Methods And Compositions For Diagnosis And Prognosis Of Renal Injury And Renal Failure
ANDERBERG; JOSEPH ; et al.
U.S. patent application number 17/153134 was filed with the patent office on 2021-05-27 for methods and compositions for diagnosis and prognosis of renal injury and renal failure. This patent application is currently assigned to ASTUTE MEDICAL, INC.. The applicant listed for this patent is ASTUTE MEDICAL, INC.. Invention is credited to JOSEPH ANDERBERG, JEFF GRAY, JAMES PATRICK KAMPF, PAUL MCPHERSON, KEVIN NAKAMURA.
| Application Number | 20210156850 17/153134 |
| Document ID | / |
| Family ID | 1000005373606 |
| Filed Date | 2021-05-27 |
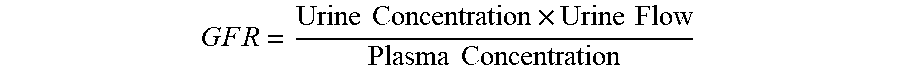









View All Diagrams
| United States Patent Application | 20210156850 |
| Kind Code | A1 |
| ANDERBERG; JOSEPH ; et al. | May 27, 2021 |
METHODS AND COMPOSITIONS FOR DIAGNOSIS AND PROGNOSIS OF RENAL INJURY AND RENAL FAILURE
Abstract
The present invention relates to methods and compositions for monitoring, diagnosis, prognosis, and determination of treatment regimens in sepsis patients . In particular, the invention relates to using assays that detect one or more biomarkers selected from the group consisting of Insulin-like growth factor-binding protein 7, Beta-2-glycoprotein 1, Metalloproteinase inhibitor 2, Alpha-1 Antitrypsin, Leukocyte elastase, Serum Amyloid P Component, C--X--C motif chemokine 6, Immunoglobulin A, Immunoglobulin G subclass I, C--C motif chemokine 24, Neutrophil collagenase, Cathepsin D, C--X--C motif chemokine 13, Involucrin, Interleukin-6 receptor subunit beta, Hepatocyte Growth Factor, CXCL-1, -2, -3, Immunoglobulin G subclass II, Metalloproteinase inhibitor 4, C--C motif chemokine 18, Matrilysin, C--X--C motif chemokine 11, and Antileukoproteinase as diagnostic and prognostic biomarker assays of renal injury in the sepsis patient.
| Inventors: | ANDERBERG; JOSEPH; (ENCINITAS, CA) ; GRAY; JEFF; (SOLANA BEACH, CA) ; MCPHERSON; PAUL; (ENCINITAS, CA) ; NAKAMURA; KEVIN; (CARDIFF BY THE SEA, CA) ; KAMPF; JAMES PATRICK; (SAN DIEGO, CA) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Assignee: | ASTUTE MEDICAL, INC. SAN DIEGO CA |
||||||||||
| Family ID: | 1000005373606 | ||||||||||
| Appl. No.: | 17/153134 | ||||||||||
| Filed: | January 20, 2021 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 14363724 | Jun 6, 2014 | 10935548 | ||
| PCT/US12/68498 | Dec 7, 2012 | |||
| 17153134 | ||||
| 61568447 | Dec 8, 2011 | |||
| 61593561 | Feb 1, 2012 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | G01N 2800/347 20130101; G01N 2800/52 20130101; G01N 33/6893 20130101; G01N 2333/8146 20130101; G01N 2800/56 20130101; G01N 33/545 20130101; G01N 2800/26 20130101 |
| International Class: | G01N 33/545 20060101 G01N033/545; G01N 33/68 20060101 G01N033/68 |
Claims
1. A method for evaluating renal status in a sepsis patient, comprising: performing one or more assays configured to detect one or more biomarkers selected from the group consisting of Insulin-like growth factor-binding protein 7, Beta-2-glycoprotein 1, Metalloproteinase inhibitor 2, Alpha-1 Antitrypsin, Leukocyte elastase, Serum Amyloid P Component, C--X--C motif chemokine 6, Immunoglobulin A, Immunoglobulin G subclass I, C--C motif chemokine 24, Neutrophil collagenase, Cathepsin D, C--X--C motif chemokine 13, Involucrin, Interleukin-6 receptor subunit beta, Hepatocyte Growth Factor, CXCL-1, -2, -3, Immunoglobulin G subclass II, Metalloproteinase inhibitor 4, C--C motif chemokine 18, Matrilysin, C--X--C motif chemokine 11, and Antileukoproteinase on a body fluid sample obtained from the sepsis patient to provide an assay result; and correlating the assay result(s) to the renal status of the sepsis patient.
2. A method according to claim 1, wherein said correlation step comprises correlating the assay result(s) to one or more of risk stratification, diagnosis, staging, prognosis, classifying and monitoring of the renal status of the sepsis patient.
3. A method according to claim 1, wherein said correlating step comprises assigning a likelihood of one or more future changes in renal status to the sepsis patient based on the assay result(s).
4. A method according to claim 3, wherein said one or more future changes in renal status comprise one or more of a future injury to renal function, future reduced renal function, future improvement in renal function, and future acute renal failure (ARF).
5. A method according to one of claims 1-4, wherein said assay results comprise at least 2, 3, or 4 of: a measured urine or plasma concentration of Insulin-like growth factor-binding protein 7, a measured urine or plasma concentration of Beta-2-glycoprotein 1, a measured urine or plasma concentration of Metalloproteinase inhibitor 2, a measured urine or plasma concentration of Alpha-1 Antitrypsin, a measured urine or plasma concentration of Leukocyte elastase, a measured urine or plasma concentration of Serum Amyloid P Component, a measured urine or plasma concentration of C--X--C motif chemokine 6, a measured urine or plasma concentration of Immunoglobulin A, a measured urine or plasma concentration of Immunoglobulin G subclass I, a measured urine or plasma concentration of C--C motif chemokine 24, a measured urine or plasma concentration of Neutrophil collagenase, a measured urine or plasma concentration of Cathepsin D, a measured urine or plasma concentration of C--X--C motif chemokine 13, a measured urine or plasma concentration of Involucrin, a measured urine or plasma concentration of Interleukin-6 receptor subunit beta, a measured urine or plasma concentration of Hepatocyte Growth Factor, a measured urine or plasma concentration of CXCL-1, a measured urine or plasma concentration of -2, a measured urine or plasma concentration of -3, a measured urine or plasma concentration of Immunoglobulin G subclass II, a measured urine or plasma concentration of Metalloproteinase inhibitor 4, a measured urine or plasma concentration of C--C motif chemokine 18, a measured urine or plasma concentration of Matrilysin, a measured urine or plasma concentration of C--X--C motif chemokine 11, and a measured urine or plasma concentration of Antileukoproteinase.
6. A method according to one of claims 1-5, wherein a plurality of assay results are combined using a function that converts the plurality of assay results into a single composite result.
7. A method according to claim 3, wherein said one or more future changes in renal status comprise a clinical outcome related to a renal injury suffered by the sepsis patient.
8. A method according to claim 3, wherein the likelihood of one or more future changes in renal status is that an event of interest is more or less likely to occur within 30 days of the time at which the body fluid sample is obtained from the sepsis patient.
9. A method according to claim 8, wherein the likelihood of one or more future changes in renal status is that an event of interest is more or less likely to occur within a period selected from the group consisting of 21 days, 14 days, 7 days, 5 days, 96 hours, 72 hours, 48 hours, 36 hours, 24 hours, and 12 hours.
10. A method according to one of claims 1-5, wherein the sepsis patient is a severe sepsis patient.
11. A method according to one of claims 1-5, wherein the sepsis patient is a septic shock patient.
12. A method according to one of claims 1-5, wherein said correlating step comprises assigning a diagnosis of the occurrence or nonoccurrence of one or more of an injury to renal function, reduced renal function, or ARF to the sepsis patient based on the assay result(s).
13. A method according to one of claims 1-5, wherein said correlating step comprises assessing whether or not renal function is improving or worsening in a sepsis patient who has suffered from an injury to renal function, reduced renal function, or ARF based on the assay result(s).
14. A method according to one of claims 1-5, wherein said method is a method of diagnosing the occurrence or nonoccurrence of an injury to renal function in said sepsis patient.
15. A method according to one of claims 1-5, wherein said method is a method of diagnosing the occurrence or nonoccurrence of reduced renal function in said sepsis patient.
16. A method according to one of claims 1-5, wherein said method is a method of diagnosing the occurrence or nonoccurrence of acute renal failure in said sepsis patient.
17. A method according to one of claims 1-5, wherein said method is a method of diagnosing the occurrence or nonoccurrence of a need for renal replacement therapy in said sepsis patient.
18. A method according to one of claims 1-5, wherein said method is a method of diagnosing the occurrence or nonoccurrence of a need for renal transplantation in said sepsis patient.
19. A method according to one of claims 1-5, wherein said method is a method of assigning a risk of the future occurrence or nonoccurrence of an injury to renal function in said sepsis patient.
20. A method according to one of claims 1-5, wherein said method is a method of assigning a risk of the future occurrence or nonoccurrence of reduced renal function in said sepsis patient.
21. A method according to one of claims 1-5, wherein said method is a method of assigning a risk of the future occurrence or nonoccurrence of acute renal failure in said sepsis patient.
22. A method according to one of claims 1-5, wherein said method is a method of assigning a risk of the future occurrence or nonoccurrence of a need for renal replacement therapy in said sepsis patient.
23. A method according to one of claims 1-5, wherein said method is a method of assigning a risk of the future occurrence or nonoccurrence of a need for renal transplantation in said sepsis patient.
24. A method according to one of claims 1-5, wherein said one or more future changes in renal status comprise one or more of a future injury to renal function, future reduced renal function, future improvement in renal function, and future acute renal failure (ARF) within 72 hours of the time at which the body fluid sample is obtained.
25. A method according to one of claims 1-5, wherein said one or more future changes in renal status comprise one or more of a future injury to renal function, future reduced renal function, future improvement in renal function, and future acute renal failure (ARF) within 48 hours of the time at which the body fluid sample is obtained.
26. A method according to one of claims 1-5, wherein said one or more future changes in renal status comprise one or more of a future injury to renal function, future reduced renal function, future improvement in renal function, and future acute renal failure (ARF) within 24 hours of the time at which the body fluid sample is obtained.
27. A method according to one of claims 1-5, wherein the sepsis patient is in RIFLE stage 0 or R.
28. A method according to claim 27, wherein the sepsis patient is in RIFLE stage 0, and said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage R, I or F within 72 hours.
29. A method according to claim 28, wherein the sepsis patient is in RIFLE stage 0, and said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage I or F within 72 hours.
30. A method according to claim 28, wherein the sepsis patient is in RIFLE stage 0, and said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage F within 72 hours.
31. A method according to claim 27, wherein the sepsis patient is in RIFLE stage 0 or R, and said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage I or F within 72 hours.
32. A method according to claim 31, wherein the sepsis patient is in RIFLE stage 0 or R, and said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage F within 72 hours.
33. A method according to claim 27, wherein the sepsis patient is in RIFLE stage R, and said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage I or F within 72 hours.
34. A method according to claim 33, wherein the sepsis patient is in RIFLE stage R, and said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage F within 72 hours.
35. A method according to one of claims 1-5, wherein the sepsis patient is in RIFLE stage 0, R, or I, and said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage F within 72 hours.
36. A method according to claim 35, wherein the sepsis patient is in RIFLE stage I, and said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage F within 72 hours.
37. A method according to claim 28, wherein said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage R, I or F within 48 hours.
38. A method according to claim 29, wherein said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage I or F within 48 hours.
39. A method according to claim 30, wherein said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage F within 48 hours.
40. A method according to claim 31, wherein said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage I or F within 48 hours.
41. A method according to claim 32, wherein said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage F within 48 hours.
42. A method according to claim 33, wherein said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage I or F within 48 hours.
43. A method according to claim 34, wherein said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage F within 48 hours.
44. A method according to claim 35, wherein said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage F within 48 hours.
45. A method according to claim 36, wherein said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage F within 48 hours.
46. A method according to claim 28, wherein said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage R, I or F within 24 hours.
47. A method according to claim 29, wherein said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage I or F within 24 hours.
48. A method according to claim 30, wherein said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage F within 24 hours.
49. A method according to claim 31, wherein said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage I or F within 24 hours.
50. A method according to claim 32, wherein said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage F within 24 hours.
51. A method according to claim 33, wherein said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage I or F within 24 hours.
52. A method according to claim 34, wherein said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage F within 24 hours.
53. A method according to claim 35, wherein said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage F within 24 hours.
54. A method according to claim 36, wherein said correlating step comprises assigning a likelihood that the sepsis patient will reach RIFLE stage F within 24 hours.
55. A method according to one of claims 1-5, wherein the sepsis patient is not in acute renal failure.
56. A method according to one of claims 1-5, wherein the sepsis patient has not experienced a 1.5-fold or greater increase in serum creatinine over a baseline value determined prior to the time at which the body fluid sample is obtained.
57. A method according to one of claims 1-5, wherein the sepsis patient has a urine output of at least 0.5 ml/kg/hr over the 6 hours preceding the time at which the body fluid sample is obtained.
58. A method according to one of claims 1-5, wherein the sepsis patient has not experienced an increase of 0.3 mg/dL or greater in serum creatinine over a baseline value determined prior to the time at which the body fluid sample is obtained.
59. A method according to one of claims 1-5, wherein the sepsis patient (i) has not experienced a 1.5-fold or greater increase in serum creatinine over a baseline value determined prior to the time at which the body fluid sample is obtained, (ii) has a urine output of at least 0.5 ml/kg/hr over the 6 hours preceding the time at which the body fluid sample is obtained, and (iii) has not experienced an increase of 0.3 mg/dL or greater in serum creatinine over a baseline value determined prior to the time at which the body fluid sample is obtained.
60. A method according to one of claims 1-5, wherein the sepsis patient has not experienced a 1.5-fold or greater increase in serum creatinine over a baseline value determined prior to the time at which the body fluid sample is obtained.
61. A method according to one of claims 1-5, wherein the sepsis patient has a urine output of at least 0.5 ml/kg/hr over the 6 hours preceding the time at which the body fluid sample is obtained.
62. A method according to one of claims 1-5, wherein the sepsis patient (i) has not experienced a 1.5-fold or greater increase in serum creatinine over a baseline value determined prior to the time at which the body fluid sample is obtained, (ii) has a urine output of at least 0.5 ml/kg/hr over the 12 hours preceding the time at which the body fluid sample is obtained, and (iii) has not experienced an increase of 0.3 mg/dL or greater in serum creatinine over a baseline value determined prior to the time at which the body fluid sample is obtained.
63. A method according to one of claims 1-5, wherein said correlating step comprises assigning one or more of: a likelihood that within 72 hours the sepsis patient will (i) experience a 1.5-fold or greater increase in serum creatinine (ii) have a urine output of less than 0.5 ml/kg/hr over a 6 hour period, or (iii) experience an increase of 0.3 mg/dL, or greater in serum creatinine.
64. A method according to claim 63, wherein said correlating step comprises assigning one or more of: a likelihood that within 48 hours the sepsis patient will (i) experience a 1.5-fold or greater increase in serum creatinine (ii) have a urine output of less than 0.5 ml/kg/hr over a 6 hour period, or (iii) experience an increase of 0.3 mg/dL or greater in serum creatinine.
65. A method according to claim 63, wherein said correlating step comprises assigning one or more of: a likelihood that within 24 hours the sepsis patient will (i) experience a 1.5-fold or greater increase in serum creatinine (ii) have a urine output of less than 0.5 ml/kg/hr over a 6 hour period, or (iii) experience an increase of 0.3 mg/dL or greater in serum creatinine.
66. A method according to claim 63, wherein said correlating step comprises assigning a likelihood that within 72 hours the sepsis patient will experience a 1.5-fold or greater increase in serum creatinine.
67. A method according to claim 63, wherein said correlating step comprises assigning a likelihood that within 72 hours the sepsis patient will have a urine output of less than 0.5 ml/kg/hr over a 6 hour period.
68. A method according to claim 63, wherein said correlating step comprises assigning a likelihood that within 72 hours the sepsis patient will experience an increase of 0.3 mg/dL or greater in serum creatinine.
69. A method according to claim 63, wherein said correlating step comprises assigning a likelihood that within 48 hours the sepsis patient will experience a 1.5-fold or greater increase in serum creatinine.
70. A method according to claim 63, wherein said correlating step comprises assigning a likelihood that within 48 hours the sepsis patient will have a urine output of less than 0.5 ml/kg/hr over a 6 hour period.
71. A method according to claim 63, wherein said correlating step comprises assigning a likelihood that within 48 hours the sepsis patient will experience an increase of 0.3 mg/dL or greater in serum creatinine.
72. A method according to claim 63, wherein said correlating step comprises assigning a likelihood that within 24 hours the sepsis patient will experience a 1.5-fold or greater increase in serum creatinine.
73. A method according to claim 63, wherein said correlating step comprises assigning a likelihood that within 24 hours the sepsis patient will have a urine output of less than 0.5 ml/kg/hr over a 6 hour period.
74. A method according to claim 63, wherein said correlating step comprises assigning a likelihood that within 24 hours the sepsis patient will experience an increase of 0.3 mg/dL or greater in serum creatinine.
75. A method according to one of claims 1-5, wherein the sepsis patient has not experienced a 2-fold or greater increase in serum creatinine over a baseline value determined prior to the time at which the body fluid sample is obtained.
76. A method according to one of claims 1-5, wherein the sepsis patient has a urine output of at least 0.5 ml/kg/hr over the 12 hours preceding the time at which the body fluid sample is obtained.
77. A method according to one of claims 1-5, wherein the sepsis patient (i) has not experienced a 2-fold or greater increase in serum creatinine over a baseline value determined prior to the time at which the body fluid sample is obtained, (ii) has a urine output of at least 0.5 ml/kg/hr over the 2 hours preceding the time at which the body fluid sample is obtained, and (iii) has not experienced an increase of 0.3 mg/dL or greater in serum creatinine over a baseline value determined prior to the time at which the body fluid sample is obtained.
78. A method according to one of claims 1-5, wherein the sepsis patient has not experienced a 3-fold or greater increase in serum creatinine over a baseline value determined prior to the time at which the body fluid sample is obtained.
79. A method according to one of claims 1-5, wherein the sepsis patient has a urine output of at least 0.3 ml/kg/hr over the 24 hours preceding the time at which the body fluid sample is obtained, or anuria over the 12 hours preceding the time at which the body fluid sample is obtained.
80. A method according to one of claims 1-5, wherein the sepsis patient (i) has not experienced a 3-fold or greater increase in serum creatinine over a baseline value determined prior to the time at which the body fluid sample is obtained, (ii) has a urine output of at least 0.3 ml/kg/hr over the 24 hours preceding the time at which the body fluid sample is obtained, or anuria over the 12 hours preceding the time at which the body fluid sample is obtained, and (iii) has not experienced an increase of 0.3 mg/dL or greater in serum creatinine over a baseline value determined prior to the time at which the body fluid sample is obtained.
81. A method according to one of claims 1-5, wherein said correlating step comprises assigning one or more of: a likelihood that within 72 hours the sepsis patient will (i) experience a 2-fold or greater increase in serum creatinine (ii) have a urine output of less than 0.5 ml/kg/hr over a 12 hour period, or (iii) experience an increase of 0.3 mg/dL or greater in serum creatinine.
82. A method according to claim 81, wherein said correlating step comprises assigning one or more of: a likelihood that within 48 hours the sepsis patient will (i) experience a 2-fold or greater increase in serum creatinine (ii) have a urine output of less than 0.5 ml/kg/hr over a 6 hour period, or (iii) experience an increase of 0.3 mg/dL or greater in serum creatinine.
83. A method according to claim 81, wherein said correlating step comprises assigning one or more of: a likelihood that within 24 hours the sepsis patient will (i) experience a 2-fold or greater increase in serum creatinine, or (ii) have a urine output of less than 0.5 ml/kg/hr over a 6 hour period.
84. A method according to claim 81, wherein said correlating step comprises assigning a likelihood that within 72 hours the sepsis patient will experience a 2-fold or greater increase in serum creatinine.
85. A method according to claim 81, wherein said correlating step comprises assigning a likelihood that within 72 hours the sepsis patient will have a urine output of less than 0.5 ml/kg/hr over a 6 hour period.
86. A method according to claim 81, wherein said correlating step comprises assigning a likelihood that within 48 hours the sepsis patient will experience a 2-fold or greater increase in serum creatinine.
87. A method according to claim 81, wherein said correlating step comprises assigning a likelihood that within 48 hours the sepsis patient will have a urine output of less than 0.5 ml/kg/hr over a 6 hour period.
88. A method according to claim 81, wherein said correlating step comprises assigning a likelihood that within 24 hours the sepsis patient will experience a 2-fold or greater increase in serum creatinine.
89. A method according to claim 81, wherein said correlating step comprises assigning a likelihood that within 24 hours the sepsis patient will have a urine output of less than 0.5 ml/kg/hr over a 6 hour period.
90. A method according to one of claims 1-5, wherein said correlating step comprises assigning one or more of: a likelihood that within 72 hours the sepsis patient will (i) experience a 3-fold or greater increase in serum creatinine, or (ii) have a urine output of less than 0.3 ml/kg/hr over a 24 hour period or anuria over a 12 hour period.
91. A method according to claim 90, wherein said correlating step comprises assigning one or more of: a likelihood that within 48 hours the sepsis patient will (i) experience a 3-fold or greater increase in serum creatinine, or (ii) have a urine output of less than 0.3 ml/kg/hr over a 24 hour period or anuria over a 12 hour period.
92. A method according to claim 90, wherein said correlating step comprises assigning one or more of: a likelihood that within 24 hours the sepsis patient will (i) experience a 3-fold or greater increase in serum creatinine, or (ii) have a urine output of less than 0.3 ml/kg/hr over a 24 hour period or anuria over a 12 hour period.
93. A method according to claim 90, wherein said correlating step comprises assigning a likelihood that within 72 hours the sepsis patient will experience a 3-fold or greater increase in serum creatinine.
94. A method according to claim 90, wherein said correlating step comprises assigning a likelihood that within 72 hours the sepsis patient will have a urine output of less than 0.3 ml/kg/hr over a 24 hour period or anuria over a 12 hour period.
95. A method according to claim 90, wherein said correlating step comprises assigning a likelihood that within 48 hours the sepsis patient will experience a 3-fold or greater increase in serum creatinine.
96. A method according to claim 90, wherein said correlating step comprises assigning a likelihood that within 48 hours the sepsis patient will have a urine output of less than 0.3 ml/kg/hr over a 24 hour period or anuria over a 12 hour period.
97. A method according to claim 90, wherein said correlating step comprises assigning a likelihood that within 24 hours the sepsis patient will experience a 3-fold or greater increase in serum creatinine.
98. A method according to claim 90, wherein said correlating step comprises assigning a likelihood that within 24 hours the sepsis patient will have a urine output of less than 0.3 ml/kg/hr over a 24 hour period or anuria over a 12 hour period.
99. A method according to one of claims 1-98, wherein the body fluid sample is a urine sample.
100. A method according to one of claims 1-99, wherein said method comprises performing assays that detect one, two or three, or more of Insulin-like growth factor-binding protein 7, Beta-2-glycoprotein 1, Metalloproteinase inhibitor 2, Alpha-1 Antitrypsin, Leukocyte elastase, Serum Amyloid P Component, C--X--C motif chemokine 6, Immunoglobulin A, Immunoglobulin G subclass I, C--C motif chemokine 24, Neutrophil collagenase, Cathepsin D, C--X--C motif chemokine 13, Involucrin, Interleukin-6 receptor subunit beta, Hepatocyte Growth Factor, CXCL-1, -2, -3, Immunoglobulin G subclass II, Metalloproteinase inhibitor 4, C--C motif chemokine 18, Matrilysin, C--X--C motif chemokine 11, and Antileukoproteinase.
101. Measurement of one or more biomarkers selected from the group consisting of Insulin-like growth factor-binding protein 7, Beta-2-glycoprotein 1, Metalloproteinase inhibitor 2, Alpha-1 Antitrypsin, Leukocyte elastase, Serum Amyloid P Component, C--X--C motif chemokine 6, Immunoglobulin A, Immunoglobulin G subclass I, C--C motif chemokine 24, Neutrophil collagenase, Cathepsin D, C--X--C motif chemokine 13, Involucrin, Interleukin-6 receptor subunit beta, Hepatocyte Growth Factor, CXCL-1, -2, -3, Immunoglobulin G subclass II, Metalloproteinase inhibitor 4, C--C motif chemokine 18, Matrilysin, C--X--C motif chemokine 11, and Antileukoproteinase for the evaluation of renal injury.
102. Measurement of one or more biomarkers selected from the group consisting of Insulin-like growth factor-binding protein 7, Beta-2-glycoprotein 1, Metalloproteinase inhibitor 2, Alpha-1 Antitrypsin, Leukocyte elastase, Serum Amyloid P Component, C--X--C motif chemokine 6, Immunoglobulin A, Immunoglobulin G subclass I, C--C motif chemokine 24, Neutrophil collagenase, Cathepsin D, C--X--C motif chemokine 13, Involucrin, Interleukin-6 receptor subunit beta, Hepatocyte Growth Factor, CXCL-1, -2, -3, Immunoglobulin G subclass 11, Metalloproteinase inhibitor 4, C--C motif chemokine 18, Matrilysin, C--X--C motif chemokine 11, and Antileukoproteinase for the evaluation of acute renal injury.
103. A kit, comprising: reagents for performing one or more assays configured to detect one or more kidney injury markers selected from the group consisting of Insulin-like growth factor-binding protein 7, Beta-2-glycoprotein 1, Metalloproteinase inhibitor 2, Alpha-1 Antitrypsin, Leukocyte elastase, Serum Amyloid P Component, C--X--C motif chemokine 6, Immunoglobulin A, Immunoglobulin G subclass I, C--C motif chemokine 24, Neutrophil collagenase, Cathepsin D, C--X--C motif chemokine 13, Involucrin, Interleukin-6 receptor subunit beta, Hepatocyte Growth Factor, CXCL-1, -2, -3, Immunoglobulin G subclass II, Metalloproteinase inhibitor 4, C--C motif chemokine 18, Matrilysin, C--X--C motif chemokine 11, and Antileukoproteinase.
104. A kit according to claim 103, wherein said reagents comprise one or more binding reagents, each of which specifically binds one of said of kidney injury markers.
105. A kit according to claim 104, wherein a plurality of binding reagents are contained in a single assay device.
106. A kit according to claim 103, wherein at least one of said assays is configured as a sandwich binding assay.
107. A kit according to claim 103, wherein at least one of said assays is configured as a competitive binding assay.
108. A kit according to one of claims 103-107, wherein said one or more assays comprise assays that detect one, two or three, or more of Insulin-like growth factor-binding protein 7, Beta-2-glycoprotein 1, Metalloproteinase inhibitor 2, Alpha-1 Antitrypsin, Leukocyte elastase, Serum Amyloid P Component, C--X--C motif chemokine 6, Immunoglobulin A, Immunoglobulin G subclass I, C--C motif chemokine 24, Neutrophil collagenase, Cathepsin D, C--X--C motif chemokine 13, Involucrin, Interleukin-6 receptor subunit beta, Hepatocyte Growth Factor, CXCL-1, -2, -3, Immunoglobulin G subclass II, Metalloproteinase inhibitor 4, C--C motif chemokine 18, Matrilysin, C--X--C motif chemokine 11, and Antileukoproteinase.
109. A method for evaluating biomarker levels in a body fluid sample, comprising: obtaining a body fluid sample from a subject selected for evaluation based on a determination that the subject has sepsis; and performing a plurality of analyte binding assays configured to detect a plurality of biomarkers, one or more of which is selected from the group consisting of Insulin-like growth factor-binding protein 7, Beta-2-glycoprotein 1, Metalloproteinase inhibitor 2, Alpha-1 Antitrypsin, Leukocyte elastase, Serum Amyloid P Component, C--X--C motif chemokine 6, Immunoglobulin A, Immunoglobulin G subclass I, C--C motif chemokine 24, Neutrophil collagenase, Cathepsin D, C--X--C motif chemokine 13, Involucrin, Interleukin-6 receptor subunit beta, Hepatocyte Growth Factor, CXCL-1, -2, -3, Immunoglobulin G subclass II, Metalloproteinase inhibitor 4, C--C motif chemokine 18, Matrilysin, C--X--C motif chemokine 11, and Antileukoproteinase by introducing the urine sample obtained from the subject into an assay instrument which (i) contacts a plurality of reagents which specifically bind for detection the plurality of biomarkers with the urine sample, and (ii) generates one or more assay results indicative of binding of each biomarker which is assayed to a respective specific binding reagent in the plurality of reagents, and (iii) correlates the one or more assay results to a likelihood of worsening or improving renal function.
110. A method according to claim 109, wherein the body fluid sample is a urine sample.
111. A method according to claim 109 or 110, wherein the correlation is to a likelihood of future acute renal injury (AKI) or acute renal failure (ARF).
112. A method according to claim 111, wherein the correlation is to a likelihood of of a future acute renal injury within a period selected from the group consisting of 21 days, 14 days, 7 days, 5 days, 96 hours, 72 hours, 48 hours, 36 hours, 24 hours, 18 hours, and 12 hours.
113. A method according to claim 111, wherein the wherein the correlation is to a likelihood of of a future acute renal failure within a period selected from the group consisting of 21 days, 14 days, 7 days, 5 days, 96 hours, 72 hours, 48 hours, 36 hours, 24 hours, 18 hours, and 12 hours.
114. A method according to one of claims 109-113, wherein the plurality of assays are immunoassays performed by (i) introducing the urine sample into an assay device comprising a plurality of antibodies, at least one of which binds to each biomarker which is assayed, and (ii) generating an assay result indicative of binding of each biomarker to its respective antibody.
115. A method according to one of claims 109-110, wherein the subject is in RIFLE stage 0 or R.
116. A method according to one of claims 109-110, wherein the subject is in RIFLE stage 0, R, or I.
117. A system for evaluating biomarker levels, comprising: a plurality of reagents which specifically bind for detection the plurality of biomarkers, one or more of which is selected from the group consisting of Insulin-like growth factor-binding protein 7, Beta-2-glycoprotein 1, Metalloproteinase inhibitor 2, Alpha-1 Antitrypsin, Leukocyte elastase, Serum Amyloid P Component, C--X--C motif chemokine 6, Immunoglobulin A, Immunoglobulin G subclass I, C--C motif chemokine 24, Neutrophil collagenase, Cathepsin D, C--X--C motif chemokine 13, Involucrin, Interleukin-6 receptor subunit beta, Hepatocyte Growth Factor, CXCL-1, -2, -3, Immunoglobulin G subclass II, Metalloproteinase inhibitor 4, C--C motif chemokine 18, Matrilysin, C--X--C motif chemokine 11, and Antileukoproteinase; an assay instrument configured to receive a urine sample and contact the plurality of reagents with the urine sample to generate one or more assay results indicative of binding of each biomarker which is assayed to a respective specific binding reagent in the plurality of reagents, and to correlate the one or more assay results to a likelihood of worsening or improving renal function
118. A system according to claim 117, wherein the reagents comprise a plurality of antibodies, at least one of which binds to each of the biomarkers which are assayed.
119. A system according to claim 118, wherein assay instrument comprises an assay device and an assay device reader, wherein the plurality of antibodies are immobilized at a plurality of predetermined locations within the assay device, wherein the assay device is configured to receive the urine sample such that the urine sample contacts the plurality of predetermined locations, and wherein the assay device reader interrogates the plurality of predetermined locations to generate the assay results.
Description
[0001] This application claims priority to U.S. Provisional Patent Application 61/568,447, filed Dec. 8, 2011, and to U.S. Provisional Patent Application 61/593,561, filed Feb. 1, 2012, each of which is hereby incorporated by reference in its entirety.
BACKGROUND OF THE INVENTION
[0002] The following discussion of the background of the invention is merely provided to aid the reader in understanding the invention and is not admitted to describe or constitute prior art to the present invention.
[0003] The term "sepsis" has been used to describe a variety of clinical conditions related to systemic manifestations of inflammation accompanied by an infection. Because of clinical similarities to inflammatory responses secondary to non-infectious etiologies, identifying sepsis has been a particularly challenging diagnostic problem. Recently, the American College of Chest Physicians and the American Society of Critical Care Medicine (Bone et al., Chest 101: 1644-53, 1992) published definitions for "Systemic Inflammatory Response Syndrome" (or "SIRS"), which refers generally to a severe systemic response to an infectious or non-infectious insult, and for the related syndromes "sepsis," "severe sepsis," and "septic shock," and extending to multiple organ dysfunction syndrome ("MODS"). These definitions, described below, are intended for each of these phrases for the purposes of the present application.
[0004] "SIRS" refers to a condition that exhibits two or more of the following:
a temperature >38.degree. C. or <36.degree. C.; a heart rate of >90 heats per minute (tachycardia); a respiratory rate of >20 breaths per minute (tachypnea) or a P.sub.aCO.sub.2<4.3 kPa; and a white blood cell count >12,000 per mm.sup.3, <4,000 per mm.sup.3, or >10% immature (band) forms.
[0005] "Sepsis" refers to SIRS, further accompanied by a clinically evident or microbiologically confirmed infection. This infection may be bacterial, fungal, parasitic, or viral.
[0006] "Severe sepsis" refers to a subset of sepsis patients, in which sepsis is further accompanied by organ hypoperfusion made evident by at least one sign of organ dysfunction such as hypoxemia, oliguria, metabolic acidosis, or altered cerebral function.
[0007] "Septic shock" refers to a subset of severe sepsis patients, in which severe sepsis is further accompanied by hypotension, made evident by a systolic blood pressure <90 mm Hg, or the requirement for pharmaceutical intervention to maintain blood pressure.
[0008] MODS (multiple organ dysfunction syndrome) is the presence of altered organ function in a patient who is acutely ill such that homeostasis cannot be maintained without intervention. Primary MODS is the direct result of a well-defined insult in which organ dysfunction occurs early and can be directly attributable to the insult itself. Secondary MODS develops as a consequence of a host response and is identified within the context of SIRS.
[0009] A systemic inflammatory response leading to a diagnosis of SIRS may be related to both infection and to numerous non-infective etiologies, including burns, pancreatitis, trauma, heat stroke, and neoplasia. While conceptually it may be relatively simple to distinguish between sepsis and non-septic SIRS, no diagnostic tools have been described to unambiguously distinguish these related conditions. See, e.g., Llewelyn and Cohen, Int. Care Med. 27: S10-S32, 200L For example, because more than 90% of sepsis cases involve bacterial infection, the "gold standard" for confirming infection has been microbial growth from blood, urine, pleural fluid, cerebrospinal fluid, peritoneal fluid, synnovial fluid, sputum, or other tissue specimens. Such culture has been reported, however, to fail to confirm 50% or more of patients exhibiting strong clinical evidence of sepsis. See, e.g., Jaimes et al., Int. Care Med 29: 1368-71, published electronically Jun. 26, 2003.
[0010] Development of acute kidney injury (AKI) during sepsis increases patient morbidity, predicts higher mortality, has a significant effect on multiple organ functions, is associated with an increased length of stay in the intensive care unit, and hence consumes considerable healthcare resources. Several authors have noted that, when compared with AKI of nonseptic origin, septic AKI is characterized by a distinct pathophysiology and therefore requires a different approach. Sepsis-related AKI has been described in terms of elevated and imbalanced pro- and anti-inflammatory mediators (the so-called "peak concentration hypothesis"), coupled with severe endothelial dysfunction and a perturbed coagulation cascade operate synergistically to induce chemically and biologically mediated kidney injury. Major impediments to progress in understanding, early diagnosis, and application of appropriate therapeutic modalities in sepsis-induced AKI include limited histopathologic information, few animal models that closely mimic human sepsis, and a relative shortage of specific diagnostic tools. See, e.g., Zarjou and Agarwal, J. Am. Soc. Nephrol. 22: 999-1006, 2011; Ronco et al., Clin. J. Am. Soc. Nephrol. 3: 531-44, 2008.
[0011] These limitations underscore the need for better methods to evaluate sepsis patients in order to identify those most at risk for AKI, particularly in the early and subclinical stages, but also in later stages when recovery and repair of the kidney can occur. Furthermore, there is a need to better identify patients who are at risk of having an AKI.
BRIEF SUMMARY OF THE INVENTION
[0012] It is an object of the invention to provide methods and compositions for evaluating renal function in a sepsis patient diagnosed with sepsis. As described herein, measurement of one or more biomarkers selected from the group consisting of Insulin-like growth factor-binding protein 7, Beta-2-glycoprotein 1, Metalloproteinase inhibitor 2, Alpha-1 Antitrypsin, Leukocyte elastase, Serum Amyloid P Component, C--X--C motif chemokine 6, Immunoglobulin A, Immunoglobulin G subclass I, C--C motif chemokine 24, Neutrophil collagenase, Cathepsin D, C--X--C motif chemokine 13, Involucrin, Interleukin-6 receptor subunit beta, Hepatocyte Growth Factor, CXCL-1, -2, -3, Immunoglobulin G subclass II, Metalloproteinase inhibitor 4, C--C motif chemokine 18, Matrilysin, C--X--C motif chemokine 11, and Antileukoproteinase (referred to herein as a "kidney injury marker") can be used for diagnosis, prognosis, risk stratification, staging, monitoring, categorizing and determination of further diagnosis and treatment regimens in sepsis patients.
[0013] The kidney injury markers of the present invention may be used, individually or in panels comprising a plurality of kidney injury markers, for risk stratification (that is, to identify sepsis patients at risk for a future injury to renal function, for future progression to reduced renal function, for future progression to ARF, for future improvement in renal function, etc.); for diagnosis of existing disease (that is, to identify sepsis patients who have suffered an injury to renal function, who have progressed to reduced renal function, who have progressed to ARF, etc.); for monitoring for deterioration or improvement of renal function; and for predicting a future medical outcome, such as improved or worsening renal function, a decreased or increased mortality risk, a decreased or increased risk that a sepsis patient will require renal replacement therapy (i.e., hemodialysis, peritoneal dialysis, hemofiltration, and/or renal transplantation, a decreased or increased risk that a sepsis patient will recover from an injury to renal function, a decreased or increased risk that a sepsis patient will recover from ARF, a decreased or increased risk that a sepsis patient will progress to end stage renal disease, a decreased or increased risk that a sepsis patient will progress to chronic renal failure, a decreased or increased risk that a sepsis patient will suffer rejection of a transplanted kidney, etc.
[0014] In a first aspect, the present invention relates to methods for evaluating renal status in a sepsis patient. These methods comprise performing an assay method that is configured to detect one or more biomarkers selected from the group consisting of Insulin-like growth factor-binding protein 7, Beta-2-glycoprotein 1, Metalloproteinase inhibitor 2, Alpha-1 Antitrypsin, Leukocyte elastase, Serum Amyloid P Component, C--X--C motif chemokine 6, Immunoglobulin A, Immunoglobulin G subclass I, C--C motif chemokine 24, Neutrophil collagenase, Cathepsin D, C--X--C motif chemokine 13, Involucrin, Interleukin-6 receptor subunit beta, Hepatocyte Growth Factor, CXCL-1, -2, -3, Immunoglobulin G subclass II, Metalloproteinase inhibitor 4, C--C motif chemokine 18, Matrilysin, C--X--C motif chemokine 11, and Antileukoproteinase in a body fluid sample obtained from the sepsis patient. The assay result(s), for example measured concentration(s) of one or more biomarkers selected from the group consisting of Insulin-like growth factor-binding protein 7, Beta-2-glycoprotein 1, Metalloproteinase inhibitor 2, Alpha-1 Antitrypsin, Leukocyte elastase, Serum Amyloid P Component, C--X--C motif chemokine 6, Immunoglobulin A, Immunoglobulin G subclass I, C--C motif chemokine 24, Neutrophil collagenase, Cathepsin D, C--X--C motif chemokine 13, Involucrin, Interleukin-6 receptor subunit beta, Hepatocyte Growth Factor, CXCL,-1, -2, -3, Immunoglobulin G subclass 11, Metalloproteinase inhibitor 4, C--C motif chemokine 18, Matrilysin, C--X--C motif chemokine 11, and Antileukoproteinase is/are then correlated to the renal status of the sepsis patient. This correlation to renal status may include correlating the assay result(s) to one or more of risk stratification, diagnosis, prognosis, staging, classifying and monitoring of the sepsis patient as described herein. Thus, the present invention utilizes one or more kidney injury markers of the present invention for the evaluation of renal injury in a sepsis patient.
[0015] In certain embodiments, the methods for evaluating renal status described herein are methods for risk stratification of the sepsis patient; that is, assigning a likelihood of one or more future changes in renal status to the sepsis patient. In these embodiments, the assay result(s) is/are correlated to one or more such future changes. The following are preferred risk stratification embodiments.
[0016] In preferred risk stratification embodiments, these methods comprise determining a sepsis patient's risk for a future injury to renal function, and the assay result(s) is/are correlated to a likelihood of such a future injury to renal function. For example, the measured concentration(s) may each be compared to a threshold value. For a "positive going" kidney injury marker, an increased likelihood of suffering a future injury to renal function is assigned to the sepsis patient when the measured concentration is above the threshold, relative to a likelihood assigned when the measured concentration is below the threshold. For a "negative going" kidney injury marker, an increased likelihood of suffering a future injury to renal function is assigned to the sepsis patient when the measured concentration is below the threshold, relative to a likelihood assigned when the measured concentration is above the threshold.
[0017] In other preferred risk stratification embodiments, these methods comprise determining a sepsis patient's risk for future reduced renal function, and the assay result(s) is/are correlated to a likelihood of such reduced renal function. For example, the measured concentrations may each be compared to a threshold value. For a "positive going" kidney injury marker, an increased likelihood of suffering a future reduced renal function is assigned to the sepsis patient when the measured concentration is above the threshold, relative to a likelihood assigned when the measured concentration is below the threshold. For a "negative going" kidney injury marker, an increased likelihood of future reduced renal function is assigned to the sepsis patient when the measured concentration is below the threshold, relative to a likelihood assigned when the measured concentration is above the threshold.
[0018] In still other preferred risk stratification embodiments, these methods comprise determining a sepsis patient's likelihood for a future improvement in renal function, and the assay result(s) is/are correlated to a likelihood of such a future improvement in renal function. For example, the measured concentration(s) may each be compared to a threshold value. For a "positive going" kidney injury marker, an increased likelihood of a future improvement in renal function is assigned to the sepsis patient when the measured concentration is below the threshold, relative to a likelihood assigned when the measured concentration is above the threshold. For a "negative going" kidney injury marker, an increased likelihood of a future improvement in renal function is assigned to the sepsis patient when the measured concentration is above the threshold, relative to a likelihood assigned when the measured concentration is below the threshold.
[0019] In yet other preferred risk stratification embodiments, these methods comprise determining a sepsis patient's risk for progression to ARF, and the result(s) is/are correlated to a likelihood of such progression to ARF. For example, the measured concentration(s) may each be compared to a threshold value. For a "positive going" kidney injury marker, an increased likelihood of progression to ARF is assigned to the sepsis patient when the measured concentration is above the threshold, relative to a likelihood assigned when the measured concentration is below the threshold. For a "negative going" kidney injury marker, an increased likelihood of progression to ARF is assigned to the sepsis patient when the measured concentration is below the threshold, relative to a likelihood assigned when the measured concentration is above the threshold.
[0020] And in other preferred risk stratification embodiments, these methods comprise determining a sepsis patient's outcome risk, and the assay result(s) is/are correlated to a likelihood of the occurrence of a clinical outcome related to a renal injury suffered by the sepsis patient. For example, the measured concentration(s) may each be compared to a threshold value. For a "positive going" kidney injury marker, an increased likelihood of one or more of: acute kidney injury, progression to a worsening stage of AKI, mortality, a requirement for renal replacement therapy, a requirement for withdrawal of renal toxins, end stage renal disease, heart failure, stroke, myocardial infarction, progression to chronic kidney disease, etc., is assigned to the sepsis patient when the measured concentration is above the threshold, relative to a likelihood assigned when the measured concentration is below the threshold. For a "negative going" kidney injury marker, an increased likelihood of one or more of: acute kidney injury, progression to a worsening stage of AKI, mortality, a requirement for renal replacement therapy, a requirement for withdrawal of renal toxins, end stage renal disease, heart failure, stroke, myocardial infarction, progression to chronic kidney disease, etc., is assigned to the sepsis patient when the measured concentration is below the threshold, relative to a likelihood assigned when the measured concentration is above the threshold.
[0021] In such risk stratification embodiments, preferably the likelihood or risk assigned is that an event of interest is more or less likely to occur within 180 days of the time at which the body fluid sample is obtained from the sepsis patient. In particularly preferred embodiments, the likelihood or risk assigned relates to an event of interest occurring within a shorter time period such as 18 months, 120 days, 90 days, 60 days, 45 days, 30 days, 21 days, 14 days, 7 days, 5 days, 96 hours, 72 hours, 48 hours, 36 hours, 24 hours, 12 hours, or less. A risk at 0 hours of the time at which the body fluid sample is obtained from the sepsis patient is equivalent to diagnosis of a current condition.
[0022] In other embodiments, the methods for evaluating renal status described herein are methods for diagnosing a renal injury in a sepsis patient; that is, assessing whether or not a sepsis patient has suffered from an injury to renal function, reduced renal function, or ARF. In these embodiments, the assay result(s), for example measured concentration(s) of one or more biomarkers selected from the group consisting of Insulin-like growth factor-binding protein 7, Beta-2-glycoprotein 1, Metalloproteinase inhibitor 2, Alpha-1 Antitrypsin, Leukocyte elastase, Serum Amyloid P Component, C--X--C motif chemokine 6, Immunoglobulin A, Immunoglobulin G subclass I, C--C motif chemokine 24, Neutrophil collagenase, Cathepsin D, C--X--C motif chemokine 13, Involucrin, Interleukin-6 receptor subunit beta, Hepatocyte Growth Factor, CXCL-1, -2, -3, Immunoglobulin G subclass II, Metalloproteinase inhibitor 4, C--C motif chemokine 18, Matrilysin, C--X--C motif chemokine 11, and Antileukoproteinase is/are correlated to the occurrence or nonoccurrence of a change in renal status. The following are preferred diagnostic embodiments.
[0023] In preferred diagnostic embodiments, these methods comprise diagnosing the occurrence or nonoccurrence of an injury to renal function, and the assay result(s) is/are correlated to the occurrence or nonoccurrence of such an injury. For example, each of the measured concentration(s) may be compared to a threshold value. For a positive going marker, an increased likelihood of the occurrence of an injury to renal function is assigned to the sepsis patient when the measured concentration is above the threshold (relative to the likelihood assigned when the measured concentration is below the threshold); alternatively, when the measured concentration is below the threshold, an increased likelihood of the nonoccurrence of an injury to renal function may be assigned to the sepsis patient (relative to the likelihood assigned when the measured concentration is above the threshold). For a negative going marker, an increased likelihood of the occurrence of an injury to renal function is assigned to the sepsis patient when the measured concentration is below the threshold (relative to the likelihood assigned when the measured concentration is above the threshold); alternatively, when the measured concentration is above the threshold, an increased likelihood of the nonoccurrence of an injury to renal function may be assigned to the sepsis patient (relative to the likelihood assigned when the measured concentration is below the threshold).
[0024] In other preferred diagnostic embodiments, these methods comprise diagnosing the occurrence or nonoccurrence of reduced renal function, and the assay result(s) is/are correlated to the occurrence or nonoccurrence of an injury causing reduced renal function. For example, each of the measured concentration(s) may be compared to a threshold value. For a positive going marker, an increased likelihood of the occurrence of an injury causing reduced renal function is assigned to the sepsis patient when the measured concentration is above the threshold (relative to the likelihood assigned when the measured concentration is below the threshold); alternatively, when the measured concentration is below the threshold, an increased likelihood of the nonoccurrence of an injury causing reduced renal function may be assigned to the sepsis patient (relative to the likelihood assigned when the measured concentration is above the threshold). For a negative going marker, an increased likelihood of the occurrence of an injury causing reduced renal function is assigned to the sepsis patient when the measured concentration is below the threshold (relative to the likelihood assigned when the measured concentration is above the threshold); alternatively, when the measured concentration is above the threshold, an increased likelihood of the nonoccurrence of an injury causing reduced renal function may be assigned to the sepsis patient (relative to the likelihood assigned when the measured concentration is below the threshold).
[0025] In yet other preferred diagnostic embodiments, these methods comprise diagnosing the occurrence or nonoccurrence of ARF, and the assay result(s) is/are correlated to the occurrence or nonoccurrence of an injury causing ARF. For example, each of the measured concentration(s) may be compared to a threshold value. For a positive going marker, an increased likelihood of the occurrence of ARF is assigned to the sepsis patient when the measured concentration is above the threshold (relative to the likelihood assigned when the measured concentration is below the threshold); alternatively, when the measured concentration is below the threshold, an increased likelihood of the nonoccurrence of ARF may be assigned to the sepsis patient (relative to the likelihood assigned when the measured concentration is above the threshold). For a negative going marker, an increased likelihood of the occurrence of ARF is assigned to the sepsis patient when the measured concentration is below the threshold (relative to the likelihood assigned when the measured concentration is above the threshold); alternatively, when the measured concentration is above the threshold, an increased likelihood of the nonoccurrence of ARF may be assigned to the sepsis patient (relative to the likelihood assigned when the measured concentration is below the threshold).
[0026] In still other preferred diagnostic embodiments, these methods comprise diagnosing a sepsis patient as being in need of renal replacement therapy, and the assay result(s) is/are correlated to a need for renal replacement therapy. For example, each of the measured concentration(s) may be compared to a threshold value. For a positive going marker, an increased likelihood of the occurrence of an injury creating a need for renal replacement therapy is assigned to the sepsis patient when the measured concentration is above the threshold (relative to the likelihood assigned when the measured concentration is below the threshold); alternatively, when the measured concentration is below the threshold, an increased likelihood of the nonoccurrence of an injury creating a need for renal replacement therapy may be assigned to the sepsis patient (relative to the likelihood assigned when the measured concentration is above the threshold). For a negative going marker, an increased likelihood of the occurrence of an injury creating a need for renal replacement therapy is assigned to the sepsis patient when the measured concentration is below the threshold (relative to the likelihood assigned when the measured concentration is above the threshold); alternatively, when the measured concentration is above the threshold, an increased likelihood of the nonoccurrence of an injury creating a need for renal replacement therapy may be assigned to the sepsis patient (relative to the likelihood assigned when the measured concentration is below the threshold).
[0027] In still other preferred diagnostic embodiments, these methods comprise diagnosing a sepsis patient as being in need of renal transplantation, and the assay result(s0 is/are correlated to a need for renal transplantation. For example, each of the measured concentration(s) may be compared to a threshold value. For a positive going marker, an increased likelihood of the occurrence of an injury creating a need for renal transplantation is assigned to the sepsis patient when the measured concentration is above the threshold (relative to the likelihood assigned when the measured concentration is below the threshold); alternatively, when the measured concentration is below the threshold, an increased likelihood of the nonoccurrence of an injury creating a need for renal transplantation may be assigned to the sepsis patient (relative to the likelihood assigned when the measured concentration is above the threshold). For a negative going marker, an increased likelihood of the occurrence of an injury creating a need for renal transplantation is assigned to the sepsis patient when the measured concentration is below the threshold (relative to the likelihood assigned when the measured concentration is above the threshold); alternatively, when the measured concentration is above the threshold, an increased likelihood of the nonoccurrence of an injury creating a need for renal transplantation may be assigned to the sepsis patient (relative to the likelihood assigned when the measured concentration is below the threshold).
[0028] In still other embodiments, the methods for evaluating renal status described herein are methods for monitoring a renal injury in a sepsis patient; that is, assessing whether or not renal function is improving or worsening in a sepsis patient who has suffered from an injury to renal function, reduced renal function, or ARF. In these embodiments, the assay result(s), for example measured concentration(s) of one or more biomarkers selected from the group consisting of Insulin-like growth factor-binding protein 7, Beta-2-glycoprotein 1, Metalloproteinase inhibitor 2, Alpha-1 Antitrypsin, Leukocyte elastase, Serum Amyloid P Component, C--X--C motif chemokine 6, Immunoglobulin A, Immunoglobulin G subclass I, C--C motif chemokine 24, Neutrophil collagenase, Cathepsin D, C--X--C motif chemokine 13, Involucrin, Interleukin-6 receptor subunit beta, Hepatocyte Growth Factor, CXCL,-1, -2, -3, Immunoglobulin G subclass IT, Metalloproteinase inhibitor 4, C--C motif chemokine 18, Matrilysin, C--X--C motif chemokine 11, and Antileukoproteinase is/are correlated to the occurrence or nonoccurrence of a change in renal status. The following are preferred monitoring embodiments.
[0029] In preferred monitoring embodiments, these methods comprise monitoring renal status in a sepsis patient suffering from an injury to renal function, and the assay result(s) is/are correlated to the occurrence or nonoccurrence of a change in renal status in the sepsis patient. For example, the measured concentration(s) may be compared to a threshold value. For a positive going marker, when the measured concentration is above the threshold, a worsening of renal function may be assigned to the sepsis patient; alternatively, when the measured concentration is below the threshold, an improvement of renal function may be assigned to the sepsis patient. For a negative going marker, when the measured concentration is below the threshold, a worsening of renal function may be assigned to the sepsis patient; alternatively, when the measured concentration is above the threshold, an improvement of renal function may be assigned to the sepsis patient.
[0030] In other preferred monitoring embodiments, these methods comprise monitoring renal status in a sepsis patient suffering from reduced renal function, and the assay result(s) is/are correlated to the occurrence or nonoccurrence of a change in renal status in the sepsis patient. For example, the measured concentration(s) may be compared to a threshold value. For a positive going marker, when the measured concentration is above the threshold, a worsening of renal function may be assigned to the sepsis patient; alternatively, when the measured concentration is below the threshold, an improvement of renal function may be assigned to the sepsis patient. For a negative going marker, when the measured concentration is below the threshold, a worsening of renal function may be assigned to the sepsis patient; alternatively, when the measured concentration is above the threshold, an improvement of renal function may be assigned to the sepsis patient.
[0031] In yet other preferred monitoring embodiments, these methods comprise monitoring renal status in a sepsis patient suffering from acute renal failure, and the assay result(s) is/are correlated to the occurrence or nonoccurrence of a change in renal status in the sepsis patient. For example, the measured concentration(s) may be compared to a threshold value. For a positive going marker, when the measured concentration is above the threshold, a worsening of renal function may be assigned to the sepsis patient; alternatively, when the measured concentration is below the threshold, an improvement of renal function may be assigned to the sepsis patient. For a negative going marker, when the measured concentration is below the threshold, a worsening of renal function may be assigned to the sepsis patient; alternatively, when the measured concentration is above the threshold, an improvement of renal function may be assigned to the sepsis patient.
[0032] In still other embodiments, the methods for evaluating renal status described herein are methods for classifying a renal injury in a sepsis patient; that is, determining whether a renal injury in a sepsis patient is prerenal, intrinsic renal, or postrenal; and/or further subdividing these classes into subclasses such as acute tubular injury, acute glomerulonephritis acute tubulointerstitial nephritis, acute vascular nephropathy, or infiltrative disease; and/or assigning a likelihood that a sepsis patient will progress to a particular RIFLE stage. In these embodiments, the assay result(s), for example measured concentration(s) of one or more biomarkers selected from the group consisting of Insulin-like growth factor-binding protein 7, Beta-2-glycoprotein 1, Metalloproteinase inhibitor 2, Alpha-1 Antitrypsin, Leukocyte elastase, Serum Amyloid P Component, C--X--C motif chemokine 6, Immunoglobulin A, Immunoglobulin G subclass I, C--C motif chemokine 24, Neutrophil collagenase, Cathepsin D, C--X--C motif chemokine 13, Involucrin, Interleukin-6 receptor subunit beta, Hepatocyte Growth Factor, CXCL-1, -2, -3, Immunoglobulin G subclass II, Metalloproteinase inhibitor 4, C--C motif chemokine 18, Matrilysin, C--X--C motif chemokine 11, and Antileukoproteinase is/are correlated to a particular class and/or subclass. The following are preferred classification embodiments.
