Silk Proteins
Sutherland; Tara D. ; et al.
U.S. patent application number 16/931710 was filed with the patent office on 2021-04-01 for silk proteins. The applicant listed for this patent is COMMONWEALTH SCIENTIFIC AND INDUSTRIAL RESEARCH ORGANISATION. Invention is credited to Peter M. Campbell, Victoria Shirley Haritos, Alagacone Sriskantha, Tara D. Sutherland, Holly Trueman, Sarah Weisman.
| Application Number | 20210094989 16/931710 |
| Document ID | / |
| Family ID | 1000005273718 |
| Filed Date | 2021-04-01 |











View All Diagrams
| United States Patent Application | 20210094989 |
| Kind Code | A1 |
| Sutherland; Tara D. ; et al. | April 1, 2021 |
Silk Proteins
Abstract
The present invention provides silk proteins, as well as nucleic acids encoding these proteins. The present invention also provides recombinant cells and/or organisms which synthesize silk proteins. Silk proteins of the invention can be used for a variety of purposes such as in the manufacture of personal care products, plastics, textiles, and biomedical products.
| Inventors: | Sutherland; Tara D.; (Watson, AU) ; Haritos; Victoria Shirley; (Kingsville, AU) ; Trueman; Holly; (Downer, AU) ; Sriskantha; Alagacone; (Nicholls, AU) ; Weisman; Sarah; (Griffith, AU) ; Campbell; Peter M.; (Cook, AU) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 1000005273718 | ||||||||||
| Appl. No.: | 16/931710 | ||||||||||
| Filed: | July 17, 2020 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 15197541 | Jun 29, 2016 | |||
| 16931710 | ||||
| 12089045 | Jun 27, 2008 | 9409959 | ||
| PCT/AU2006/001453 | Oct 4, 2006 | |||
| 15197541 | ||||
| 60723766 | Oct 5, 2005 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C12P 21/02 20130101; C07K 14/43563 20130101; C12N 15/8257 20130101; C07K 14/43572 20130101; C12N 15/70 20130101; C07K 2319/00 20130101; C07K 2319/73 20130101 |
| International Class: | C07K 14/435 20060101 C07K014/435; C12N 15/70 20060101 C12N015/70; C12N 15/82 20060101 C12N015/82; C12P 21/02 20060101 C12P021/02 |
Claims
1.-46. (canceled)
47. A recombinant host cell comprising a polynucleotide which encodes a polypeptide having a coiled coil structure, wherein the polypeptide comprises an amino acid sequence which is at least 40% identical to at least any one or more of SEQ ID NO:40, SEQ ID NO:41, SEQ ID NO:5, SEQ ID NO:6, SEQ ID NO:26, or SEQ ID NO:27; and wherein a) the polynucleotide is operably linked to a heterologous promoter, and/or b) the recombinant host cell is a bacterial, yeast or plant cell.
48. The recombinant host cell of claim 47, wherein the polypeptide comprises an amino acid sequence which is at least 50% identical to at least any one or more of SEQ ID NO:40, SEQ ID NO:41, SEQ ID NO:5, SEQ ID NO:6, SEQ ID NO:26, or SEQ ID NO:27.
49. The recombinant host cell of claim 47, wherein the polypeptide comprises an amino acid sequence which is at least 70% identical to at least any one or more of SEQ ID NO:40, SEQ ID NO:41, SEQ ID NO:5, SEQ ID NO:6, SEQ ID NO:26, or SEQ ID NO:27.
50. The recombinant host cell of claim 47, wherein the polypeptide comprises an amino acid sequence which is at least 80% identical to at least any one or more of SEQ ID NO:40, SEQ ID NO:41, SEQ ID NO:5, SEQ ID NO:6, SEQ ID NO:26, or SEQ ID NO:27.
51. The recombinant host cell of claim 47, wherein the polynucleotide comprises a nucleic acid sequence which is at least 40% identical to any one or more of SEQ ID NO:48, SEQ ID NO:49, SEQ ID NO: 15, SEQ ID NO: 16, SEQ ID NO:35, or SEQ ID NO:36.
52. The recombinant host cell of claim 47 which is a bacterial cell.
53. The recombinant host cell of claim 47, wherein the polypeptide comprises an amino acid sequence which is at least 60% identical to at least any one or more of SEQ ID NO:40, SEQ ID NO:41, SEQ ID NO:5, SEQ ID NO:6, SEQ ID NO:26, or SEQ ID NO:27.
54. The recombinant host cell of claim 47, wherein the polypeptide comprises an amino acid sequence which is at least 85% identical to at least any one or more of SEQ ID NO:40, SEQ ID NO:41, SEQ ID NO:5, SEQ ID NO:6, SEQ ID NO:26, or SEQ ID NO:27.
55. The recombinant host cell of claim 47, wherein the coiled coil structure comprises at least 10 copies of the heptad sequence abcdefg, and wherein at least 25% of the amino acids at positions a and d are alanine residues.
56. The recombinant host cell of claim 55, wherein the coiled coil structure comprises at least 10 copies of the heptad sequence abcdefg, and at least 25% of the amino acids at positions a, d and e are alanine residues.
57. The recombinant host cell of claim 47, wherein the coiled coil structure comprises at least 18 copies of the heptad sequence abcdefg, and wherein at least 25% of the amino acids at positions a and d are alanine residues.
58. The recombinant host cell of claim 57, wherein a coiled coil structure comprises at least 18 copies of the heptad sequence abcdefg, and at least 25% of the amino acids at positions a, d and e are alanine residues.
59. A process for preparing a polypeptide comprising an amino acid sequence which is at least 40% identical to at least any one or more of SEQ ID NO:40, SEQ ID NO:41, SEQ ID NO:5, SEQ ID NO:6, SEQ ID NO:26, or SEQ ID NO:27, the method comprising cultivating the recombinant host cell of claim 47, under conditions which allow expression of the polynucleotide encoding the polypeptide, and recovering the expressed polypeptide.
60. A recombinant host cell comprising a polynucleotide which encodes a polypeptide having a coiled coil structure, wherein the polypeptide comprises an amino acid sequence which is at least 40% identical to at least any one or more of SEQ ID NO:40, SEQ ID NO:41, SEQ ID NO:5, SEQ ID NO:6, SEQ ID NO:26, or SEQ ID NO:27, and wherein the coiled coil structure comprises at least 10 copies of the heptad sequence abcdefg, and wherein at least 25% of the amino acids at positions a and d are alanine residues; and wherein a) the polynucleotide is operably linked to a heterologous promoter, and/or b) the recombinant host cell is a bacterial, yeast or plant cell.
61. A recombinant host cell comprising a polynucleotide which encodes a polypeptide having a coiled coil structure, wherein the polypeptide comprises an amino acid sequence which is at least 80% identical to at least any one or more of SEQ ID NO:40, SEQ ID NO:41, SEQ ID NO:5, SEQ ID NO:6, SEQ ID NO:26, or SEQ ID NO:27; and wherein a) the polynucleotide is operably linked to a heterologous promoter, and/or b) the recombinant host cell is a bacterial, yeast or plant cell.
Description
FIELD OF THE INVENTION
[0001] The present invention relates to silk proteins, as well as nucleic acids encoding such proteins. The present invention also relates to recombinant cells and/or organisms which synthesize silk proteins. Silk proteins of the invention can be used for a variety of purposes such as in the production of personal care products, plastics, textiles, and biomedical products.
BACKGROUND OF THE INVENTION
[0002] Silks are fibrous protein secretions that exhibit exceptional strength and toughness and as such have been the target of extensive study. Silks are produced by over 30,000 species of spiders and by many insects. Very few of these silks have been characterised, with most research concentrating on the cocoon silk of the domesticated silkworm, Bombyx mori and on the dragline silk of the orb-weaving spider Nephila clavipes.
[0003] In the Lepidoptera and spider, the fibroin silk genes code for proteins that are generally large with prominent hydrophilic terminal domains at either end spanning an extensive region of alternating hydrophobic and hydrophilic blocks (Bini et al., 2004). Generally these proteins comprise different combinations of crystalline arrays of .beta.-pleated sheets loosely associated with .beta.-sheets, .beta.-spirals, .alpha.-helices and amorphous regions (see Craig and Riekel, 2002 for review).
[0004] As silk fibres represent some of the strongest natural fibres known, they have been subject to extensive research in attempts to reproduce their synthesis. However, a recurrent problem with expression of Lepidopteran and spider fibroin genes has been low expression rates in various recombinant expression systems due to the combination of the repeating nucleotide motifs in the silk gene that lead to deleterious recombination events, the large gene size and the small number of codons used for each amino acid in the gene which leads to depletion of tRNA pools in the host cells. Recombinant expression leads to difficulties during translation such as translational pauses as a result of codon preferences and codon demands and extensive recombination rates leading to truncation of the genes. Shorter, less repetitive sequences would avoid many of the problems associated with silk gene expression to date.
[0005] In contrast to the extensive knowledge that has accumulated about the Lepidopteran (in particular the cocoon silk of Bombyx mori) and spider (in particular the dragline silk of Nephila clavipes) little is known about the chemical composition and molecular organisation of other insect silks.
[0006] In the early 1960s, the silk of the aculeate Hymenopteran was shown to have an alpha-helical structure by X-ray diffraction patterns obtained from silk fibres drawn from the salivary gland of honeybee larvae (Rudall, 1962). As well as demonstrating that this silk was helical, the patterns obtained were indicative of a coiled-coil system of alpha-helical chains (Atkins, 1967). Similar X-ray diffraction patterns have been obtained for cocoon silks from other Aculeata species including the wasp Pseudopompilus humbolti (Rudall, 1962) and the bumblebee, Bombus lucorum (Lucas and Rudall, 1967).
[0007] In contrast to the alpha-helical structure described in the Aculeata silks, the silks characterised from a related clade to the aculeata, the Ichneumonoidea, have parallel-.beta. structures. X-ray diagrams for four examples of this structure have been described in the Braconidae (Cotesia(=Apenteles) glomerate; Cotesia(=Apenteles) gonopterygis; Apenteles bignelli) and three in Ichneumonidae (Dusona sp.; Phytodietris sp.; Branchus femoralis) (Lucas and Rudall, 1967). In addition the sequence of a single Braconidae (Cotesia glomerate) silk has been described (Genbank database accession number AB188680; Yamada et al., 2004). This partial protein sequence consists of a highly conserved 28 X-asparagine repeat (where X is alanine or serine) and is not predicted to contain coiled coil forming heptad repeats. Extensive analysis of the amino acid composition of the cocoon silks of the Braconidae has shown that the silks from the subfamily Microgastrinae are unique in their high asparagine and serine content (Lucas et al., 1960; Quicke et al., 2004). Related subfamilies produce silks with significantly different amino acid compositions suggesting that the Microgastrinae silks have evolved specifically in this subfamily (Yamada et al., 2004). The partial cDNA of Cotesia glomerata was isolated using PCR primers designed from sequence obtained from internal peptides derived from isolated cocoon silk proteins. The predicted amino acid composition of this partial sequence closely resembles the amino acid composition of the extensively washed silk from this species.
[0008] The structure of many of the silks within other non aculeate Apocrita and within the rest of the Hymenoptera (Symphata) are most commonly parallel-.beta. sheets, with both collagen-like and polyglycine silks produced by the Tenthredinidae (Lucas and Rudall, 1967).
[0009] Honeybee silk proteins are synthesised in the middle of the final instar and can be imaged as a mix of depolymerised silk proteins (Silva-Zacarin et al., 2003). As the instar progresses, water is removed from the gland and dehydration results in the polymerisation of the silk protein to form well-organised and insoluble silk filaments labelled tactoids (Silva-Zacarin et al., 2003). Progressive dehydration leads to further reorganisation of the tactoids (Silva-Zacarin et al., 2003) and possibly new inter-filamentary bonding between filaments (Rudall, 1962). Electron microscope images of fibrils isolated from the honeybee silk gland show structures of approximately 20-25 angstroms diameter (Flower and Kenchington, 1967). This value is consistent with three-, four-, or five-stranded coiled coils.
[0010] The amino acid composition of the silks of various aculeate Hymenopteran species was determined by Lucas and Rudall (1967) and found to contain high contents of alanine, serine, the acid residues, aspartic acid and glutamic acids, and reduced amounts of glycine in comparison to classical fibroins. It was considered that the helical content of the aculeate Hymenoptera silk was a consequence of a reduced glycine content and increased content of acidic residues (Rudall and Kenchington, 1971).
[0011] Little is known about the larval silk of the lacewings (Order: Neuroptera). The cocoon is comprised of two layers, an inner solid layer and an outer fibrous layer. Previously the cocoon was described as being comprised of a cuticulin silk (Rudall and Kenchington, 1971), a description that only related to the inner solid layer. LaMunyon (1988) described a substance excreted from the malphigian tubules that made up the outer fibres. After deposition of this layer, the solid inner wall was constructed from secretions from the epithelial cells in the highly villous lumen (LaMunyon, 1988).
[0012] It is also known that lacewing larva produce a proteinaceous adhesive substance from the malpighian tubules throughout all instars to stick the larvae to substrates, to glue items of camouflage on to the larvae's back or to entrap prey (Speilger, 1962). In the genus Lomamyia (Bethothidae), the larvae produce the silk and adhesive substance at the same time and it has been postulated that these two substances may well be the same product (Speilger, 1962). The adhesive secretion is highly soluble and is also thought to be associated with defense against predators (LaMunyon & Adams, 1987).
[0013] Considering the unique properties of silks produced by insects such as Hymenopterans and Neuropterans, there is a need for the identification of novel nucleic acids encoding silk proteins from these organisms.
SUMMARY OF THE INVENTION
[0014] The present inventors have identified numerous silk proteins from insects. These silk proteins are surprisingly different to other known silk proteins in their primary sequence, secondary structure and/or amino acid content.
[0015] Thus, in a first aspect the present invention provides a substantially purified and/or recombinant silk polypeptide, wherein at least a portion of the polypeptide has a coiled coil structure.
[0016] As known in the art, coiled coil structures of polypeptides are characterized by heptad repeats represented by the consensus sequence (abcdefg).sub.n, with generally hydrophobic residues in position a and d, and generally polar residues at the remaining positions. Surprisingly, the heptads of the polypeptides of the present invention have a novel composition when viewed collectively--with an unusually high abundance of alanine in the `hydrophobic` heptad positions a and d. Additionally, there are high levels of small polar residues in these positions. Furthermore, the e position also has high levels of alanine and small hydrophobic residues.
[0017] Accordingly, in a particularly preferred embodiment, the portion of the polypeptide that has a coiled coil structure comprises at least 10 copies of the heptad sequence abcdefg, and at least 25% of the amino acids at positions a and d are alanine residues.
[0018] In a further preferred embodiment, the portion of the polypeptide that has a coiled coil structure comprises at least 10 copies of the heptad sequence abcdefg, and at least 25% of the amino acids at positions a, d and e are alanine residues.
[0019] In a further preferred embodiment, the portion of the polypeptide that has a coiled coil structure comprises at least 10 copies of the heptad sequence abcdefg, and at least 25% of the amino acids at position a are alanine residues.
[0020] In a further preferred embodiment, the portion of the polypeptide that has a coiled coil structure comprises at least 10 copies of the heptad sequence abcdefg, and at least 25% of the amino acids at position d are alanine residues.
[0021] In a further preferred embodiment, the portion of the polypeptide that has a coiled coil structure comprises at least 10 copies of the heptad sequence abcdefg, and at least 25% of the amino acids at position e are alanine residues.
[0022] In a particularly preferred embodiment, the at least 10 copies of the heptad sequence are contiguous.
[0023] In a further preferred embodiment, the portion of the polypeptide that has a coiled coil structure comprises at least 5 copies of the heptad sequence abcdefg, and at least 15% of the amino acids at positions a and d are alanine residues.
[0024] In a further preferred embodiment, the portion of the polypeptide that has a coiled coil structure comprises at least 5 copies of the heptad sequence abcdefg, and at least 15% of the amino acids at positions a, d and e are alanine residues.
[0025] In a further preferred embodiment, the portion of the polypeptide that has a coiled coil structure comprises at least 5 copies of the heptad sequence abcdefg, and at least 15% of the amino acids at position a are alanine residues.
[0026] In a further preferred embodiment, the portion of the polypeptide that has a coiled coil structure comprises at least 5 copies of the heptad sequence abcdefg, and at least 15% of the amino acids at position d are alanine residues.
[0027] In a further preferred embodiment, the portion of the polypeptide that has a coiled coil structure comprises at least 5 copies of the heptad sequence abcdefg, and at least 15% of the amino acids at position e are alanine residues.
[0028] In a particularly preferred embodiment, the at least 5 copies of the heptad sequence are contiguous.
[0029] In one embodiment, the polypeptide comprises a sequence selected from:
[0030] i) an amino acid sequence as provided in any one of SEQ ID NO:1, SEQ ID NO:2, SEQ ID NO:22, SEQ ID NO:23, SEQ ID NO:40, SEQ ID NO:41, SEQ ID NO:56, and SEQ ID NO:57;
[0031] ii) an amino acid sequence which is at least 30% identical to any one or more of SEQ ID NO:1, SEQ ID NO:2, SEQ ID NO:22, SEQ ID NO:23, SEQ ID NO:40, SEQ ID NO:41, SEQ ID NO:56, and SEQ ID NO:57; and
[0032] iii) a biologically active fragment of i) or ii).
[0033] In another embodiment, the polypeptide comprises a sequence selected from:
[0034] i) an amino acid sequence as provided in any one of SEQ ID NO:3, SEQ ID NO:4, SEQ ID NO:24, SEQ ID NO:25, SEQ ID NO:42, SEQ ID NO:43, SEQ ID NO:58, and SEQ ID NO:59;
[0035] ii) an amino acid sequence which is at least 30% identical to any one or more of SEQ ID NO:3, SEQ ID NO:4, SEQ ID NO:24, SEQ ID NO:25, SEQ ID NO:42, SEQ ID NO:43, SEQ ID NO:58, and SEQ ID NO:59; and
[0036] iii) a biologically active fragment of i) or ii).
[0037] In another embodiment, the polypeptide comprises a sequence selected from:
[0038] i) an amino acid sequence as provided in any one of SEQ ID NO:5, SEQ ID NO:6, SEQ ID NO:26, SEQ ID NO:27, SEQ ID NO:44, SEQ ID NO:45, SEQ ID NO:60, and SEQ ID NO:61;
[0039] ii) an amino acid sequence which is at least 30% identical to any one or more of SEQ ID NO:5, SEQ ID NO:6, SEQ ID NO:26, SEQ ID NO:27, SEQ ID NO:44, SEQ ID NO:45, SEQ ID NO:60, and SEQ ID NO:61; and
[0040] iii) a biologically active fragment of i) or ii).
[0041] In another embodiment, the polypeptide comprises a sequence selected from:
[0042] i) an amino acid sequence as provided in any one of SEQ ID NO:7, SEQ ID NO:8, SEQ ID NO:28, SEQ ID NO:29, SEQ ID NO:46, SEQ ID NO:47, SEQ ID NO:62, and SEQ ID NO:63;
[0043] ii) an amino acid sequence which is at least 30% identical to any one or more of SEQ ID NO:7, SEQ ID NO:8, SEQ ID NO:28, SEQ ID NO:29, SEQ ID NO:46, SEQ ID NO:47, SEQ ID NO:62, and SEQ ID NO:63; and
[0044] iii) a biologically active fragment of i) or ii).
[0045] In a further embodiment, the polypeptide comprises a sequence selected from: i) an amino acid sequence as provided in SEQ ID NO:72 or SEQ ID NO:73;
[0046] ii) an amino acid sequence which is at least 30% identical to SEQ ID NO:72 and/or SEQ ID NO:73; and
[0047] iii) a biologically active fragment of i) or ii).
[0048] Further silk proteins which co-associate with proteins of the first aspect have been identified. One of these proteins (SEQ ID NO:10) is predicted to have 41% alpha-helical, 8% beta-sheet and 50% loop secondary structure by PROFsec, and therefore is classified as a mixed structure protein. MARCOIL analysis of this protein predicted only a short region of heptad repeats characteristic of proteins with a coiled coil structure.
[0049] Accordingly, in a second aspect, the present invention provides a substantially purified and/or recombinant silk polypeptide which comprises a sequence selected from:
[0050] i) an amino acid sequence as provided in any one of SEQ ID NO:9, SEQ ID NO:10 and SEQ ID NO:30;
[0051] ii) an amino acid sequence which is at least 30% identical to any one or more of SEQ ID NO:9, SEQ ID NO:10 and SEQ ID NO:30; and
[0052] iii) a biologically active fragment of i) or ii).
[0053] Without wishing to be limited by theory, it appears that four proteins of the first aspect become intertwined to form a bundle with helical axes almost parallel to each other, and this bundle extends axially into a fibril. Furthermore, it is predicted that in at least some species such as the honyebee and bumblebee the proteins of the second aspect act as a "glue" assisting in binding various bundles of coiled coil proteins of the first aspect together to form a fibrous protein complex. However, silk fibers and copolymers can still be formed without a polypeptide of second aspect.
[0054] In a preferred embodiment, a polypeptide of the invention can be purified from, or is a mutant of a polypeptide purified from, a species of Hymenoptera or Neuroptera. Preferably, the species of Hymenoptera is Apis mellifera, Oecophylla smaragdina, Myrmecia foricata or Bombus terrestris. Preferably, the species of Neuroptera is Mallada signata.
[0055] In another aspect, the present invention provides a polypeptide of the invention fused to at least one other polypeptide.
[0056] In a preferred embodiment, the at least one other polypeptide is selected from the group consisting of: a polypeptide that enhances the stability of a polypeptide of the present invention, a polypeptide that assists in the purification of the fusion protein, and a polypeptide which assists in the polypeptide of the invention being secreted from a cell (for example secreted from a plant cell).
[0057] In another aspect, the present invention provides an isolated and/or exogenous polynucleotide which encodes a silk polypeptide, wherein at least a portion of the polypeptide has a coiled coil structure.
[0058] In one embodiment, the polynucleotide comprises a sequence selected from:
[0059] i) a sequence of nucleotides as provided in any one of SEQ ID NO:11, SEQ ID NO:12, SEQ ID NO:31, SEQ ID NO:32, SEQ ID NO:48, SEQ ID NO:49, SEQ ID NO:64, and SEQ ID NO:65;
[0060] ii) a sequence of nucleotides encoding a polypeptide of the invention,
[0061] iii) a sequence of nucleotides which is at least 30% identical to any one or more of SEQ ID NO:11, SEQ ID NO:12, SEQ ID NO:31, SEQ ID NO:32, SEQ ID NO:48, SEQ ID NO:49, SEQ ID NO:64, and SEQ ID NO:65, and
[0062] iv) a sequence which hybridizes to any one of i) to iii) under stringent conditions.
[0063] In another embodiment, the polynucleotide comprises a sequence selected from:
[0064] i) a sequence of nucleotides as provided in any one of SEQ ID NO:13, SEQ ID NO:14, SEQ ID NO:33, SEQ ID NO:34, SEQ ID NO:50, SEQ ID NO:51, SEQ ID NO:66, and SEQ ID NO:67;
[0065] ii) a sequence of nucleotides encoding a polypeptide of the invention,
[0066] iii) a sequence of nucleotides which is at least 30% identical to any one or more of SEQ ID NO:13, SEQ ID NO:14, SEQ ID NO:33, SEQ ID NO:34, SEQ ID NO:50, SEQ ID NO:51, SEQ ID NO:66, and SEQ ID NO:67, and
[0067] iv) a sequence which hybridizes to any one of i) to iii) under stringent conditions.
[0068] In another embodiment, the polynucleotide comprises a sequence selected from:
[0069] i) a sequence of nucleotides as provided in any one of SEQ ID NO:15, SEQ ID NO:16, SEQ ID NO:35, SEQ ID NO:36, SEQ ID NO:52, SEQ ID NO:53, SEQ ID NO:68, and SEQ ID NO:69;
[0070] ii) a sequence of nucleotides encoding a polypeptide of the invention,
[0071] iii) a sequence of nucleotides which is at least 30% identical to any one or more of SEQ ID NO:15, SEQ ID NO:16, SEQ ID NO:35, SEQ ID NO:36, SEQ ID NO:52, SEQ ID NO:53, SEQ ID NO:68, and SEQ ID NO:69, and
[0072] iv) a sequence which hybridizes to any one of i) to iii) under stringent conditions.
[0073] In a further embodiment, the polynucleotide comprises a sequence selected from:
[0074] i) a sequence of nucleotides as provided in any one of SEQ ID NO:17, SEQ ID NO:18, SEQ ID NO:37, SEQ ID NO:38, SEQ ID NO:54, SEQ ID NO:55, SEQ ID NO:70, SEQ ID NO:71 and SEQ ID NO:76;
[0075] ii) a sequence of nucleotides encoding a polypeptide of the invention,
[0076] iii) a sequence of nucleotides which is at least 30% identical to any one or more of SEQ ID NO:17, SEQ ID NO:18, SEQ ID NO:37, SEQ ID NO:38, SEQ ID NO:54, SEQ ID NO:55, SEQ ID NO:70, SEQ ID NO:71 and SEQ ID NO:76, and
[0077] iv) a sequence which hybridizes to any one of i) to iii) under stringent conditions.
[0078] In another embodiment, the polynucleotide comprises a sequence selected from:
[0079] i) a sequence of nucleotides as provided in SEQ ID NO:74 or SEQ ID NO:75;
[0080] ii) a sequence of nucleotides encoding a polypeptide of the invention,
[0081] iii) a sequence of nucleotides which is at least 30% identical to SEQ ID NO:74 and/or SEQ ID NO:75, and
[0082] iv) a sequence which hybridizes to any one of i) to iii) under stringent conditions.
[0083] In a further aspect, the present invention provides an isolated and/or exogenous polynucleotide, the polynucleotide comprising a sequence selected from:
[0084] i) a sequence of nucleotides as provided in any one of SEQ ID NO:19, SEQ ID NO:20, SEQ ID NO:21, and SEQ ID NO:39;
[0085] ii) a sequence of nucleotides encoding a polypeptide of the invention,
[0086] iii) a sequence of nucleotides which is at least 30% identical to any one or more of SEQ ID NO:19, SEQ ID NO:20, SEQ ID NO:21, and SEQ ID NO:39, and
[0087] iv) a sequence which hybridizes to any one of i) to iii) under stringent conditions.
[0088] In a preferred embodiment, a polynucleotide can be isolated from, or is a mutant of a polynucleotide isolated from, a species of Hymenoptera or Neuroptera. Preferably, the species of Hymenoptera is Apis mellifera, Oecophylla smaragdina, Myrmecia foricata or Bombus terrestris. Preferably, the species of Neuroptera is Mallada signata.
[0089] In a further aspect, the present invention provides a vector comprising at least one polynucleotide of the invention.
[0090] Preferably, the vector is an expression vector.
[0091] In another aspect, the present invention provides a host cell comprising at least one polynucleotide of the invention, and/or at least one vector of the invention.
[0092] The host cell can be any type of cell. Examples include, but are not limited to, a bacterial, yeast or plant cell.
[0093] Also provided is a process for preparing a polypeptide according to the invention, the process comprising cultivating a host cell of the invention, or a vector of the invention, under conditions which allow expression of the polynucleotide encoding the polypeptide, and recovering the expressed polypeptide.
[0094] It is envisaged that transgenic plants will be particularly useful for the production of polypeptides of the invention. Thus, in yet another aspect, the present provides a transgenic plant comprising an exogenous polynucleotide, the polynucleotide encoding at least one polypeptide of the invention.
[0095] In another aspect, the present invention provides a transgenic non-human animal comprising an exogenous polynucleotide, the polynucleotide encoding at least one polypeptide of the invention.
[0096] In yet another aspect, the present invention provides an antibody which specifically binds a polypeptide of the invention.
[0097] In a further aspect, the present invention provides a silk fiber comprising at least one polypeptide of the invention.
[0098] Preferably, the polypeptide is a recombinant polypeptide.
[0099] In an embodiment, at least some of the polypeptides are crosslinked. In an embodiment, at least some of the lysine residues of the polypeptides are crosslinked.
[0100] In another aspect, the present invention provides a copolymer comprising at least two polypeptides of the invention.
[0101] Preferably, the polypeptides are recombinant polypeptides.
[0102] In an embodiment, the copolymer comprises at least four different polypeptide of the first aspect. In another embodiment, the copolymer further comprises a polypeptide of the second aspect.
[0103] In an embodiment, at least some of the polypeptides are crosslinked. In an embodiment, at least some of the lysine residues of the polypeptides are crosslinked.
[0104] As the skilled addressee will appreciate, the polypeptides of the invention have a wide variety of uses as is known in the art for other types of silk proteins. Thus, in a further aspect, the present invention provides a product comprising at least one polypeptide of the invention, a silk fiber of the invention and/or a copolymer of the invention.
[0105] Examples of products include, but are not limited to, personal care products, textiles, plastics, and biomedical products.
[0106] In yet a further aspect, the present invention provides a composition comprising at least one polypeptide of the invention, a silk fiber of the invention and/or a copolymer of the invention, and one or more acceptable carriers.
[0107] In one embodiment, the composition further comprises a drug.
[0108] In another embodiment, the composition is used as a medicine, in a medical device or a cosmetic.
[0109] In another aspect, the present invention provides a composition comprising at least one polynucleotide of the invention, and one or more acceptable carriers.
[0110] In a preferred embodiment, a composition, silk fiber, copolymer and/or product of the invention does not comprise a royal jelly protein produced by an insect.
[0111] In a further aspect, the present invention provides a method of treating or preventing a disease, the method comprising administering a composition comprising a drug for treating or preventing the disease and a pharmaceutically acceptable carrier, wherein the pharmaceutically acceptable carrier is selected from at least one polypeptide of the invention, a silk fiber of the invention and/or a copolymer of the invention.
[0112] In yet another aspect, the present invention provides for the use of at least one polypeptide of the invention, a silk fiber of the invention and/or a copolymer of the invention, and a drug, for the manufacture of a medicament for treating or preventing a disease.
[0113] In a further aspect, the present invention provides a kit comprising at least one polypeptide of the invention, at least one polynucleotide of the invention, at least one vector of the invention, at least one silk fiber of the invention and/or a copolymer of the invention.
[0114] Preferably, the kit further comprises information and/or instructions for use of the kit.
[0115] As will be apparent, preferred features and characteristics of one aspect of the invention are applicable to many other aspects of the invention.
[0116] Throughout this specification the word "comprise", or variations such as "comprises" or "comprising", will be understood to imply the inclusion of a stated element, integer or step, or group of elements, integers or steps, but not the exclusion of any other element, integer or step, or group of elements, integers or steps.
[0117] The invention is hereinafter described by way of the following non-limiting Examples and with reference to the accompanying figures.
BRIEF DESCRIPTION OF THE ACCOMPANYING DRAWINGS
[0118] FIG. 1. Fourier transform infrared spectra of the amide I and II regions of the silks: 1) honeybee silk, 2) bumblebee silk, 3) bulldog ant silk, 4) weaver ant silk 5) lacewing larval silk. All the silks have spectra expected of helical proteins. The Hymenopteran silks (ants and bees) have spectral maxima at 1645-1646 cm.sup.-1 (labelled), shifted approximately 10 cm.sup.-1 lower than a classical alpha-helical signal and broadened, as is typical of coiled-coil proteins (Heimburg et al., 1999).
[0119] FIG. 2. Comparison of amino acid composition of SDS washed honeybee brood comb silk with amino acid composition of Xenospira proteins (namely, Xenospira1, Xenospira2, Xenospira3 and Xenospira4) (equimolar amounts totalling 65%) and Xenosin (35%).
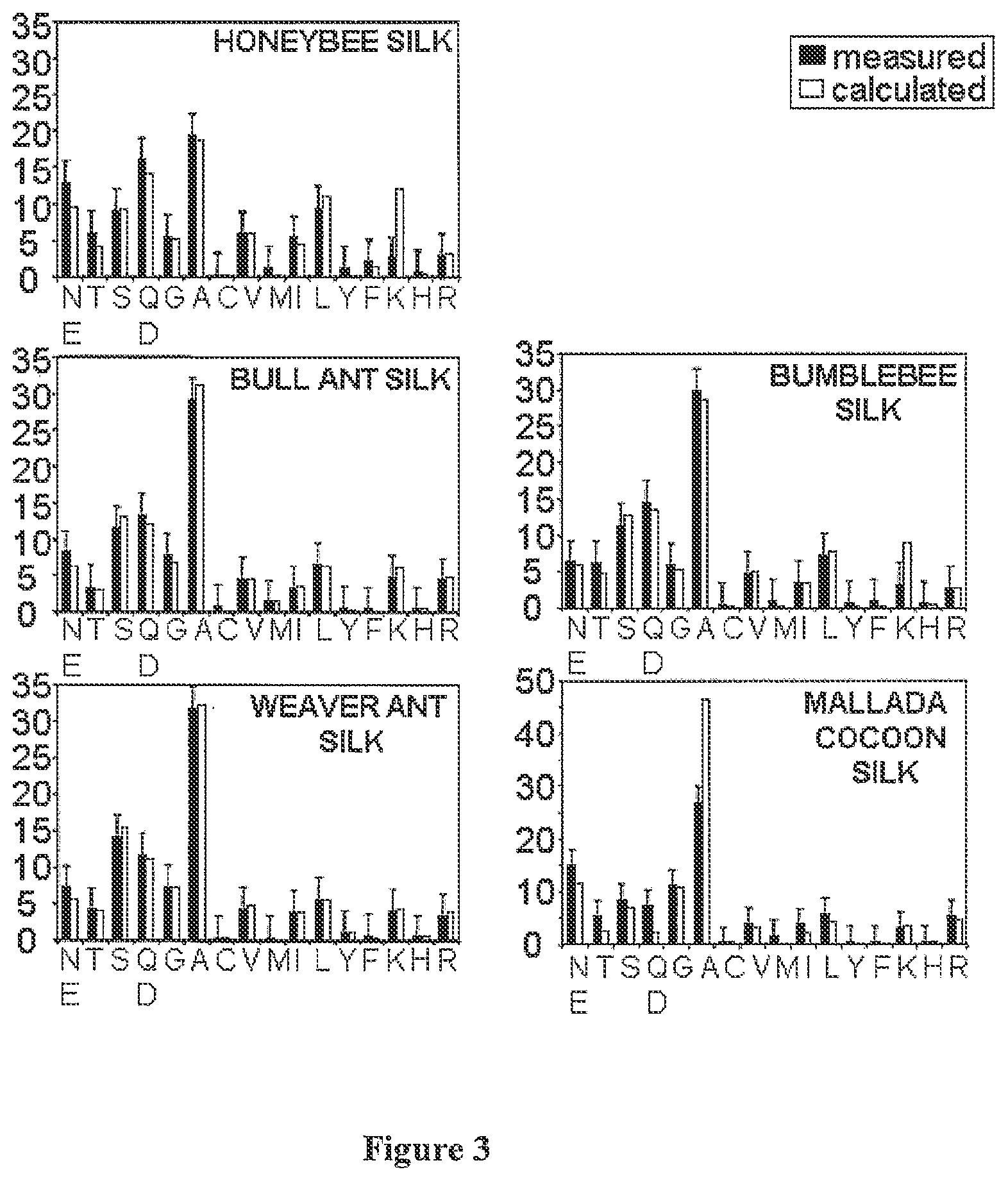
[0120] FIG. 3. Comparison of amino acid composition of silk with amino acid composition predicted from proteins encoded by silk genes.
[0121] FIG. 4. Prediction of coiled coil regions in honeybee silk proteins. COILS is a program that compares a sequence to a database of known parallel two-stranded coiled-coils and derives a similarity score. By comparing this score to the distribution of scores in globular and coiled-coil proteins, the program then calculates the probability that the sequence will adopt a coiled-coil conformation as described in Lupas et al. (1991). Using a window size of 28 this program predicts the following numbers of residues exist in each protein in coiled coil domains: Xenospira3: 77; Xenospira4: 35; Xenospira1: 28; Xenospira2: 80.
[0122] FIG. 5. Alignment of honey bee silk proteins showing MARCOIL prediction of major heptads that form a coiled-coil structure. Heptad sequences are shown above the amino acids, and alanine residues in positions a and d are highlighted.
[0123] FIG. 6. Alignment of Marciol predicted coiled coil regions of hymenopteran (bees and ants) silk proteins showing the heptad position assignment. Amel, honeybee; BB, bumblebee; BA, bulldog ant; WA, weaver ant; F1-4, silk fibroins 1-4. Heptad sequences are shown above the amino acids, and alanine residues in positions a, d and e are highlighted.
[0124] FIG. 7. The amino acid character of heptad positions in the predicted coiled coil regions of the Mallada signata larval silk protein and the orthologous clusters of the Hymenopteran silk proteins.
[0125] FIG. 8. SDS polyacrylamide gel electrophoresis of late last instar salivary gland proteins. Proteins were identified after tryptic digest and analysis of mass spectral data set using Agilent's Spectrum Mill software to match the data with predictions of protein sequences from proteins identified from cDNA sequences. The software generated scores for the quality of each match between experimentally observed sets of masses of fragments of peptides and the predictions of fragments that might be generated according to the sequences of proteins in a provided database. All the sequence matches shown here received scores greater than 20 by the Spectrum Mill software, where a score of 20 would be sufficient for automatic, confident acceptance of a valid match.
[0126] FIG. 9. Parsimony analysis of the coiled coil region of silk proteins. The relatedness of the four coiled-coil proteins suggests that the genes evolved from a common ancestor predating the divergence of the Euaculeata. The area bound by the dashed line indicates variation that occurred before the ants and wasps (Vespoidea) diverged from the bees (Apoidea) in the Late Jurassic (155 myrs; Grimaldi and Engel, 2005). Numbers indicating bootstrap values from 1000 iterations are shown.
[0127] FIG. 10. A) Apis mellifera silk proteins identified by mass spectral analysis of peptides generated from bee silk after digestion with trypsin. Shading indicates peptides identified by the mass spectral analysis. All the sequence matches shown here received scores greater than 20 by the Spectrum Mill software, where a score of 20 would be sufficient for automatic, confident acceptance of a valid match. [0128] B) Full length amino sequences of bumblebee, bulldog ant, weaver and lacewing silk proteins.
[0129] FIG. 11. Open reading frames encoding honeybee, bumblebee, bulldog ant, weaver ant and lacewing silk proteins.
[0130] FIG. 12. Sequence of gene encoding Xenosin. Entire coding sequence provided which is interrupted by a single intron (highlighted).
[0131] FIG. 13. Expression of silk protein in tobacco. Detection of histidine tagged proteins after western blot analysis of proteins from: 1. E. coli transformed with empty expression vector, 2. E. coli transformed with expression vector containing AmelF4 (Xenospira4) coding region, 3. tobacco transformed with empty expression vector, 4. tobacco transformed with expression vector containing AmelF4 coding region.
[0132] FIG. 14. Fibres made from recombinant honeybee silk proteins showing birefringent threads. Biorefringence indicates structure is present in the threads. Different recombinant honeybee threads are shown in each panel A-D, and recombinant lacewing thread is shown in panel E.
KEY TO THE SEQUENCE LISTING
[0133] SEQ ID NO:1--Honeybee silk protein termed herein Xenospira1 (also termed herein AmelF1) (minus signal peptide).
[0134] SEQ ID NO:2--Honeybee silk protein termed herein Xenospira1.
[0135] SEQ ID NO:3--Honeybee silk protein termed herein Xenospira2 (also termed herein AmelF2) (minus signal peptide).
[0136] SEQ ID NO:4--Honeybee silk protein termed herein Xenospira2.
[0137] SEQ ID NO:5--Honeybee silk protein termed herein Xenospira3 (also termed herein AmelF3) (minus signal peptide).
[0138] SEQ ID NO:6--Honeybee silk protein termed herein Xenospira3.
[0139] SEQ ID NO:7--Honeybee silk protein termed herein Xenospira4 (also termed herein AmelF4) (minus signal peptide).
[0140] SEQ ID NO:8--Honeybee silk protein termed herein Xenospira4.
[0141] SEQ ID \ NO:9--Honeybee silk protein termed herein Xenosin (also termed herein AmelSA1) (minus signal peptide).
[0142] SEQ ID NO:10--Honeybee silk protein termed herein Xenosin.
[0143] SEQ ID NO:11--Nucleotide sequence encoding honeybee silk protein Xenospira1 (minus region encoding signal peptide).
[0144] SEQ ID NO:12--Nucleotide sequence encoding honeybee silk protein Xenospira1.
[0145] SEQ ID NO:13--Nucleotide sequence encoding honeybee silk protein Xenospira2 (minus region encoding signal peptide).
[0146] SEQ ID NO:14--Nucleotide sequence encoding honeybee silk protein Xenospira2.
[0147] SEQ ID NO:15--Nucleotide sequence encoding honeybee silk protein Xenospira3 (minus region encoding signal peptide).
[0148] SEQ ID NO:16--Nucleotide sequence encoding honeybee silk protein Xenospira3.
[0149] SEQ ID NO:17--Nucleotide sequence encoding honeybee silk protein Xenospira4 (minus region encoding signal peptide).
[0150] SEQ ID NO:18--Nucleotide sequence encoding honeybee silk protein Xenospira4.
[0151] SEQ ID NO:19--Nucleotide sequence encoding honeybee silk protein Xenosin (minus region encoding signal peptide).
[0152] SEQ ID NO:20--Nucleotide sequence encoding honeybee silk protein Xenosin.
[0153] SEQ ID NO:21--Gene sequence encoding honeybee silk protein Xenosin.
[0154] SEQ ID NO:22--Bumblebee silk protein termed herein BBF1 (minus signal peptide).
[0155] SEQ ID NO:23--Bumblebee silk protein termed herein BBF1.
[0156] SEQ ID NO:24--Bumblebee silk protein termed herein BBF2 (minus signal peptide).
[0157] SEQ ID NO:25--Bumblebee silk protein termed herein BBF2.
[0158] SEQ ID NO:26--Bumblebee silk protein termed herein BBF3 (minus signal peptide).
[0159] SEQ ID NO:27--Bumblebee silk protein termed herein BBF3.
[0160] SEQ ID NO:28--Bumblebee silk protein termed herein BBF4 (minus signal peptide).
[0161] SEQ ID NO:29--Bumblebee silk protein termed herein BBF4.
[0162] SEQ ID NO:30--Partial amino acid sequence of bumblebee silk protein termed herein BBSA1.
[0163] SEQ ID NO:31--Nucleotide sequence encoding bumblebee silk protein BBF1 (minus region encoding signal peptide).
[0164] SEQ ID NO:32--Nucleotide sequence encoding bumblebee silk protein BBF1.
[0165] SEQ ID NO:33--Nucleotide sequence encoding bumblebee silk protein BBF2 (minus region encoding signal peptide).
[0166] SEQ ID NO:34--Nucleotide sequence encoding bumblebee silk protein BBF2.
[0167] SEQ ID NO:35--Nucleotide sequence encoding bumblebee silk protein BBF3 (minus region encoding signal peptide).
[0168] SEQ ID NO:36--Nucleotide sequence encoding bumblebee silk protein BBF3.
[0169] SEQ ID NO:37--Nucleotide sequence encoding bumblebee silk protein BBF4 (minus region encoding signal peptide).
[0170] SEQ ID NO:38--Nucleotide sequence encoding bumblebee silk protein BBF4.
[0171] SEQ ID NO:39--Partial nucleotide sequence encoding bumblebee silk protein BBSA1.
[0172] SEQ ID NO:40--Bulldog ant silk protein termed herein BAF1 (minus signal peptide).
[0173] SEQ ID NO:41--Bulldog ant silk protein termed herein BAF1.
[0174] SEQ ID NO:42--Bulldog ant silk protein termed herein BAF2 (minus signal peptide).
[0175] SEQ ID NO:43--Bulldog ant silk protein termed herein BAF2.
[0176] SEQ ID NO:44--Bulldog ant silk protein termed herein BAF3 (minus signal peptide).
[0177] SEQ ID NO:45--Bulldog ant silk protein termed herein BAF3.
[0178] SEQ ID NO:46--Bulldog ant silk protein termed herein BAF4 (minus signal peptide).
[0179] SEQ ID NO:47--Bulldog ant silk protein termed herein BAF4.
[0180] SEQ ID NO:48--Nucleotide sequence encoding bulldog ant silk protein BAF1 (minus region encoding signal peptide).
[0181] SEQ ID NO:49--Nucleotide sequence encoding bulldog ant silk protein BAF1.
[0182] SEQ ID NO:50--Nucleotide sequence encoding bulldog ant silk protein BAF2 (minus region encoding signal peptide).
[0183] SEQ ID NO:51--Nucleotide sequence encoding bulldog ant silk protein BAF2.
[0184] SEQ ID NO:52--Nucleotide sequence encoding bulldog ant silk protein BAF3 (minus region encoding signal peptide).
[0185] SEQ ID NO:53--Nucleotide sequence encoding bulldog ant silk protein BAF3.
[0186] SEQ ID NO:54--Nucleotide sequence encoding bulldog ant silk protein BAF4 (minus region encoding signal peptide).
[0187] SEQ ID NO:55--Nucleotide sequence encoding bulldog ant silk protein BAF4.
[0188] SEQ ID NO:56--Weaver ant silk protein termed herein GAF1 (minus signal peptide).
[0189] SEQ ID NO:57--Weaver ant silk protein termed herein GAF1.
[0190] SEQ ID NO:58--Weaver ant silk protein termed herein GAF2 (minus signal peptide).
[0191] SEQ ID NO:59--Weaver ant silk protein termed herein GAF2.
[0192] SEQ ID NO:60--Weaver ant silk protein termed herein GAF3 (minus signal peptide).
[0193] SEQ ID NO:61--Weaver ant silk protein termed herein GAF3.
[0194] SEQ ID NO:62--Weaver ant silk protein termed herein GAF4 (minus signal peptide).
