Plasmid Constructs For Heterologous Protein Expression And Methods Of Use
Campbell; Jean ; et al.
U.S. patent application number 17/032034 was filed with the patent office on 2021-03-04 for plasmid constructs for heterologous protein expression and methods of use. The applicant listed for this patent is OncoSec Medical Incorporated. Invention is credited to Jean Campbell, David A. Canton, Robert H. Pierce.
| Application Number | 20210062218 17/032034 |
| Document ID | / |
| Family ID | 1000005210028 |
| Filed Date | 2021-03-04 |







| United States Patent Application | 20210062218 |
| Kind Code | A1 |
| Campbell; Jean ; et al. | March 4, 2021 |
PLASMID CONSTRUCTS FOR HETEROLOGOUS PROTEIN EXPRESSION AND METHODS OF USE
Abstract
Provided are plasmid vector constructs encoding multiple immunomodulatory proteins where each protein or component thereof can be expressed utilizing appropriate promoters and/or translation modifiers. Additional immunomodulatory proteins and genetic adjuvants containing shared tumor antigens can be added to further therapeutic potential as well as allow tracking of therapeutic treatment. Also provides are methods of expressing the plasmid constructs.
| Inventors: | Campbell; Jean; (Seattle, WA) ; Canton; David A.; (Poway, CA) ; Pierce; Robert H.; (Seattle, WA) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 1000005210028 | ||||||||||
| Appl. No.: | 17/032034 | ||||||||||
| Filed: | September 25, 2020 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 16062983 | Jun 15, 2018 | |||
| PCT/US2016/067388 | Dec 16, 2016 | |||
| 17032034 | ||||
| 62375245 | Aug 15, 2016 | |||
| 62269702 | Dec 18, 2015 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C07K 14/57 20130101; C07K 14/52 20130101; C07K 14/7155 20130101; A61P 35/00 20180101; C07K 14/5434 20130101; G01N 33/574 20130101; C07K 14/5443 20130101; C07K 2319/00 20130101; A61K 9/0009 20130101; A61K 9/0019 20130101; C12N 15/85 20130101 |
| International Class: | C12N 15/85 20060101 C12N015/85; C07K 14/52 20060101 C07K014/52; C07K 14/54 20060101 C07K014/54; A61P 35/00 20060101 A61P035/00; A61K 9/00 20060101 A61K009/00; C07K 14/57 20060101 C07K014/57; C07K 14/715 20060101 C07K014/715; G01N 33/574 20060101 G01N033/574 |
Claims
1. A method of reducing cancer growth in a lung in a subject comprising injecting at least one pharmaceutically effective dose of at least one nucleic acid encoding IL-12 into a tumor in the subject and administering at least one electroporation pulse to the tumor.
2. The method of claim 1, wherein the nucleic acid comprises the formula: P-A-T-A' wherein: a) P is an expression promoter; b) A and A' encode IL-12 subunits; and c) T is a translation modulating element.
3. The method of claim 2, wherein A encodes IL-12p35 and A' encodes IL 12p40.
4. The method of claim 2, wherein the translation modulating element is selected from the group consisting of: a 2A ribosomal skipping modulator and an internal ribosomal entry site (IRES).
5. The method of claim 2, wherein the translation modulating element comprises a 2A ribosomal skipping modulator.
6. The method of claim 5, wherein the 2A ribosomal skipping modulator comprises a P2A ribosomal skipping modulator.
7. The method of claim 2, wherein the translation modulating element comprises an IRES.
8. The method of claim 2, wherein A encodes IL-12p35, A' encodes IL 12p40, and T encodes a P2A ribosomal skipping modulator.
9. The method of claim 2, wherein A encodes IL-12p35, A' encodes IL 12p40, and T encodes an IRES.
10. The method of claim 2, wherein P is selected from the group consisting of a human CMV promoter, a simian CMV promoter, SV-40, mPGK, and .beta.-Actin.
11. The method of claim 2, wherein the nucleic acid comprises SEQ ID NO: 5, SEQ ID NO: 6, or SEQ ID NO: 7.
12. The method of claim 1, wherein the electroporation pulse has a field strength of about 200 V/cm to about 1500 V/cm.
13. The method of claim 12, wherein the electroporation pulse has a field strength selected from the group consisting of: 350 V/cm, 400 V/cm, and 1500 V/cm.
14. The method of claim 1, wherein administering at least one electroporation pulse to the tumor comprises administering at least one electroporation pulse having a field strength of about 1500 V/cm and a pulse length of about 100 .mu.seconds.
15. The method of claim 1, wherein administering at least one electroporation pulse to the tumor comprises administering at least one electroporation pulse having a field strength of about 400 V/cm and a pulse length of about 10 mseconds.
16. The method of claim 1, wherein administering at least one electroporation pulse to the tumor comprises administering at least one electroporation pulse having a field strength of about 350 V/cm and a pulse length of about 10 mseconds.
17. The method of claim 1, wherein administering at least one electroporation pulse to the tumor comprises administering 8 electroporation pulses having a field strength of about 350 V/cm and a pulse length of about 10 mseconds.
18. The method of claim 1, wherein the subject is a human.
Description
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] The application is a continuation of U.S. application Ser. No. 16/062,983, filed Jun. 15, 2018, which is the National Stage of International Application PCT/US2016/067388, filed Dec. 16, 2016, which claims the benefit of U.S. Provisional Application No. 62/375,245, filed Aug. 15, 2016 and U.S. Provisional Application No. 62/269,702, filed Dec. 18, 2015, each of which is herein incorporated by reference in its entirety for all purposes.
SEQUENCE LISTING
[0002] The Sequence Listing written in file OM1507WO01-SEQLIST.txt is 41 kilobytes in size, was created Dec. 16, 2016, and is hereby incorporated by reference.
FIELD OF THE INVENTION
[0003] The present invention relates to recombinant expression vectors for intratumoral delivery of at least two genes encoding each chain of a therapeutically active multimeric polypeptide. Each nucleic acid chain encoding the multimer is separated by at least one translation modulating element. Additional genes encoding therapeutic polypeptides and tracking antigens can be added using additional translation modifiers to the nucleic acid chain or as a separate gene in the expression vector.
BACKGROUND OF THE INVENTION
[0004] E. coli plasmids have long been an important source of recombinant DNA molecules used by researchers and by industry. Today, plasmid DNA is becoming increasingly important as the next generation of biotechnology products (e.g., gene medicines and DNA vaccines) make their way into clinical trials, and eventually into the pharmaceutical marketplace. Expression plasmid DNA may find application as vehicles to deliver therapeutic proteins to sites on a patient where treatment is needed, e.g., tumors.
[0005] This "intratumoral delivery" often involves the delivery of immunomodulators to the tumor microenvironment. Immunotherapy has recently drawn attention as a fourth method following surgery, chemotherapy and radiation therapy for treating tumors. Since immunotherapy utilizes the immunity inherent to humans, it is said that the physical burden on patients are less in immunotherapy than those in other therapies. The therapeutic approaches known as immunotherapies include: cell transfer therapy in which cells such as lymphokine-activated cells, natural killer T-cells or .gamma..delta.T cells obtained, for example, from exogenously-induced cytotoxic T-lymphocytes (CTLs) or peripheral blood lymphocytes by expansion culture using various method are transferred; dendritic cell-transfer therapy or peptide vaccine therapy by which in vivo induction of antigen-specific CTLs is expected; Th1 cell therapy; and immune gene therapy in which genes expected to have various effects are introduced ex vivo into the above-mentioned cells to transfer them in vivo. In these immunotherapies, CD4-positive T cells and CD8-positive T cells have traditionally known to play a critical role.
[0006] In vivo electroporation is a gene delivery technique that has been used successfully for efficient delivery of plasmid DNA to many different tissues. Studies have reported the administration of in vivo electroporation for delivery of plasmid DNA to B16 melanomas and other tumor tissues. Systemic and local expression of a gene or cDNA encoded by a plasmid can be obtained with administration of in vivo electroporation. Use of in vivo electroporation enhances plasmid DNA uptake in tumor tissue, resulting in expression within the tumor, and delivers plasmids to muscle tissue, resulting in systemic cytokine expression.
[0007] It has been shown that electroporation can be used to transfect cells in vivo with plasmid DNA. Recent studies have shown that electroporation is capable of enhancing delivery of plasmid DNA as an antitumor agent. Electroporation has been administered for treatment of hepatocellular carcinomas, adenocarcinoma, breast tumors, squamous cell carcinoma and B16.F10 melanoma in rodent models. The B16.F10 murine melanoma model has been used extensively for testing potential immunotherapy protocols for the delivery of an immunomodulatory molecule including cytokines either as recombinant protein or by gene therapy.
[0008] Various protocols known in the art can be utilized for the delivery of plasmid encoding an immunomodulatory protein utilizing in vivo electroporation for the treatment of cancer. The protocols known in the art describe in vivo electroporation mediated cytokine based gene therapy, both intratumor and intramuscular, utilizing low-voltage and long-pulse currents.
[0009] Combination immunotherapies that involve various phases of the cancer-immunity cycle may enhance the ability to prevent immune escape by targeting multiple mechanisms by which tumor cells avoid elimination by the immune system, with synergistic effects that may offer improved efficacy in broader patient populations. Often these combination therapeutic immunomodulatory proteins are complex molecules involving one or more homo- or heterodimeric chains, e.g., IL-12 or IL-15/IL-15R.alpha., fusion proteins encoding genetic adjuvants and shared tumor antigens. Administration of multiple proteins as therapeutics is complex and costly. Use of intratumoral delivery of multiple encoded proteins using expression plasmids is simpler and more cost effective. However, current expression plasmid constructs do not address the need for adequate production of each immunomodulatory protein. The present invention addresses this need by providing expression plasmids encoding multiple immunomodulators with appropriately placed promoters and translation modifiers.
BRIEF DESCRIPTION OF THE DRAWINGS
[0010] FIG. 1 shows the plasmid map of human IL-12 and P2A in pOMI2A.
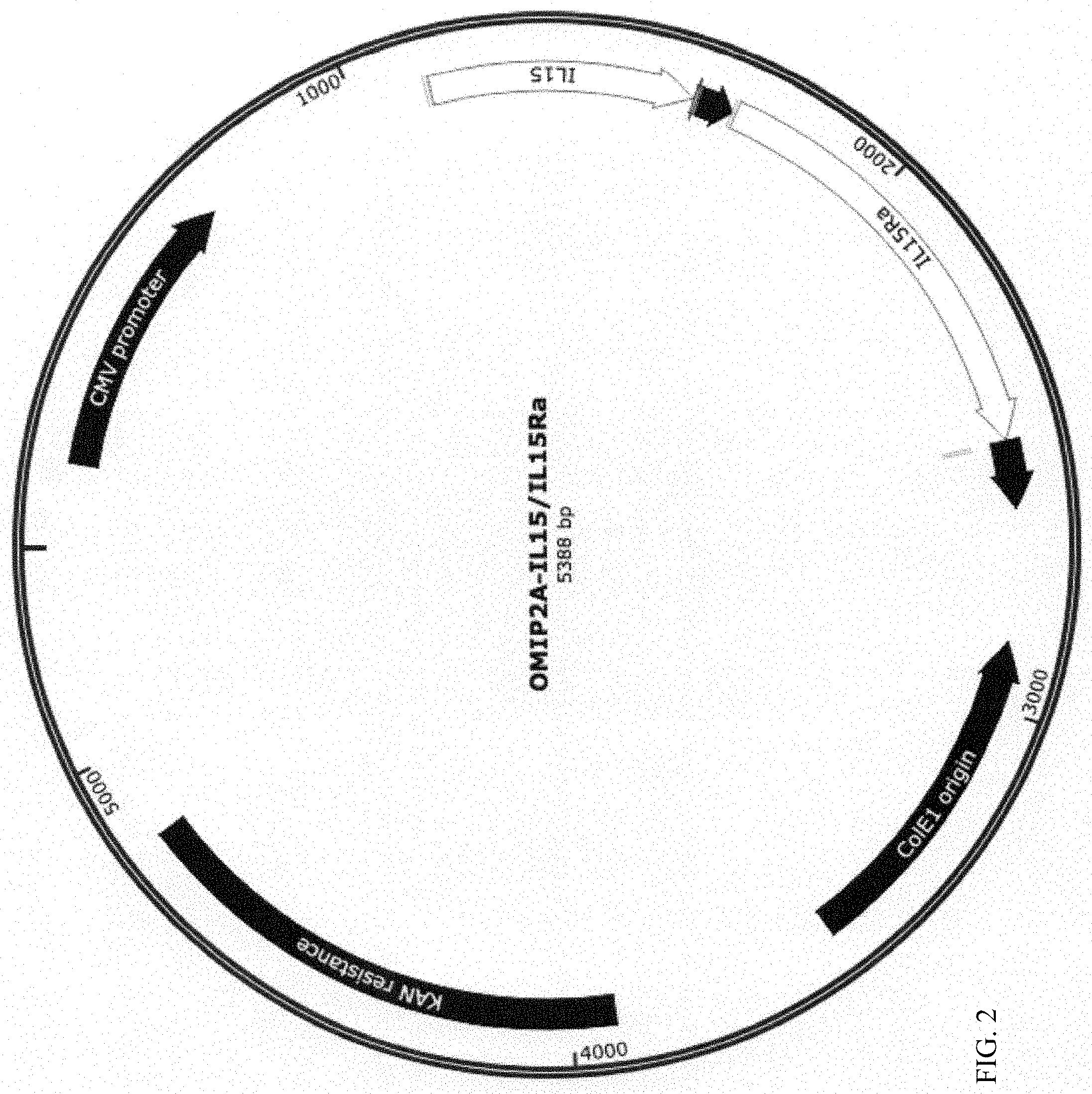
[0011] FIG. 2 shows the plasmid map of human IL-15/IL-15Ra and P2A in pOMI2A.
[0012] FIG. 3A shows the plasmid map for vectors for expression of more than two immunomodulatory gene cassettes: OMI2x2A: Promoter 1+gene cassette A+P2A+gene cassette B+P2A+gene cassette B'.
[0013] FIG. 3B shows the plasmid map for vectors for expression of more than two immunomodulatory gene cassettes: OMI2x2A': Promoter 1+gene cassette A+P2A+gene cassette A'+P2A+gene cassette B.
[0014] FIG. 4 (A) illustrates the protein expression levels of cells transfected with pOMI2A-hIL-12 and pOMIIRES-hIL-12, as measured by ELISA. (B) illustrates the proliferative activity of IL-12 produced by transfection of pOMI2A-hIL-12 in comparison to pOMIIRES-hIL-12 on peripheral blood monocyte cells (PBMC).
[0015] FIG. 5 (A) illustrates the protein expression levels of cells transfected with pOMI2A-hIL-15/hIL-15Ra and pOMI2A-IL-15/IL-15Ra-Fc, as measured by ELISA. (B) illustrates the proliferative activity on human primary CD8+ T cells of hIL-15 produced by transfection of pOMI2A-hIL-15/IL-15Ra and pOMI2A-hIL-15/IL-15Ra-Fc.
[0016] FIG. 6 illustrates the activity tissue culture cell conditioned media containing secreted IL-12 p70 heterodimers expressed from OMIP2A-IL12-Flt3L-NYESO1 vectors as measured using HEK Blue reporter cells. Controls (Addition of neutralizing anti-IL12 antibodies; conditioned media from un-transfected cells) and shown with dotted lines.
SUMMARY OF THE INVENTION
[0017] The present invention provides an expression plasmid construct comprising a plurality of expression cassettes defined by the formula: P-A-T-B where: a) P is an expression promoter; b) A and B encode immunomodulatory molecules; and c) T is a translation modification element. In certain embodiments, P is selected from the group consisting of a human CMV promoter, a simian CMV promoter, SV-40, mPGK, and .beta.-Actin, and the immunomodulatory molecules are selected from the group consisting of immunostimulatory cytokines and genetic adjuvants fused to at least one antigen.
[0018] The present invention provides an expression plasmid comprising a plurality of expression cassettes defined by the formula: P-A-T-A'-T-B where a) P is an expression promoter; b) A and A' are chains of a heterodimeric cytokine; c) B is at least one genetic adjuvant fused to at least one antigen; and d) T is a translation modification element. In certain embodiments, P is selected from the group consisting of a human CMV promoter, a simian CMV promoter, SV-40, mPGK, and .beta.-Actin; the heterodimeric cytokine is selected from the group consisting of IL-12, IL-15, IL-23, and IL-27; A is selected from the group consisting of IL-12p35, IL-23p19, EBI3, IL-15; A' is selected from the group consisting of IL-12p40, IL-27p28, and IL-15R.alpha.; the translation modification element is selected from the group consisting of a P2A family member and IRES; the genetic adjuvant is selected from the group consisting of Flt3 ligand, LAMP-1, Calreticulin, Human heat shock protein 96; GM-CSF, and CSF Receptor 1; antigen is selected from the group consisting of: NYESO-1, OVA, RNEU, MAGE-A1, MAGE-A2, Mage-A10, SSX-2, Melan-A, MART-1, Tyr, Gp100, LAGE-1, Survivin, PRS pan-DR, CEA peptide CAP-1, OVA, HCV-N53, and an HPV vaccine peptide.
[0019] The present invention provides for an expression plasmid comprising a plurality of expression cassettes defined by the formula: P-A-T-B-T-B' where P is an expression promoter; A is at least one genetic adjuvant fused to at least one antigen; B and B' are chains of a heterodimeric cytokine; and T is a translation modification element. In certain embodiments, P is selected from the group consisting of a human CMV promoter, a simian CMV promoter, SV-40, mPGK, and .beta.-Actin; the heterodimeric cytokine is selected from the group consisting of IL-12, IL-15, IL-23, and IL-27; A is selected from the group consisting of IL-12p35, IL-23p19, EBI3, IL-15; A' is selected from the group consisting of IL-12p40, IL-27p28, and IL-15R.alpha.; the translation modification element is selected from the group consisting of a P2A family member and IRES; the genetic adjuvant is selected from the group consisting of Flt3 ligand; LAMP-1; Calreticulin; Human heat shock protein 96; GM-CSF; and CSF Receptor 1; and the antigen is selected from the group consisting of: NYESO-1, OVA, RNEU, MAGE-A1, MAGE-A2, Mage-A10, SSX-2, Melan-A, MART-1, Tyr, Gp100, LAGE-1, Survivin, PRS pan-DR, CEA peptide CAP-1, OVA, HCV-NS3, and an HPV vaccine peptide.
[0020] The present invention provides a method of treating a tumor in a subject comprising delivering the expression plasmid of either of the formulas P-A-T-A'-T-B or P-A-T-B-T-B' into the tumor using at least one intratumoral electroporation pulse. In certain embodiments, the intratumoral electroporation pulse has a field strength of about 200 V/cm to 1500 V/cm; the subject is a human; the tumor is selected from the group of melanoma, triple negative breast cancer, Merkel Cell Carcinoma, CTCL, and head and neck squamous cell carcinoma; and the electroporation pulse is delivered by a generator capable of electrochemical impedance spectroscopy.
[0021] The present invention provides an expression plasmid construct comprising a plurality of expression cassettes defined by the formula: P-A-T-A' where a) P is an expression promoter; b) A, and A' encode subunits of an immunomodulatory molecule; and c) T is a translation modification sequence. In certain embodiments, P is selected from group consisting of human CMV promoter, a simian CMV promoter, SV-40, mPGK, and .beta.-Actin; A is selected from the group consisting of IL-12p35, IL-23p19, EBI3, IL-15; A' is selected from the group consisting of IL-12p40, IL-27p28, and IL-15R.alpha.; and T is selected from the group consisting of a P2A and IRES.
DETAILED DESCRIPTION
[0022] As used herein, including the appended claims, the singular forms of words such as "a," "an," and "the," include their corresponding plural references unless the context clearly dictates otherwise.
[0023] All references cited herein are incorporated by reference to the same extent as if each individual publication, patent application, or patent, was specifically and individually indicated to be incorporated by reference.
