 1985 1990 199554PRTArtificial SequenceSynthetic Polypeptide 5Gly Gly Gly Ser161333PRTArtificial SequenceSynthetic Polypeptide 6Met Tyr Pro Tyr Asp Val Pro Asp Tyr Ala Ser Pro Lys Lys Lys Arg1 5 10 15Lys Val Glu Ala Ser Asp Lys Lys Tyr Ser Ile Gly Leu Asp Ile Gly 20 25 30Thr Asn Ser Val Gly Trp Ala Val Ile Thr Asp Glu Tyr Lys Val Pro 35 40 45Ser Lys Lys Phe Lys Val Leu Gly Asn Thr Asp Arg His Ser Ile Lys 50 55 60Lys Asn Leu Ile Gly Ala Leu Leu Phe Asp Ser Gly Glu Thr Ala Glu65 70 75 80Ala Thr Arg Leu Lys Arg Thr Ala Arg Arg Arg Tyr Thr Arg Arg Lys 85 90 95Asn Arg Ile Cys Tyr Leu Gln Glu Ile Phe Ser Asn Glu Met Ala Lys 100 105 110Val Asp Asp Ser Phe Phe His Arg Leu Glu Glu Ser Phe Leu Val Glu 115 120 125Glu Asp Lys Lys His Glu Arg His Pro Ile Phe Gly Asn Ile Val Asp 130 135 140Glu Val Ala Tyr His Glu Lys Tyr Pro Thr Ile Tyr His Leu Arg Lys145 150 155 160Lys Leu Val Asp Ser Thr Asp Lys Ala Asp Leu Arg Leu Ile Tyr Leu 165 170 175Ala Leu Ala His Met Ile Lys Phe Arg Gly His Phe Leu Ile Glu Gly 180 185 190Asp Leu Asn Pro Asp Asn Ser Asp Val Asp Lys Leu Phe Ile Gln Leu 195 200 205Val Gln Thr Tyr Asn Gln Leu Phe Glu Glu Asn Pro Ile Asn Ala Ser 210 215 220Gly Val Asp Ala Lys Ala Ile Leu Ser Ala Arg Leu Ser Lys Ser Arg225 230 235 240Arg Leu Glu Asn Leu Ile Ala Gln Leu Pro Gly Glu Lys Lys Asn Gly 245 250 255Leu Phe Gly Asn Leu Ile Ala Leu Ser Leu Gly Leu Thr Pro Asn Phe 260 265 270Lys Ser Asn Phe Asp Leu Ala Glu Asp Ala Lys Leu Gln Leu Ser Lys 275 280 285Asp Thr Tyr Asp Asp Asp Leu Asp Asn Leu Leu Ala Gln Ile Gly Asp 290 295 300Gln Tyr Ala Asp Leu Phe Leu Ala Ala Lys Asn Leu Ser Asp Ala Ile305 310 315 320Leu Leu Ser Asp Ile Leu Arg Val Asn Thr Glu Ile Thr Lys Ala Pro 325 330 335Leu Ser Ala Ser Met Ile Lys Arg Tyr Asp Glu His His Gln Asp Leu 340 345 350Thr Leu Leu Lys Ala Leu Val Arg Gln Gln Leu Pro Glu Lys Tyr Lys 355 360 365Glu Ile Phe Phe Asp Gln Ser Lys Asn Gly Tyr Ala Gly Tyr Ile Asp 370 375 380Gly Gly Ala Ser Gln Glu Glu Phe Tyr Lys Phe Ile Lys Pro Ile Leu385 390 395 400Glu Lys Met Asp Gly Thr Glu Glu Leu Leu Val Lys Leu Asn Arg Glu 405 410 415Asp Leu Leu Arg Lys Gln Arg Thr Phe Asp Asn Gly Ser Ile Pro His 420 425 430Gln Ile His Leu Gly Glu Leu His Ala Ile Leu Arg Arg Gln Glu Asp 435 440 445Phe Tyr Pro Phe Leu Lys Asp Asn Arg Glu Lys Ile Glu Lys Ile Leu 450 455 460Thr Phe Arg Ile Pro Tyr Tyr Val Gly Pro Leu Ala Arg Gly Asn Ser465 470 475 480Arg Phe Ala