[0195] SEQ ID NO:63--Weaver ant silk protein termed herein GAF4.
[0196] SEQ ID NO:64--Nucleotide sequence encoding weaver ant silk protein GAF1 (minus region encoding signal peptide).
[0197] SEQ ID NO:65--Nucleotide sequence encoding weaver ant silk protein GAF1.
[0198] SEQ ID NO:66--Nucleotide sequence encoding weaver ant silk protein GAF2 (minus region encoding signal peptide).
[0199] SEQ ID NO:67--Nucleotide sequence encoding weaver ant silk protein GAF2,
[0200] SEQ ID NO:68--Nucleotide sequence encoding weaver ant silk protein GAF3 (minus region encoding signal peptide).
[0201] SEQ ID NO:69--Nucleotide sequence encoding weaver ant silk protein GAF3.
[0202] SEQ ID NO:70--Nucleotide sequence encoding weaver ant silk protein GAF4 (minus region encoding signal peptide).
[0203] SEQ ID NO:71--Nucleotide sequence encoding weaver ant silk protein GAF4.
[0204] SEQ ID NO:72--Lacewing silk protein termed herein MalF1 (minus signal peptide).
[0205] SEQ ID NO:73--Lacewing silk protein termed herein MalF1.
[0206] SEQ ID NO:74--Nucleotide sequence encoding lacewing silk protein MalF1 (minus region encoding signal peptide).
[0207] SEQ ID NO:75--Nucleotide sequence encoding lacewing silk protein MalF1.
[0208] SEQ ID NO:76--Nucleotide sequence encoding honeybee silk protein termed herein Xenospira4 codon-optimized for plant expression (before subcloning into pET14b and pVEC8).
[0209] SEQ ID NO:77--Honeybee silk protein (Xenospira4) open reading frame optimized for plant expression (without translational fusion).
DETAILED DESCRIPTION OF THE INVENTION
General Techniques and Definitions
[0210] Unless specifically defined otherwise, all technical and scientific terms used herein shall be taken to have the same meaning as commonly understood by one of ordinary skill in the art (e.g., in cell culture, molecular genetics, immunology, immunohistochemistry, protein chemistry, and biochemistry).
[0211] Unless otherwise indicated, the recombinant protein, cell culture, and immunological techniques utilized in the present invention are standard procedures, well known to those skilled in the art. Such techniques are described and explained throughout the literature in sources such as, J. Perbal, A Practical Guide to Molecular Cloning, John Wiley and Sons (1984), J. Sambrook et al., Molecular Cloning: A Laboratory Manual, Cold Spring Harbour Laboratory Press (1989), T. A. Brown (editor), Essential Molecular Biology: A Practical Approach, Volumes 1 and 2, IRL Press (1991), D. M. Glover and B. D. Hames (editors), DNA Cloning: A Practical Approach, Volumes 1-4, IRL Press (1995 and 1996), and F. M. Ausubel et al. (editors), Current Protocols in Molecular Biology, Greene Pub. Associates and Wiley-Interscience (1988, including all updates until present), Ed Harlow and David Lane (editors) Antibodies: A Laboratory Manual, Cold Spring Harbour Laboratory, (1988), and J. E. Coligan et al. (editors) Current Protocols in Immunology, John Wiley & Sons (including all updates until present), and are incorporated herein by reference.
[0212] As used herein, the terms "silk protein" and "silk polypeptide" refer to a fibrous protein/polypeptide that can be used to produce a silk fibre, and/or a fibrous protein complex. Naturally occurring silk proteins of the invention form part of the brood comb silk of insects such as honeybees, however, as described herein variants of these proteins could readily be produced which would perform the same function if expressed within an appropriate insect.
[0213] As used herein, a "silk fibre" refers to filaments comprising proteins of the invention which can be woven into various items such as textiles.
[0214] As used herein, a "copolymer" is composition comprising two or more silk proteins of the invention. This term excludes naturally occurring copolymers such as the brood comb of insects.
[0215] The term "plant" includes whole plants, vegetative structures (for example, leaves, stems), roots, floral organs/structures, seed (including embryo, endosperm, and seed coat), plant tissue (for example, vascular tissue, ground tissue, and the like), cells and progeny of the same.
[0216] A "transgenic plant" refers to a plant that contains a gene construct ("transgene") not found in a wild-type plant of the same species, variety or cultivar. A "transgene" as referred to herein has the normal meaning in the art of biotechnology and includes a genetic sequence which has been produced or altered by recombinant DNA or RNA technology and which has been introduced into the plant cell. The transgene may include genetic sequences derived from a plant cell. Typically, the transgene has been introduced into the plant by human manipulation such as, for example, by transformation but any method can be used as one of skill in the art recognizes.
[0217] "Polynucleotide" refers to an oligonucleotide, nucleic acid molecule or any fragment thereof. It may be DNA or RNA of genomic or synthetic origin, double-stranded or single-stranded, and combined with carbohydrate, lipids, protein, or other materials to perform a particular activity defined herein.
[0218] "Operably linked" as used herein refers to a functional relationship between two or more nucleic acid (e.g., DNA) segments. Typically, it refers to the functional relationship of transcriptional regulatory element to a transcribed sequence. For example, a promoter is operably linked to a coding sequence, such as a polynucleotide defined herein, if it stimulates or modulates the transcription of the coding sequence in an appropriate host cell. Generally, promoter transcriptional regulatory elements that are operably linked to a transcribed sequence are physically contiguous to the transcribed sequence, i.e., they are cis-acting. However, some transcriptional regulatory elements, such as enhancers, need not be physically contiguous or located in close proximity to the coding sequences whose transcription they enhance.
[0219] The term "signal peptide" refers to an amino terminal polypeptide preceding a secreted mature protein. The signal peptide is cleaved from and is therefore not present in the mature protein. Signal peptides have the function of directing and trans-locating secreted proteins across cell membranes. The signal peptide is also referred to as signal sequence.
[0220] As used herein, "transformation" is the acquisition of new genes in a cell by the incorporation of a polynucleotide.
[0221] As used herein, the term "drug" refers to any compound that can be used to treat or prevent a particular disease, examples of drugs which can be formulated with a silk protein of the invention include, but are not limited to, proteins, nucleic acids, anti-tumor agents, analgesics, antibiotics, anti-inflammatory compounds (both steroidal and non-steroidal), hormones, vaccines, labeled substances, and the like.
Polypeptides
[0222] By "substantially purified polypeptide" we mean a polypeptide that has generally been separated from the lipids, nucleic acids, other polypeptides, and other contaminating molecules such as wax with which it is associated in its native state. With the exception of other proteins of the invention, it is preferred that the substantially purified polypeptide is at least 60% free, more preferably at least 75% free, and more preferably at least 90% free from other components with which it is naturally associated.
[0223] The term "recombinant" in the context of a polypeptide refers to the polypeptide when produced by a cell, or in a cell-free expression system, in an altered amount or at an altered rate compared to its native state. In one embodiment the cell is a cell that does not naturally produce the polypeptide. However, the cell may be a cell which comprises a non-endogenous gene that causes an altered, preferably increased, amount of the polypeptide to be produced. A recombinant polypeptide of the invention includes polypeptides which have not been separated from other components of the transgenic (recombinant) cell, or cell-free expression system, in which it is produced, and polypeptides produced in such cells or cell-free systems which are subsequently purified away from at least some other components.
[0224] The terms "polypeptide" and "protein" are generally used interchangeably and refer to a single polypeptide chain which may or may not be modified by addition of non-amino acid groups. The terms "proteins" and "polypeptides" as used herein also include variants, mutants, modifications, analogous and/or derivatives of the polypeptides of the invention as described herein.
[0225] The % identity of a polypeptide is determined by GAP (Needleman and Wunsch, 1970) analysis (GCG program) with a gap creation penalty=5, and a gap extension penalty-0.3. The query sequence is at least 15 amino acids in length, and the GAP analysis aligns the two sequences over a region of at least 15 amino acids. More preferably, the query sequence is at least 50 amino acids in length, and the GAP analysis aligns the two sequences over a region of at least 50 amino acids. More preferably, the query sequence is at least 100 amino acids in length and the GAP analysis aligns the two sequences over a region of at least 100 amino acids. Even more preferably, the query sequence is at least 250 amino acids in length and the GAP analysis aligns the two sequences over a region of at least 250 amino acids. Even more preferably, the GAP analysis aligns the two sequences over their entire length.
[0226] As used herein a "biologically active" fragment is a portion of a polypeptide of the invention which maintains a defined activity of the full-length polypeptide, namely the ability to be used to produce silk. Biologically active fragments can be any size as long as they maintain the defined activity.
[0227] With regard to a defined polypeptide, it will be appreciated that % identity figures higher than those provided above will encompass preferred embodiments. Thus, where applicable, in light of the minimum % identity figures, it is preferred that the polypeptide comprises an amino acid sequence which is at least 40%, more preferably at least 45%, more preferably at least 50%, more preferably at least 55%, more preferably at least 60%, more preferably at least 65%, more preferably at least 70%, more preferably at least 75%, more preferably at least 80%, more preferably at least 85%, more preferably at least 90%, more preferably at least 91%, more preferably at least 92%, more preferably at least 93%, more preferably at least 94%, more preferably at least 95%, more preferably at least 96%, more preferably at least 97%, more preferably at least 98%, more preferably at least 99%, more preferably at least 99.1%, more preferably at least 99.2%, more preferably at least 99.3%, more preferably at least 99.4%, more preferably at least 99.5%, more preferably at least 99.6%, more preferably at least 99.7%, more preferably at least 99.8%, and even more preferably at least 99.9% identical to the relevant nominated SEQ ID NO.
[0228] Amino acid sequence mutants of the polypeptides of the present invention can be prepared by introducing appropriate nucleotide changes into a nucleic acid of the present invention, or by in vitro synthesis of the desired polypeptide. Such mutants include, for example, deletions, insertions or substitutions of residues within the amino acid sequence. A combination of deletion, insertion and substitution can be made to arrive at the final construct, provided that the final polypeptide product possesses the desired characteristics.
[0229] Mutant (altered) polypeptides can be prepared using any technique known in the art. For example, a polynucleotide of the invention can be subjected to in vitro mutagenesis. Such in vitro mutagenesis techniques include sub-cloning the polynucleotide into a suitable vector, transforming the vector into a "mutator" strain such as the E. coli XL-1 red (Stratagene) and propagating the transformed bacteria for a suitable number of generations. In another example, the polynucleotides of the invention are subjected to DNA shuffling techniques as broadly described by Harayama (1998). These DNA shuffling techniques may include genes of the invention possibly in addition to genes related to those of the present invention, such as silk genes from Hymenopteran or Neuroptean species other than the specific species characterized herein. Products derived from mutated/altered DNA can readily be screened using techniques described herein to determine if they can be used as silk proteins.
[0230] In designing amino acid sequence mutants, the location of the mutation site and the nature of the mutation will depend on characteristic(s) to be modified. The sites for mutation can be modified individually or in series, e.g., by (1) substituting first with conservative amino acid choices and then with more radical selections depending upon the results achieved, (2) deleting the target residue, or (3) inserting other residues adjacent to the located site.
[0231] Amino acid sequence deletions generally range from about 1 to 15 residues, more preferably about 1 to 10 residues and typically about 1 to 5 contiguous residues.
[0232] Substitution mutants have at least one amino acid residue in the polypeptide molecule removed and a different residue inserted in its place. The sites of greatest interest for substitutional mutagenesis include sites identified as important for function. Other sites of interest are those in which particular residues obtained from various strains or species are identical. These positions may be important for biological activity. These sites, especially those falling within a sequence of at least three other identically conserved sites, are preferably substituted in a relatively conservative manner. Such conservative substitutions are shown in Table 1 under the heading of "exemplary substitutions".
[0233] As outlined above, a portion of some of the polypeptides of the invention have a coiled coil structure. Coiled coil structures of polypeptides are characterized by heptad repeats represented by the consensus sequence (abcdefg).sub.n. In a preferred embodiment, the portion of the polypeptide that has a coiled coil structure comprises at least 10 copies of the heptad sequence abcdefg, and at least 25% of the amino acids at positions a and d are alanine residues.
TABLE-US-00001 TABLE 1 Exemplary substitutions Original Exemplary Residue Substitutions Ala (A) val; leu; ile; gly; cys; ser; thr Arg (R) lys Asn (N) gln; his Asp (D) glu Cys (C) Ser; thr; ala; gly; val Gln (Q) asn; his Glu (E) asp Gly (G) pro; ala; ser; val; thr His (H) asn; gln Ile (I) leu; val; ala; met Leu (L) ile; val; met; ala; phe Lys (K) arg Met (M) leu; phe Phe (F) leu; val; ala Pro (P) gly Ser (S) thr; ala; gly; val; gln Thr (T) ser; gln; ala Trp (W) tyr Tyr (Y) trp; phe Val (V) ile; leu; met; phe; ala; ser; thr
[0234] In a preferred embodiment, the polypeptide that has a coiled coil structure comprises at least 12 consecutive copies, more preferably at least 15 consecutive copies, and even more preferably at least 18 consecutive copies of the heptad. In further embodiments, the polypeptide that has a coiled coil structure can have up to at least 28 copies of the heptad. Typically, the copies of the heptad will be tandemly repeated. However, they do not necessarily have to be perfect tandem repeats, for example, as shown in FIGS. 5 and 6 a few amino acids may be found between two heptads, or a few truncated heptads may be found (see, for example, Xenospira1 in FIG. 5).
[0235] Guidance regarding amino acid substitutions which can be made to the polypeptides of the invention which have a coiled coil structure is provided in FIGS. 5 and 6, as well as Tables 6 to 10. Where a predicted useful amino acid substitution based on the experimental data provided herein is in anyway in conflict with the exemplary substitutions provided in Table 1 it is preferred that a substitution based on the experimental data is used.
[0236] Coiled coil structures of polypeptides of the invention have a high content of alanine residues, particularly at amino acid positions a, d and e of the heptad. However, positions b, c, f and g also have a high frequency of alanine residues. In a preferred embodiment, at least 15% of the amino acids at positions a, d and/or e of the heptads are alanine residues, more preferably at least 25%, more preferably at least 30%, more preferably at least 40%, and even more preferably at least 50%. In a further preferred embodiment, at least 25% of the amino acids at both positions a and d of the heptads are alanine residues, more preferably at least 30%, more preferably at least 40%, and even more preferably at least 50%. Furthermore, it is preferred that at least 15% of the amino acids at positions b, c, f and g of the heptads are alanine residues, more preferably at least 20%, and even more preferably at least 25%.
[0237] Typically, the heptads will not comprise any proline or histidine residues. Furthermore, the heptads will comprise few (1 or 2), if any, phenylalanine, methionine, tyrosine, cysteine, glycine or tryptophan residues. Apart from alanine, common (for example greater than 5%, more preferably greater than 10%) amino acids in the heptads include leucine (particularly at positions b and d), serine (particularly at positions b, e and f), glutamic acid (particularly at positions c, e and f), lysine (particularly at positions b, c, d, f and g) as well as arginine at position g.
[0238] Polypeptides (and polynucleotides) of the invention can be purified (isolated) from a wide variety of Hymenopteran and Neuropteran species. Examples of Hymenopterans include, but are not limited to, any species of the Suborder Apocrita (bees, ants and wasps), which include the following Families of insects; Chrysididae (cuckoo wasps), Formicidae (ants), Mutillidae (velvet ants), Pompilidae (spider wasps), Scoliidae, Vespidae (paper wasps, potter wasps, hornets), Agaonidae (fig wasps), Chalcididae (chalcidids), Eucharitidae (eucharitids), Eupelmidae (eupelmids), Pteromalidae (pteromalids), Evaniidae (ensign wasps), Braconidae, Ichneumonidae (ichneumons), Megachilidae, Apidae, Colletidae, Halictidae, and Melittidae (oil collecting bees). Examples of Neuropterans include species from the following insect Families: Mantispidae, Chrysopidae (lacewings), Myrmeleontidae (antlions), and Ascalaphidae (owlflies). Such further polypeptides (and polynucleotides) can be characterized using the same procedures described herein for silks from Bombus terrestris, Myrmecia forficata, Oecophylla smaragdina and Mallada signata.
[0239] Furthermore, if desired, unnatural amino acids or chemical amino acid analogues can be introduced as a substitution or addition into the polypeptides of the present invention. Such amino acids include, but are not limited to, the D-isomers of the common amino acids, 2,4-diaminobutyric acid, .alpha.-amino isobutyric acid, 4-aminobutyric acid, 2-aminobutyric acid, 6-amino hexanoic acid, 2-amino isobutyric acid, 3-amino propionic acid, ornithine, norleucine, norvaline, hydroxyproline, sarcosine, citrulline, homocitrulline, cysteic acid, t-butylglycine, t-butylalanine, phenylglycine, cyclohexylalanine, .beta.-alanine, fluoro-amino acids, designer amino acids such as .beta.-methyl amino acids, C.alpha.-methyl amino acids, N.alpha.-methyl amino acids, and amino acid analogues in general.
[0240] Also included within the scope of the invention are polypeptides of the present invention which are differentially modified during or after synthesis, e.g., by biotinylation, benzylation, glycosylation, acetylation, phosphorylation, amidation, derivatization by known protecting/blocking groups, proteolytic cleavage, linkage to an antibody molecule or other cellular ligand, etc. These modifications may serve to increase the stability and/or bioactivity of the polypeptide of the invention.
[0241] Polypeptides of the present invention can be produced in a variety of ways, including production and recovery of natural polypeptides, production and recovery of recombinant polypeptides, and chemical synthesis of the polypeptides. In one embodiment, an isolated polypeptide of the present invention is produced by culturing a cell capable of expressing the polypeptide under conditions effective to produce the polypeptide, and recovering the polypeptide. A preferred cell to culture is a recombinant cell of the present invention. Effective culture conditions include, but are not limited to, effective media, bioreactor, temperature, pH and oxygen conditions that permit polypeptide production. An effective medium refers to any medium in which a cell is cultured to produce a polypeptide of the present invention. Such medium typically comprises an aqueous medium having assimilable carbon, nitrogen and phosphate sources, and appropriate salts, minerals, metals and other nutrients, such as vitamins. Cells of the present invention can be cultured in conventional fermentation bioreactors, shake flasks, test tubes, microtiter dishes, and petri plates. Culturing can be carried out at a temperature, pH and oxygen content appropriate for a recombinant cell. Such culturing conditions are within the expertise of one of ordinary skill in the art.
Polynucleotides
[0242] By an "isolated polynucleotide", including DNA, RNA, or a combination of these, single or double stranded, in the sense or antisense orientation or a combination of both, dsRNA or otherwise, we mean a polynucleotide which is at least partially separated from the polynucleotide sequences with which it is associated or linked in its native state. Preferably, the isolated polynucleotide is at least 60% free, preferably at least 75% free, and most preferably at least 90% free from other components with which they are naturally associated. Furthermore, the term "polynucleotide" is used interchangeably herein with the term "nucleic acid".
[0243] The term "exogenous" in the context of a polynucleotide refers to the polynucleotide when present in a cell, or in a cell-free expression system, in an altered amount compared to its native state. In one embodiment, the cell is a cell that does not naturally comprise the polynucleotide. However, the cell may be a cell which comprises a non-endogenous polynucleotide resulting in an altered, preferably increased, amount of production of the encoded polypeptide. An exogenous polynucleotide of the invention includes polynucleotides which have not been separated from other components of the transgenic (recombinant) cell, or cell-free expression system, in which it is present, and polynucleotides produced in such cells or cell-free systems which are subsequently purified away from at least some other components.
[0244] The % identity of a polynucleotide is determined by GAP (Needleman and Wunsch, 1970) analysis (GCG program) with a gap creation penalty=5, and a gap extension penalty=0.3. Unless stated otherwise, the query sequence is at least 45 nucleotides in length, and the GAP analysis aligns the two sequences over a region of at least 45 nucleotides. Preferably, the query sequence is at least 150 nucleotides in length, and the GAP analysis aligns the two sequences over a region of at least 150 nucleotides. More preferably, the query sequence is at least 300 nucleotides in length and the GAP analysis aligns the two sequences over a region of at least 300 nucleotides. Even more preferably, the GAP analysis aligns the two sequences over their entire length.
[0245] With regard to the defined polynucleotides, it will be appreciated that % identity figures higher than those provided above will encompass preferred embodiments. Thus, where applicable, in light of the minimum % identity figures, it is preferred that a polynucleotide of the invention comprises a sequence which is at least 40%, more preferably at least 45%, more preferably at least 50%, more preferably at least 55%, more preferably at least 60%, more preferably at least 65%, more preferably at least 70%, more preferably at least 75%, more preferably at least 80%, more preferably at least 85%, more preferably at least 90%, more preferably at least 91%, more preferably at least 92%, more preferably at least 93%, more preferably at least 94%, more preferably at least 95%, more preferably at least 96%, more preferably at least 97%, more preferably at least 98%, more preferably at least 99%, more preferably at least 99.1%, more preferably at least 99.2%, more preferably at least 99.3%, more preferably at least 99.4%, more preferably at least 99.5%, more preferably at least 99.6%, more preferably at least 99.7%, more preferably at least 99.8%, and even more preferably at least 99.9% identical to the relevant nominated SEQ ID NO.
[0246] Polynucleotides of the present invention may possess, when compared to naturally occurring molecules, one or more mutations which are deletions, insertions, or substitutions of nucleotide residues. Mutants can be either naturally occurring (that is to say, isolated from a natural source) or synthetic (for example, by performing site-directed mutagenesis on the nucleic acid).
[0247] Oligonucleotides and/or polynucleotides of the invention hybridize to a silk gene of the present invention, or a region flanking said gene, under stringent conditions. The term "stringent hybridization conditions" and the like as used herein refers to parameters with which the art is familiar, including the variation of the hybridization temperature with length of an oligonucleotide. Nucleic acid hybridization parameters may be found in references which compile such methods, Sambrook, et al. (supra), and Ausubel, et al. (supra). For example, stringent hybridization conditions, as used herein, can refer to hybridization at 65.degree. C. in hybridization buffer (3.5.times.SSC, 0.02% Ficoll, 0.02% polyvinyl pyrrolidone, 0.02% Bovine Serum Albumin (BSA), 2.5 mM NaH.sub.2PO.sub.4 (pH7), 0.5% SDS, 2 mM EDTA), followed by one or more washes in 0.2.xSSC, 0.01% BSA at 50.degree. C. Alternatively, the nucleic acid and/or oligonucleotides (which may also be referred to as "primers" or "probes") hybridize to the region of the an insect genome of interest, such as the genome of a honeybee, under conditions used in nucleic acid amplification techniques such as PCR.
[0248] Oligonucleotides of the present invention can be RNA, DNA, or derivptives of either. Although the terms polynucleotide and oligonucleotide have overlapping meaning, oligonucleotides are typically relatively short single stranded molecules. The minimum size of such oligonucleotides is the size required for the formation of a stable hybrid between an oligonucleotide and a complementary sequence on a target nucleic acid molecule. Preferably, the oligonucleotides are at least 15 nucleotides, more preferably at least 18 nucleotides, more preferably at least 19 nucleotides, more preferably at least 20 nucleotides, even more preferably at least 25 nucleotides in length.
[0249] Usually, monomers of a polynucleotide or oligonucleotide are linked by phosphodiester bonds or analogs thereof to form oligonucleotides ranging in size from a relatively short monomeric units, e.g., 12-18, to several hundreds of monomeric units. Analogs of phosphodiester linkages include: phosphorothioate, phosphorodithioate, phosphoroselenoate, phosphorodiselenoate, phosphoroanilothioate, phosphoranilidate, phosphoramidate.
[0250] The present invention includes oligonucleotides that can be used as, for example, probes to identify nucleic acid molecules, or primers to produce nucleic acid molecules. Oligonucleotides of the present invention used as a probe are typically conjugated with a detectable label such as a radioisotope, an enzyme, biotin, a fluorescent molecule or a chemiluminescent molecule.
Recombinant Vectors
[0251] One embodiment of the present invention includes a recombinant vector, which comprises at least one isolated polynucleotide molecule of the present invention, inserted into any vector capable of delivering the polynucleotide molecule into a host cell. Such a vector contains heterologous polynucleotide sequences, that is polynucleotide sequences that are not naturally found adjacent to polynucleotide molecules of the present invention and that preferably are derived from a species other than the species from which the polynucleotide molecule(s) are derived. The vector can be either RNA or DNA, either prokaryotic or eukaryotic, and typically is a transposon (such as described in U.S. Pat. No. 5,792,294), a virus or a plasmid.
[0252] One type of recombinant vector comprises a polynucleotide molecule of the present invention operatively linked to an expression vector. The phrase operatively linked refers to insertion of a polynucleotide molecule into an expression vector in a manner such that the molecule is able to be expressed when transformed into a host cell. As used herein, an expression vector is a DNA or RNA vector that is capable of transforming a host cell and of effecting expression of a specified polynucleotide molecule. Preferably, the expression vector is also capable of replicating within the host cell. Expression vectors can be either prokaryotic or eukaryotic, and are typically viruses or plasmids. Expression vectors of the present invention include any vectors that function (i.e., direct gene expression) in recombinant cells of the present invention, including in bacterial, fungal, endoparasite, arthropod, animal, and plant cells. Particularly preferred expression vectors of the present invention can direct gene expression in plants cells. Vectors of the invention can also be used to produce the polypeptide in a cell-free expression system, such systems are well known in the art.
[0253] In particular, expression vectors of the present invention contain regulatory sequences such as transcription control sequences, translation control sequences, origins of replication, and other regulatory sequences that are compatible with the recombinant cell and that control the expression of polynucleotide molecules of the present invention. In particular, recombinant molecules of the present invention include transcription control sequences. Transcription control sequences are sequences which control the initiation, elongation, and termination of transcription. Particularly important transcription control sequences are those which control transcription initiation, such as promoter, enhancer, operator and repressor sequences. Suitable transcription control sequences include any transcription control sequence that can function in at least one of the recombinant cells of the present invention. A variety of such transcription control sequences are known to those skilled in the art. Preferred transcription control sequences include those which function in bacterial, yeast, arthropod, plant or mammalian cells, such as, but not limited to, tac, lac, trp, trc, oxy-pro, omp/lpp, rrnB, bacteriophage lambda, bacteriophage T7, T7lac, bacteriophage T3, bacteriophage SP6, bacteriophage SP01, metallothionein, alpha-mating factor, Pichia alcohol oxidase, alphavirus subgenomic promoters (such as Sindbis virus subgenomic promoters), antibiotic resistance gene, baculovirus, Heliothis zea insect virus, vaccinia virus, herpesvirus, raccoon poxvirus, other poxvirus, adenovirus, cytomegalovirus (such as intermediate early promoters), simian virus 40, retrovirus, actin, retroviral long terminal repeat, Rous sarcoma virus, heat shock, phosphate and nitrate transcription control sequences as well as other sequences capable of controlling gene expression in prokaryotic or eukaryotic cells.
[0254] Particularly preferred transcription control sequences are promoters active in directing transcription in plants, either constitutively or stage and/or tissue specific, depending on the use of the plant or parts thereof. These plant promoters include, but are not limited to, promoters showing constitutive expression, such as the 35S promoter of Cauliflower Mosaic Virus (CaMV), those for leaf-specific expression, such as the promoter of the ribulose bisphosphate carboxylase small subunit gene, those for root-specific expression, such as the promoter from the glutamine synthase gene, those for seed-specific expression, such as the cruciferin A promoter from Brassica napus, those for tuber-specific expression, such as the class-I patatin promoter from potato or those for fruit-specific expression, such as the polygalacturonase (PG) promoter from tomato.
[0255] Recombinant molecules of the present invention may also (a) contain secretory signals (i.e., signal segment nucleic acid sequences) to enable an expressed polypeptide of the present invention to be secreted from the cell that produces the polypeptide and/or (b) contain fusion sequences which lead to the expression of nucleic acid molecules of the present invention as fusion proteins. Examples of suitable signal segments include any signal segment capable of directing the secretion of a polypeptide of the present invention. Preferred signal segments include, but are not limited to, tissue plasminogen activator (t-PA), interferon, interleukin, growth hormone, viral envelope glycoprotein signal segments, Nicotiana nectarin signal peptide (U.S. Pat. No. 5,939,288), tobacco extensin signal, the soy oleosin oil body binding protein signal, Arabidopsis thaliana vacuolar basic chitinase signal peptide, as well as native signal sequences of a polypeptide of the invention. In addition, a nucleic acid molecule of the present invention can be joined to a fusion segment that directs the encoded polypeptide to the proteosome, such as a ubiquitin fusion segment. Recombinant molecules may also include intervening and/or untranslated sequences surrounding and/or within the nucleic acid sequences of the present invention.
Host Cells
[0256] Another embodiment of the present invention includes a recombinant cell comprising a host cell transformed with one or more recombinant molecules of the present invention, or progeny cells thereof. Transformation of a polynucleotide molecule into a cell can be accomplished by any method by which a polynucleotide molecule can be inserted into the cell. Transformation techniques include, but are not limited to, transfection, electroporation, microinjection, lipofection, adsorption, and protoplast fusion. A recombinant cell may remain unicellular or may grow into a tissue, organ or a multicellular organism. Transformed polynucleotide molecules of the present invention can remain extrachromosomal or can integrate into one or more sites within a chromosome of the transformed (i.e., recombinant) cell in such a manner that their ability to be expressed is retained.
[0257] Suitable host cells to transform include any cell that can be transformed with a polynucleotide of the present invention. Host cells of the present invention either can be endogenously (i.e., naturally) capable of producing polypeptides of the present invention or can be capable of producing such polypeptides after being transformed with at least one polynucleotide molecule of the present invention. Host cells of the present invention can be any cell capable of producing at least one protein of the present invention, and include bacterial, fungal (including yeast), parasite, arthropod, animal and plant cells. Examples of host cells include Salmonella, Escherichia, Bacillus, Listeria, Saccharomyces, Spodoptera, Mycobacteria, Trichoplusia, BHK (baby hamster kidney) cells, MDCK cells, CRFK cells, CV-1 cells, COS (e.g., COS-7) cells, and Vero cells. Further examples of host cells are E. coli, including E. coli K-12 derivatives; Salmonella typhi; Salmonella typhimurium, including attenuated strains; Spodoptera frugiperda; Trichoplusia ni; and non-tumorigenic mouse myoblast G8 cells (e.g., ATCC CRL 1246). Additional appropriate mammalian cell hosts include other kidney cell lines, other fibroblast cell lines (e.g., human, murine or chicken embryo fibroblast cell lines), myeloma cell lines, Chinese hamster ovary cells, mouse NIH/3T3 cells, LMTK cells and/or HeLa cells. Particularly preferred host cells are plant cells such as those available from Deutsche Sammlung von Mikroorganismen and Zellkulturen GmbH (German Collection of Microorganisms and Cell Cultures).
[0258] Recombinant DNA technologies can be used to improve expression of a transformed polynucleotide molecule by manipulating, for example, the number of copies of the polynucleotide molecule within a host cell, the efficiency with which those polynucleotide molecules are transcribed, the efficiency with which the resultant transcripts are translated, and the efficiency of post-translational modifications. Recombinant techniques useful for increasing the expression of polynucleotide molecules of the present invention include, but are not limited to, operatively linking polynucleotide molecules to high-copy number plasmids, integration of the polynucleotide molecule into one or more host cell chromosomes, addition of vector stability sequences to plasmids, substitutions or modifications of transcription control signals (e.g., promoters, operators, enhancers), substitutions or modifications of translational control signals (e.g., ribosome binding sites, Shine-Dalgarno sequences), modification of polynucleotide molecules of the present invention to correspond to the codon usage of the host cell, and the deletion of sequences that destabilize transcripts.
Transgenic Plants
[0259] The term "plant" refers to whole plants, plant organs (e.g. leaves, stems roots, etc), seeds, plant cells and the like. Plants contemplated for use in the practice of the present invention include both monocotyledons and dicotyledons. Target plants include, but are not limited to, the following: cereals (wheat, barley, rye, oats, rice, sorghum and related crops); beet (sugar beet and fodder beet); pomes, stone fruit and soft fruit (apples, pears, plums, peaches, almonds, cherries, strawberries, raspberries and black-berries); leguminous plants (beans, lentils, peas, soybeans); oil plants (rape, mustard, poppy, olives, sunflowers, coconut, castor oil plants, cocoa beans, groundnuts); cucumber plants (marrows, cucumbers, melons); fibre plants (cotton, flax, hemp, jute); citrus fruit (oranges, lemons, grapefruit, mandarins); vegetables (spinach, lettuce, asparagus, cabbages, carrots, onions, tomatoes, potatoes, paprika); lauraceae (avocados, cinnamon, camphor); or plants such as maize, tobacco, nuts, coffee, sugar cane, tea, vines, hops, turf, bananas and natural rubber plants, as well as ornamentals (flowers, shrubs, broad-leaved trees and evergreens, such as conifers).
[0260] Transgenic plants, as defined in the context of the present invention include plants (as well as parts and cells of said plants) and their progeny which have been genetically modified using recombinant techniques to cause production of at least one polypeptide of the present invention in the desired plant or plant organ. Transgenic plants can be produced using techniques known in the art, such as those generally described in A. Slater et al., Plant Biotechnology--The Genetic Manipulation of Plants, Oxford University Press (2003), and P. Christou and H. Klee, Handbook of Plant Biotechnology, John Wiley and Sons (2004).
[0261] A polynucleotide of the present invention may be expressed constitutively in the transgenic plants during all stages of development. Depending on the use of the plant or plant organs, the polypeptides may be expressed in a stage-specific manner. Furthermore, the polynucleotides may be expressed tissue-specifically.
[0262] Regulatory sequences which are known or are found to cause expression of a gene encoding a polypeptide of interest in plants may be used in the present invention. The choice of the regulatory sequences used depends on the target plant and/or target organ of interest. Such regulatory sequences may be obtained from plants or plant viruses, or may be chemically synthesized. Such regulatory sequences are well known to those skilled in the art.
[0263] Constitutive plant promoters are well known. Further to previously mentioned promoters, some other suitable promoters include but are not limited to the nopaline synthase promoter, the octopine synthase promoter, CaMV 35S promoter, the ribulose-1,5-bisphosphate carboxylase promoter, Adh1-based pEmu, Act1, the SAM synthase promoter and Ubi promoters and the promoter of the chlorophyll a/b binding protein. Alternatively it may be desired to have the transgene(s) expressed in a regulated fashion. Regulated expression of the polypeptides is possible by placing the coding sequence of the silk protein under the control of promoters that are tissue-specific, developmental-specific, or inducible. Several tissue-specific regulated genes and/or promoters have been reported in plants. These include genes encoding the seed storage proteins (such as napin, cruciferin, .beta.-conglycinin, glycinin and phaseolin), zein or oil body proteins (such as oleosin), or genes involved in fatty acid biosynthesis (including acyl carrier protein, stearoyl-ACP desaturase, and fatty acid desaturases (fad 2-1)), and other genes expressed during embryo development (such as Bce4). Particularly useful for seed-specific expression is the pea vicilin promoter. Other useful promoters for expression in mature leaves are those that are switched on at the onset of senescence, such as the SAG promoter from Arabidopsis). A class of fruit-specific promoters expressed at or during anthesis through fruit development, at least until the beginning of ripening, is discussed in U.S. Pat. No. 4,943,674. Other examples of tissue-specific promoters include those that direct expression in tubers (for example, patatin gene promoter), and in fiber cells (an example of a developmentally-regulated fiber cell protein is E6 fiber).
[0264] Other regulatory sequences such as terminator sequences and polyadenylation signals include any such sequence functioning as such in plants, the choice of which would be obvious to the skilled addressee. The termination region used in the expression cassette will be chosen primarily for convenience, since the termination regions appear to be relatively interchangeable. The termination region which is used may be native with the transcriptional initiation region, may be native with the polynucleotide sequence of interest, or may be derived from another source. The termination region may be naturally occurring, or wholly or partially synthetic. Convenient termination regions are available from the Ti-plasmid of A. tumefaciens, such as the octopine synthase and nopaline synthase termination regions or from the genes for .beta.-phaseolin, the chemically inducible plant gene, pIN.
[0265] Several techniques are available for the introduction of an expression construct containing a nucleic acid sequence encoding a polypeptide of interest into the target plants. Such techniques include but are not limited to transformation of protoplasts using the calcium/polyethylene glycol method, electroporation and microinjection or (coated) particle bombardment. In addition to these so-called direct DNA transformation methods, transformation systems involving vectors are widely available, such as viral and bacterial vectors (e.g. from the genus Agrobacterium). After selection and/or screening, the protoplasts, cells or plant parts that have been transformed can be regenerated into whole plants, using methods known in the art. The choice of the transformation and/or regeneration techniques is not critical for this invention.
[0266] To confirm the presence of the transgenes in transgenic cells and plants, a polymerase chain reaction (PCR) amplification or Southern blot analysis can be performed using methods known to those skilled in the art. Expression products of the transgenes can be detected in any of a variety of ways, depending upon the nature of the product, and include Western blot and enzyme assay. One particularly useful way to quantitate protein expression and to detect replication in different plant tissues is to use a reporter gene, such as GUS. Once transgenic plants have been obtained, they may be grown to produce plant tissues or parts having the desired phenotype. The plant tissue or plant parts, may be harvested, and/or the seed collected. The seed may serve as a source for growing additional plants with tissues or parts having the desired characteristics.
Transgenic Hon-Human Animals
[0267] Techniques for producing transgenic animals are well known in the art. A useful general textbook on this subject is Houdebine, Transgenic animals--Generation and Use (Harwood Academic, 1997).
[0268] Heterologous DNA can be introduced, for example, into fertilized mammalian ova. For instance, totipotent or pluripotent stem cells can be transformed by microinjection, calcium phosphate mediated precipitation, liposome fusion, retroviral infection or other means, the transformed cells are then introduced into the embryo, and the embryo then develops into a transgenic animal. In a highly preferred method, developing embryos are infected with a retrovirus containing the desired DNA, and transgenic animals produced from the infected embryo. In a most preferred method, however, the appropriate DNAs are coinjected into the pronucleus or cytoplasm of embryos, preferably at the single cell stage, and the embryos allowed to develop into mature transgenic animals.
[0269] Another method used to produce a transgenic animal involves microinjecting a nucleic acid into pro-nuclear stage eggs by standard methods. Injected eggs are then cultured before transfer into the oviducts of pseudopregnant recipients.
[0270] Transgenic animals may also be produced by nuclear transfer technology. Using this method, fibroblasts from donor animals are stably transfected with a plasmid incorporating the coding sequences for a binding domain or binding partner of interest under the control of regulatory sequences. Stable transfectants are then fused to enucleated oocytes, cultured and transferred into female recipients.
Recovery Methods and Production of Silk
[0271] The silk proteins of the present invention may be extracted and purified from recombinant cells, such as plant, bacteria or yeast cells, producing said protein by a variety of methods. In one embodiment, the method involves removal of native cell proteins from homogenized cells/tissues/plants etc. by lowering pH and heating, followed by ammonium sulfate fractionation. Briefly, total soluble proteins are extracted by homogenizing cells/tissues/plants. Native proteins are removed by precipitation at pH 4.7 and then at 60.degree. C. The resulting supernatant is then fractionated with ammonium sulfate at 40% saturation. The resulting protein will be of the order of 95% pure. Additional purification may be achieved with conventional gel or affinity chromatography.
[0272] In another example, cell lysates are treated with high concentrations of acid e.g. HCl or propionic acid to reduce pH to .about.1-2 for 1 hour or more which will solubilise the silk proteins but precipitate other proteins.
[0273] Fibrillar aggregates will form from solutions by spontaneous self-assembly of silk proteins of the invention when the protein concentration exceeds a critical value. The aggregates may be gathered and mechanically spun into macroscopic fibers according to the method of O'Brien et al. (I. O'Brien et al., "Design, Synthesis and Fabrication of Novel Self-Assembling Fibrillar Proteins", in Silk Polymers: Materials Science and Biotechnology, pp. 104-117, Kaplan, Adams, Fanner and Viney, eds., c. 1994 by American Chemical Society, Washington, D.C.).
[0274] By nature of the inherent coiled coil secondary structure, proteins such as Xenospira1-4, BBF1-4, BAF1-4 and GAF1-4 will spontaneously form the coiled coil secondary structure upon dehydration. As described below, the strength of the coiled coil can be enhanced through enzymatic or chemical cross-linking of lysine residues in close proximity.
[0275] Silk fibres and/or copolymers of the invention have a low processing requirement. The silk proteins of the invention require minimal processing e.g. spinning to form a strong fibre as they spontaneously forms strong coiled coils which can be reinforced with crosslinks such as lysine crosslinks. This contrasts with B. mori and spider recombinant silk polypeptides which require sophisticated spinning techniques in order to obtain the secondary structure (.beta.-sheet) and strength of the fibre.
[0276] However, fibers may be spun from solutions having properties characteristic of a liquid crystal phase. The fiber concentration at which phase transition can occur is dependent on the composition of a protein or combination of proteins present in the solution. Phase transition, however, can be detected by monitoring the clarity and birefringence of the solution. Onset of a liquid crystal phase can be detected when the solution acquires a translucent appearance and registers birefringence when viewed through crossed polarizing filters.
[0277] In one fiber-forming technique, fibers can first be extruded from the protein solution through an orifice into methanol, until a length sufficient to be picked up by a mechanical means is produced. Then a fiber can be pulled by such mechanical means through a methanol solution, collected, and dried. Methods for drawing fibers are considered well-known in the art.
[0278] Further examples of methods which may be used for producing silk fibres and/or copolymers of the present are described in US 2004/0170827 and US 2005/0054830.
[0279] In a preferred embodiment, silk fibres and/or copolymers of the invention are crosslinked. In one embodiment, the silk fibres and/or copolymers are crosslinked to a surface/article/product etc of interest using techniques known in the art. In another embodiment (or in combination with the previous embodiment), at least some silk proteins in the silk fibres and/or copolymers are crosslinked to each other. Preferably, the silk proteins are crosslinked via lysine residues in the proteins. Such crosslinking can be performed using chemical and/or enzymatic techniques known in the art. For example, enzymatic cross links can be catalysed by lysyl oxidase, whereas nonenzymatic cross links can be generated from glycated lysine residues (Reiser et al., 1992).
Antibodies
[0280] The invention also provides monoclonal or polyclonal antibodies to polypeptides of the invention or fragments thereof. Thus, the present invention further provides a process for the production of monoclonal or polyclonal antibodies to polypeptides of the invention.
[0281] The term "binds specifically" refers to the ability of the antibody to bind to at least one polypeptide of the present invention but not other known silk proteins.
[0282] As used herein, the term "epitope" refers to a region of a polypeptide of the invention which is bound by the antibody. An epitope can be administered to an animal to generate antibodies against the epitope, however, antibodies of the present invention preferably specifically bind the epitope region in the context of the entire polypeptide.
[0283] If polyclonal antibodies are desired, a selected mammal (e.g., mouse, rabbit, goat, horse, etc.) is immunised with an immunogenic polypeptide of the invention. Serum from the immunised animal is collected and treated according to known procedures. If serum containing polyclonal antibodies contains antibodies to other antigens, the polyclonal antibodies can be purified by immunoaffinity chromatography. Techniques for producing and processing polyclonal antisera are known in the art. In order that such antibodies may be made, the invention also provides polypeptides of the invention or fragments thereof haptenised to another polypeptide for use as immunogens in animals.
[0284] Monoclonal antibodies directed against polypeptides of the invention can also be readily produced by one skilled in the art. The general methodology for making monoclonal antibodies by hybridomas is well known. Immortal antibody-producing cell lines can be created by cell fusion, and also by other techniques such as direct transformation of B lymphocytes with oncogenic DNA, or transfection with Epstein-Barr virus. Panels of monoclonal antibodies produced can be screened for various properties; i.e., for isotype and epitope affinity.
[0285] An alternative technique involves screening phage display libraries where, for example the phage express scFv fragments on the surface of their coat with a large variety of complementarity determining regions (CDRs). This technique is well known in the art.
[0286] For the purposes of this invention, the term "antibody", unless specified to the contrary, includes fragments of whole antibodies which retain their binding activity for a target antigen. Such fragments include Fv, F(ab') and F(ab').sub.2 fragments, as well as single chain antibodies (scFv). Furthermore, the antibodies and fragments thereof may be humanised antibodies, for example as described in EP-A-239400.
[0287] Antibodies of the invention may be bound to a solid support and/or packaged into kits in a suitable container along with suitable reagents, controls, instructions and the like.
[0288] Preferably, antibodies of the present invention are detectably labeled. Exemplary detectable labels that allow for direct measurement of antibody binding include radiolabels, fluorophores, dyes, magnetic beads, chemiluminescers, colloidal particles, and the like. Examples of labels which permit indirect measurement of binding include enzymes where the substrate may provide for a coloured or fluorescent product. Additional exemplary detectable labels include covalently bound enzymes capable of providing a detectable product signal after addition of suitable substrate. Examples of suitable enzymes for use in conjugates include horseradish peroxidase, alkaline phosphatase, malate dehydrogenase and the like. Where not commercially available, such antibody-enzyme conjugates are readily produced by techniques known to those skilled in the art. Further exemplary detectable labels include biotin, which binds with high affinity to avidin or streptavidin; fluorochromes (e.g., phycobiliproteins, phycoerythrin and allophycocyanins; fluorescein and Texas red), which can be used with a fluorescence activated cell sorter; haptens; and the like. Preferably, the detectable label allows for direct measurement in a plate luminometer, e.g., biotin. Such labeled antibodies can be used in techniques known in the art to detect polypeptides of the invention.
Compositions
[0289] Compositions of the present invention may include an "acceptable carrier". Examples of such acceptable carriers include water, saline, Ringer's solution, dextrose solution, Hank's solution, and other aqueous physiologically balanced salt solutions. Nonaqueous vehicles, such as fixed oils, sesame oil, ethyl oleate, or triglycerides may also be used.
