Anti-pd-1 Antibodies, Compositions Comprising Anti-pd-1 Antibodies And Methods Of Using Anti-pd-1 Antibodies
YAM; Alice ; et al.
U.S. patent application number 17/068477 was filed with the patent office on 2021-04-15 for anti-pd-1 antibodies, compositions comprising anti-pd-1 antibodies and methods of using anti-pd-1 antibodies. The applicant listed for this patent is SUTRO BIOPHARMA, INC.. Invention is credited to Avinash GILL, John LEE, Aaron SATO, Ryan STAFFORD, Heather STEPHENSON, Alice YAM, Junhao YANG.
| Application Number | 20210107981 17/068477 |
| Document ID | / |
| Family ID | 1000005291731 |
| Filed Date | 2021-04-15 |



| United States Patent Application | 20210107981 |
| Kind Code | A1 |
| YAM; Alice ; et al. | April 15, 2021 |
ANTI-PD-1 ANTIBODIES, COMPOSITIONS COMPRISING ANTI-PD-1 ANTIBODIES AND METHODS OF USING ANTI-PD-1 ANTIBODIES
Abstract
Provided herein are antibodies that selectively bind to PD-1 and its isoforms and homologs, and compositions comprising the antibodies. Also provided are methods of using the antibodies, such as therapeutic and diagnostic methods.
| Inventors: | YAM; Alice; (Tiburon, CA) ; STAFFORD; Ryan; (Emeryville, CA) ; SATO; Aaron; (Burlingame, CA) ; LEE; John; (San Francisco, CA) ; GILL; Avinash; (Emeryville, CA) ; YANG; Junhao; (Palo Alto, CA) ; STEPHENSON; Heather; (San Jose, CA) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 1000005291731 | ||||||||||
| Appl. No.: | 17/068477 | ||||||||||
| Filed: | October 12, 2020 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 15525943 | May 10, 2017 | 10822414 | ||
| PCT/US2015/060033 | Nov 10, 2015 | |||
| 17068477 | ||||
| 62078115 | Nov 11, 2014 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C07K 2317/24 20130101; C07K 2317/34 20130101; C07K 2317/33 20130101; C07K 2317/92 20130101; C07K 2317/41 20130101; C07K 16/2818 20130101; A61P 37/00 20180101; A61K 2039/505 20130101; C07K 2317/76 20130101; A61P 35/00 20180101; C07K 2317/94 20130101; C07K 2317/622 20130101 |
| International Class: | C07K 16/28 20060101 C07K016/28; A61P 37/00 20060101 A61P037/00; A61P 35/00 20060101 A61P035/00 |
Claims
1.-20. (canceled)
21.-67. (canceled)
68. A method of treating disease in a subject, comprising administering an effective amount of an antibody conjugate, or a pharmaceutical composition comprising the same, wherein the antibody conjugate comprises an antibody or antibody fragment of the IgG class comprising: three heavy chain CDRs of the V.sub.H region SEQ ID NO: 252, or a variant thereof, and hree light chain CDRs of the V.sub.L region SEQ ID NO: 276, or a variant thereof.
69. A method of diagnosing a disease in a subject, comprising administering an effective amount of an antibody conjugate, or a pharmaceutical composition comprising the same, wherein the antibody conjugate comprises an antibody or antibody fragment of the IgG class comprising: three heavy chain CDRs of the V.sub.H region SEQ ID NO: 252, or a variant thereof, and three light chain CDRs of the V.sub.L region SEQ ID NO: 276, or a variant thereof.
70. The method of claim 68, wherein the disease is cancer, an autoimmune disease or condition, infection, or any combination thereof.
71. The method of claim 68, wherein the pharmaceutical composition comprises the antibody or antibody fragment of claim 68 and a pharmaceutically acceptable carrier.
72. The method of claim 68, wherein the pharmaceutical composition is substantially pure.
73. The method of claim 72, wherein the pharmaceutical composition comprises an antibody that is at least 95% by mass of the total antibody or antibody fragment mass of said composition.
74. The method of claim 68, wherein the pharmaceutical composition is administered intramuscularly, intradermally, intraperitoneally, intravenously, subcutaneously administration, or any combination thereof.
75. The method of claim 68, wherein the antibody or antibody fragment comprises: a CDR-H1 comprising SEQ ID NO: 13; a CDR-H2 comprising SEQ ID NO: 66; a CDR-H3 comprising SEQ ID NO: 119; a CDR-L1 comprising SEQ ID NO: 148; a CDR-L2 comprising SEQ ID NO: 177; and a CDR-L3 comprising SEQ ID NO: 206; or b. a CDR-H1 comprising SEQ ID NO: 37; a CDR-H2 comprising SEQ ID NO: 90; a CDR-H3 comprising SEQ ID NO: 119; a CDR-L1 comprising SEQ ID NO: 148; a CDR-L2 comprising SEQ ID NO: 177; and a CDR-L3 comprising SEQ ID NO: 206.
76. The method of claim 68, wherein the variant of the V.sub.H and V.sub.L regions have 20 or fewer amino acid substitutions, and wherein the substitutions are conservative amino acid substitutions.
77. The method of claim 68, wherein the antibody or antibody fragment further comprises at least one constant region domain.
78. The antibody of claim 77, wherein the constant region domain comprises a sequence selected from SEQ ID NOs: 224-226 and 297.
79. The method of claim 68, wherein the antibody or antibody fragment is a monoclonal antibody.
80. The method of claim 68, wherein the antibody or antibody fragment is aglycosylated.
81. The method of claim 68, wherein the antibody fragment is selected from an Fv fragment, a Fab fragment, a F(ab').sub.2 fragment, a Fab' fragment, an scFv (sFv) fragment, and an scFv-Fc fragment.
82. The antibody fragment of claim 81, wherein the antibody fragment is an scFv fragment.
83. The antibody fragment of claim 81, wherein the antibody fragment is an scFv-Fc fragment.
84. The antibody fragment of claim 83, wherein the scFv-Fc fragment comprises a sequence selected from SEQ ID NO: 243 with AAGSDQEPK (SEQ ID NO: 301) removed from the sequence.
85. The method of claim 68, wherein the antibody or antibody fragment has a k.sub.a of about 4.74.times.10.sup.4 M.sup.-1.times.sec.sup.-1 to about 1.23.times.10.sup.6 M.sup.-1.times.sec.sup.-1 when associating with human PD-1 at a temperature of 25.degree. C.
86. The method of claim 68, wherein the antibody or antibody fragment has a k.sub.d of about 1.87.times.10.sup.-2 sec.sup.-1 to about 4.17.times.10.sup.-4 sec.sup.-1 when dissociating from human PD-1 at a temperature of 25.degree. C.
87. The method of claim 68, wherein the antibody or antibody fragment has a K.sub.D of about 3.85.times.10.sup.-8 M to about 2.52.times.10.sup.-10 M when bound to human PD-1 at a temperature of 25.degree. C.
88. The method of claim 68, wherein the antibody or antibody fragment specifically binds one or more of murine PD-1 and cynomolgus PD-1.
Description
FIELD
[0001] Provided herein are antibodies with binding specificity for PD-1 and compositions comprising the antibodies, including pharmaceutical compositions, diagnostic compositions and kits. Also provided are methods of using anti-PD-1 antibodies for therapeutic and diagnostic purposes.
BACKGROUND
[0002] Programmed cell death protein 1 (PD-1, also known as CD279) is a cell surface protein molecule that belongs to the immunoglobulin superfamily. It is expressed on T and B lymphocytes and macrophages, and plays a role in cell fate and differentiation. See Ishida et al., EMBO J., 1992, 11:3887-3895, incorporated by reference in its entirety. Activation of PD-1 is thought to negatively regulate the immune response. See Blank et al., Cancer Immunol. Immunother., 2007, 56:739-745; and Freeman et al., J. Exp. Med., 2000, 192:1027-1034, each of which is incorporated by reference in its entirety.
[0003] PD-1 has two known ligands, PD-L1 and PD-L2, which are both members of the B7 family. See Freeman et al., supra; and Latchman et al., Nat. Immunol., 2001, 2:261-268, each of which is incorporated by reference in its entirety. The interaction between PD-1 and these ligands is thought to play a role in a variety of diseases, including cancer (see Ribas and Tumeh, Clin. Cancer Res., 2014, Jun. 26, PMID: 24970841 [Epub ahead of print]), autoimmune disease (see Dai et al., Cell Immunol., 2014, 290:72-79), and infection (see Day et al., Nature, 2006, 443:350-354). Each of the references cited in the preceding sentence is incorporated by reference in its entirety. In particular, the engagement of PD-1 by one of its ligands is thought to inhibit T-cell effector functions in an antigen-specific manner.
[0004] In view of the role of PD-1 in multiple disease processes, there is a need for improved methods of modulating the interaction of PD-1 with its ligands and the downstream signaling processes activated by PD-1. Moreover, given the role of PD-1 in several diseases, there is also a need for therapeutics that specifically target cells and tissues that express PD-1.
SUMMARY
[0005] Provided herein are antibodies that selectively bind PD-1. In some embodiments, the antibodies bind human PD-1. In some embodiments, the antibodies also bind homologs of human PD-1. In some aspects, the homolog is a cynomolgus monkey homolog. In some aspects, the homolog is a murine homolog. In some embodiments, the antibodies bind to human PD-1, a cynomolgus monkey homolog, and a murine homolog.
[0006] In some embodiments, the antibodies comprise at least one CDR sequence defined by a consensus sequence provided in this disclosure. In some embodiments, the antibodies comprise an illustrative CDR, V.sub.H, or V.sub.L sequence provided in this disclosure, or a variant thereof. In some aspects, the variant is a variant with one or more conservative amino acid substitutions.
[0007] Also provided are compositions and kits comprising the antibodies. In some embodiments, the compositions are pharmaceutical compositions. Any suitable pharmaceutical composition may be used. In some embodiments, the pharmaceutical composition is a composition for parenteral administration.
[0008] This disclosure also provides methods of using the anti-PD-1 antibodies provided herein. In some embodiments, the method is a method of treatment. In some embodiments, the method is a diagnostic method. In some embodiments, the method is an analytical method. In some embodiments, the method is a method of purifying and/or quantifying PD-1.
[0009] In some embodiments, the antibodies are used to treat a disease or condition. In some aspects, the disease or condition is selected from a cancer, autoimmune disease, and infection.
BRIEF DESCRIPTION OF THE DRAWINGS
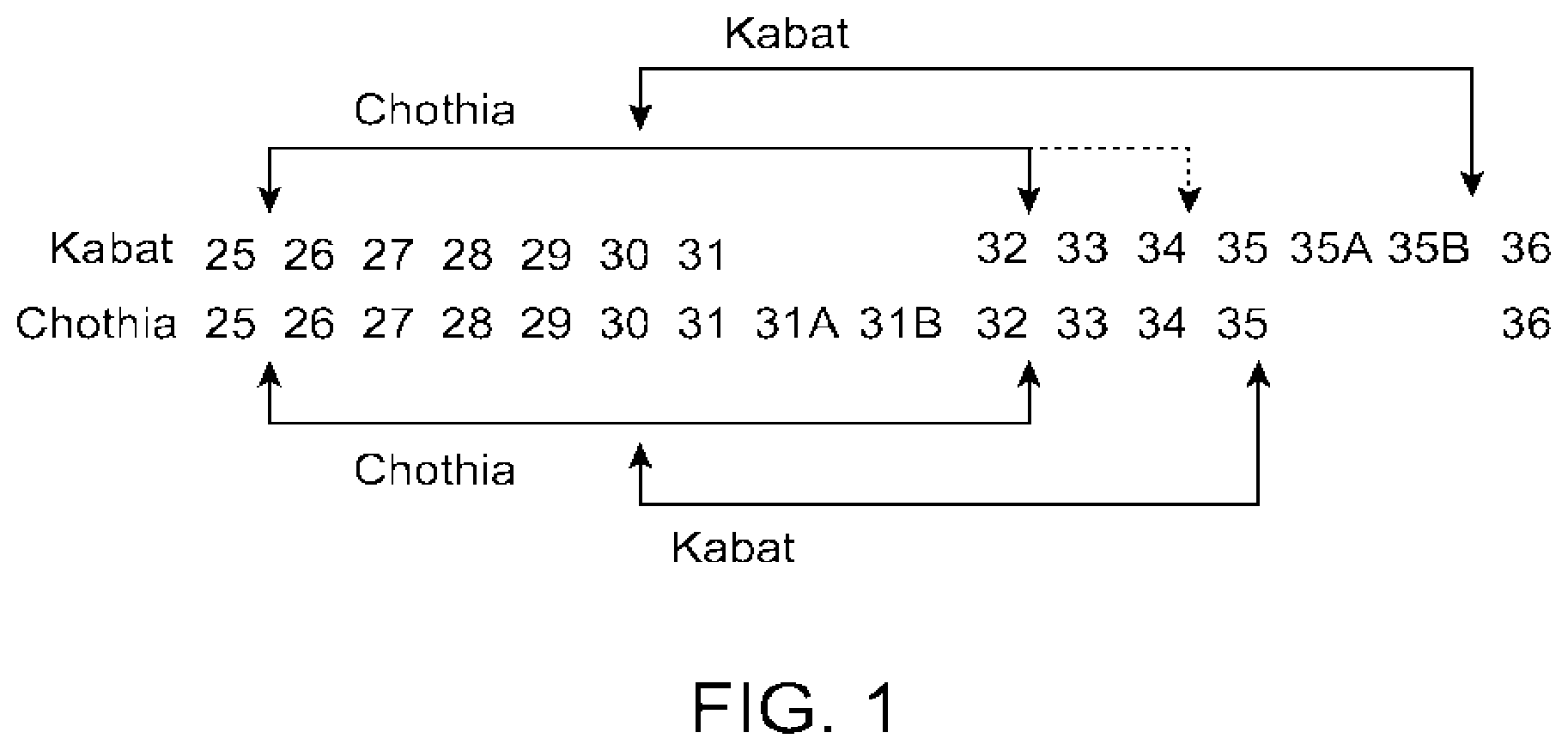
[0010] FIG. 1 provides a comparison of the Kabat and Chothia numbering systems for CDR-H1. Adapted from Martin A. C. R. (2010). Protein Sequence and Structure Analysis of Antibody Variable Domains. In R. Kontermann & S. Dubel (Eds.), Antibody Engineering vol. 2 (pp. 33-51). Springer-Verlag, Berlin Heidelberg.
[0011] FIG. 2 provides a chart of tumor volume over 17-days of treatment with various anti-PD-1 antibodies, as described in Example 15.
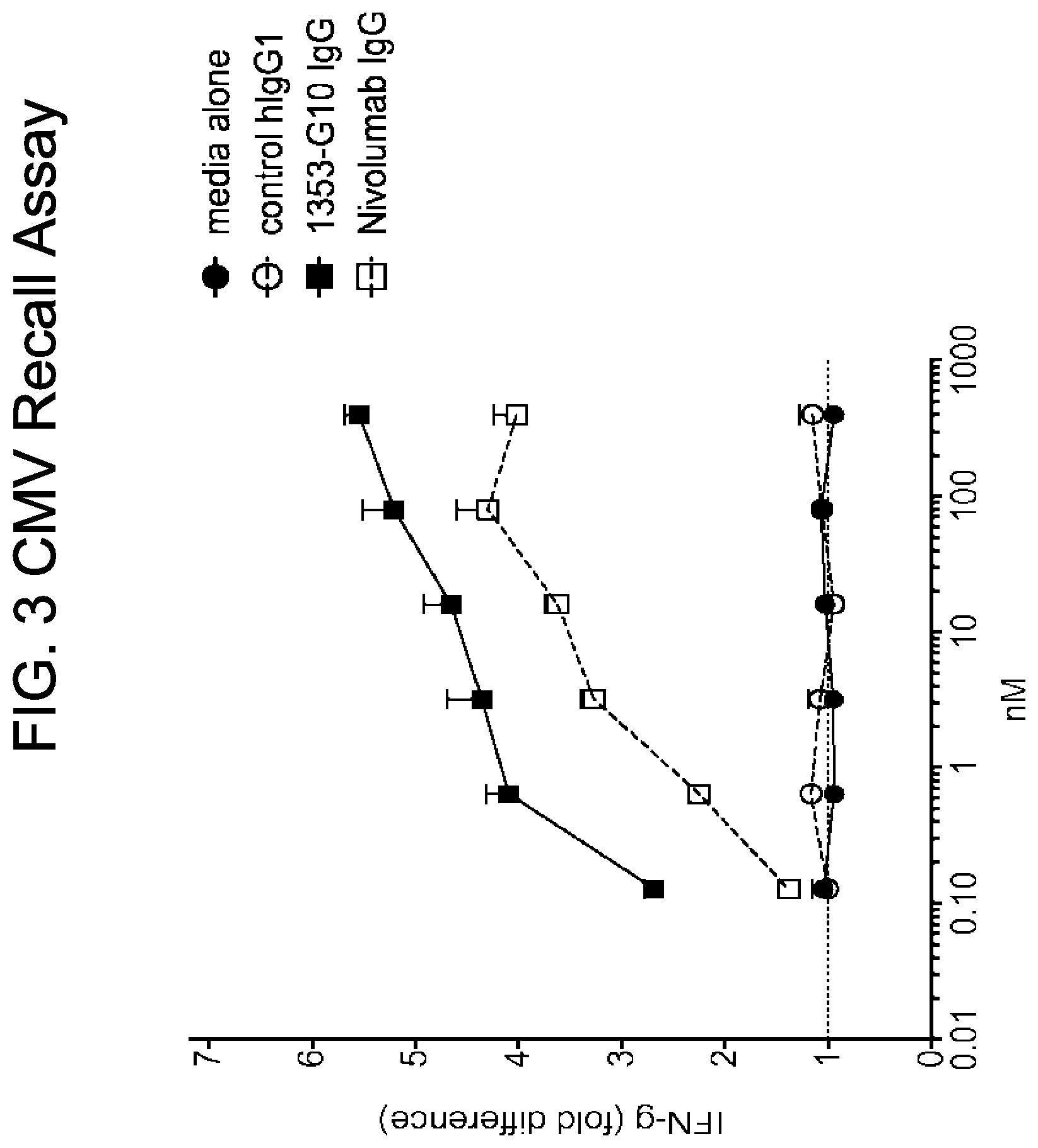
[0012] FIG. 3 provides a chart of interferon gamma (IFN-g) secretion in a cytomegalovirus (CMV) recall assay, as described in Example 16.
[0013] FIG. 4 provides a chart of interferon gamma (IFN-g) secretion in a mixed lymphocyte response (MLR) assay, as described in Example 17.
[0014] FIG. 5 provides a chart of mouse survival in a model of graft versus host disease, as described in Example 18.
DETAILED DESCRIPTION
1. Definitions
[0015] Unless otherwise defined, all terms of art, notations and other scientific terminology used herein are intended to have the meanings commonly understood by those of skill in the art to which this invention pertains. In some cases, terms with commonly understood meanings are defined herein for clarity and/or for ready reference, and the inclusion of such definitions herein should not necessarily be construed to represent a difference over what is generally understood in the art. The techniques and procedures described or referenced herein are generally well understood and commonly employed using conventional methodologies by those skilled in the art, such as, for example, the widely utilized molecular cloning methodologies described in Sambrook et al., Molecular Cloning: A Laboratory Manual 2nd ed. (1989) Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. As appropriate, procedures involving the use of commercially available kits and reagents are generally carried out in accordance with manufacturer defined protocols and/or parameters unless otherwise noted.
[0016] As used herein, the singular forms "a," "an," and "the" include the plural referents unless the context clearly indicates otherwise.
[0017] The term "about" indicates and encompasses an indicated value and a range above and below that value. In certain embodiments, the term "about" indicates the designated value.+-.10%, .+-.5%, or .+-.1%. In certain embodiments, the term "about" indicates the designated value.+-.one standard deviation of that value.
[0018] The term "combinations thereof" includes every possible combination of elements to which the term refers to. For example, a sentence stating that "if .alpha..sub.2 is A, then .alpha..sub.3 is not D; .alpha..sub.5 is not S; or .alpha..sub.6 is not S; or combinations thereof" includes the following combinations when .alpha..sub.2 is A: (1) .alpha..sub.3 is not D; (2) .alpha..sub.5 is not S; (3) .alpha..sub.6 is not S; (4) .alpha..sub.3 is not D; as is not S; and .alpha..sub.6 is not S; (5) .alpha..sub.3 is not D and .alpha..sub.5 is not S; (6) .alpha..sub.3 is not D and .alpha..sub.6 is not S; and (7) .alpha..sub.5 is not S and .alpha..sub.6 is not S.
[0019] The terms "PD-1" and "PD-1 antigen" are used interchangeably herein. Unless specified otherwise, the terms include any variants, isoforms and species homologs of human PD-1 that are naturally expressed by cells, or that are expressed by cells transfected with a PD-1 gene. PD-1 proteins include full-length PD-1 (e.g., human PD-1; GI: 167857792; SEQ ID NO: 1; extracellular domain: Pro21-Gln167), as well as alternative splice variants of PD-1, such as PD-1.DELTA.ex2, PD-1.DELTA.ex3, PD-1.DELTA.ex2,3, and PD-1.DELTA.ex2,3,4. See Nielsen et al., Cellular Immunology, 2005, 235:109-116, incorporated by reference in its entirety. In some embodiments, PD-1 proteins include murine PD-1 (e.g., SEQ ID NO: 299; extracellular domain: Leu25-Gln167). In some embodiments, PD-1 proteins include cynomolgus PD-1 (e.g., SEQ ID NO: 300; extracellular domain: Pro21-Gln167).
[0020] The term "immunoglobulin" refers to a class of structurally related proteins generally comprising two pairs of polypeptide chains: one pair of light (L) chains and one pair of heavy (H) chains. In an "intact immunoglobulin," all four of these chains are interconnected by disulfide bonds. The structure of immunoglobulins has been well characterized. See, e.g., Paul, Fundamental Immunology 7th ed., Ch. 5 (2013) Lippincott Williams & Wilkins, Philadelphia, Pa. Briefly, each heavy chain typically comprises a heavy chain variable region (V.sub.H) and a heavy chain constant region (CH). The heavy chain constant region typically comprises three domains, C.sub.H1, C.sub.H2, and C.sub.H3. Each light chain typically comprises a light chain variable region (V.sub.L) and a light chain constant region. The light chain constant region typically comprises one domain, abbreviated CL.
[0021] The term "antibody" describes a type of immunoglobulin molecule and is used herein in its broadest sense. An antibody specifically includes intact antibodies (e.g., intact immunoglobulins), and antibody fragments. Antibodies comprise at least one antigen-binding domain. One example of an antigen-binding domain is an antigen binding domain formed by a V.sub.H-V.sub.L dimer. A "PD-1 antibody," "anti-PD-1 antibody," "PD-1 Ab," "PD-1-specific antibody" or "anti-PD-1 Ab" is an antibody, as described herein, which binds specifically to the antigen PD-1. In some embodiments, the antibody binds the extracellular domain of PD-1.
[0022] The V.sub.H and V.sub.L regions may be further subdivided into regions of hypervariability ("hypervariable regions (HVRs);" also called "complementarity determining regions" (CDRs)) interspersed with regions that are more conserved. The more conserved regions are called framework regions (FRs). Each V.sub.H and V.sub.L generally comprises three CDRs and four FRs, arranged in the following order (from N-terminus to C-terminus): FR1-CDR1-FR2-CDR2-FR3-CDR3-FR4. The CDRs are involved in antigen binding, and confer antigen specificity and binding affinity to the antibody. See Kabat et al., Sequences of Proteins of Immunological Interest 5th ed. (1991) Public Health Service, National Institutes of Health, Bethesda, Md., incorporated by reference in its entirety.
[0023] The light chain from any vertebrate species can be assigned to one of two types, called kappa and lambda, based on the sequence of the constant domain.
[0024] The heavy chain from any vertebrate species can be assigned to one of five different classes (or isotypes): IgA, IgD, IgE, IgG, and IgM. These classes are also designated .alpha., .delta., .epsilon., .gamma., and .mu., respectively. The IgG and IgA classes are further divided into subclasses on the basis of differences in sequence and function. Humans express the following subclasses: IgG1, IgG2, IgG3, IgG4, IgA1, and IgA2.
[0025] The amino acid sequence boundaries of a CDR can be determined by one of skill in the art using any of a number of known numbering schemes, including those described by Kabat et al., supra ("Kabat" numbering scheme); Al-Lazikani et al., 1997, J. Mol. Biol., 273:927-948 ("Chothia" numbering scheme); MacCallum et al., 1996, J. Mol. Biol. 262:732-745 ("Contact" numbering scheme); Lefranc et al., Dev. Comp. Immunol., 2003, 27:55-77 ("IMGT" numbering scheme); and Honegge and Pluckthun, J. Mol. Biol., 2001, 309:657-70 ("AHo" numbering scheme), each of which is incorporated by reference in its entirety.
[0026] Table 1 provides the positions of CDR-L1, CDR-L2, CDR-L3, CDR-H1, CDR-H2, and CDR-H3 as identified by the Kabat and Chothia schemes. For CDR-H1, residue numbering is provided using both the Kabat and Chothia numbering schemes. FIG. 1 provides a comparison of the Kabat and Chothia numbering schemes for CDR-H1. See Martin (2010), supra.
[0027] Unless otherwise specified, the numbering scheme used for identification of a particular CDR herein is the Kabat/Chothia numbering scheme. Where the residues encompassed by these two numbering schemes diverge, the numbering scheme is specified as either Kabat or Chothia.
TABLE-US-00001 TABLE 1 Residues in CDRs according to Kabat and Chothia numbering schemes. CDR Kabat Chothia L1 L24-L34 L24-L34 L2 L50-L56 L50-L56 L3 L89-L97 L89-L97 H1 (Kabat Numbering) H31-H35B H26-H32 or H34* H1 (Chothia Numbering) H31-H35 H26-H32 H2 H50-H65 H52-H56 H3 H95-H102 H95-H102 *The C-terminus of CDR-H1, when numbered using the Kabat numbering convention, varies between H32 and H34, depending on the length of the CDR, as illustrated in FIG. 1.
[0028] The "EU numbering scheme" is generally used when referring to a residue in an antibody heavy chain constant region (e.g., as reported in Kabat et al., supra). Unless stated otherwise, the EU numbering scheme is used to refer to residues in antibody heavy chain constant regions described herein.
[0029] An "antibody fragment" comprises a portion of an intact antibody, such as the antigen binding or variable region of an intact antibody. Antibody fragments include, for example, Fv fragments, Fab fragments, F(ab').sub.2 fragments, Fab' fragments, scFv (sFv) fragments, and scFv-Fc fragments.
[0030] "Fv" fragments comprise a non-covalently-linked dimer of one heavy chain variable domain and one light chain variable domain.
[0031] "Fab" fragments comprise, in addition to the heavy and light chain variable domains, the constant domain of the light chain and the first constant domain (Cm) of the heavy chain. Fab fragments may be generated, for example, by papain digestion of a full-length antibody.
[0032] "F(ab').sub.2" fragments contain two Fab' fragments joined, near the hinge region, by disulfide bonds. F(ab').sub.2 fragments may be generated, for example, by pepsin digestion of an intact antibody. The F(ab') fragments can be dissociated, for example, by treatment with -mercaptoethanol.
[0033] "Single-chain Fv" or "sFv" or "scFv" antibody fragments comprise a VII domain and a V.sub.L domain in a single polypeptide chain. The V.sub.H and V.sub.L are generally linked by a peptide linker. See Pluckthun A. (1994). Antibodies from Escherichia coli. In Rosenberg M. & Moore G. P. (Eds.), The Pharmacology of Monoclonal Antibodies vol. 113 (pp. 269-315). Springer-Verlag, New York, incorporated by reference in its entirety. "scFv-Fc" fragments comprise an scFv attached to an Fc domain. For example, an Fc domain may be attached to the C-terminal of the scFv. The Fc domain may follow the V.sub.H or V.sub.L, depending on the orientation of the variable domains in the scFv (i.e., V.sub.H-V.sub.L or V.sub.L-V.sub.H). Any suitable Fc domain known in the art or described herein may be used. In some cases, the Fc domain is an IgG1 Fc domain (e.g., SEQ ID NO: 295). In some embodiments, the linker is (G.sub.4S).sub.3 (see SEQ ID NO: 298).
[0034] The term "monoclonal antibody" refers to an antibody from a population of substantially homogeneous antibodies. A population of substantially homogeneous antibodies comprises antibodies that are substantially similar and that bind the same epitope(s), except for variants that may normally arise during production of the monoclonal antibody. Such variants are generally present in only minor amounts. A monoclonal antibody is typically obtained by a process that includes the selection of a single antibody from a plurality of antibodies. For example, the selection process can be the selection of a unique clone from a plurality of clones, such as a pool of hybridoma clones, phage clones, yeast clones, bacterial clones, or other recombinant DNA clones. The selected antibody can be further altered, for example, to improve affinity for the target ("affinity maturation"), to humanize the antibody, to improve its production in cell culture, and/or to reduce its immunogenicity in a subject.
[0035] The term "chimeric antibody" refers to an antibody in which a portion of the heavy and/or light chain is derived from a particular source or species, while the remainder of the heavy and/or light chain is derived from a different source or species.
[0036] "Humanized" forms of non-human antibodies are chimeric antibodies that contain minimal sequence derived from the non-human antibody. A humanized antibody is generally a human immunoglobulin (recipient antibody) in which residues from one or more CDRs are replaced by residues from one or more CDRs of a non-human antibody (donor antibody). The donor antibody can be any suitable non-human antibody, such as a mouse, rat, rabbit, chicken, or non-human primate antibody having a desired specificity, affinity, or biological effect. In some instances, selected framework region residues of the recipient antibody are replaced by the corresponding framework region residues from the donor antibody. Humanized antibodies may also comprise residues that are not found in either the recipient antibody or the donor antibody. Such modifications may be made to further refine antibody function. For further details, see Jones et al., Nature, 1986, 321:522-525; Riechmann et al., Nature, 1988, 332:323-329; and Presta, Curr. Op. Struct. Biol., 1992, 2:593-596, each of which is incorporated by reference in its entirety.
[0037] A "human antibody" is one which possesses an amino acid sequence corresponding to that of an antibody produced by a human or a human cell, or derived from a non-human source that utilizes a human antibody repertoire or human antibody-encoding sequences (e.g., obtained from human sources or designed de novo). Human antibodies specifically exclude humanized antibodies.
[0038] An "isolated antibody" is one that has been separated and/or recovered from a component of its natural environment. Components of the natural environment may include enzymes, hormones, and other proteinaceous or nonproteinaceous materials. In some embodiments, an isolated antibody is purified to a degree sufficient to obtain at least 15 residues of N-terminal or internal amino acid sequence, for example by use of a spinning cup sequenator. In some embodiments, an isolated antibody is purified to homogeneity by gel electrophoresis (e.g., SDS-PAGE) under reducing or nonreducing conditions, with detection by Coomassie blue or silver stain. An isolated antibody includes an antibody in situ within recombinant cells, since at least one component of the antibody's natural environment is not present. In some aspects, an isolated antibody is prepared by at least one purification step.
[0039] In some embodiments, an isolated antibody is purified to at least 80%, 85%, 90%, 95%, or 99% by weight. In some embodiments, an isolated antibody is provided as a solution comprising at least 85%, 90%, 95%, 98%, 99% to 100% by weight of an antibody, the remainder of the weight comprising the weight of other solutes dissolved in the solvent.
[0040] "Affinity" refers to the strength of the sum total of non-covalent interactions between a single binding site of a molecule (e.g., an antibody) and its binding partner (e.g., an antigen). Unless indicated otherwise, as used herein, "binding affinity" refers to intrinsic binding affinity, which reflects a 1:1 interaction between members of a binding pair (e.g., antibody and antigen). The affinity of a molecule X for its partner Y can generally be represented by the dissociation constant (K.sub.D). Affinity can be measured by common methods known in the art, including those described herein. Affinity can be determined, for example, using surface plasmon resonance (SPR) technology, such as a Biacore.RTM. instrument.
[0041] With regard to the binding of an antibody to a target molecule, the terms "specific binding," "specifically binds to," "specific for," "selectively binds," and "selective for" a particular antigen (e.g., a polypeptide target) or an epitope on a particular antigen mean binding that is measurably different from a non-specific or non-selective interaction. Specific binding can be measured, for example, by determining binding of a molecule compared to binding of a control molecule. Specific binding can also be determined by competition with a control molecule that is similar to the target, such as an excess of non-labeled target. In that case, specific binding is indicated if the binding of the labeled target to a probe is competitively inhibited by the excess non-labeled target.
[0042] The term "k.sub.d" (sec.sup.-1), as used herein, refers to the dissociation rate constant of a particular antibody-antigen interaction. This value is also referred to as the k.sub.off value.
[0043] The term "k.sub.a" (M.sup.-1.times.sec.sup.-1), as used herein, refers to the association rate constant of a particular antibody-antigen interaction. This value is also referred to as the k.sub.on value.
[0044] The term "K.sub.D" (M), as used herein, refers to the dissociation equilibrium constant of a particular antibody-antigen interaction. K.sub.D=k.sub.d/k.sub.a.
[0045] The term "K.sub.A" (M.sup.-1), as used herein, refers to the association equilibrium constant of a particular antibody-antigen interaction. K.sub.A=k.sub.a/k.sub.d.
[0046] An "affinity matured" antibody is one with one or more alterations in one or more CDRs or FRs that result in an improvement in the affinity of the antibody for its antigen, compared to a parent antibody which does not possess the alteration(s). In one embodiment, an affinity matured antibody has nanomolar or picomolar affinity for the target antigen. Affinity matured antibodies may be produced using a variety of methods known in the art. For example, Marks et al. (Bio/Technology, 1992, 10:779-783, incorporated by reference in its entirety) describes affinity maturation by V.sub.H and V.sub.L domain shuffling. Random mutagenesis of CDR and/or framework residues is described by, for example, Barbas et al. (Proc. Nat. Acad. Sci. USA., 1994, 91:3809-3813); Schier et al., Gene, 1995, 169:147-155; Yelton et al., J. Immunol., 1995, 155:1994-2004; Jackson et al., J. Immunol., 1995, 154:3310-33199; and Hawkins et al, J. Mol. Biol., 1992, 226:889-896, each of which is incorporated by reference in its entirety.
[0047] When used herein in the context of two or more antibodies, the term "competes with" or "cross-competes with" indicates that the two or more antibodies compete for binding to an antigen (e.g., PD-1). In one exemplary assay, PD-1 is coated on a plate and allowed to bind a first antibody, after which a second, labeled antibody is added. If the presence of the first antibody reduces binding of the second antibody, then the antibodies compete. The term "competes with" also includes combinations of antibodies where one antibody reduces binding of another antibody, but where no competition is observed when the antibodies are added in the reverse order. However, in some embodiments, the first and second antibodies inhibit binding of each other, regardless of the order in which they are added. In some embodiments, one antibody reduces binding of another antibody to its antigen by at least 50%, at least 60%, at least 70%, at least 80%, or at least 90%.
[0048] The term "epitope" means a portion of an antigen capable of specific binding to an antibody. Epitopes frequently consist of surface-accessible amino acid residues and/or sugar side chains and may have specific three dimensional structural characteristics, as well as specific charge characteristics. Conformational and non-conformational epitopes are distinguished in that the binding to the former but not the latter is lost in the presence of denaturing solvents. An epitope may comprise amino acid residues that are directly involved in the binding, and other amino acid residues, which are not directly involved in the binding. The epitope to which an antibody binds can be determined using known techniques for epitope determination such as, for example, testing for antibody binding to PD-1 variants with different point-mutations.
[0049] Percent "identity" between a polypeptide sequence and a reference sequence, is defined as the percentage of amino acid residues in the polypeptide sequence that are identical to the amino acid residues in the reference sequence, after aligning the sequences and introducing gaps, if necessary, to achieve the maximum percent sequence identity. Alignment for purposes of determining percent amino acid sequence identity can be achieved in various ways that are within the skill in the art, for instance, using publicly available computer software such as BLAST, BLAST-2, ALIGN, MEGALIGN (DNASTAR), CLUSTALW, or CLUSTAL OMEGA software. Those skilled in the art can determine appropriate parameters for aligning sequences, including any algorithms needed to achieve maximal alignment over the full length of the sequences being compared.
[0050] A "conservative substitution" or a "conservative amino acid substitution," refers to the substitution of one or more amino acids with one or more chemically or functionally similar amino acids. Conservative substitution tables providing similar amino acids are well known in the art. Polypeptide sequences having such substitutions are known as "conservatively modified variants." Such conservatively modified variants are in addition to and do not exclude polymorphic variants, interspecies homologs, and alleles. By way of example, the following groups of amino acids are considered conservative substitutions for one another.
TABLE-US-00002 Acidic Residues D and E Basic Residues K, R, and H Hydrophilic Uncharged Residues S, T, N, and Q Aliphatic Uncharged Residues G, A, V, L, and I Non-polar Uncharged Residues C, M, and P Aromatic Residues F, Y, and W Alcohol Group-Containing Residues S and T Aliphatic Residues I, L, V, and M Cycloalkenyl-associated Residues F, H, W, and Y Hydrophobic Residues A, C, F, G, H, I, L, M, R, T, V, W, and Y Negatively Charged Residues D and E Polar Residues C, D, E, H, K, N, Q, R, S, and T Positively Charged Residues H, K, and R Small Residues A, C, D, G, N, P, S, T, and V Very Small Residues A, G, and S Residues Involved in A, C, D, E, G, H, K, Turn Formation N, Q, R, S, P, and T Flexible Residues Q, T, K, S, G, P, D, E, and R Group 1 A, S, and T Group 2 D and E Group 3 N and Q Group 4 R and K Group 5 I, L, and M Group 6 F, Y, and W Group A A and G Group B D and E Group C N and Q Group D R, K, and H Group E I, L, M, V Group F F, Y, and W Group G S and T Group H C and M
Additional conservative substitutions may be found, for example, in Creighton, Proteins: Structures and Molecular Properties 2nd ed. (1993) W. H. Freeman & Co., New York, N.Y. An antibody generated by making one or more conservative substitutions of amino acid residues in a parent antibody is referred to as a "conservatively modified variant."
[0051] The term "amino acid" refers to the twenty common naturally occurring amino acids. Naturally occurring amino acids include alanine (Ala; A), arginine (Arg; R), asparagine (Asn; N), aspartic acid (Asp; D), cysteine (Cys; C); glutamic acid (Glu; E), glutamine (Gln; Q), Glycine (Gly; G); histidine (His; H), isoleucine (Ile; I), leucine (Leu; L), lysine (Lys; K), methionine (Met; M), phenylalanine (Phe; F), proline (Pro; P), serine (Ser; S), threonine (Thr; T), tryptophan (Trp; W), tyrosine (Tyr; Y), and valine (Val; V).
[0052] "Treating" or "treatment" of any disease or disorder refers, in certain embodiments, to ameliorating a disease or disorder that exists in a subject. In another embodiment, "treating" or "treatment" includes ameliorating at least one physical parameter, which may be indiscernible by the subject. In yet another embodiment, "treating" or "treatment" includes modulating the disease or disorder, either physically (e.g., stabilization of a discernible symptom) or physiologically (e.g., stabilization of a physical parameter) or both. In yet another embodiment, "treating" or "treatment" includes delaying or preventing the onset of the disease or disorder.
[0053] As used herein, the term "therapeutically effective amount" or "effective amount" refers to an amount of an antibody or composition that when administered to a subject is effective to treat a disease or disorder.
[0054] As used herein, the term "subject" means a mammalian subject. Exemplary subjects include, but are not limited to humans, monkeys, dogs, cats, mice, rats, cows, horses, camels, avians, goats and sheep. In certain embodiments, the subject is a human. In some embodiments, the subject has cancer, an autoimmune disease or condition, and/or an infection that can be treated with an antibody provided herein. In some embodiments, the subject is a human that is suspected to have cancer, an autoimmune disease or condition, and/or an infection.
2. Antibodies
[0055] Provided herein are antibodies that selectively bind human PD-1. In some aspects, the antibody selectively binds to the extracellular domain of human PD-1. In some aspects, the antibody selectively binds to one or more of full-length human PD-1, PD-1.DELTA.ex2, PD-1.DELTA.ex3, PD-1.DELTA.ex2,3, and PD-1.DELTA.ex2,3,4. See Nielsen et al., Cellular Immunology, 2005, 235:109-116, incorporated by reference in its entirety.
[0056] In some embodiments, the antibody binds to homologs of human PD-1. In some aspects, the antibody binds to a homolog of human PD-1 from a species selected from monkeys, mice, dogs, cats, rats, cows, horses, goats and sheep. In some aspects, the homolog is a cynomolgus monkey homolog. In some aspects, the homolog is a murine homolog.
[0057] In some embodiments, the antibody has one or more CDRs having particular lengths, in terms of the number of amino acid residues. In some embodiments, the Chothia CDR-H1 of the antibody is 6, 7, 8, or 9 residues in length. In some embodiments, the Kabat CDR-H1 of the antibody is 4, 5, 6, or 7 residues in length. In some embodiments, the Chothia CDR-H2 of the antibody is 5, 6, or 7 residues in length. In some embodiments, the Kabat CDR-H2 of the antibody is 15, 16, 17, or 18 residues in length. In some embodiments, the Kabat/Chothia CDR-H3 of the antibody is 5, 6, 7, 8, 9, 10, 11, or 12 residues in length.
[0058] In some aspects, the Kabat/Chothia CDR-L1 of the antibody is 9, 10, 11, 12, 13, 14, 15, or 16 residues in length. In some aspects, the Kabat/Chothia CDR-L2 of the antibody is 6, 7, or 8 residues in length. In some aspects, the Kabat/Chothia CDR-L3 of the antibody is 8, 9, 10, 11, or 12 residues in length.
[0059] In some embodiments, the antibody comprises a light chain. In some aspects, the light chain is a kappa light chain. In some aspects, the light chain is a lambda light chain.
[0060] In some embodiments, the antibody comprises a heavy chain. In some aspects, the heavy chain is an IgA. In some aspects, the heavy chain is an IgD. In some aspects, the heavy chain is an IgE. In some aspects, the heavy chain is an IgG. In some aspects, the heavy chain is an IgM. In some aspects, the heavy chain is an IgG1. In some aspects, the heavy chain is an IgG2. In some aspects, the heavy chain is an IgG3. In some aspects, the heavy chain is an IgG4. In some aspects, the heavy chain is an IgA1. In some aspects, the heavy chain is an IgA2.
[0061] In some embodiments, the antibody is an antibody fragment. In some aspects, the antibody fragment is an Fv fragment. In some aspects, the antibody fragment is a Fab fragment. In some aspects, the antibody fragment is a F(ab').sub.2 fragment. In some aspects, the antibody fragment is a Fab' fragment. In some aspects, the antibody fragment is an scFv (sFv) fragment. In some aspects, the antibody fragment is an scFv-Fc fragment.
[0062] In some embodiments, the antibody is a monoclonal antibody. In some embodiments, the antibody is a polyclonal antibody.
[0063] In some embodiments, the antibody is a chimeric antibody. In some embodiments, the antibody is a humanized antibody. In some embodiments, the antibody is a human antibody.
[0064] In some embodiments, the antibody is an affinity matured antibody. In some aspects, the antibody is an affinity matured antibody derived from an illustrative sequence provided in this disclosure.
[0065] In some embodiments, the antibody inhibits the binding of PD-1 to its ligands. In some aspects, the antibody inhibits the binding of PD-1 to PD-L1. In some aspects, the antibody inhibits the binding of PD-1 to PD-L2. In some aspects, the antibody inhibits the binding of PD-1 to PD-L1 and PD-L2.
[0066] The antibodies provided herein may be useful for the treatment of a variety of diseases and conditions, including cancers, autoimmune diseases, and infections.
[0067] 2.1. CDR-H3 Sequences
[0068] In some embodiments, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 113-131 and 309-315. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 113. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 114. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 115. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 116. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 117. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 118. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 119. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 120. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 121. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 122. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 123. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 124. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 125. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 126. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 127. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 128. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 129. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 130. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 131. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 309. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 310. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 311. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 312. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 313. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 314. In some aspects, the antibody comprises a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 315.
[0069] In some aspects, the CDR-H3 sequence comprises, consists of, or consists essentially of a variant of an illustrative CDR-H3 sequence provided in this disclosure. In some aspects, the CDR-H3 sequence comprises, consists of, or consists essentially of a sequence having at least 70%, 75%, 80%, 85%, 90%, or 95% identity with any of the illustrative CDR-H3 sequences provided in this disclosure. In some aspects, the CDR-H3 sequence comprises, consists of, or consists essentially of any of the illustrative CDR-H3 sequences provided in this disclosure, with 1, 2, or 3 amino acid substitutions. In some aspects, the amino acid substitutions are conservative amino acid substitutions.
[0070] In some aspects, the CDR-H3 sequence does not comprise, consist of, or consist essentially of a sequence selected from SEQ ID NOs: 132-136. In some aspects, the CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 132. In some aspects, the CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 133. In some aspects, the CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 134. In some aspects, the CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 135. In some aspects, the CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 136.
[0071] 2.2. V.sub.H Sequences Comprising Illustrative CDRs
[0072] In some embodiments, the antibody comprises a V.sub.H sequence comprising one or more CDR-H sequences comprising, consisting of, or consisting essentially of one or more illustrative CDR-H sequences provided in this disclosure, and variants thereof.
[0073] 2.2.1. V.sub.H Sequences Comprising Illustrative Kabat CDRs
[0074] In some embodiments, the antibody comprises a V.sub.H sequence comprising one or more Kabat CDR-H sequences comprising, consisting of, or consisting essentially of one or more illustrative Kabat CDR-H sequences provided in this disclosure, and variants thereof.
[0075] 2.2.1.1. Kabat CDR-H3
[0076] In some embodiments, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 113-131 and 309-315. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 113. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 114. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 115. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 116. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 117. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 118. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 119. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 120. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 121. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 122. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 123. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 124. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 125. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 126. In some aspects, the antibody comprises a VII sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 127. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 128. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 129. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 130. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 131. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 309. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 310. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 311. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 312. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 313. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 314. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 315.
[0077] 2.2.1.2. Kabat CDR-H2
[0078] In some embodiments, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 84-102 or 331. In some aspects, the antibody comprises a VII sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 84. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 85. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 86. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 87. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 88. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 89. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 90. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 91. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 92. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 93. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 94. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 95. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 96. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 97. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 98. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 99. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 100. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 101. In some aspects, the antibody comprises a VII sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 102. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 331.
[0079] 2.2.1.3. Kabat CDR-H1
[0080] In some embodiments, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 31-49. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 31. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 32. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 33. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 34 In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 35. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 36. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 37. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 38. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 39. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 40. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 41. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 42. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 43. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 44. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 45. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 46. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 47. In some aspects, the antibody comprises a VII sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 48. In some aspects, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 49.
[0081] 2.2.1.4. Kabat CDR-H3+Kabat CDR-H2
[0082] In some embodiments, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 113-131 and 309-315, and a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 84-102 or 331. In some aspects, the Kabat CDR-H3 sequence and the Kabat CDR-H2 sequence are both from a single illustrative V.sub.H sequence provided in this disclosure. For example, in some aspects, the Kabat CDR-H3 and Kabat CDR-H2 are both from a single illustrative V.sub.H sequence selected from SEQ ID NOs: 246-264 and 316-322.
[0083] 2.2.1.5. Kabat CDR-H3+Kabat CDR-H1
[0084] In some embodiments, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 113-131 and 309-315, and a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 31-49. In some aspects, the Kabat CDR-H3 sequence and the Kabat CDR-H1 sequence are both from a single illustrative V.sub.H sequence provided in this disclosure. For example, in some aspects, the Kabat CDR-H3 and Kabat CDR-H1 are both from a single illustrative VII sequence selected from SEQ ID NOs: 246-264 and 316-322.
[0085] 2.2.1.6. Kabat CDR-H1+Kabat CDR-H2
[0086] In some embodiments, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 31-49 and a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 84-102 or 331. In some aspects, the Kabat CDR-H1 sequence and the Kabat CDR-H2 sequence are both from a single illustrative V.sub.H sequence provided in this disclosure. For example, in some aspects, the Kabat CDR-H1 and Kabat CDR-H2 are both from a single illustrative V.sub.H sequence selected from SEQ ID NOs: 246-264. 2.2.1.7. Kabat CDR-H1+Kabat CDR-H2+Kabat CDR-H3
[0087] In some embodiments, the antibody comprises a V.sub.H sequence comprising a Kabat CDR-H1 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 31-49, a Kabat CDR-H2 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 84-102 or 331, and a Kabat CDR-H3 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 113-131 and 309-315. In some aspects, the Kabat CDR-H1 sequence, Kabat CDR-H2 sequence, and Kabat CDR-H3 sequence are all from a single illustrative VII sequence provided in this disclosure. For example, in some aspects, the Kabat CDR-H1, Kabat CDR-H2, and Kabat CDR-H3 are all from a single illustrative V.sub.H sequence selected from SEQ ID NOs: 246-264 and 316-322.
[0088] 2.2.1.8. Variants of V.sub.H Sequences Comprising Illustrative Kabat CDRs
[0089] In some embodiments, the V.sub.H sequences provided herein comprise a variant of an illustrative Kabat CDR-H3, CDR-H2, and/or CDR-H1 sequence provided in this disclosure.
[0090] In some aspects, the Kabat CDR-H3 sequence comprises, consists of, or consists essentially of a variant of an illustrative Kabat CDR-H3 sequence provided in this disclosure. In some aspects, the Kabat CDR-H3 sequence comprises, consists of, or consists essentially of a sequence having at least 70%, 75%, 80%, 85%, 90%, or 95% identity with any of the illustrative Kabat CDR-H3 sequences provided in this disclosure. In some aspects, the Kabat CDR-H3 sequence comprises, consists of, or consists essentially of any of the illustrative Kabat CDR-H3 sequences provided in this disclosure, with 1, 2, or 3 amino acid substitutions. In some aspects, the amino acid substitutions are conservative amino acid substitutions.
[0091] In some aspects, the Kabat CDR-H2 sequence comprises, consists of, or consists essentially of a variant of an illustrative Kabat CDR-H2 sequence provided in this disclosure. In some aspects, the Kabat CDR-H2 sequence comprises, consists of, or consists essentially of a sequence having at least 70%, 75%, 80%, 85%, 90%, or 95% identity with any of the illustrative Kabat CDR-H2 sequences provided in this disclosure. In some aspects, the Kabat CDR-H2 sequence comprises, consists of, or consists essentially of any of the illustrative Kabat CDR-H2 sequences provided in this disclosure, with 1, 2, or 3 amino acid substitutions. In some aspects, the amino acid substitutions are conservative amino acid substitutions.
[0092] In some aspects, the Kabat CDR-H1 sequence comprises, consists of, or consists essentially of a variant of an illustrative Kabat CDR-H1 sequence provided in this disclosure. In some aspects, the Kabat CDR-H1 sequence comprises, consists of, or consists essentially of a sequence having at least 70%, 75%, 80%, 85%, 90%, or 95% identity with any of the illustrative Kabat CDR-H1 sequences provided in this disclosure. In some aspects, the Kabat CDR-H1 sequence comprises, consists of, or consists essentially of any of the illustrative Kabat CDR-H1 sequences provided in this disclosure, with 1, 2, or 3 amino acid substitutions. In some aspects, the amino acid substitutions are conservative amino acid substitutions.
[0093] 2.2.1.9. Excluded V.sub.H Sequences Comprising Kabat CDRs
[0094] In some embodiments, the V.sub.H sequences provided herein do not comprise certain Kabat CDR-H3, CDR-H2, and/or CDR-H1 sequences.
[0095] In some aspects, the Kabat CDR-H3 sequence does not comprise, consist of, or consist essentially of a sequence selected from SEQ ID NOs: 108-112 or 132-136. In some aspects, the Kabat CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 108. In some aspects, the Kabat CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 109. In some aspects, the Kabat CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 110. In some aspects, the Kabat CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 111. In some aspects, the Kabat CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 112. In some aspects, the Kabat CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 132. In some aspects, the Kabat CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 133. In some aspects, the Kabat CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 134. In some aspects, the Kabat CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 135. In some aspects, the Kabat CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 136.
[0096] In some aspects, the Kabat CDR-H2 sequence does not comprise, consist of, or consist essentially of a sequence selected from SEQ ID NOs: 55-59 or 103-107. In some aspects, the Kabat CDR-H2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 55. In some aspects, the Kabat CDR-H2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 56. In some aspects, the Kabat CDR-H2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 57. In some aspects, the Kabat CDR-H2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 58. In some aspects, the Kabat CDR-H2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 59. In some aspects, the Kabat CDR-H2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 103. In some aspects, the Kabat CDR-H2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 104. In some aspects, the Kabat CDR-H2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 105. In some aspects, the Kabat CDR-H2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 106. In some aspects, the Kabat CDR-H2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 107.
