C3b Binding Polypeptide
BISHOP; Paul ; et al.
U.S. patent application number 16/962108 was filed with the patent office on 2021-05-20 for c3b binding polypeptide. This patent application is currently assigned to THE UNIVERSITY OF MANCHESTER. The applicant listed for this patent is THE UNIVERSITY OF MANCHESTER. Invention is credited to Paul BISHOP, Simon CLARK, Richard UNWIN.
| Application Number | 20210145933 16/962108 |
| Document ID | / |
| Family ID | 1000005401836 |
| Filed Date | 2021-05-20 |


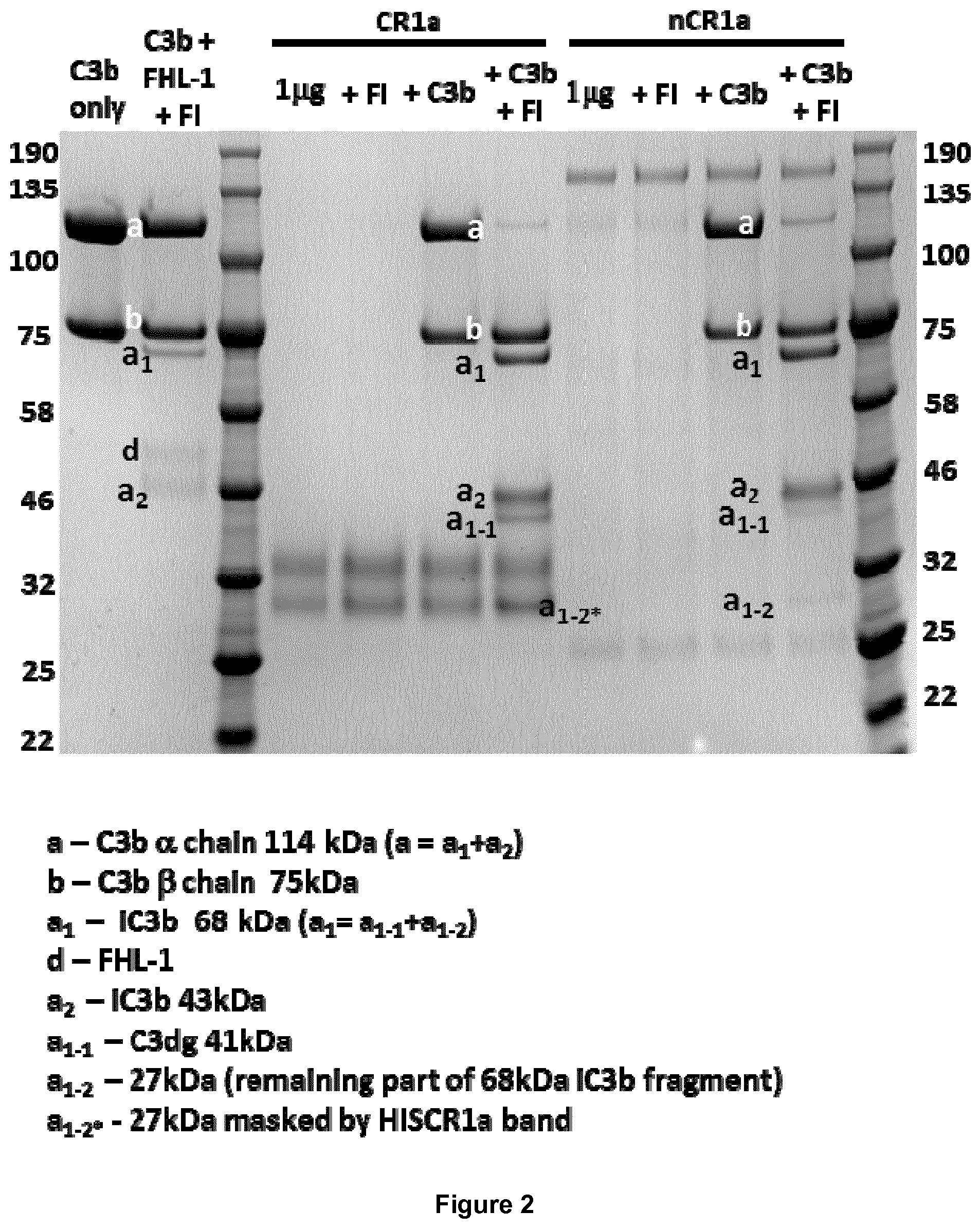


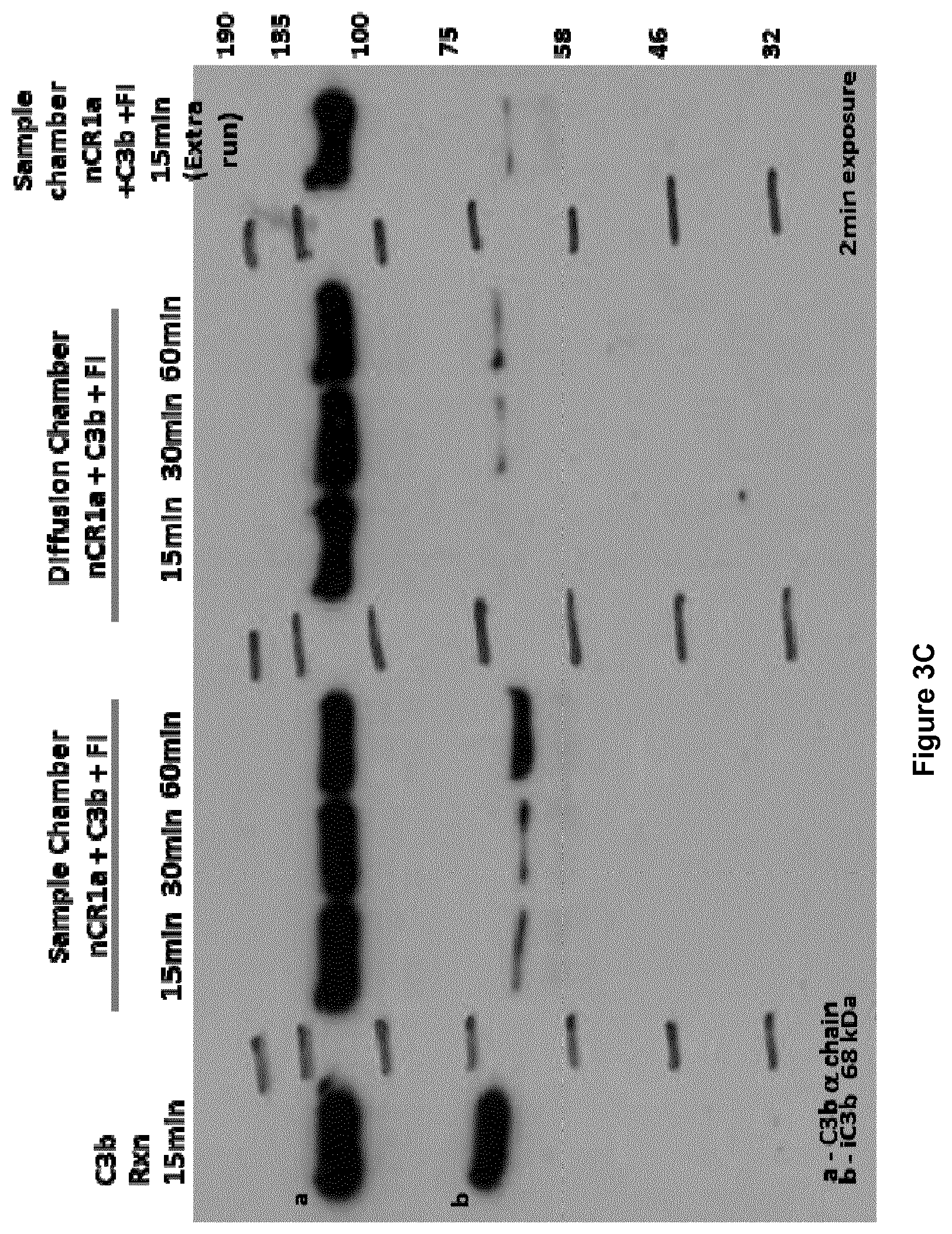


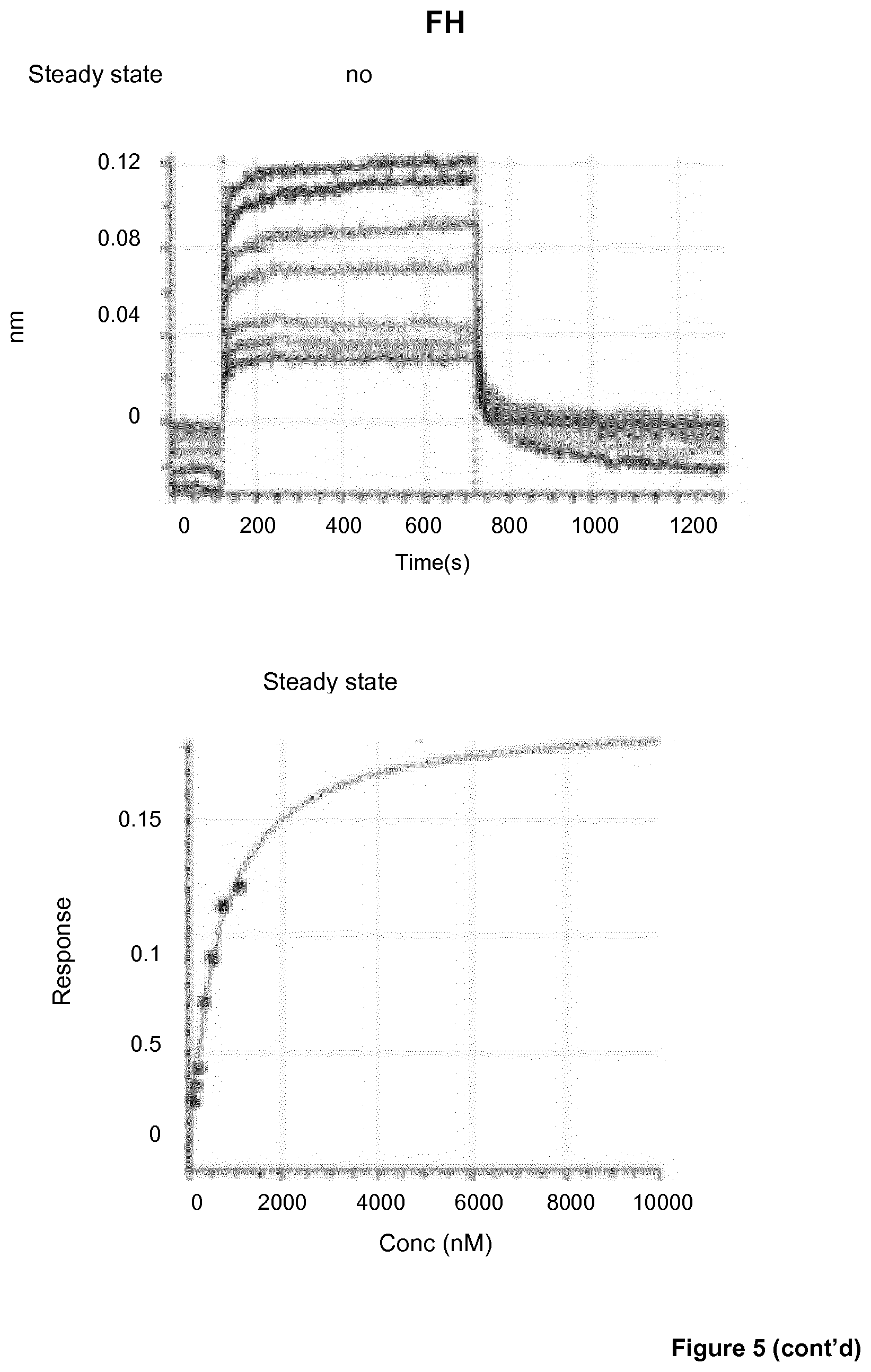
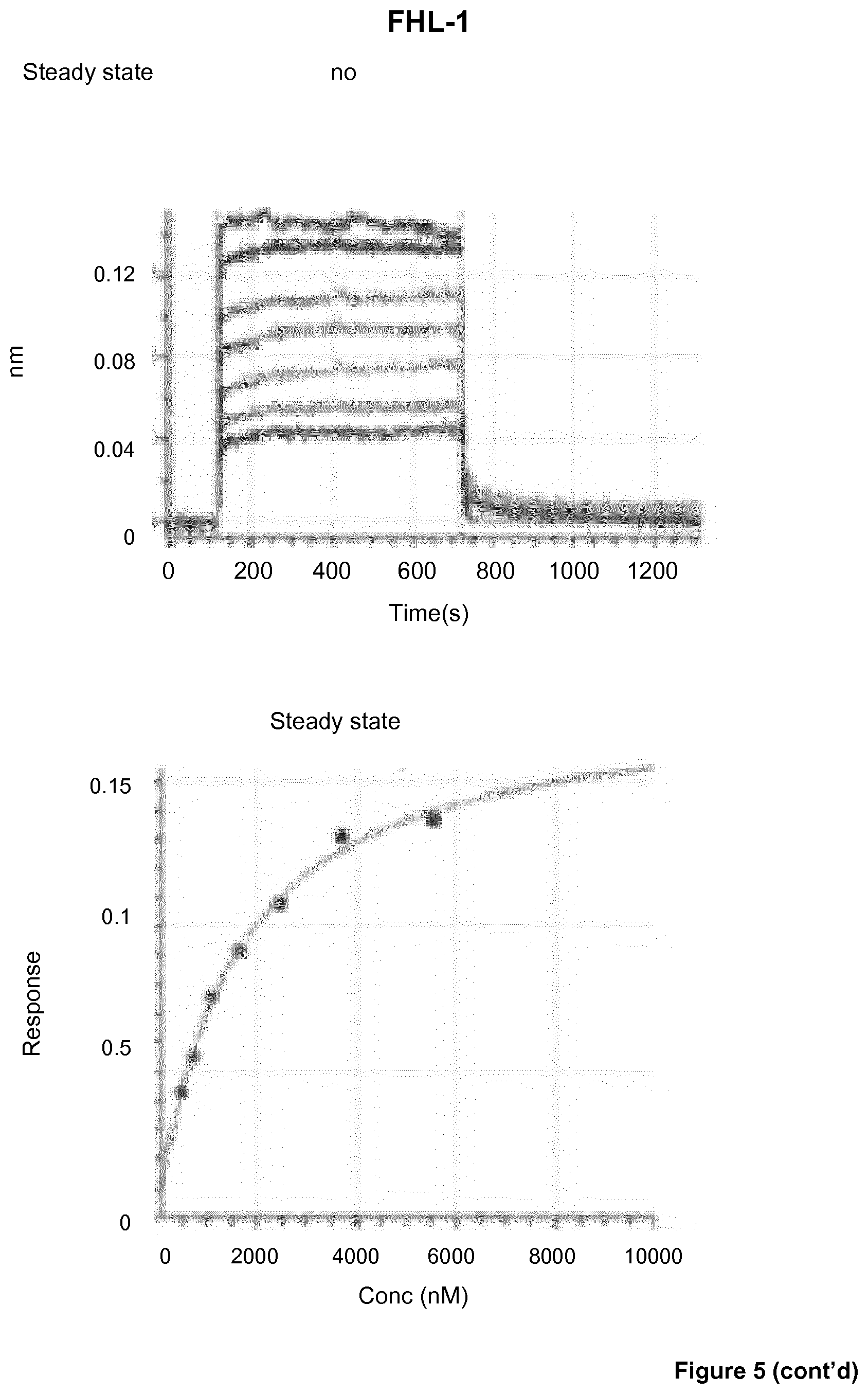


