Method For Detecting An Antigen-specific Antibody And Reagent For Use Therein
FUTAMI; Junichiro ; et al.
U.S. patent application number 17/029206 was filed with the patent office on 2021-02-04 for method for detecting an antigen-specific antibody and reagent for use therein. This patent application is currently assigned to Junichiro FUTAMI. The applicant listed for this patent is Junichiro FUTAMI, Medinet Co., Ltd.. Invention is credited to Junichiro FUTAMI, Kazuhiro KAKIMI, Ryuji MAEKAWA, Masato SHIRAKI.
| Application Number | 20210032302 17/029206 |
| Document ID | / |
| Family ID | 1000005150200 |
| Filed Date | 2021-02-04 |











View All Diagrams
| United States Patent Application | 20210032302 |
| Kind Code | A1 |
| FUTAMI; Junichiro ; et al. | February 4, 2021 |
METHOD FOR DETECTING AN ANTIGEN-SPECIFIC ANTIBODY AND REAGENT FOR USE THEREIN
Abstract
A method for detecting an antigen-specific antibody detection reagent, which includes a cationized, denatured antigenic protein immobilized on a solid-phase surface of a carrier material, is provided. The cationized, denatured antigenic protein has cationized SH groups formed from reaction with a cationizing agent. The detection reagent is capable of specifically binding to an antigen-specific antibody which binds to an epitope of the antigenic protein. The method includes contacting a sample containing the antigen-specific antibody with the detection reagent to bind the antigen-specific antibody with the detection reagent. A labeled secondary antibody is then added to allow the labeled secondary antibody to bind to the antigen-specific antibody; and the detection reagent bound with the antigen-specific antibody is detected.
| Inventors: | FUTAMI; Junichiro; (Okayama, JP) ; KAKIMI; Kazuhiro; (Tokyo, JP) ; MAEKAWA; Ryuji; (Tokyo, JP) ; SHIRAKI; Masato; (Kanagawa, JP) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Assignee: | Junichiro FUTAMI Okayama JP Medinet Co., Ltd. Kanagawa JP |
||||||||||
| Family ID: | 1000005150200 | ||||||||||
| Appl. No.: | 17/029206 | ||||||||||
| Filed: | September 23, 2020 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 14389016 | Sep 29, 2014 | 10822384 | ||
| PCT/JP2013/059692 | Mar 29, 2013 | |||
| 17029206 | ||||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | G01N 2333/47 20130101; C07K 1/113 20130101; C07K 14/4748 20130101; G01N 33/57492 20130101; G01N 33/543 20130101; G01N 33/574 20130101; G01N 2333/4704 20130101; C07K 14/4703 20130101; G01N 33/6854 20130101; G01N 33/57426 20130101 |
| International Class: | C07K 14/47 20060101 C07K014/47; G01N 33/574 20060101 G01N033/574; G01N 33/68 20060101 G01N033/68; G01N 33/543 20060101 G01N033/543; C07K 1/113 20060101 C07K001/113 |
Foreign Application Data
| Date | Code | Application Number |
|---|---|---|
| Mar 30, 2012 | JP | 2012-082735 |
Claims
1. A method for detecting an antigen-specific antibody contained in a sample, the method comprising the steps of: contacting the sample with a reagent for detection of antibody binding to an epitope of an antigenic protein to bind the antigen-specific antibody with the reagent for detection; wherein the reagent for detection comprises a carrier material and a cationized, denatured antigenic protein immobilized on a solid-phase surface of the carrier material; wherein the cationized, denatured antigenic protein has cationized SH groups formed from reaction with a cationizing agent; adding thereto a labeled secondary antibody to allow the labeled secondary antibody to bind to the antigen-specific antibody; and detecting the reagent for detection bound with the antigen-specific antibody.
2. The method of claim 1, wherein the cationized, denatured antigenic protein is a cationized, denatured full-length protein.
3. The method of claim 1, wherein the cationized, denatured antigenic protein is a cationized, denatured membrane protein.
4. The method of claim 1, wherein the cationized, denatured antigenic protein is a cationized, denatured cancer antigenic protein.
5. The method of claim 1, wherein the carrier material is a membrane, wafer, microplate or bead.
6. The method of claim 1, wherein the carrier material is a magnetic microbead.
7. The method of claim 1, wherein the solid-phase surface is a glass, nylon or semiconductor surface.
8. The method of claim 1, wherein the carrier material is a carboxylated polystyrene bead.
9. The method of claim 1, wherein the cationized, denatured antigenic protein is immobilized through a reaction of an antigenic protein amino group with an activated carboxylic acid group on the solid-phase surface.
10. The method of claim 1, wherein the cationized, denatured antigenic protein is indirectly immobilized on the solid-phase surface.
11. The method of claim 10, wherein the cationized, denatured antigenic protein is immobilized indirectly on the solid-phase surface via a biotin-avidin bond.
12. The method of claim 1, wherein the cationized, denatured antigenic protein has cationized SH groups formed from reaction with an alkyl halide cationizing agent.
13. The method of claim 12, wherein the alkyl halide cationizing agent is (3-bromopropyl)-trimethylammonium (TAP-Br).
14. The method of claim 1, wherein the cationized, denatured antigenic protein has cationized SH groups formed from reaction with a cationizing agent comprising a thiosulfonate compound, a mixed disulfide compound, a pyridyl sulfide cationizing agent, or a mixture of two of more thereof.
15. The method of claim 1, wherein the cationizing agent is trimethylammoniopropyl methanethiosulfonate (TAPS-sulfonate).
16. A reagent for detection of antibody binding to an epitope of an antigenic protein comprising a carrier material and a cationized, denatured antigenic protein immobilized on a solid-phase surface of the carrier material; wherein the cationized, denatured antigenic protein has cationized SH groups formed from reaction with a cationizing agent.
17. The reagent of claim 16, wherein the carrier material is a membrane, wafer, microplate or bead.
18. The reagent of claim 16, wherein the cationizing agent comprises a thiosulfonate compound, a mixed disulfide compound, a pyridyl sulfide cationizing agent, alkyl halide cationizing agent or a mixture of two of more thereof.
19. The reagent of claim 16, wherein the cationized, denatured antigenic protein is immobilized indirectly on the solid-phase surface via a biotin-avidin bond.
20. The reagent of claim 16, wherein the cationized, denatured antigenic protein is irreversibly immobilized and indirectly bound to the solid-phase surface via a linker molecule.
Description
REFERENCE TO RELATED APPLICATIONS
[0001] This application is a continuation of U.S. application Ser. No. 14/389,016, filed on Sep. 29, 2014; which is a U.S. national stage of international application PCT/JP2013/059692, filed on Mar. 29, 2013; which claims priority to Japanese patent application no. JP 2012-082735, filed on Mar. 30, 2012; the disclosures of which are herein incorporated by reference in their entirety.
BACKGROUND
[0002] The present application relates to a method for producing a reagent for antibody detection. The present application also relates to a reagent for antibody detection produced by the method and use thereof.
[0003] There exist a large number of methods for detecting antibodies contained in liquid samples, for example, radioimmunoassay and enzyme-linked immunosorbent assay (ELISA). ELISA is a method which involves: immobilizing particular antigens onto a microplate; after serial dilution of an antibody-containing sample, performing antigen-antibody reaction on the microplate; and detecting the bound antibody using an enzyme-labeled secondary antibody. This method requires evaluating antibody tests directed to individual antigens using separate assay plates.
[0004] As an approach for solving this problem, a multiplex technique has received attention, which can simultaneously analyze antibodies against diverse antigens by use of microbeads bearing reporter fluorescence as a carrier for immobilization.
[0005] In the application of any of these techniques, such diverse antigens must be prepared in water-soluble forms. Proteins of native structures that can be prepared in water-soluble forms or chemically synthesized polypeptide fragments have conventionally been used in most cases.
[0006] For example, a method for detecting an antibody contained in the blood of a cancer patient is described in Japanese Patent No. 3960614 (JP 3960614--Immunodia Co., Ltd.) or Japanese Patent Publication JP 2005-098877A (Hitachi Software Engineering Co., Ltd.). This method involves immobilizing an antigen epitope peptide (antigenic peptide composed of several amino acids) onto beads; contacting the beads with the blood components of a subject; and detecting antigen epitope peptide-specific antibody contained in the blood of the subject. The epitope portion to which the antibody binds, however, differs depending on the type of HLA. Use of the method described in JP 3960614 or JP 2005-098877A therefore requires clearing various conditions such as the examination of the HLA type of the subject and the identification of an epitope peptide appropriate for the HLA type of the subject.
[0007] For preparing a detection reagent for an antibody against a particular antigenic protein, the reagent to be prepared comprises all epitope portions derived from one type of antigenic protein on the surface of one type of bead and performs highly sensitive and stable detection. For this purpose, it is preferred to obtain an antigen having a water-soluble and flexible structure. Nonetheless, most of denatured proteins, poorly soluble proteins (e.g., membrane proteins), or proteins having unstable physical properties tend to aggregate. In this respect, partial peptides capable of exhibiting stable physical properties have conventionally been used in most cases.
[0008] Use of such partial peptides requires synthesizing diverse overlapping peptides for covering all epitopes and also requires preparing many types of beads. In addition, it is practically difficult to provide seamless epitope peptides. Even if a full-length antigen having a native structure can be obtained luckily, a general protein, which has a higher-order structure with a hydrophobic moiety buried in the interior, does not always expose its epitope to react with an antibody.
[0009] Even in a reagent for antibody detection prepared using solubilized proteins, thiol groups contained in the proteins might form a disulfide bond over time and thereby influence the antibody detection.
[0010] For example, Japanese Patent No. 3225468 (Dainabot Co., Ltd.) describes a method for detecting an anti-HCV antibody contained in the serum of a subject by use of the long-chain polypeptide of human hepatitis C virus (HCV). Japanese Patent No. 3225468 states that an intraprotein or interprotein disulfide bond generated over time reduces antibody detection sensitivity. The solution to this problem described therein is to dissociate the intraprotein or interprotein disulfide bond using a reducing agent before or during detection, thereby improving the antibody detection sensitivity. Since even the solubilized proteins might precipitate over time, some approach is necessary for solving this problem.
[0011] TAPS-sulfonate (trimethylammoniopropyl methanethiosulfonate; hereinafter, referred to as TAPS) is known as a compound that solubilizes proteins. TAPS can bind to thiol groups in a protein to reversibly cationize the protein (see e.g., M. Seno et al., Growth Factors, 15, 215-229 (1998) and M. Inoue et al., Biotechnol. Appl. Biochem., 28, 207-213 (1998)).
[0012] The cationized protein exhibits improved solubility in water. Since the binding of TAPS to the protein is reversible reaction, TAPS is known to dissociate from the protein upon cellular uptake so that the protein can exert its original functions as a result of refolding. Nonetheless, no attempt has been made so far on the process of preparing a reagent for antibody detection by use of solubilization using TAPS. Also, antibodies are generally known to bind to glycosylated proteins. No previous report, however, shows whether an antibody can bind to a protein bound with an artificially synthesized compound such as TAPS.
SUMMARY
[0013] The present reagent and methods have been made in light of the circumstances mentioned above. The present application provides a method for efficiently producing a reagent for the detection of an antibody present in a liquid sample, the antibody specifically binding to a poorly soluble antigenic protein. The present application also provides a reagent for antibody detection produced by the production method and use thereof.
Solution to Problem
[0014] Specifically, an object of the present application is to provide the following aspects:
[0015] (1) A method for producing a reagent for antibody detection comprising an antigenic protein and a carrier, the method comprising the steps of: solubilizing the antigenic protein by cationization; and allowing the cationized antigenic protein to bind to the carrier;
[0016] (2) The method for producing a reagent for antibody detection according to (1), wherein the antigenic protein is a full-length protein;
[0017] (3) The method for producing a reagent for antibody detection according to (1) or (2), wherein the antigenic protein is a membrane protein;
[0018] (4) The method for producing a reagent for antibody detection according to any one of (1) to (3), wherein the antigenic protein is a cancer antigenic protein;
[0019] (5) The method for producing a reagent for antibody detection according to any one of
[0020] (1) to (4), wherein the cationization is performed by the binding of a cationizing agent to thiol groups of the antigenic protein.
[0021] (6) The method for producing a reagent for antibody detection according to (5), wherein the cationizing agent is selected from any one of a thiosulfonate compound, a mixed disulfide compound, a pyridyl sulfide cationizing agent, and an alkyl halide cationizing agent, and mixtures thereof;
[0022] (7) The method for producing a reagent for antibody detection according to (6), wherein the thiosulfonate compound is a compound represented by the following formula:
##STR00001##
[0023] wherein R.sup.1 represents a linear alkylene group having 2 to 20 carbon atoms; R.sup.2 represents an alkyl group having 1 to 3 carbon atoms; and n represents any integer of 1 to 3.
[0024] (8) The method according to (7), wherein the compound is TAPS-sulfonate wherein R.sup.1 is --(CH.sub.2)3-; R.sup.2 is CH.sub.3--; and n is 1;
[0025] (9) The method for producing a reagent for antibody detection according to (7), wherein the compound is TAP3S-sulfonate wherein R.sup.1 is --(CH.sub.2).sub.3--; R.sup.2 is CH.sub.3; and n is 3;
[0026] (10) The method for producing a reagent for antibody detection according to (6), wherein the alkyl halide cationizing agent is TAP-Br;
[0027] (11) The method for producing a reagent for antibody detection according to any one of (1) to (10), wherein the carrier is magnetic beads;
[0028] (12) A reagent for antibody detection produced by a method according to any one of (I) to (11);
[0029] (13) A method for detecting an antigen-specific antibody contained in a sample, the 10 method comprising the steps of: contacting a reagent for antibody detection according to (12) with the sample; adding thereto an antibody-binding labeled secondary antibody to allow the secondary antibody to bind to the antibody; recovering the reagent for antibody detection; and detecting the reagent for antibody detection bound with the antibody; and
[0030] (14) The method according to (13), wherein the sample is an isolated body fluid.
[0031] The present application describes that a reagent for antibody detection that is intended to detect an antibody against an antigenic protein can be produced by the configuration as described above.
Advantageous Effects
[0032] A cationizing agent can be used in an antigenic protein solubilization step to thereby efficiently solubilize and recover antigenic proteins. Hence, a reagent for antibody detection comprising a large number of antigenic protein molecules bound with a carrier can be efficiently produced, compared with conventional methods.