[0097] In some aspects, the Kabat CDR-H1 sequence does not comprise, consist of, or consist essentially of a sequence selected from SEQ ID NOs: 2-6 or 50-54. In some aspects, the Kabat CDR-H1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 2. In some aspects, the Kabat CDR-H1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 3. In some aspects, the Kabat CDR-H1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 4. In some aspects, the Kabat CDR-H1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 5. In some aspects, the Kabat CDR-H1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 6. In some aspects, the Kabat CDR-H1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 50. In some aspects, the Kabat CDR-H1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 51. In some aspects, the Kabat CDR-H1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 52. In some aspects, the Kabat CDR-H1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 53. In some aspects, the Kabat CDR-H1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 54.
[0098] 2.2.2. V.sub.H Sequences Comprising Illustrative Chothia CDRs
[0099] In some embodiments, the antibody comprises a V.sub.H sequence comprising one or more Chothia CDR-H sequences comprising, consisting of, or consisting essentially of one or more illustrative Chothia CDR-H sequences provided in this disclosure, and variants thereof.
[0100] 2.2.2.1. Chothia CDR-H3
[0101] In some embodiments, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 113-131 and 309-315. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 113. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 114. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 115. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 116. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 117. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 118. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 119. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 120. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 121. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 122. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 123. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 124. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 125. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 126. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 127. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 128. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 129. In some aspects, the antibody comprises a VII sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 130. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 131. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 309. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 310. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 311. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 312. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 313. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 314. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 315.
[0102] 2.2.2.2. Chothia CDR-H2
[0103] In some embodiments, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 60-78. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 60. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 61. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 62. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 63. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 64. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 65. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 66. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 67. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 68. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 69. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 70. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 71. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 72. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 73. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 74. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 75. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 76. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 77. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 78.
[0104] 2.2.2.3. Chothia CDR-H1
[0105] In some embodiments, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 7-25. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 7. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 8. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 9. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 10 In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 11. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 12. In some aspects, the antibody comprises a VII sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 13. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 14. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 15. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 16. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 17. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 18. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 19. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 20. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 21. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 22. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 23. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 24. In some aspects, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 25.
[0106] 2.2.2.4. Chothia CDR-H3+Chothia CDR-H2
[0107] In some embodiments, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 113-131 and 309-315, and a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 60-78. In some aspects, the Chothia CDR-H3 sequence and the Chothia CDR-H2 sequence are both from a single illustrative V.sub.H sequence provided in this disclosure. For example, in some aspects, the Chothia CDR-H3 and Chothia CDR-H2 are both from a single illustrative V.sub.H sequence selected from SEQ ID NOs: 246-264 and 316-322.
[0108] 2.2.2.5. Chothia CDR-H3+Chothia CDR-H1
[0109] In some embodiments, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 113-131 and 309-315, and a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 7-25. In some aspects, the Chothia CDR-H3 sequence and the Chothia CDR-H1 sequence are both from a single illustrative V.sub.H sequence provided in this disclosure. For example, in some aspects, the Chothia CDR-H3 and Chothia CDR-H1 are both from a single illustrative VII sequence selected from SEQ ID NOs: 246-264 and 316-322.
[0110] 2.2.2.6. Chothia CDR-H1+Chothia CDR-H2
[0111] In some embodiments, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 7-25 and a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 60-78. In some aspects, the Chothia CDR-H1 sequence and the Chothia CDR-H2 sequence are both from a single illustrative V.sub.H sequence provided in this disclosure. For example, in some aspects, the Chothia CDR-H1 and Chothia CDR-H2 are both from a single illustrative V.sub.H sequence selected from SEQ ID NOs: 246-264.
[0112] 2.2.2.7. Chothia CDR-H1+Chothia CDR-H2+Chothia CDR-H3
[0113] In some embodiments, the antibody comprises a V.sub.H sequence comprising a Chothia CDR-H1 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 7-25, a Chothia CDR-H2 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 60-78, and a Chothia CDR-H3 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 113-131 and 309-315. In some aspects, the Chothia CDR-H1 sequence, Chothia CDR-H2 sequence, and Chothia CDR-H3 sequence are all from a single illustrative V.sub.H sequence provided in this disclosure. For example, in some aspects, the Chothia CDR-H1, Chothia CDR-H2, and Chothia CDR-H3 are all from a single illustrative V.sub.H sequence selected from SEQ ID NOs: 246-264 and 316-322.
[0114] 2.2.2.8. Variants of V.sub.H Sequences Comprising Illustrative Chothia CDRs
[0115] In some embodiments, the V.sub.H sequences provided herein comprise a variant of an illustrative Chothia CDR-H3, CDR-H2, and/or CDR-H1 sequence provided in this disclosure.
[0116] In some aspects, the Chothia CDR-H3 sequence comprises, consists of, or consists essentially of a variant of an illustrative Chothia CDR-H3 sequence provided in this disclosure. In some aspects, the Chothia CDR-H3 sequence comprises, consists of, or consists essentially of a sequence having at least 70%, 75%, 80%, 85%, 90%, or 95% identity with any of the illustrative Chothia CDR-H3 sequences provided in this disclosure. In some aspects, the Chothia CDR-H3 sequence comprises, consists of, or consists essentially of any of the illustrative Chothia CDR-H3 sequences provided in this disclosure, with 1, 2, or 3 amino acid substitutions. In some aspects, the amino acid substitutions are conservative amino acid substitutions.
[0117] In some aspects, the Chothia CDR-H2 sequence comprises, consists of, or consists essentially of a variant of an illustrative Chothia CDR-H2 sequence provided in this disclosure. In some aspects, the Chothia CDR-H2 sequence comprises, consists of, or consists essentially of a sequence having at least 70%, 75%, 80%, 85%, 90%, or 95% identity with any of the illustrative Chothia CDR-H2 sequences provided in this disclosure. In some aspects, the Chothia CDR-H2 sequence comprises, consists of, or consists essentially of any of the illustrative Chothia CDR-H2 sequences provided in this disclosure, with 1, 2, or 3 amino acid substitutions. In some aspects, the amino acid substitutions are conservative amino acid substitutions.
[0118] In some aspects, the Chothia CDR-H1 sequence comprises, consists of, or consists essentially of a variant of an illustrative Chothia CDR-H1 sequence provided in this disclosure. In some aspects, the Chothia CDR-H1 sequence comprises, consists of, or consists essentially of a sequence having at least 70%, 75%, 80%, 85%, 90%, or 95% identity with any of the illustrative Chothia CDR-H1 sequences provided in this disclosure. In some aspects, the Chothia CDR-H1 sequence comprises, consists of, or consists essentially of any of the illustrative Chothia CDR-H1 sequences provided in this disclosure, with 1, 2, or 3 amino acid substitutions. In some aspects, the amino acid substitutions are conservative amino acid substitutions.
[0119] 2.2.2.9. Excluded V.sub.H Sequences Comprising Chothia CDRs
[0120] In some embodiments, the V.sub.H sequences provided herein do not comprise certain Chothia CDR-H3, CDR-H2, and/or CDR-H1 sequences.
[0121] In some aspects, the Chothia CDR-H3 sequence does not comprise, consist of, or consist essentially of a sequence selected from SEQ ID NOs: 108-112 or 132-136. In some aspects, the Chothia CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 108. In some aspects, the Chothia CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 109. In some aspects, the Chothia CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 110. In some aspects, the Chothia CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 111. In some aspects, the Chothia CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 112. In some aspects, the Chothia CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 132. In some aspects, the Chothia CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 133. In some aspects, the Chothia CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 134. In some aspects, the Chothia CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 135. In some aspects, the Chothia CDR-H3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 136.
[0122] In some aspects, the Chothia CDR-H2 sequence does not comprise, consist of, or consist essentially of a sequence selected from SEQ ID NOs: 79-83. In some aspects, the Chothia CDR-H2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 79. In some aspects, the Chothia CDR-H2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 80. In some aspects, the Chothia CDR-H2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 81. In some aspects, the Chothia CDR-H2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 82. In some aspects, the Chothia CDR-H2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 83.
[0123] In some aspects, the Chothia CDR-H1 sequence does not comprise, consist of, or consist essentially of a sequence selected from SEQ ID NOs: 26-30. In some aspects, the Chothia CDR-H1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 26. In some aspects, the Chothia CDR-H1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 27. In some aspects, the Chothia CDR-H1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 28. In some aspects, the Chothia CDR-H1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 29. In some aspects, the Chothia CDR-H1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 30.
[0124] 2.3. V.sub.H Sequences
[0125] In some embodiments, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 246-264 and 316-322. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 246. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 247. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 248. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 249. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 250. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 251. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 252. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 253. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 254. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 255 (with or without a serine prepended to the sequence). In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 256. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 257. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 258. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 259. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 260. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 261. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 262. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 263. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 264. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 316. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 317. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 318. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 319. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 320. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 321. In some aspects, the antibody comprises a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 322.
[0126] 2.3.1. Variants of V.sub.H Sequences
[0127] In some embodiments, the V.sub.H sequences provided herein comprise, consist of, or consist essentially of a variant of an illustrative V.sub.H sequence provided in this disclosure.
[0128] In some aspects, the V.sub.H sequence comprises, consists of, or consists essentially of a variant of an illustrative V.sub.H sequence provided in this disclosure. In some aspects, the V.sub.H sequence comprises, consists of, or consists essentially of a sequence having at least 85%, 90%, 95%, 96%, 97%, 98%, 99%, or 99.5% identity with any of the illustrative V.sub.H sequences provided in this disclosure.
[0129] In some embodiments, the V.sub.H sequence comprises, consists of, or consists essentially of any of the illustrative V.sub.H sequences provided in this disclosure, 20 or fewer, 19 or fewer, 18 or fewer, 17 or fewer, 16 or fewer, 15 or fewer, 14 or fewer, 13 or fewer, 12 or fewer, 11 or fewer, 10 or fewer, 9 or fewer, 8 or fewer, 7 or fewer, 6 or fewer, 5 or fewer, 4 or fewer, 3 or fewer, 2 or fewer, or 1 or fewer amino acid substitutions. In some aspects, the amino acid substitutions are conservative amino acid substitutions.
[0130] 2.3.2. Excluded V.sub.H Sequences
[0131] In some embodiments, the V.sub.H sequences provided herein do not comprise certain V.sub.H sequences.
[0132] In some aspects, the V.sub.H sequence does not comprise, consist of, or consist essentially of a sequence selected from SEQ ID NOs: 265-269. In some aspects, the V.sub.H sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 265. In some aspects, the V.sub.H sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 266. In some aspects, the V.sub.H sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 267. In some aspects, the V.sub.H sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 268. In some aspects, the V.sub.H sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 269.
[0133] 2.4. CDR-L3 Sequences
[0134] In some embodiments, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 200-218. In some aspects, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 200. In some aspects, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 201. In some aspects, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 202. In some aspects, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 203. In some aspects, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 204. In some aspects, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 205. In some aspects, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 206. In some aspects, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 207. In some aspects, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 208. In some aspects, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 209. In some aspects, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 210. In some aspects, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 211. In some aspects, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 212. In some aspects, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 213. In some aspects, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 214. In some aspects, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 215. In some aspects, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 216. In some aspects, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 217. In some aspects, the antibody comprises a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 218.
[0135] In some aspects, the CDR-L3 sequence comprises, consists of, or consists essentially of a variant of an illustrative CDR-L3 sequence provided in this disclosure. In some aspects, the CDR-L3 sequence comprises, consists of, or consists essentially of a sequence having at least 70%, 75%, 80%, 85%, 90%, or 95% identity with any of the illustrative CDR-L3 sequences provided in this disclosure. In some aspects, the CDR-L3 sequence comprises, consists of, or consists essentially of any of the illustrative CDR-L3 sequences provided in this disclosure, with 1, 2, or 3 amino acid substitutions. In some aspects, the amino acid substitutions are conservative amino acid substitutions.
[0136] In some aspects, the CDR-L3 sequence does not comprise, consist of, or consist essentially of a sequence selected from SEQ ID NOs: 195-199 or 219-223. In some aspects the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 195. In some aspects the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 196. In some aspects the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 197. In some aspects the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 198. In some aspects the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 199. In some aspects the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 219. In some aspects the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 220. In some aspects the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 221. In some aspects the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 222. In some aspects the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 223.
[0137] 2.5. V.sub.L Sequences Comprising Illustrative CDRs
[0138] In some embodiments, the antibody comprises a V.sub.L sequence comprising one or more CDR-L sequences comprising, consisting of, or consisting essentially of one or more illustrative CDR-L sequences provided in this disclosure, and variants thereof.
[0139] 2.5.1. CDR-L3
[0140] In some embodiments, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 200-218. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 200. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 201. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 202. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 203. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 204. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 205. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 206. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 207. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 208. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 209. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 210. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 211. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 212. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 213. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 214. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 215. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 216. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 217. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 218.
[0141] 2.5.2. CDR-L2
[0142] In some embodiments, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 171-189. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 171. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 172. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 173. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 174. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 175. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 176. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 177. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 178. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 179. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 180. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 181. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 182. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 183. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 184. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 185. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 186. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 187. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 188. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L2 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 189.
[0143] 2.5.3. CDR-L1
[0144] In some embodiments, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 142-160. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 142. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 143. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 144. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 145. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 146. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 147. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 148. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 149. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 150. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 151. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 152. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 153. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 154. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 155. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 156. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 157. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 158. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 159. In some aspects, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 160.
[0145] 2.5.4. CDR-L3+CDR-L2
[0146] In some embodiments, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 200-218 and a CDR-L2 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 171-189. In some aspects, the CDR-L3 sequence and the CDR-L2 sequence are both from a single illustrative V.sub.L sequence provided in this disclosure. For example, in some aspects, the CDR-L3 and CDR-L2 are both from a single illustrative V.sub.L sequence selected from SEQ ID NOs: 270-288.
[0147] 2.5.5. CDR-L3+CDR-L1
[0148] In some embodiments, the antibody comprises a V.sub.L sequence comprising a CDR-L3 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 200-218 and a CDR-L1 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 142-160. In some aspects, the CDR-L3 sequence and the CDR-L1 sequence are both from a single illustrative V.sub.L sequence provided in this disclosure. For example, in some aspects, the CDR-L3 and CDR-L1 are both from a single illustrative V.sub.L sequence selected from SEQ ID NOs: 270-288.
[0149] 2.5.6. CDR-L1+CDR-L2
[0150] In some embodiments, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 142-160 and a CDR-L2 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 171-189. In some aspects, the CDR-L1 sequence and the CDR-L2 sequence are both from a single illustrative V.sub.L sequence provided in this disclosure. For example, in some aspects, the CDR-L1 and CDR-L2 are both from a single illustrative V.sub.L sequence selected from SEQ ID NOs: 270-288.
[0151] 2.5.7. CDR-L1+CDR-L2+CDR-L3
[0152] In some embodiments, the antibody comprises a V.sub.L sequence comprising a CDR-L1 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 142-160, a CDR-L2 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 171-189, and a CDR-L3 sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L1 sequence, CDR-L2 sequence, and CDR-L3 sequence are all from a single illustrative V.sub.L sequence provided in this disclosure. For example, in some aspects, the CDR-L1, CDR-L2, and CDR-L3 are all from a single illustrative V.sub.L sequence selected from SEQ ID NOs: 270-288.
[0153] 2.5.8. Variants of V.sub.L Sequences Comprising Illustrative CDR-Ls
[0154] In some embodiments, the V.sub.L sequences provided herein comprise a variant of an illustrative CDR-L3, CDR-L2, and/or CDR-L1 sequence provided in this disclosure.
[0155] In some aspects, the CDR-L3 sequence comprises, consists of, or consists essentially of a variant of an illustrative CDR-L3 sequence provided in this disclosure. In some aspects, the CDR-L3 sequence comprises, consists of, or consists essentially of a sequence having at least 70%, 75%, 80%, 85%, 90%, or 95% identity with any of the illustrative CDR-L3 sequences provided in this disclosure. In some aspects, the CDR-L3 sequence comprises, consists of, or consists essentially of any of the illustrative CDR-L3 sequences provided in this disclosure, with 1, 2, or 3 amino acid substitutions. In some aspects, the amino acid substitutions are conservative amino acid substitutions.
[0156] In some aspects, the CDR-L2 sequence comprises, consists of, or consists essentially of a variant of an illustrative CDR-L2 sequence provided in this disclosure. In some aspects, the CDR-L2 sequence comprises, consists of, or consists essentially of a sequence having at least 70%, 75%, 80%, 85%, 90%, or 95% identity with any of the illustrative CDR-L2 sequences provided in this disclosure. In some aspects, the CDR-L2 sequence comprises, consists of, or consists essentially of any of the illustrative CDR-L2 sequences provided in this disclosure, with 1, 2, or 3 amino acid substitutions. In some aspects, the amino acid substitutions are conservative amino acid substitutions.
[0157] In some aspects, the CDR-L1 sequence comprises, consists of, or consists essentially of a variant of an illustrative CDR-L1 sequence provided in this disclosure. In some aspects, the CDR-L1 sequence comprises, consists of, or consists essentially of a sequence having at least 70%, 75%, 80%, 85%, 90%, or 95% identity with any of the illustrative CDR-L1 sequences provided in this disclosure. In some aspects, the CDR-L1 sequence comprises, consists of, or consists essentially of any of the illustrative CDR-L1 sequences provided in this disclosure, with 1, 2, or 3 amino acid substitutions. In some aspects, the amino acid substitutions are conservative amino acid substitutions.
[0158] 2.5.9. Excluded V.sub.L Sequences Comprising CDR-Ls
[0159] In some embodiments, the V.sub.L sequences provided herein do not comprise certain CDR-L3, CDR-L2, and/or CDR-L1 sequences.
[0160] In some aspects, the CDR-L3 sequence does not comprise, consist of, or consist essentially of a sequence selected from SEQ ID NOs: 195-199 or 219-223. In some aspects, the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NOs: 195. In some aspects, the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NOs: 196. In some aspects, the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NOs: 197. In some aspects, the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NOs: 198. In some aspects, the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NOs: 199. In some aspects, the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NOs: 219. In some aspects, the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NOs: 220. In some aspects, the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NOs: 221. In some aspects, the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NOs: 222. In some aspects, the CDR-L3 sequence does not comprise, consist of, or consist essentially of SEQ ID NOs: 223.
[0161] In some aspects, the CDR-L2 sequence does not comprise, consist of, or consist essentially of a sequence selected from SEQ ID NOs: 166-170 or 190-194. In some aspects, the CDR-L2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 190. In some aspects, the CDR-L2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 166. In some aspects, the CDR-L2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 167. In some aspects, the CDR-L2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 168. In some aspects, the CDR-L2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 169. In some aspects, the CDR-L2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 170. In some aspects, the CDR-L2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 191. In some aspects, the CDR-L2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 192. In some aspects, the CDR-L2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 193. In some aspects, the CDR-L2 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 194.
[0162] In some aspects, the CDR-L1 sequence does not comprise, consist of, or consist essentially of a sequence selected from SEQ ID NOs: 137-141 or 161-165. In some aspects, the CDR-L1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 137. In some aspects, the CDR-L1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 138. In some aspects, the CDR-L1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 139. In some aspects, the CDR-L1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 140. In some aspects, the CDR-L1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 141. In some aspects, the CDR-L1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 161. In some aspects, the CDR-L1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 162. In some aspects, the CDR-L1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 163. In some aspects, the CDR-L1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 164. In some aspects, the CDR-L1 sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 165.
[0163] 2.6. V.sub.L Sequences
[0164] In some embodiments, the antibody comprises a V.sub.L sequence comprising, consisting of, or consisting essentially of a sequence selected from SEQ ID NOs: 270-288. In some aspects, the antibody comprises a V.sub.L sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 270. In some aspects, the antibody comprises a V.sub.L sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 281. In some aspects, the antibody comprises a V.sub.L sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 282. In some aspects, the antibody comprises a V.sub.L sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 283. In some aspects, the antibody comprises a V.sub.L sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 284. In some aspects, the antibody comprises a V.sub.L sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 285. In some aspects, the antibody comprises a V.sub.L sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 286. In some aspects, the antibody comprises a V.sub.L sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 287. In some aspects, the antibody comprises a V.sub.L sequence comprising, consisting of, or consisting essentially of SEQ ID NO: 288.
[0165] 2.6.1. Variants of V.sub.L Sequences
[0166] In some embodiments, the V.sub.L sequences provided herein comprise, consist of, or consist essentially of a variant of an illustrative V.sub.L sequence provided in this disclosure.
[0167] In some aspects, the V.sub.L sequence comprises, consists of, or consists essentially of a variant of an illustrative V.sub.L sequence provided in this disclosure. In some aspects, the V.sub.L sequence comprises, consists of, or consists essentially of a sequence having at least 85%, 90%, 95%, 96%, 97%, 98%, 99%, or 99.05% identity with any of the illustrative V.sub.L sequences provided in this disclosure.
[0168] In some embodiments, the V.sub.L sequence comprises, consists of, or consists essentially of any of the illustrative V.sub.L sequences provided in this disclosure, 20 or fewer, 19 or fewer, 18 or fewer, 17 or fewer, 16 or fewer, 15 or fewer, 14 or fewer, 13 or fewer, 12 or fewer, 11 or fewer, 10 or fewer, 9 or fewer, 8 or fewer, 7 or fewer, 6 or fewer, 5 or fewer, 4 or fewer, 3 or fewer, 2 or fewer, or 1 or fewer amino acid substitutions. In some aspects, the amino acid substitutions are conservative amino acid substitutions.
[0169] 2.6.2. Excluded V.sub.L Sequences
[0170] In some embodiments, the V.sub.L sequences provided herein do not comprise certain V.sub.L sequences.
[0171] In some aspects, the V.sub.L sequence does not comprise, consist of, or consist essentially of a sequence selected from SEQ ID NOs: 289-293. In some aspects, the V.sub.L sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 289. In some aspects, the V.sub.L sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 290. In some aspects, the V.sub.L sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 291. In some aspects, the V.sub.L sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 292. In some aspects, the V.sub.L sequence does not comprise, consist of, or consist essentially of SEQ ID NO: 293.
[0172] 2.7. Pairs
[0173] 2.7.1. CDR-H3-CDR-L3 Pairs
[0174] In some embodiments, the antibody comprises a CDR-H3 sequence and a CDR-L3 sequence. In some aspects, the CDR-H3 sequence is part of a VII and the CDR-L3 sequence is part of a V.sub.L.
[0175] In some aspects, the CDR-H3 sequence is a CDR-H3 sequence comprising, consisting of, or consisting essentially of SEQ ID NOs: 113-131 and 309-315, and the CDR-L3 sequence is a CDR-L3 sequence comprising, consisting of, or consisting essentially of SEQ ID NOs: 200-218.
[0176] In some aspects, the CDR-H3 sequence is SEQ ID NO: 113, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0177] In some aspects, the CDR-H3 sequence is SEQ ID NO: 114, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0178] In some aspects, the CDR-H3 sequence is SEQ ID NO: 115, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0179] In some aspects, the CDR-H3 sequence is SEQ ID NO: 116, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0180] In some aspects, the CDR-H3 sequence is SEQ ID NO: 117, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0181] In some aspects, the CDR-H3 sequence is SEQ ID NO: 118, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0182] In some aspects, the CDR-H3 sequence is SEQ ID NO: 119, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0183] In some aspects, the CDR-H3 sequence is SEQ ID NO: 120, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0184] In some aspects, the CDR-H3 sequence is SEQ ID NO: 121, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0185] In some aspects, the CDR-H3 sequence is SEQ ID NO: 122, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0186] In some aspects, the CDR-H3 sequence is SEQ ID NO: 123, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0187] In some aspects, the CDR-H3 sequence is SEQ ID NO: 124, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0188] In some aspects, the CDR-H3 sequence is SEQ ID NO: 125, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0189] In some aspects, the CDR-H3 sequence is SEQ ID NO: 126, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0190] In some aspects, the CDR-H3 sequence is SEQ ID NO: 127, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0191] In some aspects, the CDR-H3 sequence is SEQ ID NO: 128, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0192] In some aspects, the CDR-H3 sequence is SEQ ID NO: 129, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0193] In some aspects, the CDR-H3 sequence is SEQ ID NO: 130, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0194] In some aspects, the CDR-H3 sequence is SEQ ID NO: 131, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0195] In some aspects, the CDR-H3 sequence is SEQ ID NO: 309, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0196] In some aspects, the CDR-H3 sequence is SEQ ID NO: 310, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0197] In some aspects, the CDR-H3 sequence is SEQ ID NO: 311, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0198] In some aspects, the CDR-H3 sequence is SEQ ID NO: 312, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0199] In some aspects, the CDR-H3 sequence is SEQ ID NO: 313, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0200] In some aspects, the CDR-H3 sequence is SEQ ID NO: 314, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0201] In some aspects, the CDR-H3 sequence is SEQ ID NO: 315, and the CDR-L3 sequence is selected from SEQ ID NOs: 200-218. In some aspects, the CDR-L3 sequence is SEQ ID NO: 200. In some aspects, the CDR-L3 sequence is SEQ ID NO: 201. In some aspects, the CDR-L3 sequence is SEQ ID NO: 202. In some aspects, the CDR-L3 sequence is SEQ ID NO: 203. In some aspects, the CDR-L3 sequence is SEQ ID NO: 204. In some aspects, the CDR-L3 sequence is SEQ ID NO: 205. In some aspects, the CDR-L3 sequence is SEQ ID NO: 206. In some aspects, the CDR-L3 sequence is SEQ ID NO: 207. In some aspects, the CDR-L3 sequence is SEQ ID NO: 208. In some aspects, the CDR-L3 sequence is SEQ ID NO: 209. In some aspects, the CDR-L3 sequence is SEQ ID NO: 210. In some aspects, the CDR-L3 sequence is SEQ ID NO: 211. In some aspects, the CDR-L3 sequence is SEQ ID NO: 212. In some aspects, the CDR-L3 sequence is SEQ ID NO: 213. In some aspects, the CDR-L3 sequence is SEQ ID NO: 214. In some aspects, the CDR-L3 sequence is SEQ ID NO: 215. In some aspects, the CDR-L3 sequence is SEQ ID NO: 216. In some aspects, the CDR-L3 sequence is SEQ ID NO: 217. In some aspects, the CDR-L3 sequence is SEQ ID NO: 218.
[0202] 2.7.1.1. Variants of CDR-H3-CDR-L3 Pairs
[0203] In some embodiments, the CDR-H3-CDR-L3 pairs provided herein comprise a variant of an illustrative CDR-H3 and/or CDR-L1 sequence provided in this disclosure.
[0204] In some aspects, the CDR-H3 sequence comprises, consists of, or consists essentially of a variant of an illustrative CDR-H3 sequence provided in this disclosure. In some aspects, the CDR-H3 sequence comprises, consists of, or consists essentially of a sequence having at least 70%, 75%, 80%, 85%, 90%, or 95% identity with any of the illustrative CDR-H3 sequences provided in this disclosure. In some aspects, the CDR-H3 sequence comprises, consists of, or consists essentially of any of the illustrative CDR-H3 sequences provided in this disclosure, with 1, 2, or 3 amino acid substitutions. In some aspects, the amino acid substitutions are conservative amino acid substitutions.
[0205] In some aspects, the CDR-L3 sequence comprises, consists of, or consists essentially of a variant of an illustrative CDR-L3 sequence provided in this disclosure. In some aspects, the CDR-L3 sequence comprises, consists of, or consists essentially of a sequence having at least 70%, 75%, 80%, 85%, 90%, or 95% identity with any of the illustrative CDR-L3 sequences provided in this disclosure. In some aspects, the CDR-L3 sequence comprises, consists of, or consists essentially of any of the illustrative CDR-L3 sequences provided in this disclosure, with 1, 2, or 3 amino acid substitutions. In some aspects, the amino acid substitutions are conservative amino acid substitutions.
[0206] 2.7.1.2. Excluded CDR-H3-CDR-L3 Pairs
[0207] In some embodiments, the CDR-H3-CDR-L3 pairs provided herein do not comprise certain CDR-H3-CDR-L3 pairs.
[0208] In some aspects, the CDR-H3 sequence is not selected from SEQ ID NOs: 108-112 or 132-136, and the CDR-L3 sequence is not selected from SEQ ID NOs: 195-199 or 219-223.
[0209] In some aspects, the CDR-H3 sequence is not SEQ ID NO: 108, and the CDR-L3 sequence is not selected from SEQ ID NOs: 195-199 or 219-223. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 195. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 196. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 197. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 198. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 199. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 219. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 220. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 221. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 222. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 223.
[0210] In some aspects, the CDR-H3 sequence is not SEQ ID NO: 109, and the CDR-L3 sequence is not selected from SEQ ID NOs: 195-199 or 219-223. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 195. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 196. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 197. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 198. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 199. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 219. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 220. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 221. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 222. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 223.
[0211] In some aspects, the CDR-H3 sequence is not SEQ ID NO: 110, and the CDR-L3 sequence is not selected from SEQ ID NOs: 195-199 or 219-223. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 195. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 196. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 197. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 198. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 199. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 219. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 220. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 221. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 222. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 223.
[0212] In some aspects, the CDR-H3 sequence is not SEQ ID NO: 111, and the CDR-L3 sequence is not selected from SEQ ID NOs: 195-199 or 219-223. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 195. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 196. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 197. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 198. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 199. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 219. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 220. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 221. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 222. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 223.
[0213] In some aspects, the CDR-H3 sequence is not SEQ ID NO: 112, and the CDR-L3 sequence is not selected from SEQ ID NOs: 195-199 or 219-223. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 195. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 196. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 197. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 198. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 199. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 219. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 220. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 221. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 222. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 223.
[0214] In some aspects, the CDR-H3 sequence is not SEQ ID NO: 132, and the CDR-L3 sequence is not selected from SEQ ID NOs: 195-199 or 219-223. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 195. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 196. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 197. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 198. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 199. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 219. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 220. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 221. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 222. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 223.
[0215] In some aspects, the CDR-H3 sequence is not SEQ ID NO: 133, and the CDR-L3 sequence is not selected from SEQ ID NOs: 195-199 or 219-223. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 195. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 196. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 197. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 198. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 199. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 219. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 220. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 221. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 222. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 223.
[0216] In some aspects, the CDR-H3 sequence is not SEQ ID NO: 134, and the CDR-L3 sequence is not selected from SEQ ID NOs: 195-199 or 219-223. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 195. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 196. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 197. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 198. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 199. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 219. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 220. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 221. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 222. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 223.
[0217] In some aspects, the CDR-H3 sequence is not SEQ ID NO: 135, and the CDR-L3 sequence is not selected from SEQ ID NOs: 195-199 or 219-223. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 195. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 196. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 197. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 198. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 199. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 219. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 220. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 221. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 222. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 223.
[0218] In some aspects, the CDR-H3 sequence is not SEQ ID NO: 136, and the CDR-L3 sequence is not selected from SEQ ID NOs: 195-199 or 219-223. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 195. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 196. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 197. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 198. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 199. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 219. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 220. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 221. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 222. In some aspects, the CDR-L3 sequence is not SEQ ID NO: 223.
[0219] 2.7.2. V.sub.H-V.sub.L Pairs
[0220] In some embodiments, the antibody comprises a V.sub.H sequence and a V.sub.L sequence.
[0221] In some aspects, the V.sub.H sequence is a V.sub.H sequence comprising, consisting of, or consisting essentially of SEQ ID NOs: 246-264 and 316-322, and the V.sub.L sequence is a V.sub.L sequence comprising, consisting of, or consisting essentially of SEQ ID NOs: 270-288.
[0222] In some aspects, the V.sub.H sequence is SEQ ID NO: 246, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0223] In some aspects, the V.sub.H sequence is SEQ ID NO: 247, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0224] In some aspects, the V.sub.H sequence is SEQ ID NO: 248, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0225] In some aspects, the V.sub.H sequence is SEQ ID NO: 249, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0226] In some aspects, the V.sub.H sequence is SEQ ID NO: 250, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0227] In some aspects, the V.sub.H sequence is SEQ ID NO: 251, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0228] In some aspects, the V.sub.H sequence is SEQ ID NO: 252, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0229] In some aspects, the V.sub.H sequence is SEQ ID NO: 253, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0230] In some aspects, the V.sub.H sequence is SEQ ID NO: 254, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0231] In some aspects, the V.sub.H sequence is SEQ ID NO: 255 (with or without a serine prepended to the sequence), and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0232] In some aspects, the V.sub.H sequence is SEQ ID NO: 256, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0233] In some aspects, the V.sub.H sequence is SEQ ID NO: 257, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0234] In some aspects, the V.sub.H sequence is SEQ ID NO: 258, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0235] In some aspects, the V.sub.H sequence is SEQ ID NO: 259, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0236] In some aspects, the V.sub.H sequence is SEQ ID NO: 260, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0237] In some aspects, the V.sub.H sequence is SEQ ID NO: 261, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0238] In some aspects, the V.sub.H sequence is SEQ ID NO: 262, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0239] In some aspects, the V.sub.H sequence is SEQ ID NO: 263, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0240] In some aspects, the V.sub.H sequence is SEQ ID NO: 264, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0241] In some aspects, the V.sub.H sequence is SEQ ID NO: 316, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0242] In some aspects, the V.sub.H sequence is SEQ ID NO: 317, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0243] In some aspects, the V.sub.H sequence is SEQ ID NO: 318, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0244] In some aspects, the V.sub.H sequence is SEQ ID NO: 319, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0245] In some aspects, the V.sub.H sequence is SEQ ID NO: 320, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0246] In some aspects, the V.sub.H sequence is SEQ ID NO: 321, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0247] In some aspects, the V.sub.H sequence is SEQ ID NO: 322, and the V.sub.L sequence is selected from SEQ ID NOs: 270-288. In some aspects, the V.sub.L sequence is SEQ ID NO: 270. In some aspects, the V.sub.L sequence is SEQ ID NO: 271. In some aspects, the V.sub.L sequence is SEQ ID NO: 272. In some aspects, the V.sub.L sequence is SEQ ID NO: 273. In some aspects, the V.sub.L sequence is SEQ ID NO: 274. In some aspects, the V.sub.L sequence is SEQ ID NO: 275. In some aspects, the V.sub.L sequence is SEQ ID NO: 276. In some aspects, the V.sub.L sequence is SEQ ID NO: 277. In some aspects, the V.sub.L sequence is SEQ ID NO: 278. In some aspects, the V.sub.L sequence is SEQ ID NO: 279. In some aspects, the V.sub.L sequence is SEQ ID NO: 280. In some aspects, the V.sub.L sequence is SEQ ID NO: 281. In some aspects, the V.sub.L sequence is SEQ ID NO: 282. In some aspects, the V.sub.L sequence is SEQ ID NO: 283. In some aspects, the V.sub.L sequence is SEQ ID NO: 284. In some aspects, the V.sub.L sequence is SEQ ID NO: 285. In some aspects, the V.sub.L sequence is SEQ ID NO: 286. In some aspects, the V.sub.L sequence is SEQ ID NO: 287. In some aspects, the V.sub.L sequence is SEQ ID NO: 288.
[0248] 2.7.2.1. Variants of V.sub.H-V.sub.L Pairs
[0249] In some embodiments, the V.sub.H-V.sub.L pairs provided herein comprise a variant of an illustrative V.sub.H and/or V.sub.L sequence provided in this disclosure.
[0250] In some aspects, the V.sub.H sequence comprises, consists of, or consists essentially of a variant of an illustrative V.sub.H sequence provided in this disclosure. In some aspects, the V.sub.H sequence comprises, consists of, or consists essentially of a sequence having at least 85%, 90%, 95%, 96%, 97%, 98%, 99%, or 99.1% identity with any of the illustrative V.sub.H sequences provided in this disclosure.
[0251] In some embodiments, the V.sub.H sequence comprises, consists of, or consists essentially of any of the illustrative V.sub.H sequences provided in this disclosure, 20 or fewer, 19 or fewer, 18 or fewer, 17 or fewer, 16 or fewer, 15 or fewer, 14 or fewer, 13 or fewer, 12 or fewer, 11 or fewer, 10 or fewer, 9 or fewer, 8 or fewer, 7 or fewer, 6 or fewer, 5 or fewer, 4 or fewer, 3 or fewer, 2 or fewer, or 1 or fewer amino acid substitutions. In some aspects, the amino acid substitutions are conservative amino acid substitutions.
[0252] In some aspects, the V.sub.L sequence comprises, consists of, or consists essentially of a variant of an illustrative V.sub.L sequence provided in this disclosure. In some aspects, the V.sub.L sequence comprises, consists of, or consists essentially of a sequence having at least 85%, 90%, 95%, 96%, 97%, 98%, 99%, or 99.05% identity with any of the illustrative V.sub.L sequences provided in this disclosure.
[0253] In some embodiments, the V.sub.L sequence comprises, consists of, or consists essentially of any of the illustrative V.sub.L sequences provided in this disclosure, 20 or fewer, 19 or fewer, 18 or fewer, 17 or fewer, 16 or fewer, 15 or fewer, 14 or fewer, 13 or fewer, 12 or fewer, 11 or fewer, 10 or fewer, 9 or fewer, 8 or fewer, 7 or fewer, 6 or fewer, 5 or fewer, 4 or fewer, 3 or fewer, 2 or fewer, or 1 or fewer amino acid substitutions. In some aspects, the amino acid substitutions are conservative amino acid substitutions.
[0254] 2.7.2.2. Excluded V.sub.H-V.sub.L Pairs
[0255] In some embodiments, the V.sub.H-V.sub.L pairs provided herein do not comprise certain V.sub.H-V.sub.L pairs.
[0256] In some aspects, the V.sub.H sequence is not selected from SEQ ID NOs: 265-269, and the V.sub.L sequence is not selected from SEQ ID NOs: 289-293.
[0257] In some aspects, the V.sub.H sequence is not SEQ ID NO: 265, and the V.sub.L sequence is not selected from SEQ ID NO: 289-293. In some aspects, the V.sub.L sequence is not SEQ ID NO: 289. In some aspects, the V.sub.L sequence is not SEQ ID NO: 290. In some aspects, the V.sub.L sequence is not SEQ ID NO: 291. In some aspects, the V.sub.L sequence is not SEQ ID NO: 292. In some aspects, the V.sub.L sequence is not SEQ ID NO: 293.
[0258] In some aspects, the V.sub.H sequence is not SEQ ID NO: 266, and the V.sub.L sequence is not selected from SEQ ID NO: 289-293. In some aspects, the V.sub.L sequence is not SEQ ID NO: 289. In some aspects, the V.sub.L sequence is not SEQ ID NO: 290. In some aspects, the V.sub.L sequence is not SEQ ID NO: 291. In some aspects, the V.sub.L sequence is not SEQ ID NO: 292. In some aspects, the V.sub.L sequence is not SEQ ID NO: 293.
[0259] In some aspects, the V.sub.H sequence is not SEQ ID NO: 267, and the V.sub.L sequence is not selected from SEQ ID NO: 289-293. In some aspects, the V.sub.L sequence is not SEQ ID NO: 289. In some aspects, the V.sub.L sequence is not SEQ ID NO: 290. In some aspects, the V.sub.L sequence is not SEQ ID NO: 291. In some aspects, the V.sub.L sequence is not SEQ ID NO: 292. In some aspects, the V.sub.L sequence is not SEQ ID NO: 293.
[0260] In some aspects, the V.sub.H sequence is not SEQ ID NO: 268, and the V.sub.L sequence is not selected from SEQ ID NO: 289-293. In some aspects, the V.sub.L sequence is not SEQ ID NO: 289. In some aspects, the V.sub.L sequence is not SEQ ID NO: 290. In some aspects, the V.sub.L sequence is not SEQ ID NO: 291. In some aspects, the V.sub.L sequence is not SEQ ID NO: 292. In some aspects, the V.sub.L sequence is not SEQ ID NO: 293.
[0261] In some aspects, the V.sub.H sequence is not SEQ ID NO: 269, and the V.sub.L sequence is not selected from SEQ ID NO: 289-293. In some aspects, the V.sub.L sequence is not SEQ ID NO: 289. In some aspects, the V.sub.L sequence is not SEQ ID NO: 290. In some aspects, the V.sub.L sequence is not SEQ ID NO: 291. In some aspects, the V.sub.L sequence is not SEQ ID NO: 292. In some aspects, the V.sub.L sequence is not SEQ ID NO: 293.
[0262] 2.8. Consensus Sequences
[0263] In some embodiments, provided herein are anti-PD-1 antibodies comprising one or more sequences defined by consensus sequences. Each consensus sequence is based, at least in part, on one or more alignments of two or more useful anti-PD-1 CDR sequences provided in this disclosure. Based on such alignments, a person of skill in the art would recognize that different amino acid residues may useful in certain positions of the CDRs. Accordingly, each consensus sequence encompasses two or more useful anti-PD-1 CDR sequences.
[0264] 2.8.1. CDR-H3 Consensus Sequences
[0265] In some embodiments, the antibody comprises a CDR-H3 sequence defined by the consensus sequence D-.alpha..sub.2-.alpha..sub.3-Y-.alpha..sub.5-.alpha..sub.6-G-S-G-Y, where .alpha..sub.2 is A, V, or S; .alpha..sub.3 is D or E; .alpha..sub.5 is S or G; and .alpha..sub.6 is S, L, or T. In some embodiments, as is S, G, or R. Sequencing of individual clones isolated from the output of the antibody selection process revealed that R occurred at position as at nearly the same frequency as G.
[0266] In some aspects, if .alpha..sub.2 is A, then .alpha..sub.3 is not D; .alpha..sub.5 is not S; or .alpha..sub.6 is not S; or combinations thereof.
[0267] In some aspects, .alpha..sub.2 is V or S; .alpha..sub.3 is E; as is G; or .alpha..sub.6 is L or T; or combinations thereof.
[0268] In some aspects, .alpha..sub.2 is not A; .alpha..sub.3 is not D; .alpha..sub.5 is not S; or .alpha..sub.6 is not S; or combinations thereof.
[0269] In some embodiments, the antibody comprises a CDR-H3 sequence defined by the consensus sequence .sub.1-G-Y- .sub.4- .sub.5-Y- .sub.7-.beta..sub.8-F- .sub.10- .sub.11, where .sub.1 is not present or Q; .sub.4 is G or D; .sub.5 is N or V; .sub.7 is L or S; .sub.8 is Y or W; .sub.10 is D or A; and .sub.11 is V or Y.
[0270] 2.8.2. Chothia CDR-H1 Consensus Sequences
[0271] In some embodiments, the antibody comprises a Chothia CDR-H1 sequence defined by the consensus sequence G-.epsilon..sub.2-.epsilon..sub.3-.epsilon..sub.4-.epsilon..sub.5-.epsilo- n..sub.6-.epsilon..sub.7, where .epsilon..sub.2 is Y or F; .epsilon..sub.3 is T, R or I; .epsilon..sub.4 is F or L; .epsilon..sub.5 is S, E, T, P, or R; .epsilon..sub.6 is T, S, H, Q, R, or W; and .epsilon..sub.7 is F, Y, or Q.
[0272] In some aspects, if .epsilon..sub.5 is T, then .epsilon..sub.6 is not S.
[0273] In some aspects, .epsilon..sub.5 is S, E, P, or R; or .epsilon..sub.6 is T, H, Q, R, or W; or combinations thereof.
[0274] In some aspects, .epsilon..sub.2 is not Y; .epsilon..sub.3 is not T or R; .epsilon..sub.5 is not T; .epsilon..sub.6 is not S; or .epsilon..sub.7 is not Y; or combinations thereof.
[0275] 2.8.3. Kabat CDR-H2 Consensus Sequences
[0276] In some embodiments, the antibody comprises a Kabat CDR-H2 sequence defined by the consensus sequence W-.gamma..sub.2-S-A-.gamma..sub.5-N-G-N-T-.gamma..sub.10-Y-A-Q-K-L-Q-G, where .gamma.2 is I or V; .gamma.5 is Y or H; and .gamma.10 is K or N.
[0277] In some aspects, .gamma.2 is not I; .gamma.5 is not Y; or .gamma.10 is not N; or combinations thereof.
[0278] In some embodiments, the antibody comprises a Kabat CDR-H2 sequence defined by the consensus sequence .delta..sub.1-I-S-G-.delta..sub.5-G-.delta..sub.7-.delta..sub.8-T-Y-Y-.de- lta..sub.12-D-S-V-.delta..sub.16-G, where .delta..sub.1 is T or A; .delta..sub.5 is S or G; .delta..sub.7 is S or G; .delta..sub.8 is S, D or N; .delta..sub.12 is A, P or S; and .delta..sub.16 is K or Q.
[0279] In some aspects, if .delta..sub.1 is A, then .delta..sub.5 is not S; .delta..sub.7 is not G; .delta..sub.8 is not S; .delta..sub.12 is not A; or .delta..sub.16 is not K; or combinations thereof.
[0280] In some aspects, .delta..sub.1 is T; .delta..sub.5 is G; .delta..sub.7 is S; .delta..sub.8 is D or N; or .delta..sub.12 is P or S; or combinations thereof.
[0281] In some aspects, .delta..sub.1 is not A; .delta..sub.5 is not S; .delta..sub.7 is not G; .delta..sub.8 is not S; .delta..sub.12 is not A; or .delta..sub.16 is not K; or combinations thereof.
[0282] 2.8.4. Kabat CDR-H1 Consensus Sequences
[0283] In some embodiments, the antibody comprises a Kabat CDR-H1 sequence defined by the consensus sequence .THETA..sub.1-.THETA..sub.2-G-.THETA..sub.4-S, where .THETA..sub.1 is T, R, W, Q, H, or S; .THETA..sub.2 is Y, F, or Q; and .THETA..sub.4 is M or I.
[0284] In some aspects, .THETA..sub.1 is T, R, W, Q, or H; .THETA..sub.2 is F or Q; or .THETA..sub.4 is M; or combinations thereof.
[0285] In some aspects, .THETA..sub.1 is not S; .THETA..sub.2 is not Y; or .THETA..sub.4 is not I; or combinations thereof.
[0286] 2.8.5. CDR-L3 Consensus Sequences
[0287] In some embodiments, the antibody comprises a CDR-L3 sequence defined by the consensus sequence Q-Q-.pi..sub.3-.pi..sub.4-.pi..sub.5-.pi..sub.6-P-.pi..sub.8-T, where .pi..sub.3 is N, S, or W; .pi..sub.4 is Y, K, or I; .pi..sub.5 is N, E, or S; .pi..sub.6 is S, V, D, or T; and .pi..sub.8 is Y or W.
[0288] In some aspects, if .pi..sub.4 is Y, then .pi..sub.3 is not S; .pi..sub.5 is not S; .pi..sub.6 is not T; or .pi..sub.8 is not W; or combinations thereof.
[0289] In some aspects, .pi..sub.3 is N or W; .pi..sub.4 is K or I; .pi..sub.5 is N or E; .pi..sub.6 is S, V, or D; or .pi..sub.8 is Y; or combinations thereof.
[0290] In some aspects, .pi..sub.3 is not S; .pi..sub.4 is not Y; .pi..sub.5 is not S; .pi..sub.6 is not T; or .pi..sub.8 is not W; or combinations thereof.
[0291] 2.8.6. CDR-L1 Consensus Sequences
[0292] In some embodiments, the antibody comprises a CDR-L1 sequence defined by the consensus sequence S-G-D-A-L-.mu.6-.mu.7-Q-Y-.mu.10-Y, where .mu.6 is P, T, or S; .mu.7 is M, T, E, or K; and .mu..sub.10 is G or A.
[0293] In some aspects, if .mu..sub.6 is P, then .mu..sub.7 is not K, .mu..sub.10 is not A, or combinations thereof.
[0294] In some aspects, .mu..sub.6 is T or S; .mu..sub.7 is M, T, or E; or .mu..sub.10 is G; or combinations thereof.
[0295] In some aspects, .mu..sub.6 is not P; .mu..sub.7 is not K; or .mu..sub.10 is not A; or combinations thereof.
[0296] In some embodiments, the antibody comprises a CDR-L1 sequence defined by the consensus sequence R-A-S-E-.SIGMA..sub.5-V-D-.SIGMA..sub.8-.SIGMA..sub.9-G-.SIGMA..sub.11-S-- F-M-.SIGMA..sub.15, where .SIGMA..sub.5 is S or N; .SIGMA..sub.8 is N or D; .SIGMA..sub.9 is S or Y; .SIGMA..sub.11 is I or V; and .SIGMA..sub.15 is S or N.
3. Germline
[0297] In some embodiments, the antibody that specifically binds PD-1 is an antibody comprising a variable region that is encoded by a particular germline gene, or a variant thereof. The illustrative antibodies provided herein comprise variable regions that are encoded by the heavy chain variable region germline genes VH1-18, VH3-21, VH3-7, and VH3-15, or variants thereof; and the light chain variable region germline genes V.lamda.3-25, V.kappa.1-9, V.kappa.3-11, V.kappa.3-20, and V.kappa.4-1, or variants thereof. One of skill in the art would recognize that the CDR sequences provided herein may also be useful when combined with variable regions encoded by other variable region germline genes, or variants thereof. In particular, the CDR sequences provided herein may be useful when combined with variable regions encoded by variable region germline genes, or variants thereof, that are structurally similar to the variable region germline genes recited above. For example, in some embodiments, a CDR-H sequence provided herein may be combined with a variable region encoded by a variable region germline gene selected from the VH1 or VH3 family, or a variant thereof. In some embodiments, a CDR-L sequence provided herein may be combined with a variable region encoded by a variable region germline gene selected from the V.lamda.3, V.kappa.1, V.kappa.3, and V.kappa.4 families, or a variant thereof.
4. Affinity
[0298] In some embodiments, the affinity of the antibody for PD-1, as indicated by K.sub.D, is less than about 10.sup.-5 M, less than about 10.sup.-6 M, less than about 10.sup.-7 M, less than about 10.sup.-8 M, less than about 10.sup.-9 M, less than about 10.sup.-10 M, less than about 10.sup.-11 M, or less than about 10.sup.-12 M. In some embodiments, the affinity of the antibody is between about 10.sup.-7 M and 10.sup.-11 M. In some embodiments, the affinity of the antibody is between about 10.sup.-7 M and 10.sup.-10 M. In some embodiments, the affinity of the antibody is between about 10.sup.-7 M and 10.sup.-9 M. In some embodiments, the affinity of the antibody is between about 10.sup.-7 M and 10.sup.-8 M. In some embodiments, the affinity of the antibody is between about 10.sup.-8 M and 10.sup.-11 M. In some embodiments, the affinity of the antibody is between about 10.sup.-8 M and 10.sup.-10 M. In some embodiments, the affinity of the antibody is between about 10.sup.-9 M and 10.sup.-11 M. In some embodiments, the affinity of the antibody is between about 10.sup.-10 M and 10.sup.-11M.
[0299] In some embodiments, the affinity of the antibody for human PD-1 is between about 3.85.times.10.sup.-8 M and 2.52.times.10.sup.-10 M. In some embodiment, the affinity of the antibody for human PD-1 is about 2.55.times.10.sup.-8 M, about 1.52.times.10.sup.-8 M, about 9.52.times.10.sup.-9 M, about 1.09.times.10.sup.-8 M, about 4.50.times.10.sup.-9 M, about 1.90.times.10.sup.-9 M, about 4.76.times.10.sup.-9 M, about 4.5.times.10.sup.-9 M, about 1.04.times.10.sup.-8 M, about 9.90.times.10.sup.-9 M, about 9.13.times.10.sup.-10 M, about 2.52.times.10.sup.-10 M, about 2.58.times.10.sup.-9 M, about 3.85.times.10.sup.-8 M, about 3.66.times.10.sup.-9 M, about 3.15.times.10.sup.-9 M, about 5.14.times.10.sup.-9 M, about 2.47.times.10.sup.-9M, about 2.79.times.10.sup.-9M, about 1.20.times.10.sup.-9M, or about 1.28.times.10.sup.-8 M
[0300] In some embodiments, the affinity of the antibody for human PD-1 expressed on the surface of a cell is between about 3.2 and about 0.2 nM. In some embodiment, the affinity of the antibody for human PD-1 expressed on the surface of a cell is about 0.2 nM, about 0.4 nM, about 0.9 nM, about 1 nM, about 0.3 nM, about 0.7 nM, about 0.2 nM, about 0.8 nM, about 3.2 nM, about 2.9 nM, about 1.39 nM, or about 1.34 nM.