[0033] In preferred classification embodiments, these methods comprise determining whether a renal injury in a sepsis patient is prerenal, intrinsic renal, or postrenal; and/or further subdividing these classes into subclasses such as acute tubular injury, acute glomerulonephritis acute tubulointerstitial nephritis, acute vascular nephropathy, or infiltrative disease; and/or assigning a likelihood that a sepsis patient will progress to a particular RIFLE stage, and the assay result(s) is/are correlated to the injury classification for the sepsis patient. For example, the measured concentration may be compared to a threshold value, and when the measured concentration is above the threshold, a particular classification is assigned; alternatively, when the measured concentration is below the threshold, a different classification may be assigned to the sepsis patient.
[0034] A variety of methods may be used by the skilled artisan to arrive at a desired threshold value for use in these methods. For example, the threshold value may be determined from a population of normal sepsis patients by selecting a concentration representing the 75.sup.th, 85.sup.th, 90.sup.th, 95.sup.th, or 99.sup.th percentile of a kidney injury marker measured in such normal sepsis patients. Alternatively, the threshold value may be determined from a "diseased" population of sepsis patients, e.g., those suffering from an injury or having a predisposition for an injury (e.g., progression to ARF or some other clinical outcome such as death, dialysis, renal transplantation, etc.), by selecting a concentration representing the 75.sup.th, 85.sup.th, 90.sup.th, 95.sup.th, or 99.sup.th percentile of a kidney injury marker measured in such sepsis patients. In another alternative, the threshold value may be determined from a prior measurement of a kidney injury marker in the same sepsis patient; that is, a temporal change in the level of a kidney injury marker in the sepsis patient may be used to assign risk to the sepsis patient.
[0035] The foregoing discussion is not meant to imply, however, that the kidney injury markers of the present invention must be compared to corresponding individual thresholds. Methods for combining assay results can comprise the use of multivariate logistical regression, loglinear modeling, neural network analysis, n-of-m analysis, decision tree analysis, calculating ratios of markers, etc. This list is not meant to be limiting. In these methods, a composite result which is determined by combining individual markers may be treated as if it is itself a marker; that is, a threshold may be determined for the composite result as described herein for individual markers, and the composite result for an individual patient compared to this threshold.
[0036] The ability of a particular test to distinguish two populations can be established using ROC analysis. For example, ROC curves established from a "first" subpopulation which is predisposed to one or more future changes in renal status, and a "second" subpopulation which is not so predisposed can be used to calculate a ROC curve, and the area under the curve provides a measure of the quality of the test. Preferably, the tests described herein provide a ROC curve area greater than 0.5, preferably at least 0.6, more preferably 0.7, still more preferably at least 0.8, even more preferably at least 0.9, and most preferably at least 0.95.
[0037] In certain aspects, the measured concentration of one or more kidney injury markers, or a composite of such markers, may be treated as continuous variables. For example, any particular concentration can be converted into a corresponding probability of a future reduction in renal function for the sepsis patient, the occurrence of an injury, a classification, etc. In yet another alternative, a threshold that can provide an acceptable level of specificity and sensitivity in separating a population of sepsis patients into "bins" such as a "first" subpopulation (e.g., which is predisposed to one or more future changes in renal status, the occurrence of an injury, a classification, etc.) and a "second" subpopulation which is not so predisposed. A threshold value is selected to separate this first and second population by one or more of the following measures of test accuracy:
an odds ratio greater than 1, preferably at least about 2 or more or about 0.5 or less, more preferably at least about 3 or more or about 0.33 or less, still more preferably at least about 4 or more or about 0.25 or less, even more preferably at least about 5 or more or about 0.2 or less, and most preferably at least about 10 or more or about 0.1 or less; a specificity of greater than 0.5, preferably at least about 0.6, more preferably at least about 0.7, still more preferably at least about 0.8, even more preferably at least about 0.9 and most preferably at least about 0.95, with a corresponding sensitivity greater than 0.2, preferably greater than about 0.3, more preferably greater than about 0.4, still more preferably at least about 0.5, even more preferably about 0.6, yet more preferably greater than about 0.7, still more preferably greater than about 0.8, more preferably greater than about 0.9, and most preferably greater than about 0.95; a sensitivity of greater than 0.5, preferably at least about 0.6, more preferably at least about 0.7, still more preferably at least about 0.8, even more preferably at least about 0.9 and most preferably at least about 0.95, with a corresponding specificity greater than 0.2, preferably greater than about 0.3, more preferably greater than about 0.4, still more preferably at least about 0.5, even more preferably about 0.6, yet more preferably greater than about 0.7, still more preferably greater than about 0.8, more preferably greater than about 0.9, and most preferably greater than about 0.95; at least about 75% sensitivity, combined with at least about 75% specificity; a positive likelihood ratio (calculated as sensitivity/(1-specificity)) of greater than 1, at least about 2, more preferably at least about 3, still more preferably at least about 5, and most preferably at least about 10; or a negative likelihood ratio (calculated as (1-sensitivity)/specificity) of less than 1, less than or equal to about 0.5, more preferably less than or equal to about 0.3, and most preferably less than or equal to about 0.1.
[0038] The term "about" in the context of any of the above measurements refers to +/-5% of a given measurement.
[0039] Multiple thresholds may also be used to assess renal status in a sepsis patient. For example, a "first" subpopulation which is predisposed to one or more future changes in renal status, the occurrence of an injury, a classification, etc., and a "second" subpopulation which is not so predisposed can be combined into a single group. This group is then subdivided into three or more equal parts (known as tertiles, quartiles, quintiles, etc., depending on the number of subdivisions). An odds ratio is assigned to sepsis patients based on which subdivision they fall into. If one considers a tertile, the lowest or highest tertile can be used as a reference for comparison of the other subdivisions. This reference subdivision is assigned an odds ratio of 1. The second tertile is assigned an odds ratio that is relative to that first tertile. That is, someone in the second tertile might be 3 times more likely to suffer one or more future changes in renal status in comparison to someone in the first tertile. The third tertile is also assigned an odds ratio that is relative to that first tertile.
[0040] In certain embodiments, the assay method is an immunoassay. Antibodies for use in such assays will specifically hind a full length kidney injury marker of interest, and may also bind one or more polypeptides that are "related" thereto, as that term is defined hereinafter. Numerous immunoassay formats are known to those of skill in the art. Preferred body fluid samples are selected from the group consisting of urine, blood, serum, saliva, tears, and plasma.
[0041] The foregoing method steps should not be interpreted to mean that the kidney injury marker assay result(s) is/are used in isolation in the methods described herein. Rather, additional variables or other clinical indicia may be included in the methods described herein. For example, a risk stratification, diagnostic, classification, monitoring, etc. method may combine the assay result(s) with one or more variables measured for the sepsis patient selected from the group consisting of demographic information (e.g., weight, sex, age, race), medical history (e.g., family history, type of surgery, pre-existing disease such as aneurism, congestive heart failure, preeclampsia, eclampsia, diabetes mellitus, hypertension, coronary artery disease, proteinuria, renal insufficiency, or sepsis, type of toxin exposure such as NSAIDs, cyclosporines, tacrolimus, aminoglycosides, foscarnet, ethylene glycol, hemoglobin, myoglobin, ifosfamide, heavy metals, methotrexate, radiopaque contrast agents, or streptozotocin), clinical variables (e.g., blood pressure, temperature, respiration rate), risk scores (APACHE score, PREDICT score, TIMI Risk Score for UA/NSTEMI, Framingham Risk Score), a glomerular filtration rate, an estimated glomerular filtration rate, a urine production rate, a serum or plasma creatinine concentration, a urine creatinine concentration, a fractional excretion of sodium, a urine sodium concentration, a urine creatinine to serum or plasma creatinine ratio, a urine specific gravity, a urine osmolality, a urine urea nitrogen to plasma urea nitrogen ratio, a plasma BUN to creatnine ratio, a renal failure index calculated as urine sodium/(urine creatinine/plasma creatinine), a serum or plasma neutrophil gelatinase (NGAL) concentration, a urine NGAL concentration, a serum or plasma cystatin C concentration, a serum or plasma cardiac troponin concentration, a serum or plasma BNP concentration, a serum or plasma NTproBNP concentration, and a serum or plasma proBNP concentration. Other measures of renal function which may be combined with one or more kidney injury marker assay result(s) are described hereinafter and in Harrison's Principles of Internal Medicine, 17.sup.th Ed., McGraw Hill, New York, pages 1741-1830, and Current Medical Diagnosis & Treatment 2008, 47.sup.th Ed, McGraw Hill, New York, pages 785-815, each of which are hereby incorporated by reference in their entirety.
[0042] When more than one marker is measured, the individual markers may be measured in samples obtained at the same time, or may be determined from samples obtained at different (e.g., an earlier or later) times. The individual markers may also be measured on the same or different body fluid samples. For example, one kidney injury marker may be measured in a serum or plasma sample and another kidney injury marker may be measured in a urine sample. In addition, assignment of a likelihood may combine an individual kidney injury marker assay result with temporal changes in one or more additional variables.
[0043] In various related aspects, the present invention also relates to devices and kits for performing the methods described herein. Suitable kits comprise reagents sufficient for performing an assay for at least one of the described kidney injury markers, together with instructions for performing the described threshold comparisons.
[0044] In certain embodiments, reagents for performing such assays are provided in an assay device, and such assay devices may be included in such a kit. Preferred reagents can comprise one or more solid phase antibodies, the solid phase antibody comprising antibody that detects the intended biomarker target(s) bound to a solid support. In the case of sandwich immunoassays, such reagents can also include one or more detectably labeled antibodies, the detectably labeled antibody comprising antibody that detects the intended biomarker target(s) bound to a detectable label. Additional optional elements that may be provided as part of an assay device are described hereinafter.
[0045] Detectable labels may include molecules that are themselves detectable (e.g., fluorescent moieties, electrochemical labels, ecl (electrochemical luminescence) labels, metal chelates, colloidal metal particles, etc.) as well as molecules that may be indirectly detected by production of a detectable reaction product (e.g., enzymes such as horseradish peroxidase, alkaline phosphatase, etc.) or through the use of a specific binding molecule which itself may be detectable (e.g., a labeled antibody that binds to the second antibody, biotin, digoxigenin, maltose, oligohistidine, 2,4-dintrobenzene, phenylarsenate, ssDNA, dsDNA, etc.).
[0046] Generation of a signal from the signal development element can be performed using various optical, acoustical, and electrochemical methods well known in the art. Examples of detection modes include fluorescence, radiochemical detection, reflectance, absorbance, amperometry, conductance, impedance, interferometry, ellipsometry, etc. In certain of these methods, the solid phase antibody is coupled to a transducer (e.g., a diffraction grating, electrochemical sensor, etc) for generation of a signal, while in others, a signal is generated by a transducer that is spatially separate from the solid phase antibody (e.g., a fluorometer that employs an excitation light source and an optical detector). This list is not meant to be limiting. Antibody-based biosensors may also be employed to determine the presence or amount of analytes that optionally eliminate the need for a labeled molecule.
DETAILED DESCRIPTION OF THE INVENTION
[0047] The present invention relates to methods and compositions for diagnosis, differential diagnosis, risk stratification, monitoring, classifying and determination of treatment regimens in sepsis patients diagnosed with sepsis. In various embodiments, a measured concentration of one or more biomarkers selected from the group consisting of Insulin-like growth factor-binding protein 7, Beta-2-glycoprotein 1, Metalloproteinase inhibitor 2, Alpha-1 Antitrypsin, Leukocyte elastase, Serum Amyloid P Component, C--X--C motif chemokine 6, Immunoglobulin A, Immunoglobulin G subclass I, C--C motif chemokine 24, Neutrophil collagenase, Cathepsin D, C--X--C motif chemokine 13, Involucrin, Interleukin-6 receptor subunit beta, Hepatocyte Growth Factor, CXCL-1, -2, -3, Immunoglobulin G subclass II, Metalloproteinase inhibitor 4, C--C motif chemokine 18, Matrilysin, C--X--C motif chemokine 11, and Antileukoproteinase or one or more markers related thereto, are correlated to the renal status of the sepsis patient.
[0048] The kidney is responsible for water and solute excretion from the body. Its functions include maintenance of acid-base balance, regulation of electrolyte concentrations, control of blood volume, and regulation of blood pressure. As such, loss of kidney function through injury and/or disease results in substantial morbidity and mortality. A detailed discussion of renal injuries is provided in Harrison's Principles of Internal Medicine, 17.sup.th Ed., McGraw Hill, New York, pages 1741-1830, which are hereby incorporated by reference in their entirety. Renal disease and/or injury may be acute or chronic. Acute and chronic kidney disease are described as follows (from Current Medical Diagnosis & Treatment 2008, 47.sup.th Ed, McGraw Hill, New York, pages 785-815, which are hereby incorporated by reference in their entirety): "Acute renal failure is worsening of renal function over hours to days, resulting in the retention of nitrogenous wastes (such as urea nitrogen) and creatinine in the blood. Retention of these substances is called azotemia. Chronic renal failure (chronic kidney disease) results from an abnormal loss of renal function over months to years".
[0049] Acute renal failure (ARF, also known as acute kidney injury, or AKI) is an abrupt (typically detected within about 48 hours to 1 week)reduction in glomerular filtration. This loss of filtration capacity results in retention of nitrogenous (urea and creatinine) and non-nitrogenous waste products that are normally excreted by the kidney, a reduction in urine output, or both. It is reported that ARF complicates about 5% of hospital admissions, 4-15% of cardiopulmonary bypass surgeries, and up to 30% of intensive care admissions. ARF may be categorized as prerenal, intrinsic renal, or postrenal in causation. Intrinsic renal disease can be further divided into glomerular, tubular, interstitial, and vascular abnormalities. Major causes of ARF are described in the following table, which is adapted from the Merck Manual, 17.sup.th ed., Chapter 222, and which is hereby incorporated by reference in their entirety:
TABLE-US-00001 Type Risk Factors Prerenal ECF volume Excessive diuresis, hemorrhage, GI losses, depletion loss of intravascular fluid into the extravascular space (due to ascites, peritonitis, pancreatitis, or burns), loss of skin and mucus membranes, renal salt-and water-wasting states Low cardiac Cardiomyopathy, MI, cardiac tamponade, output pulmonary embolism, pulmonary hypertension, positive-pressure mechanical ventilation Low systemic Septic shock, liver failure, antihypertensive vascular drugs resistance Increased renal NSAIDs, cyclosporines, tacrolimus, vascular hypercalcemia, anaphylaxis, anesthetics, resistance renal artery obstruction, renal vein thrombosis, sepsis, hepatorenal syndrome Decreased ACE inhibitors or angiotensin II receptor efferent arteriolar blockers tone (leading to decreased GFR from reduced glomerular trans- capillary pressure, especially in patients with bilateral renal artery stenosis) Intrinsic Renal Acute tubular Ischemia (prolonged or severe prerenal state): injury surgery, hemorrhage, arterial or venous obstruction; Toxins: NSAIDs, cyclosporines, tacrolimus, aminoglycosides, foscarnet, ethylene glycol, hemoglobin, myoglobin, ifosfamide, heavy metals, methotrexate, radiopaque contrast agents, streptozotocin Acute ANCA-associated: Crescentic glomerulone- glomerulone- phritis, polyarteritis nodosa, Wegener's phritis granulomatosis; Anti-GBM glomerulone- phritis: Goodpasture's syndrome; Immune-complex: Lupus glomerulonephritis, postinfectious glomerulonephritis, cryoglobulinemic glomerulonephritis Acute tubuloin- Drug reaction (eg, .beta.-lactams, NSAIDs, terstitial nephritis sulfonamides, ciprofloxacin, thiazide diuretics, furosemide, phenytoin, allopurinol, pyelonephritis, papillary necrosis Acute vascular Vasculitis, malignant hypertension, nephropathy thrombotic microangiopathies, scleroderma, atheroembolism Infiltrative Lymphoma, sarcoidosis, leukemia diseases Postrenal Tubular Uric acid (tumor lysis), sulfonamides, precipitation triamterene, acyclovir, indinavir, methotrexate, ethylene glycol ingestion, myeloma protein, myoglobin Ureteral Intrinsic: Calculi, clots, sloughed renal obstruction tissue, fungus ball, edema, malignancy, congenital defects; Extrinsic: Malignancy, retroperitoneal fibrosis, ureteral trauma during surgery or high impact injury Bladder Mechanical: Benign prostatic hyperplasia, obstruction prostate cancer, bladder cancer, urethral strictures, phimosis, paraphimosis, urethral valves, obstructed indwelling urinary catheter; Neurogenic: Anticholinergic drugs, upper or lower motor neuron lesion
[0050] In the case of ischemic ARF, the course of the disease may be divided into four phases. During an initiation phase, which lasts hours to days, reduced perfusion of the kidney is evolving into injury. Glomerular ultrafiltration reduces, the flow of filtrate is reduced due to debris within the tubules, and back leakage of filtrate through injured epithelium occurs. Renal injury can be mediated during this phase by reperfusion of the kidney. Initiation is followed by an extension phase which is characterized by continued ischemic injury and inflammation and may involve endothelial damage and vascular congestion. During the maintenance phase, lasting from 1 to 2 weeks, renal cell injury occurs, and glomerular filtration and urine output reaches a minimum. A recovery phase can follow in which the renal epithelium is repaired and GFR gradually recovers. Despite this, the survival rate of sepsis patients with ARF may be as low as about 60%.
[0051] A commonly reported criteria for defining and detecting AKI is an abrupt (typically within about 2-7 days or within a period of hospitalization) elevation of serum creatinine. Although the use of serum creatinine elevation to define and detect AKI is well established, the magnitude of the serum creatinine elevation and the time over which it is measured to define AKI varies considerably among publications. Traditionally, relatively large increases in serum creatinine such as 100%, 200%, an increase of at least 100% to a value over 2 mg/dL and other definitions were used to define AKI. However, the recent trend has been towards using smaller serum creatinine rises to define AKI. The relationship between serum creatinine rise, AKI and the associated health risks are reviewed in Praught and Shlipak, Curr Opin Nephrol Hypertens 14:265-270, 2005 and Chertow et al, J Am Soc Nephrol 16: 3365-3370, 2005, which, with the references listed therein, are hereby incorporated by reference in their entirety. As described in these publications, acute worsening renal function (AKI) and increased risk of death and other detrimental outcomes are now known to be associated with very small increases in serum creatinine. These increases may be determined as a relative (percent) value or a nominal value. Relative increases in serum creatinine as small as 20% from the pre-injury value have been reported to indicate acutely worsening renal function (AKI) and increased health risk, but the more commonly reported value to define AKI and increased health risk is a relative increase of at least 25%. Nominal increases as small as 0.3 mg/dL, 0.2 mg/dL or even 0.1 mg/dL have been reported to indicate worsening renal function and increased risk of death. Various time periods for the serum creatinine to rise to these threshold values have been used to define AKI, for example, ranging from 2 days, 3 days, 7 days, or a variable period defined as the time the patient is in the hospital or intensive care unit. These studies indicate there is not a particular threshold serum creatinine rise (or time period for the rise) for worsening renal function or AKI, but rather a continuous increase in risk with increasing magnitude of serum creatinine rise.
[0052] One study (Lassnigg et all, J Am Soc Nephrol 15:1597-1605, 2004, hereby incorporated by reference in its entirety) investigated both increases and decreases in serum creatinine. Patients with a mild fall in serum creatinine of -0.1 to -0.3 mg/dL, following heart surgery had the lowest mortality rate. Patients with a larger fall in serum creatinine (more than or equal to -0.4 mg/dL) or any increase in serum creatinine had a larger mortality rate. These findings caused the authors to conclude that even very subtle changes in renal function (as detected by small creatinine changes within 48 hours of surgery) seriously effect patient's outcomes. In an effort to reach consensus on a unified classification system for using serum creatinine to define AKI in clinical trials and in clinical practice, Bellomo et al., Crit Care. 8(4):R204-12, 2004, which is hereby incorporated by reference in its entirety, proposes the following classifications for stratifying AKI patients:
"Risk": serum creatinine increased 1.5 fold from baseline OR urine production of <0.5 ml/kg body weight/hr for 6 hours; "Injury": serum creatinine increased 2.0 fold from baseline OR urine production <0.5 ml/kg/hr for 12 h; "Failure": serum creatinine increased 3.0 fold from baseline OR creatinine >355 .mu.mol/l (with a rise of >44) or urine output below 0.3 ml/kg/hr for 24 h or anuria for at least 12 hours; And included two clinical outcomes: "Loss": persistent need for renal replacement therapy for more than four weeks. "ESRD": end stage renal disease--the need for dialysis for more than 3 months.
[0053] These criteria are called the RIFLE criteria, which provide a useful clinical tool to classify renal status. As discussed in Kellum, Crit. Care Med. 36: S141-45, 2008 and Ricci et al., Kidney Int. 73, 538-546, 2008, each hereby incorporated by reference in its entirety, the RIFLE criteria provide a uniform definition of AKI which has been validated in numerous studies.
[0054] More recently, Mehta et al., Crit. Care 11:R31 (doi:10.1186.cc5713), 2007, hereby incorporated by reference in its entirety, proposes the following similar classifications for stratifying AKI patients, which have been modified from RIFLE:
"Stage I": increase in serum creatinine of more than or equal to 0.3 mg/dL (.gtoreq.26.4 .mu.mol/L) or increase to more than or equal to 150% (1.5-fold) from baseline OR urine output less than 0.5 mL/kg per hour for more than 6 hours; "Stage II": increase in serum creatinine to more than 200% (>2-fold) from baseline OR urine output less than 0.5 mL/kg per hour for more than 12 hours; "Stage III": increase in serum creatinine to more than 300% (>3-fold) from baseline OR serum creatinine .gtoreq.354 .mu.mol/L accompanied by an acute increase of at least 44 .mu.mol/L OR urine output less than 0.3 mL/kg per hour for 24 hours or anuria for 12 hours.
[0055] The CIN Consensus Working Panel (McCollough et al, Rev Cardiovasc Med. 2006; 7(4):177-197, hereby incorporated by reference in its entirety) uses a serum creatinine rise of 25% to define Contrast induced nephropathy (which is a type of AKI).Although various groups propose slightly different criteria for using serum creatinine to detect AKI, the consensus is that small changes in serum creatinine, such as 0.3 mg/dL or 25%, are sufficient to detect AKI (worsening renal function) and that the magnitude of the serum creatinine change is an indicator of the severity of the AKI and mortality risk.
[0056] Although serial measurement of serum creatinine over a period of days is an accepted method of detecting and diagnosing AKI and is considered one of the most important tools to evaluate AKI patients, serum creatinine is generally regarded to have several limitations in the diagnosis, assessment and monitoring of AKI patients. The time period for serum creatinine to rise to values (e.g., a 0.3 mg/dL or 25% rise) considered diagnostic for AKI can be 48 hours or longer depending on the definition used. Since cellular injury in AKI can occur over a period of hours, serum creatinine elevations detected at 48 hours or longer can be a late indicator of injury, and relying on serum creatinine can thus delay diagnosis of AKI. Furthermore, serum creatinine is not a good indicator of the exact kidney status and treatment needs during the most acute phases of AKI when kidney function is changing rapidly. Some patients with AKI will recover fully, some will need dialysis (either short term or long term) and some will have other detrimental outcomes including death, major adverse cardiac events and chronic kidney disease. Because serum creatinine is a marker of filtration rate, it does not differentiate between the causes of AKI (pre-renal, intrinsic renal, post-renal obstruction, atheroembolic, etc) or the category or location of injury in intrinsic renal disease (for example, tubular, glomerular or interstitial in origin). Urine output is similarly limited, Knowing these things can be of vital importance in managing and treating patients with AKI.
[0057] For purposes of this document, the following definitions apply:
[0058] As used herein, an "injury to renal function" is an abrupt (within 14 days, preferably within 7 days, more preferably within 72 hours, and still more preferably within 48 hours) measurable reduction in a measure of renal function. Such an injury may be identified, for example, by a decrease in glomerular filtration rate or estimated GFR, a reduction in urine output, an increase in serum creatinine, an increase in serum cystatin C, a requirement for renal replacement therapy, etc. "Improvement in Renal Function" is an abrupt (within 14 days, preferably within 7 days, more preferably within 72 hours, and still more preferably within 48 hours) measurable increase in a measure of renal function. Preferred methods for measuring and/or estimating GFR are described hereinafter.
[0059] As used herein, "reduced renal function" is an abrupt (within 14 days, preferably within 7 days, more preferably within 72 hours, and still more preferably within 48 hours) reduction in kidney function identified by an absolute increase in serum creatinine of greater than or equal to 0.1 mg/dL (.gtoreq.8.8 mol/L), a percentage increase in serum creatinine of greater than or equal to 20% (1.2-fold from baseline), or a reduction in urine output (documented oliguria of less than 0. 5 ml/kg per hour).
[0060] As used herein, "acute renal failure" or "ARF" is an abrupt (within 14 days, preferably within 7 days, more preferably within 72 hours, and still more preferably within 48 hours) reduction in kidney function identified by an absolute increase in serum creatinine of greater than or equal to 0.3 mg/dl (.gtoreq.26.4 .mu.map, a percentage increase in serum creatinine of greater than or equal to 50% (1. 5-fold from baseline), or a reduction in urine output (documented oliguria of less than 0.5 ml/kg per hour for at least 6 hours). This term is synonymous with "acute kidney injury" or "AKI."
[0061] As used herein, the term "Insulin-like growth factor-binding protein 7" or "IGFBP7" refers to one or more polypeptides present in a biological sample that are derived from the Insulin-like growth factor-binding protein 7 precursor (human precursor: Swiss-Prot Q16270 (SEQ ID NO: 1))
TABLE-US-00002 10 20 30 40 MERPSLRALL LGAAGLLLLL LPLSSSSSSD TCGPCEPASC 50 60 70 80 PPLPPLGCLL GETRDACGCC PMCARGEGEP CGGGGAGRGY 90 100 110 120 CAPGMECVKS RKRRKGKAGA AAGGPGVSGV CVCKSRYPVC 130 140 150 160 GSDGTTYPSG CQLRAASQRA ESRGEKAITQ VSKGTCEQGP 170 180 190 200 SIVTPPKDIW NVTGAQVYLS CEVIGIPTPV LIWNKVKRGH 210 220 230 240 YGVQRTELLP GDRDNLAIQT RGGPEKHEVT GWVLVSPLSK 250 260 270 280 EDAGEYECHA SNSQGQASAS AKITVVDALH EIPVKKGEGA EL
[0062] The following domains have been identified in Insulin-like growth factor-binding protein 7:
TABLE-US-00003 Residues Length Domain ID 1-26 26 Signal peptide 27-282 256 Insulin-like growth factor-binding protein 7
[0063] As used herein, the term "Beta-2-glycoprotein 1" refers to one or polypeptides present in a biological sample that are derived from the Beta-2-glycoprotein 1 precursor (human precursor: Swiss-Prot P02749 (SEQ ID NO: 2)).
TABLE-US-00004 10 20 30 MISPVLILFS SFLCHVAIAG RTCPKPDDLP 40 50 60 FSTVVPLKTF YEPGEEITYS CKPGYVSRGG 70 80 90 MRKFICPLTG LWPINTLKCT PRVCPFAGIL 100 110 120 ENGAVRYTTF EYPNTISFSC NTGFYLNGAD 130 140 150 SAKCTEEGKW SPELPVCAPI ICPPPSIPTF 160 170 180 ATLRVYKPSA GNNSLYRDTA VFECLPQHAM 190 200 210 FGNDTITCTT HGNWTKLPEC REVKCPFPSR 220 230 240 PDNGFVNYPA KPTLYYKDKA TFGCHDGYSL 250 260 270 DGPEEIECTK LGNWSAMPSC KASCKVPVKK 280 290 300 ATVVYQGERV KIQEKFKNGM LHGDKVSFFC 310 320 330 KNKEKKCSYT EDAQCIDGTI EVPKCFKEHS 340 SLAFWKTDAS DVKPC
[0064] The following domains have been identified in Beta-2-glycoprotein 1:
TABLE-US-00005 Residues Length Domain ID 1-19 19 Signal sequence 20-345 326 Beta-2-glycoprotein 1
[0065] In addition, several naturally occurring variants have been identified:
TABLE-US-00006 Residue Change 5 V to A 107 S to N 154 R to H 266 V to L 325 C to G 335 W to S
[0066] As used herein, the term "Metalloproteinase inhibitor 2" refers to one or more polypeptides present in a biological sample that are derived from the Metalloproteinase inhibitor 2 precursor (human precursor: Swiss-Prot P16035 (SEQ ID NO: 3)).
TABLE-US-00007 10 20 30 MGAAARTLRL ALGLLLLATL LRPADACSCS 40 50 60 PVHPQQAFCN ADVVIRAKAV SEKEVDSGND 70 80 90 IYGNPIKRIQ YEIKQIKMFK GPEKDIEFIY 100 110 120 TAPSSAVCGV SLDVGGKKEY LIAGKAEGDG 130 140 150 KMHITLCDFI VPWDTLSTTQ KKSLNHRYQM 160 170 180 GCECKITRCP MIPCYISSPD ECLWMDWVTE 190 200 210 KNINGHQAKF FACIKRSDGS CAWYRGAAPP 220 KQEFLDIEDP
[0067] The following domains have been identified in Metalloproteinase inhibitor 2:
TABLE-US-00008 Residues Length Domain ID 1-26 26 Signal peptide 27-220 194 Metalloproteinase inhibitor 2
[0068] As used herein, the term "alpha-1-antitrypsin" refers to one or more polypeptides present in a biological sample that are derived from the alpha-1 -antitrypsin precursor (human precursor: Swiss-Prot P01009 (SEQ ID NO: 4)).
TABLE-US-00009 10 20 30 MPSSVSWGIL LLAGLCCLVP VSLAEDPQGD 40 50 60 AAQKTDTSHH DQDHPTFNKI TPNLAEFAFS 70 80 90 LYRQLAHQSN STNIFFSPVS IATAFAMLSL 100 110 120 GTKADTHDEI LEGLNFNLTE IPEAQIHEGF 130 140 150 QELLRTLNQP DSQLQLTTGN GLFLSEGLKL 160 170 180 VDKFLEDVKK LYHSEAFTVN FGDTEEAKKQ 190 200 210 INDYVEKGTQ GKIVDLVKEL DRDTVFALVN 220 230 240 YIFFKGKWER PFEVKDTEEE DFHVDQVTTV 250 260 270 KVPMMKRLGM FNIQHCKKLS SWVLLMKYLG 280 290 300 NATAIFFLPD EGKLQHLENE LTHDIITKFL 310 320 330 ENEDRRSASL HLPKLSITGT YDLKSVLGQL 340 350 360 GITKVFSNGA DLSGVTEEAP LKLSKAVHKA 370 380 390 VLTIDEKGTE AAGAMFLEAT PMSIPPEVKF 400 410 NKPFVFLMIE QNTKSPLFMG KVVNPTQK
[0069] The following domains have been identified in alpha-1-antitrypsin:
TABLE-US-00010 Residues Length Domain ID 1-24 24 signal sequence 25-418 394 alpha-1-antitrypsin
[0070] As used herein, the term "leukocyte elastase" refers to one or more polypeptides present in a biological sample that are derived from the leukocyte elastase precursor (human precursor: Swiss-Prot P08246 (SEQ ID NO: 5)).
TABLE-US-00011 10 20 30 MTLGRRLACL FLACVLPALL LGGTALASEI 40 50 60 VGGRRARPHA WPFMVSLQLR GGHFCGATLI 70 80 90 APNFVMSAAH CVANVNVRAV RVVLGAHNLS 100 110 120 RREPTRQVFA VQRIFENGYD PVNLLNDIVI 130 140 150 LQLNGSATIN ANVQVAQLPA QGRRLGNGVQ 160 170 180 CLAMGWGLLG RNRGIASVLQ ELNVIVVISL 190 200 210 CRRSNVCTLV RGRQAGVCFG DSGSPLVCNG 220 230 240 LIHGIASFVR GGCASGLYPD AFAPVAQFVN 250 260 WIDSIIQRSE DNPCPHPRDP DPASRTH
[0071] The following domains have been identified in leukocyte elastase:
TABLE-US-00012 Residues Length Domain ID 1-27 315 signal sequence 28-29 2 pro-peptide 30-267 238 leukocyte elastase
[0072] As used herein, the term "Serum amyloid P-component" refers to one or more polypeptides present in a biological sample that are derived from the Serum amyloid P-component precursor (human precursor: Swiss-Prot P02743 (SEQ ID NO: 6)).
TABLE-US-00013 10 20 30 MNKPLLWISV LTSLLEAFAH TDLSGKVFVF 40 50 60 PRESVTDHVN LITPLEKPLQ NFTLCFRAYS 70 80 90 DLSRAYSLFS YNTQGRDNEL LVYKERVGEY 100 110 120 SLYIGRHKVT SKVIEKFPAP VHICVSWESS 130 140 150 SGIAEFWING TPLVKKGLRQ GYFVEAQPKI 160 170 180 VLGQEQDSYG GKFDRSQSFV GEIGDLYMWD 190 200 210 SVLPPENILS AYQGTPLPAN ILDWQALNYE 220 IRGYVIIKPL VW
[0073] The following domains have been identified in Serum amyloid P-component:
TABLE-US-00014 Residues Length Domain ID 1-19 19 Signal peptide 20-223 204 Serum amyloid P-component 20-222 203 Serum amyloid P-component (1-203)
[0074] As used herein, the term "C--X--C motif chemokine 6" refers to one or more polypeptides present in a biological sample that are derived from the C--X--C motif chemokine 6 precursor (human precursor: Swiss-Prot P80162 (SEQ ID NO: 7))
TABLE-US-00015 10 20 30 MSLPSSRAAR VPGPSGSLCA LLALLLLLTP 40 50 60 PGPLASAGPV SAVLTELRCT CLRVTLRVNP 70 80 90 KTIGKLQVFP AGPQCSKVEV VASLKNGKQV 100 110 CLDPEAPFLK KVIQKILDSG NKKN
[0075] The following domains have been identified in C--X--C motif chemokine 6:
TABLE-US-00016 Residues Length Domain ID 1-37 37 Signal peptide 38-114 77 C-X-C motif chemokine 6 40-114 75 C-X-C motif chemokine 6 (N-processed variant 1) 43-114 72 C-X-C motif chemokine 6 (N-processed variant 2) 46-114 69 C-X-C motif chemokine 6 (N-processed variant 3)
[0076] As used herein, the term "C--C motif chemokine 24" refers to one or more polypeptides present in a biological sample that are derived from the C--C motif chemokine 24 precursor (human precursor: Swiss-Prot 000175 (SEQ ID NO: 8)).
TABLE-US-00017 10 20 30 40 MAGLMTIVTS LLFLGVCAHH IIPTGSVVIP SPCCMFFVSK 50 60 70 80 RIPENRVVSY QLSSRSTCLK AGVIFTTKKG QQFCGDPKQE 90 100 110 WVQRYMKNLD AKQKKASPRA RAVAVKGPVQ RYPGNQTTC
[0077] The following domains have been identified in C--C motif chemokine 24:
TABLE-US-00018 Residues Length Domain ID 1-26 26 Signal peptide 27-119 93 C-C motif chemokine 24
[0078] As used herein, the term "Neutrophil collagenase" (also known as MMP-8 and matrix metalloproteinase 8) refers to one or more polypeptides present in a biological sample that are derived from the Neutrophil collagenase precursor (human precursor: Swiss-Prot P22894 (SEQ ID NO: 9)).
TABLE-US-00019 10 20 30 40 MFSLKTLPFL LLLHVQISKA FPVSSKEKNT KTVQDYLEKF 50 60 70 80 YQLPSNQYQS TRKNGTNVIV EKLKEMQRFF GLNVTGKPNE 90 100 110 120 ETLDMMKKPR CGVPDSGGFM LTPGNPKWER TNLTYRIRNY 130 140 150 160 TPQLSEAEVE RAIKDAFELW SVASPLIFTR ISQGEADINI 170 180 190 200 AFYQRDHGDN SPFDGPNGIL AHAFQPGQGI GGDAHFDAEE 210 220 230 240 TWINTSANYN LFLVAAHEFG HSLGLAHSSD PGALMYPNYA 250 260 270 280 FRETSNYSLP QDDIDGIQAI YGLSSNPIQP TGPSTPKPCD 290 300 310 320 PSLTFDAITT LRGEILFFKD RYFWRRHPQL QRVEMNFISL 330 340 350 360 FWPSLPTGIQ AAYEDFDRDL IFLFKGNQYW ALSGYDILQG 370 380 390 400 YPKDISNYGF PSSVQAIDAA VFYRSKTYFF VNDQFWRYDN 410 420 430 440 QRQFMEPGYP KSISGAFPGI ESKVDAVFQQ EHFFHVFSGP 450 460 RYYAFDLIAQ RVTRVARGNK WLNCRYG
[0079] The following domains have been identified in Neutrophil collagenase:
TABLE-US-00020 Residues Length Domain ID 1-20 20 Signal peptide 21-100 80 Activation peptide 101-467 367 Neutrophil collagenase
[0080] As used herein, the term "Cathepsin D" refers to one or more polypeptides present in a biological sample that are derived from the Cathepsin D precursor (human precursor: Swiss-Prot P07339 (SEQ ID NO: 10)).
TABLE-US-00021 10 20 30 40 MQPSSLLPLA LCLLAAPASA LVRIPLHKFT SIRRTMSEVG 50 60 70 80 GSVEDLIAKG PVSKYSQAVP AVTEGPIPEV LKNYMDAQYY 90 100 110 120 GEIGIGTPPQ CFTVVFDTGS SNLWVPSIHC KLLDIACWIH 130 140 150 160 HKYNSDKSST YVKNGTSFDI HYGSGSLSGY LSQDTVSVPC 170 180 190 200 QSASSASALG GVKVERQVFG EATKQPGITF IAAKFDGILG 210 220 230 240 MAYPRISVNN VLPVFDNLMQ QKLVDQNIFS FYLSRDPDAQ 250 260 270 280 PGGELMLGGT DSKYYKGSLS YLNVTRKAYW QVHLDQVEVA 290 300 310 320 SGLTLCKEGC EAIVDTGTSL MVGPVDEVRE LQKAIGAVPL 330 340 350 360 IQGEYMIPCE KVSTLPAITL KLGGKGYKLS PEDYTLKVSQ 370 380 390 400 AGKTLCLSGF MGMDIPPPSG PLWILGDVFI GRYYTVFDRD 410 NNRVGFAEAA RL
[0081] The following domains have been identified in Capthesin D:
TABLE-US-00022 Residues Length Domain ID 1-18 18 Signal peptide 19-64 46 Activation peptide 65-412 348 Cathepsin D 65-161 348 Cathepsin D light chain 169-412 348 Cathepsin D heavy chain
[0082] As used herein, the term "C--X--C Motif chemokine 13" refers to one or more polypeptides present in a biological sample that are derived from the C--X--C Motif chemokine 13 precursor (human precursor: Swiss-Prot 043927 (SEQ ID NO: 11)).
TABLE-US-00023 10 20 30 40 MKFISTSLLL MLLVSSLSPV QGVLEVYYTS LRCRCVQESS 50 60 70 80 VFIPRRFIDR IQILPRGNGC PRKEIIVWKK NKSIVCVDPQ 90 100 AFWIQRMMEV LRKRSSSTLP VPVFKRKIP
[0083] The following domains have been identified in C--X--C Motif chemokine 13:
TABLE-US-00024 Residues Length Domain ID 1-22 22 Signal peptide 23-109 87 C-X-C Motif chemokine 13
[0084] As used herein, the term "Involucrin" refers to one or more polypeptides present in a biological sample that are derived from the Involucrin precursor (human precursor: Swiss-Prot P07476 (human precursor: SEQ ID NO: 12)).
TABLE-US-00025 10 20 30 40 MSQQHTLPVT LSPALSQELL KTVPPPVNTH QEQMKQPTPL 50 60 70 80 PPPCQKVPVE LPVEVPSKQE EKHMTAVKGL PEQECEQQQK 90 100 110 120 EPQEQELQQQ HWEQHEEYQK AENPEQQLKQ EKTQRDQQLN 130 140 150 160 KQLEEEKKLL DQQLDQELVK RDEQLGMKKE QLLELPEQQE 170 180 190 200 GHLKHLEQQE GQLKHPEQQE GQLELPEQQE GQLELPEQQE 210 220 230 240 GQLELPEQQE GQLELPEQQE GQLELPEQQE GQLELPQQQE 250 260 270 280 GQLELSEQQE GQLELSEQQE GQLKHLEHQE GQLEVPEEQM 290 300 310 320 GQLKYLEQQE GQLKHLDQQE KQPELPEQQM GQLKHLEQQE 330 340 350 360 GQPKHLEQQE GQLEQLEEQE GQLKHLEQQE GQLEHLEHQE 370 380 390 400 GQLGLPEQQV LQLKQLEKQQ GQPKHLEEEE GQLKHLVQQE 410 420 430 440 GQLKHLVQQE GQLEQQERQV EHLEQQVGQL KHLEEQEGQL 450 460 470 480 KHLEQQQGQL EVPEQQVGQP KNLEQEEKQL ELPEQQEGQV 490 500 510 520 KHLEKQEAQL ELPEQQVGQP KHLEQQEKHL EHPEQQDGQL 530 540 550 560 KHLEQQEGQL KDLEQQKGQL EQPVFAPAPG QVQDIQPALP 570 580 TKGEVLLPVE HQQQKQEVQW PPKHK
[0085] As used herein, the term "Interleukin-6 receptor subunit beta" refers to one or more polypeptides present in a biological sample that are derived from the Interleukin-6 receptor subunit beta precursor (human precursor: Swiss-Prot P40189 (SEQ ID NO: 13))
TABLE-US-00026 10 20 30 40 MLTLQTWLVQ ALFIFLTTES TGELLDPCGY ISPESPVVQL 50 60 70 80 HSNFTAVCVL KEKCMDYFHV NANYIVWKTN HFTIPKEQYT 90 100 110 120 IINRTASSVT FTDIASLNIQ LTCNILTFGQ LEQNVYGITI 130 140 150 160 ISGLPPEKPK NLSCIVNEGK KMRCEWDGGR ETHLETNFTL 170 180 190 200 KSEWATHKFA DCKAKRDTPT SCTVDYSTVY FVNIEVWVEA 210 220 230 240 ENALGKVTSD HINFDPVYKV KPNPPHNLSV INSEELSSIL 250 260 270 280 KLTWTNPSIK SVIILKYNIQ YRTKDASTWS QIPPEDTAST 290 300 310 320 RSSFTVQDLK PFTEYVFRIR CMKEDGKGYW SDWSEEASGI 330 340 350 360 TYEDRPSKAP SFWYKIDPSH TQGYRTVQLV WKTLPPFEAN 370 380 390 400 GKILDYEVTL TRWKSHLQNY TVNATKLTVN LTNDRYLATL 410 420 430 440 TVRNLVGKSD AAVLTIPACD FQATHPVMDL KAFPKDNMLW 450 460 470 480 VEWTTPRESV KKYILEWCVL SDKAPCITDW QQEDGTVHRT 490 500 510 520 YLRGNLAESK CYLITVTPVY ADGPGSPESI KAYLKQAPPS 530 540 550 560 KGPTVRTKKV GKNEAVLEWD QLPVDVQNGF IRNYTIFYRT 570 580 590 600 IIGNETAVNV DSSHTEYTLS SLTSDTLYMV RMAAYTDEGG 610 620 630 640 KDGPEFTFTT PKFAQGEIEA IVVPVCLAFL LTTLLGVLFC 650 660 670 680 FNKRDLIKKH IWPNVPDPSK SHIAQWSPHT PPRHNFNSKD 690 700 710 720 QMYSDGNFTD VSVVEIEAND KKPFPEDLKS LDLFKKEKIN 730 740 750 760 TEGHSSGIGG SSCMSSSRPS ISSSDENESS QNTSSTVQYS 770 780 790 800 TVVHSGYRHQ VPSVQVFSRS ESTQPLLDSE ERPEDLQLVD 810 820 830 840 HVDGGDGILP RQQYFKQNCS QHESSPDISH FERSKQVSSV 850 860 870 880 NEEDFVRLKQ QISDHISQSC GSGQMKMFQE VSAADAFGPG 890 900 910 TEGQVERFET VGMEAATDEG MPKSYLPQTV RQGGYMPQ
[0086] Most preferably, the Interleukin-6 receptor subunit beta assay detects one or more soluble forms of Interleukin-6 receptor subunit beta. Interleukin-6 receptor subunit beta is a type I membrane protein having a large extracellular domain, most or all of which is present in soluble forms of Interleukin-6 receptor subunit beta generated either through alternative splicing event which deletes all or a portion of the transmembrane domain, or by proteolysis of the membrane-bound form. In the case of an immunoassay, one or more antibodies that hind to epitopes within this extracellular domain may be used to detect these soluble form(s). The following domains have been identified in Interleukin-6 receptor subunit beta:
TABLE-US-00027 Residues Length Domain ID 1-22 22 Signal peptide 23-918 896 Interleukin-6 receptor subunit beta 642-918 277 Cytoplasmic domain 620-641 21 transmembrane domain 23-619 597 Extracellular domain 330-918 589 Missing in isoform 2 325-329 5 RPSKA (SEQ ID NO: 14) .fwdarw. NIASF (SEQ ID NO: 15) in isoform 2
[0087] As used herein, the term "Hepatocyte growth factor" refers to one or more polypeptides present in a biological sample that are derived from the Hepatocyte growth factor precursor (human precursor: Swiss-Prot P14210 (SEQ ID NO: 16)).
TABLE-US-00028 10 20 30 40 MWVTKLLPAL LLQHVLLHLL LLPIAIPYAE GQRKRRNTIH 50 60 70 80 EFKKSAKTTL IKIDPALKIK TKKVNTADQC ANRCTRNKGL 90 100 110 120 PFTCKAFVFD KARKQCLWFP FNSMSSGVKK EFGHEFDLYE 130 140 150 160 NKDYIRNCII GKGRSYKGTV SITKSGIKCQ PWSSMIPHEH 170 180 190 200 SFLPSSYRGK DLQENYCRNP RGEEGGPWCF TSNPEVRYEV 210 220 230 240 CDIPQCSEVE CMTCNGESYR GLMDHTESGK ICQRWDHQTP 250 260 270 280 HRHKFLPERY PDKGFDDNYC RNPDGQPRPW CYTLDPHTRW 290 300 310 320 EYCAIKTCAD NTMNDTDVPL ETTECIQGQG EGYRGTVNTI 330 340 350 360 WNGIPCQRWD SQYPHEHDMT PENFKCKDLR ENYCRNPDGS 370 380 390 400 ESPWCFTTDP NIRVGYCSQI PNCDMSHGQD CYRGNGKNYM 410 420 430 440 GNLSQTRSGL TCSMWDKNME DLHRHIFWEP DASKLNENYC 450 460 470 480 RNPDDDAHGP WCYTGNPLIP WDYCPISRCE GDTTPTIVNL 490 500 510 520 DHPVISCAKT KQLRVVNGIP TRTNIGWMVS LRYRNKHICG 530 540 550 560 GSLIKESWVL TARQCFPSRD LKDYEAWLGI HDVHGRGDEK 570 580 590 600 CKQVLNVSQL VYGPEGSDLV LMKLARPAVL DDFVSTIDLP 610 620 630 640 NYGCTIPEKT SCSVYGWGYT GLINYDGLLR VAHLYIMGNE 650 660 670 680 KCSQHHRGKV TLNESEICAG AEKIGSGPCE GDYGGPLVCE 690 700 710 720 QHKMRMVLGV IVPGRGCAIP NRPGIFVRVA YYAKWIHKII LTYKVPQS
[0088] The following domains have been identified in Hepatocyte growth factor:
TABLE-US-00029 Residues Length Domain ID 1-31 31 signal sequence 32-494 463 Hepatocyte growth factor alpha chain 495-728 234 Hepatocyte growth factor beta chain
[0089] As used herein, the term "Metalloproteinase inhibitor 4" refers to one or polypeptides present in a biological sample that are derived from the Metalloproteinase inhibitor 4 precursor (human precursor: Swiss-Prot Q99727 (SEQ ID NO: 17)).
TABLE-US-00030 10 20 30 40 MPGSPRPAPS WVLLLRLLAL LRPPGLGEAC SCAPAHPQQH 50 60 70 80 ICHSALVIRA KISSEKVVPA SADPADTEKM LRYEIKQIKM 90 100 110 120 FKGFEKVKDV QYIYTPFDSS LCGVKLEANS QKQYLLTGQV 130 140 150 160 LSDGKVFIHL CNYIEPWEDL SLVQRESLNH HYHLNCGCQI 170 180 190 200 TTCYTVPCTI SAPNECLWTD WLLERKLYGY QAQHYVCMKH 210 220 VDGTCSWYRG HLPLRKEFVD IVQP
[0090] The following domains have been identified in Metalloproteinase inhibitor 4:
TABLE-US-00031 Residues Length Domain ID 1-27 27 Signal sequence 28-224 197 Metalloproteinase inhibitor 4
[0091] As used herein, the term "C--C motif chemokine 18" refers to one or more polypeptides present in a biological sample that are derived from the C--C motif chemokine 18 precursor (human precursor: Swiss-Prot P55774 (SEQ ID NO: 18)).
TABLE-US-00032 10 20 30 40 MKGLAAALLV LVCTMALCSC AQVGTNKELC CLVYTSWQIP 50 60 70 80 QKFIVDYSET SPQCPKPGVI LLTKRGRQIC ADPNKKWVQK YISDLKLNA
[0092] The following domains have been identified in C--C motif chemokine 18:
TABLE-US-00033 Residues Length Domain ID 1-20 20 Signal peptide 21-89 69 C-C motif chemokine 18 21-88 68 CCL 18 (1-68) 23-89 67 CCL 18 (3-69) 24-89 66 CCL 18 (4-69)
[0093] As used herein, the term "Matrilysin" refers to one or more polypeptides present in a biological sample that are derived from the Matrilysin precursor (Swiss-Prot P09237 (human precursor: SEQ ID NO: 19))
TABLE-US-00034 10 20 30 40 MRLTVLCAVC LLPGSLALPL PQEAGGMSEL QWEQAQDYLK 50 60 70 80 RFYLYDSETK NANSLEAKLK EMQKFFGLPI TGMLNSRVIE 90 100 110 120 IMQKPRCGVP DVAEYSLFPN SPKWTSKVVT YRIVSYTRDL 130 140 150 160 PHITVDRLVS KALNMWGKEI PLHFRKVVWG TADIMIGFAR 170 180 190 200 GAHGDSYPFD GPGNTLAHAF APGTGLGGDA HFDEDERWTD 210 220 230 240 GSSLGINFLY AATHELGHSL GMGHSSDPNA VMYPTYGNGD 250 260 PQNFKLSQDD IKGIQKLYGK RSNSRKK
[0094] The following domains have been identified in Matrilysin:
TABLE-US-00035 Residues Length Domain ID 1-17 17 signal peptide 18-94 77 activation peptide 95-267 173 Matrilysin
[0095] As used herein, the term "C--X--C motif chemokine 11" refers to one or more polypeptides present in a biological sample that are derived from the C--X--C motif chemokine 11 precursor (human precursor: Swiss-Prot 014625 (SEQ ID NO: 20))
TABLE-US-00036 10 20 30 40 MSVKGMAIAL AVILCATVVQ GFPMFKRGRC LCIGPGVKAV 50 60 70 80 KVADIEKASI MYPSNNCDKI EVIITLKENK GQRCLNPKSK 90 QARLIIKKVE RKNF
[0096] The following domains have been identified in C--X--C motif chemokine 11:
TABLE-US-00037 Residues Length Domain ID 1-21 21 signal peptide 22-94 73 C-X-C motif chemokine 11
[0097] As used herein, the term "C--X--C motif chemokines -1, -2, and -3" refers to one or more polypeptides present in a biological sample that are common to the C--X--C motif chemokines -1, -2, and -3 precursors (Swiss-Prot accession numbers of the human precursors: C--X--C motif chemokine -1 (P09341), -2 (P19875), and -3 (P19876)).
[0098] CXC motif chemokine-1 is also known as "Growth-regulated alpha protein" (human precursor Swiss-Prot P09341 (SEQ ID NO: 21)).
TABLE-US-00038 10 20 30 40 MARAALSAAP SNPRLLRVAL LLLLLVAAGR RAAGASVATE 50 60 70 80 LRCQCLQTLQ GIHPKNIQSV NVKSPGPHCA QTEVIATLKN 90 100 GRKACLNPAS PIVKKIIEKM LNSDKSN
[0099] The following domains have been identified in Growth-regulated alpha protein:
TABLE-US-00039 Residues Length Domain ID 1-34 34 Signal peptide 35-107 73 Growth-regulated alpha protein 38-107 70 GRO-alpha (4-73) 39-107 69 GRO-alpha (5-73) 40-107 68 GRO-alpha (6-73)
[0100] CXC motif chemokine-2 is also known as "Macrophage inflammatory protein 2-alpha" (human precursor Swiss-Prot P19875 (SEQ ID NO: 22)).
TABLE-US-00040 10 20 30 40 MARATLSAAP SNPRLLRVAL LLLLLVAASR RAAGAPLATE 50 60 70 80 LRCQCLQTLQ GIHLKNIQSV KVKSPGPHCA QTEVIATLKN 90 100 GQKACLNPAS PMVKKIIEKM LKNGKSN
[0101] The following domains have been identified in Macrophage inflammatory protein 2-alpha:
TABLE-US-00041 Residues Length Domain ID 1-34 34 Signal peptide 35-107 73 C-X-C motif chemokine 2 39-107 69 GRO-beta (5-73)
[0102] CXC motif chemokine-2 is also known as "Growth-regulated protein gamma" (human precursor Swiss-Prot P19876 (SEQ ID NO: 23)).
TABLE-US-00042 10 20 30 40 MAHATLSAAP SNPRLLRVAL LLLLLVAASR RAAGASVVTE 50 60 70 80 LRCQCLQTLQ GIHLKNIQSV NVRSPGPHCA QTEVIATLKN 90 100 GKKACLNPAS PMVQKIIEKI LNKGSTN
[0103] The following domains have been identified in C--X--C motif chemokine 3:
TABLE-US-00043 Residues Length Domain ID 1-34 34 Signal peptide 35-107 73 C-X-C motif chemokine 3 39-107 73 GRO-gamma (5-73)
[0104] As used herein, the term "Antileukoproteinase" refers to one or more polypeptides present in a biological sample that are derived from the Antileukoproteinase precursor (Swiss-Prot P03973 (SEQ ID NO: 24)).
TABLE-US-00044 10 20 30 40 MKSSGLFPFL VLLALGTLAP WAVEGSGKSF KAGVCPPKKS 50 60 70 80 AQCLRYKKPE CQSDWQCPGK KRCCPDTCGI KCLDPVDTPN 90 100 110 120 PTRRKPGKCP VTYGQCLMLN PPNFCEMDGQ CKRDLKCCMG 130 MCGKSCVSPV KA
[0105] The following domains have been identified in Antileukoproteinase:
TABLE-US-00045 Residues Length Domain ID 1-25 25 signal sequence 26-132 107 Antileukoproteinase
[0106] As used herein, the term "IgA" refers to an antibody having two subclasses (IgA1 and IgA2) and which can exist in a dimeric form linked by a J chain (called secretory IgA, or sIgA). In its secretory form, IgA is the main immunoglobulin found in mucous secretions, including tears, saliva, colostrum and secretions from the genito-urinary tract, gastrointestinal tractprostate and respiratory epithelium. It is also found in small amounts in blood. IgA may be measured separately from other immunoglobulins such as IgG or IgM, for example, using antibodies which bind to the IgA .alpha.-chain.
[0107] As used herein, the terms "IgG1" and "IgG subclass I" refer to subclass 1 of the glycoprotein immunoglobulin G (IgG), a major effector molecule of the humoral immune response in man. Antibodies of the IgG class express their predominant activity during a secondary antibody response. The basic immunoglobulin G molecule has a four-chain structure, comprising two identical heavy (H) chains and two identical light (L) chains, linked together by inter-chain disulfide bonds. Each heavy chain is encoded by 4 distinct types of gene segments, designated V.sub.H (variable), D (diversity), J.sub.H (joining) and C.sub.H(constant). The variable region of the heavy chain is encoded by the V.sub.H, D and J.sub.H segments. The light chains are encoded by the 3 gene segments, V.sub.L, J.sub.L and C.sub.L. The variable region of the light chains is encoded by the V.sub.L and J.sub.L segments.
[0108] As used herein, the terms "IgG2" and "IgG subclass II" refer to subclass 2 of the glycoprotein immunoglobulin G (TgG), a major effector molecule of the humoral immune response in man. Antibodies of the IgG class express their predominant activity during a secondary antibody response. The basic immunoglobulin G molecule has a four-chain structure, comprising two identical heavy (H) chains and two identical light (L) chains, linked together by inter-chain disulfide bonds. Each heavy chain is encoded by 4 distinct types of gene segments, designated V.sub.H (variable), D (diversity), J.sub.H (joining) and C.sub.H(constant). The variable region of the heavy chain is encoded by the V.sub.H, D and J.sub.H segments. The light chains are encoded by the 3 gene segments, V.sub.L, J.sub.L and C.sub.L. The variable region of the light chains is encoded by the V.sub.L and J.sub.L segments.
[0109] The length and flexibility of the hinge region varies among the IgG subclasses. The hinge region of IgG1 encompasses amino acids 216-231 and since it is freely flexible, the Fab fragments can rotate about their axes of symmetry and move within a sphere centered at the first of two inter-heavy chain disulfide bridges (23). IgG2 has a shorter hinge than IgG 1, with 12 amino acid residues and four disulfide bridges. The hinge region of IgG2 lacks a glycine residue, it is relatively short and contains a rigid poly-proline double helix, stabilised by extra inter-heavy chain disulfide bridges. These properties restrict the flexibility of the IgG2 molecule (24). IgG3 differs from the other subclasses by its unique extended hinge region (about four times as long as the IgG1 hinge), containing 62 amino acids (including 21 prolines and 11 cysteines), forming an inflexible poly-proline double helix (25,26). In IgG3 the Fab fragments are relatively far away from the Fc fragment, giving the molecule a greater flexibility. The elongated hinge in IgG3 is also responsible for its higher molecular weight compared to the other subclasses. The hinge region of IgG4 is shorter than that of IgG1 and its flexibility is intermediate between that of IgG1 and IgG2.
[0110] The four IgG subclasses also differ with respect to the number of inter-heavy chain disulfide bonds in the hinge region (26). The structural differences between the IgG subclasses are also reflected in their susceptibility to proteolytic enzymes. IgG3 is very susceptible to cleavage by these enzymes, whereas IgG2 is relatively resistant. IgG1 and IgG4 exhibit an intermediary sensitivity, depending upon the enzyme used. Since these proteolytic enzymes all cleave IgG molecules near or within the hinge region, it is likely that the high sensitivity of IgG3 to enzyme digestion is related to its accessible hinge. Another structural difference between the human IgG subclasses is the linkage of the heavy and light chain by a disulfide bond. This bond links the carboxy-terminal of the light chain with the cysteine residue at position 220 (in IgG) or at position 131 (in IgG2, IgG3 and IgG4) of the CH1 sequence of the heavy chain.
[0111] As a consequence of the structural differences, the four IgG subclasses may be distinguished from one another, for example using antibodies that are specific for differences between the isoforms. In the present application, a level of IgG1 is determined using an assay which distinguishes this subclass, relative to the other subclasses.
[0112] As used herein, the term "relating a signal to the presence or amount" of an analyte reflects the following understanding. Assay signals are typically related to the presence or amount of an analyte through the use of a standard curve calculated using known concentrations of the analyte of interest. As the term is used herein, an assay is "configured to detect" an analyte if an assay can generate a detectable signal indicative of the presence or amount of a physiologically relevant concentration of the analyte. Because an antibody epitope is on the order of 8 amino acids, an immunoassay configured to detect a marker of interest will also detect polypeptides related to the marker sequence, so long as those polypeptides contain the epitope(s) necessary to bind to the antibody or antibodies used in the assay. The term "related marker" as used herein with regard to a biomarker such as one of the kidney injury markers described herein refers to one or more fragments, variants, etc., of a particular marker or its biosynthetic parent that may be detected as a surrogate for the marker itself or as independent biomarkers. The term also refers to one or more polypeptides present in a biological sample that are derived from the biomarker precursor complexed to additional species, such as binding proteins, receptors, heparin, lipids, sugars, etc.
[0113] In this regard, the skilled artisan will understand that the signals obtained from an immunoassay are a direct result of complexes formed between one or more antibodies and the target biomolecule (i.e., the analyte) and polypeptides containing the necessary epitope(s) to which the antibodies bind. While such assays may detect the full length biomarker and the assay result be expressed as a concentration of a biomarker of interest, the signal from the assay is actually a result of all such "immunoreactive" polypeptides present in the sample. Expression of biomarkers may also be determined by means other than immunoassays, including protein measurements (such as dot blots, western blots, chromatographic methods, mass spectrometry, etc.) and nucleic acid measurements (mRNA quatitation). This list is not meant to be limiting.
[0114] The term "positive going" marker as that term is used herein refer to a marker that is determined to be elevated in sepsis patients suffering from a disease or condition, relative to sepsis patients not suffering from that disease or condition. The term "negative going" marker as that term is used herein refer to a marker that is determined to be reduced in sepsis patients suffering from a disease or condition, relative to sepsis patients not suffering from that disease or condition.
[0115] The term "sepsis patient" as used herein refers to a human or non-human organism. Thus, the methods and compositions described herein are applicable to both human and veterinary disease. Further, while a sepsis patient is preferably a living organism, the invention described herein may be used in post-mortem analysis as well. Preferred sepsis patients are humans, and most preferably "patients," which as used herein refers to living humans that are receiving medical care for a disease or condition. This includes persons with no defined illness who are being investigated for signs of pathology.
[0116] Preferably, an analyte is measured in a sample. Such a sample may be obtained from a sepsis patient, or may be obtained from biological materials intended to be provided to the sepsis patient. For example, a sample may be obtained from a kidney being evaluated for possible transplantation into a sepsis patient, and an analyte measurement used to evaluate the kidney for preexisting damage. Preferred samples are body fluid samples.
[0117] The term "body fluid sample" as used herein refers to a sample of bodily fluid obtained for the purpose of diagnosis, prognosis, classification or evaluation of a sepsis patient of interest, such as a patient or transplant donor. In certain embodiments, such a sample may be obtained for the purpose of determining the outcome of an ongoing condition or the effect of a treatment regimen on a condition. Preferred body fluid samples include blood, serum, plasma, cerebrospinal fluid, urine, saliva, sputum, and pleural effusions. In addition, one of skill in the art would realize that certain body fluid samples would be more readily analyzed following a fractionation or purification procedure, for example, separation of whole blood into serum or plasma components.
[0118] The term "diagnosis" as used herein refers to methods by which the skilled artisan can estimate and/or determine the probability ("a likelihood") of whether or not a patient is suffering from a given disease or condition. In the case of the present invention, "diagnosis" includes using the results of an assay, most preferably an immunoassay, for a kidney injury marker of the present invention, optionally together with other clinical characteristics, to arrive at a diagnosis (that is, the occurrence or nonoccurrence) of an acute renal injury or ARF for the sepsis patient from which a sample was obtained and assayed. That such a diagnosis is "determined" is not meant to imply that the diagnosis is 100% accurate. Many biomarkers are indicative of multiple conditions. The skilled clinician does not use biomarker results in an informational vacuum, but rather test results are used together with other clinical indicia to arrive at a diagnosis. Thus, a measured biomarker level on one side of a predetermined diagnostic threshold indicates a greater likelihood of the occurrence of disease in the sepsis patient relative to a measured level on the other side of the predetermined diagnostic threshold.
[0119] Similarly, a prognostic risk signals a probability ("a likelihood") that a given course or outcome will occur. A level or a change in level of a prognostic indicator, which in turn is associated with an increased probability of morbidity (e.g., worsening renal function, future ARF, or death) is referred to as being "indicative of an increased likelihood" of an adverse outcome in a patient.
[0120] Marker Assays
[0121] In general, immunoassays involve contacting a sample containing or suspected of containing a biomarker of interest with at least one antibody that specifically binds to the biomarker. A signal is then generated indicative of the presence or amount of complexes formed by the binding of polypeptides in the sample to the antibody. The signal is then related to the presence or amount of the biomarker in the sample. Numerous methods and devices are well known to the skilled artisan for the detection and analysis of biomarkers. See, e.g., U.S. Pat. Nos. 6,143,576; 6,113,855; 6,019,944; 5,985,579; 5,947,124; 5,939,272; 5,922,615; 5,885,527; 5,851,776; 5,824,799; 5,679,526; 5,525,524; and 5,480,792, and The Immunoassay Handbook, David Wild, ed. Stockton Press, New York, 1994, each of which is hereby incorporated by reference in its entirety, including all tables, figures and claims.
[0122] The assay devices and methods known in the art can utilize labeled molecules in various sandwich, competitive, or non-competitive assay formats, to generate a signal that is related to the presence or amount of the biomarker of interest. Suitable assay formats also include chromatographic, mass spectrographic, and protein "blotting" methods. Additionally, certain methods and devices, such as biosensors and optical immunoassays, may be employed to determine the presence or amount of analytes without the need for a labeled molecule. See, e.g., U.S. Pat. Nos. 5,631,171; and 5,955,377, each of which is hereby incorporated by reference in its entirety, including all tables, figures and claims. One skilled in the art also recognizes that robotic instrumentation including but not limited to Beckman ACCESS.RTM., Abbott AXSYM.RTM., Roche ELECSYS.RTM., Dade Behring STRATUS.RTM. systems are among the immunoassay analyzers that are capable of performing immunoassays. But any suitable immunoassay may be utilized, for example, enzyme-linked immunoassays (ELISA), radioimmunoassays (RIAs), competitive binding assays, and the like.
[0123] Antibodies or other polypeptides may be immobilized onto a variety of solid supports for use in assays. Solid phases that may be used to immobilize specific binding members include include those developed and/or used as solid phases in solid phase binding assays. Examples of suitable solid phases include membrane filters, cellulose-based papers, beads (including polymeric, latex and paramagnetic particles), glass, silicon wafers, microparticles, nanoparticles, TentaGels, AgroGels, PEGA gels, SPOCC gels, and multiple-well plates. An assay strip could be prepared by coating the antibody or a plurality of antibodies in an array on solid support. This strip could then be dipped into the test sample and then processed quickly through washes and detection steps to generate a measurable signal, such as a colored spot. Antibodies or other polypeptides may be bound to specific zones of assay devices either by conjugating directly to an assay device surface, or by indirect binding. In an example of the later case, antibodies or other polypeptides may be immobilized on particles or other solid supports, and that solid support immobilized to the device surface.
[0124] Biological assays require methods for detection, and one of the most common methods for quantitation of results is to conjugate a detectable label to a protein or nucleic acid that has affinity for one of the components in the biological system being studied. Detectable labels may include molecules that are themselves detectable (e.g., fluorescent moieties, electrochemical labels, metal chelates, etc.) as well as molecules that may be indirectly detected by production of a detectable reaction product (e.g., enzymes such as horseradish peroxidase, alkaline phosphatase, etc.) or by a specific binding molecule which itself may be detectable (e.g., biotin, digoxigenin, maltose, oligohistidine, 2,4-dintrobenzene, phenylarsenate, ssDNA, dsDNA, etc.).