Trp Met Thr Arg Lys Ser Glu Glu Thr Ile Thr Pro Trp 485 490 495Asn Phe Glu Glu Val Val Asp Lys Gly Ala Ser Ala Gln Ser Phe Ile 500 505 510Glu Arg Met Thr Asn Phe Asp Lys Asn Leu Pro Asn Glu Lys Val Leu 515 520 525Pro Lys His Ser Leu Leu Tyr Glu Tyr Phe Thr Val Tyr Asn Glu Leu 530 535 540Thr Lys Val Lys Tyr Val Thr Glu Gly Met Arg Lys Pro Ala Phe Leu545 550 555 560Ser Gly Glu Gln Lys Lys Ala Ile Val Asp Leu Leu Phe Lys Thr Asn 565 570 575Arg Lys Val Thr Val Lys Gln Leu Lys Glu Asp Tyr Phe Lys Lys Ile 580 585 590Glu Cys Phe Asp Ser Val Glu Ile Ser Gly Val Glu Asp Arg Phe Asn 595 600 605Ala Ser Leu Gly Thr Tyr His Asp Leu Leu Lys Ile Ile Lys Asp Lys 610 615 620Asp Phe Leu Asp Asn Glu Glu Asn Glu Asp Ile Leu Glu Asp Ile Val625 630 635 640Leu Thr Leu Thr Leu Phe Glu Asp Arg Glu Met Ile Glu Glu Arg Leu 645 650 655Lys Thr Tyr Ala His Leu Phe Asp Asp Lys Val Met Lys Gln Leu Lys 660 665 670Arg Arg Arg Tyr Thr Gly Trp Gly Arg Leu Ser Arg Lys Leu Ile Asn 675 680 685Gly Ile Arg Asp Lys Gln Ser Gly Lys Thr Ile Leu Asp Phe Leu Lys 690 695 700Ser Asp Gly Phe Ala Asn Arg Asn Phe Met Gln Leu Ile His Asp Asp705 710 715 720Ser Leu Thr Phe Lys Glu Asp Ile Gln Lys Ala Gln Val Ser Gly Gln 725 730 735Gly Asp Ser Leu His Glu His Ile Ala Asn Leu Ala Gly Ser Pro Ala 740 745 750Ile Lys Lys Gly Ile Leu Gln Thr Val Lys Val Val Asp Glu Leu Val 755 760 765Lys Val Met Gly Arg His Lys Pro Glu Asn Ile Val Ile Glu Met Ala 770 775 780Arg Glu Asn Gln Thr Thr Gln Lys Gly Gln Lys Asn Ser Arg Glu Arg785 790 795 800Met Lys Arg Ile Glu Glu Gly Ile Lys Glu Leu Gly Ser Gln Ile Leu 805 810 815Lys Glu His Pro Val Glu Asn Thr Gln Leu Gln Asn Glu Lys Leu Tyr 820 825 830Leu Tyr Tyr Leu Gln Asn Gly Arg Asp Met Tyr Val Asp Gln Glu Leu 835 840 845Asp Ile Asn Arg Leu Ser Asp Tyr Asp Val Asp His Ile Val Pro Gln 850 855 860Ser Phe Leu Lys Asp Asp Ser Ile Asp Asn Lys Val Leu Thr Arg Ser865 870 875 880Asp Lys Asn Arg Gly Lys Ser Asp Asn Val Pro Ser Glu Glu Val Val 885 890 895Lys Lys Met Lys Asn Tyr Trp Arg Gln Leu Leu Asn Ala Lys Leu Ile 900 905 910Thr Gln Arg Lys Phe Asp Asn Leu Thr Lys Ala Glu Arg Gly Gly Leu 915 920 925Ser Glu Leu Asp Lys Ala Gly Phe Ile Lys Arg Gln Leu Val Glu Thr 930 935 940Arg Gln Ile Thr Lys His Val Ala Gln Ile Leu Asp Ser Arg Met Asn945 950 955 960Thr Lys Tyr Asp Glu Asn Asp Lys Leu Ile Arg Glu Val Lys Val Ile 965 970 975Thr Leu Lys Ser Lys Leu Val Ser Asp Phe Arg Lys Asp Phe