[0290] In one embodiment, the "acceptable carrier" is a "pharmaceutically acceptable carrier". The term pharmaceutically acceptable carrier refers to molecular entities and compositions that do not produce an allergic, toxic or otherwise adverse reaction when administered to an animal, particularly a mammal, and more particularly a human. Useful examples of pharmaceutically acceptable carriers or diluents include, but are not limited to, solvents, dispersion media, coatings, stabilizers, protective colloids, adhesives, thickeners, thixotropic agents, penetration agents, sequestering agents and isotonic and absorption delaying agents that do not affect the activity of the polypeptides of the invention. The proper fluidity can be maintained, for example, by the use of a coating, such as lecithin, by the maintenance of the required particle size in the case of dispersion and by the use of surfactants. More generally, the polypeptides of the invention can be combined with any non-toxic solid or liquid additive corresponding to the usual formulating techniques.
[0291] As outlined herein, in some embodiments a polypeptide, a silk fiber and/or a copolymer of the invention is used as a pharmaceutically acceptable carrier.
[0292] Other suitable compositions are described below with specific reference to specific uses of the polypeptides of the invention.
Uses
[0293] Silk proteins are useful for the creation of new biomaterials because of their exceptional toughness and strength. However, to date the fibrous proteins of spiders and insects are large proteins (over 100 kDa) and consist of highly repetitive amino acid sequences. These proteins are encoded by large genes containing highly biased codons making them particularly difficult to produce in recombinant systems. By comparison, the silk proteins of the invention are short and non-repetitive. These properties make the genes encoding these proteins particularly attractive for recombinant production of new biomaterials.
[0294] The silk proteins, silk fibers and/or copolymers of the invention can be used for a broad and diverse array of medical, military, industrial and commercial applications. The fibers can be used in the manufacture of medical devices such as sutures, skin grafts, cellular growth matrices, replacement ligaments, and surgical mesh, and in a wide range of industrial and commercial products, such as, for example, cable, rope, netting, fishing line, clothing fabric, bullet-proof vest lining, container fabric, backpacks, knapsacks, bag or purse straps, adhesive binding material, non-adhesive binding material, strapping material, tent fabric, tarpaulins, pool covers, vehicle covers, fencing material, sealant, construction material, weatherproofing material, flexible partition material, sports equipment; and, in fact, in nearly any use of fiber or fabric for which high tensile strength and elasticity are desired characteristics. The silk proteins, silk fibers and/or copolymers of the present invention also have applications in compositions for personal care products such as cosmetics, skin care, hair care and hair colouring; and in coating of particles, such as pigments.
[0295] The silk proteins may be used in their native form or they may be modified to form derivatives, which provide a more beneficial effect. For example, the silk protein may be modified by conjugation to a polymer to reduce allergenicity as described in U.S. Pat. Nos. 5,981,718 and 5,856,451. Suitable modifying polymers include, but are not limited to, polyalkylene oxides, polyvinyl alcohol, poly-carboxylates, poly(vinylpyrolidone), and dextrans. In another example, the silk proteins may be modified by selective digestion and splicing of other protein modifiers. For example, the silk proteins may be cleaved into smaller peptide units by treatment with acid at an elevated temperature of about 60.degree. C. The useful acids include, but are not limited to, dilute hydrochloric, sulfuric or phosphoric acids. Alternatively, digestion of the silk proteins may be done by treatment with a base, such as sodium hydroxide, or enzymatic digestion using a suitable protease may be used.
[0296] The proteins may be further modified to provide performance characteristics that are beneficial in specific applications for personal care products. The modification of proteins for use in personal care products is well known in the art. For example, commonly used methods are described in U.S. Pat. Nos. 6,303,752, 6,284,246, and 6,358,501. Examples of modifications include, but are not limited to, ethoxylation to promote water-oil emulsion enhancement, siloxylation to provide lipophilic compatibility, and esterification to aid in compatibility with soap and detergent compositions. Additionally, the silk proteins may be derivatized with functional groups including, but not limited to, amines, oxiranes, cyanates, carboxylic acid esters, silicone copolyols, siloxane esters, quaternized amine aliphatics, urethanes, polyacrylamides, dicarboxylic acid esters, and halogenated esters. The silk proteins may also be derivatized by reaction with diimines and by the formation of metal salts.
[0297] Consistent with the above definitions of "polypeptide" (and "protein"), such derivatized and/or modified molecules are also referred to herein broadly as "polypeptides" and "proteins".
[0298] Silk proteins of the invention can be spun together and/or bundled or braided with other fiber types. Examples include, but are not limited to, polymeric fibers (e.g., polypropylene, nylon, polyester), fibers and silks of other plant and animal sources (e.g., cotton, wool, Bombyx mori or spider silk), and glass fibers. A preferred embodiment is silk fiber braided with 10% polypropylene fiber. The present invention contemplates that the production of such combinations of fibers can be readily practiced to enhance any desired characteristics, e.g., appearance, softness, weight, durability, water-repellant properties, improved cost-of-manufacture, that may be generally sought in the manufacture and production of fibers for medical, industrial, or commercial applications.
Personal Care Products
[0299] Cosmetic and skin care compositions may be anhydrous compositions comprising an effective amount of silk protein in a cosmetically acceptable medium. The uses of these compositions include, but are not limited to, skin care, skin cleansing, make-up, and anti-wrinkle products. An effective amount of a silk protein for cosmetic and skin care compositions is herein defined as a proportion of from about 10.sup.-4 to about 30% by weight, but preferably from about 10.sup.-3 to 15% by weight, relative to the total weight of the composition. This proportion may vary as a function of the type of cosmetic or skin care composition. Suitable compositions for a cosmetically acceptable medium are described in U.S. Pat. No. 6,280,747. For example, the cosmetically acceptable medium may contain a fatty substance in a proportion generally of from about 10 to about 90% by weight relative to the total weight of the composition, where the fatty phase containing at least one liquid, solid or semi-solid fatty substance. The fatty substance includes, but is not limited to, oils, waxes, gums, and so-called pasty fatty substances. Alternatively, the compositions may be in the form of a stable dispersion such as a water-in-oil or oil-in-water emulsion. Additionally, the compositions may contain one or more conventional cosmetic or dermatological additives or adjuvants, including but not limited to, antioxidants, preserving agents, fillers, surfactants, UVA and/or UVB sunscreens, fragrances, thickeners, wetting agents and anionic, nonionic or amphoteric polymers, and dyes or pigments.
[0300] Emulsified cosmetics and quasi drugs which are producible with the use of emulsified materials comprising at least one silk protein of the present invention include, for example, cleansing cosmetics (beauty soap, facial wash, shampoo, rinse, and the like), hair care products (hair dye, hair cosmetics, and the like), basic cosmetics (general cream, emulsion, shaving cream, conditioner, cologne, shaving lotion, cosmetic oil, facial mask, and the like), make-up cosmetics (foundation, eyebrow pencil, eye cream, eye shadow, mascara, and the like), aromatic cosmetics (perfume and the like), tanning and sunscreen cosmetics (tanning and sunscreen cream, tanning and sunscreen lotion, tanning and sunscreen oil, and the like), nail cosmetics (nail cream and the like), eyeliner cosmetics (eyeliner and the like), lip cosmetics (lipstick, lip cream, and the like), oral care products (tooth paste and the like) bath cosmetics (bath products and the like), and the like.
[0301] The cosmetic composition may also be in the form of products for nail care, such as a nail varnish. Nail varnishes are herein defined as compositions for the treatment and colouring of nails, comprising an effective amount of silk protein in a cosmetically acceptable medium. An effective amount of a silk protein for use in a nail varnish composition is herein defined as a proportion of from about 10.sup.4 to about 30% by weight relative to the total weight of the varnish. Components of a cosmetically acceptable medium for nail varnishes are described in U.S. Pat. No. 6,280,747. The nail varnish typically contains a solvent and a film forming substance, such as cellulose derivatives, polyvinyl derivatives, acrylic polymers or copolymers, vinyl copolymers and polyester polymers. The composition may also contain an organic or inorganic pigment.
[0302] Hair care compositions are herein defined as compositions for the treatment of hair, including but not limited to shampoos, conditioners, lotions, aerosols, gels, and mousses, comprising an effective amount of silk protein in a cosmetically acceptable medium. An effective amount of a silk protein for use in a hair care composition is herein defined as a proportion of from about 10.sup.0.2 to about 90% by weight relative to the total weight of the composition. Components of a cosmetically acceptable medium for hair care compositions are described in US 2004/0170590, U.S. Pat. Nos. 6,280,747, 6,139,851, and 6,013,250. For example, these hair care compositions can be aqueous, alcoholic or aqueous-alcoholic solutions, the alcohol preferably being ethanol or isopropanol, in a proportion of from about 1 to about 75% by weight relative to the total weight, for the aqueous-alcoholic solutions. Additionally, the hair care compositions may contain one or more conventional cosmetic or dermatological additives or adjuvants, as given above.
[0303] Hair colouring compositions are herein defined as compositions for the colouring, dyeing, or bleaching of hair, comprising an effective amount of silk protein in a cosmetically acceptable medium. An effective amount of a silk protein for use in a hair colouring composition is herein defined as a proportion of from about 10.sup.-4 to about 60% by weight relative to the total weight of the composition. Components of a cosmetically acceptable medium for hair colouring compositions are described in US 2004/0170590, U.S. Pat. Nos. 6,398,821 and 6,129,770. For example, hair colouring compositions generally contain a mixture of inorganic peroxygen-based dye oxidizing agent and an oxidizable coloring agent. The peroxygen-based dye oxidizing agent is most commonly hydrogen peroxide. The oxidative hair coloring agents are formed by oxidative coupling of primary intermediates (for example p-phenylenediamines, p-aminophenols, p-diaminopyridines, hydroxyindoles, aminoindoles, aminothymidines, or cyanophenols) with secondary intermediates (for example phenols, resorcinols, m-aminophenols, m-phenylenediamines, naphthols, pyrazolones, hydroxyindoles, catechols or pyrazoles). Additionally, hair colouring compositions may contain oxidizing acids, sequestrants, stabilizers, thickeners, buffers carriers, surfactants, solvents, antioxidants, polymers, non-oxidative dyes and conditioners.
[0304] The silk proteins can also be used to coat pigments and cosmetic particles in order to improve dispersibility of the particles for use in cosmetics and coating compositions. Cosmetic particles are herein defined as particulate materials such as pigments or inert particles that are used in cosmetic compositions. Suitable pigments and cosmetic particles, include, but are not limited to, inorganic color pigments, organic pigments, and inert particles. The inorganic color pigments include, but are not limited to, titanium dioxide, zinc oxide, and oxides of iron, magnesium, cobalt, and aluminium. Organic pigments include, but are not limited to, D&C Red No. 36, D&C Orange No. 17, the calcium lakes of D&C Red Nos. 7, 11, 31 and 34, the barium lake of D&C Red No. 12, the strontium lake D&C Red No. 13, the aluminium lake of FD&C Yellow No. 5 and carbon black particles. Inert particles include, but are not limited to, calcium carbonate, aluminium silicate, calcium silicate, magnesium silicate, mica, talc, barium sulfate, calcium sulfate, powdered Nylon.TM., perfluorinated alkanes, and other inert plastics.
[0305] The silk proteins may also be used in dental floss (see, for example, US 2005/0161058). The floss may be monofilament yarn or multifilament yarn, and the fibers may or may not be twisted. The dental floss may be packaged as individual pieces or in a roll with a cutter for cutting pieces to any desired length. The dental floss may be provided in a variety of shapes other than filaments, such as but not limited to, strips and sheets and the like. The floss may be coated with different materials, such as but not limited to, wax, polytetrafluoroethylene monofilament yarn for floss.
[0306] The silk proteins may also be used in soap (see, for example, US 2005/0130857).
Pigment and Cosmetic Particle Coating
[0307] The effective amount of a silk protein for use in pigment and cosmetic particle coating is herein defined as a proportion of from about 10.sup.-4 to about 50%, but preferably from about 0.25 to about 15% by weight relative to the dry weight of particle. The optimum amount of the silk protein to be used depends on the type of pigment or cosmetic particle being coated. For example, the amount of silk protein used with inorganic color pigments is preferably between about 0.01% and 20% by weight. In the case of organic pigments, the preferred amount of silk protein is between about 1% to about 15% by weight, while for inert particles, the preferred amount is between about 0.25% to about 3% by weight. Methods for the preparation of coated pigments and particles are described in U.S. Pat. No. 5,643,672. These methods include: adding an aqueous solution of the silk protein to the particles while tumbling or mixing, forming a slurry of the silk protein and the particles and drying, spray drying a solution of the silk protein onto the particles or lyophilizing a slurry of the silk protein and the particles. These coated pigments and cosmetic particles may be used in cosmetic formulations, paints, inks and the like.
Biomedical
[0308] The silk proteins may be used as a coating on a bandage to promote wound healing. For this application, the bandage material is coated with an effective amount of the silk protein. For the purpose of a wound-healing bandage, an effective amount of silk protein is herein defined as a proportion of from about e to about 30% by weight relative to the weight of the bandage material. The material to be coated may be any soft, biologically inert, porous cloth or fiber. Examples include, but are not limited to, cotton, silk, rayon, acetate, acrylic, polyethylene, polyester, and combinations thereof. The coating of the cloth or fiber may be accomplished by a number of methods known in the art. For example, the material to be coated may be dipped into an aqueous solution containing the silk protein. Alternatively, the solution containing the silk protein may be sprayed onto the surface of the material to be coated using a spray gun. Additionally, the solution containing the silk protein may be coated onto the surface using a roller coat printing process. The wound bandage may include other additives including, but not limited to, disinfectants such as iodine, potassium iodide, povidon iodine, acrinol, hydrogen peroxide, benzalkonium chloride, and chlorohexidine; cure accelerating agents such as allantoin, dibucaine hydrochloride, and chlorophenylamine malate; vasoconstrictor agents such as naphazoline hydrochloride; astringent agents such as zinc oxide; and crust generating agents such as boric acid.
[0309] The silk proteins of the present invention may also be used in the form of a film as a wound dressing material. The use of silk proteins, in the form of an amorphous film, as a wound dressing material is described in U.S. Pat. No. 6,175,053. The amorphous film comprises a dense and nonporous film of a crystallinity below 10% which contains an effective amount of silk protein. For a film for wound care, an effective amount of silk protein is herein defined as between about 1 to 99% by weight. The film may also contain other components including but not limited to other proteins such as sericin, and disinfectants, cure accelerating agents, vasoconstrictor agents, astringent agents, and crust generating agents, as described above. Other proteins such as sericin may comprise 1 to 99% by weight of the composition. The amount of the other ingredients listed is preferably below a total of about 30% by weight, more preferably between about 0.5 to 20% by weight of the composition. The wound dressing film may be prepared by dissolving the above mentioned materials in an aqueous solution, removing insolubles by filtration or centrifugation, and casting the solution on a smooth solid surface such as an acrylic plate, followed by drying.
[0310] The silk proteins of the present invention may also be used in sutures (see, for example, US 2005/0055051). Such sutures can feature a braided jacket made of ultrahigh molecular weight fibers and silk fibers. The polyethylene provides strength. Polyester fibers may be woven with the high molecular weight polyethylene to provide improved tie down properties. The silk may be provided in a contrasting color to provide a trace for improved suture recognition and identification. Silk also is more tissue compliant than other fibers, allowing the ends to be cut close to the knot without concern for deleterious interaction between the ends of the suture and surrounding tissue. Handling properties of the high strength suture also can be enhanced using various materials to coat the suture. The suture advantageously has the strength of Ethibond No. 5 suture, yet has the diameter, feel and tie-ability of No. 2 suture. As a result, the suture is ideal for most orthopedic procedures such as rotator cuff repair, Achilles tendon repair, patellar tendon repair, ACL/PCL reconstruction, hip and shoulder reconstruction procedures, and replacement for suture used in or with suture anchors. The suture can be uncoated, or coated with wax (beeswax, petroleum wax, polyethylene wax, or others), silicone (Dow Corning silicone fluid 202A or others), silicone rubbers, PBA (polybutylate acid), ethyl cellulose (Filodel) or other coatings, to improve lubricity of the braid, knot security, or abrasion resistance, for example.
[0311] The silk proteins of the present invention may also be used in stents (see, for example, US 2004/0199241). For example, a stent graft is provided that includes an endoluminal stent and a graft, wherein the stent graft includes silk. The silk induces a response in a host who receives the stent graft, where the response can lead to enhanced adhesion between the silk stent graft and the host's tissue that is adjacent to the silk of the silk stent graft. The silk may be attached to the graft by any of various means, e.g., by interweaving the silk into the graft or by adhering the silk to the graft (e.g., by means of an adhesive or by means of suture). The silk may be in the form of a thread, a braid, a sheet, powder, etc. As for the location of the silk on the stent graft, the silk may be attached only the exterior of the stent, and/or the silk may be attached to distal regions of the stent graft, in order to assist in securing those distal regions to neighbouring tissue in the host. A wide variety of stent grafts may be utilized within the context of the present invention, depending on the site and nature of treatment desired. Stent grafts may be, for example, bifurcated or tube grafts, cylindrical or tapered, self-expandable or balloon-expandable, unibody or, modular, etc.
[0312] In addition to silk, the stent graft may contain a coating on some or all of the silk, where the coating degrades upon insertion of the stent graft into a host, the coating thereby delaying contact between the silk and the host. Suitable coatings include, without limitation, gelatin, degradable polyesters (e.g., PLGA, PLA, MePEG-PLGA, PLGA-PEG-PLGA, and copolymers and blends thereof), cellulose and cellulose derivatives (e.g., hydroxypropyl cellulose), polysaccharides (e.g., hyaluronic acid, dextran, dextran sulfate, chitosan), lipids, fatty acids, sugar esters, nucleic acid esters, polyanhydrides, polyorthoesters and polyvinylalcohol (PVA). The silk-containing stent grafts may contain a biologically active agent (drug), where the agent is released from the stent graft and then induces an enhanced cellular response (e.g., cellular or extracellular matrix deposition) and/or fibrotic response in a host into which the stent graft has been inserted.
[0313] The silk proteins of the present invention may also be used in a matrix for producing ligaments and tendons ex vivo (see, for example, US 2005/0089552). A silk-fiber-based matrix can be seeded with pluripotent cells, such as bone marrow stromal cells (BMSCs). The bioengineered ligament or tendon is advantageously characterized by a cellular orientation and/or matrix crimp pattern in the direction of applied mechanical forces, and also by the production of ligament and tendon specific markers including collagen type I, collagen type III, and fibronectin proteins along the axis of mechanical load produced by the mechanical forces or stimulation, if such forces are applied. In a preferred embodiment, the ligament or tendon is characterized by the presence of fiber bundles which are arranged into a helical organization. Some examples of ligaments or tendons that can be produced include anterior cruciate ligament, posterior cruciate ligament, rotator cuff tendons, medial collateral ligament of the elbow and knee, flexor tendons of the hand, lateral ligaments of the ankle and tendons and ligaments of the jaw or temporomandibular joint. Other tissues that may be produced by methods of the present invention include cartilage (both articular and meniscal), bone, muscle, skin and blood vessels.
[0314] The silk proteins of the present invention may also be used in hydrogels (see, for example, US 2005/0266992). Silk fibroin hydrogels can be characterized by an open pore structure which allows their use as tissue engineering scaffolds, substrate for cell culture, wound and burn dressing, soft tissue substitutes, bone filler, and as well as support for pharmaceutical or biologically active compounds.
[0315] The silk proteins may also be used in dermatological compositions (see, for example, US 2005/0019297). Furthermore, the silk proteins of the invention and derivatives thereof may also be used in sustained release compositions (see, for example, US 2004/0005363).
Textiles
[0316] The silk proteins of the present invention may also be applied to the surface of fibers for subsequent use in textiles. This provides a monolayer of the protein film on the fiber, resulting in a smooth finish. U.S. Pat. Nos. 6,416,558 and 5,232,611 describe the addition of a finishing coat to fibers. The methods described in these disclosures provide examples of the versatility of finishing the fiber to provide a good feel and a smooth surface. For this application, the fiber is coated with an effective amount of the silk protein. For the purpose of fiber coating for use in textiles, an effective amount of silk protein is herein defined as a proportion of from about 1 to about 99% by weight relative to the weight of the fiber material. The fiber materials include, but are not limited to textile fibers of cotton, polyesters such as rayon and Lycra.TM., nylon, wool, and other natural fibers including native silk. Compositions suitable for applying the silk protein onto the fiber may include co-solvents such as ethanol, isopropanol, hexafluoranols, isothiocyanouranates, and other polar solvents that can be mixed with water to form solutions or microemulsions. The silk protein-containing solution may be sprayed onto the fiber or the fiber may be dipped into the solution. While not necessary, flash drying of the coated material is preferred. An alternative protocol is to apply the silk protein composition onto woven fibers. An ideal embodiment of this application is the use of silk proteins to coat stretchable weaves such as used for stockings.
Composite Materials
[0317] Silk fibres can be added to polyurethane, other resins or thermoplastic fillers to prepare panel boards and other construction material or as moulded furniture and benchtops that replace wood and particle board. The composites can be also be used in building and automotive construction especially rooftops and door panels. The silk fibres re-enforce the resin making the material much stronger and allowing lighterweight construction which is of equal or superior strength to other particle boards and composite materials. Silk fibres may be isolated and added to a synthetic composite-forming resin or be used in combination with plant-derived proteins, starch and oils to produce a biologically-based composite materials. Processes for the production of such materials are described in JP 2004284246, US 2005175825, U.S. Pat. No. 4,515,737, JP 47020312 and WO 2005/017004.
Paper Additives
[0318] The fibre properties of the silk of the invention can add strength and quality texture to paper making. Silk papers are made by mottling silk threads in cotton pulp to prepare extra smooth handmade papers is used for gift wrapping, notebook covers, carry bags. Processes for production of paper products which can include silk proteins of the invention are generally described in JP 2000139755.
Advanced Materials
[0319] Silks of the invention have considerable toughness and stands out among other silks in maintaining these properties when wet (Hepburn et al., 1979).
[0320] Areas of substantial growth in the clothing textile industry are the technical and intelligent textiles. There is a rising demand for healthy, high value functional, environmentally friendly and personalized textile products. Fibers, such as those of the invention, that do not change properties when wet and in particular maintain their strength and extensibility are useful for functional clothing for sports and leisure wear as well as work wear and protective clothing.
[0321] Developments in the weapons and surveillance technologies are prompting innovations in individual protection equipments and battle-field related systems and structures. Besides conventional requirements such as material durability to prolonged exposure, heavy wear and protection from external environment, silk textiles of the invention can be processed to resist ballistic projectiles, fire and chemicals. Processes for the production of such materials are described in WO 2005/045122 and US 2005268443.
EXAMPLES
Example 1--Preparation and Analysis of Late Last Instar Salivary Gland cDNAs
[0322] The proteins that are found in euaculeatan and neuropteran (Apis mellifera, Bombus terrestris, Myrmecia forficata, Oecophylla smaragdina, Mallada signata) silks were identified by matching ion trap consecutive mass spectral (MS/MS) fragmentation patterns of peptides obtained by trypsin digestion of the silk with the predicted mass spectral data of proteins encoded by cDNAs isolated from the salivary gland of late final instar larvae. For confirmation that no proteins were missed by this analysis for the honeybee, the peptide mass spectral data were also compared to virtual tryptic digests of Apis mellifera proteins predicted by the bee genome project and translations of the Amel3 honeybee genomic sequences in all six reading frames.
Honeybee
[0323] Apis mellifera larvae were obtained from domestic hives. Previously it was shown that silk production in Apis mellifera is confined to the salivary gland during the latter half of the final instar (Silva-Zacarin et al., 2003). During this period, RNA is more abundant in the posterior end of the gland (Flower and Kenchington, 1967). The cubical cell regions of 50 salivary glands were dissected from late fifth instar Apis mellifera immersed in phosphate buffered saline. The posterior end of the dissected gland was immediately placed into RNAlater.RTM. (Ambion, Austin, Tex., USA), to stabilise the mRNA, and subsequently stored at 4.degree. C.
[0324] Total RNA (35m) was isolated from the late final instar salivary glands using the RNAqueous for PCR kit from Ambion (Austin, Tex., USA). Message RNA was isolated from the total RNA using the Micro-FastTrack.TM. 2.0 mRNA Isolation kit from Invitrogen (Calsbad, Calif., USA) according to the manufacturer's directions with the isolated mRNA being eluted into 10 ul RNAse free water.
[0325] A cDNA library was constructed from the mRNA isolated from Apis mellifera larvae using the CloneMiner.TM. cDNA library construction kit of Invitrogen (Calsbad, Calif., USA) with the following modifications from the standard protocol: For the first strand synthesis, 0.5 .mu.l of Biotin-attB2-Oligo(dT) primer at 6 pmol.mu.l.sup.-1 and 0.5 .mu.l of dNTPs at 2 mM each was added to the 10 .mu.l mRNA. After incubation at 65.degree. C. for 5 min then 45.degree. C. for 2 min, 2 .mu.l 5.times. First strand buffer, 1 .mu.l of 0.1M DTT, and 0.5 .mu.l SuperScript.TM. II RT at 200 U.mu.l.sup.-1 were added. For second strand synthesis, the total volume of all reagents was halved and after ethanol precipitation, the cDNA was resuspended in 5 .mu.l of DEPC-treated water. The aatB1 adapter (1 .mu.l) was ligated in a total volume of 10 .mu.l to the 5 ul cDNA with 2 .mu.l 5.times. Adapter buffer, 1 ul 0.1M DTT and 1 .mu.l T4 DNA ligase (1 U.mu.l.sup.-1) at 16.degree. C. for 48 hrs with an additional 0.5 .mu.l T4 DNA ligase (1 U.mu.l.sup.-1) added after 16 hrs. The cDNA was size fractionated according, to the manufactures instructions with samples eluting between 300-500 .mu.l being precipitated with ethanol, resuspended and transformed into the provided E. coli DH10B.TM. T1 phage resistant cells as recommended. The cDNA library comprised approximately 1,200,000 colony forming units (cfu) with approximately 1% the original vector. The average insert size was 1.3.+-.1.4 kbp.
[0326] Eighty two clones were randomly selected and sequenced using the GenomeLab.TM. DTCS Quick start kit (BeckmanCoulter, Fullerton Calif. USA) and run on a CEQ8000 Biorad sequencer. These clustered into fifty four groups (Table 2). Identification of the cDNAs that encoded the silk proteins is described below.
Other Species
[0327] Total RNA was isolated from 4 bumblebee (Bombus terrestris) (2 .mu.g RNA), 4 bulldog ant (Myrmecia forficata) (3 .mu.g RNA), approximately 100 Weaver ants (Oecophylla smaragdina) (0.4 .mu.g RNA) and approximately 50 green lacewing (Mallada signata) late larval labial glands using the RNAqueous for PCR kit from Ambion (Austin, Tex., USA). mRNA was isolated from the total RNA using the Micro-FastTrack.TM. 2.0 mRNA Isolation kit from Invitrogen (Calsbad, Calif., USA) into a final volume of 10 .mu.l water. cDNA libraries were constructed from the mRNA using the CloneMiner.TM. cDNA kit of Invitrogen (Calsbad, Calif., USA) with the following modifications from the standard protocol: For the first strand synthesis, 3 pmol of Biotin-attB2-Oligo(dT) primer and 1 nmol each dNTPs were added to the 10 .mu.l mRNA. After 5 min at 65.degree. C. followed by 2 min at 45.degree. C., 2 .mu.l 5.times. First strand buffer, 50 nmol DTT, and 100 U SuperScript.TM. II RT were added.
TABLE-US-00002 TABLE 2 A. mellifera final instar salivary gland cDNAs and MS ion trap fragmentation patterns of peptides from trypsin digestion of SDS treated brood comb silk. Number of Distinct Abundance tryptic summed Coverage Number of in salivary Protein or peptides MS/MS of protein cDNA's gland gene identified search sequence Protein in cluster library (%) synonyms in the silk score (% protein) identification Proteins identified in cDNA library and in honeybee silk 10 13 Xenosin; 9 143.89 25 AC004701 GB15233-PA 8 11 Xenospiral; 10 165.13 37 No matches GB12184-PA 6 7 Xenospira4; 8 142.16 35 No matches GB19585-PA 6 7 Xenospira2; 9 145.91 28 No matches GB12348-PA 5 6 Xenospira3; 9 147.02 31 No matches GB17818-PA Proteins identified in cDNA library only 4 4 GB14261-PA 0 2 2 Contig 2504 0 2 2 GB17108-PA 0 1 1 Contig 68 0 1 1 Contig 110 0 1 1 Contig 487 0 1 1 GB14199-PA 0 1 1 GB10847-PA 0 1 1 Contig 1047 0 1 1 GB17558-PA 0 1 1 Contig 1471 0 1 1 GB16480-PA 0 1 1 Contig 1818 0 1 1 GB16911-PA 0 1 1 Contig 2046 0 1 1 Contig 2136 0 1 1 Contig 2196 0 1 1 GB11234-PA 0 1 1 GB11199-PA 0 1 1 GB18183-PA 0 1 1 Contig 2938 0 1 1 Contig 2976 0 1 1 Contig 3263 0 1 1 Contig 3527 0 1 1 GB16412-PA 0 1 1 GB18750-PA 0 1 1 GB16132-PA 0 1 1 Contig 4536 0 1 1 GB19431-PA 0 1 1 Contig 4704 0 1 1 Contig 4758 0 1 1 Contig 4830 0 1 1 Contig 4968 0 1 1 Contig 5402 0 1 1 Contig 5971 0 1 1 GB11274-PA 0 1 1 GB14693-PA 0 1 1 GB19585-PA 0 1 1 GB15606-PA 0 1 1 GB16801-PA 0 1 1 GB12085-PA 0 1 1 Contig 7704 0 1 1 Contig 8630 0 1 1 Contig 9774 0 1 1 GB16452-PA 0 1 1 GB10420-PA 0 1 1 GB14724-PA 0
[0328] For second strand synthesis, the total volume of all reagents was halved from the manufacturer's recommended amounts and after ethanol precipitation, the cDNA was resuspended in 5 .mu.l of DEPC-treated water. The aatB1 adapter (1 .mu.l) was ligated in a total volume of 10 .mu.l to the 5 .mu.l cDNA with 2 .mu.l 5.times. Adapter buffer, 50 nmol DTT and 1 U T4 DNA ligase at 16.degree. C. for 12 hrs. The cDNA libraries comprised approximately 2.4.times.10.sup.7 (bumblebee), 5.0.times.10.sup.7 (bulldog ant) and 6000 (green ant) colony forming units (cfu) with less than 1% the original vector for the bulldog ant and bumblebee libraries and greater than 80% original vector in the green ant library. The average insert size within the libraries was 1.3 Kbp.
[0329] Sequence data was obtained from more than 100 random clones from the cDNA libraries from bumblebee and bulldog ant, 82 clones from the honeybee and 60 clones from the lacewing. The technical difficulties of obtaining salivary glands from the minute green ants (approximately 1 mm in length) reduced the efficiency of the library from this species and as such only 40 sequences were examined. A summary of the silk proteins identified is provided in Table 3.
TABLE-US-00003 TABLE 3 Identification and properties of the euaculeatan silk proteins. Length of % Distinct MARCOIL protein cDNA summed MS/MS predicted coiled Protein (amino library identification % helical coil length*** Species name acids) clones .cndot. score structure** (amino acids) Honeybee AmelF1* 333 6 52 76 117 Honeybee AmelF2* 290 7 51 88 175 Honeybee AmelF3* 335 11 107 81 154 Honeybee AmelF4* 342 7 88 76 174 Honeybee AmelSA1* 578 13 40 41 45 Bumblebee BBF1 327 4 180 86 147 Bumblebee BBF2 313 14 100 84 199 Bumblebee BBF3 332 20 218 86 146 Bumblebee BBF4 357 32 137 80 188 Bumblebee BBSA1 >501 3 138 21 0 Bulldog ant BAF1 422 16 99 69 121 Bulldog ant BAF2 411 30 90 76 132 Bulldog ant BAF3 394 26 88 79 131 Bulldog ant BAF4 441 24 116 76 157 Weaver ant GAF1 391 35 228 74 177 Weaver ant GAF2 400 22 191 79 158 Weaver ant GAF3 395 13 156 72 103 Weaver ant GAF4 443 17 148 74 166 Lacewing MalF1 596 23 45 89 151 *also referred to herein as Xenospiral-4 and Xenosin respectively, **predicted by PROFsec, ***predicted by MARCOIL at 90% threshold
Example 2--Preparation and Proteomic Analysis of Native Silk
[0330] Honeybee brood comb after the removal of larvae, bumblebee cocoons after the removal of larvae, bulldog ant cocoons after the removal of larvae, or weaver ant silk sheets were washed extensively three times in warm water to remove water soluble contaminants and then washed extensively three times in chloroform to remove wax. Chloroform was removed by rinsing in distilled water and a subset of this silk was retained for analysis. A subset of the Hymenopteran (ants and bees) silk samples was further washed by boiling for 30 minutes in 0.05% sodium carbonate solution, a standard procedure for degumming silkworm silk, then rinsed in distilled water. Lacewing silk was rinsed in distilled water only. A subset of the lacewing silk samples was degummed by boiling for 30 minutes in 0.05% sodium carbonate solution.
[0331] A subset of the honeybee material was soaked overnight in 2% SDS at 95.degree. C., followed by three washes in distilled water. Extraction in hot SDS solution solubilises most proteins, but in this case the silk sheets retained their conformation.
[0332] The clean silks were analysed by liquid chromatography followed by tandem mass spectrometry (LCMS) as described below.
[0333] Pieces of cleaned silk were placed in a well of a Millipore `zipplate`, a 96 well microtitre tray containing a plug of C18 reversed phase chromatography medium through the bottom of each well to which was added 20 .mu.l 25 mM ammonium bicarbonate containing 160 ng of sequencing grade trypsin (Promega). Then the tray was incubated overnight in a humidified plastic bag at 30.degree. C.
[0334] The C18 material was wetted by pipetting acetonitrile (10 .mu.l) to the sides of each well and incubating the plate at 37.degree. C. for 15 min. Formic acid solution (130 .mu.l, 1% v/v) was added to each well and after 30 min peptides from the digested bee proteins were captured on the C18 material by slowly drawing the solutions from each well through the base of the plate under a reduced vacuum. The C18 material was washed twice by drawing through 100 .mu.l of formic acid solution. Peptides were eluted with 6 .mu.l of 1% formic acid in 70% methanol pipetted directly onto the C18 material and promptly centrifuged through the C18 plug to an underlying microtitre tray. This tray was placed under vacuum till the volume in each well was reduced about 2-fold by evaporation. Formic acid solution (10 .mu.l) was added to each well and the tray was transferred to the well plate sampler of an Agilent 1100 capillary liquid chromatography system.
[0335] Peptides (8 .mu.l) from the silk extract were bound to an Agilent Zorbax SB-C18 5 .mu.m 150.times.0.5 mm column with a flow rate of 0.1% formic acid/5% acetonitrile at 20 .mu.lmin.sup.-1 for one min then eluted with gradients of increasing acetonitrile concentration to 0.1% formic acid/20% acetonitrile over one minute at 5 .mu.lmin.sup.-1, then to 0.1% formic acid/50% acetonitrile over 28 minutes, then to 0.1% formic acid/95% acetonitrile over one minute. The column was washed with 0.1% formic acid/95%-100% acetonitrile over 5 mins at 20 .mu.lmin.sup.-1 and reequilibrated with 0.1% formic acid/5% acetonitrile for 7 mins before peptides from the next well were sampled.
[0336] Eluate from the column was introduced to an Agilent XCT ion trap mass spectrometer through the instrument's electrospray ion source fitted with a micronebuliser. Briefly, as peptides were eluting from the column, the ion trap collected full spectrum positive ion scans (100-2200 m/z) followed by two MS/MS scans of ions observed in the full spectrum avoiding the selection of ions that carried only a single charge. When an ion was selected for MS/MS analysis all others were excluded from the ion trap, the selected ion was fragmented according to the instrument's recommended "SmartFrag" and "Peptide Scan" settings. Once two fragmentation spectra were collected for any particular m/z value it was excluded from selection for analysis for a further 30 seconds to avoid collecting redundant data.
[0337] Mass spectral data sets from the entire experiment were analysed using Agilent's Spectrum Mill software to match the data with predictions of protein sequences from the cDNA libraries. The software generated scores for the quality of each match between experimentally observed sets of masses of fragments of peptides and the predictions of fragments that might be generated according to the sequences of proteins in a provided database. All the sequence matches reported here received scores greater than 20, the default setting for automatic, confident acceptance of valid matches.
[0338] This analysis identified that five proteins expressed at high levels in the labial gland matched the silk from each of the cognate bee species (shown in Tables 2 and 3) and four proteins expressed at high levels in the labial gland matched the silk from each of the cognate ant species (shown in Table 3). The abundance of message RNA encoding these proteins in the labial gland of the larvae was consistent with the proteins being abundantly produced (abundance of message shown in Table 3).
[0339] To ensure that none of the honeybee silk proteins were missed by this identification process, we also compared the honeybee silk trypsin peptide mass spectral data to a set of publicly available predicted protein sequences from the honeybee genome project, generated by a computer algorithm that tries to recognise transcribed genes in the complete genomic DNA sequences of the bee. Additionally, we generated a database of translations in the six possible reading frames of each contiguous genomic DNA sequence provided by the bee genome project (Amel3 release). These translated DNA sequences were presented to the Spectrum Mill software as if they were the sequences of very large proteins. Matching MS/MS peptide data identified open reading frames within the genomic sequences that had encoded parts of the isolated bee proteins without the need to first predict the organisation of genes. No additional proteins were identified in the silk by this analysis.
Example 3--Structural Analysis of the Native Silk
[0340] Native silk samples were prepared as described in Example 2. Silk samples were examined using a Bruker Tensor 37 Fourier transform infrared spectrometer with a Pike Miracle diamond attenuated total reflection accessory. Analysis of the amide I and II regions of the spectra of honeybee, bumblebee, green ant, bulldog ant silks and lacewing larval silk (FIG. 1) shows that all these silks have a predominantly alpha-helical secondary structure. The silks of the Euaculeatan species have dominant peaks in the FT-IR spectra at 1645-1646 cm.sup.-1, shifted approximately 10 cm.sup.-1 lower than a classical .alpha.-helical signal and broadened. This shift in the .alpha.-helical signal is typical of coiled-coil proteins (Heimburg et al., 1999). Spectra from samples that were degummed were unchanged.
Example 4--the Amino Acid Composition of Native Silks Closely Resembles that of the Identified Silk Proteins
[0341] The amino acid composition of the native silks was determined after 24 hr gas phase hydrolysis at 110.degree. C. using the Waters AccQTag chemistry by Australian Proteome Analysis Facility Ltd (Macquarie University, Sydney).
[0342] The measured amino acid composition of the SDS washed silk was similar to that predicted from the identified silks protein sequences (FIGS. 2 and 3).
Example 5--Structural Analysis of the Silk Proteins
Predicted Secretory Peptides
[0343] As expected for silk proteins, the SignalP 3.0 signal prediction program (Bendtsen et al., 2004), which uses two models to identify signal peptides predicted that all the identified silk genes encoded proteins which contain signal peptides that targeted them for secretion from a cell (data not shown). The predicted cleavage sites of the polypeptides are as follows:
[0344] Xenospira1 (AmelF1)--between pos 19 and 20 (ASA-GL),
[0345] Xenospira2 (AmelF2)--between pos 19 and 20 (AEG-RV),
[0346] Xenospira3 (AmelF3)--between pos 19 and 20 (VHA-GV),
[0347] Xenospira4 (AmelF4)--between pos 19 and 20 (ASG-AR),
[0348] Xenosin (AmelSA1)--between pos 19 and 20 (VCA-GV),
[0349] BBF1--between pos 19 and 20 (ASA-GQ),
[0350] BBF2--between pos 20 and 21 (AEG-HV),
[0351] BBF3--between pos 19 and 20 (VHA-GS),
[0352] BBF4--between pos 19 and 20 (ASA-GK),
[0353] BAF1--between pos 19 and 20 (ASA-SG),
[0354] BAF2--between pos 19 and 20 (ASG-RV),
[0355] BAF3--between pos 19 and 20 (ASG-NL),
[0356] BAF4--between pos 19 and 20 (VGA-SE),
[0357] GAF1--between pos 19 and 20 (ADA-SK),
[0358] GAF2--between pos 19 and 20 (ASG-GV),
[0359] GAF3--between pos 19 and 20 (ASG-GV),
[0360] GAF4--between pos 19 and 20 (VGA-SE),
[0361] MalF1--between pos 26 and 27 (SST-AV).
All Four of the Ant and Four of the Five Bee Silk Proteins are Helical and Formed Coiled Coils
[0362] Protein modelling and results from pattern recognition algorithms confirmed that the majority of the identified honeybee silk proteins were helical proteins that formed coiled coils.
[0363] PROFsec (Rost and Sander, 1993) and NNPredict (McClelland and Rumelhart, 1988; Kneller et al., 1990), algorithms were used to investigate the secondary structure of the identified silk genes. These algorithms identified Xenospira1 [GB12184-PA] (SEQ ID NO:1), Xenospira2 [GB12348-PA] (SEQ ID NO:3), Xenospira3 [GB17818-PA] (SEQ ID NO:5), and Xenospria4 [GB19585-PA] (SEQ ID NO:7), as highly helical proteins, with between 76-85% helical structure (Table 4). Xenosin [GB15233-PA] (SEQ ID NO:10) had significantly less helical structure.
TABLE-US-00004 TABLE 4 The secondary structure of Apis mellifera silk proteins predicted by PROFsec (Rost and Sander, 1993) showing percentages of helices, extended sheets and loops. helical extended loop Protein PROFsec NNPredict PROFsec NNPredict PROFsec NNPredict Xenospira3 77 70 3 6 20 27 Xenospira4 85 82 2 6 14 16 Xenospira1 80 73 1 4 19 26 Xenospira2 77 69 2 5 21 29 Xenosin 41 41 8 9 51 50
[0364] Further protein modelling and results from pattern recognition algorithms confirmed that the majority of the identified silk proteins were helical proteins that formed coiled coils. PredictProtein (Rost et al., 2004) algorithms were used to investigate the secondary structure of the identified silk genes. These algorithms identified Xenospira1 (SEQ ID NO:1), Xenospira2 (SEQ ID NO:3), Xenospira3 (SEQ ID NO:5), Xenospira4 (SEQ ID NO:7), BBF1 (SEQ ID NO:22), BBF2 (SEQ ID NO:24), BBF3 (SEQ ID NO:26), BBF4 (SEQ ID NO:28), BAF1 (SEQ ID NO:40), BAF2 (SEQ ID NO:42), BAF3 (SEQ ID NO:44), BAF4 (SEQ ID NO:46), GAF1 (SEQ ID NO:56), GAF2 (SEQ ID NO:58), GAF3 (SEQ ID NO:60), GAF4 (SEQ ID NO:62), and MalF1 (SEQ ID NO:72) as highly helical proteins, with between 69-88% helical structure (Table 3). AmelSA1 [GB15233-PA] (Xenosin) (SEQ ID NO:10) and BBSA1 (SEQ ID NO:30) had significantly less helical structure.
[0365] Super-coiling of helical proteins (coiled coils) arises from a characteristic heptad repeat sequence normally denoted as (abcdefg).sub.n with generally hydrophobic residues in position a and d, and generally charged or polar residues at the remaining positions. The pattern recognition programs (MARCOIL (Delorenzi and Speed, 2002), COILS (Lupas et al., 1991)) identified numerous heptad repeats typical of coiled-coils in Xenospira1 [GB12184-PA] (SEQ ID NO:1), Xenospira2 [GB12348-PA] (SEQ ID NO:3), Xenospira3 [GB17818-PA] (SEQ ID NO:5), and Xenospira4 [GB19585-PA] (SEQ ID NO:7) (MARCOIL: Table 5; COILS: FIG. 4), as well as BBF1 (SEQ ID NO:22), BBF2 (SEQ ID NO:24), BBF3 (SEQ ID NO:26), BBF4 (SEQ ID NO:28), BAF1 (SEQ ID NO:40), BAF2 (SEQ ID NO:42), BAF3 (SEQ ID NO:44), BAF4 (SEQ ID NO:46), GAF1 (SEQ ID NO:56), GAF2 (SEQ ID NO:58), GAF3 (SEQ ID NO:60), GAF4 (SEQ ID NO:62), and MalF1 (SEQ ID NO:72) (MARCOIL: Table 3).
Identification of a Novel Coiled Coil Sequence in the Honeybee Silk Proteins
[0366] The heptad repeats of amino acid residues identified in the sequences of Xenospira1 [GB 12184-PA], Xenospira2 [GB12348-PA], Xenospira3 [GB17818-PA], Xenospria4 [GB19585-PA], were each highly indicative of a coiled coil secondary structure (FIG. 5) (see Table 5 for confidence levels). The fact that the heptads are found consecutively and numerously suggests the proteins adopt a very regular structure. Overlapping heptads were identified in two of the honeybee proteins: the major coiled coil region of Xenospira1 contained overlapping heptads with a 3 residue offset followed by a space of 5 residues and then four consecutive heptads; and the entire coiled coil region of Xenospira2 had multiple overlapping heptads with a single offset and 4 residue offset (equivalent to 3 residue offset). The composition of amino acids in the various positions of the major heptad are shown in the first column in Table 6, with the positions of the overlapping heptads indicated in adjacent columns.
