Methods And Kits For Determining A Personalized Treatment Regimen For A Subject Suffering From A Pathologic Disorder
SMITH; Yoav
U.S. patent application number 17/100987 was filed with the patent office on 2021-03-18 for methods and kits for determining a personalized treatment regimen for a subject suffering from a pathologic disorder. The applicant listed for this patent is YISSUM RESEARCH DEVELOPMENT COMPANY OF THE HEBREW UNIVERSITY OF JERUSALEM LTD.. Invention is credited to Yoav SMITH.
| Application Number | 20210079475 17/100987 |
| Document ID | / |
| Family ID | 1000005237342 |
| Filed Date | 2021-03-18 |











View All Diagrams
| United States Patent Application | 20210079475 |
| Kind Code | A1 |
| SMITH; Yoav | March 18, 2021 |
METHODS AND KITS FOR DETERMINING A PERSONALIZED TREATMENT REGIMEN FOR A SUBJECT SUFFERING FROM A PATHOLOGIC DISORDER
Abstract
The invention relates to methods and kits for determining and optimizing a personalized treatment regimen for a subject suffering from a pathologic disorder based on calculating the value of M, that indicates the ability of said subject to eliminate said disorder. The invention specifically relates to optimization of interferon treatment of viral disorders.
| Inventors: | SMITH; Yoav; (Jerusalem, IL) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 1000005237342 | ||||||||||
| Appl. No.: | 17/100987 | ||||||||||
| Filed: | November 23, 2020 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 15303427 | Oct 11, 2016 | 10846371 | ||
| PCT/IL2015/050363 | Apr 2, 2015 | |||
| 17100987 | ||||
| 61977966 | Apr 10, 2014 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C12Q 1/6883 20130101; G16H 10/40 20180101; C12Q 2600/106 20130101; Y02A 90/10 20180101; G01N 2800/52 20130101; C12Q 2600/158 20130101; G16H 20/10 20180101; G16B 20/00 20190201 |
| International Class: | C12Q 1/6883 20060101 C12Q001/6883; G16B 20/00 20060101 G16B020/00; G16H 10/40 20060101 G16H010/40; G16H 20/10 20060101 G16H020/10 |
Claims
1. A method for determining a personalized treatment regimen for a subject suffering from a pathologic disorder, said method comprises the step of: a. Calculating the value of M, wherein said value indicates the ability of said subject to eliminate said disorder; b. Determining the value of M1, said value indicates the minimal ability required for eliminating said disorder; c. providing the dose A1 and number B1 of administrations of said dose to obtain an amount C1 of said medicament required for eliminating said disorder in subjects having a value of M that is equal or above said M1 value, wherein A1*B1=C1; d. Calculating the dose A and number B of administrations of said dose A to obtain an amount C1 required for said subject having said M determined/calculated in step (a), wherein said A=A1/(M1/M) and B=B1*(M1/M); thereby at least one of determining and optimizing the treatment regimen for said subject.
2. The method according to claim 1, wherein calculating the value of M is performed by the steps of any one of: I. A static analysis comprising: Ia. determining the level of expression of at least one of ISG15, IFIT1, IFIT2, IFITM3, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7, IFI6, STAT1, IFI44, EIF2AK2 and DHX58 genes in a biological sample of said subject, to obtain an expression value Ex.sub.samp in said sample; Ib. providing a standard curve of expression values of subjects suffering from the same pathologic disorder; Ic. Obtaining a maximal expression value Ex.sub.max and a minimal expression value Ex.sub.min from said standard curve of (Ib); and Id. Calculating the M value of said sample, wherein M=1-[(Ex.sub.samp-Ex.sub.min)/(Ex.sub.max-Ex.sub.min]; II. An induced dynamic analysis comprising: IIa. determining the level of expression of at least one of genes in a biological sample of said subject, to obtain an expression value in said sample; IIb. exposing or contacting said subject or at least one other sample obtained from said subject to or with an immuno-stimulant; IIc. determining the level of expression of at least one of ISG15, IFIT1, IFIT2, IFITM3, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7, IFI6, STAT1, IFI44, EIF2AK2 and DHX58 genes in said at least one other biological sample of said subject obtained in step IIb; and IId. calculating the rate of change (RC) between the expression value obtained in step (IIa), and the expression value obtained in step (IIc), thereby obtaining the rate of change in the sample RC.sub.samp; IIe. providing a standard curve of the rate of change in the expression of at least one of ISG15, IFIT1, IFIT2, IFITM3, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7, IFI6, STAT1, IFI44, EIF2AK2 and DHX58 genes in subjects treated with said immuno-stimulant; IIf. Obtaining a maximal rate of change value RC.sub.max and a minimal rate of change RC.sub.min value from said standard curve of (IIe); and IIg. Calculating the M value of said sample, wherein M=[(RC.sub.samp-RC.sub.min)/(RC.sub.max-RC.sub.min)], thereby obtaining an M value of said subject; or III. A dynamic analysis comprising: IIIa. determining the level of expression of at least one of ISG15, IFIT1, IFIT2, IFITM3, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7, IFI6, STAT1, IFI44, EIF2AK2 and DHX58 genes in a biological sample of said subject, to obtain an expression value in said sample, wherein said sample is obtained prior the initiation of said treatment with said medicament; IIIb. determining the level of expression of at least one ISG15, IFIT1, IFIT2, IFITM3, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7, IFI6, STAT1, IFI44, EIF2AK2 and DHX58 genes in at least one other biological sample of said subject, wherein said at least one other sample is obtained after the initiation of said treatment; IIIc. calculating the rate of change between the expression value obtained in step (IIIa), and the expression value obtained in step (IIIb), thereby obtaining the rate of change in the sample RC.sub.samp; IIId. providing a predetermined standard curve of the rate of change in the expression of at least one of ISG15, IFIT1, IFIT2, IFITM3, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7, IFI6, STAT1, IFI44, EIF2AK2 and DHX58 genes in subjects suffering from the same disorder that were treated with said medicament; IIIe. Obtaining a maximal rate of change value RC.sub.max and a minimal rate of change value RC.sub.min from said standard curve of (IId); and IIIf. Calculating the M value of said sample, wherein M=RC.sub.samp-RC.sub.min)/(RC.sub.max-RC.sub.min)], thereby obtaining an M value of said subject.
3. The method according to claim 2, wherein calculating the value of M1 is performed by the steps of any one of: I. Ia. Providing a K value for said disorder; Ib. calculating the M1, wherein M1.gtoreq.1-(1/k), thereby determining the M1 value; and II. IIa. Providing a standard M1 value calculated for a responder population.
4. The method according to claim 2, wherein in steps (a) and (b) the expression of OAS2, HERC5, UPS18, UBE216 and optionally of ISG15 genes is determined.
5. The method according to claim 1, wherein said method is for determining a personalized interferon treatment regimen for a subject suffering from a pathologic disorder.
6. The method according to claim 2, wherein determining the level of expression of at least one of said ISG15, IFIT1, IFIT2, IFITM3, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7, IFI6, STAT1, IFI44, EIF2AK2 and DHX58 genes in a biological sample of said subject is performed by the step of contacting detecting molecules specific for said genes with a biological sample of said subject, or with any nucleic acid or protein product obtained therefrom, and wherein said detecting molecules are selected from isolated detecting nucleic acid molecules and isolated detecting amino acid molecules.
7. The method according to claim 6, wherein said nucleic acid detecting molecule comprises isolated oligonucleotide/s, each oligonucleotide specifically hybridizes to a nucleic acid sequence of said at least one of ISG15, IFIT1, IFIT2, IFITM3, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7, IFI6, STAT1, IFI44, EIF2AK2 and DHX58 genes and optionally, to a control reference gene, and wherein said detecting molecule is at least one of a pair of primers, at least one primer, nucleotide probes or any combinations thereof.
8. The method according to claim 1, wherein said subject is suffering from an immune-related disorder, and wherein said immune-related disorder is any one of an infectious condition, an autoimmune disease, and a proliferative disorder.
9. The method according to claim 8, wherein said subject is suffering from an infectious condition caused by any one of (Hepatitis C virus) HCV, dengue virus, influenza, poliovirus, HIV (human immune deficiency virus) and West Nile virus (WNV) infection.
10. The method according to claim 8, wherein said subject is suffering from Multiple sclerosis (MS).
11. The method according to claim 8, wherein said subject is suffering from Rheumatoid Arthritis (RA), and wherein said genes are at least one of IFIT1, IFITM3, IFIT3, OAS1, OAS3, HERC5, RSAD2, MX1, IFI44L, IFI6, IFI44 and DDX58 genes.
12. The method according to claim 2, wherein said immuno-stimulant is any one of a synthetic double stranded RNA (poly ICLC), yellow fever (YF) vaccine 17D (YF17D).
13. A kit for determining and/or optimizing a personalized treatment regimen for a subject suffering from a pathologic disorder comprising: a. detecting molecules specific for determining the level of expression of at least one of ISG15, IFIT1, IFIT2, IFITM3, IFI44L, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7, IFI6, STAT1, IFI44, EIF2AK2 and DHX58 genes in a biological sample, wherein said detecting molecules are selected from isolated detecting nucleic acid molecules and isolated detecting amino acid molecules; b. means for calculating the M value of a tested subject, wherein said value indicates the ability of said subject to eliminate said disorder; c. means for calculating the value of M1 or a standard M1 value calculated for a responder population, said M1 value indicates the minimal ability required for eliminating said disorder; and d. means for calculating the dose A and number B of administrations of said dose A to obtain an amount C of said medicament required for said subject.
14. The kit according to claim 13, wherein means for calculating the value of M comprise at least one of: I. means for static analysis comprising: Ia. detecting molecules specific for determining the level of expression of ISG15, IFIT1, IFIT2, IFITM3, IFI44L, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7, IFI6, STAT1, IFI44, EIF2AK2 and DHX58 genes in a biological sample for determining an expression value Ex.sub.samp in said sample; Ib. a standard curve of expression values of subjects suffering from the same pathologic disorder or predetermined maximal expression value Ex.sub.max and a minimal expression value Ex.sub.min calculated from said standard curve; and Ic. a formula for calculating M value, wherein said formula is M=[(Ex.sub.samp-Ex.sub.min)/(Ex.sub.max-Ex.sub.min)]; II. means for an induced dynamic analysis comprising: IIa. detecting molecules specific for determining the level of expression of ISG15, IFIT1, IFIT2, IFITM3, IFI44L, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7, IFI6, STAT1, IFI44, EIF2AK2 and DHX58 genes in a biological sample for determining an expression value Ex.sub.samp in said sample before and after stimulation of said subject with an immuno-stimulant, and for calculating the rate of change RC.sub.samp in the expression value Ex.sub.samp of said sample before and after stimulation; IIb. an immuno-stimulant; IIc. a standard curve of the rate of change in the expression of at least one of ISG15, FIT1, IFIT2, IFITM3, IFI44L, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7, IFI6, STAT1, IFI44, EIF2AK2 and DHX58 genes in subjects treated with said immuno-stimulant, or predetermined maximal rate of change value RC.sub.max and a minimal rate of change RC.sub.min value calculated from said standard curve; and IId. a formula for calculating said M value, wherein said formula is M=[(RC.sub.samp-RC.sub.min)/(RC.sub.max-RC.sub.min)]; and III. means for a dynamic analysis comprising: IIIa. detecting molecules specific for determining the level of expression of ISG15, IFIT1, IFIT2, IFITM3, IFI44L, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7, IFI6, STAT1, IFI44, EIF2AK2 and DHX58 genes in a biological sample for determining an expression value Ex.sub.samp in said sample before and after treatment of said subject with said medicament, and for calculating the rate of change RC.sub.samp in the expression value Ex.sub.samp of said sample; IIIb. a predetermined standard curve of the rate of change in the expression of at least one of ISG15, IFIT1, IFIT2, IFITM3, IFI44L, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7, IFI6, STAT1, IFI44, EIF2AK2 and DHX58 genes in subjects suffering from the same disorder and treated with said medicament, or predetermined maximal rate of change value RC.sub.max and minimal rate of change value RC.sub.min calculated from said standard curve; and IIIc. a formula for calculating said M value, wherein said formula is M=[(RC.sub.samp-RC.sub.min)/(RC.sub.max-RC.sub.min)]; and wherein means for calculating the value of M1 comprise: a. a predetermined K value of said disorder; b. a formula for calculating said M1 value, wherein said formula is M1.gtoreq.1-(1/k).
15. The kit according to claim 13, wherein said nucleic acid detecting molecule comprises isolated oligonucleotides, each oligonucleotide specifically hybridizes to a nucleic acid sequence of said at least one of ISG15, IFIT1, IFIT2, IFITM3, IFI44L, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7, IFI6, STAT1, IFI44, EIF2AK2 and DHX58 genes and optionally, to a control reference gene, and wherein said detecting molecule is at least one of a pair of primers, at least one primer, nucleotide probes or any combinations thereof.
16. The kit according to claim 13, wherein said subject is suffering from an infectious condition caused by any one of HCV, dengue virus, influenza, poliovirus, HIV and WNV infection.
17. The kit according to claim 13, wherein said subject is suffering from Multiple sclerosis (MS).
18. The kit according to claim 13, wherein said subject is suffering from Rheumatoid Arthritis (RA) and wherein said kit comprises detecting molecules specific for determining the level of expression of at least one of IFIT1, IFITM3, IFIT3, OAS1, OAS3, HERC5, RSAD2, MX1, IFI44L, IFI6, IFI44 and DDX58 genes.
19. The kit according to claim 13, wherein said immuno-stimulant is any one of a synthetic double stranded RNA (poly ICLC), yellow fever (YF) vaccine 17D (YF17D).
20. A computer software product for determining and/or optimizing a personalized treatment regimen for a subject suffering from a pathologic disorder, the product comprising a computer readable medium in which program instructions are stored, which instructions, when read by a computer, cause the computer to a. Calculate and/or determine the value of M, wherein said value indicates the ability of said subject to eliminate said disorder; b. Determine the value of M1, said value indicates the minimal ability required for eliminating said disorder; c. calculate the dose A and number B of administrations of said dose A to obtain an amount C required for said subject having said M determined/calculated in step (a), from predetermined dose A1 and number B1 of administrations of said dose, using the formula of A=A/(M1/M) and B=B1*(M1/M).
Description
CROSS REFERENCE TO RELATED APPLICATIONS
[0001] This application is a continuation of U.S. patent application Ser. No. 15/303,427, filed on Oct. 11, 2016, which is a National Phase of PCT Patent Application No. PCT/IL2015/050363 having International filing date of Apr. 2, 2015, which claims the benefit of priority under 35 U. S.C. .sctn. 119(e) of U.S. Provisional Patent Application No. 61/977,966, filed on Apr. 10, 2014. The contents of the above applications are all incorporated by reference as if fully set forth herein in their entirety.
FIELD OF THE INVENTION
[0002] The invention relates to personalized medicine. More specifically, the invention provides methods and kits for determining and optimizing a treatment regimen of a medicament, for a subject suffering from a pathologic disorder.
BACKGROUND REFERENCES
[0003] References considered to be relevant as background to the presently disclosed subject matter are listed below: [0004] Chen Limin, et al., Gastroenterology 128:1437-1444 (2005). [0005] Taylor, M W, et al., Journal of Virology 81:3391-3401 (2007). [0006] van Baarsen L G, et al., PLoS ONE 3:e1927 (2008). [0007] Zeremski M, et al., J. Acquir. Immune. Defic. Syndr. 45:262-268 (2007). [0008] Tarantino G, et al., Digestive and Liver Disease 40:A1-A40 (2008). [0009] US2009/157324 [0010] WO10/076788 [0011] Sadlet A J et al, Nature Reviews Immunology 8: 559 (2008) [0012] Grinde B, et al, Virol J. 4: 24 (2007) [0013] David Stifflerl J. et al., PLoS ONE 4(8) e6661 (2009)
[0014] Acknowledgement of the above references herein is not to be inferred as meaning that these are in any way relevant to the patentability of the presently disclosed subject matter.
BACKGROUND OF THE INVENTION
[0015] Determining treatment protocols that may be suitable for each individual or a subset of individuals is highly desirable. Clinical diagnosis and management has been long focused on clinical sign and symptoms of a patient in order to treat specific diseases. Recently along with the advances in genetic profiling, it became possible to understand the impact of genetic variability as measured in individuals or subsets of individuals on the disease progression.
[0016] Personalized medicine is therefore aimed at enabling decisions and practices to the individual patient by use for example of genetic information.
[0017] It has been recently shown that evaluating the differences in the genetic profile of the two or more groups of patients can provide valuable insight into resistant to treatment.
[0018] For example, interferon therapy is widely used in the treatment of a variety of diseases including for example, multiple sclerosis (MS), hepatitis B, hepatitis C, inflammatory diseases and many cancers types. However, not all subjects treated with interferon equally respond to this therapy and moreover, responsive subjects experience relapse of the disease after remission periods. In fact, in both MS and type 1 hepatitis C Virus (HCV) the success of treatment is only about 50%, namely about half of the patients administered with interferon will not benefit but rather experience only related side effects.
[0019] Chen et al. 2005, compared the gene expression levels in liver specimens taken before treatment from 15 non-responders and 16 responders to Pegylated interferon (IFN-alpha), identified 18 genes that have a significantly different expression between all responders and all non-responders and concluded that up-regulation of a specific set of interferon-responsive gens predict non response to exogenous treatment.
[0020] Taylor M., et al. 2007, found that the induced levels of known interferon-stimulated genes such as the OAS1, OAS2, MX1, IRF-7 and TLR-7 genes is lower in poor-response patients than in marked- or intermediate-response patients.
[0021] Van Baarsen et al., 2008 show that the expression level of interferon response genes in the peripheral blood of multiple sclerosis patients prior to treatment can serve a role as a biomarker for the differential clinical response to interferon beta.
[0022] Zeremaki M, et al., 2007 showed that PEG-interferon induced elevations in IP-10 are greater in responders than in non-responders after the first PEG-interferon dose.
[0023] Tarantino et al., 2008 described that serum levels of B-Lymphocyes stimulator (BLyS) have a potential role as a predictor of outcome in patients with acute hepatitis C.
[0024] The Inventor previous US Patent Application, US2009157324 describes a computational method for selecting a group of genes from a predetermined group of genes whose expression level is significantly different among a first group of individuals (being for example responders to a treatment) and comparing their expression in a second group of individuals (for example not responders). The statistical significance of each group of genes is determined in both up regulated genes or down regulated genes, namely their expression in the first group is higher or lower than in the second group, respectively. The genes in both groups (up regulated and down regulated) are ranked according to number of times each gene was ranked in the highest statistical significant score. A subset of genes having the highest score, either up regulated or down regulated are then selected as biomarkers.
[0025] In another application by the Inventor, International Patent Publication WO10076788, computational and experimental methods are provided for predicting the responsiveness of a subject to interferon therapy by measuring the expression level of various genes such as OAS3, IF16, ISG15, OAS2, IFIT1, KIR3DL3, KIR3DL2, KIR3DL1, KIR2DL1, KIR2DL2, KIR2DL3, KLRG1, KIR3DS1, CD160, HLA-A, HLA-B, HLA-C, HLA-F, HLA-G and IFI27. Specifically, the inventor has found that OAS3, IF16, ISG15, OAS2 and IFIT1 are up-regulated in patients that do not respond to interferon treatment as compared to patients that respond to interferon therapy or compared to healthy controls.
[0026] Thus, the correlations between genetic profiling and personalized medicine, namely treatment regimens, needs to be considered for predicting response to therapy, predicting treatment success and monitoring disease prognosis and pathogenesis, specifically chances for disease relapse.
SUMMARY OF THE INVENTION
[0027] A first aspect of the invention relates to a method for determining and optimizing a personalized treatment regimen for a subject suffering from a pathologic disorder. In certain embodiments, the method of the invention comprises the step of:
[0028] First step (a) involves calculating and determining the value of M. The value M indicates the ability, capability of a specific subject, in this case, the examined subject, to eliminate the specific disorder.
[0029] More specifically, the value of the individual's M reflects the efficiency of the specific tested subject in cellular elements that are required for challenging and eliminating a specific disorder. In certain embodiments the M value indicates the strength of the individual's innate immunity, and may be used for predicting it's ability to eliminate a specific disorder.
[0030] The next step (b), involves determining the value of M1 that indicates the minimal ability required for eliminating said disorder.
[0031] In the nest step (c), providing the dose A1 and number B1 of administrations of such dose to obtain an amount C1 of a specific medicament required for eliminating a specific disorder in subjects having a value of M that is equal or above the optimal M1 value, wherein A1*B1=C1.
[0032] The next step (d) involves calculating the dose A and number B of administrations of such specific dose A to obtain an amount C1 required for the examined subject having the specific M value determined and calculated in step (a). More specifically, the specific optimal dose required for a successful treatment for the tested subject would be A=A1/(M1/M). The specific number of administrations of such dose may be calculated using the formula B=B1*(M1/M); thereby determining and optimizing the treatment regimen for the specific tested subject.
[0033] A further aspect of the invention relates to a kit for determining and optimizing a personalized treatment regimen for a subject suffering from a pathologic disorder.
[0034] In certain embodiments, such kit may comprise elements required for performing any of the methods described above. More specifically, such kit may comprise:
[0035] (a) detecting molecules specific for determining the level of expression of at least one of ISG15, IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7 and IFI6 genes in a biological sample. In certain embodiments the kits of the invention may further comprise detecting molecules for the STAT1, IFI44, EIF2AK2 and DHX58 genes and any combinations thereof with any of the marker genes of the invention.
[0036] The kit of the invention further comprises (b), means for calculating the M value of a tested subject. As noted above, the M value indicates the ability of said subject to eliminate said disorder. The kit of the invention further comprises (c) means for calculating the value of M1 or a standard M1 value calculated for a responder population. As indicated above, the M1 value indicates the minimal ability, or in other words, the optimal M1 value required for a successful elimination of the disorder. Finally, the kit of the invention comprises (d) means for calculating the dose A and number B of administrations of said dose A to obtain an amount C of said medicament required for said subject.
[0037] In yet a further aspect, the invention provides a computer software product for determining and optimizing a personalized treatment regimen for a subject suffering from a pathologic disorder. Such product comprising a computer readable medium in which program instructions are stored, which instructions, when read by a computer, cause the computer to: (a) calculate and determine the value of M that indicates the ability of said subject to eliminate said disorder; (b) determine the value of M1, that indicates the minimal ability required for eliminating said disorder. (c) calculate the dose A and number B of administrations of said dose A to obtain an amount C required for said subject having said M determined/calculated in step (a), from predetermined dose A1 and number B1 of administrations of said dose, using the formula of A=A1/(M1/M) and B=B1*(M1/M).
BRIEF DESCRIPTION OF THE DRAWINGS
[0038] In order to understand the disclosure and to see how it may be carried out in practice, embodiments will now be described, by way of non-limiting example only, with reference to the accompanying drawings, in which:
[0039] FIG. 1 shows a schematic representation at the cellular level of a cell infected with a virus that is multiplied by a rate K followed by the regulation of specific genes caused either by the immune system or by an external treatment that may lead to viral elimination by rate M. The virus or other viruses may penetrate other cells by a certain rate. As schematically illustrated here, the virus may be destroyed by the activity of a set of proteins encoded by a set of genes for example, ISG15, USP18, HERC5 and OAS, in the UPS (Ubiquitin Proteasome System).
[0040] FIG. 2 shows MATLAB simulation of the model with k=2 and p=0 as constant parameters and M being varied from 0 to 1. The lower line represents 0.1 of initial load and provides information whether the down regulation was more than one tenth of the initial virus load.
[0041] FIG. 3 shows MATLAB simulation of the model with k=3 and p=0 as constant parameters and M being varied from 0 to 1. The lower line represents 0.1 of initial load and provides information whether the down regulation was more than one tenth of the initial virus load.
[0042] FIG. 4 shows MATLAB simulation of the model including PI administration with k=2, p=0 and M=0.44 as constant parameters and M1 being varied from 0 to 0.44. As shown by the figure, the optimal M1 should be over 0.3 using the PI treatment.
[0043] FIG. 5 is a volcano plot showing the significant changes in the expression level of different genes in West Nile virus (WNV) infected retinal pigment epithelial (RPE). Expression data was obtained Gene Expression Omnibus Accession No. GSE30719. The "X"-axis represents log 2 of ratio between gene expression measured after 24 hours after infection and a baseline level of the same gene measured before infection, the points present to the right of the right vertical line (shown at a value of 1 on the x-axis), represent genes that were up regulated by more than 2 folds. The "Y" axis shows the p value assigned to each point. The horizontal line corresponds to p-value of 0.05, with points above this line correspond to a p values lower than 0.05 (namely, more significant).
[0044] FIG. 6 is a volcano plot showing the significant changes in the expression level of different genes in H1N1 (left) compare to H5N1 (right). Expression data was obtained from Gene Expression Omnibus Accession No. GSE18816. The "X"-axis represents log 2 of ratio between gene expression measured after 6 hours after infection and a baseline level of the same gene measured before infection, the points present to the right of the right vertical line (shown at a value of 1 on the x-axis), represent genes that were up regulated by more than 2 folds. The "Y" axis shows the p value assigned to each point. The horizontal line corresponds to p-value of 0.05, with points above this line correspond to a p values lower than 0.05 (namely, more significant).
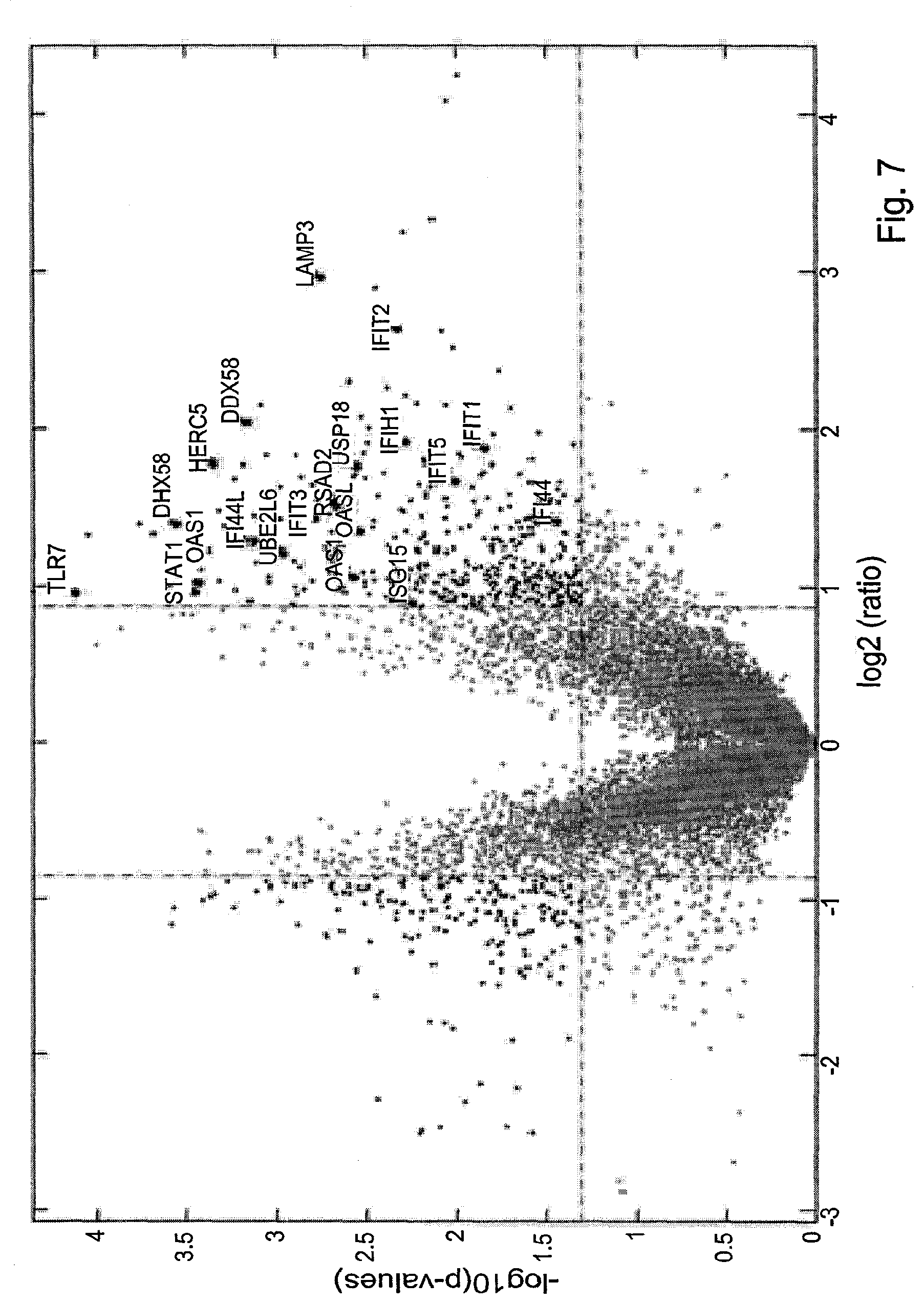
[0045] FIG. 7 is a volcano plot showing the significant changes in the expression level of different genes in blood of children infected with dengue virus. Expression data was obtained from Gene Expression Omnibus Accession No. GSE13052. The "X"-axis represents log 2 of ratio between gene expression measured 4 days after infection in 9 acute dengue shock patients and a baseline level of the same gene measured in 9 acute uncomplicated dengue patients, the points present to the right of the right vertical line (shown at a value of 1 on the x-axis), represent genes that were up regulated by more than 2 folds. The "Y" axis shows the p value assigned to each point. The horizontal line corresponds to p-value of 0.05, with points above this line correspond to a p values lower than 0.05 (namely, more significant).
[0046] FIG. 8 is a volcano plot showing the significant changes in the expression level of different genes in liver biopsies of ten responders and ten non-responders HCV patients before treatment (left) and after one week of IFN and Rib treatment (right). Expression data was obtained from Gene Expression Omnibus Accession No. GSE17183. The "X"-axis represents log 2 of ratio between gene expression measured in responders vs. non-responders, the points present to the right of the right vertical line (shown at a value of 1 on the x-axis), represent genes that were up regulated by more than 2 folds whereas the points present to the left of the left vertical line (shown at a value of -0.75 on the x-axis), represent genes that were down regulated by more than 2 folds. The "Y" axis shows the p value assigned to each point. The horizontal line corresponds to p-value of 0.05, with points above this line correspond to a p values lower than 0.05 (namely, more significant).
[0047] FIG. 9 is a volcano plot showing the significant changes in the expression level of different genes in MS patients three months after treatment with IFN-.beta.. Expression data was obtained from Gene Expression Omnibus Accession No GSE16214. The "X"-axis represents log 2 of ratio between gene expression measured in after treatment, the points present to the right of the right vertical line (shown at a value of 1 on the x-axis), represent genes that were up regulated by more than 2 folds. The "Y" axis shows the p value assigned to each point. The horizontal line corresponds to p-value of 0.05, with points above this line correspond to a p values lower than 0.05 (namely, more significant).
[0048] FIGS. 10A and 10B are graphs showing the expression of IFI27, IFI44L, IFI6, MX1 and ISG15 genes measured in PBMCs of MS patients before (FIG. 10A) and three month after treatment with interferon alpha (IFN-.alpha.) (FIG. 10B). Expression data was downloaded from Gene Expression Omnibus Accession No GSE16214. The "X"-axis represents the subject number and the "Y" axis represents the normalized expression level of the genes.
[0049] FIG. 11 is a graph showing the sum of the expression of the USP18, IFI44, MX1, IFI44L, OAS3, HERC5 and RSAD2 genes (square) and the relapse rate (diamond) of 50 MS patients (patients are indicated in the X-axis.
[0050] FIG. 12 is a graph illustrating the differential expression as calculated from the sum of the common genes, MX1, IFITM3, IFI44L, HERC5, IFI44, IFI6, OAS1, OAS3, RSAD2, IFIT1, IFIT3 and DDX58 in RA patients that are responders and non-responders to infliximab (influx) treatment, and responders and non-responders to RTX treatment (Rituxi=Rituximab).
[0051] FIG. 13 is a graph showing the sum of expression of the IFI27, ISG15, IFIH1, IFI44L, OAS2, DDX58, IFIT1 and IFI6 genes in 44 HIV patients treated with HAART (squares) and the virus load (diamonds).
[0052] FIG. 14 is a graph showing the expression of the ISG15, HERC5, USP10 and UBE2L6 genes, in the ferret experimental groups as indicated therein.
[0053] FIG. 15 is a graph showing the clustering of genes measured for a population of 15 healthy individuals. Expression data was downloaded from Gene Expression Omnibus Accession No GSE838. The "X" axis denotes the tested individual and the "Y" axis represents the measured genes.
[0054] FIG. 16 is a graph showing the expression of IGS15, IFIT1, OAS2 and USP18 genes measured in healthy individuals. Expression data was downloaded from Gene Expression Omnibus Accession No GSE838 the "X"-axis represents the subject number (patients 1-15) and the "Y" axis represents the normalized expression level of the genes ranging from 0 to 1.
[0055] FIG. 17 is a graph showing the clustering of genes measured for a population of 145 healthy individuals. Expression data was downloaded from Gene Expression Omnibus Accession No GSE3649. The "X" axis denotes the tested individual and the "Y" axis represents the measured genes
[0056] FIG. 18 is a graph showing the expression IGS15 and IFIT1 gene measured in healthy individuals. Expression data was downloaded from Gene Expression Omnibus Accession No. GSE3649. The "X"-axis represents the subject number and the "Y" axis represents the normalized expression level of the genes ranging from 0 to 1.
[0057] FIG. 19 is a volcano plot showing the significant changes in the expression level of different genes in healthy individuals 24 hours following injection of poly ICLC. Expression data was obtained from Gene Expression Omnibus Accession No GSE32862. The "X"-axis represents log 2 of ratio between gene expression measured 24 hours as compared to base line level before administration, the points present to the right of the right vertical line (shown at a value of 1 on the x-axis), represent genes that were up regulated by more than 2 folds. The "Y" axis shows the p value assigned to each point. Abbreviations: val. (value); rat. (ratio).
[0058] FIG. 20 is a volcano plot showing the significant changes in the expression level of different genes in healthy individuals 24 hours following injection of poly ICLC. Expression data was obtained from Gene Expression Omnibus Accession No GSE32862. The "X"-axis represents log 2 of ratio between gene expression measured 24 hours as compared to base line level before administration, the points present to the right of the right vertical line (shown at a value of 1 on the x-axis), represent genes that were up regulated by more than 2 folds. The "Y" axis shows the p value assigned to each point. Abbreviations: val. (value); rat. (ratio). The genes ISG15, HERC5 and UBE2L6 are given in squares, IFI44 set (in triangle pointing right), IFIT set (in circles), OAS set (in triangles pointing left), triggers DDX58, TLR7, IFIH1, MYd88.
[0059] FIG. 21 is a graph showing principal component analysis (PCA) of the expression level of IFIT1, IFI44L, IFI6 and ISG15. The data was obtained from PBMC of healthy donors 24 hours after administration of poly ICLC as compared to baseline levels.
[0060] FIG. 22 are graphs showing timing of genes dynamics. Dashed lines correspond to non responders and full lines to responders. X-axis represents time and the Y-axis represents expression of the indicated genes.
[0061] FIG. 23 are graphs showing timing of genes dynamics for longer periods. Dashed lines correspond to non responders and full lines to responders. X-axis represents time and the Y-axis represents expression of the indicated genes.
[0062] FIG. 24 is a histogram graph showing the changes in genes expression after 24 hours following PolyC treatment in healthy donors divided by baseline level of the same gene.
[0063] FIG. 25 is a graph showing the calculation of M from the model equations.
[0064] FIGS. 26A-26B is a graph showing simulation of replication vs. immune defense, per different M. As can be seen for the same individual with an M value suitable for K=3, that is calculated as follows M=1-1/3=0.66 being infected by a variety of viruses with varying K (multiplication rate). FIG. 26A shows that at K rate higher than 3, the virus progresses. FIG. 26B shows situation where K smaller than M, attenuation of the virus is achieved. The X-axis represents time from initial infection different k values and the Y-axis represents the virus load per the different k values.
[0065] FIGS. 27A-27B is a graph showing simulation instructing how much PI is needed per each individuals M and virus K. The PI effectively increases the individuals M, FIG. 27A shows an individual with M=0.6, FIG. 27B shows an individual with M=0.8 both are affected by the same range of PI injections. The better M the quicker an individual to become a responder with the same PI.
[0066] FIG. 28 is a graph showing sum of the expression of the ubiquitin genes, ISG15, USP18, HERC5, UBE2L6, as measured in A549 cells at 2, 4, 6, 8 and 10 hours post infection with the three different influenza strains.
[0067] FIG. 29 is a graph showing the sum of the expression of the ubiquitin genes, ISG15, USP18, HERC5 and UBE2L6 in different time points up to 120 hr (X-axis) post infection of H3N2, in nine different individuals. Each individual is represented in one panel numbered 1 to 9.
[0068] FIG. 30 is a graph showing the simulation results, in the upper panel the virus load of a virus having replication rate of 1.93, the lower panel shows the sum of the expression of the 4 genes, in individual 6 as presented in FIG. 29.
[0069] FIG. 31 is a graph showing the simulation results, in the upper panel the virus load of a virus having replication rate of 1.94, the lower panel shows the sum of the expression of the 4 genes, in individual 6 as presented in FIG. 29.
[0070] FIG. 32 is a bar graph showing the normalized and scaled sum expression of the genes UBE2L6, USP18, HERC5, OAS2 and ISG15 in each one of the tested patients and the amount of reduction in virus load.
[0071] FIG. 33 is a bar graph showing the normalized and scaled (0-1) sum expression of the five genes UBE2L6, USP18, HERC5 and OAS2 in each one of the tested patients and the scaled M values of each patient as calculated using the simulation.
[0072] FIGS. 34A and 34B are bar graphs showing the sum expression of HERC5 and UBE2L6 genes (FIG. 34A) and the M value (FIG. 34B) in IFN responsive and non-responsive HCV patients. The data was obtained from PBMC of HCV patients.
[0073] FIG. 35 is a bar graph showing the expression of HERC5 and the viral load in responsive and non-responsive HCV patients. The data was obtained from PBMC of HCV patients.
[0074] FIG. 36 is a bar graph showing the normalized expression of HERC5 gene (open box) and normalized M value (black box) obtained from model simulation using viral load data in responsive and non-responsive HCV patients. The data was obtained from PBMC of HCV patients.
[0075] FIGS. 37A to 37C are graphs showing correlation between gene expression and M value, with FIG. 37A showing the sum of normalized and scaled expression of the five genes UBE2L6, USP18, HERC5, OAS2 and ISG15, FIG. 37B showing the M value calculated from the gene expression. The data was obtained from liver samples of HCV patients. FIG. 37C shows MATLAB simulation of the model with k=1.92 and p=0 as described herein.
[0076] FIG. 38 is a volcano plot showing the significant changes in the expression level of different genes in liver biopsies in responders and non-responders HCV patients before treatment (left) and after one week of IFN and Rib treatment (right). Expression data was obtained from Masao H. et al. The "X"-axis represents log 2 of ratio between gene expression measured in responders vs. non-responders, the points present to the right of the right vertical line (shown at a value of 1 on the x-axis), represent genes that were up regulated by more than 2 folds whereas the points present to the left of the left vertical line (shown at a value of -0.75 on the x-axis), represent genes that were down regulated by more than 2 folds. The "Y" axis shows the p value assigned to each point. The horizontal line corresponds to p-value of 0.05, with points above this line correspond to a p values lower than 0.05 (namely, more significant).
[0077] FIGS. 39A and 39B are graphs showing the expression of HERC5 gene expression before initiation of IFN and Rib treatment (FIG. 39A) and ratio between HERC5 gene expression measured after one week of treatment and a baseline level of the same gene measured before infection (FIG. 39B). The data was obtained from liver samples of HCV patients.
[0078] FIG. 40 is a graph showing normalized M value obtained from the model simulation for each one of the patients using the virus load data.
[0079] FIGS. 41A and 41B are model simulations predicting viral progression in a non responsive (FIG. 41A) and responsive (FIG. 41B) HCV patients.
[0080] FIG. 42 is a model simulation predicting treatment regimen in HCV patients having an M value of 0.82.
[0081] FIG. 43 is a model simulation predicting treatment regimen in HCV patient having an M value of 0.7995 (patient p18).
DETAILED DESCRIPTION OF THE INVENTION
[0082] The importance of adjusting suitable treatment protocols is highly valuable and clinically desired in view of the fact that a large number of treatment protocols are often associated with some extent of undesired side effects, and moreover, may be unsuccessful. Thus, optimizing a treatment protocol before and/or at early stages after initiation of treatment and/or throughout or after a treatment period may avoid inadequate treatments, reduce unnecessary side effects and improve chance of success. Interferon is widely clinically used for treatment of a variety of diseases including for example inflammatory diseases such as hepatitis C infections, autoimmune diseases such as multiple sclerosis and different types of proliferative disorders. Significant therapeutic advances were made in the treatment of interferon associated diseases however, it is still difficult to determine at the time of disease diagnosis and treatment adjustments, which patients will respond to treatment and which would eventually relapse. Surprisingly, although interferon is considered as a state of art therapy in treatment of these diseases, many of the treated patients do not respond to the therapy and even if they do, many of the patients experience a relapse of the disease.
[0083] Thus, there is a critical need for reliable tailor-made optimization methods that will provide gaudiness and identification of treatment success and failure, breakthrough point and predict inadequate treatments, providing efficient dosing regimens of interferon.
[0084] Thus, a first aspect of the invention relates to a method for determining and optimizing a personalized treatment regimen for a subject suffering from a pathologic disorder. In certain embodiments, the method of the invention comprises the step of:
[0085] First step (a) involves calculating and determining the value of M. It should be further noted that the value M indicates the ability of a specific subject, in this case, the examined subject, to eliminate the specific disorder. More specifically, the value of the individual's M reflects the efficiency of the specific tested subject in requiting cellular elements that are required for challenging and eliminating a specific disorder. In certain embodiments the M value indicates the strength of the individual's innate immunity, and may be used for predicting the individual's ability to eliminate a specific disorder.
[0086] The next step (b), involves determining the value of M1 that indicates the minimal ability required for eliminating the specific disorder. Moreover, the value M1 reflects the optimal threshold of the ability and efficiency of requiting elements required for eliminating a specific disorder. It should be noted that this value is calculated for populations of subjects that perform successful recovery in response to a certain treatment. In some embodiments, this group of subjects may be considered a "responders".
[0087] In the next step (c), providing the dose A1 and number B1 of administrations of such dose to obtain an amount C1 of a specific medicament required for eliminating a specific disorder in subjects having a value of M that is equal or above the optimal M1 value, wherein A1*B1=C1.
[0088] The next step (d) involves calculating the dose A and number B of administrations of such specific dose A to obtain an amount C1 required for the examined subject having the specific M value determined and calculated in step (a). More specifically, the specific optimal dose required for a successful treatment for the tested subject would be A=A1/(M1/M). The specific number of administrations of such dose may be calculated using the formula B=B1*(M1/M); thereby determining and optimizing the treatment regimen for the specific tested subject.
[0089] According to more specific embodiments of the method of the invention, calculating the value of M of the tested individual may be performed using different approaches. It should be noted that such determination may be performed using any of the approaches of the invention or any combination thereof.
[0090] More specifically, determination of the specific M value of the tested individual may be performed by (I) using a static analysis. More specifically, "static" analysis means that the M value may be calculated for a specific individual even before starting a treatment with the particular medicament, and would not reflect any change occurring in response to such treatment.
[0091] In some embodiments, such approach may comprise the steps of:
[0092] First (Ia), determining the level of expression of at least one of ISG15, IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7 and IFI6 genes in a biological sample of the tested subject, to obtain an expression value Ex.sub.samp in the tested sample. In certain embodiments of the method of the invention the level of expression of at least one of STAT1, IFI44, EIF2AK2 and DHX58 genes may be also determined.
[0093] In the next step (Ib), providing a standard curve, specifically, predetermined standard curve of expression values of subjects suffering from the same pathologic disorder.
[0094] In the next step (Ic) Obtaining a maximal expression value Ex.sub.max and a minimal expression value Exon from the standard curve of (Ib), indicating the variance in the gene expression of a certain marker gene in a predetermined population; and finally, step (Id) Calculating the M value of the tested sample. Such calculation is based on using the following formula wherein M=1-[(Ex.sub.samp-Ex.sub.min)/(Ex.sub.max-Ex.sub.min)].
[0095] In yet another embodiment, as an approach for determining the individual's M value, an induced dynamic analysis (II) may be used. It should be appreciated that such approach is based on pre measurements of the M value for an individual, specifically, before such individual was affected by a certain pathologic disorder. More specifically, using such approach, the specific M value of a specific individual may be predetermined, providing information that may be used in the future in case such subject may be affected by any pathologic disorder. More specifically, such predetermined individual value may serve as valuable information that may be used for optimizing treatment regimen for such individual. Moreover, the method of the invention provides the use of such M value for specifically optimized treatment regimen suitable for a certain pathologic disorder.
[0096] In more specific embodiment, the induced dynamic analysis (II) comprises the steps of:
[0097] First (IIa), determining the level of expression of at least one of ISG15, IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7 and IFI6 genes, and optionally of STAT1, IFI44, EIF2AK2 and DHX58 genes in a biological sample of said subject, to obtain an expression value in the tested sample.
[0098] In the second step (IIb) exposing the tested subject to an immuno-stimulant. Alternatively, this step may be performed in vitro, more specifically, a sample of the examined subject may be contacted with an immuno-stimulant.
[0099] The next step (IIc) involves determining the level of expression of at least one of ISG15, IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7 and IFI6 genes, and optionally of STAT1, IFI44, EIF2AK2 and DHX58 genes in a sample of said individual that has been exposed to said immuno-stimulant. In case of the alternative in vitro analysis, determining the level of at least one of these marker genes in at least one other biological sample of the tested subject that has been contacted in vitro with the immuno-stimulant, as indicated in step (IIb).
[0100] In the next step (IId) calculating the rate of change between the expression value obtained in step (IIa), and the expression value obtained in step (IIc), thereby obtaining the rate of change in the sample RC.sub.samp, more specifically, the rate of change in the expression of at least one of the marker genes of the invention, in response to such immuno-stimulant. Such change of expression reflects the intrinsic ability of the tested subject in requiting elements that may be involved in eliminating of any disorder, and therefore reflects the specific ability a certain subject to challenge disorders.
[0101] In the next step (IIe), providing a standard curve, specifically, predetermined standard curve of the rate of change in the expression of at least one of ISG15, IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7 and IFI6 genes, and optionally of STAT1, IFI44, EIF2AK2 and DHX58 genes or any combinations thereof in subjects treated with said immuno-stimulant. It should be noted that such predetermined standard curve is based on a population of healthy subjects (or non-diseased subjects) treated with the same immuno-stimulant.
[0102] In step (IIf) obtaining a maximal rate of change value RC.sub.max and a minimal rate of change RC.sub.min value from said standard curve of (IIe); and
[0103] In final step (IIg), calculating the M value of the tested sample using the formula: wherein M=[(RC.sub.samp-RC.sub.min)/(RC.sub.max-RC.sub.min)], thereby obtaining an M value of said subject.
[0104] In yet another alternative approach, were predetermined M values of an individual are not available, the invention provides a method for optimizing treatment regimen for a subject that has been already started a certain treatment, using a dynamic analysis (III) comprising:
[0105] In the first step (IIIa), determining the level of expression of at least one of ISG15, IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7 and IFI6 genes, and optionally of STAT1, IFI44, EIF2AK2 and DHX58 genes in a biological sample of the tested subject, to obtain an expression value in the tested sample. It should be noted that such sample should be obtained prior the initiation of the specific treatment with said medicament.
[0106] In the next step (IIIb) determining the level of expression of at least one of ISG15, IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7 and IFI6 genes, and optionally of STAT1, IFI44, EIF2AK2 and DHX58 genes in at least one other biological sample of the tested subject. Such at least one other sample should be obtained after the initiation of the specific treatment.
[0107] In step (IIIc) calculating the rate of change between the expression value obtained in step (IIIa), and the expression value obtained in step (IIIb), thereby obtaining the rate of change in the sample RC.sub.samp, in response to such treatment.
[0108] In the next step (IId) providing a standard curve, specifically, predetermined standard curve of the rate of change in the expression of at least one of ISG15, IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7 and IFI6 genes, and optionally of STAT1, IFI44, EIF2AK2 and DHX58 genes in subjects suffering from the same disorder that were treated with the same medicament.
[0109] In step (IIe) obtaining a maximal rate of change value RC.sub.max and a minimal rate of change value RC.sub.min from the standard curve, specifically, predetermined standard curve of (IId); and
[0110] Finally in (IIf), calculating the M value of the tested sample using the following formula: wherein M=[(RC.sub.samp-RC.sub.min)/(RC.sub.max-RC.sub.min)], thereby obtaining an M value of said subject.
[0111] As indicated above, an essential step in the method of the invention is the determination of the expression level of several specific marker genes provided herein. In certain embodiments, these marker genes include at least one of ISG15, IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7 and IFI6 or any combinations thereof.
[0112] In certain embodiments the methods of the invention may further use the STAT1, IFI44, EIF2AK2 and DHX58 genes and any combinations thereof with any of the marker genes of the invention.
[0113] It should be therefore appreciated that the method of the invention may use at least one, at least two, at least three, at least four, at least five, at least six, at least seven, at least eight, at least nine, at least ten, at least eleven, at least twelve, at least thirteen, at least fourteen, at least fifteen, at least sixteen, at least seventeen, at least eighteen, at least nineteen, at least twenty, at least twenty one, at least twenty two, at least twenty three, at least twenty four, at least twenty five or at least twenty six of said marker genes, specifically of any one of ISG15, IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7, IFI6, STAT1, IFI44, EIF2AK2 and DHX58 genes and any combinations thereof. In yet some other embodiments the methods of the invention may use 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 and 26 or more or any combination of the marker genes of the invention. In yet further embodiments, the methods and kits of the invention may use any of the marker genes of the invention with any combination thereof with additional 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100 and more, specifically, 110, 120, 130, 140, 150, 160, 170, 180, 190, 200 and more, specifically, 300, 350 or 400 further marker genes or control reference genes or any combinations thereof. In certain embodiments, such control reference gene (having an equal expression in samples of responsive and non-responsive subjects) may be a house keeping gene, for example, GAPDH or actin.
[0114] As mentioned above, the method and kits of the invention may use the marker genes provided herein, ISG15, IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7, IFI6, STAT1, IFI44, EIF2AK2 and DHX58 genes and any combination thereof.
[0115] More specifically, ISG15 ubiquitin-like modifier (ISG15) gene (GenBank Accession No. NM_005101; SEQ ID NO: 1) encodes the ISG15 protein (GenBank Accession No. NP_005092.1; SEQ ID NO: 2). ISG15 is reported to be an ubiquitin-like protein that is conjugated to intracellular target proteins after IFN-alpha or IFN-beta stimulation. Its enzymatic pathway is partially distinct from that of ubiquitin, differing in substrate specificity and interaction with ligating enzymes. ISG15 conjugation pathway uses a dedicated E1 enzyme, but seems to converge with the ubiquitin conjugation pathway at the level of a specific E2 enzyme. Targets include STAT1, SERPINA3G/SPI2A, JAK1, MAPK3/ERK1, PLCG1, EIF2AK2/PKR, MX1/MxA, and RIG-1. It undergoes deconjugation by USP18/UBP43. It shows specific chemotactic activity towards neutrophils and activates them to induce release of eosinophil chemotactic factors. It was suggested to serve as a trans-acting binding factor directing the association of ligated target proteins to intermediate filaments.
[0116] Interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) gene (GenBank Accession No. NM_001548; SEQ ID NO: 3) encodes the IRF1 protein (GenBank Accession No. NP_001539; SEQ ID NO: 4).
[0117] Interferon-induced protein with tetratricopeptide repeats 2 (IFIT2) gene (GenBank Accession No. NM_001547; SEQ ID NO: 5) encodes the IFIT2 protein (GenBank Accession No. NP_001538; SEQ ID NO: 6).
[0118] Interferon-induced protein with tetratricopeptide repeats 3 (IFIT3) gene (GenBank Accession Nos. NM_001031683; SEQ ID NO: 7, NM_001549; SEQ ID NO: 9) encodes the IFIT3 protein (GenBank Accession Nos. NP_001026853; SEQ ID NO: 8, NP_001540; SEQ ID NO: 10).
[0119] Interferon-induced protein with tetratricopeptide repeats 5 (IFIT5) gene (GenBank Accession No. NM_012420; SEQ ID NO: 11) encodes the IFIT5 protein (GenBank Accession No. NP_036552; SEQ ID NO: 12).
[0120] 2'-5'-oligoadenylate synthetase 1 (OAS1) gene (GenBank Accession No. NM_016816 SEQ ID NO:13, NM_002534 SEQ ID NO:15, NM_001032409 SEQ ID NO:17) encodes the OAS1 protein (GenBank Accession No. NP_058132 SEQ ID NO:14, NP_002525 SEQ ID NO:16, NP_001027581 SEQ ID NO:18). OAS1 encodes a member of the 2-5A synthetase family, essential proteins involved in the innate immune response to viral infection. The encoded protein is induced by interferons and uses adenosine triphosphate in 2'-specific nucleotidyl transfer reactions to synthesize 2', 5'-oligoadenylates (2-5As). These molecules activate latent RNase L, which results in viral RNA degradation and the inhibition of viral replication. The three known members of this gene family are located in a cluster on chromosome 12. Mutations in this gene have been associated with host susceptibility to viral infection. Alternatively spliced transcript variants encoding different isoforms have been described.
[0121] 2'-5'-oligoadenylate synthetase 2 (OAS2) gene (GenBank Accession No. NM_016817 SEQ ID NO:19, NM_002535 SEQ ID NO:21, NM_001032731 SEQ ID NO:23) encodes the OAS2 protein (GenBank Accession No. NP_058197 SEQ ID NO:20, NP_002526 SEQ ID NO:22, NP_001027903 SEQ ID NO:24).
[0122] 2'-5'-oligoadenylate synthetase 3 (OAS3) gene (GenBank Accession No. NM_006187 SEQ ID NO:25) encodes the OAS3 protein (GenBank Accession No. NP_006178.2 SEQ ID NO:26). OAS3 may play a role in mediating resistance to virus infection, control of cell growth, differentiation, and apoptosis. OAS3 synthesizes preferentially dimeric 2', 5'-oligoadenylate molecules. GTP can be an alternative substrate.
[0123] 2'-5'-oligoadenylate synthetase-like (OASL) gene (GenBank Accession Nos. NM_003733; SEQ ID NO: 27, NM_198213; SEQ ID NO: 29) encodes the OASL protein (GenBank Accession Nos. NP_003724; SEQ ID NO: 28, NP_937856; SEQ ID NO: 30).
[0124] HECT and RLD domain containing E3 ubiquitin protein ligase 5 (HERC5) gene (GenBank Accession No. NM_016323; SEQ ID NO: 31) encodes the HERC5 protein (GenBank Accession No. NP_057407 SEQ ID NO: 32). HERC5 gene is a member of the HERC family of ubiquitin ligases and encodes a protein with a HECT domain and five RCC1 repeats. Pro-inflammatory cytokines up regulate expression of this gene in endothelial cells. The HERC5 protein localizes to the cytoplasm and perinuclear region and functions as an interferon-induced E3 protein ligase that mediates ISGylation of protein targets. It is a major E3 ligase for ISG15 conjugation. HERC5 Acts as a positive regulator of innate antiviral response in cells induced by interferon. Makes part of the ISGylation machinery that recognizes target proteins in a broad and relatively non-specific manner.
[0125] Ubiquitin specific peptidase 18 (USP18) gene (GenBank Accession No. NM_017414; SEQ ID NO: 33) encodes the USP18 protein (GenBank Accession No. NP_059110 SEQ ID NO: 34). The protein encoded by this gene belongs to the ubiquitin-specific proteases (UBP) family of enzymes that cleave ubiquitin from ubiquitinated protein substrates. It is highly expressed in liver and thymus, and is localized to the nucleus. USP18 protein efficiently cleaves only ISG15 (a ubiquitin-like protein) fusions, and deletion of this gene in mice results in a massive increase of ISG15 conjugates in tissues, indicating that this protein is a major ISG15-specific protease. Mice lacking this gene are also hypersensitive to interferon, suggesting a function of this protein in downregulating interferon responses, independent of its isopeptidase activity towards ISG15. USP18 can efficiently cleave only ISG15 fusions including native ISG15 conjugates linked via isopeptide bonds. Necessary to maintain a critical cellular balance of ISG15-conjugated proteins in both healthy and stressed organisms.
[0126] Radical S-adenosyl methionine domain containing 2 (RSAD2) gene (GenBank Accession No. NM_080657; SEQ ID NO: 35) encodes the RSAD2 protein (GenBank Accession No. NP_542388; SEQ ID NO: 36). RSAD2 is reported to be involved in antiviral defense. It was suggested to impair virus budding by disrupting lipid rafts at the plasma membrane, a feature which is essential for the budding process of many viruses. In addition, it was reported to act through binding with and inactivating FPPS, an enzyme involved in synthesis of cholesterol, farnesylated and geranylated proteins, ubiquinones dolichol and heme. Moreover, it is considered to play a major role in the cell antiviral state induced by type I and type II interferon. Finally, it was reported to display antiviral effect against HIV-1 virus, hepatitis C virus, human cytomegalovirus, and aphaviruses, but not vesiculovirus.
[0127] Myxovirus (influenza virus) resistance 1 (MX) gene (GenBank Accession No. NM_002462 SEQ ID NO:37, NM_001178046 SEQ ID NO:39, NM_001144925 SEQ ID NO:41) encodes the MX1 protein (GenBank Accession No. NP_002453 SEQ ID NO:38, NP_001171517 SEQ ID NO:40, NP_001138397 SEQ ID NO:42). In mouse, the interferon-inducible Mx protein is responsible for a specific antiviral state against influenza virus infection. The protein encoded by this gene is similar to the mouse protein as determined by its antigenic relatedness, induction conditions, physicochemical properties, and amino acid analysis. This cytoplasmic protein is a member of both the dynamin family and the family of large GTPases. Two transcript variants encoding the same protein have been found for this gene. MX1 may regulate the calcium channel activity of TRPCs. Ring-like assemblies may induce membrane tabulation.
[0128] Interferon-inducedprotein 44-like (IFI44L) gene (GenBank Accession No. NM_006820.3; SEQ ID NO: 43) encodes the IFI44L protein (GenBank Accession No. NP_006811; SEQ ID NO: 44) that belongs to the IFI44 family of proteins is located in the cytoplasm and exhibits a low antiviral activity against hepatitis C. The expression of the protein is induced by type I interferon.
[0129] DEAD (Asp-Glu-Ala-Asp) boxpolypeptide 58 (DDX58) gene (GenBank Accession No. NM_014314; SEQ ID NO: 45) encodes the DDX58 protein (GenBank Accession No. NP_055129; SEQ ID NO: 46). DEAD box proteins, characterized by the conserved motif Asp-Glu-Ala-Asp (DEAD), are putative RNA helicases which are implicated in a number of cellular processes involving RNA binding and alteration of RNA secondary structure. This gene encodes a protein containing RNA helicase-DEAD box protein motifs and a caspase recruitment domain (CARD). It is involved in viral double-stranded (ds) RNA recognition and the regulation of immune response. It is an innate immune receptor which acts as a cytoplasmic sensor of viral nucleic acids and plays a major role in sensing viral infection and in the activation of a cascade of antiviral responses including the induction of type I interferons and pro-inflammatory cytokines. Upon ligand binding it associates with mitochondria antiviral signaling protein (MAVS/IPS1) which activates the IKK-related kinases: TBK1 and IKBKE which phosphorylate interferon regulatory factors: IRF3 and IRF7 which in turn activate transcription of antiviral immunological genes, including interferons (IFNs); IFN-alpha and IFN-beta. Detects both positive and negative strand RNA viruses including members of the families Paramyxoviridae: Human respiratory synctial virus and measles virus (MeV), Rhabdoviridae: vesicular stomatitis virus (VSV), Orthomyxoviridae: influenza A and B virus, Flaviviridae: Japanese encephalitis virus (JEV), hepatitis C virus (HCV), dengue virus (DENV) and west Nile virus (WNV).
[0130] E1-like ubiquitin-activating enzyme (UBE1L) gene (GenBank Accession No. AF294032; SEQ ID NO: 79) encodes the UBE1L protein (GenBank Accession No. AAG49557; SEQ ID NO: 80). UBE1L is the E1-like ubiquitin-activating enzyme for the IFN-stimulated gene, 15-kDa protein (ISG15).
[0131] Ubiquitin-conjugating enzyme E2L 6 (UBE2L6) gene (GenBank Accession No. NM_198183 SEQ ID NO: 81; GenBank Accession No. NM_004223 SEQ ID NO: 83) encodes the UBE2L6 protein (GenBank Accession No. NP_937826 SEQ ID NO: 82; GenBank Accession No. NP_004214 SEQ ID NO: 84). The UBE2L6 gene encodes a member of the E2 ubiquitin-conjugating enzyme family. This enzyme is highly similar in primary structure to the enzyme encoded by the UBE2L3 gene. UBE2L6 catalyzes the covalent attachment of ubiquitin or ISG15 to other proteins. UBE2L6 functions in the E6/E6-AP-induced ubiquitination of p53/TP53. It also promotes ubiquitination and subsequent proteasomal degradation of FLT3.
[0132] Interferon alpha-inducible protein 27 (IFI27) gene (GenBank Accession Nos. NM_001130080 and NM_005532; SEQ ID NOs: 85, 87, respectively) encodes the IFI27 protein (GenBank Accession Nos. NP_001123552 and NP_005523; SEQ ID NOs: 86, 88, respectively). The IFI27 protein was reported to promote cell death and mediate IFN-induced apoptosis characterized by a rapid and robust release of cytochrome C from the mitochondria and activation of BAX and caspases 2, 3, 6, 8 and 9.
[0133] Interferon induced with helicase C domain 1 (IFIH1) gene (GenBank Accession No. NM_022168 SEQ ID NO: 89) encodes the IFIH1 protein (GenBank Accession No. NP_071451 SEQ ID NO: 90). IFIH1 is an innate immune receptor which acts as a cytoplasmic sensor of viral nucleic acids and plays a major role in sensing viral infection and in the activation of a cascade of antiviral responses including the induction of type I interferons and proinflammatory cytokines. Its ligands include mRNA lacking 2'-O-methylation at their 5' cap and long-dsRNA (>1 kb in length). Upon ligand binding it associates with mitochondria antiviral signaling protein (MAVS/IPS1) which activates the IKK-related kinases.
[0134] Toll-like receptor 7 (TLR-7) gene (GenBank Accession No. NM_016562 SEQ ID NO: 91) encodes the TLR-7 protein (GenBank Accession No. NP_057646 SEQ ID NO: 92). The protein encoded by this gene is a member of the Toll-like receptor (TLR) family which plays a fundamental role in pathogen recognition and activation of innate immunity. TLRs are highly conserved from Drosophila to humans and share structural and functional similarities. They recognize pathogen-associated molecular patterns (PAMPs) that are expressed on infectious agents, and mediate the production of cytokines necessary for the development of effective immunity. The various TLRs exhibit different patterns of expression. This gene is predominantly expressed in lung, placenta, and spleen, and lies in close proximity to another family member, TLR8, on chromosome X.
[0135] Interferon regulatory factor 7 (IRF7) gene (GenBank Accession Nos. NM_001572 SEQ ID NO: 93; NM_004029 SEQ ID NO: 95) encodes the IRF7 protein (GenBank Accession Nos. NP_001563 SEQ ID NO: 94; NP_004020 SEQ ID NO: 96). IFR7 is reported to be a transcriptional activator. It binds to the interferon-stimulated response element (ISRE) in IFN promoters and in the Q promoter (Qp) of EBV nuclear antigen 1 (EBNA1). It is also reported to function as a molecular switch for antiviral activity. It is reported to be activated by phosphorylation in response to infection. The activation leads to nuclear retention, DNA binding, and depression of transactivation ability.
[0136] Interferon, alpha-inducible protein 6 (IFI6) gene (GenBank Accession Nos. NM_022873, SEQ ID NO:97; NM_022872, SEQ ID NO:99; NM_002038, SEQ ID NO:101) encodes the IFI6 protein (GenBank Accession Nos. NP_075011, SEQ ID NO:98; NP_075010, SEQ ID NO:100; NP_002029, SEQ ID NO:102). IFI6 gene was first identified as one of the many genes induced by interferon. The encoded IFI6 protein may play a critical role in the regulation of apoptosis.
[0137] In yet another embodiment, the methods, kits and compositions of the invention may further include detecting molecules for the STAT1 gene. Signal transducer and activator of transcription 1 (STAT) gene (GenBank Accession No. NM_007315 SEQ ID NO:103, NM_139266 SEQ ID NO:104) encodes the STAT1 protein (GenBank Accession No. NP_009330 SEQ ID NO:105, NP 644671 SEQ ID NO: 106). Signal transducer and transcription activator that mediates cellular responses to interferons (IFNs), cytokine KITLG/SCF and other cytokines and growth factors.
[0138] Interferon-induced protein 44 (IFI44) gene (GenBank Accession No. NM_006417; SEQ ID NO: 107) encodes the IFI44 protein (GenBank Accession No. NP_006408; SEQ ID NO: 108), that was reported to aggregate to form microtubular structures.
[0139] EIF2AK2 eukaryotic translation initiation factor 2-alpha kinase 2 (EIF2AK2) gene (GenBank Accession No. NM_002759.1; SEQ ID NO: 109) encodes the EIF2AK2 protein (GenBank Accession No. NC_000002.11; SEQ ID NO: 110). The protein encoded by this gene is a serine/threonine protein kinase that is activated by autophosphorylation after binding to dsRNA. The activated form of the encoded protein can phosphorylate translation initiation factor EIF2S1, which in turn inhibits protein synthesis. This protein is also activated by manganese ions and heparin. Three transcript variants encoding two different isoforms have been found for this gene.
[0140] DHX58 DHX58 (DEXH (Asp-Glu-X-His) box polypeptide 58), gene (GenBank Accession No. NM_024119; SEQ ID NO: 111) encodes the DHX58 protein (GenBank Accession No. NC_000017.10; SEQ ID NO: 112). DHX58 acts as a regulator of DDX58/RIG-I and IFIH1/MDA5 mediated antiviral signaling. Cannot initiateantiviral signaling as it lacks the CARD domain required for activating MAVS/IPS1-dependent signaling
[0141] The terms "level of expression" or "expression level" are used interchangeably and generally refer to a numerical representation of the amount (quantity) of a polynucleotide which encodes an amino acid product or protein in a biological sample.
[0142] "Expression" generally refers to the process by which gene-encoded information is converted into the structures present and operating in the cell. For example, biomarker gene expression values measured in Real-Time Polymerase Chain Reaction, sometimes also referred to as RT-PCR or quantitative PCR (qPCR), represent luminosity measured in a tested sample, where an intercalating fluorescent dye is integrated into double-stranded DNA products of the qPCR reaction performed on reverse-transcribed sample RNA, i.e., test sample RNA converted into DNA for the purpose of the assay. The luminosity is captured by a detector that converts the signal intensity into a numerical representation which is said expression value, in terms of miRNA. Therefore, according to the invention "expression" of a gene, specifically, a gene encoding the biomarker genes of the invention may refer to transcription into a polynucleotide. Fragments of the transcribed polynucleotide, the translated protein, or the post-translationally modified protein shall also be regarded as expressed whether they originate from a transcript generated by alternative splicing or a degraded transcript, or from a post-translational processing of the protein, e.g., by proteolysis. Methods for determining the level of expression of the biomarkers of the invention will be described in more detail herein after.
[0143] In certain and specific embodiments, the step of determining the level of expression to obtain an expression value by the method of the invention further comprises an additional and optional step of normalization. According to this embodiment, in addition to determination of the level of expression of the biomarkers of the invention, the level of expression of at least one suitable control reference gene (e.g., housekeeping genes) is being determined in the same sample. According to such embodiment, the expression level of the biomarkers of the invention obtained in step (a) is normalized according to the expression level of said at least one reference control gene obtained in the additional optional step in said test sample, thereby obtaining a normalized expression value. Optionally, similar normalization is performed also in at least one control sample or a representing standard when applicable.
[0144] The term "expression value" thus refers to the result of a calculation, that uses as an input the "level of expression" or "expression level" obtained experimentally and by normalizing the "level of expression" or "expression level" by at least one normalization step as detailed herein, the calculated value termed herein "expression value" is obtained.