[0033] Furthermore, this reagent for antibody detection is much more stable than reagents produced by the conventional methods and can thus be stored for a long period.
[0034] In one aspect of the present application, poorly soluble antigenic proteins can be used as antigens for antibody detection. Use of such antigenic proteins permits detection of antibodies even if the antigenic proteins, because of their difference in HLA type, differ in epitope portions which can be recognized by the antibodies. This eliminates the need of producing a plurality of reagents according to the HLA type of a subject and can provide an efficient production method, compared with the methods involving the immobilization of epitope peptides on beads.
[0035] In addition, the cationizing agent bound with thiol groups in an antigenic protein can inhibit the time-dependent generation of an intraprotein or interprotein disulfide bond. This can be expected to be effective for preventing reagents for antibody detection from aggregating over time through interprotein disulfide bonds. Thus, the reagents for antibody detection can maintain their functions, compared with the conventional methods, even when poorly soluble antigenic proteins are bound with a carrier or even after long-term storage at room temperature or at 4.degree. C. or -20.degree. C.
[0036] On the other hand, such artificially synthesized compounds bound with antigenic proteins might hinder the antigenic proteins from binding to antibodies. However, the present application reveals that antibodies can be detected using antigenic proteins even bound with cationizing agents.
[0037] According to these features, a reagent for antibody detection can be efficiently produced by use of the production method of the present application. The reagent for antibody detection produced by the production method of the present application can detect an antibody (against an antigenic protein) present in a liquid sample and as such, can detect a cancer antigenic protein-specific antibody from a serum sample, for example, regardless of the HLA type of a cancer patient.
[0038] TAPS-sulfonate or TAP-Br, in particular, has a low molecular weight. This compound can therefore minimize steric hindrance that inhibits protein-antibody reaction, while maintaining its high solubilizing ability. This low molecular weight also facilitates the binding of a plurality of its molecules to a protein. As a result, all SH groups contained in the protein can be cationized. This can prevent beads from aggregating during long-term storage. By virtue of these features, the method of the present application using TAPS-sulfonate or TAP-Br is more effective than the conventional methods.
BRIEF DESCRIPTION OF DRAWINGS
[0039] FIG. 1 is a diagram illustrating protein cationization.
[0040] FIG. 2 is a diagram showing results of SDS-PAGE analysis after cationization of WT-1 with TAPS.
[0041] FIG. 3 is a diagram showing results of SDS-PAGE analysis after cationization of MAGE-A4 with TAPS.
[0042] FIG. 4 is a diagram showing results of HPLC purification of TAPS-bound WT-1.
[0043] FIG. 5 is a diagram showing results of HPLC purification of TAPS-bound MAGE-A4.
[0044] FIG. 6 is a diagram showing results of SDS-PAGE analysis after HPLC purification of TAPS-bound WT-1 and MAGE-A4. Min represents an elution time in the HPLC purification (FIG. 4 or 5). For example, the lane indicated by "49 min" depicts the migration of fractions collected for 1 minute from the elution time of 49 minutes.
[0045] FIG. 7 is a diagram showing results of SDS-PAGE analysis of prepared antigenic proteins. Each lane was charged with 2 .mu.g of each antigenic protein.
[0046] FIG. 8 is a diagram showing results of detecting antibodies against 6 types of cancer antigenic proteins contained in the serum of cancer patients using a reagent for antibody detection produced by the production method of the present application.
[0047] FIG. 9 is a diagram showing results of detecting antibodies against 3 types of cancer antigenic proteins contained in the serum of cancer patients using a reagent for antibody detection produced by the production method of the present application.
[0048] FIG. 10 is a diagram showing results of detecting antibodies against 3 types of cancer antigenic proteins contained in the serum of cancer patients using a reagent for antibody detection produced by the production method of the present application.
[0049] FIG. 11 is a diagram showing results of reverse-phase HPLC (high-performance liquid chromatography) performed on TAPS-bound, TAP3 S-bound, native, and reduced lysozymes.
[0050] FIG. 12 is a diagram showing the percentages of bead aggregates contained in suspensions of TAPS-bound, TAP3S-bound, native, and reduced MAGE-A4-immobilized beads stored at 4.degree. C. for 5 days or at 37.degree. C. for 10 days or 41 days.
[0051] FIG. 13 is a diagram showing results of Western blotting performed on storage solutions of TAPS-bound MAGE-A4-immobilized beads (lane 2), TAP3S-bound MAGE-A4-immobilized beads (lane 3), and native MAGE-A4-immobilized beads (lane 4) stored for 104 days. Lane 1 depicts the migration of 100 ng of the MAGE-A4 protein as a control.
[0052] FIG. 14 provides images showing the detection of proteins bound with the surface of Bio-Plex COOH beads, TAPS-bound MAGE-A4-immobilized beads stored for 104 days, TAP3S-bound MAGE-A4-immobilized beads stored for 104 days, and native MAGE-A4-immobilized beads stored for 104 days. The left images are images taken by a confocal laser scanning fluorescence microscope. The right images are differential interference images (bright field) taken at the same time therewith.
DESCRIPTION OF EMBODIMENTS
[0053] Hereinafter, embodiments of the present reagent and methods will be described.
[0054] First, the method for producing a reagent for antibody detection according to the present application (hereinafter, also referred to as the production method of the present application) will be described.
[0055] The production method of the present application is a method for producing a reagent for antibody detection, comprising the steps of: solubilizing the antigenic protein by cationization; and allowing the cationized antigenic protein to bind to the carrier. The antigenic protein may be a poorly soluble protein.
[0056] In the present specification, the term "poorly soluble" is provided merely for illustrating a property of the protein and refers to the property of not forming a uniform mixed solution by dissolution in a liquid, particularly, the property of being impossible or difficult to dissolve in water or a physiological solvent. The poorly soluble proteins of types described herein can be defined as poorly soluble proteins when substantially the majorities thereof are recovered into precipitated fractions after centrifugation at 15,100.times.g for 1 hour of the proteins in water, in water and a salt, or in water and a physiological solvent that does not denature the proteins.
[0057] The "protein" described herein includes peptides, polypeptides, and the like. The protein is not limited to naturally occurring proteins that are found in nature and also encompasses recombinant proteins derived from cells transformed by gene transfer or the like, proteins expressed using an in vitro cell-free protein expression system, and synthetic proteins prepared in a synthetic organic chemistry manner. Alternatively, a functional group may be added to a portion or the whole of amino acids constituting the protein in such a way that the amino acid(s) is acetylated, phosphorylated, or methylated, or a portion or the whole of amino acids constituting the protein may be modified with a sugar, a protein, a lipid, or the like.
[0058] The "poorly soluble protein" described herein refers to a protein that is impossible or difficult to dissolve even by stirring in water or a physiological solvent at room temperature and may be dissolved by use of, for example, a denaturant but forms precipitates as a result of the replacement of the denaturant with a physiological solvent. Even in the case of a protein that is soluble in nature, the protein of interest is also expressed as a poorly soluble protein when this protein of interest is recovered in the form of an inclusion body by the expression thereof as a recombinant protein using an organism of different species (e.g., by the construction of an expression system in E. coli using a gene recombination technique). Examples of the poorly soluble protein include, but are not limited to, full-length proteins, membrane proteins, and cancer antigenic proteins.
[0059] In the present specification, the term "solubilization" refers to the dissolution of a protein in a physiological solvent with its amino acid sequence maintained. The term "solubilization" means that when a solution containing a protein dissolved with a denaturant is centrifuged after replacement with a physiological solvent, the amount of the protein recovered into a supernatant is increased after the centrifugation.
[0060] The full-length proteins mean not only natural proteins confirmed to exist in vivo but a protein encoded by the largest open reading frame (ORF) predicted from the genomic sequence. The amino acid sequences of such full-length proteins can be obtained from, but not limited to, database, for example, The ORFeome Collaboration (http://www.orfeomecollaboration.org/) or GeneCards(R) (http://www.genecards.org/).
[0061] The membrane proteins refer to proteins having a transmembrane structure. These proteins are present at the surface of cell membranes, nuclear envelopes, and other intracellular organelles. One protein molecule contains a hydrophilic moiety which is within the cell or is in contact with the outside of the cell, and a hydrophobic moiety which is buried in the cell membrane. For this reason, these membrane proteins, when obtained by expression in E. coli or the like, rarely form a three-dimensional structure in an aqueous solution and tend to form an inclusion body.
[0062] The cancer antigenic proteins refer to antigenic proteins that are expressed in tumors and induce immune response or antigenic proteins that can serve as an index for the presence of tumors. Examples of the cancer antigenic proteins include proteins that are expressed at increased levels by the malignant transformation of cells, and proteins having one or some amino acids thereof mutated as a result of the malignant transformation of cells.
[0063] A feature of the production method of the present invention is to cationize a protein. More preferably, a feature of the production method of the present invention is to cationize a poorly soluble protein.
[0064] The cationization of the protein refers to the addition of excessive positive charges to the protein. The cationized protein exhibits improved solubility in water owing to charge repulsion. Examples of an approach for the protein cationization include the binding of a cationizing agent to a protein.
[0065] The "carrier" refers to a material having solid-phase surface to which the antigenic protein is to be bound. Specific examples thereof include, but are not limited to, glass, nylon membranes, semiconductor wafers, and microbeads.
[0066] The binding of the antigenic protein to the carrier means that the "antigenic protein" is immobilized directly onto the surface of the carrier using a technique known in the art. Alternatively, the antigenic protein may be immobilized indirectly thereonto via, for example, a biotin-avidin bond or via a linker molecule.
[0067] The "sample" refers to a test piece containing the antibody (including subtypes such as IgG, IgA, IgM, IgD, and IgE) and an active fragment thereof (including e.g., Fab and F(ab').sub.2 fragments) to be detected by the reagent for antibody detection according to the present application.
[0068] The "body fluid" refers to a sample in a liquid state that can be collected from an organism. The body fluid corresponds to peripheral blood, bone marrow fluid, cord blood, pleural fluid, ascitic fluid, urine, and the like. The body fluid also corresponds to samples obtained by the treatment of these body fluids according to methods well known to those skilled in the art (e.g., plasma or serum obtained from a supernatant by the centrifugation of peripheral blood).
[0069] The "secondary antibody" refers to an antibody and an active fragment thereof that recognizes the antibody (including subtypes such as IgG, IgA, IgM, IgD, and IgE) and an active fragment thereof (including e.g., Fab and F(ab'h fragments) to be detected by the reagent for antibody detection according to the present invention. The "labeled secondary antibody" refers to a secondary antibody bound with a label such as a radioisotope, a luminescent agent, or a fluorophore. Alternatively, a protein such as an enzyme (such as luciferase or peroxidase), biotin or green fluorescent protein (GFP) may be used as the "label". In the case of such a "label" derived from the protein, the "labeled secondary antibody" may be prepared in a genetic engineering manner as one recombinant protein in the form of a fusion protein.
[0070] The cationizing agent that can be used in the production method of the present invention can be any of various compounds that can add positive charges to the protein via disulfide bonds (FIG. 1). For example, a thiosulfonate compound, a mixed disulfide compound, a pyridyl disulfide cationizing agent, or an alkyl halide cationizing agent can be used.
[0071] The thiosulfonate compound used in the production method of the present invention is a compound represented by Formula 2 given below. In this formula, X represents a group having a cation. One group having a cation represented by X may be used, or a linkage of the groups represented by X may be used. In the formula, R.sup.2 represents a lower alkyl group having 1 to 3 carbon atoms. Specifically, the thiosulfonate compound used in the production method of the present application is a thiosulfonate compound having one or more cations derived from X in one molecule. Examples of the group represented by X include quaternary ammonium groups.
##STR00002##
[0072] The thiosulfonate compound used in the production method of the present invention is a thiosulfonate compound represented by Formula 3 given below having one or more quaternary ammonium group-derived cations in one molecule.
##STR00003##
wherein R.sup.1 represents a linear or branched alkylene group having 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 carbon atoms; R.sup.2 represents a lower alkyl group having 1, 2, or 3 carbon atoms; and n represents any integer of 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10, more preferably any integer of 1, 2, and 3.
[0073] Examples of features of the thiosulfonate compound include an inert dissociable group formed as the dissociable group R.sup.2 after protein cationization reaction as illustrated in FIG. 1.
[0074] Examples of the compound containing one quaternary ammonium group-derived cation in one molecule include trimethylammoniopropyl methanethiosulfonate (TAPS-sulfonate; hereinafter, referred to as TAPS). Examples of the compound containing three quaternary ammonium group-derived cations in one molecule include TAP3S-sulfonate (hereinafter, referred to as TAP3S).
[0075] TAPS refers to a compound represented by [Formula 4] given below. TAPS has a strongly positively charged quaternary amine in the molecule and cationizes a protein through cysteine residue-mediated binding to the protein. The cationized protein exhibits improved solubility and contains stably protected SH groups. TAPS can be synthesized with reference to, for example, Biotechnol. Appl. Biochem. (1998) 28, 207-213 or purchased as a reagent in the form of Br salt (represented by Formula 5 given below) (molecular weight: 292.26) (from Wako Pure Chemical Industries, Ltd., Katayama Chemical, Ltd., etc.).
##STR00004##
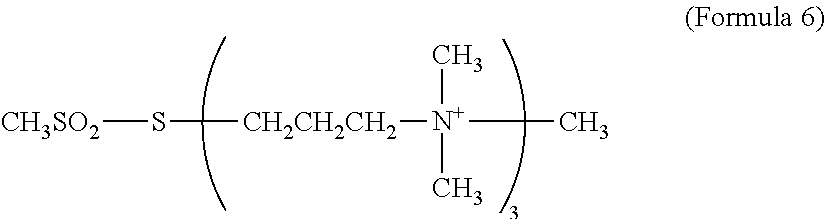
[0076] TAP3S refers to a molecular-weight compound represented by Formula 6 given below. TAP3 S has 3 strongly positively charged quaternary amines in the molecule and can more strongly cationize a protein than TAPS. TAP3S can be synthesized with reference to, for example, International Publication No. WO 2011/118731.
##STR00005##
[0077] Examples of the mixed disulfide compound used in the production method of the present invention include cystamine as represented by Formula 7 given below. In the case of cystamine, NH.sub.3.sup.+ is the group having a cation.