I. Definitions
[0024] "Activity" of a molecule may describe or refer to the binding of the molecule to a ligand or to a receptor, to catalytic activity, to the ability to stimulate gene expression, to antigenic activity, to the modulation of activities of other molecules, and the like. "Activity" of a molecule may also refer to activity in modulating or maintaining cell-to-cell interactions, e.g., adhesion, or activity in maintaining a structure of a cell, e.g., cell membranes or cytoskeleton. "Activity" may also mean specific activity, e.g., [catalytic activity]/[mg protein], or [immunological activity]/[mg protein], or the like.
[0025] "Translation modulating element" or "translation modifier" as used herein, means a specific translation initiator or ribosomal skipping modulator wherein a picornavirus-derived sequence in the nascent polypeptide chain prevents covalent amide linkage with the next amino acid. Incorporation of this sequence results in co-expression of each chain of a heterodimeric protein with equal molar levels of the translated polypeptides. Contemplated are: the 2A family of ribosomal skipping modulators that include, but are not limited to, P2A, T2A, E2A or F2A, all of which share the PG/P cleavage site (See Table 5); and internal ribosomal entry sites (IRES).
[0026] In accordance with the present invention there may be employed conventional molecular biology, microbiology, and recombinant DNA techniques within the skill of the art. Such techniques are explained in the literature. See, e.g., Sambrook, Fritsch & Maniatis, Molecular Cloning: A Laboratory Manual, Second Edition (1989) Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. (herein "Sambrook, et al., 1989"); DNA Cloning: A Practical Approach, Volumes I and II (D. N. Glover ed. 1985); Oligonucleotide Synthesis (M. J. Gait ed. 1984); Nucleic Acid Hybridization (B. D. Hames & S. J. Higgins eds. (1985)); Transcription And Translation (B. D. Hames & S. J. Higgins, eds. (1984)); Animal Cell Culture (R. I. Freshney, ed. (1986)); Immobilized Cells And Enzymes (IRL Press, (1986)); B. Perbal, A Practical Guide To Molecular Cloning (1984); F. M. Ausubel, et al. (eds.), Current Protocols in Molecular Biology, John Wiley & Sons, Inc. (1994).
[0027] A "polynucleotide," "nucleic acid" or "nucleic acid molecule" includes DNA or RNA. For example, in an embodiment of the invention, the polynucleotide is the circular plasmid pOMI2A.
[0028] A "polynucleotide sequence," "nucleic acid sequence" or "nucleotide sequence" is a series of nucleotides in a nucleic acid, such as DNA or RNA, and means any chain of two or more nucleotides.
[0029] A "coding sequence" or a sequence "encoding" an expression product such as a RNA or peptide (e.g., an immunoglobulin chain), is a nucleotide sequence that, when expressed, results in production of the product.
[0030] As used herein, the term "oligonucleotide" refers to a nucleic acid, generally of no more than about 300 nucleotides (e.g., 30, 40, 50, 60, 70, 80, 90, 150, 175, 200, 250 or 300), that may be hybridizable to a genomic DNA molecule, a cDNA molecule, or an mRNA molecule encoding a gene, mRNA, cDNA, or other nucleic acid of interest. Oligonucleotides are usually single-stranded, but may be double-stranded. Oligonucleotides can be labeled, e.g., by incorporation of 32P-nucleotides, 3H-nucleotides, 14C-nucleotides, 35S-nucleotides or nucleotides to which a label, such as biotin, has been covalently conjugated. In one embodiment, a labeled oligonucleotide can be used as a probe to detect the presence of a nucleic acid. In another embodiment, oligonucleotides (one or both of which may be labeled) can be used as PCR primers, either for cloning full length or a fragment of the gene, or to detect the presence of nucleic acids. Generally, oligonucleotides are prepared synthetically, e.g., on a nucleic acid synthesizer.
[0031] A "protein sequence," "peptide sequence" or "polypeptide sequence," or "amino acid sequence" refers to a series of two or more amino acids in a protein, peptide or polypeptide.
[0032] "Protein," "peptide" or "polypeptide" includes a contiguous string of two or more amino acids.
[0033] The term "isolated polynucleotide" or "isolated polypeptide" includes a polynucleotide (e.g., RNA or DNA molecule, or a mixed polymer) or a polypeptide, respectively, which is partially or fully separated from other components that are normally found in cells or in recombinant DNA expression systems or any other contaminant. These components include, but are not limited to, cell membranes, cell walls, ribosomes, polymerases, serum components and extraneous genomic sequences.
[0034] An isolated polynucleotide (e.g., pOMI2A) or polypeptide will, preferably, be an essentially homogeneous composition of molecules but may contain some heterogeneity.
[0035] The term "host cell" includes any cell of any organism that is selected, modified, transfected, transformed, grown, or used or manipulated in any way, for the production of a substance by the cell, for example the expression or replication, by the cell, of a gene, a polynucleotide such as a circular plasmid (e.g., pOMI2A) or RNA or a protein. For example, a host cell may be a mammalian cell or bacterial cell (e.g., E. coli) or any isolated cell capable of maintaining pOMI2A plasmid and, in an embodiment of the invention, promoting expression of a polypeptide encoded by a polynucleotide in the plasmid, e.g., an immunoglobulin chain.
[0036] Vectors of the invention, such as pOMI2A, may be introduced into host cells according to any of the many techniques known in the art, e.g., dextran-mediated transfection, polybrene-mediated transfection, protoplast fusion, electroporation, calcium phosphate co-precipitation, lipofection, direct microinjection of the vector into nuclei, or any other means appropriate for a given host cell type.
[0037] A "cassette" or an "expression cassette" refers to a DNA coding sequence or segment of DNA that codes for an expression product (e.g., peptide or RNA) that can be inserted into a vector, e.g., at defined restriction sites. The expression cassette may comprise a promoter and/or a terminator and/or polyA signal operably linked to the DNA coding sequence.
[0038] In general, a "promoter" or "promoter sequence" is a DNA regulatory region capable of binding an RNA polymerase in a cell (e.g., directly or through other promoter-bound proteins or substances) and initiating transcription of a coding sequence. A promoter sequence is, in general, bounded at its 3' terminus by the transcription initiation site and extends upstream (5' direction) to include the minimum number of bases or elements necessary to initiate transcription at any level. Within the promoter sequence may be found a transcription initiation site (conveniently defined, for example, by mapping with nuclease 51), as well as protein binding domains (consensus sequences) responsible for the binding of RNA polymerase. The promoter may be operably associated with or operably linked to other expression control sequences, including enhancer and repressor sequences or with a nucleic acid to be expressed. An expression control sequence is operably associated with or operably linked to a promoter if it regulates expression from said promoter.
[0039] Promoters which may be used to control gene expression include, but are not limited to, SR.alpha. promoter (Takebe et al., Molec. and Cell. Bio. 8:466-472 (1988)), the human CMV immediate early promoter (Boshart et al., Cell 41:521-530 (1985); Foecking et al., Gene 45:101-105 (1986)), the mouse CMV immediate early promoter, the SV40 early promoter region (Benoist et al., Nature 290:304-310 (1981)), the Orgyia pseudotsugata immediate early promoter, the herpes thymidine kinase promoter (Wagner et al., Proc. Natl. Acad. Sci. USA 78:1441-1445 (1981)), the regulatory sequences of the metallothionein gene (Brinster et al., Nature 296:39-42 (1982)); prokaryotic expression vectors such as the .beta.-lactamase promoter (Villa-Komaroff et al., Proc. Natl. Acad. Sci. USA 75:3727-3731 (1978)), or the tac promoter (DeBoer et al., Proc. Natl. Acad. Sci. USA 80:21-25 (1983)); and promoter elements from yeast or other fungi such as the GAL1, GAL4 or GAL10 promoter, the ADH (alcohol dehydrogenase) promoter, PGK (phosphoglycerol kinase) promoter or the alkaline phosphatase promoter.
[0040] Viral long terminal repeat promoters such as the mouse mammary tumor virus long terminal repeat (MMTV-LTR) (Fasel et al., EMBO J. 1(1):3-7 (1982)), the moloney murine sarcoma virus long terminal repeat (Reddy et al., Proc. Natl. Acad. Sci. USA 77(9): 5234-5238 (1980)), the moloney murine leukemia virus long terminal repeat (Van Beveren et al., Proc. Natl. Acad. Sci. USA 77(6): 3307-3311 (1980)), the HIV LTR (Genbank Accession No. AB100245), the bovine foamy virus LTR (Genbank Accession No. NC-001831), RSV 5'-LTR (Genbank Accession No. K00087), the HIV-2 LTR (Genbank Accession No. NC-001722), an avian retroviral LTR (Ju et al., Cell 22: 379-386 (1980)) and the human herpesvirus LTR (Genbank Accession No. NC-001806) may be included in the vectors of the present invention.
[0041] Other acceptable promoters include the human and simian CMV5 promoter, the murine CMV promoter, the EF1.alpha. promoter, the SV40 promoter, a hybrid CMV promoter for liver specific expression (e.g., made by conjugating CMV immediate early promoter with the transcriptional promoter elements of either human .alpha.1-antitrypsin (HAT) or albumin (HAL) promoter), or promoters for hepatoma specific expression (e.g., wherein the transcriptional promoter elements of either human albumin (HAL; about 1000 bp) or human .alpha.1-antitrypsin (HAT, about 2000 bp) are combined with a 145 bp long enhancer element of human .alpha.1-microglobulin and bikunin precursor gene (AMBP); HAL-AMBP and HAT-AMBP). Table 1 provides examples of promoters that may be utilized.
TABLE-US-00001 TABLE 1 Transcriptional Promoter/Enhancer DNA element Structure Nucleotide sequence Human CMV promoter/enhancer Seq ID 1 Simian CMV promoter/enhancer Seq ID 2 SV-40 promoter/enhancer Seq ID 3 mPGK promoter/enhancer Seq ID 4
[0042] One or more promoters on a single plasmid construct may be employed to drive expression of one or more expression cassettes.
[0043] In addition, bacterial promoters, such as the T7 RNA Polymerase promoter or the tac promoter, may be used to control expression.
[0044] In one embodiment, the promoter is the human CMV (hCMV) promoter. The hCMV promoter provides a high level of expression in a variety of mammalian cell types.
[0045] A coding sequence is "under the control of", "functionally associated with", "operably linked to" or "operably associated with" transcriptional and translational control sequences in a cell when the sequences direct or regulate expression of the sequence. For example, a promoter operably linked to a gene will direct RNA polymerase mediated transcription of the coding sequence into RNA, preferably mRNA, which may then be spliced (if it contains introns) and, optionally, translated into a protein encoded by the coding sequence. A terminator/polyA signal operably linked to a gene terminates transcription of the gene into RNA and directs addition of a polyA signal onto the RNA.
[0046] The terms "express" and "expression" mean allowing or causing the information in a gene, RNA or DNA sequence to become manifest; for example, producing a protein by activating the cellular functions involved in transcription and translation of a corresponding gene. "Express" and "expression" include transcription of DNA to RNA and of RNA to protein. A DNA sequence is expressed in or by a cell to form an "expression product" such as an RNA (e.g., mRNA) or a protein. The expression product itself may also be said to be "expressed" by the cell.
[0047] The term "transformation" means the introduction of a nucleic acid into a cell. The introduced gene or sequence may be called a "clone". A host cell that receives the introduced DNA or RNA has been "transformed" and is a "transformant" or a "clone." The DNA or RNA introduced to a host cell can come from any source, including cells of the same genus or species as the host cell, or from cells of a different genus or species. Examples of transformation methods which are very well known in the art include liposome delivery, electroporation, CaPO.sub.4 transformation, DEAE-Dextran transformation, microinjection and viral infection.
[0048] The present invention includes vectors which comprise polynucleotides of the invention. The term "vector" may refer to a vehicle (e.g., a plasmid) by which a DNA or RNA sequence can be introduced into a host cell, so as to transform the host and, optionally, promote expression and/or replication of the introduced sequence.
[0049] The polynucleotides of the invention may be expressed in an expression system. The term "expression system" means a host cell and compatible vector which, under suitable conditions, can express a protein or nucleic acid which is carried by the vector and introduced to the host cell. Common expression systems include E. coli host cells and plasmid vectors, insect host cells and baculovirus vectors, and mammalian host cells and vectors such as plasmids, cosmids, BACs, YACs and viruses such as adenovirus and adenovirus associated virus (AAV).
[0050] The terms "immunostimulatory cytokine" or "immunostimulatory cytokines" refer to protein naturally secreted by cells involved in immunity that have the capacity to stimulate an immune response. Examples of immunostimulatory cytokines are provided in Table 2A and 2B.
[0051] The phrase "genetic adjuvants containing shared tumor antigens" as used herein refers to fusion proteins of receptor tyrosine kinases and known tumor antigens as described in Table 4.
II. General
[0052] The present invention provides expression vectors that allow adequate expression of multiple proteins following transfection of an in vivo cell, particularly a tumor cell.
[0053] Vectors are provided that contain some or all of the modifications described herein designed to improve their efficacy and safety. The optimization of the vectors includes the incorporation of sequences encoding appropriate peptides and the tailoring of sites to maximize gene expression. A peptide is understood to be any translation product regardless of size, and whether or not post-translationally modified, as, for example, in glycosylation and phosphorylation.
[0054] The present invention provides expression vectors comprising the translation control element, e.g., P2A, operatively linked to gene sequences to be expressed. In certain embodiments, the expression vector comprises at least two nucleic acid sequences to be translated and the translation control element is operatively linked to at least one of the sequences to be translated. Vectors are known or can be constructed by those skilled in the art and contain all expression elements necessary to achieve the desired transcription of the sequences in addition to the sequence of the present invention as shown in the Examples herein below. The vectors contain elements for use in either prokaryotic or eukaryotic host systems depending on their use. One of ordinary skill in the art will know which host systems are compatible with a particular vector.
[0055] Recombinant gene expression depends upon transcription of the appropriate gene and efficient translation of the message. A failure to perform correctly either one of these processes can result in the failure of a given gene to be expressed. This is further complicated when more than one gene needs to be expressed from a single plasmid. Traditionally, internal ribosomal entry sites (IRES's) were used between the genes to be expressed. IRES's have limitations because of their size and the translation efficiency of the second gene is much lower than the first. Recent studies have found that the use of picornavirus polyprotein 2A ("P2A") peptide results in stoichiometric expression of multiple proteins flanking the P2A peptide (see, e.g., Kim et al (2011) PloS One 6:318556).
[0056] Adequate recombinant expression of diverse immunomodulators including, e.g., heterodimeric proteins such as IL-12, IL-15/IL-15R.alpha., IL-23, IL-27; and genetic adjuvants containing shared tumor antigens, e.g., Flt3L-NYESO-1 fusion protein, in expression plasmids. This is especially true when the plasmid is delivered to a tumor (intratumoral delivery) via in vivo electroporation.
[0057] Examples of immunostimulatory cytokines are provided in Table 2A.
TABLE-US-00002 TABLE 2A Immunostimulatory cytokines. Sequence Gene Structure nucleotide Protein IL-12 p35 and p40 SEQ ID 5 NP_000873.2, subunits NP_002178.2 heterodimer IL-12 p35 and p40 SEQ ID 6 NP_001152896.1, (mouse) subunits NP_001290173.1 heterodimer IL-12 p35 and p40 SEQ ID 7 XP_013965819.1, (canine) subunits NP_001003292.1 heterodimer IL-15/ IL15 and soluble SEQ ID 8 SEQ ID 9, IL-15 IL15 receptor SEQ ID 10 SEQ ID 11_(IL- receptor heterodimer (IL-15Ra- 15Ra-Fc fusion) Fc fusion) IL-23 p19 and p40 XM_011538477.2 XP_011536779.1 subunits NM_002187.2 NP_002178.2 heterodimer IL-27 p28 and IL27B NM_145659.3; NP_663634.2; subunits NM_005755.2 NP_005746.2 heterodimer IFN.alpha. Full length protein NM_006900.3. NP_008831.3 NM_024013.2. NP_076918.1 IFN.beta. Full length protein NM_002176.3. NP_002167.1 INF.gamma. Full length protein SEQ ID 12 NP_000610.2 TNF.alpha. Full length protein X02910 ADV31546 IL-4 Full length protein NM_000589.3 NP_000580.1 IL-7 Full length protein NM_001199886.1 NP_001186815.1 IL-9 Full length protein NM_000590.1 NP_000581.1 IL-21 Full length protein NM_021803.3 NP_068575.1 IL-2 Full length protein NM_000586.3. NP_000577.2
[0058] Also contemplated for immunostimulation are innate immunity regulators as described in Table 2B.
TABLE-US-00003 TABLE 2B Innate immunity regulators Gene Structure Reference IL-33 Recombinant protein: Gao et al., J. Immunol. amino acid 109 to 266 2015; 194: 438 Flagellin TLR5 binding domain Hayashi et al., Nature 2001; 410: 1099 IL-10 Receptor Recombinant soluble, Marchi et al., Cancer secreted protein Gene Therapy 2011, 18: 110 Sting Receptor Dominant-active mutant pUNO1-hSTING-M155 (InvivoGen) IRF3 Dominant-active mutant pUNO1-hsaIRF3 (invivoGen)
TABLE-US-00004 TABLE 3 Genetic Adjuvants Gene Structure Reference Flt3 ligand Extralcellular domain XM_017026533.1 LAMP-1 (ECD) XM_011537494.1 Calreticulin Full length protein NM_004343; Cheng et al., 2001, J Clin Invest. 108: 669 Human heat shock Full length protein Rivoltini et al., 2003. J. protein 96 Immunol. 171: 3467 GM-CSF Full length protein NM_000758.3 CSF Receptor 1 NM_001288705.2
TABLE-US-00005 TABLE 4 Genetic Adjuvants fused to shared tumor antigens or viral antigens (Flt3L protein fusions) Gene Structure Reference NY-ESO-1 Fusion of full length protein to SEQ ID 13 (DNA); SEQ ID 14 ECD of Flt3L (protein):, Gnjatic et al., Advances in Cancer Res. 2006 NY-ESO-1 Fusion of amino acid# 80-180 to SEQ ID 15 (DNA); SEQ ID 16 ECD of Flt3L (protein):, Sabado-R L, Cancer Immunol Res 2015 MARCH; 3(3) NY-ESO-1 Fusion of overlapping peptides: SEQ ID 17 (DNA); SEQ ID 18 Amino acid# 81-100, 87-111, (protein): 157-165, 157-170, 161-180 to ECD of Flt3L NY-ESO-1 Fusion of amino acid # 157-165 RAPOPORT-A P, NATURE to ECD of Flt3L MEDICINE, 2015 AUGUST 21(8) MAGE-A1 Fusion of full legth protein or Almeida et al., Nuc, Acids Res antigenic peptides to ECD of 2009; database url: Flt3L http://www.cta.lncc.br/index.php MAGE-A2 Fusion of full legth protein or Almeida et al., Nuc, Acids Res antigenic peptides to ECD of 2009; database url: Flt3L http://www.cta.lncc.br/index.php MAGE-A3 Fusion of full legth protein or Almeida et al., Nuc, Acids Res antigenic peptides to ECD of 2009; database url: Flt3L http://www.cta.lncc.br/index.php MAGE-A10 Fusion of full legth protein or Almeida et al., Nuc, Acids Res antigenic peptides to ECD of 2009; database url: Flt3L http://www.cta.lncc.br/index.php SSX-2 Fusion of full legth protein or Almeida et al., Nuc, Acids Res antigenic peptides to ECD of 2009; database url: Flt3L http://www.cta.lncc.br/index.php MART-1 Fusion of full length protein or Li et al., J. Immunol. 2010, antigenic peptide ELAGIGILTV 184: 452 to ECD of Flt3L Tyrosinase Fusion of antigenic peptide Skipper et al., J. Exp. Med YMDGTMSQV to ECD of Flt3L 1996, 183: 527 Gp100 Fusion of full legth protein or Bakker et al., J. Exp. Med. antigenic peptides to ECD of 1994, 179: 1005 Flt3L Survivin Fusion of full legth protein or Schmidt et al., Blood 2002, antigenic peptide ELTLGEFLKL 102: 571 to ECD of Flt3L hTERT Fusion of full legth protein or Vonderheide et al., Nature antigenic peptides to ECD of 2002, 21: 674 Flt3L PRS pan- Fusion of full legth protein or Almeida et al., Nuc, Acids Res DR antigenic peptides to ECD of 2009; database url: Flt3L http://www.cta.lncc.br/index.php B7-H6 Full length protein or fusion of Brandt et al., J. Exp Med. 2009, full legth protein to ECD of Flt3L 206: 1495 HPV E7 Full length protein or fusion of Huang et al., Cancer Res. 2001 full legth protein to ECD of Flt3L 61: 1080; Seo et al., Vaccine 2009 27: 5906; Lin et al., HPV16 1-85 aa E6, 1-65 aa E7, 71-158 Kim et al, Nature 2014 5: 5317 E6/E7 aa E6, 51-98 aa E7 fused to ECD of Flt3L HPV16 E6 mutant L50A; E6 mutant Wieking et al., 2012, Cancer E6/E7 ETNL146-151AAAA; E7 mutant Gene Ther. 19: 667 H2P; E7 mutant C24G; E7 mutant E46A; E7 mutant L67R HPV11 E6 44-51 aa E6 Peng et al., 2010, Larynoscope 120: 504 HPV6b/11E7 21-29 aa E7, 82-90 aa E7 Peng et al., 2016, Cancer Immunol. Immunother. 65: 261 HCV-NS3 Fusion of full legth protein or Grubor-Bauk et al., 2016, Gene antigenic peptides fused to ECD Ther. 23: 26 of Flt3L Influenza Fusion of full legth protein or Chow et al., 1979. Infect HA and NA antigenic peptides to ECD of Immun. 25: 103 Flt3L Polyoma- MCPyV LTA aa1-258, aa136- Zeng et al., Vaccine 2012 virus 160; various other peptides from 30: 1322; Lyngaa et al., 2014, VP1, LTA, and STA Clin Can Res 2014, 20: 1768
[0059] Several studies have shown that the translation modifiers can efficiently drive translation of genes encoding multimeric proteins (see, e.g., Kim, et al. (2011) PloS ONE 6:1-8; Ibrahimi, et al. (2009) Human Gene Ther. 20:845-860; Szymczak, et al. (2004) Nat. Biotechnol. 22:589-594). Table 5 provides examples of translational modifiers.
