Increasing Productivity Of E. Coli Host Cells That Functionally Express P450 Enzymes
DONALD; Jason ; et al.
U.S. patent application number 16/990424 was filed with the patent office on 2020-12-03 for increasing productivity of e. coli host cells that functionally express p450 enzymes. The applicant listed for this patent is Manus Bio, Inc.. Invention is credited to Jason DONALD, Ajikumar Parayil KUMARAN, Liwei LI, Huey-Ming MAK, Christopher PIRIE, Srishti TIBREWALA.
| Application Number | 20200377865 16/990424 |
| Document ID | / |
| Family ID | 1000005022875 |
| Filed Date | 2020-12-03 |





View All Diagrams
| United States Patent Application | 20200377865 |
| Kind Code | A1 |
| DONALD; Jason ; et al. | December 3, 2020 |
INCREASING PRODUCTIVITY OF E. COLI HOST CELLS THAT FUNCTIONALLY EXPRESS P450 ENZYMES
Abstract
The present invention relates to the production of chemical species in bacterial host cells. Particularly, the present invention provides for the production of chemical species in Escherichia coli (E. coli) host cells that functionally express engineered P450 enzymes.
| Inventors: | DONALD; Jason; (Cambridge, MA) ; PIRIE; Christopher; (Cambridge, MA) ; LI; Liwei; (Cambridge, MA) ; MAK; Huey-Ming; (Cambridge, MA) ; TIBREWALA; Srishti; (Cambridge, MA) ; KUMARAN; Ajikumar Parayil; (Cambridge, MA) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 1000005022875 | ||||||||||
| Appl. No.: | 16/990424 | ||||||||||
| Filed: | August 11, 2020 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 15754105 | Feb 21, 2018 | 10774314 | ||
| PCT/US2016/047692 | Aug 19, 2016 | |||
| 16990424 | ||||
| 62208166 | Aug 21, 2015 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C12P 19/56 20130101; C12N 9/00 20130101; C12N 9/0079 20130101; C12N 15/70 20130101; C12N 15/52 20130101; C07K 2319/03 20130101; C12P 5/007 20130101 |
| International Class: | C12N 9/02 20060101 C12N009/02; C12N 15/52 20060101 C12N015/52; C12P 5/00 20060101 C12P005/00; C12N 9/00 20060101 C12N009/00; C12P 19/56 20060101 C12P019/56; C12N 15/70 20060101 C12N015/70 |
Claims
1. A method for biosynthesis of one or more chemical species in E. coli, comprising: expressing one or more biosynthetic pathways in E. coli, the one or more biosynthetic pathways comprising at least one membrane-anchored P450 enzyme having a transmembrane domain derived from an E. coli inner membrane cytoplasmic C-terminus protein, and culturing the E. coli to produce the one or more chemical species from the biosynthetic pathway(s).
2. The method of claim 1, wherein the E. coli does not exhibit a substantially stressed phenotype during the culturing.
3. The method of claim 1 or 2, wherein the E. coli expresses at least two, at least three, or at least four recombinant enzymes.
4. The method of claim 3, wherein the biosynthetic pathway(s) produce a secondary metabolite through the overexpression of at least two foreign genes.
5. The method of claim 4, wherein the biosynthetic pathway(s) produce a secondary metabolite through the overexpression of at least three foreign genes, at least four foreign genes, or at least five foreign genes.
6. The method of any one of claims 1 to 5, wherein the E. coli contains an overexpression of at leak two E. coli genes.
7. The method of claim 6, wherein the E. coli overexpresses at least one gene in the MEP pathway.
8. The method of claim 6 or 7, wherein at least one gene is expressed by a strong promoter.
9. The method of any one of claims 6 to 8, wherein at least one gene is expressed from a plasmid.
10. The method of any one of claims 6 to 9, wherein at least one gene is chromosomally integrated.
11. The method of any one of claims 1 to 10, wherein at least one P450 enzyme is not strongly expressed.
12. The method of any one of claims 1 to 11, wherein the E. coli expresses at least two P450 enzymes, which are optionally derived from plant P450 enzymes.
13. The method of claim 12, wherein the E. coli expresses a membrane-anchored P450 selected from CiVO, HmPO, LsGAO, BsGAO, NtEAO, SrKO, SrKAH, AtKAEI, ZzHO, CpVO, MsL6OH, NtVO, StVO, AtKO, Ci2VO, AaAO, and Taxus 5-alpha hydroxylase, or derivative thereof.
14. The method of any one of claims 1 to 13, wherein the biosynthetic pathway produces a secondary metabolite selected from a terpenoid, alkaloid, cannabinoid, steroid, saponin, glycoside, stilbenoid, polyphenol, antibiotic, polyketide, fatty acid, or non-ribosomal peptide.
15. The method of claim 14, wherein the biosynthetic pathway produces a terpenoid selected from a monoterpenoid, a sesquiterpenoid, diterpenoid, a sesterpenoid, or a triterpenoid.
16. The method of claim 15, wherein the biosynthetic pathway involves overexpression of a geranyl diphosphate synthase (GPS), a gernanylgeranyl diphosphate synthase (GGPS), a farnsesyl diphosphate synthase (FPS), or a farnesyl geranyl diphosphate synthase (FGPPS).
17. The method of any one of claims 14 to 16, wherein the biosynthetic pathway(s) produce at least one terpenoid selected from alpha-sinensal, beta-Thujone, Camphor, Carveol, Carvone, Cineole, Citral, Citronellal, Cubebol, Geraniol, Limonene, Menthol, Menthone, Myrcene, Nootkatone, Nootkatoi, Patchouli, Piperitone, Sabinene, Steviol, Steviol glycoside, Taxadiene, Thymol, and Valencene, or derivative thereof.
18. The method of claim 17, wherein the biosynthetic pathway(s) produce steviol or steviol glycoside.
19. The method of claim 18, wherein the biosynthetic pathway comprises a geranylgeranyl pyrophosphate synthase (GPPS), a copalyl diphosphate synthase (CPPS), and a kaurene synthase (KS), as well as a kaurene oxidase (KO) and a kaureneoic acid hydroxylase (KAH) having the transmembrane domain derived from an E. coli gene.
20. The method of claim 18 or 19, wherein the biosynthetic pathway further comprises one or more uridine diphosphate dependent glycosyltransferase enzymes (UGT).
21. The method of claim 17, wherein the biosynthetic pathway produces Valencene and/or Nootkatone.
22. The method of claim 21, wherein the biosynthetic pathway comprises a farnesyl pyrophosphate synthase, a Valencene Synthase, and a Valencene Oxidase.
23. The method of any one of claims 1 to 22, wherein IbpA is not overexpressed during the culturing.
24. The method of any one of claims 1 to 23, wherein the culturing is conducted at 30.degree. C. or greater.
25. The method of claim 24, wherein the culturing is conducted at 32.degree. C. or greater.
26. The method of claim 24, wherein the culturing is conducted at 34.degree. C. or greater.
27. The method of any one of claims 1 to 26, wherein the size of the culture is at least 100 L.
28. The method of claim 27, wherein the size of the culture is at least 1000 L.
29. The method of claim 28, wherein the culturing is conducted in batch culture.
30. The method of claim 28, wherein the culturing is conducted in continuous culture or semi-continuous culture.
31. The method of claims 23 to 30, wherein the E. coli expresses one or more CPR enzymes as a translational fusion or operon with the P450 enzymes.
32. The method of any one of claims 1 to 31, wherein the P450 and the CPR are expressed separately, and the level of expression of the P450 enzyme and the CPR are approximately 2:1 to 1:2.
33. The method of any one of claims 1 to 32, wherein the cell expresses a single CPR protein.
34. The method of any one of claims 1 to 33, wherein at least one CPR partner comprises a membrane-anchor having a single pass transmembrane domain derived from an E. coli gene.
35. The method of any one of claims 1 to 34, wherein at least one membrane anchor is a single pass transmembrane domain derived from an E. coli gene selected from waaA, vpfN, yhcB, yhbM, yhhm, zipA, ycgG, djlA, sohB, lpxK, F11O, motA, htpx, pgaC, ygdD, hemr, and ycls, or derivative thereof.
36. The method of claim 35, wherein at least one membrane anchor is a single pass transmembrane domain derived from yhcB or zipA, or a derivative thereof.
37. The method of any one of claims 1 to 36, wherein the P450 enzyme has a deletion of part or all of its native N-terminal transmembrane domain.
38. The method of claim 37, wherein the P450 enzyme has an N-terminal truncation of from about 10 to about 50 amino acids with respect to the wildtype enzyme.
39. The method of claim 37, wherein the P450 enzyme has a N-terminal truncation of from about 15 to about 45 amino acids with respect to the wildtype enzyme.
40. The method of claim 37, wherein the P450 enzyme has an N-terminal truncation of from about 20 to about 40 amino acids with respect to the wildtype enzyme.
41. The method of claim 37, wherein the P450 enzyme has an N-terminal truncation of from about 20 to about 35 amino acids with respect to the wildtype enzyme.
42. The method of claim 37, wherein the P450 enzyme has an N-terminal truncation of about 29 or 30 amino acids with respect to the wildtype enzyme.
43. The method of any one of claims 1 to 42, wherein the membrane anchor is from about 8 to about 75 amino acids in length.
44. The method of claim 43, wherein the membrane anchor is from about 15 to about 50 amino acids in length.
45. The method of claim 43, wherein the membrane anchor is from about 20 to about 40 amino acids in length.
46. The method of claim 43, wherein the membrane anchor is from about 20 to about 30 amino acids in length.
47. The method of claim 43, wherein at least one membrane anchor is selected from: about the N-terminal 20 to 22 amino acids of yhcB, about the N-terminal 19 to 21 amino acids of yhhM, about the N-terminal 24 to 26 amino acids of zipA, about the N-terminal 21 to 23 amino acids of ypfN, about the N-terminal 27 to 29 amino acids of SohB, and about the N-terminal 20-22 amino acids of waaA, or derivative thereof.
48. The method of any one of claims 1 to 47, wherein the membrane anchor has from 1 to about 8 deletions, insertions, or substitutions relative to the wildtype E. coli sequence.
49. The method of any one of claims 37 to 48, wherein the length of the truncation and selection of anchor sequence is by testing ibpA expression in cultures.
50. The method of any one of claims 1 to 49, further comprising recovering the chemical species from the culture.
51. The method of any one of claims 1 to 49, wherein the culturing produces at least 25 mg/L of the chemical species.
52. The method of claim 51, wherein the culturing produces at least 50 mg/L of the chemical species or at least 100 mg/L of the chemical species.
53. The method of any one of claims 1 to 52, further comprising, incorporating the chemical species into a product.
54. A method for producing a product comprising one or more terpenoid compounds, comprising: expressing a terpenoid biosynthetic pathway in E. coli, the biosynthetic pathway comprising at least one membrane-anchored P450 enzyme having a transmembrane domain derived from an E. coli inner membrane cytoplasmic C-terminus protein; and culturing the E. coli to produce the one or more terpenoids from the biosynthetic pathway; recovering the terpenoid(s) from the culture; and incorporating the terpenoid into a product.
55. The method of claim 54, wherein the E. coli does not exhibit a substantially stressed phenotype during the culturing.
56. The method of claim 54 or 55, wherein the E. coli expresses at least two, at least three, or at least four recombinant enzymes.
57. The method of claim 56, wherein the terpenoid biosynthetic pathway comprises the overexpression of at least two foreign genes.
58. The method of claim 57, wherein the terpenoid biosynthetic pathway comprises the overexpression of at least three foreign genes, at least four foreign genes, or at least five foreign genes.
59. The method of any one of claims 54 to 58, wherein the E. coli contains an overexpression of at least two E. coli genes.
60. The method of claim 59, wherein the E. coli overexpresses at least one gene in the MEP pathway.
61. The method of claim 59 or 60, wherein at least one gene is expressed by a strong promoter.
62. The method of any one of claims 59 to 61, wherein at least one gene is expressed from a plasmid.
63. The method of any one of claims 59 to 62, wherein at least one gene is chromosomally integrated.
64. The method of any one of claims 54 to 63, wherein at least one P450 enzyme is not strongly expressed.
65. The method of any one of claims 54 to 64, wherein the E. coli expresses at least two P450 enzymes, which are optionally derived from plant P450 enzymes.
66. The method of claim 65, wherein the E. coli expresses a membrane-anchored P450 selected from CiVO, HmPO, LsGAO, BsGAO, NtEAO, SrKO, SrKAH, AtKAH, ZzHO, CpVO, MsL6OH, NtVO, StVO, AtKO, Ci2VO, AaAO, and Taxus 5-alpha hydroxylase, or derivative thereof.
67. The method of any one of claims 54 to 66, wherein the biosynthetic pathway produces a terpenoid selected from a monoterpenoid, a sesquiterpenoid, diterpenoid, a sesterpenoid, or a triterpenoid.
68. The method of claim 67, wherein the biosynthetic pathway involves overexpression of a geranyl diphosphate synthase (GPS), a gernanylgeranyl diphosphate synthase (GGPS), a farnsesyl diphosphate synthase (FPS), or a famesyl geranyl diphosphate synthase (FGPPS).
69. The method of any one of claims 54 to 68, wherein the biosynthetic pathway produces at least one terpenoid selected from alpha-sinensal, beta-Thujone, Camphor, Carveol, Carvone, Cineole, Citral, Citronellal, Cubebol, Geraniol, Limonene, Menthol, Menthone, Myrcene, Nootkatone, Nootkatol, Patchouli, Piperitone, Sabinene, Steviol, Steviol glycoside, Taxadiene, Thymol, and Valencene, or derivative thereof.
70. The method of claim 69, wherein the biosynthetic pathway(s) produce steviol or steviol glycoside.
71. The method of claim 70, wherein the biosynthetic pathway comprises a geranylgeranyl pyrophosphate synthase (GPPS), a copalyl diphosphate synthase (CPPS), and a kaurene synthase (KS), as well as a kaurene oxidase (KO) and a kaureneoic acid hydroxylase (KAH) having the transmembrane domain derived from an E. coli gene.
72. The method of claim 71, wherein the biosynthetic pathway further comprises one or more uridine diphosphate dependent glycosyltransferase enzymes (UGT).
73. The method of claim 69, wherein the biosynthetic pathway produces Valencene and/or Nootkatone.
74. The method of claim 73, wherein the biosynthetic pathway comprises a farnesyl pyrophosphate synthase, a Valencene Synthase, and a Valencene Oxidase.
75. The method of any one of claims 54 to 74, wherein IbpA is not overexpressed during the culturing.
76. The method of any one of claims 54 to 75, wherein the culturing is conducted at 30.degree. C. or greater.
77. The method of claim 76, wherein the culturing is conducted at 32.degree. C. or greater.
78. The method of claim 76, wherein the culturing is conducted at 34.degree. C. or greater.
79. The method of any one of claims 54 to 78, wherein the size of the culture is at least 100 L.
80. The method of claim 79, wherein the size of the culture is at least 1000 L.
81. The method of claim 79 or 80, wherein the culturing is conducted in batch culture.
82. The method of any one of claims 76 to 81, wherein the culturing is conducted in continuous culture or semi-continuous culture.
83. The method of claims 76 to 82, wherein the E. coli expresses one or more CPR enzymes as a translational fusion or operon with the P450 enzymes.
84. The method of any one of claims 54 to 83, wherein the P450 and the CPR are expressed separately, and the level of expression of the P450 enzyme and the CPR are approximately 2:1 to 1:2.
85. The method of any one of claims 54 to 84, wherein the cell expresses a single CPR protein.
86. The method of any one of claims 54 to 85, wherein at least one CPR partner comprises a membrane-anchor having a single pass transmembrane domain derived from an E. coli gene.
87. The method of any one of claims 54 to 86, wherein at least one membrane anchor is a single pass transmembrane domain derived from an E. coli gene selected from waaA, ypfN, yhcB, yhbM, yhhm, zipA, ycgG, djlA, sohB, lpxK, F11O, motA, htpx, pgaC, ygdD, hemr, and ycls, or derivative thereof.
88. The method of claim 87, wherein at least one membrane anchor is a single pass transmembrane domain derived from yhcB or zipA, or derivative thereof.
89. The method of any one of claims 54 to 88, wherein the P450 enzyme has a deletion of part or all of its native N-terminal transmembrane domain.
90. The method of claim 89, wherein the P450 enzyme has an N-terminal truncation of from about 10 to about 50 amino acids with respect to the wildtype enzyme.
91. The method of claim 89, wherein the P450 enzyme has a N-terminal truncation of from about 15 to about 45 amino acids with respect to the wildtype enzyme.
92. The method of claim 89, wherein the P450 enzyme has an N-terminal truncation of from about 20 to about 40 amino acids with respect to the wildtype enzyme.
93. The method of claim 89, wherein the P450 enzyme has an N-terminal truncation of from about 20 to about 35 amino acids with respect to the wildtype enzyme.
94. The method of claim 89, wherein the 1450 enzyme has an N-terminal truncation of about 29 or 30 amino acids with respect to the wildtype enzyme.
95. The method of any one of claims 54 to 94, wherein the membrane anchor is from about 8 to about 75 amino acids in length.
96. The method of claim 95, wherein the membrane anchor is from about 15 to about 50 amino acids in length.
97. The method of claim 95, wherein the membrane anchor is from about 20 to about 40 amino acids in length.
98. The method of claim 95, wherein the membrane anchor is from about 20 to about 30 amino acids in length.
99. The method of claim 95, wherein at least one membrane anchor is selected from: about the N-terminal 20 to 22 amino acids of yhcB, about the N-terminal 19 to 21 amino acids of yhhM, about the N-terminal 24 to 26 amino acids of zipA, about the N-terminal 21 to 23 amino acids of ypfN, about the N-terminal 27 to 29 amino acids of SohB, and about the N-terminal 20-22 amino acids of waaA, or derivative thereof.
100. The method of any one of claims 54 to 99, wherein the membrane anchor has from 1 to about 8 deletions, insertions, or substitutions relative to the wildtype E. coli sequence.
101. The method of claim 100, wherein the length of the truncation and selection of anchor sequence is by testing ibpA expression in cultures.
102. The method of any one of claims 54 to 101, further comprising recovering the chemical species from the culture.
103. The method of claim 102, wherein the culturing produces at least 25 mg/L of the chemical species.
104. The method of claim 102, wherein the culturing produces at least 50 mg/L of the chemical species or at least 100 mg/L of the chemical species.
105. An E. coli host cell expressing one or more recombinant biosynthetic pathways, where the biosynthetic pathways comprise at least one membrane-anchored P450 protein having a transmembrane domain derived from an E. coli inner membrane cytoplasmic C-terminus protein.
106. The host cell of claim 105, wherein the E. coli does not exhibit a substantially stressed phenotype during culturing.
107. The host cell of claim 105 or 106, wherein the E. coli expresses at least two, at least three, or at least four recombinant enzymes.
108. The host cell of claim 107, wherein the biosynthetic pathway(s) produce a secondary metabolite through the overexpression of at least two foreign genes.
109. The host cell of claim 108, wherein the biosynthetic pathway(s) produce a secondary metabolite through the overexpression of at least three foreign genes, at least four foreign genes, or at least five foreign genes.
110. The host cell of any one of claims 105 to 109, wherein the E. coli contains an overexpression of at least two E. coli genes.
111. The host cell of claim 110, wherein the E. coli overexpresses at least one gene in the MFP pathway.
112. The host cell of claim 110 or 111, wherein at least one gene is expressed by a strong promoter.
113. The host cell of any one of claims 110 to 112, wherein at least one gene is expressed from a plasmid.
114. The host cell of any one of claims 110 to 113, wherein at least one gene is chromosomally integrated.
115. The host cell of any one of claims 105 to 114, wherein at least one P450 enzyme is not strongly expressed.
116. The host cell of any one of claims 105 to 114, wherein the E. coli expresses at least two P450 enzymes, which are optionally derived from plant P450 enzymes.
117. The host cell of claim 116, wherein the E. coli expresses a membrane-anchored P450 selected from CiVO, HmPO, LsGAO, BsGAO, NtEAO, SrKO, SrKAH, AtKAH, ZzHO, CpVO, MsL6OH, NtVO, StVO, AtKO, Ci2VO, AaAO, and Taxus 5-alpha hydroxylase, or derivative thereof.
118. The host cell of any one of claims 105 to 117, wherein the biosynthetic pathway produces a secondary metabolite selected from a terpenoid, alkaloid, cannabinoid, steroid, saponin, glycoside, stilbenoid, polyphenol, antibiotic, polyketide, fatty acid, or non-ribosomal peptide.
119. The host cell of claim 118, wherein the biosynthetic pathway produces a terpenoid selected from a monoterpenoid, a sesquiterpenoid, diterpenoid, a sesterpenoid, or a triterpenoid.
120. The host cell of claim 119, wherein the biosynthetic pathway involves overexpression of a geranyl diphosphate synthase (GPS), a gernanylgeranyl diphosphate synthase (GGPS), a farnsesyl diphosphate synthase (FPS), or a farnesyl geranyl diphosphate synthase (FGPPS).
121. The host cell of any one of claims 118 to 120, wherein the biosynthetic pathway(s) produce at least one terpenoid selected from alpha-sinensal, beta-Thujone, Camphor, Carveol, Carvone, Cineole, Citral, Citronellal, Cubebol, Geraniol, Limonene, Menthol, Menthone, Myrcene, Nootkatone, Nootkatol, Patchouli, Piperitone, Sabinene, Steviol, Steviol glycoside, Taxadiene, Thymol, and Valencene, or derivative thereof.