[0301] In some embodiments, the affinity of the antibody for murine PD-1 is between about 6.09.times.10.sup.-8 M and 9.08.times.10.sup.-9 M. In some embodiment, the affinity of the antibody for murine PD-1 is about 6.09.times.10.sup.-8 M, about 6.22.times.10.sup.-8 M, or about 9.08.times.10.sup.-9 M.
[0302] In some embodiments, the affinity of the antibody for cynomolgus PD-1 is between about 2.43.times.10.sup.-8 M and 1.95.times.10.sup.-10 M. In some embodiment, the affinity of the antibody for cynomolgus PD-1 is about 2.43.times.10.sup.-8 M, about 1.55.times.10.sup.-8 M, about 2.22.times.10.sup.-8 M, about 2.56.times.10.sup.-9M, about 2.54.times.10.sup.-9M, about 5.61.times.10.sup.-10 M, or about 1.95.times.10.sup.-10 M
[0303] In some embodiments the antibody has a k.sub.a of at least about 10.sup.4 M.sup.-1.times.sec.sup.-1. In some embodiments the antibody has a k.sub.a of at least about 10.sup.5 M.sup.-1.times.sec.sup.-1. In some embodiments the antibody has a k.sub.a of at least about 10.sup.6 M.sup.-1.times.sec.sup.-1. In some embodiments the antibody has a k.sub.a of between about 10.sup.4 M.sup.-1.times.sec.sup.-1 and about 10.sup.5 M.sup.-1.times.sec.sup.-1. In some embodiments the antibody has a k.sub.a of between about 10.sup.5 M.sup.-1.times.sec.sup.-1 and about 10.sup.6 M.sup.-1.times.sec.sup.-1.
[0304] In some embodiments the antibody has a k.sub.a when associating with human PD-1 of between about 4.74.times.10.sup.4 M.sup.-1.times.sec.sup.-1 and about 1.23.times.10.sup.6 M.sup.-1.times.sec.sup.-1. In some embodiments the antibody has a k.sub.a when associating with human PD-1 of about 4.88.times.10.sup.5 M.sup.-1.times.sec.sup.-1, about 1.23.times.10.sup.6 M.sup.-1.times.sec.sup.-1, about 7.37.times.10.sup.5 M.sup.-1.times.sec.sup.-1, about 6.87.times.10.sup.5 M.sup.-1.times.sec.sup.-1, about 5.63.times.10.sup.5 M.sup.-1.times.sec.sup.-1, about 5.16.times.10.sup.5 M.sup.-1.times.sec.sup.-1, about 2.48.times.10.sup.5 M.sup.-1.times.sec.sup.-1, about 7.98.times.10.sup.5 M.sup.-1.times.sec.sup.-1, about 1.82.times.10.sup.5 M.sup.-1.times.sec.sup.-1, about 4.74.times.10.sup.4 M.sup.-1.times.sec.sup.-1, about 1.85.times.10.sup.5 M.sup.-1.times.sec.sup.-1, about 2.00.times.10.sup.5 M.sup.-1.times.sec.sup.-1, about 8.12.times.10.sup.4 M.sup.-1.times.sec.sup.-1, about 1.21.times.10.sup.6 M.sup.-1.times.sec.sup.-1, about 1.16.times.10.sup.6 M.sup.-1.times.sec.sup.-1, about 5.13.times.10.sup.5 M.sup.-1.times.sec.sup.-1, or about 1.86.times.10.sup.5 M.sup.-1.times.sec.sup.-1.
[0305] In some embodiments the antibody has a k.sub.d of about 10.sup.-5 sec.sup.-1 or less. In some embodiments the antibody has a k.sub.d of about 10.sup.4 sec.sup.-1 or less. In some embodiments the antibody has a k.sub.d of about 10.sup.-3 sec.sup.-1 or less. In some embodiments the antibody has a k.sub.d of between about 10.sup.-2 sec.sup.-1 and about 10.sup.-5 sec.sup.-1. In some embodiments the antibody has a k.sub.d of between about 10.sup.-2 sec.sup.-1 and about 10.sup.-4 sec.sup.-1. In some embodiments the antibody has a k.sub.d of between about 10.sup.-3 sec.sup.-1 and about 10.sup.-5 sec.sup.-1.
[0306] In some embodiments the antibody has a k.sub.d when dissociating from human PD-1 of between about 1.87.times.10.sup.-2 sec.sup.-1 and about 4.17.times.10.sup.-4 sec.sup.-1. In some embodiments the antibody has a k.sub.d when dissociating from human PD-1 of about 1.24.times.10.sup.-2 sec.sup.-1, about 1.87.times.10.sup.-2 sec.sup.-1, about 7.01.times.10.sup.-3 sec.sup.-1, about 7.74.times.10.sup.-3 sec.sup.-1, about 2.54.times.10.sup.-3 sec.sup.-1, about 9.80.times.10.sup.-4 sec.sup.-1, about 1.18.times.10.sup.-3 sec.sup.-1, about 3.59.times.10.sup.-3 sec.sup.-1, about 4.68.times.10.sup.-4 sec.sup.-1, about 1.82.times.10.sup.-3 sec.sup.-1, about 6.79.times.10.sup.-4 sec.sup.-1, about 6.28.times.10.sup.-4 sec.sup.-1, about 4.17.times.10.sup.-4 sec.sup.-1, about 2.99.times.10.sup.-3 sec.sup.-1, about 3.24.times.10.sup.-3 sec.sup.-1, about 6.17.times.10.sup.-4 sec.sup.-1, or about 2.39.times.10.sup.-3 sec.sup.-1.
[0307] In some aspects, the K.sub.D, k.sub.a, and k.sub.d are determined at 25.degree. C. In some embodiments, the K.sub.D, k.sub.a, and k.sub.d are determined by surface plasmon resonance. In some embodiments, the K.sub.D, k.sub.a, and k.sub.d are determined according to the methods described in Examples 4 and 6.
5. Inhibition of PD-L1 and PD-L2 Binding
[0308] In some embodiments, the antibody inhibits binding of one or more of PD-L1 and PD-L2 to PD-1.
[0309] In some embodiments, the antibody inhibits binding of PD-L1 to PD-1 with an IC50 of about 1 to about 7 nM. In some aspects, the antibody inhibits binding of PD-L1 to PD-1 with an IC50 of about 1.99, about 2.53, about 5.86, or about 5.96 nM.
[0310] In some embodiments, the antibody inhibits binding of PD-L2 to PD-1 with an IC.sub.50 of about 0.01 to about 1 nM. In some aspects, the antibody inhibits binding of PD-L2 to PD-1 with an IC.sub.50 of about 0.01, about 0.18, about 0.56, or about 0.58 nM.
[0311] In some aspects, the antibody inhibits binding of PD-L1 to PD-1 with an IC.sub.50 of about 5.96 nM, and inhibits binding of PD-L2 to PD-1 with an IC.sub.50 of about 0.56 nM. In some aspects, the antibody inhibits binding of PD-L1 to PD-1 with an IC.sub.50 of about 5.86 nM, and inhibits binding of PD-L2 to PD-1 with an IC.sub.50 of about 0.58 nM. In some aspects, the antibody inhibits binding of PD-L1 to PD-1 with an IC.sub.50 of about 1.99 nM, and inhibits binding of PD-L2 to PD-1 with an IC.sub.50 of about 0.01 nM. In some aspects, the antibody inhibits binding of PD-L1 to PD-1 with an IC.sub.50 of about 2.53 nM, and inhibits binding of PD-L2 to PD-1 with an IC.sub.50 of about 0.18 nM.
6. PD-1 Assays
[0312] In some embodiments, the anti-PD-1 antibodies induce the secretion of interferon gamma when added to a peripheral blood mononuclear cell (PBMC) two-way mixed lymphocyte reaction (MLR) assay, as described in Examples 8 and 16.
[0313] In some embodiments, the anti-PD-1 antibodies induce the secretion of interferon gamma when added to a PBMC cytomegalovirus recall assay, as described in Example 16.
[0314] In some embodiments, the anti-PD-1 antibodies accelerate the onset of graft versus host disease, as shown in Example 18.
7. Glycosylation Variants
[0315] In certain embodiments, an antibody may be altered to increase, decrease or eliminate the extent to which it is glycosylated. Glycosylation of polypeptides is typically either "N-linked" or "O-linked."
[0316] "N-linked" glycosylation refers to the attachment of a carbohydrate moiety to the side chain of an asparagine residue. The tripeptide sequences asparagine-X-serine and asparagine-X-threonine, where X is any amino acid except proline, are the recognition sequences for enzymatic attachment of the carbohydrate moiety to the asparagine side chain. Thus, the presence of either of these tripeptide sequences in a polypeptide creates a potential glycosylation site.
[0317] "O-linked" glycosylation refers to the attachment of one of the sugars N-acetylgalactosamine, galactose, or xylose to a hydroxyamino acid, most commonly serine or threonine, although 5-hydroxyproline or 5-hydroxylysine may also be used.
[0318] Addition or deletion of N-linked glycosylation sites to the antibody may be accomplished by altering the amino acid sequence such that one or more of the above-described tripeptide sequences is created or removed. Addition or deletion of O-linked glycosylation sites may be accomplished by addition, deletion, or substitution of one or more serine or threonine residues in or to (as the case may be) the sequence of an antibody.
8. Fc Variants
[0319] In certain embodiments, amino acid modifications may be introduced into the Fc region of an antibody provided herein to generate an Fc region variant. In certain embodiments, the Fc region variant possesses some, but not all, effector functions. Such antibodies may be useful, for example, in applications in which the half-life of the antibody in vivo is important, yet certain effector functions are unnecessary or deleterious. Examples of effector functions include complement-dependent cytotoxicity (CDC) and antibody-directed complement-mediated cytotoxicity (ADCC). Numerous substitutions or substitutions or deletions with altered effector function are known in the art.
[0320] An alteration in in CDC and/or ADCC activity can be confirmed using in vitro and/or in vivo assays. For example, Fc receptor (FcR) binding assays can be conducted to measure Fc.gamma.R binding. The primary cells for mediating ADCC, NK cells, express Fc.gamma.RIII only, whereas monocytes express Fc.gamma.RI, Fc.gamma.RII and Fc.gamma.RIII. FcR expression on hematopoietic cells is summarized in Ravetch and Kinet, Ann. Rev. Immunol., 1991, 9:457-492.
[0321] Non-limiting examples of in vitro assays to assess ADCC activity of a molecule of interest are provided in U.S. Pat. Nos. 5,500,362 and 5,821,337; Hellstrom et al., Proc. Natl. Acad. Sci. USA., 1986, 83:7059-7063; Hellstrom et al., Proc. Natl. Acad. Sci. USA., 1985, 82:1499-1502; and Bruggemann et al., J. Exp. Med., 1987, 166:1351-1361. Useful effector cells for such assays include peripheral blood mononuclear cells (PBMC) and Natural Killer (NK) cells. Alternatively, or additionally, ADCC activity of the molecule of interest may be assessed in vivo, using an animal model such as that disclosed in Clynes et al. Proc. Natl. Acad. Sci. USA., 1998, 95:652-656.
[0322] C1q binding assays may also be carried out to confirm that the antibody is unable to bind C1q and hence lacks CDC activity. Examples of C1q binding assays include those described in WO 2006/029879 and WO 2005/100402.
[0323] Complement activation assays include those described, for example, in Gazzano-Santoro et al., J. Immunol. Methods, 1996, 202:163-171; Cragg et al., Blood, 2003, 101:1045-1052; and Cragg and Glennie, Blood, 2004, 103:2738-2743.
[0324] FcRn binding and in vivo clearance (half-life determination) can also be measured, for example, using the methods described in Petkova et al., Intl. Immunol., 2006, 18:1759-1769.
9. Preparation of Antibodies
[0325] 9.1. Antigen Preparation
[0326] The PD-1 antigen to be used for production of antibodies may be intact PD-1 or a fragment of PD-1. The intact PD-1, or fragment of PD-1, may be in the form of an isolated protein or expressed by a cell. Other forms of PD-1 useful for generating antibodies will be apparent to those skilled in the art.
[0327] 9.2. Monoclonal Antibodies
[0328] Monoclonal antibodies may be obtained, for example, using the hybridoma method first described by Kohler et al., Nature, 1975, 256:495-497, and/or by recombinant DNA methods (see e.g., U.S. Pat. No. 4,816,567). Monoclonal antibodies may also be obtained, for example, using phage or yeast-based libraries. See e.g., U.S. Pat. Nos. 8,258,082 and 8,691,730.
[0329] In the hybridoma method, a mouse or other appropriate host animal is immunized to elicit lymphocytes that produce or are capable of producing antibodies that will specifically bind to the protein used for immunization. Alternatively, lymphocytes may be immunized in vitro. Lymphocytes are then fused with myeloma cells using a suitable fusing agent, such as polyethylene glycol, to form a hybridoma cell. See Goding J. W., Monoclonal Antibodies: Principles and Practice 3.sup.rd ed. (1986) Academic Press, San Diego, Calif.
[0330] The hybridoma cells are seeded and grown in a suitable culture medium that contains one or more substances that inhibit the growth or survival of the unfused, parental myeloma cells. For example, if the parental myeloma cells lack the enzyme hypoxanthine guanine phosphoribosyl transferase (HGPRT or HPRT), the culture medium for the hybridomas typically will include hypoxanthine, aminopterin, and thymidine (HAT medium), which substances prevent the growth of HGPRT-deficient cells.
[0331] Useful myeloma cells are those that fuse efficiently, support stable high-level production of antibody by the selected antibody-producing cells, and are sensitive media conditions, such as the presence or absence of HAT medium. Among these, preferred myeloma cell lines are murine myeloma lines, such as those derived from MOP-21 and MC-11 mouse tumors (available from the Salk Institute Cell Distribution Center, San Diego, Calif.), and SP-2 or X63-Ag8-653 cells (available from the American Type Culture Collection, Rockville, Md.). Human myeloma and mouse-human heteromyeloma cell lines also have been described for the production of human monoclonal antibodies. See e.g., Kozbor, J. Immunol., 1984, 133:3001.
[0332] After the identification of hybridoma cells that produce antibodies of the desired specificity, affinity, and/or biological activity, selected clones may be subcloned by limiting dilution procedures and grown by standard methods. See Goding, supra. Suitable culture media for this purpose include, for example, D-MEM or RPMI-1640 medium. In addition, the hybridoma cells may be grown in vivo as ascites tumors in an animal.
[0333] DNA encoding the monoclonal antibodies may be readily isolated and sequenced using conventional procedures (e.g., by using oligonucleotide probes that are capable of binding specifically to genes encoding the heavy and light chains of the monoclonal antibodies). Thus, the hybridoma cells can serve as a useful source of DNA encoding antibodies with the desired properties. Once isolated, the DNA may be placed into expression vectors, which are then transfected into host cells such as bacteria (e.g., E. coli), yeast (e.g., Saccharomyces or Pichia sp.), COS cells, Chinese hamster ovary (CHO) cells, or myeloma cells that do not otherwise produce antibody, to produce the monoclonal antibodies.
[0334] 9.3. Humanized Antibodies
[0335] Humanized antibodies may be generated by replacing most, or all, of the structural portions of a monoclonal antibody with corresponding human antibody sequences. Consequently, a hybrid molecule is generated in which only the antigen-specific variable, or CDR, is composed of non-human sequence. Methods to obtain humanized antibodies include those described in, for example, Winter and Milstein, Nature, 1991, 349:293-299; Rader et al., Proc. Nat. Acad. Sci. USA., 1998, 95:8910-8915; Steinberger et al., J. Biol. Chem., 2000, 275:36073-36078; Queen et al., Proc. Natl. Acad. Sci. USA., 1989, 86:10029-10033; and U.S. Pat. Nos. 5,585,089, 5,693,761, 5,693,762, and 6,180,370.
[0336] 9.4. Human Antibodies
[0337] Human antibodies can be generated by a variety of techniques known in the art, for example by using transgenic animals (e.g., humanized mice). See, e.g., Jakobovits et al., Proc. Natl. Acad. Sci. USA., 1993, 90:2551; Jakobovits et al., Nature, 1993, 362:255-258; Bruggermann et al., Year in Immuno., 1993, 7:33; and U.S. Pat. Nos. 5,591,669, 5,589,369 and 5,545,807. Human antibodies can also be derived from phage-display libraries (see e.g., Hoogenboom et al., J. Mol. Biol., 1991, 227:381-388; Marks et al., J. Mol. Biol., 1991, 222:581-597; and U.S. Pat. Nos. 5,565,332 and 5,573,905). Human antibodies may also be generated by in vitro activated B cells (see e.g., U.S. Pat. Nos. 5,567,610 and 5,229,275). Human antibodies may also be derived from yeast-based libraries (see e.g., U.S. Pat. No. 8,691,730).
10. Vectors, Host Cells, and Recombinant Methods
[0338] The invention also provides isolated nucleic acids encoding anti-PD-1 antibodies, vectors and host cells comprising the nucleic acids, and recombinant techniques for the production of the antibodies.
[0339] For recombinant production of the antibody, the nucleic acid encoding it may be isolated and inserted into a replicable vector for further cloning (i.e., amplification of the DNA) or expression. In some aspects, the nucleic acid may be produced by homologous recombination, for example as described in U.S. Pat. No. 5,204,244.
[0340] Many different vectors are known in the art. The vector components generally include, but are not limited to, one or more of the following: a signal sequence, an origin of replication, one or more marker genes, an enhancer element, a promoter, and a transcription termination sequence, for example as described in U.S. Pat. No. 5,534,615.
[0341] Illustrative examples of suitable host cells are provided below, these host cells are not meant to be limiting.
[0342] Suitable host cells include any prokaryotic (e.g., bacterial), lower eukaryotic (e.g., yeast), or higher eukaryotic (e.g., mammalian) cells. Suitable prokaryotes include eubacteria, such as Gram-negative or Gram-positive organisms, for example, Enterobacteriaceae such as Escherichia (E. coli), Enterobacter, Erwinia, Klebsiella, Proteus, Salmonella (S. typhimurium), Serratia (S. marcescans), Shigella, Bacilli (B. subtilis and B. licheniformis), Pseudomonas (P. aeruginosa), and Streptomyces. One useful E. coli cloning host is E. coli 294, although other strains such as E. coli B, E. coli X1776, and E. coli W3110 are suitable.
[0343] In addition to prokaryotes, eukaryotic microbes such as filamentous fungi or yeast are also suitable cloning or expression hosts for anti-PD-1 antibody-encoding vectors. Saccharomyces cerevisiae, or common baker's yeast, is a commonly used lower eukaryotic host microorganism. However, a number of other genera, species, and strains are available and useful, such as Schizosaccharomyces pombe, Kluyveromyces (K. lactis, K. fragilis, K. bulgaricus K. wickeramii, K. waltii, K. drosophilarum, K. thermotolerans, and K. marxianus), Yarrowia, Pichia pastoris, Candida (C. albicans), Trichoderma reesia, Neurospora crassa, Schwanniomyces (S. occidentalis), and filamentous fungi such as, for example Penicillium, Tolypocladium, and Aspergillus (A. nidulans and A. niger).
[0344] Useful mammalian host cells include COS-7 cells, HEK293 cells; baby hamster kidney (BHK) cells; Chinese hamster ovary (CHO); mouse sertoli cells; African green monkey kidney cells (VERO-76), and the like.
[0345] The host cells used to produce the anti-PD-1 antibody of this invention may be cultured in a variety of media. Commercially available media such as, for example, Ham's F10, Minimal Essential Medium (MEM), RPMI-1640, and Dulbecco's Modified Eagle's Medium (DMEM) are suitable for culturing the host cells. In addition, any of the media described in Ham et al., Meth. Enz., 1979, 58:44; Barnes et al., Anal. Biochem., 1980, 102:255; and U.S. Pat. Nos. 4,767,704, 4,657,866, 4,927,762, 4,560,655, and 5,122,469, or WO 90/03430 and WO 87/00195 may be used.
[0346] Any of these media may be supplemented as necessary with hormones and/or other growth factors (such as insulin, transferrin, or epidermal growth factor), salts (such as sodium chloride, calcium, magnesium, and phosphate), buffers (such as HEPES), nucleotides (such as adenosine and thymidine), antibiotics, trace elements (defined as inorganic compounds usually present at final concentrations in the micromolar range), and glucose or an equivalent energy source. Any other necessary supplements may also be included at appropriate concentrations that would be known to those skilled in the art.
[0347] The culture conditions, such as temperature, pH, and the like, are those previously used with the host cell selected for expression, and will be apparent to the ordinarily skilled artisan.
[0348] When using recombinant techniques, the antibody can be produced intracellularly, in the periplasmic space, or directly secreted into the medium. If the antibody is produced intracellularly, as a first step, the particulate debris, either host cells or lysed fragments, is removed, for example, by centrifugation or ultrafiltration. For example, Carter et al. (Bio/Technology, 1992, 10:163-167) describes a procedure for isolating antibodies which are secreted to the periplasmic space of E. coli. Briefly, cell paste is thawed in the presence of sodium acetate (pH 3.5), EDTA, and phenylmethylsulfonylfluoride (PMSF) over about 30 min. Cell debris can be removed by centrifugation.
[0349] In some embodiments, the antibody is produced in a cell-free system. In some aspects, the cell-free system is an in vitro transcription and translation system as described in Yin et al., mAbs, 2012, 4:217-225, incorporated by reference in its entirety. In some aspects, the cell-free system utilizes a cell-free extract from a eukaryotic cell or from a prokaryotic cell. In some aspects, the prokaryotic cell is E. coli. Cell-free expression of the antibody may be useful, for example, where the antibody accumulates in a cell as an insoluble aggregate, or where yields from periplasmic expression are low.
[0350] Where the antibody is secreted into the medium, supernatants from such expression systems are generally first concentrated using a commercially available protein concentration filter, for example, an Amicon.RTM. or Millipore.RTM. Pellcon.RTM. ultrafiltration unit. A protease inhibitor such as PMSF may be included in any of the foregoing steps to inhibit proteolysis and antibiotics may be included to prevent the growth of adventitious contaminants.
[0351] The antibody composition prepared from the cells can be purified using, for example, hydroxylapatite chromatography, gel electrophoresis, dialysis, and affinity chromatography, with affinity chromatography being a particularly useful purification technique. The suitability of protein A as an affinity ligand depends on the species and isotype of any immunoglobulin Fc domain that is present in the antibody. Protein A can be used to purify antibodies that are based on human .gamma..sub.1, .gamma..sub.2, or .gamma..sub.4 heavy chains (Lindmark et al., J. Immunol. Meth., 1983, 62:1-13). Protein G is useful for all mouse isotypes and for human .gamma.3 (Guss et al., EMBO J., 1986, 5:1567-1575).
[0352] The matrix to which the affinity ligand is attached is most often agarose, but other matrices are available. Mechanically stable matrices such as controlled pore glass or poly(styrenedivinyl)benzene allow for faster flow rates and shorter processing times than can be achieved with agarose. Where the antibody comprises a C.sub.H3 domain, the BakerBond ABX.RTM. resin is useful for purification.
[0353] Other techniques for protein purification, such as fractionation on an ion-exchange column, ethanol precipitation, Reverse Phase HPLC, chromatography on silica, chromatography on heparin Sepharose.RTM., chromatofocusing, SDS-PAGE, and ammonium sulfate precipitation are also available, and can be applied by one of skill in the art.
[0354] Following any preliminary purification step(s), the mixture comprising the antibody of interest and contaminants may be subjected to low pH hydrophobic interaction chromatography using an elution buffer at a pH between about 2.5 to about 4.5, generally performed at low salt concentrations (e.g., from about 0 to about 0.25 M salt).
11. Pharmaceutical Compositions and Methods of Administration
[0355] Any of the antibodies provided herein can be provided in any appropriate pharmaceutical composition and be administered by any suitable route of administration. Suitable routes of administration include, but are not limited to, the inhalation, intraarterial, intradermal, intramuscular, intraperitoneal, intravenous, nasal, parenteral, pulmonary, and subcutaneous routes.
[0356] The pharmaceutical composition may comprise one or more pharmaceutical excipients. Any suitable pharmaceutical excipient may be used, and one of ordinary skill in the art is capable of selecting suitable pharmaceutical excipients. Accordingly, the pharmaceutical excipients provided below are intended to be illustrative, and not limiting. Additional pharmaceutical excipients include, for example, those described in the Handbook of Pharmaceutical Excipients, Rowe et al. (Eds.) 6th Ed. (2009), incorporated by reference in its entirety.
[0357] In some embodiments, the pharmaceutical composition comprises an anti-foaming agent. Any suitable anti-foaming agent may be used. In some aspects, the anti-foaming agent is selected from an alcohol, an ether, an oil, a wax, a silicone, a surfactant, and combinations thereof. In some aspects, the anti-foaming agent is selected from a mineral oil, a vegetable oil, ethylene bis stearamide, a paraffin wax, an ester wax, a fatty alcohol wax, a long chain fatty alcohol, a fatty acid soap, a fatty acid ester, a silicon glycol, a fluorosilicone, a polyethylene glycol-polypropylene glycol copolymer, polydimethylsiloxane-silicon dioxide, ether, octyl alcohol, capryl alcohol, sorbitan trioleate, ethyl alcohol, 2-ethyl-hexanol, dimethicone, oleyl alcohol, simethicone, and combinations thereof.
[0358] In some embodiments, the pharmaceutical composition comprises a cosolvent. Illustrative examples of cosolvents include ethanol, poly(ethylene) glycol, butylene glycol, dimethylacetamide, glycerin, and propylene glycol.
[0359] In some embodiments, the pharmaceutical composition comprises a buffer. Illustrative examples of buffers include acetate, borate, carbonate, lactate, malate, phosphate, citrate, hydroxide, diethanolamine, monoethanolamine, glycine, methionine, guar gum, and monosodium glutamate.
[0360] In some embodiments, the pharmaceutical composition comprises a carrier or filler. Illustrative examples of carriers or fillers include lactose, maltodextrin, mannitol, sorbitol, chitosan, stearic acid, xanthan gum, and guar gum.
[0361] In some embodiments, the pharmaceutical composition comprises a surfactant. Illustrative examples of surfactants include d-alpha tocopherol, benzalkonium chloride, benzethonium chloride, cetrimide, cetylpyridinium chloride, docusate sodium, glyceryl behenate, glyceryl monooleate, lauric acid, macrogol 15 hydroxystearate, myristyl alcohol, phospholipids, polyoxyethylene alkyl ethers, polyoxyethylene sorbitan fatty acid esters, polyoxyethylene stearates, polyoxylglycerides, sodium lauryl sulfate, sorbitan esters, and vitamin E polyethylene(glycol) succinate.
[0362] In some embodiments, the pharmaceutical composition comprises an anti-caking agent. Illustrative examples of anti-caking agents include calcium phosphate (tribasic), hydroxymethyl cellulose, hydroxypropyl cellulose, and magnesium oxide.
[0363] Other excipients that may be used with the pharmaceutical compositions include, for example, albumin, antioxidants, antibacterial agents, antifungal agents, bioabsorbable polymers, chelating agents, controlled release agents, diluents, dispersing agents, dissolution enhancers, emulsifying agents, gelling agents, ointment bases, penetration enhancers, preservatives, solubilizing agents, solvents, stabilizing agents, and sugars. Specific examples of each of these agents are described, for example, in the Handbook of Pharmaceutical Excipients, Rowe et al. (Eds.) 6th Ed. (2009), The Pharmaceutical Press, incorporated by reference in its entirety.
[0364] In some embodiments, the pharmaceutical composition comprises a solvent. In some aspects, the solvent is saline solution, such as a sterile isotonic saline solution or dextrose solution. In some aspects, the solvent is water for injection.
[0365] In some embodiments, the pharmaceutical compositions are in a particulate form, such as a microparticle or a nanoparticle. Microparticles and nanoparticles may be formed from any suitable material, such as a polymer or a lipid. In some aspects, the microparticles or nanoparticles are micelles, liposomes, or polymersomes. In certain embodiments, a composition provided herein is a pharmaceutical composition or a single unit dosage form. Pharmaceutical compositions and single unit dosage forms provided herein comprise a prophylactically or therapeutically effective amount of one or more prophylactic or therapeutic antibodies.
[0366] Further encompassed herein are anhydrous pharmaceutical compositions and dosage forms comprising an antibody, since water can facilitate the degradation of some antibodies.
[0367] Anhydrous pharmaceutical compositions and dosage forms provided herein can be prepared using anhydrous or low moisture containing ingredients and low moisture or low humidity conditions. Pharmaceutical compositions and dosage forms that comprise lactose and at least one active ingredient that comprises a primary or secondary amine can be anhydrous if substantial contact with moisture and/or humidity during manufacturing, packaging, and/or storage is expected.
[0368] An anhydrous pharmaceutical composition should be prepared and stored such that its anhydrous nature is maintained. Accordingly, anhydrous compositions can be packaged using materials known to prevent exposure to water such that they can be included in suitable formulary kits. Examples of suitable packaging include, but are not limited to, hermetically sealed foils, plastics, unit dose containers (e.g., vials), blister packs, and strip packs.
[0369] 11.1. Parenteral Dosage Forms
[0370] In certain embodiments, provided are parenteral dosage forms. Parenteral dosage forms can be administered to subjects by various routes including, but not limited to, subcutaneous, intravenous (including bolus injection), intramuscular, and intraarterial. Because their administration typically bypasses subjects' natural defenses against contaminants, parenteral dosage forms are typically, sterile or capable of being sterilized prior to administration to a subject. Examples of parenteral dosage forms include, but are not limited to, solutions ready for injection, dry products ready to be dissolved or suspended in a pharmaceutically acceptable vehicle for injection, suspensions ready for injection, and emulsions.
[0371] Suitable vehicles that can be used to provide parenteral dosage forms are well known to those skilled in the art. Examples include, but are not limited to: Water for Injection USP; aqueous vehicles such as, but not limited to, Sodium Chloride Injection, Ringer's Injection, Dextrose Injection, Dextrose and Sodium Chloride Injection, and Lactated Ringer's Injection; water miscible vehicles such as, but not limited to, ethyl alcohol, polyethylene glycol, and polypropylene glycol; and non-aqueous vehicles such as, but not limited to, corn oil, cottonseed oil, peanut oil, sesame oil, ethyl oleate, isopropyl myristate, and benzyl benzoate.
[0372] Excipients that increase the solubility of one or more of the antibodies disclosed herein can also be incorporated into the parenteral dosage forms.
[0373] 11.2. Dosage and Unit Dosage Forms
[0374] In human therapeutics, the doctor will determine the posology which he considers most appropriate according to a preventive or curative treatment and according to the age, weight, condition and other factors specific to the subject to be treated.
[0375] The amount of the antibody or composition which will be effective in the prevention or treatment of a disorder or one or more symptoms thereof will vary with the nature and severity of the disease or condition, and the route by which the antibody is administered. The frequency and dosage will also vary according to factors specific for each subject depending on the specific therapy (e.g., therapeutic or prophylactic agents) administered, the severity of the disorder, disease, or condition, the route of administration, as well as age, body, weight, response, and the past medical history of the subject. Effective doses may be extrapolated from dose-response curves derived from in vitro or animal model test systems.
[0376] In certain embodiments, exemplary doses of a composition include milligram or microgram amounts of the antibody per kilogram of subject or sample weight (e.g., about 10 micrograms per kilogram to about 50 milligrams per kilogram, about 100 micrograms per kilogram to about 25 milligrams per kilogram, or about 100 microgram per kilogram to about 10 milligrams per kilogram). In certain embodiment, the dosage of the antibody provided herein, based on weight of the antibody, administered to prevent, treat, manage, or ameliorate a disorder, or one or more symptoms thereof in a subject is 0.1 mg/kg, 1 mg/kg, 2 mg/kg, 3 mg/kg, 4 mg/kg, 5 mg/kg, 6 mg/kg, 10 mg/kg, or 15 mg/kg or more of a subject's body weight. In another embodiment, the dosage of the composition or a composition provided herein administered to prevent, treat, manage, or ameliorate a disorder, or one or more symptoms thereof in a subject is 0.1 mg to 200 mg, 0.1 mg to 100 mg, 0.1 mg to 50 mg, 0.1 mg to 25 mg, 0.1 mg to 20 mg, 0.1 mg to 15 mg, 0.1 mg to 10 mg, 0.1 mg to 7.5 mg, 0.1 mg to 5 mg, 0.1 to 2.5 mg, 0.25 mg to 20 mg, 0.25 to 15 mg, 0.25 to 12 mg, 0.25 to 10 mg, 0.25 mg to 7.5 mg, 0.25 mg to 5 mg, 0.25 mg to 2.5 mg, 0.5 mg to 20 mg, 0.5 to 15 mg, 0.5 to 12 mg, 0.5 to 10 mg, 0.5 mg to 7.5 mg, 0.5 mg to 5 mg, 0.5 mg to 2.5 mg, 1 mg to 20 mg, 1 mg to 15 mg, 1 mg to 12 mg, 1 mg to 10 mg, 1 mg to 7.5 mg, 1 mg to 5 mg, or 1 mg to 2.5 mg.
[0377] The dose can be administered according to a suitable schedule, for example, once, two times, three times, or for times weekly. It may be necessary to use dosages of the antibody outside the ranges disclosed herein in some cases, as will be apparent to those of ordinary skill in the art. Furthermore, it is noted that the clinician or treating physician will know how and when to interrupt, adjust, or terminate therapy in conjunction with subject response.
[0378] Different therapeutically effective amounts may be applicable for different diseases and conditions, as will be readily known by those of ordinary skill in the art. Similarly, amounts sufficient to prevent, manage, treat or ameliorate such disorders, but insufficient to cause, or sufficient to reduce, adverse effects associated with the antibodies provided herein are also encompassed by the herein described dosage amounts and dose frequency schedules. Further, when a subject is administered multiple dosages of a composition provided herein, not all of the dosages need be the same. For example, the dosage administered to the subject may be increased to improve the prophylactic or therapeutic effect of the composition or it may be decreased to reduce one or more side effects that a particular subject is experiencing.
[0379] In certain embodiments, treatment or prevention can be initiated with one or more loading doses of an antibody or composition provided herein followed by one or more maintenance doses.
[0380] In certain embodiments, a dose of an antibody or composition provided herein can be administered to achieve a steady-state concentration of the antibody in blood or serum of the subject. The steady-state concentration can be determined by measurement according to techniques available to those of skill or can be based on the physical characteristics of the subject such as height, weight and age.
[0381] In certain embodiments, administration of the same composition may be repeated and the administrations may be separated by at least 1 day, 2 days, 3 days, 5 days, 10 days, 15 days, 30 days, 45 days, 2 months, 75 days, 3 months, or 6 months. In other embodiments, administration of the same prophylactic or therapeutic agent may be repeated and the administration may be separated by at least 1 day, 2 days, 3 days, 5 days, 10 days, 15 days, 30 days, 45 days, 2 months, 75 days, 3 months, or 6 months.
12. Therapeutic Applications
[0382] For therapeutic applications, the antibodies of the invention are administered to a mammal, generally a human, in a pharmaceutically acceptable dosage form such as those known in the art and those discussed above. For example, the antibodies of the invention may be administered to a human intravenously as a bolus or by continuous infusion over a period of time, by intramuscular, intraperitoneal, intra-cerebrospinal, subcutaneous, intra-articular, intrasynovial, intrathecal, or intratumoral routes. The antibodies also are suitably administered by peritumoral, intralesional, or perilesional routes, to exert local as well as systemic therapeutic effects. The intraperitoneal route may be particularly useful, for example, in the treatment of ovarian tumors.
[0383] The antibodies provided herein may be useful for the treatment of any disease or condition involving PD-1, such as cancer, autoimmune disease, and infection.
[0384] Any suitable cancer may be treated with the antibodies provided herein. Illustrative suitable cancers include, for example, acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), adrenocortical carcinoma, anal cancer, appendix cancer, astrocytoma, basal cell carcinoma, brain tumor, bile duct cancer, bladder cancer, bone cancer, breast cancer, bronchial tumor, Burkitt Lymphoma, carcinoma of unknown primary origin, cardiac tumor, cervical cancer, chordoma, chronic lymphocytic leukemia (CLL), chronic myelogenous leukemia (CML), chronic myeloproliferative neoplasm, colon cancer, colorectal cancer, craniopharyngioma, cutaneous T-cell lymphoma, ductal carcinoma, embryonal tumor, endometrial cancer, ependymoma, esophageal cancer, esthesioneuroblastoma, fibrous histiocytoma, Ewing sarcoma, eye cancer, germ cell tumor, gallbladder cancer, gastric cancer, gastrointestinal carcinoid tumor, gastrointestinal stromal tumor, gestational trophoblastic disease, glioma, head and neck cancer, hairy cell leukemia, hepatocellular cancer, histiocytosis, Hodgkin lymphoma, hypopharyngeal cancer, intraocular melanoma, islet cell tumor, Kaposi sarcoma, kidney cancer, Langerhans cell histiocytosis, laryngeal cancer, leukemia, lip and oral cavity cancer, liver cancer, lobular carcinoma in situ, lung cancer, lymphoma, macroglobulinemia, malignant fibrous histiocytoma, melanoma, Merkel cell carcinoma, mesothelioma, metastatic squamous neck cancer with occult primary, midline tract carcinoma involving NUT gene, mouth cancer, multiple endocrine neoplasia syndrome, multiple myeloma, mycosis fungoides, myelodysplastic syndrome, myelodysplastic/myeloproliferative neoplasm, nasal cavity and par nasal sinus cancer, nasopharyngeal cancer, neuroblastoma, non-Hodgkin lymphoma, non-small cell lung cancer, oropharyngeal cancer, osteosarcoma, ovarian cancer, pancreatic cancer, papillomatosis, paraganglioma, parathyroid cancer, penile cancer, pharyngeal cancer, pheochromocytomas, pituitary tumor, pleuropulmonary blastoma, primary central nervous system lymphoma, prostate cancer, rectal cancer, renal cell cancer, renal pelvis and ureter cancer, retinoblastoma, rhabdoid tumor, salivary gland cancer, Sezary syndrome, skin cancer, small cell lung cancer, small intestine cancer, soft tissue sarcoma, spinal cord tumor, stomach cancer, T-cell lymphoma, teratoid tumor, testicular cancer, throat cancer, thymoma and thymic carcinoma, thyroid cancer, urethral cancer, uterine cancer, vaginal cancer, vulvar cancer, and Wilms tumor.
[0385] Any suitable autoimmune disease may be treated with the antibodies provided herein. Illustrative suitable autoimmune diseases, or diseases with an autoimmune component, include, for example, acute disseminated encephalomyelitis (ADEM), acute necrotizing hemorrhagic leukoencephalitis, Addison's disease, agammaglobulinemia, alopecia areata, amyloidosis, ankylosing spondylitis, anti-GBM/anti-TBM nephritis, antiphospholipid syndrome (APS), autoimmune angioedema, autoimmune aplastic anemia, autoimmune dysautonomia, autoimmune hepatitis, autoimmune hyperlipidemia, autoimmune immunodeficiency, autoimmune inner ear disease (AIED), autoimmune myocarditis, autoimmune oophoritis, autoimmune pancreatitis, autoimmune retinopathy, autoimmune thrombocytopenic purpura (ATP), autoimmune thyroid disease, autoimmune urticarial, axonal & neuronal neuropathies, Balo disease, Behcet's disease, bullous pemphigoid, cardiomyopathy, Castleman disease, Celiac disease, Chagas disease, chronic fatigue syndrome, chronic inflammatory demyelinating polyneuropathy (CIDP), chronic recurrent multifocal ostomyelitis (CRMO), Churg-Strauss syndrome, cicatricial pemphigoid/benign mucosal pemphigoid, Crohn's disease, Cogans syndrome, cold agglutinin disease, congenital heart block, coxsackie myocarditis, CREST disease, essential mixed cryoglobulinemia, demyelinating neuropathies, dermatitis herpetiformis, dermatomyositis, Devic's disease (neuromyelitis optica), discoid lupus, Dressler's syndrome, endometriosis, eosinophilic esophagitis, eosinophilic fasciitis, erythema nodosum, experimental allergic encephalomyelitis, Evans syndrome, fibromyalgia, fibrosing alveolitis, giant cell arteritis (temporal arteritis), giant cell myocarditis, glomerulonephritis, Goodpasture's syndrome, granulomatosis with polyangiitis (GPA) (formerly called Wegener's Granulomatosis), Graves' disease, Guillain-Barre syndrome, Hashimoto's encephalitis, Hashimoto's thyroiditis, hemolytic anemia, Henoch-Schonlein purpura, herpes gestationis, hypogammaglobulinemia, idiopathic thrombocytopenic purpura (ITP), IgA nephropathy, IgG4-related sclerosing disease, immunoregulatory lipoproteins, inclusion body myositis, interstitial cystitis, juvenile arthritis, juvenile diabetes (Type 1 diabetes), juvenile myositis, Kawasaki syndrome, Lambert-Eaton syndrome, leukocytoclastic vasculitis, lichen planus, lichen sclerosus, ligneous conjunctivitis, linear IgA disease (LAD), lupus (SLE), Lyme disease (chronic), Meniere's disease, microscopic polyangiitis, mixed connective tissue disease (MCTD), Mooren's ulcer, Mucha-Habermann disease, multiple sclerosis, myasthenia gravis, myositis, narcolepsy, neuromyelitis optica (Devic's), neutropenia, ocular cicatricial pemphigoid, optic neuritis, palindromic rheumatism, PANDAS (Pediatric Autoimmune Neuropsychiatric Disorders Associated with Streptococcus), paraneoplastic cerebellar degeneration, paroxysmal nocturnal hemoglobinuria (PNH), Parry Romberg syndrome, Parsonnage-Turner syndrome, pars planitis (peripheral uveitis), pemphigus, peripheral neuropathy, perivenous encephalomyelitis, pernicious anemia, POEMS syndrome, polyarteritis nodosa, type I, II, & III autoimmune polyglandular syndromes, polymyalgia rheumatic, polymyositis, postmyocardial infarction syndrome, postpericardiotomy syndrome, progesterone dermatitis, primary biliary cirrhosis, rimary sclerosing cholangitis, psoriasis, psoriatic arthritis, idiopathic pulmonary fibrosis, pyoderma gangrenosum, pure red cell aplasia, Raynauds phenomenon, reactive arthritis, reflex sympathetic dystrophy, Reiter's syndrome, relapsing polychondritis, restless legs syndrome, retroperitoneal fibrosis, rheumatic fever, rheumatoid arthritis, sarcoidosis, Schmidt syndrome, scleritis, scleroderma, Sjogren's syndrome, sperm & testicular autoimmunity, stiff person syndrome, subacute bacterial endocarditis (SBE), Susac's syndrome, sympathetic ophthalmia, Takayasu's arteritis, temporal arteritis/giant cell arteritis, thrombocytopenic purpura (TTP), Tolosa-Hunt syndrome, transverse myelitis, type 1 diabetes, ulcerative colitis, undifferentiated connective tissue disease (UCTD), uveitis, vasculitis, vesiculobullous dermatosis, vitiligo, and Wegener's granulomatosis (now termed Granulomatosis with Polyangiitis (GPA).
[0386] Any suitable infection may be treated with the antibodies provided herein. Illustrative suitable infections include, for example, hepatitis A virus, hepatitis B virus, hepatitis C virus (HCV), human immunodeficiency virus (HIV), and other viral infections.
13. Diagnostic Applications
[0387] In some embodiments, the antibodies provided herein are used in diagnostic applications. For example, an ant-PD-1 antibody may be useful in assays for PD-1 protein. In some aspects the antibody can be used to detect the expression of PD-1 in various cells and tissues. These assays may be useful, for example, evaluating cancer and autoimmune disease.
[0388] In some diagnostic applications, the antibody may be labeled with a detectable moiety. Suitable detectable moieties include, but are not limited to radioisotopes, fluorescent labels, and enzyme-substrate labels. In another embodiment of the invention, the anti-PD-1 antibody need not be labeled, and the presence thereof can be detected using a labeled antibody which specifically binds to the anti-PD-1 antibody.
14. Affinity Purification Reagents
[0389] The antibodies of the invention may be used as affinity purification agents. In this process, the antibodies may be immobilized on a solid phase such a resin or filter paper, using methods well known in the art. The immobilized antibody is contacted with a sample containing the PD-1 protein (or fragment thereof) to be purified, and thereafter the support is washed with a suitable solvent that will remove substantially all the material in the sample except the PD-1 protein, which is bound to the immobilized antibody. Finally, the support is washed with another suitable solvent, such as glycine buffer, pH 5.0, that will release the PD-1 protein from the antibody.
15. Kits
[0390] In some embodiments, an anti-PD-1 antibody provided herein is provided in the form of a kit, i.e., a packaged combination of reagents in predetermined amounts with instructions for performing a procedure. In some embodiments, the procedure is a diagnostic assay. In other embodiments, the procedure is a therapeutic procedure.
[0391] In some embodiments, the kit further comprises a solvent for the reconstitution of the anti-PD-1 antibody. In some embodiments, the anti-PD-1 antibody is provided in the form of a pharmaceutical composition.
EXAMPLES
Example 1: Generation and Primary Screening of Anti-PD-1 Antibodies
[0392] Antibody Fab or scFv libraries were constructed using a standard overlap extension PCR protocol with mutagenic primers targeting CDRs. See Heckman and Pease, Nat. Protoc., 2007, 2:924-932, incorporated by reference in its entirety. Selections for novel antibodies were performed using standard ribosome display protocols. See Dreir and Pluckthun, Methods Mol. Biol., 2011, Clifton, N.J., 687:283-306, incorporated by reference in its entirety. Specifically, scFv-based selection formats were performed according to published protocols. See Hanes and Pluckthun, Proc. Natl. Acad. Sci. USA., 1997, 94:4937-4942, incorporated by reference in its entirety. After multiple rounds of selection, the DNA from RT-PCR output was cloned into an optimized vector for cell-free expression using standard molecular biology techniques. See Yin et al., mAbs, 2012, 4:217-225, incorporated by reference in its entirety. All constructs were HIS- and FLAG-tagged to streamline purification and testing during screening.
[0393] Libraries of antibody variants isolated by the selections were transformed into E. coli and grown on agar plates with antibiotic (kanamycin). Individual colonies were picked and grown in liquid broth (TB+kanamycin), and used as a template for DNA amplification via rolling circle amplification (RCA). The variants were then expressed in a cell-free protein synthesis reaction as described in Zawada et al. (Biotechnol. Bioeng., 2011, 108:1570-1578, incorporated by reference in its entirety).
[0394] Briefly, cell-free extracts were treated with 50 .mu.M iodoacetamide for 30 minutes at room temperature (RT; 20.degree. C.) and added to a premix containing cell-free reaction components (see Groff et al., mAbs, 2014, 6:671-678, incorporated by reference in its entirety) and 10% (v/v) RCA DNA template (approximately 10 .mu.g/mL DNA) for variants of interest. Cell free reactions, at a final volume of 604, were incubated at 30.degree. C. for 12 h on a shaker at 650 rpm in 96-well plates. Four hundred to one-thousand-five-hundred colonies were screened, depending on the predicted diversity of the libraries used in the different selection campaigns. Following synthesis, each reaction was diluted 1:50 into PBST (PBS at pH 7.4 with 0.2% Tween-20+0.2% BSA) and the variants expressed in each reaction were tested for functional activity via ELISA-based binding to recombinant human PD-1 (ACROBiosystems, Inc., Catalog No. PD1-H5221 or SINO Biological Inc. Catalog No. 10377-H08H).
[0395] Standard ELISA-based methods were employed. Specifically, 384-well plates were coated with 2 .mu.g/mL recombinant PD-1 diluted in bicarbonate buffer, and then blocked with BSA. Antibody variants of interest were allowed to bind to the PD-1-coated plates, and detected with secondary antibodies (e.g., HRP-conjugated anti-human Fc or anti-FLAG) and then detected with chemiluminescent substrate (Pierce ELISA SuperSignal.TM. Substrate). Plates were analyzed on a Molecular Devices SpectraMax.RTM. M5 plate reader. Top hits were selected based on ELISA signal or signal/noise ratio and sequenced. Based on functional activity and sequence analysis, a subset of variants was selected for further scale-up and characterization.
Example 2: Secondary Screening of Antibody Variants
[0396] The top leads from the initial round of screening were cultured and plasmids encoding the antibody genes of interest were isolated using a QIAprep 96 Turbo.RTM. Miniprep Kit (Qiagen) according to the manufacturer's instructions. DNA was added to 4 mL of cell-free reaction medium to achieve a final concentration of 10 .mu.g/mL. The cell-free reaction medium was then incubated overnight for 12 hr at 30.degree. C., at 650 rpm.
[0397] The expressed variants from clarified cell-free reactions were purified via immobilized metal ion affinity chromatography (IMAC) using a semi-automated high throughput batch purification method. Briefly, purifications were performed in a 96-well plate format where 50 .mu.L/well of IMAC resin (Ni Sepharose.RTM. High Performance, GE Healthcare) was equilibrated in IMAC binding buffer (50 mM Tris pH 8.0, 300 mM NaCl, 10 mM imidazole), incubated with 1 mL cell-free reaction for 15 minutes, followed by two washes in IMAC binding buffer. His-tagged antibody variants were then eluted using 200 .mu.L IMAC elution buffer (50 mM Tris pH 8.0, 300 mM NaCl, 500 mM imidazole), and buffer exchanged into PBS using a 96-well Zeba.TM. plate (7 kDa MWCO, Thermo Fisher). Purified antibodies were quantified via high throughput capillary electrophoresis using the LabChip GXII.RTM. (Perkin Elmer) against a trastuzumab standard curve, according to the manufacturer's instructions.
Example 3: Hybridoma Generation
[0398] Balb/C mice were immunized with the extracellular domain of human PD-1 fused with human Fc (R&D Systems, supra) using standard immunization methods. The spleens and/or lymph nodes of the mice were harvested and fused with P3X cells to generate the hybridomas (Aragen Biosciences, Morgan Hill, Calif.), similar to what has been previously described. See Chronopoulou et al., Methods Mol. Biol., 2014, 1131:47-70; and Kim et al., Methods Mol. Biol., 2014, 1131:33-45, each of which is incorporated by reference in its entirety.
[0399] Total RNA was extracted from hybridoma cells using an RNeasy.RTM. Mini Kit (Qiagen) and converted to cDNA using a SMARTer.TM. RACE cDNA Amplification Kit (Clontech). Positive clones were identified by gel electrophoresis, cloned using a TOPO.RTM. kit (Invitrogen), and sequenced using standard Sanger methods.
[0400] Mouse single-chain antibodies were constructed by using total gene synthesis using codons optimized for E. coli. The genes encoding the antibodies were cloned into a standard cell-free expression vector. See Yin et al., mAbs, 2012, 4:217-225, incorporated by reference in its entirety.
[0401] The CDRs from m1E9 were grafted onto human antibody frameworks VH3-21, VH3-7, V.kappa.1-9, and V.kappa.3-11 by standard methodology to yield humanized antibodies h1E9-1, h1E9-2, h1E9-4, and h1E9-5. See Kuramochi et al., Methods Mole. Biol., 2014, 1060:123-137, incorporated by reference in its entirety. The same method was used to graft the CDRs from m4B10 onto human antibody frameworks VH3-15, V.kappa.3-11, V.kappa.3-20, and V.kappa.4-1 to yield humanized antibodies h4B10-1, h4B10-2, and h4B10-3.
Example 4: Kinetic Analysis of Selected Antibody Variants
[0402] Human PD-1 (ACROBiosystems, Inc., Catalog No. PD1-H5221), cynomolgus PD-1 (ACROBiosystems, Inc., Catalog No. PD1-H5254), and murine PD-1 (R&D Systems Inc., Catalog No. 1021-PD-100) were used, as indicated, for kinetic analysis.
[0403] Monoclonal anti-FLAG M2 IgG (Sigma-Aldrich #F9291) was immobilized onto a CMS chip (GE Life Sciences) using amine coupling chemistry (from Amine Coupling Kit, GE Life Sciences). The immobilization steps were carried out at a flow rate of 25 .mu.l/min in 1.times.HBS-EP+ buffer (GE Life Sciences; 10.times. Stock diluted before use). The sensor surfaces were activated for 7 min with a mixture of NHS (0.05 M) and EDC (0.2 M). The Anti-FLAG M2 IgG was injected over all 4 flow cells at a concentration of 25 .mu.g/ml in 10 mM sodium acetate, pH 4.5, for 7 min. Ethanolamine (1 M, pH 8.5) was injected for 7 min to block any remaining activated groups. An average of 12,000 response units (RU) of capture antibody was immobilized on each flow cell.
