Chimeric Antigen Receptor
Pule; Martin ; et al.
U.S. patent application number 17/077782 was filed with the patent office on 2021-04-08 for chimeric antigen receptor. The applicant listed for this patent is AUTOLUS LIMITED. Invention is credited to Ben Draper, Lydia Lee, Martin Pule, Kwee Yong.
| Application Number | 20210101959 17/077782 |
| Document ID | / |
| Family ID | 1000005277982 |
| Filed Date | 2021-04-08 |





View All Diagrams
| United States Patent Application | 20210101959 |
| Kind Code | A1 |
| Pule; Martin ; et al. | April 8, 2021 |
CHIMERIC ANTIGEN RECEPTOR
Abstract
The present invention provides a chimeric antigen recept- or (CAR) comprising: (i) a B cell maturation antigen (BCMA)-binding domain which comprises at least part of a proliferation-inducing ligand (APRIL); (ii) a spacer domain (iii) a transmembrane domain; and (iv) an intracellular T cell signaling domain. The invention also provides the use of such a T-cell expressing such a CAR in the treatment of plasma-cell mediated diseases, such as multiple myeloma.
| Inventors: | Pule; Martin; (London, GB) ; Yong; Kwee; (London, GB) ; Lee; Lydia; (London, GB) ; Draper; Ben; (London, GB) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 1000005277982 | ||||||||||
| Appl. No.: | 17/077782 | ||||||||||
| Filed: | October 22, 2020 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 16162747 | Oct 17, 2018 | 10919951 | ||
| 17077782 | ||||
| 15028064 | Apr 8, 2016 | 10160794 | ||
| PCT/GB2014/053058 | Oct 10, 2014 | |||
| 16162747 | ||||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C07K 16/18 20130101; A61K 48/00 20130101; C07K 14/7051 20130101; C07K 14/70575 20130101; C07K 14/70578 20130101; C12N 15/85 20130101; A61K 35/17 20130101 |
| International Class: | C07K 14/705 20060101 C07K014/705; A61K 35/17 20060101 A61K035/17; C07K 14/725 20060101 C07K014/725; C07K 16/18 20060101 C07K016/18; C12N 15/85 20060101 C12N015/85 |
Foreign Application Data
| Date | Code | Application Number |
|---|---|---|
| Oct 10, 2013 | GB | 1317929.6 |
Claims
1-18. (canceled)
19. A nucleic acid encoding a chimeric antigen receptor (CAR) which comprises: (i) truncated proliferation-inducing ligand (APRIL) that (a) binds B cell maturation antigen (BCMA) and (b) binds transmembrane activator and calcium modulator and cyclophilin ligand interactor (TACI); (ii) a spacer domain; and (ii) a transmembrane domain; and (iii) an intracellular T cell signaling domain.
20. The nucleic acid according to claim 19, wherein the truncated APRIL lacks the amino terminal portion of APRIL responsible for proteoglycan binding.
21. The nucleic acid according to claim 20, which comprises the sequence shown as SEQ ID NO: 14.
22. The nucleic acid according to claim 19, wherein the transmembrane and intracellular T-cell signalling domain comprise the sequence shown as SEQ ID NO: 7.
23. The nucleic acid according to claim 19, wherein the spacer comprises one of the following: a human IgG1 spacer, an IgG1 hinge, or a CD8 stalk.
24. The nucleic acid according to claim 23, wherein the spacer comprises a CD8 stalk.
25. The nucleic acid according to claim 19 which encodes the sequence shown as SEQ ID NO: 1, 2, 3, 4, 5 or 6.
26. A nucleic acid sequence according to claim 19 which comprises the sequence shown as SEQ ID NO: 15, 16, 17, 18, 19 or 20.
27. A vector which comprises a nucleic acid sequence according to claim 19.
28. A T cell or NK cell which expresses a CAR according to claim 19.
29. A method for making a T cell or NK cell, which comprises the step of introducing a nucleic acid according to claim 19 into a T cell or NK cell.
30. A pharmaceutical composition which comprises a vector according to claim 27 together with a pharmaceutically acceptable carrier, diluent or excipient.
31. A pharmaceutical composition which comprises a cell according to claim 28 together with a pharmaceutically acceptable carrier, diluent or excipient.
Description
FIELD OF THE INVENTION
[0001] The present invention relates to chimeric antigen receptor (CAR) which binds the B cell maturation antigen (BCMA). T cells expressing such a CAR are useful in the treatment of plasma cell diseases such as multiple myeloma.
BACKGROUND TO THE INVENTION
[0002] Multiple Myeloma
[0003] Multiple Myeloma (myeloma) is a bone-marrow malignancy of plasma cells. Collections of abnormal plasma cells accumulate in the bone marrow, where they interfere with the production of normal blood cells. Myeloma is the second most common hematological malignancy in the U.S. (after non-Hodgkin lymphoma), and constitutes 13% of haematologic malignancies and 1% of all cancers. The disease is burdensome in terms of suffering as well as medical expenditure since it causes pathological fractures, susceptibility to infection, renal and then bone-marrow failure before death.
[0004] Unlike many lymphomas, myeloma is currently incurable. Standard chemotherapy agents used in lymphoma are largely ineffective for myeloma. In addition, since CD20 expression is lost in plasma cells, Rituximab cannot be used against this disease. New agents such as Bortezamib and Lenolidomide are partially effective, but fail to lead to long-lasting remissions.
[0005] There is thus a need for alternative agents for the treatment of myeloma which have increased efficacy and improved long-term effects.
[0006] Chimeric Antigen Receptors (CARs)
[0007] Chimeric antigen receptors are proteins which, in their usual format, graft the specificity of a monoclonal antibody (mAb) to the effector function of a T-cell. Their usual form is that of a type I transmembrane domain protein with an antigen recognizing amino terminus, a spacer, a transmembrane domain all connected to a compound endodomain which transmits T-cell survival and activation signals (see FIG. 3).
[0008] The most common form of these molecules use single-chain variable fragments (scFv) derived from monoclonal antibodies to recognize a target antigen. The scFv is fused via a spacer and a transmembrane domain to a signaling endodomain. Such molecules result in activation of the T-cell in response to recognition by the scFv of its target. When T cells express such a CAR, they recognize and kill target cells that express the target antigen. Several CARs have been developed against tumour associated antigens, and adoptive transfer approaches using such CAR-expressing T cells are currently in clinical trial for the treatment of various cancers. Carpenter et al (2013, Clin Cancer Res 19(8) 2048-60) describe a CAR which incorporates a scFv against the B-cell maturation antigen (BCMA).
[0009] BCMA is a transmembrane protein that is preferentially expressed in mature lymphocytes, i.e. memory B cells, plasmablasts and bone marrow plasma cells. BCMA is also expressed on multiple myeloma cells.
[0010] Carpenter eta/demonstrate that T cells transduced to express the anti-BCMA CAR are capable of specifically killing myeloma cells from a plasmacytoma of a myeloma patient.
[0011] Although CAR approaches using anti-BCMA antibodies show promise, a particular consideration when targeting this antigen is the particularly low density of BCMA on myeloma cells, in comparison for instance with CD19 on a lymphoma cell. Hence there is a need to increase the sensitivity of target cell recognition of an anti-BCMA CAR T cell.
DESCRIPTION OF THE FIGURES
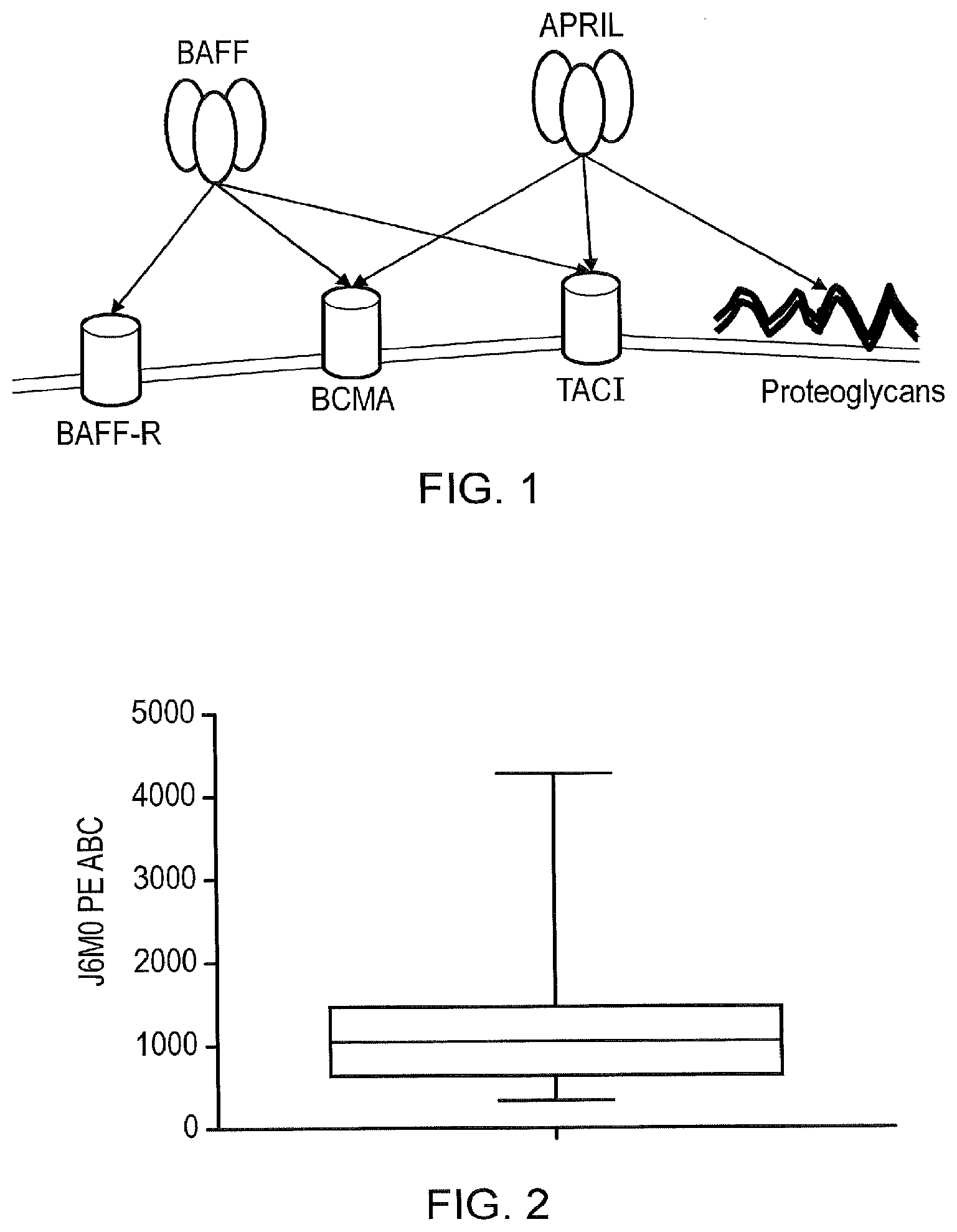
[0012] FIG. 1--Ligand Specificity and Function Assignment of APRIL and BAFF
[0013] B-cell-activating factor (BAFF, TNFSF13B) interacts with BAFF-Receptor (BAFF-R, TNFRSF13C), B-cell membrane antigen (BCMA, TNFRSF17) and transmembrane activator and calcium modulator and cyclophilin ligand interactor (TACI, TNFRSF13B) while A proliferation-inducing ligand (APRIL, TNFSF13) interacts with BCMA, TACI and proteoglycans. BAFF-R activation affects peripheral B-cell survival, while BCMA may affect plasma cell survival. APRIL interaction with proteoglycans involves acidic sulphated glycol-saminoglycan side-chain containing amino-terminus of APRIL.
[0014] FIG. 2--Expression Data of BCMA on Myeloma
[0015] Myeloma cells from bone marrow samples from 39 multiple myeloma patients were isolated by a CD138+ magnetic bead selection. These cells were stained with the anti-BCMA monoclonal antibody J6MO conjugated with PE (GSK). Antigen copy number was quantified using PE Quantibrite beads (Becton Dickenson) as per the manufacturer's instructions. A box and whiskers plot of antigen copy number is presented along with the range, interquartile and median values plotted. We found the range is 348.7-4268.4 BCMA copies per cell with a mean of 1181 and a median of 1084.9.
[0016] FIG. 3--Standard Design of a Chimeric Antigen Receptor
[0017] The typical format of a chimeric antigen receptor is shown. These are type I transmembrane proteins. An ectodomain recognizes antigen. This is composed of an antibody derived single-chain variable fragment (scFv) which is attached to a spacer domain. This in turn is connected to a transmembrane domain which acts to anchor the molecule in the membrane. Finally, this is connected to an endodomain which acts to transmits intracellular signals to the cell. This is composed of one or more signalling domains.
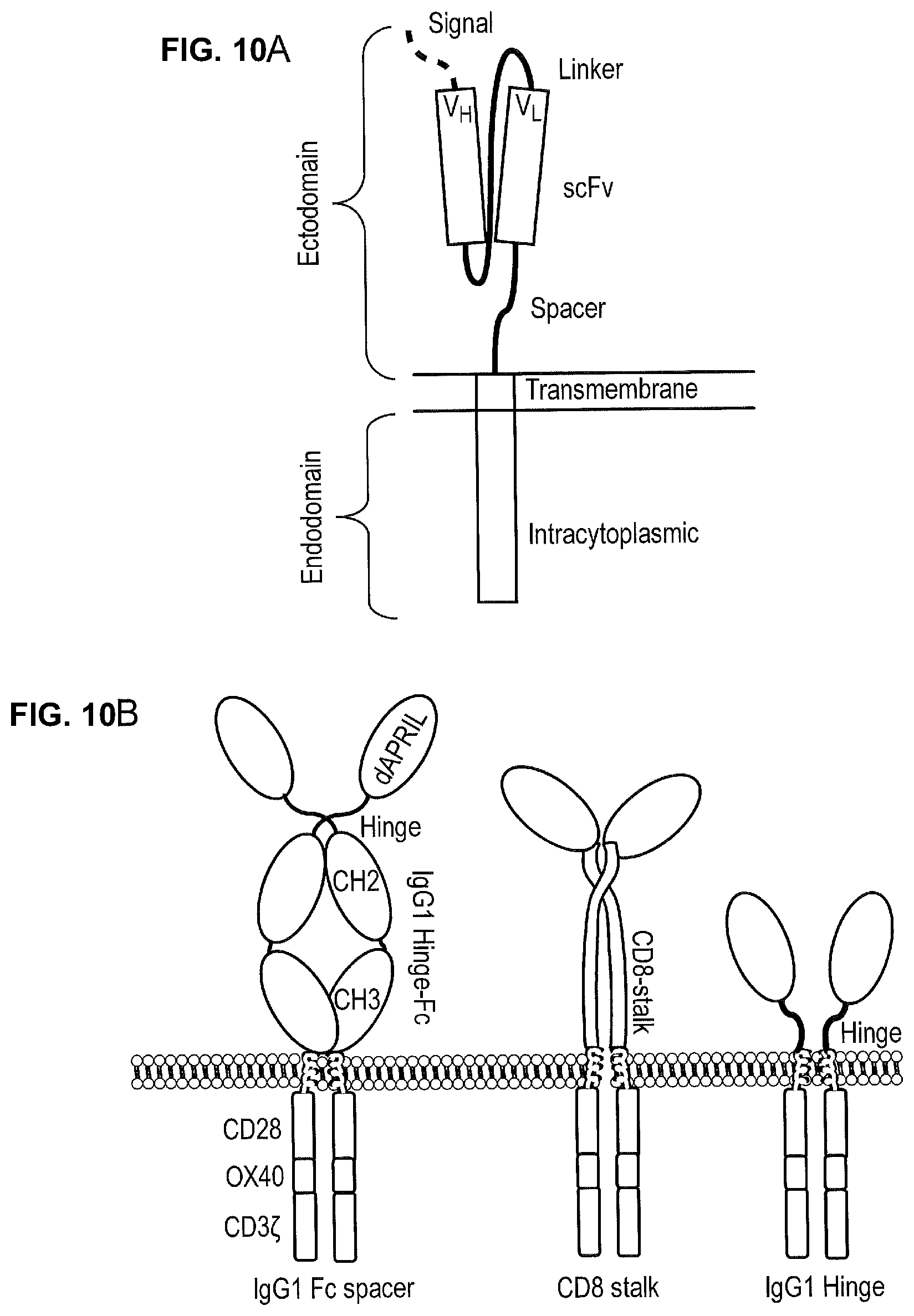
[0018] FIGS. 4A-4C--Design of the Different APRIL-Based CARs Generated.
[0019] The CAR design as shown in FIG. 3 was modified so that the scFv was replaced with a modified form of APRIL to act as an antigen binding domain: APRIL was truncated so that the proteoglycan binding amino-terminus is absent. A signal peptide was then attached to truncated APRIL amino-terminus to direct the protein to the cell surface. Three CARs were generated with this APRIL based binding domain: FIG. 4A. In the first CAR, the human CD8 stalk domain was used as a spacer domain. FIG. 4B. In the second CAR, the hinge from IgG1 was used as a spacer domain. FIG. 4C. In the third CAR, the hinge, CH2 and CH3 domains of human IgG1 modified with the pva/a mutations described by Hombach et al (2010 Gene Ther. 17:1206-1213) to reduce Fc Receptor binding was used as a spacer (henceforth referred as Fc-pvaa). In all CARs, these spacers were connected to the CD28 transmembrane domain and then to a tripartite endodomain containing a fusion of the CD28, OX40 and the CD3-Zeta endodomain (Pule et al, Molecular therapy, 2005: Volume 12; Issue 5; Pages 933-41).
[0020] FIGS. 5A-5C--Annotated Amino Acid Sequence of the Above Three APRIL-CARS
[0021] FIG. 5A: Shows the annotated amino acid sequence of the CD8 stalk APRIL CAR; FIG. 5B: Shows the annotated amino acid sequence of the APRIL IgG1 hinge based CAR; FIG. 5C: Shows the annotated amino acid sequence of the APRIL Fc-pvaa based CAR.