TABLE-US-00005 TABLE 5 Percent of residues in the identified silk proteins predicted to exist as coiled coil by the MARCOIL (Delorenzi and Speed, 2002) pattern recognition algorithm. Length of mature Percent protein protein that exists as coiled coil (amino 50% 90% 99% Protein acids) threshold threshold threshold Xenospira3 315 64% 34% 20% (residues (residues (residues 68 to 268) 128-223 and 149-211) 235-246) Xenospira4 290 73% 60% 27% (residues (residues (residues 83-293) 98-168 and 113-154 and 182-285) 212-247) Xenospira1 316 69% 49% 18% (residues (residues (residues 67-282) 103-256) 113-169) Xenospira2 328 65% 54% 45% (residues (residues (residues 89-298) 110-283) 127-270) Xenosin 350 26% 9% 2% (residues (residues (residues 32-127) 42-75) 59-67)
[0367] Surprisingly the major heptads have a novel composition when viewed collectively--with an unusually high abundance of alanine in the `hydrophobic` heptad positions a and d (see Table 6 and FIG. 5). Additionally, a high proportion of heptads have alanine at both a and d positions within the same heptad (33% in Xenospira1 [GB12184-PA]; 36% in Xenospira2 [GB12348-PA]; 27% in Xenospira3 [GB17818-PA]; and 38% in Xenospira4 [GB19585-PA]; see Tables 6 and 7).
TABLE-US-00006 TABLE 6 Summary of the number of each amino acid residues in the various heptad positions in coiled coil regions of honeybee silk proteins. A I R L K T E V F S Q N D G M Y W Total Xenospira4 a 23 0 1 1 0 1 1 1 0 1 0 0 0 0 0 0 0 29 b 12 0 0 2 2 2 3 1 0 3 1 1 1 1 0 0 0 29 c 12 0 0 1 5 1 3 1 0 3 1 1 0 1 0 0 0 29 d 17 0 0 5 1 0 1 2 0 2 1 0 0 0 0 0 0 29 e 12 0 1 0 0 2 4 2 0 5 2 1 0 0 0 0 0 29 f 13 1 0 1 2 0 7 1 0 1 1 2 0 0 0 0 0 29 g 9 3 4 0 2 1 2 1 0 2 0 1 2 2 0 0 0 29 Xenospira3 a 19 0 0 1 0 4 2 0 0 1 1 1 0 0 0 1 0 30 b 8 0 0 5 1 2 2 0 0 5 4 2 1 0 0 0 0 30 c 13 0 1 0 3 2 2 3 0 1 2 0 1 1 0 0 1 30 d 13 3 0 2 2 0 2 2 0 4 0 1 1 0 0 0 0 30 e 8 0 0 2 2 2 4 0 0 7 4 0 0 1 0 0 0 30 f 7 0 2 3 4 2 4 0 0 4 1 2 1 0 0 0 0 30 g 9 0 5 2 3 0 1 2 0 5 0 2 1 0 0 0 0 30 Xenospira2 a 20 0 0 1 0 3 1 1 1 1 0 0 0 0 0 0 0 28 b 7 2 2 2 2 2 2 4 0 1 1 3 0 0 0 0 0 28 c 9 0 2 0 4 1 2 4 0 1 3 2 0 0 1 1 1 28 d 16 0 0 3 3 1 0 1 0 1 2 0 1 0 0 0 0 28 e 11 0 1 3 0 3 4 1 0 2 2 0 1 0 0 0 0 28 f 10 2 1 0 1 2 6 1 0 3 1 1 0 0 0 0 0 28 g 8 4 1 0 1 1 5 0 0 0 2 4 0 1 1 0 0 28 Xenospira1 a 13 3 0 1 2 0 1 1 0 2 1 1 0 2 0 0 0 27 b 7 1 1 1 6 0 2 1 0 3 1 0 4 0 0 0 0 27 c 8 1 2 1 1 1 7 2 0 1 1 1 0 1 0 0 0 27 d 18 0 0 2 1 2 1 0 2 1 0 0 0 0 0 0 27 e 11 1 2 1 1 2 3 2 0 4 0 0 0 0 0 0 0 27 f 7 0 3 0 2 1 3 3 0 7 0 1 0 0 0 0 0 27 g 13 0 0 3 3 0 2 1 0 3 1 0 0 0 1 0 0 27
TABLE-US-00007 TABLE 7 Summary of alanine residues in heptads of honeybee silk proteins. Amount of Amount of Amount of Ala in Ala in Amount of Ala Amount of Number protein Amount of position a position d in position a helical of major in major Ala in major of major of major and d of major Protein structure (%).sup.1 heptads heptad (%) heptads (%) heptads (%) heptads (%) heptads (%) Xenospira1 77 (70) 27 41 44 74 33 Xenospira2 85 (82) 28 37 71 57 36 Xenospira3 80 (73) 30 37 63 43 27 Xenospira4 77 (69) 29 48 79 58 38 Xenosin 41 (41) n/a n/a n/a n/a .sup.1PROFsec predictions with NNPredict predictions shown in brackets.
[0368] The composition of amino acids in the various heptad positions in the coiled coil region of the hymenopteran silks are summarised in FIGS. 6 and 7. As noted above, the positions within the heptads have a novel composition--the `hydrophobic` heptad positions a and d of the bee and ant silks contain very high levels of alanine (average 58%) and high levels of small polar residues (average 21%) in comparison to other coiled coils. Additionally, position e is unusually small and hydrophobic (Table 8, FIG. 7). Topographically this position is located adjacent to the a residues within the helices. Its compositional similarity with the a and d residues suggest that the silks adopt a coiled coil structure with three core residues per .alpha.-helix. Three residue cores contribute a larger hydrophobic interface than two residues in the core (Deng et al., 2006)--a feature that would assist coiled coil formation and stability.
[0369] In addition, when viewed collectively the positions b, c, e, f and g within the heptad are generally more hydrophobic, less polar and less charged than protein coiled coil regions previously characterised (see FIG. 7, and Tables 8 and 9). Therefore, although historically it was regarded that the helical content of the aculeate Hymenopteran silk was a consequence of a reduced glycine content and increased content of acidic residues (Rudall and Kenchington, 1971), we have discovered that it is not the glycine/acid residues that are responsible for the novel silk structure but rather the position of the alanine residues within the polypeptide chains.
TABLE-US-00008 TABLE 8 Average size and hydrophobicity at each heptad position of the orthologous hymenopteran silk proteins and of the green lacewing silk protein (MalF1) showing that a, d, and e positions (core) are smaller and more hydrophobic than other positions. In some cases the b position (partially submerged) is also small and hydrophobic. Heptad position a b c d e f g Amel F1 orthologs Average residue 0.36 0.20 0.20 0.30 0.26 -0.16 0.03 side chain hydrophobicity Average residue 1.7 2.5 2.5 2.1 2.3 3.0 2.6 side chain length Amel F2 orthologs Average residue 0.53 0.20 0.03 0.36 0.24 0.05 0.12 side chain hydrophobicity Average residue 1.5 2.6 2.6 2.0 2.2 2.5 3.0 side chain length Amel F3 orthologs Average residue 0.44 0.36 0.06 0.41 0.27 -0.10 0.00 side chain hydrophobicity Average residue 1.9 2.3 2.4 2.1 2.3 2.8 2.8 side chain length Amel F4 orthologs Average residue 0.46 0.17 -0.13 0.61 0.04 0.06 0.06 side chain hydrophobicity Average residue 1.4 2.2 2.6 2.04 2.3 2.6 2.7 side chain length MalF1 Average residue -0.05 0.14 -0.61 0.27 0.59 0.23 -0.22 side chain hydrophobicity Average residue 2.1 1.7 2.5 1.4 1.5 1.7 3.5 side chain length
Example 6--the Bee Silk Proteins are Likely to be Extensively Cross-Linked
[0370] The bee silk proteins all contain a high proportion of lysine (6.5%-16.3%). A comparison between the measured amino acid composition of bee silk and the sequences of the identified silk proteins reveals a substantial mismatch in the number of lysine residues, with much less lysine detected in the silk than expected (FIGS. 2 and 3). This suggests that lysine residues in the silk have been modified, so are not being identified by standard amino acid analysis. Lysine is known to form a variety of cross-links: either enzymatic cross links catalysed by lysyl oxidase or nonenzymatic cross links generated from glycated lysine residues (Reiser et al., 1992). The under-representation of lysine in the honeybee and bumblebee silk amino acid analysis is consistent with the presence of lysine cross-linking
TABLE-US-00009 TABLE 9 Number of residues in each class of amino acids at various heptad positions in coiled coil regions of silk proteins. Heptad Nonpolar Polar Charged Small Medium Large position Xenospira4 25 2 2 26 2 1 a 16 7 6 19 10 0 b 15 6 8 18 11 0 c 24 3 2 21 8 0 d 14 10 5 21 7 1 e 16 4 9 15 14 0 f 15 4 10 15 10 4 g Xenospira3 20 8 2 24 5 1 a 13 13 4 15 15 0 b 17 6 7 20 8 2 c 20 5 5 19 11 0 d 11 13 6 18 12 0 e 10 9 11 13 15 2 f 13 7 10 16 9 5 g Xenospira2 23 4 1 25 2 1 a 15 7 6 14 12 2 b 13 7 8 15 11 2 c 20 4 4 19 9 0 d 15 7 6 17 10 1 e 13 7 8 16 11 1 f 14 7 7 10 17 1 g Xenospira1 20 4 3 18 9 0 a 10 4 13 11 15 1 b 13 4 10 13 12 2 c 20 5 2 22 5 0 d 15 6 6 19 6 2 e 10 9 8 18 6 3 f 18 4 5 17 10 0 g
[0371] Covalently cross-linked proteins subjected to SDS polyacrylamide gel electrophoresis (PAGE) are expected to migrate according to the molecular weight of the cross-linked complex. We subjected late last instar honeybee labial gland proteins to SDS PAGE and measured the migration of the silk proteins in relation to standard protein markers. Bands were observed corresponding to monomers of each of the identified silk proteins, however higher molecular weight bands containing these proteins were also present, as expected in a cross-linked system (FIG. 8).
[0372] As described above, the honeybee labial gland contains a mixture of organised and disorganised silk proteins. The cross-linked proteins observed probably correspond to the protein population of the anterior region of the gland, where the silk is prepared for extrusion. It is reasonable to assume that extracellular honeybee silk contains a substantially higher proportion of cross-linked proteins than is observed in a heterogenous mixture of all stages of salivary gland silk proteins. The bonds are unlikely to be cysteine cross-links, as the silk was unaffected by reductive treatment, and the identified silk proteins contain few or no cysteine residues.
Example 7--the Euaculeatan Silk Proteins Differ Significantly from the Other Silk Proteins
[0373] The euaculeatan silk is significantly different from other described silk genes in relation to amino acid composition (Table 10), molecular weight of the proteins involved, secondary structure and physical properties (Tables 11 and 12). The lepidopteran silks are primarily composed of the small amino acid residues alanine, serine and glycine (for example the silk of Bombyx mori, Table 10) and are dominated by extended beta sheet secondary structure. The Cotesia glomerata silk protein is high in asparagine and serine--the abundance of the latter residue being characteristic of Lepidopteran silk sericins (glues) (Table 10). Modelling of the Cotesia glomerata silk protein does not identify helices or coiled coils in the secondary structure. In contrast, the bee, ant and lacewing silks are high in alanine (Table 10) and are comprised of a high level of helical secondary structure that forms coiled coils.
TABLE-US-00010 TABLE 10 Amino acid composition of silk from various Insects with most abundant residues shown in boldface. Euacu- Honey- leatan Mallada Cotesia Bombyx bee silk silk glomerata mori Alanine 22.6 27.5 26.9 12.5 29.3 Glutamic 16.1 13.9 7.4 0.6 0.9 acid + Glutamine Aspartic 13.2 8.6 15.0 37.6 1.2 acid + (Asn 33.7) Asparagine Serine 10.4 11.5 8.5 37.1 11.3 Leucine 9.0 7.2 5.9 0.4 0.4 Valine 6.6 4.8 4.1 0.3 2.1 Glycine 5.7 6.6 11.2 5.5 46.0 Isoleucine 5.6 4.0 3.9 0.4 0.6 Threonine 5.1 4.9 5.3 0.5 0.8 Lysine 3.7 3.7 3.2 0.1 0.3 Phenylalanine 2.0 1.0 0.5 0.5 0.6 Tyrosine 0 0.9 0.5 3.1 5.3 Proline 0 0 0 0.7 0.4 Histidine 0 0.5 0.5 0.4 0.2 Arginine 0 3.3 5.4 0.2 0.4 Methionine 0 1.0 1.6 0 0.1 Tryptophan 0 Not Not Not 0.2 reported reported reported Cysteine 0 0.4 0.3 Not 0.1 reported
TABLE-US-00011 TABLE 11 Differences between insect silks. Ant Lepidoptera and bee Mallada Cotesia For example silk silk sp. Bombyx mori Most Ala Ala Ser, Asn Gly, Ala abundant amino acids Size of 25-35 57 Approx >100 KDa fibroin kDa KDa 500 KDa proteins Secondary Coiled Coiled Most likely beta-pleated structure coil coil beta sheets. sheets Secondary loosely structure associated prediction with beta- programs sheets, PROFsec and beta- MARCOIL spirals, do not alpha recognise helices and any helical amorphous structure or regions coiled coil regions.
TABLE-US-00012 TABLE 12 Solubility of insect silks. Ant and bee silk Mallada silk Cotesia sp. Bombyx mori Solvent 20.degree. C. 95.degree. C. 20.degree. C. 95.degree. C. 20.degree. C. 95.degree. C. 20.degree. C. 95.degree. C. LiBr 54% -- -- -- -- -- part -- LiSCN saturated -- -- -- -- -- part -- 8M urea -- -- -- -- -- -- -- part 6M guanidine HCl -- -- -- -- -- -- -- part 1M NaOH -- part ? ? -- part part 6M HCl -- part part -- part -- 3M HCl/50% -- part ? ? -- part part propanoic acid
[0374] Cladistic analysis of the coiled coil regions of the silk proteins of the four Hymenopteran species (FIG. 9) suggests that the genes evolved in a common ancestor that predates the divergence of the Euaculeata from the parasitic wasps. The sequences of the silk have diverged extensively and we were only able to align the 210 amino acids that comprise the coiled coil region of each protein. The amino acid sequence identity between the coiled coil regions of each of the silk proteins provided herein is shown in Table 13 and DNA identity in the corresponding region is shown in Table 14. Whilst the proteins have similar amino acid contents (especially high levels of alanine) and tertiary structure, the primary amino acid sequence identity is very low. In fact, the gene encoding the Mallada silk protein has evolved independently and as such the silk protein sequence cannot be aligned to the Hymenopteran sequences. This indicates that considerable variety in the identity of the amino acids can occur, whilst not affecting the biological function of the proteins.
[0375] The cladistic analysis predicts that silk of euaculeatan wasps is comprised of related proteins to the silk of ants and bees and that although these proteins will have similar composition and architecture to the proteins described here, they will have highly diverged primary sequence.
[0376] The amino acid sequences of the silk proteins provided herein (FIG. 10) were subjected to comparisons with protein databases, however, no prior art proteins were identified with any reasonable level of sequence identity (for example, none greater than 30% identical over the length of the silk protein sequence).
TABLE-US-00013 TABLE 13 Percent identity between protein sequences of the coiled coil region of the fibre proteins in ants and bees. Honeybee Bumblebee Bulldog ant Green ant F1 F2 F3 F4 F1 F2 F3 F4 F1 F2 F3 F4 F1 F2 F3 F4 beeF1 100 beeF2 26.7 100 beeF3 23.3 31.4 100 beeF4 34.8 32.4 30.0 100 BBF1 65.7 28.1 24.8 35.7 100 BBF2 28.6 71.4 28.6 31.9 31.0 100 BBF3 25.2 31.0 65.7 27.6 27.1 29.5 100 BBF4 33.3 31.0 29.5 64.8 34.8 31.4 28.1 100 BAF1 37.1 20.0 20.0 32.4 39.5 21.4 21.4 29.1 100 BAF2 25.2 44.3 29.5 33.8 28.1 38.1 28.6 27.6 27.1 100 BAF3 23.8 26.2 36.7 28.1 24.8 25.2 36.7 28.1 21.0 27.6 100 BAF4 28.1 33.8 24.8 45.2 28.6 33.8 23.3 43.8 26.1 27.6 25.2 100 GAF1 33.8 20.0 23.8 32.9 36.2 22.9 23.8 29.1 66.7 28.1 25.2 28.6 100 GAF2 24.8 41.9 27.6 29.5 28.1 39.5 29.0 26.7 21.9 66.2 23.8 26.7 23.8 100 GAF3 26.9 28.8 40.1 31.6 25.5 28.3 38.2 30.2 24.0 28.3 62.7 27.4 27.4 26.4 100 GAF4 24.7 32.4 24.3 37.6 27.1 32.4 24.8 38.1 23.9 29.5 21.0 63.3 24.8 27.6 24.1 100
TABLE-US-00014 TABLE 14 Percent identity between nucleotide sequences encoding coiled coil region of the fibre proteins in ants and bees. Honeybee Bumblebee Bulldog ant Green ant F1 F2 F3 F4 F1 F2 F3 F4 F1 F2 F3 F4 F1 F2 F3 F4 beeF1 100 beeF2 39.4 100 beeF3 37.0 40.2 100 beeF4 45.1 44.8 41.0 100 BBF1 68.9 40.9 37.5 45.2 100 BBF2 42.5 72.9 42.5 44.9 42.2 100 BBF3 40.6 40.0 67.6 40.5 38.4 41.0 100 BBF4 45.4 41.0 41.7 66.0 45.9 43.6 40.0 100 BAF1 45.7 35.1 35.9 41.1 47.9 36.5 36.0 38.7 100 BAF2 38.1 49.8 41.4 44.6 38.7 47.3 40.0 41.0 40.6 100 BAF3 33.3 36.7 45.4 40.3 36.3 36.8 46.2 39.4 36.0 40.5 100 BAF4 39.5 43.3 41.4 46.8 43.0 47.6 39.8 49.4 42.5 41.7 40.3 100 GAF1 45.6 35.1 37.3 42.4 47.6 38.5 37.8 41.4 68.9 41.7 36.7 43.0 100 GAF2 38.5 47.8 38.4 43.2 38.1 46.5 41.4 40.0 37.5 69.7 38.9 40.6 39.4 100 GAF3 39.0 40.1 46.1 41.8 37.7 39.3 46.1 40.0 37.7 41.7 65.1 41.2 40.0 41.7 100 GAF4 38.9 42.4 38.1 44.9 38.9 43.8 38.4 44.3 37.3 42.7 36.7 67.8 38.2 40.3 37.7 100
[0377] The open reading frames encoding the silk proteins (provided on FIG. 11) were subjected to similar database searching as that described above. The only related molecules that were identified have been published as part of the honeybee genome project (www.ncbi.nlm.nih.gov/genome/guide/bee). The open reading frames had been predicted by the bee genome project, however, the function of the encoded proteins had not been suggested. Furthermore, there is no evidence that a polynucleotide comprising the open reading frame of the mRNA had ever been produced for any of these molecules.
[0378] The genes encoding Xenospira1, Xenospira2, Xenospira3 and Xenospira4 comprise an exon covering the entire single open reading frame, whereas the gene encoding Xenosin comprises at least one intron (see FIG. 12).
Example 8--Expression of Silk Proteins in Transgenic Plants
[0379] A plant expression vector encoding a silk protein of the invention may consist of a recombinant nucleic acid molecule coding for said protein (for example a polynucleotide provided in any one of SEQ ID NO's:11 to 21, 31 to 39, 48 to 55, 64 to 71, 74 or 75) placed downstream of the CaMV 35S promoter in a binary vector backbone containing a kanamycin-resistance gene (NptII).
[0380] For the polynucleotides comprising any one of SEQ ID NO's 11, 13, 15, 17, 19, 31, 33, 35, 37, 48, 50, 52, 54, 64, 66, 68, 70 or 74 the construct further may comprise a signal peptide encoding region such as Arabidopsis thaliana vacuolar basic chitinase signal peptide, which is placed in-frame and upstream of the sequence encoding the silk protein.
[0381] The construct carrying a silk protein encoding polypeptide is transformed separately into Agrobacterium tumefaciens by electroporation prior to transformation into Arabidopsis thaliana. The hypocotyl method of transformation can be used to transform A. thaliana which can be selected for survival on selective media comprising kanamycin media. After roots are formed on the regenerates they are transferred to soil to establish primary transgenic plants.
[0382] Verification of the transformation process can be achieved via PCR screening. Incorporation and expression of polynucleotide can be measured using PCR, Southern blot analysis and/or LC/MS of trypsin-digested expressed proteins.
[0383] Two or more different silk protein encoding constructs can be provided in the same vector, or numerous different vectors can be transformed into the plant each encoding a different protein.
[0384] As an experimental example of plant expression, a codon-optimised version of AmelF4 (Xenospira4) (SEQ ID NO:76) was cloned into pET14b (Novagen), generating pET14b-6.times.His:F4op, forming an in-frame translational fusion with a 6.times.histidine at the N-terminal of the protein. The sequence encoding the protein "6.times.Histidine:F4op" was cloned into pVEC8 (Wang et al., 1992) under the control of the CaMV 35S promoter and ocs polyadenylation regulatory apparatus, generating pVEC8-35S-6.times.His:F4op-ocs. pET14b-6.times.His:F4op was transformed into chemically-competent E. coli and pVEC8-35S-6.times.His:F4op was transformed into tobacco leaf discs by Agrobacterium mediated transformation. Proteins from antibiotic resistant E. coli (induced expression) and tobacco leaves were isolated and subjected to western blot analysis using the Tetra-Histidine antibody (Qiagen, Karlsrule, Germany) for detection. The empty vectors pET14b and pVEC8-35S-ocs were used as negative controls in there respective host backgrounds. As shown in FIG. 13, these experiments resulted in the plant producing the Xenospira4 (AmelF4) protein.
Example 9--Fermentation and Purification of Silk Proteins
[0385] Expression constructs were constructed after the silk coding regions of honeybee genes AmelF1-F4 (Xenospira1 to 4 respectively) and lacewing MalF1 genes were amplified by PCR and cloned into pET14b expression vectors (Novagen, Madison, Wis.). The resultant expression plasmids were then electroporated into E. coli BL21 (DE3) Rosetta cells and grown overnight on LB agar containing ampicilin. A single colony was then used to inoculate LB broth containing ampicilin then grown at 37.degree. C. overnight. Cells were harvested by centrifugation and lysed with detergent (Bugbuster, Novagen). Inclusion bodies were washed extensively and re-solubilised in 6M guanidinium.
[0386] This procedure yielded proteins mixtures with greater than 95% purity of the honeybee proteins and greater than 50% purity of the lacewing MalF1 protein. Yields of up to 50% of the wet weight of the E. coli cell pellet were regularly obtained, indicating that the proteins are easy to express in this manner.
[0387] The solubilised honeybee recombinant proteins were applied to a Talon resin column prepared according to manufactures directions. They were then eluted off the column in 100 mM Tris.HCL, 150 mM imidazole pH 8.
Example 10--Processing of Silk Proteins into Threads
[0388] The honeybee and lacewing silk proteins have been readily made into threads using a variety of methods (see FIG. 14) using the following procedure.
[0389] The anterior segment of the salivary gland from late final instar Apis mellifera was dissected under phosphate buffered saline and removed to a flat surface in a droplet of buffer. Forceps were used to grasp either end of the segment. One end was raised out of the droplet and away from the other at a steady rate. This enabled the drawing of a fine thread that rapidly solidified in air.
[0390] The honeybee and lacewing larval recombinant silk proteins formed threads or sheets after dehydration or concentration. For example, by dropping soluble protein into a butanol solution or by concentrating proteins on the Talon resin column.
[0391] Threads were also obtained after honeybee or lacewing recombinant silk proteins were mixed with an organic solvent (such as hexane) to concentrate them at the interface in the correct conformation, and then addition of a reagent to exclude them from the interface (such as butanol). The threads formed by this procedure had similar FT-IR spectra to the native silk indicating that they were comprised of the same coiled coil structure.
[0392] Silk proteins from other species described herein can also be processed by this procedure.
[0393] It will be appreciated by persons skilled in the art that numerous variations and/or modifications may be made to the invention as shown in the specific embodiments without departing from the spirit or scope of the invention as broadly described. The present embodiments are, therefore, to be considered in all respects as illustrative and not restrictive.
[0394] All publications discussed above are incorporated herein in their entirety.
[0395] Any discussion of documents, acts, materials, devices, articles or the like which has been included in the present specification is solely for the purpose of providing a context for the present invention. It is not to be taken as an admission that any or all of these matters form part of the prior art base or were common general knowledge in the field relevant to the present invention as it existed before the priority date of each claim of this application.
REFERENCES
[0396] Atkins E. D. T. (1967) J Mol Biol 24:139-141. [0397] Bendtsen J. D., Nielsen H., von Heijne G. and Brunak S. (2004) J. Mol. Biol. 340:783-795. [0398] Bini E., Knight D. P. and Kaplan D. L. (2004) J. Mol. Biol. 335:27-40. [0399] Craig C. L. and Riekel C. (2002) Comparative Biochemistry and Physiology Part B 133:493-507. [0400] Delorenzi M. and Speed T. (2002) Bioinformatics 18:617-625. [0401] Deng Y., Liu J., Zheng Q., Eliezer D., Kallenbach N. R. and Lu M. (2006) Structure 14:247-255. [0402] Flower N. E. and Kenchington W. R. (1967) Journal of the Royal Microscopical Society 86:297. [0403] Grimaldi D. and Engel M. S. (2005) Evolution of insects. Cambridge University Press, New York. [0404] Harayama S. (1998) Trends Biotech., 16; 76-82. [0405] Heimburg T, Schunemann J., Weber K., and Geisler N. (1999) Biochemistry 38:12727-12734. [0406] Hepburn H. R., Chandler H. D. and Davidoff M. R. (1979) Insect Biochem. 9:66. [0407] Kneller D. G., Cohen F. E. and Langridge R. (1990) J. Mol. Biol. 214:171-182. [0408] LaMunyon C. W. (1988) Psyche 95:203-209. [0409] LaMunyon C. W. and Adams P. A. (1987) Annals of the Entomological Society of America 80:804-808. [0410] Lucas F. Shaw J. T. B. and Smith S. G. (1960) J. Mol. Biol. 2:339-349. [0411] Lucas F. and Rudall K. M. (1967) In Comprehensive Biochemistry (Ed. Florkin M and Stotz H) Vol 26B pp 475-559 Elsevier Amsterdam. [0412] Lupas A., Van Dyke M. and Stock J. (1991) Science 252:1162-1164. [0413] McClelland J. L. and Rumelhart D. E. (1988) Explorations in Parallel Distributed Processing vol 13. pp 318-362. MITPress, Cambridge Mass. [0414] Needleman, S. B. and Wunsch, C. D. (1970) J. Mol. Biol., 48; 443-453. [0415] Quicke D. L. J., Shaw M. R., Takahashi M. and Yanechin B. (2004) Journal of Natural History 38:2167-2181. [0416] Reiser K., McCormick, Rucker R. B. (1992) The FASEB Journal 6:2439-2449. [0417] Rost B. and Sander C. (1993) J. Molecular Biology 232:584-599. [0418] Rost B., Yachdav G. and Liu J. (2004) Nucleic Acids Research 32(Web Server issue):W321-W326. [0419] Rudall K. M. (1962) In Comparative Biochemistry (Ed. By Florkin M and Mason HS) Vol 4, pp. 297-435. Academic Press, New York. [0420] Rudall K. M. and Kenchington W. (1971) Annual Reviews in Entomology 16:73-96. [0421] Silva-Zacarin E. C. M., Silva De Moraes R. L. M. and Taboga S. R. (2003) J. Biosci. 6:753-764. [0422] Speilger P. E. (1962) Annals of the Entomological Society of America. 55: 69-77. [0423] Wang M. B., Li Z. Y. et al. (1998). Acta Hort. 461: 401-407. [0424] Yamada H., Shigesada K., Igarashi Y., Takasu Y., Tsubouchi K. and Kato Y. (2004) Int. J. Wild Silkmoth and Silk 9:61-66.
Sequence CWU 1
1
771314PRTApis mellifera 1Gly Leu Glu Gly Pro Gly Asn Ser Leu Pro
Glu Leu Val Lys Gly Ser1 5 10 15Ala Ser Ala Thr Ala Ser Thr Ala Val
Thr Ala Arg Ser Gly Leu Arg 20 25 30Ala Gly Gln Val Ala Leu Ala Ser
Gln Lys Asp Ala Val Leu Gln Ala 35 40 45Gln Ala Ala Ala Ser Ala Ala
Ser Glu Ala Arg Ala Ala Ala Asp Leu 50 55 60Thr Ala Lys Leu Ser Gln
Glu Ser Ala Ser Val Gln Ser Gln Ala Ala65 70 75 80Ala Lys Gly Lys
Glu Thr Glu Glu Ala Ala Val Gly Gln Ala Arg Ala 85 90 95Gly Leu Glu
Ser Val Ser Met Ala Ala Ser Ala Thr Ser Ala Ala Lys 100 105 110Glu
Ala Ser Thr Ala Ala Lys Ala Ala Ala Ser Ala Leu Ser Thr Ala 115 120
125Val Val Gln Ala Lys Ile Ala Glu Arg Ala Ala Lys Ala Glu Ala Val
130 135 140Ala Ser Asp Glu Ala Lys Ala Lys Ala Ile Ala Ala Ala Asn
Leu Ala145 150 155 160Ala Glu Ala Ser Val Ala Ala Glu Ala Ala Leu
Lys Ala Glu Lys Val 165 170 175Ala Glu Glu Ala Ile Ala Arg Ala Ala
Ser Ala Lys Ala Ala Ala Arg 180 185 190Ala Ala Ala Ala Ala Leu Ala
Ser Ser Lys Glu Ala Ala Thr Ala Ser 195 200 205Ala Arg Asn Ala Ala
Glu Ser Glu Ala Arg Asn Glu Val Ala Val Leu 210 215 220Ile Ala Glu
Ile Asp Lys Lys Ser Arg Glu Ile Asp Ala Ala Ser Ser225 230 235
240Leu Asn Ala Arg Ala Ala Ala Lys Ala Ser Ser Arg Asn Val Glu Thr
245 250 255Ala Thr Ile Gly Ala Asn Ile Asn Ser Ser Lys Gln Val Val
Ser Ile 260 265 270Pro Val Glu Ile Lys Lys Phe Ser Glu Pro Glu Val
Ser Thr Ser Trp 275 280 285Arg Glu Asp Glu Glu Val Thr Lys Glu Lys
Lys Glu His Ile Asn Leu 290 295 300Asn Asp Phe Asp Leu Lys Ser Asn
Val Phe305 3102333PRTApis mellifera 2Met Lys Ile Pro Val Leu Leu
Ala Thr Cys Leu Tyr Leu Cys Gly Phe1 5 10 15Ala Ser Ala Gly Leu Glu
Gly Pro Gly Asn Ser Leu Pro Glu Leu Val 20 25 30Lys Gly Ser Ala Ser
Ala Thr Ala Ser Thr Ala Val Thr Ala Arg Ser 35 40 45Gly Leu Arg Ala
Gly Gln Val Ala Leu Ala Ser Gln Lys Asp Ala Val 50 55 60Leu Gln Ala
Gln Ala Ala Ala Ser Ala Ala Ser Glu Ala Arg Ala Ala65 70 75 80Ala
Asp Leu Thr Ala Lys Leu Ser Gln Glu Ser Ala Ser Val Gln Ser 85 90
95Gln Ala Ala Ala Lys Gly Lys Glu Thr Glu Glu Ala Ala Val Gly Gln
100 105 110Ala Arg Ala Gly Leu Glu Ser Val Ser Met Ala Ala Ser Ala
Thr Ser 115 120 125Ala Ala Lys Glu Ala Ser Thr Ala Ala Lys Ala Ala
Ala Ser Ala Leu 130 135 140Ser Thr Ala Val Val Gln Ala Lys Ile Ala
Glu Arg Ala Ala Lys Ala145 150 155 160Glu Ala Val Ala Ser Asp Glu
Ala Lys Ala Lys Ala Ile Ala Ala Ala 165 170 175Asn Leu Ala Ala Glu
Ala Ser Val Ala Ala Glu Ala Ala Leu Lys Ala 180 185 190Glu Lys Val
Ala Glu Glu Ala Ile Ala Arg Ala Ala Ser Ala Lys Ala 195 200 205Ala
Ala Arg Ala Ala Ala Ala Ala Leu Ala Ser Ser Lys Glu Ala Ala 210 215
220Thr Ala Ser Ala Arg Asn Ala Ala Glu Ser Glu Ala Arg Asn Glu
Val225 230 235 240Ala Val Leu Ile Ala Glu Ile Asp Lys Lys Ser Arg
Glu Ile Asp Ala 245 250 255Ala Ser Ser Leu Asn Ala Arg Ala Ala Ala
Lys Ala Ser Ser Arg Asn 260 265 270Val Glu Thr Ala Thr Ile Gly Ala
Asn Ile Asn Ser Ser Lys Gln Val 275 280 285Val Ser Ile Pro Val Glu
Ile Lys Lys Phe Ser Glu Pro Glu Val Ser 290 295 300Thr Ser Trp Arg
Glu Asp Glu Glu Val Thr Lys Glu Lys Lys Glu His305 310 315 320Ile
Asn Leu Asn Asp Phe Asp Leu Lys Ser Asn Val Phe 325 3303290PRTApis
mellifera 3Arg Val Ile Asn His Glu Ser Leu Lys Thr Ser Glu Asp Ile
Gln Gly1 5 10 15Gly Tyr Ser Ala Gly Ile Val Gly Asp Gly Ser Asp Ala
Leu Gly Ser 20 25 30Ser Ile Glu Asn Ala Gln Lys Val Ala Arg Ala Ala
Glu Asn Val Gly 35 40 45Leu Asn Leu Glu Leu Gly Ala Gly Ala Arg Ala
Ala Ser Val Ala Ala 50 55 60Ala Ala Gln Ala Lys Asn Thr Glu Ala Ala
Glu Ala Gly Ala Asn Ala65 70 75 80Ala Leu Ala Ala Ala Ile Ala Lys
Arg Glu Glu Ala Ile Lys Ala Ser 85 90 95Glu Ile Ala Asn Gln Leu Leu
Thr Asn Ala Ala Lys Ala Ala Glu Ala 100 105 110Thr Val Ser Ala Thr
Lys Arg Ala Ala Gln Leu Thr Ala Ala Ala Lys 115 120 125Glu Ala Thr
Arg Ala Ser Ala Ala Ala Ala Glu Ala Ala Thr Glu Ala 130 135 140Gln
Val Lys Ala Asn Ala Asp Ser Ile Ile Thr Lys Arg Ala Ala Ile145 150
155 160Ala Glu Ala Gln Ala Ala Ala Glu Ala Gln Val Lys Ala Ala Ile
Ala 165 170 175Arg Lys Ser Ala Ala Asn Phe Leu Ala Lys Ala Gln Ile