TABLE-US-00006 TABLE 5 Translational modifiers DNA element Structure Nucleotide sequence P2A Exon skipping motif in mRNA Seq ID 19 T2A Exon skipping motif in mRNA Seq ID 20 E2A Exon skipping motif in mRNA Seq ID 21 F2A Exon skipping motif in mRNA Seq ID 21 IRES Internal Ribosome Entry Site Seq ID 23
III. Devices and Uses
[0060] The invention finds use in intratumoral gene electrotransfer. In particular the current plasmid constructs can be used to generate adequate concentrations of several recombinantly expressed immunomodulatory molecules such as, multimeric cytokines or combination of multimeric cytokines, co-stimulatory molecules in native or engineered forms, genetic adjuvants containing shared tumor antigens, etc. To achieve transfer of the instant plasmid constructs into a tissue, e.g., a tumor, an electroporation device is employed.
[0061] The devices and methods of the present embodiment work to treat cancerous tumors by delivering electrical therapy continuously and/or in pulses for a period of time ranging from a fraction of a second to several days, weeks, and/or months to tumors. In a preferred embodiment, electrical therapy is direct current electrical therapy.
[0062] The term "electroporation" (i.e., rendering cellular membranes permeable) as used herein may be caused by any amount of coulombs, voltage, and/or current delivered to a patient in any period of time sufficient to open holes in cellular membranes (e.g., to allow diffusion of molecules such as pharmaceuticals, solutions, genes, and other agents into a viable cell).
[0063] Delivering electrical therapy to tissue causes a series of biological and electrochemical reactions. At a high enough voltage, cellular structures and cellular metabolism are severely disturbed by the application of electrical therapy. Although both cancerous and non-cancerous cells are destroyed at certain levels of electrical therapy tumor cells are more sensitive to changes in their microenvironment than are non-cancerous cells. Distributions of macroelements and microelements are changed as a result of electrical therapy. Destruction of cells in the vicinity of the electroporation is known as irreversible electroporation.
[0064] The use of reversible electroporation is also contemplated. Reversible electroporation occurs when the electricity applied with the electrodes is below the electric field threshold of the target tissue. Because the electricity applied is below the cells' threshold, cells are able to repair their phospholipid bilayer and continue on with their normal cell functions. Reversible electroporation is typically done with treatments that involve getting a drug or gene (or other molecule that is not normally permeable to the cell membrane) into the cell. (Garcia, et al. (2010) "Non-thermal irreversible electroporation for deep intracranial disorders". 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology: 2743-6.)
[0065] In a single electrode configuration, voltage may be applied for fractions of seconds to hours between a lead electrode and the generator housing, to begin destruction of cancerous tissue. Application of a given voltage may be in a series of pulses, with each pulse lasting fractions of a second to several minutes. In certain embodiments, the pulse duration or width can be from about. Low voltage may also be applied for of a duration of fractions of seconds to minutes, which may attract white blood cells to the tumor site. In this way, the cell mediated immune system may remove dead tumor cells and may develop antibodies against tumor cells. Furthermore, the stimulated immune system may attack borderline tumor cells and metastases.
[0066] Various adjuvants may be used to increase any immunological response, depending on the host species, including but not limited to Freund's adjuvant (complete and incomplete), mineral salts such as aluminum hydroxide or aluminum phosphate, various cytokines, surface active substances such as lysolecithin, pluronic polyols, polyanions, peptides, oil emulsions, and potentially useful human adjuvants such as BCG (bacille Calmette-Guerin) and Corynebacterium parvum. Alternatively, the immune response could be enhanced by combination and or coupling with molecules such as keyhole limpet hemocyanin, tetanus toxoid, diphtheria toxoid, ovalbumin, cholera toxin or fragments thereof.
[0067] U.S. Pat. No. 7,245,963 by Draghia-Akli, et al. describes modular electrode systems and their use for facilitating the introduction of a biomolecule into cells of a selected tissue in a body or plant. The modular electrode systems comprise a plurality of needle electrodes; a hypodermic needle; an electrical connector that provides a conductive link from a programmable constant-current pulse controller to the plurality of needle electrodes; and a power source. An operator can grasp the plurality of needle electrodes that are mounted on a support structure and firmly insert them into the selected tissue in a body or plant. The biomolecules are then delivered via the hypodermic needle into the selected tissue. The programmable constant-current pulse controller is activated and constant-current electrical pulse is applied to the plurality of needle electrodes. The applied constant-current electrical pulse facilitates the introduction of the biomolecule into the cell between the plurality of electrodes. The entire content of U.S. Pat. No. 7,245,963 is hereby incorporated by reference.
[0068] U.S. Patent Pub. 2005/0052630 describes an electroporation device which may be used to effectively facilitate the introduction of a biomolecule into cells of a selected tissue in a body or plant. The electroporation device comprises an electro-kinetic device ("EKD device") whose operation is specified by software or firmware. The EKD device produces a series of programmable constant-current pulse patterns between electrodes in an array based on user control and input of the pulse parameters, and allows the storage and acquisition of current waveform data. The electroporation device also comprises a replaceable electrode disk having an array of needle electrodes, a central injection channel for an injection needle, and a removable guide disk (see, e.g., U.S. Patent Pub. 2005/0052630) is hereby incorporated by reference.
[0069] The electrode arrays and methods described in U.S. Pat. No. 7,245,963 and U.S. Patent Pub. 2005/0052630 are adapted for deep penetration into not only tissues such as muscle, but also other tissues or organs. Because of the configuration of the electrode array, the injection needle (to deliver the biomolecule of choice) is also inserted completely into the target organ, and the injection is administered perpendicular to the target issue, in the area that is pre-delineated by the electrodes.
[0070] Also encompassed are electroporation devices incorporating electrochemical impedance spectroscopy ("EIS"). Such devices provide real-time information on in vivo, in particular, intratumoral electroporation efficiency, allowing for the optimization of conditions. Examples of electroporation devices incorporating EIS can be found, e.g., in WO2016161201, which is hereby incorporated by reference.
[0071] Other alternative electroporation technologies are also contemplated. In vivo plasmid delivery can also be performed using cold plasma. Plasma is one of the four fundamental states of matter, the others being solid, liquid, and gas. Plasma is an electrically neutral medium of unbound positive and negative particles (i.e., the overall charge of a plasma is roughly zero). A plasma can be created by heating a gas or subjecting it to a strong electromagnetic field, applied with a laser or microwave generator. This decreases or increases the number of electrons, creating positive or negative charged particles called ions (Luo, et al. (1998) Phys. Plasma 5:2868-2870) and is accompanied by the dissociation of molecular bonds, if present.
[0072] Cold plasmas (i.e., non-thermal plasmas) are produced by the delivery of pulsed high voltage signals to a suitable electrode. Cold plasma devices may take the form of a gas jet device or a dielectric barrier discharge (DBD) device. Cold temperature plasmas have attracted a great deal of enthusiasm and interest by virtue of their provision of plasmas at relatively low gas temperatures. The provision of plasmas at such a temperature is of interest to a variety of applications, including wound healing, anti-bacterial processes, various other medical therapies and sterilization. As noted earlier, cold plasmas (i.e., non-thermal plasmas) are produced by the delivery of pulsed high voltage signals to a suitable electrode. Cold plasma devices may take the form of a gas jet device, a dielectric barrier discharge (DBD) device or multi-frequency harmonic-rich power supply.
[0073] Dielectric barrier discharge device, relies on a different process to generate the cold plasma. A dielectric barrier discharge (DBD) device contains at least one conductive electrode covered by a dielectric layer. The electrical return path is formed by the ground that can be provided by the target substrate undergoing the cold plasma treatment or by providing an in-built ground for the electrode. Energy for the dielectric barrier discharge device can be provided by a high voltage power supply, such as that mentioned above. More generally, energy is input to the dielectric barrier discharge device in the form of pulsed DC electrical voltage to form the plasma discharge. By virtue of the dielectric layer, the discharge is separated from the conductive electrode and electrode etching and gas heating is reduced. The pulsed DC electrical voltage can be varied in amplitude and frequency to achieve varying regimes of operation. Any device incorporating such a principle of cold plasma generation (e.g., a DBD electrode device) falls within the scope of various embodiments of the present invention.
[0074] Cold plasma has been employed to transfect cells with foreign nucleic acids. In particular, transfection of tumor cells (see, e.g., Connolly, et al. (2012) Human Vaccines & Immunotherapeutics 8:1729-1733; and Connolly et al (2015) Bioelectrochemistry 103: 15-21).
[0075] The devices are contemplated for use in patients afflicted with cancer or other non-cancerous (benign) growths. These growths may manifest themselves as any of a lesion, polyp, neoplasm (e.g., papillary urothelial neoplasm), papilloma, malignancy, tumor (e.g., Klatskin tumor, hilar tumor, noninvasive papillary urothelial tumor, germ cell tumor, Ewing's tumor, Askin's tumor, primitive neuroectodermal tumor, Leydig cell tumor, Wilms' tumor, Sertoli cell tumor), sarcoma, carcinoma (e.g., squamous cell carcinoma, cloacogenic carcinoma, adenocarcinoma, adenosquamous carcinoma, cholangiocarcinoma, hepatocellular carcinoma, invasive papillary urothelial carcinoma, flat urothelial carcinoma), lump, or any other type of cancerous or non-cancerous growth. Tumors treated with the devices and methods of the present embodiment may be any of noninvasive, invasive, superficial, papillary, flat, metastatic, localized, unicentric, multicentric, low grade, and high grade.
[0076] The devices are contemplated for use in numerous types of malignant tumors (i.e., cancer) and benign tumors. For example, the devices and methods described herein are contemplated for use in adrenal cortical cancer, anal cancer, bile duct cancer (e.g., periphilar cancer, distal bile duct cancer, intrahepatic bile duct cancer) bladder cancer, benign and cancerous bone cancer (e.g., osteoma, osteoid osteoma, osteoblastoma, osteochrondroma, hemangioma, chondromyxoid fibroma, osteosarcoma, chondrosarcoma, fibrosarcoma, malignant fibrous histiocytoma, giant cell tumor of the bone, chordoma, lymphoma, multiple myeloma), brain and central nervous system cancer (e.g., meningioma, astocytoma, oligodendrogliomas, ependymoma, gliomas, medulloblastoma, ganglioglioma, Schwannoma, germinoma, craniopharyngioma), breast cancer (e.g., ductal carcinoma in situ, infiltrating ductal carcinoma, infiltrating lobular carcinoma, lobular carcinoma in situ, gynecomastia), Castleman disease (e.g., giant lymph node hyperplasia, angiofollicular lymph node hyperplasia), cervical cancer, colorectal cancer, endometrial cancer (e.g., endometrial adenocarcinoma, adenocanthoma, papillary serous adenocarcinoma, clear cell) esophagus cancer, gallbladder cancer (mucinous adenocarcinoma, small cell carcinoma), gastrointestinal carcinoid tumors (e.g., choriocarcinoma, chorioadenoma destruens), Hodgkin's disease, non-Hodgkin's lymphoma, Cutaneous T-Cell Lymphoma (CTCL), Kaposi's sarcoma, kidney cancer (e.g., renal cell cancer), laryngeal and hypopharyngeal cancer, liver cancer (e.g., hemangioma, hepatic adenoma, focal nodular hyperplasia, hepatocellular carcinoma), lung cancer (e.g., small cell lung cancer, non-small cell lung cancer), mesothelioma, plasmacytoma, nasal cavity and paranasal sinus cancer (e.g., esthesioneuroblastoma, midline granuloma), nasopharyngeal cancer, neuroblastoma, oral cavity and oropharyngeal cancer, ovarian cancer, pancreatic cancer, penile cancer, pituitary cancer, prostate cancer, retinoblastoma, rhabdomyosarcoma (e.g., embryonal rhabdomyosarcoma, alveolar rhabdomyosarcoma, pleomorphic rhabdomyosarcoma), salivary gland cancer, skin cancer, both melanoma and non-melanoma skin cancer (including Merkel Cell Carcinoma), stomach cancer, testicular cancer (e.g., seminoma, nonseminoma germ cell cancer), thymus cancer, thyroid cancer (e.g., follicular carcinoma, anaplastic carcinoma, poorly differentiated carcinoma, medullary thyroid carcinoma, thyroid lymphoma), vaginal cancer, vulvar cancer, and uterine cancer (e.g., uterine leiomyosarcoma).
IV. Combination Therapies
[0077] It is contemplated that intratumoral electroporation of DNA encoding immune-modulatory proteins can be administered with other therapeutic entities. Table 6 provides possible combinations. Administration of the combination therapies can be achieved by electroporation alone or a combination of electroporation and systemic delivery.
TABLE-US-00007 TABLE 6 Combination Therapies Combination Proposed delivery method Reference IT-pOMI-2A/EP + Anti- Intratumoral i.e. Quetglas et al. Can, PD1 antagonist Ab Electroporation (`IT-EP") Immol, Res. 2015, 3: 449 of plasmids encoding cytokines, co-stimulators, immune-directors in pOMI-2A plus systemic anti-PD-1 Ab treatment 1. co-administration 2. Administration of IT-EP, followed by systemic anti-PD-1 inhibitor IT-pOMI-2A/EP + anti- IT-EP of pOMI-2A/EP PDL1 antagonist Ab plus systemic anti-PDL-1 Ab treatment 1. co-administration 2. sequential administration of IT-EP, followed by systemic anti-PDL-1 inhibitor IT-pOMI-2A/EP + CTLA4 IT-EP of pOMI-2A/EP Vom Berg et al., 2013, J. agonist antibody ("Ab") plus systemic delivery of Exp. Med. 210: 2803 or ligand CTLA4 antagonist Abs 1. co-administration 2. sequential administration of IT-EP, followed by systemic anti- CTLA4 antagonist Ab IT-pOMI-2A/EP + tumor 1. EP of IT-pOMI-2A + Vergati et al., 2010. J. vaccine cytotoxic agent Biomed. Biotechnol. (separately) to 2010: Article ID 596432 create local tumor antigen pool 2. EP of IT-pOMI-2A + system delivery of tumor vaccine (i.e gp100 peptide vaccine for melanoma) IT-pOMI-2A/EP + 1. intratumoral EP of i.e. Zhang et al., 2015, J. Bleomycin, Gemzar, drug + IT-pOMI- Immunother. 38: 137 Cytozan, 5-fluoro-uracil, 2A Adriamycin or other 2. EP of IT-pOMI-2A + chemotherapeutic agent system delivery of drug IT-pOMI-2A/EP + small 1. EP of IT-pOMI-2A Hu-Lieskovan et al., (2014) molecule inhibitors (i.e. combined with J. Clin. Oncol. 32(21 ): 2248-54 Sunitiinib, Imatinib, local drug delivery Vanneman and Dranoff Vemurafenib, 2. EP of IT-pOMI-2A (2014) Nat. Rev. Cancer Trastuzumab, combined with 12(4): 237-251 Bevacizumab , systemic drug Cetuximb, rapamycin, treatment Bortezomib, PI3K-AKT inhibitors, IAP inhibitors IT-pOMI-2A/EP + Sublethal radiation dose Almo S C, Guha C. (2014) targeted radiation locally at tumor site, Radiation Res. 182(2): 230- followed by IT-pOMI- 238. 2A/EP
[0078] The broad scope of this invention is best understood with reference to the following examples, which are not intended to limit the inventions to the specific embodiments.
Examples
I. General Methods
[0079] Standard methods in molecular biology are described. Maniatis et al. (1982) Molecular Cloning, A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.; Sambrook and Russell (2001) Molecular Cloning, 3rd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.; Wu (1993) Recombinant DNA, Vol. 217, Academic Press, San Diego, Calif. Standard methods also appear in Ausbel et al. (2001) Current Protocols in Molecular Biology, Vols. 1-4, John Wiley and Sons, Inc. New York, N.Y., which describes cloning in bacterial cells and DNA mutagenesis (Vol. 1), cloning in mammalian cells and yeast (Vol. 2), glycoconjugates and protein expression (Vol. 3), and bioinformatics (Vol. 4).
[0080] Methods for protein purification including immunoprecipitation, chromatography, electrophoresis, centrifugation, and crystallization are described. Coligan et al. (2000) Current Protocols in Protein Science, Vol. 1, John Wiley and Sons, Inc., New York. Chemical analysis, chemical modification, post-translational modification, production of fusion proteins, glycosylation of proteins are described. See, e.g., Coligan et al. (2000) Current Protocols in Protein Science, Vol. 2, John Wiley and Sons, Inc., New York; Ausubel et al. (2001) Current Protocols in Molecular Biology, Vol. 3, John Wiley and Sons, Inc., NY, NY, pp. 16.0.5-16.22.17; Sigma-Aldrich, Co. (2001) Products for Life Science Research, St. Louis, Mo.; pp. 45-89; Amersham Pharmacia Biotech (2001) BioDirectory, Piscataway, N.J., pp. 384-391. Production, purification, and fragmentation of polyclonal and monoclonal antibodies are described. Coligan et al. (2001) Current Protocols in Immunology, Vol. 1, John Wiley and Sons, Inc., New York; Harlow and Lane (1999) Using Antibodies, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.; Harlow and Lane, supra. Standard techniques for characterizing ligand/receptor interactions are available. See, e.g., Coligan et al. (2001) Current Protocols in Immunology, Vol. 4, John Wiley, Inc., New York.