[0145] More specifically, as used herein, "normalized values" are the quotient of raw expression values of marker genes, divided by the expression value of a control reference gene from the same sample, such as a stably-expressed housekeeping control gene. Any assayed sample may contain more or less biological material than is intended, due to human error and equipment failures. Importantly, the same error or deviation applies to both the marker genes of the invention and to the control reference gene, whose expression is essentially constant. Thus, division of the marker gene raw expression value by the control reference gene raw expression value yields a quotient which is essentially free from any technical failures or inaccuracies (except for major errors which destroy the sample for testing purposes) and constitutes a normalized expression value of said marker gene. This normalized expression value may then be compared with normalized cutoff values, i.e., cutoff values calculated from normalized expression values. In certain embodiments, the control reference gene may be a gene that maintains stable in all samples analyzed in the microarray analysis.
[0146] It should be noted that normalized biomarker genes expression level values that are higher (positive) or lower (negative) in comparison with a corresponding predetermined standard expression value or a cut-off value in a control sample predict to which population of patients the tested sample belongs.
[0147] It should be appreciated that in some embodiments an important step in determining the expression level is to examine whether the normalized expression value of any one of the biomarker genes of the tested sample is within the range of the expression value of a standard population or a cutoff value for such population.
[0148] More specifically, the specific expression values of the tested samples are compared to a predetermined cutoff value. As used herein the term "comparing" denotes any examination of the expression level and/or expression values obtained in the samples of the invention as detailed throughout in order to discover similarities or differences between at least two different samples. It should be noted that comparing according to the present invention encompasses the possibility to use a computer based approach.
[0149] As described hereinabove, the method of the invention refers to a predetermined cutoff value. It should be noted that a "cutoff value", sometimes referred to simply as "cutoff" herein, is a value that meets the requirements for both high diagnostic sensitivity (true positive rate) and high diagnostic specificity (true negative rate).
[0150] It should be noted that the terms "sensitivity" and "specificity" are used herein with respect to the ability of one or more markers, to correctly classify a sample as belonging to a pre-established population associated with responsiveness to treatment with a certain medicament.
[0151] In certain alternative embodiments, a control sample may be used (instead of, or in addition to, pre-determined cutoff values). Accordingly, the normalized expression values of the biomarker genes used by the invention in the test sample are compared to the expression values in the control sample. In certain embodiments, such control sample may be obtained from at least one of a healthy subject, a subject suffering from the same pathologic disorder, a subject that responds to treatment with said medicament and a non-responder subject.
[0152] The term "response" or "responsiveness" to a certain treatment refers to an improvement in at least one relevant clinical parameter as compared to an untreated subject diagnosed with the same pathology (e.g., the same type, stage, degree and/or classification of the pathology), or as compared to the clinical parameters of the same subject prior to interferon treatment with said medicament.
[0153] The term "non responder" to treatment with a specific medicament, refers to a patient not experiencing an improvement in at least one of the clinical parameter and is diagnosed with the same condition as an untreated subject diagnosed with the same pathology (e.g., the same type, stage, degree and/or classification of the pathology), or experiencing the clinical parameters of the same subject prior to treatment with the specific medicament.
[0154] In case the method of the invention uses the dynamic approaches for determining the M value of the tested individual, at least two samples may be obtained from the subjects. These samples should be obtained from different time points, before and after the treatment, and therefore may be considered as "temporally separated samples". As indicated above, in accordance with some embodiments of the invention, in order to asses response and determine the rate of change in the expression of the marker genes of the invention upon treatment with a specific medicament, at least two "temporally-separated" test samples must be collected from the treated patient and compared thereafter in order to obtain the rate of expression change in the biomarker genes. In practice, to detect a change in the biomarker genes expression, at least two "temporally-separated" test samples and preferably more must be collected from the patient.
[0155] The expression of at least one of the markers is then determined using the method of the invention, applied for each sample. As detailed above, the rate of change in marker expression is calculated by determining the ratio between the two expression values, obtained from the same patient in different time-points or time intervals.
[0156] This period of time, also referred to as "time interval", or the difference between time points (wherein each time point is the time when a specific sample was collected) may be any period deemed appropriate by medical staff and modified as needed according to the specific requirements of the patient and the clinical state he or she may be in. For example, this interval may be at least one day, at least three days, at least three days, at least one week, at least two weeks, at least three weeks, at least one month, at least two months, at least three months, at least four months, at least five months, at least one year, or even more.
[0157] More specifically, one sample should be obtained prior to treatment with the specific medicament. Prior as used herein is meant the first time point is at any time before initiation of treatment, ideally several minutes before initiation of treatment. However, it should be noted that any time point before initiation of the treatment, including hours, days, weeks, months or years, may be useful for this method and is therefore encompassed by the invention. The second time point is collected from the same patient after hours, days, weeks, months or even years after initiation of treatment. More specifically, at least 3 hours, at least 4 hours, at least 6 hours, at least 10 hours, at least 12 hours, at least 24 hours, at least 1 day, at least 2 days, at least 3 days, at least 4 days, at least 5 days, at least 6 days, at least 7 days, at least 8 days, at least 9 days, at least 10 days, at least 11 days, at least 12 days, at least 13 days, at least 14 days, at least 15 days, at least 16 days, at least 17 days, at least 18 days, at least 19 days, at least 20 days, at least 21 days, at least 22 days, at least 23 days, at least 24 days, at least 25 days, at least 26 days, at least 27 days, at least 28 days, at least 29 days, at least 30 days, at least 31 days, at least 32 days, at least 33 days, at least 40 days, at least 50 days, at least 60 days, at least 70 days, at least 78 days, at least 80, at least 90 days, at least 100 days, at least 110, at least 120 days, at least 130 days, at least 140 days or at least 150 days after initiation of treatment.
[0158] In some embodiments, the second time point is obtained between 1 hour to 24 month after initiation of the treatment. In some other embodiments, the second time point is between 1 hour to 6 hours after initiation of the treatment. In yet some other embodiments, the second time point is between 1 month to 3 month after initiation of the treatment.
[0159] In practice, for assessing response to a specific treatment, at least two test samples (before and after treatment) must be collected from the treated patient, and preferably more. The expression level of the genes is then determined using the method of the invention, applied for each sample. As detailed above, the expression value is obtained from the experimental expression level. The rate of change of each biomarker expression, namely at least one of the genes indicated by the invention, is then calculated and determined by dividing the two expression values obtained from the same patient in different time-points or time intervals one by the other.
[0160] It should be appreciated that in some embodiments, the term "before treatment" may also encompass samples that are obtained from a treated subject, between two treatments. More specifically, in cases wherein the interval between treatments is once a day, a week, a month, a year or every several days, months or years, "before treatment" may be obtained right before the next treatment. The second "after treatment" sample may be taken after several hours or days of the treatment as indicated above.
[0161] It should be noted that it is possible to divide the prior-treatment expression value by the after treatment expression value and vise versa. For the sake of clarity, as used herein, the rate of change is referred as the ratio obtained when dividing the expression value obtained at the later time point of the time interval by the expression value obtained at the earlier time point (for example before initiation of treatment).
[0162] For example, this interval may be at least one day, at least three days, at least three days, at least one week, at least two weeks, at least three weeks, at least one month, at least two months, at least three months, at least four months, at least five months, at least one year, or even more. Permeably the second point is obtained at the earlier time point that can provide valuable information regarding assessing response of the patient to interferon treatment or to treatment with any other drug, medicament or any other combination of drugs or medicaments.
[0163] The rate of change in the expression value of the different marker genes of the invention may reflect either reduction or elevation of expression. More specifically, "reduction" or "down-regulation" of the marker genes as a result of interferon treatment includes any "decrease", "inhibition", "moderation", "elimination" or "attenuation" in the expression of said genes and relate to the retardation, restraining or reduction of the biomarker genes expression or levels by any one of about 1% to 99.9%, specifically, about 1% to about 5%, about 5% to 10%, about 10% to 15%, about 15% to 20%, about 20% to 25%, about 25% to 30%, about 30% to 35%, about 35% to 40%, about 40% to 45%, about 45% to 50%, about 50% to 55%, about 55% to 60%, about 60% to 65%, about 65% to 70%, about 75% to 80%, about 80% to 85% about 85% to 90%, about 90% to 95%, about 95% to 99%, or about 99% to 99.9%.
[0164] Alternatively, "up-regulation" of any one of the biomarker genes as a result of interferon or any other drug treatment includes any "increase", "elevation", "enhancement" or "elevation" in the expression of said genes and relate to the enhancement and increase of at least one of the biomarker genes expression or levels by any one of about 1% to 99.9%, specifically, about 1% to about 5%, about 5% to 10%, about 10% to 15%, about 15% to 20%, about 20% to 25%, about 25% to 30%, about 30% to 35%, about 35% to 40%, about 40% to 45%, about 45% to 50%, about 50% to 55%, about 55% to 60%, about 60% to 65%, about 65% to 70%, about 75% to 80%, about 80% to 85% about 85% to 90%, about 90% to 95%, about 95% to 99%, or about 99% to 99.9%.
[0165] As appreciated, a predetermined rate of change calculated for a pre-established population as detailed above for example encompasses a range for the rate of change having a low value and a high value, as obtained from a population of individuals including healthy controls, responders and non-responders to said medicament. Thus a subgroup of responsive patients can be obtained from the entire tested population. In this pre-established responsive population, the low value may be characterized by a low response whereas the high value may be associated with a high response as indicated by regular clinical evaluation. Therefore, in addition to assessing responsiveness to treatment, the rate of change may provide insight into the degree of responsiveness. For example, a calculated rate of change that is closer in its value to the low value may be indicative of a low response and thus although the patient is considered responsive, increasing dosing or frequency of administration may be considered. Alternatively, a calculated rate of change that is closer in its value to the high value may be indicative of a high response, even at times leading to remission and thus lowering the administration dosage may be considered.
[0166] For clarity, when referring to a pre-established population associated with responsiveness, or the ability to eradicate pathogens, it is meant that a statistically-meaningful group of patients treated with a specific medicament was analyzed as disclosed herein, and the correlations between the biomarker gene/s expression values (and optionally other patient clinical parameters) and responsiveness to such treatment was calculated. The population may optionally be further divided into sub-populations according to other patient parameters, for example gender and age.
[0167] Another embodiment of the method of the invention defines the step of calculating the value of M1, the optimal threshold required for successful elimination of the pathologic disorder. In such embodiment, this optimal M1 value may be determined using two alternative approaches:
[0168] In one embodiment, determination of the M1 value may be performed by the steps of:
[0169] First (Ia) Providing a K value for the specific disorder. It should be noted that the K value reflects the severity of the disorder. For example, disorders caused by a viral infection, the K value may be the multiplicity rate of such virus, in other words, the pathogen growth rate. Methods for obtaining the multiplicity rate of a virus (the K value of the present application) are described for example in Ruy M. Ribeiro et al., PLOS Pathogens 8 (8):e1002881 (2012); Deborah Cromer et al., Journal of Virology 87: 3376-3381 (2013); Ying Fang et al., J. Virol. Methods. 173(2): 251-258 (2011) and Stiffler J D, et al. PLoS ONE 4(8): e6661. doi:10.1371/journal.pone.0006661 (2009).
[0170] In the next step (Ib) involves calculating the M1 using the formula, wherein M1>1-(1/k), thereby determining the M1 value.
[0171] Another alternative approach for determining the M1 value, may involve the use of standard curve, specifically, predetermined standard curve of a responder population thereby calculating for such curve, the optimal M1 value.
[0172] According to some embodiments, the method of the invention may be specifically practiced using 4 or 5 marker genes. More specifically, in some embodiments, the method of the invention may use the expression value of OAS2, HERC5, UPS18, UBE216 and optionally of ISG15 genes. In some embodiments, the method of the invention may use OAS2, HERC5, UPS18 and UBE216 as markers for calculating M. In yet further embodiments, the method of the invention may use OAS2, HERC5, UPS18, UBE216 and ISG15 genes.
[0173] According to certain embodiments, the method of the invention may be specifically suitable for determining and optimizing a personalized interferon treatment regimen for a subject suffering from a pathologic disorder.
[0174] More specifically, the methods of the invention described herein, relate to interferon treatment, specifically, to optimize interferon treatment regimen to a specific individual, as a personalized medicine approach. As used herein the term "interferon" or "IFN" which is interchangeably used herein, refers to a synthetic, recombinant or purified interferon, and encompasses interferon type I that binds to the cell surface receptor complex IFN-.alpha. receptor (IFNAR) consisting of IFNAR and IFNAR2 chains; interferon type II that binds to the IFNGR receptor; and interferon type III, that binds to a receptor complex consisting of IL10R2 (also called CRF2-4) and IFNLR1 (also called CRF2-12).
[0175] Interferon type I in human includes interferon alpha 1 (GenBank Accession No. NM_024013 and NP_076918; SEQ ID NOs: 47 and 48 respectively), interferon alpha 2 (GenBank Accession No. NM_000605 and NP_000596; SEQ ID NO: 49 and 50, respectively), Interferon alpha-4 (GenBank Accession No. NM_021068 and NP_066546; SEQ ID NO: 51 and 52, respectively), Interferon alpha-5 (GenBank Accession No. NM_002169 and NP_002160; SEQ ID NO: 53 and 54, respectively), Interferon alpha-6 (GenBank Accession No. NM_021002 and NP_066282; SEQ ID NO: 55 and 56, respectively), Interferon alpha-7 (GenBank Accession No. NM_021057 and NP_066401; SEQ ID NO: 57 and 58, respectively), Interferon alpha-8 (GenBank Accession No. NM_002170 and NP_002161; SEQ ID NO: 59 and 60, respectively), Interferon alpha-10 (GenBank Accession No. NM_002171 and NP_002162; SEQ ID NO: 61 and 62, respectively), Interferon alpha-1/13 (GenBank Accession No. NM_006900 and NP_008831; SEQ ID NO: 63 and 64, respectively), Interferon alpha-14 (GenBank Accession No. NM_002172 and NP_002163; SEQ ID NO: 65 and 66, respectively), Interferon alpha-16 (GenBank Accession No. NM_002173 and NP_002164; SEQ ID NO: 67 and 68, respectively), Interferon alpha-17 (GenBank Accession No. NM_021268 and NP_067091; SEQ ID NO: 69 and 70, respectively) and Interferon alpha-21 (GenBank Accession No. NM_002175 and NP_002166; SEQ ID NO: 71 and 72, respectively), Interferon, beta 1 (GenBank Accession No. NM_002176 and NP_002167; SEQ ID NO: 73 and 74, respectively), and Interferon omega-1 (GenBank Accession No. NM_002177 and NP_002168; SEQ ID NOs: 75 and 76 respectively)].
[0176] Interferon type II in humans is Interferon-gamma (GenBank Accession No. NM_000619 and NP_000610; SEQ ID NOs: 77 and 78 respectively).
[0177] As used herein the phrase "interferon treatment" refers to administration of interferon into a subject in need thereof. It should be noted that administration of interferon may comprise a single or multiple dosages, as well as a continuous administration, depending on the pathology to be treated and a clinical assessment of the subject receiving the treatment.
[0178] Various modes of interferon administration are known in the art. These include, but are not limited to, injection (e.g., using a subcutaneous, intramuscular, intravenous, or intradermal injection), intranasal administration and oral administration.
[0179] According to some embodiments of the invention, interferon treatment is provided to the subject in doses matching his weight, at a frequency of once a week, for a period of up to 48 weeks.
[0180] Non-limiting examples of interferon treatment and representative diseases includes the following interferon beta-la (multiple sclerosis), interferon beta-Ib (multiple sclerosis), recombinant IFN-.alpha.2b (various cancers).
[0181] As appreciated in the art, interferon alfa-2a treatment is known as Roferon. Interferon alpha 2b treatment is by Intron A or Reliferon or Uniferon. Interferon beta-1a is sold under the trade names Avonex and Rebif. CinnaGen is a biosimilar compound. Interferon beta-1b is sold under trade names Betaferon, Betaseron, Extavia and ZIFERON.
[0182] Interferon treatment may comprise PEGylated interferon i.e., conjugated to a polyethylene glycol (PEG) polymer. For example, PEGylated interferon alpha 2a is sold under the trade name Pegasys. PEGylated interferon alpha 2a in Egypt is sold under the trade name Reiferon Retard. PEGylated interferon alpha 2b is sold under the trade name PegIntron.
[0183] The interferon treatment can also comprise a combination of interferon and ribavirin. For example, PEGylated interferon alpha 2b plus ribavirin is sold under the trade name Pegetron.
[0184] In yet another specific embodiment, determining the level of expression of at least one of said ISG15, IFIT1-5, OAS1-3L, HERC5, USP18, IFIT2, RSAD2, ISIT1, MX1, IFIT3, IFI44L, OASL, OAS1, OAS2, OAS3, DIX5B, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7 and IFI6 genes, and optionally of STAT1, IFI44, EIF2AK2 and DHX58 genes in a biological sample of the tested subject in order to calculate the specific M value of the individual as described herein above, may be performed by the step of contacting detecting molecules specific for said genes with a biological sample of said subject, or with any nucleic acid or protein product obtained therefrom.
[0185] The term "contacting" means to bring, put, incubate or mix together. As such, a first item is contacted with a second item when the two items are brought or put together, e.g., by touching them to each other or combining them. In the context of the present invention, the term "contacting" includes all measures or steps which allow interaction between the at least one of the detection molecules for the biomarker genes and optionally one suitable control reference gene and the nucleic acid or amino acid molecules of the tested sample. The contacting is performed in a manner so that the at least one of detecting molecule of the genes and at least one suitable control reference gene can interact with or bind to the nucleic acid molecules or alternatively, a protein product of the at least one biomarker gene, in the tested sample. The binding will preferably be non-covalent, reversible binding, e.g., binding via salt bridges, hydrogen bonds, hydrophobic interactions or a combination thereof.
[0186] In certain embodiments, the detection step further involves detecting a signal from the detecting molecules that correlates with the expression level of said genes or any product thereof in the sample from the subject, by a suitable means. According to some embodiments, the signal detected from the sample by any one of the experimental methods detailed herein below reflects the expression level of said genes or product thereof. Such signal-to-expression level data may be calculated and derived from a calibration curve. Thus, in certain embodiments, the method of the invention may optionally further involve the use of a calibration curve created by detecting a signal for each one of increasing pre-determined concentrations of said genes or product. Obtaining such a calibration curve may be indicative to evaluate the range at which the expression levels correlate linearly with the concentrations of said genes or product. It should be noted in this connection that at times when no change in expression level of genes or product is observed, the calibration curve should be evaluated in order to rule out the possibility that the measured expression level is not exhibiting a saturation type curve, namely a range at which increasing concentrations exhibit the same signal.
[0187] It must be appreciated that in certain embodiments such calibration curve as described above may by also part or component in any of the kits provided by the invention herein after.
[0188] In more specific embodiments, the detecting molecules used by the method of the invention for determining the expression level of the marker genes, may be selected from isolated detecting nucleic acid molecules and isolated detecting amino acid molecules.
[0189] According to certain embodiments, the method of the invention may use nucleic acid detecting molecules that may comprise isolated oligonucleotide/s, each oligonucleotide specifically hybridizes to a nucleic acid sequence of said at least one of ISG15, IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7 and IFI6 genes (optionally further of STAT1, IFI44, EIF2AK2 and DHX58 genes) and optionally, to a control reference gene. More specifically, such detecting molecule may be at least one of, a pair of primers, at least one primer and/or nucleotide probe/s or any combination thereof. It should be noted that in some embodiments, each of said oligonucleotides is specifically directed against a specific marker gene or against a specific control gene (e.g., house keeping genes).
[0190] As used herein, "nucleic acid molecules" or "nucleic acid sequence" are interchangeable with the term "polynucleotide(s)" and it generally refers to any polyribonucleotide or poly-deoxyribonucleotide, which may be unmodified RNA or DNA or modified RNA or DNA or any combination thereof. "Nucleic acids" include, without limitation, single- and double-stranded nucleic acids. As used herein, the term "nucleic acid(s)" also includes DNAs or RNAs as described above that contain one or more modified bases. Thus, DNAs or RNAs with backbones modified for stability or for other reasons are "nucleic acids". The term "nucleic acids" as it is used herein embraces such chemically, enzymatically or metabolically modified forms of nucleic acids, as well as the chemical forms of DNA and RNA characteristic of viruses and cells, including for example, simple and complex cells. A "nucleic acid" or "nucleic acid sequence" may also include regions of single- or double-stranded RNA or DNA or any combinations.
[0191] As used herein, the term "oligonucleotide" is defined as a molecule comprised of two or more deoxyribonucleotides and/or ribonucleotides, and preferably more than three. Its exact size will depend upon many factors which in turn, depend upon the ultimate function and use of the oligonucleotide. The oligonucleotides may be from about 3 to about 1,000 nucleotides long. Although oligonucleotides of 5 to 100 nucleotides are useful in the invention, preferred oligonucleotides range from about 5 to about 15 bases in length, from about 5 to about 20 bases in length, from about 5 to about 25 bases in length, from about 5 to about 30 bases in length, from about 5 to about 40 bases in length or from about 5 to about 50 bases in length. More specifically, the detecting oligonucleotides molecule used by the composition of the invention may comprise any one of 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 35, 40, 45, 50 bases in length. It should be further noted that the term "oligonucleotide" refers to a single stranded or double stranded oligomer or polymer of ribonucleic acid (RNA) or deoxyribonucleic acid (DNA) or mimetics thereof. This term includes oligonucleotides composed of naturally-occurring bases, sugars and covalent internucleoside linkages (e.g., backbone) as well as oligonucleotides having non-naturally-occurring portions which function similarly.
[0192] As indicated throughout, in certain embodiments when the detecting molecules used are nucleic acid based molecules, specifically, oligonucleotides. It should be noted that the oligonucleotides used in here specifically hybridize to nucleic acid sequences of the biomarker genes of the invention. Optionally, where also the expression of at least one of the biomarker genes is being examined, the method of the invention may use as detecting molecules oligonucleotides that specifically hybridize to a nucleic acid sequence of said at least one of the genes. As used herein, the term "hybridize" refers to a process where two complementary nucleic acid strands anneal to each other under appropriately stringent conditions. Hybridizations are typically and preferably conducted with probe-length nucleic acid molecules, for example, 5-100 nucleotides in length, 5-50, 5-40, 5-30 or 5-20.
[0193] As used herein "selective or specific hybridization" in the context of this invention refers to a hybridization which occurs between a polynucleotide encompassed by the invention as detecting molecules, and the specific biomarker gene and/or any control reference gene, wherein the hybridization is such that the polynucleotide binds to the gene or any control reference gene preferentially to any other RNA in the tested sample. In a specific embodiment a polynucleotide which "selectively hybridizes" is one which hybridizes with a selectivity of greater than 60 percent, greater than 70 percent, greater than 80 percent, greater than 90 percent and most preferably on 100 percent (i.e. cross hybridization with other RNA species preferably occurs at less than 40 percent, less than 30 percent, less than 20 percent, less than 10 percent). As would be understood to a person skilled in the art, a detecting polynucleotide which "selectively hybridizes" to the biomarker genes or any control reference gene can be designed taking into account the length and composition.
[0194] The measuring of the expression of any one of the biomarker genes and any control reference gene or any combination thereof can be done by using those polynucleotides as detecting molecules, which are specific and/or selective for the biomarker genes of the invention to quantitate the expression of said biomarker genes or any control reference gene. In a specific embodiment of the invention, the polynucleotides which are specific and/or selective for said genes may be probes or a pair of primers.
[0195] It should be further appreciated that the methods, as well as the compositions and kits of the invention may comprise, as an oligonucleotide-based detection molecule, both primers and probes.
[0196] The term, "primer", as used herein refers to an oligonucleotide, whether occurring naturally as in a purified restriction digest, or produced synthetically, which is capable of acting as a point of initiation of synthesis when placed under conditions in which synthesis of a primer extension product, which is complementary to a nucleic acid strand, is induced, i.e., in the presence of nucleotides and an inducing agent such as a DNA polymerase and at a suitable temperature and pH. The primer may be single-stranded or double-stranded and must be sufficiently long to prime the synthesis of the desired extension product in the presence of the inducing agent. The exact length of the primer will depend upon many factors, including temperature, source of primer and the method used. For example, for diagnostic applications, depending on the complexity of the target sequence, the oligonucleotide primer typically contains 10-30 or more nucleotides, although it may contain fewer nucleotides. More specifically, the primer used by the methods, as well as the compositions and kits of the invention may comprise 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29 or 30 nucleotides or more. In certain embodiments, such primers may comprise 30, 40, 50, 60, 70, 80, 90, 100 nucleotides or more. In specific embodiments, the primers used by the method of the invention may have a stem and loop structure. The factors involved in determining the appropriate length of primer are known to one of ordinary skill in the art and information regarding them is readily available.
[0197] As used herein, the term "probe" means oligonucleotides and analogs thereof and refers to a range of chemical species that recognize polynucleotide target sequences through hydrogen bonding interactions with the nucleotide bases of the target sequences. The probe or the target sequences may be single- or double-stranded RNA or single- or double-stranded DNA or a combination of DNA and RNA bases. A probe is at least 5 or preferably, 8 nucleotides in length. A probe may be 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29 and up to 30 nucleotides in length as long as it is less than the full length of the target marker gene. Probes can include oligonucleotides modified so as to have a tag which is detectable by fluorescence, chemiluminescence and the like. The probe can also be modified so as to have both a detectable tag and a quencher molecule, for example TaqMan.RTM. and Molecular Beacon.RTM. probes, that will be described in detail below.
[0198] The oligonucleotides and analogs thereof may be RNA or DNA, or analogs of RNA or DNA, commonly referred to as antisense oligomers or antisense oligonucleotides. Such RNA or DNA analogs comprise, but are not limited to, 2-'0-alkyl sugar modifications, methylphosphonate, phosphorothiate, phosphorodithioate, formacetal, 3-thioformacetal, sulfone, sulfamate, and nitroxide backbone modifications, and analogs, for example, LNA analogs, wherein the base moieties have been modified. In addition, analogs of oligomers may be polymers in which the sugar moiety has been modified or replaced by another suitable moiety, resulting in polymers which include, but are not limited to, morpholino analogs and peptide nucleic acid (PNA) analogs. Probes may also be mixtures of any of the oligonucleotide analog types together or in combination with native DNA or RNA. At the same time, the oligonucleotides and analogs thereof may be used alone or in combination with one or more additional oligonucleotides or analogs thereof.
[0199] Thus, according to one embodiment, such oligonucleotides are any one of a pair of primers or nucleotide probes, and wherein the level of expression of at least one of the biomarker genes is determined using a nucleic acid amplification assay selected from the group consisting of: a Real-Time PCR, micro array, PCR, in situ hybridization and comparative genomic hybridization.
[0200] The term "amplification assay", with respect to nucleic acid sequences, refers to methods that increase the representation of a population of nucleic acid sequences in a sample. Nucleic acid amplification methods, such as PCR, isothermal methods, rolling circle methods, etc., are well known to the skilled artisan. More specifically, as used herein, the term "amplified", when applied to a nucleic acid sequence, refers to a process whereby one or more copies of a particular nucleic acid sequence is generated from a template nucleic acid, preferably by the method of polymerase chain reaction.
[0201] "Polymerase chain reaction" or "PCR" refers to an in vitro method for amplifying a specific nucleic acid template sequence. The PCR reaction involves a repetitive series of temperature cycles and is typically performed in a volume of 50-100 microliter. The reaction mix comprises dNTPs (each of the four deoxynucleotides dATP, dCTP, dGTP, and dTTP), primers, buffers, DNA polymerase, and nucleic acid template. The PCR reaction comprises providing a set of polynucleotide primers wherein a first primer contains a sequence complementary to a region in one strand of the nucleic acid template sequence and primes the synthesis of a complementary DNA strand, and a second primer contains a sequence complementary to a region in a second strand of the target nucleic acid sequence and primes the synthesis of a complementary DNA strand, and amplifying the nucleic acid template sequence employing a nucleic acid polymerase as a template-dependent polymerizing agent under conditions which are permissive for PCR cycling steps of (i) annealing of primers required for amplification to a target nucleic acid sequence contained within the template sequence, (ii) extending the primers wherein the nucleic acid polymerase synthesizes a primer extension product. "A set of polynucleotide primers", "a set of PCR primers" or "pair of primers" can comprise two, three, four or more primers.
[0202] Real time nucleic acid amplification and detection methods are efficient for sequence identification and quantification of a target since no pre-hybridization amplification is required. Amplification and hybridization are combined in a single step and can be performed in a fully automated, large-scale, closed-tube format.
[0203] Methods that use hybridization-triggered fluorescent probes for real time PCR are based either on a quench-release fluorescence of a probe digested by DNA Polymerase (e.g., methods using TaqMan.RTM., MGB-TaqMan.RTM.), or on a hybridization-triggered fluorescence of intact probes (e.g., molecular beacons, and linear probes). In general, the probes are designed to hybridize to an internal region of a PCR product during annealing stage (also referred to as amplicon). For those methods utilizing TaqMan.RTM. and MGB-TaqMan.RTM. the 5'-exonuclease activity of the approaching DNA Polymerase cleaves a probe between a fluorophore and a quencher, releasing fluorescence.
[0204] Thus, a "real time PCR" or "RT-PCT" assay provides dynamic fluorescence detection of amplified genes or any control reference gene produced in a PCR amplification reaction. During PCR, the amplified products created using suitable primers hybridize to probe nucleic acids (TaqMan.RTM. probe, for example), which may be labeled according to some embodiments with both a reporter dye and a quencher dye. When these two dyes are in close proximity, i.e. both are present in an intact probe oligonucleotide, the fluorescence of the reporter dye is suppressed. However, a polymerase, such as AmpliTaq Gold.TM., having 5'-3' nuclease activity can be provided in the PCR reaction. This enzyme cleaves the fluorogenic probe if it is bound specifically to the target nucleic acid sequences between the priming sites. The reporter dye and quencher dye are separated upon cleavage, permitting fluorescent detection of the reporter dye. Upon excitation by a laser provided, e.g., by a sequencing apparatus, the fluorescent signal produced by the reporter dye is detected and/or quantified. The increase in fluorescence is a direct consequence of amplification of target nucleic acids during PCR. The method and hybridization assays using self-quenching fluorescence probes with and/or without internal controls for detection of nucleic acid application products are known in the art, for example, U.S. Pat. Nos. 6,258,569; 6,030,787; 5,952,202; 5,876,930; 5,866,336; 5,736,333; 5,723,591; 5,691,146; and 5,538,848.
[0205] More particularly, QRT-PCR or "qPCR" (Quantitative RT-PCR), which is quantitative in nature, can also be performed to provide a quantitative measure of gene expression levels. In QRT-PCR reverse transcription and PCR can be performed in two steps, or reverse transcription combined with PCR can be performed. One of these techniques, for which there are commercially available kits such as TaqMan.RTM. (Perkin Elmer, Foster City, Calif.), is performed with a transcript-specific antisense probe.
[0206] This probe is specific for the PCR product (e.g. a nucleic acid fragment derived from a gene, or in this case, from a pre-miRNA) and is prepared with a quencher and fluorescent reporter probe attached to the 5' end of the oligonucleotide. Different fluorescent markers are attached to different reporters, allowing for measurement of at least two products in one reaction.
[0207] When Taq DNA polymerase is activated, it cleaves off the fluorescent reporters of the probe bound to the template by virtue of its 5-to-3' exonuclease activity. In the absence of the quenchers, the reporters now fluoresce. The color change in the reporters is proportional to the amount of each specific product and is measured by a fluorometer; therefore, the amount of each color is measured and the PCR product is quantified. The PCR reactions can be performed in any solid support, for example, slides, microplates, 96 well plates, 384 well plates and the like so that samples derived from many individuals are processed and measured simultaneously. The TaqMan.RTM. system has the additional advantage of not requiring gel electrophoresis and allows for quantification when used with a standard curve.
[0208] A second technique useful for detecting PCR products quantitatively without is to use an intercalating dye such as the commercially available QuantiTect SYBR Green PCR (Qiagen, Valencia Calif.).
[0209] RT-PCR is performed using SYBR green as a fluorescent label which is incorporated into the PCR product during the PCR stage and produces fluorescence proportional to the amount of PCR product.
[0210] Both TaqMan.RTM. and QuantiTect SYBR systems can be used subsequent to reverse transcription of RNA. Reverse transcription can either be performed in the same reaction mixture as the PCR step (one-step protocol) or reverse transcription can be performed first prior to amplification utilizing PCR (two-step protocol).
[0211] Additionally, other known systems to quantitatively measure mRNA expression products include Molecular Beacons.RTM. which uses a probe having a fluorescent molecule and a quencher molecule, the probe capable of forming a hairpin structure such that when in the hairpin form, the fluorescence molecule is quenched, and when hybridized, the fluorescence increases giving a quantitative measurement of gene expression.
[0212] According to this embodiment, the detecting molecule may be in the form of probe corresponding and thereby hybridizing to any region or part of the biomarker genes or any control reference gene. More particularly, it is important to choose regions which will permit hybridization to the target nucleic acids. Factors such as the Tm of the oligonucleotide, the percent GC content, the degree of secondary structure and the length of nucleic acid are important factors.
[0213] It should be further noted that a standard Northern blot assay can also be used to ascertain an RNA transcript size and the relative amounts of the biomarker genes or any control gene product, in accordance with conventional Northern hybridization techniques known to those persons of ordinary skill in the art.
[0214] The invention further contemplates the use of amino acid based molecules such as proteins or polypeptides as detecting molecules disclosed herein and would be known by a person skilled in the art to measure the protein products of the marker genes of the invention. Techniques known to persons skilled in the art (for example, techniques such as Western Blotting, Immunoprecipitation, ELISAs, protein microarray analysis, Flow cytometry and the like) can then be used to measure the level of protein products corresponding to the biomarker of the invention. As would be understood to a person skilled in the art, the measure of the level of expression of the protein products of the biomarker of the invention requires a protein, which specifically and/or selectively binds to the biomarker genes of the invention.
[0215] As indicated above, the detecting molecules of the invention may be amino acid based molecules that may be referred to as protein/s or polypeptide/s. As used herein, the terms "protein" and "polypeptide" are used interchangeably to refer to a chain of amino acids linked together by peptide bonds. In a specific embodiment, a protein is composed of less than 200, less than 175, less than 150, less than 125, less than 100, less than 50, less than 45, less than 40, less than 35, less than 30, less than 25, less than 20, less than 15, less than 10, or less than 5 amino acids linked together by peptide bonds. In another embodiment, a protein is composed of at least 200, at least 250, at least 300, at least 350, at least 400, at least 450, at least 500 or more amino acids linked together by peptide bonds. It should be noted that peptide bond as described herein is a covalent amid bond formed between two amino acid residues.
[0216] In specific embodiments, the detecting amino acid molecules are isolated antibodies, with specific binding selectively to the proteins encoded by the biomarker genes as detailed above. Using these antibodies, the level of expression of proteins encoded by the genes may be determined using an immunoassay which is selected from the group consisting of FACS, a Western blot, an ELISA, a RIA, a slot blot, a dot blot, immunohistochemical assay and a radio-imaging assay.
[0217] In yet other specific embodiments, the method of the invention may use any sample. In more specific embodiment, such sample may be any one of peripheral blood mononuclear cells and biopsies of organs or tissues.
[0218] It should be noted that any of the detecting molecules used by the methods, compositions and kits of the invention are isolated and purified. Still further, it must be understood that any of the detecting molecules (for example, primers and/or probes) or reagents used by the compositions, kits, arrays and in any step of the methods of the invention are non-naturally occurring products or compounds, As such, none of the detecting molecules of the invention are directed to naturally occurring compounds or products.
[0219] According to certain embodiments, the sample examined by the method of the invention may be any one of peripheral blood mononuclear cells and biopsies of organs or tissues.
[0220] Still further, according to certain embodiments, the method of the invention uses any appropriate biological sample. The term "biological sample" in the present specification and claims is meant to include samples obtained from a mammal subject.
[0221] It should be recognized that in certain embodiments a biological sample may be for example, bone marrow, lymph fluid, blood cells, blood, serum, plasma, urine, sputum, saliva, faeces, semen, spinal fluid or CSF, the external secretions of the skin, respiratory, intestinal, and genitourinary tracts, tears, milk, any human organ or tissue, any sample obtained by lavage, optionally of the breast ducal system, plural effusion, sample of in vitro or ex vivo cell culture and cell culture constituents. More specific embodiments, the sample may be any one of peripheral blood mononuclear cells and biopsies of organs or tissues.
[0222] According to an embodiment of the invention, the sample is a cell sample. More specifically, the cell is a blood cell (e.g., white blood cells, macrophages, B- and T-lymphocytes, monocytes, neutrophiles, eosinophiles, and basophiles) which can be obtained using a syringe needle from a vein of the subject. It should be noted that the cell may be isolated from the subject (e.g., for in vitro detection) or may optionally comprise a cell that has not been physically removed from the subject (e.g., in vivo detection).
[0223] According to a specific embodiment, the sample used by the method of the invention is a sample of peripheral blood mononuclear cells (PBMCs).
[0224] The phrase, "peripheral blood mononuclear cells (PBMCs)" as used herein, refers to a mixture of monocytes and lymphocytes. Several methods for isolating white blood cells are known in the art. For example, PBMCs can be isolated from whole blood samples using density gradient centrifugation procedures. Typically, anticoagulated whole blood is layered over the separating medium. At the end of the centrifugation step, the following layers are visually observed from top to bottom: plasma/platelets, PBMCs, separating medium and erythrocytes/granulocytes. The PBMC layer is then removed and washed to remove contaminants (e.g., red blood cells) prior to determining the expression level of the polynucleotide (s) bio-markers of the invention.
[0225] In yet another embodiment, the sample may be a biopsy of human organs or tissue, specifically, liver biopsy.
[0226] According to some embodiments, the sample may be biopsies of organs or tissues. The biopsies may be obtained by a surgical operation from an organ or tissue of interest, for example liver biopsy, cerebrospinal fluid (CSF), brain biopsy, skin biopsy.
[0227] The term biopsy used herein refers to a medical test commonly performed by a surgeon or an interventional radiologist involving sampling of cells or tissues for examination. It is the medical removal of tissue from a living subject to determine the presence or extent of a disease. The tissue is generally examined under a microscope by a pathologist, and can also be analyzed chemically. When an entire lump or suspicious area is removed, the procedure is called an excisional biopsy. When only a sample of tissue is removed with preservation of the histological architecture of the tissue's cells, the procedure is called an incisional biopsy or core biopsy. When a sample of tissue or fluid is removed with a needle in such a way that cells are removed without preserving the histological architecture of the tissue cells, the procedure is called a needle aspiration biopsy.
[0228] According to some embodiments of the invention, the cell is a liver cell.
[0229] It should be noted that liver cells (hepatic cell) can be obtained by a liver biopsy (e.g., using a surgical tool or a needle). It should be noted that certain embodiments of the invention contemplate the use of different biological samples.
[0230] According to certain embodiments, the method of the invention may be specifically suitable for optimizing personalized treatment regimen for a subject suffering from an immune-related disorder.
[0231] It should be noted that an "Immune-related disorder" is a condition that is associated with the immune system of a subject, either through activation or inhibition of the immune system, or that can be treated, prevented or diagnosed by targeting a certain component of the immune response in a subject, such as the adaptive or innate immune response.
[0232] In more specific embodiments, the immune-related disorder may be any one of an infectious condition, autoimmune disease and a proliferative disorder.
[0233] It should be appreciated that the method of the invention may be applicable for determining the appropriate treatment regimen for a specific individual affected with any disorder, for example, any disorder caused by any pathogenic agent. Pathogenic agents include prokaryotic microorganisms, lower eukaryotic microorganisms, complex eukaryotic organisms, viruses, fungi, prions, parasites, yeasts, toxins and venoms.
[0234] A prokaryotic microorganism includes bacteria such as Gram positive, Gram negative and Gram variable bacteria and intracellular bacteria. Examples of bacteria contemplated herein include the species of the genera Treponema sp., Borrelia sp., Neisseria sp., Legionella sp., Bordetella sp., Escherichia sp., Salmonella sp., Shigella sp., Klebsiella sp., Yersinia sp., Vibrio sp., Hemophilus sp., Rickettsia sp., Chlamydia sp., Mycoplasma sp., Staphylococcus sp., Streptococcus sp., Bacillus sp., Clostridium sp., Corynebacterium sp., Proprionibacterium sp., Mycobacterium sp., Ureaplasma sp. and Listeria sp.
[0235] Particular species include Treponema pallidum, Borrelia burgdorferi, Neisseria gonorrhea, Neisseria meningitidis, Legionella pneumophila, Bordetella pertussis, Escherichia coli, Salmonella typhi, Salmonella typhimurium, Shigella dysenteriae, Klebsiella pneumoniae, Yersinia pestis, Vibrio cholerae, Hemophilus influenzae, Rickettsia rickettsii, Chlamydia trachomatis, Mycoplasma pneumoniae, Staphylococcus aureus, Streptococcus pneumoniae, Streptococcus pyogenes, Bacillus anthracis, Clostridium botulinum, Clostridium tetani, Clostridium perfringens, Corynebacterium diphtheriae, Proprionibacterium acnes, Mycobacterium tuberculosis, Mycobacterium leprae and Listeria monocytogenes.
[0236] A lower eukaryotic organism includes a yeast or fungus such as but not limited to Pneumocystis carinii, Candida albicans, Aspergillus, Histoplasma capsulatum, Blastomyces dermatitidis, Cryptococcus neoformans, Trichophyton and Microsporum.
[0237] A complex eukaryotic organism includes worms, insects, arachnids, nematodes, aemobe, Entamoeba histolytica, Giardia lamblia, Trichomonas vaginalis, Trypanosoma brucei gambiense, Trypanosoma cruzi, Balantidium coli, Toxoplasma gondii, Cryptosporidium or Leishmania.
[0238] The term "fungi" includes for example, fungi that cause diseases such as ringworm, histoplasmosis, blastomycosis, aspergillosis, cryptococcosis, sporotrichosis, coccidioidomycosis, paracoccidio-idoinycosis, and candidiasis.
[0239] The term parasite includes, but not limited to, infections caused by somatic tapeworms, blood flukes, tissue roundworms, ameba, and Plasmodium, Trypanosoma, Leishmania, and Toxoplasma species.
[0240] The term "viruses" is used in its broadest sense to include viruses of the families adenoviruses, papovaviruses, herpesviruses: simplex, varicella-zoster, Epstein-Barr, CMV, pox viruses: smallpox, vaccinia, hepatitis B, rhinoviruses, hepatitis A, poliovirus, rubella virus, hepatitis C, arboviruses, rabies virus, influenza viruses A and B, flaviviruses, measles virus, mumps virus, HIV, HTLV I and II.
[0241] As shown by the following Examples, the method of the invention may be particularly useful for optimizing treatment for HCV infected subjects. Therefore, the method of the invention may be used for optimizing treatment in subjects suffering from viral infections, for example, Hepatitis C virus infection (type 1, 2, 3 or 4), or HCV or influenza infections.
[0242] According to a particular embodiment, the subject is suffering from an infectious condition caused by hepatitis C virus (HCV).
[0243] As used herein the term "HCV" refers to hepatitis C virus having genotype 1 (also known as HCV Type 1), genotype 2 (also known as HCV Type 2), genotype 3 (also known as HCV Type 3), genotype 4 (also known as HCV Type 4), genotype 5 (also known as HCV Type 5) or genotype 6 (also known as HCV Type 6).
[0244] The phrase "HCV infection" encompasses acute (refers to the first 6 months after infection) and chronic (refers to infection with hepatitis C virus which persists more than 6 month) infection with the hepatitis C virus. Thus, according to some embodiments of the invention, the subject is diagnosed with chronic HCV infection.
[0245] According to some embodiments of the invention, the subject is infected with HCV type 1. According to some embodiments of the invention, the subject is infected with HCV type 2, 3 or 4. More specifically, Hepatitis C virus (HCV or sometimes HVC) is a small (55-65 nm in size), enveloped, positive-sense single-stranded RNA virus of the family Flaviviridae and as indicated herein, is the cause of hepatitis C in humans. The hepatitis C virus particle consists of a core of RNA, surrounded by an icosahedral protective shell of protein, and further encased in a lipid (fatty) envelope of cellular origin. The Hepatitis C virus has a positive sense single-stranded RNA genome consisting of a single open reading frame that is 9600 nucleotide bases long.
[0246] Hepatitis C is an infectious disease affecting primarily the liver, is caused by the hepatitis C virus (HCV). The infection is often asymptomatic, but chronic infection can lead to scarring of the liver and ultimately to cirrhosis, which is generally apparent after many years. In some cases, those with cirrhosis will go on to develop liver failure, liver cancer, or life-threatening esophageal and gastric varices. The invention in some embodiments thereof provides methods, kits and compositions for predicting responsiveness of HCV patients to treatment, specifically, interferon.
[0247] Still further, in certain embodiments the method of the invention may be particularly suitable for optimizing treatment regimen for subjects suffering from an infectious condition caused by any one of HCV, dengue virus, influenza, poliovirus, HIV (human immuno deficiency virus), West Nile virus (WNV) infection and Middle East respiratory syndrome coronavirus (MERS-CoV).
[0248] According to some particular embodiments, the subject may be a subject suffering from an infectious condition caused by a CMV (cytomegalovirus). In more specific embodiments, the virus may be Human cytomegalovirus (HCMV). CMV belongs to the Herpesviridae family that may be also referred to herein as herpesviruses. HCMV may be also referred to as Human herpesvirus 5 (HHV-5). HCMV infections are frequently associated with the salivary glands. HCMV infection is typically unnoticed in healthy people, but can be life-threatening for the immunocompromised, such as HIV-infected persons, organ transplant recipients, or new born infants. It should be therefore appreciated that the method if the invention may be applicable for determining treatment regimen also for subjects infected by CMV.
[0249] A subset of immune-mediated diseases is known as autoimmune diseases. As used herein autoimmune diseases arise from an inappropriate immune response of the body against substances and tissues normally present in the body. In other words, the immune system mistakes some part of the body as a pathogen and attacks its own cells. This may be restricted to certain organs (e.g. in autoimmune thyroiditis) or involve a particular tissue in different places (e.g. Goodpasture's disease which may affect the basement membrane in both the lung and the kidney). Autoimmune disease are categorized by Witebsky's postulates (first formulated by Ernst Witebsky and colleagues in 1957) and include (i) direct evidence from transfer of pathogenic antibody or pathogenic T cells, (ii) indirect evidence based on reproduction of the autoimmune disease in experimental animals and (iii) circumstantial evidence from clinical clues. The treatment of autoimmune diseases is typically done by compounds that decrease the immune response.
[0250] Non-limiting examples for autoimmune disorders include Multiple Sclerosis (MS), inflammatory arthritis. rheumatoid arthritis (RA), Eaton-Lambert syndrome, Goodpasture's syndrome, Greave's disease, Guillain-Barr syndrome, autoimmune hemolytic anemia (AIHA), hepatitis, insulin-dependent diabetes mellitus (IDDM) and NIDDM, systemic lupus erythematosus (SLE), myasthenia gravis, plexus disorders e.g. acute brachial neuritis, polyglandular deficiency syndrome, primary biliary cirrhosis, rheumatoid arthritis, scleroderma, thrombocytopenia, thyroiditis e.g. Hashimoto's disease, Sjogren's syndrome, allergic purpura, psoriasis, mixed connective tissue disease, polymyositis, dermatomyositis, vasculitis, polyarteritis nodosa, arthritis, alopecia areata, polymyalgia rheumatica, Wegener's granulomatosis, Reiter's syndrome, Behget's syndrome, ankylosing spondylitis, pemphigus, bullous pemphigoid, dermatitis herpetiformis, inflammatory bowel disease, ulcerative colitis and Crohn's disease and fatty liver disease.
[0251] In yet another embodiment, the subject is suffering from Multiple sclerosis (MS).
[0252] Thus, in more specific embodiment, the method of the invention may be particularly useful for optimizing treatment, specifically, interferon treatment for a subject suffering from an autoimmune disorder, specifically, Multiple sclerosis (MS).
[0253] As used herein the phrase "multiple sclerosis" (abbreviated MS, formerly known as disseminated sclerosis or encephalomyelitis disseminata) is a chronic, inflammatory, demyelinating disease that affects the central nervous system (CNS). Disease onset usually occurs in young adults, is more common in women, and has a prevalence that ranges between 2 and 150 per 100,000 depending on the country or specific population.
[0254] MS is characterized by presence of at least two neurological attacks affecting the central nervous system (CNS) and accompanied by demyelinating lesions on brain magnetic resonance imaging (MRI). MS takes several forms, with new symptoms occurring either in discrete episodes (relapsing forms) or slowly accumulating over time (progressive forms). Most people are first diagnosed with relapsing-remitting MS (RRMS) but develop secondary-progressive MS (SPMS) after a number of years. Between episodes or attacks, symptoms may go away completely, but permanent neurological problems often persist, especially as the disease advances.
[0255] Relapsing-remitting multiple sclerosis (RRMS) occurring in 85 percent of the patients and a progressive multiple sclerosis occurring in 15 percent of the patients.
[0256] According to some embodiments of the invention, the method of the invention may be particularly applicable for subjects diagnosed with RRMS, where early diagnosis of relapse may improve the treatment.
[0257] In certain embodiments, the methods of the invention may be also useful for determining and optimizing treatment regimen for subjects suffering from Rehumatoid arthritis (RA). It should be appreciated that there are different forms of arthritis that may be generally grouped into two main categories, inflammatory arthritis, and degenerative arthritis, each with different causes. Therefore, according to one specific embodiments the method of the invention may be specifically applicable for inflammatory arthritis. It should be noted that inflammatory arthritis is characterized by synovitis, bone erosions, osteopenia, soft-tissue swelling, and uniform joint space narrowing. More specifically, the hallmarks of joint inflammation are synovitis and erosion of bone. The latter will initially appear as a focal discontinuity of the thin, white, subchondral bone plate. Normally, this subchondral bone plate can be seen even in cases of severe osteopenia, whereas its discontinuity indicates erosion. Still further, the method of the invention may be applicable for determining the most effective personally tailored treatment regimen for a subject suffering from a malignant disorder.
[0258] As used herein to describe the present invention, "cancer", "tumor" and "malignancy" all relate equivalently to a hyperplasia of a tissue or organ. If the tissue is a part of the lymphatic or immune systems, malignant cells may include non-solid tumors of circulating cells. Malignancies of other tissues or organs may produce solid tumors. In general, the methods of the present invention may be applicable for non-solid and solid tumors.
[0259] Malignancy, as contemplated in the present invention may be selected from the group consisting of carcinomas, melanomas, lymphomas and sarcomas. Malignancies that may find utility in the present invention can comprise but are not limited to hematological malignancies (including leukemia, lymphoma and myeloproliferative disorders), hypoplastic and aplastic anemia (both virally induced and idiopathic), myelodysplastic syndromes, all types of paraneoplastic syndromes (both immune mediated and idiopathic) and solid tumors (including lung, liver, breast, colon, prostate GI tract, pancreas and Karposi). More particularly, the malignant disorder may be hepaotcellular carcinoma, colon cancer, melanoma, myeloma, acute or chronic leukemia.
[0260] In certain embodiments, the methods of the invention may be also useful for determining and optimizing treatment regimen for subjects suffering from a proliferative disorder, specifically a cancer, even in cases the medicament is used only as an adjuvant treatment for cell therapy. More specifically, the methods and kits of the invention may be used for optimizing interferon treatment regimen in cases that interferon is being used as an adjuvant for cell therapy, for example in melanoma patients.
[0261] It should be noted that in certain embodiments, were the method of the invention uses an induced dynamic approach for determining the M value of the tested individual, an immuno-stimulant suitable for such method may be any one of a synthetic double stranded RNA (poly ICLC), yellow fever (YF) vaccine 17D (YF17D), TLR stimulants such as double-strand RNA or GC.
[0262] In some specific embodiments, Poly ICLC as used herein is an immunostimulant comprising a synthetic complex of carboxymethylcellulose, polyinosinic-polycytidylic acid, and poly-L-lysine double-stranded RNA. Poly ICLC may stimulate the release of cytotoxic cytokines and induce interferon-gamma production.
[0263] A further aspect of the invention relates to a kit for determining and optimizing a personalized treatment regimen for a subject suffering from a pathologic disorder.
[0264] In certain embodiments, such kit may comprise elements required for performing any of the methods described above. More specifically, such kit may comprise:
[0265] (a) detecting molecules specific for determining the level of expression of at least one of ISG15, IFIT1-5, OAS1-3L, HERC5, USP18, IFIT2, RSAD2, ISIT1, MX1, IFIT3, IFI44L, OASL, OAS1, OAS2, OAS3, DIX5B, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7 and IFI6 genes in a biological sample.
[0266] The kit of the invention further comprises (b), means for calculating the M value of a tested subject. As noted above, the M value indicates the ability of said subject to eliminate said disorder.
[0267] The Kit of the invention further comprises (c) means for calculating the value of M1 or a standard M1 value calculated for a responder population. As indicated above, the M1 value indicates the minimal ability, or specifically, the optimal M1 value required for a successful elimination of the disorder.
[0268] Finally, the kit of the invention comprises (d) means for calculating the dose A and number B of administrations of said dose A to obtain an amount C of said medicament required for said subject.
[0269] According to some specific embodiments, means for calculating the value of M comprised within the kit of the invention should enable determination of the M value by any of the different approaches mentioned by the invention. More specifically, the kit of the invention may comprise at least one of:
[0270] (I) means for performing static analysis for measuring the individual's M value, comprising:
[0271] (Ia) detecting molecules specific for determining the level of expression of ISG15, IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7 and IFI6 genes in a biological sample for determining an expression value Ex.sub.samp in said sample;
[0272] (Ib) a standard curve, specifically, a predetermined standard curve of expression values of subjects suffering from the same pathologic disorder. Alternatively, the kit of the invention may comprise predetermined values, specifically, maximal expression value Ex.sub.max and a minimal expression value Ex.sub.min calculated from such standard curve. In yet another embodiment, the kit of the invention may comprise control samples of at least one individual having a Ex.sub.max expression value and at least one individual having an Ex.sub.min expression value; and
[0273] (Ic) a formula for calculating M value, more specifically, such formula is M=[(Ex.sub.samp-Ex.sub.min)/(Ex.sub.max-Ex.sub.min]. It should be further noted that the kit of the invention may further comprise instructions for determining the expression of any one of the marker genes used by the invention. Moreover, the kit of the invention may further comprise instructions for calculating the required values from the standard curve as well as instructions for calculating the M value using the formula provided.
[0274] In yet another alternative or additional embodiment, the kit of the invention may comprise means for performing an induced dynamic analysis (II). It should be noted that such analysis should be performed on healthy individuals. In more specific embodiments, such means comprise:
[0275] (IIa) detecting molecules specific for determining the level of expression of ISG15, IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7 and IFI6 genes in a biological sample for determining an expression value Ex.sub.samp in the tested sample before and after stimulation of the subject (or in case of in vitro stimulation of a sample of said subject) with an immuno-stimulant. The kit of the invention further comprises means for calculating the rate of change RC.sub.samp in the expression value Ex.sub.samp of the sample before and after stimulation;
[0276] (IIb) an immuno-stimulant;
[0277] (IIc) a standard curve, specifically, predetermined standard curve of the rate of change in the expression of at least one of ISG15, IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7 and IFI6 genes (and optionally of STAT1, IFI44, EIF2AK2 and DHX58 genes) in subjects (specifically, healthy subjects) treated with said immuno-stimulant. Alternatively, the kit of the invention may comprise predetermined maximal rate of change value RC.sub.max and a minimal rate of change RC.sub.min value calculated from such standard curve; and
[0278] (IId) the kit further comprises a formula for calculating said M value. Such formula is M=[(RC.sub.samp-RC.sub.min)/(RC.sub.max-RC.sub.min>]. The kit of the invention further comprises instructions for determining the expression of any one of the marker genes used by the invention. Moreover, the kit of the invention may further comprise instructions for calculating the required values from the standard curve as well as instructions for calculating the M value using the formula provided.
[0279] In yet another embodiment, the kit of the invention may comprise means for a dynamic analysis (III) comprising:
[0280] (IIIa) detecting molecules specific for determining the level of expression of ISG15, IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7 and IFI6 genes (and optionally of STAT1, IFI44, EIF2AK2 and DHX58 genes) in a biological sample for determining an expression value Ex.sub.samp in the examined sample before and after treatment of the tested subject with said specific medicament, and for calculating the rate of change RC.sub.samp in the expression value Ex.sub.samp of the tested sample.
[0281] The kit of the invention further comprises (IIb) a standard curve, specifically, predetermined standard curve of the rate of change in the expression of at least one of ISG15, IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7 and IFI6 (and optionally of STAT1, IFI44, EIF2AK2 and DHX58 genes) genes in subjects suffering from the same disorder and treated with the same medicament. Alternatively, the kit of the invention may comprise predetermined maximal rate of change value RC.sub.max and minimal rate of change value RC.sub.min calculated from such standard curve; and
[0282] The kit of the invention further comprises (IIc) a formula for calculating the M value/More specifically, such formula is M=[(RC.sub.samp-RC.sub.min)/(RC.sub.max-RC.sub.min)].
[0283] It should be further noted that the kit of the invention may further comprise instructions for determining the expression of any one of the marker genes used by the invention. Moreover, the kit of the invention may further comprise instructions for calculating the required values from the standard curve as well as instructions for calculating the M value using the formula provided.
[0284] According to certain embodiments, means for calculating the value of M1 comprised within the kit of the invention may comprise:
[0285] (a) a standard curve, specifically, predetermined K value of the specific disorder;
[0286] (b) a formula for calculating said M1 value. More specifically, such formula is M1>1-(1/k).
[0287] In yet another embodiment, the kit of the invention comprises means for calculating the dose A and number B of administrations of said dose A to obtain an amount C of said medicament required for said subject. It should be note that these means include for example, a predetermined dose A1 and predetermined number B1 of administrations of said dose to obtain a predetermined amount C1 of said medicament required for eliminating said disorder in subjects having a value of M that is equal or above said M1 value. These means further comprise the formulas A=A1/(M1/M) and B=B1*(M1/M); that are required for calculating the dose required for the tested subject.
[0288] According to some embodiments, the kit of the invention may be specifically practiced using 4 or 5 marker genes. More specifically, in some embodiments, the kit of the invention may comprise detecting molecule specific for determining the expression value of OAS2, HERC5, UPS18, UBE216 and optionally of ISG15 genes. In some embodiments, the kit of the invention may comprise detecting molecules specific for OAS2, HERC5, UPS18 and UBE216. In yet further embodiments, the kit of the invention may comprise detecting molecules specific for OAS2, HERC5, UPS18, UBE216 and ISG15 genes.
[0289] According to one specific embodiment, the kit of the invention comprises detecting molecules that are isolated oligonucleotides, each oligonucleotide specifically hybridize to a nucleic acid sequence of at least one of genes and optionally, to a control reference gene. More specifically, such detecting molecules may be at least one of pair of primer/s at least one primer, and/or nucleotide probes.
[0290] According to specific embodiments, the kit of the invention may further comprise at least one reagent for conducting a nucleic acid amplification based assay selected from the group consisting of a Real-Time PCR, micro arrays, PCR, in situ Hybridization and Comparative Genomic Hybridization.
[0291] According to some specific embodiments, the kit of the invention may be specifically suitable for determining and optimizing a personalized interferon treatment regimen for a subject suffering from a pathologic disorder.
[0292] In more specific embodiments, the detecting molecules comprised within the kit of the invention are selected from isolated detecting nucleic acid molecules and isolated detecting amino acid molecules.
[0293] In more specific embodiments, such nucleic acid detecting molecule comprises isolated oligonucleotides, each oligonucleotide specifically hybridizes to a nucleic acid sequence of said at least one of ISG15, IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7 and IFI6 genes (and optionally of STAT1, IFI44, EIF2AK2 and DHX58 genes) and optionally, to a control reference gene.
[0294] Still further, such detecting molecule may be at least one of a pair of primers or nucleotide probes.
[0295] In one embodiment, the polynucleotide-based detection molecules of the invention may be in the form of nucleic acid probes which can be spotted onto an array to measure RNA from the sample of a subject to be diagnosed.
[0296] As defined herein, a "nucleic acid array" refers to a plurality of nucleic acids (or "nucleic acid members"), optionally attached to a support where each of the nucleic acid members is attached to a support in a unique pre-selected and defined region. These nucleic acid sequences are used herein as detecting nucleic acid molecules. In one embodiment, the nucleic acid member attached to the surface of the support is DNA. In a preferred embodiment, the nucleic acid member attached to the surface of the support is either cDNA or oligonucleotides. In another embodiment, the nucleic acid member attached to the surface of the support is cDNA synthesized by polymerase chain reaction (PCR). In another embodiment, a "nucleic acid array" refers to a plurality of unique nucleic acid detecting molecules attached to nitrocellulose or other membranes used in Southern and/or Northern blotting techniques. For oligonucleotide-based arrays, the selection of oligonucleotides corresponding to the gene of interest which are useful as probes is well understood in the art.