122. The host cell of claim 121, wherein the biosynthetic pathway(s) produce steviol or steviol glycoside.
123. The host cell of claim 122, wherein the biosynthetic pathway comprises a geranylgeranyl pyrophosphate synthase (GPPS), a copalyl diphosphate synthase (CPPS), and a kaurene synthase (KS), as well as a kaurene oxidase (KO) and a kaureneoic acid hydroxylase (KAH) having the transmembrane domain derived from an E. coli gene.
124. The host cell of claim 122 or 123, wherein the biosynthetic pathway further comprises one or more uridine diphosphate dependent glycosyltransferase enzymes (UGT).
125. The host cell of claim 121, wherein the biosynthetic pathway produces Valencene and/or Nootkatone.
126. The host cell of claim 125, wherein the biosynthetic pathway comprises a farnesyl pyrophosphate synthase, a Valencene Synthase, and a Valencene Oxidase.
127. The host cell of any one of claims 105 to 126, wherein IbpA is not overexpressed during culturing.
128. The host cell of claims 105 to 127, wherein the E. coli expresses one or more CPR enzymes as a translational fusion or operon with the P450 enzymes.
129. The host cell of any one of claims 105 to 128, wherein the P450 and the CPR are expressed separately, and the level of expression of the P450 enzyme and the CPR are approximately 2:1 to 1:2.
130. The host cell of any one of claims 105 to 129, wherein the cell expresses a single CPR protein.
131. The host cell of any one of claims 105 to 130, wherein at least one CPR partner comprises a membrane-anchor having a single pass transmembrane domain derived from an E. coli gene.
132. The host cell of any one of claims 105 to 131, wherein at least one membrane anchor is a single pass transmembrane domain derived from an E. coli gene selected from waaA, ypfN, yhcB, yhbM, yhhm, zipA, ycgG, djlA, sohB, lpxK, F11O, motA, htpx, pgaC, ygdD, hemr, and ycls, or derivative thereof.
133. The host cell of claim 132, wherein at least one membrane anchor is a single pass transmembrane domain derived from yhcB or zipA, or derivative thereof.
134. The host cell of any one of claims 105 to 133, wherein the P450 enzyme has a deletion of part or all of its native N-terminal transmembrane domain.
135. The host cell of claim 134, wherein the P450 enzyme has an N-terminal truncation of from about 10 to about 50 amino acids with respect to the wildtype enzyme.
136. The host cell of claim 134, wherein the P450 enzyme has a N-terminal truncation of from about 15 to about 45 amino acids with respect to the wildtype enzyme.
137. The host cell of claim 134, wherein the P450 enzyme has an N-terminal truncation of from about 20 to about 40 amino acids with respect to the wildtype enzyme.
138. The host cell of claim 134, wherein the P450 enzyme has an N-terminal truncation of from about 20 to about 35 amino acids with respect to the wildtype enzyme.
139. The host cell of claim 134, wherein the P450 enzyme has an N-terminal truncation of about 29 or 30 amino acids with respect to the wildtype enzyme.
140. The host cell of any one of claims 105 to 139, wherein the membrane anchor is from about 8 to about 75 amino acids in length.
141. The host cell of claim 140, wherein the membrane anchor is from about 15 to about 50 amino acids in length.
142. The host cell of claim 140, wherein the membrane anchor is from about 20 to about 40 amino acids in length.
143. The host cell of claim 140, wherein the membrane anchor is from about 20 to about 30 amino acids in length.
144. The host cell of claim 140, wherein at least one membrane anchor is selected from: about the N-terminal 20 to 22 amino acids of yhcB, about the N-terminal 19 to 21 amino acids of yhhM, about the N-terminal 24 to 26 amino acids of zipA, about the N-terminal 21 to 23 amino acids of ypfN, about the N-terminal 27 to 29 amino acids of SohB, and about the N-terminal 20-22 amino acids of waaA, or derivatives thereof.
145. The host cell of any one of claims 105 to 144, wherein the membrane anchor has from 1 to about 8 deletions, insertions, or substitutions relative to the wildtype E. coli sequence.
146. The host cell of claim 145, wherein the length of the truncation and selection of anchor sequence is by testing ibpA expression in cultures.
147. A plant P450 enzyme comprising an N-terminal truncation and a single-pass transmembrane region derived from an E. coli inner membrane cytoplasmic C-terminus protein.
148. The enzyme of claim 147, wherein the membrane-anchored P450 is selected from CiVO, HmPO, LsGAO, BsGAO, NtEAO, SrKO, SrKAH, AtKAH, ZzHO, CpVO, MsL6OH, NtVO, StVO, AtKO, Ci2VO, AaAO, and Taxus 5-alpha hydroxylase, or derivative thereof.
149. The enzyme of claim 148, wherein at least one membrane anchor is a single pass transmembrane domain derived from an E. coli gene selected from waaA, ypfN, yhcB, yhbM, yhhm, zipA, ycgG, djlA, sohB, lpxK, F11O, motA, htpx, pgaC, ygdD, hemr, and ycls, or derivative thereof.
150. The enzyme of claim 149, wherein at least one membrane anchor is a single pass transmembrane domain derived from yhcB or zipA, or derivative thereof.
151. The enzyme of any one of claims 147 to 150, wherein the P450 enzyme has a deletion of part or all of its native N-terminal transmembrane domain.
152. The enzyme of claim 151, wherein the P450 enzyme has an N-terminal truncation of from about 10 to about 50 amino acids with respect to the wildtype enzyme.
153. The enzyme of claim 151, wherein the P450 enzyme has a N-terminal truncation of from about 15 to about 45 amino acids with respect to the wildtype enzyme.
154. The enzyme of claim 151, wherein the P450 enzyme has an N-terminal truncation of from about 20 to about 40 amino acids with respect to the wildtype enzyme.
155. The enzyme of claim 151, wherein the P450 enzyme has an N-terminal truncation of from about 20 to about 35 amino acids with respect to the wildtype enzyme.
156. The enzyme of claim 151, wherein the P450 enzyme has an N-terminal truncation of about 29 or 30 amino acids with respect to the wildtype enzyme.
157. The enzyme of any one of claims 147 to 156, wherein the membrane anchor is from about 8 to about 75 amino acids in length.
158. The enzyme of claim 157, wherein the membrane anchor is from about 15 to about 50 amino acids in length.
159. The enzyme of claim 157, wherein the membrane anchor is from about 20 to about 40 amino acids in length.
160. The enzyme of claim 157, wherein the membrane anchor is from about 20 to about 30 amino acids in length.
161. The enzyme of claim 157, wherein at least one membrane anchor is selected from: about the N-terminal 20 to 22 amino acids of yhcB, about the N-terminal 19 to 21 amino acids of yhhM, about the N-terminal 24 to 26 amino acids of zipA, about the N-terminal 21 to 23 amino acids of ypfN, about the N-terminal 27 to 29 amino acids of SohB, and about the N-terminal 20-22 amino acids of waaA, or derivative thereof.
162. The enzyme of any one of claims 147 to 161, wherein the membrane anchor has from 1 to about 8 deletions, insertions, or substitutions relative to the wildtype E. coli sequence.
163. A polynucleotide encoding the enzyme of any one of claims 147 to 162.
Description
FIELD OF THE INVENTION
[0001] The present invention relates to production of chemical species through oxidative chemistry in bacterial host cells. Particularly, the present invention provides P450 enzymes engineered for functional expression in bacterial host cells such as E. coli.
BACKGROUND OF THE INVENTION
[0002] E. coli is widely used for production of chemicals by the recombinant expression of biosynthetic pathways, which can involve overexpression of several native and/or foreign genes. However, where the biosynthetic pathway involves recombinant expression of one or more cytochrome P450 enzymes (e.g., to perform oxidative chemistry), other host organisms such as yeast are generally preferred. See Chang & Keasling, Nat. Chem. Bio. 2006. The perceived limitations of the bacterial system for oxidative chemistry include the absence of electron transfer machinery and P450-reductases (CPRs), and translational incompatibility of the membrane signal modules of P450 enzymes due to the lack of an endoplasmic reticulum. Thus, it remains a commonly held belief in the scientific community that E. coli is a generally unsuitable host for P450 expression and expression of biosynthetic pathways incorporating the same. See Tippman et al., Biotech. Journal (2013); Thodey et al., Nat. Chem. Bio. (2014).
[0003] It is an object of the invention to improve productivity of P450 enzymes in bacterial platforms such as E. coli, to thereby expand the utility of these platforms for P450 chemistry.
SUMMARY OF THE INVENTION
[0004] In various aspects, the invention provides P450 enzymes engineered for functional expression in bacterial cells (e.g., E. coli), and polynucleotides encoding the same. The invention further provides bacterial host cells expressing the engineered P450 enzymes, and methods for producing chemical species through recombinant expression of biosynthetic pathways involving P450 enzymes.
[0005] The engineered P450 enzymes described herein have a deletion of all or part of the wild type P450 N-terminal transmembrane region, and the addition of a transmembrane domain derived from an E. coli inner membrane, cytoplasmic C-terminus protein. It is believed that the transmembrane domain acts to anchor the P450 in the E. coli inner membrane. In various embodiments, the transmembrane domain is a single-pass transmembrane domain.
[0006] In various embodiments, the transmembrane domain (or "N-terminal anchor") is derived from an E. coli gene selected from waaA, yptN, yhcB, yhbM, yhhm, zipA, ycgG, djlA, sohB, lpxK, F11O, motA, htpx, pgaC, ygdD, hemr, and ycls. These genes were identified as inner membrane, cytoplasmic C-terminus proteins through bioinformatic prediction as well as experimental validation. The invention may employ an N-terminal anchor sequence that is a derivative of the E. coli wild-type transmembrane domain, that is, having one or more mutations with respect to the wild-type sequence.
[0007] In some aspects, the invention provides methods for the production of chemical species by expressing in E. coli cells one or more biosynthetic pathways including at least one membrane-anchored P450 (CYP) enzyme, and culturing the E. coli cells to produce the chemical species. At least one membrane-anchored P450 enzyme contains a transmembrane domain derived from an E. coli inner membrane, cytoplasmic C-terminus protein. As demonstrated herein, previous methods for expressing P450 proteins in E. coli can result in a substantial stress response, which limits productivity of the host cell. E. coli cells expressing the engineered P450 enzymes described herein do not exhibit a substantially stressed phenotype in some embodiments, thereby improving pathway productivity.
[0008] The invention in various aspects is applicable to various P450 enzymes, including plant-derived P450 enzymes, which can be further engineered fur productivity in a bacterial host cell system. These engineered P450 enzymes can be used in the production of a variety of chemical species through recombinant pathway expression, including but not limited to production of terpenoid compounds. Terpenoids represent a diverse class of molecules that provide beneficial health and nutritional attributes, as well as numerous other commercial applications. For example, terpenoids find use in the food and beverage industries as well as the perfume, cosmetic and health care industries.
[0009] In various embodiments, the E. coli cell is used for the production of chemicals by the recombinant expression of biosynthetic pathways, which can involve overexpression of several native and/or foreign genes. Often, expression of several foreign genes in E. coli and/or overexpression of native E. coli genes can induce a substantial stress response, which limits productivity. Conventional expression of P450 enzymes in E. coli, together with cytochrome P450 reductase (CPR) partners to regenerate the cofactor, can substantially add to this stress response, as exhibited for example by overexpression of IbpA, a protein that is overexpressed in E. coli under conditions of high protein aggregation and stress. It is critical that the P450 enzyme expression induce as little cell stress as possible to avoid limits on pathway productivity. Accordingly, the invention helps minimize cellular stress in a host E. coli cell to increase productivity of the host cell for production of chemical species.
[0010] Other aspects and embodiments of the invention will be apparent from the following detailed disclosure.
DESCRIPTION OF THE FIGURES
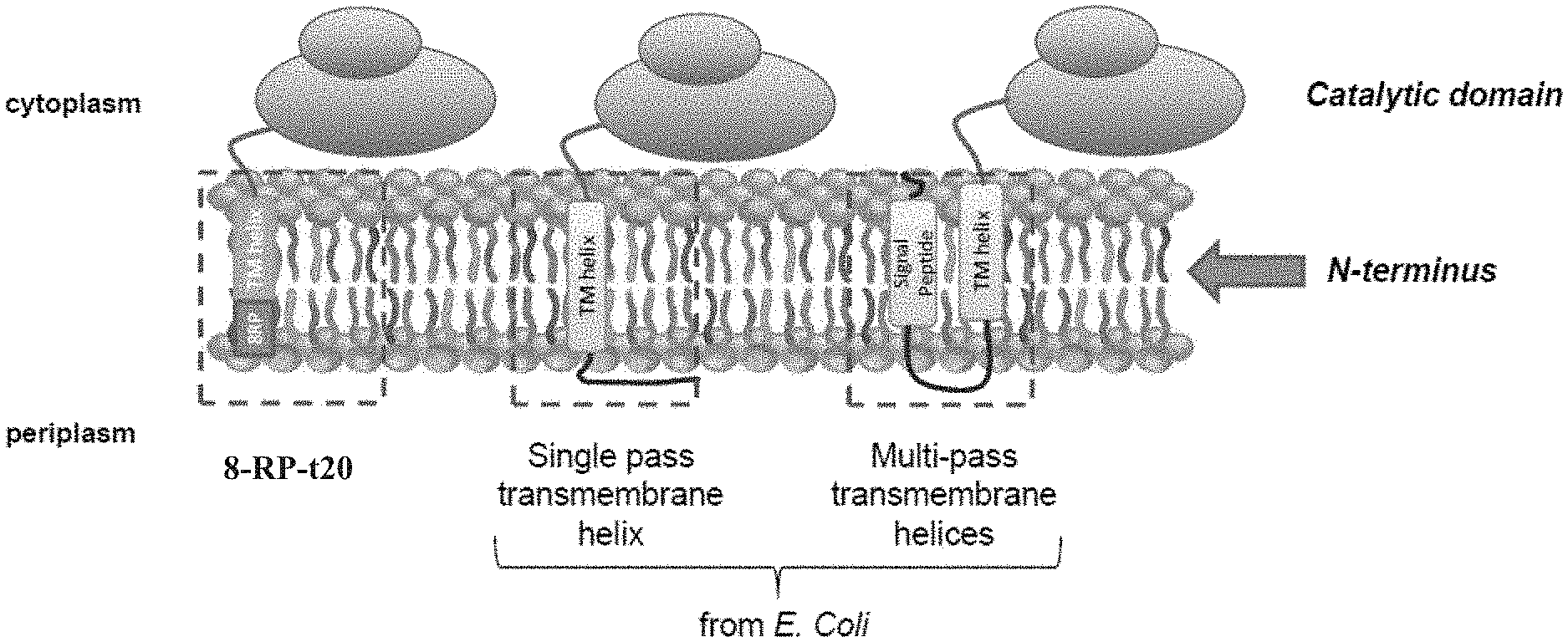
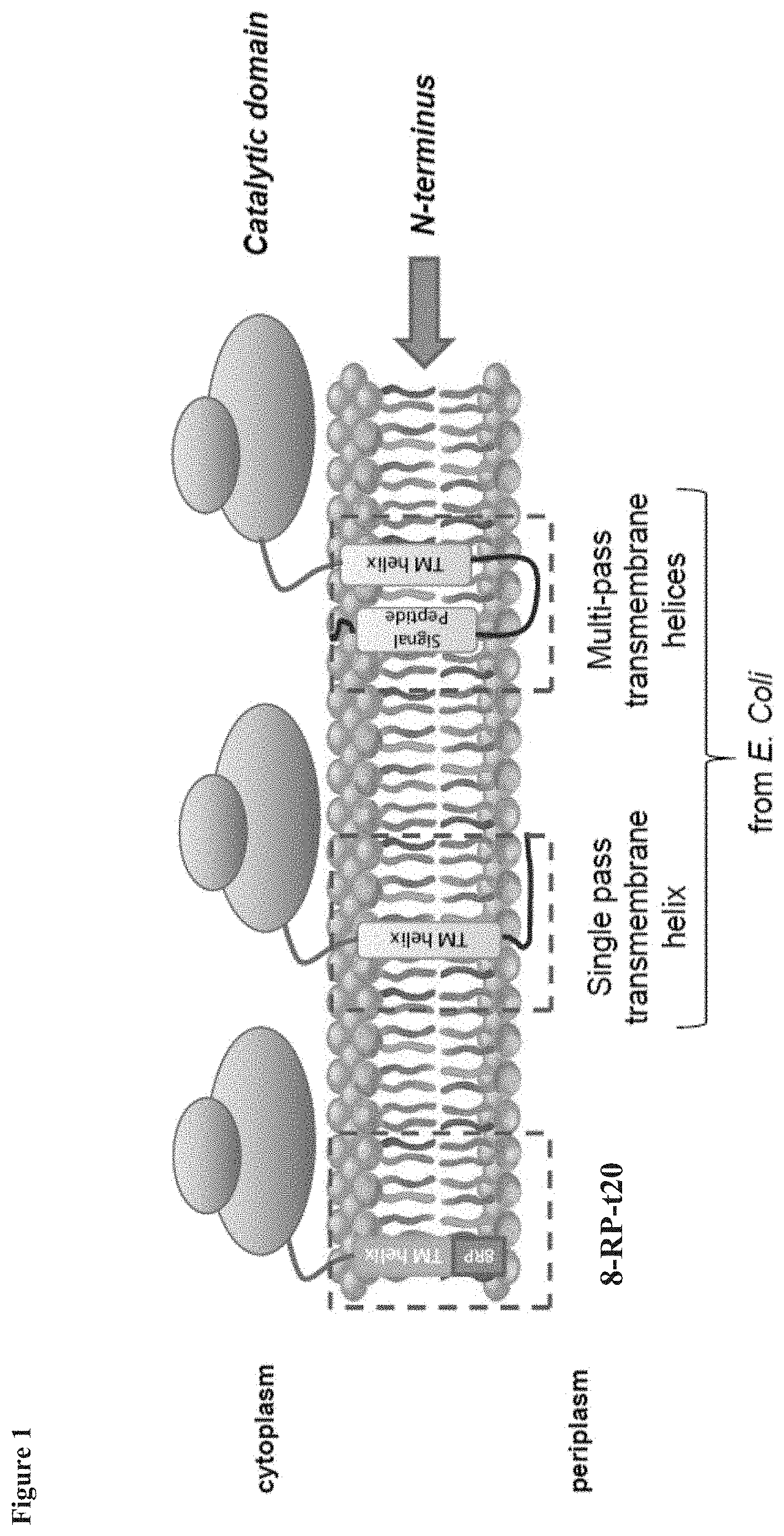
[0011] FIG. 1 illustrates N-terminal anchors for expressing P450 proteins in E. coli, including the previous designs based on truncation of the P450 transmembrane helix with the addition of an 8-amino acid peptide (8RP), and the use of single-pass and multi-pass transmembrane helices from E. coli proteins as described herein.
[0012] FIG. 2 shows a computational prediction of signal peptides and/or transmembrane helices using the Phobius predictive tool. SrKO and AtKAH are predicted to have an N-terminal transmembrane region. Bovine P450-C17 N-terminus is predicted as a signal peptide.
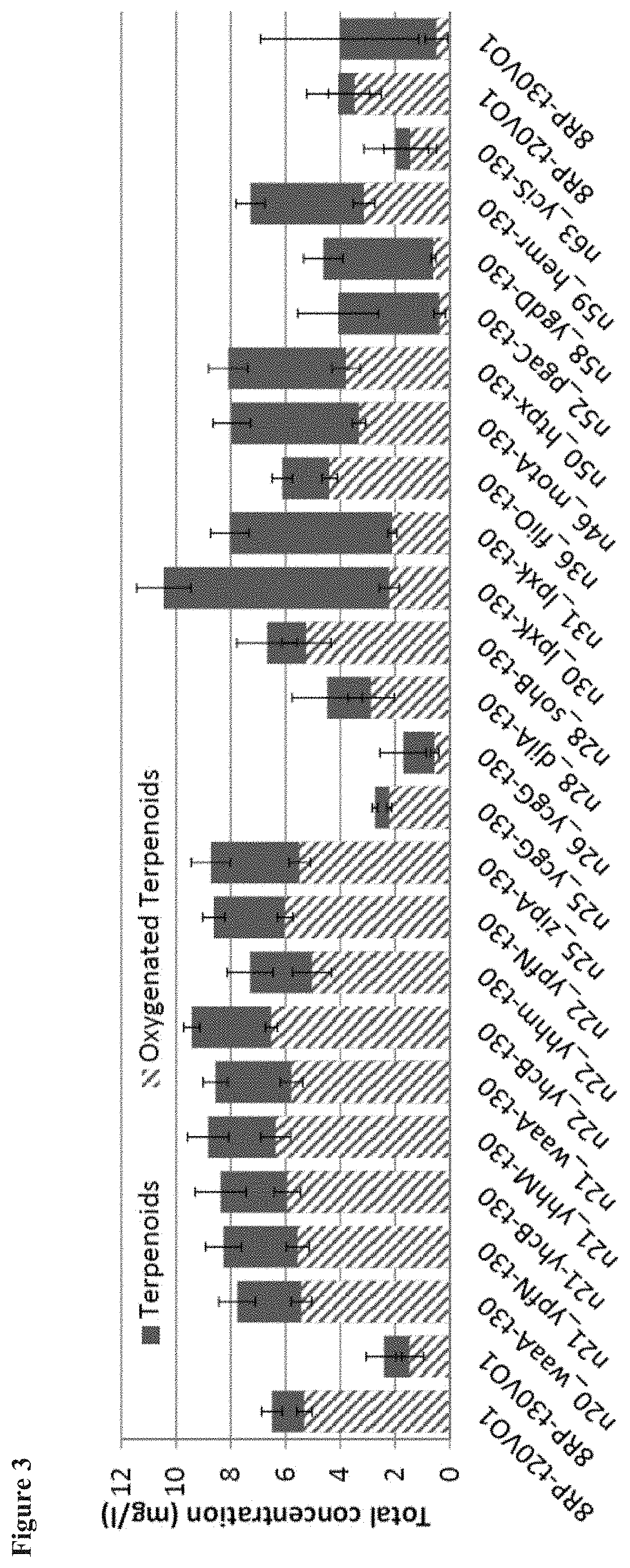
[0013] FIG. 3 shows the total terpenoid flux and oxygenated terpenoid formation upon expression in E. coli of Valencene Oxidase (VO) enzymes truncated at residue 30 and having various E. coli anchors. The E. coli cells express a valencene synthesis pathway, producing the valencene substrate. Results with control VO enzymes having the 8rp signal peptide with truncation of 20 or 30 residues are also shown. Enzymes include a translationally coupled CPR.