Gln Phe 980 985 990Tyr Lys Val Arg Glu Ile Asn Asn Tyr His His Ala His Asp Ala Tyr 995 1000 1005Leu Asn Ala Val Val Gly Thr Ala Leu Ile Lys Lys Tyr Pro Lys 1010 1015 1020Leu Glu Ser Glu Phe Val Tyr Gly Asp Tyr Lys Val Tyr Asp Val 1025 1030 1035Arg Lys Met Ile Ala Lys Ser Glu Gln Glu Ile Gly Lys Ala Thr 1040 1045 1050Ala Lys Tyr Phe Phe Tyr Ser Asn Ile Met Asn Phe Phe Lys Thr 1055 1060 1065Glu Ile Thr Leu Ala Asn Gly Glu Ile Arg Lys Arg Pro Leu Ile 1070 1075 1080Glu Thr Asn Gly Glu Thr Gly Glu Ile Val Trp Asp Lys Gly Arg 1085 1090 1095Asp Phe Ala Thr Val Arg Lys Val Leu Ser Met Pro Gln Val Asn 1100 1105 1110Ile Val Lys Lys Thr Glu Val Gln Thr Gly Gly Phe Ser Lys Glu 1115 1120 1125Ser Ile Leu Pro Lys Arg Asn Ser Asp Lys Leu Ile Ala Arg Lys 1130 1135 1140Lys Asp Trp Asp Pro Lys Lys Tyr Gly Gly Phe Asp Ser Pro Thr 1145 1150 1155Val Ala Tyr Ser Val Leu Val Val Ala Lys Val Glu Lys Gly Lys 1160 1165 1170Ser Lys Lys Leu Lys Ser Val Lys Glu Leu Leu Gly Ile Thr Ile 1175 1180 1185Met Glu Arg Ser Ser Phe Glu Lys Asn Pro Ile Asp Phe Leu Glu 1190 1195 1200Ala Lys Gly Tyr Lys Glu Val Lys Lys Asp Leu Ile Ile Lys Leu 1205 1210 1215Pro Lys Tyr Ser Leu Phe Glu Leu Glu Asn Gly Arg Lys Cys Leu 1220 1225 1230Ser Tyr Glu Thr Glu Ile Leu Thr Val Glu Tyr Gly Leu Leu Pro 1235 1240 1245Ile Gly Lys Ile Val Glu Lys Arg Ile Glu Cys Thr Val Tyr Ser 1250 1255 1260Val Asp Asn Asn Gly Asn Ile Tyr Thr Gln Pro Val Ala Gln Trp 1265 1270 1275His Asp Arg Gly Glu Gln Glu Val Phe Glu Tyr Cys Leu Glu Asp 1280 1285 1290Gly Ser Leu Ile Arg Ala Thr Lys Asp His Lys Phe Met Thr Val 1295 1300 1305Asp Gly Gln Met Leu Pro Ile Asp Glu Ile Phe Glu Arg Glu Leu 1310 1315 1320Asp Leu Met Arg Val Asp Asn Leu Pro Asn 1325 13307803PRTArtificial SequenceSynthetic Polypeptide 7Met Ile Lys Ile Ala Thr Arg Lys Tyr Leu Gly Lys Gln Asn Val Tyr1 5 10 15Asp Ile Gly Val Glu Arg Asp His Asn Phe Ala Leu Lys Asn Gly Phe 20 25 30Ile Ala Ser Asn Cys Met Leu Ala Ser Ala Gly Glu Leu Gln Lys Gly 35 40 45Asn Glu Leu Ala Leu Pro Ser Lys Tyr Val Asn Phe Leu Tyr Leu Ala 50 55 60Ser His Tyr Glu Lys Leu Lys Gly Ser Pro Glu Asp Asn Glu Gln Lys65 70 75 80Gln Leu Phe Val Glu Gln His Lys His Tyr Leu Asp Glu Ile Ile Glu 85 90 95Gln Ile Ser Glu Phe Ser Lys Arg Val Ile Leu Ala Asp Ala Asn Leu 100 105 110Asp Lys Val Leu Ser Ala Tyr Asn Lys His Arg Asp Lys Pro Ile Arg 115 120 125Glu Gln Ala Glu Asn Ile Ile His Leu Phe Thr Leu Thr Asn Leu