[0081] Methods for flow cytometry, including fluorescence activated cell sorting detection systems (FACS.RTM.), are available. See, e.g., Owens et al. (1994) Flow Cytometry Principles for Clinical Laboratory Practice, John Wiley and Sons, Hoboken, N.J.; Givan (2001) Flow Cytometry, 2nd ed.; Wiley-Liss, Hoboken, N.J.; Shapiro (2003) Practical Flow Cytometry, John Wiley and Sons, Hoboken, N.J. Fluorescent reagents suitable for modifying nucleic acids, including nucleic acid primers and probes, polypeptides, and antibodies, for use, e.g., as diagnostic reagents, are available. Molecular Probes (2003) Catalogue, Molecular Probes, Inc., Eugene, Oreg.; Sigma-Aldrich (2003) Catalogue, St. Louis, Mo.
[0082] Standard methods of histology of the immune system are described. See, e.g., Muller-Harmelink (ed.) (1986) Human Thymus: Histopathology and Pathology, Springer Verlag, New York, N.Y.; Hiatt, et al. (2000) Color Atlas of Histology, Lippincott, Williams, and Wilkins, Phila, Pa.; Louis, et al. (2002) Basic Histology: Text and Atlas, McGraw-Hill, New York, N.Y.
[0083] Software packages and databases for determining, e.g., antigenic fragments, leader sequences, protein folding, functional domains, glycosylation sites, and sequence alignments, are available. See, e.g., GenBank, Vector NTI.RTM. Suite (Informax, Inc, Bethesda, Md.); GCG Wisconsin Package (Accelrys, Inc., San Diego, Calif.); DeCypher.RTM. (TimeLogic Corp., Crystal Bay, Nev.); Menne et al. (2000) Bioinformatics 16: 741-742; Menne et al. (2000) Bioinformatics Applications Note 16:741-742; Wren et al. (2002) Comput. Methods Programs Biomed. 68:177-181; von Heijne (1983) Eur. J. Biochem. 133:17-21; von Heijne (1986) Nucleic Acids Res. 14:4683-4690.
II. Subcloning of Human IL-12 p35 and p40 Subunits into pOMI2A
[0084] A pUMVC3 backbone was purchased from Aldevron (Fargo, N. Dak.). A 1071 bp DNA fragment (gene block) encoding the translation modulating element P2A linked in-frame to hIL12p40 (P2A-hIL12p40) was purchased from IDT (Coralville, Iowa). The p40 geneblock was PCR amplified using Phusion polymerase (NEB, Ipswich Mass., cat. #M0530S) and ligated into pUMVC3 downstream of the CMV promoter/enhancer using standard restriction enzyme pairing and T4 DNA ligase (Life Technologies, Grand Island N.Y., cat. #15224-017). Positives clones of P2A-hIL12p40/pOMI2A were identified via restriction enzyme digests and verified with DNA sequencing.
[0085] Human p35 was ordered as a 789 bp geneblock from IDT (Coralville Iowa) with internal BamH1, BglII and Xba1 sites removed to facilitate cloning. The p35 geneblock was PCR amplified as described above and ligated upstream of the p40 geneblock in P2A-hIL12p40/pOMI2A. Positives clones of hIL12p35-P2A-p40/pOMI2A were identified via restriction enzyme digests and verified with DNA sequencing.
[0086] Other heterodimeric cytokines, single chain cytokines, or innate immune-regulators (Tables 2A, 2B) are cloned into pOMI2A vectors similar to IL-12.
III. Subcloning of IL-15-P2A-IL-15R.alpha. into pOMI2A
[0087] A 1384 bp geneblock was ordered from IDT encoding hIL15 and hIL15R.alpha., linked together in-frame with the translation-modulating element P2A. The geneblock was PCR amplified as described above and ligated into pOMI2A. Positives clones were identified via restriction enzyme digests and verified with DNA sequencing.
[0088] A mutant form of IL-15 showing increased activity was also subcloned into the pOMI2A vector as above (see, e.g., Zhu, et al. (2009) J. Immunol. 183:3598).
IV. Subcloning of IL-15-P2A-IL-15R.alpha.-IgG1Fc into pOMI2A
[0089] A 708 bp DNA geneblock was ordered from IDT encoding the human IgG1 Fc sequence. The geneblock was PCR amplified as described above and ligated downstream of IL-15-P2A-IL-15R.alpha. in pOMI2A. The stop site between IL15Ra and Fc was then removed via a QuikChange mutagenesis reaction (Agilent Technologies, La Jolla Calif., cat. #200521). Finally, the complete IL15-P2A-IL15R.alpha.-IgG1Fc sequence was PCR amplified and ligated back into pOMI2A.
V. Subcloning of INF.gamma. into pOMI2A
[0090] A 501 bp DNA geneblock was ordered from IDT encoding the full-length human INF gamma coding sequence. The geneblock was PCR amplified as described above and ligated into pUMVC3 (Aldevron). Positives clones were identified via restriction enzyme digests and verified with DNA sequencing. Finally, the IFN.gamma. insert was PCR amplified and ligated into various pOMI2A vectors.
VI. Generation of FLT3L-Antigen Fusion Protein Constructs
[0091] The FMS-like tyrosine kinase 3 ligand (Flt3L) has been shown to direct antigen to antigen presenting cells (APC) for preferential presentation to T cells (Kim et al. Nat Comm. 2014, Kreiter et al., Cancer Res. 2011, 71:6132). A soluble, secreted form of Flt3L is fused to a variety of protein or peptide antigens (Table 4; Kim et al. Nat Comm. 2014). An example protocol is given for generating a FLT3L-NY-ESO-1 fusion protein construct.
[0092] Three gene blocks were obtained from IDT that each contained the IgK signal peptide sequence followed by the ECD of Flt3L, a short hinge region, and three different segments of the NY-ESO-1 antigen. PCR was used to add flanking restriction sites and introduce these three fusion protein constructs into pUMVC3 (Sequence ID Nos. 17-22). Flt3L was also fused to a concatamer of 3 peptides containing the SIINFEKL peptide antigen from the ovalbumin gene (Seq ID 24) for pre-clinical studies in mice. From pUMVC3, these fusion constructs are introduced into pOMI-2x2A (described below).
[0093] An alternative fusion protein using viral antigens (Table 4) is constructed using the same method.
[0094] An alternative fusion protein with full length calreticulin (Table 3) is constructed using the same method.
[0095] In addition to identified shared tumor antigens, patient-specific neoantigens could be identified and immunogenic peptide antigens tailored to that patient can be fused to Flt3L for personalized therapy via intratumoral electroporation, (see, e.g., Beckhove et al., J. Clin. Invest. 2010, 120:2230).
[0096] Versions of all immune-modulatory proteins are constructed in parallel using mouse homolog sequences and are used in pre-clinical studies.
VII. Generation of OMI-2x2A for Expression of Three Proteins from a Single Transcript.
[0097] A schematic diagram of the vector is shown in FIG. 3. All three genes are expressed from the same promoter, with intervening exon skipping motifs to allow all three proteins to be expressed from a single polycistronic message.
[0098] An example subcloning protocol is given for IL-12 heterodimeric cytokine, and Flt3L-NY-ESO-1. A DNA geneblock (IDT) encoding FLT3L-NYESO-1 was PCR-amplified with an upstream P2A site and flanking restriction sites and ligated downstream of hIL-12p40. Quikchange mutagenesis (Agilent, Santa Clara, USA) was performed to delete the stop site 3' of p40. Positives clones were identified via restriction enzyme digests and verified with DNA sequencing.
[0099] A forth gene can be added either upstream or downstream of the three genes already in the polycistronic message using the same methods.
VIII. ELISA
[0100] Clones of OMI2A-IL-12 and OMI2A-IL-15/IL-15R, and OMI2x2A-IL12-Flt3L-NY-ESO-1 were transfected into HEK293 cells using TransIT LT-1 (Mirus, Madison Wis., cat. #MIR 2300) according to the manufacturer's recommendations. Two days later, supernatants were collected and spun for 5 minutes at 3000 rpm to remove any cell debris. Cleared supernatants were transferred to new tubes, aliquoted and frozen at -86.degree. C. The levels of hIL-12p70 and hIL15-IL15R.alpha. heterodimeric proteins in the conditioned media were quantitated using an ELISA that specifically detects the complexes (R&D Systems, Minneapolis Minn. cat. #DY1270, DY6924). The level of FLT3L-NYESO-1 fusion protein were quantified by ELISA with anti-Flt3L antibodies (R&D Systems, Minneapolis Minn. cat. #DY308).
[0101] Comparison of hIL-12p70 expression and secretion from cells transfected with pOMI2A-hIL-12 and pOMIIRES-hIL-12 revealed that pOMI2A-hIL-12 generated higher expression levels of the mature heterodimeric p70 protein secreted by transfected cells as measured by ELISA (FIG. 4A). Expression and secretion from cells transfected with pOMI2A-hIL-15/IL-15R.alpha. and pOMI2A-hIL-15/IL-15R.alpha.Fc domain were measured by ELISA and are shown in FIG. 5A.
TABLE-US-00008 TABLE 7 Expression and secretion of IL-12 p70 and Flt3L-NY-ESO-1 fusion protein from cells transfected with OMI2x2A-IL- 12-Flt3L-NY-ESO-1 were measured by ELISA and are shown. Secreted protein ng/ml; Mean .+-. SEM IL-12 p70 1364 .+-. 5.5 Flt3L-NY-ESO-1 fusion protein 25.1 .+-. 3.1
IX. Protein Detection by Western Blots
[0102] For Western Blotting, Laemmli SDS sample buffer NuPAGE 4.times.LDS, ThermoFisher Scientific) was added to each sample and boiled at 100.degree. C. for 10 minutes and samples were centrifuged. 23 .mu.l of protein+sample buffer was loaded per well and gel was run at 150 volts for about an hour until the smallest standard reached the bottom of the gel. Gel proteins were transferred to PVDF membranes at 100 volts for 1 hour, rinsed with 1.times.TBST, and then blocked for 1 hour at room temperature on a rocker with 5% BSA in TBST. Rinsed membranes were incubated overnight with rabbit anti-2A peptide antibody (EMD Millipore ABS031) or anti-HA antibody (Cell Signaling, cat #3724) diluted in TBST+5% nonfat dry milk. Blots were incubated for 1 hour at room temperature with donkey anti-rabbit secondary antibody conjugated to horseradish peroxidase (BioRad, Hercules, Calif.). Blots were developed with enhanced chemiluminescence reagents (SuperSignal West Pico, ThermoFisher Scientific) and captured on a digital imaging system (Protein Simple, San Jose, Calif.). Western Blots on HEK 293 conditioned supernatants probed for Flt3L-OVA and Flt3L-NY-ESO-1 revealed that these fusion proteins were stable, secreted and had the predicted molecular weight.
X. In Vitro Functional Assays
[0103] Frozen human PBMCs were purchased from ATCC (Manassas Va., cat. #PCS-800-011) thawed and pre-stimulated for 5 days in RPMI 1640 supplemented with 10% FBS, 1% P/S, 50 ng/mL recombinant human IL-2 and 10 .mu.g/mL PHA-L. Cells (2.times.10.sup.4) were then seeded into triplicate wells of opaque white 96-well plates and cultured for 72 hours in growth media (RPMI 1640 containing 10% FBS and 1% P/S) with increasing amounts of IL-12p35/p40 heterodimer-containing HEK293 cell culture supernatant, protein concentration was determined via ELISA as described above. Supernatants from un-transfected cells were used as negative controls. CellTiter-Glo (Promega, Madison Wis., cat. #G7570) was diluted to 1.times. as described by the manufacturer and 100 .mu.L was pipetted into each well. The plates were gently shaken for 10 min at room temperature, then the luminescence was read on a SpectraMax plate reader (Molecular Devices, Sunnyvale, Calif.) with a is integration time.
[0104] Culture supernatants from transfected HEK293 cells expressing and secreting IL-12 expression plasmids were added to the cells and proliferative responses were measured. The half-maximal response for PBMC proliferation was achieved with a 3-fold higher dilution factor for OMIP2A-hIL-12 as compared to OMIIRES-hIL-12 (69244 vs. 19548). When relative p70 protein concentrations were normalized, IL-12p70 expressed from the two vectors had comparable ability to stimulate cell proliferation in human PBMCs (FIG. 4B).
[0105] This result indicated that pOMI2A-IL-12 can generate 3 times more IL-12 mediated T cell proliferation from a given dose of plasmid.
[0106] Human CD8+ T cells were purchased fresh from AllCells (Alameda Calif., cat. #PB009-3), resuspended in RPMI 1640 containing 10% FBS and 1% P/S, and then seeded in triplicate wells of a black 96-well plate (2.times.10.sup.4 cells per well). Increasing amounts of IL15/IL15Ra-containing HEK293 cell culture supernatant (determined via ELISA as described above) were added and the cells were cultured for 3 days at 37.degree. C., 5% CO2. CellTiter-Blue (Promega, Madison Wis., cat. #G8080) was then added to the wells followed by a 4 hr incubation at 37.degree. C., The resulting fluorescence signal (Ex 560/Em 590 nm) was read on a Cytation 3 plate-reader (Biotek, Winooski Vt.).
[0107] Protein expressed from cells transfected with pOMI2A-IL-15/IL15Ra and pOMI2A-IL-15/IL15Ra-Fc both stimulated cell proliferation in human primary CD8+ T cells (FIG. 5B).
[0108] Tissue culture supernatants from cells expressing pOMIP2A-IL12-Flt3L-NY-ESO-1 were tested for the expression of functional IL-12 p70 using HEK-Blue cells. These cells are engineered to express human IL-12 receptors, and a STAT4-driven secreted form of alkaline phosphatase.
[0109] This reporter assay was performed according to the manufacturer protocol (HEK-Blue IL-12 cells, InvivoGen catalog #hkb-il12). Expression of secreted alkaline phosphatase (SEAP) was measured according to the manufacturer's protocol (Quanti-Blue, InvivoGen catalog #rep-qbl).
[0110] IL-12 p70 protein expressed and secreted from the OMIP2A polycistronic vector demonstrated strong activity in the induction of SEAP protein (FIG. 6). This activity was comparable to rhIL-12 protein controls, and was blocked by a neutralizing IL-12 antibody (R&D systems; AB-219-NA) (FIG. 6).
[0111] Human Flt3L and Flt3L-NY-ESO-1 fusion protein expressed from pOMIP2A vectors and secreted into the culture medium of HEK 293 cells were tested for binding to FLT3 receptors expressed on the surface THP-1 monocytic cells.
[0112] HEK cells were transfected with pOMIP2A-hFlt3L or pOMIP2A-hFlt3L-NYESO-1 (80-180aa) using Minis TransIT LT-1. Supernatants were collected after 72 hours. The amount of secreted FLT3L proteins was quantified using hFlt3L ELISA (R&D Systems cat. #DY308).
[0113] The THP-1 monocyte cell line was cultured in RPMI+10% FBS+1% P/S (ATCC, cat. #TIB-202). For each experiment, 750,000 THP-1 cells were washed in Fc buffer (PBS+5% filtered FBS+0.1% NaN3), preincubated with human Fc block (TruStain FcX, Biolegend 422301) for 10 minutes and then incubated with 150 ng of recombinant hFlt3L-Fc (R&D Systems, cat. #AAA17999.1) or HEK 293 conditioned media containing 150 ng hFlt3L or hFlt3L-NYESO-1 protein and incubated for 1 hour at 4.degree. C. Cells were then washed in Fc buffer and incubated with biotinylated anti-hFlt3L antibodies (R&D Systems, cat. #BAF308) for 1 hour. Cells were then washed in Fc buffer and incubated with streptactin-AlexaFluor-647 2.degree. Ab for 1 hr (ThermoFisher, #S32357). Cell were washed again and analyzed by flow cytometry using a Guava 12HT cytometer (Millipore) on the Red-R channel. HEK 293 cells which do not express Flt3 receptors were also tested as a negative control.
TABLE-US-00009 TABLE 8 Secreted recombinant Flt3 ligand proteins bind to Flt3 receptors of the surface of THP-1 monocytes Mean fluorescence intensity Cell line unstained Control super hFlt3L h-Flt3L-NYESO1 THP-1 9.0 9.7 32.2 52.2 HEK293 9.0 7.5 8.4 8.8
[0114] Over 90% of THP-1 cells showed an increase in mean fluorescence intensity with both hFlt3L and hFLT3L-NY-ESO-1 fusion proteins expressed from pOMIP2A vectors indicating that these recombinant proteins bind efficiently to Flt3 receptors on the cell surface.
[0115] In order to further test the functionality of the recombinant Flt3L proteins, HEK 293 conditioned media were used to test for induction of dendritic cell maturation in mouse splenocytes.
[0116] Spleens were excised from a B16-F10 tumor bearing C58/BL6 mice. Under sterile conditions, spleens were placed in DMEM media into the 70 micron cell strainer (Miltenyi) and mechanically dissociated using the rubber tip of the plunger from a 3 ml syringe. Once the spleen is completely dissociated, 10 mls of HBSS with 10% FBS (PFB) wad used to wash the strainer. Flow-though was spun in a centrifuge at 300.times.g for 10 mins. to pellet cells. Cells were washed once with PFB. Red blood cells were lysed with ACK lysis buffer according to the manufacturer's instructions (Thermo Fisher A1049201). Cells were filtered through a 40-micron cell strainer into a 15 ml conical tube and spun in a centrifuge at 300.times.g. Single cell suspension from the spleens were resuspended in complete RPMI-10 media. 1.5 million splenocytes were plated in a 12 well plate and allowed to adhere to the plate approximately 3 hrs. Non-adherent cells were removed and 2 mls of complete RPMI-10 media containing murine GMCSF (100 ng/ml) and murine IL4 (50 ng/ml) were added. The media was changed every 2 days for a week. The adherent dendritic cells were treated in triplicate wells with 1 ml of HEK 293 conditioned supernatants (containing 100 ng/ml Flt3L-NY-ESO-1 fusion protein) for 7 days. 100 ng Recombinant Human Flt-3 Ligand Protein was compared as a positive control (R&D systems, AAA17999.1). Cells were gently scraped from a plate and the number of CD11c+ cells was determined by flow cytometric analysis.
[0117] When the number of CD3(-)CD11c(+) dendritic cells was tabulated, conditioned media from cells transfected with pOMIP2A-Flt3L-NYESO1 plasmid generated a significant increase in the number of these cells as compared to splenocytes incubated with conditioned media from un-transfected cells.
[0118] This result indicated that the FLT3L-NY-ESO-1 fusion protein can function to stimulate Flt3 receptor mediated dendritic cell maturation ex-vivo in mouse splenocytes.
XI. Tumors and Mice
[0119] Female C57BL/6J or Balb/c mice, 6-8 weeks of age were obtained from Jackson Laboratories and housed in accordance with AALAM guidelines.
[0120] B16.F10 cells were cultured with McCoy's 5A medium (2 mM L-Glutamine) supplemented with 10% FBS and 50 .mu.g/ml gentamicin. Cells were harvested by trypsinizing with 0.25% trypsin and resuspended in Hank's balanced salt solution (HBSS). Anesthetized mice were subcutaneously injected with 1 million cells in a total volume of 0.1 ml into the right flank of each mouse. 0.5 million cells in a total volume of 0.1 ml were injected subcutaneously into the left flank of each mouse Tumor growth was monitored by digital caliper measurements starting day 8 until average tumor volume reaches .about.100 mm.sup.3. Once tumors are staged to the desired volume, mice with very large or small tumors were culled. Remaining mice were divided into groups of 10 mice each, randomized by tumor volume implanted on right flank.
[0121] Additional tumor cell types were tested including B16OVA in C57Bl/6J mice as well as CT26 and 4T1 in Balb/c mice.
[0122] This protocol was used as a standard model to test simultaneously for the effect on the treated tumor (primary) and untreated (contralateral). Lung metastases were also quantified in Balb/c mice bearing 4T1 tumors.
XII. Intratumoral Treatment
[0123] Mice were anesthetized with isoflurane for treatment. Circular plasmid DNA was diluted to 1 .mu.g/.mu.L in sterile 0.9% saline. 50 .mu.L of plasmid DNA was injected centrally into primary tumors using a 1 ml syringe with a 26 Ga needle. Electroporation was performed immediately after injection. Electroporation of DNA was achieved using a Medpulser with clinical electroporation parameters of 1500 V/cm, 100 .mu.s pulses, 0.5 cm, 6 needle electrode. Alternative parameters used were 400 V/cm, 10-msec pulses, using either a BTX generator or a generator incorporating impedance spectroscopy, as described above. Tumor volumes were measured twice weekly. Mice were euthanized when the total tumor burden of the primary and contralateral reached 2000 mm.sup.3.