[0125] Preparation of solid phases and detectable label conjugates often comprise the use of chemical cross-linkers. Cross-linking reagents contain at least two reactive groups, and are divided generally into homofunctional cross-linkers (containing identical reactive groups) and heterofunctional cross-linkers (containing non-identical reactive groups). Homobifunctional cross-linkers that couple through amines, sulfhydryls or react non-specifically are available from many commercial sources. Maleimides, alkyl and aryl halides, alpha-haloacyls and pyridyl disulfides are thiol reactive groups. Maleimides, alkyl and aryl halides, and alpha-haloacyls react with sulfhydryls to form thiol ether bonds, while pyridyl disulfides react with sulfhydryls to produce mixed disulfides. The pyridyl disulfide product is cleavable. Imidoesters are also very useful for protein-protein cross-links. A variety of heterobifunctional cross-linkers, each combining different attributes for successful conjugation, are commercially available.
[0126] In certain aspects, the present invention provides kits for the analysis of the described kidney injury markers. The kit comprises reagents for the analysis of at least one test sample which comprise at least one antibody that a kidney injury marker. The kit can also include devices and instructions for performing one or more of the diagnostic and/or prognostic correlations described herein. Preferred kits will comprise an antibody pair for performing a sandwich assay, or a labeled species for performing a competitive assay, for the analyte. Preferably, an antibody pair comprises a first antibody conjugated to a solid phase and a second antibody conjugated to a detectable label, wherein each of the first and second antibodies that bind a kidney injury marker. Most preferably each of the antibodies are monoclonal antibodies. The instructions for use of the kit and performing the correlations can be in the form of labeling, which refers to any written or recorded material that is attached to, or otherwise accompanies a kit at any time during its manufacture, transport, sale or use. For example, the term labeling encompasses advertising leaflets and brochures, packaging materials, instructions, audio or video cassettes, computer discs, as well as writing imprinted directly on kits.
[0127] Antibodies
[0128] The term "antibody" as used herein refers to a peptide or polypeptide derived from, modeled after or substantially encoded by an immunoglobulin gene or immunoglobulin genes, or fragments thereof, capable of specifically binding an antigen or epitope. See, e.g. Fundamental Immunology, 3rd Edition, W. E. Paul, ed., Raven Press, New York (1993); Wilson (1994; J. Immunol. Methods 175:267-273; Yarmush (1992) J. Biochem. Biophys. Methods 25:85-97. The term antibody includes antigen-binding portions, i.e., "antigen binding sites," (e.g., fragments, subsequences, complementarity determining regions (CDRs)) that retain capacity to bind antigen, including (i) a Fab fragment, a monovalent fragment consisting of the VL, VH, CL and CH1 domains; (ii) a F(ab')2 fragment, a bivalent fragment comprising two Fab fragments linked by a disulfide bridge at the hinge region; (iii) a Fd fragment consisting of the VH and CH1 domains; (iv) a Fv fragment consisting of the VL and VH domains of a single arm of an antibody, (v) a dAb fragment (Ward et al., (1989) Nature 341:544-546), which consists of a VII domain; and (vi) an isolated complementarity determining region (CDR). Single chain antibodies are also included by reference in the term "antibody."
[0129] Antibodies used in the immunoassays described herein preferably specifically bind to a kidney injury marker of the present invention. The term "specifically binds" is not intended to indicate that an antibody binds exclusively to its intended target since, as noted above, an antibody binds to any polypeptide displaying the epitope(s) to which the antibody binds. Rather, an antibody "specifically binds" if its affinity for its intended target is about 5-fold greater when compared to its affinity for a non-target molecule which does not display the appropriate epitope(s). Preferably the affinity of the antibody will be at least about 5 fold, preferably 10 fold, more preferably 25-fold, even more preferably 50-fold, and most preferably 100-fold or more, greater for a target molecule than its affinity for a non-target molecule. In preferred embodiments, Preferred antibodies bind with affinities of at least about 10.sup.7 M.sup.-1, and preferably between about 10.sup.8 M.sup.-1 to about 10.sup.9 M.sup.-1, about 10.sup.9 M.sup.-1 to about 10.sup.10M.sup.-1, or about 10.sup.10 M.sup.-1 to about 10.sup.12 M.sup.-1 .
[0130] Affinity is calculated as K.sub.d=k.sub.off/K.sub.on (k.sub.off is the dissociation rate constant, K.sub.on is the association rate constant and K.sub.d is the equilibrium constant). Affinity can be determined at equilibrium by measuring the fraction bound (r) of labeled ligand at various concentrations (c). The data are graphed using the Scatchard equation: r/c=K(n-r): where r=moles of bound ligand/mole of receptor at equilibrium; c=free ligand concentration at equilibrium; K=equilibrium association constant; and n=number of ligand binding sites per receptor molecule. By graphical analysis, r/c is plotted on the Y-axis versus r on the X-axis, thus producing a Scatchard plot. Antibody affinity measurement by Scatchard analysis is well known in the art. See, e.g., van Erp et al., J. Immunoassay 12: 425-43, 1991; Nelson and Griswold, Comput. Methods Programs Biomed. 27: 65-8, 1988.
[0131] The term "epitope" refers to an antigenic determinant capable of specific binding to an antibody. Epitopes usually consist of chemically active surface groupings of molecules such as amino acids or sugar side chains and usually have specific three dimensional structural characteristics, as well as specific charge characteristics. Conformational and nonconformational epitopes are distinguished in that the binding to the former but not the latter is lost in the presence of denaturing solvents.
[0132] Numerous publications discuss the use of phage display technology to produce and screen libraries of polypeptides for binding to a selected analyte. See, e.g, Cwirla et al., Proc. Natl. Acad. Sci. USA 87, 6378-82, 1990; Devlin et al., Science 249, 404-6, 1990, Scott and Smith, Science 249, 386-88, 1990; and Ladner et al., U.S. Pat. No. 5,571,698. A basic concept of phage display methods is the establishment of a physical association between DNA encoding a polypeptide to be screened and the polypeptide. This physical association is provided by the phage particle, which displays a polypeptide as part of a capsid enclosing the phage genome which encodes the polypeptide. The establishment of a physical association between polypeptides and their genetic material allows simultaneous mass screening of very large numbers of phage bearing different polypeptides. Phage displaying a polypeptide with affinity to a target bind to the target and these phage are enriched by affinity screening to the target. The identity of polypeptides displayed from these phage can be determined from their respective genomes. Using these methods a polypeptide identified as having a binding affinity for a desired target can then be synthesized in bulk by conventional means. See, e.g., U.S. Pat. No. 6,057,098, which is hereby incorporated in its entirety, including all tables, figures, and claims.
[0133] The antibodies that are generated by these methods may then be selected by first screening for affinity and specificity with the purified polypeptide of interest and, if required, comparing the results to the affinity and specificity of the antibodies with polypeptides that are desired to be excluded from binding. The screening procedure can involve immobilization of the purified polypeptides in separate wells of microtiter plates. The solution containing a potential antibody or groups of antibodies is then placed into the respective microtiter wells and incubated for about 30 min to 2 h. The microtiter wells are then washed and a labeled secondary antibody (for example, an anti-mouse antibody conjugated to alkaline phosphatase if the raised antibodies are mouse antibodies) is added to the wells and incubated for about 30 min and then washed. Substrate is added to the wells and a color reaction will appear where antibody to the immobilized polypeptide(s) are present.
[0134] The antibodies so identified may then he further analyzed for affinity and specificity in the assay design selected. In the development of immunoassays for a target protein, the purified target protein acts as a standard with which to judge the sensitivity and specificity of the immunoassay using the antibodies that have been selected. Because the binding affinity of various antibodies may differ; certain antibody pairs (e.g., in sandwich assays) may interfere with one another sterically, etc., assay performance of an antibody may be a more important measure than absolute affinity and specificity of an antibody.
[0135] While the present application describes antibody-based binding assays in detail, alternatives to antibodies as binding species in assays are well known in the art. These include receptors for a particular target, aptamers, etc. Aptamers are oligonucleic acid or peptide molecules that bind to a specific target molecule. Aptamers are usually created by selecting them from a large random sequence pool, but natural aptamers also exist. High-affinity aptamers containing modified nucleotides conferring improved characteristics on the ligand, such as improved in vivo stability or improved delivery characteristics. Examples of such modifications include chemical substitutions at the ribose and/or phosphate and/or base positions, and may include amino acid side chain functionalities.
[0136] Assay Correlations
[0137] The term "correlating" as used herein in reference to the use of biomarkers refers to comparing the presence or amount of the biomarker(s) in a patient to its presence or amount in persons known to suffer from, or known to be at risk of, a given condition; or in persons known to be free of a given condition. Often, this takes the form of comparing an assay result in the form of a biomarker concentration to a predetermined threshold selected to be indicative of the occurrence or nonoccurrence of a disease or the likelihood of some future outcome.
[0138] Selecting a diagnostic threshold involves, among other things, consideration of the probability of disease, distribution of true and false diagnoses at different test thresholds, and estimates of the consequences of treatment (or a failure to treat) based on the diagnosis. For example, when considering administering a specific therapy which is highly efficacious and has a low level of risk, few tests are needed because clinicians can accept substantial diagnostic uncertainty. On the other hand, in situations where treatment options are less effective and more risky, clinicians often need a higher degree of diagnostic certainty. Thus, cost/benefit analysis is involved in selecting a diagnostic threshold.
[0139] Suitable thresholds may be determined in a variety of ways. For example, one recommended diagnostic threshold for the diagnosis of acute myocardial infarction using cardiac troponin is the 97.5th percentile of the concentration seen in a normal population. Another method may be to look at serial samples from the same patient, where a prior "baseline" result is used to monitor for temporal changes in a biomarker level.
[0140] Population studies may also be used to select a decision threshold. Reciever Operating Characteristic ("ROC") arose from the field of signal dectection therory developed during World War II for the analysis of radar images, and ROC analysis is often used to select a threshold able to best distinguish a "diseased" subpopulation from a "nondiseased" subpopulation. A false positive in this case occurs when the person tests positive, but actually does not have the disease. A false negative, on the other hand, occurs when the person tests negative, suggesting they are healthy, when they actually do have the disease. To draw a ROC curve, the true positive rate (TPR) and false positive rate (FPR) are determined as the decision threshold is varied continuously. Since TPR is equivalent with sensitivity and FPR is equal to 1--specificity, the ROC graph is sometimes called the sensitivity vs (1--specificity) plot. A perfect test will have an area under the ROC curve of 1.0; a random test will have an area of 0.5. A threshold is selected to provide an acceptable level of specificity and sensitivity.
[0141] In this context, "diseased" is meant to refer to a population having one characteristic (the presence of a disease or condition or the occurrence of some outcome) and "nondiseased" is meant to refer to a population lacking the characteristic. While a single decision threshold is the simplest application of such a method, multiple decision thresholds may be used. For example, below a first threshold, the absence of disease may be assigned with relatively high confidence, and above a second threshold the presence of disease may also be assigned with relatively high confidence. Between the two thresholds may be considered indeterminate. This is meant to be exemplary in nature only.
[0142] In addition to threshold comparisons, other methods for correlating assay results to a patient classification (occurrence or nonoccurrence of disease, likelihood of an outcome, etc.) include decision trees, rule sets, Bayesian methods, and neural network methods. These methods can produce probability values representing the degree to which a sepsis patient belongs to one classification out of a plurality of classifications.
[0143] Measures of test accuracy may be obtained as described in Fischer et al., Intensive Care Med. 29: 1043-51 2003, and used to determine the effectiveness of a given biomarker. These measures include sensitivity and specificity, predictive values, likelihood ratios, diagnostic odds ratios, and ROC curve areas. The area under the curve ("AUC") of a ROC plot is equal to the probability that a classifier will rank a randomly chosen positive instance higher than a randomly chosen negative one. The area under the ROC curve may be thought of as equivalent to the Mann-Whitney U test, which tests for the median difference between scores obtained in the two groups considered if the groups are of continuous data, or to the Wilcoxon test of ranks.
[0144] As discussed above, suitable tests may exhibit one or more of the following results on these various measures: a specificity of greater than 0.5, preferably at least 0.6, more preferably at least 0.7, still more preferably at least 0.8, even more preferably at least 0.9 and most preferably at least 0.95, with a corresponding sensitivity greater than 0.2, preferably greater than 0.3, more preferably greater than 0.4, still more preferably at least 0.5, even more preferably 0.6, yet more preferably greater than 0.7, still more preferably greater than 0.8, more preferably greater than 0.9, and most preferably greater than 0.95; a sensitivity of greater than 0.5, preferably at least 0.6, more preferably a least 0.7, still more preferably at least 0.8, even more preferably at least 0.9 and most preferably at least 0.95, with a corresponding specificity greater than 0.2, preferably greater than 0.3, more preferably greater than 0.4, still more preferably at least 0.5, even more preferably 0.6, yet more preferably greater than 0.7, still more preferably greater than 0.8, more preferably greater than 0.9, and most preferably greater than 0.95; at least 75% sensitivity, combined with at least 75% specificity; a ROC curve area of greater than 0.5, preferably at least 0.6, more preferably 0.7, still more preferably at least 0.8, even more preferably at least 0.9, and most preferably at least 0.95; an odds ratio different from 1, preferably at least about 2 or more or about 0.5 or less, more preferably at least about 3 or more or about 0.33 or less, still more preferably at least about 4 or more or about 0.25 or less, even more preferably at least about 5 or more or about 0.2 or less, and most preferably at least about 10 or more or about 0.1 or less; a positive likelihood ratio (calculated as sensitivity/(1-specificity)) of greater than 1, at least 2, more preferably at least 3, still more preferably at least 5, and most preferably at least 10; and or a negative likelihood ratio (calculated as (1-sensitivity)/specificity) of less than 1, less than or equal to 0.5, more preferably less than or equal to 0.3, and most preferably less than or equal to 0.1
[0145] Additional clinical indicia may be combined with the kidney injury marker assay result(s) of the present invention. These include other biomarkers related to renal status. Other clinical indicia which may be combined with the kidney injury marker assay result(s) of the present invention includes demographic information (e.g., weight, sex, age, race), medical history (e.g., family history, type of surgery, pre-existing disease such as aneurism, congestive heart failure, preeclampsia, eclampsia, diabetes mellitus, hypertension, coronary artery disease, proteinuria, renal insufficiency, or sepsis, type of toxin exposure such as NSAIDs, cyclosporines, tacrolimus, aminoglycosides, foscarnet, ethylene glycol, hemoglobin, myoglobin, ifosfamide, heavy metals, methotrexate, radiopaque contrast agents, or streptozotocin), clinical variables (e.g., blood pressure, temperature, respiration rate), risk scores (APACHE score, PREDICT score, TIMI Risk Score for UA/NSTEMI, Framingham Risk Score), a urine total protein measurement, a glomerular filtration rate, an estimated glomerular filtration rate, a urine production rate, a serum or plasma creatinine concentration, a renal papillary antigen 1 (RPA1) measurement; a renal papillary antigen 2 (RPA2) measurement; a urine creatinine concentration, a fractional excretion of sodium, a urine sodium concentration, a urine creatinine to serum or plasma creatinine ratio, a urine specific gravity, a urine osmolality, a urine urea nitrogen to plasma urea nitrogen ratio, a plasma BUN to creatnine ratio, and/or a renal failure index calculated as urine sodium/(urine creatinine/plasma creatinine). Other measures of renal function which may be combined with the kidney injury marker assay result(s) are described hereinafter and in Harrison's Principles of Internal Medicine, 17.sup.th Ed., McGraw Hill, New York, pages 1741-1830, and Current Medical Diagnosis & Treatment 2008, 47.sup.th Ed, McGraw Hill, New York, pages 785-815, each of which arc hereby incorporated by reference in their entirety.
[0146] Combining assay results/clinical indicia in this manner can comprise the use of multivariate logistical regression, loglinear modeling, neural network analysis, n-of-m analysis, decision tree analysis, etc. This list is not meant to be limiting.
[0147] Diagnosis of Acute Renal Failure
[0148] As noted above, the terms "acute renal (or kidney) injury" and "acute renal (or kidney) failure" as used herein are defined in part in terms of changes in serum creatinine from a baseline value. Most definitions of ARF have common elements, including the use of serum creatinine and, often, urine output. Patients may present with renal dysfunction without an available baseline measure of renal function for use in this comparison. In such an event, one may estimate a baseline serum creatinine value by assuming the patient initially had a normal GFR. Glomerular filtration rate (GFR) is the volume of fluid filtered from the renal (kidney) glomerular capillaries into the Bowman's capsule per unit time. Glomerular filtration rate (GFR) can be calculated by measuring any chemical that has a steady level in the blood, and is freely filtered but neither reabsorbed nor secreted by the kidneys. GFR is typically expressed in units of ml/min:
G F R = Urine Concentration .times. Urine Flow Plasma Concentration ##EQU00001##
[0149] By normalizing the GFR to the body surface area, a GFR of approximately 75-100 ml/min per 1.73 m.sup.2 can be assumed. The rate therefore measured is the quantity of the substance in the urine that originated from a calculable volume of blood.
[0150] There are several different techniques used to calculate or estimate the glomerular filtration rate (GFR or eGFR). In clinical practice, however, creatinine clearance is used to measure GFR. Creatinine is produced naturally by the body (creatinine is a metabolite of creatine, which is found in muscle). It is freely filtered by the glomerulus, but also actively secreted by the renal tubules in very small amounts such that creatinine clearance overestimates actual GFR by 10-20%. This margin of error is acceptable considering the ease with which creatinine clearance is measured.
[0151] Creatinine clearance (CCr) can be calculated if values for creatinine's urine concentration (U.sub.Cr), urine flow rate (V), and creatinine's plasma concentration (P.sub.Cr) are known. Since the product of urine concentration and urine flow rate yields creatinine's excretion rate, creatinine clearance is also said to be its excretion rate (U.sub.Cr.times.V) divided by its plasma concentration. This is commonly represented mathematically as:
C Cr = U Cr .times. V P Cr ##EQU00002##
[0152] Commonly a 24 hour urine collection is undertaken, from empty-bladder one morning to the contents of the bladder the following morning, with a comparative blood test then taken:
C Cr = U Cr .times. 24 - hour volume P Cr .times. 24 .times. 60 mins ##EQU00003##
[0153] To allow comparison of results between people of different sizes, the CCr is often corrected for the body surface area (BSA) and expressed compared to the average sized man as ml/min/1.73 m2. While most adults have a BSA that approaches 1.7 (1.6-1.9), extremely obese or slim patients should have their CCr corrected for their actual BSA:
C Cr - corrected = C Cr .times. 1.73 B S A ##EQU00004##
[0154] The accuracy of a creatinine clearance measurement (even when collection is complete) is limited because as glomerular filtration rate (GFR) falls creatinine secretion is increased, and thus the rise in serum creatinine is less. Thus, creatinine excretion is much greater than the filtered load, resulting in a potentially large overestimation of the GFR (as much as a twofold difference). However, for clinical purposes it is important to determine whether renal function is stable or getting worse or better. This is often determined by monitoring serum creatinine alone. Like creatinine clearance, the serum creatinine will not be an accurate reflection of GFR in the non-steady-state condition of ARF. Nonetheless, the degree to which serum creatinine changes from baseline will reflect the change in GFR. Serum creatinine is readily and easily measured and it is specific for renal function.
[0155] For purposes of determining urine output on a Urine output on a mL/kg/hr basis, hourly urine collection and measurement is adequate. In the case where, for example, only a cumulative 24-h output was available and no patient weights are provided, minor modifications of the RIFLE urine output criteria have been described. For example, Bagshaw et al., Nephrol. Dial. Transplant. 23: 1203-1210, 2008, assumes an average patient weight of 70 kg, and patients are assigned a RIFLE classification based on the following: <35 mL/h (Risk), <21 mL/h (Injury) or <4 mL/h (Failure).
[0156] Selecting a Treatment Regimen
[0157] Once a diagnosis is obtained, the clinician can readily select a treatment regimen that is compatible with the diagnosis, such as initiating renal replacement therapy, withdrawing delivery of compounds that are known to be damaging to the kidney, kidney transplantation, delaying or avoiding procedures that are known to be damaging to the kidney, modifying diuretic administration, initiating goal directed therapy, etc. The skilled artisan is aware of appropriate treatments for numerous diseases discussed in relation to the methods of diagnosis described herein. See, e.g., Merck Manual of Diagnosis and Therapy, 17th Ed. Merck Research Laboratories, Whitehouse Station, N.J., 1999. In addition, since the methods and compositions described herein provide prognostic information, the markers of the present invention may be used to monitor a course of treatment. For example, improved or worsened prognostic state may indicate that a particular treatment is or is not efficacious.
[0158] One skilled in the art readily appreciates that the present invention is well adapted to carry out the objects and obtain the ends and advantages mentioned, as well as those inherent therein. The examples provided herein are representative of preferred embodiments, are exemplary, and are not intended as limitations on the scope of the invention.
Example 1: Septic Sepsis Patient Sample Collection
[0159] The objective of this study was to collect samples from patients expected to be in the ICU for at least 48 hours were enrolled. To be enrolled in the study, each patient must meet all of the following inclusion criteria and none of the following exclusion criteria:
Inclusion Criteria: males and females 18 years of age or older and which either acquire sepsis or have sepsis on admission.
Exclusion Criteria
[0160] known pregnancy; institutionalized individuals; previous renal transplantation; known acutely worsening renal function prior to enrollment (e.g., any category of RIFLE criteria); received dialysis (either acute or chronic) within 5 days prior to enrollment or in imminent need of dialysis at the time of enrollment; known infection with human immunodeficiency virus (HIV) or a hepatitis virus; meets only the SBP <90 mmHg inclusion criterion set forth above, and does not have shock in the attending physician's or principal investigator's opinion.
[0161] After providing informed consent, an EDTA anti-coagulated blood sample (10 mL) and a urine sample (25-30 mL) are collected from each patient. Blood and urine samples are then collected at 4 (.+-.0.5) and 8 (.+-.1) hours after contrast administration (if applicable); at 12 (.+-.1), 24 (.+-.2), and 48 (.+-.2) hours after enrollment, and thereafter daily up to day 7 to day 14 while the sepsis patient is hospitalized. Blood is collected via direct venipuncture or via other available venous access, such as an existing femoral sheath, central venous line, peripheral intravenous line or hep-lock. These study blood samples are processed to plasma at the clinical site, frozen and shipped to Astute Medical, Inc., San Diego, Calif. The study urine samples are frozen and shipped to Astute Medical, Inc.
Example 2. Immunoassay Format
[0162] Analytes are measured using standard sandwich enzyme immunoassay techniques. A first antibody which binds the analyte is immobilized in wells of a 96 well polystyrene microplate. Analyte standards and test samples are pipetted into the appropriate wells and any analyte present is hound by the immobilized antibody. After washing away any unbound substances, a horseradish peroxidase-conjugated second antibody which binds the analyte is added to the wells, thereby forming sandwich complexes with the analyte (if present) and the first antibody. Following a wash to remove any unbound antibody-enzyme reagent, a substrate solution comprising tetramethylbenzidine and hydrogen peroxide is added to the wells. Color develops in proportion to the amount of analyte present in the sample. The color development is stopped and the intensity of the color is measured at 540 nm or 570 nm. An analyte concentration is assigned to the test sample by comparison to a standard curve determined from the analyte standards.
[0163] Concentrations for the various markers are reported as follows:
Insulin-like growth factor-binding protein 7 ng/ml Beta-2-glycoprotein 1 ng/ml Metalloproteinase inhibitor 2 pg/ml Alpha-1 Antitrypsin ng/ml Neutrophil Elastase ng/ml Serum Amyloid P Component ng/ml C--X--C motif chemokine 6 pg/ml Immunoglobulin A ng/ml Immunoglobulin G, subclass I ng/ml C--C motif chemokine 24 pg/ml Neutrophil collagenase pg/ml Cathepsin D pg/ml C--X--C motif chemokine 13 pg/ml Involucrin ng/ml Interleukin-6 receptor subunit beta pg/ml Hepatocyte Growth Factor pg/ml CXCL-1, -2, -3 mix pg/ml Immunoglobulin G, subclass II ng/ml Metalloproteinase inhibitor 4 pg/ml C--C motif chemokine 18 ng/ml Matrilysin pg/ml C--X--C motif chemokine 11 pg/ml Antileukoproteinase (WAP4) pg/ml
Example 3. Use of Kidney Injury Markers for Evaluating Sepsis Patients
[0164] Patients from the sepsis study (Example 1) were classified by kidney status as non-injury (0), risk of injury (R), injury (I), and failure (F) according to the maximum stage reached within 7 days of enrollment as determined by the RIFLE criteria. EDTA anti-coagulated blood samples (10 mL) and a urine samples (25-30 mL) were collected from each patient at enrollment, 4 (.+-.0.5) and 8 (.+-.1) hours after contrast administration (if applicable); at 12 (.+-.1), 24 (.+-.2), and 48 (.+-.2) hours after enrollment, and thereafter daily up to day 7 to day 14 while the sepsis patient is hospitalized. Markers were each measured by standard immunoassay methods using commercially available assay reagents in the urine samples and the plasma component of the blood samples collected.
[0165] Two cohorts were defined to represent a "diseased" and a "normal" population. While these terms are used for convenience, "diseased" and "normal" simply represent two cohorts for comparison (say RIFLE 0 vs RIFLE R, I and F; RIFLE 0 vs RIFLE R; RIFLE 0 and R vs RIFLE I and F; etc.). The time "prior max stage" represents the time at which a sample is collected, relative to the time a particular patient reaches the lowest disease stage as defined for that cohort, binned into three groups which are +/-12 hours. For example, "24 hr prior" which uses 0 vs R, I, F as the two cohorts would mean 24 hr (+/-12 hours) prior to reaching stage R (or I if no sample at R, or F if no sample at R or I).
[0166] A receiver operating characteristic (ROC) curve was generated for each biomarker measured and the area under each ROC curve (AUC) is determined. Patients in Cohort 2 were also separated according to the reason for adjudication to cohort 2 as being based on serum creatinine measurements (sCr), being based on urine output (UO), or being based on either serum creatinine measurements or urine output. Using the same example discussed above (0 vs R, I, F), for those patients adjudicated to stage R, I, or F on the basis of serum creatinine measurements alone, the stage 0 cohort may include patients adjudicated to stage R, I, or F on the basis of urine output; for those patients adjudicated to stage R, I, or F on the basis of urine output alone, the stage 0 cohort may include patients adjudicated to stage R, I, or F on the basis of serum creatinine measurements; and for those patients adjudicated to stage R, I, or F on the basis of serum creatinine measurements or urine output, the stage 0 cohort contains only patients in stage 0 for both serum creatinine measurements and urine output. Also, in the data for patients adjudicated on the basis of serum creatinine measurements or urine output, the adjudication method which yielded the most severe RIFLE stage is used.
[0167] The ability to distinguish cohort 1 from Cohort 2 was determined using ROC analysis. SE is the standard error of the AUC, n is the number of sample or individual patients ("pts," as indicated). Standard errors are calculated as described in Hanley, J. A., and McNeil, B. J., The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology (1982) 143: 29-36; p values are calculated with a two-tailed Z-test. An AUC <0.5 is indicative of a negative going marker for the comparison, and an AUC >0.5 is indicative of a positive going marker for the comparison.
[0168] Various threshold (or "cutoff") concentrations were selected, and the associated sensitivity and specificity for distinguishing cohort 1 from cohort 2 are determined. OR is the odds ratio calculated for the particular cutoff concentration, and 95% CI is the confidence interval for the odds ratio.
[0169] In the following tables 1-12, a population which either acquire sepsis days 1-7 or have sepsis on admission are used as the disease cohort; in tables 13-24, only those patients with sepsis on admission were included.
[0170] Table 1: Comparison of marker levels in urine samples collected from Cohort 1 (patients that did not progress beyond RIFLE stage 0) and in urine samples collected from subjects at 0, 24 hours, and 48 hours prior to reaching stage R, I or F in Cohort 2.
TABLE-US-00046 Lengthy table referenced here US20210156850A1-20210527-T00001 Please refer to the end of the specification for access instructions.
[0171] While the invention has been described and exemplified in sufficient detail for those skilled in this art to make and use it, various alternatives, modifications, and improvements should be apparent without departing from the spirit and scope of the invention. The examples provided herein are representative of preferred embodiments, are exemplary, and are not intended as limitations on the scope of the invention. Modifications therein and other uses will occur to those skilled in the art. These modifications are encompassed within the spirit of the invention and are defined by the scope of the claims.
[0172] It will be readily apparent to a person skilled in the art that varying substitutions and modifications may be made to the invention disclosed herein without departing from the scope and spirit of the invention.
[0173] All patents and publications mentioned in the specification are indicative of the levels of those of ordinary skill in the art to which the invention pertains. All patents and publications are herein incorporated by reference to the same extent as if each individual publication was specifically and individually indicated to be incorporated by reference.
[0174] The invention illustratively described herein suitably may be practiced in the absence of any element or elements, limitation or limitations which is not specifically disclosed herein. Thus, for example, in each instance herein any of the terms "comprising", "consisting essentially of" and "consisting of" may be replaced with either of the other two terms. The terms and expressions which have been employed are used as terms of description and not of limitation, and there is no intention that in the use of such terms and expressions of excluding any equivalents of the features shown and described or portions thereof, but it is recognized that various modifications are possible within the scope of the invention claimed. Thus, it should be understood that although the present invention has been specifically disclosed by preferred embodiments and optional features, modification and variation of the concepts herein disclosed may be resorted to by those skilled in the art, and that such modifications and variations are considered to be within the scope of this invention as defined by the appended claims.
[0175] Other embodiments are set forth within the following claims.
TABLE-US-LTS-00001 LENGTHY TABLES The patent application contains a lengthy table section. A copy of the table is available in electronic form from the USPTO web site (https://seqdata.uspto.gov/?pageRequest=docDetail&DocID=US20210156850A1). An electronic copy of the table will also be available from the USPTO upon request and payment of the fee set forth in 37 CFR 1.19(b)(3).
Sequence CWU 1
1
241282PRTHomo sapiens 1Met Glu Arg Pro Ser Leu Arg Ala Leu Leu Leu
Gly Ala Ala Gly Leu1 5 10 15Leu Leu Leu Leu Leu Pro Leu Ser Ser Ser
Ser Ser Ser Asp Thr Cys 20 25 30Gly Pro Cys Glu Pro Ala Ser Cys Pro
Pro Leu Pro Pro Leu Gly Cys 35 40 45Leu Leu Gly Glu Thr Arg Asp Ala
Cys Gly Cys Cys Pro Met Cys Ala 50 55 60Arg Gly Glu Gly Glu Pro Cys
Gly Gly Gly Gly Ala Gly Arg Gly Tyr65 70 75 80Cys Ala Pro Gly Met
Glu Cys Val Lys Ser Arg Lys Arg Arg Lys Gly 85 90 95Lys Ala Gly Ala
Ala Ala Gly Gly Pro Gly Val Ser Gly Val Cys Val 100 105 110Cys Lys
Ser Arg Tyr Pro Val Cys Gly Ser Asp Gly Thr Thr Tyr Pro 115 120
125Ser Gly Cys Gln Leu Arg Ala Ala Ser Gln Arg Ala Glu Ser Arg Gly
130 135 140Glu Lys Ala Ile Thr Gln Val Ser Lys Gly Thr Cys Glu Gln
Gly Pro145 150 155 160Ser Ile Val Thr Pro Pro Lys Asp Ile Trp Asn
Val Thr Gly Ala Gln 165 170 175Val Tyr Leu Ser Cys Glu Val Ile Gly
Ile Pro Thr Pro Val Leu Ile 180 185 190Trp Asn Lys Val Lys Arg Gly
His Tyr Gly Val Gln Arg Thr Glu Leu 195 200 205Leu Pro Gly Asp Arg
Asp Asn Leu Ala Ile Gln Thr Arg Gly Gly Pro 210 215 220Glu Lys His
Glu Val Thr Gly Trp Val Leu Val Ser Pro Leu Ser Lys225 230 235
240Glu Asp Ala Gly Glu Tyr Glu Cys His Ala Ser Asn Ser Gln Gly Gln
245 250 255Ala Ser Ala Ser Ala Lys Ile Thr Val Val Asp Ala Leu His
Glu Ile 260 265 270Pro Val Lys Lys Gly Glu Gly Ala Glu Leu 275
2802345PRTHomo sapiens 2Met Ile Ser Pro Val Leu Ile Leu Phe Ser Ser
Phe Leu Cys His Val1 5 10 15Ala Ile Ala Gly Arg Thr Cys Pro Lys Pro
Asp Asp Leu Pro Phe Ser 20 25 30Thr Val Val Pro Leu Lys Thr Phe Tyr
Glu Pro Gly Glu Glu Ile Thr 35 40 45Tyr Ser Cys Lys Pro Gly Tyr Val
Ser Arg Gly Gly Met Arg Lys Phe 50 55 60Ile Cys Pro Leu Thr Gly Leu
Trp Pro Ile Asn Thr Leu Lys Cys Thr65 70 75 80Pro Arg Val Cys Pro
Phe Ala Gly Ile Leu Glu Asn Gly Ala Val Arg 85 90 95Tyr Thr Thr Phe
Glu Tyr Pro Asn Thr Ile Ser Phe Ser Cys Asn Thr 100 105 110Gly Phe
Tyr Leu Asn Gly Ala Asp Ser Ala Lys Cys Thr Glu Glu Gly 115 120
125Lys Trp Ser Pro Glu Leu Pro Val Cys Ala Pro Ile Ile Cys Pro Pro
130 135 140Pro Ser Ile Pro Thr Phe Ala Thr Leu Arg Val Tyr Lys Pro
Ser Ala145 150 155 160Gly Asn Asn Ser Leu Tyr Arg Asp Thr Ala Val
Phe Glu Cys Leu Pro 165 170 175Gln His Ala Met Phe Gly Asn Asp Thr
Ile Thr Cys Thr Thr His Gly 180 185 190Asn Trp Thr Lys Leu Pro Glu
Cys Arg Glu Val Lys Cys Pro Phe Pro 195 200 205Ser Arg Pro Asp Asn
Gly Phe Val Asn Tyr Pro Ala Lys Pro Thr Leu 210 215 220Tyr Tyr Lys
Asp Lys Ala Thr Phe Gly Cys His Asp Gly Tyr Ser Leu225 230 235
240Asp Gly Pro Glu Glu Ile Glu Cys Thr Lys Leu Gly Asn Trp Ser Ala
245 250 255Met Pro Ser Cys Lys Ala Ser Cys Lys Val Pro Val Lys Lys
Ala Thr 260 265 270Val Val Tyr Gln Gly Glu Arg Val Lys Ile Gln Glu
Lys Phe Lys Asn 275 280 285Gly Met Leu His Gly Asp Lys Val Ser Phe
Phe Cys Lys Asn Lys Glu 290 295 300Lys Lys Cys Ser Tyr Thr Glu Asp
Ala Gln Cys Ile Asp Gly Thr Ile305 310 315 320Glu Val Pro Lys Cys
Phe Lys Glu His Ser Ser Leu Ala Phe Trp Lys 325 330 335Thr Asp Ala
Ser Asp Val Lys Pro Cys 340 3453220PRTHomo sapiens 3Met Gly Ala Ala
Ala Arg Thr Leu Arg Leu Ala Leu Gly Leu Leu Leu1 5 10 15Leu Ala Thr
Leu Leu Arg Pro Ala Asp Ala Cys Ser Cys Ser Pro Val 20 25 30His Pro
Gln Gln Ala Phe Cys Asn Ala Asp Val Val Ile Arg Ala Lys 35 40 45Ala
Val Ser Glu Lys Glu Val Asp Ser Gly Asn Asp Ile Tyr Gly Asn 50 55
60Pro Ile Lys Arg Ile Gln Tyr Glu Ile Lys Gln Ile Lys Met Phe Lys65
70 75 80Gly Pro Glu Lys Asp Ile Glu Phe Ile Tyr Thr Ala Pro Ser Ser
Ala 85 90 95Val Cys Gly Val Ser Leu Asp Val Gly Gly Lys Lys Glu Tyr
Leu Ile 100 105 110Ala Gly Lys Ala Glu Gly Asp Gly Lys Met His Ile
Thr Leu Cys Asp 115 120 125Phe Ile Val Pro Trp Asp Thr Leu Ser Thr
Thr Gln Lys Lys Ser Leu 130 135 140Asn His Arg Tyr Gln Met Gly Cys
Glu Cys Lys Ile Thr Arg Cys Pro145 150 155 160Met Ile Pro Cys Tyr
Ile Ser Ser Pro Asp Glu Cys Leu Trp Met Asp 165 170 175Trp Val Thr
Glu Lys Asn Ile Asn Gly His Gln Ala Lys Phe Phe Ala 180 185 190Cys
Ile Lys Arg Ser Asp Gly Ser Cys Ala Trp Tyr Arg Gly Ala Ala 195 200
205Pro Pro Lys Gln Glu Phe Leu Asp Ile Glu Asp Pro 210 215
2204418PRTHomo sapiens 4Met Pro Ser Ser Val Ser Trp Gly Ile Leu Leu
Leu Ala Gly Leu Cys1 5 10 15Cys Leu Val Pro Val Ser Leu Ala Glu Asp
Pro Gln Gly Asp Ala Ala 20 25 30Gln Lys Thr Asp Thr Ser His His Asp
Gln Asp His Pro Thr Phe Asn 35 40 45Lys Ile Thr Pro Asn Leu Ala Glu
Phe Ala Phe Ser Leu Tyr Arg Gln 50 55 60Leu Ala His Gln Ser Asn Ser
Thr Asn Ile Phe Phe Ser Pro Val Ser65 70 75 80Ile Ala Thr Ala Phe
Ala Met Leu Ser Leu Gly Thr Lys Ala Asp Thr 85 90 95His Asp Glu Ile
Leu Glu Gly Leu Asn Phe Asn Leu Thr Glu Ile Pro 100 105 110Glu Ala
Gln Ile His Glu Gly Phe Gln Glu Leu Leu Arg Thr Leu Asn 115 120
125Gln Pro Asp Ser Gln Leu Gln Leu Thr Thr Gly Asn Gly Leu Phe Leu
130 135 140Ser Glu Gly Leu Lys Leu Val Asp Lys Phe Leu Glu Asp Val
Lys Lys145 150 155 160Leu Tyr His Ser Glu Ala Phe Thr Val Asn Phe
Gly Asp Thr Glu Glu 165 170 175Ala Lys Lys Gln Ile Asn Asp Tyr Val
Glu Lys Gly Thr Gln Gly Lys 180 185 190Ile Val Asp Leu Val Lys Glu
Leu Asp Arg Asp Thr Val Phe Ala Leu 195 200 205Val Asn Tyr Ile Phe
Phe Lys Gly Lys Trp Glu Arg Pro Phe Glu Val 210 215 220Lys Asp Thr
Glu Glu Glu Asp Phe His Val Asp Gln Val Thr Thr Val225 230 235
240Lys Val Pro Met Met Lys Arg Leu Gly Met Phe Asn Ile Gln His Cys
245 250 255Lys Lys Leu Ser Ser Trp Val Leu Leu Met Lys Tyr Leu Gly
Asn Ala 260 265 270Thr Ala Ile Phe Phe Leu Pro Asp Glu Gly Lys Leu
Gln His Leu Glu 275 280 285Asn Glu Leu Thr His Asp Ile Ile Thr Lys
Phe Leu Glu Asn Glu Asp 290 295 300Arg Arg Ser Ala Ser Leu His Leu
Pro Lys Leu Ser Ile Thr Gly Thr305 310 315 320Tyr Asp Leu Lys Ser
Val Leu Gly Gln Leu Gly Ile Thr Lys Val Phe 325 330 335Ser Asn Gly
Ala Asp Leu Ser Gly Val Thr Glu Glu Ala Pro Leu Lys 340 345 350Leu
Ser Lys Ala Val His Lys Ala Val Leu Thr Ile Asp Glu Lys Gly 355 360
365Thr Glu Ala Ala Gly Ala Met Phe Leu Glu Ala Ile Pro Met Ser Ile
370 375 380Pro Pro Glu Val Lys Phe Asn Lys Pro Phe Val Phe Leu Met
Ile Glu385 390 395 400Gln Asn Thr Lys Ser Pro Leu Phe Met Gly Lys
Val Val Asn Pro Thr 405 410 415Gln Lys5267PRTHomo sapiens 5Met Thr
Leu Gly Arg Arg Leu Ala Cys Leu Phe Leu Ala Cys Val Leu1 5 10 15Pro
Ala Leu Leu Leu Gly Gly Thr Ala Leu Ala Ser Glu Ile Val Gly 20 25
30Gly Arg Arg Ala Arg Pro His Ala Trp Pro Phe Met Val Ser Leu Gln
35 40 45Leu Arg Gly Gly His Phe Cys Gly Ala Thr Leu Ile Ala Pro Asn
Phe 50 55 60Val Met Ser Ala Ala His Cys Val Ala Asn Val Asn Val Arg
Ala Val65 70 75 80Arg Val Val Leu Gly Ala His Asn Leu Ser Arg Arg
Glu Pro Thr Arg 85 90 95Gln Val Phe Ala Val Gln Arg Ile Phe Glu Asn
Gly Tyr Asp Pro Val 100 105 110Asn Leu Leu Asn Asp Ile Val Ile Leu
Gln Leu Asn Gly Ser Ala Thr 115 120 125Ile Asn Ala Asn Val Gln Val
Ala Gln Leu Pro Ala Gln Gly Arg Arg 130 135 140Leu Gly Asn Gly Val
Gln Cys Leu Ala Met Gly Trp Gly Leu Leu Gly145 150 155 160Arg Asn
Arg Gly Ile Ala Ser Val Leu Gln Glu Leu Asn Val Thr Val 165 170
175Val Thr Ser Leu Cys Arg Arg Ser Asn Val Cys Thr Leu Val Arg Gly
180 185 190Arg Gln Ala Gly Val Cys Phe Gly Asp Ser Gly Ser Pro Leu
Val Cys 195 200 205Asn Gly Leu Ile His Gly Ile Ala Ser Phe Val Arg
Gly Gly Cys Ala 210 215 220Ser Gly Leu Tyr Pro Asp Ala Phe Ala Pro
Val Ala Gln Phe Val Asn225 230 235 240Trp Ile Asp Ser Ile Ile Gln
Arg Ser Glu Asp Asn Pro Cys Pro His 245 250 255Pro Arg Asp Pro Asp
Pro Ala Ser Arg Thr His 260 2656223PRTHomo sapiens 6Met Asn Lys Pro
Leu Leu Trp Ile Ser Val Leu Thr Ser Leu Leu Glu1 5 10 15Ala Phe Ala
His Thr Asp Leu Ser Gly Lys Val Phe Val Phe Pro Arg 20 25 30Glu Ser
Val Thr Asp His Val Asn Leu Ile Thr Pro Leu Glu Lys Pro 35 40 45Leu
Gln Asn Phe Thr Leu Cys Phe Arg Ala Tyr Ser Asp Leu Ser Arg 50 55
60Ala Tyr Ser Leu Phe Ser Tyr Asn Thr Gln Gly Arg Asp Asn Glu Leu65
70 75 80Leu Val Tyr Lys Glu Arg Val Gly Glu Tyr Ser Leu Tyr Ile Gly
Arg 85 90 95His Lys Val Thr Ser Lys Val Ile Glu Lys Phe Pro Ala Pro
Val His 100 105 110Ile Cys Val Ser Trp Glu Ser Ser Ser Gly Ile Ala
Glu Phe Trp Ile 115 120 125Asn Gly Thr Pro Leu Val Lys Lys Gly Leu
Arg Gln Gly Tyr Phe Val 130 135 140Glu Ala Gln Pro Lys Ile Val Leu
Gly Gln Glu Gln Asp Ser Tyr Gly145 150 155 160Gly Lys Phe Asp Arg
Ser Gln Ser Phe Val Gly Glu Ile Gly Asp Leu 165 170 175Tyr Met Trp
Asp Ser Val Leu Pro Pro Glu Asn Ile Leu Ser Ala Tyr 180 185 190Gln
Gly Thr Pro Leu Pro Ala Asn Ile Leu Asp Trp Gln Ala Leu Asn 195 200
205Tyr Glu Ile Arg Gly Tyr Val Ile Ile Lys Pro Leu Val Trp Val 210
215 2207114PRTHomo sapiens 7Met Ser Leu Pro Ser Ser Arg Ala Ala Arg
Val Pro Gly Pro Ser Gly1 5 10 15Ser Leu Cys Ala Leu Leu Ala Leu Leu
Leu Leu Leu Thr Pro Pro Gly 20 25 30Pro Leu Ala Ser Ala Gly Pro Val
Ser Ala Val Leu Thr Glu Leu Arg 35 40 45Cys Thr Cys Leu Arg Val Thr
Leu Arg Val Asn Pro Lys Thr Ile Gly 50 55 60Lys Leu Gln Val Phe Pro
Ala Gly Pro Gln Cys Ser Lys Val Glu Val65 70 75 80Val Ala Ser Leu
Lys Asn Gly Lys Gln Val Cys Leu Asp Pro Glu Ala 85 90 95Pro Phe Leu
Lys Lys Val Ile Gln Lys Ile Leu Asp Ser Gly Asn Lys 100 105 110Lys
Asn8119PRTHomo sapiens 8Met Ala Gly Leu Met Thr Ile Val Thr Ser Leu
Leu Phe Leu Gly Val1 5 10 15Cys Ala His His Ile Ile Pro Thr Gly Ser
Val Val Ile Pro Ser Pro 20 25 30Cys Cys Met Phe Phe Val Ser Lys Arg
Ile Pro Glu Asn Arg Val Val 35 40 45Ser Tyr Gln Leu Ser Ser Arg Ser
Thr Cys Leu Lys Ala Gly Val Ile 50 55 60Phe Thr Thr Lys Lys Gly Gln
Gln Phe Cys Gly Asp Pro Lys Gln Glu65 70 75 80Trp Val Gln Arg Tyr
Met Lys Asn Leu Asp Ala Lys Gln Lys Lys Ala 85 90 95Ser Pro Arg Ala
Arg Ala Val Ala Val Lys Gly Pro Val Gln Arg Tyr 100 105 110Pro Gly
Asn Gln Thr Thr Cys 1159467PRTHomo sapiens 9Met Phe Ser Leu Lys Thr
Leu Pro Phe Leu Leu Leu Leu His Val Gln1 5 10 15Ile Ser Lys Ala Phe
Pro Val Ser Ser Lys Glu Lys Asn Thr Lys Thr 20 25 30Val Gln Asp Tyr
Leu Glu Lys Phe Tyr Gln Leu Pro Ser Asn Gln Tyr 35 40 45Gln Ser Thr
Arg Lys Asn Gly Thr Asn Val Ile Val Glu Lys Leu Lys 50 55 60Glu Met
Gln Arg Phe Phe Gly Leu Asn Val Thr Gly Lys Pro Asn Glu65 70 75
80Glu Thr Leu Asp Met Met Lys Lys Pro Arg Cys Gly Val Pro Asp Ser
85 90 95Gly Gly Phe Met Leu Thr Pro Gly Asn Pro Lys Trp Glu Arg Thr
Asn 100 105 110Leu Thr Tyr Arg Ile Arg Asn Tyr Thr Pro Gln Leu Ser
Glu Ala Glu 115 120 125Val Glu Arg Ala Ile Lys Asp Ala Phe Glu Leu
Trp Ser Val Ala Ser 130 135 140Pro Leu Ile Phe Thr Arg Ile Ser Gln
Gly Glu Ala Asp Ile Asn Ile145 150 155 160Ala Phe Tyr Gln Arg Asp
His Gly Asp Asn Ser Pro Phe Asp Gly Pro 165 170 175Asn Gly Ile Leu
Ala His Ala Phe Gln Pro Gly Gln Gly Ile Gly Gly 180 185 190Asp Ala
His Phe Asp Ala Glu Glu Thr Trp Thr Asn Thr Ser Ala Asn 195 200
205Tyr Asn Leu Phe Leu Val Ala Ala His Glu Phe Gly His Ser Leu Gly
210 215 220Leu Ala His Ser Ser Asp Pro Gly Ala Leu Met Tyr Pro Asn
Tyr Ala225 230 235 240Phe Arg Glu Thr Ser Asn Tyr Ser Leu Pro Gln
Asp Asp Ile Asp Gly 245 250 255Ile Gln Ala Ile Tyr Gly Leu Ser Ser
Asn Pro Ile Gln Pro Thr Gly 260 265 270Pro Ser Thr Pro Lys Pro Cys
Asp Pro Ser Leu Thr Phe Asp Ala Ile 275 280 285Thr Thr Leu Arg Gly
Glu Ile Leu Phe Phe Lys Asp Arg Tyr Phe Trp 290 295 300Arg Arg His
Pro Gln Leu Gln Arg Val Glu Met Asn Phe Ile Ser Leu305 310 315
320Phe Trp Pro Ser Leu Pro Thr Gly Ile Gln Ala Ala Tyr Glu Asp Phe
325 330 335Asp Arg Asp Leu Ile Phe Leu Phe Lys Gly Asn Gln Tyr Trp
Ala Leu 340 345 350Ser Gly Tyr Asp Ile Leu Gln Gly Tyr Pro Lys Asp
Ile Ser Asn Tyr 355 360 365Gly Phe Pro Ser Ser Val Gln Ala Ile Asp
Ala Ala Val Phe Tyr Arg 370 375 380Ser Lys Thr Tyr Phe Phe Val Asn
Asp Gln Phe Trp Arg Tyr Asp Asn385 390 395 400Gln Arg Gln Phe Met
Glu Pro Gly Tyr Pro Lys Ser Ile Ser Gly Ala 405 410 415Phe Pro Gly
Ile Glu Ser Lys Val Asp Ala Val Phe Gln Gln Glu His 420 425 430Phe
Phe His Val Phe Ser Gly Pro Arg Tyr Tyr Ala Phe Asp Leu Ile 435 440
445Ala Gln Arg Val Thr Arg Val Ala Arg Gly Asn Lys Trp Leu Asn Cys
450 455 460Arg Tyr Gly46510412PRTHomo sapiens 10Met Gln
Pro Ser Ser Leu Leu Pro Leu Ala Leu Cys Leu Leu Ala Ala1 5 10 15Pro
Ala Ser Ala Leu Val Arg Ile Pro Leu His Lys Phe Thr Ser Ile 20 25
30Arg Arg Thr Met Ser Glu Val Gly Gly Ser Val Glu Asp Leu Ile Ala
35 40 45Lys Gly Pro Val Ser Lys Tyr Ser Gln Ala Val Pro Ala Val Thr
Glu 50 55 60Gly Pro Ile Pro Glu Val Leu Lys Asn Tyr Met Asp Ala Gln
Tyr Tyr65 70 75 80Gly Glu Ile Gly Ile Gly Thr Pro Pro Gln Cys Phe
Thr Val Val Phe 85 90 95Asp Thr Gly Ser Ser Asn Leu Trp Val Pro Ser
Ile His Cys Lys Leu 100 105 110Leu Asp Ile Ala Cys Trp Ile His His
Lys Tyr Asn Ser Asp Lys Ser 115 120 125Ser Thr Tyr Val Lys Asn Gly
Thr Ser Phe Asp Ile His Tyr Gly Ser 130 135 140Gly Ser Leu Ser Gly
Tyr Leu Ser Gln Asp Thr Val Ser Val Pro Cys145 150 155 160Gln Ser
Ala Ser Ser Ala Ser Ala Leu Gly Gly Val Lys Val Glu Arg 165 170
175Gln Val Phe Gly Glu Ala Thr Lys Gln Pro Gly Ile Thr Phe Ile Ala
180 185 190Ala Lys Phe Asp Gly Ile Leu Gly Met Ala Tyr Pro Arg Ile
Ser Val 195 200 205Asn Asn Val Leu Pro Val Phe Asp Asn Leu Met Gln
Gln Lys Leu Val 210 215 220Asp Gln Asn Ile Phe Ser Phe Tyr Leu Ser
Arg Asp Pro Asp Ala Gln225 230 235 240Pro Gly Gly Glu Leu Met Leu
Gly Gly Thr Asp Ser Lys Tyr Tyr Lys 245 250 255Gly Ser Leu Ser Tyr
Leu Asn Val Thr Arg Lys Ala Tyr Trp Gln Val 260 265 270His Leu Asp
Gln Val Glu Val Ala Ser Gly Leu Thr Leu Cys Lys Glu 275 280 285Gly
Cys Glu Ala Ile Val Asp Thr Gly Thr Ser Leu Met Val Gly Pro 290 295
300Val Asp Glu Val Arg Glu Leu Gln Lys Ala Ile Gly Ala Val Pro
Leu305 310 315 320Ile Gln Gly Glu Tyr Met Ile Pro Cys Glu Lys Val
Ser Thr Leu Pro 325 330 335Ala Ile Thr Leu Lys Leu Gly Gly Lys Gly
Tyr Lys Leu Ser Pro Glu 340 345 350Asp Tyr Thr Leu Lys Val Ser Gln
Ala Gly Lys Thr Leu Cys Leu Ser 355 360 365Gly Phe Met Gly Met Asp
Ile Pro Pro Pro Ser Gly Pro Leu Trp Ile 370 375 380Leu Gly Asp Val
Phe Ile Gly Arg Tyr Tyr Thr Val Phe Asp Arg Asp385 390 395 400Asn
Asn Arg Val Gly Phe Ala Glu Ala Ala Arg Leu 405 41011109PRTHomo
sapiens 11Met Lys Phe Ile Ser Thr Ser Leu Leu Leu Met Leu Leu Val
Ser Ser1 5 10 15Leu Ser Pro Val Gln Gly Val Leu Glu Val Tyr Tyr Thr
Ser Leu Arg 20 25 30Cys Arg Cys Val Gln Glu Ser Ser Val Phe Ile Pro
Arg Arg Phe Ile 35 40 45Asp Arg Ile Gln Ile Leu Pro Arg Gly Asn Gly
Cys Pro Arg Lys Glu 50 55 60Ile Ile Val Trp Lys Lys Asn Lys Ser Ile
Val Cys Val Asp Pro Gln65 70 75 80Ala Glu Trp Ile Gln Arg Met Met
Glu Val Leu Arg Lys Arg Ser Ser 85 90 95Ser Thr Leu Pro Val Pro Val
Phe Lys Arg Lys Ile Pro 100 10512585PRTHomo sapiens 12Met Ser Gln
Gln His Thr Leu Pro Val Thr Leu Ser Pro Ala Leu Ser1 5 10 15Gln Glu
Leu Leu Lys Thr Val Pro Pro Pro Val Asn Thr His Gln Glu 20 25 30Gln
Met Lys Gln Pro Thr Pro Leu Pro Pro Pro Cys Gln Lys Val Pro 35 40
45Val Glu Leu Pro Val Glu Val Pro Ser Lys Gln Glu Glu Lys His Met
50 55 60Thr Ala Val Lys Gly Leu Pro Glu Gln Glu Cys Glu Gln Gln Gln
Lys65 70 75 80Glu Pro Gln Glu Gln Glu Leu Gln Gln Gln His Trp Glu
Gln His Glu 85 90 95Glu Tyr Gln Lys Ala Glu Asn Pro Glu Gln Gln Leu
Lys Gln Glu Lys 100 105 110Thr Gln Arg Asp Gln Gln Leu Asn Lys Gln
Leu Glu Glu Glu Lys Lys 115 120 125Leu Leu Asp Gln Gln Leu Asp Gln
Glu Leu Val Lys Arg Asp Glu Gln 130 135 140Leu Gly Met Lys Lys Glu
Gln Leu Leu Glu Leu Pro Glu Gln Gln Glu145 150 155 160Gly His Leu
Lys His Leu Glu Gln Gln Glu Gly Gln Leu Lys His Pro 165 170 175Glu
Gln Gln Glu Gly Gln Leu Glu Leu Pro Glu Gln Gln Glu Gly Gln 180 185
190Leu Glu Leu Pro Glu Gln Gln Glu Gly Gln Leu Glu Leu Pro Glu Gln
195 200 205Gln Glu Gly Gln Leu Glu Leu Pro Glu Gln Gln Glu Gly Gln
Leu Glu 210 215 220Leu Pro Glu Gln Gln Glu Gly Gln Leu Glu Leu Pro
Gln Gln Gln Glu225 230 235 240Gly Gln Leu Glu Leu Ser Glu Gln Gln
Glu Gly Gln Leu Glu Leu Ser 245 250 255Glu Gln Gln Glu Gly Gln Leu
Lys His Leu Glu His Gln Glu Gly Gln 260 265 270Leu Glu Val Pro Glu
Glu Gln Met Gly Gln Leu Lys Tyr Leu Glu Gln 275 280 285Gln Glu Gly
Gln Leu Lys His Leu Asp Gln Gln Glu Lys Gln Pro Glu 290 295 300Leu
Pro Glu Gln Gln Met Gly Gln Leu Lys His Leu Glu Gln Gln Glu305 310
315 320Gly Gln Pro Lys His Leu Glu Gln Gln Glu Gly Gln Leu Glu Gln
Leu 325 330 335Glu Glu Gln Glu Gly Gln Leu Lys His Leu Glu Gln Gln
Glu Gly Gln 340 345 350Leu Glu His Leu Glu His Gln Glu Gly Gln Leu
Gly Leu Pro Glu Gln 355 360 365Gln Val Leu Gln Leu Lys Gln Leu Glu
Lys Gln Gln Gly Gln Pro Lys 370 375 380His Leu Glu Glu Glu Glu Gly
Gln Leu Lys His Leu Val Gln Gln Glu385 390 395 400Gly Gln Leu Lys
His Leu Val Gln Gln Glu Gly Gln Leu Glu Gln Gln 405 410 415Glu Arg
Gln Val Glu His Leu Glu Gln Gln Val Gly Gln Leu Lys His 420 425
430Leu Glu Glu Gln Glu Gly Gln Leu Lys His Leu Glu Gln Gln Gln Gly
435 440 445Gln Leu Glu Val Pro Glu Gln Gln Val Gly Gln Pro Lys Asn
Leu Glu 450 455 460Gln Glu Glu Lys Gln Leu Glu Leu Pro Glu Gln Gln
Glu Gly Gln Val465 470 475 480Lys His Leu Glu Lys Gln Glu Ala Gln
Leu Glu Leu Pro Glu Gln Gln 485 490 495Val Gly Gln Pro Lys His Leu
Glu Gln Gln Glu Lys His Leu Glu His 500 505 510Pro Glu Gln Gln Asp
Gly Gln Leu Lys His Leu Glu Gln Gln Glu Gly 515 520 525Gln Leu Lys
Asp Leu Glu Gln Gln Lys Gly Gln Leu Glu Gln Pro Val 530 535 540Phe
Ala Pro Ala Pro Gly Gln Val Gln Asp Ile Gln Pro Ala Leu Pro545 550
555 560Thr Lys Gly Glu Val Leu Leu Pro Val Glu His Gln Gln Gln Lys
Gln 565 570 575Glu Val Gln Trp Pro Pro Lys His Lys 580
58513918PRTHomo sapiens 13Met Leu Thr Leu Gln Thr Trp Leu Val Gln
Ala Leu Phe Ile Phe Leu1 5 10 15Thr Thr Glu Ser Thr Gly Glu Leu Leu
Asp Pro Cys Gly Tyr Ile Ser 20 25 30Pro Glu Ser Pro Val Val Gln Leu
His Ser Asn Phe Thr Ala Val Cys 35 40 45Val Leu Lys Glu Lys Cys Met
Asp Tyr Phe His Val Asn Ala Asn Tyr 50 55 60Ile Val Trp Lys Thr Asn
His Phe Thr Ile Pro Lys Glu Gln Tyr Thr65 70 75 80Ile Ile Asn Arg
Thr Ala Ser Ser Val Thr Phe Thr Asp Ile Ala Ser 85 90 95Leu Asn Ile
Gln Leu Thr Cys Asn Ile Leu Thr Phe Gly Gln Leu Glu 100 105 110Gln
Asn Val Tyr Gly Ile Thr Ile Ile Ser Gly Leu Pro Pro Glu Lys 115 120
125Pro Lys Asn Leu Ser Cys Ile Val Asn Glu Gly Lys Lys Met Arg Cys
130 135 140Glu Trp Asp Gly Gly Arg Glu Thr His Leu Glu Thr Asn Phe
Thr Leu145 150 155 160Lys Ser Glu Trp Ala Thr His Lys Phe Ala Asp
Cys Lys Ala Lys Arg 165 170 175Asp Thr Pro Thr Ser Cys Thr Val Asp
Tyr Ser Thr Val Tyr Phe Val 180 185 190Asn Ile Glu Val Trp Val Glu
Ala Glu Asn Ala Leu Gly Lys Val Thr 195 200 205Ser Asp His Ile Asn
Phe Asp Pro Val Tyr Lys Val Lys Pro Asn Pro 210 215 220Pro His Asn
Leu Ser Val Ile Asn Ser Glu Glu Leu Ser Ser Ile Leu225 230 235
240Lys Leu Thr Trp Thr Asn Pro Ser Ile Lys Ser Val Ile Ile Leu Lys
245 250 255Tyr Asn Ile Gln Tyr Arg Thr Lys Asp Ala Ser Thr Trp Ser
Gln Ile 260 265 270Pro Pro Glu Asp Thr Ala Ser Thr Arg Ser Ser Phe
Thr Val Gln Asp 275 280 285Leu Lys Pro Phe Thr Glu Tyr Val Phe Arg
Ile Arg Cys Met Lys Glu 290 295 300Asp Gly Lys Gly Tyr Trp Ser Asp
Trp Ser Glu Glu Ala Ser Gly Ile305 310 315 320Thr Tyr Glu Asp Arg
Pro Ser Lys Ala Pro Ser Phe Trp Tyr Lys Ile 325 330 335Asp Pro Ser
His Thr Gln Gly Tyr Arg Thr Val Gln Leu Val Trp Lys 340 345 350Thr
Leu Pro Pro Phe Glu Ala Asn Gly Lys Ile Leu Asp Tyr Glu Val 355 360
365Thr Leu Thr Arg Trp Lys Ser His Leu Gln Asn Tyr Thr Val Asn Ala
370 375 380Thr Lys Leu Thr Val Asn Leu Thr Asn Asp Arg Tyr Leu Ala
Thr Leu385 390 395 400Thr Val Arg Asn Leu Val Gly Lys Ser Asp Ala
Ala Val Leu Thr Ile 405 410 415Pro Ala Cys Asp Phe Gln Ala Thr His
Pro Val Met Asp Leu Lys Ala 420 425 430Phe Pro Lys Asp Asn Met Leu
Trp Val Glu Trp Thr Thr Pro Arg Glu 435 440 445Ser Val Lys Lys Tyr
Ile Leu Glu Trp Cys Val Leu Ser Asp Lys Ala 450 455 460Pro Cys Ile
Thr Asp Trp Gln Gln Glu Asp Gly Thr Val His Arg Thr465 470 475
480Tyr Leu Arg Gly Asn Leu Ala Glu Ser Lys Cys Tyr Leu Ile Thr Val
485 490 495Thr Pro Val Tyr Ala Asp Gly Pro Gly Ser Pro Glu Ser Ile
Lys Ala 500 505 510Tyr Leu Lys Gln Ala Pro Pro Ser Lys Gly Pro Thr
Val Arg Thr Lys 515 520 525Lys Val Gly Lys Asn Glu Ala Val Leu Glu
Trp Asp Gln Leu Pro Val 530 535 540Asp Val Gln Asn Gly Phe Ile Arg
Asn Tyr Thr Ile Phe Tyr Arg Thr545 550 555 560Ile Ile Gly Asn Glu
Thr Ala Val Asn Val Asp Ser Ser His Thr Glu 565 570 575Tyr Thr Leu
Ser Ser Leu Thr Ser Asp Thr Leu Tyr Met Val Arg Met 580 585 590Ala
Ala Tyr Thr Asp Glu Gly Gly Lys Asp Gly Pro Glu Phe Thr Phe 595 600
605Thr Thr Pro Lys Phe Ala Gln Gly Glu Ile Glu Ala Ile Val Val Pro
610 615 620Val Cys Leu Ala Phe Leu Leu Thr Thr Leu Leu Gly Val Leu
Phe Cys625 630 635 640Phe Asn Lys Arg Asp Leu Ile Lys Lys His Ile
Trp Pro Asn Val Pro 645 650 655Asp Pro Ser Lys Ser His Ile Ala Gln
Trp Ser Pro His Thr Pro Pro 660 665 670Arg His Asn Phe Asn Ser Lys
Asp Gln Met Tyr Ser Asp Gly Asn Phe 675 680 685Thr Asp Val Ser Val
Val Glu Ile Glu Ala Asn Asp Lys Lys Pro Phe 690 695 700Pro Glu Asp
Leu Lys Ser Leu Asp Leu Phe Lys Lys Glu Lys Ile Asn705 710 715
720Thr Glu Gly His Ser Ser Gly Ile Gly Gly Ser Ser Cys Met Ser Ser
725 730 735Ser Arg Pro Ser Ile Ser Ser Ser Asp Glu Asn Glu Ser Ser
Gln Asn 740 745 750Thr Ser Ser Thr Val Gln Tyr Ser Thr Val Val His
Ser Gly Tyr Arg 755 760 765His Gln Val Pro Ser Val Gln Val Phe Ser
Arg Ser Glu Ser Thr Gln 770 775 780Pro Leu Leu Asp Ser Glu Glu Arg
Pro Glu Asp Leu Gln Leu Val Asp785 790 795 800His Val Asp Gly Gly
Asp Gly Ile Leu Pro Arg Gln Gln Tyr Phe Lys 805 810 815Gln Asn Cys
Ser Gln His Glu Ser Ser Pro Asp Ile Ser His Phe Glu 820 825 830Arg
Ser Lys Gln Val Ser Ser Val Asn Glu Glu Asp Phe Val Arg Leu 835 840
845Lys Gln Gln Ile Ser Asp His Ile Ser Gln Ser Cys Gly Ser Gly Gln
850 855 860Met Lys Met Phe Gln Glu Val Ser Ala Ala Asp Ala Phe Gly
Pro Gly865 870 875 880Thr Glu Gly Gln Val Glu Arg Phe Glu Thr Val
Gly Met Glu Ala Ala 885 890 895Thr Asp Glu Gly Met Pro Lys Ser Tyr
Leu Pro Gln Thr Val Arg Gln 900 905 910Gly Gly Tyr Met Pro Gln
915145PRTHomo sapiens 14Arg Pro Ser Lys Ala1 5155PRTHomo sapiens
15Asn Ile Ala Ser Phe1 516728PRTHomo sapiens 16Met Trp Val Thr Lys
Leu Leu Pro Ala Leu Leu Leu Gln His Val Leu1 5 10 15Leu His Leu Leu
Leu Leu Pro Ile Ala Ile Pro Tyr Ala Glu Gly Gln 20 25 30Arg Lys Arg
Arg Asn Thr Ile His Glu Phe Lys Lys Ser Ala Lys Thr 35 40 45Thr Leu
Ile Lys Ile Asp Pro Ala Leu Lys Ile Lys Thr Lys Lys Val 50 55 60Asn
Thr Ala Asp Gln Cys Ala Asn Arg Cys Thr Arg Asn Lys Gly Leu65 70 75
80Pro Phe Thr Cys Lys Ala Phe Val Phe Asp Lys Ala Arg Lys Gln Cys
85 90 95Leu Trp Phe Pro Phe Asn Ser Met Ser Ser Gly Val Lys Lys Glu
Phe 100 105 110Gly His Glu Phe Asp Leu Tyr Glu Asn Lys Asp Tyr Ile
Arg Asn Cys 115 120 125Ile Ile Gly Lys Gly Arg Ser Tyr Lys Gly Thr
Val Ser Ile Thr Lys 130 135 140Ser Gly Ile Lys Cys Gln Pro Trp Ser
Ser Met Ile Pro His Glu His145 150 155 160Ser Phe Leu Pro Ser Ser
Tyr Arg Gly Lys Asp Leu Gln Glu Asn Tyr 165 170 175Cys Arg Asn Pro
Arg Gly Glu Glu Gly Gly Pro Trp Cys Phe Thr Ser 180 185 190Asn Pro
Glu Val Arg Tyr Glu Val Cys Asp Ile Pro Gln Cys Ser Glu 195 200
205Val Glu Cys Met Thr Cys Asn Gly Glu Ser Tyr Arg Gly Leu Met Asp
210 215 220His Thr Glu Ser Gly Lys Ile Cys Gln Arg Trp Asp His Gln
Thr Pro225 230 235 240His Arg His Lys Phe Leu Pro Glu Arg Tyr Pro
Asp Lys Gly Phe Asp 245 250 255Asp Asn Tyr Cys Arg Asn Pro Asp Gly
Gln Pro Arg Pro Trp Cys Tyr 260 265 270Thr Leu Asp Pro His Thr Arg
Trp Glu Tyr Cys Ala Ile Lys Thr Cys 275 280 285Ala Asp Asn Thr Met
Asn Asp Thr Asp Val Pro Leu Glu Thr Thr Glu 290 295 300Cys Ile Gln
Gly Gln Gly Glu Gly Tyr Arg Gly Thr Val Asn Thr Ile305 310 315
320Trp Asn Gly Ile Pro Cys Gln Arg Trp Asp Ser Gln Tyr Pro His Glu
325 330 335His Asp Met Thr Pro Glu Asn Phe Lys Cys Lys Asp Leu Arg
Glu Asn 340 345 350Tyr Cys Arg Asn Pro Asp Gly Ser Glu Ser Pro Trp
Cys Phe Thr Thr 355 360 365Asp Pro Asn Ile Arg Val Gly Tyr Cys Ser
Gln Ile Pro Asn Cys Asp 370 375 380Met Ser His Gly Gln Asp Cys Tyr
Arg Gly Asn Gly Lys Asn Tyr Met385 390 395 400Gly Asn Leu Ser Gln
Thr Arg Ser Gly Leu Thr Cys Ser Met Trp Asp 405 410 415Lys Asn Met
Glu Asp Leu His Arg His Ile Phe Trp Glu Pro Asp Ala 420 425
430Ser Lys Leu Asn Glu Asn Tyr Cys Arg Asn Pro Asp Asp Asp Ala His
435 440 445Gly Pro Trp Cys Tyr Thr Gly Asn Pro Leu Ile Pro Trp Asp
Tyr Cys 450 455 460Pro Ile Ser Arg Cys Glu Gly Asp Thr Thr Pro Thr
Ile Val Asn Leu465 470 475 480Asp His Pro Val Ile Ser Cys Ala Lys
Thr Lys Gln Leu Arg Val Val 485 490 495Asn Gly Ile Pro Thr Arg Thr
Asn Ile Gly Trp Met Val Ser Leu Arg 500 505 510Tyr Arg Asn Lys His
Ile Cys Gly Gly Ser Leu Ile Lys Glu Ser Trp 515 520 525Val Leu Thr
Ala Arg Gln Cys Phe Pro Ser Arg Asp Leu Lys Asp Tyr 530 535 540Glu
Ala Trp Leu Gly Ile His Asp Val His Gly Arg Gly Asp Glu Lys545 550
555 560Cys Lys Gln Val Leu Asn Val Ser Gln Leu Val Tyr Gly Pro Glu
Gly 565 570 575Ser Asp Leu Val Leu Met Lys Leu Ala Arg Pro Ala Val
Leu Asp Asp 580 585 590Phe Val Ser Thr Ile Asp Leu Pro Asn Tyr Gly
Cys Thr Ile Pro Glu 595 600 605Lys Thr Ser Cys Ser Val Tyr Gly Trp
Gly Tyr Thr Gly Leu Ile Asn 610 615 620Tyr Asp Gly Leu Leu Arg Val
Ala His Leu Tyr Ile Met Gly Asn Glu625 630 635 640Lys Cys Ser Gln
His His Arg Gly Lys Val Thr Leu Asn Glu Ser Glu 645 650 655Ile Cys
Ala Gly Ala Glu Lys Ile Gly Ser Gly Pro Cys Glu Gly Asp 660 665
670Tyr Gly Gly Pro Leu Val Cys Glu Gln His Lys Met Arg Met Val Leu
675 680 685Gly Val Ile Val Pro Gly Arg Gly Cys Ala Ile Pro Asn Arg
Pro Gly 690 695 700Ile Phe Val Arg Val Ala Tyr Tyr Ala Lys Trp Ile
His Lys Ile Ile705 710 715 720Leu Thr Tyr Lys Val Pro Gln Ser
72517224PRTHomo sapiens 17Met Pro Gly Ser Pro Arg Pro Ala Pro Ser
Trp Val Leu Leu Leu Arg1 5 10 15Leu Leu Ala Leu Leu Arg Pro Pro Gly
Leu Gly Glu Ala Cys Ser Cys 20 25 30Ala Pro Ala His Pro Gln Gln His
Ile Cys His Ser Ala Leu Val Ile 35 40 45Arg Ala Lys Ile Ser Ser Glu
Lys Val Val Pro Ala Ser Ala Asp Pro 50 55 60Ala Asp Thr Glu Lys Met
Leu Arg Tyr Glu Ile Lys Gln Ile Lys Met65 70 75 80Phe Lys Gly Phe
Glu Lys Val Lys Asp Val Gln Tyr Ile Tyr Thr Pro 85 90 95Phe Asp Ser
Ser Leu Cys Gly Val Lys Leu Glu Ala Asn Ser Gln Lys 100 105 110Gln
Tyr Leu Leu Thr Gly Gln Val Leu Ser Asp Gly Lys Val Phe Ile 115 120
125His Leu Cys Asn Tyr Ile Glu Pro Trp Glu Asp Leu Ser Leu Val Gln
130 135 140Arg Glu Ser Leu Asn His His Tyr His Leu Asn Cys Gly Cys
Gln Ile145 150 155 160Thr Thr Cys Tyr Thr Val Pro Cys Thr Ile Ser
Ala Pro Asn Glu Cys 165 170 175Leu Trp Thr Asp Trp Leu Leu Glu Arg
Lys Leu Tyr Gly Tyr Gln Ala 180 185 190Gln His Tyr Val Cys Met Lys
His Val Asp Gly Thr Cys Ser Trp Tyr 195 200 205Arg Gly His Leu Pro
Leu Arg Lys Glu Phe Val Asp Ile Val Gln Pro 210 215 2201889PRTHomo
sapiens 18Met Lys Gly Leu Ala Ala Ala Leu Leu Val Leu Val Cys Thr
Met Ala1 5 10 15Leu Cys Ser Cys Ala Gln Val Gly Thr Asn Lys Glu Leu
Cys Cys Leu 20 25 30Val Tyr Thr Ser Trp Gln Ile Pro Gln Lys Phe Ile
Val Asp Tyr Ser 35 40 45Glu Thr Ser Pro Gln Cys Pro Lys Pro Gly Val
Ile Leu Leu Thr Lys 50 55 60Arg Gly Arg Gln Ile Cys Ala Asp Pro Asn
Lys Lys Trp Val Gln Lys65 70 75 80Tyr Ile Ser Asp Leu Lys Leu Asn
Ala 8519267PRTHomo sapiens 19Met Arg Leu Thr Val Leu Cys Ala Val
Cys Leu Leu Pro Gly Ser Leu1 5 10 15Ala Leu Pro Leu Pro Gln Glu Ala
Gly Gly Met Ser Glu Leu Gln Trp 20 25 30Glu Gln Ala Gln Asp Tyr Leu
Lys Arg Phe Tyr Leu Tyr Asp Ser Glu 35 40 45Thr Lys Asn Ala Asn Ser
Leu Glu Ala Lys Leu Lys Glu Met Gln Lys 50 55 60Phe Phe Gly Leu Pro
Ile Thr Gly Met Leu Asn Ser Arg Val Ile Glu65 70 75 80Ile Met Gln
Lys Pro Arg Cys Gly Val Pro Asp Val Ala Glu Tyr Ser 85 90 95Leu Phe
Pro Asn Ser Pro Lys Trp Thr Ser Lys Val Val Thr Tyr Arg 100 105
110Ile Val Ser Tyr Thr Arg Asp Leu Pro His Ile Thr Val Asp Arg Leu
115 120 125Val Ser Lys Ala Leu Asn Met Trp Gly Lys Glu Ile Pro Leu
His Phe 130 135 140Arg Lys Val Val Trp Gly Thr Ala Asp Ile Met Ile
Gly Phe Ala Arg145 150 155 160Gly Ala His Gly Asp Ser Tyr Pro Phe
Asp Gly Pro Gly Asn Thr Leu 165 170 175Ala His Ala Phe Ala Pro Gly
Thr Gly Leu Gly Gly Asp Ala His Phe 180 185 190Asp Glu Asp Glu Arg
Trp Thr Asp Gly Ser Ser Leu Gly Ile Asn Phe 195 200 205Leu Tyr Ala
Ala Thr His Glu Leu Gly His Ser Leu Gly Met Gly His 210 215 220Ser
Ser Asp Pro Asn Ala Val Met Tyr Pro Thr Tyr Gly Asn Gly Asp225 230
235 240Pro Gln Asn Phe Lys Leu Ser Gln Asp Asp Ile Lys Gly Ile Gln
Lys 245 250 255Leu Tyr Gly Lys Arg Ser Asn Ser Arg Lys Lys 260
2652094PRTHomo sapiens 20Met Ser Val Lys Gly Met Ala Ile Ala Leu
Ala Val Ile Leu Cys Ala1 5 10 15Thr Val Val Gln Gly Phe Pro Met Phe
Lys Arg Gly Arg Cys Leu Cys 20 25 30Ile Gly Pro Gly Val Lys Ala Val
Lys Val Ala Asp Ile Glu Lys Ala 35 40 45Ser Ile Met Tyr Pro Ser Asn
Asn Cys Asp Lys Ile Glu Val Ile Ile 50 55 60Thr Leu Lys Glu Asn Lys
Gly Gln Arg Cys Leu Asn Pro Lys Ser Lys65 70 75 80Gln Ala Arg Leu
Ile Ile Lys Lys Val Glu Arg Lys Asn Phe 85 9021107PRTHomo sapiens
21Met Ala Arg Ala Ala Leu Ser Ala Ala Pro Ser Asn Pro Arg Leu Leu1
5 10 15Arg Val Ala Leu Leu Leu Leu Leu Leu Val Ala Ala Gly Arg Arg
Ala 20 25 30Ala Gly Ala Ser Val Ala Thr Glu Leu Arg Cys Gln Cys Leu
Gln Thr 35 40 45Leu Gln Gly Ile His Pro Lys Asn Ile Gln Ser Val Asn
Val Lys Ser 50 55 60Pro Gly Pro His Cys Ala Gln Thr Glu Val Ile Ala
Thr Leu Lys Asn65 70 75 80Gly Arg Lys Ala Cys Leu Asn Pro Ala Ser
Pro Ile Val Lys Lys Ile 85 90 95Ile Glu Lys Met Leu Asn Ser Asp Lys
Ser Asn 100 10522107PRTHomo sapiens 22Met Ala Arg Ala Thr Leu Ser
Ala Ala Pro Ser Asn Pro Arg Leu Leu1 5 10 15Arg Val Ala Leu Leu Leu
Leu Leu Leu Val Ala Ala Ser Arg Arg Ala 20 25 30Ala Gly Ala Pro Leu
Ala Thr Glu Leu Arg Cys Gln Cys Leu Gln Thr 35 40 45Leu Gln Gly Ile
His Leu Lys Asn Ile Gln Ser Val Lys Val Lys Ser 50 55 60Pro Gly Pro
His Cys Ala Gln Thr Glu Val Ile Ala Thr Leu Lys Asn65 70 75 80Gly
Gln Lys Ala Cys Leu Asn Pro Ala Ser Pro Met Val Lys Lys Ile 85 90
95Ile Glu Lys Met Leu Lys Asn Gly Lys Ser Asn 100 10523107PRTHomo
sapiens 23Met Ala His Ala Thr Leu Ser Ala Ala Pro Ser Asn Pro Arg
Leu Leu1 5 10 15Arg Val Ala Leu Leu Leu Leu Leu Leu Val Ala Ala Ser
Arg Arg Ala 20 25 30Ala Gly Ala Ser Val Val Thr Glu Leu Arg Cys Gln
Cys Leu Gln Thr 35 40 45Leu Gln Gly Ile His Leu Lys Asn Ile Gln Ser
Val Asn Val Arg Ser 50 55 60Pro Gly Pro His Cys Ala Gln Thr Glu Val
Ile Ala Thr Leu Lys Asn65 70 75 80Gly Lys Lys Ala Cys Leu Asn Pro
Ala Ser Pro Met Val Gln Lys Ile 85 90 95Ile Glu Lys Ile Leu Asn Lys
Gly Ser Thr Asn 100 10524132PRTHomo sapiens 24Met Lys Ser Ser Gly
Leu Phe Pro Phe Leu Val Leu Leu Ala Leu Gly1 5 10 15Thr Leu Ala Pro
Trp Ala Val Glu Gly Ser Gly Lys Ser Phe Lys Ala 20 25 30Gly Val Cys
Pro Pro Lys Lys Ser Ala Gln Cys Leu Arg Tyr Lys Lys 35 40 45Pro Glu
Cys Gln Ser Asp Trp Gln Cys Pro Gly Lys Lys Arg Cys Cys 50 55 60Pro
Asp Thr Cys Gly Ile Lys Cys Leu Asp Pro Val Asp Thr Pro Asn65 70 75
80Pro Thr Arg Arg Lys Pro Gly Lys Cys Pro Val Thr Tyr Gly Gln Cys
85 90 95Leu Met Leu Asn Pro Pro Asn Phe Cys Glu Met Asp Gly Gln Cys
Lys 100 105 110Arg Asp Leu Lys Cys Cys Met Gly Met Cys Gly Lys Ser
Cys Val Ser 115 120 125Pro Val Lys Ala 130
References
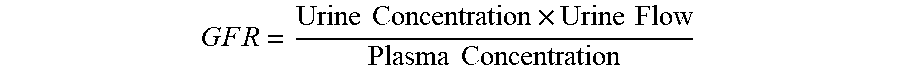