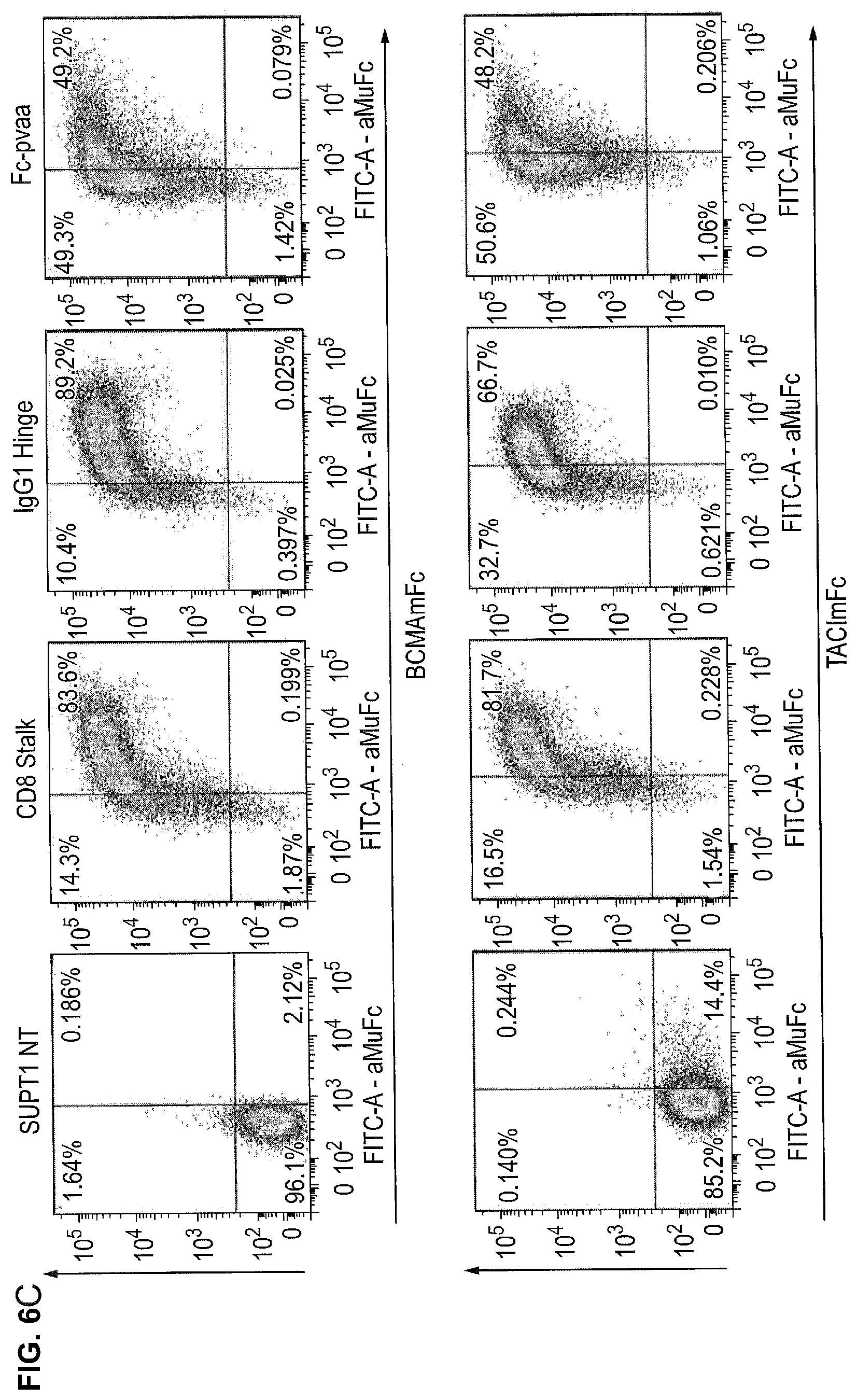
[0022] FIGS. 6A-6C--Expression and Ligand Binding of Different APRIL Based CARs
[0023] FIG. 6A. The receptors were co-expressed with a marker gene truncated CD34 in a retroviral gene vector. Expression of the marker gene on transduced cells allows confirmation of transduction. FIG. 6B. T-cells were transduced with APRIL based CARs with either the CD8 stalk spacer, IgG1 hinge or Fc spacer. To test whether these receptors could be stably expressed on the cell surface, T-cells were then stained with anti-APRIL-biotin/Streptavidin APC and anti-CD34. Flow-cytometric analysis was performed. APRIL was equally detected on the cell surface in the three CARs suggesting they are equally stably expressed. FIG. 6C. Next, the capacity of the CARs to recognize TACI and BCMA was determined. The transduced T-cells were stained with either recombinant BCMA or TACI fused to mouse IgG2a Fc fusion along with an anti-mouse secondary and anti-CD34. All three receptor formats showed binding to both BCMA and TACI. A surprising finding was that binding to BCMA seemed greater than to TACI. A further surprising finding was that although all three CARs were equally expressed, the CD8 stalk and IgG1 hinge CARs appeared better at recognizing BCMA and TACI than that with the Fc spacer.
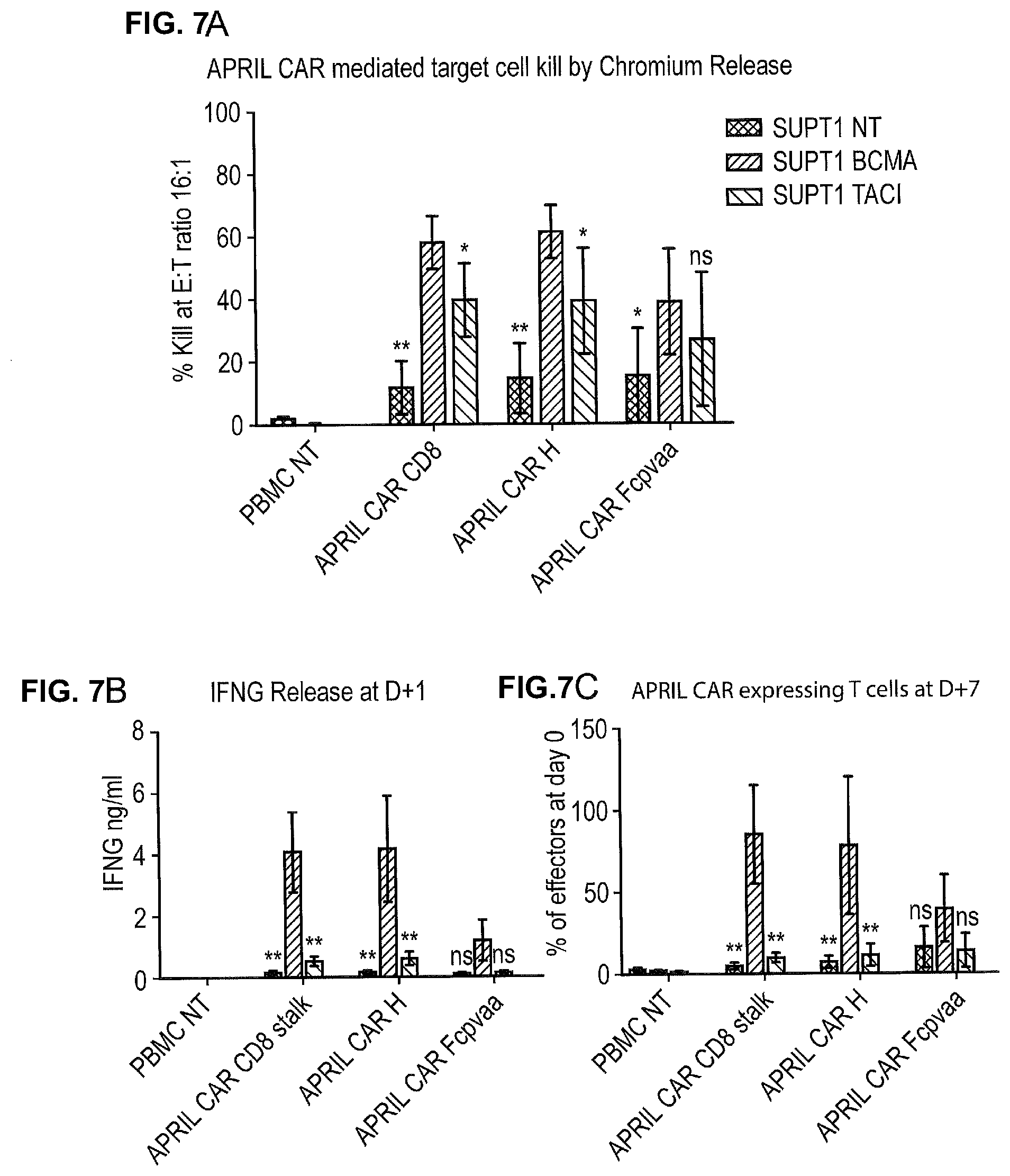
[0024] FIGS. 7A-7C--Function of the Different CAR Constructs.
[0025] Functional assays were performed of the three different APRIL based CARs. Normal donor peripheral blood T-cells either non-transduced (NT), or transduced to express the different CARs. Transduction was performed using equal titer supernatant. These T-cells were then CD56 depleted to remove non-specific NK activity and used as effectors. SupT1 cells either non-transduced (NT), or transduced to express BCMA or TACI were used as targets. Data shown is mean and standard deviation from 5 independent experiments. FIG. 7A. Specific killing of BCMA and TACI expressing T-cells was determined using Chromium release. FIG. 7B. Interferon-.gamma. release was also determined. Targets and effectors were co-cultured at a ratio of 1:1. After 24 hours, Interferon-.gamma. in the supernatant was assayed by ELISA. FIG. 7C. Proliferation/survival of CAR T-cells were also determined by counting number of CAR T-cells in the same co-culture incubated for a further 6 days. All 3 CARs direct responses against BCMA and TACI expressing targets. The responses to BCMA were greater than for TACI.
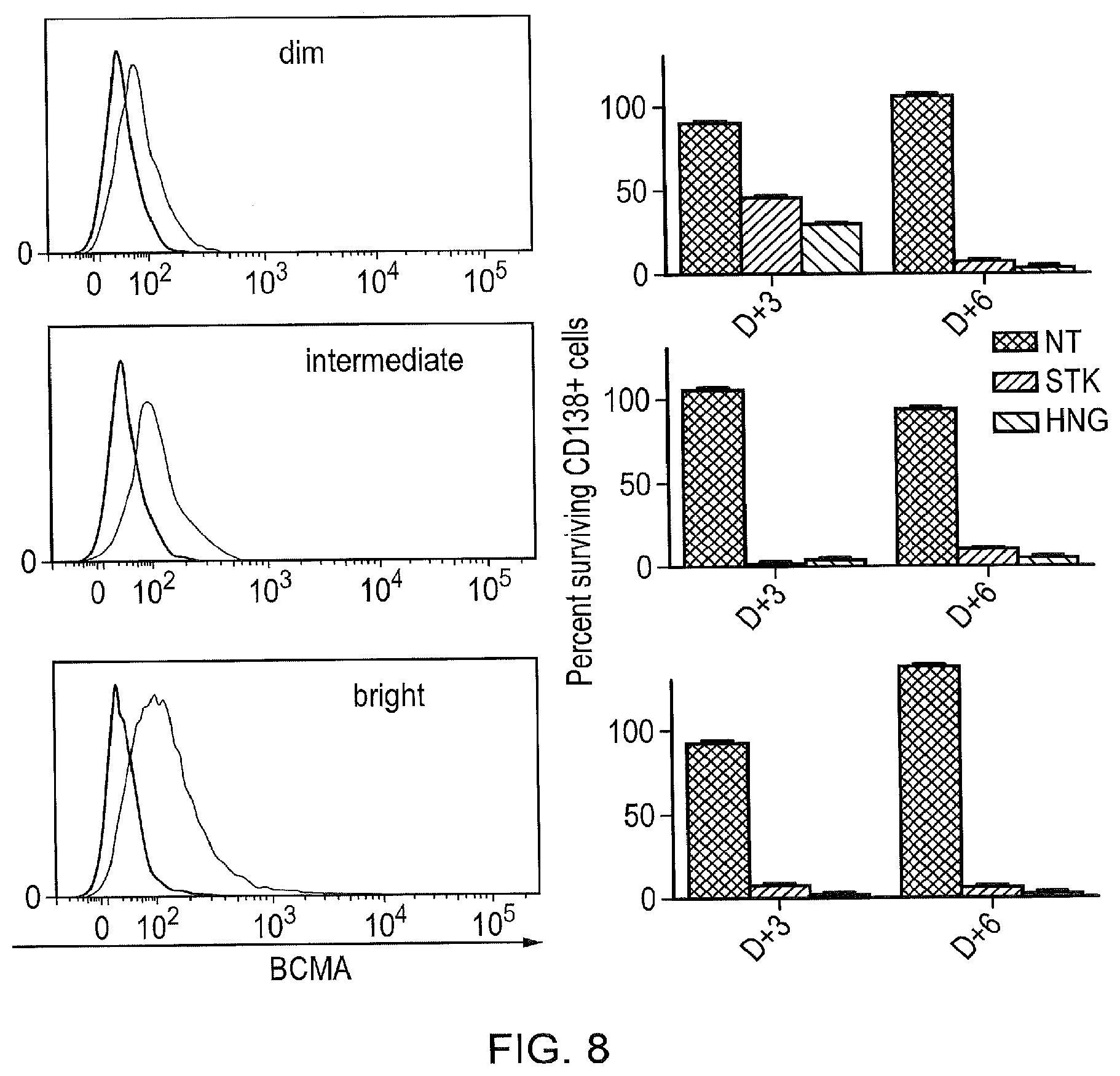
[0026] FIG. 8--Killing of Primary Myeloma Cells by APRIL CAR T-Cells
[0027] Since most primary myeloma cells express a low number of BCMA molecules on their surface, it was investigated whether killing of primary myeloma cells occurs despite low-density expression. Three cases were selected which represented the range of BCMA expression described in FIG. 2: the first had dim expression (lower than mean); the second case had intermediate expression (approximately mean expression) and the third had bright (above mean expression). A histogram of BCMA staining against isotype control for all three cases is shown on the left. In this assay, only the CD8 stalk and hinge APRIL CARs were tested. On the left, survival of myeloma cells compared with starting numbers is shown at day 3 and day 6 after a 1:1 co-culture of myeloma cells and CAR T-cells. By day 6, >95% of the myeloma cells were eliminated, including those with dim BCMA expression.
[0028] FIG. 9--Vector Co-Expressing APRIL Based CAR with Truncated CD34
[0029] A cell line expressing the vector used for screening was incubated with either BCMA-Fc or TACI-Fc and stained with both anti-CD34 and anti-human-Fc PE and FITC conjugated mAbs. The cells were then studied by flow-cytometery. This shows a typical pattern of binding of BCMA and TACI relative to the marker gene CD34.
[0030] FIGS. 10A-10B--FIG. 10A Schematic Diagram Illustrating a Classical CAR FIG. 10B Design of the Different APRIL-Based CARs Generated.
[0031] A signal peptide is attached to truncated APRIL amino-terminus. This was fused to different spacers: either the hinge, CH2 and CH3 domains of human IgG1 modified with the pvaa mutation described by Hombach et al (2010 Gene Ther. 17:1206-1213) to reduce Fc Receptor binding; the stalk of human CD8.alpha.; and the hinge of IgG1. These spacers were connected to a tripartite endodomain containing CD28 transmembrane domain, the OX40 endodomain and the CD3-Zeta endodomain.
[0032] FIG. 11--Expression of Different CARs
[0033] The receptors were co-expressed with enhanced blue fluorescence protein 2 (eBFP2) using an IRES sequence. Primary human T-cells were transduced and stained with anti-APRIL-biotin/Streptavidin APC. Flow-cytometric analysis was performed. eBFP2 signal is shown against APRIL detection. All three CARs are stably expressed (representative experiment of 3 independent experiments performed using 3 different normal donor T-cells).
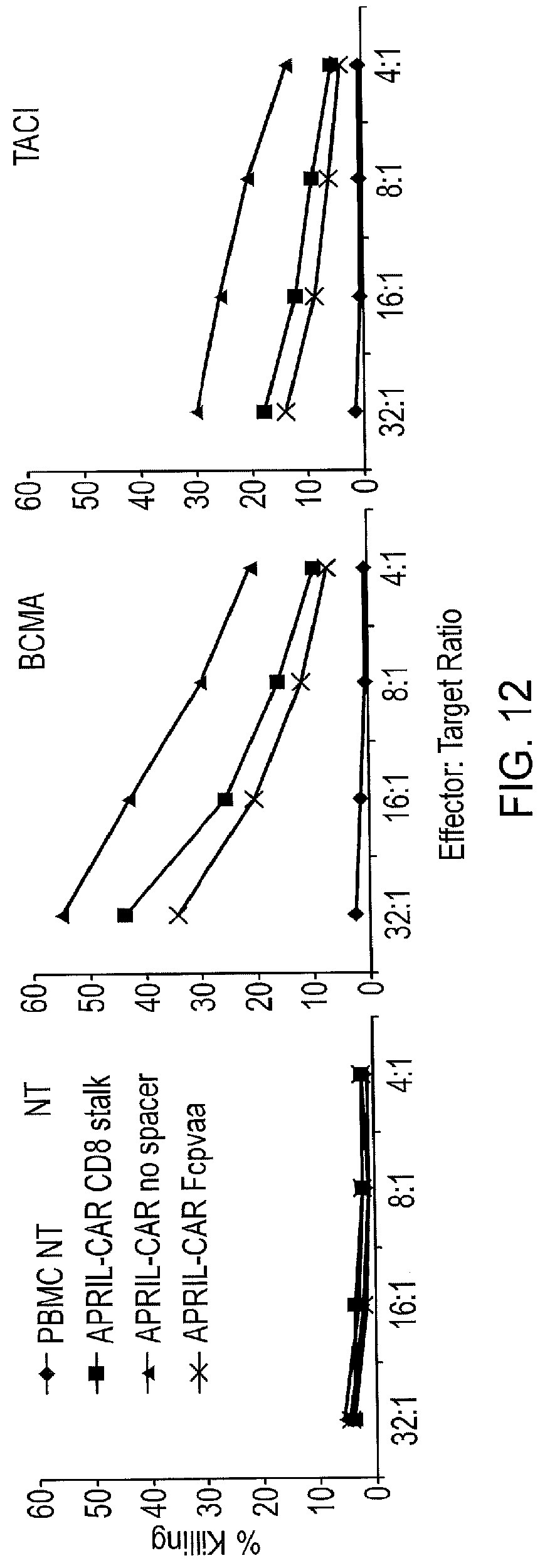
[0034] FIG. 12--Chromium Release Assay
[0035] Using normal donor peripheral blood T-cells either non-transduced (NT), or transduced to express different spacer CARs as effectors, and SupT1 cells either non-transduced (NT), or transduced to express BCMA or TACI as targets. The T-cells were CD56 depleted to reduce NK activity. This is a representative of three independent experiments and is shown as an example. Cumulative killing data is shown in FIG. 7A. Specific killing of BCMA and TACI expressing T-cells is seen with no activity against negative target cells.