[0297] As indicated above, assay based on micro array or RT-PCR may involve attaching or spotting of the probes in a solid support. As used herein, the terms "attaching" and "spotting" refer to a process of depositing a nucleic acid onto a substrate to form a nucleic acid array such that the nucleic acid is stably bound to the substrate via covalent bonds, hydrogen bonds or ionic interactions.
[0298] As used herein, "stably associated" or "stably bound" refers to a nucleic acid that is stably bound to a solid substrate to form an array via covalent bonds, hydrogen bonds or ionic interactions such that the nucleic acid retains its unique pre-selected position relative to all other nucleic acids that are stably associated with an array, or to all other pre-selected regions on the solid substrate under conditions in which an array is typically analyzed (i.e., during one or more steps of hybridization, washes, and/or scanning, etc.).
[0299] As used herein, "substrate" or "support" or "solid support", when referring to an array, refers to a material having a rigid or semi-rigid surface. The support may be biological, non-biological, organic, inorganic, or a combination of any of these, existing as particles, strands, precipitates, gels, sheets, tubing, spheres, beads, containers, capillaries, pads, slices, films, plates, slides, chips, etc. Often, the substrate is a silicon or glass surface, (poly)tetrafluoroethylene, (poly) vinylidendifmoride, polystyrene, polycarbonate, a charged membrane, such as nylon or nitrocellulose, or combinations thereof. Preferably, at least one surface of the substrate will be substantially flat. The support may optionally contain reactive groups, including, but not limited to, carboxyl, amino, hydroxyl, thiol, and the like. In one embodiment, the support may be optically transparent. As noted above, the solid support may include polymers, such as polystyrene, agarose, sepharose, cellulose, glass, glass beads and magnetizable particles of cellulose or other polymers. The solid-support can be in the form of large or small beads, chips or particles, tubes, plates, or other forms.
[0300] The method of the invention may be used for personalized medicine, namely adjusting and customizing healthcare with decisions and practices being suitable to the individual patient by use of genetic information and any additional information collected at different stages of the disease.
[0301] According to specific embodiments, the biological sample may be a blood sample. Specifically, the biological sample is a sample of peripheral blood mononuclear cells (PBMCs). The kit of the invention may therefore optionally comprise suitable mans for obtaining said sample. More specifically, for using the kit of the invention, one must first obtain samples from the tested subjects. To do so, means for obtaining such samples may be required. Such means for obtaining a sample from the mammalian subject can be by any means for obtaining a sample from the subject known in the art. Examples for obtaining e.g. blood or bone marrow samples are known in the art and could be any kind of finger or skin prick or lancet based device, which basically pierces the skin and results in a drop of blood being released from the skin. In addition, aspirating or biopsy needles may be also used for obtaining spleen lymph nodes tissue samples. Samples may of course be taken from any other living tissue, or body secretions comprising viable cells, such as biopsies, saliva or even urine.
[0302] The inventors consider the kit of the invention in compartmental form. It should be therefore noted that the detecting molecules used for detecting the expression levels of the biomarker genes may be provided in a kit attached to an array. As defined herein, a "detecting molecule array" refers to a plurality of detection molecules that may be nucleic acids based or protein based detecting molecules (specifically, probes, primers and antibodies), optionally attached to a support where each of the detecting molecules is attached to a support in a unique pre-selected and defined region.
[0303] For example, an array may contain different detecting molecules, such as specific antibodies or primers. As indicated herein before, in case a combined detection of the biomarker genes expression level, the different detecting molecules for each target may be spatially arranged in a predetermined and separated location in an array. For example, an array may be a plurality of vessels (test tubes), plates, micro-wells in a micro-plate, each containing different detecting molecules, specifically, probes, primers and antibodies, against polypeptides encoded by the marker genes used by the invention. An array may also be any solid support holding in distinct regions (dots, lines, columns) different and known, predetermined detecting molecules.
[0304] As used herein, "solid support" is defined as any surface to which molecules may be attached through either covalent or non-covalent bonds. Thus, useful solid supports include solid and semi-solid matrixes, such as aero gels and hydro gels, resins, beads, biochips (including thin film coated biochips), micro fluidic chip, a silicon chip, multi-well plates (also referred to as microtiter plates or microplates), membranes, filters, conducting and no conducting metals, glass (including microscope slides) and magnetic supports. More specific examples of useful solid supports include silica gels, polymeric membranes, particles, derivative plastic films, glass beads, cotton, plastic beads, alumina gels, polysaccharides such as Sepharose, nylon, latex bead, magnetic bead, paramagnetic bead, super paramagnetic bead, starch and the like. This also includes, but is not limited to, microsphere particles such as Lumavidin.TM.. Or LS-beads, magnetic beads, charged paper, Langmuir-Blodgett films, functionalized glass, germanium, silicon, PTFE, polystyrene, gallium arsenide, gold, and silver. Any other material known in the art that is capable of having functional groups such as amino, carboxyl, thiol or hydroxyl incorporated on its surface, is also contemplated. This includes surfaces with any topology, including, but not limited to, spherical surfaces and grooved surfaces.
[0305] It should be further appreciated that any of the reagents, substances or ingredients included in any of the methods and kits of the invention may be provided as reagents embedded, linked, connected, attached, placed or fused to any of the solid support materials described above.
[0306] According to other embodiments, the kit of the invention may be suitable for examining samples such as peripheral blood mononuclear cells and biopsies of organs or tissues.
[0307] According to some embodiments, the kit of the invention is specifically suitable for optimizing a treatment regimen for subjects suffering from an immune-related disorder.
[0308] In more specific embodiments, such immune-related disorder may be any one of an infectious condition, an autoimmune disease, and a proliferative disorder.
[0309] In certain embodiments, the kit of the invention is suitable for optimizing treatment regimen to a subject suffering from an infectious condition caused by any one of HCV, dengue virus, influenza, poliovirus, HIV (human immune-deficiency virus) and West Nile virus (WNV) infection.
[0310] In yet other embodiments, the kit of the invention may be suitable for optimizing treatment regimen for a subject suffering from Multiple sclerosis (MS).
[0311] In yet other embodiments, the kit of the invention may be suitable for optimizing treatment regimen for a subject suffering from Rheumatoid Arthritis (RA).
[0312] In more specific embodiments, were the kit of the invention comprises means for determining the M value using the induced dynamic approach, the kit of the invention may comprises at least one immuno-stimulant that may be any one of a synthetic double stranded RNA (poly ICLC), yellow fever (YF) vaccine 17D (YF17D).
[0313] In yet a further aspect, the invention provides a computer software product for determining and optimizing a personalized treatment regimen for a subject suffering from a pathologic disorder. Such product comprising a computer readable medium in which program instructions are stored, which instructions, when read by a computer, cause the computer to:
[0314] (a) calculate and determine the value of M that indicates the ability of said subject to eliminate said disorder;
[0315] (b) determine the value of M1, that indicates the minimal ability required for eliminating said disorder.
[0316] (c) calculate the dose A and number B of administrations of said dose A to obtain an amount C required for said subject having said M determined/calculated in step (a), from predetermined dose A1 and number B1 of administrations of said dose, using the formula of A=A1/(M1/M) and B=B1*(M1/M).
[0317] Still further, it must be understood that in certain embodiments, the invention further provides a prognostic composition comprising (a) detecting molecules specific for determining the level of expression of ISG15, IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OAS2, OAS3, OASL, HERC5, USP18, RSAD2, MX1, IFI44L, DDX58, UBE1L, UBE2L6, IFI27, IFIH1, TLR7, IRF7 and IFI6 genes (and optionally of STAT1, IFI44, EIF2AK2 and DHX58 genes) and (b) a biological sample. In certain embodiments, the biological sample may be obtained from the subject that is to be prognosed. In some embodiments, the sample may be a control sample, as discussed herein before. In an optional embodiment, the detecting molecules may be attached to a solid support. As such, the composition of the invention may be specifically suitable for performing any of the prognostic methods disclosed by the invention.
[0318] As used herein, "disease", "disorder", "condition" and the like, as they relate to a subject's health, are used interchangeably and have meanings ascribed to each and all of such terms.
[0319] The present invention relates to the treatment of subjects, or patients, in need thereof. By "patient", "individual" or "subject in need" it is meant any organism who may be affected by the above-mentioned conditions, and to whom the treatment and diagnosis methods herein described is desired, including humans. More specifically, the composition of the invention is intended for mammals. By "mammalian subject" is meant any mammal for which the proposed therapy is desired, including human, equine, canine, and feline subjects, most specifically humans.
[0320] It should be noted that specifically in cases of non-human subjects, the method of the invention may be performed using administration via injection, drinking water, feed, spraying, oral gavages and directly into the digestive tract of subjects in need thereof.
[0321] The term "treatment or prevention" refers to the complete range of therapeutically positive effects of administrating to a subject including inhibition, reduction of, alleviation of, and relief from, a condition known to be treated with interferon, for example an immune-related disorder as detailed herein. More specifically, treatment or prevention of relapse or recurrence of the disease includes the prevention or postponement of development of the disease, prevention or postponement of development of symptoms and/or a reduction in the severity of such symptoms that will or are expected to develop. These further include ameliorating existing symptoms, preventing-additional symptoms and ameliorating or preventing the underlying metabolic causes of symptoms. It should be appreciated that the terms "inhibition", "moderation", "reduction" or "attenuation" as referred to herein, relate to the retardation, restraining or reduction of a process by any one of about 1% to 99.9%, specifically, about 1% to about 5%, about 5% to 10%, about 10% to 15%, about 15% to 20%, about 20% to 25%, about 25% to 30%, about 30% to 35%, about 35% to 40%, about 40% to 45%, about 45% to 50%, about 50% to 55%, about 55% to 60%, about 60% to 65%, about 65% to 70%, about 75% to 80%, about 80% to 85% about 85% to 90%, about 90% to 95%, about 95% to 99%, or about 99% to 99.9%.
[0322] With regards to the above, it is to be understood that, where provided, percentage values such as, for example, 10%, 50%, 120%, 500%, etc., are interchangeable with "fold change" values, i.e., 0.1, 0.5, 1.2, 5, etc., respectively.
[0323] All scientific and technical terms used herein have meanings commonly used in the art unless otherwise specified. The definitions provided herein are to facilitate understanding of certain terms used frequently herein and are not meant to limit the scope of the present disclosure.
[0324] As used herein the term "about" refers to 10% The terms "comprises", "comprising", "includes", "including", "having" and their conjugates mean "including but not limited to". The term "consisting essentially of" means that the composition, method or structure may include additional ingredients, steps and/or parts, but only if the additional ingredients, steps and/or parts do not materially alter the basic and novel characteristics of the claimed composition, method or structure.
[0325] The term "about" as used herein indicates values that may deviate up to 1%, more specifically 5%, more specifically 10%, more specifically 15%, and in some cases up to 20% higher or lower than the value referred to, the deviation range including integer values, and, if applicable, non-integer values as well, constituting a continuous range.
[0326] As used herein the term "about" refers to 10%. The terms "comprises", "comprising", "includes", "including", "having" and their conjugates mean "including but not limited to". This term encompasses the terms "consisting of" and "consisting essentially of". The phrase "consisting essentially of" means that the composition or method may include additional ingredients and/or steps, but only if the additional ingredients and/or steps do not materially alter the basic and novel characteristics of the claimed composition or method. Throughout this specification and the Examples and claims which follow, unless the context requires otherwise, the word "comprise", and variations such as "comprises" and "comprising", will be understood to imply the inclusion of a stated integer or step or group of integers or steps but not the exclusion of any other integer or step or group of integers or steps.
[0327] As used herein the term "method" refers to manners, means, techniques and procedures for accomplishing a given task including, but not limited to, those manners, means, techniques and procedures either known to, or readily developed from known manners, means, techniques and procedures by practitioners of the chemical, pharmacological, biological, biochemical and medical arts.
[0328] The term "about" as used herein indicates values that may deviate up to 1 percent, more specifically 5 percent, more specifically 10 percent, more specifically 15 percent, and in some cases up to 20 percent higher or lower than the value referred to, the deviation range including integer values, and, if applicable, non-integer values as well, constituting a continuous range.
[0329] It must be noted that, as used in this specification and the appended claims, the singular forms "a", "an" and "the" include plural referents unless the content clearly dictates otherwise.
EXAMPLES
Experimental Procedures
[0330] The expression levels of the genes of interest were obtained from publicly available data bases [http://www.ncbi.nlm.nih.gov/geo/] using the following Gene Expression Omnibus Accession Nos:
[0331] Gene Expression Omnibus Accession No. GSE30719 (described in Example 2A) describes genetic data from retinal pigment epithelial (RPE) infected with immunopathogenic West Nile virus (WNV). RNA was extracted after 24 hours and analyzed using Affymetrix arrays.
[0332] Gene Expression Omnibus Accession No. GSE18816 (described in Example 2B) describes peripheral-blood leucocytes that were separated from buffy coats of three healthy blood donors and were differentiated for 14 days before use. Differentiated macrophages infected with H1N1 and H5N1 viruses at a multiplicity of infection (MOD of two were analyzed. Total RNA was extracted from cells after 1, 3, and 6h post-infection, and the gene expression profiling was performed using an Affymetrix Human Gene 1.0 ST microarray platform.
[0333] Gene Expression Omnibus Accession No. GSE13052 (described in Example 2C) describes studies from whole blood transcriptional profiles of children infected 4 days with dengue virus with different clinical outcomes. The tested subjects included 9 acute dengue shock samples, 9 acute uncomplicated dengue samples, 6 autologous follow up dengue samples and 6 autologous follow up uncomplicated dengue patients. Microarray data was normalized using Gene spring GX7 software, statistical analysis was performed in Multiexperiment viewer software. Pathway analysis was performed using Ingenuity Pathway analysis online software.
[0334] Gene Expression Omnibus Accession No. GSE17183 (described in Example 3A) provides data from liver biopsy from 30 patients before and one week after starting combination therapy with IFN+Rib. Hepatocytes and liver-infiltrating lymphocytes (LILs) were obtained from 12 patients using laser capture micro dissection (LCM).
[0335] Gene Expression Omnibus Accession No. GSE16214 (described in Example 3B) provides data from PBMC samples that were collected from relapsing-remitting MS subjects and CIS subjects. The first time point was chosen for each subject with multiple measurements based on an at least three months of treatment criteria. We thereafter analyzed the data for each treatment category.
[0336] Gene Expression Omnibus Accession No. GSE 5549 (described in Example 3C) provides gene expression microarrays data obtained from embryonic fibroblast cell line was synchronously infected with poliovirus in the absence or presence of interferon-.alpha., or with vacciniavirus, a virus that is not inhibited by interferon. The cells were incubated for 1 h with either poliovirus or vacciniavirus, washed and incubated for another 4 to 16 h. Total RNA from three parallel cell cultures were used for each time point and compared with mock infected cells.
[0337] Gene Expression Omnibus Accession No. Gene Expression Omnibus Accession No. GSE15245 (described in Example 4)
[0338] Gene Expression Omnibus Accession No. GSE 37107 and GEO 42296, disclosed gene profiling of RA patients treated with RTX or infliximab, respectively (described in Example 5).
[0339] Gene Expression Omnibus Accession No. GSE18464 provides gene expression data of CD 14+ monocytes isolated from 55 subjects, 22 with HIV HVL, 22 with HIV LVL and 11 HIV seronegative controls (described in Example 6).
[0340] Gene Expression Omnibus Accession No. GSE27248 provides gene expression of Ferrets (3 ferrets in each group) immunized with different adjuvant human seasonal vaccines of CFA plus vaccine, CpG plus vaccine, pegylated IFN-alpha plus vaccine and vaccine alone (PBS plus vaccine) (described in Example 7).
[0341] Gene Expression Omnibus Accession No. GSE31518, GSE31471 and GSE31472 (described in Example 10) provide gene expression data obtained at 2, 4, 6, 8 and 10 hours post infection of three different host cell lines (A549, MDCK and CEF) with three different Influenza A virus strains, pH1N1 (A/Singapore/478/2009), H9N2 and H5N2.
[0342] Gene Expression Omnibus Accession No. GSE52428 (described in Example 10) provide gene expression data obtained from microarrays assay of peripheral blood at baseline and every 8 hours for 7 days following intranasal influenza A H1N1 or H3N2 inoculation in healthy volunteers.
[0343] Gene Expression Omnibus Accession No. GSE838 (described in Example 8A) provides gene expression data in peripheral blood leukocytes (PBL) from normal individuals sampled multiple times over periods ranging from several weeks up to 6 months.
[0344] Gene Expression Omnibus Accession No. GSE3649 (described in Example 8B) provides data of variation in gene expression patterns in the blood of healthy individuals, by using cDNA microarrays.
[0345] Gene Expression Omnibus Accession No. GSE32862 (described in Example 9) provides data from synthetic double stranded RNA that induces innate immunity similar to a live viral vaccine in humans.
[0346] The innate immune response in humans to synthetic double stranded RNA (poly ICLC), a ligand for TLR3 and MDA-5 cytosolic RNA helicase was studied. Transcriptional analysis of blood samples from eight volunteers, after subcutaneous administration of poly ICLC were obtained and analyzed.
[0347] Gene Expression Omnibus Accession No. GSE13699 (described in Example 9) provides data of the immune response to the yellow fever vaccine 17D.
[0348] The data was downloaded from the each one of these selected Gene Expression Omnibus Accession and was analyzed using custom programs written in MATLAB.
[0349] Specifically, after verifying normalization of data (such as RMA quantile on Affymetrix arrays) and averaging multiple probes per gene, MATLAB mattest is carried out with permutations to calculate pvals. In brief, mattest perform two-sample t-test to evaluate differential expression of genes from two experimental conditions or phenotypes.
Example 1
[0350] A Mathematical Model for Determining a Treatment Regimen
[0351] The model developed in here is based on a biological situation in which an animal cell is being infected with a virus having a multiplicity rate K. The cell may be subjected to additional viruses penetrating the cytoplasm by a rate P. The viral infection is reduced or terminated at a rate of M (A person's M is considered to be from 0 to 1. For example, a value of M=0.5 means half of existing viruses will be destroyed).
[0352] The viral infection may be terminated by the immune system of a subject that upon infection is induced and thus capable of destroying the virus by itself with no external therapy. Alternately, the viral infection may be terminated by injection of an appropriate treatment for example with IFN that leads to distraction of the virus. After the IFN effect is diminished, infection may occur again.
[0353] The following set of equations was designed generally to describe the above situation:
[0354] The period at which the cell is being infected with a virus starts at time X(N-1) and ends at time X(N), the virus load at both time points should be therefore described as follows:
X(N).dbd.X(N-1)*K+P*X(N-1);
[0355] The period at which the virus is destroyed either by the immune system, by administration of treatment for example interferon or combination of both starts at time (N) and ends at time (N+1). The virus load (X) at the start point X(N) and the end time point X(N+1) is described as follows:
X(N+1)=X(N)-M*X(N);
[0356] As shown above, the virus load at a certain time point depends on the ability of said individual to eliminate and reduce said virus, as reflected by the M value. The following equations were used in MATLAB for simulation purposes.
[0357] FIG. 1 [based on schematics from Sadler A J. et al. Nature Reviews Immunology 8:559-568 (July 2008)], shows a schematic representation of such a model at a cellular level and emphasizes the ongoing balance between invading virus that is multiplying in the cytoplasm at a rate K and the effect of defending genes that are participating and assisting to diminish the virus. As detailed above, these genes are regulated either by the immune response, external treatment or combination of the two.
[0358] FIG. 2 shows simulations of the above model equations by assuming the following: P=0 (namely, no additional virus from neighboring cells is penetrating to the cell) and K=2 (namely, the virus population doubles itself. The value of M was varied throughout the simulation from 0 to 1. The results in FIG. 2 show the virus amount (virus load) as a function of the rate of M during time. As can be seen at lower M values of 0.04 to 0.48, the virus is capable of multiplying and hence the disease is progressing, as the immune system is not succeeding in elimination thereof. The effect of M is observed as follows: upon increasing the value of M for example from 0.04 to 0.36, the amount of virus is reduced, indicating that that the viral load is reduced to some extend as a function of M. Increasing M to a value higher than 0.6 shows that the viral load is reduced, namely the disease is eliminated. This may indicate that the immune system is succeeding in stopping the virus.
[0359] The simulation shown in FIG. 2, shows that a person having at least an M value of 0.6 will reach this limit using 12 shots (3 month), or he can reach this limit with 4 shots if he has M greater than 0.8. On the x axis the peaks represent IFN cycles assuming they are given once a week.
[0360] The model clearly shows that X(0) the initial load, has no impact on response rate as setting the initial load of the virus to different values does not change the curves, namely there is a dependency only on the relations between M and K, specifically, the ability of the individual and the virulence of the pathogen.
[0361] FIG. 3 shown similar simulations, however, in this simulation the virus is simulated to multiply faster and k is set to 3. The results show that the response curves for the different M's in this new situation are different (FIG. 3). For example, the virus amount is reduced at a large M value of about 0.68. In other words, for eliminating more virulent viruses, a larger M is required.
[0362] The assumption for P=0 is based on virological consideration for example once the virus penetrates the cell penetrations of other viruses from the exterior are blocked.
[0363] These two simulation results can be interpreted as follows:
[0364] First--assuming that an individual is infected with a virus, initially the immune system responds to this infection in an attempt to destroy the virus. The virus is multiplied by a rate K and the immune system of the tested subject may be viewed as a component having the rate M. The "decision" or determination if a person will be able to fight the viral infection on its own depends on the interplay between M and K as shown in FIGS. 2 and 3. The simulation using different M values may be viewed as a heterogeneous population having varying immune system capabilities.
[0365] Second--assuming that an individual is infected with a virus and is being treated with an anti viral treatment directed to distraction of the virus. The virus is multiplied by a rate K and the antiviral treatment may be viewed as a component having the rate M. The interplay between M and K as shown in FIGS. 2 and 3 may be used for determining if a person will cure from the viral infection using the treatment. The simulation with different M values may be viewed as different treatments regimens (dosing and timing), or a heterogeneous population having varying response to treatment (possibly because of immune response) or combinations of the two. In such a situation, the time unit (X-axis) may be considered as the number of treatment administrations (for example injections) to be used.
[0366] The clinical implications of this model suggest that if an intrinsic M of an individual can be measured, it may be possible to precisely predict if an individual will be able to use its own immune system to fight, eradicate and/or eliminate a viral infection (assuming K is known). This model may be further used to predict if an individual is a responder or non responder to treatment with a specific therapeutic agent, for example, IFN treatment. This can be used to predict the outcome of the response, namely what will be the viral amount (viral load) at the end of the treatment and as such, may help in determining the required dosage regimen accordingly. Moreover, this model may be used for calculating and determining the rate of the elevation in the M value required for eliminating of a specific virus having a specific K rate in a specific viral load. Such elevation of M may be achieved by designing an appropriate treatment regimen.
[0367] For example, a person that is characterized by having a M value of 0.6 is being administered with IFN in order to treat a viral infection of a virus that has a K value of 2 (namely, doubles itself every week), will need 12 injections to reach one tenth of his initial load. The results may also serve for prediction of the interval time between treatments, namely, if the time between the injections is shortened from a week to 3 days, an individual may reach the targeted virus load in half the time.
[0368] As known in the art, occasionally infectious disease/s are also treated with a new generation of compounds known as Protease Inhibitors (PI), or with a combination of interferon with any "new generation" drug. Thus, the model incorporates a further possibility that after a subject is being infected with a virus, interferon is administered together with a Protease Inhibitor.
[0369] In such a case the virus is in fact affected by interferon and the protease inhibitors, reflected by M1. Accordingly, the equation
X(N+1)=X(N)-M*X(N);
is now represented as:
X(N+1)=X(N)-M*X(N)-M1*X(N);
[0370] FIG. 4 shows a simulation of a situation with K of 2, P of 0 and M is 0.44. In such case, the tuning should be M1 greater than 0.3 to shift an original non-responder person having a basic rate of M=0.44 to a responder. In fact, having a value of M1 of 0.14 seems to be sufficient to be considered as a responder.
[0371] Turning to FIG. 2, a person with M=0.44 being infected with a virus of k=2 and assume D p=0, seems not to recover from the infection. However, the use of the correct M1 value that may be achieved in this case using combined treatment with Protease Inhibitors might help as seen in FIG. 4.
[0372] M may be considered as a result of the amount of IFN genes that are released upon infection and/or upon treatment (such as for example ISG15, HERC5, USP18, OAS2, OAS 3, OASL, IFI44L). Thus, in case treatment includes PI, the M1 value is added to the value of M that may be now considered as a total amount required for eliminating existing viruses. Similarly, the methods of the invention provide any specific regimen of treatment, being personally adapted to achieve the required M1 value in a person having a particular M value that is required to achieve the desired result of virus elimination, or elimination of any of the disease symptoms.
[0373] The following examples provide results obtained from statistical analysis of data base information and are aimed at showing the importance of combining genetic data from different groups of individuals, and uses thereof in determining a suitable Taylor-made treatment for each individual. Specifically, the inventor has used data from healthy individuals, healthy individuals after stimulation of the immune system, individuals infected with different types of viruses (having different lethality potential) and patients treated with appropriate treatments. The complete data presented in the Examples below provide a comprehensive analysis and a unique understanding on a representative arsenal of genes and the degree to which each gene can be up regulated.
[0374] Moreover, the data presented herein clearly indicate that the value of M is an individual value for each subject and determination thereof (prior to the occurrence of any pathologic disorder), may reflect the ability of a given subject to overcome pathologic disorders, specifically, viral infections. Furthermore, predetermination of such characterizing M value for each individual enables determination, optimization and fine tuning of treatment regimen specifically suitable and effective for such individual.
Example 2
[0375] Gene Profiling after Viral Infections
[0376] The purpose of this example was to find a minimal representative set of genes that are regulated after infection and that measuring their expression will enable determination of the capability of an individual to overcome a viral infection. More specifically, a set of genes enabling the determination of the specific M value of a given individual.
Example 2A
[0377] Genes Expression in West Nile Virus (WNV) Infected RPE
[0378] FIG. 5 shows a representation of genes, each depicted by a different point, such that each point represents the ratio of the specific gene between its expression 24 hours after infection and its base line value. Each point corresponds to an average value of the ratio of the specific gene calculated for all the tested individuals. Each gene (point) is assigned with a value along the X axis that corresponds to the regulation fold (either up regulation or down regulation) and with a value along the Y axis corresponding to the significant of the regulation. Thus, this analysis provides a quantitative indication for the dominating genes that are regulated in infected individuals with respect to a baseline level determined before infection.
[0379] The analysis was obtained by averaging all probes per gene analyzed using volcano analysis on their RMA affymetrix normalized data, at 24h post infection.
[0380] The results indicate that in individuals who were infected with the virus, a high number of genes were up regulated. Specifically, IFIT1-5, OAS1-3L, ISG15, HERC5, USP18 and triggering genes like TLR3, IFIH1, DDX58(RIG-I)
Example 2B
[0381] Gene Expression in Macrophages Infected with H1N1 and H5N1 Viruses
[0382] As appreciated, human disease caused by highly pathogenic avian influenza (HPAI) H5N1 can lead to a rapidly progressive viral pneumonia leading to acute respiratory distress syndrome.
[0383] Table 1 shows a list of genes that were found to be up regulated 6 hours post infection in the more challenging H5N1 compare to H1N1. FIG. 6 is a volcano plot showing the genes that are up regulated six hours post infection with both viruses.
[0384] As shown in Table 1 and FIG. 6, the group of genes including the ISG15, HERC5, USP18, OAS, IFIT and IFI44 show an enhanced up regulation pattern after infection with H5N1.
[0385] As can be seen, the degree of up regulation for each gene is increased after infection with H5N1 compared to H1N1.
[0386] These results suggest that an increased immune response is provided by the host upon infection with H5N1 that is considered more lethal. This enhanced host response may be mediated for example by IFIH1 (MDA5) and DDX58 (RIG-I) gene products.
TABLE-US-00001 TABLE 1 IFN Genes up regulated in the more challenging H5N1 compare to H1N1 at 6 hours post infection. Gene Name Fold Change LAMP3 2.019927 OASL 1.740487 HERC5 1.66951 IFNA13///IFNA1 1.605767 RSAD2 1.257327 ISG15 1.22565 DDX58 1.22122 IFIT1 1.173913 IFIH1 1.141153 IFNA8 1.098233 IFIT2 0.884427 DHX58 0.843863 IFIT3 0.7848 IFI44 0.77259 OAS2 0.745073 USP41///USP18 0.739487 IFIT5 0.722987 MX1 0.65857
Example 2C
[0387] Gene expression in whole blood transcriptional profiles of children infected 4 days with dengue virus Genetic expression from blood obtained from children infected with dengue virus (dengue fever) that developed to dengue shock syndrome (DSS) was compared to well-matched patients with uncomplicated dengue.
[0388] FIG. 7 is a volcano plot showing the group of genes that are regulated for children presenting with dengue shock syndrome (DSS) and well-matched patients with uncomplicated dengue. The plot shows uncomplicated vs. DSS.
[0389] The genes include ISG15, HERC5, UBE2L6, USP18, OAS, IFIT, and IFI44. These results show that this set of genes is regulated to protect the host from the infection. Without being bound by any theory, the inventors suggest that this is mediated by IFIH1 (MDA5), DDX58 (RIG-I) and TLR7.
[0390] Taken together the results shown in all the individuals infected with different viruses having different lethality potential, that a representative set of genes that is being unregulated. The set of genes include for example IFIT2, RSAD2, ISIT1, HERC5, MX1, IFIT3, IFI44L, OASL, OAS1, OAS2, OAS3, DDX58, DHX58, ISG15, USP18, and UBE2L.
[0391] These genes that are involved in innate immunity may be used as an indicator for the ability of a specific individual to eliminate that pathologic disorder.
Example 3
[0392] Gene Profiling after Interferon Treatment
Example 3A
[0393] Gene Analysis in HCV Patients Treated with Interferon
[0394] The differential genetic expression in liver biopsies of responders and non-responders HCV patients after combined therapy is shown in FIG. 8. Specifically, the left hand side shows the ratio of expression level of genes in responders vs. non-responders at day 0 (namely, before treatment), whereas the right hand side shows the results obtained after one week following treatment of IFN and RBV. The results provide a representative set of genes having a low expression value before treatment in patients who are referred to as responders. One week after treatment, these genes were clearly up regulated in the responders group.
[0395] A non-limiting example is the results of ISG15 expression as specifically shown in Table 2. The initial ISG15 expression level in responders is low compared to non-responders. As shown by the Table, a week after treatment, a clear elevation in the expression of said gene appears in the responder group, whereas the non-responders show a clear reduction. Based on these results it may be suggested that the expression level of this gene in responders is close to its saturation level, and therefore interferon cannot induce elevation in the expression of these genes.
[0396] The same results were obtained for HERC5, USP18, OAS, IFIT and IFI44.
TABLE-US-00002 TABLE 2 Expression level of ISG15 in biopsies of ten responders and ten non-responders HCV patients before treatment and after one week of IFN and Rib treatment. non responders responders log2 expression before IFN 9.016884851 7.682601881 log2 change after 1 week of 0.399427605 2.694511557 treatment
[0397] The results obtained here are in accordance with previous results shown in International Patent Application WO10076788 that is a previous application by the inventor, which describes five signature genes that are up regulated before interferon treatment in patients that are considered non-responders to interferon treatment. Thus, based on the expression of the five signature genes before treatment, one can assess the probability to respond to treatment.
[0398] These finding were also repeated in additional data sets. Chen et al which gene expression from tissue taken from HCV patients before treatment in Gene Expression Omnibus Accession No. GSE 11190 provides data on tissue before and 4 hours after IFN injection. The same behavior as explained here was shown in these data sets.
Example 3B
[0399] MS Patients Treated with Interferon
[0400] Analysis of a cohort of 90 patients from PBMC samples of relapsing-remitting MS subjects and Clinically Isolated Demyelinating Syndrome (CIS) subjects.
[0401] Samples were selected at the first time point for each subject with multiple measurements based on an at least three months of treatment criteria Data was obtained Gene Expression Omnibus Accession No. GSE16214.
[0402] Both the volcano (FIG. 9) and the dynamic analysis disclosed in FIGS. 10A and 10B show the same observation of the determination of available dynamic range to predict the outcome of the treatment.
[0403] The volcano plot in FIG. 9 shows the genes are up regulated following treatment with IFN .beta. compared to not-treated patients.
[0404] FIGS. 10A and 10B show the level of genes expression in MS patients before and after three month of treatment, respectively. The expression level of the genes is lower in most patients before treatment. Specifically, about 70 patients show low levels of expression and only about 20 patients show higher levels of expression (FIG. 10A). After treatment, as shown in FIG. 10B, patients who had a low gene expression before treatment (namely patients 1 to 70), show an increased expression after treatment, whereas those patients who had a high gene expression before treatment (namely patients 71 to 90), show a reduced expression after treatment.
[0405] The patients exhibiting an increase in the expression level may be considered as responders whereas the patients showing a decrease in the expression level or lack of elevation, may be considered as non-responders.
Example 3C
[0406] Gene Analysis in Poliovirus Infected Cells and Treated with Interferon
[0407] A human embryonic fibroblast cell line was synchronously infected with poliovirus in the absence or presence of interferon-.alpha., or with vaccinia virus, a virus that is not inhibited by interferon. The samples were washed and incubated for another 4 to 16 h. Total RNA from three parallel cell cultures were used for each time point and compared with mock infected cells.
[0408] Interferon-alpha, at a concentration sufficient to inhibit poliovirus replication, was used to define genes that might be involved in viral defense.
[0409] Analysis of GSE 5549 database shows that the top genes of the Interferon are up regulated when Embryonic cells are injected with IFN (Table 3 shown 16h following injection), and Table 4, shows the elevation of gene expression at 16 hr after IFN was added to culture infected with polio virus (Table 4). Tables 3 and 4 show data from Grinde B, et al. (2007).
TABLE-US-00003 TABLE 3 HE cells, interferon, Name of gene 16 h Interferon. alpha-inducible protein (clone IFI-15K) 5.11 Interferon. alpha-inducible protein (clone IFI-6-16) 3.09 Major histocompatibility complex. class I. C 2.71 Interferon induced transmembrane protein 2 (1-8D) 2.63 H300000271 2.6 Lymphocyte antigen 6 complex. locus E 2.56 HLA class I histocompatibility antigen. A-3 alpha 2.55 chain precursor (MHC class I antigen A*3). [Source: Uniprot/SWISSPROT:Acc:P04439] Signal transducer and activator of transcription 2.483333 1. 91kDa Interferon-induced protein 44-like 2.45 Major histocompatibility complex. class I. B 2.4 Interferon-induced protein with tetratricopeptide 2.29 repeats 3 Tripartite motif-containing 22 2.2 HLA-G histocompatibility antigen. class I. G 2.145 Ubiquitin-conjugating enzyme E2L 6 2.14 HLA class I histocompatibility antigen. alpha 2.136667 chain G precursor (HLA G antigen). [Source: Uniprot/SWISSPROT:Acc:P17693] Major histocompatibility complex. class I. F 2.12 Beta-2-microglobulin 2.08 HLA class I histocompatibility antigen. B-7 alpha 2.05 chain precursor (MHC class I antigen B*7). [Source: Uniprot/SWISSPROT:Acc:P01889] Bone marrow stromal cell antigen 2 2.02 Epithelial stromal interaction 1 (breast) 2.01 2'.5'-oligoadenylate synthetase 1. 40/46kDa 1.98
TABLE-US-00004 TABLE 4 HE cells, interferon + poliovirus, Name of gene 16h Interferon, alpha-inducible protein (clone IFI-15K) 4.91 Interferon, alpha-inducible protein (clone IFI-6-16) 2.89 Major histocompatibility complex. class I. C 2.8 Major histocompatibility complex. class I. C 2.73 Interferon induced transmembrane protein 2 (1-8D) 2.71 Interferon-induced protein 44-like 2.7 Signal transducer and activator of transcription 2.6 1. 91kDa Major histocompatibility complex. class I. C 2.58 Major histocompatibility complex. class I. B 2.47 Signal transducer and activator of transcription 2.44 1. 91kDa Major histocompatibility complex. class I. A 2.43 H300000271 2.38 HLA class I histocompatibility antigen. B-7 alpha 2.37 chain precursor (MHC class I antigen B*7). [Source: Uniprot/SWISSPROT:Acc:P01889] major histocompatibility complex. class I. B 2.35 2'.5'-oligoadenylate synthetase 1. 40/46kDa 2.29 Interferon-induced protein with tetratricopeptide 2.28 repeats 3 Signal transducer and activator of transcription 1. 2.26 91kDa HLA class I histocompatibility antigen. A-3 alpha 2.25 chain precursor (MHC class I antigen A*3). [Source: Uniprot/SWISSPROT:Acc:P04439] HLA-G histocompatibility antigen. class I. G 2.19 Tripartite motif-containing 22 2.18 HLA-G histocompatibility antigen. class I. G 2.16 Beta-2-microglobulin 2.15 Ubiquitin-conjugating enzyme E2L 6 2.15
[0410] The results show that a set of genes can be detected in all the studied cases regardless of the viral origin, namely the set is reproducible and universal. In this connection, the M value, that reflects the ability of the specific individual to eliminate the disease symptoms (viral infection, for example), can be considered as a phenotype.
[0411] Taken together the results shown above suggest that there is a dynamic range for each gene that controls the extent to which a gene can be up regulated and down regulated. This dynamic range of a given gene is required and accordingly the protein encoded by said gene is recruited by the host for eliminating a variety of viral infections. For example, the more pathogenic virus H5N1 compared to H1N1, dengue virus in children and western Nile virus. This dynamic range varies between people as evident from the differences between responders and non responders and from the differences between people that mange to fight viral infection on their own and those who do not. The results presented herein clearly suggested that M as defined in the model above is a phenotypic properties of a specific individual.
Example 4
[0412] Inverse Correlation Between the Expression of the Genes of the Invention and Relapse in MS Patients
[0413] The inventors used gene expression data of Gene Expression Omnibus Accession No. GSE15245, to determine whether the expression of the genes of the invention, namely, USP18, IFI44, MX1, IFI44L, OAS3, HERC5 and RSAD2, can distinguish between MS patients experiencing relapse and patients that respond to interferon treatment and therefore do not experience relapse. As shown in FIG. 11, sum of the expression values of these genes was inversely correlated with relapse rate of fifty MS patients. More specifically, patients 27 to 50 that displayed low initial expression level of the genes of the invention, showed no relapse, whereas patients exhibiting high expression level of the genes of the invention showed enhanced relapse rate.
[0414] The inventors have further analyzed data obtained from GSE5574 that provides gene expression data of MS patients treated with Avonex (0-interferon once a week). The expression of the ISG15, UPS18, UBE2L6 and HERC5 genes of the invention was examined before and during treatment (6 points including 2 reading naive prior to treatment, 2 readings 24 hr following first treatment, 2 readings 6 month following treatment and a week after last IFNB Avonex treatment. The expression level was compared with the following clinical parameters, wherein nonresponsive patients experienced clinical exacerbations including optic neuritis and ataxia requiring steroid treatment, none of the other patients reported any progression of symptoms during the course of the study. The non-responsive patients could not elevate the expression of the genes of the invention (data not shown). Therefore, follow-up of the expression of the signatory genes of the invention during treatment reflects the responsiveness of the patient.
Example 5
[0415] Determination of Treatment Regimen in Rheumatoid Arthritis (RA) Patients
[0416] B cell depletion therapy, for example, by using Rituximab, a chimeric monoclonal antibody against the protein CD20 which is primarily found on the surface of immune system B cells, is efficacious in rheumatoid arthritis (RA) patients that do not respond to tumor necrosis factor (TNF) blocking agents. However, approximately 40% to 50% of rituximab (RTX) treated RA patients display a poor response. The inventors therefore next explored the possibility of using the method of the invention as a tool for determining an appropriate treatment regimen for RA patients. More specifically, the invention provides for any specific individual, a molecular tool to determine whether a RTX treatment is appropriate, or alternatively, treatment with TNF blockers, such as Infliximab (INN; trade name Remicade), that is a chimeric monoclonal antibody specific for tumor necrosis factor alpha (TNF-.alpha.), may be more appropriate. Therefore, the inventors analyzed gene expression data provided by GSE 37107 and GEO 42296 that disclosed gene profiling of RA patients treated with RTX or infliximab, respectively.
[0417] More specifically, Gene Expression Omnibus Accession No. GSE 37107 provides expression profiling data of on whole peripheral blood RNA obtained from 14 RA patients treated with RTX. Expression data of 6 non responders were compared to 8 responders. Responsiveness has been determined 6 months after treatment, using disease activity score (.DELTA.DAS28<1.2) and European League against Rheumatism (EULAR). The samples were obtained and examined prior to treatment.
[0418] Gene Expression Omnibus Accession No. GEO 42296 provides expression profiling data of whole peripheral blood RNA obtained from 29 individuals treated with infliximab and compares the gene expression profiling of 13 non-responders with 6 responders. The samples were obtained and examined prior to treatment.
[0419] The inventors have found that the genes presented in Table 5, are common to both groups and are differentially expressed in the RTX and the infliximab treatment. More specifically, the genes of Table 5, were found to be up-regulated in infliximab responders and down regulated in non-responders. In contrast, the very same genes were found to be down-regulated RTX responders, and up-regulated in RTX non-responders.
[0420] FIG. 12 illustrates the differential expression as calculated from the sum of the common genes, indicating that an individual displaying a high initial expression of these genes will benefit infliximab treatment, whereas an individual displaying a low initial level of expression of these genes will benefit RTX treatment.
TABLE-US-00005 TABLE 5 common signatory genes MXI IFITM3 IFI44L HERC5 IFI44 IFI6 OAS1 OAS3 RSAD2 IFIT1 IFIT3 DDX58
Example 6
[0421] Inverse Correlation Between the Expression of the Signatory Genes of the Invention and Responsiveness to HARRT Treatment of HIV Infected Patient
[0422] To examine whether the signatory genes of the invention may have a predictive value on further viral infections and treatment of patients with other therapeutic agents, The inventors next examined whether the signatory genes of the invention, namely, IFI27, ISG15, IFIH1, IFI44L, OAS2, DDX58, IFIT1 and IFI6, may correlate with responsiveness of HIV infected patients to HAART treatment. More specifically, HIV infected patients that were treated with highly active antiviral therapies (HAART) that is a combination of multiple drugs that act on different viral targets, as reflected by the reduction in virus load.
[0423] The inventors used gene expression data of Gene Expression Omnibus Accession No. GSE18464 that provides gene expression data of high-density cDNA microarrays was performed on CD 14+ monocytes isolated from 55 subjects, 22 with HIV HVL, 22 with HIV LVL and 11 HIV seronegative controls. The examined patients were evaluated for virus load. The categorization of high or low viral load was based on clinical criteria with LVL <10,000 RNA copies/ml and HVL as >10,000 RNA copies/ml. Subjects in the study were males between 30 and 66 years of age and the cohort was comprised of white (62%), black (19%), Hispanic (12%), Asian (4%) and other (3%) individuals. At the time of the study individuals in the LVL group were on highly active antiretroviral therapies (HAART), while subjects with HVL fell into one of three categories: on HAART (15); scheduled treatment interruption (6) or HAART naive (1).
[0424] FIG. 13 presents the correlation between reduced virus load of the HIV patients and the initial expression of the genes of the invention in all 44 examined patients. As shown by the figure, a low expression rate of the genes of the invention is associated with a low virus load that reflects responsiveness to HAART treatment in patients 22-44 (that are the LVL group).
Example 7
[0425] In Vivo Adjuvant Activity in Ferrets Vaccinated Against Influenza Virus
[0426] To examine whether the signatory genes of the invention may be applicable for prediction of responsiveness in other mammals, the gene expression profile of Ferrets vaccinated against influenza virus was next analyzed. The inventors used gene expression data of Gene Expression Omnibus Accession No. GSE27248 that provides gene expression of Ferrets (3 ferrets in each group) immunized with different adjuvant human seasonal vaccines of CFA plus vaccine, CpG plus vaccine, pegylated IFN-alpha plus vaccine and vaccine alone (PBS plus vaccine). The control group comprised 4 ferrets received PBS only. The whole blood was collected for RNA extraction and subsequent gene expression analysis was performed with Affymetrix GeneChip Canine Genome 2.0 Array. The inventors analyzed the expression of the genes of the invention ISG15, HERC5, USP18 and UBE2L6, in all experimental groups. As shown in FIG. 14, a clear correlation of elevated expression of the genes of the invention is exhibited in response to treatment with CpG adjuvant. It should be noted that CpG clearly enhanced activation and antibody production, indicating that dynamic analysis of the expression of the genes of the invention may serve as a tool for evaluating successful treatment.
Example 8
[0427] Genetic Data Obtained from Healthy Populations
[0428] The purpose of these examples was to study variations of gene expression in peripheral blood leukocytes of healthy individuals and thus to obtain an individual specific finger printing.
Example 8A
[0429] PBL Samples of Healthy Individuals
[0430] The data used herein was obtained from peripheral blood leukocytes (PBL) of normal individuals sampled multiple times over periods ranging from several weeks up to 6 months. The genetic data obtained after the first reading for each individual was clustered using k-mean clustering algorithm. FIG. 15 shows the clustering results of the tested individuals, as can be seen, the genes that are clustered within one group include for example ISG15, HERC5, USP18 OAS and IFIT and their triggering elements RIG-I and DDX60. The expression of these genes is correlated and changes together in healthy individuals between high and low levels of expression.
[0431] The close ties between these genes can be better appreciated by looking at Table 5 showing the measured correlation to each of these genes.
TABLE-US-00006 TABLE 6 correlation between ISG15 and all other genes p-value Column # Column ID r correlation Lower CI Upper CI N 3694 FLJ11354 0.930123 5.11E-07 0.79815 0.976917 15 8622 MX1 0.925057 7.95E-07 0.784585 0.9752 15 7251 KPTN 0.92249 9.83E-07 0.777762 0.974328 15 1522 cig5 0.919924 1.21E-06 0.770975 0.973454 15 5764 IFIT4 0.917704 1.43E-06 0.76513 0.972697 15 12247 TREX1 0.903768 3.83E-06 0.72901 0.967918 15 2307 DKFZp434J0310 0.893967 7.02E-06 0.704176 0.964529 15 12478 USP18 0.893168 7.36E-06 0.702173 0.964252 15 8030 LY6E 0.891366 8.17E-06 0.697663 0.963625 15 6185 KIAA0082 0.887221 1.03E-05 0.687348 0.962182 15 5763 IFIT1 0.884509 1.20E-05 0.680644 0.961236 15 924 BST2 0.883635 1.25E-05 0.67849 0.960931 15 12390 UBE2L6 0.882725 1.31E-05 0.67625 0.960612 15 9071 OAS1 0.871427 2.32E-05 0.648773 0.956645 15 7574 LOC51191 0.870744 2.40E-05 0.647129 0.956404 15 9942 PRKR 0.868662 2.65E-05 0.642136 0.955669 15 8572 MTAP44 0.86321 3.40E-05 0.629145 0.95374 15 5758 IFI27 0.860744 3.80E-05 0.623314 0.952865 15 4966 GS3686 0.855596 4.75E-05 0.611221 0.951032 15 5760 IF135 0.853527 5.18E-05 0.606395 0.950294 15 9158 OS4 0.852918 5.32E-05 0.604976 0.950076 15 9131 OR1F1 0.847821 6.55E-05 0.593175 0.948253 15 47 ABCC1 0.828275 0.000137 0.548906 0.941196 15 7728 LOC51667 0.827948 0.000138 0.548179 0.941077 15 10972 SCO2 0.813388 0.000226 0.516209 0.935754 15 2505 DKFZP586A0522 0.805792 0.000287 0.499852 0.932954 15 5654 HSXIAPAF1 0.800053 0.000342 0.487637 0.930829 15 10571 REC8 0.798916 0.000354 0.485231 0.930407 15 4554 G1P3 0.794898 0.000398 0.47677 0.928913 15 3532 FLJ10783 0.785073 0.000526 0.456321 0.92524 15 8997 NRGN 0.782342 0.000567 0.450698 0.924214 15 5961 IRF7 0.781768 0.000576 0.44952 0.923999 15 3907 FLJ20037 0.779139 0.000618 0.444136 0.923009 15 5509 HSPC018 0.767953 0.000827 0.421502 0.918777 15 9558 PIK3R2 0.765603 0.000878 0.416799 0.917883 15 3811 FLJ13102 0.758369 0.00105 0.402444 0.915123 15 4012 FLJ20281 0.755838 0.001116 0.397462 0.914154 15 6423 KIAA0456 0.75358 0.001177 0.393035 0.913288 15 5970 ISG20 0.749655 0.001291 0.385382 0.911779 15
[0432] FIG. 16 shows the same expression but specially demonstrates the expression of IGS15, IFIT1, OAS2 and USP18 in the tested healthy individuals.
[0433] Using the clustering data (FIG. 15) and the specific genes expression graph (FIG. 16), it may be concluded that individuals 2, 7, 8 and 14 who express high levels of the genes at base line namely healthy individuals, will not benefit from IFN treatment if required.
[0434] The other individuals that are characterized by a low expression level would probably respond to IFN and in addition may have a better immune-response.
Example 8B
[0435] A similar approach was used to analyze large cohort of 145 healthy individuals.
[0436] More specifically, large dataset of 145 individuals was used to observe variation in gene expression patterns in blood, by using cDNA microarrays. Again in this group the correlated pattern of the IFN genes reappears as shown by the gene expression correlation in Table 7.
TABLE-US-00007 TABLE 7 correlation of genes with ISG15. p-value Column # Column ID r (correlation) Lower CI Upper CI N 7036 OAS3 0.744385 7.40E-27 0.661661 0.809205 145 4648 IFIT1 0.72467 6.83E-25 0.636894 0.793899 145 8999 SERPING1 0.708683 2.03E-23 0.616943 0.781422 145 4647 IFI6 0.701859 8.06E-23 0.608461 0.776078 145 5868 LY6E 0.694095 3.69E-22 0.598839 0.769986 145 4646 IFI44L 0.693568 4.09E-22 0.598186 0.769572 145 7035 OAS2 0.688079 1.16E-21 0.591402 0.765256 145 2079 CMPK2 0.668012 4.39E-20 0.566711 0.749417 145 6487 MX2 0.640268 4.28E-18 0.532872 0.727364 145 6486 MX1 0.633634 1.20E-17 0.524832 0.722064 145 7202 PARP14 0.625737 3.93E-17 0.515285 0.715741 145 3839 GBP1 0.603309 9.68E-16 0.488322 0.697704 145 9739 STAT1 0.593401 3.68E-15 0.47648 0.689698 145 9204 SLC22A23 0.578805 2.43E-14 0.459111 0.67786 145 4649 IFIT2 0.575336 3.76E-14 0.454997 0.675039 145 7209 PARP9 0.560641 2.25E-13 0.437624 0.663056 145 6230 MLC1 0.536979 3.34E-12 0.409841 0.643649 145 8743 RTP4 0.526616 1.21E-11 0.397258 0.635454 144 3517 FCGR1A 0.520442 1.95E-11 0.390565 0.630006 145 3840 GBP2 0.50984 5.76E-11 0.378266 0.621222 145 10725 UBE2L6 0.503062 1.13E-10 0.370427 0.615592 145 4845 ISG20 0.500953 1.39E-10 0.367991 0.613838 145 4651 IFIT5 0.49784 1.88E-10 0.3644 0.611247 145 10495 TRIM22 0.497346 1.97E-10 0.363831 0.610836 145 4644 IFI35 0.494534 2.58E-10 0.360591 0.608492 145 1645 CCR1 0.493976 2.72E-10 0.359949 0.608028 145 4640 IFI16 0.492824 3.04E-10 0.358622 0.607066 145
[0437] Analysis of the data by clustering is shown in FIG. 17 demonstrating the genes clustering together in one group include for example LY6E, SLC22A23, IFI44L, ISG15, SERPING1, MX2, OAS3, IFIT1 and CMPK2.
[0438] FIG. 18 shows an expression graph showing the expression level of ISG15 and IFIT1, where the individuals were sorted by the ISG15 expression level and not by the individual numbering. It can be suggested that the individuals in the right hand side of the graph (provided in the rectangular) would be non responsive to IFN as the initiation level of the genes is high before any treatment or pathological infection.
Example 9
[0439] Dynamic Analysis of Stimulated Healthy Individuals
[0440] As noted above, changes in the expression levels of genes are observed between cohorts of populations (healthy, infected but not treated yet and treated individuals) and well as between individuals that are responsive or non-responsive. One of the challenges was to quantify the dynamic range of a gene, namely what is the possible expression level a gene can exhibit.
[0441] In fact, it can be suggested that understanding and measuring this dynamic range of a set of known genes may help in determine the capabilities of an individual to use its own immune system in response to infection and/or the predicting if an individual will respond to interferon treatment, namely should interferon be administered to a patients as part of any medical treatment.
[0442] The data provided in Gene Expression Omnibus Accession No. Gse32862 provides information on the innate immune response in humans in response to synthetic double stranded RNA (poly ICLC), a ligand for TLR3 and MDA-5 cytosolic RNA helicase. poly ICLC is an immuno-stimulant and may be considered to have the same effect as interferon.
[0443] The study included transcriptional analysis of blood samples obtained from eight volunteers at different time points, after subcutaneous administration of poly ICLC.
[0444] The data analysis showed a peak in gene expression 24 hr following injection of poly ICLC (FIG. 19). FIG. 20 is a magnified volcano showing ISG15, IFIT1, IFI44, OASL and the triggering of IFIH1 and DDX58.
[0445] Principal component analysis (PCA) was applied using the data for ISG15, IFIT, IFI44, IFIT6 in order to evaluate the importance of the genes in predicting behavior of individuals and is shown in FIG. 21. As can be seen, three groups were obtained, one group including the majority of the individuals and two additional groups each one including one individual. It was suggested that the group with the majority of the individuals corresponds to responders or to individuals that will be able to fight the viral infection on their own. The two individuals were suggested to be non-responders.
[0446] Further, the volcano analysis shows that the up regulation following the poly ICLC mimics exactly the model simulation seen in all previous cases.
[0447] FIG. 22 shows the dynamics of gene expression with time in responders and non-responders.
[0448] As can be seen in FIG. 22, the dynamics of the expression is different in responders and non-responders, specifically, the expression in the non responders (dashed line is high at the beginning (time "0"--healthy) and thus the magnitude of change in the expression is limited and narrow. The expression in the responders, on the other hand, is low at the beginning (time 0) and this enables a large change in their future expression. FIG. 23 shows the same results obtained in longer time intervals with the dashed line corresponding to data obtained from non responders.
[0449] FIG. 24 shows the average increase in each gene expression as measured 24 hours after poly ICLC administration compared to baseline.
[0450] The fold increases are shown in Table 8.
TABLE-US-00008 TABLE 8 fold increase of genes Individual IFIT1 ISG15 IF144L IFIT6 i302 5.23 4.54 4.78 4.68 i304 4.99 4.08 3.68 3.58 i305 4.84 4.55 4.42 4.51 i307 1.99 1.12 2.78 2.35 i308 1.30 1.69 4.41 0.86 i309 4.42 4.47 3.98 3.96 i310 3.49 4.06 3.45 3.53
[0451] Since the observed changes in the expression level of the genes are a result of stimulation of the immune system, the results obtained here may be used to assess the capability of an immune system to react for example to a viral infection. In addition, these results may be also used to obtain information on the magnitude of possible changes in genes expression following interferon administration. Thus, taken together the magnitude of change observed in the expression level of the genes provides a range of the M values in the tested population. Namely, an individual having the highest increase in the expression, for example i302 is characterized with an M value of 1 and is expected to be able to use the immune system and to respond to treatment. On the contrary, an individual having the lowest increase in the expression, for example i308 is characterized with an M value of 0.2 and is expected to fail in inducing the immune system and not to respond to treatment.
[0452] M may be calculated from the area under the curve from baseline to its peak (around 24 hr) and back to baseline should be calculated (FIG. 25). The area of the triangle represents added amount of the fighting gene used to combat the virus in the next cycle (.about.48 hr). Thus the maximal reached pick can represent M=1 and other M's can be derived from the ratio of the triangle area to this maximum triangle area.
[0453] Table 9 shows the change in gene expression obtained after administrating poly ICLC in vivo and in vitro. In vitro samples were obtained from PBMCs isolated from blood of healthy donors via density gradient centrifugation. In vivo samples were obtained from subjects randomized to either 1.6 mg poly ICLC or placebo (sterile saline) in a 2:1 ratio, administered s.c.
[0454] As can be seen, similar patterns are observed both in vivo and in vitro suggesting that in vitro data can be used to calculate M values.
TABLE-US-00009 TABLE 9 changes in gene expression after administrating poly ICLC in vivo and in vitro. logFC logFC Marker in vivo in vitro gene pICLC pIC IFIT3 4.473713385 4.741052 IFIT1 4.384596081 5.420633 IFI44L 4.171223257 4.799476 IFI6 4.016294707 2.691117 HERC5 3.871166452 3.248894 IFIT3 3.75211871 4.750157 ISG15 3.726288606 4.480175 IFIT2 3.702681715 4.218502 RSAD2 3.628919044 4.957172 MX1 3.440496672 2.968754 IFIT3 3.438007383 2.896002 OASL 3.437579624 3.757009 IFITM3 3.301297669 3.898404 EPSTI1 3.288106942 3.001505 IFI44 3.233595065 3.578491 OAS1 3.122394636 3.803166 LAMP3 2.958338552 1.798831 MT2A 2.951819567 1.502288 HES4 2.904583684 2.542752 GBP1 2.885817399 2.094531 IRF7 2.818183268 2.601032 FCGR1B 2.815964332 0.34899 OAS2 2.744736242 2.835379 TNFSF10 2.708541769 3.520021 GBP5 2.707972339 1.841636 MT1A 2.696990876 1.162177 CXCL10 2.673387432 4.427627 OAS1 2.669871256 3.51157 IFI35 2.653402642 2.562161 LAP3 2.647847669 1.792573 SERPING1 2.629612763 2.03007 XAF1 2.627899914 3.065335 IFI27 2.616810145 3.207039 LY6E 2.578162512 2.557162 ZBP1 2.572024498 1.786611 SAMD9L 2.559958622 2.829755 OASL 2.488510388 2.512218
[0455] An exact pattern is shown by Gaucher et al in Gene Expression Omnibus Accession No. GSE13699, who examined the signature of the immune response to the yellow fever (YF) vaccine 17D (YF17D) in a cohort of forty volunteers.
[0456] Table 10 shows the top ranking genes in two tested in vivo vaccination studies in two different locations (Montreal, Canada and Lausanne, Switzerland). Both groups received YF17D (ratio measured 1 week after administration) and the polyI CLC, averaged on all participants.
TABLE-US-00010 TABLE 10 Top ranked genes expressed in poly ICLC administered individuals and in individuals administered with yellow fever (YF) vaccine 17D (YF17D) in two tested groups. Group A Group B polyc (yf Canada) (yf Swiss) ratio 24 hr/ ratio 1 week/ ratio 1 week/ baseline baseline baseline RSAD2 IFI44L IFI44L IFI44L IFIT1 RSAD2 ISG15 ISG15 IFI44 IFI44 RSAD2 ISG15 OA53 HERC5 IFI27 HERC5 IFITM3 OAS3 LY6E IFI6 IFIT3 EPSTI1 IFIT3 EPSTI1 IFI27 LAMP3 HES4 IFITM3 IFI27 HERC5
[0457] Table 10 shows that there is a representative set of genes that is being regulated after administration of immune response stimulants. This suggests that the arsenal of observed genes may be regarded as the genes related to the M phenotype in a person.
[0458] Table 11 shows that among the cohort of individuals tested in Group A, a phenotype can be seen as provided in bold. The individuals that show a marked increase in the genes expression following stimulation of the immune response are suggested to correspond to individuals that will be able to use their immune system or to respond to therapy or both. The individuals that show a low increase in the genes expression are given in plain numbers and correspond to individuals that will not be able to use their own immune system, will not respond to treatment or both.
TABLE-US-00011 TABLE 11 Top ranking genes that are up regulated and extent of regulation. Marker gene diff diff diff diff diff diff diff diff diff diff diff avg IFI44L 1.9 3.25 5.7 3.2 4.1 5.5 3.7 5.2 4.7 0.8 3 3.73 RSAD2 1.9 3.47 5.6 3.4 4.2 4.5 3.9 5.4 3.9 1.2 3.5 3.72 IFI44 2.1 2.79 5.4 3.8 3.6 5.1 3.7 4.8 4 0.7 3 3.55 ISG15 1.8 3.16 4.6 2.5 3.5 4.2 3.3 4.4 3.2 1.3 2.7 3.16 IFI27 5 3.39 2.7 2.6 3.2 4.7 4.5 3.8 3.1 0 2.96 OAS3 1.2 2.22 4.6 2.2 3.2 3.5 2.8 4 2.5 0.4 2.4 2.64 IFIT3 2.3 2.61 3.7 2.6 2.6 2.9 2.8 3.5 2.1 0.8 1.8 2.52 EPSTI1 1.4 2.43 4 2.23 2.9 3.6 2.1 3.6 2.9 0.5 1.5 2.47 HES4 1.7 3.2 3.3 2.02 2.1 3.1 2.9 3.5 1.9 0.5 2.2 2.4 HERC5 0.9 2.03 3.7 1.53 2.7 3.2 2.4 3.4 1.6 1 2.4 2.26
[0459] FIG. 26 is a graph showing simulation of replication vs. immune defense, per different M. As can be seen for the same individual with an M value suitable for K=3, that is calculated as follows M=1-1/3=0.66 being infected by a variety of viruses with varying K (multiplication rate). FIG. 26A shows that at K rate higher than 3, the virus progresses. FIG. 26B shows situation where K smaller than M, attenuation of the virus is achieved.
[0460] FIGS. 27A-B is a graph showing simulation instructing how much PI is needed per each individuals M and virus K. The PI effectively increases the individuals M, FIG. 27A shows an individual with M=0.6, FIG. 27B shows an individual with M=0.8 both are affected by the same range of PI injections. The better M the quicker an individual to become a responder with the same PI. The results shown here suggest that if M is measured (as shown above) and K is known for each type of virus, a simulator may be used to guide any clinical decision on the frequency of treatment (by simulation that changes each point on the graph 2 days or 3 days instead of one week). In addition, such a simulator may also indicate if and how much further combined therapy, for example protease inhibitors (PI) is required.
Example 10
[0461] Correlation Between Virus Load and Induction of the Ubiquitin Genes Expression
[0462] As shown in Example 2B, infection of macrophages with influenza H1N1 and H5N1 strains, led to a significant elevation in the expression of the ubiquitin genes. The virulent strain, H5N1 showed a clear enhanced induction of the expression of said genes. To further establish the hypothesis that the virulence of the pathogen, as determined by measuring the increase in virus load, is correlated with the extent of the induced expression of the ubiquitin genes of the invention, specifically, ISG15, USP18, HERC5 and UBE2L6, the inventors have analyzed data of different host cells infected with three strains of influenza virus. Gene Expression Omnibus Accession No. GSE31518, GSE 31471 and GSE31472 provide gene expression data obtained at 2, 4, 6, 8 and 10 hours post infection of three different host cell lines (A549, MDCK and CEF) with three different Influenza A virus strains, pH1N1 (A/Singapore/478/2009), H9N2 and H5N2. Table 12, presents virus load (as indicated by measuring the vRNA copy number) of the three influenza strains 10 hr post infection of each of the three different host cell lines. FIG. 28 shows sum of the expression of the ubiquitin genes, ISG15, USP18, HERC5, UBE2L6, as measured in A549 cells at 2, 4, 6, 8 and 10 hours post infection with the three different influenza strains. A significant correlation between viral load in the infected A549 and the Ubiquitin genes sum expression is clearly observed.
[0463] The inventors have next examined the feasibility of using data of virus load and gene expression data to evaluate the ability of a specific individual (having a specific M value), to overcome an infection of a specific pathogen having a specific replication rate. Therefore, gene expression data of healthy human volunteers inoculated with intranasal influenza A H1N1 and H3N2 strains was analyzed by the inventors. Gene Expression Omnibus Accession No. GSE52428 provide gene expression data obtained from microarrays assay of peripheral blood at baseline and every 8 hours for 7 days following intranasal influenza A H1N1 or H3N2 inoculation in healthy volunteers.
[0464] FIG. 29 shows the sum of the expression of the 4 ubiquitin genes of the invention in different time points up to 120 hr post infection of H3N2 in all nine individuals (numbered as 1 to 9).
[0465] Simulation based on the model of the invention as described in Example 1 was performed using the data of all 9 infected individuals based on the calculated rate of induced expression of the ubiquitin genes, ISG15. USP18, HERC5 and UBE2L6.
[0466] FIGS. 30 and 31, present the result of such simulation using the data of individual 6. Assuming that individual I is infected with a virus having a replication rate of 1.93 every 6 hrs, as presented in FIG. 30, the sum of the expression of the 4 genes, is maximal between 12 to 18 hrs post infection, and reduced after 36 hrs, as shown in the lower panel of the figure. The rate of expression of these genes is correlated to the replication of the virus, that is maximal between 12 to 18 hrs post infection and is significantly reduced after 36 hrs, indicating that the specific individual may successfully reduce the virus load of a virus having replication rate of 1.93, and therefore overcome the infection. FIG. 31 presents simulation of the same individual infected with a virus having a little higher replication rate, of 1.94. As clearly shown in the figure, the same individual, when confronted with said virus, shows increase in the gene expression of the signatory genes of the invention (bottom panel), however, 24 hrs post infection, the replication rate of the virus increases with no corresponding increase in the expression of the signatory genes, indicating that said individual may experience failure of overcoming an infection of virus having a rate of replication of 1.94 each 6 hrs. It should be appreciated that M as it is measured for the whole range (min to max) or alternatively, as it is made of the segmented parts measured for the individual.
[0467] This example clearly demonstrates the correlation between the expression of the signatory genes of the invention and the ability of the virus to propagate in a specific individual having a specific ability of increasing expression of the signatory genes, or in other words, having a specific M value.