[0404] Off-rate and kinetic binding experiments were performed at 25.degree. C. using 1.times.HBS-EP+ buffer. Test and control antibodies were injected over the Anti-FLAG surface at concentrations of 5-10 .mu.g/mL for 12 seconds at a flow rate of 10 .mu.l/min on flow cells 2, 3 and 4, followed by a buffer wash for 30 seconds at the same flow rate. Kinetic characterization of antibody samples was carried out with a single concentration of antigen (for off-rate ranking) or a 1:2 dilution series of antigen (for kinetic characterization) and 1 injection of 0 nM antigen (i.e., buffer alone). After capturing ligand (anti-PD-1 antibody) on the anti-FLAG surface, the analyte (huPD1-His) was bound at 50, 25, 12.5, 6.25 and 0 nM for 180 seconds, followed by a 600 second dissociation phase at a flow rate of 50 .mu.l/min. Between each ligand capture and analyte binding cycle, regeneration was carried out using 2 injections of 10 mM glycine pH 2.0 for 30 seconds at 30 .mu.L/min, followed by a 30 second buffer wash step.
[0405] The data was fit with the Biacore T200 Evaluation software, using a 1:1 Langmuir binding model. K.sub.D (affinity, nM) was determined as a ratio of the kinetic rate constants calculated from the fits of the association and dissociation phases.
Example 5: PD-1-PD-L1 Competition ELISA
[0406] Anti-PD1 antibodies were tested for their ability to block a PD-1/PD-L1 interaction. PD-1 (ACROBiosystems, Inc.) was adsorbed on 384-well white Maxisorp.RTM. plates (Nunc) at 2 .mu.g/mL in sodium bicarbonate buffer (pH 8.9) and incubated at 30.degree. C. for 1 hour or overnight at 4.degree. C. The plate was washed 3 times with PBS pH 7.4 with 0.05% Tween20 and blocked with 2% bovine serum albumin (BSA) in PBS pH 7.4+0.1% Tween20 for 1 hour at 30.degree. C.
[0407] The blocking solution was aspirated, and a dilution series of antibody was mixed with 100 nM PD-L1-Fc (ACROBiosystems, Inc.) in 0.2% BSA in PBS pH 7.4+0.1% Tween20 (diluent buffer) and incubated at 30.degree. C. for 1 hour. The plate was washed, and 10 nM anti-PD-L1 antibody (BioLegend, clone 29E.2A3) in diluent buffer was added to all wells. After a 1 hour incubation at 30.degree. C., the plate was washed and incubated with HRP-conjugated anti-mouse Fc (Jackson Laboratories), followed by detection with SuperSignal.TM. Pico Chemiluminescent Substrate (Thermo Pierce). Luminescence was detected on a SpectraMax.RTM. M5 plate reader (Molecular Devices).
Example 6: Cell Binding Experiments
[0408] Antibodies with expression levels >250 nM and mouse IgGs from hybridomas were tested in a fluorescence-activated cell sorting (FACS) cell-binding assay. Chinese Hamster Ovary (or CHO) Cells stably expressing the human target molecule PD-1 on the cell surface (CHO-PD1) were used to screen for binding. Parental CHO cells were used as a negative control to determine background-binding levels. Parental CHO cells and CHO-PD1 cells were cultured in RPMI w/10% FCS penicillin/streptomycin (Pen/Strep) and glutamine (or Gln) and split every 3-4 days at 10.sup.5 cells/mL.
[0409] A mix of parental CHO cells and CHO-PD1 cells was prepared as follows: Parental CHO cells were washed 2.times. in PBS then incubated in PBS containing 1 .mu.M CellTrace.TM. Oregon Green488.RTM. (Life Technologies) at 37.degree. C. for 30 minutes. Cells were then washed 2.times. with RPMI w/10% fetal calf serum (FCS), washed 2.times. with FACS buffer (PBS w/2% FCS), suspended thoroughly in ice-cold FACS buffer at a final concentration of 2.times.10.sup.6 cells/mL and kept on ice. CHO-PD1 cells were similarly washed with FACS buffer and kept on ice at 2.times.10.sup.6 cells/mL. Parental CHO cells and CHO-PD1 cells were then mixed to obtain a 1:1 cell suspension and seeded at 100 .mu.L per well on 96 well polypropylene plates. Plates were spun at 1500 rpm for 5 minutes and cell pellets were suspended in 50 .mu.L FACS buffer containing 6-12 point dilutions of anti-PD-1 variants starting from concentrations of -100-200 nM antibody, dispensed using BioMek FX (Beckman Coulter). Cells were then incubated on ice for 1 hr, washed with FACS buffer and incubated for 1 hr on ice with 50 .mu.L FACS buffer containing 2.5 .mu.g/ml R-phycoerythrin-conjugated goat anti-Human IgG (Jackson ImmunoResearch) or AF647-conjugated goat anti-mouse IgG (Life Technologies) dispensed using BioMek FX (Beckman Coulter). Cells were then washed 2.times. with FACS buffer and fixed for 10 minutes in 200 .mu.L PBS with 2% paraformaldehyde (PFA) prior to fluorescence detection. Samples were acquired using a Becton Dickinson LSRII FACS. Mean Fluorescence Intensity of PD-1 antibody binding was analyzed using FlowJo.RTM. software (Tree Star, Inc.).
Example 7: Cell-Based Ligand Competition Experiments
[0410] Variants that showed cell-binding activity were tested in a fluorescence-activated cell sorting (FACS) cell-based competition assay. CHO cells stably expressing the human target molecule PD-1 on the cell surface (CHO-PD1) were used to screen for antibodies that compete with hFc-tagged recombinant human PD-L1 or PD-L2 proteins (R&D systems) for binding to PD-L1 expressed on the cell surface.
[0411] CHO-PD1 cells were cultured in RPMI with 10% FCS Pen/Strep and Gln and split every 3-4 days at 10.sup.5 cells/ml. Cells were washed 2.times. with FACS buffer (PBS w/2% FCS), thoroughly in ice-cold FACS buffer at a final concentration of 1.times.10.sup.6 cells/ml and seeded at 1004 per well on 96 well polypropylene plates. Plates were spun at 1500 rpm for 5 minutes and cell pellets were suspended in 50 .mu.L FACS buffer containing 8 point 1:3 dilutions (2.times. concentrated) of anti-PD-1 antibody variants, starting from high concentration of -200 nM. 50 .mu.L FACS buffer containing a fixed amount of either 6 .mu.g/ml rhPDL2-Fc or 50 .mu.g/ml rhPDL1-Fc proteins were then added to the cells. Cell were then incubated on ice for 1 hr, washed with FACS buffer and incubated for 1 hr on ice with 50 .mu.l FACS buffer containing 2.5 .mu.g/ml R-phycoerythrin-conjugated anti-human IgG (Jackson ImmunoResearch). Cells were then washed 2.times. with FACS buffer and fixed for 10 minutes in 200 .mu.l PBS with 2% PFA prior to acquisition. Samples were acquired using a Becton Dickinson LSRII FACS. Mean Fluorescence Intensity of rhPDL1 or rhPDL2 protein binding was analyzed using FlowJo.RTM. software (Tree Star, Inc.).
Example 8: Evaluating the Effect of Anti-PD-1 Antibodies on Interferon Gamma Production in a Mixed Lymphocyte Reaction
[0412] Anti-PD-1 antibodies were functionally tested for potency in blocking the PD-1 pathway in a peripheral blood mononuclear cell (PBMC) two-way mixed lymphocyte reaction (MLR) assay by measuring interferon gamma (IFN-g) secretion in cell culture medium. 1.times.10.sup.5 human PBMC from 2 allogeneic donors were co-cultured in RPMI media+10% FBS in a total volume of 150 .mu.l in a 96-well U-bottom plate. Anti-PD-1 antibodies were added at specific concentrations to each well. Isotype control antibody, non-PD-1 targeting antibody, or nothing were used as a negative controls. Cells were cultured for 5 days at 37.degree. C. At day 5, conditioned media was collected and levels of IFN-g were measured using DuoSet.RTM. ELISA kits (R&D Systems).
Example 9: Characteristics of Antibodies Isolated from Primary and Secondary Screen
[0413] Table 1 shows the characteristics of scFv-Fc antibodies (VH1-18/V.lamda.3-25) isolated as described in Examples 1-2, and characterized as described above.
TABLE-US-00003 TABLE 1 Characteristics of antibodies isolated as described in Examples 1-2, and characterized as described above. Cell Binding, K.sub.D Murine Cynomolgus MLR k.sub.a (1/Ms) k.sub.d (1/s) K.sub.D (M) (nM) PD-1 PD-1 Activity Human Human Human PD-L1 Human Binding, Binding, K.sub.D (IFNg Clone ID PD-1 PD-1 PD-1 Competition PD-1 K.sub.D (M) (M) release) 1353-A09 4.88E+05 1.24E-02 2.55E-08 yes, ++ 0.2 Not Not tested Not (SEQ ID NO: 238) tested tested 1353-C07 1.23E+06 1.87E-02 1.52E-08 yes 0.4 Not Not tested Not (SEQ ID NO: 239) tested tested 1353-E07 7.37E+05 7.01E-03 9.52E-09 yes 0.9 Not Not tested Not (SEQ ID NO: 240) tested tested 1353-F09 6.87E+05 7.47E-03 1.09E-08 yes 1 Not Not tested Not (SEQ ID NO: 241) tested tested 1353-G08 5.63E+05 2.54E-03 4.50E-09 yes, +++ 0.3 6.09E-08 2.43E-08 Not (SEQ ID NO: 242) tested 1353-G10 5.16E+05 9.80E-04 1.90E-09 yes, ++ 0.7 6.22E-08 1.55E-08 positive (SEQ ID NO: 243) 1353-H08 2.48E+05 1.18E-03 4.76E-09 yes, +++ 0.2 Not Not tested Not (SEQ ID NO: 244) tested tested 1353-H09 7.98E+05 3.59E-03 4.50E-09 yes, ++ 0.8 9.08E-09 2.22E-08 positive (SEQ ID NO: 245)
Example 10: Characteristics of Murine Hybridoma Antibodies
[0414] Table 2 shows the characteristics of IgG antibodies isolated as described in Example 3, and characterized as described above.
TABLE-US-00004 TABLE 2 Characteristics of murine hybridoma antibodies, characterized as described above. PD-L1 PD-L2 Biacore Biacore Cell, Competition, Competition, Human Cynomolgus MLR, EC50.sup.1 IC50.sup.2 Binding, IC.sub.50.sup.4 IC.sub.50.sup.4 PD1 K.sub.D K.sub.D IFNg Clone ID (nM) (nM) K.sub.D.sup.3 (nM) (nM) (nM) (M) (M) secretion 1B10 1.71 9.47 3.2 5.96 0.56 1.04E-08 2.56E-09 positive VH: SEQ ID NO: 255, with serine prepended to the sequence VL: SEQ ID NO: 279 1E9 0.33 0.89 2.9 5.86 0.58 9.90E-09 2.54E-09 positive VH: SEQ ID NO: 256 VL: SEQ ID NO: 280 4B10 0.47 1.43 1.39 1.99 0.01 9.13E-10 5.61E-10 positive VH: SEQ ID NO: 257 VL: SEQ ID NO: 281 10B4 0.59 1.32 1.34 2.53 0.18 2.52E-10 1.95E-10 positive VH: SEQ ID NO: 258 VL: SEQ ID NO: 282 .sup.1Binding of human PD-1 (ACROBiosystems, Inc., Cat. No. PD1-H5221) via ELISA. .sup.2Competition against PD-L1 (ACROBiosystems, Inc., Cat No. PD1-H5258) .sup.3CHO cell line overexpressing human PD-1. .sup.4Cell-based competition. i.e., Inhibition of PD-L1 or PD-L2 binding to CHO cells overexpressing human PD-1 is inhibited by IgG.
Example 11: Characteristics of Humanized Antibodies
[0415] Table 3 shows the characteristics of humanized scFv antibodies derived from the murine hybridoma antibodies of Example 10, and characterized as described above.
TABLE-US-00005 TABLE 3 Characteristics of humanized antibodies, characterized as described above. k.sub.a (1/Ms) k.sub.d (1/s) K.sub.D (M) Clone Human PD-1 Human PD-1 Human PD-1 h1E9-1 (VH3-21-V.kappa.1-9) 1.82E+05 4.68E-04 2.58E-09 scFv (SEQ ID NO: 227) h1E9-2 (VH3-21-V.kappa.3-11) 4.74E+04 1.82E-03 3.85E-08 scFv (SEQ ID NO: 228) h1E9-4 (VH3-7-V.kappa.1-9) 1.85E+05 6.79E-04 3.66E-09 scFv (SEQ ID NO: 229) h1E9-5 (VH3-7-V.kappa.3-11) 2.00E+05 6.28E-04 3.15E-09 scFv (SEQ ID NO: 230) m4B10 scFv 8.12E+04 4.17E-04 5.14E-09 (SEQ ID NO: 237) h4B10-1 (VH3-15-V.kappa.3- 1.21E+06 2.99E-03 2.47E-09 11) scFv (SEQ ID NO: 231) h4B10-2 (VH3-15-V.kappa.3- 1.16E+06 3.24E-03 2.79E-09 20) scFv (SEQ ID NO: 232) h4B10-3 (VH3-15-V.kappa.4-1) 5.13E+05 6.17E-04 1.20E-09 scFv (SEQ ID NO: 233) m1B10 scFv 1.86E+05 2.39E-03 1.28E-08 (SEQ ID NO: 235)
Example 12: Thermal Stability Data
[0416] Table 4 provides thermal stability of selected antibodies, as determined by differential scanning fluorimetry (DSF).
TABLE-US-00006 TABLE 4 Thermal stability of selected antibodies, as determined by DSF. Tm1 Tm2 Variant ID Target Scaffold (.degree. C.) (.degree. C.) 1353-G12 PD1 scFv-Fc 48.2 1353-G08 PD1 scFv-Fc 45.3 1353-G10 PD1 scFv-Fc 48.6 1353-H09 PD1 scFv-Fc 49.4 m1B10 PD1 scFv 55.3 h1E9-1 PD1 scFv 52.5 h1E9-2 PD1 scFv 50.9 h1E9-4 PD1 scFv 55.2 h1E9-5 PD1 scFv 51.7 h4b10-1 PD1 scFv 45.5 59.4 h4b10-2 PD1 scFv 45 61.7 h4b10-3 PD1 scFv 52.1 59.3
Example 13: Construction and Evaluation of h1E9-4 and h1E9-5 IgGs
[0417] Variable domains from h1E9-4 scFv (V.sub.H: SEQ ID NO: 260; V.sub.L: SEQ ID NO: 284), and h1E9-5 scFv (V.sub.H: SEQ ID NO: 261; V.sub.L: SEQ ID NO: 285) were grafted onto human antibody constant domains to generate human IgG1 antibodies based on these scFvs.
[0418] Specifically, the V.sub.H sequences were grafted onto C.sub.H1-C.sub.H2-C.sub.H3 constant domains to yield full-length IgG HCs with C-terminal FlagHis tags (GSGDYKDDDDKGSGHHHHHH; SEQ ID NO: 294) for ease of purification and assay development. The V.sub.L sequences were grafted onto CL domains to yield full-length IgG LCs. The sequence for both the h1E9-4 and h1E9-5 HCs, with C-terminal FlagHis tag, is provided in SEQ ID NO: 302. The sequence for the h1E9-4 LC is provided in SEQ ID NO: 304. The sequence for the h1E9-5 LC is provided in SEQ ID NO: 303.
[0419] The IgGs were expressed in a cell-free reaction, as described in Example 1, and purified using the FlagHis tags. The affinity of the IgGs for PD-1 was measured by surface plasmon resonance (Biacore.RTM.) and determined to be essentially equivalent to that of the parent scFvs. Affinity data is provided in Table 5.
TABLE-US-00007 TABLE 5 Affinity of h1E9-4 and h1E9-5 IgGs for PD-1 antigen. Sample Ligand k.sub.a (1/Ms) k.sub.d (1/s) K.sub.D (M) PD-1- h1E9-4 IgG 3.56E+04 4.42E-04 1.24E-08 his PD-1- h1E9-5 IgG 2.43E+04 4.71E-04 1.94E-08 his
Example 14: Construction and Evaluation of h1E9 Humanized IgGs
[0420] The CDRs for m1E9 were grafted onto human antibody frameworks V.sub.H3-23, VH3-30, Vk4-1, Vk3-20, Vk2-28, and Vk1-33 by standard methodology (T. Kuramochi, T. Igawa, H. Tsunoda, and K. Hattori, Method in Molecular Biology, Human Monoclonal Antibodies: Methods and Protocols, 1060, 123-137) to yield humanized antibodies h1E9-HC1, h1E9-HC2, h1E9-HC3, h1E9-LC1, h1E9-LC2, h1E9-LC3, and h1E9-LC4. The HC constructs included C-terminal FlagHis tags (GSGDYKDDDDKGSGHHHHHH; SEQ ID NO: 294) for ease of purification and assay development. The resulting h1E9 heavy chains were: h1E9-HC3 (SEQ ID NO:324), h1E9-HC2 (SEQ ID NO:325), and h1E9-HC3 (SEQ ID NO:326). The resulting light chains were: h1E9-LC4 (SEQ ID NO:327), h1E9-LC3 (SEQ ID NO:328), h1E9-LC2 (SEQ ID NO:329), and h1E9-LC1 (SEQ ID NO:330).
[0421] The humanized heavy chains h1E9-HC1, h1E9-HC2, h1E9-HC3, and h1E9-5 were paired with humanized IgG light chains h1E9-LC1, h1E9-LC2, h1E9-LC3, and h1E9-LC4. These 16 combinations were then assessed for binding to human PD1 in vitro by surface plasmon resonance (Biacore.RTM.) and subsequently human and cynomolgus PD1 binding on CHO cells using FACS as described below.
[0422] Surface plasmon resonance (Biacore data is provided in Table 6.
TABLE-US-00008 TABLE 6 Affinity of h1E9 humanized IgGs for PD-1 antigen. Sample Ligand SEQ ID NOS K.sub.D (M) PD-1-his h1E9-HC1 .times. h1E9-LC1 326 330 5.08E-09 PD-1-his h1E9-HC1 .times. h1E9-LC2 326 329 1.90E-08 PD-1-his h1E9-HC1 .times. h1E9-LC3 326 328 6.38E-09 PD-1-his h1E9-HC1 .times. h1E9-LC4 326 327 6.33E-09 PD-1-his h1E9-HC2 .times. h1E9-LC1 325 330 3.11E-09 PD-1-his h1E9-HC2 .times. h1E9-LC2 325 329 1.23E-08 PD-1-his h1E9-HC2 .times. h1E9-LC3 325 328 1.09E-08 PD-1-his h1E9-HC2 .times. h1E9-LC4 325 327 4.96E-09 PD-1-his h1E9-HC3 .times. h1E9-LC1 324 330 3.60E-09 PD-1-his h1E9-HC3 .times. h1E9-LC2 324 329 3.19E-09 PD-1-his h1E9-HC3 .times. h1E9-LC3 324 328 1.69E-08 PD-1-his h1E9-HC3 .times. h1E9-LC4 324 327 9.40E-09 PD-1-his h1E9-5-IgG-HC-FLAG-HIS .times. 261 330 3.70E-09 h1E9-LC1 PD-1-his h1E9-5-IgG-HC-FLAG-HIS .times. 261 329 1.62E-08 h1E9-LC2 PD-1-his h1E9-5-IgG-HC-FLAG-HIS .times. 261 328 1.63E-08 h1E9-LC3 PD-1-his h1E9-5-IgG-HC-FLAG-HIS .times. 261 327 7.31E-09 h1E9-LC4
[0423] The affinity of the IgGs for cell-surface expressed human PD-1 were measured according to Example 6, above. Affinity data is provided in Table 7.
TABLE-US-00009 TABLE 7 Affinity of h1E9 humanized IgGs for Cell-Surface Human PD-1 antigen (nM). h1E9-HC1 h1E9-HC2 h1E9-HC3 h1E9-5 HC SEQ ID SEQ ID SEQ ID SEQ ID NO: 326 NO: 325 NO: 324 NO: 261 h1E9-LC1 SEQ ID 6 7 8 9 NO: 330 h1E9-LC2 SEQ ID 12 11 19 16 NO: 329 h1E9-LC3 SEQ ID 12 9 12 11 NO: 328 h1E9-LC4 SEQ ID 8 4 9 14 NO: 327
[0424] The affinity of the IgGs for cell-surface expressed cynomolgus PD-1 were measured according to Example 6, above. Affinity data is provided in Table 8.
TABLE-US-00010 TABLE 8 Affinity of h1E9 humanized IgGs for Cell-Surface Cynomolgus PD-1 antigen (nM). h1E9-HC1 h1E9-HC2 h1E9-HC3 h1E9-5 HC SEQ ID SEQ ID SEQ ID SEQ ID NO: 326 NO: 325 NO: 324 NO: 261 h1E9-LC1 SEQ ID 7 6 4 4 NO: 330 h1E9-LC2 SEQ ID 19 9 11 11 NO: 329 h1E9-LC3 SEQ ID 15 9 8 9 NO: 328 h1E9-LC4 SEQ ID 11 6 5 12 NO: 327
Example 15: In Vivo Efficacy of Anti-PD-1 Antibodies on Tumor Establishment and Growth
[0425] MC38 colorectal cancer cells (2.times.10.sup.6 cells in 0.1 mL PBS) were implanted subcutaneously on the right flank of C57BL/6 mice (Charles River Laboratories). On day 2 post-cell implantation (day 0), mice were treated with anti-PD-1 or control antibody at a dose of 5 mg/kg intraperitoneally. Animals were dosed on dosing days 0, 4, 8, 11, and 14.
[0426] The treatment groups were as follows:
[0427] (1) PBS vehicle;
[0428] (2) control rat IgG2a, clone 2A3;
[0429] (3) anti-PD-1 rat IgG2a, clone RMP1-14;
[0430] (4) anti-PD-1 human scFv-Fc, clone PD1-F2 (see SEQ ID NOs: 306 and 293 for V.sub.H and V.sub.L sequences, respectively, and SEQ ID NO: 305 for the scFv-Fc);
[0431] (5) anti-PD-1 human scFv-Fc, clone 1353-G08 (see SEQ ID NOs: 251 and 275 for V.sub.H and V.sub.L sequences, respectively, and SEQ ID NO: 242 for the scFv-Fc);
[0432] (6) anti-PD-1 human scFv-Fc, clone 1353-G10 (see SEQ ID NOs: 252 and 276 for V.sub.H and V.sub.L sequences, respectively, and SEQ ID NO: 243 for the scFv-Fc); and
[0433] (7) anti-PD-1 human IgG, clone PD1-F2v (see SEQ ID NOs: 603 and 293 for V.sub.H and V.sub.L sequences, respectively, SEQ ID NO: 307 for the HC sequence, and SEQ ID NO: 308 for the LC sequence).
[0434] Antibodies (2) and (3) were obtained commercially from Bio X Cell. Antibodies (4)-(6) were produced using the cell-free expression methods described in Example 1. Antibody 7 was produced by mammalian cell expression using HEK293 cells and standard techniques.
[0435] Tumors were measured using an electronic caliper and tumor volumes were calculated using the formula, volume=(width.sup.2.times.length)/2. Statistical analysis was performed via a two-tailed Mann-Whitney test at day 17 post-treatment.
[0436] At day 17 post-treatment, all anti-PD-1 antibodies with strong mouse PD-1 reactivity (i.e., antibodies RMP1-14, PD1-F2, and 1353-G08) resulted in a significant delay in tumor establishment and growth (p<0.01) compared to the PBS and control rat IgG2a treatment groups. In contrast, antibody 1353-G10, which binds human PD-1 but has weaker reactivity with mouse PD-1, showed no significant effect on tumor establishment and growth (p>0.05).
[0437] FIG. 2 provides a chart of the tumor volume over the 17-days of treatment with the various antibodies.
Example 16: Cytomegalovirus (CMV) Recall Assay
[0438] Anti-PD-1 antibodies were functionally tested for potency in blocking the PD-pathway in restimulating peripheral blood mononuclear cells (PBMC) from cytomegalovirus-positive (CMV+) human donors by measuring IFN-g secretion in cell culture medium.
[0439] CD14+ monocytes and CD3+ T cells were obtained from peripheral blood mononuclear cells (PBMC) isolated from CMV+ human donors (AllCells, Alameda, Calif.) using MACS Cell Separation kits (Miltenyi Biotec).
[0440] CD14+ monocytes were differentiated into immature dendritic cells (DC) by culturing cells at 1.times.10.sup.6 cells/ml for 7 days in presence of GM-CSF and IL-4 (Peprotech) in X-Vivo 15 media (Lonza) containing 2% human AB serum (Sigma-Aldrich), penicillin-streptomycin (Corning Mediatech) and GlutaMAX (Life Technologies). Following differentiation, DCs were matured by culturing in X-Vivo 15+2% human AB serum media at 1.times.10.sup.6 cells/ml for 2 days in the presence of GM-CSF, IL-4, TNF-.alpha., IL-1b, IL-6 (Peprotech) and prostaglandin E2 (Sigma-Aldrich).
[0441] To set-up the CMV recall assay, mature DCs were collected and washed. 10,000 DCs and 100,000 pan CD3+ T cells were plated per well in a 96-well U-bottom plate in a total volume of 100 .mu.l media containing peptide pools for the CMV IE-1 and CMV pp65 proteins (Miltenyi Biotec).
[0442] Anti-PD-1 or control human IgG antibody (50 final volume of 150 .mu.l per well) were added starting at a final concentration of 400 nM with 5-fold serial dilutions. Cells were co-cultured with peptides and antibodies for 5-6 days. Conditioned media was collected and tested for human IFN-g levels by ELISA (BD Biosciences).
[0443] As shown in FIG. 3, anti-PD-1 human IgG 1353-G10 (SEQ ID NOs: 252 and 276) showed significant fold-increase in IFN-g levels (closed circles). The IFN-g levels were substantial compared to Nivolumab IgG (open circles).
Example 17: DC/CD4+ T Cell Mixed Lymphocyte Reaction (MLR) Assay
[0444] Anti-PD-1 antibodies were functionally tested for potency in blocking the PD-1 pathway in a peripheral blood mononuclear cell (PBMC) two-way mixed lymphocyte reaction (MLR) assay by measuring interferon gamma (IFN-g) secretion in cell culture medium.
[0445] CD14+ monocytes and CD4+ T cells were obtained from PBMC isolated from human donors using MACS Cell Separation kits. CD14+ monocytes were differentiated into immature DC by culturing cells at 1.times.10.sup.6 cells/ml cell density for 7 days in presence of GM-CSF and IL-4 in RPMI media containing 10% fetal bovine serum, penicillin-streptomycin and GlutaMAX.
[0446] Following differentiation, DCs were matured by culturing in RPMI+10% FBS media at 1.times.10.sup.6 cells/ml cell density for 2 days in the presence of GM-CSF, IL-4, TNF-a, IL-1b, IL-6 and prostaglandin E2.
[0447] To set-up the DC/CD4+ T cell MLR, mature DCs were collected and washed. 10,000 DCs and 100,000 CD4+ T cells were plated per well in a 96-well U-bottom plate in a total volume of 100 .mu.l media. Anti-PD-1 or control human IgG antibody (50 .mu.l, final volume of 150 .mu.l per well) were added starting at a final concentration of 400 nM with 5-fold serial dilutions. Cells were co-cultured with peptides and antibodies for 5-6 days. Conditioned media was collected and tested for human IFN-g levels by ELISA.
[0448] As shown in FIG. 4, anti-PD-1 human IgG 1353-G10 (SEQ ID NOs: 252 and 276) showed significant fold-increase in IFN-g levels (closed circles). The IFN-g levels were substantial compared to Nivolumab IgG (open circles).
Example 18: Effect of PD-1 Blockade on Graft Versus Host Disease (GVHD) Response
[0449] Anti-PD-1 antibodies described herein were evaluated for acceleration of graft versus host disease (GVHD) in a mouse model. Since PD-1 negatively regulates immune response, blockade of PD-1 by an effective antibody should accelerate the immune and GVHD response.
[0450] 40 female NSG mice aged 7 weeks and weighing approximately 19-22 g were used as human PBMC recipients. Groups were randomized into four groups of ten. On day 0, all animals received 2.times.10.sup.7 human PBMC injected via the tail vein. Prior to PBMC injection, animals were dosed intraperitoneally (IP) with either control anti-GFP, Nivolumab anti-PD-1 antibody, or 1353-G10 anti-PD1 antibody (SEQ ID NOs: 252 and 276) as detailed in Table 9. Treatment was administered 2.times. per week until study conclusion.
TABLE-US-00011 TABLE 9 Treatment groups huPBMC Total con- Test Dosing number Group centration article Dose frequency of doses Route N 1 2 .times. 10.sup.7 Control 10 2.times./week 10 IP 10 Anti-GFP mg/kg 2 2 .times. 10.sup.7 Anti-PD1 10 2.times./week 10 IP 10 Nivolumab mg/kg 3 2 .times. 10.sup.7 Anti-PD1 10 2.times./week 10 IP 10 1353-G10 mg/kg 4 2 .times. 10.sup.7 Anti-PD1 3 2.times./week 10 IP 10 1353-G10 mg/kg
[0451] Animals were weighed and observed at least two times per week for 5 weeks. Monitoring increased to daily when 10% body weight loss (compared to initial mouse weight) occurred. Animals were euthanized with CO.sub.2 at the end of five weeks (end of study), when they exhibited weight loss greater than 20%, or were unable to right themselves, cold to the touch, and moribund (severe hair ruffling, body weight loss, hunched posture, or decreased activity).
[0452] Upon sacrifice (after 5 weeks) or pre-mature euthanasia (due to body weight loss or moribundity), the spleen, liver, and serum were collected for further analyses. The liver and spleen were placed in formalin for 24 hours then transferred to 70% ethanol for storage. The serum was flash frozen and stored at -80.degree. C. All procedures were conducted according to the guidelines of the Sutro Institutional Animal Care and Use Committee (IACUC) and Sutro IACUC protocol SU-001-2013
[0453] As shown in FIG. 5, anti-PD-1 treatment with human IgG 1353-G10 (SEQ ID NOs: 252 and 276) showed accelerated mouse morbidity by approximately two weeks (median survival of 13 days for 10 mg/kg 1353-G10 versus 27 days for anti-GFP control). The onset of GVHD severity was similar in mice treated with high (10 mg/kg) and low (3 mg/kg) 1353-G10. Body weight loss was also faster with both doses of 1353-G10 compared to control and Nivolumab (data not shown). Median survival with both doses of 1353-G10 was less compared to Nivolumab, indicating greater PD-1 blockade potency for 1353-G10 in vivo.
Example 19: Sequences
[0454] Table 10 provides sequences referred to herein.
TABLE-US-00012 TABLE 10 Sequences. SEQ ID NO Molecule Region Scheme Sequence 1 hPD-1 MQIPQAPWPVVWAVLQLGWRPGWFLDSPDR PWNPPTFSPALLVVTEGDNATFTCSFSNTS ESFVLNWYRMSPSNQTDKLAAFPEDRSQPG QDCRFRVTQLPNGRDFHMSVVRARRNDSGT YLCGAISLAPKAQIKESLRAELRVTERRAE VPTAHPSPSPRPAGQFQTLVVGVVGGLLGS LVLLVWVLAVICSRAARGTIGARRTGQPLK EDPSAVPVFSVDYGELDFQWREKTPEPPVP CVPEQTEYATIVFPSGMGTSSPARRGSADG PRSAQPLRPEDGHCSWPL 2 PD1-17 CDR-H1 SGGSIRSTRWWS 3 PD1-28 CDR-H1 SYGIS 4 PD1-33 CDR-H1 SYYIH 5 PD1-35 CDR-H1 SGAYYWS 6 PD1-F2 CDR-H1 SSYWMS 7 10B4 CDR-H1 Chothia GYIFSSY 8 1353-A09 CDR-H1 Chothia GYRFTWY 9 1353-C07 CDR-H1 Chothia GYRFSTF 10 1353-E07 CDR-H1 Chothia GYRFETY 11 1353-F09 CDR-H1 Chothia GYRFRQY 12 1353-G08 CDR-H1 Chothia GYRFTRY 13 1353-G10 CDR-H1 Chothia GYRFPHY 14 1353-H08 CDR-H1 Chothia GYRFTRQ 15 1353-H09 CDR-H1 Chothia GYRFPHY 16 1B10 CDR-H1 Chothia GHSITSDY 17 1E9 CDR-H1 Chothia GFTFSTF 18 4B10 CDR-H1 Chothia GFTFSTY 19 h1E9-1 CDR-H1 Chothia GFTFSTF 20 h1E9-2 CDR-H1 Chothia GFTFSTF 21 h1E9-4 CDR-H1 Chothia GFTFSTF 22 h1E9-5 CDR-H1 Chothia GFTFSTF 23 h4B10-1 CDR-H1 Chothia GFTFSTY 24 h4B10-2 CDR-H1 Chothia GFTFSTY 25 h4B10-3 CDR-H1 Chothia GFTFSTY 26 PD1-17 CDR-H1 Chothia GGSIGSGGSIRSTR 27 PD1-28 CDR-H1 Chothia GYRFTSY 28 PD1-33 CDR-H1 Chothia GYTLTSY 29 PD1-35 CDR-H1 Chothia GGSISSGAY 30 PD1-F2 CDR-H1 Chothia GFTFSSYWCD 31 10B4 CDR-H1 Kabat SYWIG 32 1353-A09 CDR-H1 Kabat WYGIS 33 1353-C07 CDR-H1 Kabat TFGIS 34 1353-E07 CDR-H1 Kabat TYGIS 35 1353-F09 CDR-H1 Kabat QYGIS 36 1353-G08 CDR-H1 Kabat RYGIS 37 1353-G10 CDR-H1 Kabat HYGIS 38 1353-H08 CDR-H1 Kabat RQGIS 39 1353-H09 CDR-H1 Kabat HYGIS 40 1B10 CDR-H1 Kabat SDYAWN 41 1E9 CDR-H1 Kabat TFGMS 42 4B10 CDR-H1 Kabat TYGMS 43 h1E9-1 CDR-H1 Kabat TFGMS 44 h1E9-2 CDR-H1 Kabat TFGMS 45 h1E9-4 CDR-H1 Kabat TFGMS 46 h1E9-5 CDR-H1 Kabat TFGMS 47 h4B10-1 CDR-H1 Kabat TYGMS 48 h4B10-2 CDR-H1 Kabat TYGMS 49 h4B10-3 CDR-H1 Kabat TYGMS 50 PD1-17 CDR-H1 Kabat SGGSIRSTRWWS 51 PD1-28 CDR-H1 Kabat SYGIS 52 PD1-33 CDR-H1 Kabat SYYIH 53 PD1-35 CDR-H1 Kabat SGAYYWS 54 PD1-F2 CDR-H1 Kabat SYWCDRMS 55 PD1-17 CDR-H2 EIYHSGSTNYNPSLKS 56 PD1-28 CDR-H2 WISAYNGNTNYAQKLQG 57 PD1-33 CDR-H2 IINPRGATISYAQKFQG 58 PD1-35 CDR-H2 YIYYNGNTYYNPSLRS 59 PD1-F2 CDR-H2 AISGSGGSTYYADSVKG 60 10B4 CDR-H2 Chothia FPGSGS 61 1353-A09 CDR-H2 Chothia SAYNGN 62 1353-C07 CDR-H2 Chothia SAYNGN 63 1353-E07 CDR-H2 Chothia SAYNGN 64 1353-F09 CDR-H2 Chothia SAYNGN 65 1353-G08 CDR-H2 Chothia SAHNGN 66 1353-G10 CDR-H2 Chothia SAYNGN 67 1353-H08 CDR-H2 Chothia SAYNGN 68 1353-H09 CDR-H2 Chothia SAYNGN 69 1B10 CDR-H2 Chothia SYSGR 70 1E9 CDR-H2 Chothia SGGGSD 71 4B10 CDR-H2 Chothia SGGGSN 72 h1E9-1 CDR-H2 Chothia SGGGSD 73 h1E9-2 CDR-H2 Chothia SGGGSD 74 h1E9-4 CDR-H2 Chothia SGGGSD 75 h1E9-5 CDR-H2 Chothia SGGGSD 76 h4B10-1 CDR-H2 Chothia SGGGSN 77 h4B10-2 CDR-H2 Chothia SGGGSN 78 h4B10-3 CDR-H2 Chothia SGGGSN 79 PD1-17 CDR-H2 Chothia YHSGS 80 PD1-28 CDR-H2 Chothia SAYNGN 81 PD1-33 CDR-H2 Chothia NPRGAT 82 PD1-35 CDR-H2 Chothia YYNGN 83 PD1-F2 CDR-H2 Chothia SGSGGS 84 10B4 CDR-H2 Kabat KIFPGSGSADYNENFKG 85 1353-A09 CDR-H2 Kabat WISAYNGNTNYAQKLQG 86 1353-C07 CDR-H2 Kabat WISAYNGNTNYAQKLQG 87 1353-E07 CDR-H2 Kabat WISAYNGNTNYAQKLQG 88 1353-F09 CDR-H2 Kabat WISAYNGNTNYAQKLQG 89 1353-G08 CDR-H2 Kabat WVSAHNGNTNYAQKLQG 90 1353-G10 CDR-H2 Kabat WISAYNGNTNYAQKLQG 91 1353-H08 CDR-H2 Kabat WISAYNGNTKYAQKLQG 92 1353-H09 CDR-H2 Kabat WISAYNGNTNYAQKLQG 93 1B10 CDR-H2 Kabat YISYSGRTSYNPSLTS 94 1E9 CDR-H2 Kabat TISGGGSDTYYPDSVQG 95 4B10 CDR-H2 Kabat TISGGGSNTYYSDSVKG 96 h1E9-1 CDR-H2 Kabat TISGGGSDTYYPDSVQG 97 h1E9-2 CDR-H2 Kabat TISGGGSDTYYPDSVQG 98 h1E9-4 CDR-H2 Kabat TISGGGSDTYYPDSVQG 99 h1E9-5 CDR-H2 Kabat TISGGGSDTYYPDSVQG 100 h4B10-1 CDR-H2 Kabat TISGGGSNTYYSDSVKG 101 h4B10-2 CDR-H2 Kabat TISGGGSNTYYSDSVKG 102 h4B10-3 CDR-H2 Kabat TISGGGSNTYYSDSVKG 103 PD1-17 CDR-H2 Kabat EIYHSGSTNYNPSLKS 104 PD1-28 CDR-H2 Kabat WISAYNGNTNYAQKLQG 105 PD1-33 CDR-H2 Kabat IINPRGATISYAQKFQG 106 PD1-35 CDR-H2 Kabat YIYYNGNTYYNPSLRS 107 PD1-F2 CDR-H2 Kabat AISGSGGSTYYADSVKG 108 PD1-17 CDR-H3 QDYGDSGDWYFDL 109 PD1-28 CDR-H3 DADYSSGSGY 110 PD1-33 CDR-H3 AGIYGFDFDY 111 PD1-35 CDR-H3 ASDYVWGGYRYMDAFDI 112 PD1-F2 CDR-H3 ENWGSYFDL 113 10B4 CDR-H3 GYGNYLYFDV 114 1353-A09 CDR-H3 DSEYSSGSGY 115 1353-C07 CDR-H3 DVDYSSGSGY 116 1353-E07 CDR-H3 DAEYSLGSGY 117 1353-F09 CDR-H3 DAEYGSGSGY 118 1353-G08 CDR-H3 DADYGSGSGY