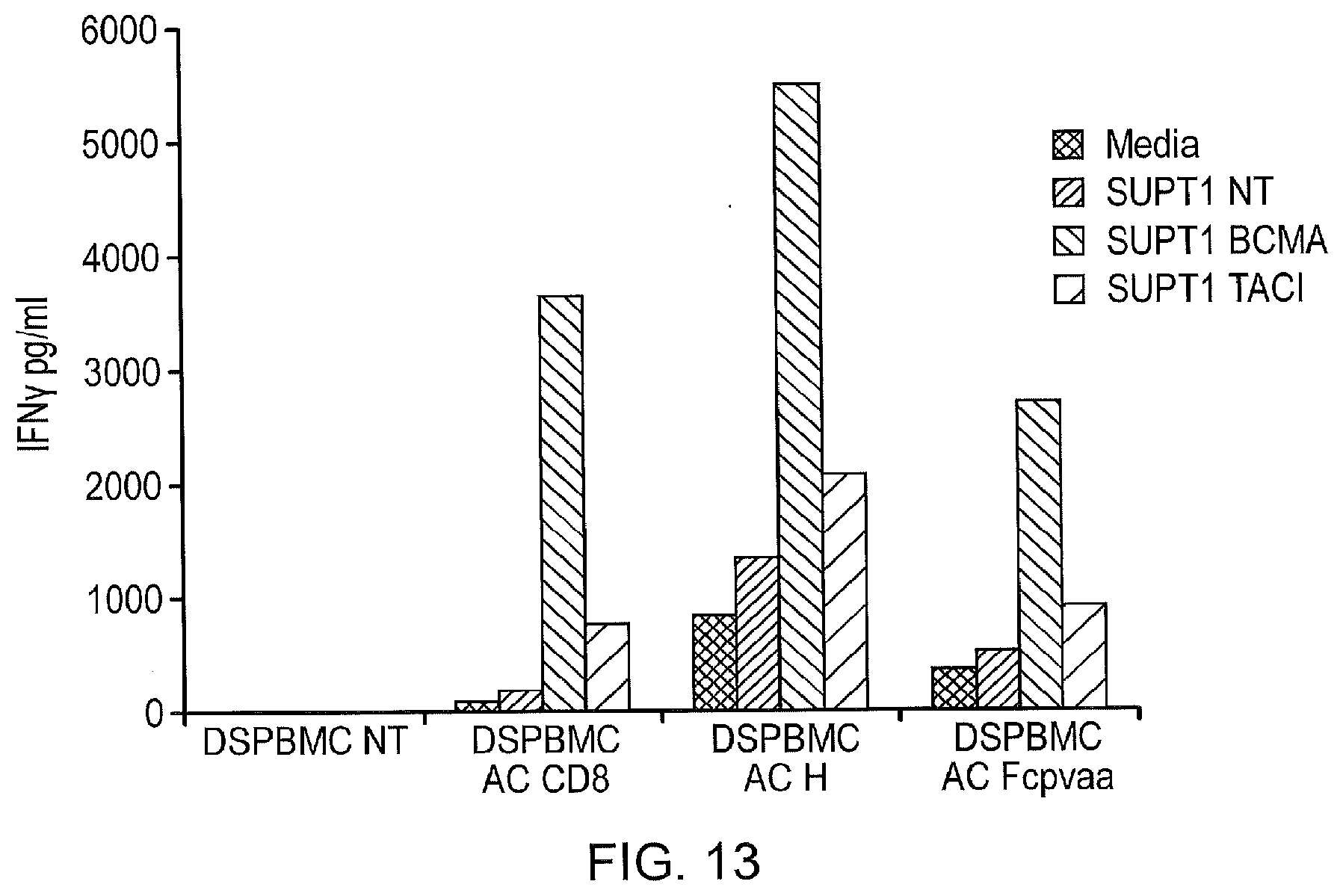
[0036] FIG. 13--Interferon-Gamma Release
[0037] From a 1:1 co-culture of effectors and targets is measured by ELISA. The CD8 stalk construct appears to have the best specificity while the hinge construct results in the most Interferon release demonstrates some non-specific activity. This is representative of 3 independent experiments and is shown as an example. Cumulative interferon-gamma release data is shown in FIG. 7B.
[0038] FIG. 14--Examples of BCMA Expression on Primary Myelomas
[0039] Four examples of myeloma samples stained with the rat anti-human BCMA mAb Vicky1 is shown. The first panel shows bright BCMA staining in a patient with a plasma cell leukemia (an unusual, advanced and aggressive form of myeloma). The other three cases are clinically and morphologically typical myelomas. They show the intermediate or dim staining typically seen. Staining with isotype control (grey) is superimposed. These are examples of cumulative BCMA expression data shown in FIG. 2.
[0040] FIGS. 15A-15C--Amino Acid Sequence of APRIL-CARS with a V5 Epitope Tag.
[0041] FIG. 15A: dAPRIL-HCH2CH3pvaa-CD28OXZ
[0042] FIG. 15B: dAPRIL-CD8STK-CD28OXZ
[0043] FIG. 15C: dAPRIL-HNG-CD28OXZ
[0044] Sequences in this figure differ from those in FIG. 5 have a different signal peptide and no V5 tag.
[0045] FIG. 16--Demonstration of In Vivo Function of APRIL CAR T-cells
[0046] Six 3 month old female NSG mice received 1.times.10.sup.7 MM1.s.FLuc cells vial tail-vein injection. Mice were imaged with bioluminescence at day 8 and day 13. After imaging on day 13, four mice received 5.times.10.sup.6 APRIL CAR T-cells via tail vein injection. Mice were imaged on day 13 and day 18. Mice which received CAR T-cells are indicated with (*). Remission of Myeloma could be observed by Day 18 in all treated mice, while disease in untreated mice progressed.
SUMMARY OF ASPECTS OF THE INVENTION
[0047] B-cell membrane antigen (BCMA) is a surface protein expressed on nearly all Multiple Myeloma (MM). BCMA is only otherwise expressed on plasma cells hence targeting this antigen may prove an effective treatment of myeloma. However, the low-level expression of BCMA (See FIG. 2), is a consideration when targeting this antigen.
[0048] The present inventors have surprisingly found that if a binding domain is used based on A proliferation-inducing ligand (APRIL), rather than a BCMA-binding antibody, in a CAR-type molecule, T cells expressing such CARs cause very efficient killing of BCMA-expressing target cells, even those with low-levels of expression.
[0049] Without wishing to be bound by theory, the present inventors predict that this is because the three-fold symmetry inherent in the binding of BCMA with APRIL. This means that every interaction between the CAR and BCMA will involve 3 CARs, approximating 3 endodomains on the T-cell surface. Since T-cell activation is triggered by close approximation of signalling endodomains in an immunological synapse, the CAR design of the present invention is highly sensitive and specific. As BCMA is expressed at a very low density on primary myeloma cells (see FIGS. 2 and 7), this receptor design is particularly suited to this target.
[0050] Thus, in a first aspect the present invention provides a chimeric antigen receptor (CAR) comprising:
[0051] (i) a B cell maturation antigen (BCMA)-binding domain which comprises at least part of a proliferation-inducing ligand (APRIL);
[0052] (ii) a spacer domain
[0053] (iii) a transmembrane domain; and
[0054] (iv) an intracellular T cell signaling domain.
[0055] The BCMA-binding domain may comprise a truncated APRIL which comprises the BCMA binding site but lacks the amino terminal portion of APRIL responsible for proteoglycan binding. Such a molecule may comprise the sequence shown as SEQ ID No. 14. Alternatively the molecule may comprise a variant of that sequence having at least 80% sequence identity which binds BCMA.
[0056] The transmembrane and intracellular T-cell signalling domain may comprise the sequence shown as SEQ ID No. 7 or a variant thereof having at least 80% sequence identity.
[0057] The BCMA-binding domain and the transmembrane domain may be connected by a spacer. The spacer may comprise one of the following: a human IgG1 spacer; an IgG1 hinge; or a CD8 stalk.
[0058] The CAR of the first aspect of the invention may comprise the sequence shown as SEQ ID No. 1, 2, 3, 4, 5 or 6 or a variant thereof which has at least 80% sequence identity but retains the capacity to i) bind BCMA and ii) induce T cell signalling.
[0059] The CAR of the first aspect of the invention may bind to BCMA as a trimer.
[0060] In a second aspect, the present invention provides a nucleic acid sequence which encodes a CAR according to any preceding claim.
[0061] The nucleic acid sequence may comprise the sequence shown as SEQ ID No 15, 16, 17, 18, 19 or 20 or a variant thereof having at least 80% sequence identity.
[0062] In a third aspect, the present invention provides a vector which comprises a nucleic acid sequence according to the second aspect of the invention.
[0063] In a fourth aspect, the present invention provides a T cell or an NK cell which expresses a CAR according to the first aspect of the invention.
[0064] In a fifth aspect, the present invention provides a method for making a T cell or an NK cell according to the fourth aspect of the invention, which comprises the step of introducing a nucleic acid according to the second aspect of the invention into a T cell or an NK cell.
[0065] In a sixth aspect, the present invention provides a pharmaceutical composition which comprises a vector according to the third aspect of the invention or T cell/NK cell according to the fourth aspect of the invention, together with a pharmaceutically acceptable carrier, diluent or excipient.
[0066] In a seventh aspect, the present invention provides a method for treating a plasma cell disorder which comprises the step of administering a vector according to the third aspect of the invention or T cell/NK cell according to the fourth aspect of the invention to a subject.
[0067] The plasma cell disorder may be selected from plasmacytoma, plasma cell leukemia, multiple myeloma, macroglobulinemia, amyloidosis, Waldenstrom's macroglobulinemia, solitary bone plasmacytoma, extramedullary plasmacytoma, osteosclerotic myeloma, heavy chain diseases, monoclonal gammopathy of undetermined significance and smoldering multiple myeloma.
[0068] The plasma cell disorder may be multiple myeloma.
[0069] In an eighth aspect, the present invention provides a vector according to the third aspect of the invention or T cell/NK cell according to the fourth aspect of the invention for use in treating a plasma cell disorder.
[0070] In a ninth aspect, the present invention provides use of a vector according to the third aspect of the invention or T cell/NK cell according to the fourth aspect of the invention in the manufacture of a medicament for treating a plasma cell disorder.
DETAILED DESCRIPTION
[0071] Chimeric Antigen Receptors (CARS)
[0072] Chimeric antigen receptors (CARs), also known as chimeric T cell receptors, artificial T cell receptors and chimeric immunoreceptors, are engineered receptors, which graft an arbitrary specificity onto an immune effector cell. In a classical CAR (FIG. 3), the specificity of a monoclonal antibody is grafted on to a T cell or NK cell. CAR-encoding nucleic acids may be introduced into T cells or NK cells using, for example, retroviral vectors. In this way, a large number of cancer-specific T cells or NK cells can be generated for adoptive cell transfer. Early clinical studies of this approach have shown efficacy in some cancers, primarily when targeting the pan-B-cell antigen CD19 to treat B-cell malignancies.
[0073] The target-antigen binding domain of a CAR is commonly fused via a spacer and transmembrane domain to a signaling endodomain. When the CAR binds the target-antigen, this results in the transmission of an activating signal to the T-cell it is expressed on.
[0074] The CAR of the present invention comprises:
[0075] (i) a B cell maturation antigen (BCMA)-binding domain which comprises at least part of a proliferation-inducing ligand (APRIL), which is discussed in more detail below;
[0076] (ii) a spacer
[0077] (iii) a transmembrane domain; and
[0078] (iv) an intracellular T cell signaling domain
[0079] The CAR of the present invention may comprise one of the following amino acid sequences:
TABLE-US-00001 (dAPRIL-HCH2CH3pvaa-CD28OXZ) SEQ ID No. 1 METDTLLLWVLLLWVPGSTGSVLHLVPINATSKDDSDVTEVMWQPALRRGRGLQAQGYGVRIQ DAGVYLLYSQVLFQDVTFTMGQVVSREGQGRQETLFRCIRSMPSHPDRAYNSCYSAGVFHLHQ GDILSVIIPRARAKLNLSPHGTFLGFVKLSGGGSDPAEPKSPDKTHTCPPCPAPPVAGPSVFL FPPKPKDTLMIARTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSV LTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCL VKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEA LHNHYTQKSLSLSPGKKDPKFWVLVVVGGVLACYSLLVTVAFIIFWVRSKRSRLLHSDYMNMT PRRPGPTRKHYQPYAPPRDFAAYRSRDQRLPPDAHKPPGGGSFRTPIQEEQADAHSTLAKIRV KFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKD KMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPR (dAPRIL-CD8STK-CD28OXZ) SEQ ID No. 2 METDTLLLWVLLLWVPGSTGSVLHLVPINATSKDDSDVTEVMWQPALRRGRGLQAQGYGVRIQ DAGVYLLYSQVLFQDVTFTMGQVVSREGQGRQETLFRCIRSMPSHPDRAYNSCYSAGVFHLHQ GDILSVIIPRARAKLNLSPHGTFLGFVKLSGGGSDPTTTPAPRPPTPAPTIASQPLSLRPEAC RPAAGGAVHTRGLDFACDIFWVLVVVGGVLACYSLLVTVAFIIFWVRSKRSRLLHSDYMNMTP RRPGPTRKHYQPYAPPRDFAAYRSRDQRLPPDAHKPPGGGSFRTPIQEEQADAHSTLAKIRVK FSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDK MAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPR (dAPRIL-HNG-CD28OXZ) SEQ ID No. 3 METDTLLLWVLLLWVPGSTGSVLHLVPINATSKDDSDVTEVMWQPALRRGRGLQAQGYGVRIQ DAGVYLLYSQVLFQDVTFTMGQVVSREGQGRQETLFRCIRSMPSHPDRAYNSCYSAGVFHLHQ GDILSVIIPRARAKLNLSPHGTFLGFVKLSGGGSDPAEPKSPDKTHTCPPCPKDPKFWVLVVV GGVLACYSLLVTVAFIIFWVRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRSRD QRLPPDAHKPPGGGSFRTPIQEEQADAHSTLAKIRVKFSRSADAPAYQQGQNQLYNELNLGRR EEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQG LSTATKDTYDALHMQALPPR (dAPRIL-HCH2CH3pvaa-CD28OXZ) SEQ ID No. 4 MGTSLLCWMALCLLGADHADGKPIPNPLLGLDSTSGGGGSVLHLVPINATSKDDSDVTEVMWQ PALRRGRGLQAQGYGVRIQDAGVYLLYSQVLFQDVTFTMGQVVSREGQGRQETLFRCIRSMPS HPDRAYNSCYSAGVFHLHQGDILSVIIPRARAKLNLSPHGTFLGFVKLSGGGSDPAEPKSPDK THTCPPCPAPPVAGPSVFLFPPKPKDTLMIARTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHN AKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVY TLPPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTV DKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGKKDPKFWVLVVVGGVLACYSLLVTVAFII FWVRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRSRDQRLPPDAHKPPGGGSFR TPIQEEQADAHSTLAKIRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMG GKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQAL PPR (dAPRIL-CD8STK-CD28OXZ) SEQ ID No. 5 MGTSLLCWMALCLLGADHADGKPIPNPLLGLDSTSGGGGSVLHLVPINATSKDDSDVTEVMWQ PALRRGRGLQAQGYGVRIQDAGVYLLYSQVLFQDVTFTMGQVVSREGQGRQETLFRCIRSMPS HPDRAYNSCYSAGVFHLHQGDILSVIIPRARAKLNLSPHGTFLGFVKLSGGGSDPTTTPAPRP PTPAPTIASQPLSLRPEACRPAAGGAVHTRGLDFACDIFWVLVVVGGVLACYSLLVTVAFIIF WVRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRSRDQRLPPDAHKPPGGGSFRT PIQEEQADAHSTLAKIRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGG KPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALP PR (dAPRIL-HNG-CD28OXZ) SEQ ID No. 6 MGTSLLCWMALCLLGADHADGKPIPNPLLGLDSTSGGGGSVLHLVPINATSKDDSDVTEVMWQ PALRRGRGLQAQGYGVRIQDAGVYLLYSQVLFQDVTFTMGQVVSREGQGRQETLFRCIRSMPS HPDRAYNSCYSAGVFHLHQGDILSVIIPRARAKLNLSPHGTFLGFVKLSGGGSDPAEPKSPDK THTCPPCPKDPKFWVLVVVGGVLACYSLLVTVAFIIFWVRSKRSRLLHSDYMNMTPRRPGPTR KHYQPYAPPRDFAAYRSRDQRLPPDAHKPPGGGSFRTPIQEEQADAHSTLAKIRVKFSRSADA PAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSE IGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPR
[0080] The molecule of the invention may comprise a variant of the sequence shown as SEQ ID No. 1, 2, 3, 4, 5 or 6 having at least 80, 85, 90, 95, 98 or 99% sequence identity, provided that the variant sequence is a molecule as defined in the first aspect of the invention, i.e. a CAR which comprises:
[0081] (i) a BCMA-binding domain;
[0082] (ii) a spacer domain
[0083] (iii) a transmembrane domain; and
[0084] (iv) an intracellular T cell signaling domain.
[0085] The percentage identity between two polypeptide sequences may be readily determined by programs such as BLAST which is freely available at http://blast.ncbi.nlm.nih.gov.
[0086] Transmembrane Domain
[0087] The transmembrane domain is the sequence of the CAR that spans the membrane. It may comprise a hydrophobic alpha helix. The transmembrane domain may be derived from CD28, which gives good receptor stability. The transmembrane domain may be derived from any type I transmembrane protein. The transmembrane domain may be a synthetic sequence predicted to form a hydrophobic helix.
[0088] Intracellular T Cell Signaling Domain (Endodomain)
[0089] The endodomain is the signal-transmission portion of the CAR. After antigen recognition, receptors cluster and a signal is transmitted to the cell. The most commonly used endodomain component is that of CD3-zeta which contains 3 ITAMs. This transmits an activation signal to the T cell after antigen is bound. CD3-zeta may not provide a fully competent activation signal and additional co-stimulatory signaling may be needed. For example, chimeric CD28 and OX40 can be used with CD3-Zeta to transmit a proliferative/survival signal, or all three can be used together (Pule et al, Molecular therapy, 2005: Volume 12; Issue 5; Pages 933-41). The CAR endodomain may also be derived from other signaling domains either individually or in combination, derived from signaling proteins found in nature or artificial ones constructed by those skilled in the art such that the CAR transmits a suitable signal to for an effective CAR therapeutic.
[0090] The endodomain of the CAR of the present invention may comprise the CD28 endodomain and OX40 and CD3-Zeta endodomain.
[0091] The transmembrane and intracellular T-cell signalling domain (endodomain) of the CAR of the present invention may comprise the sequence shown as SEQ ID No. 7 or a variant thereof having at least 80% sequence identity.
TABLE-US-00002 SEQ ID No. 7 FWVLVVVGGVLACYSLLVTVAFIIFWVRSKRSRLLHSDYMNMTPR RPGPTRKHYQPYAPPRDFAAYRSRDQRLPPDAHKPPGGGSFRTPI QEEQADAHSTLAKIRVKFSRSADAPAYQQGQNQLYNELNLGRREE YDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGM KGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPR
[0092] A variant sequence may have at least 80%, 85%, 90%, 95%, 98% or 99% sequence identity to SEQ ID No. 7, provided that the sequence provides an effective transmembrane domain and an effective intracellular T cell signaling domain.
[0093] Signal Peptide
[0094] The CAR of the present invention may comprise a signal peptide so that when the CAR is expressed inside a cell, such as a T-cell, the nascent protein is directed to the endoplasmic reticulum and subsequently to the cell surface, where it is expressed.
[0095] The core of the signal peptide may contain a long stretch of hydrophobic amino acids that has a tendency to form a single alpha-helix. The signal peptide may begin with a short positively charged stretch of amino acids, which helps to enforce proper topology of the polypeptide during translocation. At the end of the signal peptide there is typically a stretch of amino acids that is recognized and cleaved by signal peptidase. Signal peptidase may cleave either during or after completion of translocation to generate a free signal peptide and a mature protein. The free signal peptides are then digested by specific proteases.
[0096] The signal peptide may be at the amino terminus of the molecule.
[0097] The CAR of the invention may have the general formula:
[0098] Signal peptide--BCMA-binding domain spacer domain--transmembrane domain--intracellular T cell signaling domain.
[0099] The signal peptide may comprise the SEQ ID No. 8 or 9 or a variant thereof having 5, 4, 3, 2 or 1 amino acid mutations (insertions, substitutions or additions) provided that the signal peptide still functions to cause cell surface expression of the CAR.