TABLE-US-00012 TABLE 12 Comparison of the M gene vRNA levels at 10 hpi in influenza virus-infected A549, MDCK and CEF vRNA (copy numbers) Virus A549 MDCK CEF H1N1 4.43 .+-. 0.07 5.74 .+-. 0.07 6.91 .+-. 0.05 H9N2 1.71 .+-. 0.04 3.42 .+-. 0.08 7.34 .+-. 0.07 H5N2(F118) 5.71 .+-. 0.04 4.91 .+-. 0.02 7.47 .+-. 0.10 H4N2(F189) 5.70 .+-. 0.08 5.01 .+-. 0.20 7.28 .+-. 0.06 H5N3 6.83 .+-. 0.04 5.93 .+-. 0.03 8.29 .+-. 0.06 pH1N1/276 1.92 .+-. 0.06 2.11 .+-. 0.26 5.10 .+-. 0.52 pH1N1/471 1.06 .+-. 0.09 2.69 .+-. 0.20 5.27 .+-. 0.11 pH1N1/478 1.67 .+-. 0.12 2.53 .+-. 0.20 4.92 .+-. 0.33 pH1N1/527 1.32 .+-. 0.11 2.46 .+-. 0.18 5.70 .+-. 0.21
Example 11
[0468] Prediction of Response to Treatment of IFN-.alpha. in Blood Samples and Liver Tissue of HCV Patients
[0469] To further establish the model of the invention, the inventors next evaluated the ability of calculating the M parameter of an individual (that reflects the ability of a specific individual to overcome a pathologic disorder), infected by HCV in this case, from the measured data of the expression of the signatory genes of the invention, namely, UBE2L6, USP18, HERC5, OAS2 and ISG15, and the reduction in virus load as measured 4 weeks after treatment with Interferon alpha. Therefore, RT-PCR analysis of the genetic profile in Peripheral Blood Mononucleated Cell (PBMC) and liver tissue of HCV patients was performed on samples obtained before initiation of IFN-.alpha. treatment, and one month after.
[0470] The expression levels of the following genes: UBE2L6, USP18, HERC5, OAS2 and ISG1 (using 3 probes) in each patient was measured by RT-PCR and normalized to a control gene GAPDH. In addition, in each one of the eight patients, the virus load was determined before treatment and 4 weeks after treatment with IFN-.alpha. using commercial kits.
[0471] Based on the sum expression of the five genes, an experimental M was calculated as follows:
M=1-[(Ex.sub.samp-Ex.sub.min)/(Ex.sub.max-Ex.sub.min)].
[0472] Wherein Ex.sub.max is a maximal measured sum expression value of the five genes and Ex.sub.min is a minimal measured sum expression value of the five genes within a population and Ex.sub.samp is the measured sum expression value of the five genes for a specific patient within this population, to whom the M is calculated.
[0473] Based on the results of the change in virus load measured before treatment and after 4 weeks of treatment, two populations of HCV patients were defined: responders and non-responders.
[0474] A responder was considered as a patient that the amount of viral load was reduced by more than 100 within 4 weeks, (2 in log 10). A non-responder was considered as a patient that the amount of viral load was reduced by less than 100 within 4 weeks, (2 in log 10).
[0475] Experiments were conducted on different populations of HCV patients from samples obtained from PBMC and from liver tissue samples.
[0476] Experiments on Blood Samples:
[0477] As can be seen in FIG. 32, the patients denoted as p2, p1, p3, p5, p4, and p8 experienced an amount of down regulation of virus load higher than 100 (observed as 2 in log 10 scale) and are thus considered responders to IFN-.alpha. treatment in line with the definition above.
[0478] In contrast, patients denoted as p6 and p7 experienced an amount of down regulation of virus load lower than 100 (observed as 2 in log 10 scale) and are thus considered non-responders to IFN-.alpha. treatment in line with the definition above.
[0479] The results in FIG. 32 demonstrated that the sum of normalized and scaled expression of the five genes UBE2L6, USP18, HERC5, OAS2 and ISG15 was significantly reduced in patients that were considered as responders (p2, p1, p3, p5, p4, and p8) compared to the expression in patients considered as non-responders (p6 and p7).
[0480] As indicated above, the virus load of these 8 HCV patients was recorded before and one month (4 injection cycles) after Pegylated Interferon-alpha Treatment. Gene signature of the UBE2L6, USP18, HERC5 and OAS2 genes expression was derived and scaled (0-1) before treatment. Simulation is carried out for different M values per 4 cycles of treatment.
[0481] Table 13 shows M values and their corresponding virus load decline in each of the 4 weeks of treatment with Peg Interferon as calculated by the model of the invention as described in Example 1.
TABLE-US-00013 TABLE 13 Simulation of calculated M values vs. reduction of virus load after treatment M value treatment_w1 treatment_w2 treatment_w3 treatment_w4 0.87 1 1.538461538 2.366863905 3.641329085 0.88 1 1.666666667 2.777777778 4.62962963 0.89 1 1.818181818 3.305785124 0.89 0.9 1 2 4 8 0.91 1 2.222222222 4.938271605 10.9739369 0.92 1 2.5 6.25 15.625 0.93 1 2.857142857 8.163265306 23.32361516 0.94 1 3.333333333 11.11111111 37.03703704 0.95 1 4 16 64 0.96 1 5 25 125 0.97 1 6.666666667 44.44444444 296.2962963 0.98 1 10 100 1000 0.985 1 13.33333333 177.7777778 2370.37037 0.998 1 100 10000 1000000 0.999 1 200 40000 8000000
[0482] As shown by the table, assuming that the initial measured virus load is 1 (treatment w1), different calculated M values indicated in the table, result in the indicated reduction (folds of reduction) in virus load.
[0483] The virus load and expression values of the UBE2L6, USP18, HERC5 and OAS2 genes obtained for 7 of the HCV patients analyzed above, were now calculated using the simulation values of Table 13, and are presented in Table 14.
[0484] More specifically, Table 14 shows the correlation between the measured fold of virus load reduction after 1 month of interferon treatment (second column from left), and the measured expression of the 4 genes of the invention (the third column from left presents sum of the expression values of all 4 genes, each value scaled between 0 to 1).
TABLE-US-00014 TABLE 14 Calculated M values vs. reduction of measured HCV virus load after IFN treatment fold decline in VL after 1 scaled_expression scaled simulation patient month simulation M scaled_expression (0-1) M (0_1) p2 14535.3 0.9918 0 0 0.950819672 p1 16760.7865 0.9921 0.96137233 0.251731718 0.953161593 p3 51955.95 0.9948 1.032753453 0.270422596 0.974238876 p4 20120.4 0.9928 1.144285694 0.299626892 0.958626073 p8 2061843.2 0.9981 2.057379621 0.538717093 1 p6 3.48570259 0.87 3.525351059 0.923099876 0 p7 52.4136592 0.948 3.81903535 1 0.608899297
[0485] As shown in the table, for patient p1, for example, the measured reduction in virus load was about 16,000 folds, going back to the simulation of reduction in virus load as a parameter of M value, as presented in Table 13, an M value of about 0.992 is correlated with reduction of about 15,625 folds in the measured virus load. This predicted M value (shown in the second column from left), is correlated to a scaled expression value of 0.25. In contrast, patient p6 that showed only 3.48 folds reduction in virus load, was correlated with a scaled expression of 0.9, and a low M value of 0.87 (in the simulation Table 13, reduction of about 3.6 folds is correlated with an M value of 0.87). The right column of Table 14 shows scaled M values, of between 0 to 1, were the lowest M value, 0.87, was considered as 0 (as shown for patient p6), and the higher M value in the simulation, 0.9981, is considered as 1 (as shown for patient p8). When correlating to the scaled expression values (sum of the expression values of the 4 signatory genes), it seems that patients having scaled expression value of below 0.5, efficiently reduce the virus load and are therefore considered as responsive to interferon treatment, whereas patients presenting an initial scaled expression value of above 0.5, and a low M value, show poor response that is reflected in low ability to reduce virus load.
[0486] FIG. 33 clearly shows that the four responders have M values between 0.95 to 1 while their expression value of the signatory genes is below 0.5. The two non responders (p6 and p7 having low M values and a corresponding high levels of initial expression of the signature genes, of above 0.9.
[0487] This example clearly demonstrate the feasibility of using the measured initial expression of the signatory genes of the invention, before starting any treatment, to evaluate the personal M value that distinguish between responders and non-responders and also indicate the extent of predicted responsiveness of a specific individual. The method of the invention thereby provides a powerful tool for personalized medicine.
[0488] Further analysis of the sum of the expression of two genes, HERC5 and UBE2L6 in HCV patients is shown in FIGS. 34A and 34B. In the analysis shown in these figures, patients denoted as p2, p1, p3, p4, p8, p11, p101 and p12 experienced clear reduction of virus load that is more than by 100 (observed as 2 in log 10 scale) and are thus considered responders to IFN-.alpha. treatment in line with the definition above, whereas patients denoted as p6 and p7 experienced reduction of virus load lower than by 100 (observed as 2 in log 10 scale) and are thus considered non-responders to IFN-.alpha. treatment in line with the definition above.
[0489] Patient p12 experienced a reduction in virus load of 2.02 and thus theoretically should be considered as responder. However, in the following analysis, this patient was not categorized to any one of the groups since the value of 2.02 is in the border between responders and non-responders.
[0490] Analysis of the gene as described above is shown in FIG. 34A. As shown in FIG. 34A, a strong correlation was observed between the sum expression of the five genes and the patient's response to IFN treatment. A lower expression value was measured in patients p2, p1, p3, p4, p8, p11, p101 and p12 who were found responsive to IFN treatment. On the other hand, a high expression value was measured in patients p6 and p7 who were found not responsive.
[0491] Based on the experimental data of the expression of the two genes detailed above, the M value was calculated for each one of the patients. FIG. 34B shows the experimental M value calculated for each one of the tested patients presented in FIG. 34A.
[0492] As can be seen, a correlation exists between the M value and the patient's responsiveness to treatment. Patients having a high M value were found to be responsive to IFN treatment, whereas patients having a lower M value were found to be not responsive to treatment.
[0493] The effect of HCV in liver tissue may be considered different than the effect in blood samples. In HCV liver the battle is occurring inside hepatocytes and the inventor assume k=5 based on previous publications [Ruy M. Ribeiro et al., (2012)]. In PBMC the specificity of the cells is not as clear and the inventors assume it's in a close range to the shaded amount transferred to the blood from the source hepatocytes.
[0494] The cutoff value may be calculated by using the following equation:
M.sub.cutoff=I1-/K
[0495] Thus for a virus characterized by a K value of 5, the theoretical M.sub.cutoff value is 0.8. As can be seen in FIG. 34B, the inventor assumes for this example the range of M to be between 7.9 to 8.9 and scaled M accordingly. Once more viral loads data points in time were received the final range for the group was narrower (0.815 to 0.862). It should be noted that there is an inverse correlation between the calculated M value and the sum of the genes expression.
[0496] The inventors have also performed an analysis using the expression of a single gene, HERC5 which is considered as a predictive gene. FIG. 35 shows the patients denoted as p208, p213, p102, p201, p211, p203, p204, p202 and p101 experienced a reduction of virus load by more than 100 (observed as 2 in log 10 scale) and are thus considered responders to IFN-.alpha. treatment in line with the definition above.
[0497] In contrast, patients denoted as p206 and p207 experienced reduction of virus load that is lower than by 100 (observed as 2 in log 10 scale) and are thus considered non-responders to IFN-.alpha. treatment in line with the definition above.
[0498] Patient denoted as p212 experienced reduction regulation of virus load of 2.02 and is thus considered on the boarder between responder and non-responder to IFN-.alpha. treatment in line with the definition above.
[0499] The results in FIG. 35 demonstrated that the expression of the gene HERC5 was significantly reduced in patients that were considered as responders compared to the expression in patients considered as non-responders. Interestingly, the expression of the gene HERC5 was significantly reduced also in patient denoted as p212, which experienced a reduction of virus load by about 100 (observed as 2 in log 10 scale).
[0500] The inventors have used the virus load measured or each patient at the beginning and at the end of the experiment and used these parameters for the model simulation to obtain a value of M for each patient, taking into account that K for HCV in blood samples is 5 [Ruy M. Ribeiro et al., (2012)].
TABLE-US-00015 TABLE 15 The expression of HERC5 gene (arbitrary units -2{circumflex over ( )}-dct rt-pcr_) and the calculated M for each one of the tested HCV patients. M calculated from Patient Expression of simulation # HERC5 assuming 4 weeks p202 0.00144 0.8635 p204 0.042563 0.863 p201 0.047145 0.862 p203 0.076972 0.867 p212 0.100299 0.836 p211 0.108098 0.872 p208 0.111671 0.883 p102 0.181633 0.8785 p213 0.188302 0.875 p101 0.28833 0.861 p207 1.801784 0.832 p206 3.157128 0.816
[0501] As shown in Table 15, the M values of all patients varied between 0.816 to 0.883.
[0502] The expression value of HERC5 and the M value obtained from the simulation were normalized with respect to the patient's population. FIG. 36, shows for each patient the normalized simulated M value (black box) and the normalized expression of HERC5 gene (open box). As can be seen, an inverse correlation is observed between the M value and the expression of HERC5 gene, with the patients being considered as responsive having considerably higher M value and the patients being considered as non-responsive having considerably lower M value.
[0503] Interestingly, the patient denoted as p212 that was considered on the boarder with respect to the virus load and responsive with respect to the HERC5 expression, has an intermediate M value. Simulation of the data of the patient denoted as p212 for a long time period of three month resulted in a M value correlating to responsiveness (data not shown).
[0504] Experiments on Tissue Samples
[0505] The data of liver tissue analysis of HCV patients is shown in FIGS. 37A and 37B. The patients denoted as p25, s12, p24, p26, s6, p22, s5 and s13 experienced reduction of virus load higher than by 100 (observed as 2 in log 10 scale) and are thus considered responders to IFN-.alpha. treatment in line with the definition above.
[0506] In contrast, patients denoted as p27, s20, p23, s18, s15, p21, s16, cts17 and sb11 experienced an amount of down regulation of virus load lower than 100 (observed as 2 in log 10 scale) and are thus considered non-responders to IFN-.alpha. treatment in line with the definition above.
[0507] The results presented in FIG. 37A demonstrate that the sum of normalized and scaled expression of the five genes UBE2L6, USP18, HERC5, OAS2 and ISG15 was significantly lower in patients that were considered as responders (patients p25, s12, p24, p26, s6, p22, s5 and s13) compared to the sum of the normalized expression in patients considered as non-responders (patients p25, s12, p24, p26, s6, p22, s5 and s13).
[0508] As indicated above, the M value was calculated for each patient using the experimental data obtained for the five genes. FIG. 37B shows the experimental M value for each one of the tested patients as in FIG. 37A.
[0509] As can be seen in FIG. 37B, there exists a strong correlation between the M value of a patient and the patient's response to treatment. The patients who were found to be responsive to IFN treatment (namely, patients p25, s12, p24, p26, s6, p22, s5 and s13) were characterized by a high M value (ranging between 0.86 to 0.88, whereas patients who were found to be non-responders to IFN treatment (namely, patients p25, s12, p24, p26, s6, p22, s5 and s13) were characterized by a low M value (ranges between 0.79 to 0.834).
[0510] As shown in FIG. 37B, the M value (that may be considered as a cut off to distinguish between responders and non-responders may range between 00.835 to 0.855. In this illustration example as in the PBMC case we assumed a distribution of M between 0.79 to 0.89.
[0511] A clear link between the experimental data obtained from tissue samples and the mathematical model described herein indicate that there is a strong correlation between the simulation of M and the experimental data. HCV in tissue samples is characterized by a doubling time of 5, namely K=5. As shown herein in FIG. 37C, a simulations of the above model equations by taking K as 4, showed that at lower M values of 0 to 0.72, namely in those patients being characterized by M values of up to 0.72, the virus is capable of multiplying and hence the disease is progressing, as the immune system or IFN treatment fail to eradicate the virus.
[0512] An M value that is higher than 0.72, clearly reduce virus load thereby eliminating the disease caused by a virus with k=4. This may indicate that either the immune system or IFN treatment regimen or both succeed in eliminating the virus.
[0513] Similarly, HCV patients characterized by M values of above 0.8 will most likely be able to reduce or eliminate the disease.
[0514] These results clearly indicate that there is the model simulation of the invention predicts with a high accuracy a patient's behavior for a particular virus. Namely, for a given virus an accurate cutoff value of M can be determined, and such M value distinguishes patients that will be able to "fight" the disease by responding to treatment, and those who will still suffer from the disease, namely, patients that are not responsive to treatment.
Example 12
[0515] Calculation of M Using Model Simulation in HCV Patients Treated with Combination Therapy of IFN-.alpha. and Ribavirin (Rib)
[0516] Data from the publication by Honda M. et al. [Journal of Hepatology 53: 817-826 (2010)] was used for correlation analysis with the mathematical model. Superficially, the inventors used virus load measured in thirty HCV patients before and after administration of IFN-.alpha. 2b at different time points. In accordance with the response to treatment, Honda M. et al. have defined treatment outcomes according to as follows: sustained viral response (SVR) --clearance of HCV viremia at 24 weeks after initiation of therapy; transient response (TR)--no detectable HCV viremia at 24 weeks but relapse during the follow-up period; and non-response (NR).
[0517] FIG. 38 shows a differential genetic expression obtained in liver biopsies of responders and non-responders HCV patients after combined therapy as described in Masao H. et al. Specifically, the left hand side shows the ratio of expression level of genes in responders vs. non-responders at day 0 (namely, before treatment), whereas the right hand side shows the results obtained after one week following treatment of IFN and RBV. The results provide a representative set of genes having a low expression value before treatment in patients who are referred to as responders. One week after treatment, these genes were clearly up regulated in the responders group.
[0518] Among the genes shown in the volcano plot, the inventors have used the expression of HERC5 for further analysis as this gene was the predominant gene and obtained the best p-value in the analysis. FIG. 39A shows the expression of HERC5 before treatment and FIG. 39B shows the expression of HERC5 after one week of treatment relative to the expression before treatment.
[0519] The results in FIG. 39A show that the initial HERC5 expression level in responders (including the patients defined as TR is low compared to non-responders. As shown in FIG. 39B, a week after treatment, a clear elevation in the expression of HERC5 gene appears in the responder group (and in the patients defined as TR), whereas the non-responders show a clear reduction. Based on these results it may be suggested that the expression level of this gene in responders is close to its saturation level, and therefore interferon cannot induce elevation in the expression of these genes.
[0520] The inventors of the present application then used the virus load data measured for each patient at different time points in a model simulation as described herein to obtain an M value for each one of the tested patients (assuming that k is 5).
[0521] FIG. 40 show normalized M value obtained from the model simulation for each one of the patients. The results indicate that the patients being considered as non-responsive have considerably lower M values, whereas the patients that show response (defined as SVR or TR) have considerably higher M values.
[0522] Interestingly, in two of the responsive patients that were characterized with the heights M values in FIG. 40 (0.97 and 0.99), no virus was detected after 48 hours of treatment. These results suggest that for patients characterized with higher M value, a short treatment period is sufficient to reduce/eliminate the virus and there is no need to treat these patients using long-term treatment.
[0523] The M values calculated from the model simulation described herein were correlated to normalized expression of HERC5 in order to obtain a "calibration data" of M values.
[0524] Table 16 shows for each patient, the normalized expression of HERC5 before treatment and the model calculated M value. Such calibration data may be further used for derivation of M. The patients category NR, SVR or TR is as defined above.
TABLE-US-00016 TABLE 16 The expression of HERC5 gene and the simulated M for each one of the tested HCV patients. Normalized Expression of Patient HERC5 Simulated category before treatment M value NR 0.480499 0.7995 NR 1 0.8002 NR 0.780031 0.8004 NR 0.533021 0.803 NR 0.704628 0.804 NR 0.689028 0.805 SVR 0.694228 0.81 SVR 0.161206 0.81 TR 0.460218 0.81 SVR 0.330213 0.82 TR 0 0.824 SVR 0.25741 0.85 SVR 0.0078 0.97 SVR 0.01014 0.99
Example 13
[0525] Determining Treatment Duration by Calculating M Using Genetic Expression in HCV Patients Treated with IFN-.alpha.
[0526] Seventeen HCV patients were examined in this study as also presented in Example 11 and FIGS. 37A and 37B. The expression of HERC5 was determined for each one of the patients before initiation of treatment using RT-PCR.
[0527] Based on the response to treatment, the patients were categorized into responders or non-responders as shown in FIG. 37.
[0528] Using the calibration curve prepared in Example 12, the inventors have determined for each one of the patients, an M value based on the experimental normalized value of expression of HERC5. It should be noted that the inventor considers treatment with every day using IFN plus ribavirin, as the best way for calculating an accurate M and therefore approximates the treatment with PegIFN during the week.
[0529] Table 17 shows the normalized expression of HERC5 as measured by RT-PCT and the M value determined using the calibration data described above.
TABLE-US-00017 TABLE 17 The expression of HERC5 gene and the derived M (from the calibration data) for each one of the tested HCV patients. Normalized Expression of Patient HERC5 derived category before treatment M value NR s18 0.40773 0.7995 NR p21 0.413093 0.7995 NR p23 0.448118 0.7995 NR p27 0.494851 0.7995 NR cts17 0.589386 0.803 NR s20 0.654809 0.805 NR s16 0.804371 0.8004 NR s15 0.873918 0.8004 NRsb11 1 0.8002 Responsive p25 0 0.95 Responsive p12 0.030833 0.95 Responsive p26 0.076105 0.95 Responsive p24 0.113371 0.84 Responsive s6 0.120362 0.84 Responsive p22 0.170882 0.84 Responsive p13 0.250061 0.85 Responsive s5 0.276657 0.85
[0530] These results show that the patients that were experimentally categorized as non-responders have lower M value compared with the patients that were experimentally categorized as responders. This suggest that measuring the expression of a single gene before treatment in a given patient and using this expression to obtain the corresponding M value for this patient, may predict if the patient will respond to treatment.
[0531] For prediction of treatment regimen, data from two patients were used denoted as s18 (non-responder) and p25 (responder). As shown in Table 17, using the initial virus load and the derived M for each one of these two patients, the viral load after 4 weeks was measured as presented in Table 18. The figure also discloses the normalized expression of HERC5 marker gene calculated for both patients.
TABLE-US-00018 TABLE 18 virus load and M values of HCV patients Baseline Virus load HERC5 Derived virus after Patient expression M load 4 weeks S18 0.40773 0.7995 79986/07 168,162 P25 0 0.95 4539 HCV Not detected
[0532] Based on the model described herein, FIGS. 41A and 41B show results of model simulation for s18 and p25, respectively providing calculated predicted virus load.
[0533] As shown in FIG. 41A, patient denoted as s18 having a measured initial virus load of 79986.07 (Table 18) and a calculated derived M value of 0.7995, exhibits an increase in virus load after a month up to a value of about 170,000. This simulation strongly correlates with the virus load measured in this patient after four weeks of treatment, which is about 168,162, as presented in Table 18.
[0534] In addition, as shown in FIG. 41B, patient denoted as p25 having an initial virus load of 4,539 and a derived M value of 0.95, exhibits a decrease in virus load after a month up to a value basically to baseline level. This strongly correlates with the fact that no virus load was measured in this patient after four weeks of treatment as presented in Table 18.
[0535] Thus, for a patient having an M value of 0.795 (p18), IFN treatment would not reduce viral load and therefore should be avoided. However, for a patient having an M value of about 0.95, eradication of the virus is achieved within several days (less than a week) of treatment. The data shown herein therefore also provide means to determine the treatment duration and also type of treatment.
Example 14
[0536] Predicting Treatment Regimen for Patient Suffering from HCV and HIV
[0537] Data from the publication of Murphya, Alison A. et al, AIDS 2011, 25:1179-1187 was used to study the ability of the model to predict treatment regimen. Specifically, the data of average virus load obtained from all patients at different time points was used in the simulation to obtain a M value of 0.82 (K was set at 5).
[0538] FIG. 42 shows the model simulation (right curve) of the data providing a M value of 0.82.
[0539] The simulated M value (of 0.82) was then used together with the initial average virus load in a further simulation that used different dosing regimen, twice a week instead of once weekly. As can be seen in the left curve of FIG. 42, treatment twice a week was more efficient.
[0540] Further, the patient denoted as s18 (non responder) was further used in a model simulation of treatment regimen. Using the M value of 0.7995 and the initial virus load, the treatment outcome was simulated. As shown in the right curve in FIG. 43, there was an increase in virus load indicating that the patient was not responsive to treatment. Interestingly, as shown in left curve of FIG. 43, increasing the dosing regimen from once a week to twice a week did not result in a response to treatment. This data suggest that even treating this patient with higher amount and/or different regimen is not efficient and new medications need to be used.
TABLE-US-00019 TABLE 19 List of Sequences SEQ ID NO: Details 1 DNA sequence of ISG15 ubiquitin-like modifier (ISG15) 2 Protein sequence of ISG15 ubiquitin-like modifier (ISG15) 3 DNA sequence of Interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) 4 Protein sequence of Interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) 5 DNA sequence of Interferon-induced protein with tetratricopeptide repeats 2 (IFIT2) 6 Protein sequence of Interferon-induced protein with tetratricopeptide repeats 2 (IFIT2) 7 DNA sequence of Interferon-induced protein with tetratricopeptide repeats 3 (IFIT3) 8 Protein sequence of Interferon-induced protein with tetratricopeptide repeats 3 (IFIT3) 9 DNA sequence of Interferon-induced protein with tetratricopeptide repeats 3 (IFIT3) 10 Protein sequence of Interferon-induced protein with tetratricopeptide repeats 3 (IFIT3) 11 DNA sequence of Interferon-induced protein with tetratricopeptide repeats 5 (IFIT5) 12 Protein sequence of Interferon-induced protein with tetratricopeptide repeats 5 (IFIT5) 13 DNA sequence of 2'-5'-oligoadenylate synthetase 1 (OAS1) 14 Protein sequence of 2'-5'-oligoadenylate synthetase 1 (OAS1) 15 DNA sequence of 2'-5'-oligoadenylate synthetase 1 (OAS1) 16 Protein sequence of 2'-5'-oligoadenylate synthetase 1 (OAS1) 17 DNA sequence of 2'-5'-oligoadenylate synthetase 1 (OAS1) 18 Protein sequence of 2'-5'-oligoadenylate synthetase 1 (OAS1) 19 DNA sequence of 2'-5'-oligoadenylate synthetase 2 (OAS2) 20 Protein sequence of 2'-5'-oligoadenylate synthetase 2 (OAS2) 21 DNA sequence of 2'-5'-oligoadenylate synthetase 2 (OAS2) 22 Protein sequence of 2'-5'-oligoadenylate synthetase 2 (OAS2) 23 DNA sequence of 2'-5'-oligoadenylate synthetase 2 (OAS2) 24 Protein sequence of 2'-5'-oligoadenylate synthetase 2 (OAS2) 25 DNA sequence of 2'-5'-oligoadenylate synthetase 3 (OAS3) 26 Protein sequence of 2'-5'-oligoadenylate synthetase 3 (OAS3) 27 DNA sequence of 2'-5'-oligoadenylate synthetase-like (OASL) 28 Protein sequence of 2'-5'-oligoadenylate synthetase-like (OASL) 29 DNA sequence of 2'-5'-oligoadenylate synthetase-like (OASL) 30 Protein sequence of 2'-5'-oligoadenylate synthetase-like (OASL) 31 DNA sequence of HECT and RLD domain containing E3 ubiquitin protein ligase 5 (HERC5) 32 Protein sequence of HECT and RLD domain containing E3 ubiquitin protein ligase 5 (HERC5) 33 DNA sequence of ubiquitin specific peptidase 18 (USP18) 34 Protein sequence of ubiquitin specific peptidase 18 (USP18) 35 DNA sequence of Radical S-adenosyl methionine domain containing 2 (RSAD2) 36 Protein sequence of Radical S-adenosyl methionine domain containing 2 (RSAD2) 37 DNA sequence of myxovirus (influenza virus) resistance 1 (MX1) 38 Protein sequence of myxovirus (influenza virus) resistance 1 (MX1) 39 DNA sequence of myxovirus (influenza virus) resistance 1 (MX1) 40 Protein sequence of myxovirus (influenza virus) resistance 1 (MX1) 41 DNA sequence of myxovirus (influenza virus) resistance 1 (MX1) 42 Protein sequence of myxovirus (influenza virus) resistance 1 (MX1) 43 DNA sequence of Interferon-induced protein 44-like (IFI44L) 44 Protein sequence of Interferon-induced protein 44-like (IFI44L) 45 DNA sequence of DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 (DDX58) 46 Protein sequence of DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 (DDX58) 47 DNA sequence of interferon alpha 1 48 Protein sequence interferon alpha 1 49 DNA sequence of interferon alpha 2 50 Protein sequence of interferon alpha 2 51 DNA sequence of Interferon alpha-4 52 Protein sequence of Interferon alpha-4 53 DNA sequence of Interferon alpha-5 54 Protein sequence of Interferon alpha-5 55 DNA sequence of Interferon alpha-6 56 Protein sequence of Interferon alpha-6 57 DNA sequence of Interferon alpha-7 58 Protein sequence of Interferon alpha-7 59 DNA sequence of Interferon alpha-8 60 Protein sequence of Interferon alpha-8 61 DNA sequence of Interferon alpha-10 62 Protein sequence of Interferon alpha-10 63 DNA sequence of Interferon alpha-1/13 64 Protein sequence of Interferon alpha-1/13 65 DNA sequence of Interferon alpha-14 66 Protein sequence of Interferon alpha-14 67 DNA sequence of Interferon alpha-16 68 Protein sequence of Interferon alpha-16 69 DNA sequence of Interferon alpha-17 70 Protein sequence of Interferon alpha-17 71 DNA sequence of Interferon alpha-21 72 Protein sequence of Interferon alpha-21 73 DNA sequence of Interferon, beta 1 74 Protein sequence of Interferon, beta 1 75 DNA sequence of Interferon omega-1 76 Protein sequence of Interferon omega-1 77 DNA sequence of Interferon-gamma 78 Protein sequence of Interferon-gamma 79 DNA sequence of E1-like ubiquitin-activating enzyme (UBElL) 80 Protein sequence of E1-like ubiquitin-activating enzyme (UBElL) 81 DNA sequence of Ubiquitin-conjugating enzyme E2L 6 (UBE2L6) 82 Protein sequence of Ubiquitin-conjugating enzyme E2L 6 (UBE2L6) 83 DNA sequence of Ubiquitin-conjugating enzyme E2L 6 (UBE2L6) 84 Protein sequence of Ubiquitin-conjugating enzyme E2L 6 (UBE2L6) 85 DNA sequence of Interferon alpha-inducible protein 27 (IFI27) 86 protein sequence of Interferon alpha-inducible protein 27 (IFI27) 87 DNA sequence of Interferon alpha-inducible protein 27 (IFI27) 88 Protein sequence of Interferon alpha-inducible protein 27 (IFI27) 89 DNA sequence of Interferon induced with helicase C domain 1 (IFIH1) 90 Protein sequence of Interferon induced with helicase C domain 1 (IFIH1) 91 DNA sequence of Toll-like receptor 7 (TLR-7) 92 Protein sequence of Toll-like receptor 7 (TLR-7) 93 DNA sequence of Interferon regulatory factor 7 (IRF7) 94 Protein sequence of Interferon regulatory factor 7 (IRF7) 95 DNA sequence of Interferon regulatory factor 7 (IRF7) 96 Protein sequence of Interferon regulatory factor 7 (IRF7) 97 DNA sequence of Interferon, alpha-inducible protein 6 (IFI6) 98 Protein sequence of Interferon, alpha-inducible protein 6 (IFI6) 99 DNA sequence of Interferon, alpha-inducible protein 6 (IFI6) 100 Protein sequence of Interferon, alpha-inducible protein 6 (IFI6) 101 DNA sequence of Interferon, alpha-inducible protein 6 (IFI6) 102 Protein sequence of Interferon, alpha-inducible protein 6 (IFI6) 103 DNA sequence of Signal transducer and activator of transcription 1 (STAT1) 104 DNA sequence of Signal transducer and activator of transcription 1 (STAT1) 105 Protein sequence of Signal transducer and activator of transcription 1 (STAT1) 106 Protein sequence of Signal transducer and activator of transcription 1 (STAT1) 107 DNA sequence of Interferon-induced protein 44 (IFI44) gene 108 Protein sequence of Interferon-induced protein 44 (IFI44) gene 109 DNA sequence of eukaryotic translation initiation factor 2-alpha kinase 2 (EIF2AK2) 110 Protein sequence of eukaryotic translation initiation factor 2-alpha kinase 2 (EIF2AK2) 111 DNA sequence of DEXH (Asp-Glu-X-His) box polypeptide 58 (DHX58) 112 Protein sequence of DEXH (Asp-Glu-X-His) box polypeptide 58 (DHX58)
Sequence CWU 1
1
1121685DNAHomo sapiens 1ataatagggc cggtgctgcc tgccgaagcc ggcggctgag
aggcagcgaa ctcatctttg 60ccagtacagg agcttgtgcc gtggcccaca gcccacagcc
cacagccatg ggctgggacc 120tgacggtgaa gatgctggcg ggcaacgaat
tccaggtgtc cctgagcagc tccatgtcgg 180tgtcagagct gaaggcgcag
atcacccaga agatcggcgt gcacgccttc cagcagcgtc 240tggctgtcca
cccgagcggt gtggcgctgc aggacagggt cccccttgcc agccagggcc
300tgggccccgg cagcacggtc ctgctggtgg tggacaaatg cgacgaacct
ctgagcatcc 360tggtgaggaa taacaagggc cgcagcagca cctacgaggt
acggctgacg cagaccgtgg 420cccacctgaa gcagcaagtg agcgggctgg
agggtgtgca ggacgacctg ttctggctga 480ccttcgaggg gaagcccctg
gaggaccagc tcccgctggg ggagtacggc ctcaagcccc 540tgagcaccgt
gttcatgaat ctgcgcctgc ggggaggcgg cacagagcct ggcgggcgga
600gctaagggcc tccaccagca tccgagcagg atcaagggcc ggaaataaag
gctgttgtaa 660agagaaaaaa aaaaaaaaaa aaaaa 6852165PRTHomo sapiens
2Met Gly Trp Asp Leu Thr Val Lys Met Leu Ala Gly Asn Glu Phe Gln1 5
10 15Val Ser Leu Ser Ser Ser Met Ser Val Ser Glu Leu Lys Ala Gln
Ile 20 25 30Thr Gln Lys Ile Gly Val His Ala Phe Gln Gln Arg Leu Ala
Val His 35 40 45Pro Ser Gly Val Ala Leu Gln Asp Arg Val Pro Leu Ala
Ser Gln Gly 50 55 60Leu Gly Pro Gly Ser Thr Val Leu Leu Val Val Asp
Lys Cys Asp Glu65 70 75 80Pro Leu Ser Ile Leu Val Arg Asn Asn Lys
Gly Arg Ser Ser Thr Tyr 85 90 95Glu Val Arg Leu Thr Gln Thr Val Ala
His Leu Lys Gln Gln Val Ser 100 105 110Gly Leu Glu Gly Val Gln Asp
Asp Leu Phe Trp Leu Thr Phe Glu Gly 115 120 125Lys Pro Leu Glu Asp
Gln Leu Pro Leu Gly Glu Tyr Gly Leu Lys Pro 130 135 140Leu Ser Thr
Val Phe Met Asn Leu Arg Leu Arg Gly Gly Gly Thr Glu145 150 155
160Pro Gly Gly Arg Ser 16534396DNAHomo sapiens 3gcaaggacac
acccacagct tacaccattg gctgctgttt agctccctta tataacactg 60tcttggggtt
taaacgtaac tgaaaatcca caagacagaa tagccagatc tcagaggagc
120ctggctaagc aaaaccctgc agaacggctg cctaatttac agcaaccatg
agtacaaatg 180gtgatgatca tcaggtcaag gatagtctgg agcaattgag
atgtcacttt acatgggagt 240tatccattga tgacgatgaa atgcctgatt
tagaaaacag agtcttggat cagattgaat 300tcctagacac caaatacagt
gtgggaatac acaacctact agcctatgtg aaacacctga 360aaggccagaa
tgaggaagcc ctgaagagct taaaagaagc tgaaaactta atgcaggaag
420aacatgacaa ccaagcaaat gtgaggagtc tggtgacctg gggcaacttt
gcctggatgt 480attaccacat gggcagactg gcagaagccc agacttacct
ggacaaggtg gagaacattt 540gcaagaagct ttcaaatccc ttccgctata
gaatggagtg tccagaaata gactgtgagg 600aaggatgggc cttgctgaag
tgtggaggaa aaaattatga acgggccaag gcctgctttg 660aaaaggtgct
tgaagtggac cctgaaaacc ctgaatccag cgctgggtat gcgatctctg
720cctatcgcct ggatggcttt aaattagcca caaaaaatca caagccattt
tctttgcttc 780ccctaaggca ggctgtccgc ttaaatccag acaatggata
tattaaggtt ctccttgccc 840tgaagcttca ggatgaagga caggaagctg
aaggagaaaa gtacattgaa gaagctctag 900ccaacatgtc ctcacagacc
tatgtctttc gatatgcagc caagttttac cgaagaaaag 960gctctgtgga
taaagctctt gagttattaa aaaaggcctt gcaggaaaca cccacttctg
1020tcttactgca tcaccagata gggctttgct acaaggcaca aatgatccaa
atcaaggagg 1080ctacaaaagg gcagcctaga gggcagaaca gagaaaagct
agacaaaatg ataagatcag 1140ccatatttca ttttgaatct gcagtggaaa
aaaagcccac atttgaggtg gctcatctag 1200acctggcaag aatgtatata
gaagcaggca atcacagaaa agctgaagag aattttcaaa 1260aattgttatg
catgaaacca gtggtagaag aaacaatgca agacatacat ttccactatg
1320gtcggtttca ggaatttcaa aagaaatctg acgtcaatgc aattatccat
tatttaaaag 1380ctataaaaat agaacaggca tcattaacaa gggataaaag
tatcaattct ttgaagaaat 1440tggttttaag gaaacttcgg agaaaggcat
tagatctgga aagcttgagc ctccttgggt 1500tcgtctacaa attggaagga
aatatgaatg aagccctgga gtactatgag cgggccctga 1560gactggctgc
tgactttgag aactctgtga gacaaggtcc ttaggcaccc agatatcagc
1620cactttcaca tttcatttca ttttatgcta acatttacta atcatctttt
ctgcttactg 1680ttttcagaaa cattataatt cactgtaatg atgtaattct
tgaataataa atctgacaaa 1740atattagttg tgttcaacaa ttagtgaaac
agaatgtgtg tatgcatgta agaaagagaa 1800atcatttgta tgagtgctat
gtagtagaga aaaaatgtta gttaactttg taggaaataa 1860aacattggac
ttacactaaa tgtttaattc attcatttta ttgtgaaata aaaataaaat
1920ccttagctcc tccaccaact gaacagaccc tcttggccaa ggagacccca
gaaaccttaa 1980aaactaagtt tcccaaccat gacaagatga gagatcattc
acacctcatt atattccctc 2040ccttgctaac tgccattgga ctttttccac
tgagttaaac agaaacccat ggaaaacaaa 2100gaacagaaga ctcactcctt
ggctgacttc acctagctca ctccacgtag cgccacagcc 2160agactcccct
cccctcttgc ggtttccaca tgacaactga tcagccttcc ctcctgataa
2220gtgaccactg cccacagact ggttctggcc agtccatgga ggctgcacac
agggtgcctc 2280tatgtccttt gtttcacctt ttgatataga aaggctaatt
ttgctgtatt ttaatgttaa 2340gtctccacca cagagtgaac acagaatgca
tgtgacatac atgtttacat accactattg 2400tgtgactgcc cctcatgaat
attcatagcc ccccataacc tgttaactat gtgtgtctag 2460ccaatccacc
aaccataaaa cttctgtaat accctccctt cctccaagag cctgcttttg
2520gttgctgtgg taggctctgc ttcccaggct gcaggttgca ggagaggagg
ctgcagtggc 2580tcacgcctgt aatctcagca cttcgatggg acgaggcagg
cagatcacct gaacccagga 2640gttcgagagc agccttggca atggcaaaac
caaccgtctc tacaaaaaat gcaaaaactt 2700agctgggtgt ggtggcatgc
acctgtagct tcagttccag ctactcagga ggctgaggtg 2760agtggactgc
tggagccagg gagttcgagg ctgcagtgtc gagatcttgc cactgcactc
2820cattctggat gatagaacga gaccccatct caaaaaaaaa aaaagttctc
tccaattgta 2880tatagcttgt gattttatgt caacactatc aataaatagc
tttcagtgca agaaaccaaa 2940aatactgtaa taaacaggca catattcttc
ccaaacctca tgcagtttac aatctagtga 3000gagacacaga tagcagtaca
gagtcaatta aaggttagtt ttcttcatga agatgtttta 3060attttaattc
aatgtgaaag ggttccaagg agtttatctt gttttatgcc attttatttg
3120aagcactact tactaagtca tttgctgata ttaatctagt taaatcaaga
aatattacat 3180gaaaatgttg ctaaatcaga gatcatgggt aacaatcacc
tttgattatg aataatcata 3240ttttattgaa aggcaaggca caacaaataa
taagaaggaa aaaataaata agcaatgtta 3300ttgatctttc attctgtata
tgttttgggg ggaatatact agtttctttt agtggctgta 3360acaaattacc
acaaacttgg tgacttaaaa tttcacagat ttactctttc ttacagttct
3420ggaggtcaga agtctgaaat gggtttcaat gagccaaagt caaggtattg
atgacgctac 3480actcctccgg aggctctagg cagatagcct tttccagctt
ccagaggctg cctgaattct 3540ttcatccatc ttaaaaacca acagtgtagt
agcctcaaat ctctctctct gcttccttct 3600tcacatctcc ttctctcctc
tgactctttt gcctctttct tctaaggacg caccaggtcc 3660acctgcataa
tccagaataa ttgccccatc cgcaaatcct taatttaata acatctgcaa
3720agtccctttt gctatgtaaa gtagcatgtt cacaggttct ggagacttgg
ccatggatac 3780gattgcgggg ggggcattat tcttaccaca gagcacccca
agaaaatctc caaattttgg 3840gcttccaatc cattttgctt caattattta
atatttttac tccttccagt agatactgat 3900ttcatccatt gcccttaaga
aggtaggaca gagattatgg cacatctcac attaaatgct 3960atattttcgt
tggaaataca ttttttgctt caacttttat tttaaattca agggtacatg
4020tgcaggatgt tcaggtttgt tacacaggta aacgtgtgcc atggcggttt
gctgaacaga 4080tcatcccatc accaacagat catcccattg agaggtgaag
ccggctgggc ttctgggttg 4140ggtggggact tggagaactt ttctgtctag
ctaaagtatt gtaaaatgga ccagtcaaca 4200ctctgtaaaa tggaccaatc
agctctctgt aaaatggacc aatcagcagg atgtgggtgg 4260ggccaagtaa
gggaataaaa gcaggccacc cgagctggca gcggcaaccc gctcgggtcc
4320ccttccatgc tgtggaagtt ttgttctttc gctctttcaa taaatcttgc
tgctgctcaa 4380aaaaaaaaaa aaaaaa 43964478PRTHomo sapiens 4Met Ser
Thr Asn Gly Asp Asp His Gln Val Lys Asp Ser Leu Glu Gln1 5 10 15Leu
Arg Cys His Phe Thr Trp Glu Leu Ser Ile Asp Asp Asp Glu Met 20 25
30Pro Asp Leu Glu Asn Arg Val Leu Asp Gln Ile Glu Phe Leu Asp Thr
35 40 45Lys Tyr Ser Val Gly Ile His Asn Leu Leu Ala Tyr Val Lys His
Leu 50 55 60Lys Gly Gln Asn Glu Glu Ala Leu Lys Ser Leu Lys Glu Ala
Glu Asn65 70 75 80Leu Met Gln Glu Glu His Asp Asn Gln Ala Asn Val
Arg Ser Leu Val 85 90 95Thr Trp Gly Asn Phe Ala Trp Met Tyr Tyr His
Met Gly Arg Leu Ala 100 105 110Glu Ala Gln Thr Tyr Leu Asp Lys Val
Glu Asn Ile Cys Lys Lys Leu 115 120 125Ser Asn Pro Phe Arg Tyr Arg
Met Glu Cys Pro Glu Ile Asp Cys Glu 130 135 140Glu Gly Trp Ala Leu
Leu Lys Cys Gly Gly Lys Asn Tyr Glu Arg Ala145 150 155 160Lys Ala
Cys Phe Glu Lys Val Leu Glu Val Asp Pro Glu Asn Pro Glu 165 170
175Ser Ser Ala Gly Tyr Ala Ile Ser Ala Tyr Arg Leu Asp Gly Phe Lys
180 185 190Leu Ala Thr Lys Asn His Lys Pro Phe Ser Leu Leu Pro Leu
Arg Gln 195 200 205Ala Val Arg Leu Asn Pro Asp Asn Gly Tyr Ile Lys
Val Leu Leu Ala 210 215 220Leu Lys Leu Gln Asp Glu Gly Gln Glu Ala
Glu Gly Glu Lys Tyr Ile225 230 235 240Glu Glu Ala Leu Ala Asn Met
Ser Ser Gln Thr Tyr Val Phe Arg Tyr 245 250 255Ala Ala Lys Phe Tyr
Arg Arg Lys Gly Ser Val Asp Lys Ala Leu Glu 260 265 270Leu Leu Lys
Lys Ala Leu Gln Glu Thr Pro Thr Ser Val Leu Leu His 275 280 285His
Gln Ile Gly Leu Cys Tyr Lys Ala Gln Met Ile Gln Ile Lys Glu 290 295
300Ala Thr Lys Gly Gln Pro Arg Gly Gln Asn Arg Glu Lys Leu Asp
Lys305 310 315 320Met Ile Arg Ser Ala Ile Phe His Phe Glu Ser Ala
Val Glu Lys Lys 325 330 335Pro Thr Phe Glu Val Ala His Leu Asp Leu
Ala Arg Met Tyr Ile Glu 340 345 350Ala Gly Asn His Arg Lys Ala Glu
Glu Asn Phe Gln Lys Leu Leu Cys 355 360 365Met Lys Pro Val Val Glu
Glu Thr Met Gln Asp Ile His Phe His Tyr 370 375 380Gly Arg Phe Gln
Glu Phe Gln Lys Lys Ser Asp Val Asn Ala Ile Ile385 390 395 400His
Tyr Leu Lys Ala Ile Lys Ile Glu Gln Ala Ser Leu Thr Arg Asp 405 410
415Lys Ser Ile Asn Ser Leu Lys Lys Leu Val Leu Arg Lys Leu Arg Arg
420 425 430Lys Ala Leu Asp Leu Glu Ser Leu Ser Leu Leu Gly Phe Val
Tyr Lys 435 440 445Leu Glu Gly Asn Met Asn Glu Ala Leu Glu Tyr Tyr
Glu Arg Ala Leu 450 455 460Arg Leu Ala Ala Asp Phe Glu Asn Ser Val
Arg Gln Gly Pro465 470 47553505DNAHomo sapiens 5agtttcactt
tcccttttgt aacgtcagct gaagggaaac aaacaaaaag gaaccagagg 60ccacttgtat
atataggtct cttcagcatt tattggtggc agaagaggaa gatttctgaa
120gagtgcagct gcctgaaccg agccctgccg aacagctgag aattgcactg
caaccatgag 180tgagaacaat aagaattcct tggagagcag cctacggcaa
ctaaaatgcc atttcacctg 240gaacttgatg gagggagaaa actccttgga
tgattttgaa gacaaagtat tttaccggac 300tgagtttcag aatcgtgaat
tcaaagccac aatgtgcaac ctactggcct atctaaagca 360cctcaaaggg
caaaacgagg cagccctgga atgcttacgt aaagctgaag agttaatcca
420gcaagagcat gctgaccagg cagaaatcag aagtctggtc acctggggaa
actatgcctg 480ggtctactat cacatgggcc gactctcaga cgttcagatt
tatgtagaca aggtgaaaca 540tgtctgtgag aagttttcca gtccctatag
aattgagagt ccagagcttg actgtgagga 600agggtggaca cggttaaagt
gtggaggaaa ccaaaatgaa agagcgaagg tgtgctttga 660gaaggctctg
gaaaagaagc caaagaaccc agaattcacc tctggactgg caatagcaag
720ctaccgtctg gacaactggc caccatctca gaacgccatt gaccctctga
ggcaagccat 780tcggctgaat cctgacaacc agtaccttaa agtcctcctg
gctctgaagc ttcataagat 840gcgtgaagaa ggtgaagagg aaggtgaagg
agagaagtta gttgaagaag ccttggagaa 900agccccaggt gtaacagatg
ttcttcgcag tgcagccaag ttttatcgaa gaaaagatga 960gccagacaaa
gcgattgaac tgcttaaaaa ggctttagaa tacataccaa acaatgccta
1020cctgcattgc caaattgggt gctgctatag ggcaaaagtc ttccaagtaa
tgaatctaag 1080agagaatgga atgtatggga aaagaaagtt actggaacta
ataggacacg ctgtggctca 1140tctgaagaaa gctgatgagg ccaatgataa
tctcttccgt gtctgttcca ttcttgccag 1200cctccatgct ctagcagatc
agtatgaaga cgcagagtat tacttccaaa aggaattcag 1260taaagagctt
actcctgtag cgaaacaact gctccatctg cggtatggca actttcagct
1320gtaccaaatg aagtgtgaag acaaggccat ccaccacttt atagagggtg
taaaaataaa 1380ccagaaatca agggagaaag aaaagatgaa agacaaactg
caaaaaattg ccaaaatgcg 1440actttctaaa aatggagcag attctgaggc
tttgcatgtc ttggcattcc ttcaggagct 1500gaatgaaaaa atgcaacaag
cagatgaaga ctctgagagg ggtttggagt ctggaagcct 1560catcccttca
gcatcaagct ggaatgggga atgaagaata gagatgtggt gcccactagg
1620ctactgctga aagggagctg aaattcctcc accaagttgg tattcaaaat
atgtaatgac 1680tggtatggca aaagattgga ctaagacact ggccatacca
ctggacaggg ttatgttaac 1740acctgaattg ctgggtcttg agagagccca
aggagttctg ggagagggac cagattgggg 1800ggtaggtcca cgggcttggt
gatagaatta tttctcgatt gacttcttga gtgcaatttg 1860aactgtaaca
tttgcttagt cacctttagt ggagtaatct actgggcttg tttctatatt
1920tatataaagc agccaaatcc ttcatgtaat attgaagtcc atttttgcaa
tgttgttcca 1980tacttggagt cattttgcat cccatagagg ttagtcctgc
atagccagta atgtgctaag 2040ttcatccaaa agctggcgga ccaaagtcta
aatagggctc agtatccccc atcgcttatc 2100tctgcctcct tcctcctcct
tcccagtcta tcatcaacct tgagtattct acacaatgtg 2160aattcaagtg
cctgattaat tgaggtggca acatagtttg agacgagggc agagaacagg
2220aagatacata gctagaagcg acgggtacaa aaagcaatgt gtacaagaag
actttcagca 2280agtatacaga gagttcacct ctactctgcc ctcctcatag
tcataatgta gcaagtaaag 2340aatgagaatg gattctgtac aatacactag
aaaccaacat aatgtatttc tttaaaacct 2400gtgtgaaaaa ataaatgttc
caccagtagg gataggggaa aagtaaccaa aagagagaaa 2460gagaaaggaa
tgctggttta tctttgtaga ttgtaatcga atggagaaat ttgcagtatt
2520ttagccacta ttaggaattt tttttttttg taaaatgaag actgaactct
gttcaaatgc 2580tttcatgaac ctggtttgag acggtaggaa agcaacaaaa
cgtgggaacc tggtgactaa 2640gggcctggtg caaggacttg ggaaatgtca
ttgataatag atggtggggt tttcccccct 2700ttagaaatgt tggatattaa
gtgatataaa cacttctttt aactccgaaa atcttctgag 2760aaatcacaaa
attcacggta tgcttggaac gattgagatt ttctaggtag atgctgaata
2820gcctagacat caaagttggt gtgaaccaaa atagagtcag ctgacccagc
atcagccaca 2880ctctgggttg gaaaatgttt gcctgttgga attaatttaa
gcttaagtat atatcaacat 2940tattttattg tgcaattaaa acaatacaaa
ttcatggttt tttaaagtta aaaattctaa 3000ccactgtaac aacagttttt
gtgttatttt ctgtattaaa catcttgttg cacgcatttg 3060aggtcatcag
ggtgcaaaat ttgtattcct gaaaatgtca tatattttca ttaataaata
3120acctaaatat gataaaacat aaagcagtgt tctggttcat ctggaatttt
gctgtacttt 3180aaatctttca gactcagcta ctgataaatg aaacgttaca
caggtgtgaa ccaaatccaa 3240ataacctcga ctggtctact atcataatca
cctgaacaga acaaaacttt ttcctcagct 3300ttaagagtcc agggcttcgg
ataacagctg ccatctgcca cctgctacca ttgacctacg 3360tgaacacaga
cattctgtct ccaccttgat ggtgggtggg ctgctcccct tttctttgtt
3420aaattttgtg ctttcatcac attttctcta ttctgacctc tgttatgaga
aataaaagtc 3480actgattcca ttttaaaaaa aaaaa 35056472PRTHomo sapiens
6Met Ser Glu Asn Asn Lys Asn Ser Leu Glu Ser Ser Leu Arg Gln Leu1 5
10 15Lys Cys His Phe Thr Trp Asn Leu Met Glu Gly Glu Asn Ser Leu
Asp 20 25 30Asp Phe Glu Asp Lys Val Phe Tyr Arg Thr Glu Phe Gln Asn
Arg Glu 35 40 45Phe Lys Ala Thr Met Cys Asn Leu Leu Ala Tyr Leu Lys
His Leu Lys 50 55 60Gly Gln Asn Glu Ala Ala Leu Glu Cys Leu Arg Lys
Ala Glu Glu Leu65 70 75 80Ile Gln Gln Glu His Ala Asp Gln Ala Glu
Ile Arg Ser Leu Val Thr 85 90 95Trp Gly Asn Tyr Ala Trp Val Tyr Tyr
His Met Gly Arg Leu Ser Asp 100 105 110Val Gln Ile Tyr Val Asp Lys
Val Lys His Val Cys Glu Lys Phe Ser 115 120 125Ser Pro Tyr Arg Ile
Glu Ser Pro Glu Leu Asp Cys Glu Glu Gly Trp 130 135 140Thr Arg Leu
Lys Cys Gly Gly Asn Gln Asn Glu Arg Ala Lys Val Cys145 150 155
160Phe Glu Lys Ala Leu Glu Lys Lys Pro Lys Asn Pro Glu Phe Thr Ser
165 170 175Gly Leu Ala Ile Ala Ser Tyr Arg Leu Asp Asn Trp Pro Pro
Ser Gln 180 185 190Asn Ala Ile Asp Pro Leu Arg Gln Ala Ile Arg Leu
Asn Pro Asp Asn 195 200 205Gln Tyr Leu Lys Val Leu Leu Ala Leu Lys
Leu His Lys Met Arg Glu 210 215 220Glu Gly Glu Glu Glu Gly Glu Gly
Glu Lys Leu Val Glu Glu Ala Leu225 230 235 240Glu Lys Ala Pro Gly
Val Thr Asp Val Leu Arg Ser Ala Ala Lys Phe 245 250 255Tyr Arg Arg
Lys Asp Glu Pro Asp Lys Ala Ile Glu Leu Leu Lys Lys 260 265 270Ala
Leu Glu Tyr Ile Pro Asn Asn Ala Tyr Leu His Cys Gln Ile Gly 275 280
285Cys Cys Tyr Arg Ala Lys Val Phe Gln Val Met Asn Leu Arg Glu Asn
290 295 300Gly Met Tyr Gly Lys Arg Lys Leu Leu Glu Leu Ile Gly His
Ala Val305 310 315 320Ala His Leu Lys Lys Ala Asp Glu Ala Asn Asp
Asn Leu Phe Arg Val 325 330 335Cys Ser Ile Leu Ala Ser Leu His Ala
Leu Ala Asp Gln Tyr Glu Asp 340 345 350Ala Glu Tyr Tyr Phe Gln Lys
Glu Phe Ser Lys Glu Leu Thr Pro Val 355 360 365Ala Lys Gln Leu Leu
His Leu Arg Tyr Gly Asn Phe Gln Leu Tyr Gln 370 375 380Met Lys Cys
Glu Asp Lys Ala Ile His His
Phe Ile Glu Gly Val Lys385 390 395 400Ile Asn Gln Lys Ser Arg Glu
Lys Glu Lys Met Lys Asp Lys Leu Gln 405 410 415Lys Ile Ala Lys Met
Arg Leu Ser Lys Asn Gly Ala Asp Ser Glu Ala 420 425 430Leu His Val
Leu Ala Phe Leu Gln Glu Leu Asn Glu Lys Met Gln Gln 435 440 445Ala
Asp Glu Asp Ser Glu Arg Gly Leu Glu Ser Gly Ser Leu Ile Pro 450 455
460Ser Ala Ser Ser Trp Asn Gly Glu465 47072464DNAHomo sapiens
7actttccttt cccctttcat aaaagcacag acctaacagc accctgggtg gaaacctctt
60cagcatttgc ttggaatcag taagctaaaa acaaaatcaa ccgggacccc agcttttcag
120aactgcaggg aaacagccat catgagtgag gtcaccaaga attccctgga
gaaaatcctt 180ccacagctga aatgccattt cacctggaac ttattcaagg
aagacagtgt ctcaagggat 240ctagaagata gagtgtgtaa ccagattgaa
tttttaaaca ctgagttcaa agctacaatg 300tacaacttgt tggcctacat
aaaacaccta gatggtaaca acgaggcagc cctggaatgc 360ttacggcaag
ctgaagagtt aatccagcaa gaacatgctg accaagcaga aatcagaagt
420ctagtcactt ggggaaacta cgcctgggtc tactatcact tgggcagact
ctcagatgct 480cagatttatg tagataaggt gaaacaaacc tgcaagaaat
tttcaaatcc atacagtatt 540gagtattctg aacttgactg tgaggaaggg
tggacacaac tgaagtgtgg aagaaatgaa 600agggcgaagg tgtgttttga
gaaggctctg gaagaaaagc ccaacaaccc agaattctcc 660tctggactgg
caattgcgat gtaccatctg gataatcacc cagagaaaca gttctctact
720gatgttttga agcaggccat tgagctgagt cctgataacc aatacgtcaa
ggttctcttg 780ggcctgaaac tgcagaagat gaataaagaa gctgaaggag
agcagtttgt tgaagaagcc 840ttggaaaagt ctccttgcca aacagatgtc
ctccgcagtg cagccaaatt ttacagaaga 900aaaggtgacc tagacaaagc
tattgaactg tttcaacggg tgttggaatc cacaccaaac 960aatggctacc
tctatcacca gattgggtgc tgctacaagg caaaagtaag acaaatgcag
1020aatacaggag aatctgaagc tagtggaaat aaagagatga ttgaagcact
aaagcaatat 1080gctatggact attcgaataa agctcttgag aagggactga
atcctctgaa tgcatactcc 1140gatctcgctg agttcctgga gacggaatgt
tatcagacac cattcaataa ggaagtccct 1200gatgctgaaa agcaacaatc
ccatcagcgc tactgcaacc ttcagaaata taatgggaag 1260tctgaagaca
ctgctgtgca acatggttta gagggtttgt ccataagcaa aaaatcaact
1320gacaaggaag agatcaaaga ccaaccacag aatgtatctg aaaatctgct
tccacaaaat 1380gcaccaaatt attggtatct tcaaggatta attcataagc
agaatggaga tctgctgcaa 1440gcagccaaat gttatgagaa ggaactgggc
cgcctgctaa gggatgcccc ttcaggcata 1500ggcagtattt tcctgtcagc
atctgagctt gaggatggta gtgaggaaat gggccagggc 1560gcagtcagct
ccagtcccag agagctcctc tctaactcag agcaactgaa ctgagacaga
1620ggaggaaaac agagcatcag aagcctgcag tggtggttgt gacgggtagg
acgataggaa 1680gacagggggc cccaacctgg gattgctgag cagggaagct
ttgcatgttg ctctaaggta 1740catttttaaa gagttgtttt ttggccgggc
gcagtggctc atgcctgtaa tcccagcact 1800ttgggaggcc gaggtgggcg
gatcacgagg tctggagttt gagaccatcc tggctaacac 1860agtgaaatcc
cgtctctact aaaaatacaa aaaattagcc aggcgtggtg gctggcacct
1920gtagtcccag ctacttggga ggctgaggca ggagaatggc gtgaacctgg
aaggaagagg 1980ttgcagtgag ccaagattgc gcccctgcac tccagcctgg
gcaacagagc aagactccat 2040ctcaaaaaaa aaaaaaaaaa aaaaaaagag
ttgttttctc atgttcatta tagttcatta 2100cagttacata gtccgaaggt
cttacaacta atcactggta gcaataaatg cttcaggccc 2160acatgatgct
gattagttct cagttttcat tcagttcaca atataaccac cattcctgcc
2220ctccctgcca agggtcataa atggtgactg cctaacaaca aaatttgcag
tctcatctca 2280ttttcatcca gacttctgga actcaaagat taacttttga
ctaaccctgg aatatctctt 2340atctcactta tagcttcagg catgtattta
tatgtattct tgatagcaat accataatca 2400atgtgtattc ctgatagtaa
tgctacaata aatccaaaca tttcaactct gttaaaaaaa 2460aaaa
24648490PRTHomo sapiens 8Met Ser Glu Val Thr Lys Asn Ser Leu Glu
Lys Ile Leu Pro Gln Leu1 5 10 15Lys Cys His Phe Thr Trp Asn Leu Phe
Lys Glu Asp Ser Val Ser Arg 20 25 30Asp Leu Glu Asp Arg Val Cys Asn
Gln Ile Glu Phe Leu Asn Thr Glu 35 40 45Phe Lys Ala Thr Met Tyr Asn
Leu Leu Ala Tyr Ile Lys His Leu Asp 50 55 60Gly Asn Asn Glu Ala Ala
Leu Glu Cys Leu Arg Gln Ala Glu Glu Leu65 70 75 80Ile Gln Gln Glu
His Ala Asp Gln Ala Glu Ile Arg Ser Leu Val Thr 85 90 95Trp Gly Asn
Tyr Ala Trp Val Tyr Tyr His Leu Gly Arg Leu Ser Asp 100 105 110Ala
Gln Ile Tyr Val Asp Lys Val Lys Gln Thr Cys Lys Lys Phe Ser 115 120
125Asn Pro Tyr Ser Ile Glu Tyr Ser Glu Leu Asp Cys Glu Glu Gly Trp
130 135 140Thr Gln Leu Lys Cys Gly Arg Asn Glu Arg Ala Lys Val Cys
Phe Glu145 150 155 160Lys Ala Leu Glu Glu Lys Pro Asn Asn Pro Glu
Phe Ser Ser Gly Leu 165 170 175Ala Ile Ala Met Tyr His Leu Asp Asn
His Pro Glu Lys Gln Phe Ser 180 185 190Thr Asp Val Leu Lys Gln Ala
Ile Glu Leu Ser Pro Asp Asn Gln Tyr 195 200 205Val Lys Val Leu Leu
Gly Leu Lys Leu Gln Lys Met Asn Lys Glu Ala 210 215 220Glu Gly Glu
Gln Phe Val Glu Glu Ala Leu Glu Lys Ser Pro Cys Gln225 230 235
240Thr Asp Val Leu Arg Ser Ala Ala Lys Phe Tyr Arg Arg Lys Gly Asp
245 250 255Leu Asp Lys Ala Ile Glu Leu Phe Gln Arg Val Leu Glu Ser
Thr Pro 260 265 270Asn Asn Gly Tyr Leu Tyr His Gln Ile Gly Cys Cys
Tyr Lys Ala Lys 275 280 285Val Arg Gln Met Gln Asn Thr Gly Glu Ser
Glu Ala Ser Gly Asn Lys 290 295 300Glu Met Ile Glu Ala Leu Lys Gln
Tyr Ala Met Asp Tyr Ser Asn Lys305 310 315 320Ala Leu Glu Lys Gly
Leu Asn Pro Leu Asn Ala Tyr Ser Asp Leu Ala 325 330 335Glu Phe Leu
Glu Thr Glu Cys Tyr Gln Thr Pro Phe Asn Lys Glu Val 340 345 350Pro
Asp Ala Glu Lys Gln Gln Ser His Gln Arg Tyr Cys Asn Leu Gln 355 360
365Lys Tyr Asn Gly Lys Ser Glu Asp Thr Ala Val Gln His Gly Leu Glu
370 375 380Gly Leu Ser Ile Ser Lys Lys Ser Thr Asp Lys Glu Glu Ile
Lys Asp385 390 395 400Gln Pro Gln Asn Val Ser Glu Asn Leu Leu Pro
Gln Asn Ala Pro Asn 405 410 415Tyr Trp Tyr Leu Gln Gly Leu Ile His
Lys Gln Asn Gly Asp Leu Leu 420 425 430Gln Ala Ala Lys Cys Tyr Glu
Lys Glu Leu Gly Arg Leu Leu Arg Asp 435 440 445Ala Pro Ser Gly Ile
Gly Ser Ile Phe Leu Ser Ala Ser Glu Leu Glu 450 455 460Asp Gly Ser
Glu Glu Met Gly Gln Gly Ala Val Ser Ser Ser Pro Arg465 470 475
480Glu Leu Leu Ser Asn Ser Glu Gln Leu Asn 485 49092552DNAHomo
sapiens 9attttcctcc tcccaacgat tttaaattag tttcactttc cagtttcctc
ttccttcccc 60taaaagcaat tactcaaaaa cggagaaaac atcagctgat gcgtgcccta
ctctcccacc 120cctttatata gttccttcag tatttacttg aggcagacag
gaagacttct gaagaacaaa 180tcagcctggt caccagcttt tcggaacagc
agagacacag agggcagtca tgagtgaggt 240caccaagaat tccctggaga
aaatccttcc acagctgaaa tgccatttca cctggaactt 300attcaaggaa
gacagtgtct caagggatct agaagataga gtgtgtaacc agattgaatt
360tttaaacact gagttcaaag ctacaatgta caacttgttg gcctacataa
aacacctaga 420tggtaacaac gaggcagccc tggaatgctt acggcaagct
gaagagttaa tccagcaaga 480acatgctgac caagcagaaa tcagaagtct
agtcacttgg ggaaactacg cctgggtcta 540ctatcacttg ggcagactct
cagatgctca gatttatgta gataaggtga aacaaacctg 600caagaaattt
tcaaatccat acagtattga gtattctgaa cttgactgtg aggaagggtg
660gacacaactg aagtgtggaa gaaatgaaag ggcgaaggtg tgttttgaga
aggctctgga 720agaaaagccc aacaacccag aattctcctc tggactggca
attgcgatgt accatctgga 780taatcaccca gagaaacagt tctctactga
tgttttgaag caggccattg agctgagtcc 840tgataaccaa tacgtcaagg
ttctcttggg cctgaaactg cagaagatga ataaagaagc 900tgaaggagag
cagtttgttg aagaagcctt ggaaaagtct ccttgccaaa cagatgtcct
960ccgcagtgca gccaaatttt acagaagaaa aggtgaccta gacaaagcta
ttgaactgtt 1020tcaacgggtg ttggaatcca caccaaacaa tggctacctc
tatcaccaga ttgggtgctg 1080ctacaaggca aaagtaagac aaatgcagaa
tacaggagaa tctgaagcta gtggaaataa 1140agagatgatt gaagcactaa
agcaatatgc tatggactat tcgaataaag ctcttgagaa 1200gggactgaat
cctctgaatg catactccga tctcgctgag ttcctggaga cggaatgtta
1260tcagacacca ttcaataagg aagtccctga tgctgaaaag caacaatccc
atcagcgcta 1320ctgcaacctt cagaaatata atgggaagtc tgaagacact
gctgtgcaac atggtttaga 1380gggtttgtcc ataagcaaaa aatcaactga
caaggaagag atcaaagacc aaccacagaa 1440tgtatctgaa aatctgcttc
cacaaaatgc accaaattat tggtatcttc aaggattaat 1500tcataagcag
aatggagatc tgctgcaagc agccaaatgt tatgagaagg aactgggccg
1560cctgctaagg gatgcccctt caggcatagg cagtattttc ctgtcagcat
ctgagcttga 1620ggatggtagt gaggaaatgg gccagggcgc agtcagctcc
agtcccagag agctcctctc 1680taactcagag caactgaact gagacagagg
aggaaaacag agcatcagaa gcctgcagtg 1740gtggttgtga cgggtaggac
gataggaaga cagggggccc caacctggga ttgctgagca 1800gggaagcttt
gcatgttgct ctaaggtaca tttttaaaga gttgtttttt ggccgggcgc
1860agtggctcat gcctgtaatc ccagcacttt gggaggccga ggtgggcgga
tcacgaggtc 1920tggagtttga gaccatcctg gctaacacag tgaaatcccg
tctctactaa aaatacaaaa 1980aattagccag gcgtggtggc tggcacctgt
agtcccagct acttgggagg ctgaggcagg 2040agaatggcgt gaacctggaa
ggaagaggtt gcagtgagcc aagattgcgc ccctgcactc 2100cagcctgggc
aacagagcaa gactccatct caaaaaaaaa aaaaaaaaaa aaaaagagtt
2160gttttctcat gttcattata gttcattaca gttacatagt ccgaaggtct
tacaactaat 2220cactggtagc aataaatgct tcaggcccac atgatgctga
ttagttctca gttttcattc 2280agttcacaat ataaccacca ttcctgccct
ccctgccaag ggtcataaat ggtgactgcc 2340taacaacaaa atttgcagtc
tcatctcatt ttcatccaga cttctggaac tcaaagatta 2400acttttgact
aaccctggaa tatctcttat ctcacttata gcttcaggca tgtatttata
2460tgtattcttg atagcaatac cataatcaat gtgtattcct gatagtaatg
ctacaataaa 2520tccaaacatt tcaactctgt taaaaaaaaa aa 255210490PRTHomo
sapiens 10Met Ser Glu Val Thr Lys Asn Ser Leu Glu Lys Ile Leu Pro
Gln Leu1 5 10 15Lys Cys His Phe Thr Trp Asn Leu Phe Lys Glu Asp Ser
Val Ser Arg 20 25 30Asp Leu Glu Asp Arg Val Cys Asn Gln Ile Glu Phe
Leu Asn Thr Glu 35 40 45Phe Lys Ala Thr Met Tyr Asn Leu Leu Ala Tyr
Ile Lys His Leu Asp 50 55 60Gly Asn Asn Glu Ala Ala Leu Glu Cys Leu
Arg Gln Ala Glu Glu Leu65 70 75 80Ile Gln Gln Glu His Ala Asp Gln
Ala Glu Ile Arg Ser Leu Val Thr 85 90 95Trp Gly Asn Tyr Ala Trp Val
Tyr Tyr His Leu Gly Arg Leu Ser Asp 100 105 110Ala Gln Ile Tyr Val
Asp Lys Val Lys Gln Thr Cys Lys Lys Phe Ser 115 120 125Asn Pro Tyr
Ser Ile Glu Tyr Ser Glu Leu Asp Cys Glu Glu Gly Trp 130 135 140Thr
Gln Leu Lys Cys Gly Arg Asn Glu Arg Ala Lys Val Cys Phe Glu145 150
155 160Lys Ala Leu Glu Glu Lys Pro Asn Asn Pro Glu Phe Ser Ser Gly
Leu 165 170 175Ala Ile Ala Met Tyr His Leu Asp Asn His Pro Glu Lys
Gln Phe Ser 180 185 190Thr Asp Val Leu Lys Gln Ala Ile Glu Leu Ser
Pro Asp Asn Gln Tyr 195 200 205Val Lys Val Leu Leu Gly Leu Lys Leu
Gln Lys Met Asn Lys Glu Ala 210 215 220Glu Gly Glu Gln Phe Val Glu
Glu Ala Leu Glu Lys Ser Pro Cys Gln225 230 235 240Thr Asp Val Leu
Arg Ser Ala Ala Lys Phe Tyr Arg Arg Lys Gly Asp 245 250 255Leu Asp
Lys Ala Ile Glu Leu Phe Gln Arg Val Leu Glu Ser Thr Pro 260 265
270Asn Asn Gly Tyr Leu Tyr His Gln Ile Gly Cys Cys Tyr Lys Ala Lys
275 280 285Val Arg Gln Met Gln Asn Thr Gly Glu Ser Glu Ala Ser Gly
Asn Lys 290 295 300Glu Met Ile Glu Ala Leu Lys Gln Tyr Ala Met Asp
Tyr Ser Asn Lys305 310 315 320Ala Leu Glu Lys Gly Leu Asn Pro Leu
Asn Ala Tyr Ser Asp Leu Ala 325 330 335Glu Phe Leu Glu Thr Glu Cys
Tyr Gln Thr Pro Phe Asn Lys Glu Val 340 345 350Pro Asp Ala Glu Lys
Gln Gln Ser His Gln Arg Tyr Cys Asn Leu Gln 355 360 365Lys Tyr Asn
Gly Lys Ser Glu Asp Thr Ala Val Gln His Gly Leu Glu 370 375 380Gly
Leu Ser Ile Ser Lys Lys Ser Thr Asp Lys Glu Glu Ile Lys Asp385 390
395 400Gln Pro Gln Asn Val Ser Glu Asn Leu Leu Pro Gln Asn Ala Pro
Asn 405 410 415Tyr Trp Tyr Leu Gln Gly Leu Ile His Lys Gln Asn Gly
Asp Leu Leu 420 425 430Gln Ala Ala Lys Cys Tyr Glu Lys Glu Leu Gly
Arg Leu Leu Arg Asp 435 440 445Ala Pro Ser Gly Ile Gly Ser Ile Phe
Leu Ser Ala Ser Glu Leu Glu 450 455 460Asp Gly Ser Glu Glu Met Gly
Gln Gly Ala Val Ser Ser Ser Pro Arg465 470 475 480Glu Leu Leu Ser
Asn Ser Glu Gln Leu Asn 485 490114034DNAHomo sapiens 11agtttctgag
cgctcggcat ctgattcaat ctccagtttc ctgttcttgc tggggctggg 60gtctctcctt
taacaaagac acgccgcgcg gccgagtcca ggggctgcag aggcctggcg
120cgcgcacgcg cacgcgcacg cccaccgcgc ggcttcccgc ggtccccggt
gctgaggaga 180gagcgatccg agggactgcg ccgcccggac ggcctgcaga
gcgctgccat catgagtgaa 240attcgtaagg acaccttgaa ggccattctg
ttggagttag aatgtcattt tacatggaat 300ttacttaagg aagacattga
tctgtttgag gtagaagata caattgggca acagcttgaa 360tttcttacca
caaaatctag acttgctctt tataacctat tggcctatgt gaaacaccta
420aaaggccaaa ataaagacgc ccttgagtgc ttggaacaag cagaagaaat
aatccagcaa 480gaacactcag acaaagaaga agtacgaagc ctggtcactt
ggggaaacta tgcctgggtg 540tattatcaca tggaccagct tgaagaagct
cagaagtata caggtaagat agggaatgtc 600tgtaagaaat tgtccagtcc
ttctaactac aagttggagt gtcctgagac tgactgtgag 660aaaggctggg
cactcttgaa atttggagga aagtattatc aaaaggctaa agcggctttt
720gagaaggctc tggaagtgga gcctgacaat ccagaattta acatcggcta
tgctatcaca 780gtgtatcggc tggatgattc tgatagagaa gggtctgtaa
agagcttttc tctggggcct 840ttgagaaagg ctgttaccct gaacccagat
aacagctata ttaaggtttt tctggcactg 900aagcttcaag atgtacatgc
agaagctgaa ggggaaaagt atattgaaga aatcctggac 960caaatatcat
cccagcctta cgtccttcgt tatgcagcca agttctatag gagaaaaaat
1020tcctggaaca aagctctcga acttttaaaa aaggccttgg aggtgacacc
aacttcttct 1080ttcctgcatc accagatggg actttgctac agggcacaaa
tgatccaaat caagaaggcc 1140acacacaaca gacctaaagg aaaggataaa
ctaaaggttg atgagctgat ttcatctgct 1200atatttcatt tcaaagcagc
catggaacga gactctatgt ttgcatttgc ctacacagac 1260ctggccaaca
tgtacgctga aggaggccag tatagcaatg ctgaggacat tttccggaaa
1320gctcttcgtc tggagaacat aaccgatgat cacaaacatc agatccatta
ccactatggc 1380cgctttcagg aatttcaccg taaatcagaa aatactgcca
tccatcatta tttagaagcc 1440ttaaaggtca aagacagatc accccttcgc
accaaactga caagtgctct gaagaaattg 1500tctaccaaga gactttgtca
caatgcttta gatgtgcaga gtttaagtgc cctagggttt 1560gtttacaagc
tggaaggaga aaagaggcaa gctgctgagt actatgagaa ggcacaaaag
1620atagatccag aaaatgcaga attcctgact gctctctgtg agctccgact
ttccatttaa 1680atacatactc taggaaatta gctctaagtt tttcccttca
ttttgggttc tcctgtttgt 1740ttttttttta ttattttaat cccttgttta
ttatagagct aatatttatt gaatagttat 1800tgtgtaccaa gcattgtgct
aaatacttta tatgcattat gatgaatctt gtgcggtttt 1860ctttcttttt
ttctttttaa ttaaaatact ataatccatt gagaaatagc aatattctag
1920ctattgtaac ttctaaaaat ggtatggcca ttagatctgt gctttttatc
tctgctcttt 1980gaatttctca tattatatag taaatatatt cctacgtaaa
cctttgatac ctagatcagg 2040aatactcttc caggagtaca aaattacatt
attgatagtt aagctcttaa ttgtgtagct 2100tgcaaaagac agcacttttt
agttacagat gttttgactt tgatgaggat atttagctat 2160caatctaata
gtcacctaaa atatcttttt tgttggaaaa aagtttataa taaaaaagtt
2220tgtcatctct agtgacttca ataaagaaaa aactagaaga ggagaaaaag
gatttcctca 2280aattttaaat atgtaacttc agggattcaa tccccaaatg
tttattaagt agctagaaat 2340aattatgtgg aaaaaaatga ataatggaaa
atagtgagtc tcaaattgtt ctcttttttt 2400ttttaactaa aacaaatctg
caatgaatct agatgcaatt aattttattc cttccaacta 2460aaattacaat
atttttaggt taaaattatt gagatataaa gcagccattg ggaaattggg
2520agaaatgata aacaaatgga aaaagaagat gtccctaacc tacacccata
gattaccaag 2580gtttcagtgt actagttttg aatctgttct gaatggagtt
tttataccct caatttctgg 2640cctttggcta ttttagcatt tcaaagtgac
ttctatgaag cttttttttt aatgtgaaat 2700tttcagaatg ttgttttttt
catgtagata ctccaggaag agttaagcac tgctttcagt 2760tttaatatcc
accttgaggg gtcgctgctt gagggctctt atcccagggg actttttaat
2820tcggatgtta cttaatgtgg cttctctaat gtagtttctt tgattaccga
ctacacaatt 2880atgtaccatc acagtattag tggaaaagta ccatgtgatt
taattctcca ttcctccaat 2940gtaactctta aaattattat gtatgtgtgt
gtgttttact ttttgttttt tatcatcttt 3000aaaatttcta ttatggtttg
attattataa aaataatgaa ttctcactgt aaatttcaaa 3060aaaaaattac
aaaagtatgt gaatttaaaa atgagagcag tcctctcacc ctaccacagt
3120tccacaccct caaggtaaac ttataactta taatttgata tgtaaacttc
cagatctttt 3180ttctatgcgt aatcagacat acatatatac tgcagtgtat
ctcacgtatt aatttttaaa 3240aatcttttgt tttacttaat tctgttttta
ttattattat tattttgttt gatctattaa 3300ggaagaacaa ggaagggaat
gatctttact caagaatttc agaaagtcag cactgaagtc 3360ctgacctatc
agtagacaca tttgtccctt tcagatattt taggatattc tagcaaagca
3420ggccatttct cccacctgaa agtacataac ttctatcact tgccacataa
ttaaaagaac 3480tcacattaag cggttactca gacagttaat catagaaaag
attatttgct tcatcagttc 3540atagaaaaga ttatttgctt catcagttaa
cttgttttta taaatcaggg ctgtgttcat 3600acacagaagg ggcctgagat
ttctgcactt taaacaagct cctcctaggt gaggatgctg 3660tggctgttct
aattacattt tgagtagtaa ggtctacagc attgttcctc aaacttggct
3720acgtattgga atcacctaaa aagttaaaac aaaacatgga tgtctgggtc
ccgccccata 3780gagaatgact taattggcat ggggtgcagt ccaggcatca
tgatttttag atttcccagt 3840tggaacttgt gcagcaaagt ttgggagcta
ctgatggaca tgtgaaaagt aagtataaat 3900ggaataaaat taattaggct
aataggctta acccaggaaa tcctaagttc cttgaatatc 3960cagtttgcat
ttggactcct catcatatac ttggtatata atactctaat aaaagctgcc
4020tgagttgaat tgta 403412482PRTHomo sapiens 12Met Ser Glu Ile Arg
Lys Asp Thr Leu Lys Ala Ile Leu Leu Glu Leu1 5 10 15Glu Cys His Phe
Thr Trp Asn Leu Leu Lys Glu Asp Ile Asp Leu Phe 20 25 30Glu Val Glu
Asp Thr Ile Gly Gln Gln Leu Glu Phe Leu Thr Thr Lys 35 40 45Ser Arg
Leu Ala Leu Tyr Asn Leu Leu Ala Tyr Val Lys His Leu Lys 50 55 60Gly
Gln Asn Lys Asp Ala Leu Glu Cys Leu Glu Gln Ala Glu Glu Ile65 70 75
80Ile Gln Gln Glu His Ser Asp Lys Glu Glu Val Arg Ser Leu Val Thr
85 90 95Trp Gly Asn Tyr Ala Trp Val Tyr Tyr His Met Asp Gln Leu Glu
Glu 100 105 110Ala Gln Lys Tyr Thr Gly Lys Ile Gly Asn Val Cys Lys
Lys Leu Ser 115 120 125Ser Pro Ser Asn Tyr Lys Leu Glu Cys Pro Glu
Thr Asp Cys Glu Lys 130 135 140Gly Trp Ala Leu Leu Lys Phe Gly Gly
Lys Tyr Tyr Gln Lys Ala Lys145 150 155 160Ala Ala Phe Glu Lys Ala
Leu Glu Val Glu Pro Asp Asn Pro Glu Phe 165 170 175Asn Ile Gly Tyr
Ala Ile Thr Val Tyr Arg Leu Asp Asp Ser Asp Arg 180 185 190Glu Gly
Ser Val Lys Ser Phe Ser Leu Gly Pro Leu Arg Lys Ala Val 195 200
205Thr Leu Asn Pro Asp Asn Ser Tyr Ile Lys Val Phe Leu Ala Leu Lys
210 215 220Leu Gln Asp Val His Ala Glu Ala Glu Gly Glu Lys Tyr Ile
Glu Glu225 230 235 240Ile Leu Asp Gln Ile Ser Ser Gln Pro Tyr Val
Leu Arg Tyr Ala Ala 245 250 255Lys Phe Tyr Arg Arg Lys Asn Ser Trp
Asn Lys Ala Leu Glu Leu Leu 260 265 270Lys Lys Ala Leu Glu Val Thr
Pro Thr Ser Ser Phe Leu His His Gln 275 280 285Met Gly Leu Cys Tyr
Arg Ala Gln Met Ile Gln Ile Lys Lys Ala Thr 290 295 300His Asn Arg
Pro Lys Gly Lys Asp Lys Leu Lys Val Asp Glu Leu Ile305 310 315
320Ser Ser Ala Ile Phe His Phe Lys Ala Ala Met Glu Arg Asp Ser Met
325 330 335Phe Ala Phe Ala Tyr Thr Asp Leu Ala Asn Met Tyr Ala Glu
Gly Gly 340 345 350Gln Tyr Ser Asn Ala Glu Asp Ile Phe Arg Lys Ala
Leu Arg Leu Glu 355 360 365Asn Ile Thr Asp Asp His Lys His Gln Ile
His Tyr His Tyr Gly Arg 370 375 380Phe Gln Glu Phe His Arg Lys Ser
Glu Asn Thr Ala Ile His His Tyr385 390 395 400Leu Glu Ala Leu Lys
Val Lys Asp Arg Ser Pro Leu Arg Thr Lys Leu 405 410 415Thr Ser Ala
Leu Lys Lys Leu Ser Thr Lys Arg Leu Cys His Asn Ala 420 425 430Leu
Asp Val Gln Ser Leu Ser Ala Leu Gly Phe Val Tyr Lys Leu Glu 435 440
445Gly Glu Lys Arg Gln Ala Ala Glu Tyr Tyr Glu Lys Ala Gln Lys Ile
450 455 460Asp Pro Glu Asn Ala Glu Phe Leu Thr Ala Leu Cys Glu Leu
Arg Leu465 470 475 480Ser Ile131673DNAHomo sapiens 13tcccttctga
ggaaacgaaa ccaacagcag tccaagctca gtcagcagaa gagataaaag 60caaacaggtc
tgggaggcag ttctgttgcc actctctctc ctgtcaatga tggatctcag
120aaatacccca gccaaatctc tggacaagtt cattgaagac tatctcttgc
cagacacgtg 180tttccgcatg caaatcaacc atgccattga catcatctgt
gggttcctga aggaaaggtg 240cttccgaggt agctcctacc ctgtgtgtgt
gtccaaggtg gtaaagggtg gctcctcagg 300caagggcacc accctcagag
gccgatctga cgctgacctg gttgtcttcc tcagtcctct 360caccactttt
caggatcagt taaatcgccg gggagagttc atccaggaaa ttaggagaca
420gctggaagcc tgtcaaagag agagagcatt ttccgtgaag tttgaggtcc
aggctccacg 480ctggggcaac ccccgtgcgc tcagcttcgt actgagttcg
ctccagctcg gggagggggt 540ggagttcgat gtgctgcctg cctttgatgc
cctgggtcag ttgactggcg gctataaacc 600taacccccaa atctatgtca
agctcatcga ggagtgcacc gacctgcaga aagagggcga 660gttctccacc
tgcttcacag aactacagag agacttcctg aagcagcgcc ccaccaagct
720caagagcctc atccgcctag tcaagcactg gtaccaaaat tgtaagaaga
agcttgggaa 780gctgccacct cagtatgccc tggagctcct gacggtctat
gcttgggagc gagggagcat 840gaaaacacat ttcaacacag cccagggatt
tcggacggtc ttggaattag tcataaacta 900ccagcaactc tgcatctact
ggacaaagta ttatgacttt aaaaacccca ttattgaaaa 960gtacctgaga
aggcagctca cgaaacccag gcctgtgatc ctggacccgg cggaccctac
1020aggaaacttg ggtggtggag acccaaaggg ttggaggcag ctggcacaag
aggctgaggc 1080ctggctgaat tacccatgct ttaagaattg ggatgggtcc
ccagtgagct cctggattct 1140gctggctgaa agcaacagtg cagacgatga
gaccgacgat cccaggaggt atcagaaata 1200tggttacatt ggaacacatg
agtaccctca tttctctcat agacccagca cactccaggc 1260agcatccacc
ccacaggcag aagaggactg gacctgcacc atcctctgaa tgccagtgca
1320tcttggggga aagggctcca gtgttatctg gaccagttcc ttcattttca
ggtgggactc 1380ttgatccaga gaggacaaag ctcctcagtg agctggtgta
taatccagga cagaacccag 1440gtctcctgac tcctggcctt ctatgccctc
tatcctatca tagataacat tctccacagc 1500ctcacttcat tccacctatt
ctctgaaaat attccctgag agagaacaga gagatttaga 1560taagagaatg
aaattccagc cttgactttc ttctgtgcac ctgatgggag ggtaatgtct
1620aatgtattat caataacaat aaaaataaag caaataccat ttaaaaaaaa aaa
167314400PRTHomo sapiens 14Met Met Asp Leu Arg Asn Thr Pro Ala Lys
Ser Leu Asp Lys Phe Ile1 5 10 15Glu Asp Tyr Leu Leu Pro Asp Thr Cys
Phe Arg Met Gln Ile Asn His 20 25 30Ala Ile Asp Ile Ile Cys Gly Phe
Leu Lys Glu Arg Cys Phe Arg Gly 35 40 45Ser Ser Tyr Pro Val Cys Val
Ser Lys Val Val Lys Gly Gly Ser Ser 50 55 60Gly Lys Gly Thr Thr Leu
Arg Gly Arg Ser Asp Ala Asp Leu Val Val65 70 75 80Phe Leu Ser Pro
Leu Thr Thr Phe Gln Asp Gln Leu Asn Arg Arg Gly 85 90 95Glu Phe Ile
Gln Glu Ile Arg Arg Gln Leu Glu Ala Cys Gln Arg Glu 100 105 110Arg
Ala Phe Ser Val Lys Phe Glu Val Gln Ala Pro Arg Trp Gly Asn 115 120
125Pro Arg Ala Leu Ser Phe Val Leu Ser Ser Leu Gln Leu Gly Glu Gly
130 135 140Val Glu Phe Asp Val Leu Pro Ala Phe Asp Ala Leu Gly Gln
Leu Thr145 150 155 160Gly Gly Tyr Lys Pro Asn Pro Gln Ile Tyr Val
Lys Leu Ile Glu Glu 165 170 175Cys Thr Asp Leu Gln Lys Glu Gly Glu
Phe Ser Thr Cys Phe Thr Glu 180 185 190Leu Gln Arg Asp Phe Leu Lys
Gln Arg Pro Thr Lys Leu Lys Ser Leu 195 200 205Ile Arg Leu Val Lys
His Trp Tyr Gln Asn Cys Lys Lys Lys Leu Gly 210 215 220Lys Leu Pro
Pro Gln Tyr Ala Leu Glu Leu Leu Thr Val Tyr Ala Trp225 230 235
240Glu Arg Gly Ser Met Lys Thr His Phe Asn Thr Ala Gln Gly Phe Arg
245 250 255Thr Val Leu Glu Leu Val Ile Asn Tyr Gln Gln Leu Cys Ile
Tyr Trp 260 265 270Thr Lys Tyr Tyr Asp Phe Lys Asn Pro Ile Ile Glu
Lys Tyr Leu Arg 275 280 285Arg Gln Leu Thr Lys Pro Arg Pro Val Ile
Leu Asp Pro Ala Asp Pro 290 295 300Thr Gly Asn Leu Gly Gly Gly Asp
Pro Lys Gly Trp Arg Gln Leu Ala305 310 315 320Gln Glu Ala Glu Ala
Trp Leu Asn Tyr Pro Cys Phe Lys Asn Trp Asp 325 330 335Gly Ser Pro
Val Ser Ser Trp Ile Leu Leu Ala Glu Ser Asn Ser Ala 340 345 350Asp
Asp Glu Thr Asp Asp Pro Arg Arg Tyr Gln Lys Tyr Gly Tyr Ile 355 360
365Gly Thr His Glu Tyr Pro His Phe Ser His Arg Pro Ser Thr Leu Gln
370 375 380Ala Ala Ser Thr Pro Gln Ala Glu Glu Asp Trp Thr Cys Thr
Ile Leu385 390 395 400151470DNAHomo sapiens 15tcccttctga ggaaacgaaa
ccaacagcag tccaagctca gtcagcagaa gagataaaag 60caaacaggtc tgggaggcag
ttctgttgcc actctctctc ctgtcaatga tggatctcag 120aaatacccca
gccaaatctc tggacaagtt cattgaagac tatctcttgc cagacacgtg
180tttccgcatg caaatcaacc atgccattga catcatctgt gggttcctga
aggaaaggtg 240cttccgaggt agctcctacc ctgtgtgtgt gtccaaggtg
gtaaagggtg gctcctcagg 300caagggcacc accctcagag gccgatctga
cgctgacctg gttgtcttcc tcagtcctct 360caccactttt caggatcagt
taaatcgccg gggagagttc atccaggaaa ttaggagaca 420gctggaagcc
tgtcaaagag agagagcatt ttccgtgaag tttgaggtcc aggctccacg
480ctggggcaac ccccgtgcgc tcagcttcgt actgagttcg ctccagctcg
gggagggggt 540ggagttcgat gtgctgcctg cctttgatgc cctgggtcag
ttgactggcg gctataaacc 600taacccccaa atctatgtca agctcatcga
ggagtgcacc gacctgcaga aagagggcga 660gttctccacc tgcttcacag
aactacagag agacttcctg aagcagcgcc ccaccaagct 720caagagcctc
atccgcctag tcaagcactg gtaccaaaat tgtaagaaga agcttgggaa
780gctgccacct cagtatgccc tggagctcct gacggtctat gcttgggagc
gagggagcat 840gaaaacacat ttcaacacag cccagggatt tcggacggtc
ttggaattag tcataaacta 900ccagcaactc tgcatctact ggacaaagta
ttatgacttt aaaaacccca ttattgaaaa 960gtacctgaga aggcagctca
cgaaacccag gcctgtgatc ctggacccgg cggaccctac 1020aggaaacttg
ggtggtggag acccaaaggg ttggaggcag ctggcacaag aggctgaggc
1080ctggctgaat tacccatgct ttaagaattg ggatgggtcc ccagtgagct
cctggattct 1140gctggtgaga cctcctgctt cctccctgcc attcatccct
gcccctctcc atgaagcttg 1200agacatatag ctggagacca ttctttccaa
agaacttacc tcttgccaaa ggccatttat 1260attcatatag tgacaggctg
tgctccatat tttacagtca ttttggtcac aatcgagggt 1320ttctggaatt
ttcacatccc ttgtccagaa ttcattcccc taagagtaat aataaataat
1380ctctaacacc atttattgac tgtctgcttc gggctcaggt tctgtcctaa
gccctttaat 1440atgcactctc tcattaaata gtcacaacaa 147016364PRTHomo
sapiens 16Met Met Asp Leu Arg Asn Thr Pro Ala Lys Ser Leu Asp Lys
Phe Ile1 5 10 15Glu Asp Tyr Leu Leu Pro Asp Thr Cys Phe Arg Met Gln
Ile Asn His 20 25 30Ala Ile Asp Ile Ile Cys Gly Phe Leu Lys Glu Arg
Cys Phe Arg Gly 35 40 45Ser Ser Tyr Pro Val Cys Val Ser Lys Val Val
Lys Gly Gly Ser Ser 50 55 60Gly Lys Gly Thr Thr Leu Arg Gly Arg Ser
Asp Ala Asp Leu Val Val65 70 75 80Phe Leu Ser Pro Leu Thr Thr Phe
Gln Asp Gln Leu Asn Arg Arg Gly 85 90 95Glu Phe Ile Gln Glu Ile Arg
Arg Gln Leu Glu Ala Cys Gln Arg Glu 100 105 110Arg Ala Phe Ser Val
Lys Phe Glu Val Gln Ala Pro Arg Trp Gly Asn 115 120 125Pro Arg Ala
Leu Ser Phe Val Leu Ser Ser Leu Gln Leu Gly Glu Gly 130 135 140Val
Glu Phe Asp Val Leu Pro Ala Phe Asp Ala Leu Gly Gln Leu Thr145 150
155 160Gly Gly Tyr Lys Pro Asn Pro Gln Ile Tyr Val Lys Leu Ile Glu
Glu 165 170 175Cys Thr Asp Leu Gln Lys Glu Gly Glu Phe Ser Thr Cys
Phe Thr Glu 180 185 190Leu Gln Arg Asp Phe Leu Lys Gln Arg Pro Thr
Lys Leu Lys Ser Leu 195 200 205Ile Arg Leu Val Lys His Trp Tyr Gln
Asn Cys Lys Lys Lys Leu Gly 210 215 220Lys Leu Pro Pro Gln Tyr Ala
Leu Glu Leu Leu Thr Val Tyr Ala Trp225 230 235 240Glu Arg Gly Ser
Met Lys Thr His Phe Asn Thr Ala Gln Gly Phe Arg 245 250 255Thr Val
Leu Glu Leu Val Ile Asn Tyr Gln Gln Leu Cys Ile Tyr Trp 260 265
270Thr Lys Tyr Tyr Asp Phe Lys Asn Pro Ile Ile Glu Lys Tyr Leu Arg