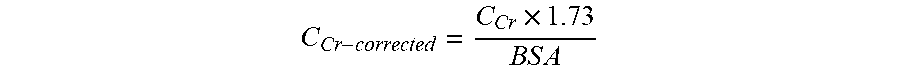
S00001
T00001










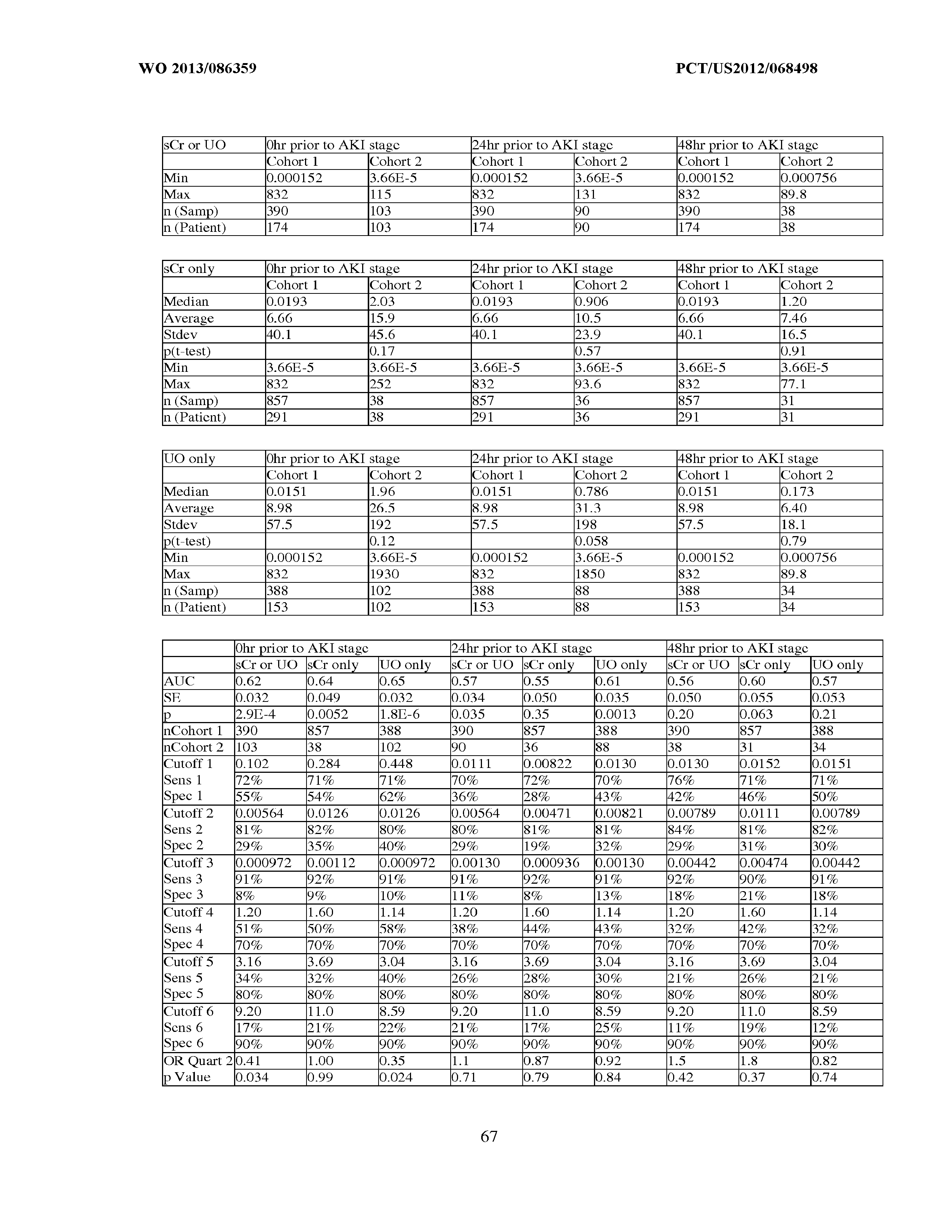










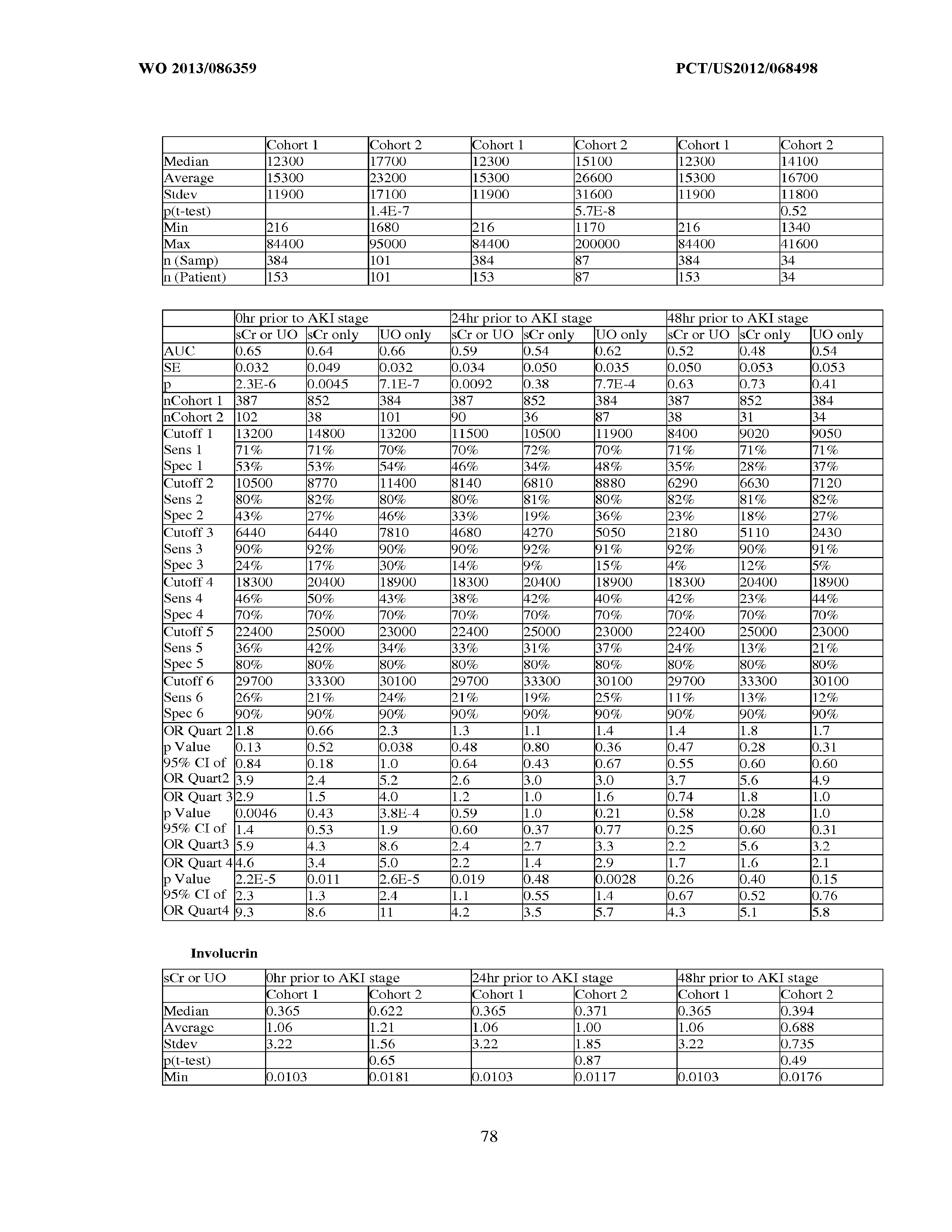







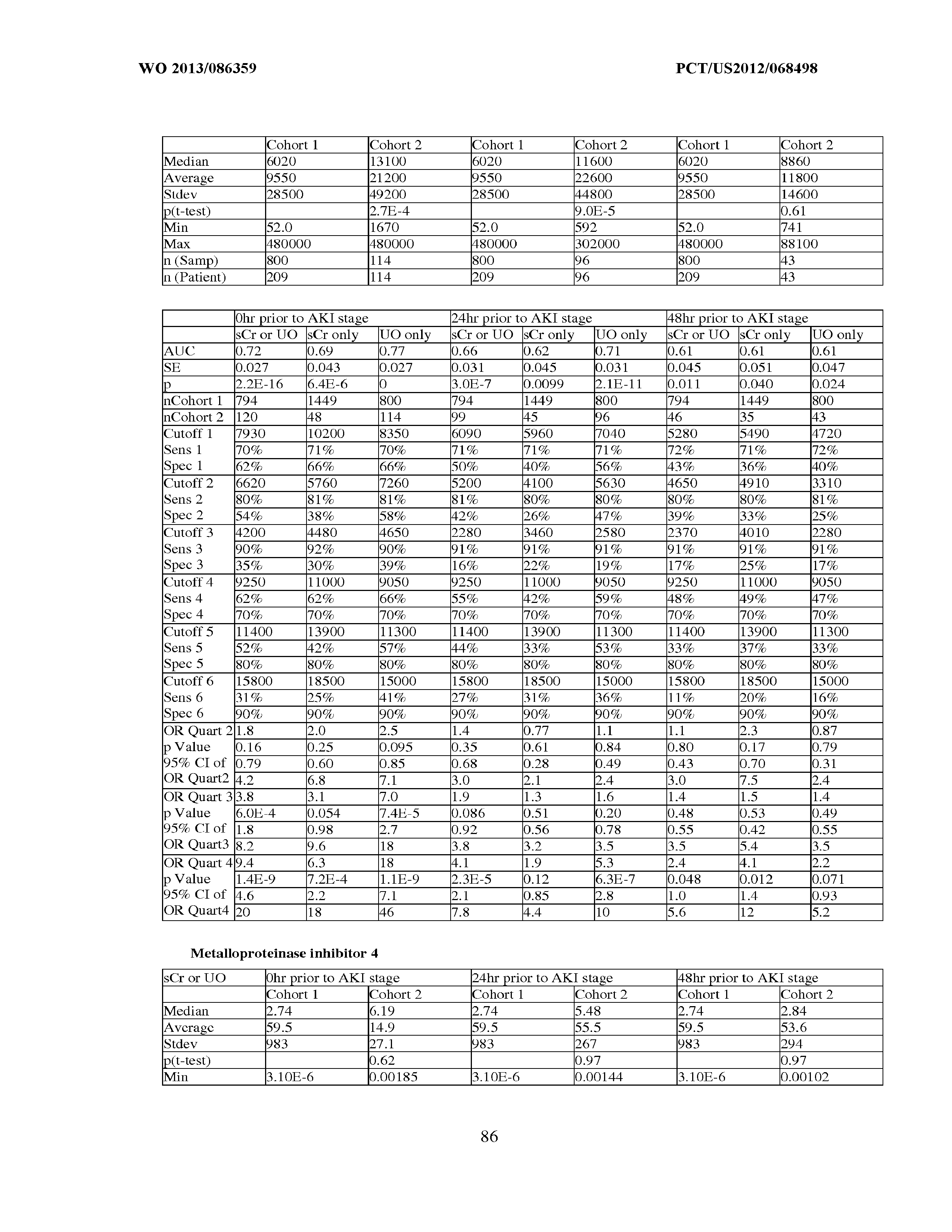
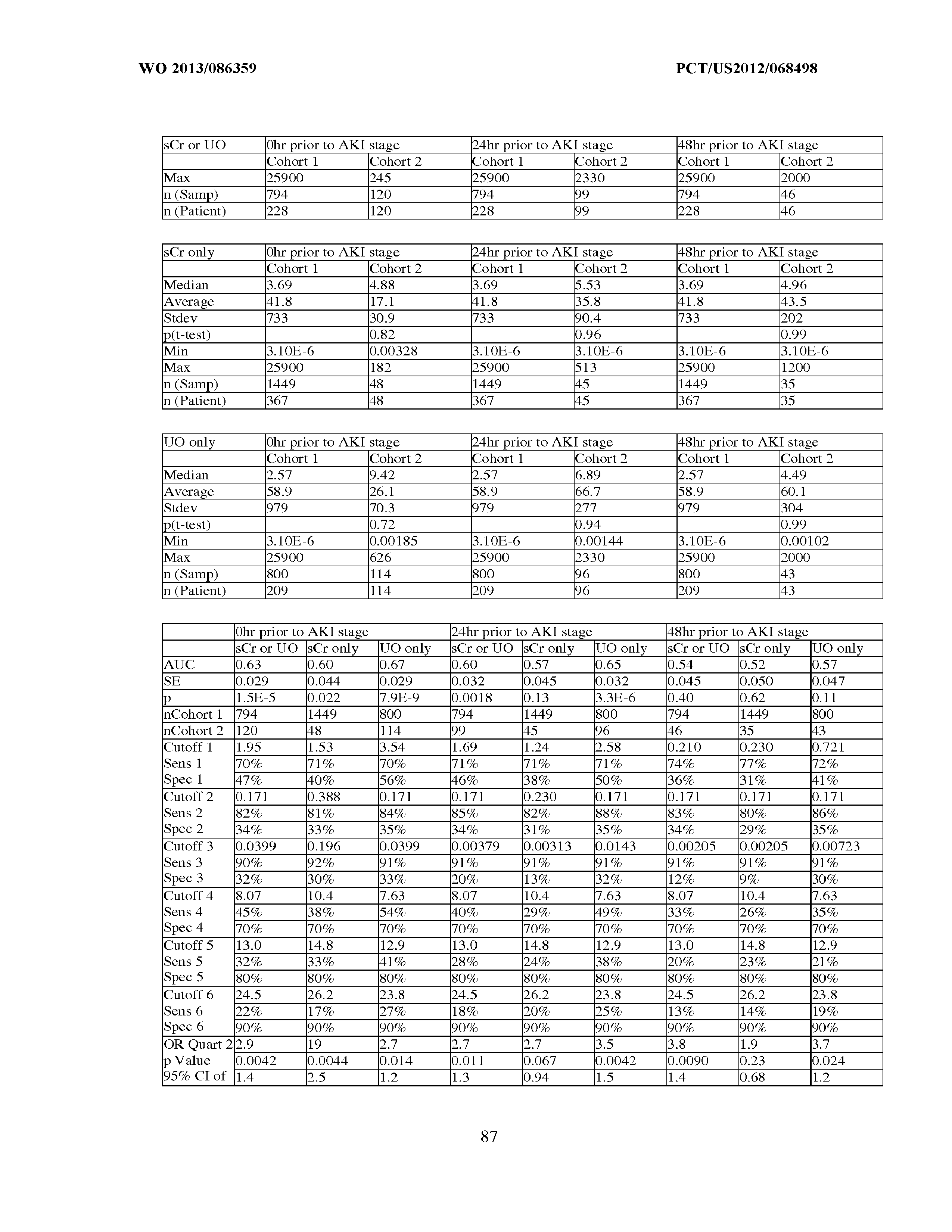















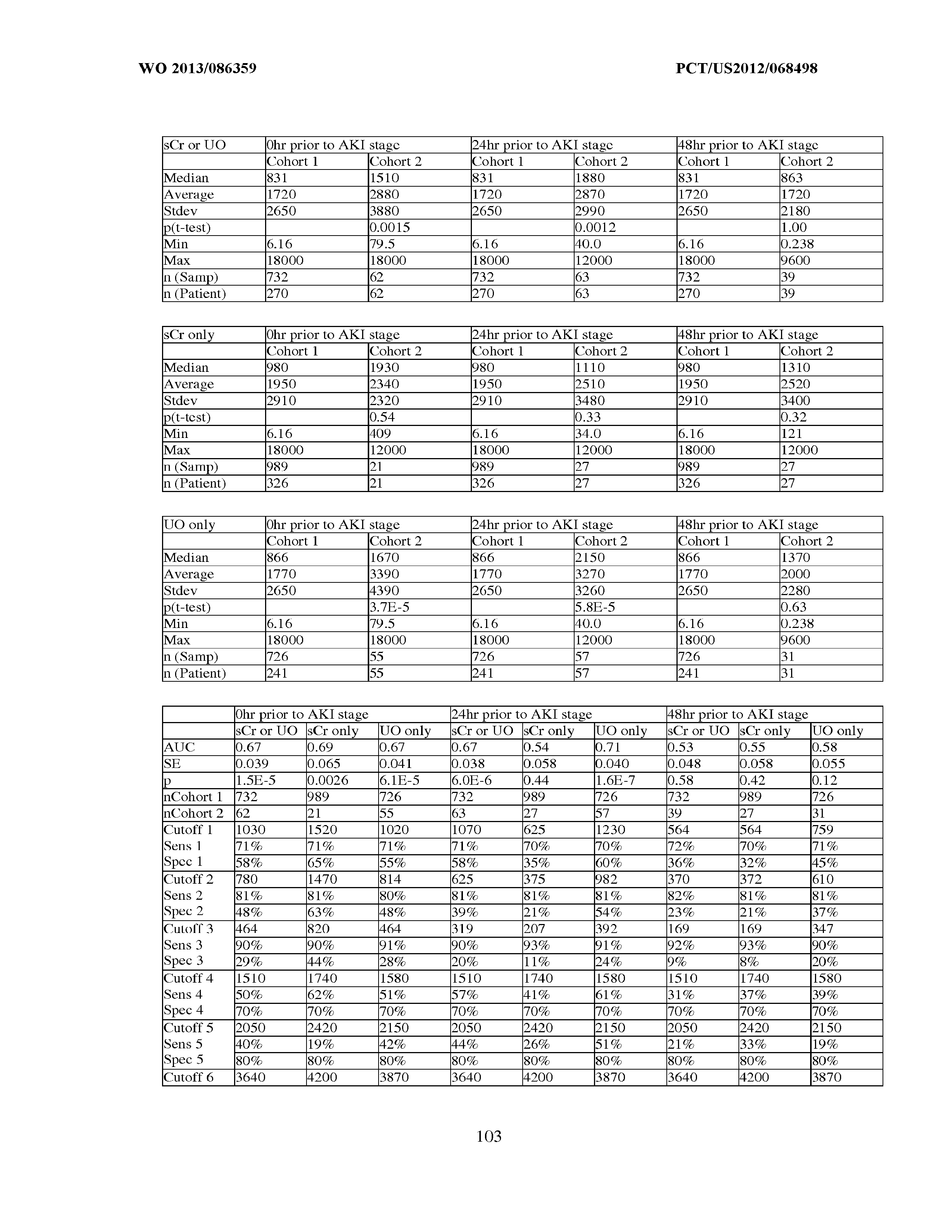






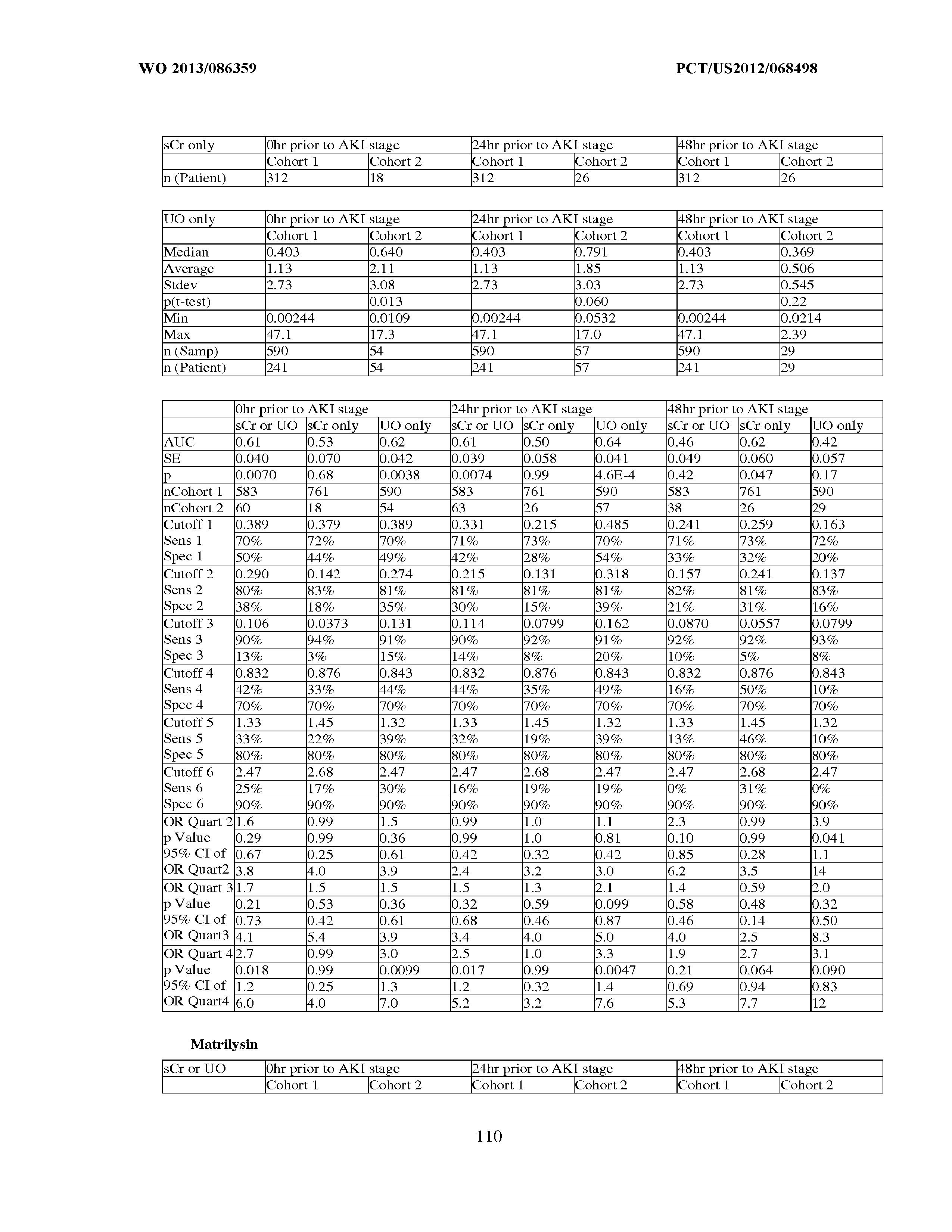









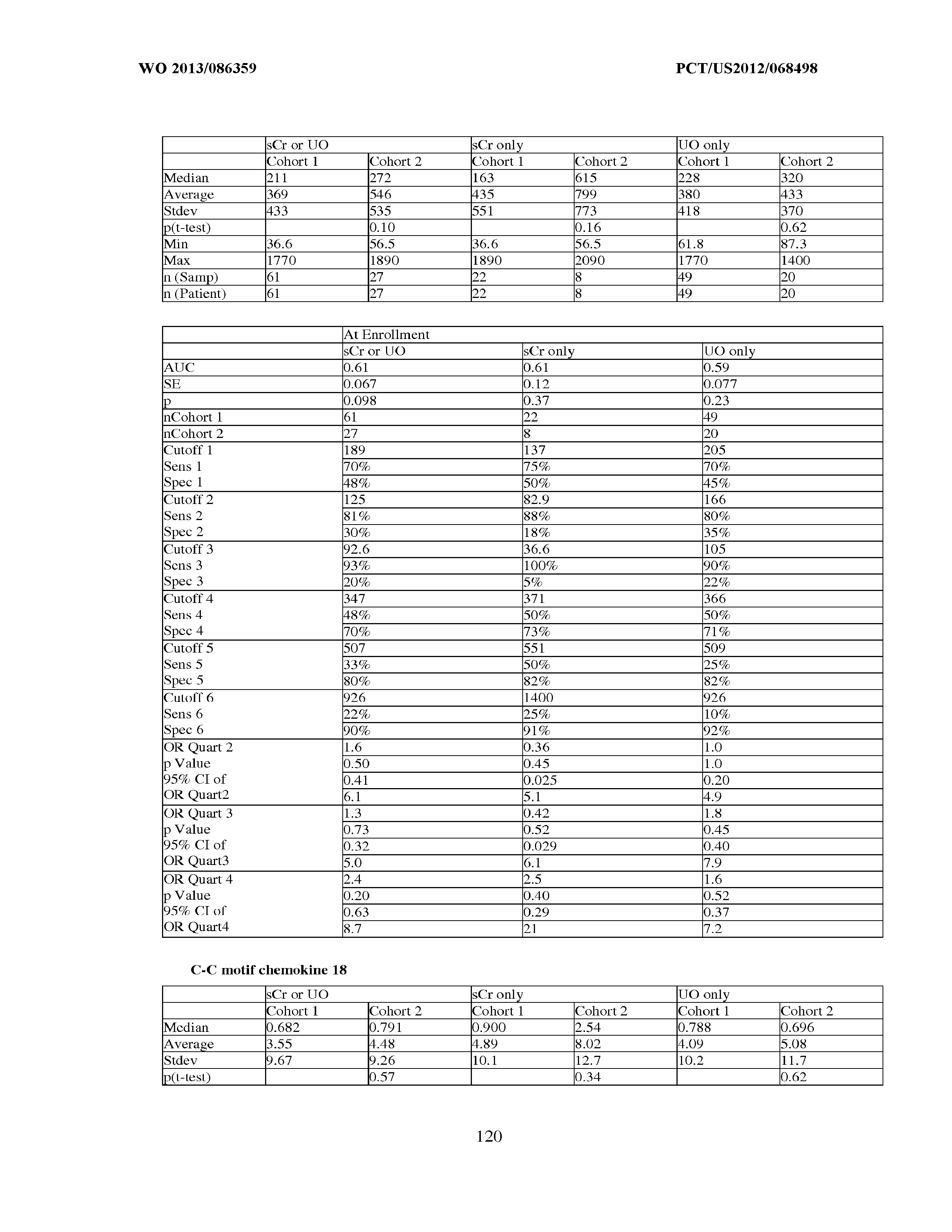

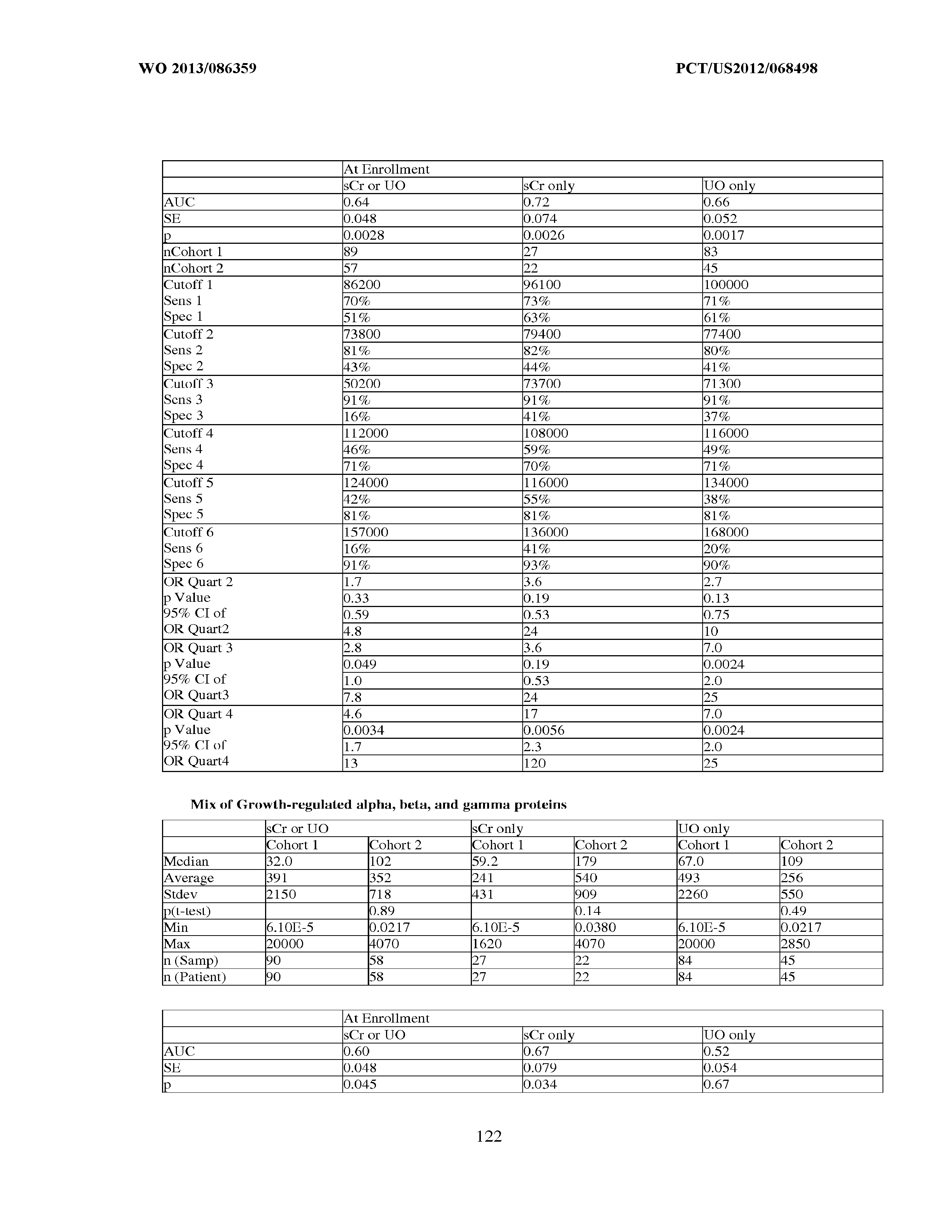




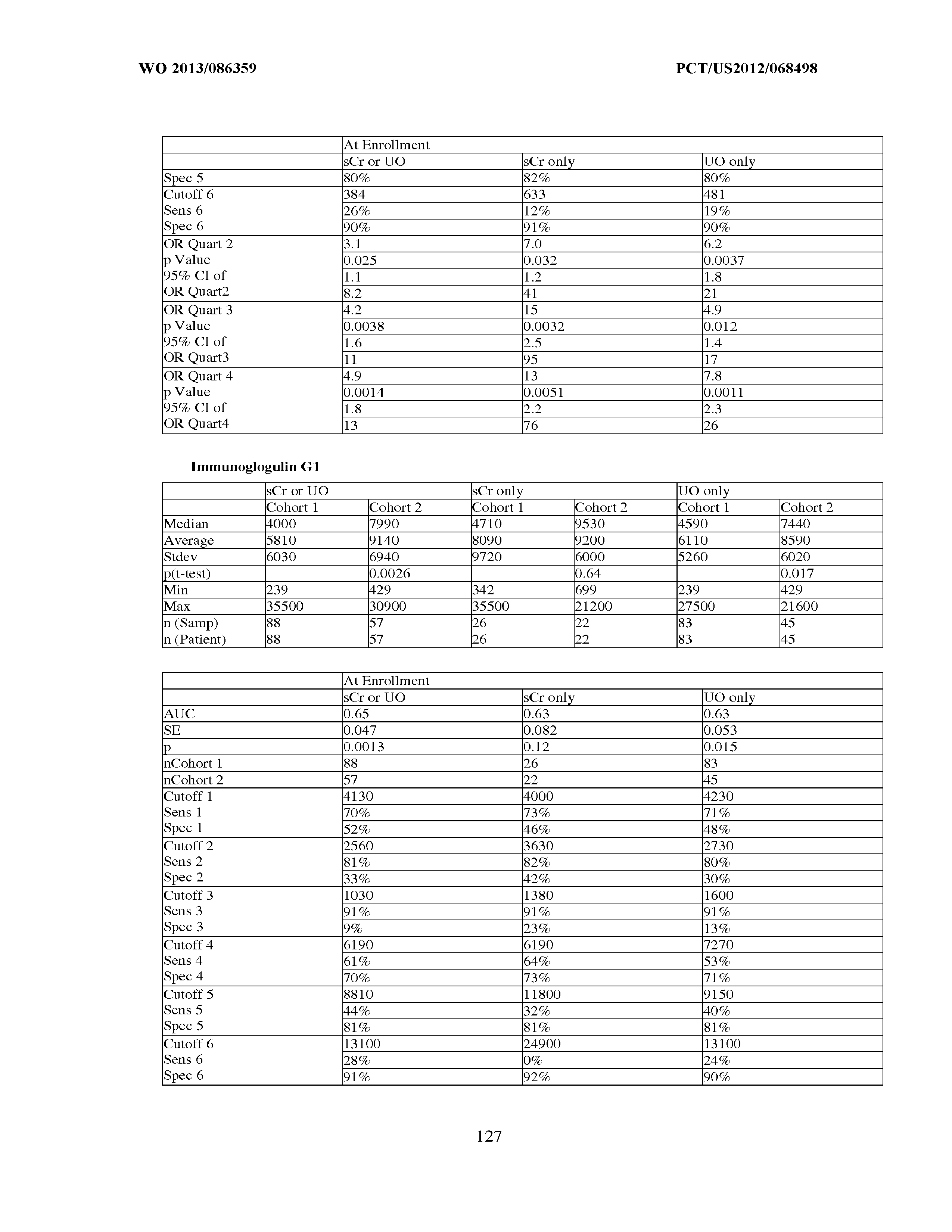





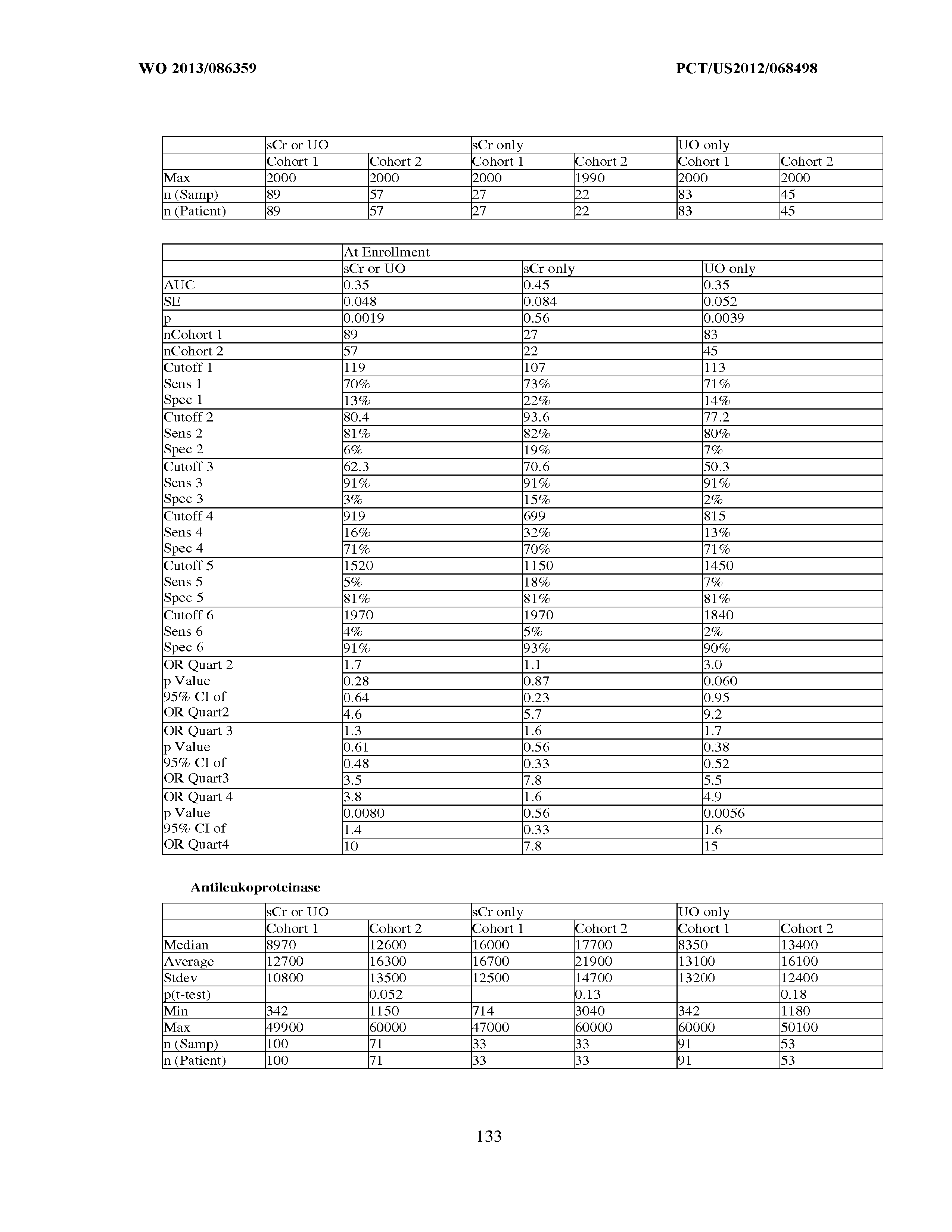























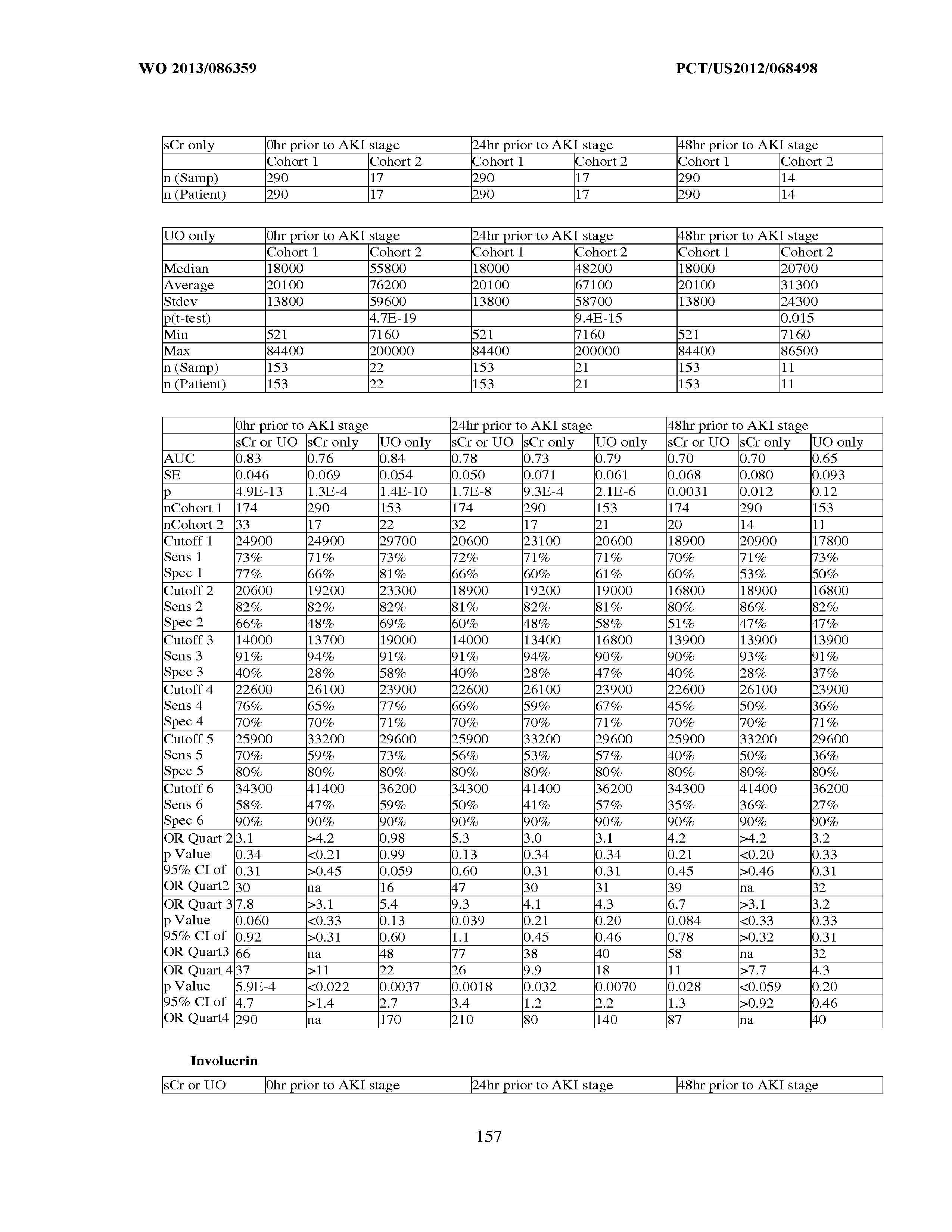

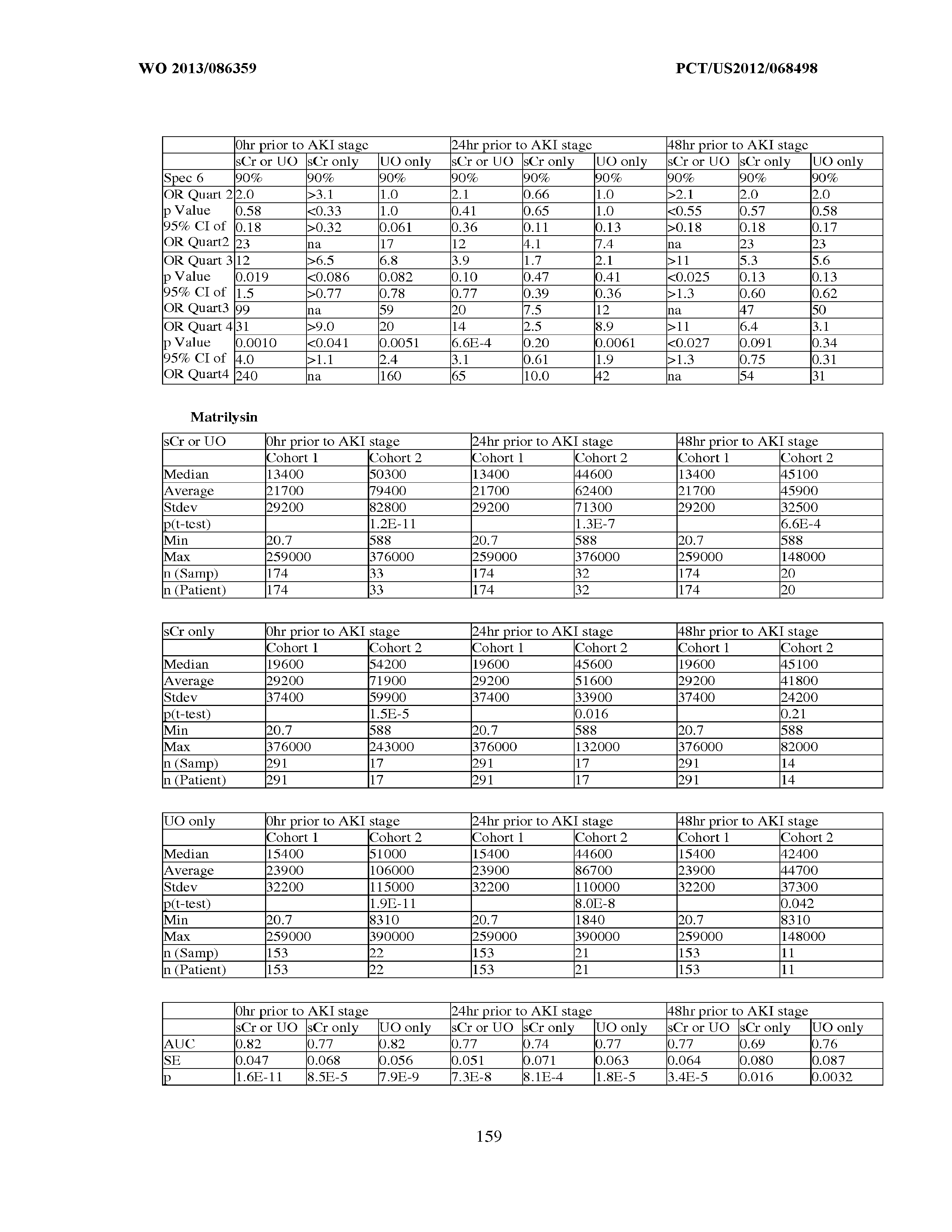
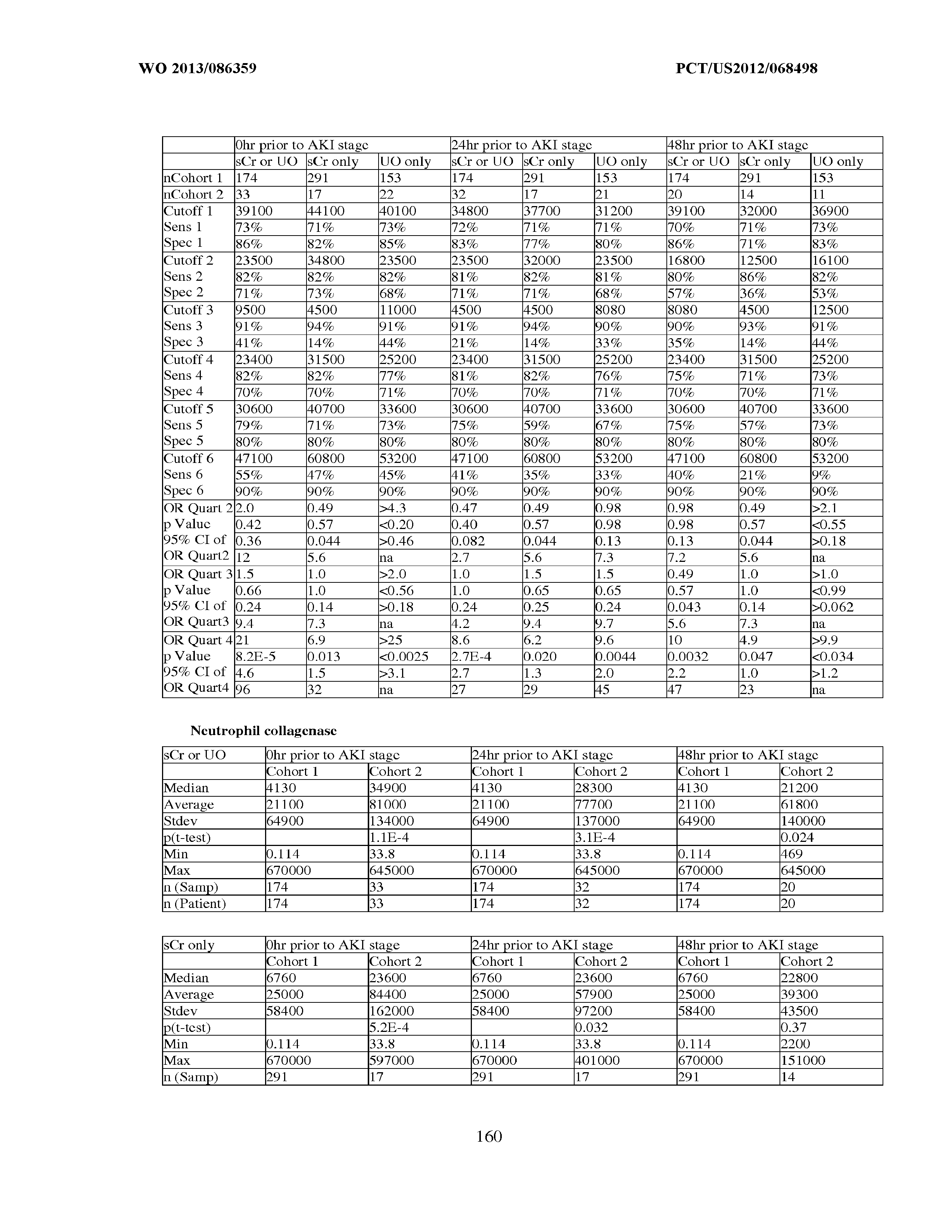