TABLE-US-00003 SEQ ID No. 8: MGTSLLCWMALCLLGADHADG SEQ ID No. 9: METDTLLLWVLLLWVPGSTG
[0100] The signal peptide of SEQ ID No. 8 and SEQ ID No 9 is compact and highly efficient. It is predicted to give about 95% cleavage after the terminal glycine, giving efficient removal by signal peptidase.
[0101] Spacer
[0102] The CAR of the present invention may comprise a spacer sequence to connect the BCMA-binding domain with the transmembrane domain and spatially separate the BCMA-binding domain from the endodomain. A flexible spacer allows to the BCMA-binding domain to orient in different directions to enable BCMA binding.
[0103] The spacer sequence may, for example, comprise an IgG1 Fc region, an IgG1 hinge or a CD8 stalk. The linker may alternatively comprise an alternative linker sequence which has similar length and/or domain spacing properties as an IgG1 Fc region, an IgG1 hinge or a CD8 stalk.
[0104] The spacer may be a short spacer, for example a spacer which comprises less than 100, less than 80, less than 60 or less than 45 amino acids. The spacer may be or comprise an IgG1 hinge or a CD8 stalk or a modified version thereof.
[0105] A human IgG1 spacer may be altered to remove Fc binding motifs.
[0106] Examples of amino acid sequences for these spacers are given below:
TABLE-US-00004 (hinge-CH2CH3 of human IgG1) SEQ ID No. 10 AEPKSPDKTHTCPPCPAPPVAGPSVFLFPPKPKDTLMIARTPEVT CVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSV LTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYT LPPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTP PVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKS LSLSPGKKD (human CD8 stalk): SEQ ID No. 11 TTTPAPRPPTPAPTIASQPLSLRPEACRPAAGGAVHTRGLDFACDI (human IgG1 hinge): SEQ ID No. 12 AEPKSPDKTHTCPPCPKDPK
[0107] B-Cell Membrane Antigen (BCMA)
[0108] The CAR of the first aspect of the invention comprises a domain which binds BCMA.
[0109] BCMA, also known as TNFRSF17, is a plasma cell specific surface antigen which is expressed exclusively on B-lineage haemopoietic cells or dendritic cells. It is a member of the TNF receptor family. BCMA is not expressed on naive B cells but is up-regulated during B-cell differentiation into plasmablasts, and is brightly expressed on memory B cells, plasmablasts and bone marrow plasma cells. BCMA is also expressed on the majority of primary myeloma cells. Unlike other CAR targets such as CD19, BCMA is expressed at low density (FIG. 2).
[0110] BCMA functions within a network of interconnected ligands and receptors which is shown schematically in FIG. 1. Two other TNF receptors share the ligands APRIL and BAFF with BCMA-TACI (TNFRSF13B), which is found on activated T-cells and all B-cells and BAFF-R (TNFRSF13C) which is predominantly expressed on B-lymphocytes. Multiple myeloma cells express TACI in some cases and BCMA in most cases, but never BAFF-R.
[0111] April
[0112] The BCMA-binding domain of the CAR of the invention and comprises at least part of a proliferation-inducing ligand (APRIL). APRIL is also known as TNFSF13.
[0113] The wild-type sequence of APRIL is available at UNIPROT/O75888 and is show below (SEQ ID No. 13). It is not a classical secreted protein in that it has no signal peptide. It has a furin cleavage site "KQKKQK" (underlined in SEQ ID No. 13). The amino terminus is involved in proteoglycan binding.
[0114] The BCMA-binding domain may comprise the BCMA-binding site of APRIL. The BCMA-binding domain may comprise a fragment of APRIL which comprises the BCMA-binding site.
[0115] The BCMA-binding domain may comprise a truncated APRIL, which lacks the amino terminal end of the molecule. The truncated APRIL may retain BCMA and TACI binding but lose proteoglycan binding. Truncated APRIL can be cleaved at or immediately after the furin cleavage site. Truncated APRIL may lack the amino terminal 116 amino acids from the wild-type APRIL molecule shown as SEQ ID No. 13. Truncated APRIL may comprise the sequence shown as SEQ ID No. 14 (which corresponds to the portion of SEQ ID No. 13 shown in bold) or a variant thereof. This corresponds to the portion of the molecule which is needed for BCMA and TACI binding.
TABLE-US-00005 SEQ ID No. 13 10 20 30 40 MPASSPFLLA PKGPPGNMGG PVREPALSVA LWLSWGAALG 50 60 70 80 AVACAMALLT QQTELQSLRR EVSRLQGTGG PSQNGEGYPW 90 100 110 120 QSLPEQSSDA LEAWENGERS RKRRAVLTQK QKKQHSVLHL 130 140 150 160 VPINATSKDD SDVTEVMWQP ALRRGRGLQA QGYGVRIQDA 170 180 190 200 GVYLLYSQVL FQDVTFTMGQ VVSREGQGRQ ETLFRCIRSM 210 220 230 240 PSHPDRAYNS CYSAGVFHLH QGDILSVIIP RARAKLNLSP 250 HGTFLGFVKL SEQ ID No. 14 VLHLVPINATSKDDSDVTEVMWQPALRRGRGLQAQGYGVRIQDAGVY LLYSQVLFQDVTFTMGQVVSREGQGRQETLFRCIRSMPSHPDRAYNS CYSAGVFHLHQGDILSVIIPRARAKLNLSPHGTFLGFVKL
[0116] The CAR of the present invention may comprise a variant of the truncated APRIL molecule shown as SEQ ID No. 14 which has at least 80% amino acid sequence identity and which has the same or improved BCMA binding capabilities. The variant sequence may have at least 80%, 85%, 90%, 95%, 98% or 99% sequence identity to SEQ ID No. 14.
[0117] Nucleic Acid Sequence
[0118] The second aspect of the invention relates to a nucleic acid sequence which codes for a CAR of the first aspect of the invention.
[0119] The nucleic acid sequence may be or comprise one of the following sequences:
TABLE-US-00006 (dAPRIL-HCH2CH3pvaa-CD28OXZ) SEQ ID No. 15 ATGGAGACCGACACCCTGCTGCTGTGGGTGCTGCTGCTGTGGGTGCCAGGCAGCACCGGCAGC GTGCTCCACCTGGTGCCCATCAACGCCACCAGCAAGGACGACTCTGATGTGACCGAGGTGATG TGGCAGCCAGCCCTGAGACGGGGCAGAGGCCTGCAGGCCCAGGGCTACGGCGTGAGAATCCAG GACGCTGGCGTGTACCTGCTGTACTCCCAGGTGCTGTTCCAGGACGTGACCTTCACAATGGGC CAGGTGGTGAGCCGGGAGGGCCAGGGCAGACAGGAGACCCTGTTCCGGTGCATCCGGAGCATG CCCAGCCACCCCGACAGAGCCTACAACAGCTGCTACAGCGCTGGCGTGTTTCACCTGCACCAG GGCGACATCCTGAGCGTGATCATCCCCAGAGCCAGAGCCAAGCTGAACCTGTCCCCCCACGGC ACCTTTCTGGGCTTCGTGAAGCTGTCTGGAGGCGGCTCGGATCCCGCCGAGCCCAAATCTCCT GACAAAACTCACACATGCCCACCGTGCCCAGCACCTCCCGTGGCCGGCCCGTCAGTCTTCCTC TTCCCCCCAAAACCCAAGGACACCCTCATGATCGCCCGGACCCCTGAGGTCACATGCGTGGTG GTGGACGTGAGCCACGAAGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGGAGGTG CATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTCAGCGTC CTCACCGTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAA GCCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACAG GTGTACACCCTGCCCCCATCCCGGGATGAGCTGACCAAGAACCAGGTCAGCCTGACCTGCCTG GTCAAAGGCTTCTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAACCGGAGAAC AACTACAAGACCACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCTACAGCAAGCTC ACCGTGGACAAGAGCAGGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGTGATGCATGAGGCT CTGCACAACCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGTAAAAAAGATCCCAAATTT TGGGTGCTGGTGGTGGTTGGTGGAGTCCTGGCTTGCTATAGCTTGCTAGTAACAGTGGCCTTT ATTATTTTCTGGGTGAGGAGTAAGAGGAGCAGGCTCCTGCACAGTGACTACATGAACATGACT CCCCGCCGCCCCGGGCCCACCCGCAAGCATTACCAGCCCTATGCCCCACCACGCGACTTCGCA GCCTATCGCTCCAGGGACCAGAGGCTGCCCCCCGATGCCCACAAGCCCCCTGGGGGAGGCAGT TTCCGGACCCCCATCCAAGAGGAGCAGGCCGACGCCCACTCCACCCTGGCCAAGATCAGAGTG AAGTTCAGCAGGAGCGCAGACGCCCCCGCGTACCAGCAGGGCCAGAACCAGCTCTATAACGAG CTCAATCTAGGACGAAGAGAGGAGTACGATGTTTTGGACAAGAGACGTGGCCGGGACCCTGAG ATGGGGGGAAAGCCGAGAAGGAAGAACCCTCAGGAAGGCCTGTACAATGAACTGCAGAAAGAT AAGATGGCGGAGGCCTACAGTGAGATTGGGATGAAAGGCGAGCGCCGGAGGGGCAAGGGGCAC GATGGCCTTTACCAGGGTCTCAGTACAGCCACCAAGGACACCTACGACGCCCTTCACATGCAG GCCCTGCCTCCTCGCTAA (dAPRIL-CD8STK-CD28OXZ) SEQ ID No. 16 ATGGAGACCGACACCCTGCTGCTGTGGGTGCTGCTGCTGTGGGTGCCAGGCAGCACCGGCAGC GTGCTCCACCTGGTGCCCATCAACGCCACCAGCAAGGACGACTCTGATGTGACCGAGGTGATG TGGCAGCCAGCCCTGAGACGGGGCAGAGGCCTGCAGGCCCAGGGCTACGGCGTGAGAATCCAG GACGCTGGCGTGTACCTGCTGTACTCCCAGGTGCTGTTCCAGGACGTGACCTTCACAATGGGC CAGGTGGTGAGCCGGGAGGGCCAGGGCAGACAGGAGACCCTGTTCCGGTGCATCCGGAGCATG CCCAGCCACCCCGACAGAGCCTACAACAGCTGCTACAGCGCTGGCGTGTTTCACCTGCACCAG GGCGACATCCTGAGCGTGATCATCCCCAGAGCCAGAGCCAAGCTGAACCTGTCCCCCCACGGC ACCTTTCTGGGCTTCGTGAAGCTGTCTGGAGGCGGCTCGGATCCCACCACGACGCCAGCGCCG CGACCACCAACACCGGCGCCCACCATCGCGTCGCAGCCCCTGTCCCTGCGCCCAGAGGCGTGC CGGCCAGCGGCGGGGGGCGCAGTGCACACGAGGGGGCTGGACTTCGCCTGTGATATCTTTTGG GTGCTGGTGGTGGTTGGTGGAGTCCTGGCTTGCTATAGCTTGCTAGTAACAGTGGCCTTTATT ATTTTCTGGGTGAGGAGTAAGAGGAGCAGGCTCCTGCACAGTGACTACATGAACATGACTCCC CGCCGCCCCGGGCCCACCCGCAAGCATTACCAGCCCTATGCCCCACCACGCGACTTCGCAGCC TATCGCTCCAGGGACCAGAGGCTGCCCCCCGATGCCCACAAGCCCCCTGGGGGAGGCAGTTTC CGGACCCCCATCCAAGAGGAGCAGGCCGACGCCCACTCCACCCTGGCCAAGATCAGAGTGAAG TTCAGCAGGAGCGCAGACGCCCCCGCGTACCAGCAGGGCCAGAACCAGCTCTATAACGAGCTC AATCTAGGACGAAGAGAGGAGTACGATGTTTTGGACAAGAGACGTGGCCGGGACCCTGAGATG GGGGGAAAGCCGAGAAGGAAGAACCCTCAGGAAGGCCTGTACAATGAACTGCAGAAAGATAAG ATGGCGGAGGCCTACAGTGAGATTGGGATGAAAGGCGAGCGCCGGAGGGGCAAGGGGCACGAT GGCCTTTACCAGGGTCTCAGTACAGCCACCAAGGACACCTACGACGCCCTTCACATGCAGGCC CTGCCTCCTCGCTAA (dAPRIL-HNG-CD28OXZ) SEQ ID No. 17 ATGGAGACCGACACCCTGCTGCTGTGGGTGCTGCTGCTGTGGGTGCCAGGCAGCACCGGCAGC GTGCTCCACCTGGTGCCCATCAACGCCACCAGCAAGGACGACTCTGATGTGACCGAGGTGATG TGGCAGCCAGCCCTGAGACGGGGCAGAGGCCTGCAGGCCCAGGGCTACGGCGTGAGAATCCAG GACGCTGGCGTGTACCTGCTGTACTCCCAGGTGCTGTTCCAGGACGTGACCTTCACAATGGGC CAGGTGGTGAGCCGGGAGGGCCAGGGCAGACAGGAGACCCTGTTCCGGTGCATCCGGAGCATG CCCAGCCACCCCGACAGAGCCTACAACAGCTGCTACAGCGCTGGCGTGTTTCACCTGCACCAG GGCGACATCCTGAGCGTGATCATCCCCAGAGCCAGAGCCAAGCTGAACCTGTCCCCCCACGGC ACCTTTCTGGGCTTCGTGAAGCTGTCTGGAGGCGGCTCGGATCCCGCCGAGCCCAAATCTCCT GACAAAACTCACACATGCCCACCGTGCCCAAAAGATCCCAAATTTTGGGTGCTGGTGGTGGTT GGTGGAGTCCTGGCTTGCTATAGCTTGCTAGTAACAGTGGCCTTTATTATTTTCTGGGTGAGG AGTAAGAGGAGCAGGCTCCTGCACAGTGACTACATGAACATGACTCCCCGCCGCCCCGGGCCC ACCCGCAAGCATTACCAGCCCTATGCCCCACCACGCGACTTCGCAGCCTATCGCTCCAGGGAC CAGAGGCTGCCCCCCGATGCCCACAAGCCCCCTGGGGGAGGCAGTTTCCGGACCCCCATCCAA GAGGAGCAGGCCGACGCCCACTCCACCCTGGCCAAGATCAGAGTGAAGTTCAGCAGGAGCGCA GACGCCCCCGCGTACCAGCAGGGCCAGAACCAGCTCTATAACGAGCTCAATCTAGGACGAAGA GAGGAGTACGATGTTTTGGACAAGAGACGTGGCCGGGACCCTGAGATGGGGGGAAAGCCGAGA AGGAAGAACCCTCAGGAAGGCCTGTACAATGAACTGCAGAAAGATAAGATGGCGGAGGCCTAC AGTGAGATTGGGATGAAAGGCGAGCGCCGGAGGGGCAAGGGGCACGATGGCCTTTACCAGGGT CTCAGTACAGCCACCAAGGACACCTACGACGCCCTTCACATGCAGGCCCTGCCTCCTCGCTAA (dAPRIL-HCH2CH3pvaa-CD28OXZ) SEQ ID No. 18 ATGGGCACCTCCCTGCTGTGCTGGATGGCCCTGTGCCTGCTGGGAGCCGACCACGCCGACGGC AAGCCCATTCCCAACCCCCTGCTGGGCCTGGACTCCACCTCTGGCGGAGGCGGCAGCGTGCTG CACCTGGTGCCCATCAACGCCACCAGCAAGGACGACTCTGATGTGACCGAGGTGATGTGGCAG CCAGCCCTGAGACGGGGCAGAGGCCTGCAGGCCCAGGGCTACGGCGTGAGAATCCAGGACGCT GGCGTGTACCTGCTGTACTCCCAGGTGCTGTTCCAGGACGTGACCTTCACAATGGGCCAGGTG GTGAGCCGGGAGGGCCAGGGCAGACAGGAGACCCTGTTCCGGTGCATCCGGAGCATGCCCAGC CACCCCGACAGAGCCTACAACAGCTGCTACAGCGCTGGCGTGTTTCACCTGCACCAGGGCGAC ATCCTGAGCGTGATCATCCCCAGAGCCAGAGCCAAGCTGAACCTGTCCCCCCACGGCACCTTT CTGGGCTTCGTGAAGCTGTCTGGAGGCGGCTCGGATCCCGCCGAGCCCAAATCTCCTGACAAA ACTCACACATGCCCACCGTGCCCAGCACCTCCCGTGGCCGGCCCGTCAGTCTTCCTCTTCCCC CCAAAACCCAAGGACACCCTCATGATCGCCCGGACCCCTGAGGTCACATGCGTGGTGGTGGAC GTGAGCCACGAAGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGGAGGTGCATAAT GCCAAGACAAAGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTCAGCGTCCTCACC GTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAGCCCTC CCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACAGGTGTAC ACCCTGCCCCCATCCCGGGATGAGCTGACCAAGAACCAGGTCAGCCTGACCTGCCTGGTCAAA GGCTTCTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAACCGGAGAACAACTAC AAGACCACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCTACAGCAAGCTCACCGTG GACAAGAGCAGGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCAC AACCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGTAAAAAAGATCCCAAATTTTGGGTG CTGGTGGTGGTTGGTGGAGTCCTGGCTTGCTATAGCTTGCTAGTAACAGTGGCCTTTATTATT TTCTGGGTGAGGAGTAAGAGGAGCAGGCTCCTGCACAGTGACTACATGAACATGACTCCCCGC CGCCCCGGGCCCACCCGCAAGCATTACCAGCCCTATGCCCCACCACGCGACTTCGCAGCCTAT CGCTCCAGGGACCAGAGGCTGCCCCCCGATGCCCACAAGCCCCCTGGGGGAGGCAGTTTCCGG ACCCCCATCCAAGAGGAGCAGGCCGACGCCCACTCCACCCTGGCCAAGATCAGAGTGAAGTTC AGCAGGAGCGCAGACGCCCCCGCGTACCAGCAGGGCCAGAACCAGCTCTATAACGAGCTCAAT CTAGGACGAAGAGAGGAGTACGATGTTTTGGACAAGAGACGTGGCCGGGACCCTGAGATGGGG GGAAAGCCGAGAAGGAAGAACCCTCAGGAAGGCCTGTACAATGAACTGCAGAAAGATAAGATG GCGGAGGCCTACAGTGAGATTGGGATGAAAGGCGAGCGCCGGAGGGGCAAGGGGCACGATGGC CTTTACCAGGGTCTCAGTACAGCCACCAAGGACACCTACGACGCCCTTCACATGCAGGCCCTG CCTCCTCGCTAA (dAPRIL-CD8STK-CD28OXZ) SEQ ID No. 19 ATGGGCACCTCCCTGCTGTGCTGGATGGCCCTGTGCCTGCTGGGAGCCGACCACGCCGACGGC AAGCCCATTCCCAACCCCCTGCTGGGCCTGGACTCCACCTCTGGCGGAGGCGGCAGCGTGCTG CACCTGGTGCCCATCAACGCCACCAGCAAGGACGACTCTGATGTGACCGAGGTGATGTGGCAG CCAGCCCTGAGACGGGGCAGAGGCCTGCAGGCCCAGGGCTACGGCGTGAGAATCCAGGACGCT GGCGTGTACCTGCTGTACTCCCAGGTGCTGTTCCAGGACGTGACCTTCACAATGGGCCAGGTG GTGAGCCGGGAGGGCCAGGGCAGACAGGAGACCCTGTTCCGGTGCATCCGGAGCATGCCCAGC CACCCCGACAGAGCCTACAACAGCTGCTACAGCGCTGGCGTGTTTCACCTGCACCAGGGCGAC ATCCTGAGCGTGATCATCCCCAGAGCCAGAGCCAAGCTGAACCTGTCCCCCCACGGCACCTTT CTGGGCTTCGTGAAGCTGTCTGGAGGCGGCTCGGATCCCACCACGACGCCAGCGCCGCGACCA CCAACACCGGCGCCCACCATCGCGTCGCAGCCCCTGTCCCTGCGCCCAGAGGCGTGCCGGCCA GCGGCGGGGGGCGCAGTGCACACGAGGGGGCTGGACTTCGCCTGTGATATCTTTTGGGTGCTG GTGGTGGTTGGTGGAGTCCTGGCTTGCTATAGCTTGCTAGTAACAGTGGCCTTTATTATTTTC TGGGTGAGGAGTAAGAGGAGCAGGCTCCTGCACAGTGACTACATGAACATGACTCCCCGCCGC CCCGGGCCCACCCGCAAGCATTACCAGCCCTATGCCCCACCACGCGACTTCGCAGCCTATCGC TCCAGGGACCAGAGGCTGCCCCCCGATGCCCACAAGCCCCCTGGGGGAGGCAGTTTCCGGACC CCCATCCAAGAGGAGCAGGCCGACGCCCACTCCACCCTGGCCAAGATCAGAGTGAAGTTCAGC AGGAGCGCAGACGCCCCCGCGTACCAGCAGGGCCAGAACCAGCTCTATAACGAGCTCAATCTA GGACGAAGAGAGGAGTACGATGTTTTGGACAAGAGACGTGGCCGGGACCCTGAGATGGGGGGA AAGCCGAGAAGGAAGAACCCTCAGGAAGGCCTGTACAATGAACTGCAGAAAGATAAGATGGCG
GAGGCCTACAGTGAGATTGGGATGAAAGGCGAGCGCCGGAGGGGCAAGGGGCACGATGGCCTT TACCAGGGTCTCAGTACAGCCACCAAGGACACCTACGACGCCCTTCACATGCAGGCCCTGCCT CCTCGCTAA (dAPRIL-HNG-CD28OXZ) SEQ ID No. 20 ATGGGCACCTCCCTGCTGTGCTGGATGGCCCTGTGCCTGCTGGGAGCCGACCACGCCGACGGC AAGCCCATTCCCAACCCCCTGCTGGGCCTGGACTCCACCTCTGGCGGAGGCGGCAGCGTGCTG CACCTGGTGCCCATCAACGCCACCAGCAAGGACGACTCTGATGTGACCGAGGTGATGTGGCAG CCAGCCCTGAGACGGGGCAGAGGCCTGCAGGCCCAGGGCTACGGCGTGAGAATCCAGGACGCT GGCGTGTACCTGCTGTACTCCCAGGTGCTGTTCCAGGACGTGACCTTCACAATGGGCCAGGTG GTGAGCCGGGAGGGCCAGGGCAGACAGGAGACCCTGTTCCGGTGCATCCGGAGCATGCCCAGC CACCCCGACAGAGCCTACAACAGCTGCTACAGCGCTGGCGTGTTTCACCTGCACCAGGGCGAC ATCCTGAGCGTGATCATCCCCAGAGCCAGAGCCAAGCTGAACCTGTCCCCCCACGGCACCTTT CTGGGCTTCGTGAAGCTGTCTGGAGGCGGCTCGGATCCCGCCGAGCCCAAATCTCCTGACAAA ACTCACACATGCCCACCGTGCCCAAAAGATCCCAAATTTTGGGTGCTGGTGGTGGTTGGTGGA GTCCTGGCTTGCTATAGCTTGCTAGTAACAGTGGCCTTTATTATTTTCTGGGTGAGGAGTAAG AGGAGCAGGCTCCTGCACAGTGACTACATGAACATGACTCCCCGCCGCCCCGGGCCCACCCGC AAGCATTACCAGCCCTATGCCCCACCACGCGACTTCGCAGCCTATCGCTCCAGGGACCAGAGG CTGCCCCCCGATGCCCACAAGCCCCCTGGGGGAGGCAGTTTCCGGACCCCCATCCAAGAGGAG CAGGCCGACGCCCACTCCACCCTGGCCAAGATCAGAGTGAAGTTCAGCAGGAGCGCAGACGCC CCCGCGTACCAGCAGGGCCAGAACCAGCTCTATAACGAGCTCAATCTAGGACGAAGAGAGGAG TACGATGTTTTGGACAAGAGACGTGGCCGGGACCCTGAGATGGGGGGAAAGCCGAGAAGGAAG AACCCTCAGGAAGGCCTGTACAATGAACTGCAGAAAGATAAGATGGCGGAGGCCTACAGTGAG ATTGGGATGAAAGGCGAGCGCCGGAGGGGCAAGGGGCACGATGGCCTTTACCAGGGTCTCAGT ACAGCCACCAAGGACACCTACGACGCCCTTCACATGCAGGCCCTGCCTCCTCGCTAA
[0120] The nucleic acid sequence may encode the same amino acid sequence as that encoded by SEQ ID No. 15, 16, 17, 18 19 or 20 but may have a different nucleic acid sequence, due to the degeneracy of the genetic code. The nucleic acid sequence may have at least 80, 85, 90, 95, 98 or 99% identity to the sequence shown as SEQ ID No. 15, 16, 17, 18 19 or 20 provided that it encodes a CAR as defined in the first aspect of the invention.