View All Diagrams
| United States Patent Application | 20210145933 |
| Kind Code | A1 |
| BISHOP; Paul ; et al. | May 20, 2021 |
C3B BINDING POLYPEPTIDE
Abstract
Polypeptides comprising a C3b binding region are disclosed, as well as nucleic acids and vectors encoding such polypeptides, and cells and compositions comprising such polypeptides. Also disclosed are uses and methods using the polypeptides for treating and preventing diseases and conditions.
| Inventors: | BISHOP; Paul; (Manchester, GB) ; CLARK; Simon; (Manchester, GB) ; UNWIN; Richard; (Manchester, GB) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Assignee: | THE UNIVERSITY OF
MANCHESTER Manchester GB |
||||||||||
| Family ID: | 1000005401836 | ||||||||||
| Appl. No.: | 16/962108 | ||||||||||
| Filed: | January 15, 2019 | ||||||||||
| PCT Filed: | January 15, 2019 | ||||||||||
| PCT NO: | PCT/EP2019/050949 | ||||||||||
| 371 Date: | July 14, 2020 |
| Current U.S. Class: | 1/1 |
| Current CPC Class: | A61P 27/02 20180101; A61K 38/1709 20130101 |
| International Class: | A61K 38/17 20060101 A61K038/17; A61P 27/02 20060101 A61P027/02 |
Foreign Application Data
| Date | Code | Application Number |
|---|---|---|
| Jan 15, 2018 | GB | 1800620.5 |
Claims
1. A method of treating or preventing a complement-related disease or condition, the method comprising administering a polypeptide which is capable of binding C3b, the polypeptide comprising an amino acid sequence having at least 85% identity to SEQ ID NO:4 and wherein the polypeptide has a total length of 450 amino acids or fewer.
2. (canceled)
3. (canceled)
4. (canceled)
5. A method according to claim 1, wherein the complement-related disease or condition is an ocular disease or condition.
6. A method or use according to claim 5, wherein the treatment or prevention of an ocular disease or condition comprises modifying at least one ocular cell of a subject to express or comprise the polypeptide.
7. A method according to claim 5, wherein the treatment or prevention of an ocular disease or condition comprises modifying at least one ocular cell of a subject to express or comprise a nucleic acid encoding the polypeptide.
8. A method according to claim 5, wherein the treatment or prevention of an ocular disease or condition comprises administering a vector comprising a nucleic acid encoding the polypeptide to at least one ocular cell of a subject.
9. A method according to claim 6, wherein the at least one ocular cell is a retinal pigment epithelial (RPE) cell.
10. A method according to claim 1, wherein the disease or condition is a disease or condition in which C3b or a C3b-containing complex, an activity/response associated with C3b or a C3b-containing complex, or a product of an activity/response associated with C3b or a C3b-containing complex is pathologically implicated.
11. (canceled)
12. A method according to claim 1, wherein the disease or condition is selected from one or more of: macular degeneration, age-related macular degeneration (AMD), early AMD, intermediate AMD, late AMD, geographic atrophy (`dry` AMD), `wet` (neovascular) AMD, choroidal neovascularisation (CNV), glaucoma, autoimmune uveitis, diabetic retinopathy, and early-onset macular degeneration (EOMD).
13. A method according to claim 1, wherein the polypeptide comprises an amino acid sequence having at least 95% identity to SEQ ID NO:4.
14. A method according to claim 1, wherein X.sub.1 is A or T, X.sub.2 is P or L, and/or X.sub.3 is G or R.
15. A method according to claim 1, wherein the polypeptide has a total length of 50 to 250 amino acids.
16. A method according to claim 1, wherein the polypeptide comprises, or consists of, SEQ ID NO:2 SEQ ID NO:3 or SEQ ID NO:13.
17. (canceled)
18. A method according to claim 1, wherein the polypeptide is capable of acting as a co-factor for Complement Factor I.
19. A method according to claim 1, wherein the polypeptide is capable of being expressed functionally by RPE cells and/or is capable of diffusing across Bruch's membrane (BrM).
20. A method according to claim 1, wherein the polypeptide binds to C3b in the region bound by a co-factor for Complement Factor I, or wherein the polypeptide binds to C3b in the region bound by Complement Receptor 1 (CR1).
21. (canceled)
22. A method according to claim 1, wherein the polypeptide comprises a secretory pathway sequence.
23. A method or use according to claim 22, wherein the secretory pathway sequence comprises, or consists of, SEQ ID NO:7, or wherein the polypeptide comprises, or consists of, SEQ ID NO:47, 49, or 51.
24. (canceled)
25. A method according to claim 22, wherein the polypeptide comprises a cleavage site for removing the secretory pathway sequence.
26. A polypeptide which is capable of binding to C3b, the polypeptide comprising an amino acid sequence having at least 85% sequence identity to SEQ ID NO:4 and wherein the polypeptide has a total length of 450 amino acids or fewer.
27. A polypeptide according to claim 26, wherein the polypeptide comprises an amino acid sequence having at least 95% identity to SEQ ID NO:4.
28. A polypeptide according to any claim 26, wherein X.sub.1 is A or T, X.sub.2 is P or L, and/or X.sub.3 is G or R.
29. A polypeptide according to claim 26, wherein the polypeptide has a total length of 50 to 250 amino acids.
30. A polypeptide according to claim 26, comprising, or consisting of, SEQ ID NO:2 SEQ ID NO:3, or SEQ ID NO:13.
31. (canceled)
32. A polypeptide according to claim 26, wherein the polypeptide is capable of acting as a co-factor for Complement Factor I.
33. A polypeptide according to claim 26, wherein the polypeptide is capable of being expressed functionally by RPE cells and/or is capable of diffusing across Bruch's membrane (BrM).
34. A polypeptide according to claim 26, which binds to C3b in the region bound by a co-factor for Complement Factor I or which binds to C3b in the region bound by Complement Receptor 1 (CR1).
35. (canceled)
36. A polypeptide according to claim 26, wherein the polypeptide comprises a secretory pathway sequence.
37. A polypeptide according to claim 36, wherein the secretory pathway sequence comprises, or consists of, SEQ ID NO:7, or wherein the polypeptide comprises, or consists of, SEQ ID NO:47, 49 or 51.
38. (canceled)
39. The polypeptide according to claim 36, wherein the polypeptide comprises a cleavage site for removing the secretory pathway sequence.
40. A nucleic acid encoding the polypeptide according to claim 36.
41. (canceled)
42. (canceled)
43. (canceled)
44. (canceled)
45. (canceled)
46. (canceled)
47. (canceled)
48. (canceled)
49. A method according to claim 1, wherein the treatment or prevention comprises administering a nucleic acid encoding the polypeptide.
50. A polypeptide according to claim 26, wherein the polypeptide is functional on the RPE side of Bruch's membrane and in the choroid and/or choriocapillaris.
Description
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] This application is a National Phase of International Application No. PCT/EP2019/050949 filed Jan. 15, 2019, which designated the U.S. and that International Application was published under PCT Article 21(2) in English. This application also includes a claim of priority under 35 U.S.C. .sctn. 119(a) and .sctn. 365(b) to British patent application No. GB 1800620.5 filed Jan. 15, 2018, the entirety of which is hereby incorporated by reference.
REFERENCE TO A SEQUENCE LISTING
[0002] This application contains a sequence listing named "SequenceListing" in ASCII text file format, created on Jan. 20, 2021, being 148 kb in size, which is herein incorporated by reference as though fully disclosed.
FIELD OF THE INVENTION
[0003] The present invention relates to the fields of molecular biology, immunology, and medicine. More specifically, the present invention relates to a polypeptide that binds to C3b.
BACKGROUND TO THE INVENTION
[0004] Age-related macular degeneration (AMD) is the leading cause of blindness in the developed world: AMD is currently responsible for 8.7% of all global blind registrations and it is estimated that 196 million people will be affected by 2020 (Wong et al. Lancet Glob Heal (2014) 2:e106-16). AMD manifests as the progressive destruction of the macula, the central part of the retina at the back of the eye, leading to loss of central visual acuity. Early stages of the disease see morphological changes in the macula, including first the loss of blood vessels in the choriocapillaris (Whitmore et al., Prog Retin Eye Res (2015) 45:1-29) which are fenestrated blood vessels found in the choroid (a highly vascularized layer that supplies oxygen and nutrition to the outer retina).
[0005] AMD is largely a genetic disease. Mutations in genes of the complement system, part of our immune system, are highly associated with increased risk of AMD. Indeed, it has become clear that over-activation of complement is a main driver of disease pathogenesis and many examples of complement over-activation can be seen in the choriocapillaris. The role of complement in AMD is reviewed, for example, by Zipfel et al. Chapter 2, in Lambris and Adamis (eds.), Inflammation and Retinal Disease: Complement Biology and Pathology, Advances in Experimental Medicine and Biology 703, Springer Science+Business Media, LLC (2010), which is hereby incorporated by reference in its entirety. Complement is activated by the deposition onto a surface of protein C3b, a pro-inflammatory breakdown product of immune system protein C3. C3b associates with other proteins to form convertase enzyme complexes for activating and amplifying complement responses, and initiates the amplification loop of the complement cascade, ultimately leading to cell/tissue destruction and a local inflammatory response (all characteristics of AMD).
[0006] The choriocapillaris is separated from the metabolically active retinal pigment epithelium (RPE) by Bruch's membrane (BrM), a thin (2-4 .mu.m), acellular, five-layered, extracellular matrix. The BrM serves two major functions: the substratum of the RPE and a blood vessel wall. The structure and function of BrM is reviewed e.g. in Curcio and Johnson, Structure, Function and Pathology of Bruch's Membrane, In: Ryan et al. (2013), Retina, Vol. 1, Part 2: Basic Science and Translation to Therapy. 5th ed. London: Elsevier, pp 466-481, which is hereby incorporated by reference in its entirety.
[0007] C3b activation of complement on acellular structures, such as BrM and the intercapillary septa (extracellular matrix filling the spaces between capillaries in the choriocapillaris), is regulated by proteins `complement factor H` (FH) and `complement factor I` (FI). FI prevents complement activation by cleaving C3b to an proteolytically-inactive form, designated iC3b, which is unable to participate in convertase assembly. However, iC3b is an opsonin and therefore a mediator of leucocyte recruitment with a subsequent inflammatory response, whereas the further breakdown products of C3b, iC3dg and C3d, are poor opsonins. In order to cleave C3b, FI requires the presence of a cofactor, examples of which include the blood borne FH protein and the membrane-bound surface co-factor `complement factor 1` (CR1; CD35). CR1 is a membrane receptor expressed on a wide range of cells and is involved in immune complex clearance, phagocytosis, and complement regulation. As well as serving as a co-factor in the FI-mediated cleavage of C3b, CR1 acts as a regulator of complement by accelerating the decay of C3 and C5 convertases. CR1 structure and function is reviewed e.g. in Khera and Das, Mol Immunol (2009) 46(5): 761-772 and Jacquet et al., J Immunol (2013) 190(7): 3721-3731, both of which are hereby incorporated by reference in their entirety.
[0008] Hallmark lesions of early AMD, termed drusen, develop within BrM adjacent to the RPE layer (Bird et al, Surv Ophthalmol (1995) 39(5):367-374). Drusen are formed from the accumulation of lipids and cellular debris, and include a swathe of complement activation products (Anderson et al., Prog Retin Eye Res (2009) 29:95-112; Whitcup et al., Int J Inflam (2013) 1-10). The presence of drusen within BrM disrupts the flow of nutrients from the choroid across this extracellular matrix to the RPE cells, which leads to cell dysfunction and eventual death. As the RPE cell monolayer supports the rod and cone cells of the neurosensory retina by providing nutrients and removing waste, their cell death causes dysfunction of photoreceptor cells and subsequent loss of visual acuity.
[0009] This represents one of the late stages of AMD, known as `dry` AMD and also as geographic atrophy, which represents around 90% of AMD cases. In the remaining percentage of cases of late-stage AMD, the presence of drusen promotes choroidal neovascularisation (CNV), where the increased synthesis of vascular endothelial growth factor (VEGF) by RPE cells promotes new blood vessel growth from the choroid/choriocapillaris that breaks through BrM into the retina. These new blood vessels leak and eventually form scar tissue; this is referred to as `wet` AMD. `Wet` AMD, while only representing 10% of cases, is the most virulent form of late-stage AMD and has different disease characteristics to `dry` AMD. There are treatments for wet AMD, where for example the injection of anti-VEGF agents into the vitreous of the eye can slow or reverse the growth of these blood vessels, although it cannot prevent their formation in the first place. Geographic atrophy (`dry` AMD) remains untreatable.
SUMMARY OF THE INVENTION
[0010] The present invention provides polypeptides that bind to C3b which are useful for treating or preventing complement-related diseases or conditions.
[0011] In one aspect, the present invention provides a polypeptide which is capable of binding C3b, the polypeptide comprising an amino acid sequence having at least 85% identity to SEQ ID NO:4 and wherein the polypeptide has a total length of 450 amino acids or fewer, for use in a method of treating or preventing a complement-related disease or condition.
[0012] Also provided is a nucleic acid encoding a polypeptide which is capable of binding C3b, the polypeptide comprising an amino acid sequence having at least 85% identity to SEQ ID NO:4 and wherein the polypeptide has a total length of 450 amino acids or fewer, for use in a method of treating or preventing a complement-related disease or condition.
[0013] In another aspect, the present invention provides the use of a polypeptide which is capable of binding C3b, the polypeptide comprising an amino acid sequence having at least 85% identity to SEQ ID NO:4 and wherein the polypeptide has a total length of 450 amino acids or fewer, in the manufacture of a medicament for treating or preventing a complement-related disease or condition.
[0014] Also provided is the use of a nucleic acid encoding a polypeptide which is capable of binding C3b, the polypeptide comprising an amino acid sequence having at least 85% identity to SEQ ID NO:4 and wherein the polypeptide has a total length of 450 amino acids or fewer, in the manufacture of a medicament for treating or preventing a complement-related disease or condition.
[0015] In another aspect, provided is a method of treating or preventing a complement-related disease or condition, comprising administering to a subject a polypeptide which is capable of binding C3b, the polypeptide comprising an amino acid sequence having at least 85% identity to SEQ ID NO:4 and wherein the polypeptide has a total length of 450 amino acids or fewer.
[0016] In another aspect, provided is a method of treating or preventing a complement-related disease or condition in a subject, comprising modifying at least one cell of the subject to express or comprise a polypeptide capable of binding C3b, the polypeptide comprising an amino acid sequence having at least 85% identity to SEQ ID NO:4 and wherein the polypeptide has a total length of 450 amino acids or fewer.
[0017] In some embodiments the complement-related disease or condition is an ocular disease or condition.
[0018] In some embodiments the treatment or prevention of an ocular disease or condition comprises modifying at least one ocular cell of a subject to express or comprise the polypeptide. In some embodiments the treatment or prevention of an ocular disease or condition comprises modifying at least one ocular cell of a subject to express or comprise a nucleic acid encoding the polypeptide. In some embodiments the treatment or prevention of an ocular disease or condition comprises administering a vector comprising a nucleic acid encoding the polypeptide to at least one ocular cell of a subject. In some embodiments the at least one ocular cell is a retinal pigment epithelial (RPE) cell.
[0019] In some embodiments the disease or condition is a disease or condition in which C3b or a C3b-containing complex, an activity/response associated with C3b or a C3b-containing complex, or a product of an activity/response associated with C3b or a C3b-containing complex is pathologically implicated.
[0020] In some embodiments the disease or condition is macular degeneration. In some embodiments the disease or condition is selected from one or more of: age-related macular degeneration (AMD), early AMD, intermediate AMD, late AMD, geographic atrophy (`dry` AMD), `wet` (neovascular) AMD, choroidal neovascularisation (CNV), glaucoma, autoimmune uveitis, diabetic retinopathy, and early-onset macular degeneration (EOMD).
[0021] In some embodiments the polypeptide comprises an amino acid sequence having at least 95% identity to SEQ ID NO:4. In some embodiments, X.sub.1 is A or T, X.sub.2 is P or L, and/or X.sub.3 is G or R of SEQ ID NO:4.
[0022] In some embodiments the polypeptide has a total length of 50 to 250 amino acids. In some embodiments the polypeptide comprises, or consists of, SEQ ID NO:2 or SEQ ID NO:3.
[0023] In some embodiments the polypeptide comprises, or consists of, SEQ ID NO:13.
[0024] In some embodiments the polypeptide is capable of acting as a co-factor for Complement Factor I. In some embodiments the polypeptide is capable of diffusing across Bruch's membrane (BrM). In some embodiments the polypeptide binds to C3b in the region bound by a co-factor for Complement Factor I. In some embodiments the polypeptide binds to C3b in the region bound by Complement Receptor 1 (CR1).
[0025] In some embodiments the polypeptide comprises a secretory pathway sequence. In some embodiments the secretory pathway sequence comprises, or consists of, SEQ ID NO:7. In some embodiments the polypeptide comprises, or consists of, SEQ ID NO:47, 49, or 51. In some embodiments the polypeptide comprises a cleavage site for removing the secretory pathway sequence.
[0026] In one aspect, the present invention provides a polypeptide having at least 80% sequence identity to SEQ ID NO:4, wherein the polypeptide has a length of 700 amino acids or fewer.
[0027] In some embodiments, the polypeptide has a length of 50 to 700 amino acids. In some embodiments, the polypeptide has at least 80% sequence identity to SEQ ID NO:4 wherein X.sub.1 is A or T, X.sub.2 is P or L, and/or X.sub.3 is G or R.
[0028] Also provided is a polypeptide which is capable of binding to C3b, the polypeptide comprising an amino acid sequence having at least 85% sequence identity to SEQ ID NO:4 and wherein the polypeptide has a total length of 450 amino acids or fewer.
[0029] In some embodiments the polypeptide comprises an amino acid sequence having at least 95% identity to SEQ ID NO:4. In some embodiments X.sub.1 is A or T, X.sub.2 is P or L, and/or X.sub.3 is G or R.
[0030] In some embodiments the polypeptide has a total length of 50 to 250 amino acids.
[0031] In some embodiments, the polypeptide comprises, or consists of, an amino acid sequence according to SEQ ID NO:2. In some embodiments, the polypeptide comprises, or consists of, an amino acid sequence according to SEQ ID NO:3. In some embodiments, the polypeptide comprises, or consists of, an amino acid sequence according to SEQ ID NO:13.
[0032] In some embodiments, the polypeptide is capable of binding to C3b. In some embodiments, the polypeptide binds to C3b in the region bound by a co-factor for Complement Factor I. In some embodiments, the polypeptide binds to C3b in the region bound by Complement Receptor 1 (CR1).
[0033] In some embodiments, the polypeptide acts as a co-factor for Complement Factor I.
[0034] In some embodiments, the polypeptide is capable of diffusing across Bruch's membrane (BrM). In some embodiments, the polypeptide is not glycosylated or is partially glycosylated. In some embodiments, the polypeptide comprises at least one amino acid substitution, e.g. one, two, three or four substitutions, at position 509, 578, 959 and/or 1028 (numbered according to Uniprot: P17927 (SEQ ID NO:1)). In some embodiments, the at least one amino acid substitution is one or more of N509Q, N578Q, N959Q and/or N1028Q (numbered according to Uniprot: P17927 (SEQ ID NO:1)). In some embodiments, the polypeptide comprises, or consists of, SEQ ID NO:5, SEQ ID NO:6, and/or SEQ ID NO:15.
[0035] In some embodiments, the polypeptide additionally comprises a secretory pathway sequence. In some embodiments, the secretory pathway sequence comprises, or consists of, SEQ ID NO:7. In some embodiments, the polypeptide additionally comprises a cleavage site for removing the secretory pathway sequence. In some embodiments the polypeptide comprises, or consists of, SEQ ID NO:47, 49 or 51.
[0036] In another aspect, the present invention provides a nucleic acid encoding a polypeptide according to the present invention.
[0037] In another aspect, the present invention provides a vector comprising a nucleic acid of the present invention.
[0038] In another aspect, the present invention provides a cell comprising a polypeptide, nucleic acid, or vector according to the present invention.
[0039] In another aspect, the present invention provides a method for producing a polypeptide, comprising introducing into a cell a nucleic acid or a vector according to the present invention, and culturing the cell under conditions suitable for expression of the polypeptide.
[0040] In another aspect, the present invention provides a cell, which is obtained or obtainable by the method for producing a polypeptide according to the present invention.
[0041] In another aspect, the present invention provides a pharmaceutical composition comprising a polypeptide, nucleic acid, vector or cell according to the present invention. In some embodiments, the pharmaceutical composition comprises a pharmaceutically acceptable carrier, adjuvant, excipient, or diluent.
[0042] In another aspect, the present invention provides a polypeptide, nucleic acid, vector, cell or pharmaceutical composition according to the present invention, for use in a method of treating or preventing a disease or condition.
[0043] In another aspect, the present invention provides the use of a polypeptide, nucleic acid, vector, cell or pharmaceutical composition according to the present invention, in the manufacture of a medicament for treating or preventing a disease or condition.
[0044] In another aspect, the present invention provides a method of treating or preventing a disease or condition, comprising administering to a subject a polypeptide, nucleic acid, vector, cell or pharmaceutical composition according to the present invention.
[0045] In another aspect, the present invention provides a method of treating or preventing a disease or condition in a subject, comprising modifying at least one cell of the subject to express or comprise a nucleic acid, vector or polypeptide according to the present invention.
[0046] In some embodiments in accordance with various aspects of the present invention, the disease or condition is a disease or condition in which C3b or a C3b-containing complex, an activity/response associated with C3b or a C3b-containing complex, or a product of an activity/response associated with C3b or a C3b-containing complex is pathologically implicated. In some embodiments, the disease or condition is macular degeneration. In some embodiments, the disease or condition is age-related macular degeneration (AMD). In some embodiments, the method for treating or preventing a disease or condition comprises modifying at least one retinal pigment epithelial (RPE) cell of the subject to express or comprise a nucleic acid, vector, or polypeptide according to the present invention.
[0047] In another aspect, the present invention provides a kit of parts comprising a predetermined quantity of a polypeptide, nucleic acid, vector, cell, or pharmaceutical composition according to the present invention.
DESCRIPTION
[0048] Complement-based therapies for AMD have thus far concentrated upon injecting complement regulating antibodies into the eye. Such therapies have provided little to no therapeutic benefit as these proteins cannot reach the target area, i.e. the BrM and its underlying vasculature, the choriocapillaris at all, or in effective concentrations.
[0049] Complement Factor I (FI)-mediated regulation of complement, i.e. the cleavage of C3b to iC3b (proteolytically-inactive C3b), requires cofactors such as membrane-anchored CR1. However, the present inventors have discovered that it is not necessary to provide full-length membrane-bound CR1, nor even a soluble version of CR1 merely lacking the transmembrane domain, for successful cofactor activity. Instead, the inventors have discovered that short CR1 fragments comprising the CR1 C3b-binding domains are sufficient to enable efficient FI-mediated C3b cleavage.
[0050] Thus, the present invention relates to soluble, truncated polypeptides derived from the FI cofactor CR1. The polypeptides comprise domains that are capable of binding to C3b, such that they can act as essential FI cofactors for the regulation of complement activation. A key advantage of the soluble, truncated polypeptides is their ability to pass through BrM, and thus they are able to reach all regions associated with AMD, i.e. the RPE/BrM interface, BrM and the choroid, including the intercapillary septa (the extracellular matrix between the blood vessels of the choriocapillaris). The present invention also provides non-glycosylated polypeptides derived from CR1, which may aid polypeptide passage through BrM. The polypeptides are expressed and secreted easily, enabling in situ expression by cells local to the affected sites and targeting of the polypeptides to areas affected by complement over-activation. In situ expression of the polypeptides may be achieved using gene therapy techniques. In situ expression provides targeted therapy to areas of need without disrupting functioning complement regulation elsewhere in the body.
[0051] The present invention enables supplementation of a deteriorating complement regulation system without replacing the endogenous complement regulation currently in place, or interfering in the rest of the complement cascade.
[0052] Polypeptides
[0053] A polypeptide according to the present invention may comprise, or consist of, one or more C3b binding regions.
[0054] A polypeptide according to the present invention has at least 80% sequence identity to SEQ ID NO:4, wherein the polypeptide has a length of 700 amino acids or fewer. In some embodiments, the polypeptide comprises, or consists of, an amino acid sequence having at least 80% sequence identity to SEQ ID NO:4, wherein the polypeptide has a length of 700 amino acids or fewer.
[0055] A polypeptide according to the present invention may comprise, or consist of, an amino acid sequence of 700 amino acids or fewer having at least 80% sequence identity to SEQ ID NO:4.
[0056] In some embodiments, the polypeptide comprises, or consists of, an amino acid sequence of 650, 600, 550, 500, 450, 400, 350, 300, 250, or 200 amino acids or fewer. In some embodiments, the polypeptide comprises, or consists of, an amino acid sequence having 1 to 200 amino acids, 1 to 250 amino acids, 1 to 300 amino acids, 1 to 350 amino acids, 1 to 400 amino acids, 1 to 450 amino acids, 1 to 500 amino acids, 1 to 550 amino acids, 1 to 600 amino acids, 1 to 650 amino acids, or 1 to 700 amino acids. In some embodiments, the polypeptide has a length of 50 to 700 amino acids. In some embodiments, the polypeptide has a length of 100 to 650 amino acids. In some embodiments, the polypeptide has a length of 100 to 550 amino acids. In some embodiments, the polypeptide has a length of 150 to 450 amino acids. In some embodiments, the polypeptide has a length of 400 to 700 amino acids. In some embodiments, the polypeptide has a length of 700 to 1000 or greater than 1000 amino acids.
[0057] "Length" as used herein refers to the total length of the polypeptide; that is, "length" refers to the measurement or extent of the entire polypeptide from end to end, i.e. from the N-terminus to the C-terminus. "Length" as used herein is measured by the number of amino acid residues within the polypeptide.
[0058] In some embodiments the polypeptide is a detached/discrete/separate/individual molecule. In some embodiments, the polypeptide is a single contiguous amino acid sequence that is unconnected, i.e. not joined, fused or attached, to another amino acid sequence. In some embodiments the polypeptide is not attached by an amino acid linker or a non-amino acid linker to another polypeptide or amino acid sequence. In some embodiments the polypeptide is not a section, part or region of a longer amino acid sequence, i.e. it is not part of an amino acid sequence that exceeds the maximum, specified, polypeptide length. In some embodiments the polypeptide is not part of, or does not form a section of, a fusion protein. In some embodiments the polypeptide may comprise a sequence provided herein and one or more additional amino acids, as long as the maximum length of the polypeptide is not exceeded. The short length of the polypeptides described herein enables the polypeptides to pass through the BrM and reach sites of complement activation.
[0059] In some embodiments the polypeptide has a total length of 700, 650, 600, 550, 500, 450, 400, 350, 300, 250, or 200 amino acids or fewer. In some embodiments the polypeptide has a total length of 450, 440, 430, 420, 410, 400, 390, 380, 370, 360, 350, 340, 330, 320, 310, 300, 290, 280, 270, 260, 250, 240, 230, 220, 210, 200, 190, 180, 170, 160, 150, 140, 130, 120, 110, 100, 90, 80, or 70 amino acids or fewer. In some embodiments the polypeptide has a total length of 1 to 70 amino acids, 1 to 80 amino acids, 1 to 90 amino acids, 1 to 100 amino acids, 1 to 110 amino acids, 1 to 120 amino acids, 1 to 130 amino acids, 1 to 140 amino acids, 1 to 150 amino acids, 1 to 160 amino acids, 1 to 170 amino acids, 1 to 180 amino acids, 1 to 190 amino acids, 1 to 200 amino acids, 1 to 210 amino acids, 1 to 220 amino acids, 1 to 230 amino acids, 1 to 240 amino acids, 1 to 250 amino acids, 1 to 260 amino acids, 1 to 270 amino acids, 1 to 280 amino acids, 1 to 290 amino acids, 1 to 300 amino acids, 1 to 310 amino acids, 1 to 320 amino acids, 1 to 330 amino acids, 1 to 340 amino acids, 1 to 350 amino acids, 1 to 360 amino acids, 1 to 370 amino acids, 1 to 380 amino acids, 1 to 390 amino acids, 1 to 400 amino acids, 1 to 410 amino acids, 1 to 420 amino acids, 1 to 430 amino acids, 1 to 440 amino acids, or 1 to 450 amino acids. In some embodiments the polypeptide has a total length of 50 to 450 amino acids, 50 to 400 amino acids, 50 to 350 amino acids, 50 to 300 amino acids, 50 to 250 amino acids, 50 to 200 amino acids, 100 to 250 amino acids, 100 to 200 amino acids, 150 to 250 amino acids, or 150 to 200 amino acids. In some embodiments the polypeptide has a total length of one of 61, 72, 194, 212, 231, 388, 406, or 644 amino acids.
[0060] In some embodiments a polypeptide of the present invention has a maximum molecular weight of 80 kDa, whether the polypeptide is covalently/non-covalently bonded to a larger complex, part of a larger complex, or is not part of a larger complex. In some embodiments a polypeptide of the present invention has a molecular weight of 75 kDa or less, 70 kDa or less, 65 kDa or less, 60 kDa or less, 55 kDa or less, 50 kDa or less, 45 kDa or less, 40 kDa or less, 35 kDa or less, 30 kDa or less, 29 kDa or less, 28 kDa or less, 27 kDa or less, 26 kDa or less, 25 kDa or less, 24 kDa or less, 23 kDa or less, 22 kDa or less, 21 kDa or less, 20 kDa or less, 19 kDa or less, 18 kDa or less, 17 kDa or less, 16 kDa or less, 15 kDa or less, 14 kDa or less, 13 kDa or less, 12 kDa or less, 11 kDa or less, or 10 kDa or less. In some embodiments the polypeptide has a maximum molecular weight of 50 kDa, i.e. 50 kDa or less. In some embodiments the polypeptide has a maximum molecular weight of 26 kDa, i.e. 26 kDa or less. In some embodiments the polypeptide has a maximum molecular weight of 24 kDa, i.e. 24 kDa or less. In some embodiments the polypeptide has a maximum molecular weight of 22 kDa, i.e. 22 kDa or less. In some embodiments the polypeptide has a maximum molecular weight of 20 kDa, i.e. 20 kDa or less.
[0061] In some embodiments, a polypeptide of the present invention comprises, or consists of, an amino acid sequence having at least 70%, 75%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID NO:4. In some embodiments, X.sub.1 is A or T, X.sub.2 is P or L, and/or X.sub.3 is G or R. In some embodiments, X.sub.1 is A, X.sub.2 is P, and/or X.sub.3 is G. In some embodiments, X.sub.1 is A, X.sub.2 is L, and/or X.sub.3 is R. In some embodiments, X.sub.1 is A, X.sub.2 is P, and/or X.sub.3 is R. In some embodiments, X.sub.1 is A, X.sub.2 is L, and/or X.sub.3 is G. In some embodiments, X.sub.1 is T, X.sub.2 is L, and/or X.sub.3 is R. In some embodiments, X.sub.1 is T, X.sub.2 is P, and/or X.sub.3 is G. In some embodiments, X.sub.1 is T, X.sub.2 is L, and/or X.sub.3 is G. In some embodiments, X.sub.1 is T, X.sub.2 is P, and/or X.sub.3 is R.
[0062] In some embodiments the polypeptide comprises, or consists of, an amino acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID NO:4, wherein the polypeptide has a length provided herein. For example, in some embodiments the polypeptide comprises, or consists of, an amino acid sequence having 85% sequence identity to SEQ ID NO:4, wherein the polypeptide has a total length of 450, 440, 430, 420, 410, 400, 390, 380, 370, 360, 350, 340, 330, 320, 310, 300, 290, 280, 270, 260, 250, 240, 230, 220, 210, 200, 190, 180, 170, or 160 amino acids or fewer; in some embodiments the polypeptide comprises, or consists of, an amino acid sequence having 90% sequence identity to SEQ ID NO:4, wherein the polypeptide has a total length of 450, 440, 430, 420, 410, 400, 390, 380, 370, 360, 350, 340, 330, 320, 310, 300, 290, 280, 270, 260, 250, 240, 230, 220, 210, 200, 190, or 180 amino acids or fewer; in some embodiments the polypeptide comprises, or consists of, an amino acid sequence having 95% sequence identity to SEQ ID NO:4, wherein the polypeptide has a total length of 450, 440, 430, 420, 410, 400, 390, 380, 370, 360, 350, 340, 330, 320, 310, 300, 290, 280, 270, 260, 250, 240, 230, 220, 210, 200, 190, or 180 amino acids or fewer; in some embodiments the polypeptide comprises, or consists of, an amino acid sequence having 98% sequence identity to SEQ ID NO:4, wherein the polypeptide has a total length of 450, 440, 430, 420, 410, 400, 390, 380, 370, 360, 350, 340, 330, 320, 310, 300, 290, 280, 270, 260, 250, 240, 230, 220, 210, or 200 amino acids or fewer.
[0063] In some embodiments the polypeptide comprises, or consists of, an amino acid sequence having at least 85% sequence identity to SEQ ID NO:4, wherein the polypeptide has a total length of 450 amino acids or fewer. In some embodiments the polypeptide comprises, or consists of, an amino acid sequence having at least 85% sequence identity to SEQ ID NO:4, wherein the polypeptide has a total length of 50 to 450 amino acids. In some embodiments the polypeptide comprises, or consists of, an amino acid sequence having at least 85% sequence identity to SEQ ID NO:4, wherein the polypeptide has a total length of 250 amino acids or fewer. In some embodiments the polypeptide comprises, or consists of, an amino acid sequence having at least 85% sequence identity to SEQ ID NO:4, wherein the polypeptide has a total length of 50 to 250 amino acids. In some embodiments the polypeptide comprises, or consists of, an amino acid sequence having at least 95% sequence identity to SEQ ID NO:4, wherein the polypeptide has a total length of 450 amino acids or fewer. In some embodiments the polypeptide comprises, or consists of, an amino acid sequence having at least 95% sequence identity to SEQ ID NO:4, wherein the polypeptide has a total length of 50 to 450 amino acids. In some embodiments the polypeptide comprises, or consists of, an amino acid sequence having at least 95% sequence identity to SEQ ID NO:4, wherein the polypeptide has a total length of 250 amino acids or fewer. In some embodiments the polypeptide comprises, or consists of, an amino acid sequence having at least 95% sequence identity to SEQ ID NO:4, wherein the polypeptide has a total length of 50 to 250 amino acids.
[0064] Human CR1 (UniProt: P17927 (Entry version 181 (25 Oct. 2017), Sequence version 3 (2 Mar. 2010)); SEQ ID NO:1) has a 2,039 amino acid sequence (including an N-terminal, 41 amino acid signal peptide), and comprises 30 complement control protein (CCP) domains (also known as sushi domains or short consensus repeats (SCRs)), with the N-terminal 28 CCPs organised into four long homologous repeat (LHR) domains each comprising 7 CCPs: LHR-A, LHR-B, LHR-C and LHR-D. The C3b binding region of CR1 is found in CCPs 8-10 in LHR-B (UniProt: P17927 positions 491 to 684; SEQ ID NO:2), and CCPs 15-17 in LHR-C (UniProt: P17927 positions 941 to 1134; SEQ ID NO:3). CCPs 8-10 and 15-17 differ in sequence by three amino acid residues, as shown in consensus sequence SEQ ID NO:4.
[0065] A polypeptide according to the present invention may comprise, or consist of, an amino acid sequence corresponding to CCPs 8-10 (SEQ ID NO:2) and/or CCPs 15-17 (SEQ ID NO:3). In some embodiments, a polypeptide of the present invention comprises, or consists of, an amino acid sequence having at least 70%, 75%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID NO:2 and/or SEQ ID NO:3. Such polypeptides may have any length provided herein.
[0066] A polypeptide according to the present invention may comprise, or consist of, an amino acid sequence corresponding to CCPs 8-10 and 15-17. The polypeptide may comprise or consist of CCPs 8-10 and 15-17 in their native CR1 sequence (SEQ ID NO:30). The polypeptide may comprise or consist of CCPs 8-10 joined to CCPs 15-17. This may be in a contiguous sequence (SEQ ID NO:13), or achieved by a linker between CCPs 8-10 and 15-17 (e.g. SEQ ID NO:14). In some embodiments, a polypeptide of the present invention comprises, or consists of, an amino acid sequence having at least 70%, 75%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID NO:13, SEQ ID NO:14 and/or SEQ ID NO:30. Such polypeptides may have any length provided herein.
[0067] A polypeptide according to the present invention may comprise, or consist of, an amino acid sequence corresponding to one or more of sequence `A` (SEQ ID NO:8), sequence `B` (SEQ ID NO:16) and/or `Sequence C` (SEQ ID NO:17). In some embodiments, the polypeptide consists of a sequence selected from sequence `A` (SEQ ID NO:8), sequence `B` (SEQ ID NO:16) and `Sequence C` (SEQ ID NO:17). In some embodiments, a polypeptide of the present invention comprises, or consists of, an amino acid sequence having at least 70%, 75%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to one or more of sequences `A` (SEQ ID NO:8), `B` (SEQ ID NO:16) and/or `C` (SEQ ID NO:17).
[0068] In some embodiments, where a polypeptide according to the present invention comprises, or consists of, sequence `B` (SEQ ID NO:16) or an amino acid sequence having at least 70%, 75%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to sequence `B` (SEQ ID NO:16), X.sub.1 is A or T. In some embodiments, X.sub.1 is A. In some embodiments, X.sub.1 is T.
[0069] In some embodiments, where a polypeptide according to the present invention comprises, or consists of, sequence `C` (SEQ ID NO:17) or an amino acid sequence having at least 70%, 75%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to sequence `C` (SEQ ID NO:17), X.sub.2 is P or L and/or X.sub.3 is G or R. In some embodiments, X.sub.2 is P and/or X.sub.3 is G. In some embodiments, X.sub.2 is P and/or X.sub.3 is R. In some embodiments, X.sub.2 is L and/or X.sub.3 is G. In some embodiments, X.sub.2 is L and/or X.sub.3 is R.
[0070] In some embodiments sequence `B` corresponds to SEQ ID NO:9 or SEQ ID NO:11. In some embodiments sequence `C` corresponds to SEQ ID NO:10 or SEQ ID NO:12.
[0071] The present invention includes polypeptides comprising sequences `A`, `B`, and/or `C` as described herein, and combinations thereof, including at least the following combinations (organised from N-terminus to C-terminus): [0072] A+B [0073] B+C [0074] A+C [0075] C+A [0076] A+B+C [0077] B+C+A [0078] C+A+B [0079] A+B+C+A [0080] B+C+A+B [0081] C+A+B+C [0082] A+B+C+A+B [0083] B+C+A+B+C [0084] A+B+C+A+B+C [0085] A+B+C+A+B+C+Y (where Y=one or more of A, B and/or C).
[0086] In some embodiments, the polypeptide comprises, or consists of, an amino acid sequence having at least 70%, 75%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to any combination of CCP domains or any combination of sequences `A`, `B` and/or `C` described herein.