275 280 285Arg Gln Leu Thr Lys Pro Arg Pro Val Ile Leu Asp Pro Ala
Asp Pro 290 295 300Thr Gly Asn Leu Gly Gly Gly Asp Pro Lys Gly Trp
Arg Gln Leu Ala305 310 315 320Gln Glu Ala Glu Ala Trp Leu Asn Tyr
Pro Cys Phe Lys Asn Trp Asp 325 330 335Gly Ser Pro Val Ser Ser Trp
Ile Leu Leu Val Arg Pro Pro Ala Ser 340 345 350Ser Leu Pro Phe Ile
Pro Ala Pro Leu His Glu Ala 355 360171575DNAHomo sapiens
17tcccttctga ggaaacgaaa ccaacagcag tccaagctca gtcagcagaa gagataaaag
60caaacaggtc tgggaggcag ttctgttgcc actctctctc ctgtcaatga tggatctcag
120aaatacccca gccaaatctc tggacaagtt cattgaagac tatctcttgc
cagacacgtg 180tttccgcatg caaatcaacc atgccattga catcatctgt
gggttcctga aggaaaggtg 240cttccgaggt agctcctacc ctgtgtgtgt
gtccaaggtg gtaaagggtg gctcctcagg 300caagggcacc accctcagag
gccgatctga cgctgacctg gttgtcttcc tcagtcctct 360caccactttt
caggatcagt taaatcgccg gggagagttc atccaggaaa ttaggagaca
420gctggaagcc tgtcaaagag agagagcatt ttccgtgaag tttgaggtcc
aggctccacg 480ctggggcaac ccccgtgcgc tcagcttcgt actgagttcg
ctccagctcg gggagggggt 540ggagttcgat gtgctgcctg cctttgatgc
cctgggtcag ttgactggcg gctataaacc 600taacccccaa atctatgtca
agctcatcga ggagtgcacc gacctgcaga aagagggcga 660gttctccacc
tgcttcacag aactacagag agacttcctg aagcagcgcc ccaccaagct
720caagagcctc atccgcctag tcaagcactg gtaccaaaat tgtaagaaga
agcttgggaa 780gctgccacct cagtatgccc tggagctcct gacggtctat
gcttgggagc gagggagcat 840gaaaacacat ttcaacacag cccagggatt
tcggacggtc ttggaattag tcataaacta 900ccagcaactc tgcatctact
ggacaaagta ttatgacttt aaaaacccca ttattgaaaa 960gtacctgaga
aggcagctca cgaaacccag gcctgtgatc ctggacccgg cggaccctac
1020aggaaacttg ggtggtggag acccaaaggg ttggaggcag ctggcacaag
aggctgaggc 1080ctggctgaat tacccatgct ttaagaattg ggatgggtcc
ccagtgagct cctggattct 1140gctgacccag cacactccag gcagcatcca
ccccacaggc agaagaggac tggacctgca 1200ccatcctctg aatgccagtg
catcttgggg gaaagggctc cagtgttatc tggaccagtt 1260ccttcatttt
caggtgggac tcttgatcca gagaggacaa agctcctcag tgagctggtg
1320tataatccag gacagaaccc aggtctcctg actcctggcc ttctatgccc
tctatcctat 1380catagataac attctccaca gcctcacttc attccaccta
ttctctgaaa atattccctg 1440agagagaaca gagagattta gataagagaa
tgaaattcca gccttgactt tcttctgtgc 1500acctgatggg agggtaatgt
ctaatgtatt atcaataaca ataaaaataa agcaaatacc 1560atttaaaaaa aaaaa
157518414PRTHomo sapiens 18Met Met Asp Leu Arg Asn Thr Pro Ala Lys
Ser Leu Asp Lys Phe Ile1 5 10 15Glu Asp Tyr Leu Leu Pro Asp Thr Cys
Phe Arg Met Gln Ile Asn His 20 25 30Ala Ile Asp Ile Ile Cys Gly Phe
Leu Lys Glu Arg Cys Phe Arg Gly 35 40 45Ser Ser Tyr Pro Val Cys Val
Ser Lys Val Val Lys Gly Gly Ser Ser 50 55 60Gly Lys Gly Thr Thr Leu
Arg Gly Arg Ser Asp Ala Asp Leu Val Val65 70 75 80Phe Leu Ser Pro
Leu Thr Thr Phe Gln Asp Gln Leu Asn Arg Arg Gly 85 90 95Glu Phe Ile
Gln Glu Ile Arg Arg Gln Leu Glu Ala Cys Gln Arg Glu 100 105 110Arg
Ala Phe Ser Val Lys Phe Glu Val Gln Ala Pro Arg Trp Gly Asn 115 120
125Pro Arg Ala Leu Ser Phe Val Leu Ser Ser Leu Gln Leu Gly Glu Gly
130 135 140Val Glu Phe Asp Val Leu Pro Ala Phe Asp Ala Leu Gly Gln
Leu Thr145 150 155 160Gly Gly Tyr Lys Pro Asn Pro Gln Ile Tyr Val
Lys Leu Ile Glu Glu 165 170 175Cys Thr Asp Leu Gln Lys Glu Gly Glu
Phe Ser Thr Cys Phe Thr Glu 180 185 190Leu Gln Arg Asp Phe Leu Lys
Gln Arg Pro Thr Lys Leu Lys Ser Leu 195 200 205Ile Arg Leu Val Lys
His Trp Tyr Gln Asn Cys Lys Lys Lys Leu Gly 210 215 220Lys Leu Pro
Pro Gln Tyr Ala Leu Glu Leu Leu Thr Val Tyr Ala Trp225 230 235
240Glu Arg Gly Ser Met Lys Thr His Phe Asn Thr Ala Gln Gly Phe Arg
245 250 255Thr Val Leu Glu Leu Val Ile Asn Tyr Gln Gln Leu Cys Ile
Tyr Trp 260 265 270Thr Lys Tyr Tyr Asp Phe Lys Asn Pro Ile Ile Glu
Lys Tyr Leu Arg 275 280 285Arg Gln Leu Thr Lys Pro Arg Pro Val Ile
Leu Asp Pro Ala Asp Pro 290 295 300Thr Gly Asn Leu Gly Gly Gly Asp
Pro Lys Gly Trp Arg Gln Leu Ala305
310 315 320Gln Glu Ala Glu Ala Trp Leu Asn Tyr Pro Cys Phe Lys Asn
Trp Asp 325 330 335Gly Ser Pro Val Ser Ser Trp Ile Leu Leu Thr Gln
His Thr Pro Gly 340 345 350Ser Ile His Pro Thr Gly Arg Arg Gly Leu
Asp Leu His His Pro Leu 355 360 365Asn Ala Ser Ala Ser Trp Gly Lys
Gly Leu Gln Cys Tyr Leu Asp Gln 370 375 380Phe Leu His Phe Gln Val
Gly Leu Leu Ile Gln Arg Gly Gln Ser Ser385 390 395 400Ser Val Ser
Trp Cys Ile Ile Gln Asp Arg Thr Gln Val Ser 405 410193539DNAHomo
sapiens 19caagagttgg taagctcgct gcagtgggtg gagagaggcc tctagacttc
agtttcagtt 60tcctggctct gggcagcagc aagaattcct ctgcctccca tcctaccatt
cactgtcttg 120ccggcagcca gctgagagca atgggaaatg gggagtccca
gctgtcctcg gtgcctgctc 180agaagctggg ttggtttatc caggaatacc
tgaagcccta cgaagaatgt cagacactga 240tcgacgagat ggtgaacacc
atctgtgacg tcctgcagga acccgaacag ttccccctgg 300tgcagggagt
ggccataggt ggctcctatg gacggaaaac agtcttaaga ggcaactccg
360atggtaccct tgtcctcttc ttcagtgact taaaacaatt ccaggatcag
aagagaagcc 420aacgtgacat cctcgataaa actggggata agctgaagtt
ctgtctgttc acgaagtggt 480tgaaaaacaa tttcgagatc cagaagtccc
ttgatgggtt caccatccag gtgttcacaa 540aaaatcagag aatctctttc
gaggtgctgg ccgccttcaa cgctctgagc ttaaatgata 600atcccagccc
ctggatctat cgagagctca aaagatcctt ggataagaca aatgccagtc
660ctggtgagtt tgcagtctgc ttcactgaac tccagcagaa gttttttgac
aaccgtcctg 720gaaaactaaa ggatttgatc ctcttgataa agcactggca
tcaacagtgc cagaaaaaaa 780tcaaggattt accctcgctg tctccgtatg
ccctggagct gcttacggtg tatgcctggg 840aacaggggtg cagaaaagac
aactttgaca ttgctgaagg cgtcagaacc gtactggagc 900tgatcaaatg
ccaggagaag ctgtgtatct attggatggt caactacaac tttgaagatg
960agaccatcag gaacatcctg ctgcaccagc tccaatcagc gaggccagta
atcttggatc 1020cagttgaccc aaccaataat gtgagtggag ataaaatatg
ctggcaatgg ctgaaaaaag 1080aagctcaaac ctggttgact tctcccaacc
tggataatga gttacctgca ccatcttgga 1140atgttctgcc tgcaccactc
ttcacgaccc caggccacct tctggataag ttcatcaagg 1200agtttctcca
gcccaacaaa tgcttcctag agcagattga cagtgctgtt aacatcatcc
1260gtacattcct taaagaaaac tgcttccgac aatcaacagc caagatccag
attgtccggg 1320gaggatcaac cgccaaaggc acagctctga agactggctc
tgatgccgat ctcgtcgtgt 1380tccataactc acttaaaagc tacacctccc
aaaaaaacga gcggcacaaa atcgtcaagg 1440aaatccatga acagctgaaa
gccttttgga gggagaagga ggaggagctt gaagtcagct 1500ttgagcctcc
caagtggaag gctcccaggg tgctgagctt ctctctgaaa tccaaagtcc
1560tcaacgaaag tgtcagcttt gatgtgcttc ctgcctttaa tgcactgggt
cagctgagtt 1620ctggctccac acccagcccc gaggtttatg cagggctcat
tgatctgtat aaatcctcgg 1680acctcccggg aggagagttt tctacctgtt
tcacagtcct gcagcgaaac ttcattcgct 1740cccggcccac caaactaaag
gatttaattc gcctggtgaa gcactggtac aaagagtgtg 1800aaaggaaact
gaagccaaag gggtctttgc ccccaaagta tgccttggag ctgctcacca
1860tctatgcctg ggagcagggg agtggagtgc cggattttga cactgcagaa
ggtttccgga 1920cagtcctgga gctggtcaca caatatcagc agctctgcat
cttctggaag gtcaattaca 1980actttgaaga tgagaccgtg aggaagtttc
tactgagcca gttgcagaaa accaggcctg 2040tgatcttgga cccagccgaa
cccacaggtg acgtgggtgg aggggaccgt tggtgttggc 2100atcttctggc
aaaagaagca aaggaatggt tatcctctcc ctgcttcaag gatgggactg
2160gaaacccaat accaccttgg aaagtgccga caatgcagac accaggaagt
tgtggagcta 2220ggatccatcc tattgtcaat gagatgttct catccagaag
ccatagaatc ctgaataata 2280attctaaaag aaacttctag agatcatctg
gcaatcgctt ttaaagactc ggctcaccgt 2340gagaaagagt cactcacatc
cattcttccc ttgatggtcc ctattcctcc ttcccttgct 2400tcttggactt
cttgaaatca atcaagactg caaacccttt cataaagtct tgccttgctg
2460aactccctct ctgcaggcag cctgccttta aaaatagttg ctgtcatcca
ctttatgtgc 2520atcttatttc tgtcaacttg tatttttttt cttgtatttt
tccaattagc tcctcctttt 2580tccttccagt ctaaaaaagg aatcctctgt
gtcttcaaag caaagctctt tactttcccc 2640ttggttctca taactctgtg
atcttgctct cggtgcttcc aactcatcca cgtcctgtct 2700gtttcctctg
tatacaaaac cctttctgcc cctgctgaca cagacatcct ctatgccagc
2760agccagccaa ccctttcatt agaacttcaa gctctccaaa ggctcagatt
ataactgttg 2820tcatatttat atgaggctgt tgtcttttcc ttctgagcct
gcctttctcc cccccaccca 2880ggagtatcct cttgccaaat caaaagactt
tttccttggg ctttagcctt aaagatactt 2940gaaggtctag gtgctttaac
ctcacatacc ctcacttaaa cttttatcac tgttgcatat 3000accagttgtg
atacaataaa gaatgtatct ggattttgtg cctagttcct agcacacagc
3060ttcaaaaatt ctagagtttc ctgataggag tgtcttttgt attcataaca
agcccttttc 3120acccatgcct gggtttatgc taacaaggtt acccatggtg
ggcccttagt ttcaaggaag 3180gagttggcca agccagaaag accaagcatg
tggttaaagc attggaattt tcagccccat 3240cccaccccca atctccaagg
aggtgatggg gctggaaatt gagttcaatt ttaacatggc 3300cagtgattta
agcaatgctg cctatgtaaa gaaaccccaa taaaaactct ggacagtgag
3360gcttggggag cttcctgatt ggcagacatt ccaatgtact aggaaggtag
cgcatcttga 3420ttccacaggg acaaaggctc ctgagctctg ggcccttcca
gtgcttgcca ccctacatac 3480tctttgtctg gctcttcatt tgtattcttt
ataataaaat ggtgattgta agtagagca 353920719PRTHomo sapiens 20Met Gly
Asn Gly Glu Ser Gln Leu Ser Ser Val Pro Ala Gln Lys Leu1 5 10 15Gly
Trp Phe Ile Gln Glu Tyr Leu Lys Pro Tyr Glu Glu Cys Gln Thr 20 25
30Leu Ile Asp Glu Met Val Asn Thr Ile Cys Asp Val Leu Gln Glu Pro
35 40 45Glu Gln Phe Pro Leu Val Gln Gly Val Ala Ile Gly Gly Ser Tyr
Gly 50 55 60Arg Lys Thr Val Leu Arg Gly Asn Ser Asp Gly Thr Leu Val
Leu Phe65 70 75 80Phe Ser Asp Leu Lys Gln Phe Gln Asp Gln Lys Arg
Ser Gln Arg Asp 85 90 95Ile Leu Asp Lys Thr Gly Asp Lys Leu Lys Phe
Cys Leu Phe Thr Lys 100 105 110Trp Leu Lys Asn Asn Phe Glu Ile Gln
Lys Ser Leu Asp Gly Phe Thr 115 120 125Ile Gln Val Phe Thr Lys Asn
Gln Arg Ile Ser Phe Glu Val Leu Ala 130 135 140Ala Phe Asn Ala Leu
Ser Leu Asn Asp Asn Pro Ser Pro Trp Ile Tyr145 150 155 160Arg Glu
Leu Lys Arg Ser Leu Asp Lys Thr Asn Ala Ser Pro Gly Glu 165 170
175Phe Ala Val Cys Phe Thr Glu Leu Gln Gln Lys Phe Phe Asp Asn Arg
180 185 190Pro Gly Lys Leu Lys Asp Leu Ile Leu Leu Ile Lys His Trp
His Gln 195 200 205Gln Cys Gln Lys Lys Ile Lys Asp Leu Pro Ser Leu
Ser Pro Tyr Ala 210 215 220Leu Glu Leu Leu Thr Val Tyr Ala Trp Glu
Gln Gly Cys Arg Lys Asp225 230 235 240Asn Phe Asp Ile Ala Glu Gly
Val Arg Thr Val Leu Glu Leu Ile Lys 245 250 255Cys Gln Glu Lys Leu
Cys Ile Tyr Trp Met Val Asn Tyr Asn Phe Glu 260 265 270Asp Glu Thr
Ile Arg Asn Ile Leu Leu His Gln Leu Gln Ser Ala Arg 275 280 285Pro
Val Ile Leu Asp Pro Val Asp Pro Thr Asn Asn Val Ser Gly Asp 290 295
300Lys Ile Cys Trp Gln Trp Leu Lys Lys Glu Ala Gln Thr Trp Leu
Thr305 310 315 320Ser Pro Asn Leu Asp Asn Glu Leu Pro Ala Pro Ser
Trp Asn Val Leu 325 330 335Pro Ala Pro Leu Phe Thr Thr Pro Gly His
Leu Leu Asp Lys Phe Ile 340 345 350Lys Glu Phe Leu Gln Pro Asn Lys
Cys Phe Leu Glu Gln Ile Asp Ser 355 360 365Ala Val Asn Ile Ile Arg
Thr Phe Leu Lys Glu Asn Cys Phe Arg Gln 370 375 380Ser Thr Ala Lys
Ile Gln Ile Val Arg Gly Gly Ser Thr Ala Lys Gly385 390 395 400Thr
Ala Leu Lys Thr Gly Ser Asp Ala Asp Leu Val Val Phe His Asn 405 410
415Ser Leu Lys Ser Tyr Thr Ser Gln Lys Asn Glu Arg His Lys Ile Val
420 425 430Lys Glu Ile His Glu Gln Leu Lys Ala Phe Trp Arg Glu Lys
Glu Glu 435 440 445Glu Leu Glu Val Ser Phe Glu Pro Pro Lys Trp Lys
Ala Pro Arg Val 450 455 460Leu Ser Phe Ser Leu Lys Ser Lys Val Leu
Asn Glu Ser Val Ser Phe465 470 475 480Asp Val Leu Pro Ala Phe Asn
Ala Leu Gly Gln Leu Ser Ser Gly Ser 485 490 495Thr Pro Ser Pro Glu
Val Tyr Ala Gly Leu Ile Asp Leu Tyr Lys Ser 500 505 510Ser Asp Leu
Pro Gly Gly Glu Phe Ser Thr Cys Phe Thr Val Leu Gln 515 520 525Arg
Asn Phe Ile Arg Ser Arg Pro Thr Lys Leu Lys Asp Leu Ile Arg 530 535
540Leu Val Lys His Trp Tyr Lys Glu Cys Glu Arg Lys Leu Lys Pro
Lys545 550 555 560Gly Ser Leu Pro Pro Lys Tyr Ala Leu Glu Leu Leu
Thr Ile Tyr Ala 565 570 575Trp Glu Gln Gly Ser Gly Val Pro Asp Phe
Asp Thr Ala Glu Gly Phe 580 585 590Arg Thr Val Leu Glu Leu Val Thr
Gln Tyr Gln Gln Leu Cys Ile Phe 595 600 605Trp Lys Val Asn Tyr Asn
Phe Glu Asp Glu Thr Val Arg Lys Phe Leu 610 615 620Leu Ser Gln Leu
Gln Lys Thr Arg Pro Val Ile Leu Asp Pro Ala Glu625 630 635 640Pro
Thr Gly Asp Val Gly Gly Gly Asp Arg Trp Cys Trp His Leu Leu 645 650
655Ala Lys Glu Ala Lys Glu Trp Leu Ser Ser Pro Cys Phe Lys Asp Gly
660 665 670Thr Gly Asn Pro Ile Pro Pro Trp Lys Val Pro Thr Met Gln
Thr Pro 675 680 685Gly Ser Cys Gly Ala Arg Ile His Pro Ile Val Asn
Glu Met Phe Ser 690 695 700Ser Arg Ser His Arg Ile Leu Asn Asn Asn
Ser Lys Arg Asn Phe705 710 715213647DNAHomo sapiens 21caagagttgg
taagctcgct gcagtgggtg gagagaggcc tctagacttc agtttcagtt 60tcctggctct
gggcagcagc aagaattcct ctgcctccca tcctaccatt cactgtcttg
120ccggcagcca gctgagagca atgggaaatg gggagtccca gctgtcctcg
gtgcctgctc 180agaagctggg ttggtttatc caggaatacc tgaagcccta
cgaagaatgt cagacactga 240tcgacgagat ggtgaacacc atctgtgacg
tcctgcagga acccgaacag ttccccctgg 300tgcagggagt ggccataggt
ggctcctatg gacggaaaac agtcttaaga ggcaactccg 360atggtaccct
tgtcctcttc ttcagtgact taaaacaatt ccaggatcag aagagaagcc
420aacgtgacat cctcgataaa actggggata agctgaagtt ctgtctgttc
acgaagtggt 480tgaaaaacaa tttcgagatc cagaagtccc ttgatgggtt
caccatccag gtgttcacaa 540aaaatcagag aatctctttc gaggtgctgg
ccgccttcaa cgctctgagc ttaaatgata 600atcccagccc ctggatctat
cgagagctca aaagatcctt ggataagaca aatgccagtc 660ctggtgagtt
tgcagtctgc ttcactgaac tccagcagaa gttttttgac aaccgtcctg
720gaaaactaaa ggatttgatc ctcttgataa agcactggca tcaacagtgc
cagaaaaaaa 780tcaaggattt accctcgctg tctccgtatg ccctggagct
gcttacggtg tatgcctggg 840aacaggggtg cagaaaagac aactttgaca
ttgctgaagg cgtcagaacc gtactggagc 900tgatcaaatg ccaggagaag
ctgtgtatct attggatggt caactacaac tttgaagatg 960agaccatcag
gaacatcctg ctgcaccagc tccaatcagc gaggccagta atcttggatc
1020cagttgaccc aaccaataat gtgagtggag ataaaatatg ctggcaatgg
ctgaaaaaag 1080aagctcaaac ctggttgact tctcccaacc tggataatga
gttacctgca ccatcttgga 1140atgttctgcc tgcaccactc ttcacgaccc
caggccacct tctggataag ttcatcaagg 1200agtttctcca gcccaacaaa
tgcttcctag agcagattga cagtgctgtt aacatcatcc 1260gtacattcct
taaagaaaac tgcttccgac aatcaacagc caagatccag attgtccggg
1320gaggatcaac cgccaaaggc acagctctga agactggctc tgatgccgat
ctcgtcgtgt 1380tccataactc acttaaaagc tacacctccc aaaaaaacga
gcggcacaaa atcgtcaagg 1440aaatccatga acagctgaaa gccttttgga
gggagaagga ggaggagctt gaagtcagct 1500ttgagcctcc caagtggaag
gctcccaggg tgctgagctt ctctctgaaa tccaaagtcc 1560tcaacgaaag
tgtcagcttt gatgtgcttc ctgcctttaa tgcactgggt cagctgagtt
1620ctggctccac acccagcccc gaggtttatg cagggctcat tgatctgtat
aaatcctcgg 1680acctcccggg aggagagttt tctacctgtt tcacagtcct
gcagcgaaac ttcattcgct 1740cccggcccac caaactaaag gatttaattc
gcctggtgaa gcactggtac aaagagtgtg 1800aaaggaaact gaagccaaag
gggtctttgc ccccaaagta tgccttggag ctgctcacca 1860tctatgcctg
ggagcagggg agtggagtgc cggattttga cactgcagaa ggtttccgga
1920cagtcctgga gctggtcaca caatatcagc agctctgcat cttctggaag
gtcaattaca 1980actttgaaga tgagaccgtg aggaagtttc tactgagcca
gttgcagaaa accaggcctg 2040tgatcttgga cccagccgaa cccacaggtg
acgtgggtgg aggggaccgt tggtgttggc 2100atcttctggc aaaagaagca
aaggaatggt tatcctctcc ctgcttcaag gatgggactg 2160gaaacccaat
accaccttgg aaagtgccgg taaaagtcat ctaaaggagg cgttgtctgg
2220aaatagccct gtaacaggct tgaatcaaag aacttctcct actgtagcaa
cctgaaatta 2280actcagacac aaataaagga aacccagctc acaggagctt
aaacagctgg tcagccccct 2340aagcccccac tacaagtgat cctcaggcag
gtaaccccag attcatgcac tgtagggtgc 2400tgcgcagcat ccctagtctc
tacccagtag atgccactag ccctcctctc ccagtgacaa 2460ccaaaagtct
tcagacattg tcaaacgttc ccctgggttc acagatcttt ctgcctttgg
2520cttttggctc caccctcttt agctgttaat ttgagtactt atggccctga
aagcggccac 2580ggtgcctcca gatggcaggt ttgcaatcca agcaggaaga
aggaaaagat acccaaaggt 2640caagaacaca gtgattttat tagaagtttc
atccgcaaat tttcttccat ttcattgctc 2700agaaatgtca tgtggctacc
tgtaacttga aggtggctac aaagatgact gtggacgtgg 2760gttgcactgg
ccacccaagg atgtctgcca cacctctcca aagccctccc tacctaccaa
2820gatatacctg atatattcca ccaggatatc ctccctccag atatacttgg
ttctctccac 2880caggttcttt ctttaaagca ggatttctca actttgatac
ttactcacat ttggggctag 2940acagttcttt gtttggaggc tctcttgtgc
attgtaggat gttgagcagc atctctggcc 3000tgtacccagt agatgccacc
cagttgtgac aattaaaagt gtcttgagac tttatcatgt 3060gtcttctgcc
ctaggtgaga acccttgcac tagaggaacc ctacacccca accctggggg
3120gaatgtaggg aagaggtggc caagccaacc gtggggttag ctctaattat
taagatatgc 3180attataaata aataccaaaa aattgtctct ggcaatagtt
accttcccag atacaggtcc 3240cccctttttt cccctaactc ttttaagcaa
tgattgtaac tattaggaga cattgctctc 3300ccacgtatgt ttttcttttt
agacaatgca gacaccagga agttgtggag ctaggatcca 3360tcctattgtc
aatgagatgt tctcatccag aagccataga atcctgaata ataattctaa
3420aagaaacttc tagagatcat ctggcaatcg cttttaaaga ctcggctcac
cgtgagaaag 3480agtcactcac atccattctt cccttgatgg tccctattcc
tccttccctt gcttcttgga 3540cttcttgaaa tcaatcaaga ctgcaaaccc
tttcataaag tcttgccttg ctgaactccc 3600tctctgcagg cagcctgcct
ttaaaaatag ttgctgtcat ccacttt 364722687PRTHomo sapiens 22Met Gly
Asn Gly Glu Ser Gln Leu Ser Ser Val Pro Ala Gln Lys Leu1 5 10 15Gly
Trp Phe Ile Gln Glu Tyr Leu Lys Pro Tyr Glu Glu Cys Gln Thr 20 25
30Leu Ile Asp Glu Met Val Asn Thr Ile Cys Asp Val Leu Gln Glu Pro
35 40 45Glu Gln Phe Pro Leu Val Gln Gly Val Ala Ile Gly Gly Ser Tyr
Gly 50 55 60Arg Lys Thr Val Leu Arg Gly Asn Ser Asp Gly Thr Leu Val
Leu Phe65 70 75 80Phe Ser Asp Leu Lys Gln Phe Gln Asp Gln Lys Arg
Ser Gln Arg Asp 85 90 95Ile Leu Asp Lys Thr Gly Asp Lys Leu Lys Phe
Cys Leu Phe Thr Lys 100 105 110Trp Leu Lys Asn Asn Phe Glu Ile Gln
Lys Ser Leu Asp Gly Phe Thr 115 120 125Ile Gln Val Phe Thr Lys Asn
Gln Arg Ile Ser Phe Glu Val Leu Ala 130 135 140Ala Phe Asn Ala Leu
Ser Leu Asn Asp Asn Pro Ser Pro Trp Ile Tyr145 150 155 160Arg Glu
Leu Lys Arg Ser Leu Asp Lys Thr Asn Ala Ser Pro Gly Glu 165 170
175Phe Ala Val Cys Phe Thr Glu Leu Gln Gln Lys Phe Phe Asp Asn Arg
180 185 190Pro Gly Lys Leu Lys Asp Leu Ile Leu Leu Ile Lys His Trp
His Gln 195 200 205Gln Cys Gln Lys Lys Ile Lys Asp Leu Pro Ser Leu
Ser Pro Tyr Ala 210 215 220Leu Glu Leu Leu Thr Val Tyr Ala Trp Glu
Gln Gly Cys Arg Lys Asp225 230 235 240Asn Phe Asp Ile Ala Glu Gly
Val Arg Thr Val Leu Glu Leu Ile Lys 245 250 255Cys Gln Glu Lys Leu
Cys Ile Tyr Trp Met Val Asn Tyr Asn Phe Glu 260 265 270Asp Glu Thr
Ile Arg Asn Ile Leu Leu His Gln Leu Gln Ser Ala Arg 275 280 285Pro
Val Ile Leu Asp Pro Val Asp Pro Thr Asn Asn Val Ser Gly Asp 290 295
300Lys Ile Cys Trp Gln Trp Leu Lys Lys Glu Ala Gln Thr Trp Leu
Thr305 310 315 320Ser Pro Asn Leu Asp Asn Glu Leu Pro Ala Pro Ser
Trp Asn Val Leu 325 330 335Pro Ala Pro Leu Phe Thr Thr Pro Gly His
Leu Leu Asp Lys Phe Ile 340 345 350Lys Glu Phe Leu Gln Pro Asn Lys
Cys Phe Leu Glu Gln Ile Asp Ser 355 360 365Ala Val Asn Ile Ile Arg
Thr Phe Leu Lys Glu Asn Cys Phe Arg Gln 370 375 380Ser Thr Ala Lys
Ile Gln Ile Val Arg Gly Gly Ser Thr Ala Lys Gly385 390 395 400Thr
Ala Leu Lys Thr Gly Ser Asp Ala Asp Leu Val Val Phe His Asn 405 410
415Ser Leu Lys Ser Tyr Thr Ser Gln Lys Asn Glu Arg His Lys Ile Val
420 425 430Lys Glu Ile His Glu Gln Leu Lys Ala Phe Trp Arg Glu Lys
Glu Glu 435 440 445Glu Leu Glu Val Ser Phe Glu Pro Pro Lys Trp Lys
Ala Pro Arg Val 450 455
460Leu Ser Phe Ser Leu Lys Ser Lys Val Leu Asn Glu Ser Val Ser
Phe465 470 475 480Asp Val Leu Pro Ala Phe Asn Ala Leu Gly Gln Leu
Ser Ser Gly Ser 485 490 495Thr Pro Ser Pro Glu Val Tyr Ala Gly Leu
Ile Asp Leu Tyr Lys Ser 500 505 510Ser Asp Leu Pro Gly Gly Glu Phe
Ser Thr Cys Phe Thr Val Leu Gln 515 520 525Arg Asn Phe Ile Arg Ser
Arg Pro Thr Lys Leu Lys Asp Leu Ile Arg 530 535 540Leu Val Lys His
Trp Tyr Lys Glu Cys Glu Arg Lys Leu Lys Pro Lys545 550 555 560Gly
Ser Leu Pro Pro Lys Tyr Ala Leu Glu Leu Leu Thr Ile Tyr Ala 565 570
575Trp Glu Gln Gly Ser Gly Val Pro Asp Phe Asp Thr Ala Glu Gly Phe
580 585 590Arg Thr Val Leu Glu Leu Val Thr Gln Tyr Gln Gln Leu Cys
Ile Phe 595 600 605Trp Lys Val Asn Tyr Asn Phe Glu Asp Glu Thr Val
Arg Lys Phe Leu 610 615 620Leu Ser Gln Leu Gln Lys Thr Arg Pro Val
Ile Leu Asp Pro Ala Glu625 630 635 640Pro Thr Gly Asp Val Gly Gly
Gly Asp Arg Trp Cys Trp His Leu Leu 645 650 655Ala Lys Glu Ala Lys
Glu Trp Leu Ser Ser Pro Cys Phe Lys Asp Gly 660 665 670Thr Gly Asn
Pro Ile Pro Pro Trp Lys Val Pro Val Lys Val Ile 675 680
685232123DNAHomo sapiens 23caagagttgg taagctcgct gcagtgggtg
gagagaggcc tctagacttc agtttcagtt 60tcctggctct gggcagcagc aagaattcct
ctgcctccca tcctaccatt cactgtcttg 120ccggcagcca gctgagagca
atgggaaatg gggagtccca gctgtcctcg gtgcctgctc 180agaagctggg
ttggtttatc caggaatacc tgaagcccta cgaagaatgt cagacactga
240tcgacgagat ggtgaacacc atctgtgacg tcctgcagga acccgaacag
ttccccctgg 300tgcagggagt ggccataggt ggctcctatg gacggaaaac
agtcttaaga ggcaactccg 360atggtaccct tgtcctcttc ttcagtgact
taaaacaatt ccaggatcag aagagaagcc 420aacgtgacat cctcgataaa
actggggata agctgaagtt ctgtctgttc acgaagtggt 480tgaaaaacaa
tttcgagatc cagaagtccc ttgatgggtt caccatccag gtgttcacaa
540aaaatcagag aatctctttc gaggtgctgg ccgccttcaa cgctctgagt
aagcattgct 600gggtgtcagg agagaaaagc caaagaagcg ggtgccagac
agctctgtgc aacctctagg 660ccatgagtgg gatagatacc actgctgctt
taaaaaatgg gagaccatag accctcagga 720gagaagaatc ccttctaccc
tggactcgct ctcttctctg gaactaactt ctcccccata 780ccctgattgt
ctttggagaa aatgttctgg attctagaat ctaaggcaga gccttttaag
840ccatactgta cacataaatc acctggaacc ttgttaaaat gcagatcctg
actcaggagg 900tctgagttag agcccaggat ttcatatttc tagccagctc
catgatgagc tgctggtccg 960cagatcatgc ttgcaggttt tgaccagagt
cagtgttggt tagagtaaga ggatgaggca 1020gacatctggg aaaagtccag
ctggggcaag catttgaagt ctgccttcct accaggtcaa 1080aatcaaggca
acgaccttcc atagataact atcaaagctt gagggggtgc cttgaaccca
1140actcctaaat ccctaagacc tgcccacctc ttgtgtctcc tgtctcagca
aacattccca 1200cactcttgca tattgttaaa gtaacctctg cttaccaggc
ttctggttta ataaaagatg 1260gctagagtga ctccatctta aagcaagtag
ctaggcactc aaaaggaacc tacaggctta 1320atacttgggt ctgaaaatag
ccacagtcta agctgaccac caattataat tgcagaatat 1380ttaaggccat
acaaaacatc tcccactaag cctacaaaat gtccaggtgt cctaaaagtt
1440cagcccactt aaaggcagca ttaatgagca ggtttaggtt gaaggattaa
tggtcatcaa 1500taccactgtt aagaagaaaa ttcttggcca aattgaattt
aatggagttt aactgagcag 1560acaattcaca aatctagaag cctcctgagc
cagagtaggt tcagagagtc ttgaacacag 1620ccacgtggtg gaagaagatt
tatggacagg aaaaggaaaa tgatgtactg aaaatgaaag 1680tgaggtacag
aaacagccag actggttata gctcagcatt ggccttattt gaacgagatt
1740tgaacagttg gccacctttg attggccgaa actcagtgat tggcacaaga
gtaggttgca 1800gtctgtttac acatcctttt aggttatagt tcaccatgta
cagagaaatt ttaggccaaa 1860cttaaaatat gtaaggaggc agctttaggc
taaacttgat ttaacagcac caataccccc 1920tacctttagt gagcacatct
gcacattcca attttaatga cagctcctta gaatttctta 1980tcaacgaaga
cactaacaaa gaatggcgca ttcctccttc tcctttctga ggatgcccta
2040ccctgtaaca aagtcgtttc taataaattt gcttctttca ccataaaaaa
aaaaaaaaaa 2100aaaaaaaaaa aaaaaaaaaa aaa 212324172PRTHomo sapiens
24Met Gly Asn Gly Glu Ser Gln Leu Ser Ser Val Pro Ala Gln Lys Leu1
5 10 15Gly Trp Phe Ile Gln Glu Tyr Leu Lys Pro Tyr Glu Glu Cys Gln
Thr 20 25 30Leu Ile Asp Glu Met Val Asn Thr Ile Cys Asp Val Leu Gln
Glu Pro 35 40 45Glu Gln Phe Pro Leu Val Gln Gly Val Ala Ile Gly Gly
Ser Tyr Gly 50 55 60Arg Lys Thr Val Leu Arg Gly Asn Ser Asp Gly Thr
Leu Val Leu Phe65 70 75 80Phe Ser Asp Leu Lys Gln Phe Gln Asp Gln
Lys Arg Ser Gln Arg Asp 85 90 95Ile Leu Asp Lys Thr Gly Asp Lys Leu
Lys Phe Cys Leu Phe Thr Lys 100 105 110Trp Leu Lys Asn Asn Phe Glu
Ile Gln Lys Ser Leu Asp Gly Phe Thr 115 120 125Ile Gln Val Phe Thr
Lys Asn Gln Arg Ile Ser Phe Glu Val Leu Ala 130 135 140Ala Phe Asn
Ala Leu Ser Lys His Cys Trp Val Ser Gly Glu Lys Ser145 150 155
160Gln Arg Ser Gly Cys Gln Thr Ala Leu Cys Asn Leu 165
170256646DNAHomo sapiens 25gttcggagag ccgggcggga aaacgaaacc
agaaatccga aggccgcgcc agagccctgc 60ttccccttgc acctgcgccg ggcggccatg
gacttgtaca gcaccccggc cgctgcgctg 120gacaggttcg tggccagaag
gctgcagccg cggaaggagt tcgtagagaa ggcgcggcgc 180gctctgggcg
ccctggccgc tgccctgagg gagcgcgggg gccgcctcgg tgctgctgcc
240ccgcgggtgc tgaaaactgt caagggaggc tcctcgggcc ggggcacagc
tctcaagggt 300ggctgtgatt ctgaacttgt catcttcctc gactgcttca
agagctatgt ggaccagagg 360gcccgccgtg cagagatcct cagtgagatg
cgggcatcgc tggaatcctg gtggcagaac 420ccagtccctg gtctgagact
cacgtttcct gagcagagcg tgcctggggc cctgcagttc 480cgcctgacat
ccgtagatct tgaggactgg atggatgtta gcctggtgcc tgccttcaat
540gtcctgggtc aggccggctc cggcgtcaaa cccaagccac aagtctactc
taccctcctc 600aacagtggct gccaaggggg cgagcatgcg gcctgcttca
cagagctgcg gaggaacttt 660gtgaacattc gcccagccaa gttgaagaac
ctaatcttgc tggtgaagca ctggtaccac 720caggtgtgcc tacaggggtt
gtggaaggag acgctgcccc cggtctatgc cctggaattg 780ctgaccatct
tcgcctggga gcagggctgt aagaaggatg ctttcagcct agccgaaggc
840ctccgaactg tcctgggcct gatccaacag catcagcacc tgtgtgtttt
ctggactgtc 900aactatggct tcgaggaccc tgcagttggg cagttcttgc
agcggcagct taagagaccc 960aggcctgtga tcctggaccc agctgacccc
acatgggacc tggggaatgg ggcagcctgg 1020cactgggatt tgctagccca
ggaggcagca tcctgctatg accacccatg ctttctgagg 1080gggatggggg
acccagtgca gtcttggaag gggccgggcc ttccacgtgc tggatgctca
1140ggtttgggcc accccatcca gctagaccct aaccagaaga cccctgaaaa
cagcaagagc 1200ctcaatgctg tgtacccaag agcagggagc aaacctccct
catgcccagc tcctggcccc 1260actggggcag ccagcatcgt cccctctgtg
ccgggaatgg ccttggacct gtctcagatc 1320cccaccaagg agctggaccg
cttcatccag gaccacctga agccgagccc ccagttccag 1380gagcaggtga
aaaaggccat cgacatcatc ttgcgctgcc tccatgagaa ctgtgttcac
1440aaggcctcaa gagtcagtaa agggggctca tttggccggg gcacagacct
aagggatggc 1500tgtgatgttg aactcatcat cttcctcaac tgcttcacgg
actacaagga ccaggggccc 1560cgccgcgcag agatccttga tgagatgcga
gcgcagctag aatcctggtg gcaggaccag 1620gtgcccagcc tgagccttca
gtttcctgag cagaatgtgc ctgaggctct gcagttccag 1680ctggtgtcca
cagccctgaa gagctggacg gatgttagcc tgctgcctgc cttcgatgct
1740gtggggcagc tcagttctgg caccaaacca aatccccagg tctactcgag
gctcctcacc 1800agtggctgcc aggagggcga gcataaggcc tgcttcgcag
agctgcggag gaacttcatg 1860aacattcgcc ctgtcaagct gaagaacctg
attctgctgg tgaagcactg gtaccgccag 1920gttgcggctc agaacaaagg
aaaaggacca gcccctgcct ctctgccccc agcctatgcc 1980ctggagctcc
tcaccatctt tgcctgggag cagggctgca ggcaggattg tttcaacatg
2040gcccaaggct tccggacggt gctggggctc gtgcaacagc atcagcagct
ctgtgtctac 2100tggacggtca actatagcac tgaggaccca gccatgagaa
tgcaccttct tggccagctt 2160cgaaaaccca gacccctggt cctggacccc
gctgatccca cctggaacgt gggccacggt 2220agctgggagc tgttggccca
ggaagcagca gcgctgggga tgcaggcctg ctttctgagt 2280agagacggga
catctgtgca gccctgggat gtgatgccag ccctccttta ccaaacccca
2340gctggggacc ttgacaagtt catcagtgaa tttctccagc ccaaccgcca
gttcctggcc 2400caggtgaaca aggccgttga taccatctgt tcatttttga
aggaaaactg cttccggaat 2460tctcccatca aagtgatcaa ggtggtcaag
ggtggctctt cagccaaagg cacagctctg 2520cgaggccgct cagatgccga
cctcgtggtg ttcctcagct gcttcagcca gttcactgag 2580cagggcaaca
agcgggccga gatcatctcc gagatccgag cccagctgga ggcatgtcaa
2640caggagcggc agttcgaggt caagtttgaa gtctccaaat gggagaatcc
ccgcgtgctg 2700agcttctcac tgacatccca gacgatgctg gaccagagtg
tggactttga tgtgctgcca 2760gcctttgacg ccctaggcca gctggtctct
ggctccaggc ccagctctca agtctacgtc 2820gacctcatcc acagctacag
caatgcgggc gagtactcca cctgcttcac agagctacaa 2880cgggacttca
tcatctctcg ccctaccaag ctgaagagcc tgatccggct ggtgaagcac
2940tggtaccagc agtgtaccaa gatctccaag gggagaggct ccctaccccc
acagcacggg 3000ctggaactcc tgactgtgta tgcctgggag cagggcggga
aggactccca gttcaacatg 3060gctgagggct tccgcacggt cctggagctg
gtcacccagt accgccagct ctgtatctac 3120tggaccatca actacaacgc
caaggacaag actgttggag acttcctgaa acagcagctt 3180cagaagccca
ggcctatcat cctggatccg gctgacccga caggcaacct gggccacaat
3240gcccgctggg acctgctggc caaggaagct gcagcctgca catctgccct
gtgctgcatg 3300ggacggaatg gcatccccat ccagccatgg ccagtgaagg
ctgctgtgtg aagttgagaa 3360aatcagcggt cctactggat gaagagaaga
tggacaccag ccctcagcat gaggaaattc 3420agggtcccct accagatgag
agagattgtg tacatgtgtg tgtgagcaca tgtgtgcatg 3480tgtgtgcaca
cgtgtgcatg tgtgtgtttt agtgaatctg ctctcccagc tcacacactc
3540ccctgcctcc catggcttac acactaggat ccagactcca tggtttgaca
ccagcctgcg 3600tttgcagctt ctctgtcact tccatgactc tatcctcata
ccaccactgc tgcttcccac 3660ccagctgaga atgccccctc ctccctgact
cctctctgcc catgcaaatt agctcacatc 3720tttcctcctg ctgcaatcca
tcccttcctc ccattggcct ctccttgcca aatctaaata 3780gtttatatag
ggatggcaga gagttcccat ctcatctgtc agccacagtc atttggtact
3840ggctacctgg agccttatct tctgaagggt tttaaagaat ggccaattag
ctgagaagaa 3900ttatctaatc aattagtgat gtctgccatg gatgcagtag
aggaaagtgg tggtacaagt 3960gccatgattg attagcaatg tctgcactgg
atacggaaaa aagaaggtgc ttgcaggttt 4020acagtgtata tgtgggctat
tgaagagccc tctgagctcg gttgctagca ggagagcatg 4080cccatattgg
cttactttgt ctgccacaga cacagacaga gggagttggg acatgcatgc
4140tatggggacc ctcttgttgg acacctaatt ggatgcctct tcatgagagg
cctccttttc 4200ttcacctttt atgctgcact cctcccctag tttacacatc
ttgatgctgt ggctcagttt 4260gccttcctga atttttattg ggtccctgtt
ttctctccta acatgctgag attctgcatc 4320cccacagcct aaactgagcc
agtggccaaa caaccgtgct cagcctgttt ctctctgccc 4380tctagagcaa
ggcccaccag gtccatccag gaggctctcc tgacctcaag tccaacaaca
4440gtgtccacac tagtcaaggt tcagcccaga aaacagaaag cactctagga
atcttaggca 4500gaaagggatt ttatctaaat cactggaaag gctggaggag
cagaaggcag aggccaccac 4560tggactattg gtttcaatat tagaccactg
tagccgaatc agaggccaga gagcagccac 4620tgctactgct aatgccacca
ctacccctgc catcactgcc ccacatggac aaaactggag 4680tcgagaccta
ggttagattc ctgcaaccac aaacatccat cagggatggc cagctgccag
4740agctgcggga agacggatcc cacctccctt tcttagcaga atctaaatta
cagccagacc 4800tctggctgca gaggagtctg agacatgtat gattgaatgg
gtgccaagtg ccagggggcg 4860gagtccccag cagatgcatc ctggccatct
gttgcgtgga tgagggagtg ggtctatctc 4920agaggaagga acaggaaaca
aagaaaggaa gccactgaac atcccttctc tgctccacag 4980gagtgcctta
gacagcctga ctctccacaa accactgtta aaacttacct gctaggaatg
5040ctagattgaa tgggatggga agagccttcc ctcattattg tcattcttgg
agagaggtga 5100gcaaccaagg gaagctcctc tgattcacct agaacctgtt
ctctgccgtc tttggctcag 5160cctacagaga ctagagtagg tgaagggaca
gaggacaggg cttctaatac ctgtgccata 5220ttgacagcct ccatccctgt
cccccatctt ggtgctgaac caacgctaag ggcaccttct 5280tagactcacc
tcatcgatac tgcctggtaa tccaaagcta gaactctcag gaccccaaac
5340tccacctctt ggattggccc tggctgctgc cacacacata tccaagagct
cagggccagt 5400tctggtgggc agcagagacc tgctctgcca agttgtccag
cagcagagtg gccctggcct 5460gggcatcaca agccagtgat gctcctggga
agaccaggtg gcaggtcgca gttgggtacc 5520ttccattccc accacacaga
ctctgggcct ccccgcaaaa tggctccaga attagagtaa 5580ttatgagatg
gtgggaacca gagcaactca ggtgcatgat acaaggagag gttgtcatct
5640gggtagggca gagaggaggg cttgctcatc tgaacagggg tgtatttcat
tccaggccct 5700cagtctttgg caatggccac cctggtgttg gcatattggc
cccactgtaa cttttggggg 5760cttcccggtc tagccacacc ctcggatgga
aagacttgac tgcataaaga tgtcagttct 5820ccctgagttg attgataggc
ttaatggtca ccctaaaaac acccacatat gcttttcgat 5880ggaaccaggt
aagttgacgc taaagttctt atggaaaaat acacacgcaa tagctaggaa
5940aacacaggga aagaagagtt ctgagcaggg cctagtctta gccaatatta
aaacatacta 6000tgaagcctct gatacttaaa cagcatggcg ctggtacgta
aatagaccaa tgcagttagg 6060tggctctttc caagactctg gggaaaaaag
tagtaaaaag ctaaatgcaa tcaatcagca 6120attgaaagct aagtgagaga
gccagagggc ctccttggtg gtaaaagagg gttgcatttc 6180ttgcagccag
aaggcagaga aagtgaagac caagtccaga actgaatcct aagaaatgca
6240ggactgcaaa gaaattggtg tgtgtgtgtg tgtgtgtgtg tgtgtgtgtg
tttaattttt 6300aaaaagtttt tattgagata caagtcaata ccataaagct
ctcacccttc taaagtgtac 6360aattcagtgg tgtgagtata ttcataagat
ttatacttgg tgtctattca taagacttat 6420atccagcata ttcataacta
gagccatatc acagatgcat tcatcataat aattccagac 6480attttcatca
ccctaaaagg aaaccctgaa acccattagc agtcattccc cattcctcca
6540acccattctc tccctaatcc ctagaaacca ccaatctgct gtgtatttca
tctattgcca 6600acatttcata taaatggcat catacaaaaa aaaaaaaaaa aaaaaa
6646261087PRTHomo sapiens 26Met Asp Leu Tyr Ser Thr Pro Ala Ala Ala
Leu Asp Arg Phe Val Ala1 5 10 15Arg Arg Leu Gln Pro Arg Lys Glu Phe
Val Glu Lys Ala Arg Arg Ala 20 25 30Leu Gly Ala Leu Ala Ala Ala Leu
Arg Glu Arg Gly Gly Arg Leu Gly 35 40 45Ala Ala Ala Pro Arg Val Leu
Lys Thr Val Lys Gly Gly Ser Ser Gly 50 55 60Arg Gly Thr Ala Leu Lys
Gly Gly Cys Asp Ser Glu Leu Val Ile Phe65 70 75 80Leu Asp Cys Phe
Lys Ser Tyr Val Asp Gln Arg Ala Arg Arg Ala Glu 85 90 95Ile Leu Ser
Glu Met Arg Ala Ser Leu Glu Ser Trp Trp Gln Asn Pro 100 105 110Val
Pro Gly Leu Arg Leu Thr Phe Pro Glu Gln Ser Val Pro Gly Ala 115 120
125Leu Gln Phe Arg Leu Thr Ser Val Asp Leu Glu Asp Trp Met Asp Val
130 135 140Ser Leu Val Pro Ala Phe Asn Val Leu Gly Gln Ala Gly Ser
Gly Val145 150 155 160Lys Pro Lys Pro Gln Val Tyr Ser Thr Leu Leu
Asn Ser Gly Cys Gln 165 170 175Gly Gly Glu His Ala Ala Cys Phe Thr
Glu Leu Arg Arg Asn Phe Val 180 185 190Asn Ile Arg Pro Ala Lys Leu
Lys Asn Leu Ile Leu Leu Val Lys His 195 200 205Trp Tyr His Gln Val
Cys Leu Gln Gly Leu Trp Lys Glu Thr Leu Pro 210 215 220Pro Val Tyr
Ala Leu Glu Leu Leu Thr Ile Phe Ala Trp Glu Gln Gly225 230 235
240Cys Lys Lys Asp Ala Phe Ser Leu Ala Glu Gly Leu Arg Thr Val Leu
245 250 255Gly Leu Ile Gln Gln His Gln His Leu Cys Val Phe Trp Thr
Val Asn 260 265 270Tyr Gly Phe Glu Asp Pro Ala Val Gly Gln Phe Leu
Gln Arg Gln Leu 275 280 285Lys Arg Pro Arg Pro Val Ile Leu Asp Pro
Ala Asp Pro Thr Trp Asp 290 295 300Leu Gly Asn Gly Ala Ala Trp His
Trp Asp Leu Leu Ala Gln Glu Ala305 310 315 320Ala Ser Cys Tyr Asp
His Pro Cys Phe Leu Arg Gly Met Gly Asp Pro 325 330 335Val Gln Ser
Trp Lys Gly Pro Gly Leu Pro Arg Ala Gly Cys Ser Gly 340 345 350Leu
Gly His Pro Ile Gln Leu Asp Pro Asn Gln Lys Thr Pro Glu Asn 355 360
365Ser Lys Ser Leu Asn Ala Val Tyr Pro Arg Ala Gly Ser Lys Pro Pro
370 375 380Ser Cys Pro Ala Pro Gly Pro Thr Gly Ala Ala Ser Ile Val
Pro Ser385 390 395 400Val Pro Gly Met Ala Leu Asp Leu Ser Gln Ile
Pro Thr Lys Glu Leu 405 410 415Asp Arg Phe Ile Gln Asp His Leu Lys
Pro Ser Pro Gln Phe Gln Glu 420 425 430Gln Val Lys Lys Ala Ile Asp
Ile Ile Leu Arg Cys Leu His Glu Asn 435 440 445Cys Val His Lys Ala
Ser Arg Val Ser Lys Gly Gly Ser Phe Gly Arg 450 455 460Gly Thr Asp
Leu Arg Asp Gly Cys Asp Val Glu Leu Ile Ile Phe Leu465 470 475
480Asn Cys Phe Thr Asp Tyr Lys Asp Gln Gly Pro Arg Arg Ala Glu Ile
485 490 495Leu Asp Glu Met Arg Ala Gln Leu Glu Ser Trp Trp Gln Asp
Gln Val 500 505 510Pro Ser Leu Ser Leu Gln Phe Pro Glu Gln Asn Val
Pro Glu Ala Leu 515 520 525Gln Phe Gln Leu Val Ser Thr Ala Leu Lys
Ser Trp Thr Asp Val Ser 530 535 540Leu Leu Pro Ala Phe Asp Ala Val
Gly Gln Leu Ser Ser Gly Thr Lys545 550 555 560Pro Asn Pro Gln Val
Tyr Ser Arg Leu Leu Thr Ser Gly Cys Gln Glu 565 570 575Gly Glu His
Lys Ala Cys Phe Ala Glu Leu Arg Arg Asn Phe Met Asn 580 585 590Ile
Arg Pro Val Lys Leu Lys Asn Leu Ile Leu Leu Val Lys His Trp 595 600
605Tyr Arg Gln Val Ala Ala Gln Asn Lys Gly Lys Gly
Pro Ala Pro Ala 610 615 620Ser Leu Pro Pro Ala Tyr Ala Leu Glu Leu
Leu Thr Ile Phe Ala Trp625 630 635 640Glu Gln Gly Cys Arg Gln Asp
Cys Phe Asn Met Ala Gln Gly Phe Arg 645 650 655Thr Val Leu Gly Leu
Val Gln Gln His Gln Gln Leu Cys Val Tyr Trp 660 665 670Thr Val Asn
Tyr Ser Thr Glu Asp Pro Ala Met Arg Met His Leu Leu 675 680 685Gly
Gln Leu Arg Lys Pro Arg Pro Leu Val Leu Asp Pro Ala Asp Pro 690 695
700Thr Trp Asn Val Gly His Gly Ser Trp Glu Leu Leu Ala Gln Glu
Ala705 710 715 720Ala Ala Leu Gly Met Gln Ala Cys Phe Leu Ser Arg
Asp Gly Thr Ser 725 730 735Val Gln Pro Trp Asp Val Met Pro Ala Leu
Leu Tyr Gln Thr Pro Ala 740 745 750Gly Asp Leu Asp Lys Phe Ile Ser
Glu Phe Leu Gln Pro Asn Arg Gln 755 760 765Phe Leu Ala Gln Val Asn
Lys Ala Val Asp Thr Ile Cys Ser Phe Leu 770 775 780Lys Glu Asn Cys
Phe Arg Asn Ser Pro Ile Lys Val Ile Lys Val Val785 790 795 800Lys
Gly Gly Ser Ser Ala Lys Gly Thr Ala Leu Arg Gly Arg Ser Asp 805 810
815Ala Asp Leu Val Val Phe Leu Ser Cys Phe Ser Gln Phe Thr Glu Gln
820 825 830Gly Asn Lys Arg Ala Glu Ile Ile Ser Glu Ile Arg Ala Gln
Leu Glu 835 840 845Ala Cys Gln Gln Glu Arg Gln Phe Glu Val Lys Phe
Glu Val Ser Lys 850 855 860Trp Glu Asn Pro Arg Val Leu Ser Phe Ser
Leu Thr Ser Gln Thr Met865 870 875 880Leu Asp Gln Ser Val Asp Phe
Asp Val Leu Pro Ala Phe Asp Ala Leu 885 890 895Gly Gln Leu Val Ser
Gly Ser Arg Pro Ser Ser Gln Val Tyr Val Asp 900 905 910Leu Ile His
Ser Tyr Ser Asn Ala Gly Glu Tyr Ser Thr Cys Phe Thr 915 920 925Glu
Leu Gln Arg Asp Phe Ile Ile Ser Arg Pro Thr Lys Leu Lys Ser 930 935
940Leu Ile Arg Leu Val Lys His Trp Tyr Gln Gln Cys Thr Lys Ile
Ser945 950 955 960Lys Gly Arg Gly Ser Leu Pro Pro Gln His Gly Leu
Glu Leu Leu Thr 965 970 975Val Tyr Ala Trp Glu Gln Gly Gly Lys Asp
Ser Gln Phe Asn Met Ala 980 985 990Glu Gly Phe Arg Thr Val Leu Glu
Leu Val Thr Gln Tyr Arg Gln Leu 995 1000 1005Cys Ile Tyr Trp Thr
Ile Asn Tyr Asn Ala Lys Asp Lys Thr Val 1010 1015 1020Gly Asp Phe
Leu Lys Gln Gln Leu Gln Lys Pro Arg Pro Ile Ile 1025 1030 1035Leu
Asp Pro Ala Asp Pro Thr Gly Asn Leu Gly His Asn Ala Arg 1040 1045
1050Trp Asp Leu Leu Ala Lys Glu Ala Ala Ala Cys Thr Ser Ala Leu
1055 1060 1065Cys Cys Met Gly Arg Asn Gly Ile Pro Ile Gln Pro Trp
Pro Val 1070 1075 1080Lys Ala Ala Val 1085272103DNAHomo sapiens
27tgactagacg gccagcctgt taaggtggcc ccagatattc cagcctcagc ccagagtcct
60cctgtgcccc tactgcagca agggtgtctc caagaagggg gacctggagt cagcccgtca
120cacctggttt cctctctgct agggtccctc ctcccacaga gcactggagg
gcagctgagg 180aggagctacc ttaaaaaagg aggtgtgtgc cagggagctg
ggtaggagcc tggctatata 240tctgcccagc agcggtactc tcgggacaga
gatggcactg atgcaggaac tgtatagcac 300accagcctcc aggctggact
ccttcgtggc tcagtggctg cagccccacc gggagtggaa 360ggaagaggtg
ctagacgctg tgcggaccgt ggaggagttt ctgaggcagg agcatttcca
420ggggaagcgt gggctggacc aggatgtgcg ggtgctgaag gtagtcaagg
tgggctcctt 480cgggaatggc acggttctca ggagcaccag agaggtggag
ctggtggcgt ttctgagctg 540tttccacagc ttccaggagg cagccaagca
tcacaaagat gttctgaggc tgatatggaa 600aaccatgtgg caaagccagg
acctgctgga cctcgggctc gaggacctga ggatggagca 660gagagtcccc
gatgctctcg tcttcaccat ccagaccagg gggactgcgg agcccatcac
720ggtcaccatt gtgcctgcct acagagccct ggggccttct cttcccaact
cccagccacc 780ccctgaggtc tatgtgagcc tgatcaaggc ctgcggtggt
cctggaaatt tctgcccatc 840cttcagcgag ctgcagagaa atttcgtgaa
acatcggcca actaagctga agagcctcct 900gcgcctggtg aaacactggt
accagcagta tgtgaaagcc aggtccccca gagccaatct 960gccccctctc
tatgctcttg aacttctaac catctatgcc tgggaaatgg gtactgaaga
1020agacgagaat ttcatgttgg acgaaggctt caccactgtg atggacctgc
tcctggagta 1080tgaagtcatc tgtatctact ggaccaagta ctacacactc
cacaatgcaa tcattgagga 1140ttgtgtcaga aaacagctca aaaaagagag
gcccatcatc ctggatccgg ccgaccccac 1200cctcaacgtg gcagaagggt
acagatggga catcgttgct cagagggcct cccagtgcct 1260gaaacaggac
tgttgctatg acaacaggga gaaccccatc tccagctgga acgtgaagag
1320ggcacgagac atccacttga cagtggagca gaggggttac ccagatttca
acctcatcgt 1380gaacccttat gagcccataa ggaaggttaa agagaaaatc
cggaggacca ggggctactc 1440tggcctgcag cgtctgtcct tccaggttcc
tggcagtgag aggcagcttc tcagcagcag 1500gtgctcctta gccaaatatg
ggatcttctc ccacactcac atctatctgc tggagaccat 1560cccctccgag
atccaggtct tcgtgaagaa tcctgatggt gggagctacg cctatgccat
1620caaccccaac agcttcatcc tgggtctgaa gcagcagatt gaagaccagc
aggggcttcc 1680taaaaagcag cagcagctgg aattccaagg ccaagtcctg
caggactggt tgggtctggg 1740gatctatggc atccaagaca gtgacactct
catcctctcg aagaagaaag gagaggctct 1800gtttccagcc agttagtttt
ctctgggaga cttctctgta catttctgcc atgtactcca 1860gaactcatcc
tgtcaatcac tctgtcccat tgtctactgg gaaggtccca ggtcttcacc
1920agttttacaa tgagttatcc caggccagac gtggtagctc acacctgtaa
tcccagaact 1980ttgggaggcc gaggtgggag gagcgcttga gccgaggagt
tcaagaccag cctgggtatc 2040acagggagac cccgtctcta caaaataaaa
aaataattca ctgggaaaaa aaaaaaaaaa 2100aaa 210328514PRTHomo sapiens
28Met Ala Leu Met Gln Glu Leu Tyr Ser Thr Pro Ala Ser Arg Leu Asp1
5 10 15Ser Phe Val Ala Gln Trp Leu Gln Pro His Arg Glu Trp Lys Glu
Glu 20 25 30Val Leu Asp Ala Val Arg Thr Val Glu Glu Phe Leu Arg Gln
Glu His 35 40 45Phe Gln Gly Lys Arg Gly Leu Asp Gln Asp Val Arg Val
Leu Lys Val 50 55 60Val Lys Val Gly Ser Phe Gly Asn Gly Thr Val Leu
Arg Ser Thr Arg65 70 75 80Glu Val Glu Leu Val Ala Phe Leu Ser Cys
Phe His Ser Phe Gln Glu 85 90 95Ala Ala Lys His His Lys Asp Val Leu
Arg Leu Ile Trp Lys Thr Met 100 105 110Trp Gln Ser Gln Asp Leu Leu
Asp Leu Gly Leu Glu Asp Leu Arg Met 115 120 125Glu Gln Arg Val Pro
Asp Ala Leu Val Phe Thr Ile Gln Thr Arg Gly 130 135 140Thr Ala Glu
Pro Ile Thr Val Thr Ile Val Pro Ala Tyr Arg Ala Leu145 150 155
160Gly Pro Ser Leu Pro Asn Ser Gln Pro Pro Pro Glu Val Tyr Val Ser
165 170 175Leu Ile Lys Ala Cys Gly Gly Pro Gly Asn Phe Cys Pro Ser
Phe Ser 180 185 190Glu Leu Gln Arg Asn Phe Val Lys His Arg Pro Thr
Lys Leu Lys Ser 195 200 205Leu Leu Arg Leu Val Lys His Trp Tyr Gln
Gln Tyr Val Lys Ala Arg 210 215 220Ser Pro Arg Ala Asn Leu Pro Pro
Leu Tyr Ala Leu Glu Leu Leu Thr225 230 235 240Ile Tyr Ala Trp Glu
Met Gly Thr Glu Glu Asp Glu Asn Phe Met Leu 245 250 255Asp Glu Gly
Phe Thr Thr Val Met Asp Leu Leu Leu Glu Tyr Glu Val 260 265 270Ile
Cys Ile Tyr Trp Thr Lys Tyr Tyr Thr Leu His Asn Ala Ile Ile 275 280
285Glu Asp Cys Val Arg Lys Gln Leu Lys Lys Glu Arg Pro Ile Ile Leu
290 295 300Asp Pro Ala Asp Pro Thr Leu Asn Val Ala Glu Gly Tyr Arg
Trp Asp305 310 315 320Ile Val Ala Gln Arg Ala Ser Gln Cys Leu Lys
Gln Asp Cys Cys Tyr 325 330 335Asp Asn Arg Glu Asn Pro Ile Ser Ser
Trp Asn Val Lys Arg Ala Arg 340 345 350Asp Ile His Leu Thr Val Glu
Gln Arg Gly Tyr Pro Asp Phe Asn Leu 355 360 365Ile Val Asn Pro Tyr
Glu Pro Ile Arg Lys Val Lys Glu Lys Ile Arg 370 375 380Arg Thr Arg
Gly Tyr Ser Gly Leu Gln Arg Leu Ser Phe Gln Val Pro385 390 395
400Gly Ser Glu Arg Gln Leu Leu Ser Ser Arg Cys Ser Leu Ala Lys Tyr
405 410 415Gly Ile Phe Ser His Thr His Ile Tyr Leu Leu Glu Thr Ile
Pro Ser 420 425 430Glu Ile Gln Val Phe Val Lys Asn Pro Asp Gly Gly
Ser Tyr Ala Tyr 435 440 445Ala Ile Asn Pro Asn Ser Phe Ile Leu Gly
Leu Lys Gln Gln Ile Glu 450 455 460Asp Gln Gln Gly Leu Pro Lys Lys
Gln Gln Gln Leu Glu Phe Gln Gly465 470 475 480Gln Val Leu Gln Asp
Trp Leu Gly Leu Gly Ile Tyr Gly Ile Gln Asp 485 490 495Ser Asp Thr
Leu Ile Leu Ser Lys Lys Lys Gly Glu Ala Leu Phe Pro 500 505 510Ala
Ser291861DNAHomo sapiens 29tgactagacg gccagcctgt taaggtggcc
ccagatattc cagcctcagc ccagagtcct 60cctgtgcccc tactgcagca agggtgtctc
caagaagggg gacctggagt cagcccgtca 120cacctggttt cctctctgct
agggtccctc ctcccacaga gcactggagg gcagctgagg 180aggagctacc
ttaaaaaagg aggtgtgtgc cagggagctg ggtaggagcc tggctatata
240tctgcccagc agcggtactc tcgggacaga gatggcactg atgcaggaac
tgtatagcac 300accagcctcc aggctggact ccttcgtggc tcagtggctg
cagccccacc gggagtggaa 360ggaagaggtg ctagacgctg tgcggaccgt
ggaggagttt ctgaggcagg agcatttcca 420ggggaagcgt gggctggacc
aggatgtgcg ggtgctgaag gtagtcaagg tgggctcctt 480cgggaatggc
acggttctca ggagcaccag agaggtggag ctggtggcgt ttctgagctg
540tttccacagc ttccaggagg cagccaagca tcacaaagat gttctgaggc
tgatatggaa 600aaccatgtgg caaagccagg acctgctgga cctcgggctc
gaggacctga ggatggagca 660gagagtcccc gatgctctcg tcttcaccat
ccagaccagg gggactgcgg agcccatcac 720ggtcaccatt gtgcctgcct
acagagccct ggggccttct cttcccaact cccagccacc 780ccctgaggtc
tatgtgagcc tgatcaaggc ctgcggtggt cctggaaatt tctgcccatc
840cttcagcgag ctgcagagaa atttcgtgaa acatcggcca actaagctga
agagcctcct 900gcgcctggtg aaacactggt accagcaggc ccatcatcct
ggatccggcc gaccccaccc 960tcaacgtggc agaagggtac agatgggaca
tcgttgctca gagggcctcc cagtgcctga 1020aacaggactg ttgctatgac
aacagggaga accccatctc cagctggaac gtgaagaggg 1080cacgagacat
ccacttgaca gtggagcaga ggggttaccc agatttcaac ctcatcgtga
1140acccttatga gcccataagg aaggttaaag agaaaatccg gaggaccagg
ggctactctg 1200gcctgcagcg tctgtccttc caggttcctg gcagtgagag
gcagcttctc agcagcaggt 1260gctccttagc caaatatggg atcttctccc
acactcacat ctatctgctg gagaccatcc 1320cctccgagat ccaggtcttc
gtgaagaatc ctgatggtgg gagctacgcc tatgccatca 1380accccaacag
cttcatcctg ggtctgaagc agcagattga agaccagcag gggcttccta
1440aaaagcagca gcagctggaa ttccaaggcc aagtcctgca ggactggttg
ggtctgggga 1500tctatggcat ccaagacagt gacactctca tcctctcgaa
gaagaaagga gaggctctgt 1560ttccagccag ttagttttct ctgggagact
tctctgtaca tttctgccat gtactccaga 1620actcatcctg tcaatcactc
tgtcccattg tctactggga aggtcccagg tcttcaccag 1680ttttacaatg
agttatccca ggccagacgt ggtagctcac acctgtaatc ccagaacttt
1740gggaggccga ggtgggagga gcgcttgagc cgaggagttc aagaccagcc
tgggtatcac 1800agggagaccc cgtctctaca aaataaaaaa ataattcact
gggaaaaaaa aaaaaaaaaa 1860a 186130255PRTHomo sapiens 30Met Ala Leu
Met Gln Glu Leu Tyr Ser Thr Pro Ala Ser Arg Leu Asp1 5 10 15Ser Phe
Val Ala Gln Trp Leu Gln Pro His Arg Glu Trp Lys Glu Glu 20 25 30Val
Leu Asp Ala Val Arg Thr Val Glu Glu Phe Leu Arg Gln Glu His 35 40
45Phe Gln Gly Lys Arg Gly Leu Asp Gln Asp Val Arg Val Leu Lys Val
50 55 60Val Lys Val Gly Ser Phe Gly Asn Gly Thr Val Leu Arg Ser Thr
Arg65 70 75 80Glu Val Glu Leu Val Ala Phe Leu Ser Cys Phe His Ser
Phe Gln Glu 85 90 95Ala Ala Lys His His Lys Asp Val Leu Arg Leu Ile
Trp Lys Thr Met 100 105 110Trp Gln Ser Gln Asp Leu Leu Asp Leu Gly
Leu Glu Asp Leu Arg Met 115 120 125Glu Gln Arg Val Pro Asp Ala Leu
Val Phe Thr Ile Gln Thr Arg Gly 130 135 140Thr Ala Glu Pro Ile Thr
Val Thr Ile Val Pro Ala Tyr Arg Ala Leu145 150 155 160Gly Pro Ser
Leu Pro Asn Ser Gln Pro Pro Pro Glu Val Tyr Val Ser 165 170 175Leu
Ile Lys Ala Cys Gly Gly Pro Gly Asn Phe Cys Pro Ser Phe Ser 180 185
190Glu Leu Gln Arg Asn Phe Val Lys His Arg Pro Thr Lys Leu Lys Ser
195 200 205Leu Leu Arg Leu Val Lys His Trp Tyr Gln Gln Ala His His
Pro Gly 210 215 220Ser Gly Arg Pro His Pro Gln Arg Gly Arg Arg Val
Gln Met Gly His225 230 235 240Arg Cys Ser Glu Gly Leu Pro Val Pro
Glu Thr Gly Leu Leu Leu 245 250 255311240DNAHomo sapiens
31ttatgggttt catttagtgg agaaattggg tatgacctcg gaggagtcaa gaaagagttc
60ttctactgtc tgtttgcaga gatgatccag ccggaatatg ggatgttcat gtatcctgaa
120ggggcttcct gcatgtggtt tcctgtcaag cctaaatttg agaagaaaag
atacttcttt 180tttggggttc tatgtggact ttccctgttc aattgcaatg
ttgccaacct tcctttccca 240ctggcactgt ttaagaaact tttggaccaa
atgccatcat tggaagactt gaaagaactc 300agtcctgatt tgggaaagaa
tttgcaaaca cttctggatg atgaaggtga taactttgag 360gaagtatttt
acatccattt taatgtgcac tgggacagaa acgacacaaa cttaattcct
420aatggaagta gcataactgt caaccagact aacaagagag actatgtttc
taagtatatc 480aattacattt tcaacgactc tgtaaaggcg gtttatgaag
aatttcggag aggattttat 540aaaatgtgcg acgaagacat tatcaaatta
ttccaccccg aagaactgaa ggatgtgatt 600gttggaaata cagattatga
ttggaaaaca tttgaaaaga atgcacgtta tgaaccagga 660tataacagtt
cacatcccac catagtgatg ttttggaagg ctttccacaa attgactctg
720gaagaaaaga aaaaattcct tgtatttctt acaggaactg acagactaca
aatgaaagat 780ttaaataata tgaaaataac attttgctgt cctgaaagtt
ggaatgaaag agaccctata 840agagcactga catgtttcag tgtcctcttc
ctccctaaat attctacaat ggaaacagtt 900gaagaagcgc ttcaagaagc
catcaacaac aacagaggat ttggctgacc agcttgcttg 960tccaacagcc
ttattttgtt gttgttatcg ttgttgttgt tgttgttgtt gttgtttctc
1020tactttgttt tgttttaggc ttttagcagc ctgaagccat ggtttttcat
ttctgtctct 1080agtgataagc aggaaagagg gatgaagaag agggtttact
ggccggttag aacccgtgac 1140tgtattctct cccttggata cccctatgcc
tacatcatat tccttacctc ttttgggaaa 1200tatttttcaa aaataaaata
accgaaaaat taacataaaa 1240321024PRTHomo sapiens 32Met Glu Arg Arg
Ser Arg Arg Lys Ser Arg Arg Asn Gly Arg Ser Thr1 5 10 15Ala Gly Lys
Ala Ala Ala Thr Gln Pro Ala Lys Ser Pro Gly Ala Gln 20 25 30Leu Trp
Leu Phe Pro Ser Ala Ala Gly Leu His Arg Ala Leu Leu Arg 35 40 45Arg
Val Glu Val Thr Arg Gln Leu Cys Cys Ser Pro Gly Arg Leu Ala 50 55
60Val Leu Glu Arg Gly Gly Ala Gly Val Gln Val His Gln Leu Leu Ala65
70 75 80Gly Ser Gly Gly Ala Arg Thr Pro Lys Cys Ile Lys Leu Gly Lys
Asn 85 90 95Met Lys Ile His Ser Val Asp Gln Gly Ala Glu His Met Leu
Ile Leu 100 105 110Ser Ser Asp Gly Lys Pro Phe Glu Tyr Asp Asn Tyr
Ser Met Lys His 115 120 125Leu Arg Phe Glu Ser Ile Leu Gln Glu Lys
Lys Ile Ile Gln Ile Thr 130 135 140Cys Gly Asp Tyr His Ser Leu Ala
Leu Ser Lys Gly Gly Glu Leu Phe145 150 155 160Ala Trp Gly Gln Asn
Leu His Gly Gln Leu Gly Val Gly Arg Lys Phe 165 170 175Pro Ser Thr
Thr Thr Pro Gln Ile Val Glu His Leu Ala Gly Val Pro 180 185 190Leu
Ala Gln Ile Ser Ala Gly Glu Ala His Ser Met Ala Leu Ser Met 195 200
205Ser Gly Asn Ile Tyr Ser Trp Gly Lys Asn Glu Cys Gly Gln Leu Gly
210 215 220Leu Gly His Thr Glu Ser Lys Asp Asp Pro Ser Leu Ile Glu
Gly Leu225 230 235 240Asp Asn Gln Lys Val Glu Phe Val Ala Cys Gly
Gly Ser His Ser Ala 245 250 255Leu Leu Thr Gln Asp Gly Leu Leu Phe
Thr Phe Gly Ala Gly Lys His 260 265 270Gly Gln Leu Gly His Asn Ser
Thr Gln Asn Glu Leu Arg Pro Cys Leu 275 280 285Val Ala Glu Leu Val
Gly Tyr Arg Val Thr Gln Ile Ala Cys Gly Arg 290 295 300Trp His Thr
Leu Ala Tyr Val Ser Asp Leu Gly Lys Val Phe Ser Phe305 310 315
320Gly Ser Gly Lys Asp Gly Gln Leu Gly Asn Gly Gly Thr Arg Asp Gln
325 330
335Leu Met Pro Leu Pro Val Lys Val Ser Ser Ser Glu Glu Leu Lys Leu
340 345 350Glu Ser His Thr Ser Glu Lys Glu Leu Ile Met Ile Ala Gly
Gly Asn 355 360 365Gln Ser Ile Leu Leu Trp Ile Lys Lys Glu Asn Ser
Tyr Val Asn Leu 370 375 380Lys Arg Thr Ile Pro Thr Leu Asn Glu Gly
Thr Val Lys Arg Trp Ile385 390 395 400Ala Asp Val Glu Thr Lys Arg
Trp Gln Ser Thr Lys Arg Glu Ile Gln 405 410 415Glu Ile Phe Ser Ser
Pro Ala Cys Leu Thr Gly Ser Phe Leu Arg Lys 420 425 430Arg Arg Thr
Thr Glu Met Met Pro Val Tyr Leu Asp Leu Asn Lys Ala 435 440 445Arg
Asn Ile Phe Lys Glu Leu Thr Gln Lys Asp Trp Ile Thr Asn Met 450 455
460Ile Thr Thr Cys Leu Lys Asp Asn Leu Leu Lys Arg Leu Pro Phe
His465 470 475 480Ser Pro Pro Gln Glu Ala Leu Glu Ile Phe Phe Leu
Leu Pro Glu Cys 485 490 495Pro Met Met His Ile Ser Asn Asn Trp Glu
Ser Leu Val Val Pro Phe 500 505 510Ala Lys Val Val Cys Lys Met Ser
Asp Gln Ser Ser Leu Val Leu Glu 515 520 525Glu Tyr Trp Ala Thr Leu
Gln Glu Ser Thr Phe Ser Lys Leu Val Gln 530 535 540Met Phe Lys Thr
Ala Val Ile Cys Gln Leu Asp Tyr Trp Asp Glu Ser545 550 555 560Ala
Glu Glu Asn Gly Asn Val Gln Ala Leu Leu Glu Met Leu Lys Lys 565 570
575Leu His Arg Val Asn Gln Val Lys Cys Gln Leu Pro Glu Ser Ile Phe
580 585 590Gln Val Asp Glu Leu Leu His Arg Leu Asn Phe Phe Val Glu
Val Cys 595 600 605Arg Arg Tyr Leu Trp Lys Met Thr Val Asp Ala Ser
Glu Asn Val Gln 610 615 620Cys Cys Val Ile Phe Ser His Phe Pro Phe
Ile Phe Asn Asn Leu Ser625 630 635 640Lys Ile Lys Leu Leu His Thr
Asp Thr Leu Leu Lys Ile Glu Ser Lys 645 650 655Lys His Lys Ala Tyr
Leu Arg Ser Ala Ala Ile Glu Glu Glu Arg Glu 660 665 670Ser Glu Phe
Ala Leu Arg Pro Thr Phe Asp Leu Thr Val Arg Arg Asn 675 680 685His
Leu Ile Glu Asp Val Leu Asn Gln Leu Ser Gln Phe Glu Asn Glu 690 695
700Asp Leu Arg Lys Glu Leu Trp Val Ser Phe Ser Gly Glu Ile Gly
Tyr705 710 715 720Asp Leu Gly Gly Val Lys Lys Glu Phe Phe Tyr Cys
Leu Phe Ala Glu 725 730 735Met Ile Gln Pro Glu Tyr Gly Met Phe Met
Tyr Pro Glu Gly Ala Ser 740 745 750Cys Met Trp Phe Pro Val Lys Pro
Lys Phe Glu Lys Lys Arg Tyr Phe 755 760 765Phe Phe Gly Val Leu Cys
Gly Leu Ser Leu Phe Asn Cys Asn Val Ala 770 775 780Asn Leu Pro Phe
Pro Leu Ala Leu Phe Lys Lys Leu Leu Asp Gln Met785 790 795 800Pro
Ser Leu Glu Asp Leu Lys Glu Leu Ser Pro Asp Leu Gly Lys Asn 805 810
815Leu Gln Thr Leu Leu Asp Asp Glu Gly Asp Asn Phe Glu Glu Val Phe
820 825 830Tyr Ile His Phe Asn Val His Trp Asp Arg Asn Asp Thr Asn
Leu Ile 835 840 845Pro Asn Gly Ser Ser Ile Thr Val Asn Gln Thr Asn
Lys Arg Asp Tyr 850 855 860Val Ser Lys Tyr Ile Asn Tyr Ile Phe Asn
Asp Ser Val Lys Ala Val865 870 875 880Tyr Glu Glu Phe Arg Arg Gly
Phe Tyr Lys Met Cys Asp Glu Asp Ile 885 890 895Ile Lys Leu Phe His
Pro Glu Glu Leu Lys Asp Val Ile Val Gly Asn 900 905 910Thr Asp Tyr
Asp Trp Lys Thr Phe Glu Lys Asn Ala Arg Tyr Glu Pro 915 920 925Gly
Tyr Asn Ser Ser His Pro Thr Ile Val Met Phe Trp Lys Ala Phe 930 935
940His Lys Leu Thr Leu Glu Glu Lys Lys Lys Phe Leu Val Phe Leu
Thr945 950 955 960Gly Thr Asp Arg Leu Gln Met Lys Asp Leu Asn Asn
Met Lys Ile Thr 965 970 975Phe Cys Cys Pro Glu Ser Trp Asn Glu Arg
Asp Pro Ile Arg Ala Leu 980 985 990Thr Cys Phe Ser Val Leu Phe Leu
Pro Lys Tyr Ser Thr Met Glu Thr 995 1000 1005Val Glu Glu Ala Leu
Gln Glu Ala Ile Asn Asn Asn Arg Gly Phe 1010 1015
1020Gly332037DNAHomo sapiens 33gggaagctcg ggccggcagg gtttccccgc
acgctggcgc ccagctcccg gcgcggaggc 60cgctgtaagt ttcgctttcc attcagtgga
aaacgaaagc tgggcggggt gccacgagcg 120cggggccaga ccaaggcggg
cccggagcgg aacttcggtc ccagctcggt ccccggctca 180gtcccgacgt
ggaactcagc agcggaggct ggacgcttgc atggcgcttg agagattcca
240tcgtgcctgg ctcacataag cgcttcctgg aagtgaagtc gtgctgtcct
gaacgcgggc 300caggcagctg cggcctgggg gttttggagt gatcacgaat
gagcaaggcg tttgggctcc 360tgaggcaaat ctgtcagtcc atcctggctg
agtcctcgca gtccccggca gatcttgaag 420aaaagaagga agaagacagc
aacatgaaga gagagcagcc cagagagcgt cccagggcct 480gggactaccc
tcatggcctg gttggtttac acaacattgg acagacctgc tgccttaact
540ccttgattca ggtgttcgta atgaatgtgg acttcaccag gatattgaag
aggatcacgg 600tgcccagggg agctgacgag cagaggagaa gcgtcccttt
ccagatgctt ctgctgctgg 660agaagatgca ggacagccgg cagaaagcag
tgcggcccct ggagctggcc tactgcctgc 720agaagtgcaa cgtgcccttg
tttgtccaac atgatgctgc ccaactgtac ctcaaactct 780ggaacctgat
taaggaccag atcactgatg tgcacttggt ggagagactg caggccctgt
840atacgatccg ggtgaaggac tccttgattt gcgttgactg tgccatggag
agtagcagaa 900acagcagcat gctcaccctc ccactttctc tttttgatgt
ggactcaaag cccctgaaga 960cactggagga cgccctgcac tgcttcttcc
agcccaggga gttatcaagc aaaagcaagt 1020gcttctgtga gaactgtggg
aagaagaccc gtgggaaaca ggtcttgaag ctgacccatt 1080tgccccagac
cctgacaatc cacctcatgc gattctccat caggaattca cagacgagaa
1140agatctgcca ctccctgtac ttcccccaga gcttggattt cagccagatc
cttccaatga 1200agcgagagtc ttgtgatgct gaggagcagt ctggagggca
gtatgagctt tttgctgtga 1260ttgcgcacgt gggaatggca gactccggtc
attactgtgt ctacatccgg aatgctgtgg 1320atggaaaatg gttctgcttc
aatgactcca atatttgctt ggtgtcctgg gaagacatcc 1380agtgtaccta
cggaaatcct aactaccact ggcaggaaac tgcatatctt ctggtttaca
1440tgaagatgga gtgctaatgg aaatgcccaa aaccttcaga gattgacacg
ctgtcatttt 1500ccatttccgt tcctggatct acggagtctt ctaagagatt
ttgcaatgag gagaagcatt 1560gttttcaaac tatataactg agccttattt
ataattaggg atattatcaa aatatgtaac 1620catgaggccc ctcaggtcct
gatcagtcag aatggatgct ttcaccagca gacccggcca 1680tgtggctgct
cggtcctggg tgctcgctgc tgtgcaagac attagccctt tagttatgag
1740cctgtgggaa cttcaggggt tcccagtggg gagagcagtg gcagtgggag
gcatctgggg 1800gccaaaggtc agtggcaggg ggtatttcag tattatacaa
ctgctgtgac cagacttgta 1860tactggctga atatcagtgc tgtttgtaat
ttttcacttt gagaaccaac attaattcca 1920tatgaatcaa gtgttttgta
actgctattc atttattcag caaatattta ttgatcatct 1980cttctccata
agatagtgtg ataaacacag tcatgaataa agttattttc cacaaaa
203734372PRTHomo sapiens 34Met Ser Lys Ala Phe Gly Leu Leu Arg Gln
Ile Cys Gln Ser Ile Leu1 5 10 15Ala Glu Ser Ser Gln Ser Pro Ala Asp
Leu Glu Glu Lys Lys Glu Glu 20 25 30Asp Ser Asn Met Lys Arg Glu Gln
Pro Arg Glu Arg Pro Arg Ala Trp 35 40 45Asp Tyr Pro His Gly Leu Val
Gly Leu His Asn Ile Gly Gln Thr Cys 50 55 60Cys Leu Asn Ser Leu Ile
Gln Val Phe Val Met Asn Val Asp Phe Thr65 70 75 80Arg Ile Leu Lys
Arg Ile Thr Val Pro Arg Gly Ala Asp Glu Gln Arg 85 90 95Arg Ser Val
Pro Phe Gln Met Leu Leu Leu Leu Glu Lys Met Gln Asp 100 105 110Ser
Arg Gln Lys Ala Val Arg Pro Leu Glu Leu Ala Tyr Cys Leu Gln 115 120
125Lys Cys Asn Val Pro Leu Phe Val Gln His Asp Ala Ala Gln Leu Tyr
130 135 140Leu Lys Leu Trp Asn Leu Ile Lys Asp Gln Ile Thr Asp Val
His Leu145 150 155 160Val Glu Arg Leu Gln Ala Leu Tyr Thr Ile Arg
Val Lys Asp Ser Leu 165 170 175Ile Cys Val Asp Cys Ala Met Glu Ser
Ser Arg Asn Ser Ser Met Leu 180 185 190Thr Leu Pro Leu Ser Leu Phe
Asp Val Asp Ser Lys Pro Leu Lys Thr 195 200 205Leu Glu Asp Ala Leu
His Cys Phe Phe Gln Pro Arg Glu Leu Ser Ser 210 215 220Lys Ser Lys
Cys Phe Cys Glu Asn Cys Gly Lys Lys Thr Arg Gly Lys225 230 235
240Gln Val Leu Lys Leu Thr His Leu Pro Gln Thr Leu Thr Ile His Leu
245 250 255Met Arg Phe Ser Ile Arg Asn Ser Gln Thr Arg Lys Ile Cys
His Ser 260 265 270Leu Tyr Phe Pro Gln Ser Leu Asp Phe Ser Gln Ile
Leu Pro Met Lys 275 280 285Arg Glu Ser Cys Asp Ala Glu Glu Gln Ser
Gly Gly Gln Tyr Glu Leu 290 295 300Phe Ala Val Ile Ala His Val Gly
Met Ala Asp Ser Gly His Tyr Cys305 310 315 320Val Tyr Ile Arg Asn
Ala Val Asp Gly Lys Trp Phe Cys Phe Asn Asp 325 330 335Ser Asn Ile
Cys Leu Val Ser Trp Glu Asp Ile Gln Cys Thr Tyr Gly 340 345 350Asn
Pro Asn Tyr His Trp Gln Glu Thr Ala Tyr Leu Leu Val Tyr Met 355 360
365Lys Met Glu Cys 370353512DNAHomo sapiens 35aactcagctg agtgttagtc
aaagaaggtg tgtcctgctc cccaatgaca ggttgctcag 60agactgctga tttccatccc
tatataaaga gagtccctgg catacagaga ctgctctgct 120ccaggcatct
gccacaatgt gggtgcttac acctgctgct tttgctggga agctcttgag
180tgtgttcagg caacctctga gctctctgtg gaggagcctg gtcccgctgt
tctgctggct 240gagggcaacc ttctggctgc tagctaccaa gaggagaaag
cagcagctgg tcctgagagg 300gccagatgag accaaagagg aggaagagga
ccctcctctg cccaccaccc caaccagcgt 360caactatcac ttcactcgcc
agtgcaacta caaatgcggc ttctgtttcc acacagccaa 420aacatccttt
gtgctgcccc ttgaggaagc aaagagagga ttgcttttgc ttaaggaagc
480tggtatggag aagatcaact tttcaggtgg agagccattt cttcaagacc
ggggagaata 540cctgggcaag ttggtgaggt tctgcaaagt agagttgcgg
ctgcccagcg tgagcatcgt 600gagcaatgga agcctgatcc gggagaggtg
gttccagaat tatggtgagt atttggacat 660tctcgctatc tcctgtgaca
gctttgacga ggaagtcaat gtccttattg gccgtggcca 720aggaaagaag
aaccatgtgg aaaaccttca aaagctgagg aggtggtgta gggattatag
780agtcgctttc aagataaatt ctgtcattaa tcgtttcaac gtggaagagg
acatgacgga 840acagatcaaa gcactaaacc ctgtccgctg gaaagtgttc
cagtgcctct taattgaggg 900tgagaattgt ggagaagatg ctctaagaga
agcagaaaga tttgttattg gtgatgaaga 960atttgaaaga ttcttggagc
gccacaaaga agtgtcctgc ttggtgcctg aatctaacca 1020gaagatgaaa
gactcctacc ttattctgga tgaatatatg cgctttctga actgtagaaa
1080gggacggaag gacccttcca agtccatcct ggatgttggt gtagaagaag
ctataaaatt 1140cagtggattt gatgaaaaga tgtttctgaa gcgaggagga
aaatacatat ggagtaaggc 1200tgatctgaag ctggattggt agagcggaaa
gtggaacgag acttcaacac accagtggga 1260aaactcctag agtaactgcc
attgtctgca atactatccc gttggtattt cccagtggct 1320gaaaacctga
ttttctgctg cacgtggcat ctgattacct gtggtcactg aacacacgaa
1380taacttggat agcaaatcct gagacaatgg aaaaccatta actttacttc
attggcttat 1440aaccttgttg ttattgaaac agcacttctg tttttgagtt
tgttttagct aaaaagaagg 1500aatacacaca ggaataatga ccccaaaaat
gcttagataa ggcccctata cacaggacct 1560gacatttagc tcaatgatgc
gtttgtaaga aataagctct agtgatatct gtgggggcaa 1620aatttaattt
ggatttgatt ttttaaaaca atgtttactg cgatttctat atttccattt
1680tgaaactatt tcttgttcca ggtttgttca tttgacagag tcagtatttt
ttgccaaata 1740tccagataac cagttttcac atctgagaca ttacaaagta
tctgcctcaa ttatttctgc 1800tggttataat gctttttttt ttttgccttt
atgccattgc agtcttgtac tttttactgt 1860gatgtacaga aatagtcaac
agatgtttcc aagaacatat gatatgataa tcctaccaat 1920tttcaagaag
tctctagaaa gagataacac atggaaagac ggtgtggtgc agcccagccc
1980acggtggctg ttccatgaat gctggctacc tatgtgtgtg gtacctgttg
tgtccctttc 2040tcttcaaaga tcctgagcaa aacaaagata cgctttccat
ttgatgatgg agttgacatg 2100gaggcagtgc ttgcattgct ttgttcgcct
atcatctggc cacatgaggc tgtcaagcaa 2160aagaatagga gtgtagttga
gtagctggtt ggccctacat ctctgagaag tgacggcaca 2220ctgggttggc
ataagatatc ctaaaatcac gctggaacct tgggcaagga agaatgtgag
2280caagagtaga gagagtgcct ggatttcatg tcagtgaagc caagtcacca
tatcatattt 2340ttgaatgaac tctgagtcag ttgaaatagg gtaccatcta
ggtcagttta agaagagtca 2400gctcagagaa agcaagcata agggaaaatg
tcacgtaaac tagatcaggg aacaaaatcc 2460tctccttgtg gaaatatccc
atgcagtttg ttgatacaac ttagtatctt attgcctaaa 2520aaaaaatttc
ttatcattgt ttcaaaaaag caaaatcatg gaaaattttt gttgtccagg
2580caaataaaag gtcattttaa tttagctgca atttcagtgt tcctcactag
gtggcattta 2640aatgtcgcct gatgtcatta agcaccatcc aaaaagtctg
cttcataatc tattttcaag 2700acttggtgat tctgaaagtt ttggtttttg
tgactttgtt tctcaggaaa aaaaatattc 2760ctacttaaat tttaagtcta
taattcaatt taaatatgtg tgtgtctcat ccaggatagg 2820ataggttgtc
ttctattttc cattttacct atttactttt tttgtaagaa aagagaaaaa
2880tgaattctaa agatgttccc catgggtttt gattgtgtct aagctatgat
gaccttcata 2940taatcagcat aaacataaaa caaatttttt acttaacatg
agtgcacttt actaatcctc 3000atggcacagt ggctcacgcc tgtaatccca
gcacttggga ggacaatgtg ggtggatcac 3060gaggtcagga gttcgagaac
agcctggcca acatggtgaa accccgtctc cactaaaaat 3120acaaaaatta
gccaggcatg gtggcgtaca cttgtaattc cagctactca agaggctgag
3180gcaggaggat tgcttgaacc ctgaaggcag aggttacaga gccaagatag
cgccactgca 3240ctccagcctg gatgacagag caagactccg tctcaaaaaa
aaaaaaaaaa aaaagcaaga 3300gagttcaact aagaaaggtc acatatgtga
aagcccaagg acactgtttg atatacagca 3360ggtattcaat cagtgttatt
tgaaaccaaa tctgaatttg aagtttgaat cttctgagtt 3420ggaatgaatt
tttttctagc tgagggaaac tgtatttttc tttccccaaa gaggaatgta
3480atgtaaagtg aaataaaact ataagctatg tt 351236361PRTHomo sapiens
36Met Trp Val Leu Thr Pro Ala Ala Phe Ala Gly Lys Leu Leu Ser Val1
5 10 15Phe Arg Gln Pro Leu Ser Ser Leu Trp Arg Ser Leu Val Pro Leu
Phe 20 25 30Cys Trp Leu Arg Ala Thr Phe Trp Leu Leu Ala Thr Lys Arg
Arg Lys 35 40 45Gln Gln Leu Val Leu Arg Gly Pro Asp Glu Thr Lys Glu
Glu Glu Glu 50 55 60Asp Pro Pro Leu Pro Thr Thr Pro Thr Ser Val Asn
Tyr His Phe Thr65 70 75 80Arg Gln Cys Asn Tyr Lys Cys Gly Phe Cys
Phe His Thr Ala Lys Thr 85 90 95Ser Phe Val Leu Pro Leu Glu Glu Ala
Lys Arg Gly Leu Leu Leu Leu 100 105 110Lys Glu Ala Gly Met Glu Lys
Ile Asn Phe Ser Gly Gly Glu Pro Phe 115 120 125Leu Gln Asp Arg Gly
Glu Tyr Leu Gly Lys Leu Val Arg Phe Cys Lys 130 135 140Val Glu Leu
Arg Leu Pro Ser Val Ser Ile Val Ser Asn Gly Ser Leu145 150 155
160Ile Arg Glu Arg Trp Phe Gln Asn Tyr Gly Glu Tyr Leu Asp Ile Leu
165 170 175Ala Ile Ser Cys Asp Ser Phe Asp Glu Glu Val Asn Val Leu
Ile Gly 180 185 190Arg Gly Gln Gly Lys Lys Asn His Val Glu Asn Leu
Gln Lys Leu Arg 195 200 205Arg Trp Cys Arg Asp Tyr Arg Val Ala Phe
Lys Ile Asn Ser Val Ile 210 215 220Asn Arg Phe Asn Val Glu Glu Asp
Met Thr Glu Gln Ile Lys Ala Leu225 230 235 240Asn Pro Val Arg Trp
Lys Val Phe Gln Cys Leu Leu Ile Glu Gly Glu 245 250 255Asn Cys Gly
Glu Asp Ala Leu Arg Glu Ala Glu Arg Phe Val Ile Gly 260 265 270Asp
Glu Glu Phe Glu Arg Phe Leu Glu Arg His Lys Glu Val Ser Cys 275 280
285Leu Val Pro Glu Ser Asn Gln Lys Met Lys Asp Ser Tyr Leu Ile Leu
290 295 300Asp Glu Tyr Met Arg Phe Leu Asn Cys Arg Lys Gly Arg Lys
Asp Pro305 310 315 320Ser Lys Ser Ile Leu Asp Val Gly Val Glu Glu
Ala Ile Lys Phe Ser 325 330 335Gly Phe Asp Glu Lys Met Phe Leu Lys
Arg Gly Gly Lys Tyr Ile Trp 340 345 350Ser Lys Ala Asp Leu Lys Leu
Asp Trp 355 360372983DNAHomo sapiens 37gttcgggccc gagaacctgc
gtctcccgcg agttcccgcg aggcaagtgc tgcaggtgcg 60gggccaggag ctaggtttcg
tttctgcgcc cggagccgcc ctcagcacag ggtctgtgag 120tttcatttct
tcgcggcgcg gggcggggct gggcgcgggg tgaaagaggc gaagcgagag
180cggaggccgc actccagcac tgcgcaggga ccgccttgga ccgcagttgc
cggccaggaa 240tcccagtgtc acggtggaca cgcctccctc gcgcccttgc
cgcccacctg ctcacccagc 300tcaggggctt tggaattctg tggccacact
gcgaggagat cggttctggg tcggaggcta 360caggaagact cccactccct
gaaatctgga gtgaagaacg ccgccatcca gccaccattc 420caaggaggtg
caggagaaca gctctgtgat accatttaac ttgttgacat tacttttatt
480tgaaggaacg tatattagag cttactttgc aaagaaggaa gatggttgtt
tccgaagtgg 540acatcgcaaa agctgatcca gctgctgcat cccaccctct
attactgaat ggagatgcta 600ctgtggccca gaaaaatcca ggctcggtgg
ctgagaacaa cctgtgcagc cagtatgagg 660agaaggtgcg cccctgcatc
gacctcattg actccctgcg ggctctaggt gtggagcagg
720acctggccct gccagccatc gccgtcatcg gggaccagag ctcgggcaag
agctccgtgt 780tggaggcact gtcaggagtt gcccttccca gaggcagcgg
gatcgtgacc agatgcccgc 840tggtgctgaa actgaagaaa cttgtgaacg
aagataagtg gagaggcaag gtcagttacc 900aggactacga gattgagatt
tcggatgctt cagaggtaga aaaggaaatt aataaagccc 960agaatgccat
cgccggggaa ggaatgggaa tcagtcatga gctaatcacc ctggagatca
1020gctcccgaga tgtcccggat ctgactctaa tagaccttcc tggcataacc
agagtggctg 1080tgggcaatca gcctgctgac attgggtata agatcaagac
actcatcaag aagtacatcc 1140agaggcagga gacaatcagc ctggtggtgg
tccccagtaa tgtggacatc gccaccacag 1200aggctctcag catggcccag
gaggtggacc ccgagggaga caggaccatc ggaatcttga 1260cgaagcctga
tctggtggac aaaggaactg aagacaaggt tgtggacgtg gtgcggaacc
1320tcgtgttcca cctgaagaag ggttacatga ttgtcaagtg ccggggccag
caggagatcc 1380aggaccagct gagcctgtcc gaagccctgc agagagagaa
gatcttcttt gagaaccacc 1440catatttcag ggatctgctg gaggaaggaa
aggccacggt tccctgcctg gcagaaaaac 1500ttaccagcga gctcatcaca
catatctgta aatctctgcc cctgttagaa aatcaaatca 1560aggagactca
ccagagaata acagaggagc tacaaaagta tggtgtcgac ataccggaag
1620acgaaaatga aaaaatgttc ttcctgatag ataaagttaa tgcctttaat
caggacatca 1680ctgctctcat gcaaggagag gaaactgtag gggaggaaga
cattcggctg tttaccagac 1740tccgacacga gttccacaaa tggagtacaa
taattgaaaa caattttcaa gaaggccata 1800aaattttgag tagaaaaatc
cagaaatttg aaaatcagta tcgtggtaga gagctgccag 1860gctttgtgaa
ttacaggaca tttgagacaa tcgtgaaaca gcaaatcaag gcactggaag
1920agccggctgt ggatatgcta cacaccgtga cggatatggt ccggcttgct
ttcacagatg 1980tttcgataaa aaattttgaa gagtttttta acctccacag
aaccgccaag tccaaaattg 2040aagacattag agcagaacaa gagagagaag
gtgagaagct gatccgcctc cacttccaga 2100tggaacagat tgtctactgc
caggaccagg tatacagggg tgcattgcag aaggtcagag 2160agaaggagct
ggaagaagaa aagaagaaga aatcctggga ttttggggct ttccagtcca
2220gctcggcaac agactcttcc atggaggaga tctttcagca cctgatggcc
tatcaccagg 2280aggccagcaa gcgcatctcc agccacatcc ctttgatcat
ccagttcttc atgctccaga 2340cgtacggcca gcagcttcag aaggccatgc
tgcagctcct gcaggacaag gacacctaca 2400gctggctcct gaaggagcgg
agcgacacca gcgacaagcg gaagttcctg aaggagcggc 2460ttgcacggct
gacgcaggct cggcgccggc ttgcccagtt ccccggttaa ccacactctg
2520tccagccccg tagacgtgca cgcacactgt ctgcccccgt tcccgggtag
ccactggact 2580gacgacttga gtgctcagta gtcagactgg atagtccgtc
tctgcttatc cgttagccgt 2640ggtgatttag caggaagctg tgagagcagt
ttggtttcta gcatgaagac agagccccac 2700cctcagatgc acatgagctg
gcgggattga aggatgctgt cttcgtactg ggaaagggat 2760tttcagccct
cagaatcgct ccaccttgca gctctcccct tctctgtatt cctagaaact
2820gacacatgct gaacatcaca gcttatttcc tcatttttat aatgtccctt
cacaaaccca 2880gtgttttagg agcatgagtg ccgtgtgtgt gcgtcctgtc
ggagccctgt ctcctctctc 2940tgtaataaac tcatttctag cagacaaaaa
aaaaaaaaaa aaa 298338662PRTHomo sapiens 38Met Val Val Ser Glu Val
Asp Ile Ala Lys Ala Asp Pro Ala Ala Ala1 5 10 15Ser His Pro Leu Leu
Leu Asn Gly Asp Ala Thr Val Ala Gln Lys Asn 20 25 30Pro Gly Ser Val
Ala Glu Asn Asn Leu Cys Ser Gln Tyr Glu Glu Lys 35 40 45Val Arg Pro
Cys Ile Asp Leu Ile Asp Ser Leu Arg Ala Leu Gly Val 50 55 60Glu Gln
Asp Leu Ala Leu Pro Ala Ile Ala Val Ile Gly Asp Gln Ser65 70 75
80Ser Gly Lys Ser Ser Val Leu Glu Ala Leu Ser Gly Val Ala Leu Pro
85 90 95Arg Gly Ser Gly Ile Val Thr Arg Cys Pro Leu Val Leu Lys Leu
Lys 100 105 110Lys Leu Val Asn Glu Asp Lys Trp Arg Gly Lys Val Ser
Tyr Gln Asp 115 120 125Tyr Glu Ile Glu Ile Ser Asp Ala Ser Glu Val
Glu Lys Glu Ile Asn 130 135 140Lys Ala Gln Asn Ala Ile Ala Gly Glu
Gly Met Gly Ile Ser His Glu145 150 155 160Leu Ile Thr Leu Glu Ile
Ser Ser Arg Asp Val Pro Asp Leu Thr Leu 165 170 175Ile Asp Leu Pro
Gly Ile Thr Arg Val Ala Val Gly Asn Gln Pro Ala 180 185 190Asp Ile
Gly Tyr Lys Ile Lys Thr Leu Ile Lys Lys Tyr Ile Gln Arg 195 200
205Gln Glu Thr Ile Ser Leu Val Val Val Pro Ser Asn Val Asp Ile Ala
210 215 220Thr Thr Glu Ala Leu Ser Met Ala Gln Glu Val Asp Pro Glu
Gly Asp225 230 235 240Arg Thr Ile Gly Ile Leu Thr Lys Pro Asp Leu
Val Asp Lys Gly Thr 245 250 255Glu Asp Lys Val Val Asp Val Val Arg
Asn Leu Val Phe His Leu Lys 260 265 270Lys Gly Tyr Met Ile Val Lys
Cys Arg Gly Gln Gln Glu Ile Gln Asp 275 280 285Gln Leu Ser Leu Ser
Glu Ala Leu Gln Arg Glu Lys Ile Phe Phe Glu 290 295 300Asn His Pro
Tyr Phe Arg Asp Leu Leu Glu Glu Gly Lys Ala Thr Val305 310 315
320Pro Cys Leu Ala Glu Lys Leu Thr Ser Glu Leu Ile Thr His Ile Cys
325 330 335Lys Ser Leu Pro Leu Leu Glu Asn Gln Ile Lys Glu Thr His
Gln Arg 340 345 350Ile Thr Glu Glu Leu Gln Lys Tyr Gly Val Asp Ile
Pro Glu Asp Glu 355 360 365Asn Glu Lys Met Phe Phe Leu Ile Asp Lys
Val Asn Ala Phe Asn Gln 370 375 380Asp Ile Thr Ala Leu Met Gln Gly
Glu Glu Thr Val Gly Glu Glu Asp385 390 395 400Ile Arg Leu Phe Thr
Arg Leu Arg His Glu Phe His Lys Trp Ser Thr 405 410 415Ile Ile Glu
Asn Asn Phe Gln Glu Gly His Lys Ile Leu Ser Arg Lys 420 425 430Ile
Gln Lys Phe Glu Asn Gln Tyr Arg Gly Arg Glu Leu Pro Gly Phe 435 440
445Val Asn Tyr Arg Thr Phe Glu Thr Ile Val Lys Gln Gln Ile Lys Ala
450 455 460Leu Glu Glu Pro Ala Val Asp Met Leu His Thr Val Thr Asp
Met Val465 470 475 480Arg Leu Ala Phe Thr Asp Val Ser Ile Lys Asn
Phe Glu Glu Phe Phe 485 490 495Asn Leu His Arg Thr Ala Lys Ser Lys
Ile Glu Asp Ile Arg Ala Glu 500 505 510Gln Glu Arg Glu Gly Glu Lys
Leu Ile Arg Leu His Phe Gln Met Glu 515 520 525Gln Ile Val Tyr Cys
Gln Asp Gln Val Tyr Arg Gly Ala Leu Gln Lys 530 535 540Val Arg Glu
Lys Glu Leu Glu Glu Glu Lys Lys Lys Lys Ser Trp Asp545 550 555
560Phe Gly Ala Phe Gln Ser Ser Ser Ala Thr Asp Ser Ser Met Glu Glu
565 570 575Ile Phe Gln His Leu Met Ala Tyr His Gln Glu Ala Ser Lys
Arg Ile 580 585 590Ser Ser His Ile Pro Leu Ile Ile Gln Phe Phe Met
Leu Gln Thr Tyr 595 600 605Gly Gln Gln Leu Gln Lys Ala Met Leu Gln
Leu Leu Gln Asp Lys Asp 610 615 620Thr Tyr Ser Trp Leu Leu Lys Glu
Arg Ser Asp Thr Ser Asp Lys Arg625 630 635 640Lys Phe Leu Lys Glu
Arg Leu Ala Arg Leu Thr Gln Ala Arg Arg Arg 645 650 655Leu Ala Gln
Phe Pro Gly 660392683DNAHomo sapiens 39tttcttcgcg gcgcggggcg
gggctgggcg cggggtgaaa gaggcgaagc gagagcggag 60gccgcactcc agcactgcgc
agggaccgga attctgtggc cacactgcga ggagatcggt 120tctgggtcgg
aggctacagg aagactccca ctccctgaaa tctggagtga agaacgccgc
180catccagcca ccattccaag cttactttgc aaagaaggaa gatggttgtt
tccgaagtgg 240acatcgcaaa agctgatcca gctgctgcat cccaccctct
attactgaat ggagatgcta 300ctgtggccca gaaaaatcca ggctcggtgg
ctgagaacaa cctgtgcagc cagtatgagg 360agaaggtgcg cccctgcatc
gacctcattg actccctgcg ggctctaggt gtggagcagg 420acctggccct
gccagccatc gccgtcatcg gggaccagag ctcgggcaag agctccgtgt
480tggaggcact gtcaggagtt gcccttccca gaggcagcgg gatcgtgacc
agatgcccgc 540tggtgctgaa actgaagaaa cttgtgaacg aagataagtg
gagaggcaag gtcagttacc 600aggactacga gattgagatt tcggatgctt
cagaggtaga aaaggaaatt aataaagccc 660agaatgccat cgccggggaa
ggaatgggaa tcagtcatga gctaatcacc ctggagatca 720gctcccgaga
tgtcccggat ctgactctaa tagaccttcc tggcataacc agagtggctg
780tgggcaatca gcctgctgac attgggtata agatcaagac actcatcaag
aagtacatcc 840agaggcagga gacaatcagc ctggtggtgg tccccagtaa
tgtggacatc gccaccacag 900aggctctcag catggcccag gaggtggacc
ccgagggaga caggaccatc ggaatcttga 960cgaagcctga tctggtggac
aaaggaactg aagacaaggt tgtggacgtg gtgcggaacc 1020tcgtgttcca
cctgaagaag ggttacatga ttgtcaagtg ccggggccag caggagatcc
1080aggaccagct gagcctgtcc gaagccctgc agagagagaa gatcttcttt
gagaaccacc 1140catatttcag ggatctgctg gaggaaggaa aggccacggt
tccctgcctg gcagaaaaac 1200ttaccagcga gctcatcaca catatctgta
aatctctgcc cctgttagaa aatcaaatca 1260aggagactca ccagagaata
acagaggagc tacaaaagta tggtgtcgac ataccggaag 1320acgaaaatga
aaaaatgttc ttcctgatag ataaagttaa tgcctttaat caggacatca
1380ctgctctcat gcaaggagag gaaactgtag gggaggaaga cattcggctg
tttaccagac 1440tccgacacga gttccacaaa tggagtacaa taattgaaaa
caattttcaa gaaggccata 1500aaattttgag tagaaaaatc cagaaatttg
aaaatcagta tcgtggtaga gagctgccag 1560gctttgtgaa ttacaggaca
tttgagacaa tcgtgaaaca gcaaatcaag gcactggaag 1620agccggctgt
ggatatgcta cacaccgtga cggatatggt ccggcttgct ttcacagatg
1680tttcgataaa aaattttgaa gagtttttta acctccacag aaccgccaag
tccaaaattg 1740aagacattag agcagaacaa gagagagaag gtgagaagct
gatccgcctc cacttccaga 1800tggaacagat tgtctactgc caggaccagg
tatacagggg tgcattgcag aaggtcagag 1860agaaggagct ggaagaagaa
aagaagaaga aatcctggga ttttggggct ttccagtcca 1920gctcggcaac
agactcttcc atggaggaga tctttcagca cctgatggcc tatcaccagg
1980aggccagcaa gcgcatctcc agccacatcc ctttgatcat ccagttcttc
atgctccaga 2040cgtacggcca gcagcttcag aaggccatgc tgcagctcct
gcaggacaag gacacctaca 2100gctggctcct gaaggagcgg agcgacacca
gcgacaagcg gaagttcctg aaggagcggc 2160ttgcacggct gacgcaggct
cggcgccggc ttgcccagtt ccccggttaa ccacactctg 2220tccagccccg
tagacgtgca cgcacactgt ctgcccccgt tcccgggtag ccactggact
2280gacgacttga gtgctcagta gtcagactgg atagtccgtc tctgcttatc
cgttagccgt 2340ggtgatttag caggaagctg tgagagcagt ttggtttcta
gcatgaagac agagccccac 2400cctcagatgc acatgagctg gcgggattga
aggatgctgt cttcgtactg ggaaagggat 2460tttcagccct cagaatcgct
ccaccttgca gctctcccct tctctgtatt cctagaaact 2520gacacatgct
gaacatcaca gcttatttcc tcatttttat aatgtccctt cacaaaccca
2580gtgttttagg agcatgagtg ccgtgtgtgt gcgtcctgtc ggagccctgt
ctcctctctc 2640tgtaataaac tcatttctag cagacaaaaa aaaaaaaaaa aaa
268340662PRTHomo sapiens 40Met Val Val Ser Glu Val Asp Ile Ala Lys
Ala Asp Pro Ala Ala Ala1 5 10 15Ser His Pro Leu Leu Leu Asn Gly Asp
Ala Thr Val Ala Gln Lys Asn 20 25 30Pro Gly Ser Val Ala Glu Asn Asn
Leu Cys Ser Gln Tyr Glu Glu Lys 35 40 45Val Arg Pro Cys Ile Asp Leu
Ile Asp Ser Leu Arg Ala Leu Gly Val 50 55 60Glu Gln Asp Leu Ala Leu
Pro Ala Ile Ala Val Ile Gly Asp Gln Ser65 70 75 80Ser Gly Lys Ser
Ser Val Leu Glu Ala Leu Ser Gly Val Ala Leu Pro 85 90 95Arg Gly Ser
Gly Ile Val Thr Arg Cys Pro Leu Val Leu Lys Leu Lys 100 105 110Lys
Leu Val Asn Glu Asp Lys Trp Arg Gly Lys Val Ser Tyr Gln Asp 115 120
125Tyr Glu Ile Glu Ile Ser Asp Ala Ser Glu Val Glu Lys Glu Ile Asn
130 135 140Lys Ala Gln Asn Ala Ile Ala Gly Glu Gly Met Gly Ile Ser
His Glu145 150 155 160Leu Ile Thr Leu Glu Ile Ser Ser Arg Asp Val
Pro Asp Leu Thr Leu 165 170 175Ile Asp Leu Pro Gly Ile Thr Arg Val
Ala Val Gly Asn Gln Pro Ala 180 185 190Asp Ile Gly Tyr Lys Ile Lys
Thr Leu Ile Lys Lys Tyr Ile Gln Arg 195 200 205Gln Glu Thr Ile Ser
Leu Val Val Val Pro Ser Asn Val Asp Ile Ala 210 215 220Thr Thr Glu
Ala Leu Ser Met Ala Gln Glu Val Asp Pro Glu Gly Asp225 230 235
240Arg Thr Ile Gly Ile Leu Thr Lys Pro Asp Leu Val Asp Lys Gly Thr
245 250 255Glu Asp Lys Val Val Asp Val Val Arg Asn Leu Val Phe His
Leu Lys 260 265 270Lys Gly Tyr Met Ile Val Lys Cys Arg Gly Gln Gln
Glu Ile Gln Asp 275 280 285Gln Leu Ser Leu Ser Glu Ala Leu Gln Arg
Glu Lys Ile Phe Phe Glu 290 295 300Asn His Pro Tyr Phe Arg Asp Leu
Leu Glu Glu Gly Lys Ala Thr Val305 310 315 320Pro Cys Leu Ala Glu
Lys Leu Thr Ser Glu Leu Ile Thr His Ile Cys 325 330 335Lys Ser Leu
Pro Leu Leu Glu Asn Gln Ile Lys Glu Thr His Gln Arg 340 345 350Ile
Thr Glu Glu Leu Gln Lys Tyr Gly Val Asp Ile Pro Glu Asp Glu 355 360
365Asn Glu Lys Met Phe Phe Leu Ile Asp Lys Val Asn Ala Phe Asn Gln
370 375 380Asp Ile Thr Ala Leu Met Gln Gly Glu Glu Thr Val Gly Glu
Glu Asp385 390 395 400Ile Arg Leu Phe Thr Arg Leu Arg His Glu Phe
His Lys Trp Ser Thr 405 410 415Ile Ile Glu Asn Asn Phe Gln Glu Gly
His Lys Ile Leu Ser Arg Lys 420 425 430Ile Gln Lys Phe Glu Asn Gln
Tyr Arg Gly Arg Glu Leu Pro Gly Phe 435 440 445Val Asn Tyr Arg Thr
Phe Glu Thr Ile Val Lys Gln Gln Ile Lys Ala 450 455 460Leu Glu Glu
Pro Ala Val Asp Met Leu His Thr Val Thr Asp Met Val465 470 475
480Arg Leu Ala Phe Thr Asp Val Ser Ile Lys Asn Phe Glu Glu Phe Phe
485 490 495Asn Leu His Arg Thr Ala Lys Ser Lys Ile Glu Asp Ile Arg
Ala Glu 500 505 510Gln Glu Arg Glu Gly Glu Lys Leu Ile Arg Leu His
Phe Gln Met Glu 515 520 525Gln Ile Val Tyr Cys Gln Asp Gln Val Tyr
Arg Gly Ala Leu Gln Lys 530 535 540Val Arg Glu Lys Glu Leu Glu Glu
Glu Lys Lys Lys Lys Ser Trp Asp545 550 555 560Phe Gly Ala Phe Gln
Ser Ser Ser Ala Thr Asp Ser Ser Met Glu Glu 565 570 575Ile Phe Gln
His Leu Met Ala Tyr His Gln Glu Ala Ser Lys Arg Ile 580 585 590Ser
Ser His Ile Pro Leu Ile Ile Gln Phe Phe Met Leu Gln Thr Tyr 595 600
605Gly Gln Gln Leu Gln Lys Ala Met Leu Gln Leu Leu Gln Asp Lys Asp
610 615 620Thr Tyr Ser Trp Leu Leu Lys Glu Arg Ser Asp Thr Ser Asp
Lys Arg625 630 635 640Lys Phe Leu Lys Glu Arg Leu Ala Arg Leu Thr
Gln Ala Arg Arg Arg 645 650 655Leu Ala Gln Phe Pro Gly
660413409DNAHomo sapiens 41aaattcgcgg tgggggcgga gagcgcaggg
agaagtaagc ccagtgcagg atcctgaggc 60ccgtgtttgc aggaccaggg ccggccttcc
gattccccat tcattccaga agcaccgaac 120cacgctgtgc ccggatccca
agtgcagcgg cacccagcgt gggcctgggg ttgccggttg 180acccggtcct
cagcctggta gcagaggcca ggccagtgcc acaaggcacc taagtccacc
240tgggcctgga gcaggacagg ttgcaaaaga aaatatctcg ggacccccaa
actccttatg 300ctaagggaaa catcgagcct gggaactgag ccatcaacgc
tgccattctt tttcccaaac 360agaaccctgt tgtcagaggt acacccagag
caactccaca ccgggtgcat gccacagcaa 420ctccatctta aataggagct
ggtaaaacga ggctgatacc tactgggctg cattcccaga 480cggcatagcg
aggaggtgct gaagagcgca ggtttggaga atgatcacct ggattggaac
540catagctcta ccaatatgga acccagctcc ttaggcctcg gtcttctcat
ggagaacatg 600gtgtgataat cctactcctc tgggagggtg gctgttaagc
cttggaccgc agttgccggc 660caggaatccc agtgtcacgg tggacacgcc
tccctcgcgc ccttgccgcc cacctgctca 720cccagctcag gggctttgga
attctgtggc cacactgcga ggagatcggt tctgggtcgg 780aggctacagg
aagactccca ctccctgaaa tctggagtga agaacgccgc catccagcca
840ccattccaag gaggtgcagg agaacagctc tgtgatacca tttaacttgt
tgacattact 900tttatttgaa ggaacgtata ttagagctta ctttgcaaag
aaggaagatg gttgtttccg 960aagtggacat cgcaaaagct gatccagctg
ctgcatccca ccctctatta ctgaatggag 1020atgctactgt ggcccagaaa
aatccaggct cggtggctga gaacaacctg tgcagccagt 1080atgaggagaa
ggtgcgcccc tgcatcgacc tcattgactc cctgcgggct ctaggtgtgg
1140agcaggacct ggccctgcca gccatcgccg tcatcgggga ccagagctcg
ggcaagagct 1200ccgtgttgga ggcactgtca ggagttgccc ttcccagagg
cagcgggatc gtgaccagat 1260gcccgctggt gctgaaactg aagaaacttg
tgaacgaaga taagtggaga ggcaaggtca 1320gttaccagga ctacgagatt
gagatttcgg atgcttcaga ggtagaaaag gaaattaata 1380aagcccagaa
tgccatcgcc ggggaaggaa tgggaatcag tcatgagcta atcaccctgg
1440agatcagctc ccgagatgtc ccggatctga ctctaataga ccttcctggc