[0121] Vector
[0122] The present invention also provides a vector which comprises a nucleic acid sequence according to the present invention. Such a vector may be used to introduce the nucleic acid sequence into a host cell so that it expresses and produces a molecule according to the first aspect of the invention.
[0123] The vector may, for example, be a plasmid or synthetic mRNA or a viral vector, such as a retroviral vector or a lentiviral vector.
[0124] The vector may be capable of transfecting or transducing an effector cell.
[0125] Host Cell
[0126] The invention also provides a host cell which comprises a nucleic acid according to the invention. The host cell may be capable of expressing a CAR according to the first aspect of the invention.
[0127] The host cell may be human T cell or a human NK cell.
[0128] A T-cell capable of expressing a CAR according to the invention may be made by transducing or transfecting a T cell with CAR-encoding nucleic acid.
[0129] The T-cell may be an ex vivo T cell. The T cell may be from a peripheral blood mononuclear cell (PBMC) sample. T cells may be activated and/or expanded prior to being transduced with CAR-encoding nucleic acid, for example by treatment with a anti-CD3 monoclonal antibody.
[0130] Pharmaceutical Composition
[0131] The present invention also relates to a pharmaceutical composition containing a vector or a CAR-expressing T cell of the invention together with a pharmaceutically acceptable carrier, diluent or excipient, and optionally one or more further pharmaceutically active polypeptides and/or compounds. Such a formulation may, for example, be in a form suitable for intravenous infusion).
[0132] Method of Treatment
[0133] T cells expressing a CAR molecule of the present invention are capable of killing cancer cells, such as multiple myeloma cells. CAR-expressing T cells may either be created ex vivo either from a patient's own peripheral blood (1.sup.st party), or in the setting of a haematopoietic stem cell transplant from donor peripheral blood (2.sup.nd party), or peripheral blood from an unconnected donor (3.sup.rd party). Alternatively, CAR T-cells may be derived from ex-vivo differentiation of inducible progenitor cells or embryonic progenitor cells to T-cells. In these instances, CAR T-cells are generated by introducing DNA or RNA coding for the CAR by one of many means including transduction with a viral vector, transfection with DNA or RNA.
[0134] T cells expressing a CAR molecule of the present invention may be used for the treatment of a cancerous disease, in particular a plasma cell disorder or a B cell disorder which correlates with enhanced BCMA expression.
[0135] Plasma cell disorders include plasmacytoma, plasma cell leukemia, multiple myeloma, macroglobulinemia, amyloidosis, Waldenstrom's macroglobulinemia, solitary bone plasmacytoma, extramedullary plasmacytoma, osteosclerotic myeloma (POEMS Syndrome) and heavy chain diseases as well as the clinically unclear monoclonal gammopathy of undetermined significance/smoldering multiple myeloma.
[0136] The disease may be multiple myeloma.
[0137] Examples for B cell disorders which correlate with elevated BCMA expression levels are CLL (chronic lymphocytic leukemia) and non-Hodgkins lymphoma (NHL). The bispecific binding agents of the invention may also be used in the therapy of autoimmune diseases like Systemic Lupus Erythematosus (SLE), multiple sclerosis (MS) and rheumatoid arthritis (RA).
[0138] The method of the present invention may be for treating a cancerous disease, in particular a plasma cell disorder or a B cell disorder which correlates with enhanced BCMA expression.
[0139] A method for the treatment of disease relates to the therapeutic use of a vector or T cell of the invention. In this respect, the vector or T cell may be administered to a subject having an existing disease or condition in order to lessen, reduce or improve at least one symptom associated with the disease and/or to slow down, reduce or block the progression of the disease. The method of the invention may cause or promote T-cell mediated killing of BCMA-expressing cells, such as plasma cells.
[0140] The invention will now be further described by way of Examples, which are meant to serve to assist one of ordinary skill in the art in carrying out the invention and are not intended in any way to limit the scope of the invention.
EXAMPLES
Example 1
Characterisation of BCMA as a Target for Myeloma
[0141] Primary myeloma cells were isolated by performing a CD138 immunomagnetic selection on fresh bone marrow samples from Multiple myeloma patients that were known to have frank disease. These cells were stained with the BCMA specific J6MO mAb (GSK) which was conjugated to PE. At the same time, a standard of beads with known numbers of binding sites was generated using the PE Quantibrite bead kit (Becton Dickenson) as per the manufacturer's instructions. The BCMA copy number on myeloma cells could be derived by correlating the mean-fluorescent intensity from the myeloma cells with the standard curve derived from the beads. It was found that the range of BCMA copy number on a myeloma cell surface is low: at 348.7-4268.4 BCMA copies per cell with a mean of 1181 and a median of 1084.9 (FIG. 2). This is considerably lower than e.g. CD19 and GD2, classic targets for CARs. Presence of BCMA expression on primary myeloma cells was also confirmed with the Vicky-1 antibody (Abcam Ab17323), examples of which are shown in FIG. 14.
Example 2
Design and Construction of APRIL Based CARs
[0142] APRIL in its natural form is a secreted type II protein. The use of APRIL as a BCMA binding domain for a CAR requires conversion of this type II secreted protein to a type I membrane bound protein and for this protein to be stable and to retain binding to BCMA in this form. To generate candidate molecules, the extreme amino-terminus of APRIL was deleted to remove binding to proteoglycans. Next, a signal peptide was added to direct the nascent protein to the endoplasmic reticulum and hence the cell surface. Also, because the nature of spacer used can alter the function of a CAR, three different spacer domains were tested: an APRIL based CAR was generated comprising (i) a human IgG1 spacer altered to remove Fc binding motifs; (ii) a CD8 stalk; and (iii) the IgG1 hinge alone (cartoon in FIGS. 4A-4C and amino acid sequences in FIGS. 5A-5C, and also amino acid sequences in FIG. 19 which differ from the sequences in FIGS. 5A-5C by having a different signal peptide and the V5 epitope tag). These CARs were expressed in a bicistronic retroviral vector (FIG. 6A) so that a marker protein-truncated CD34 could be co-expressed as a convenient marker gene.
Example 3
Expression and Function of APRIL Based CARs
[0143] The aim of this study was to test whether the APRIL based CARs which had been constructed were expressed on the cell surface and whether APRIL had folded to form the native protein. T-cells were transduced with these different CAR constructs and stained using a commercially available anti-APRIL mAb, along with staining for the marker gene and analysed by flow-cytometry. The results of this experiment are shown in FIG. 6B where APRIL binding is plotting against marker gene fluorescence. These data show that in this format, the APRIL based CARs are expressed on the cell surface and APRIL folds sufficiently to be recognized by an anti-APRIL mAb.
[0144] Next, it was determined whether APRIL in this format could recognize BCMA and TACI. Recombinant BCMA and TACI were generated as fusions with mouse IgG2a-Fc. These recombinant proteins were incubated with the transduced T-cells. After this, the cells were washed and stained with an anti-mouse fluorophore conjugated antibody and an antibody to detect the marker gene conjugated to a different fluorophore. The cells were analysed by flow cytometry and the results are presented in FIG. 6C. The different CARs were able to bind both BCMA and TACI. Surprisingly, the CARs were better able to bind BCMA than TACI. Also, surprisingly CARs with a CD8 stalk or IgG1 hinge spacer were better able to bind BCMA and TACI than CAR with an Fc spacer.
Example 4
APRIL Based Chimeric Antigen Receptors are Active Against BCMA Expressing Cells
[0145] T-cells from normal donors were transduced with the different APRIL CARs and tested against SupT1 cells either wild-type, or engineered to express BCMA and TACI. Several different assays were used to determine function. A classical chromium release assay was performed. Here, the target cells (the SupT1 cells) were labelled with .sup.51Cr and mixed with effectors (the transduced T-cells) at different ratio. Lysis of target cells was determined by counting .sup.51Cr in the co-culture supernatant (FIG. 6A shows the cumulative data, example data from a single assay with different effector:target ratios is shown in FIG. 12).
[0146] In addition, supernatant from T-cells cultured 1:1 with SupT1 cells was assayed by ELISA for Interferon-gamma (FIG. 6B shows cumulative data, example data from a single assay is shown in FIG. 13). Measurement of T-cell expansion after one week of co-culture with SupT1 cells was also performed (FIG. 6C). T-cells were counted by flow-cytometry calibrated with counting beads. These experimental data show that APRIL based CARs can kill BCMA expressing targets. Further, these data show that CARs based on the CD8 stalk or IgG1 hinge performed better than the Fc-pvaa based CAR.
Example 5
APRIL Based CARs are Able to Kill Primary Myeloma Cells
[0147] The above data are encouraging since they demonstrate that it in principle, it is possible to make an APRIL based CAR. However, since most primary myeloma cells express a low number of BCMA molecules on their surface, it was investigated whether such an APRIL based CAR would cause killing of primary myeloma cells, particularly in cases with low-density expression. Three cases were selected which represented the range of BCMA expression described in FIG. 2: the first had dim expression (lower than mean); the second case had intermediate expression (approximately mean expression) and the third had bright (above mean expression). FIG. 8 shows a histogram of BCMA staining against isotype control for all three cases on the left to illustrate BCMA expression. Since when comparing APRIL based CARs with different spacers it had been determined that CARs with CD8 stalk spacer and IgG1 hinge spacer performed better than the Fc-pvaa spacered CAR, in this assay, only the CD8 stalk and hinge APRIL CARs were tested. On the left, survival of myeloma cells compared with starting numbers is shown at day 3 and day 6 after a 1:1 co-culture of myeloma cells and CAR T-cells. By day 6, >95% of the myeloma cells were eliminated, including those with dim BCMA expression. Dim BCMA expressing myeloma cells can be targeted by the APRIL CARs albeit with a slower tempo of killing than higher expressers.
Example 6
Secreted and Truncated APRIL Fused to an Fc Spacer Recognizes BCMA and TACI
[0148] In order to investigate whether truncated APRIL in a CAR format (i.e. fused to a trans-membrane domain and anchored to a cell membrane) could bind BCMA and TACI, a basic CAR was engineered in frame with the self-cleaving foot and mouth disease 2A peptide with truncated CD34, as a convenient marker gene. A stable SUPT1 cell line was established which expresses this construct. Secreted truncated BCMA and TACI fused to human (and other species, not shown) Ig Fc domain was also generated and recombinant protein produced. It was shown that both BCMA-Fc and TACI-Fc bind the engineered SUPT1 cell line. Only cells expressing the CD34 marker gene were found to bind BCMA-Fc and TACI-Fc (FIG. 9).
Example 7
APRIL Based Chimeric Antigen Receptors are Stably Expressed on the Surface of T-Cells
[0149] The CAR spacer domain can alter sensitivity and specificity. Three versions of an APRIL-based CAR were generated with three spacer domains: (i) a human IgG1 spacer altered to remove Fc binding motifs; (ii) a CD8 stalk; and (iii) the IgG1 hinge alone (FIG. 10B). Primary human T-cells were transduced with these different CARs and stained using a commercially available anti-APRIL mAb (FIG. 11).
Example 8
APRIL Based Chimeric Antigen Receptors are Active Against Cognate Target Expressing Cells
[0150] T-cells from normal donors were transduced with the different APRIL CARs and tested against SupT1 cells either wild-type, or engineered to express BCMA and TACI. Several different assays were used to determine function. A classical chromium release assay was performed. Here, the target cells (the SupT1 cells) were labelled with .sup.51Cr and mixed with effectors (the transduced T-cells) at different ratio. Lysis of target cells was determined by counting .sup.51Cr in the co-culture supernatant (FIG. 12).
[0151] In addition, supernatant from T-cells cultured 1:1 with SupT1 cells was assayed by ELISA for Interferon-gamma (FIG. 13).
[0152] Measurement of T-cell expansion after one week of co-culture with SupT1 cells was also performed. T-cells were counted by flow-cytometry calibrated with counting beads. Initial data (not shown) appears to indicate that the CD8 stalk based construct results in more T-cell proliferation than the other constructs.
Example 9
Demonstration of In Vivo Function of APRIL CAR T-Cells
[0153] In order to demonstrate APRIL CAR T-cell function in vivo, APRIL CAR T-cells were tested in a human/mouse chimeric model. MM1.s (ATCC CRL-2974) is a human myeloma cell line which expresses intermediate levels of BCMA. The inventors engineered this cell line to express firefly Luciferase to derive the cell-line MM1.s.FLuc.
[0154] NOD scid gamma (NSG: NOD.Cg-Prkdc.sup.scid II2rgtm1.sup.Wjl/SzJ) mice are profoundly immunosuppressed mice capable of engrafting several human cell lines and human peripheral blood lymphocytes. Three month old female NSG mice received 1.times.10.sup.7 MM1.s.FLuc cells vial tail-vein injection without any preparative therapy. Engraftment was determined by serial bioluminescence imaging (FIG. 16). Robust and increasing intramedullary engraftment was observed in all mice. At day 13, 5.times.10.sup.6 APRIL-HNG-CD28OXZ CAR T-cells were administered via tail vein injection. Serial bioluminescence was performed which showed rapid decrease in burden of MM1.s (FIG. 16) in all treated mice to a complete remission. This response to CAR therapy was confirmed by flow-cytometry and immunohistochemistry.
[0155] All publications mentioned in the above specification are herein incorporated by reference. Various modifications and variations of the described methods and system of the invention will be apparent to those skilled in the art without departing from the scope and spirit of the invention. Although the invention has been described in connection with specific preferred embodiments, it should be understood that the invention as claimed should not be unduly limited to such specific embodiments. Indeed, various modifications of the described modes for carrying out the invention which are obvious to those skilled in molecular biology, cellular immunology or related fields are intended to be within the scope of the following claims.