Ala Ala Ala 180 185 190Ala Glu Ser Glu Ala Thr Lys Leu Ala Ala Glu
Ala Val Val Ala Leu 195 200 205Thr Asn Ala Glu Val Ala Val Asn Gln
Ala Arg Asn Ala Gln Ala Asn 210 215 220Ala Ser Thr Gln Ala Ser Met
Ala Val Arg Val Asp Ser Gln Ala Ala225 230 235 240Asn Ala Glu Ala
Ala Ala Val Ala Gln Ala Glu Thr Leu Leu Val Thr 245 250 255Ala Glu
Ala Val Ala Ala Ala Glu Ala Glu Val Ala Asn Lys Ala Ala 260 265
270Thr Phe Ala Lys Gln Ile Val Asn Glu Lys Lys Ile His Val Ala Lys
275 280 285Leu Glu 2904309PRTApis mellifera 4Met Lys Ile Pro Ala
Ile Phe Val Thr Ser Leu Leu Val Trp Gly Leu1 5 10 15Ala Glu Gly Arg
Val Ile Asn His Glu Ser Leu Lys Thr Ser Glu Asp 20 25 30Ile Gln Gly
Gly Tyr Ser Ala Gly Ile Val Gly Asp Gly Ser Asp Ala 35 40 45Leu Gly
Ser Ser Ile Glu Asn Ala Gln Lys Val Ala Arg Ala Ala Glu 50 55 60Asn
Val Gly Leu Asn Leu Glu Leu Gly Ala Gly Ala Arg Ala Ala Ser65 70 75
80Val Ala Ala Ala Ala Gln Ala Lys Asn Thr Glu Ala Ala Glu Ala Gly
85 90 95Ala Asn Ala Ala Leu Ala Ala Ala Ile Ala Lys Arg Glu Glu Ala
Ile 100 105 110Lys Ala Ser Glu Ile Ala Asn Gln Leu Leu Thr Asn Ala
Ala Lys Ala 115 120 125Ala Glu Ala Thr Val Ser Ala Thr Lys Arg Ala
Ala Gln Leu Thr Ala 130 135 140Ala Ala Lys Glu Ala Thr Arg Ala Ser
Ala Ala Ala Ala Glu Ala Ala145 150 155 160Thr Glu Ala Gln Val Lys
Ala Asn Ala Asp Ser Ile Ile Thr Lys Arg 165 170 175Ala Ala Ile Ala
Glu Ala Gln Ala Ala Ala Glu Ala Gln Val Lys Ala 180 185 190Ala Ile
Ala Arg Lys Ser Ala Ala Asn Phe Leu Ala Lys Ala Gln Ile 195 200
205Ala Ala Ala Ala Glu Ser Glu Ala Thr Lys Leu Ala Ala Glu Ala Val
210 215 220Val Ala Leu Thr Asn Ala Glu Val Ala Val Asn Gln Ala Arg
Asn Ala225 230 235 240Gln Ala Asn Ala Ser Thr Gln Ala Ser Met Ala
Val Arg Val Asp Ser 245 250 255Gln Ala Ala Asn Ala Glu Ala Ala Ala
Val Ala Gln Ala Glu Thr Leu 260 265 270Leu Val Thr Ala Glu Ala Val
Ala Ala Ala Glu Ala Glu Val Ala Asn 275 280 285Lys Ala Ala Thr Phe
Ala Lys Gln Ile Val Asn Glu Lys Lys Ile His 290 295 300Val Ala Lys
Leu Glu3055316PRTApis mellifera 5Gly Val Glu Glu Phe Lys Ser Ser
Ala Thr Glu Glu Val Ile Ser Lys1 5 10 15Asn Leu Glu Val Asp Leu Leu
Lys Asn Val Asp Thr Ser Ala Lys Arg 20 25 30Arg Glu Asn Gly Ala Pro
Val Leu Gly Lys Asn Thr Leu Gln Ser Leu 35 40 45Glu Lys Ile Lys Thr
Ser Ala Ser Val Asn Ala Lys Ala Ala Ala Val 50 55 60Val Lys Ala Ser
Ala Leu Ala Leu Ala Glu Ala Tyr Leu Arg Ala Ser65 70 75 80Ala Leu
Ser Ala Ala Ala Ser Ala Lys Ala Ala Ala Ala Leu Lys Asn 85 90 95Ala
Gln Gln Ala Gln Leu Asn Ala Gln Glu Lys Ser Leu Ala Ala Leu 100 105
110Lys Ala Gln Ser Glu Glu Glu Ala Ala Ser Ala Arg Ala Asn Ala Ala
115 120 125Thr Ala Ala Thr Gln Ser Ala Leu Glu Arg Ala Gln Ala Ser
Ser Arg 130 135 140Leu Ala Thr Val Ala Gln Asn Val Ala Ser Asp Leu
Gln Lys Arg Thr145 150 155 160Ser Thr Lys Ala Ala Ala Glu Ala Ala
Ala Thr Leu Arg Gln Leu Gln 165 170 175Asp Ala Glu Arg Thr Lys Trp
Ser Ala Asn Ala Ala Leu Glu Val Ser 180 185 190Ala Ala Ala Ala Ala
Ala Glu Thr Lys Thr Thr Ala Ser Ser Glu Ala 195 200 205Ala Asn Ala
Ala Ala Lys Lys Ala Ala Ala Ile Ala Ser Asp Ala Asp 210 215 220Gly
Ala Glu Arg Ser Ala Ser Thr Glu Ala Gln Ser Ala Ala Lys Ile225 230
235 240Glu Ser Val Ala Ala Ala Glu Gly Ser Ala Asn Ser Ala Ser Glu
Asp 245 250 255Ser Arg Ala Ala Gln Leu Glu Ala Ser Thr Ala Ala Arg
Ala Asn Val 260 265 270Ala Ala Ala Val Gly Asp Gly Ala Ile Ile Gly
Leu Gly Glu Glu Ala 275 280 285Gly Ala Ala Ala Gln Leu Leu Ala Gln
Ala Lys Ala Leu Ala Glu Val 290 295 300Ser Ser Lys Ser Glu Asn Ile
Glu Asp Lys Lys Phe305 310 3156335PRTApis mellifera 6Met Gln Ile
Pro Thr Phe Val Ala Ile Cys Leu Leu Thr Ser Gly Leu1 5 10 15Val His
Ala Gly Val Glu Glu Phe Lys Ser Ser Ala Thr Glu Glu Val 20 25 30Ile
Ser Lys Asn Leu Glu Val Asp Leu Leu Lys Asn Val Asp Thr Ser 35 40
45Ala Lys Arg Arg Glu Asn Gly Ala Pro Val Leu Gly Lys Asn Thr Leu
50 55 60Gln Ser Leu Glu Lys Ile Lys Thr Ser Ala Ser Val Asn Ala Lys
Ala65 70 75 80Ala Ala Val Val Lys Ala Ser Ala Leu Ala Leu Ala Glu
Ala Tyr Leu 85 90 95Arg Ala Ser Ala Leu Ser Ala Ala Ala Ser Ala Lys
Ala Ala Ala Ala 100 105 110Leu Lys Asn Ala Gln Gln Ala Gln Leu Asn
Ala Gln Glu Lys Ser Leu 115 120 125Ala Ala Leu Lys Ala Gln Ser Glu
Glu Glu Ala Ala Ser Ala Arg Ala 130 135 140Asn Ala Ala Thr Ala Ala
Thr Gln Ser Ala Leu Glu Arg Ala Gln Ala145 150 155 160Ser Ser Arg
Leu Ala Thr Val Ala Gln Asn Val Ala Ser Asp Leu Gln 165 170 175Lys
Arg Thr Ser Thr Lys Ala Ala Ala Glu Ala Ala Ala Thr Leu Arg 180 185
190Gln Leu Gln Asp Ala Glu Arg Thr Lys Trp Ser Ala Asn Ala Ala Leu
195 200 205Glu Val Ser Ala Ala Ala Ala Ala Ala Glu Thr Lys Thr Thr
Ala Ser 210 215 220Ser Glu Ala Ala Asn Ala Ala Ala Lys Lys Ala Ala
Ala Ile Ala Ser225 230 235 240Asp Ala Asp Gly Ala Glu Arg Ser Ala
Ser Thr Glu Ala Gln Ser Ala 245 250 255Ala Lys Ile Glu Ser Val Ala
Ala Ala Glu Gly Ser Ala Asn Ser Ala 260 265 270Ser Glu Asp Ser Arg
Ala Ala Gln Leu Glu Ala Ser Thr Ala Ala Arg 275 280 285Ala Asn Val
Ala Ala Ala Val Gly Asp Gly Ala Ile Ile Gly Leu Gly 290 295 300Glu
Glu Ala Gly Ala Ala Ala Gln Leu Leu Ala Gln Ala Lys Ala Leu305 310
315 320Ala Glu Val Ser Ser Lys Ser Glu Asn Ile Glu Asp Lys Lys Phe
325 330 3357323PRTApis mellifera 7Ala Arg Glu Glu Val Glu Thr Arg
Asp Lys Thr Lys Thr Ser Thr Val1 5 10 15Val Lys Ser Glu Lys Val Glu
Val Val Ala Pro Ala Lys Asp Glu Leu 20 25 30Lys Leu Thr Ser Glu Pro
Ile Phe Gly Arg Arg Val Gly Thr Gly Ala 35 40 45Ser Glu Val Ala Ser
Ser Ser Gly Glu Ala Ile Ala Ile Ser Leu Gly 50 55 60Ala Gly Gln Ser
Ala Ala Glu Ser Gln Ala Leu Ala Ala Ser Gln Ser65 70 75 80Lys Thr
Ala Ala Asn Ala Ala Ile Gly Ala Ser Glu Leu Thr Asn Lys 85 90 95Val
Ala Ala Leu Val Ala Gly Ala Thr Gly Ala Gln Ala Arg Ala Thr 100 105
110Ala Ala Ser Ser Ser Ala Leu Lys Ala Ser Leu Ala Thr Glu Glu Ala
115 120 125Ala Glu Glu Ala Glu Ala Ala Val Ala Asp Ala Lys Ala Ala
Ala Glu 130 135 140Lys Ala Glu Ser Leu Ala Lys Asn Leu Ala Ser Ala
Ser Ala Arg Ala145 150 155 160Ala Leu Ser Ser Glu Arg Ala Asn Glu
Leu Ala Gln Ala Glu Ser Ala 165 170 175Ala Ala Ala Glu Ala Gln Ala
Lys Thr Ala Ala Ala Ala Lys Ala Ala 180 185 190Glu Ile Ala Leu Lys
Val Ala Glu Ile Ala Val Lys Ala Glu Ala Asp 195 200 205Ala Ala Ala
Ala Ala Val Ala Ala Ala Lys Ala Arg Ala Val Ala Asp 210 215 220Ala
Ala Ala Ala Arg Ala Ala Ala Val Asn Ala Ile Ala Lys Ala Glu225 230
235 240Glu Glu Ala Ser Ala Gln Ala Glu Asn Ala Ala Gly Val Leu Gln
Ala 245 250 255Ala Ala Ser Ala Ala Ala Glu Ser Arg Ala Ala Ala Ala
Ala Ala Ala 260 265 270Ala Thr Ser Glu Ala Ala Ala Glu Ala Gly Pro
Leu Ala Gly Glu Met 275 280 285Lys Pro Pro His Trp Lys Trp Glu Arg
Ile Pro Val Lys Lys Glu Glu 290 295 300Trp Lys Thr Ser Thr Lys Glu
Glu Trp Lys Thr Thr Asn Glu Glu Trp305 310 315 320Glu Val
Lys8342PRTApis mellifera 8Met Lys Ile Pro Ser Ile Leu Ala Val Ser
Leu Leu Ile Trp Gly Leu1 5 10 15Ala Ser Gly Ala Arg Glu Glu Val Glu
Thr Arg Asp Lys Thr Lys Thr 20 25 30Ser Thr Val Val Lys Ser Glu Lys
Val Glu Val Val Ala Pro Ala Lys 35 40 45Asp Glu Leu Lys Leu Thr Ser
Glu Pro Ile Phe Gly Arg Arg Val Gly 50 55 60Thr Gly Ala Ser Glu Val
Ala Ser Ser Ser Gly Glu Ala Ile Ala Ile65 70 75 80Ser Leu Gly Ala
Gly Gln Ser Ala Ala Glu Ser Gln Ala Leu Ala Ala 85 90 95Ser Gln Ser
Lys Thr Ala Ala Asn Ala Ala Ile Gly Ala Ser Glu Leu 100 105 110Thr
Asn Lys Val Ala Ala Leu Val Ala Gly Ala Thr Gly Ala Gln Ala 115 120
125Arg Ala Thr Ala Ala Ser Ser Ser Ala Leu Lys Ala Ser Leu Ala Thr
130 135 140Glu Glu Ala Ala Glu Glu Ala Glu Ala Ala Val Ala Asp Ala
Lys Ala145 150 155 160Ala Ala Glu Lys Ala Glu Ser Leu Ala Lys Asn
Leu Ala Ser Ala Ser 165 170 175Ala Arg Ala Ala Leu Ser Ser Glu Arg
Ala Asn Glu Leu Ala Gln Ala 180 185 190Glu Ser Ala Ala Ala Ala Glu
Ala Gln Ala Lys Thr Ala Ala Ala Ala 195 200 205Lys Ala Ala Glu Ile
Ala Leu Lys Val Ala Glu Ile Ala Val Lys Ala 210 215 220Glu Ala Asp
Ala Ala Ala Ala Ala Val Ala Ala Ala Lys Ala Arg Ala225 230 235
240Val Ala Asp Ala
Ala Ala Ala Arg Ala Ala Ala Val Asn Ala Ile Ala 245 250 255Lys Ala
Glu Glu Glu Ala Ser Ala Gln Ala Glu Asn Ala Ala Gly Val 260 265
270Leu Gln Ala Ala Ala Ser Ala Ala Ala Glu Ser Arg Ala Ala Ala Ala
275 280 285Ala Ala Ala Ala Thr Ser Glu Ala Ala Ala Glu Ala Gly Pro
Leu Ala 290 295 300Gly Glu Met Lys Pro Pro His Trp Lys Trp Glu Arg
Ile Pro Val Lys305 310 315 320Lys Glu Glu Trp Lys Thr Ser Thr Lys
Glu Glu Trp Lys Thr Thr Asn 325 330 335Glu Glu Trp Glu Val Lys
3409353PRTApis mellifera 9Gly Val Asn Thr Glu Leu Lys Lys Asp Gly
Glu Leu Lys Glu Glu Ser1 5 10 15Tyr Glu Lys Ser Glu Ser Lys Ser Leu
Lys Glu Ile Lys Glu Glu Arg 20 25 30Ala Ser Lys Ser Lys Ser Glu Arg
Leu Lys Ile Arg Glu Glu Lys Arg 35 40 45Glu Glu Glu Glu Lys Ser Lys
Ser Leu Asn Leu Val Val Val Arg Glu 50 55 60Lys Ile Thr Lys Leu Ser
Ser Trp Leu Lys Glu Glu Lys Asp Ile Ser65 70 75 80Pro Leu Leu Glu
Glu Lys Asn Gly Lys Gly Leu Leu Gly Leu Glu Asp 85 90 95Val Thr Asp
Glu Leu Asn Ile Ala Leu Lys Ser Leu Lys Glu Gly Lys 100 105 110Lys
Phe Asp Thr Trp Lys Phe Glu Lys Gly Ser Glu Asp Val Arg Ser 115 120
125Leu Glu Glu Leu Asp Thr Ser Val Val Glu Leu Leu Lys Leu Ile Lys
130 135 140Glu Gly Lys Thr Asp His Gly Ala Ile Asp Leu Glu Lys Asn
Gly Lys145 150 155 160Val Leu Val Asp Leu Glu Lys Ile Ser Glu Asn
Ile Leu Glu Thr Cys 165 170 175Gly Ser Gln Lys Lys Thr Val Glu Val
Val Asp Asp Lys Asp Lys Lys 180 185 190Trp Asn Lys Glu Ser Gly Trp
Lys Lys Asn Leu Asn Asp Leu Asp Trp 195 200 205Lys Lys Asp Leu Asp
Lys Asp Lys Val Gly Gly Gly Leu Leu Gly Gly 210 215 220Leu Ser Gly
Leu Leu Asn Ser Leu Lys Ser Glu Lys Gly Leu Leu Gly225 230 235
240Leu Leu Asn Lys Asn Gln Ile Glu Leu Leu Ile Pro Leu Ile Ser Glu
245 250 255Ile Lys Lys Lys Asn Ile Asp Phe Asn Leu Phe Asp Ser Val
Asp Ser 260 265 270Val Glu Arg Asn Leu Asp Leu Lys Leu Phe Thr Ser
Ser Val Ser Lys 275 280 285Val Thr Glu Leu Leu Asn Lys Gly Ile Asp
Ile Gln Thr Ile Leu Asn 290 295 300Ala Lys Asn Gly Asp Glu Phe Asp
Leu Ser Gly Lys Glu Leu Lys Asn305 310 315 320Val Lys Gly Ile Phe
Gly Leu Ile Gly Ser Leu Lys Arg Ser Leu Gly 325 330 335Leu Glu Asn
Ile Leu Asn Leu Pro Phe Lys Arg Ile Pro Leu Leu Lys 340 345
350Leu10372PRTApis mellifera 10Met Lys Tyr Met Leu Leu Leu Leu Ser
Ile Phe Ile Cys Ala His Ile1 5 10 15Val Cys Ala Gly Val Asn Thr Glu
Leu Lys Lys Asp Gly Glu Leu Lys 20 25 30Glu Glu Ser Tyr Glu Lys Ser
Glu Ser Lys Ser Leu Lys Glu Ile Lys 35 40 45Glu Glu Arg Ala Ser Lys
Ser Lys Ser Glu Arg Leu Lys Ile Arg Glu 50 55 60Glu Lys Arg Glu Glu
Glu Glu Lys Ser Lys Ser Leu Asn Leu Val Val65 70 75 80Val Arg Glu
Lys Ile Thr Lys Leu Ser Ser Trp Leu Lys Glu Glu Lys 85 90 95Asp Ile
Ser Pro Leu Leu Glu Glu Lys Asn Gly Lys Gly Leu Leu Gly 100 105
110Leu Glu Asp Val Thr Asp Glu Leu Asn Ile Ala Leu Lys Ser Leu Lys
115 120 125Glu Gly Lys Lys Phe Asp Thr Trp Lys Phe Glu Lys Gly Ser
Glu Asp 130 135 140Val Arg Ser Leu Glu Glu Leu Asp Thr Ser Val Val
Glu Leu Leu Lys145 150 155 160Leu Ile Lys Glu Gly Lys Thr Asp His
Gly Ala Ile Asp Leu Glu Lys 165 170 175Asn Gly Lys Val Leu Val Asp
Leu Glu Lys Ile Ser Glu Asn Ile Leu 180 185 190Glu Thr Cys Gly Ser
Gln Lys Lys Thr Val Glu Val Val Asp Asp Lys 195 200 205Asp Lys Lys
Trp Asn Lys Glu Ser Gly Trp Lys Lys Asn Leu Asn Asp 210 215 220Leu
Asp Trp Lys Lys Asp Leu Asp Lys Asp Lys Val Gly Gly Gly Leu225 230
235 240Leu Gly Gly Leu Ser Gly Leu Leu Asn Ser Leu Lys Ser Glu Lys
Gly 245 250 255Leu Leu Gly Leu Leu Asn Lys Asn Gln Ile Glu Leu Leu
Ile Pro Leu 260 265 270Ile Ser Glu Ile Lys Lys Lys Asn Ile Asp Phe
Asn Leu Phe Asp Ser 275 280 285Val Asp Ser Val Glu Arg Asn Leu Asp
Leu Lys Leu Phe Thr Ser Ser 290 295 300Val Ser Lys Val Thr Glu Leu
Leu Asn Lys Gly Ile Asp Ile Gln Thr305 310 315 320Ile Leu Asn Ala
Lys Asn Gly Asp Glu Phe Asp Leu Ser Gly Lys Glu 325 330 335Leu Lys
Asn Val Lys Gly Ile Phe Gly Leu Ile Gly Ser Leu Lys Arg 340 345
350Ser Leu Gly Leu Glu Asn Ile Leu Asn Leu Pro Phe Lys Arg Ile Pro
355 360 365Leu Leu Lys Leu 37011942DNAApis mellifera 11ggtttggagg
ggccgggcaa ctcgttgccc gagctcgtga aaggtagcgc atcggccacc 60gcgtcgaccg
ctgtgaccgc tagatcagga cttagagccg gacaagtagc tttagcttcg
120cagaaggatg ccgtactcca agctcaagct gctgcatccg ccgcgtcaga
ggcgcgcgct 180gctgccgatc tgacggctaa acttagccaa gaatcggcat
cagtgcaatc gcaggctgcc 240gccaaaggga aggaaacgga ggaggcagct
gttggtcaag ctagggctgg cctcgagtcg 300gtgtccatgg ccgcatcagc
cacatctgct gccaaagaag catcgaccgc cgccaaagcc 360gcagcatccg
cactatccac agccgtggtg caagcgaaaa tagctgagag ggcagccaaa
420gctgaagctg ttgcctcgga cgaagccaag gccaaggcga ttgcagcagc
caacttggcg 480gctgaggcca gtgtagccgc agaagcagct ctcaaggccg
agaaagtggc cgaagaagcc 540atcgcaagag cggcctctgc aaaggctgcc
gcaagagctg ctgctgccgc tctagcctcc 600tcgaaggaag cagccacggc
cagcgcaaga aacgccgcgg aatccgaggc caggaacgaa 660gtagctgtat
tgatcgccga gattgataaa aagagtaggg aaatcgacgc agccagttcg
720cttaatgcgc gtgccgctgc caaggcaagc tccaggaacg tagaaacggc
gacaatcggg 780gccaacatca actcttcgaa acaagtcgtg tcaattccag
tggaaataaa gaaattctcg 840gagccggaag tgtcaacatc atggagagaa
gatgaagagg ttacgaaaga gaagaaggag 900cacataaatc tgaacgactt
cgacttgaag agcaacgtat tt 94212999DNAApis mellifera 12atgaagattc
cagtattgct tgcaacgtgc ctctaccttt gcggatttgc gtccgccggt 60ttggaggggc
cgggcaactc gttgcccgag ctcgtgaaag gtagcgcatc ggccaccgcg
120tcgaccgctg tgaccgctag atcaggactt agagccggac aagtagcttt
agcttcgcag 180aaggatgccg tactccaagc tcaagctgct gcatccgccg
cgtcagaggc gcgcgctgct 240gccgatctga cggctaaact tagccaagaa
tcggcatcag tgcaatcgca ggctgccgcc 300aaagggaagg aaacggagga
ggcagctgtt ggtcaagcta gggctggcct cgagtcggtg 360tccatggccg
catcagccac atctgctgcc aaagaagcat cgaccgccgc caaagccgca
420gcatccgcac tatccacagc cgtggtgcaa gcgaaaatag ctgagagggc
agccaaagct 480gaagctgttg cctcggacga agccaaggcc aaggcgattg
cagcagccaa cttggcggct 540gaggccagtg tagccgcaga agcagctctc
aaggccgaga aagtggccga agaagccatc 600gcaagagcgg cctctgcaaa
ggctgccgca agagctgctg ctgccgctct agcctcctcg 660aaggaagcag
ccacggccag cgcaagaaac gccgcggaat ccgaggccag gaacgaagta
720gctgtattga tcgccgagat tgataaaaag agtagggaaa tcgacgcagc
cagttcgctt 780aatgcgcgtg ccgctgccaa ggcaagctcc aggaacgtag
aaacggcgac aatcggggcc 840aacatcaact cttcgaaaca agtcgtgtca
attccagtgg aaataaagaa attctcggag 900ccggaagtgt caacatcatg
gagagaagat gaagaggtta cgaaagagaa gaaggagcac 960ataaatctga
acgacttcga cttgaagagc aacgtattt 99913870DNAApis mellifera
13cgcgtgatta atcacgagtc cctgaagacg agcgaggata ttcaaggagg atattcagca
60ggaatagtcg gtgatggatc tgacgcgctt ggctcctcca tagaaaacgc ccaaaaagtc
120gctcgagcgg ctgaaaacgt gggcttgaat ctggaattgg gcgcaggcgc
gcgtgctgcc 180agtgttgccg ctgctgccca ggccaaaaac acagaggctg
cggaagcagg agcaaacgcc 240gctctggccg ccgccattgc caaacgggag
gaagcgatta aagccagcga gatagcaaac 300caattgttga ccaatgcagc
aaaagcggca gaagcgactg tatcggcaac gaagagggca 360gcacaattga
cggctgcagc gaaagaagca accagagctt ctgcagccgc tgctgaagct
420gctacggagg cccaggtaaa ggctaacgcc gattcaatca tcacgaagag
ggctgcgatt 480gccgaggctc aagctgcggc ggaagctcaa gttaaggcgg
caatcgccag aaaatcggca 540gcgaattttt tggctaaggc tcaaatagcg
gctgccgcgg aatccgaggc cacgaaactc 600gcggccgaag ctgtagtggc
actaacaaac gccgaagtcg ccgtgaacca ggctagaaac 660gcacaggcaa
acgcctcgac tcaagcttcc atggctgtta gggtagattc tcaagcagcg
720aacgctgaag cagccgctgt agcgcaagcc gaaactctct tggttacggc
agaagctgtc 780gcagctgcgg aggctgaggt tgcgaacaaa gccgccacat
ttgcaaaaca gatcgtcaac 840gagaagaaaa tacatgtagc aaagttggaa
87014927DNAApis mellifera 14atgaagattc cagcaatatt cgtcacgtct
ctgctggtct ggggattggc cgagggccgc 60gtgattaatc acgagtccct gaagacgagc
gaggatattc aaggaggata ttcagcagga 120atagtcggtg atggatctga
cgcgcttggc tcctccatag aaaacgccca aaaagtcgct 180cgagcggctg
aaaacgtggg cttgaatctg gaattgggcg caggcgcgcg tgctgccagt
240gttgccgctg ctgcccaggc caaaaacaca gaggctgcgg aagcaggagc
aaacgccgct 300ctggccgccg ccattgccaa acgggaggaa gcgattaaag
ccagcgagat agcaaaccaa 360ttgttgacca atgcagcaaa agcggcagaa
gcgactgtat cggcaacgaa gagggcagca 420caattgacgg ctgcagcgaa
agaagcaacc agagcttctg cagccgctgc tgaagctgct 480acggaggccc
aggtaaaggc taacgccgat tcaatcatca cgaagagggc tgcgattgcc
540gaggctcaag ctgcggcgga agctcaagtt aaggcggcaa tcgccagaaa
atcggcagcg 600aattttttgg ctaaggctca aatagcggct gccgcggaat
ccgaggccac gaaactcgcg 660gccgaagctg tagtggcact aacaaacgcc
gaagtcgccg tgaaccaggc tagaaacgca 720caggcaaacg cctcgactca
agcttccatg gctgttaggg tagattctca agcagcgaac 780gctgaagcag
ccgctgtagc gcaagccgaa actctcttgg ttacggcaga agctgtcgca
840gctgcggagg ctgaggttgc gaacaaagcc gccacatttg caaaacagat
cgtcaacgag 900aagaaaatac atgtagcaaa gttggaa 92715949DNAApis
mellifera 15ggcgtcgagg aattcaagtc ctcggcaacc gaggaggtga tcagcaaaaa
cttagaagtc 60gacctgttga aaaatgtgga cactagcgcg aaacgaagag agaacggcgc
cccggtgctc 120ggcaagaaca cacttcaatc cctggagaag atcaagacgt
cggcgagcgt gaatgccaaa 180gcagcagccg tggtgaaagc gtccgctctg
gctcttgcag aggcctattt gcgagcgtcc 240gcattgtcag ccgccgcttc
agccaaggca gccgccgccc tgaaaaatgc tcaacaagcg 300caattaaacg
cccaggaaaa gtctttggcc gcgttgaaag ctcagtccga ggaagaggca
360gcttctgctc gtgcaaacgc agcaaccgcc gcgacacagt cggcactgga
acgcgctcaa 420gcctcctcca ggttagcaac ggtcgcccaa aacgtagcca
gcgacttgca gaaacggacc 480agcaccaagg ccgcggctga agccgctgcc
accctcagac aattacagga cgcggaacga 540acgaaatgga gtgccaacgc
tgccttagaa gtctccgccg ctgcagctgc cgcagaaacc 600aagaccactg
cctcctcgga ggccgccaac gccgccgcca aaaaggcggc cgcgatagct
660tctgacgcgg acggcgcgga aaggtcggca tctaccgagg cacaatcagc
tgcgaagatc 720gagagtgtgg cagccgccga gggatccgcc aactcggcct
ctgaggattc ccgggccgct 780caattggaag cctccaccgc ggcgagagcc
aacgtggccg cagctgtcgg ggatggagcg 840attataggac ttggagagga
agcgggtgcc gcggctcagt tgcttgcaca ggcgaaggca 900ttggccgaag
ttagctcgaa atccgaaaat attgaggata aaaaatttt 949161006DNAApis
mellifera 16atgcagatcc caacgtttgt cgccatatgc ttgctcacat cgggcttggt
gcacgcaggc 60gtcgaggaat tcaagtcctc ggcaaccgag gaggtgatca gcaaaaactt
agaagtcgac 120ctgttgaaaa atgtggacac tagcgcgaaa cgaagagaga
acggcgcccc ggtgctcggc 180aagaacacac ttcaatccct ggagaagatc
aagacgtcgg cgagcgtgaa tgccaaagca 240gcagccgtgg tgaaagcgtc
cgctctggct cttgcagagg cctatttgcg agcgtccgca 300ttgtcagccg
ccgcttcagc caaggcagcc gccgccctga aaaatgctca acaagcgcaa
360ttaaacgccc aggaaaagtc tttggccgcg ttgaaagctc agtccgagga
agaggcagct 420tctgctcgtg caaacgcagc aaccgccgcg acacagtcgg
cactggaacg cgctcaagcc 480tcctccaggt tagcaacggt cgcccaaaac
gtagccagcg acttgcagaa acggaccagc 540accaaggccg cggctgaagc
cgctgccacc ctcagacaat tacaggacgc ggaacgaacg 600aaatggagtg
ccaacgctgc cttagaagtc tccgccgctg cagctgccgc agaaaccaag
660accactgcct cctcggaggc cgccaacgcc gccgccaaaa aggcggccgc
gatagcttct 720gacgcggacg gcgcggaaag gtcggcatct accgaggcac
aatcagctgc gaagatcgag 780agtgtggcag ccgccgaggg atccgccaac
tcggcctctg aggattcccg ggccgctcaa 840ttggaagcct ccaccgcggc
gagagccaac gtggccgcag ctgtcgggga tggagcgatt 900ataggacttg
gagaggaagc gggtgccgcg gctcagttgc ttgcacaggc gaaggcattg
960gccgaagtta gctcgaaatc cgaaaatatt gaggataaaa aatttt
100617969DNAApis mellifera 17gcaagggaag aggtggagac acgggacaag
accaagacct cgacagtggt gaaaagcgag 60aaagtggaag tcgttgctcc cgctaaggat
gaacttaaat taacgagcga gcctatcttt 120ggaagaagag tgggaactgg
agcatccgag gtggcatcta gcagcggtga agccatcgcg 180ataagtcttg
gagcagggca gtcagcggca gagtctcagg ccttggccgc ctcgcaatcc
240aaaacggcag cgaacgccgc cataggcgcg agcgagctta ccaacaaagt
tgctgctcta 300gttgctggcg cgactggtgc gcaggcgaga gctacggccg
cctcctcgag cgcgttgaag 360gccagcttgg cgaccgaaga agcggcggaa
gaggccgagg cggccgtggc tgacgccaag 420gctgccgcgg aaaaggccga
atccctggcg aaaaatctcg cgtcggcgag cgctcgcgcg 480gccctctcct
ccgaaagggc gaacgaattg gctcaagctg agagcgctgc agcggccgag
540gcgcaggcca agacagcagc cgccgccaaa gcagcggaaa tcgcccttaa
ggtcgctgag 600atagcggtga aggcggaagc ggacgcagca gctgccgccg
tggcagctgc aaaggcaaga 660gccgtggcag acgcggccgc tgcccgtgcc
gcagccgtga acgccatcgc caaggcggaa 720gaggaggcct cggcccaagc
agagaacgcc gccggtgttt tgcaagcagc cgcctccgcc 780gcggcggaat
cgcgagccgc tgcagctgcc gccgctgcta cctcggaggc agcggctgaa
840gctggcccgt tggcaggtga gatgaaacca ccgcactgga aatgggaacg
gattcctgtg 900aagaaggagg agtggaaaac gtcaacgaag gaagaatgga
aaacgacgaa tgaagaatgg 960gaggtgaag 969181026DNAApis mellifera
18atgaagatcc catccatact cgcggtttcc ctgctgatct ggggtttggc aagcggcgca
60agggaagagg tggagacacg ggacaagacc aagacctcga cagtggtgaa aagcgagaaa
120gtggaagtcg ttgctcccgc taaggatgaa cttaaattaa cgagcgagcc
tatctttgga 180agaagagtgg gaactggagc atccgaggtg gcatctagca
gcggtgaagc catcgcgata 240agtcttggag cagggcagtc agcggcagag
tctcaggcct tggccgcctc gcaatccaaa 300acggcagcga acgccgccat
aggcgcgagc gagcttacca acaaagttgc tgctctagtt 360gctggcgcga
ctggtgcgca ggcgagagct acggccgcct cctcgagcgc gttgaaggcc
420agcttggcga ccgaagaagc ggcggaagag gccgaggcgg ccgtggctga
cgccaaggct 480gccgcggaaa aggccgaatc cctggcgaaa aatctcgcgt
cggcgagcgc tcgcgcggcc 540ctctcctccg aaagggcgaa cgaattggct
caagctgaga gcgctgcagc ggccgaggcg 600caggccaaga cagcagccgc
cgccaaagca gcggaaatcg cccttaaggt cgctgagata 660gcggtgaagg
cggaagcgga cgcagcagct gccgccgtgg cagctgcaaa ggcaagagcc
720gtggcagacg cggccgctgc ccgtgccgca gccgtgaacg ccatcgccaa
ggcggaagag 780gaggcctcgg cccaagcaga gaacgccgcc ggtgttttgc
aagcagccgc ctccgccgcg 840gcggaatcgc gagccgctgc agctgccgcc
gctgctacct cggaggcagc ggctgaagct 900ggcccgttgg caggtgagat
gaaaccaccg cactggaaat gggaacggat tcctgtgaag 960aaggaggagt
ggaaaacgtc aacgaaggaa gaatggaaaa cgacgaatga agaatgggag 1020gtgaag
1026191059DNAApis mellifera 19ggcgtaaata cagaattaaa aaaagatggt
gaactaaagg aagagtctta tgagaaaagc 60gagtcaaaga gtttaaaaga aattaaagaa
gaacgtgctt caaaatcaaa aagtgaacgt 120ttgaagattc gtgaagaaaa
acgcgaagag gaagaaaaat ccaagagtct gaatctggtc 180gtggtcagag
aaaagattac caaactttct tcatggctca aagaagagaa agatatcagt
240cctcttttgg aagaaaaaaa tggcaaaggt ctattgggtt tggaagatgt
cacggacgag 300ttaaatatcg ctcttaaatc gttgaaggag ggcaaaaagt
ttgatacttg gaaattcgag 360aaaggtagcg aagacgttcg ttctttggaa
gaacttgata cgagcgtcgt tgaactttta 420aaattaataa aggaaggaaa
aactgaccat ggtgctatag atttggagaa gaatggtaag 480gtacttgtag
atttggaaaa aatctcagaa aacatacttg aaacttgtgg atcacaaaag
540aagactgtgg aagttgtaga tgataaagac aaaaaatgga ataaagaatc
aggttggaaa 600aaaaatctaa atgatctaga ttggaaaaaa gatttagata
aagataaagt tggtggcggt 660ttgcttggcg gtttaagtgg cctcttaaat
agtttaaaat cagaaaaagg tcttctaggt 720cttttgaata agaatcaaat
tgagttatta attcctttaa tcagtgagat aaaaaagaaa 780aatatagatt
ttaatctctt cgattctgtt gattctgtcg aaagaaattt agacttgaaa
840cttttcacaa gttctgtttc aaaagttact gaattattaa ataaaggaat
cgatattcaa 900acaattttga atgcgaaaaa tggagatgaa ttcgatttaa
gcggcaaaga attgaaaaac 960gtcaaaggga tatttggttt gattggaagt
ttgaaacgct cattaggatt agaaaatata 1020ttgaacttac cgtttaaacg
tatacctctg cttaaatta 1059201116DNAApis mellifera 20atgaaataca
tgctcttgtt gctatctata ttcatctgtg cacatattgt atgcgcaggc 60gtaaatacag
aattaaaaaa agatggtgaa ctaaaggaag agtcttatga gaaaagcgag
120tcaaagagtt taaaagaaat taaagaagaa cgtgcttcaa aatcaaaaag
tgaacgtttg 180aagattcgtg aagaaaaacg cgaagaggaa gaaaaatcca
agagtctgaa tctggtcgtg 240gtcagagaaa agattaccaa actttcttca
tggctcaaag aagagaaaga tatcagtcct 300cttttggaag aaaaaaatgg
caaaggtcta ttgggtttgg aagatgtcac ggacgagtta 360aatatcgctc
ttaaatcgtt gaaggagggc aaaaagtttg atacttggaa attcgagaaa
420ggtagcgaag acgttcgttc tttggaagaa cttgatacga gcgtcgttga
acttttaaaa 480ttaataaagg aaggaaaaac tgaccatggt gctatagatt
tggagaagaa tggtaaggta 540cttgtagatt tggaaaaaat ctcagaaaac
atacttgaaa cttgtggatc acaaaagaag 600actgtggaag ttgtagatga
taaagacaaa aaatggaata aagaatcagg ttggaaaaaa 660aatctaaatg
atctagattg gaaaaaagat ttagataaag ataaagttgg tggcggtttg
720cttggcggtt taagtggcct cttaaatagt ttaaaatcag aaaaaggtct
tctaggtctt
780ttgaataaga atcaaattga gttattaatt cctttaatca gtgagataaa
aaagaaaaat 840atagatttta atctcttcga ttctgttgat tctgtcgaaa
gaaatttaga cttgaaactt 900ttcacaagtt ctgtttcaaa agttactgaa
ttattaaata aaggaatcga tattcaaaca 960attttgaatg cgaaaaatgg
agatgaattc gatttaagcg gcaaagaatt gaaaaacgtc 1020aaagggatat
ttggtttgat tggaagtttg aaacgctcat taggattaga aaatatattg
1080aacttaccgt ttaaacgtat acctctgctt aaatta 1116211660DNAApis
mellifera 21atgaaataca tgctcttgtt gctatctata ttcatctgtg cacatattgt
atgcgcaggc 60gtaaatacag aattaaaaaa agatggtgaa ctaaaggaag agtcttatga
gaaaagcgag 120tcaaagagtt taaaagaaat taaagaagaa cgtgcttcaa
aatcaaaaag tgaacgtttg 180aagattcgtg aaggtaattc gtgagattca
agattcaaat caattaaatt tgaaaattat 240gaaagtagta ttgttaaatt
ataagataga agattttatc taaaaaataa taaattaagc 300tttttgtatt
tttggatatt gtagatattt ttaatataga attcttataa agttaaaaaa
360tattttataa attaaacaac tttttattat ttttatgatc taaaaattaa
aaatttcaag 420ttaaagttca aattaaaaat ttgtaaaaaa tatggaaaaa
acataaaaat tgaatttgtt 480gtaatttaaa aaggattttt attatttatt
gattaattat gaatataagt tcgaaaaatc 540ctaaatatta atgtttaaaa
ttttaattct taacaaaata tatttaattt aattcttaac 600aaagatacat
ttaaagaatt tcgcaaattt aaaaattagg tttttaattt taagaatcaa
660atggtaaaaa acattttaaa tttgaaatat ataaaagtaa atcttttaat
cgacaaacgg 720atgaatttat tgattagaaa aacgcgaaga ggaagaaaaa
tccaagagtc tgaatctggt 780cgtggtcaga gaaaagatta ccaaactttc
ttcatggctc aaagaagaga aagatatcag 840tcctcttttg gaagaaaaaa
atggcaaagg tctattgggt ttggaagatg tcacggacga 900gttaaatatc
gctcttaaat cgttgaagga gggcaaaaag tttgatactt ggaaattcga
960gaaaggtagc gaagacgttc gttctttgga agaacttgat acgagcgtcg
ttgaactttt 1020aaaattaata aaggaaggaa aaactgacca tggtgctata
gatttggaga agaatggtaa 1080ggtacttgta gatttggaaa aaatctcaga
aaacatactt gaaacttgtg gatcacaaaa 1140gaagactgtg gaagttgtag
atgataaaga caaaaaatgg aataaagaat caggttggaa 1200aaaaaatcta
aatgatctag attggaaaaa agatttagat aaagataaag ttggtggcgg
1260tttgcttggc ggtttaagtg gcctcttaaa tagtttaaaa tcagaaaaag
gtcttctagg 1320tcttttgaat aagaatcaaa ttgagttatt aattccttta
atcagtgaga taaaaaagaa 1380aaatatagat tttaatctct tcgattctgt
tgattctgtc gaaagaaatt tagacttgaa 1440acttttcaca agttctgttt
caaaagttac tgaattatta aataaaggaa tcgatattca 1500aacaattttg
aatgcgaaaa atggagatga attcgattta agcggcaaag aattgaaaaa
1560cgtcaaaggg atatttggtt tgattggaag tttgaaacgc tcattaggat
tagaaaatat 1620attgaactta ccgtttaaac gtatacctct gcttaaatta
166022308PRTBombus terrestris 22Gly Gln Ser Ser Pro Leu Leu Glu Ile
Val Gln Gly Ser Ala Ser Ala1 5 10 15Thr Ala Ser Thr Ala Val Thr Ala
Arg Ser Gly Leu Arg Ala Gly Gln 20 25 30Val Ala Val Ala Ser Gln Lys
Asp Ala Thr Leu Gln Ala Asp Ala Ser 35 40 45Ala Ala Ala Ala Ala Ala
Ala Arg Ala Ser Ala Asp Gln Ser Ala Ser 50 55 60Leu Ala Gln Gln Ser
Ala Ser Leu Gln Ser Lys Ala Ala Ala Arg Ala65 70 75 80Lys Ser Ala
Glu Glu Ser Ala Ala Ala Thr Ala Lys Ala Glu Leu Gln 85 90 95Ala Glu
Ser Ile Ala Ala Ser Ala Ser Ser Asn Ala Arg Glu Ala Ala 100 105
110Ala Ser Ala Lys Ala Ser Ala Ser Ala Met Ser Ser Ala Ala Val Gln
115 120 125Ala Lys Leu Ala Glu Lys Thr Ala Lys Asn Gln Ala Leu Ala
Ser Glu 130 135 140Glu Ala Lys Leu Lys Ala Ala Ala Ala Ala Ser Ala
Ala Ala Ala Ala145 150 155 160Ser Ala Ala Ala Glu Ala Ala Leu Lys
Ala Glu Arg Ile Ala Glu Glu 165 170 175Ala Ile Ala Lys Ala Ala Ala
Ala Lys Ala Ala Ala Arg Ala Ala Ala 180 185 190Ala Ala Leu Asn Ser
Ala Lys Glu Ala Ala Thr Ser Ser Ala Arg Ser 195 200 205Ala Ala Glu
Ala Glu Ala Lys Ser Glu Val Ala Ile Leu Ile Ser Glu 210 215 220Leu
Asp Lys Lys Ser Arg Glu Val Ala Ala Ser Ala Ser Ala Lys Ala225 230
235 240Arg Ala Ala Ala Ala Ala Ser Ser Arg Asn Ala Glu Thr Ala Val
Ile 245 250 255Gly Ala Asn Ile Asn Val Ala Lys Glu Val Leu Ala Ile
Pro Ile Glu 260 265 270Pro Lys Lys Leu Pro Glu Pro Glu Leu Ala Leu
Lys Glu Glu Asn Val 275 280 285Ala Val Ala Ser Ser Glu Ser Glu Val
Lys Val Glu Thr Ser Ser Glu 290 295 300Ala Trp Ser
Ile30523327PRTBombus terrestris 23Met Lys Ile Pro Ala Leu Leu Val
Thr Cys Leu Tyr Leu Trp Gly Phe1 5 10 15Ala Ser Ala Gly Gln Ser Ser
Pro Leu Leu Glu Ile Val Gln Gly Ser 20 25 30Ala Ser Ala Thr Ala Ser
Thr Ala Val Thr Ala Arg Ser Gly Leu Arg 35 40 45Ala Gly Gln Val Ala
Val Ala Ser Gln Lys Asp Ala Thr Leu Gln Ala 50 55 60Asp Ala Ser Ala
Ala Ala Ala Ala Ala Ala Arg Ala Ser Ala Asp Gln65 70 75 80Ser Ala
Ser Leu Ala Gln Gln Ser Ala Ser Leu Gln Ser Lys Ala Ala 85 90 95Ala
Arg Ala Lys Ser Ala Glu Glu Ser Ala Ala Ala Thr Ala Lys Ala 100 105
110Glu Leu Gln Ala Glu Ser Ile Ala Ala Ser Ala Ser Ser Asn Ala Arg
115 120 125Glu Ala Ala Ala Ser Ala Lys Ala Ser Ala Ser Ala Met Ser
Ser Ala 130 135 140Ala Val Gln Ala Lys Leu Ala Glu Lys Thr Ala Lys
Asn Gln Ala Leu145 150 155 160Ala Ser Glu Glu Ala Lys Leu Lys Ala
Ala Ala Ala Ala Ser Ala Ala 165 170 175Ala Ala Ala Ser Ala Ala Ala
Glu Ala Ala Leu Lys Ala Glu Arg Ile 180 185 190Ala Glu Glu Ala Ile
Ala Lys Ala Ala Ala Ala Lys Ala Ala Ala Arg 195 200 205Ala Ala Ala
Ala Ala Leu Asn Ser Ala Lys Glu Ala Ala Thr Ser Ser 210 215 220Ala
Arg Ser Ala Ala Glu Ala Glu Ala Lys Ser Glu Val Ala Ile Leu225 230
235 240Ile Ser Glu Leu Asp Lys Lys Ser Arg Glu Val Ala Ala Ser Ala
Ser 245 250 255Ala Lys Ala Arg Ala Ala Ala Ala Ala Ser Ser Arg Asn
Ala Glu Thr 260 265 270Ala Val Ile Gly Ala Asn Ile Asn Val Ala Lys
Glu Val Leu Ala Ile 275 280 285Pro Ile Glu Pro Lys Lys Leu Pro Glu
Pro Glu Leu Ala Leu Lys Glu 290 295 300Glu Asn Val Ala Val Ala Ser
Ser Glu Ser Glu Val Lys Val Glu Thr305 310 315 320Ser Ser Glu Ala
Trp Ser Ile 32524293PRTBombus terrestris 24His Val Val Lys Arg Asp
Lys Glu Leu Lys Ala Pro Ala Leu Pro Glu1 5 10 15Leu Leu Gly Asp Gly
Ser Asp Thr Leu Gly Ala Ser Met Glu Asn Gly 20 25 30Ile Lys Val Ala
Arg Ala Ser Gln Asn Val Gly Leu Arg Thr Glu Leu 35 40 45Asn Ala Ala
Ala Arg Ala Ala Ala Ala Ala Ala Thr Lys Gln Ala Lys 50 55 60Asp Thr
Glu Ala Ala Glu Ala Gly Ala Ala Ala Ala Ile Ala Ile Ala65 70 75
80Ile Ala Lys Arg Glu Glu Ala Ile Lys Ala Ser Glu Leu Ala Ser Lys
85 90 95Leu Leu Thr Ala Ala Ala Gly Ser Ser Glu Ala Ala Val Ser Ala
Thr 100 105 110Val Arg Ala Ala Gln Leu Thr Ala Ala Ala Ser Ala Ala
Ala Lys Ala 115 120 125Ser Ala Ser Ala Ser Glu Ala Ser Ala Glu Ala
Gln Val Arg Ala Asn 130 135 140Ala Glu Ala Asn Ile Ala Lys Lys Ala
Ser Ala Ala Glu Ala Lys Ala145 150 155 160Ala Ala Glu Ala Gln Val
Lys Ala Glu Leu Ala Lys Lys Ala Ala Ala 165 170 175Gly Phe Leu Ala