Gly 130 135 140Ala Pro Ala Ala Phe Lys Tyr Phe Asp Thr Thr Ile Asp Arg Lys Arg145 150 155 160Tyr Thr Ser Thr Lys Glu Val Leu Asp Ala Thr Leu Ile His Gln Ser 165 170 175Ile Thr Gly Leu Tyr Glu Thr Arg Ile Asp Leu Ser Gln Leu Gly Gly 180 185 190Asp Ser Pro Lys Lys Lys Arg Lys Val Glu Ala Ser Gly Ser Ala Pro 195 200 205Gly Lys Lys Arg Pro Val Glu His Ser Pro Val Glu Pro Asp Ser Ser 210 215 220Ser Gly Thr Gly Lys Ala Gly Gln Gln Pro Ala Arg Lys Arg Leu Asn225 230 235 240Phe Gly Gln Thr Gly Asp Ala Asp Ser Val Pro Asp Pro Gln Pro Leu 245 250 255Gly Gln Pro Pro Ala Ala Pro Ser Gly Leu Gly Thr Asn Thr Met Ala 260 265 270Thr Gly Ser Gly Ala Pro Met Ala Asp Asn Asn Glu Gly Ala Asp Gly 275 280 285Val Gly Asn Ser Ser Gly Asn Trp His Cys Asp Ser Thr Trp Met Gly 290 295 300Asp Arg Val Ile Thr Thr Ser Thr Arg Thr Trp Ala Leu Pro Thr Tyr305 310 315 320Asn Asn His Leu Tyr Lys Gln Ile Ser Ser Gln Ser Gly Ala Ser Asn 325 330 335Asp Asn His Tyr Phe Gly Tyr Ser Thr Pro Trp Gly Tyr Phe Asp Phe 340 345 350Asn Arg Phe His Cys His Phe Ser Pro Arg Asp Trp Gln Arg Leu Ile 355 360 365Asn Asn Asn Trp Gly Phe Arg Pro Lys Arg Leu Asn Phe Lys Leu Phe 370 375 380Asn Ile Gln Val Lys Glu Val Thr Gln Asn Asp Gly Thr Thr Thr Ile385 390 395 400Ala Asn Asn Leu Thr Ser Thr Val Gln Val Phe Thr Asp Ser Glu Tyr 405 410 415Gln Leu Pro Tyr Val Leu Gly Ser Ala His Gln Gly Cys Leu Pro Pro 420 425 430Phe Pro Ala Asp Val Phe Met Val Pro Gln Tyr Gly Tyr Leu Thr Leu 435 440 445Asn Asn Gly Ser Gln Ala Val Gly Arg Ser Ser Phe Tyr Cys Leu Glu 450 455 460Tyr Phe Pro Ser Gln Met Leu Arg Thr Gly Asn Asn Phe Thr Phe Ser465 470 475 480Tyr Thr Phe Glu Asp Val Pro Phe His Ser Ser Tyr Ala His Ser Gln 485 490 495Ser Leu Asp Arg Leu Met Asn Pro Leu Ile Asp Gln Tyr Leu Tyr Tyr 500 505 510Leu Ser Arg Thr Asn Thr Pro Ser Gly Thr Thr Thr Gln Ser Arg Leu 515 520 525Gln Phe Ser Gln Ala Gly Ala Ser Asp Ile Arg Asp Gln Ser Arg Asn 530 535 540Trp Leu Pro Gly Pro Cys Tyr Arg Gln Gln Arg Val Ser Lys Thr Ser545 550 555 560Ala Asp Asn Asn Asn Ser Glu Tyr Ser Trp Thr Gly Ala Thr Lys Tyr 565 570 575His Leu Asn Gly Arg Asp Ser Leu Val Asn Pro Gly Pro Ala Met Ala 580 585 590Ser His Lys Asp Asp Glu Glu Lys Phe Phe Pro Gln Ser Gly Val Leu 595 600 605Ile Phe Gly Lys Gln Gly Ser Glu Lys Thr Asn Val Asp Ile Glu Lys 610 615 620Val Met Ile Thr Asp Glu Glu Glu Ile Arg Thr Thr Asn Pro Val Ala625 630 635 640Thr Glu Gln Tyr Gly Ser Val