[0014] FIG. 4 shows the total terpenoid flux and oxygenated terpenoid formation upon expression in E. coli of VO enzymes truncated at residue 30 with candidate E. coli anchors (sequences from yhcB, yhhM, zipA, ypfN, sohB, and waaA).
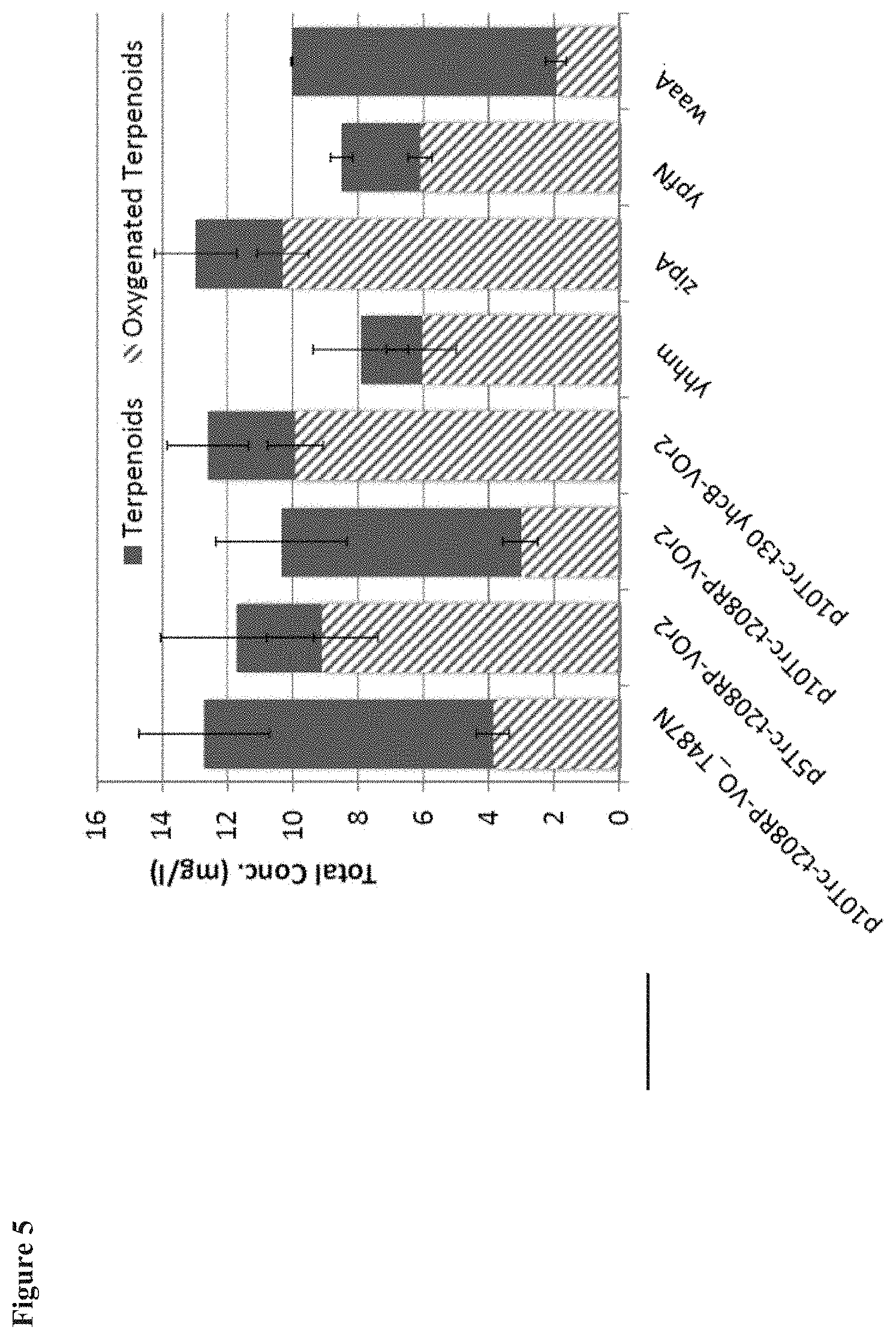
[0015] FIG. 5 shows the total terpenoid flux and oxygenated terpenoid formation upon expression of VO enzymes from a p10 expression plasmid or a p5 expression plasmid. While the 8RP anchor shows markedly decreased productivity at the higher expression level provided by a p10 plasmid, the higher expression level has little impact on productivity with the VO enzymes engineered with the E. coli anchor sequences.
[0016] FIG. 6 shows the total terpenoid flux and oxygenated terpenoid titers upon expression of the VO enzymes in a valencene-producing E. coli strain with a cytochrome P450 reductase (CPR) partner. The VO enzymes were expressed with the CPR either as separate proteins from the same operon or were translationally coupled.
[0017] FIG. 7 shows the level of VO protein expression with candidate N-terminal E. coli anchors (i.e., yhcB, yhhm, zipA, and ypfN), in comparison to t20-8RP. All four of the native E. coli anchors show lower total VO expression compared to 8RP, which results in a significantly lower relative VO/CPR ratio.
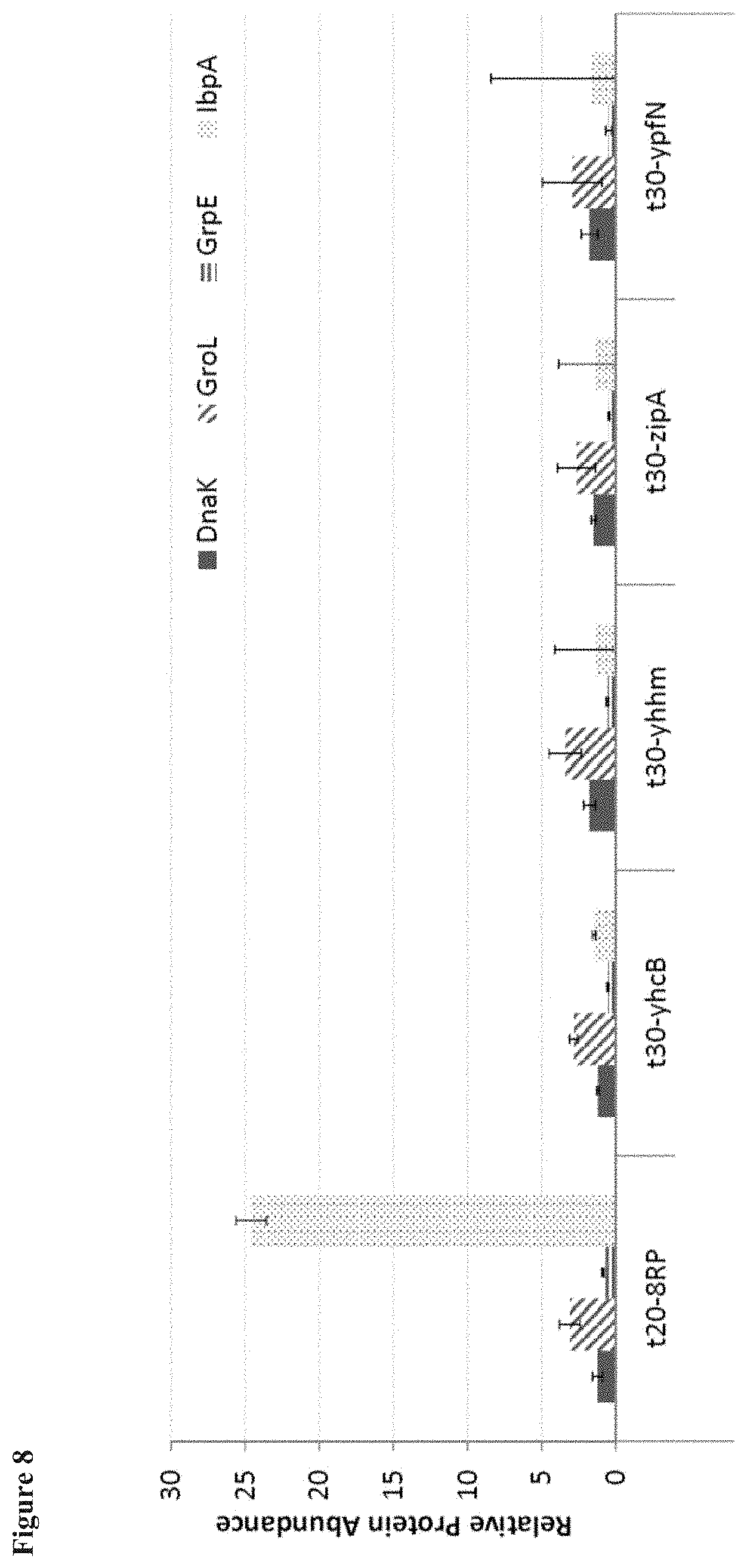
[0018] FIG. 8 shows the cellular stress response to VO expression with different N-terminal E. coli anchors, by assessing known E. coli stress response proteins. The IbpA protein, which is overexpressed in E. coli under conditions of high protein aggregation, was highly expressed in response to t20-8RP expression, but not with the native E. coli anchors.
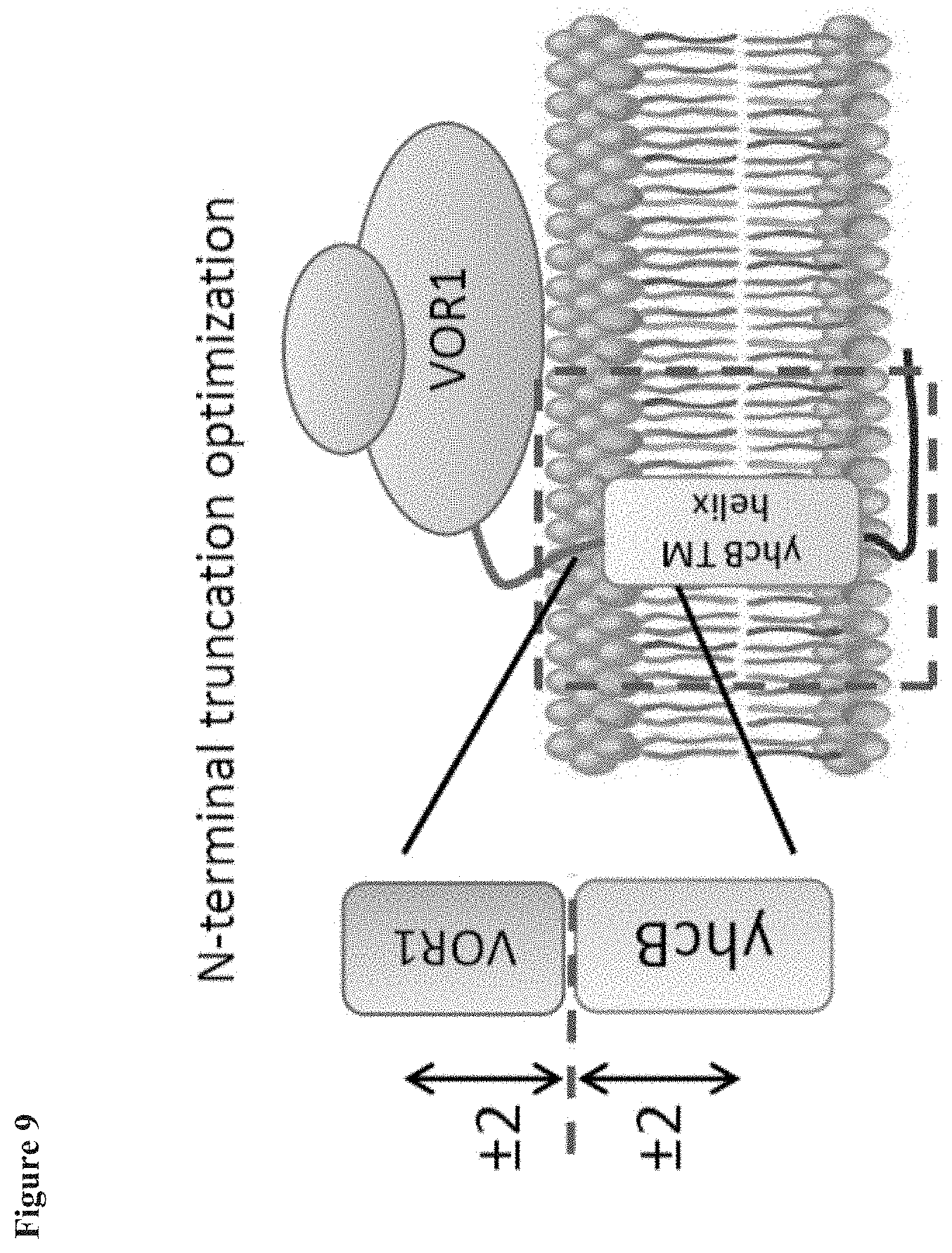
[0019] FIG. 9 is a diagram showing the optimization of truncation length and anchor length of yhcB anchored VO enzymes.
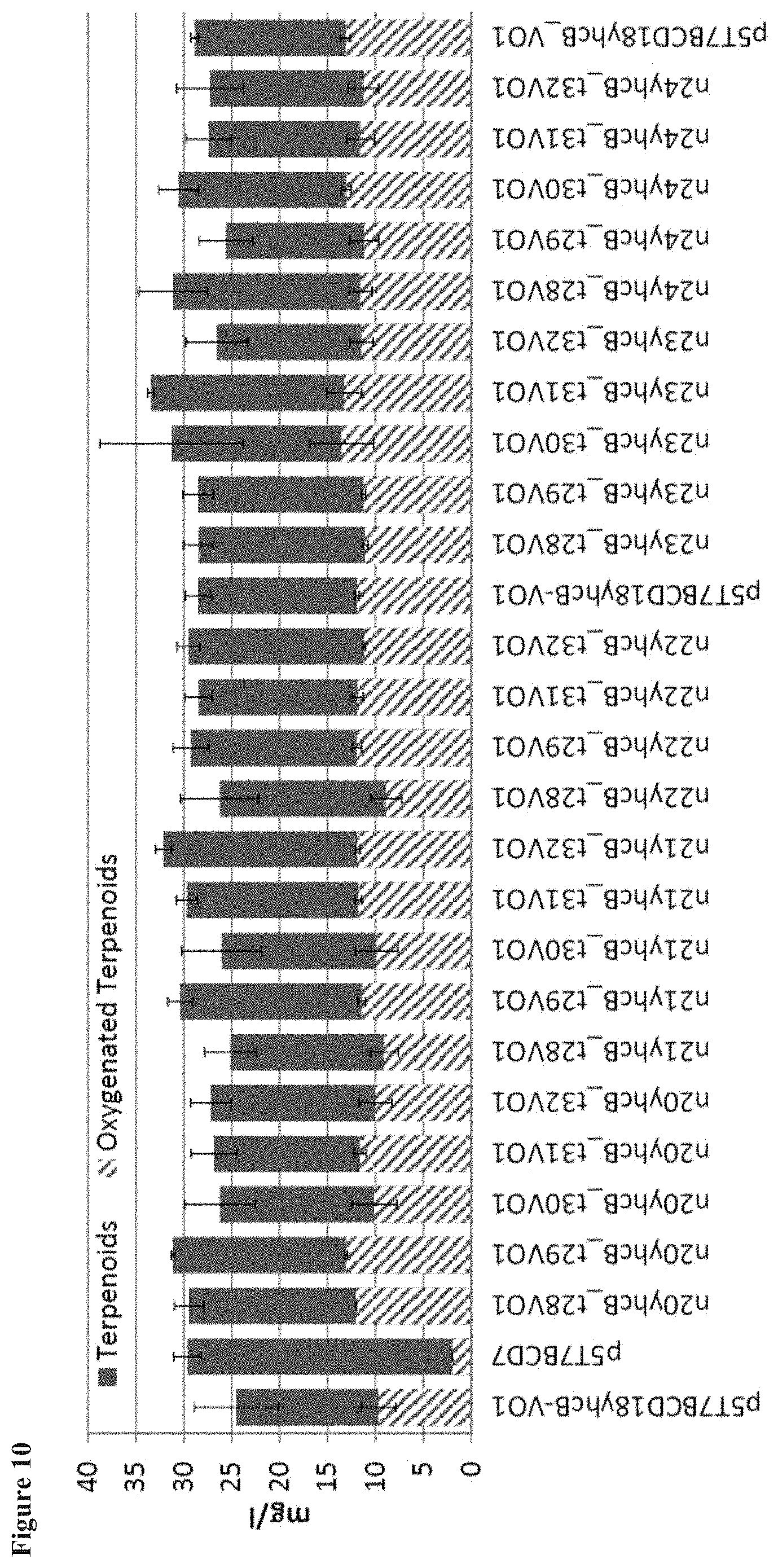
[0020] FIG. 10 shows the total terpenoid flux and oxygenated terpenoid production with various truncated and yhcB anchored VO enzymes. Truncations varied from 28 to 32 amino acids of the Valencene Oxidase, and the anchored VO included from 20 to 24 amino acids of the yhcB N-terminus.
[0021] FIG. 11 shows the E(z) score of AtKAH enzymes truncated at residue 26 with various E. coli anchors. Wild-type (wt) is the score in the wild-type E. coli enzyme, nXX is the number of residues taken from the protein for a swap with the E. coli anchor, and the name of the source protein. Asterisks show truncations not tested with VO. E(z) score estimates the suitability of a protein sequence for insertion into a cell membrane based on statistics from solved transmembrane crystal structures.
[0022] FIG. 12A shows kaurenoic acid formation upon expression in kaurene-producing strains of SrKO enzymes engineered by truncation at residue 30, and with addition of E. coli anchors. The enzymes were expressed from a p5 plasmid along with a cytochrome P450 reductase (CPR) in the same operon. Kaurenoic acid formation is shown at 30.degree. C. and 34.degree. C. FIG. 12B shows the kaurenoic acid formation/OD of the E. coli strains.
[0023] FIG. 13 shows a detailed proteomic analysis of SrKO expressing cells. The relative abundance of various pathway and stress response proteins is assessed. SrKO is significantly over expressed when paired with non-native anchors (E. coli or 8rp) although the increased expression is dampened at higher temperatures (34.degree. C. vs. 30.degree. C.). IbpA stress response is also significantly increased at the higher temperature.
DETAILED DESCRIPTION OF THE INVENTION
[0024] The invention provides for improved functional expression of P450 enzymes in bacterial host cells, and in particular, bacterial cells that do not naturally possess P450 enzymes, such as E. coli. While these bacterial platforms are widely used for production of a wide variety of chemicals, they are generally considered insufficient when P450 chemistry is required. The perceived limitations of bacterial systems such as E. coli for oxidative chemistry include the absence of electron transfer machinery and P450-reductases (CPRs), and translational incompatibility of the membrane signal modules of P450 enzymes due to the lack of an endoplasmic reticulum.
[0025] Basic P450 expression in E. coli has been obtained by co-expression of the P450 with a CPR. The P450 enzyme contained a truncation of at least part of the native P450 transmembrane region, which was replaced with the 8 amino acid tag MALLLAVF (which is derived from bovine P45017.alpha.) at the N-terminus. The present invention demonstrates that this tag is far from optimal, and results in a substantial cell stress response.
[0026] The invention provides P450 enzymes engineered for functional expression in bacterial platforms such as E. coli. The P450 enzymes comprise an N-terminal membrane anchor sequence derived from a native E. coli inner membrane protein having a cytoplasmic C-terminus. The N-terminal membrane anchor sequence replaces some or the entire native P450 N-terminal transmembrane region, where present in the wild-type enzyme. Expression of the engineered P450 enzymes in bacteria induces less cell stress than previous attempts to functionally express P450 enzymes in E. coli, for example. The invention allows for increases in biosynthetic productivity in bacterial host platforms, due in-part, to substantial improvements in P450 efficiency and to minimizing the cell stress response.
[0027] In one aspect, the present invention relates to a P450 enzyme having a transmembrane domain derived from an E. coli inner membrane protein having a cytoplasmic C-terminus. The E. coli transmembrane domain (or derivative thereof) replaces part or the entire native P450 N-terminal transmembrane region. In some embodiments, the transmembrane domain is a single-pass transmembrane domain, or in other embodiments, is a multi-pass (e.g., 2, 3, or more transmembrane helices) transmembrane domain.