119 1353-G10 CDR-H3 DVDYGTGSGY 120 1353-H08 CDR-H3 DVDYGSGSGY 121 1353-H09 CDR-H3 DAEYGSGSGY 122 1B10 CDR-H3 GYALDY 123 1E9 CDR-H3 QGYDVYSWFAY 124 4B10 CDR-H3 QRDSAWFAS 125 h1E9-1 CDR-H3 QGYDVYSWFAY 126 h1E9-2 CDR-H3 QGYDVYSWFAY 127 h1E9-4 CDR-H3 QGYDVYSWFAY 128 h1E9-5 CDR-H3 QGYDVYSWFAY 129 h4B10-1 CDR-H3 QRDSAWFAS 130 h4B10-2 CDR-H3 QRDSAWFAS 131 h4B10-3 CDR-H3 QRDSAWFAS 132 PD1-17 CDR-H3 QDYGDSGDWYFDL 133 PD1-28 CDR-H3 DADYSSGSGY 134 PD1-33 CDR-H3 AGIYGFDFDY 135 PD1-35 CDR-H3 ASDYVWGGYRYMDAFDI 136 PD1-F2 CDR-H3 ENWGSYFDL 137 PD1-17 CDR-L1 TRSSGSIASNSVQ 138 PD1-28 CDR-L1 SGDALPKQYAY 139 PD1-33 CDR-L1 TGTSNDVGGYNYVS 140 PD1-35 CDR-L1 SGSNSNIGSNSVN 141 PD1-F2 CDR-L1 RASQGISSWLA 142 10B4 CDR-L1 KASQSVSDDVA 143 1353-A09 CDR-L1 SGDALTTQYAY 144 1353-C07 CDR-L1 SGDALSEQYAY 145 1353-E07 CDR-L1 SGDALPKQYAY 146 1353-F09 CDR-L1 SGDALPKQYAY 147 1353-G08 CDR-L1 SGDALPMQYGY 148 1353-G10 CDR-L1 SGDALPKQYAY 149 1353-H08 CDR-L1 SGDALPKQYAY 150 1353-H09 CDR-L1 SGDALPKQYAY 151 1B10 CDR-L1 RTSSSVNYMH 152 1E9 CDR-L1 RASESVDNSGISFMS 153 4B10 CDR-L1 RASENVDDYGVSFMN 154 h1E9-1 CDR-L1 RASESVDNSGISFMS 155 h1E9-2 CDR-L1 RASESVDNSGISFMS 156 h1E9-4 CDR-L1 RASESVDNSGISFMS 157 h1E9-5 CDR-L1 RASESVDNSGISFMS 158 h4B10-1 CDR-L1 RASENVDDYGVSFMN 159 h4B10-2 CDR-L1 RASENVDDYGVSFMN 160 h4B10-3 CDR-L1 RASENVDDYGVSFMN 161 PD1-17 CDR-L1 TRSSGSIASNSVQ 162 PD1-28 CDR-L1 SGDALPKQYAY 163 PD1-33 CDR-L1 TGTSNDVGGYNYVS 164 PD1-35 CDR-L1 SGSNSNIGSNSVN 165 PD1-F2 CDR-L1 RASQGISSWLA 166 PD1-17 CDR-L2 EDNQRPS 167 PD1-28 CDR-L2 KDTERPS 168 PD1-33 CDR-L2 DVTNRPS 169 PD1-35 CDR-L2 GNNQRPS 170 PD1-F2 CDR-L2 KASTLES 171 10B4 CDR-L2 YAFKRYI 172 1353-A09 CDR-L2 KDTERPS 173 1353-C07 CDR-L2 KDTERPS 174 1353-E07 CDR-L2 KDTERPS 175 1353-F09 CDR-L2 KDTERPS 176 1353-G08 CDR-L2 KDTERPS 177 1353-G10 CDR-L2 KDTERPS 178 1353-H08 CDR-L2 KDTERPS 179 1353-H09 CDR-L2 KDTERPS 180 1B10 CDR-L2 ATSKLAS 181 1E9 CDR-L2 TASNQGS 182 4B10 CDR-L2 PASNQGS 183 h1E9-1 CDR-L2 TASNQGS 184 h1E9-2 CDR-L2 TASNQGS 185 h1E9-4 CDR-L2 TASNQGS 186 h1E9-5 CDR-L2 TASNQGS 187 h4B10-1 CDR-L2 PASNQGS 188 h4B10-2 CDR-L2 PASNQGS 189 h4B10-3 CDR-L2 PASNQGS 190 PD1-17 CDR-L2 EDNQRPS 191 PD1-28 CDR-L2 KDTERPS 192 PD1-33 CDR-L2 DVTNRPS 193 PD1-35 CDR-L2 GNNQRPS 194 PD1-F2 CDR-L2 KASTLES 195 PD1-17 CDR-L3 QSSDSSAVV 196 PD1-28 CDR-L3 QSADNSITYRV 197 PD1-33 CDR-L3 SSYTIVTNFEVL 198 PD1-35 CDR-L3 AAWDDSLNGPV 199 PD1-F2 CDR-L3 QQSYSTPWT 200 10B4 CDR-L3 QQNYNSPYT 201 1353-A09 CDR-L3 QSADNSITYRV 202 1353-C07 CDR-L3 QSADNSITYRV 203 1353-E07 CDR-L3 QSADNSITYRV 204 1353-F09 CDR-L3 QSADNSITYRV 205 1353-G08 CDR-L3 QSADNSITYRV 206 1353-G10 CDR-L3 QSADNSITYRV 207 1353-H08 CDR-L3 QSADNSITYRV 208 1353-H09 CDR-L3 QSADNSITYRV 209 1B10 CDR-L3 QQWISDPWT 210 1E9 CDR-L3 QQSKEVPWT 211 4B10 CDR-L3 QQSKEVPWT 212 h1E9-1 CDR-L3 QQSKEVPWT 213 h1E9-2 CDR-L3 QQSKEVPWT 214 h1E9-4 CDR-L3 QQSKEVPWT 215 h1E9-5 CDR-L3 QQSKEVPWT 216 h4B10-1 CDR-L3 QQSKEVPWT 217 h4B10-2 CDR-L3 QQSKEVPWT 218 h4B10-3 CDR-L3 QQSKEVPWT 219 PD1-17 CDR-L3 QSSDSSAVV 220 PD1-28 CDR-L3 QSADNSITYRV 221 PD1-33 CDR-L3 SSYTIVTNFEVL 222 PD1-35 CDR-L3 AAWDDSLNGPV 223 PD1-F2 CDR-L3 QQSYSTPWT 224 HC ASTKGPSVFPLAPSSKSTSGGTAALGCLVK Constant DYFPEPVTVSWNSGALTSGVHTFPAVLQSS GLYSLSSVVTVPSSSLGTQTYICNVNHKPS NTKVDKKVEPKSCDKTHTCPPCPAPELLGG PSVFLEPPKPKDTLMISRTPEVTCVVVDVS HEDPEVKFNWYVDGVEVHNAKTKPREEQYN STYRVVSVLTVLHQDWLNGKEYKCKVSNKA LPAPIEKTISKAKGQPREPQVYTLPPSREE MTKNQVSLTCLVKGFYPSDIAVEWESNGQP ENNYKTTPPVLDSDGSFFLYSKLTVDKSRW QQGNVFSCSVMHEALHNHYTQKSLSLSPGK 225 Kappa LC HMTVAAPSVFIFPPSDEQLKSGTASVVCLL NNFYPREAKVQWKVDNALQSGNSQESVTEQ DSKDSTYSLSSTLTLSKADYEKHKVYACEV THQGLSSPVTKSFNRGEC 226 Lambda GQPKAAPSVTLFPPSSEELQANKATLVCLI LD SDFYPGAVTVAWKADSSPVKAGVETTTPSK QSNNKYAASSYLSLTPEQWKSHRSYSCQVT HEGSTVEKTVAPTECS 227 h1E9-1 scFv MEVQLVESGGGLVKPGGSLRLSCAASGFTF STFGMSWVRQAPGKGLEWVSTISGGGSDTY YPDSVQGRFTISRDNAKNSLYLQMNSLRAE DTAVYYCARQGYDVYSWFAYWGQGTLVTVS SGGGGSGGGGSGGGGSDIQLTQSPSFLSAS VGDRVTITCRASESVDNSGISFMSWYQQKP GKAPKLLIYTASNQGSGVPSRFSGSGSGTE FTLTISSLQPEDFATYYCQQSKEVPWTFGQ GTKVEIKGSGDYKDDDDKGSGHHHHHH 228 h1E9-2 scFv MEVQLVESGGGLVKPGGSLRLSCAASGFTF STFGMSWVRQAPGKGLEWVSTISGGGSDTY YPDSVQGRFTISRDNAKNSLYLQMNSLRAE DTAVYYCARQGYDVYSWFAYWGQGTLVTVS SGGGGSGGGGSGGGGSEIVLTQSPATLSLS PGERATLSCRASESVDNSGISFMSWYQQKP GQAPRLLIYTASNQGSGIPARFSGSGSGTD FTLTISSLEPEDFAVYYCQQSKEVPWTFGQ
GTKVEIKGSGDYKDDDDKGSGHHHHHH 229 h1E9-4 scFv MEVQLVESGGGLVQPGGSLRLSCAASGFTF STFGMSWVRQAPGKGLEWVATISGGGSDTY YPDSVQGRFTISRDNAKNSLYLQMNSLRAE DTAVYYCARQGYDVYSWFAYWGQGTLVTVS SGGGGSGGGGSGGGGSDIQLTQSPSFLSAS VGDRVTITCRASESVDNSGISFMSWYQQKP GKAPKLLIYTASNQGSGVPSRFSGSGSGTE FTLTISSLQPEDFATYYCQQSKEVPWTFGQ GTKVEIKGSGDYKDDDDKGSGHHHHHH 230 h1E9-5 scFv MEVQLVESGGGLVQPGGSLRLSCAASGFTF STFGMSWVRQAPGKGLEWVATISGGGSDTY YPDSVQGRFTISRDNAKNSLYLQMNSLRAE DTAVYYCARQGYDVYSWFAYWGQGTLVTVS SGGGGSGGGGSGGGGSEIVLTQSPATLSLS PGERATLSCRASESVDNSGISFMSWYQQKP GQAPRLLIYTASNQGSGIPARFSGSGSGTD FTLTISSLEPEDFAVYYCQQSKEVPWTFGQ GTKVEIKGSGDYKDDDDKGSGHHHHHH 231 h4B10-1 scFv MEVQLVESGGGLVKPGGSLRLSCAASGFTF TSYGMSWVRQAPGKGLEWVATISGGGSNTY YSDSVKGRFTISRDDSKNTLYLQMNSLKTE DTAVYYCARQRDSAWFASWGQGTLVTVSSG GGGSGGGGSGGGGSEIVLTQSPATLSLSPG ERATLSCRASENVDDYGVSFMNWYQQKPGQ APRLLIYPASNQGSGIPARFSGSGSGTDFT LTISSLEPEDFAVYYCQQSKEVPWTFGQGT KVEIKGSGDYKDDDDKGSGHHHHHH 232 h4B10-2 scFv MEVQLVESGGGLVKPGGSLRLSCAASGFTF STYGMSWVRQAPGKGLEWVATISGGGSNTY YSDSVKGRFTISRDDSKNTLYLQMNSLKTE DTAVYYCARQRDSAWFASWGQGTLVTVSSG GGGSGGGGSGGGGSEIVLTQSPGTLSLSPG ERATLSCRASENVDDYGVSFMNWYQQKPGQ APRLLIYPASNQGSGIPDRFSGSGSGTDFT LTISRLEPEDFAVYYCQQSKEVPWTFGQGT KVEIKGSGDYKDDDDKGSGHHHHHH 233 h4B10-3 scFv MEVQLVESGGGLVKPGGSLRLSCAASGFTF STYGMSWVRQAPGKGLEWVATISGGGSNTY YSDSVKGRFTISRDDSKNTLYLQMNSLKTE DTAVYYCARQRDSAWFASWGQGTLVTVSSG GGGSGGGGSGGGGSDIVMTQSPDSLAVSLG ERATINCRASENVDDYGVSFMNWYQQKPGQ PPKLLIYPASNQGSGVPDRFSGSGSGTDFT LTISSLQAEDVAVYYCQQSKEVPWTFGGGT KLEIKGSGDYKDDDDKGSGHHHHHH 234 m10B4 scFv MQVQLQQSGAELMKPGASVKMSCKTTGYIF SSYWIGWVKQRPGHGLEWIGKIFPGSGSAD YNENFKGKATFTVDTSSNTAYMQLSSLTSE DSAVYYCARGYGNYLYFDVWGAGTTVTVSS GGGGSGGGGSGGGGSNIVMTQTPKFLLVSA GDRITITCKASQSVSDDVAWYQQKPGQSPK LLISYAFKRYIGVPDRFTGSGYGTDFTFTI STVQAEDLAVYFCQQNYNSPYTFGGGTKLE LKGSGDYKDDDDKGSGHHHHHH 235 m1B10 scFv MSDVQLQESGPGLVKPSQSLSLTCTVTGHS ITSDYAWNWIRQFPGNKLEWMGYISYSGRT SYNPSLTSRISITRDTSKNQFFLQLNSVTT EDTATYYCARGYALDYWGQGTSVTVSSGGG GSGGGGSGGGGSQIVLSQSPAILSASPGEK VTMTCRTSSSVNYMHWFQQKPGSSPKPWIY ATSKLASGVPARFSGSGSGTSYSLTISRVE AEDAATYFCQQWISDPWTFGGGTKLEIKGS GDYKDDDDKGSGHHHHHH 236 m1E9 scFv MEVKLVESGGGLVSPGGSLKLSCAASGFTF STFGMSWVRQTPEKRLEWVATISGGGSDTY YPDSVQGRFIISRYNAKNNLYLQMNSLRPE DTALYYCARQGYDVYSWFAYWGQGTLVTVS AGGGGSGGGGSGGGGSDIILTQSPASLAVS LGQRAAISCRASESVDNSGISFMSWFQQKP GQPPKLLIYTASNQGSGVPARFSGSGSGTE FSLNIHPMEEDDTAMYFCQQSKEVPWTFGG GTKLEIRGSGDYKDDDDKGSGHHHHHH 237 m4B10 scFv MEVKLVESGGGLVKPGGSLKLSCAASGFTF STYGMSWVRQTPEKRLQWVATISGGGSNTY YSDSVKGRFTISRDNAKNNLYLQMSSLRSE DTALYYCARQRDSAWFASWGQGTLVTVSAG GGGSGGGGSGGGGSDIVLTQSPASLAVSLG QRATISCRASENVDDYGVSFMNWFQQKPGQ PPKLLIYPASNQGSGVPARFSGSGSGTDFS LNIHPMEEDDTAMYFCQQSKEVPWTEGGGT KLEIKGSGDYKDDDDKGSGHHHHHH 238 1353-A09 scFv-Fc MEVQLVQSGAEVKKPGASVKVSCKASGYRF TWYGISWVRQAPGQGLEWMGWISAYNGNTN YAQKLQGRVTMTTDTSTNTAYMELRSLRSD DTAVYYCARDSEYSSGSGYWGQGTLVTVSS GGGGSGGGGSGGGGSSYELTQPPSVSVSPG QTARTTCSGDALTTQYAYWYQQKPGQAPVM VIYKDTERPSGIPERFSGSSSGTKVTLTIS GVQAEDEADYYCQSADNSITYRVFGGGTKV TVLAAGSDQEPKKLAAGSDQEPKSSDKTHT CPPCSAPELLGGSSVFLFPPKPKDTLMISR TPEVTCVVVDVSHEDPEVKFNWYVDGVEVH NAKTKPREEQYNSTYRVVSVLTVLHQDWLN GKEYKCKVSNKALPAPIEKTISKAKGQPRE PQVYTLPPSRDELTKNQVSLTCLVKGFYPS DIAVEWESNGQPENNYKTTPPVLDSDGSFE LYSKLTVDKSRWQQGNVFSCSVMHEALHNH YTQKSLSLSPGKGSGDYKDDDDKGSGHHHH HH 239 1353-C07 scFv-Fc MEVQLVQSGAEVKKPGASVKVSCKASGYRF STFGISWVRQAPGQGLEWMGWISAYNGNTN YAQKLQGRVTMTTDTSTNTAYMELRSLRSD DTAVYYCARDVDYSSGSGYWGQGTLVTVSS GGGGSGGGGSGGGGSSYELTQPPSVSVSPG QTARTTCSGDALSEQYAYWYQQKPGQAPVM VIYKDTERPSGIPERFSGSSSGTKVTLTIS GVQAEDEADYYCQSADNSITYRVFGGGTKV TVLAAGSDQEPKKLAAGSDQEPKSSDKTHT CPPCSAPELLGGSSVFLEPPKPKDTLMISR TPEVTCVVVDVSHEDPEVKFNWYVDGVEVH NAKTKPREEQYNSTYRVVSVLTVLHQDWLN GKEYKCKVSNKALPAPIEKTISKAKGQPRE PQVYTLPPSRDELTKNQVSLTCLVKGFYPS DIAVEWESNGQPENNYKTTPPVLDSDGSFF LYSKLTVDKSRWQQGNVFSCSVMHEALHNH YTQKSLSLSPGKGSGDYKDDDDKGSGHHHH HH 240 1353-E07 scFv-Fc MEVQLVQSGAEVKKPGASVKVSCKASGYRF ETYGISWVRQAPGQGLEWMGWISAYNGNTN YAQKLQGRVTMTTDTSTNTAYMELRSLRSD DTAVYYCARDAEYSLGSGYWGQGTLVTVSS GGGGSGGGGSGGGGSSYELTQPPSVSVSPG QTARTTCSGDALPKQYAYWYQQKPGQAPVM VIYKDTERPSGIPERFSGSSSGTKVTLTIS GVQAEDEADYYCQSADNSITYRVFGGGTKV TVLAAGSDQEPKKLAAGSDQEPKSSDKTHT CPPCSAPELLGGSSVFLEPPKPKDTLMISR TPEVTCVVVDVSHEDPEVKFNWYVDGVEVH NAKTKPREEQYNSTYRVVSVLTVLHQDWLN GKEYKCKVSNKALPAPIEKTISKAKGQPRE PQVYTLPPSRDELTKNQVSLTCLVKGFYPS DIAVEWESNGQPENNYKTTPPVLDSDGSFF YLSKLTVDKSRWQQGNVFSCSVMHEALHNH YTQKSLSLSPGKGSGDYKDDDDKGSGHHHH HH 241 1353-F09 scFv-Fc MEVQLVQSGAEVKKPGASVKVSCKASGYRF RQYGISWVRQAPGQGLEWMGWISAYNGNTN YAQKLQGRVTMTTDTSTNTAYMELRSLRSD DTAVYYCARDAEYGSGSGYWGQGTLVTVSS GGGGSGGGGSGGGGSSYELTQPPSVSVSPG QTARTTCSGDALPKQYAYWYQQKPGQAPVM VLYKDTERPSGIPERFSGSSSGTKVTLTIS GVQAEDEADYYCQSADNSITYRVFGGGTKV TVLAAGSDQEPKKLAAGSDQEPKSSDKTHT CPPCSAPELLGGSSVFLEPPKPKDTLMISR TPEVTCVVVDVSHEDPEVKFNWYVDGVEVH NAKTKPREEQYNSTYRVVSVLTVLHQDWLN GKEYKCKVSNKALPAPIEKTISKAKGQPRE PQVYTLPPSRDELTKNQVSLTCLVKGFYPS DIAVEWESNGQPENNYKTTPPVLDSDGSFF LYSKLTVDKSRWQQGNVFSCSVMHEALHNH YTQKSLSLSPGKGSGDYKDDDDKGSGHHHH HH 242 1353-G08 scFv-Fc MEVQLVQSGAEVKKPGASVKVSCKASGYRF TRYGISWVRQAPGQGLEWMGWVSAHNGNTN YAQKLQGRVTMTTDTSTNTAYMELRSLRSD DTAVYYCARDADYGSGSGYWGQGTLVTVSS GGGVSGGGGSGGGGSSYELTQPPSVSVSPG QTARTTCSGDALPMQYGYWYQQKPGQAPVM VIYKDTERPSGIPERFSGSSSGTKVTLTIS GVQAEDEADYYCQSADNSITYRVFGGGTKV TVLAAGSDQEPKKLAAGSDQEPKSSDKTHT CPPCSAPELLGGSSVFLEPPKPKDTLMISR TPEVTCVVVDVSHEDPEVKFNWYVDGVEVH NAKTKPREEQYNSTYRVVSVLTVLHQDWLN GKEYKCKVSNKALPAPIEKTISKAKGQPRE PQVYTLPPSRDELTKNQVSLTCLVKGFYPS DIAVEWESNGQPENNYKTTPPVLDSDGSFF LYSKLTVDKSRWQQGNVFSCSVMHEALHNH YTQKSLSLSPGKGSGDYKDDDDKGSGHHHH HH 243 1353-G10 scFv-Fc MEVQLVQSGAEVKKPGASVKVSCKASGYRF PHYGISWVRQAPGQGLEWMGWISAYNGNTN YAQKLQGRVTMTTDTSTNTAYMELRSLRSD DTAVYYCARDVDYGTGSGYWGQGTLVTVSS GGGGSGGGGSGGGGSSYELTQPPSVSVSPG QTARTTCSGDALPKQYAYWYQQKPGQAPVM VIYKDTERPSGIPERFSGSSSGTKVTLTIS GVQAEDEADYYCQSADNSITYRVFGGGTKV TVLAAGSDQEPKKLAAGSDQEPKSSDKTHT CPPCSAPELLGGSSVFLEPPKPKDTLMISR TPEVTCVVVDVSHEDPEVKFNWYVDGVEVH NAKTKPREEQYNSTYRVVSVLTVLHQDWLN GKEYKCKVSNKALPAPIEKTISKAKGQPRE PQVYTLPPSRDELTKNQVSLTCLVKGFYPS DIAVEWESNGQPENNYKTTPPVLDSDGSFF LYSKLTVDKSRWQQGNVFSCSVMHEALHNH YTQKSLSLSPGKGSGDYKDDDDKGSGHHHH HH 244 1353-H08 scFv-Fc MEVQLVQSGAEVKKPGASVKVSCKASGYRF TRQGISWVRQAPGQGLEWMGWISAYNGNTK YAQKLQGRVTMTTDTSTNTAYMELRSLRSD DTAVYYCARDVDYGSGSGYWGQGTLVTVSS GGGGSGGGGSGGGGSSYELTQPPSVSVSPG QTARTTCSGDALPKQYAYWYQQKPGQAPVM VIYKDTERPSGIPERFSGSSSGTKVTLTIS GVQAEDEADYYCQSADNSITYRVFGGGTKV TVLAAGSDQEPKKLAAGSDQEPKSSDKTHT CPPCSAPELLGGSSVFLEPPKPKDTLMISR TPEVTCVVVDVSHEDPEVKFNWYVDGVEVH NAKTKPREEQYNSTYRVVSVLTVLHQDWLN GKEYKCKVSNKALPAPIEKTISKAKGQPRE PQVYTLPPSRDELTKNQVSLTCLVKGFYPS DIAVEWESNGQPENNYKTTPPVLDSDGSFF LYSKLTVDKSRWQQGNVFSCSVMHEALHNH YTQKSLSLSPGKGSGDYKDDDDKGSGHHHH HH 245 1353-H09 scFv-Fc MEVQLVQSGAEVKKPGASVKVSCKASGYRF PHYGISWVRQAPGQGLEWMGWISAYNGNTN YAQKLQGRVTMTTDTSTNTAYMELRSLRSD DTAVYYCARDAEYGSGSGYWGQGTLVTVSS GGGGSGGGGSGGGGSSYELTQPPSVSVSPG QTARTTCSGDALPKQYAYWYQQKPGQAPVM VIYKDTERPSGIPERFSGSSSGTKVTLTIS GVQAEDEADYYCQSADNSITYRVFGGGTKV TVLAAGSDQEPKKLAAGSDQEPKSSDKTHT PCPCSAPELLGGSSVFLEPPKPKDTLMISR TPEVTCVVVDVSHEDPEVKFNWYVDGVEVH NAKTKPREEQYNSTYRVVSVLTVLHQDWLN GKEYKCKVSNKALPAPIEKTISKAKGQPRE PQVYTLPPSRDELTKNQVSLTCLVKGFYPS DIAVEWESNGQPENNYKTTPPVLDSDGSFF LYSKLTVDKSRWQQGNVFSCSVMHEALHNH YTQKSLSLSPGKGSGDYKDDDDKGSGHHHH HH 246 10B4 VH QVQLQQSGAELMKPGASVKMSCKTTGYIFS SYWIGWVKQRPGHGLEWIGKIFPGSGSADY NENFKGKATFTVDTSSNTAYMQLSSLTSED SAVYYCARGYGNYLYFDVWGAGTTVTVSS 247 1353-A09 VH EVQLVQSGAEVKKPGASVKVSCKASGYRFT WYGISWVRQAPGQGLEWMGWISAYNGNTNY
AQKLQGRVTMTTDTSTNTAYMELRSLRSDD TAVYYCARDSEYSSGSGYWGQGTLVTVSS 248 1353-C07 VH EVQLVQSGAEVKKPGASVKVSCKASGYRFS TFGISWVRQAPGQGLEWMGWISAYNGNTNY AQKLQGRVTMTTDTSTNTAYMELRSLRSDD TAVYYCARDVDYSSGSGYWGQGTLVTVSS 249 1353-E07 VH EVQLVQSGAEVKKPGASVKVSCKASGYRFE TYGISWVRQAPGQGLEWMGWISAYNGNTNY AQKLQGRVTMTTDTSTNTAYMELRSLRSDD TAVYYCARDAEYSLGSGYWGQGTLVTVSS 250 1353-F09 VH EVQLVQSGAEVKKPGASVKVSCKASGYRFR QYGISWVRQAPGQGLEWMGWISAYNGNTNY AQKLQGRVTMTTDTSTNTAYMELRSLRSDD TAVYYCARDAEYGSGSGYWGQGTLVTVSS 251 1353-G08 VH EVQLVQSGAEVKKPGASVKVSCKASGYRFT RYGISWVRQAPGQGLEWMGWVSAHNGNTNY AQKLQGRVTMTTDTSTNTAYMELRSLRSDD TAVYYCARDADYGSGSGYWGQGTLVTVSS 252 1353-G10 VH EVQLVQSGAEVKKPGASVKVSCKASGYRFP HYGISWVRQAPGQGLEWMGWISAYNGNTNY AQKLQGRVTMTTDTSTNTAYMELRSLRSDD TAVYYCARDVDYGTGSGYWGQGTLVTVSS 253 1353-H08 VH EVQLVQSGAEVKKPGASVKVSCKASGYRFT RQGISWVRQAPGQGLEWMGWISAYNGNTKY AQKLQGRVTMTTDTSTNTAYMELRSLRSDD TAVYYCARDVDYGSGSGYWGQGTLVTVSS 254 1353-H09 VH EVQLVQSGAEVKKPGASVKVSCKASGYRFP HYGISWVRQAPGQGLEWMGWISAYNGNTNY AQKLQGRVTMTTDTSTNTAYMELRSLRSDD TAVYYCARDAEYGSGSGYWGQGTLVTVSS 255 1B10 VH DVQLQESGPGLVKPSQSLSLTCTVTGHSIT SDYAWNWIRQFPGNKLEWMGYISYSGRTSY NPSLTSRISITRDTSKNQFFLQLNSVTTED TATYYCARGYALDYWGQGTSVTVSS 256 1E9 VH EVKLVESGGGLVSPGGSLKLSCAASGFTFS TFGMSWVRQTPEKRLEWVATISGGGSDTYY PDSVQGRFIISRYNAKNNLYLQMNSLRPED TALYYCARQGYDVYSWFAYWGQGTLVTVSA 257 4B10 VH EVKLVESGGGLVKPGGSLKLSCAASGFTFS TYGMSWVRQTPEKRLQWVATISGGGSNTYY SDSVKGRFTISRDNAKNNLYLQMSSLRSED TALYYCARQRDSAWFASWGQGTLVTVSA 258 h1E9-1 VH EVQLVESGGGLVKPGGSLRLSCAASGFTFS TFGMSWVRQAPGKGLEWVSTISGGGSDTYY PDSVQGRFTISRDNAKNSLYLQMNSLRAED TAVYYCARQGYDVYSWFAYWGQGTLVTVSS 259 h1E9-2 VH EVQLVESGGGLVKPGGSLRLSCAASGFTFS TFGMSWVRQAPGKGLEWVSTISGGGSDTYY PDSVQGRFTISRDNAKNSLYLQMNSLRAED TAVYYCARQGYDVYSWFAYWGQGTLVTVSS 260 h1E9-4 VH EVQLVESGGGLVQPGGSLRLSCAASGFTFS TFGMSWVRQAPGKGLEWVATISGGGSDTYY PDSVQGRFTISRDNAKNSLYLQMNSLRAED TAVYYCARQGYDVYSWFAYWGQGTLVTVSS 261 h1E9-5 VH EVQLVESGGGLVQPGGSLRLSCAASGFTFS TFGMSWVRQAPGKGLEWVATISGGGSDTYY PDSVQGRFTISRDNAKNSLYLQMNSLRAED TAVYYCARQGYDVYSWFAYWGQGTLVTVSS 262 h4B10-1 VH EVQLVESGGGLVKPGGSLRLSCAASGFTFS TYGMSWVRQAPGKGLEWVATISGGGSNTYY SDSVKGRFTISRDDSKNTLYLQMNSLKTED TAVYYCARQRDSAWFASWGQGTLVTVSS 263 h4B10-2 VH EVQLVESGGGLVKPGGSLRLSCAASGFTFS TYGMSWVRQAPGKGLEWVATISGGGSNTYY SDSVKGRFTISRDDSKNTLYLQMNSLKTED TAVYYCARQRDSAWFASWGQGTLVTVSS 264 h4B10-3 VH EVQLVESGGGLVKPGGSLRLSCAASGFTFS TYGMSWVRQAPGKGLEWVATISGGGSNTYY SDSVKGRFTISRDDSKNTLYLQMNSLKTED TAVYYCARQRDSAWFASWGQGTLVTVSS 265 PD1-17 VH QVQLQESGPGVVKPSGTLSLTCAISGGSIG SGGSIRSTRWWSWVRQSPGKGLEWIGEIYH SGSTNYNPSLKSRVTISLDKSRNHFSLRLN SVTAADTAVYYCARQDYGDSGDWYFDLWGK GTMVTVSS 266 PD1-28 VH EVQLVQSGAEVKKPGASVKVSCKASGYRFT SYGISWVRQAPGQGLEWMGWISAYNGNTNY AQKLQGRVTMTTDTSTNTAYMELRSLRSDD TAVYYCARDADYSSGSGYWGQGTLVTVSS 267 PD1-33 VH QVQLVQSGAEVKKPGASVRVSCKASGYTLT SYYIHWVRQAPGQGLEWMGIINPRGATISY AQKFQGRVTMTRDTSTSTVYMELRNLKSED TALYYCATAGIYGFDFDYWGRGTLVTVSS 268 PD1-35 VH QVQLQESGPGLVKPSQTLSLTCTVSGGSIS SGAYYWSWIRQHPGKGLEWIGYIYYNGNTY YNPSLRSLVTISVDASKNQFSLKLSSVTAA DTAVYYCARASDYVWGGYRYMDAFDIWGRG TLITVSS 269 PD1-F2 VH EVQLVQSGGGVVQPGRSLRLSCAASGFTFS SYWCDRMSWVRQAPGKGLEWVSAISGSGGS TYYADSVKGRFTISRDNSKNTLYLQMNSLR AEDTAVYYCAKENWGSYFDLWGQGTTVTVS S 270 10B4 VL NIVMTQTPKFLLVSAGDRITITCKASQSVS DDVAWYQQKPGQSPKLLISYAFKRYIGVPD RFTGSGYGTDFTFTISTVQAEDLAVYFCQQ NYNSPYTFGGGTKLELKR 271 1353-A09 VL SYELTQPPSVSVSPGQTARITCSGDALTTQ YAYWYQQKPGQAPVMVIYKDTERPSGIPER FSGSSSGTKVTLTISGVQAEDEADYYCQSA DNSITYRVFGGGTKVTVL 272 1353-C07 VL SYELTQPPSVSVSPGQTARITCSGDALSEQ YAYWYQQKPGQAPVMVIYKDTERPSGIPER FSGSSSGTKVTLTISGVQAEDEADYYCQSA DNSITYRVFGGGTKVTVL 273 1353-E07 VL SYELTQPPSVSVSPGQTARITCSGDALPKQ YAYWYQQKPGQAPVMVIYKDTERPSGIPER FSGSSSGTKVTLTISGVQAEDEADYYCQSA DNSITYRVFGGGTKVTVL 274 1353-F09 VL SYELTQPPSVSVSPGQTARITCSGDALPKQ YAYWYQQKPGQAPVMVLYKDTERPSGIPER FSGSSSGTKVTLTISGVQAEDEADYYCQSA DNSITYRVFGGGTKVTVL 275 1353-G08 VL SYELTQPPSVSVSPGQTARITCSGDALPMQ YGYWYQQKPGQAPVMVIYKDTERPSGIPER FSGSSSGTKVTLTISGVQAEDEADYYCQSA DNSITYRVFGGGTKVTVL 276 1353-G10 VL SYELTQPPSVSVSPGQTARITCSGDALPKQ YAYWYQQKPGQAPVMVIYKDTERPSGIPER FSGSSSGTKVTLTISGVQAEDEADYYCQSA DNSITYRVFGGGTKVTVL 277 1353-H08 VL SYELTQPPSVSVSPGQTARITCSGDALPKQ YAYWYQQKPGQAPVMVIYKDTERPSGIPER FSGSSSGTKVTLTISGVQAEDEADYYCQSA DNSITYRVFGGGTKVTVL 278 1353-H09 VL SYELTQPPSVSVSPGQTARITCSGDALPKQ YAYWYQQKPGQAPVMVIYKDTERPSGIPER FSGSSSGTKVTLTISGVQAEDEADYYCQSA DNSITYRVFGGGTKVTVL 279 1B10 VL QIVLSQSPAILSASPGEKVTMTCRTSSSVN YMHWFQQKPGSSPKPWIYATSKLASGVPAR FSGSGSGTSYSLTISRVEAEDAATYFCQQW ISDPWTFGGGTKLEIK 280 1E9 VL DIILTQSPASLAVSLGQRAAISCRASESVD NSGISFMSWFQQKPGQPPKLLIYTASNQGS GVPARFSGSGSGTEFSLNIHPMEEDDTAMY FCQQSKEVPWTFGGGTKLEIR 281 4B10 VL DIVLTQSPASLAVSLGQRATISCRASENVD DYGVSFMNWFQQKPGQPPKLLIYPASNQGS GVPARFSGSGSGTDFSLNIHPMEEDDTAMY FCQQSKEVPWTFGGGTKLEIK 282 h1E9-1 VL DIQLTQSPSFLSASVGDRVTITCRASESVD NSGISFMSWYQQKPGKAPKLLIYTASNQGS GVPSRFSGSGSGTEFTLTISSLQPEDFATY YCQQSKEVPWTFGQGTKVEIK 283 h1E9-2 VL EIVLTQSPATLSLSPGERATLSCRASESVD NSGISFMSWYQQKPGQAPRLLIYTASNQGS GIPARFSGSGSGTDFTLTISSLEPEDFAVY YCQQSKEVPWTFGQGTKVEIK 284 h1E9-4 VL DIQLTQSPSFLSASVGDRVTITCRASESVD NSGISFMSWYQQKPGKAPKLLIYTASNQGS GVPSRFSGSGSGTEFTLTISSLQPEDFATY YCQQSKEVPWTFGQGTKVEIK 285 h1E9-5 VL EIVLTQSPATLSLSPGERATLSCRASESVD NSGISFMSWYQQKPGQAPRLLIYTASNQGS GIPARFSGSGSGTDFTLTISSLEPEDFAVY YCQQSKEVPWTFGQGTKVEIK 286 h4B10-1 VL EIVLTQSPATLSLSPGERATLSCRASENVD DYGVSFMNWYQQKPGQAPRLLTYPASNQGS GIPARFSGSGSGTDFTLTISSLEPEDFAVY YCQQSKEVPWTFGQGTKVEIK 287 h4B10-2 VL EIVLTQSPGTLSLSPGERATLSCRASENVD DYGVSFMNWYQQKPGQAPRLLTYPASNQGS GIPDRFSGSGSGTDFTLTISRLEPEDFAVY YCQQSKEVPWTFGQGTKVEIK 288 h4B10-3 VL DIVMTQSPDSLAVSLGERATINCRASENVD DYGVSFMNWYQQKPGQPPKLLTYPASNQGS GVPDRFSGSGSGTDFTLTISSLQAEDVAVY YCQQSKEVPWTFGGGTKLEIK 289 PD1-17 VL NFMLTQPHSVSESPGKTVTISCTRSSGSIA SNSVQWYQQRPGSSPTTVIYEDNQRPSGVP DRFSGSIDSSSNSASLTVSGLKTEDEADYY CQSSDSSAVVFGSGTKLTVL 290 PD1-28 VL SYELTQPPSVSVSPGQTARITCSGDALPKQ YAYWYQQKPGQAPVMVIYKDTERPSGIPER FSGSSSGTKVTLTISGVQAEDEADYYCQSA DNSITYRVFGGGTKVTVL 291 PD1-33 VL QSALTQPASVSGSPGQSITISCTGTSNDVG GYNYVSWYQHHPGKAPKLIIYDVTNRPSGV SDRFSGSKSGNTASLTISGLLAEDEGDYYC SSYTIVTNFEVLEGGGTKLTV 292 PD1-35 VL QSVLTQPPSASGTPGQRVTISCSGSNSNIG SNSVNWYQQLPGTAPKLLIYGNNQRPSGVP DRFSGSKSGTSASLAISGLQSENEADYYCA AWDDSLNGPVFGRGTKVTVL 293 PD1-F2 VL DIVMTQSPSTLSASVGDRVTITCRASQGIS SWLAWYQQKPGRAPKVLIYKASTLESGVPS RFSGSGSGTDFTLTISSLQPEDFATYYCQQ SYSTPWTFGQGTKLEIK 294 FlagHis GSGDYKDDDDKGSGHHHHHH Tag 295 IgG1 Fc AAGSDQEPKSSDKTHTCPPCSAPELLGGSS from VFLFPPKPKDTLMISRTPEVTCVVVDVSHE scFv-Fc DPEVKFNWYVDGVEVHNAKTKPREEQYNST YRVVSVLTVLHQDWLNGKEYKCKVSNKALP APIEKTISKAKGQPREPQVYTLPPSRDELT KNQVSLTCLVKGFYPSDIAVEWESNGQPEN NYKTTPPVLDSDGSFFLYSKLTVDKSRWQQ GNVFSCSVMHEALHNHYTQKSLSLSPGKGS GDYKDDDDKGSG 296 HC ASTKGPSVFPLAPSSKSTSGGTAALGCLVK Constant DYFPEPVTVSWNSGALTSGVHTFPAVLQSS
GLYSLSSVVTVPSSSLGTQTYICNVNHKPS NTKVDKKVEPKSCDKTHTCPPCPAPELLGG PSVFLFPPKPKDTLMISRTPEVTCVVVDVS HEDPEVKFNWYVDGVEVHNAKTKPREEQYN STYRVVSVLTVLHQDWLNGKEYKCKVSNKA LPAPIEKTISKAKGQPREPQVYTLPPSREE MTKNQVSLTCLVKGFYPSDIAVEWESNGQP ENNYKTTPPVLDSDGSFFLYSKLTVDKSRW QQGNVFSCSVMHEALHNHYTQKSLSLSPGK 297 LC RTVAAPSVFIFPPSDEQLKSGTASVVCLLN Constant NFYPREAKVQWKVDNALQSGNSQESVTEQD SKDSTYSLSSTLTLSKADYEKHKVYACEVT HQGLSSPVTKSFNRGEC 298 Linker GGGGSGGGGSGGGGS 299 murine MWVRQVPWSFTWAVLQLSWQSGWLLEVPNG PD-1 PWRSLTFYPAWLTVSEGANATFTCSLSNWS EDLMLNWNRLSPSNQTEKQAAFCNGLSQPV QDARFQITQLPNRHDEHMNILDTRRNDSGI YLCGAISLHPKAKIEESPGAELVVTERILE TSTRYPSPSPKPEGRFQGMVIGIMSALVGI PVLLLLAWALAVFCSTSMSEARGAGSKDDT LKEEPSAAPVPSVAYEELDFQGREKTPELP TACVHTEYATIVFTEGLGASAMGRRGSADG LQGPRPPRHEDGHCSWPL 300 cyno MWVRQVPWSFTWAVLQLSWQSGWLLEVPNG PD-1 PWRSLTFYPAWLTVSEGANATFTCSLSNWS EDLMLNWNRLSPSNQTEKQAAFCNGLSQPV QDARFQITQLPNRHDEHMNILDTRRNDSGI YLCGAISLHPKAKIEESPGAELVVTERILE TSTRYPSPSPKPEGRFQGMVIGIMSALVGI PVLLLLAWALAVFCSTSMSEARGAGSKDDT LKEEPSAAPVPSVAYEELDFQGREKTPELP TACVHTEYATIVFTEGLGASAMGRRGSADG LQGPRPPRHEDGHCSWPL 301 Linker AAGSDQEPK 302 h1E9-4 & HC- MEVQLVESGGGLVQPGGSLRLSCAASGFTF h1E9-5 FlagHis STFGMSWVRQAPGKGLEWVATISGGGSDTY YPDSVQGRFTISRDNAKNSLYLQMNSLRAE DTAVYYCARQGYDVYSWFAYWGQGTLVTVS SASTKGPSVFPLAPSSKSTSGGTAALGCLV KDYFPEPVTVSWNSGALTSGVHTFPAVLQS SGLYSLSSVVTVPSSSLGTQTYICNVNHKP SNTKVDKKVEPKSCDKTHTCPPCPAPELLG GPSVFLEPPKPKDTLMISRTPEVTCVVVDV SHEDPEVKFNWYVDGVEVHNAKTKPREEQY NSTYRVVSVLTVLHQDWLNGKEYKCKVSNK ALPAPIEKTISKAKGQPREPQVYTLPPSRE EMTKNQVSLTCLVKGFYPSDIAVEWESNGQ PENNYKTTPPVLDSDGSFFLYSKLTVDKSR WQQGNVFSCSVMHEALHNHYTQKSLSLSPG KGSGDYKDDDDKGSGHHHHHH 303 h1E9-5 LC MEIVLTQSPATLSLSPGERATLSCRASESV DNSGISFMSWYQQKPGQAPRLLIYTASNQG SGIPARFSGSGSGTDFTLTISSLEPEDFAV YYCQQSKEVPWTFGQGTKVEIKRTVAAPSV FIFPPSDEQLKSGTASVVCLLNNFYPREAK VQWKVDNALQSGNSQESVTEQDSKDSTYSL SSTLTLSKADYEKHKVYACEVTHQGLSSPV TKSFNRGEC 304 h1E9-4 LC MDIQLTQSPSFLSASVGDRVTITCRASESV DNSGISFMSWYQQKPGKAPKLLIYTASNQG SGVPSRFSGSGSGTEFTLTISSLQPEDFAT YYCQQSKEVPWTFGQGTKVEIKRTVAAPSV FIFPPSDEQLKSGTASVVCLLNNFYPREAK VQWKVDNALQSGNSQESVTEQDSKDSTYSL SSTLTLSKADYEKHKVYACEVTHQGLSSPV TKSFNRGEC 305 PD1-F2v- scFv-Fc MGAHSEVQLVQSGGGVVQPGRSLRLSCAAS scFvFcFlag GFTFSSYWMSWVRQAPGKGLEWVSAISGSG His GSTYYADSVKGRFTISRDNSKNTLYLQMNS LRAEDTAVYYCAKENWGSYFDLWGQGTTVT VSSGGGGSGGGGSGGGGSGVHSDIVMTQSP STLSASVGDRVTITCRASQGISSWLAWYQQ KPGRAPKVLTYKASTLESGVPSRFSGSGSG TDFTLTISSLQPEDFATYYCQQSYSTPWTF GQGTKLEIKAAGSDQEPKSSDKTHTCPPCS APELLGGSSVFLEPPKPKDTLMISRTPEVT CVVVDVSHEDPEVKFNWYVDGVEVHNAKTK PREEQYNSTYRVVSVLTVLHQDWLNGKEYK CKVSNKALPAPIEKTISKAKGQPREPQVYT LPPSRDELTKNQVSLTCLVKGFYPSDIAVE WESNGQPENNYKTTPPVLDSDGSFFLYSKL TVDKSRWQQGNVFSCSVMHEALHNHYTQKS LSLSPGKGSGDYKDDDDKGSGHHHHHH 306 PD1-F2v VH EVQLVQSGGGVVQPGRSLRLSCAASGFTES SYWMSWVRQAPGKGLEWVSAISGSGGSTYY ADSVKGRFTISRDNSKNTLYLQMNSLRAED TAVYYCAKENWGSYFDLWGQGTTVTVSS 307 PD1-F2v HC GAHSEVQLVQSGGGVVQPGRSLRLSCAASG FTESSYWMSWVRQAPGKGLEWVSAISGSGG STYYADSVKGRFTISRDNSKNTLYLQMNSL RAEDTAVYYCAKENWGSYFDLWGQGTTVTV SSASTKGPSVFPLAPSSKSTSGGTAALGCL VKDYFPEPVTVSWNSGALTSGVHTFPAVLQ SSGLYSLSSVVTVPSSSLGTQTYICNVNHK PSNTKVDKKVEPKSCDKTHTCPPCPAPELL GGPSVFLFPPKPKDTLMISRTPEVTCVVVD VSHEDPEVKFNWYVDGVEVHNAKTKPREEQ YNSTYRVVSVLTVLHQDWLNGKEYKCKVSN KALPAPIEKTISKAKGQPREPQVYTLPPSR EEMTKNQVSLTCLVKGFYPSDIAVEWESNG QPENNYKTTPPVLDSDGSFFLYSKLTVDKS RWQQGNVFSCSVMHEALHNHYTQKSLSLSP GK 308 PD1-F2v LC GVHSDIVMTQSPSTLSASVGDRVTITCRAS QGISSWLAWYQQKPGRAPKVLIYKASTLES GVPSRFSGSGSGTDFTLTISSLQPEDFATY YCQQSYSTPWTFGQGTKLEIKRHMTVAAPS VFIFPPSDEQLKSGTASVVCLLNNFYPREA KVQWKVDNALQSGNSQESVTEQDSKDSTYS LSSTLTLSKADYEKHKVYACEVTHQGLSSP VTKSFNRGEC 309 1353-E07- CDR-H3 DAEYRLGSGY R5 310 1353-A09- CDR-H3 DSEYRSGSGY R5 311 1353-F09- CDR-H3 DAEYRSGSGY R5 312 1353-G08- CDR-H3 DADYRSGSGY R5 313 1353-G10- CDR-H3 DVDYRTGSGY R5 314 1353-C07- CDR-H3 DVDYRSGSGY R5 315 1353-H08- CDR-H3 DVDYRSGSGY R5 316 1353-E07- VH EVQLVQSGAEVKKPGASVKVSCKASGYRFE R5 TYGISWVRQAPGQGLEWMGWISAYNGNTNY AQKLQGRVTMTTDTSTNTAYMELRSLRSDD TAVYYCARDAEYRLGSGYWGQGTLVTVSS 317 1353-A09- VH EVQLVQSGAEVKKPGASVKVSCKASGYRFT R5 WYGISWVRQAPGQGLEWMGWISAYNGNTNY AQKLQGRVTMTTDTSTNTAYMELRSLRSDD TAVYYCARDSEYRSGSGYWGQGTLVTVSS 318 1353-F09- VH EVQLVQSGAEVKKPGASVKVSCKASGYRFR R5 QYGISWVRQAPGQGLEWMGWISAYNGNTNY AQKLQGRVTMTTDTSTNTAYMELRSLRSDD TAVYYCARDAEYRSGSGYWGQGTLVTVSS 319 1353-G08- VH EVQLVQSGAEVKKPGASVKVSCKASGYRFT R5 RYGISWVRQAPGQGLEWMGWVSAHNGNTNY AQKLQGRVTMTTDTSTNTAYMELRSLRSDD TAVYYCARDADYRSGSGYWGQGTLVTVSS 320 1353-G10- VH EVQLVQSGAEVKKPGASVKVSCKASGYRFP R5 HYGISWVRQAPGQGLEWMGWISAYNGNTNY AQKLQGRVTMTTDTSTNTAYMELRSLRSDD TAVYYCARDVDYRTGSGYWGQGTLVTVSS 321 1353-C07- VH EVQLVQSGAEVKKPGASVKVSCKASGYRFS R5 TFGISWVRQAPGQGLEWMGWISAYNGNTNY AQKLQGRVTMTTDTSTNTAYMELRSLRSDD TAVYYCARDVDYRSGSGYWGQGTLVTVSS 322 1353-H08- VH EVQLVQSGAEVKKPGASVKVSCKASGYRFT R5 RQGISWVRQAPGQGLEWMGWISAYNGNTKY AQKLQGRVTMTTDTSTNTAYMELRSLRSDD TAVYYCARDVDYRSGSGYWGQGTLVTVSS 323 1353-H09- VH EVQLVQSGAEVKKPGASVKVSCKASGYRFP R5 HYGISWVRQAPGQGLEWMGWISAYNGNTNY AQKLQGRVTMTTDTSTNTAYMELRSLRSDD TAVYYCARDAEYRSGSGYWGQGTLVTVSS 324 H1E9-HC3 HC- MEVQLVESGGGLVQPGGSLRLSCAASGFTF FlagHis STFGMSWVRQAPGKGLEWVATISGGGSDTY YPDSVKGRFTISRDNAKNSLYLQMNSLRAE DTAVYYCARQGYDVYSWFAYWGQGTLVTVS SASTKGPSVFPLAPSSKSTSGGTAALGCLV KDYFPEPVTVSWNSGALTSGVHTFPAVLQS SGLYSLSSVVTVPSSSLGTQTYICNVNHKP SNTKVDKKVEPKSCDKTHTCPPCPAPELLG GPSVFLFPPKPKDTLMISRTPEVTCVVVDV SHEDPEVKFNWYVDGVEVHNAKTKPREEQY NSTYRVVSVLTVLHQDWLNGKEYKCKVSNK ALPAPIEKTISKAKGQPREPQVYTLPPSRE EMTKNQVSLTCLVKGFYPSDIAVEWESNGQ PENNYKTTPPVLDSDGSFFLYSKLTVDKSR WQQGNVFSCSVMHEALHNHYTQKSLSLSPG KGSGDYKDDDDKGSGHHHHHH 325 H1E9-HC2 HC- MQVQLVESGGGVVQPGRSLRLSCAASGFTF FlagHis STFGMSWVRQAPGKGLEWVATISGGGSDTY YPDSVKGRFTISRDNSKNTLYLQMNSLRAE DTAVYYCARQGYDVYSWFAYWGQGTLVTVS SASTKGPSVFPLAPSSKSTSGGTAALGCLV KDYFPEPVTVSWNSGALTSGVHTFPAVLQS SGLYSLSSVVTVPSSSLGTQTYICNVNHKP SNTKVDKKVEPKSCDKTHTCPPCPAPELLG GPSVFLFPPKPKDTLMISRTPEVTCVVVDV SHEDPEVKFNWYVDGVEVHNAKTKPREEQY NSTYRVVSVLTVLHQDWLNGKEYKCKVSNK ALPAPIEKTISKAKGQPREPQVYTLPPSRE EMTKNQVSLTCLVKGFYPSDIAVEWESNGQ PENNYKTTPPVLDSDGSFFLYSKLTVDKSR WQQGNVFSCSVMHEALHNHYTQKSLSLSPG KGSGDYKDDDDKGSGHHHHHH 326 H1E9-HC1 HC- MEVQLLESGGGLVQPGGSLRLSCAASGFTF FlagHis STFGMSWVRQAPGKGLEWVATISGGGSDTY YPDSVKGRFTISRDNSKNTLYLQMNSLRAE DTAVYYCARQGYDVYSWFAYWGQGTLVTVS SASTKGPSVFPLAPSSKSTSGGTAALGCLV KDYFPEPVTVSWNSGALTSGVHTFPAVLQS SGLYSLSSVVTVPSSSLGTQTYICNVNHKP SNTKVDKKVEPKSCDKTHTCPPCPAPELLG GPSVFLFPPKPKDTLMISRTPEVTCVVVDV SHEDPEVKFNWYVDGVEVHNAKTKPREEQY NSTYRVVSVLTVLHQDWLNGKEYKCKVSNK ALPAPIEKTISKAKGQPREPQVYTLPPSRE EMTKNQVSLTCLVKGFYPSDIAVEWESNGQ PENNYKTTPPVLDSDGSFFLYSKLTVDKSR WQQGNVFSCSVMHEALHNHYTQKSLSLSPG KGSGDYKDDDDKGSGHHHHHH 327 H1E9-LC4 LC MDIVLTQSPDSLAVSLGERATINCRASESV DNSGISFMSWYQQKPGQPPKLLIYTASNQG SGVPDRFSGSGSGTDFTLTISSLQAEDVAV YYCQQSKEVPWTFGQGTKVEIKRTVAAPSV FIFPPSDEQLKSGTASVVCLLNNFYPREAK VQWKVDNALQSGNSQESVTEQDSKDSTYSL SSTLTLSKADYEKHKVYACEVTHQGLSSPV TKSFNRGEC 328 H1E9-LC3 LC MEIVLTQSPGTLSLSPGERATLSCRASESV DNSGISFMSWYQQKPGQAPRLLIYTASNQG SGIPDRFSGSGSGTDFTLTISRLEPEDFAV YYCQQSKEVPWTFGQGTKVEIKRTVAAPSV FIFPPSDEQLKSGTASVVCLLNNFYPREAK
VQWKVDNALQSGNSQESVTEQDSKDSTYSL SSTLTLSKADYEKHKVYACEVTHQGLSSPV TKSFNRGEC 329 H1E9-LC2 LC MDIVLTQSPLSLPVTPGEPASISCRASESV DNSGISFMSWYLQKPGQSPQLLIYTASNQG SGVPDRFSGSGSGTDFTLKISRVEAEDVGV YYCQQSKEVPWTFGQGTKVEIKRTVAAPSV FIFPPSDEQLKSGTASVVCLLNNFYPREAK VQWKVDNALQSGNSQESVTEQDSKDSTYSL SSTLTLSKADYEKHKVYACEVTHQGLSSPV TKSFNRGEC 330 H1E9-LC1 LC MDIQLTQSPSSLSASVGDRVTITCRASESV DNSGISFMSWYQQKPGKAPKLLIYTASNQG SGVPSRFSGSGSGTDFTFTISSLQPEDIAT YYCQQSKEVPWTFGQGTKVEIKRTVAAPSV FIFPPSDEQLKSGTASVVCLLNNFYPREAK VQWKVDNALQSGNSQESVTEQDSKDSTYSL SSTLTLSKADYEKHKVYACEVTHQGLSSPV TKSFNRGEC 331 CDR-H2 Kabat TISGGGSDTYYPDSVQG
EQUIVALENTS
[0455] The disclosure set forth above may encompass multiple distinct inventions with independent utility. Although each of these inventions has been disclosed in its preferred form(s), the specific embodiments thereof as disclosed and illustrated herein are not to be considered in a limiting sense, because numerous variations are possible. The subject matter of the inventions includes all novel and nonobvious combinations and subcombinations of the various elements, features, functions, and/or properties disclosed herein. The following claims particularly point out certain combinations and subcombinations regarded as novel and nonobvious. Inventions embodied in other combinations and subcombinations of features, functions, elements, and/or properties may be claimed in this application, in applications claiming priority from this application, or in related applications. Such claims, whether directed to a different invention or to the same invention, and whether broader, narrower, equal, or different in scope in comparison to the original claims, also are regarded as included within the subject matter of the inventions of the present disclosure.