ataaccagag 1500tggctgtggg caatcagcct gctgacattg ggtataagat
caagacactc atcaagaagt 1560acatccagag gcaggagaca atcagcctgg
tggtggtccc cagtaatgtg gacatcgcca 1620ccacagaggc tctcagcatg
gcccaggagg tggaccccga gggagacagg accatcggaa 1680tcttgacgaa
gcctgatctg gtggacaaag gaactgaaga caaggttgtg gacgtggtgc
1740ggaacctcgt gttccacctg aagaagggtt acatgattgt caagtgccgg
ggccagcagg 1800agatccagga ccagctgagc ctgtccgaag ccctgcagag
agagaagatc ttctttgaga 1860accacccata tttcagggat ctgctggagg
aaggaaaggc cacggttccc tgcctggcag 1920aaaaacttac cagcgagctc
atcacacata
tctgtaaatc tctgcccctg ttagaaaatc 1980aaatcaagga gactcaccag
agaataacag aggagctaca aaagtatggt gtcgacatac 2040cggaagacga
aaatgaaaaa atgttcttcc tgatagataa agttaatgcc tttaatcagg
2100acatcactgc tctcatgcaa ggagaggaaa ctgtagggga ggaagacatt
cggctgttta 2160ccagactccg acacgagttc cacaaatgga gtacaataat
tgaaaacaat tttcaagaag 2220gccataaaat tttgagtaga aaaatccaga
aatttgaaaa tcagtatcgt ggtagagagc 2280tgccaggctt tgtgaattac
aggacatttg agacaatcgt gaaacagcaa atcaaggcac 2340tggaagagcc
ggctgtggat atgctacaca ccgtgacgga tatggtccgg cttgctttca
2400cagatgtttc gataaaaaat tttgaagagt tttttaacct ccacagaacc
gccaagtcca 2460aaattgaaga cattagagca gaacaagaga gagaaggtga
gaagctgatc cgcctccact 2520tccagatgga acagattgtc tactgccagg
accaggtata caggggtgca ttgcagaagg 2580tcagagagaa ggagctggaa
gaagaaaaga agaagaaatc ctgggatttt ggggctttcc 2640agtccagctc
ggcaacagac tcttccatgg aggagatctt tcagcacctg atggcctatc
2700accaggaggc cagcaagcgc atctccagcc acatcccttt gatcatccag
ttcttcatgc 2760tccagacgta cggccagcag cttcagaagg ccatgctgca
gctcctgcag gacaaggaca 2820cctacagctg gctcctgaag gagcggagcg
acaccagcga caagcggaag ttcctgaagg 2880agcggcttgc acggctgacg
caggctcggc gccggcttgc ccagttcccc ggttaaccac 2940actctgtcca
gccccgtaga cgtgcacgca cactgtctgc ccccgttccc gggtagccac
3000tggactgacg acttgagtgc tcagtagtca gactggatag tccgtctctg
cttatccgtt 3060agccgtggtg atttagcagg aagctgtgag agcagtttgg
tttctagcat gaagacagag 3120ccccaccctc agatgcacat gagctggcgg
gattgaagga tgctgtcttc gtactgggaa 3180agggattttc agccctcaga
atcgctccac cttgcagctc tccccttctc tgtattccta 3240gaaactgaca
catgctgaac atcacagctt atttcctcat ttttataatg tcccttcaca
3300aacccagtgt tttaggagca tgagtgccgt gtgtgtgcgt cctgtcggag
ccctgtctcc 3360tctctctgta ataaactcat ttctagcaga caaaaaaaaa
aaaaaaaaa 340942662PRTHomo sapiens 42Met Val Val Ser Glu Val Asp
Ile Ala Lys Ala Asp Pro Ala Ala Ala1 5 10 15Ser His Pro Leu Leu Leu
Asn Gly Asp Ala Thr Val Ala Gln Lys Asn 20 25 30Pro Gly Ser Val Ala
Glu Asn Asn Leu Cys Ser Gln Tyr Glu Glu Lys 35 40 45Val Arg Pro Cys
Ile Asp Leu Ile Asp Ser Leu Arg Ala Leu Gly Val 50 55 60Glu Gln Asp
Leu Ala Leu Pro Ala Ile Ala Val Ile Gly Asp Gln Ser65 70 75 80Ser
Gly Lys Ser Ser Val Leu Glu Ala Leu Ser Gly Val Ala Leu Pro 85 90
95Arg Gly Ser Gly Ile Val Thr Arg Cys Pro Leu Val Leu Lys Leu Lys
100 105 110Lys Leu Val Asn Glu Asp Lys Trp Arg Gly Lys Val Ser Tyr
Gln Asp 115 120 125Tyr Glu Ile Glu Ile Ser Asp Ala Ser Glu Val Glu
Lys Glu Ile Asn 130 135 140Lys Ala Gln Asn Ala Ile Ala Gly Glu Gly
Met Gly Ile Ser His Glu145 150 155 160Leu Ile Thr Leu Glu Ile Ser
Ser Arg Asp Val Pro Asp Leu Thr Leu 165 170 175Ile Asp Leu Pro Gly
Ile Thr Arg Val Ala Val Gly Asn Gln Pro Ala 180 185 190Asp Ile Gly
Tyr Lys Ile Lys Thr Leu Ile Lys Lys Tyr Ile Gln Arg 195 200 205Gln
Glu Thr Ile Ser Leu Val Val Val Pro Ser Asn Val Asp Ile Ala 210 215
220Thr Thr Glu Ala Leu Ser Met Ala Gln Glu Val Asp Pro Glu Gly
Asp225 230 235 240Arg Thr Ile Gly Ile Leu Thr Lys Pro Asp Leu Val
Asp Lys Gly Thr 245 250 255Glu Asp Lys Val Val Asp Val Val Arg Asn
Leu Val Phe His Leu Lys 260 265 270Lys Gly Tyr Met Ile Val Lys Cys
Arg Gly Gln Gln Glu Ile Gln Asp 275 280 285Gln Leu Ser Leu Ser Glu
Ala Leu Gln Arg Glu Lys Ile Phe Phe Glu 290 295 300Asn His Pro Tyr
Phe Arg Asp Leu Leu Glu Glu Gly Lys Ala Thr Val305 310 315 320Pro
Cys Leu Ala Glu Lys Leu Thr Ser Glu Leu Ile Thr His Ile Cys 325 330
335Lys Ser Leu Pro Leu Leu Glu Asn Gln Ile Lys Glu Thr His Gln Arg
340 345 350Ile Thr Glu Glu Leu Gln Lys Tyr Gly Val Asp Ile Pro Glu
Asp Glu 355 360 365Asn Glu Lys Met Phe Phe Leu Ile Asp Lys Val Asn
Ala Phe Asn Gln 370 375 380Asp Ile Thr Ala Leu Met Gln Gly Glu Glu
Thr Val Gly Glu Glu Asp385 390 395 400Ile Arg Leu Phe Thr Arg Leu
Arg His Glu Phe His Lys Trp Ser Thr 405 410 415Ile Ile Glu Asn Asn
Phe Gln Glu Gly His Lys Ile Leu Ser Arg Lys 420 425 430Ile Gln Lys
Phe Glu Asn Gln Tyr Arg Gly Arg Glu Leu Pro Gly Phe 435 440 445Val
Asn Tyr Arg Thr Phe Glu Thr Ile Val Lys Gln Gln Ile Lys Ala 450 455
460Leu Glu Glu Pro Ala Val Asp Met Leu His Thr Val Thr Asp Met
Val465 470 475 480Arg Leu Ala Phe Thr Asp Val Ser Ile Lys Asn Phe
Glu Glu Phe Phe 485 490 495Asn Leu His Arg Thr Ala Lys Ser Lys Ile
Glu Asp Ile Arg Ala Glu 500 505 510Gln Glu Arg Glu Gly Glu Lys Leu
Ile Arg Leu His Phe Gln Met Glu 515 520 525Gln Ile Val Tyr Cys Gln
Asp Gln Val Tyr Arg Gly Ala Leu Gln Lys 530 535 540Val Arg Glu Lys
Glu Leu Glu Glu Glu Lys Lys Lys Lys Ser Trp Asp545 550 555 560Phe
Gly Ala Phe Gln Ser Ser Ser Ala Thr Asp Ser Ser Met Glu Glu 565 570
575Ile Phe Gln His Leu Met Ala Tyr His Gln Glu Ala Ser Lys Arg Ile
580 585 590Ser Ser His Ile Pro Leu Ile Ile Gln Phe Phe Met Leu Gln
Thr Tyr 595 600 605Gly Gln Gln Leu Gln Lys Ala Met Leu Gln Leu Leu
Gln Asp Lys Asp 610 615 620Thr Tyr Ser Trp Leu Leu Lys Glu Arg Ser
Asp Thr Ser Asp Lys Arg625 630 635 640Lys Phe Leu Lys Glu Arg Leu
Ala Arg Leu Thr Gln Ala Arg Arg Arg 645 650 655Leu Ala Gln Phe Pro
Gly 660435889DNAHomo sapiens 43gctgccagct gagttttttt gctgctttga
gtctcagttt tctttctttc ctagagtctc 60tgaagccaca gatctcttaa gaactttctg
tctccaaacc gtggctgctc gataaatcag 120acagaacagt taatcctcaa
tttaagcctg atctaacccc tagaaacaga tatagaacaa 180tggaagtgac
aacaagattg acatggaatg atgaaaatca tctgcgcaag ctgcttggaa
240atgtttcttt gagtcttctc tataagtcta gtgttcatgg aggtagcatt
gaagatatgg 300ttgaaagatg cagccgtcag ggatgtacta taacaatggc
ttacattgat tacaatatga 360ttgtagcctt tatgcttgga aattatatta
atttacatga aagttctaca gagccaaatg 420attccctatg gttttcactt
caaaagaaaa atgacaccac tgaaatagaa actttactct 480taaatacagc
accaaaaatt attgatgagc aactggtgtg tcgtttatcg aaaacggata
540ttttcattat atgtcgagat aataaaattt atctagataa aatgataaca
agaaacttga 600aactaaggtt ttatggccac cgtcagtatt tggaatgtga
agtttttcga gttgaaggaa 660ttaaggataa cctagacgac ataaagagga
taattaaagc cagagagcac agaaataggc 720ttctagcaga catcagagac
tataggccct atgcagactt ggtttcagaa attcgtattc 780ttttggtggg
tccagttggg tctggaaagt ccagtttttt caattcagtc aagtctattt
840ttcatggcca tgtgactggc caagccgtag tggggtctga tatcaccagc
ataaccgagc 900ggtataggat atattctgtt aaagatggaa aaaatggaaa
atctctgcca tttatgttgt 960gtgacactat ggggctagat ggggcagaag
gagcaggact gtgcatggat gacattcccc 1020acatcttaaa aggttgtatg
ccagacagat atcagtttaa ttcccgtaaa ccaattacac 1080ctgagcattc
tacttttatc acctctccat ctctgaagga caggattcac tgtgtggctt
1140atgtcttaga catcaactct attgacaatc tctactctaa aatgttggca
aaagtgaagc 1200aagttcacaa agaagtatta aactgtggta tagcatatgt
ggccttgctt actaaagtgg 1260atgattgcag tgaggttctt caagacaact
ttttaaacat gagtagatct atgacttctc 1320aaagccgggt catgaatgtc
cataaaatgc taggcattcc tatttccaat attttgatgg 1380ttggaaacta
tgcttcagat ttggaactgg accccatgaa ggatattctc atcctctctg
1440cactgaggca gatgctgcgg gctgcagatg attttttaga agatttgcct
cttgaggaaa 1500ctggtgcaat tgagagagcg ttacagccct gcatttgaga
taagttgcct tgattctgac 1560atttggccca gcctgtactg gtgtgccgca
atgagagtca atctctattg acagcctgct 1620tcagattttg cttttgttcg
ttttgccttc tgtccttgga acagtcatat ctcaagttca 1680aaggccaaaa
cctgagaagc ggtgggctaa gataggtcct actgcaaacc acccctccat
1740atttccgtac catttacaat tcagtttctg tgacatcttt ttaaaccact
ggaggaaaaa 1800tgagatattc tctaatttat tcttctataa cactctatat
agagctatgt gagtactaat 1860cacattgaat aatagttata aaattattgt
atagacatct gcttcttaaa cagattgtga 1920gttctttgag aaacagcgtg
gattttactt atctgtgtat tcacagagct tagcacagtg 1980cctggtaatg
agcaagcata cttgccatta cttttccttc ccactctctc caacatcaca
2040ttcactttaa atttttctgt atatagaaag gaaaactagc ctgggcaaca
tgatgaaacc 2100ccatctccac tgcaaaaaaa aaaaaaaaaa ataagaaaga
acaaaacaaa ccccacaaaa 2160attagctggg tatgatggca cgtgcctgta
gtcccagtta ctcaggatga ttgattgagc 2220cttggaggtg gaggctacag
tgagctgaga ttgtgccact gtactctagc cagggagaaa 2280gagtgagatc
ctggctcaaa aaaaccaaat aaaacaaaac aaacaaacga aaaacagaaa
2340ggaagactga aagagaatga aaagctgggg agaggaaata aaaataaaga
aggaagagtg 2400tttcatttat atctgaatga aaatatgaat gactctaagt
aattgaatta attaaaatga 2460gccaactttt ttttaacaat ttacatttta
tttctatggg aaaaaataaa tattcctctt 2520ctaacaaacc catgcttgat
tttcattaat tgaattccaa atcatcctag ccatgtgtcc 2580ttccatttag
gttactgggg caaatcagta agaaagttct tatatttatg ctccaaataa
2640ttctgaagtc ctcttactag ctgtgaaagc tagtactatt aagaaagaaa
acaaaattcc 2700caaaagatag ctttcacttt tttttttcct taaagacttc
ctaattctct tctccaaatt 2760cttagtcttc ttcaaaataa tatgctttgg
ttcaatagtt atccacattc tgacagtcta 2820atttagtttt aatcagaatt
atactcatct tttgggtagt catagatatt aagaaagcaa 2880gagtttctta
tgtccagtta tggaatattt cctaaagcaa ggctgcaggt gaagttgtgc
2940tcaagtgaat gttcaggaga cacaattcag tggaagaaat taagtcttta
aaaaagacct 3000aggaatagga gaaccatgga aattgaggag gtaggcctac
aagtagatat tgggaacaaa 3060attagagagg caaccagaaa aagttatttt
aggctcacca gagttgttct tattgcacag 3120taacacacca atataccaaa
acagcaggta ttgcagtaga gaaagagttt aataattgaa 3180tggcagaaaa
atgaggaagg ttgaggaaac ctcaaatcta cctccctgct gagtctaagt
3240ttaggatttt taagagaaag gcaggtaagg tgctgaaggt ctggagctgc
tgatttgttg 3300gggtataggg aatgaaatga aacatacaga gatgaaaact
ggaagttttt ttttgtttgt 3360tttgtttttt ttttgttgtt gttttttttt
ttttttgttt ttttgctgag tcaattcctt 3420ggagggggtc ttcagactga
ctggtgtcag cagacccatg ggattccaag atctggaaaa 3480ctttttagat
agaaacttga tgtttcttaa cgttacatat attatcttat agaaataact
3540aagggaagtt agtgccttgt gaccacatct atgtgacttt taggcagtaa
gaaactataa 3600ggaaaggagc taacagtcat gctgtaagta gctacaggga
attggcttaa agggcaagtt 3660ggttagtact tagctgtgtt tttattcaaa
gtctacattt tatgtagtgg ttaatgtttg 3720ctgttcatta ggatggtttc
acagttacca tacaaatgta gaagcaacag gtccaaaaag 3780tagggcatga
ttttctccat gtaatccagg gagaaaacaa gccatgacca ttgttggttg
3840ggagactgaa ggtgattgaa ggttcaccat catcctcacc aacttttggg
ccataattca 3900cccaaccctt tggtggagcc tgaaaaaaat ctgggcagaa
tgtaggactt ctttattttg 3960tttaaagggg taacacagag tgcccttatg
aaggagttgg agatcctgca aggaagagaa 4020ggagtgaagg agagatcaag
agagagaaac aatgaggaac atttcatttg acccaacatc 4080ctttaggagc
ataaatgttg acactaagtt atcccttttg tgctaaaatg gacagtattg
4140gcaaaatgat accacaactt cttattctct ggctctatat tgctttggaa
acacttaaac 4200atcaaatgga gttaaataca tatttgaaat ttaggttagg
aaatattggt gaggaggcct 4260caaaaagggg gaaacatctt ttgtctggga
ggatattttc cattttgtgg atttccctga 4320tctttttcta ccaccctgag
gggtggtggg aattatcatt ttgctacatt ttagaggtca 4380tccaggattt
ttgaaacttt acattcttta cggttaagca agatgtacag ctcagtcaaa
4440gacactaaat tcttcttaga aaaatagtgc taaggagtat agcagatgac
ctatatgtgt 4500gttggctggg agaatatcat cttaaagtga gagtgatgtt
gtggagacag ttgaaatgtc 4560aatgctagag cctctgtggt gtgaatgggc
acgttaggtt gttgcattag aaagtgactg 4620tttctgacag aaatttgtag
ctttgtgcaa actcacccac catctacctc aataaaatat 4680agagaaaaga
aaaatagagc agtttgagtt ctatgaggta tgcaggccca gagagacata
4740agtatgttcc tttagtcttg cttcctgtgt gccacactgc ccctccacaa
ccatagctgg 4800gggcaattgt ttaaagtcat tttgttcccg actagctgcc
ttgcacatta tcttcatttt 4860cctggaattt gatacagaga gcaatttata
gccaattgat agcttatgct gtttcaatgt 4920aaattcgtgg taaataactt
aggaactgcc tcttcttttt ctttgaaaac ctacttataa 4980ctgttgctaa
taagaatgtg tattgttcag gacaacttgt ctccatacag ttgggttgta
5040accctcatgc ttggcccaaa taaactctct acttatatca gtttttccta
cacttcttcc 5100ttttaggtca acaataccaa gaggggttac tgtgctgggt
aatgtgtaaa cttgtgtctt 5160gtttagaaag ataaatttaa agactatcac
attgcttttt cataaaacaa gacaggtcta 5220caattaattt attttgacgc
aaattgatag gggggccaag taagccccat atgcttaatg 5280atcagctgat
gaataatcat ctcctagcaa cataactcaa tctaatgcta aggtacccac
5340aagatggcaa ggctgatcaa agtcgtcatg gaatcctgca accaaaagcc
atgggaattt 5400ggaagccctc aaatcccatt cctaatctga tgagtctatg
gaccaatttg tggaggacag 5460tagattaaat agatctgatt tttgccatca
atgtaaggag gataaaaact tgcataccaa 5520ttgtacaccc ttgcaaaatc
tttctctgat gttggagaaa atgggccagt gagatcatgg 5580atatagaagt
acagtcaatg ttcagctgta ccctcccaca atcccacttc cttcctcaac
5640acaattcaaa caaatagact cagactgttt caggctccag gacaggaagt
gcagtgtagg 5700caaaattgca aaaattgagg gcacaggggt ggaggtgggg
gggttgaata acaagctgtg 5760ctaaataatt acgtgtaaat atattttttc
atttttaaaa attgatttct tttgcacatt 5820ccatgacaat atatgtcaca
tttttaaaat aaatgcaaag aagcatacat ccaaaaaaaa 5880aaaaaaaaa
588944452PRTHomo sapiens 44Met Glu Val Thr Thr Arg Leu Thr Trp Asn
Asp Glu Asn His Leu Arg1 5 10 15Lys Leu Leu Gly Asn Val Ser Leu Ser
Leu Leu Tyr Lys Ser Ser Val 20 25 30His Gly Gly Ser Ile Glu Asp Met
Val Glu Arg Cys Ser Arg Gln Gly 35 40 45Cys Thr Ile Thr Met Ala Tyr
Ile Asp Tyr Asn Met Ile Val Ala Phe 50 55 60Met Leu Gly Asn Tyr Ile
Asn Leu His Glu Ser Ser Thr Glu Pro Asn65 70 75 80Asp Ser Leu Trp
Phe Ser Leu Gln Lys Lys Asn Asp Thr Thr Glu Ile 85 90 95Glu Thr Leu
Leu Leu Asn Thr Ala Pro Lys Ile Ile Asp Glu Gln Leu 100 105 110Val
Cys Arg Leu Ser Lys Thr Asp Ile Phe Ile Ile Cys Arg Asp Asn 115 120
125Lys Ile Tyr Leu Asp Lys Met Ile Thr Arg Asn Leu Lys Leu Arg Phe
130 135 140Tyr Gly His Arg Gln Tyr Leu Glu Cys Glu Val Phe Arg Val
Glu Gly145 150 155 160Ile Lys Asp Asn Leu Asp Asp Ile Lys Arg Ile
Ile Lys Ala Arg Glu 165 170 175His Arg Asn Arg Leu Leu Ala Asp Ile
Arg Asp Tyr Arg Pro Tyr Ala 180 185 190Asp Leu Val Ser Glu Ile Arg
Ile Leu Leu Val Gly Pro Val Gly Ser 195 200 205Gly Lys Ser Ser Phe
Phe Asn Ser Val Lys Ser Ile Phe His Gly His 210 215 220Val Thr Gly
Gln Ala Val Val Gly Ser Asp Ile Thr Ser Ile Thr Glu225 230 235
240Arg Tyr Arg Ile Tyr Ser Val Lys Asp Gly Lys Asn Gly Lys Ser Leu
245 250 255Pro Phe Met Leu Cys Asp Thr Met Gly Leu Asp Gly Ala Glu
Gly Ala 260 265 270Gly Leu Cys Met Asp Asp Ile Pro His Ile Leu Lys
Gly Cys Met Pro 275 280 285Asp Arg Tyr Gln Phe Asn Ser Arg Lys Pro
Ile Thr Pro Glu His Ser 290 295 300Thr Phe Ile Thr Ser Pro Ser Leu
Lys Asp Arg Ile His Cys Val Ala305 310 315 320Tyr Val Leu Asp Ile
Asn Ser Ile Asp Asn Leu Tyr Ser Lys Met Leu 325 330 335Ala Lys Val
Lys Gln Val His Lys Glu Val Leu Asn Cys Gly Ile Ala 340 345 350Tyr
Val Ala Leu Leu Thr Lys Val Asp Asp Cys Ser Glu Val Leu Gln 355 360
365Asp Asn Phe Leu Asn Met Ser Arg Ser Met Thr Ser Gln Ser Arg Val
370 375 380Met Asn Val His Lys Met Leu Gly Ile Pro Ile Ser Asn Ile
Leu Met385 390 395 400Val Gly Asn Tyr Ala Ser Asp Leu Glu Leu Asp
Pro Met Lys Asp Ile 405 410 415Leu Ile Leu Ser Ala Leu Arg Gln Met
Leu Arg Ala Ala Asp Asp Phe 420 425 430Leu Glu Asp Leu Pro Leu Glu
Glu Thr Gly Ala Ile Glu Arg Ala Leu 435 440 445Gln Pro Cys Ile
450454759DNAHomo sapiens 45tagttattaa agttcctatg cagctccgcc
tcgcgtccgg cctcatttcc tcggaaaatc 60cctgctttcc ccgctcgcca cgccctcctc
ctacccggct ttaaagctag tgaggcacag 120cctgcgggga acgtagctag
ctgcaagcag aggccggcat gaccaccgag cagcgacgca 180gcctgcaagc
cttccaggat tatatccgga agaccctgga ccctacctac atcctgagct
240acatggcccc ctggtttagg gaggaagagg tgcagtatat tcaggctgag
aaaaacaaca 300agggcccaat ggaggctgcc acactttttc tcaagttcct
gttggagctc caggaggaag 360gctggttccg tggctttttg gatgccctag
accatgcagg ttattctgga ctttatgaag 420ccattgaaag ttgggatttc
aaaaaaattg aaaagttgga ggagtataga ttacttttaa 480aacgtttaca
accagaattt aaaaccagaa ttatcccaac cgatatcatt tctgatctgt
540ctgaatgttt aattaatcag gaatgtgaag aaattctaca gatttgctct
actaagggga 600tgatggcagg tgcagagaaa ttggtggaat gccttctcag
atcagacaag gaaaactggc 660ccaaaacttt gaaacttgct ttggagaaag
aaaggaacaa gttcagtgaa ctgtggattg 720tagagaaagg tataaaagat
gttgaaacag aagatcttga ggataagatg gaaacttctg 780acatacagat
tttctaccaa
gaagatccag aatgccagaa tcttagtgag aattcatgtc 840caccttcaga
agtgtctgat acaaacttgt acagcccatt taaaccaaga aattaccaat
900tagagcttgc tttgcctgct atgaaaggaa aaaacacaat aatatgtgct
cctacaggtt 960gtggaaaaac ctttgtttca ctgcttatat gtgaacatca
tcttaaaaaa ttcccacaag 1020gacaaaaggg gaaagttgtc ttttttgcga
atcagatccc agtgtatgaa cagcagaaat 1080ctgtattctc aaaatacttt
gaaagacatg ggtatagagt tacaggcatt tctggagcaa 1140cagctgagaa
tgtcccagtg gaacagattg ttgagaacaa tgacatcatc attttaactc
1200cacagattct tgtgaacaac cttaaaaagg gaacgattcc atcactatcc
atctttactt 1260tgatgatatt tgatgaatgc cacaacacta gtaaacaaca
cccgtacaat atgatcatgt 1320ttaattatct agatcagaaa cttggaggat
cttcaggccc actgccccag gtcattgggc 1380tgactgcctc ggttggtgtt
ggggatgcca aaaacacaga tgaagccttg gattatatct 1440gcaagctgtg
tgcttctctt gatgcgtcag tgatagcaac agtcaaacac aatctggagg
1500aactggagca agttgtttat aagccccaga agtttttcag gaaagtggaa
tcacggatta 1560gcgacaaatt taaatacatc atagctcagc tgatgaggga
cacagagagt ctggcaaaga 1620gaatctgcaa agacctcgaa aacttatctc
aaattcaaaa tagggaattt ggaacacaga 1680aatatgaaca atggattgtt
acagttcaga aagcatgcat ggtgttccag atgccagaca 1740aagatgaaga
gagcaggatt tgtaaagccc tgtttttata cacttcacat ttgcggaaat
1800ataatgatgc cctcattatc agtgagcatg cacgaatgaa agatgctctg
gattacttga 1860aagacttctt cagcaatgtc cgagcagcag gattcgatga
gattgagcaa gatcttactc 1920agagatttga agaaaagctg caggaactag
aaagtgtttc cagggatccc agcaatgaga 1980atcctaaact tgaagacctc
tgcttcatct tacaagaaga gtaccactta aacccagaga 2040caataacaat
tctctttgtg aaaaccagag cacttgtgga cgctttaaaa aattggattg
2100aaggaaatcc taaactcagt tttctaaaac ctggcatatt gactggacgt
ggcaaaacaa 2160atcagaacac aggaatgacc ctcccggcac agaagtgtat
attggatgca ttcaaagcca 2220gtggagatca caatattctg attgccacct
cagttgctga tgaaggcatt gacattgcac 2280agtgcaatct tgtcatcctt
tatgagtatg tgggcaatgt catcaaaatg atccaaacca 2340gaggcagagg
aagagcaaga ggtagcaagt gcttccttct gactagtaat gctggtgtaa
2400ttgaaaaaga acaaataaac atgtacaaag aaaaaatgat gaatgactct
attttacgcc 2460ttcagacatg ggacgaagca gtatttaggg aaaagattct
gcatatacag actcatgaaa 2520aattcatcag agatagtcaa gaaaaaccaa
aacctgtacc tgataaggaa aataaaaaac 2580tgctctgcag aaagtgcaaa
gccttggcat gttacacagc tgacgtaaga gtgatagagg 2640aatgccatta
cactgtgctt ggagatgctt ttaaggaatg ctttgtgagt agaccacatc
2700ccaagccaaa gcagttttca agttttgaaa aaagagcaaa gatattctgt
gcccgacaga 2760actgcagcca tgactgggga atccatgtga agtacaagac
atttgagatt ccagttataa 2820aaattgaaag ttttgtggtg gaggatattg
caactggagt tcagacactg tactcgaagt 2880ggaaggactt tcattttgag
aagataccat ttgatccagc agaaatgtcc aaatgatatc 2940aggtcctcaa
tcttcagcta cagggaatga gtaactttga gtggagaaga aacaaacata
3000gtgggtataa tcatggatcg cttgtacccc tgtgaaaata tattttttaa
aaatatcttt 3060agcagtttgt actatattat atatgcaaag cacaaatgag
tgaatcacag cactgagtat 3120tttgtaggcc aacagagctc atagtacttg
ggaaaaatta aaaagcctca tttctagcct 3180tctttttaga gtcaactgcc
aacaaacaca cagtaatcac tctgtacaca ctgggataga 3240tgaatgaatg
gaatgttggg aatttttatc tccctttgtc tccttaacct actgtaaact
3300ggcttttgcc cttaacaatc tactgaaatt gttcttttga aggttaccag
tgactctggt 3360tgccaaatcc actgggcact tcttaacctt ctatttgacc
tctgcgcatt tggccctgtt 3420gagcactctt cttgaagctc tccctgggct
tctctctctt ctagttctat tctagtcttt 3480ttttattgag tcctcctctt
tgctgatccc ttccaagggt tcaatatata tacatgtata 3540tactgtacat
atgtatatgt aactaatata catacataca ggtatgtata tgtaatggtt
3600atatgtactc atgttcctgg tgtagcaacg tgtggtatgg ctacacagag
aacatgagaa 3660cataaagcca tttttatgct tactactaaa agctgtccac
tgtagagttg ctgtatgtag 3720caatgtgtat ccactctaca gtggtcagct
tttagtagag agcataaaaa tgataaaata 3780cttcttgaaa acttagttta
ctatacatct tgccctatta atatgttctc ttaacgtgtg 3840ccattgttct
ctttgaccat tttcctataa tgatgttgat gttcaacacc tggactgaat
3900gtctgttctc agatcccttg gatgttacag atgaggcagt ctgactgtcc
tttctacttg 3960aaagattaga atatgtatcc aaatggcatt cacgtgtcac
ttagcaaggt ttgctgatgc 4020ttcaaagagc ttagtttgcg gtttcctgga
cgtggaaaca agtatctgag ttccctggag 4080atcaacggga tgaggtgtta
cagctgcctc cctcttcatg caatctggtg agcagtggtg 4140caggcgggga
gccagagaaa cttgccagtt atataacttc tctttggctt ttcttcatct
4200gtaaaacaag gataatactg aactgtaagg gttagtggag agtttttaat
taaaagaatg 4260tgtgaaaagt acatgacaca gtagttgctt gataatagtt
actagtagta gtattcttac 4320taagacccaa tacaaatgga ttatttaaac
caagtttatg agttggtttt ttttcatttt 4380ctatttgtat tttattaaga
gtgtcttttc ttatgtgatt ttttttaatt gctatttgat 4440atggtttggc
tatatgtccc cacccaaatc tcatcttgaa ttataatccc catgtgtcaa
4500gggagggacc tgacgggagg tgattggatc acgggggcag ttgtccccat
gctgttcttg 4560ggatagtgag ttagttctca tgagatctga tggttttata
agtgtttgac aattcctcct 4620ttacacacac tctctctctc atctgctgcc
atgtaagact tgcctgcttc cccttctgcc 4680atgattgtaa gtttcctgag
gcctcctcag ccatgtggaa ctgtgaatct attaagcctc 4740ttttctttat
aaatgaaaa 475946925PRTHomo sapiens 46Met Thr Thr Glu Gln Arg Arg
Ser Leu Gln Ala Phe Gln Asp Tyr Ile1 5 10 15Arg Lys Thr Leu Asp Pro
Thr Tyr Ile Leu Ser Tyr Met Ala Pro Trp 20 25 30Phe Arg Glu Glu Glu
Val Gln Tyr Ile Gln Ala Glu Lys Asn Asn Lys 35 40 45Gly Pro Met Glu
Ala Ala Thr Leu Phe Leu Lys Phe Leu Leu Glu Leu 50 55 60Gln Glu Glu
Gly Trp Phe Arg Gly Phe Leu Asp Ala Leu Asp His Ala65 70 75 80Gly
Tyr Ser Gly Leu Tyr Glu Ala Ile Glu Ser Trp Asp Phe Lys Lys 85 90
95Ile Glu Lys Leu Glu Glu Tyr Arg Leu Leu Leu Lys Arg Leu Gln Pro
100 105 110Glu Phe Lys Thr Arg Ile Ile Pro Thr Asp Ile Ile Ser Asp
Leu Ser 115 120 125Glu Cys Leu Ile Asn Gln Glu Cys Glu Glu Ile Leu
Gln Ile Cys Ser 130 135 140Thr Lys Gly Met Met Ala Gly Ala Glu Lys
Leu Val Glu Cys Leu Leu145 150 155 160Arg Ser Asp Lys Glu Asn Trp
Pro Lys Thr Leu Lys Leu Ala Leu Glu 165 170 175Lys Glu Arg Asn Lys
Phe Ser Glu Leu Trp Ile Val Glu Lys Gly Ile 180 185 190Lys Asp Val
Glu Thr Glu Asp Leu Glu Asp Lys Met Glu Thr Ser Asp 195 200 205Ile
Gln Ile Phe Tyr Gln Glu Asp Pro Glu Cys Gln Asn Leu Ser Glu 210 215
220Asn Ser Cys Pro Pro Ser Glu Val Ser Asp Thr Asn Leu Tyr Ser
Pro225 230 235 240Phe Lys Pro Arg Asn Tyr Gln Leu Glu Leu Ala Leu
Pro Ala Met Lys 245 250 255Gly Lys Asn Thr Ile Ile Cys Ala Pro Thr
Gly Cys Gly Lys Thr Phe 260 265 270Val Ser Leu Leu Ile Cys Glu His
His Leu Lys Lys Phe Pro Gln Gly 275 280 285Gln Lys Gly Lys Val Val
Phe Phe Ala Asn Gln Ile Pro Val Tyr Glu 290 295 300Gln Gln Lys Ser
Val Phe Ser Lys Tyr Phe Glu Arg His Gly Tyr Arg305 310 315 320Val
Thr Gly Ile Ser Gly Ala Thr Ala Glu Asn Val Pro Val Glu Gln 325 330
335Ile Val Glu Asn Asn Asp Ile Ile Ile Leu Thr Pro Gln Ile Leu Val
340 345 350Asn Asn Leu Lys Lys Gly Thr Ile Pro Ser Leu Ser Ile Phe
Thr Leu 355 360 365Met Ile Phe Asp Glu Cys His Asn Thr Ser Lys Gln
His Pro Tyr Asn 370 375 380Met Ile Met Phe Asn Tyr Leu Asp Gln Lys
Leu Gly Gly Ser Ser Gly385 390 395 400Pro Leu Pro Gln Val Ile Gly
Leu Thr Ala Ser Val Gly Val Gly Asp 405 410 415Ala Lys Asn Thr Asp
Glu Ala Leu Asp Tyr Ile Cys Lys Leu Cys Ala 420 425 430Ser Leu Asp
Ala Ser Val Ile Ala Thr Val Lys His Asn Leu Glu Glu 435 440 445Leu
Glu Gln Val Val Tyr Lys Pro Gln Lys Phe Phe Arg Lys Val Glu 450 455
460Ser Arg Ile Ser Asp Lys Phe Lys Tyr Ile Ile Ala Gln Leu Met
Arg465 470 475 480Asp Thr Glu Ser Leu Ala Lys Arg Ile Cys Lys Asp
Leu Glu Asn Leu 485 490 495Ser Gln Ile Gln Asn Arg Glu Phe Gly Thr
Gln Lys Tyr Glu Gln Trp 500 505 510Ile Val Thr Val Gln Lys Ala Cys
Met Val Phe Gln Met Pro Asp Lys 515 520 525Asp Glu Glu Ser Arg Ile
Cys Lys Ala Leu Phe Leu Tyr Thr Ser His 530 535 540Leu Arg Lys Tyr
Asn Asp Ala Leu Ile Ile Ser Glu His Ala Arg Met545 550 555 560Lys
Asp Ala Leu Asp Tyr Leu Lys Asp Phe Phe Ser Asn Val Arg Ala 565 570
575Ala Gly Phe Asp Glu Ile Glu Gln Asp Leu Thr Gln Arg Phe Glu Glu
580 585 590Lys Leu Gln Glu Leu Glu Ser Val Ser Arg Asp Pro Ser Asn
Glu Asn 595 600 605Pro Lys Leu Glu Asp Leu Cys Phe Ile Leu Gln Glu
Glu Tyr His Leu 610 615 620Asn Pro Glu Thr Ile Thr Ile Leu Phe Val
Lys Thr Arg Ala Leu Val625 630 635 640Asp Ala Leu Lys Asn Trp Ile
Glu Gly Asn Pro Lys Leu Ser Phe Leu 645 650 655Lys Pro Gly Ile Leu
Thr Gly Arg Gly Lys Thr Asn Gln Asn Thr Gly 660 665 670Met Thr Leu
Pro Ala Gln Lys Cys Ile Leu Asp Ala Phe Lys Ala Ser 675 680 685Gly
Asp His Asn Ile Leu Ile Ala Thr Ser Val Ala Asp Glu Gly Ile 690 695
700Asp Ile Ala Gln Cys Asn Leu Val Ile Leu Tyr Glu Tyr Val Gly
Asn705 710 715 720Val Ile Lys Met Ile Gln Thr Arg Gly Arg Gly Arg
Ala Arg Gly Ser 725 730 735Lys Cys Phe Leu Leu Thr Ser Asn Ala Gly
Val Ile Glu Lys Glu Gln 740 745 750Ile Asn Met Tyr Lys Glu Lys Met
Met Asn Asp Ser Ile Leu Arg Leu 755 760 765Gln Thr Trp Asp Glu Ala
Val Phe Arg Glu Lys Ile Leu His Ile Gln 770 775 780Thr His Glu Lys
Phe Ile Arg Asp Ser Gln Glu Lys Pro Lys Pro Val785 790 795 800Pro
Asp Lys Glu Asn Lys Lys Leu Leu Cys Arg Lys Cys Lys Ala Leu 805 810
815Ala Cys Tyr Thr Ala Asp Val Arg Val Ile Glu Glu Cys His Tyr Thr
820 825 830Val Leu Gly Asp Ala Phe Lys Glu Cys Phe Val Ser Arg Pro
His Pro 835 840 845Lys Pro Lys Gln Phe Ser Ser Phe Glu Lys Arg Ala
Lys Ile Phe Cys 850 855 860Ala Arg Gln Asn Cys Ser His Asp Trp Gly
Ile His Val Lys Tyr Lys865 870 875 880Thr Phe Glu Ile Pro Val Ile
Lys Ile Glu Ser Phe Val Val Glu Asp 885 890 895Ile Ala Thr Gly Val
Gln Thr Leu Tyr Ser Lys Trp Lys Asp Phe His 900 905 910Phe Glu Lys
Ile Pro Phe Asp Pro Ala Glu Met Ser Lys 915 920 92547863DNAHomo
sapiens 47caaggttcag agtcacccat ctcagcaagc ccagaagtat ctgcaatatc
tacgatggcc 60tcgccctttg ctttactgat ggtcctggtg gtgctcagct gcaagtcaag
ctgctctctg 120ggctgtgatc tccctgagac ccacagcctg gataacagga
ggaccttgat gctcctggca 180caaatgagca gaatctctcc ttcctcctgt
ctgatggaca gacatgactt tggatttccc 240caggaggagt ttgatggcaa
ccagttccag aaggctccag ccatctctgt cctccatgag 300ctgatccagc
agatcttcaa cctctttacc acaaaagatt catctgctgc ttgggatgag
360gacctcctag acaaattctg caccgaactc taccagcagc tgaatgactt
ggaagcctgt 420gtgatgcagg aggagagggt gggagaaact cccctgatga
atgcggactc catcttggct 480gtgaagaaat acttccgaag aatcactctc
tatctgacag agaagaaata cagcccttgt 540gcctgggagg ttgtcagagc
agaaatcatg agatccctct ctttatcaac aaacttgcaa 600gaaagattaa
ggaggaagga ataacatctg gtccaacatg aaaacaattc ttattgactc
660atacaccagg tcacgctttc atgaattctg tcatttcaaa gactctcacc
cctgctataa 720ctatgaccat gctgataaac tgatttatct atttaaatat
ttatttaact attcataaga 780tttaaattat ttttgttcat ataacgtcat
gtgcaccttt acactgtggt tagtgtaata 840aaacatgttc cttatattta ctc
86348189PRTHomo sapiens 48Met Ala Ser Pro Phe Ala Leu Leu Met Val
Leu Val Val Leu Ser Cys1 5 10 15Lys Ser Ser Cys Ser Leu Gly Cys Asp
Leu Pro Glu Thr His Ser Leu 20 25 30Asp Asn Arg Arg Thr Leu Met Leu
Leu Ala Gln Met Ser Arg Ile Ser 35 40 45Pro Ser Ser Cys Leu Met Asp
Arg His Asp Phe Gly Phe Pro Gln Glu 50 55 60Glu Phe Asp Gly Asn Gln
Phe Gln Lys Ala Pro Ala Ile Ser Val Leu65 70 75 80His Glu Leu Ile
Gln Gln Ile Phe Asn Leu Phe Thr Thr Lys Asp Ser 85 90 95Ser Ala Ala
Trp Asp Glu Asp Leu Leu Asp Lys Phe Cys Thr Glu Leu 100 105 110Tyr
Gln Gln Leu Asn Asp Leu Glu Ala Cys Val Met Gln Glu Glu Arg 115 120
125Val Gly Glu Thr Pro Leu Met Asn Ala Asp Ser Ile Leu Ala Val Lys
130 135 140Lys Tyr Phe Arg Arg Ile Thr Leu Tyr Leu Thr Glu Lys Lys
Tyr Ser145 150 155 160Pro Cys Ala Trp Glu Val Val Arg Ala Glu Ile
Met Arg Ser Leu Ser 165 170 175Leu Ser Thr Asn Leu Gln Glu Arg Leu
Arg Arg Lys Glu 180 185491143DNAHomo sapiens 49gagaacctgg
agcctaaggt ttaggctcac ccatttcaac cagtctagca gcatctgcaa 60catctacaat
ggccttgacc tttgctttac tggtggccct cctggtgctc agctgcaagt
120caagctgctc tgtgggctgt gatctgcctc aaacccacag cctgggtagc
aggaggacct 180tgatgctcct ggcacagatg aggagaatct ctcttttctc
ctgcttgaag gacagacatg 240actttggatt tccccaggag gagtttggca
accagttcca aaaggctgaa accatccctg 300tcctccatga gatgatccag
cagatcttca atctcttcag cacaaaggac tcatctgctg 360cttgggatga
gaccctccta gacaaattct acactgaact ctaccagcag ctgaatgacc
420tggaagcctg tgtgatacag ggggtggggg tgacagagac tcccctgatg
aaggaggact 480ccattctggc tgtgaggaaa tacttccaaa gaatcactct
ctatctgaaa gagaagaaat 540acagcccttg tgcctgggag gttgtcagag
cagaaatcat gagatctttt tctttgtcaa 600caaacttgca agaaagttta
agaagtaagg aatgaaaact ggttcaacat ggaaatgatt 660ttcattgatt
cgtatgccag ctcacctttt tatgatctgc catttcaaag actcatgttt
720ctgctatgac catgacacga tttaaatctt ttcaaatgtt tttaggagta
ttaatcaaca 780ttgtattcag ctcttaaggc actagtccct tacagaggac
catgctgact gatccattat 840ctatttaaat atttttaaaa tattatttat
ttaactattt ataaaacaac ttatttttgt 900tcatattatg tcatgtgcac
ctttgcacag tggttaatgt aataaaatat gttctttgta 960tttggtaaat
ttattttgtg ttgttcattg aacttttgct atggaaactt ttgtacttgt
1020ttattcttta aaatgaaatt ccaagcctaa ttgtgcaacc tgattacaga
ataactggta 1080cacttcattt atccatcaat attatattca agatataagt
aaaaataaac tttctgtaaa 1140cca 114350188PRTHomo sapiens 50Met Ala
Leu Thr Phe Ala Leu Leu Val Ala Leu Leu Val Leu Ser Cys1 5 10 15Lys
Ser Ser Cys Ser Val Gly Cys Asp Leu Pro Gln Thr His Ser Leu 20 25
30Gly Ser Arg Arg Thr Leu Met Leu Leu Ala Gln Met Arg Arg Ile Ser
35 40 45Leu Phe Ser Cys Leu Lys Asp Arg His Asp Phe Gly Phe Pro Gln
Glu 50 55 60Glu Phe Gly Asn Gln Phe Gln Lys Ala Glu Thr Ile Pro Val
Leu His65 70 75 80Glu Met Ile Gln Gln Ile Phe Asn Leu Phe Ser Thr
Lys Asp Ser Ser 85 90 95Ala Ala Trp Asp Glu Thr Leu Leu Asp Lys Phe
Tyr Thr Glu Leu Tyr 100 105 110Gln Gln Leu Asn Asp Leu Glu Ala Cys
Val Ile Gln Gly Val Gly Val 115 120 125Thr Glu Thr Pro Leu Met Lys
Glu Asp Ser Ile Leu Ala Val Arg Lys 130 135 140Tyr Phe Gln Arg Ile
Thr Leu Tyr Leu Lys Glu Lys Lys Tyr Ser Pro145 150 155 160Cys Ala
Trp Glu Val Val Arg Ala Glu Ile Met Arg Ser Phe Ser Leu 165 170
175Ser Thr Asn Leu Gln Glu Ser Leu Arg Ser Lys Glu 180
18551982DNAHomo sapiens 51agaaaaccta gaggccgaag ttcaaggtta
tccatctcaa gtagcctagc aatatttgca 60acatcccaat ggccctgtcc ttttctttac
tgatggccgt gctggtgctc agctacaaat 120ccatctgttc tctgggctgt
gatctgcctc agacccacag cctgggtaat aggagggcct 180tgatactcct
ggcacaaatg ggaagaatct ctcatttctc ctgcctgaag gacagacatg
240atttcggatt ccccgaggag gagtttgatg gccaccagtt ccagaaggct
caagccatct 300ctgtcctcca tgagatgatc cagcagacct tcaatctctt
cagcacagag gactcatctg 360ctgcttggga acagagcctc ctagaaaaat
tttccactga actttaccag caactgaatg 420acctggaagc atgtgtgata
caggaggttg gggtggaaga gactcccctg atgaatgagg 480actccatcct
ggctgtgagg aaatacttcc aaagaatcac tctttatcta acagagaaga
540aatacagccc ttgtgcctgg gaggttgtca gagcagaaat catgagatcc
ctctcgtttt 600caacaaactt gcaaaaaaga ttaaggagga aggattgaaa
cctggttcaa catggaaatg 660atcctgattg actaatacat tatctcacac
tttcatgagt tcttccattt caaagactca 720cttctataac caccacgagt
tgaatcaaaa ttttcaaatg ttttcagcag tgtgaagaag 780cttggtgtat
acctgtgcag gcactagtcc tttacagatg acaatgctga tgtctctgtt
840catctattta tttaaatatt tatttatttt taaaatttaa attatttttt
atgtgatatc 900atgagtacct
ttacattgtg gtgaatgtaa caatatatgt tcttcatatt tagccaatat
960attaatttcc tttttcatta aa 98252189PRTHomo sapiens 52Met Ala Leu
Ser Phe Ser Leu Leu Met Ala Val Leu Val Leu Ser Tyr1 5 10 15Lys Ser
Ile Cys Ser Leu Gly Cys Asp Leu Pro Gln Thr His Ser Leu 20 25 30Gly
Asn Arg Arg Ala Leu Ile Leu Leu Ala Gln Met Gly Arg Ile Ser 35 40
45His Phe Ser Cys Leu Lys Asp Arg His Asp Phe Gly Phe Pro Glu Glu
50 55 60Glu Phe Asp Gly His Gln Phe Gln Lys Ala Gln Ala Ile Ser Val
Leu65 70 75 80His Glu Met Ile Gln Gln Thr Phe Asn Leu Phe Ser Thr
Glu Asp Ser 85 90 95Ser Ala Ala Trp Glu Gln Ser Leu Leu Glu Lys Phe
Ser Thr Glu Leu 100 105 110Tyr Gln Gln Leu Asn Asp Leu Glu Ala Cys
Val Ile Gln Glu Val Gly 115 120 125Val Glu Glu Thr Pro Leu Met Asn
Glu Asp Ser Ile Leu Ala Val Arg 130 135 140Lys Tyr Phe Gln Arg Ile
Thr Leu Tyr Leu Thr Glu Lys Lys Tyr Ser145 150 155 160Pro Cys Ala
Trp Glu Val Val Arg Ala Glu Ile Met Arg Ser Leu Ser 165 170 175Phe
Ser Thr Asn Leu Gln Lys Arg Leu Arg Arg Lys Asp 180 18553700DNAHomo
sapiens 53gcccaaggtt cagggtcact caatctcaac agcccagaag catctgcaac
ctccccaatg 60gccttgccct ttgttttact gatggccctg gtggtgctca actgcaagtc
aatctgttct 120ctgggctgtg atctgcctca gacccacagc ctgagtaaca
ggaggacttt gatgataatg 180gcacaaatgg gaagaatctc tcctttctcc
tgcctgaagg acagacatga ctttggattt 240cctcaggagg agtttgatgg
caaccagttc cagaaggctc aagccatctc tgtcctccat 300gagatgatcc
agcagacctt caatctcttc agcacaaagg actcatctgc tacttgggat
360gagacacttc tagacaaatt ctacactgaa ctttaccagc agctgaatga
cctggaagcc 420tgtatgatgc aggaggttgg agtggaagac actcctctga
tgaatgtgga ctctatcctg 480actgtgagaa aatactttca aagaatcacc
ctctatctga cagagaagaa atacagccct 540tgtgcatggg aggttgtcag
agcagaaatc atgagatcct tctctttatc agcaaacttg 600caagaaagat
taaggaggaa ggaatgaaaa ctggttcaac atcgaaatga ttctcattga
660ctagtacacc atttcacact tcttgagttc tgccgtttca 70054189PRTHomo
sapiens 54Met Ala Leu Pro Phe Val Leu Leu Met Ala Leu Val Val Leu
Asn Cys1 5 10 15Lys Ser Ile Cys Ser Leu Gly Cys Asp Leu Pro Gln Thr
His Ser Leu 20 25 30Ser Asn Arg Arg Thr Leu Met Ile Met Ala Gln Met
Gly Arg Ile Ser 35 40 45Pro Phe Ser Cys Leu Lys Asp Arg His Asp Phe
Gly Phe Pro Gln Glu 50 55 60Glu Phe Asp Gly Asn Gln Phe Gln Lys Ala
Gln Ala Ile Ser Val Leu65 70 75 80His Glu Met Ile Gln Gln Thr Phe
Asn Leu Phe Ser Thr Lys Asp Ser 85 90 95Ser Ala Thr Trp Asp Glu Thr
Leu Leu Asp Lys Phe Tyr Thr Glu Leu 100 105 110Tyr Gln Gln Leu Asn
Asp Leu Glu Ala Cys Met Met Gln Glu Val Gly 115 120 125Val Glu Asp
Thr Pro Leu Met Asn Val Asp Ser Ile Leu Thr Val Arg 130 135 140Lys
Tyr Phe Gln Arg Ile Thr Leu Tyr Leu Thr Glu Lys Lys Tyr Ser145 150
155 160Pro Cys Ala Trp Glu Val Val Arg Ala Glu Ile Met Arg Ser Phe
Ser 165 170 175Leu Ser Ala Asn Leu Gln Glu Arg Leu Arg Arg Lys Glu
180 18555570DNAHomo sapiens 55atggctttgc cttttgcttt actgatggcc
ctggtggtgc tcagctgcaa gtcaagctgc 60tctctggact gtgatctgcc tcagacccac
agcctgggtc acaggaggac catgatgctc 120ctggcacaaa tgaggagaat
ctctcttttc tcctgtctga aggacagaca tgacttcaga 180tttccccagg
aggagtttga tggcaaccag ttccagaagg ctgaagccat ctctgtcctc
240catgaggtga ttcagcagac cttcaacctc ttcagcacaa aggactcatc
tgttgcttgg 300gatgagaggc ttctagacaa actctatact gaactttacc
agcagctgaa tgacctggaa 360gcctgtgtga tgcaggaggt gtgggtggga
gggactcccc tgatgaatga ggactccatc 420ctggctgtga gaaaatactt
ccaaagaatc actctctacc tgacagagaa aaagtacagc 480ccttgtgcct
gggaggttgt cagagcagaa atcatgagat ccttctcttc atcaagaaac
540ttgcaagaaa ggttaaggag gaaggaataa 57056189PRTHomo sapiens 56Met
Ala Leu Pro Phe Ala Leu Leu Met Ala Leu Val Val Leu Ser Cys1 5 10
15Lys Ser Ser Cys Ser Leu Asp Cys Asp Leu Pro Gln Thr His Ser Leu
20 25 30Gly His Arg Arg Thr Met Met Leu Leu Ala Gln Met Arg Arg Ile
Ser 35 40 45Leu Phe Ser Cys Leu Lys Asp Arg His Asp Phe Arg Phe Pro
Gln Glu 50 55 60Glu Phe Asp Gly Asn Gln Phe Gln Lys Ala Glu Ala Ile
Ser Val Leu65 70 75 80His Glu Val Ile Gln Gln Thr Phe Asn Leu Phe
Ser Thr Lys Asp Ser 85 90 95Ser Val Ala Trp Asp Glu Arg Leu Leu Asp
Lys Leu Tyr Thr Glu Leu 100 105 110Tyr Gln Gln Leu Asn Asp Leu Glu
Ala Cys Val Met Gln Glu Val Trp 115 120 125Val Gly Gly Thr Pro Leu
Met Asn Glu Asp Ser Ile Leu Ala Val Arg 130 135 140Lys Tyr Phe Gln
Arg Ile Thr Leu Tyr Leu Thr Glu Lys Lys Tyr Ser145 150 155 160Pro
Cys Ala Trp Glu Val Val Arg Ala Glu Ile Met Arg Ser Phe Ser 165 170
175Ser Ser Arg Asn Leu Gln Glu Arg Leu Arg Arg Lys Glu 180
18557736DNAHomo sapiens 57tacccacctc aggtagccta gtgatatttg
caaaatccca atggcccggt ccttttcttt 60actgatggtc gtgctggtac tcagctacaa
atccatctgc tctctgggct gtgatctgcc 120tcagacccac agcctgcgta
ataggagggc cttgatactc ctggcacaaa tgggaagaat 180ctctcctttc
tcctgcttga aggacagaca tgaattcaga ttcccagagg aggagtttga
240tggccaccag ttccagaaga ctcaagccat ctctgtcctc catgagatga
tccagcagac 300cttcaatctc ttcagcacag aggactcatc tgctgcttgg
gaacagagcc tcctagaaaa 360attttccact gaactttacc agcaactgaa
tgacctggaa gcatgtgtga tacaggaggt 420tggggtggaa gagactcccc
tgatgaatga ggacttcatc ctggctgtga ggaaatactt 480ccaaagaatc
actctttatc taatggagaa gaaatacagc ccttgtgcct gggaggttgt
540cagagcagaa atcatgagat ccttctcttt ttcaacaaac ttgaaaaaag
gattaaggag 600gaaggattga aaactggttc atcatggaaa tgattctcat
tgactaatgc atcatctcac 660actttcatga gttcttccat ttcaaagact
cacttctata accaccacaa gttaatcaaa 720atttccaaat gttttc
73658189PRTHomo sapiens 58Met Ala Arg Ser Phe Ser Leu Leu Met Val
Val Leu Val Leu Ser Tyr1 5 10 15Lys Ser Ile Cys Ser Leu Gly Cys Asp
Leu Pro Gln Thr His Ser Leu 20 25 30Arg Asn Arg Arg Ala Leu Ile Leu
Leu Ala Gln Met Gly Arg Ile Ser 35 40 45Pro Phe Ser Cys Leu Lys Asp
Arg His Glu Phe Arg Phe Pro Glu Glu 50 55 60Glu Phe Asp Gly His Gln
Phe Gln Lys Thr Gln Ala Ile Ser Val Leu65 70 75 80His Glu Met Ile
Gln Gln Thr Phe Asn Leu Phe Ser Thr Glu Asp Ser 85 90 95Ser Ala Ala
Trp Glu Gln Ser Leu Leu Glu Lys Phe Ser Thr Glu Leu 100 105 110Tyr
Gln Gln Leu Asn Asp Leu Glu Ala Cys Val Ile Gln Glu Val Gly 115 120
125Val Glu Glu Thr Pro Leu Met Asn Glu Asp Phe Ile Leu Ala Val Arg
130 135 140Lys Tyr Phe Gln Arg Ile Thr Leu Tyr Leu Met Glu Lys Lys
Tyr Ser145 150 155 160Pro Cys Ala Trp Glu Val Val Arg Ala Glu Ile
Met Arg Ser Phe Ser 165 170 175Phe Ser Thr Asn Leu Lys Lys Gly Leu
Arg Arg Lys Asp 180 185591039DNAHomo sapiens 59accagctcag
cagcatccac aacatctaca atggccttga ctttttattt actggtggcc 60ctagtggtgc
tcagctacaa gtcattcagc tctctgggct gtgatctgcc tcagactcac
120agcctgggta acaggagggc cttgatactc ctggcacaaa tgcgaagaat
ctctcctttc 180tcctgcctga aggacagaca tgactttgaa ttcccccagg
aggagtttga tgataaacag 240ttccagaagg ctcaagccat ctctgtcctc
catgagatga tccagcagac cttcaacctc 300ttcagcacaa aggactcatc
tgctgctttg gatgagaccc ttctagatga attctacatc 360gaacttgacc
agcagctgaa tgacctggag tcctgtgtga tgcaggaagt gggggtgata
420gagtctcccc tgatgtacga ggactccatc ctggctgtga ggaaatactt
ccaaagaatc 480actctatatc tgacagagaa gaaatacagc tcttgtgcct
gggaggttgt cagagcagaa 540atcatgagat ccttctcttt atcaatcaac
ttgcaaaaaa gattgaagag taaggaatga 600gacctggtac aacacggaaa
tgattcttat agactaatac agcagctcac acttcgacaa 660gttgtgctct
ttcaaagacc cttgtttctg ccaaaaccat gctatgaatt gaatcaaatg
720tgtcaagtgt tttcaggagt gttaagcaac atcctgttca gctgtatggg
cactagtccc 780ttacagatga ccatgctgat ggatctattc atctatttat
ttaaatcttt atttagttaa 840ctatctatag ggcttaaatt agttttgttc
atattatatt atgtgaactt ttacattgtg 900aattgtgtaa caaaaacatg
ttctttatat ttattatttt gccttgttta ttaaattttt 960actatagaaa
aattctttat ttattcttta aaattgaact ccaaccctga ttgtgcaaac
1020tgattaaaga atggatggt 103960189PRTHomo sapiens 60Met Ala Leu Thr
Phe Tyr Leu Leu Val Ala Leu Val Val Leu Ser Tyr1 5 10 15Lys Ser Phe
Ser Ser Leu Gly Cys Asp Leu Pro Gln Thr His Ser Leu 20 25 30Gly Asn
Arg Arg Ala Leu Ile Leu Leu Ala Gln Met Arg Arg Ile Ser 35 40 45Pro
Phe Ser Cys Leu Lys Asp Arg His Asp Phe Glu Phe Pro Gln Glu 50 55
60Glu Phe Asp Asp Lys Gln Phe Gln Lys Ala Gln Ala Ile Ser Val Leu65
70 75 80His Glu Met Ile Gln Gln Thr Phe Asn Leu Phe Ser Thr Lys Asp
Ser 85 90 95Ser Ala Ala Leu Asp Glu Thr Leu Leu Asp Glu Phe Tyr Ile
Glu Leu 100 105 110Asp Gln Gln Leu Asn Asp Leu Glu Ser Cys Val Met
Gln Glu Val Gly 115 120 125Val Ile Glu Ser Pro Leu Met Tyr Glu Asp
Ser Ile Leu Ala Val Arg 130 135 140Lys Tyr Phe Gln Arg Ile Thr Leu
Tyr Leu Thr Glu Lys Lys Tyr Ser145 150 155 160Ser Cys Ala Trp Glu
Val Val Arg Ala Glu Ile Met Arg Ser Phe Ser 165 170 175Leu Ser Ile
Asn Leu Gln Lys Arg Leu Lys Ser Lys Glu 180 18561963DNAHomo sapiens
61caaggttatc catctcaagt agcctagcaa tatttgcaac atcccaatgg ccctgtcctt
60ttctttactt atggccgtgc tggtgctcag ctacaaatcc atctgttctc tgggctgtga
120tctgcctcag acccacagcc tcggtaatag gagggccttg atactcctgg
gacaaatggg 180aagaatctct cctttctcct gcctgaagga cagacatgat
ttccgaatcc cccaggagga 240gtttgatggc aaccagttcc agaaggctca
agccatctct gtcctccatg agatgatcca 300gcagaccttc aatctcttca
gcacagagga ctcatctgct gcttgggaac agagcctcct 360agaaaaattt
tccactgaac tttaccagca actgaatgac ctggaagcat gtgtgataca
420ggaggttggg gtggaagaga ctcccctgat gaatgaggac tccatcctgg
ctgtgaggaa 480atacttccaa agaatcactc tttatctaat agagaggaaa
tacagccctt gtgcctggga 540ggttgtcaga gcagaaatca tgagatccct
ctcgttttca acaaacttgc aaaaaagatt 600aaggaggaag gattgaaaac
tggttcaaca tggcaatgat cctgattgac taatacatta 660tctcacactt
tcatgagttc ttccatttca aagactcact tctataacca cgacgcgttg
720aatcaaaatt ttcaaatgtt ttcagcagtg taaagaagtg tcgtgtatac
ctgtgcaggc 780actagtcctt tacagatgac cattctgatg tctctgttca
tcttttgttt aaatatttat 840ttaattattt ttaaaattta tgtaatatca
tgagtcgctt tacattgtgg ttaatgtaac 900aatatatgtt cttcatattt
agccaatata ttaatttcct ttttcattaa atttttacta 960tac 96362189PRTHomo
sapiens 62Met Ala Leu Ser Phe Ser Leu Leu Met Ala Val Leu Val Leu
Ser Tyr1 5 10 15Lys Ser Ile Cys Ser Leu Gly Cys Asp Leu Pro Gln Thr
His Ser Leu 20 25 30Gly Asn Arg Arg Ala Leu Ile Leu Leu Gly Gln Met
Gly Arg Ile Ser 35 40 45Pro Phe Ser Cys Leu Lys Asp Arg His Asp Phe
Arg Ile Pro Gln Glu 50 55 60Glu Phe Asp Gly Asn Gln Phe Gln Lys Ala
Gln Ala Ile Ser Val Leu65 70 75 80His Glu Met Ile Gln Gln Thr Phe
Asn Leu Phe Ser Thr Glu Asp Ser 85 90 95Ser Ala Ala Trp Glu Gln Ser
Leu Leu Glu Lys Phe Ser Thr Glu Leu 100 105 110Tyr Gln Gln Leu Asn
Asp Leu Glu Ala Cys Val Ile Gln Glu Val Gly 115 120 125Val Glu Glu
Thr Pro Leu Met Asn Glu Asp Ser Ile Leu Ala Val Arg 130 135 140Lys
Tyr Phe Gln Arg Ile Thr Leu Tyr Leu Ile Glu Arg Lys Tyr Ser145 150
155 160Pro Cys Ala Trp Glu Val Val Arg Ala Glu Ile Met Arg Ser Leu
Ser 165 170 175Phe Ser Thr Asn Leu Gln Lys Arg Leu Arg Arg Lys Asp
180 18563705DNAHomo sapiens 63agagaaccta gagcccaagg ttcagagtca
cccatctcag caagcccaga agcatctgca 60atatctatga tggcctcgcc ctttgcttta
ctgatggccc tggtggtgct cagctgcaag 120tcaagctgct ctctgggctg
tgatctccct gagacccaca gcctggataa caggaggacc 180ttgatgctcc
tggcacaaat gagcagaatc tctccttcct cctgtctgat ggacagacat
240gactttggat ttccccagga ggagtttgat ggcaaccagt tccagaaggc
tccagccatc 300tctgtcctcc atgagctgat ccagcagatc ttcaacctct
ttaccacaaa agattcatct 360gctgcttggg atgaggacct cctagacaaa
ttctgcaccg aactctacca gcagctgaat 420gacttggaag cctgtgtgat
gcaggaggag agggtgggag aaactcccct gatgaatgcg 480gactccatct
tggctgtgaa gaaatacttc cgaagaatca ctctctatct gacagagaag
540aaatacagcc cttgtgcctg ggaggttgtc agagcagaaa tcatgagatc
cctctcttta 600tcaacaaact tgcaagaaag attaaggagg aaggaataac
acctggtcca acatgaaaca 660attcttattg actcatatac caggtcacgc
tttcatgaat tctgc 70564190PRTHomo sapiens 64Met Met Ala Ser Pro Phe
Ala Leu Leu Met Ala Leu Val Val Leu Ser1 5 10 15Cys Lys Ser Ser Cys
Ser Leu Gly Cys Asp Leu Pro Glu Thr His Ser 20 25 30Leu Asp Asn Arg
Arg Thr Leu Met Leu Leu Ala Gln Met Ser Arg Ile 35 40 45Ser Pro Ser
Ser Cys Leu Met Asp Arg His Asp Phe Gly Phe Pro Gln 50 55 60Glu Glu
Phe Asp Gly Asn Gln Phe Gln Lys Ala Pro Ala Ile Ser Val65 70 75
80Leu His Glu Leu Ile Gln Gln Ile Phe Asn Leu Phe Thr Thr Lys Asp
85 90 95Ser Ser Ala Ala Trp Asp Glu Asp Leu Leu Asp Lys Phe Cys Thr
Glu 100 105 110Leu Tyr Gln Gln Leu Asn Asp Leu Glu Ala Cys Val Met
Gln Glu Glu 115 120 125Arg Val Gly Glu Thr Pro Leu Met Asn Ala Asp
Ser Ile Leu Ala Val 130 135 140Lys Lys Tyr Phe Arg Arg Ile Thr Leu
Tyr Leu Thr Glu Lys Lys Tyr145 150 155 160Ser Pro Cys Ala Trp Glu
Val Val Arg Ala Glu Ile Met Arg Ser Leu 165 170 175Ser Leu Ser Thr
Asn Leu Gln Glu Arg Leu Arg Arg Lys Glu 180 185 19065778DNAHomo
sapiens 65gttacccctc atcaaccagc ccagcagcat cttcgggatt cccaatggca
ttgccctttg 60ctttaatgat ggccctggtg gtgctcagct gcaagtcaag ctgctctctg
ggctgtaatc 120tgtctcaaac ccacagcctg aataacagga ggactttgat
gctcatggca caaatgagga 180gaatctctcc tttctcctgc ctgaaggaca
gacatgactt tgaatttccc caggaggaat 240ttgatggcaa ccagttccag
aaagctcaag ccatctctgt cctccatgag atgatgcagc 300agaccttcaa
tctcttcagc acaaagaact catctgctgc ttgggatgag accctcctag
360aaaaattcta cattgaactt ttccagcaaa tgaatgacct ggaagcctgt
gtgatacagg 420aggttggggt ggaagagact cccctgatga atgaggactc
catcctggct gtgaagaaat 480acttccaaag aatcactctt tatctgatgg
agaagaaata cagcccttgt gcctgggagg 540ttgtcagagc agaaatcatg
agatccctct ctttttcaac aaacttgcaa aaaagattaa 600ggaggaagga
ttgaaaactg gttcatcatg gaaatgattc tcattgacta atacatcatc
660tcacactttc atgagttctt ccatttcaaa gactcacttc tcctataacc
accacaagtt 720gaatcaaaat tttcaaatgt tttcaggagt gtaaagaagc
atcatgtata cctgtgca 77866189PRTHomo sapiens 66Met Ala Leu Pro Phe
Ala Leu Met Met Ala Leu Val Val Leu Ser Cys1 5 10 15Lys Ser Ser Cys
Ser Leu Gly Cys Asn Leu Ser Gln Thr His Ser Leu 20 25 30Asn Asn Arg
Arg Thr Leu Met Leu Met Ala Gln Met Arg Arg Ile Ser 35 40 45Pro Phe
Ser Cys Leu Lys Asp Arg His Asp Phe Glu Phe Pro Gln Glu 50 55 60Glu
Phe Asp Gly Asn Gln Phe Gln Lys Ala Gln Ala Ile Ser Val Leu65 70 75
80His Glu Met Met Gln Gln Thr Phe Asn Leu Phe Ser Thr Lys Asn Ser
85 90 95Ser Ala Ala Trp Asp Glu Thr Leu Leu Glu Lys Phe Tyr Ile Glu
Leu 100 105 110Phe Gln Gln Met Asn Asp Leu Glu Ala Cys Val Ile Gln
Glu Val Gly 115 120 125Val Glu Glu Thr Pro Leu Met Asn Glu Asp Ser
Ile Leu Ala Val Lys 130 135 140Lys Tyr Phe Gln Arg Ile Thr Leu Tyr
Leu Met Glu Lys Lys Tyr Ser145 150
155 160Pro Cys Ala Trp Glu Val Val Arg Ala Glu Ile Met Arg Ser Leu
Ser 165 170 175Phe Ser Thr Asn Leu Gln Lys Arg Leu Arg Arg Lys Asp
180 18567939DNAHomo sapiens 67atcccaatgg ccctgtcctt ttctttactg
atggccgtgc tggtgctcag ctacaaatcc 60atctgttctc tgggctgtga tctgcctcag
actcacagcc tgggtaatag gagggccttg 120atactcctgg cacaaatggg
aagaatctct catttctcct gcctgaagga cagatatgat 180ttcggattcc
cccaggaggt gtttgatggc aaccagttcc agaaggctca agccatctct
240gccttccatg agatgatcca gcagaccttc aatctcttca gcacaaagga
ttcatctgct 300gcttgggatg agaccctcct agacaaattc tacattgaac
ttttccagca actgaatgac 360ctagaagcct gtgtgacaca ggaggttggg
gtggaagaga ttgccctgat gaatgaggac 420tccatcctgg ctgtgaggaa
atactttcaa agaatcactc tttatctgat ggggaagaaa 480tacagccctt
gtgcctggga ggttgtcaga gcagaaatca tgagatcctt ctctttttca
540acaaacttgc aaaaaggatt aagaaggaag gattgaaaac tcattcaaca
tggaaatgat 600cctcattgat taatacatca tctcacactt tcatgagttc
ttccatttca aagactcact 660tctataacca ccacaagttg aatcaaaatt
tcaaaatgtt ttcaggagtg taaagaagca 720tcgtgtttac ctgtgcaggc
actagtcctt tacagatgac catgctgatg tctctattca 780tctatttatt
taaatattta tttatttaac tatttttaag gtttaaatca tgttttatgt
840aatatcatgt gtacctttac attttgctta atgtaacaat atatgttctt
catatttagt 900taatatatta acttcctttt cattaaattt ttactatac
93968189PRTHomo sapiens 68Met Ala Leu Ser Phe Ser Leu Leu Met Ala
Val Leu Val Leu Ser Tyr1 5 10 15Lys Ser Ile Cys Ser Leu Gly Cys Asp
Leu Pro Gln Thr His Ser Leu 20 25 30Gly Asn Arg Arg Ala Leu Ile Leu
Leu Ala Gln Met Gly Arg Ile Ser 35 40 45His Phe Ser Cys Leu Lys Asp
Arg Tyr Asp Phe Gly Phe Pro Gln Glu 50 55 60Val Phe Asp Gly Asn Gln
Phe Gln Lys Ala Gln Ala Ile Ser Ala Phe65 70 75 80His Glu Met Ile
Gln Gln Thr Phe Asn Leu Phe Ser Thr Lys Asp Ser 85 90 95Ser Ala Ala
Trp Asp Glu Thr Leu Leu Asp Lys Phe Tyr Ile Glu Leu 100 105 110Phe
Gln Gln Leu Asn Asp Leu Glu Ala Cys Val Thr Gln Glu Val Gly 115 120
125Val Glu Glu Ile Ala Leu Met Asn Glu Asp Ser Ile Leu Ala Val Arg
130 135 140Lys Tyr Phe Gln Arg Ile Thr Leu Tyr Leu Met Gly Lys Lys
Tyr Ser145 150 155 160Pro Cys Ala Trp Glu Val Val Arg Ala Glu Ile
Met Arg Ser Phe Ser 165 170 175Phe Ser Thr Asn Leu Gln Lys Gly Leu
Arg Arg Lys Asp 180 18569980DNAHomo sapiens 69gttcaaggtt acccatctca
agtagcctag caacatttgc aacatcccaa tggccctgtc 60cttttcttta ctgatggccg
tgctggtgct cagctacaaa tccatctgtt ctctaggctg 120tgatctgcct
cagacccaca gcctgggtaa taggagggcc ttgatactcc tggcacaaat
180gggaagaatc tctcctttct cctgcctgaa ggacagacat gactttggac
ttccccagga 240ggagtttgat ggcaaccagt tccagaagac tcaagccatc
tctgtcctcc atgagatgat 300ccagcagacc ttcaatctct tcagcacaga
ggactcatct gctgcttggg aacagagcct 360cctagaaaaa ttttccactg
aactttacca gcaactgaat aacctggaag catgtgtgat 420acaggaggtt
gggatggaag agactcccct gatgaatgag gactccatcc tggctgtgag
480gaaatacttc caaagaatca ctctttatct aacagagaag aaatacagcc
cttgtgcctg 540ggaggttgtc agagcagaaa tcatgagatc tctctctttt
tcaacaaact tgcaaaaaat 600attaaggagg aaggattgaa aactggttca
acatggcaat gatcctgatt gactaataca 660ttatctcaca ctttcatgag
ttcctccatt tcaaagactc acttctataa ccaccacgag 720ttgaatcaaa
attttcaaat gttttcagca gtgtaaagaa gcgtcgtgta tacctgtgca
780ggcactagta ctttacagat gaccatgctg atgtctctgt tcatctattt
atttaaatat 840ttatttaatt atttttaaga tttaaattat ttttttatgt
aatatcatgt gtacctttac 900attgtggtga atgtaacaat atatgttctt
catatttagc caatatatta atttcctttt 960tcattaaatt tttactatac
98070189PRTHomo sapiens 70Met Ala Leu Ser Phe Ser Leu Leu Met Ala
Val Leu Val Leu Ser Tyr1 5 10 15Lys Ser Ile Cys Ser Leu Gly Cys Asp
Leu Pro Gln Thr His Ser Leu 20 25 30Gly Asn Arg Arg Ala Leu Ile Leu
Leu Ala Gln Met Gly Arg Ile Ser 35 40 45Pro Phe Ser Cys Leu Lys Asp
Arg His Asp Phe Gly Leu Pro Gln Glu 50 55 60Glu Phe Asp Gly Asn Gln
Phe Gln Lys Thr Gln Ala Ile Ser Val Leu65 70 75 80His Glu Met Ile
Gln Gln Thr Phe Asn Leu Phe Ser Thr Glu Asp Ser 85 90 95Ser Ala Ala
Trp Glu Gln Ser Leu Leu Glu Lys Phe Ser Thr Glu Leu 100 105 110Tyr
Gln Gln Leu Asn Asn Leu Glu Ala Cys Val Ile Gln Glu Val Gly 115 120
125Met Glu Glu Thr Pro Leu Met Asn Glu Asp Ser Ile Leu Ala Val Arg
130 135 140Lys Tyr Phe Gln Arg Ile Thr Leu Tyr Leu Thr Glu Lys Lys
Tyr Ser145 150 155 160Pro Cys Ala Trp Glu Val Val Arg Ala Glu Ile
Met Arg Ser Leu Ser 165 170 175Phe Ser Thr Asn Leu Gln Lys Ile Leu
Arg Arg Lys Asp 180 185711024DNAHomo sapiens 71ttcaaggtta
cccatctcaa gtagcctagc aatattggca acatcccaat ggccctgtcc 60ttttctttac
tgatggccgt gctggtgctc agctacaaat ccatctgttc tctgggctgt
120gatctgcctc agacccacag cctgggtaat aggagggcct tgatactcct
ggcacaaatg 180ggaagaatct ctcctttctc ctgcctgaag gacagacatg
actttggatt cccccaggag 240gagtttgatg gcaaccagtt ccagaaggct
caagccatct ctgtcctcca tgagatgatc 300cagcagacct tcaatctctt
cagcacaaag gactcatctg ctacttggga acagagcctc 360ctagaaaaat
tttccactga acttaaccag cagctgaatg acctggaagc ctgcgtgata
420caggaggttg gggtggaaga gactcccctg atgaatgtgg actccatcct
ggctgtgaag 480aaatacttcc aaagaatcac tctttatctg acagagaaga
aatacagccc ttgtgcctgg 540gaggttgtca gagcagaaat catgagatcc
ttctctttat caaaaatttt tcaagaaaga 600ttaaggagga aggaatgaaa
cctgtttcaa catggaaatg atctgtattg actaatacac 660cagtccacac
ttctatgact tctgccattt caaagactca tttctcctat aaccaccgca
720tgagttgaat caaaattttc agatcttttc aggagtgtaa ggaaacatca
tgtttacctg 780tgcaggcact agtcctttac agatgaccat gctgatagat
ctaattatct atctattgaa 840atatttattt atttattaga tttaaattat
ttttgtccat gtaatattat gtgtactttt 900acattgtgtt atatcaaaat
atgttattta tatttagtca atatattatt ttctttttat 960taatttttac
tattaaaact tcttatatta tttgtttatt ctttaataaa gaaataccaa 1020gccc
102472189PRTHomo sapiens 72Met Ala Leu Ser Phe Ser Leu Leu Met Ala
Val Leu Val Leu Ser Tyr1 5 10 15Lys Ser Ile Cys Ser Leu Gly Cys Asp
Leu Pro Gln Thr His Ser Leu 20 25 30Gly Asn Arg Arg Ala Leu Ile Leu
Leu Ala Gln Met Gly Arg Ile Ser 35 40 45Pro Phe Ser Cys Leu Lys Asp
Arg His Asp Phe Gly Phe Pro Gln Glu 50 55 60Glu Phe Asp Gly Asn Gln
Phe Gln Lys Ala Gln Ala Ile Ser Val Leu65 70 75 80His Glu Met Ile
Gln Gln Thr Phe Asn Leu Phe Ser Thr Lys Asp Ser 85 90 95Ser Ala Thr
Trp Glu Gln Ser Leu Leu Glu Lys Phe Ser Thr Glu Leu 100 105 110Asn
Gln Gln Leu Asn Asp Leu Glu Ala Cys Val Ile Gln Glu Val Gly 115 120
125Val Glu Glu Thr Pro Leu Met Asn Val Asp Ser Ile Leu Ala Val Lys
130 135 140Lys Tyr Phe Gln Arg Ile Thr Leu Tyr Leu Thr Glu Lys Lys
Tyr Ser145 150 155 160Pro Cys Ala Trp Glu Val Val Arg Ala Glu Ile
Met Arg Ser Phe Ser 165 170 175Leu Ser Lys Ile Phe Gln Glu Arg Leu
Arg Arg Lys Glu 180 18573840DNAHomo sapiens 73acattctaac tgcaaccttt
cgaagccttt gctctggcac aacaggtagt aggcgacact 60gttcgtgttg tcaacatgac
caacaagtgt ctcctccaaa ttgctctcct gttgtgcttc 120tccactacag
ctctttccat gagctacaac ttgcttggat tcctacaaag aagcagcaat
180tttcagtgtc agaagctcct gtggcaattg aatgggaggc ttgaatactg
cctcaaggac 240aggatgaact ttgacatccc tgaggagatt aagcagctgc
agcagttcca gaaggaggac 300gccgcattga ccatctatga gatgctccag
aacatctttg ctattttcag acaagattca 360tctagcactg gctggaatga
gactattgtt gagaacctcc tggctaatgt ctatcatcag 420ataaaccatc
tgaagacagt cctggaagaa aaactggaga aagaagattt caccagggga
480aaactcatga gcagtctgca cctgaaaaga tattatggga ggattctgca
ttacctgaag 540gccaaggagt acagtcactg tgcctggacc atagtcagag
tggaaatcct aaggaacttt 600tacttcatta acagacttac aggttacctc
cgaaactgaa gatctcctag cctgtgcctc 660tgggactgga caattgcttc
aagcattctt caaccagcag atgctgttta agtgactgat 720ggctaatgta
ctgcatatga aaggacacta gaagattttg aaatttttat taaattatga
780gttattttta tttatttaaa ttttattttg gaaaataaat tatttttggt
gcaaaagtca 84074187PRTHomo sapiens 74Met Thr Asn Lys Cys Leu Leu
Gln Ile Ala Leu Leu Leu Cys Phe Ser1 5 10 15Thr Thr Ala Leu Ser Met
Ser Tyr Asn Leu Leu Gly Phe Leu Gln Arg 20 25 30Ser Ser Asn Phe Gln
Cys Gln Lys Leu Leu Trp Gln Leu Asn Gly Arg 35 40 45Leu Glu Tyr Cys
Leu Lys Asp Arg Met Asn Phe Asp Ile Pro Glu Glu 50 55 60Ile Lys Gln
Leu Gln Gln Phe Gln Lys Glu Asp Ala Ala Leu Thr Ile65 70 75 80Tyr
Glu Met Leu Gln Asn Ile Phe Ala Ile Phe Arg Gln Asp Ser Ser 85 90
95Ser Thr Gly Trp Asn Glu Thr Ile Val Glu Asn Leu Leu Ala Asn Val
100 105 110Tyr His Gln Ile Asn His Leu Lys Thr Val Leu Glu Glu Lys
Leu Glu 115 120 125Lys Glu Asp Phe Thr Arg Gly Lys Leu Met Ser Ser
Leu His Leu Lys 130 135 140Arg Tyr Tyr Gly Arg Ile Leu His Tyr Leu
Lys Ala Lys Glu Tyr Ser145 150 155 160His Cys Ala Trp Thr Ile Val
Arg Val Glu Ile Leu Arg Asn Phe Tyr 165 170 175Phe Ile Asn Arg Leu
Thr Gly Tyr Leu Arg Asn 180 185751514DNAHomo sapiens 75gatctggtaa
acctgaagca aatatagaaa cctatagggc ctgacttcct acataaagta 60aggagggtaa
aaatggaggc tagaataagg gttaaaattt tgcttctaga acagagaaaa
120tgattttttt catatatata tgaatatata ttatatatac acatatatac
atatattcac 180tatagtgtgt atacataaat atataatata tatattgtta
gtgtagtgtg tgtctgatta 240tttacatgca tatagtatat acacttatga
ctttagtacc cagacgtttt tcatttgatt 300aagcattcat ttgtattgac
acagctgaag tttactggag tttagctgaa gtctaatgca 360aaattaatag
attgttgtca tcctcttaag gtcataggga gaacacacaa atgaaaacag
420taaaagaaac tgaaagtaca gagaaatgtt cagaaaatga aaaccatgtg
tttcctatta 480aaagccatgc atacaagcaa tgtcttcaga aaacctaggg
tccaaggtta agccatatcc 540cagctcagta aagccaggag catcctcatt
tcccaatggc cctcctgttc cctctactgg 600cagccctagt gatgaccagc
tatagccctg ttggatctct gggctgtgat ctgcctcaga 660accatggcct
acttagcagg aacaccttgg tgcttctgca ccaaatgagg agaatctccc
720ctttcttgtg tctcaaggac agaagagact tcaggttccc ccaggagatg
gtaaaaggga 780gccagttgca gaaggcccat gtcatgtctg tcctccatga
gatgctgcag cagatcttca 840gcctcttcca cacagagcgc tcctctgctg
cctggaacat gaccctccta gaccaactcc 900acactggact tcatcagcaa
ctgcaacacc tggagacctg cttgctgcag gtagtgggag 960aaggagaatc
tgctggggca attagcagcc ctgcactgac cttgaggagg tacttccagg
1020gaatccgtgt ctacctgaaa gagaagaaat acagcgactg tgcctgggaa
gttgtcagaa 1080tggaaatcat gaaatccttg ttcttatcaa caaacatgca
agaaagactg agaagtaaag 1140atagagacct gggctcatct tgaaatgatt
ctcattgatt aatttgccat ataacacttg 1200cacatgtgac tctggtcaat
tcaaaagact cttatttcgg ctttaatcac agaattgact 1260gaattagttc
tgcaaatact ttgtcggtat attaagccag tatatgttaa aaagacttag
1320gttcaggggc atcagtccct aagatgttat ttatttttac tcatttattt
attcttacat 1380tttatcatat ttatactatt tatattctta tataacaaat
gtttgccttt acattgtatt 1440aagataacaa aacatgttca gctttccatt
tggttaaata ttgtattttg ttatttatta 1500aattattttc aaac
151476195PRTHomo sapiens 76Met Ala Leu Leu Phe Pro Leu Leu Ala Ala
Leu Val Met Thr Ser Tyr1 5 10 15Ser Pro Val Gly Ser Leu Gly Cys Asp
Leu Pro Gln Asn His Gly Leu 20 25 30Leu Ser Arg Asn Thr Leu Val Leu
Leu His Gln Met Arg Arg Ile Ser 35 40 45Pro Phe Leu Cys Leu Lys Asp
Arg Arg Asp Phe Arg Phe Pro Gln Glu 50 55 60Met Val Lys Gly Ser Gln
Leu Gln Lys Ala His Val Met Ser Val Leu65 70 75 80His Glu Met Leu
Gln Gln Ile Phe Ser Leu Phe His Thr Glu Arg Ser 85 90 95Ser Ala Ala
Trp Asn Met Thr Leu Leu Asp Gln Leu His Thr Gly Leu 100 105 110His
Gln Gln Leu Gln His Leu Glu Thr Cys Leu Leu Gln Val Val Gly 115 120
125Glu Gly Glu Ser Ala Gly Ala Ile Ser Ser Pro Ala Leu Thr Leu Arg
130 135 140Arg Tyr Phe Gln Gly Ile Arg Val Tyr Leu Lys Glu Lys Lys
Tyr Ser145 150 155 160Asp Cys Ala Trp Glu Val Val Arg Met Glu Ile
Met Lys Ser Leu Phe 165 170 175Leu Ser Thr Asn Met Gln Glu Arg Leu
Arg Ser Lys Asp Arg Asp Leu 180 185 190Gly Ser Ser 195771240DNAHomo
sapiens 77cacattgttc tgatcatctg aagatcagct attagaagag aaagatcagt
taagtccttt 60ggacctgatc agcttgatac aagaactact gatttcaact tctttggctt
aattctctcg 120gaaacgatga aatatacaag ttatatcttg gcttttcagc
tctgcatcgt tttgggttct 180cttggctgtt actgccagga cccatatgta
aaagaagcag aaaaccttaa gaaatatttt 240aatgcaggtc attcagatgt
agcggataat ggaactcttt tcttaggcat tttgaagaat 300tggaaagagg
agagtgacag aaaaataatg cagagccaaa ttgtctcctt ttacttcaaa
360ctttttaaaa actttaaaga tgaccagagc atccaaaaga gtgtggagac
catcaaggaa 420gacatgaatg tcaagttttt caatagcaac aaaaagaaac
gagatgactt cgaaaagctg 480actaattatt cggtaactga cttgaatgtc
caacgcaaag caatacatga actcatccaa 540gtgatggctg aactgtcgcc
agcagctaaa acagggaagc gaaaaaggag tcagatgctg 600tttcgaggtc
gaagagcatc ccagtaatgg ttgtcctgcc tgcaatattt gaattttaaa
660tctaaatcta tttattaata tttaacatta tttatatggg gaatatattt
ttagactcat 720caatcaaata agtatttata atagcaactt ttgtgtaatg
aaaatgaata tctattaata 780tatgtattat ttataattcc tatatcctgt
gactgtctca cttaatcctt tgttttctga 840ctaattaggc aaggctatgt
gattacaagg ctttatctca ggggccaact aggcagccaa 900cctaagcaag
atcccatggg ttgtgtgttt atttcacttg atgatacaat gaacacttat
960aagtgaagtg atactatcca gttactgccg gtttgaaaat atgcctgcaa
tctgagccag 1020tgctttaatg gcatgtcaga cagaacttga atgtgtcagg
tgaccctgat gaaaacatag 1080catctcagga gatttcatgc ctggtgcttc
caaatattgt tgacaactgt gactgtaccc 1140aaatggaaag taactcattt
gttaaaatta tcaatatcta atatatatga ataaagtgta 1200agttcacaac
aaaaaaaaaa aaaaaaaaaa aaaaaaaaaa 124078166PRTHomo sapiens 78Met Lys
Tyr Thr Ser Tyr Ile Leu Ala Phe Gln Leu Cys Ile Val Leu1 5 10 15Gly
Ser Leu Gly Cys Tyr Cys Gln Asp Pro Tyr Val Lys Glu Ala Glu 20 25
30Asn Leu Lys Lys Tyr Phe Asn Ala Gly His Ser Asp Val Ala Asp Asn
35 40 45Gly Thr Leu Phe Leu Gly Ile Leu Lys Asn Trp Lys Glu Glu Ser
Asp 50 55 60Arg Lys Ile Met Gln Ser Gln Ile Val Ser Phe Tyr Phe Lys
Leu Phe65 70 75 80Lys Asn Phe Lys Asp Asp Gln Ser Ile Gln Lys Ser
Val Glu Thr Ile 85 90 95Lys Glu Asp Met Asn Val Lys Phe Phe Asn Ser
Asn Lys Lys Lys Arg 100 105 110Asp Asp Phe Glu Lys Leu Thr Asn Tyr
Ser Val Thr Asp Leu Asn Val 115 120 125Gln Arg Lys Ala Ile His Glu
Leu Ile Gln Val Met Ala Glu Leu Ser 130 135 140Pro Ala Ala Lys Thr
Gly Lys Arg Lys Arg Ser Gln Met Leu Phe Arg145 150 155 160Gly Arg
Arg Ala Ser Gln 1657912979DNAHomo
sapiensmisc_feature(5427)..(5427)n is a, c, g, or
tmisc_feature(5460)..(5461)n is a, c, g, or
tmisc_feature(12190)..(12190)n is a, c, g, or
tmisc_feature(12192)..(12192)n is a, c, g, or
tmisc_feature(12237)..(12240)n is a, c, g, or t 79gaattccttg
agcccaggaa gctgcagtga gccacgtttg taccattgca ctccagcttg 60ggagatagag
tgagaccctg tctcaaaaaa aaaaaaaaaa aaaaaaagaa aaatttattt
120ggcttacaga tgtctgggct gtacaagaag tgtggcagca gcatctgctt
ccagtgaagg 180cattaggctc cttctactca gcagaagaca acatgcagct
ggcatgtgca gagaccacat 240ggtaagagaa gaagccaatg ggggaatgca
atggggggac ggggagacac acactttttt 300tttttttttt gagatggagt
ctcgctctgt cacccaggct ggagtgcagt ggcgcgatct 360cggctcactg
caagctctgc ctcccgggtt cacgccattc tcctgcctca gccgcccgag
420tagctgggac tacaggtgcc caccaccacg ccccgctgat tttttgtatt
tttagtagag 480atggggtttc accatgttag ccaggatggt ctcgatctcc
tgacctcgtg atctgcctgc 540cttggcttcc caaagtgctg ggattacagg
catgagccac catgcctgtc cctttttttt 600tttttttttt tgagacaggg
gctgtcaccc aggctggagt gcactggcgt ggtcatggct 660cattgcacct
ttcacctcag gctcaagaga ccctcctacc tcagcctcct cagtagctgg
720gactaaggaa gtacaccacc atgcccagct aattattttt tttaagatgc
aatctcactc 780tgttgcccag gctgcactgc agtggtacaa tctcagctca
ctgcaacctc cgcctcctgg 840gttcaagcga ttctcttgct
tcagcctcct gagtagctgg gattacagat gcctgccacc 900aggcccggct
aatttttgta tttttagtag atacagggtt tcgccatgtt ggccaggctg
960gtctcgaact cctaacctaa ggtgatccgc ccatctcggc ctcccaaagt
gctgggatta 1020caggcatgag ccaccgtgcc tggcccatgc ccagctaagt
taaaaaaatt tttggctggg 1080caaggtgatt catacctgta attccaacaa
tttgggaagc taaggcaggc agatcccatt 1140tgagctcagg aattggagac
catcttggac aatatggcga aatcctgtct ctacaaaata 1200tacaaaaatt
agctgggtgt ggtggcgctg tgcctgtagt cccagctact ccagaggctg
1260aggtgggagg atggcttgag cccgggaggc agaggttgca gtgagccaaa
atggtgacat 1320tgcactccag cctgggcaac agagccagac cctgtctcaa
aaaaaaaaaa aaaaagctct 1380aaaaatagat agtggtgata gttagttaca
caacattgtg aatgtactta aatgtcactg 1440aattatacac ttaaatgttt
aaatggtaaa ttctatctta tgtatatttt accacaattt 1500aaaaatttta
tctatttcta ttttaatgag agttttaaaa agcaggaatg gatattgaat
1560ttccttaaat actctttgga gtctattaaa gatagcattt tacttcaaaa
tccagccctg 1620gtttctgtac ctagtacttc tgtcacacca gtaaatgtta
ttgaatgaaa acaaacaaaa 1680gaccataaag acatacctct tcacgttcac
taggattggt gtcatttttt taaaaaaagg 1740aaagaaaaat aaccagtatt
ggcaacagtg tggatatata gaaatttttg tatattgctg 1800gtaggaatat
aaaataggga aaacggtttg gtggttcctc aaaaagttca acataaaccc
1860aggcgcggtg gctcatgcct gtaatcctag cactttggga ggctgaggtg
ggagaactgt 1920ttgatcccag gagtttgaga ccagcctagg caatatggtg
agacctcatc tctacaaaaa 1980attagctagg catggtggcg catgcctgta
gtcccagcta cctgggaggc taagacagaa 2040agatcacttg agcctggaag
ggagtttgag gctgcagtga gcccagattg cgccactgca 2100ctccagcctg
ggtgacagag tgatatcctg tctcaaaata aataaataaa taataaacaa
2160acacagaatt accatatgac ccagcaactc tactcctgtg tatatgccca
aaaaagctga 2220aagcccaggc actgtggctt acacctgtaa tcccagcact
ttaggaggcc gaggcacttg 2280accccaggat tttgagacca gcctgggcaa
catagtactt gttcaagaat caggctgggc 2340atggtggctc acacctgtaa
tcccagcact ttgggagggc aaggtgggag gatcacttga 2400ggccaggcat
tcgagaccag cctgggcaag atagcaatac ccacccccaa tctctacaaa
2460agcaagtaat taattaggaa attagccaaa gccaggtgcg gtggctcacg
cctgtagtcc 2520cagcactttg ggaggctgag gtgggcagat tgcttgagtc
cagagattcg agaccagcct 2580aggcaacata tcaaaacccc gtctctacta
aaagtaaaaa aagaaaaatt gcactttggg 2640aggccaaggc ggggggatca
cctgatgtca ggagttcaag accagcctgg ccaacatggt 2700gaaaccccat
ctctacaaaa atacaaaagt tagccgggca tgatggcagt gtctgtaatc
2760ccagctactc aggaggctgg gtcggaagaa tcatttgaat ccaggaggcg
gaggttgcag 2820tgaaccaaga ttgtgccatt gcattccagc ctgggtgaca
gagcaagcct ccgtctcaaa 2880aaaaaaaaaa aaaagaaaaa ttagccagat
gtggtggtgc atgcctgtgg tcccagctac 2940tcaggaggct gtggtgggag
gattgcttga gcccaagaca ctgaggctgc agtgagccat 3000gatcacacca
ctgcactcca gcctaggcaa ccagagtgag accctgtctc aaaaacaaac
3060aaacaaacaa aaaccacttt aacagggtat ggtggtgcac acctctagtc
ccagctactt 3120gggaggttga ggcagccgga tcacttgacc ccaggagatc
gaggctgcag tccagcctgg 3180gcaaccgagt gagactgtct caaaaagaaa
aaaaaaaaaa aaaggacata gcagcactat 3240tcacaaatcc aaaagttaga
aataactcag atgtccatca acagatgaat ggataaacga 3300attgtggtat
atacatataa tggactatta ttcagccatt aaaaggaatg aaatattgat
3360acaggctata aactctatga acattgaaaa cattctaagt gaaaggaaat
agacataaga 3420ggtcacattt tgcaattctt tttttttttt tttttttttt
tgagactgag tctcactctg 3480ttgcccaggc tggagcgcag tggctcgatc
tcagctcact gcaacctcca tctcccgaat 3540tcaagcaatt cttctgcctc
agcctcccga gtagctggga ttacatgtag gcatgcacca 3600ccatgcctgg
ctaatttttg tacttttagt agagatgggg tttcaccatg ttggtcaggc
3660tggtcttgaa ctccagacct caggtggtcc acccgccttg gcctcccaaa
gtgctaggat 3720tacaggtgtg agccaccatg cccggccaca tgtatggtaa
ttattgaatg tgtttggtat 3780gttcgttgtg ggtgatgaaa tattttggaa
ctagatagac gtgatggttg aacaacactg 3840tggatgcact aaatgccact
gaattgtaca ctttaaaatt gttaacttta tgttacatga 3900atttcaccta
aattaacaac aacaacaaaa aagaacttaa gacagcactt ggtttggcta
3960ttacgtagtt tcgtgacaaa cagtggtcca tctcccagag aactggcccc
aggttcctaa 4020gaaggcaaaa ggagacacag gacctctctg cactattttt
tttgcaactt cttatgagtc 4080tataattatt tcaaaataaa agtctaaaag
gaaaataaga acatgtgtga atgtggctgc 4140cccatgcctc ccaccctcag
gtctgacact cagagactga tcacctcttg agagtcctgg 4200aactcatccc
aggttttaga ccctgaatgg cctgtctggg gctggcgtct ggaggcagga
4260tcaggagcca gctcagagca tagtttaact ttcacttttc ttttctccag
aggagccagg 4320aagagagctg tgaccagcag cgtcccttat tcgcttggcc
ttggttcctg tttgcactgg 4380ctacagcagg gcactggccc ctactgtcac
cgccacctac acaaagaccc tatctctgag 4440cgctgcagcc tactgttcag
ccccaggttt gaggatggat gccctggacg cttcgaagct 4500actggatgag
gagctgtatt caagacagct gtgaggcccg aggtgggggg tggagagtgg
4560gatggtcttc agaccttgat ctacaactgc ttgccttctg cttcccatcc
acaggtatgt 4620gctgggctca cctgccatgc agaggattca gggagccagg
gtcctggtgt caggcctgca 4680gggcctgggg gccgaggtgg ccaagaactt
ggttctgatg ggtgtgggca gcctcactct 4740gcatgatccc caccccacct
gctggtccga cctggctgcc caggtaagtg tcctggggct 4800atgggctgcc
agaccaagtg gggcacggcc caagaggagt gtctttgctc aggctgcact
4860ggctctctcc ctagtttctc ctctcagagc aggacttgga aaggagcaga
gccgaggcct 4920ctcaagagct cttggctcag ctcaacagag ctgtccaggt
cgtcgtgcac acgggtgaca 4980tcactgagga cctgctgttg gacttccagg
tcagctcagg cctgcagccc tcaagagcag 5040gaagggctgg gcaatggttt
tggccctgct gatcactgtg tccacccagg tggtggtgct 5100gactgctgca
aagctggagg agcagctgaa ggtgggcacc ttgtgtcata agcatggagt
5160ttgctttctg gcggctgaca cccggggcct cgtggggtga gtaagactgc
ctgcccagcc 5220taccatatta cagccagcaa ctggcctcat gctgtcctca
gctccaggct tgctccagtg 5280cccctccaac cagcctcagg tctatcccag
catgcctttc tgattctggt ccccagtcct 5340gccctctggt tcctccaacc
tagcctccag acctgctcca gtaaccctct caaattctag 5400ttcccaagcc
tctccttgca ttccttnccc aattctggcc ctctggccct gccctagtan
5460nccctatcct taagtacaat ctgtaagcca cctcagtgac ccctaccacc
ccatctcagg 5520cagttgttct gtgactttgg tgaggacttc actgtgcagg
accccacaga ggcagaaccc 5580ctgacagctg ccatccagca catctcccag
gtgggtgctg agctgtaggc attcacccgc 5640tgaccaagga gaggctgcca
gggcctgtgg aaggcaggtc caggcaaccc tgagccaagc 5700ctcctcctac
ccagggctcc cctggcattc tcactctgag gaaaggggcc aatacccact
5760acttccgtga tggagacttg gtgactttct cgggaattga gggaatggtt
gagctcaacg 5820actgtgatcc ccggtctatc cacgtgcggg gtaagccaat
cccattccaa ttccaggtgc 5880agggcccaag cctccactgg aagtgagcac
agcctggccc ttgggatggg tttttctccc 5940tccaccttct acaaggtgca
gcaaggtttg ggacacagat gcaagatagg atggggtgtg 6000ggaactactc
aggctcaagg atcattactg actagactgg aactccctca gaggatgggt
6060ccctggagat tggagacaca acaactttct ctcggtactt gcgtggtggg
gctatcactg 6120aagtcaagag acccaagact gtgagacatg tgagtgcaag
tccatctgag gtaggggagc 6180ttggtcgcct tgaggggccc atagcattct
ggactagacc ctgagccagg tgcccttgca 6240gaagtccctg gacacagccc
tgctccagcc ccatgtggtg gcccagagct cccaggaagt 6300tcaccatgcc
cactgcctgc atcaggcctt ctgtgcactg cacaagttcc agcacctcca
6360tggccggcca ccccagccct gggatcctgt gagtagtcct gttgctccca
cccccagcct 6420ctgtcattta ttggggtccc acctgccaga ggcaacaatg
accattcaca aatccaagtc 6480tgatctccca acactgcagc ctttagagta
gagactggtt ccatggaagt gccaggcaca 6540catcctgggg actcctgcta
caccccgacc cctcagatct gtgctggaag ctgcactcag 6600attagtgaag
cctcctggac tgctgtctgg tactgggcat cctctggtgg tgctgtgcag
6660gctggcagca gggccaggcc ttcccaccca ggcttctgct tcctcttctg
tggaacaggg 6720tggatggagg gtggctggaa ggatttgagt caggagtaga
gctcaggctg gggctactat 6780gcccacagag tcctaccaac aggttgatgc
agagactgtg gtgggcctgg cccgggacct 6840ggaaccactg aagcggacag
aggaagagcc actggaagag ccactggatg aggccctagt 6900gcggacagtc
gccctaagca gtgcaaggtg tcttgagcct atggtggcat gctgggtcag
6960tagctgccca ggaagtgctg aaggtgggca gaggcatagg tgtggggggt
actgggaaga 7020tgtggagatc agtgtgtgtg tcagagggca cccagcgcta
gagagcagcc ctggagcctt 7080caccaacctg ggtgaagcct ccagccagga
tctgaggggg gtcaggaggt ggcaggagtg 7140cccagcctga agtgctgccc
ctaggcaatc tccagaagtt catgcctctg gaccagtggc 7200tttactttga
tgccctcgat tgtcttccgg aagatgggga gctccttccc agtcctgagg
7260actgtgccct gagaggcagc cgctatgatg ggcaaattgc agtgtttggg
gctggttttc 7320aggagaaact gagacgccag cactacctcc tggtgagctg
tggggtgaga ctgggggtgc 7380ctttgggaga gccagcccag cccctctggc
taaggctgtt cctgccaaca ggtgggcgct 7440ggtgccattg gttgtgagct
gctcaaagtc tttgccctag tgggactggg ggccgggaac 7500agcgggggct
tgactgttgt tgacatggac cacatagagc gctccaatct cagccgtcag
7560ttcctcttca ggtcccagga cgttggtgtg agtgctgacc cctctccaca
ctcctgcatc 7620ccagaccgtc ctcccataca gcttcccacc caacatcttc
ctgccttctt cccagagacc 7680caaggcagag gtggctgcag cagctgcccg
gggcctgaac ccagacttac aggtgatccc 7740gctcacctac ccactggatc
ccaccacaga gcacatctat ggggataact ttttctcccg 7800tgtggatggt
gtggctgctg ccctggacag tttccaggcc cgtgagtgct tgacttcgga
7860ggtcagtccc ttgcccacag ctgtgccagt cccacttctg acccactgct
cccctgccag 7920ggcgctatgt ggctgctcgt tgcacccact atctgaagcc
actgctggag gcaggcacat 7980cgggcacctg gggcagtgct acagtattca
tgccacatgt gactgaggcc tacagagccc 8040ctgcctcagc tgcagcttct
gaggatgccc cctaccctgt ctgtaccgtg cggtacttcc 8100ctagcacagc
cgagcacacc ctgcaggtag gaagcaccct ggagactccc accccaccca
8160gctcagccct cagctgcaga cctgttctcc acctgatacc tcattcttcc
tccctcctcc 8220acagtgggcc cggcatgagt ttgaagaact cttccgactg
tctgcagaga ccatcaacca 8280ccaccaacag taaggccacc aacagaggca
gatgggagtc cagggctcca agcatgagtc 8340tgcaggactc agtctcacac
ttcctcctct ctctgcaggg cacacacctc cctggcagac 8400atggatgagc
cacagacact caccttactg aagccagtgc ttggggtcct gagagtgcgt
8460ccacagaact ggcaagactg tgtggcgtgg gctcttggcc actggaaact
ctgctttcat 8520tatggcatca aacagctgct gaggcacttc ccacctaata
aagtgtgtgg ctaggggttg 8580ggacgctggg ggctcagggg gaccagactg
agcccagcag cttctactta cctacctagg 8640tgcttgagga tggaactccc
ttctggtcag gtcccaaaca gtgtccccag cccttggagt 8700ttgacaccaa
ccaagtgagt gggattctgt agggagctcc aagatagaga tgtggcccct
8760cagagcagag gtaggcattt ctgcattctg cagagatgca cagatgccca
gagagagcca 8820tgcttgtgca tatatgggtg tctacatgtg aggcaaaggc
aggcactcaa acagatccac 8880aaatggacag tgaccccacc catgcaccat
gcctctctgt tctgctctct gctcttggtc 8940tggctgcagg acacacacct
cctctacgta ctggcagctg ccaacctgta tgcccagatg 9000catgggctgc
ctggctcaca ggactggact gcactcaggg agctgctgaa gctgctgcca
9060cagcctgacc cccaacagat ggcccccatc tttgctagta atctagagct
ggcttcggct 9120tctgctgagt ttggtgaggc tcctggccct ggcccctcat
gctgtctttc aaaggcctga 9180acctgtcctg tcctcagcct gtgctgcaga
aggaagatag ggcctagggg atctacagcc 9240aatttgctac ctctcaggcc
tcctaacctc actcctccat agtttcaggc ttatcctctg 9300gtccctcagt
aggtcttctc cctgctgcct accccacatc ccagttcttg tggcagattc
9360ttggcaaaat aaataagtaa ataaataaag tccattggtt cctggggagt
gtctagctat 9420ggcctgcagg tgaggacagg gtcacagagg tcatgagcac
acatgggtga agactggggc 9480ttctagaggg gagattgtag cattaattaa
gggggcttct tgatttgatc agggaatagt 9540aagtgacagg cttggcaaag
accaagaata ggcacagggc tccaagaaga gtgcaggaga 9600caggggctaa
ggactgcctc aacatcccct tccctgacag gccctgagca gcagaaggaa
9660ctgaacaaag ccctggaagt ctggagtgtg ggccctcccc tgaagcctct
gatgtttgag 9720aaggtgggtg cccaagtggc agtgaggagt ggggctgggg
agtttgtgga gaaaggtcag 9780gagctaataa ggtagttttg gagccccttg
gcctgaattc cacagctgca gtgttaacac 9840tactttgact tgggccttac
aggatgatga cagcaacttc catgtggact ttgtggtagc 9900ggcagctagc
ctgagatgtc agaactacgg gattccaccg gtcaaccgtg cccaggtaac
9960cccacccctt gaggcttggg cctggaggtg gagggcaaac cctggcccta
cgccttgggc 10020ccagaccaaa tctcttgtcc ttggcagagc aagcgaattg
tgggccagat tatcccagcc 10080attgccacca ctacagcagc tgtggcaggc
ctgttgggcc tggagctgta taaggtggtg 10140agtgggccac ggcctcgtag
tgcctttcgc cacagctacc tacatctggc tgaaaactac 10200ctcatccgct
atatgccttt tgccccagcc atccagacgg ttgagcccat gataccccac
10260ccttagccct actaggcctg ggtttcccct gcacctgccc atacaggccc
caatctagct 10320gccggctctc actgaaactc agactgtgca gaagtcctga
agactccctc cagccctctt 10380cctgctatga agccaggctg ggacctgtca
gacacaggaa gcagccgtca gccatcccca 10440cccccaatcc tccaaagccc
aggatctggg ggagctgcag ctttaactca ttagtggagc 10500cagacatccc
ccacagtccc ctccttcctt ggagtacccc tgagggtagg tagtggggag
10560gggaccaggg catcagccca gaaagagtct agcttccccc tgcatggtac
gggggccctg 10620gcctacctcc tacaagctga gttaaaaggt aataggcttg
ccactagagg tgtggggtct 10680gcagccctga gtgtatttgt gtcacagttg
tgagtgcaac tggggtctgg gcatccccgg 10740agtgtgggta tggaaggagc
aagtttgcac atctgtgcat cagatgggag tgcagggctc 10800ccatcttcct
gtgtcctcaa tgtgggcatg catggagatg catgtgggca cacatgtgtg
10860tgtttcctgg gcttgtgggc atttgtgttc ctgtgactgc agcgctgtga
atgcctccag 10920ccttgtgccc aggctctgca gttggcattt ccttggggca
aggatgaggg tagaaggaat 10980gccctcatga ggggagaggg cagaggtcat
gggccagcac tgggtttctg ctgagcagcc 11040tggggtccct ctggactagc
acacagagcc cctttgtgag gcaccctgcc tctaaccagc 11100atacaggctg
cctttgtcca cagagtgaag caggtgaagg agaagccccc atctcacctc
11160tagcctagtc tcagcttgac cccagtgtgt tttctcaggg gctgataagc
accccctgtc 11220ctggtctcta gatacctgcc agccatcctt ctgtccttag
ctctaggtcc cagaacccca 11280agactcttgg aaggaaggaa gggacaggag
gaggaagcct ctatgcattg tatccctgct 11340ggggtctctg ggacagtggg
gccctggtgg tcactgtgcc ctttgcccct gtgtcccaat 11400gagtagccca
aggcactggc agtacataca agattggggg acaggatgtg ccccccagct
11460cccagccttg tctttgagga acaagcagct ttatcagagg ctgcaggggc
cctgctctgg 11520gtttcctcag gaagcaccac cgccgcatcc cccactctca
caactggccc atgtgatgga 11580tcgtctgttt ccctgtgcgg gccccatagc
cccatttcct gtgctggccc cggcctggac 11640ggggaggggg ctgagactct
gggcccagat cccacctccc ccccaccccc caccccctgg 11700ctcctgtttc
ctgctagtcc agctcttccc ctaggaaagg ctgctggtaa ctgggatggg
11760ggttgggggg aggtaagaag tctctgactc ctcctctacc tcatcccagt
tccatcacct 11820gaagtggacc tcttgggacc gtctgaaggt accagctggg
cagcctgaga ggaccctgga 11880gtcgctgctg gctcatcttc aggagcagca
cgggttgagg gtgaggatcc tgctgcacgg 11940ctcagccctg ctctatgcgg
ccggatggtc acctgaaaag caggcccagc acctgcccct 12000caggtgagcc
cacttgggct ttagacaggc cccaccagtc cctggaggct ggggctaggg
12060accacactgc cttttgtctt cccagcccca ttctgggccc ctcacacctt
cccaagcatt 12120ctttccccaa atggagccag caaacaggct ggaggtgggg
tgagggccga gagctgagga 12180ggagtcttcn anggagctcg tatttggcca
gcccatggct cccacatgct gcacagnnnn 12240ttcacagcca ctcctaagga
cccatagctt cctgcctcct gcttggcctc atcagctgct 12300cctaaaatag
tttcagatgt ttcctgtctt gagcagctcc tgctcctggc ttgggctcct
12360gacggcctgc cagcaccctc tctagtccat gccaggctgc cttctgcttg
ccatggctca 12420cctctccaat ctcccctaaa cccaccccta ccagggtgac
agaactggtt cagcagctga 12480caggccaggc acctgctcct gggcagcggg
tgttggtgct agagctgagc tgtgagggtg 12540acgacgagga cactgccttc
ccacctctgc actatgagct gtgacaaggc agccaccctg 12600tcacctagct
caatggagcc ccggatccca agccctgcat tgtaagccca cagtaggcac
12660tcaataattg cttgttaaag gaaggcattg cagagaggac ggacgataga
aaacagtgca 12720ctaatgcaca cgggtgtgac atgggcatga cagggacctt
cacacagaga aaaaaagctc 12780ttcagaagat ttgtctccct gggcagtgct
cacagggctg gggctgcctc ttagtgcctc 12840aggggtatgg agccaggaca
gtctagaaaa aaggctttta ttgtcccagg ctggagggca 12900gggtcagagg
tagctgacat cattgcagat gatgggctgg cggctacgac agctcatgag
12960agctgcaaag ctgagacat 12979801011PRTHomo sapiens 80Met Asp Ala
Leu Asp Ala Ser Lys Leu Leu Asp Glu Glu Leu Tyr Ser1 5 10 15Arg Gln
Leu Tyr Val Leu Gly Ser Pro Ala Met Gln Arg Ile Gln Gly 20 25 30Ala
Arg Val Leu Val Ser Gly Leu Gln Gly Leu Gly Ala Glu Val Ala 35 40
45Lys Asn Leu Val Leu Met Gly Val Gly Ser Leu Thr Leu His Asp Pro
50 55 60His Pro Thr Cys Trp Ser Asp Leu Ala Ala Gln Phe Leu Leu Ser
Glu65 70 75 80Gln Asp Leu Glu Arg Ser Arg Ala Glu Ala Ser Gln Glu
Leu Leu Ala 85 90 95Gln Leu Asn Arg Ala Val Gln Val Val Val His Thr
Gly Asp Ile Thr 100 105 110Glu Asp Leu Leu Leu Asp Phe Gln Val Val
Val Leu Thr Ala Ala Lys 115 120 125Leu Glu Glu Gln Leu Lys Val Gly
Thr Leu Cys His Lys His Gly Val 130 135 140Cys Phe Leu Ala Ala Asp
Thr Arg Gly Leu Val Gly Gln Leu Phe Cys145 150 155 160Asp Phe Gly
Glu Asp Phe Thr Val Gln Asp Pro Thr Glu Ala Glu Pro 165 170 175Leu
Thr Ala Ala Ile Gln His Ile Ser Gln Gly Ser Pro Gly Ile Leu 180 185
190Thr Leu Arg Lys Gly Ala Asn Thr His Tyr Phe Arg Asp Gly Asp Leu
195 200 205Val Thr Phe Ser Gly Ile Glu Gly Met Val Glu Leu Asn Asp
Cys Asp 210 215 220Pro Arg Ser Ile His Val Arg Glu Asp Gly Ser Leu
Glu Ile Gly Asp225 230 235 240Thr Thr Thr Phe Ser Arg Tyr Leu Arg
Gly Gly Ala Ile Thr Glu Val 245 250 255Lys Arg Pro Lys Thr Val Arg
His Lys Ser Leu Asp Thr Ala Leu Leu 260 265 270Gln Pro His Val Val
Ala Gln Ser Ser Gln Glu Val His His Ala His 275 280 285Cys Leu His
Gln Ala Phe Cys Ala Leu His Lys Phe Gln His Leu His 290 295 300Gly
Arg Pro Pro Gln Pro Trp Asp Pro Val Asp Ala Glu Thr Val Val305 310
315 320Gly Leu Ala Arg Asp Leu Glu Pro Leu Lys Arg Thr Glu Glu Glu
Pro 325 330 335Leu Glu Glu Pro Leu Asp Glu Ala Leu Val Arg Thr Val
Ala Leu Ser 340 345 350Ser Ala Arg Cys Leu Glu Pro Met Val Ala Cys
Trp Val Ser Ser Cys 355 360 365Pro Gly Ser Ala Glu Gly Asn Leu Gln
Lys Phe Met Pro Leu Asp Gln 370 375 380Trp Leu Tyr Phe Asp Ala Leu
Asp Cys Leu Pro Glu Asp Gly Glu Leu385 390 395 400Leu Pro Ser Pro
Glu Asp Cys Ala Leu Arg Gly Ser Arg Tyr Asp Gly 405 410 415Gln Ile
Ala Val Phe Gly Ala Gly Phe Gln Glu Lys Leu Arg Arg Gln 420 425
430His Tyr Leu Leu Val Gly Ala Gly Ala Ile Gly Cys Glu Leu Leu Lys
435 440 445Val Phe Ala Leu Val Gly Leu Gly Ala Gly Asn Ser Gly Gly
Leu Thr 450 455 460Val Val Asp Met Asp His Ile Glu Arg Ser Asn Leu
Ser Arg Gln Phe465
470 475 480Leu Phe Arg Ser Gln Asp Val Gly Arg Pro Lys Ala Glu Val
Ala Ala 485 490 495Ala Ala Ala Arg Gly Leu Asn Pro Asp Leu Gln Val
Ile Pro Leu Thr 500 505 510Tyr Pro Leu Asp Pro Thr Thr Glu His Ile
Tyr Gly Asp Asn Phe Phe 515 520 525Ser Arg Val Asp Gly Val Ala Ala
Ala Leu Asp Ser Phe Gln Ala Arg 530 535 540Arg Tyr Val Ala Ala Arg
Cys Thr His Tyr Leu Lys Pro Leu Leu Glu545 550 555 560Ala Gly Thr
Ser Gly Thr Trp Gly Ser Ala Thr Val Phe Met Pro His 565 570 575Val
Thr Glu Ala Tyr Arg Ala Pro Ala Ser Ala Ala Ala Ser Glu Asp 580 585
590Ala Pro Tyr Pro Val Cys Thr Val Arg Tyr Phe Pro Ser Thr Ala Glu
595 600 605His Thr Leu Gln Trp Ala Arg His Glu Phe Glu Glu Leu Phe