[0087] In some embodiments, the combination of CCP domains is a combination found in native CR1. In some embodiments, the combination of CCP domains is not a combination found in native CR1.
[0088] In some embodiments, the polypeptide comprises, or consists of, an amino acid sequence having multiple copies of sequence `A`, multiple copies of sequence `B`, and/or multiple copies of sequence `C`. In some embodiments, the polypeptide comprises 1, 2, 3, 4, 5, 6, 7, 8 or more copies of one or more of sequences `A`, `B` and/or `C`. In some embodiments, the polypeptide comprises 1, 2, 3, 4, 5, 6, 7, 8 or more copies of sequence `A`; 1, 2, 3, 4, 5, 6, 7, 8 or more copies of sequence `B`; and/or 1, 2, 3, 4, 5, 6, 7, 8 or more copies of sequence `C`. In some embodiments, the polypeptide comprises 9, 10, or more copies of one or more of sequences `A`, `B` and/or `C`.
[0089] In some embodiments, a polypeptide according to the present invention lacks substantial sequence identity to one or more of amino acid sequences 1-490, 685-940 and/or 1135-2039 of human CR1 (SEQ ID NO:1). A polypeptide that lacks substantial sequence identity as described herein may have less than 80%, less than 75%, less than 70%, less than 65%, less than 60%, less than 55%, less than 50%, less than 45%, less than 40%, less than 35%, less than 30%, less than 25%, less than 20%, less than 15%, less than 10%, or less than 5% sequence identity to one or more of amino acid sequences 1-490, 685-940 and/or 1135-2039 of human CR1 (SEQ ID NO:1). In some embodiments a polypeptide according to the present invention lacks amino acid sequence having substantial sequence identity to CR1 long homologous repeat (LHR) domains LHR-A and/or LHR-D. In some embodiments a polypeptide according to the present invention lacks amino acid sequence having substantial sequence identity to CR1 CCP domains 1-7, 11-14 and/or 18-30. Amino acid residues of SEQ ID NO:1 are numbered according to Uniprot P17927; Entry version 181 (25 Oct. 2017), Sequence version 3 (2 Mar. 2010).
[0090] A polypeptide according to the present invention, and/or described herein, may be isolated and/or substantially purified.
[0091] Further Features of the Polypeptide
[0092] Polypeptides according to the present invention may comprise modifications and/or additional amino acid sequences. The modifications and/or additional amino acid sequences may be included in the length limitation of a polypeptide provided herein such that the length limitation of that polypeptide is not exceeded.
[0093] In some embodiments, an additional amino acid sequence comprises, or consists of, no more than 25, 50, 100, 150, or 200 amino acids, i.e. an additional amino acid sequence comprises, or consists of, 1-25, 1-50, 1-100, 1-150, or 1-200 amino acids. In some embodiments, an additional amino acid sequence comprises more than 200 amino acids. In some embodiments, an additional amino acid sequence comprises no more than 100 amino acids at the C-terminus of a polypeptide according to the present invention, and/or no more than 100 amino acids at the N-terminus of a polypeptide according to the present invention.
[0094] In some embodiments, an additional amino acid sequence results in a polypeptide longer than 700 amino acids. In some embodiments, a polypeptide according to the present invention comprises, or consists of, 700 or more amino acids. For example, the polypeptide may comprise, or consist of, 700-750, 750-800, 800-850, 850-900, 900-950, 950-100, or more than 1000 amino acids.
[0095] In some embodiments, an additional amino acid sequence described herein lacks substantial sequence identity to one or more of amino acid sequences 1-490, 685-940 and/or 1135-2039 of human CR1 (SEQ ID NO:1, numbered according to Uniprot P17927; Entry version 181 (25 Oct. 2017), Sequence version 3 (2 Mar. 2010)). In some embodiments, the additional amino acid sequence lacks substantial sequence identity to CR1 CCP domains 1-7, 11-14 and/or 18-30. In some embodiments, the additional amino acid sequence has less than 80%, less than 75%, less than 70%, less than 65%, less than 60%, less than 55%, less than 50%, less than 45%, less than 40%, less than 35%, less than 30%, less than 25%, less than 20%, less than 15%, less than 10%, or less than 5% sequence identity to one or more of amino acid sequences 1-490, 685-940 and/or 1135-2039 of human CR1 (SEQ ID NO:1). In some embodiments the additional amino acid sequence lacks substantial sequence identity to CR1 long homologous repeat (LHR) domains LHR-A and/or LHR-D.
[0096] In some embodiments, a polypeptide may lack amino acid sequence having substantial sequence identity to a region of a co-factor for Complement Factor I (e.g. CR1) other than in the C3b binding region. For example, the polypeptide may lack amino acid sequence having substantial sequence identity to CR1 other than in CR1 CCP domains 8-10 and/or 15-17 (residues 491 to 684 and/or 941 to 1134, respectively, of SEQ ID NO:1). In some embodiments, the polypeptide may lack amino acid sequence having substantial sequence identity to CR1 CCP domains 1-7, 11-14 and/or 18-30. A polypeptide lacking amino acid sequence having substantial sequence identity as described herein may have less than 80%, less than 75%, less than 70%, less than 65%, less than 60%, less than 55%, less than 50%, less than 45%, less than 40%, less than 35%, less than 30%, less than 25%, less than 20%, less than 15%, less than 10%, or less than 5% sequence identity to one or more of amino acid sequences 1-490, 685-940 and/or 1135-2039 of human CR1 (SEQ ID NO:1).
[0097] In some embodiments, a polypeptide according to the present invention may comprise a secretory pathway sequence. As used herein, a secretory pathway sequence is an amino acid sequence which directs secretion of polypeptide. The secretory pathway sequence may be cleaved from the mature protein once export of the polypeptide chain across the rough endoplasmic reticulum is initiated. Polypeptides secreted by mammalian cells generally have a signal peptide fused to the N-terminus of the polypeptide, which is cleaved from the translated polypeptide to produce a "mature" form of the polypeptide.
[0098] In some embodiments, the secretory pathway sequence may comprise or consist of a leader sequence (also known as a signal peptide or signal sequence). Leader sequences normally consist of a sequence of 5-30 hydrophobic amino acids, which form a single alpha helix. Secreted proteins and proteins expressed at the cell surface often comprise leader sequences. The leader sequence may be present in the newly-translated polypeptide (e.g. prior to processing to remove the leader sequence). Leader sequences are known for many proteins, and are recorded in databases such as GenBank, UniProt, Swiss-Prot, TrEMBL, Protein Information Resource, Protein Data Bank, Ensembl, and InterPro, and/or can be identified/predicted e.g. using amino acid sequence analysis tools such as SignalP (Petersen et al., 2011 Nature Methods 8: 785-786) or Signal-BLAST (Frank and Sippl, 2008 Bioinformatics 24: 2172-2176).
[0099] In some embodiments, the secretory pathway sequence is derived from Complement Factor H (FH). In some embodiments, the secretory pathway sequence comprises or consists of SEQ ID NO:7. In some embodiments, the secretory pathway sequence of the polypeptide of the present invention comprises, or consists of, an amino acid sequence having at least 80%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to the amino acid sequence of SEQ ID NO:7.
[0100] In some embodiments a polypeptide according to the present invention comprises, or consists of, an amino acid sequence corresponding to SEQ ID NO:47, 49, and/or 51. In some embodiments the polypeptide comprises, or consists of, an amino acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID NO:47, 49 and/or 51. Such polypeptides may have any length provided herein.
[0101] In some embodiments, a polypeptide according to the present invention may additionally comprise a cleavage site for removing the secretory pathway sequence from the polypeptide. In some embodiments, the cleavage site for removing the secretory pathway sequence from the polypeptide is a cleavage site for an endoprotease. In some embodiments, the cleavage site is for an endoprotease expressed by the cell in which the polypeptide is expressed. In some embodiments, the cleavage site is a signal peptidase cleavage site. In some embodiments, the cleavage site is a protease cleavage site, e.g. a cleavage site for an endoprotease expressed by cells expressing the polypeptide. In some embodiments, the cleavage site is a cleavage site for an endoprotease expressed by RPE cells.
[0102] A polypeptide according to the present invention may comprise one or more linker sequences between amino acid sequences. A linker sequence may be provided between any two or more of sequences `A`, `B` and/or `C`. In some embodiments, a polypeptide according to the present invention comprises, or consists of, an amino acid sequence A+B+C-[LINKER]-A+B+C. In some embodiments, a polypeptide comprises, or consists of, SEQ ID NO:14.
[0103] Linker sequences are known to the skilled person, and are described, for example in Chen et al., Adv Drug Deliv Rev (2013) 65(10): 1357-1369, which is hereby incorporated by reference in its entirety. In some embodiments, a linker sequence may be a flexible linker sequence. Flexible linker sequences allow for relative movement of the amino acid sequences which are linked by the linker sequence. Flexible linkers are known to the skilled person, and several are identified in Chen et al., Adv Drug Deliv Rev (2013) 65(10): 1357-1369. Flexible linker sequences often comprise high proportions of glycine and/or serine residues.
[0104] In some embodiments, the linker sequence comprises at least one glycine residue and/or at least one serine residue. In some embodiments the linker sequence consists of glycine and serine residues. In some embodiments, the linker sequence has a length of 1-2, 1-3, 1-4, 1-5, 1-10, 1-15, 1-20, 1-25, 1-30 or 1-35 amino acids.
[0105] In some embodiments, a polypeptide according to the present invention comprises a non-amino acid linker. In some embodiments, a polypeptide according to the present invention may comprise two or more polypeptides linked by conjugation, e.g. by nucleophilic substitutions (e.g., reactions of amines and alcohols with acyl halides, active esters), electrophilic substitutions (e.g., enamine reactions) and additions to carbon-carbon and carbon-heteroatom multiple bonds (e.g., Michael reaction, Diels-Alder addition). These and other useful reactions are discussed in, for example, March, Advanced Organic Chemistry, 3rd Ed., John Wiley & Sons, New York, 1985; Hermanson, Bioconjugate Techniques, Academic Press, San Diego, 1996; and Feeney et al., Modification of Proteins; Advances in Chemistry Series, Vol. 198, American Chemical Society, Washington, D.C., 1982.
[0106] In some embodiments, a polypeptide according to the present invention comprises a cleavable linker.
[0107] It may be desirable for a polypeptide according to the present invention to lack certain properties of CR1. For example, it may be desirable for the polypeptide to lack regions that would otherwise inhibit diffusion through Bruch's membrane (BrM) or that would interfere with the action of native co-factor family proteins.
[0108] A polypeptide according to the present invention lacks the CR1 transmembrane domain (SEQ ID NO:32). A polypeptide according to the present invention may lack the CR1 cytoplasmic tail (SEQ ID NO:33). In a preferred embodiment, a polypeptide according to the present invention is soluble.
[0109] A polypeptide according to the present invention may lack regions which could otherwise be exploited by pathogenic bacteria to subvert the host immune system. Bacteria have developed molecules on their surface that can bind and recruit soluble complement factor H from the blood. This enables the bacteria to effectively coat themselves in a complement regulator and evade a host immune response. Polypeptides according to the present invention may lack bacterial binding sites such that they cannot be used by invading pathogens to evade an immune response.
[0110] In some embodiments a polypeptide according to the present invention comprises one or more sites for glycosylation. In some embodiments a polypeptide according to the present invention is glycosylated.
[0111] In some embodiments, a polypeptide according to the present invention is not glycosylated. In some embodiments, a polypeptide according to the present invention lacks one or more sites for glycosylation. In some embodiments, the polypeptide of the present invention lacks one or more sites for N-linked glycosylation. In some embodiments, a polypeptide according to the present invention lacks N-linked glycans. In some embodiments, a polypeptide according to the present invention is expressed and/or secreted by cells that are unable to glycosylate or fully glycosylate polypeptides. For example, cells may lack functional glycosyl transferase enzymes. In some embodiments, the polypeptide is aglycosyl (i.e. is not glycosylated). In some embodiments, the polypeptide has been deglycosylated, e.g. by treatment with a glycosidase (e.g. Peptide N-Glycosidase). Deglycosylation is preferably non-denaturing. In some embodiments a polypeptide according to the present invention is partially glycosylated, non-glycosylated or de-glycosylated.
[0112] In some embodiments, a polypeptide according to the present invention lacks sequence conforming to the consensus sequence of SEQ ID NO:27. In some embodiments, the polypeptide according to the present invention comprises one or more sequences conforming to the consensus sequence of SEQ ID NO:27 that have been mutated to remove sites for N-glycosylation. In some embodiments, the Asn (N) residue in one or more consensus sequences according to SEQ ID NO:27 is substituted with another amino acid residue, e.g. a residue selected from: Ala (A), Cys (C), Asp (D), Glu (E), Phe (F), Gly (G), His (H), Ile (I), Lys (K), Leu (L), Met (M), Pro (P), Gln (Q), Arg (R), Ser (S), Thr (T), Val (V), Trp (W) or Tyr (Y). In some embodiments, the Asn (N) residue in one or more consensus sequences according to SEQ ID NO:27 is substituted with a Gln (Q) residue. In some embodiments, residue X.sub.2 of SEQ ID NO:27 is, or is mutated to be, an amino acid that is not Ser (S) or Thr (T).
[0113] In some embodiments, a polypeptide comprising, or consisting of, an amino acid sequence having at least 70%, 75%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID NO:2 or SEQ ID NO:4 comprises one or more amino acid substitutions at position 509 and/or position 578 (numbered according to Uniprot: P17927). In some embodiments, the one or more amino acid substitutions are selected from N509Q and/or N578Q. In some embodiments, a polypeptide comprising, or consisting of, an amino acid sequence having at least 70%, 75%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID NO:3 comprises one or more amino acid substitutions at position 959 and/or position 1028 (numbered according to Uniprot: P17927). In some embodiments, the one or more amino acid substitutions are selected from N959Q and/or N1028Q. In some embodiments, a polypeptide comprising, or consisting of, an amino acid sequence having at least 70%, 75%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID NO:13 comprises one or more amino acid substitutions at position 509, 578, 959 and/or position 1028 (numbered according to Uniprot: P17927). In some embodiments, the one or more amino acid substitutions are selected from N509Q, N578Q, N959Q and/or N1028Q. Such polypeptides may have any length provided herein.
[0114] In some embodiments, the polypeptide comprises, or consists, of an amino acid sequence having at least 70%, 75%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID NO:5, SEQ ID NO:6, SEQ ID NO:15, and/or SEQ ID NO:31. Such polypeptides may have any length provided herein.
[0115] In some embodiments a polypeptide according to the present invention comprises a secretory pathway sequence and one or more sequences conforming to the consensus sequence of SEQ ID NO:27 that have been mutated to remove sites for N-glycosylation. In some embodiments a polypeptide according to the present invention comprises, or consists of, an amino acid sequence corresponding to SEQ ID NO:48, 50, 52, 53, and/or 54. In some embodiments the polypeptide comprises, or consists, of an amino acid sequence having at least 70%, 75%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID NO:48, 50, 52, 53, and/or 54. Such polypeptides may have any length provided herein.
[0116] In some embodiments, a polypeptide according to the present invention may comprise amino acid sequence(s) to facilitate expression, folding, trafficking, processing, purification or detection of the polypeptide. For example, the polypeptide may comprise a sequence encoding a protein tag, e.g. a His, (e.g. 6.times.His), FLAG, Myc, GST, MBP, HA, E, or Biotin tag, optionally: at the N- or C-terminus of the polypeptide; in a linker; or at the N- or C-terminus of a linker. In some embodiments the polypeptide comprises a detectable moiety, e.g. a fluorescent, luminescent, immuno-detectable, radio, chemical, nucleic acid or enzymatic label. In some embodiments, the detectable moiety facilitates detection of the polypeptide in a sample obtained from a subject, e.g. following administration to the subject of the polypeptide, nucleic acid, vector, cell or pharmaceutical composition according to the present invention. The sample may be any biological sample obtained from a subject. In some embodiments the sample is a liquid biopsy, such as ocular fluid (tear fluid, aqueous humour, or vitreous), blood, plasma, etc. In some embodiments the sample is a cytological sample or a tissue sample such as a surgical sample, e.g. of ocular cells/tissue.
[0117] In some embodiments, the polypeptide according to the present invention may be detected and/or distinguished from endogenous CR1 by Western blotting, mass spectrometry and/or enzyme digestion, e.g. by a specific peptidase. In some embodiments, the polypeptide may comprise a point mutation to generate peptides by enzyme digestion that are distinct from post-digestion peptides from endogenous CR1.
[0118] In some embodiments, a polypeptide according to the present invention may additionally comprise a cleavage site for removing a protein tag. For example, it may be desired to remove a tag used for purification of the polypeptide following purification. In some embodiments the cleavage site may e.g. be a Tobacco Etch Virus (TEV) protease cleavage site, for example as shown in SEQ ID NO:34.
[0119] In some embodiments a polypeptide according to the present invention comprises, or consists of, an amino acid sequence corresponding to SEQ ID NO:40, 42, 44, and/or 46. In some embodiments the polypeptide comprises, or consists of, an amino acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID NO:40, 42, 44, and/or 46. Such polypeptides may have any length provided herein.
[0120] As used herein, a "polypeptide" includes molecules comprising more than one polypeptide chain, which may be associated (e.g. covalently or non-covalently) into a complex. That is, a "polypeptide" within the meaning of the present invention encompasses molecules comprising one or more polypeptide chains. The polypeptide of the invention may in various different embodiments and at different stages of expression/production in vitro or in vivo comprise e.g. a signal peptide, protein tag, cleavage sites for removal thereof, etc. The polypeptide of the invention may comprise any CR1 CCP sequence described herein, or any combination of CR1 CCP domains 8 (SEQ ID NO:8), 9 (SEQ ID NO:9), 10 (SEQ ID NO:10), 15 (SEQ ID NO:8), 16 (SEQ ID NO:11), and/or 17 (SEQ ID NO:12), or any combination of sequences `A`, `B` and/or `C` described herein, optionally in combination with one or more of any of the further features of the polypeptide of the invention described herein (e.g. signal peptide, linker, detection sequence, lack of glycosylation site, substituted amino acid residue, protein tag, cleavage site for removing a protein tag, secretory pathway sequence, cleavage site for removing a secretory pathway sequence).
[0121] Sequence Identity
[0122] As used herein, an amino acid sequence which corresponds to a reference amino acid sequence may comprise at least 60%, e.g. one of at least 65%, 70%, 75%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% sequence identity to the reference sequence.
[0123] Pairwise and multiple sequence alignment for the purposes of determining percent identity between two or more amino acid or nucleic acid sequences can be achieved in various ways known to a person of skill in the art, for instance, using publicly available computer software such as ClustalOmega (Sdding, J. 2005, Bioinformatics 21, 951-960), T-coffee (Notredame et al. 2000, J. Mol. Biol. (2000) 302, 205-217), Kalign (Lassmann and Sonnhammer 2005, BMC Bioinformatics, 6(298)) and MAFFT (Katoh and Standley 2013, Molecular Biology and Evolution, 30(4) 772-780 software. When using such software, the default parameters, e.g. for gap penalty and extension penalty, are preferably used.
TABLE-US-00001 Sequences SEQ ID NO: Description Sequence 1 Human Complement MGASSPRSPEPVGPPAPGLPFCCGGSLLAVVVLLALPVAWGQCNAPEW Receptor 1 (UniProt: LPFARPTNLTDEFEFPIGTYLNYECRPGYSGRPFSIICLKNSVWTGAK P17927; Entry version DRCRRKSCRNPPDPVNGMVHVIKGIQFGSQIKYSCTKGYRLIGSSSAT 181 (25 Oct. 2017), CIISGDTVIWDNETPICDRIPCGLPPTITNGDFISTNRENFHYGSVVT Sequence version 3 (02 YRCNPGSGGRKVFELVGEPSIYCTSNDDQVGIWSGPAPQCIIPNKCTP Mar. 2010)); residues 1- PNVENGILVSDNRSLFSLNEVVEFRCQPGFVMKGPRRVKCQALNKWEP 2039 ELPSCSRVCQPPPDVLHAERTQRDKDNFSPGQEVFYSCEPGYDLRGAA Including signal sequence SMRCTPQGDWSPAAPTCEVKSCDDFMGQLLNGRVLFPVNLQLGAKVDF [CCP domains '8-10' and VCDEGFQLKGSSASYCVLAGMESLWNSSVPVCEQIFCPSPPVIPNGRH 15-17 indicated by single TGKPLEVFPFGKTVNYTCDPHPDRGTSFDLIGESTIRCTSDPQGNGVW underline, individual CCP SSPAPRCGILGHCQAPDHFLFAKLKTQTNASDFPIGTSLKYECRPEYY domains 8, 9, 10, 15, 16, GRPFSITCLDNLVWSSPKDVCKRKSCKTPPDPVNGMVHVITDIQVGSR 17 indicated by double INYSCTTGHRLIGHSSAECILSGN AHWSTKPPICQRIPCGLPPTIAN underline; GDFISTNRENFHYGSVVTYRCN GS GRKVFELVGEPSIYCTSNDDQV amino acid differences GIWSGPAPQCIIPNKCTPPNVENGILVSDNRSLFSLNEVVEFRCQPGF between CCPs 8-10 and VMKGPRRVKCQALNKWEPELPSCSRVCQPPPDVLHAERTQRDKDNFSP 15-17 indicated with wavy GQEVFYSCEPGYDLRGAASMRCTPQGDWSPAAPTCEVKSCDDFMGQLL underline] NGRVLFPVNLQLGAKVDFVCDEGFQLKGSSASYCVLAGMESLWNSSVP VCEQIFCPSPPVIPNGRHTGKPLEVFPFGKAVNYTCDPHPDRGTSFDL IGESTIRCTSDPQGNGVWSSPAPRCGILGHCQAPDHFLFAKLKTQTNA SDFPIGTSLKYECRPEYYGRPFSITCLDNLVWSSPKDVCKRKSCKTPP DPVNGMVHVITDIQVGSRINYSCTTGHRLIGHSSAECILSGN AHWST KPPICQRIPCGLPPTIANGDFISTNRENFHYGSVVTYRCN GS GRKV FELVGEPSIYCTSNDDQVGIWSGPAPQCIIPNKCTPPNVENGILVSDN RSLFSLNEVVEFRCQPGFVMKGPRRVKCQALNKWEPELPSCSRVCQPP PEILHGEHTPSHQDNFSPGQEVFYSCEPGYDLRGAASLHCTPQGDWSP EAPRCAVKSCDDFLGQLPHGRVLFPLNLQLGAKVSFVCDEGFRLKGSS VSHCVLVGMRSLWNNSVPVCEHIFCPNPPAILNGRHTGTPSGDIPYGK EISYTCDPHPDRGMTFNLIGESTIRCTSDPHGNGVWSSPAPRCELSVR AGHCKTPEQFPFASPTIPINDFEFPVGTSLNYECRPGYFGKMFSISCL ENLVWSSVEDNCRRKSCGPPPEPFNGMVHINTDTQFGSTVNYSCNEGF RLIGSPSTTCLVSGNNVTWDKKAPICEIISCEPPPTISNGDFYSNNRT SFHNGTVVTYQCHTGPDGEQLFELVGERSIYCTSKDDQVGVWSSPPPR CISTNKCTAPEVENAIRVPGNRSFFSLTEIIRFRCQPGFVMVGSHTVQ CQTNGRWGPKLPHCSRVCQPPPEILHGEHTLSHQDNFSPGQEVFYSCE PSYDLRGAASLHCTPQGDWSPEAPRCTVKSCDDFLGQLPHGRVLLPLN LQLGAKVSFVCDEGFRLKGRSASHCVLAGMKALWNSSVPVCEQIFCPN PPAILNGRHTGTPFGDIPYGKEISYACDTHPDRGMTFNLIGESSIRCT SDPQGNGVWSSPAPRCELSVPAACPHPPKIQNGHYIGGHVSLYLPGMT ISYICDPGYLLVGKGFIFCTDQGIWSQLDHYCKEVNCSFPLFMNGISK ELEMKKVYHYGDYVTLKCEDGYTLEGSPWSQCQADDRWDPPLAKCTSR THDALIVGTLSGTIFFILLIIFLSWIILKHRKGNNAHENPKEVAIHLH SQGGSSVHPRTLQTNEENSRVLP 2 Human Complement GHCQAPDHFLFAKLKTQTNASDFPIGTSLKYECRPEYYGRPFSITCLD Receptor 1 CCPs 8-10 NLVWSSPKDVCKRKSCKTPPDPVNGMVHVITDIQVGSRINYSCTTGHR (UniProt: P17927 LIGHSSAECILSGNAAHWSTKPPICQRIPCGLPPTIANGDFISTNREN residues 491 to 684) FHYGSVVTYRCNPGSGGRKVFELVGEPSIYCTSNDDQVGIWSGPAPQC Without leader sequence II 3 Human Complement GHCQAPDHFLFAKLKTQTNASDFPIGTSLKYECRPEYYGRPFSITCLD Receptor 1 CCPs 15-17 NLVWSSPKDVCKRKSCKTPPDPVNGMVHVITDIQVGSRINYSCTTGHR (UniProt: P17927 LIGHSSAECILSGNTAHWSTKPPICQRIPCGLPPTIANGDFISTNREN residues 941 to 1134) FHYGSVVTYRCNLGSRGRKVFELVGEPSIYCTSNDDQVGIWSGPAPQC Without leader sequence II 4 Human Complement GHCQAPDHFLFAKLKTQTNASDFPIGTSLKYECRPEYYGRPFSITCLD Receptor 1; consensus NLVWSSPKDVCKRKSCKTPPDPVNGMVHVITDIQVGSRINYSCTIGHR sequence for CCPs 8-10, LIGHSSAECILSGNX.sub.1AHWSTKPPICQRIPCGLPPTIANGDFISTNRE 15-17 NFHYGSVVTYRCNX.sub.2GSX.sub.3GRKVFELVGEPSIYCTSNDDQVGIWSGPAP (UniProt: P17927 QCII residues 491 to 684; residues 941 to 1134) Without leader sequence 5 Human Complement GHCQAPDHFLFAKLKTQTQASDFPIGTSLKYECRPEYYGRPFSITCLD Receptor 1 CCPs 8-10 NLVWSSPKDVCKRKSCKTPPDPVNGMVHVITDIQVGSRIQYSCTIGHR (UniProt: P17927 LIGHSSAECILSGNAAHWSTKPPICQRIPCGLPPTIANGDFISTNREN residues 491 to 684) FHYGSVVTYRCNPGSGGRKVFELVGEPSIYCTSNDDQVGIWSGPAPQC Non-glycosylated; II Without leader sequence [mutated glycosylation sites underlined; substitutions in bold correspond to positions 509 and 578 of UniProt: P17927] 6 Human Complement GHCQAPDHFLFAKLKTQTQASDFPIGTSLKYECRPEYYGRPFSITCLD Receptor 1 CCPs 15-17 NLVWSSPKDVCKRKSCKTPPDPVNGMVHVITDIQVGSRIQYSCTIGHR (UniProt: P17927 LIGHSSAECILSGNTAHWSTKPPICQRIPCGLPPTIANGDFISTNREN residues 941 to 1134) FHYGSVVTYRCNLGSRGRKVFELVGEPSIYCTSNDDQVGIWSGPAPQC Non-glycosylated; II Without leader sequence [mutated glycosylation sites underlined; substitutions in bold correspond to positions 959 and 1028 of UniProt: P17927] 7 Native signal/leader MRLLAKIICLMLWAICVA sequence from Factor H 8 Human Complement GHCQAPDHFLFAKLKTQTNASDFPIGTSLKYECRPEYYGRPFSITCLD Receptor 1 CCP8/ NLVWSSPKDVCKR CCP15 (UniProt: P17927 residues 491-550/941- 1000) 'Sequence A' 9 Human Complement KSCKTPPDPVNGMVHVITDIQVGSRINYSCTIGHRLIGHSSAECILSG Receptor 1 N AHWSTKPPICQ CCP9 (UniProt: P17927 residues 552-612) 10 Human Complement RIPCGLPPTIANGDFISTNRENFHYGSVVTYRCN GS GRKVFELVGE Receptor 1 PSIYCTSNDDQVGIWSGPAPQCII CCP10 (UniProt: P17927 residues 613-684 11 Human Complement KSCKTPPDPVNGMVHVITDIQVGSRINYSCTIGHRLIGHSSAECILSG Receptor 1 N AHWSTKPPICQ CCP16 (UniProt: P17927 residues 1002-1062) 12 Human Complement RIPCGLPPTIANGDFISTNRENFHYGSVVTYRCN GS GRKVFELVGE Receptor 1 PSIYCTSNDDQVGIWSGPAPQCII CCP17 (UniProt: P17927 residues 1063-1134 13 Human Complement GHCQAPDHFLFAKLKTQTNASDFPIGTSLKYECRPEYYGRPFSITCLD Receptor 1 NLVWSSPKDVCKRKSCKIPPDPVNGMVHVITDIQVGSRINYSCTIGHR CCPs 8-10 and 15-17 LIGHSSAECILSGN AHWSTKPPICQRIPCGLPPTIANGDFISTNREN (contiguous; without FHYGSVVTYRCN GS GRKVFELVGEPSIYCTSNDDQVGIWSGPAPQC leader sequence) IIGHCQAPDHFLFAKLKTQTNASDFPIGTSLKYECRPEYYGRPFSITC [amino acid differences LDNLVWSSPKDVCKRKSCKTPPDPVNGMVHVITDIQVGSRINYSCITG between CCPs 8-10 and HRLIGHSSAECILSGN AHWSTKPPICQRIPCGLPPTIANGDFISTNR 15-17 indicated with wavy ENFHYGSVVTYRCN GS GRKVFELVGEPSIYCTSNDDQVGIWSGPAP underline] QCII 14 Human Complement GHCQAPDHFLFAKLKTQTNASDFPIGTSLKYECRPEYYGRPFSITCLD Receptor 1 NLVWSSPKDVCKRKSCKIPPDPVNGMVHVITDIQVGSRINYSCTIGHR CCPs 8-10 (linker) LIGHSSAECILSGNAAHWSTKPPICQRIPCGLPPTIANGDFISTNREN 15-17 FHYGSVVTYRCNPGSGGRKVFELVGEPSIYCTSNDDQVGIWSGPAPQC II-[linker]- GHCQAPDHFLFAKLKTQTNASDFPIGTSLKYECRPEYYGRPFSITCLD NLVWSSPKDVCKRKSCKIPPDPVNGMVHVITDIQVGSRINYSCTIGHR LIGHSSAECILSGNTAHWSTKPPICQRIPCGLPPTIANGDFISTNREN FHYGSVVTYRCNLGSRGRKVFELVGEPSIYCTSNDDQVGIWSGPAPQC II 15 Human Complement GHCQAPDHELFAKLKTQTQASDEPIGTSLKYECRPEYYGRPFSITCLD Receptor 1 NLVWSSPKDVCKRKSCKIPPDPVNGMVHVITDIQVGSRIQYSCTIGHR CCPs 8-10 and 15-17 LIGHSSAECILSGNAAHWSTKPPICQRIPCGLPPTIANGDFISTNREN (contiguous) FHYGSVVTYRCNPGSGGRKVFELVGEPSIYCTSNDDQVGIWSGPAPQC Non-glycosylated; IIGHCQAPDHFLFAKLKTQTQASDFPIGTSLKYECRPEYYGRPFSITC Without leader sequence LDNLVWSSPKDVCKRKSCKIPPDPVNGMVHVITDIQVGSRIQYSCITG [Mutated glycosylation HRLIGHSSAECILSGNTAHWSTKPPICQRIPCGLPPTIANGDFISTNR sites underlined; ENFHYGSVVTYRCNLGSRGRKVFELVGEPSIYCTSNDDQVGIWSGPAP substitutions in bold QCII correspond to positions 509, 578, 959 and 1028 of UniProt: P17927] 16 Human Complement KSCKIPPDPVNGMVHVITDIQVGSRINYSCTIGHRLIGHSSAECILSG Receptor 1; Consensus NX.sub.1AHWSTKPPICQ sequence for CCPs 9, 16 (corresponding to UniProt: P17927 residues 552-612 and 1002-1062) 'Sequence B' 17 Human Complement RIPCGLPPTIANGDFISTNRENFHYGSVVTYRCNX.sub.2GSX.sub.3GRKVFELVG Receptor 1; Consensus EPSIYCTSNDDQVGIWSGPAPQCII sequence for CCPs 10, 17 (corresponding to (UniProt: P17927 residues 613-684 and 1063-1134) 'Sequence C' 18 Human C3 (UniProt: MGPTSGPSLLLLLLTHLPLALGSPMYSIITPNILRLESEETMVLEAHD P01024; Entry version AQGDVPVIVIVHDFPGKKLVLSSEKTVLIPAINHMGNVIFTIPANREF 221 (20 Dec. 2017); KSEKGRNKFVTVQATFGTQVVEKVVLVSLQSGYLFIQTDKTIYIPGST Sequence version 2 (12 VLYRIFTVNHKLLPVGRIVMVNIENPEGIPVKQDSLSSQNQLGVLPLS Dec. 2006)) WDIPELVNMGQWKIRAYYENSPQQVFSTEFEVKEYVLPSFEVIVEPTE including signal peptide KFYYIYNEKGLEVTITARFLYGKKVEGTAFVIFGIQDGEQRISLPESL KRIPIEDGSGEVVLSRKVLLDGVQNPRAEDLVGKSLYVSATVILHSGS DMVQAERSGIPIVISPYQIHFIKTPKYFKPGMPFDLMVFVINPDGSPA YRVPVAVQGEDTVQSLIQGDGVAKLSINTHPSQKPLSITVRIKKQELS EAEQATRTMQALPYSTVGNSNNYLHLSVLRTELRPGETLNVNFLLRMD RAHEAKIRYYTYLIMNKGRLLKAGRQVREPGQDLVVLPLSITTDFIPS FRLVAYYTLIGASGQREVVADSVWVDVKDSCVGSLVVKSGQSEDRQPV PGQQMTLKIEGDHGARVVLVAVDKGVFVLNKKNKLTQSKIWDVVEKAD IGCTPGSGKDYAGVFSDAGLTFTSSSGQQTAQRAELQCPQPAARRRRS VQLTEKRMDKVGKYPKELRKCCEDGMRENPMRFSCQRRTRFISLGEAC KKVFLDCCNYITELRRQHARASHLGLARSNLDEDIIAEENIVSRSEFP ESWLWNVEDLKEPPKNGISTKLMNIFLKDSITTWEILAVSMSDKKGIC VADPFEVTVMQDFFIDLRLPYSVVRNEQVEIRAVLYNYRQNQELKVRV ELLHNPAFCSLATTKRRHQQTVTIPPKSSLSVPYVIVPLKTGLQEVEV KAAVYHHFISDGVRKSLKVVPEGIRMNKTVAVRTLDPERLGREGVQKE DIPPADLSDQVPDTESETRILLQGTPVAQMTEDAVDAERLKHLIVTPS GCGEQNMIGMTPTVIAVHYLDETEQWEKFGLEKRQGALELIKKGYTQQ LAFRQPSSAFAAFVKRAPSTWLTAYVVKVFSLAVNLIAIDSQVLCGAV KWLILEKQKPDGVFQEDAPVIHQEMIGGLRNNNEKDMALTAFVLISLQ EAKDICEEQVNSLPGSITKAGDFLEANYMNLQRSYTVAIAGYALAQMG RLKGPLLNKFLTTAKDKNRWEDPGKQLYNVEATSYALLALLQLKDFDF VPPVVRWLNEQRYYGGGYGSTQATFMVFQALAQYQKDAPDHQELNLDV SLQLPSRSSKITHRIHWESASLLRSEETKENEGFTVTAEGKGQGTLSV VTMYHAKAKDQLTCNKFDLKVTIKPAPETEKRPQDAKNTMILEICTRY RGDQDATMSILDISMMTGFAPDTDDLKQLANGVDRYISKYELDKAFSD RNTLIIYLDKVSHSEDDCLAFKVHQYFNVELIQPGAVKVYAYYNLEES CTRFYHPEKEDGKLNKLCRDELCRCAEENCFIQKSDDKVTLEERLDKA CEPGVDYVYKTRLVKVQLSNDFDEYIMAIEQTIKSGSDEVQVGQQRTF ISPIKCREALKLEEKKHYLMWGLSSDFWGEKPNLSYIIGKDTWVEHWP EEDECQDEENQKQCQDLGAFTESMVVFGCPN 19 Human C3 .beta. chain SPMYSIITPNILRLESEETMVLEAHDAQGDVPVTVTVHDFPGKKLVLS (UniProt: P01024; Entry SEKTVLTPATNHMGNVTFTIPANREFKSEKGRNKFVTVQATFGTQVVE version 221 (20 Dec. KVVLVSLQSGYLFIQTDKTIYTPGSTVLYRIFTVNHKLLPVGRTVMVN 2017); IENPEGIPVKQDSLSSQNQLGVLPLSWDIPELVNMGQWKIRAYYENSP Sequence version 2 (12 QQVFSTEFEVKEYVLPSFEVIVEPTEKFYYIYNEKGLEVTITARFLYG Dec. 2006); residues 23- KKVEGTAFVIFGIQDGEQRISLPESLKRIPIEDGSGEVVLSRKVLLDG 667) VQNPRAEDLVGKSLYVSATVILHSGSDMVQAERSGIPIVTSPYQIHFT KTPKYFKPGMPFDLMVFVTNPDGSPAYRVPVAVQGEDTVQSLTQGDGV AKLSINTHPSQKPLSITVRTKKQELSEAEQATRTMQALPYSTVGNSNN YLHLSVLRTELRPGETLNVNFLLRMDRAHEAKIRYYTYLIMNKGRLLK AGRQVREPGQDLVVLPLSITTDFIPSFRLVAYYTLIGASGQREVVADS VWVDVKDSCVGSLVVKSGQSEDRQPVPGQQMTLKIEGDHGARVVLVAV DKGVFVLNKKNKLTQSKIWDVVEKADIGCTPGSGKDYAGVFSDAGLTF TSSSGQQTAQRAELQCPQPAA 20 Human C3 .alpha.' chain SNLDEDIIAEENIVSRSEFPESWLWNVEDLKEPPKNGISTKLMNIFLK (UniProt: P01024; Entry DSITTWEILAVSMSDKKGICVADPFEVTVMQDFFIDLRLPYSVVRNEQ version 221 (20 Dec. VEIRAVLYNYRQNQELKVRVELLHNPAFCSLATTKRRHQQTVTIPPKS 2017); SLSVPYVIVPLKTGLQEVEVKAAVYHHFISDGVRKSLKVVPEGIRMNK Sequence version 2 (12 TVAVRTLDPERLGREGVQKEDIPPADLSDQVPDTESETRILLQGTPVA Dec. 2006); residues 749- QMTEDAVDAERLKHLIVTPSGCGEQNMIGMTPTVIAVHYLDETEQWEK 1663) FGLEKRQGALELIKKGYTQQLAFRQPSSAFAAFVKRAPSTWLTAYVVK
VFSLAVNLIAIDSQVLCGAVKWLILEKQKPDGVFQEDAPVIHQEMIGG LRNNNEKDMALTAFVLISLQEAKDICEEQVNSLPGSITKAGDFLEANY MNLQRSYTVAIAGYALAQMGRLKGPLLNKFLTTAKDKNRWEDPGKQLY NVEATSYALLALLQLKDFDFVPPVVRWLNEQRYYGGGYGSTQATFMVF QALAQYQKDAPDHQELNLDVSLQLPSRSSKITHRIHWESASLLRSEET KENEGFTVTAEGKGQGTLSVVTMYHAKAKDQLTCNKFDLKVTIKPAPE TEKRPQDAKNTMILEICTRYRGDQDATMSILDISMMTGFAPDTDDLKQ LANGVDRYISKYELDKAFSDRNTLIIYLDKVSHSEDDCLAFKVHQYFN VELIQPGAVKVYAYYNLEESCTRFYHPEKEDGKLNKLCRDELCRCAEE NCFIQKSDDKVTLEERLDKACEPGVDYVYKTRLVKVQLSNDFDEYIMA IEQTIKSGSDEVQVGQQRTFISPIKCREALKLEEKKHYLMWGLSSDFW GEKPNLSYIIGKDTWVEHWPEEDECQDEENQKQCQDLGAFTESMVVFG CPN 21 Human C3a (UniProt: SVQLTEKRMDKVGKYPKELRKCCEDGMRENPMRFSCQRRTRFISLGEA P01024; Entry version CKKVFLDCCNYITELRRQHARASHLGLAR 221 (20 Dec. 2017); Sequence version 2 (12 Dec. 2006); residues 672- 748) 22 Human C3 .alpha.' chain SNLDEDIIAEENIVSRSEFPESWLWNVEDLKEPPKNGISTKLMNIFLK fragment 1 DSITTWEILAVSMSDKKGICVADPFEVTVMQDFFIDLRLPYSVVRNEQ (UniProt: P01024; Entry VEIRAVLYNYRQNQELKVRVELLHNPAFCSLATTKRRHQQTVTIPPKS version 221 (20 Dec. SLSVPYVIVPLKTGLQEVEVKAAVYHHFISDGVRKSLKVVPEGIRMNK 2017); TVAVRTLDPERLGREGVQKEDIPPADLSDQVPDTESETRILLQGTPVA Sequence version 2 (12 QMTEDAVDAERLKHLIVTPSGCGEQNMIGMTPTVIAVHYLDETEQWEK Dec. 2006); residues 749- FGLEKRQGALELIKKGYTQQLAFRQPSSAFAAFVKRAPSTWLTAYVVK 1303) VFSLAVNLIAIDSQVLCGAVKWLILEKQKPDGVFQEDAPVIHQEMIGG LRNNNEKDMALTAFVLISLQEAKDICEEQVNSLPGSITKAGDFLEANY MNLQRSYTVAIAGYALAQMGRLKGPLLNKFLTTAKDKNRWEDPGKQLY NVEATSYALLALLQLKDFDFVPPVVRWLNEQRYYGGGYGSTQATFMVF QALAQYQKDAPDHQELNLDVSLQLPSR 23 Human C3 .alpha.' chain SEETKENEGFTVTAEGKGQGTLSVVTMYHAKAKDQLTCNKFDLKVTIK fragment 2 (UniProt: PAPETEKRPQDAKNTMILEICTRYRGDQDATMSILDISMMTGFAPDTD P01024;; Entry version DLKQLANGVDRYISKYELDKAFSDRNTLIIYLDKVSHSEDDCLAFKVH 221 (20 Dec. 2017); QYFNVELIQPGAVKVYAYYNLEESCTRFYHPEKEDGKLNKLCRDELCR Sequence version 2 (12 CAEENCFIQKSDDKVTLEERLDKACEPGVDYVYKTRLVKVQLSNDFDE Dec. 2006); residues YIMAIEQTIKSGSDEVQVGQQRTFISPIKCREALKLEEKKHYLMWGLS 1321-1663) SDFWGEKPNLSYIIGKDTWVEHWPEEDECQDEENQKQCQDLGAFTESM Also known as C3c VVFGCPN fragment 2 24 Human C3f (UniProt: SSKITHRIHWESASLLR P01024;; Entry version 221 (20 Dec. 2017); Sequence version 2 (12 Dec. 2006); residues 1304-1320) 25 Human Complement MKLLHVFLLFLCFHLRFCKVTYTSQEDLVEKKCLAKKYTHLSCDKVFC Factor I (UniProt: QPWQRCIEGTCVCKLPYQCPKNGTAVCATNRRSFPTYCQQKSLECLHP P05156; Entry version GTKFLNNGTCTAEGKFSVSLKHGNTDSEGIVEVKLVDQDKTMFICKSS 192 (20 Dec. 2017) WSMREANVACLDLGFQQGADTQRRFKLSDLSINSTECLHVHCRGLETS Sequence version 2 (11 LAECTFTKRRTMGYQDFADVVCYTQKADSPMDDFFQCVNGKYISQMKA Jan. 2011)) CDGINDCGDQSDELCCKACQGKGFHCKSGVCIPSQYQCNGEVDCITGE DEVGCAGFASVTQEETEILTADMDAERRRIKSLLPKLSCGVKNRMHIR RKRIVGGKRAQLGDLPWQVAIKDASGITCGGIYIGGCWILTAAHCLRA SKTHRYQIWTTVVDWIHPDLKRIVIEYVDRIIFHENYNAGTYQNDIAL IEMKKDGNKKDCELPRSIPACVPWSPYLFQPNDTCIVSGWGREKDNER VFSLQWGEVKLISNCSKFYGNRFYEKEMECAGTYDGSIDACKGDSGGP LVCMDANNVTYVWGVVSWGENCGKPEFPGVYTKVANYFDWISYHVGRP FISQYNV 26 Human Complement IVGGKRAQLGDLPWQVAIKDASGITCGGIYIGGCWILTAAHCLRASKT Factor I proteolytic HRYQIWTTVVDWIHPDLKRIVIEYVDRIIFHENYNAGTYQNDIALIEM domain (UniProt: P05156; KKDGNKKDCELPRSIPACVPWSPYLFQPNDTCIVSGWGREKDNERVFS Entry version 192 (20 LQWGEVKLISNCSKFYGNRFYEKEMECAGTYDGSIDACKGDSGGPLVC Dec. 2017) MDANNVTYVWGVVSWGENCGKPEFPGVYTKVANYFDWISYHVG Sequence version 2 (11 Jan. 2011); residues 340- 574) 27 Consensus sequence for NX.sub.1X.sub.2 N-linked glycosylation wherein X.sub.1 = any amino acid except for P X.sub.2 = S or T 28 Complement Factor H MRLLAKIICLMLWAICVAEDCNELPPRRNTEILTGSWSDQTYPEGTQA isoform FHL-1 (UniProt: IYKCRPGYRSLGNVIMVCRKGEWVALNPLRKCQKRPCGHPGDTPFGTF P08603-2) TLTGGNVFEYGVKAVYTCNEGYQLLGEINYRECDTDGWTNDIPICEVV Including leader KCLPVTAPENGKIVSSAMEPDREYHFGQAVRFVCNSGYKIEGDEEMHC sequence (underlined) SDDGFWSKEKPKCVEISCKSPDVINGSPISQKIIYKENERFQYKCNMG SEQ ID No. 7 YEYSERGDAVCTESGWRPLPSCEEKSCDNPYIPNGDYSPLRIKHRTGD EITYQCRNGFYPATRGNTAKCTSTGWIPAPRCTLKPCDYPDIKHGGLY HENMRRPYFPVAVGKYYSYYCDEHFETPSGSYWDHIHCTQDGWSPAVP CLRKCYFPYLENGYNQNYGRKFVQGKSIDVACHPGYALPKAQTTVTCM ENGWSPTPRCIRVSFTL 29 Human Complement MRLLAKIICLMLWAICVAEDCNELPPRRNTEILTGSWSDQTYPEGTQA Factor H (UniProt: TYKCRPGYRSLGNVIMVCRKGEWVALNPLRKCQKRPCGHPGDTPFGTF P08603; Entry version TLTGGNVFEYGVKAVYTCNEGYQLLGEINYRECDTDGWTNDIPICEVV 214 (25 Oct. 2017) KCLPVTAPENGKIVSSAMEPDREYHFGQAVREVCNSGYKIEGDEEMHC Sequence version 4(11 SDDGEWSKEKPKCVEISCKSPDVINGSPISQKITYKENERFQYKCNMG Sep. 2007)) YEYSERGDAVCTESGWRPLPSCEEKSCDNPYIPNGDYSPLRIKHRTGD Including leader EITYQCRNGFYPATRGNTAKCTSTGWIPAPRCTLKPCDYPDIKHGGLY sequence (underlined) HENMRRPYFPVAVGKYYSYYCDEHFETPSGSYWDHIHCTQDGWSPAVP SEQ ID No. 7 CLRKCYFPYLENGYNQNYGRKFVQGKSIDVACHPGYALPKAQTTVTCM ENGWSPTPRCIRVKTCSKSSIDIENGFISESQYTYALKEKAKYQCKLG YVTADGETSGSITCGKDGWSAQPTCIKSCDIPVFMNARTKNDFTWFKL NDTLDYECHDGYESNTGSTTGSIVCGYNGWSDLPICYERECELPKIDV HLVPDRKKDQYKVGEVLKESCKPGFTIVGPNSVQCYHEGLSPDLPICK EQVQSCGPPPELLNGNVKEKTKEEYGHSEVVEYYCNPRFLMKGPNKIQ CVDGEWTTLPVCIVEESTCGDIPELEHGWAQLSSPPYYYGDSVEFNCS ESFTMIGHRSITCIHGVWTQLPQCVAIDKLKKCKSSNLIILEEHLKNK KEEDHNSNIRYRCRGKEGWIHTVCINGRWDPEVNCSMAQIQLCPPPPQ IPNSHNMTTTLNYRDGEKVSVLCQENYLIQEGEEITCKDGRWQSIPLC VEKIPCSQPPQIEHGTINSSRSSQESYAHGTKLSYTCEGGFRISEENE TTCYMGKWSSPPQCEGLPCKSPPEISHGVVAHMSDSYQYGEEVTYKCF EGEGIDGPAIAKCLGEKWSHPPSCIKTDCLSLPSFENAIPMGEKKDVY KAGEQVTYTCATYYKMDGASNVTCINSRWTGRPTCRDTSCVNPPTVQN AYIVSRQMSKYPSGERVRYQCRSPYEMFGDEEVMCLNGNWTEPPQCKD STGKCGPPPPIDNGDITSFPLSVYAPASSVEYQCQNLYQLEGNKRITC RNGQWSEPPKCLHPCVISREIMENYNIALRWTAKQKLYSRTGESVEFV CKRGYRLSSRSHTLRTTCWDGKLEYPTCAKR 30 Human Complement GHCQAPDHELFAKLKTQTNASDEPIGTSLKYECRPEYYGRPFSITCLD Receptor 1 (UniProt: NLVWSSPKDVCKRKSCKTPPDPVNGMVHVITDIQVGSRINYSCTTGHR P17927 residues 491 to LIGHSSAECILSGN AHWSTKPPICQRIPCGLPPTIANGDFISTNREN 1134, from CCP8 to FHYGSVVTYRCN GS GRKVFELVGEPSIYCTSNDDQVGIWSGPAPQC CCP17) IIPNKCTPPNVENGILVSDNRSLFSLNEVVEFRCQPGFVMKGPRRVKC [CCP domains '8-10' and QALNKWEPELPSCSRVCQPPPDVLHAERTQRDKDNESPGQEVEYSCEP '15-17' indicated by single GYDLRGAASMRCTPQGDWSPAAPTCEVKSCDDFMGQLLNGRVLFPVNL underline, individual CCP QLGAKVDFVCDEGFQLKGSSASYCVLAGMESLWNSSVPVCEQIFCPSP domains 8, 9, 10, 15, 16, PVIPNGRHTGKPLEVFPFGKAVNYTCDPHPDRGTSFDLIGESTIRCTS 17 indicated by double DPQGNGVWSSPAPRCGILGHCQAPDHELFAKLKTQTNASDEPIGTSLK underline; YECRPEYYGRPFSITCLDNLVWSSPKDVCKRKSCKTPPDPVNGMVHVI amino acid differences TDIQVGSRINYSCTTGHRLIGHSSAECILSGN AHWSTKPPICQRIPC between CCPs 8-10 and GLPPTIANGDFISTNRENFHYGSVVTYRCN GS GRKVFELVGEPSIY 15-17 indicated in CTSNDDQVGIWSGPAPQCII bold/wavy underline] 31 Human Complement GHCQAPDHELFAKLKTQT ASDEPIGTSLKYECRPEYYGRPFSITCLD Receptor 1 (UniProt: NLVWSSPKDVCKRKSCKTPPDPVNGMVHVITDIQVGSRI YSCTTGHR P17927 residues 491 to LIGHSSAECILSGN AHWSTKPPICQRIPCGLPPTIANGDFISTNREN 1134, CCP8 to CCP17) FHYGSVVTYRCN GS GRKVFELVGEPSIYCTSNDDQVGIWSGPAPQC Non-glycosylated IIPNKCTPPNVENGILVSDNRSLFSLNEVVEFRCQPGFVMKGPRRVKC [CCP domains '8-10' and QALNKWEPELPSCSRVCQPPPDVLHAERTQRDKDNESPGQEVEYSCEP '15-17' indicated by single GYDLRGAASMRCTPQGDWSPAAPTCEVKSCDDFMGQLLNGRVLFPVNL underline, individual CCP QLGAKVDFVCDEGFQLKGSSASYCVLAGMESLWNSSVPVCEQIFCPSP domains 8, 9, 10, 15, 16, PVIPNGRHTGKPLEVFPFGKAVNYTCDPHPDRGTSFDLIGESTIRCTS 17 indicated by double DPQGNGVWSSPAPRCGILGHCQAPDHELFAKLKTQT ASDEPIGTSLK underline; YECRPEYYGRPFSITCLDNLVWSSPKDVCKRKSCKTPPDPVNGMVHVI amino acid differences TDIQVGSRI YSCTTGHRLIGHSSAECILSGN AHWSTKPPICQRIPC between CCPs 8-10 and GLPPTIANGDFISTNRENFHYGSVVTYRCN GS GRKVFELVGEPSIY 15-17 indicated in CTSNDDQVGIWSGPAPQCII bold/wavy underline; substitutions in bold/dotted underline correspond to positions 509, 578, 959 and 1028 of UniProt: P17927]] 32 Human Complement ALIVGTLSGTIFFILLIIFLSWITL Receptor 1 (UniProt: P17927; Entry version 181 (25 Oct. 2017), Sequence version 3 (02 Mar. 2010)) Transmembrane domain (residues 1972-1996 33 Human Complement KHRKGNNAHENPKEVAIHLHSQGGSSVHPRTLQTNEENSRVLP Receptor 1 (UniProt: P17927; Entry version 181 (25 Oct. 2017), Sequence version 3 (02 Mar. 2010)) Cytoplasmic tail (residues 1997-2039) 34 Tobacco Etch Virus ENLYFQGS (TEV) cleavage site 35 Human Complement AAGCTTGCCACCATGAGACTGCTGGCCAAGATCATCTGCCTGATGCTG Receptor 1 CCPs 8-10 TGGGCCATCTGCGTGGCCGGACATTGTCAGGCCCCTGACCACTTCCTG protein coding sequence TTCGCCAAGCTGAAAACCCAGACCAACGCCAGCGACTTCCCTATCGGC Including leader ACCAGCCTGAAGTACGAGTGCAGACCCGAGTACTACGGCAGACCCTTC sequence (double AGCATCACCTGTCTGGACAACCTCGTGTGGTCTAGCCCCAAGGACGTG underlined) TGCAAGAGAAAGAGCTGCAAGACCCCTCCTGATCCTGTGAACGGCATG Including HindIII GTGCACGTGATCACCGACATCCAAGTGGGCAGCAGAATCAACTACAGC restriction enzyme TGCACCACCGGCCACAGACTGATCGGACACTCTAGCGCCGAGTGTATC cleavage site (AAGCTT), CTGAGCGGCAATGCCGCACACTGGTCCACCAAGCCTCCAATCTGCCAG Kozak consensus AGAATCCCTTGCGGCCTGCCTCCTACAATCGCCAACGGCGATTTCATC sequence (GCCACC), AGCACCAACAGAGAGAACTTCCACTACGGCTCCGTGGTCACCTACAGA stop codon (TAA) and TGCAATCCTGGCAGCGGCGGCAGAAAGGTGTTCGAACTTGTGGGCGAG EcoRV restriction enzyme CCCAGCATCTACTGCACCAGCAACGATGACCAAGTCGGCATTTGGAGC cleavage site (GATATC) GGCCCTGCTCCTCAGTGCATCATCTAAGATATC 36 Human Complement AAGCTTGCCACCATGAGACTGCTGGCCAAGATCATCTGCCTGATGCTG Receptor 1 CCPs 8-10 TGGGCCATCTGCGTGGCCGGACATTGTCAGGCCCCTGACCACTTCCTG protein coding sequence TTCGCCAAGCTGAAAACCCAGACACAGGCCAGCGACTTCCCTATCGGC (underlined) ACCAGCCTGAAGTACGAGTGCAGACCCGAGTACTACGGCAGACCCTTC Non-glycosylated AGCATCACCTGTCTGGACAACCTCGTGTGGTCTAGCCCCAAGGACGTG Including leader TGCAAGAGAAAGAGCTGCAAGACCCCTCCTGATCCTGTGAACGGCATG sequence (double GTGCACGTGATCACCGACATCCAAGTGGGCAGCAGAATCCAGTACAGC underlined) TGCACCACAGGCCACAGACTGATCGGCCACTCTAGCGCCGAGTGTATC Including HindIII CTGTCTGGCAATGCCGCTCACTGGTCCACCAAGCCTCCAATCTGCCAG restriction enzyme AGAATCCCTTGCGGCCTGCCTCCTACAATCGCCAACGGCGATTTCATC cleavage site (AAGCTT), AGCACCAACAGAGAGAACTTCCACTACGGCTCCGTGGTCACCTACAGA Kozak consensus TGCAATCCTGGCAGCGGCGGCAGAAAGGTGTTCGAACTTGTGGGCGAG sequence (GCCACC), CCCAGCATCTACTGCACCAGCAACGATGACCAAGTCGGCATTTGGAGC stop codon (TAA) and GGCCCTGCTCCTCAGTGCATCATCTAAGATATC EcoRV restriction enzyme cleavage site (GATATC) 37 Human Complement AAGCTTGCCACCATGAGACTGCTGGCCAAGATCATCTGCCTGATGCTG Receptor 1 CCPs 15-17 TGGGCCATCTGCGTGGCCGGCCACTGTCAGGCCCCTGATCACTTCCTG protein coding sequence TTCGCCAAGCTGAAAACCCAGACCAACGCCAGCGACTTCCCTATCGGC (underlined) ACCAGCCTGAAGTACGAGTGCAGACCCGAGTACTACGGCAGACCCTTC Including leader AGCATCACCTGTCTGGACAACCTCGTGTGGTCTAGCCCCAAGGACGTG sequence (double TGCAAGAGAAAGAGCTGCAAGACCCCTCCTGATCCTGTGAACGGCATG underlined) GTGCACGTGATCACCGACATCCAAGTGGGCAGCAGAATCAACTACAGC Including HindIII TGCACCACCGGCCACAGACTGATCGGACACTCTAGCGCCGAGTGTATC restriction enzyme CTGAGCGGCAATGCCGCACACTGGTCCACCAAGCCTCCAATCTGCCAG cleavage site (AAGCTT), AGAATCCCTTGCGGCCTGCCTCCTACAATCGCCAACGGCGATTTCATC Kozak consensus AGCACCAACAGAGAGAACTTCCACTACGGCTCCGTGGTCACCTACAGA sequence (GCCACC), TGCAATCCTGGCAGCGGCGGCAGAAAGGTGTTCGAACTTGTGGGCGAG stop codon (TAA) and CCCAGCATCTACTGCACCAGCAACGATGACCAAGTCGGCATTTGGAGC EcoRV restriction enzyme GGCCCTGCTCCTCAGTGCATCATCCCTAAGATATC cleavage site (GATATC) 38 Human Complement AAGCTTGCCACCATGAGACTGCTGGCCAAGATCATCTGCCTGATGCTG Receptor 1 CCPs 15-17 TGGGCCATCTGCGTGGCCGGCCACTGTCAGGCCCCTGATCACTTCCTG protein coding sequence TTCGCCAAGCTGAAAACCCAGACACAGGCCAGCGACTTCCCTATCGGC (underlined) ACCAGCCTGAAGTACGAGTGCAGACCCGAGTACTACGGCAGACCCTTC Non-glycosylated AGCATCACCTGTCTGGACAACCTCGTGTGGTCTAGCCCCAAGGACGTG Including leader TGCAAGAGAAAGAGCTGCAAGACCCCTCCTGATCCTGTGAACGGCATG sequence (double GTGCACGTGATCACCGACATCCAAGTGGGCAGCAGAATCCAGTACAGC underlined) TGCACCACAGGCCACAGACTGATCGGCCACTCTAGCGCCGAGTGTATC Including HindIII CTGAGCGGAAACACAGCCCACTGGTCCACCAAGCCTCCAATCTGCCAG restriction enzyme AGAATCCCTTGCGGCCTGCCTCCTACAATCGCCAACGGCGATTTCATC cleavage site (AAGCTT), AGCACCAACAGAGAGAACTTCCACTACGGCTCCGTGGTCACCTACAGA Kozak consensus TGCAACCTGGGCTCCAGAGGCCGGAAGGTGTTCGAACTTGTGGGCGAG sequence (GCCACC), CCTAGCATCTACTGCACCAGCAACGACGACCAAGTCGGCATTTGGAGC stop codon (TAA) and GGACCTGCTCCTCAGTGCATCATCCCTAAGATATC EcoRV restriction enzyme cleavage site (GATATC)