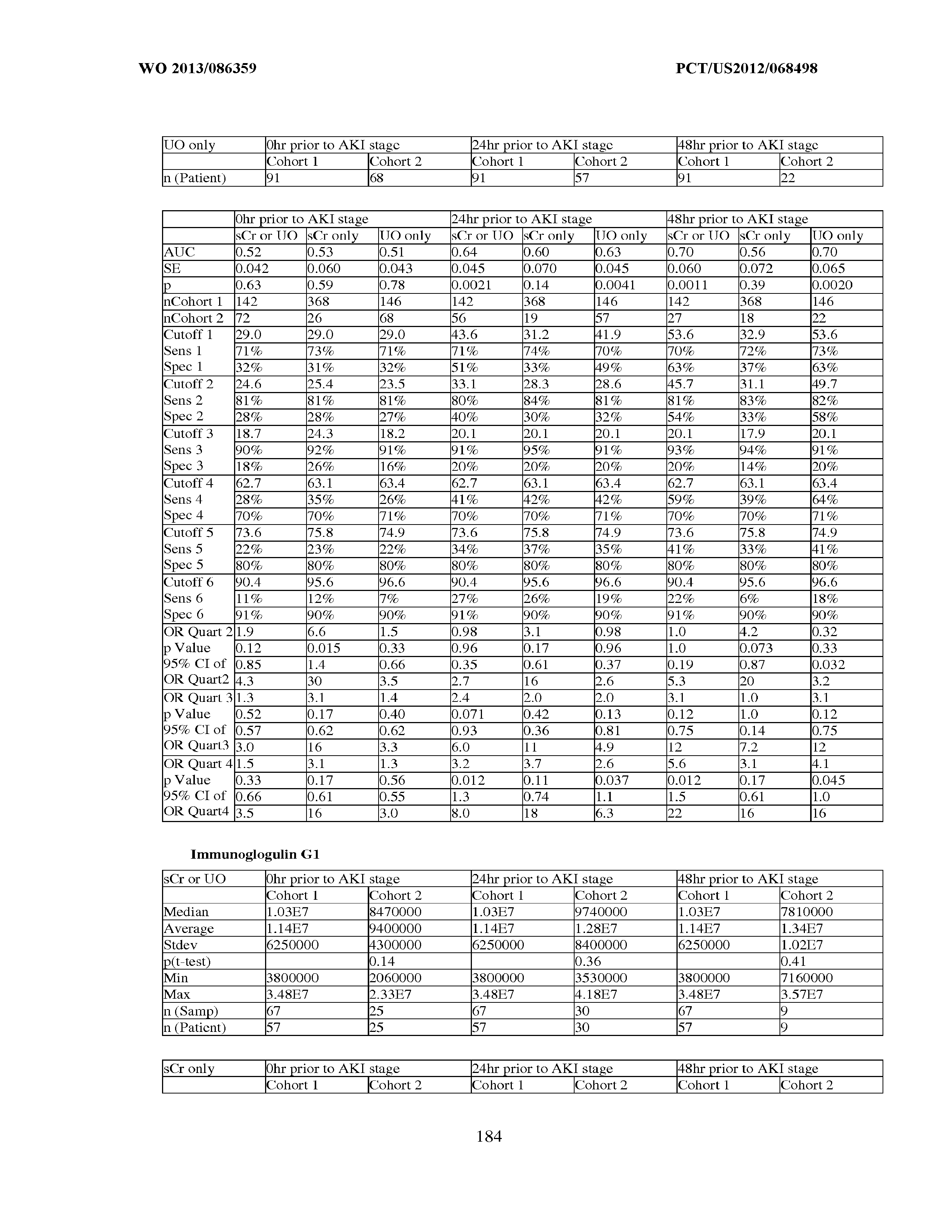










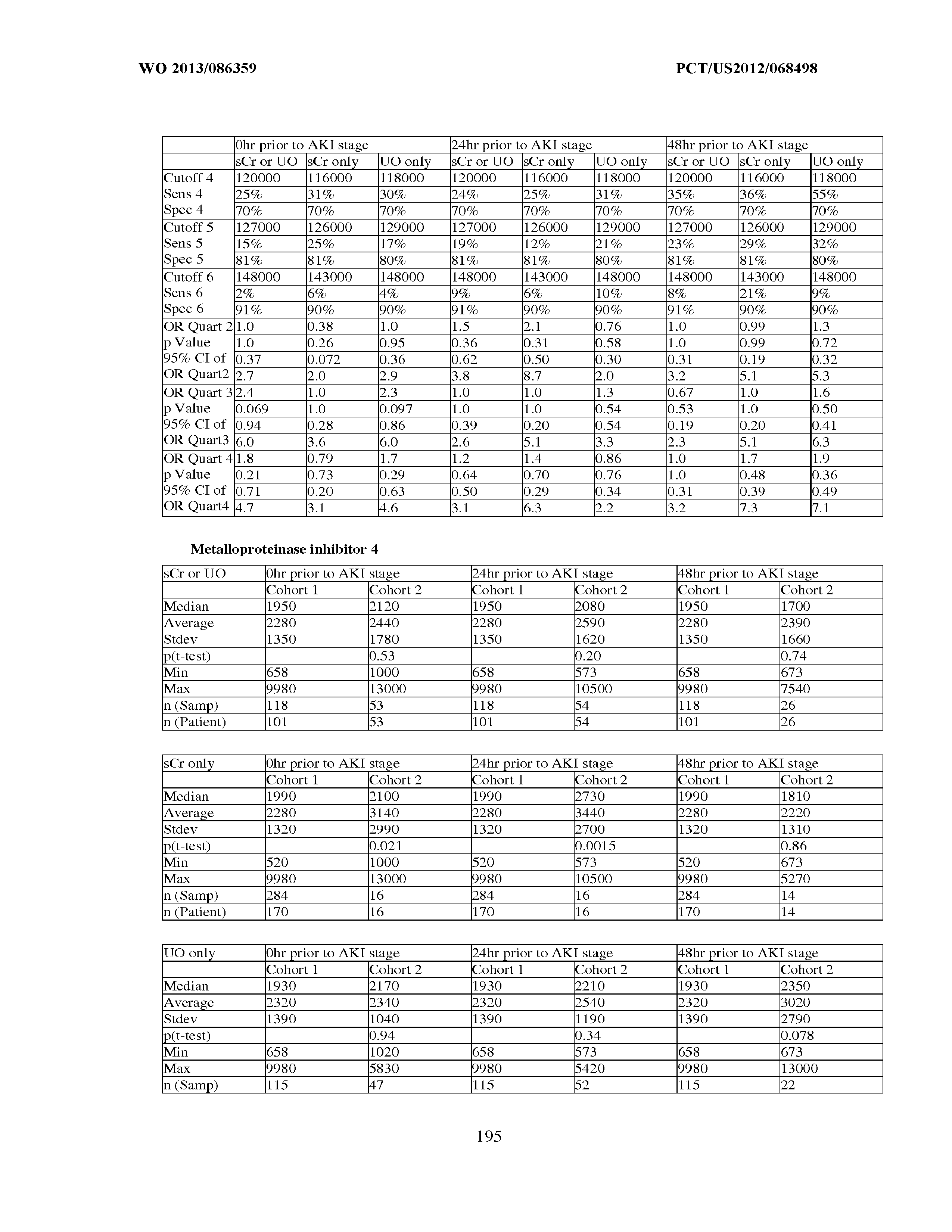



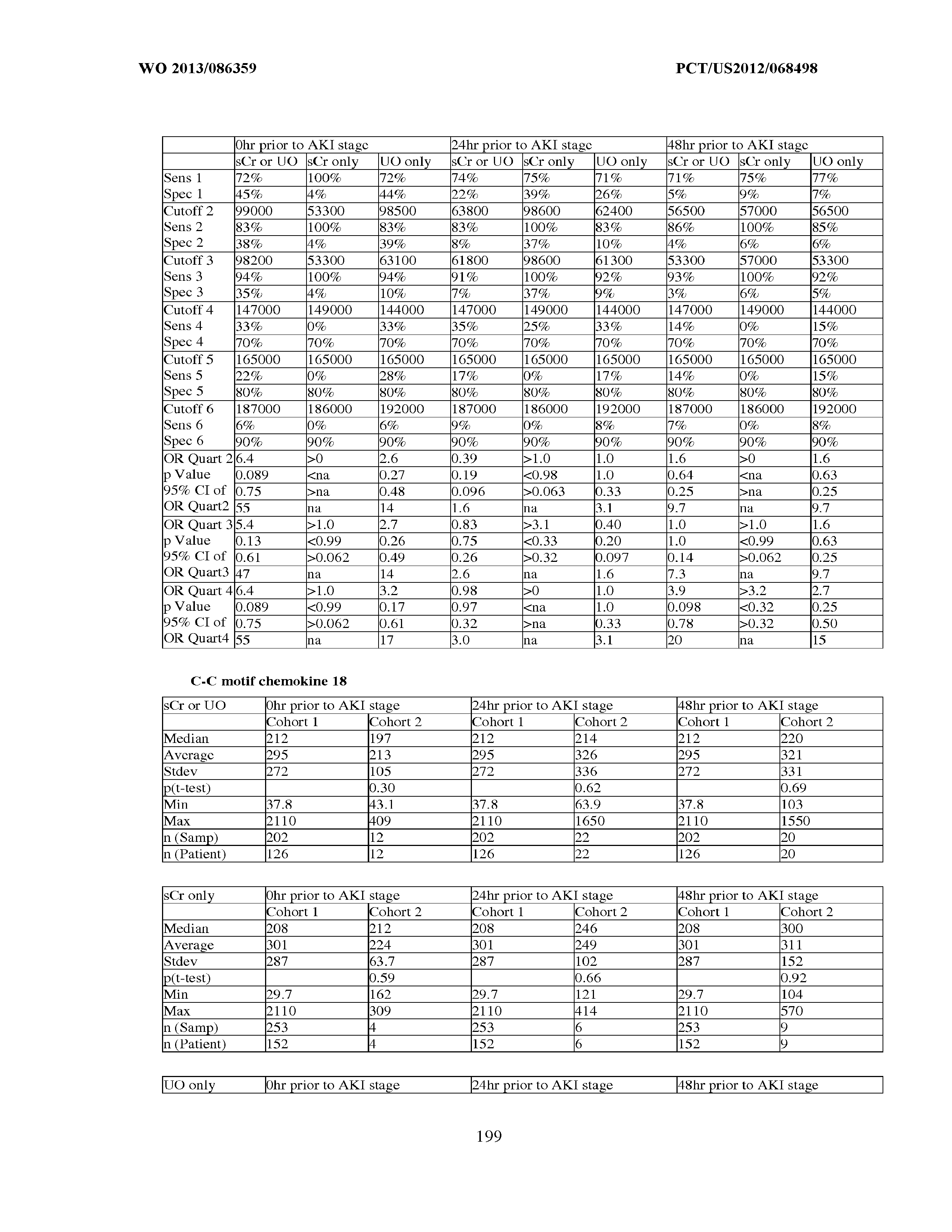





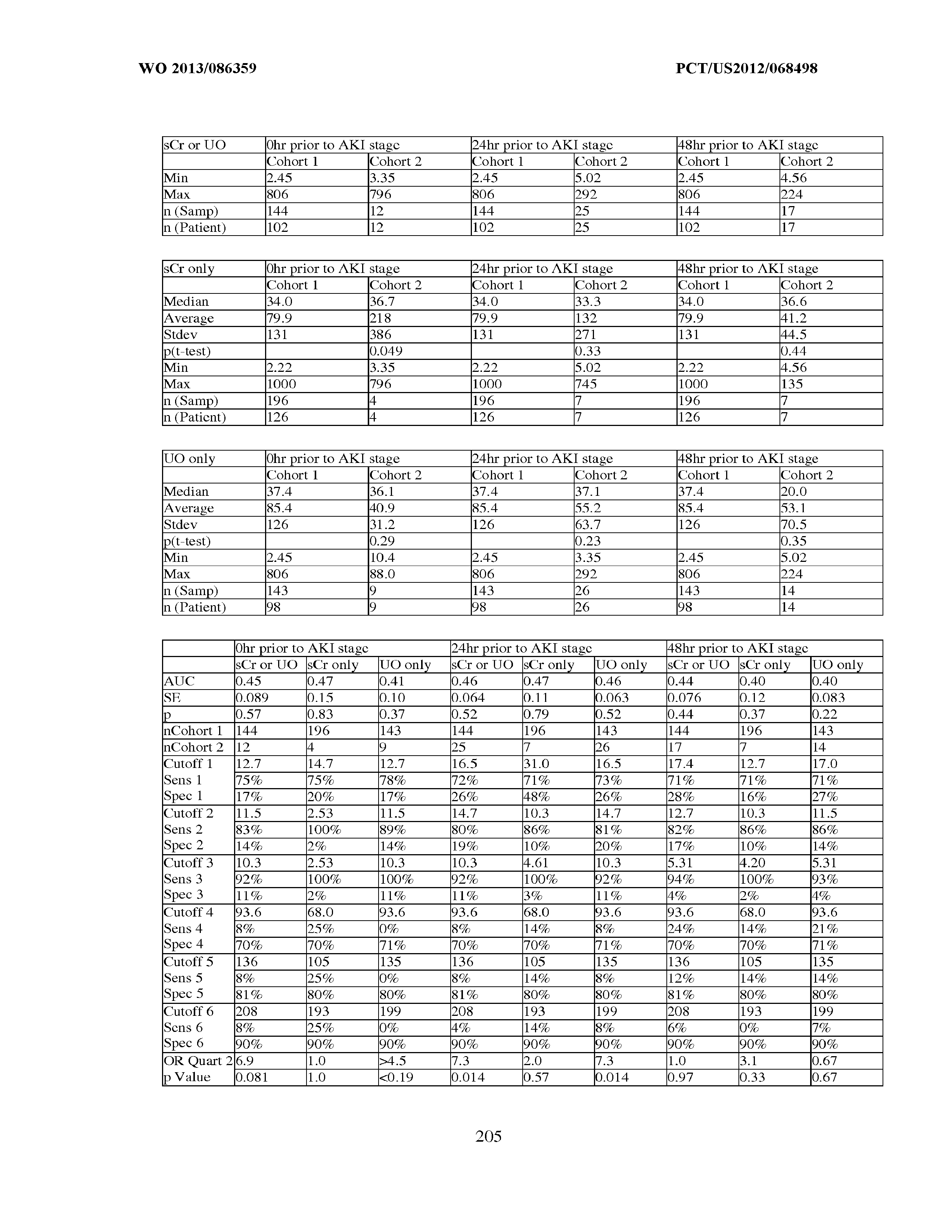










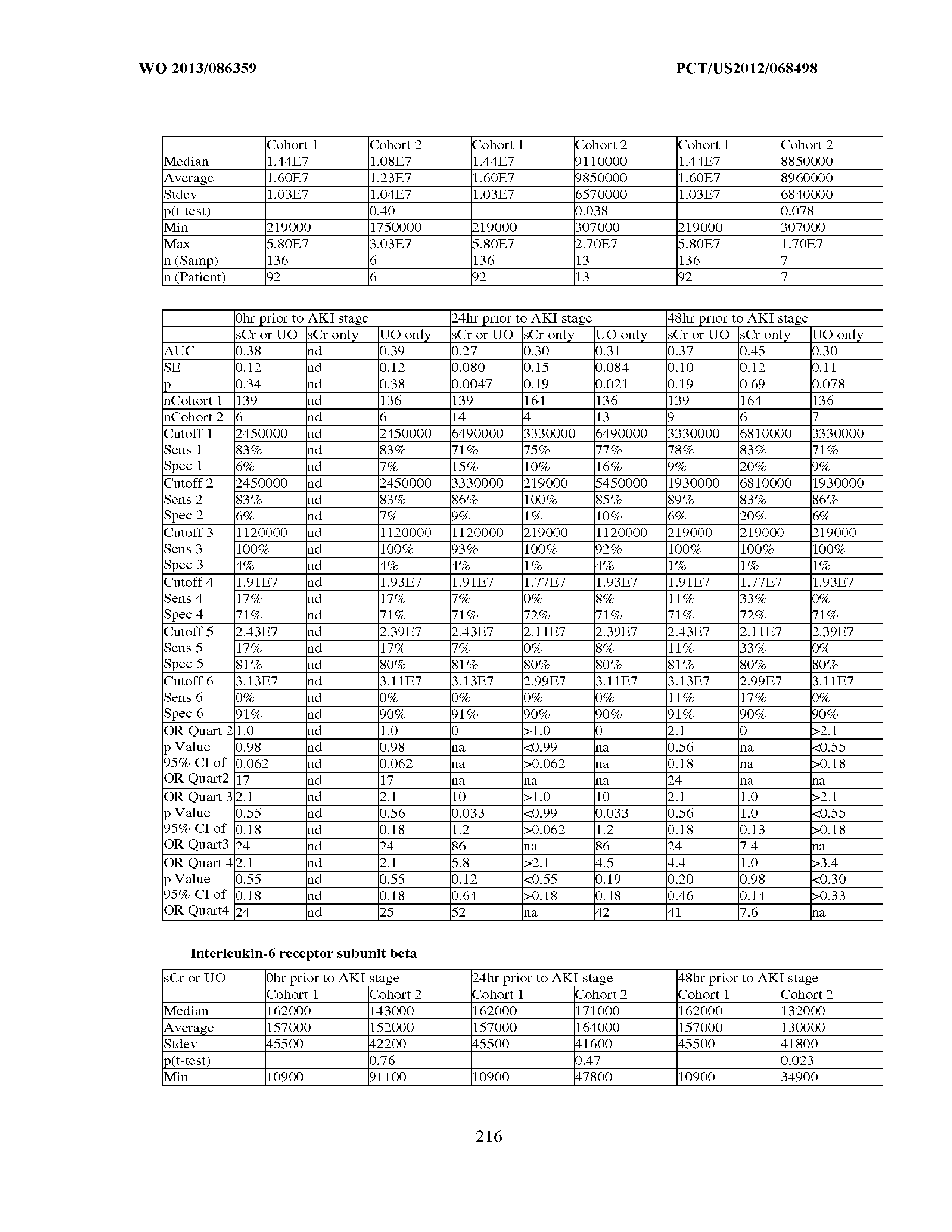













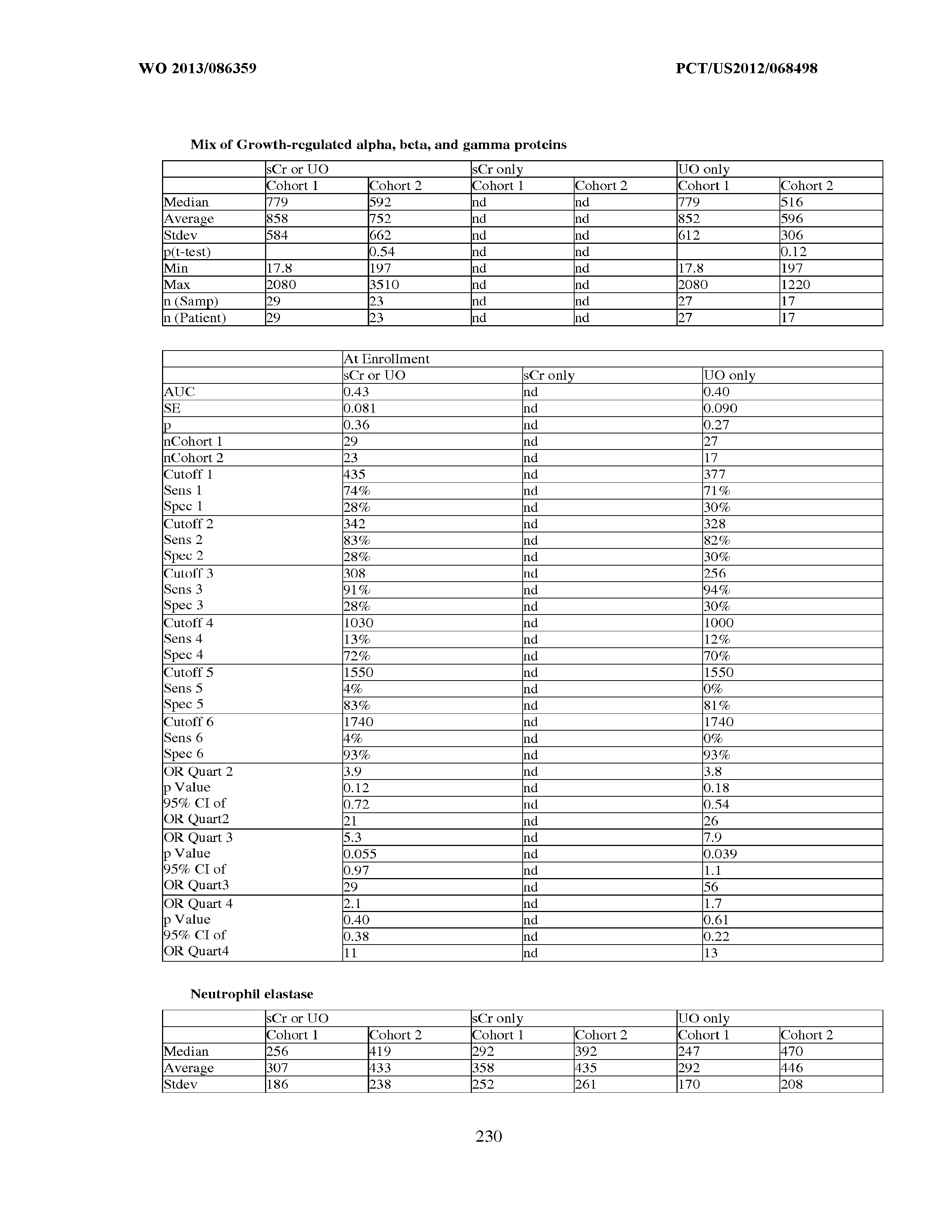




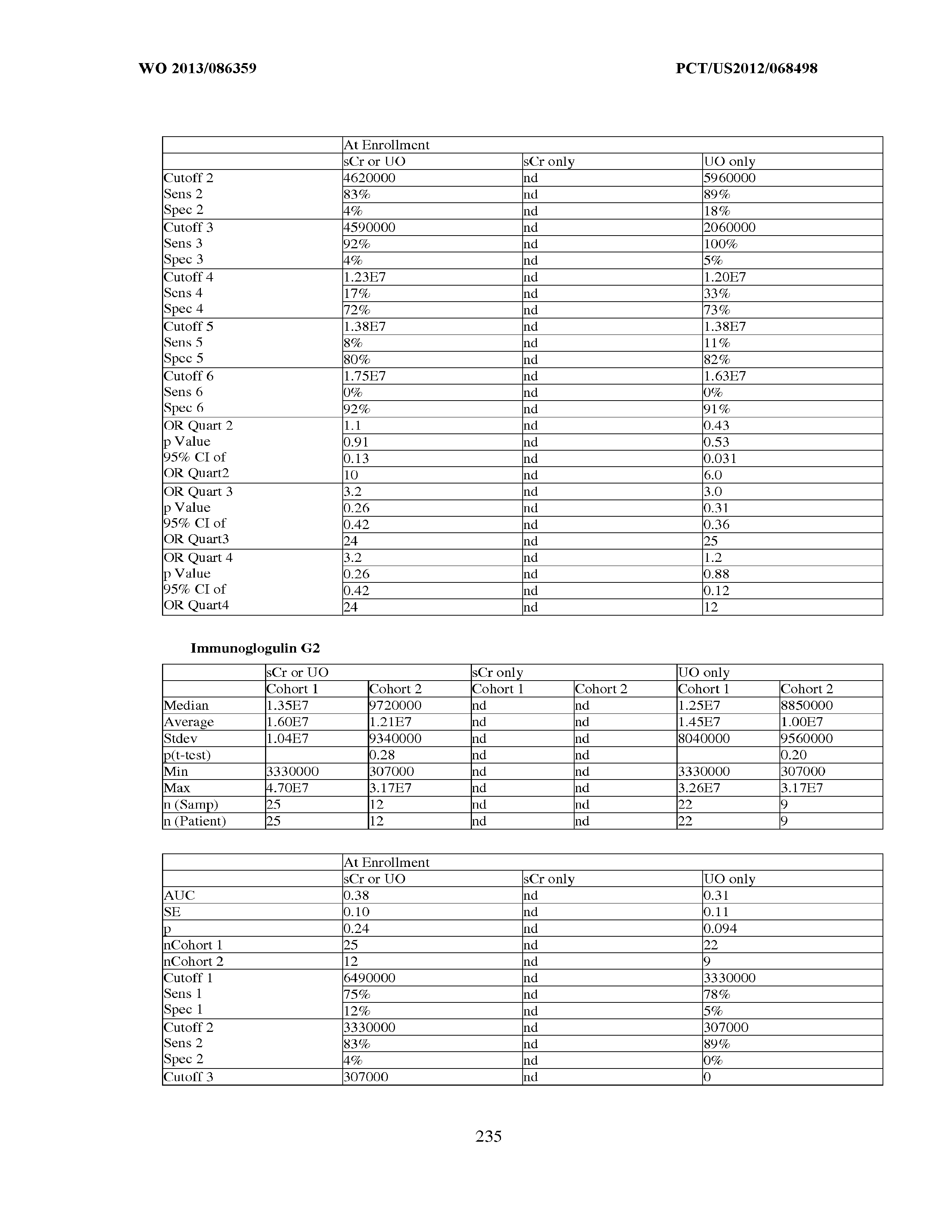



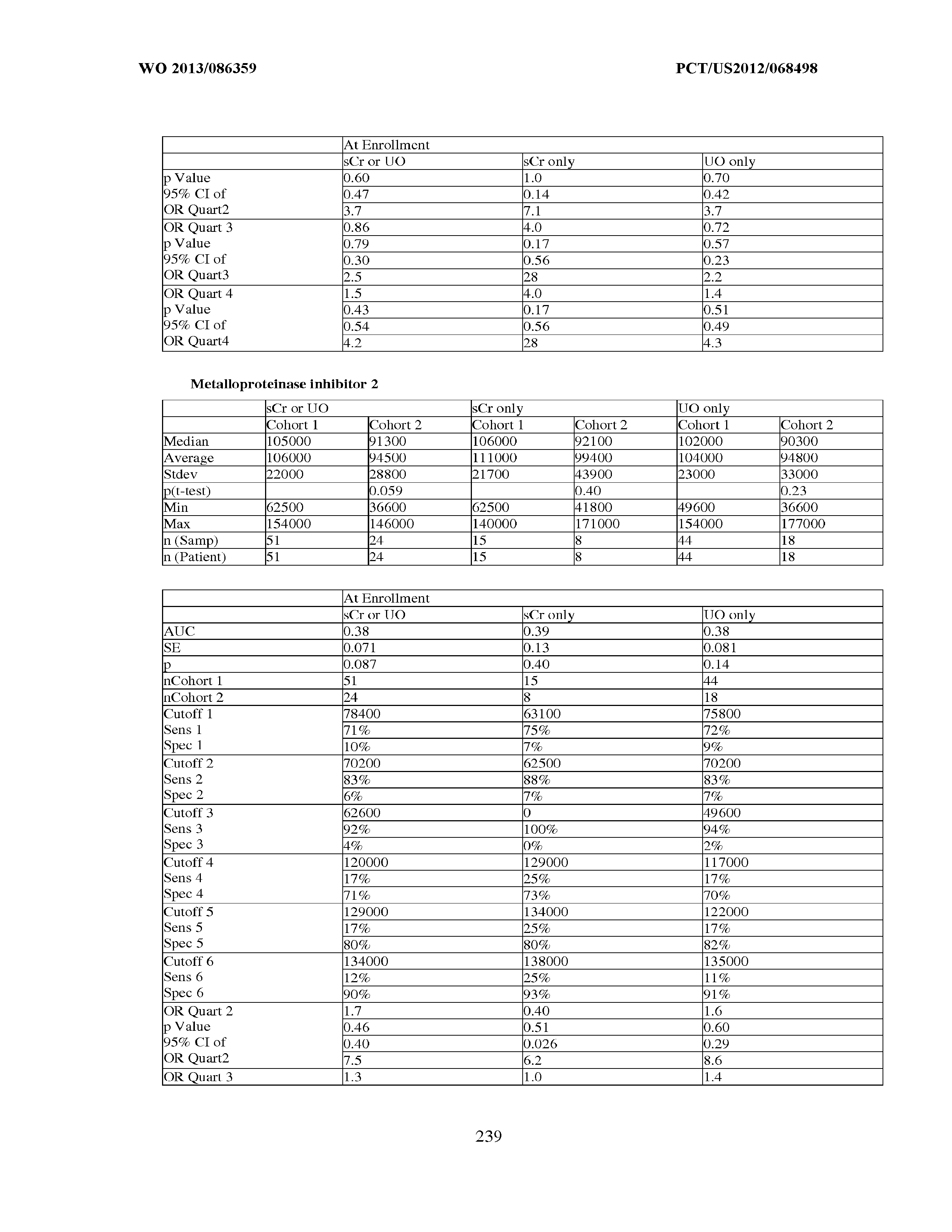















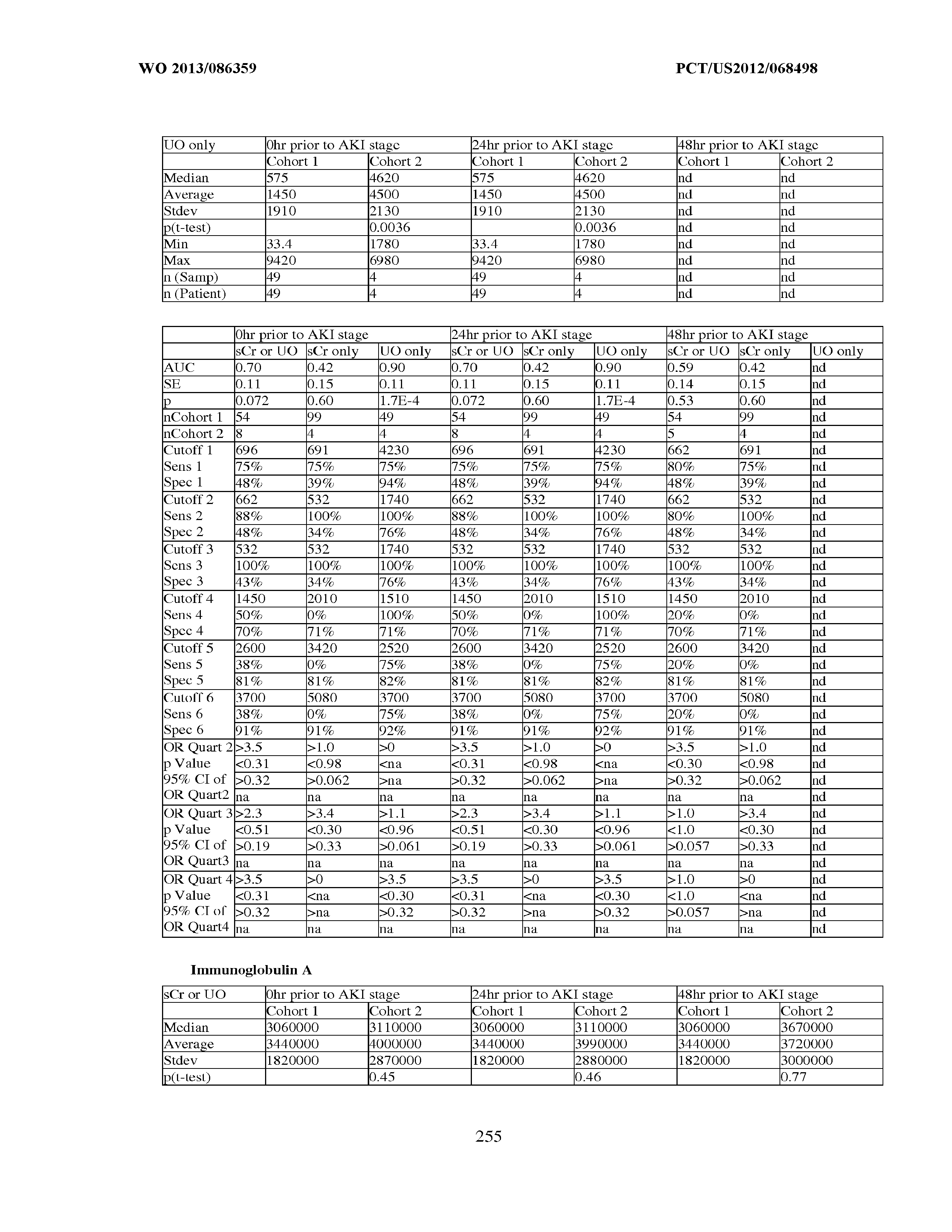


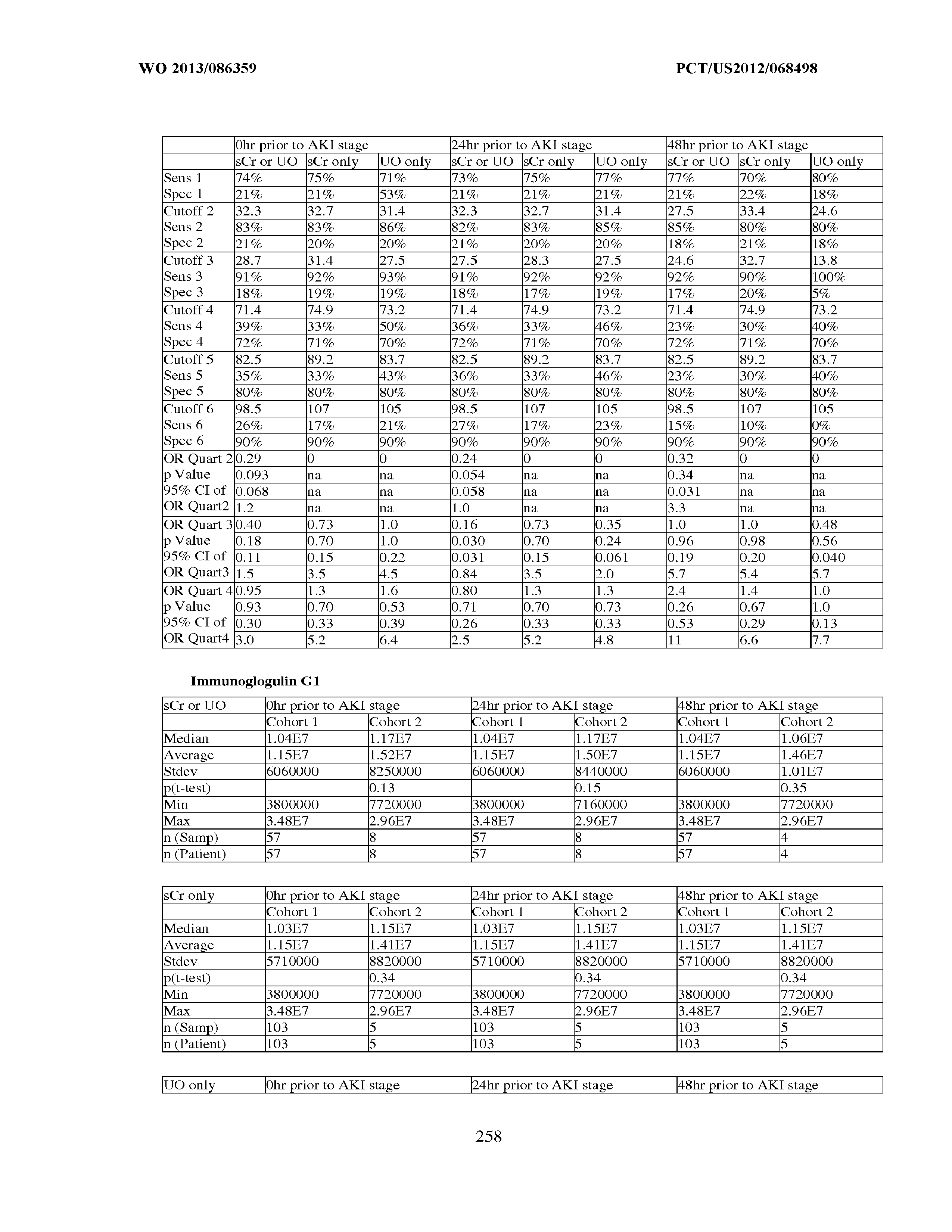


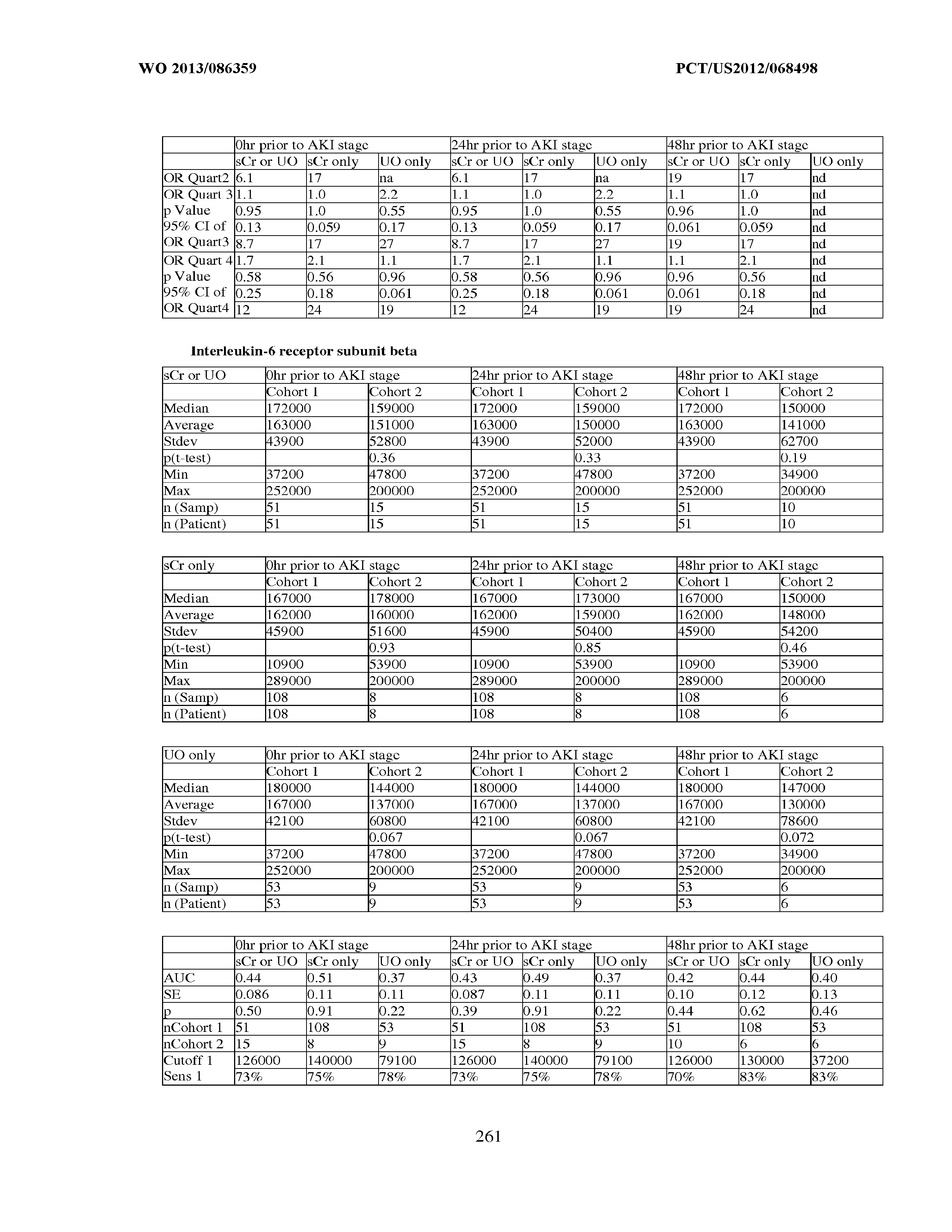

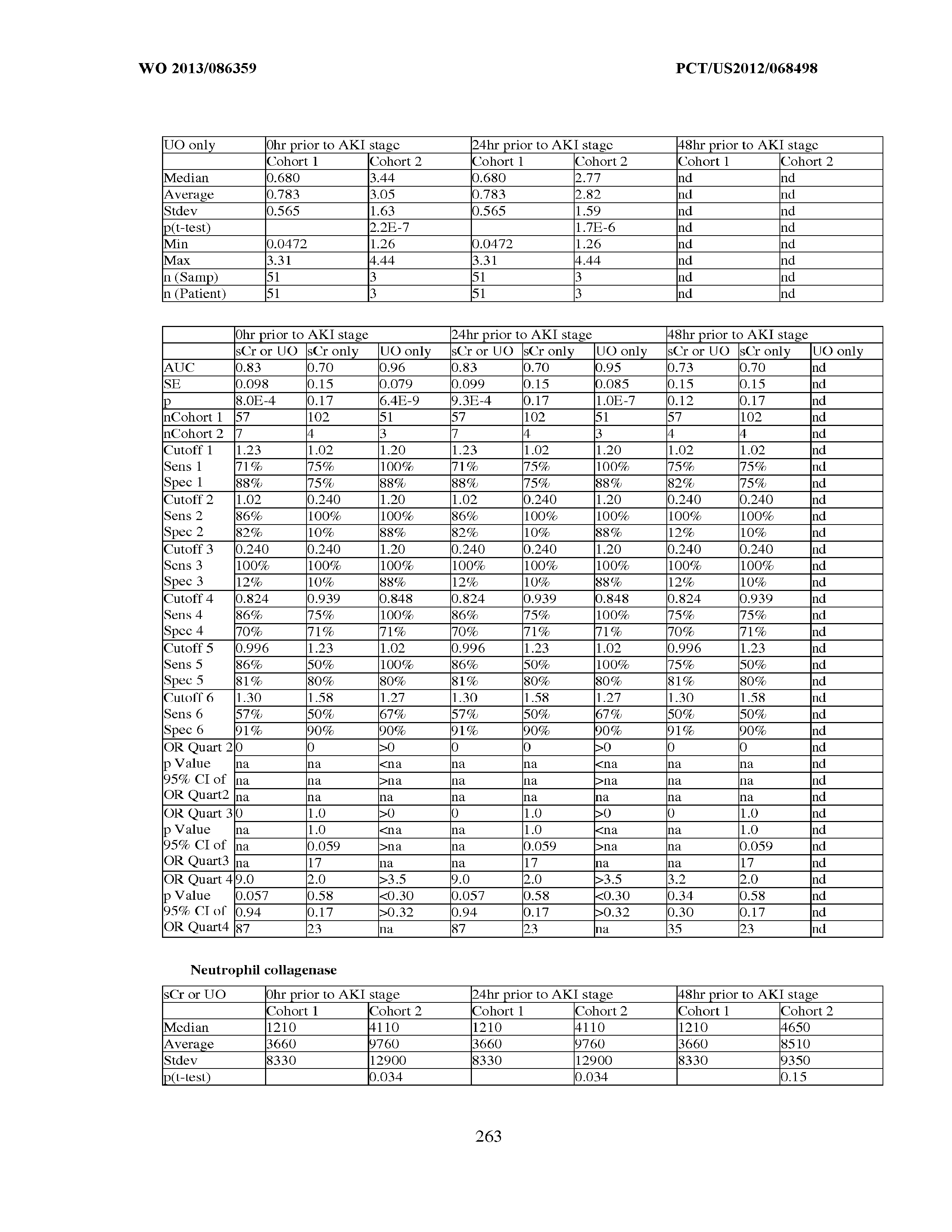
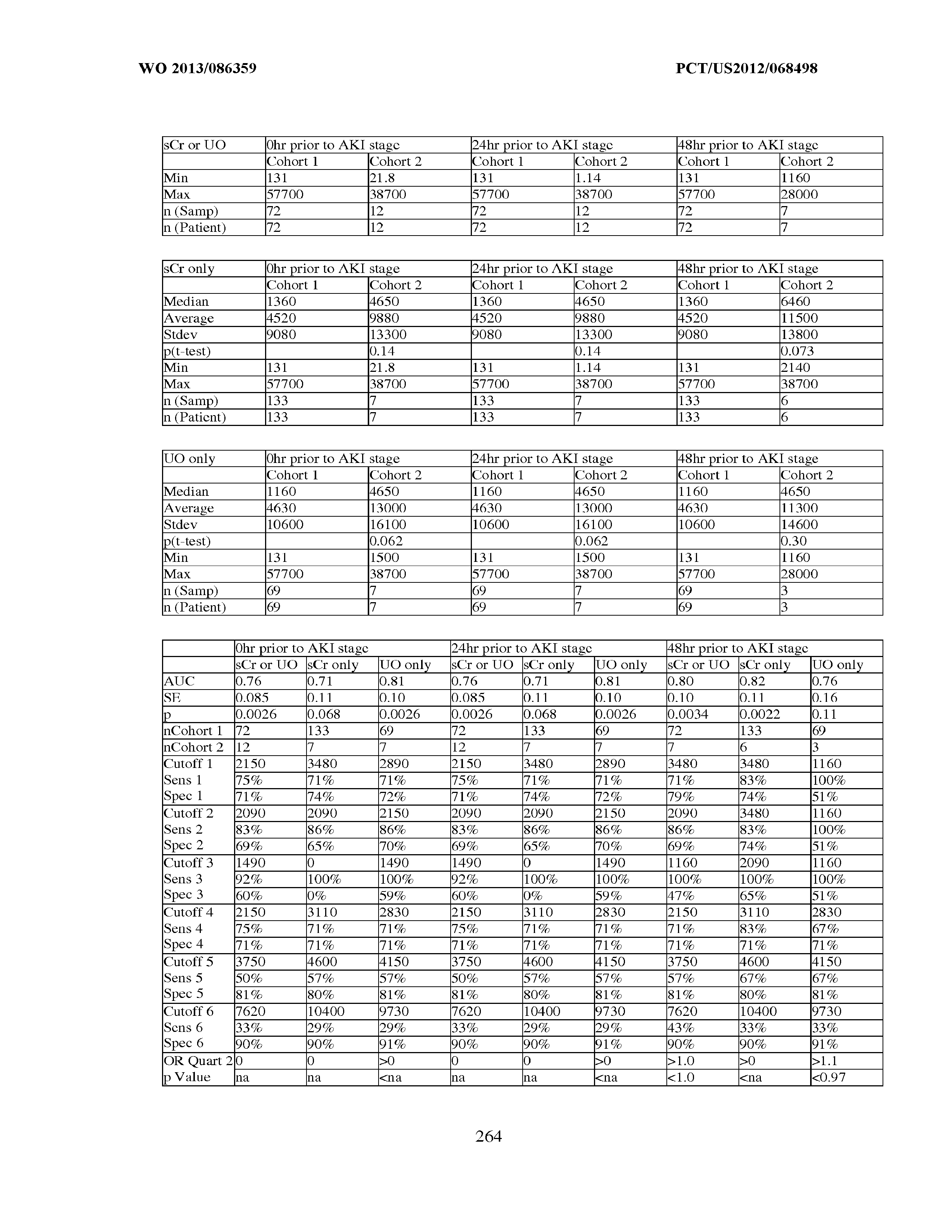


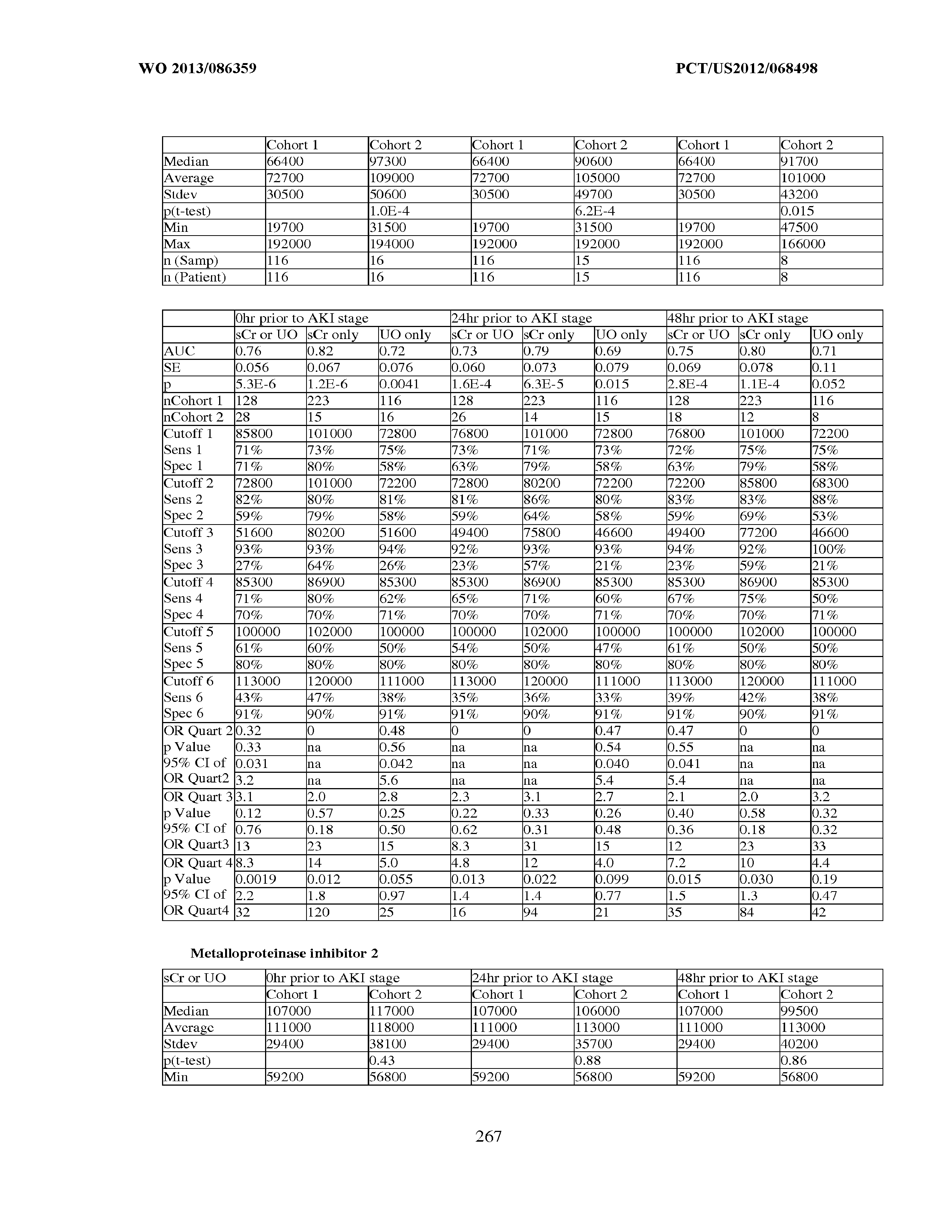
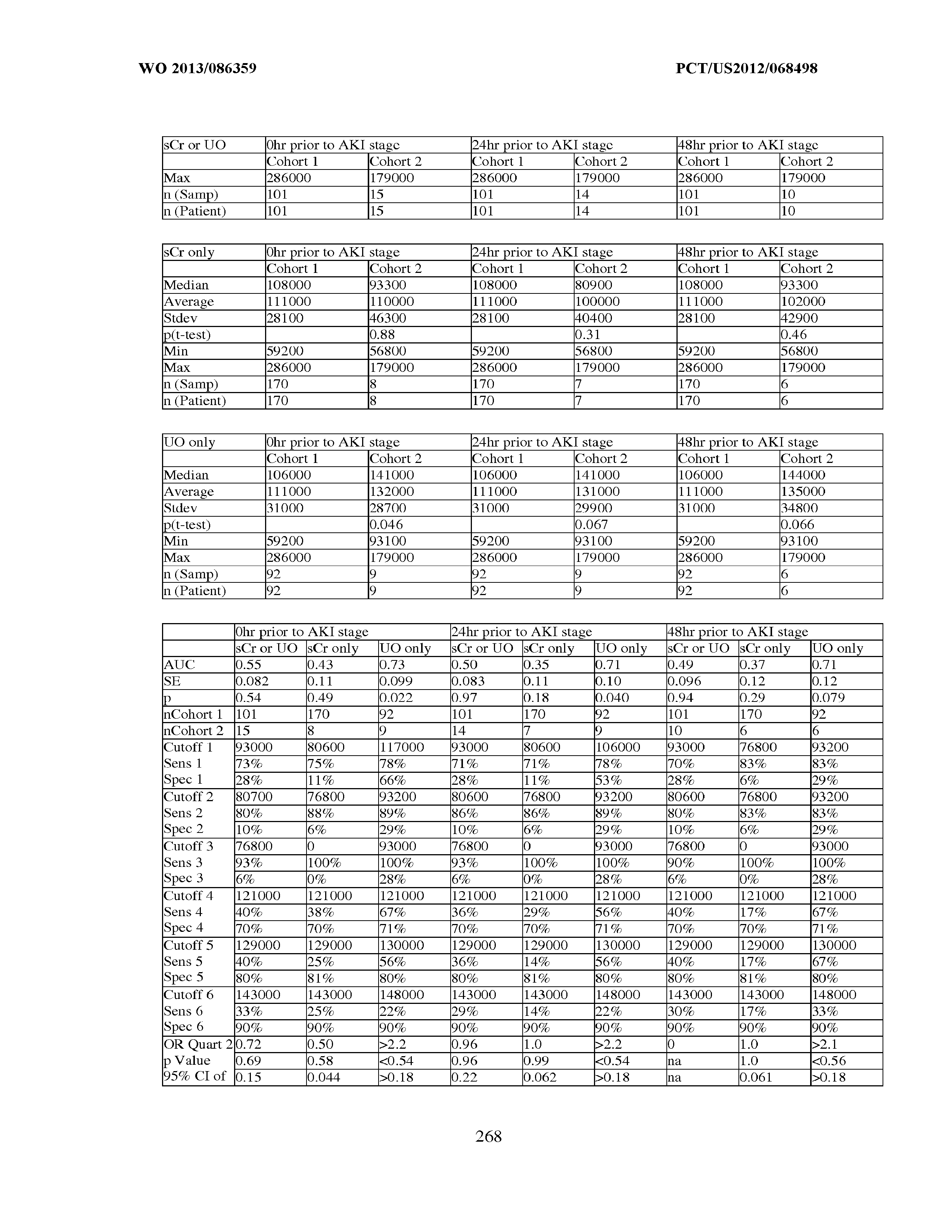









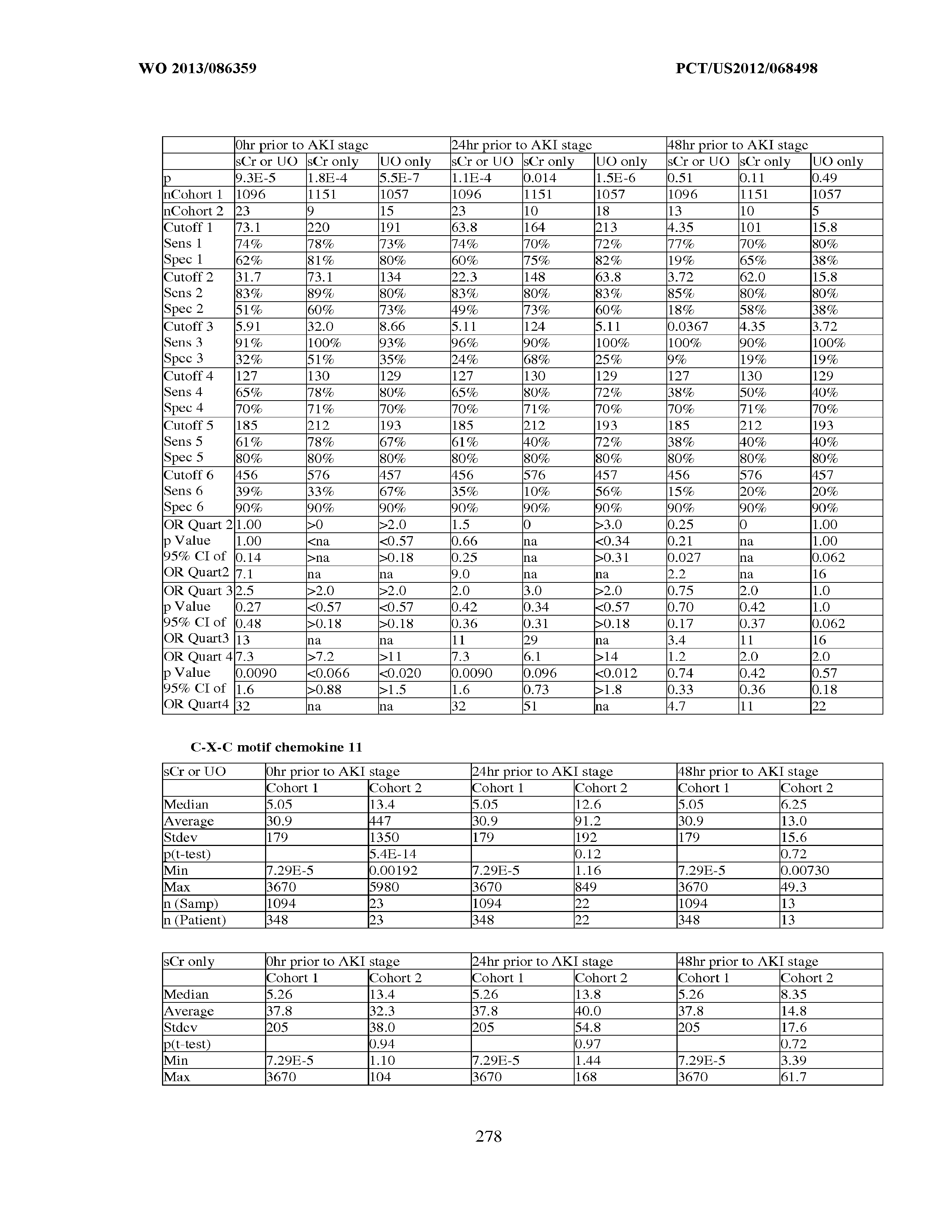

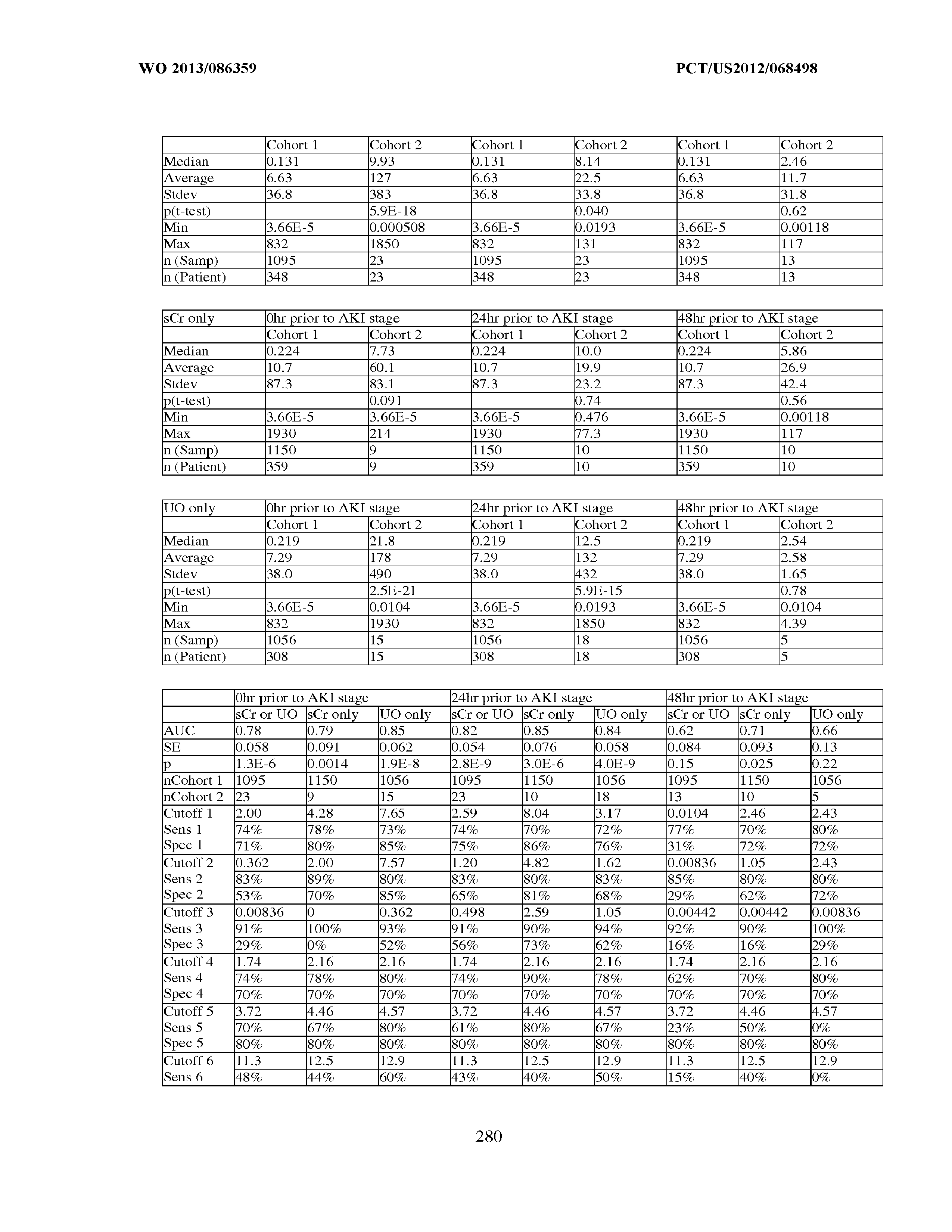





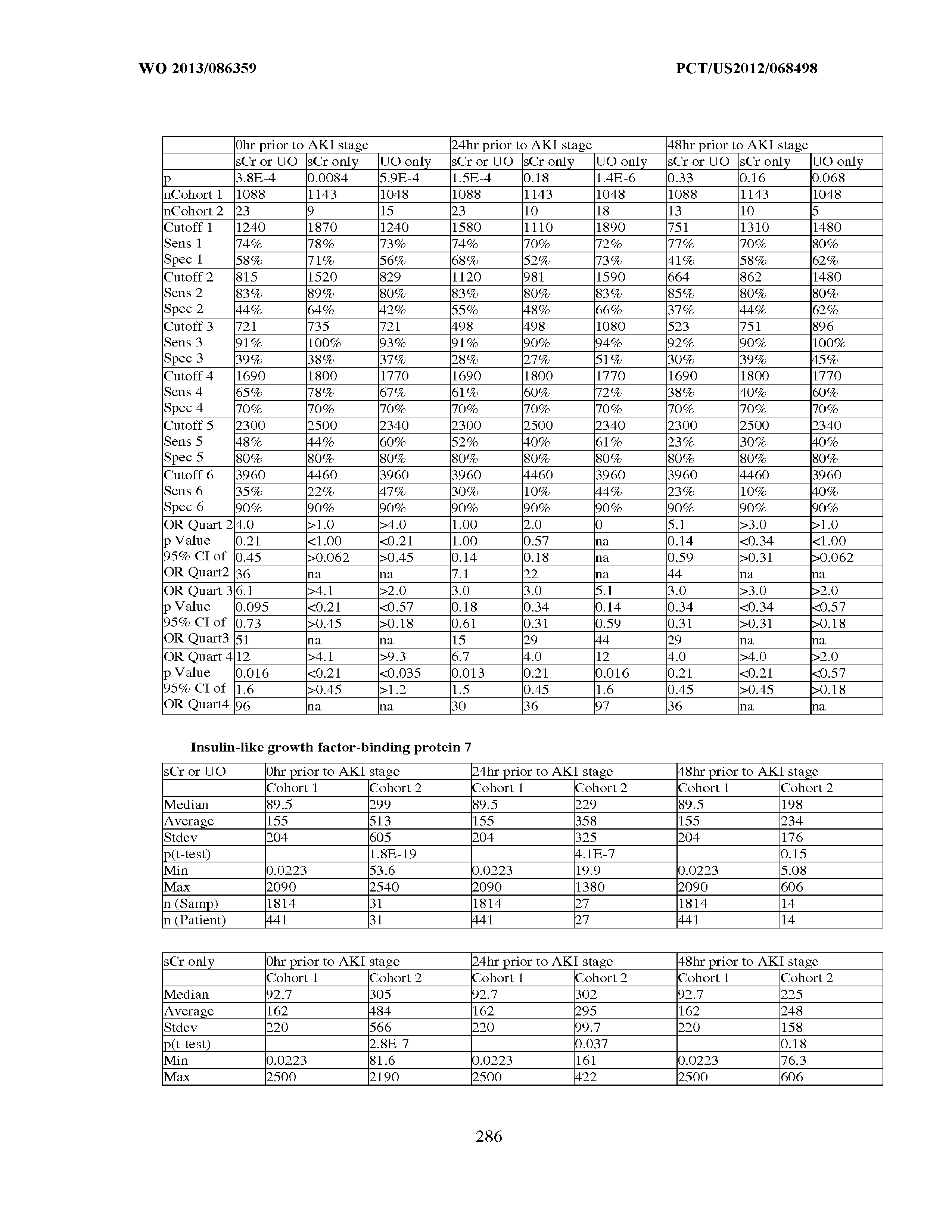
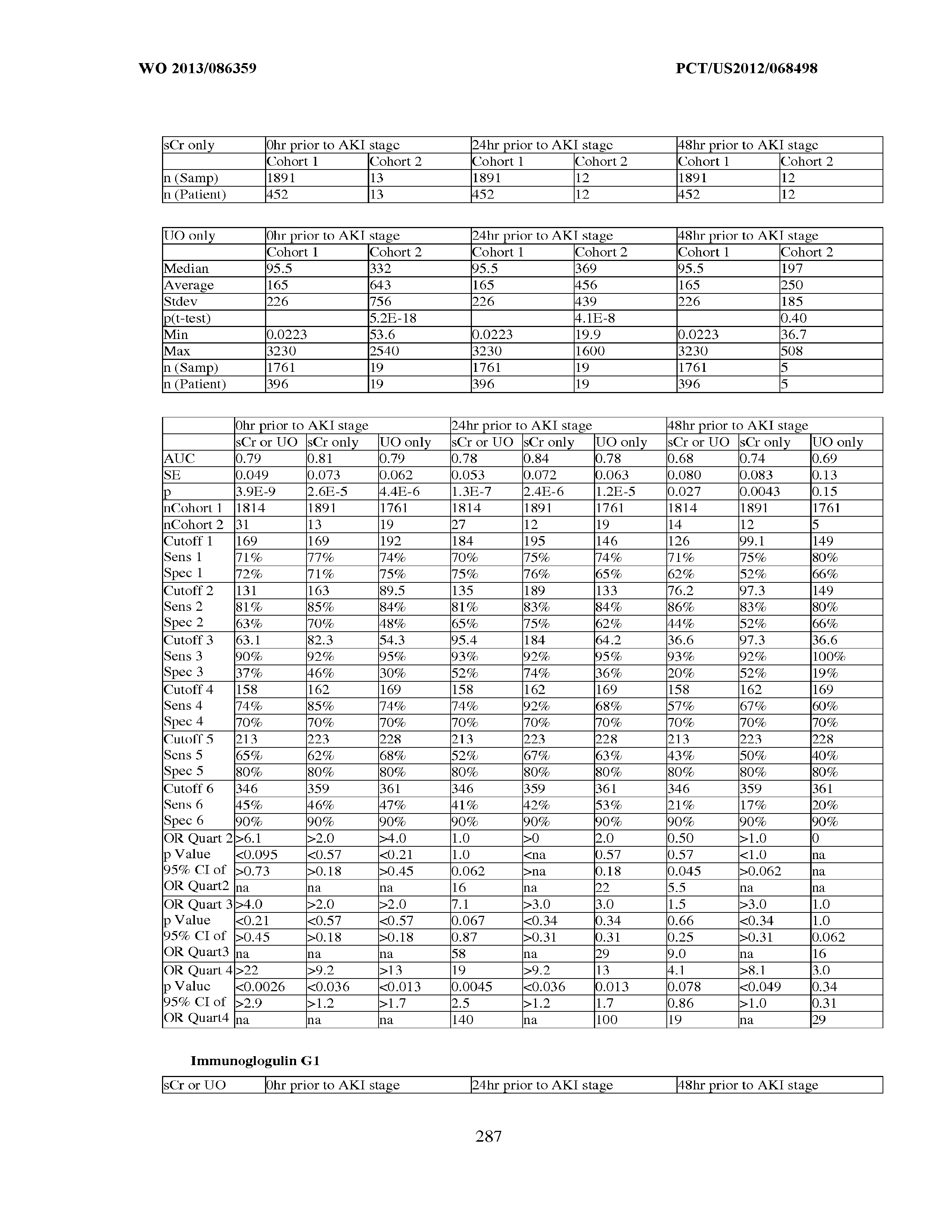