Sequence CWU 1
1
211614PRTArtificial SequenceChimeric antigen receptor (CAR) dAPRIL-
HCH2CH3pvaa-CD28OXZ 1Met Glu Thr Asp Thr Leu Leu Leu Trp Val Leu
Leu Leu Trp Val Pro1 5 10 15Gly Ser Thr Gly Ser Val Leu His Leu Val
Pro Ile Asn Ala Thr Ser 20 25 30Lys Asp Asp Ser Asp Val Thr Glu Val
Met Trp Gln Pro Ala Leu Arg 35 40 45Arg Gly Arg Gly Leu Gln Ala Gln
Gly Tyr Gly Val Arg Ile Gln Asp 50 55 60Ala Gly Val Tyr Leu Leu Tyr
Ser Gln Val Leu Phe Gln Asp Val Thr65 70 75 80Phe Thr Met Gly Gln
Val Val Ser Arg Glu Gly Gln Gly Arg Gln Glu 85 90 95Thr Leu Phe Arg
Cys Ile Arg Ser Met Pro Ser His Pro Asp Arg Ala 100 105 110Tyr Asn
Ser Cys Tyr Ser Ala Gly Val Phe His Leu His Gln Gly Asp 115 120
125Ile Leu Ser Val Ile Ile Pro Arg Ala Arg Ala Lys Leu Asn Leu Ser
130 135 140Pro His Gly Thr Phe Leu Gly Phe Val Lys Leu Ser Gly Gly
Gly Ser145 150 155 160Asp Pro Ala Glu Pro Lys Ser Pro Asp Lys Thr
His Thr Cys Pro Pro 165 170 175Cys Pro Ala Pro Pro Val Ala Gly Pro
Ser Val Phe Leu Phe Pro Pro 180 185 190Lys Pro Lys Asp Thr Leu Met
Ile Ala Arg Thr Pro Glu Val Thr Cys 195 200 205Val Val Val Asp Val
Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp 210 215 220Tyr Val Asp
Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu225 230 235
240Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu
245 250 255His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val
Ser Asn 260 265 270Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser
Lys Ala Lys Gly 275 280 285Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu
Pro Pro Ser Arg Asp Glu 290 295 300Leu Thr Lys Asn Gln Val Ser Leu
Thr Cys Leu Val Lys Gly Phe Tyr305 310 315 320Pro Ser Asp Ile Ala
Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn 325 330 335Asn Tyr Lys
Thr Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe 340 345 350Leu
Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn 355 360
365Val Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr
370 375 380Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys Lys Asp Pro Lys
Phe Trp385 390 395 400Val Leu Val Val Val Gly Gly Val Leu Ala Cys
Tyr Ser Leu Leu Val 405 410 415Thr Val Ala Phe Ile Ile Phe Trp Val
Arg Ser Lys Arg Ser Arg Leu 420 425 430Leu His Ser Asp Tyr Met Asn
Met Thr Pro Arg Arg Pro Gly Pro Thr 435 440 445Arg Lys His Tyr Gln
Pro Tyr Ala Pro Pro Arg Asp Phe Ala Ala Tyr 450 455 460Arg Ser Arg
Asp Gln Arg Leu Pro Pro Asp Ala His Lys Pro Pro Gly465 470 475
480Gly Gly Ser Phe Arg Thr Pro Ile Gln Glu Glu Gln Ala Asp Ala His
485 490 495Ser Thr Leu Ala Lys Ile Arg Val Lys Phe Ser Arg Ser Ala
Asp Ala 500 505 510Pro Ala Tyr Gln Gln Gly Gln Asn Gln Leu Tyr Asn
Glu Leu Asn Leu 515 520 525Gly Arg Arg Glu Glu Tyr Asp Val Leu Asp
Lys Arg Arg Gly Arg Asp 530 535 540Pro Glu Met Gly Gly Lys Pro Arg
Arg Lys Asn Pro Gln Glu Gly Leu545 550 555 560Tyr Asn Glu Leu Gln
Lys Asp Lys Met Ala Glu Ala Tyr Ser Glu Ile 565 570 575Gly Met Lys
Gly Glu Arg Arg Arg Gly Lys Gly His Asp Gly Leu Tyr 580 585 590Gln
Gly Leu Ser Thr Ala Thr Lys Asp Thr Tyr Asp Ala Leu His Met 595 600
605Gln Ala Leu Pro Pro Arg 6102424PRTArtificial SequenceChimeric
antigen receptor (CAR) dAPRIL-CD8STK- CD28OXZ 2Met Glu Thr Asp Thr
Leu Leu Leu Trp Val Leu Leu Leu Trp Val Pro1 5 10 15Gly Ser Thr Gly
Ser Val Leu His Leu Val Pro Ile Asn Ala Thr Ser 20 25 30Lys Asp Asp
Ser Asp Val Thr Glu Val Met Trp Gln Pro Ala Leu Arg 35 40 45Arg Gly
Arg Gly Leu Gln Ala Gln Gly Tyr Gly Val Arg Ile Gln Asp 50 55 60Ala
Gly Val Tyr Leu Leu Tyr Ser Gln Val Leu Phe Gln Asp Val Thr65 70 75
80Phe Thr Met Gly Gln Val Val Ser Arg Glu Gly Gln Gly Arg Gln Glu
85 90 95Thr Leu Phe Arg Cys Ile Arg Ser Met Pro Ser His Pro Asp Arg
Ala 100 105 110Tyr Asn Ser Cys Tyr Ser Ala Gly Val Phe His Leu His
Gln Gly Asp 115 120 125Ile Leu Ser Val Ile Ile Pro Arg Ala Arg Ala
Lys Leu Asn Leu Ser 130 135 140Pro His Gly Thr Phe Leu Gly Phe Val
Lys Leu Ser Gly Gly Gly Ser145 150 155 160Asp Pro Thr Thr Thr Pro
Ala Pro Arg Pro Pro Thr Pro Ala Pro Thr 165 170 175Ile Ala Ser Gln
Pro Leu Ser Leu Arg Pro Glu Ala Cys Arg Pro Ala 180 185 190Ala Gly
Gly Ala Val His Thr Arg Gly Leu Asp Phe Ala Cys Asp Ile 195 200
205Phe Trp Val Leu Val Val Val Gly Gly Val Leu Ala Cys Tyr Ser Leu
210 215 220Leu Val Thr Val Ala Phe Ile Ile Phe Trp Val Arg Ser Lys
Arg Ser225 230 235 240Arg Leu Leu His Ser Asp Tyr Met Asn Met Thr
Pro Arg Arg Pro Gly 245 250 255Pro Thr Arg Lys His Tyr Gln Pro Tyr
Ala Pro Pro Arg Asp Phe Ala 260 265 270Ala Tyr Arg Ser Arg Asp Gln
Arg Leu Pro Pro Asp Ala His Lys Pro 275 280 285Pro Gly Gly Gly Ser
Phe Arg Thr Pro Ile Gln Glu Glu Gln Ala Asp 290 295 300Ala His Ser
Thr Leu Ala Lys Ile Arg Val Lys Phe Ser Arg Ser Ala305 310 315
320Asp Ala Pro Ala Tyr Gln Gln Gly Gln Asn Gln Leu Tyr Asn Glu Leu
325 330 335Asn Leu Gly Arg Arg Glu Glu Tyr Asp Val Leu Asp Lys Arg
Arg Gly 340 345 350Arg Asp Pro Glu Met Gly Gly Lys Pro Arg Arg Lys
Asn Pro Gln Glu 355 360 365Gly Leu Tyr Asn Glu Leu Gln Lys Asp Lys
Met Ala Glu Ala Tyr Ser 370 375 380Glu Ile Gly Met Lys Gly Glu Arg
Arg Arg Gly Lys Gly His Asp Gly385 390 395 400Leu Tyr Gln Gly Leu
Ser Thr Ala Thr Lys Asp Thr Tyr Asp Ala Leu 405 410 415His Met Gln
Ala Leu Pro Pro Arg 4203398PRTArtificial SequenceChimeric antigen
receptor (CAR) dAPRIL-HNG- CD28OXZ 3Met Glu Thr Asp Thr Leu Leu Leu
Trp Val Leu Leu Leu Trp Val Pro1 5 10 15Gly Ser Thr Gly Ser Val Leu
His Leu Val Pro Ile Asn Ala Thr Ser 20 25 30Lys Asp Asp Ser Asp Val
Thr Glu Val Met Trp Gln Pro Ala Leu Arg 35 40 45Arg Gly Arg Gly Leu
Gln Ala Gln Gly Tyr Gly Val Arg Ile Gln Asp 50 55 60Ala Gly Val Tyr
Leu Leu Tyr Ser Gln Val Leu Phe Gln Asp Val Thr65 70 75 80Phe Thr
Met Gly Gln Val Val Ser Arg Glu Gly Gln Gly Arg Gln Glu 85 90 95Thr
Leu Phe Arg Cys Ile Arg Ser Met Pro Ser His Pro Asp Arg Ala 100 105
110Tyr Asn Ser Cys Tyr Ser Ala Gly Val Phe His Leu His Gln Gly Asp
115 120 125Ile Leu Ser Val Ile Ile Pro Arg Ala Arg Ala Lys Leu Asn
Leu Ser 130 135 140Pro His Gly Thr Phe Leu Gly Phe Val Lys Leu Ser
Gly Gly Gly Ser145 150 155 160Asp Pro Ala Glu Pro Lys Ser Pro Asp
Lys Thr His Thr Cys Pro Pro 165 170 175Cys Pro Lys Asp Pro Lys Phe
Trp Val Leu Val Val Val Gly Gly Val 180 185 190Leu Ala Cys Tyr Ser
Leu Leu Val Thr Val Ala Phe Ile Ile Phe Trp 195 200 205Val Arg Ser
Lys Arg Ser Arg Leu Leu His Ser Asp Tyr Met Asn Met 210 215 220Thr
Pro Arg Arg Pro Gly Pro Thr Arg Lys His Tyr Gln Pro Tyr Ala225 230
235 240Pro Pro Arg Asp Phe Ala Ala Tyr Arg Ser Arg Asp Gln Arg Leu
Pro 245 250 255Pro Asp Ala His Lys Pro Pro Gly Gly Gly Ser Phe Arg
Thr Pro Ile 260 265 270Gln Glu Glu Gln Ala Asp Ala His Ser Thr Leu
Ala Lys Ile Arg Val 275 280 285Lys Phe Ser Arg Ser Ala Asp Ala Pro
Ala Tyr Gln Gln Gly Gln Asn 290 295 300Gln Leu Tyr Asn Glu Leu Asn
Leu Gly Arg Arg Glu Glu Tyr Asp Val305 310 315 320Leu Asp Lys Arg
Arg Gly Arg Asp Pro Glu Met Gly Gly Lys Pro Arg 325 330 335Arg Lys
Asn Pro Gln Glu Gly Leu Tyr Asn Glu Leu Gln Lys Asp Lys 340 345
350Met Ala Glu Ala Tyr Ser Glu Ile Gly Met Lys Gly Glu Arg Arg Arg
355 360 365Gly Lys Gly His Asp Gly Leu Tyr Gln Gly Leu Ser Thr Ala
Thr Lys 370 375 380Asp Thr Tyr Asp Ala Leu His Met Gln Ala Leu Pro
Pro Arg385 390 3954633PRTArtificial SequenceChimeric antigen
receptor (CAR) dAPRIL- HCH2CH3pvaa-CD28OXZ 4Met Gly Thr Ser Leu Leu
Cys Trp Met Ala Leu Cys Leu Leu Gly Ala1 5 10 15Asp His Ala Asp Gly
Lys Pro Ile Pro Asn Pro Leu Leu Gly Leu Asp 20 25 30Ser Thr Ser Gly
Gly Gly Gly Ser Val Leu His Leu Val Pro Ile Asn 35 40 45Ala Thr Ser
Lys Asp Asp Ser Asp Val Thr Glu Val Met Trp Gln Pro 50 55 60Ala Leu
Arg Arg Gly Arg Gly Leu Gln Ala Gln Gly Tyr Gly Val Arg65 70 75
80Ile Gln Asp Ala Gly Val Tyr Leu Leu Tyr Ser Gln Val Leu Phe Gln
85 90 95Asp Val Thr Phe Thr Met Gly Gln Val Val Ser Arg Glu Gly Gln
Gly 100 105 110Arg Gln Glu Thr Leu Phe Arg Cys Ile Arg Ser Met Pro
Ser His Pro 115 120 125Asp Arg Ala Tyr Asn Ser Cys Tyr Ser Ala Gly
Val Phe His Leu His 130 135 140Gln Gly Asp Ile Leu Ser Val Ile Ile
Pro Arg Ala Arg Ala Lys Leu145 150 155 160Asn Leu Ser Pro His Gly
Thr Phe Leu Gly Phe Val Lys Leu Ser Gly 165 170 175Gly Gly Ser Asp
Pro Ala Glu Pro Lys Ser Pro Asp Lys Thr His Thr 180 185 190Cys Pro
Pro Cys Pro Ala Pro Pro Val Ala Gly Pro Ser Val Phe Leu 195 200
205Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ala Arg Thr Pro Glu
210 215 220Val Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu
Val Lys225 230 235 240Phe Asn Trp Tyr Val Asp Gly Val Glu Val His
Asn Ala Lys Thr Lys 245 250 255Pro Arg Glu Glu Gln Tyr Asn Ser Thr
Tyr Arg Val Val Ser Val Leu 260 265 270Thr Val Leu His Gln Asp Trp
Leu Asn Gly Lys Glu Tyr Lys Cys Lys 275 280 285Val Ser Asn Lys Ala
Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys 290 295 300Ala Lys Gly
Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser305 310 315
320Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys
325 330 335Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn
Gly Gln 340 345 350Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu
Asp Ser Asp Gly 355 360 365Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val
Asp Lys Ser Arg Trp Gln 370 375 380Gln Gly Asn Val Phe Ser Cys Ser
Val Met His Glu Ala Leu His Asn385 390 395 400His Tyr Thr Gln Lys
Ser Leu Ser Leu Ser Pro Gly Lys Lys Asp Pro 405 410 415Lys Phe Trp
Val Leu Val Val Val Gly Gly Val Leu Ala Cys Tyr Ser 420 425 430Leu
Leu Val Thr Val Ala Phe Ile Ile Phe Trp Val Arg Ser Lys Arg 435 440
445Ser Arg Leu Leu His Ser Asp Tyr Met Asn Met Thr Pro Arg Arg Pro
450 455 460Gly Pro Thr Arg Lys His Tyr Gln Pro Tyr Ala Pro Pro Arg
Asp Phe465 470 475 480Ala Ala Tyr Arg Ser Arg Asp Gln Arg Leu Pro
Pro Asp Ala His Lys 485 490 495Pro Pro Gly Gly Gly Ser Phe Arg Thr
Pro Ile Gln Glu Glu Gln Ala 500 505 510Asp Ala His Ser Thr Leu Ala
Lys Ile Arg Val Lys Phe Ser Arg Ser 515 520 525Ala Asp Ala Pro Ala
Tyr Gln Gln Gly Gln Asn Gln Leu Tyr Asn Glu 530 535 540Leu Asn Leu
Gly Arg Arg Glu Glu Tyr Asp Val Leu Asp Lys Arg Arg545 550 555
560Gly Arg Asp Pro Glu Met Gly Gly Lys Pro Arg Arg Lys Asn Pro Gln
565 570 575Glu Gly Leu Tyr Asn Glu Leu Gln Lys Asp Lys Met Ala Glu
Ala Tyr 580 585 590Ser Glu Ile Gly Met Lys Gly Glu Arg Arg Arg Gly
Lys Gly His Asp 595 600 605Gly Leu Tyr Gln Gly Leu Ser Thr Ala Thr