39 Human Complement AAGCTTGCCACCATGAGACTGCTGGCCAAGATCATCTGCCTGATGCTG Receptor 1 CCPs 8-10 TGGGCCATCTGCGTGGCC GGCAGCAGC protein coding sequence CTGGCGGCCACTGTCAGGCCCCTGAT (underlined) CACTTCCTGTTCGCCAAGCTGAAAACCCAGACCAACGCCAGCGACTTC Including leader CCTATCGGCACCAGCCTGAAGTACGAGTGCAGACCCGAGTACTACGGC sequence (doubled AGACCCTTCAGCATCACCTGTCTGGACAACCTCGTGTGGTCTAGCCCC underlined); AAGGACGTGTGCAAGAGAAAGAGCTGCAAGACCCCTCCTGATCCTGTG His tag (dotted line); AACGGCATGGTGCACGTGATCACCGACATCCAAGTGGGCAGCAGAATC TEV cleavage site AACTACAGCTGCACCACCGGCCACAGACTGATCGGACACTCTAGCGCC (dashed line) GAGTGTATCCTGAGCGGCAATGCCGCACACTGGTCCACCAAGCCTCCA Including HindIII ATCTGCCAGAGAATCCCTTGCGGCCTGCCTCCTACAATCGCCAACGGC restriction enzyme GATTTCATCAGCACCAACAGAGAGAACTTCCACTACGGCTCCGTGGTC cleavage site (AAGCTT), ACCTACAGATGCAATCCTGGCAGCGGCGGCAGAAAGGTGTTCGAACTT Kozak consensus GTGGGCGAGCCCAGCATCTACTGCACCAGCAACGATGACCAAGTCGGC sequence (GCCACC), ATTTGGAGCGGCCCTGCTCCTCAGTGCATCATCTAAGATATC stop codon (TAA) and EcoRV restriction enzyme cleavage site (GATATC) 40 Human Complement MRLLAKIICLMLWAICVA GSS SGGHCQAPDHFLF Receptor 1 CCPs 8-10 AKLKTQTNASDFPIGTSLKYECRPEYYGRPFSITCLDNLVWSSPKDVC Including leader KRKSCKTPPDPVNGMVHVITDIQVGSRINYSCTTGHRLIGHSSAECIL sequence (doubled SGNAAHWSTKPPICQRIPCGLPPTIANGDFISTNRENFHYGSVVTYRC underlined); NPGSGGRKVFELVGEPSIYCTSNDDQVGIWSGPAPQCII His tag (dotted line); TEV cleavage site (dashed line) 41 Human Complement AAGCTTGCCACCATGAGACTGCTGGCCAAGATCATCTGCCTGATGCTG Receptor 1 CCPs 8-10 TGGGCCATCTGCGTGGCC GGCAGCAGC protein coding sequence TTCTGGCGGCCACTGTCAGGCCCCTGAT (underlined) CACTTCCTGTTCGCCAAGCTGAAAACCCAGACACAGGCCAGCGACTTC Non-glycosylated CCTATCGGCACCAGCCTGAAGTACGAGTGCAGACCCGAGTACTACGGC Including leader AGACCCTTCAGCATCACCTGTCTGGACAACCTCGTGTGGTCTAGCCCC sequence (doubled AAGGACGTGTGCAAGAGAAAGAGCTGCAAGACCCCTCCTGATCCTGTG underlined); AACGGCATGGTGCACGTGATCACCGACATCCAAGTGGGCAGCAGAATC His tag (dotted line); CAGTACAGCTGCACCACAGGCCACAGACTGATCGGCCACTCTAGCGCC TEV cleavage site GAGTGTATCCTGTCTGGCAATGCCGCTCACTGGTCCACCAAGCCTCCA (dashed line) ATCTGCCAGAGAATCCCTTGCGGCCTGCCTCCTACAATCGCCAACGGC Including HindIII GATTTCATCAGCACCAACAGAGAGAACTTCCACTACGGCTCCGTGGTC restriction enzyme ACCTACAGATGCAATCCTGGCAGCGGCGGCAGAAAGGTGTTCGAACTT cleavage site (AAGCTT), GTGGGCGAGCCCAGCATCTACTGCACCAGCAACGATGACCAAGTCGGC Kozak consensus ATTTGGAGCGGCCCTGCTCCTCAGTGCATCATCTAAGATATC sequence (GCCACC), stop codon (TAA) and EcoRV restriction enzyme cleavage site (GATATC) 42 Human Complement MRLLAKIICLMLWAICVA GSS SGGHCQAPDHFLF Receptor 1 CCPs 8-10 AKLKTQTQASDFPIGTSLKYECRPEYYGRPFSITCLDNLVWSSPKDVC Non-glycosylated KRKSCKTPPDPVNGMVHVITDIQVGSRIQYSCTTGHRLIGHSSAECIL Including leader SGNAAHWSTKPPICQRIPCGLPPTIANGDFISTNRENFHYGSVVTYRC sequence (doubled NPGSGGRKVFELVGEPSIYCTSNDDQVGIWSGPAPQCII underlined); His tag (dotted line); TEV cleavage site (dashed line) 43 Human Complement AAGCTTGCCACCATGAGACTGCTGGCCAAGATCATCTGCCTGATGCTG Receptor 1 CCPs 15-17 TGGGCCATCTGCGTGGCC GGCAGCAGC protein coding sequence TCTGGCGGCCACTGTCAGGCCCCTGAT (underlined) CACTTCCTGTTCGCCAAGCTGAAAACCCAGACCAACGCCAGCGACTTC Including leader CCTATCGGCACCAGCCTGAAGTACGAGTGCAGACCCGAGTACTACGGC sequence (doubled AGACCCTTCAGCATCACCTGTCTGGACAACCTCGTGTGGTCTAGCCCC underlined); AAGGACGTGTGCAAGAGAAAGAGCTGCAAGACCCCTCCTGATCCTGTG His tag (dotted line); AACGGCATGGTGCACGTGATCACCGACATCCAAGTGGGCAGCAGAATC TEV cleavage site AACTACAGCTGCACCACCGGCCACAGACTGATCGGACACTCTAGCGCC (dashed line) GAGTGTATCCTGAGCGGCAATGCCGCACACTGGTCCACCAAGCCTCCA Including HindIII ATCTGCCAGAGAATCCCTTGCGGCCTGCCTCCTACAATCGCCAACGGC restriction enzyme GATTTCATCAGCACCAACAGAGAGAACTTCCACTACGGCTCCGTGGTC cleavage site (AAGCTT), ACCTACAGATGCAATCCTGGCAGCGGCGGCAGAAAGGTGTTCGAACTT Kozak consensus GTGGGCGAGCCCAGCATCTACTGCACCAGCAACGATGACCAAGTCGGC sequence (GCCACC), ATTTGGAGCGGCCCTGCTCCTCAGTGCATCATCCCTAAGATATC stop codon (TAA) and EcoRV restriction enzyme cleavage site (GATATC) 44 Human Complement MRLLAKIICLMLWAICVA GSS SGGHCQAPDHFLF Receptor 1 CCPs 15-17 AKLKTQTNASDFPIGTSLKYECRPEYYGRPFSITCLDNLVWSSPKDVC Including leader KRKSCKTPPDPVNGMVHVITDIQVGSRINYSCTTGHRLIGHSSAECIL sequence (doubled SGNTAHWSTKPPICQRIPCGLPPTIANGDFISTNRENFHYGSVVTYRC underlined); NLGSRGRKVFELVGEPSIYCTSNDDQVGIWSGPAPQCII His tag (dotted line); TEV cleavage site (dashed line) 45 Human Complement AAGCTTGCCACCATGAGACTGCTGGCCAAGATCATCTGCCTGATGCTG Receptor 1 CCPs 15-17 TGGGCCATCTGCGTGGCC GGCAGCAGC protein coding sequence TCTGGCGGCCACTGTCAGGCCCCTGAT (underlined) CACTTCCTGTTCGCCAAGCTGAAAACCCAGACACAGGCCAGCGACTTC Non-glycosylated CCTATCGGCACCAGCCTGAAGTACGAGTGCAGACCCGAGTACTACGGC Including leader AGACCCTTCAGCATCACCTGTCTGGACAACCTCGTGTGGTCTAGCCCC sequence (doubled AAGGACGTGTGCAAGAGAAAGAGCTGCAAGACCCCTCCTGATCCTGTG underlined); AACGGCATGGTGCACGTGATCACCGACATCCAAGTGGGCAGCAGAATC His tag (dotted line); CAGTACAGCTGCACCACAGGCCACAGACTGATCGGCCACTCTAGCGCC TEV cleavage site GAGTGTATCCTGAGCGGAAACACAGCCCACTGGTCCACCAAGCCTCCA (dashed line) ATCTGCCAGAGAATCCCTTGCGGCCTGCCTCCTACAATCGCCAACGGC Including HindIII GATTTCATCAGCACCAACAGAGAGAACTTCCACTACGGCTCCGTGGTC restriction enzyme ACCTACAGATGCAACCTGGGCTCCAGAGGCCGGAAGGTGTTCGAACTT cleavage site (AAGCTT), GTGGGCGAGCCTAGCATCTACTGCACCAGCAACGACGACCAAGTCGGC Kozak consensus ATTTGGAGCGGACCTGCTCCTCAGTGCATCATCCCTAAGATATC sequence (GCCACC), stop codon (TAA) and EcoRV restriction enzyme cleavage site (GATATC) 46 Human Complement MRLLAKIICLMLWAICVA GSS SGGHCQAPDHFLF Receptor 1 CCPs 15-17 AKLKTQTQASDFPIGTSLKYECRPEYYGRPFSITCLDNLVWSSPKDVC Non-glycosylated KRKSCKTPPDPVNGMVHVITDIQVGSRIQYSCTTGHRLIGHSSAECIL Including leader SGNAAHWSTKPPICQRIPCGLPPTIANGDFISTNRENFHYGSVVTYRC sequence (doubled NPGSGGRKVFELVGEPSIYCTSNDDQVGIWSGPAPQCII underlined); His tag (dotted line); TEV cleavage site (dashed line) 47 CR1a MRLLAKIICLMLWAICVAGHCQAPDHFLFAKLKTQTNASDFPIGTSLK Including leader YECRPEYYGRPFSITCLDNLVWSSPKDVCKRKSCKTPPDPVNGMVHVI sequence (double TDIQVGSRINYSCTTGHRLIGHSSAECILSGNAAHWSTKPPICQRIPC underline); CCPs 8-10 GLPPTIANGDFISTNRENFHYGSVVTYRCNPGSGGRKVFELVGEPSIY CTSNDDQVGIWSGPAPQCII 48 nCR1a MRLLAKIICLMLWAICVAGHCQAPDHFLFAKLKTQTQASDFPIGTSLK Including leader YECRPEYYGRPFSITCLDNLVWSSPKDVCKRKSCKTPPDPVNGMVHVI sequence (double TDIQVGSRIQYSCTTGHRLIGHSSAECILSGNAAHWSTKPPICQRIPC underline); CCPs 8-10; GLPPTIANGDFISTNRENFHYGSVVTYRCNPGSGGRKVFELVGEPSIY N509Q and N578Q CTSNDDQVGIWSGPAPQCII (numbered according to UniProt: P17927) 49 CR1b MRLLAKIICLMLWAICVAGHCQAPDHFLFAKLKTQTNASDFPIGTSLK Including leader YECRPEYYGRPFSITCLDNLVWSSPKDVCKRKSCKTPPDPVNGMVHVI sequence (double TDIQVGSRINYSCTTGHRLIGHSSAECILSGNTAHWSTKPPICQRIPC underline); CCPs 15-17 GLPPTIANGDFISTNRENFHYGSVVTYRCNLGSRGRKVFELVGEPSIY CTSNDDQVGIWSGPAPQCII 50 nCR1b MRLLAKIICLMLWAICVAGHCQAPDHFLFAKLKTQTQASDFPIGTSLK Including leader YECRPEYYGRPFSITCLDNLVWSSPKDVCKRKSCKTPPDPVNGMVHVI sequence (double TDIQVGSRIQYSCTTGHRLIGHSSAECILSGNTAHWSTKPPICQRIPC underline); CCPs 15-17; GLPPTIANGDFISTNRENFHYGSVVTYRCNLGSRGRKVFELVGEPSIY N959Q and N1028Q CTSNDDQVGIWSGPAPQCII (numbered according to UniProt: P17927) 51 CR1a + CR1b MRLLAKIICLMLWAICVAGHCQAPDHFLFAKLKTQTNASDFPIGTSLK Including leader YECRPEYYGRPFSITCLDNLVWSSPKDVCKRKSCKTPPDPVNGMVHVI sequence (double TDIQVGSRINYSCTTGHRLIGHSSAECILSGNAAHWSTKPPICQRIPC underline); CCPs 8-10 +30 GLPPTIANGDFISTNRENFHYGSVVTYRCNPGSGGRKVFELVGEPSIY 15-17 CTSNDDQVGIWSGPAPQCIIGHCQAPDHFLFAKLKTQTNASDFPIGTS LKYECRPEYYGRPFSITCLDNLVWSSPKDVCKRKSCKTPPDPVNGMVH VITDIQVGSRINYSCTIGHRLIGHSSAECILSGNTAHWSTKPPICQRI PCGLPPTIANGDFISTNRENFHYGSVVTYRCNLGSRGRKVFELVGEPS IYCTSNDDQVGIWSGPAPQCII 52 nCR1a + nCR1b MRLLAKIICLMLWAICVAGHCQAPDHFLFAKLKTQTQASDFPIGTSLK Including leader YECRPEYYGRPFSITCLDNLVWSSPKDVCKRKSCKTPPDPVNGMVHVI sequence (double TDIQVGSRIQYSCTIGHRLIGHSSAECILSGNAAHWSTKPPICQRIPC underline); CCPs 8-10 +30 GLPPTIANGDFISTNRENFHYGSVVTYRCNPGSGGRKVFELVGEPSIY 15-17; N509Q and CTSNDDQVGIWSGPAPQCIIGHCQAPDHFLFAKLKTQTQASDFPIGTS N578Q , N959Q and LKYECRPEYYGRPFSITCLDNLVWSSPKDVCKRKSCKTPPDPVNGMVH N1028Q (numbered VITDIQVGSRIQYSCTIGHRLIGHSSAECILSGNTAHWSTKPPICQRI according to UniProt: PCGLPPTIANGDFISTNRENFHYGSVVTYRCNLGSRGRKVFELVGEPS P17927) IYCTSNDDQVGIWSGPAPQCII 53 CR1a + nCR1b MRLLAKIICLMLWAICVAGHCQAPDHFLFAKLKTQTNASDFPIGTSLK Including leader YECRPEYYGRPFSITCLDNLVWSSPKDVCKRKSCKTPPDPVNGMVHVI sequence (double TDIQVGSRINYSCTIGHRLIGHSSAECILSGNAAHWSTKPPICQRIPC underline); CCPs 8-10 +30 GLPPTIANGDFISTNRENFHYGSVVTYRCNPGSGGRKVFELVGEPSIY 15-17; N959Q and CTSNDDQVGIWSGPAPQCIIGHCQAPDHFLFAKLKTQTQASDFPIGTS N1028Q (numbered LKYECRPEYYGRPFSITCLDNLVWSSPKDVCKRKSCKTPPDPVNGMVH according to UniProt: VITDIQVGSRIQYSCTIGHRLIGHSSAECILSGNTAHWSTKPPICQRI P17927) PCGLPPTIANGDFISTNRENFHYGSVVTYRCNLGSRGRKVFELVGEPS IYCTSNDDQVGIWSGPAPQCII 54 nCR1a + CR1b MRLLAKIICLMLWAICVAGHCQAPDHFLFAKLKTQTQASDFPIGTSLK Including leader YECRPEYYGRPFSITCLDNLVWSSPKDVCKRKSCKTPPDPVNGMVHVI sequence (double TDIQVGSRIQYSCTIGHRLIGHSSAECILSGNAAHWSTKPPICQRIPC underline); CCPs 8-10 +30 GLPPTIANGDFISTNRENFHYGSVVTYRCNPGSGGRKVFELVGEPSIY 15-17; N509Q and CTSNDDQVGIWSGPAPQCIIGHCQAPDHFLFAKLKTQTNASDFPIGTS N578Q (numbered LKYECRPEYYGRPFSITCLDNLVWSSPKDVCKRKSCKTPPDPVNGMVH according to UniProt: VITDIQVGSRINYSCTIGHRLIGHSSAECILSGNTAHWSTKPPICQRI P17927) PCGLPPTIANGDFISTNRENFHYGSVVTYRCNLGSRGRKVFELVGEPS IYCTSNDDQVGIWSGPAPQCII
[0124] Functional Properties of the Polypeptide
[0125] The polypeptide according to the present invention may be characterised by reference to one or more functional properties.
[0126] In particular, a polypeptide according to the present invention may possess one or more of the following properties (as determined by analysis in an appropriate assay for said property): [0127] Binds to C3b; [0128] Binds to C3b with an affinity of binding which is similar to the affinity of binding to C3b displayed by a co-factor for Complement Factor I (or a fragment thereof; [0129] Binds to C3b with an affinity of binding which is higher than the affinity of binding to C3b displayed by a co-factor for Complement Factor I (or a fragment thereof; [0130] Binds to C3b with an affinity of binding which is similar to the affinity of binding to C3b displayed by Complement Receptor 1 (or a fragment thereof; [0131] Binds to C3b in the region of C3b bound by a co-factor for Complement Factor I (or a fragment thereof); [0132] Binds to C3b in the region of C3b bound by Complement Receptor 1 (or a fragment thereof); [0133] Binds to C3b in the region of C3b bound by Complement Receptor 1 CCP domains 8-10 and/or 15-17 (or a fragment thereof); [0134] Acts as a co-factor to enable Complement Factor I-mediated inactivation of C3b; [0135] Acts as a co-factor to enable Complement Factor I-mediated reduction/prevention of the formation of a functional C3bBb-type C3 convertase; [0136] Acts as a co-factor to enable Complement Factor I-mediated reduction/prevention of the formation of a functional C3bBb3b-type C5 convertase; [0137] Acts as a co-factor to enable Complement Factor I-mediated reduction/prevention of the formation of a functional C4b2a3b-type C5 convertase; [0138] Acts as a co-factor to enable Complement Factor I-mediated reduction of C3bBb-type C3 convertase activity; [0139] Acts as a co-factor to enable Complement Factor I-mediated reduction of C3bBb3b-type C5 convertase activity; [0140] Acts as a co-factor to enable Complement Factor I-mediated reduction of C4b2a3b-type C5 convertase activity; [0141] Acts as a co-factor to enable Complement Factor I-mediated reduction of the amount of C3bBb-type C3 convertase; [0142] Acts as a co-factor to enable Complement Factor I-mediated reduction of the amount of C3bBb3b-type C5 convertase; [0143] Acts as a co-factor to enable Complement Factor I-mediated reduction of the amount of C4b2a3b-type C5 convertase; [0144] Reduces the amount of C3b via Complement Factor I; [0145] Increases the amount of iC3b via Complement Factor I; [0146] Increases the amount of C3dg via Complement Factor I; [0147] Increases the amount of C3d via Complement Factor I; [0148] Increases the amount of C3f via Complement Factor I; [0149] Reduces the amount of C5b via Complement Factor I; [0150] Reduces the amount of C5a via Complement Factor I; [0151] Decreases the amount of iC3b via Complement Factor I compared to the amount of iC3b produced by FH and/or FHL-1 via Complement Factor I; [0152] Increases the ratio of C3dg to iC3b via Complement Factor I; [0153] Is capable of inhibiting complement activation; [0154] Diffuses through Bruch's membrane (BrM); [0155] Displays superior ability to diffuse through BrM compared to Complement Factor I; [0156] Displays superior ability to diffuse through BrM compared to a co-factor for Complement Factor I (or a fragment thereof); [0157] Displays similar ability to diffuse through BrM compared to a co-factor for Complement Factor I (or a fragment thereof; [0158] Displays superior ability to diffuse through BrM compared to Complement Factor H; [0159] Displays similar ability to diffuse through BrM compared to Complement Factor H isoform FHL-1; [0160] Displays superior ability to diffuse through BrM compared to Complement Factor H isoform FHL-1; [0161] Displays similar ability to diffuse through BrM compared to soluble Complement Receptor 1; [0162] Displays superior ability to diffuse through BrM compared to soluble Complement Receptor 1.
[0163] Whether a given polypeptide possesses the functional properties referred to in the previous paragraph can be analysed, for example, as described herein.
[0164] A polypeptide according to the present invention may be capable of binding to C3b. In some embodiments, a polypeptide according to the present invention may comprise or consist of a C3b binding region. In some embodiments, the C3b binding region of the polypeptide according to the present invention comprises or consists of a C3b binding region of CR1, e.g. CCP domains 8-10 and/or 15-17.
[0165] As used herein, a "C3b binding region" refers to a region capable of binding to C3b. In some embodiments, the C3b binding region is capable of specific binding to C3b. Binding to C3b may be mediated by non-covalent interactions such as Van der Waals forces, electrostatic interactions, hydrogen bonding, and hydrophobic interactions formed between the C3b binding region and C3b. In some embodiments, the C3b binding region binds to C3b with greater affinity, and/or with greater duration than it binds to molecules other than C3b.
[0166] The ability of a polypeptide according to the present invention or a putative C3b binding region to bind to C3b can be analysed using techniques well known to the person skilled in the art, including ELISA, Surface Plasmon Resonance (SPR; see e.g. Hearty et al., Methods Mol Biol (2012) 907:411-442; or Rich et al., Anal Biochem. 2008 Feb. 1; 373(1):112-20), Bio-Layer Interferometry (see e.g. Lad et al., (2015) J Biomol Screen 20(4): 498-507; or Concepcion et al., Comb Chem High Throughput Screen. 2009 September; 12(8):791-800), MicroScale Thermophoresis (MST) analysis (see e.g. Jerabek-Willemsen et al., Assay Drug Dev Technol. 2011 August; 9(4): 342-353), or by a radiolabelled antigen binding assay (RIA). Through such analysis binding to a given target can be determined and quantified. In some embodiments, the binding may be the response detected in a given assay.
[0167] In some embodiments, a polypeptide according to the present invention displays binding to C3b in such an assay which is greater than 1 times, e.g. one of >1.01, >1.02, >1.03, >1.04, >1.05, >1.06, >1.07, >1.08, >1.09, >1.1, >1.2, >1.3, >1.4, >1.5, >1.6, >1.7, >1.8, >1.9, >2, >3, >4, >5, >6, >7, >8, >9, >10, >15, >20, >25, >30, >35, >40, >45, >50, >60, >70, >80, >90, or >100 times the level of binding signal detected in such an assay to a negative control molecule to which the C3b binding region does not bind.
[0168] In some embodiments, a polypeptide according to the present invention is capable of binding to C3b with an affinity of binding which is similar to the affinity of binding to C3b displayed by CR1 or another co-factor for Complement Factor I (or a fragment thereof) in a given assay. An affinity of binding which is similar to a reference affinity of binding can be e.g. .+-.40% of the level of binding, e.g. one of 35%, .+-.30%, 25%, 20%, .+-.15%, .+-.10% or .+-.5% of the level of binding to C3b displayed by reference CR1 or the reference co-factor for Complement Factor I in a comparable assay.
[0169] In some embodiments a polypeptide according to the present invention is capable of binding to C3b with an affinity of binding which is higher than the affinity of binding to C3b displayed by a co-factor for Complement Factor I (or a fragment thereof) in a given assay. In some embodiments a polypeptide according to the present invention is capable of binding to C3b with an affinity of binding which is 1.5 times, 2 times, 2.5 times, 3 times, 3.5 times, 4 times, 4.5 times, 5 times, 5.5 times, 6 times, 6.5 times, 7 times, 7.5 times, 8 times, 8.5 times, 9 times, 9.5 times, 10 times, 15 times, 20 times, 25 times, 30 times, 35 times, 40 times, 45 times, 50 times, 75 times, 100 times, 150 times, 200 times, 250 times, 300 times, 350 times, 400 times, 450 times, 500 times, 550 times, 600 times, 650 times, 700 times, 750 times, 800 times, 850 times, 900 times, 950 times, or 1000 times the affinity of binding to C3b displayed by a co-factor for Complement Factor I (or a fragment thereof) in a given assay. In some embodiments a polypeptide according to the present invention is capable of binding to C3b with an affinity of binding which is 10000 times, 100000 times, or 1000000 times the affinity of binding to C3b displayed by a co-factor for Complement Factor I (or a fragment thereof) in a given assay. In some embodiments a polypeptide according to the present invention is capable of binding to C3b with an affinity of binding which is 2, 3, 4, 5, 6, 7, 8, 9 or 10 order(s) of magnitude greater than the affinity of binding to C3b displayed by a co-factor for Complement Factor I (or a fragment thereof) in a given assay. In some embodiments the co-factor for Complement Factor I is Complement Factor H or truncated FH isoform FHL-1. The co-factor for Complement Factor I may be CR1.
[0170] In some embodiments, a polypeptide according to the present invention is capable of binding to C3b in the region of C3b that is bound by a co-factor for Complement Factor I (i.e. binds to the same region or an overlapping region). In some embodiments, the polypeptide is capable of binding to C3b in the region bound by CR1 (or a fragment thereof). In some embodiments, the polypeptide is capable of binding to C3b in the region bound by CR1 CCP domains 8-10 and/or 15-17. In some embodiments, the polypeptide is capable of binding to C3b in the region bound by one or more of Complement Factor I co-factors Complement Factor H, CD46, CD55, C4BP, SPICE, VCP, or MOPICE (or fragments thereof).
[0171] Whether a polypeptide according to the present invention binds to C3b in the region of C3b bound by a given co-factor for Complement Factor I (or a fragment thereof) can be determined by various methods known to the skilled person, including ELISA, and surface plasmon resonance (SPR) analysis. An example of a suitable assay to determine whether a C3b binding region binds to C3b in the region bound by a given co-factor for Complement Factor I (or a fragment thereof is a competition ELISA assay.
[0172] For example, whether a polypeptide according to the present invention binds to C3b in the region of C3b bound by a given co-factor for Complement Factor I (or a fragment thereof can be determined by analysis of interaction of the co-factor/fragment with C3b in the presence of, or following incubation of one or both of the co-factor/fragment and C3b with the polypeptide according to the present invention. A C3b binding region which binds to C3b in the region of C3b bound by a given co-factor/fragment is identified by the observation of a reduction/decrease in the level of interaction between the co-factor/fragment and C3b in the presence of--or following incubation of one or both of the interaction partners with--the polypeptide according to the present invention, as compared to the level of interaction in the absence of the polypeptide according to the present invention (or in the presence of an appropriate control peptide/polypeptide). Suitable analysis can be performed in vitro, e.g. using recombinant interaction partners. For the purposes of such assays, one or both of the interaction partners and/or the polypeptide according to the present invention may be labelled, or used in conjunction with a detectable entity for the purposes of detecting and/or measuring the level of interaction.
[0173] In some embodiments, the polypeptide according to the present invention acts as a co-factor for Complement Factor I. For example, the polypeptide may potentiate cleavage of C3b by Complement Factor I, and/or present C3b in a favourable orientation for proteolytic cleavage by Complement Factor I. The polypeptide preferably does not inhibit proteolytic cleavage of C3b by Complement Factor I. In some embodiments, the Complement Factor I is endogenous. In some embodiments, the Complement Factor I is exogenous.
[0174] As used herein, an `endogenous` protein/peptide refers to a protein/peptide which is encoded/expressed by the relevant cell type, tissue, or subject (prior to treatment with a polypeptide, nucleic acid, vector, cell or pharmaceutical composition according to the present invention). A `non-endogenous` or `exogenous` protein/peptide refers to a protein/peptide which is not encoded/expressed by, the relevant cell type, tissue, or subject (prior to treatment with a polypeptide, nucleic acid, vector, cell or pharmaceutical composition according to the present invention).
[0175] A polypeptide according to the present invention that acts as a co-factor for Complement Factor I can be determined by e.g. by analysis of the level or rate of proteolytic cleavage of C3b by Complement Factor I in a suitable assay in the presence of (or after incubation with) a polypeptide according to the present invention as compared to the level or rate of proteolytic cleavage of C3b by Complement Factor I in the absence of the polypeptide according to the present invention (or in the presence of an appropriate control peptide/polypeptide). A C3b binding region which acts as a co-factor for Complement Factor I is identified by the detection of an increased level or rate of proteolytic cleavage of C3b by Complement Factor I in the presence of (or after incubation with) a polypeptide according to the present invention as compared to the level or rate of proteolytic cleavage of C3b by Complement Factor I in the absence of the polypeptide according to the present invention (or in the presence of an appropriate control peptide/polypeptide). The level or rate of proteolytic cleavage of C3b by Complement Factor I can be determined e.g. by detection of one or more products of cleavage of C3b by Complement Factor I, e.g. iC3b, C3dg, C3d or C3f. The level or rate of proteolytic cleavage of C3b by Complement Factor I can be determined e.g. by detection of a reduction in the presence of C3b. In some embodiments a polypeptide according to the present invention that acts as a co-factor for Complement Factor I produces a smaller amount of iC3b overall compared to an amount of iC3b produced by FH/FHL-1 via Complement Factor I. In some embodiments a polypeptide according to the present invention that acts as a co-factor for Complement Factor I increases the ratio of C3dg to iC3b via Complement Factor I compared to the ratio of C3dg to iC3b produced by FH/FHL-1 via Complement Factor I. For example, the polypeptide may not increase the overall amount of iC3b, but may instead increase the amount of C3dg, C3f and/or C3d via Complement Factor I.
[0176] In some embodiments a polypeptide according to the present invention is capable of inhibiting or reducing complement activation. A polypeptide may inhibit or reduce complement over-activation. The level of complement activation/over-activation may be determined by the assays described herein, e.g. abnormal levels of complement components, or by tests/assays that are known by one skilled in the art, e.g. as described in Shih and Murali Am. J. Hematol. 2015, 90: 1180-1186; Kirschfink and Mollnes, Clin Diagn Lab Immunol. 2003, 10(6): 982-989; Nilsson and Ekdahl, Clinical and Developmental Immunology, 2012, Article ID 962702; which are hereby incorporated by reference in their entirety.
[0177] In some embodiments, the polypeptide according to the present invention possesses the ability to diffuse through, i.e. pass through, Bruch's Membrane (BrM), as determined by analysis in an appropriate assay for said property.
[0178] The ability of a given polypeptide to diffuse through BrM can be analysed e.g. in vitro, e.g. as described in Clark et al J. Immunol (2014) 193, 4962-4970. Briefly, BrM can be isolated from donor eyes as described in McHarg et al., J Vis Exp (2015) 1-7, and the macular area can be mounted in an Ussing chamber. Once mounted, the 5 mm diameter macular area is the only barrier between two identical compartments. Both sides of BrM can be washed with PBS, and human serum can be diluted 1:1 with PBS and added to the Ussing compartment on one side of the BrM (the sample chamber). The polypeptide to be analysed can be added to the sample chamber in PBS, and PBS alone can be added to the compartment on the other side of the BrM (the diffusate chamber), and the Ussing chamber can be incubated at room temperature for 24 hours with gentle stirring in both the sample and diffusate chambers. Samples from each chamber can subsequently be analysed for the presence of the polypeptide, e.g. using antibody based detection methods such as ELISA analysis or western blot. Detection of the polypeptide in the diffusate chamber indicates that the polypeptide is capable of diffusing through BrM. Suitable positive and negative control proteins known to be able to/not to be able to diffuse through BrM can be included in such experiments.
[0179] In some embodiments, a polypeptide according to the present invention displays superior ability to diffuse through BrM than Complement Factor I. In some embodiments, a polypeptide according to the present invention displays superior ability to diffuse through BrM than Complement Factor H. FH, consisting of 20 CCP domains, is a large molecule and does not pass through BrM. In some embodiments, a polypeptide according to the present invention displays similar ability to diffuse through BrM as compared to the truncated Complement Factor H isoform FHL-1 (UniProt: P08603-2; SEQ ID NO:28). In some embodiments, a polypeptide according to the present invention displays superior ability to diffuse through BrM as compared to Complement Factor H isoform FHL-1. In some embodiments, a polypeptide according to the present invention displays similar ability to diffuse through BrM as compared to full length soluble CR1 (30 CCP domains; SEQ ID NO:1 lacking SEQ ID NO:32 and 33). In some embodiments, a polypeptide according to the present invention displays superior ability to diffuse through BrM as compared to full length soluble CR1. A polypeptide according to the present invention that is able to diffuse though BrM preferably remains functionally active, i.e. acts as a cofactor for Complement Factor I, after diffusing though BrM.
[0180] A polypeptide of the present invention displaying superior ability to diffuse through BrM as compared to a given reference polypeptide can be identified by analysing diffusion through BrM as described above. The diffusion through BrM may be detected by measuring the rate of diffusion through to the diffusate chamber and/or detecting the proportion of polypeptide present in the diffusate chamber at the end of the experiment. A polypeptide of the present invention displaying similar ability to diffuse through BrM as compared to a given reference polypeptide can be identified by analysing diffusion through BrM as described above. A similar ability to diffuse through BrM may be indicated by detecting a rate of diffusion through to the diffusate chamber which is within 30%, e.g. within one of 25%, 20%, 15%, or 10% of the rate of diffusion for a reference polypeptide, and/or by detecting a proportion of the polypeptide of the present invention present in the diffusate chamber at the end of the experiment that is within 30%, e.g. within one of 25%, 20%, 15%, or 10% of the proportion of a reference polypeptide present in the diffusate chamber.
[0181] As a result of the ability of the polypeptides to diffuse through the BrM, the polypeptides of the present invention are also able to diffuse away from the site of C3b inactivation if/once they have performed their cofactor role with FI. In other words, the polypeptides of the present invention may be capable of being present transiently at areas of complement activation. This is advantageous because accumulation of complement-related debris is undesirable, particularly in the context of macular degeneration where cellular debris accumulation can lead to the formation of drusen.
[0182] A polypeptide of the present invention may be capable of being expressed in a cell, e.g. a cell as described herein. A polypeptide of the present invention may be capable of being secreted by a cell, e.g. a cell as described herein. In some embodiments the cell is an ocular cell, e.g. an RPE cell, as described herein.
[0183] Nucleic Acids, Cells, Compositions and Kits
[0184] The present invention provides a nucleic acid encoding a polypeptide according to the present invention. In some embodiments, the nucleic acid is purified or isolated, e.g. from other nucleic acid, or naturally-occurring biological material. In some embodiments the nucleic acid(s) comprise or consist of DNA and/or RNA.
[0185] Provided herein are nucleic acid sequences that encode a polypeptide comprising or consisting of SEQ ID NO:2, 3, 5, 6, 13, 14, 15, 30, 31, 40, 42, 44, 46, 47, 48, 49, 50, 51, 52, 53 or 54. The encoded polypeptide may be produced with or without a leader sequence e.g. a secretory pathway sequence. The encoded polypeptide may be produced together with a leader sequence which is then subsequently removed from said polypeptide.
[0186] In some embodiments, a nucleic acid according to the present invention comprises, or consists of, one or more of SEQ ID NO:35, 36, 37, 38, 39, 41, 43, and/or 45, or equivalent nucleic acid sequences thereof which would be translated into the same respective polypeptides due to codon degeneracy.
[0187] The present invention also provides a vector comprising nucleic acid encoding a polypeptide according to the present invention.
[0188] The nucleotide sequence may be contained in a vector, e.g. an expression vector. A "vector" as used herein is a nucleic acid molecule used as a vehicle to transfer exogenous nucleic acid into a cell. The vector may be a vector for expression of the nucleic acid in the cell. Such vectors may include a promoter sequence operably linked to the nucleotide sequence encoding the sequence to be expressed. A vector may also include a termination codon and expression enhancers. A vector may include regulatory elements, such as a polyadenylation site. Any suitable vectors, promoters, enhancers and termination codons known in the art may be used to express a peptide or polypeptide from a vector according to the invention. Nucleic acid sequences described herein may be codon optimised for optimised expression in a desired cell or organism.
[0189] The term "operably linked" may include the situation where a selected nucleic acid sequence and regulatory nucleic acid sequence (e.g. promoter and/or enhancer) are covalently linked in such a way as to place the expression of nucleic acid sequence under the influence or control of the regulatory sequence (thereby forming an expression cassette). Thus a regulatory sequence is operably linked to the selected nucleic acid sequence if the regulatory sequence is capable of effecting transcription of the nucleic acid sequence. The resulting transcript(s) may then be translated into a desired peptide(s)/polypeptide(s).
[0190] The nucleic acid and/or vector according to the present invention is preferably provided for introduction into a cell, e.g. a human cell. Suitable vectors include plasmids, binary vectors, DNA vectors, mRNA vectors, viral vectors (e.g. gammaretroviral vectors (e.g. murine Leukemia virus (MLV)-derived vectors), lentiviral vectors, retroviral vectors, adenovirus vectors, adeno-associated virus (AAV) vectors, vaccinia virus vectors and herpesvirus vectors, e.g. Herpes Simplex Virus vectors), transposon-based vectors, and artificial chromosomes (e.g. yeast artificial chromosomes), e.g. as described in Maus et al., Annu Rev Immunol (2014) 32:189-225 or Morgan and Boyerinas, Biomedicines 2016 4, 9, which are both hereby incorporated by reference in its entirety. In some embodiments, the lentiviral vector may be pELNS, or may be derived from pELNS. In some embodiments, the vector may be a vector encoding CRISPR/Cas9. In some embodiments, the adeno-associated virus (AAV) vector is selected from AAV serotype 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or 11, or hybrids and/or mutants thereof. In some embodiments, the AAV vector is an AAV serotype 2 (AAV-2) vector, or a hybrid and/or mutant thereof. Viral and non-viral delivery systems for introducing genetic material into cells are reviewed, for example, in Nayerossadat et al., Adv Biomed Res. 2012; 1: 27; MacLaren et al. Ophthalmology. 2016, 123(10 Suppl): S98-S106; Petit and Punzo, Discov Med. 2016, 22(121): 221-229; Aguirre, Invest Ophthalmol Vis Sci. 2017, 58(12): 5399-5411; Lundstrom, Diseases. 2018, 6(2): 42; which are hereby incorporated by reference in their entirety. Any suitable nucleotide or vector delivery method can be used in the context of the present invention.
[0191] In some embodiments the expression of a nucleic acid or a nucleic acid contained in a vector, according to the present invention, is driven by a promoter that drives expression in a specific retinal cell type, e.g. rods, cones, RPE, or ganglion cells, as described for example in Beltran W A, et al. Gene Ther. 2010; 17:1162-74 and Boye S E, et al. Hum Gene Ther. 2012; 23:1101-15, which are hereby incorporated by reference in their entirety.
[0192] In some embodiments, the expression of a nucleic acid or a nucleic acid contained in a vector, according to the present invention, is driven by a promoter that drives expression of that nucleic acid in retinal pigment epithelial (RPE) cells. In some embodiments, the promoter is an RPE65 or VMD2 promoter, or modified version thereof. In some embodiments the promoter is a chicken .beta. actin promoter.
[0193] In some embodiments, the vector may be a eukaryotic vector, e.g. a vector comprising the elements necessary for expression of protein from the vector in a eukaryotic cell. In some embodiments, the vector may be a mammalian vector, e.g. comprising a cytomegalovirus (CMV) or SV40 promoter to drive protein expression.
[0194] In some embodiments the vector comprises an inducible promoter, i.e. gene expression is activated by the promoter only in the presence or absence of a particular molecule. Suitable inducible promoters will be known to the skilled person. Examples of inducible promoters are described in e.g. Le at al. Invest Ophthalmol Vis Sci. 2008, 49(3): 1248-1253 and McGee Sanftner et al. Mol Ther. 2001. 3(5): 688-696; which are hereby incorporated by reference in their entirety.
[0195] In some embodiments, the nucleic acid comprises, or consists of, a nucleic acid sequence encoding a polypeptide having an amino acid sequence having at least 70%, 75%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID NO:2, 3, 4, 5, 6, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 30, 31, 40, 42, 44, 46, 47, 48, 49, 50, 51, 52, 53, or 54, or any combination of sequences A, B, and/or C as described hereinabove.
[0196] The present invention also provides a cell comprising or expressing a polypeptide according to the present invention. Also provided is a cell comprising or expressing a nucleic acid or vector according to the invention. The cell comprising or expressing polypeptide, nucleic acid or vector according to the present invention may secrete a polypeptide according to the present invention. That is, expression of the polypeptide, nucleic acid or vector by the cell may result in the soluble production of a polypeptide according of the present invention from the cell.
[0197] The cell may be a eukaryotic cell, e.g. a mammalian cell. The mammal may be a human, or a non-human mammal (e.g. rabbit, guinea pig, rat, mouse or other rodent (including any animal in the order Rodentia), cat, dog, pig, sheep, goat, cattle (including cows, e.g. dairy cows, or any animal in the order Bos), horse (including any animal in the order Equidae), donkey, and non-human primate). In some embodiments, the cell may be from, or may have been obtained from, a human subject.
[0198] In some embodiments, the cell is a cell of the eye. In some embodiments, the cell is a cell of the neurosensory retina, retinal pigment epithelium (RPE), choroid or macula. In some embodiments, the cell is a retinal cell. In some embodiments, the cell is a retinal pigment epithelial cell. In some embodiments, the cell is a human retinal pigment epithelial cell (RPE). In some embodiments the cell is a photoreceptor cell.
[0199] The present invention also provides a method for producing a cell comprising a nucleic acid or vector according to the present invention, comprising introducing a nucleic acid or vector according to the present invention into a cell. In some embodiments, introducing an isolated nucleic acid(s) or vector(s) according to the invention into a cell comprises transformation, transfection, electroporation or transduction (e.g. retroviral transduction). Methods for producing a cell according to the present invention may be performed according to methods known to the skilled person for the production of cells comprising nucleic acid/vectors.
[0200] The present invention also provides a method for producing a cell comprising or expressing a polypeptide according to the present invention, comprising introducing a nucleic acid or vector according to the present invention into a cell. In some embodiments, the methods additionally comprise culturing the cell under conditions suitable for expression of the nucleic acid or vector by the cell. In some embodiments, the methods are performed in vitro or ex vivo. In some embodiments, the methods are performed in vivo.
[0201] The present invention also provides cells obtained or obtainable by the methods according to the present invention.
[0202] The present invention also provides compositions comprising a polypeptide, nucleic acid, vector, or cell according to the present invention.
[0203] Polypeptides, nucleic acids, vectors and cells according to the present invention may be formulated as pharmaceutical compositions for clinical use and may comprise a pharmaceutically acceptable carrier, diluent, excipient or adjuvant.
[0204] In accordance with the present invention methods are also provided for the production of pharmaceutically useful compositions, such methods of production may comprise one or more steps selected from: isolating a polypeptide, cell, nucleic acid or vector as described herein; and/or mixing a polypeptide, cell, nucleic acid or vector as described herein with a pharmaceutically acceptable carrier, adjuvant, excipient or diluent.
[0205] A kit of parts is also provided. In some embodiments the kit may have at least one container having a predetermined quantity of a polypeptide, nucleic acid, vector, cell, and/or composition according to the present invention.