Ser Thr Asn Leu Gln Arg Gly Asn Arg 645 650 655Gln Ala Ala Thr Ala Asp Val Asn Thr Gln Gly Val Leu Pro Gly Met 660 665 670Val Trp Gln Asp Arg Asp Val Tyr Leu Gln Gly Pro Ile Trp Ala Lys 675 680 685Ile Pro His Thr Asp Gly His Phe His Pro Ser Pro Leu Met Gly Gly 690 695 700Phe Gly Leu Lys His Pro Pro Pro Gln Ile Leu Ile Lys Asn Thr Pro705 710 715 720Val Pro Ala Asn Pro Ser Thr Thr Phe Ser Ala Ala Lys Phe Ala Ser 725 730 735Phe Ile Thr Gln Tyr Ser Thr Gly Gln Val Ser Val Glu Ile Glu Trp 740 745 750Glu Leu Gln Lys Glu Asn Ser Lys Arg Trp Asn Pro Glu Ile Gln Tyr 755 760 765Thr Ser Asn Tyr Asn Lys Ser Val Asn Val Asp Phe Thr Val Asp Thr 770 775 780Asn Gly Val Tyr Ser Glu Pro Arg Pro Ile Gly Thr Arg Tyr Leu Thr785 790 795 800Arg Asn Leu8816PRTArtificial SequenceSynthetic Polypeptide 8Met Ile Lys Ile Ala Thr Arg Lys Tyr Leu Gly Lys Gln Asn Val Tyr1 5 10 15Asp Ile Gly Val Glu Arg Asp His Asn Phe Ala Leu Lys Asn Gly Phe 20 25 30Ile Ala Ser Asn Cys Met Leu Ala Ser Ala Gly Glu Leu Gln Lys Gly 35 40 45Asn Glu Leu Ala Leu Pro Ser Lys Tyr Val Asn Phe Leu Tyr Leu Ala 50 55 60Ser His Tyr Glu Lys Leu Lys Gly Ser Pro Glu Asp Asn Glu Gln Lys65 70 75 80Gln Leu Phe Val Glu Gln His Lys His Tyr Leu Asp Glu Ile Ile Glu 85 90 95Gln Ile Ser Glu Phe Ser Lys Arg Val Ile Leu Ala Asp Ala Asn Leu 100 105 110Asp Lys Val Leu Ser Ala Tyr Asn Lys His Arg Asp Lys Pro Ile Arg 115 120
125Glu Gln Ala Glu Asn Ile Ile His Leu Phe Thr Leu Thr Asn Leu Gly 130 135 140Ala Pro Ala Ala Phe Lys Tyr Phe Asp Thr Thr Ile Asp Arg Lys Arg145 150 155 160Tyr Thr Ser Thr Lys Glu Val Leu Asp Ala Thr Leu Ile His Gln Ser 165 170 175Ile Thr Gly Leu Tyr Glu Thr Arg Ile Asp Leu Ser Gln Leu Gly Gly 180 185 190Asp Ser Pro Lys Lys Lys Arg Lys Val Glu Ala Ser Gly Gly Gly Gly 195 200 205Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Ala Pro Gly Lys Lys 210 215 220Arg Pro Val Glu His Ser Pro Val Glu Pro Asp Ser Ser Ser Gly Thr225 230 235 240Gly Lys Ala Gly Gln Gln Pro Ala Arg Lys Arg Leu Asn Phe Gly Gln 245 250 255Thr Gly Asp Ala Asp Ser Val Pro Asp Pro Gln Pro Leu Gly Gln Pro 260 265 270Pro Ala Ala Pro Ser Gly Leu Gly Thr Asn Thr Met Ala Thr Gly Ser 275 280 285Gly Ala Pro Met Ala Asp Asn Asn Glu Gly Ala Asp Gly Val Gly Asn 290 295 300Ser Ser Gly Asn Trp His Cys Asp Ser Thr Trp Met Gly Asp Arg Val305 310 315 320Ile Thr Thr Ser Thr Arg Thr Trp Ala Leu Pro Thr Tyr Asn Asn His 325 330 335Leu Tyr Lys Gln Ile Ser