Lys Ala Arg Leu Ala Ala Ser Ala Glu Ser Glu Ala 180 185 190Thr Lys
Leu Ala Ala Glu Ala Glu Val Ala Leu Ala Lys Ala Arg Val 195 200
205Ala Val Asp Gln Ser Gln Ser Ala Gln Ala Thr Ala Thr Ala Gln Ala
210 215 220Ala Thr Ala Val Gln Leu Gln Ser Gln Ala Ala Asn Ala Glu
Ala Ser225 230 235 240Ala Val Ala Gln Ala Glu Thr Leu Leu Val Thr
Ala Glu Ala Val Ser 245 250 255Ala Ala Glu Ala Glu Ala Ala Thr Lys
Ala Thr Ser Trp Gly Glu Glu 260 265 270Cys His Gln Arg Glu Lys Val
Thr Phe Ser Glu Asp Arg Leu Asn Glu 275 280 285Arg Gln Asp Asn Trp
29025313PRTBombus terrestris 25Met Lys Ile Pro Ala Ile Leu Val Thr
Ser Leu Leu Val Trp Gly Gly1 5 10 15Leu Ala Glu Gly His Val Val Lys
Arg Asp Lys Glu Leu Lys Ala Pro 20 25 30Ala Leu Pro Glu Leu Leu Gly
Asp Gly Ser Asp Thr Leu Gly Ala Ser 35 40 45Met Glu Asn Gly Ile Lys
Val Ala Arg Ala Ser Gln Asn Val Gly Leu 50 55 60Arg Thr Glu Leu Asn
Ala Ala Ala Arg Ala Ala Ala Ala Ala Ala Thr65 70 75 80Lys Gln Ala
Lys Asp Thr Glu Ala Ala Glu Ala Gly Ala Ala Ala Ala 85 90 95Ile Ala
Ile Ala Ile Ala Lys Arg Glu Glu Ala Ile Lys Ala Ser Glu 100 105
110Leu Ala Ser Lys Leu Leu Thr Ala Ala Ala Gly Ser Ser Glu Ala Ala
115 120 125Val Ser Ala Thr Val Arg Ala Ala Gln Leu Thr Ala Ala Ala
Ser Ala 130 135 140Ala Ala Lys Ala Ser Ala Ser Ala Ser Glu Ala Ser
Ala Glu Ala Gln145 150 155 160Val Arg Ala Asn Ala Glu Ala Asn Ile
Ala Lys Lys Ala Ser Ala Ala 165 170 175Glu Ala Lys Ala Ala Ala Glu
Ala Gln Val Lys Ala Glu Leu Ala Lys 180 185 190Lys Ala Ala Ala Gly
Phe Leu Ala Lys Ala Arg Leu Ala Ala Ser Ala 195 200 205Glu Ser Glu
Ala Thr Lys Leu Ala Ala Glu Ala Glu Val Ala Leu Ala 210 215 220Lys
Ala Arg Val Ala Val Asp Gln Ser Gln Ser Ala Gln Ala Thr Ala225 230
235 240Thr Ala Gln Ala Ala Thr Ala Val Gln Leu Gln Ser Gln Ala Ala
Asn 245 250 255Ala Glu Ala Ser Ala Val Ala Gln Ala Glu Thr Leu Leu
Val Thr Ala 260 265 270Glu Ala Val Ser Ala Ala Glu Ala Glu Ala Ala
Thr Lys Ala Thr Ser 275 280 285Trp Gly Glu Glu Cys His Gln Arg Glu
Lys Val Thr Phe Ser Glu Asp 290 295 300Arg Leu Asn Glu Arg Gln Asp
Asn Trp305 31026313PRTBombus terrestris 26Gly Ser Val Glu Leu Gly
Ala Pro Lys Gln Glu Ser Val Leu Val Glu1 5 10 15Gln Leu Leu Leu Lys
Asn Val Glu Thr Ser Ala Lys Arg Lys Glu Asn 20 25 30Gly Ala Pro Lys
Leu Gly Glu Ser Thr Ala Ala Ala Leu Ala Ser Thr 35 40 45Lys Ala Thr
Ala Ala Ala Glu Ala Lys Ala Ser Ala Lys Val Lys Ala 50 55 60Ser Ala
Leu Ala Leu Ala Glu Ala Phe Leu Arg Ala Ser Ala Ala Phe65 70 75
80Ala Ala Ala Ser Ala Lys Ala Ala Ala Ala Val Lys Glu Ala Thr Gln
85 90 95Ala Gln Leu Leu Ala Gln Glu Lys Ala Leu Ile Ala Leu Lys Thr
Gln 100 105 110Ser Glu Gln Gln Ala Ala Ser Ala Arg Ala Asp Ala Ala
Ala Ala Ala 115 120 125Ala Val Ser Ala Leu Glu Arg Ala Gln Ala Ser
Ser Arg Ala Ala Thr 130 135 140Thr Ala Gln Asp Ile Ser Ser Asp Leu
Glu Lys Arg Val Ala Thr Ser145 150 155 160Ala Ala Ala Glu Ala Gly
Ala Thr Leu Arg Ala Glu Gln Ser Ala Ala 165 170 175Gln Ser Lys Trp
Ser Ala Ala Leu Ala Ala Gln Thr Ala Ala Ala Ala 180 185 190Ala Ala
Ile Glu Ala Lys Ala Thr Ala Ser Ser Glu Ser Thr Ala Ala 195 200
205Ala Thr Ser Lys Ala Ala Val Leu Thr Ala Asp Thr Ser Ser Ala Glu
210 215 220Ala Ala Ala Ala Ala Glu Ala Gln Ser Ala Ser Arg Ile Ala
Gly Thr225 230 235 240Ala Ala Thr Glu Gly Ser Ala Asn Trp Ala Ser
Glu Asn Ser Arg Thr 245 250 255Ala Gln Leu Glu Ala Ser Ala Ser Ala
Lys Ala Thr Ala Ala Ala Ala 260 265 270Val Gly Asp Gly Ala Ile Ile
Gly Leu Ala Arg Asp Ala Ser Ala Ala 275 280 285Ala Gln Ala Ala Ala
Glu Val Lys Ala Leu Ala Glu Ala Ser Ala Ser 290 295 300Leu Gly Ala
Ser Glu Lys Asp Lys Lys305 31027332PRTBombus terrestris 27Met Gln
Ile Pro Ala Ile Phe Val Thr Cys Leu Leu Thr Trp Gly Leu1 5 10 15Val
His Ala Gly Ser Val Glu Leu Gly Ala Pro Lys Gln Glu Ser Val 20 25
30Leu Val Glu Gln Leu Leu Leu Lys Asn Val Glu Thr Ser Ala Lys Arg
35 40 45Lys Glu Asn Gly Ala Pro Lys Leu Gly Glu Ser Thr Ala Ala Ala
Leu 50 55 60Ala Ser Thr Lys Ala Thr Ala Ala Ala Glu Ala Lys Ala Ser
Ala Lys65 70 75 80Val Lys Ala Ser Ala Leu Ala Leu Ala Glu Ala Phe
Leu Arg Ala Ser 85 90 95Ala Ala Phe Ala Ala Ala Ser Ala Lys Ala Ala
Ala Ala Val Lys Glu 100 105 110Ala Thr Gln Ala Gln Leu Leu Ala Gln
Glu Lys Ala Leu Ile Ala Leu 115 120 125Lys Thr Gln Ser Glu Gln Gln
Ala Ala Ser Ala Arg Ala Asp Ala Ala 130 135 140Ala Ala Ala Ala Val
Ser Ala Leu Glu Arg Ala Gln Ala Ser Ser Arg145 150 155 160Ala Ala
Thr Thr Ala Gln Asp Ile Ser Ser Asp Leu Glu Lys Arg Val 165 170
175Ala Thr Ser Ala Ala Ala Glu Ala Gly Ala Thr Leu Arg Ala Glu Gln
180 185 190Ser Ala Ala Gln Ser Lys Trp Ser Ala Ala Leu Ala Ala Gln
Thr Ala 195 200 205Ala Ala Ala Ala Ala Ile Glu Ala Lys Ala Thr Ala
Ser Ser Glu Ser 210 215 220Thr Ala Ala Ala Thr Ser Lys Ala Ala Val
Leu Thr Ala Asp Thr Ser225 230 235 240Ser Ala Glu Ala Ala Ala Ala
Ala Glu Ala Gln Ser Ala Ser Arg Ile 245 250 255Ala Gly Thr Ala Ala
Thr Glu Gly Ser Ala Asn Trp Ala Ser Glu Asn 260 265 270Ser Arg Thr
Ala Gln Leu Glu Ala Ser Ala Ser Ala Lys Ala Thr Ala 275 280 285Ala
Ala Ala Val Gly Asp Gly Ala Ile Ile Gly Leu Ala Arg Asp Ala 290 295
300Ser Ala Ala Ala Gln Ala Ala Ala Glu Val Lys Ala Leu Ala Glu
Ala305 310 315 320Ser Ala Ser Leu Gly Ala Ser Glu Lys Asp Lys Lys
325 33028338PRTBombus terrestris 28Gly Lys Pro Leu Ile Ala Asn Ala
Gln Ile Gly Lys Val Lys Thr Glu1 5 10 15Thr Ser Ser Ser Ser Glu Ile
Glu Thr Leu Val Ser Gly Ser Gln Thr 20 25 30Leu Val Ala Gly Ser Glu
Thr Leu Ala Ser Glu Ser Glu Ala Leu Ala 35 40 45Ser Lys Ser Glu Ala
Leu Thr Ser Glu Ala Glu Ile Ala Ser Val Thr 50 55 60Thr Lys Asp Glu
Leu Ile Leu Lys Gly Glu Ala Ile Thr Gly Lys Lys65 70 75 80Leu Gly
Thr Gly Ala Ser Glu Val Ala Ala Ala Ser Gly Glu Ala Ile 85 90 95Ala
Thr Thr Leu Gly Ala Gly Gln Ala Ala Ala Glu Ala Gln Ala Ala 100 105
110Ala Ala Ala Gln Ala Lys Ser Ala Ala Ala Ala Ala Ala Asn Ala Gly
115 120 125Glu Ser Ser Asn Ser Ala Ala Ala Leu Val Ala Ala Ala Ala
Ala Ala 130 135 140Gln Gly Lys Ala Ala Ala Ala Ala Ala Ala Ala Thr
Lys Ala Ser Leu145 150 155 160Glu Ala Ala Asp Ala Ala Glu Glu Ala
Glu Ser Ala Val Ala Leu Ala 165 170 175Arg Ala Ala Ser Ala Lys Ala
Glu Ala Leu Ala Ser Thr Ala Ala Ala 180 185 190Ala Asn Thr Arg Ala
Ala Leu Gln Ala Glu Lys Ser Asn Glu Leu Ala 195 200 205Gln Ala Glu
Ala Ala Ala Ala Ala Glu Ala Gln Ala Lys Ala Ala Ala 210 215 220Ala
Ala Lys Ala Thr Gln Leu Ala Leu Lys Val Ala Glu Thr Ala Val225
230 235 240Lys Thr Glu Ala Asp Ala Ala Ala Ala Ala Val Ala Ala Ala
Lys Ala 245 250 255Arg Ala Val Ala Asp Ala Ala Ala Ser Arg Ala Thr
Ala Val Asn Ala 260 265 270Ile Ala Glu Ala Glu Glu Arg Asp Ser Ala
Gln Ala Glu Asn Thr Ala 275 280 285Gly Val Ala Gln Ala Ala Leu Ala
Ala Ala Glu Ala Gln Asp Ser Cys 290 295 300Ile Gly Ala Ala Ala Thr
Pro Arg His Ser Ser Ser Tyr Ala Trp Trp305 310 315 320Lys Leu Arg
Ile Thr Ser Leu Ile Val Ile Leu Ser Pro Arg Asn Arg 325 330 335Arg
Thr29357PRTBombus terrestris 29Met Lys Ile Pro Ser Ile Leu Ala Val
Ser Leu Leu Val Trp Gly Leu1 5 10 15Ala Ser Ala Gly Lys Pro Leu Ile
Ala Asn Ala Gln Ile Gly Lys Val 20 25 30Lys Thr Glu Thr Ser Ser Ser
Ser Glu Ile Glu Thr Leu Val Ser Gly 35 40 45Ser Gln Thr Leu Val Ala
Gly Ser Glu Thr Leu Ala Ser Glu Ser Glu 50 55 60Ala Leu Ala Ser Lys
Ser Glu Ala Leu Thr Ser Glu Ala Glu Ile Ala65 70 75 80Ser Val Thr
Thr Lys Asp Glu Leu Ile Leu Lys Gly Glu Ala Ile Thr 85 90 95Gly Lys
Lys Leu Gly Thr Gly Ala Ser Glu Val Ala Ala Ala Ser Gly 100 105
110Glu Ala Ile Ala Thr Thr Leu Gly Ala Gly Gln Ala Ala Ala Glu Ala
115 120 125Gln Ala Ala Ala Ala Ala Gln Ala Lys Ser Ala Ala Ala Ala
Ala Ala 130 135 140Asn Ala Gly Glu Ser Ser Asn Ser Ala Ala Ala Leu
Val Ala Ala Ala145 150 155 160Ala Ala Ala Gln Gly Lys Ala Ala Ala
Ala Ala Ala Ala Ala Thr Lys 165 170 175Ala Ser Leu Glu Ala Ala Asp
Ala Ala Glu Glu Ala Glu Ser Ala Val 180 185 190Ala Leu Ala Arg Ala
Ala Ser Ala Lys Ala Glu Ala Leu Ala Ser Thr 195 200 205Ala Ala Ala
Ala Asn Thr Arg Ala Ala Leu Gln Ala Glu Lys Ser Asn 210 215 220Glu
Leu Ala Gln Ala Glu Ala Ala Ala Ala Ala Glu Ala Gln Ala Lys225 230
235 240Ala Ala Ala Ala Ala Lys Ala Thr Gln Leu Ala Leu Lys Val Ala
Glu 245 250 255Thr Ala Val Lys Thr Glu Ala Asp Ala Ala Ala Ala Ala
Val Ala Ala 260 265 270Ala Lys Ala Arg Ala Val Ala Asp Ala Ala Ala
Ser Arg Ala Thr Ala 275 280 285Val Asn Ala Ile Ala Glu Ala Glu Glu
Arg Asp Ser Ala Gln Ala Glu 290 295 300Asn Thr Ala Gly Val Ala Gln
Ala Ala Leu Ala Ala Ala Glu Ala Gln305 310 315 320Asp Ser Cys Ile
Gly Ala Ala Ala Thr Pro Arg His Ser Ser Ser Tyr 325 330 335Ala Trp
Trp Lys Leu Arg Ile Thr Ser Leu Ile Val Ile Leu Ser Pro 340 345
350Arg Asn Arg Arg Thr 35530422PRTBombus terrestris 30Gly Asn Ser
Glu Ser Gly Glu Asn Trp Lys Asn Gly Glu Ser Ser Glu1 5 10 15Ser Gly
Lys Asn Trp Arg Asn Ser Gly Ser Ser Glu Ser Gly Lys Asn 20 25 30Trp
Lys Asn Gly Gly Ser Ser Glu Ser Asn Lys His Trp Lys Asn Gly 35 40
45Gly Ser Ser Glu Ser Gly Glu Lys Trp Lys Asn Ser Glu Ser Ser Glu
50 55 60Ser Gly Lys Asn Trp Lys Asn Ser Gly Ser Ser Glu Ser Gly Lys
Asn65 70 75 80Trp Lys Asn Gly Gly Ser Ser Glu Ser Asn Lys His Trp
Lys Ser Gly 85 90 95Gly Ser Ser Glu Ser Gly Glu Lys Trp Lys Asn Ser
Glu Ser Gly Asn 100 105 110Lys Gly Lys Ser Ser Lys Ser Ser Glu Ser
Trp Lys Ser Asn Glu Asn 115 120 125Ser Lys Asn Asp Gly Ser Trp Lys
Ser Ser Glu Glu Ser Glu Lys Trp 130 135 140Lys Asp Gly Lys Ala Val
Ala Glu Asp Ser Val Ser Ile Asn Trp Ala145 150 155 160Asp Val Lys
Glu Gln Ile Ser Asn Ile Ala Thr Ser Leu Glu Lys Gly 165 170 175Gly
Asn Leu Glu Ala Val Leu Lys Ile Lys Lys Gly Glu Lys Lys Ile 180 185
190Ser Ser Leu Glu Glu Ile Lys Glu Lys Ile Ser Val Leu Leu Lys Trp
195 200 205Ile Gln Glu Gly Lys Asp Thr Ser Ser Leu Leu Asp Leu Lys
Glu Gly 210 215 220Ser Lys Asp Ile Ala Ser Leu Lys Glu Ile Lys Gly
Lys Ile Leu Leu225 230 235 240Ile Val Lys Leu Val Asn Glu Gly Lys
Asp Thr Ser Gly Leu Leu Asp 245 250 255Leu Glu Ala Ser Gly Lys Val
Ile Leu Glu Leu Gln Ser Ala Ile Glu 260 265 270Lys Val Leu Val Lys
Ser Glu Lys Val Thr Lys Val Ser Glu Val Ser 275 280 285Gly Leu Val
Lys Ser Lys Thr Val Ser Asp Ile Lys Pro Leu Gln Ala 290 295 300Val
Ile Pro Leu Ile Leu Glu Leu Gln Lys Thr Asp Ile Asn Leu Ser305 310
315 320Thr Leu Asn Lys Trp Ser Thr Val Asn Val Asn Ser Ile Asp Lys
Glu 325 330 335Arg Val Thr Lys Thr Val Pro Val Leu Leu Gln Ser Met
Lys Gly Gly 340 345 350Glu Asp Ile Gln Asn Leu Leu Ser Ala Lys Gly
Ala Lys Lys Leu Gly 355 360 365Ile Ser Ala Leu Asp Leu Gln Ala Val
Gln Gly Ala Leu Gly Val Val 370 375 380Gly Lys Leu Ser Ser Gly Gly
Ala Leu Asn Ser Lys Gly Leu Leu Asn385 390 395 400Leu Lys Asp Gly
Ala Ser Val Leu Gly Ala Gly Lys Ile Gly Gly Leu 405 410 415Ile Pro
Leu Pro Lys Leu 42031927DNABombus terrestris 31ggccagagct
cacctctgct cgagatcgtg cagggtagcg cgtcggccac cgcatccacc 60gctgtgaccg
ctagatccgg acttcgtgcc ggtcaggtag ccgtggcctc gcagaaggat
120gccacacttc aggcagatgc ctcagcggcc gccgcggccg ctgcacgcgc
ttccgccgac 180cagtcggcca gtctagccca acagtcggcg tctttgcagt
ccaaagctgc cgccagagca 240aaatcagccg aggagtcagc ggcagctacg
gccaaagccg agttgcaggc agaatccatt 300gctgcatctg ccagttccaa
tgccagagag gctgcagcgt ccgcaaaagc ctccgcatcc 360gcgatgtcat
cggctgccgt gcaggcgaaa ctcgctgaaa agacggccaa gaatcaagct
420ctggcttccg aagaagccaa actcaaggct gccgccgctg ccagcgcagc
agcagcagcc 480agcgccgccg ccgaggcagc cctgaaagct gagagaatag
cggaagaagc catcgccaag 540gcggccgctg ccaaagcagc cgccagagcc
gctgcagccg cgttaaactc cgcgaaggaa 600gccgccacga gcagcgcaag
gagcgccgcc gaagccgaag ctaagagcga agtcgctata 660ctgatcagcg
aactcgacaa gaagagcagg gaagtcgccg cttccgcgtc cgccaaggca
720cgcgctgctg ctgcggctag ctccagaaac gcagaaacgg ctgttatcgg
agctaacatc 780aatgtggcca aagaggtctt ggcgattccc atcgagccaa
agaaacttcc ggagccagag 840ctggcgttga aagaagagaa tgtcgcggtc
gcgagctcag agagtgaagt gaaggtagaa 900acgagcagcg aagcatggtc aatttaa
92732984DNABombus terrestris 32atgaagattc cagcactgct cgtaacgtgc
ctctaccttt ggggcttcgc gtccgccggc 60cagagctcac ctctgctcga gatcgtgcag
ggtagcgcgt cggccaccgc atccaccgct 120gtgaccgcta gatccggact
tcgtgccggt caggtagccg tggcctcgca gaaggatgcc 180acacttcagg
cagatgcctc agcggccgcc gcggccgctg cacgcgcttc cgccgaccag
240tcggccagtc tagcccaaca gtcggcgtct ttgcagtcca aagctgccgc
cagagcaaaa 300tcagccgagg agtcagcggc agctacggcc aaagccgagt
tgcaggcaga atccattgct 360gcatctgcca gttccaatgc cagagaggct
gcagcgtccg caaaagcctc cgcatccgcg 420atgtcatcgg ctgccgtgca
ggcgaaactc gctgaaaaga cggccaagaa tcaagctctg 480gcttccgaag
aagccaaact caaggctgcc gccgctgcca gcgcagcagc agcagccagc
540gccgccgccg aggcagccct gaaagctgag agaatagcgg aagaagccat
cgccaaggcg 600gccgctgcca aagcagccgc cagagccgct gcagccgcgt
taaactccgc gaaggaagcc 660gccacgagca gcgcaaggag cgccgccgaa
gccgaagcta agagcgaagt cgctatactg 720atcagcgaac tcgacaagaa
gagcagggaa gtcgccgctt ccgcgtccgc caaggcacgc 780gctgctgctg
cggctagctc cagaaacgca gaaacggctg ttatcggagc taacatcaat
840gtggccaaag aggtcttggc gattcccatc gagccaaaga aacttccgga
gccagagctg 900gcgttgaaag aagagaatgt cgcggtcgcg agctcagaga
gtgaagtgaa ggtagaaacg 960agcagcgaag catggtcaat ttaa
98433882DNABombus terrestris 33cacgtggtga agcgcgacaa ggagctcaag
gccccggctt taccggaact actcggtgat 60gggtctgaca cgctcggtgc ctcgatggag
aacgggatca aagtcgccag agcatcgcag 120aatgtgggtc tgagaacaga
gttgaatgca gccgcgcggg ctgcagccgc tgctgcgacc 180aagcaggcca
aagacacaga ggccgcggaa gctggagcgg ccgctgcgat tgccatcgct
240atcgccaagc gtgaagaagc tatcaaagca agcgaattag ccagcaagtt
gttgacagcc 300gcggctgggt ccagcgaagc tgccgtgtca gcgacggtga
gggcggcgca attgacggcc 360gcagctagcg cagctgccaa agcttctgca
tccgcctctg aggcttctgc cgaagcccag 420gtgagggcca acgccgaagc
aaacatcgcc aagaaagctt cggcagctga agcaaaagcc 480gcagccgaag
cccaggttaa ggcggaactc gccaagaaag cggccgccgg tttcttagct
540aaggctagac tagcggccag cgccgaatcc gaggccacta aactcgcagc
cgaagctgaa 600gtagcactgg ctaaggccag agtcgccgtc gaccagtcgc
agagcgcaca ggcaaccgct 660accgctcaag ctgccacagc cgttcagctg
cagtctcaag cagctaacgc ggaagcctcc 720gctgtagcac aggctgaaac
tctgctggtc acggcggaag ccgtctctgc cgcggaagcc 780gaagccgcga
ccaaagctac cagttggggc gaagaatgtc atcaacgaga aaaagttacg
840tttagcgaag atcgattaaa cgagagacaa gacaattggt ag 88234942DNABombus
terrestris 34atgaagattc cagcaatact ggttacgtct ctgctggtct ggggtggtct
ggccgagggc 60cacgtggtga agcgcgacaa ggagctcaag gccccggctt taccggaact
actcggtgat 120gggtctgaca cgctcggtgc ctcgatggag aacgggatca
aagtcgccag agcatcgcag 180aatgtgggtc tgagaacaga gttgaatgca
gccgcgcggg ctgcagccgc tgctgcgacc 240aagcaggcca aagacacaga
ggccgcggaa gctggagcgg ccgctgcgat tgccatcgct 300atcgccaagc
gtgaagaagc tatcaaagca agcgaattag ccagcaagtt gttgacagcc
360gcggctgggt ccagcgaagc tgccgtgtca gcgacggtga gggcggcgca
attgacggcc 420gcagctagcg cagctgccaa agcttctgca tccgcctctg
aggcttctgc cgaagcccag 480gtgagggcca acgccgaagc aaacatcgcc
aagaaagctt cggcagctga agcaaaagcc 540gcagccgaag cccaggttaa
ggcggaactc gccaagaaag cggccgccgg tttcttagct 600aaggctagac
tagcggccag cgccgaatcc gaggccacta aactcgcagc cgaagctgaa
660gtagcactgg ctaaggccag agtcgccgtc gaccagtcgc agagcgcaca
ggcaaccgct 720accgctcaag ctgccacagc cgttcagctg cagtctcaag
cagctaacgc ggaagcctcc 780gctgtagcac aggctgaaac tctgctggtc
acggcggaag ccgtctctgc cgcggaagcc 840gaagccgcga ccaaagctac
cagttggggc gaagaatgtc atcaacgaga aaaagttacg 900tttagcgaag
atcgattaaa cgagagacaa gacaattggt ag 94235942DNABombus terrestris
35ggtagcgtgg aactcggtgc ccccaagcag gagtctgtcc tcgtggagca gctcctattg
60aagaacgtgg agactagtgc gaagcgaaag gagaacggcg caccgaaact cggcgagagc
120acagctgcgg ctctggctag taccaaggca actgcagccg cagaggctaa
ggcatccgcc 180aaagtgaaag cttctgcctt ggccctcgct gaggctttct
tgcgtgcgtc ggcagcgttt 240gctgctgctt cagccaaagc tgctgccgct
gtaaaggaag caacgcaggc acagttgctg 300gcacaggaga aggctttgat
agcgttgaaa actcaatctg agcaacaagc tgcctctgct 360cgcgcggacg
ccgcggctgc cgcagccgta tccgcgctag aacgcgccca ggcctcctcc
420agagcagcca cgaccgccca agacatctcc agcgatctgg agaaacgtgt
cgccacctca 480gccgctgctg aagcaggtgc caccctcaga gcggaacaat
ccgccgcgca atcgaaatgg 540tccgccgcac tggccgccca aaccgccgct
gctgcagccg ctatagaagc aaaggccacc 600gcttcctcag aaagcaccgc
tgccgctact agtaaggccg ccgtgttgac cgctgacact 660agcagcgcag
aagctgccgc tgcagcggag gcacaatccg cttcgcggat cgcaggtaca
720gcagccaccg agggatccgc caactgggct agcgagaact cgcgtaccgc
acaactggaa 780gcttccgcct cagcgaaggc caccgcagcc gcagctgtcg
gagatggagc tattatagga 840cttgcacggg acgctagtgc cgcagctcag
gcagccgcag aagttaaagc cttagctgaa 900gctagtgcca gcttaggtgc
ttcagaaaag gacaagaaat ga 94236999DNABombus terrestris 36atgcagatcc
cagcgatttt cgtcacgtgc ctgctcacat ggggcctggt gcacgcaggt 60agcgtggaac
tcggtgcccc caagcaggag tctgtcctcg tggagcagct cctattgaag
120aacgtggaga ctagtgcgaa gcgaaaggag aacggcgcac cgaaactcgg
cgagagcaca 180gctgcggctc tggctagtac caaggcaact gcagccgcag
aggctaaggc atccgccaaa 240gtgaaagctt ctgccttggc cctcgctgag
gctttcttgc gtgcgtcggc agcgtttgct 300gctgcttcag ccaaagctgc
tgccgctgta aaggaagcaa cgcaggcaca gttgctggca 360caggagaagg
ctttgatagc gttgaaaact caatctgagc aacaagctgc ctctgctcgc
420gcggacgccg cggctgccgc agccgtatcc gcgctagaac gcgcccaggc
ctcctccaga 480gcagccacga ccgcccaaga catctccagc gatctggaga
aacgtgtcgc cacctcagcc 540gctgctgaag caggtgccac cctcagagcg
gaacaatccg ccgcgcaatc gaaatggtcc 600gccgcactgg ccgcccaaac
cgccgctgct gcagccgcta tagaagcaaa ggccaccgct 660tcctcagaaa
gcaccgctgc cgctactagt aaggccgccg tgttgaccgc tgacactagc
720agcgcagaag ctgccgctgc agcggaggca caatccgctt cgcggatcgc
aggtacagca 780gccaccgagg gatccgccaa ctgggctagc gagaactcgc
gtaccgcaca actggaagct 840tccgcctcag cgaaggccac cgcagccgca
gctgtcggag atggagctat tataggactt 900gcacgggacg ctagtgccgc
agctcaggca gccgcagaag ttaaagcctt agctgaagct 960agtgccagct
taggtgcttc agaaaaggac aagaaatga 999371017DNABombus terrestris
37ggcaaaccac tcattgccaa tgcgcaaata gggaaggtca agaccgaaac gtcatcgtct
60tcagagattg agacgttggt atcaggaagc cagacattgg tggcaggaag tgagacattg
120gcttcagaaa gcgaggcatt ggcgtcaaaa agcgaggcat tgacgtcaga
agccgagata 180gcgagcgtga caacgaagga cgagctcata ctaaagggcg
aagctatcac tggaaagaaa 240ctaggaaccg gggcgtcgga agtagcggcg
gcctctgggg aggctatcgc aactaccctt 300ggcgcgggac aagctgcagc
agaggcacaa gcagccgccg ccgcgcaagc aaaatcagca 360gcggcagctg
ccgcgaatgc aggtgaatcc agcaacagtg ctgctgcgtt ggttgctgct
420gcagctgcag cacaaggaaa agcggctgcc gccgcagcag ccgcgacgaa
ggctagctta 480gaggccgcag acgctgctga ggaagctgag tcggccgtgg
ccttggctag ggctgcctcc 540gcaaaggcgg aagcgctcgc atcgaccgcc
gctgctgcga atacccgtgc tgctctccaa 600gcggaaaaat cgaacgagct
ggcgcaagct gaggctgcag ccgccgccga agcccaggct 660aaagccgccg
ctgctgccaa ggcaacacaa ctcgccctta aagttgccga aactgcggtg
720aaaacggaag cagatgcagc agctgccgcc gttgcggccg caaaagccag
agcagtcgca 780gacgcagccg cgtctcgtgc gaccgcagtg aacgccattg
ctgaagcgga agaaagagac 840tctgcacagg cggagaacac cgctggtgta
gcacaagcag cgctcgctgc tgcggaagca 900caagactcct gcatcggcgc
tgccgcgact cctaggcatt cgtcgagcta tgcatggtgg 960aagcttagga
taacatcctt gatcgtcatt ctatcgccac gcaatcgacg tacttaa
1017381074DNABombus terrestris 38atgaagattc catcgatact cgcggtgtcc
ctgctggttt ggggtctggc cagcgcaggc 60aaaccactca ttgccaatgc gcaaataggg
aaggtcaaga ccgaaacgtc atcgtcttca 120gagattgaga cgttggtatc
aggaagccag acattggtgg caggaagtga gacattggct 180tcagaaagcg
aggcattggc gtcaaaaagc gaggcattga cgtcagaagc cgagatagcg
240agcgtgacaa cgaaggacga gctcatacta aagggcgaag ctatcactgg
aaagaaacta 300ggaaccgggg cgtcggaagt agcggcggcc tctggggagg
ctatcgcaac tacccttggc 360gcgggacaag ctgcagcaga ggcacaagca
gccgccgccg cgcaagcaaa atcagcagcg 420gcagctgccg cgaatgcagg
tgaatccagc aacagtgctg ctgcgttggt tgctgctgca 480gctgcagcac
aaggaaaagc ggctgccgcc gcagcagccg cgacgaaggc tagcttagag
540gccgcagacg ctgctgagga agctgagtcg gccgtggcct tggctagggc
tgcctccgca 600aaggcggaag cgctcgcatc gaccgccgct gctgcgaata
cccgtgctgc tctccaagcg 660gaaaaatcga acgagctggc gcaagctgag
gctgcagccg ccgccgaagc ccaggctaaa 720gccgccgctg ctgccaaggc
aacacaactc gcccttaaag ttgccgaaac tgcggtgaaa 780acggaagcag
atgcagcagc tgccgccgtt gcggccgcaa aagccagagc agtcgcagac
840gcagccgcgt ctcgtgcgac cgcagtgaac gccattgctg aagcggaaga
aagagactct 900gcacaggcgg agaacaccgc tggtgtagca caagcagcgc
tcgctgctgc ggaagcacaa 960gactcctgca tcggcgctgc cgcgactcct
aggcattcgt cgagctatgc atggtggaag 1020cttaggataa catccttgat
cgtcattcta tcgccacgca atcgacgtac ttaa 1074391411DNABombus
terrestris 39ggaaattcgg aaagcggcga aaattggaag aacggtgaaa gctccgaaag
cggcaaaaat 60tggaggaaca gcggaagctc cgaaagcggc aaaaattgga agaatggcgg
aagctcagaa 120agcaacaaac attggaagaa cggtggaagc tcggaaagcg
gcgagaaatg gaaaaacagt 180gaaagctccg aaagcggcaa aaattggaag
aacagcggaa gctccgaaag cggcaaaaat 240tggaaaaacg gcggaagctc
ggaaagcaac aaacattgga agagcggtgg aagctcggaa 300agtggcgaga
aatggaaaaa cagtgaaagc ggaaataaag gcaaaagctc aaaaagcagc
360gaaagttgga agagcaacga aaactcgaag aacgacggca gctggaagag
cagtgaagaa 420tcagaaaagt ggaaagatgg taaagcagtg gcggaagaca
gcgttagtat aaactgggca 480gatgtcaaag agcagattag caacattgct
acatccttag aaaagggtgg taacctcgag 540gctgtattga aaataaagaa
aggagaaaag aaaatttcaa gtttggagga aatcaaggag 600aaaatctctg
tcctactgaa atggattcaa gaaggcaaag atactagcag cctattagat
660ttgaaagagg gtagcaagga tattgcgtcg ttgaaagaaa tcaaaggaaa
gatccttttg 720attgttaaat tagtgaacga agggaaagac actagtggtc
ttttagattt agaagcgagt 780ggcaaagtaa ttttagaatt gcaaagcgcc
atagaaaagg ttctcgtaaa gtcagaaaag 840gtaaccaaag tatctgaagt
ttccggttta gtaaaaagca aaactgtctc ggacataaaa 900ccgcttcaag
cagtaattcc tttaatcctt gaattgcaaa aaacagacat taaccttagt
960accttaaaca agtggtccac tgttaacgta aattctatag ataaagaacg
cgtcacgaaa 1020acggttccag tgctccttca atccatgaaa ggaggcgaag
atattcagaa ccttttgagt 1080gcgaaaggtg caaagaaact tggcattagt
gctttggact tacaggcagt tcaaggagct 1140cttggcgtgg ttggaaagct
aagttcaggt ggtgcgttga actcaaaagg cttgttgaac 1200ttgaaagacg
gcgctagtgt gttaggtgca ggaaaaatcg gaggattaat tcctttaccg
1260aaactttaag agatagaccg ataaaggcag atatactctc ggaagatttt
tttggaagtt 1320gaatagtccg caaaaaaatt atctctgatt attataattt
agcctaaaat attaaataaa 1380atggagaaat aacgttgaaa tatataaata a
141140403PRTMyrmecia forficata 40Ser Gly Pro Arg Leu Leu Gly Gly
Arg Ser Ala Ala Ser Ala Ser Ala1 5 10 15Ser Ala Ser Ala Glu Ala Ser
Ala Gly Gly Trp Arg Lys Ser Gly Ala 20 25 30Ser Ala Ser Ala Ser Ala
Lys Ala Gly Ser Ser Asn Ile Leu Ser Arg 35 40 45Val Gly Ala Ser Arg
Ala Ala Ala Thr Leu Val Ala Ser Ala Ala Val 50 55 60Glu Ala Lys Ala
Gly Leu Arg Ala Gly Lys Ala Thr Ala Glu Glu Gln65 70 75 80Arg Glu
Ala Leu Glu Met Leu Thr Leu Ser Ala Asp Lys Asn Ala Glu 85 90 95Ala
Arg Ile Leu Ala Asp Asp Thr Ala Val Leu Val Gln Gly Ser Ala 100 105
110Glu Ala Gln Ser Val Ala Ala Ala Lys Thr Val Ala Val Glu Glu Glu
115 120 125Ser Ala Ser Leu Asp Ala Ala Ala Val Glu Ala Glu Val Ala
Ala Ala 130 135 140Thr Ser Lys Ser Ser Ala Gly Gln Ala Leu Gln Ser
Ala Gln Thr Ala145 150 155 160Ala Ser Ala Leu Arg Thr Ser Ala Arg
Ser Ala Leu Thr Ala Leu Lys 165 170 175Leu Ala Arg Leu Gln Gly Ala
Ala Ser Ser Asn Ala Ala Arg Met Met 180 185 190Glu Lys Ala Leu Ala
Ala Thr Gln Asp Ala Asn Ala Ala Ala Gln Gln 195 200 205Ala Met Ala
Ala Glu Ser Ala Ala Ala Glu Ala Ala Ala Ile Ala Ala 210 215 220Ala
Lys Gln Ser Glu Ala Arg Asp Ala Gly Ala Glu Ala Lys Ala Ala225 230
235 240Met Ala Ala Leu Ile Thr Ala Gln Arg Asn Leu Val Gln Ala Asn
Ala 245 250 255Arg Ala Glu Met Ala Ser Glu Glu Ala Glu Leu Asp Ser
Lys Ser Arg 260 265 270Ala Ser Asp Ala Lys Val Asn Ala Val Ala Arg
Ala Ala Ser Lys Ser 275 280 285Ser Ile Arg Arg Asp Glu Leu Ile Glu
Ile Gly Ala Glu Phe Gly Lys 290 295 300Ala Ser Gly Glu Val Ile Ser
Thr Gly Thr Arg Ser Asn Gly Gly Gln305 310 315 320Asp Ala Ile Ala
Thr Ala Glu Ala Ser Ser Ser Ala Ser Ala Val Gly 325 330 335Ile Lys
Lys Thr Ser Gly His Trp Gly Ser Gly Lys Trp Ser Arg Val 340 345
350Ser Lys Gly Lys Gly Trp Ala Ser Ser Asn Ala Asp Ala Asp Ala Ser
355 360 365Ser Ser Ser Ile Ile Ile Gly Gly Leu Lys Arg Gly Gly Leu
Gly Ser 370 375 380Glu Ala Ser Ala Ala Ala Ser Ala Glu Ala Glu Ala
Ser Ala Gly Thr385 390 395 400Leu Leu Leu41422PRTMyrmecia forficata
41Met Lys Ile Pro Ala Ile Ile Ala Thr Ser Leu Leu Leu Trp Gly Phe1
5 10 15Ala Ser Ala Ser Gly Pro Arg Leu Leu Gly Gly Arg Ser Ala Ala
Ser 20 25 30Ala Ser Ala Ser Ala Ser Ala Glu Ala Ser Ala Gly Gly Trp
Arg Lys 35 40 45Ser Gly Ala Ser Ala Ser Ala Ser Ala Lys Ala Gly Ser
Ser Asn Ile 50 55 60Leu Ser Arg Val Gly Ala Ser Arg Ala Ala Ala Thr
Leu Val Ala Ser65 70 75 80Ala Ala Val Glu Ala Lys Ala Gly Leu Arg
Ala Gly Lys Ala Thr Ala 85 90 95Glu Glu Gln Arg Glu Ala Leu Glu Met
Leu Thr Leu Ser Ala Asp Lys 100 105 110Asn Ala Glu Ala Arg Ile Leu
Ala Asp Asp Thr Ala Val Leu Val Gln 115 120 125Gly Ser Ala Glu Ala
Gln Ser Val Ala Ala Ala Lys Thr Val Ala Val 130 135 140Glu Glu Glu
Ser Ala Ser Leu Asp Ala Ala Ala Val Glu Ala Glu Val145 150 155
160Ala Ala Ala Thr Ser Lys Ser Ser Ala Gly Gln Ala Leu Gln Ser Ala
165 170 175Gln Thr Ala Ala Ser Ala Leu Arg Thr Ser Ala Arg Ser Ala
Leu Thr 180 185 190Ala Leu Lys Leu Ala Arg Leu Gln Gly Ala Ala Ser
Ser Asn Ala Ala 195 200 205Arg Met Met Glu Lys Ala Leu Ala Ala Thr
Gln Asp Ala Asn Ala Ala 210 215 220Ala Gln Gln Ala Met Ala Ala Glu
Ser Ala Ala Ala Glu Ala Ala Ala225 230 235 240Ile Ala Ala Ala Lys
Gln Ser Glu Ala Arg Asp Ala Gly Ala Glu Ala 245 250 255Lys Ala Ala
Met Ala Ala Leu Ile Thr Ala Gln Arg Asn Leu Val Gln 260 265 270Ala
Asn Ala Arg Ala Glu Met Ala Ser Glu Glu Ala Glu Leu Asp Ser 275 280
285Lys Ser Arg Ala Ser Asp Ala Lys Val Asn Ala Val Ala Arg Ala Ala
290 295 300Ser Lys Ser Ser Ile Arg Arg Asp Glu Leu Ile Glu Ile Gly
Ala Glu305 310 315 320Phe Gly Lys Ala Ser Gly Glu Val Ile Ser Thr
Gly Thr Arg Ser Asn 325 330 335Gly Gly Gln Asp Ala Ile Ala Thr Ala
Glu Ala Ser Ser Ser Ala Ser 340 345 350Ala Val Gly Ile Lys Lys Thr
Ser Gly His Trp Gly Ser Gly Lys Trp 355 360 365Ser Arg Val Ser Lys
Gly Lys Gly Trp Ala Ser Ser Asn Ala Asp Ala 370 375 380Asp Ala Ser
Ser Ser Ser Ile Ile Ile Gly Gly Leu Lys Arg Gly Gly385 390 395
400Leu Gly Ser Glu Ala Ser Ala Ala Ala Ser Ala Glu Ala Glu Ala Ser
405 410 415Ala Gly Thr Leu Leu Leu 42042392PRTMyrmecia forficata
42Arg Val Ile Glu Ser Ser Ser Ser Ala Ser Ala Gln Ala Ser Ala Ser1
5 10 15Ala Gly Ser Arg Gly Leu Leu Gly Lys Arg Pro Ile Gly Lys Leu
Glu 20 25 30Trp Gly Lys Glu Glu Lys Lys Leu Glu Glu Leu Asp Glu Glu
Ser Leu 35 40 45Asn Glu Ala Ala Leu Lys Val Gly Ile Lys Asn Gly Gly
Leu Asp Val 50 55 60Ala Lys Gly Ala Ala Val Leu Glu Ala Ala Met Ser
Asp Val Ala Thr65 70 75 80Leu Thr Asp Gln Arg Ser Leu Val Asp Leu
Gly Leu Gly Pro Val Ala 85 90 95Asn Glu Ala Glu Ile Leu Ala Glu Ala
Gln Ala Ala Thr Ser Ala Gln 100 105 110Ala Gly Ala Val Ala Asn Ser
Ala Ala Glu Arg Ala Ile Ala Ala Met 115 120 125Glu Met Ala Asp Arg
Thr Glu Tyr Ile Ala Ala Leu Val Thr Thr Lys 130 135 140Ala Ala Lys
Ala Ala Glu Ala Thr Met Ala Ala Thr Ala Arg Ala Thr145 150 155
160Ala Ala Ala Ser Ala Ser Lys Ile Ser Ser Gln Glu Ser Ala Ala Ser
165 170 175Ala Ala Asn Ala Ala Asn Ala Glu Ala Lys Ala Asn Ala Ala
Ser Ile 180 185 190Ile Ala Asn Lys Ala Asn Ala Val Leu Ala Glu Ala
Ala Ala Val Leu 195 200 205Ala Ala Thr Ala Ala Lys Ala Lys Glu Ser
Ala Met Lys Ser Leu Ser 210 215 220Ala Ala Gln Ala Ala Ala Lys Ala
Gln Ala Arg Asn Ala Glu Ala Ser225 230 235 240Ala Glu Ala Gln Ile
Lys Leu Ser Gln Ala Arg Ala Ala Val Ala Arg 245 250 255Ala Ala Ala
Asp Gln Ala Val Cys Ser Ser Gln Ala Gln Ala Ala Ser 260 265 270Gln
Ile Gln Ser Arg Ala Ser Ala Ser Glu Ser Ala Ala Ser Ala Gln 275 280
285Ser Glu Thr Asn Thr Ala Ala Ala Glu Ala Val Ala Thr Ala Asp Ala
290 295 300Glu Ala Ala Ala Gln Ala Glu Ala Trp Val Met Ser Leu Lys
Asn Asp305 310 315 320Leu Trp Leu His Leu Asn Met Lys Gly Glu Ala
Lys Ala Glu Gly Glu 325 330 335Ala Val Ser Ile Ser Lys Gly His Arg
Gly Gly Ile Arg Ser Gly Ser 340 345 350Ile Ser Glu Ala Ser Ala Glu
Ala Ser Ser Asn Val Ser Met Gly Gly 355 360 365Arg His Gly Arg Lys
Asp Leu Val Ser Glu Ala Leu Ala Gly Ala Ser 370 375 380Ala Gly Ser
Ser Ala Asp Ser Leu385 39043411PRTMyrmecia forficata 43Met Lys Ile
Pro Ala Ile Leu Val Thr Ser Leu Leu Ala Trp Gly Leu1 5 10 15Ala Ser
Gly Arg Val Ile Glu Ser Ser Ser Ser Ala Ser Ala Gln Ala 20 25 30Ser
Ala Ser Ala Gly Ser Arg Gly Leu Leu Gly Lys Arg Pro Ile Gly 35 40
45Lys Leu Glu Trp Gly Lys Glu Glu Lys Lys Leu Glu Glu Leu Asp Glu
50 55 60Glu Ser Leu Asn Glu Ala Ala Leu Lys Val Gly Ile Lys Asn Gly
Gly65 70 75 80Leu Asp Val Ala Lys Gly Ala Ala Val Leu Glu Ala Ala
Met Ser Asp 85 90 95Val Ala Thr Leu Thr Asp Gln Arg Ser Leu Val Asp
Leu Gly Leu Gly 100 105 110Pro Val Ala Asn Glu Ala Glu Ile Leu Ala
Glu Ala Gln Ala Ala Thr 115 120 125Ser Ala Gln Ala Gly Ala Val Ala
Asn Ser Ala Ala Glu Arg Ala Ile 130 135 140Ala Ala Met Glu Met Ala
Asp Arg Thr Glu Tyr Ile Ala Ala Leu Val145 150 155 160Thr Thr Lys
Ala Ala Lys Ala Ala Glu Ala Thr Met Ala Ala Thr Ala 165 170 175Arg
Ala Thr Ala Ala Ala Ser Ala Ser Lys Ile Ser Ser Gln Glu Ser 180 185
190Ala Ala Ser Ala Ala Asn Ala Ala Asn Ala Glu Ala Lys Ala Asn Ala
195 200 205Ala Ser Ile Ile Ala Asn Lys Ala Asn Ala Val Leu Ala Glu
Ala Ala 210 215 220Ala Val Leu Ala Ala Thr Ala Ala Lys Ala Lys Glu
Ser Ala Met Lys225 230 235 240Ser Leu Ser Ala Ala Gln Ala Ala Ala
Lys Ala Gln Ala Arg Asn Ala 245 250 255Glu Ala Ser Ala Glu Ala Gln
Ile Lys Leu Ser Gln Ala Arg Ala Ala 260 265 270Val Ala Arg Ala Ala
Ala Asp Gln Ala Val Cys Ser Ser Gln Ala Gln 275 280 285Ala Ala Ser
Gln Ile Gln Ser Arg Ala Ser Ala Ser Glu Ser Ala Ala 290 295 300Ser
Ala Gln Ser Glu Thr Asn Thr Ala Ala Ala Glu Ala Val Ala Thr305 310
315 320Ala Asp Ala Glu Ala Ala Ala Gln Ala Glu Ala Trp Val Met Ser
Leu 325 330 335Lys Asn Asp Leu Trp Leu His Leu Asn Met Lys Gly Glu
Ala Lys Ala 340 345 350Glu Gly Glu Ala Val Ser Ile Ser Lys Gly His
Arg Gly Gly Ile Arg 355 360 365Ser Gly Ser Ile Ser Glu Ala Ser Ala
Glu Ala Ser Ser Asn Val Ser 370 375 380Met Gly Gly Arg His Gly Arg
Lys Asp Leu Val Ser Glu Ala Leu Ala385 390 395 400Gly Ala Ser Ala
Gly Ser Ser Ala Asp Ser Leu 405 41044375PRTMyrmecia forficata 44Asn
Leu Leu Lys Glu Ser Lys Ala Ser Ala Ser Ala Ser Ala Ser Ala1 5 10
15Ser Ala Arg Ala Ser Gly Lys Lys Asn Leu His Val Leu Pro Leu Pro