[0206] The kit may provide the polypeptide, nucleic acid, vector, cell or composition together with instructions for administration to a subject in order to treat a specified disease/condition. The polypeptide, nucleic acid, vector, cell or composition may be formulated so as to be suitable for injection or infusion. In some embodiments, the polypeptide, nucleic acid, vector, cell or composition may be formulated so as to be suitable for intravenous, intraocular, sub-retinal, suprachoroidal or intraconjunctival injection, administration as an eye drop (i.e. ophthalmic administration), or oral administration.
[0207] In some embodiments the kit may comprise materials for producing a cell according to the present invention. For example, the kit may comprise materials for modifying a cell to express or comprise a polypeptide, nucleic acid or vector according to the present invention, or materials for introducing into a cell the nucleic acid or vector according to the present invention.
[0208] In some embodiments the kit may further comprise at least one container having a predetermined quantity of another therapeutic agent (e.g. a therapeutic agent for the treatment of AMD). In such embodiments, the kit may also comprise a second medicament or pharmaceutical composition such that the two medicaments or pharmaceutical compositions may be administered simultaneously or separately such that they provide a combined treatment for the specific disease or condition. In some embodiments, the second medicament or pharmaceutical composition comprises Complement Factor I.
[0209] Producing Polypeptides
[0210] The present invention also provides a method for producing a polypeptide according to the present invention, the method comprising introducing into a cell a nucleic acid or vector according to the present invention, and culturing the cell under conditions suitable for expression of the polypeptide. The polypeptide may be a fusion protein. The polypeptide may be subsequently isolated and/or substantially purified.
[0211] Polypeptides according to the present invention may be prepared according to methods for the production of polypeptides known to the skilled person.
[0212] Polypeptides may be prepared by chemical synthesis, e.g. liquid or solid phase synthesis. For example, peptides/polypeptides can by synthesised using the methods described in, for example, Chandrudu et al., Molecules (2013), 18: 4373-4388, which is hereby incorporated by reference in its entirety.
[0213] Alternatively, polypeptides may be produced by recombinant expression. Molecular biology techniques suitable for recombinant production of polypeptides are well known in the art, such as those set out in Green and Sambrook, Molecular Cloning: A Laboratory Manual (4th Edition), Cold Spring Harbor Press, 2012, and in Nat Methods. (2008); 5(2): 135-146 both of which are hereby incorporated by reference in their entirety.
[0214] For recombinant production according to the invention, any cell suitable for the expression of polypeptides may be used. The cell may be a prokaryote or eukaryote. In some embodiments the cell is a prokaryotic cell, such as a cell of archaea or bacteria. In some embodiments the bacteria may be Gram-negative bacteria such as bacteria of the family Enterobacteriaceae, for example Escherichia coli. In some embodiments, the cell is a eukaryotic cell such as a yeast cell, a plant cell, insect cell or a mammalian cell, e.g. CHO, HEK (e.g. HEK293), HeLa or COS cells.
[0215] In some cases the cell is not a prokaryotic cell because some prokaryotic cells do not allow for the same folding or post-translational modifications as eukaryotic cells. In addition, very high expression levels are possible in eukaryotes and proteins can be easier to purify from eukaryotes using appropriate tags. Specific plasmids may also be utilised which enhance secretion of the protein into the media.
[0216] In some embodiments polypeptides may be prepared by cell-free-protein synthesis (CFPS), e.g. according using a system described in Zemella et al. Chembiochem (2015) 16(17): 2420-2431, which is hereby incorporated by reference in its entirety.
[0217] Production may involve culture or fermentation of a eukaryotic cell modified to express the polypeptide(s) of interest. The culture or fermentation may be performed in a bioreactor provided with an appropriate supply of nutrients, air/oxygen and/or growth factors. Secreted proteins can be collected by partitioning culture media/fermentation broth from the cells, extracting the protein content, and separating individual proteins to isolate secreted polypeptide(s). Culture, fermentation and separation techniques are well known to those of skill in the art, and are described, for example, in Green and Sambrook, Molecular Cloning: A Laboratory Manual (4th Edition; incorporated by reference herein above).
[0218] Bioreactors include one or more vessels in which cells may be cultured. Culture in the bioreactor may occur continuously, with a continuous flow of reactants into, and a continuous flow of cultured cells from, the reactor. Alternatively, the culture may occur in batches. The bioreactor monitors and controls environmental conditions such as pH, oxygen, flow rates into and out of, and agitation within the vessel such that optimum conditions are provided for the cells being cultured.
[0219] Following culturing the cells that express the antigen-binding molecule/polypeptide(s), the polypeptide(s) of interest may be isolated. Any suitable method for separating proteins from cells known in the art may be used. In order to isolate the polypeptide it may be necessary to separate the cells from nutrient medium. If the polypeptide(s) are secreted from the cells, the cells may be separated by centrifugation from the culture media that contains the secreted polypeptide(s) of interest. If the polypeptide(s) of interest collect within the cell, protein isolation may comprise centrifugation to separate cells from cell culture medium, treatment of the cell pellet with a lysis buffer, and cell disruption e.g. by sonication, rapid freeze-thaw or osmotic lysis.
[0220] It may then be desirable to isolate the polypeptide(s) of interest from the supernatant or culture medium, which may contain other protein and non-protein components. A common approach to separating protein components from a supernatant or culture medium is by precipitation. Proteins of different solubilities are precipitated at different concentrations of precipitating agent such as ammonium sulfate. For example, at low concentrations of precipitating agent, water soluble proteins are extracted. Thus, by adding different increasing concentrations of precipitating agent, proteins of different solubilities may be distinguished. Dialysis may be subsequently used to remove ammonium sulfate from the separated proteins.
[0221] Other methods for distinguishing different proteins are known in the art, for example ion exchange chromatography and size chromatography. The polypeptide may also be affinity-purified using an appropriate binding partner for a molecular tag on the polypeptide (e.g. a His, FLAG, Myc, GST, MBP, HA, E, or Biotin tag). These may be used as an alternative to precipitation, or may be performed subsequently to precipitation.
[0222] In some cases it may further be desired to process the polypeptide, e.g. to remove a sequence of amino acids, molecular tag, moiety, etc.
[0223] In some embodiments, treatment is with an appropriate endopeptidase for the cleavage and removal of an amino acid sequence.
[0224] In some embodiments, treatment is with an enzyme to remove the moiety of interest. In some embodiments, the polypeptide is treated to remove glycans (i.e. the polypeptide is degylcosylated), e.g. by treatment with a glycosidase such as with a Peptide:N-glycosidase (PNGase).
[0225] Once the polypeptide(s) of interest have been isolated from culture it may be desired or necessary to concentrate the polypeptide(s). A number of methods for concentrating proteins are known in the art, such as ultrafiltration or lyophilisation.
[0226] In some embodiments, the production of the polypeptide occurs in vivo, e.g. after introduction to the host of a cell comprising a nucleic acid or vector encoding a polypeptide of the present invention, or following introduction into a cell of the host of a nucleic acid or vector encoding a polypeptide of the present invention. In such embodiments, the polypeptide is transcribed, translated and post-translationally processed to the mature polypeptide. In some embodiments, the polypeptide is produced in situ at the desired location in the host. In some embodiments, the desired location is the eye, e.g. in a cell of the retina, choroid, retinal pigment epithelium (RPE) or macula. In some embodiments, the desired location is at or in a retinal cell. In some embodiments, the desired location is at or in a RPE cell.
[0227] Therapeutic Applications
[0228] Any of the polypeptides, nucleic acids, vectors, cells and pharmaceutical compositions according to the present invention find use in therapeutic and prophylactic methods.
[0229] The present invention provides a polypeptide, nucleic acid, vector, cell, or pharmaceutical composition according to the present invention, for use in a method of medical treatment or prophylaxis. The present invention also provides the use of a polypeptide, nucleic acid, vector, cell or pharmaceutical composition according to the present invention in the manufacture of a medicament for treating or preventing a disease or condition. The present invention also provides a method of treating or preventing a disease or condition, comprising administering to a subject a therapeutically or prophylactically effective amount of a polypeptide, nucleic acid, vector, cell or pharmaceutical composition according to the present invention.
[0230] In particular, the polypeptides, nucleic acids, vectors, cells and pharmaceutical compositions according to the present invention find use to treat or prevent diseases/conditions associated with complement dysregulation, in particular overactive complement response. In some embodiments, the overactive complement response is linked to the presence of C3b. In some embodiments the disease/condition to be treated or prevented is a complement-related disease. In some embodiments the disease/condition to be treated or prevented is pathologically associated with complement activation. In some embodiments the disease/condition to be treated or prevented is pathologically associated with complement over-activation. In some embodiments the disease/condition to be treated or prevented is driven by complement activation or over-activation. In some embodiments the disease/condition is complement activation or over-activation.
[0231] The polypeptides, nucleic acids, vectors, cells and pharmaceutical compositions find use to treat or prevent diseases/conditions which would benefit from one or more of: a reduction in the level or activity of C3bBb-type C3 convertase, C3bBb3b-type C5 convertase or C4b2a3b-type C5 convertase; a reduction in the level of C3b, C5b or C5a; an increase in the level of iC3b, C3f, C3dg or C3d; or a reduction in the level or activity of iC3b and an increase in the level of C3f, C3dg or C3d.
[0232] `Treatment` may, for example, be reduction in the development or progression of a disease/condition, alleviation of the symptoms of a disease/condition or reduction in the pathology of a disease/condition. Treatment or alleviation of a disease/condition may be effective to prevent progression of the disease/condition, e.g. to prevent worsening of the condition or to slow the rate of development. In some embodiments treatment or alleviation may lead to an improvement in the disease/condition, e.g. a reduction in the symptoms of the disease/condition or reduction in some other correlate of the severity/activity of the disease/condition. Prevention/prophylaxis of a disease/condition may refer to prevention of a worsening of the condition or prevention of the development of the disease/condition, e.g. preventing an early stage disease/condition developing to a later, chronic, stage.
[0233] In some embodiments, the disease or condition to be treated or prevented may be a disease/condition associated with C3b or a C3b-containing complex, an activity/response associated with C3b or a C3b-containing complex, or a product of an activity/response associated with C3b or a C3b-containing complex. That is, in some embodiments, the disease or condition to be treated or prevented is a disease/condition in which C3b, a C3b-containing complex, an activity/response associated with C3b or a C3b-containing complex, or the product of said activity/response is pathologically implicated. In some embodiments, the disease/condition may be associated with an increased level of C3b or a C3b-containing complex, an increased level of an activity/response associated with C3b or a C3b-containing complex, or increased level of a product of an activity/response associated with C3b or a C3b-containing complex as compared to the control state.
[0234] The treatment may be aimed at reducing the level of C3b or a C3b-containing complex, an activity/response associated with C3b or a C3b-containing complex, or a product of an activity/response associated with C3b or a C3b-containing complex. In some embodiments, the treatment is aimed at: reducing the level or activity of C3bBb-type C3 convertase, C3bBb3b-type C5 convertase or C4b2a3b-type C5 convertase; reducing the level of C3b, C5b or C5a; increasing the level of iC3b, C3f, C3dg or C3d, or reducing the level of iC3b and increasing the level of C3f, C3dg or C3d.
[0235] Administration of the polypeptides, nucleic acids, vectors, cells and compositions of the present invention may cause a reduction in the level of C3b or a C3b-containing complex, an activity/response associated with C3b or a C3b-containing complex, or a product of an activity/response associated with C3b or a C3b-containing complex through cleavage of C3b.
[0236] In some embodiments, the treatment may be aimed at reducing the level of C3b or a C3b-containing complex, an activity/response associated with C3b or a C3b-containing complex, or a product of an activity/response associated with C3b or a C3b-containing complex in a subject, e.g. at a particular location, in a particular organ, tissue, structure or cell type. In some embodiments, the treatment may be aimed at reducing the level of C3b or a C3b-containing complex, an activity/response associated with C3b or a C3b-containing complex, or a product of an activity/response associated with C3b or a C3b-containing complex in the eye, e.g. in the retina, choroid, RPE, macula and/or at the BrM/RPE interface.
[0237] In some embodiments, the treatment may comprise modifying a cell or population of cells to comprise/express a polypeptide, nucleic acid or vector of the present invention. In some embodiments, the treatment may comprise modification of the cell/population in vivo, for in situ production of the polypeptide of the invention. In some embodiments the cell/population of cells is/are an ocular cell/cells. In some embodiments, the cell/population of cells is a RPE cell and/or population of RPE cells. In some embodiments the cell/population of cells is a photoreceptor cell and/or population of photoreceptor cells.
[0238] In some embodiments, the present invention provides a nucleic acid or vector of the present invention for use in gene therapy. In some embodiments, the treatment comprises administering the nucleic acid and/or vector to a subject. In some embodiments, the treatment comprises introducing the nucleic acid and/or vector into a cell of a subject, using techniques described herein or well known in the art, see e.g. MacLaren et al. Ophthalmology. 2016, 123(10 Suppl): S98-S106; Aguirre, Invest Ophthalmol Vis Sci. 2017, 58(12): 5399-5411; Lundstrom, Diseases. 2018, 6(2): 42; which are hereby incorporated by reference in their entirety. In some embodiments the cell is an ocular cell or cells. In some embodiments, the cell is an RPE cell or cells. In some embodiments the cell is a photoreceptor cell or cells.
[0239] In some embodiments, the treatment may comprise administering to a subject a cell or population of cells modified to comprise/express a polypeptide, nucleic acid or vector of the present invention. In some embodiments, the treatment may comprise modification of the cell/population ex vivo or in vitro.
[0240] In some embodiments, the treatment is aimed at providing the subject with a cell or population of cells which produce and/or will produce the polypeptide of the invention, e.g. by administering a cell according to the present invention, or by generating a cell according to the present invention.
[0241] In some embodiments, the cell referred to herein is a cell of the eye i.e. an ocular cell. In some embodiments, the cell is a cell of the retina, choroid, retinal pigment epithelium (RPE) or macula. In some embodiments, the cell is a retinal cell. In some embodiments, the cell is an RPE cell. In some embodiments the cell is a photoreceptor cell.
[0242] The present invention provides a method of treating or preventing a disease or condition in a subject, the method comprising modifying at least one cell to express or comprise a polypeptide, nucleic acid or vector according to the present invention. In some embodiments the at least one cell is an ocular cell. In some embodiments, the at least one cell is an RPE cell.
[0243] The at least one cell modified according to the present invention can be modified according to methods well known to the skilled person. The modification may comprise nucleic acid transfer for permanent or transient expression of the transferred nucleic acid. Any suitable genetic engineering platform may be used to modify a cell according to the present invention. Suitable methods for modifying a cell include the use of genetic engineering platforms such as gammaretroviral vectors, lentiviral vectors, adenovirus vectors, adeno-associated virus (AAV) vectors, DNA transfection, transposon-based gene delivery and RNA transfection, for example as described in Maus et al., Annu Rev Immunol (2014) 32:189-225, incorporated by reference hereinabove.
[0244] The subject to be treated may be any animal or human. The subject is preferably mammalian, more preferably human. The subject may be a non-human mammal, but is more preferably human. The subject may be male or female. The subject may be a patient. A subject may have been diagnosed with a disease or condition requiring treatment, or be suspected of having such a disease or condition.
[0245] The subject to be treated may display an elevated level of C3b or a C3b-containing complex, an activity/response associated with C3b or a C3b-containing complex, or a product of an activity/response associated with C3b or a C3b-containing complex, e.g. as determined by analysis of the subject, or a sample (e.g. a cell, tissue, blood sample) obtained from the subject, using an appropriate assay.
[0246] The subject may have an increased level of expression or activity of a positive regulator/effector of C3b or a C3b-containing complex or of an activity/response associated with C3b or a C3b-containing complex, or may have an increased level of expression or activity of a product of an activity/response associated with C3b or a C3b-containing complex. The subject may have an increased level of an activity upregulated by C3b or a C3b-containing complex.
[0247] The subject may have a reduced level of expression or activity of a negative regulator of C3b or a C3b-containing complex or of an activity/response associated with C3b or a C3b-containing complex, or may have a reduced level of expression or activity a factor downregulated by C3b or a C3b-containing complex. The subject may have a reduced level of an activity downregulated by C3b or a C3b-containing complex.
[0248] The increase/reduction may be relative to the level of expression/activity in the absence of the relevant disease/condition, e.g. the level of expression/activity in a healthy control subject or sample obtained from a healthy control subject.
[0249] In some embodiments, the subject may be at risk of developing/contracting a disease or condition. In some embodiments, the subject may possess one or more predisposing factors increasing risk of developing/contracting a disease or condition.
[0250] In some embodiments, the subject may possess one or more risk factors for Age-related Macular Degeneration (AMD). In some embodiments, the subject may possess one or more of AMD-associated genetic variants. AMD-associated genetic variants are described e.g. in Clark et al., J Clin Med (2015) 4(1):18-31, which is hereby incorporated by reference in its entirety. In some embodiments, the subject may possess one or more of the following AMD-associated genetic variants (or a variant having LD=r.sup.2.gtoreq.0.8 with such variant): Y402H in CFH (i.e. rs1061170.sup.c), rs1410996c, 162V in CFH, R53C in CFH, D90G in CFH, R1210C in CFH, or rs6685931T in CFHR4.
[0251] In some embodiments the subject may possess one or more risk factors for early-onset macular degeneration (EOMD). EOMD is thought to be caused by monogenic inheritance of rare variants of the CFH gene (see e.g. Boon C J et al. Am J Hum Genet 2008; 82(2):516-23; van de Ven J P, et al. Arch Ophthalmol 2012; 130(8):1038-47; Yu Y et al. Hum Mol Genet 2014; 23(19):5283-93; Duvvari M R, et al. Mol Vis 2015; 21:285-92; Hughes A E, et al. Acta Ophthalmol 2016; 94(3):e247-8; Wagner et al. Sci Rep 2016; 6:31531). In some embodiments, the subject may possess one or more of EOMD-associated genetic variants. EOMD-associated genetic variants are described in e.g. Servais A et al. Kidney Int, 2012; 82(4):454-64 and Dragon-Durey M A, et al. J Am Soc Nephrol 2004; 15(3):787-95; which are hereby incorporated by reference in their entirety. In some embodiments, the subject may possess one or more of the following EOMD-associated genetic variants: CFH c.1243del, p.(Ala415Profs*39) het; CFH c.350+1G>T het; CFH c.619+1G>A het; CFH c.380G>A, p.(Arg127His); CFH c.694C>T, p.(Arg232Ter); or CFH c.1291T>A, p.(Cys431Ser).
[0252] In some embodiments, the subject is selected for therapeutic or prophylactic treatment with the polypeptide, nucleic acid, vector, cell or composition of the present invention based on their being determined to possess one or more risk factors for AMD and/or EOMD, e.g. one or more AMD/EOMD-associated genetic variants. In some embodiments, the subject has been determined to have one or more such risk factors. In some embodiments, the methods of the present invention involving determining whether a subject possesses one or more such risk factors.
[0253] In some embodiments, the disease or condition to be treated or prevented may be an ocular disease/condition. In some embodiments, the disease or condition to be treated or prevented is a complement-related ocular disease. In some embodiments, the disease or condition to be treated or prevented is macular degeneration. In some embodiments, the disease or condition to be treated or prevented is age-related macular degeneration (AMD). AMD is commonly-defined as causing vision loss in subjects age 50 and older.
[0254] In some embodiments, the disease or condition to be treated or prevented is selected from age-related macular degeneration (AMD), early AMD, intermediate AMD, late AMD, geographic atrophy (`dry` (i.e. non-exudative) AMD), `wet` (neovascular or exudative) AMD, choroidal neovascularisation (CNV), glaucoma, autoimmune uveitis, and diabetic retinopathy. In some embodiments, the disease or condition to be treated or prevented is AMD. In some embodiments, the disease or condition to be treated or prevented is geographic atrophy (`dry` AMD). In some embodiments, the disease or condition to be treated or prevented is `wet` AMD. In some embodiments the disease or condition to be treated or prevented is a combination of the diseases/conditions above, e.g. `dry` and `wet` AMD. In some embodiments the disease or condition to be treated or prevented is not `wet` AMD or choroidal neovascularisation. In some embodiments a subject to be treated is age 50 or older, i.e. is at least 50 years old.
[0255] As used herein "early AMD" refers to a stage of AMD characterised by the presence of medium-sized drusen, commonly having a width of up to .about.200 .mu.m, within the Bruch's membrane adjacent to the RPE layer. Subjects with early AMD typically do not present significant vision loss. As used herein "intermediate AMD" refers to a stage of AMD characterised by large drusen and/or pigment changes in the retina. Intermediate AMD may be accompanied by some vision loss. As used herein "late AMD" refers to a stage of AMD characterised by the presence of drusen and vision loss due to damage to the macula. In all stages of AMD, `reticular pseudodrusen` (RPD) or `reticular drusen` may be present, referring to the accumulation of extracellular material in the subretinal space between the neurosensory retina and RPE. "Late AMD" encompasses `dry` and `wet` AMD. In `dry` AMD (also known as geographic atrophy), there is a gradual breakdown of the light-sensitive cells in the macula that convey visual information to the brain and of the supporting tissue beneath the macula. In `wet` AMD (also known as choroidal neovascularization and exudative AMD), abnormal blood vessels grow underneath and into the retina. These vessels can leak fluid and blood which can lead to swelling and damage of the macula and subsequent scar formation. The damage may be rapid and severe.
[0256] In some embodiments the disease or condition to be treated or prevented is early-onset macular degeneration (EOMD). As used herein "EOMD" refers to a phenotypically severe sub-type of macular degeneration that demonstrates a much earlier age of onset than classical AMD and results in many more years of substantial visual loss. The EOMD subset is described in e.g. Boon C J et al. Am J Hum Genet 2008; 82(2):516-23 and van de Ven J P, et al. Arch Ophthalmol 2012; 130(8):1038-47. In some embodiments a subject to be treated is age 49 or younger. In some embodiments a subject to be treated is between ages 15 and 49, i.e. is between 15 and 49 years old.
[0257] In some embodiments, the disease or condition to be treated or prevented is a disease/condition driven by complement over-activation. In some embodiments, the disease or condition to be treated or prevented may be selected from atypical Haemolytic Uremic Syndrome (aHUS), Membranoproliferative Glomerulonephritis Type II (MPGN II), sepsis, and Paroxysmal nocturnal hemoglobinuria (PNH).
[0258] Methods of medical treatment may also involve in vivo, ex vivo, and adoptive immunotherapies, including those using autologous and/or heterologous cells or immortalized cell lines.
[0259] Administration of a polypeptide described herein is preferably in a "therapeutically effective amount", this being sufficient to show benefit to the individual. The actual amount administered, and rate and time-course of administration, will depend on the nature and severity of the disease being treated. Prescription of treatment, e.g. decisions on dosage etc., is within the responsibility of general practitioners and other medical doctors, and typically takes account of the condition to be treated, the condition of the individual patient, the site of delivery, the method of administration and other factors known to practitioners. Examples of the techniques and protocols mentioned above can be found in Remington's Pharmaceutical Sciences, 20th Edition, 2000, pub. Lippincott, Williams & Wilkins.
[0260] Polypeptides, nucleic acids, vectors and cells according to the present invention may be formulated as pharmaceutical compositions or medicaments for clinical use and may comprise a pharmaceutically acceptable carrier, diluent, excipient or adjuvant. The composition may be formulated for topical, parenteral, systemic, intracavitary, intravenous, intra-arterial, intramuscular, intrathecal, intraocular, intraconjunctival, subretinal, suprachoroidal, subcutaneous, intradermal, intrathecal, oral or transdermal routes of administration which may include injection or infusion, or administration as an eye drop (i.e. ophthalmic administration). Suitable formulations may comprise the polypeptide, nucleic acid, vector, or cell in a sterile or isotonic medium. Medicaments and pharmaceutical compositions may be formulated in fluid, including gel, form. Fluid formulations may be formulated for administration by injection or infusion (e.g. via catheter) to a selected organ or region of the human or animal body. In some embodiments the polypeptides, nucleic acids, vector, cells and compositions of the invention are formulated for intravitreal routes of administration e.g. by intravitreal injection. In some embodiments polypeptides, nucleic acids, vector, cells and compositions of the invention are formulated for submacular delivery i.e. placing the therapeutic molecules in direct contact with the target cell layers.
[0261] The particular mode and/or site of administration may be selected in accordance with the location where the C3b inactivation is desired. In some embodiments, the polypeptides, nucleic acids, vectors, or pharmaceutical compositions of the present invention are formulated for administration and/or administered into the subretinal space between the photoreceptor cells and the retinal pigment epithelium (RPE) in the eye. In some embodiments, the polypeptides, nucleic acids, vectors, or pharmaceutical compositions of the present invention are formulated for administration and/or administered into the retinal pigment epithelium (RPE).
[0262] In accordance with the present invention methods are also provided for the production of pharmaceutically useful compositions, such methods of production may comprise one or more steps selected from: isolating a polypeptide, nucleic acid, vector, or cell as described herein; and/or mixing a polypeptide, nucleic acid, vector, or cell as described herein with a pharmaceutically acceptable carrier, adjuvant, excipient or diluent.
[0263] For example, a further aspect of the present invention relates to a method of formulating or producing a medicament or pharmaceutical composition for use in a method of medical treatment, the method comprising formulating a pharmaceutical composition or medicament by mixing polypeptide, nucleic acid, vector, or cell as described herein with a pharmaceutically acceptable carrier, adjuvant, excipient or diluent.
[0264] Administration may be alone or in combination with other treatments (e.g. other therapeutic or prophylactic intervention), either simultaneously or sequentially dependent upon the condition to be treated. The polypeptide, nucleic acid, vector, cell or composition according to the present invention and a therapeutic agent may be administered simultaneously or sequentially.
[0265] Simultaneous administration refers to administration of the polypeptide, nucleic acid, vector, cell or composition and therapeutic agent together, for example as a pharmaceutical composition containing both agents (combined preparation), or immediately after each other and optionally via the same route of administration, e.g. to the same tissue, artery, vein or other blood vessel. Sequential administration refers to administration of one of the polypeptide, nucleic acid, vector, cell or composition or therapeutic agent followed after a given time interval by separate administration of the other agent. It is not required that the two agents are administered by the same route, although this is the case in some embodiments. The time interval may be any time interval. In some embodiments, the polypeptide, nucleic acid, vector, cell or composition and therapeutic agent are administered separately, simultaneously or sequentially to the eye.
[0266] In some embodiments, the other treatment/therapeutic agent is a therapeutically effective amount of Complement Factor I. In some embodiments, Complement Factor I is administered to the subject simultaneously or sequentially with administration of a polypeptide, nucleic acid, vector, cell, or pharmaceutical composition according to the present invention. In some embodiments, the treatment may comprise modifying a cell or population of cells in vitro, ex vivo or in vivo to express and/or secrete Complement Factor I. The cell or population of cells may be the same cell or population of cells as a cell or population of cells modified to comprise/express a polypeptide, nucleic acid or vector according to the present invention, for example the treatment may comprise modifying a cell or population of cells in vitro, ex vivo or in vivo to express and/or secrete a polypeptide, nucleic acid or vector according to the present invention, and Complement Factor I. In some embodiments, Complement Factor I is administered to a subject, wherein the subject comprises a cell or population of cells modified to comprise/express a polypeptide, nucleic acid or vector of the present invention. In some embodiments, Complement Factor I is administered to a subject wherein the subject has expressed in situ or is expressing in situ a polypeptide, nucleic acid or vector of the present invention.
[0267] Complement Factor I, or a composition comprising Complement Factor I, may be formulated for topical, parenteral, systemic, intracavitary, intravenous, intravitreal, intra-arterial, intramuscular, intrathecal, intraocular, intraconjunctival, subretinal, suprachoroidal, subcutaneous, intradermal, intrathecal, oral or transdermal routes of administration which may include injection or infusion, or administration as an eye drop (i.e. ophthalmic administration). Suitable formulations may comprise a sterile or isotonic medium. Medicaments and pharmaceutical compositions may be formulated in fluid, including gel, form. Fluid formulations may be formulated for administration by injection or infusion (e.g. via catheter) to a selected organ or region of the human or animal body.
[0268] In some embodiments, the other treatment/therapeutic agent is a therapeutically effective amount of an anti-VEGF therapy (e.g. ranibizumab (Lucentis; Genentech/Novartis), bevacizumab (off label Avastin; Genentech), aflibercept (Eylea/VEGF Trap-Eye; Regeneron/Bayer)), pegaptanib (Macugen), laser photocoagulation, or photodynamic therapy (PDT) e.g. with Visudyne.TM. (verteporfin).
[0269] Multiple doses of the polypeptide, nucleic acid, vector, cell or composition may be provided. One or more, or each, of the doses may be accompanied by simultaneous or sequential administration of another therapeutic agent.
[0270] Multiple doses may be separated by a predetermined time interval, which may be selected to be one of 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, or 31 days, or 1, 2, 3, 4, 5, or 6 months. By way of example, doses may be given once every 7, 14, 21 or 28 days (plus or minus 3, 2, or 1 days).
[0271] A polypeptide, nucleic acid, vector or composition according to the present invention may be formulated in a sustained release delivery system, in order to release the polypeptide, nucleic acid, vector or composition at a predetermined rate. Sustained release delivery systems may maintain a constant drug/therapeutic concentration for a specified period of time. In some embodiments, a polypeptide, nucleic acid, vector or composition according to the present invention is formulated in a liposome, gel, implant, device, or drug-polymer conjugate e.g. hydrogel.
[0272] Genetic Factors in Age-Related Macular Degeneration (AMD)
[0273] Complement Factor H (encoded by the CFH gene) is another co-factor for Complement Factor I. Complement Factor H structure and function is reviewed e.g. in Wu et al., Nat Immunol (2009) 10(7): 728-733, which is hereby incorporated by reference in its entirety. Human Complement Factor H (UniProt: P08603; SEQ ID NO:29) has a 1,233 amino acid sequence (including an N-terminal, 18 amino acid signal peptide), and comprises 20 complement control protein (CCP) domains. The first four CCP domains (i.e. CCP1 to CCP4) of Complement Factor H are necessary for Complement Factor I co-factor activity for cleavage of C3b to iC3b. CCPs 19 to 20 have also been shown to engage with C3b and C3d (Morgan et al., Nat Struct Mol Biol (2011) 18(4): 463-470), whilst CCP7 and CCPs 19 to 20 bind to glycosaminoglycans (GAGs) and sialic acid, and are involved in discrimination between self and non-self (Schmidt et al., J Immunol (2008) 181(4): 2610-2619; Kajander et al., PNAS (2011) 108(7): 2897-2902).
[0274] One of the major SNPs associated with genetic risk of developing AMD is found in the CFH gene and leads to the Y402H polymorphism in Complement Factor H (see e.g. Haines et al., Science (2005) 308:419-21), and it's alternative splice variant factor H-like protein 1 (FHL-1). Around 30% of individuals of white European heritage have at least one copy of this polymorphism, whilst being a heterozygote increases the risk of AMD by .about.3-fold (Sofat et al., Int J Epidemiol (2012) 41:250-262). The Y402H polymorphism, which manifests in the seventh complement control protein (CCP) domain, reduces the binding of FH/FHL-1 to BrM, leading to perturbations in the binding of these blood-borne complement regulators and dampened complement regulation on this surface (Clark et al., J Biol Chem (2010) 285:30192-202).
[0275] The binding of FH/FHL-1 to BrM is mediated by sulphated sugars including the glycosaminoglycans (GAGs) heparan sulphate (HS) and dermatan sulphate (DS). The family of GAG sequences found in BrM appears to have greater tissue specificity than previously thought, as they are able to recruit FH/FHL-1 through their CCP7 domains and not FH's secondary anchoring site found in CCPs19-20 (Clark et al., J Immunol (2013) 190:2049-2057). This is likely to be an evolutionary twist, as it has been discovered that the main regulator of complement within BrM is the truncated FHL-1 protein (Clark et al J. Immunol (2014) 193, 4962-4970), which only has the one surface anchoring site in CCP7 and lacks CCPs19-20. In contrast, the Y402H polymorphism is not associated with kidney disease where the CCP19-20 domain of FH is known to be the main GAG-mediated anchoring site (Clark et al., J Immunol (2013) 190:2049-2057). Age-related changes in the BrM expression levels of HS and DS, themselves considered part of the normal ageing process, have also been associated with AMD, and may go some way as to explain the age-related nature of the genetically driven AMD.
[0276] A rare mutation (R1210C) in the C-terminal CCP19-20 region of FH, which does not bind to BrM, has a very high level of association with AMD, and FH protein carrying this mutation is found covalently bound to albumin (Senchez-Corral et al., Am J Hum Genet (2002) 71:1285-1295) preventing the FH protein from leaving the circulation and entering eye tissue. Some research suggests that the large confluent drusen that precede geographic atrophy and the associated pigmentary changes in the RPE indicate that dry AMD results firstly from dysfunction of the RPE with secondary effects within the choroid (Bhutto and Lutty Mol Aspects Med (2012) 33:295-317). In contrast, Whitmore et al. reported changes in the choriocapillaris preceding all forms of late-stage AMD including the deposition of the terminal complement membrane attack complex (MAC), and argue that excessive complement activation in the choriocapillaris is the primary event with RPE atrophy being secondary (Whitmore et al., Prog Retin Eye Res (2015) 45:1-29). These data imply that a genetic predisposition conferred by alterations in complement genes is tolerated until changes in both BrM and the underlying choriocapillaris come to the fore. Whether these changes are age-related, driven by oxidative stress or a result of RPE cell dysfunction remains to be seen, but naturally occurring changes in these structures are known to be age-related.
[0277] C3 and C3b
[0278] Complement component 3 (C3) is an immune system protein having a central role in innate immunity and the complement system. Processing of C3 is described, for example, in Foley et al. J Thromb Haemostasis (2015) 13: 610-618, which is hereby incorporated by reference in its entirety. Human C3 (UniProt: P01024; SEQ ID NO:18) comprises a 1,663 amino acid sequence (including an N-terminal, 22 amino acid signal peptide). Amino acids 23 to 667 encode C3 .beta. chain (SEQ ID NO:19), and amino acids 749 to 1,663 encode C3 .alpha.' chain (SEQ ID NO:20). C3 .beta. chain and C3 .alpha.' chain associate through interchain disulphide bonds (formed between cysteine 559 of C3 .beta. chain, and cysteine 816 of the C3 .alpha.' chain) to form C3b. C3a is a 77 amino acid fragment corresponding to amino acid positions 672 to 748 of C3 (SEQ ID NO:21), generated by proteolytic cleavage of C3 following activation through the classical complement pathway and the lectin pathways.
[0279] C3b is a potent opsonin, targeting pathogens, antibody-antigen immune complexes and apoptotic cells for phagocytosis by phagocytes and NK cells. C3b is also involved in the formation of convertase enzyme complexes for activating and amplifying complement responses. C3b associates with Factor B to form the C3bBb-type C3 convertase (alternative complement pathway), and can associate with C4b and C2a to form the C4b2a3b-type C5 convertase (classical pathway), or with C3bBb to form the C3bBb3b-type C5 convertase (alternative pathway).