XIII. Intratumoral Expression
[0124] One, 2 or 7 days after IT-EP (350 v/cm, 8 10-msec pulses), tumor tissue was isolated from sacrificed mice at various time point to determine expression of the transgenes. Tumor were dissected from mice and transferred to a cryotube in liquid nitrogen. The frozen tumor was transferred to a 4 ml tube containing 300 .mu.l of tumor lysis buffer (50 mM TRIS pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, Protease inhibitor cocktail) and placed on ice and homogenized for 30 seconds (LabGen 710 homogenizer). Lysates were transferred to 1.5 ml centrifuge tube and spun at 10,000.times.g for 10 minutes at 4.degree. C. Supernatants were to a new tube. Spin and transfer procedure was repeated three times. Tumor extracts were analyzed immediately according to manufacturer's instruction (Mouse Cytokine/Chemokine Magnetic Bead Panel MCYTOMAG-70K, Millepore) or frozen at -80.degree. C. Recombinant Flt3L-OVA were done by standard ELISA protocols (R&D systems) using anti-Flt3L antibody for capture (R&D Systems, Minneapolis Minn. cat. #DY308) and the following antibodies for detection (ThermoFisher, cat. #PA1-196).
TABLE-US-00010 TABLE 9 Intratumoral expression of cytokines after electroporation of a polycistronic plasmid encoding hIL-12, hIL-15/hIL-15R.alpha. and hIFN.gamma. Untreated EP/pOMI-hIL12/hIL15/hINF.gamma. [Protein] pg/mg [Protein] pg/mg Recombinant protein Mean .+-. SEM n = 2 Mean .+-. SEM n = 3 detected Day 1 Day 2 Day 7 Day 1 Day 2 Day 7 IL-12 p70 0 0 0 3000.5 .+-. 1872.7 2874.7 .+-. 1459.1 19.1 .+-. 4.2 IL-15/IL-15R.alpha. 0 0 0 1.19 .+-. 0.22 0.41 .+-. 0.29 0.09 .+-. 0.05 INF.gamma. 0 0 0 36.6 .+-. 6.4 45.0 .+-. 12.8 1.0 .+-. 0.4
[0125] To test for expression and function of our Flt3L-tracking antigen-fusion protein, we constructed a fusion of mouse Flt3L (extracellular domain) and peptides from the ovalbumin gene (Seq ID 25) in OMI2A vectors and electroporated intratumorally as above.
TABLE-US-00011 TABLE 10 Intratumoral expression of Flt3L-OVA fusion protein (genetic adjuvant with shared tumor antigen) 2 days after electroporation as analyzed by ELISA (n = 8). EP/recombinant Recombinant protein EP/pUMVC3 control protein construct construct Mean .+-. SEM pg/ml Mean .+-. SEM pg/ml Flt3L-OVA fusion 30.6 .+-. 1.4 441 .+-. 102
[0126] After intratumoral electroporation of pOMI2A vectors containing mouse homologs of the various immunomodulatory proteins, significant levels of IL-12p70, IL-15/IL-15R.alpha., INF.gamma. (Table 9), and Flt3L-OVA recombinant proteins (Table x10) were all detectable in tumor homogenates by ELISA.
XIV(a). Tumor Regression
TABLE-US-00012 [0127] TABLE 11 B16F0 tumor regression for treated and untreated tumors after intratumoral electroporation (IT-EP) of OMI vectors encoding mouse IL-12. Electroporation with the parameters of 1500 V/cm, 100 .mu.s, 0.5 cm, 6 needle electrode was performed 8, 12, and 15 days after implantation. Tumor volume measurements shown were taken 16 days after implantation. Tumor volume (mm.sup.3), Mean .+-. SEM, n = 10 Treated Untreated Treatment tumor tumor Untreated 1005.2 .+-. 107.4 626.6 .+-. 71.8 EP/pUMVC3 control 345.2 .+-. 130.5 951.1 .+-. 77.0 EP/pOMIIRES-mIL-12 140.3 .+-. 49.8 441.0 .+-. 80.8 EP/pOMI2A-mIL-12 92.1 .+-. 38.7 283.3 .+-. 87.2
[0128] Comparison of tumor regression after electroporation of pOMI2A-IL-12 vs. pOMIIRES-IL-12 demonstrated that using P2A exon skipping motif for expression of p35 and p40 subunits not only gave higher p70 IL-12 expression (FIG. 4A), but also better efficacy for tumor regression in vivo.
TABLE-US-00013 TABLE 12 B16F10 tumor regression for treated and untreated tumors after IT-EP with different doses of OMIP2A-IL-12. Electroporation with the parameters of 350 V/cm, 8 10-msec pulses using acupuncture needles was performed once, 8 days after implantation. Tumor volume (mm.sup.3), Mean .+-. SEM, n = 10 Plasmid dose introduced Treated Untreated by IT-EP tumor tumor pUMVC3 control 50 ug 556.4 .+-. 59.0 211.3 .+-. 46.5 pOMI2A-mIL-12 1 ug 546.1 .+-. 92.5 158.4 .+-. 47.1 pOMI2A-mIL-12 10 ug 398.6 .+-. 78.4 79.7 .+-. 18.7 pOMI2A-mIL-12 50 ug 373.6 .+-. 46.3 74.3 .+-. 12.1
[0129] The extent of regression of both treated and untreated tumors increased with electroporation of increasing dose of OMIP2A-mIL-12 plasmid.
[0130] The ability of IT-EP of pOMIP2A-mIL12 to affect 4T1 primary tumor growth and lung metastases in Balb/c mice was also tested. One million 4T1 cells were injected subcutaneously on the right flank of the mice and 0.25 million 4T1 cells were injected into the left flank. Larger tumors on the right flank were subject to IT-EP with empty vector (pUMVC3, Aldevron) or with pOMIP2A-mIL12. Tumor volumes were measured every two days and on Day 19, mice were sacrificed, and the lungs were excised and weighed.
TABLE-US-00014 TABLE 13 Primary tumor growth and post-mortem weight of lungs of mice electroporated with 350 V/cm, 8 10-msec pulses with acupuncture needles on day 8, and day 15 post-implantation. Primary tumor volumes were measured on Day 17, and lung weights on Day 18. Primary tumor Lung weight volume (mm.sup.3) (grams) Mean .+-. SEM, Mean .+-. SEM, Treatment n = 5 n = 5 Untreated 897 .+-. 131 0.252 .+-. 0.019 EP/pUMVC3 593 .+-. 27 0.228 .+-. 0.006 EP/pOMIP2A-mIL12 356 .+-. 80 0.184 .+-. 0.004
[0131] It has been previously reported that systemic IL-12 treatment can reduce lung metastases in mice with 4T1 tumors (Shi et al., J Immunol. 2004, 172:4111). Our finding indicate that local IT-EP treatment of the tumors also reduced metastasis of these tumor cells to the lung in this model.
[0132] In addition to Bl6F10 tumors, electroporation of pOMIP2A-mIL12 also resulting in regression of both primary (treated) and contralateral (untreated) B16OVA and CT26 tumors. In the 4T1 tumor model, the primary tumor regressed after EP/pOMI-mIL12, and the mice demonstrated a significant reduction in lung weight, indicating a reduction in lung metastases. We show that IT-EP of OMIP2A-mIL12 can reduce tumor burden in 4 different tumor models in two different strains of mice.
TABLE-US-00015 TABLE 14 B16F10 tumor regression for treated and untreated tumors after intratumoral electroporation of pOMI2A plasmids containing genes encoding mIL-12 and Flt3L-OVA using 350 V/cm, and 8 10-msec pulses on day 7 and 14 after tumor cell inoculation; tumors measurements shown from Day 16. Tumor volume (mm.sup.3), Mean .+-. SEM, n = 10 Treated Untreated Treatment tumor tumor EP/pUMVC3 control 600.7 .+-. 113.3 383.4 .+-. 75.9 EP/OMI2A_IL12/ 94.2 .+-. 31.7 115.7 .+-. 42.3 OMI2A_Flt3LOVA
TABLE-US-00016 TABLE 15 B16F10 tumor regression for treated tumors after IT-EP of pOMIP2A-mIL12-Flt3L-NYESO1 using 350 V/cm, and 8 10-msec pulses on day 7 after tumor cell inoculation; tumors measurements shown from Day 14. Tumor volume (mm.sup.3), Treatment Mean .+-. SEM, n= Untreated 230.0 .+-. 67.5 EP/pUMVC3 empty vector 170.8 .+-. 20.8 EP/pOMIP2A-mIL12-Flt3L-NYESO1 4.0 .+-. 0.0
[0133] Electroporation of plasmid expressing both mouse IL-12 p70 and human Flt3L-NY-ESO-1 fusion protein caused complete regression of the treated tumor in 7 days.
[0134] The volume of both primary and contralateral tumors is significantly reduced in mice where immunomodulatory genes were introduced by electroporation as compared with electroporation of empty vector control, indicating not only a local effect within the treated tumor microenvironment, but an increase in systemic immunity as well.
XIV(b). Flow Cytometry
[0135] At various time points after IT-pIL12-EP treatment, mice were sacrificed and tumor and spleen tissue were surgically removed.
[0136] Splenocytes were isolated by pressing spleens through a 70 micron filter, followed by red blood cell lysis (RBC lysis buffer, VWR, 420301OBL), and lympholyte (Cedarlane CL5035) fractionation. Lymphocytes were stained with SIINFEKL-tetramers (MBL International T03002), followed by staining with antibody cocktails containing: anti-CD3 (Biolegend 100225), anti-CD4 (Biolegend 100451), anti-CD8a (Biolegend 100742), anti-CD19 (Biolegend 115546), and vital stain (live-dead Aqua; Thermo-Fisher L-34966). Cells were fixed and analyzed on an LSR II flow cytometer (Beckman).
[0137] Tumors were dissociated using Gentle-MACS for tumors (Miltenyi tumor dissociation kit 130-096-730, C-tubes, 130-093-237) and homogenized using an Miltenyi gentleMACS.TM. Octo Dissociator with Heaters (130-096-427). Cells were pelleted at 800.times.g for 5 min at 4'C and re-suspended in 5 mL of PBS+2% FBS+1 mM EDTA (PFB) and overlaid onto 5 mL of Lympholyte-M (Cedarlane). Lympholyte columns were spun in centrifuge at 1500.times.g for 20 min at room temperature with no brake. Lymphocyte layer was washed with PBF. Cell pellets were gently re-suspended in 500 .mu.L of PFB with Fc block (BD Biosciences 553142). In 96-well plate, cells were mixed with a solution of SIINFEKL tetramer (MBL), representing the immunodominant antigen in B16OVA tumors, according to the manufacturers instruction and incubated for 10 minutes at room temperature. Antibody staining cocktails containing the following: Anti-CD45-AF488 (Biolegend 100723), anti-CD3-BV785 (Biolegend 100232), Anti-CD4-PE (eBioscience 12-0041), anti-CD8a-APC (eBioscience 17-0081), anti-CD44-APC-Cy7 (Biolegend 103028), anti-CD19-BV711 (Biolegend 11555), anti-CD127 (135010), anti-KLRG1 (138419), were added and incubated at room temperature for 30 minutes. Cells were washed 3 times with PFB. Cells were fixed in PFB with 1% paraformaldehyde for 1 minutes on ice. Cells were washed twice with PFB and stored at 4.degree. C. in the dark. Samples were analyzed on an LSR II flow cytometer (Beckman).
TABLE-US-00017 TABLE 16 IT-pIL12-EP increased SIINFEKL-tetramer-binding CD8+ T cells in the spleens of treated, B16OVA tumor-bearing mice. Mice were electroporated intratumorally (IT-EP) once on Day 0 using 350 V/cm, 10-msec pulses, 300 ms pulse frequency, with 0.5 cm acupuncture needles. Percent of CD3.sup.+CD8.sup.+CD44.sup.+ T cells that are Treatment SIINFEKL-tetramer positive on Day 13, n = 6 IT-pIL12-EP 2.36 .+-. 0.75 IT-pUMVC3-EP 0.24 .+-. 0.04 untreated 0.10 .+-. 0.04
[0138] IT-pIL12-EP induces an increase in circulating CD8.sup.+ T cells directed against the SIINFEKL peptide from ovalbumin, the dominant antigen in B16OVA tumors. These data indicate that local IL-12 therapy can lead to system tumor immunity in mice.
TABLE-US-00018 TABLE 17 Intratumoral electroporation of pIL12 alters the immune environment in B16OVA contralateral tumors. Mice were electroporated intratumorally (IT-EP) once on Day 0 using 350 V/cm, 10-msec pulses, 300 ms pulse frequency, with 0.5 cm acupuncture needles. The composition of infiltrating lymphocytes (TIL) in untreated tumors measured 18 days after treatment is shown. Composition of TIL in untreated tumors Mean .+-. SEM, n = 6 % CD3.sup.+CD8.sup.+ % SLEC CD8.sup.+/T.sub.reg Treatment T cells T cells T cell ratio IT-pIL12-EP 14.8 .+-. 2.7 1.0 .+-. 0.1 1892 .+-. 602 IT-pUMVC3-EP 3.6 .+-. 1.1 0.2 .+-. 0.07 659 .+-. 129 Untreated 2.9 .+-. 0.9 0.09 .+-. 0.03 753 .+-. 288
[0139] Electroporation of OMI2A-pIL-12 into the primary tumor can significantly alter the composition of TILs within the contralateral, untreated tumor. These results show that intratumoral treatment with pOMI2A-IL-12 can affect the immune environment in untreated tumors indicating that local treatment leads to a systemic anti-tumor immune response. This conclusion is corroborated by increased detection of tumor antigen-specific CD8.sup.+ T cells in the spleen (Table 16), contralateral tumor regression (Table 12), and reduction in lung metastases (Table 13).
XV(a). Analysis of Mouse Gene Expression
[0140] NanoString was used for analysis of changes in gene expression in treated and untreated tumors induced by IT-EP of pOMIP2A-IL12 and pOMI-Flt3L-NYESO1 plasmids. Tumor tissue was carefully harvested from mice using scalpel and flash frozen in liquid nitrogen. Tissues were weighed using a balance (Mettler Toledo, Model ML54). 1 ml of Trizol (Thermo Fisher Scientific, Waltham, Mass.) was added to the tissue and homogenized using a probe homogenizer on ice. RNA was extracted from Trizol using manufacturer's instructions. Contaminating DNA was removed by Dnase (Thermo Fisher, Cat no: EN0525) treatment. Total RNA concentrations were determined using the NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific). Gene expression profiling was performed using NanoString technology. In brief, 50 ng of Total RNA was hybridized at 96.degree. C. overnight with the nCounter.RTM. (Mouse immune `v1` Expression Panel NanoString.RTM. Technologies). This panel profiles 561 immunology-related mouse gene as well as two types of built-in controls: positive controls (spiked RNA at various concentrations to evaluate the overall assay performance) and 15 negative controls (to normalize for differences in total RNA input). Hybridized samples were then digitally analyzed for frequency of each RNA species using the nCounter SPRINT.TM. profiler. Raw mRNA abundance frequencies were analyzed using the nSolver.TM. analysis software 2.5 pack. In this process, normalization factors derived from the geometric mean of housekeeping genes, mean of negative controls and geometric mean of positive controls were used.
TABLE-US-00019 TABLE 18 IT-EP of pOM1P2A-IL12 caused an increase in intratumoral levels of lymphocyte and monocyte cell surface markers in both primary and contralateral tumors. Fold change of treated vs. untreated mice values are shown Immune IT-pIL12-EP IT-pUMVC3-EP Untreated Checkpoint Mean .+-. SEM n = 5 Mean .+-. SEM n = 4 Mean .+-. SEM n = 3 Protein RNA Primary Contralateral Primary Contralateral Primary Contralateral CD45 11.54 .+-. 1.65 3.55 .+-. 0.40 1.70 .+-. 0.72 1.26 .+-. 0.51 1.00 .+-. 0.38 1.00 .+-. 0.50 CD3 13.16 .+-. 2.95 5.30 .+-. 0.72 1.26 .+-. 0.38 1.09 .+-. 0.32 1.00 .+-. 0.22 1.00 .+-. 0.40 CD4 2.35 .+-. 0.39 2.74 .+-. 0.44 0.73 .+-. 0.18 1.00 .+-. 0.22 1.00 .+-. 0.20 1.00 .+-. 0.09 CD8 16.28 .+-. 3.10 4.60 .+-. 0.50 1.23 .+-. 0.32 1.00 .+-. 0.15 1.00 .+-. 0.14 1.00 .+-. 0.45 KLRC1 14.03 .+-. 2.73 5.62 .+-. 0.23 1.16 .+-. 0.45 1.28 .+-. 0.44 1.00 .+-. 0.07 1.00 .+-. 0.43 KLRD1 4.64 .+-. 1.00 4.17 .+-. 0.33 1.05 .+-. 0.27 1.65 .+-. 0.45 1.00 .+-. 0.20 1.00 .+-. 0.30 CD11b 11.13 .+-. 2.39 4.17 .+-. 0.48 1.55 .+-. 0.52 1.11 .+-. 0.40 1.00 .+-. 0.42 1.00 .+-. 0.34
TABLE-US-00020 TABLE 19 IT-EP of pOM1P2A-IL12 caused an increase in intratumoral levels of INF-y regulated genes in both primary and contralateral tumors. Fold change of treated vs. untreated mice values are shown. IT-pIL12-EP IT-pUMVC3 -EP Untreated IFN-7 related Mean .+-. SEM n = 5 Mean .+-. SEM n = 4 Mean .+-. SEM n = 3 RNA Primary Contralateral Primary Contralateral Primary Contralateral IFNy 8.63 .+-. 1.38 1.80 .+-. 0.44 0.76 .+-. 0.22 0.98 .+-. 0.43 1.00 .+-. 0.15 1.00 .+-. 0.29 CD274 12.47 .+-. 2.24 7.03 .+-. 2.30 1.00 .+-. 0.30 1.18 .+-. 0.83 1.00 .+-. 0.48 1.00 .+-. 0.84 (PD-L1) CXCL10 3.18 .+-. 0.58 2.26 .+-. 0.42 0.99 .+-. 0.30 1.44 .+-. 0.85 1.00 .+-. 0.43 1.00 .+-. 0.73 CXCL11 5.02 .+-. 0.74 3.14 .+-. 0.41 0.74 .+-. 0.10 1.38 .+-. 0.82 1.00 .+-. 0.16 1.00 .+-. 0.55 CXCL9 5.92 .+-. 0.60 3.75 .+-. 0.57 1.03 .+-. 0.31 1.67 .+-. 1.37 1.00 .+-. 0.50 1.00 .+-. 0.85 H2A-a 9.21 .+-. 1.86 6.63 .+-. 2.21 1.26 .+-. 0.36 1.52 .+-. 0.99 1.00 .+-. 0.61 1.00 .+-. 1.28 H2k-1 4.23 .+-. 1.02 3.71 .+-. 0.68 1.06 .+-. 0.19 1.42 .+-. 0.52 1.00 .+-. 0.54 1.00 .+-. 0.87 IRF 1 4.18 .+-. 0.28 2.72 .+-. 0.46 1.09 .+-. 0.28 1.28 .+-. 0.93 1.00 .+-. 0.45 1.00 .+-. 0.78 PDCD1 (PD-1) 3.80 .+-. 0.48 2.78 .+-. 0.84 1.13 .+-. 0.25 1.18 .+-. 0.37 1.00 .+-. 0.28 1.00 .+-. 0.56 Stat 1 3.51 .+-. 0.28 3.47 .+-. 0.68 1.04 .+-. 0.26 1.36 .+-. 0.79 1.00 .+-. 0.48 1.00 .+-. 0.79 TAP 1 3.80 .+-. 0.48 2.84 .+-. 0.37 1.17 .+-. 0.27 1.36 .+-. 0.85 1.00 .+-. 0.50 1.00 .+-. 0.97 CCL5 24.47 .+-. 7.81 14.59 .+-. 2.97 2.21 .+-. 0.72 1.48 .+-. 0.40 1.00 .+-. 0.29 1.00 .+-. 0.40 CCR5 11.29 .+-. 2.72 3.70 .+-. 0.70 1.31 .+-. 0.42 1.21 .+-. 0.42 1.00 .+-. 0.27 1.00 .+-. 0.40 GZMA 11.08 .+-. 1.18 4.60 .+-. 0.96 1.43 .+-. 0.53 2.05 .+-. 0.91 1.00 .+-. 0.23 1.00 .+-. 0.22 GZMB 3.11 .+-. 0.83 2.11 .+-. 0.10 0.68 .+-. 0.22 1.47 .+-. 0.67 1.00 .+-. 0.33 1.00 .+-. 0.47 PRF1 8.21 .+-. 2.27 2.06 .+-. 0.26 1.0 .+-. 0.32 1.13 .+-. 0.45 1.00 .+-. 0.23 1.00 .+-. 0.39
[0141] Additional NanoString gene expression analysis of extracts from treated and untreated tumors in the 4T1 and MC-38 tumor models after pOMIP2A-IL12 electroporation revealed similar upregulation of lymphocyte and monocyte cell surface markers as well as INF.gamma.-regulated genes, indicating that these effects of IL-12 on the tumor microenvironment are generalizable to multiple mouse tumor models.