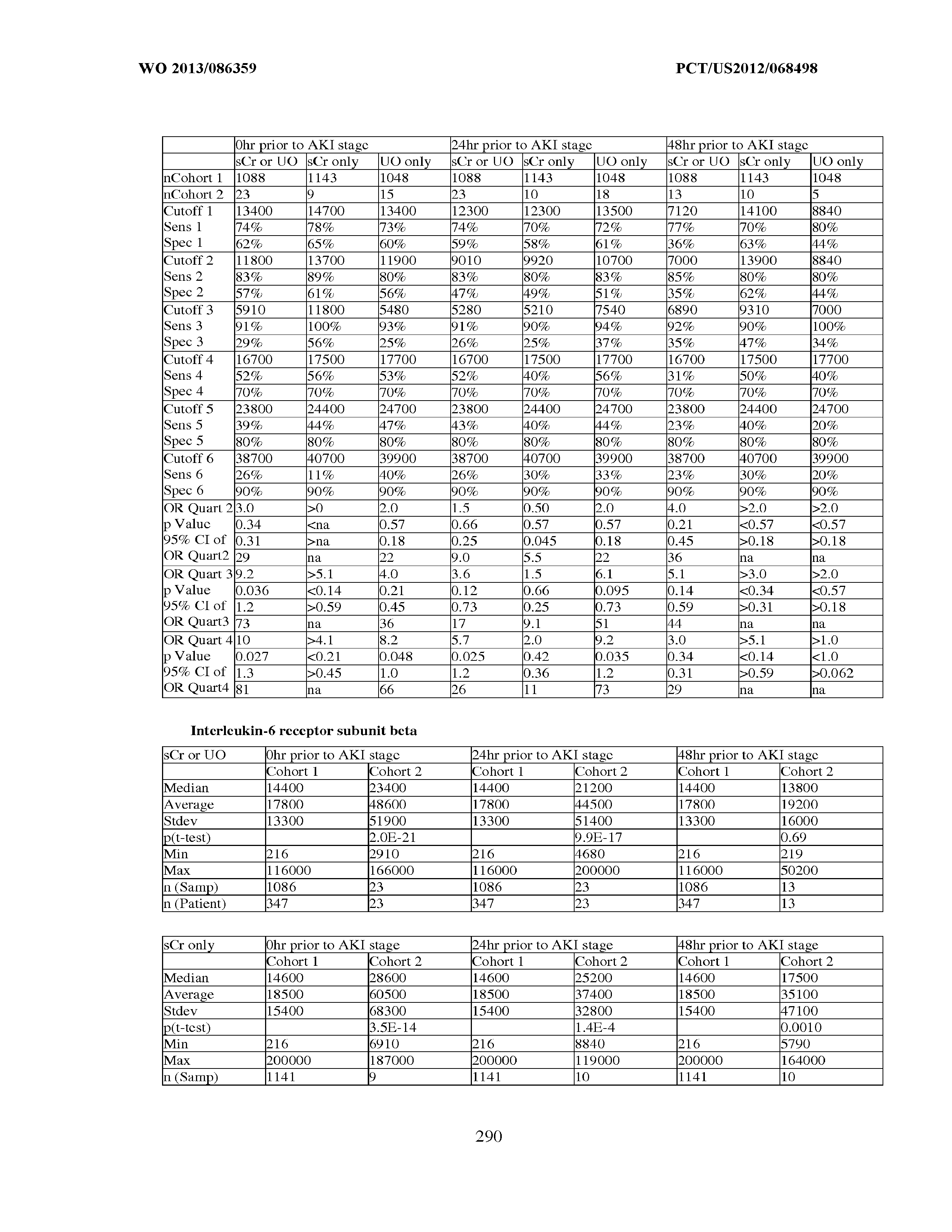



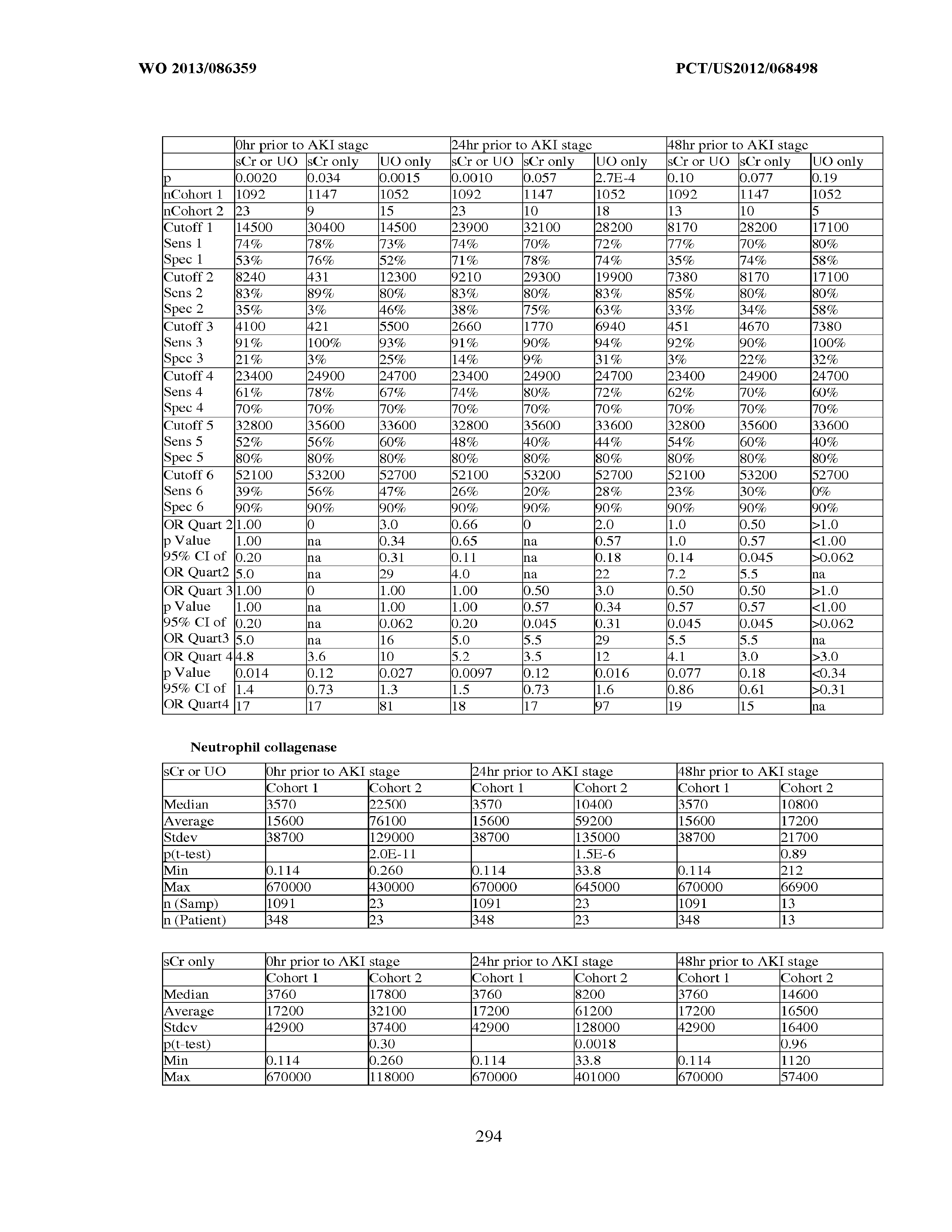








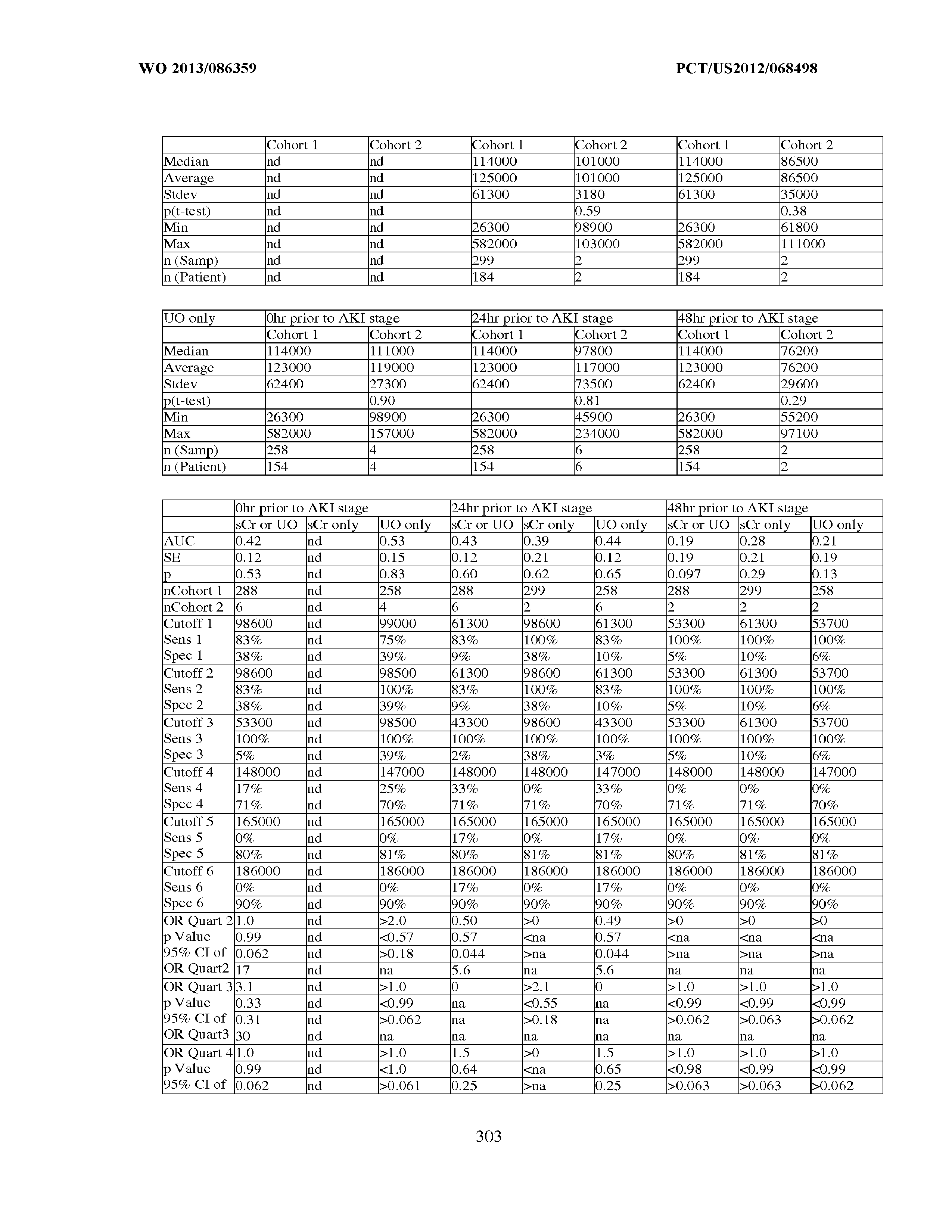
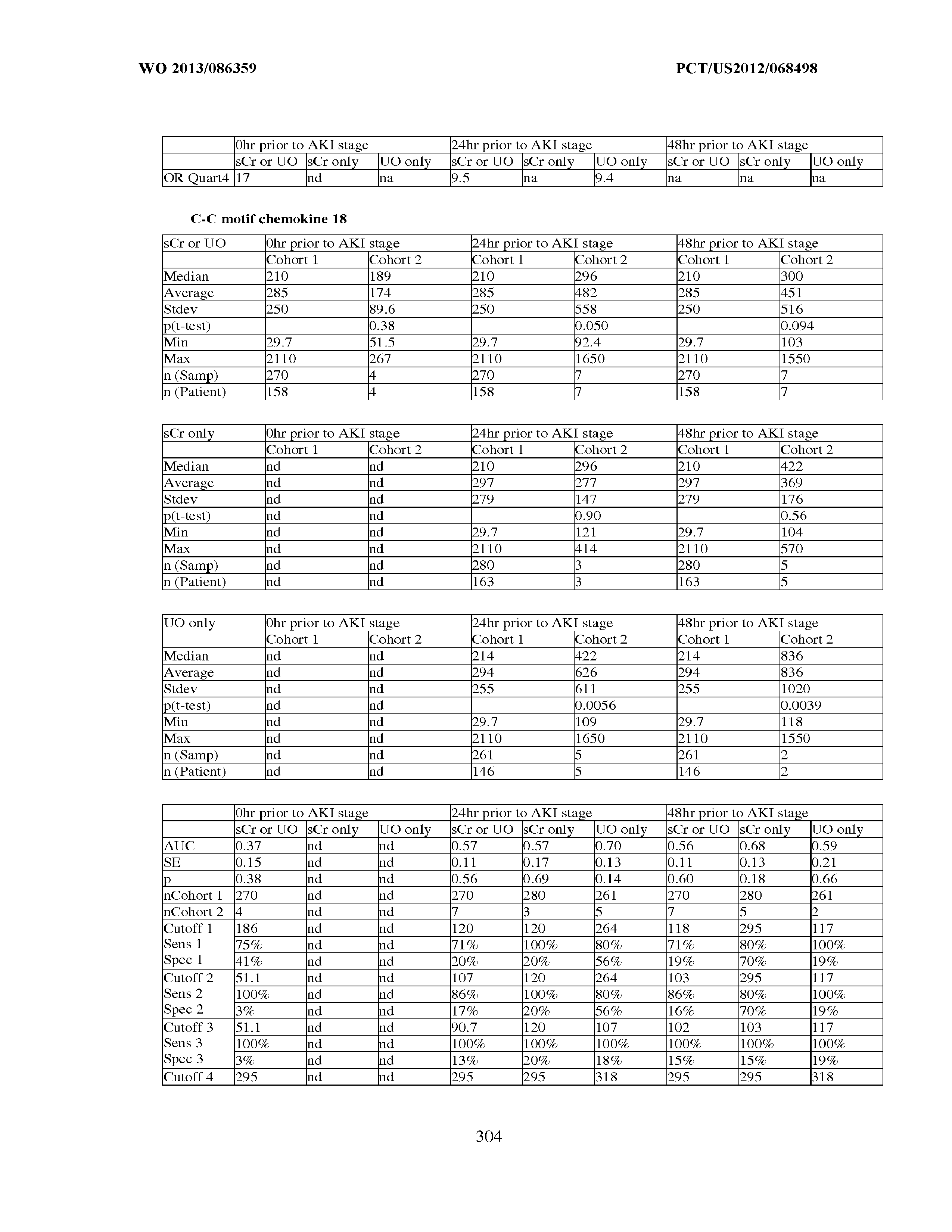

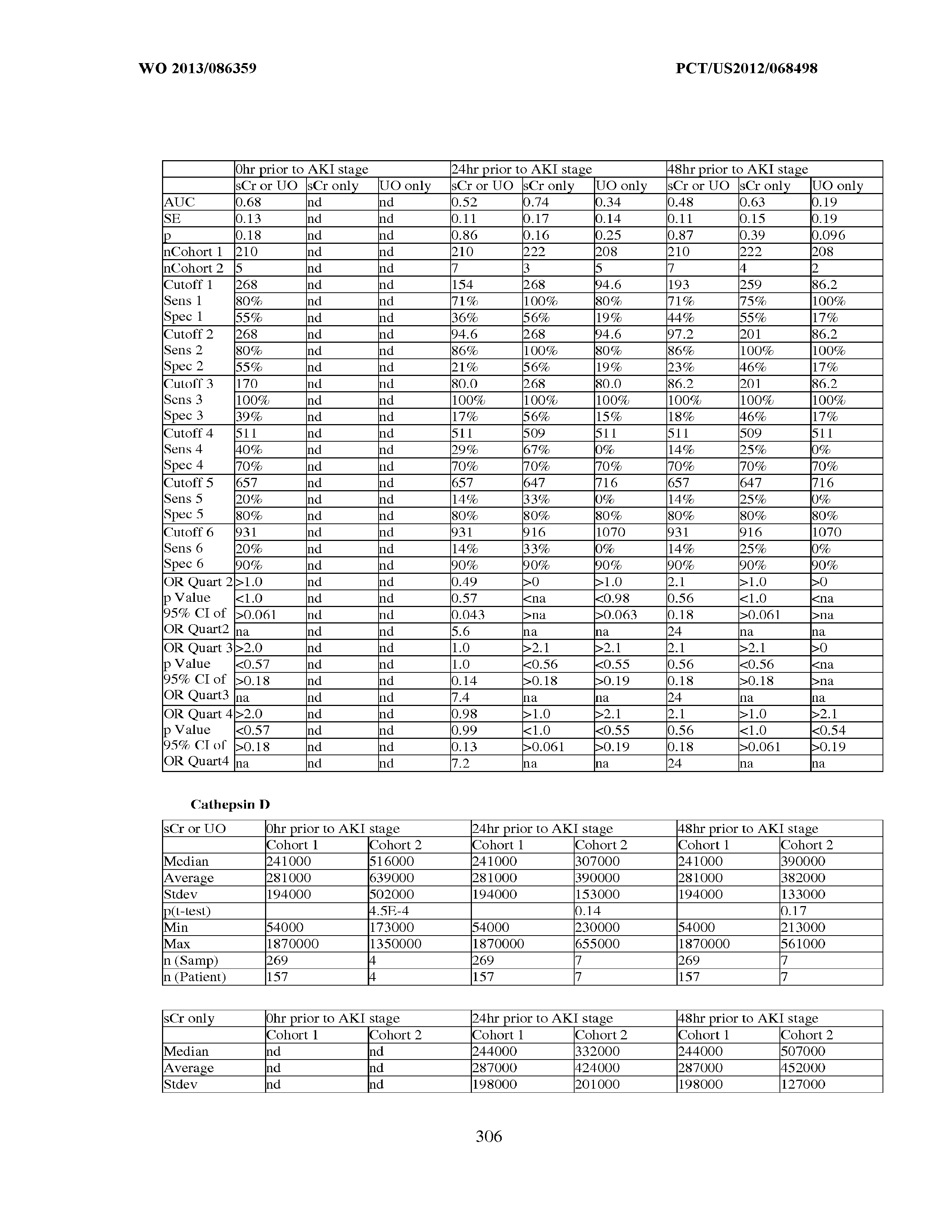

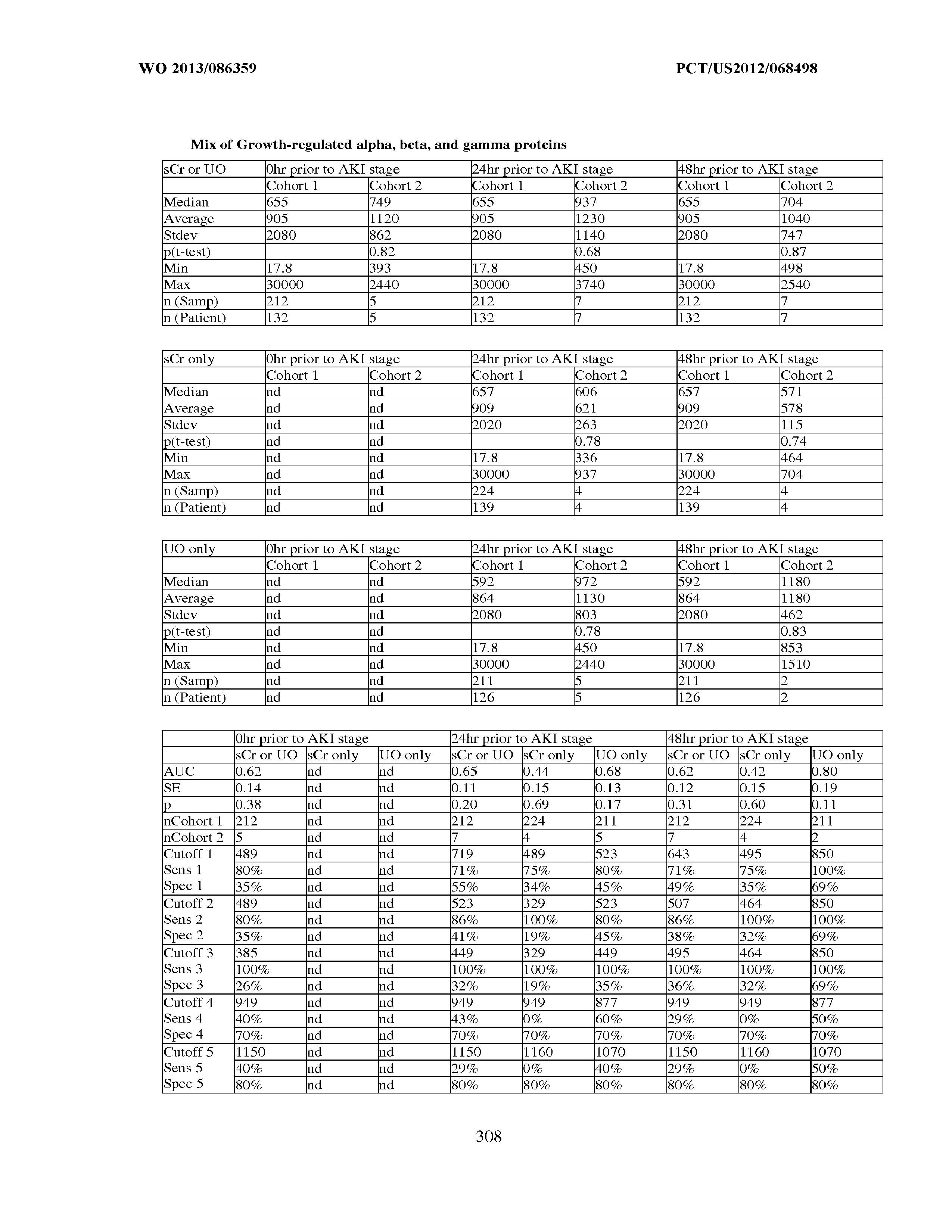

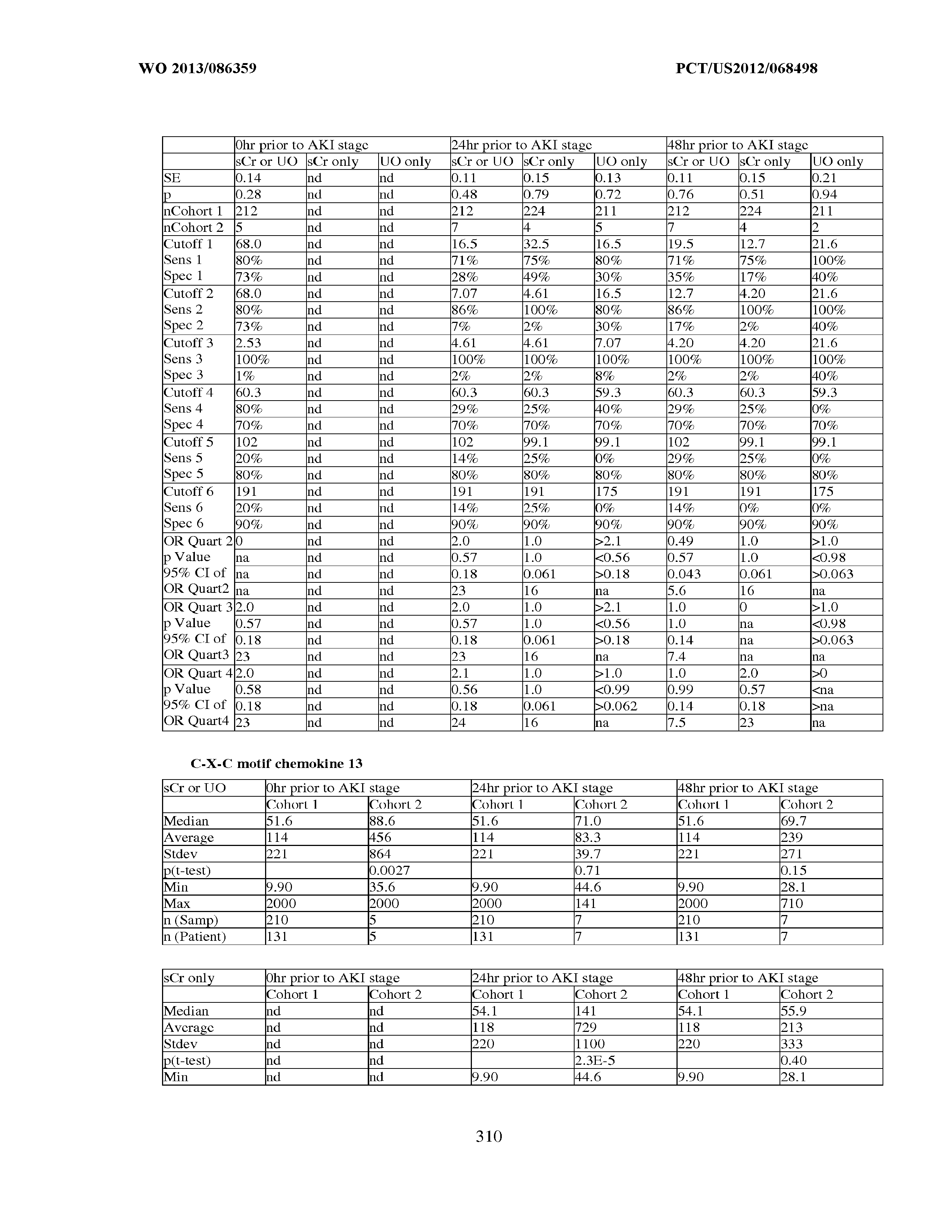













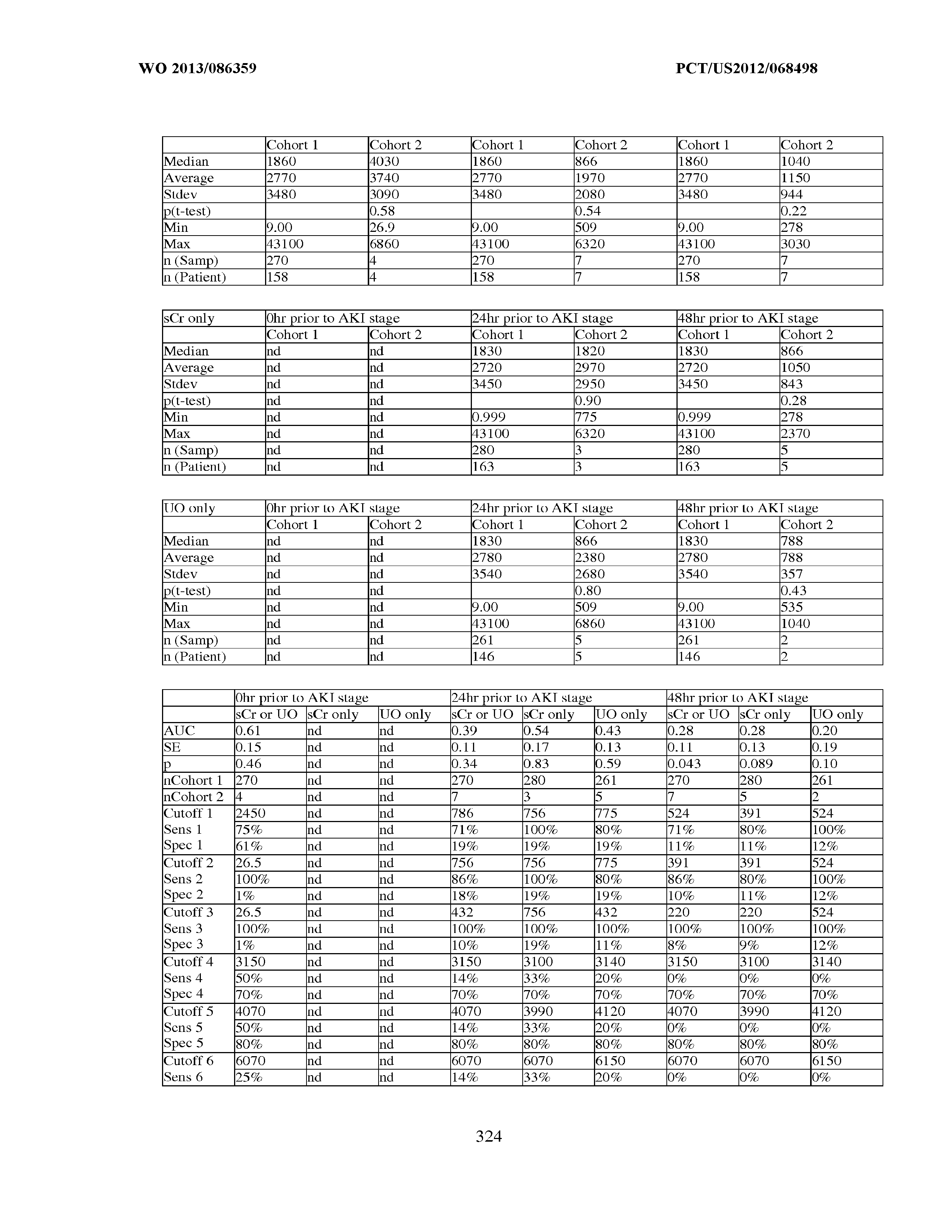
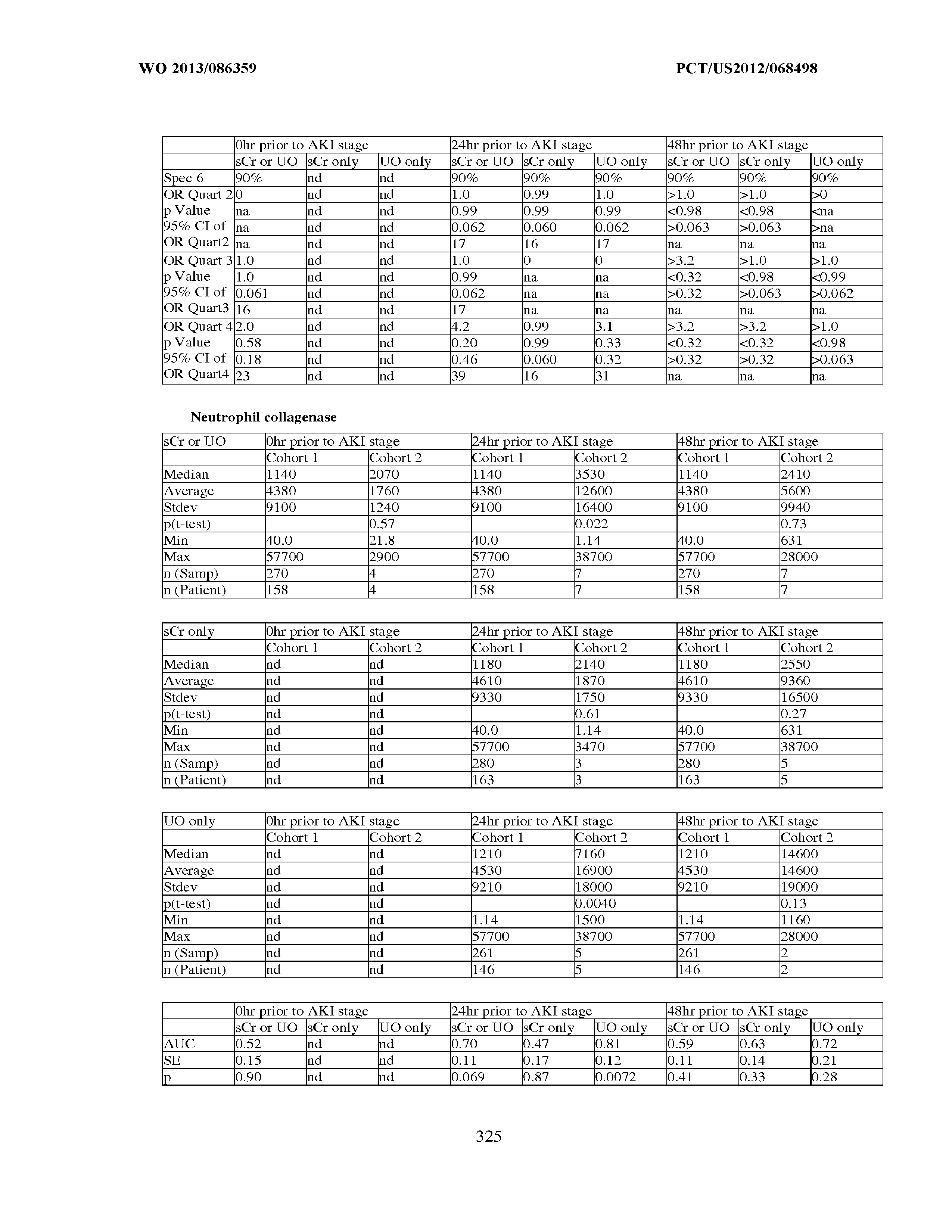