[0280] Processing of C3b to the form iC3b, which is proteolytically inactive and which cannot itself promote further complement amplification, involves proteolytic cleavage of the C3b .alpha.' chain at amino acid positions 1303 and 1320 to form an .alpha.' chain fragment 1 (corresponding to amino acid positions 672 to 748 of C3; SEQ ID NO:22), an .alpha.' chain fragment 2 (corresponding to amino acid positions 1321 to 1,663 of C3; SEQ ID NO:23). Thus, iC3b comprises the C3 .beta. chain, C3 .alpha.' chain fragment 1 and C3 .alpha.' chain fragment 2 (associated via disulphide bonds). Cleavage of the .alpha.' chain also liberates C3f, which corresponds to amino acid positions 1304 to 1320 of C3 (SEQ ID NO:24).
[0281] As used herein "C3" refers to C3 from any species and include isoforms, fragments, variants or homologues of C3 from any species. In some embodiments, the C3 is mammalian C3 (e.g. cynomolgous, human and/or rodent (e.g. rat and/or murine) C3). Isoforms, fragments, variants or homologues of C3 may optionally be characterised as having at least 70%, preferably one of 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% amino acid sequence identity to the amino acid sequence of immature or mature C3 from a given species, e.g. human C3 (SEQ ID NO:18).
[0282] As used herein "C3b" refers to and includes isoforms, fragments, variants or homologues of C3b from any species. In some embodiments, the C3b is mammalian C3b (e.g. cynomolgous, human and/or rodent (e.g. rat and/or murine) C3b).
[0283] Isoforms, fragments, variants or homologues of C3b may optionally be characterised as comprising a C3 .alpha.' chain fragment 1, C3 .alpha.' chain fragment 2 and a C3 .beta. having at least 70%, preferably one of 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% amino acid sequence identity to the amino acid sequences of the respective polypeptides from a given species, e.g. human. That is, the C3b may comprise: a C3 .alpha.' chain fragment 1 having at least 70%, preferably one of 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% amino acid sequence identity to SEQ ID NO:22; a C3 .alpha.' chain fragment 2 having at least 70%, preferably one of 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% amino acid sequence identity to SEQ ID NO:23; and a C3 .beta. chain having at least 70%, preferably one of 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% amino acid sequence identity to SEQ ID NO:19.
[0284] Isoforms, fragments, variants or homologues of C3b may optionally be functional isoforms, fragments, variants or homologues, e.g. having a functional property/activity of the reference C3b, as determined by analysis by a suitable assay for the functional property/activity. For example, Isoforms, fragments, variants or homologues of C3b may be characterised by the ability to act as an opsonin, and/or to form functional C3/C5 convertase.
[0285] Complement Factor I
[0286] Processing of C3b to iC3b is performed by Complement Factor I (encoded in humans by the gene CF). Human Complement Factor I (UniProt: P05156; SEQ ID NO:25) has a 583 amino acid sequence (including an N-terminal, 18 amino acid signal peptide). The precursor polypeptide is cleaved by furin to yield the mature Complement Factor I, comprising a heavy chain (amino acids 19 to 335), and light chain (amino acids 340 to 583) linked by interchain disulphide bonds. Amino acids 340 to 574 of the light chain encode the proteolytic domain of Complement Factor I (SEQ ID NO:26), which is a serine protease containing the catalytic triad responsible for cleaving C3b to produce iC3b (Ekdahl et al., J Immunol (1990) 144 (11): 4269-74).
[0287] As used herein "Complement Factor I (FI)" refers to Complement Factor I from any species and includes isoforms, fragments, variants or homologues of Complement Factor I from any species. In some embodiments, the Complement Factor I is mammalian Complement Factor I (e.g. cynomolgous, human and/or rodent (e.g. rat and/or murine) Complement Factor I).
[0288] Isoforms, fragments, variants or homologues of Complement Factor I may optionally be characterised as having at least 70%, preferably one of 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% amino acid sequence identity to the amino acid sequence of immature or mature Complement Factor I from a given species, e.g. human Complement Factor I (SEQ ID NO:25). Isoforms, fragments, variants or homologues of Complement Factor I may optionally be functional isoforms, fragments, variants or homologues, e.g. having a functional property/activity of the reference Complement Factor I (e.g. full-length human Complement Factor I), as determined by analysis by a suitable assay for the functional property/activity. For example, an isoform, fragment, variant or homologue of Complement Factor I may display serine protease activity and/or may be capable of inactivating C3b.
[0289] Proteolytic cleavage of C3b by Complement Factor I to yield iC3b is facilitated by co-factors for Complement Factor I. Co-factors for Complement Factor I typically bind to C3b and/or Complement Factor I, and potentiate processing of C3b to iC3b by Complement Factor I.
[0290] Complement Receptor 1
[0291] Complement Receptor 1 (CR1) acts as a cofactor for Complement Factor I, enabling cleavage of C3b to iC3b and downstream products.
[0292] iC3b does not amplify or activate the complement system, but it can still act as an opsonin to target pathogens for phagocytosis. iC3b production therefore results in local immune system activation and inflammatory effects. This can have negative consequences for sufferers of complement-related disease and may contribute to the development or worsening of an existing complement-related disease/condition.
[0293] Factor H (FH) and truncated FH isoform FHL-1 act as co-factors for FI to produce iC3b, but they cannot promote further degradation of iC3b which can lead to undesirable iC3b accumulation. In addition, the accumulation of iC3b may contribute to further debris in the affected area, leading to e.g. the (further) development of drusen in macular degeneration.
[0294] In contrast, CR1 and the polypeptides according to the present invention, see e.g. FIGS. 2, 3B, 7A, can act in combination with FI to promote further breakdown of iC3b into advantageous downstream products such as C3c, C3dg and C3b. These molecules are not opsonins and thus avoid recruiting immune system components. Their presence in an affected area is preferable to iC3b accumulation. As used herein, "Complement Receptor 1 (CR1)" refers to CR1 from any species and includes isoforms, fragments, variants or homologues of CR1 from any species. In some embodiments, the CR1 is mammalian CR1 (e.g. cynomolgous, human and/or rodent (e.g. rat and/or murine) CR1).
[0295] Aspects and embodiments of the present invention will now be illustrated, by way of example, with reference to the accompanying figures. Further aspects and embodiments will be apparent to those skilled in the art. All documents mentioned in this text are incorporated herein by reference.
[0296] The invention includes the combination of the aspects and preferred features described except where such a combination is clearly impermissible or expressly avoided.
[0297] The section headings used herein are for organizational purposes only and are not to be construed as limiting the subject matter described.
[0298] The features disclosed in the foregoing description, or in the following claims, or in the accompanying drawings, expressed in their specific forms or in terms of a means for performing the disclosed function, or a method or process for obtaining the disclosed results, as appropriate, may, separately, or in any combination of such features, be utilised for realising the invention in diverse forms thereof.
[0299] While the invention has been described in conjunction with the exemplary embodiments described above, many equivalent modifications and variations will be apparent to those skilled in the art when given this disclosure. Accordingly, the exemplary embodiments of the invention set forth above are considered to be illustrative and not limiting. Various changes to the described embodiments may be made without departing from the spirit and scope of the invention.
[0300] For the avoidance of any doubt, any theoretical explanations provided herein are provided for the purposes of improving the understanding of a reader. The inventors do not wish to be bound by any of these theoretical explanations.
[0301] Throughout this specification, including the claims which follow, unless the context requires otherwise, the word "comprise," and variations such as "comprises" and "comprising," will be understood to imply the inclusion of a stated integer or step or group of integers or steps but not the exclusion of any other integer or step or group of integers or steps.
[0302] It must be noted that, as used in the specification and the appended claims, the singular forms "a," "an," and "the" include plural referents unless the context clearly dictates otherwise. Ranges may be expressed herein as from "about" one particular value, and/or to "about" another particular value. When such a range is expressed, another embodiment includes from the one particular value and/or to the other particular value. Similarly, when values are expressed as approximations, by the use of the antecedent "about," it will be understood that the particular value forms another embodiment.
[0303] Where a nucleic acid sequence in disclosed the reverse complement thereof is also expressly contemplated.
[0304] The following numbered paragraphs (paras) describe particular aspects and embodiments of the present invention:
[0305] 1. A polypeptide having at least 80% sequence identity to SEQ ID NO:4, wherein the polypeptide has a length of 700 amino acids or fewer.
[0306] 2. The polypeptide according to para 1, wherein the polypeptide has a length of 50 to 700 amino acids.
[0307] 3. The polypeptide according to para 1 or 2, wherein X.sub.1 is A or T, X.sub.2 is P or L, and/or X.sub.3 is G or R.
[0308] 4. The polypeptide according to any one of paras 1 to 3, comprising SEQ ID NO:2, SEQ ID NO:3 or SEQ ID NO:13.
[0309] 5. The polypeptide according to any one of paras 1 to 4, consisting of SEQ ID NO:2, SEQ ID NO:3, or SEQ ID NO:13.
[0310] 6. The polypeptide according to any one of paras 1 to 5, which is capable of binding to C3b.
[0311] 7. The polypeptide according to any one of paras 1 to 6, which binds to C3b in the region bound by a co-factor for Complement Factor I.
[0312] 8. The polypeptide according to any one of paras 1 to 7, which binds to C3b in the region bound by Complement Receptor 1 (CR1).
[0313] 9. The polypeptide according to any one of paras 1 to 8, which acts as a co-factor for Complement Factor I.
[0314] 10. The polypeptide according to any one of paras 1 to 9, which is capable of diffusing across Bruch's membrane (BrM).
[0315] 11. The polypeptide according to any one of paras 1 to 10, which is not glycosylated or is partially glycosylated.
[0316] 12. The polypeptide according to any one of paras 1 to 11, wherein the amino acid sequence comprises one or more amino acid substitutions at position 509, 578, 959 and/or 1028 (numbered according to Uniprot: P17927).
[0317] 13. The polypeptide according to para 12, wherein the one or more amino acid substitutions are selected from N509Q, N578Q, N959Q and/or N1028Q (numbered according to Uniprot: P17927).
[0318] 14. The polypeptide according to any one of paras 1 to 13, comprising, or consisting, of SEQ ID NO:5, SEQ ID NO:6, and/or SEQ ID NO:15.
[0319] 15. The polypeptide according to any one of paras 1 to 14, additionally comprising a secretory pathway sequence.
[0320] 16. The polypeptide according to para 15, wherein the secretory pathway sequence comprises SEQ ID NO:7.
[0321] 17. The polypeptide according to para 15 or para 16, wherein the polypeptide additionally comprises a cleavage site for removing the secretory pathway sequence.
[0322] 18. A nucleic acid encoding the polypeptide according to any one of paras 1 to 17.
[0323] 19. A vector comprising the nucleic acid of para 18.
[0324] 20. A cell comprising the polypeptide according to any one of paras 1 to 17, the nucleic acid according to para 18, or the vector according to para 19.
[0325] 21. A method for producing a polypeptide, comprising introducing into a cell a nucleic acid according to para 18 or a vector according to para 19, and culturing the cell under conditions suitable for expression of the polypeptide.
[0326] 22. A cell which is obtained or obtainable by the method according to para 21.
[0327] 23. A pharmaceutical composition comprising the polypeptide according to any one of paras 1 to 17, the nucleic acid according to para 18, the vector according to para 19, or the cell according to para 20 or 22, optionally comprising a pharmaceutically acceptable carrier, adjuvant, excipient, or diluent.
[0328] 24. The polypeptide according to any one of paras 1 to 17, the nucleic acid according to para 18, the vector according to para 19, the cell according to para 20 or 22, or the pharmaceutical composition according to para 23, for use in a method of treating or preventing a disease or condition.
[0329] 25. Use of the polypeptide according to any one of paras 1 to 17, the nucleic acid according to para 18, the vector according to para 19, the cell according to para 20 or 22, or the pharmaceutical composition according to para 23, in the manufacture of a medicament for treating or preventing a disease or condition.
[0330] 26. A method of treating or preventing a disease or condition, comprising administering to a subject the polypeptide according to any one of paras 1 to 17, the nucleic acid according to para 18, the vector according to para 19, the cell according to para 20 or 22, or the pharmaceutical composition according to para 23.
[0331] 27. A method of treating or preventing a disease or condition in a subject, comprising modifying at least one cell of the subject to express or comprise a polypeptide according to any one of paras 1 to 17, a nucleic acid according to para 18, or a vector according to para 19.
[0332] 28. The polypeptide, nucleic acid, vector, cell, or pharmaceutical composition for use according to para 24, the use according to para 25, or the method according to para 26 or para 27, wherein the disease or condition is a disease or condition in which C3b or a C3b-containing complex, an activity/response associated with C3b or a C3b-containing complex, or a product of an activity/response associated with C3b or a C3b-containing complex is pathologically implicated.
[0333] 29. The polypeptide, nucleic acid, vector, cell, or pharmaceutical composition for use, the use, or the method according to any one of paras 24 to 28, wherein the disease or condition is age-related macular degeneration (AMD).
[0334] 30. The polypeptide, nucleic acid, vector, cell, or pharmaceutical composition for use, the use, or the method according to any one of paras 24 to 29, wherein the method for treating or preventing a disease or condition comprises modifying at least one retinal pigment epithelial (RPE) cell of the subject to express or comprise a polypeptide according to any one of paras 1 to 17, a nucleic acid according to para 18, or a vector according to para 19.
[0335] 31. A kit of parts comprising a predetermined quantity of the polypeptide according to any one of paras 1 to 17, the nucleic acid according to para 18, the vector according to para 19, the cell according to para 20 or para 22 or the pharmaceutical composition according to para 23.
BRIEF DESCRIPTION OF THE FIGURES
[0336] Embodiments and experiments illustrating the principles of the invention will now be discussed with reference to the accompanying figures.
[0337] FIGS. 1A and 1B. CR1a and nCR1a protein expression from human HEK293 cells. (1A) CR1a is expressed as two glyco-forms that are reduced to one lower MW band after deglycosylation treatment with PNGase. (1B) nCR1a is expressed as single form of same MW as deglycosylated CR1a.
[0338] FIG. 2. The ability of CR1a and nCR1a expressed and secreted by human cells to act as cofactors for Factor I-mediated breakdown of C3b. CR1a/nCR1a+FI+C3b reactions produced iC3b and C3dg (lanes 7 and 11).
[0339] FIGS. 3A to 3C. (3A) Presence of CR1a in the diffusion chamber of an Ussing chamber after 24 hours, showing ability to diffuse through Bruch's membrane (BrM). (3B, 3C) The ability of (3B) CR1a and (3C) nCR1a to act as cofactors for Factor I to break down C3b after polypeptide diffusion through Bruch's membrane (BrM).
[0340] FIG. 4. Schematic of an Ussing chamber used in diffusion experiments, in which (a) is enriched Bruch's membrane from human donor eyes, covering a 5 mm aperture and representing the only passage of liquid from one chamber to another; (b) are sampling access points; and (c) are magnetic stirrer bars.
[0341] FIG. 5. Binding kinetics of CR1a, FH and FHL-1 for C3b measured by Biolayer interferometry (BLI).
[0342] FIG. 6. Detection of secreted CR1a (AAV-CR1a media) from the tissue culture media of AAV-CR1a transduced ARPE-19 cells using anti-CR1a antibody. Recombinant CR1a protein was used as a positive control. Media alone and media from culture of AAV-GFP transduced ARPE-19 cells were used as negative controls.
[0343] FIGS. 7A and 7B. Ability of secreted CR1a from human ARPE-19 cells to act as a cofactor for Factor I-mediated breakdown of C3b. (7A) Secreted CR1a acts as FI cofactor to produce iC3b (product e) and C3dg (product 0. CR1a secreted from human HEK293 cells was provided as a control. (7B) Cultured AAV-CR1a transduced RPE cells showed increased ability to break down C3b (product a) to iC3b (product b) compared to media containing non-transduced cells.
[0344] FIG. 8. Schematic of the macular region of the eye, showing photoreceptors, retinal pigment epithelium, Bruch's membrane, and the choriocapillaris and intercapillary septa in the choroid.
[0345] FIG. 9. Representative fluorescence microscopy image of retinal tissue from Forest et al. (2015) Dis. Mod. Mech. 8, 421-427 (FIG. 1). Shown are drusen deposits underlying degraded RPE cells and areas of complement activation. Scale bar 20 .mu.m.
[0346] FIG. 10. Schematic of the macular region of the eye showing key stages of localised expression of effective complement therapeutic.
[0347] FIG. 11. Example of an expression vector comprising nucleic acid encoding a polypeptide according to the present invention (e.g. CR1a), promoter elements, replication elements and selection elements.
EXAMPLES
[0348] In the following Example(s), the inventors describe the design of recombinant CR1 proteins comprising the C3b binding co-factor regions of Complement Receptor 1. Also described is the ability of these proteins to be expressed by human cells, diffuse through Bruch's membrane enriched from human donor eyes, and confer regulatory activity, i.e. facilitate the FI-mediated breakdown of C3b into iC3b and further breakdown products.
Example 1
[0349] DNA inserts encoding the amino acid sequences shown in SEQ ID NOs:2 and 5 were prepared by recombinant DNA techniques, and cloned into a vector to generate constructs for recombinant expression of CR1 peptides. The amino acid sequences and features thereof are shown below:
[0350] CR1a (SEQ ID NO:47)
[0351] MRLLAKIICLMLWAICVAGHCQAPDHFLFAKLKTQTNASDFPIGTSLKYECRPEYYGRPFSITCLDN- LV WSSPKDVCKRKSCKTPPDPVNGMVHVITDIQVGSRINYSCTTGHRLIGHSSAECILSGNAAHWSTKP PICQRIPCGLPPTIANGDFISTNRENFHYGSVVTYRCNPGSGGRKVFELVGEPSIYCTSNDDQVGIWS GPAPQCII
[0352] Signal peptide (SEQ ID NO:7); CCPs8-10 of CR1 (SEQ ID NO:2) (UniProt: P17927, residues 491-685)
[0353] nCR1a (SEQ ID NO:48)
[0354] MRLLAKIICLMLWAICVAGHCQAPDHFLFAKLKTQTQASDFPIGTSLKYECRPEYYGRPFSITCLDN- LV WSSPKDVCKRKSCKTPPDPVNGMVHVITDIQVGSRIQYSCTTGHRLIGHSSAECILSGNAAHWSTKP PICQRIPCGLPPTIANGDFISTNRENFHYGSVVTYRCNPGSGGRKVFELVGEPSIYCTSNDDQVGIWS GPAPQCII
[0355] Signal peptide (SEQ ID NO:7); CCPs8-10 of CR1 (SEQ ID NO:5) (UniProt: P17927, residues 491-685) comprising substitutions N509Q and N578Q
[0356] The 18-amino acid signal peptide is designed to be cleaved from the polypeptides upon secretion.
[0357] In some experiments, HIS-tagged CR1a and nCR1a were used, e.g. as shown in SEQ ID NO:40 and 21, respectively.
[0358] Expression of Protein from Human Cells
[0359] HEK 293T cells (7.times.10.sup.6 cell per plate) were grown in 15 cm culture plates overnight in 17 ml of Dulbecco's Modified Eagle's Medium with high glucose (DMEM, Sigma, catalogue number D469) supplemented with 10% Foetal Bovine Serum (FBS, Sigma, catalogue number F9665) in 5% CO.sub.2 incubator at 37.degree. C. Once the cells reached 60% confluence they were transiently transfected with 14.4 .mu.g plasmid expressing either CR1a (SEQ ID NO:2) or nCR1a (SEQ ID NO:5) linked to the signal peptide (SEQ ID NO:7) with 86.4 .mu.l of 7.5 mM Polyethylenimine (PEI, Polysciences, catalogue number 24765-2) 150 mM NaCl (Fisher Scientific UK Ltd, catalogue number 1073592). For the negative control 14.4 .mu.l of Tris-EDTA buffer was used instead of the plasmid DNA. Five hours after transfection, the transfection medium containing 10% FBS was replaced with 17.5 ml fresh DMEM with high glucose supplemented with 2% FBS (referred to henceforth as expression media). Expression media was collected 24, 48, 72 and 140 hours after transfection. 80 .mu.l of 0.5 M Phenylmethanesulfonyl fluoride (PMSF, Sigma, Catalogue number P7626) was added to every 100 ml of expression media collected and stored at 4.degree. C. Expression media collected after 24 hrs was used for diffusion and function studies described below.
[0360] Characterisation of Secreted Protein from Human Cells
[0361] Protein expression from human cells is shown in FIGS. 1A and 1B. Recombinantly expressed and purified CR1a protein was found to be secreted from the cells as two glyco-forms. Treatment with PNGaseF, an enzyme that removes glycosylation, reduced the two bands to one band of lower apparent molecular weight (1A). A non-glycosylated form of the protein (nCR1a) was expressed. Western blotting demonstrated that nCR1a produces a single band that migrates to the same position as the enzymatically de-glycosylated protein (1B).
[0362] C3b Breakdown Activity
[0363] CR1a and nCR1a expressed and secreted from human HEK293 cells were tested for their ability to act as cofactors for Factor I-mediated breakdown of C3b.
[0364] 1 .mu.g of recombinant CR1a or nCR1a protein was mixed with 2 .mu.g of pure C3b protein and 0.04 .mu.g of pure complement Factor I (FI; VWR International, catalogue no. 341280) for 15 minutes at 37.degree. C. FHL-1 was provided as a co-factor control for FI. CR1a/nCR1a, CR1a/nCR1a+FI, CR1a/nCR1a+C3b and C3b alone were also provided as controls. Reactions were stopped with the addition of 4.times.SDS-loading buffer and heating to 100.degree. C. for 5 minutes. Samples were then run on 4-12% NuPAGE Bis-Tris gels, run at 200V for 60 minutes. Samples were transferred onto nitrocellulose membranes at 80 mA for 1.5 hours using semi-dry transfer apparatus in transfer buffer (25 mM Tris, 192 mM glycine, 10% (v/v) Methanol). The membranes were the blocked in PBS, 10% (w/v) milk, 0.2% (w/v) BSA for 16 hours at 4.degree. C. before the addition of anti-C3b antibody (HycultBiotech, catalogue no. HM2287), at 100 .mu.g/ml, in PBS, 0.2% (v/v) Tween-20 (PBS-T) for 1 hours at room temperature. Membranes were washed 2.times.30 min in PBS-T before the addition of a 1:2500 dilution of HRP conjugated goat anti-mouse for 1 hour at room temperature, protected from light. Membranes were washed 2.times.30 min in PBS-T before the addition of SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific, catalogue no. 34080) for 3 min at room temperature. Reactive bands were detected by exposing Super RX-N X-ray film (FujiFilm, catalogue no. PPB5080) to the treated membrane for 2 min at room temperature, and developed on an automated X-ray film developer.
[0365] The results are shown in FIG. 2. Both secreted proteins were found to be able to act as cofactors for Factor I, leading to the breakdown of C3b to firstly iC3b (a.sub.1), and further to C3dg (a.sub.1-1). The C3b breakdown using CR1a/nCR1a continues further than the normal native breakdown of C3b observed using FI+FHL-1 (second lane), which only produces iC3b (a).
[0366] Ussing Chamber Diffusion Experiments and C3b Breakdown Activity
[0367] The macular region of enriched Bruch's membrane isolated from donor eyes was mounted in an Ussing chamber (Harvard Apparatus, Hamden, Conn.) as described in McHarg et al., J Vis Exp (2015) 1-7, supra. Once mounted, the 5-mm-diameter macular area was the only barrier between two identical compartments, i.e. liquid must pass through Bruch's membrane (FIG. 4). Both sides of Bruch's membrane were washed with 2 ml PBS for 5 min at room temperature. The structural integrity of Bruch's membrane was tested prior to the experiments by its ability to retain 2 ml of liquid in one chamber without leaking into the next. 2 ml of expression media containing recombinant protein (CR1a and/or nCR1a, see above) was added to a chamber (henceforth designated the sample chamber) and 2 ml of fresh PBS was added to the other chamber (henceforth referred to as the diffusate/diffusion chamber). The Ussing chamber was left at room temperature for 24 hours with gentle stirring in each compartment with magnetic stirrer bars to avoid generating gradients of diffusing protein.
[0368] CR1a was added to the sample chamber. After 24 hours, samples from each chamber (original sample chamber and diffusion chamber) were analysed for the presence of CR1a.
[0369] The results are shown in FIG. 3A. CR1a was found to be present in the diffusion chamber after 24 hours.
[0370] Separately, CR1a and nCR1a were added to sample chambers and samples from both chambers were analysed for C3b-breakdown activity. After 24 hours, 18.6 .mu.l samples were taken from each chamber and mixed with 1 .mu.l (1 .mu.g) of pure C3b protein and 0.4 .mu.l of pure complement factor I (0.04 .mu.g, VWR International, catalogue no. 341280) for one of 15, 30, or 60 minutes at 37.degree. C. Reactions were stopped with the addition of 4.times.SDS-loading buffer and heating to 100.degree. C. for 5 minutes. Samples were then run on 4-12% NuPAGE Bis-Tris gels, run at 200V for 60 minutes.
[0371] The results are shown in FIGS. 3B and 3C. Breakdown of C3b in the diffusion chamber demonstrates that both glycosylated CR1a (3B) and non-glycosylated nCR1a (3C) were able to cross Bruch's membrane from the sample chamber and remain functionally active. CR1a and nCR1a from both chambers were found to act successfully as co-factors for FI to break down C3b into proteolytically-inactive C3b (iC3b) and further products, as demonstrated by the presence of bands representing C3b breakdown products iC3b and C3dg. Thus, CR1a and nCR1a are able to diffuse through BrM and retain the ability to confer C3b breakdown in the presence of Factor I.
Example 2
[0372] The expression levels of polypeptides according to the present invention are compared to assess if there is an optimal formulation, i.e. whether glycosylated polypeptides express at higher levels than non-glycosylated polypeptides. It is anticipated that the glycosylation state of the polypeptides will have a minimal effect on level of expression.
[0373] CR1a was found to be expressed well by human cells. nCR1a was found to be expressed at lower levels than CR1a but both polypeptides were found to be functionally active, see e.g. FIG. 2.
Example 3
[0374] The binding kinetics of purified polypeptides according to the present invention for C3b is tested using Surface Plasmon Resonance (SPR), whereby C3b is immobilised onto SPR chips and the polypeptides used in the fluid phase. The association and dissociation constants are directly measured and a kD value for the interaction inferred.
[0375] Whilst no differences are observed in the binding of the CR1a (CCPs 8-10) or CR1b (CCPs 15-17) species to C3b, glycosylated polypeptides bind more strongly to C3b than non-glycosylated polypeptides.
[0376] Binding Kinetics Measured by Bio-Layer Interferometry (BLI)
[0377] The affinity of CR1a for C3b protein was measured using OctetRed96 System (ForteBio, Pall Corp., USA). Biotinylated C3b protein was diluted in 0.2% PBST to final concentration of 0.4 .mu.g/mL and loaded onto High Precision Streptavidin (SAX) Biosensors (ForteBio, Pall Corp., USA) for 600 s, previously hydrated with the same buffer for 20 min. C3b-loaded sensors were then washed with 0.2% PBST for 150 s (baseline) and dipped into wells containing CR1a in different concentrations ranging from 30.0 .mu.g/mL to 2.6 .mu.g/mL for 600 s (association) followed by wash with 0.2% PBST for 600 s (dissociation). Association and dissociation profiles were recorded and analysed with ForeBio Data Analysis v9 (ForteBio, Pall Corp., USA). The negative control i.e. 0.2% PBST containing well was used in parallel to subtract binding resulting from nonspecific interactions with the sensors. Experiments were performed using the kinetics mode, at 25.degree. C. and sample plates were pre-agitated for 3 min. The binding profiles were globally fitted to 1:1 (one analyte in solution to one binding site on the surface). The KD was determined using data of the association (from 0 s to 600 s) and dissociation (from 0 s to 100 s) phases from four the lowest available analyte concentration by steady-state analysis. Binding affinities were also determined for native soluble C3b-binding complement regulators Factor H (FH) and FHL-1 to immobilised C3b.
[0378] The results are shown in FIG. 5. The binding affinity of CR1a for C3b was found to be 21 nM. For comparison, the binding affinity for C3b of FH was 580 nM and of FHL-1 was 1.2 mM. CR1a therefore binds significantly more strongly to C3b than either native soluble complement regulator. The strong binding affinity of CR1a for C3b means that CR1a is a more effective agent to promote C3b break down than agents based on FH or FHL-1.
[0379] A strong binding affinity also enables CR1a to promote degradation of iC3b into desirable further downstream products e.g. C3dg (see FIG. 2). In contrast, FH and FHL-1 cannot cause degradation of C3b beyond iC3b. iC3b is a pro-inflammatory molecule which acts to recruit immune cells to the site of complement activation, which in turn cause negative inflammatory effects. Further breakdown of iC3b to C3dg by CR1a is thus advantageous and avoids further damage caused by the immune system.
Example 4
[0380] The diffusion rates of polypeptides according to the present invention are compared using Ussing chamber experiments (described above) to see if any differences arise due to the formulation of the protein.
[0381] Experiments include using Bruch's membranes derived from donors with AMD to determine whether the material deposited in the Bruch's membrane in this condition, including drusen and basal linear deposits, compromises the ability of the polypeptide to cross Bruch's membrane. It is anticipated that the optimal polypeptide will cross Bruch's membrane efficiently even in the presence of AMD changes.
Example 5
[0382] Nucleic acid according to the present invention, optionally encoding the preferred signal peptide, a polypeptide having at least 80% sequence identity to SEQ ID NO:4 and a termination codon, is inserted into an AAV vector. The resultant expression vector is used to transfect cultured RPE cells. The expression and secretion of the encoded polypeptide is evaluated. It is anticipated that the polypeptide having at least 80% sequence identity to SEQ ID NO:4 is secreted by the RPE cells and that the signal peptide has been cleaved from the secreted polypeptide.
[0383] Adeno-Associated Virus (AAV) Transduction
[0384] Nucleic acid encoding the CR1a polypeptide described in Example 1 was transfected into human APRE-19 cells (ATCC, USA) derived from retinal pigment epithelium.
[0385] AAV2 serotype viral particles were pre-packed with CR1a plasmid. ARPE-19 cells were seeded on six well cell culture plate (Corning) in 2 ml of DMEM/F12 growth medium (ATCC, USA) supplemented with 10% (v/v) fetal bovine serum (ATCC, USA) at the density of 300,000 cells per well. Cells were then left for incubation at 37.degree. C. in humidified atmosphere of 5% CO.sub.2 for 24 hrs. After incubation, cells were washed twice with 2 ml serum free DMEM/F12 growth medium. AAV2-CR1a at multiplicity of infection (MOI) 100,000 in serum free DMEM/F12 growth medium in total volume of 1 ml was added. AAV2-CR1a containing medium was incubated with cells for 24 hrs (37.degree. C., 5% CO.sub.2) followed by replacement with 2 ml of fresh serum free DMEM/F12 growth medium the next day. Control cells transduced with AAV-GFP were grown in parallel. Transduction efficiency was assessed 14 days post-infection. Secretion of CR1a by ARPE-19 cells was detected by dot blot: conditioned media from CR1a-transduced RPE cells was contacted with in-house polyclonal anti-CR1a antibody.
[0386] The results are shown in FIG. 6. Immunoreactivity with the anti-CR1a antibody was seen in both samples tested (AAV-CR1a media), compared to media from RPE cells transduced with AAV-GFP (Ctrl media) used as a negative control. Purified recombinant CR1a protein was included as a positive control.
[0387] Recombinant CR1a polypeptides secreted from human APRE-19 and HEK293 cells were assessed for their ability to break down C3b to iC3b and C3dg.
[0388] The results are shown in FIG. 7A. Functional CR1a polypeptide secreted from human APRE-19 cells was found to act as a cofactor for Factor I leading to breakdown of C3b into iC3b (product e) and C3dg (product 0. Reaction containing FHL-1+FI+C3b provided a MW control for C3b and product iC3b.
[0389] C3b break down was assessed in tissue culture media of human RPE cells (ARPE-19) transduced with AAV-delivered CR1a. AAV-GFP transduced RPE cells were used as a negative control. Transduced cells were supplemented with purified C3b and Factor 114 days after transduction. C3b and iC3b levels in tissue culture media were detected by Western blot.
[0390] The results are shown in FIG. 7B. RPE cells produce their own complement components and were found to have a low native turnover of C3b (product a) into iC3b (product b; lane 2). However, RPE cells expressing AAV-delivered CR1a were found to have increased C3b breakdown capacity to iC3b (product b; lane 3) compared to the native turnover rate.
[0391] This indicates that CR1a is secreted successfully from eye cells, e.g. retinal pigment epithelium cells, is functionally active as a FI cofactor, enhances C3b break down to downstream products, is capable of complement regulation, and would provide therapeutic benefit for conditions involving over-activation of complement e.g. an excess of C3b.
Example 6
[0392] The polypeptides of the present invention find use in methods of treatment or prevention of complement-related disorders. One example of a complement related disorder is macular degeneration in the eye, e.g. AMD.
[0393] FIG. 8 provides a schematic of the eye macular region. The retinal pigment epithelium (RPE) is a continuous monolayer of cuboidal/columnar epithelial cells between the neurosensory retina and the vascular choroid. The cells have physical, optical, metabolic/biochemical and transport functions and play a critical role in the normal visual process. The RPE is separated from the choroid by Bruch's membrane (BrM): a thin (2-4 .mu.m), acellular, five-layered, extracellular matrix. The BrM serves two major functions: the substratum of the RPE and a blood vessel wall. Immediately adjacent to the BrM, and within the choroid, is a layer of capillaries termed the choriocapillaris. Complement activation centres on the extracellular matrix of the choriocapillaris, termed the intercapillary septa.
[0394] The hallmark lesions of AMD, drusen, are formed from the accumulation of lipids and cellular debris, including many complement activation products. Drusen develop within the BrM adjacent to the RPE layer and disrupt the flow of nutrients from the choroid to the RPE, leading to cell dysfunction and death. The death of RPE cells also causes dysfunction of photoreceptor cells and subsequent loss of visual acuity.
[0395] A representative fluorescence microscopy image of drusen deposits is provided in FIG. 9, taken from Forest et al. (2015) Dis. Mod. Mech. 8, 421-427 (FIG. 1). FIG. 9 shows retinal tissue from an 82-year-old female with AMD. A cell membrane marker shows degraded RPE cells overlying drusen. Areas of complement activation within the drusen and around the blood vessels are indicated by the terminal complement complex marker C5b-9. Nuclei are stained. Scale bar 20 .mu.m.
[0396] Systemic administration of a complement-regulating molecule would require a high dose and carries a substantial risk of detrimental off-target effects on functioning complement systems. Localised administration, e.g. expression from RPE cells, is a safer and more effective delivery method. However, for an RPE-expressed molecule to be effective in treating/preventing a complement-related disorder in the intercapillary septa of the choriocapillaris, i.e. the site of complement over-activation, the molecule will have to cross Bruch's membrane (see FIG. 10). That is, an effective complement-inhibiting therapy must meet the requirements of all three stages in FIG. 10.
[0397] As demonstrated herein, CR1a is: [0398] 1. Secreted from human RPE cells (see e.g. FIG. 6); [0399] 2. Able to passively diffuse across human BrM (see e.g. FIG. 3); and [0400] 3. Able to mediate the breakdown of C3b into iC3b, and further desirable break down products, in the presence of complement factor I (see e.g. FIGS. 2, 3, 7).
[0401] Therefore, CR1a is able to be expressed by RPE cells, is able to reach the areas of complement activation where C3b regulation is necessary, and can act as an effective therapeutic agent to treat complement over-activation in the eye, e.g. in AMD.
Example 7
[0402] Laser-Induced Choroidal Neovascularisation Model
[0403] This model applies laser burns to the retinas of mice or rats to induce choroidal neovascularization, for example as described in Schnabolk et al, Mol Ther Methods Clin Dev. 2018; 9: 1-11. The size of the choroidal neovascular complexes can be measured by fluorescein angiography or by histology.
[0404] Rodents are given a subretinal injection of AAV vector, e.g. AAV2, containing CR1a cDNA, or empty AAV vector as a control. Rodents receiving AAV vector containing CR1a cDNA secrete CR1a protein from their retinal pigment epithelial cells. When maximal CR1a protein excretion is achieved, laser burns are applied to the rodent retinas and the size of the choroidal neovascular complexes is measured at a pre-specified time after the laser burns. The complement inhibitory effect of CR1a polypeptide is found to decrease the size of the choroidal neovascular complexes compared to rodents administered the empty AAV vector.
[0405] Sodium Iodate Induced Retinal Degeneration
[0406] Mice are injected intravenously with sodium iodate which induces a degeneration of the retinal pigment epithelium that is partially dependent upon complement activation, for example as described in Katschke et al., Sci Rep. 2018; 8(1):7348. CR1a is delivered by intravitreal injection as a recombinant protein or by subretinal delivery using AAV vector, e.g. AAV2, containing CR1a cDNA. Empty AAV vector is administered as a control. CR1a treated mice are found to have less retinal degeneration than the control mice that did not receive CR1a treatment.
Sequence CWU 1
1
5412039PRTHomo sapiens 1Met Gly Ala Ser Ser Pro Arg Ser Pro Glu Pro
Val Gly Pro Pro Ala1 5 10 15Pro Gly Leu Pro Phe Cys Cys Gly Gly Ser
Leu Leu Ala Val Val Val 20 25 30Leu Leu Ala Leu Pro Val Ala Trp Gly
Gln Cys Asn Ala Pro Glu Trp 35 40 45Leu Pro Phe Ala Arg Pro Thr Asn
Leu Thr Asp Glu Phe Glu Phe Pro 50 55 60Ile Gly Thr Tyr Leu Asn Tyr
Glu Cys Arg Pro Gly Tyr Ser Gly Arg65 70 75 80Pro Phe Ser Ile Ile
Cys Leu Lys Asn Ser Val Trp Thr Gly Ala Lys 85 90 95Asp Arg Cys Arg
Arg Lys Ser Cys Arg Asn Pro Pro Asp Pro Val Asn 100 105 110Gly Met
Val His Val Ile Lys Gly Ile Gln Phe Gly Ser Gln Ile Lys 115 120
125Tyr Ser Cys Thr Lys Gly Tyr Arg Leu Ile Gly Ser Ser Ser Ala Thr
130 135 140Cys Ile Ile Ser Gly Asp Thr Val Ile Trp Asp Asn Glu Thr
Pro Ile145 150 155 160Cys Asp Arg Ile Pro Cys Gly Leu Pro Pro Thr
Ile Thr Asn Gly Asp 165 170 175Phe Ile Ser Thr Asn Arg Glu Asn Phe
His Tyr Gly Ser Val Val Thr 180 185 190Tyr Arg Cys Asn Pro Gly Ser
Gly Gly Arg Lys Val Phe Glu Leu Val 195 200 205Gly Glu Pro Ser Ile
Tyr Cys Thr Ser Asn Asp Asp Gln Val Gly Ile 210 215 220Trp Ser Gly
Pro Ala Pro Gln Cys Ile Ile Pro Asn Lys Cys Thr Pro225 230 235
240Pro Asn Val Glu Asn Gly Ile Leu Val Ser Asp Asn Arg Ser Leu Phe
245 250 255Ser Leu Asn Glu Val Val Glu Phe Arg Cys Gln Pro Gly Phe
Val Met 260 265 270Lys Gly Pro Arg Arg Val Lys Cys Gln Ala Leu Asn
Lys Trp Glu Pro 275 280 285Glu Leu Pro Ser Cys Ser Arg Val Cys Gln
Pro Pro Pro Asp Val Leu 290 295 300His Ala Glu Arg Thr Gln Arg Asp
Lys Asp Asn Phe Ser Pro Gly Gln305 310 315 320Glu Val Phe Tyr Ser
Cys Glu Pro Gly Tyr Asp Leu Arg Gly Ala Ala 325 330 335Ser Met Arg