##STR00006##
[0078] Examples of the pyridyl sulfide cationizing agent used in the production method of the present invention include compounds as represented by Formula 8 given below. X represents a group having a cation. One group having a cation represented by X may be used, or a linkage of the groups represented by X may be used.
##STR00007##
[0079] The cationizing agent that can be used m the production method of the present application may be an alkyl halide cationizing agent that can add positive charges to a protein through S-alkylation. For example, (3-bromopropyl)trimethylammonium (TAP-Br) can be used. TAP-Br irreversibly binds SH groups in a protein and thereby improves its protein-solubilizing ability. TAP-Br (molecular weight: 261.00) can be purchased, for example, as a reagent in the form of Br salt (represented by Formula 9) (e.g., from Sigma-Aldrich Corp.).
##STR00008##
[0080] Various cationizing agents can be used in the production method of the present invention. TAPS-sulfonate or TAP-Br, in particular, has a low molecular weight. This compound can therefore minimize steric hindrance that inhibits protein-antibody reaction, while maintaining its high solubilizing ability. This low molecular weight also facilitates the binding of a plurality of its molecules to a protein. As a result, all SH groups contained in the protein can be cationized. This can prevent beads from aggregating during long-term storage.
[0081] Hereinafter, the steps of preparing an antigenic protein and preparing a TAPS-bound antigenic protein using TAPS will be described as one example of the obtainment of a cationized antigenic protein.
[0082] A gene of the antigenic protein is transferred to E. coli, which is then cultured. The cultured E. coli is homogenized to recover the antigenic protein. In this recovery, the antigenic protein may be in the form of an inclusion body.
[0083] The recovered inclusion body is solubilized with a denaturant. Examples of the denaturant used include, but are not limited to, urea, guanidine hydrochloride, and surfactants.
[0084] The antigenic protein thus denatured is treated with a reducing agent to cleave the SS bonds. Examples of the reducing agent include, but are not limited to, dithiothreitol (DTT) and 2-mercaptoethanol (2ME).
[0085] TAPS is added to the solution containing the denatured antigenic protein, and the mixture is left standing at room temperature for 30 minutes. The amount of TAPS added is preferably a TAPS molar concentration of I to 10 times, more preferably 1.1 to 1.2 times the molar concentration of thiol groups contained in the antigenic protein and the solution.
[0086] After 30 minutes, polyethyleneimine having an average molecular weight of 600 is added at a final concentration of 0.2% to the solution containing the TAPS-bound antigenic protein. Then, 10% acetic acid is added thereto in 4 times the amount of the resulting solution. The solution containing the TAPS-bound antigenic protein is centrifuged to recover a supernatant.
[0087] Subsequently, the TAPS-bound antigenic protein contained in the supernatant is purified. In the purification, a method such as dialysis or column chromatography can be used. For example, the supernatant is transferred to a dialysis tube and dialyzed against pure water or up to 0.5% acetic acid at 4.degree. C. to remove the denaturant. The dialyzed liquid is centrifuged at 12,000 rpm at 4.degree. C. to room temperature for 15 minutes to recover a supernatant.
[0088] The cationized antigenic protein is obtained through these steps.
[0089] The cationized antigenic protein exhibits high water solubility, is easily prepared, and exhibits very high stability under weakly acidic conditions (preferably pH 6 or lower, more preferably pH 2 to 5). The cationized antigenic protein therefore can maintain its water solubility in a state where epitopes in its full-length protein are seamlessly exposed. For this reason, all epitopes contained in one antigenic protein can be immobilized on one type of bead so as to facilitate the reaction of the epitopes with antibodies.
[0090] Next, the TAPS-bound protein is allowed to bind to a carrier.
[0091] In one aspect, magnetic beads, non-magnetic beads, a microplate, or the like can be used as the carrier. Magnetic beads are preferably used in terms of convenient analysis operation. Hereinafter, a case using the beads as the carrier will be described.
[0092] The beads are suspended in a solution for binding. In the case where the beads have already been modified to help the beads bind to the protein, the beads are bound directly to the TAPS-bound protein. In the absence of such modification to help the beads bind to the protein, the beads are modified with a material, such as skimmed milk, which does not inhibit the reaction of the TAPS-bound protein with an antibody during antibody detection.
[0093] The solution of the TAPS-bound protein is mixed with the suspension of the beads. For example, 2 to 16 hours are preferred as the mixing time of beads activated into a state reactive with amino groups.
[0094] The beads are recovered, then suspended in a solution for washing, and washed by centrifugation. Examples of the solution for washing include phosphate buffer solutions. For storing the beads thus washed, the beads are suspended in a buffer for storage and stored. Examples of the buffer for storage include a storage buffer available from Bio-Rad Laboratories, Inc. A weakly acidic (preferably pH 6 or lower, more preferably pH 2 to 5) buffer for storage more stably maintains the solubility of the TAPS-bound protein.
[0095] The reagent for antibody detection can be produced through these steps.
[0096] Next, an antibody detection method will be described in detail as one aspect using the reagent for antibody detection produced by the production method of the present application (hereinafter, referred to as the reagent of the present application).
[0097] The reagent of the present invention may be a reagent for antibody detection comprising a cationized poorly soluble antigenic protein and beads.
[0098] A sample presumed to contain an antibody is obtained from a subject. A body fluid can be used as the sample. Peripheral blood, bone marrow fluid, cord blood, pleural fluid, ascitic fluid, urine, or the like can be used as the body fluid. Peripheral blood is preferably used in consideration of easy collection and a small burden on the subject. The amount of the sample collected is preferably an amount that puts no heavy burden on the subject. The peripheral blood can be collected by use of a whole blood collection method using a vacuum blood collection tube, a blood collection bag, or the like. For blood collection, heparin or the like may be added in order to prevent the coagulation of blood.
[0099] Plasma is collected from the collected blood by centrifugation. Alternatively, the plasma may be obtained during the partial collection of blood by use of an apheresis apparatus.
[0100] Also, peripheral blood may be collected, and serum can be obtained by the removal of blood cell components and blood clotting components and also used as the sample.
[0101] For antibody detection, the plasma or the serum is diluted, if necessary.
[0102] The reagent of the present invention is mixed with the plasma. The mixing time is preferably approximately 2 hours, for example, for reaction at room temperature or overnight reaction at 4.degree. C. The reagent of the present invention is recovered using a centrifugation method or a magnetic apparatus and washed.
[0103] There may be apprehension that the cationization influences antigen-antibody reaction. In such a case, the influence can be easily tested, because the cationization can be canceled by treatment with a reducing agent such as dithiothreitol.
[0104] For antibody detection, a secondary antibody is allowed to bind to the antibody. In the case of, for example, a human subject, the antibody bound with the reagent of the present invention is a human antibody. Thus, a labeled anti-human antibody is used as the secondary antibody for labeling. Examples thereof include biotinylated anti-human IgG mouse monoclonal antibodies.
[0105] The beads bound with the secondary antibody are detected using a flow cytometer. Bio-Plex beads (Bio-Rad Laboratories, Inc.), in which beads themselves are stained, may be used in the detection. In such a case, plural types of beads can be applied simultaneously to an apparatus. Thus, plural types of antibodies can be analyzed by one operation. This can shorten the analysis time.
[0106] An antibody can also be detected from other liquid samples using the reagent of the present invention in the same way as described in the above paragraphs except that the plasma is changed to the liquid samples such as serum.
[0107] The analysis of an antibody contained in the blood of a subject according to these embodiments presumably achieves the following situations: [0108] The allergenic reactivity of the subject is determined by the analysis of in vivo antibodies in the subject; [0109] An antibody that correlates with therapeutic effects or a progress after treatment is found by antibody analysis conducted before and after the treatment. The antibody thus found is used as an index to decide a therapeutic strategy or to predict or determine prognosis; [0110] For immune cell therapy, an antigen whose specific antibody has been detected in the serum of a patient is used in the treatment to thereby improve therapeutic effects; [0111] Before and after radiotherapy, antibodies are analyzed to thereby predict therapeutic effects reportedly involving immune functions, such as abscopal effects.
[0112] Although the effects mentioned above may be attained by a reagent for antibody detection using an epitope peptide, the reagent of the present invention is superior in that proteins can be used. This is because use of the proteins allows antibodies to be detected without being limited by the HLA type of a subject. As a result, even an HLA type that is generally regarded as being minor and thus less understood in epitope peptide analysis can be analyzed.
[0113] Hereinafter, the present invention will be described in detail with reference to Examples. However, the present invention is not intended to be limited by these Examples by any means, as a matter of course.
Example 1
[0114] <Study on Large-Scale Culture and Preparation Methods for MAGE-A4 and WT-1>
[0115] First, each antigenic protein for use in antibody detection was prepared.
[0116] E. coli T7 Express was transformed with each of 2 types of plasmid DNAs of His-tag-fused MAGE-A4 (SEQ ID NOs: 16 and 17, antigenic protein sequence: SEQ ID NOs: 3 and 4) and His-tag-fused WT-1 (SEQ ID NOs: 14 and 15, antigenic protein sequence: SEQ ID NOs: 1 and 2) cloned into pET28b vectors. Several colonies formed on a plate were picked up, then added to IO mL of an LB/Kan25 medium, and shake-cultured for 2 hours.
[0117] Next, a small amount of the cultures was added to 400 mL of a TB medium and further cultured at 37.degree. C. When the bacterial cell concentration reached OD600=0.7 to 0.8, IPTG was added thereto at a final concentration of 0.5 mM. The cells were further cultured at 37.degree. C. for 3 hours.
[0118] The bacterial cells were dispersed using a sonicator in 40 mL of a Iysis butter (20 mM Tris-HCl (pH 8.0), 50 mM NaCl, 5 mM MgSO4, and 0.2% Tween 20) and further homogenized by 1 minute.times.3 sets.
[0119] To the solution containing the bacterial cell homogenates, 1 .mu.L of a strong nuclease Benzonase (HC) was added, and the mixture was left standing at room temperature for approximately 15 minutes.
[0120] For WT-1, precipitates (inclusion body) were recovered by centrifugation (8 krpm, 10 to 15 min, room temperature), while the supernatant was removed. To the recovered precipitates, 50 to 100 mL of RO water was added, and the precipitates were resuspended using a sonicator and recovered by centrifugation (8 krpm, 10 to 15 min, room temperature).
[0121] For MAGE-A4, a supernatant was recovered by centrifugation (8 krpm, 10 to 15 min, room temperature).
<Solubilization of WT-I by Reversible Cationization Method of Denatured Precipitated Fraction>
[0122] The precipitates were dissolved in 5 to 10 mL of 6 M guanidine and 0.1 M Tris-HCl (pH 8)+1 mM EDTA. After deaeration and nitrogen substitution, 30 mM DTT (solid) was added thereto, followed by treatment at 37.degree. C. for approximately I hour.
[0123] To the solution containing WT-1, 70 mM TAPS-sulfonate was added, followed by treatment at 37.degree. C. for approximately 30 minutes.
[0124] To the solution containing WT-1 and TAPS-sulfonate, acetic acid in an amount of 1/10 thereof and 0.1% PEI600 were added, and the mixture was then well dialyzed against Milli-Q water at 4.degree. C. After the dialysis, SDS-PAGE was performed. The results are shown in FIG. 2. The lane indicated by sup depicts the migration of the solution thus dialyzed. The lane indicated by ppt depicts the migration of a sample obtained as an insoluble fraction after the dialysis. The lane indicated by "Reduced" depicts the migration of the protein separated from TAPS by the addition of a reducing agent to the sample. The lane indicated by "Non-reduced" depicts the migration of the TAPS-bound protein without the addition of a reducing agent to the sample. The SDS-PAGE analysis results shown in FIG. 2 demonstrated that WT-1 was successfully TAPS-bound and solubilized.
<His-Tag Purification of MAGE-A4 Using Co.sup.2 Column>
[0125] A Co.sup.2 column was equilibrated with solution A(20 mM Tris-HCl (pH 8.0), 50 mM NaCl, and 0.2% Tween 20). Then, the supernatant of MAGE-A4 was adsorbed onto the resin. Then, the resin was thoroughly washed with solution A Next, 30 mL of solution A+5 mM imidazole was injected thereto. After addition of 4 mL of solution A+200 mM imidazole, the column was left standing for 5 minutes, followed by protein elution. This operation was repeated five times to recover purified MAGE-A4.
<Binding of TAPS to MAGE-A4>
[0126] MAGE-A4 thus affinity-purified was added to an eggplant-shaped flask, then dehydrated using a freeze dryer, and dissolved in 3 mL of 6 M guanidine and 0.1 M Tris-Hct (pH 8). After deaeration and nitrogen substitution, 30 mM DTT (solid) was added thereto, followed by treatment at 37.degree. C. for approximately I hour. To the solution containing MAGE-A4, 70 mM TAPS-sulfonate was added, followed by treatment at 37.degree. C. for approximately 30 minutes.
[0127] To the solution containing MAGE-A4 and TAPS-sulfonate, acetic acid in an amount of 1/10 thereof and 0.1% PEI600 were added, and the mixture was then well dialyzed against Milli-Q water at 4.degree. C. After the dialysis, SDS-PAGE was performed. The results are shown in FIG. 3.
[0128] <HPLC Purification>
[0129] TAPS-bound MAGE-A4 and WT-1 were purified using reverse-phase HPLC. The chromatograms are shown in FIGS. 4 and 5.
[0130] After HPLC, TAPS-bound MAGE-A4 and WT-I were analyzed by SDS-PAGE. The results are shown in FIG. 6. The analysis results demonstrated that a highly pure antigenic protein can be prepared by the reverse-phase HPLC purification of the TAPS-bound protein. Fractions were recovered as MAGE-A4 at elution times of 50 minutes to 51 minutes (lane "S0min" in FIG. 6), while fractions were recovered as WT-1 at elution times of 52 minutes to 53 minutes (lane "52 min" in FIG. 6). These fractions were used in the subsequent steps.