[0142] Gene expression analysis of tissue from treated and untreated tumors corroborate flow cytometric analysis showing a robust increase in tumor TIL with IT-EP of pOMIP2A-IL12. In addition, an increase in interferon gamma-regulated genes suggest induction of an immunostimulatory environment within the tumors. A significant increase in expression of checkpoint proteins indicate that IT-pIL12-EP could increase the substrate for the action of checkpoint inhibitors used in combination.
[0143] Intratumoral electroporation of an OMI plasmid encoding human Flt3L-NY-ESO-1 fusion protein alone also had effects on tumor regression and changes to the immune phenotype of tumor TIL.
TABLE-US-00021 TABLE 20 IT-EP of pOMI-Flt3L-NYESO1 plasmid reduced tumor growth. Subcutaneous 4T1 tumors were electroporated once at 350 V/cm, 8 10 msec pulses with acupuncture needles after plamdis injection. Tumor measurements on Day 6 after treatment are shown. Tumor volume (mm.sup.3) Treatment Mean .+-. SEM n = 5 Untreated 273.8 .+-. 35.7 EP/pUMVC3 (empty vector) 380.4 .+-. 84.7 EP/pOMI-Flt3L-NYESO1 127.1 .+-. 13.2 EP/pOMIP2A-IL12 69.4 .+-. 16.4
TABLE-US-00022 TABLE 21 Changes INF.gamma. related gene expression in treated tumors after IT-EP of pOMI-Flt3L-NYESO1 as measured by NanoString in tumor extracts. Fold change of treated vs. untreated mice values are shown. IFN-.gamma. related IT-EP pUMVC3 IT-EP pOMI-Flt3L-NYESO1 RNA Mean .+-. SEM n = 3 Mean .+-. SEM n = 5 Cxcl9 1.00 .+-. 0.07 3.68 .+-. 0.42 Cxcl10 1.00 .+-. 0.02 1.80 .+-. 0.17 Cxcl11 1.00 .+-. 0.35 2.29 .+-. 0.41 Cd274 1.00 .+-. 0.28 3.31 .+-. 0.55 Irf1 1.00 .+-. 0.07 2.31 .+-. 0.16 Stat1 1.00 .+-. 0.13 2.46 .+-. 0.25
TABLE-US-00023 TABLE 22 Changes in antigen presentation machinery (APM) gene expression in treated tumors after IT-EP of pOMI-Flt3L-NYESO1 as measured by NanoString in tumor extracts. Fold change of treated vs. untreated mice values are shown. IT-EP pUMVC3 IT-EP pOMI-Flt3L-NYESO1 APM RNA Mean .+-. SEM n = 3 Mean .+-. SEM n = 5 H2-Ob 1.00 .+-. 0.24 2.09 .+-. 0.48 H2-Aa 1.00 .+-. 0.29 4.41 .+-. 0.78 H2-K1 1.00 .+-. 0.21 2.20 .+-. 0.16 H2-Ab1 1.00 .+-. 0.22 4.78 .+-. 0.82 H2-Eb1 1.00 .+-. 0.22 3.74 .+-. 0.50 Tap1 1.00 .+-. 0.08 2.63 .+-. 0.25 Tapbp 1.00 .+-. 0.11 2.61 .+-. 0.23 Cd74 1.00 .+-. 0.22 4.71 .+-. 0.81 Ccr7 1.00 .+-. 0.09 2.08 .+-. 0.33 Cd11b 1.00 .+-. 0.18 2.22 .+-. 0.27
TABLE-US-00024 TABLE 23 Changes in co-stimulatory gene expression in treated tumors after IT-EP of pOMI-Flt3L-NYESO1 as measured by NanoString in tumor extracts. Fold change of treated vs. untreated mice values are shown. Co-stimulatory IT-EP pUMVC3 IT-EP pOMI-Flt3L-NYESO1 RNA Mean .+-. SEM n = 3 Mean .+-. SEM n = 5 Cd80 1.00 .+-. 0.12 2.01 .+-. 0.35 Cd40 1.00 .+-. 0.18 3.15 .+-. 0.52 Ctla4 1.00 .+-. 0.06 3.11 .+-. 0.47 Cd274 1.00 .+-. 0.28 3.31 .+-. 0.55 Icam1 1.00 .+-. 0.33 2.67 .+-. 0.55
TABLE-US-00025 TABLE 24 Changes in T cell and Natural Killer (NK) cell gene expression in treated tumors after IT-EP of pOMI-Flt3L-NYESO1 as measured by NanoString in tumor extracts. Fold change of treated vs. untreated mice values are shown. T and NK cell IT-EP pUMVC3 IT-EP pOMI-Flt3L-NYESO1 RNA Mean .+-. SEM n = 3 Mean .+-. SEM n = 5 Klrc1 1.00 .+-. 0.37 2.84 .+-. 0.40 Klrd1 1.00 .+-. 0.11 3.91 .+-. 0.74 Cd3e 1.00 .+-. 0.38 3.57 .+-. 0.70 Cd8a 1.00 .+-. 0.36 2.03 .+-. 0.38 Cd4 1.00 .+-. 0.10 2.08 .+-. 0.36
[0144] In order to test for host response to electroporation of plasmids encoding a tracking antigen fused to Flt3L, Bl6F10 tumors were electroporated with pOMI-Flt3L-OVA and the host response to the OVA antigen was measured. Mice were injected with 1 million Bl6F10 cells on the right flank. Seven days later, tumors were electroporated with pOMI-mIL12-mFlt3L-OVA, empty vector, or left untreated. Electroporation was done using the Genesis generator, 400 V/cm, 8 10-msec pulses. Tumors regression was observed with pOMI-mIL12-mFLT3-OVA (Table 15).
[0145] Detection of tracking antigen-specific CD8+ T cells in mouse was tested inguinal lymph nodes 7 days after introduction of plasmid encoding FLt3L-OVA fusion proteins into tumors.
[0146] Lymph nodes were isolated 7 days after electroporation treatment. Mice were sacrificed; inguinal lymph nodes were excised, mashed in PBS+2% FBS+1 mM EDTA (PFB) and then strained through a 70 micro filter. Cells were pelleted in a centrifuge at 300.times.g at 4.degree. C. and washed in PFB, and counted on a Cellometer (Nexcelom).
[0147] Lymph node cell pellets were gently re-suspended in PFB with Fc block (BD Biosciences 553142). Cells were then mixed with a solution of SIINFEKL tetramer (MBL), according to the manufacturers instruction and incubated for 10 minutes at room temperature. Antibody staining cocktails containing the following: Live/Dead Aqua (Thermo Fisher L34966), Anti-CD3 (Biolegend 100228), anti-CD19 (Biolegend 115555), anti-CD127 (Biolegend), anti-CD8a (MBL D271-4), anti-CD44 (Biolegend 103028), anti-PD-1 (Biolegend 109110), anti-CD4 (Biolegend 100547), anti-KLRG1 (138419), anti-CD62L (Biolegend 104448) were added and incubated at 4'C for 30 minutes. Cells were washed with PFB. Cells were fixed in PFB with 1% paraformaldehyde for 1 minute on ice. Cells were washed 3 times with PFB, and analyzed by flow cytometry (LSR Fortessa X-20).
TABLE-US-00026 TABLE 25 Detection of host immune cells displaying tracking antigen and T cells reactive to tracking antigen after IT-EP of pOMI-mIL12-hFlt3L-OVA as compared to pUMVC3 empty vector into B16F10 subcutaneous tumors. Plasmid Frequency of introduced by CD44.sup.+SIINFEKLtetramer.sup.+CD8 Frequency of IT-EP T cells SIINFEKLtetramer.sup.+CD8.sup.+T cells Untreated n = 3 0.0003 .+-. 0.0003 0.0067 .+-. 0.0018 pUMVC3 n = 4 0.0026 .+-. 0.0003 0.0100 .+-. 0.0027 pOMI-mIL12-hFlt3L-OVA 0.4050 .+-. 0.2457 0.2958 .+-. 0.0582 n = 6
[0148] Using OVA as a surrogate tracking antigen in mice, we demonstrate that we can readily detect circulating T cells directed against the tracking antigen which was electroporated into tumor as a Flt3L-fusion protein.
XV(b). Introduction of Plasmids by Hydrodynamic Injection into Mouse Tail Vein
[0149] The in vivo activity of Flt3L fusion proteins expressed from OMI plasmids was tested by hydrodynamic injection of 5 .mu.g of plasmids into the tail vein of C57Bl/6J mice. Seven days later, mice were sacrificed, the spleens were excised, weighed, and dissociated for analysis of changes in cell composition by flow cytometry.
[0150] Splenocytes were isolated as described above, washed with PFB and re-suspended in PFB with Fc block (BD Biosciences 553142) and incubated for 10 minutes at room temp. Antibody cocktails containing the following were added: Anti NK1.1 (Biolegend 108731), Live/Dead Aqua ( ), anti-CD4 (Biolegend 100547), anti-F4/80 (Biolegend 123149), anti-CD19 (Biolegend 115555), Anti-I-A/I-E (Biolegend 107645), Anti-CD8 (MBL International D271-4), anti-CD80 (Biolegend 104722), anti-CD3 (Biolegend 117308), anti-CD40 (Biolegend 124630), anti-GR-1 (Biolegend 108424), anti-CD11c (Biolegend 117324), anti-CD86 (Biolegend 105024, anti-CD11b (Biolegend 101212). Incubate at 37.degree. C. Cells were washed 3 times with PFB, and analyzed by flow cytometry (LSR Fortessa X-20).
TABLE-US-00027 TABLE 26 Effect of systemic exposure to pOMIP2A-FLt3L and pOMIP2A- Flt3LNYESO1 plasmids introduced by tail vein injection. CD11c.sup.+ frequency Spleen weight Absolute CD11c.sup.+ of parent CD3.sup.- (grams) cell number; Mean .times. CD19.sup.-NK1.1.sup.-; Mean .+-. SEM, 10.sup.6 .+-. Mean percent .+-. Injected Plasmid n = 6 SEM, n = 6 SEM, n = 6 None 0.085 .+-. 0.005 1.82 7.68 .+-. 0.66 pUMVC3 empty vector 0.090 .+-. 0.006 2.75 12.11 .+-. 0.08 OMI-Flt3LNYESO1 0.123 .+-. 0.009 5.26 31.75 .+-. 2.88 OMI-Flt3L 0.141 .+-. 0.011 5.42 37.60 .+-. 3.22
[0151] Introduction of plasmids encoding human Flt3L or human FLt3L fused to a portion of the NY-ESO-1 proteins (80-180 aa) lead to an increase in CD11+ dendritic cells (DC) in the spleen. Moreover, the majority of these DC demonstrated high levels of MEW Class II indicating that they are mature DCs. In addition, a portion of these DCs demonstrated higher levels of cell surface CD86, indicating they were activated.
[0152] These data are consistent with exposure to active Flt3 ligand being expressed from these plasmids and leading to DC maturation and activation in the mice (Maraskovsky et al., 2000. Blood 96:878)
Sequence Identifiers
TABLE-US-00028 [0153] TABLE 27 Sequence Identifier Table SEQ ID NO Description 1 Promoter/enhancer: Human CMV 2 Promoter/enhancer: Simian CMV 3 Promoter/enhancer: SV-40 4 Promoter/enhancer: mPGK 5 Human IL-12p35-P2A-IL-12p40 (DNA) 6 Mouse IL-12p35-P2A-IL-12p40 (DNA) 7 Canine IL-12p35-P2A-IL-12p40 (DNA) 8 Human IL-15/IL-15Ra with P2A (DNA) 9 Human IL-15/IL-15Ra with P2A (protein) 10 Human IL-15/IL-15Ra-Fc with P2A (DNA) 11 Human IL-15/IL-15Ra-Fc with P2A (protein) 12 Human interferon gamma (DNA) 13 Flt3L- NY-ESO-1 (full length) fusion protein (DNA) 14 Flt3L- NY-ESO-1 (full length) fusion protein (protein) 15 Flt3L- NY-ESO-1 (amino acids 80-180) fusion protein (DNA) 16 Flt3L- NY-ESO-1 (amino acids 80-180) fusion protein (protein) 17 Flt3L- NY-ESO-1 (fusion of peptides) fusion protein (DNA) 18 Flt3L- NY-ESO-1 (fusion of peptides) fusion protein (protein) 19 Translation Modulator, exon-skipping motif P2A 20 Translation Modulator, exon-skipping motif T2A 21 Translation Modulator, exon-skipping motif E2A 22 Translation Modulator, exon-skipping motif F2A 23 Translation modifier: Internal Ribosomal Entry Site (IRES) 24 Ovalbumin aa 241-270 (DNA) 25 Ovalbumin aa 241-270 (protein)
Sequence CWU 1
1
251576DNAArtificial SequenceHuman CMV promoter/enhancer 1tagtaatcaa
ttacggggtc attagttcat agcccatata tggagttccg cgttacataa 60cttacggtaa
atggcccgcc tggctgaccg cccaacgacc cccgcccatt gacgtcaata
120atgacgtatg ttcccatagt aacgccaata gggactttcc attgacgtca
atgggtggag 180tatttacggt aaactgccca cttggcagta catcaagtgt
atcatatgcc aagtacgccc 240cctattgacg tcaatgacgg taaatggccc
gcctggcatt atgcccagta catgacctta 300tgggactttc ctacttggca
gtacatctac gtattagtca tcgctattac catggtgatg 360cggttttggc
agtacatcaa tgggcgtgga tagcggtttg actcacgggg atttccaagt
420ctccacccca ttgacgtcaa tgggagtttg ttttggcacc aaaatcaacg
ggactttcca 480aaatgtcgta acaactccgc cccattgacg caaatgggcg
gtaggcgtgt acggtgggag 540gtctatataa gcagagctcg tttagtgaac cgtcag
5762593DNAArtificial SequenceSimian CMV promoter/enhancer
2atagtattcc atatatgggt tttcctattg acgtagatag cccctcccaa tgggcggtcc
60catataccat atatggggct tcctaatacc gcccatagcc actcccccat tgacgtcaat
120ggtctctata tatggtcttt ttcgaaccta ttgacgtcat atgggcggtc
ctattgacgt 180atatggcgcc tcccccattg acgtcaatta cggtaaatgg
cccgcctggc tcaatgccca 240ttgacgtcaa taggaccacc caccattgac
gtcaatggga tggctcattg cccattcata 300tccgttctca cgccccctat
tgacgtcaat gacggtaaat ggcccacttg gcagtacatc 360aatatctatt
aatagtaact tggcaagtac attactattg gaagtacgcc agggtacatt
420ggcagtactc ccattgacgt caatggcggt aaatggcccg cgatggctgc
caagtacatc 480cccattgacg tcaatgggga ggggcaatga cgcaaatggg
cgttccattg acgtaaatgg 540gcggtaggcg tgcctaatgg gaggtctata
taagcaatgc tcgtttaggg aac 5933511DNAMus musculus 3ttctaccggg
taggggaggc gcttttccca aggcagtctg gagcatgcgc tttagcagcc 60ccgctgggca
cttggcgcta cacaagtggc ctctggcctc gcacacattc cacatccacc
120ggtaggcgcc aaccggctcc gttctttggt ggccccttcg cgccaccttc
tactcctccc 180ctagtcagga agttcccccc cgccccgcag ctcgcgtcgt
gcaggacgtg acaaatggaa 240gtagcacgtc tcactagtct cgtgcagatg
gacagcaccg ctgagcaatg gaagcgggta 300ggcctttggg gcagcggcca
atagcagctt tgctccttcg ctttctgggc tcagaggctg 360ggaaggggtg
ggtccggggg cgggctcagg ggcgggctca ggggcggggc gggcgcccga
420aggtcctccg gaggcccggc attctgcacg cttcaaaagc gcacgtctgc
cgcgctgttc 480tcctcttcct catctccggg cctttcgacc t
5114344DNAArtificial SequenceSV-40 promoter 4ctgtggaatg tgtgtcagtt
agggtgtgga aagtccccag gctccccagc aggcagaagt 60atgcaaagca tgcatctcaa
ttagtcagca accaggtgtg gaaagtcccc aggctcccca 120gcaggcagaa
gtatgcaaag catgcatctc aattagtcag caaccatagt cccgccccta
180actccgccca tcccgcccct aactccgccc agttccgccc attctccgcc
ccatggctga 240ctaatttttt ttatttatgc agaggccgag gccgcctctg
cctctgagct attccagaag 300tagtgaggag gcttttttgg aggcctaggc
ttttgcaaaa agct 34451824DNAArtificial SequenceHuman
IL12p35-P2A-IL12p40 5atgtggcccc ctgggtcagc ctcccagcca ccgccctcac
ctgccgcggc cacaggtctg 60catccagcgg ctcgccctgt gtccctgcag tgccggctca
gcatgtgtcc agcgcgcagc 120ctcctccttg tggctaccct ggtcctcctg
gaccacctca gtttggccag aaacctcccc 180gtggccactc cagacccagg
aatgttccca tgccttcacc actcccaaaa cctgctgagg 240gccgtcagca
acatgctcca gaaggccaga caaactctcg aattttaccc ttgcacttct
300gaagagattg atcatgaaga tatcacaaaa gataaaacca gcacagtgga
ggcctgttta 360ccattggaat taaccaagaa tgagagttgc ctaaattcca
gagagacctc tttcataact 420aatgggagtt gcctggcctc cagaaagacc
tcttttatga tggccctgtg ccttagtagt 480atttatgaag acttgaagat
gtaccaggtg gagttcaaga ccatgaatgc aaagcttctg 540atggacccta
agaggcaaat cttcctagat caaaacatgc tggcagttat tgatgagctg
600atgcaggccc tgaatttcaa cagtgagact gtgccacaaa aatcctccct