Lys Asp Thr Tyr Asp Ala 610 615 620Leu His Met Gln Ala Leu Pro Pro
Arg625 6305443PRTArtificial SequenceChimeric antigen receptor (CAR)
dAPRIL-CD8STK- CD28OXZ 5Met Gly Thr Ser Leu Leu Cys Trp Met Ala Leu
Cys Leu Leu Gly Ala1 5 10 15Asp His Ala Asp Gly Lys Pro Ile Pro Asn
Pro Leu Leu Gly Leu Asp 20 25 30Ser Thr Ser Gly Gly Gly Gly Ser Val
Leu His Leu Val Pro Ile Asn 35 40 45Ala Thr Ser Lys Asp Asp Ser Asp
Val Thr Glu Val Met Trp Gln Pro 50 55 60Ala Leu Arg Arg Gly Arg Gly
Leu Gln Ala Gln Gly Tyr Gly Val Arg65 70 75 80Ile Gln Asp Ala Gly
Val Tyr Leu Leu Tyr Ser Gln Val Leu Phe Gln 85 90 95Asp Val Thr Phe
Thr Met Gly Gln Val Val Ser Arg Glu Gly Gln Gly 100 105 110Arg Gln
Glu Thr Leu Phe Arg Cys Ile Arg Ser Met Pro Ser His Pro 115 120
125Asp Arg Ala Tyr Asn Ser Cys Tyr Ser Ala Gly Val Phe His Leu His
130 135 140Gln Gly Asp Ile Leu Ser Val Ile Ile Pro Arg Ala Arg Ala
Lys Leu145 150 155 160Asn Leu Ser Pro His Gly Thr Phe Leu Gly Phe
Val Lys Leu Ser Gly 165 170 175Gly Gly Ser Asp Pro Thr Thr Thr Pro
Ala Pro Arg Pro Pro Thr Pro 180 185 190Ala Pro Thr Ile Ala Ser Gln
Pro Leu Ser Leu Arg Pro Glu Ala Cys 195 200 205Arg Pro Ala Ala Gly
Gly Ala Val His Thr Arg Gly Leu Asp Phe Ala 210 215 220Cys Asp Ile
Phe Trp Val Leu Val Val Val Gly Gly Val Leu Ala Cys225 230 235
240Tyr Ser Leu Leu Val Thr Val Ala Phe Ile Ile Phe Trp Val Arg Ser
245 250 255Lys Arg Ser Arg Leu Leu His Ser Asp Tyr Met Asn Met Thr
Pro Arg 260 265 270Arg Pro Gly Pro Thr Arg Lys His Tyr Gln Pro Tyr
Ala Pro Pro Arg 275 280 285Asp Phe Ala Ala Tyr Arg Ser Arg Asp Gln
Arg Leu Pro Pro Asp Ala 290 295 300His Lys Pro Pro Gly Gly Gly Ser
Phe Arg Thr Pro Ile Gln Glu Glu305 310 315 320Gln Ala Asp Ala His
Ser Thr Leu Ala Lys Ile Arg Val Lys Phe Ser 325 330 335Arg Ser Ala
Asp Ala Pro Ala Tyr Gln Gln Gly Gln Asn Gln Leu Tyr 340 345
350Asn Glu Leu Asn Leu Gly Arg Arg Glu Glu Tyr Asp Val Leu Asp Lys
355 360 365Arg Arg Gly Arg Asp Pro Glu Met Gly Gly Lys Pro Arg Arg
Lys Asn 370 375 380Pro Gln Glu Gly Leu Tyr Asn Glu Leu Gln Lys Asp
Lys Met Ala Glu385 390 395 400Ala Tyr Ser Glu Ile Gly Met Lys Gly
Glu Arg Arg Arg Gly Lys Gly 405 410 415His Asp Gly Leu Tyr Gln Gly
Leu Ser Thr Ala Thr Lys Asp Thr Tyr 420 425 430Asp Ala Leu His Met
Gln Ala Leu Pro Pro Arg 435 4406417PRTArtificial SequenceChimeric
antigen receptor (CAR) dAPRIL-HNG- CD28OXZ 6Met Gly Thr Ser Leu Leu
Cys Trp Met Ala Leu Cys Leu Leu Gly Ala1 5 10 15Asp His Ala Asp Gly
Lys Pro Ile Pro Asn Pro Leu Leu Gly Leu Asp 20 25 30Ser Thr Ser Gly
Gly Gly Gly Ser Val Leu His Leu Val Pro Ile Asn 35 40 45Ala Thr Ser
Lys Asp Asp Ser Asp Val Thr Glu Val Met Trp Gln Pro 50 55 60Ala Leu
Arg Arg Gly Arg Gly Leu Gln Ala Gln Gly Tyr Gly Val Arg65 70 75
80Ile Gln Asp Ala Gly Val Tyr Leu Leu Tyr Ser Gln Val Leu Phe Gln
85 90 95Asp Val Thr Phe Thr Met Gly Gln Val Val Ser Arg Glu Gly Gln
Gly 100 105 110Arg Gln Glu Thr Leu Phe Arg Cys Ile Arg Ser Met Pro
Ser His Pro 115 120 125Asp Arg Ala Tyr Asn Ser Cys Tyr Ser Ala Gly
Val Phe His Leu His 130 135 140Gln Gly Asp Ile Leu Ser Val Ile Ile
Pro Arg Ala Arg Ala Lys Leu145 150 155 160Asn Leu Ser Pro His Gly
Thr Phe Leu Gly Phe Val Lys Leu Ser Gly 165 170 175Gly Gly Ser Asp
Pro Ala Glu Pro Lys Ser Pro Asp Lys Thr His Thr 180 185 190Cys Pro
Pro Cys Pro Lys Asp Pro Lys Phe Trp Val Leu Val Val Val 195 200
205Gly Gly Val Leu Ala Cys Tyr Ser Leu Leu Val Thr Val Ala Phe Ile
210 215 220Ile Phe Trp Val Arg Ser Lys Arg Ser Arg Leu Leu His Ser
Asp Tyr225 230 235 240Met Asn Met Thr Pro Arg Arg Pro Gly Pro Thr
Arg Lys His Tyr Gln 245 250 255Pro Tyr Ala Pro Pro Arg Asp Phe Ala
Ala Tyr Arg Ser Arg Asp Gln 260 265 270Arg Leu Pro Pro Asp Ala His
Lys Pro Pro Gly Gly Gly Ser Phe Arg 275 280 285Thr Pro Ile Gln Glu
Glu Gln Ala Asp Ala His Ser Thr Leu Ala Lys 290 295 300Ile Arg Val
Lys Phe Ser Arg Ser Ala Asp Ala Pro Ala Tyr Gln Gln305 310 315
320Gly Gln Asn Gln Leu Tyr Asn Glu Leu Asn Leu Gly Arg Arg Glu Glu
325 330 335Tyr Asp Val Leu Asp Lys Arg Arg Gly Arg Asp Pro Glu Met
Gly Gly 340 345 350Lys Pro Arg Arg Lys Asn Pro Gln Glu Gly Leu Tyr
Asn Glu Leu Gln 355 360 365Lys Asp Lys Met Ala Glu Ala Tyr Ser Glu
Ile Gly Met Lys Gly Glu 370 375 380Arg Arg Arg Gly Lys Gly His Asp
Gly Leu Tyr Gln Gly Leu Ser Thr385 390 395 400Ala Thr Lys Asp Thr
Tyr Asp Ala Leu His Met Gln Ala Leu Pro Pro 405 410
415Arg7216PRTArtificial SequenceTransmembrane and intracellular
T-cell signalling domain (endodomain) 7Phe Trp Val Leu Val Val Val
Gly Gly Val Leu Ala Cys Tyr Ser Leu1 5 10 15Leu Val Thr Val Ala Phe
Ile Ile Phe Trp Val Arg Ser Lys Arg Ser 20 25 30Arg Leu Leu His Ser
Asp Tyr Met Asn Met Thr Pro Arg Arg Pro Gly 35 40 45Pro Thr Arg Lys
His Tyr Gln Pro Tyr Ala Pro Pro Arg Asp Phe Ala 50 55 60Ala Tyr Arg
Ser Arg Asp Gln Arg Leu Pro Pro Asp Ala His Lys Pro65 70 75 80Pro
Gly Gly Gly Ser Phe Arg Thr Pro Ile Gln Glu Glu Gln Ala Asp 85 90
95Ala His Ser Thr Leu Ala Lys Ile Arg Val Lys Phe Ser Arg Ser Ala
100 105 110Asp Ala Pro Ala Tyr Gln Gln Gly Gln Asn Gln Leu Tyr Asn
Glu Leu 115 120 125Asn Leu Gly Arg Arg Glu Glu Tyr Asp Val Leu Asp
Lys Arg Arg Gly 130 135 140Arg Asp Pro Glu Met Gly Gly Lys Pro Arg
Arg Lys Asn Pro Gln Glu145 150 155 160Gly Leu Tyr Asn Glu Leu Gln
Lys Asp Lys Met Ala Glu Ala Tyr Ser 165 170 175Glu Ile Gly Met Lys
Gly Glu Arg Arg Arg Gly Lys Gly His Asp Gly 180 185 190Leu Tyr Gln
Gly Leu Ser Thr Ala Thr Lys Asp Thr Tyr Asp Ala Leu 195 200 205His
Met Gln Ala Leu Pro Pro Arg 210 215821PRTArtificial SequenceSignal
peptide 8Met Gly Thr Ser Leu Leu Cys Trp Met Ala Leu Cys Leu Leu
Gly Ala1 5 10 15Asp His Ala Asp Gly 20920PRTArtificial
SequenceSignal peptide 9Met Glu Thr Asp Thr Leu Leu Leu Trp Val Leu
Leu Leu Trp Val Pro1 5 10 15Gly Ser Thr Gly 2010234PRTArtificial
SequenceSpacer (hinge-CH2CH3 of human IgG1) 10Ala Glu Pro Lys Ser
Pro Asp Lys Thr His Thr Cys Pro Pro Cys Pro1 5 10 15Ala Pro Pro Val
Ala Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro 20 25 30Lys Asp Thr
Leu Met Ile Ala Arg Thr Pro Glu Val Thr Cys Val Val 35 40 45Val Asp
Val Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val 50 55 60Asp
Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln65 70 75
80Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln
85 90 95Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys
Ala 100 105 110Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys
Gly Gln Pro 115 120 125Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser
Arg Asp Glu Leu Thr 130 135 140Lys Asn Gln Val Ser Leu Thr Cys Leu
Val Lys Gly Phe Tyr Pro Ser145 150 155 160Asp Ile Ala Val Glu Trp
Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr 165 170 175Lys Thr Thr Pro
Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr 180 185 190Ser Lys
Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe 195 200
205Ser Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys
210 215 220Ser Leu Ser Leu Ser Pro Gly Lys Lys Asp225
2301146PRTArtificial SequenceSpacer (human CD8 stalk) 11Thr Thr Thr
Pro Ala Pro Arg Pro Pro Thr Pro Ala Pro Thr Ile Ala1 5 10 15Ser Gln
Pro Leu Ser Leu Arg Pro Glu Ala Cys Arg Pro Ala Ala Gly 20 25 30Gly
Ala Val His Thr Arg Gly Leu Asp Phe Ala Cys Asp Ile 35 40
451220PRTArtificial SequenceSpacer (human IgG1 hinge) 12Ala Glu Pro
Lys Ser Pro Asp Lys Thr His Thr Cys Pro Pro Cys Pro1 5 10 15Lys Asp
Pro Lys 2013250PRTHomo sapiens 13Met Pro Ala Ser Ser Pro Phe Leu
Leu Ala Pro Lys Gly Pro Pro Gly1 5 10 15Asn Met Gly Gly Pro Val Arg
Glu Pro Ala Leu Ser Val Ala Leu Trp 20 25 30Leu Ser Trp Gly Ala Ala
Leu Gly Ala Val Ala Cys Ala Met Ala Leu 35 40 45Leu Thr Gln Gln Thr
Glu Leu Gln Ser Leu Arg Arg Glu Val Ser Arg 50 55 60Leu Gln Gly Thr
Gly Gly Pro Ser Gln Asn Gly Glu Gly Tyr Pro Trp65 70 75 80Gln Ser
Leu Pro Glu Gln Ser Ser Asp Ala Leu Glu Ala Trp Glu Asn 85 90 95Gly
Glu Arg Ser Arg Lys Arg Arg Ala Val Leu Thr Gln Lys Gln Lys 100 105
110Lys Gln His Ser Val Leu His Leu Val Pro Ile Asn Ala Thr Ser Lys
115 120 125Asp Asp Ser Asp Val Thr Glu Val Met Trp Gln Pro Ala Leu
Arg Arg 130 135 140Gly Arg Gly Leu Gln Ala Gln Gly Tyr Gly Val Arg
Ile Gln Asp Ala145 150 155 160Gly Val Tyr Leu Leu Tyr Ser Gln Val
Leu Phe Gln Asp Val Thr Phe 165 170 175Thr Met Gly Gln Val Val Ser
Arg Glu Gly Gln Gly Arg Gln Glu Thr 180 185 190Leu Phe Arg Cys Ile
Arg Ser Met Pro Ser His Pro Asp Arg Ala Tyr 195 200 205Asn Ser Cys
Tyr Ser Ala Gly Val Phe His Leu His Gln Gly Asp Ile 210 215 220Leu
Ser Val Ile Ile Pro Arg Ala Arg Ala Lys Leu Asn Leu Ser Pro225 230
235 240His Gly Thr Phe Leu Gly Phe Val Lys Leu 245 25014134PRTHomo
sapiens 14Val Leu His Leu Val Pro Ile Asn Ala Thr Ser Lys Asp Asp
Ser Asp1 5 10 15Val Thr Glu Val Met Trp Gln Pro Ala Leu Arg Arg Gly
Arg Gly Leu 20 25 30Gln Ala Gln Gly Tyr Gly Val Arg Ile Gln Asp Ala
Gly Val Tyr Leu 35 40 45Leu Tyr Ser Gln Val Leu Phe Gln Asp Val Thr
Phe Thr Met Gly Gln 50 55 60Val Val Ser Arg Glu Gly Gln Gly Arg Gln
Glu Thr Leu Phe Arg Cys65 70 75 80Ile Arg Ser Met Pro Ser His Pro
Asp Arg Ala Tyr Asn Ser Cys Tyr 85 90 95Ser Ala Gly Val Phe His Leu
His Gln Gly Asp Ile Leu Ser Val Ile 100 105 110Ile Pro Arg Ala Arg
Ala Lys Leu Asn Leu Ser Pro His Gly Thr Phe 115 120 125Leu Gly Phe
Val Lys Leu 130151845DNAArtificial SequenceNucleic acid sequences
coding for a CAR (dAPRIL-HCH2CH3pvaa-CD28OXZ) 15atggagaccg
acaccctgct gctgtgggtg ctgctgctgt gggtgccagg cagcaccggc 60agcgtgctcc
acctggtgcc catcaacgcc accagcaagg acgactctga tgtgaccgag
120gtgatgtggc agccagccct gagacggggc agaggcctgc aggcccaggg
ctacggcgtg 180agaatccagg acgctggcgt gtacctgctg tactcccagg
tgctgttcca ggacgtgacc 240ttcacaatgg gccaggtggt gagccgggag
ggccagggca gacaggagac cctgttccgg 300tgcatccgga gcatgcccag
ccaccccgac agagcctaca acagctgcta cagcgctggc 360gtgtttcacc
tgcaccaggg cgacatcctg agcgtgatca tccccagagc cagagccaag
420ctgaacctgt ccccccacgg cacctttctg ggcttcgtga agctgtctgg
aggcggctcg 480gatcccgccg agcccaaatc tcctgacaaa actcacacat
gcccaccgtg cccagcacct 540cccgtggccg gcccgtcagt cttcctcttc
cccccaaaac ccaaggacac cctcatgatc 600gcccggaccc ctgaggtcac
atgcgtggtg gtggacgtga gccacgaaga ccctgaggtc 660aagttcaact
ggtacgtgga cggcgtggag gtgcataatg ccaagacaaa gccgcgggag
720gagcagtaca acagcacgta ccgtgtggtc agcgtcctca ccgtcctgca
ccaggactgg 780ctgaatggca aggagtacaa gtgcaaggtc tccaacaaag