Cys Thr Pro Gln Gly Asp Trp Ser Pro Ala Ala Pro Thr 340 345 350Cys
Glu Val Lys Ser Cys Asp Asp Phe Met Gly Gln Leu Leu Asn Gly 355 360
365Arg Val Leu Phe Pro Val Asn Leu Gln Leu Gly Ala Lys Val Asp Phe
370 375 380Val Cys Asp Glu Gly Phe Gln Leu Lys Gly Ser Ser Ala Ser
Tyr Cys385 390 395 400Val Leu Ala Gly Met Glu Ser Leu Trp Asn Ser
Ser Val Pro Val Cys 405 410 415Glu Gln Ile Phe Cys Pro Ser Pro Pro
Val Ile Pro Asn Gly Arg His 420 425 430Thr Gly Lys Pro Leu Glu Val
Phe Pro Phe Gly Lys Thr Val Asn Tyr 435 440 445Thr Cys Asp Pro His
Pro Asp Arg Gly Thr Ser Phe Asp Leu Ile Gly 450 455 460Glu Ser Thr
Ile Arg Cys Thr Ser Asp Pro Gln Gly Asn Gly Val Trp465 470 475
480Ser Ser Pro Ala Pro Arg Cys Gly Ile Leu Gly His Cys Gln Ala Pro
485 490 495Asp His Phe Leu Phe Ala Lys Leu Lys Thr Gln Thr Asn Ala
Ser Asp 500 505 510Phe Pro Ile Gly Thr Ser Leu Lys Tyr Glu Cys Arg
Pro Glu Tyr Tyr 515 520 525Gly Arg Pro Phe Ser Ile Thr Cys Leu Asp
Asn Leu Val Trp Ser Ser 530 535 540Pro Lys Asp Val Cys Lys Arg Lys
Ser Cys Lys Thr Pro Pro Asp Pro545 550 555 560Val Asn Gly Met Val
His Val Ile Thr Asp Ile Gln Val Gly Ser Arg 565 570 575Ile Asn Tyr
Ser Cys Thr Thr Gly His Arg Leu Ile Gly His Ser Ser 580 585 590Ala
Glu Cys Ile Leu Ser Gly Asn Ala Ala His Trp Ser Thr Lys Pro 595 600
605Pro Ile Cys Gln Arg Ile Pro Cys Gly Leu Pro Pro Thr Ile Ala Asn
610 615 620Gly Asp Phe Ile Ser Thr Asn Arg Glu Asn Phe His Tyr Gly
Ser Val625 630 635 640Val Thr Tyr Arg Cys Asn Pro Gly Ser Gly Gly
Arg Lys Val Phe Glu 645 650 655Leu Val Gly Glu Pro Ser Ile Tyr Cys
Thr Ser Asn Asp Asp Gln Val 660 665 670Gly Ile Trp Ser Gly Pro Ala
Pro Gln Cys Ile Ile Pro Asn Lys Cys 675 680 685Thr Pro Pro Asn Val
Glu Asn Gly Ile Leu Val Ser Asp Asn Arg Ser 690 695 700Leu Phe Ser
Leu Asn Glu Val Val Glu Phe Arg Cys Gln Pro Gly Phe705 710 715
720Val Met Lys Gly Pro Arg Arg Val Lys Cys Gln Ala Leu Asn Lys Trp
725 730 735Glu Pro Glu Leu Pro Ser Cys Ser Arg Val Cys Gln Pro Pro
Pro Asp 740 745 750Val Leu His Ala Glu Arg Thr Gln Arg Asp Lys Asp
Asn Phe Ser Pro 755 760 765Gly Gln Glu Val Phe Tyr Ser Cys Glu Pro
Gly Tyr Asp Leu Arg Gly 770 775 780Ala Ala Ser Met Arg Cys Thr Pro
Gln Gly Asp Trp Ser Pro Ala Ala785 790 795 800Pro Thr Cys Glu Val
Lys Ser Cys Asp Asp Phe Met Gly Gln Leu Leu 805 810 815Asn Gly Arg
Val Leu Phe Pro Val Asn Leu Gln Leu Gly Ala Lys Val 820 825 830Asp
Phe Val Cys Asp Glu Gly Phe Gln Leu Lys Gly Ser Ser Ala Ser 835 840
845Tyr Cys Val Leu Ala Gly Met Glu Ser Leu Trp Asn Ser Ser Val Pro
850 855 860Val Cys Glu Gln Ile Phe Cys Pro Ser Pro Pro Val Ile Pro
Asn Gly865 870 875 880Arg His Thr Gly Lys Pro Leu Glu Val Phe Pro
Phe Gly Lys Ala Val 885 890 895Asn Tyr Thr Cys Asp Pro His Pro Asp
Arg Gly Thr Ser Phe Asp Leu 900 905 910Ile Gly Glu Ser Thr Ile Arg
Cys Thr Ser Asp Pro Gln Gly Asn Gly 915 920 925Val Trp Ser Ser Pro
Ala Pro Arg Cys Gly Ile Leu Gly His Cys Gln 930 935 940Ala Pro Asp
His Phe Leu Phe Ala Lys Leu Lys Thr Gln Thr Asn Ala945 950 955
960Ser Asp Phe Pro Ile Gly Thr Ser Leu Lys Tyr Glu Cys Arg Pro Glu
965 970 975Tyr Tyr Gly Arg Pro Phe Ser Ile Thr Cys Leu Asp Asn Leu
Val Trp 980 985 990Ser Ser Pro Lys Asp Val Cys Lys Arg Lys Ser Cys
Lys Thr Pro Pro 995 1000 1005Asp Pro Val Asn Gly Met Val His Val
Ile Thr Asp Ile Gln Val 1010 1015 1020Gly Ser Arg Ile Asn Tyr Ser
Cys Thr Thr Gly His Arg Leu Ile 1025 1030 1035Gly His Ser Ser Ala
Glu Cys Ile Leu Ser Gly Asn Thr Ala His 1040 1045 1050Trp Ser Thr
Lys Pro Pro Ile Cys Gln Arg Ile Pro Cys Gly Leu 1055 1060 1065Pro
Pro Thr Ile Ala Asn Gly Asp Phe Ile Ser Thr Asn Arg Glu 1070 1075
1080Asn Phe His Tyr Gly Ser Val Val Thr Tyr Arg Cys Asn Leu Gly
1085 1090 1095Ser Arg Gly Arg Lys Val Phe Glu Leu Val Gly Glu Pro
Ser Ile 1100 1105 1110Tyr Cys Thr Ser Asn Asp Asp Gln Val Gly Ile
Trp Ser Gly Pro 1115 1120 1125Ala Pro Gln Cys Ile Ile Pro Asn Lys
Cys Thr Pro Pro Asn Val 1130 1135 1140Glu Asn Gly Ile Leu Val Ser
Asp Asn Arg Ser Leu Phe Ser Leu 1145 1150 1155Asn Glu Val Val Glu
Phe Arg Cys Gln Pro Gly Phe Val Met Lys 1160 1165 1170Gly Pro Arg
Arg Val Lys Cys Gln Ala Leu Asn Lys Trp Glu Pro 1175 1180 1185Glu
Leu Pro Ser Cys Ser Arg Val Cys Gln Pro Pro Pro Glu Ile 1190 1195
1200Leu His Gly Glu His Thr Pro Ser His Gln Asp Asn Phe Ser Pro
1205 1210 1215Gly Gln Glu Val Phe Tyr Ser Cys Glu Pro Gly Tyr Asp
Leu Arg 1220 1225 1230Gly Ala Ala Ser Leu His Cys Thr Pro Gln Gly
Asp Trp Ser Pro 1235 1240 1245Glu Ala Pro Arg Cys Ala Val Lys Ser
Cys Asp Asp Phe Leu Gly 1250 1255 1260Gln Leu Pro His Gly Arg Val
Leu Phe Pro Leu Asn Leu Gln Leu 1265 1270 1275Gly Ala Lys Val Ser
Phe Val Cys Asp Glu Gly Phe Arg Leu Lys 1280 1285 1290Gly Ser Ser
Val Ser His Cys Val Leu Val Gly Met Arg Ser Leu 1295 1300 1305Trp
Asn Asn Ser Val Pro Val Cys Glu His Ile Phe Cys Pro Asn 1310 1315
1320Pro Pro Ala Ile Leu Asn Gly Arg His Thr Gly Thr Pro Ser Gly
1325 1330 1335Asp Ile Pro Tyr Gly Lys Glu Ile Ser Tyr Thr Cys Asp
Pro His 1340 1345 1350Pro Asp Arg Gly Met Thr Phe Asn Leu Ile Gly
Glu Ser Thr Ile 1355 1360 1365Arg Cys Thr Ser Asp Pro His Gly Asn
Gly Val Trp Ser Ser Pro 1370 1375 1380Ala Pro Arg Cys Glu Leu Ser
Val Arg Ala Gly His Cys Lys Thr 1385 1390 1395Pro Glu Gln Phe Pro
Phe Ala Ser Pro Thr Ile Pro Ile Asn Asp 1400 1405 1410Phe Glu Phe
Pro Val Gly Thr Ser Leu Asn Tyr Glu Cys Arg Pro 1415 1420 1425Gly
Tyr Phe Gly Lys Met Phe Ser Ile Ser Cys Leu Glu Asn Leu 1430 1435
1440Val Trp Ser Ser Val Glu Asp Asn Cys Arg Arg Lys Ser Cys Gly
1445 1450 1455Pro Pro Pro Glu Pro Phe Asn Gly Met Val His Ile Asn
Thr Asp 1460 1465 1470Thr Gln Phe Gly Ser Thr Val Asn Tyr Ser Cys
Asn Glu Gly Phe 1475 1480 1485Arg Leu Ile Gly Ser Pro Ser Thr Thr
Cys Leu Val Ser Gly Asn 1490 1495 1500Asn Val Thr Trp Asp Lys Lys
Ala Pro Ile Cys Glu Ile Ile Ser 1505 1510 1515Cys Glu Pro Pro Pro
Thr Ile Ser Asn Gly Asp Phe Tyr Ser Asn 1520 1525 1530Asn Arg Thr
Ser Phe His Asn Gly Thr Val Val Thr Tyr Gln Cys 1535 1540 1545His
Thr Gly Pro Asp Gly Glu Gln Leu Phe Glu Leu Val Gly Glu 1550 1555
1560Arg Ser Ile Tyr Cys Thr Ser Lys Asp Asp Gln Val Gly Val Trp
1565 1570 1575Ser Ser Pro Pro Pro Arg Cys Ile Ser Thr Asn Lys Cys
Thr Ala 1580 1585 1590Pro Glu Val Glu Asn Ala Ile Arg Val Pro Gly
Asn Arg Ser Phe 1595 1600 1605Phe Ser Leu Thr Glu Ile Ile Arg Phe
Arg Cys Gln Pro Gly Phe 1610 1615 1620Val Met Val Gly Ser His Thr
Val Gln Cys Gln Thr Asn Gly Arg 1625 1630 1635Trp Gly Pro Lys Leu
Pro His Cys Ser Arg Val Cys Gln Pro Pro 1640 1645 1650Pro Glu Ile
Leu His Gly Glu His Thr Leu Ser His Gln Asp Asn 1655 1660 1665Phe
Ser Pro Gly Gln Glu Val Phe Tyr Ser Cys Glu Pro Ser Tyr 1670 1675
1680Asp Leu Arg Gly Ala Ala Ser Leu His Cys Thr Pro Gln Gly Asp
1685 1690 1695Trp Ser Pro Glu Ala Pro Arg Cys Thr Val Lys Ser Cys
Asp Asp 1700 1705 1710Phe Leu Gly Gln Leu Pro His Gly Arg Val Leu
Leu Pro Leu Asn 1715 1720 1725Leu Gln Leu Gly Ala Lys Val Ser Phe
Val Cys Asp Glu Gly Phe 1730 1735 1740Arg Leu Lys Gly Arg Ser Ala
Ser His Cys Val Leu Ala Gly Met 1745 1750 1755Lys Ala Leu Trp Asn
Ser Ser Val Pro Val Cys Glu Gln Ile Phe 1760 1765 1770Cys Pro Asn
Pro Pro Ala Ile Leu Asn Gly Arg His Thr Gly Thr 1775 1780 1785Pro
Phe Gly Asp Ile Pro Tyr Gly Lys Glu Ile Ser Tyr Ala Cys 1790 1795
1800Asp Thr His Pro Asp Arg Gly Met Thr Phe Asn Leu Ile Gly Glu
1805 1810 1815Ser Ser Ile Arg Cys Thr Ser Asp Pro Gln Gly Asn Gly
Val Trp 1820 1825 1830Ser Ser Pro Ala Pro Arg Cys Glu Leu Ser Val
Pro Ala Ala Cys 1835 1840 1845Pro His Pro Pro Lys Ile Gln Asn Gly
His Tyr Ile Gly Gly His 1850 1855 1860Val Ser Leu Tyr Leu Pro Gly
Met Thr Ile Ser Tyr Ile Cys Asp 1865 1870 1875Pro Gly Tyr Leu Leu
Val Gly Lys Gly Phe Ile Phe Cys Thr Asp 1880 1885 1890Gln Gly Ile
Trp Ser Gln Leu Asp His Tyr Cys Lys Glu Val Asn 1895 1900 1905Cys
Ser Phe Pro Leu Phe Met Asn Gly Ile Ser Lys Glu Leu Glu 1910 1915
1920Met Lys Lys Val Tyr His Tyr Gly Asp Tyr Val Thr Leu Lys Cys
1925 1930 1935Glu Asp Gly Tyr Thr Leu Glu Gly Ser Pro Trp Ser Gln
Cys Gln 1940 1945 1950Ala Asp Asp Arg Trp Asp Pro Pro Leu Ala Lys
Cys Thr Ser Arg 1955 1960 1965Thr His Asp Ala Leu Ile Val Gly Thr
Leu Ser Gly Thr Ile Phe 1970 1975 1980Phe Ile Leu Leu Ile Ile Phe
Leu Ser Trp Ile Ile Leu Lys His 1985 1990 1995Arg Lys Gly Asn Asn
Ala His Glu Asn Pro Lys Glu Val Ala Ile 2000 2005 2010His Leu His
Ser Gln Gly Gly Ser Ser Val His Pro Arg Thr Leu 2015 2020 2025Gln
Thr Asn Glu Glu Asn Ser Arg Val Leu Pro 2030 20352194PRTHomo
sapiens 2Gly His Cys Gln Ala Pro Asp His Phe Leu Phe Ala Lys Leu
Lys Thr1 5 10 15Gln Thr Asn Ala Ser Asp Phe Pro Ile Gly Thr Ser Leu
Lys Tyr Glu 20 25 30Cys Arg Pro Glu Tyr Tyr Gly Arg Pro Phe Ser Ile
Thr Cys Leu Asp 35 40 45Asn Leu Val Trp Ser Ser Pro Lys Asp Val Cys
Lys Arg Lys Ser Cys 50 55 60Lys Thr Pro Pro Asp Pro Val Asn Gly Met
Val His Val Ile Thr Asp65 70 75 80Ile Gln Val Gly Ser Arg Ile Asn
Tyr Ser Cys Thr Thr Gly His Arg 85 90 95Leu Ile Gly His Ser Ser Ala
Glu Cys Ile Leu Ser Gly Asn Ala Ala 100 105 110His Trp Ser Thr Lys
Pro Pro Ile Cys Gln Arg Ile Pro Cys Gly Leu 115 120 125Pro Pro Thr
Ile Ala Asn Gly Asp Phe Ile Ser Thr Asn Arg Glu Asn 130 135 140Phe
His Tyr Gly Ser Val Val Thr Tyr Arg Cys Asn Pro Gly Ser Gly145 150
155 160Gly Arg Lys Val Phe Glu Leu Val Gly Glu Pro Ser Ile Tyr Cys
Thr 165 170 175Ser Asn Asp Asp Gln Val Gly Ile Trp Ser Gly Pro Ala
Pro Gln Cys 180 185 190Ile Ile3194PRTHomo sapiens 3Gly His Cys Gln
Ala Pro Asp His Phe Leu Phe Ala Lys Leu Lys Thr1 5 10 15Gln Thr Asn
Ala Ser Asp Phe Pro Ile Gly Thr Ser Leu Lys Tyr Glu 20 25 30Cys Arg
Pro Glu Tyr Tyr Gly Arg Pro Phe Ser Ile Thr Cys Leu Asp 35 40 45Asn
Leu Val Trp Ser Ser Pro Lys Asp Val Cys Lys Arg Lys Ser Cys 50 55
60Lys Thr Pro Pro Asp Pro Val Asn Gly Met Val His Val Ile Thr Asp65
70 75 80Ile Gln Val Gly Ser Arg Ile Asn Tyr Ser Cys Thr Thr Gly His
Arg 85 90 95Leu Ile Gly His Ser Ser Ala Glu Cys Ile Leu Ser Gly Asn
Thr Ala 100 105 110His Trp Ser Thr Lys Pro Pro Ile Cys Gln Arg Ile
Pro Cys Gly Leu 115 120 125Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile
Ser Thr Asn Arg Glu Asn 130 135 140Phe His Tyr Gly Ser Val Val Thr
Tyr Arg Cys Asn Leu Gly Ser Arg145 150 155 160Gly Arg Lys Val Phe
Glu Leu Val Gly Glu Pro Ser Ile Tyr Cys Thr 165 170 175Ser Asn Asp
Asp Gln Val Gly Ile Trp Ser Gly Pro Ala Pro Gln Cys 180 185 190Ile
Ile4194PRTHomo sapiensMISC_FEATURE(111)..(111)X = A or
TMISC_FEATURE(157)..(157)X = P or LMISC_FEATURE(160)..(160)X = G or
R 4Gly His Cys Gln Ala Pro Asp His Phe Leu Phe Ala Lys Leu Lys
Thr1
5 10 15Gln Thr Asn Ala Ser Asp Phe Pro Ile Gly Thr Ser Leu Lys Tyr
Glu 20 25 30Cys Arg Pro Glu Tyr Tyr Gly Arg Pro Phe Ser Ile Thr Cys
Leu Asp 35 40 45Asn Leu Val Trp Ser Ser Pro Lys Asp Val Cys Lys Arg
Lys Ser Cys 50 55 60Lys Thr Pro Pro Asp Pro Val Asn Gly Met Val His
Val Ile Thr Asp65 70 75 80Ile Gln Val Gly Ser Arg Ile Asn Tyr Ser
Cys Thr Thr Gly His Arg 85 90 95Leu Ile Gly His Ser Ser Ala Glu Cys
Ile Leu Ser Gly Asn Xaa Ala 100 105 110His Trp Ser Thr Lys Pro Pro
Ile Cys Gln Arg Ile Pro Cys Gly Leu 115 120 125Pro Pro Thr Ile Ala
Asn Gly Asp Phe Ile Ser Thr Asn Arg Glu Asn 130 135 140Phe His Tyr
Gly Ser Val Val Thr Tyr Arg Cys Asn Xaa Gly Ser Xaa145 150 155
160Gly Arg Lys Val Phe Glu Leu Val Gly Glu Pro Ser Ile Tyr Cys Thr
165 170 175Ser Asn Asp Asp Gln Val Gly Ile Trp Ser Gly Pro Ala Pro
Gln Cys 180 185 190Ile Ile5194PRTHomo sapiens 5Gly His Cys Gln Ala
Pro Asp His Phe Leu Phe Ala Lys Leu Lys Thr1 5 10 15Gln Thr Gln Ala
Ser Asp Phe Pro Ile Gly Thr Ser Leu Lys Tyr Glu 20 25 30Cys Arg Pro
Glu Tyr Tyr Gly Arg Pro Phe Ser Ile Thr Cys Leu Asp 35 40 45Asn Leu
Val Trp Ser Ser Pro Lys Asp Val Cys Lys Arg Lys Ser Cys 50 55 60Lys
Thr Pro Pro Asp Pro Val Asn Gly Met Val His Val Ile Thr Asp65 70 75
80Ile Gln Val Gly Ser Arg Ile Gln Tyr Ser Cys Thr Thr Gly His Arg
85 90 95Leu Ile Gly His Ser Ser Ala Glu Cys Ile Leu Ser Gly Asn Ala
Ala 100 105 110His Trp Ser Thr Lys Pro Pro Ile Cys Gln Arg Ile Pro
Cys Gly Leu 115 120 125Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile Ser
Thr Asn Arg Glu Asn 130 135 140Phe His Tyr Gly Ser Val Val Thr Tyr
Arg Cys Asn Pro Gly Ser Gly145 150 155 160Gly Arg Lys Val Phe Glu
Leu Val Gly Glu Pro Ser Ile Tyr Cys Thr 165 170 175Ser Asn Asp Asp
Gln Val Gly Ile Trp Ser Gly Pro Ala Pro Gln Cys 180 185 190Ile
Ile6194PRTHomo sapiens 6Gly His Cys Gln Ala Pro Asp His Phe Leu Phe
Ala Lys Leu Lys Thr1 5 10 15Gln Thr Gln Ala Ser Asp Phe Pro Ile Gly
Thr Ser Leu Lys Tyr Glu 20 25 30Cys Arg Pro Glu Tyr Tyr Gly Arg Pro
Phe Ser Ile Thr Cys Leu Asp 35 40 45Asn Leu Val Trp Ser Ser Pro Lys
Asp Val Cys Lys Arg Lys Ser Cys 50 55 60Lys Thr Pro Pro Asp Pro Val
Asn Gly Met Val His Val Ile Thr Asp65 70 75 80Ile Gln Val Gly Ser
Arg Ile Gln Tyr Ser Cys Thr Thr Gly His Arg 85 90 95Leu Ile Gly His
Ser Ser Ala Glu Cys Ile Leu Ser Gly Asn Thr Ala 100 105 110His Trp
Ser Thr Lys Pro Pro Ile Cys Gln Arg Ile Pro Cys Gly Leu 115 120
125Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile Ser Thr Asn Arg Glu Asn
130 135 140Phe His Tyr Gly Ser Val Val Thr Tyr Arg Cys Asn Leu Gly
Ser Arg145 150 155 160Gly Arg Lys Val Phe Glu Leu Val Gly Glu Pro
Ser Ile Tyr Cys Thr 165 170 175Ser Asn Asp Asp Gln Val Gly Ile Trp
Ser Gly Pro Ala Pro Gln Cys 180 185 190Ile Ile718PRTArtificial
SequenceSynthetic construct 7Met Arg Leu Leu Ala Lys Ile Ile Cys
Leu Met Leu Trp Ala Ile Cys1 5 10 15Val Ala861PRTHomo sapiens 8Gly
His Cys Gln Ala Pro Asp His Phe Leu Phe Ala Lys Leu Lys Thr1 5 10
15Gln Thr Asn Ala Ser Asp Phe Pro Ile Gly Thr Ser Leu Lys Tyr Glu
20 25 30Cys Arg Pro Glu Tyr Tyr Gly Arg Pro Phe Ser Ile Thr Cys Leu
Asp 35 40 45Asn Leu Val Trp Ser Ser Pro Lys Asp Val Cys Lys Arg 50
55 60961PRTHomo sapiens 9Lys Ser Cys Lys Thr Pro Pro Asp Pro Val
Asn Gly Met Val His Val1 5 10 15Ile Thr Asp Ile Gln Val Gly Ser Arg
Ile Asn Tyr Ser Cys Thr Thr 20 25 30Gly His Arg Leu Ile Gly His Ser
Ser Ala Glu Cys Ile Leu Ser Gly 35 40 45Asn Ala Ala His Trp Ser Thr
Lys Pro Pro Ile Cys Gln 50 55 601072PRTHomo sapiens 10Arg Ile Pro
Cys Gly Leu Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile1 5 10 15Ser Thr
Asn Arg Glu Asn Phe His Tyr Gly Ser Val Val Thr Tyr Arg 20 25 30Cys
Asn Pro Gly Ser Gly Gly Arg Lys Val Phe Glu Leu Val Gly Glu 35 40
45Pro Ser Ile Tyr Cys Thr Ser Asn Asp Asp Gln Val Gly Ile Trp Ser
50 55 60Gly Pro Ala Pro Gln Cys Ile Ile65 701161PRTHomo sapiens
11Lys Ser Cys Lys Thr Pro Pro Asp Pro Val Asn Gly Met Val His Val1
5 10 15Ile Thr Asp Ile Gln Val Gly Ser Arg Ile Asn Tyr Ser Cys Thr
Thr 20 25 30Gly His Arg Leu Ile Gly His Ser Ser Ala Glu Cys Ile Leu
Ser Gly 35 40 45Asn Thr Ala His Trp Ser Thr Lys Pro Pro Ile Cys Gln
50 55 601272PRTHomo sapiens 12Arg Ile Pro Cys Gly Leu Pro Pro Thr
Ile Ala Asn Gly Asp Phe Ile1 5 10 15Ser Thr Asn Arg Glu Asn Phe His
Tyr Gly Ser Val Val Thr Tyr Arg 20 25 30Cys Asn Leu Gly Ser Arg Gly
Arg Lys Val Phe Glu Leu Val Gly Glu 35 40 45Pro Ser Ile Tyr Cys Thr
Ser Asn Asp Asp Gln Val Gly Ile Trp Ser 50 55 60Gly Pro Ala Pro Gln
Cys Ile Ile65 7013388PRTHomo sapiens 13Gly His Cys Gln Ala Pro Asp
His Phe Leu Phe Ala Lys Leu Lys Thr1 5 10 15Gln Thr Asn Ala Ser Asp
Phe Pro Ile Gly Thr Ser Leu Lys Tyr Glu 20 25 30Cys Arg Pro Glu Tyr
Tyr Gly Arg Pro Phe Ser Ile Thr Cys Leu Asp 35 40 45Asn Leu Val Trp
Ser Ser Pro Lys Asp Val Cys Lys Arg Lys Ser Cys 50 55 60Lys Thr Pro
Pro Asp Pro Val Asn Gly Met Val His Val Ile Thr Asp65 70 75 80Ile
Gln Val Gly Ser Arg Ile Asn Tyr Ser Cys Thr Thr Gly His Arg 85 90
95Leu Ile Gly His Ser Ser Ala Glu Cys Ile Leu Ser Gly Asn Ala Ala
100 105 110His Trp Ser Thr Lys Pro Pro Ile Cys Gln Arg Ile Pro Cys
Gly Leu 115 120 125Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile Ser Thr
Asn Arg Glu Asn 130 135 140Phe His Tyr Gly Ser Val Val Thr Tyr Arg
Cys Asn Pro Gly Ser Gly145 150 155 160Gly Arg Lys Val Phe Glu Leu
Val Gly Glu Pro Ser Ile Tyr Cys Thr 165 170 175Ser Asn Asp Asp Gln
Val Gly Ile Trp Ser Gly Pro Ala Pro Gln Cys 180 185 190Ile Ile Gly
His Cys Gln Ala Pro Asp His Phe Leu Phe Ala Lys Leu 195 200 205Lys
Thr Gln Thr Asn Ala Ser Asp Phe Pro Ile Gly Thr Ser Leu Lys 210 215
220Tyr Glu Cys Arg Pro Glu Tyr Tyr Gly Arg Pro Phe Ser Ile Thr
Cys225 230 235 240Leu Asp Asn Leu Val Trp Ser Ser Pro Lys Asp Val
Cys Lys Arg Lys 245 250 255Ser Cys Lys Thr Pro Pro Asp Pro Val Asn
Gly Met Val His Val Ile 260 265 270Thr Asp Ile Gln Val Gly Ser Arg
Ile Asn Tyr Ser Cys Thr Thr Gly 275 280 285His Arg Leu Ile Gly His
Ser Ser Ala Glu Cys Ile Leu Ser Gly Asn 290 295 300Thr Ala His Trp
Ser Thr Lys Pro Pro Ile Cys Gln Arg Ile Pro Cys305 310 315 320Gly
Leu Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile Ser Thr Asn Arg 325 330
335Glu Asn Phe His Tyr Gly Ser Val Val Thr Tyr Arg Cys Asn Leu Gly
340 345 350Ser Arg Gly Arg Lys Val Phe Glu Leu Val Gly Glu Pro Ser
Ile Tyr 355 360 365Cys Thr Ser Asn Asp Asp Gln Val Gly Ile Trp Ser
Gly Pro Ala Pro 370 375 380Gln Cys Ile Ile38514423PRTHomo
sapiensmisc_feature(195)..(195)Xaa can be any naturally occurring
amino acidMISC_FEATURE(196)..(229)Xaa can be any naturally
occurring amino acid or absent 14Gly His Cys Gln Ala Pro Asp His
Phe Leu Phe Ala Lys Leu Lys Thr1 5 10 15Gln Thr Asn Ala Ser Asp Phe
Pro Ile Gly Thr Ser Leu Lys Tyr Glu 20 25 30Cys Arg Pro Glu Tyr Tyr
Gly Arg Pro Phe Ser Ile Thr Cys Leu Asp 35 40 45Asn Leu Val Trp Ser
Ser Pro Lys Asp Val Cys Lys Arg Lys Ser Cys 50 55 60Lys Thr Pro Pro
Asp Pro Val Asn Gly Met Val His Val Ile Thr Asp65 70 75 80Ile Gln
Val Gly Ser Arg Ile Asn Tyr Ser Cys Thr Thr Gly His Arg 85 90 95Leu
Ile Gly His Ser Ser Ala Glu Cys Ile Leu Ser Gly Asn Ala Ala 100 105
110His Trp Ser Thr Lys Pro Pro Ile Cys Gln Arg Ile Pro Cys Gly Leu
115 120 125Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile Ser Thr Asn Arg
Glu Asn 130 135 140Phe His Tyr Gly Ser Val Val Thr Tyr Arg Cys Asn
Pro Gly Ser Gly145 150 155 160Gly Arg Lys Val Phe Glu Leu Val Gly
Glu Pro Ser Ile Tyr Cys Thr 165 170 175Ser Asn Asp Asp Gln Val Gly
Ile Trp Ser Gly Pro Ala Pro Gln Cys 180 185 190Ile Ile Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 195 200 205Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 210 215 220Xaa
Xaa Xaa Xaa Xaa Gly His Cys Gln Ala Pro Asp His Phe Leu Phe225 230
235 240Ala Lys Leu Lys Thr Gln Thr Asn Ala Ser Asp Phe Pro Ile Gly
Thr 245 250 255Ser Leu Lys Tyr Glu Cys Arg Pro Glu Tyr Tyr Gly Arg
Pro Phe Ser 260 265 270Ile Thr Cys Leu Asp Asn Leu Val Trp Ser Ser
Pro Lys Asp Val Cys 275 280 285Lys Arg Lys Ser Cys Lys Thr Pro Pro
Asp Pro Val Asn Gly Met Val 290 295 300His Val Ile Thr Asp Ile Gln
Val Gly Ser Arg Ile Asn Tyr Ser Cys305 310 315 320Thr Thr Gly His
Arg Leu Ile Gly His Ser Ser Ala Glu Cys Ile Leu 325 330 335Ser Gly
Asn Thr Ala His Trp Ser Thr Lys Pro Pro Ile Cys Gln Arg 340 345
350Ile Pro Cys Gly Leu Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile Ser
355 360 365Thr Asn Arg Glu Asn Phe His Tyr Gly Ser Val Val Thr Tyr
Arg Cys 370 375 380Asn Leu Gly Ser Arg Gly Arg Lys Val Phe Glu Leu
Val Gly Glu Pro385 390 395 400Ser Ile Tyr Cys Thr Ser Asn Asp Asp
Gln Val Gly Ile Trp Ser Gly 405 410 415Pro Ala Pro Gln Cys Ile Ile
42015388PRTHomo sapiens 15Gly His Cys Gln Ala Pro Asp His Phe Leu
Phe Ala Lys Leu Lys Thr1 5 10 15Gln Thr Gln Ala Ser Asp Phe Pro Ile
Gly Thr Ser Leu Lys Tyr Glu 20 25 30Cys Arg Pro Glu Tyr Tyr Gly Arg
Pro Phe Ser Ile Thr Cys Leu Asp 35 40 45Asn Leu Val Trp Ser Ser Pro
Lys Asp Val Cys Lys Arg Lys Ser Cys 50 55 60Lys Thr Pro Pro Asp Pro
Val Asn Gly Met Val His Val Ile Thr Asp65 70 75 80Ile Gln Val Gly
Ser Arg Ile Gln Tyr Ser Cys Thr Thr Gly His Arg 85 90 95Leu Ile Gly
His Ser Ser Ala Glu Cys Ile Leu Ser Gly Asn Ala Ala 100 105 110His
Trp Ser Thr Lys Pro Pro Ile Cys Gln Arg Ile Pro Cys Gly Leu 115 120
125Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile Ser Thr Asn Arg Glu Asn
130 135 140Phe His Tyr Gly Ser Val Val Thr Tyr Arg Cys Asn Pro Gly
Ser Gly145 150 155 160Gly Arg Lys Val Phe Glu Leu Val Gly Glu Pro
Ser Ile Tyr Cys Thr 165 170 175Ser Asn Asp Asp Gln Val Gly Ile Trp
Ser Gly Pro Ala Pro Gln Cys 180 185 190Ile Ile Gly His Cys Gln Ala
Pro Asp His Phe Leu Phe Ala Lys Leu 195 200 205Lys Thr Gln Thr Gln
Ala Ser Asp Phe Pro Ile Gly Thr Ser Leu Lys 210 215 220Tyr Glu Cys
Arg Pro Glu Tyr Tyr Gly Arg Pro Phe Ser Ile Thr Cys225 230 235
240Leu Asp Asn Leu Val Trp Ser Ser Pro Lys Asp Val Cys Lys Arg Lys
245 250 255Ser Cys Lys Thr Pro Pro Asp Pro Val Asn Gly Met Val His
Val Ile 260 265 270Thr Asp Ile Gln Val Gly Ser Arg Ile Gln Tyr Ser
Cys Thr Thr Gly 275 280 285His Arg Leu Ile Gly His Ser Ser Ala Glu
Cys Ile Leu Ser Gly Asn 290 295 300Thr Ala His Trp Ser Thr Lys Pro
Pro Ile Cys Gln Arg Ile Pro Cys305 310 315 320Gly Leu Pro Pro Thr
Ile Ala Asn Gly Asp Phe Ile Ser Thr Asn Arg 325 330 335Glu Asn Phe
His Tyr Gly Ser Val Val Thr Tyr Arg Cys Asn Leu Gly 340 345 350Ser
Arg Gly Arg Lys Val Phe Glu Leu Val Gly Glu Pro Ser Ile Tyr 355 360
365Cys Thr Ser Asn Asp Asp Gln Val Gly Ile Trp Ser Gly Pro Ala Pro
370 375 380Gln Cys Ile Ile3851661PRTHomo
sapiensMISC_FEATURE(50)..(50)Xaa = Ala or Thr 16Lys Ser Cys Lys Thr
Pro Pro Asp Pro Val Asn Gly Met Val His Val1 5 10 15Ile Thr Asp Ile
Gln Val Gly Ser Arg Ile Asn Tyr Ser Cys Thr Thr 20 25 30Gly His Arg
Leu Ile Gly His Ser Ser Ala Glu Cys Ile Leu Ser Gly 35 40 45Asn Xaa
Ala His Trp Ser Thr Lys Pro Pro Ile Cys Gln 50 55 601772PRTHomo
sapiensMISC_FEATURE(35)..(35)Xaa = Pro or
LeuMISC_FEATURE(38)..(38)Xaa = Gly or Arg 17Arg Ile Pro Cys Gly Leu
Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile1 5 10 15Ser Thr Asn Arg Glu
Asn Phe His Tyr Gly Ser Val Val Thr Tyr Arg 20 25 30Cys Asn Xaa Gly
Ser Xaa Gly Arg Lys Val Phe Glu Leu Val Gly Glu 35 40 45Pro Ser Ile
Tyr Cys Thr Ser Asn Asp Asp Gln Val Gly Ile Trp Ser 50 55 60Gly Pro
Ala Pro Gln Cys Ile Ile65 70181663PRTHomo sapiens 18Met Gly Pro Thr
Ser Gly Pro Ser Leu Leu Leu Leu Leu Leu Thr His1 5 10 15Leu Pro Leu
Ala Leu Gly Ser Pro Met Tyr Ser Ile Ile Thr Pro Asn 20 25 30Ile Leu
Arg Leu Glu Ser Glu Glu Thr Met Val Leu Glu Ala His Asp 35 40 45Ala
Gln Gly Asp Val Pro Val Thr Val Thr Val His Asp Phe Pro Gly 50 55
60Lys Lys Leu Val Leu Ser Ser Glu Lys Thr Val Leu Thr Pro Ala Thr65
70 75 80Asn His Met Gly Asn Val Thr Phe Thr Ile Pro Ala Asn Arg Glu
Phe 85 90 95Lys Ser Glu Lys Gly Arg Asn Lys Phe Val Thr Val Gln Ala
Thr Phe 100 105 110Gly Thr Gln Val Val Glu Lys Val Val Leu Val Ser
Leu Gln Ser Gly 115 120 125Tyr Leu Phe Ile Gln Thr Asp Lys Thr Ile
Tyr Thr Pro Gly Ser Thr 130 135 140Val Leu Tyr Arg Ile Phe Thr Val
Asn His Lys Leu Leu Pro Val Gly145 150 155 160Arg Thr Val Met Val
Asn
Ile Glu Asn Pro Glu Gly Ile Pro Val Lys 165 170 175Gln Asp Ser Leu
Ser Ser Gln Asn Gln Leu Gly Val Leu Pro Leu Ser 180 185 190Trp Asp
Ile Pro Glu Leu Val Asn Met Gly Gln Trp Lys Ile Arg Ala 195 200
205Tyr Tyr Glu Asn Ser Pro Gln Gln Val Phe Ser Thr Glu Phe Glu Val
210 215 220Lys Glu Tyr Val Leu Pro Ser Phe Glu Val Ile Val Glu Pro
Thr Glu225 230 235 240Lys Phe Tyr Tyr Ile Tyr Asn Glu Lys Gly Leu
Glu Val Thr Ile Thr 245 250 255Ala Arg Phe Leu Tyr Gly Lys Lys Val
Glu Gly Thr Ala Phe Val Ile 260 265 270Phe Gly Ile Gln Asp Gly Glu
Gln Arg Ile Ser Leu Pro Glu Ser Leu 275 280 285Lys Arg Ile Pro Ile
Glu Asp Gly Ser Gly Glu Val Val Leu Ser Arg 290 295 300Lys Val Leu
Leu Asp Gly Val Gln Asn Pro Arg Ala Glu Asp Leu Val305 310 315
320Gly Lys Ser Leu Tyr Val Ser Ala Thr Val Ile Leu His Ser Gly Ser
325 330 335Asp Met Val Gln Ala Glu Arg Ser Gly Ile Pro Ile Val Thr
Ser Pro 340 345 350Tyr Gln Ile His Phe Thr Lys Thr Pro Lys Tyr Phe
Lys Pro Gly Met 355 360 365Pro Phe Asp Leu Met Val Phe Val Thr Asn
Pro Asp Gly Ser Pro Ala 370 375 380Tyr Arg Val Pro Val Ala Val Gln
Gly Glu Asp Thr Val Gln Ser Leu385 390 395 400Thr Gln Gly Asp Gly
Val Ala Lys Leu Ser Ile Asn Thr His Pro Ser 405 410 415Gln Lys Pro
Leu Ser Ile Thr Val Arg Thr Lys Lys Gln Glu Leu Ser 420 425 430Glu
Ala Glu Gln Ala Thr Arg Thr Met Gln Ala Leu Pro Tyr Ser Thr 435 440
445Val Gly Asn Ser Asn Asn Tyr Leu His Leu Ser Val Leu Arg Thr Glu
450 455 460Leu Arg Pro Gly Glu Thr Leu Asn Val Asn Phe Leu Leu Arg
Met Asp465 470 475 480Arg Ala His Glu Ala Lys Ile Arg Tyr Tyr Thr
Tyr Leu Ile Met Asn 485 490 495Lys Gly Arg Leu Leu Lys Ala Gly Arg
Gln Val Arg Glu Pro Gly Gln 500 505 510Asp Leu Val Val Leu Pro Leu
Ser Ile Thr Thr Asp Phe Ile Pro Ser 515 520 525Phe Arg Leu Val Ala
Tyr Tyr Thr Leu Ile Gly Ala Ser Gly Gln Arg 530 535 540Glu Val Val
Ala Asp Ser Val Trp Val Asp Val Lys Asp Ser Cys Val545 550 555
560Gly Ser Leu Val Val Lys Ser Gly Gln Ser Glu Asp Arg Gln Pro Val
565 570 575Pro Gly Gln Gln Met Thr Leu Lys Ile Glu Gly Asp His Gly
Ala Arg 580 585 590Val Val Leu Val Ala Val Asp Lys Gly Val Phe Val
Leu Asn Lys Lys 595 600 605Asn Lys Leu Thr Gln Ser Lys Ile Trp Asp
Val Val Glu Lys Ala Asp 610 615 620Ile Gly Cys Thr Pro Gly Ser Gly
Lys Asp Tyr Ala Gly Val Phe Ser625 630 635 640Asp Ala Gly Leu Thr
Phe Thr Ser Ser Ser Gly Gln Gln Thr Ala Gln 645 650 655Arg Ala Glu
Leu Gln Cys Pro Gln Pro Ala Ala Arg Arg Arg Arg Ser 660 665 670Val
Gln Leu Thr Glu Lys Arg Met Asp Lys Val Gly Lys Tyr Pro Lys 675 680
685Glu Leu Arg Lys Cys Cys Glu Asp Gly Met Arg Glu Asn Pro Met Arg
690 695 700Phe Ser Cys Gln Arg Arg Thr Arg Phe Ile Ser Leu Gly Glu
Ala Cys705 710 715 720Lys Lys Val Phe Leu Asp Cys Cys Asn Tyr Ile
Thr Glu Leu Arg Arg 725 730 735Gln His Ala Arg Ala Ser His Leu Gly
Leu Ala Arg Ser Asn Leu Asp 740 745 750Glu Asp Ile Ile Ala Glu Glu
Asn Ile Val Ser Arg Ser Glu Phe Pro 755 760 765Glu Ser Trp Leu Trp
Asn Val Glu Asp Leu Lys Glu Pro Pro Lys Asn 770 775 780Gly Ile Ser
Thr Lys Leu Met Asn Ile Phe Leu Lys Asp Ser Ile Thr785 790 795
800Thr Trp Glu Ile Leu Ala Val Ser Met Ser Asp Lys Lys Gly Ile Cys
805 810 815Val Ala Asp Pro Phe Glu Val Thr Val Met Gln Asp Phe Phe
Ile Asp 820 825 830Leu Arg Leu Pro Tyr Ser Val Val Arg Asn Glu Gln
Val Glu Ile Arg 835 840 845Ala Val Leu Tyr Asn Tyr Arg Gln Asn Gln
Glu Leu Lys Val Arg Val 850 855 860Glu Leu Leu His Asn Pro Ala Phe
Cys Ser Leu Ala Thr Thr Lys Arg865 870 875 880Arg His Gln Gln Thr
Val Thr Ile Pro Pro Lys Ser Ser Leu Ser Val 885 890 895Pro Tyr Val
Ile Val Pro Leu Lys Thr Gly Leu Gln Glu Val Glu Val 900 905 910Lys
Ala Ala Val Tyr His His Phe Ile Ser Asp Gly Val Arg Lys Ser 915 920
925Leu Lys Val Val Pro Glu Gly Ile Arg Met Asn Lys Thr Val Ala Val
930 935 940Arg Thr Leu Asp Pro Glu Arg Leu Gly Arg Glu Gly Val Gln
Lys Glu945 950 955 960Asp Ile Pro Pro Ala Asp Leu Ser Asp Gln Val
Pro Asp Thr Glu Ser 965 970 975Glu Thr Arg Ile Leu Leu Gln Gly Thr
Pro Val Ala Gln Met Thr Glu 980 985 990Asp Ala Val Asp Ala Glu Arg
Leu Lys His Leu Ile Val Thr Pro Ser 995 1000 1005Gly Cys Gly Glu
Gln Asn Met Ile Gly Met Thr Pro Thr Val Ile 1010 1015 1020Ala Val
His Tyr Leu Asp Glu Thr Glu Gln Trp Glu Lys Phe Gly 1025 1030
1035Leu Glu Lys Arg Gln Gly Ala Leu Glu Leu Ile Lys Lys Gly Tyr
1040 1045 1050Thr Gln Gln Leu Ala Phe Arg Gln Pro Ser Ser Ala Phe
Ala Ala 1055 1060 1065Phe Val Lys Arg Ala Pro Ser Thr Trp Leu Thr
Ala Tyr Val Val 1070 1075 1080Lys Val Phe Ser Leu Ala Val Asn Leu
Ile Ala Ile Asp Ser Gln 1085 1090 1095Val Leu Cys Gly Ala Val Lys
Trp Leu Ile Leu Glu Lys Gln Lys 1100 1105 1110Pro Asp Gly Val Phe
Gln Glu Asp Ala Pro Val Ile His Gln Glu 1115 1120 1125Met Ile Gly
Gly Leu Arg Asn Asn Asn Glu Lys Asp Met Ala Leu 1130 1135 1140Thr
Ala Phe Val Leu Ile Ser Leu Gln Glu Ala Lys Asp Ile Cys 1145 1150
1155Glu Glu Gln Val Asn Ser Leu Pro Gly Ser Ile Thr Lys Ala Gly
1160 1165 1170Asp Phe Leu Glu Ala Asn Tyr Met Asn Leu Gln Arg Ser
Tyr Thr 1175 1180 1185Val Ala Ile Ala Gly Tyr Ala Leu Ala Gln Met
Gly Arg Leu Lys 1190 1195 1200Gly Pro Leu Leu Asn Lys Phe Leu Thr
Thr Ala Lys Asp Lys Asn 1205 1210 1215Arg Trp Glu Asp Pro Gly Lys
Gln Leu Tyr Asn Val Glu Ala Thr 1220 1225 1230Ser Tyr Ala Leu Leu
Ala Leu Leu Gln Leu Lys Asp Phe Asp Phe 1235 1240 1245Val Pro Pro
Val Val Arg Trp Leu Asn Glu Gln Arg Tyr Tyr Gly 1250 1255 1260Gly
Gly Tyr Gly Ser Thr Gln Ala Thr Phe Met Val Phe Gln Ala 1265 1270
1275Leu Ala Gln Tyr Gln Lys Asp Ala Pro Asp His Gln Glu Leu Asn
1280 1285 1290Leu Asp Val Ser Leu Gln Leu Pro Ser Arg Ser Ser Lys
Ile Thr 1295 1300 1305His Arg Ile His Trp Glu Ser Ala Ser Leu Leu
Arg Ser Glu Glu 1310 1315 1320Thr Lys Glu Asn Glu Gly Phe Thr Val
Thr Ala Glu Gly Lys Gly 1325 1330 1335Gln Gly Thr Leu Ser Val Val
Thr Met Tyr His Ala Lys Ala Lys 1340 1345 1350Asp Gln Leu Thr Cys
Asn Lys Phe Asp Leu Lys Val Thr Ile Lys 1355 1360 1365Pro Ala Pro
Glu Thr Glu Lys Arg Pro Gln Asp Ala Lys Asn Thr 1370 1375 1380Met
Ile Leu Glu Ile Cys Thr Arg Tyr Arg Gly Asp Gln Asp Ala 1385 1390
1395Thr Met Ser Ile Leu Asp Ile Ser Met Met Thr Gly Phe Ala Pro
1400 1405 1410Asp Thr Asp Asp Leu Lys Gln Leu Ala Asn Gly Val Asp
Arg Tyr 1415 1420 1425Ile Ser Lys Tyr Glu Leu Asp Lys Ala Phe Ser
Asp Arg Asn Thr 1430 1435 1440Leu Ile Ile Tyr Leu Asp Lys Val Ser
His Ser Glu Asp Asp Cys 1445 1450 1455Leu Ala Phe Lys Val His Gln
Tyr Phe Asn Val Glu Leu Ile Gln 1460 1465 1470Pro Gly Ala Val Lys
Val Tyr Ala Tyr Tyr Asn Leu Glu Glu Ser 1475 1480 1485Cys Thr Arg
Phe Tyr His Pro Glu Lys Glu Asp Gly Lys Leu Asn 1490 1495 1500Lys
Leu Cys Arg Asp Glu Leu Cys Arg Cys Ala Glu Glu Asn Cys 1505 1510
1515Phe Ile Gln Lys Ser Asp Asp Lys Val Thr Leu Glu Glu Arg Leu
1520 1525 1530Asp Lys Ala Cys Glu Pro Gly Val Asp Tyr Val Tyr Lys
Thr Arg 1535 1540 1545Leu Val Lys Val Gln Leu Ser Asn Asp Phe Asp
Glu Tyr Ile Met 1550 1555 1560Ala Ile Glu Gln Thr Ile Lys Ser Gly
Ser Asp Glu Val Gln Val 1565 1570 1575Gly Gln Gln Arg Thr Phe Ile
Ser Pro Ile Lys Cys Arg Glu Ala 1580 1585 1590Leu Lys Leu Glu Glu
Lys Lys His Tyr Leu Met Trp Gly Leu Ser 1595 1600 1605Ser Asp Phe
Trp Gly Glu Lys Pro Asn Leu Ser Tyr Ile Ile Gly 1610 1615 1620Lys
Asp Thr Trp Val Glu His Trp Pro Glu Glu Asp Glu Cys Gln 1625 1630
1635Asp Glu Glu Asn Gln Lys Gln Cys Gln Asp Leu Gly Ala Phe Thr
1640 1645 1650Glu Ser Met Val Val Phe Gly Cys Pro Asn 1655
166019645PRTHomo sapiens 19Ser Pro Met Tyr Ser Ile Ile Thr Pro Asn
Ile Leu Arg Leu Glu Ser1 5 10 15Glu Glu Thr Met Val Leu Glu Ala His
Asp Ala Gln Gly Asp Val Pro 20 25 30Val Thr Val Thr Val His Asp Phe
Pro Gly Lys Lys Leu Val Leu Ser 35 40 45Ser Glu Lys Thr Val Leu Thr
Pro Ala Thr Asn His Met Gly Asn Val 50 55 60Thr Phe Thr Ile Pro Ala
Asn Arg Glu Phe Lys Ser Glu Lys Gly Arg65 70 75 80Asn Lys Phe Val
Thr Val Gln Ala Thr Phe Gly Thr Gln Val Val Glu 85 90 95Lys Val Val
Leu Val Ser Leu Gln Ser Gly Tyr Leu Phe Ile Gln Thr 100 105 110Asp
Lys Thr Ile Tyr Thr Pro Gly Ser Thr Val Leu Tyr Arg Ile Phe 115 120
125Thr Val Asn His Lys Leu Leu Pro Val Gly Arg Thr Val Met Val Asn
130 135 140Ile Glu Asn Pro Glu Gly Ile Pro Val Lys Gln Asp Ser Leu
Ser Ser145 150 155 160Gln Asn Gln Leu Gly Val Leu Pro Leu Ser Trp
Asp Ile Pro Glu Leu 165 170 175Val Asn Met Gly Gln Trp Lys Ile Arg
Ala Tyr Tyr Glu Asn Ser Pro 180 185 190Gln Gln Val Phe Ser Thr Glu
Phe Glu Val Lys Glu Tyr Val Leu Pro 195 200 205Ser Phe Glu Val Ile
Val Glu Pro Thr Glu Lys Phe Tyr Tyr Ile Tyr 210 215 220Asn Glu Lys
Gly Leu Glu Val Thr Ile Thr Ala Arg Phe Leu Tyr Gly225 230 235
240Lys Lys Val Glu Gly Thr Ala Phe Val Ile Phe Gly Ile Gln Asp Gly
245 250 255Glu Gln Arg Ile Ser Leu Pro Glu Ser Leu Lys Arg Ile Pro
Ile Glu 260 265 270Asp Gly Ser Gly Glu Val Val Leu Ser Arg Lys Val
Leu Leu Asp Gly 275 280 285Val Gln Asn Pro Arg Ala Glu Asp Leu Val
Gly Lys Ser Leu Tyr Val 290 295 300Ser Ala Thr Val Ile Leu His Ser
Gly Ser Asp Met Val Gln Ala Glu305 310 315 320Arg Ser Gly Ile Pro
Ile Val Thr Ser Pro Tyr Gln Ile His Phe Thr 325 330 335Lys Thr Pro
Lys Tyr Phe Lys Pro Gly Met Pro Phe Asp Leu Met Val 340 345 350Phe
Val Thr Asn Pro Asp Gly Ser Pro Ala Tyr Arg Val Pro Val Ala 355 360
365Val Gln Gly Glu Asp Thr Val Gln Ser Leu Thr Gln Gly Asp Gly Val
370 375 380Ala Lys Leu Ser Ile Asn Thr His Pro Ser Gln Lys Pro Leu
Ser Ile385 390 395 400Thr Val Arg Thr Lys Lys Gln Glu Leu Ser Glu
Ala Glu Gln Ala Thr 405 410 415Arg Thr Met Gln Ala Leu Pro Tyr Ser
Thr Val Gly Asn Ser Asn Asn 420 425 430Tyr Leu His Leu Ser Val Leu
Arg Thr Glu Leu Arg Pro Gly Glu Thr 435 440 445Leu Asn Val Asn Phe
Leu Leu Arg Met Asp Arg Ala His Glu Ala Lys 450 455 460Ile Arg Tyr
Tyr Thr Tyr Leu Ile Met Asn Lys Gly Arg Leu Leu Lys465 470 475
480Ala Gly Arg Gln Val Arg Glu Pro Gly Gln Asp Leu Val Val Leu Pro
485 490 495Leu Ser Ile Thr Thr Asp Phe Ile Pro Ser Phe Arg Leu Val
Ala Tyr 500 505 510Tyr Thr Leu Ile Gly Ala Ser Gly Gln Arg Glu Val
Val Ala Asp Ser 515 520 525Val Trp Val Asp Val Lys Asp Ser Cys Val
Gly Ser Leu Val Val Lys 530 535 540Ser Gly Gln Ser Glu Asp Arg Gln
Pro Val Pro Gly Gln Gln Met Thr545 550 555 560Leu Lys Ile Glu Gly
Asp His Gly Ala Arg Val Val Leu Val Ala Val 565 570 575Asp Lys Gly
Val Phe Val Leu Asn Lys Lys Asn Lys Leu Thr Gln Ser 580 585 590Lys
Ile Trp Asp Val Val Glu Lys Ala Asp Ile Gly Cys Thr Pro Gly 595 600
605Ser Gly Lys Asp Tyr Ala Gly Val Phe Ser Asp Ala Gly Leu Thr Phe
610 615 620Thr Ser Ser Ser Gly Gln Gln Thr Ala Gln Arg Ala Glu Leu
Gln Cys625 630 635 640Pro Gln Pro Ala Ala 64520915PRTHomo sapiens
20Ser Asn Leu Asp Glu Asp Ile Ile Ala Glu Glu Asn Ile Val Ser Arg1
5 10 15Ser Glu Phe Pro Glu Ser Trp Leu Trp Asn Val Glu Asp Leu Lys
Glu 20 25 30Pro Pro Lys Asn Gly Ile Ser Thr Lys Leu Met Asn Ile Phe
Leu Lys 35 40 45Asp Ser Ile Thr Thr Trp Glu Ile Leu Ala Val Ser Met
Ser Asp Lys 50 55 60Lys Gly Ile Cys Val Ala Asp Pro Phe Glu Val Thr
Val Met Gln Asp65 70 75 80Phe Phe Ile Asp Leu Arg Leu Pro Tyr Ser
Val Val Arg Asn Glu Gln 85 90 95Val Glu Ile Arg Ala Val Leu Tyr Asn
Tyr Arg Gln Asn Gln Glu Leu 100 105 110Lys Val Arg Val Glu Leu Leu
His Asn Pro Ala Phe Cys Ser Leu Ala 115 120 125Thr Thr Lys Arg Arg
His Gln Gln Thr Val Thr Ile Pro Pro Lys Ser 130 135 140Ser Leu Ser
Val Pro Tyr Val Ile Val Pro Leu Lys Thr Gly Leu Gln145 150 155
160Glu Val Glu Val Lys Ala Ala Val Tyr His His Phe Ile Ser Asp Gly
165 170 175Val Arg Lys Ser Leu Lys Val Val Pro Glu Gly Ile Arg Met
Asn Lys 180 185 190Thr Val Ala Val Arg Thr Leu Asp Pro Glu Arg Leu
Gly Arg Glu Gly 195 200 205Val Gln Lys Glu Asp Ile Pro Pro Ala Asp
Leu Ser Asp Gln Val Pro 210 215 220Asp Thr Glu Ser Glu Thr Arg Ile
Leu Leu Gln Gly Thr Pro Val Ala225 230 235 240Gln Met Thr Glu Asp
Ala Val Asp Ala Glu Arg Leu Lys His Leu Ile 245 250 255Val Thr Pro
Ser Gly Cys Gly Glu Gln Asn Met Ile Gly Met Thr Pro 260 265 270Thr
Val Ile Ala Val His Tyr Leu Asp Glu Thr Glu Gln Trp Glu Lys 275 280
285Phe Gly Leu Glu Lys Arg Gln Gly Ala Leu Glu Leu Ile Lys Lys Gly
290 295 300Tyr Thr Gln Gln Leu Ala Phe Arg Gln Pro Ser Ser Ala Phe
Ala Ala305
310 315 320Phe Val Lys Arg Ala Pro Ser Thr Trp Leu Thr Ala Tyr Val
Val Lys 325 330 335Val Phe Ser Leu Ala Val Asn Leu Ile Ala Ile Asp
Ser Gln Val Leu 340 345 350Cys Gly Ala Val Lys Trp Leu Ile Leu Glu
Lys Gln Lys Pro Asp Gly 355 360 365Val Phe Gln Glu Asp Ala Pro Val
Ile His Gln Glu Met Ile Gly Gly 370 375 380Leu Arg Asn Asn Asn Glu
Lys Asp Met Ala Leu Thr Ala Phe Val Leu385 390 395 400Ile Ser Leu
Gln Glu Ala Lys Asp Ile Cys Glu Glu Gln Val Asn Ser 405 410 415Leu
Pro Gly Ser Ile Thr Lys Ala Gly Asp Phe Leu Glu Ala Asn Tyr 420 425
430Met Asn Leu Gln Arg Ser Tyr Thr Val Ala Ile Ala Gly Tyr Ala Leu
435 440 445Ala Gln Met Gly Arg Leu Lys Gly Pro Leu Leu Asn Lys Phe
Leu Thr 450 455 460Thr Ala Lys Asp Lys Asn Arg Trp Glu Asp Pro Gly
Lys Gln Leu Tyr465 470 475 480Asn Val Glu Ala Thr Ser Tyr Ala Leu
Leu Ala Leu Leu Gln Leu Lys 485 490 495Asp Phe Asp Phe Val Pro Pro
Val Val Arg Trp Leu Asn Glu Gln Arg 500 505 510Tyr Tyr Gly Gly Gly
Tyr Gly Ser Thr Gln Ala Thr Phe Met Val Phe 515 520 525Gln Ala Leu
Ala Gln Tyr Gln Lys Asp Ala Pro Asp His Gln Glu Leu 530 535 540Asn
Leu Asp Val Ser Leu Gln Leu Pro Ser Arg Ser Ser Lys Ile Thr545 550
555 560His Arg Ile His Trp Glu Ser Ala Ser Leu Leu Arg Ser Glu Glu
Thr 565 570 575Lys Glu Asn Glu Gly Phe Thr Val Thr Ala Glu Gly Lys
Gly Gln Gly 580 585 590Thr Leu Ser Val Val Thr Met Tyr His Ala Lys
Ala Lys Asp Gln Leu 595 600 605Thr Cys Asn Lys Phe Asp Leu Lys Val
Thr Ile Lys Pro Ala Pro Glu 610 615 620Thr Glu Lys Arg Pro Gln Asp
Ala Lys Asn Thr Met Ile Leu Glu Ile625 630 635 640Cys Thr Arg Tyr
Arg Gly Asp Gln Asp Ala Thr Met Ser Ile Leu Asp 645 650 655Ile Ser
Met Met Thr Gly Phe Ala Pro Asp Thr Asp Asp Leu Lys Gln 660 665
670Leu Ala Asn Gly Val Asp Arg Tyr Ile Ser Lys Tyr Glu Leu Asp Lys
675 680 685Ala Phe Ser Asp Arg Asn Thr Leu Ile Ile Tyr Leu Asp Lys
Val Ser 690 695 700His Ser Glu Asp Asp Cys Leu Ala Phe Lys Val His
Gln Tyr Phe Asn705 710 715 720Val Glu Leu Ile Gln Pro Gly Ala Val
Lys Val Tyr Ala Tyr Tyr Asn 725 730 735Leu Glu Glu Ser Cys Thr Arg
Phe Tyr His Pro Glu Lys Glu Asp Gly 740 745 750Lys Leu Asn Lys Leu
Cys Arg Asp Glu Leu Cys Arg Cys Ala Glu Glu 755 760 765Asn Cys Phe
Ile Gln Lys Ser Asp Asp Lys Val Thr Leu Glu Glu Arg 770 775 780Leu
Asp Lys Ala Cys Glu Pro Gly Val Asp Tyr Val Tyr Lys Thr Arg785 790
795 800Leu Val Lys Val Gln Leu Ser Asn Asp Phe Asp Glu Tyr Ile Met
Ala 805 810 815Ile Glu Gln Thr Ile Lys Ser Gly Ser Asp Glu Val Gln
Val Gly Gln 820 825 830Gln Arg Thr Phe Ile Ser Pro Ile Lys Cys Arg
Glu Ala Leu Lys Leu 835 840 845Glu Glu Lys Lys His Tyr Leu Met Trp
Gly Leu Ser Ser Asp Phe Trp 850 855 860Gly Glu Lys Pro Asn Leu Ser
Tyr Ile Ile Gly Lys Asp Thr Trp Val865 870 875 880Glu His Trp Pro
Glu Glu Asp Glu Cys Gln Asp Glu Glu Asn Gln Lys 885 890 895Gln Cys
Gln Asp Leu Gly Ala Phe Thr Glu Ser Met Val Val Phe Gly 900 905
910Cys Pro Asn 9152177PRTHomo sapiens 21Ser Val Gln Leu Thr Glu Lys
Arg Met Asp Lys Val Gly Lys Tyr Pro1 5 10 15Lys Glu Leu Arg Lys Cys
Cys Glu Asp Gly Met Arg Glu Asn Pro Met 20 25 30Arg Phe Ser Cys Gln
Arg Arg Thr Arg Phe Ile Ser Leu Gly Glu Ala 35 40 45Cys Lys Lys Val
Phe Leu Asp Cys Cys Asn Tyr Ile Thr Glu Leu Arg 50 55 60Arg Gln His
Ala Arg Ala Ser His Leu Gly Leu Ala Arg65 70 7522555PRTHomo sapiens
22Ser Asn Leu Asp Glu Asp Ile Ile Ala Glu Glu Asn Ile Val Ser Arg1
5 10 15Ser Glu Phe Pro Glu Ser Trp Leu Trp Asn Val Glu Asp Leu Lys
Glu 20 25 30Pro Pro Lys Asn Gly Ile Ser Thr Lys Leu Met Asn Ile Phe
Leu Lys 35 40 45Asp Ser Ile Thr Thr Trp Glu Ile Leu Ala Val Ser Met
Ser Asp Lys 50 55 60Lys Gly Ile Cys Val Ala Asp Pro Phe Glu Val Thr
Val Met Gln Asp65 70 75 80Phe Phe Ile Asp Leu Arg Leu Pro Tyr Ser
Val Val Arg Asn Glu Gln 85 90 95Val Glu Ile Arg Ala Val Leu Tyr Asn
Tyr Arg Gln Asn Gln Glu Leu 100 105 110Lys Val Arg Val Glu Leu Leu
His Asn Pro Ala Phe Cys Ser Leu Ala 115 120 125Thr Thr Lys Arg Arg
His Gln Gln Thr Val Thr Ile Pro Pro Lys Ser 130 135 140Ser Leu Ser
Val Pro Tyr Val Ile Val Pro Leu Lys Thr Gly Leu Gln145 150 155
160Glu Val Glu Val Lys Ala Ala Val Tyr His His Phe Ile Ser Asp Gly
165 170 175Val Arg Lys Ser Leu Lys Val Val Pro Glu Gly Ile Arg Met
Asn Lys 180 185 190Thr Val Ala Val Arg Thr Leu Asp Pro Glu Arg Leu
Gly Arg Glu Gly 195 200 205Val Gln Lys Glu Asp Ile Pro Pro Ala Asp
Leu Ser Asp Gln Val Pro 210 215 220Asp Thr Glu Ser Glu Thr Arg Ile
Leu Leu Gln Gly Thr Pro Val Ala225 230 235 240Gln Met Thr Glu Asp
Ala Val Asp Ala Glu Arg Leu Lys His Leu Ile 245 250 255Val Thr Pro
Ser Gly Cys Gly Glu Gln Asn Met Ile Gly Met Thr Pro 260 265 270Thr
Val Ile Ala Val His Tyr Leu Asp Glu Thr Glu Gln Trp Glu Lys 275 280
285Phe Gly Leu Glu Lys Arg Gln Gly Ala Leu Glu Leu Ile Lys Lys Gly
290 295 300Tyr Thr Gln Gln Leu Ala Phe Arg Gln Pro Ser Ser Ala Phe
Ala Ala305 310 315 320Phe Val Lys Arg Ala Pro Ser Thr Trp Leu Thr
Ala Tyr Val Val Lys 325 330 335Val Phe Ser Leu Ala Val Asn Leu Ile
Ala Ile Asp Ser Gln Val Leu 340 345 350Cys Gly Ala Val Lys Trp Leu
Ile Leu Glu Lys Gln Lys Pro Asp Gly 355 360 365Val Phe Gln Glu Asp
Ala Pro Val Ile His Gln Glu Met Ile Gly Gly 370 375 380Leu Arg Asn
Asn Asn Glu Lys Asp Met Ala Leu Thr Ala Phe Val Leu385 390 395
400Ile Ser Leu Gln Glu Ala Lys Asp Ile Cys Glu Glu Gln Val Asn Ser
405 410 415Leu Pro Gly Ser Ile Thr Lys Ala Gly Asp Phe Leu Glu Ala
Asn Tyr 420 425 430Met Asn Leu Gln Arg Ser Tyr Thr Val Ala Ile Ala
Gly Tyr Ala Leu 435 440 445Ala Gln Met Gly Arg Leu Lys Gly Pro Leu
Leu Asn Lys Phe Leu Thr 450 455 460Thr Ala Lys Asp Lys Asn Arg Trp
Glu Asp Pro Gly Lys Gln Leu Tyr465 470 475 480Asn Val Glu Ala Thr
Ser Tyr Ala Leu Leu Ala Leu Leu Gln Leu Lys 485 490 495Asp Phe Asp
Phe Val Pro Pro Val Val Arg Trp Leu Asn Glu Gln Arg 500 505 510Tyr
Tyr Gly Gly Gly Tyr Gly Ser Thr Gln Ala Thr Phe Met Val Phe 515 520
525Gln Ala Leu Ala Gln Tyr Gln Lys Asp Ala Pro Asp His Gln Glu Leu
530 535 540Asn Leu Asp Val Ser Leu Gln Leu Pro Ser Arg545 550
55523343PRTHomo sapiens 23Ser Glu Glu Thr Lys Glu Asn Glu Gly Phe
Thr Val Thr Ala Glu Gly1 5 10 15Lys Gly Gln Gly Thr Leu Ser Val Val
Thr Met Tyr His Ala Lys Ala 20 25 30Lys Asp Gln Leu Thr Cys Asn Lys
Phe Asp Leu Lys Val Thr Ile Lys 35 40 45Pro Ala Pro Glu Thr Glu Lys
Arg Pro Gln Asp Ala Lys Asn Thr Met 50 55 60Ile Leu Glu Ile Cys Thr
Arg Tyr Arg Gly Asp Gln Asp Ala Thr Met65 70 75 80Ser Ile Leu Asp
Ile Ser Met Met Thr Gly Phe Ala Pro Asp Thr Asp 85 90 95Asp Leu Lys
Gln Leu Ala Asn Gly Val Asp Arg Tyr Ile Ser Lys Tyr 100 105 110Glu
Leu Asp Lys Ala Phe Ser Asp Arg Asn Thr Leu Ile Ile Tyr Leu 115 120
125Asp Lys Val Ser His Ser Glu Asp Asp Cys Leu Ala Phe Lys Val His
130 135 140Gln Tyr Phe Asn Val Glu Leu Ile Gln Pro Gly Ala Val Lys
Val Tyr145 150 155 160Ala Tyr Tyr Asn Leu Glu Glu Ser Cys Thr Arg
Phe Tyr His Pro Glu 165 170 175Lys Glu Asp Gly Lys Leu Asn Lys Leu
Cys Arg Asp Glu Leu Cys Arg 180 185 190Cys Ala Glu Glu Asn Cys Phe
Ile Gln Lys Ser Asp Asp Lys Val Thr 195 200 205Leu Glu Glu Arg Leu
Asp Lys Ala Cys Glu Pro Gly Val Asp Tyr Val 210 215 220Tyr Lys Thr
Arg Leu Val Lys Val Gln Leu Ser Asn Asp Phe Asp Glu225 230 235
240Tyr Ile Met Ala Ile Glu Gln Thr Ile Lys Ser Gly Ser Asp Glu Val
245 250 255Gln Val Gly Gln Gln Arg Thr Phe Ile Ser Pro Ile Lys Cys
Arg Glu 260 265 270Ala Leu Lys Leu Glu Glu Lys Lys His Tyr Leu Met
Trp Gly Leu Ser 275 280 285Ser Asp Phe Trp Gly Glu Lys Pro Asn Leu
Ser Tyr Ile Ile Gly Lys 290 295 300Asp Thr Trp Val Glu His Trp Pro
Glu Glu Asp Glu Cys Gln Asp Glu305 310 315 320Glu Asn Gln Lys Gln
Cys Gln Asp Leu Gly Ala Phe Thr Glu Ser Met 325 330 335Val Val Phe
Gly Cys Pro Asn 3402417PRTHomo sapiens 24Ser Ser Lys Ile Thr His
Arg Ile His Trp Glu Ser Ala Ser Leu Leu1 5 10 15Arg25583PRTHomo
sapiens 25Met Lys Leu Leu His Val Phe Leu Leu Phe Leu Cys Phe His
Leu Arg1 5 10 15Phe Cys Lys Val Thr Tyr Thr Ser Gln Glu Asp Leu Val
Glu Lys Lys 20 25 30Cys Leu Ala Lys Lys Tyr Thr His Leu Ser Cys Asp
Lys Val Phe Cys 35 40 45Gln Pro Trp Gln Arg Cys Ile Glu Gly Thr Cys