tgaagaaccg 660gatttctaca agactaaaat caagctctgc atacttcttc
atgctttcag aattcgggca 720gtgactattg atagagtgat gagctatctg
aatgcttccg cggccgcagg atctggggcc 780accaactttt cattgctcaa
gcaggcgggc gatgtggagg aaaaccctgg ccccggatcc 840tgtcaccagc
agttggtcat ctcttggttt tccctggttt ttctggcatc tcccctcgtg
900gccatatggg aactgaagaa agatgtttat gtcgtagaat tggattggta
tccggatgcc 960cctggagaaa tggtggtcct cacctgtgac acccctgaag
aagatggtat cacctggacc 1020ttggaccaga gcagtgaggt cttaggctct
ggcaaaaccc tgaccatcca agtcaaagag 1080tttggagatg ctggccagta
cacctgtcac aaaggaggcg aggttctaag ccattcgctc 1140ctgctgcttc
acaaaaagga agatggaatt tggtccactg atattttaaa ggaccagaaa
1200gaacccaaaa ataagacctt tctaagatgc gaggccaaga attattctgg
acgtttcacc 1260tgctggtggc tgacgacaat cagtactgat ttgacattca
gtgtcaaaag cagcagaggc 1320tcttctgacc cccaaggggt gacgtgcgga
gctgctacac tctctgcaga gagagtcaga 1380ggggacaaca aggagtatga
gtactcagtg gagtgccagg aggacagtgc ctgcccagct 1440gctgaggaga
gtctgcccat tgaggtcatg gtggatgccg ttcacaagct caagtatgaa
1500aactacacca gcagcttctt catcagggac atcatcaaac ctgacccacc
caagaacttg 1560cagctgaagc cattaaagaa ttctcggcag gtggaggtca
gctgggagta ccctgacacc 1620tggagtactc cacattccta cttctccctg
acattctgcg ttcaggtcca gggcaagagc 1680aagagagaaa agaaagatag
agtcttcacg gacaagacct cagccacggt catctgccgc 1740aaaaatgcca
gcattagcgt gcgggcccag gaccgctact atagctcatc ttggagcgaa
1800tgggcatctg tgccctgcag ttag 182461794DNAArtificial SequenceMouse
IL12p35-P2A-IL12p40 6atggtcagcg ttccaacagc ctcaccctcg gcatccagca
gctcctctca gtgccggtcc 60agcatgtgtc aatcacgcta cctcctcttt ttggccaccc
ttgccctcct aaaccacctc 120agtttggcca gggtcattcc agtctctgga
cctgccaggt gtcttagcca gtcccgaaac 180ctgctgaaga ccacagatga
catggtgaag acggccagag aaaaactgaa acattattcc 240tgcactgctg
aagacatcga tcatgaagac atcacacggg accaaaccag cacattgaag
300acctgtttac cactggaact acacaagaac gagagttgcc tggctactag
agagacttct 360tccacaacaa gagggagctg cctgccccca cagaagacgt
ctttgatgat gaccctgtgc 420cttggtagca tctatgagga cttgaagatg
taccagacag agttccaggc catcaacgca 480gcacttcaga atcacaacca
tcagcagatc attcttgaca agggcatgct ggtggccatc 540gatgagctga
tgcagtctct gaatcataat ggcgagactc tgcgccagaa acctcctgtg
600ggagaagcag acccttacag agtgaaaatg aagctctgca tcctgcttca
cgccttcagc 660acccgcgtcg tgaccatcaa cagggtgatg ggctatctga
gctccgccgc ggccgcagga 720tctggggcca ccaacttttc attgctcaag
caggcgggcg atgtggagga aaaccctggc 780cccggatcct gtcctcagaa
gctaaccatc tcctggtttg ccatcgtttt gctggtgtct 840ccactcatgg
ccatgtggga gctggagaaa gacgtttatg ttgtagaggt ggactggact
900cccgatgccc ctggagaaac agtgaacctc acctgtgaca cgcctgaaga
agatgacatc 960acctggacct cagaccagag acatggagtc ataggctctg
gaaagaccct gaccatcact 1020gtcaaagagt ttcttgatgc tggccagtac
acctgccaca aaggaggcga gactctgagc 1080cactcacatc tgctgctcca
caagaaggaa aatggaattt ggtccactga aattttaaag 1140aatttcaaga
acaagacttt cctgaagtgt gaagcaccaa attactccgg acggttcacg
1200tgctcatggc tggtgcaaag aaacatggac ttgaagttca acatcaagag
cagtagcagt 1260tcccctgact ctcgggcagt gacatgtgga atggcgtctc
tgtctgcaga gaaggtcaca 1320ctggaccaaa gggactatga gaagtattca
gtgtcctgcc aggaggatgt cacctgccca 1380actgccgagg agaccctgcc
cattgaactg gcgttggaag cacggcagca gaataaatat 1440gagaactaca
gcaccagctt cttcatcagg gacatcatca aaccagaccc gcccaagaac
1500ttgcagatga agcctttgaa gaactcacag gtggaggtca gctgggagta
ccctgactcc 1560tggagcactc cccattccta cttctccctc aagttctttg
ttcgaatcca gcgcaagaaa 1620gaaaagatga aggagacaga ggaggggtgt
aaccagaaag gtgcgttcct cgtagagaag 1680acatctaccg aagtccaatg
caaaggcggg aatgtctgcg tgcaagctca ggatcgctat 1740tacaattcct
catgcagcaa gtgggcatgt gttccctgca gggtccgatc ctag
179471734DNAArtificial SequenceCanine IL12p35-P2A-IL12p40
7atgtgcccgc cgcgcggcct cctccttgtg accatcctgg tcctgctaag ccacctggac
60caccttactt gggccaggag cctccccaca gcctcaccga gcccaggaat attccagtgc
120ctcaaccact cccaaaacct gctgagagcc gtcagcaaca cgcttcagaa
ggccagacaa 180actctcgatt atattccctg cacttccgaa gagattgatc
atgaagatat cacaaaggat 240aaaaccagca cagtggaggc ctgcttacca
ctggaattaa ccatgaatga gagttgcctg 300gcttccagag agatttcttt
gataactaac gggagttgcc tggcctctgg aaaggcctct 360tttatgacgg
tcctgtgcct tagcagcatc tatgaggact tgaagatgta ccagatggaa
420ttcaaggcca tgaacgcaaa gcttttaatg gaccccaaga ggcagatttt
tctggatcaa 480aacatgttga cagctatcga tgagctgtta caggccctga
atttcaacag tgtgactgtg 540ccacagaaat cctcccttga agagccggac
ttctacaaga ctaaaatcaa gctctgcata 600cttcttcatg ctttcagaat
tcgtgcggtg accatcgata gaatgatgag ttatctgaat 660tcttccgcgg
ccgcaggatc tggggccacc aacttttcat tgctcaagca ggcgggcgat
720gtggaggaaa accctggccc cggatcccat cctcagcagt tggtcatctc
ctggttttcc 780ctcgttttgc tggcgtcttc cctcatgacc atatgggaac
tggagaaaga tgtttatgtt 840gtagagttgg actggcaccc tgatgccccc
ggagaaatgg tggtcctcac ctgccatacc 900cctgaagaag atgacatcac
ttggacctca gcgcagagca gtgaagtcct aggttctggt 960aaaactctga
ccatccaagt caaagaattt ggagatgctg gccagtatac ctgccataaa
1020ggaggcaagg ttctgagccg ctcactcctg ttgattcaca aaaaagaaga
tggaatttgg 1080tccactgata tcttaaagga acagaaagaa tccaaaaata
agatttttct gaaatgtgag 1140gcaaagaatt attctggacg tttcacatgc
tggtggctga cggcaatcag tactgatttg 1200aaattcagtg tcaaaagtag
cagaggcttc tctgaccccc aaggggtgac atgtggagca 1260gtgacacttt
cagcagagag ggtcagagtg gacaacaggg attataagaa gtacacagtg
1320gagtgtcagg aaggcagtgc ctgcccctct gccgaggaga gcctacccat
cgaggtcgtg 1380gtggatgcta ttcacaagct caagtatgaa aactacacca
gcagcttctt catcagagac 1440atcatcaaac cagacccacc cacaaacctg
cagctgaagc cattgaaaaa ttctcggcac 1500gtggaggtca gctgggaata
ccccgacacc tggagcaccc cacattccta cttctccctg 1560acattttgcg
tacaggccca gggcaagaac aatagagaaa agaaagatag actctgcgtg
1620gacaagacct cagccaaggt cgtgtgccac aaggatgcca agatccgcgt
gcaagcccga 1680gaccgctact atagttcatc ctggagcgac tgggcatctg
tgtcctgcag ttag 173481368DNAArtificial SequenceHuman IL-15/IL-15Ra
with P2A 8atgagaattt cgaaaccaca tttgagaagt atttccatcc agtgctactt
gtgtttactt 60ctaaacagtc attttctaac tgaagctggc attcatgtct tcattttggg
ctgtttcagt 120gcagggcttc ctaaaacaga agccaactgg gtgaatgtaa
taagtgattt gaaaaaaatt 180gaggatctta ttcaatctat gcatattgat
gctactttat atacggaaag tgatgttcac 240cccagttgca aagtaacagc
aatgaagtgc tttctcttgg agttacaagt tatttcactt 300gagtccggag
atgcaagtat tcatgataca gtagaaaatc tgatcatcct agcaaacaac
360agtttgtctt ctaatgggaa tgtaacagaa tctggatgca aagaatgtga
ggaactggag 420gaaaagaata ttaaagagtt tttgcagagt tttgtacata
ttgtccaaat gttcatcaac 480acttctgcgg ccgcaggatc tggggccacc
aacttttcat tgctcaagca ggcgggcgat 540gtggaggaaa accctggccc
cggatccgct ccccgccgcg cgagagggtg tcgaacgctt 600ggtcttcctg
cgttgctgct gttgttgttg ctcaggccgc ctgccacccg cggcataacc
660tgccccccac cgatgagcgt ggagcacgca gacatctggg tcaaaagcta
tagtctgtac 720tctagggaaa ggtacatttg taacagtggg ttcaaaagaa
aggctggcac atcatccctt 780acagaatgcg tccttaacaa ggccaccaac
gtcgcgcatt ggactacccc ttccctgaag 840tgcatcagag atcctgcgct
ggttcaccaa cgcccggccc ccccatctac agttacaacc 900gctggtgtta
cacctcagcc cgaatctctg tcaccttctg gcaaagaacc agcagcctcc
960tccccaagtt ccaacaatac cgccgccact accgctgcta tcgttccagg
ctcacaactg 1020atgccgagta aatctcccag caccggtaca acagaaatct
ctagtcatga gtcctcccat 1080gggaccccgt cccagaccac tgctaaaaac
tgggagctga ccgcctccgc tagccatcag 1140ccgcccgggg tgtacccaca
ggggcactct gacaccactg tcgcgattag tactagcaca 1200gtgctgttgt
gcggactgtc tgccgtcagc ttgttggctt gctatctgaa atcccggcag
1260acgccaccac tggccagcgt ggagatggaa gcaatggaag ctttgcctgt
aacctggggg 1320acgagctcac gggatgagga tctggagaac tgctcacatc acctgtaa
13689455PRTArtificial SequenceHuman IL-15/IL-15Ra Fc P2A 9Met Arg
Ile Ser Lys Pro His Leu Arg Ser Ile Ser Ile Gln Cys Tyr1 5 10 15Leu
Cys Leu Leu Leu Asn Ser His Phe Leu Thr Glu Ala Gly Ile His 20 25
30Val Phe Ile Leu Gly Cys Phe Ser Ala Gly Leu Pro Lys Thr Glu Ala
35 40 45Asn Trp Val Asn Val Ile Ser Asp Leu Lys Lys Ile Glu Asp Leu
Ile 50 55 60Gln Ser Met His Ile Asp Ala Thr Leu Tyr Thr Glu Ser Asp
Val His65 70 75 80Pro Ser Cys Lys Val Thr Ala Met Lys Cys Phe Leu
Leu Glu Leu Gln 85 90 95Val Ile Ser Leu Glu Ser Gly Asp Ala Ser Ile
His Asp Thr Val Glu 100 105 110Asn Leu Ile Ile Leu Ala Asn Asn Ser
Leu Ser Ser Asn Gly Asn Val 115 120 125Thr Glu Ser Gly Cys Lys Glu
Cys Glu Glu Leu Glu Glu Lys Asn Ile 130 135 140Lys Glu Phe Leu Gln
Ser Phe Val His Ile Val Gln Met Phe Ile Asn145 150 155 160Thr Ser
Ala Ala Ala Gly Ser Gly Ala Thr Asn Phe Ser Leu Leu Lys 165 170
175Gln Ala Gly Asp Val Glu Glu Asn Pro Gly Pro Gly Ser Ala Pro Arg
180 185 190Arg Ala Arg Gly Cys Arg Thr Leu Gly Leu Pro Ala Leu Leu
Leu Leu 195 200 205Leu Leu Leu Arg Pro Pro Ala Thr Arg Gly Ile Thr
Cys Pro Pro Pro 210 215 220Met Ser Val Glu His Ala Asp Ile Trp Val
Lys Ser Tyr Ser Leu Tyr225 230 235 240Ser Arg Glu Arg Tyr Ile Cys
Asn Ser Gly Phe Lys Arg Lys Ala Gly 245 250 255Thr Ser Ser Leu Thr
Glu Cys Val Leu Asn Lys Ala Thr Asn Val Ala 260 265 270His Trp Thr
Thr Pro Ser Leu Lys Cys Ile Arg Asp Pro Ala Leu Val 275 280 285His
Gln Arg Pro Ala Pro Pro Ser Thr Val Thr Thr Ala Gly Val Thr 290 295
300Pro Gln Pro Glu Ser Leu Ser Pro Ser Gly Lys Glu Pro Ala Ala
Ser305 310 315 320Ser Pro Ser Ser Asn Asn Thr Ala Ala Thr Thr Ala
Ala Ile Val Pro 325 330 335Gly Ser Gln Leu Met Pro Ser Lys Ser Pro
Ser Thr Gly Thr Thr Glu 340 345 350Ile Ser Ser His Glu Ser Ser His
Gly Thr Pro Ser Gln Thr Thr Ala 355 360 365Lys Asn Trp Glu Leu Thr
Ala Ser Ala Ser His Gln Pro Pro Gly Val 370 375 380Tyr Pro Gln Gly
His Ser Asp Thr Thr Val Ala Ile Ser Thr Ser Thr385 390 395 400Val
Leu Leu Cys Gly Leu Ser Ala Val Ser Leu Leu Ala Cys Tyr Leu 405 410
415Lys Ser Arg Gln Thr Pro Pro Leu Ala Ser Val Glu Met Glu Ala Met
420 425 430Glu Ala Leu Pro Val Thr Trp Gly Thr Ser Ser Arg Asp Glu
Asp Leu 435 440 445Glu Asn Cys Ser His His Leu 450
455102070DNAArtificial SequenceHuman IL-15/IL-15Ra-Fc with P2A
10atgagaattt cgaaaccaca tttgagaagt atttccatcc agtgctactt gtgtttactt
60ctaaacagtc attttctaac tgaagctggc attcatgtct tcattttggg ctgtttcagt
120gcagggcttc ctaaaacaga agccaactgg gtgaatgtaa taagtgattt
gaaaaaaatt 180gaggatctta ttcaatctat gcatattgat gctactttat
atacggaaag tgatgttcac 240cccagttgca aagtaacagc aatgaagtgc
tttctcttgg agttacaagt tatttcactt 300gagtccggag atgcaagtat
tcatgataca gtagaaaatc tgatcatcct agcaaacaac 360agtttgtctt
ctaatgggaa tgtaacagaa tctggatgca aagaatgtga ggaactggag
420gaaaagaata ttaaagagtt tttgcagagt tttgtacata ttgtccaaat
gttcatcaac 480acttctgcgg ccgcaggatc tggggccacc aacttttcat
tgctcaagca ggcgggcgat 540gtggaggaaa accctggccc cggatccgct
ccccgccgcg cgagagggtg tcgaacgctt 600ggtcttcctg cgttgctgct
gttgttgttg ctcaggccgc ctgccacccg cggcataacc 660tgccccccac
cgatgagcgt ggagcacgca gacatctggg tcaaaagcta tagtctgtac
720tctagggaaa ggtacatttg taacagtggg ttcaaaagaa aggctggcac
atcatccctt 780acagaatgcg tccttaacaa ggccaccaac gtcgcgcatt
ggactacccc ttccctgaag 840tgcatcagag atcctgcgct ggttcaccaa
cgcccggccc ccccatctac agttacaacc 900gctggtgtta cacctcagcc
cgaatctctg tcaccttctg gcaaagaacc agcagcctcc 960tccccaagtt
ccaacaatac cgccgccact accgctgcta tcgttccagg ctcacaactg
1020atgccgagta aatctcccag caccggtaca acagaaatct ctagtcatga
gtcctcccat 1080gggaccccgt cccagaccac tgctaaaaac tgggagctga
ccgcctccgc tagccatcag 1140ccgcccgggg tgtacccaca ggggcactct
gacaccactg tcgcgattag tactagcaca 1200gtgctgttgt gcggactgtc
tgccgtcagc ttgttggctt gctatctgaa atcccggcag 1260acgccaccac
tggccagcgt ggagatggaa gcaatggaag ctttgcctgt aacctggggg
1320acgagctcac gggatgagga tctggagaac tgctcacatc acctgggaag
atctggcagt 1380tctggggaca aaactcacac atgcccaccg tgcccagcac
ctgaactcct ggggggaccg 1440tcagtcttcc tcttcccccc aaaacccaag
gacaccctca tgatctcccg gacccctgag 1500gtcacatgcg tggtggtgga
cgtgagccac gaagaccctg aggtcaagtt caactggtac 1560gtggacggcg
tggaggtgca taatgccaag acaaagccgc gggaggagca gtacaacagc
1620acgtaccgtg tggtcagcgt cctcaccgtc ctgcaccagg actggctgaa
tggcaaggag 1680tacaagtgca aggtctccaa caaagccctc ccagccccca
tcgagaaaac catctccaaa 1740gccaaagggc agccccgaga accacaggtc
tacaccctgc ccccatcccg ggaggagatg 1800accaagaacc aggtcagcct
gacctgcctg gtcaaaggct tctatcccag cgacatcgcc 1860gtggagtggg
agagcaatgg gcagccggag aacaactaca agaccacgcc tcccgtgctg
1920gactccgacg gctccttctt cctctacagc aagctcaccg tggacaagag
caggtggcag 1980caggggaacg tcttctcatg ctccgtgatg cacgaggctc
tgcacaacca ctacacgcag 2040aagagcctct ccctgtctcc gggtaaatag
207011689PRTArtificial SequenceHuman IL-15/IL-15Ra-Fc with P2A
11Met Arg Ile Ser Lys Pro His Leu Arg Ser Ile Ser Ile Gln Cys Tyr1
5 10 15Leu Cys Leu Leu Leu Asn Ser His Phe Leu Thr Glu Ala Gly Ile
His 20 25 30Val Phe Ile Leu Gly Cys Phe Ser Ala Gly Leu Pro Lys Thr
Glu Ala 35 40 45Asn Trp Val Asn Val Ile Ser Asp Leu Lys Lys Ile Glu
Asp Leu Ile 50 55 60Gln Ser Met His Ile Asp Ala Thr Leu Tyr Thr Glu
Ser Asp Val His65 70 75 80Pro Ser Cys Lys Val Thr Ala Met Lys Cys
Phe Leu Leu Glu Leu Gln 85 90 95Val Ile Ser Leu Glu Ser Gly Asp Ala
Ser Ile His Asp Thr Val Glu 100 105 110Asn Leu Ile Ile Leu Ala Asn
Asn Ser Leu Ser Ser Asn Gly
Asn Val 115 120 125Thr Glu Ser Gly Cys Lys Glu Cys Glu Glu Leu Glu
Glu Lys Asn Ile 130 135 140Lys Glu Phe Leu Gln Ser Phe Val His Ile
Val Gln Met Phe Ile Asn145 150 155 160Thr Ser Ala Ala Ala Gly Ser
Gly Ala Thr Asn Phe Ser Leu Leu Lys 165 170 175Gln Ala Gly Asp Val
Glu Glu Asn Pro Gly Pro Gly Ser Ala Pro Arg 180 185 190Arg Ala Arg
Gly Cys Arg Thr Leu Gly Leu Pro Ala Leu Leu Leu Leu 195 200 205Leu
Leu Leu Arg Pro Pro Ala Thr Arg Gly Ile Thr Cys Pro Pro Pro 210 215
220Met Ser Val Glu His Ala Asp Ile Trp Val Lys Ser Tyr Ser Leu