Ser Gln Ser Gly Ala Ser Asn Asp Asn His 340 345 350Tyr Phe Gly Tyr Ser Thr Pro Trp Gly Tyr Phe Asp Phe Asn Arg Phe 355 360 365His Cys His Phe Ser Pro Arg Asp Trp Gln Arg Leu Ile Asn Asn Asn 370 375 380Trp Gly Phe Arg Pro Lys Arg Leu Asn Phe Lys Leu Phe Asn Ile Gln385 390 395 400Val Lys Glu Val Thr Gln Asn Asp Gly Thr Thr Thr Ile Ala Asn Asn 405 410 415Leu Thr Ser Thr Val Gln Val Phe Thr Asp Ser Glu Tyr Gln Leu Pro 420 425 430Tyr Val Leu Gly Ser Ala His Gln Gly Cys Leu Pro Pro Phe Pro Ala 435 440 445Asp Val Phe Met Val Pro Gln Tyr Gly Tyr Leu Thr Leu Asn Asn Gly 450 455 460Ser Gln Ala Val Gly Arg Ser Ser Phe Tyr Cys Leu Glu Tyr Phe Pro465 470 475 480Ser Gln Met Leu Arg Thr Gly Asn Asn Phe Thr Phe Ser Tyr Thr Phe 485 490 495Glu Asp Val Pro Phe His Ser Ser Tyr Ala His Ser Gln Ser Leu Asp 500 505 510Arg Leu Met Asn Pro Leu Ile Asp Gln Tyr Leu Tyr Tyr Leu Ser Arg 515 520 525Thr Asn Thr Pro Ser Gly Thr Thr Thr Gln Ser Arg Leu Gln Phe Ser 530 535 540Gln Ala Gly Ala Ser Asp Ile Arg Asp Gln Ser Arg Asn Trp Leu Pro545 550 555 560Gly Pro Cys Tyr Arg Gln Gln Arg Val Ser Lys Thr Ser Ala Asp Asn 565 570 575Asn Asn Ser Glu Tyr Ser Trp Thr Gly Ala Thr Lys Tyr His Leu Asn 580 585 590Gly Arg Asp Ser Leu Val Asn Pro Gly Pro Ala Met Ala Ser His Lys 595 600 605Asp Asp Glu Glu Lys Phe Phe Pro Gln Ser Gly Val Leu Ile Phe Gly 610 615 620Lys Gln Gly Ser Glu Lys Thr Asn Val Asp Ile Glu Lys Val Met Ile625 630 635 640Thr Asp Glu Glu Glu Ile Arg Thr Thr Asn Pro Val Ala Thr Glu Gln 645 650 655Tyr Gly Ser Val Ser Thr Asn Leu Gln Arg Gly Asn Arg Gln Ala Ala 660 665 670Thr Ala Asp Val Asn Thr Gln Gly Val Leu Pro Gly Met Val Trp Gln 675 680 685Asp Arg Asp Val Tyr Leu Gln Gly Pro Ile Trp Ala Lys Ile Pro His 690 695 700Thr Asp Gly His Phe His Pro Ser Pro Leu Met Gly Gly Phe Gly Leu705 710 715 720Lys His Pro Pro Pro Gln Ile Leu Ile Lys Asn Thr Pro Val Pro Ala 725 730 735Asn Pro Ser Thr Thr Phe Ser Ala Ala Lys Phe Ala Ser Phe Ile Thr 740 745 750Gln Tyr Ser Thr Gly Gln Val Ser Val Glu Ile Glu Trp Glu Leu Gln 755 760 765Lys Glu Asn Ser Lys Arg Trp Asn Pro Glu Ile Gln Tyr Thr Ser Asn 770 775 780Tyr Asn Lys Ser Val Asn Val Asp Phe Thr Val Asp Thr Asn Gly Val785 790 795 800Tyr Ser Glu Pro Arg Pro Ile Gly Thr Arg Tyr Leu Thr Arg Asn Leu 805 810 8159816PRTArtificial SequenceSynthetic Polypeptide 9Met Ile Lys Ile Ala Thr Arg Lys Tyr Leu Gly Lys Gln Asn Val Tyr1 5 10 15Asp Ile Gly Val Glu Arg