[0028] The P450 enzyme may be derived from any source, including plants, animals, or microbes. The P450 enzyme may be a CYP70, CYP71, CYP73, CYP76 (e.g., CYP76F), CYP82 or CYP92 family P450. The P450 may be an enzyme disclosed in U.S. Pat. No. 8,722,363, which is hereby incorporated by reference. In some embodiments, the P450 is a plant P450 enzyme. Plant cytochrome P450s are involved in a wide range of biosynthetic reactions, leading to various fatty acid conjugates, plant hormones, defensive compounds, or medically and commercially important compounds, including terpenoids. Terpenoids represent the largest class of characterized natural plant compounds and are often substrates for plant P450 enzymes. In some embodiments, the P450 is derived from a species selected from Zingiber sp., Barnadesia sp., Hyoscyamus sp., Latuca sp., Nicotiana sp., Citrus sp., Artemesia sp., Arabidopsis sp, Stevia sp., Bacillus sp., Pleurotus sp., Cichorium sp., Helianthus sp., and Physcomitrella sp., Taxus sp., Rosa sp., Cymbopogon sp., Humulus sp., Pogostemon sp., and Cannabis sp. Wild-type P450 enzyme sequences are known and publically available, and/or can be obtained by genetic analysis of select plants based on well-known P450 motifs. See, for example, Saxena A. et al., Identification of cytochrome P450 heme motif in plants proteome, Plant Omics (2013); Chapple C., Molecular-genetic analysis of plant cytochrome p450-dependent monooxygenases, Annual Review of Plant Physiology and Plant Molecular Biology Vol. 49:311-343 (1998).
[0029] Table 1 provides a list of exemplary P450 enzymes that may be engineered in accordance with the invention:
TABLE-US-00001 TABLE 1 Native Reaction Species Name Native Substrate Product Zingiber zzHO .alpha.-humulene 8-hydroxy-.alpha.- zerumbet humulene Barnadesia BsGAO germacrene A germacra- spinosa 1(10),4,11(13)- trien-12-ol Hyoscyamus HmPO premnaspirodiene solavetivol muticus Latuca spicata LsGAO germacrene A germacra- 1(10),4,11(13)- trien-12-ol Nicotiana NtEAO 5-epi-aristolochene capsidiol tabacum Citrus .times. CpVO valencene nootkatol paradisi Artemesia AaAO amorphadiene artemisinic acid annua Arabidopsis AtKO kaurene kaurenoic acid thaliana Stevia SrKO kaurene kaurenoic acid rebaudiana Physcomitrella PpKO kaurene kaurenoic acid patens Bacillus BmVO fatty acids hydroxylated megaterium FAs Pleurotus PsVO valencene nootkatone sapidus Pleurotus PoLO unknown unknown ostreatus Cichorium CiVO valencene nootkatone intybus Helianthus HaGAO germacrene A germacrene anmius A acid
[0030] Thus, the engineered P450 enzyme may be based on wild-type sequences of ZzHO (SEQ ID NO: 1), BsGAO (SEQ ID NO: 2), HmPO (SEQ ID NO: 3), LsGAO (SEQ ID NO: 4), NtEAO (SEQ ID NO: 5), CpVO (SEQ ID NO: 6), AaAO (SEQ ID NO: 7), AtKO (SEQ ID NO: 8), SrKO (SEQ ID NO: 9), PpKO (SEQ ID NO:10), BmVO (SEQ ID NO: 11), PsVO (SEQ ID NO:12), PoLO (SEQ ID NO: 13), CiVO (SEQ ID NO: 14), or HaGAO (SEQ ID NO: 15).
[0031] Additional P450 enzymes that can be engineered in accordance with the invention include limonene-6-hydroxylase (AAQ18706.1, AAD44150.1), (-)-limonene-3-hydroxylase (EF426464, AY622319), kaurenoic acid 13-hydroxylase (EU722415.1), carotenoid cleavage dioxygenase (ABY60886.1, BAJ05401.1), beta-carotene hydroxylase (AAA64983.1), amorpha-4,11-diene monoxygenase (DQ315671), taxadiene 5-alpha hydroxylase (AY289209.2), 5-alpha-taxadienol-10-beta-hydroxylase (AF318211.1), taxoid 10-beta hydroxylase (AY563635.1), taxane 13-alpha-hydroxylase (AY056019.1), taxane 14b-hydroxylase (AY188177.1), taxoid 7-beta-hydroxylase (AY307951.1). The amino acid and encoding nucleotide sequences of these enzymes are hereby incorporated by reference. Derivatives of these P450s may be constructed in accordance with this disclosure.
[0032] The particular P450 enzyme scaffold can be selected based on the desired substrate specificity, which may be its natural substrate, or a non-natural substrate similar to the natural substrate, or otherwise determined experimentally. P450's can have varying substrate specificities, and thus can be engineered for chemistry on non-natural substrates. See, for example, Wu et al., Expansion of substrate specificity of cytochrome P450 2A6 by random and site-directed mutagenesis, J. Biol. Chem 280(49): 41090-100 (2005). Exemplary substrates for P450 chemistry include various secondary metabolites such as, without limitation, terpenoids, alkaloids, cannabinoids, steroids, saponins, glycosides, stilbenoids, polyphenols, antibiotics, polyketides, fatty acids, and non-ribosomal peptides. Exemplary products that may be produced through P450 chemistry include, without limitation, lutein, tocopherol, abietic acid, mogroside, forskolin, amyrin, lupeol, butyrospermol, quillic acid, triterpenoid saponins, oleanic acid, betulinic acid, boswellic acid, gymnemic acid, banaba/corosolic acid, cissas keto-steroid, curcurbitane triterpenoid, santalol, marrubiin, montbretin A, tropolone, sclareol, pseudolaric acid, grindelic acid, kauralexin, viteagnusin, diterpenoid epoxide triptolide, quinone triterpene celastrol, gibberellic acid, pseudolaric acid, carveol, carvone, nootkatol, nootkatone, piperitone, steviol, perillaldehyde, tagetone, verbenone, menthol, thymol, 3-oxo-alpha-lonone, zeanthin, artemisinin, taxol, gingkolide, gossypol, pseudoterosin, crotophorbolone, englerin, psiguadial, stemodinone, maritimol, cyclopamine, veratramine, aplyviolene, macfarlandin E, betulinic acid, oleanolic acid, ursoloic acid, dolichol, lupeol, euphol, cassaic acid, erthroxydiol, trisporic acid, podocarpic acid, retene, dehydroleucodine, phorbol, cafestol, kahweol, tetrahydrocannabinol, androstenol, tanshinone IIA or IIB or VI, cryptotanshinone, 15,16-dihydrotanshinone, trijuganone A or B, dihydrotanshinone I, miltirone, ferruginol, hydrotanshinone IIA, and 1,2-dihydromitotanshinone.
[0033] Exemplary terpenoid products that may be produced in accordance with the invention are described in U.S. Pat. No. 8,927,241, which is hereby incorporated by reference, and include: alpha-sinensal, beta-Thujone, Camphor, Carveol, Carvone, Cineole, Citral, Citronellal, Cubebol, Geraniol, Limonene, Menthol, Menthone, Nootkatone, Nootkatol, Patchouli, Piperitone, Sabinene, Steviol, Steviol glycoside, Taxadiene, and Thymol.
[0034] In various embodiments, the engineered P450 enzyme comprises an amino acid sequence that has at least about 30% sequence identity, at least about 40% sequence identity, or at least about 50% sequence identity to any one of SECS ID NOS: 1-15, or other P450 enzyme described herein. While the P450 need not display high sequence identity to these exemplary P450 enzymes in some embodiments, the P450 exhibits well-known P450 motifs and/or secondary structure. Generally, P450 sequence identity is determined by alignment of the full amino acid sequences, except for the N-terminal transmembrane regions (e.g., the alignment does not include about the first 30 amino acids of the wild-type sequence). In some embodiments, the engineered P450 enzyme comprises an amino acid sequence that has at least about 60% identity, at least about 70% identity, at least about 75% identity, at least about 80% identity, at least about 85% identity, at least about 90% identity, at least about 95% identity, at least about 96% identity, at least about 97% identity, or at least about 98% identity to any one of SEQ ID NOS: 48 to 60. SEQ ID NOS: 48 to 60 show P450 enzymes without the predicted transmembrane region (txx is the length of the N-terminal truncation): t22ZzHO (SEQ ID NO:48), t20BsGAO (SEQ ID NO:49), t16HmPO (SEQ NO:50), t19LsGAO (SEQ ID NO:51), t16NtEAO (SEQ ID NO:52), t26CpVO (SEQ ID NO:53), t23AaAO (SEQ ID NO:54), t21AtKO (SEQ ID NO:55), t30SrKO (SEQ ID NO:56), t52PpKO (SEQ ID NO:57), t15PsVO (SEQ ID NO:58), t20CiVO (SEQ ID NO:59), t20HaGAO (SEQ ID NO:60).
[0035] The similarity of nucleotide and amino acid sequences, i.e. the percentage of sequence identity, can be determined via sequence alignments. Such alignments can be carried out with several art-known algorithms, such as with the mathematical algorithm of Karlin and Altschul (Karlin & Altschul (1993) Proc. Nail. Acad. Sci. USA 90: 5873-5877), with hmmalign (HMMER package, http://hmmer.wustl.edu/) or with the CLUSTAL algorithm (Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-80). The grade of sequence identity (sequence matching) may be calculated using e.g. BLAST, BLAT or BlastZ (or BlastX). A similar algorithm is incorporated into the BLASTN and BLASTP programs of Altschul et al (1990) J. Mol. Biol. 215: 403-410. BLAST polynucleotide searches can be performed with the BLASTN program, score=100, word length=12.
[0036] BLAST protein searches may be performed with the BLASTP program, score=50, word length=3. To obtain gapped alignments for comparative purposes, Gapped BLAST is utilized as described in Altschul et al (1997) Nucleic Acids Res. 25: 3389-3402. When utilizing BLAST and Gapped BLAST programs, the default parameters of the respective programs are used. Sequence matching analysis may be supplemented by established homology mapping techniques like Shuffle-LAGAN (Brudno M., Bioinformatics 2003b, 19 Suppl 1:154-162) or Markov random fields.
[0037] In various embodiments, the engineered P450 enzyme may comprise an amino acid sequence having one or more amino acid mutations relative to the wild-type sequence, not including the modifications to the N-terminal transmembrane region (e.g., about the first 18 to 30 amino acids). For example, the P450 enzyme may comprise an amino acid sequence having from 1 to about 50, or from 1 to about 40, or from 1 to about 30, or from 1 to about 25, or from 1 to about 20, or from 1 to about 15, or from 1 to about 10, or from 1 to about 5 mutations relative to the wild- type sequence (e.g., any one of SEQ ID NOS: 48 to 60). In some embodiments, the one or more amino acid mutations may be independently selected from substitutions, insertions, deletions, and truncations. In some embodiments, the amino acid mutations are amino acid substitutions, and may include conservative and/or non-conservative substitutions.
[0038] "Conservative substitutions" may be made, for instance, on the basis of similarity in polarity, charge, size, solubility, hydrophobicity, hydrophilicity, and/or the amphipathic nature of the amino acid residues involved. The 20 naturally occurring amino acids can be grouped into the following six standard amino acid groups:
[0039] (1) hydrophobic: Met, Ala, Val, Leu, Ile;
[0040] (2) neutral hydrophilic: Cys, Ser, Thr; Asn, Gin;
[0041] (3) acidic: Asp, Glu;
[0042] (4) basic: His, Lys, Arg;
[0043] (5) residues that influence chain orientation: Gly, Pro; and
[0044] (6) aromatic: Trp, Tyr, Phe.
[0045] As used herein, "conservative substitutions" are defined as exchanges of an amino acid by another amino acid listed within the same group of the six standard amino acid groups shown above. For example, the exchange of Asp by Glu retains one negative charge in the so modified polypeptide. In addition, glycine and proline may be substituted for one another based on their ability to disrupt .alpha.-helices. Some preferred conservative substitutions within the above six groups are exchanges within the following sub-groups: (i) Ala, Val, Leu and Ile; (ii) Ser and Thr; (ii) Asn and Gin; (iv) Lys and Arg; and (v) Tyr and Phe.
[0046] As used herein, "non-conservative substitutions" are defined as exchanges of an amino acid by another amino acid listed in a different group of the six standard amino acid groups (1) to (6) shown above.
[0047] In various embodiments, the engineered P450 enzyme has a deletion or truncation of part or all of it native transmembrane domain. Generally, the deletion or truncation is about the first 15 to 30 amino acids, and the desired length can be determined based on the present disclosure and using predictive tools known in the art (e.g., PHOBIUS, http://phobius.sbc.su.se/). See Lukas K, et al., A Combined Transmembrane Topology and Signal Peptide Prediction Method, Journal of Molecular Biology, 338(5):1027-1036 (2004); Lukas K, et al., An HMM posterior decoder for sequence feature prediction that includes homology information, Bioinformatics, 21 (Suppl 1):i251-i257 (2005); Lukas K, et al., Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server, Nucleic Acids Res., 35:W429-32 (2007).
[0048] In various embodiments, the engineered P450 enzyme may have an N-terminal truncation of from about 10 to about 55 amino acids, or from about 15 to about 45 amino acids, or from about 15 to about 40 amino acids, or from about 15 to about 35 amino acids with respect to the wild-type enzyme. In various embodiments, the engineered P450 enzyme may have an N-terminal truncation of about 10, about 11, about 12, about 13, about 14, about 15, about 16, about 17, about 18, about 19, about 20, about 21, about 22, about 23 about 24, about 25, about 26, about 27, about 28, about 29, about 30, about 31, about 32, about 33, about 34, about 35, about 36, about 37, about 38, about 39, or about 40 amino acids with respect to the wild-type enzyme.
[0049] The wild-type transmembrane region is replaced with a membrane anchor sequence derived from an E. coli protein. The E. coli protein is an inner membrane protein with its C-terminus in the cytoplasm. The membrane anchor derived from E. coli may be a single-pass transmembrane domain or a multiple-pass transmembrane domain. In some embodiments, the membrane anchor is a single-pass transmembrane domain. Exemplary single-pass transmembrane domains derived from E. coli include, but are not limited to, N-terminal domains from the following genes: waaA (SEQ ID NO: 16), ypfN (SEQ ID NO: 17), yhcB (SEQ :ID NO: 18), yhbM (SEQ :ID NO: 19), yhhm (SEQ ID NO: 20), zipA (SEQ ID NO: 21), ycgG (SEQ ID NO: 22), djlA (SEQ ID NO: 23), sohB (SEQ ID NO: 24), lpxK (SEQ NO: 25), F11O (SEQ NO: 26), motA (SEQ NO: 27), htpx (SEQ ID NO: 28), pgaC (SEQ ID NO: 29), ygdD (SEQ ID NO: 30), hemr (SEQ ID NO: 31), and ycls (SEQ ID NO: 32). In an embodiment, the transmembrane domain is derived from yhcB, yhhm, zipA, sohB, and waaA. The transmembrane regions can likewise be determined by predictive tools known in the art (including PHOBIUS).
[0050] In various embodiments, the membrane anchor sequence is from about 8 to about 75 amino acids in length. For example, the membrane anchor may be from about 15 to about 50, or from about 15 to about 40, or from about 15 to about 30, or from about 20 to about 40, or from about 20 to about 30 amino acids in length. In various embodiments, the membrane anchor is about 8, about 9, about 10, about 11, about 12, about 13, about 14, about 15, about 16, about 17, about 18, about 19, about 20, about 21, about 22, about 23, about 24, about 25, about 26, about 27, about 28, about 29, about 30, about 31, about 32, about 33, about 34, about 35, about 36, about 37, about 38, about 39, about 40, about 41, about 42, about 43, about 44, about 45, about 46, about 47, about 48, about 49, about 50, about 51, about 52, about 53, about 54, about 55, about 56, about 57, about 58, about 59, about 60, about 61, about 62, about 63, about 64, about 65, about 66, about 67, about 68, about 69, about 70, about , about 72, about 73, about 74, or about 75 amino acids in length.
[0051] In an embodiment, the transmembrane domain is a yhcB transmembrane domain or a derivative thereof. For example, the transmembrane domain may include the N-terminal 15 to 30 amino acids of yhcB, such as the N-terminal 20 to 22 amino acids of yhcB. For example, the transmembrane domain may include the N-terminal 20, 21, or 22 amino acids of yhcB. The transmembrane domain may have one or more amino acid mutations relative to the wild-type yhcB domain (SEQ ID NO: 18). In some embodiments, the transmembrane domain may have from about 1 to about 8, or from about 1 to about 7, or from about 1 to about 6, or from about 1 to about 5, or from 1 to about 3 mutations relative to the wild-type yhcB sequence. The one or more amino acid mutations may be independently selected from substitutions, insertions, or deletions. In some embodiments, the amino acid mutations are amino acid substitutions. In some embodiments, mutations are selected based on their predicted score as a transmembrane region, using known predictive tools.
[0052] In an embodiment, the transmembrane domain is a yhhM transmembrane domain or derivative thereof. For example, the transmembrane domain may include the N-terminal 15 to 30, or 19 to 21, amino acids of yhhM. For example, the transmembrane domain may include the N-terminal 19, 20, or 21 amino acids of yhhM. The transmembrane domain may have one or more amino acid mutations relative to the wild type yhhM domain (SEQ ID NO: 20). In some embodiments, the transmembrane domain may have from about 1 to about 8, or from about 1 to about 7, or from about 1 to about 6, or from about 1 to about 5, or from 1 to about 3 mutations relative to the wild-type yhhM sequence. The one or more amino acid mutations may be independently selected from substitutions, insertions, or deletions. In some embodiments, the amino acid mutations are amino acid substitutions. In some embodiments, mutations are selected based on their predicted score as a transmembrane region, using known predictive tools.
[0053] In an embodiment, the transmembrane domain is a zipA transmembrane domain or derivative thereof. In such an embodiment, the transmembrane domain may include the N-terminal 15 to 30, or 24 to 26, amino acids of zipA. For example, the transmembrane domain may include the N-terminal 24, 25, or 26 amino acids of zipA. The transmembrane domain may have one or more amino acid mutations relative to the wild type zipA domain (SEQ ID NO: 21). In some embodiments, the transmembrane domain may have from about 1 to about 8, or from about 1 to about 7, or from about 1 to about 6, or from about 1 to about 5, or from 1 to about 3 mutations relative to the wild-type zipA sequence. The one or more amino acid mutations may be independently selected from substitutions, insertions, or deletions. In some embodiments, the amino acid mutations are amino acid substitutions. In some embodiments, mutations are selected based on their predicted score as a transmembrane region, using known predictive tools.
[0054] In an embodiment, the transmembrane domain is a ypfN transmembrane domain or derivative thereof. In such an embodiment, the transmembrane domain may include the N-terminal 15 to 30, or 21 to 23, amino acids of ypfN. For example, the transmembrane domain may include the N-terminal 21, 22, or 23 amino acids of ypfN. The transmembrane domain may have one or more amino acid mutations relative to the wild-type ypfN domain (SEQ ID NO: 17). In some embodiments, the transmembrane domain may have from about 1 to about 8, or from about 1 to about 7, or from about 1 to about 6, or from about 1 to about 5, or from 1 to about 3 mutations relative to the wild-type ypfN sequence. The one or more amino acid mutations may be independently selected from substitutions, insertions, or deletions. In some embodiments, the amino acid mutations are amino acid substitutions. In some embodiments, mutations are selected based on their predicted score as a transmembrane region, using known predictive tools.
[0055] In an embodiment, the transmembrane domain is a sohB transmembrane domain or derivative. In such an embodiment, the transmembrane domain may include the N-terminal 20 to 35, or 27 to 29, amino acids, of sohB. For example, the transmembrane domain may include the N-terminal 27, 28, or 29 amino acids of sohB. The transmembrane domain may have one or more amino acid mutations relative to the wild-type sohB domain (SEQ ID NO: 24). In some embodiments, the transmembrane domain may have from about 1 to about 8, or from about 1 to about 7, or from about 1 to about 6, or from about 1 to about 5, or from 1 to about 3 mutations relative to the wild-type sohB sequence. The one or more amino acid mutations may be independently selected from substitutions, insertions, or deletions. In some embodiments, the amino acid mutations are amino acid substitutions. In some embodiments, mutations are selected based on their predicted score as a transmembrane region, using known predictive tools.
[0056] In an embodiment, the transmembrane domain is a waaA transmembrane domain or derivative thereof. In such an embodiment, the transmembrane domain may include the N-terminal 15 to 30, or 20 to 22, amino acids of waaA. For example, the transmembrane domain may include the N-terminal 20, 21, or 22 amino acids of waaA. The transmembrane domain may have one or more amino acid mutations relative to the wild-type waaA domain (SEQ ID NO: 16). In some embodiments, the transmembrane domain may have from about 1 to about 8, or from about 1 to about 7, or from about 1 to about 6, or from about 1 to about 5, or from 1 to about 3 mutations relative to the wild-type waaA sequence. The one or more amino acid mutations may be independently selected from substitutions, insertions, or deletions. In some embodiments, the amino acid mutations are amino acid substitutions. In some embodiments, mutations are selected based on their predicted score as a transmembrane region, using known predictive tools.
[0057] In still other embodiments, the transmembrane domain is a transmembrane domain of yhbM (SEQ ID NO: 19), ycgG (SEQ ID NO: 22), djlA (SEQ ID NO: 23), lpxK (SEQ ID NO: 25), F11O (SEQ ID NO: 26), motA (SEQ ID NO: 27), htpx (SEQ ID NO: 28), pgaC (SEQ ID NO: 29), ygdD (SEQ ID NO: 30), hemr (SEQ ID NO: 31), or ycls (SEQ ID NO: 32), or a derivative thereof. The derivative may have one or more amino acid mutations relative to the wild-type E. coli sequence. In some embodiments, the transmembrane domain may have from about 1 to about 10, or from about 1 to about 8, or from about 1 to about 5, or from about 1 to about 3 mutations relative to the wild-type E. coli sequence. In some embodiments, the one or more amino acid mutations may be independently selected from substitutions, insertions, and deletions. In some embodiments, the amino acid mutations are amino acid substitutions. In some embodiments, mutations are selected based on their predicted score as a transmembrane region, using known predictive tools.