Tyr225 230 235 240Ser Arg Glu Arg Tyr Ile Cys Asn Ser Gly Phe Lys
Arg Lys Ala Gly 245 250 255Thr Ser Ser Leu Thr Glu Cys Val Leu Asn
Lys Ala Thr Asn Val Ala 260 265 270His Trp Thr Thr Pro Ser Leu Lys
Cys Ile Arg Asp Pro Ala Leu Val 275 280 285His Gln Arg Pro Ala Pro
Pro Ser Thr Val Thr Thr Ala Gly Val Thr 290 295 300Pro Gln Pro Glu
Ser Leu Ser Pro Ser Gly Lys Glu Pro Ala Ala Ser305 310 315 320Ser
Pro Ser Ser Asn Asn Thr Ala Ala Thr Thr Ala Ala Ile Val Pro 325 330
335Gly Ser Gln Leu Met Pro Ser Lys Ser Pro Ser Thr Gly Thr Thr Glu
340 345 350Ile Ser Ser His Glu Ser Ser His Gly Thr Pro Ser Gln Thr
Thr Ala 355 360 365Lys Asn Trp Glu Leu Thr Ala Ser Ala Ser His Gln
Pro Pro Gly Val 370 375 380Tyr Pro Gln Gly His Ser Asp Thr Thr Val
Ala Ile Ser Thr Ser Thr385 390 395 400Val Leu Leu Cys Gly Leu Ser
Ala Val Ser Leu Leu Ala Cys Tyr Leu 405 410 415Lys Ser Arg Gln Thr
Pro Pro Leu Ala Ser Val Glu Met Glu Ala Met 420 425 430Glu Ala Leu
Pro Val Thr Trp Gly Thr Ser Ser Arg Asp Glu Asp Leu 435 440 445Glu
Asn Cys Ser His His Leu Gly Arg Ser Gly Ser Ser Gly Asp Lys 450 455
460Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly
Pro465 470 475 480Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr
Leu Met Ile Ser 485 490 495Arg Thr Pro Glu Val Thr Cys Val Val Val
Asp Val Ser His Glu Asp 500 505 510Pro Glu Val Lys Phe Asn Trp Tyr
Val Asp Gly Val Glu Val His Asn 515 520 525Ala Lys Thr Lys Pro Arg
Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val 530 535 540Val Ser Val Leu
Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu545 550 555 560Tyr
Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys 565 570
575Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr
580 585 590Leu Pro Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser
Leu Thr 595 600 605Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala
Val Glu Trp Glu 610 615 620Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys
Thr Thr Pro Pro Val Leu625 630 635 640Asp Ser Asp Gly Ser Phe Phe
Leu Tyr Ser Lys Leu Thr Val Asp Lys 645 650 655Ser Arg Trp Gln Gln
Gly Asn Val Phe Ser Cys Ser Val Met His Glu 660 665 670Ala Leu His
Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly 675 680
685Lys12501DNAArtificial SequenceHuman interferon gamma
12atgaaatata caagttatat cttggctttt cagctctgca tcgttttggg ttctcttggc
60tgttactgcc aggacccata tgtaaaagaa gcagaaaacc ttaagaaata ttttaatgca
120ggtcattcag atgtagcgga taatggaact cttttcttag gcattttgaa
gaattggaaa 180gaggagagtg acagaaaaat aatgcagagc caaattgtct
ccttttactt caaacttttt 240aaaaacttta aagatgacca gagcatccaa
aagagtgtgg agaccatcaa ggaagacatg 300aatgtcaagt ttttcaatag
caacaaaaag aaacgagatg acttcgaaaa gctgactaat 360tattcggtaa
ctgacttgaa tgtccaacgc aaagcaatac atgaactcat ccaagtgatg
420gctgaactgt cgccagcagc taaaacaggg aagcgaaaaa ggagtcagat
gctgtttcga 480ggtcgaagag catcccagta a 501131101DNAArtificial
SequencehFlt3L-NY-ESO-1 full length 13atggatgcta tgaaacgagg
actgtgttgt gtgttgcttc tgtgtggagc ggtttttgtg 60agtcccactc aggattgcag
cttccagcat tcacccatat catcagattt tgcagtaaag 120atcagggaac
tctccgatta tctccttcaa gactaccccg taacagtggc ctccaatttg
180caagacgaag agctttgtgg tgccctctgg cggctcgttt tggcccaaag
gtggatggaa 240cggcttaaga cagtcgctgg cagcaagatg caagggttgc
tcgaacgagt caatacagag 300atccattttg taaccaagtg tgcatttcaa
ccgccgccaa gctgccttcg ctttgttcag 360acgaatataa gtagactgtt
gcaggaaacc tccgagcaac tcgtagccct gaagccctgg 420attacacggc
aaaatttcag tcggtgcctt gagcttcagt gtcagcctga tagtagtacc
480ttgcctccgc catggtcccc caggcctctt gaagctacag ctccgacagc
ccctcagccg 540ggcagtagtg gtagttctgg acaagcggag ggaagaggga
cggggggctc aacgggagac 600gccgatggac ctggcggtcc tggtatcccg
gacggcccag gtggcaatgc tggcggtcca 660ggcgaggctg gtgcaactgg
tggacgcggg ccgcggggcg cgggtgctgc acgggcaagt 720ggccctgggg
gcggcgctcc tcggggtccg catgggggtg cagccagtgg actcaatggg
780tgctgcagat gcggcgcgag aggtccggaa agtcgattgc ttgaatttta
cctcgcaatg 840cccttcgcaa cgccgatgga ggctgaactg gccaggcgca
gtcttgccca ggacgcgcct 900cccttgccag tgcccggtgt gctgcttaaa
gagttcactg tttctggtaa catcttgacg 960attcgcctga cggcagcaga
ccatcgacaa ctccagctct ccatctcttc atgtcttcag 1020cagctctcac
tgttgatgtg gatcacacag tgtttcttgc ccgtgtttct cgcacagcct
1080ccatctggac agagaaggta a 110114366PRTArtificial
SequencehFlt3L-NY-ESO-1 full length 14Met Asp Ala Met Lys Arg Gly
Leu Cys Cys Val Leu Leu Leu Cys Gly1 5 10 15Ala Val Phe Val Ser Pro
Thr Gln Asp Cys Ser Phe Gln His Ser Pro 20 25 30Ile Ser Ser Asp Phe
Ala Val Lys Ile Arg Glu Leu Ser Asp Tyr Leu 35 40 45Leu Gln Asp Tyr
Pro Val Thr Val Ala Ser Asn Leu Gln Asp Glu Glu 50 55 60Leu Cys Gly
Ala Leu Trp Arg Leu Val Leu Ala Gln Arg Trp Met Glu65 70 75 80Arg
Leu Lys Thr Val Ala Gly Ser Lys Met Gln Gly Leu Leu Glu Arg 85 90
95Val Asn Thr Glu Ile His Phe Val Thr Lys Cys Ala Phe Gln Pro Pro
100 105 110Pro Ser Cys Leu Arg Phe Val Gln Thr Asn Ile Ser Arg Leu
Leu Gln 115 120 125Glu Thr Ser Glu Gln Leu Val Ala Leu Lys Pro Trp
Ile Thr Arg Gln 130 135 140Asn Phe Ser Arg Cys Leu Glu Leu Gln Cys
Gln Pro Asp Ser Ser Thr145 150 155 160Leu Pro Pro Pro Trp Ser Pro
Arg Pro Leu Glu Ala Thr Ala Pro Thr 165 170 175Ala Pro Gln Pro Gly
Ser Ser Gly Ser Ser Gly Gln Ala Glu Gly Arg 180 185 190Gly Thr Gly
Gly Ser Thr Gly Asp Ala Asp Gly Pro Gly Gly Pro Gly 195 200 205Ile
Pro Asp Gly Pro Gly Gly Asn Ala Gly Gly Pro Gly Glu Ala Gly 210 215
220Ala Thr Gly Gly Arg Gly Pro Arg Gly Ala Gly Ala Ala Arg Ala
Ser225 230 235 240Gly Pro Gly Gly Gly Ala Pro Arg Gly Pro His Gly
Gly Ala Ala Ser 245 250 255Gly Leu Asn Gly Cys Cys Arg Cys Gly Ala
Arg Gly Pro Glu Ser Arg 260 265 270Leu Leu Glu Phe Tyr Leu Ala Met
Pro Phe Ala Thr Pro Met Glu Ala 275 280 285Glu Leu Ala Arg Arg Ser
Leu Ala Gln Asp Ala Pro Pro Leu Pro Val 290 295 300Pro Gly Val Leu
Leu Lys Glu Phe Thr Val Ser Gly Asn Ile Leu Thr305 310 315 320Ile
Arg Leu Thr Ala Ala Asp His Arg Gln Leu Gln Leu Ser Ile Ser 325 330
335Ser Cys Leu Gln Gln Leu Ser Leu Leu Met Trp Ile Thr Gln Cys Phe
340 345 350Leu Pro Val Phe Leu Ala Gln Pro Pro Ser Gly Gln Arg Arg
355 360 36515867DNAArtificial SequencehFlt3L-NY-ESO-1 80-180aa
15atggatgcta tgaaacgagg actgtgttgt gtgttgcttc tgtgtggagc ggtttttgtg
60agtcccactc aggattgcag cttccagcat tcacccatat catcagattt tgcagtaaag
120atcagggaac tctccgatta tctccttcaa gactaccccg taacagtggc
ctccaatttg 180caagacgaag agctttgtgg tgccctctgg cggctcgttt
tggcccaaag gtggatggaa 240cggcttaaga cagtcgctgg cagcaagatg
caagggttgc tcgaacgagt caatacagag 300atccattttg taaccaagtg
tgcatttcaa ccgccgccaa gctgccttcg ctttgttcag 360acgaatataa
gtagactgtt gcaggaaacc tccgagcaac tcgtagccct gaagccctgg
420attacacggc aaaatttcag tcggtgcctt gagcttcagt gtcagcctga
tagtagtacc 480ttgcctccgc catggtcccc caggcctctt gaagctacag
ctccgacagc ccctcagccg 540ggcagtagtg gtagttctgg agccaggggg
ccggagagcc gcctgcttga gttctacctc 600gccatgcctt tcgcgacacc
catggaagca gagctggccc gcaggagcct ggcccaggat 660gccccaccgc
ttcccgtgcc aggggtgctt ctgaaggagt tcactgtgtc cggcaacata
720ctgactatcc gactgactgc tgcagaccac cgccaactgc agctctccat
cagctcctgt 780ctccagcagc tttccctgtt gatgtggatc acgcagtgct
ttctgcccgt gtttttggct 840cagcctccct cagggcagag gcgctaa
86716288PRTArtificial SequencehFlt3L-NY-ESO-1 80-180aa 16Met Asp
Ala Met Lys Arg Gly Leu Cys Cys Val Leu Leu Leu Cys Gly1 5 10 15Ala
Val Phe Val Ser Pro Thr Gln Asp Cys Ser Phe Gln His Ser Pro 20 25
30Ile Ser Ser Asp Phe Ala Val Lys Ile Arg Glu Leu Ser Asp Tyr Leu
35 40 45Leu Gln Asp Tyr Pro Val Thr Val Ala Ser Asn Leu Gln Asp Glu
Glu 50 55 60Leu Cys Gly Ala Leu Trp Arg Leu Val Leu Ala Gln Arg Trp
Met Glu65 70 75 80Arg Leu Lys Thr Val Ala Gly Ser Lys Met Gln Gly
Leu Leu Glu Arg 85 90 95Val Asn Thr Glu Ile His Phe Val Thr Lys Cys
Ala Phe Gln Pro Pro 100 105 110Pro Ser Cys Leu Arg Phe Val Gln Thr
Asn Ile Ser Arg Leu Leu Gln 115 120 125Glu Thr Ser Glu Gln Leu Val
Ala Leu Lys Pro Trp Ile Thr Arg Gln 130 135 140Asn Phe Ser Arg Cys
Leu Glu Leu Gln Cys Gln Pro Asp Ser Ser Thr145 150 155 160Leu Pro
Pro Pro Trp Ser Pro Arg Pro Leu Glu Ala Thr Ala Pro Thr 165 170
175Ala Pro Gln Pro Gly Ser Ser Gly Ser Ser Gly Ala Arg Gly Pro Glu
180 185 190Ser Arg Leu Leu Glu Phe Tyr Leu Ala Met Pro Phe Ala Thr
Pro Met 195 200 205Glu Ala Glu Leu Ala Arg Arg Ser Leu Ala Gln Asp
Ala Pro Pro Leu 210 215 220Pro Val Pro Gly Val Leu Leu Lys Glu Phe
Thr Val Ser Gly Asn Ile225 230 235 240Leu Thr Ile Arg Leu Thr Ala
Ala Asp His Arg Gln Leu Gln Leu Ser 245 250 255Ile Ser Ser Cys Leu
Gln Gln Leu Ser Leu Leu Met Trp Ile Thr Gln 260 265 270Cys Phe Leu
Pro Val Phe Leu Ala Gln Pro Pro Ser Gly Gln Arg Arg 275 280
28517828DNAArtificial SequencehFlt3L-NY-ESO various peptides
17atggatgcta tgaaacgagg actgtgttgt gtgttgcttc tgtgtggagc ggtttttgtg
60agtcccactc aggattgcag cttccagcat tcacccatat catcagattt tgcagtaaag
120atcagggaac tctccgatta tctccttcaa gactaccccg taacagtggc
ctccaatttg 180caagacgaag agctttgtgg tgccctctgg cggctcgttt
tggcccaaag gtggatggaa 240cggcttaaga cagtcgctgg cagcaagatg
caagggttgc tcgaacgagt caatacagag 300atccattttg taaccaagtg
tgcatttcaa ccgccgccaa gctgccttcg ctttgttcag 360acgaatataa
gtagactgtt gcaggaaacc tccgagcaac tcgtagccct gaagccctgg
420attacacggc aaaatttcag tcggtgcctt gagcttcagt gtcagcctga
tagtagtacc 480ttgcctccgc catggtcccc caggcctctt gaagctacag
ctccgacagc ccctcagccg 540ggcagtagtg gtagttctgg aaggggacct
gagtcaaggc tgctcgaatt ctacttggca 600atgccttttg cgacccctat
gctgcttgaa ttttaccttg caatgccctt cgccaccccc 660atggaagcag
aactcgcccg gcgatccttg gctcagagcc tcttgatgtg gataactcaa
720tgcagtttgt tgatgtggat aactcaatgc tttctgcccg ttttttggat
aacgcaatgc 780tttctcccgg tgtttctcgc tcaaccccct agcgggcaaa ggcgctaa
82818275PRTArtificial SequencehFlt3L-NY-ESO various peptides 18Met
Asp Ala Met Lys Arg Gly Leu Cys Cys Val Leu Leu Leu Cys Gly1 5 10
15Ala Val Phe Val Ser Pro Thr Gln Asp Cys Ser Phe Gln His Ser Pro
20 25 30Ile Ser Ser Asp Phe Ala Val Lys Ile Arg Glu Leu Ser Asp Tyr
Leu 35 40 45Leu Gln Asp Tyr Pro Val Thr Val Ala Ser Asn Leu Gln Asp
Glu Glu 50 55 60Leu Cys Gly Ala Leu Trp Arg Leu Val Leu Ala Gln Arg
Trp Met Glu65 70 75 80Arg Leu Lys Thr Val Ala Gly Ser Lys Met Gln
Gly Leu Leu Glu Arg 85 90 95Val Asn Thr Glu Ile His Phe Val Thr Lys
Cys Ala Phe Gln Pro Pro 100 105 110Pro Ser Cys Leu Arg Phe Val Gln
Thr Asn Ile Ser Arg Leu Leu Gln 115 120 125Glu Thr Ser Glu Gln Leu
Val Ala Leu Lys Pro Trp Ile Thr Arg Gln 130 135 140Asn Phe Ser Arg
Cys Leu Glu Leu Gln Cys Gln Pro Asp Ser Ser Thr145 150 155 160Leu
Pro Pro Pro Trp Ser Pro Arg Pro Leu Glu Ala Thr Ala Pro Thr 165 170
175Ala Pro Gln Pro Gly Ser Ser Gly Ser Ser Gly Arg Gly Pro Glu Ser
180 185 190Arg Leu Leu Glu Phe Tyr Leu Ala Met Pro Phe Ala Thr Pro
Met Leu 195 200 205Leu Glu Phe Tyr Leu Ala Met Pro Phe Ala Thr Pro
Met Glu Ala Glu 210 215 220Leu Ala Arg Arg Ser Leu Ala Gln Ser Leu
Leu Met Trp Ile Thr Gln225 230 235 240Cys Ser Leu Leu Met Trp Ile
Thr Gln Cys Phe Leu Pro Val Phe Trp 245 250 255Ile Thr Gln Cys Phe
Leu Pro Val Phe Leu Ala Gln Pro Pro Ser Gly 260 265 270Gln Arg Arg
2751966DNAArtificial SequenceTranslational modifier Exon skipping
motif P2A 19ggatctgggg ccaccaactt ttcattgctc aagcaggcgg gcgatgtgga
ggaaaaccct 60ggcccc 662063DNAArtificial SequenceTranslational
modifier Exon skipping motif T2A 20ggaagcggag agggcagagg aagtctgcta
acatgcggtg acgtcgagga gaatcctggc 60cca 632169DNAArtificial
SequenceTranslational modifier Exon skipping motif E2A 21ggaagcggac
agtgtactaa ttatgctctc ttgaaattgg ctggagatgt tgagagcaac 60cctggacct
692275DNAArtificial SequenceTranslational modifier Exon skipping
motif F2A 22ggcagcggcg tgaagcagac cctgaacttc gacctgctga agctggccgg
cgacgtggag 60agcaaccccg gcccc 7523568DNAArtificial
SequenceTranslational modifier Internal Ribosomal Entry Sequence
(IRES) 23tccgcccccc cccctaacgt tactggccga agccgcttgg aataaggccg
gtgtgcgttt 60gtctatatgt tattttccac catattgccg tcttttggca atgtgagggc
ccggaaacct 120ggccctgtct tcttgacgag cattcctagg ggtctttccc
ctctcgccaa aggaatgcaa 180ggtctgttga atgtcgtgaa ggaagcagtt
cctctggaag cttcttgaag acaaacaacg 240tctgtagcga ccctttgcag
gcagcggaac cccccacctg gcgacaggtg cctctgcggc 300caaaagccac
gtgtataaga tacacctgca aaggcggcac aaccccagtg ccacgttgtg
360agttggatag ttgtggaaag agtcaaatgg ctctcctcaa gcgtattcaa
caaggggctg 420aaggatgccc agaaggtacc ccattgtatg ggatctgatc
tggggcctcg gtgcacatgc 480tttacatgtg tttagtcgag gttaaaaaaa
cgtctaggcc ccccgaacca cggggacgtg 540gttttccttt gaaaaacacg atgataat
5682490DNAArtificial SequenceOvalbumin aa 241-270 24tccatgcttg
tgttgttgcc tgacgaggtg tccgggttgg agcagttgga aagcataata 60aacttcgaga
agttgactga atggacataa 902529PRTArtificial SequenceOvalbumin aa
241-270 25Ser Met Leu Val Leu Leu Pro Asp Glu Val Ser Gly Leu Glu
Gln Leu1 5 10 15Glu Ser Ile Ile Asn Phe Glu Lys Leu Thr Glu Trp Thr
20 25
References
-
cta.lncc.br/index.phpMAGE-A2FusionoffulllegthproteinorAlmeidaetal
-
cta.lncc.br/index.phpMAGE-A3FusionoffulllegthproteinorAlmeidaetal
-
cta.lncc.br/index.phpMAGE-A10FusionoffulllegthproteinorAlmeidaetal
-
cta.lncc.br/index.phpSSX-2FusionoffulllegthproteinorAlmeidaetal
-
cta.lncc.br/index.phpMART-1FusionoffulllengthproteinorLietal
-
cta.lncc.br/index.phpB7-H6FulllengthproteinorfusionofBrandtetal
D00000

D00001

D00002

D00003

D00004

D00005

D00006

D00007

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.