Val Cys Lys Leu Pro 50 55 60Tyr Gln Cys Pro Lys Asn Gly Thr Ala Val
Cys Ala Thr Asn Arg Arg65 70 75 80Ser Phe Pro Thr Tyr Cys Gln Gln
Lys Ser Leu Glu Cys Leu His Pro 85 90 95Gly Thr Lys Phe Leu Asn Asn
Gly Thr Cys Thr Ala Glu Gly Lys Phe 100 105 110Ser Val Ser Leu Lys
His Gly Asn Thr Asp Ser Glu Gly Ile Val Glu 115 120 125Val Lys Leu
Val Asp Gln Asp Lys Thr Met Phe Ile Cys Lys Ser Ser 130 135 140Trp
Ser Met Arg Glu Ala Asn Val Ala Cys Leu Asp Leu Gly Phe Gln145 150
155 160Gln Gly Ala Asp Thr Gln Arg Arg Phe Lys Leu Ser Asp Leu Ser
Ile 165 170 175Asn Ser Thr Glu Cys Leu His Val His Cys Arg Gly Leu
Glu Thr Ser 180 185 190Leu Ala Glu Cys Thr Phe Thr Lys Arg Arg Thr
Met Gly Tyr Gln Asp 195 200 205Phe Ala Asp Val Val Cys Tyr Thr Gln
Lys Ala Asp Ser Pro Met Asp 210 215 220Asp Phe Phe Gln Cys Val Asn
Gly Lys Tyr Ile Ser Gln Met Lys Ala225 230 235 240Cys Asp Gly Ile
Asn Asp Cys Gly Asp Gln Ser Asp Glu Leu Cys Cys 245 250 255Lys Ala
Cys Gln Gly Lys Gly Phe His Cys Lys Ser Gly Val Cys Ile 260 265
270Pro Ser Gln Tyr Gln Cys Asn Gly Glu Val Asp Cys Ile Thr Gly Glu
275 280 285Asp Glu Val Gly Cys Ala Gly Phe Ala Ser Val Thr Gln Glu
Glu Thr 290 295 300Glu Ile Leu Thr Ala Asp Met Asp Ala Glu Arg Arg
Arg Ile Lys Ser305 310 315 320Leu Leu Pro Lys Leu Ser Cys Gly Val
Lys Asn Arg Met His Ile Arg 325 330 335Arg Lys Arg Ile Val Gly Gly
Lys Arg Ala Gln Leu Gly Asp Leu Pro 340 345 350Trp Gln Val Ala Ile
Lys Asp Ala Ser Gly Ile Thr Cys Gly Gly Ile 355 360 365Tyr Ile Gly
Gly Cys Trp Ile Leu Thr Ala Ala His Cys Leu Arg Ala 370 375 380Ser
Lys Thr His Arg Tyr Gln Ile Trp Thr Thr Val Val Asp Trp Ile385 390
395 400His Pro Asp Leu Lys Arg Ile Val Ile Glu Tyr Val Asp Arg Ile
Ile 405 410 415Phe His Glu Asn Tyr Asn Ala Gly Thr Tyr Gln Asn Asp
Ile Ala Leu 420 425 430Ile Glu Met Lys Lys Asp Gly Asn Lys Lys Asp
Cys Glu Leu Pro Arg 435 440 445Ser Ile Pro Ala Cys Val Pro Trp Ser
Pro Tyr Leu Phe Gln Pro Asn 450 455 460Asp Thr Cys Ile Val Ser Gly
Trp Gly Arg Glu Lys Asp Asn Glu Arg465 470 475 480Val Phe Ser Leu
Gln Trp Gly Glu Val Lys Leu Ile Ser Asn Cys Ser 485 490 495Lys Phe
Tyr Gly Asn Arg Phe Tyr Glu Lys Glu Met Glu Cys Ala Gly 500 505
510Thr Tyr Asp Gly Ser Ile Asp Ala Cys Lys Gly Asp Ser Gly Gly Pro
515 520 525Leu Val Cys Met Asp Ala Asn Asn Val Thr Tyr Val Trp Gly
Val Val 530 535 540Ser Trp Gly Glu Asn Cys Gly Lys Pro Glu Phe Pro
Gly Val Tyr Thr545 550 555 560Lys Val Ala Asn Tyr Phe Asp Trp Ile
Ser Tyr His Val Gly Arg Pro 565 570 575Phe Ile Ser Gln Tyr Asn Val
58026235PRTHomo sapiens 26Ile Val Gly Gly Lys Arg Ala Gln Leu Gly
Asp Leu Pro Trp Gln Val1 5 10 15Ala Ile Lys Asp Ala Ser Gly Ile Thr
Cys Gly Gly Ile Tyr Ile Gly 20 25 30Gly Cys Trp Ile Leu Thr Ala Ala
His Cys Leu Arg Ala Ser Lys Thr 35 40 45His Arg Tyr Gln Ile Trp Thr
Thr Val Val Asp Trp Ile His Pro Asp 50 55 60Leu Lys Arg Ile Val Ile
Glu Tyr Val Asp Arg Ile Ile Phe His Glu65 70 75 80Asn Tyr Asn Ala
Gly Thr Tyr Gln Asn Asp Ile Ala Leu Ile Glu Met 85 90 95Lys Lys Asp
Gly Asn Lys Lys Asp Cys Glu Leu Pro Arg Ser Ile Pro 100 105 110Ala
Cys Val Pro Trp Ser Pro Tyr Leu Phe Gln Pro Asn Asp Thr Cys 115 120
125Ile Val Ser Gly Trp Gly Arg Glu Lys Asp Asn Glu Arg Val Phe Ser
130 135 140Leu Gln Trp Gly Glu Val Lys Leu Ile Ser Asn Cys Ser Lys
Phe Tyr145 150 155 160Gly Asn Arg Phe Tyr Glu Lys Glu Met Glu Cys
Ala Gly Thr Tyr Asp 165 170 175Gly Ser Ile Asp Ala Cys Lys Gly Asp
Ser Gly Gly Pro Leu Val Cys 180 185 190Met Asp Ala Asn Asn Val Thr
Tyr Val Trp Gly Val Val Ser Trp Gly 195 200 205Glu Asn Cys Gly Lys
Pro Glu Phe Pro Gly Val Tyr Thr Lys Val Ala 210 215 220Asn Tyr Phe
Asp Trp Ile Ser Tyr His Val Gly225 230 235273PRTArtificial
SequenceSynthetic constructMISC_FEATURE(2)..(2)Xaa = any amino acid
except for ProMISC_FEATURE(3)..(3)Xaa = Ser or Thr 27Asn Xaa
Xaa128449PRTArtificial SequenceSynthetic construct 28Met Arg Leu
Leu Ala Lys Ile Ile Cys Leu Met Leu Trp Ala Ile Cys1 5 10 15Val Ala
Glu Asp Cys Asn Glu Leu Pro Pro
Arg Arg Asn Thr Glu Ile 20 25 30Leu Thr Gly Ser Trp Ser Asp Gln Thr
Tyr Pro Glu Gly Thr Gln Ala 35 40 45Ile Tyr Lys Cys Arg Pro Gly Tyr
Arg Ser Leu Gly Asn Val Ile Met 50 55 60Val Cys Arg Lys Gly Glu Trp
Val Ala Leu Asn Pro Leu Arg Lys Cys65 70 75 80Gln Lys Arg Pro Cys
Gly His Pro Gly Asp Thr Pro Phe Gly Thr Phe 85 90 95Thr Leu Thr Gly
Gly Asn Val Phe Glu Tyr Gly Val Lys Ala Val Tyr 100 105 110Thr Cys
Asn Glu Gly Tyr Gln Leu Leu Gly Glu Ile Asn Tyr Arg Glu 115 120
125Cys Asp Thr Asp Gly Trp Thr Asn Asp Ile Pro Ile Cys Glu Val Val
130 135 140Lys Cys Leu Pro Val Thr Ala Pro Glu Asn Gly Lys Ile Val
Ser Ser145 150 155 160Ala Met Glu Pro Asp Arg Glu Tyr His Phe Gly
Gln Ala Val Arg Phe 165 170 175Val Cys Asn Ser Gly Tyr Lys Ile Glu
Gly Asp Glu Glu Met His Cys 180 185 190Ser Asp Asp Gly Phe Trp Ser
Lys Glu Lys Pro Lys Cys Val Glu Ile 195 200 205Ser Cys Lys Ser Pro
Asp Val Ile Asn Gly Ser Pro Ile Ser Gln Lys 210 215 220Ile Ile Tyr
Lys Glu Asn Glu Arg Phe Gln Tyr Lys Cys Asn Met Gly225 230 235
240Tyr Glu Tyr Ser Glu Arg Gly Asp Ala Val Cys Thr Glu Ser Gly Trp
245 250 255Arg Pro Leu Pro Ser Cys Glu Glu Lys Ser Cys Asp Asn Pro
Tyr Ile 260 265 270Pro Asn Gly Asp Tyr Ser Pro Leu Arg Ile Lys His
Arg Thr Gly Asp 275 280 285Glu Ile Thr Tyr Gln Cys Arg Asn Gly Phe
Tyr Pro Ala Thr Arg Gly 290 295 300Asn Thr Ala Lys Cys Thr Ser Thr
Gly Trp Ile Pro Ala Pro Arg Cys305 310 315 320Thr Leu Lys Pro Cys
Asp Tyr Pro Asp Ile Lys His Gly Gly Leu Tyr 325 330 335His Glu Asn
Met Arg Arg Pro Tyr Phe Pro Val Ala Val Gly Lys Tyr 340 345 350Tyr
Ser Tyr Tyr Cys Asp Glu His Phe Glu Thr Pro Ser Gly Ser Tyr 355 360
365Trp Asp His Ile His Cys Thr Gln Asp Gly Trp Ser Pro Ala Val Pro
370 375 380Cys Leu Arg Lys Cys Tyr Phe Pro Tyr Leu Glu Asn Gly Tyr
Asn Gln385 390 395 400Asn Tyr Gly Arg Lys Phe Val Gln Gly Lys Ser
Ile Asp Val Ala Cys 405 410 415His Pro Gly Tyr Ala Leu Pro Lys Ala
Gln Thr Thr Val Thr Cys Met 420 425 430Glu Asn Gly Trp Ser Pro Thr
Pro Arg Cys Ile Arg Val Ser Phe Thr 435 440 445Leu291231PRTHomo
sapiens 29Met Arg Leu Leu Ala Lys Ile Ile Cys Leu Met Leu Trp Ala
Ile Cys1 5 10 15Val Ala Glu Asp Cys Asn Glu Leu Pro Pro Arg Arg Asn
Thr Glu Ile 20 25 30Leu Thr Gly Ser Trp Ser Asp Gln Thr Tyr Pro Glu
Gly Thr Gln Ala 35 40 45Ile Tyr Lys Cys Arg Pro Gly Tyr Arg Ser Leu
Gly Asn Val Ile Met 50 55 60Val Cys Arg Lys Gly Glu Trp Val Ala Leu
Asn Pro Leu Arg Lys Cys65 70 75 80Gln Lys Arg Pro Cys Gly His Pro
Gly Asp Thr Pro Phe Gly Thr Phe 85 90 95Thr Leu Thr Gly Gly Asn Val
Phe Glu Tyr Gly Val Lys Ala Val Tyr 100 105 110Thr Cys Asn Glu Gly
Tyr Gln Leu Leu Gly Glu Ile Asn Tyr Arg Glu 115 120 125Cys Asp Thr
Asp Gly Trp Thr Asn Asp Ile Pro Ile Cys Glu Val Val 130 135 140Lys
Cys Leu Pro Val Thr Ala Pro Glu Asn Gly Lys Ile Val Ser Ser145 150
155 160Ala Met Glu Pro Asp Arg Glu Tyr His Phe Gly Gln Ala Val Arg
Phe 165 170 175Val Cys Asn Ser Gly Tyr Lys Ile Glu Gly Asp Glu Glu
Met His Cys 180 185 190Ser Asp Asp Gly Phe Trp Ser Lys Glu Lys Pro
Lys Cys Val Glu Ile 195 200 205Ser Cys Lys Ser Pro Asp Val Ile Asn
Gly Ser Pro Ile Ser Gln Lys 210 215 220Ile Ile Tyr Lys Glu Asn Glu
Arg Phe Gln Tyr Lys Cys Asn Met Gly225 230 235 240Tyr Glu Tyr Ser
Glu Arg Gly Asp Ala Val Cys Thr Glu Ser Gly Trp 245 250 255Arg Pro
Leu Pro Ser Cys Glu Glu Lys Ser Cys Asp Asn Pro Tyr Ile 260 265
270Pro Asn Gly Asp Tyr Ser Pro Leu Arg Ile Lys His Arg Thr Gly Asp
275 280 285Glu Ile Thr Tyr Gln Cys Arg Asn Gly Phe Tyr Pro Ala Thr
Arg Gly 290 295 300Asn Thr Ala Lys Cys Thr Ser Thr Gly Trp Ile Pro
Ala Pro Arg Cys305 310 315 320Thr Leu Lys Pro Cys Asp Tyr Pro Asp
Ile Lys His Gly Gly Leu Tyr 325 330 335His Glu Asn Met Arg Arg Pro
Tyr Phe Pro Val Ala Val Gly Lys Tyr 340 345 350Tyr Ser Tyr Tyr Cys
Asp Glu His Phe Glu Thr Pro Ser Gly Ser Tyr 355 360 365Trp Asp His
Ile His Cys Thr Gln Asp Gly Trp Ser Pro Ala Val Pro 370 375 380Cys
Leu Arg Lys Cys Tyr Phe Pro Tyr Leu Glu Asn Gly Tyr Asn Gln385 390
395 400Asn Tyr Gly Arg Lys Phe Val Gln Gly Lys Ser Ile Asp Val Ala
Cys 405 410 415His Pro Gly Tyr Ala Leu Pro Lys Ala Gln Thr Thr Val
Thr Cys Met 420 425 430Glu Asn Gly Trp Ser Pro Thr Pro Arg Cys Ile
Arg Val Lys Thr Cys 435 440 445Ser Lys Ser Ser Ile Asp Ile Glu Asn
Gly Phe Ile Ser Glu Ser Gln 450 455 460Tyr Thr Tyr Ala Leu Lys Glu
Lys Ala Lys Tyr Gln Cys Lys Leu Gly465 470 475 480Tyr Val Thr Ala
Asp Gly Glu Thr Ser Gly Ser Ile Thr Cys Gly Lys 485 490 495Asp Gly
Trp Ser Ala Gln Pro Thr Cys Ile Lys Ser Cys Asp Ile Pro 500 505
510Val Phe Met Asn Ala Arg Thr Lys Asn Asp Phe Thr Trp Phe Lys Leu
515 520 525Asn Asp Thr Leu Asp Tyr Glu Cys His Asp Gly Tyr Glu Ser
Asn Thr 530 535 540Gly Ser Thr Thr Gly Ser Ile Val Cys Gly Tyr Asn
Gly Trp Ser Asp545 550 555 560Leu Pro Ile Cys Tyr Glu Arg Glu Cys
Glu Leu Pro Lys Ile Asp Val 565 570 575His Leu Val Pro Asp Arg Lys
Lys Asp Gln Tyr Lys Val Gly Glu Val 580 585 590Leu Lys Phe Ser Cys
Lys Pro Gly Phe Thr Ile Val Gly Pro Asn Ser 595 600 605Val Gln Cys
Tyr His Phe Gly Leu Ser Pro Asp Leu Pro Ile Cys Lys 610 615 620Glu
Gln Val Gln Ser Cys Gly Pro Pro Pro Glu Leu Leu Asn Gly Asn625 630
635 640Val Lys Glu Lys Thr Lys Glu Glu Tyr Gly His Ser Glu Val Val
Glu 645 650 655Tyr Tyr Cys Asn Pro Arg Phe Leu Met Lys Gly Pro Asn
Lys Ile Gln 660 665 670Cys Val Asp Gly Glu Trp Thr Thr Leu Pro Val
Cys Ile Val Glu Glu 675 680 685Ser Thr Cys Gly Asp Ile Pro Glu Leu
Glu His Gly Trp Ala Gln Leu 690 695 700Ser Ser Pro Pro Tyr Tyr Tyr
Gly Asp Ser Val Glu Phe Asn Cys Ser705 710 715 720Glu Ser Phe Thr
Met Ile Gly His Arg Ser Ile Thr Cys Ile His Gly 725 730 735Val Trp
Thr Gln Leu Pro Gln Cys Val Ala Ile Asp Lys Leu Lys Lys 740 745
750Cys Lys Ser Ser Asn Leu Ile Ile Leu Glu Glu His Leu Lys Asn Lys
755 760 765Lys Glu Phe Asp His Asn Ser Asn Ile Arg Tyr Arg Cys Arg
Gly Lys 770 775 780Glu Gly Trp Ile His Thr Val Cys Ile Asn Gly Arg
Trp Asp Pro Glu785 790 795 800Val Asn Cys Ser Met Ala Gln Ile Gln
Leu Cys Pro Pro Pro Pro Gln 805 810 815Ile Pro Asn Ser His Asn Met
Thr Thr Thr Leu Asn Tyr Arg Asp Gly 820 825 830Glu Lys Val Ser Val
Leu Cys Gln Glu Asn Tyr Leu Ile Gln Glu Gly 835 840 845Glu Glu Ile
Thr Cys Lys Asp Gly Arg Trp Gln Ser Ile Pro Leu Cys 850 855 860Val
Glu Lys Ile Pro Cys Ser Gln Pro Pro Gln Ile Glu His Gly Thr865 870
875 880Ile Asn Ser Ser Arg Ser Ser Gln Glu Ser Tyr Ala His Gly Thr
Lys 885 890 895Leu Ser Tyr Thr Cys Glu Gly Gly Phe Arg Ile Ser Glu
Glu Asn Glu 900 905 910Thr Thr Cys Tyr Met Gly Lys Trp Ser Ser Pro
Pro Gln Cys Glu Gly 915 920 925Leu Pro Cys Lys Ser Pro Pro Glu Ile
Ser His Gly Val Val Ala His 930 935 940Met Ser Asp Ser Tyr Gln Tyr
Gly Glu Glu Val Thr Tyr Lys Cys Phe945 950 955 960Glu Gly Phe Gly
Ile Asp Gly Pro Ala Ile Ala Lys Cys Leu Gly Glu 965 970 975Lys Trp
Ser His Pro Pro Ser Cys Ile Lys Thr Asp Cys Leu Ser Leu 980 985
990Pro Ser Phe Glu Asn Ala Ile Pro Met Gly Glu Lys Lys Asp Val Tyr
995 1000 1005Lys Ala Gly Glu Gln Val Thr Tyr Thr Cys Ala Thr Tyr
Tyr Lys 1010 1015 1020Met Asp Gly Ala Ser Asn Val Thr Cys Ile Asn
Ser Arg Trp Thr 1025 1030 1035Gly Arg Pro Thr Cys Arg Asp Thr Ser
Cys Val Asn Pro Pro Thr 1040 1045 1050Val Gln Asn Ala Tyr Ile Val
Ser Arg Gln Met Ser Lys Tyr Pro 1055 1060 1065Ser Gly Glu Arg Val
Arg Tyr Gln Cys Arg Ser Pro Tyr Glu Met 1070 1075 1080Phe Gly Asp
Glu Glu Val Met Cys Leu Asn Gly Asn Trp Thr Glu 1085 1090 1095Pro
Pro Gln Cys Lys Asp Ser Thr Gly Lys Cys Gly Pro Pro Pro 1100 1105
1110Pro Ile Asp Asn Gly Asp Ile Thr Ser Phe Pro Leu Ser Val Tyr
1115 1120 1125Ala Pro Ala Ser Ser Val Glu Tyr Gln Cys Gln Asn Leu
Tyr Gln 1130 1135 1140Leu Glu Gly Asn Lys Arg Ile Thr Cys Arg Asn
Gly Gln Trp Ser 1145 1150 1155Glu Pro Pro Lys Cys Leu His Pro Cys
Val Ile Ser Arg Glu Ile 1160 1165 1170Met Glu Asn Tyr Asn Ile Ala
Leu Arg Trp Thr Ala Lys Gln Lys 1175 1180 1185Leu Tyr Ser Arg Thr
Gly Glu Ser Val Glu Phe Val Cys Lys Arg 1190 1195 1200Gly Tyr Arg
Leu Ser Ser Arg Ser His Thr Leu Arg Thr Thr Cys 1205 1210 1215Trp
Asp Gly Lys Leu Glu Tyr Pro Thr Cys Ala Lys Arg 1220 1225
123030644PRTHomo sapiens 30Gly His Cys Gln Ala Pro Asp His Phe Leu
Phe Ala Lys Leu Lys Thr1 5 10 15Gln Thr Asn Ala Ser Asp Phe Pro Ile
Gly Thr Ser Leu Lys Tyr Glu 20 25 30Cys Arg Pro Glu Tyr Tyr Gly Arg
Pro Phe Ser Ile Thr Cys Leu Asp 35 40 45Asn Leu Val Trp Ser Ser Pro
Lys Asp Val Cys Lys Arg Lys Ser Cys 50 55 60Lys Thr Pro Pro Asp Pro
Val Asn Gly Met Val His Val Ile Thr Asp65 70 75 80Ile Gln Val Gly
Ser Arg Ile Asn Tyr Ser Cys Thr Thr Gly His Arg 85 90 95Leu Ile Gly
His Ser Ser Ala Glu Cys Ile Leu Ser Gly Asn Ala Ala 100 105 110His
Trp Ser Thr Lys Pro Pro Ile Cys Gln Arg Ile Pro Cys Gly Leu 115 120
125Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile Ser Thr Asn Arg Glu Asn
130 135 140Phe His Tyr Gly Ser Val Val Thr Tyr Arg Cys Asn Pro Gly
Ser Gly145 150 155 160Gly Arg Lys Val Phe Glu Leu Val Gly Glu Pro
Ser Ile Tyr Cys Thr 165 170 175Ser Asn Asp Asp Gln Val Gly Ile Trp
Ser Gly Pro Ala Pro Gln Cys 180 185 190Ile Ile Pro Asn Lys Cys Thr
Pro Pro Asn Val Glu Asn Gly Ile Leu 195 200 205Val Ser Asp Asn Arg
Ser Leu Phe Ser Leu Asn Glu Val Val Glu Phe 210 215 220Arg Cys Gln
Pro Gly Phe Val Met Lys Gly Pro Arg Arg Val Lys Cys225 230 235
240Gln Ala Leu Asn Lys Trp Glu Pro Glu Leu Pro Ser Cys Ser Arg Val
245 250 255Cys Gln Pro Pro Pro Asp Val Leu His Ala Glu Arg Thr Gln
Arg Asp 260 265 270Lys Asp Asn Phe Ser Pro Gly Gln Glu Val Phe Tyr
Ser Cys Glu Pro 275 280 285Gly Tyr Asp Leu Arg Gly Ala Ala Ser Met
Arg Cys Thr Pro Gln Gly 290 295 300Asp Trp Ser Pro Ala Ala Pro Thr
Cys Glu Val Lys Ser Cys Asp Asp305 310 315 320Phe Met Gly Gln Leu
Leu Asn Gly Arg Val Leu Phe Pro Val Asn Leu 325 330 335Gln Leu Gly
Ala Lys Val Asp Phe Val Cys Asp Glu Gly Phe Gln Leu 340 345 350Lys
Gly Ser Ser Ala Ser Tyr Cys Val Leu Ala Gly Met Glu Ser Leu 355 360
365Trp Asn Ser Ser Val Pro Val Cys Glu Gln Ile Phe Cys Pro Ser Pro
370 375 380Pro Val Ile Pro Asn Gly Arg His Thr Gly Lys Pro Leu Glu
Val Phe385 390 395 400Pro Phe Gly Lys Ala Val Asn Tyr Thr Cys Asp
Pro His Pro Asp Arg 405 410 415Gly Thr Ser Phe Asp Leu Ile Gly Glu
Ser Thr Ile Arg Cys Thr Ser 420 425 430Asp Pro Gln Gly Asn Gly Val
Trp Ser Ser Pro Ala Pro Arg Cys Gly 435 440 445Ile Leu Gly His Cys
Gln Ala Pro Asp His Phe Leu Phe Ala Lys Leu 450 455 460Lys Thr Gln
Thr Asn Ala Ser Asp Phe Pro Ile Gly Thr Ser Leu Lys465 470 475
480Tyr Glu Cys Arg Pro Glu Tyr Tyr Gly Arg Pro Phe Ser Ile Thr Cys
485 490 495Leu Asp Asn Leu Val Trp Ser Ser Pro Lys Asp Val Cys Lys
Arg Lys 500 505 510Ser Cys Lys Thr Pro Pro Asp Pro Val Asn Gly Met
Val His Val Ile 515 520 525Thr Asp Ile Gln Val Gly Ser Arg Ile Asn
Tyr Ser Cys Thr Thr Gly 530 535 540His Arg Leu Ile Gly His Ser Ser
Ala Glu Cys Ile Leu Ser Gly Asn545 550 555 560Thr Ala His Trp Ser
Thr Lys Pro Pro Ile Cys Gln Arg Ile Pro Cys 565 570 575Gly Leu Pro
Pro Thr Ile Ala Asn Gly Asp Phe Ile Ser Thr Asn Arg 580 585 590Glu
Asn Phe His Tyr Gly Ser Val Val Thr Tyr Arg Cys Asn Leu Gly 595 600
605Ser Arg Gly Arg Lys Val Phe Glu Leu Val Gly Glu Pro Ser Ile Tyr
610 615 620Cys Thr Ser Asn Asp Asp Gln Val Gly Ile Trp Ser Gly Pro
Ala Pro625 630 635 640Gln Cys Ile Ile31644PRTHomo sapiens 31Gly His
Cys Gln Ala Pro Asp His Phe Leu Phe Ala Lys Leu Lys Thr1 5 10 15Gln
Thr Gln Ala Ser Asp Phe Pro Ile Gly Thr Ser Leu Lys Tyr Glu 20 25
30Cys Arg Pro Glu Tyr Tyr Gly Arg Pro Phe Ser Ile Thr Cys Leu Asp
35 40 45Asn Leu Val Trp Ser Ser Pro Lys Asp Val Cys Lys Arg Lys Ser
Cys 50 55 60Lys Thr Pro Pro Asp Pro Val Asn Gly Met Val His Val Ile
Thr Asp65 70 75 80Ile Gln Val Gly Ser Arg Ile Gln Tyr Ser Cys Thr
Thr Gly His Arg 85 90 95Leu Ile Gly His Ser Ser Ala Glu Cys Ile Leu
Ser Gly Asn Ala Ala 100 105 110His Trp Ser Thr Lys Pro Pro Ile Cys
Gln Arg Ile Pro Cys Gly Leu 115 120 125Pro Pro Thr Ile Ala Asn Gly
Asp Phe Ile Ser Thr Asn Arg Glu Asn 130 135 140Phe His Tyr Gly Ser
Val Val Thr Tyr Arg Cys Asn Pro Gly Ser Gly145 150 155 160Gly Arg
Lys Val Phe Glu Leu Val Gly Glu
Pro Ser Ile Tyr Cys Thr 165 170 175Ser Asn Asp Asp Gln Val Gly Ile
Trp Ser Gly Pro Ala Pro Gln Cys 180 185 190Ile Ile Pro Asn Lys Cys
Thr Pro Pro Asn Val Glu Asn Gly Ile Leu 195 200 205Val Ser Asp Asn
Arg Ser Leu Phe Ser Leu Asn Glu Val Val Glu Phe 210 215 220Arg Cys
Gln Pro Gly Phe Val Met Lys Gly Pro Arg Arg Val Lys Cys225 230 235
240Gln Ala Leu Asn Lys Trp Glu Pro Glu Leu Pro Ser Cys Ser Arg Val
245 250 255Cys Gln Pro Pro Pro Asp Val Leu His Ala Glu Arg Thr Gln
Arg Asp 260 265 270Lys Asp Asn Phe Ser Pro Gly Gln Glu Val Phe Tyr
Ser Cys Glu Pro 275 280 285Gly Tyr Asp Leu Arg Gly Ala Ala Ser Met
Arg Cys Thr Pro Gln Gly 290 295 300Asp Trp Ser Pro Ala Ala Pro Thr
Cys Glu Val Lys Ser Cys Asp Asp305 310 315 320Phe Met Gly Gln Leu
Leu Asn Gly Arg Val Leu Phe Pro Val Asn Leu 325 330 335Gln Leu Gly
Ala Lys Val Asp Phe Val Cys Asp Glu Gly Phe Gln Leu 340 345 350Lys
Gly Ser Ser Ala Ser Tyr Cys Val Leu Ala Gly Met Glu Ser Leu 355 360
365Trp Asn Ser Ser Val Pro Val Cys Glu Gln Ile Phe Cys Pro Ser Pro
370 375 380Pro Val Ile Pro Asn Gly Arg His Thr Gly Lys Pro Leu Glu
Val Phe385 390 395 400Pro Phe Gly Lys Ala Val Asn Tyr Thr Cys Asp
Pro His Pro Asp Arg 405 410 415Gly Thr Ser Phe Asp Leu Ile Gly Glu
Ser Thr Ile Arg Cys Thr Ser 420 425 430Asp Pro Gln Gly Asn Gly Val
Trp Ser Ser Pro Ala Pro Arg Cys Gly 435 440 445Ile Leu Gly His Cys
Gln Ala Pro Asp His Phe Leu Phe Ala Lys Leu 450 455 460Lys Thr Gln
Thr Gln Ala Ser Asp Phe Pro Ile Gly Thr Ser Leu Lys465 470 475
480Tyr Glu Cys Arg Pro Glu Tyr Tyr Gly Arg Pro Phe Ser Ile Thr Cys
485 490 495Leu Asp Asn Leu Val Trp Ser Ser Pro Lys Asp Val Cys Lys
Arg Lys 500 505 510Ser Cys Lys Thr Pro Pro Asp Pro Val Asn Gly Met
Val His Val Ile 515 520 525Thr Asp Ile Gln Val Gly Ser Arg Ile Gln
Tyr Ser Cys Thr Thr Gly 530 535 540His Arg Leu Ile Gly His Ser Ser
Ala Glu Cys Ile Leu Ser Gly Asn545 550 555 560Thr Ala His Trp Ser
Thr Lys Pro Pro Ile Cys Gln Arg Ile Pro Cys 565 570 575Gly Leu Pro
Pro Thr Ile Ala Asn Gly Asp Phe Ile Ser Thr Asn Arg 580 585 590Glu
Asn Phe His Tyr Gly Ser Val Val Thr Tyr Arg Cys Asn Leu Gly 595 600
605Ser Arg Gly Arg Lys Val Phe Glu Leu Val Gly Glu Pro Ser Ile Tyr
610 615 620Cys Thr Ser Asn Asp Asp Gln Val Gly Ile Trp Ser Gly Pro
Ala Pro625 630 635 640Gln Cys Ile Ile3225PRTHomo sapiens 32Ala Leu
Ile Val Gly Thr Leu Ser Gly Thr Ile Phe Phe Ile Leu Leu1 5 10 15Ile
Ile Phe Leu Ser Trp Ile Ile Leu 20 253343PRTHomo sapiens 33Lys His
Arg Lys Gly Asn Asn Ala His Glu Asn Pro Lys Glu Val Ala1 5 10 15Ile
His Leu His Ser Gln Gly Gly Ser Ser Val His Pro Arg Thr Leu 20 25
30Gln Thr Asn Glu Glu Asn Ser Arg Val Leu Pro 35 40348PRTArtificial
SequenceSynthetic construct 34Glu Asn Leu Tyr Phe Gln Gly Ser1
535657DNAHomo sapiens 35aagcttgcca ccatgagact gctggccaag atcatctgcc
tgatgctgtg ggccatctgc 60gtggccggac attgtcaggc ccctgaccac ttcctgttcg
ccaagctgaa aacccagacc 120aacgccagcg acttccctat cggcaccagc
ctgaagtacg agtgcagacc cgagtactac 180ggcagaccct tcagcatcac
ctgtctggac aacctcgtgt ggtctagccc caaggacgtg 240tgcaagagaa
agagctgcaa gacccctcct gatcctgtga acggcatggt gcacgtgatc
300accgacatcc aagtgggcag cagaatcaac tacagctgca ccaccggcca
cagactgatc 360ggacactcta gcgccgagtg tatcctgagc ggcaatgccg
cacactggtc caccaagcct 420ccaatctgcc agagaatccc ttgcggcctg
cctcctacaa tcgccaacgg cgatttcatc 480agcaccaaca gagagaactt
ccactacggc tccgtggtca cctacagatg caatcctggc 540agcggcggca
gaaaggtgtt cgaacttgtg ggcgagccca gcatctactg caccagcaac
600gatgaccaag tcggcatttg gagcggccct gctcctcagt gcatcatcta agatatc
65736657DNAHomo sapiens 36aagcttgcca ccatgagact gctggccaag
atcatctgcc tgatgctgtg ggccatctgc 60gtggccggac attgtcaggc ccctgaccac
ttcctgttcg ccaagctgaa aacccagaca 120caggccagcg acttccctat
cggcaccagc ctgaagtacg agtgcagacc cgagtactac 180ggcagaccct
tcagcatcac ctgtctggac aacctcgtgt ggtctagccc caaggacgtg
240tgcaagagaa agagctgcaa gacccctcct gatcctgtga acggcatggt
gcacgtgatc 300accgacatcc aagtgggcag cagaatccag tacagctgca
ccacaggcca cagactgatc 360ggccactcta gcgccgagtg tatcctgtct
ggcaatgccg ctcactggtc caccaagcct 420ccaatctgcc agagaatccc
ttgcggcctg cctcctacaa tcgccaacgg cgatttcatc 480agcaccaaca
gagagaactt ccactacggc tccgtggtca cctacagatg caatcctggc
540agcggcggca gaaaggtgtt cgaacttgtg ggcgagccca gcatctactg
caccagcaac 600gatgaccaag tcggcatttg gagcggccct gctcctcagt
gcatcatcta agatatc 65737659DNAHomo sapiens 37aagcttgcca ccatgagact
gctggccaag atcatctgcc tgatgctgtg ggccatctgc 60gtggccggcc actgtcaggc
ccctgatcac ttcctgttcg ccaagctgaa aacccagacc 120aacgccagcg
acttccctat cggcaccagc ctgaagtacg agtgcagacc cgagtactac
180ggcagaccct tcagcatcac ctgtctggac aacctcgtgt ggtctagccc
caaggacgtg 240tgcaagagaa agagctgcaa gacccctcct gatcctgtga
acggcatggt gcacgtgatc 300accgacatcc aagtgggcag cagaatcaac
tacagctgca ccaccggcca cagactgatc 360ggacactcta gcgccgagtg
tatcctgagc ggcaatgccg cacactggtc caccaagcct 420ccaatctgcc
agagaatccc ttgcggcctg cctcctacaa tcgccaacgg cgatttcatc
480agcaccaaca gagagaactt ccactacggc tccgtggtca cctacagatg
caatcctggc 540agcggcggca gaaaggtgtt cgaacttgtg ggcgagccca
gcatctactg caccagcaac 600gatgaccaag tcggcatttg gagcggccct
gctcctcagt gcatcatccc taagatatc 65938659DNAHomo sapiens
38aagcttgcca ccatgagact gctggccaag atcatctgcc tgatgctgtg ggccatctgc
60gtggccggcc actgtcaggc ccctgatcac ttcctgttcg ccaagctgaa aacccagaca
120caggccagcg acttccctat cggcaccagc ctgaagtacg agtgcagacc
cgagtactac 180ggcagaccct tcagcatcac ctgtctggac aacctcgtgt
ggtctagccc caaggacgtg 240tgcaagagaa agagctgcaa gacccctcct
gatcctgtga acggcatggt gcacgtgatc 300accgacatcc aagtgggcag
cagaatccag tacagctgca ccacaggcca cagactgatc 360ggccactcta
gcgccgagtg tatcctgagc ggaaacacag cccactggtc caccaagcct
420ccaatctgcc agagaatccc ttgcggcctg cctcctacaa tcgccaacgg
cgatttcatc 480agcaccaaca gagagaactt ccactacggc tccgtggtca
cctacagatg caacctgggc 540tccagaggcc ggaaggtgtt cgaacttgtg
ggcgagccta gcatctactg caccagcaac 600gacgaccaag tcggcatttg
gagcggacct gctcctcagt gcatcatccc taagatatc 65939714DNAHomo sapiens
39aagcttgcca ccatgagact gctggccaag atcatctgcc tgatgctgtg ggccatctgc
60gtggcccacc accatcacca tcacggcagc agcgagaacc tgtacttcca aggatcttct
120ggcggccact gtcaggcccc tgatcacttc ctgttcgcca agctgaaaac
ccagaccaac 180gccagcgact tccctatcgg caccagcctg aagtacgagt
gcagacccga gtactacggc 240agacccttca gcatcacctg tctggacaac
ctcgtgtggt ctagccccaa ggacgtgtgc 300aagagaaaga gctgcaagac
ccctcctgat cctgtgaacg gcatggtgca cgtgatcacc 360gacatccaag
tgggcagcag aatcaactac agctgcacca ccggccacag actgatcgga
420cactctagcg ccgagtgtat cctgagcggc aatgccgcac actggtccac
caagcctcca 480atctgccaga gaatcccttg cggcctgcct cctacaatcg
ccaacggcga tttcatcagc 540accaacagag agaacttcca ctacggctcc
gtggtcacct acagatgcaa tcctggcagc 600ggcggcagaa aggtgttcga
acttgtgggc gagcccagca tctactgcac cagcaacgat 660gaccaagtcg
gcatttggag cggccctgct cctcagtgca tcatctaaga tatc 71440231PRTHomo
sapiens 40Met Arg Leu Leu Ala Lys Ile Ile Cys Leu Met Leu Trp Ala
Ile Cys1 5 10 15Val Ala His His His His His His Gly Ser Ser Glu Asn
Leu Tyr Phe 20 25 30Gln Gly Ser Ser Gly Gly His Cys Gln Ala Pro Asp
His Phe Leu Phe 35 40 45Ala Lys Leu Lys Thr Gln Thr Asn Ala Ser Asp
Phe Pro Ile Gly Thr 50 55 60Ser Leu Lys Tyr Glu Cys Arg Pro Glu Tyr
Tyr Gly Arg Pro Phe Ser65 70 75 80Ile Thr Cys Leu Asp Asn Leu Val
Trp Ser Ser Pro Lys Asp Val Cys 85 90 95Lys Arg Lys Ser Cys Lys Thr
Pro Pro Asp Pro Val Asn Gly Met Val 100 105 110His Val Ile Thr Asp
Ile Gln Val Gly Ser Arg Ile Asn Tyr Ser Cys 115 120 125Thr Thr Gly
His Arg Leu Ile Gly His Ser Ser Ala Glu Cys Ile Leu 130 135 140Ser
Gly Asn Ala Ala His Trp Ser Thr Lys Pro Pro Ile Cys Gln Arg145 150
155 160Ile Pro Cys Gly Leu Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile
Ser 165 170 175Thr Asn Arg Glu Asn Phe His Tyr Gly Ser Val Val Thr
Tyr Arg Cys 180 185 190Asn Pro Gly Ser Gly Gly Arg Lys Val Phe Glu
Leu Val Gly Glu Pro 195 200 205Ser Ile Tyr Cys Thr Ser Asn Asp Asp
Gln Val Gly Ile Trp Ser Gly 210 215 220Pro Ala Pro Gln Cys Ile
Ile225 23041714DNAHomo sapiens 41aagcttgcca ccatgagact gctggccaag
atcatctgcc tgatgctgtg ggccatctgc 60gtggcccacc accatcacca tcacggcagc
agcgagaacc tgtacttcca aggatcttct 120ggcggccact gtcaggcccc
tgatcacttc ctgttcgcca agctgaaaac ccagacacag 180gccagcgact
tccctatcgg caccagcctg aagtacgagt gcagacccga gtactacggc
240agacccttca gcatcacctg tctggacaac ctcgtgtggt ctagccccaa
ggacgtgtgc 300aagagaaaga gctgcaagac ccctcctgat cctgtgaacg
gcatggtgca cgtgatcacc 360gacatccaag tgggcagcag aatccagtac
agctgcacca caggccacag actgatcggc 420cactctagcg ccgagtgtat
cctgtctggc aatgccgctc actggtccac caagcctcca 480atctgccaga
gaatcccttg cggcctgcct cctacaatcg ccaacggcga tttcatcagc
540accaacagag agaacttcca ctacggctcc gtggtcacct acagatgcaa
tcctggcagc 600ggcggcagaa aggtgttcga acttgtgggc gagcccagca
tctactgcac cagcaacgat 660gaccaagtcg gcatttggag cggccctgct
cctcagtgca tcatctaaga tatc 71442231PRTHomo sapiens 42Met Arg Leu
Leu Ala Lys Ile Ile Cys Leu Met Leu Trp Ala Ile Cys1 5 10 15Val Ala
His His His His His His Gly Ser Ser Glu Asn Leu Tyr Phe 20 25 30Gln
Gly Ser Ser Gly Gly His Cys Gln Ala Pro Asp His Phe Leu Phe 35 40
45Ala Lys Leu Lys Thr Gln Thr Gln Ala Ser Asp Phe Pro Ile Gly Thr
50 55 60Ser Leu Lys Tyr Glu Cys Arg Pro Glu Tyr Tyr Gly Arg Pro Phe
Ser65 70 75 80Ile Thr Cys Leu Asp Asn Leu Val Trp Ser Ser Pro Lys
Asp Val Cys 85 90 95Lys Arg Lys Ser Cys Lys Thr Pro Pro Asp Pro Val
Asn Gly Met Val 100 105 110His Val Ile Thr Asp Ile Gln Val Gly Ser
Arg Ile Gln Tyr Ser Cys 115 120 125Thr Thr Gly His Arg Leu Ile Gly
His Ser Ser Ala Glu Cys Ile Leu 130 135 140Ser Gly Asn Ala Ala His
Trp Ser Thr Lys Pro Pro Ile Cys Gln Arg145 150 155 160Ile Pro Cys
Gly Leu Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile Ser 165 170 175Thr
Asn Arg Glu Asn Phe His Tyr Gly Ser Val Val Thr Tyr Arg Cys 180 185
190Asn Pro Gly Ser Gly Gly Arg Lys Val Phe Glu Leu Val Gly Glu Pro
195 200 205Ser Ile Tyr Cys Thr Ser Asn Asp Asp Gln Val Gly Ile Trp
Ser Gly 210 215 220Pro Ala Pro Gln Cys Ile Ile225 23043716DNAHomo
sapiens 43aagcttgcca ccatgagact gctggccaag atcatctgcc tgatgctgtg
ggccatctgc 60gtggcccacc accatcacca tcacggcagc agcgagaacc tgtacttcca
aggatcttct 120ggcggccact gtcaggcccc tgatcacttc ctgttcgcca
agctgaaaac ccagaccaac 180gccagcgact tccctatcgg caccagcctg
aagtacgagt gcagacccga gtactacggc 240agacccttca gcatcacctg
tctggacaac ctcgtgtggt ctagccccaa ggacgtgtgc 300aagagaaaga
gctgcaagac ccctcctgat cctgtgaacg gcatggtgca cgtgatcacc
360gacatccaag tgggcagcag aatcaactac agctgcacca ccggccacag
actgatcgga 420cactctagcg ccgagtgtat cctgagcggc aatgccgcac
actggtccac caagcctcca 480atctgccaga gaatcccttg cggcctgcct
cctacaatcg ccaacggcga tttcatcagc 540accaacagag agaacttcca
ctacggctcc gtggtcacct acagatgcaa tcctggcagc 600ggcggcagaa
aggtgttcga acttgtgggc gagcccagca tctactgcac cagcaacgat
660gaccaagtcg gcatttggag cggccctgct cctcagtgca tcatccctaa gatatc
71644231PRTHomo sapiens 44Met Arg Leu Leu Ala Lys Ile Ile Cys Leu
Met Leu Trp Ala Ile Cys1 5 10 15Val Ala His His His His His His Gly
Ser Ser Glu Asn Leu Tyr Phe 20 25 30Gln Gly Ser Ser Gly Gly His Cys
Gln Ala Pro Asp His Phe Leu Phe 35 40 45Ala Lys Leu Lys Thr Gln Thr
Asn Ala Ser Asp Phe Pro Ile Gly Thr 50 55 60Ser Leu Lys Tyr Glu Cys
Arg Pro Glu Tyr Tyr Gly Arg Pro Phe Ser65 70 75 80Ile Thr Cys Leu
Asp Asn Leu Val Trp Ser Ser Pro Lys Asp Val Cys 85 90 95Lys Arg Lys
Ser Cys Lys Thr Pro Pro Asp Pro Val Asn Gly Met Val 100 105 110His
Val Ile Thr Asp Ile Gln Val Gly Ser Arg Ile Asn Tyr Ser Cys 115 120
125Thr Thr Gly His Arg Leu Ile Gly His Ser Ser Ala Glu Cys Ile Leu
130 135 140Ser Gly Asn Thr Ala His Trp Ser Thr Lys Pro Pro Ile Cys
Gln Arg145 150 155 160Ile Pro Cys Gly Leu Pro Pro Thr Ile Ala Asn
Gly Asp Phe Ile Ser 165 170 175Thr Asn Arg Glu Asn Phe His Tyr Gly
Ser Val Val Thr Tyr Arg Cys 180 185 190Asn Leu Gly Ser Arg Gly Arg
Lys Val Phe Glu Leu Val Gly Glu Pro 195 200 205Ser Ile Tyr Cys Thr
Ser Asn Asp Asp Gln Val Gly Ile Trp Ser Gly 210 215 220Pro Ala Pro
Gln Cys Ile Ile225 23045716DNAHomo sapiens 45aagcttgcca ccatgagact
gctggccaag atcatctgcc tgatgctgtg ggccatctgc 60gtggcccacc accatcacca
tcacggcagc agcgagaacc tgtacttcca aggatcttct 120ggcggccact
gtcaggcccc tgatcacttc ctgttcgcca agctgaaaac ccagacacag
180gccagcgact tccctatcgg caccagcctg aagtacgagt gcagacccga
gtactacggc 240agacccttca gcatcacctg tctggacaac ctcgtgtggt
ctagccccaa ggacgtgtgc 300aagagaaaga gctgcaagac ccctcctgat
cctgtgaacg gcatggtgca cgtgatcacc 360gacatccaag tgggcagcag
aatccagtac agctgcacca caggccacag actgatcggc 420cactctagcg
ccgagtgtat cctgagcgga aacacagccc actggtccac caagcctcca
480atctgccaga gaatcccttg cggcctgcct cctacaatcg ccaacggcga
tttcatcagc 540accaacagag agaacttcca ctacggctcc gtggtcacct
acagatgcaa cctgggctcc 600agaggccgga aggtgttcga acttgtgggc
gagcctagca tctactgcac cagcaacgac 660gaccaagtcg gcatttggag
cggacctgct cctcagtgca tcatccctaa gatatc 71646231PRTHomo sapiens
46Met Arg Leu Leu Ala Lys Ile Ile Cys Leu Met Leu Trp Ala Ile Cys1
5 10 15Val Ala His His His His His His Gly Ser Ser Glu Asn Leu Tyr
Phe 20 25 30Gln Gly Ser Ser Gly Gly His Cys Gln Ala Pro Asp His Phe
Leu Phe 35 40 45Ala Lys Leu Lys Thr Gln Thr Gln Ala Ser Asp Phe Pro
Ile Gly Thr 50 55 60Ser Leu Lys Tyr Glu Cys Arg Pro Glu Tyr Tyr Gly
Arg Pro Phe Ser65 70 75 80Ile Thr Cys Leu Asp Asn Leu Val Trp Ser
Ser Pro Lys Asp Val Cys 85 90 95Lys Arg Lys Ser Cys Lys Thr Pro Pro
Asp Pro Val Asn Gly Met Val 100 105 110His Val Ile Thr Asp Ile Gln
Val Gly Ser Arg Ile Gln Tyr Ser Cys 115 120 125Thr Thr Gly His Arg
Leu Ile Gly His Ser Ser Ala Glu Cys Ile Leu 130 135 140Ser Gly Asn
Ala Ala His Trp Ser Thr Lys Pro Pro Ile Cys Gln Arg145 150 155
160Ile Pro Cys Gly Leu Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile Ser
165 170 175Thr Asn Arg Glu Asn Phe His Tyr Gly Ser Val Val Thr Tyr
Arg Cys 180 185 190Asn Pro Gly Ser Gly Gly Arg Lys Val Phe Glu Leu
Val Gly Glu Pro 195 200 205Ser Ile Tyr Cys Thr Ser Asn Asp Asp Gln
Val Gly Ile Trp Ser Gly 210 215 220Pro Ala Pro Gln Cys Ile Ile225
23047212PRTArtificial SequenceSynthetic construct 47Met Arg Leu Leu
Ala Lys Ile Ile Cys Leu Met Leu Trp Ala Ile Cys1 5 10 15Val Ala Gly
His Cys Gln Ala Pro Asp His Phe Leu Phe Ala Lys Leu 20 25 30Lys Thr
Gln Thr Asn Ala Ser Asp Phe Pro
Ile Gly Thr Ser Leu Lys 35 40 45Tyr Glu Cys Arg Pro Glu Tyr Tyr Gly
Arg Pro Phe Ser Ile Thr Cys 50 55 60Leu Asp Asn Leu Val Trp Ser Ser
Pro Lys Asp Val Cys Lys Arg Lys65 70 75 80Ser Cys Lys Thr Pro Pro
Asp Pro Val Asn Gly Met Val His Val Ile 85 90 95Thr Asp Ile Gln Val
Gly Ser Arg Ile Asn Tyr Ser Cys Thr Thr Gly 100 105 110His Arg Leu
Ile Gly His Ser Ser Ala Glu Cys Ile Leu Ser Gly Asn 115 120 125Ala
Ala His Trp Ser Thr Lys Pro Pro Ile Cys Gln Arg Ile Pro Cys 130 135
140Gly Leu Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile Ser Thr Asn
Arg145 150 155 160Glu Asn Phe His Tyr Gly Ser Val Val Thr Tyr Arg
Cys Asn Pro Gly 165 170 175Ser Gly Gly Arg Lys Val Phe Glu Leu Val
Gly Glu Pro Ser Ile Tyr 180 185 190Cys Thr Ser Asn Asp Asp Gln Val
Gly Ile Trp Ser Gly Pro Ala Pro 195 200 205Gln Cys Ile Ile
21048212PRTArtificial SequenceSynthetic construct 48Met Arg Leu Leu
Ala Lys Ile Ile Cys Leu Met Leu Trp Ala Ile Cys1 5 10 15Val Ala Gly
His Cys Gln Ala Pro Asp His Phe Leu Phe Ala Lys Leu 20 25 30Lys Thr
Gln Thr Gln Ala Ser Asp Phe Pro Ile Gly Thr Ser Leu Lys 35 40 45Tyr
Glu Cys Arg Pro Glu Tyr Tyr Gly Arg Pro Phe Ser Ile Thr Cys 50 55
60Leu Asp Asn Leu Val Trp Ser Ser Pro Lys Asp Val Cys Lys Arg Lys65
70 75 80Ser Cys Lys Thr Pro Pro Asp Pro Val Asn Gly Met Val His Val
Ile 85 90 95Thr Asp Ile Gln Val Gly Ser Arg Ile Gln Tyr Ser Cys Thr
Thr Gly 100 105 110His Arg Leu Ile Gly His Ser Ser Ala Glu Cys Ile
Leu Ser Gly Asn 115 120 125Ala Ala His Trp Ser Thr Lys Pro Pro Ile
Cys Gln Arg Ile Pro Cys 130 135 140Gly Leu Pro Pro Thr Ile Ala Asn
Gly Asp Phe Ile Ser Thr Asn Arg145 150 155 160Glu Asn Phe His Tyr
Gly Ser Val Val Thr Tyr Arg Cys Asn Pro Gly 165 170 175Ser Gly Gly
Arg Lys Val Phe Glu Leu Val Gly Glu Pro Ser Ile Tyr 180 185 190Cys
Thr Ser Asn Asp Asp Gln Val Gly Ile Trp Ser Gly Pro Ala Pro 195 200
205Gln Cys Ile Ile 21049212PRTArtificial SequenceSynthetic
construct 49Met Arg Leu Leu Ala Lys Ile Ile Cys Leu Met Leu Trp Ala
Ile Cys1 5 10 15Val Ala Gly His Cys Gln Ala Pro Asp His Phe Leu Phe
Ala Lys Leu 20 25 30Lys Thr Gln Thr Asn Ala Ser Asp Phe Pro Ile Gly
Thr Ser Leu Lys 35 40 45Tyr Glu Cys Arg Pro Glu Tyr Tyr Gly Arg Pro
Phe Ser Ile Thr Cys 50 55 60Leu Asp Asn Leu Val Trp Ser Ser Pro Lys
Asp Val Cys Lys Arg Lys65 70 75 80Ser Cys Lys Thr Pro Pro Asp Pro
Val Asn Gly Met Val His Val Ile 85 90 95Thr Asp Ile Gln Val Gly Ser
Arg Ile Asn Tyr Ser Cys Thr Thr Gly 100 105 110His Arg Leu Ile Gly
His Ser Ser Ala Glu Cys Ile Leu Ser Gly Asn 115 120 125Thr Ala His
Trp Ser Thr Lys Pro Pro Ile Cys Gln Arg Ile Pro Cys 130 135 140Gly
Leu Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile Ser Thr Asn Arg145 150
155 160Glu Asn Phe His Tyr Gly Ser Val Val Thr Tyr Arg Cys Asn Leu
Gly 165 170 175Ser Arg Gly Arg Lys Val Phe Glu Leu Val Gly Glu Pro
Ser Ile Tyr 180 185 190Cys Thr Ser Asn Asp Asp Gln Val Gly Ile Trp
Ser Gly Pro Ala Pro 195 200 205Gln Cys Ile Ile
21050212PRTArtificial SequenceSynthetic construct 50Met Arg Leu Leu
Ala Lys Ile Ile Cys Leu Met Leu Trp Ala Ile Cys1 5 10 15Val Ala Gly
His Cys Gln Ala Pro Asp His Phe Leu Phe Ala Lys Leu 20 25 30Lys Thr
Gln Thr Gln Ala Ser Asp Phe Pro Ile Gly Thr Ser Leu Lys 35 40 45Tyr
Glu Cys Arg Pro Glu Tyr Tyr Gly Arg Pro Phe Ser Ile Thr Cys 50 55
60Leu Asp Asn Leu Val Trp Ser Ser Pro Lys Asp Val Cys Lys Arg Lys65
70 75 80Ser Cys Lys Thr Pro Pro Asp Pro Val Asn Gly Met Val His Val
Ile 85 90 95Thr Asp Ile Gln Val Gly Ser Arg Ile Gln Tyr Ser Cys Thr
Thr Gly 100 105 110His Arg Leu Ile Gly His Ser Ser Ala Glu Cys Ile
Leu Ser Gly Asn 115 120 125Thr Ala His Trp Ser Thr Lys Pro Pro Ile
Cys Gln Arg Ile Pro Cys 130 135 140Gly Leu Pro Pro Thr Ile Ala Asn
Gly Asp Phe Ile Ser Thr Asn Arg145 150 155 160Glu Asn Phe His Tyr
Gly Ser Val Val Thr Tyr Arg Cys Asn Leu Gly 165 170 175Ser Arg Gly
Arg Lys Val Phe Glu Leu Val Gly Glu Pro Ser Ile Tyr 180 185 190Cys
Thr Ser Asn Asp Asp Gln Val Gly Ile Trp Ser Gly Pro Ala Pro 195 200
205Gln Cys Ile Ile 21051406PRTArtificial SequenceSynthetic
construct 51Met Arg Leu Leu Ala Lys Ile Ile Cys Leu Met Leu Trp Ala
Ile Cys1 5 10 15Val Ala Gly His Cys Gln Ala Pro Asp His Phe Leu Phe
Ala Lys Leu 20 25 30Lys Thr Gln Thr Asn Ala Ser Asp Phe Pro Ile Gly
Thr Ser Leu Lys 35 40 45Tyr Glu Cys Arg Pro Glu Tyr Tyr Gly Arg Pro
Phe Ser Ile Thr Cys 50 55 60Leu Asp Asn Leu Val Trp Ser Ser Pro Lys
Asp Val Cys Lys Arg Lys65 70 75 80Ser Cys Lys Thr Pro Pro Asp Pro
Val Asn Gly Met Val His Val Ile 85 90 95Thr Asp Ile Gln Val Gly Ser
Arg Ile Asn Tyr Ser Cys Thr Thr Gly 100 105 110His Arg Leu Ile Gly
His Ser Ser Ala Glu Cys Ile Leu Ser Gly Asn 115 120 125Ala Ala His
Trp Ser Thr Lys Pro Pro Ile Cys Gln Arg Ile Pro Cys 130 135 140Gly
Leu Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile Ser Thr Asn Arg145 150
155 160Glu Asn Phe His Tyr Gly Ser Val Val Thr Tyr Arg Cys Asn Pro
Gly 165 170 175Ser Gly Gly Arg Lys Val Phe Glu Leu Val Gly Glu Pro
Ser Ile Tyr 180 185 190Cys Thr Ser Asn Asp Asp Gln Val Gly Ile Trp
Ser Gly Pro Ala Pro 195 200 205Gln Cys Ile Ile Gly His Cys Gln Ala
Pro Asp His Phe Leu Phe Ala 210 215 220Lys Leu Lys Thr Gln Thr Asn
Ala Ser Asp Phe Pro Ile Gly Thr Ser225 230 235 240Leu Lys Tyr Glu
Cys Arg Pro Glu Tyr Tyr Gly Arg Pro Phe Ser Ile 245 250 255Thr Cys
Leu Asp Asn Leu Val Trp Ser Ser Pro Lys Asp Val Cys Lys 260 265
270Arg Lys Ser Cys Lys Thr Pro Pro Asp Pro Val Asn Gly Met Val His
275 280 285Val Ile Thr Asp Ile Gln Val Gly Ser Arg Ile Asn Tyr Ser
Cys Thr 290 295 300Thr Gly His Arg Leu Ile Gly His Ser Ser Ala Glu
Cys Ile Leu Ser305 310 315 320Gly Asn Thr Ala His Trp Ser Thr Lys
Pro Pro Ile Cys Gln Arg Ile 325 330 335Pro Cys Gly Leu Pro Pro Thr
Ile Ala Asn Gly Asp Phe Ile Ser Thr 340 345 350Asn Arg Glu Asn Phe
His Tyr Gly Ser Val Val Thr Tyr Arg Cys Asn 355 360 365Leu Gly Ser
Arg Gly Arg Lys Val Phe Glu Leu Val Gly Glu Pro Ser 370 375 380Ile
Tyr Cys Thr Ser Asn Asp Asp Gln Val Gly Ile Trp Ser Gly Pro385 390
395 400Ala Pro Gln Cys Ile Ile 40552406PRTArtificial
SequenceSynthetic construct 52Met Arg Leu Leu Ala Lys Ile Ile Cys
Leu Met Leu Trp Ala Ile Cys1 5 10 15Val Ala Gly His Cys Gln Ala Pro
Asp His Phe Leu Phe Ala Lys Leu 20 25 30Lys Thr Gln Thr Gln Ala Ser
Asp Phe Pro Ile Gly Thr Ser Leu Lys 35 40 45Tyr Glu Cys Arg Pro Glu
Tyr Tyr Gly Arg Pro Phe Ser Ile Thr Cys 50 55 60Leu Asp Asn Leu Val
Trp Ser Ser Pro Lys Asp Val Cys Lys Arg Lys65 70 75 80Ser Cys Lys
Thr Pro Pro Asp Pro Val Asn Gly Met Val His Val Ile 85 90 95Thr Asp
Ile Gln Val Gly Ser Arg Ile Gln Tyr Ser Cys Thr Thr Gly 100 105
110His Arg Leu Ile Gly His Ser Ser Ala Glu Cys Ile Leu Ser Gly Asn
115 120 125Ala Ala His Trp Ser Thr Lys Pro Pro Ile Cys Gln Arg Ile
Pro Cys 130 135 140Gly Leu Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile
Ser Thr Asn Arg145 150 155 160Glu Asn Phe His Tyr Gly Ser Val Val
Thr Tyr Arg Cys Asn Pro Gly 165 170 175Ser Gly Gly Arg Lys Val Phe
Glu Leu Val Gly Glu Pro Ser Ile Tyr 180 185 190Cys Thr Ser Asn Asp
Asp Gln Val Gly Ile Trp Ser Gly Pro Ala Pro 195 200 205Gln Cys Ile
Ile Gly His Cys Gln Ala Pro Asp His Phe Leu Phe Ala 210 215 220Lys
Leu Lys Thr Gln Thr Gln Ala Ser Asp Phe Pro Ile Gly Thr Ser225 230
235 240Leu Lys Tyr Glu Cys Arg Pro Glu Tyr Tyr Gly Arg Pro Phe Ser
Ile 245 250 255Thr Cys Leu Asp Asn Leu Val Trp Ser Ser Pro Lys Asp
Val Cys Lys 260 265 270Arg Lys Ser Cys Lys Thr Pro Pro Asp Pro Val
Asn Gly Met Val His 275 280 285Val Ile Thr Asp Ile Gln Val Gly Ser
Arg Ile Gln Tyr Ser Cys Thr 290 295 300Thr Gly His Arg Leu Ile Gly
His Ser Ser Ala Glu Cys Ile Leu Ser305 310 315 320Gly Asn Thr Ala
His Trp Ser Thr Lys Pro Pro Ile Cys Gln Arg Ile 325 330 335Pro Cys
Gly Leu Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile Ser Thr 340 345
350Asn Arg Glu Asn Phe His Tyr Gly Ser Val Val Thr Tyr Arg Cys Asn
355 360 365Leu Gly Ser Arg Gly Arg Lys Val Phe Glu Leu Val Gly Glu
Pro Ser 370 375 380Ile Tyr Cys Thr Ser Asn Asp Asp Gln Val Gly Ile
Trp Ser Gly Pro385 390 395 400Ala Pro Gln Cys Ile Ile
40553406PRTArtificial SequenceSynthetic construct 53Met Arg Leu Leu
Ala Lys Ile Ile Cys Leu Met Leu Trp Ala Ile Cys1 5 10 15Val Ala Gly
His Cys Gln Ala Pro Asp His Phe Leu Phe Ala Lys Leu 20 25 30Lys Thr
Gln Thr Asn Ala Ser Asp Phe Pro Ile Gly Thr Ser Leu Lys 35 40 45Tyr
Glu Cys Arg Pro Glu Tyr Tyr Gly Arg Pro Phe Ser Ile Thr Cys 50 55
60Leu Asp Asn Leu Val Trp Ser Ser Pro Lys Asp Val Cys Lys Arg Lys65
70 75 80Ser Cys Lys Thr Pro Pro Asp Pro Val Asn Gly Met Val His Val
Ile 85 90 95Thr Asp Ile Gln Val Gly Ser Arg Ile Asn Tyr Ser Cys Thr
Thr Gly 100 105 110His Arg Leu Ile Gly His Ser Ser Ala Glu Cys Ile
Leu Ser Gly Asn 115 120 125Ala Ala His Trp Ser Thr Lys Pro Pro Ile
Cys Gln Arg Ile Pro Cys 130 135 140Gly Leu Pro Pro Thr Ile Ala Asn
Gly Asp Phe Ile Ser Thr Asn Arg145 150 155 160Glu Asn Phe His Tyr
Gly Ser Val Val Thr Tyr Arg Cys Asn Pro Gly 165 170 175Ser Gly Gly
Arg Lys Val Phe Glu Leu Val Gly Glu Pro Ser Ile Tyr 180 185 190Cys
Thr Ser Asn Asp Asp Gln Val Gly Ile Trp Ser Gly Pro Ala Pro 195 200
205Gln Cys Ile Ile Gly His Cys Gln Ala Pro Asp His Phe Leu Phe Ala
210 215 220Lys Leu Lys Thr Gln Thr Gln Ala Ser Asp Phe Pro Ile Gly
Thr Ser225 230 235 240Leu Lys Tyr Glu Cys Arg Pro Glu Tyr Tyr Gly
Arg Pro Phe Ser Ile 245 250 255Thr Cys Leu Asp Asn Leu Val Trp Ser
Ser Pro Lys Asp Val Cys Lys 260 265 270Arg Lys Ser Cys Lys Thr Pro
Pro Asp Pro Val Asn Gly Met Val His 275 280 285Val Ile Thr Asp Ile
Gln Val Gly Ser Arg Ile Gln Tyr Ser Cys Thr 290 295 300Thr Gly His
Arg Leu Ile Gly His Ser Ser Ala Glu Cys Ile Leu Ser305 310 315
320Gly Asn Thr Ala His Trp Ser Thr Lys Pro Pro Ile Cys Gln Arg Ile
325 330 335Pro Cys Gly Leu Pro Pro Thr Ile Ala Asn Gly Asp Phe Ile
Ser Thr 340 345 350Asn Arg Glu Asn Phe His Tyr Gly Ser Val Val Thr
Tyr Arg Cys Asn 355 360 365Leu Gly Ser Arg Gly Arg Lys Val Phe Glu
Leu Val Gly Glu Pro Ser 370 375 380Ile Tyr Cys Thr Ser Asn Asp Asp
Gln Val Gly Ile Trp Ser Gly Pro385 390 395 400Ala Pro Gln Cys Ile
Ile 40554406PRTArtificial SequenceSynthetic construct 54Met Arg Leu
Leu Ala Lys Ile Ile Cys Leu Met Leu Trp Ala Ile Cys1 5 10 15Val Ala
Gly His Cys Gln Ala Pro Asp His Phe Leu Phe Ala Lys Leu 20 25 30Lys
Thr Gln Thr Gln Ala Ser Asp Phe Pro Ile Gly Thr Ser Leu Lys 35 40
45Tyr Glu Cys Arg Pro Glu Tyr Tyr Gly Arg Pro Phe Ser Ile Thr Cys
50 55 60Leu Asp Asn Leu Val Trp Ser Ser Pro Lys Asp Val Cys Lys Arg
Lys65 70 75 80Ser Cys Lys Thr Pro Pro Asp Pro Val Asn Gly Met Val
His Val Ile 85 90 95Thr Asp Ile Gln Val Gly Ser Arg Ile Gln Tyr Ser
Cys Thr Thr Gly 100 105 110His Arg Leu Ile Gly His Ser Ser Ala Glu
Cys Ile Leu Ser Gly Asn 115 120 125Ala Ala His Trp Ser Thr Lys Pro
Pro Ile Cys Gln Arg Ile Pro Cys 130 135 140Gly Leu Pro Pro Thr Ile
Ala Asn Gly Asp Phe Ile Ser Thr Asn Arg145 150 155 160Glu Asn Phe
His Tyr Gly Ser Val Val Thr Tyr Arg Cys Asn Pro Gly 165 170 175Ser
Gly Gly Arg Lys Val Phe Glu Leu Val Gly Glu Pro Ser Ile Tyr 180 185
190Cys Thr Ser Asn Asp Asp Gln Val Gly Ile Trp Ser Gly Pro Ala Pro
195 200 205Gln Cys Ile Ile Gly His Cys Gln Ala Pro Asp His Phe Leu
Phe Ala 210 215 220Lys Leu Lys Thr Gln Thr Asn Ala Ser Asp Phe Pro
Ile Gly Thr Ser225 230 235 240Leu Lys Tyr Glu Cys Arg Pro Glu Tyr
Tyr Gly Arg Pro Phe Ser Ile 245 250 255Thr Cys Leu Asp Asn Leu Val
Trp Ser Ser Pro Lys Asp Val Cys Lys 260 265 270Arg Lys Ser Cys Lys
Thr Pro Pro Asp Pro Val Asn Gly Met Val His 275 280 285Val Ile Thr
Asp Ile Gln Val Gly Ser Arg Ile Asn Tyr Ser Cys Thr 290 295 300Thr
Gly His Arg Leu Ile Gly His Ser Ser Ala Glu Cys Ile Leu Ser305 310
315 320Gly Asn Thr Ala His Trp Ser Thr Lys Pro Pro Ile Cys Gln Arg
Ile 325 330 335Pro Cys Gly Leu Pro Pro Thr Ile Ala Asn Gly Asp Phe
Ile Ser Thr 340 345 350Asn Arg Glu Asn Phe His Tyr Gly Ser Val Val
Thr Tyr Arg Cys Asn 355 360 365Leu Gly Ser Arg Gly Arg Lys Val Phe
Glu Leu Val Gly Glu Pro Ser 370 375 380Ile Tyr Cys Thr Ser Asn Asp
Asp Gln Val Gly Ile Trp Ser Gly Pro385 390 395 400Ala Pro Gln Cys
Ile Ile 405
D00000

D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014

D00015

D00016

D00017

P00001

P00002
P00003
P00004
P00005
P00006
P00007

P00008
P00009

P00010

P00011

P00012

P00013

P00014
P00015

P00016

P00017

P00018

P00019

P00020

P00021
P00022

P00023
P00024
P00025

P00026

P00027
P00028

P00029
P00030

P00031
P00032
P00033
P00034
P00035

P00036

P00037

P00038

P00039
P00040

P00041

P00042

P00043

P00044

P00045

P00046

P00047

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.