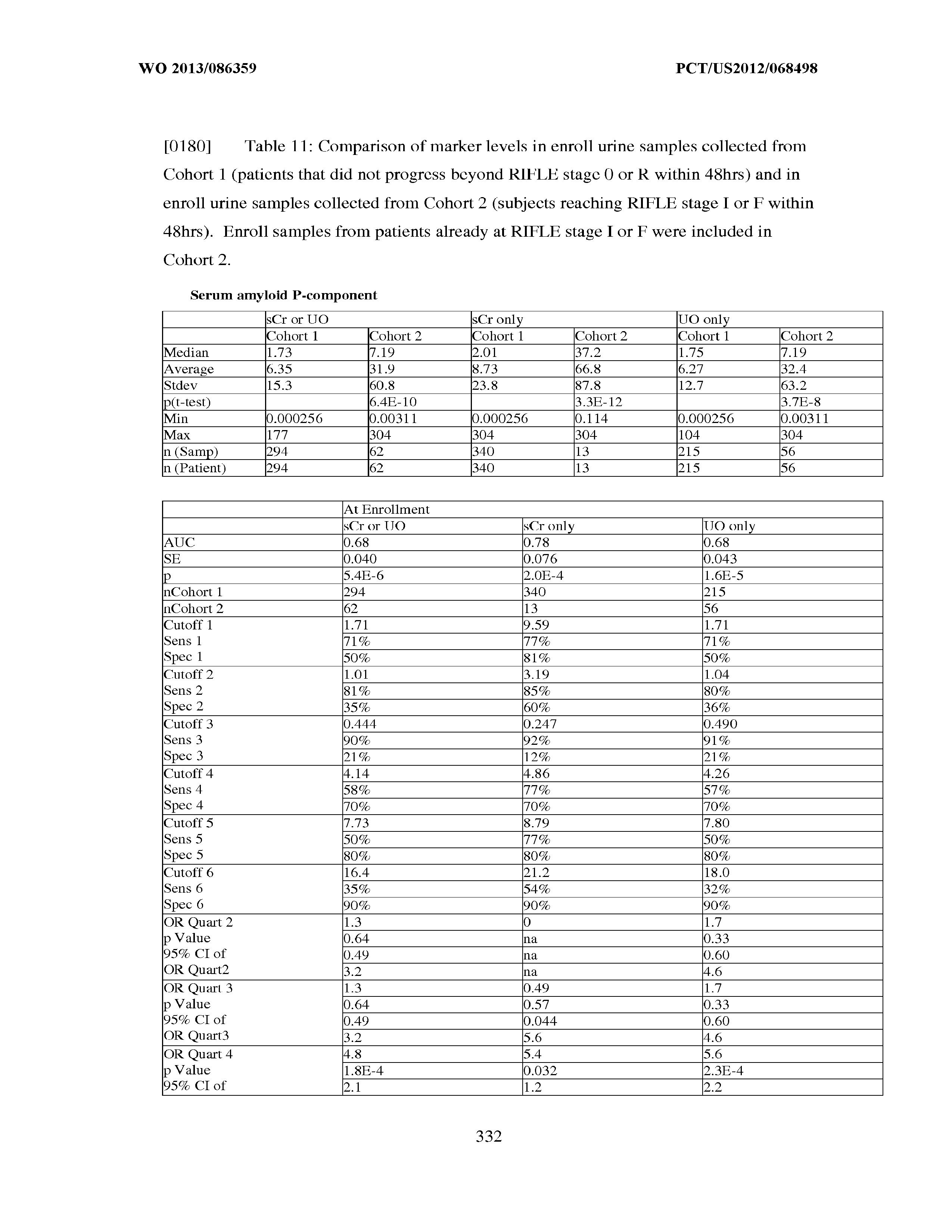








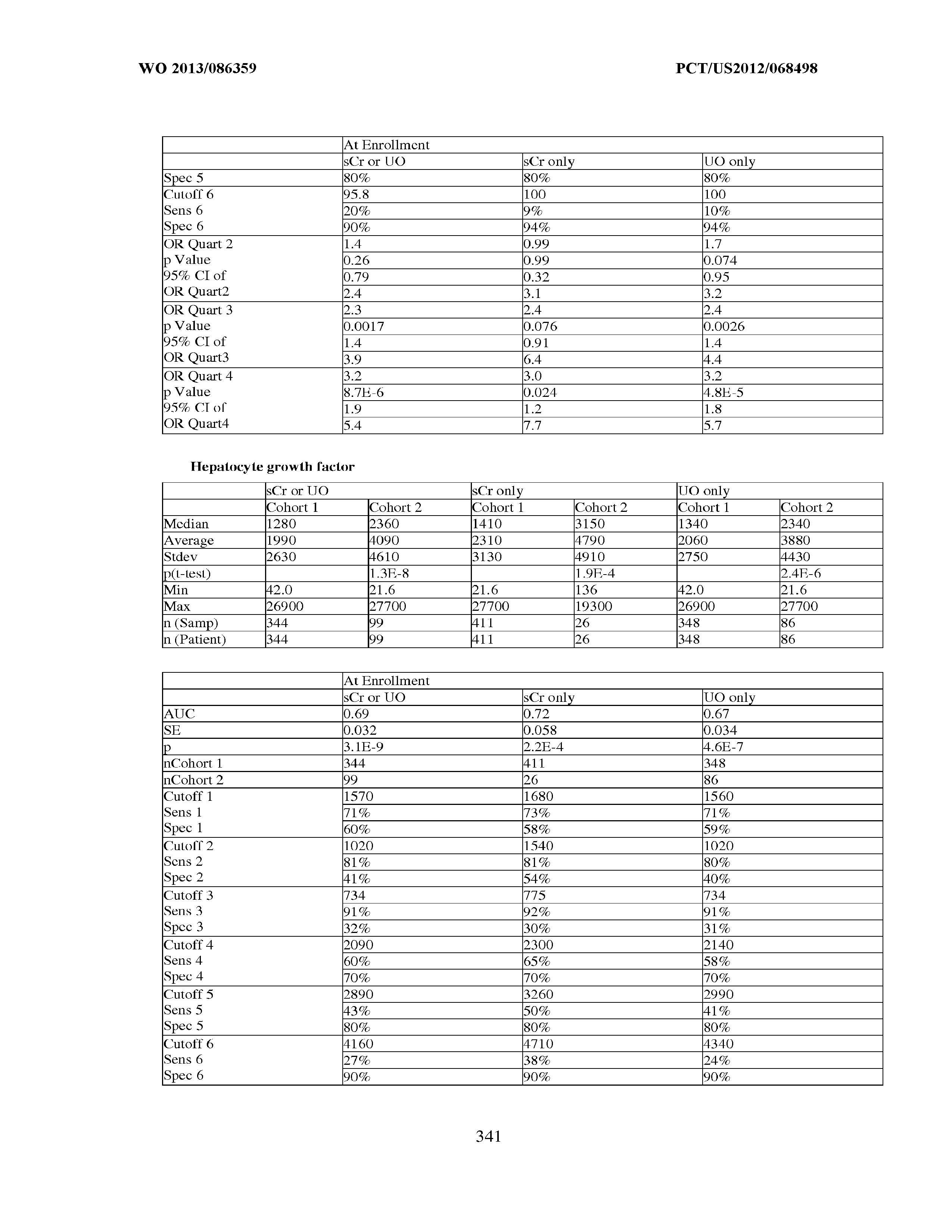
























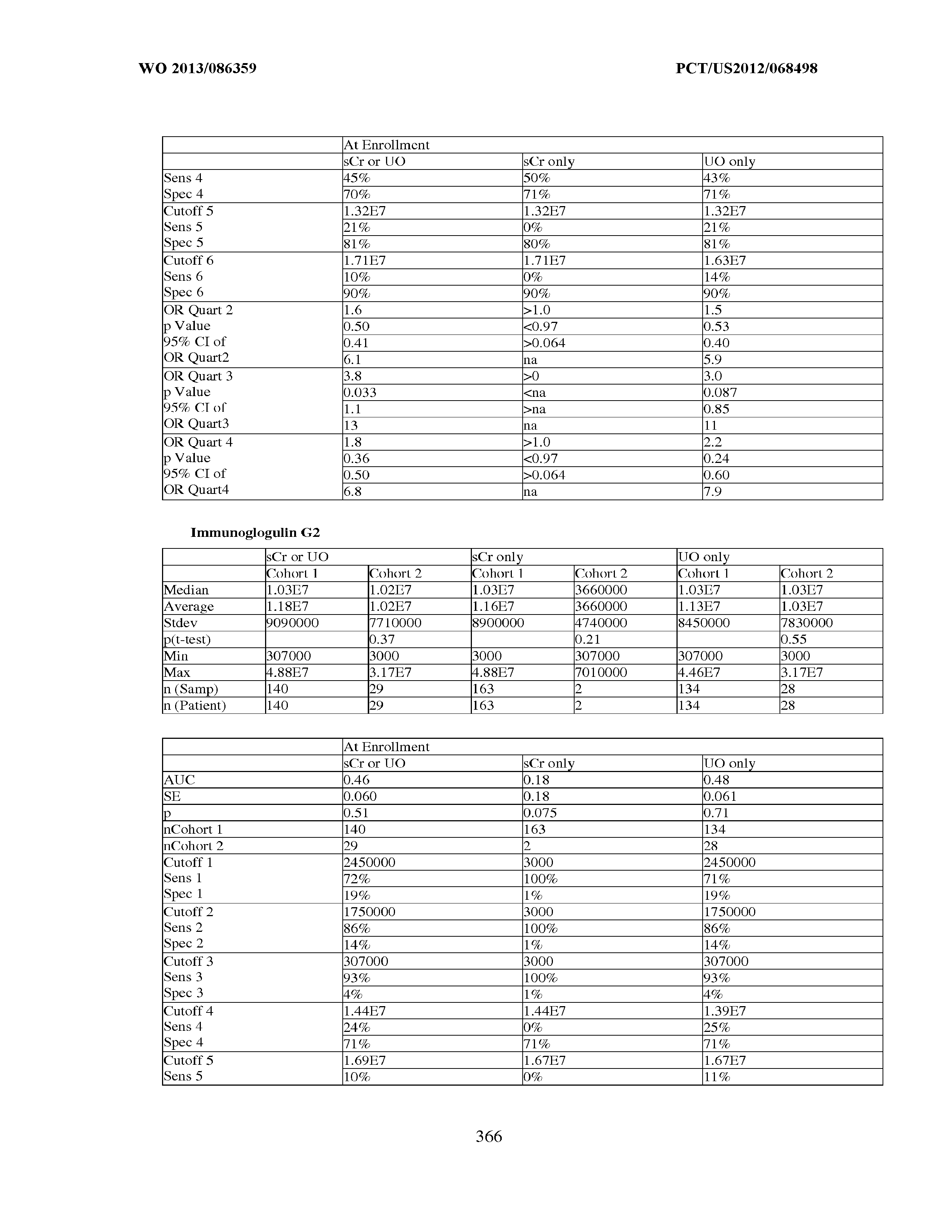


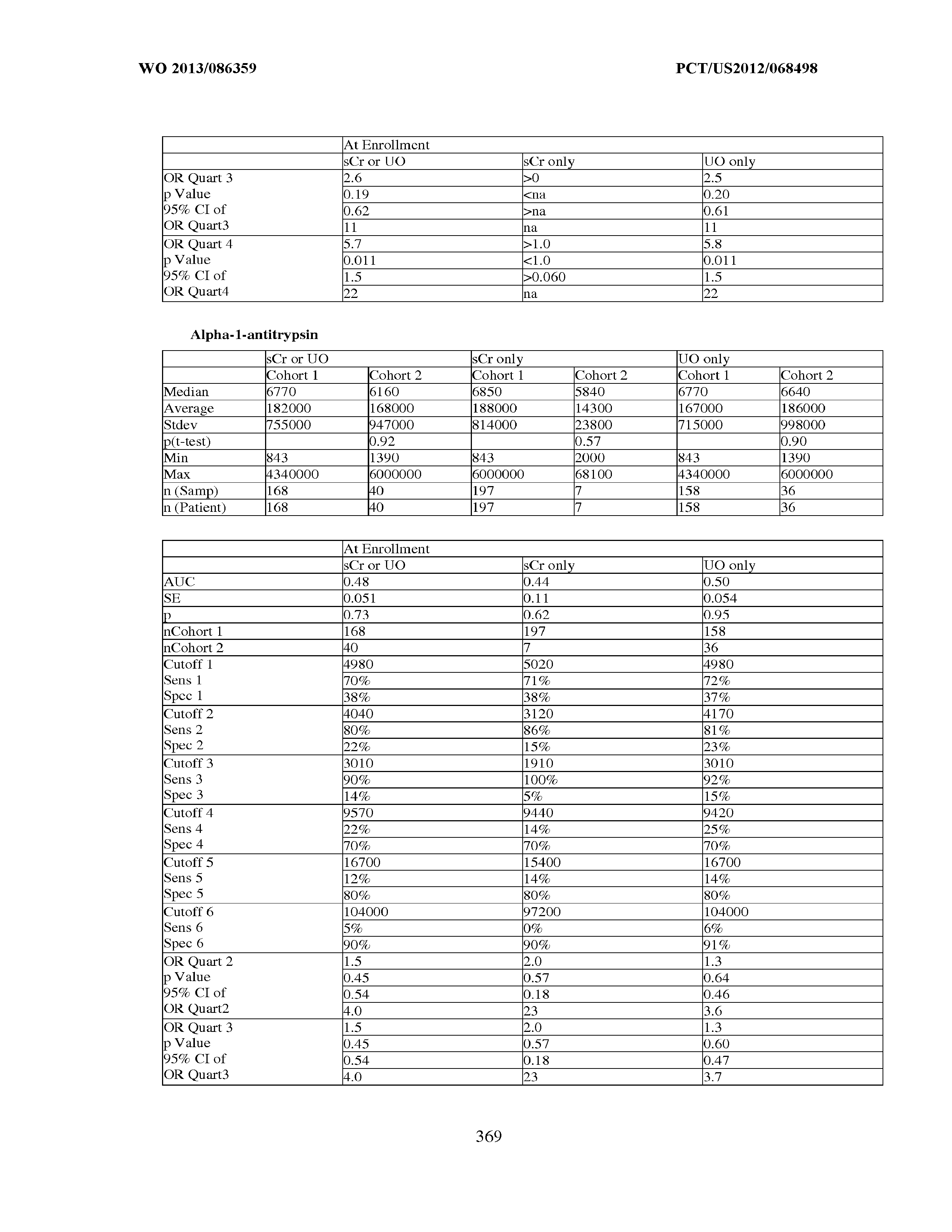





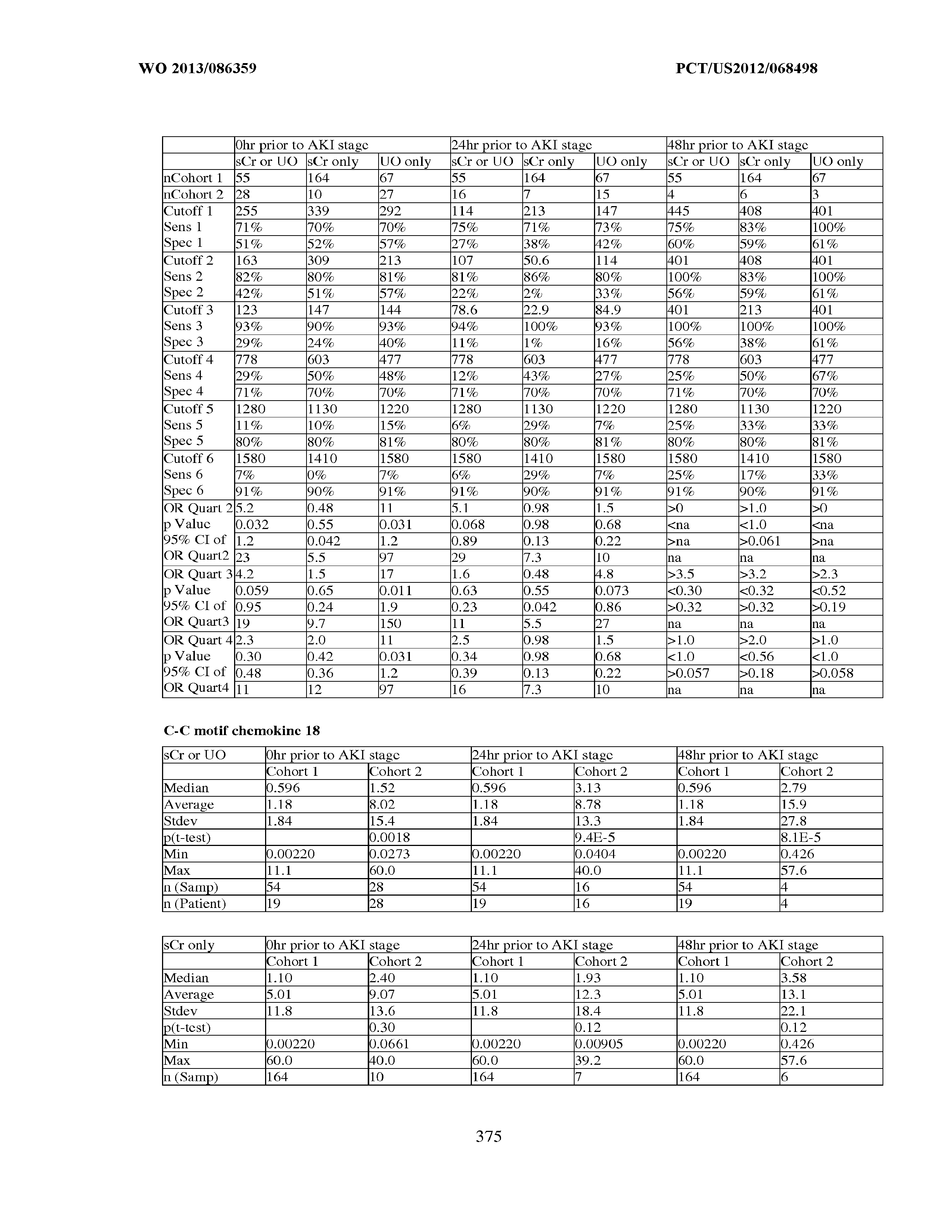




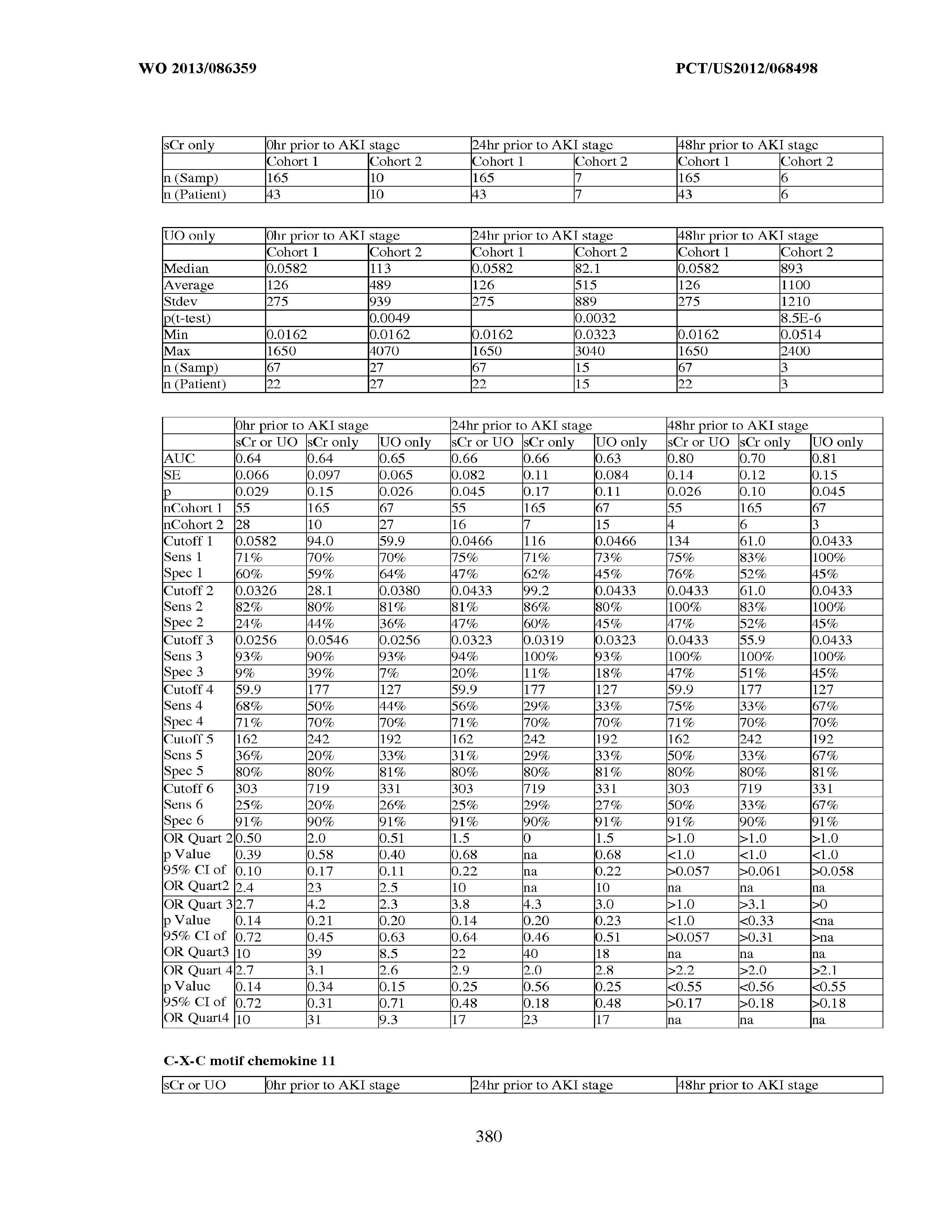

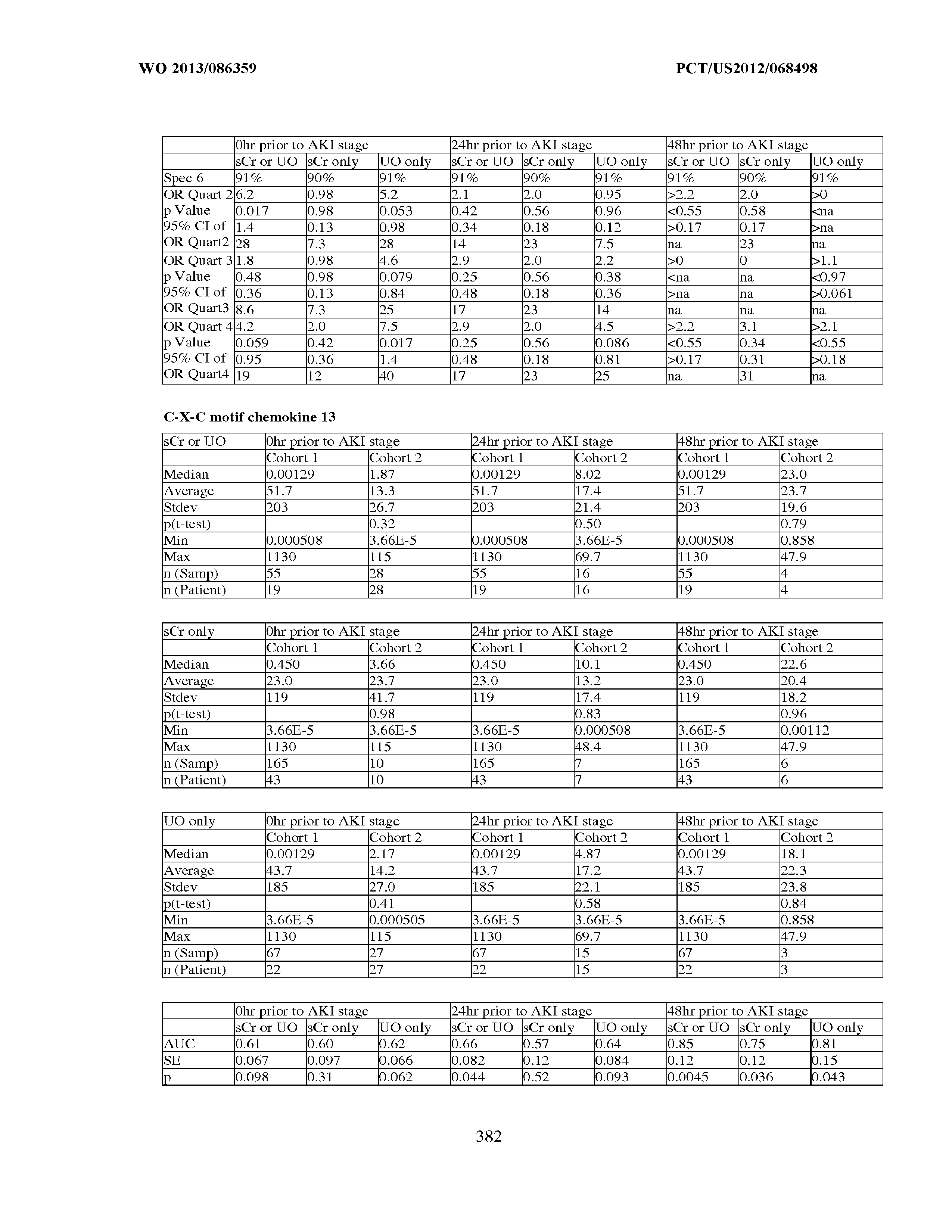

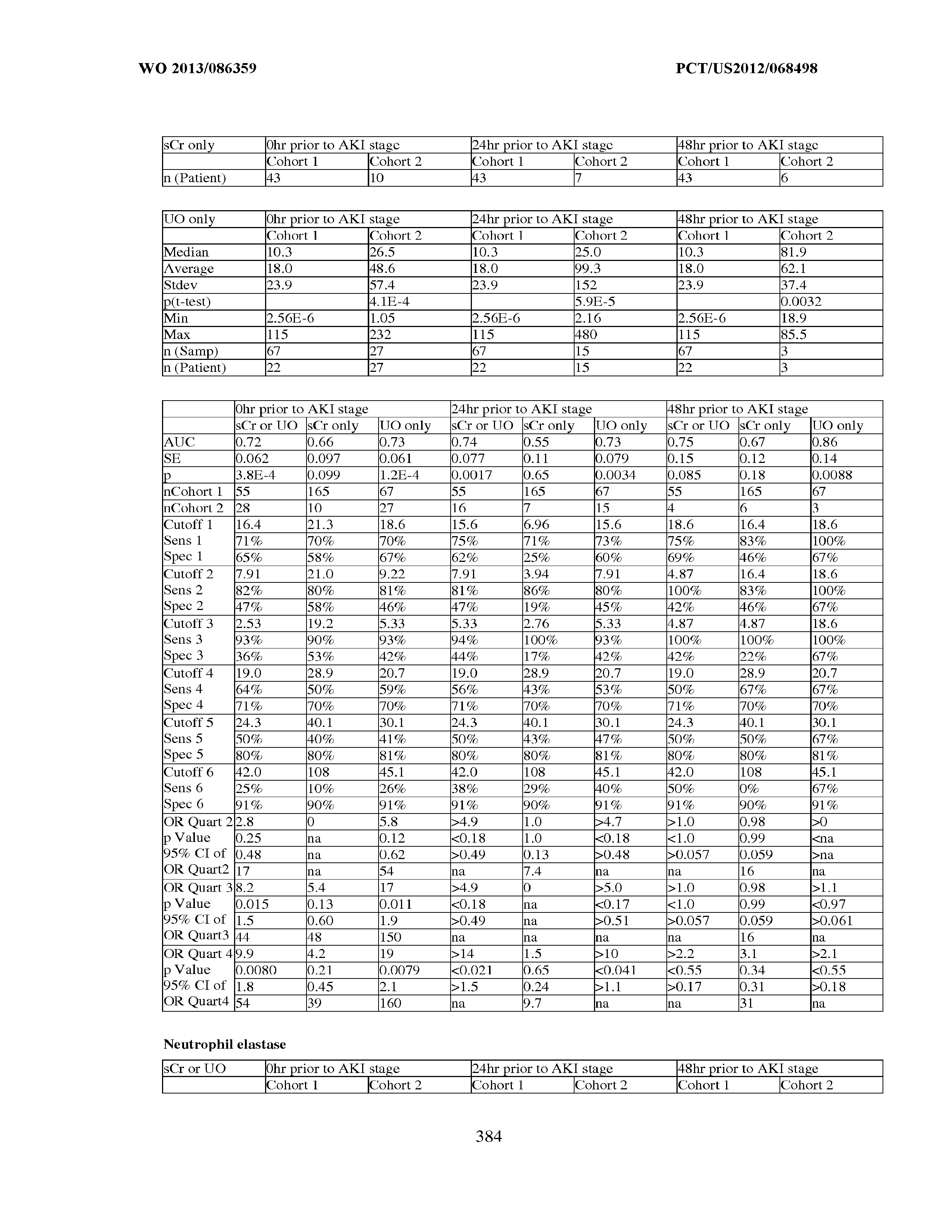


















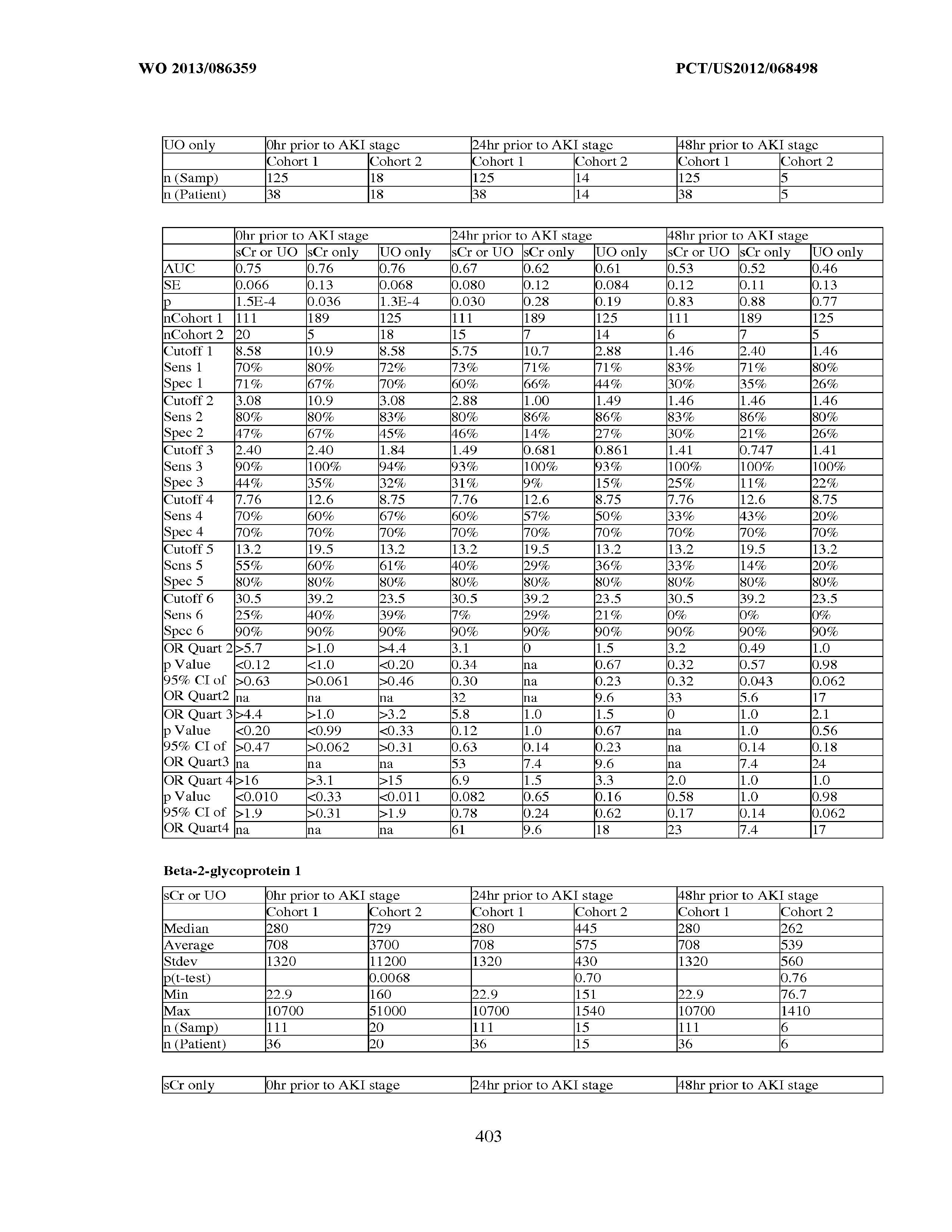


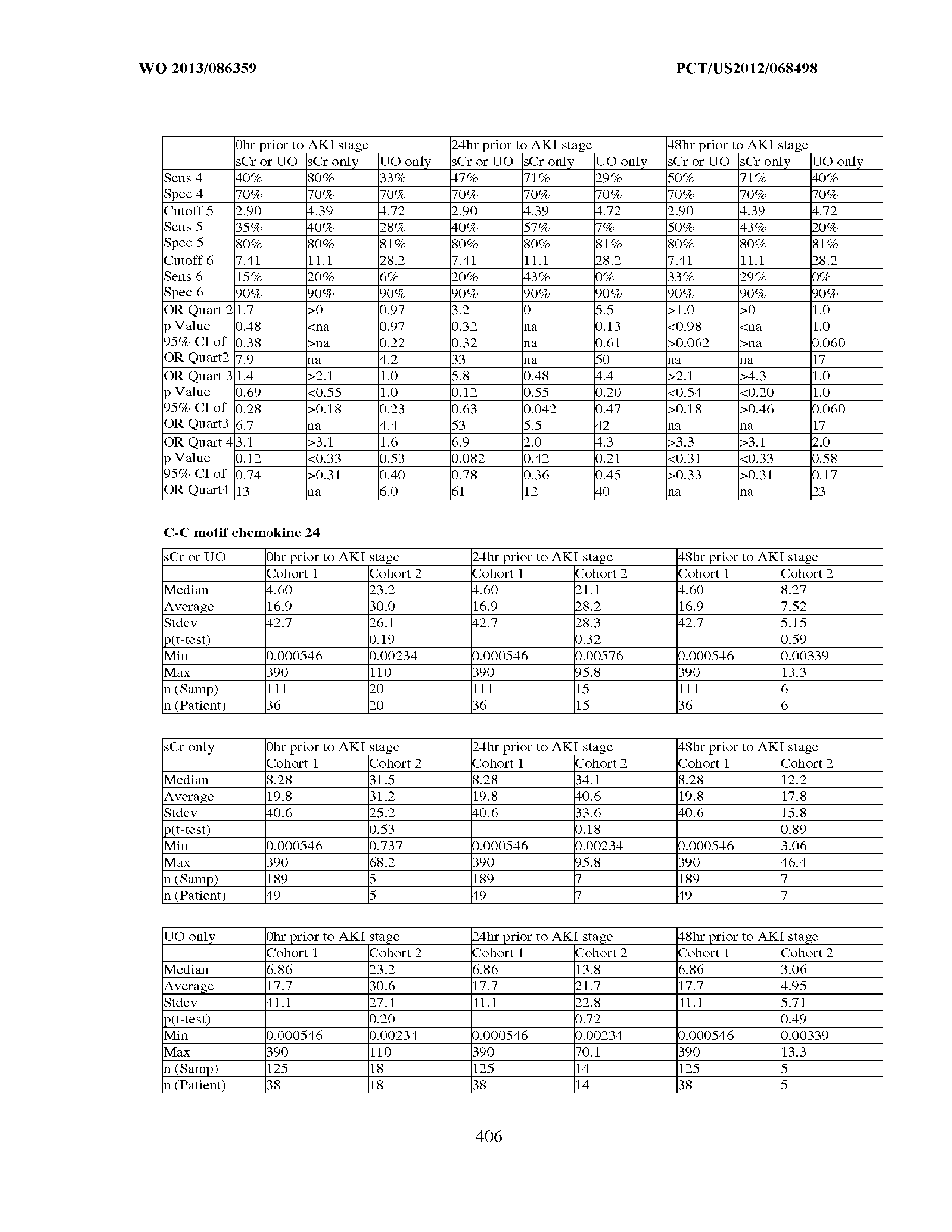







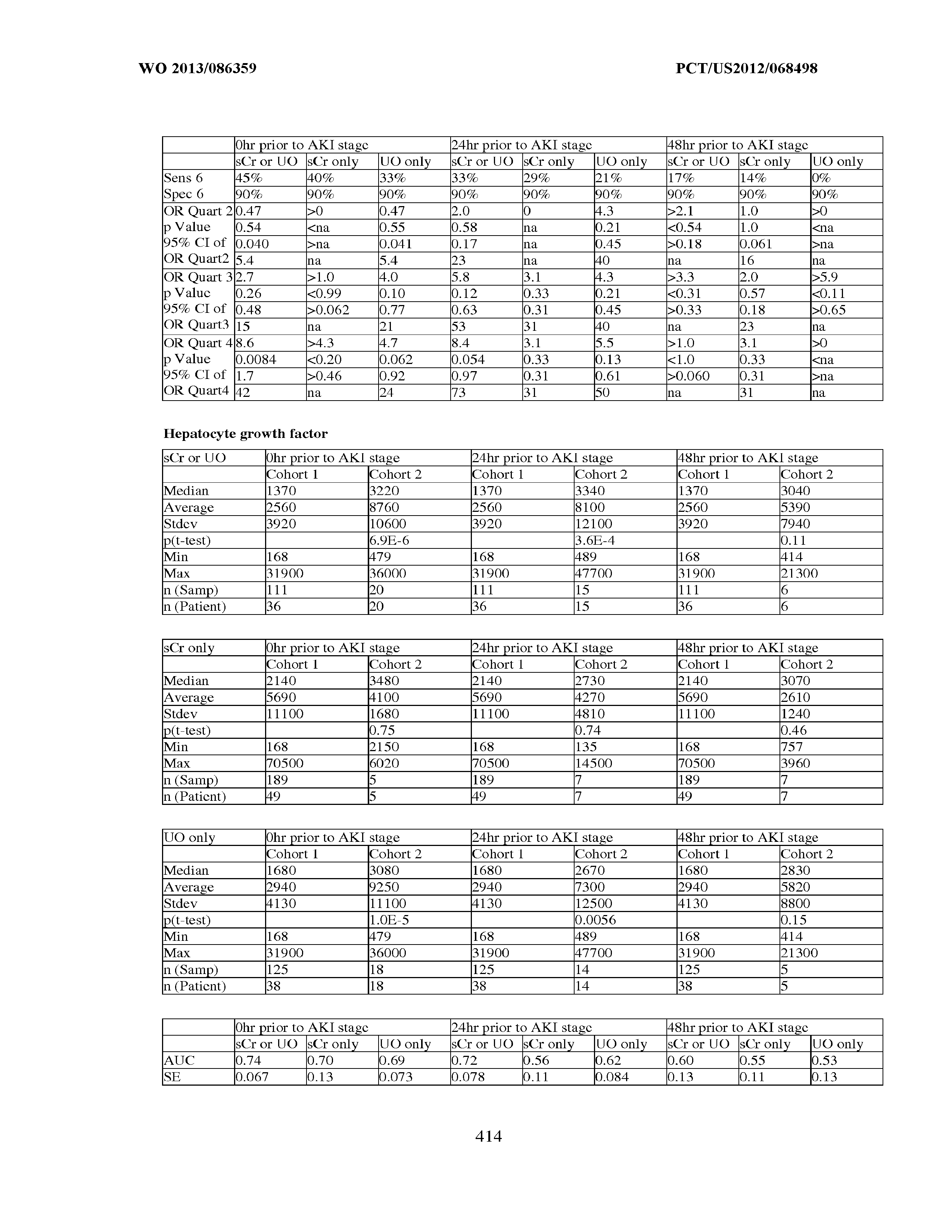
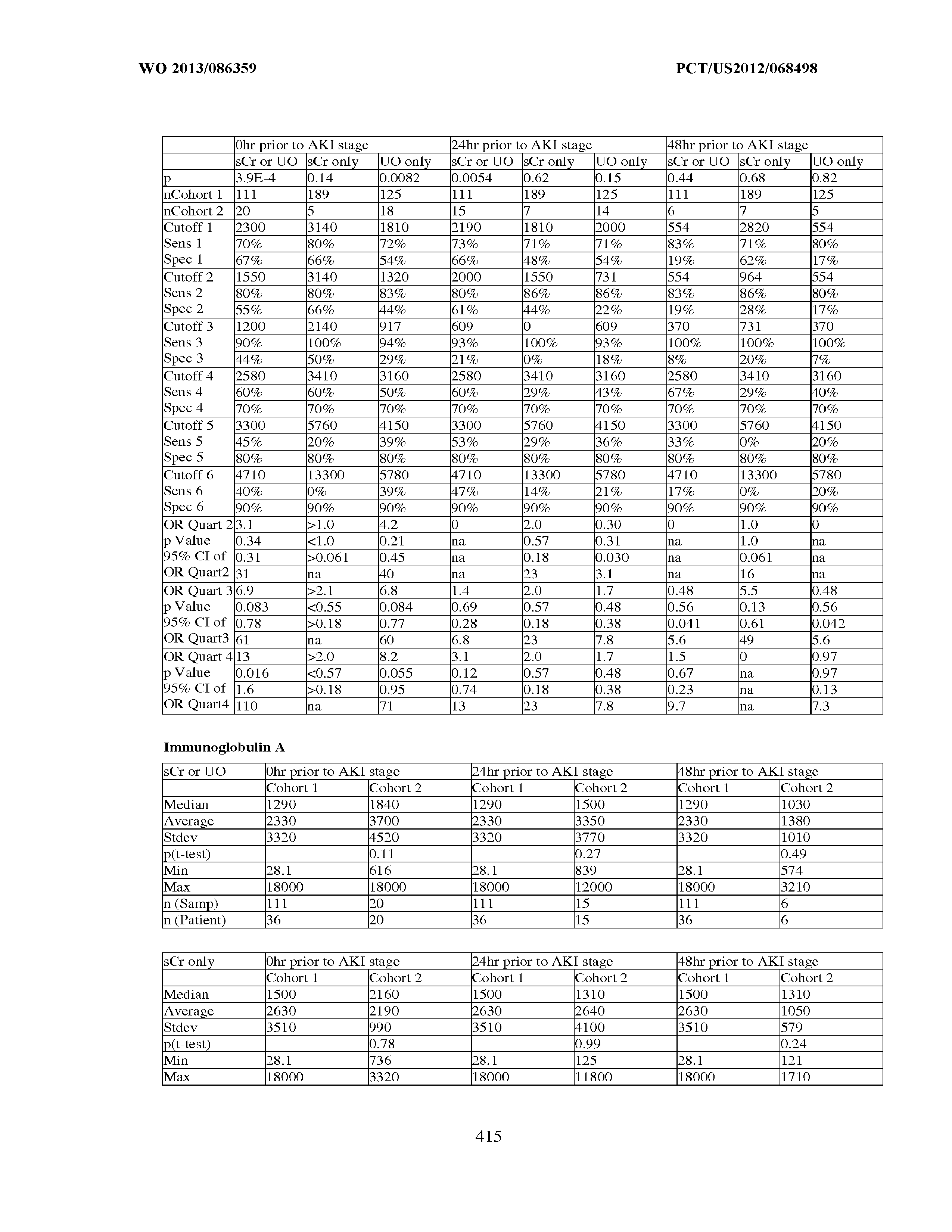





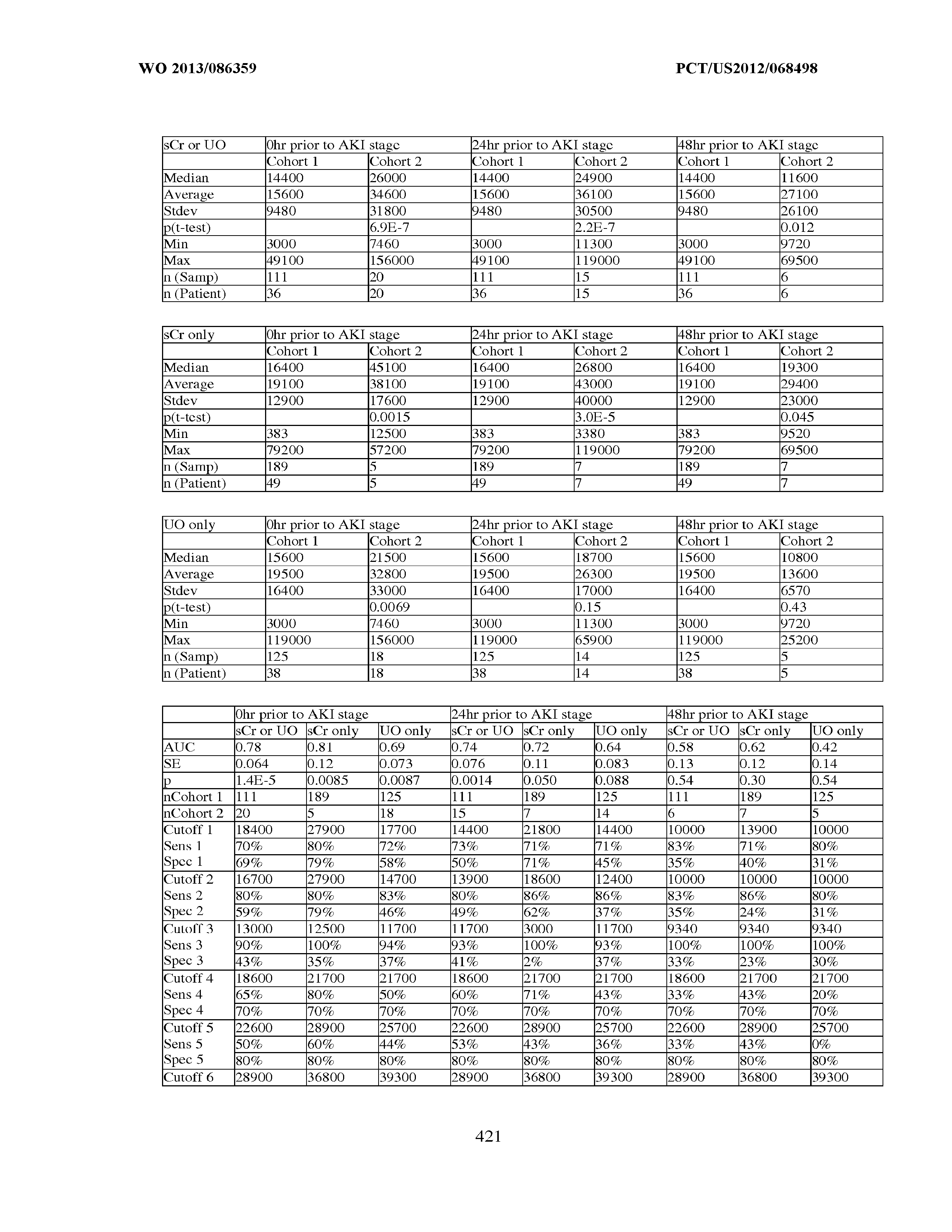


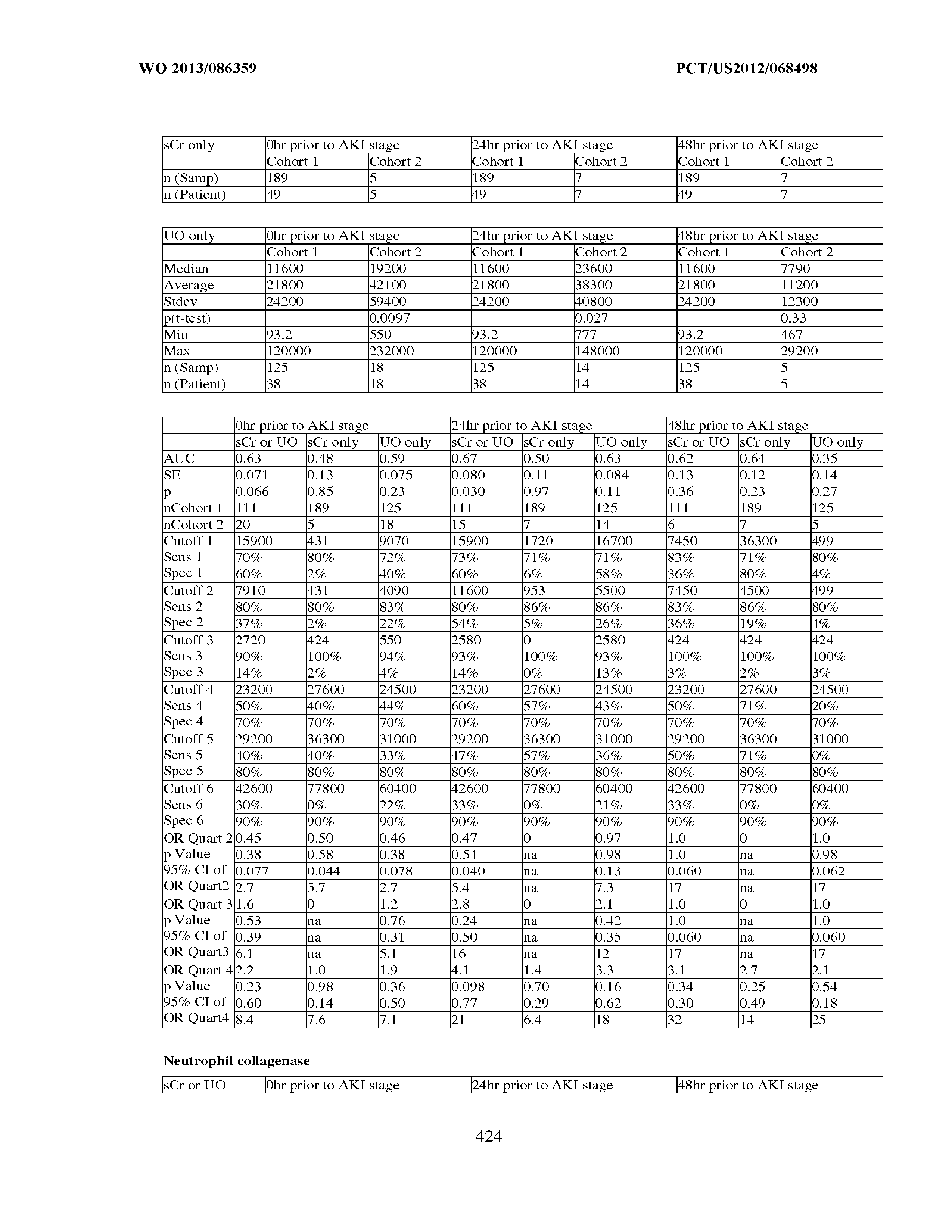
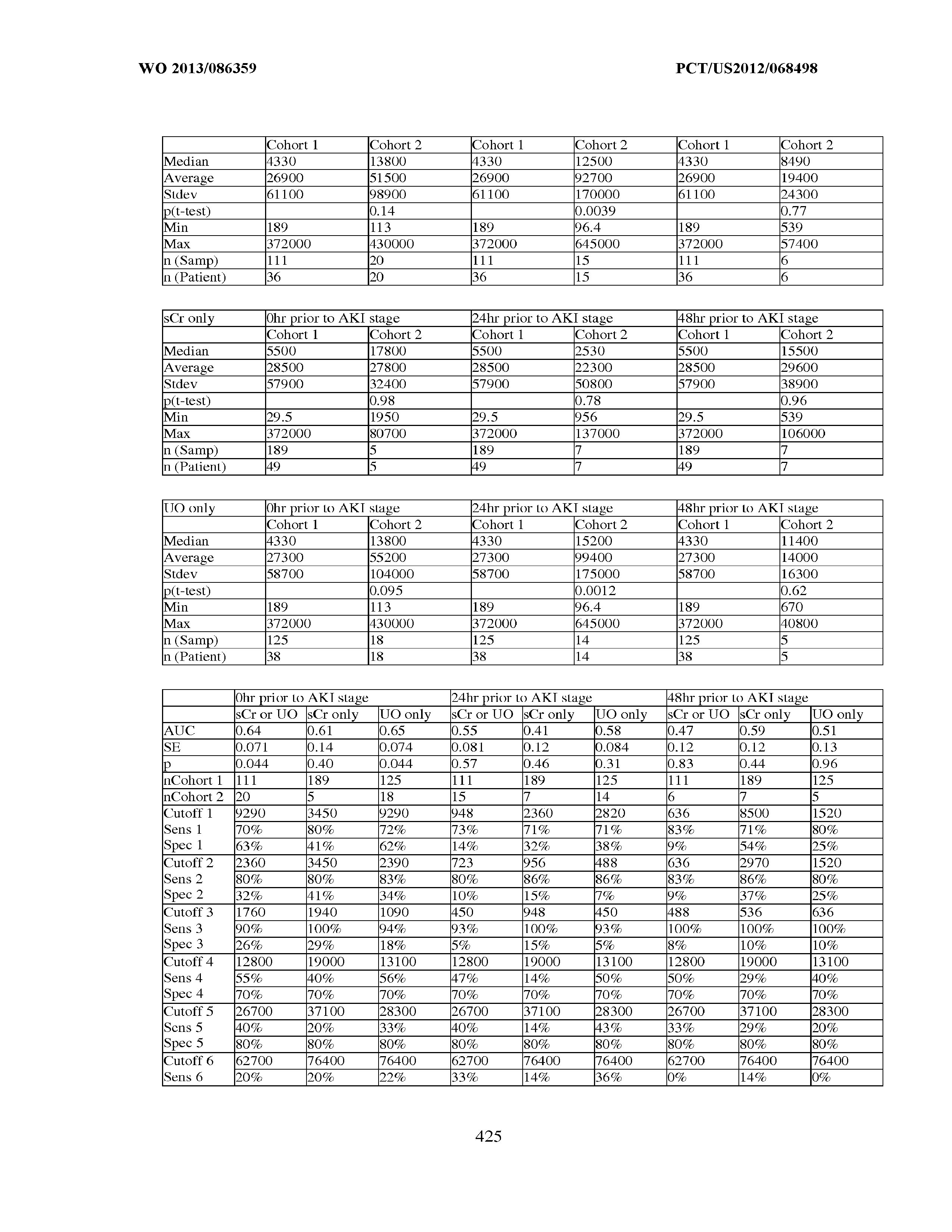
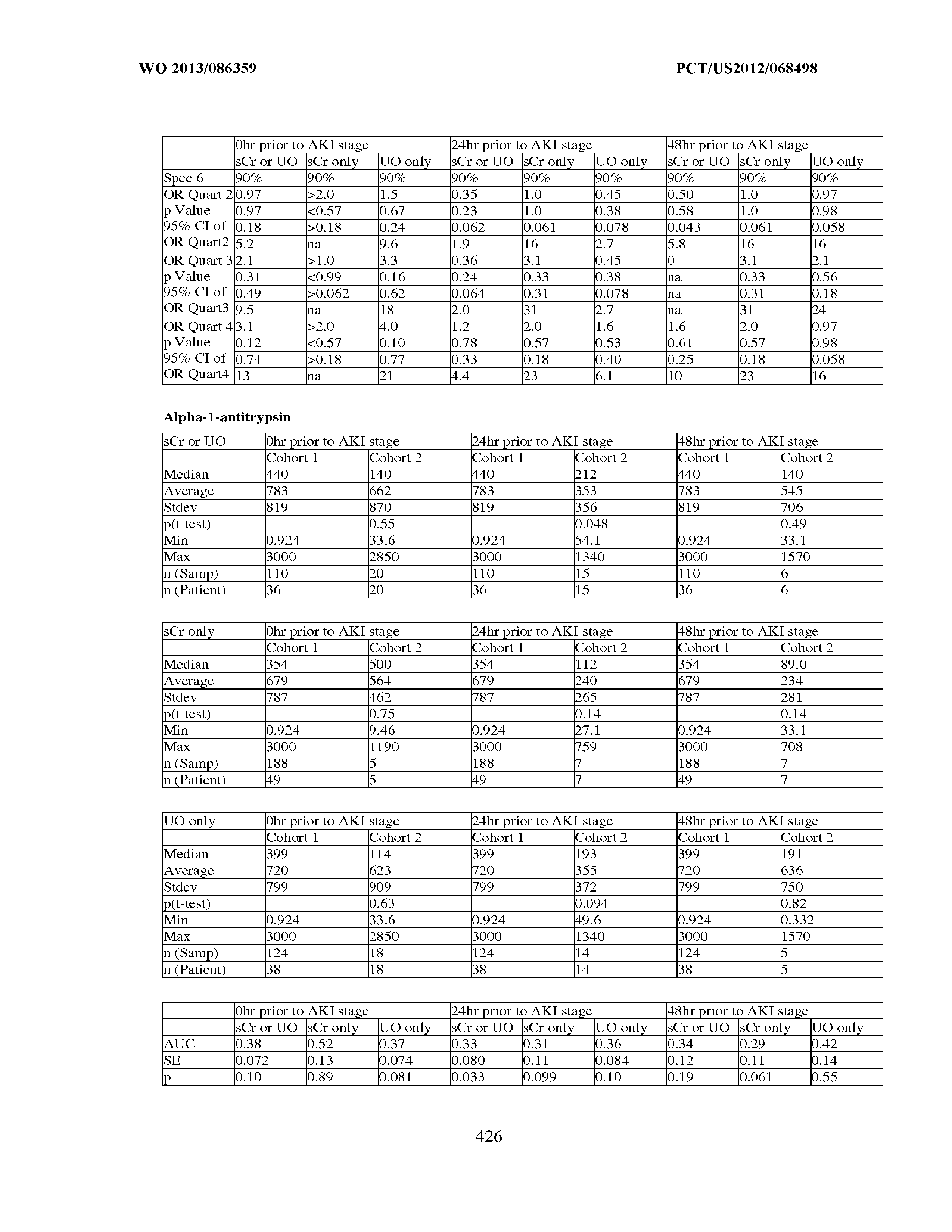















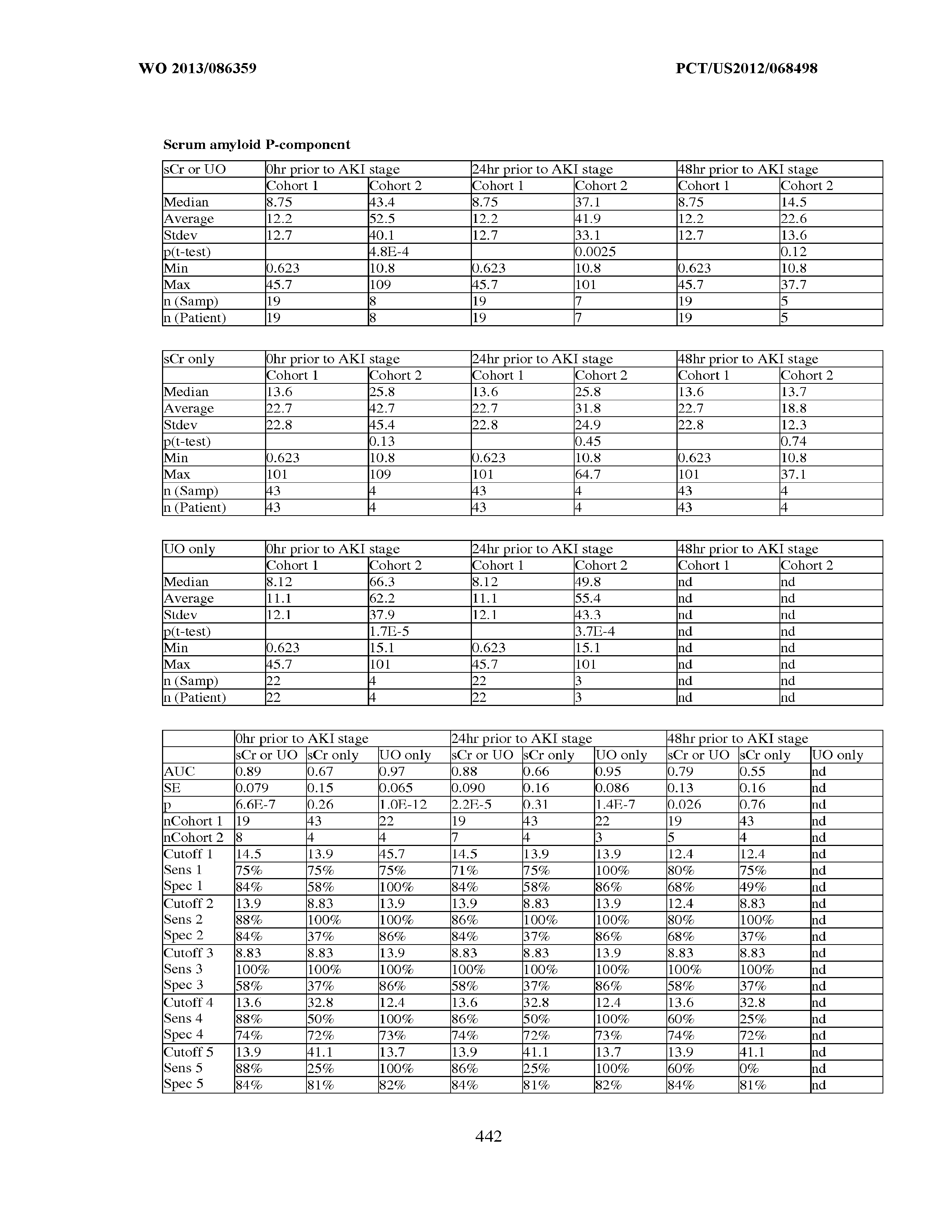

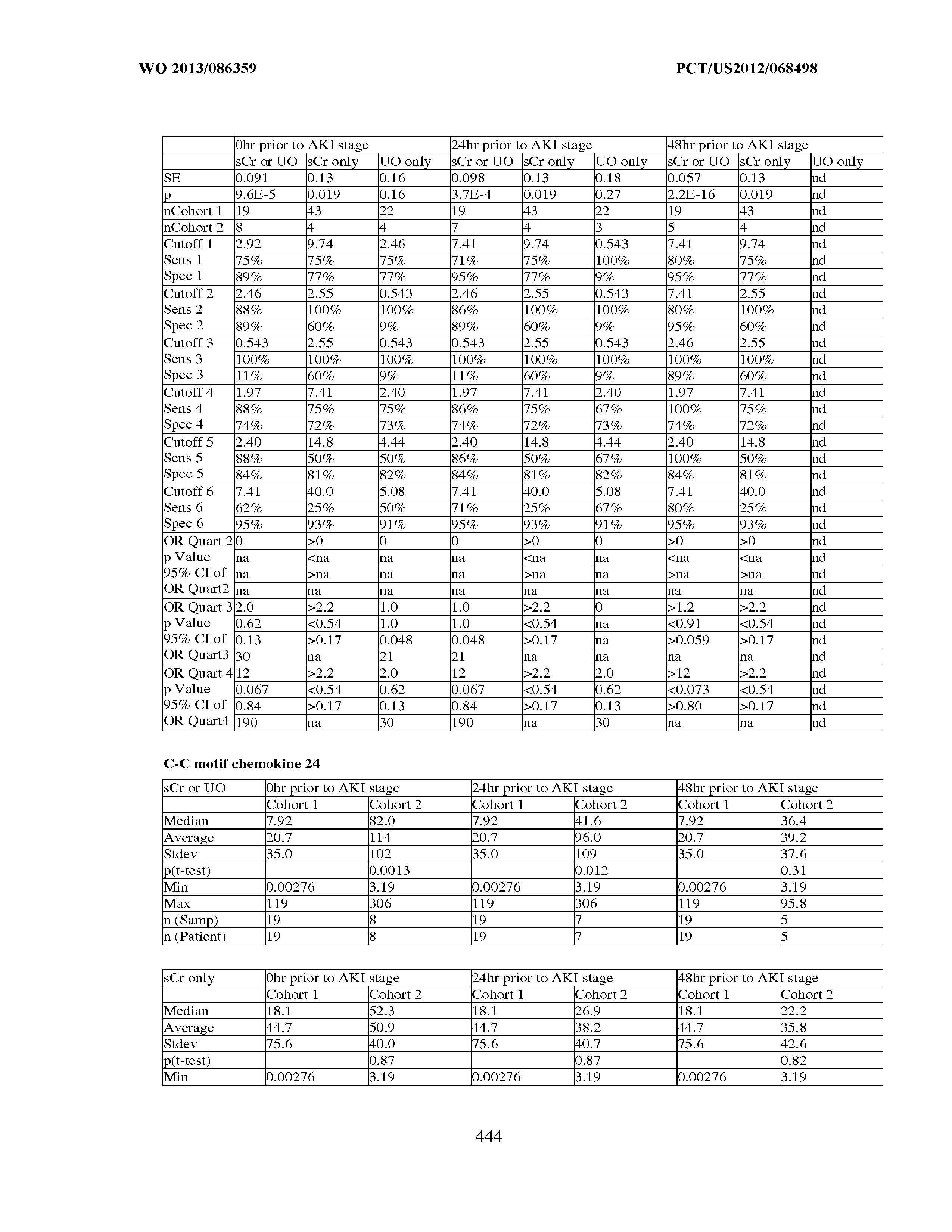




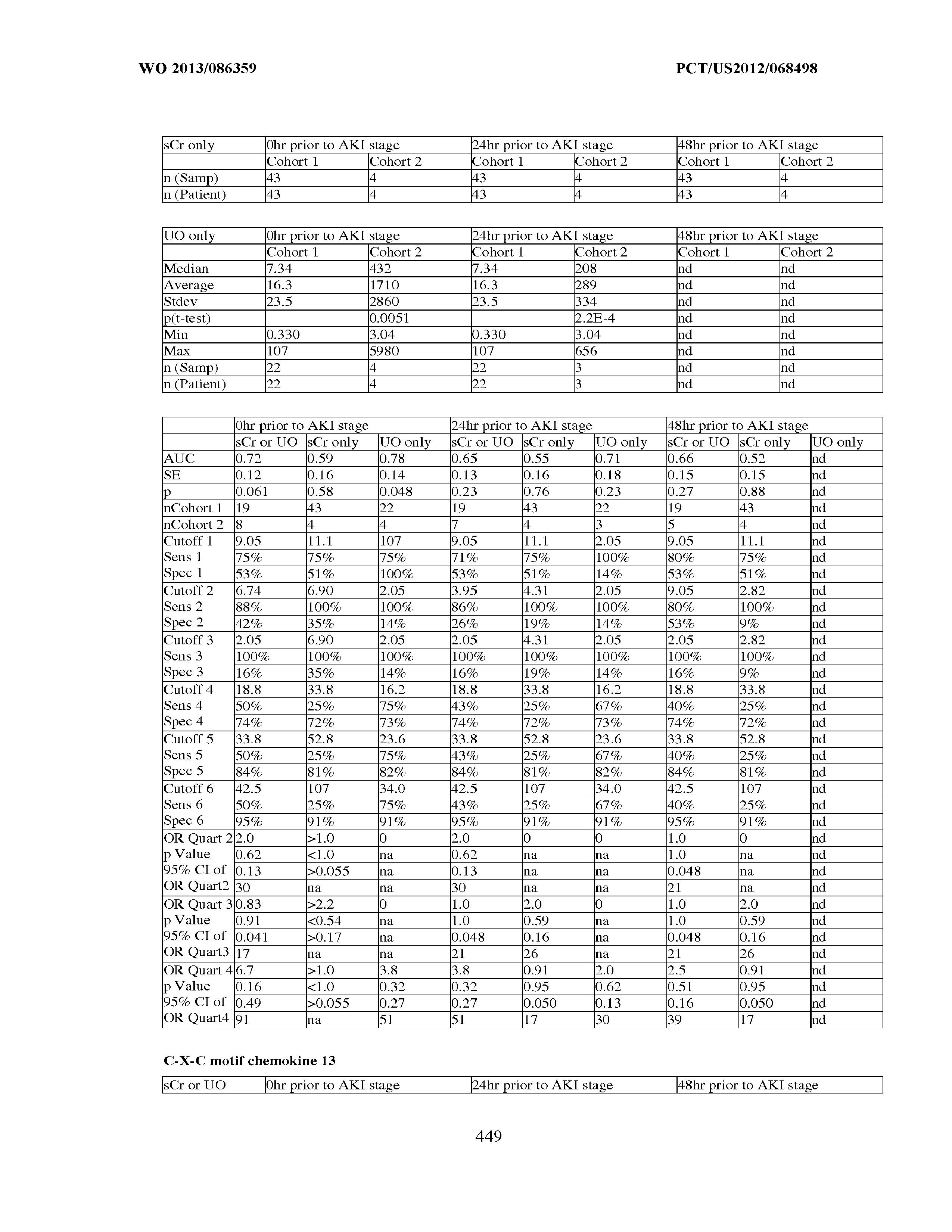





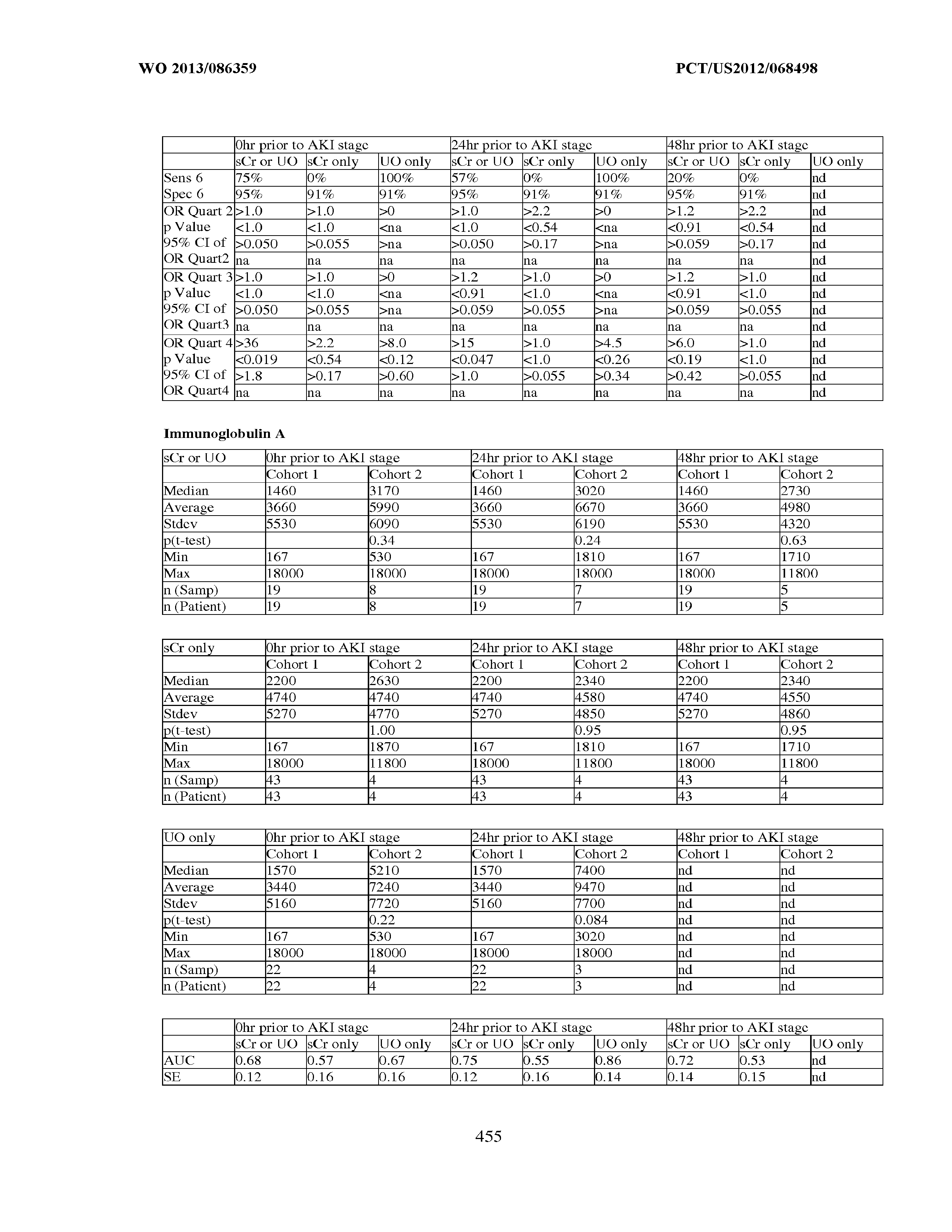
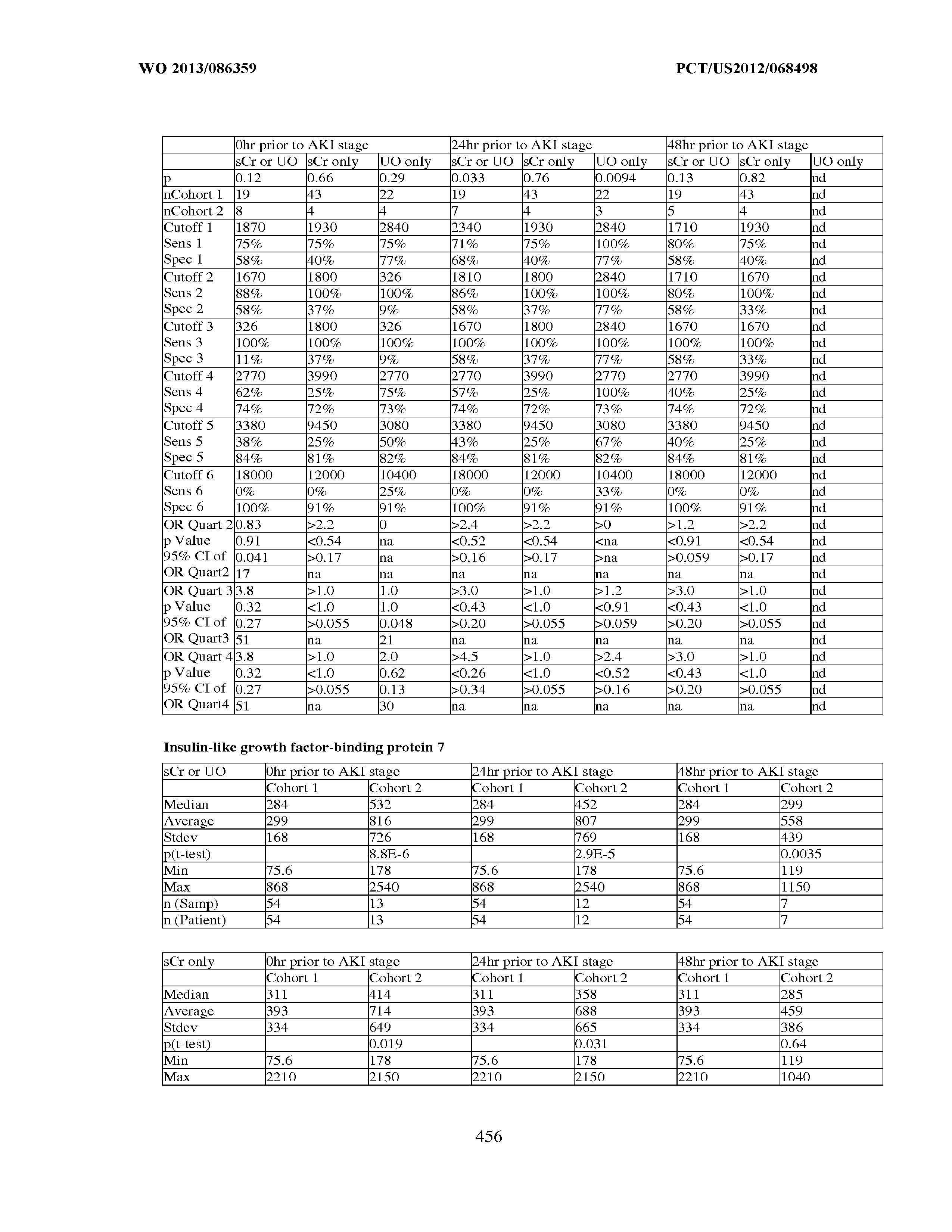