[0131] His-tag-fused NY-ESO-1 (SEQ ID NOs: 18 and 19, antigenic protein sequence: SEQ ID NOs: 5 and 6), XAGElb (SEQ ID NOs: 20 and 21, antigenic protein sequence: SEQ ID NOs: 7 and 8), gp100 (SEQ ID NOs: 22 and 23, antigenic protein sequence: SEQ ID NOs: 9 and 10), Survivin-2B (SEQ ID NOs: 24 and 25, antigenic protein sequence: SEQ ID NOs: 11 and 12) were prepared by the same method as that for WT-1. The prepared proteins were analyzed by SDS-PAGE. The results are shown in FIG. 7.
<Binding to Bio-Plex COOH Beads (Manufactured by Bio-Rad Laboratories, Inc.)>
[0132] The antigens were immobilized onto 6 types of Bio-Plex COOH beads (amount of 1/10 (1.25.times.10.sup.6 beads)) using Bio-Plex amine coupling kit (manufactured by Bio-Rad Laboratories, Inc.). All of the antigens used were antigens in a denatured state solubilized by TAPS binding (11 .mu.g each).
[0133] Each solubilized protein was dissolved in a buffer and left standing on ice.
[0134] The container of the COOH beads was shaken for 30 seconds with a vortex mixer, and the beads were suspended by sonication for 30 seconds.
[0135] The COOH bead suspension was centrifuged at 14,000.times.g for 4 minutes to remove a supernatant.
[0136] To the recovered precipitates, 100 .mu.L of a bead wash buffer was added. The mixture was shaken for 10 seconds with a vortex mixer, and the beads were suspended by sonication for 10 seconds. The COOH bead suspension was centrifuged at 14,000.times.g for 4 minutes to remove a supernatant.
[0137] To the recovered precipitates, 80 .mu.L of a bead activation buffer was added. The mixture was well shaken for 30 seconds with a vortex mixer, and the beads were suspended by sonication for 30 seconds.
[0138] To the COOH bead suspension, 10% of 50 mg/mL EDAC was added, and then, 10 .mu.L of 50 mg/mL S-NHS (N-hydroxysulfosuccinimide) was added. The COOH bead suspension was shaken for 30 seconds with a vortex mixer.
[0139] While the beads were shielded from light, the COOH bead suspension was shaken for 20 minutes in a rotary incubator.
[0140] To the COOH bead suspension, 150 .mu.L of PBS was added, and the mixture was shaken for 10 seconds with a vortex mixer. The COOH bead suspension was centrifuged at 14,000.times.g for 4 minutes to remove a supernatant. This operation was repeated again.
[0141] To the recovered precipitates, 100 .mu.L of PBS was added. The mixture was shaken for 30 seconds with a vortex mixer, and the beads were suspended by sonication for 15 seconds.
[0142] The COOH bead suspension was mixed with the TAPS-bound protein suspension. The total amount of the mixture was adjusted to 500 .mu.L by the addition of PBS.
[0143] While the beads were shielded from light, the COOH bead suspension was shaken at room temperature for 2 hours in a rotary incubator.
[0144] The suspension containing the COOR beads and the TAPS-bound protein was centrifuged at 14,000.times.g for 4 minutes to remove a supernatant.
[0145] The COOH beads (on which the TAPS-bound protein was immobilized) recovered as precipitates were washed by the addition of 500 .mu.L of PBS. The COOH bead suspension was centrifuged at 14,000.times.g for 4 minutes to remove a supernatant.
[0146] The COOH beads recovered as precipitates were resuspended by the addition of 250 .mu.L of a blocking buffer. The COOR bead suspension was shaken for 15 seconds with a vortex mixer.
[0147] While the COOH beads were shielded from light using an aluminum foil, the COOH bead suspension was shaken at room temperature for 30 minutes in a rotary incubator. The COOH bead suspension was centrifuged at 14,000.times.g for 4 minutes to remove a supernatant.
[0148] The COOH beads recovered as precipitates were resuspended by the addition of 500 .mu.L of a buffer for storage. The COOH bead suspension was centrifuged at 16,000.times.g for 6 minutes to remove a supernatant.
[0149] The TAPS-bound protein-immobilized beads thus obtained were suspended in 100 .mu.L of a storage buffer and stored therein. Table 1 shows the concentration of each type of bead.
TABLE-US-00001 TABLE 1 Bead concentration Cancer antigenic protein Bead No. (the number of beads/mL) NY-ESO-1 26 4.33 .times. 10.sup.6 XAGE1b 28 5.59 .times. 10.sup.6 MAGE-A4 43 3.68 .times. 10.sup.6 WT-1 45 1.95 .times. 10.sup.6 gp100 62 6.09 .times. 10.sup.6 Survivin-2B 64 1.61 .times. 10.sup.6
Example 2
<Analysis of Antibody Contained in Serum of Cancer Patient-1>
[0150] Blood was collected from two cancer patients (Donor 1 and Donor 2), and serum was obtained therefrom.
[0151] Six types of magnetic beads on which the antigenic proteins (NY-ESO-1, WT-I, MAGE-A4, XAGElb, gp100, and Survivin-2B) were respectively immobilized were mixed at the same concentrations and dispensed to the wells of a 96-well plate (Bio-Rad Laboratories, Inc., #171-025001). The serum of each patient was added to each well. While shielded from light using an aluminum foil, the plate was shaken at room temperature for 1 hour for reaction. The beads were washed with Wash Station (Bio-Rad Laboratories, Inc.). For antibody detection, biotinylated anti-human IgG (H+L) (manufactured by Vector Laboratories, Inc., BA-3000) was added as a secondary antibody to each well. While shielded from light using an aluminum foil, the plate was shaken at room temperature for 30 minutes for reaction. The beads were washed with Wash Station (Bio-Rad Laboratories, Inc.). (R-)Phycoerythrin (PE)-labeled streptavidin (Vector Laboratories, Inc.) was added to each well. While shielded from light using an aluminum foil, the plate was shaken at room temperature for 10 minutes for reaction. The beads were washed with Wash Station (Bio-Rad Laboratories, Inc.) and analyzed using Bio-Plex (Bio-Rad Laboratories, Inc.). The serum of each patient used was diluted 400-fold, 1600-fold, and 6400-fold.
[0152] As shown in FIG. 8, the sample derived from Donor 1 was confirmed to respond to NY-ES0-1.
[0153] This result suggested that: the blood of Donor 1 contained an antibody recognizing NY-ES0-1; cancer in Donor 1 expressed NY-ES0-1; and peptide vaccine or DC vaccine therapy using NY-ES0-1 was possibly effective for Donor I.
[0154] As a result of this test, the sample derived from Donor 2 was confirmed to respond to XAGElb.
[0155] This result suggested that: the blood of Donor 2 contained an antibody recognizing XAGEib; cancer in Donor 2 expressed XAGElb; and peptide vaccine or DC vaccine therapy using XAGElb was possibly effective for Donor 2.
Example 3
<Analysis of Antibody Contained in Serum of Cancer Patient-2>
[0156] Serum was obtained from 8 renal cell cancer patients (Donor 2 to Donor 9) before and after EP-DC therapy.
[0157] The EP-DC therapy refers to a treatment method which involves: preparing lysates by the freeze-thaw method or the like from tumor tissues removed by surgery from a cancer patient; electroloading the lysates into dendritic cells; and administering the dendritic cell vaccine thus prepared to the patient.
[0158] Six types of magnetic beads on which the antigenic proteins were respectively immobilized were mixed at the same concentrations and dispensed to the wells of a 96-well plate (Bio-Rad Laboratories, Inc., #I 71-025001). The serum of each patient was added to each well. While shielded from light using an aluminum foil, the plate was shaken at room temperature for 1 hour for reaction. The beads were washed with Wash Station (Bio-Rad Laboratories, Inc.). For antibody detection, biotinylated anti-human IgG (H+L) (manufactured by Vector Laboratories, Inc., BA-3000) was added as a secondary antibody to each well. While shielded from light using an aluminum foil, the plate was shaken at room temperature for 30 minutes for reaction. The beads were washed with Wash Station (Bio-Rad Laboratories, Inc.). PE-labeled streptavidin (Vector Laboratories, Inc.) was added to each well. While shielded from light using an aluminum foil, the plate was shaken at room temperature for 10 minutes for reaction. The beads were washed with Wash Station (Bio-Rad Laboratories, Inc.) and analyzed using an assay apparatus of Bio-Plex (Bio-Rad Laboratories, Inc.). The serum of each patient used was diluted 400-fold, 1600-fold, and 6400-fold.
[0159] As shown in FIG. 9, the serum of Donor 6 and Donor 8 was confirmed to respond to MAGE-A4. Accordingly, the blood of Donor 6 and Donor 8 contained an antibody recognizing MAGE-A4, suggesting that the in vivo tumor tissues of the patients Donor 6 and Donor 8 possibly expressed MAGE-A4. In this case, peptide vaccine or DC vaccine therapy using MAGE-A4 was presumably appropriate for these patients.
[0160] As shown in FIG. 10, the serum of Donor 3 was confirmed to respond more highly to WT-1 after DC vaccine therapy compared with before the therapy. Accordingly, the blood of Donor 3 contained an antibody recognizing WT-1, suggesting that the tumor tissues of Donor 3 possibly expressed WT-1. In this case, peptide vaccine or DC vaccine therapy using WT-1 was presumably appropriate for this patient.
Example 4
[0161] <Evaluation of Cationizing Reagent for its Ability to Protect SH Group>
[0162] Chicken egg white lysozyme (SEQ ID NO: 13; hereinafter, also simply referred to as lysozyme; manufactured by Kewpie Corp.) was used as a sample to evaluate the ability of a cationizing reagent to protect SH groups.
[0163] 15 mg of the lysozyme was weighed into a 100-mL pear-shaped flask and dissolved in 2 mL of 6 M guanidine hydrochloride, 0.1 M Tris-HCl (pH 8.5), and 2 mM EDTA. After deaeration and nitrogen substitution of the obtained solution, dithiothreitol (DTT) was added thereto at a final concentration of 30 mM, and the mixture was reacted for 2 hours in a thermostat bath of 37.degree. C. To the obtained reduced lysozyme, each cationizing reagent (TAPS-sulfonate (manufactured by Katayama Chemical, Ltd.) or TAP3 S-sulfonate) was added at a final concentration of 70 mM, and the mixture was further reacted at room temperature for 2 hours. The reaction was stopped by the addition of acetic acid in 1/10 of the amount of the obtained solution. Then, the solution was dialyzed against Milli-Q water at 4.degree. C. for 1 day to obtain TAPS-bound lysozyme and TAP3S-bound lysozyme. The TAP3S-sulfonate used was synthesized according to the approach described in Japanese Patent Application No. 2010-070804.
[0164] TAPS-bound, TAP3S-bound, native, and reduced lysozymes were subjected to reverse--phase HPLC (high-performance liquid chromatography). The native lysozyme contains four SS bonds in one molecule, and has hydrophilicity to some extent because its hydrophobic groups are not exposed. The reduced lysozyme is a denatured protein with its SS bonds reduced and has the highest hydrophobicity (lowest solubility in water). The column used was COSMOSIL Protein-R (manufactured by Nacalai Tesque, Inc.). The solvent used was acetonitrile diluted with 0.1% hydrochloric acid, and the acetonitrile concentration was set to 1% to 50%.
[0165] The results summarizing reverse-phase HPLC charts are shown in FIG. 11. The straight line represents the concentrations of acetonitrile. Peaks positioned closer to the left side mean lower hydrophobicity (higher solubility in water) of samples. The TAP3S-bound lysozyme was more hydrophilic than the native and reduced lysozymes and was eluted with a low-concentration acetonitrile solvent. TAP3S, however, has the difficulty in protecting and cationizing all of the 8 SH groups in the lysozyme and showed peak dispersion due to contamination by imperfect products in which a portion of SH groups was unprotected. By contrast, in the TAPS-bound lysozyme, all of the 8 SH groups were protected and cationized, demonstrating high homogeneity. Such a difference in the ability to protect SH groups between TAPS and TAP3S is attributed to the sizes of their molecules. This is presumably because, in the case of protecting somewhat dense SH groups on a protein molecule, the large TAP3S molecule causes steric hindrance on the protein molecule to be cationized, resulting in a trace amount of residual SH groups incapable of binding to TAP3S; thus, the subsequent SH/SS exchange reaction proceeds slowly.
Example 5
<Evaluation of Storage Stability of Beads>
[0166] When a reagent for antibody detection is prepared and stored in a state where the SH groups of the protein are incompletely protected, it is possible that unprotected residual SH groups form SS bonds to cause the aggregation of proteins or the aggregation of reagents for antibody detection. Thus, the storage stability of the reagent for antibody detection was confirmed by the following procedures.
[0167] According to the procedures described in Example 1, TAPS-bound, TAP3S-bound, or native MAGE-A4 was prepared, and beads on which each protein was immobilized were prepared.
[0168] The prepared beads were suspended in a buffer for storage and stored under conditions of 4.degree. C. or 37.degree. C. After a lapse of given time, the abundance of bead aggregates among the beads in the suspension was measured using a hemocytometer. The percentage of bead aggregates shown in the results was calculated according to the number of aggregates each involving 3 or more beads/the total number of beads x 100.
[0169] FIG. 12 shows the percentage of bead aggregates contained in each suspension stored at 4.degree. C. for 5 days or at 37.degree. C. for 10 days or 41 days. In the case of storage at 4.degree. C., the percentage of bead aggregates was decreased in the stored TAPS-bound or TAP3S-bound MAGE-A4-immobilized beads compared with the stored native MAGE-A4-immobilized beads. In the case of storage at 37.degree. C., the percentage of bead aggregates was decreased in the stored TAPS-bound MAGE-A4-immobilized beads compared with the stored native MAGE-A4-immobilized beads.
[0170] Next, the bead storage solution was subjected to SOS-PAGE in order to confirm that this decrease in the percentage of bead aggregates was not attributed to the liberation of the immobilized antigenic protein from the beads.
[0171] MAGE-A4 immobilized on the beads was N-terminally His-tagged. Thus, liberated MAGE-A4 proteins in the storage solution of the MAGE-A4-immobilized beads were detected by Western blotting using an anti-His-tag antibody (OGHis, manufactured by Medical & Biological Laboratories Co., Ltd. (MBL)).
[0172] The results are shown in FIG. 13. The storage solutions of the TAPS-bound MAGE-A4-immobilized beads, the TAP3 S-bound MAGE-A4-immobilized beads, and the native MAGE-A4-immobilized beads stored for I 04 days were used as the samples migrated in lanes 2, 3, and 4, respectively. Free proteins were detected at the same levels among all of these samples, demonstrating the absence of the phenomenon in which cationization facilitates the liberation of antigenic proteins from beads.