[0058] In other aspects, the invention provides polynucleotides comprising a nucleotide sequence encoding an engineered P450 enzyme described above. The polynucleotide may be codon optimized for expression in E. coli in some embodiments. In another example, the polynucleotide may comprise a nucleotide sequence encoding at least one engineered P450 enzyme with one or more cytochrome P450 reductase (CPR) enzymes described herein as a translational fusion or operon. Such polynucleotides may further comprise, in addition to sequences encoding the engineered P450 enzyme, one or more expression control elements. For example, the polynucleotide may comprise one or more promoters or transcriptional enhancers, ribosomal binding sites, transcription termination signals, as expression control elements. The polynucleotide may be inserted within any suitable vector, including an expression vector, and which may be contained within any suitable host cell for expression. The polynucleotide may be designed for introduction and/or protein expression in any suitable host cell, including bacterial cells such as E. coli cells.
[0059] In other aspects, the invention provides E. coli host cells expressing the engineered P450 enzyme, either integrated into the genome, or extrachromosomally (e.g., on a plasmid). In some embodiments, the P450 enzyme is expressed by a strong promoter, such as T7, T5, T3, or Trc, or a promoter having promoter strength in E. coli equal to or more than T7, T5, T3, or Trc. The promoter may be a strong constitutive E. coli promoter or a coliphage promoter, or a variant thereof. Deuschle et al., Promoters of Escherichia coil: a hierarchy of in vivo strength indicates alternate structures, EMBO J. 5(11): 2987-2994 (1986); http://parts.igem.org/Promoters/Catalog/Ecoli/Constitutive. When expressed from a plasmid, the plasmid may be a low or high copy number plasmid (e.g., p5, p10, p20). In another embodiment, the P450 gene is chromosomally integrated into the genome of the host E. coli cell, which may further include tandem repeats of the gene to increase the expression level. See US 20110236927, which is hereby incorporated by reference in its entirety.
[0060] The E. coli cells may be fed the desired P450 substrate for chemical transformation, or biochemical pathways may be expressed in the host cell to generate the substrate in vivo.
[0061] In some embodiments the bacterial host expresses one or more recombinant biosynthetic pathways. For example, an E. coli host cell may express one or more recombinant biosynthetic pathways that include at least 1, at least 2, at least 3, at least 4, or at least 5 recombinant enzymes. The biosynthetic pathways may produce a secondary metabolite through the overexpression of at least 1, at least 2, at least 3, at least 4, or at least 5 foreign genes. In these or other embodiments, the E. coli host cell may overexpress at least 1, at least 2, at least 3, at least 4, or at least 5 E. coli genes. Overexpression of several E. coli genes and/or foreign genes can produce substantial cell stress responses. Where these pathways include one or more P450 enzymes, these stress responses can be substantially higher. For example, as shown herein, P450 enzymes can induce substantial overexpression of the IbpA protein, a protein that is overexpressed under conditions of protein aggregation and cell stress. For example, overexpression of native E. coli genes as well as foreign genes can result in conditions of protein aggregation that induce a cell stress response (e.g., as observed by overexpression of IbpA).
[0062] In various embodiments, the invention results in reduced cell stress such that the E. coli cell does not exhibit a substantially stressed phenotype during culturing, or the cell stress is minimized. Cell stress may be assessed by measuring the expression of various cell stress proteins including, but not limited to, IbpA, DnaK, GrpE, and GroL. In some embodiments, methods of the invention do not result in overexpression of the cell stress protein IbpA. In this context, overexpression refers to at least two times the IbpA expression level of the parent strain. In some embodiments, the engineering of the P450 enzyme of the invention may be guided by testing IbpA expression in cultures. For example, a determination of the length of the truncation of the P450 enzyme and/or the anchor size and/or sequence may be guided by IbpA expression levels in the culture.
[0063] In some embodiments, at least one foreign gene is expressed by a strong promoter, such as T7, T5, T3, or Trc, or a promoter having promoter strength in E. coli equal to or more than T7, T5, T3, or Trc. The promoter may be a strong constitutive E. coli promoter or a coliphage promoter, or a variant thereof. Deuschle et al., Promoters of Escherichia coli: a hierarchy of in vivo strength indicates alternate structures, EMBO J. 5(11): 2987-2994 (1986); http://parts.igem.org/Protnoters/Catalog/Ecoli/Constitutive. In an embodiment, the genes are expressed from a plasmid, which may be a low or high copy number plasmid p5, p10, p20). In another embodiment, the genes are chromosomally integrated into the genome of the host E. coli cell, which may further include tandem repeats of the gene to increase the expression level. See US 20110236927, which is hereby incorporated by reference in its entirety.
[0064] In some embodiments, the E. coli produces a compound from isopentyl pyrophosphate (IPP) and/or dimethylallyl pyrophosphate DMAPP, such as a terpene or terpenoid compound. In an exemplary embodiment, the E. coli cell may overexpress at least one gene in the MEP pathway, which is endogenous to E. coli. The MEP (2-C-methyl-D-erythritol 4-phosphate) pathway, also called the MEP/DOXP (2-C-methyl-D-erythritol 4-phosphated-deoxy-D-xylulose 5-phosphate) pathway or the non-mevalonate pathway or the mevalonic acid-independent pathway refers to the pathway that converts glyceraldehyde-3-phosphate and pyruvate to IPP and DMAPP. In the MEP pathway, pyruvate and D-glyceraldehyde-3-phosphate are converted via a series of reactions to IPP and DMAPP. The pathway typically involves action of the following enzymes: 1-deoxy-D-xylulose-5-phosphate synthase (Dxs), 1-deoxy-D-xylulose-5-phosphate reductoisomerase (IspC), 4-diphosphocytidyl-2-C-methyl-D-erythritol synthase (IspD), 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (IspE), 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (IspF), 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase (IspG), and isopentenyl diphosphate isomerase (IspH). The MEP pathway, and the genes and enzymes that make up the MEP pathway, are described in U.S. Pat. No. 8,512,988, which is hereby incorporated by reference in its entirety. For example, genes that make up the MEP pathway include dxs, ispC, ispD, ispE, ispF, ispG, idi, ispA, and ispB. In some embodiments, one or more terpenoid compounds are produced at least in part by metabolic flux through an MEP pathway. In an embodiment, the E. coli host cell may express at least one additional copy of a dxs and/or idi gene. In some embodiments, the E. coli host as at least one additional copy of dxs, ispD, ispF, and/or idi gene, so as to overexpress these gene products.
[0065] In various embodiments, the E. coli cell expresses one or more biosynthetic pathways that include at least one membrane-anchored engineered P450 enzyme as described herein. In some embodiments, the P450 enzyme is not strongly expressed, but the more efficient membrane anchoring in accordance with the invention allows for sufficient activity without stronger expression. In various embodiments, the E. coli cell expresses at least 1, at least 2, at least 3, at least 4, or at least 5 P450 enzymes, which may operate in serial fashion in a biosynthetic pathway.
[0066] In some embodiments, the P450 enzyme is expressed from a strong (e.g., constitutive or inducible) E. coli or coliphage promoter, or variant thereof (e.g., Trc, T7, T5, or T3, or variant thereof). While overexpression of P450 enzymes can induce significant cell stress, the membrane anchoring system in accordance with the invention renders the P450-membrane association more productive, with less protein misfolding and/or aggregation, which would otherwise induce cell stress.
[0067] In various embodiments, the E. coli host cell expresses the engineered P450 enzyme alongside one or more cytochrome P450 reductase (CPR) partner that regenerates the P450 enzyme. As used herein, the term "cytochrome P450 reductase partner" or "CPR partner" refers to a cytochrome P450 reductase capable of regenerating the cofactor component of the cytochrome P450 oxidase of interest for oxidative chemistry. The CPR may be a natural CPR partner for the P450 enzyme, and in other embodiments, the CPR partner is not the natural CPR partner for the P450 enzyme. In nature, cytochrome P450 reductase is a membrane protein generally found in the endoplasmic reticulum. It catalyzes pyridine nucleotide dehydration and electron transfer to membrane bound cytochrome P450s. CPRs may be derived from any species that naturally employs P450 biochemistry, including: Zingiber sp., Barnadesia sp., Hyoscyamus sp., Latuca sp., Nicotiana sp., Citrus sp., Artemesia sp., Arabidopsis sp, Stevia sp., Bacillus sp., Pleurotus sp., Cichorium sp., Helianthus sp., and Physcomitrella sp., Taxus sp., Rosa sp., Cymbopogon sp., Humulus sp., Pogostenion sp., and Cannabis sp. Exemplary CPRs include those from Stevia rebaudiana (e.g., SEQ ID NO: 33, 40, 41, and 42), Arabidopsis thaliana (SEQ ID NO: 34, 37, 38, and 39), Taxus cuspidata (SEQ ID NO: 35), Atemisia annua (SEQ ID NO:36), and Pelargonium graveolans (SEQ ID NO: 43). In various embodiments, the wild-type CPR or derivative thereof is expressed separately from the P450 enzymes (e.g., from the same or different operon), or in some embodiments as a translational fusion with the P450 enzyme. Generally, CPR derivatives comprise amino acid sequences having at least 70%, or at least 80%, or at least 90%, or at least 95% identity to the wild-type sequence (e.g., SEQ ID NOS: 33-43), and which can be employed in the various embodiments.
[0068] In an embodiment, the CPR may be expressed as a translational fusion protein with an engineered P450 enzyme. The CPR may be fused to the P450 enzyme through a linker. Exemplary linker sequences can be predominantly serine, glycine, and/or alanine, and may be from three to one hundred amino acids in various embodiments. Linker sequences include, for example, GSG, GSGGGGS (SEQ ID NO: 44), GSGEAAAK (SEQ ID NO: 45), GSGEAAAKEAAAK (SEQ ID NO: 46), and GSGMGSSSN (SEQ ID NO: 47).
[0069] In some embodiments, the invention allows for better control of P450 enzyme efficiency, by allowing for efficient ratios of expression of P450 enzymes in relation to the CPR partner (when expressed separately). In some embodiments, the ratio of the expression levels of the P450 enzyme(s) and the CPR partners may range from about 5:1 to about 1:5, for example, about 5:1, or about 4:1, or about 3:1, or about 2:1, or about 1:1, or about 1:2, or about 1:3, or about 1:4, or about 1:5. For example, the ratio of the expression levels of the P450 enzyme(s) and the CPR partner may be from about 2:1 to about 1:2. In various embodiments, the CPR may also be modified to include at least one membrane anchor sequence derived from an E. coli protein as described herein. In an embodiment, the E. coli cell expresses a single CPR protein, and optionally expresses more than one P450 enzyme.
[0070] In various embodiments, the E. coli host cell expresses a biosynthetic pathway that produces a secondary metabolite selected from a terpenoid, alkaloid, cannabinoid, steroid, saponin, glycoside, stilbenoid, polyphenol, antibiotic, polyketide, fatty acid, or non-ribosomal peptide. In certain embodiments, the E. coli cell produces one or more terpenoid compounds. A terpenoid, also referred to as an isoprenoid, is an organic chemical derived from a five-carbon isoprene unit (C5). Several non-limiting examples of terpenoids, classified based on the number of isoprene units that they contain, include: hemiterpenoids (1 isoprene unit), monoterpenoids (2 isoprene units), sesquiterpenoids (3 isoprene units), diterpenoids (4 isoprene units), sesterterpenoids (5 isoprene units), triterpenoids (6 isoprene units), tetraterpenoids (8 isoprene units), and polyterpenoids with a larger number of isoprene units. In an embodiment, the E. coli cell produces a terpenoid selected from a monoterpenoid, a sesquiterpenoid, diterpenoid, a sesterpenoid, or a triterpenoid. Terpenoids represent a diverse class of molecules that provide numerous commercial applications, including in the food and beverage industries as well as the perfume, cosmetic and health care industries. By way of example, terpenoid compounds find use in perfumery (e.g. patchoulol), in the flavor industry (e.g., nootkatone), as sweeteners (e.g., steviol), or therapeutic agents (e.g., taxol) and many are conventionally extracted from plants. Nevertheless, terpenoid molecules are found in ppm levels in nature, and therefore require massive harvesting to obtain sufficient amounts for commercial applications.
[0071] Where the chemical species is a terpenoid, the host cell will generally contain a recombinant downstream pathway that produces the terpenoid from IPP and DMAPP precursors. Terpenes such as Monoterpenes (C10), Sesquiterpenes (C15) and Diterpenes (C20) are derived from the prenyl diphosphate substrates, geranyl diphosphate (GPP), farnesyl diphosphate (FPP) and geranylgeranyl diphosphate (GGPP) respectively through the action of a very large group of enzymes called the terpene (terpenoid) synthases. These enzymes are often referred to as terpene cyclases since the product of the reactions are cyclized to various monoterpene, sesquiterpene and diterpene carbon skeleton products. Many of the resulting carbon skeletons undergo subsequence oxygenation by cytochrome P450 hydrolysase enzymes to give rise to large families of derivatives. In various embodiments, the E. coli cell expresses a biosynthetic pathway involving the overexpression of a geranyl diphosphate synthase (GPS), a gernanylgeranyl diphosphate synthase (GGPS), a farnsesyl diphosphate synthase (FPS), or a farnesyl geranyl diphosphate synthase (FGPPS).
[0072] The product of the invention in some embodiments is one or more oxygenated terpenoids. As used herein, the term "oxygenated terpenoid" refers to a terpene scaffold having one or more oxygenation events, producing a corresponding alcohol, aldehyde, carboxylic acid and/or ketone. In some embodiments, the E. coli cell produces at least one terpenoid selected from alpha-sinensal, beta-Thujone, Camphor, Carveol, Carvone, Cineole, Citral, Citronellal, Cubebol, Geraniol, Limonene, Menthol, Menthone, Myrcene, Nootkatone, Nootkatol, Patchouli, Piperitone, Sabinene, Steviol, Steviol glycoside, Taxadiene, Thymol, and Valencene, either as a P450 oxygenated product or as a substrate for P450 chemistry.
[0073] In another embodiment, the E. coli cell produces Valencene and/or Nootkatone. In such an embodiment, the E. coli cell may express a biosynthetic pathway that further includes a farnesyl pyrophosphate synthase, a Valencene Synthase, and a Valencene Oxidase (the VO comprising the membrane anchor described herein). Farnesyl pyrophosphate synthases (FPPS) produce farnesyl pyrophosphates from iso-pentyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMPP). An exemplary farnesyl pyrophosphate synthase is ERG20 of Saccharomyces cerevisiae (NCBI accession P08524) and E. coli ispA. Valencene synthase produces sesquiterpene scaffolds and are described in, for example, US 2012/0107893, US 2012/0246767, and U.S. Pat. No. 7,273,735, which are hereby incorporated by reference in their entireties.
[0074] In an embodiment, the E. coli cell produces steviol or steviol glycoside (e.g., RebM). Steviol is produced from kaurene by the action of two P450 enzymes, kaurene oxidase (KO) and kaurenoic acid hydroxylase (KAH). After production of steviol, various steviol glycoside products may be produced through a series of glycosylation reactions, which can take place in vitro or in vivo. Pathways and enzymes for production of steviol and steviol glycosides are disclosed in US 2013/0171328, US 2012/0107893, WO 2012/075030, WO 2014/122328, which are hereby incorporated by reference in their entireties.
[0075] In various embodiments, the E. coli cell may express a biosynthetic pathway involving a geranylgeranyl pyrophosphate synthase (GPPS), a copalyl diphosphate synthase (CPPS), and a kaurene synthase (KS), as well as a kaurene oxidase (KO) and a kaureneoic acid hydroxylase (KAH) having a single pass transmembrane domain derived from an E. coli gene. In some embodiments, the biosynthetic pathway may further include or more uridine diphosphate dependent glycosyltransferase enzymes (UGT).
[0076] Other biosynthetic pathways for production of terpenoid compounds are disclosed in U.S. Pat. No. 8,927,241, which is hereby incorporated by reference in its entirety.
[0077] In various embodiments, E. coli cell is cultured to produce the one or more chemical species. For example, the E. coli cell may be cultured to produce one or more terpenoid compounds.
[0078] While commercial biosynthesis in E. coli can be limited by the temperature at which overexpressed and/or foreign enzymes are stable, the substantial improvements in stability of the P450 enzymes described herein, may allow for cultures to be maintained at higher temperatures, resulting in higher yields and higher overall productivity. In some embodiments, the culturing is conducted at about 30.degree. C. or greater, about 31.degree. C. or greater, about 32.degree. C. or greater, about 33.degree. C. or greater, about 34.degree. C. or greater, about 35.degree. C. or greater, about 36.degree. C. or greater, or about 37.degree. C. or greater.
[0079] The host cells are further suitable for commercial production of chemical species, and therefor can be productive at commercial scale. In some embodiments, the size of the culture is at least about 100 L, at least about 200 L, at least about 500 L, at least about 1,000 L, or at least about 10,000 L. In an embodiment, the culturing may be conducted in batch culture, continuous culture, or semi-continuous culture.
[0080] In various embodiments, methods of the invention further include recovering the one or more chemical species such as one or more terpenoid compounds from the cell culture or from cell lysates. In some embodiments, the culture produces at least about 20 mg/L, at least about 25 mg/L, at least about 30 mg/L, at least about 35 mg/L, at least about 40 mg/L, at least about 45 mg/L, at least about 50 mg/L, at least about 55 mg/L, at least about 60 mg/L, at least about 65 mg/L, at least about 70 mg/L, at least about 75 mg/L, at least about 80 mg/L, at least about 85 mg/L, at least about 90 mg/L, at least about 95 mg/L, at least about 100 mg/L, at least about 150 mg/L, or at least about 200 mg/L of the chemical species.
[0081] In some embodiments involving the production of a terpenoid compound, the production of indole is used as a surrogate marker for terpenoid production, and/or the accumulation of indole in the culture is controlled to increase terpenoid production. For example, in various embodiments, accumulation of indole in the culture is controlled to below about 100 mg/L, or below about 75 mg/L, or below about 50 mg/L, or below about 25 mg/L, or below about 10 mg/L. The accumulation of indole can be controlled by balancing protein expression and activity using the multivariate modular approach as described in U.S. Pat. No. 8,927,241 (which is hereby incorporated by reference), and/or is controlled by chemical means.
[0082] The oxidized product can be recovered by any suitable process, including partitioning the desired product into an organic phase. The production of the desired product can be determined and/or quantified, for example, by gas chromatography (e.g., GC-MS). The desired product can be produced in batch or continuous bioreactor systems. Production of product, recovery, and/or analysis of the product can be done as described in US 2012/0246767, which is hereby incorporated by reference in its entirety. For example, in some embodiments, particularly in relation to terpenoids, oxidized oil is extracted from aqueous reaction medium using an organic solvent, such as an alkane such as heptane, followed by fractional distillation. Terpenoid components of fractions may be measured quantitatively by GC/MS, followed by blending of fractions to generate a desired product profile.
[0083] In various embodiments, the recovered chemical species such as one or more terpenoid compounds are incorporated into a product (e.g., a consumer or industrial product). For example, the product may be a flavor product, a fragrance product, a sweetener, a cosmetic, a cleaning product, a detergent or soap, or a pest control product. In some embodiments, the oxygenated product recovered is nootkatol and/or nootkatone, and the product is a flavor product selected from a beverage, a chewing gum, a candy, or a flavor additive, or the product is an insect repellant. in some embodiments, the oxygenated product is steviol or a steviol glycoside, which is provided as a sweetener, or is incorporated into beverages or food products.
[0084] The invention further provides methods of making products such as foods, beverages, texturants starches, fibers, gums, fats and fat mimetics, and emulsifiers), pharmaceutical products, tobacco products, nutraceutical products, oral hygiene products, and cosmetic products, by incorporating the chemical species produced herein. The higher yields of such species produced in embodiments of the invention can provide significant cost advantages as well as sustainability.
EXAMPLES
Example 1. Identification of Candidate E. coli Genes with Membrane Anchor Sequences
[0085] FIG. 1 illustrates the N-terminal membrane anchor concept. Previously, P450 enzymes were expressed in E. coli by truncation of at least a portion of the P450 N-terminal transmembrane region, with the addition of an 8 amino acid peptide (MALLLAVF) derived from bovine P45017.alpha.. See, for example, Barnes H J, et al., Expression and enzymatic activity of recombinant cytochrome P450 17.alpha.-hydroxylase in Escherichia coli, PNAS 88: 5597-5601 (1991); Ajikumar P K, et al., Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coil. Science 330(6000): 70-74 (2010). However, activity of these P450 constructs in E. coli was generally limiting, making eukaryotes such as yeast the preferred hosts for oxidative transformations involving P450 enzymes. In part, the lack of P450 productivity in E. coli could result from a cell stress response triggered by the non-native membrane anchoring. By engineering P450 proteins to contain an N-terminal anchor derived from an E. coli protein that is natively anchored in the inner membrane by its N-terminus (either by single pass or multi-pass transmembrane helices), this cell stress response could be minimized, and other benefits for the functional expression of P450 enzymes might be identified.