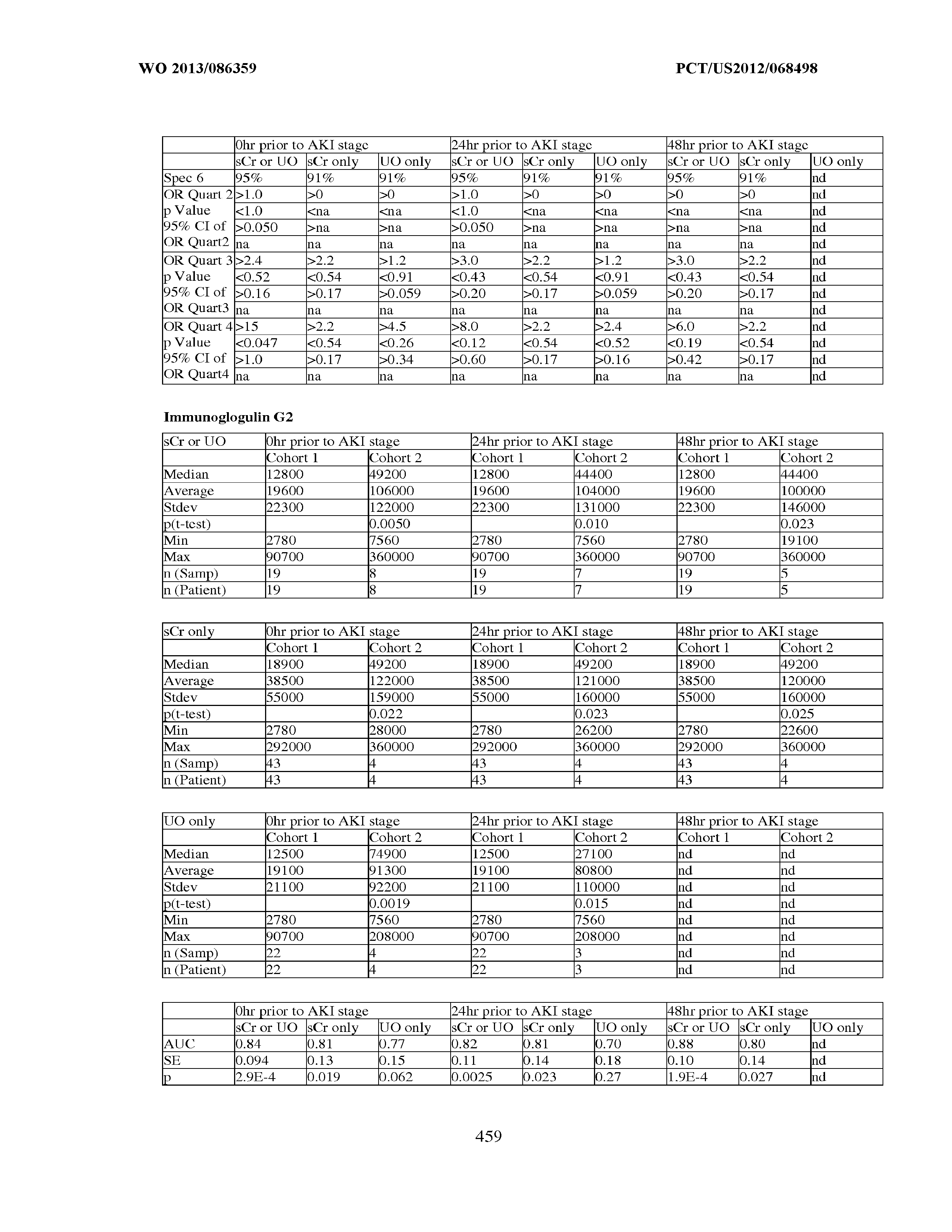



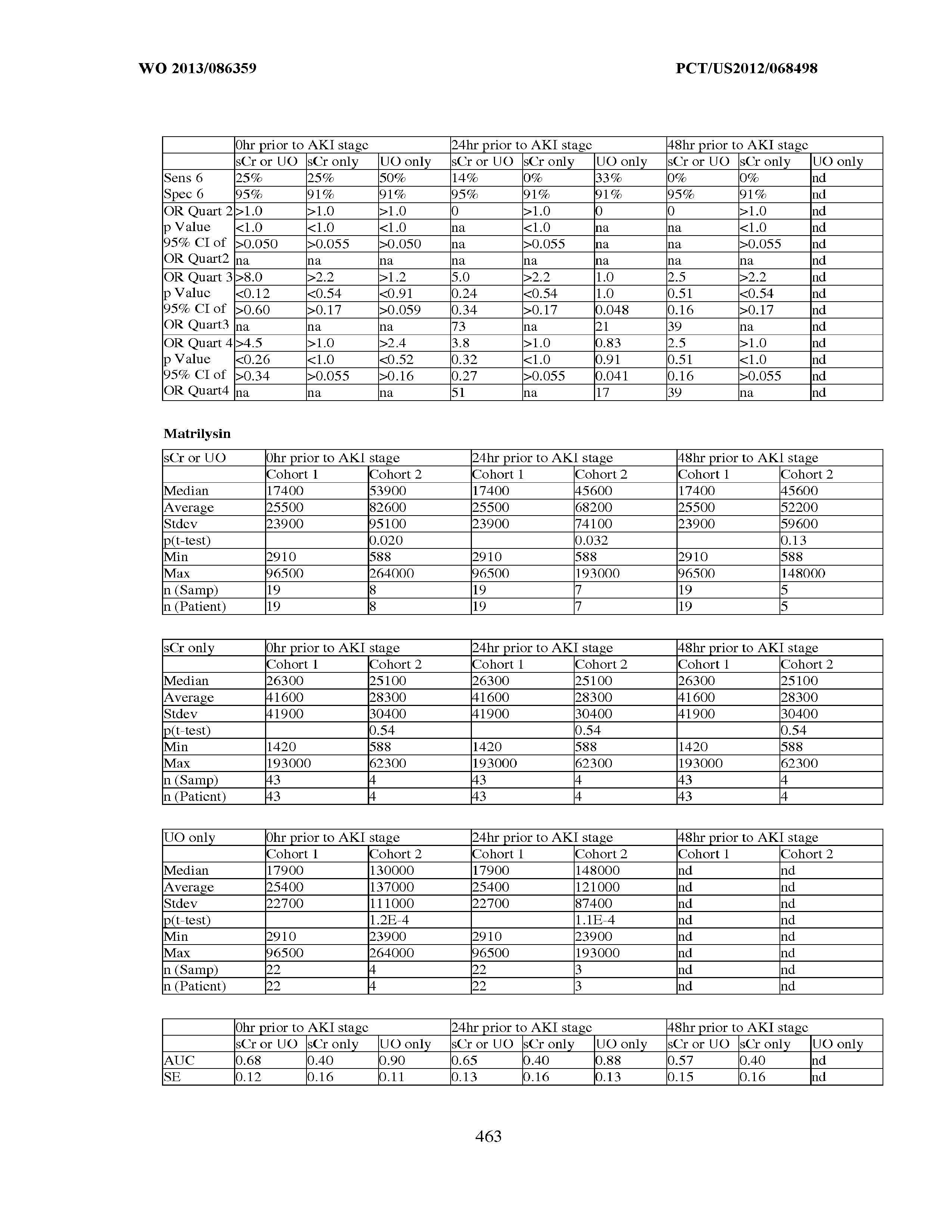



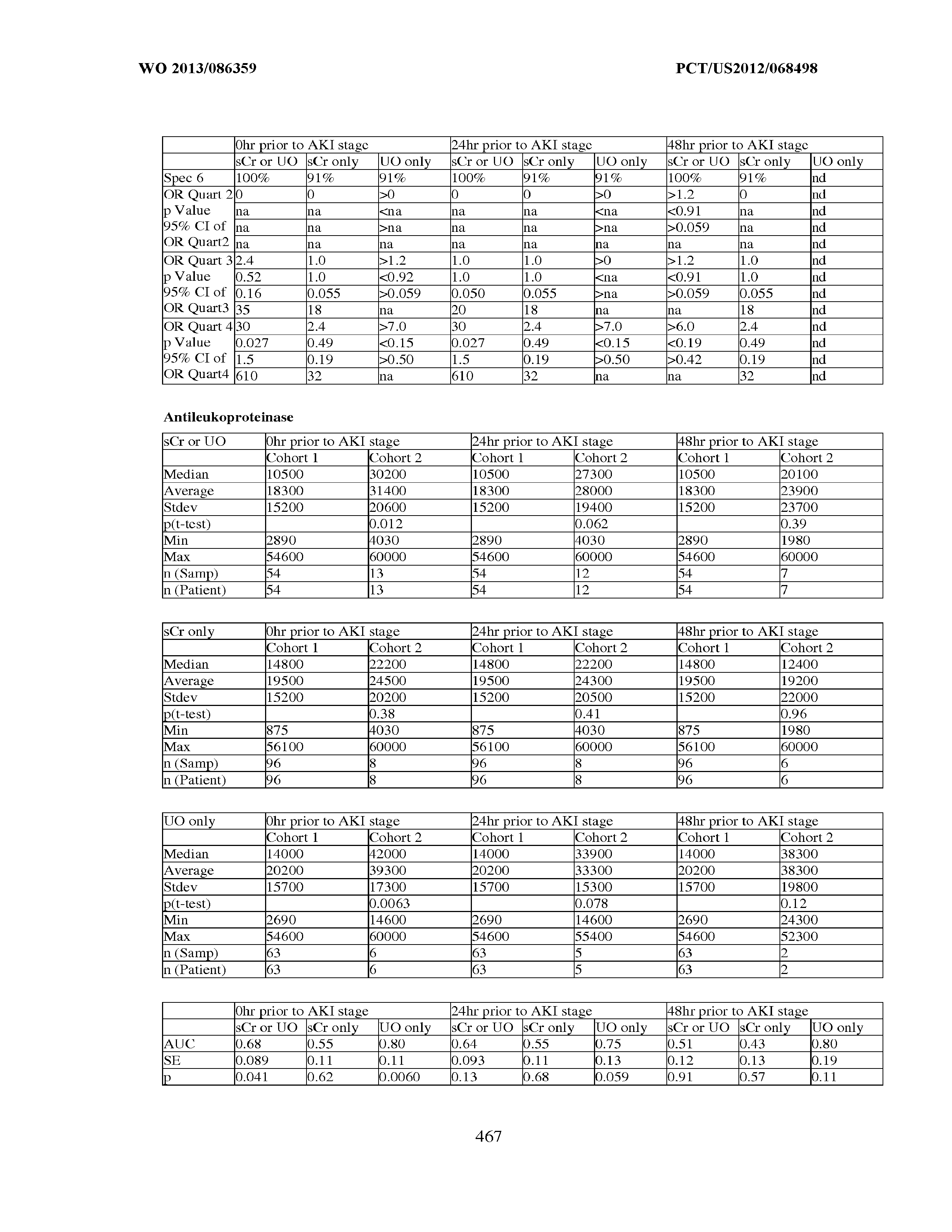



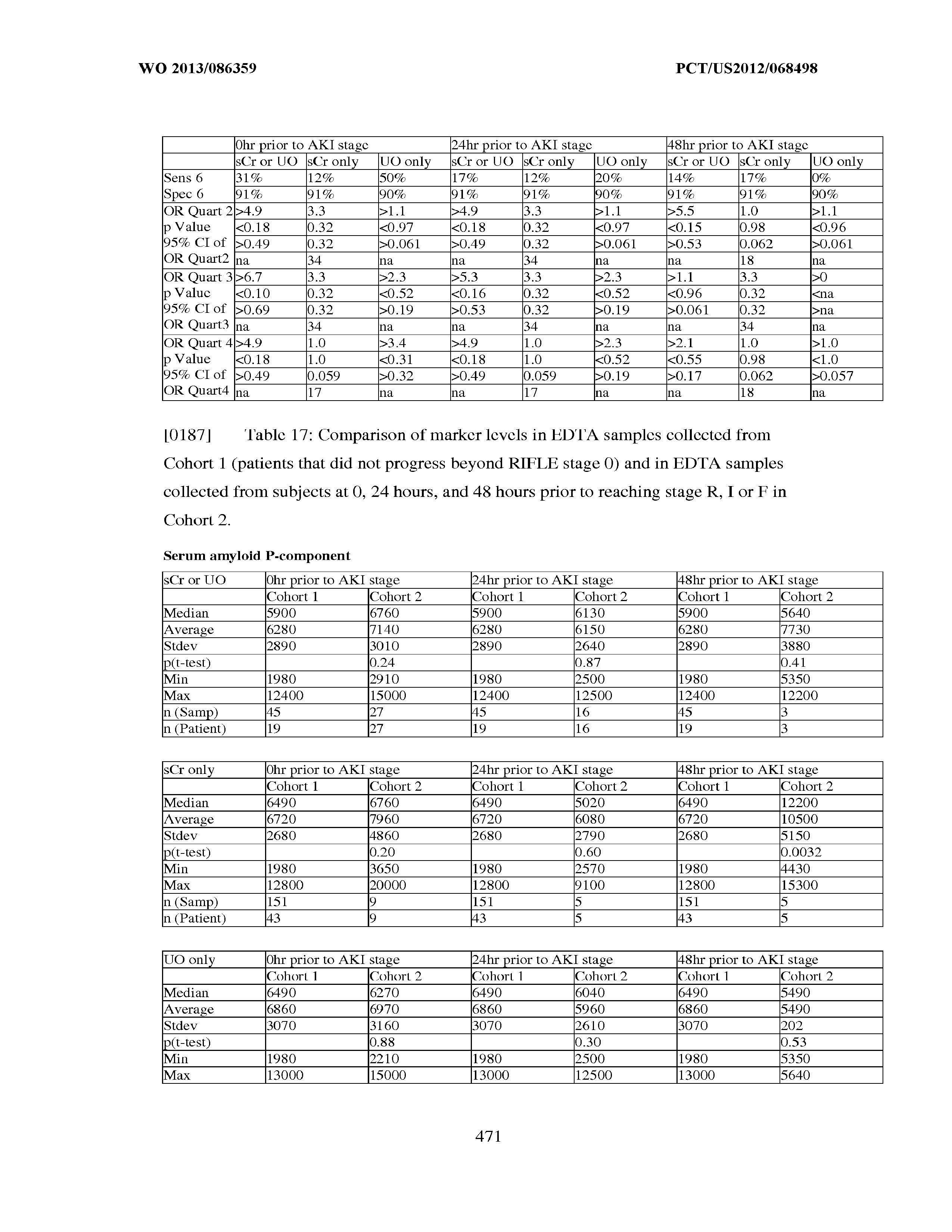



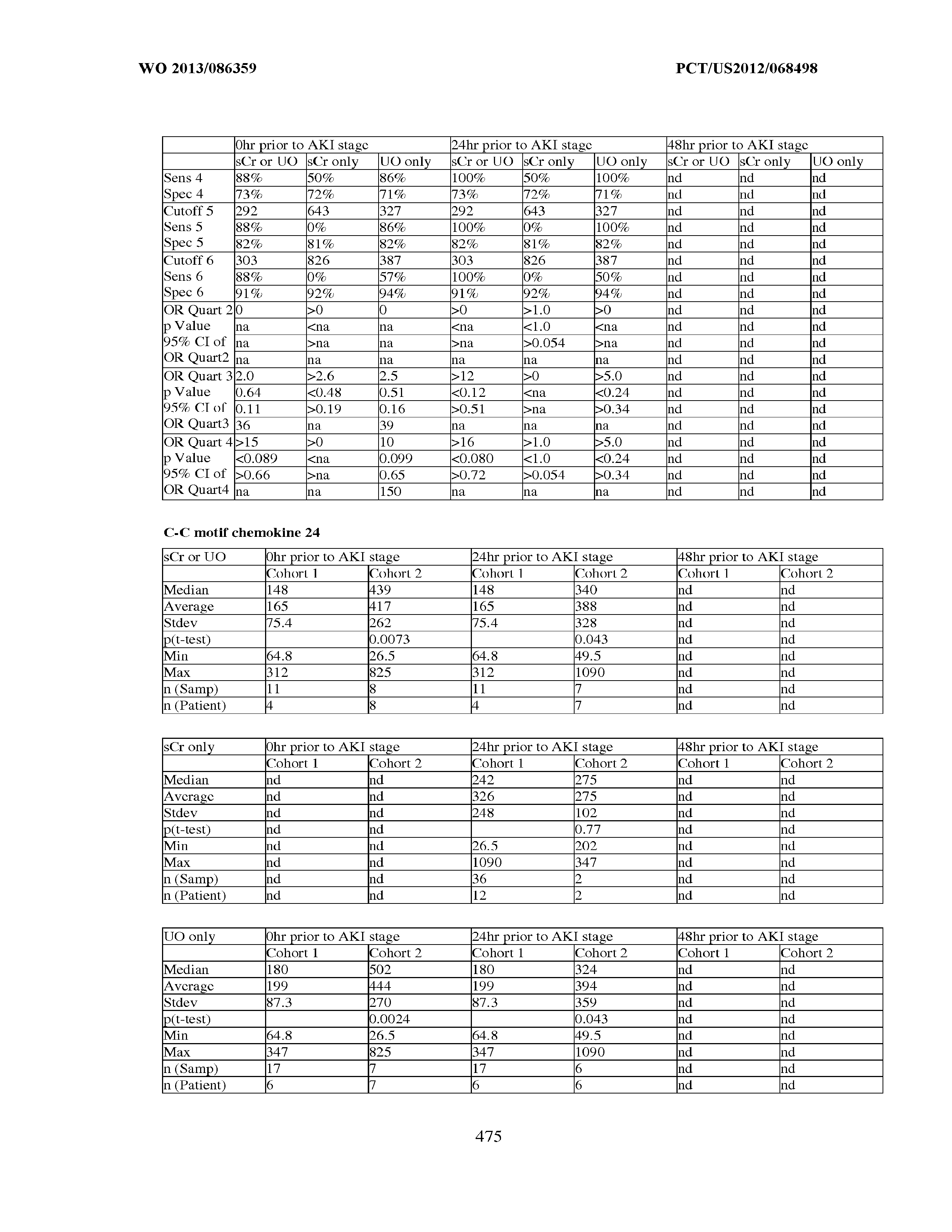


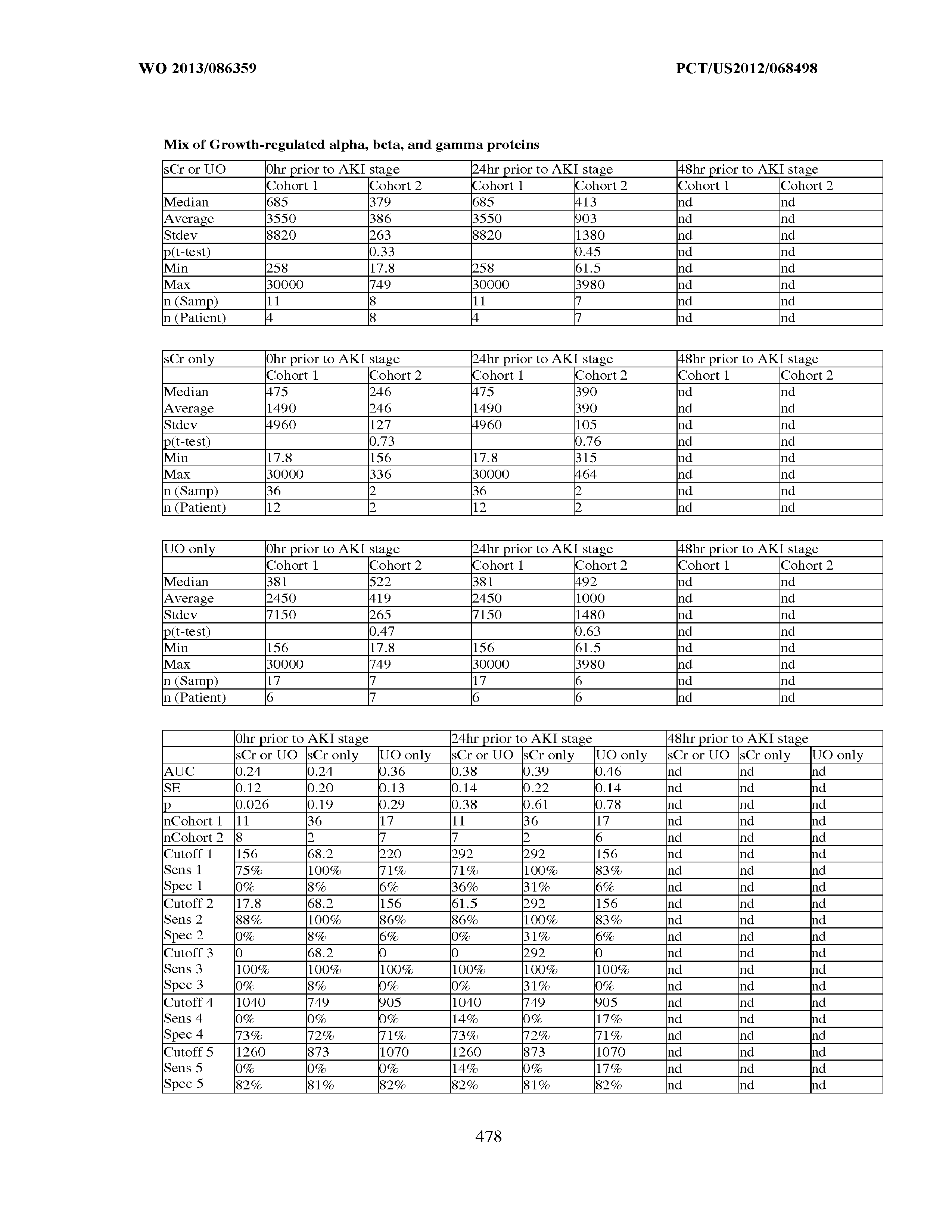
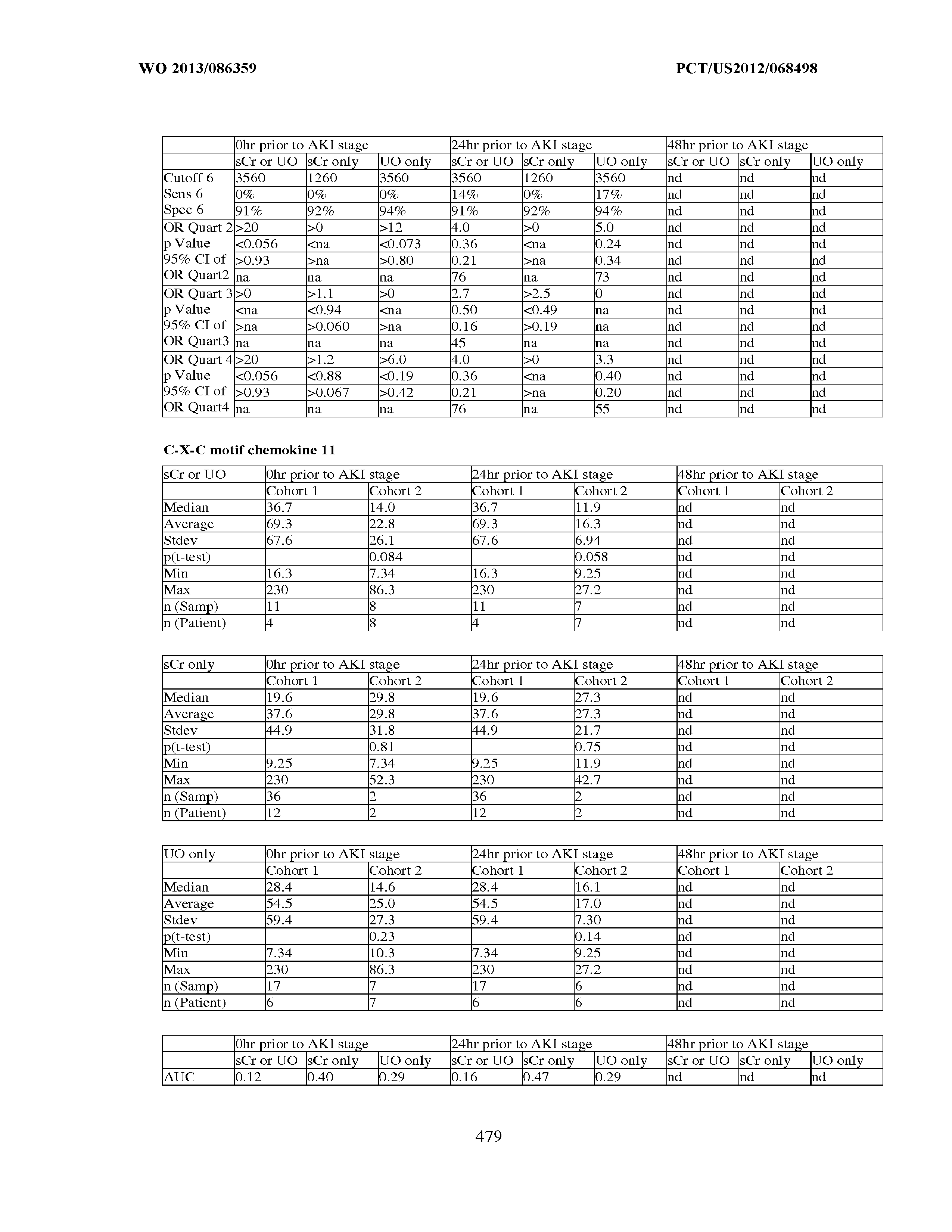


















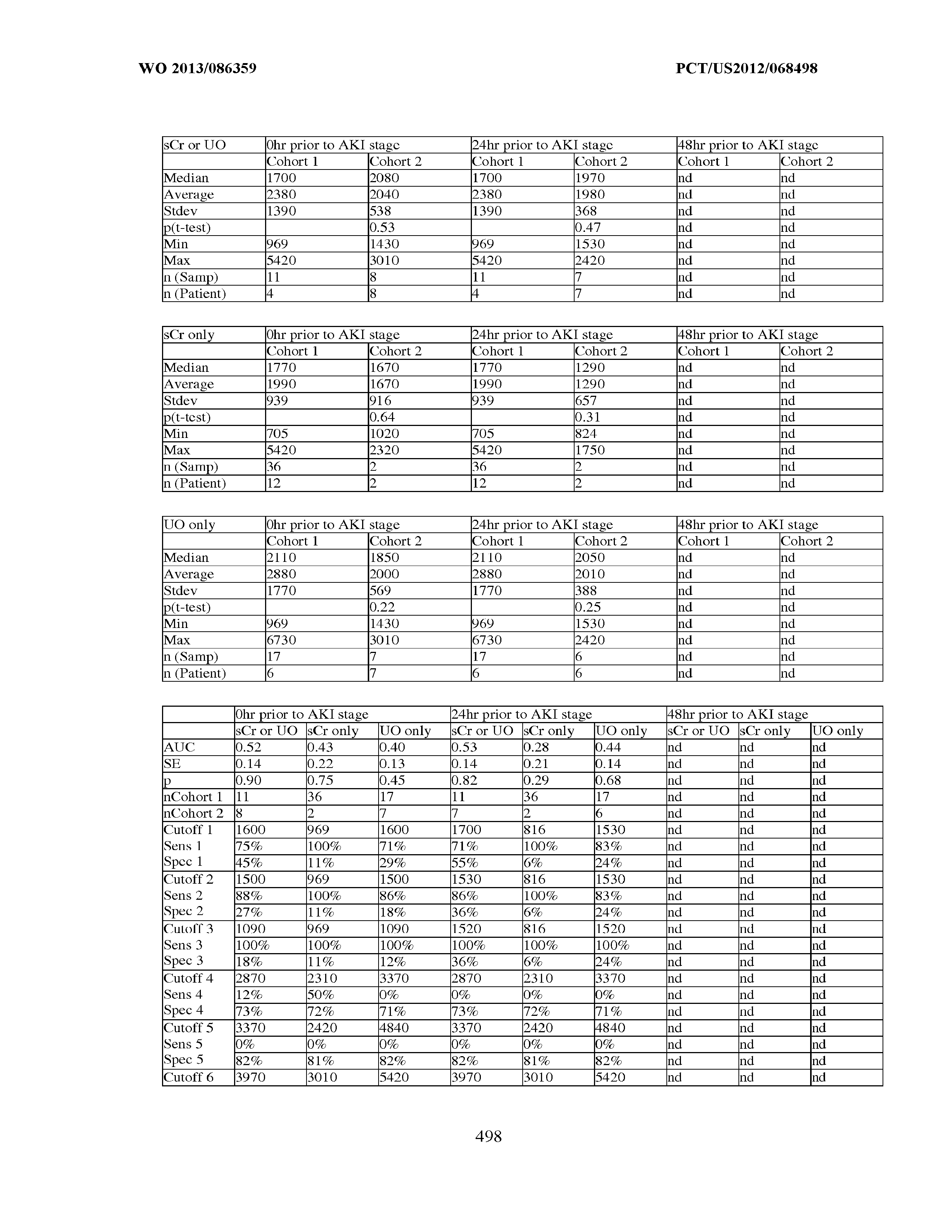









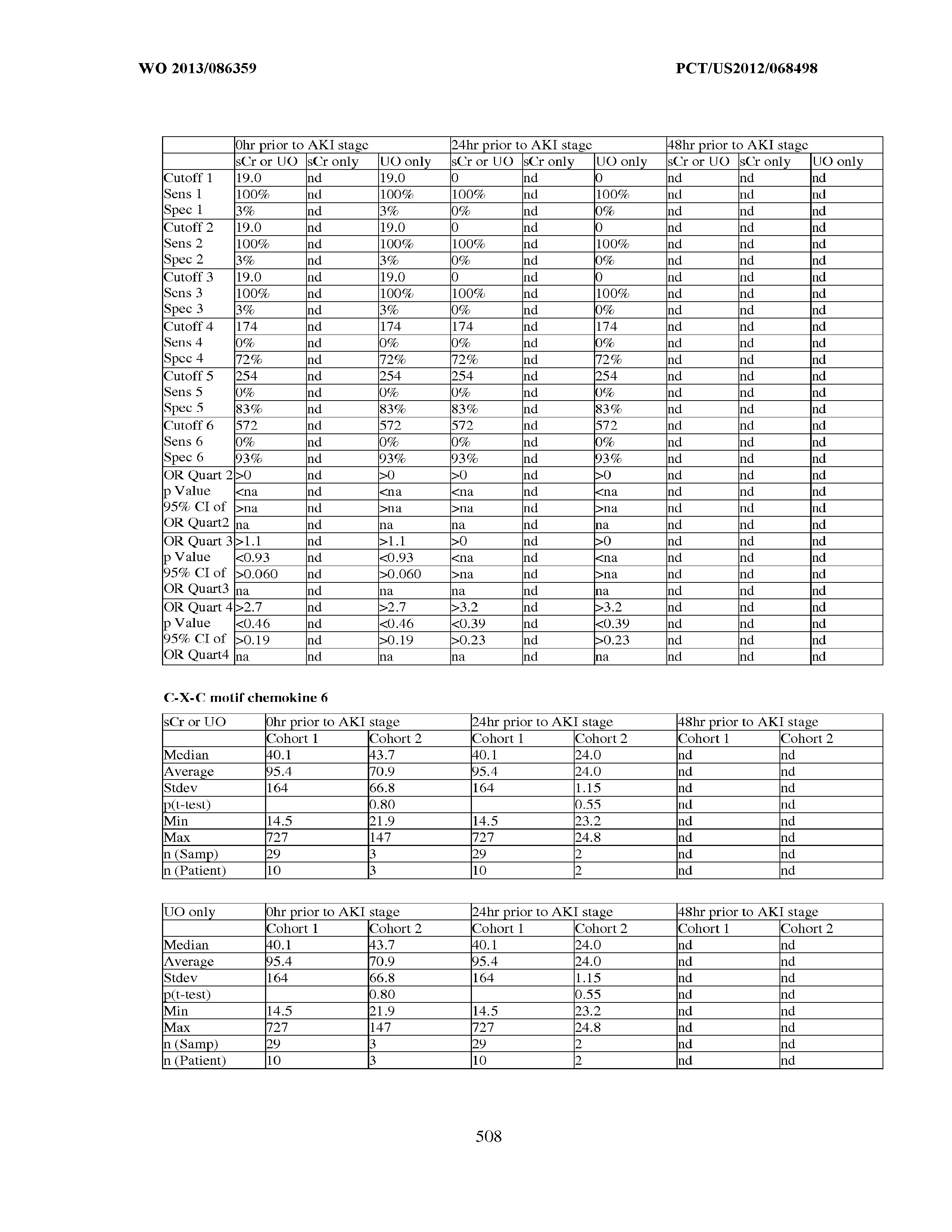





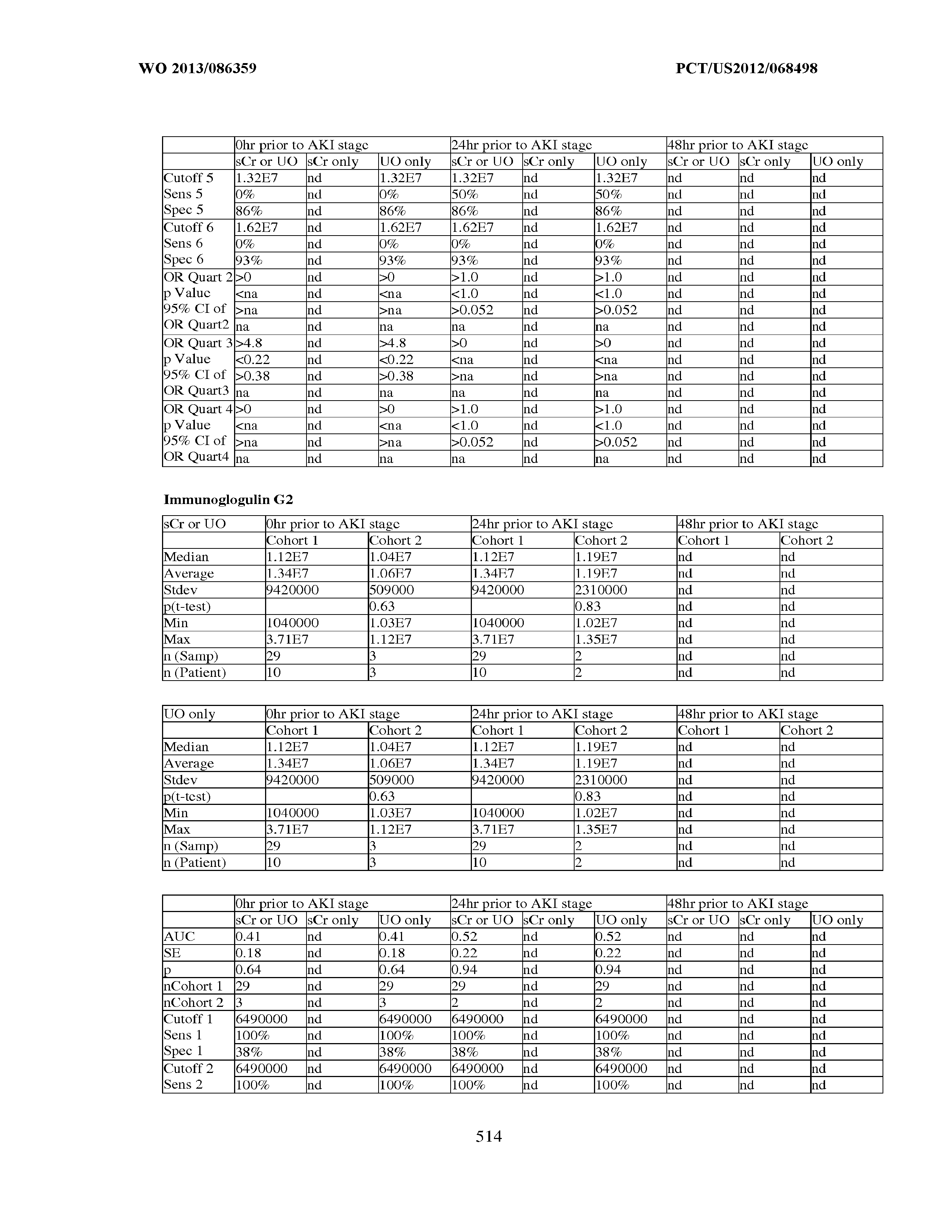

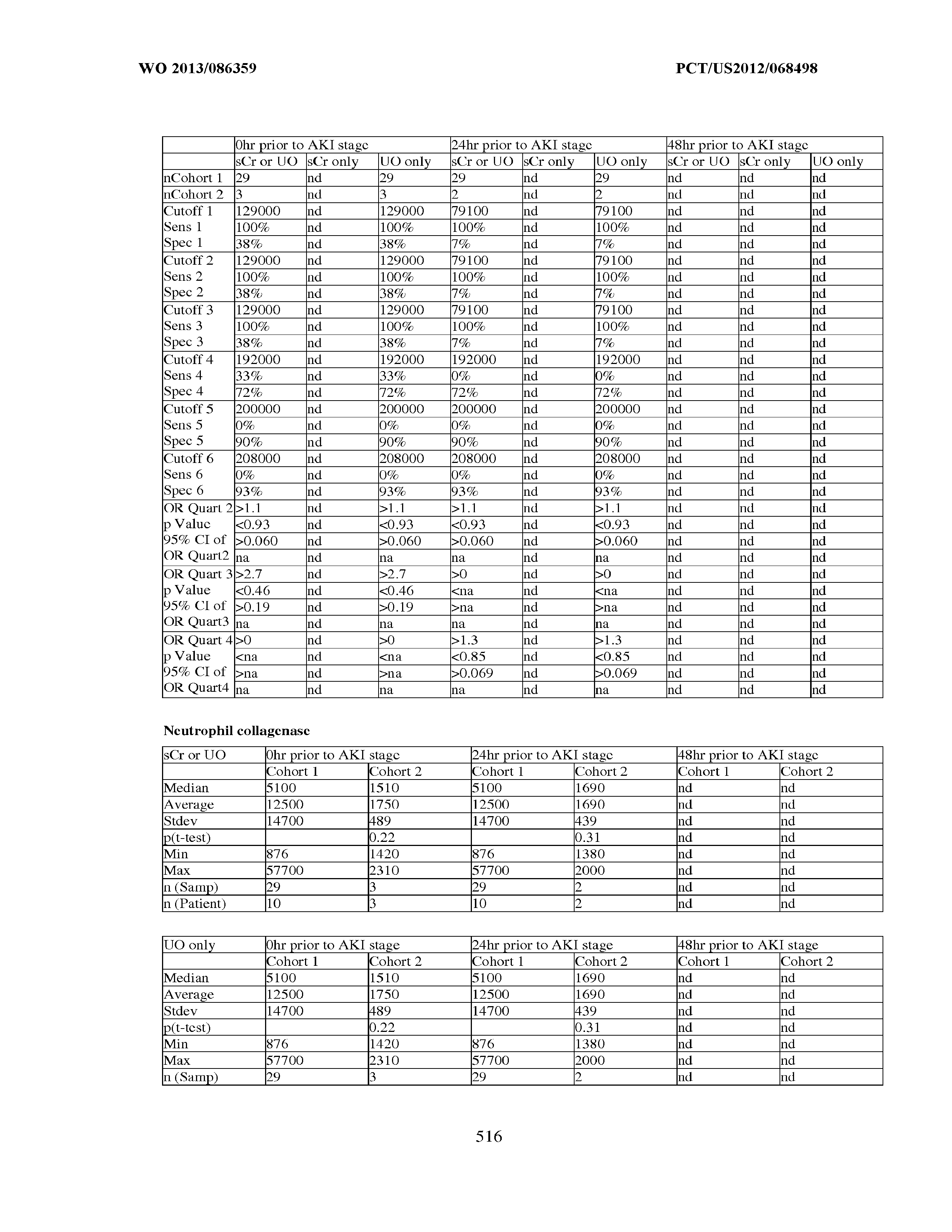












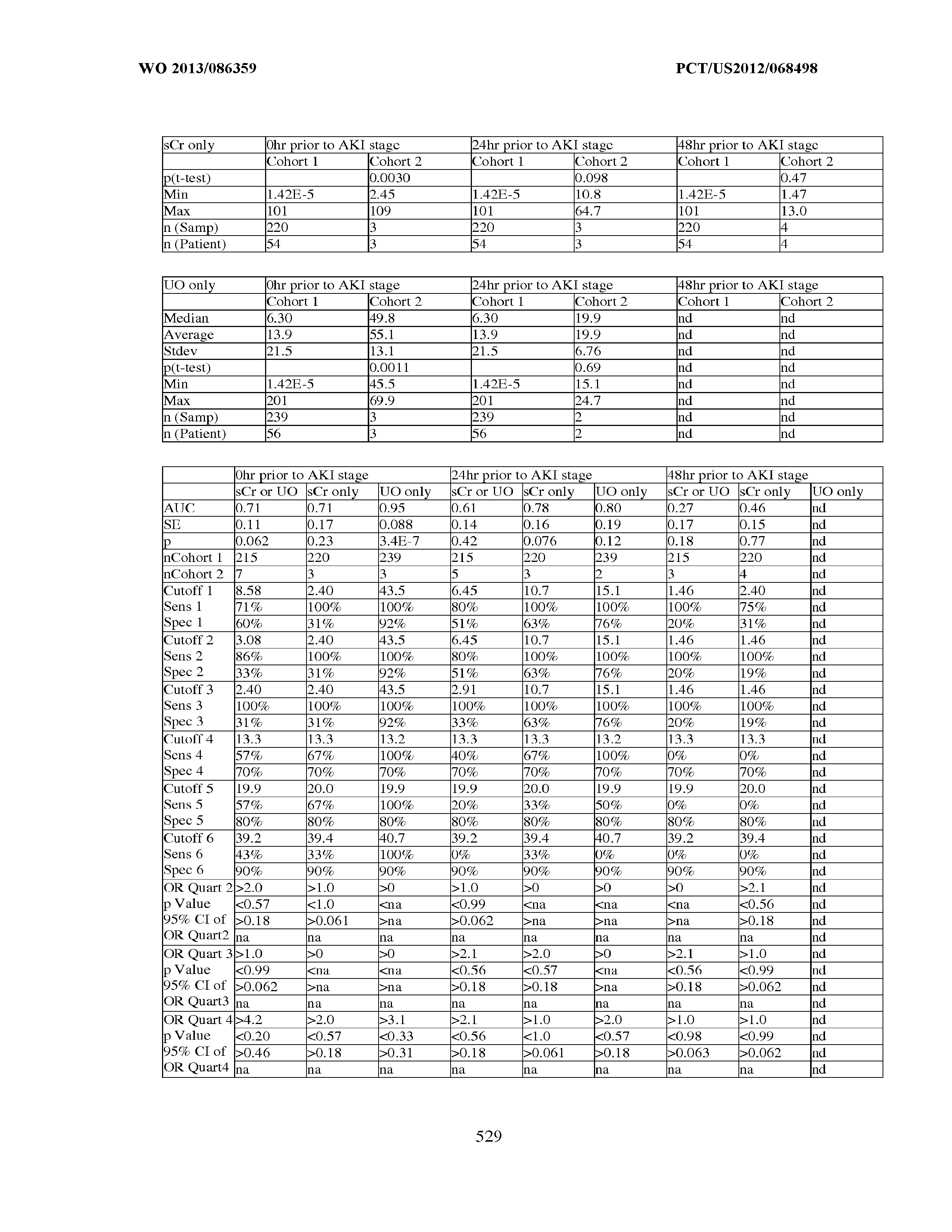










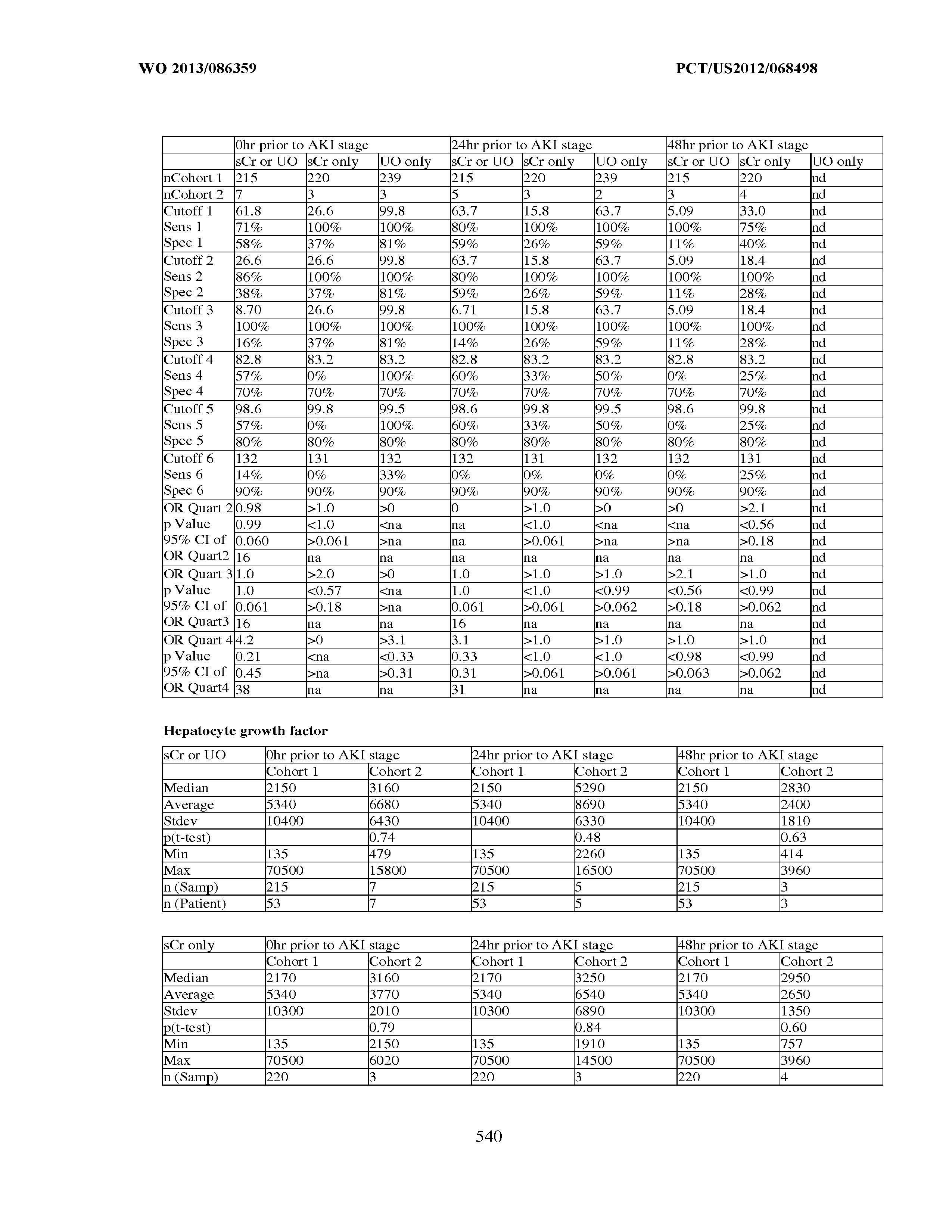











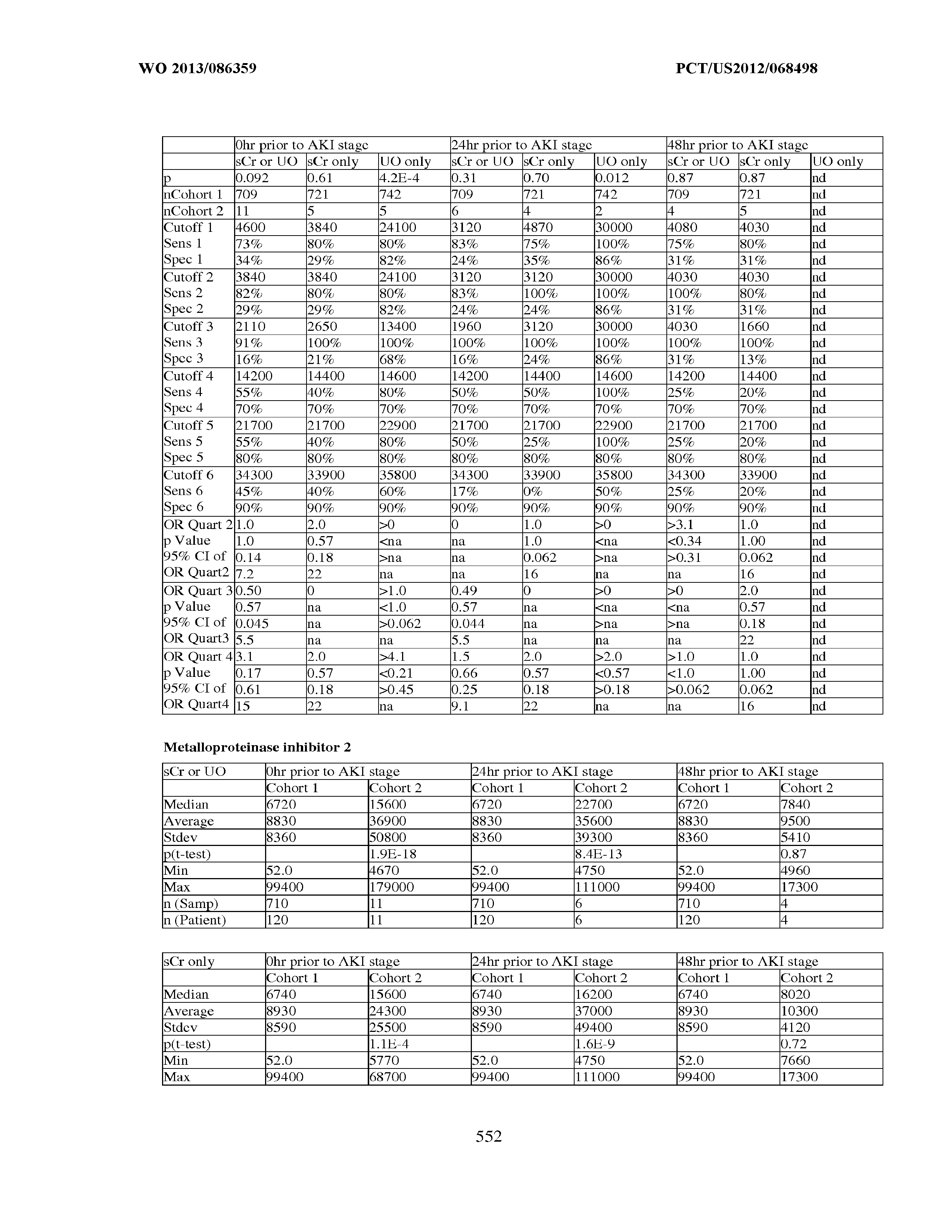



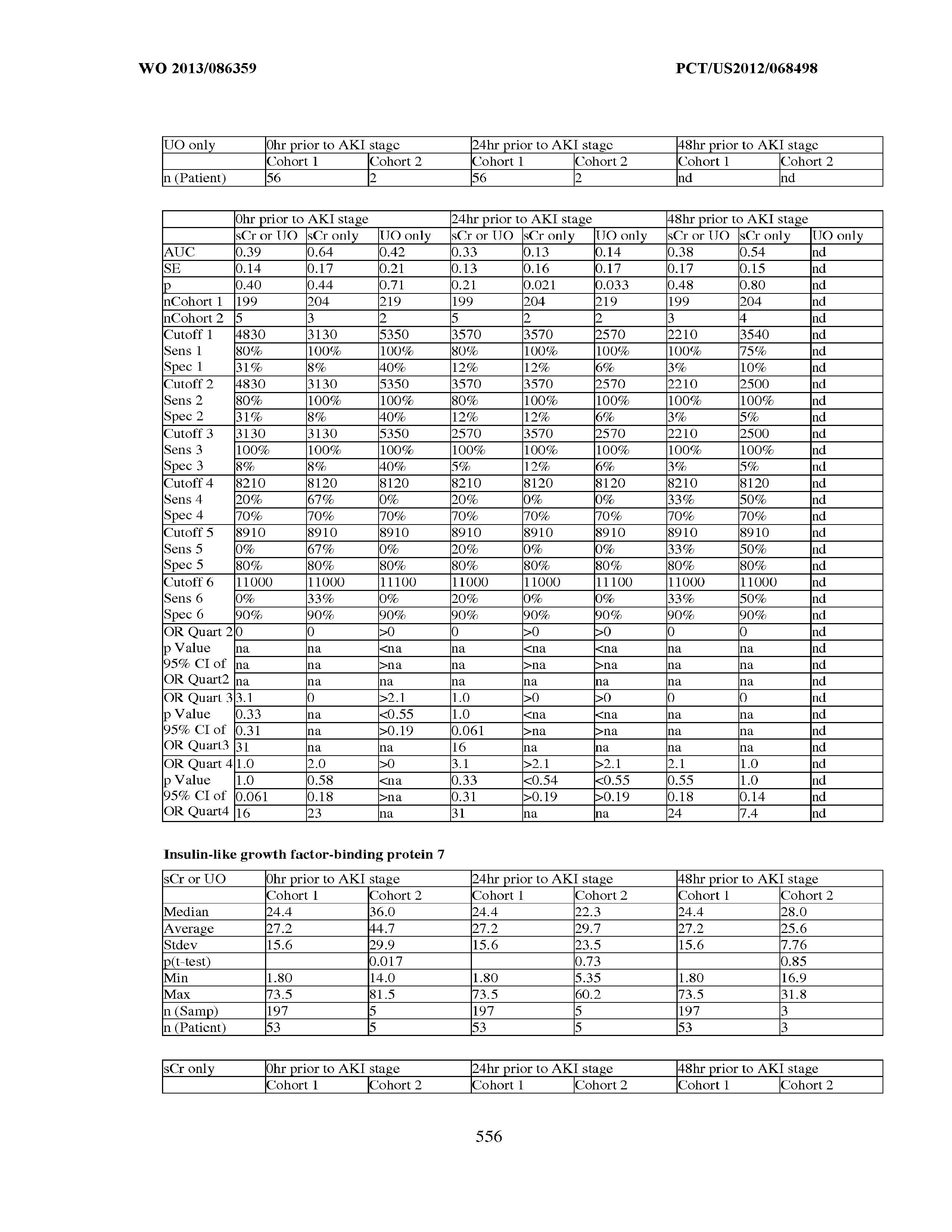







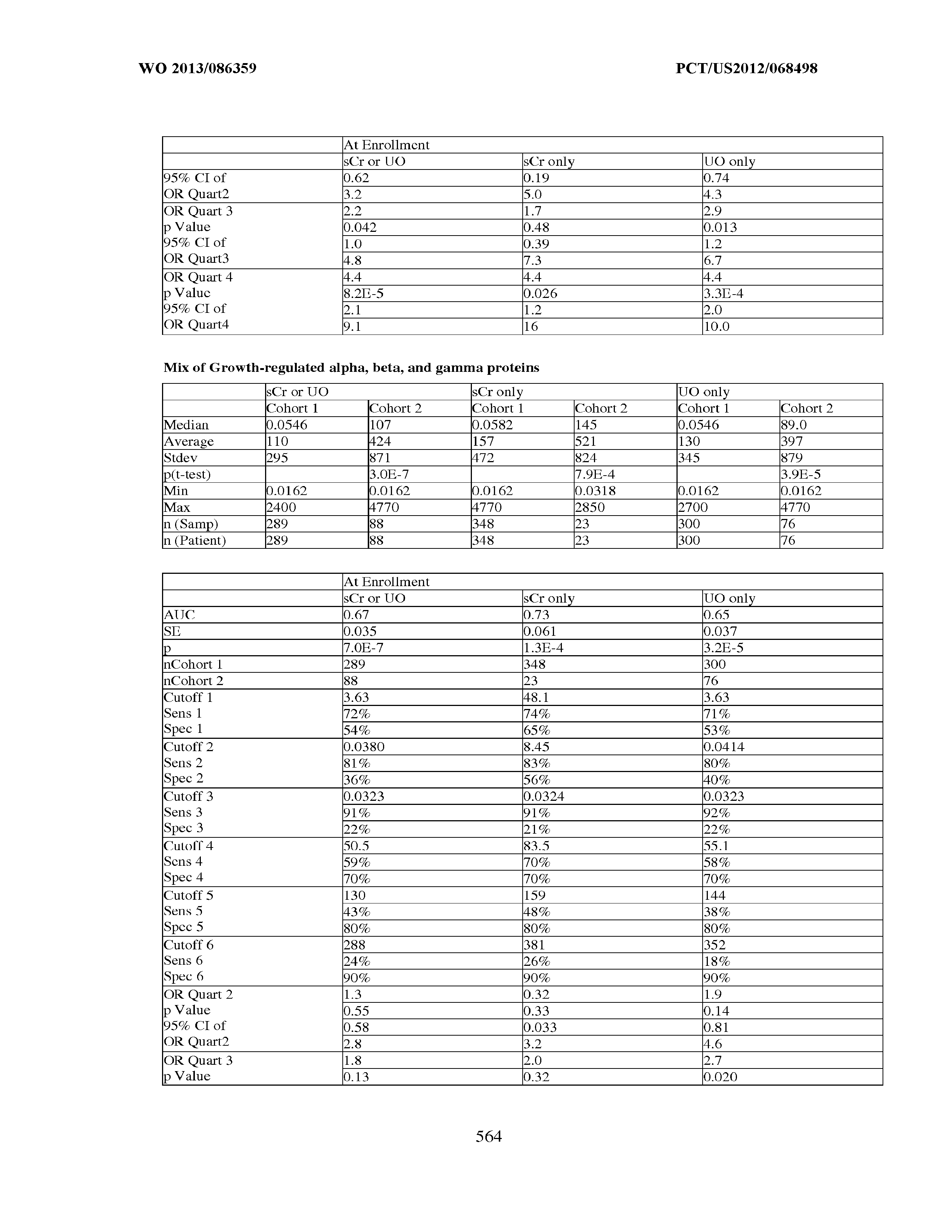






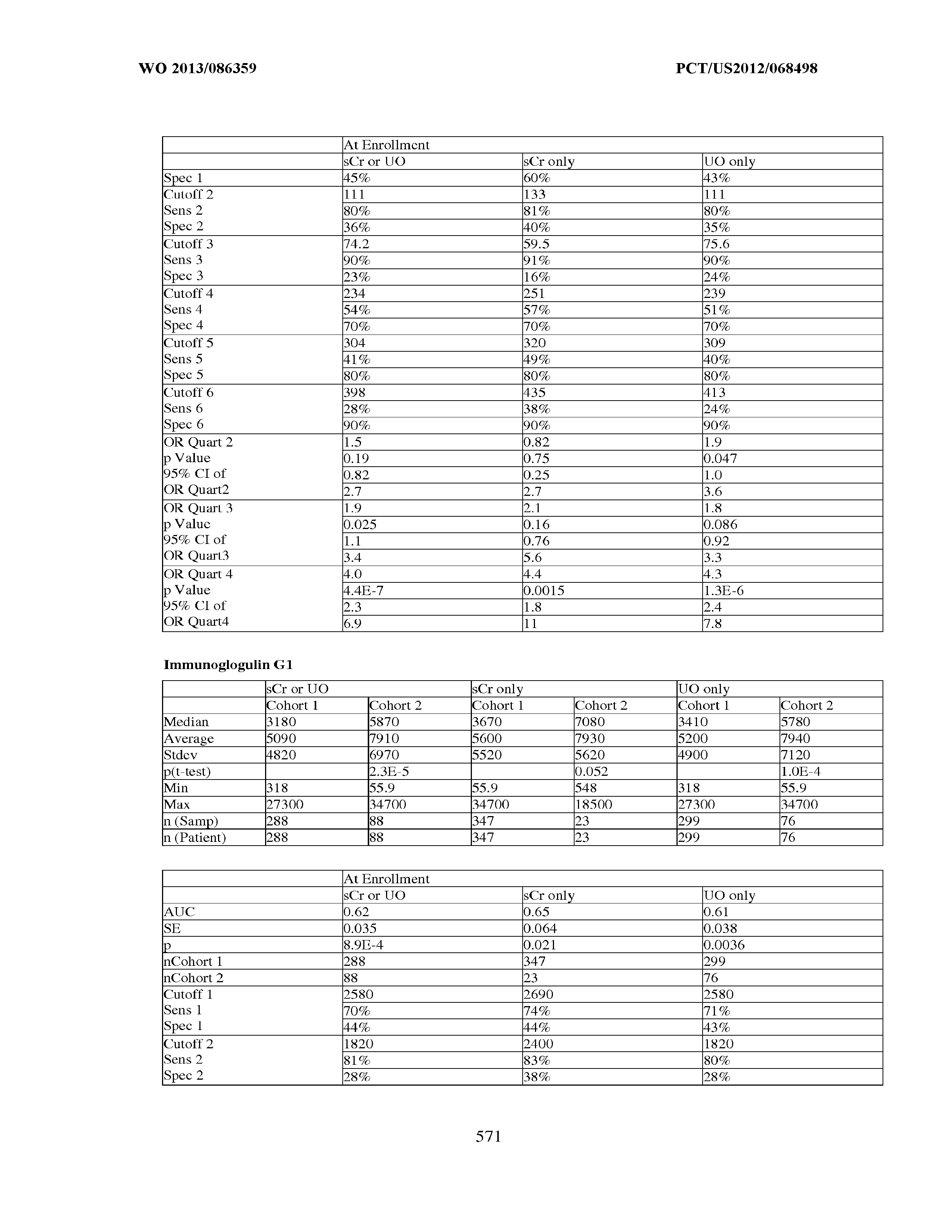
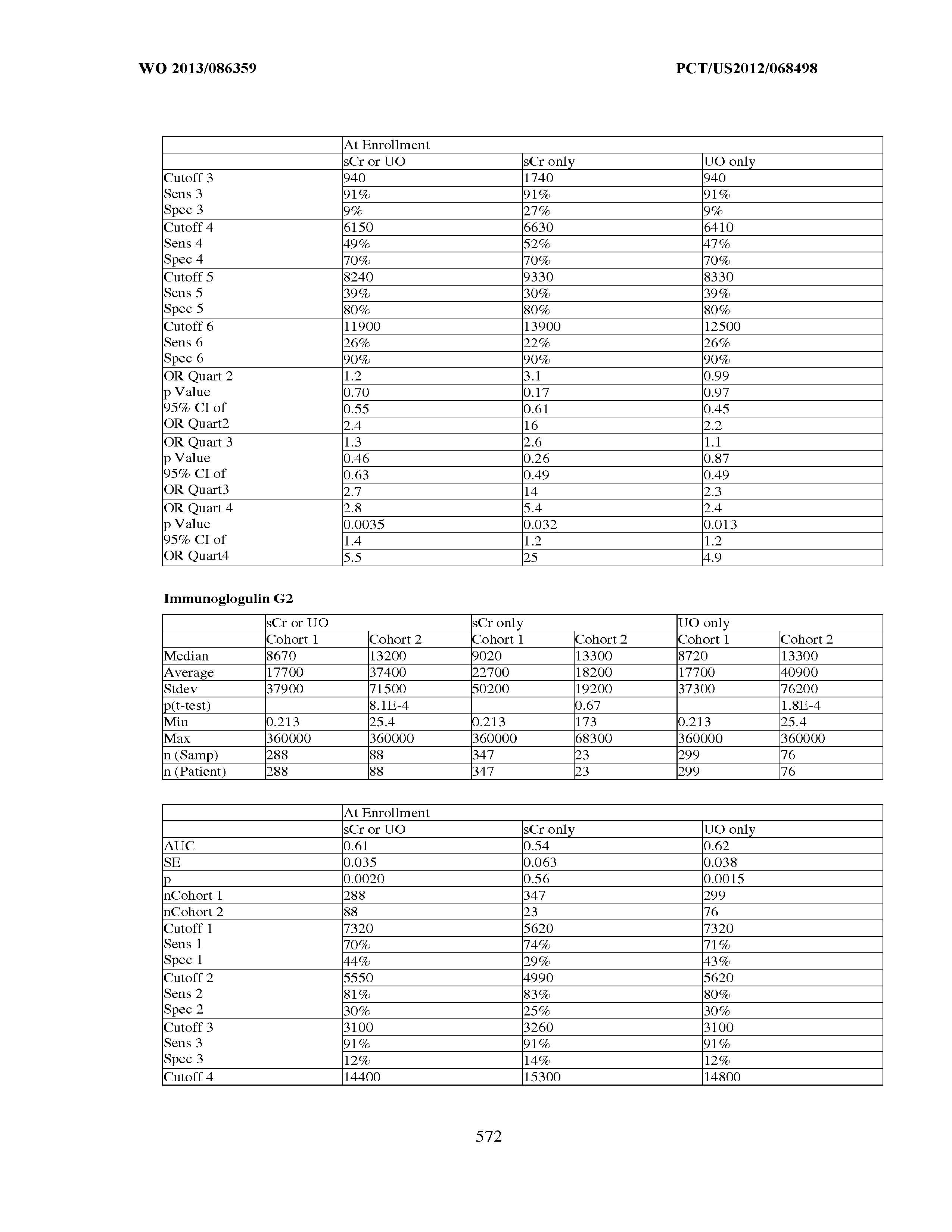










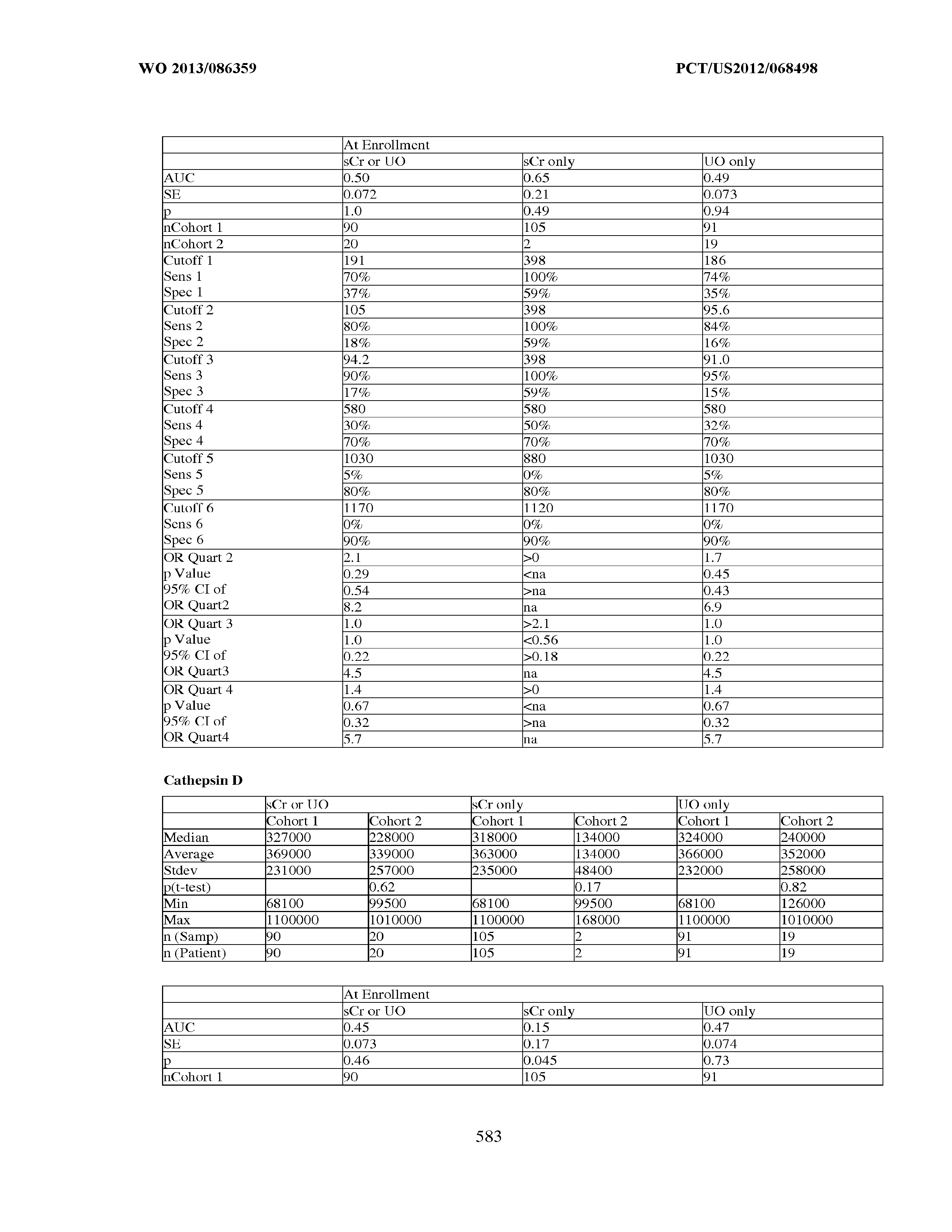
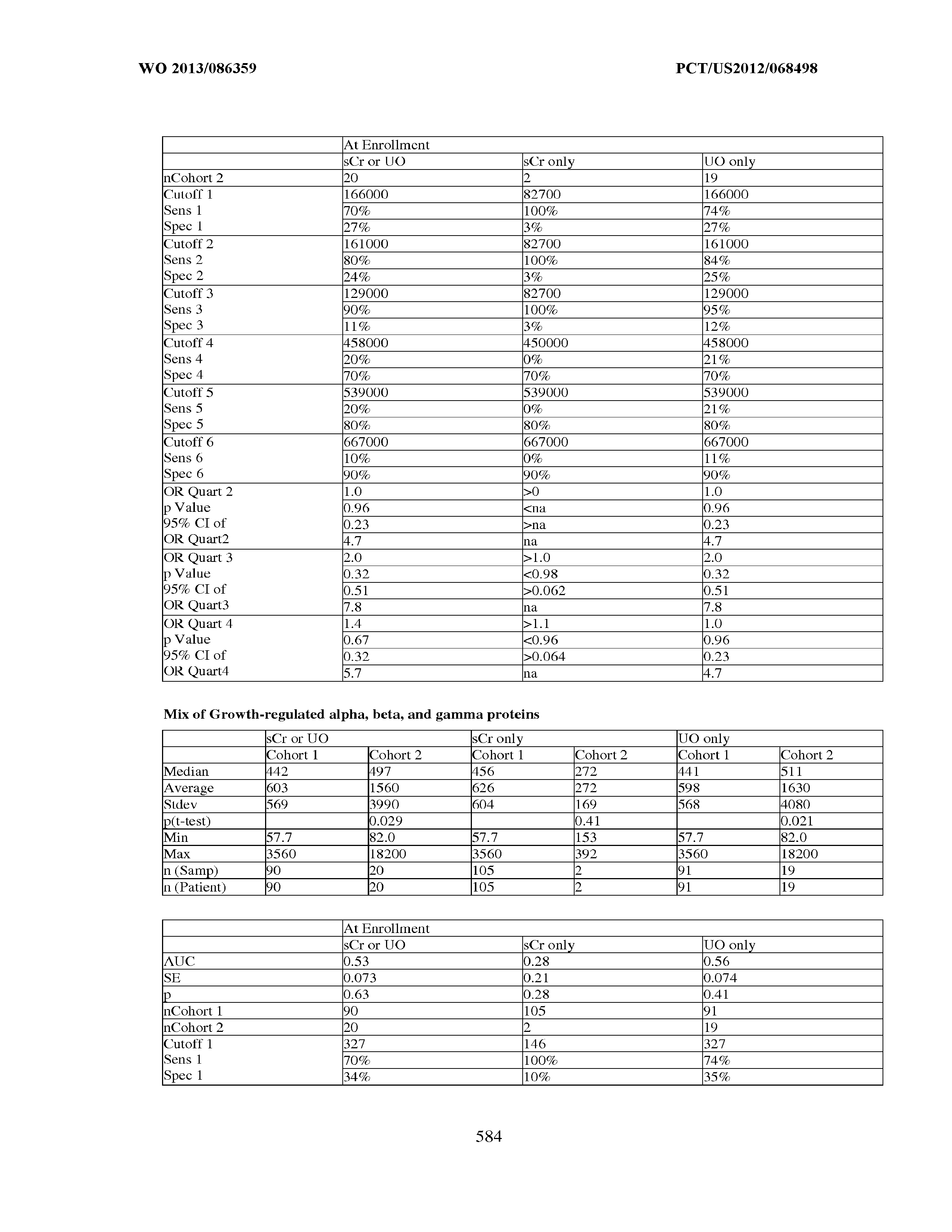








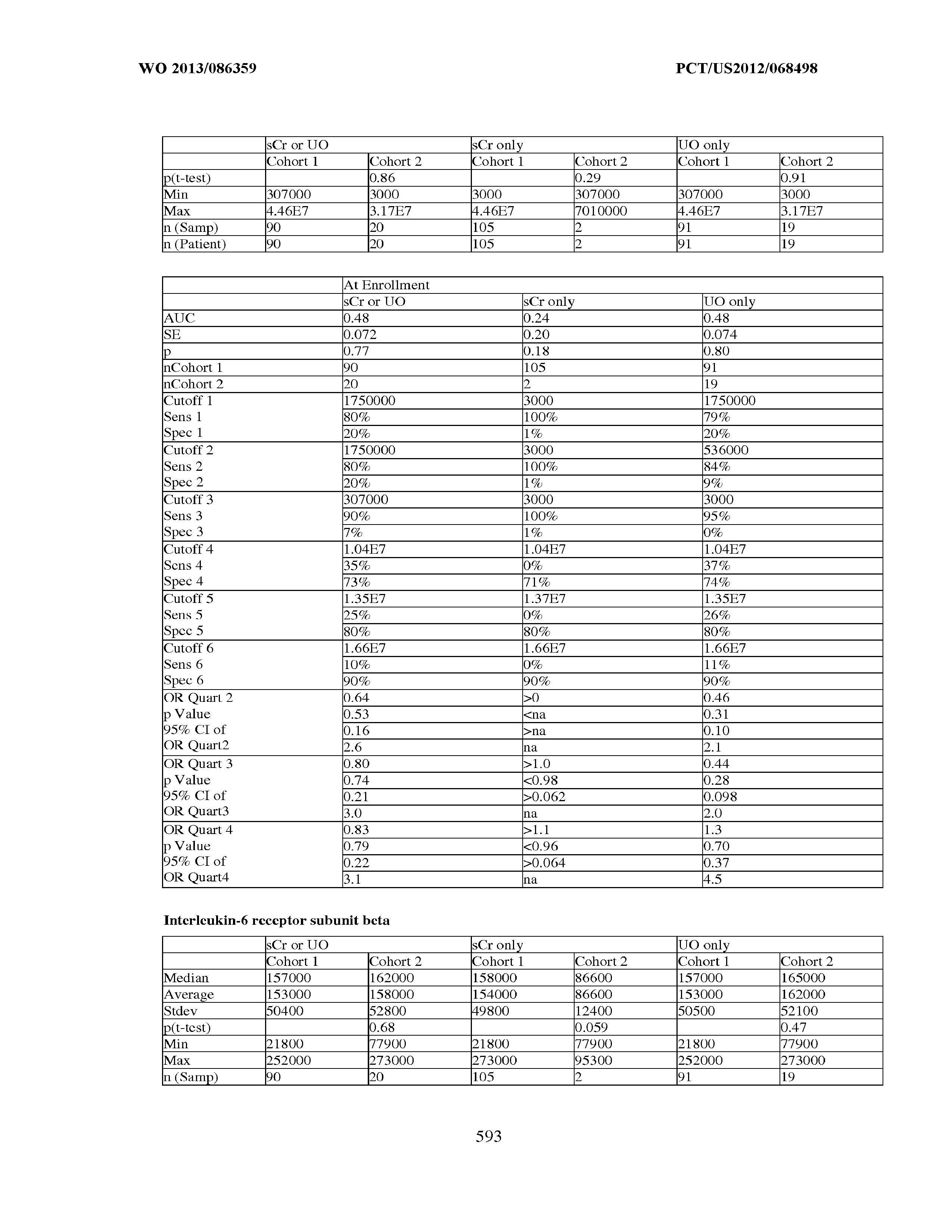






XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.