[0173] In addition, bound proteins were detected as to Bio-Plex COOH beads, the TAPS-bound MAGE-A4-immobilized beads stored for 104 days, the TAP3S-bound MAGE-A4-immobilized beads stored for 104 days, and the native MAGE-A4-immobilized beads stored for 104 days.
[0174] An anti-His-tag antibody (OGHis, manufactured by Medical & Biological Laboratories Co., Ltd. (MBL)) diluted to 200 ng/mL in PBS was mixed with the beads of each type at room temperature for 30 minutes. Then, the beads were washed twice with PBS. Next, an anti-mouse IgG-Alexa 488 (secondary antibody; Invitrogen Corp.) diluted to 200 ng/mL in PBS was mixed with the beads of each type at room temperature for 30 minutes. Then, the beads were washed twice with PBS. These beads were observed under a confocal laser scanning fluorescence microscope (excitation light: 488 nm, fluorescence filter LP505) to visualize the presence of the His-tagged MAGE-A4 protein on the surface of the beads. The taken images are shown in FIG. 14. All of the images were taken with the same detection sensitivity. As shown in FIG. 14, the MAGE-A4 protein was confirmed to be bound with the surface of all of the beads.
[0175] The results of Example 5 demonstrated that the beads prepared by the method for producing a reagent for antibody detection disclosed in the present invention are more effective for inhibiting the formation of aggregates than beads prepared using a native protein. This inhibition of the formation of aggregates was also confirmed to be not attributed to the liberation of the antigenic protein from the bead surface. TAP3S is generally known to be superior in protein-solubilizing ability to TAPS. Unlike this solubilizing ability, however, the inhibitory effects on the aggregation of prepared beads were shown to be higher in TAPS.
Example 6
[0176] <Protein Cationization Using TAP-Br>
[0177] XAGEl b or NY-ES0-1 affinity-purified as described above was added to an eggplant-shaped flask, then dehydrated using a freeze dryer, and dissolved in 3 mL of 6 M guanidine and 0.1 M Tris-HCl (pH 8). After deaeration and nitrogen substitution, 30 mM OTT (solid) was added thereto, followed by treatment at 37.degree. C. for approximately 1 hour. To the solution containing XAGElb or NY-ESO-1, 70 mM TAP-Br was added, followed by treatment at 37.degree. C. for approximately 60 minutes.
[0178] To the solution containing XAGElb or NY-ESO-1 and TAP-Br, acetic acid in an amount of 1/10 thereof was added, and the mixture was then well dialyzed against Milli-Q water at 4.degree. C. TAP-bound XAGE lb or NY-ESO-1 was purified by reverse-phase HPLC.
[0179] Beads on which TAP-bound XAGElb, TAPS-bound XAGElb, TAP-bound NY-ESO-1, and TAPS-bound NY-ESO-1 were respectively immobilized were prepared by the method described in Example 1. The TAP-bound antigenic protein was also confirmed to be successfully immobilized on the beads, as with the TAPS-bound antigenic protein.
[0180] All publications and patents cited herein are incorporated herein by reference in their entirety.
INDUSTRIAL APPLICABILITY
[0181] As described above, a reagent for antibody detection comprising an antigenic protein can be efficiently produced by use of the production method of the present invention. The reagent for antibody detection produced by the production method of the present invention was confirmed to be highly stable and also capable of efficiently detecting an antibody in a liquid sample. Use of the reagent of the present invention can provide an antibody analysis test that is free from the constraints of the HLA type of a subject. This test can probably be carried out, for example, to thereby decide a therapeutic strategy for a disease involving the immunity or to thereby predict or determine therapeutic effects on the disease.
Sequence CWU 1
1
2511554DNAHomo sapiens 1atgcaggacc cggcttccac gtgtgtcccg gagccggcgt
ctcagcacac gctccgctcc 60gggcctgggt gcctacagca gccagagcag cagggagtcc
gggacccggg cggcatctgg 120gccaagttag gcgccgccga ggccagcgct
gaacgtctcc agggccggag gagccgcggg 180gcgtccgggt ctgagccgca
gcaaatgggc tccgacgtgc gggacctgaa cgcgctgctg 240cccgccgtcc
cctccctggg tggcggcggc ggctgtgccc tgcctgtgag cggcgcggcg
300cagtgggcgc cggtgctgga ctttgcgccc ccgggcgctt cggcttacgg
gtcgttgggc 360ggccccgcgc cgccaccggc tccgccgcca cccccgccgc
cgccgcctca ctccttcatc 420aaacaggagc cgagctgggg cggcgcggag
ccgcacgagg agcagtgcct gagcgccttc 480actgtccact tttccggcca
gttcactggc acagccggag cctgtcgcta cgggcccttc 540ggtcctcctc
cgcccagcca ggcgtcatcc ggccaggcca ggatgtttcc taacgcgccc
600tacctgccca gctgcctcga gagccagccc gctattcgca atcagggtta
cagcacggtc 660accttcgacg ggacgcccag ctacggtcac acgccctcgc
accatgcggc gcagttcccc 720aaccactcat tcaagcatga ggatcccatg
ggccagcagg gctcgctggg tgagcagcag 780tactcggtgc cgcccccggt
ctatggctgc cacaccccca ccgacagctg caccggcagc 840caggctttgc
tgctgaggac gccctacagc agtgacaatt tataccaaat gacatcccag
900cttgaatgca tgacctggaa tcagatgaac ttaggagcca ccttaaaggg
agttgctgct 960gggagctcca gctcagtgaa atggacagaa gggcagagca
accacagcac agggtacgag 1020agcgataacc acacaacgcc catcctctgc
ggagcccaat acagaataca cacgcacggt 1080gtcttcagag gcattcagga
tgtgcgacgt gtgcctggag tagccccgac tcttgtacgg 1140tcggcatctg
agaccagtga gaaacgcccc ttcatgtgtg cttacccagg ctgcaataag
1200agatatttta agctgtccca cttacagatg cacagcagga agcacactgg
tgagaaacca 1260taccagtgtg acttcaagga ctgtgaacga aggttttctc
gttcagacca gctcaaaaga 1320caccaaagga gacatacagg tgtgaaacca
ttccagtgta aaacttgtca gcgaaagttc 1380tcccggtccg accacctgaa
gacccacacc aggactcata caggtaaaac aagtgaaaag 1440cccttcagct
gtcggtggcc aagttgtcag aaaaagtttg cccggtcaga tgaattagtc
1500cgccatcaca acatgcatca gagaaacatg accaaactcc agctggcgct ttga
15542517PRTHomo sapiens 2Met Gln Asp Pro Ala Ser Thr Cys Val Pro
Glu Pro Ala Ser Gln His1 5 10 15Thr Leu Arg Ser Gly Pro Gly Cys Leu
Gln Gln Pro Glu Gln Gln Gly 20 25 30Val Arg Asp Pro Gly Gly Ile Trp
Ala Lys Leu Gly Ala Ala Glu Ala 35 40 45Ser Ala Glu Arg Leu Gln Gly
Arg Arg Ser Arg Gly Ala Ser Gly Ser 50 55 60Glu Pro Gln Gln Met Gly
Ser Asp Val Arg Asp Leu Asn Ala Leu Leu65 70 75 80Pro Ala Val Pro
Ser Leu Gly Gly Gly Gly Gly Cys Ala Leu Pro Val 85 90 95Ser Gly Ala
Ala Gln Trp Ala Pro Val Leu Asp Phe Ala Pro Pro Gly 100 105 110Ala
Ser Ala Tyr Gly Ser Leu Gly Gly Pro Ala Pro Pro Pro Ala Pro 115 120
125Pro Pro Pro Pro Pro Pro Pro Pro His Ser Phe Ile Lys Gln Glu Pro
130 135 140Ser Trp Gly Gly Ala Glu Pro His Glu Glu Gln Cys Leu Ser
Ala Phe145 150 155 160Thr Val His Phe Ser Gly Gln Phe Thr Gly Thr
Ala Gly Ala Cys Arg 165 170 175Tyr Gly Pro Phe Gly Pro Pro Pro Pro
Ser Gln Ala Ser Ser Gly Gln 180 185 190Ala Arg Met Phe Pro Asn Ala
Pro Tyr Leu Pro Ser Cys Leu Glu Ser 195 200 205Gln Pro Ala Ile Arg
Asn Gln Gly Tyr Ser Thr Val Thr Phe Asp Gly 210 215 220Thr Pro Ser
Tyr Gly His Thr Pro Ser His His Ala Ala Gln Phe Pro225 230 235
240Asn His Ser Phe Lys His Glu Asp Pro Met Gly Gln Gln Gly Ser Leu
245 250 255Gly Glu Gln Gln Tyr Ser Val Pro Pro Pro Val Tyr Gly Cys
His Thr 260 265 270Pro Thr Asp Ser Cys Thr Gly Ser Gln Ala Leu Leu
Leu Arg Thr Pro 275 280 285Tyr Ser Ser Asp Asn Leu Tyr Gln Met Thr
Ser Gln Leu Glu Cys Met 290 295 300Thr Trp Asn Gln Met Asn Leu Gly
Ala Thr Leu Lys Gly Val Ala Ala305 310 315 320Gly Ser Ser Ser Ser
Val Lys Trp Thr Glu Gly Gln Ser Asn His Ser 325 330 335Thr Gly Tyr
Glu Ser Asp Asn His Thr Thr Pro Ile Leu Cys Gly Ala 340 345 350Gln
Tyr Arg Ile His Thr His Gly Val Phe Arg Gly Ile Gln Asp Val 355 360
365Arg Arg Val Pro Gly Val Ala Pro Thr Leu Val Arg Ser Ala Ser Glu
370 375 380Thr Ser Glu Lys Arg Pro Phe Met Cys Ala Tyr Pro Gly Cys
Asn Lys385 390 395 400Arg Tyr Phe Lys Leu Ser His Leu Gln Met His
Ser Arg Lys His Thr 405 410 415Gly Glu Lys Pro Tyr Gln Cys Asp Phe
Lys Asp Cys Glu Arg Arg Phe 420 425 430Ser Arg Ser Asp Gln Leu Lys
Arg His Gln Arg Arg His Thr Gly Val 435 440 445Lys Pro Phe Gln Cys
Lys Thr Cys Gln Arg Lys Phe Ser Arg Ser Asp 450 455 460His Leu Lys
Thr His Thr Arg Thr His Thr Gly Lys Thr Ser Glu Lys465 470 475
480Pro Phe Ser Cys Arg Trp Pro Ser Cys Gln Lys Lys Phe Ala Arg Ser
485 490 495Asp Glu Leu Val Arg His His Asn Met His Gln Arg Asn Met
Thr Lys 500 505 510Leu Gln Leu Ala Leu 5153954DNAHomo sapiens
3atgtcttctg agcagaagag tcagcactgc aagcctgagg aaggcgttga ggcccaagaa
60gaggccctgg gcctggtggg tgcgcaggct cctactactg aggagcagga ggctgctgtc
120tcctcctcct ctcctctggt ccctggcacc ctggaggaag tgcctgctgc
tgagtcagca 180ggtcctcccc agagtcctca gggagcctct gccttaccca
ctaccatcag cttcacttgc 240tggaggcaac ccaatgaggg ttccagcagc
caagaagagg aggggccaag cacctcgcct 300gacgcagagt ccttgttccg
agaagcactc agtaacaagg tggatgagtt ggctcatttt 360ctgctccgca
agtatcgagc caaggagctg gtcacaaagg cagaaatgct ggagagagtc
420atcaaaaatt acaagcgctg ctttcctgtg atcttcggca aagcctccga
gtccctgaag 480atgatctttg gcattgacgt gaaggaagtg gaccccacca
gcaacaccta cacccttgtc 540acctgcctgg gcctttccta tgatggcctg
ctgggtaata atcagatctt tcccaagaca 600ggccttctga taatcgtcct
gggcacaatt gcaatggagg gcgacagcgc ctctgaggag 660gaaatctggg
aggagctggg tgtgatgggg gtgtatgatg ggagggagca cactgtctat
720ggggagccca ggaaactgct cacccaagat tgggtgcagg aaaactacct
ggagtaccgg 780caggtacccg gcagtaatcc tgcgcgctat gagttcctgt