Arg Leu 610 615 620Ser Ala Glu Thr Ile Asn His His Gln Gln Ala His
Thr Ser Leu Ala625 630 635 640Asp Met Asp Glu Pro Gln Thr Leu Thr
Leu Leu Lys Pro Val Leu Gly 645 650 655Val Leu Arg Val Arg Pro Gln
Asn Trp Gln Asp Cys Val Ala Trp Ala 660 665 670Leu Gly His Trp Lys
Leu Cys Phe His Tyr Gly Ile Lys Gln Leu Leu 675 680 685Arg His Phe
Pro Pro Asn Lys Val Leu Glu Asp Gly Thr Pro Phe Trp 690 695 700Ser
Gly Pro Lys Gln Cys Pro Gln Pro Leu Glu Phe Asp Thr Asn Gln705 710
715 720Asp Thr His Leu Leu Tyr Val Leu Ala Ala Ala Asn Leu Tyr Ala
Gln 725 730 735Met His Gly Leu Pro Gly Ser Gln Asp Trp Thr Ala Leu
Arg Glu Leu 740 745 750Leu Lys Leu Leu Pro Gln Pro Asp Pro Gln Gln
Met Ala Pro Ile Phe 755 760 765Ala Ser Asn Leu Glu Leu Ala Ser Ala
Ser Ala Glu Phe Gly Pro Glu 770 775 780Gln Gln Lys Glu Leu Asn Lys
Ala Leu Glu Val Trp Ser Val Gly Pro785 790 795 800Pro Leu Lys Pro
Leu Met Phe Glu Lys Asp Asp Asp Ser Asn Phe His 805 810 815Val Asp
Phe Val Val Ala Ala Ala Ser Leu Arg Cys Gln Asn Tyr Gly 820 825
830Ile Pro Pro Val Asn Arg Ala Gln Ser Lys Arg Ile Val Gly Gln Ile
835 840 845Ile Pro Ala Ile Ala Thr Thr Thr Ala Ala Val Ala Gly Leu
Leu Gly 850 855 860Leu Glu Leu Tyr Lys Val Val Ser Gly Pro Arg Pro
Arg Ser Ala Phe865 870 875 880Arg His Ser Tyr Leu His Leu Ala Glu
Asn Tyr Leu Ile Arg Tyr Met 885 890 895Pro Phe Ala Pro Ala Ile Gln
Thr Phe His His Leu Lys Trp Thr Ser 900 905 910Trp Asp Arg Leu Lys
Val Pro Ala Gly Gln Pro Glu Arg Thr Leu Glu 915 920 925Ser Leu Leu
Ala His Leu Gln Glu Gln His Gly Leu Arg Val Arg Ile 930 935 940Leu
Leu His Gly Ser Ala Leu Leu Tyr Ala Ala Gly Trp Ser Pro Glu945 950
955 960Lys Gln Ala Gln His Leu Pro Leu Arg Val Thr Glu Leu Val Gln
Gln 965 970 975Leu Thr Gly Gln Ala Pro Ala Pro Gly Gln Arg Val Leu
Val Leu Glu 980 985 990Leu Ser Cys Glu Gly Asp Asp Glu Asp Thr Ala
Phe Pro Pro Leu His 995 1000 1005Tyr Glu Leu 1010811642DNAHomo
sapiens 81aagccacggg gagaaacgtt gcagcccgcg ccgaacgccg ggcagcacaa
aggatccccg 60actgccgggg agcggtgctc ggagggcaca ggtctacgcc atcccccacg
cagtttcgga 120gatggagcgc tgggcccatg gagggaaggc ggcaggctcg
gcggctccgg cagcttgctg 180gggcaggggc tcaaggcggc agtccgatag
tggaggccgc tgagaactgt cacggagctg 240cgtctgtaca gcgagcatcc
cttatttatt cagggcgagt gtgtatttgg ggcggcgtgc 300agggggctga
caaagaccgg agagctcccg gtgcggccgc cggcggagcg aagactggaa
360cccgtatgag cgccccccag cgcccctgag cgctcgccgc cggtgcacgg
cgcaccccgc 420gggaggcagg gatcagcaaa gccgtgcgcc ccgaggcccg
cccccgtctc cgcacaaaga 480ccgagctgga ggatcttcag aagaagcctc
ccccatacct gcggaacctg tccagcgatg 540atgccaatgt cctggtgtgg
cacgctctcc tcctacccga ccaacctccc taccacctga 600aagccttcaa
cctgcgcatc agcttcccgc cggagtatcc gttcaagcct cccatgatca
660aattcacaac caagatctac caccccaacg tggacgagaa cggacagatt
tgcctgccca 720tcatcagcag tgagaactgg aagccttgca ccaagacttg
ccaagtcctg gaggccctca 780atgtgctggt gaatagaccg aatatcaggg
agcccctgcg gatggacctc gctgacctgc 840tgacacagaa tccggagctg
ttcagaaaga atgccgaaga gttcaccctc cgattcggag 900tggaccggcc
ctcctaactc atgttctgac cctctgtgca ctggatcctc ggcatagcgg
960acggacacac ctcatggact gaggccagag ccccctgtgg cccattcccc
attcattttt 1020cccttcttag gttgttagtc attagtttgt gtgtgtgtgt
ggtggaggga agggagctat 1080gagtgtgtgt gttgtgtatg gactcactcc
caggttcacc tggccacagg tgcacccttc 1140ccacaccctt tacattcccc
agagccaagg gagtttaagt ttgcagttac aggccagttc 1200tccagctctc
catcttagag agacaggtca ccttgcaggc ctgcttgcag gaaatgaatc
1260cagcagccaa ctcgaatccc cctagggctc aggcactgag ggcctgggga
cagtggagca 1320tatgggtggg agacagatgg agggtaccct atttacaact
gagtcagcca agccactgat 1380gggaatatac agatttaggt gctaaaccat
ttattttcca cggatgagtc acaatctgaa 1440gaatcaaact tccatcctga
aaatctatat gtttcaaaac cacttgccat cctgttagat 1500tgccagttcc
tgggaccagg cctcagactg tgaagtatat atcctccagc attcagtcca
1560gggggagcca cggaaaccat gttcttgctt aagccattaa agtcagagat
gaattctgga 1620aaaaaaaaaa aaaaaaaaaa aa 16428287PRTHomo sapiens
82Met Ile Lys Phe Thr Thr Lys Ile Tyr His Pro Asn Val Asp Glu Asn1
5 10 15Gly Gln Ile Cys Leu Pro Ile Ile Ser Ser Glu Asn Trp Lys Pro
Cys 20 25 30Thr Lys Thr Cys Gln Val Leu Glu Ala Leu Asn Val Leu Val
Asn Arg 35 40 45Pro Asn Ile Arg Glu Pro Leu Arg Met Asp Leu Ala Asp
Leu Leu Thr 50 55 60Gln Asn Pro Glu Leu Phe Arg Lys Asn Ala Glu Glu
Phe Thr Leu Arg65 70 75 80Phe Gly Val Asp Arg Pro Ser
85831283DNAHomo sapiens 83gggggtgggg tccccggggc ggggcggggc
gcgctgtgtc gcgggtcgga gctcggtcct 60gctggaggcc acgggtgcca cacactcggt
cccgacatga tggcgagcat gcgagtggtg 120aaggagctgg aggatcttca
gaagaagcct cccccatacc tgcggaacct gtccagcgat 180gatgccaatg
tcctggtgtg gcacgctctc ctcctacccg accaacctcc ctaccacctg
240aaagccttca acctgcgcat cagcttcccg ccggagtatc cgttcaagcc
tcccatgatc 300aaattcacaa ccaagatcta ccaccccaac gtggacgaga
acggacagat ttgcctgccc 360atcatcagca gtgagaactg gaagccttgc
accaagactt gccaagtcct ggaggccctc 420aatgtgctgg tgaatagacc
gaatatcagg gagcccctgc ggatggacct cgctgacctg 480ctgacacaga
atccggagct gttcagaaag aatgccgaag agttcaccct ccgattcgga
540gtggaccggc cctcctaact catgttctga ccctctgtgc actggatcct
cggcatagcg 600gacggacaca cctcatggac tgaggccaga gccccctgtg
gcccattccc cattcatttt 660tcccttctta ggttgttagt cattagtttg
tgtgtgtgtg tggtggaggg aagggagcta 720tgagtgtgtg tgttgtgtat
ggactcactc ccaggttcac ctggccacag gtgcaccctt 780cccacaccct
ttacattccc cagagccaag ggagtttaag tttgcagtta caggccagtt
840ctccagctct ccatcttaga gagacaggtc accttgcagg cctgcttgca
ggaaatgaat 900ccagcagcca actcgaatcc ccctagggct caggcactga
gggcctgggg acagtggagc 960atatgggtgg gagacagatg gagggtaccc
tatttacaac tgagtcagcc aagccactga 1020tgggaatata cagatttagg
tgctaaacca tttattttcc acggatgagt cacaatctga 1080agaatcaaac
ttccatcctg aaaatctata tgtttcaaaa ccacttgcca tcctgttaga
1140ttgccagttc ctgggaccag gcctcagact gtgaagtata tatcctccag
cattcagtcc 1200agggggagcc acggaaacca tgttcttgct taagccatta
aagtcagaga tgaattctgg 1260aaaaaaaaaa aaaaaaaaaa aaa
128384153PRTHomo sapiens 84Met Met Ala Ser Met Arg Val Val Lys Glu
Leu Glu Asp Leu Gln Lys1 5 10 15Lys Pro Pro Pro Tyr Leu Arg Asn Leu
Ser Ser Asp Asp Ala Asn Val 20 25 30Leu Val Trp His Ala Leu Leu Leu
Pro Asp Gln Pro Pro Tyr His Leu 35 40 45Lys Ala Phe Asn Leu Arg Ile
Ser Phe Pro Pro Glu Tyr Pro Phe Lys 50 55 60Pro Pro Met Ile Lys Phe
Thr Thr Lys Ile Tyr His Pro Asn Val Asp65 70 75 80Glu Asn Gly Gln
Ile Cys Leu Pro Ile Ile Ser Ser Glu Asn Trp Lys 85 90 95Pro Cys Thr
Lys Thr Cys Gln Val Leu Glu Ala Leu Asn Val Leu Val 100 105 110Asn
Arg Pro Asn Ile Arg Glu Pro Leu Arg Met Asp Leu Ala Asp Leu 115 120
125Leu Thr Gln Asn Pro Glu Leu Phe Arg Lys Asn Ala Glu Glu Phe Thr
130 135 140Leu Arg Phe Gly Val Asp Arg Pro Ser145 15085665DNAHomo
sapiens 85gggaacacat ccaagcttaa gacggtgagg tcagcttcac attctcagga
actctccttc 60tttgggtctg gctgaagttg aggatctctt actctctagg ccacggaatt
aacccgagca 120ggcatggagg cctctgctct cacctcatca gcagtgacca
gtgtggccaa agtggtcagg 180gtggcctctg gctctgccgt agttttgccc
ctggccagga ttgctacagt tgtgattgga 240ggagttgtgg ccatggcggc
tgtgcccatg gtgctcagtg ccatgggctt cactgcggcg 300ggaatcgcct
cgtcctccat agcagccaag atgatgtccg cggcggccat tgccaatggg
360ggtggagttg cctcgggcag ccttgtggct actctgcagt cactgggagc
aactggactc 420tccggattga ccaagttcat cctgggctcc attgggtctg
ccattgcggc tgtcattgcg 480aggttctact agctccctgc ccctcgccct
gcagagaaga gaaccatgcc aggggagaag 540gcacccagcc atcctgaccc
agcgaggagc caactatccc aaatatacct ggggtgaaat 600ataccaaatt
ctgcatctcc agaggaaaat aagaaataaa gatgaattgt tgcaactctt 660caaaa
66586122PRTHomo sapiens 86Met Glu Ala Ser Ala Leu Thr Ser Ser Ala
Val Thr Ser Val Ala Lys1 5 10 15Val Val Arg Val Ala Ser Gly Ser Ala
Val Val Leu Pro Leu Ala Arg 20 25 30Ile Ala Thr Val Val Ile Gly Gly
Val Val Ala Met Ala Ala Val Pro 35 40 45Met Val Leu Ser Ala Met Gly
Phe Thr Ala Ala Gly Ile Ala Ser Ser 50 55 60Ser Ile Ala Ala Lys Met
Met Ser Ala Ala Ala Ile Ala Asn Gly Gly65 70 75 80Gly Val Ala Ser
Gly Ser Leu Val Ala Thr Leu Gln Ser Leu Gly Ala 85 90 95Thr Gly Leu
Ser Gly Leu Thr Lys Phe Ile Leu Gly Ser Ile Gly Ser 100 105 110Ala
Ile Ala Ala Val Ile Ala Arg Phe Tyr 115 12087656DNAHomo sapiens
87gggaacacat ccaagcttaa gacggtgagg tcagcttcac attctcagga actctccttc
60tttgggtctg gctgaagttg aggatctctt actctctagg ccacggaatt aacccgagca
120ggcatggagg cctctgctct cacctcatca gcagtgacca gtgtggccaa
agtggtcagg 180gtggcctctg gctctgccgt agttttgccc ctggccagga
ttgctacagt tgtgattgga 240ggagttgtgg ctgtgcccat ggtgctcagt
gccatgggct tcactgcggc gggaatcgcc 300tcgtcctcca tagcagccaa
gatgatgtcc gcggcggcca ttgccaatgg gggtggagtt 360gcctcgggca
gccttgtggc tactctgcag tcactgggag caactggact ctccggattg
420accaagttca tcctgggctc cattgggtct gccattgcgg ctgtcattgc
gaggttctac 480tagctccctg cccctcgccc tgcagagaag agaaccatgc
caggggagaa ggcacccagc 540catcctgacc cagcgaggag ccaactatcc
caaatatacc tggggtgaaa tataccaaat 600tctgcatctc cagaggaaaa
taagaaataa agatgaattg ttgcaactct tcaaaa 65688119PRTHomo sapiens
88Met Glu Ala Ser Ala Leu Thr Ser Ser Ala Val Thr Ser Val Ala Lys1
5 10 15Val Val Arg Val Ala Ser Gly Ser Ala Val Val Leu Pro Leu Ala
Arg 20 25 30Ile Ala Thr Val Val Ile Gly Gly Val Val Ala Val Pro Met
Val Leu 35 40 45Ser Ala Met Gly Phe Thr Ala Ala Gly Ile Ala Ser Ser
Ser Ile Ala 50 55 60Ala Lys Met Met Ser Ala Ala Ala Ile Ala Asn Gly
Gly Gly Val Ala65 70 75 80Ser Gly Ser Leu Val Ala Thr Leu Gln Ser
Leu Gly Ala Thr Gly Leu 85 90 95Ser Gly Leu Thr Lys Phe Ile Leu Gly
Ser Ile Gly Ser Ala Ile Ala 100 105 110Ala Val Ile Ala Arg Phe Tyr
115893617DNAHomo sapiens 89atcgaaacag aaaccaaagt caggcaaact
ctgtaagaac tgcctgacag aaagctggac 60tcaaagctcc tacccgagtg tgcagcagga
tcgccccggt ccgggacccc aggcgcacac 120cgcagagtcc aaagtgccgc
gcctgccggc cgcacctgcc tgccgcggcc ccgcgcgccg 180ccccgctgcc
cacctgcccg cctgcccacc tgcccaggtg cgagtgcagc cccgcgcgcc
240ggcctgagag ccctgtggac aacctcgtca ttgtcaggca cagagcggta
gaccctgctt 300ctctaagtgg gcagcggaca gcggcacgca catttcacct
gtcccgcaga caacagcacc 360atctgcttgg gagaaccctc tcccttctct
gagaaagaaa gatgtcgaat gggtattcca 420cagacgagaa tttccgctat
ctcatctcgt gcttcagggc cagggtgaaa atgtacatcc 480aggtggagcc
tgtgctggac tacctgacct ttctgcctgc agaggtgaag gagcagattc
540agaggacagt cgccacctcc gggaacatgc aggcagttga actgctgctg
agcaccttgg 600agaagggagt ctggcacctt ggttggactc gggaattcgt
ggaggccctc cggagaaccg 660gcagccctct ggccgcccgc tacatgaacc
ctgagctcac ggacttgccc tctccatcgt 720ttgagaacgc tcatgatgaa
tatctccaac tgctgaacct ccttcagccc actctggtgg 780acaagcttct
agttagagac gtcttggata agtgcatgga ggaggaactg ttgacaattg
840aagacagaaa ccggattgct gctgcagaaa acaatggaaa tgaatcaggt
gtaagagagc 900tactaaaaag gattgtgcag aaagaaaact ggttctctgc
atttctgaat gttcttcgtc 960aaacaggaaa caatgaactt gtccaagagt
taacaggctc tgattgctca gaaagcaatg 1020cagagattga gaatttatca
caagttgatg gtcctcaagt ggaagagcaa cttctttcaa 1080ccacagttca
gccaaatctg gagaaggagg tctggggcat ggagaataac tcatcagaat
1140catcttttgc agattcttct gtagtttcag aatcagacac aagtttggca
gaaggaagtg 1200tcagctgctt agatgaaagt cttggacata acagcaacat
gggcagtgat tcaggcacca 1260tgggaagtga ttcagatgaa gagaatgtgg
cagcaagagc atccccggag ccagaactcc 1320agctcaggcc ttaccaaatg
gaagttgccc agccagcctt ggaagggaag aatatcatca 1380tctgcctccc
tacagggagt ggaaaaacca gagtggctgt ttacattgcc aaggatcact
1440tagacaagaa gaaaaaagca tctgagcctg gaaaagttat agttcttgtc
aataaggtac 1500tgctagttga acagctcttc cgcaaggagt tccaaccatt
tttgaagaaa tggtatcgtg 1560ttattggatt aagtggtgat acccaactga
aaatatcatt tccagaagtt gtcaagtcct 1620gtgatattat tatcagtaca
gctcaaatcc ttgaaaactc cctcttaaac ttggaaaatg 1680gagaagatgc
tggtgttcaa ttgtcagact tttccctcat tatcattgat gaatgtcatc
1740acaccaacaa agaagcagtg tataataaca tcatgaggca ttatttgatg
cagaagttga 1800aaaacaatag actcaagaaa gaaaacaaac cagtgattcc
ccttcctcag atactgggac 1860taacagcttc acctggtgtt ggaggggcca
cgaagcaagc caaagctgaa gaacacattt 1920taaaactatg tgccaatctt
gatgcattta ctattaaaac tgttaaagaa aaccttgatc 1980aactgaaaaa
ccaaatacag gagccatgca agaagtttgc cattgcagat gcaaccagag
2040aagatccatt taaagagaaa cttctagaaa taatgacaag gattcaaact
tattgtcaaa 2100tgagtccaat gtcagatttt ggaactcaac cctatgaaca
atgggccatt caaatggaaa 2160aaaaagctgc aaaagaagga aatcgcaaag
aacgtgtttg tgcagaacat ttgaggaagt 2220acaatgaggc cctacaaatt
aatgacacaa ttcgaatgat agatgcgtat actcatcttg 2280aaactttcta
taatgaagag aaagataaga agtttgcagt catagaagat gatagtgatg
2340agggtggtga tgatgagtat tgtgatggtg atgaagatga ggatgattta
aagaaacctt 2400tgaaactgga tgaaacagat agatttctca tgactttatt
ttttgaaaac aataaaatgt 2460tgaaaaggct ggctgaaaac ccagaatatg
aaaatgaaaa gctgaccaaa ttaagaaata 2520ccataatgga gcaatatact
aggactgagg aatcagcacg aggaataatc tttacaaaaa 2580cacgacagag
tgcatatgcg ctttcccagt ggattactga aaatgaaaaa tttgctgaag
2640taggagtcaa agcccaccat ctgattggag ctggacacag cagtgagttc
aaacccatga 2700cacagaatga acaaaaagaa gtcattagta aatttcgcac
tggaaaaata aatctgctta 2760tcgctaccac agtggcagaa gaaggtctgg
atattaaaga atgtaacatt gttatccgtt 2820atggtctcgt caccaatgaa
atagccatgg tccaggcccg tggtcgagcc agagctgatg 2880agagcaccta
cgtcctggtt gctcacagtg gttcaggagt tatcgaacat gagacagtta
2940atgatttccg agagaagatg atgtataaag ctatacattg tgttcaaaat
atgaaaccag 3000aggagtatgc tcataagatt ttggaattac agatgcaaag
tataatggaa aagaaaatga 3060aaaccaagag aaatattgcc aagcattaca
agaataaccc atcactaata actttccttt 3120gcaaaaactg cagtgtgcta
gcctgttctg gggaagatat ccatgtaatt gagaaaatgc 3180atcacgtcaa
tatgacccca gaattcaagg aactttacat tgtaagagaa aacaaagcac
3240tgcaaaagaa gtgtgccgac tatcaaataa atggtgaaat catctgcaaa
tgtggccagg 3300cttggggaac aatgatggtg cacaaaggct tagatttgcc
ttgtctcaaa ataaggaatt 3360ttgtagtggt tttcaaaaat aattcaacaa
agaaacaata caaaaagtgg gtagaattac 3420ctatcacatt tcccaatctt
gactattcag aatgctgttt atttagtgat gaggattagc 3480acttgattga
agattctttt aaaatactat cagttaaaca tttaatatga ttatgattaa
3540tgtattcatt atgctacaga actgacataa gaatcaataa aatgattgtt
ttactctgca 3600aaaaaaaaaa aaaaaaa 3617901025PRTHomo sapiens 90Met
Ser Asn Gly Tyr Ser Thr Asp Glu Asn Phe Arg Tyr Leu Ile Ser1 5 10
15Cys Phe Arg Ala Arg Val Lys Met Tyr Ile Gln Val Glu Pro Val Leu
20 25 30Asp Tyr Leu Thr Phe Leu Pro Ala Glu Val Lys Glu Gln Ile Gln
Arg 35 40 45Thr Val Ala Thr Ser Gly Asn Met Gln Ala Val Glu Leu Leu
Leu Ser 50 55 60Thr Leu Glu Lys Gly Val Trp His Leu Gly Trp Thr Arg
Glu Phe Val65 70 75 80Glu Ala Leu Arg Arg Thr Gly Ser Pro Leu Ala
Ala Arg Tyr Met Asn 85 90 95Pro Glu Leu Thr Asp Leu Pro Ser Pro Ser
Phe Glu Asn Ala His Asp
100 105 110Glu Tyr Leu Gln Leu Leu Asn Leu Leu Gln Pro Thr Leu Val
Asp Lys 115 120 125Leu Leu Val Arg Asp Val Leu Asp Lys Cys Met Glu
Glu Glu Leu Leu 130 135 140Thr Ile Glu Asp Arg Asn Arg Ile Ala Ala
Ala Glu Asn Asn Gly Asn145 150 155 160Glu Ser Gly Val Arg Glu Leu
Leu Lys Arg Ile Val Gln Lys Glu Asn 165 170 175Trp Phe Ser Ala Phe
Leu Asn Val Leu Arg Gln Thr Gly Asn Asn Glu 180 185 190Leu Val Gln
Glu Leu Thr Gly Ser Asp Cys Ser Glu Ser Asn Ala Glu 195 200 205Ile
Glu Asn Leu Ser Gln Val Asp Gly Pro Gln Val Glu Glu Gln Leu 210 215
220Leu Ser Thr Thr Val Gln Pro Asn Leu Glu Lys Glu Val Trp Gly
Met225 230 235 240Glu Asn Asn Ser Ser Glu Ser Ser Phe Ala Asp Ser
Ser Val Val Ser 245 250 255Glu Ser Asp Thr Ser Leu Ala Glu Gly Ser
Val Ser Cys Leu Asp Glu 260 265 270Ser Leu Gly His Asn Ser Asn Met
Gly Ser Asp Ser Gly Thr Met Gly 275 280 285Ser Asp Ser Asp Glu Glu
Asn Val Ala Ala Arg Ala Ser Pro Glu Pro 290 295 300Glu Leu Gln Leu
Arg Pro Tyr Gln Met Glu Val Ala Gln Pro Ala Leu305 310 315 320Glu
Gly Lys Asn Ile Ile Ile Cys Leu Pro Thr Gly Ser Gly Lys Thr 325 330
335Arg Val Ala Val Tyr Ile Ala Lys Asp His Leu Asp Lys Lys Lys Lys
340 345 350Ala Ser Glu Pro Gly Lys Val Ile Val Leu Val Asn Lys Val
Leu Leu 355 360 365Val Glu Gln Leu Phe Arg Lys Glu Phe Gln Pro Phe
Leu Lys Lys Trp 370 375 380Tyr Arg Val Ile Gly Leu Ser Gly Asp Thr
Gln Leu Lys Ile Ser Phe385 390 395 400Pro Glu Val Val Lys Ser Cys
Asp Ile Ile Ile Ser Thr Ala Gln Ile 405 410 415Leu Glu Asn Ser Leu
Leu Asn Leu Glu Asn Gly Glu Asp Ala Gly Val 420 425 430Gln Leu Ser
Asp Phe Ser Leu Ile Ile Ile Asp Glu Cys His His Thr 435 440 445Asn
Lys Glu Ala Val Tyr Asn Asn Ile Met Arg His Tyr Leu Met Gln 450 455
460Lys Leu Lys Asn Asn Arg Leu Lys Lys Glu Asn Lys Pro Val Ile
Pro465 470 475 480Leu Pro Gln Ile Leu Gly Leu Thr Ala Ser Pro Gly
Val Gly Gly Ala 485 490 495Thr Lys Gln Ala Lys Ala Glu Glu His Ile
Leu Lys Leu Cys Ala Asn 500 505 510Leu Asp Ala Phe Thr Ile Lys Thr
Val Lys Glu Asn Leu Asp Gln Leu 515 520 525Lys Asn Gln Ile Gln Glu
Pro Cys Lys Lys Phe Ala Ile Ala Asp Ala 530 535 540Thr Arg Glu Asp
Pro Phe Lys Glu Lys Leu Leu Glu Ile Met Thr Arg545 550 555 560Ile
Gln Thr Tyr Cys Gln Met Ser Pro Met Ser Asp Phe Gly Thr Gln 565 570
575Pro Tyr Glu Gln Trp Ala Ile Gln Met Glu Lys Lys Ala Ala Lys Glu
580 585 590Gly Asn Arg Lys Glu Arg Val Cys Ala Glu His Leu Arg Lys
Tyr Asn 595 600 605Glu Ala Leu Gln Ile Asn Asp Thr Ile Arg Met Ile
Asp Ala Tyr Thr 610 615 620His Leu Glu Thr Phe Tyr Asn Glu Glu Lys
Asp Lys Lys Phe Ala Val625 630 635 640Ile Glu Asp Asp Ser Asp Glu
Gly Gly Asp Asp Glu Tyr Cys Asp Gly 645 650 655Asp Glu Asp Glu Asp
Asp Leu Lys Lys Pro Leu Lys Leu Asp Glu Thr 660 665 670Asp Arg Phe
Leu Met Thr Leu Phe Phe Glu Asn Asn Lys Met Leu Lys 675 680 685Arg
Leu Ala Glu Asn Pro Glu Tyr Glu Asn Glu Lys Leu Thr Lys Leu 690 695
700Arg Asn Thr Ile Met Glu Gln Tyr Thr Arg Thr Glu Glu Ser Ala
Arg705 710 715 720Gly Ile Ile Phe Thr Lys Thr Arg Gln Ser Ala Tyr
Ala Leu Ser Gln 725 730 735Trp Ile Thr Glu Asn Glu Lys Phe Ala Glu
Val Gly Val Lys Ala His 740 745 750His Leu Ile Gly Ala Gly His Ser
Ser Glu Phe Lys Pro Met Thr Gln 755 760 765Asn Glu Gln Lys Glu Val
Ile Ser Lys Phe Arg Thr Gly Lys Ile Asn 770 775 780Leu Leu Ile Ala
Thr Thr Val Ala Glu Glu Gly Leu Asp Ile Lys Glu785 790 795 800Cys
Asn Ile Val Ile Arg Tyr Gly Leu Val Thr Asn Glu Ile Ala Met 805 810
815Val Gln Ala Arg Gly Arg Ala Arg Ala Asp Glu Ser Thr Tyr Val Leu
820 825 830Val Ala His Ser Gly Ser Gly Val Ile Glu His Glu Thr Val
Asn Asp 835 840 845Phe Arg Glu Lys Met Met Tyr Lys Ala Ile His Cys
Val Gln Asn Met 850 855 860Lys Pro Glu Glu Tyr Ala His Lys Ile Leu
Glu Leu Gln Met Gln Ser865 870 875 880Ile Met Glu Lys Lys Met Lys
Thr Lys Arg Asn Ile Ala Lys His Tyr 885 890 895Lys Asn Asn Pro Ser
Leu Ile Thr Phe Leu Cys Lys Asn Cys Ser Val 900 905 910Leu Ala Cys
Ser Gly Glu Asp Ile His Val Ile Glu Lys Met His His 915 920 925Val
Asn Met Thr Pro Glu Phe Lys Glu Leu Tyr Ile Val Arg Glu Asn 930 935
940Lys Ala Leu Gln Lys Lys Cys Ala Asp Tyr Gln Ile Asn Gly Glu
Ile945 950 955 960Ile Cys Lys Cys Gly Gln Ala Trp Gly Thr Met Met
Val His Lys Gly 965 970 975Leu Asp Leu Pro Cys Leu Lys Ile Arg Asn
Phe Val Val Val Phe Lys 980 985 990Asn Asn Ser Thr Lys Lys Gln Tyr
Lys Lys Trp Val Glu Leu Pro Ile 995 1000 1005Thr Phe Pro Asn Leu
Asp Tyr Ser Glu Cys Cys Leu Phe Ser Asp 1010 1015 1020Glu
Asp1025914992DNAHomo sapiens 91gaagactcca gatataggat cactccatgc
catcaagaaa gttgatgcta ttgggcccat 60ctcaagctga tcttggcacc tctcatgctc
tgctctcttc aaccagacct ctacattcca 120ttttggaaga agactaaaaa
tggtgtttcc aatgtggaca ctgaagagac aaattcttat 180cctttttaac
ataatcctaa tttccaaact ccttggggct agatggtttc ctaaaactct
240gccctgtgat gtcactctgg atgttccaaa gaaccatgtg atcgtggact
gcacagacaa 300gcatttgaca gaaattcctg gaggtattcc cacgaacacc
acgaacctca ccctcaccat 360taaccacata ccagacatct ccccagcgtc
ctttcacaga ctggaccatc tggtagagat 420cgatttcaga tgcaactgtg
tacctattcc actggggtca aaaaacaaca tgtgcatcaa 480gaggctgcag
attaaaccca gaagctttag tggactcact tatttaaaat ccctttacct
540ggatggaaac cagctactag agataccgca gggcctcccg cctagcttac
agcttctcag 600ccttgaggcc aacaacatct tttccatcag aaaagagaat
ctaacagaac tggccaacat 660agaaatactc tacctgggcc aaaactgtta
ttatcgaaat ccttgttatg tttcatattc 720aatagagaaa gatgccttcc
taaacttgac aaagttaaaa gtgctctccc tgaaagataa 780caatgtcaca
gccgtcccta ctgttttgcc atctacttta acagaactat atctctacaa
840caacatgatt gcaaaaatcc aagaagatga ttttaataac ctcaaccaat
tacaaattct 900tgacctaagt ggaaattgcc ctcgttgtta taatgcccca
tttccttgtg cgccgtgtaa 960aaataattct cccctacaga tccctgtaaa
tgcttttgat gcgctgacag aattaaaagt 1020tttacgtcta cacagtaact
ctcttcagca tgtgccccca agatggttta agaacatcaa 1080caaactccag
gaactggatc tgtcccaaaa cttcttggcc aaagaaattg gggatgctaa
1140atttctgcat tttctcccca gcctcatcca attggatctg tctttcaatt
ttgaacttca 1200ggtctatcgt gcatctatga atctatcaca agcattttct
tcactgaaaa gcctgaaaat 1260tctgcggatc agaggatatg tctttaaaga
gttgaaaagc tttaacctct cgccattaca 1320taatcttcaa aatcttgaag
ttcttgatct tggcactaac tttataaaaa ttgctaacct 1380cagcatgttt
aaacaattta aaagactgaa agtcatagat ctttcagtga ataaaatatc
1440accttcagga gattcaagtg aagttggctt ctgctcaaat gccagaactt
ctgtagaaag 1500ttatgaaccc caggtcctgg aacaattaca ttatttcaga
tatgataagt atgcaaggag 1560ttgcagattc aaaaacaaag aggcttcttt
catgtctgtt aatgaaagct gctacaagta 1620tgggcagacc ttggatctaa
gtaaaaatag tatatttttt gtcaagtcct ctgattttca 1680gcatctttct
ttcctcaaat gcctgaatct gtcaggaaat ctcattagcc aaactcttaa
1740tggcagtgaa ttccaacctt tagcagagct gagatatttg gacttctcca
acaaccggct 1800tgatttactc cattcaacag catttgaaga gcttcacaaa
ctggaagttc tggatataag 1860cagtaatagc cattattttc aatcagaagg
aattactcat atgctaaact ttaccaagaa 1920cctaaaggtt ctgcagaaac
tgatgatgaa cgacaatgac atctcttcct ccaccagcag 1980gaccatggag
agtgagtctc ttagaactct ggaattcaga ggaaatcact tagatgtttt
2040atggagagaa ggtgataaca gatacttaca attattcaag aatctgctaa
aattagagga 2100attagacatc tctaaaaatt ccctaagttt cttgccttct
ggagtttttg atggtatgcc 2160tccaaatcta aagaatctct ctttggccaa
aaatgggctc aaatctttca gttggaagaa 2220actccagtgt ctaaagaacc
tggaaacttt ggacctcagc cacaaccaac tgaccactgt 2280ccctgagaga
ttatccaact gttccagaag cctcaagaat ctgattctta agaataatca
2340aatcaggagt ctgacgaagt attttctaca agatgccttc cagttgcgat
atctggatct 2400cagctcaaat aaaatccaga tgatccaaaa gaccagcttc
ccagaaaatg tcctcaacaa 2460tctgaagatg ttgcttttgc atcataatcg
gtttctgtgc acctgtgatg ctgtgtggtt 2520tgtctggtgg gttaaccata
cggaggtgac tattccttac ctggccacag atgtgacttg 2580tgtggggcca
ggagcacaca agggccaaag tgtgatctcc ctggatctgt acacctgtga
2640gttagatctg actaacctga ttctgttctc actttccata tctgtatctc
tctttctcat 2700ggtgatgatg acagcaagtc acctctattt ctgggatgtg
tggtatattt accatttctg 2760taaggccaag ataaaggggt atcagcgtct
aatatcacca gactgttgct atgatgcttt 2820tattgtgtat gacactaaag
acccagctgt gaccgagtgg gttttggctg agctggtggc 2880caaactggaa
gacccaagag agaaacattt taatttatgt ctcgaggaaa gggactggtt
2940accagggcag ccagttctgg aaaacctttc ccagagcata cagcttagca
aaaagacagt 3000gtttgtgatg acagacaagt atgcaaagac tgaaaatttt
aagatagcat tttacttgtc 3060ccatcagagg ctcatggatg aaaaagttga
tgtgattatc ttgatatttc ttgagaagcc 3120ctttcagaag tccaagttcc
tccagctccg gaaaaggctc tgtgggagtt ctgtccttga 3180gtggccaaca
aacccgcaag ctcacccata cttctggcag tgtctaaaga acgccctggc
3240cacagacaat catgtggcct atagtcaggt gttcaaggaa acggtctagc
ccttctttgc 3300aaaacacaac tgcctagttt accaaggaga ggcctggctg
tttaaattgt tttcatatat 3360atcacaccaa aagcgtgttt tgaaattctt
caagaaatga gattgcccat atttcagggg 3420agccaccaac gtctgtcaca
ggagttggaa agatggggtt tatataatgc atcaagtctt 3480ctttcttatc
tctctgtgtc tctatttgca cttgagtctc tcacctcagc tcctgtaaaa
3540gagtggcaag taaaaaacat ggggctctga ttctcctgta attgtgataa
ttaaatatac 3600acacaatcat gacattgaga agaactgcat ttctaccctt
aaaaagtact ggtatataca 3660gaaatagggt taaaaaaaac tcaagctctc
tctatatgag accaaaatgt actagagtta 3720gtttagtgaa ataaaaaacc
agtcagctgg ccgggcatgg tggctcatgc ttgtaatccc 3780agcactttgg
gaggccgagg caggtggatc acgaggtcag gagtttgaga ccagtctggc
3840caacatggtg aaaccccgtc tgtactaaaa atacaaaaat tagctgggcg
tggtggtggg 3900tgcctgtaat cccagctact tgggaggctg aggcaggaga
atcgcttgaa cccgggaggt 3960ggaggtggca gtgagccgag atcacgccac
tgcaatgcag cccgggcaac agagctagac 4020tgtctcaaaa gaacaaaaaa
aaaaaaacac aaaaaaactc agtcagcttc ttaaccaatt 4080gcttccgtgt
catccagggc cccattctgt gcagattgag tgtgggcacc acacaggtgg
4140ttgctgcttc agtgcttcct gctctttttc cttgggcctg cttctgggtt
ccatagggaa 4200acagtaagaa agaaagacac atccttacca taaatgcata
tggtccacct acaaatagaa 4260aaatatttaa atgatctgcc tttatacaaa
gtgatattct ctacctttga taatttacct 4320gcttaaatgt ttttatctgc
actgcaaagt actgtatcca aagtaaaatt tcctcatcca 4380atatctttca
aactgttttg ttaactaatg ccatatattt gtaagtatct gcacacttga
4440tacagcaacg ttagatggtt ttgatggtaa accctaaagg aggactccaa
gagtgtgtat 4500ttatttatag ttttatcaga gatgacaatt atttgaatgc
caattatatg gattcctttc 4560attttttgct ggaggatggg agaagaaacc
aaagtttata gaccttcaca ttgagaaagc 4620ttcagttttg aacttcagct
atcagattca aaaacaacag aaagaaccaa gacattctta 4680agatgcctgt
actttcagct gggtataaat tcatgagttc aaagattgaa acctgaccaa
4740tttgctttat ttcatggaag aagtgatcta caaaggtgtt tgtgccattt
ggaaaacagc 4800gtgcatgtgt tcaagcctta gattggcgat gtcgtatttt
cctcacgtgt ggcaatgcca 4860aaggctttac tttacctgtg agtacacact
atatgaatta tttccaacgt acatttaatc 4920aataagggtc acaaattccc
aaatcaatct ctggaataaa tagagaggta attaaattgc 4980tggagccaac ta
4992921049PRTHomo sapiens 92Met Val Phe Pro Met Trp Thr Leu Lys Arg
Gln Ile Leu Ile Leu Phe1 5 10 15Asn Ile Ile Leu Ile Ser Lys Leu Leu
Gly Ala Arg Trp Phe Pro Lys 20 25 30Thr Leu Pro Cys Asp Val Thr Leu
Asp Val Pro Lys Asn His Val Ile 35 40 45Val Asp Cys Thr Asp Lys His
Leu Thr Glu Ile Pro Gly Gly Ile Pro 50 55 60Thr Asn Thr Thr Asn Leu
Thr Leu Thr Ile Asn His Ile Pro Asp Ile65 70 75 80Ser Pro Ala Ser
Phe His Arg Leu Asp His Leu Val Glu Ile Asp Phe 85 90 95Arg Cys Asn
Cys Val Pro Ile Pro Leu Gly Ser Lys Asn Asn Met Cys 100 105 110Ile
Lys Arg Leu Gln Ile Lys Pro Arg Ser Phe Ser Gly Leu Thr Tyr 115 120
125Leu Lys Ser Leu Tyr Leu Asp Gly Asn Gln Leu Leu Glu Ile Pro Gln
130 135 140Gly Leu Pro Pro Ser Leu Gln Leu Leu Ser Leu Glu Ala Asn
Asn Ile145 150 155 160Phe Ser Ile Arg Lys Glu Asn Leu Thr Glu Leu
Ala Asn Ile Glu Ile 165 170 175Leu Tyr Leu Gly Gln Asn Cys Tyr Tyr
Arg Asn Pro Cys Tyr Val Ser 180 185 190Tyr Ser Ile Glu Lys Asp Ala
Phe Leu Asn Leu Thr Lys Leu Lys Val 195 200 205Leu Ser Leu Lys Asp
Asn Asn Val Thr Ala Val Pro Thr Val Leu Pro 210 215 220Ser Thr Leu
Thr Glu Leu Tyr Leu Tyr Asn Asn Met Ile Ala Lys Ile225 230 235
240Gln Glu Asp Asp Phe Asn Asn Leu Asn Gln Leu Gln Ile Leu Asp Leu
245 250 255Ser Gly Asn Cys Pro Arg Cys Tyr Asn Ala Pro Phe Pro Cys
Ala Pro 260 265 270Cys Lys Asn Asn Ser Pro Leu Gln Ile Pro Val Asn
Ala Phe Asp Ala 275 280 285Leu Thr Glu Leu Lys Val Leu Arg Leu His
Ser Asn Ser Leu Gln His 290 295 300Val Pro Pro Arg Trp Phe Lys Asn
Ile Asn Lys Leu Gln Glu Leu Asp305 310 315 320Leu Ser Gln Asn Phe
Leu Ala Lys Glu Ile Gly Asp Ala Lys Phe Leu 325 330 335His Phe Leu
Pro Ser Leu Ile Gln Leu Asp Leu Ser Phe Asn Phe Glu 340 345 350Leu
Gln Val Tyr Arg Ala Ser Met Asn Leu Ser Gln Ala Phe Ser Ser 355 360
365Leu Lys Ser Leu Lys Ile Leu Arg Ile Arg Gly Tyr Val Phe Lys Glu
370 375 380Leu Lys Ser Phe Asn Leu Ser Pro Leu His Asn Leu Gln Asn
Leu Glu385 390 395 400Val Leu Asp Leu Gly Thr Asn Phe Ile Lys Ile
Ala Asn Leu Ser Met 405 410 415Phe Lys Gln Phe Lys Arg Leu Lys Val
Ile Asp Leu Ser Val Asn Lys 420 425 430Ile Ser Pro Ser Gly Asp Ser
Ser Glu Val Gly Phe Cys Ser Asn Ala 435 440 445Arg Thr Ser Val Glu
Ser Tyr Glu Pro Gln Val Leu Glu Gln Leu His 450 455 460Tyr Phe Arg
Tyr Asp Lys Tyr Ala Arg Ser Cys Arg Phe Lys Asn Lys465 470 475
480Glu Ala Ser Phe Met Ser Val Asn Glu Ser Cys Tyr Lys Tyr Gly Gln
485 490 495Thr Leu Asp Leu Ser Lys Asn Ser Ile Phe Phe Val Lys Ser
Ser Asp 500 505 510Phe Gln His Leu Ser Phe Leu Lys Cys Leu Asn Leu
Ser Gly Asn Leu 515 520 525Ile Ser Gln Thr Leu Asn Gly Ser Glu Phe
Gln Pro Leu Ala Glu Leu 530 535 540Arg Tyr Leu Asp Phe Ser Asn Asn
Arg Leu Asp Leu Leu His Ser Thr545 550 555 560Ala Phe Glu Glu Leu
His Lys Leu Glu Val Leu Asp Ile Ser Ser Asn 565 570 575Ser His Tyr
Phe Gln Ser Glu Gly Ile Thr His Met Leu Asn Phe Thr 580 585 590Lys
Asn Leu Lys Val Leu Gln Lys Leu Met Met Asn Asp Asn Asp Ile 595 600
605Ser Ser Ser Thr Ser Arg Thr Met Glu Ser Glu Ser Leu Arg Thr Leu
610 615 620Glu Phe Arg Gly Asn His Leu Asp Val Leu Trp Arg Glu Gly
Asp Asn625 630 635 640Arg Tyr Leu Gln Leu Phe Lys Asn Leu Leu Lys
Leu Glu Glu Leu Asp 645 650 655Ile Ser Lys Asn Ser Leu Ser Phe Leu
Pro Ser Gly Val Phe Asp Gly 660 665 670Met Pro Pro Asn Leu Lys Asn
Leu Ser Leu Ala Lys Asn Gly Leu Lys 675 680 685Ser Phe Ser Trp Lys
Lys Leu Gln Cys Leu Lys Asn Leu Glu Thr Leu 690 695 700Asp Leu Ser
His Asn Gln Leu Thr Thr Val Pro Glu Arg Leu Ser Asn705 710 715
720Cys Ser Arg Ser Leu Lys Asn Leu Ile Leu Lys Asn Asn Gln Ile Arg
725 730 735Ser Leu Thr Lys Tyr Phe Leu Gln Asp Ala Phe Gln Leu Arg
Tyr Leu 740 745 750Asp Leu Ser Ser Asn Lys Ile Gln Met Ile Gln Lys
Thr Ser Phe Pro 755 760 765Glu Asn Val Leu Asn Asn Leu Lys Met Leu
Leu Leu His His Asn Arg 770 775 780Phe Leu Cys Thr Cys Asp Ala Val
Trp Phe Val Trp Trp Val Asn His785 790 795 800Thr Glu Val Thr Ile
Pro Tyr Leu Ala Thr Asp Val Thr Cys Val Gly 805 810 815Pro Gly Ala
His Lys Gly Gln Ser Val Ile Ser Leu Asp Leu Tyr Thr 820 825 830Cys
Glu Leu Asp Leu Thr Asn Leu Ile Leu Phe Ser Leu Ser Ile Ser 835 840
845Val Ser Leu Phe Leu Met Val Met Met Thr Ala Ser His Leu Tyr Phe
850 855 860Trp Asp Val Trp Tyr Ile Tyr His Phe Cys Lys Ala Lys Ile
Lys Gly865 870 875 880Tyr Gln Arg Leu Ile Ser Pro Asp Cys Cys Tyr
Asp Ala Phe Ile Val 885 890 895Tyr Asp Thr Lys Asp Pro Ala Val Thr
Glu Trp Val Leu Ala Glu Leu 900 905 910Val Ala Lys Leu Glu Asp Pro
Arg Glu Lys His Phe Asn Leu Cys Leu 915 920 925Glu Glu Arg Asp Trp
Leu Pro Gly Gln Pro Val Leu Glu Asn Leu Ser 930 935 940Gln Ser Ile
Gln Leu Ser Lys Lys Thr Val Phe Val Met Thr Asp Lys945 950 955
960Tyr Ala Lys Thr Glu Asn Phe Lys Ile Ala Phe Tyr Leu Ser His Gln
965 970 975Arg Leu Met Asp Glu Lys Val Asp Val Ile Ile Leu Ile Phe
Leu Glu 980 985 990Lys Pro Phe Gln Lys Ser Lys Phe Leu Gln Leu Arg
Lys Arg Leu Cys 995 1000 1005Gly Ser Ser Val Leu Glu Trp Pro Thr
Asn Pro Gln Ala His Pro 1010 1015 1020Tyr Phe Trp Gln Cys Leu Lys
Asn Ala Leu Ala Thr Asp Asn His 1025 1030 1035Val Ala Tyr Ser Gln
Val Phe Lys Glu Thr Val 1040 1045931972DNAHomo sapiens 93gaaactcccg
cctggccacc ataaaagcgc cggccctccg cttccccgcg agacgaaact 60tcccgtcccg
gcggctctgg cacccagggt ccggcctgcg ccttcccgcc aggcctggac
120actggttcaa cacctgtgac ttcatgtgtg cgcgccggcc acacctgcag
tcacacctgt 180agccccctct gccaagagat ccataccgag gcagcgtcgg
tggctacaag ccctcagtcc 240acacctgtgg acacctgtga cacctggcca
cacgacctgt ggccgcggcc tggcgtctgc 300tgcgacagga gcccttacct
cccctgttat aacacctgac cgccacctaa ctgcccctgc 360agaaggagca
atggccttgg ctcctgagag ggcagcccca cgcgtgctgt tcggagagtg
420gctccttgga gagatcagca gcggctgcta tgaggggctg cagtggctgg
acgaggcccg 480cacctgtttc cgcgtgccct ggaagcactt cgcgcgcaag
gacctgagcg aggccgacgc 540gcgcatcttc aaggcctggg ctgtggcccg
cggcaggtgg ccgcctagca gcaggggagg 600tggcccgccc cccgaggctg
agactgcgga gcgcgccggc tggaaaacca acttccgctg 660cgcactgcgc
agcacgcgtc gcttcgtgat gctgcgggat aactcggggg acccggccga
720cccgcacaag gtgtacgcgc tcagccggga gctgtgctgg cgagaaggcc
caggcacgga 780ccagactgag gcagaggccc ccgcagctgt cccaccacca
cagggtgggc ccccagggcc 840attcctggca cacacacatg ctggactcca
agccccaggc cccctccctg ccccagctgg 900tgacaagggg gacctcctgc
tccaggcagt gcaacagagc tgcctggcag accatctgct 960gacagcgtca
tggggggcag atccagtccc aaccaaggct cctggagagg gacaagaagg
1020gcttcccctg actggggcct gtgctggagg cccagggctc cctgctgggg
agctgtacgg 1080gtgggcagta gagacgaccc ccagccccgg gccccagccc
gcggcactaa cgacaggcga 1140ggccgcggcc ccagagtccc cgcaccaggc
agagccgtac ctgtcaccct ccccaagcgc 1200ctgcaccgcg gtgcaagagc
ccagcccagg ggcgctggac gtgaccatca tgtacaaggg 1260ccgcacggtg
ctgcagaagg tggtgggaca cccgagctgc acgttcctat acggcccccc
1320agacccagct gtccgggcca cagaccccca gcaggtagca ttccccagcc
ctgccgagct 1380cccggaccag aagcagctgc gctacacgga ggaactgctg
cggcacgtgg cccctgggtt 1440gcacctggag cttcgggggc cacagctgtg
ggcccggcgc atgggcaagt gcaaggtgta 1500ctgggaggtg ggcggacccc
caggctccgc cagcccctcc accccagcct gcctgctgcc 1560tcggaactgt
gacaccccca tcttcgactt cagagtcttc ttccaagagc tggtggaatt
1620ccgggcacgg cagcgccgtg gctccccacg ctataccatc tacctgggct
tcgggcagga 1680cctgtcagct gggaggccca aggagaagag cctggtcctg
gtgaagctgg aaccctggct 1740gtgccgagtg cacctagagg gcacgcagcg
tgagggtgtg tcttccctgg atagcagcag 1800cctcagcctc tgcctgtcca
gcgccaacag cctctatgac gacatcgagt gcttccttat 1860ggagctggag
cagcccgcct agaacccagt ctaatgagaa ctccagaaag ctggagcagc
1920ccacctagag ctggccgcgg ccgcccagtc taataaaaag aactccagaa ca
197294503PRTHomo sapiens 94Met Ala Leu Ala Pro Glu Arg Ala Ala Pro
Arg Val Leu Phe Gly Glu1 5 10 15Trp Leu Leu Gly Glu Ile Ser Ser Gly
Cys Tyr Glu Gly Leu Gln Trp 20 25 30Leu Asp Glu Ala Arg Thr Cys Phe
Arg Val Pro Trp Lys His Phe Ala 35 40 45Arg Lys Asp Leu Ser Glu Ala
Asp Ala Arg Ile Phe Lys Ala Trp Ala 50 55 60Val Ala Arg Gly Arg Trp
Pro Pro Ser Ser Arg Gly Gly Gly Pro Pro65 70 75 80Pro Glu Ala Glu
Thr Ala Glu Arg Ala Gly Trp Lys Thr Asn Phe Arg 85 90 95Cys Ala Leu
Arg Ser Thr Arg Arg Phe Val Met Leu Arg Asp Asn Ser 100 105 110Gly
Asp Pro Ala Asp Pro His Lys Val Tyr Ala Leu Ser Arg Glu Leu 115 120
125Cys Trp Arg Glu Gly Pro Gly Thr Asp Gln Thr Glu Ala Glu Ala Pro
130 135 140Ala Ala Val Pro Pro Pro Gln Gly Gly Pro Pro Gly Pro Phe
Leu Ala145 150 155 160His Thr His Ala Gly Leu Gln Ala Pro Gly Pro
Leu Pro Ala Pro Ala 165 170 175Gly Asp Lys Gly Asp Leu Leu Leu Gln
Ala Val Gln Gln Ser Cys Leu 180 185 190Ala Asp His Leu Leu Thr Ala
Ser Trp Gly Ala Asp Pro Val Pro Thr 195 200 205Lys Ala Pro Gly Glu
Gly Gln Glu Gly Leu Pro Leu Thr Gly Ala Cys 210 215 220Ala Gly Gly
Pro Gly Leu Pro Ala Gly Glu Leu Tyr Gly Trp Ala Val225 230 235
240Glu Thr Thr Pro Ser Pro Gly Pro Gln Pro Ala Ala Leu Thr Thr Gly
245 250 255Glu Ala Ala Ala Pro Glu Ser Pro His Gln Ala Glu Pro Tyr
Leu Ser 260 265 270Pro Ser Pro Ser Ala Cys Thr Ala Val Gln Glu Pro
Ser Pro Gly Ala 275 280 285Leu Asp Val Thr Ile Met Tyr Lys Gly Arg
Thr Val Leu Gln Lys Val 290 295 300Val Gly His Pro Ser Cys Thr Phe
Leu Tyr Gly Pro Pro Asp Pro Ala305 310 315 320Val Arg Ala Thr Asp
Pro Gln Gln Val Ala Phe Pro Ser Pro Ala Glu 325 330 335Leu Pro Asp
Gln Lys Gln Leu Arg Tyr Thr Glu Glu Leu Leu Arg His 340 345 350Val
Ala Pro Gly Leu His Leu Glu Leu Arg Gly Pro Gln Leu Trp Ala 355 360
365Arg Arg Met Gly Lys Cys Lys Val Tyr Trp Glu Val Gly Gly Pro Pro
370 375 380Gly Ser Ala Ser Pro Ser Thr Pro Ala Cys Leu Leu Pro Arg
Asn Cys385 390 395 400Asp Thr Pro Ile Phe Asp Phe Arg Val Phe Phe
Gln Glu Leu Val Glu 405 410 415Phe Arg Ala Arg Gln Arg Arg Gly Ser
Pro Arg Tyr Thr Ile Tyr Leu 420 425 430Gly Phe Gly Gln Asp Leu Ser
Ala Gly Arg Pro Lys Glu Lys Ser Leu 435 440 445Val Leu Val Lys Leu
Glu Pro Trp Leu Cys Arg Val His Leu Glu Gly 450 455 460Thr Gln Arg
Glu Gly Val Ser Ser Leu Asp Ser Ser Ser Leu Ser Leu465 470 475
480Cys Leu Ser Ser Ala Asn Ser Leu Tyr Asp Asp Ile Glu Cys Phe Leu
485 490 495Met Glu Leu Glu Gln Pro Ala 500951885DNAHomo sapiens
95gaaactcccg cctggccacc ataaaagcgc cggccctccg cttccccgcg agacgaaact
60tcccgtcccg gcggctctgg cacccagggt ccggcctgcg ccttcccgcc aggcctggac
120actggttcaa cacctgtgac ttcatgtgtg cgcgccggcc acacctgcag
tcacacctgt 180agccccctct gccaagagat ccataccgag gcagcgtcgg
tggctacaag ccctcagtcc 240acacctgtgg acacctgtga cacctggcca
cacgacctgt ggccgcggcc tggcgtctgc 300tgcgacagga gcccttacct
cccctgttat aacacctgac cgccacctaa ctgcccctgc 360agaaggagca
atggccttgg ctcctgagag ggcagcccca cgcgtgctgt tcggagagtg
420gctccttgga gagatcagca gcggctgcta tgaggggctg cagtggctgg
acgaggcccg 480cacctgtttc cgcgtgccct ggaagcactt cgcgcgcaag
gacctgagcg aggccgacgc 540gcgcatcttc aaggcctggg ctgtggcccg
cggcaggtgg ccgcctagca gcaggggagg 600tggcccgccc cccgaggctg
agactgcgga gcgcgccggc tggaaaacca acttccgctg 660cgcactgcgc
agcacgcgtc gcttcgtgat gctgcgggat aactcggggg acccggccga
720cccgcacaag gtgtacgcgc tcagccggga gctgtgctgg cgagaaggcc
caggcacgga 780ccagactgag gcagaggccc ccgcagctgt cccaccacca
cagggtgggc ccccagggcc 840attcctggca cacacacatg ctggactcca
agccccaggc cccctccctg ccccagctgg 900tgacaagggg gacctcctgc
tccaggcagt gcaacagagc tgcctggcag accatctgct 960gacagcgtca
tggggggcag atccagtccc aaccaaggct cctggagagg gacaagaagg
1020gcttcccctg actggggcct gtgctggagg cgaggccgcg gccccagagt
ccccgcacca 1080ggcagagccg tacctgtcac cctccccaag cgcctgcacc
gcggtgcaag agcccagccc 1140aggggcgctg gacgtgacca tcatgtacaa
gggccgcacg gtgctgcaga aggtggtggg 1200acacccgagc tgcacgttcc
tatacggccc cccagaccca gctgtccggg ccacagaccc 1260ccagcaggta
gcattcccca gccctgccga gctcccggac cagaagcagc tgcgctacac
1320ggaggaactg ctgcggcacg tggcccctgg gttgcacctg gagcttcggg
ggccacagct 1380gtgggcccgg cgcatgggca agtgcaaggt gtactgggag
gtgggcggac ccccaggctc 1440cgccagcccc tccaccccag cctgcctgct
gcctcggaac tgtgacaccc ccatcttcga 1500cttcagagtc ttcttccaag
agctggtgga attccgggca cggcagcgcc gtggctcccc 1560acgctatacc
atctacctgg gcttcgggca ggacctgtca gctgggaggc ccaaggagaa
1620gagcctggtc ctggtgaagc tggaaccctg gctgtgccga gtgcacctag
agggcacgca 1680gcgtgagggt gtgtcttccc tggatagcag cagcctcagc
ctctgcctgt ccagcgccaa 1740cagcctctat gacgacatcg agtgcttcct
tatggagctg gagcagcccg cctagaaccc 1800agtctaatga gaactccaga
aagctggagc agcccaccta gagctggccg cggccgccca 1860gtctaataaa
aagaactcca gaaca 188596474PRTHomo sapiens 96Met Ala Leu Ala Pro Glu
Arg Ala Ala Pro Arg Val Leu Phe Gly Glu1 5 10 15Trp Leu Leu Gly Glu
Ile Ser Ser Gly Cys Tyr Glu Gly Leu Gln Trp 20 25 30Leu Asp Glu Ala
Arg Thr Cys Phe Arg Val Pro Trp Lys His Phe Ala 35 40 45Arg Lys Asp
Leu Ser Glu Ala Asp Ala Arg Ile Phe Lys Ala Trp Ala 50 55 60Val Ala
Arg Gly Arg Trp Pro Pro Ser Ser Arg Gly Gly Gly Pro Pro65 70 75
80Pro Glu Ala Glu Thr Ala Glu Arg Ala Gly Trp Lys Thr Asn Phe Arg
85 90 95Cys Ala Leu Arg Ser Thr Arg Arg Phe Val Met Leu Arg Asp Asn
Ser 100 105 110Gly Asp Pro Ala Asp Pro His Lys Val Tyr Ala Leu Ser
Arg Glu Leu 115 120 125Cys Trp Arg Glu Gly Pro Gly Thr Asp Gln Thr
Glu Ala Glu Ala Pro 130 135 140Ala Ala Val Pro Pro Pro Gln Gly Gly
Pro Pro Gly Pro Phe Leu Ala145 150 155 160His Thr His Ala Gly Leu
Gln Ala Pro Gly Pro Leu Pro Ala Pro Ala 165 170 175Gly Asp Lys Gly
Asp Leu Leu Leu Gln Ala Val Gln Gln Ser Cys Leu 180 185 190Ala Asp
His Leu Leu Thr Ala Ser Trp Gly Ala Asp Pro Val Pro Thr 195 200
205Lys Ala Pro Gly Glu Gly Gln Glu Gly Leu Pro Leu Thr Gly Ala Cys
210 215 220Ala Gly Gly Glu Ala Ala Ala Pro Glu Ser Pro His Gln Ala
Glu Pro225 230 235 240Tyr Leu Ser Pro Ser Pro Ser Ala Cys Thr Ala
Val Gln Glu Pro Ser 245 250 255Pro Gly Ala Leu Asp Val Thr Ile Met
Tyr Lys Gly Arg Thr Val Leu 260 265 270Gln Lys Val Val Gly His Pro
Ser Cys Thr Phe Leu Tyr Gly Pro Pro 275 280 285Asp Pro Ala Val Arg
Ala Thr Asp Pro Gln Gln Val Ala Phe Pro Ser 290 295 300Pro Ala Glu
Leu Pro Asp Gln Lys Gln Leu Arg Tyr Thr Glu Glu Leu305 310 315
320Leu Arg His Val Ala Pro Gly Leu His Leu Glu Leu Arg Gly Pro Gln
325 330 335Leu Trp Ala Arg Arg Met Gly Lys Cys Lys Val Tyr Trp Glu
Val Gly 340 345 350Gly Pro Pro Gly Ser Ala Ser Pro Ser Thr Pro Ala
Cys Leu Leu Pro 355 360 365Arg Asn Cys Asp Thr Pro Ile Phe Asp Phe
Arg Val Phe Phe Gln Glu 370 375 380Leu Val Glu Phe Arg Ala Arg Gln
Arg Arg Gly Ser Pro Arg Tyr Thr385 390 395 400Ile Tyr Leu Gly Phe
Gly Gln Asp Leu Ser Ala Gly Arg Pro Lys Glu 405 410 415Lys Ser Leu
Val Leu Val Lys Leu Glu Pro Trp Leu Cys Arg Val His 420 425 430Leu
Glu Gly Thr Gln Arg Glu Gly Val Ser Ser Leu Asp Ser Ser Ser 435 440
445Leu Ser Leu Cys Leu Ser Ser Ala Asn Ser Leu Tyr Asp Asp Ile Glu
450 455 460Cys Phe Leu Met Glu Leu Glu Gln Pro Ala465
47097860DNAHomo sapiens 97ccagccttca gccggagaac cgtttactcg
ctgctgtgcc catctatcag caggctccgg 60gctgaagatt gcttctcttc tctcctccaa
ggtctagtga cggagcccgc gcgcggcgcc 120accatgcggc agaaggcggt
atcgcttttc ttgtgctacc tgctgctctt cacttgcagt 180ggggtggagg
caggtgagaa tgcgggtaag gatgcaggta agaaaaagtg ctcggagagc
240tcggacagcg gctccgggtt ctggaaggcc ctgaccttca tggccgtcgg
aggaggactc 300gcagtcgccg ggctgcccgc gctgggcttc accggcgccg
gcatcgcggc caactcggtg 360gctgcctcgc tgatgagctg gtctgcgatc
ctgaatgggg gcggcgtgcc cgccgggggg 420ctagtggcca cgctgcagag
cctcggggct ggtggcagca gcgtcgtcat aggtaatatt 480ggtgccctga
tgggctacgc cacccacaag tatctcgata gtgaggagga tgaggagtag
540ccagcagctc ccagaacctc ttcttccttc ttggcctaac tcttccagtt
aggatctaga 600actttgcctt tttttttttt tttttttttt tgagatgggt
tctcactata ttgtccaggc 660tagagtgcag tggctattca cagatgcgaa
catagtacac tgcagcctcc aactcctagc 720ctcaagtgat cctcctgtct
caacctccca agtaggatta caagcatgcg ccgacgatgc 780ccagaatcca
gaactttgtc tatcactctc cccaacaacc tagatgtgaa aacagaataa
840acttcaccca gaaaacactt 86098138PRTHomo sapiens 98Met Arg Gln Lys
Ala Val Ser Leu Phe Leu Cys Tyr Leu Leu Leu Phe1 5 10 15Thr Cys Ser
Gly Val Glu Ala Gly Glu Asn Ala Gly Lys Asp Ala Gly 20 25 30Lys Lys
Lys Cys Ser Glu Ser Ser Asp Ser Gly Ser Gly Phe Trp Lys 35 40 45Ala
Leu Thr Phe Met Ala Val Gly Gly Gly Leu Ala Val Ala Gly Leu 50 55
60Pro Ala Leu Gly Phe Thr Gly Ala Gly Ile Ala Ala Asn Ser Val Ala65
70 75 80Ala Ser Leu Met Ser Trp Ser Ala Ile Leu Asn Gly Gly Gly Val
Pro 85 90 95Ala Gly Gly Leu Val Ala Thr Leu Gln Ser Leu Gly Ala Gly
Gly Ser 100 105 110Ser Val Val Ile Gly Asn Ile Gly Ala Leu Met Gly
Tyr Ala Thr His 115 120 125Lys Tyr Leu Asp Ser Glu Glu Asp Glu Glu
130 13599848DNAHomo sapiens 99ccagccttca gccggagaac cgtttactcg
ctgctgtgcc catctatcag caggctccgg 60gctgaagatt gcttctcttc tctcctccaa
ggtctagtga cggagcccgc gcgcggcgcc 120accatgcggc agaaggcggt
atcgcttttc ttgtgctacc tgctgctctt cacttgcagt 180ggggtggagg
caggtgagaa tgcgggtaag aaaaagtgct cggagagctc ggacagcggc
240tccgggttct ggaaggccct gaccttcatg gccgtcggag gaggactcgc
agtcgccggg 300ctgcccgcgc tgggcttcac cggcgccggc atcgcggcca
actcggtggc tgcctcgctg 360atgagctggt ctgcgatcct gaatgggggc
ggcgtgcccg ccggggggct agtggccacg 420ctgcagagcc tcggggctgg
tggcagcagc gtcgtcatag gtaatattgg tgccctgatg 480ggctacgcca
cccacaagta tctcgatagt gaggaggatg aggagtagcc agcagctccc
540agaacctctt cttccttctt ggcctaactc ttccagttag gatctagaac
tttgcctttt 600tttttttttt tttttttttg agatgggttc tcactatatt
gtccaggcta gagtgcagtg 660gctattcaca gatgcgaaca tagtacactg
cagcctccaa ctcctagcct caagtgatcc 720tcctgtctca acctcccaag
taggattaca agcatgcgcc gacgatgccc agaatccaga 780actttgtcta
tcactctccc caacaaccta gatgtgaaaa cagaataaac ttcacccaga 840aaacactt
848100134PRTHomo sapiens 100Met Arg Gln Lys Ala Val Ser Leu Phe Leu
Cys Tyr Leu Leu Leu Phe1 5 10 15Thr Cys Ser Gly Val Glu Ala Gly Glu
Asn Ala Gly Lys Lys Lys Cys 20 25 30Ser Glu Ser Ser Asp Ser Gly Ser
Gly Phe Trp Lys Ala Leu Thr Phe 35 40 45Met Ala Val Gly Gly Gly Leu
Ala Val Ala Gly Leu Pro Ala Leu Gly 50 55 60Phe Thr Gly Ala Gly Ile
Ala Ala Asn Ser Val Ala Ala Ser Leu Met65
70 75 80Ser Trp Ser Ala Ile Leu Asn Gly Gly Gly Val Pro Ala Gly Gly
Leu 85 90 95Val Ala Thr Leu Gln Ser Leu Gly Ala Gly Gly Ser Ser Val
Val Ile 100 105 110Gly Asn Ile Gly Ala Leu Met Gly Tyr Ala Thr His
Lys Tyr Leu Asp 115 120 125Ser Glu Glu Asp Glu Glu 130101836DNAHomo
sapiens 101ccagccttca gccggagaac cgtttactcg ctgctgtgcc catctatcag
caggctccgg 60gctgaagatt gcttctcttc tctcctccaa ggtctagtga cggagcccgc
gcgcggcgcc 120accatgcggc agaaggcggt atcgcttttc ttgtgctacc
tgctgctctt cacttgcagt 180ggggtggagg caggtaagaa aaagtgctcg
gagagctcgg acagcggctc cgggttctgg 240aaggccctga ccttcatggc
cgtcggagga ggactcgcag tcgccgggct gcccgcgctg 300ggcttcaccg
gcgccggcat cgcggccaac tcggtggctg cctcgctgat gagctggtct
360gcgatcctga atgggggcgg cgtgcccgcc ggggggctag tggccacgct
gcagagcctc 420ggggctggtg gcagcagcgt cgtcataggt aatattggtg
ccctgatggg ctacgccacc 480cacaagtatc tcgatagtga ggaggatgag
gagtagccag cagctcccag aacctcttct 540tccttcttgg cctaactctt
ccagttagga tctagaactt tgcctttttt tttttttttt 600tttttttgag
atgggttctc actatattgt ccaggctaga gtgcagtggc tattcacaga
660tgcgaacata gtacactgca gcctccaact cctagcctca agtgatcctc
ctgtctcaac 720ctcccaagta ggattacaag catgcgccga cgatgcccag
aatccagaac tttgtctatc 780actctcccca acaacctaga tgtgaaaaca
gaataaactt cacccagaaa acactt 836102130PRTHomo sapiens 102Met Arg
Gln Lys Ala Val Ser Leu Phe Leu Cys Tyr Leu Leu Leu Phe1 5 10 15Thr
Cys Ser Gly Val Glu Ala Gly Lys Lys Lys Cys Ser Glu Ser Ser 20 25
30Asp Ser Gly Ser Gly Phe Trp Lys Ala Leu Thr Phe Met Ala Val Gly
35 40 45Gly Gly Leu Ala Val Ala Gly Leu Pro Ala Leu Gly Phe Thr Gly
Ala 50 55 60Gly Ile Ala Ala Asn Ser Val Ala Ala Ser Leu Met Ser Trp
Ser Ala65 70 75 80Ile Leu Asn Gly Gly Gly Val Pro Ala Gly Gly Leu
Val Ala Thr Leu 85 90 95Gln Ser Leu Gly Ala Gly Gly Ser Ser Val Val
Ile Gly Asn Ile Gly 100 105 110Ala Leu Met Gly Tyr Ala Thr His Lys
Tyr Leu Asp Ser Glu Glu Asp 115 120 125Glu Glu 1301034326DNAHomo
sapiens 103gctgagcgcg gagccgcccg gtgattggtg ggggcggaag ggggccgggc
gccagcgctg 60ccttttctcc tgccgggtag tttcgctttc ctgcgcagag tctgcggagg
ggctcggctg 120caccgggggg atcgcgcctg gcagacccca gaccgagcag
aggcgaccca gcgcgctcgg 180gagaggctgc accgccgcgc ccccgcctag
cccttccgga tcctgcgcgc agaaaagttt 240catttgctgt atgccatcct
cgagagctgt ctaggttaac gttcgcactc tgtgtatata 300acctcgacag
tcttggcacc taacgtgctg tgcgtagctg ctcctttggt tgaatcccca
360ggcccttgtt ggggcacaag gtggcaggat gtctcagtgg tacgaacttc
agcagcttga 420ctcaaaattc ctggagcagg ttcaccagct ttatgatgac
agttttccca tggaaatcag 480acagtacctg gcacagtggt tagaaaagca
agactgggag cacgctgcca atgatgtttc 540atttgccacc atccgttttc
atgacctcct gtcacagctg gatgatcaat atagtcgctt 600ttctttggag
aataacttct tgctacagca taacataagg aaaagcaagc gtaatcttca
660ggataatttt caggaagacc caatccagat gtctatgatc atttacagct
gtctgaagga 720agaaaggaaa attctggaaa acgcccagag atttaatcag
gctcagtcgg ggaatattca 780gagcacagtg atgttagaca aacagaaaga
gcttgacagt aaagtcagaa atgtgaagga 840caaggttatg tgtatagagc
atgaaatcaa gagcctggaa gatttacaag atgaatatga 900cttcaaatgc
aaaaccttgc agaacagaga acacgagacc aatggtgtgg caaagagtga
960tcagaaacaa gaacagctgt tactcaagaa gatgtattta atgcttgaca
ataagagaaa 1020ggaagtagtt cacaaaataa tagagttgct gaatgtcact
gaacttaccc agaatgccct 1080gattaatgat gaactagtgg agtggaagcg
gagacagcag agcgcctgta ttggggggcc 1140gcccaatgct tgcttggatc
agctgcagaa ctggttcact atagttgcgg agagtctgca 1200gcaagttcgg
cagcagctta aaaagttgga ggaattggaa cagaaataca cctacgaaca
1260tgaccctatc acaaaaaaca aacaagtgtt atgggaccgc accttcagtc
ttttccagca 1320gctcattcag agctcgtttg tggtggaaag acagccctgc
atgccaacgc accctcagag 1380gccgctggtc ttgaagacag gggtccagtt
cactgtgaag ttgagactgt tggtgaaatt 1440gcaagagctg aattataatt
tgaaagtcaa agtcttattt gataaagatg tgaatgagag 1500aaatacagta
aaaggattta ggaagttcaa cattttgggc acgcacacaa aagtgatgaa
1560catggaggag tccaccaatg gcagtctggc ggctgaattt cggcacctgc
aattgaaaga 1620acagaaaaat gctggcacca gaacgaatga gggtcctctc
atcgttactg aagagcttca 1680ctcccttagt tttgaaaccc aattgtgcca
gcctggtttg gtaattgacc tcgagacgac 1740ctctctgccc gttgtggtga
tctccaacgt cagccagctc ccgagcggtt gggcctccat 1800cctttggtac
aacatgctgg tggcggaacc caggaatctg tccttcttcc tgactccacc
1860atgtgcacga tgggctcagc tttcagaagt gctgagttgg cagttttctt
ctgtcaccaa 1920aagaggtctc aatgtggacc agctgaacat gttgggagag
aagcttcttg gtcctaacgc 1980cagccccgat ggtctcattc cgtggacgag
gttttgtaag gaaaatataa atgataaaaa 2040ttttcccttc tggctttgga
ttgaaagcat cctagaactc attaaaaaac acctgctccc 2100tctctggaat
gatgggtgca tcatgggctt catcagcaag gagcgagagc gtgccctgtt
2160gaaggaccag cagccgggga ccttcctgct gcggttcagt gagagctccc
gggaaggggc 2220catcacattc acatgggtgg agcggtccca gaacggaggc
gaacctgact tccatgcggt 2280tgaaccctac acgaagaaag aactttctgc
tgttactttc cctgacatca ttcgcaatta 2340caaagtcatg gctgctgaga
atattcctga gaatcccctg aagtatctgt atccaaatat 2400tgacaaagac
catgcctttg gaaagtatta ctccaggcca aaggaagcac cagagccaat
2460ggaacttgat ggccctaaag gaactggata tatcaagact gagttgattt
ctgtgtctga 2520agttcaccct tctagacttc agaccacaga caacctgctc
cccatgtctc ctgaggagtt 2580tgacgaggtg tctcggatag tgggctctgt
agaattcgac agtatgatga acacagtata 2640gagcatgaat ttttttcatc
ttctctggcg acagttttcc ttctcatctg tgattccctc 2700ctgctactct
gttccttcac atcctgtgtt tctagggaaa tgaaagaaag gccagcaaat
2760tcgctgcaac ctgttgatag caagtgaatt tttctctaac tcagaaacat
cagttactct 2820gaagggcatc atgcatctta ctgaaggtaa aattgaaagg
cattctctga agagtgggtt 2880tcacaagtga aaaacatcca gatacaccca
aagtatcagg acgagaatga gggtcctttg 2940ggaaaggaga agttaagcaa
catctagcaa atgttatgca taaagtcagt gcccaactgt 3000tataggttgt
tggataaatc agtggttatt tagggaactg cttgacgtag gaacggtaaa
3060tttctgtggg agaattctta catgttttct ttgctttaag tgtaactggc
agttttccat 3120tggtttacct gtgaaatagt tcaaagccaa gtttatatac
aattatatca gtcctctttc 3180aaaggtagcc atcatggatc tggtaggggg
aaaatgtgta ttttattaca tctttcacat 3240tggctattta aagacaaaga
caaattctgt ttcttgagaa gagaatatta gctttactgt 3300ttgttatggc
ttaatgacac tagctaatat caatagaagg atgtacattt ccaaattcac
3360aagttgtgtt tgatatccaa agctgaatac attctgcttt catcttggtc
acatacaatt 3420atttttacag ttctcccaag ggagttaggc tattcacaac
cactcattca aaagttgaaa 3480ttaaccatag atgtagataa actcagaaat
ttaattcatg tttcttaaat gggctacttt 3540gtcctttttg ttattagggt
ggtatttagt ctattagcca caaaattggg aaaggagtag 3600aaaaagcagt
aactgacaac ttgaataata caccagagat aatatgagaa tcagatcatt
3660tcaaaactca tttcctatgt aactgcattg agaactgcat atgtttcgct
gatatatgtg 3720tttttcacat ttgcgaatgg ttccattctc tctcctgtac
tttttccaga cacttttttg 3780agtggatgat gtttcgtgaa gtatactgta
tttttacctt tttccttcct tatcactgac 3840acaaaaagta gattaagaga
tgggtttgac aaggttcttc ccttttacat actgctgtct 3900atgtggctgt
atcttgtttt tccactactg ctaccacaac tatattatca tgcaaatgct
3960gtattcttct ttggtggaga taaagatttc ttgagttttg ttttaaaatt
aaagctaaag 4020tatctgtatt gcattaaata taatatgcac acagtgcttt
ccgtggcact gcatacaatc 4080tgaggcctcc tctctcagtt tttatataga
tggcgagaac ctaagtttca gttgatttta 4140caattgaaat gactaaaaaa
caaagaagac aacattaaaa caatattgtt tctaattgct 4200gaggtttagc
tgtcagttct ttttgccctt tgggaattcg gcatggtttc attttactgc
4260actagccaag agactttact tttaagaagt attaaaattc taaaattcaa
aaaaaaaaaa 4320aaaaaa 43261042798DNAHomo sapiens 104gctgagcgcg
gagccgcccg gtgattggtg ggggcggaag ggggccgggc gccagcgctg 60ccttttctcc
tgccgggtag tttcgctttc ctgcgcagag tctgcggagg ggctcggctg
120caccgggggg atcgcgcctg gcagacccca gaccgagcag aggcgaccca
gcgcgctcgg 180gagaggctgc accgccgcgc ccccgcctag cccttccgga
tcctgcgcgc agaaaagttt 240catttgctgt atgccatcct cgagagctgt
ctaggttaac gttcgcactc tgtgtatata 300acctcgacag tcttggcacc
taacgtgctg tgcgtagctg ctcctttggt tgaatcccca 360ggcccttgtt
ggggcacaag gtggcaggat gtctcagtgg tacgaacttc agcagcttga
420ctcaaaattc ctggagcagg ttcaccagct ttatgatgac agttttccca
tggaaatcag 480acagtacctg gcacagtggt tagaaaagca agactgggag
cacgctgcca atgatgtttc 540atttgccacc atccgttttc atgacctcct
gtcacagctg gatgatcaat atagtcgctt 600ttctttggag aataacttct
tgctacagca taacataagg aaaagcaagc gtaatcttca 660ggataatttt
caggaagacc caatccagat gtctatgatc atttacagct gtctgaagga
720agaaaggaaa attctggaaa acgcccagag atttaatcag gctcagtcgg
ggaatattca 780gagcacagtg atgttagaca aacagaaaga gcttgacagt
aaagtcagaa atgtgaagga 840caaggttatg tgtatagagc atgaaatcaa
gagcctggaa gatttacaag atgaatatga 900cttcaaatgc aaaaccttgc
agaacagaga acacgagacc aatggtgtgg caaagagtga 960tcagaaacaa
gaacagctgt tactcaagaa gatgtattta atgcttgaca ataagagaaa
1020ggaagtagtt cacaaaataa tagagttgct gaatgtcact gaacttaccc
agaatgccct 1080gattaatgat gaactagtgg agtggaagcg gagacagcag
agcgcctgta ttggggggcc 1140gcccaatgct tgcttggatc agctgcagaa
ctggttcact atagttgcgg agagtctgca 1200gcaagttcgg cagcagctta
aaaagttgga ggaattggaa cagaaataca cctacgaaca 1260tgaccctatc
acaaaaaaca aacaagtgtt atgggaccgc accttcagtc ttttccagca
1320gctcattcag agctcgtttg tggtggaaag acagccctgc atgccaacgc
accctcagag 1380gccgctggtc ttgaagacag gggtccagtt cactgtgaag
ttgagactgt tggtgaaatt 1440gcaagagctg aattataatt tgaaagtcaa
agtcttattt gataaagatg tgaatgagag 1500aaatacagta aaaggattta
ggaagttcaa cattttgggc acgcacacaa aagtgatgaa 1560catggaggag
tccaccaatg gcagtctggc ggctgaattt cggcacctgc aattgaaaga
1620acagaaaaat gctggcacca gaacgaatga gggtcctctc atcgttactg
aagagcttca 1680ctcccttagt tttgaaaccc aattgtgcca gcctggtttg
gtaattgacc tcgagacgac 1740ctctctgccc gttgtggtga tctccaacgt
cagccagctc ccgagcggtt gggcctccat 1800cctttggtac aacatgctgg
tggcggaacc caggaatctg tccttcttcc tgactccacc 1860atgtgcacga
tgggctcagc tttcagaagt gctgagttgg cagttttctt ctgtcaccaa
1920aagaggtctc aatgtggacc agctgaacat gttgggagag aagcttcttg
gtcctaacgc 1980cagccccgat ggtctcattc cgtggacgag gttttgtaag
gaaaatataa atgataaaaa 2040ttttcccttc tggctttgga ttgaaagcat
cctagaactc attaaaaaac acctgctccc 2100tctctggaat gatgggtgca
tcatgggctt catcagcaag gagcgagagc gtgccctgtt 2160gaaggaccag
cagccgggga ccttcctgct gcggttcagt gagagctccc gggaaggggc
2220catcacattc acatgggtgg agcggtccca gaacggaggc gaacctgact
tccatgcggt 2280tgaaccctac acgaagaaag aactttctgc tgttactttc
cctgacatca ttcgcaatta 2340caaagtcatg gctgctgaga atattcctga
gaatcccctg aagtatctgt atccaaatat 2400tgacaaagac catgcctttg
gaaagtatta ctccaggcca aaggaagcac cagagccaat 2460ggaacttgat
ggccctaaag gaactggata tatcaagact gagttgattt ctgtgtctga
2520agtgtaagtg aacacagaag agtgacatgt ttacaaacct caagccagcc
ttgctcctgg 2580ctggggcctg ttgaagatgc ttgtatttta cttttccatt
gtaattgcta tcgccatcac 2640agctgaactt gttgagatcc ccgtgttact
gcctatcagc attttactac tttaaaaaaa 2700aaaaaaaagc caaaaaccaa
atttgtattt aaggtatata aattttccca aaactgatac 2760cctttgaaaa
agtataaata aaatgagcaa aagttgat 2798105750PRTHomo sapiens 105Met Ser
Gln Trp Tyr Glu Leu Gln Gln Leu Asp Ser Lys Phe Leu Glu1 5 10 15Gln
Val His Gln Leu Tyr Asp Asp Ser Phe Pro Met Glu Ile Arg Gln 20 25
30Tyr Leu Ala Gln Trp Leu Glu Lys Gln Asp Trp Glu His Ala Ala Asn
35 40 45Asp Val Ser Phe Ala Thr Ile Arg Phe His Asp Leu Leu Ser Gln
Leu 50 55 60Asp Asp Gln Tyr Ser Arg Phe Ser Leu Glu Asn Asn Phe Leu
Leu Gln65 70 75 80His Asn Ile Arg Lys Ser Lys Arg Asn Leu Gln Asp
Asn Phe Gln Glu 85 90 95Asp Pro Ile Gln Met Ser Met Ile Ile Tyr Ser
Cys Leu Lys Glu Glu 100 105 110Arg Lys Ile Leu Glu Asn Ala Gln Arg
Phe Asn Gln Ala Gln Ser Gly 115 120 125Asn Ile Gln Ser Thr Val Met
Leu Asp Lys Gln Lys Glu Leu Asp Ser 130 135 140Lys Val Arg Asn Val
Lys Asp Lys Val Met Cys Ile Glu His Glu Ile145 150 155 160Lys Ser
Leu Glu Asp Leu Gln Asp Glu Tyr Asp Phe Lys Cys Lys Thr 165 170
175Leu Gln Asn Arg Glu His Glu Thr Asn Gly Val Ala Lys Ser Asp Gln
180 185 190Lys Gln Glu Gln Leu Leu Leu Lys Lys Met Tyr Leu Met Leu
Asp Asn 195 200 205Lys Arg Lys Glu Val Val His Lys Ile Ile Glu Leu
Leu Asn Val Thr 210 215 220Glu Leu Thr Gln Asn Ala Leu Ile Asn Asp
Glu Leu Val Glu Trp Lys225 230 235 240Arg Arg Gln Gln Ser Ala Cys
Ile Gly Gly Pro Pro Asn Ala Cys Leu 245 250 255Asp Gln Leu Gln Asn
Trp Phe Thr Ile Val Ala Glu Ser Leu Gln Gln 260 265 270Val Arg Gln
Gln Leu Lys Lys Leu Glu Glu Leu Glu Gln Lys Tyr Thr 275 280 285Tyr
Glu His Asp Pro Ile Thr Lys Asn Lys Gln Val Leu Trp Asp Arg 290 295
300Thr Phe Ser Leu Phe Gln Gln Leu Ile Gln Ser Ser Phe Val Val
Glu305 310 315 320Arg Gln Pro Cys Met Pro Thr His Pro Gln Arg Pro
Leu Val Leu Lys 325 330 335Thr Gly Val Gln Phe Thr Val Lys Leu Arg
Leu Leu Val Lys Leu Gln 340 345 350Glu Leu Asn Tyr Asn Leu Lys Val
Lys Val Leu Phe Asp Lys Asp Val 355 360 365Asn Glu Arg Asn Thr Val
Lys Gly Phe Arg Lys Phe Asn Ile Leu Gly 370 375 380Thr His Thr Lys
Val Met Asn Met Glu Glu Ser Thr Asn Gly Ser Leu385 390 395 400Ala
Ala Glu Phe Arg His Leu Gln Leu Lys Glu Gln Lys Asn Ala Gly 405 410
415Thr Arg Thr Asn Glu Gly Pro Leu Ile Val Thr Glu Glu Leu His Ser
420 425 430Leu Ser Phe Glu Thr Gln Leu Cys Gln Pro Gly Leu Val Ile
Asp Leu 435 440 445Glu Thr Thr Ser Leu Pro Val Val Val Ile Ser Asn
Val Ser Gln Leu 450 455 460Pro Ser Gly Trp Ala Ser Ile Leu Trp Tyr
Asn Met Leu Val Ala Glu465 470 475 480Pro Arg Asn Leu Ser Phe Phe
Leu Thr Pro Pro Cys Ala Arg Trp Ala 485 490 495Gln Leu Ser Glu Val
Leu Ser Trp Gln Phe Ser Ser Val Thr Lys Arg 500 505 510Gly Leu Asn
Val Asp Gln Leu Asn Met Leu Gly Glu Lys Leu Leu Gly 515 520 525Pro
Asn Ala Ser Pro Asp Gly Leu Ile Pro Trp Thr Arg Phe Cys Lys 530 535
540Glu Asn Ile Asn Asp Lys Asn Phe Pro Phe Trp Leu Trp Ile Glu
Ser545 550 555 560Ile Leu Glu Leu Ile Lys Lys His Leu Leu Pro Leu
Trp Asn Asp Gly 565 570 575Cys Ile Met Gly Phe Ile Ser Lys Glu Arg
Glu Arg Ala Leu Leu Lys 580 585 590Asp Gln Gln Pro Gly Thr Phe Leu
Leu Arg Phe Ser Glu Ser Ser Arg 595 600 605Glu Gly Ala Ile Thr Phe
Thr Trp Val Glu Arg Ser Gln Asn Gly Gly 610 615 620Glu Pro Asp Phe
His Ala Val Glu Pro Tyr Thr Lys Lys Glu Leu Ser625 630 635 640Ala
Val Thr Phe Pro Asp Ile Ile Arg Asn Tyr Lys Val Met Ala Ala 645 650
655Glu Asn Ile Pro Glu Asn Pro Leu Lys Tyr Leu Tyr Pro Asn Ile Asp
660 665 670Lys Asp His Ala Phe Gly Lys Tyr Tyr Ser Arg Pro Lys Glu
Ala Pro 675 680 685Glu Pro Met Glu Leu Asp Gly Pro Lys Gly Thr Gly
Tyr Ile Lys Thr 690 695 700Glu Leu Ile Ser Val Ser Glu Val His Pro
Ser Arg Leu Gln Thr Thr705 710 715 720Asp Asn Leu Leu Pro Met Ser
Pro Glu Glu Phe Asp Glu Val Ser Arg 725 730 735Ile Val Gly Ser Val
Glu Phe Asp Ser Met Met Asn Thr Val 740 745 750106712PRTHomo
sapiens 106Met Ser Gln Trp Tyr Glu Leu Gln Gln Leu Asp Ser Lys Phe
Leu Glu1 5 10 15Gln Val His Gln Leu Tyr Asp Asp Ser Phe Pro Met Glu
Ile Arg Gln 20 25 30Tyr Leu Ala Gln Trp Leu Glu Lys Gln Asp Trp Glu
His Ala Ala Asn 35 40 45Asp Val Ser Phe Ala Thr Ile Arg Phe His Asp
Leu Leu Ser Gln Leu 50 55 60Asp Asp Gln Tyr Ser Arg Phe Ser Leu Glu
Asn Asn Phe Leu Leu Gln65 70 75 80His Asn Ile Arg Lys Ser Lys Arg
Asn Leu Gln Asp Asn Phe Gln Glu 85 90 95Asp Pro Ile Gln Met Ser Met
Ile Ile Tyr Ser Cys Leu Lys Glu Glu 100 105 110Arg Lys Ile Leu Glu
Asn Ala Gln Arg Phe Asn Gln Ala Gln Ser Gly 115 120 125Asn Ile Gln
Ser Thr Val Met Leu Asp Lys Gln Lys Glu Leu Asp Ser 130 135 140Lys
Val Arg Asn Val Lys Asp Lys Val Met Cys Ile Glu His Glu Ile145 150
155 160Lys Ser Leu Glu Asp Leu Gln Asp Glu Tyr Asp Phe Lys Cys Lys
Thr 165 170 175Leu Gln Asn Arg Glu His Glu Thr Asn Gly Val Ala Lys
Ser Asp Gln 180 185 190Lys
Gln Glu Gln Leu Leu Leu Lys Lys Met Tyr Leu Met Leu Asp Asn 195 200
205Lys Arg Lys Glu Val Val His Lys Ile Ile Glu Leu Leu Asn Val Thr
210 215 220Glu Leu Thr Gln Asn Ala Leu Ile Asn Asp Glu Leu Val Glu
Trp Lys225 230 235 240Arg Arg Gln Gln Ser Ala Cys Ile Gly Gly Pro
Pro Asn Ala Cys Leu 245 250 255Asp Gln Leu Gln Asn Trp Phe Thr Ile
Val Ala Glu Ser Leu Gln Gln 260 265 270Val Arg Gln Gln Leu Lys Lys
Leu Glu Glu Leu Glu Gln Lys Tyr Thr 275 280 285Tyr Glu His Asp Pro
Ile Thr Lys Asn Lys Gln Val Leu Trp Asp Arg 290 295 300Thr Phe Ser
Leu Phe Gln Gln Leu Ile Gln Ser Ser Phe Val Val Glu305 310 315
320Arg Gln Pro Cys Met Pro Thr His Pro Gln Arg Pro Leu Val Leu Lys
325 330 335Thr Gly Val Gln Phe Thr Val Lys Leu Arg Leu Leu Val Lys
Leu Gln 340 345 350Glu Leu Asn Tyr Asn Leu Lys Val Lys Val Leu Phe
Asp Lys Asp Val 355 360 365Asn Glu Arg Asn Thr Val Lys Gly Phe Arg
Lys Phe Asn Ile Leu Gly 370 375 380Thr His Thr Lys Val Met Asn Met
Glu Glu Ser Thr Asn Gly Ser Leu385 390 395 400Ala Ala Glu Phe Arg
His Leu Gln Leu Lys Glu Gln Lys Asn Ala Gly 405 410 415Thr Arg Thr
Asn Glu Gly Pro Leu Ile Val Thr Glu Glu Leu His Ser 420 425 430Leu
Ser Phe Glu Thr Gln Leu Cys Gln Pro Gly Leu Val Ile Asp Leu 435 440
445Glu Thr Thr Ser Leu Pro Val Val Val Ile Ser Asn Val Ser Gln Leu
450 455 460Pro Ser Gly Trp Ala Ser Ile Leu Trp Tyr Asn Met Leu Val
Ala Glu465 470 475 480Pro Arg Asn Leu Ser Phe Phe Leu Thr Pro Pro
Cys Ala Arg Trp Ala 485 490 495Gln Leu Ser Glu Val Leu Ser Trp Gln
Phe Ser Ser Val Thr Lys Arg 500 505 510Gly Leu Asn Val Asp Gln Leu
Asn Met Leu Gly Glu Lys Leu Leu Gly 515 520 525Pro Asn Ala Ser Pro
Asp Gly Leu Ile Pro Trp Thr Arg Phe Cys Lys 530 535 540Glu Asn Ile
Asn Asp Lys Asn Phe Pro Phe Trp Leu Trp Ile Glu Ser545 550 555
560Ile Leu Glu Leu Ile Lys Lys His Leu Leu Pro Leu Trp Asn Asp Gly
565 570 575Cys Ile Met Gly Phe Ile Ser Lys Glu Arg Glu Arg Ala Leu
Leu Lys 580 585 590Asp Gln Gln Pro Gly Thr Phe Leu Leu Arg Phe Ser
Glu Ser Ser Arg 595 600 605Glu Gly Ala Ile Thr Phe Thr Trp Val Glu
Arg Ser Gln Asn Gly Gly 610 615 620Glu Pro Asp Phe His Ala Val Glu
Pro Tyr Thr Lys Lys Glu Leu Ser625 630 635 640Ala Val Thr Phe Pro
Asp Ile Ile Arg Asn Tyr Lys Val Met Ala Ala 645 650 655Glu Asn Ile
Pro Glu Asn Pro Leu Lys Tyr Leu Tyr Pro Asn Ile Asp 660 665 670Lys
Asp His Ala Phe Gly Lys Tyr Tyr Ser Arg Pro Lys Glu Ala Pro 675 680
685Glu Pro Met Glu Leu Asp Gly Pro Lys Gly Thr Gly Tyr Ile Lys Thr
690 695 700Glu Leu Ile Ser Val Ser Glu Val705 7101071742DNAHomo
sapiens 107tctttgaagc ttcaaggctg ctgaataatt tccttctccc attttgtgcc
tgcctagcta 60tccagacaga gcagctaccc tcagctctag ctgatactac agacagtaca
acagatcaag 120aagtatggca gtgacaactc gtttgacatg gttgcacgaa
aagatcctgc aaaatcattt 180tggagggaag cggcttagcc ttctctataa
gggtagtgtc catggattcc gtaatggagt 240tttgcttgac agatgttgta
atcaagggcc tactctaaca gtgatttata gtgaagatca 300tattattgga
gcatatgcag aagagagtta ccaggaagga aagtatgctt ccatcatcct
360ttttgcactt caagatacta aaatttcaga atggaaacta ggactatgta
caccagaaac 420actgttttgt tgtgatgtta caaaatataa ctccccaact
aatttccaga tagatggaag 480aaatagaaaa gtgattatgg acttaaagac
aatggaaaat cttggacttg ctcaaaattg 540tactatctct attcaggatt
atgaagtttt tcgatgcgaa gattcactgg atgaaagaaa 600gataaaaggg
gtcattgagc tcaggaagag cttactgtct gccttgagaa cttatgaacc
660atatggatcc ctggttcaac aaatacgaat tctgctgctg ggtccaattg
gagctgggaa 720gtccagcttt ttcaactcag tgaggtctgt tttccaaggg
catgtaacgc atcaggcttt 780ggtgggcact aatacaactg ggatatctga
gaagtatagg acatactcta ttagagacgg 840gaaagatggc aaatacctgc
cgtttattct gtgtgactca ctggggctga gtgagaaaga 900aggcggcctg
tgcagggatg acatattcta tatcttgaac ggtaacattc gtgatagata
960ccagtttaat cccatggaat caatcaaatt aaatcatcat gactacattg
attccccatc 1020gctgaaggac agaattcatt gtgtggcatt tgtatttgat
gccagctcta ttcaatactt 1080ctcctctcag atgatagtaa agatcaaaag
aattcgaagg gagttggtaa acgctggtgt 1140ggtacatgtg gctttgctca
ctcatgtgga tagcatggat ttgattacaa aaggtgacct 1200tatagaaata
gagagatgtg agcctgtgag gtccaagcta gaggaagtcc aaagaaaact
1260tggatttgct ctttctgaca tctcggtggt tagcaattat tcctctgagt
gggagctgga 1320ccctgtaaag gatgttctaa ttctttctgc tctgagacga
atgctatggg ctgcagatga 1380cttcttagag gatttgcctt ttgagcaaat
agggaatcta agggaggaaa ttatcaactg 1440tgcacaagga aaaaaataga
tatgtgaaag gttcacgtaa atttcctcac atcacagaag 1500attaaaattc
agaaaggaga aaacacagac caaagagaag tatctaagac caaagggatg
1560tgttttatta atgtctagga tgaagaaatg catagaacat tgtagtactt
gtaaataact 1620agaaataaca tgatttagtc ataattgtga aaaataataa
taatttttct tggatttatg 1680ttctgtatct gtgaaaaaat aaatttctta
taaaactcgg gtctaaaaaa aaaaaaaaaa 1740aa 1742108444PRTHomo sapiens
108Met Ala Val Thr Thr Arg Leu Thr Trp Leu His Glu Lys Ile Leu Gln1
5 10 15Asn His Phe Gly Gly Lys Arg Leu Ser Leu Leu Tyr Lys Gly Ser
Val 20 25 30His Gly Phe Arg Asn Gly Val Leu Leu Asp Arg Cys Cys Asn
Gln Gly 35 40 45Pro Thr Leu Thr Val Ile Tyr Ser Glu Asp His Ile Ile
Gly Ala Tyr 50 55 60Ala Glu Glu Ser Tyr Gln Glu Gly Lys Tyr Ala Ser
Ile Ile Leu Phe65 70 75 80Ala Leu Gln Asp Thr Lys Ile Ser Glu Trp
Lys Leu Gly Leu Cys Thr 85 90 95Pro Glu Thr Leu Phe Cys Cys Asp Val
Thr Lys Tyr Asn Ser Pro Thr 100 105 110Asn Phe Gln Ile Asp Gly Arg
Asn Arg Lys Val Ile Met Asp Leu Lys 115 120 125Thr Met Glu Asn Leu
Gly Leu Ala Gln Asn Cys Thr Ile Ser Ile Gln 130 135 140Asp Tyr Glu
Val Phe Arg Cys Glu Asp Ser Leu Asp Glu Arg Lys Ile145 150 155
160Lys Gly Val Ile Glu Leu Arg Lys Ser Leu Leu Ser Ala Leu Arg Thr
165 170 175Tyr Glu Pro Tyr Gly Ser Leu Val Gln Gln Ile Arg Ile Leu
Leu Leu 180 185 190Gly Pro Ile Gly Ala Gly Lys Ser Ser Phe Phe Asn
Ser Val Arg Ser 195 200 205Val Phe Gln Gly His Val Thr His Gln Ala
Leu Val Gly Thr Asn Thr 210 215 220Thr Gly Ile Ser Glu Lys Tyr Arg
Thr Tyr Ser Ile Arg Asp Gly Lys225 230 235 240Asp Gly Lys Tyr Leu
Pro Phe Ile Leu Cys Asp Ser Leu Gly Leu Ser 245 250 255Glu Lys Glu
Gly Gly Leu Cys Arg Asp Asp Ile Phe Tyr Ile Leu Asn 260 265 270Gly
Asn Ile Arg Asp Arg Tyr Gln Phe Asn Pro Met Glu Ser Ile Lys 275 280
285Leu Asn His His Asp Tyr Ile Asp Ser Pro Ser Leu Lys Asp Arg Ile
290 295 300His Cys Val Ala Phe Val Phe Asp Ala Ser Ser Ile Gln Tyr
Phe Ser305 310 315 320Ser Gln Met Ile Val Lys Ile Lys Arg Ile Arg
Arg Glu Leu Val Asn 325 330 335Ala Gly Val Val His Val Ala Leu Leu
Thr His Val Asp Ser Met Asp 340 345 350Leu Ile Thr Lys Gly Asp Leu
Ile Glu Ile Glu Arg Cys Glu Pro Val 355 360 365Arg Ser Lys Leu Glu
Glu Val Gln Arg Lys Leu Gly Phe Ala Leu Ser 370 375 380Asp Ile Ser
Val Val Ser Asn Tyr Ser Ser Glu Trp Glu Leu Asp Pro385 390 395
400Val Lys Asp Val Leu Ile Leu Ser Ala Leu Arg Arg Met Leu Trp Ala
405 410 415Ala Asp Asp Phe Leu Glu Asp Leu Pro Phe Glu Gln Ile Gly
Asn Leu 420 425 430Arg Glu Glu Ile Ile Asn Cys Ala Gln Gly Lys Lys
435 4401092808DNAHomo sapiens 109gcggcggcgg cggcgcagtt tgctcatact
ttgtgacttg cggtcacagt ggcattcagc 60tccacacttg gtagaaccac aggcacgaca
agcatagaaa catcctaaac aatcttcatc 120gaggcatcga ggtccatccc
aataaaaatc aggagaccct ggctatcata gaccttagtc 180ttcgctggta
tactcgctgt ctgtcaacca gcggttgact ttttttaagc cttctttttt
240ctcttttacc agtttctgga gcaaattcag tttgccttcc tggatttgta
aattgtaatg 300acctcaaaac tttagcagtt cttccatctg actcaggttt
gcttctctgg cggtcttcag 360aatcaacatc cacacttccg tgattatctg
cgtgcatttt ggacaaagct tccaaccagg 420atacgggaag aagaaatggc
tggtgatctt tcagcaggtt tcttcatgga ggaacttaat 480acataccgtc
agaagcaggg agtagtactt aaatatcaag aactgcctaa ttcaggacct
540ccacatgata ggaggtttac atttcaagtt ataatagatg gaagagaatt
tccagaaggt 600gaaggtagat caaagaagga agcaaaaaat gccgcagcca
aattagctgt tgagatactt 660aataaggaaa agaaggcagt tagtccttta
ttattgacaa caacgaattc ttcagaagga 720ttatccatgg ggaattacat
aggccttatc aatagaattg cccagaagaa aagactaact 780gtaaattatg
aacagtgtgc atcgggggtg catgggccag aaggatttca ttataaatgc
840aaaatgggac agaaagaata tagtattggt acaggttcta ctaaacagga
agcaaaacaa 900ttggccgcta aacttgcata tcttcagata ttatcagaag
aaacctcagt gaaatctgac 960tacctgtcct ctggttcttt tgctactacg
tgtgagtccc aaagcaactc tttagtgacc 1020agcacactcg cttctgaatc
atcatctgaa ggtgacttct cagcagatac atcagagata 1080aattctaaca
gtgacagttt aaacagttct tcgttgctta tgaatggtct cagaaataat
1140caaaggaagg caaaaagatc tttggcaccc agatttgacc ttcctgacat
gaaagaaaca 1200aagtatactg tggacaagag gtttggcatg gattttaaag
aaatagaatt aattggctca 1260ggtggatttg gccaagtttt caaagcaaaa
cacagaattg acggaaagac ttacgttatt 1320aaacgtgtta aatataataa
cgagaaggcg gagcgtgaag taaaagcatt ggcaaaactt 1380gatcatgtaa
atattgttca ctacaatggc tgttgggatg gatttgatta tgatcctgag
1440accagtgatg attctcttga gagcagtgat tatgatcctg agaacagcaa
aaatagttca 1500aggtcaaaga ctaagtgcct tttcatccaa atggaattct
gtgataaagg gaccttggaa 1560caatggattg aaaaaagaag aggcgagaaa
ctagacaaag ttttggcttt ggaactcttt 1620gaacaaataa caaaaggggt
ggattatata cattcaaaaa aattaattca tagagatctt 1680aagccaagta
atatattctt agtagataca aaacaagtaa agattggaga ctttggactt
1740gtaacatctc tgaaaaatga tggaaagcga acaaggagta agggaacttt
gcgatacatg 1800agcccagaac agatttcttc gcaagactat ggaaaggaag
tggacctcta cgctttgggg 1860ctaattcttg ctgaacttct tcatgtatgt
gacactgctt ttgaaacatc aaagtttttc 1920acagacctac gggatggcat
catctcagat atatttgata aaaaagaaaa aactcttcta 1980cagaaattac
tctcaaagaa acctgaggat cgacctaaca catctgaaat actaaggacc
2040ttgactgtgt ggaagaaaag cccagagaaa aatgaacgac acacatgtta
gagcccttct 2100gaaaaagtat cctgcttctg atatgcagtt ttccttaaat
tatctaaaat ctgctaggga 2160atatcaatag atatttacct tttattttaa
tgtttccttt aattttttac tatttttact 2220aatctttctg cagaaacaga
aaggttttct tctttttgct tcaaaaacat tcttacattt 2280tactttttcc
tggctcatct ctttattctt tttttttttt ttaaagacag agtctcgctc
2340tgttgcccag gctggagtgc aatgacacag tcttggctca ctgcaacttc
tgcctcttgg 2400gttcaagtga ttctcctgcc tcagcctcct gagtagctgg
attacaggca tgtgccaccc 2460acccaactaa tttttgtgtt tttaataaag
acagggtttc accatgttgg ccaggctggt 2520ctcaaactcc tgacctcaag
taatccacct gcctcggcct cccaaagtgc tgggattaca 2580gggatgagcc
accgcgccca gcctcatctc tttgttctaa agatggaaaa accaccccca
2640aattttcttt ttatactatt aatgaatcaa tcaattcata tctatttatt
aaatttctac 2700cgcttttagg ccaaaaaaat gtaagatcgt tctctgcctc
acatagctta caagccagct 2760ggagaaatat ggtactcatt aaaaaaaaaa
aaaaagtgat gtacaacc 2808110551PRTHomo sapiens 110Met Ala Gly Asp
Leu Ser Ala Gly Phe Phe Met Glu Glu Leu Asn Thr1 5 10 15Tyr Arg Gln
Lys Gln Gly Val Val Leu Lys Tyr Gln Glu Leu Pro Asn 20 25 30Ser Gly
Pro Pro His Asp Arg Arg Phe Thr Phe Gln Val Ile Ile Asp 35 40 45Gly
Arg Glu Phe Pro Glu Gly Glu Gly Arg Ser Lys Lys Glu Ala Lys 50 55
60Asn Ala Ala Ala Lys Leu Ala Val Glu Ile Leu Asn Lys Glu Lys Lys65
70 75 80Ala Val Ser Pro Leu Leu Leu Thr Thr Thr Asn Ser Ser Glu Gly
Leu 85 90 95Ser Met Gly Asn Tyr Ile Gly Leu Ile Asn Arg Ile Ala Gln
Lys Lys 100 105 110Arg Leu Thr Val Asn Tyr Glu Gln Cys Ala Ser Gly
Val His Gly Pro 115 120 125Glu Gly Phe His Tyr Lys Cys Lys Met Gly
Gln Lys Glu Tyr Ser Ile 130 135 140Gly Thr Gly Ser Thr Lys Gln Glu
Ala Lys Gln Leu Ala Ala Lys Leu145 150 155 160Ala Tyr Leu Gln Ile
Leu Ser Glu Glu Thr Ser Val Lys Ser Asp Tyr 165 170 175Leu Ser Ser
Gly Ser Phe Ala Thr Thr Cys Glu Ser Gln Ser Asn Ser 180 185 190Leu
Val Thr Ser Thr Leu Ala Ser Glu Ser Ser Ser Glu Gly Asp Phe 195 200
205Ser Ala Asp Thr Ser Glu Ile Asn Ser Asn Ser Asp Ser Leu Asn Ser
210 215 220Ser Ser Leu Leu Met Asn Gly Leu Arg Asn Asn Gln Arg Lys
Ala Lys225 230 235 240Arg Ser Leu Ala Pro Arg Phe Asp Leu Pro Asp
Met Lys Glu Thr Lys 245 250 255Tyr Thr Val Asp Lys Arg Phe Gly Met
Asp Phe Lys Glu Ile Glu Leu 260 265 270Ile Gly Ser Gly Gly Phe Gly
Gln Val Phe Lys Ala Lys His Arg Ile 275 280 285Asp Gly Lys Thr Tyr
Val Ile Lys Arg Val Lys Tyr Asn Asn Glu Lys 290 295 300Ala Glu Arg
Glu Val Lys Ala Leu Ala Lys Leu Asp His Val Asn Ile305 310 315
320Val His Tyr Asn Gly Cys Trp Asp Gly Phe Asp Tyr Asp Pro Glu Thr
325 330 335Ser Asp Asp Ser Leu Glu Ser Ser Asp Tyr Asp Pro Glu Asn
Ser Lys 340 345 350Asn Ser Ser Arg Ser Lys Thr Lys Cys Leu Phe Ile
Gln Met Glu Phe 355 360 365Cys Asp Lys Gly Thr Leu Glu Gln Trp Ile
Glu Lys Arg Arg Gly Glu 370 375 380Lys Leu Asp Lys Val Leu Ala Leu
Glu Leu Phe Glu Gln Ile Thr Lys385 390 395 400Gly Val Asp Tyr Ile
His Ser Lys Lys Leu Ile His Arg Asp Leu Lys 405 410 415Pro Ser Asn
Ile Phe Leu Val Asp Thr Lys Gln Val Lys Ile Gly Asp 420 425 430Phe
Gly Leu Val Thr Ser Leu Lys Asn Asp Gly Lys Arg Thr Arg Ser 435 440
445Lys Gly Thr Leu Arg Tyr Met Ser Pro Glu Gln Ile Ser Ser Gln Asp
450 455 460Tyr Gly Lys Glu Val Asp Leu Tyr Ala Leu Gly Leu Ile Leu
Ala Glu465 470 475 480Leu Leu His Val Cys Asp Thr Ala Phe Glu Thr
Ser Lys Phe Phe Thr 485 490 495Asp Leu Arg Asp Gly Ile Ile Ser Asp
Ile Phe Asp Lys Lys Glu Lys 500 505 510Thr Leu Leu Gln Lys Leu Leu
Ser Lys Lys Pro Glu Asp Arg Pro Asn 515 520 525Thr Ser Glu Ile Leu
Arg Thr Leu Thr Val Trp Lys Lys Ser Pro Glu 530 535 540Lys Asn Glu
Arg His Thr Cys545 5501112613DNAHomo sapiens 111agtttcagtt
tccatttctg atttctgctc tctgcgctga gcacagcggc accaggctga 60gctaagcagg
gccgccttgg gcaggcctac gtggtggtgc aggcgagacc caggctgggc
120aaggcgcagt ttcagtttcc atcttgggtc tctgagctga gcagagtggc
accaggctga 180gttaagtggg actgccctgg gcagacctac ctactagagc
agaatggagc ttcggtccta 240ccaatgggag gtgatcatgc ctgccctgga
gggcaagaat atcatcatct ggctgcccac 300gggtgccggg aagacccggg
cggctgctta tgtggccaag cggcacctag agactgtgga 360tggagccaag
gtggttgtat tggtcaacag ggtgcacctg gtgacccagc atggtgaaga
420gttcaggcgc atgctggatg gacgctggac cgtgacaacc ctgagtgggg
acatgggacc 480acgtgctggc tttggccacc tggcccggtg ccatgacctg
ctcatctgca cagcagagct 540tctgcagatg gcactgacca gccccgagga
ggaggagcac gtggagctca ctgtcttctc 600cctgatcgtg gtggatgagt
gccaccacac gcacaaggac accgtctaca acgtcatcat 660gagccagtac
ctagaactta aactccagag ggcacagccg ctaccccagg tgctgggtct
720cacagcctcc ccaggcactg gcggggcctc caaactcgat ggggccatca
accacgtcct 780gcagctctgt gccaacttgg acacgtggtg catcatgtca
ccccagaact gctgccccca 840gctgcaggag cacagccaac agccttgcaa
acagtacaac ctctgccaca ggcgcagcca 900ggatccgttt ggggacttgc
tgaagaagct catggaccaa atccatgacc acctggagat 960gcctgagttg
agccggaaat ttgggacgca aatgtatgag cagcaggtgg tgaagctgag
1020tgaggctgcg gctttggctg ggcttcagga gcaacgggtg tatgcgcttc
acctgaggcg 1080ctacaatgac gcgctgctca tccatgacac cgtccgcgcc
gtggatgcct tggctgcgct
1140gcaggatttc tatcacaggg agcacgtcac taaaacccag atcctgtgtg
ccgagcgccg 1200gctgctggcc ctgttcgatg accgcaagaa tgagctggcc
cacttggcaa ctcatggccc 1260agagaatcca aaactggaga tgctggaaaa
gatcctgcaa aggcagttca gtagctctaa 1320cagccctcgg ggtatcatct
tcacccgcac ccgccaaagc gcacactccc tcctgctctg 1380gctccagcag
caacagggcc tgcagactgt ggacatccgg gcccagctac tgattggggc
1440tgggaacagc agccagagca cccacatgac ccagagggac cagcaagaag
tgatccagaa 1500gttccaagat ggaaccctga accttctggt ggccacgagt
gtggcggagg aggggctgga 1560catcccacat tgcaatgtgg tggtgcgtta
tgggctcttg accaatgaaa tctccatggt 1620ccaggccagg ggccgtgcct
gggccgatca gagtgtatac gcgtttgtag caactgaagg 1680tagccgggag
ctgaagcggg agctgatcaa cgaggcgctg gagacgctaa tggagcaggc
1740agtggctgct gtgcagaaaa tggaccaggc cgagtaccag gccaagatcc
gggatctgca 1800gcaggcagcc ttgaccaagc gggcggccca ggcagcccag
cgggagaacc agcggcagca 1860gttcccagtg gagcacgtgc agctactctg
catcaactgc atggtggctg tgggccatgg 1920cagcgacctg cggaaggtgg
agggcaccca ccatgtcaat gtgaacccca acttctcgaa 1980ctactataat
gtctccaggg atcctgtggt catcaacaaa gtcttcaagg actggaagcc
2040tgggggtgtc atcagctgca ggaactgtgg ggaggtctgg ggtctgcaga
tgatctacaa 2100gtcagtgaag ctgccagtgc tcaaagtccg cagcatgctg
ctggagaccc ctcaggggcg 2160gatccaggcc aaaaagtggt cccgcgtgcc
cttctccgtg cctgactttg acttcctgca 2220gcattgtgcc gagaacttgt
cggacctctc cctggactga ccacctcatt gctgcagtgc 2280ccggtttggg
ctgtaggggg cgggagagtc tgcagcagac tccaggcccc tccttcctga
2340atcatcagct gtgggcatca ggcccaccag ccacacagga gtcctgggca
ccctggctta 2400ggctcccgca atgggaaaac aaccggaggg ccagagctta
gtccagacct accttgtacg 2460cacatagaca ttttcatatg cactggatgg
agttagggaa actgaggcaa aagaatttgc 2520catactgtac tcagaatcac
gacattcctt ccctaccaag gccacttcta ttttttgagg 2580ctcctcataa
aaataaatga aaaaatggga tag 2613112678PRTHomo sapiens 112Met Glu Leu
Arg Ser Tyr Gln Trp Glu Val Ile Met Pro Ala Leu Glu1 5 10 15Gly Lys
Asn Ile Ile Ile Trp Leu Pro Thr Gly Ala Gly Lys Thr Arg 20 25 30Ala
Ala Ala Tyr Val Ala Lys Arg His Leu Glu Thr Val Asp Gly Ala 35 40
45Lys Val Val Val Leu Val Asn Arg Val His Leu Val Thr Gln His Gly
50 55 60Glu Glu Phe Arg Arg Met Leu Asp Gly Arg Trp Thr Val Thr Thr
Leu65 70 75 80Ser Gly Asp Met Gly Pro Arg Ala Gly Phe Gly His Leu
Ala Arg Cys 85 90 95His Asp Leu Leu Ile Cys Thr Ala Glu Leu Leu Gln
Met Ala Leu Thr 100 105 110Ser Pro Glu Glu Glu Glu His Val Glu Leu
Thr Val Phe Ser Leu Ile 115 120 125Val Val Asp Glu Cys His His Thr
His Lys Asp Thr Val Tyr Asn Val 130 135 140Ile Met Ser Gln Tyr Leu
Glu Leu Lys Leu Gln Arg Ala Gln Pro Leu145 150 155 160Pro Gln Val
Leu Gly Leu Thr Ala Ser Pro Gly Thr Gly Gly Ala Ser 165 170 175Lys
Leu Asp Gly Ala Ile Asn His Val Leu Gln Leu Cys Ala Asn Leu 180 185
190Asp Thr Trp Cys Ile Met Ser Pro Gln Asn Cys Cys Pro Gln Leu Gln
195 200 205Glu His Ser Gln Gln Pro Cys Lys Gln Tyr Asn Leu Cys His
Arg Arg 210 215 220Ser Gln Asp Pro Phe Gly Asp Leu Leu Lys Lys Leu
Met Asp Gln Ile225 230 235 240His Asp His Leu Glu Met Pro Glu Leu
Ser Arg Lys Phe Gly Thr Gln 245 250 255Met Tyr Glu Gln Gln Val Val
Lys Leu Ser Glu Ala Ala Ala Leu Ala 260 265 270Gly Leu Gln Glu Gln
Arg Val Tyr Ala Leu His Leu Arg Arg Tyr Asn 275 280 285Asp Ala Leu
Leu Ile His Asp Thr Val Arg Ala Val Asp Ala Leu Ala 290 295 300Ala
Leu Gln Asp Phe Tyr His Arg Glu His Val Thr Lys Thr Gln Ile305 310
315 320Leu Cys Ala Glu Arg Arg Leu Leu Ala Leu Phe Asp Asp Arg Lys
Asn 325 330 335Glu Leu Ala His Leu Ala Thr His Gly Pro Glu Asn Pro
Lys Leu Glu 340 345 350Met Leu Glu Lys Ile Leu Gln Arg Gln Phe Ser
Ser Ser Asn Ser Pro 355 360 365Arg Gly Ile Ile Phe Thr Arg Thr Arg
Gln Ser Ala His Ser Leu Leu 370 375 380Leu Trp Leu Gln Gln Gln Gln
Gly Leu Gln Thr Val Asp Ile Arg Ala385 390 395 400Gln Leu Leu Ile
Gly Ala Gly Asn Ser Ser Gln Ser Thr His Met Thr 405 410 415Gln Arg
Asp Gln Gln Glu Val Ile Gln Lys Phe Gln Asp Gly Thr Leu 420 425
430Asn Leu Leu Val Ala Thr Ser Val Ala Glu Glu Gly Leu Asp Ile Pro
435 440 445His Cys Asn Val Val Val Arg Tyr Gly Leu Leu Thr Asn Glu
Ile Ser 450 455 460Met Val Gln Ala Arg Gly Arg Ala Arg Ala Asp Gln
Ser Val Tyr Ala465 470 475 480Phe Val Ala Thr Glu Gly Ser Arg Glu
Leu Lys Arg Glu Leu Ile Asn 485 490 495Glu Ala Leu Glu Thr Leu Met
Glu Gln Ala Val Ala Ala Val Gln Lys 500 505 510Met Asp Gln Ala Glu
Tyr Gln Ala Lys Ile Arg Asp Leu Gln Gln Ala 515 520 525Ala Leu Thr
Lys Arg Ala Ala Gln Ala Ala Gln Arg Glu Asn Gln Arg 530 535 540Gln
Gln Phe Pro Val Glu His Val Gln Leu Leu Cys Ile Asn Cys Met545 550
555 560Val Ala Val Gly His Gly Ser Asp Leu Arg Lys Val Glu Gly Thr
His 565 570 575His Val Asn Val Asn Pro Asn Phe Ser Asn Tyr Tyr Asn
Val Ser Arg 580 585 590Asp Pro Val Val Ile Asn Lys Val Phe Lys Asp
Trp Lys Pro Gly Gly 595 600 605Val Ile Ser Cys Arg Asn Cys Gly Glu
Val Trp Gly Leu Gln Met Ile 610 615 620Tyr Lys Ser Val Lys Leu Pro
Val Leu Lys Val Arg Ser Met Leu Leu625 630 635 640Glu Thr Pro Gln
Gly Arg Ile Gln Ala Lys Lys Trp Ser Arg Val Pro 645 650 655Phe Ser
Val Pro Asp Phe Asp Phe Leu Gln His Cys Ala Glu Asn Leu 660 665
670Ser Asp Leu Ser Leu Asp 675
References
D00000