[0086] A proteomic analysis was carried out to identify candidate E. coli anchor sequences for functional expression in E. coli. Specifically, the EcoGene 3.0 software was used to identify E. coli inner membrane proteins that include a C-terminus in the cytoplasm. The Phobius predictive tool was used to assess candidate E. coli genes for the presence of signal peptides and/or transmembrane regions. The analysis identified 395 genes which were experimentally determined to be inner membrane proteins. In addition, 85 predicted inner membrane genes were identified. The prediction of transmembrane helices or signal peptides is illustrated in FIG. 2 for three known membrane-anchored P450 enzymes. Stevia rebaudiana Kaurene Oxidase (SrKO) and Arabidopsis thaliana kaurenoic acid 13-hydroxylase (AtKAH) were predicted to include transmembrane domains. The bovine P450-C17 enzyme was predicted to have a N-terminal signal peptide -8rp.
Example 2. Construction and Functional Analysis of Engineered Valencene Oxidase (VO) Enzymes
[0087] Various N-terminal anchors based on single pass E. coli transmembrane regions were constructed and incorporated into Valencene Oxidase (VO) truncated at residue 30. The Valencene Oxidase is derived from Stevia rebaudiana Kaurene Oxidase (SrKO), which has been shown to be functional for oxidative transformation of Valencene to Nootkatone and Nootkatol, which are sesquiterpene compounds. Constructs were cloned into the p5Trc-tc-CPR plasmid which includes a translationally coupled cytochrome P450 reductase (CPR). As a control, the 8rp signal peptide was also incorporated into VO truncated at either residue 20 or residue 30 (8rp-t20 or 8rp-t30).
[0088] The engineered VO enzymes with candidate E. coli anchor sequences were expressed in host E. coli cells that produce valencene through a recombinant valencene synthesis pathway. The cells were incubated at 30.degree. C. for 48 hours in 96 dispensing well plates with a dodecane overlayer. As shown in FIG. 3, many of the E. coli anchored enzymes outperformed the 8rp-t20 and 8rp-t30 constructs in both total terpenoid flux and. oxygenated product formation.
[0089] Lead VO constructs with E. coli anchor sequences were assessed. The constructs were expressed in host E. coli cells which were incubated at 30.degree. C. for 48 hours in 24 dispensing well plates with a vegetable oil overlay. Consistent with previous results, the E. coli anchored enzymes outperformed the 8rp-t20 and 8rp-t30 constructs in both total terpenoid flux and oxygenated product formation (FIG. 4).
[0090] The effects of increased expression of engineered P450 constructs on E. coli cells were evaluated. Specifically, VO constructs with lead E. coli anchor sequences were cloned into a p10 plasmid and expressed in valencene-producing coil cells which were incubated at 30.degree. C. for 48 hours in 24 dispensing well plates with a vegetable oil overlay. As shown in FIG. 5, overexpression of 8rp anchored P450 enzymes (from the p10 plasmid) resulted in reduced activity including a three-fold loss in the formation of oxygenated terpenoids compared to a weaker expressed enzyme (from a p5 plasmid). In contrast, the P450 enzymes with E. coli anchors (particularly yhcB and ZipA) showed only a modest loss of oxygenated product when expressed from a p10 plasmid. Further, P450 enzymes with E. coli anchors expressed from a p10 plasmid produced as much product as the 8rp anchored enzyme expressed from a p5 plasmid. Accordingly, the E. coli anchored P450 enzymes allowed for high functional expression in E. coli cells.
[0091] Functional expression with the CPR partner (from Stevia) was also assessed. In particular, the CPR enzyme was expressed in E. coli cells with the P450 enzymes either in the same operon or translationally coupled from one plasmid. The cells were incubated at 30.degree. C. for 48 hours in 24 dispensing well plates with a vegetable oil overlay. It was observed that membrane anchors based on native E. coli sequences (e.g., yhcB) provided increased oxygenated titer when translationally coupled to a CPR partner or when expressed separately (FIG. 6). In addition, cells with the E. coli anchors grew to a higher final OD and with less emulsion, suggesting that the cells are less stressed, despite overexpressing an upstream MEP pathway and downstream terpenoid biosynthesis (Nootkatone) path way.
[0092] During translation initiation, N-terminal sequences can significantly affect the expression of a protein. As such, a comparison was conducted to examine VO expression and VO/CPR ratio with different N-terminal anchor regions. As shown in FIG. 7, four anchors tested (yhcB, yhhm, zipA, and ypfN) showed lower total VO expression compared to the control 8rp anchor, allowing for a five-fold lower expression relative to CPR expression. This ratio is important for efficient P450 electron transfer without deleterious effects on coupling efficiency.
[0093] A proteomics approach was undertaken to assess cellular stress response to variable P450 expression. For example, IbpA, which is overexpressed in E. coli under conditions of high protein aggregation and stress, is strongly expressed with the 8rp anchored P450 enzyme, but not with the enzymes with E. coli anchors (FIG. 8).
[0094] The yhcB-anchored VO enzyme was further analyzed to determine the impact of anchor and P450 truncation length on overall productivity (FIG. 9). Variants with truncations in anchor and P450 sequences were generated using the GeneArt technology and subcloned into a p5T7-BCD18 screening plasmid. A list of the variants is provided in Table 2.
TABLE-US-00002 TABLE 2 Truncation Optimization n20yhcB_t28VOR1 n20yhcB_t29VOR1 n20yhcB_t30VOR1 n20yhcB_t31VOR1 n20yhcB_t32VOR1 n21yhcB_t28VOR1 n21yhcB_t29VOR1 n21yhcB_t30VOR1 n21yhcB_t31VOR1 n21yhcB_t32VOR1 n22yhcB_t28VOR1 n22yhcB_t29VOR1 n22yhcB_t31VOR1 n22yhcB_t32VOR1 n23yhcB_t28VOR1 n23yhcB_t29VOR1 n23yhcB_t30VOR1 n23yhcB_t31VOR1 n23yhcB_t32VOR1 n24yhcB_t28VOR1 n24yhcB_t29VOR1 n24yhcB_t30VOR1 n24yhcB_t31VOR1 n24yhcB_t32VOR1
The variants were assayed for product formation in a MEP/FPPS/VS integrated E. coli strain. The E. coli cells were incubated at 30.degree. C. for 48 hours in 96 dispensing well plates with a dodecane overlay. Products were subsequently extracted with ethyl acetate. As shown in FIG. 10, modification of the anchor and P450 truncation lengths has only modest impact on overall productivity. For example, the n20yhcB-t29VO1 construct yielded an approximately 1.2-fold increase in total terpenoid flux and oxygenated products. These results suggest that the E. coli anchored P450 enzymes are relatively robust to changes in anchor and truncation length.
Example 3. Construction and Functional Analysis of Additional Engineered P450 Enzymes
[0095] Modified Arabidopsis thaliana kaurenoic acid 13-hydroxylase (AtKAH) and Stevia rebaudiana Kaurene Oxidase (SrKO) enzymes with E. coli anchor sequences were generated.
[0096] Engineered enzymes were produced with the AtKAH enzymes truncated at position 26. The N-terminus of the truncated AtKAH enzyme is shown below:
TABLE-US-00003 t25 AtKAH (SEQ ID NO: 61) HVYGRAVVEQWR t26 AtKAH (SEQ ID NO: 62) VYGRAVVEQWR
The E(z) scoring system was used to guide truncation of the E. coli anchor sequence by similarity of scoring to the parent protein (FIG. 11). Eight combinations of E. coli anchored N-terminal segments and AtKAH truncation lengths were selected for in vivo screening.
[0097] E. coli membrane anchors (e.g., zipA, yhhm, and yhcB) were also incorporated into SrKO for production of kaurenoic acid in E. coli. The N-terminus of the truncated SrKO enzyme is shown below:
TABLE-US-00004 T29 SrVO (SEQ ID NO: 63) SWYLKSYTSARR T30 SrVO (SEQ ID NO: 64) WYLKSYTSARR
[0098] The constructs were cloned into the p5Trc plasmid which include an operon expressed CPR. The E. coli cells were incubated at either 30.degree. C. or 34.degree. C. for 72 hours in 96 dispensing well plates. Similar to results obtained with the engineered VO enzymes, variant SrKO enzymes with E. coli anchor sequences also showed improved activity compared to enzymes with 8rp anchor sequences. Particularly, as shown in FIG. 12A, the zipA anchored P450 enzyme produced the highest kaurenoic acid titer (about 5 mg/L). In addition, all SrKO variants showed a drop in activity at 34.degree. C. FIG. 12B shows that the final OD is higher in all variants except zipA when incubated at 34.degree. C.
[0099] A proteomics analysis was undertaken to examine the relative abundance of various pathway and stress response proteins in cells expressing SrKO. Both soluble and insoluble cell fractions were analyzed by tryptic digest proteomics. As shown in FIG. 13, SrKO enzymes with either E. coli anchors zipA and yhcB or control anchor 8rP all showed higher expressions than full-length SrKO at 30.degree. C. and 34.degree. C. Similar results were seen in the insoluble fractions, including higher expression of the upstream pathway enzyme dxs. At the higher temperature of 34.degree. C., there was also an increase in the stress response gene IbpA in the insoluble fractions.
[0100] In summary, P450 enzymes with E. coli anchor sequences showed improvement in total terpenoid flux and oxygenated terpenoid formation, and provided higher catalytic efficiency and reduced stress response, as compared to 8rp anchored P450 enzymes. This N-terminal engineering approach is broadly applicable to E. coli cells having several overexpressed genes, including complete biosynthetic pathways, and including one or multiple P450 enzymes, where managing cell stress is crucial to obtaining commercial yields.
TABLE-US-00005 SEQUENCES P450 ENZYMES >ZzHO [Zingiber zerumbet] (SEQ ID NO: 1) MEAISLFSPFFFITLFLGFFITLLIKRSSRSSVHKQQVLLASLPPSPPRLPLIGNIHQLVGGNPHR ILLQLARTHGPLICLRLGQVDQVVASSVEAVEEIIKRHDLKFADRPRDLTFSRIFFYDGNAVVMTP YGGEWKQMRKIYAMELLNSRRVKSFAAIREDVARKLTGEIAHKAFAQTPVINLSEMVMSMINAIVI RVAFGDKCKQQAYFLHLVKEAMSYVSSFSVADMYPSLKFLDTLTGLKSKLEGVHGKLDKVFDEIIA QRQAALAAEQAEEDLIIDVLLKLKDEGNQEFPITYTSVKAIVMEIFLAGTETSSSVIDWVMSELIK NPKAMEKVQKEMREAMQGKTKLEESDIPKFSYLNLVIKETLRLHPPGPLLFPRECRETCEVMGYRV PAGARLLINAFALSRDEKYWGEDAESFKPERFEGISVDFKGSNFEFMPFGAGRRICPGMTFGISSV EVALAHLLFHFDWQLPQGMKIEDLDMMEVSGMSATRRSPLLVLAKLIIPLP >BsGAO [Barnadesia spinosa] (SEQ ID NO: 2) MELTLTTSLGLAVFVFILFKLLTGSKSTKNSLPEAWRLPIIGHMHHLVGTLPHRGVTDMARKYGSL MHLQLGEVSTIVVSSPRWAKEVLTTYDITFANRPETLTGEIVAYHNTDIVLSPYGEYWRQLRKLCT LELLSAKKVKSFQSLREEECWNLVKEVRSSGSGSPVDLSESIFKLIATILSRAAFGKGIKDQREFT EIVKEILRLTGGFDVADIFPSKKILHHLSGKRAKLTNIHNKLDSLINNIVSEHPGSRTSSSQESLL DVLLRLKDSAELPLTSDNVKAVILDMFGAGTDTSSATIEWAISELIRCPRAMEKVQTELRQALNGK ERIQEEDIQELSYLKLVIKETLRLHPPLPLVMPRECREPCVLAGYEIPTKTKLIVNVFAINRDPEY WKDAETFMPERFENSPINIMGSEYEYLPFGAGRRMCPGAALGLANVELPLAHILYYFNWKLPNGAR LDELDMSECFGATVQRKSELLLVPTAYKTANNSA >HmPO [Hyoscyamus muticus] (SEQ ID NO: 3) MQFFSLVSIFLFLSFLFLLRKWKNSNSQSKKLPPGPWKLPLLGSMLHMVGGLPHHVLRDLAKKYGP LMHLQLGEVSAVVVTSPDMAKEVLKTHDIAFASRPKLLAPEIVCYNRSDIAFCPYGDYWRQMRKIC VLEVLSAKNVRSFSSIRRDEVLRLVNFVRSSTSEPVNFTERLFLFTSSMTCRSAFGKVFKEQETFI QLIKEVIGLAGGFDVADIFPSLKFLHVLTGMEGKIMKAHHKVDAIVEDVINEHKKNLAMGKTNGAL GGEDLIDVLLRLMNDGGLQFPITNDNIKAIIFDMFAAGTETSSSTLVWAMVQMMRNPTILAKAQAE VREAFKGKETFDENDVEELKYLKLVIKETLRLHPPVPLLVPRECREETEINGYTIPVKTKVMVNVW ALGRDPKYWDDADNFKPERFEQCSVDFIGNNFFYLPFGGGRRICPGISFGLANVYLPLAQLLYHFD WKLPTGMEPKDLDLTELVGVTAARKSDLMLVATPYQPSRE >LsGAO [Lactuca sativa] (SEQ ID NO: 4) MELSITTSIALATIVFFLYKLATRPKSTKKQLPEASRLPIIGHMHHLIGTMPHRGVMDLARKHGSL MHLQLGEVSTIVVSSPKWAKEILTTYDITFANRPETLTGEIIAYHNTDIVLAPYGEYWRQLRKLCT LELLSVKKVKSFQSIREEECWNLVKEVKESGSGKPINLSESIFTMIATILSRAAFGKGIKDQREFT EIVKEILRQTGGFDVADIFPSKKFLHHLSGKRARLTSIHKKLDNLINNIVAEHHVSTSSKANETLL