ggggtccaag ggctctggct 840gaaaccagct atgtgaaagt cctggagcat
gtggtcaggg tcaatgcaag agttcgcatt 900gcctacccat ccctgcgtga
agcagctttg ttagaggagg aagagggagt ctga 9544317PRTHomo sapiens 4Met
Ser Ser Glu Gln Lys Ser Gln His Cys Lys Pro Glu Glu Gly Val1 5 10
15Glu Ala Gln Glu Glu Ala Leu Gly Leu Val Gly Ala Gln Ala Pro Thr
20 25 30Thr Glu Glu Gln Glu Ala Ala Val Ser Ser Ser Ser Pro Leu Val
Pro 35 40 45Gly Thr Leu Glu Glu Val Pro Ala Ala Glu Ser Ala Gly Pro
Pro Gln 50 55 60Ser Pro Gln Gly Ala Ser Ala Leu Pro Thr Thr Ile Ser
Phe Thr Cys65 70 75 80Trp Arg Gln Pro Asn Glu Gly Ser Ser Ser Gln
Glu Glu Glu Gly Pro 85 90 95Ser Thr Ser Pro Asp Ala Glu Ser Leu Phe
Arg Glu Ala Leu Ser Asn 100 105 110Lys Val Asp Glu Leu Ala His Phe
Leu Leu Arg Lys Tyr Arg Ala Lys 115 120 125Glu Leu Val Thr Lys Ala
Glu Met Leu Glu Arg Val Ile Lys Asn Tyr 130 135 140Lys Arg Cys Phe
Pro Val Ile Phe Gly Lys Ala Ser Glu Ser Leu Lys145 150 155 160Met
Ile Phe Gly Ile Asp Val Lys Glu Val Asp Pro Thr Ser Asn Thr 165 170
175Tyr Thr Leu Val Thr Cys Leu Gly Leu Ser Tyr Asp Gly Leu Leu Gly
180 185 190Asn Asn Gln Ile Phe Pro Lys Thr Gly Leu Leu Ile Ile Val
Leu Gly 195 200 205Thr Ile Ala Met Glu Gly Asp Ser Ala Ser Glu Glu
Glu Ile Trp Glu 210 215 220Glu Leu Gly Val Met Gly Val Tyr Asp Gly
Arg Glu His Thr Val Tyr225 230 235 240Gly Glu Pro Arg Lys Leu Leu
Thr Gln Asp Trp Val Gln Glu Asn Tyr 245 250 255Leu Glu Tyr Arg Gln
Val Pro Gly Ser Asn Pro Ala Arg Tyr Glu Phe 260 265 270Leu Trp Gly
Pro Arg Ala Leu Ala Glu Thr Ser Tyr Val Lys Val Leu 275 280 285Glu
His Val Val Arg Val Asn Ala Arg Val Arg Ile Ala Tyr Pro Ser 290 295
300Leu Arg Glu Ala Ala Leu Leu Glu Glu Glu Glu Gly Val305 310
3155543DNAHomo sapiens 5atgcaggccg aaggccgggg cacagggggt tcgacgggcg
atgctgatgg cccaggaggc 60cctggcattc ctgatggccc agggggcaat gctggcggcc
caggagaggc gggtgccacg 120ggcggcagag gtccccgggg cgcaggggca
gcaagggcct cggggccggg aggaggcgcc 180ccgcggggtc cgcatggcgg
cgcggcttca gggctgaatg gatgctgcag atgcggggcc 240agggggccgg
agagccgcct gcttgagttc tacctcgcca tgcctttcgc gacacccatg
300gaagcagagc tggcccgcag gagcctggcc caggatgccc caccgcttcc
cgtgccaggg 360gtgcttctga aggagttcac tgtgtccggc aacatactga
ctatccgact gactgctgca 420gaccaccgcc aactgcagct ctccatcagc
tcctgtctcc agcagctttc cctgttgatg 480tggatcacgc agtgctttct
gcccgtgttt ttggctcagc ctccctcagg gcagaggcgc 540taa 5436180PRTHomo
sapiens 6Met Gln Ala Glu Gly Arg Gly Thr Gly Gly Ser Thr Gly Asp
Ala Asp1 5 10 15Gly Pro Gly Gly Pro Gly Ile Pro Asp Gly Pro Gly Gly
Asn Ala Gly 20 25 30Gly Pro Gly Glu Ala Gly Ala Thr Gly Gly Arg Gly
Pro Arg Gly Ala 35 40 45Gly Ala Ala Arg Ala Ser Gly Pro Gly Gly Gly
Ala Pro Arg Gly Pro 50 55 60His Gly Gly Ala Ala Ser Gly Leu Asn Gly
Cys Cys Arg Cys Gly Ala65 70 75 80Arg Gly Pro Glu Ser Arg Leu Leu
Glu Phe Tyr Leu Ala Met Pro Phe 85 90 95Ala Thr Pro Met Glu Ala Glu
Leu Ala Arg Arg Ser Leu Ala Gln Asp 100 105 110Ala Pro Pro Leu Pro
Val Pro Gly Val Leu Leu Lys Glu Phe Thr Val 115 120 125Ser Gly Asn
Ile Leu Thr Ile Arg Leu Thr Ala Ala Asp His Arg Gln 130 135 140Leu
Gln Leu Ser Ile Ser Ser Cys Leu Gln Gln Leu Ser Leu Leu Met145 150
155 160Trp Ile Thr Gln Cys Phe Leu Pro Val Phe Leu Ala Gln Pro Pro
Ser 165 170 175Gly Gln Arg Arg 1807246DNAHomo sapiens 7atggagagcc
ccaaaaagaa gaaccagcag ctgaaagtcg ggatcctaca cctgggcagc 60agacagaaga
agatcaggat acagctgaga tcccagtgcg cgacatggaa ggtgatctgc
120aagagctgca tcagtcaaac accggggata aatctggatt tgggttccgg
cgtcaaggtg 180aagataatac ctaaagagga acactgtaaa atgccagaag
caggtgaaga gcaaccacaa 240gtttaa 246881PRTHomo sapiens 8Met Glu Ser
Pro Lys Lys Lys Asn Gln Gln Leu Lys Val Gly Ile Leu1 5 10 15His Leu
Gly Ser Arg Gln Lys Lys Ile Arg Ile Gln Leu Arg Ser Gln 20 25 30Cys
Ala Thr Trp Lys Val Ile Cys Lys Ser Cys Ile Ser Gln Thr Pro 35 40
45Gly Ile Asn Leu Asp Leu Gly Ser Gly Val Lys Val Lys Ile Ile Pro
50 55 60Lys Glu Glu His Cys Lys Met Pro Glu Ala Gly Glu Glu Gln Pro
Gln65 70 75 80Val91698DNAHomo sapiens 9atgaaagtac ccagaaacca
ggactggctt ggtgtctcaa ggcaactcag aaccaaagcc 60tggaacaggc agctgtatcc
agagtggaca gaagcccaga gacttgactg ctggagaggt 120ggtcaagtgt
ccctcaaggt cagtaatgat gggcctacac tgattggtgc aaatgcctcc
180ttctctattg ccttgaactt ccctggaagc caaaaggtat tgccagatgg
gcaggttatc 240tgggtcaaca ataccatcat caatgggagc caggtgtggg
gaggacagcc agtgtatccc 300caggaaactg acgatgcctg catcttccct
gatggtggac cttgcccatc tggctcttgg 360tctcagaaga gaagctttgt
ttatgtctgg aagacctggg gccaatactg gcaagttcta 420gggggcccag
tgtctgggct gagcattggg acaggcaggg caatgctggg cacacacacc
480atggaagtga ctgtctacca tcgccgggga tcccggagct atgtgcctct
tgctcattcc 540agctcagcct tcaccattac tgaccaggtg cctttctccg
tgagcgtgtc ccagttgcgg 600gccttggatg gagggaacaa gcacttcctg
agaaatcagc ctctgacctt tgccctccag 660ctccatgacc ctagtggcta
tctggctgaa gctgacctct cctacacctg ggactttgga 720gacagtagtg
gaaccctgat ctctcgggca cttgtggtca ctcatactta cctggagcct
780ggcccagtca ctgcccaggt ggtcctgcag gctgccattc ctctcacctc
ctgtggctac 840tccccagttc caggcaccac agatgggcac aggccaactg
cagaggcccc taacaccaca 900gctggccaag tgcctactac agaagttgtg
ggtactacac ctggtcaggc gccaactgca 960gagccctctg gaaccacatc
tgtgcaggtg ccaaccactg aagtcataag cactgcacct 1020gtgcagatgc
caactgcaga gagcacaggt atgacacctg agaaggtgcc agtttcagag
1080gtcatgggta ccacactggc agagatgtca actccagagg ctacaggtat
gacacctgca 1140gaggtatcaa ttgtggtgct ttctggaacc acagctgcac
aggtaacaac tacagagtgg 1200gtggagacca cagctagaga gctacctatc
cctgagcctg aaggtccaga tgccagctca 1260atcatgtcta cggaaagtat
tacaggttcc ctgggccccc tgctggatgg tacagccacc 1320ttaaggctgg
tgaagagaca agtccccctg gattgtgttc tgtatcgata tggttccttt
1380tccgtcaccc tggacattgt ccagggtatt gaaagtgccg agatcctgca
ggctgtgccg 1440tccggtgagg gggatgcatt tgagctgact gtgtcctgcc
aaggcgggct gcccaaggaa 1500gcctgcatgg agatctcatc gccagggtgc
cagccccctg cccagcggct gtgccagcct 1560gtgctaccca gcccagcctg
ccagctggtt ctgcaccaga tactgaaggg tggctcgggg 1620acatactgcc
tcaatgtgtc tctggctgat accaacagcc tggcagtggt cagcacccag
1680cttatcatgc ctggttaa 169810565PRTHomo sapiens 10Met Lys Val Pro
Arg Asn Gln Asp Trp Leu Gly Val Ser Arg Gln Leu1 5 10 15Arg Thr Lys
Ala Trp Asn Arg Gln Leu Tyr Pro Glu Trp Thr Glu Ala 20 25 30Gln Arg
Leu Asp Cys Trp Arg Gly Gly Gln Val Ser Leu Lys Val Ser 35 40 45Asn
Asp Gly Pro Thr Leu Ile Gly Ala Asn Ala Ser Phe Ser Ile Ala 50 55
60Leu Asn Phe Pro Gly Ser Gln Lys Val Leu Pro Asp Gly Gln Val Ile65
70 75 80Trp Val Asn Asn Thr Ile Ile Asn Gly Ser Gln Val Trp Gly Gly
Gln 85 90 95Pro Val Tyr Pro Gln Glu Thr Asp Asp Ala Cys Ile Phe Pro
Asp Gly 100 105 110Gly Pro Cys Pro Ser Gly Ser Trp Ser Gln Lys Arg
Ser Phe Val Tyr 115 120 125Val Trp Lys Thr Trp Gly Gln Tyr Trp Gln
Val Leu Gly Gly Pro Val 130 135 140Ser Gly Leu Ser Ile Gly Thr Gly
Arg Ala Met Leu Gly Thr His Thr145 150 155 160Met Glu Val Thr Val
Tyr His Arg Arg Gly Ser Arg Ser Tyr Val Pro 165 170 175Leu Ala His
Ser Ser Ser Ala Phe Thr Ile Thr Asp Gln Val Pro Phe 180 185 190Ser
Val Ser Val Ser Gln Leu Arg Ala Leu Asp Gly Gly Asn Lys His 195 200
205Phe Leu Arg Asn Gln Pro Leu Thr Phe Ala Leu Gln Leu His Asp Pro
210 215 220Ser Gly Tyr Leu Ala Glu Ala Asp Leu Ser Tyr Thr Trp Asp
Phe Gly225 230 235 240Asp Ser Ser Gly Thr Leu Ile Ser Arg Ala Leu
Val Val Thr His Thr 245 250 255Tyr Leu Glu Pro Gly Pro Val Thr Ala
Gln Val Val Leu Gln Ala Ala 260 265 270Ile Pro Leu Thr Ser Cys Gly
Tyr Ser Pro Val Pro Gly Thr Thr Asp 275 280 285Gly His Arg Pro Thr
Ala Glu Ala Pro Asn Thr Thr Ala Gly Gln Val 290 295 300Pro Thr Thr
Glu Val Val Gly Thr Thr Pro Gly Gln Ala Pro Thr Ala305 310 315
320Glu Pro Ser Gly Thr Thr Ser Val Gln Val Pro Thr Thr Glu Val Ile
325 330 335Ser Thr Ala Pro Val Gln Met Pro Thr Ala Glu Ser Thr Gly
Met Thr 340 345 350Pro Glu Lys Val Pro Val Ser Glu Val Met Gly Thr
Thr Leu Ala Glu 355 360 365Met Ser Thr Pro Glu Ala Thr Gly Met Thr
Pro Ala Glu Val Ser Ile 370 375 380Val Val Leu Ser Gly Thr Thr Ala
Ala Gln Val Thr Thr Thr Glu Trp385 390 395 400Val Glu Thr Thr Ala
Arg Glu Leu Pro Ile Pro Glu Pro Glu Gly Pro 405 410 415Asp Ala Ser
Ser Ile Met Ser Thr Glu Ser Ile Thr Gly Ser Leu Gly 420 425 430Pro
Leu Leu Asp Gly Thr Ala Thr Leu Arg Leu Val Lys Arg Gln Val 435 440
445Pro Leu Asp Cys Val Leu Tyr Arg Tyr Gly Ser Phe Ser Val Thr Leu
450 455 460Asp Ile Val Gln Gly Ile Glu Ser Ala Glu Ile Leu Gln Ala
Val Pro465 470 475 480Ser Gly Glu Gly Asp Ala Phe Glu Leu Thr Val
Ser Cys Gln Gly Gly 485 490 495Leu Pro Lys Glu Ala Cys Met Glu Ile
Ser Ser Pro Gly Cys Gln Pro
500 505 510Pro Ala Gln Arg Leu Cys Gln Pro Val Leu Pro Ser Pro Ala
Cys Gln 515 520 525Leu Val Leu His Gln Ile Leu Lys Gly Gly Ser Gly
Thr Tyr Cys Leu 530 535 540Asn Val Ser Leu Ala Asp Thr Asn Ser Leu
Ala Val Val Ser Thr Gln545 550 555 560Leu Ile Met Pro Gly
56511429DNAHomo sapiens 11atgggtgccc cgacgttgcc ccctgcctgg
cagccctttc tcaaggacca ccgcatctct 60acattcaaga actggccctt cttggagggc
tgcgcctgca ccccggagcg gatggccgag 120gctggcttca tccactgccc
cactgagaac gagccagact tggcccagtg tttcttctgc 180ttcaaggagc
tggaaggctg ggagccagat gacgacccca tagaggaaca taaaaagcat
240tcgtccggtt gcgctttcct ttctgtcaag aagcagtttg aagaattaac
ccttggtgaa 300tttttgaaac tggacagaga aagagccaag aacaaaattg
caaaggaaac caacaataag 360aagaaagaat ttgaggaaac tgcggagaaa
gtgcgccgtg ccatcgagca gctggctgcc 420atggattga 42912142PRTHomo