20 25 30Lys Lys Ser Glu His Gly Ile Val Ile Asp Lys Ser Val Phe Asp
Ile 35 40 45Lys Asp Val Val Leu Ser Ala Val Asp Glu Ile Asn Gly Ala
Pro Lys 50 55 60Leu Gly Leu Gly Trp Lys Lys Val Ser Met Gly Val Glu
Arg Ala Glu65 70 75 80Ala Asn Ala Ala Ala Ala Ala Glu Ala Leu Ala
Met Ile Lys Lys Ile 85 90 95Ala Met Ala Arg Ser Ser Ala Tyr Val Gln
Ala Ala Trp Ala Ser Ala 100 105 110Gln Ala Ser Ala Asp Ala Leu Ala
Ser Ala Arg Val Ala Gln Ala Ser 115 120 125Gln Glu Ala Ala Glu Ala
Lys Gly Arg Ala Ala Ser Glu Ala Leu Ser 130 135 140Arg Ala Ile Glu
Ala Ser Ser Arg Ala Asp Ala Ala Ala Ala Ala Thr145 150 155 160Leu
Asp Ala Met Asp Arg Thr Met Glu Asn Ala Arg Ala Ala Asn Ala 165 170
175Ala Gln Thr Gln Ala Ser Gly Gln Ala Glu Asn Ala Asn Arg Ser Ala
180 185 190Ala Ala Ile Leu Ala Ala Leu Leu Arg Ile Ala Glu Ala Ser
Ala Leu 195 200 205Asn Asn Glu Ala Ala Val Asn Ala Ala Ala Ala Ala
Ala Ala Ala Ser 210 215 220Ala Leu Gln Ala Lys Ala Asn Ala Ala Ser
Gln Ala Thr Ala Arg Ala225 230 235 240Ala Gly Gln Ala Ser Thr Ala
Ala Glu Glu Ala Gln Ser Ala Gln Glu 245 250 255Ala Ala Asp Lys Asn
Ala Glu Leu Thr Thr Val Met Leu Glu Lys Ala 260 265 270Ser Ala Asp
Gln Gln Ala Ala Ser Ala Arg Ala Asp Tyr Tyr Thr Ala 275 280 285Ser
Thr Glu Ala Glu Ala Ala Ala Gln Ala Ser Ala Ile Asn Ala Leu 290 295
300Arg Asp Gly Ile Val Val Gly Met Gly Asn Asp Ala Gly Ala Ser
Ala305 310 315 320Gln Ala Met Ala Gln Val Glu Ala Leu Ala Arg Ala
Ser Glu His Lys 325 330 335Ala Leu Gly Glu Lys Lys Lys Gly Leu Val
Trp Gly Tyr Gly Ser Lys 340 345 350Gly Ser Ser Ser Ala Ser Ala Ser
Ala Ser Ala Ser Ala Glu Ala Ser 355 360 365Ser Arg Leu Gly Lys Asp
Trp 370 37545394PRTMyrmecia forficata 45Met Lys Ile Pro Ala Ile Leu
Val Thr Ser Phe Leu Ala Trp Gly Leu1 5 10 15Ala Ser Gly Asn Leu Leu
Lys Glu Ser Lys Ala Ser Ala Ser Ala Ser 20 25 30Ala Ser Ala Ser Ala
Arg Ala Ser Gly Lys Lys Asn Leu His Val Leu 35 40 45Pro Leu Pro Lys
Lys Ser Glu His Gly Ile Val Ile Asp Lys Ser Val 50 55 60Phe Asp Ile
Lys Asp Val Val Leu Ser Ala Val Asp Glu Ile Asn Gly65 70 75 80Ala
Pro Lys Leu Gly Leu Gly Trp Lys Lys Val Ser Met Gly Val Glu 85 90
95Arg Ala Glu Ala Asn Ala Ala Ala Ala Ala Glu Ala Leu Ala Met Ile
100 105 110Lys Lys Ile Ala Met Ala Arg Ser Ser Ala Tyr Val Gln Ala
Ala Trp 115 120 125Ala Ser Ala Gln Ala Ser Ala Asp Ala Leu Ala Ser
Ala Arg Val Ala 130 135 140Gln Ala Ser Gln Glu Ala Ala Glu Ala Lys
Gly Arg Ala Ala Ser Glu145 150 155 160Ala Leu Ser Arg Ala Ile Glu
Ala Ser Ser Arg Ala Asp Ala Ala Ala 165 170 175Ala Ala Thr Leu Asp
Ala Met Asp Arg Thr Met Glu Asn Ala Arg Ala 180 185 190Ala Asn Ala
Ala Gln Thr Gln Ala Ser Gly Gln Ala Glu Asn Ala Asn 195 200 205Arg
Ser Ala Ala Ala Ile Leu Ala Ala Leu Leu Arg Ile Ala Glu Ala 210 215
220Ser Ala Leu Asn Asn Glu Ala Ala Val Asn Ala Ala Ala Ala Ala
Ala225 230 235 240Ala Ala Ser Ala Leu Gln Ala Lys Ala Asn Ala Ala
Ser Gln Ala Thr 245 250 255Ala Arg Ala Ala Gly Gln Ala Ser Thr Ala
Ala Glu Glu Ala Gln Ser 260 265 270Ala Gln Glu Ala Ala Asp Lys Asn
Ala Glu Leu Thr Thr Val Met Leu 275 280 285Glu Lys Ala Ser Ala Asp
Gln Gln Ala Ala Ser Ala Arg Ala Asp Tyr 290 295 300Tyr Thr Ala Ser
Thr Glu Ala Glu Ala Ala Ala Gln Ala Ser Ala Ile305 310 315 320Asn
Ala Leu Arg Asp Gly Ile Val Val Gly Met Gly Asn Asp Ala Gly 325 330
335Ala Ser Ala Gln Ala Met Ala Gln Val Glu Ala Leu Ala Arg Ala Ser
340 345 350Glu His Lys Ala Leu Gly Glu Lys Lys Lys Gly Leu Val Trp
Gly Tyr 355 360 365Gly Ser Lys Gly Ser Ser Ser Ala Ser Ala Ser Ala
Ser Ala Ser Ala 370 375 380Glu Ala Ser Ser Arg Leu Gly Lys Asp
Trp385 39046422PRTMyrmecia forficata 46Ser Glu Leu Glu Ser Glu Ala
Ser Ala Ala Ala Ser Ala Gln Ala Glu1 5 10 15Ala Ser Ser Ser Gly Arg
Ser Gly Lys Leu Ser Ala Ser Gln Ala Ser 20 25 30Ala Ser Ala Ser Ala
Ser Ala Ser Ala Gly Ser Arg Gly Gly Ser Lys 35 40 45Gly Gly Trp Gly
Gln Leu Arg Arg Gly Asp Val Lys Ser Glu Ala Lys 50
55 60Ser Ala Ala Ala Ile Ala Val Glu Gly Ala Lys Ile Gly Thr Gly
Ile65 70 75 80Gly Asn Thr Ala Ser Ala Ser Ala Glu Ala Leu Ser Arg
Gly Leu Gly 85 90 95Ile Gly Gln Ala Ala Ala Glu Ala Gln Ala Ala Ala
Ala Gly Gln Ala 100 105 110Glu Val Ala Ala Lys Ser Cys Glu Leu Ala
Asp Lys Thr Thr Ala Lys 115 120 125Ala Val Ala Met Val Glu Ala Ala
Ala Glu Ala Glu Ile Glu Val Ala 130 135 140Asn Gln Glu Val Ala Ala
Val Lys Leu Ser Thr Trp Ala Ala Lys Ala145 150 155 160Ala Arg Ile
Val Glu Glu Asp Ser Ala Ala Val Arg Ala Ala Ala Gly 165 170 175Lys
Leu Leu Leu Ala Ala Arg Ala Ala Ala Ala Ala Glu Arg Arg Ala 180 185
190Asn Glu Glu Ser Glu Ala Ala Asn Glu Leu Ala Gln Ala Ser Ser Ala
195 200 205Ala Ala Ala Glu Ala Glu Ala Lys Ala Asn Ala Gly Arg Glu
Ala Ala 210 215 220Ala Ala Ala Leu Ala Ile Ala Glu Ala Ala Val Ala
Ile Glu Gln Glu225 230 235 240Ala Val Ile Leu Ala Arg Lys Ala Gln
Asp Ala Arg Leu Asn Ala Glu 245 250 255Ala Ala Ala Ala Ala Ala Met
Asn Ala Arg Val Ile Ala Ser Ala Glu 260 265 270Ser Glu Ala Ser Glu
Asp Leu Glu Asn Arg Ala Ser Val Ala Arg Ala 275 280 285Ser Ala Ala
Gly Ala Ala Glu Ala Lys Ala Ile Ala Thr Asp Ala Gly 290 295 300Ala
Thr Ala Glu Ile Ala Ala Tyr Ser Trp Ala Lys Lys Gly Glu Leu305 310
315 320Ile Asn Pro Gly Pro Leu Pro Lys Ile Ile Ser Val Asn Ala Asp
Leu 325 330 335Ser Lys Ser Glu Val Glu Ala Met Lys Ile Thr Arg Gly
Gln Val Gln 340 345 350Glu Val Lys Lys Ile Ser Thr His Lys Gly Gly
Trp Gly Trp Gly Lys 355 360 365Glu Gly Arg Ser Lys Val Ser Ser Asn
Ala Ser Ala Arg Ala Ser Ala 370 375 380Ser Ala Asn Ala Ala Ala Gly
Ser Leu Gly Ser Lys Trp Gly Arg Gln385 390 395 400Leu Ser Ala Ser
Ser Ala Ser Ala Asp Ala Asn Ala Glu Ala Asp Ser 405 410 415Gln Leu
Leu Lys Val Trp 42047441PRTMyrmecia forficata 47Met Lys Ile Pro Ala
Ile Leu Ala Thr Ser Leu Leu Ile Trp Gly Leu1 5 10 15Val Gly Ala Ser
Glu Leu Glu Ser Glu Ala Ser Ala Ala Ala Ser Ala 20 25 30Gln Ala Glu
Ala Ser Ser Ser Gly Arg Ser Gly Lys Leu Ser Ala Ser 35 40 45Gln Ala
Ser Ala Ser Ala Ser Ala Ser Ala Ser Ala Gly Ser Arg Gly 50 55 60Gly
Ser Lys Gly Gly Trp Gly Gln Leu Arg Arg Gly Asp Val Lys Ser65 70 75
80Glu Ala Lys Ser Ala Ala Ala Ile Ala Val Glu Gly Ala Lys Ile Gly
85 90 95Thr Gly Ile Gly Asn Thr Ala Ser Ala Ser Ala Glu Ala Leu Ser
Arg 100 105 110Gly Leu Gly Ile Gly Gln Ala Ala Ala Glu Ala Gln Ala
Ala Ala Ala 115 120 125Gly Gln Ala Glu Val Ala Ala Lys Ser Cys Glu
Leu Ala Asp Lys Thr 130 135 140Thr Ala Lys Ala Val Ala Met Val Glu
Ala Ala Ala Glu Ala Glu Ile145 150 155 160Glu Val Ala Asn Gln Glu
Val Ala Ala Val Lys Leu Ser Thr Trp Ala 165 170 175Ala Lys Ala Ala
Arg Ile Val Glu Glu Asp Ser Ala Ala Val Arg Ala 180 185 190Ala Ala
Gly Lys Leu Leu Leu Ala Ala Arg Ala Ala Ala Ala Ala Glu 195 200
205Arg Arg Ala Asn Glu Glu Ser Glu Ala Ala Asn Glu Leu Ala Gln Ala
210 215 220Ser Ser Ala Ala Ala Ala Glu Ala Glu Ala Lys Ala Asn Ala
Gly Arg225 230 235 240Glu Ala Ala Ala Ala Ala Leu Ala Ile Ala Glu
Ala Ala Val Ala Ile 245 250 255Glu Gln Glu Ala Val Ile Leu Ala Arg
Lys Ala Gln Asp Ala Arg Leu 260 265 270Asn Ala Glu Ala Ala Ala Ala
Ala Ala Met Asn Ala Arg Val Ile Ala 275 280 285Ser Ala Glu Ser Glu
Ala Ser Glu Asp Leu Glu Asn Arg Ala Ser Val 290 295 300Ala Arg Ala
Ser Ala Ala Gly Ala Ala Glu Ala Lys Ala Ile Ala Thr305 310 315
320Asp Ala Gly Ala Thr Ala Glu Ile Ala Ala Tyr Ser Trp Ala Lys Lys
325 330 335Gly Glu Leu Ile Asn Pro Gly Pro Leu Pro Lys Ile Ile Ser
Val Asn 340 345 350Ala Asp Leu Ser Lys Ser Glu Val Glu Ala Met Lys
Ile Thr Arg Gly 355 360 365Gln Val Gln Glu Val Lys Lys Ile Ser Thr
His Lys Gly Gly Trp Gly 370 375 380Trp Gly Lys Glu Gly Arg Ser Lys
Val Ser Ser Asn Ala Ser Ala Arg385 390 395 400Ala Ser Ala Ser Ala
Asn Ala Ala Ala Gly Ser Leu Gly Ser Lys Trp 405 410 415Gly Arg Gln
Leu Ser Ala Ser Ser Ala Ser Ala Asp Ala Asn Ala Glu 420 425 430Ala
Asp Ser Gln Leu Leu Lys Val Trp 435 440481212DNAMyrmecia forficata
48agcgggccgc gcttactcgg cggcagatcg gccgcgtccg cgtcggcttc cgcttcggct
60gaggcgtcgg cgggcggttg gaggaaaagc ggcgcatccg cttccgcttc cgctaaggct
120ggtagcagca acatcctcag ccgcgtggga gcttcgaggg cggccgcgac
gttggtcgct 180tccgccgcgg tggaggccaa ggcgggtctc cgtgccggca
aggcaaccgc cgaggagcag 240agggaggctt tggaaatgct caccttgtcc
gccgacaaga atgccgaggc gcgtatcctg 300gccgacgaca cggccgttct
ggttcaaggc agcgccgagg cacagtcggt cgccgccgcg 360aagaccgtcg
cggtcgagga agagtccgct tccttggatg cggccgcagt tgaagcggag
420gtcgcagccg ccacgtcgaa atcgtcggct ggccaagcac tccagtccgc
acagaccgcc 480gcatctgctc tcagaacttc cgccaggagc gccttgacgg
ccctcaagct ggcacgcctc 540caaggcgcgg cttctagcaa cgctgccagg
atgatggaaa aggcgctggc cgccacccag 600gacgcaaatg ccgccgccca
gcaagctatg gcggccgaga gtgcagccgc agaagcagcg 660gctatcgcgg
cagcgaaaca atcggaggcg agagacgccg gcgccgaggc caaggccgcc
720atggcagcac tcatcaccgc ccagaggaat ctcgtgcagg ccaatgccag
ggcggaaatg 780gcaagcgagg aagccgaatt ggattcgaag tctagagcgt
ccgacgccaa ggtgaacgcc 840gttgctcgtg cggcctccaa gtccagcata
cgcagagatg aacttatcga gatcggcgct 900gagttcggca aggccagcgg
cgaggtgatt tccaccggca cgcgttccaa cggcggtcaa 960gacgccatcg
ccaccgccga ggcatcgagt agcgcgtccg ccgtcggcat caagaaaaca
1020agcggacact gggggagcgg aaaatggagt cgtgtctcca agggtaaagg
atgggcttcc 1080tcgaatgcgg acgctgacgc cagcagcagc agcatcatca
tcggcggtct caaacgcggc 1140ggcctcggtt cggaagcctc tgcggcagct
tccgcagaag cggaagcttc cgccggcaca 1200ctcctgctgt aa
1212491269DNAMyrmecia forficata 49atgaagatcc cagcgataat cgcaacgtcc
cttctcctct ggggtttcgc cagcgccagc 60gggccgcgct tactcggcgg cagatcggcc
gcgtccgcgt cggcttccgc ttcggctgag 120gcgtcggcgg gcggttggag
gaaaagcggc gcatccgctt ccgcttccgc taaggctggt 180agcagcaaca
tcctcagccg cgtgggagct tcgagggcgg ccgcgacgtt ggtcgcttcc
240gccgcggtgg aggccaaggc gggtctccgt gccggcaagg caaccgccga
ggagcagagg 300gaggctttgg aaatgctcac cttgtccgcc gacaagaatg
ccgaggcgcg tatcctggcc 360gacgacacgg ccgttctggt tcaaggcagc
gccgaggcac agtcggtcgc cgccgcgaag 420accgtcgcgg tcgaggaaga
gtccgcttcc ttggatgcgg ccgcagttga agcggaggtc 480gcagccgcca
cgtcgaaatc gtcggctggc caagcactcc agtccgcaca gaccgccgca
540tctgctctca gaacttccgc caggagcgcc ttgacggccc tcaagctggc
acgcctccaa 600ggcgcggctt ctagcaacgc tgccaggatg atggaaaagg
cgctggccgc cacccaggac 660gcaaatgccg ccgcccagca agctatggcg
gccgagagtg cagccgcaga agcagcggct 720atcgcggcag cgaaacaatc
ggaggcgaga gacgccggcg ccgaggccaa ggccgccatg 780gcagcactca
tcaccgccca gaggaatctc gtgcaggcca atgccagggc ggaaatggca
840agcgaggaag ccgaattgga ttcgaagtct agagcgtccg acgccaaggt
gaacgccgtt 900gctcgtgcgg cctccaagtc cagcatacgc agagatgaac
ttatcgagat cggcgctgag 960ttcggcaagg ccagcggcga ggtgatttcc
accggcacgc gttccaacgg cggtcaagac 1020gccatcgcca ccgccgaggc
atcgagtagc gcgtccgccg tcggcatcaa gaaaacaagc 1080ggacactggg
ggagcggaaa atggagtcgt gtctccaagg gtaaaggatg ggcttcctcg
1140aatgcggacg ctgacgccag cagcagcagc atcatcatcg gcggtctcaa
acgcggcggc 1200ctcggttcgg aagcctctgc ggcagcttcc gcagaagcgg
aagcttccgc cggcacactc 1260ctgctgtaa 1269501179DNAMyrmecia forficata
50cgggtcatcg agtccagctc gtcggcttcc gcacaggcgt cggcatcggc cggctcgaga
60ggcctgctcg gtaaacggcc gattggcaag ctcgagtggg gcaaggagga gaagaaactc
120gaagaactcg acgaggaatc gctcaatgag gccgctctga aggtcggcat
caagaacggc 180ggattggatg tcgcgaaggg cgcggcagtc ctcgaggcag
cgatgagcga cgtcgcgacc 240cttacggatc agcgttctct tgtggatctc
ggtctcggcc cggtcgcgaa cgaggccgag 300atcctggcgg aggcgcaggc
cgccacgagc gcccaagctg gcgctgtcgc taatagcgcc 360gcggagcgtg
cgatcgcggc gatggagatg gccgacagaa ccgaatatat tgcggcactt
420gtcaccacca aagccgccaa agctgccgag gccactatgg ccgctactgc
ccgtgccacc 480gccgccgcct cagcctccaa gatatccagt caggaatcag
ccgcatcggc cgctaacgcc 540gccaacgccg aagccaaggc caacgccgct
tccataatcg ctaacaaggc gaacgccgtc 600ctggctgagg ccgccgccgt
actcgcagcc actgctgcca aggccaagga atcggcgatg 660aaatcgctta
gcgccgctca ggccgccgcc aaggcacaag ccaggaacgc cgaggcctcc
720gccgaagctc agatcaaact ttcccaggcc agggccgccg tggcacgcgc
tgcagccgat 780caggccgtct gttcctccca ggctcaggcc gcaagtcaga
tacaatcgag ggcatccgca 840tccgaatccg cggcatcggc acaatcagag
accaacaccg ccgcggccga agcggtcgcc 900accgctgacg ccgaagcggc
cgcgcaagct gaagcgtggg tcatgtcgct gaagaacgat 960ctgtggctgc
atctcaacat gaagggtgag gccaaggccg aaggcgaggc cgtttcgatc
1020agcaaaggac atcgcggcgg tatcaggtcg ggcagcatct cggaagccag
cgccgaggca 1080agcagcaacg tttccatggg cggacgtcat ggacggaagg
acctcgtctc tgaagcgtta 1140gcgggagcat cagcgggcag cagtgccgac
tccctttga 1179511236DNAMyrmecia forficata 51atgaagattc cagcgatact
cgtgacgtct ctcctcgcct ggggattagc cagcggccgg 60gtcatcgagt ccagctcgtc
ggcttccgca caggcgtcgg catcggccgg ctcgagaggc 120ctgctcggta
aacggccgat tggcaagctc gagtggggca aggaggagaa gaaactcgaa
180gaactcgacg aggaatcgct caatgaggcc gctctgaagg tcggcatcaa
gaacggcgga 240ttggatgtcg cgaagggcgc ggcagtcctc gaggcagcga
tgagcgacgt cgcgaccctt 300acggatcagc gttctcttgt ggatctcggt
ctcggcccgg tcgcgaacga ggccgagatc 360ctggcggagg cgcaggccgc
cacgagcgcc caagctggcg ctgtcgctaa tagcgccgcg 420gagcgtgcga
tcgcggcgat ggagatggcc gacagaaccg aatatattgc ggcacttgtc
480accaccaaag ccgccaaagc tgccgaggcc actatggccg ctactgcccg
tgccaccgcc 540gccgcctcag cctccaagat atccagtcag gaatcagccg
catcggccgc taacgccgcc 600aacgccgaag ccaaggccaa cgccgcttcc
ataatcgcta acaaggcgaa cgccgtcctg 660gctgaggccg ccgccgtact
cgcagccact gctgccaagg ccaaggaatc ggcgatgaaa 720tcgcttagcg
ccgctcaggc cgccgccaag gcacaagcca ggaacgccga ggcctccgcc
780gaagctcaga tcaaactttc ccaggccagg gccgccgtgg cacgcgctgc
agccgatcag 840gccgtctgtt cctcccaggc tcaggccgca agtcagatac
aatcgagggc atccgcatcc 900gaatccgcgg catcggcaca atcagagacc
aacaccgccg cggccgaagc ggtcgccacc 960gctgacgccg aagcggccgc
gcaagctgaa gcgtgggtca tgtcgctgaa gaacgatctg 1020tggctgcatc
tcaacatgaa gggtgaggcc aaggccgaag gcgaggccgt ttcgatcagc
1080aaaggacatc gcggcggtat caggtcgggc agcatctcgg aagccagcgc
cgaggcaagc 1140agcaacgttt ccatgggcgg acgtcatgga cggaaggacc
tcgtctctga agcgttagcg 1200ggagcatcag cgggcagcag tgccgactcc ctttga
1236521128DNAMyrmecia forficata 52aatctcctta aggagtcgaa agcttccgcg
tccgcgtccg cgtccgcttc cgcgagggcc 60agcggcaaga agaatcttca cgtgttgcca
ttaccgaaga aaagcgagca tggcatcgtg 120atcgacaagt cggtgttcga
catcaaggat gtagtgctga gcgcggtcga cgagatcaac 180ggcgccccga
aactcggcct gggatggaag aaggtcagca tgggggtgga gcgcgccgag
240gcgaacgcag ccgctgccgc cgaggcattg gcgatgatca agaagattgc
catggcccgc 300agcagtgcat acgtccaggc ggcctgggca tcggcccagg
catcagctga cgcattggct 360agcgccaggg tggcacaggc gtctcaggag
gctgcggagg caaagggtag agcggcttcc 420gaggcgctct ccagagccat
cgaagcatcc tcgcgagccg atgcggcagc cgctgcgacg 480ctggacgcga
tggaccgcac catggagaac gcgagggcgg caaatgccgc gcaaacgcag
540gccagcggcc aagctgagaa cgcaaatcgc agcgctgctg ccatcctcgc
agctctgcta 600cgtatcgcgg aggcatccgc gttgaacaac gaggccgcgg
tcaacgcggc cgcggccgca 660gccgcagcgt ctgcccttca ggccaaggct
aacgcggctt ctcaagcaac cgccagagcc 720gcaggacagg cgtcgacggc
cgccgaagag gcgcaatccg cccaagaagc cgccgataag 780aacgcggagc
tgaccacggt catgctcgaa aaggctagtg ctgatcaaca ggcggcatcc
840gctagggctg actactacac cgcctcaacc gaggccgaag ccgctgcaca
ggcgtctgct 900atcaacgcac tcagggacgg aatagttgtc ggaatgggaa
atgacgctgg cgcatcggcc 960caagcgatgg cacaggtaga agctctcgct
cgcgccagcg agcacaaggc gttaggcgag 1020aagaagaagg gcctggtttg
gggctacgga agcaagggca gtagctccgc cagcgcatcc 1080gccagcgcct
ccgccgaagc atcctcgaga ctcggaaagg actggtag 1128531185DNAMyrmecia
forficata 53atgaagatac cagcgatact cgtgacgtcc ttcctcgcct ggggactggc
cagcgggaat 60ctccttaagg agtcgaaagc ttccgcgtcc gcgtccgcgt ccgcttccgc
gagggccagc 120ggcaagaaga atcttcacgt gttgccatta ccgaagaaaa
gcgagcatgg catcgtgatc 180gacaagtcgg tgttcgacat caaggatgta
gtgctgagcg cggtcgacga gatcaacggc 240gccccgaaac tcggcctggg
atggaagaag gtcagcatgg gggtggagcg cgccgaggcg 300aacgcagccg
ctgccgccga ggcattggcg atgatcaaga agattgccat ggcccgcagc
360agtgcatacg tccaggcggc ctgggcatcg gcccaggcat cagctgacgc
attggctagc 420gccagggtgg cacaggcgtc tcaggaggct gcggaggcaa
agggtagagc ggcttccgag 480gcgctctcca gagccatcga agcatcctcg
cgagccgatg cggcagccgc tgcgacgctg 540gacgcgatgg accgcaccat
ggagaacgcg agggcggcaa atgccgcgca aacgcaggcc 600agcggccaag
ctgagaacgc aaatcgcagc gctgctgcca tcctcgcagc tctgctacgt
660atcgcggagg catccgcgtt gaacaacgag gccgcggtca acgcggccgc
ggccgcagcc 720gcagcgtctg cccttcaggc caaggctaac gcggcttctc
aagcaaccgc cagagccgca 780ggacaggcgt cgacggccgc cgaagaggcg
caatccgccc aagaagccgc cgataagaac 840gcggagctga ccacggtcat
gctcgaaaag gctagtgctg atcaacaggc ggcatccgct 900agggctgact
actacaccgc ctcaaccgag gccgaagccg ctgcacaggc gtctgctatc
960aacgcactca gggacggaat agttgtcgga atgggaaatg acgctggcgc
atcggcccaa 1020gcgatggcac aggtagaagc tctcgctcgc gccagcgagc
acaaggcgtt aggcgagaag 1080aagaagggcc tggtttgggg ctacggaagc
aagggcagta gctccgccag cgcatccgcc 1140agcgcctccg ccgaagcatc
ctcgagactc ggaaaggact ggtag 1185541269DNAMyrmecia forficata
54agcgagctcg aatcggaagc gagtgcggcg gcgtctgcgc aagcggaagc gtcctcgtct
60ggtcgctccg gcaaactgtc cgcgtctcag gcttccgcca gcgcgtccgc cagcgcgtca
120gccggcagca gaggtggcag caaaggtggc tggggccagc tccgccgtgg
tgatgttaag 180agcgaggcga agagcgccgc cgcgatcgcg gtcgaaggag
ctaaaatcgg caccggaatc 240ggaaataccg cgtccgcatc cgcggaggcg
ctctcacgag gactcggcat cggacaggcg 300gccgcggagg cgcaagccgc
agccgcaggt caggcagagg tcgccgcgaa atcgtgcgaa 360cttgccgaca
agaccaccgc caaagcggtc gccatggtcg aagcggcagc cgaggccgaa
420atcgaggtgg ccaatcagga ggtcgcagcc gtcaaattat cgacttgggc
cgctaaagca 480gcaaggatag tcgaggaaga cagcgccgcc gtgagggcgg
ctgccggcaa attgcttttg 540gccgcgagag ctgccgccgc cgccgagaga
cgcgccaacg aggaatccga ggcggccaac 600gaacttgctc aagcgtcatc
tgccgctgcc gccgaggccg aagccaaagc gaacgccggc 660cgtgaggccg
ctgccgctgc cttggctatc gccgaggccg ccgtcgccat cgaacaagaa
720gccgtcattt tggctcgcaa ggcacaagat gcccgtttga atgctgaagc
cgcagccgcc 780gctgcgatga acgcccgtgt catcgcttcc gccgaatccg
aggccagtga agatctggag 840aatcgcgcta gtgtggcgcg tgccagtgcg
gccggtgccg ctgaggcaaa ggctatcgcc 900accgatgccg gcgccactgc
cgagatcgcg gcctacagtt gggccaagaa gggcgaactg 960atcaaccccg
gcccgttgcc gaagatcatc agcgtcaacg ccgatctgtc caagagcgag
1020gtcgaggcca tgaagatcac ccggggtcaa gtacaggaag tcaagaaaat
cagcactcac 1080aaaggtggct ggggatgggg aaaggaagga aggtcgaagg
tatcttccaa cgctagtgcc 1140agagctagtg ccagcgccaa tgcagccgcc
ggtagcctcg gcagcaaatg gggaagacaa 1200ctatccgcat catccgcgtc
ggctgacgcc aacgccgaag ccgacagcca gttgctgaaa 1260gtgtggtga
1269551326DNAMyrmecia forficata 55atgaagattc cagcgatact tgcgacgtcc
ctcctcatct ggggtcttgt cggcgccagc 60gagctcgaat cggaagcgag tgcggcggcg
tctgcgcaag cggaagcgtc ctcgtctggt 120cgctccggca aactgtccgc
gtctcaggct tccgccagcg cgtccgccag cgcgtcagcc 180ggcagcagag
gtggcagcaa aggtggctgg ggccagctcc gccgtggtga tgttaagagc
240gaggcgaaga gcgccgccgc gatcgcggtc gaaggagcta aaatcggcac
cggaatcgga 300aataccgcgt ccgcatccgc ggaggcgctc tcacgaggac
tcggcatcgg acaggcggcc 360gcggaggcgc aagccgcagc cgcaggtcag
gcagaggtcg ccgcgaaatc gtgcgaactt 420gccgacaaga ccaccgccaa
agcggtcgcc atggtcgaag cggcagccga ggccgaaatc 480gaggtggcca
atcaggaggt cgcagccgtc aaattatcga cttgggccgc taaagcagca
540aggatagtcg aggaagacag cgccgccgtg agggcggctg ccggcaaatt
gcttttggcc 600gcgagagctg ccgccgccgc cgagagacgc gccaacgagg
aatccgaggc ggccaacgaa 660cttgctcaag cgtcatctgc cgctgccgcc
gaggccgaag ccaaagcgaa cgccggccgt 720gaggccgctg ccgctgcctt
ggctatcgcc gaggccgccg tcgccatcga acaagaagcc 780gtcattttgg
ctcgcaaggc acaagatgcc cgtttgaatg ctgaagccgc agccgccgct
840gcgatgaacg cccgtgtcat cgcttccgcc gaatccgagg ccagtgaaga
tctggagaat 900cgcgctagtg tggcgcgtgc cagtgcggcc ggtgccgctg
aggcaaaggc tatcgccacc 960gatgccggcg ccactgccga gatcgcggcc
tacagttggg ccaagaaggg cgaactgatc 1020aaccccggcc cgttgccgaa
gatcatcagc gtcaacgccg atctgtccaa gagcgaggtc 1080gaggccatga
agatcacccg gggtcaagta caggaagtca agaaaatcag cactcacaaa
1140ggtggctggg gatggggaaa ggaaggaagg tcgaaggtat cttccaacgc
tagtgccaga 1200gctagtgcca gcgccaatgc agccgccggt
agcctcggca gcaaatgggg aagacaacta 1260tccgcatcat ccgcgtcggc
tgacgccaac gccgaagccg acagccagtt gctgaaagtg 1320tggtga
132656372PRTOecophylla smaragdina 56Ser Lys Ser Tyr Leu Leu Gly Ser
Ser Ala Ser Ala Ser Ala Ser Ala1 5 10 15Ser Ala Ser Ala Ser Ala Gly
Gly Ser Thr Gly Gly Val Gly Val Gly 20 25 30Ser Val Ile Ser Gly Gly
Asn Asn Ile Ile Arg Gly Ala Ser Thr Thr 35 40 45Ser Val Thr Leu Ala
Ala Ala Ala Ala Glu Ala Lys Ala Ala Leu Asn 50 55 60Ala Gly Lys Ala
Thr Val Glu Glu Gln Arg Glu Ala Leu Gln Leu Leu65 70 75 80Thr Ala
Ser Ala Glu Lys Asn Ala Glu Ala Arg Ser Leu Ala Asp Asp 85 90 95Ala
Ala Val Leu Val Gln Gly Ala Ala Glu Ala Gln Ser Val Ala Ala 100 105
110Ala Lys Thr Val Ala Val Glu Gln Gly Ser Asn Ser Leu Asp Ala Ala
115 120 125Ala Ala Glu Ala Glu Ala Ala Ala Ala Ala Ser Arg Val Ser
Ala Gln 130 135 140Gln Ala Leu Gln Ala Ala Gln Thr Ser Ala Ala Ala
Ile Gln Thr Ala145 150 155 160Ala Gly Ser Ala Leu Thr Ala Leu Lys
Leu Ala Arg Lys Gln Glu Ala 165 170 175Glu Ser Asn Asn Ala Ala Glu
Gln Ala Asn Lys Ala Leu Ala Leu Ser 180 185 190Arg Ala Ala Ser Ala
Ala Thr Gln Arg Ala Val Ala Ala Gln Asn Ala 195 200 205Ala Ala Ala
Ser Ala Ala Ser Ala Gly Ala Ala Gln Ala Glu Ala Arg 210 215 220Asn
Ala Tyr Ala Lys Ala Lys Ala Ala Ile Ala Ala Leu Thr Ala Ala225 230
235 240Gln Arg Asn Tyr Ala Ala Ala Lys Ala Ser Ala Ser Ala Gly Ser
Val 245 250 255Val Ala Glu Gln Asp Ala Gln Ser Arg Ala Ala Asp Ala
Glu Val Asn 260 265 270Ala Val Ala Gln Ala Ala Ala Arg Ala Ser Val
Arg Asn Gln Glu Ile 275 280 285Val Glu Ile Gly Ala Glu Phe Gly Asn
Ala Ser Gly Gly Val Ile Ser 290 295 300Thr Gly Thr Arg Ser Ser Gly
Gly Lys Gly Val Ser Val Thr Ala Gly305 310 315 320Ala Gln Ala Ser
Ala Ser Ala Ser Ala Thr Ser Ser Ser Ser Ser Ser 325 330 335Ser Gly
Ile Asn Lys Gly His Pro Arg Trp Gly His Asn Trp Gly Leu 340 345
350Gly Ser Ser Glu Ala Ser Ala Asn Ala Glu Ala Glu Ser Ser Ala Ser
355 360 365Ser Tyr Ser Ser 37057391PRTOecophylla smaragdina 57Met
Lys Ile Pro Ala Ile Ile Ala Thr Thr Leu Leu Leu Trp Gly Phe1 5 10
15Ala Asp Ala Ser Lys Ser Tyr Leu Leu Gly Ser Ser Ala Ser Ala Ser
20 25 30Ala Ser Ala Ser Ala Ser Ala Ser Ala Gly Gly Ser Thr Gly Gly
Val 35 40 45Gly Val Gly Ser Val Ile Ser Gly Gly Asn Asn Ile Ile Arg
Gly Ala 50 55 60Ser Thr Thr Ser Val Thr Leu Ala Ala Ala Ala Ala Glu
Ala Lys Ala65 70 75 80Ala Leu Asn Ala Gly Lys Ala Thr Val Glu Glu
Gln Arg Glu Ala Leu 85 90 95Gln Leu Leu Thr Ala Ser Ala Glu Lys Asn
Ala Glu Ala Arg Ser Leu 100 105 110Ala Asp Asp Ala Ala Val Leu Val
Gln Gly Ala Ala Glu Ala Gln Ser 115 120 125Val Ala Ala Ala Lys Thr
Val Ala Val Glu Gln Gly Ser Asn Ser Leu 130 135 140Asp Ala Ala Ala
Ala Glu Ala Glu Ala Ala Ala Ala Ala Ser Arg Val145 150 155 160Ser
Ala Gln Gln Ala Leu Gln Ala Ala Gln Thr Ser Ala Ala Ala Ile 165 170
175Gln Thr Ala Ala Gly Ser Ala Leu Thr Ala Leu Lys Leu Ala Arg Lys
180 185 190Gln Glu Ala Glu Ser Asn Asn Ala Ala Glu Gln Ala Asn Lys
Ala Leu 195 200 205Ala Leu Ser Arg Ala Ala Ser Ala Ala Thr Gln Arg
Ala Val Ala Ala 210 215 220Gln Asn Ala Ala Ala Ala Ser Ala Ala Ser
Ala Gly Ala Ala Gln Ala225 230 235 240Glu Ala Arg Asn Ala Tyr Ala
Lys Ala Lys Ala Ala Ile Ala Ala Leu 245 250 255Thr Ala Ala Gln Arg
Asn Tyr Ala Ala Ala Lys Ala Ser Ala Ser Ala 260 265 270Gly Ser Val
Val Ala Glu Gln Asp Ala Gln Ser Arg Ala Ala Asp Ala 275 280 285Glu
Val Asn Ala Val Ala Gln Ala Ala Ala Arg Ala Ser Val Arg Asn 290 295
300Gln Glu Ile Val Glu Ile Gly Ala Glu Phe Gly Asn Ala Ser Gly
Gly305 310 315 320Val Ile Ser Thr Gly Thr Arg Ser Ser Gly Gly Lys
Gly Val Ser Val 325 330 335Thr Ala Gly Ala Gln Ala Ser Ala Ser Ala
Ser Ala Thr Ser Ser Ser 340 345 350Ser Ser Ser Ser Gly Ile Asn Lys
Gly His Pro Arg Trp Gly His Asn 355 360 365Trp Gly Leu Gly Ser Ser
Glu Ala Ser Ala Asn Ala Glu Ala Glu Ser 370 375 380Ser Ala Ser Ser
Tyr Ser Ser385 39058381PRTOecophylla smaragdina 58Gly Val Ile Gly
Pro Asp Thr Ser Ser Ser Ser Gln Ala Ser Ala Ser1 5 10 15Ala Ser Ala
Ser Ala Ser Ala Ser Ala Ser Ser Ser Ala Ser Ile Gly 20 25 30Tyr Asn
Glu Leu His Lys Ser Ile Asn Ala Pro Ala Leu Ala Val Gly 35 40 45Val
Lys Asn Gly Gly Val Asp Val Ala Lys Gly Ala Ala Val Val Glu 50 55
60Ser Ala Ile Ser Asp Val Ser Thr Leu Thr Asp Asp Arg Thr Leu Asn65
70 75 80Gly Leu Ala Ile Ile Gly Asn Ser Ala Glu Ser Leu Ala Arg Ala
Gln 85 90 95Ala Ser Ser Ser Ala Ser Ala Gly Ala Lys Ala Asn Ala Leu
Ile Lys 100 105 110Gln Ser Ile Ala Ala Ile Glu Ile Thr Glu Lys Ala
Glu Tyr Leu Ala 115 120 125Ser Ile Val Ala Thr Lys Ala Ala Lys Ala
Ala Glu Ala Thr Ala Ala 130 135 140Ala Thr Ala Arg Ala Thr Ala Val
Ala Glu Ala Ala Lys Val Ser Ser145 150 155 160Glu Gln Phe Ala Ala
Glu Ala Arg Ala Ala Ala Asp Ala Glu Ala Lys 165 170 175Ala Asn Ala
Ala Ser Ile Ile Ala Asn Lys Ala Asn Ala Val Leu Ala 180 185 190Glu
Ala Ala Thr Gly Leu Ser Ala Ser Ala Gly Lys Ala Gln Gln Ser 195 200
205Ala Thr Arg Ala Leu Gln Ala Ala Arg Ala Ala Ala Lys Ala Gln Ala
210 215 220Glu Leu Thr Gln Lys Ala Ala Gln Ile Leu Val Leu Ile Ala
Glu Ala225 230 235 240Lys Ala Ala Val Ser Arg Ala Ser Ala Asp Gln
Ser Val Cys Thr Ser 245 250 255Gln Ala Gln Ala Ala Ser Gln Ile Gln
Ser Arg Ala Ser Ala Ala Glu 260 265 270Ser Ala Ala Ser Ala Gln Ser
Glu Ala Asn Thr Ile Ala Ala Glu Ala 275 280 285Val Ala Arg Ala Asp
Ala Glu Ala Ala Ser Gln Ala Gln Ala Trp Ala 290 295 300Glu Ser Phe
Lys Arg Glu Leu Ser Ser Val Val Leu Glu Ala Glu Ala305 310 315
320Asn Ala Ser Ala Ser Ala Ser Ala Gly Ala Leu Ala Ser Gly Ser Ser
325 330 335Ser Ser Gly Ala Ser Ser Ser Ala Asp Ala Ser Ala Gly Ala
Ser Ser 340 345 350Tyr Gly Ser Leu Gly Gly Tyr Arg His Gly Gly Ser
Phe Ser Glu Ala 355 360 365Ser Ala Ala Ala Ser Ala Ala Ser Arg Ala
Glu Ala Ala 370 375 38059400PRTOecophylla smaragdina 59Met Lys Ile
Pro Ala Ile Phe Val Thr Ser Leu Leu Ala Trp Gly Leu1 5 10 15Ala Ser
Gly Gly Val Ile Gly Pro Asp Thr Ser Ser Ser Ser Gln Ala 20 25 30Ser
Ala Ser Ala Ser Ala Ser Ala Ser Ala Ser Ala Ser Ser Ser Ala 35 40
45Ser Ile Gly Tyr Asn Glu Leu His Lys Ser Ile Asn Ala Pro Ala Leu
50 55 60Ala Val Gly Val Lys Asn Gly Gly Val Asp Val Ala Lys Gly Ala
Ala65 70 75 80Val Val Glu Ser Ala Ile Ser Asp Val Ser Thr Leu Thr
Asp Asp Arg 85 90 95Thr Leu Asn Gly Leu Ala Ile Ile Gly Asn Ser Ala
Glu Ser Leu Ala 100 105 110Arg Ala Gln Ala Ser Ser Ser Ala Ser Ala
Gly Ala Lys Ala Asn Ala 115 120 125Leu Ile Lys Gln Ser Ile Ala Ala
Ile Glu Ile Thr Glu Lys Ala Glu 130 135 140Tyr Leu Ala Ser Ile Val
Ala Thr Lys Ala Ala Lys Ala Ala Glu Ala145 150 155 160Thr Ala Ala
Ala Thr Ala Arg Ala Thr Ala Val Ala Glu Ala Ala Lys 165 170 175Val
Ser Ser Glu Gln Phe Ala Ala Glu Ala Arg Ala Ala Ala Asp Ala 180 185
190Glu Ala Lys Ala Asn Ala Ala Ser Ile Ile Ala Asn Lys Ala Asn Ala
195 200 205Val Leu Ala Glu Ala Ala Thr Gly Leu Ser Ala Ser Ala Gly
Lys Ala 210 215 220Gln Gln Ser Ala Thr Arg Ala Leu Gln Ala Ala Arg
Ala Ala Ala Lys225 230 235 240Ala Gln Ala Glu Leu Thr Gln Lys Ala
Ala Gln Ile Leu Val Leu Ile 245 250 255Ala Glu Ala Lys Ala Ala Val
Ser Arg Ala Ser Ala Asp Gln Ser Val 260 265 270Cys Thr Ser Gln Ala
Gln Ala Ala Ser Gln Ile Gln Ser Arg Ala Ser 275 280 285Ala Ala Glu
Ser Ala Ala Ser Ala Gln Ser Glu Ala Asn Thr Ile Ala 290 295 300Ala
Glu Ala Val Ala Arg Ala Asp Ala Glu Ala Ala Ser Gln Ala Gln305 310
315 320Ala Trp Ala Glu Ser Phe Lys Arg Glu Leu Ser Ser Val Val Leu
Glu 325 330 335Ala Glu Ala Asn Ala Ser Ala Ser Ala Ser Ala Gly Ala
Leu Ala Ser 340 345 350Gly Ser Ser Ser Ser Gly Ala Ser Ser Ser Ala
Asp Ala Ser Ala Gly 355 360 365Ala Ser Ser Tyr Gly Ser Leu Gly Gly
Tyr Arg His Gly Gly Ser Phe 370 375 380Ser Glu Ala Ser Ala Ala Ala
Ser Ala Ala Ser Arg Ala Glu Ala Ala385 390 395
40060376PRTOecophylla smaragdina 60Gly Val Pro Lys Glu Leu Gly Thr
Ser Ile Ser Ser Ala Ser Ala Ser1 5 10 15Ala Ser Ala Ser Ala Ser Ala
Thr Ala Ser Ser Ser Ser Lys Asn Val 20 25 30His Leu Leu Pro Leu Lys
Ser Glu His Gly Ile Val Ile Asp Lys Ser 35 40 45Lys Phe Asn Ile Arg
Lys Val Val Leu Ser Ala Ile Asp Glu Ile Asn 50 55 60Gly Ala Pro Asn
Ile Gly Leu Gly Leu Lys Gln Val Ser Leu Ala Leu65 70 75 80Ala Lys
Ala Gln Ala Ser Ala Gln Ser Ser Ala Glu Ala Leu Ala Ile 85 90 95Ile
Lys Lys Ile Val Ala Leu Leu Ile Ser Ala Tyr Val Arg Ala Ala 100 105
110Glu Ala Ala Ala Arg Ala Ser Ala Glu Ala Leu Ala Thr Val Arg Ala
115 120 125Ala Glu Gln Ala Gln Lys Ile Ala Glu Ala Lys Gly Arg Ala
Ala Ala 130 135 140Glu Ala Leu Ser Glu Leu Val Glu Ala Ser Gln Lys
Ala Asp Ala Ala145 150 155 160Ala Ala Gly Thr Thr Asp Ala Ile Glu
Arg Thr Tyr Gln Asp Ala Arg 165 170 175Ala Ala Thr Ser Ala Gln Thr
Lys Ala Ser Gly Glu Ala Glu Asn Ala 180 185 190Asn Arg Asn Ala Ala
Ala Thr Leu Ala Ala Val Leu Ser Ile Ala Lys 195 200 205Ala Ala Ser
Gly Gln Gly Gly Thr Arg Ala Ala Val Asp Ala Ala Ala 210 215 220Ala
Ala Ala Ala Ala Ala Ala Leu His Ala Lys Ala Asn Ala Val Ser225 230
235 240Gln Ala Thr Ser Lys Ala Ala Ala Glu Ala Arg Val Ala Ala Glu
Glu 245 250 255Ala Ala Ser Ala Gln Ala Ser Ala Ser Ala Ser Ala Gln
Leu Thr Ala 260 265 270Gln Leu Glu Glu Lys Val Ser Ala Asp Gln Gln
Ala Ala Ser Ala Ser 275 280 285Thr Asp Thr Ser Ala Ala Ile Ala Glu
Ala Glu Ala Ala Ala Leu Ala 290 295 300Ser Thr Val Asn Ala Ile Asn
Asp Gly Val Val Ile Gly Leu Gly Asn305 310 315 320Thr Ala Ser Ser
Ser Ala Gln Ala Ser Ala Gln Ala Ser Ala Leu Ala 325 330 335Arg Ala
Lys Asn Ala Arg Pro Lys Ile Lys Gly Trp Tyr Lys Ile Gly 340 345
350Gly Ala Thr Ser Ala Ser Ala Ser Ala Ser Ala Ser Ala Ser Ala Gln
355 360 365Ser Ser Ser Gln Gly Leu Val Tyr 370
37561395PRTOecophylla smaragdina 61Met Lys Ile Pro Ala Ile Leu Val
Thr Ser Phe Leu Ala Trp Gly Leu1 5 10 15Ala Ser Gly Gly Val Pro Lys
Glu Leu Gly Thr Ser Ile Ser Ser Ala 20 25 30Ser Ala Ser Ala Ser Ala
Ser Ala Ser Ala Thr Ala Ser Ser Ser Ser 35 40 45Lys Asn Val His Leu
Leu Pro Leu Lys Ser Glu His Gly Ile Val Ile 50 55 60Asp Lys Ser Lys
Phe Asn Ile Arg Lys Val Val Leu Ser Ala Ile Asp65 70 75 80Glu Ile
Asn Gly Ala Pro Asn Ile Gly Leu Gly Leu Lys Gln Val Ser 85 90 95Leu
Ala Leu Ala Lys Ala Gln Ala Ser Ala Gln Ser Ser Ala Glu Ala 100 105
110Leu Ala Ile Ile Lys Lys Ile Val Ala Leu Leu Ile Ser Ala Tyr Val
115 120 125Arg Ala Ala Glu Ala Ala Ala Arg Ala Ser Ala Glu Ala Leu
Ala Thr 130 135 140Val Arg Ala Ala Glu Gln Ala Gln Lys Ile Ala Glu
Ala Lys Gly Arg145 150 155 160Ala Ala Ala Glu Ala Leu Ser Glu Leu