Asp His Asn Phe Ala Leu Lys Asn Gly Phe 20 25 30Ile Ala Ser Asn Cys Met Leu Ala Ser Ala Gly Glu Leu Gln Lys Gly 35 40 45Asn Glu Leu Ala Leu Pro Ser Lys Tyr Val Asn Phe Leu Tyr Leu Ala 50 55 60Ser His Tyr Glu Lys Leu Lys Gly Ser Pro Glu Asp Asn Glu Gln Lys65 70 75 80Gln Leu Phe Val Glu Gln His Lys His Tyr Leu Asp Glu Ile Ile Glu 85 90 95Gln Ile Ser Glu Phe Ser Lys Arg Val Ile Leu Ala Asp Ala Asn Leu 100 105 110Asp Lys Val Leu Ser Ala Tyr Asn Lys His Arg Asp Lys Pro Ile Arg 115 120 125Glu Gln Ala Glu Asn Ile Ile His Leu Phe Thr Leu Thr Asn Leu Gly 130 135 140Ala Pro Ala Ala Phe Lys Tyr Phe Asp Thr Thr Ile Asp Arg Lys Arg145 150 155 160Tyr Thr Ser Thr Lys Glu Val Leu Asp Ala Thr Leu Ile His Gln Ser 165 170 175Ile Thr Gly Leu Tyr Glu Thr Arg Ile Asp Leu Ser Gln Leu Gly Gly 180 185 190Asp Ser Pro Lys Lys Lys Arg Lys Val Glu Ala Ser Glu Ala Ala Ala 195 200 205Lys Glu Ala Ala Ala Lys Glu Ala Ala Ala Lys Ala Pro Gly Lys Lys 210 215 220Arg Pro Val Glu His Ser Pro Val Glu Pro Asp Ser Ser Ser Gly Thr225 230 235 240Gly Lys Ala Gly Gln Gln Pro Ala Arg Lys Arg Leu Asn Phe Gly Gln 245 250 255Thr Gly Asp Ala Asp Ser Val Pro Asp Pro Gln Pro Leu Gly Gln Pro 260 265 270Pro Ala Ala Pro Ser Gly Leu Gly Thr Asn Thr Met Ala Thr Gly Ser 275 280 285Gly Ala Pro Met Ala Asp Asn Asn Glu Gly Ala Asp Gly Val Gly Asn 290 295 300Ser Ser Gly Asn Trp His Cys Asp Ser Thr Trp Met Gly Asp Arg Val305 310 315 320Ile Thr Thr Ser Thr Arg Thr Trp Ala Leu Pro Thr Tyr Asn Asn His 325 330 335Leu Tyr Lys Gln Ile Ser Ser Gln Ser Gly Ala Ser Asn Asp Asn His 340 345 350Tyr Phe Gly Tyr Ser Thr Pro Trp Gly Tyr Phe Asp Phe Asn Arg Phe 355 360 365His Cys His Phe Ser Pro Arg Asp Trp Gln Arg Leu Ile Asn Asn Asn 370 375 380Trp Gly Phe Arg Pro Lys Arg Leu Asn Phe Lys Leu Phe Asn Ile Gln385 390 395 400Val Lys Glu Val Thr Gln Asn Asp Gly Thr Thr Thr Ile Ala Asn Asn 405 410 415Leu Thr Ser Thr Val Gln Val Phe Thr Asp Ser Glu Tyr Gln Leu Pro 420 425 430Tyr Val Leu Gly Ser Ala His Gln Gly Cys Leu Pro Pro Phe Pro Ala 435 440 445Asp Val Phe Met Val Pro Gln Tyr Gly Tyr Leu Thr Leu Asn Asn Gly 450 455 460Ser Gln Ala Val Gly Arg Ser Ser Phe Tyr Cys Leu Glu Tyr Phe Pro465 470 475 480Ser Gln Met Leu Arg Thr Gly Asn Asn Phe Thr Phe Ser Tyr Thr Phe 485 490 495Glu Asp Val Pro Phe His Ser Ser Tyr Ala His Ser Gln Ser Leu Asp 500 505 510Arg Leu Met Asn Pro
References
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.