sapiens 12Met Gly Ala Pro Thr Leu Pro Pro Ala Trp Gln Pro Phe Leu
Lys Asp1 5 10 15His Arg Ile Ser Thr Phe Lys Asn Trp Pro Phe Leu Glu
Gly Cys Ala 20 25 30Cys Thr Pro Glu Arg Met Ala Glu Ala Gly Phe Ile
His Cys Pro Thr 35 40 45Glu Asn Glu Pro Asp Leu Ala Gln Cys Phe Phe
Cys Phe Lys Glu Leu 50 55 60Glu Gly Trp Glu Pro Asp Asp Asp Pro Ile
Glu Glu His Lys Lys His65 70 75 80Ser Ser Gly Cys Ala Phe Leu Ser
Val Lys Lys Gln Phe Glu Glu Leu 85 90 95Thr Leu Gly Glu Phe Leu Lys
Leu Asp Arg Glu Arg Ala Lys Asn Lys 100 105 110Ile Ala Lys Glu Thr
Asn Asn Lys Lys Lys Glu Phe Glu Glu Thr Ala 115 120 125Glu Lys Val
Arg Arg Ala Ile Glu Gln Leu Ala Ala Met Asp 130 135
14013129PRTGallus gallus 13Lys Val Phe Gly Arg Cys Glu Leu Ala Ala
Ala Met Lys Arg His Gly1 5 10 15Leu Asp Asn Tyr Arg Gly Tyr Ser Leu
Gly Asn Trp Val Cys Ala Ala 20 25 30Lys Phe Glu Ser Asn Phe Asn Thr
Gln Ala Thr Asn Arg Asn Thr Asp 35 40 45Gly Ser Thr Asp Tyr Gly Ile
Leu Gln Ile Asn Ser Arg Trp Trp Cys 50 55 60Asn Asp Gly Arg Thr Pro
Gly Ser Arg Asn Leu Cys Asn Ile Pro Cys65 70 75 80Ser Ala Leu Leu
Ser Ser Asp Ile Thr Ala Ser Val Asn Cys Ala Lys 85 90 95Lys Ile Val
Ser Asp Gly Asn Gly Met Asn Ala Trp Val Ala Trp Arg 100 105 110Asn
Arg Cys Lys Gly Thr Asp Val Gln Ala Trp Ile Arg Gly Cys Arg 115 120
125Leu141618DNAArtificialHis-tag fusion protein 14atgggcagca
gccatcatca tcatcatcac agcagcggcc tggtgccgcg cggcagccat 60atgcaggacc
cggcttccac gtgtgtcccg gagccggcgt ctcagcacac gctccgctcc
120gggcctgggt gcctacagca gccagagcag cagggagtcc gggacccggg
cggcatctgg 180gccaagttag gcgccgccga ggccagcgct gaacgtctcc
agggccggag gagccgcggg 240gcgtccgggt ctgagccgca gcaaatgggc
tccgacgtgc gggacctgaa cgcgctgctg 300cccgccgtcc cctccctggg
tggcggcggc ggctgtgccc tgcctgtgag cggcgcggcg 360cagtgggcgc
cggtgctgga ctttgcgccc ccgggcgctt cggcttacgg gtcgttgggc
420ggccccgcgc cgccaccggc tccgccgcca cccccgccgc cgccgcctca
ctccttcatc 480aaacaggagc cgagctgggg cggcgcggag ccgcacgagg
agcagtgcct gagcgccttc 540actgtccact tttccggcca gttcactggc
acagccggag cctgtcgcta cgggcccttc 600ggtcctcctc cgcccagcca
ggcgtcatcc ggccaggcca ggatgtttcc taacgcgccc 660tacctgccca
gctgcctcga gagccagccc gctattcgca atcagggtta cagcacggtc
720accttcgacg ggacgcccag ctacggtcac acgccctcgc accatgcggc
gcagttcccc 780aaccactcat tcaagcatga ggatcccatg ggccagcagg
gctcgctggg tgagcagcag 840tactcggtgc cgcccccggt ctatggctgc
cacaccccca ccgacagctg caccggcagc 900caggctttgc tgctgaggac
gccctacagc agtgacaatt tataccaaat gacatcccag 960cttgaatgca
tgacctggaa tcagatgaac ttaggagcca ccttaaaggg agttgctgct
1020gggagctcca gctcagtgaa atggacagaa gggcagagca accacagcac
agggtacgag 1080agcgataacc acacaacgcc catcctctgc ggagcccaat
acagaataca cacgcacggt 1140gtcttcagag gcattcagga tgtgcgacgt
gtgcctggag tagccccgac tcttgtacgg 1200tcggcatctg agaccagtga
gaaacgcccc ttcatgtgtg cttacccagg ctgcaataag 1260agatatttta
agctgtccca cttacagatg cacagcagga agcacactgg tgagaaacca
1320taccagtgtg acttcaagga ctgtgaacga aggttttctc gttcagacca
gctcaaaaga 1380caccaaagga gacatacagg tgtgaaacca ttccagtgta
aaacttgtca gcgaaagttc 1440tcccggtccg accacctgaa gacccacacc
aggactcata caggtaaaac aagtgaaaag 1500cccttcagct gtcggtggcc
aagttgtcag aaaaagtttg cccggtcaga tgaattagtc 1560cgccatcaca
acatgcatca gagaaacatg accaaactcc agctggcgct ttgaattc
161815537PRTArtificialHis-tag fusion protein 15Met Gly Ser Ser His
His His His His His Ser Ser Gly Leu Val Pro1 5 10 15Arg Gly Ser His
Met Gln Asp Pro Ala Ser Thr Cys Val Pro Glu Pro 20 25 30Ala Ser Gln
His Thr Leu Arg Ser Gly Pro Gly Cys Leu Gln Gln Pro 35 40 45Glu Gln
Gln Gly Val Arg Asp Pro Gly Gly Ile Trp Ala Lys Leu Gly 50 55 60Ala
Ala Glu Ala Ser Ala Glu Arg Leu Gln Gly Arg Arg Ser Arg Gly65 70 75
80Ala Ser Gly Ser Glu Pro Gln Gln Met Gly Ser Asp Val Arg Asp Leu
85 90 95Asn Ala Leu Leu Pro Ala Val Pro Ser Leu Gly Gly Gly Gly Gly
Cys 100 105 110Ala Leu Pro Val Ser Gly Ala Ala Gln Trp Ala Pro Val
Leu Asp Phe 115 120 125Ala Pro Pro Gly Ala Ser Ala Tyr Gly Ser Leu
Gly Gly Pro Ala Pro 130 135 140Pro Pro Ala Pro Pro Pro Pro Pro Pro
Pro Pro Pro His Ser Phe Ile145 150 155 160Lys Gln Glu Pro Ser Trp
Gly Gly Ala Glu Pro His Glu Glu Gln Cys 165 170 175Leu Ser Ala Phe
Thr Val His Phe Ser Gly Gln Phe Thr Gly Thr Ala 180 185 190Gly Ala
Cys Arg Tyr Gly Pro Phe Gly Pro Pro Pro Pro Ser Gln Ala 195 200
205Ser Ser Gly Gln Ala Arg Met Phe Pro Asn Ala Pro Tyr Leu Pro Ser
210 215 220Cys Leu Glu Ser Gln Pro Ala Ile Arg Asn Gln Gly Tyr Ser
Thr Val225 230 235 240Thr Phe Asp Gly Thr Pro Ser Tyr Gly His Thr
Pro Ser His His Ala 245 250 255Ala Gln Phe Pro Asn His Ser Phe Lys
His Glu Asp Pro Met Gly Gln 260 265 270Gln Gly Ser Leu Gly Glu Gln
Gln Tyr Ser Val Pro Pro Pro Val Tyr 275 280 285Gly Cys His Thr Pro
Thr Asp Ser Cys Thr Gly Ser Gln Ala Leu Leu 290 295 300Leu Arg Thr
Pro Tyr Ser Ser Asp Asn Leu Tyr Gln Met Thr Ser Gln305 310 315
320Leu Glu Cys Met Thr Trp Asn Gln Met Asn Leu Gly Ala Thr Leu Lys
325 330 335Gly Val Ala Ala Gly Ser Ser Ser Ser Val Lys Trp Thr Glu
Gly Gln 340 345 350Ser Asn His Ser Thr Gly Tyr Glu Ser Asp Asn His
Thr Thr Pro Ile 355 360 365Leu Cys Gly Ala Gln Tyr Arg Ile His Thr
His Gly Val Phe Arg Gly 370 375 380Ile Gln Asp Val Arg Arg Val Pro
Gly Val Ala Pro Thr Leu Val Arg385 390 395 400Ser Ala Ser Glu Thr
Ser Glu Lys Arg Pro Phe Met Cys Ala Tyr Pro 405 410 415Gly Cys Asn
Lys Arg Tyr Phe Lys Leu Ser His Leu Gln Met His Ser 420 425 430Arg
Lys His Thr Gly Glu Lys Pro Tyr Gln Cys Asp Phe Lys Asp Cys 435 440
445Glu Arg Arg Phe Ser Arg Ser Asp Gln Leu Lys Arg His Gln Arg Arg
450 455 460His Thr Gly Val Lys Pro Phe Gln Cys Lys Thr Cys Gln Arg
Lys Phe465 470 475 480Ser Arg Ser Asp His Leu Lys Thr His Thr Arg
Thr His Thr Gly Lys 485 490 495Thr Ser Glu Lys Pro Phe Ser Cys Arg
Trp Pro Ser Cys Gln Lys Lys 500 505 510Phe Ala Arg Ser Asp Glu Leu
Val Arg His His Asn Met His Gln Arg 515 520 525Asn Met Thr Lys Leu
Gln Leu Ala Leu 530 535161020DNAArtificialHis-tag fusion protein
16atgggcagca gccatcatca tcatcatcac agcagcggcc tggtgccgcg cggcagccat
60atgtcttctg agcagaagag tcagcactgc aagcctgagg aaggcgttga ggcccaagaa
120gaggccctgg gcctggtggg tgcgcaggct cctactactg aggagcagga
ggctgctgtc 180tcctcctcct ctcctctggt ccctggcacc ctggaggaag
tgcctgctgc tgagtcagca 240ggtcctcccc agagtcctca gggagcctct
gccttaccca ctaccatcag cttcacttgc 300tggaggcaac ccaatgaggg
ttccagcagc caagaagagg aggggccaag cacctcgcct 360gacgcagagt
ccttgttccg agaagcactc agtaacaagg tggatgagtt ggctcatttt
420ctgctccgca agtatcgagc caaggagctg gtcacaaagg cagaaatgct
ggagagagtc 480atcaaaaatt acaagcgctg ctttcctgtg atcttcggca
aagcctccga gtccctgaag 540atgatctttg gcattgacgt gaaggaagtg
gaccccacca gcaacaccta cacccttgtc 600acctgcctgg gcctttccta
tgatggcctg ctgggtaata atcagatctt tcccaagaca 660ggccttctga
taatcgtcct gggcacaatt gcaatggagg gcgacagcgc ctctgaggag
720gaaatctggg aggagctggg tgtgatgggg gtgtatgatg ggagggagca
cactgtctat 780ggggagccca ggaaactgct cacccaagat tgggtgcagg
aaaactacct ggagtaccgg 840caggtacccg gcagtaatcc tgcgcgctat
gagttcctgt ggggtccaag ggctctggct 900gaaaccagct atgtgaaagt
cctggagcat gtggtcaggg tcaatgcaag agttcgcatt 960gcctacccat
ccctgcgtga agcagctttg ttagaggagg aagagggagt ctgaggatcc
102017337PRTArtificialHis-tag fusion protein 17Met Gly Ser Ser His
His His His His His Ser Ser Gly Leu Val Pro1 5 10 15Arg Gly Ser His
Met Ser Ser Glu Gln Lys Ser Gln His Cys Lys Pro 20 25 30Glu Glu Gly
Val Glu Ala Gln Glu Glu Ala Leu Gly Leu Val Gly Ala 35 40 45Gln Ala
Pro Thr Thr Glu Glu Gln Glu Ala Ala Val Ser Ser Ser Ser 50 55 60Pro
Leu Val Pro Gly Thr Leu Glu Glu Val Pro Ala Ala Glu Ser Ala65 70 75
80Gly Pro Pro Gln Ser Pro Gln Gly Ala Ser Ala Leu Pro Thr Thr Ile
85 90 95Ser Phe Thr Cys Trp Arg Gln Pro Asn Glu Gly Ser Ser Ser Gln
Glu 100 105 110Glu Glu Gly Pro Ser Thr Ser Pro Asp Ala Glu Ser Leu
Phe Arg Glu 115 120 125Ala Leu Ser Asn Lys Val Asp Glu Leu Ala His
Phe Leu Leu Arg Lys 130 135 140Tyr Arg Ala Lys Glu Leu Val Thr Lys
Ala Glu Met Leu Glu Arg Val145 150 155 160Ile Lys Asn Tyr Lys Arg
Cys Phe Pro Val Ile Phe Gly Lys Ala Ser 165 170 175Glu Ser Leu Lys
Met Ile Phe Gly Ile Asp Val Lys Glu Val Asp Pro 180 185 190Thr Ser
Asn Thr Tyr Thr Leu Val Thr Cys Leu Gly Leu Ser Tyr Asp 195 200
205Gly Leu Leu Gly Asn Asn Gln Ile Phe Pro Lys Thr Gly Leu Leu Ile
210 215 220Ile Val Leu Gly Thr Ile Ala Met Glu Gly Asp Ser Ala Ser
Glu Glu225 230 235 240Glu Ile Trp Glu Glu Leu Gly Val Met Gly Val
Tyr Asp Gly Arg Glu 245 250 255His Thr Val Tyr Gly Glu Pro Arg Lys
Leu Leu Thr Gln Asp Trp Val 260 265 270Gln Glu Asn Tyr Leu Glu Tyr
Arg Gln Val Pro Gly Ser Asn Pro Ala 275 280 285Arg Tyr Glu Phe Leu
Trp Gly Pro Arg Ala Leu Ala Glu Thr Ser Tyr 290 295 300Val Lys Val
Leu Glu His Val Val Arg Val Asn Ala Arg Val Arg Ile305 310 315
320Ala Tyr Pro Ser Leu Arg Glu Ala Ala Leu Leu Glu Glu Glu Glu Gly
325 330 335Val18735DNAArtificialHis-tag fusion protein 18atgggcagca
gccatcatca tcatcatcac agcagcggcc tggtgccgcg cggcagccat 60atggctagca
tgactggtgg acagcaaatg ggtcgcggat ccatgcaggc cgaaggccgg
120ggcacagggg gttcgacggg cgatgctgat ggcccaggag gccctggcat
tcctgatggc 180ccagggggca atgctggcgg cccaggagag gcgggtgcca
cgggcggcag aggtccccgg 240ggcgcagggg cagcaagggc ctcggggccg
ggaggaggcg ccccgcgggg tccgcatggc 300ggcgcggctt cagggctgaa