Val Glu Ala Ser Gln Lys Ala 165 170 175Asp Ala Ala Ala Ala Gly Thr
Thr Asp Ala Ile Glu Arg Thr Tyr Gln 180 185 190Asp Ala Arg Ala Ala
Thr Ser Ala Gln Thr Lys Ala Ser Gly Glu Ala 195 200 205Glu Asn Ala
Asn Arg Asn Ala Ala Ala Thr Leu Ala Ala Val Leu Ser 210 215 220Ile
Ala Lys Ala Ala Ser Gly Gln Gly Gly Thr Arg Ala Ala Val Asp225 230
235 240Ala Ala Ala Ala Ala Ala Ala Ala Ala Ala Leu His Ala Lys Ala
Asn 245 250 255Ala Val Ser Gln Ala Thr Ser Lys Ala Ala Ala Glu Ala
Arg Val Ala 260 265 270Ala Glu Glu Ala Ala Ser Ala Gln Ala Ser Ala
Ser Ala Ser Ala Gln 275 280 285Leu Thr Ala Gln Leu Glu Glu Lys Val
Ser Ala Asp Gln Gln Ala Ala 290 295 300Ser Ala Ser Thr Asp Thr Ser
Ala Ala Ile Ala Glu Ala Glu Ala Ala305 310 315 320Ala Leu Ala Ser
Thr Val Asn Ala Ile Asn Asp Gly Val Val Ile Gly 325 330 335Leu Gly
Asn Thr Ala Ser Ser Ser Ala Gln Ala Ser Ala Gln Ala Ser 340 345
350Ala Leu Ala Arg Ala Lys Asn Ala Arg Pro Lys Ile Lys Gly Trp Tyr
355 360 365Lys Ile Gly Gly Ala Thr Ser Ala Ser Ala Ser Ala Ser Ala
Ser Ala 370 375 380Ser Ala Gln Ser Ser Ser Gln Gly Leu Val Tyr385
390 39562424PRTOecophylla smaragdina 62Ser Glu Leu Val Gly Ser Asp
Ala Ser Ala Thr Ala Ser Ala Glu Ala1 5 10 15Ser Ala Ser Ser Ser Ala
Tyr Gly Ser Lys Tyr Gly Ile Gly Ser Gly 20 25 30Ala Val Ser Gly Ala
Ser Ala Ser Ala Ser Ala Ser Ala Ser Ala Ser 35 40 45Ala Ser Ala Ser
Ser Ala Pro Ala Ile Glu Gly Val Asn Val Gly Thr 50 55 60Gly Val Ser
Asn Thr Ala Ser Ala Ser Ala Glu Ala Leu Ser Arg Gly65 70 75 80Leu
Gly Ile Gly Gln Ala Ala Ala Glu Ala Gln Ala Ala Ala Ala Gly 85 90
95Gln Ala Ala Ile Ala Ala Lys Ser Cys Ala Leu Ala Ala Lys Ser Thr
100 105
110Ala Gln Ala Val Ala Leu Val Glu Lys Val Ala Arg Ala Glu Val Asp
115 120 125Leu Ala Glu Ser Ala Arg Lys Ala Thr Arg Leu Ser Ala Glu
Ala Ala 130 135 140Lys Ala Ala Ala Glu Val Glu Lys Asp Leu Val Gly
Leu Arg Gly Ala145 150 155 160Ala Gly Lys Leu Asn Leu Ala Ala Arg
Ala Gly Ser Lys Ala Gln Glu 165 170 175Arg Ala Asn Glu Asp Ser Ile
Glu Ala Asn Glu Leu Ala Gln Ala Thr 180 185 190Ala Ala Ala Gly Ala
Glu Ala Glu Ala Lys Ala Asn Ala Ala Gln Glu 195 200 205Ala Gly Ala
Ser Ala Leu Ala Ile Ala Gln Ala Ala Leu Asn Ile Glu 210 215 220Gln
Glu Thr Val Lys Leu Thr Arg Gln Ala Gln Asn Thr Arg Leu Arg225 230
235 240Ser Glu Asn Ile Leu Ala Ala Ala Ser Asn Ala Arg Ala Ile Ala
Ser 245 250 255Ala Glu Ala Glu Ala Ser Ser Asp Leu Asn Asn Arg Ala
Asn Ala Ala 260 265 270Arg Ser Asn Ala Arg Ala Ala Ala Glu Thr Arg
Ala Val Ala Thr Glu 275 280 285Ala Ala Ser Thr Ala Glu Ile Ala Ala
Tyr Ser Ser Ser Glu Lys Gly 290 295 300Glu Ile Thr Asn Pro Gly Pro
Leu Pro Lys Ile Val Ser Val Thr Ala305 310 315 320Gly Leu Thr Gln
Asn Glu Ile Ala Gly Ser Gly Ala Ala Ala Ser Ala 325 330 335Ser Ala
Ser Ala Leu Ala Ser Ala Ser Ala Gly Ala Gly Ala Gly Ala 340 345
350Gly Ala Gly Ala Gly Ala Ser Ala Gly Ala Gly Ala Val Ala Gly Ala
355 360 365Gly Ala Gly Ala Gly Ala Gly Ala Ser Ala Gly Ala Ser Ala
Gly Ala 370 375 380Asn Ala Gly Ala Gly Ala Ser Ser Leu Leu Leu Pro
Gln Ser Lys Leu385 390 395 400His Pro Ile Ser Arg Ser Ser Ala Ser
Ala Ser Ala Ser Ala Glu Ala 405 410 415Glu Ala Asn Ser Ser Ala Tyr
Ala 42063443PRTOecophylla smaragdina 63Met Lys Ile Pro Ala Ile Leu
Ala Thr Ser Leu Phe Val Trp Gly Leu1 5 10 15Val Gly Ala Ser Glu Leu
Val Gly Ser Asp Ala Ser Ala Thr Ala Ser 20 25 30Ala Glu Ala Ser Ala
Ser Ser Ser Ala Tyr Gly Ser Lys Tyr Gly Ile 35 40 45Gly Ser Gly Ala
Val Ser Gly Ala Ser Ala Ser Ala Ser Ala Ser Ala 50 55 60Ser Ala Ser
Ala Ser Ala Ser Ser Ala Pro Ala Ile Glu Gly Val Asn65 70 75 80Val
Gly Thr Gly Val Ser Asn Thr Ala Ser Ala Ser Ala Glu Ala Leu 85 90
95Ser Arg Gly Leu Gly Ile Gly Gln Ala Ala Ala Glu Ala Gln Ala Ala
100 105 110Ala Ala Gly Gln Ala Ala Ile Ala Ala Lys Ser Cys Ala Leu
Ala Ala 115 120 125Lys Ser Thr Ala Gln Ala Val Ala Leu Val Glu Lys
Val Ala Arg Ala 130 135 140Glu Val Asp Leu Ala Glu Ser Ala Arg Lys
Ala Thr Arg Leu Ser Ala145 150 155 160Glu Ala Ala Lys Ala Ala Ala
Glu Val Glu Lys Asp Leu Val Gly Leu 165 170 175Arg Gly Ala Ala Gly
Lys Leu Asn Leu Ala Ala Arg Ala Gly Ser Lys 180 185 190Ala Gln Glu
Arg Ala Asn Glu Asp Ser Ile Glu Ala Asn Glu Leu Ala 195 200 205Gln
Ala Thr Ala Ala Ala Gly Ala Glu Ala Glu Ala Lys Ala Asn Ala 210 215
220Ala Gln Glu Ala Gly Ala Ser Ala Leu Ala Ile Ala Gln Ala Ala
Leu225 230 235 240Asn Ile Glu Gln Glu Thr Val Lys Leu Thr Arg Gln
Ala Gln Asn Thr 245 250 255Arg Leu Arg Ser Glu Asn Ile Leu Ala Ala
Ala Ser Asn Ala Arg Ala 260 265 270Ile Ala Ser Ala Glu Ala Glu Ala
Ser Ser Asp Leu Asn Asn Arg Ala 275 280 285Asn Ala Ala Arg Ser Asn
Ala Arg Ala Ala Ala Glu Thr Arg Ala Val 290 295 300Ala Thr Glu Ala
Ala Ser Thr Ala Glu Ile Ala Ala Tyr Ser Ser Ser305 310 315 320Glu
Lys Gly Glu Ile Thr Asn Pro Gly Pro Leu Pro Lys Ile Val Ser 325 330
335Val Thr Ala Gly Leu Thr Gln Asn Glu Ile Ala Gly Ser Gly Ala Ala
340 345 350Ala Ser Ala Ser Ala Ser Ala Leu Ala Ser Ala Ser Ala Gly
Ala Gly 355 360 365Ala Gly Ala Gly Ala Gly Ala Gly Ala Ser Ala Gly
Ala Gly Ala Val 370 375 380Ala Gly Ala Gly Ala Gly Ala Gly Ala Gly
Ala Ser Ala Gly Ala Ser385 390 395 400Ala Gly Ala Asn Ala Gly Ala
Gly Ala Ser Ser Leu Leu Leu Pro Gln 405 410 415Ser Lys Leu His Pro
Ile Ser Arg Ser Ser Ala Ser Ala Ser Ala Ser 420 425 430Ala Glu Ala
Glu Ala Asn Ser Ser Ala Tyr Ala 435 440641119DNAOecophylla
smaragdina 64agcaagtcgt acctcttagg ctcatccgcg tctgcttccg cttccgcttc
cgcctcggca 60tcagcgggag gaagcaccgg cggcgtcggc gtcggatctg taatatccgg
tggcaacaac 120atcatcagag gagcttcgac cacatccgtg acattggcag
ccgccgcagc ggaggccaag 180gcagctctga atgctggaaa agcgactgtc
gaagagcaaa gggaagcgtt acagttgctc 240accgcgtccg ctgaaaaaaa
cgccgaggcg cgttccttgg ccgacgatgc ggccgttcta 300gttcagggtg
ccgctgaggc gcaatcggtc gccgccgcga agacggtcgc ggtcgagcaa
360ggatccaact ctctggatgc agctgcagcc gaagcggaag ccgccgccgc
cgcatccagg 420gtatcggccc agcaggcact ccaggccgcg cagacctccg
ccgccgctat tcaaaccgct 480gccggtagcg ccctgacggc tctcaaattg
gcacgcaaac aggaagcgga atccaataat 540gccgccgaac aggcaaataa
agcattggcc ttaagtcgcg cagccagcgc tgccactcaa 600cgagccgtgg
cagctcagaa cgcggctgcc gcatcagcgg cttcggctgg agccgcacaa
660gctgaggcaa ggaacgccta cgccaaagcc aaagcagcga tagctgctct
tacggccgcc 720caaagaaatt acgccgcggc caaggctagc gcaagcgcgg
gtagcgtggt ggccgaacaa 780gatgctcaat ctagagcggc cgatgccgag
gtgaacgccg ttgcccaagc cgctgcccga 840gccagcgttc gcaatcagga
gatcgttgaa atcggcgcgg aattcggcaa cgccagcggc 900ggagtgatct
cgaccggcac acgttcttcc ggaggcaagg gtgtctccgt taccgctgga
960gctcaggcta gcgcgtccgc ttccgcgacc tcctcctcct cctcctcctc
cggcatcaac 1020aaaggacatc ccagatgggg gcacaattgg ggtttaggtt
cttcggaagc gtcagcaaac 1080gctgaagccg aaagcagcgc ttcctcttat
tcatcttaa 1119651176DNAOecophylla smaragdina 65atgaagatcc
cagcgataat cgcaacgacc ctccttctct ggggtttcgc cgacgccagc 60aagtcgtacc
tcttaggctc atccgcgtct gcttccgctt ccgcttccgc ctcggcatca
120gcgggaggaa gcaccggcgg cgtcggcgtc ggatctgtaa tatccggtgg
caacaacatc 180atcagaggag cttcgaccac atccgtgaca ttggcagccg
ccgcagcgga ggccaaggca 240gctctgaatg ctggaaaagc gactgtcgaa
gagcaaaggg aagcgttaca gttgctcacc 300gcgtccgctg aaaaaaacgc
cgaggcgcgt tccttggccg acgatgcggc cgttctagtt 360cagggtgccg
ctgaggcgca atcggtcgcc gccgcgaaga cggtcgcggt cgagcaagga
420tccaactctc tggatgcagc tgcagccgaa gcggaagccg ccgccgccgc
atccagggta 480tcggcccagc aggcactcca ggccgcgcag acctccgccg
ccgctattca aaccgctgcc 540ggtagcgccc tgacggctct caaattggca
cgcaaacagg aagcggaatc caataatgcc 600gccgaacagg caaataaagc
attggcctta agtcgcgcag ccagcgctgc cactcaacga 660gccgtggcag
ctcagaacgc ggctgccgca tcagcggctt cggctggagc cgcacaagct
720gaggcaagga acgcctacgc caaagccaaa gcagcgatag ctgctcttac
ggccgcccaa 780agaaattacg ccgcggccaa ggctagcgca agcgcgggta
gcgtggtggc cgaacaagat 840gctcaatcta gagcggccga tgccgaggtg
aacgccgttg cccaagccgc tgcccgagcc 900agcgttcgca atcaggagat
cgttgaaatc ggcgcggaat tcggcaacgc cagcggcgga 960gtgatctcga
ccggcacacg ttcttccgga ggcaagggtg tctccgttac cgctggagct
1020caggctagcg cgtccgcttc cgcgacctcc tcctcctcct cctcctccgg
catcaacaaa 1080ggacatccca gatgggggca caattggggt ttaggttctt
cggaagcgtc agcaaacgct 1140gaagccgaaa gcagcgcttc ctcttattca tcttaa
1176661146DNAOecophylla smaragdina 66ggagtcatag gtcccgacac
gtcctcatcg tcccaggcat cggcatcggc atcggcgtca 60gcatcggcgt cggcatcatc
gtcggcatcg atcggttaca acgaactcca taaatcgatc 120aatgcgcccg
ccttggcggt cggcgtcaag aacggcggag tggatgtcgc caagggcgcg
180gccgttgtcg aatcagcgat atccgacgta tcgactctaa ccgatgatcg
tacgttgaac 240ggtctcgcta tcatcgggaa tagcgccgag agtctggcaa
gagcacaggc ttcctcgagc 300gccagcgccg gcgcaaaagc caatgctctc
atcaaacaat cgatagcggc tatagagatc 360accgaaaagg cagagtacct
tgcgtcgatc gtcgccacca aggcagcgaa ggccgccgag 420gccacagcgg
ccgcgaccgc tcgcgccact gccgtcgccg aggctgccaa ggtttccagc
480gagcaattcg cggccgaggc acgcgcggcc gccgacgccg aagccaaggc
caacgccgct 540tccatcatcg ccaacaaagc gaacgccgtc ctcgcggagg
cagccaccgg acttagcgcc 600agcgctggca aagcccaaca atcggcgacc
agggcgttgc aagccgcacg agctgccgct 660aaggctcaag ccgaacttac
ccagaaagcc gctcaaatct tagtcctcat tgctgaagcc 720aaagccgccg
tgagccgagc aagcgccgat caatccgtct gtacgtccca ggcacaagcc
780gccagtcaga ttcaatcgag agcctccgcg gccgaatccg cggcatcggc
tcaatcggaa 840gccaacacca ttgcggccga ggcggtcgct agagctgacg
ccgaggcggc cagtcaagct 900caagcgtggg ccgaatcctt caaacgcgaa
ctctcgagtg tcgttttgga ggccgaggcc 960aatgcctcgg ctagtgcctc
ggctggtgcc ctggccagtg gtagcagcag ctcgggcgcg 1020agttccagcg
cggatgccag cgccggagcg agcagctatg gatccttggg cggatatcga
1080cacggcggaa gcttcagcga ggcatcggca gccgcgtcag cggccagtcg
cgccgaggct 1140gcgtaa 1146671203DNAOecophylla smaragdina
67atgaagattc cagcgatatt cgtgacgtct ctgctcgcct ggggactcgc cagcggcgga
60gtcataggtc ccgacacgtc ctcatcgtcc caggcatcgg catcggcatc ggcgtcagca
120tcggcgtcgg catcatcgtc ggcatcgatc ggttacaacg aactccataa
atcgatcaat 180gcgcccgcct tggcggtcgg cgtcaagaac ggcggagtgg
atgtcgccaa gggcgcggcc 240gttgtcgaat cagcgatatc cgacgtatcg
actctaaccg atgatcgtac gttgaacggt 300ctcgctatca tcgggaatag
cgccgagagt ctggcaagag cacaggcttc ctcgagcgcc 360agcgccggcg
caaaagccaa tgctctcatc aaacaatcga tagcggctat agagatcacc
420gaaaaggcag agtaccttgc gtcgatcgtc gccaccaagg cagcgaaggc
cgccgaggcc 480acagcggccg cgaccgctcg cgccactgcc gtcgccgagg
ctgccaaggt ttccagcgag 540caattcgcgg ccgaggcacg cgcggccgcc
gacgccgaag ccaaggccaa cgccgcttcc 600atcatcgcca acaaagcgaa
cgccgtcctc gcggaggcag ccaccggact tagcgccagc 660gctggcaaag
cccaacaatc ggcgaccagg gcgttgcaag ccgcacgagc tgccgctaag
720gctcaagccg aacttaccca gaaagccgct caaatcttag tcctcattgc
tgaagccaaa 780gccgccgtga gccgagcaag cgccgatcaa tccgtctgta
cgtcccaggc acaagccgcc 840agtcagattc aatcgagagc ctccgcggcc
gaatccgcgg catcggctca atcggaagcc 900aacaccattg cggccgaggc
ggtcgctaga gctgacgccg aggcggccag tcaagctcaa 960gcgtgggccg
aatccttcaa acgcgaactc tcgagtgtcg ttttggaggc cgaggccaat
1020gcctcggcta gtgcctcggc tggtgccctg gccagtggta gcagcagctc
gggcgcgagt 1080tccagcgcgg atgccagcgc cggagcgagc agctatggat
ccttgggcgg atatcgacac 1140ggcggaagct tcagcgaggc atcggcagcc
gcgtcagcgg ccagtcgcgc cgaggctgcg 1200taa 1203681131DNAOecophylla
smargdina 68ggtgtcccta aagagttggg aacttccatt tcttccgcgt ccgcatccgc
atccgcatcc 60gcatccgcga ccgcgtcctc cagtagcaag aatgttcact tattaccatt
gaaaagcgag 120catggcatcg taattgacaa gtcaaaattc aacatcagaa
aggtagtgtt gagcgcaatc 180gatgagatca acggcgcgcc caacatcggt
ctgggattga aacaggtcag tttggcgctc 240gcaaaagccc aggctagtgc
tcaatcgagc gccgaggcat tggcaatcat caagaaaatc 300gtcgcgctcc
tcatctcggc ctacgtcaga gcagccgagg ccgcggctcg agcatccgcc
360gaagctttag ctaccgttag ggctgcggaa caagcgcaaa aaattgctga
agcgaagggt 420agagcggctg ctgaggcgct ctccgagtta gtcgaggcgt
cccagaaggc cgatgcggcg 480gccgcgggaa cgacggacgc gatcgaacgc
acctaccagg atgccagagc ggccacttcc 540gcacagacca aggccagcgg
cgaagccgag aatgctaatc gcaatgctgc cgccaccctc 600gcggcggtct
tgagcatcgc taaggccgcc tccggtcaag gaggcactcg agccgctgtc
660gatgcagctg ctgccgctgc cgccgcagcc gctctgcatg ctaaagctaa
cgcggtttcg 720caagctacca gcaaagcagc cgctgaagct agagtcgcgg
ctgaggaggc agcatccgcc 780caggcatccg cctcagcaag cgcacagctg
accgcacaat tagaggagaa agtcagcgcc 840gatcaacaag cagcctccgc
cagtactgat acctccgctg ctatagccga ggctgaagct 900gccgcgttag
cgtccaccgt caacgcgatc aacgacggag tggtcatcgg attaggaaat
960accgccagtt cttctgccca agcttccgca caggccagtg ctctcgctcg
cgcaaaaaat 1020gcgcgcccta aaataaaggg ctggtacaaa atcggaggcg
cgacttccgc ttctgcaagc 1080gcatcggcca gcgcttccgc ccagtcatcc
tcgcaaggac tggtatacta g 1131691188DNAOecophylla smaragdina
69atgaagattc cagcgatact cgtgacgtcc ttcctcgcct ggggactggc cagcgggggt
60gtccctaaag agttgggaac ttccatttct tccgcgtccg catccgcatc cgcatccgca
120tccgcgaccg cgtcctccag tagcaagaat gttcacttat taccattgaa
aagcgagcat 180ggcatcgtaa ttgacaagtc aaaattcaac atcagaaagg
tagtgttgag cgcaatcgat 240gagatcaacg gcgcgcccaa catcggtctg
ggattgaaac aggtcagttt ggcgctcgca 300aaagcccagg ctagtgctca
atcgagcgcc gaggcattgg caatcatcaa gaaaatcgtc 360gcgctcctca
tctcggccta cgtcagagca gccgaggccg cggctcgagc atccgccgaa
420gctttagcta ccgttagggc tgcggaacaa gcgcaaaaaa ttgctgaagc
gaagggtaga 480gcggctgctg aggcgctctc cgagttagtc gaggcgtccc
agaaggccga tgcggcggcc 540gcgggaacga cggacgcgat cgaacgcacc
taccaggatg ccagagcggc cacttccgca 600cagaccaagg ccagcggcga
agccgagaat gctaatcgca atgctgccgc caccctcgcg 660gcggtcttga
gcatcgctaa ggccgcctcc ggtcaaggag gcactcgagc cgctgtcgat
720gcagctgctg ccgctgccgc cgcagccgct ctgcatgcta aagctaacgc
ggtttcgcaa 780gctaccagca aagcagccgc tgaagctaga gtcgcggctg
aggaggcagc atccgcccag 840gcatccgcct cagcaagcgc acagctgacc
gcacaattag aggagaaagt cagcgccgat 900caacaagcag cctccgccag
tactgatacc tccgctgcta tagccgaggc tgaagctgcc 960gcgttagcgt
ccaccgtcaa cgcgatcaac gacggagtgg tcatcggatt aggaaatacc
1020gccagttctt ctgcccaagc ttccgcacag gccagtgctc tcgctcgcgc
aaaaaatgcg 1080cgccctaaaa taaagggctg gtacaaaatc ggaggcgcga
cttccgcttc tgcaagcgca 1140tcggccagcg cttccgccca gtcatcctcg
caaggactgg tatactag 1188701275DNAOecophylla smaragdina 70agcgaactcg
tcggatcgga cgcgagcgcg acggcatctg ctgaagcgtc agcatcgtca 60tccgcatacg
gtagcaagta tggtattggt agtggtgctg tctccggtgc atcagccagc
120gcctctgcca gcgcgtctgc tagcgcatca gccagcagtg ctcccgcgat
cgaaggagta 180aacgttggca ccggagtcag taacaccgct tccgcgtccg
cagaagctct ctcccgtgga 240ctcggcatcg gacaagcggc tgccgaagcg
caagccgctg ccgctggcca agcggcgatc 300gctgcgaaat cgtgcgcgct
agcggccaag agcaccgctc aagcggttgc cctggttgag 360aaagtggccc
gcgccgaggt agatctggcc gaaagcgcga gaaaggctac aagattatcg
420gcagaagcag ccaaggcagc ggcggaagtc gagaaggacc tcgtcggtct
gagaggggct 480gccggtaaac tgaatctggc tgcgagagcc ggttctaaag
cccaagaacg cgccaacgaa 540gactctatag aggctaacga acttgcccaa
gcaacggccg ccgccggtgc cgaggctgaa 600gccaaggcga atgccgccca
ggaggcaggc gcctccgctt tggccatcgc ccaagccgcc 660cttaacatcg
agcaagagac tgttaaattg acccgccagg cccagaatac tcgtctcaga
720tctgaaaata ttctcgccgc ggccagcaat gcccgcgcca tcgcttccgc
tgaggccgag 780gccagtagtg atttgaataa tcgtgcgaat gcagcgcgtt
ccaatgcccg agctgctgcc 840gagaccagag ccgtagctac cgaagccgct
tctaccgccg agatcgcagc ttatagttca 900tccgagaaag gcgagatcac
caatcccggt cctctgccca agatcgtcag tgttaccgca 960ggtctgaccc
agaacgaaat agcgggatca ggagcggccg ctagtgctag tgccagtgct
1020cttgccagtg ccagtgccgg tgccggtgcc ggtgcaggtg caggagccgg
tgcaagtgca 1080ggagccggtg cagttgcagg tgcaggagcc ggtgcaggag
ccggtgctag tgccggagcg 1140agtgccggag cgaatgccgg tgccggtgcc
agcagtttac tcttgccgca gagtaaactc 1200catccaatct ccaggtcttc
cgcctctgcc tccgcttccg ccgaggccga agctaacagt 1260tcggcgtatg cgtaa
1275711332DNAOecophylla smaragdina 71atgaagattc cagcgatact
tgcgacgtcc cttttcgtct ggggtcttgt cggcgccagc 60gaactcgtcg gatcggacgc
gagcgcgacg gcatctgctg aagcgtcagc atcgtcatcc 120gcatacggta
gcaagtatgg tattggtagt ggtgctgtct ccggtgcatc agccagcgcc
180tctgccagcg cgtctgctag cgcatcagcc agcagtgctc ccgcgatcga
aggagtaaac 240gttggcaccg gagtcagtaa caccgcttcc gcgtccgcag
aagctctctc ccgtggactc 300ggcatcggac aagcggctgc cgaagcgcaa
gccgctgccg ctggccaagc ggcgatcgct 360gcgaaatcgt gcgcgctagc
ggccaagagc accgctcaag cggttgccct ggttgagaaa 420gtggcccgcg
ccgaggtaga tctggccgaa agcgcgagaa aggctacaag attatcggca
480gaagcagcca aggcagcggc ggaagtcgag aaggacctcg tcggtctgag
aggggctgcc 540ggtaaactga atctggctgc gagagccggt tctaaagccc
aagaacgcgc caacgaagac 600tctatagagg ctaacgaact tgcccaagca
acggccgccg ccggtgccga ggctgaagcc 660aaggcgaatg ccgcccagga
ggcaggcgcc tccgctttgg ccatcgccca agccgccctt 720aacatcgagc
aagagactgt taaattgacc cgccaggccc agaatactcg tctcagatct
780gaaaatattc tcgccgcggc cagcaatgcc cgcgccatcg cttccgctga
ggccgaggcc 840agtagtgatt tgaataatcg tgcgaatgca gcgcgttcca
atgcccgagc tgctgccgag 900accagagccg tagctaccga agccgcttct
accgccgaga tcgcagctta tagttcatcc 960gagaaaggcg agatcaccaa
tcccggtcct ctgcccaaga tcgtcagtgt taccgcaggt 1020ctgacccaga
acgaaatagc gggatcagga gcggccgcta gtgctagtgc cagtgctctt
1080gccagtgcca gtgccggtgc cggtgccggt gcaggtgcag gagccggtgc
aagtgcagga 1140gccggtgcag ttgcaggtgc aggagccggt gcaggagccg
gtgctagtgc cggagcgagt 1200gccggagcga atgccggtgc cggtgccagc
agtttactct tgccgcagag taaactccat 1260ccaatctcca ggtcttccgc
ctctgcctcc gcttccgccg aggccgaagc taacagttcg 1320gcgtatgcgt aa
133272562PRTMallada signata 72Ala Val Leu Ile Ser Gly Ser Ala Ala
Gly Ala Ser Ser His Asn Ala1 5 10 15Ala Gly Ala Ala Ala Ala Ala Arg
Ala Ala Leu Gly Ala Ser Gly Ala 20 25 30Ala Gly Leu Gly Ala Ala Ser
Gly Ala Ala Arg Arg Asn Val Ala Val 35 40 45Gly Ala Asn Gly Ala Ala
Ala Ala Ser Ala Ala Ala Ala Ala Ala Arg 50
55 60Arg Ala Gly Ala Ile Gly Leu Asn Gly Ala Ala Gly Ala Asn Val
Ala65 70 75 80Val Ala Gly Gly Lys Lys Gly Gly Ala Ala Gly Leu Asn
Ala Gly Ala 85 90 95Gly Ala Ser Leu Val Ser Ala Ala Ala Arg Arg Asn
Gly Ala Leu Gly 100 105 110Leu Asn Gly Ala Ala Gly Ala Asn Leu Ala
Ala Ala Gly Gly Lys Lys 115 120 125Gly Gly Ala Ile Gly Leu Asn Ala
Gly Ala Ser Ala Asn Val Gly Ala 130 135 140Ala Ala Ala Lys Lys Asn
Gly Ala Ile Gly Leu Asn Ser Ala Ala Ser145 150 155 160Ala Asn Ala
Ala Ala Ala Ala Ala Lys Lys Gly Gly Ala Ile Gly Leu 165 170 175Asn
Ala Gly Ala Ser Ala Asn Ala Ala Ala Ala Ala Ala Lys Lys Ser 180 185
190Gly Ala Val Gly Leu Asn Ala Gly Ala Ser Ala Asn Ala Ala Ala Ala
195 200 205Ala Ala Lys Lys Ser Gly Ala Val Ala Ala Asn Ser Ala Ala
Ser Ala 210 215 220Asn Ala Ala Ala Ala Ala Gln Lys Lys Ala Ala Ala
Asp Ala Ala Asn225 230 235 240Ala Ala Ala Ser Glu Ser Ala Ala Ala
Ala Ala Ala Lys Lys Ala Ala 245 250 255Ala Val Ala Glu Asn Ala Ala
Ala Thr Ala Asn Ala Ala Ser Ala Leu 260 265 270Arg Lys Asn Ala Leu
Ala Ile Ala Ser Asp Ala Ala Ala Val Arg Ala 275 280 285Asp Ala Ala
Ala Ala Ala Ala Asp Asp Ala Ala Lys Ala Asn Asn Ala 290 295 300Ala
Ser Arg Gly Ser Asp Gly Leu Thr Ala Arg Ala Asn Ala Ala Thr305 310
315 320Leu Ala Ser Asp Ala Ala Arg Arg Ala Ser Asn Ala Ala Thr Ala
Ala 325 330 335Ser Asp Ala Ala Thr Asp Arg Leu Asn Ala Ala Thr Ala
Ala Ser Asn 340 345 350Ala Ala Thr Ala Arg Ala Asn Ala Ala Thr Arg
Ala Asp Asp Ala Ala 355 360 365Thr Asp Ala Asp Asn Ala Ala Ser Lys
Ala Ser Asp Val Ser Ala Ile 370 375 380Glu Ala Asp Asn Ala Ala Arg
Ala Ala Asp Ala Asp Ala Ile Ala Thr385 390 395 400Asn Arg Ala Ala
Glu Ala Ser Asp Ala Ala Ala Ile Ala Ala Asp Ala 405 410 415Ala Ala
Asn Ala Ala Asp Ala Ala Ala Gln Cys Asn Asn Lys Val Ala 420 425
430Arg Val Ser Asp Ala Leu Ala Leu Ala Ala Asn Ala Ala Ala Arg Gly
435 440 445Ser Asp Ala Ala Ala Glu Ala Gln Asp Ala Val Ala Arg Ala
Ser Asp 450 455 460Ala Ala Ala Ala Gln Ala Asp Gly Val Ala Ile Ala
Val Asn Gly Ala465 470 475 480Thr Ala Arg Asp Ser Ala Ile Glu Ala
Ala Ala Thr Ala Gly Ala Ala 485 490 495Gln Ala Lys Ala Ala Gly Arg
Ala Gly Ala Ala Ala Ala Gly Leu Arg 500 505 510Ala Gly Ala Ala Arg
Gly Ala Ala Ala Gly Ser Ala Arg Gly Leu Ala 515 520 525Gly Gly Leu
Ala Ala Gly Ser Asn Ala Gly Ile Ala Ala Gly Ala Ala 530 535 540Ser
Gly Leu Ala Arg Gly Ala Ala Ala Glu Val Cys Ala Ala Arg Ile545 550
555 560Ala Leu73588PRTMallada signata 73Met Ala Ala Ser Asn Lys Ile
Ile Phe Ser Phe Leu Ala Ile Val Leu1 5 10 15Leu Gln Leu Ala Thr His
Cys Ser Ser Thr Ala Val Leu Ile Ser Gly 20 25 30Ser Ala Ala Gly Ala
Ser Ser His Asn Ala Ala Gly Ala Ala Ala Ala 35 40 45Ala Arg Ala Ala
Leu Gly Ala Ser Gly Ala Ala Gly Leu Gly Ala Ala 50 55 60Ser Gly Ala
Ala Arg Arg Asn Val Ala Val Gly Ala Asn Gly Ala Ala65 70 75 80Ala
Ala Ser Ala Ala Ala Ala Ala Ala Arg Arg Ala Gly Ala Ile Gly 85 90
95Leu Asn Gly Ala Ala Gly Ala Asn Val Ala Val Ala Gly Gly Lys Lys
100 105 110Gly Gly Ala Ala Gly Leu Asn Ala Gly Ala Gly Ala Ser Leu
Val Ser 115 120 125Ala Ala Ala Arg Arg Asn Gly Ala Leu Gly Leu Asn
Gly Ala Ala Gly 130 135 140Ala Asn Leu Ala Ala Ala Gly Gly Lys Lys
Gly Gly Ala Ile Gly Leu145 150 155 160Asn Ala Gly Ala Ser Ala Asn
Val Gly Ala Ala Ala Ala Lys Lys Asn 165 170 175Gly Ala Ile Gly Leu
Asn Ser Ala Ala Ser Ala Asn Ala Ala Ala Ala 180 185 190Ala Ala Lys
Lys Gly Gly Ala Ile Gly Leu Asn Ala Gly Ala Ser Ala 195 200 205Asn
Ala Ala Ala Ala Ala Ala Lys Lys Ser Gly Ala Val Gly Leu Asn 210 215
220Ala Gly Ala Ser Ala Asn Ala Ala Ala Ala Ala Ala Lys Lys Ser
Gly225 230 235 240Ala Val Ala Ala Asn Ser Ala Ala Ser Ala Asn Ala
Ala Ala Ala Ala 245 250 255Gln Lys Lys Ala Ala Ala Asp Ala Ala Asn
Ala Ala Ala Ser Glu Ser 260 265 270Ala Ala Ala Ala Ala Ala Lys Lys
Ala Ala Ala Val Ala Glu Asn Ala 275 280 285Ala Ala Thr Ala Asn Ala
Ala Ser Ala Leu Arg Lys Asn Ala Leu Ala 290 295 300Ile Ala Ser Asp
Ala Ala Ala Val Arg Ala Asp Ala Ala Ala Ala Ala305 310 315 320Ala
Asp Asp Ala Ala Lys Ala Asn Asn Ala Ala Ser Arg Gly Ser Asp 325 330
335Gly Leu Thr Ala Arg Ala Asn Ala Ala Thr Leu Ala Ser Asp Ala Ala
340 345 350Arg Arg Ala Ser Asn Ala Ala Thr Ala Ala Ser Asp Ala Ala
Thr Asp 355 360 365Arg Leu Asn Ala Ala Thr Ala Ala Ser Asn Ala Ala
Thr Ala Arg Ala 370 375 380Asn Ala Ala Thr Arg Ala Asp Asp Ala Ala
Thr Asp Ala Asp Asn Ala385 390 395 400Ala Ser Lys Ala Ser Asp Val
Ser Ala Ile Glu Ala Asp Asn Ala Ala 405 410 415Arg Ala Ala Asp Ala
Asp Ala Ile Ala Thr Asn Arg Ala Ala Glu Ala 420 425 430Ser Asp Ala
Ala Ala Ile Ala Ala Asp Ala Ala Ala Asn Ala Ala Asp 435 440 445Ala
Ala Ala Gln Cys Asn Asn Lys Val Ala Arg Val Ser Asp Ala Leu 450 455
460Ala Leu Ala Ala Asn Ala Ala Ala Arg Gly Ser Asp Ala Ala Ala
Glu465 470 475 480Ala Gln Asp Ala Val Ala Arg Ala Ser Asp Ala Ala
Ala Ala Gln Ala 485 490 495Asp Gly Val Ala Ile Ala Val Asn Gly Ala
Thr Ala Arg Asp Ser Ala 500 505 510Ile Glu Ala Ala Ala Thr Ala Gly
Ala Ala Gln Ala Lys Ala Ala Gly 515 520 525Arg Ala Gly Ala Ala Ala
Ala Gly Leu Arg Ala Gly Ala Ala Arg Gly 530 535 540Ala Ala Ala Gly
Ser Ala Arg Gly Leu Ala Gly Gly Leu Ala Ala Gly545 550 555 560Ser
Asn Ala Gly Ile Ala Ala Gly Ala Ala Ser Gly Leu Ala Arg Gly 565 570
575Ala Ala Ala Glu Val Cys Ala Ala Arg Ile Ala Leu 580
585741689DNAMallada signata 74gctgtattga tttctggttc ggctgctggt
gcttcctcac acaatgctgc tggtgcagct 60gcagcagcca gagctgcctt aggcgcttct
ggggctgcag gtttaggtgc tgcatctggt 120gctgcaagaa gaaacgtagc
agttggtgct aacggtgccg ccgccgctag tgctgcagct 180gcagctgcca
gacgagctgg cgctattggc ctaaatggag cagctggagc taatgtagct
240gtcgctggtg gcaaaaaagg aggtgctgct ggattaaatg ctggcgctgg
tgcttcttta 300gtatctgcag ctgcaagacg aaatggagcc cttggactta
acggtgcagc tggagcaaat 360ctcgcagcag ctggtggcaa aaaaggaggt
gctattggat taaacgctgg agcatcagcc 420aatgttggtg ccgctgctgc
caagaaaaat ggagccatag gacttaactc agctgcttca 480gctaatgctg
ccgctgccgc tgctaaaaaa ggtggagcca ttggattgaa tgctggagct
540tcagcaaatg ctgctgctgc cgctgccaag aagagtggag ctgttggatt
aaatgctgga 600gcttctgcta acgctgctgc tgctgctgcc aagaaaagtg
gagctgttgc tgccaattcc 660gctgcttcag caaatgcagc tgctgctgca
caaaagaaag ccgctgctga tgccgcaaat 720gctgctgctt ctgaaagtgc
tgctgctgct gcagccaaga aagccgccgc tgttgctgaa 780aatgcagctg
ccaccgccaa tgccgcttca gctttacgta aaaatgcatt agccattgcc
840agtgatgcag cagctgtccg tgctgatgcc gctgccgccg ccgctgacga
tgctgctaaa 900gctaacaacg ctgcttcccg tggaagtgat ggtttaactg
cccgcgccaa tgccgccact 960ttagccagtg atgctgcccg tagagctagc
aatgcagcaa cagctgccag cgatgctgcc 1020actgaccgat tgaacgccgc
caccgctgct agcaacgctg ccactgctcg tgcaaatgcc 1080gccacacgtg
ccgatgatgc cgccactgat gccgacaatg ctgcttcaaa ggccagtgat
1140gtatcagcta ttgaagccga caacgctgca cgagctgctg atgctgatgc
tatcgctacc 1200aaccgtgccg ctgaagcaag cgatgctgct gctattgccg
ctgatgccgc tgccaatgct 1260gctgatgccg ctgcccaatg taataacaaa
gttgcccgag taagtgatgc cttagctctc 1320gccgctaatg ctgctgcccg
aggatctgat gccgccgctg aagctcaaga tgctgttgcc 1380agagcaagtg
acgctgccgc tgcccaagct gatggtgttg ccattgccgt aaatggagct
1440actgcgagag actcagcaat tgaagccgct gctactgctg gagctgccca
agctaaagcc 1500gctggacgtg ctggagctgc tgcagctggt ttaagagctg
gtgccgctag aggtgctgcc 1560gctggtagtg cccgcggtct agctggagga
ttagctgcag gttccaatgc tggaatcgcg 1620gctggtgcag cttctggatt
agcaagaggc gcagctgctg aagtttgcgc agctagaata 1680gcattgtaa
1689751767DNAMallada signata 75atggcagcgt cgaacaaaat catcttcagc
tttttagcta ttgttctatt acaacttgcc 60acacactgtt catcaacagc tgtattgatt
tctggttcgg ctgctggtgc ttcctcacac 120aatgctgctg gtgcagctgc
agcagccaga gctgccttag gcgcttctgg ggctgcaggt 180ttaggtgctg
catctggtgc tgcaagaaga aacgtagcag ttggtgctaa cggtgccgcc
240gccgctagtg ctgcagctgc agctgccaga cgagctggcg ctattggcct
aaatggagca 300gctggagcta atgtagctgt cgctggtggc aaaaaaggag
gtgctgctgg attaaatgct 360ggcgctggtg cttctttagt atctgcagct
gcaagacgaa atggagccct tggacttaac 420ggtgcagctg gagcaaatct
cgcagcagct ggtggcaaaa aaggaggtgc tattggatta 480aacgctggag
catcagccaa tgttggtgcc gctgctgcca agaaaaatgg agccatagga
540cttaactcag ctgcttcagc taatgctgcc gctgccgctg ctaaaaaagg
tggagccatt 600ggattgaatg ctggagcttc agcaaatgct gctgctgccg
ctgccaagaa gagtggagct 660gttggattaa atgctggagc ttctgctaac
gctgctgctg ctgctgccaa gaaaagtgga 720gctgttgctg ccaattccgc
tgcttcagca aatgcagctg ctgctgcaca aaagaaagcc 780gctgctgatg
ccgcaaatgc tgctgcttct gaaagtgctg ctgctgctgc agccaagaaa
840gccgccgctg ttgctgaaaa tgcagctgcc accgccaatg ccgcttcagc
tttacgtaaa 900aatgcattag ccattgccag tgatgcagca gctgtccgtg
ctgatgccgc tgccgccgcc 960gctgacgatg ctgctaaagc taacaacgct
gcttcccgtg gaagtgatgg tttaactgcc 1020cgcgccaatg ccgccacttt
agccagtgat gctgcccgta gagctagcaa tgcagcaaca 1080gctgccagcg
atgctgccac tgaccgattg aacgccgcca ccgctgctag caacgctgcc
1140actgctcgtg caaatgccgc cacacgtgcc gatgatgccg ccactgatgc
cgacaatgct 1200gcttcaaagg ccagtgatgt atcagctatt gaagccgaca
acgctgcacg agctgctgat 1260gctgatgcta tcgctaccaa ccgtgccgct
gaagcaagcg atgctgctgc tattgccgct 1320gatgccgctg ccaatgctgc
tgatgccgct gcccaatgta ataacaaagt tgcccgagta 1380agtgatgcct
tagctctcgc cgctaatgct gctgcccgag gatctgatgc cgccgctgaa
1440gctcaagatg ctgttgccag agcaagtgac gctgccgctg cccaagctga
tggtgttgcc 1500attgccgtaa atggagctac tgcgagagac tcagcaattg
aagccgctgc tactgctgga 1560gctgcccaag ctaaagccgc tggacgtgct
ggagctgctg cagctggttt aagagctggt 1620gccgctagag gtgctgccgc
tggtagtgcc cgcggtctag ctggaggatt agctgcaggt 1680tccaatgctg
gaatcgcggc tggtgcagct tctggattag caagaggcgc agctgctgaa
1740gtttgcgcag ctagaatagc attgtaa 176776975DNAArtificialHoneybee
silk protein (Xenospira4) open reading frame optimized for plant
expression (before sub-cloned into pET14b and pVEC8) 76atggctagag
aagaggttga gactagggat aagactaaga cttctactgt ggtgaagtct 60gagaaggttg
aagttgtggc tccagctaag gatgagctta agttgacttc tgagccaatt
120ttcggaagaa gagtgggaac tggagcttct gaagtggctt cttctagtgg
agaggctatt 180gctatttctc ttggagctgg acaatcagca gcagagtctc
aagctcttgc tgcttctcag 240tctaagactg ctgctaacgc tgctattggt
gcttctgagc ttactaacaa ggtggcagct 300cttgttgctg gtgctactgg
tgctcaagct agagctactg ctgcttcttc ttctgctctt 360aaggcttctc
ttgctactga agaggctgct gaagaagctg aagctgctgt tgcagatgct
420aaagcagctg ctgagaaggc tgagtctctt gctaagaacc ttgcttctgc
tagtgctaga 480gctgctcttt cttctgagag ggctaatgag cttgctcagg
ctgaaagtgc tgcagctgct 540gaagctcaag ctaagaccgc tgctgctgcc
aaagcagctg agattgctct taaggtggca 600gagattgctg taaaagctga
ggcagatgct gccgccgcag ccgtggcagc tgcaaaagct 660agagctgtgg
ctgatgcagc agccgccagg gctgctgctg ttaacgctat tgctaaggct
720gaagaagagg cttcagctca agctgagaac gcagctggtg ttcttcaagc
agctgcaagt 780gctgctgctg agtcaagagc agcagcagcc gctgccgcag
ctacttctga agcagcagct 840gaagcaggac cacttgctgg tgaaatgaag
ccaccacatt ggaagtggga gaggattcca 900gtgaagaaag aagagtggaa
aacttctaca aaagaggaat ggaaaactac taacgaagag 960tgggaggtga agtga
97577324PRTArtificialHoneybee silk protein (Xenospira4) encoded by
open reading frame optimized for plant expression (without
translational fusion) 77Met Ala Arg Glu Glu Val Glu Thr Arg Asp Lys
Thr Lys Thr Ser Thr1 5 10 15Val Val Lys Ser Glu Lys Val Glu Val Val
Ala Pro Ala Lys Asp Glu 20 25 30Leu Lys Leu Thr Ser Glu Pro Ile Phe
Gly Arg Arg Val Gly Thr Gly 35 40 45Ala Ser Glu Val Ala Ser Ser Ser
Gly Glu Ala Ile Ala Ile Ser Leu 50 55 60Gly Ala Gly Gln Ser Ala Ala
Glu Ser Gln Ala Leu Ala Ala Ser Gln65 70 75 80Ser Lys Thr Ala Ala
Asn Ala Ala Ile Gly Ala Ser Glu Leu Thr Asn 85 90 95Lys Val Ala Ala
Leu Val Ala Gly Ala Thr Gly Ala Gln Ala Arg Ala 100 105 110Thr Ala
Ala Ser Ser Ser Ala Leu Lys Ala Ser Leu Ala Thr Glu Glu 115 120
125Ala Ala Glu Glu Ala Glu Ala Ala Val Ala Asp Ala Lys Ala Ala Ala
130 135 140Glu Lys Ala Glu Ser Leu Ala Lys Asn Leu Ala Ser Ala Ser
Ala Arg145 150 155 160Ala Ala Leu Ser Ser Glu Arg Ala Asn Glu Leu
Ala Gln Ala Glu Ser 165 170 175Ala Ala Ala Ala Glu Ala Gln Ala Lys
Thr Ala Ala Ala Ala Lys Ala 180 185 190Ala Glu Ile Ala Leu Lys Val
Ala Glu Ile Ala Val Lys Ala Glu Ala 195 200 205Asp Ala Ala Ala Ala
Ala Val Ala Ala Ala Lys Ala Arg Ala Val Ala 210 215 220Asp Ala Ala
Ala Ala Arg Ala Ala Ala Val Asn Ala Ile Ala Lys Ala225 230 235
240Glu Glu Glu Ala Ser Ala Gln Ala Glu Asn Ala Ala Gly Val Leu Gln
245 250 255Ala Ala Ala Ser Ala Ala Ala Glu Ser Arg Ala Ala Ala Ala
Ala Ala 260 265 270Ala Ala Thr Ser Glu Ala Ala Ala Glu Ala Gly Pro
Leu Ala Gly Glu 275 280 285Met Lys Pro Pro His Trp Lys Trp Glu Arg
Ile Pro Val Lys Lys Glu 290 295 300Glu Trp Lys Thr Ser Thr Lys Glu
Glu Trp Lys Thr Thr Asn Glu Glu305 310 315 320Trp Glu Val Lys
D00000

D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014

D00015

D00016

D00017

D00018

D00019

D00020

D00021

D00022

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.