ccctcccagc ccccatcgag 840aaaaccatct ccaaagccaa agggcagccc
cgagaaccac aggtgtacac cctgccccca 900tcccgggatg agctgaccaa
gaaccaggtc agcctgacct gcctggtcaa aggcttctat 960cccagcgaca
tcgccgtgga gtgggagagc aatgggcaac cggagaacaa ctacaagacc
1020acgcctcccg tgctggactc cgacggctcc ttcttcctct acagcaagct
caccgtggac 1080aagagcaggt ggcagcaggg gaacgtcttc tcatgctccg
tgatgcatga ggctctgcac 1140aaccactaca cgcagaagag cctctccctg
tctccgggta aaaaagatcc caaattttgg 1200gtgctggtgg tggttggtgg
agtcctggct tgctatagct tgctagtaac agtggccttt 1260attattttct
gggtgaggag taagaggagc aggctcctgc acagtgacta catgaacatg
1320actccccgcc gccccgggcc cacccgcaag cattaccagc cctatgcccc
accacgcgac 1380ttcgcagcct atcgctccag ggaccagagg ctgccccccg
atgcccacaa gccccctggg 1440ggaggcagtt tccggacccc catccaagag
gagcaggccg acgcccactc caccctggcc 1500aagatcagag tgaagttcag
caggagcgca gacgcccccg cgtaccagca gggccagaac 1560cagctctata
acgagctcaa tctaggacga agagaggagt acgatgtttt ggacaagaga
1620cgtggccggg accctgagat ggggggaaag ccgagaagga agaaccctca
ggaaggcctg 1680tacaatgaac tgcagaaaga taagatggcg gaggcctaca
gtgagattgg gatgaaaggc 1740gagcgccgga ggggcaaggg gcacgatggc
ctttaccagg gtctcagtac agccaccaag 1800gacacctacg acgcccttca
catgcaggcc ctgcctcctc gctaa 1845161275DNAArtificial SequenceNucleic
acid sequences coding for a CAR (dAPRIL-CD8STK-CD28OXZ)
16atggagaccg acaccctgct gctgtgggtg ctgctgctgt gggtgccagg cagcaccggc
60agcgtgctcc acctggtgcc catcaacgcc accagcaagg acgactctga tgtgaccgag
120gtgatgtggc agccagccct gagacggggc agaggcctgc aggcccaggg
ctacggcgtg 180agaatccagg acgctggcgt gtacctgctg tactcccagg
tgctgttcca ggacgtgacc 240ttcacaatgg gccaggtggt gagccgggag
ggccagggca gacaggagac cctgttccgg 300tgcatccgga gcatgcccag
ccaccccgac agagcctaca acagctgcta cagcgctggc 360gtgtttcacc
tgcaccaggg cgacatcctg agcgtgatca tccccagagc cagagccaag
420ctgaacctgt ccccccacgg cacctttctg ggcttcgtga agctgtctgg
aggcggctcg 480gatcccacca cgacgccagc gccgcgacca ccaacaccgg
cgcccaccat cgcgtcgcag 540cccctgtccc tgcgcccaga ggcgtgccgg
ccagcggcgg ggggcgcagt gcacacgagg 600gggctggact tcgcctgtga
tatcttttgg gtgctggtgg tggttggtgg agtcctggct 660tgctatagct
tgctagtaac agtggccttt attattttct gggtgaggag taagaggagc
720aggctcctgc acagtgacta catgaacatg actccccgcc gccccgggcc
cacccgcaag 780cattaccagc cctatgcccc accacgcgac ttcgcagcct
atcgctccag ggaccagagg 840ctgccccccg atgcccacaa gccccctggg
ggaggcagtt tccggacccc catccaagag 900gagcaggccg acgcccactc
caccctggcc aagatcagag tgaagttcag caggagcgca 960gacgcccccg
cgtaccagca gggccagaac cagctctata acgagctcaa tctaggacga
1020agagaggagt acgatgtttt ggacaagaga cgtggccggg accctgagat
ggggggaaag 1080ccgagaagga agaaccctca ggaaggcctg tacaatgaac
tgcagaaaga taagatggcg 1140gaggcctaca gtgagattgg gatgaaaggc
gagcgccgga ggggcaaggg gcacgatggc 1200ctttaccagg gtctcagtac
agccaccaag gacacctacg acgcccttca catgcaggcc 1260ctgcctcctc gctaa
1275171197DNAArtificial SequenceNucleic acid sequences coding for a
CAR (dAPRIL-HNG-CD28OXZ) 17atggagaccg acaccctgct gctgtgggtg
ctgctgctgt gggtgccagg cagcaccggc 60agcgtgctcc acctggtgcc catcaacgcc
accagcaagg acgactctga tgtgaccgag 120gtgatgtggc agccagccct
gagacggggc agaggcctgc aggcccaggg ctacggcgtg 180agaatccagg
acgctggcgt gtacctgctg tactcccagg tgctgttcca ggacgtgacc
240ttcacaatgg gccaggtggt gagccgggag ggccagggca gacaggagac
cctgttccgg 300tgcatccgga gcatgcccag ccaccccgac agagcctaca
acagctgcta cagcgctggc 360gtgtttcacc tgcaccaggg cgacatcctg
agcgtgatca tccccagagc cagagccaag 420ctgaacctgt ccccccacgg
cacctttctg ggcttcgtga agctgtctgg aggcggctcg 480gatcccgccg
agcccaaatc tcctgacaaa actcacacat gcccaccgtg cccaaaagat
540cccaaatttt gggtgctggt ggtggttggt ggagtcctgg cttgctatag
cttgctagta 600acagtggcct ttattatttt ctgggtgagg agtaagagga
gcaggctcct gcacagtgac 660tacatgaaca tgactccccg ccgccccggg
cccacccgca agcattacca gccctatgcc 720ccaccacgcg acttcgcagc
ctatcgctcc agggaccaga ggctgccccc cgatgcccac 780aagccccctg
ggggaggcag tttccggacc cccatccaag aggagcaggc cgacgcccac
840tccaccctgg ccaagatcag agtgaagttc agcaggagcg cagacgcccc
cgcgtaccag 900cagggccaga accagctcta taacgagctc aatctaggac
gaagagagga gtacgatgtt 960ttggacaaga gacgtggccg ggaccctgag
atggggggaa agccgagaag gaagaaccct 1020caggaaggcc tgtacaatga
actgcagaaa gataagatgg cggaggccta cagtgagatt 1080gggatgaaag
gcgagcgccg gaggggcaag gggcacgatg gcctttacca gggtctcagt
1140acagccacca aggacaccta cgacgccctt cacatgcagg ccctgcctcc tcgctaa
1197181902DNAArtificial SequenceNucleic acid sequences coding for a
CAR (dAPRIL-HCH2CH3pvaa-CD28OXZ) 18atgggcacct ccctgctgtg ctggatggcc
ctgtgcctgc tgggagccga ccacgccgac 60ggcaagccca ttcccaaccc cctgctgggc
ctggactcca cctctggcgg aggcggcagc 120gtgctgcacc tggtgcccat
caacgccacc agcaaggacg actctgatgt gaccgaggtg 180atgtggcagc
cagccctgag acggggcaga ggcctgcagg cccagggcta cggcgtgaga
240atccaggacg ctggcgtgta cctgctgtac tcccaggtgc tgttccagga
cgtgaccttc 300acaatgggcc aggtggtgag ccgggagggc cagggcagac
aggagaccct gttccggtgc 360atccggagca tgcccagcca ccccgacaga
gcctacaaca gctgctacag cgctggcgtg 420tttcacctgc accagggcga
catcctgagc gtgatcatcc ccagagccag agccaagctg 480aacctgtccc
cccacggcac ctttctgggc ttcgtgaagc tgtctggagg cggctcggat
540cccgccgagc ccaaatctcc tgacaaaact cacacatgcc caccgtgccc
agcacctccc 600gtggccggcc cgtcagtctt cctcttcccc ccaaaaccca
aggacaccct catgatcgcc 660cggacccctg aggtcacatg cgtggtggtg
gacgtgagcc acgaagaccc tgaggtcaag 720ttcaactggt acgtggacgg
cgtggaggtg cataatgcca agacaaagcc gcgggaggag 780cagtacaaca
gcacgtaccg tgtggtcagc gtcctcaccg tcctgcacca ggactggctg
840aatggcaagg agtacaagtg caaggtctcc aacaaagccc tcccagcccc
catcgagaaa 900accatctcca aagccaaagg gcagccccga gaaccacagg
tgtacaccct gcccccatcc 960cgggatgagc tgaccaagaa ccaggtcagc
ctgacctgcc tggtcaaagg cttctatccc 1020agcgacatcg ccgtggagtg
ggagagcaat
gggcaaccgg agaacaacta caagaccacg 1080cctcccgtgc tggactccga
cggctccttc ttcctctaca gcaagctcac cgtggacaag 1140agcaggtggc
agcaggggaa cgtcttctca tgctccgtga tgcatgaggc tctgcacaac
1200cactacacgc agaagagcct ctccctgtct ccgggtaaaa aagatcccaa
attttgggtg 1260ctggtggtgg ttggtggagt cctggcttgc tatagcttgc
tagtaacagt ggcctttatt 1320attttctggg tgaggagtaa gaggagcagg
ctcctgcaca gtgactacat gaacatgact 1380ccccgccgcc ccgggcccac
ccgcaagcat taccagccct atgccccacc acgcgacttc 1440gcagcctatc
gctccaggga ccagaggctg ccccccgatg cccacaagcc ccctggggga
1500ggcagtttcc ggacccccat ccaagaggag caggccgacg cccactccac
cctggccaag 1560atcagagtga agttcagcag gagcgcagac gcccccgcgt
accagcaggg ccagaaccag 1620ctctataacg agctcaatct aggacgaaga
gaggagtacg atgttttgga caagagacgt 1680ggccgggacc ctgagatggg
gggaaagccg agaaggaaga accctcagga aggcctgtac 1740aatgaactgc
agaaagataa gatggcggag gcctacagtg agattgggat gaaaggcgag
1800cgccggaggg gcaaggggca cgatggcctt taccagggtc tcagtacagc
caccaaggac 1860acctacgacg cccttcacat gcaggccctg cctcctcgct aa
1902191332DNAArtificial SequenceNucleic acid sequences coding for a
CAR (dAPRIL-CD8STK-CD28OXZ) 19atgggcacct ccctgctgtg ctggatggcc
ctgtgcctgc tgggagccga ccacgccgac 60ggcaagccca ttcccaaccc cctgctgggc
ctggactcca cctctggcgg aggcggcagc 120gtgctgcacc tggtgcccat
caacgccacc agcaaggacg actctgatgt gaccgaggtg 180atgtggcagc
cagccctgag acggggcaga ggcctgcagg cccagggcta cggcgtgaga
240atccaggacg ctggcgtgta cctgctgtac tcccaggtgc tgttccagga
cgtgaccttc 300acaatgggcc aggtggtgag ccgggagggc cagggcagac
aggagaccct gttccggtgc 360atccggagca tgcccagcca ccccgacaga
gcctacaaca gctgctacag cgctggcgtg 420tttcacctgc accagggcga
catcctgagc gtgatcatcc ccagagccag agccaagctg 480aacctgtccc
cccacggcac ctttctgggc ttcgtgaagc tgtctggagg cggctcggat
540cccaccacga cgccagcgcc gcgaccacca acaccggcgc ccaccatcgc
gtcgcagccc 600ctgtccctgc gcccagaggc gtgccggcca gcggcggggg
gcgcagtgca cacgaggggg 660ctggacttcg cctgtgatat cttttgggtg
ctggtggtgg ttggtggagt cctggcttgc 720tatagcttgc tagtaacagt
ggcctttatt attttctggg tgaggagtaa gaggagcagg 780ctcctgcaca
gtgactacat gaacatgact ccccgccgcc ccgggcccac ccgcaagcat
840taccagccct atgccccacc acgcgacttc gcagcctatc gctccaggga
ccagaggctg 900ccccccgatg cccacaagcc ccctggggga ggcagtttcc
ggacccccat ccaagaggag 960caggccgacg cccactccac cctggccaag
atcagagtga agttcagcag gagcgcagac 1020gcccccgcgt accagcaggg
ccagaaccag ctctataacg agctcaatct aggacgaaga 1080gaggagtacg
atgttttgga caagagacgt ggccgggacc ctgagatggg gggaaagccg
1140agaaggaaga accctcagga aggcctgtac aatgaactgc agaaagataa
gatggcggag 1200gcctacagtg agattgggat gaaaggcgag cgccggaggg
gcaaggggca cgatggcctt 1260taccagggtc tcagtacagc caccaaggac
acctacgacg cccttcacat gcaggccctg 1320cctcctcgct aa
1332201254DNAArtificial SequenceNucleic acid sequences coding for a
CAR (dAPRIL-HNG-CD28OXZ) 20atgggcacct ccctgctgtg ctggatggcc
ctgtgcctgc tgggagccga ccacgccgac 60ggcaagccca ttcccaaccc cctgctgggc
ctggactcca cctctggcgg aggcggcagc 120gtgctgcacc tggtgcccat
caacgccacc agcaaggacg actctgatgt gaccgaggtg 180atgtggcagc
cagccctgag acggggcaga ggcctgcagg cccagggcta cggcgtgaga
240atccaggacg ctggcgtgta cctgctgtac tcccaggtgc tgttccagga
cgtgaccttc 300acaatgggcc aggtggtgag ccgggagggc cagggcagac
aggagaccct gttccggtgc 360atccggagca tgcccagcca ccccgacaga
gcctacaaca gctgctacag cgctggcgtg 420tttcacctgc accagggcga
catcctgagc gtgatcatcc ccagagccag agccaagctg 480aacctgtccc
cccacggcac ctttctgggc ttcgtgaagc tgtctggagg cggctcggat
540cccgccgagc ccaaatctcc tgacaaaact cacacatgcc caccgtgccc
aaaagatccc 600aaattttggg tgctggtggt ggttggtgga gtcctggctt
gctatagctt gctagtaaca 660gtggccttta ttattttctg ggtgaggagt
aagaggagca ggctcctgca cagtgactac 720atgaacatga ctccccgccg
ccccgggccc acccgcaagc attaccagcc ctatgcccca 780ccacgcgact
tcgcagccta tcgctccagg gaccagaggc tgccccccga tgcccacaag
840ccccctgggg gaggcagttt ccggaccccc atccaagagg agcaggccga
cgcccactcc 900accctggcca agatcagagt gaagttcagc aggagcgcag
acgcccccgc gtaccagcag 960ggccagaacc agctctataa cgagctcaat
ctaggacgaa gagaggagta cgatgttttg 1020gacaagagac gtggccggga
ccctgagatg gggggaaagc cgagaaggaa gaaccctcag 1080gaaggcctgt
acaatgaact gcagaaagat aagatggcgg aggcctacag tgagattggg
1140atgaaaggcg agcgccggag gggcaagggg cacgatggcc tttaccaggg
tctcagtaca 1200gccaccaagg acacctacga cgcccttcac atgcaggccc
tgcctcctcg ctaa 1254216PRTArtificial SequenceFurin cleavage site
21Lys Gln Lys Lys Gln Lys1 5
References
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013
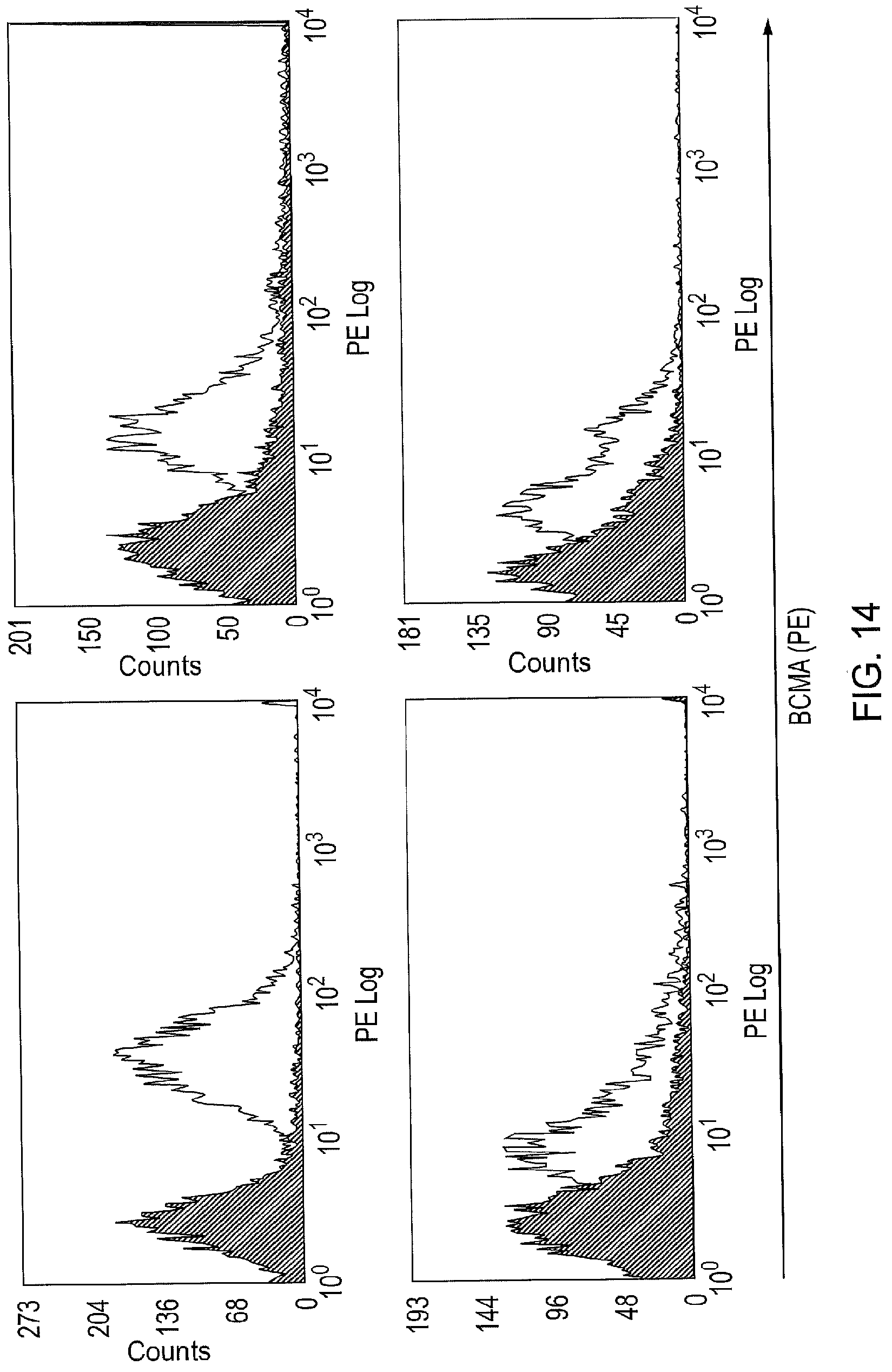
D00014
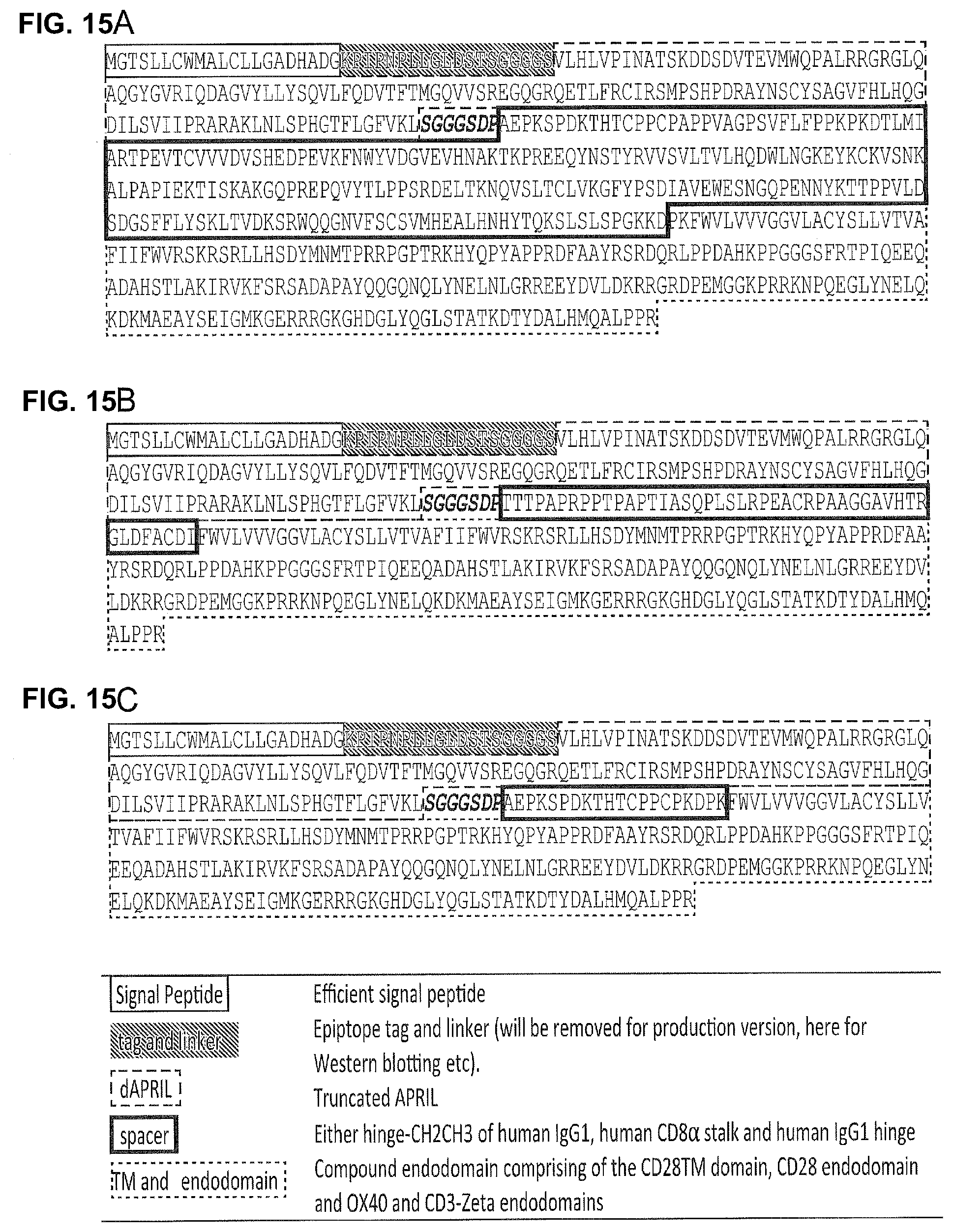
D00015

D00016
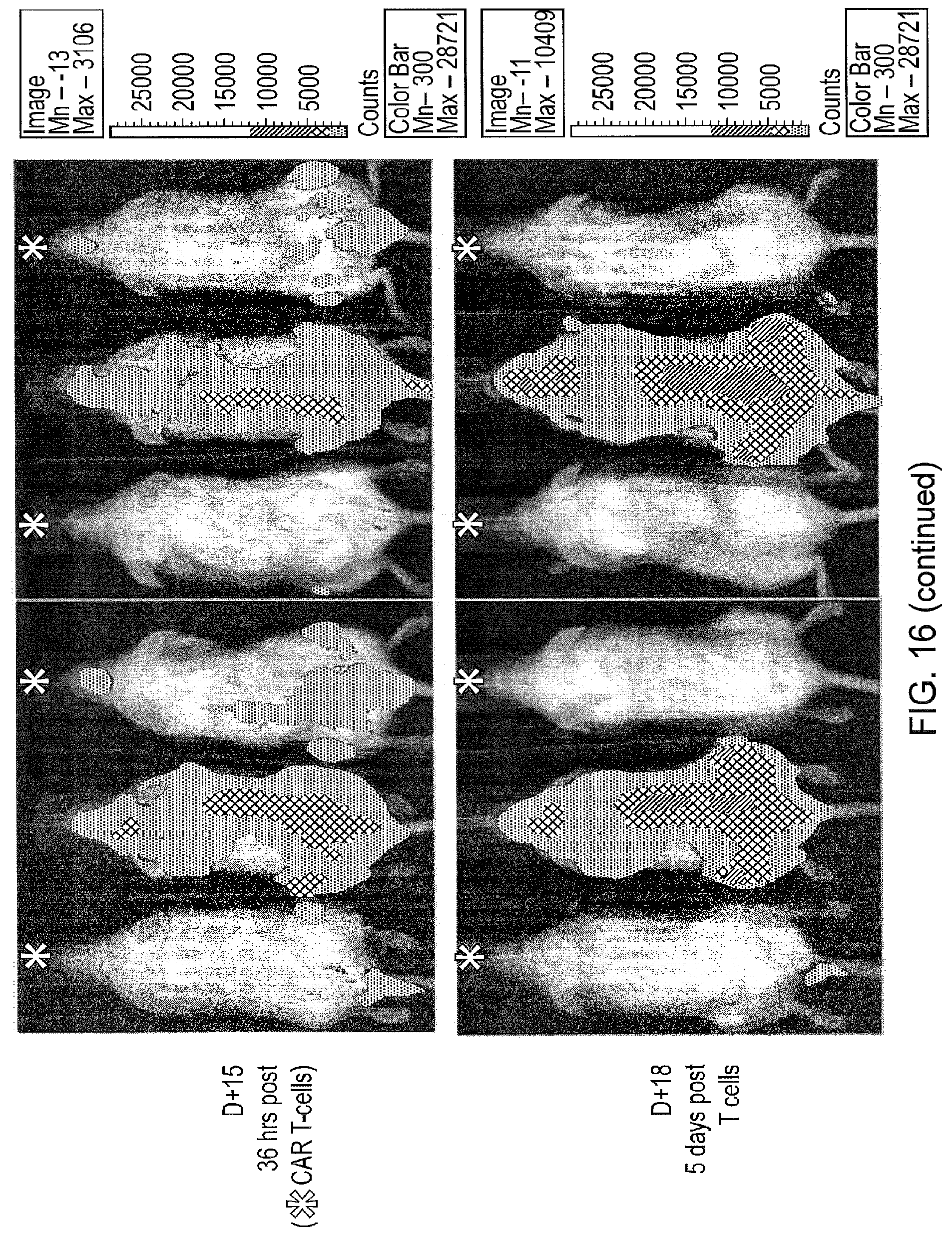
S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.