DVLLRLKDSAEFPLTADNVKAIILDMFGAGTDTSSATVEWAISELIRCPRAMEKVQAELRQALNGK EKIQEEDIQDLAYLNLVIRETLRLHPPLPLVMPRECREPVNLAGYEIANKTKLIVNVFAINRDPEY WKDAEAFIPERFENNPNNIMGADYEYLPFGAGRRMCPGAALGLANVQLPLANILYHFNWKLPNGAS HDQLDMTESFGATVQRKTELLLVPSF >NtEAO [Nicotiani tabacum] (SEQ ID NO: 5) MQFFSLVSIFLFLSFLFLLRKWKNSNSQSKKLPPGPWKIPILGSMLHMIGGEPHHVLRDLAKKYGP LMHLQLGEISAVVVTSRDMAKEVLKTHDVVFASRPKIVAMDIICYNQSDIAFSPYGDHWRQMRKIC VMELLNAKNVRSFSSIRRDEVVRLIDSIRSDSSSGELVNFTQRIIWFASSMTCRSAFGQVLKGQDI FAKKIREVIGLAEGFDVVDIFPTYKFLHVLSGMKRKLLNAHLKVDAIVEDVINEHKKNLAAGKSNG ALGGEDLIDVLLRLMNDTSLQFPITNDNIKAVIVDMFAAGTETSSTTTVWAMAEMMKNPSVFTKAQ AEVREAFRDKVSFDENDVEELKYLKLVIKETLRLHPPSPLLVPRECREDTDINGYTIPAKTKVMVN VWALGRDPKYWDDAESFKPERFEQCSVDFFGNNFEFLPFGGGRRICPGMSFGLANLYLPLAQLLYH FDWKLPTGIMPRDLDLTELSGITIARKGGLYLNATPYQPSRE >CpVO [Citrus x paradisi] (SEQ ID NO: 6) MELPLKSIALTIVIVTVLTWAWRVLNWVWLRPKKLEKFLRQQGLKGNSYRLLFGDLKENSIELKEA KARPLSLDDDIAIRVNPFLHKLVNDYGKNSFMWFGPTPRVNIMNPDQIKAIFTKINDFQKVNSIPL ARLLIVGLATLEGEKWAKHRKLINPAFHQEKLKLMLPAFYLSCIEIITKWEKQMSVEGSSELDVWP YLANLTSDVISRTAFGSSYEEGRRIFQLQAELAELTMQVFRSVHIPGWRFLPTKRNRRMKEIDKEI RASLMGIIKNREKAMRAGEAANNDLLGILMETSFREIEEHGNNKNVGFSMNDVIEECKLFYFAGQE TTSVLLNWTMVLLSKHQDWQERARQEVLQVFGNNKPDYDGLNHLKIVQMILYEVLRLYPPVTVLSR AVFKETKLGNLTLPAGVQIGLPMILVHQDPELWGDDAVEFKPERFAEGISKAAKNQVSYFPFALGP RICVGQNFALVEAKMATAMILQNYSFELSPSYVHAPTAVPTLHPELGTQLILRKLWCKNN >AaAO [Artemesia annua] (SEQ ID NO: 7) MKSILKAMALSLTTSIALATILLFVYKFATRSKSTKKSLPEPWRLPIIGHMHHLIGTTPHRGVRDL ARKYGSLMHLQLGEVPTIVVSSPKWAKEILTTYDITFANRPETLTGEIVLYHNTDVVLAPYGEYWR QLRKICTLELLSVKKVKSFQSLREEECWNLVQEIKASGSGRPVNLSENVFKLIATILSRAAFGKGI KDQKELTEIVKEILRQTGGFDVADIFPSKKFLHHLSGKRARLTSLRKKIDNLIDNLVAEHTVNTSS KTNETLLDVLLRLKDSAEFPLTSDNIKAIILDMFGAGTDTSSSTIEWAISELIKCPKAMEKVQAEL RKALNGKEKIHEEDIQELSYLNMVIKETLRLHPPLPLVLPRECRQPVNLAGYNIPNKTKLIVNVFA INRDPEYWKDAEAFIPERFENSSATVMGAEYEYLPFGAGRRMCPGAALGLANVQLPLANILYHFNW KLPNGVSYDQIDMTESSGATMQRKTELLLVPSF >AtKO [Arabidopsis thaliana] (SEQ ID NO: 8) MAFFSMISILLGFVISSFIFIFFFKKLLSFSRKNMSEVSTLPSVPVVPGFPVIGNLLQLKEKKPHK TFTRWSEIYGPIYSIKMGSSSLIVLNSTETAKEAMVTRFSSISTRKLSNALTVLTCDKSMVATSDY DDFHKLVKRCLLNGLLGANAQKRKRHYRDALIENVSSKLHAHARDHPQEPVNFRAIFEHELFGVAL KQAFGKDVESIYVKELGVTLSKDEIFKVLVHDMMEGAIDVDWRDFFPYLKWIPNKSFEARIQQKHK RRLAVMNALIQDRLKQNGSESDDDCYLNFLMSEAKTLTKEQIAILVWETIIETADTTLVTTEWAIY ELAKHPSVQDRLCKEIQNVCGGEKFKEEQLSQVPYLNGVFHETLRKYSPAPLVPIRYAHEDTQIGG YHVPAGSEIAINIYGCNMDKKRWERPEDWWPERFLDDGKYETSDLHKTMAFGAGKRVCAGALQASL MAGIAIGRLVQEFEWKLRDGEEENVDTYGLTSQKLYPLMAIINPRRS >SrKO [Stevia rebaudiana] (SEQ ID NO: 9) MDAVTGLLIVPATAITIGGTAVALAVALIFWYLKSYTSARRSQSNHLPRVPEVPGVPLLGNLLQLK EKKPYMTFTRWAATYGPIYSIKTGATSMVVVSSNEIAKEALVTRFQSISTRNLSkALKVLTADKTM VAMSDYDDYHKTVKRHILTAVLGPNAQKKHRIHRDIMMDNISTQLHEFVKNNPEQEEVDLRKIFQS ELFGLAMRQALGKDVESLYVEDLKITMNRDEIFQVLVVDPMMGAIDVDWRDFFPYLKWVPNKKFEN TIQQMYIRREAVMKSLIKEHKKRIASGEKLNSYIDYLLSEAQTLTDQQLLMSLWEPIIESSDTTMV TTEWAMYELAKNPKLQDRLYRDIKSVCGSEKITEEHLSQLPYITAIFHETLRRHSPVPIIPLRHVH EDTVLGGYHVPAGTELAVNIYGCNMDKNVWENPEEWNPERFMKENETIDFQKTMAFGGGKRVCAGS LQALLTASIGIGRMVQEFEWKLKDMTQEEVNTIGLTTQMLRPLRAIIKPRI >PpKO [Physcomitrella patens] (SEQ ID NO: 10) MAKHLATQLLQQWNEALKTMPPGFRTAGKILVWEELASNKVLITIALAWVLLFVARTCLRNKKRLP PAIPGGLPVLGNLLQLTEKKPHRTFTAWSKEHGPIFTIKVGSVPQAVVNNSEIAKEVLVTKFASIS KRQMPMALRVLTRDKTMVAMSDYGEEHRMLKKLVMTNLLGPTTQNKNRSLRDDALIGMIEGVLAEL KASPTSPKVVNVRDYVQRSLFPFALQQVFGYIPDQVEVLELGTCVSTWDMFDALVVAPLSAVINVD WRDFFPALRWIPNRSVEDLVRTVDFKRNSIMKALIRAQRMRLANLKEPPRCYADIALTEATHLTEK QLEMSLWEPIIESADTTLVTSEWAMYEIAKNPDCQDRLYREIVSVAGTERMVTEDDLPNMPYLGAI IKETLRKYTPVPLIPSRFVEEDITLGGYDIPKGYQILVNLFAIANDPAVWSNPEKWDPERMLANKK VDMGFRDFSLMPFGAGKRMCAGITQAMFIIPMNVAALVQHCEWRLSPQEISNINNKIEDVVYLTTH KLSPLSCEATPRISHRLP >BmVO [Bacillus megaterium] (SEQ ID NO: 11) MTIKEMPQPKTFGELKNLPLLNTDKPVQALMKIADELGEIFKFEAPGRVTRYLSSQRLIKEACDES RFDKNLSQALKFVRDFAGDGLATSWTHEKNWKKAHNILLPSFSQQAMKGYHAMMVDIAVQLVQKWE RLNADEHIEVPEDMTRLTLDTIGLCGFNYRFNSFYRDQPHPFITSMVRALDEAMNKLQRANPDDPA YDENKRQFQEDIKVMNDLVDKIIADRKASGEQSDDLLTHMLNGKDPETGEPLDDENIRYQIITFLI AGHETTSGLLSFALYFLVKNPHVLQKAAEEAARVLVDPVPSYKQVKQLKYVGMVLNEALRLWPTIP AFSLYAKEDTVLGGEYPLEKGDELMVLIPQLHRDKTIWGDDVEEFRPERFENPSAIPQHAFKPFGN GQRACIGQQFALHEATLVLGMMLKHFDFEDHTNYELDIKETLTLKPEGFVVKAKSKKIPLGGIPSP STEQSAKKVRKKAENAHNTPLLVLYGSNMGTAEGTARDLADIAMSKGFAPQVATLDSHAGNLPREG AVLIVTASYNGHPPDNAKQFVDWLDQASADEVKGVRYSVFGCGDKNWATTYQKVPAFIDETLAAKG AENIADRGEADASDDFEGTYEEWREHMWSDVAAYFNLDIENSEDNKSTLSLQFVDSAADMPLAKMH GAFSTNVVASKELQQPGSARSTRHLEIELPKEASYQEGDHLGVIPRNYEGIVNRVTARFGLDASQQ IRLEAEEEKLAHLPLAKTVSVEELLQYVELQDPVTRTQLRAMAAKTVCPPHKVELEALLEKQAYKE QVLAKRLTMLELLEKYPACEMKFSEFIALLPSIRPRYYSISSSPRVDEKQASITVSVVSGEAWSGY GEYKGIASNYLAELQEGDTITCFISTPQSEFTLPKDPETPLIMVGPGTGVAPFRGFVQARKQLKEQ GQSLGEAHLYFGCRSPHEDYLYQEELENAQSEGIITLHTAFSRMPNQPKTYVQHVMEQDGKKLIEL LDQGAHFYICGDSQMAPAVEATLMKSYADVHQVSEADARLWLQQLEEKGRYAKDVWAG >PsVO [Pleurotus sapidus] (SEQ ID NO: 12) MRYGCAAVALFYLTAMGKLHPLAIIPDYKGSMAASVTIFNKRTNPLDISVNQANDWPWRYAKTCVL SSDWALHEMIIHLNNTHLVEEAVIVAAQRKLSPSHIVFRLLEPHWVVTLSLNALARSVLIPEVIVP IAGFSAPHIFQFIRESFTNFDWKSLYVPADLESRGFPVDQLNSPKFHNYAYARDINDMWTTLKKFV SSVLQDAQYYPDDASVAGDTQIQAWCDEMRSGMGAGMTNFPESITTVDDLVNMVTMCIHIAAPQHT AVNYLQQYYQTFVSNKPSALFSPLPTSIAQLQKYTESDLMAALPLNAKRQWLLMAQIPYLLSMQVQ EDENIVTYAANASTDKDPIIASAGRQLAADLKKLAAVFLVNSAQLDDQNTPYDVLAPEQLANAIVI >PoLO [Pleurotus ostreatus] (SEQ ID NO: 13) MAPTMSLSRSALKNVHLPYMVQHPEPTDCSTAMKHAAEGYDRARQMIAFLFDILDYESSVPQKFTP EEKKEKYTWSHSDKFPPHLAIIPEDIDVPAYIIFSIVRLVQTLSIMSGIQCNERLAPGPEQNTMEK LTKWNAERHKNQGWVKDMFNEPNIGLRNDWYTDAVFAQQFFTGPNPTTITLASDTWMKAFTEEAAS QGKRDLISLFRSAPPNSFYVQDFSDFRARMGAKPDEELCATSDGGVTRYGCAAVALFYLPPTGELH PLAIVPDYKGSMAASITLFNKRVDPSDASVDQANDWPWRYAKTCVLSADWVLHEMIIHLNNTHLVQ EAVIVAVQRTLPDSHIVFRLLKPHWVVILSLNAQARSVLIPEVIVPIAGFSELRIFQFVGHAFTNF DWKALYVPTDLEFRGFPLDRLDDDKFHNYAYAKDIKDMWMALRKFVSSVLKDGKYYPDDSAVAADA QIQDWCDEMRSEKGAGMKKFPESISTLDDLIDMVTMCIHIAAPQHTAVNYLQQYYQTFVPNKPSAL
FSPLPTLLSQLESYTESDLMAALPLGAKQEWLLMAQVPYLLSKEVEQDGNIVTYAGTASNNEDPII AAAGKELSADLVILAGVFLKNSEKLDDQNTAYNVLAPDQLANAIVI >CiVO [Cichorium intybus] (SEQ ID NO: 14) MEISIPTTLGLAVIIFIIFKLLTRTTSKKNLLPEPWRLPIIGHMHHLIGTMPHRGVMELARKHGSL MHLQLGEVSTIVVSSPRWAKEVLTTYDITFANRPETLTGEIVAYHNTDIVLAPYGEYWRQLRKLCT LELLSNKKVKSFQSLREEECWNLVKDIRSTGQGSPINLSENIFKMIATILSRAAFGKGIKDQMKFT ELVKEILRLTGGFDVADIFPSKKLLHHLSGKRAKLTNIHNKLDNLINNIIAEHPGNRTSSSQETLL DVLLRLKESAEFPLTADNVKAVILDMFGAGTDTSSATIEWAISELIRCPRAMEKVQTELRQALNGK ERIQEEDLQELNYLKLVIKETLRLHPPLPLVMPRECREPCVLGGYDIPSKTKLIVNVFAINRDPEY WKDAETFMPERFENSPITVMGSEYEYLPFGAGRRMCPGAALGLANVELPLAHILYFNWKLPNGKTF EDLDMTESFGATVQRKTELLLVPTDFQTLTAST >HaGAO [Helianthus annuus] (SEQ ID NO: 15) MEVSLTTSIALATIVFFLYKLLTRPTSSKNRLPEPWRLPIIGHMHHLIGTMPHRGVMDLARKYGSL MHLQLGEVSAIVVSSPKWAKEILTTYDIPFANRPETLTGEIIAYHNTDIVLAPYGEYWRQLRKLCT LELLSNKKVKSFQSLREEECWNLVQEIKASGSGTPFNLSEGIFKVIATVLSRAAFGKGIKDQKQFT EIVKEILRETGGFDVADIFPSKKFLHHLSGKRGRLTSIHNKLDSLINNLVAEHTVSKSSKVNETLL DVLLRLKNSEEFPLTADNVKAIILDMFGAGTDTSSATVEWAISELIRCPRAMEKVQAELRQALNGK ERIKEEEIQDLPYLNLVIRETLRLHPPLPLVMPRECRQAMNLAGYDVANKTKLIVNVFAINRDPEY WKDAESFNPERFENSNTTIMGADYEYLPFGAGRRMCPGSALGLANVQLPLANILYYFKWKLPNGAS HDQLDMTESFGATVQRKTELMLVPSF E. coli INNER MEMBRANE PROTEINS >waaA [Escherichia coli] (SEQ ID NO: 16) MLELLYTALLYLIQPLIWIRLWVRGRKAPAYRKRWGERYGFYRHPLKPGGIMLHSVSVGETLAAIP LVRALRHRYPDLPITVTTMTPTGSERVQSAFGKDVQHVYLPYDLPDALNRFLNKVDPKLVLIMETE LWPNLIAALHKRKIPLVIANARLSARSAAGYAKLGKFVRRLLRRITLIAAQNEEDGARFVALGAKN NQVTVTGSLKFDISVTPQLAAKAVTLRRQWAPHRPVWIATSTHEGEESVVIAAHQALLQQFPNLLL ILVPRHPERFPDAINLVRQAGLSYITRSSGEVPSTSTQVVVGDTMGELMLLYGIADLAFVGGSLVE RGGHNPLEAAAHAIPVLMGPHTFNFKDICARLEQASGLITVTDATTLAKEVSSLLTDADYRSFYGR HAVEVLYQNQGALQRLLQLLEPYLPPKTH >ypfN [Escherichia coli] (SEQ ID NO: 17) MDWLAKYWWILVIVFLVGVLLNVIKDLKRVDHKKFLANKPELPPHRDFNDKWDDDDDWPKKDQPKK >yhcB [Escherichia coli] (SEQ ID NO: 18) MTWEYALIGLVVGIIIGAVAMRFGNRKLRQQQALQYELEKNKAELDEYREELVSHFARSAELLDTM AHDYRQLYQHMAKSSSSLLPELSAEANPFRNRLAESEASNDQAPVQMPRDYSEGASGLLRTGAKRD >yhbM [Escherichia coli] (SEQ ID NO: 19) MKPFLRWCFVATALTLAGCSNTSWRKSEVLAVPLQPTLQQEVILARMEQILASRALTDDERAQLLY ERGVLYDSLGLRALARNDFSQALAIRPDMPEVFNYLGIYLTQAGNFDAAYEAFDSVLELDPTYNYA HLNRGIALYYGGRDKLAQDDLLAFYQDDPNDPFRSLWLYLAEQKLDEKQAKEVLKQHFEKSDKEQW GWNIVEFYLGNISEQTLMERLKADATDNTSLAEHLSETNFYLGKYYLSLGDLDSATALFKLAVANN VHNFVEHRYALLELSLLGQDQDDLAESDQQ >yhhm [Escherichia coli] (SEQ ID NO: 20) MSKPPLFFIVIIGLIVVAASFRFMQQRREKADNDMAPLQQKLVVVSNKREKPINDRRSRQQEVTPA GTSIRYEASFKPQSGGMEQTFRLDAQQYHALTVGDKGTLSYKGTRFVSFVGEQ >zipA [Escherichia coli] (SEQ ID NO: 21) MMQDLRLILIIVGAIAIIALLVHGFWTSRKERSSMFRDRPLKRMKSKRDDDSYDEDVEDDEGVGEV RVHRVNHAPANAQEHEAARPSPQHQYQPPYASAQPRQPVQQPPEAQVPPQHAPHPAQPVQQPAYQP QPEQPLQQPVSPQVAPAPQPVHSAPQPAQQAFQPAEPVAAPQPEPVAEPAPVMDKPKRKEAVIIMN VAAHHGSELNGELLLNSIQQAGFIFGDMNIYHRHLSPDGSGPALFSLANMVKPGTFDPEMKDFTTP GVTIFMQVPSYGDELQNFKLMLQSAQHIADEVGGVVLDDQRRMMTPQKLREYQDIIREVKDANA >ycgG [Escherichia coli] (SEQ ID NO: 22) MRNTLIPILVAICLFITGVAILNIQLWYSAKAEYLAGARYAANNINHILEEASQATQTAVNIAGKE CNLEEQYQLGTEAALKPHLRTIIILKQGIVWCTSLPGNRVLLSRIPVFPDSNLLLAPAIDTVNRLP ILLYQNQFADTRILVTISDQHIRGALNVPLKGVRYVLRVADDIIGPTGDVMTLNGHYPYTEKVHST KYHFTIIFNPPPLFSFYRLIDKGFGILIFILLIACAAAFLLDRYFNKSATPEEILRRAINNGEIVP FYQPVVNGREGTLRGVEVLARWKQPHGGYISPAAFIPLAEKSGLIVPLTQSLMNQVARQMNAIASK LPEGFHIGINFSASHIISPTFVDECLNFRDSFTRRDLNLVLEVTEREPLNVDESLVQRLNILHENG FVIALDDFGTGYSGLSYLHDLHIDYIKIDKSFVGRVNADPESTRILDCVLDLARKLSISIVAEGVE TKEQLDYLNQNYITFQQGYYFYKPVTYIDLVKIILSKPKVKVVVE >djlA [Escherichia coli] (SEQ ID NO: 23) MQYWGKIIGVAVALLMGGGFWGVVLGLLIGHMFDKARSRKMAWFANQRERQALFFATTFEVMGHLT KSKGRVTEADIHIASQLMDRMNLHGASRTAAQNAFRVGKSDNYPLREKMRQFRSVCFGRFDLIRMF LEIQIQAAFADGSLHPNERAVLYVIAEELGISRAQFDQFLRMMQGGAQFGGGYQQQTGGGMWQQAQ RGPTLEDACNVLGVKPTDDATTIKRAYRKLMSEHHPDKLVAKGLPPEMMEMAKQKAQEIQQAYELI KQQKGFK >sohB [Escherichia coli] (SEQ ID NO: 24) MELLSEYGLFLAKIVTVVLAIAAIAAIIVNVAQRNKRQRGELRVNNLSEQYKEMKEELAAALMDSH QQKQWHKAQKKKHKQEAKAAKAKAKLGEVATDSKPRVWVLDFKGSMDAHEVNSLREEITAVLAAFK PQDQVVLRLESPGGMVHGYGLAASQLQRLRDKNIPLTVTVDKVAASGGYMMACVADKIVSAPFAIV GSIGVVAQMPNFNRFLKSKDIDIELHTAGQYKRTLTLLGENTEEGREKFREELNETHQLFKDFVKR MRPSLDIEQVATGEHWYGQQAVEKGLVDEINTSDEVILSLMEGREVVNVRYMQRKRLIDRFTGSAA ESADRLLLRWWQRGQKPLM >lpxK [Escherichia coli] (SEQ ID NO: 25) MIEKIWSGESPLWRLLLPLSWLYGLVSGAIRLCYKLKLKRAWRAPVPVVVVGNLTAGGNGKTPVVV WLVEQLQQRGIRVGVVSRGYGGKAESYPLLLSADTTTAQAGDEPVLIYQRTDAPVAVSPVRSDAVK AILAQHPDVQIIVTDDGLQHYRLARDVEIVVIDGVRRFGNGWWLPAGPMRERAGRLKSVDAVIVNG GVPRSGEIPMHLLPGQAVNLRTGTRCDVAQLEHVVAMAGIGHPPRFFATLKMCGVQPEKCVPLADH QSLNHADVSALVSAGQTLVMTEKDAVKCRAFAEENWWYLPVDAQLSGDEPAKLLTQLTLLASGN >fliO [Escherichia coli] (SEQ ID NO: 26) MNNHATVQSSAPVSAAPLLQVSGALIAIIALILAAAWLVKRLGFAPKRTGVNGLKISASASLGARE RVVVVDVEDARLVLGVTAGQINLLHKLPPSAPTEEIPQTDFQSVMKNLLKRSGRS >motA [Escherichia coli] (SEQ ID NO: 27) MLILLGYLVVLGTVFGGYLMTGGSLGALYQPAELVIIAGAGIGSFIVGNNGKAIKGTLKALPLLFR RSKYTKAMYMDLLALLYRLMAKSRQMGMFSLERDIENPRESEIFASYPRILADSVMLDFIVDYLRL IISGHMNTFEIEALMDEEIETHESEAEVPANSLALVGDSLPAFGIVAAVMGVVHALGSADRPAAEL GALIAHAMVGTFLGILLAYGFISPLATVLRQKSAETSKMMQCVKVTLLSNLNGYAPPIAVEFGRKT LYSSERPSFIELEEHVRAVKNPQQQTTTEEA >htpx [Escherichia coli] (SEQ ID NO: 28) MMRIALFLLTNLAVMVVFGLVLSLTGIQSSSVQGLMIMALLFGFGGSFVSLLMSKWMALRSVGGEV IEQPRNERERWLVNTVATQARQAGIAMPQVAIYHAPDINAFATGARRDASLVAVSTGLLQNMSPDE AEAVIAHEISHIANGDMVTMTLIQGVVNTFVIFISRILAQLAAGFMGGNRDEGEESNGNPLIYFAV ATVLELVFGILASIITMWFSRHREFHADAGSAKLVGREKMIAALQRLKTSYEPQEATSMMALCING KSKSLSELFMTHPPLDKRIEALRTGEYLK >pgaC [Escherichia coli] (SEQ ID NO: 29) MINRIVSFFILCLVLCIPLCVAYFHSGELMMRFVFFWPFFMSIMWIVGGVYFWVYRERHWPWGENA PAPQLKDNPSISIIIPCFNEEKNVEETIHAALAQRYENIEVIAVNDGSTDKTRAILDRMAAQIPHL RVIHLAQNQGKAIALKTGAAAAKSEYLVCIDGDALLDRDAAAYIVEPMLYNPRVGAVTGNPRIRTR STLVGKIQVGEYSSIIGLIKRTQRIYGNVFTVSGVIAAFRRSALAEVGYWSDDMITEDIDISWKLQ LNQWTIFYEPRALCWILMPETLKGLWKQRLRWAQGGAEVFLKNMTRLWRKENFRMWPLFFEYCLTT IWAFTCLVGFIIYAVQLAGVPLNIELTHIAATHTAGILLCTLCLLQFIVSLMIENRYEHNLTSSLF WIIWFPVIFWMLSLATTLVSFTRVMLMPKKQRARWVSPDRGILRG >ygdD [Escherichia coli] (SEQ ID NO: 30) MTSRFMLIFAAISGFIFVALGAFGAHVLSKTMGAVEMGWIQTGLEYQAFHTLAILGLAVAMQRRIS IWFYWSSVFLALGTVLFSGSLYCLALSHLRLWAFVTPVGGVSFLAGWALMLVGAIRLKRKGVSHE >hemr [Escherichia coli] (SEQ ID NO: 31) MNVIKTAICTLITLPVGLQAATSHSSSMTKDTITVVATGNQNTVFETPSMVSVVTNDTPWSKNAVT SAGMLRGVAGLSQTGAGRTNGQTFNLRGYDKSGVLVLVDGVRQLSDMAKSSGTYLDPALVKRIEVV RGPNSSLYGSGGLGGVVDFRTADAADFLPPGETNGVSLWGNIASGDHSTGSGLTWFGKTEKTDALL SVIMRKRGSIYQSDGERAPNKEKPAALFAKGSVSITDSNKAGASLRLYRNSTTEPGNPTLTHGDSG LRDRKTAQNDMQFWYQYAPADNSLINVKSTLYLSDITVKTNGHNKTAEWRNNRTSGVNVVNRSHSL IFPGAHQLSYGAEYYRQQQKPEGTATLYPEGHIDFTSLYFQDEMTMESYPVNIIVGSRYDRYNSFN ARAGELNAERLSPRAAMSVSPTDWLMMYGSISSAFRAPTMAEMYRDDVHFYRKGKPNYWVPNLNLK PENNTTREIGAGIQLDSLLTDNDRLQLKGGYFGTDARNYIATRVDMKRMRSYSYNVSRARIWGWDI QGNYQSDYVDWMLSYNRTESMDASSREWLGSGNPDTLISDISIPVGHRGVYAGWRAELSAPATHVK KGDPCQDGYAIHSFSLSYKPVSVKGFEASVTLDNAFNKLAMNGKGVPLSGRTVNLYTRYQW >yciS [Escherichia coli] (SEQ ID NO: 32) MKYLLIFLLVLAIFVISVTLGAQNDQQVTFNYLLAQGEYRISTLLAVLFAAGFATGWLICGLFWLR VRVSLARAERKIKRLENQLSPATDVAVVPHSSAAKE CYTOCHROME P450 REDUCTASE PARTNERS >SrCPR [Stevia rebaudiana] (SEQ ID NO: 33) MQSDSVKVSPFDLVSAAMNGKAMEKLNASESEDPTTLPALKMLVENRELLTLFTTSFAVLIGCLVF LMWRRSSSKKLVQDPVPQVIVVKKKEKESEVDDGKKKVSIFYGTQTGTAEGFAKALVEEAKVRYEK TSFKVIDLDDYAADDDEYEEKLKKESLAFFFLATYGDGEPTDNAANFYKWFTEGDDKGEWLKKLQY GVFGLGNRQYEHFNKIAIVVDDKLTEMGAKRLVPVGLGDDDQCIEDDFTAWKELVWPELDQLLRDE DDTSVTTPYTAAVLEYRVVYHDKPADSYAEDQTHTNGHVVHDAQHPSRSNVAFKKELHTSQSDRSC THLEFDISHTGLSYETGDHVGVYSENLSEVVDEALKLLGLSPDTYFSVHADKEDGTPIGGASLPPP FPPCTLRDALTRYADVLSSPKKVALLALAAHASDPSEADRLKFLASPAGKDEYAQWIVANQRSLLE VMQSFPSAKPPLGVFFAAVAPRLQPRYYSISSSPKMSPNRIHVTCALVYETTPAGRIHRGLCSTWM