tggatgctgc agatgcgggg ccagggggcc ggagagccgc 360ctgcttgagt
tctacctcgc catgcctttc gcgacaccca tggaagcaga gctggcccgc
420aggagcctgg cccaggatgc cccaccgctt cccgtgccag gggtgcttct
gaaggagttc 480actgtgtccg gcaacatact gactatccga ctgactgctg
cagaccaccg ccaactgcag 540ctctccatca gctcctgtct ccagcagctt
tccctgttga tgtggatcac gcagtgcttt 600ctgcccgtgt ttttggctca
gcctccctca gggcagaggc gctaacagct ttccctgttg 660atgtggatca
cgcagtgctt tctgcccgtg ttttggctca gcctcccctc agggcagagg
720cgctaagcca agctt 73519214PRTArtificialHis-tag fusion protein
19Met Gly Ser Ser His His His His His His Ser Ser Gly Leu Val Pro1
5 10 15Arg Gly Ser His Met Ala Ser Met Thr Gly Gly Gln Gln Met Gly
Arg 20 25 30Gly Ser Met Gln Ala Glu Gly Arg Gly Thr Gly Gly Ser Thr
Gly Asp 35 40 45Ala Asp Gly Pro Gly Gly Pro Gly Ile Pro Asp Gly Pro
Gly Gly Asn 50 55 60Ala Gly Gly Pro Gly Glu Ala Gly Ala Thr Gly Gly
Arg Gly Pro Arg65 70 75 80Gly Ala Gly Ala Ala Arg Ala Ser Gly Pro
Gly Gly Gly Ala Pro Arg 85 90 95Gly Pro His Gly Gly Ala Ala Ser Gly
Leu Asn Gly Cys Cys Arg Cys 100 105 110Gly Ala Arg Gly Pro Glu Ser
Arg Leu Leu Glu Phe Tyr Leu Ala Met 115 120 125Pro Phe Ala Thr Pro
Met Glu Ala Glu Leu Ala Arg Arg Ser Leu Ala 130 135 140Gln Asp Ala
Pro Pro Leu Pro Val Pro Gly Val Leu Leu Lys Glu Phe145 150 155
160Thr Val Ser Gly Asn Ile Leu Thr Ile Arg Leu Thr Ala Ala Asp His
165 170 175Arg Gln Leu Gln Leu Ser Ile Ser Ser Cys Leu Gln Gln Leu
Ser Leu 180 185 190Leu Met Trp Ile Thr Gln Cys Phe Leu Pro Val Phe
Leu Ala Gln Pro 195 200 205Pro Ser Gly Gln Arg Arg
21020377DNAArtificialHis-tag fusion protein 20atgggcagca gccatcatca
tcatcatcac agcagcggcc tggtgccgcg cggcagccat 60atggctagca tgactggtgg
acagcaaatg ggtcgcggat ccgaattccg catggagagc 120cccaaaaaga
agaaccagca gctgaaagtc gggatcctac acctgggcag cagacagaag
180aagatcagga tacagctgag atcccagtgc gcgacatgga aggtgatctg
caagagctgc 240atcagtcaaa caccggggat aaatctggat ttgggttccg
gcgtcaaggt gaagataata 300cctaaagagg aacactgtaa aatgccagaa
gcaggtgaag agcaaccaca agtttaaatg 360aagacaagct gctcgag
37721118PRTArtificialHis-tag fusion protein 21Met Gly Ser Ser His
His His His His His Ser Ser Gly Leu Val Pro1 5 10 15Arg Gly Ser His
Met Ala Ser Met Thr Gly Gly Gln Gln Met Gly Arg 20 25 30Gly Ser Glu
Phe Arg Met Glu Ser Pro Lys Lys Lys Asn Gln Gln Leu 35 40 45Lys Val
Gly Ile Leu His Leu Gly Ser Arg Gln Lys Lys Ile Arg Ile 50 55 60Gln
Leu Arg Ser Gln Cys Ala Thr Trp Lys Val Ile Cys Lys Ser Cys65 70 75
80Ile Ser Gln Thr Pro Gly Ile Asn Leu Asp Leu Gly Ser Gly Val Lys
85 90 95Val Lys Ile Ile Pro Lys Glu Glu His Cys Lys Met Pro Glu Ala
Gly 100 105 110Glu Glu Gln Pro Gln Val
115221766DNAArtificialHis-tag fusion protein 22atgggcagca
gccatcatca tcatcatcac agcagcggcc tggtgccgcg cggcagccat 60atgaaagtac
ccagaaacca ggactggctt ggtgtctcaa ggcaactcag aaccaaagcc
120tggaacaggc agctgtatcc agagtggaca gaagcccaga gacttgactg
ctggagaggt 180ggtcaagtgt ccctcaaggt cagtaatgat gggcctacac
tgattggtgc aaatgcctcc 240ttctctattg ccttgaactt ccctggaagc
caaaaggtat tgccagatgg gcaggttatc 300tgggtcaaca ataccatcat
caatgggagc caggtgtggg gaggacagcc agtgtatccc 360caggaaactg
acgatgcctg catcttccct gatggtggac cttgcccatc tggctcttgg
420tctcagaaga gaagctttgt ttatgtctgg aagacctggg gccaatactg
gcaagttcta 480gggggcccag tgtctgggct gagcattggg acaggcaggg
caatgctggg cacacacacc 540atggaagtga ctgtctacca tcgccgggga
tcccggagct atgtgcctct tgctcattcc 600agctcagcct tcaccattac
tgaccaggtg cctttctccg tgagcgtgtc ccagttgcgg 660gccttggatg
gagggaacaa gcacttcctg agaaatcagc ctctgacctt tgccctccag
720ctccatgacc ctagtggcta tctggctgaa gctgacctct cctacacctg
ggactttgga 780gacagtagtg gaaccctgat ctctcgggca cttgtggtca
ctcatactta cctggagcct 840ggcccagtca ctgcccaggt ggtcctgcag
gctgccattc ctctcacctc ctgtggctac 900tccccagttc caggcaccac
agatgggcac aggccaactg cagaggcccc taacaccaca 960gctggccaag
tgcctactac agaagttgtg ggtactacac ctggtcaggc
gccaactgca 1020gagccctctg gaaccacatc tgtgcaggtg ccaaccactg
aagtcataag cactgcacct 1080gtgcagatgc caactgcaga gagcacaggt
atgacacctg agaaggtgcc agtttcagag 1140gtcatgggta ccacactggc
agagatgtca actccagagg ctacaggtat gacacctgca 1200gaggtatcaa
ttgtggtgct ttctggaacc acagctgcac aggtaacaac tacagagtgg
1260gtggagacca cagctagaga gctacctatc cctgagcctg aaggtccaga
tgccagctca 1320atcatgtcta cggaaagtat tacaggttcc ctgggccccc
tgctggatgg tacagccacc 1380ttaaggctgg tgaagagaca agtccccctg
gattgtgttc tgtatcgata tggttccttt 1440tccgtcaccc tggacattgt
ccagggtatt gaaagtgccg agatcctgca ggctgtgccg 1500tccggtgagg
gggatgcatt tgagctgact gtgtcctgcc aaggcgggct gcccaaggaa
1560gcctgcatgg agatctcatc gccagggtgc cagccccctg cccagcggct
gtgccagcct 1620gtgctaccca gcccagcctg ccagctggtt ctgcaccaga
tactgaaggg tggctcgggg 1680acatactgcc tcaatgtgtc tctggctgat
accaacagcc tggcagtggt cagcacccag 1740cttatcatgc ctggttaagc ggccgc
176623585PRTArtificialHis-tag fusion protein 23Met Gly Ser Ser His
His His His His His Ser Ser Gly Leu Val Pro1 5 10 15Arg Gly Ser His
Met Lys Val Pro Arg Asn Gln Asp Trp Leu Gly Val 20 25 30Ser Arg Gln
Leu Arg Thr Lys Ala Trp Asn Arg Gln Leu Tyr Pro Glu 35 40 45Trp Thr
Glu Ala Gln Arg Leu Asp Cys Trp Arg Gly Gly Gln Val Ser 50 55 60Leu
Lys Val Ser Asn Asp Gly Pro Thr Leu Ile Gly Ala Asn Ala Ser65 70 75
80Phe Ser Ile Ala Leu Asn Phe Pro Gly Ser Gln Lys Val Leu Pro Asp
85 90 95Gly Gln Val Ile Trp Val Asn Asn Thr Ile Ile Asn Gly Ser Gln
Val 100 105 110Trp Gly Gly Gln Pro Val Tyr Pro Gln Glu Thr Asp Asp
Ala Cys Ile 115 120 125Phe Pro Asp Gly Gly Pro Cys Pro Ser Gly Ser
Trp Ser Gln Lys Arg 130 135 140Ser Phe Val Tyr Val Trp Lys Thr Trp
Gly Gln Tyr Trp Gln Val Leu145 150 155 160Gly Gly Pro Val Ser Gly
Leu Ser Ile Gly Thr Gly Arg Ala Met Leu 165 170 175Gly Thr His Thr
Met Glu Val Thr Val Tyr His Arg Arg Gly Ser Arg 180 185 190Ser Tyr
Val Pro Leu Ala His Ser Ser Ser Ala Phe Thr Ile Thr Asp 195 200
205Gln Val Pro Phe Ser Val Ser Val Ser Gln Leu Arg Ala Leu Asp Gly
210 215 220Gly Asn Lys His Phe Leu Arg Asn Gln Pro Leu Thr Phe Ala
Leu Gln225 230 235 240Leu His Asp Pro Ser Gly Tyr Leu Ala Glu Ala
Asp Leu Ser Tyr Thr 245 250 255Trp Asp Phe Gly Asp Ser Ser Gly Thr
Leu Ile Ser Arg Ala Leu Val 260 265 270Val Thr His Thr Tyr Leu Glu
Pro Gly Pro Val Thr Ala Gln Val Val 275 280 285Leu Gln Ala Ala Ile
Pro Leu Thr Ser Cys Gly Tyr Ser Pro Val Pro 290 295 300Gly Thr Thr
Asp Gly His Arg Pro Thr Ala Glu Ala Pro Asn Thr Thr305 310 315
320Ala Gly Gln Val Pro Thr Thr Glu Val Val Gly Thr Thr Pro Gly Gln
325 330 335Ala Pro Thr Ala Glu Pro Ser Gly Thr Thr Ser Val Gln Val
Pro Thr 340 345 350Thr Glu Val Ile Ser Thr Ala Pro Val Gln Met Pro
Thr Ala Glu Ser 355 360 365Thr Gly Met Thr Pro Glu Lys Val Pro Val
Ser Glu Val Met Gly Thr 370 375 380Thr Leu Ala Glu Met Ser Thr Pro
Glu Ala Thr Gly Met Thr Pro Ala385 390 395 400Glu Val Ser Ile Val
Val Leu Ser Gly Thr Thr Ala Ala Gln Val Thr 405 410 415Thr Thr Glu
Trp Val Glu Thr Thr Ala Arg Glu Leu Pro Ile Pro Glu 420 425 430Pro
Glu Gly Pro Asp Ala Ser Ser Ile Met Ser Thr Glu Ser Ile Thr 435 440
445Gly Ser Leu Gly Pro Leu Leu Asp Gly Thr Ala Thr Leu Arg Leu Val
450 455 460Lys Arg Gln Val Pro Leu Asp Cys Val Leu Tyr Arg Tyr Gly
Ser Phe465 470 475 480Ser Val Thr Leu Asp Ile Val Gln Gly Ile Glu
Ser Ala Glu Ile Leu 485 490 495Gln Ala Val Pro Ser Gly Glu Gly Asp
Ala Phe Glu Leu Thr Val Ser 500 505 510Cys Gln Gly Gly Leu Pro Lys
Glu Ala Cys Met Glu Ile Ser Ser Pro 515 520 525Gly Cys Gln Pro Pro
Ala Gln Arg Leu Cys Gln Pro Val Leu Pro Ser 530 535 540Pro Ala Cys
Gln Leu Val Leu His Gln Ile Leu Lys Gly Gly Ser Gly545 550 555
560Thr Tyr Cys Leu Asn Val Ser Leu Ala Asp Thr Asn Ser Leu Ala Val
565 570 575Val Ser Thr Gln Leu Ile Met Pro Gly 580
58524537DNAArtificialHis-tag fusion protein 24atgggcagca gccatcatca
tcatcatcac agcagcggcc tggtgccgcg cggcagccat 60atggctagca tgactggtgg
acagcaaatg ggtcgcggat ccatgggtgc cccgacgttg 120ccccctgcct
ggcagccctt tctcaaggac caccgcatct ctacattcaa gaactggccc
180ttcttggagg gctgcgcctg caccccggag cggatggccg aggctggctt
catccactgc 240cccactgaga acgagccaga cttggcccag tgtttcttct
gcttcaagga gctggaaggc 300tgggagccag atgacgaccc catagaggaa
cataaaaagc attcgtccgg ttgcgctttc 360ctttctgtca agaagcagtt
tgaagaatta acccttggtg aatttttgaa actggacaga 420gaaagagcca
agaacaaaat tgcaaaggaa accaacaata agaagaaaga atttgaggaa
480actgcggaga aagtgcgccg tgccatcgag cagctggctg ccatggattg actcgag
53725176PRTArtificialHis-tag fusion protein 25Met Gly Ser Ser His
His His His His His Ser Ser Gly Leu Val Pro1 5 10 15Arg Gly Ser His
Met Ala Ser Met Thr Gly Gly Gln Gln Met Gly Arg 20 25 30Gly Ser Met
Gly Ala Pro Thr Leu Pro Pro Ala Trp Gln Pro Phe Leu 35 40 45Lys Asp
His Arg Ile Ser Thr Phe Lys Asn Trp Pro Phe Leu Glu Gly 50 55 60Cys
Ala Cys Thr Pro Glu Arg Met Ala Glu Ala Gly Phe Ile His Cys65 70 75
80Pro Thr Glu Asn Glu Pro Asp Leu Ala Gln Cys Phe Phe Cys Phe Lys
85 90 95Glu Leu Glu Gly Trp Glu Pro Asp Asp Asp Pro Ile Glu Glu His
Lys 100 105 110Lys His Ser Ser Gly Cys Ala Phe Leu Ser Val Lys Lys
Gln Phe Glu 115 120 125Glu Leu Thr Leu Gly Glu Phe Leu Lys Leu Asp
Arg Glu Arg Ala Lys 130 135 140Asn Lys Ile Ala Lys Glu Thr Asn Asn
Lys Lys Lys Glu Phe Glu Glu145 150 155 160Thr Ala Glu Lys Val Arg
Arg Ala Ile Glu Gln Leu Ala Ala Met Asp 165 170 175
References








D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.