Sequence CWU 1
1
3311288PRTHomo sapiensmisc_feature(1)..(288)Human PD-1 1Met Gln Ile
Pro Gln Ala Pro Trp Pro Val Val Trp Ala Val Leu Gln1 5 10 15Leu Gly
Trp Arg Pro Gly Trp Phe Leu Asp Ser Pro Asp Arg Pro Trp 20 25 30Asn
Pro Pro Thr Phe Ser Pro Ala Leu Leu Val Val Thr Glu Gly Asp 35 40
45Asn Ala Thr Phe Thr Cys Ser Phe Ser Asn Thr Ser Glu Ser Phe Val
50 55 60Leu Asn Trp Tyr Arg Met Ser Pro Ser Asn Gln Thr Asp Lys Leu
Ala65 70 75 80Ala Phe Pro Glu Asp Arg Ser Gln Pro Gly Gln Asp Cys
Arg Phe Arg 85 90 95Val Thr Gln Leu Pro Asn Gly Arg Asp Phe His Met
Ser Val Val Arg 100 105 110Ala Arg Arg Asn Asp Ser Gly Thr Tyr Leu
Cys Gly Ala Ile Ser Leu 115 120 125Ala Pro Lys Ala Gln Ile Lys Glu
Ser Leu Arg Ala Glu Leu Arg Val 130 135 140Thr Glu Arg Arg Ala Glu
Val Pro Thr Ala His Pro Ser Pro Ser Pro145 150 155 160Arg Pro Ala
Gly Gln Phe Gln Thr Leu Val Val Gly Val Val Gly Gly 165 170 175Leu
Leu Gly Ser Leu Val Leu Leu Val Trp Val Leu Ala Val Ile Cys 180 185
190Ser Arg Ala Ala Arg Gly Thr Ile Gly Ala Arg Arg Thr Gly Gln Pro
195 200 205Leu Lys Glu Asp Pro Ser Ala Val Pro Val Phe Ser Val Asp
Tyr Gly 210 215 220Glu Leu Asp Phe Gln Trp Arg Glu Lys Thr Pro Glu
Pro Pro Val Pro225 230 235 240Cys Val Pro Glu Gln Thr Glu Tyr Ala
Thr Ile Val Phe Pro Ser Gly 245 250 255Met Gly Thr Ser Ser Pro Ala
Arg Arg Gly Ser Ala Asp Gly Pro Arg 260 265 270Ser Ala Gln Pro Leu
Arg Pro Glu Asp Gly His Cys Ser Trp Pro Leu 275 280
285212PRTArtificial SequenceSynthetic PD1-17, CDR-H1 2Ser Gly Gly
Ser Ile Arg Ser Thr Arg Trp Trp Ser1 5 1035PRTArtificial
SequenceSynthetic PD1-28, CDR-H1 3Ser Tyr Gly Ile Ser1
545PRTArtificial SequenceSynthetic PD1-33, CDR-H1 4Ser Tyr Tyr Ile
His1 557PRTArtificial SequenceSynthetic PD1-35, CDR-H1 5Ser Gly Ala
Tyr Tyr Trp Ser1 566PRTArtificial SequenceSynthetic PD1-F2, CDR-H1
6Ser Ser Tyr Trp Met Ser1 577PRTArtificial SequenceSynthetic 10B4,
CDR-H1 7Gly Tyr Ile Phe Ser Ser Tyr1 587PRTArtificial
SequenceSynthetic 1353-A09, CDR-H1 8Gly Tyr Arg Phe Thr Trp Tyr1
597PRTArtificial SequenceSynthetic 1353-C07, CDR-H1 9Gly Tyr Arg
Phe Ser Thr Phe1 5107PRTArtificial SequenceSynthetic 1353-E07,
CDR-H1 10Gly Tyr Arg Phe Glu Thr Tyr1 5117PRTArtificial
SequenceSynthetic 1353-F09, CDR-H1 11Gly Tyr Arg Phe Arg Gln Tyr1
5127PRTArtificial SequenceSynthetic 1353-G08, CDR-H1 12Gly Tyr Arg
Phe Thr Arg Tyr1 5137PRTArtificial SequenceSynthetic 1353-G10,
CDR-H1 13Gly Tyr Arg Phe Pro His Tyr1 5147PRTArtificial
SequenceSynthetic 1353-H08, CDR-H1 14Gly Tyr Arg Phe Thr Arg Gln1
5157PRTArtificial SequenceSynthetic 1353-H09, CDR-H1 15Gly Tyr Arg
Phe Pro His Tyr1 5168PRTArtificial SequenceSynthetic 1B10, CDR-H1
16Gly His Ser Ile Thr Ser Asp Tyr1 5177PRTArtificial
SequenceSynthetic 1E9, CDR-H1 17Gly Phe Thr Phe Ser Thr Phe1
5187PRTArtificial SequenceSynthetic 4B10, CDR-H1 18Gly Phe Thr Phe
Ser Thr Tyr1 5197PRTArtificial SequenceSynthetic h1E9-1, CDR-H1
19Gly Phe Thr Phe Ser Thr Phe1 5207PRTArtificial SequenceSynthetic
h1E9-2, CDR-H1 20Gly Phe Thr Phe Ser Thr Phe1 5217PRTArtificial
SequenceSynthetic h1E9-4, CDR-H1 21Gly Phe Thr Phe Ser Thr Phe1
5227PRTArtificial SequenceSynthetic h1E9-5, CDR-H1 22Gly Phe Thr
Phe Ser Thr Phe1 5237PRTArtificial SequenceSynthetic h4B10-1,
CDR-H1 23Gly Phe Thr Phe Ser Thr Tyr1 5247PRTArtificial
SequenceSynthetic h4B10-2, CDR-H1 24Gly Phe Thr Phe Ser Thr Tyr1
5257PRTArtificial SequenceSynthetic h4B10-3, CDR-H1 25Gly Phe Thr
Phe Ser Thr Tyr1 52614PRTArtificial SequenceSynthetic PD1-17,
CDR-H1 26Gly Gly Ser Ile Gly Ser Gly Gly Ser Ile Arg Ser Thr Arg1 5
10277PRTArtificial SequenceSynthetic PD1-28, CDR-H1 27Gly Tyr Arg
Phe Thr Ser Tyr1 5287PRTArtificial SequenceSynthetic PD1-33, CDR-H1
28Gly Tyr Thr Leu Thr Ser Tyr1 5299PRTArtificial SequenceSynthetic
PD1-35, CDR-H1 29Gly Gly Ser Ile Ser Ser Gly Ala Tyr1
53010PRTArtificial SequenceSynthetic PD1-F2, CDR-H1 30Gly Phe Thr
Phe Ser Ser Tyr Trp Cys Asp1 5 10315PRTArtificial SequenceSynthetic
10B4, CDR-H1 31Ser Tyr Trp Ile Gly1 5325PRTArtificial
SequenceSynthetic 1353-A09, CDR-H1 32Trp Tyr Gly Ile Ser1
5335PRTArtificial SequenceSynthetic 1353-C07, CDR-H1 33Thr Phe Gly
Ile Ser1 5345PRTArtificial SequenceSynthetic 1353-E07, CDR-H1 34Thr
Tyr Gly Ile Ser1 5355PRTArtificial SequenceSynthetic 1353-F09,
CDR-H1 35Gln Tyr Gly Ile Ser1 5365PRTArtificial SequenceSynthetic
1353-G08, CDR-H1 36Arg Tyr Gly Ile Ser1 5375PRTArtificial
SequenceSynthetic 1353-G10, CDR-H1 37His Tyr Gly Ile Ser1
5385PRTArtificial SequenceSynthetic 1353-H08, CDR-H1 38Arg Gln Gly
Ile Ser1 5395PRTArtificial SequenceSynthetic 1353-H09, CDR-H1 39His
Tyr Gly Ile Ser1 5406PRTArtificial SequenceSynthetic 1B10, CDR-H1
40Ser Asp Tyr Ala Trp Asn1 5415PRTArtificial SequenceSynthetic 1E9,
CDR-H1 41Thr Phe Gly Met Ser1 5425PRTArtificial SequenceSynthetic
4B10, CDR-H1 42Thr Tyr Gly Met Ser1 5435PRTArtificial
SequenceSynthetic h1E9-1, CDR-H1 43Thr Phe Gly Met Ser1
5445PRTArtificial SequenceSynthetic h1E9-2, CDR-H1 44Thr Phe Gly
Met Ser1 5455PRTArtificial SequenceSynthetic h1E9-4, CDR-H1 45Thr
Phe Gly Met Ser1 5465PRTArtificial SequenceSynthetic h1E9-5, CDR-H1
46Thr Phe Gly Met Ser1 5475PRTArtificial SequenceSynthetic h4B10-1,
CDR-H1 47Thr Tyr Gly Met Ser1 5485PRTArtificial SequenceSynthetic
h4B10-2, CDR-H1 48Thr Tyr Gly Met Ser1 5495PRTArtificial
SequenceSynthetic h4B10-3, CDR-H1 49Thr Tyr Gly Met Ser1
55012PRTArtificial SequenceSynthetic PD1-17, CDR-H1 50Ser Gly Gly
Ser Ile Arg Ser Thr Arg Trp Trp Ser1 5 10515PRTArtificial
SequenceSynthetic PD1-28, CDR-H1 51Ser Tyr Gly Ile Ser1
5525PRTArtificial SequenceSynthetic PD1-33, CDR-H1 52Ser Tyr Tyr
Ile His1 5537PRTArtificial SequenceSynthetic PD1-35, CDR-H1 53Ser
Gly Ala Tyr Tyr Trp Ser1 5548PRTArtificial SequenceSynthetic
PD1-F2, CDR-H1 54Ser Tyr Trp Cys Asp Arg Met Ser1
55516PRTArtificial SequenceSynthetic PD1-17, CDR-H2 55Glu Ile Tyr
His Ser Gly Ser Thr Asn Tyr Asn Pro Ser Leu Lys Ser1 5 10
155617PRTArtificial SequenceSynthetic PD1-28, CDR-H2 56Trp Ile Ser
Ala Tyr Asn Gly Asn Thr Asn Tyr Ala Gln Lys Leu Gln1 5 10
15Gly5717PRTArtificial SequenceSynthetic PD1-33, CDR-H2 57Ile Ile
Asn Pro Arg Gly Ala Thr Ile Ser Tyr Ala Gln Lys Phe Gln1 5 10
15Gly5816PRTArtificial SequenceSynthetic PD1-35, CDR-H2 58Tyr Ile
Tyr Tyr Asn Gly Asn Thr Tyr Tyr Asn Pro Ser Leu Arg Ser1 5 10
155917PRTArtificial SequenceSynthetic PD1-F2, CDR-H2 59Ala Ile Ser
Gly Ser Gly Gly Ser Thr Tyr Tyr Ala Asp Ser Val Lys1 5 10
15Gly606PRTArtificial SequenceSynthetic 10B4, CDR-H2 60Phe Pro Gly
Ser Gly Ser1 5616PRTArtificial SequenceSynthetic 1353-A09, CDR-H2
61Ser Ala Tyr Asn Gly Asn1 5626PRTArtificial SequenceSynthetic
1353-C07, CDR-H2 62Ser Ala Tyr Asn Gly Asn1 5636PRTArtificial
SequenceSynthetic 1353-E07, CDR-H2 63Ser Ala Tyr Asn Gly Asn1
5646PRTArtificial SequenceSynthetic 1353-F09, CDR-H2 64Ser Ala Tyr
Asn Gly Asn1 5656PRTArtificial SequenceSynthetic 1353-G08, CDR-H2
65Ser Ala His Asn Gly Asn1 5666PRTArtificial SequenceSynthetic
1353-G10, CDR-H2 66Ser Ala Tyr Asn Gly Asn1 5676PRTArtificial
SequenceSynthetic 1353-H08, CDR-H2 67Ser Ala Tyr Asn Gly Asn1
5686PRTArtificial SequenceSynthetic 1353-H09, CDR-H2 68Ser Ala Tyr
Asn Gly Asn1 5695PRTArtificial SequenceSynthetic 1B10, CDR-H2 69Ser
Tyr Ser Gly Arg1 5706PRTArtificial SequenceSynthetic 1E9, CDR-H2
70Ser Gly Gly Gly Ser Asp1 5716PRTArtificial SequenceSynthetic
4B10, CDR-H2 71Ser Gly Gly Gly Ser Asn1 5726PRTArtificial
SequenceSynthetic h1E9-1, CDR-H2 72Ser Gly Gly Gly Ser Asp1
5736PRTArtificial SequenceSynthetic h1E9-2, CDR-H2 73Ser Gly Gly
Gly Ser Asp1 5746PRTArtificial SequenceSynthetic h1E9-4, CDR-H2
74Ser Gly Gly Gly Ser Asp1 5756PRTArtificial SequenceSynthetic
h1E9-5, CDR-H2 75Ser Gly Gly Gly Ser Asp1 5766PRTArtificial
SequenceSynthetic h4B10-1, CDR-H2 76Ser Gly Gly Gly Ser Asn1
5776PRTArtificial SequenceSynthetic h4B10-2, CDR-H2 77Ser Gly Gly
Gly Ser Asn1 5786PRTArtificial SequenceSynthetic h4B10-3, CDR-H2
78Ser Gly Gly Gly Ser Asn1 5795PRTArtificial SequenceSynthetic
PD1-17, CDR-H2 79Tyr His Ser Gly Ser1 5806PRTArtificial
SequenceSynthetic PD1-28, CDR-H2 80Ser Ala Tyr Asn Gly Asn1
5816PRTArtificial SequenceSynthetic PD1-33, CDR-H2 81Asn Pro Arg
Gly Ala Thr1 5825PRTArtificial SequenceSynthetic PD1-35, CDR-H2
82Tyr Tyr Asn Gly Asn1 5836PRTArtificial SequenceSynthetic PD1-F2,
CDR-H2 83Ser Gly Ser Gly Gly Ser1 58417PRTArtificial
SequenceSynthetic 10B4, CDR-H2 84Lys Ile Phe Pro Gly Ser Gly Ser
Ala Asp Tyr Asn Glu Asn Phe Lys1 5 10 15Gly8517PRTArtificial
SequenceSynthetic 1353-A09, CDR-H2 85Trp Ile Ser Ala Tyr Asn Gly
Asn Thr Asn Tyr Ala Gln Lys Leu Gln1 5 10 15Gly8617PRTArtificial
SequenceSynthetic 1353-C07, CDR-H2 86Trp Ile Ser Ala Tyr Asn Gly
Asn Thr Asn Tyr Ala Gln Lys Leu Gln1 5 10 15Gly8717PRTArtificial
SequenceSynthetic 1353-E07, CDR-H2 87Trp Ile Ser Ala Tyr Asn Gly
Asn Thr Asn Tyr Ala Gln Lys Leu Gln1 5 10 15Gly8817PRTArtificial
SequenceSynthetic 1353-F09, CDR-H2 88Trp Ile Ser Ala Tyr Asn Gly
Asn Thr Asn Tyr Ala Gln Lys Leu Gln1 5 10 15Gly8917PRTArtificial
SequenceSynthetic 1353-G08, CDR-H2 89Trp Val Ser Ala His Asn Gly
Asn Thr Asn Tyr Ala Gln Lys Leu Gln1 5 10 15Gly9017PRTArtificial
SequenceSynthetic 1353-G10, CDR-H2 90Trp Ile Ser Ala Tyr Asn Gly
Asn Thr Asn Tyr Ala Gln Lys Leu Gln1 5 10 15Gly9117PRTArtificial
SequenceSynthetic 1353-H08, CDR-H2 91Trp Ile Ser Ala Tyr Asn Gly
Asn Thr Lys Tyr Ala Gln Lys Leu Gln1 5 10 15Gly9217PRTArtificial
SequenceSynthetic 1353-H09, CDR-H2 92Trp Ile Ser Ala Tyr Asn Gly
Asn Thr Asn Tyr Ala Gln Lys Leu Gln1 5 10 15Gly9316PRTArtificial
SequenceSynthetic 1B10, CDR-H2 93Tyr Ile Ser Tyr Ser Gly Arg Thr
Ser Tyr Asn Pro Ser Leu Thr Ser1 5 10 159417PRTArtificial
SequenceSynthetic 1E9, CDR-H2 94Thr Ile Ser Gly Gly Gly Ser Asp Thr
Tyr Tyr Pro Asp Ser Val Gln1 5 10 15Gly9517PRTArtificial
SequenceSynthetic 4B10, CDR-H2 95Thr Ile Ser Gly Gly Gly Ser Asn
Thr Tyr Tyr Ser Asp Ser Val Lys1 5 10 15Gly9617PRTArtificial
SequenceSynthetic h1E9-1, CDR-H2 96Thr Ile Ser Gly Gly Gly Ser Asp
Thr Tyr Tyr Pro Asp Ser Val Gln1 5 10 15Gly9717PRTArtificial
SequenceSynthetic h1E9-2, CDR-H2 97Thr Ile Ser Gly Gly Gly Ser Asp
Thr Tyr Tyr Pro Asp Ser Val Gln1 5 10 15Gly9817PRTArtificial
SequenceSynthetic h1E9-4, CDR-H2 98Thr Ile Ser Gly Gly Gly Ser Asp
Thr Tyr Tyr Pro Asp Ser Val Gln1 5 10 15Gly9917PRTArtificial
SequenceSynthetic h1E9-5, CDR-H2 99Thr Ile Ser Gly Gly Gly Ser Asp
Thr Tyr Tyr Pro Asp Ser Val Gln1 5 10 15Gly10017PRTArtificial
SequenceSynthetic h4B10-1, CDR-H2 100Thr Ile Ser Gly Gly Gly Ser
Asn Thr Tyr Tyr Ser Asp Ser Val Lys1 5 10 15Gly10117PRTArtificial
SequenceSynthetic h4B10-2, CDR-H2 101Thr Ile Ser Gly Gly Gly Ser
Asn Thr Tyr Tyr Ser Asp Ser Val Lys1 5 10 15Gly10217PRTArtificial
SequenceSynthetic h4B10-3, CDR-H2 102Thr Ile Ser Gly Gly Gly Ser
Asn Thr Tyr Tyr Ser Asp Ser Val Lys1 5 10 15Gly10316PRTArtificial
SequenceSynthetic PD1-17, CDR-H2 103Glu Ile Tyr His Ser Gly Ser Thr
Asn Tyr Asn Pro Ser Leu Lys Ser1 5 10 1510417PRTArtificial
SequenceSynthetic PD1-28, CDR-H2 104Trp Ile Ser Ala Tyr Asn Gly Asn
Thr Asn Tyr Ala Gln Lys Leu Gln1 5 10 15Gly10517PRTArtificial
SequenceSynthetic PD1-33, CDR-H2 105Ile Ile Asn Pro Arg Gly Ala Thr
Ile Ser Tyr Ala Gln Lys Phe Gln1 5 10 15Gly10616PRTArtificial
SequenceSynthetic PD1-35, CDR-H2 106Tyr Ile Tyr Tyr Asn Gly Asn Thr
Tyr Tyr Asn Pro Ser Leu Arg Ser1 5 10 1510717PRTArtificial
SequenceSynthetic PD1-F2, CDR-H2 107Ala Ile Ser Gly Ser Gly Gly Ser
Thr Tyr Tyr Ala Asp Ser Val Lys1 5 10 15Gly10813PRTArtificial
SequenceSynthetic PD1-17, CDR-H3 108Gln Asp Tyr Gly Asp Ser Gly Asp
Trp Tyr Phe Asp Leu1 5 1010910PRTArtificial SequenceSynthetic
PD1-28, CDR-H3 109Asp Ala Asp Tyr Ser Ser Gly Ser Gly Tyr1 5
1011010PRTArtificial SequenceSynthetic PD1-33, CDR-H3 110Ala Gly
Ile Tyr Gly Phe Asp Phe Asp Tyr1 5 1011117PRTArtificial
SequenceSynthetic PD1-35, CDR-H3 111Ala Ser Asp Tyr Val Trp Gly Gly
Tyr Arg Tyr Met Asp Ala Phe Asp1 5 10 15Ile1129PRTArtificial
SequenceSynthetic PD1-F2, CDR-H3 112Glu Asn Trp Gly Ser Tyr Phe Asp
Leu1 511310PRTArtificial SequenceSynthetic 10B4, CDR-H3 113Gly Tyr
Gly Asn Tyr Leu Tyr Phe Asp Val1 5 1011410PRTArtificial
SequenceSynthetic 1353-A09, CDR-H3 114Asp Ser Glu Tyr Ser Ser Gly
Ser Gly Tyr1 5 1011510PRTArtificial SequenceSynthetic 1353-C07,
CDR-H3 115Asp Val Asp Tyr Ser Ser Gly Ser Gly Tyr1 5
1011610PRTArtificial SequenceSynthetic 1353-E07, CDR-H3 116Asp Ala
Glu Tyr Ser Leu Gly Ser Gly Tyr1 5 1011710PRTArtificial
SequenceSynthetic 1353-F09, CDR-H3 117Asp Ala Glu Tyr Gly Ser Gly
Ser Gly Tyr1 5 1011810PRTArtificial SequenceSynthetic 1353-G08,
CDR-H3 118Asp Ala Asp Tyr Gly Ser Gly Ser Gly Tyr1 5
1011910PRTArtificial SequenceSynthetic 1353-G10, CDR-H3 119Asp Val
Asp Tyr Gly Thr Gly Ser Gly Tyr1 5 1012010PRTArtificial
SequenceSynthetic 1353-H08, CDR-H3 120Asp Val Asp Tyr Gly Ser Gly
Ser Gly Tyr1 5 1012110PRTArtificial SequenceSynthetic 1353-H09,
CDR-H3 121Asp Ala Glu Tyr Gly Ser Gly Ser Gly Tyr1 5
101226PRTArtificial SequenceSynthetic 1B10, CDR-H3 122Gly Tyr Ala
Leu Asp Tyr1 512311PRTArtificial SequenceSynthetic 1E9, CDR-H3
123Gln Gly Tyr Asp Val Tyr Ser Trp Phe Ala Tyr1 5
101249PRTArtificial SequenceSynthetic 4B10, CDR-H3 124Gln Arg Asp
Ser Ala Trp Phe Ala Ser1 512511PRTArtificial SequenceSynthetic
h1E9-1, CDR-H3 125Gln Gly Tyr Asp Val Tyr Ser Trp Phe Ala Tyr1 5
1012611PRTArtificial SequenceSynthetic h1E9-2, CDR-H3 126Gln Gly
Tyr Asp Val Tyr Ser Trp Phe Ala Tyr1 5 1012711PRTArtificial
SequenceSynthetic h1E9-4, CDR-H3 127Gln Gly Tyr Asp Val Tyr Ser Trp
Phe Ala Tyr1 5 1012811PRTArtificial SequenceSynthetic h1E9-5,
CDR-H3 128Gln Gly Tyr Asp Val Tyr Ser Trp Phe Ala Tyr1 5
101299PRTArtificial SequenceSynthetic h4B10-1, CDR-H3 129Gln Arg
Asp Ser Ala Trp Phe Ala Ser1 51309PRTArtificial SequenceSynthetic
h4B10-2, CDR-H3 130Gln Arg Asp Ser Ala Trp Phe Ala Ser1
51319PRTArtificial SequenceSynthetic h4B10-3, CDR-H3 131Gln Arg Asp
Ser Ala Trp Phe Ala Ser1 513213PRTArtificial SequenceSynthetic
PD1-17, CDR-H3 132Gln Asp Tyr Gly Asp Ser Gly Asp Trp Tyr Phe Asp
Leu1 5 1013310PRTArtificial SequenceSynthetic PD1-28, CDR-H3 133Asp
Ala Asp Tyr Ser Ser Gly Ser Gly Tyr1 5 1013410PRTArtificial
SequenceSynthetic PD1-33, CDR-H3 134Ala Gly Ile Tyr Gly Phe Asp Phe
Asp Tyr1 5 1013517PRTArtificial SequenceSynthetic PD1-35, CDR-H3
135Ala Ser Asp Tyr Val Trp Gly Gly Tyr Arg Tyr Met Asp Ala Phe Asp1
5 10 15Ile1369PRTArtificial SequenceSynthetic PD1-F2, CDR-H3 136Glu
Asn Trp Gly Ser Tyr Phe Asp Leu1 513713PRTArtificial
SequenceSynthetic PD1-17, CDR-L1 137Thr Arg Ser Ser Gly Ser Ile Ala
Ser Asn Ser Val Gln1 5 1013811PRTArtificial SequenceSynthetic
PD1-28, CDR-L1 138Ser Gly Asp Ala Leu Pro Lys Gln Tyr Ala Tyr1 5
1013914PRTArtificial SequenceSynthetic PD1-33, CDR-L1 139Thr Gly
Thr Ser Asn Asp Val Gly Gly Tyr Asn Tyr Val Ser1 5
1014013PRTArtificial SequenceSynthetic PD1-35, CDR-L1 140Ser Gly
Ser Asn Ser Asn Ile Gly Ser Asn Ser Val Asn1 5 1014111PRTArtificial
SequenceSynthetic PD1-F2, CDR-L1 141Arg Ala Ser Gln Gly Ile Ser Ser
Trp Leu Ala1 5 1014211PRTArtificial SequenceSynthetic 10B4, CDR-L1
142Lys Ala Ser Gln Ser Val Ser Asp Asp Val Ala1 5
1014311PRTArtificial SequenceSynthetic 1353-A09, CDR-L1 143Ser Gly
Asp Ala Leu Thr Thr Gln Tyr Ala Tyr1 5 1014411PRTArtificial
SequenceSynthetic 1353-C07, CDR-L1 144Ser Gly Asp Ala Leu Ser Glu
Gln Tyr Ala Tyr1 5 1014511PRTArtificial SequenceSynthetic 1353-E07,
CDR-L1 145Ser Gly Asp Ala Leu Pro Lys Gln Tyr Ala Tyr1 5
1014611PRTArtificial SequenceSynthetic 1353-F09, CDR-L1 146Ser Gly
Asp Ala Leu Pro Lys Gln Tyr Ala Tyr1 5 1014711PRTArtificial
SequenceSynthetic 1353-G08, CDR-L1 147Ser Gly Asp Ala Leu Pro Met
Gln Tyr Gly Tyr1 5 1014811PRTArtificial SequenceSynthetic 1353-G10,
CDR-L1 148Ser Gly Asp Ala Leu Pro Lys Gln Tyr Ala Tyr1 5
1014911PRTArtificial SequenceSynthetic 1353-H08, CDR-L1 149Ser Gly
Asp Ala Leu Pro Lys Gln Tyr Ala Tyr1 5 1015011PRTArtificial
SequenceSynthetic 1353-H09, CDR-L1 150Ser Gly Asp Ala Leu Pro Lys
Gln Tyr Ala Tyr1 5 1015110PRTArtificial SequenceSynthetic 1B10,
CDR-L1 151Arg Thr Ser Ser Ser Val Asn Tyr Met His1 5
1015215PRTArtificial SequenceSynthetic 1E9, CDR-L1 152Arg Ala Ser
Glu Ser Val Asp Asn Ser Gly Ile Ser Phe Met Ser1 5 10
1515315PRTArtificial SequenceSynthetic 4B10, CDR-L1 153Arg Ala Ser
Glu Asn Val Asp Asp Tyr Gly Val Ser Phe Met Asn1 5 10
1515415PRTArtificial SequenceSynthetic h1E9-1, CDR-L1 154Arg Ala
Ser Glu Ser Val Asp Asn Ser Gly Ile Ser Phe Met Ser1 5 10
1515515PRTArtificial SequenceSynthetic h1E9-2, CDR-L1 155Arg Ala
Ser Glu Ser Val Asp Asn Ser Gly Ile Ser Phe Met Ser1 5 10
1515615PRTArtificial SequenceSynthetic h1E9-4, CDR-L1 156Arg Ala
Ser Glu Ser Val Asp Asn Ser Gly Ile Ser Phe Met Ser1 5 10
1515715PRTArtificial SequenceSynthetic h1E9-5, CDR-L1 157Arg Ala
Ser Glu Ser Val Asp Asn Ser Gly Ile Ser Phe Met Ser1 5 10
1515815PRTArtificial SequenceSynthetic h4B10-1, CDR-L1 158Arg Ala
Ser Glu Asn Val Asp Asp Tyr Gly Val Ser Phe Met Asn1 5 10
1515915PRTArtificial SequenceSynthetic h4B10-2, CDR-L1 159Arg Ala
Ser Glu Asn Val Asp Asp Tyr Gly Val Ser Phe Met Asn1 5 10
1516015PRTArtificial SequenceSynthetic h4B10-3, CDR-L1 160Arg Ala
Ser Glu Asn Val Asp Asp Tyr Gly Val Ser Phe Met Asn1 5 10
1516113PRTArtificial SequenceSynthetic PD1-17, CDR-L1 161Thr Arg
Ser Ser Gly Ser Ile Ala Ser Asn Ser Val Gln1 5 1016211PRTArtificial
SequenceSynthetic PD1-28, CDR-L1 162Ser Gly Asp Ala Leu Pro Lys Gln
Tyr Ala Tyr1 5 1016314PRTArtificial SequenceSynthetic PD1-33,
CDR-L1 163Thr Gly Thr Ser Asn Asp Val Gly Gly Tyr Asn Tyr Val Ser1
5 1016413PRTArtificial SequenceSynthetic PD1-35, CDR-L1 164Ser Gly
Ser Asn Ser Asn Ile Gly Ser Asn Ser Val Asn1 5 1016511PRTArtificial
SequenceSynthetic PD1-F2, CDR-L1 165Arg Ala Ser Gln Gly Ile Ser Ser
Trp Leu Ala1 5 101667PRTArtificial SequenceSynthetic PD1-17, CDR-L2
166Glu Asp Asn Gln Arg Pro Ser1 51677PRTArtificial
SequenceSynthetic PD1-28, CDR-L2 167Lys Asp Thr Glu Arg Pro Ser1
51687PRTArtificial SequenceSynthetic PD1-33, CDR-L2 168Asp Val Thr
Asn Arg Pro Ser1 51697PRTArtificial SequenceSynthetic PD1-35,
CDR-L2 169Gly Asn Asn Gln Arg Pro Ser1 51707PRTArtificial
SequenceSynthetic PD1-F2, CDR-L2 170Lys Ala Ser Thr Leu Glu Ser1
51717PRTArtificial SequenceSynthetic 10B4, CDR-L2 171Tyr Ala Phe
Lys Arg Tyr Ile1 51727PRTArtificial SequenceSynthetic 1353-A09,
CDR-L2 172Lys Asp Thr Glu Arg Pro Ser1 51737PRTArtificial
SequenceSynthetic 1353-C07, CDR-L2 173Lys Asp Thr Glu Arg Pro Ser1
51747PRTArtificial SequenceSynthetic 1353-E07, CDR-L2 174Lys Asp
Thr Glu Arg Pro Ser1 51757PRTArtificial SequenceSynthetic 1353-F09,
CDR-L2 175Lys Asp Thr Glu Arg Pro Ser1 51767PRTArtificial
SequenceSynthetic 1353-G08, CDR-L2 176Lys Asp Thr Glu Arg Pro Ser1
51777PRTArtificial SequenceSynthetic 1353-G10, CDR-L2 177Lys Asp
Thr Glu Arg Pro Ser1 51787PRTArtificial SequenceSynthetic 1353-H08,
CDR-L2 178Lys Asp Thr Glu Arg Pro Ser1 51797PRTArtificial
SequenceSynthetic 1353-H09, CDR-L2 179Lys Asp Thr Glu Arg Pro Ser1
51807PRTArtificial SequenceSynthetic 1B10, CDR-L2 180Ala Thr Ser
Lys Leu Ala Ser1 51817PRTArtificial SequenceSynthetic 1E9, CDR-L2
181Thr Ala Ser Asn Gln Gly Ser1 51827PRTArtificial
SequenceSynthetic 4B10, CDR-L2 182Pro Ala Ser Asn Gln Gly Ser1
51837PRTArtificial SequenceSynthetic h1E9-1, CDR-L2 183Thr Ala Ser
Asn Gln Gly Ser1 51847PRTArtificial SequenceSynthetic h1E9-2,
CDR-L2 184Thr Ala Ser Asn Gln Gly Ser1 51857PRTArtificial
SequenceSynthetic h1E9-4, CDR-L2 185Thr Ala Ser Asn Gln Gly Ser1
51867PRTArtificial SequenceSynthetic h1E9-5, CDR-L2 186Thr Ala Ser
Asn Gln Gly Ser1 51877PRTArtificial SequenceSynthetic h4B10-1,
CDR-L2 187Pro Ala Ser Asn Gln Gly Ser1 51887PRTArtificial
SequenceSynthetic h4B10-2, CDR-L2 188Pro Ala Ser Asn Gln Gly Ser1
51897PRTArtificial SequenceSynthetic h4B10-3, CDR-L2 189Pro Ala Ser
Asn Gln Gly Ser1 51907PRTArtificial SequenceSynthetic PD1-17,
CDR-L2 190Glu Asp Asn Gln Arg Pro Ser1 51917PRTArtificial
SequenceSynthetic PD1-28, CDR-L2 191Lys Asp Thr Glu Arg Pro Ser1
51927PRTArtificial SequenceSynthetic PD1-33, CDR-L2 192Asp Val Thr
Asn Arg Pro Ser1 51937PRTArtificial SequenceSynthetic PD1-35,
CDR-L2 193Gly Asn Asn Gln Arg Pro Ser1 51947PRTArtificial
SequenceSynthetic PD1-F2, CDR-L2 194Lys Ala Ser Thr Leu Glu Ser1
51959PRTArtificial SequenceSynthetic PD1-17, CDR-L3 195Gln Ser Ser
Asp Ser Ser Ala Val Val1 519611PRTArtificial SequenceSynthetic
PD1-28, CDR-L3 196Gln Ser Ala Asp Asn Ser Ile Thr Tyr Arg Val1 5
1019712PRTArtificial SequenceSynthetic PD1-33, CDR-L3 197Ser Ser
Tyr Thr Ile Val Thr Asn Phe Glu Val Leu1 5 1019811PRTArtificial
SequenceSynthetic PD1-35, CDR-L3 198Ala Ala Trp Asp Asp Ser Leu Asn
Gly Pro Val1 5 101999PRTArtificial SequenceSynthetic PD1-F2, CDR-L3
199Gln Gln Ser Tyr Ser Thr Pro Trp Thr1 52009PRTArtificial
SequenceSynthetic 10B4, CDR-L3 200Gln Gln Asn Tyr Asn Ser Pro Tyr
Thr1 520111PRTArtificial SequenceSynthetic 1353-A09, CDR-L3 201Gln
Ser Ala Asp Asn Ser Ile Thr Tyr Arg Val1 5 1020211PRTArtificial
SequenceSynthetic 1353-C07, CDR-L3 202Gln Ser Ala Asp Asn Ser Ile
Thr Tyr Arg Val1 5 1020311PRTArtificial SequenceSynthetic 1353-E07,
CDR-L3 203Gln Ser Ala Asp Asn Ser Ile Thr Tyr Arg Val1 5
1020411PRTArtificial SequenceSynthetic 1353-F09, CDR-L3 204Gln Ser
Ala Asp Asn Ser Ile Thr Tyr Arg Val1 5 1020511PRTArtificial
SequenceSynthetic 1353-G08, CDR-L3 205Gln Ser Ala Asp Asn Ser Ile
Thr Tyr Arg Val1 5 1020611PRTArtificial SequenceSynthetic 1353-G10,
CDR-L3 206Gln Ser Ala Asp Asn Ser Ile Thr Tyr Arg Val1 5
1020711PRTArtificial SequenceSynthetic 1353-H08, CDR-L3 207Gln Ser
Ala Asp Asn Ser Ile Thr Tyr Arg Val1 5 1020811PRTArtificial
SequenceSynthetic 1353-H09, CDR-L3 208Gln Ser Ala Asp Asn Ser Ile
Thr Tyr Arg Val1 5 102099PRTArtificial SequenceSynthetic 1B10,
CDR-L3 209Gln Gln Trp Ile Ser Asp Pro Trp Thr1 52109PRTArtificial
SequenceSynthetic 1E9, CDR-L3 210Gln Gln Ser Lys Glu Val Pro Trp
Thr1 52119PRTArtificial SequenceSynthetic 4B10, CDR-L3 211Gln Gln
Ser Lys Glu Val Pro Trp Thr1 52129PRTArtificial SequenceSynthetic
h1E9-1, CDR-L3 212Gln Gln Ser Lys Glu Val Pro Trp Thr1
52139PRTArtificial SequenceSynthetic h1E9-2, CDR-L3 213Gln Gln Ser
Lys Glu Val Pro Trp Thr1 52149PRTArtificial SequenceSynthetic
h1E9-4, CDR-L3 214Gln Gln Ser Lys Glu Val Pro Trp Thr1
52159PRTArtificial SequenceSynthetic h1E9-5, CDR-L3 215Gln Gln Ser
Lys Glu Val Pro Trp Thr1 52169PRTArtificial SequenceSynthetic
h4B10-1, CDR-L3 216Gln Gln Ser Lys Glu Val Pro Trp Thr1
52179PRTArtificial SequenceSynthetic h4B10-2, CDR-L3 217Gln Gln Ser
Lys Glu Val Pro Trp Thr1 52189PRTArtificial SequenceSynthetic
h4B10-3, CDR-L3 218Gln Gln Ser Lys Glu Val Pro Trp Thr1
52199PRTArtificial SequenceSynthetic PD1-17, CDR-L3 219Gln Ser Ser
Asp Ser Ser Ala Val Val1 522011PRTArtificial SequenceSynthetic
PD1-28, CDR-L3 220Gln Ser Ala Asp Asn Ser Ile Thr Tyr Arg Val1 5
1022112PRTArtificial SequenceSynthetic PD1-33, CDR-L3 221Ser Ser
Tyr Thr Ile Val Thr Asn Phe Glu Val Leu1 5 1022211PRTArtificial
SequenceSynthetic PD1-35, CDR-L3 222Ala Ala Trp Asp Asp Ser Leu Asn
Gly Pro Val1 5 102239PRTArtificial SequenceSynthetic PD1-F2, CDR-L3
223Gln Gln Ser Tyr Ser Thr Pro Trp Thr1 5224330PRTArtificial
SequenceSynthetic HC Constant 224Ala Ser Thr Lys Gly Pro Ser Val
Phe Pro Leu Ala Pro Ser Ser Lys1 5 10 15Ser Thr Ser Gly Gly Thr Ala
Ala Leu Gly Cys Leu Val Lys Asp Tyr 20 25 30Phe Pro Glu Pro Val Thr
Val Ser Trp Asn Ser Gly Ala Leu Thr Ser 35 40 45Gly Val His Thr Phe
Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser 50 55 60Leu Ser Ser Val
Val Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr65 70 75 80Tyr Ile
Cys Asn Val Asn His Lys Pro Ser Asn Thr Lys Val Asp Lys 85 90 95Lys
Val Glu Pro Lys Ser Cys Asp Lys Thr His Thr Cys Pro Pro Cys 100 105
110Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro
115 120 125Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val
Thr Cys 130 135 140Val Val Val Asp Val Ser His Glu Asp Pro Glu Val
Lys Phe Asn Trp145 150 155 160Tyr Val Asp Gly Val Glu Val His Asn
Ala Lys Thr Lys Pro Arg Glu 165 170 175Glu Gln Tyr Asn Ser Thr Tyr
Arg Val Val Ser Val Leu Thr Val Leu 180 185 190His Gln Asp Trp Leu
Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn 195 200 205Lys Ala Leu
Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly 210 215 220Gln
Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu225 230
235 240Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly Phe
Tyr 245 250 255Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln
Pro Glu Asn 260 265 270Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser
Asp Gly Ser Phe Phe 275 280 285Leu Tyr Ser Lys Leu Thr Val Asp Lys
Ser Arg Trp Gln Gln Gly Asn 290 295 300Val Phe Ser Cys Ser Val Met
His Glu Ala Leu His Asn His Tyr Thr305 310 315 320Gln Lys Ser Leu
Ser Leu Ser Pro Gly Lys 325 330225108PRTArtificial
SequenceSynthetic Kappa LC 225His Met Thr Val Ala Ala Pro Ser Val
Phe Ile Phe Pro Pro Ser Asp1 5 10 15Glu Gln Leu Lys Ser Gly Thr Ala
Ser Val Val Cys Leu Leu Asn Asn 20 25 30Phe Tyr Pro Arg Glu Ala Lys
Val Gln Trp Lys Val Asp Asn Ala Leu 35 40 45Gln Ser Gly Asn Ser Gln
Glu Ser Val Thr Glu Gln Asp Ser Lys Asp 50 55 60Ser Thr Tyr Ser Leu
Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr65 70 75 80Glu Lys His
Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser 85 90 95Ser Pro
Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 100 105226106PRTArtificial
SequenceSynthetic Lambda LD 226Gly Gln Pro Lys Ala Ala Pro Ser Val
Thr Leu Phe Pro Pro Ser Ser1 5 10 15Glu Glu Leu Gln Ala Asn Lys Ala
Thr Leu Val Cys Leu Ile Ser Asp 20 25 30Phe Tyr Pro Gly Ala Val Thr
Val Ala Trp Lys Ala Asp Ser Ser Pro 35 40 45Val Lys Ala Gly Val Glu
Thr Thr Thr Pro Ser Lys Gln Ser Asn Asn 50 55 60Lys Tyr Ala Ala Ser
Ser Tyr Leu Ser Leu Thr Pro Glu Gln Trp Lys65 70 75 80Ser His Arg
Ser Tyr Ser Cys Gln Val Thr His Glu Gly Ser Thr Val 85 90 95Glu Lys
Thr Val Ala Pro Thr Glu Cys Ser 100 105227267PRTArtificial
SequenceSynthetic h1E9-1, scFv 227Met Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Lys Pro Gly1 5 10 15Gly Ser Leu Arg Leu Ser Cys
Ala Ala Ser Gly Phe Thr Phe Ser Thr 20 25 30Phe Gly Met Ser Trp Val
Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp 35 40 45Val Ser Thr Ile Ser
Gly Gly Gly Ser Asp Thr Tyr Tyr Pro Asp Ser 50 55 60Val Gln Gly Arg
Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn Ser Leu65 70 75 80Tyr Leu
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr 85 90 95Cys
Ala Arg Gln Gly Tyr Asp Val Tyr Ser Trp Phe Ala Tyr Trp Gly 100 105
110Gln Gly Thr Leu Val Thr Val Ser Ser Gly Gly Gly Gly Ser Gly Gly
115 120 125Gly Gly Ser Gly Gly Gly Gly Ser Asp Ile Gln Leu Thr Gln
Ser Pro 130 135 140Ser Phe Leu Ser Ala Ser Val Gly Asp Arg Val Thr
Ile Thr Cys Arg145 150 155 160Ala Ser Glu Ser Val Asp Asn Ser Gly
Ile Ser Phe Met Ser Trp Tyr 165 170 175Gln Gln Lys Pro Gly Lys Ala
Pro Lys Leu Leu Ile Tyr Thr Ala Ser 180 185 190Asn Gln Gly Ser Gly
Val Pro Ser Arg Phe Ser Gly Ser Gly Ser Gly 195 200 205Thr Glu Phe
Thr Leu Thr Ile Ser Ser Leu Gln Pro Glu Asp Phe Ala 210 215 220Thr
Tyr Tyr Cys Gln Gln Ser Lys Glu Val Pro Trp Thr Phe Gly Gln225 230
235 240Gly Thr Lys Val Glu Ile Lys Gly Ser Gly Asp Tyr Lys Asp Asp
Asp 245 250 255Asp Lys Gly Ser Gly His His His His His His 260
265228267PRTArtificial SequenceSynthetic h1E9-2, scFv 228Met Glu
Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Lys Pro Gly1 5 10 15Gly
Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr 20 25
30Phe Gly Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu
Trp
35 40 45Val Ser Thr Ile Ser Gly Gly Gly Ser Asp Thr Tyr Tyr Pro Asp
Ser 50 55 60Val Gln Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn
Ser Leu65 70 75 80Tyr Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr
Ala Val Tyr Tyr 85 90 95Cys Ala Arg Gln Gly Tyr Asp Val Tyr Ser Trp
Phe Ala Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val Ser Ser
Gly Gly Gly Gly Ser Gly Gly 115 120 125Gly Gly Ser Gly Gly Gly Gly
Ser Glu Ile Val Leu Thr Gln Ser Pro 130 135 140Ala Thr Leu Ser Leu
Ser Pro Gly Glu Arg Ala Thr Leu Ser Cys Arg145 150 155 160Ala Ser
Glu Ser Val Asp Asn Ser Gly Ile Ser Phe Met Ser Trp Tyr 165 170
175Gln Gln Lys Pro Gly Gln Ala Pro Arg Leu Leu Ile Tyr Thr Ala Ser
180 185 190Asn Gln Gly Ser Gly Ile Pro Ala Arg Phe Ser Gly Ser Gly
Ser Gly 195 200 205Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Glu Pro
Glu Asp Phe Ala 210 215 220Val Tyr Tyr Cys Gln Gln Ser Lys Glu Val
Pro Trp Thr Phe Gly Gln225 230 235 240Gly Thr Lys Val Glu Ile Lys
Gly Ser Gly Asp Tyr Lys Asp Asp Asp 245 250 255Asp Lys Gly Ser Gly
His His His His His His 260 265229267PRTArtificial
SequenceSynthetic h1E9-4, scFv 229Met Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly1 5 10 15Gly Ser Leu Arg Leu Ser Cys
Ala Ala Ser Gly Phe Thr Phe Ser Thr 20 25 30Phe Gly Met Ser Trp Val
Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp 35 40 45Val Ala Thr Ile Ser
Gly Gly Gly Ser Asp Thr Tyr Tyr Pro Asp Ser 50 55 60Val Gln Gly Arg
Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn Ser Leu65 70 75 80Tyr Leu
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr 85 90 95Cys
Ala Arg Gln Gly Tyr Asp Val Tyr Ser Trp Phe Ala Tyr Trp Gly 100 105
110Gln Gly Thr Leu Val Thr Val Ser Ser Gly Gly Gly Gly Ser Gly Gly
115 120 125Gly Gly Ser Gly Gly Gly Gly Ser Asp Ile Gln Leu Thr Gln
Ser Pro 130 135 140Ser Phe Leu Ser Ala Ser Val Gly Asp Arg Val Thr
Ile Thr Cys Arg145 150 155 160Ala Ser Glu Ser Val Asp Asn Ser Gly
Ile Ser Phe Met Ser Trp Tyr 165 170 175Gln Gln Lys Pro Gly Lys Ala
Pro Lys Leu Leu Ile Tyr Thr Ala Ser 180 185 190Asn Gln Gly Ser Gly
Val Pro Ser Arg Phe Ser Gly Ser Gly Ser Gly 195 200 205Thr Glu Phe
Thr Leu Thr Ile Ser Ser Leu Gln Pro Glu Asp Phe Ala 210 215 220Thr
Tyr Tyr Cys Gln Gln Ser Lys Glu Val Pro Trp Thr Phe Gly Gln225 230
235 240Gly Thr Lys Val Glu Ile Lys Gly Ser Gly Asp Tyr Lys Asp Asp
Asp 245 250 255Asp Lys Gly Ser Gly His His His His His His 260
265230267PRTArtificial SequenceSynthetic h1E9-5, scFv 230Met Glu
Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly1 5 10 15Gly
Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr 20 25
30Phe Gly Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
35 40 45Val Ala Thr Ile Ser Gly Gly Gly Ser Asp Thr Tyr Tyr Pro Asp
Ser 50 55 60Val Gln Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn
Ser Leu65 70 75 80Tyr Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr
Ala Val Tyr Tyr 85 90 95Cys Ala Arg Gln Gly Tyr Asp Val Tyr Ser Trp
Phe Ala Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val Ser Ser
Gly Gly Gly Gly Ser Gly Gly 115 120 125Gly Gly Ser Gly Gly Gly Gly
Ser Glu Ile Val Leu Thr Gln Ser Pro 130 135 140Ala Thr Leu Ser Leu
Ser Pro Gly Glu Arg Ala Thr Leu Ser Cys Arg145 150 155 160Ala Ser
Glu Ser Val Asp Asn Ser Gly Ile Ser Phe Met Ser Trp Tyr 165 170
175Gln Gln Lys Pro Gly Gln Ala Pro Arg Leu Leu Ile Tyr Thr Ala Ser
180 185 190Asn Gln Gly Ser Gly Ile Pro Ala Arg Phe Ser Gly Ser Gly
Ser Gly 195 200 205Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Glu Pro
Glu Asp Phe Ala 210 215 220Val Tyr Tyr Cys Gln Gln Ser Lys Glu Val
Pro Trp Thr Phe Gly Gln225 230 235 240Gly Thr Lys Val Glu Ile Lys
Gly Ser Gly Asp Tyr Lys Asp Asp Asp 245 250 255Asp Lys Gly Ser Gly
His His His His His His 260 265231265PRTArtificial
SequenceSynthetic h4B10-1, scFv 231Met Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Lys Pro Gly1 5 10 15Gly Ser Leu Arg Leu Ser Cys
Ala Ala Ser Gly Phe Thr Phe Ser Thr 20 25 30Tyr Gly Met Ser Trp Val
Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp 35 40 45Val Ala Thr Ile Ser
Gly Gly Gly Ser Asn Thr Tyr Tyr Ser Asp Ser 50 55 60Val Lys Gly Arg
Phe Thr Ile Ser Arg Asp Asp Ser Lys Asn Thr Leu65 70 75 80Tyr Leu
Gln Met Asn Ser Leu Lys Thr Glu Asp Thr Ala Val Tyr Tyr 85 90 95Cys
Ala Arg Gln Arg Asp Ser Ala Trp Phe Ala Ser Trp Gly Gln Gly 100 105
110Thr Leu Val Thr Val Ser Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly
115 120 125Ser Gly Gly Gly Gly Ser Glu Ile Val Leu Thr Gln Ser Pro
Ala Thr 130 135 140Leu Ser Leu Ser Pro Gly Glu Arg Ala Thr Leu Ser
Cys Arg Ala Ser145 150 155 160Glu Asn Val Asp Asp Tyr Gly Val Ser
Phe Met Asn Trp Tyr Gln Gln 165 170 175Lys Pro Gly Gln Ala Pro Arg
Leu Leu Ile Tyr Pro Ala Ser Asn Gln 180 185 190Gly Ser Gly Ile Pro
Ala Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp 195 200 205Phe Thr Leu
Thr Ile Ser Ser Leu Glu Pro Glu Asp Phe Ala Val Tyr 210 215 220Tyr
Cys Gln Gln Ser Lys Glu Val Pro Trp Thr Phe Gly Gln Gly Thr225 230
235 240Lys Val Glu Ile Lys Gly Ser Gly Asp Tyr Lys Asp Asp Asp Asp
Lys 245 250 255Gly Ser Gly His His His His His His 260
265232265PRTArtificial SequenceSynthetic h4B10-2, scFv 232Met Glu
Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Lys Pro Gly1 5 10 15Gly
Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr 20 25
30Tyr Gly Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
35 40 45Val Ala Thr Ile Ser Gly Gly Gly Ser Asn Thr Tyr Tyr Ser Asp
Ser 50 55 60Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp Ser Lys Asn
Thr Leu65 70 75 80Tyr Leu Gln Met Asn Ser Leu Lys Thr Glu Asp Thr
Ala Val Tyr Tyr 85 90 95Cys Ala Arg Gln Arg Asp Ser Ala Trp Phe Ala
Ser Trp Gly Gln Gly 100 105 110Thr Leu Val Thr Val Ser Ser Gly Gly
Gly Gly Ser Gly Gly Gly Gly 115 120 125Ser Gly Gly Gly Gly Ser Glu
Ile Val Leu Thr Gln Ser Pro Gly Thr 130 135 140Leu Ser Leu Ser Pro
Gly Glu Arg Ala Thr Leu Ser Cys Arg Ala Ser145 150 155 160Glu Asn
Val Asp Asp Tyr Gly Val Ser Phe Met Asn Trp Tyr Gln Gln 165 170
175Lys Pro Gly Gln Ala Pro Arg Leu Leu Ile Tyr Pro Ala Ser Asn Gln
180 185 190Gly Ser Gly Ile Pro Asp Arg Phe Ser Gly Ser Gly Ser Gly
Thr Asp 195 200 205Phe Thr Leu Thr Ile Ser Arg Leu Glu Pro Glu Asp
Phe Ala Val Tyr 210 215 220Tyr Cys Gln Gln Ser Lys Glu Val Pro Trp
Thr Phe Gly Gln Gly Thr225 230 235 240Lys Val Glu Ile Lys Gly Ser
Gly Asp Tyr Lys Asp Asp Asp Asp Lys 245 250 255Gly Ser Gly His His
His His His His 260 265233265PRTArtificial SequenceSynthetic
h4B10-3, scFv 233Met Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu
Val Lys Pro Gly1 5 10 15Gly Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Ser Thr 20 25 30Tyr Gly Met Ser Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Trp 35 40 45Val Ala Thr Ile Ser Gly Gly Gly Ser
Asn Thr Tyr Tyr Ser Asp Ser 50 55 60Val Lys Gly Arg Phe Thr Ile Ser
Arg Asp Asp Ser Lys Asn Thr Leu65 70 75 80Tyr Leu Gln Met Asn Ser
Leu Lys Thr Glu Asp Thr Ala Val Tyr Tyr 85 90 95Cys Ala Arg Gln Arg
Asp Ser Ala Trp Phe Ala Ser Trp Gly Gln Gly 100 105 110Thr Leu Val
Thr Val Ser Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly 115 120 125Ser
Gly Gly Gly Gly Ser Asp Ile Val Met Thr Gln Ser Pro Asp Ser 130 135
140Leu Ala Val Ser Leu Gly Glu Arg Ala Thr Ile Asn Cys Arg Ala
Ser145 150 155 160Glu Asn Val Asp Asp Tyr Gly Val Ser Phe Met Asn
Trp Tyr Gln Gln 165 170 175Lys Pro Gly Gln Pro Pro Lys Leu Leu Ile
Tyr Pro Ala Ser Asn Gln 180 185 190Gly Ser Gly Val Pro Asp Arg Phe
Ser Gly Ser Gly Ser Gly Thr Asp 195 200 205Phe Thr Leu Thr Ile Ser
Ser Leu Gln Ala Glu Asp Val Ala Val Tyr 210 215 220Tyr Cys Gln Gln
Ser Lys Glu Val Pro Trp Thr Phe Gly Gly Gly Thr225 230 235 240Lys
Leu Glu Ile Lys Gly Ser Gly Asp Tyr Lys Asp Asp Asp Asp Lys 245 250
255Gly Ser Gly His His His His His His 260 265234262PRTArtificial
SequenceSynthetic m10B4, scFv 234Met Gln Val Gln Leu Gln Gln Ser
Gly Ala Glu Leu Met Lys Pro Gly1 5 10 15Ala Ser Val Lys Met Ser Cys
Lys Thr Thr Gly Tyr Ile Phe Ser Ser 20 25 30Tyr Trp Ile Gly Trp Val
Lys Gln Arg Pro Gly His Gly Leu Glu Trp 35 40 45Ile Gly Lys Ile Phe
Pro Gly Ser Gly Ser Ala Asp Tyr Asn Glu Asn 50 55 60Phe Lys Gly Lys
Ala Thr Phe Thr Val Asp Thr Ser Ser Asn Thr Ala65 70 75 80Tyr Met
Gln Leu Ser Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Tyr 85 90 95Cys
Ala Arg Gly Tyr Gly Asn Tyr Leu Tyr Phe Asp Val Trp Gly Ala 100 105
110Gly Thr Thr Val Thr Val Ser Ser Gly Gly Gly Gly Ser Gly Gly Gly
115 120 125Gly Ser Gly Gly Gly Gly Ser Asn Ile Val Met Thr Gln Thr
Pro Lys 130 135 140Phe Leu Leu Val Ser Ala Gly Asp Arg Ile Thr Ile
Thr Cys Lys Ala145 150 155 160Ser Gln Ser Val Ser Asp Asp Val Ala
Trp Tyr Gln Gln Lys Pro Gly 165 170 175Gln Ser Pro Lys Leu Leu Ile
Ser Tyr Ala Phe Lys Arg Tyr Ile Gly 180 185 190Val Pro Asp Arg Phe
Thr Gly Ser Gly Tyr Gly Thr Asp Phe Thr Phe 195 200 205Thr Ile Ser
Thr Val Gln Ala Glu Asp Leu Ala Val Tyr Phe Cys Gln 210 215 220Gln
Asn Tyr Asn Ser Pro Tyr Thr Phe Gly Gly Gly Thr Lys Leu Glu225 230
235 240Leu Lys Gly Ser Gly Asp Tyr Lys Asp Asp Asp Asp Lys Gly Ser
Gly 245 250 255His His His His His His 260235258PRTArtificial
SequenceSynthetic m1B10, scFv 235Met Ser Asp Val Gln Leu Gln Glu
Ser Gly Pro Gly Leu Val Lys Pro1 5 10 15Ser Gln Ser Leu Ser Leu Thr
Cys Thr Val Thr Gly His Ser Ile Thr 20 25 30Ser Asp Tyr Ala Trp Asn
Trp Ile Arg Gln Phe Pro Gly Asn Lys Leu 35 40 45Glu Trp Met Gly Tyr
Ile Ser Tyr Ser Gly Arg Thr Ser Tyr Asn Pro 50 55 60Ser Leu Thr Ser
Arg Ile Ser Ile Thr Arg Asp Thr Ser Lys Asn Gln65 70 75 80Phe Phe
Leu Gln Leu Asn Ser Val Thr Thr Glu Asp Thr Ala Thr Tyr 85 90 95Tyr
Cys Ala Arg Gly Tyr Ala Leu Asp Tyr Trp Gly Gln Gly Thr Ser 100 105
110Val Thr Val Ser Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly
115 120 125Gly Gly Gly Ser Gln Ile Val Leu Ser Gln Ser Pro Ala Ile
Leu Ser 130 135 140Ala Ser Pro Gly Glu Lys Val Thr Met Thr Cys Arg
Thr Ser Ser Ser145 150 155 160Val Asn Tyr Met His Trp Phe Gln Gln
Lys Pro Gly Ser Ser Pro Lys 165 170 175Pro Trp Ile Tyr Ala Thr Ser
Lys Leu Ala Ser Gly Val Pro Ala Arg 180 185 190Phe Ser Gly Ser Gly
Ser Gly Thr Ser Tyr Ser Leu Thr Ile Ser Arg 195 200 205Val Glu Ala
Glu Asp Ala Ala Thr Tyr Phe Cys Gln Gln Trp Ile Ser 210 215 220Asp
Pro Trp Thr Phe Gly Gly Gly Thr Lys Leu Glu Ile Lys Gly Ser225 230
235 240Gly Asp Tyr Lys Asp Asp Asp Asp Lys Gly Ser Gly His His His
His 245 250 255His His236267PRTArtificial SequenceSynthetic m1E9,
scFv 236Met Glu Val Lys Leu Val Glu Ser Gly Gly Gly Leu Val Ser Pro
Gly1 5 10 15Gly Ser Leu Lys Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe
Ser Thr 20 25 30Phe Gly Met Ser Trp Val Arg Gln Thr Pro Glu Lys Arg
Leu Glu Trp 35 40 45Val Ala Thr Ile Ser Gly Gly Gly Ser Asp Thr Tyr
Tyr Pro Asp Ser 50 55 60Val Gln Gly Arg Phe Ile Ile Ser Arg Tyr Asn
Ala Lys Asn Asn Leu65 70 75 80Tyr Leu Gln Met Asn Ser Leu Arg Pro
Glu Asp Thr Ala Leu Tyr Tyr 85 90 95Cys Ala Arg Gln Gly Tyr Asp Val
Tyr Ser Trp Phe Ala Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr
Val Ser Ala Gly Gly Gly Gly Ser Gly Gly 115 120 125Gly Gly Ser Gly
Gly Gly Gly Ser Asp Ile Ile Leu Thr Gln Ser Pro 130 135 140Ala Ser
Leu Ala Val Ser Leu Gly Gln Arg Ala Ala Ile Ser Cys Arg145 150 155
160Ala Ser Glu Ser Val Asp Asn Ser Gly Ile Ser Phe Met Ser Trp Phe
165 170 175Gln Gln Lys Pro Gly Gln Pro Pro Lys Leu Leu Ile Tyr Thr
Ala Ser 180 185 190Asn Gln Gly Ser Gly Val Pro Ala Arg Phe Ser Gly
Ser Gly Ser Gly 195 200 205Thr Glu Phe Ser Leu Asn Ile His Pro Met
Glu Glu Asp Asp Thr Ala 210 215 220Met Tyr Phe Cys Gln Gln Ser Lys
Glu Val Pro Trp Thr Phe Gly Gly225 230 235 240Gly Thr Lys Leu Glu
Ile Arg Gly Ser Gly Asp Tyr Lys Asp Asp Asp 245 250 255Asp Lys Gly
Ser Gly His His His His His His 260 265237265PRTArtificial
SequenceSynthetic m4B10, scFv 237Met Glu Val Lys Leu Val Glu Ser
Gly Gly Gly Leu Val Lys Pro Gly1 5 10 15Gly Ser Leu Lys Leu Ser Cys
Ala Ala Ser Gly Phe Thr Phe Ser Thr 20 25 30Tyr Gly Met Ser Trp Val
Arg Gln Thr Pro Glu Lys Arg Leu Gln Trp 35 40 45Val Ala Thr Ile Ser
Gly Gly Gly Ser Asn Thr Tyr Tyr Ser Asp Ser 50 55
60Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn Asn Leu65
70 75 80Tyr Leu Gln Met Ser Ser Leu Arg Ser Glu Asp Thr Ala Leu Tyr
Tyr 85 90 95Cys Ala Arg Gln Arg Asp Ser Ala Trp Phe Ala Ser Trp Gly
Gln Gly 100 105 110Thr Leu Val Thr Val Ser Ala Gly Gly Gly Gly Ser
Gly Gly Gly Gly 115 120 125Ser Gly Gly Gly Gly Ser Asp Ile Val Leu
Thr Gln Ser Pro Ala Ser 130 135 140Leu Ala Val Ser Leu Gly Gln Arg
Ala Thr Ile Ser Cys Arg Ala Ser145 150 155 160Glu Asn Val Asp Asp
Tyr Gly Val Ser Phe Met Asn Trp Phe Gln Gln 165 170 175Lys Pro Gly
Gln Pro Pro Lys Leu Leu Ile Tyr Pro Ala Ser Asn Gln 180 185 190Gly
Ser Gly Val Pro Ala Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp 195 200
205Phe Ser Leu Asn Ile His Pro Met Glu Glu Asp Asp Thr Ala Met Tyr
210 215 220Phe Cys Gln Gln Ser Lys Glu Val Pro Trp Thr Phe Gly Gly
Gly Thr225 230 235 240Lys Leu Glu Ile Lys Gly Ser Gly Asp Tyr Lys
Asp Asp Asp Asp Lys 245 250 255Gly Ser Gly His His His His His His
260 265238512PRTArtificial SequenceSynthetic 1353-A09, scFv-Fc
238Met Glu Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly1
5 10 15Ala Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Arg Phe Thr
Trp 20 25 30Tyr Gly Ile Ser Trp Val Arg Gln Ala Pro Gly Gln Gly Leu
Glu Trp 35 40 45Met Gly Trp Ile Ser Ala Tyr Asn Gly Asn Thr Asn Tyr
Ala Gln Lys 50 55 60Leu Gln Gly Arg Val Thr Met Thr Thr Asp Thr Ser
Thr Asn Thr Ala65 70 75 80Tyr Met Glu Leu Arg Ser Leu Arg Ser Asp
Asp Thr Ala Val Tyr Tyr 85 90 95Cys Ala Arg Asp Ser Glu Tyr Ser Ser