D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014

D00015

D00016

D00017

D00018

D00019

D00020

D00021

D00022

D00023

D00024

D00025

D00026
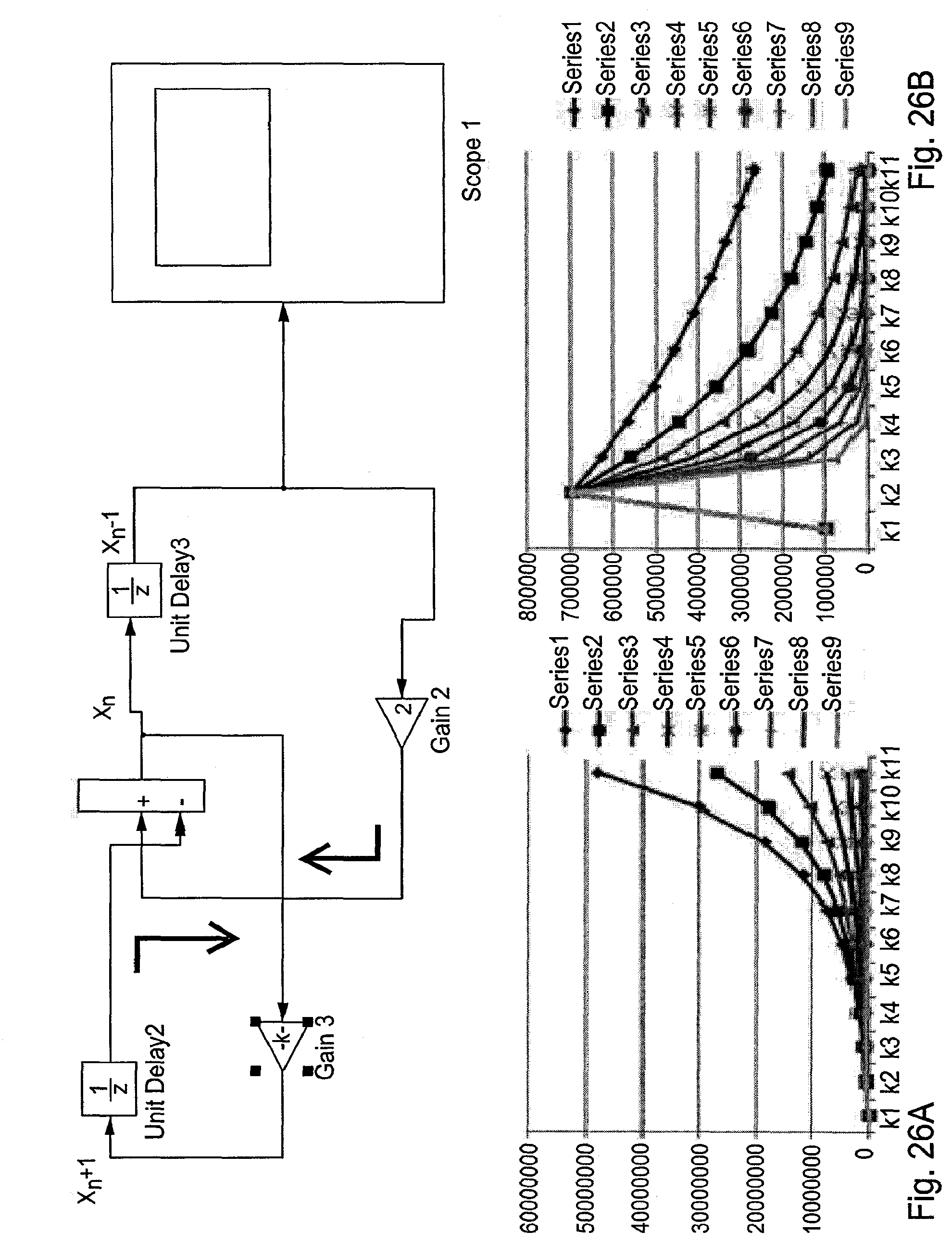
D00027

D00028

D00029

D00030

D00031

D00032

D00033

D00034

D00035

D00036

D00037

D00038

D00039

D00040

D00041

D00042

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.