Gly Ser Gly Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser
Ser Gly Gly Gly Gly Ser Gly Gly Gly 115 120 125Gly Ser Gly Gly Gly
Gly Ser Ser Tyr Glu Leu Thr Gln Pro Pro Ser 130 135 140Val Ser Val
Ser Pro Gly Gln Thr Ala Arg Ile Thr Cys Ser Gly Asp145 150 155
160Ala Leu Thr Thr Gln Tyr Ala Tyr Trp Tyr Gln Gln Lys Pro Gly Gln
165 170 175Ala Pro Val Met Val Ile Tyr Lys Asp Thr Glu Arg Pro Ser
Gly Ile 180 185 190Pro Glu Arg Phe Ser Gly Ser Ser Ser Gly Thr Lys
Val Thr Leu Thr 195 200 205Ile Ser Gly Val Gln Ala Glu Asp Glu Ala
Asp Tyr Tyr Cys Gln Ser 210 215 220Ala Asp Asn Ser Ile Thr Tyr Arg
Val Phe Gly Gly Gly Thr Lys Val225 230 235 240Thr Val Leu Ala Ala
Gly Ser Asp Gln Glu Pro Lys Lys Leu Ala Ala 245 250 255Gly Ser Asp
Gln Glu Pro Lys Ser Ser Asp Lys Thr His Thr Cys Pro 260 265 270Pro
Cys Ser Ala Pro Glu Leu Leu Gly Gly Ser Ser Val Phe Leu Phe 275 280
285Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val
290 295 300Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu Val
Lys Phe305 310 315 320Asn Trp Tyr Val Asp Gly Val Glu Val His Asn
Ala Lys Thr Lys Pro 325 330 335Arg Glu Glu Gln Tyr Asn Ser Thr Tyr
Arg Val Val Ser Val Leu Thr 340 345 350Val Leu His Gln Asp Trp Leu
Asn Gly Lys Glu Tyr Lys Cys Lys Val 355 360 365Ser Asn Lys Ala Leu
Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala 370 375 380Lys Gly Gln
Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg385 390 395
400Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly
405 410 415Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly
Gln Pro 420 425 430Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp
Ser Asp Gly Ser 435 440 445Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp
Lys Ser Arg Trp Gln Gln 450 455 460Gly Asn Val Phe Ser Cys Ser Val
Met His Glu Ala Leu His Asn His465 470 475 480Tyr Thr Gln Lys Ser
Leu Ser Leu Ser Pro Gly Lys Gly Ser Gly Asp 485 490 495Tyr Lys Asp
Asp Asp Asp Lys Gly Ser Gly His His His His His His 500 505
510239512PRTArtificial SequenceSynthetic 1353-C07, scFv-Fc 239Met
Glu Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly1 5 10
15Ala Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Arg Phe Ser Thr
20 25 30Phe Gly Ile Ser Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp 35 40 45Met Gly Trp Ile Ser Ala Tyr Asn Gly Asn Thr Asn Tyr Ala
Gln Lys 50 55 60Leu Gln Gly Arg Val Thr Met Thr Thr Asp Thr Ser Thr
Asn Thr Ala65 70 75 80Tyr Met Glu Leu Arg Ser Leu Arg Ser Asp Asp
Thr Ala Val Tyr Tyr 85 90 95Cys Ala Arg Asp Val Asp Tyr Ser Ser Gly
Ser Gly Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser
Gly Gly Gly Gly Ser Gly Gly Gly 115 120 125Gly Ser Gly Gly Gly Gly
Ser Ser Tyr Glu Leu Thr Gln Pro Pro Ser 130 135 140Val Ser Val Ser
Pro Gly Gln Thr Ala Arg Ile Thr Cys Ser Gly Asp145 150 155 160Ala
Leu Ser Glu Gln Tyr Ala Tyr Trp Tyr Gln Gln Lys Pro Gly Gln 165 170
175Ala Pro Val Met Val Ile Tyr Lys Asp Thr Glu Arg Pro Ser Gly Ile
180 185 190Pro Glu Arg Phe Ser Gly Ser Ser Ser Gly Thr Lys Val Thr
Leu Thr 195 200 205Ile Ser Gly Val Gln Ala Glu Asp Glu Ala Asp Tyr
Tyr Cys Gln Ser 210 215 220Ala Asp Asn Ser Ile Thr Tyr Arg Val Phe
Gly Gly Gly Thr Lys Val225 230 235 240Thr Val Leu Ala Ala Gly Ser
Asp Gln Glu Pro Lys Lys Leu Ala Ala 245 250 255Gly Ser Asp Gln Glu
Pro Lys Ser Ser Asp Lys Thr His Thr Cys Pro 260 265 270Pro Cys Ser
Ala Pro Glu Leu Leu Gly Gly Ser Ser Val Phe Leu Phe 275 280 285Pro
Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val 290 295
300Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Lys
Phe305 310 315 320Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala
Lys Thr Lys Pro 325 330 335Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg
Val Val Ser Val Leu Thr 340 345 350Val Leu His Gln Asp Trp Leu Asn
Gly Lys Glu Tyr Lys Cys Lys Val 355 360 365Ser Asn Lys Ala Leu Pro
Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala 370 375 380Lys Gly Gln Pro
Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg385 390 395 400Asp
Glu Leu Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly 405 410
415Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln Pro
420 425 430Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser Asp
Gly Ser 435 440 445Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser
Arg Trp Gln Gln 450 455 460Gly Asn Val Phe Ser Cys Ser Val Met His
Glu Ala Leu His Asn His465 470 475 480Tyr Thr Gln Lys Ser Leu Ser
Leu Ser Pro Gly Lys Gly Ser Gly Asp 485 490 495Tyr Lys Asp Asp Asp
Asp Lys Gly Ser Gly His His His His His His 500 505
510240512PRTArtificial SequenceSynthetic 1353-E07, scFv-Fc 240Met
Glu Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly1 5 10
15Ala Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Arg Phe Glu Thr
20 25 30Tyr Gly Ile Ser Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp 35 40 45Met Gly Trp Ile Ser Ala Tyr Asn Gly Asn Thr Asn Tyr Ala
Gln Lys 50 55 60Leu Gln Gly Arg Val Thr Met Thr Thr Asp Thr Ser Thr
Asn Thr Ala65 70 75 80Tyr Met Glu Leu Arg Ser Leu Arg Ser Asp Asp
Thr Ala Val Tyr Tyr 85 90 95Cys Ala Arg Asp Ala Glu Tyr Ser Leu Gly
Ser Gly Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser
Gly Gly Gly Gly Ser Gly Gly Gly 115 120 125Gly Ser Gly Gly Gly Gly
Ser Ser Tyr Glu Leu Thr Gln Pro Pro Ser 130 135 140Val Ser Val Ser
Pro Gly Gln Thr Ala Arg Ile Thr Cys Ser Gly Asp145 150 155 160Ala
Leu Pro Lys Gln Tyr Ala Tyr Trp Tyr Gln Gln Lys Pro Gly Gln 165 170
175Ala Pro Val Met Val Ile Tyr Lys Asp Thr Glu Arg Pro Ser Gly Ile
180 185 190Pro Glu Arg Phe Ser Gly Ser Ser Ser Gly Thr Lys Val Thr
Leu Thr 195 200 205Ile Ser Gly Val Gln Ala Glu Asp Glu Ala Asp Tyr
Tyr Cys Gln Ser 210 215 220Ala Asp Asn Ser Ile Thr Tyr Arg Val Phe
Gly Gly Gly Thr Lys Val225 230 235 240Thr Val Leu Ala Ala Gly Ser
Asp Gln Glu Pro Lys Lys Leu Ala Ala 245 250 255Gly Ser Asp Gln Glu
Pro Lys Ser Ser Asp Lys Thr His Thr Cys Pro 260 265 270Pro Cys Ser
Ala Pro Glu Leu Leu Gly Gly Ser Ser Val Phe Leu Phe 275 280 285Pro
Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val 290 295
300Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Lys
Phe305 310 315 320Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala
Lys Thr Lys Pro 325 330 335Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg
Val Val Ser Val Leu Thr 340 345 350Val Leu His Gln Asp Trp Leu Asn
Gly Lys Glu Tyr Lys Cys Lys Val 355 360 365Ser Asn Lys Ala Leu Pro
Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala 370 375 380Lys Gly Gln Pro
Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg385 390 395 400Asp
Glu Leu Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly 405 410
415Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln Pro
420 425 430Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser Asp
Gly Ser 435 440 445Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser
Arg Trp Gln Gln 450 455 460Gly Asn Val Phe Ser Cys Ser Val Met His
Glu Ala Leu His Asn His465 470 475 480Tyr Thr Gln Lys Ser Leu Ser
Leu Ser Pro Gly Lys Gly Ser Gly Asp 485 490 495Tyr Lys Asp Asp Asp
Asp Lys Gly Ser Gly His His His His His His 500 505
510241512PRTArtificial SequenceSynthetic 1353-F09, scFv-Fc 241Met
Glu Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly1 5 10
15Ala Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Arg Phe Arg Gln
20 25 30Tyr Gly Ile Ser Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp 35 40 45Met Gly Trp Ile Ser Ala Tyr Asn Gly Asn Thr Asn Tyr Ala
Gln Lys 50 55 60Leu Gln Gly Arg Val Thr Met Thr Thr Asp Thr Ser Thr
Asn Thr Ala65 70 75 80Tyr Met Glu Leu Arg Ser Leu Arg Ser Asp Asp
Thr Ala Val Tyr Tyr 85 90 95Cys Ala Arg Asp Ala Glu Tyr Gly Ser Gly
Ser Gly Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser
Gly Gly Gly Gly Ser Gly Gly Gly 115 120 125Gly Ser Gly Gly Gly Gly
Ser Ser Tyr Glu Leu Thr Gln Pro Pro Ser 130 135 140Val Ser Val Ser
Pro Gly Gln Thr Ala Arg Ile Thr Cys Ser Gly Asp145 150 155 160Ala
Leu Pro Lys Gln Tyr Ala Tyr Trp Tyr Gln Gln Lys Pro Gly Gln 165 170
175Ala Pro Val Met Val Leu Tyr Lys Asp Thr Glu Arg Pro Ser Gly Ile
180 185 190Pro Glu Arg Phe Ser Gly Ser Ser Ser Gly Thr Lys Val Thr
Leu Thr 195 200 205Ile Ser Gly Val Gln Ala Glu Asp Glu Ala Asp Tyr
Tyr Cys Gln Ser 210 215 220Ala Asp Asn Ser Ile Thr Tyr Arg Val Phe
Gly Gly Gly Thr Lys Val225 230 235 240Thr Val Leu Ala Ala Gly Ser
Asp Gln Glu Pro Lys Lys Leu Ala Ala 245 250 255Gly Ser Asp Gln Glu
Pro Lys Ser Ser Asp Lys Thr His Thr Cys Pro 260 265 270Pro Cys Ser
Ala Pro Glu Leu Leu Gly Gly Ser Ser Val Phe Leu Phe 275 280 285Pro
Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val 290 295
300Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Lys
Phe305 310 315 320Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala
Lys Thr Lys Pro 325 330 335Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg
Val Val Ser Val Leu Thr 340 345 350Val Leu His Gln Asp Trp Leu Asn
Gly Lys Glu Tyr Lys Cys Lys Val 355 360 365Ser Asn Lys Ala Leu Pro
Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala 370 375 380Lys Gly Gln Pro
Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg385 390 395 400Asp
Glu Leu Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly 405 410
415Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln Pro
420 425 430Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser Asp
Gly Ser 435 440 445Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser
Arg Trp Gln Gln 450 455 460Gly Asn Val Phe Ser Cys Ser Val Met His
Glu Ala Leu His Asn His465 470 475 480Tyr Thr Gln Lys Ser Leu Ser
Leu Ser Pro Gly Lys Gly Ser Gly Asp 485 490 495Tyr Lys Asp Asp Asp
Asp Lys Gly Ser Gly His His His His His His 500 505
510242512PRTArtificial SequenceSynthetic 1353-G08, scFv-Fc 242Met
Glu Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly1 5 10
15Ala Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Arg Phe Thr Arg
20 25 30Tyr Gly Ile Ser Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp 35 40 45Met Gly Trp Val Ser Ala His Asn Gly Asn Thr Asn Tyr Ala
Gln Lys 50 55 60Leu Gln Gly Arg Val Thr Met Thr Thr Asp Thr Ser Thr
Asn Thr Ala65 70 75 80Tyr Met Glu Leu Arg Ser Leu Arg Ser Asp Asp
Thr Ala Val Tyr Tyr 85 90 95Cys Ala Arg Asp Ala Asp Tyr Gly Ser Gly
Ser Gly Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser
Gly Gly Gly Val Ser Gly Gly Gly 115 120 125Gly Ser Gly Gly Gly Gly
Ser Ser Tyr Glu Leu Thr Gln Pro Pro Ser 130 135 140Val Ser Val Ser
Pro Gly Gln Thr Ala Arg Ile Thr Cys Ser Gly Asp145 150 155 160Ala
Leu Pro Met Gln Tyr Gly Tyr Trp Tyr Gln Gln Lys Pro Gly Gln 165 170
175Ala Pro Val Met Val Ile Tyr Lys Asp Thr Glu Arg
Pro Ser Gly Ile 180 185 190Pro Glu Arg Phe Ser Gly Ser Ser Ser Gly
Thr Lys Val Thr Leu Thr 195 200 205Ile Ser Gly Val Gln Ala Glu Asp
Glu Ala Asp Tyr Tyr Cys Gln Ser 210 215 220Ala Asp Asn Ser Ile Thr
Tyr Arg Val Phe Gly Gly Gly Thr Lys Val225 230 235 240Thr Val Leu
Ala Ala Gly Ser Asp Gln Glu Pro Lys Lys Leu Ala Ala 245 250 255Gly
Ser Asp Gln Glu Pro Lys Ser Ser Asp Lys Thr His Thr Cys Pro 260 265
270Pro Cys Ser Ala Pro Glu Leu Leu Gly Gly Ser Ser Val Phe Leu Phe
275 280 285Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro
Glu Val 290 295 300Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro
Glu Val Lys Phe305 310 315 320Asn Trp Tyr Val Asp Gly Val Glu Val
His Asn Ala Lys Thr Lys Pro 325 330 335Arg Glu Glu Gln Tyr Asn Ser
Thr Tyr Arg Val Val Ser Val Leu Thr 340 345 350Val Leu His Gln Asp
Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val 355 360 365Ser Asn Lys
Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala 370 375 380Lys
Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg385 390
395 400Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys
Gly 405 410 415Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn
Gly Gln Pro 420 425 430Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu
Asp Ser Asp Gly Ser 435 440 445Phe Phe Leu Tyr Ser Lys Leu Thr Val
Asp Lys Ser Arg Trp Gln Gln 450 455 460Gly Asn Val Phe Ser Cys Ser
Val Met His Glu Ala Leu His Asn His465 470 475 480Tyr Thr Gln Lys
Ser Leu Ser Leu Ser Pro Gly Lys Gly Ser Gly Asp 485 490 495Tyr Lys
Asp Asp Asp Asp Lys Gly Ser Gly His His His His His His 500 505
510243512PRTArtificial SequenceSynthetic 1353-G10, scFv-Fc 243Met
Glu Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly1 5 10
15Ala Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Arg Phe Pro His
20 25 30Tyr Gly Ile Ser Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp 35 40 45Met Gly Trp Ile Ser Ala Tyr Asn Gly Asn Thr Asn Tyr Ala
Gln Lys 50 55 60Leu Gln Gly Arg Val Thr Met Thr Thr Asp Thr Ser Thr
Asn Thr Ala65 70 75 80Tyr Met Glu Leu Arg Ser Leu Arg Ser Asp Asp
Thr Ala Val Tyr Tyr 85 90 95Cys Ala Arg Asp Val Asp Tyr Gly Thr Gly
Ser Gly Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser
Gly Gly Gly Gly Ser Gly Gly Gly 115 120 125Gly Ser Gly Gly Gly Gly
Ser Ser Tyr Glu Leu Thr Gln Pro Pro Ser 130 135 140Val Ser Val Ser
Pro Gly Gln Thr Ala Arg Ile Thr Cys Ser Gly Asp145 150 155 160Ala
Leu Pro Lys Gln Tyr Ala Tyr Trp Tyr Gln Gln Lys Pro Gly Gln 165 170
175Ala Pro Val Met Val Ile Tyr Lys Asp Thr Glu Arg Pro Ser Gly Ile
180 185 190Pro Glu Arg Phe Ser Gly Ser Ser Ser Gly Thr Lys Val Thr
Leu Thr 195 200 205Ile Ser Gly Val Gln Ala Glu Asp Glu Ala Asp Tyr
Tyr Cys Gln Ser 210 215 220Ala Asp Asn Ser Ile Thr Tyr Arg Val Phe
Gly Gly Gly Thr Lys Val225 230 235 240Thr Val Leu Ala Ala Gly Ser
Asp Gln Glu Pro Lys Lys Leu Ala Ala 245 250 255Gly Ser Asp Gln Glu
Pro Lys Ser Ser Asp Lys Thr His Thr Cys Pro 260 265 270Pro Cys Ser
Ala Pro Glu Leu Leu Gly Gly Ser Ser Val Phe Leu Phe 275 280 285Pro
Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val 290 295
300Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Lys
Phe305 310 315 320Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala
Lys Thr Lys Pro 325 330 335Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg
Val Val Ser Val Leu Thr 340 345 350Val Leu His Gln Asp Trp Leu Asn
Gly Lys Glu Tyr Lys Cys Lys Val 355 360 365Ser Asn Lys Ala Leu Pro
Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala 370 375 380Lys Gly Gln Pro
Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg385 390 395 400Asp
Glu Leu Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly 405 410
415Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln Pro
420 425 430Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser Asp
Gly Ser 435 440 445Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser
Arg Trp Gln Gln 450 455 460Gly Asn Val Phe Ser Cys Ser Val Met His
Glu Ala Leu His Asn His465 470 475 480Tyr Thr Gln Lys Ser Leu Ser
Leu Ser Pro Gly Lys Gly Ser Gly Asp 485 490 495Tyr Lys Asp Asp Asp
Asp Lys Gly Ser Gly His His His His His His 500 505
510244512PRTArtificial SequenceSynthetic 1353-H08, scFv-Fc 244Met
Glu Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly1 5 10
15Ala Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Arg Phe Thr Arg
20 25 30Gln Gly Ile Ser Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp 35 40 45Met Gly Trp Ile Ser Ala Tyr Asn Gly Asn Thr Lys Tyr Ala
Gln Lys 50 55 60Leu Gln Gly Arg Val Thr Met Thr Thr Asp Thr Ser Thr
Asn Thr Ala65 70 75 80Tyr Met Glu Leu Arg Ser Leu Arg Ser Asp Asp
Thr Ala Val Tyr Tyr 85 90 95Cys Ala Arg Asp Val Asp Tyr Gly Ser Gly
Ser Gly Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser
Gly Gly Gly Gly Ser Gly Gly Gly 115 120 125Gly Ser Gly Gly Gly Gly
Ser Ser Tyr Glu Leu Thr Gln Pro Pro Ser 130 135 140Val Ser Val Ser
Pro Gly Gln Thr Ala Arg Ile Thr Cys Ser Gly Asp145 150 155 160Ala
Leu Pro Lys Gln Tyr Ala Tyr Trp Tyr Gln Gln Lys Pro Gly Gln 165 170
175Ala Pro Val Met Val Ile Tyr Lys Asp Thr Glu Arg Pro Ser Gly Ile
180 185 190Pro Glu Arg Phe Ser Gly Ser Ser Ser Gly Thr Lys Val Thr
Leu Thr 195 200 205Ile Ser Gly Val Gln Ala Glu Asp Glu Ala Asp Tyr
Tyr Cys Gln Ser 210 215 220Ala Asp Asn Ser Ile Thr Tyr Arg Val Phe
Gly Gly Gly Thr Lys Val225 230 235 240Thr Val Leu Ala Ala Gly Ser
Asp Gln Glu Pro Lys Lys Leu Ala Ala 245 250 255Gly Ser Asp Gln Glu
Pro Lys Ser Ser Asp Lys Thr His Thr Cys Pro 260 265 270Pro Cys Ser
Ala Pro Glu Leu Leu Gly Gly Ser Ser Val Phe Leu Phe 275 280 285Pro
Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val 290 295
300Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Lys
Phe305 310 315 320Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala
Lys Thr Lys Pro 325 330 335Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg
Val Val Ser Val Leu Thr 340 345 350Val Leu His Gln Asp Trp Leu Asn
Gly Lys Glu Tyr Lys Cys Lys Val 355 360 365Ser Asn Lys Ala Leu Pro
Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala 370 375 380Lys Gly Gln Pro
Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg385 390 395 400Asp
Glu Leu Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly 405 410
415Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln Pro
420 425 430Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser Asp
Gly Ser 435 440 445Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser
Arg Trp Gln Gln 450 455 460Gly Asn Val Phe Ser Cys Ser Val Met His
Glu Ala Leu His Asn His465 470 475 480Tyr Thr Gln Lys Ser Leu Ser
Leu Ser Pro Gly Lys Gly Ser Gly Asp 485 490 495Tyr Lys Asp Asp Asp
Asp Lys Gly Ser Gly His His His His His His 500 505
510245512PRTArtificial SequenceSynthetic 1353-H09, scFv-Fc 245Met
Glu Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly1 5 10
15Ala Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Arg Phe Pro His
20 25 30Tyr Gly Ile Ser Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp 35 40 45Met Gly Trp Ile Ser Ala Tyr Asn Gly Asn Thr Asn Tyr Ala
Gln Lys 50 55 60Leu Gln Gly Arg Val Thr Met Thr Thr Asp Thr Ser Thr
Asn Thr Ala65 70 75 80Tyr Met Glu Leu Arg Ser Leu Arg Ser Asp Asp
Thr Ala Val Tyr Tyr 85 90 95Cys Ala Arg Asp Ala Glu Tyr Gly Ser Gly
Ser Gly Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser
Gly Gly Gly Gly Ser Gly Gly Gly 115 120 125Gly Ser Gly Gly Gly Gly
Ser Ser Tyr Glu Leu Thr Gln Pro Pro Ser 130 135 140Val Ser Val Ser
Pro Gly Gln Thr Ala Arg Ile Thr Cys Ser Gly Asp145 150 155 160Ala
Leu Pro Lys Gln Tyr Ala Tyr Trp Tyr Gln Gln Lys Pro Gly Gln 165 170
175Ala Pro Val Met Val Ile Tyr Lys Asp Thr Glu Arg Pro Ser Gly Ile
180 185 190Pro Glu Arg Phe Ser Gly Ser Ser Ser Gly Thr Lys Val Thr
Leu Thr 195 200 205Ile Ser Gly Val Gln Ala Glu Asp Glu Ala Asp Tyr
Tyr Cys Gln Ser 210 215 220Ala Asp Asn Ser Ile Thr Tyr Arg Val Phe
Gly Gly Gly Thr Lys Val225 230 235 240Thr Val Leu Ala Ala Gly Ser
Asp Gln Glu Pro Lys Lys Leu Ala Ala 245 250 255Gly Ser Asp Gln Glu
Pro Lys Ser Ser Asp Lys Thr His Thr Cys Pro 260 265 270Pro Cys Ser
Ala Pro Glu Leu Leu Gly Gly Ser Ser Val Phe Leu Phe 275 280 285Pro
Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val 290 295
300Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Lys
Phe305 310 315 320Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala
Lys Thr Lys Pro 325 330 335Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg
Val Val Ser Val Leu Thr 340 345 350Val Leu His Gln Asp Trp Leu Asn
Gly Lys Glu Tyr Lys Cys Lys Val 355 360 365Ser Asn Lys Ala Leu Pro
Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala 370 375 380Lys Gly Gln Pro
Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg385 390 395 400Asp
Glu Leu Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly 405 410
415Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln Pro
420 425 430Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser Asp
Gly Ser 435 440 445Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser
Arg Trp Gln Gln 450 455 460Gly Asn Val Phe Ser Cys Ser Val Met His
Glu Ala Leu His Asn His465 470 475 480Tyr Thr Gln Lys Ser Leu Ser
Leu Ser Pro Gly Lys Gly Ser Gly Asp 485 490 495Tyr Lys Asp Asp Asp
Asp Lys Gly Ser Gly His His His His His His 500 505
510246119PRTArtificial SequenceSynthetic 10B4, VH 246Gln Val Gln
Leu Gln Gln Ser Gly Ala Glu Leu Met Lys Pro Gly Ala1 5 10 15Ser Val
Lys Met Ser Cys Lys Thr Thr Gly Tyr Ile Phe Ser Ser Tyr 20 25 30Trp
Ile Gly Trp Val Lys Gln Arg Pro Gly His Gly Leu Glu Trp Ile 35 40
45Gly Lys Ile Phe Pro Gly Ser Gly Ser Ala Asp Tyr Asn Glu Asn Phe
50 55 60Lys Gly Lys Ala Thr Phe Thr Val Asp Thr Ser Ser Asn Thr Ala
Tyr65 70 75 80Met Gln Leu Ser Ser Leu Thr Ser Glu Asp Ser Ala Val
Tyr Tyr Cys 85 90 95Ala Arg Gly Tyr Gly Asn Tyr Leu Tyr Phe Asp Val
Trp Gly Ala Gly 100 105 110Thr Thr Val Thr Val Ser Ser
115247119PRTArtificial SequenceSynthetic 1353-A09, VH 247Glu Val
Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser
Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Arg Phe Thr Trp Tyr 20 25
30Gly Ile Ser Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Met
35 40 45Gly Trp Ile Ser Ala Tyr Asn Gly Asn Thr Asn Tyr Ala Gln Lys
Leu 50 55 60Gln Gly Arg Val Thr Met Thr Thr Asp Thr Ser Thr Asn Thr
Ala Tyr65 70 75 80Met Glu Leu Arg Ser Leu Arg Ser Asp Asp Thr Ala
Val Tyr Tyr Cys 85 90 95Ala Arg Asp Ser Glu Tyr Ser Ser Gly Ser Gly
Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val Thr Val Ser Ser
115248119PRTArtificial SequenceSynthetic 1353-C07, VH 248Glu Val
Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser
Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Arg Phe Ser Thr Phe 20 25
30Gly Ile Ser Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Met
35 40 45Gly Trp Ile Ser Ala Tyr Asn Gly Asn Thr Asn Tyr Ala Gln Lys
Leu 50 55 60Gln Gly Arg Val Thr Met Thr Thr Asp Thr Ser Thr Asn Thr
Ala Tyr65 70 75 80Met Glu Leu Arg Ser Leu Arg Ser Asp Asp Thr Ala
Val Tyr Tyr Cys 85 90 95Ala Arg Asp Val Asp Tyr Ser Ser Gly Ser Gly
Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val Thr Val Ser Ser
115249119PRTArtificial SequenceSynthetic 1353-E07, VH 249Glu Val
Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser
Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Arg Phe Glu Thr Tyr 20 25
30Gly Ile Ser Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Met
35 40 45Gly Trp Ile Ser Ala Tyr Asn Gly Asn Thr Asn Tyr Ala Gln Lys
Leu 50 55 60Gln Gly Arg Val Thr Met Thr Thr Asp Thr Ser Thr Asn Thr
Ala Tyr65 70 75 80Met Glu Leu Arg Ser Leu Arg Ser Asp Asp Thr Ala
Val Tyr Tyr Cys 85 90 95Ala Arg Asp Ala Glu Tyr Ser Leu Gly Ser Gly
Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val Thr Val Ser Ser
115250119PRTArtificial SequenceSynthetic 1353-F09, VH 250Glu Val
Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser
Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Arg Phe Arg Gln Tyr 20 25
30Gly Ile Ser Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Met
35 40 45Gly Trp Ile Ser Ala Tyr Asn Gly Asn Thr Asn Tyr Ala Gln Lys
Leu 50 55 60Gln Gly Arg Val Thr Met Thr Thr Asp Thr Ser Thr Asn Thr
Ala Tyr65 70 75
80Met Glu Leu Arg Ser Leu Arg Ser Asp Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Arg Asp Ala Glu Tyr Gly Ser Gly Ser Gly Tyr Trp Gly Gln
Gly 100 105 110Thr Leu Val Thr Val Ser Ser 115251119PRTArtificial
SequenceSynthetic 1353-G08, VH 251Glu Val Gln Leu Val Gln Ser Gly
Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys
Ala Ser Gly Tyr Arg Phe Thr Arg Tyr 20 25 30Gly Ile Ser Trp Val Arg
Gln Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Trp Val Ser Ala
His Asn Gly Asn Thr Asn Tyr Ala Gln Lys Leu 50 55 60Gln Gly Arg Val
Thr Met Thr Thr Asp Thr Ser Thr Asn Thr Ala Tyr65 70 75 80Met Glu
Leu Arg Ser Leu Arg Ser Asp Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala
Arg Asp Ala Asp Tyr Gly Ser Gly Ser Gly Tyr Trp Gly Gln Gly 100 105
110Thr Leu Val Thr Val Ser Ser 115252119PRTArtificial
SequenceSynthetic 1353-G10, VH 252Glu Val Gln Leu Val Gln Ser Gly
Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys
Ala Ser Gly Tyr Arg Phe Pro His Tyr 20 25 30Gly Ile Ser Trp Val Arg
Gln Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Trp Ile Ser Ala
Tyr Asn Gly Asn Thr Asn Tyr Ala Gln Lys Leu 50 55 60Gln Gly Arg Val
Thr Met Thr Thr Asp Thr Ser Thr Asn Thr Ala Tyr65 70 75 80Met Glu
Leu Arg Ser Leu Arg Ser Asp Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala
Arg Asp Val Asp Tyr Gly Thr Gly Ser Gly Tyr Trp Gly Gln Gly 100 105
110Thr Leu Val Thr Val Ser Ser 115253119PRTArtificial
SequenceSynthetic 1353-H08, VH 253Glu Val Gln Leu Val Gln Ser Gly
Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys
Ala Ser Gly Tyr Arg Phe Thr Arg Gln 20 25 30Gly Ile Ser Trp Val Arg
Gln Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Trp Ile Ser Ala
Tyr Asn Gly Asn Thr Lys Tyr Ala Gln Lys Leu 50 55 60Gln Gly Arg Val
Thr Met Thr Thr Asp Thr Ser Thr Asn Thr Ala Tyr65 70 75 80Met Glu
Leu Arg Ser Leu Arg Ser Asp Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala
Arg Asp Val Asp Tyr Gly Ser Gly Ser Gly Tyr Trp Gly Gln Gly 100 105
110Thr Leu Val Thr Val Ser Ser 115254119PRTArtificial
SequenceSynthetic 1353-H09, VH 254Glu Val Gln Leu Val Gln Ser Gly
Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys
Ala Ser Gly Tyr Arg Phe Pro His Tyr 20 25 30Gly Ile Ser Trp Val Arg
Gln Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Trp Ile Ser Ala
Tyr Asn Gly Asn Thr Asn Tyr Ala Gln Lys Leu 50 55 60Gln Gly Arg Val
Thr Met Thr Thr Asp Thr Ser Thr Asn Thr Ala Tyr65 70 75 80Met Glu
Leu Arg Ser Leu Arg Ser Asp Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala
Arg Asp Ala Glu Tyr Gly Ser Gly Ser Gly Tyr Trp Gly Gln Gly 100 105
110Thr Leu Val Thr Val Ser Ser 115255115PRTArtificial
SequenceSynthetic 1B10, VH 255Asp Val Gln Leu Gln Glu Ser Gly Pro
Gly Leu Val Lys Pro Ser Gln1 5 10 15Ser Leu Ser Leu Thr Cys Thr Val
Thr Gly His Ser Ile Thr Ser Asp 20 25 30Tyr Ala Trp Asn Trp Ile Arg
Gln Phe Pro Gly Asn Lys Leu Glu Trp 35 40 45Met Gly Tyr Ile Ser Tyr
Ser Gly Arg Thr Ser Tyr Asn Pro Ser Leu 50 55 60Thr Ser Arg Ile Ser
Ile Thr Arg Asp Thr Ser Lys Asn Gln Phe Phe65 70 75 80Leu Gln Leu
Asn Ser Val Thr Thr Glu Asp Thr Ala Thr Tyr Tyr Cys 85 90 95Ala Arg
Gly Tyr Ala Leu Asp Tyr Trp Gly Gln Gly Thr Ser Val Thr 100 105
110Val Ser Ser 115256120PRTArtificial SequenceSynthetic 1E9, VH
256Glu Val Lys Leu Val Glu Ser Gly Gly Gly Leu Val Ser Pro Gly Gly1
5 10 15Ser Leu Lys Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr
Phe 20 25 30Gly Met Ser Trp Val Arg Gln Thr Pro Glu Lys Arg Leu Glu
Trp Val 35 40 45Ala Thr Ile Ser Gly Gly Gly Ser Asp Thr Tyr Tyr Pro
Asp Ser Val 50 55 60Gln Gly Arg Phe Ile Ile Ser Arg Tyr Asn Ala Lys
Asn Asn Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Pro Glu Asp
Thr Ala Leu Tyr Tyr Cys 85 90 95Ala Arg Gln Gly Tyr Asp Val Tyr Ser
Trp Phe Ala Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser
Ala 115 120257118PRTArtificial SequenceSynthetic 4B10, VH 257Glu
Val Lys Leu Val Glu Ser Gly Gly Gly Leu Val Lys Pro Gly Gly1 5 10
15Ser Leu Lys Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr Tyr
20 25 30Gly Met Ser Trp Val Arg Gln Thr Pro Glu Lys Arg Leu Gln Trp
Val 35 40 45Ala Thr Ile Ser Gly Gly Gly Ser Asn Thr Tyr Tyr Ser Asp
Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn
Asn Leu Tyr65 70 75 80Leu Gln Met Ser Ser Leu Arg Ser Glu Asp Thr
Ala Leu Tyr Tyr Cys 85 90 95Ala Arg Gln Arg Asp Ser Ala Trp Phe Ala
Ser Trp Gly Gln Gly Thr 100 105 110Leu Val Thr Val Ser Ala
115258120PRTArtificial SequenceSynthetic h1E9-1, VH 258Glu Val Gln
Leu Val Glu Ser Gly Gly Gly Leu Val Lys Pro Gly Gly1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr Phe 20 25 30Gly
Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ser Thr Ile Ser Gly Gly Gly Ser Asp Thr Tyr Tyr Pro Asp Ser Val
50 55 60Gln Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn Ser Leu
Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val
Tyr Tyr Cys 85 90 95Ala Arg Gln Gly Tyr Asp Val Tyr Ser Trp Phe Ala
Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser 115
120259120PRTArtificial SequenceSynthetic h1E9-2, VH 259Glu Val Gln
Leu Val Glu Ser Gly Gly Gly Leu Val Lys Pro Gly Gly1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr Phe 20 25 30Gly
Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ser Thr Ile Ser Gly Gly Gly Ser Asp Thr Tyr Tyr Pro Asp Ser Val
50 55 60Gln Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn Ser Leu
Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val
Tyr Tyr Cys 85 90 95Ala Arg Gln Gly Tyr Asp Val Tyr Ser Trp Phe Ala
Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser 115
120260120PRTArtificial SequenceSynthetic h1E9-4, VH 260Glu Val Gln
Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr Phe 20 25 30Gly
Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ala Thr Ile Ser Gly Gly Gly Ser Asp Thr Tyr Tyr Pro Asp Ser Val
50 55 60Gln Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn Ser Leu
Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val
Tyr Tyr Cys 85 90 95Ala Arg Gln Gly Tyr Asp Val Tyr Ser Trp Phe Ala
Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser 115
120261120PRTArtificial SequenceSynthetic h1E9-5, VH 261Glu Val Gln
Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr Phe 20 25 30Gly
Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ala Thr Ile Ser Gly Gly Gly Ser Asp Thr Tyr Tyr Pro Asp Ser Val
50 55 60Gln Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn Ser Leu
Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val
Tyr Tyr Cys 85 90 95Ala Arg Gln Gly Tyr Asp Val Tyr Ser Trp Phe Ala
Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser 115
120262118PRTArtificial SequenceSynthetic h4B10-1, VH 262Glu Val Gln
Leu Val Glu Ser Gly Gly Gly Leu Val Lys Pro Gly Gly1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr Tyr 20 25 30Gly
Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ala Thr Ile Ser Gly Gly Gly Ser Asn Thr Tyr Tyr Ser Asp Ser Val
50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp Ser Lys Asn Thr Leu
Tyr65 70 75 80Leu Gln Met Asn Ser Leu Lys Thr Glu Asp Thr Ala Val
Tyr Tyr Cys 85 90 95Ala Arg Gln Arg Asp Ser Ala Trp Phe Ala Ser Trp
Gly Gln Gly Thr 100 105 110Leu Val Thr Val Ser Ser
115263118PRTArtificial SequenceSynthetic h4B10-2, VH 263Glu Val Gln
Leu Val Glu Ser Gly Gly Gly Leu Val Lys Pro Gly Gly1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr Tyr 20 25 30Gly
Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ala Thr Ile Ser Gly Gly Gly Ser Asn Thr Tyr Tyr Ser Asp Ser Val
50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp Ser Lys Asn Thr Leu
Tyr65 70 75 80Leu Gln Met Asn Ser Leu Lys Thr Glu Asp Thr Ala Val
Tyr Tyr Cys 85 90 95Ala Arg Gln Arg Asp Ser Ala Trp Phe Ala Ser Trp
Gly Gln Gly Thr 100 105 110Leu Val Thr Val Ser Ser
115264118PRTArtificial SequenceSynthetic h4B10-3, VH 264Glu Val Gln
Leu Val Glu Ser Gly Gly Gly Leu Val Lys Pro Gly Gly1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr Tyr 20 25 30Gly
Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ala Thr Ile Ser Gly Gly Gly Ser Asn Thr Tyr Tyr Ser Asp Ser Val
50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp Ser Lys Asn Thr Leu
Tyr65 70 75 80Leu Gln Met Asn Ser Leu Lys Thr Glu Asp Thr Ala Val
Tyr Tyr Cys 85 90 95Ala Arg Gln Arg Asp Ser Ala Trp Phe Ala Ser Trp
Gly Gln Gly Thr 100 105 110Leu Val Thr Val Ser Ser
115265128PRTArtificial SequenceSynthetic PD1-17, VH 265Gln Val Gln
Leu Gln Glu Ser Gly Pro Gly Val Val Lys Pro Ser Gly1 5 10 15Thr Leu
Ser Leu Thr Cys Ala Ile Ser Gly Gly Ser Ile Gly Ser Gly 20 25 30Gly
Ser Ile Arg Ser Thr Arg Trp Trp Ser Trp Val Arg Gln Ser Pro 35 40
45Gly Lys Gly Leu Glu Trp Ile Gly Glu Ile Tyr His Ser Gly Ser Thr
50 55 60Asn Tyr Asn Pro Ser Leu Lys Ser Arg Val Thr Ile Ser Leu Asp
Lys65 70 75 80Ser Arg Asn His Phe Ser Leu Arg Leu Asn Ser Val Thr
Ala Ala Asp 85 90 95Thr Ala Val Tyr Tyr Cys Ala Arg Gln Asp Tyr Gly
Asp Ser Gly Asp 100 105 110Trp Tyr Phe Asp Leu Trp Gly Lys Gly Thr
Met Val Thr Val Ser Ser 115 120 125266119PRTArtificial
SequenceSynthetic PD1-28, VH 266Glu Val Gln Leu Val Gln Ser Gly Ala
Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala
Ser Gly Tyr Arg Phe Thr Ser Tyr 20 25 30Gly Ile Ser Trp Val Arg Gln
Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Trp Ile Ser Ala Tyr
Asn Gly Asn Thr Asn Tyr Ala Gln Lys Leu 50 55 60Gln Gly Arg Val Thr
Met Thr Thr Asp Thr Ser Thr Asn Thr Ala Tyr65 70 75 80Met Glu Leu
Arg Ser Leu Arg Ser Asp Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg
Asp Ala Asp Tyr Ser Ser Gly Ser Gly Tyr Trp Gly Gln Gly 100 105
110Thr Leu Val Thr Val Ser Ser 115267119PRTArtificial
SequenceSynthetic PD1-33, VH 267Gln Val Gln Leu Val Gln Ser Gly Ala
Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Arg Val Ser Cys Lys Ala
Ser Gly Tyr Thr Leu Thr Ser Tyr 20 25 30Tyr Ile His Trp Val Arg Gln
Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Ile Ile Asn Pro Arg
Gly Ala Thr Ile Ser Tyr Ala Gln Lys Phe 50 55 60Gln Gly Arg Val Thr
Met Thr Arg Asp Thr Ser Thr Ser Thr Val Tyr65 70 75 80Met Glu Leu
Arg Asn Leu Lys Ser Glu Asp Thr Ala Leu Tyr Tyr Cys 85 90 95Ala Thr
Ala Gly Ile Tyr Gly Phe Asp Phe Asp Tyr Trp Gly Arg Gly 100 105
110Thr Leu Val Thr Val Ser Ser 115268127PRTArtificial
SequenceSynthetic PD1-35, VH 268Gln Val Gln Leu Gln Glu Ser Gly Pro
Gly Leu Val Lys Pro Ser Gln1 5 10 15Thr Leu Ser Leu Thr Cys Thr Val
Ser Gly Gly Ser Ile Ser Ser Gly 20 25 30Ala Tyr Tyr Trp Ser Trp Ile
Arg Gln His Pro Gly Lys Gly Leu Glu 35 40 45Trp Ile Gly Tyr Ile Tyr
Tyr Asn Gly Asn Thr Tyr Tyr Asn Pro Ser 50 55 60Leu Arg Ser Leu Val
Thr Ile Ser Val Asp Ala Ser Lys Asn Gln Phe65 70 75 80Ser Leu Lys
Leu Ser Ser Val Thr Ala Ala Asp Thr Ala Val Tyr Tyr 85 90 95Cys Ala
Arg Ala Ser Asp Tyr Val Trp Gly Gly Tyr Arg Tyr Met Asp 100 105
110Ala Phe Asp Ile Trp Gly Arg Gly Thr Leu Ile Thr Val Ser Ser 115
120 125269121PRTArtificial SequenceSynthetic PD1-F2, VH 269Glu Val
Gln Leu Val Gln Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser
Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25
30Trp Cys Asp Arg Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu
35 40 45Glu Trp Val Ser Ala Ile Ser Gly Ser Gly Gly Ser Thr Tyr Tyr
Ala 50 55 60Asp Ser Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser
Lys Asn65 70 75 80Thr Leu Tyr Leu Gln Met Asn Ser Leu Arg Ala Glu
Asp Thr Ala Val 85 90 95Tyr Tyr Cys Ala Lys Glu Asn Trp Gly Ser Tyr
Phe Asp Leu Trp Gly 100 105 110Gln Gly Thr Thr Val Thr Val Ser Ser
115 120270108PRTArtificial SequenceSynthetic 10B4, VL 270Asn Ile
Val Met Thr Gln Thr Pro Lys Phe Leu Leu Val Ser Ala Gly1 5 10 15Asp
Arg Ile Thr Ile Thr Cys Lys
Ala Ser Gln Ser Val Ser Asp Asp 20 25 30Val Ala Trp Tyr Gln Gln Lys
Pro Gly Gln Ser Pro Lys Leu Leu Ile 35 40 45Ser Tyr Ala Phe Lys Arg
Tyr Ile Gly Val Pro Asp Arg Phe Thr Gly 50 55 60Ser Gly Tyr Gly Thr
Asp Phe Thr Phe Thr Ile Ser Thr Val Gln Ala65 70 75 80Glu Asp Leu
Ala Val Tyr Phe Cys Gln Gln Asn Tyr Asn Ser Pro Tyr 85 90 95Thr Phe
Gly Gly Gly Thr Lys Leu Glu Leu Lys Arg 100 105271108PRTArtificial
SequenceSynthetic 1353-A09, VL 271Ser Tyr Glu Leu Thr Gln Pro Pro
Ser Val Ser Val Ser Pro Gly Gln1 5 10 15Thr Ala Arg Ile Thr Cys Ser
Gly Asp Ala Leu Thr Thr Gln Tyr Ala 20 25 30Tyr Trp Tyr Gln Gln Lys
Pro Gly Gln Ala Pro Val Met Val Ile Tyr 35 40 45Lys Asp Thr Glu Arg
Pro Ser Gly Ile Pro Glu Arg Phe Ser Gly Ser 50 55 60Ser Ser Gly Thr
Lys Val Thr Leu Thr Ile Ser Gly Val Gln Ala Glu65 70 75 80Asp Glu
Ala Asp Tyr Tyr Cys Gln Ser Ala Asp Asn Ser Ile Thr Tyr 85 90 95Arg
Val Phe Gly Gly Gly Thr Lys Val Thr Val Leu 100
105272108PRTArtificial SequenceSynthetic 1353-C07, VL 272Ser Tyr
Glu Leu Thr Gln Pro Pro Ser Val Ser Val Ser Pro Gly Gln1 5 10 15Thr
Ala Arg Ile Thr Cys Ser Gly Asp Ala Leu Ser Glu Gln Tyr Ala 20 25
30Tyr Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro Val Met Val Ile Tyr
35 40 45Lys Asp Thr Glu Arg Pro Ser Gly Ile Pro Glu Arg Phe Ser Gly
Ser 50 55 60Ser Ser Gly Thr Lys Val Thr Leu Thr Ile Ser Gly Val Gln
Ala Glu65 70 75 80Asp Glu Ala Asp Tyr Tyr Cys Gln Ser Ala Asp Asn
Ser Ile Thr Tyr 85 90 95Arg Val Phe Gly Gly Gly Thr Lys Val Thr Val
Leu 100 105273108PRTArtificial SequenceSynthetic 1353-E07, VL
273Ser Tyr Glu Leu Thr Gln Pro Pro Ser Val Ser Val Ser Pro Gly Gln1
5 10 15Thr Ala Arg Ile Thr Cys Ser Gly Asp Ala Leu Pro Lys Gln Tyr
Ala 20 25 30Tyr Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro Val Met Val
Ile Tyr 35 40 45Lys Asp Thr Glu Arg Pro Ser Gly Ile Pro Glu Arg Phe
Ser Gly Ser 50 55 60Ser Ser Gly Thr Lys Val Thr Leu Thr Ile Ser Gly
Val Gln Ala Glu65 70 75 80Asp Glu Ala Asp Tyr Tyr Cys Gln Ser Ala
Asp Asn Ser Ile Thr Tyr 85 90 95Arg Val Phe Gly Gly Gly Thr Lys Val
Thr Val Leu 100 105274108PRTArtificial SequenceSynthetic 1353-F09,
VL 274Ser Tyr Glu Leu Thr Gln Pro Pro Ser Val Ser Val Ser Pro Gly
Gln1 5 10 15Thr Ala Arg Ile Thr Cys Ser Gly Asp Ala Leu Pro Lys Gln
Tyr Ala 20 25 30Tyr Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro Val Met
Val Leu Tyr 35 40 45Lys Asp Thr Glu Arg Pro Ser Gly Ile Pro Glu Arg
Phe Ser Gly Ser 50 55 60Ser Ser Gly Thr Lys Val Thr Leu Thr Ile Ser
Gly Val Gln Ala Glu65 70 75 80Asp Glu Ala Asp Tyr Tyr Cys Gln Ser
Ala Asp Asn Ser Ile Thr Tyr 85 90 95Arg Val Phe Gly Gly Gly Thr Lys
Val Thr Val Leu 100 105275108PRTArtificial SequenceSynthetic
1353-G08, VL 275Ser Tyr Glu Leu Thr Gln Pro Pro Ser Val Ser Val Ser
Pro Gly Gln1 5 10 15Thr Ala Arg Ile Thr Cys Ser Gly Asp Ala Leu Pro
Met Gln Tyr Gly 20 25 30Tyr Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro
Val Met Val Ile Tyr 35 40 45Lys Asp Thr Glu Arg Pro Ser Gly Ile Pro
Glu Arg Phe Ser Gly Ser 50 55 60Ser Ser Gly Thr Lys Val Thr Leu Thr
Ile Ser Gly Val Gln Ala Glu65 70 75 80Asp Glu Ala Asp Tyr Tyr Cys
Gln Ser Ala Asp Asn Ser Ile Thr Tyr 85 90 95Arg Val Phe Gly Gly Gly
Thr Lys Val Thr Val Leu 100 105276108PRTArtificial
SequenceSynthetic 1353-G10, VL 276Ser Tyr Glu Leu Thr Gln Pro Pro
Ser Val Ser Val Ser Pro Gly Gln1 5 10 15Thr Ala Arg Ile Thr Cys Ser
Gly Asp Ala Leu Pro Lys Gln Tyr Ala 20 25 30Tyr Trp Tyr Gln Gln Lys
Pro Gly Gln Ala Pro Val Met Val Ile Tyr 35 40 45Lys Asp Thr Glu Arg
Pro Ser Gly Ile Pro Glu Arg Phe Ser Gly Ser 50 55 60Ser Ser Gly Thr
Lys Val Thr Leu Thr Ile Ser Gly Val Gln Ala Glu65 70 75 80Asp Glu
Ala Asp Tyr Tyr Cys Gln Ser Ala Asp Asn Ser Ile Thr Tyr 85 90 95Arg
Val Phe Gly Gly Gly Thr Lys Val Thr Val Leu 100
105277108PRTArtificial SequenceSynthetic 1353-H08, VL 277Ser Tyr
Glu Leu Thr Gln Pro Pro Ser Val Ser Val Ser Pro Gly Gln1 5 10 15Thr
Ala Arg Ile Thr Cys Ser Gly Asp Ala Leu Pro Lys Gln Tyr Ala 20 25
30Tyr Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro Val Met Val Ile Tyr
35 40 45Lys Asp Thr Glu Arg Pro Ser Gly Ile Pro Glu Arg Phe Ser Gly
Ser 50 55 60Ser Ser Gly Thr Lys Val Thr Leu Thr Ile Ser Gly Val Gln
Ala Glu65 70 75 80Asp Glu Ala Asp Tyr Tyr Cys Gln Ser Ala Asp Asn
Ser Ile Thr Tyr 85 90 95Arg Val Phe Gly Gly Gly Thr Lys Val Thr Val
Leu 100 105278108PRTArtificial SequenceSynthetic 1353-H09, VL
278Ser Tyr Glu Leu Thr Gln Pro Pro Ser Val Ser Val Ser Pro Gly Gln1
5 10 15Thr Ala Arg Ile Thr Cys Ser Gly Asp Ala Leu Pro Lys Gln Tyr
Ala 20 25 30Tyr Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro Val Met Val
Ile Tyr 35 40 45Lys Asp Thr Glu Arg Pro Ser Gly Ile Pro Glu Arg Phe
Ser Gly Ser 50 55 60Ser Ser Gly Thr Lys Val Thr Leu Thr Ile Ser Gly
Val Gln Ala Glu65 70 75 80Asp Glu Ala Asp Tyr Tyr Cys Gln Ser Ala
Asp Asn Ser Ile Thr Tyr 85 90 95Arg Val Phe Gly Gly Gly Thr Lys Val
Thr Val Leu 100 105279106PRTArtificial SequenceSynthetic 1B10, VL
279Gln Ile Val Leu Ser Gln Ser Pro Ala Ile Leu Ser Ala Ser Pro Gly1
5 10 15Glu Lys Val Thr Met Thr Cys Arg Thr Ser Ser Ser Val Asn Tyr
Met 20 25 30His Trp Phe Gln Gln Lys Pro Gly Ser Ser Pro Lys Pro Trp
Ile Tyr 35 40 45Ala Thr Ser Lys Leu Ala Ser Gly Val Pro Ala Arg Phe
Ser Gly Ser 50 55 60Gly Ser Gly Thr Ser Tyr Ser Leu Thr Ile Ser Arg
Val Glu Ala Glu65 70 75 80Asp Ala Ala Thr Tyr Phe Cys Gln Gln Trp
Ile Ser Asp Pro Trp Thr 85 90 95Phe Gly Gly Gly Thr Lys Leu Glu Ile
Lys 100 105280111PRTArtificial SequenceSynthetic 1E9, VL 280Asp Ile
Ile Leu Thr Gln Ser Pro Ala Ser Leu Ala Val Ser Leu Gly1 5 10 15Gln
Arg Ala Ala Ile Ser Cys Arg Ala Ser Glu Ser Val Asp Asn Ser 20 25
30Gly Ile Ser Phe Met Ser Trp Phe Gln Gln Lys Pro Gly Gln Pro Pro
35 40 45Lys Leu Leu Ile Tyr Thr Ala Ser Asn Gln Gly Ser Gly Val Pro
Ala 50 55 60Arg Phe Ser Gly Ser Gly Ser Gly Thr Glu Phe Ser Leu Asn
Ile His65 70 75 80Pro Met Glu Glu Asp Asp Thr Ala Met Tyr Phe Cys
Gln Gln Ser Lys 85 90 95Glu Val Pro Trp Thr Phe Gly Gly Gly Thr Lys
Leu Glu Ile Arg 100 105 110281111PRTArtificial SequenceSynthetic
4B10, VL 281Asp Ile Val Leu Thr Gln Ser Pro Ala Ser Leu Ala Val Ser
Leu Gly1 5 10 15Gln Arg Ala Thr Ile Ser Cys Arg Ala Ser Glu Asn Val
Asp Asp Tyr 20 25 30Gly Val Ser Phe Met Asn Trp Phe Gln Gln Lys Pro
Gly Gln Pro Pro 35 40 45Lys Leu Leu Ile Tyr Pro Ala Ser Asn Gln Gly
Ser Gly Val Pro Ala 50 55 60Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp
Phe Ser Leu Asn Ile His65 70 75 80Pro Met Glu Glu Asp Asp Thr Ala
Met Tyr Phe Cys Gln Gln Ser Lys 85 90 95Glu Val Pro Trp Thr Phe Gly
Gly Gly Thr Lys Leu Glu Ile Lys 100 105 110282111PRTArtificial
SequenceSynthetic h1E9-1, VL 282Asp Ile Gln Leu Thr Gln Ser Pro Ser
Phe Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg
Ala Ser Glu Ser Val Asp Asn Ser 20 25 30Gly Ile Ser Phe Met Ser Trp
Tyr Gln Gln Lys Pro Gly Lys Ala Pro 35 40 45Lys Leu Leu Ile Tyr Thr
Ala Ser Asn Gln Gly Ser Gly Val Pro Ser 50 55 60Arg Phe Ser Gly Ser
Gly Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser65 70 75 80Ser Leu Gln
Pro Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Ser Lys 85 90 95Glu Val
Pro Trp Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys 100 105
110283111PRTArtificial SequenceSynthetic h1E9-2, VL 283Glu Ile Val
Leu Thr Gln Ser Pro Ala Thr Leu Ser Leu Ser Pro Gly1 5 10 15Glu Arg
Ala Thr Leu Ser Cys Arg Ala Ser Glu Ser Val Asp Asn Ser 20 25 30Gly
Ile Ser Phe Met Ser Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro 35 40
45Arg Leu Leu Ile Tyr Thr Ala Ser Asn Gln Gly Ser Gly Ile Pro Ala
50 55 60Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile
Ser65 70 75 80Ser Leu Glu Pro Glu Asp Phe Ala Val Tyr Tyr Cys Gln
Gln Ser Lys 85 90 95Glu Val Pro Trp Thr Phe Gly Gln Gly Thr Lys Val
Glu Ile Lys 100 105 110284111PRTArtificial SequenceSynthetic
h1E9-4, VL 284Asp Ile Gln Leu Thr Gln Ser Pro Ser Phe Leu Ser Ala
Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Glu Ser
Val Asp Asn Ser 20 25 30Gly Ile Ser Phe Met Ser Trp Tyr Gln Gln Lys
Pro Gly Lys Ala Pro 35 40 45Lys Leu Leu Ile Tyr Thr Ala Ser Asn Gln
Gly Ser Gly Val Pro Ser 50 55 60Arg Phe Ser Gly Ser Gly Ser Gly Thr
Glu Phe Thr Leu Thr Ile Ser65 70 75 80Ser Leu Gln Pro Glu Asp Phe
Ala Thr Tyr Tyr Cys Gln Gln Ser Lys 85 90 95Glu Val Pro Trp Thr Phe
Gly Gln Gly Thr Lys Val Glu Ile Lys 100 105 110285111PRTArtificial
SequenceSynthetic h1E9-5, VL 285Glu Ile Val Leu Thr Gln Ser Pro Ala
Thr Leu Ser Leu Ser Pro Gly1 5 10 15Glu Arg Ala Thr Leu Ser Cys Arg
Ala Ser Glu Ser Val Asp Asn Ser 20 25 30Gly Ile Ser Phe Met Ser Trp
Tyr Gln Gln Lys Pro Gly Gln Ala Pro 35 40 45Arg Leu Leu Ile Tyr Thr
Ala Ser Asn Gln Gly Ser Gly Ile Pro Ala 50 55 60Arg Phe Ser Gly Ser
Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser65 70 75 80Ser Leu Glu
Pro Glu Asp Phe Ala Val Tyr Tyr Cys Gln Gln Ser Lys 85 90 95Glu Val
Pro Trp Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys 100 105
110286111PRTArtificial SequenceSynthetic h4B10-1, VL 286Glu Ile Val
Leu Thr Gln Ser Pro Ala Thr Leu Ser Leu Ser Pro Gly1 5 10 15Glu Arg
Ala Thr Leu Ser Cys Arg Ala Ser Glu Asn Val Asp Asp Tyr 20 25 30Gly
Val Ser Phe Met Asn Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro 35 40
45Arg Leu Leu Ile Tyr Pro Ala Ser Asn Gln Gly Ser Gly Ile Pro Ala
50 55 60Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile
Ser65 70 75 80Ser Leu Glu Pro Glu Asp Phe Ala Val Tyr Tyr Cys Gln
Gln Ser Lys 85 90 95Glu Val Pro Trp Thr Phe Gly Gln Gly Thr Lys Val
Glu Ile Lys 100 105 110287111PRTArtificial SequenceSynthetic
h4B10-2, VL 287Glu Ile Val Leu Thr Gln Ser Pro Gly Thr Leu Ser Leu
Ser Pro Gly1 5 10 15Glu Arg Ala Thr Leu Ser Cys Arg Ala Ser Glu Asn
Val Asp Asp Tyr 20 25 30Gly Val Ser Phe Met Asn Trp Tyr Gln Gln Lys
Pro Gly Gln Ala Pro 35 40 45Arg Leu Leu Ile Tyr Pro Ala Ser Asn Gln
Gly Ser Gly Ile Pro Asp 50 55 60Arg Phe Ser Gly Ser Gly Ser Gly Thr
Asp Phe Thr Leu Thr Ile Ser65 70 75 80Arg Leu Glu Pro Glu Asp Phe
Ala Val Tyr Tyr Cys Gln Gln Ser Lys 85 90 95Glu Val Pro Trp Thr Phe
Gly Gln Gly Thr Lys Val Glu Ile Lys 100 105 110288111PRTArtificial
SequenceSynthetic h4B10-3, VL 288Asp Ile Val Met Thr Gln Ser Pro
Asp Ser Leu Ala Val Ser Leu Gly1 5 10 15Glu Arg Ala Thr Ile Asn Cys
Arg Ala Ser Glu Asn Val Asp Asp Tyr 20 25 30Gly Val Ser Phe Met Asn
Trp Tyr Gln Gln Lys Pro Gly Gln Pro Pro 35 40 45Lys Leu Leu Ile Tyr
Pro Ala Ser Asn Gln Gly Ser Gly Val Pro Asp 50 55 60Arg Phe Ser Gly
Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser65 70 75 80Ser Leu
Gln Ala Glu Asp Val Ala Val Tyr Tyr Cys Gln Gln Ser Lys 85 90 95Glu
Val Pro Trp Thr Phe Gly Gly Gly Thr Lys Leu Glu Ile Lys 100 105
110289110PRTArtificial SequenceSynthetic PD1-17, VL 289Asn Phe Met
Leu Thr Gln Pro His Ser Val Ser Glu Ser Pro Gly Lys1 5 10 15Thr Val
Thr Ile Ser Cys Thr Arg Ser Ser Gly Ser Ile Ala Ser Asn 20 25 30Ser
Val Gln Trp Tyr Gln Gln Arg Pro Gly Ser Ser Pro Thr Thr Val 35 40
45Ile Tyr Glu Asp Asn Gln Arg Pro Ser Gly Val Pro Asp Arg Phe Ser
50 55 60Gly Ser Ile Asp Ser Ser Ser Asn Ser Ala Ser Leu Thr Val Ser
Gly65 70 75 80Leu Lys Thr Glu Asp Glu Ala Asp Tyr Tyr Cys Gln Ser
Ser Asp Ser 85 90 95Ser Ala Val Val Phe Gly Ser Gly Thr Lys Leu Thr
Val Leu 100 105 110290108PRTArtificial SequenceSynthetic PD1-28, VL
290Ser Tyr Glu Leu Thr Gln Pro Pro Ser Val Ser Val Ser Pro Gly Gln1
5 10 15Thr Ala Arg Ile Thr Cys Ser Gly Asp Ala Leu Pro Lys Gln Tyr
Ala 20 25 30Tyr Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro Val Met Val
Ile Tyr 35 40 45Lys Asp Thr Glu Arg Pro Ser Gly Ile Pro Glu Arg Phe
Ser Gly Ser 50 55 60Ser Ser Gly Thr Lys Val Thr Leu Thr Ile Ser Gly
Val Gln Ala Glu65 70 75 80Asp Glu Ala Asp Tyr Tyr Cys Gln Ser Ala
Asp Asn Ser Ile Thr Tyr 85 90 95Arg Val Phe Gly Gly Gly Thr Lys Val
Thr Val Leu 100 105291111PRTArtificial SequenceSynthetic PD1-33, VL
291Gln Ser Ala Leu Thr Gln Pro Ala Ser Val Ser Gly Ser Pro Gly Gln1
5 10 15Ser Ile Thr Ile Ser Cys Thr Gly Thr Ser Asn Asp Val Gly Gly
Tyr 20 25 30Asn Tyr Val Ser Trp Tyr Gln His His Pro Gly Lys Ala Pro
Lys Leu 35 40 45Ile Ile Tyr Asp Val Thr Asn Arg Pro Ser Gly Val Ser
Asp
Arg Phe 50 55 60Ser Gly Ser Lys Ser Gly Asn Thr Ala Ser Leu Thr Ile
Ser Gly Leu65 70 75 80Leu Ala Glu Asp Glu Gly Asp Tyr Tyr Cys Ser
Ser Tyr Thr Ile Val 85 90 95Thr Asn Phe Glu Val Leu Phe Gly Gly Gly
Thr Lys Leu Thr Val 100 105 110292110PRTArtificial
SequenceSynthetic PD1-35, VL 292Gln Ser Val Leu Thr Gln Pro Pro Ser
Ala Ser Gly Thr Pro Gly Gln1 5 10 15Arg Val Thr Ile Ser Cys Ser Gly
Ser Asn Ser Asn Ile Gly Ser Asn 20 25 30Ser Val Asn Trp Tyr Gln Gln
Leu Pro Gly Thr Ala Pro Lys Leu Leu 35 40 45Ile Tyr Gly Asn Asn Gln
Arg Pro Ser Gly Val Pro Asp Arg Phe Ser 50 55 60Gly Ser Lys Ser Gly
Thr Ser Ala Ser Leu Ala Ile Ser Gly Leu Gln65 70 75 80Ser Glu Asn
Glu Ala Asp Tyr Tyr Cys Ala Ala Trp Asp Asp Ser Leu 85 90 95Asn Gly
Pro Val Phe Gly Arg Gly Thr Lys Val Thr Val Leu 100 105
110293107PRTArtificial SequenceSynthetic PD1-F2, VL 293Asp Ile Val
Met Thr Gln Ser Pro Ser Thr Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg
Val Thr Ile Thr Cys Arg Ala Ser Gln Gly Ile Ser Ser Trp 20 25 30Leu
Ala Trp Tyr Gln Gln Lys Pro Gly Arg Ala Pro Lys Val Leu Ile 35 40
45Tyr Lys Ala Ser Thr Leu Glu Ser Gly Val Pro Ser Arg Phe Ser Gly
50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln
Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Ser Tyr Ser
Thr Pro Trp 85 90 95Thr Phe Gly Gln Gly Thr Lys Leu Glu Ile Lys 100
10529420PRTArtificial SequenceSynthetic FlagHis Tag 294Gly Ser Gly
Asp Tyr Lys Asp Asp Asp Asp Lys Gly Ser Gly His His1 5 10 15His His
His His 20295252PRTArtificial SequenceSynthetic IgG1 Fc from
scFv-Fc 295Ala Ala Gly Ser Asp Gln Glu Pro Lys Ser Ser Asp Lys Thr
His Thr1 5 10 15Cys Pro Pro Cys Ser Ala Pro Glu Leu Leu Gly Gly Ser
Ser Val Phe 20 25 30Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile
Ser Arg Thr Pro 35 40 45Glu Val Thr Cys Val Val Val Asp Val Ser His
Glu Asp Pro Glu Val 50 55 60Lys Phe Asn Trp Tyr Val Asp Gly Val Glu
Val His Asn Ala Lys Thr65 70 75 80Lys Pro Arg Glu Glu Gln Tyr Asn
Ser Thr Tyr Arg Val Val Ser Val 85 90 95Leu Thr Val Leu His Gln Asp
Trp Leu Asn Gly Lys Glu Tyr Lys Cys 100 105 110Lys Val Ser Asn Lys
Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser 115 120 125Lys Ala Lys
Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro 130 135 140Ser
Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val145 150
155 160Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn
Gly 165 170 175Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu
Asp Ser Asp 180 185 190Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val
Asp Lys Ser Arg Trp 195 200 205Gln Gln Gly Asn Val Phe Ser Cys Ser
Val Met His Glu Ala Leu His 210 215 220Asn His Tyr Thr Gln Lys Ser
Leu Ser Leu Ser Pro Gly Lys Gly Ser225 230 235 240Gly Asp Tyr Lys
Asp Asp Asp Asp Lys Gly Ser Gly 245 250296330PRTArtificial
SequenceSynthetic HC Constant 296Ala Ser Thr Lys Gly Pro Ser Val
Phe Pro Leu Ala Pro Ser Ser Lys1 5 10 15Ser Thr Ser Gly Gly Thr Ala
Ala Leu Gly Cys Leu Val Lys Asp Tyr 20 25 30Phe Pro Glu Pro Val Thr
Val Ser Trp Asn Ser Gly Ala Leu Thr Ser 35 40 45Gly Val His Thr Phe
Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser 50 55 60Leu Ser Ser Val
Val Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr65 70 75 80Tyr Ile
Cys Asn Val Asn His Lys Pro Ser Asn Thr Lys Val Asp Lys 85 90 95Lys
Val Glu Pro Lys Ser Cys Asp Lys Thr His Thr Cys Pro Pro Cys 100 105
110Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro
115 120 125Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val
Thr Cys 130 135 140Val Val Val Asp Val Ser His Glu Asp Pro Glu Val
Lys Phe Asn Trp145 150 155 160Tyr Val Asp Gly Val Glu Val His Asn
Ala Lys Thr Lys Pro Arg Glu 165 170 175Glu Gln Tyr Asn Ser Thr Tyr
Arg Val Val Ser Val Leu Thr Val Leu 180 185 190His Gln Asp Trp Leu
Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn 195 200 205Lys Ala Leu
Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly 210 215 220Gln
Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu225 230
235 240Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly Phe
Tyr 245 250 255Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln
Pro Glu Asn 260 265 270Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser
Asp Gly Ser Phe Phe 275 280 285Leu Tyr Ser Lys Leu Thr Val Asp Lys
Ser Arg Trp Gln Gln Gly Asn 290 295 300Val Phe Ser Cys Ser Val Met
His Glu Ala Leu His Asn His Tyr Thr305 310 315 320Gln Lys Ser Leu
Ser Leu Ser Pro Gly Lys 325 330297107PRTArtificial
SequenceSynthetic LC Constant 297Arg Thr Val Ala Ala Pro Ser Val
Phe Ile Phe Pro Pro Ser Asp Glu1 5 10 15Gln Leu Lys Ser Gly Thr Ala
Ser Val Val Cys Leu Leu Asn Asn Phe 20 25 30Tyr Pro Arg Glu Ala Lys
Val Gln Trp Lys Val Asp Asn Ala Leu Gln 35 40 45Ser Gly Asn Ser Gln
Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser 50 55 60Thr Tyr Ser Leu
Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu65 70 75 80Lys His
Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser 85 90 95Pro
Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 100 10529815PRTArtificial
SequenceSynthetic Linker 298Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser
Gly Gly Gly Gly Ser1 5 10 15299288PRTArtificial SequenceSynthetic
murine PD-1 299Met Trp Val Arg Gln Val Pro Trp Ser Phe Thr Trp Ala
Val Leu Gln1 5 10 15Leu Ser Trp Gln Ser Gly Trp Leu Leu Glu Val Pro
Asn Gly Pro Trp 20 25 30Arg Ser Leu Thr Phe Tyr Pro Ala Trp Leu Thr
Val Ser Glu Gly Ala 35 40 45Asn Ala Thr Phe Thr Cys Ser Leu Ser Asn
Trp Ser Glu Asp Leu Met 50 55 60Leu Asn Trp Asn Arg Leu Ser Pro Ser
Asn Gln Thr Glu Lys Gln Ala65 70 75 80Ala Phe Cys Asn Gly Leu Ser
Gln Pro Val Gln Asp Ala Arg Phe Gln 85 90 95Ile Ile Gln Leu Pro Asn
Arg His Asp Phe His Met Asn Ile Leu Asp 100 105 110Thr Arg Arg Asn
Asp Ser Gly Ile Tyr Leu Cys Gly Ala Ile Ser Leu 115 120 125His Pro
Lys Ala Lys Ile Glu Glu Ser Pro Gly Ala Glu Leu Val Val 130 135
140Thr Glu Arg Ile Leu Glu Thr Ser Thr Arg Tyr Pro Ser Pro Ser
Pro145 150 155 160Lys Pro Glu Gly Arg Phe Gln Gly Met Val Ile Gly
Ile Met Ser Ala 165 170 175Leu Val Gly Ile Pro Val Leu Leu Leu Leu
Ala Trp Ala Leu Ala Val 180 185 190Phe Cys Ser Thr Ser Met Ser Glu
Ala Arg Gly Ala Gly Ser Lys Asp 195 200 205Asp Thr Leu Lys Glu Glu
Pro Ser Ala Ala Pro Val Pro Ser Val Ala 210 215 220Tyr Glu Glu Leu
Asp Phe Gln Gly Arg Glu Lys Thr Pro Glu Leu Pro225 230 235 240Thr
Ala Cys Val His Thr Glu Tyr Ala Thr Ile Val Phe Thr Glu Gly 245 250
255Leu Gly Ala Ser Ala Met Gly Arg Arg Gly Ser Ala Asp Gly Leu Gln
260 265 270Gly Pro Arg Pro Pro Arg His Glu Asp Gly His Cys Ser Trp
Pro Leu 275 280 285300288PRTArtificial SequenceSynthetic cyno PD-1
300Met Trp Val Arg Gln Val Pro Trp Ser Phe Thr Trp Ala Val Leu Gln1
5 10 15Leu Ser Trp Gln Ser Gly Trp Leu Leu Glu Val Pro Asn Gly Pro
Trp 20 25 30Arg Ser Leu Thr Phe Tyr Pro Ala Trp Leu Thr Val Ser Glu
Gly Ala 35 40 45Asn Ala Thr Phe Thr Cys Ser Leu Ser Asn Trp Ser Glu
Asp Leu Met 50 55 60Leu Asn Trp Asn Arg Leu Ser Pro Ser Asn Gln Thr
Glu Lys Gln Ala65 70 75 80Ala Phe Cys Asn Gly Leu Ser Gln Pro Val
Gln Asp Ala Arg Phe Gln 85 90 95Ile Ile Gln Leu Pro Asn Arg His Asp
Phe His Met Asn Ile Leu Asp 100 105 110Thr Arg Arg Asn Asp Ser Gly
Ile Tyr Leu Cys Gly Ala Ile Ser Leu 115 120 125His Pro Lys Ala Lys
Ile Glu Glu Ser Pro Gly Ala Glu Leu Val Val 130 135 140Thr Glu Arg
Ile Leu Glu Thr Ser Thr Arg Tyr Pro Ser Pro Ser Pro145 150 155
160Lys Pro Glu Gly Arg Phe Gln Gly Met Val Ile Gly Ile Met Ser Ala
165 170 175Leu Val Gly Ile Pro Val Leu Leu Leu Leu Ala Trp Ala Leu
Ala Val 180 185 190Phe Cys Ser Thr Ser Met Ser Glu Ala Arg Gly Ala
Gly Ser Lys Asp 195 200 205Asp Thr Leu Lys Glu Glu Pro Ser Ala Ala
Pro Val Pro Ser Val Ala 210 215 220Tyr Glu Glu Leu Asp Phe Gln Gly
Arg Glu Lys Thr Pro Glu Leu Pro225 230 235 240Thr Ala Cys Val His
Thr Glu Tyr Ala Thr Ile Val Phe Thr Glu Gly 245 250 255Leu Gly Ala
Ser Ala Met Gly Arg Arg Gly Ser Ala Asp Gly Leu Gln 260 265 270Gly
Pro Arg Pro Pro Arg His Glu Asp Gly His Cys Ser Trp Pro Leu 275 280
2853019PRTArtificial SequenceSynthetic Linker 301Ala Ala Gly Ser
Asp Gln Glu Pro Lys1 5302471PRTArtificial SequenceSynthetic h1E9-4
& h1E9-5, HC-FlagHis 302Met Glu Val Gln Leu Val Glu Ser Gly Gly
Gly Leu Val Gln Pro Gly1 5 10 15Gly Ser Leu Arg Leu Ser Cys Ala Ala
Ser Gly Phe Thr Phe Ser Thr 20 25 30Phe Gly Met Ser Trp Val Arg Gln
Ala Pro Gly Lys Gly Leu Glu Trp 35 40 45Val Ala Thr Ile Ser Gly Gly
Gly Ser Asp Thr Tyr Tyr Pro Asp Ser 50 55 60Val Gln Gly Arg Phe Thr
Ile Ser Arg Asp Asn Ala Lys Asn Ser Leu65 70 75 80Tyr Leu Gln Met
Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr 85 90 95Cys Ala Arg
Gln Gly Tyr Asp Val Tyr Ser Trp Phe Ala Tyr Trp Gly 100 105 110Gln
Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120
125Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala
130 135 140Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val
Thr Val145 150 155 160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val
His Thr Phe Pro Ala 165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser
Leu Ser Ser Val Val Thr Val 180 185 190Pro Ser Ser Ser Leu Gly Thr
Gln Thr Tyr Ile Cys Asn Val Asn His 195 200 205Lys Pro Ser Asn Thr
Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys 210 215 220Asp Lys Thr
His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly225 230 235
240Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met
245 250 255Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val
Ser His 260 265 270Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp
Gly Val Glu Val 275 280 285His Asn Ala Lys Thr Lys Pro Arg Glu Glu
Gln Tyr Asn Ser Thr Tyr 290 295 300Arg Val Val Ser Val Leu Thr Val
Leu His Gln Asp Trp Leu Asn Gly305 310 315 320Lys Glu Tyr Lys Cys
Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile 325 330 335Glu Lys Thr
Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val 340 345 350Tyr
Thr Leu Pro Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser 355 360
365Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu
370 375 380Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr
Pro Pro385 390 395 400Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr
Ser Lys Leu Thr Val 405 410 415Asp Lys Ser Arg Trp Gln Gln Gly Asn
Val Phe Ser Cys Ser Val Met 420 425 430His Glu Ala Leu His Asn His
Tyr Thr Gln Lys Ser Leu Ser Leu Ser 435 440 445Pro Gly Lys Gly Ser
Gly Asp Tyr Lys Asp Asp Asp Asp Lys Gly Ser 450 455 460Gly His His
His His His His465 470303219PRTArtificial SequenceSynthetic h1E9-5,
LC 303Met Glu Ile Val Leu Thr Gln Ser Pro Ala Thr Leu Ser Leu Ser
Pro1 5 10 15Gly Glu Arg Ala Thr Leu Ser Cys Arg Ala Ser Glu Ser Val
Asp Asn 20 25 30Ser Gly Ile Ser Phe Met Ser Trp Tyr Gln Gln Lys Pro
Gly Gln Ala 35 40 45Pro Arg Leu Leu Ile Tyr Thr Ala Ser Asn Gln Gly
Ser Gly Ile Pro 50 55 60Ala Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp
Phe Thr Leu Thr Ile65 70 75 80Ser Ser Leu Glu Pro Glu Asp Phe Ala
Val Tyr Tyr Cys Gln Gln Ser 85 90 95Lys Glu Val Pro Trp Thr Phe Gly
Gln Gly Thr Lys Val Glu Ile Lys 100 105 110Arg Thr Val Ala Ala Pro
Ser Val Phe Ile Phe Pro Pro Ser Asp Glu 115 120 125Gln Leu Lys Ser
Gly Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe 130 135 140Tyr Pro
Arg Glu Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln145 150 155
160Ser Gly Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser
165 170 175Thr Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp
Tyr Glu 180 185 190Lys His Lys Val Tyr Ala Cys Glu Val Thr His Gln
Gly Leu Ser Ser 195 200 205Pro Val Thr Lys Ser Phe Asn Arg Gly Glu
Cys 210 215304219PRTArtificial SequenceSynthetic h1E9-4, LC 304Met
Asp Ile Gln Leu Thr Gln Ser Pro Ser Phe Leu Ser Ala Ser Val1 5 10
15Gly Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Glu Ser Val Asp Asn
20 25 30Ser Gly Ile Ser Phe Met Ser Trp Tyr Gln Gln Lys Pro Gly Lys
Ala 35 40 45Pro Lys Leu Leu Ile Tyr Thr Ala Ser Asn Gln Gly Ser Gly
Val Pro 50 55 60Ser Arg Phe Ser Gly Ser Gly Ser Gly Thr Glu Phe Thr
Leu Thr Ile65 70 75 80Ser Ser Leu Gln Pro Glu Asp Phe Ala Thr Tyr
Tyr Cys Gln Gln Ser 85 90 95Lys Glu Val Pro Trp Thr Phe Gly Gln Gly
Thr Lys Val Glu Ile Lys 100 105 110Arg Thr Val Ala Ala
Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu 115 120 125Gln Leu Lys
Ser Gly Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe 130 135 140Tyr
Pro Arg Glu Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln145 150
155 160Ser Gly Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp
Ser 165 170 175Thr Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala
Asp Tyr Glu 180 185 190Lys His Lys Val Tyr Ala Cys Glu Val Thr His
Gln Gly Leu Ser Ser 195 200 205Pro Val Thr Lys Ser Phe Asn Arg Gly
Glu Cys 210 215305507PRTArtificial SequenceSynthetic
PD1-F2v-scFvFcFlagHis, scFv-Fc 305Met Gly Ala His Ser Glu Val Gln
Leu Val Gln Ser Gly Gly Gly Val1 5 10 15Val Gln Pro Gly Arg Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe 20 25 30Thr Phe Ser Ser Tyr Trp
Met Ser Trp Val Arg Gln Ala Pro Gly Lys 35 40 45Gly Leu Glu Trp Val
Ser Ala Ile Ser Gly Ser Gly Gly Ser Thr Tyr 50 55 60Tyr Ala Asp Ser
Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser65 70 75 80Lys Asn
Thr Leu Tyr Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr 85 90 95Ala
Val Tyr Tyr Cys Ala Lys Glu Asn Trp Gly Ser Tyr Phe Asp Leu 100 105
110Trp Gly Gln Gly Thr Thr Val Thr Val Ser Ser Gly Gly Gly Gly Ser
115 120 125Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly Val His Ser
Asp Ile 130 135 140Val Met Thr Gln Ser Pro Ser Thr Leu Ser Ala Ser
Val Gly Asp Arg145 150 155 160Val Thr Ile Thr Cys Arg Ala Ser Gln
Gly Ile Ser Ser Trp Leu Ala 165 170 175Trp Tyr Gln Gln Lys Pro Gly
Arg Ala Pro Lys Val Leu Ile Tyr Lys 180 185 190Ala Ser Thr Leu Glu
Ser Gly Val Pro Ser Arg Phe Ser Gly Ser Gly 195 200 205Ser Gly Thr
Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro Glu Asp 210 215 220Phe
Ala Thr Tyr Tyr Cys Gln Gln Ser Tyr Ser Thr Pro Trp Thr Phe225 230
235 240Gly Gln Gly Thr Lys Leu Glu Ile Lys Ala Ala Gly Ser Asp Gln
Glu 245 250 255Pro Lys Ser Ser Asp Lys Thr His Thr Cys Pro Pro Cys
Ser Ala Pro 260 265 270Glu Leu Leu Gly Gly Ser Ser Val Phe Leu Phe
Pro Pro Lys Pro Lys 275 280 285Asp Thr Leu Met Ile Ser Arg Thr Pro
Glu Val Thr Cys Val Val Val 290 295 300Asp Val Ser His Glu Asp Pro
Glu Val Lys Phe Asn Trp Tyr Val Asp305 310 315 320Gly Val Glu Val
His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr 325 330 335Asn Ser
Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp 340 345
350Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu
355 360 365Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln
Pro Arg 370 375 380Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg Asp
Glu Leu Thr Lys385 390 395 400Asn Gln Val Ser Leu Thr Cys Leu Val
Lys Gly Phe Tyr Pro Ser Asp 405 410 415Ile Ala Val Glu Trp Glu Ser
Asn Gly Gln Pro Glu Asn Asn Tyr Lys 420 425 430Thr Thr Pro Pro Val
Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser 435 440 445Lys Leu Thr
Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser 450 455 460Cys
Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser465 470
475 480Leu Ser Leu Ser Pro Gly Lys Gly Ser Gly Asp Tyr Lys Asp Asp
Asp 485 490 495Asp Lys Gly Ser Gly His His His His His His 500
505306118PRTArtificial SequenceSynthetic PD1-F2v, VH 306Glu Val Gln
Leu Val Gln Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30Trp
Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ser Ala Ile Ser Gly Ser Gly Gly Ser Thr Tyr Tyr Ala Asp Ser Val
50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu
Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val
Tyr Tyr Cys 85 90 95Ala Lys Glu Asn Trp Gly Ser Tyr Phe Asp Leu Trp
Gly Gln Gly Thr 100 105 110Thr Val Thr Val Ser Ser
115307452PRTArtificial SequenceSynthetic PD1-F2v, HC 307Gly Ala His
Ser Glu Val Gln Leu Val Gln Ser Gly Gly Gly Val Val1 5 10 15Gln Pro
Gly Arg Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr 20 25 30Phe
Ser Ser Tyr Trp Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly 35 40
45Leu Glu Trp Val Ser Ala Ile Ser Gly Ser Gly Gly Ser Thr Tyr Tyr
50 55 60Ala Asp Ser Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser
Lys65 70 75 80Asn Thr Leu Tyr Leu Gln Met Asn Ser Leu Arg Ala Glu
Asp Thr Ala 85 90 95Val Tyr Tyr Cys Ala Lys Glu Asn Trp Gly Ser Tyr
Phe Asp Leu Trp 100 105 110Gly Gln Gly Thr Thr Val Thr Val Ser Ser
Ala Ser Thr Lys Gly Pro 115 120 125Ser Val Phe Pro Leu Ala Pro Ser
Ser Lys Ser Thr Ser Gly Gly Thr 130 135 140Ala Ala Leu Gly Cys Leu
Val Lys Asp Tyr Phe Pro Glu Pro Val Thr145 150 155 160Val Ser Trp
Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro 165 170 175Ala
Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr 180 185
190Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn
195 200 205His Lys Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro
Lys Ser 210 215 220Cys Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala
Pro Glu Leu Leu225 230 235 240Gly Gly Pro Ser Val Phe Leu Phe Pro
Pro Lys Pro Lys Asp Thr Leu 245 250 255Met Ile Ser Arg Thr Pro Glu
Val Thr Cys Val Val Val Asp Val Ser 260 265 270His Glu Asp Pro Glu
Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu 275 280 285Val His Asn
Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr 290 295 300Tyr
Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn305 310
315 320Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala
Pro 325 330 335Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg
Glu Pro Gln 340 345 350Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu Met
Thr Lys Asn Gln Val 355 360 365Ser Leu Thr Cys Leu Val Lys Gly Phe
Tyr Pro Ser Asp Ile Ala Val 370 375 380Glu Trp Glu Ser Asn Gly Gln
Pro Glu Asn Asn Tyr Lys Thr Thr Pro385 390 395 400Pro Val Leu Asp
Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr 405 410 415Val Asp
Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val 420 425
430Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu
435 440 445Ser Pro Gly Lys 450308220PRTArtificial SequenceSynthetic
PD1-F2v, LC 308Gly Val His Ser Asp Ile Val Met Thr Gln Ser Pro Ser
Thr Leu Ser1 5 10 15Ala Ser Val Gly Asp Arg Val Thr Ile Thr Cys Arg
Ala Ser Gln Gly 20 25 30Ile Ser Ser Trp Leu Ala Trp Tyr Gln Gln Lys
Pro Gly Arg Ala Pro 35 40 45Lys Val Leu Ile Tyr Lys Ala Ser Thr Leu
Glu Ser Gly Val Pro Ser 50 55 60Arg Phe Ser Gly Ser Gly Ser Gly Thr
Asp Phe Thr Leu Thr Ile Ser65 70 75 80Ser Leu Gln Pro Glu Asp Phe
Ala Thr Tyr Tyr Cys Gln Gln Ser Tyr 85 90 95Ser Thr Pro Trp Thr Phe
Gly Gln Gly Thr Lys Leu Glu Ile Lys Arg 100 105 110His Met Thr Val
Ala Ala Pro Ser Val Phe Ile Phe Pro Pro Ser Asp 115 120 125Glu Gln
Leu Lys Ser Gly Thr Ala Ser Val Val Cys Leu Leu Asn Asn 130 135
140Phe Tyr Pro Arg Glu Ala Lys Val Gln Trp Lys Val Asp Asn Ala
Leu145 150 155 160Gln Ser Gly Asn Ser Gln Glu Ser Val Thr Glu Gln
Asp Ser Lys Asp 165 170 175Ser Thr Tyr Ser Leu Ser Ser Thr Leu Thr
Leu Ser Lys Ala Asp Tyr 180 185 190Glu Lys His Lys Val Tyr Ala Cys
Glu Val Thr His Gln Gly Leu Ser 195 200 205Ser Pro Val Thr Lys Ser
Phe Asn Arg Gly Glu Cys 210 215 22030910PRTArtificial
SequenceSynthetic 1353-E07-R5, CDR-H3 309Asp Ala Glu Tyr Arg Leu
Gly Ser Gly Tyr1 5 1031010PRTArtificial SequenceSynthetic
1353-A09-R5, CDR-H3 310Asp Ser Glu Tyr Arg Ser Gly Ser Gly Tyr1 5
1031110PRTArtificial SequenceSynthetic 1353-F09-R5, CDR-H3 311Asp
Ala Glu Tyr Arg Ser Gly Ser Gly Tyr1 5 1031210PRTArtificial
SequenceSynthetic 1353-G08-R5, CDR-H3 312Asp Ala Asp Tyr Arg Ser
Gly Ser Gly Tyr1 5 1031310PRTArtificial SequenceSynthetic
1353-G10-R5, CDR-H3 313Asp Val Asp Tyr Arg Thr Gly Ser Gly Tyr1 5
1031410PRTArtificial SequenceSynthetic 1353-C07-R5, CDR-H3 314Asp
Val Asp Tyr Arg Ser Gly Ser Gly Tyr1 5 1031510PRTArtificial
SequenceSynthetic 1353-H08-R5, CDR-H3 315Asp Val Asp Tyr Arg Ser
Gly Ser Gly Tyr1 5 10316119PRTArtificial SequenceSynthetic
1353-E07-R5, VH 316Glu Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Arg Phe Glu Thr Tyr 20 25 30Gly Ile Ser Trp Val Arg Gln Ala Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Trp Ile Ser Ala Tyr Asn Gly Asn
Thr Asn Tyr Ala Gln Lys Leu 50 55 60Gln Gly Arg Val Thr Met Thr Thr
Asp Thr Ser Thr Asn Thr Ala Tyr65 70 75 80Met Glu Leu Arg Ser Leu
Arg Ser Asp Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Ala Glu
Tyr Arg Leu Gly Ser Gly Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val
Thr Val Ser Ser 115317119PRTArtificial SequenceSynthetic
1353-A09-R5, VH 317Glu Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Arg Phe Thr Trp Tyr 20 25 30Gly Ile Ser Trp Val Arg Gln Ala Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Trp Ile Ser Ala Tyr Asn Gly Asn
Thr Asn Tyr Ala Gln Lys Leu 50 55 60Gln Gly Arg Val Thr Met Thr Thr
Asp Thr Ser Thr Asn Thr Ala Tyr65 70 75 80Met Glu Leu Arg Ser Leu
Arg Ser Asp Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Ser Glu
Tyr Arg Ser Gly Ser Gly Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val
Thr Val Ser Ser 115318119PRTArtificial SequenceSynthetic
1353-F09-R5, VH 318Glu Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Arg Phe Arg Gln Tyr 20 25 30Gly Ile Ser Trp Val Arg Gln Ala Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Trp Ile Ser Ala Tyr Asn Gly Asn
Thr Asn Tyr Ala Gln Lys Leu 50 55 60Gln Gly Arg Val Thr Met Thr Thr
Asp Thr Ser Thr Asn Thr Ala Tyr65 70 75 80Met Glu Leu Arg Ser Leu
Arg Ser Asp Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Ala Glu
Tyr Arg Ser Gly Ser Gly Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val
Thr Val Ser Ser 115319119PRTArtificial SequenceSynthetic
1353-G08-R5, VH 319Glu Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Arg Phe Thr Arg Tyr 20 25 30Gly Ile Ser Trp Val Arg Gln Ala Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Trp Val Ser Ala His Asn Gly Asn
Thr Asn Tyr Ala Gln Lys Leu 50 55 60Gln Gly Arg Val Thr Met Thr Thr
Asp Thr Ser Thr Asn Thr Ala Tyr65 70 75 80Met Glu Leu Arg Ser Leu
Arg Ser Asp Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Ala Asp
Tyr Arg Ser Gly Ser Gly Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val
Thr Val Ser Ser 115320119PRTArtificial SequenceSynthetic
1353-G10-R5, VH 320Glu Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Arg Phe Pro His Tyr 20 25 30Gly Ile Ser Trp Val Arg Gln Ala Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Trp Ile Ser Ala Tyr Asn Gly Asn
Thr Asn Tyr Ala Gln Lys Leu 50 55 60Gln Gly Arg Val Thr Met Thr Thr
Asp Thr Ser Thr Asn Thr Ala Tyr65 70 75 80Met Glu Leu Arg Ser Leu
Arg Ser Asp Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Val Asp
Tyr Arg Thr Gly Ser Gly Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val
Thr Val Ser Ser 115321119PRTArtificial SequenceSynthetic
1353-C07-R5, VH 321Glu Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Arg Phe Ser Thr Phe 20 25 30Gly Ile Ser Trp Val Arg Gln Ala Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Trp Ile Ser Ala Tyr Asn Gly Asn
Thr Asn Tyr Ala Gln Lys Leu 50 55 60Gln Gly Arg Val Thr Met Thr Thr
Asp Thr Ser Thr Asn Thr Ala Tyr65 70 75 80Met Glu Leu Arg Ser Leu
Arg Ser Asp Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Val Asp
Tyr Arg Ser Gly Ser Gly Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val
Thr Val Ser Ser 115322119PRTArtificial SequenceSynthetic
1353-H08-R5, VH 322Glu Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Arg Phe Thr Arg Gln 20 25 30Gly Ile Ser Trp Val Arg Gln Ala Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Trp Ile Ser Ala Tyr Asn Gly Asn
Thr Lys Tyr Ala Gln Lys Leu 50 55 60Gln Gly Arg Val Thr Met Thr Thr
Asp Thr Ser Thr Asn Thr Ala Tyr65 70 75 80Met Glu Leu Arg Ser Leu
Arg Ser Asp Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Val Asp
Tyr Arg Ser Gly Ser Gly Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val
Thr Val Ser Ser 115323119PRTArtificial SequenceSynthetic
1353-H09-R5, VH 323Glu Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Arg Phe Pro His
Tyr 20 25 30Gly Ile Ser Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Trp Ile Ser Ala Tyr Asn Gly Asn Thr Asn Tyr Ala
Gln Lys Leu 50 55 60Gln Gly Arg Val Thr Met Thr Thr Asp Thr Ser Thr
Asn Thr Ala Tyr65 70 75 80Met Glu Leu Arg Ser Leu Arg Ser Asp Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Ala Glu Tyr Arg Ser Gly
Ser Gly Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val Thr Val Ser Ser
115324471PRTArtificial SequenceSynthetic H1E9-HC3, HC-FlagHis
324Met Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly1
5 10 15Gly Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser
Thr 20 25 30Phe Gly Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu
Glu Trp 35 40 45Val Ala Thr Ile Ser Gly Gly Gly Ser Asp Thr Tyr Tyr
Pro Asp Ser 50 55 60Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala
Lys Asn Ser Leu65 70 75 80Tyr Leu Gln Met Asn Ser Leu Arg Ala Glu
Asp Thr Ala Val Tyr Tyr 85 90 95Cys Ala Arg Gln Gly Tyr Asp Val Tyr
Ser Trp Phe Ala Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135 140Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150 155
160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala
165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
Thr Val 180 185 190Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys
Asn Val Asn His 195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys Lys
Val Glu Pro Lys Ser Cys 210 215 220Asp Lys Thr His Thr Cys Pro Pro
Cys Pro Ala Pro Glu Leu Leu Gly225 230 235 240Gly Pro Ser Val Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 245 250 255Ile Ser Arg
Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His 260 265 270Glu
Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val 275 280
285His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr
290 295 300Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu
Asn Gly305 310 315 320Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala
Leu Pro Ala Pro Ile 325 330 335Glu Lys Thr Ile Ser Lys Ala Lys Gly
Gln Pro Arg Glu Pro Gln Val 340 345 350Tyr Thr Leu Pro Pro Ser Arg
Glu Glu Met Thr Lys Asn Gln Val Ser 355 360 365Leu Thr Cys Leu Val
Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu 370 375 380Trp Glu Ser
Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro385 390 395
400Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val
405 410 415Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser
Val Met 420 425 430His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser
Leu Ser Leu Ser 435 440 445Pro Gly Lys Gly Ser Gly Asp Tyr Lys Asp
Asp Asp Asp Lys Gly Ser 450 455 460Gly His His His His His His465
470325471PRTArtificial SequenceSynthetic H1E9-HC2, HC-FlagHis
325Met Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly1
5 10 15Arg Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser
Thr 20 25 30Phe Gly Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu
Glu Trp 35 40 45Val Ala Thr Ile Ser Gly Gly Gly Ser Asp Thr Tyr Tyr
Pro Asp Ser 50 55 60Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser
Lys Asn Thr Leu65 70 75 80Tyr Leu Gln Met Asn Ser Leu Arg Ala Glu
Asp Thr Ala Val Tyr Tyr 85 90 95Cys Ala Arg Gln Gly Tyr Asp Val Tyr
Ser Trp Phe Ala Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135 140Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150 155
160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala
165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
Thr Val 180 185 190Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys
Asn Val Asn His 195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys Lys
Val Glu Pro Lys Ser Cys 210 215 220Asp Lys Thr His Thr Cys Pro Pro
Cys Pro Ala Pro Glu Leu Leu Gly225 230 235 240Gly Pro Ser Val Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 245 250 255Ile Ser Arg
Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His 260 265 270Glu
Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val 275 280
285His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr
290 295 300Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu
Asn Gly305 310 315 320Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala
Leu Pro Ala Pro Ile 325 330 335Glu Lys Thr Ile Ser Lys Ala Lys Gly
Gln Pro Arg Glu Pro Gln Val 340 345 350Tyr Thr Leu Pro Pro Ser Arg
Glu Glu Met Thr Lys Asn Gln Val Ser 355 360 365Leu Thr Cys Leu Val
Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu 370 375 380Trp Glu Ser
Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro385 390 395
400Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val
405 410 415Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser
Val Met 420 425 430His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser
Leu Ser Leu Ser 435 440 445Pro Gly Lys Gly Ser Gly Asp Tyr Lys Asp
Asp Asp Asp Lys Gly Ser 450 455 460Gly His His His His His His465
470326471PRTArtificial SequenceSynthetic H1E9-HC1, HC-FlagHis
326Met Glu Val Gln Leu Leu Glu Ser Gly Gly Gly Leu Val Gln Pro Gly1
5 10 15Gly Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser
Thr 20 25 30Phe Gly Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu
Glu Trp 35 40 45Val Ala Thr Ile Ser Gly Gly Gly Ser Asp Thr Tyr Tyr
Pro Asp Ser 50 55 60Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser
Lys Asn Thr Leu65 70 75 80Tyr Leu Gln Met Asn Ser Leu Arg Ala Glu
Asp Thr Ala Val Tyr Tyr 85 90 95Cys Ala Arg Gln Gly Tyr Asp Val Tyr
Ser Trp Phe Ala Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135 140Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150 155
160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala
165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
Thr Val 180 185 190Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys
Asn Val Asn His 195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys Lys
Val Glu Pro Lys Ser Cys 210 215 220Asp Lys Thr His Thr Cys Pro Pro
Cys Pro Ala Pro Glu Leu Leu Gly225 230 235 240Gly Pro Ser Val Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 245 250 255Ile Ser Arg
Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His 260 265 270Glu
Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val 275 280
285His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr
290 295 300Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu
Asn Gly305 310 315 320Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala
Leu Pro Ala Pro Ile 325 330 335Glu Lys Thr Ile Ser Lys Ala Lys Gly
Gln Pro Arg Glu Pro Gln Val 340 345 350Tyr Thr Leu Pro Pro Ser Arg
Glu Glu Met Thr Lys Asn Gln Val Ser 355 360 365Leu Thr Cys Leu Val
Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu 370 375 380Trp Glu Ser
Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro385 390 395
400Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val
405 410 415Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser
Val Met 420 425 430His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser
Leu Ser Leu Ser 435 440 445Pro Gly Lys Gly Ser Gly Asp Tyr Lys Asp
Asp Asp Asp Lys Gly Ser 450 455 460Gly His His His His His His465
470327219PRTArtificial SequenceSynthetic H1E9-LC4, LC 327Met Asp
Ile Val Leu Thr Gln Ser Pro Asp Ser Leu Ala Val Ser Leu1 5 10 15Gly
Glu Arg Ala Thr Ile Asn Cys Arg Ala Ser Glu Ser Val Asp Asn 20 25
30Ser Gly Ile Ser Phe Met Ser Trp Tyr Gln Gln Lys Pro Gly Gln Pro
35 40 45Pro Lys Leu Leu Ile Tyr Thr Ala Ser Asn Gln Gly Ser Gly Val
Pro 50 55 60Asp Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu
Thr Ile65 70 75 80Ser Ser Leu Gln Ala Glu Asp Val Ala Val Tyr Tyr
Cys Gln Gln Ser 85 90 95Lys Glu Val Pro Trp Thr Phe Gly Gln Gly Thr
Lys Val Glu Ile Lys 100 105 110Arg Thr Val Ala Ala Pro Ser Val Phe
Ile Phe Pro Pro Ser Asp Glu 115 120 125Gln Leu Lys Ser Gly Thr Ala
Ser Val Val Cys Leu Leu Asn Asn Phe 130 135 140Tyr Pro Arg Glu Ala
Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln145 150 155 160Ser Gly
Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser 165 170
175Thr Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu
180 185 190Lys His Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu
Ser Ser 195 200 205Pro Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 210
215328219PRTArtificial SequenceSynthetic H1E9-LC3, LC 328Met Glu
Ile Val Leu Thr Gln Ser Pro Gly Thr Leu Ser Leu Ser Pro1 5 10 15Gly
Glu Arg Ala Thr Leu Ser Cys Arg Ala Ser Glu Ser Val Asp Asn 20 25
30Ser Gly Ile Ser Phe Met Ser Trp Tyr Gln Gln Lys Pro Gly Gln Ala
35 40 45Pro Arg Leu Leu Ile Tyr Thr Ala Ser Asn Gln Gly Ser Gly Ile
Pro 50 55 60Asp Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu
Thr Ile65 70 75 80Ser Arg Leu Glu Pro Glu Asp Phe Ala Val Tyr Tyr
Cys Gln Gln Ser 85 90 95Lys Glu Val Pro Trp Thr Phe Gly Gln Gly Thr
Lys Val Glu Ile Lys 100 105 110Arg Thr Val Ala Ala Pro Ser Val Phe
Ile Phe Pro Pro Ser Asp Glu 115 120 125Gln Leu Lys Ser Gly Thr Ala
Ser Val Val Cys Leu Leu Asn Asn Phe 130 135 140Tyr Pro Arg Glu Ala
Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln145 150 155 160Ser Gly
Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser 165 170
175Thr Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu
180 185 190Lys His Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu
Ser Ser 195 200 205Pro Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 210
215329219PRTArtificial SequenceSynthetic H1E9-LC2, LC 329Met Asp
Ile Val Leu Thr Gln Ser Pro Leu Ser Leu Pro Val Thr Pro1 5 10 15Gly
Glu Pro Ala Ser Ile Ser Cys Arg Ala Ser Glu Ser Val Asp Asn 20 25
30Ser Gly Ile Ser Phe Met Ser Trp Tyr Leu Gln Lys Pro Gly Gln Ser
35 40 45Pro Gln Leu Leu Ile Tyr Thr Ala Ser Asn Gln Gly Ser Gly Val
Pro 50 55 60Asp Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu
Lys Ile65 70 75 80Ser Arg Val Glu Ala Glu Asp Val Gly Val Tyr Tyr
Cys Gln Gln Ser 85 90 95Lys Glu Val Pro Trp Thr Phe Gly Gln Gly Thr
Lys Val Glu Ile Lys 100 105 110Arg Thr Val Ala Ala Pro Ser Val Phe
Ile Phe Pro Pro Ser Asp Glu 115 120 125Gln Leu Lys Ser Gly Thr Ala
Ser Val Val Cys Leu Leu Asn Asn Phe 130 135 140Tyr Pro Arg Glu Ala
Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln145 150 155 160Ser Gly
Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser 165 170
175Thr Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu
180 185 190Lys His Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu
Ser Ser 195 200 205Pro Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 210
215330219PRTArtificial SequenceSynthetic H1E9-LC1, LC 330Met Asp
Ile Gln Leu Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val1 5 10 15Gly
Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Glu Ser Val Asp Asn 20 25
30Ser Gly Ile Ser Phe Met Ser Trp Tyr Gln Gln Lys Pro Gly Lys Ala
35 40 45Pro Lys Leu Leu Ile Tyr Thr Ala Ser Asn Gln Gly Ser Gly Val
Pro 50 55 60Ser Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Phe
Thr Ile65 70 75 80Ser Ser Leu Gln Pro Glu Asp Ile Ala Thr Tyr Tyr
Cys Gln Gln Ser 85 90 95Lys Glu Val Pro Trp Thr Phe Gly Gln Gly Thr
Lys Val Glu Ile Lys 100 105 110Arg Thr Val Ala Ala Pro Ser Val Phe
Ile Phe Pro Pro Ser Asp Glu 115 120 125Gln Leu Lys Ser Gly Thr Ala
Ser Val Val Cys Leu Leu Asn Asn Phe 130 135 140Tyr Pro Arg Glu Ala
Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln145 150 155 160Ser Gly
Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser 165 170
175Thr Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu
180 185 190Lys His Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu
Ser Ser 195 200 205Pro Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 210
21533117PRTArtificial SequenceSynthetic CDR-H2 331Thr Ile Ser Gly
Gly Gly Ser Asp Thr Tyr Tyr Pro Asp Ser Val Gln1 5 10 15Gly
D00001

D00002

D00003

D00004

D00005

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.