KNAVPLTESPDCSQASIFVRTSNFRLPVDPKVPVIMIGPGTGLAPFRGFLQERLALKESGTELGSS IFFFGCRNRKVDFIYEDELNNFVETGALSELIVAFSREGTAKEYVQHKMSQKASDIWKLLSEGAYL YVCGDAKGMAKDVHRTLHTIVQEQGSLDSSKAELYVKNLQMSGRYLRDVW >AtCPR [Arabidopsis thaliana] (SEQ ID NO: 34) MTSALYASDLFKQLKSIMGTDSLSDDVVLVIATTSLALVAGFVVLLWKKTTADRSGELKPLMIPKS LMAKDEDDDLDLGSGKTRVSIFFGTQTGTAEGFAKALSEEIKARYEKAAVKVIDLDDYAADDDQYE EKLKKETLAFFCVATYGDGEPTDNAARFSKWFTEENERDIKLQQLAYGVFALGNRQYEHFNKIGIV LDEELCKKGAKRLIEVGLGDDDQSIEDDFNAWKESLWSELDKLLKDEDDKSVATPYTAVIPEYRVV THDPRFTTQKSMESNVANGNTTIDIHHPCRVDVAVQKELHTHESDRSCIHLEFDISRTGITYETGD HVGVYAENHVEIVEEAGKLLGHSLDLVFSIHADKEDGSPLESAVPPPPPGPCTLGTGLARYADLLN PPRKSALVALAAYATEPSEAEKLKHLTSPDGKDEYSQWIVASQRSLLEVMAAFPSAKPPLGVFFAA IAPRLQPRYYSISSCQDWAPSRVHVTSALVYGPTPTGRIHKGVCSTWMKNAVPAEKSHECSGAPIF IRASNFKLPSNPSTPIVMVGPGTGLAPFRGFLQERMALKEDGEELGSSLLFFGCRNRQMDFIYEDE LNNFVDQGVISELIMAFSREGAQKEYVQHKMMEKAAQVWDLIKEEGYLYVCGDAKGMARDVHRTLH TIVQEQEGVSSSEAEAIVKKLQTEGRYLRDVW >TcCPR [Taxus cuspidata] (SEQ ID NO: 35) MQANSNTVEGASQGKSLLDISRLDHIFALLLNGKGGDLGAMTGSALILTENSQNLMILTTALAVLV ACVFFFVWRRGGSDTQKPAVRPTPLVKEEDEEEEDDSAKKKVTIFFGTQTGTAEGFAKALAEEAKA RYEKAVFKVVDLDNYAADDEQYEEKLKKEKLAFFMLATYGDGEPTDNAARFYKWFLEGKEREPWLS DLTYGVFGLGNRQYEHFNKVAKAVDEVLIEQGAKRLVPVGLGDDDQCIEDDFTAWREQVWPELDQL LRDEDDEPTSATPYTAAIPEYRVEIYDSVVSVYEETHALKQNGQAVYDIHHPCRSNVAVRRELHTP LSDRSCIHLEFDISDTGLIYETGDHVGVHTENSIETVEEAAKLLGYQLDTIFSVHGDKEDGTPLGG SSLPPPFPGPCTLRTALARYADLLNPPRKAAFLALAAHASDPAEAERLKFLSSPAGKDEYSQWVTA SQRSLLEIMAEFPSAKPPLGVFFAAIAPRLQPRYYSISSSPRFAPSRIHVTCALVYGPSPTGRIHK GVCSNWMKNSLPSEETHDCSWAPVFVRQSNFKLPADSTTPIVMVGPGTGFAPFRGFLQERAKLQEA GEKLGPAVLFFGCRNRQMDYIYEDELKGYVEKGILTNLIVAFSREGATKEYVQHKMLEKASDTWSL IAQGGYLYVCGDAKGMARDVHRTLHTIVQEQESVDSSKAEFLVKKLQMDGRYLRDIW >AaCPR [Artemisia annua] (SEQ ID NO: 36) MAQSTTSVKLSPFDLMTALLNGKVSFDTSNTSDTNIPLAVFMENRELLMILTTSVAVLIGCVVVLV WRRSSSAAKKAAESPVIVVPKKVTEDEVDDGRKKVTVFFGTQTGTAEGFAKALVEEAKARYEKAVF KVIDLDDYAAEDDEYEEKLKKESLAFFFLATYGDGEPTDNAARFYKWFTEGEEKGEWLDKLQYAVF GLGNRQYEHFNKIAKVVDEKLVEQGAKRLVPVGMGDDDQCIEDDFTAWKELVWPELDQLLRDEDDT SVATPYTAAVAEYRVVFHDKPETYDQDQLTNGHAVHDAQHPCRSNVAVKKELHSPLSDRSCTHLEF DISNTGLSYETGDHVGVYVENLSEVVDEAEKLIGLPPHTYFSVHADNEDGTPLGGASLPPPFPPCT LRKALASYADVLSSPKKSALLALAAHATDSTEADRLKFLASPAGKDEYAQWIVASHRSLLEVMEAF PSAKPPLGVFFASVAPRLQPRYYSISSSPRFAPNRIHVTCALVYEQTPSGRVHKGVCSTWMKNAVP MTESQDCSWAPIYVRTSNFRLPSDPKVPVIMIGPGTGLAPFRGFLQERLAQKEAGTELGTAILFFG CRNRKVDFIYEDELNNFVETGALSELVTAFSREGATKEYVQHKMTQKASDIWNLLSEGAYLYVCGD AKGMAKDVHRTLHTIVQEQGSLDSSKAELYVKNLQMAGRYLRDVW >AtCPR1 [Arabidopsis thaliana] (SEQ ID NO: 37) MATSALYASDLFKQLKSIMGTDSLSDDVVLVIATTSLALVAGFVVLLWKKTTADRSGELKPLMIPK SLMAKDEDDDLDLGSGKTRVSIFFGTQTGTAEGFAKALSEEIKARYEKAAVKVIDLDDYAADDDQY EEKLKKETLAFFCVATYGDGEPTDNAARFYKWFTEENERDIKLQQLAYGVFALGNRQYEHFNKIGI VLDEELCKKGAKRLIEVGLGDDDQSIEDDFNAWKESLWSELDKLLKDEDDKSVATPYTAVIPEYRV VTHDPRFTTQKSMESNVANGNTTIDIHHPCRVDVAVQKELHTHESDRSCIHLEFDISRTGITYETG DHVGVYAENHVEIVEEAGKLLGHSLDLVFSIHADKEDGSPLESAVPPPFPGPCTLGTGLARYADLL NPPRKSALVALAAYATEPSEAEKLKHLTSPDGKDEYSQWIVASQRSLLEVMAAFPSAKPPLGVFFA AIAPRLQPRYYSISSSPRLAPSRVHVTSALVYGPTPTGRIHKGVCSTWMKNAVPAEKSHECSGAPI FIRASNFKLPSNPSTPIVMVGPGTGLAPFRGFLQERMALKEDGEELGSSLLFFGCRNRQMDFIYED ELNNFVDQGVISELIMAFSREGAQKEYVQHKMMEKAAQVWDLIKEEGYLYVCGDAKGMARDVHRTL HTIVQEQEGVSSSEAEAIVKKLQTEGRYLRDVW >AtCPR2 [Arabidopsis thaliana] (SEQ ID NO: 38) MASSSSSSSTSMIDLMAAIIKGEPVIVSDPANASAYESVAAELSSMLIENRQFAMIVTTSIAVLIG CIVMLVWRRSGSGNSKRVEPLKPLVIKPREEEIDDGRKKVTIFFGTQTGTAEGFAKALGEEAKARY EKTRFKIVDLDDYAADDDEYEEKLKKEDVAFFFLATYGDGEPTDNAARFYKWFTEGNDRGEWLKNL KYGVFGLGNRQYEHFNKVAKVVDDILVEQGAQRLVQVGLGDDDQCIEDDFTAWREALWPELDTILR EEGDTAVATPYTAAVLEYRVSIHDSEDAKFNDINMANGNGYTVFDAQHPYKANVAVKRELHTPESD RSCIHLEFDIAGSGLTYETGDHVGVLCDNLSETVDEALRLLDMSPDTYFSLHAEKEDGTPISSSLP PPFPPCNLRTALTRYACLLSSPKKSALVALAAHASDPTEAERLKHLASPAGKDEYSKWVVESQRSL LEVMAEFPSAKPPLGVFFAGVAPRLQPRFYSISSSPKIAETRIHVTCALVYEKMPTGRIHKGVCST WMKNAVPYEKSENCSSAPIFVRQSNFKLPSDSKVPIIMIGPGTGLAPFRGFLQERLALVESGVELG PSVLFFGCRNRRMDFIYEEELQRFVESGALAELSVAFSREGPTKEYVQHKMMDKASDIWNMISQGA YLYVCGDAKGMARDVHRSLHTIAQEQGSMDSTKAEGFVKNLQTSGRYLRDVW >ATR2 [Arabidopsis thaliana] (SEQ ID NO: 39) MASSSSSSSTSMIDLMAAIIKGEPVIVSDPANASAYESVAAELSSMLIENRQFAMIVTTSIAVLIG CIVMLVWRRSGSGNSKRVEPLKPLVIKPREEEIDDGRKKVTIFFGTQTGTAEGFAKALGEEAKARY EKTRFKIVDLDDYAADDDEYEEKLKKEDVAFFFLATYGDGEPTDNAARFYKWFTEGNDRGEWLKNL KYGVFGLGNRQYEHFNKVAKVVDDILVEQGAQRLVQVGLGDDDQCIEDDFTAWREALWPELDTILR EEGDTAVATPYTAAVLEYRVSIHDSEDAKFNDITLANGNGYTVFDAQHPYKANVAVKRELKTPESD RSCIHLEFDIAGSGLTMKLGDHVGVLCDNLSETVDEALRLLDMSPDTYFSLHAEKEDGTPISSSLP PPFPPCNLRTALTRYACLLSSPKKSALVALAAHASDPTEAERLKHLASPAGKDEYSKWVVESQRSL LEVMAEFPSAKPPLGVFFAGVAPRLQPRFYSISSSPKIAETRIHVTCALVYEKMPTGRIHKGVCST WMKNAVPYEKSEKLFLGRPIFVRQSNFKLPSDSKVPIIMIGPGTGLAPFRGFLQERLALVESGVEL GPSVLFFGCRNRRMDFIYEEELQRFVESGALAELSVAFSREGPTKEYVQHKMMDKASDIWNMISQG AYLYVCGDAKGMARDVHRSLHTIAQEQGSMDSTKAEGFVKNLQTSGRYLRDVW >SrCPR1 [Slevia rebaudiana] (SEQ ID NO: 40) MAQSDSVKVSPFDLVSAAMNGKAMEKLNASESEDPTTLPALKMLVENRELLTLFTTSFAVLIGCLV FLMWRRSSSKKLVQDPVPQVIVVKKKEKESEVDDGKKKVSIFYGTQTGTAEGFAKALVEEAKVRYE KTSFKVIDLDDYAADDDEYEEKLKKESLAFFFLATYGDGEPTDNAANFYKWFTEGDDKGELLKKLQ YGVFGLGNRQYEHFNKIAIVVDDKLTEMGAKRLVPVGLGDDDQCIEDDFTAWKELVWPELDQLLRD EDDTSVTTPYTAAVLEYRVVYHDKPADSYAEDQTHTNGHVVHDAQHPSRSNVAFKKELHTSQSDRS CTHLEFDISHTGLSYETGDHVGVYSENLSEVVDEALKLLGLSPDTYFSVHADKEDGTPIGGASLPP PFPPCTLRDALTRYADVLSSPKKVALLALAAHASDPSEADRLKFLASPAGKDEYAQWIVANQRSLL EVMQSFPSAKPPLGVFFAAVAPRLQPRYYSISSSPKMSPNRIHVTCALVYETTPAGRIKRGLCSTW MKNAVPLTESPDCSQASIFVRTSNFRLPVDPKVPVIMIGPGTGLAPFRGFLQERLALKESGTELGS SIFFFGCRNRKVDFIYEDELNNFVETGALSELIVAFSREGTAKEYVQHKMSQKASDIWKLLSEGAY LYVCGDAKGMAKDVHRTLHTIVQEQGSLDSSKAELYVKNLQMSGRYLRDVW >SrCPR2 [Stevia rebaudiana] (SEQ ID NO: 41) MAQSESVEASTIDLMTAVLKDTVIDTANASDNGDSKMPPALAMMFEIRDLLLILTTSVAVLVGCFV VLVWKRSSGKKSGKELEPPKIVVPKRRLEQEVDDGKKKVTIFFGTQTGTAEGFAKALFEEAKARYE KAAFKVIDLDDYAADLDEYAEKLKKETYAFFFLATYGDGEPTDNAAKFYKWFTEGDEKGVWLQKLQ YGVFGLGNRQYEHFNKIGIVVDDGLTEQGAKRIVPVGLGDDDQSIEDDFSAWKELVWPELDLLLRD EDDKAAATPYTAAIPEYRVVFHDKPDAFSDDHTQTNGHAVHDAQHPCRSNVAVKKELHTPESDRSC THLEFDISHTGLSYETGDHVGVYCENLIEVVEEAGKLLGLSTDTYFSLHIDNEDGSPLGGPSLQPP FPPCTLRKALTNYADLLSSPKKSTLLALAAHASDPTEADRLRFLASREGKDEYAEWVVANQRSLLE VMEAFPSARPPLGVFFAAVAPRLQPRYYSISSSPKMEPNRIHVTCALVYEKTPAGRIHKGICSTWM KNAVPLTESQDCSWAPIFVRTSNFRLPIDPKVPVIMIGPGTGLAPFRGFLQERLALKESGTELGSS ILFFGCRNRKVDYIYENELNNFVENGALSELDVAFSRDGPTKEYVQHKMTQKASEIWNMLSEGAYL YVCGDAKGMAKDVHRTLHTIVQEQGSLDSSKAELYVKNLQMSGRYLRDVW >SrCPR3 [Stevia rebaudiana] (SEQ ID NO: 42) MAQSNSVKISPLDLVTALFSGKVLDTSNASESGESAMLPTIAMIMENRELLMILTTSVAVLIGCVV VLVWRRSSTKKSALEPPVIVVPKRVQEEEVDDGKKKVTVFFGTQTGTAEGFAKALVEEAKARYEKA VFKVIDLDDYAADDDEYEEKLKKESLAFFFLATYGDGEPTDNAARFYKWFTEGDAKGEWLNKLQYG VFGLGNRQYEHFNKIAKVVDDGLVEQGAKRLVPVGLGDDDQCIEDDFTAWKELVWPELDQLLRDED DTTVATPYTAAVAEYRVVFHEKPDALSEDYSYTNGHAVKDAQHPCRSNVAVKKELHSPESDRSCTH LEFDISNTGLSYETGDHVGVYCENLSEVVNDAERLVGLPPDTYFSIHTDSEDGSPLGGASLPPPFP PCTLRKALTCYADVLSSPKKSALLALAAHATDPSEADRLKFLASPAGKDEYSQWIVASQRSLLEVM EAFPSAKPSLGVFFASVAPRLQPRYYSISSSPKMAPDRIHVTCALVYEKTPAGRIHKGVCSTWMKN AVPMTESQDCSWAPIYVRTSNFRLPSDPKVPVIMIGPGTGLAPFRGFLQERLALKEAGTDLGLSIL FFGCRNRKVDFIYENELNNFVETGALSELIVAFSREGPTKEYVQHKMSEKASDIWNLLSEGAYLYV CGDAKGMAKDVHRTLHTIVQEQGSLDSSKAELYVKNLQMSGRYLRDVW >PgCPR [Pelargonium graveolens] (SEQ ID NO: 43) MAQSSSGSMSPFDFMTAIIKGKMEPSNASLGAAGEVTAMILDNRELVMILTTSIAVLIGCVVVFIW RRSSSQTPTAVQPLKPLLAKETESEVDDGKQKVTIFFGTQTGTAEGFAKALADEAKARYDKVTFKV VDLDDYAADDEEYEEKLKKETLAFFFLATYGDGEPTDNAARFYKWFLEGKERGEWLQNLKFGVFGL GNRQYEHFNKIAIVVDEILAEQGGKRLISVGLGDDDQCIEDDFTAWRESLWPELDQLLRDEDDTTV STPYTAAVLEYRVVFHDPADAPTLEKSYSNANGHSVVDAQHPLRANVAVRRELHTPASDRSCTHLE FDISGTGIAYETGDHVGVYCENLAETVEEALELLGLSPDTYFSVHADKEDGTPLSGSSLPPPFPPC TLRTALTLHADLLSSPKKSALLALAAHASDPTEADRLRHLASPAGKDEYAQWIVASQRSLLEVMAE FPSAKPPLGVFFASVAPRLQPRYYSISSSPRIAPSRIHVTCALVYEKTPTGRVHKGVCSTWMKNSV PSEKSDECSWAPIFVRQSNFKLPADAKVPIIMIGPGTGLAPFRGFLQERLALKEAGTELGPSILFF GCRNSKMDYIYEDELDNFVQNGALSELVLAFSREGPTKEYVQHKMMEKASDIWNLISQGAYLYVCG DAKGMARDVHRTLHTIAQEQGSLDSSKAESMVKNLQMSGRYLRDVW LINKER SEQUENCES (SEQ ID NO: 44) GSGGGGS
(SEQ ID NO: 45) GSGEAAAK (SEQ ID NO: 46) GSGEAAAKEAAAK (SEQ ID NO: 47) GSGMGSSSN. P450 ENZYMES WITH TRANSMEMBRANE DOMAIN DELETED >t22ZzHO [Zingiber zerumbet] (SEQ ID NO: 48) LLIKRSSRSSVHKQQVLLASLPPSPPRLPLIGNIHQLVGGMPHRILLQLARTHGPLICLRLGQVDQ VVASSVEAVEEIIKRHDLKFADRPRDLTFSRIFFYDGNAVVMTPYGGEWKQMRKIYAMELLNSRRV KSFAAIREDVARKLTGEIAHKAFAQTPVINLSEMVMSMINAIVIRVAFGDKCKQQAYFLHLVKEAM SYVSSFSVADMYPSLKFLDTLTGLKSKLEGVHGKLDKVFDEIIAQRQAALAAEQAEEDLIIDVLLK LKDEGNQEFPITYTSVKAIVMEIFLAGTETSSSVIDWVMSELIKNPKAMEKVQKEMREAMQGKTKL EESDIPKFSYLNLVIKETLRLHPPGPLLFPRECRETCEVMGYRVPAGARLLINAFALSRDEKYWGS DAESFKPERFEGISVDFKGSNFEFMPFGAGRRICPGMTFGISSVEVALAHLLFHFDWQLPQGMKIE DLDMMEVSGMSATRRSPLLVLAKLIIPLP >t20BsGAO [Barnadesia spinosa] (SEQ ID NO: 49) LLTGSKSTKNSLPEAWRLPIIGHMHHLVGTLPHRGVTDMARKYGSLMHLQLGEVSTIVVSSPRWAK EVLTTYDITFANRPETLTGEIVAYHNTDIVLSPYGEYWRQLRKLCTLELLSAKKVKSFQSLREEEC WNLVKEVRSSGSGSPVDLSESIFKLIATILSRAAFGKGIKDQREFTEIVKEILRLTGGFDVADIFP SKKILHHLSGKRAKLTNIHNKLDSLINNIVSEHPGSRTSSSQESLLDVLLRLKDSAELPLTSDNVK AVILDMFGAGTDTSSATIEWAISELIRCPRAMEKVQTELRQALNGKERIQEEDIQELSYLKLVIKE TLRLHPPLPLVMPRECREPCVLAGYEIPTKTKLIVNVFAINRDPEYWKDAETFMPERFENSPINIM GSEYEYLPFGAGRRMCPGAALGLANVELPLAHILYYFNWKLPNGARLDELDMSECFGATVQRKSEL LLVPTAYKTANNSA >t16HmPO [Hyoscyamus muticus] (SEQ ID NO: 50) FLLRKWKNSNSQSKKLPPGPWKLPLLGSMLHMVGGLPHHVLRDLAKKYGPLMKLQLGEVSAVVVTS PDMAKEVLKTHDIAFASRPKLLAPEIVCYNRSDIAFCPYGDYWRQMRKICVLEVLSAKNVRSFSSI RRDEVLRLVNFVRSSTSEPVNFTERLFLFTSSMTCRSAFGKVFKEQETFIQLIKEVIGLAGGFDVA DIFPSLKFLHVLTGMEGKIMKAHHKVDAIVEDVINEHKKNLAMGKTNGALGGEDLIDVLLRLMNDG GLQFPITNDNIKAIIFDMFAAGTETSSSTLVWAMVQMMRNPTILAKAQAEVREAFKGKETFDENDV EELKYLKLVIKETLRLHPPVPLLVPRECREETEINGYTIPVKTKVMVNVWALGRDPKYWDDADNFK PERFEQCSVDFIGNNFEYLPFGGGRRICPGISFGLANVYLPLAQLLYHFDWKLPTGMEPKDLDLTE LVGVTAARKSDLMLVATPYQPSRE >t19LsGAO [Lactuca sativa] (SEQ ID NO: 51) KLATRPKSTKKQLPEASRLPIIGHMHHLIGTMPHRGVMDLARKHGSLMHLQLGEVSTIVVSSPKWA KEILTTYDITFANRPETLTGEIIAYHNTDIVLAPYGEYWRQLRKLCTLELLSVKKVKSFQSIREEE CWNLVKEVKESGSGKPINLSESIFTMIATILSRAAFGKGIKDQREFTEIVKEILRQTGGFDVADIF PSKKFLHHLSGKRARLTSIHKKLDNLINNIVAEHHVSTSSKANETLLDVLLRLKDSAEFPLTADNV KAIILDMFGAGTDTSSATVEWAISELIRCPRAMEKVQAELRQALNGKEKIQEEDIQDLAYLNLVIR ETLRLHPPLPLVMPRECREPVNLAGYEIANKTKLIVNVFAINRDPEYWKDAEAFIPERFEMNPNNI MGADYEYLPFGAGRRMCPGAALGLANVQLPLANILYHFNWKLPNGASHDQLDMTESFGATVQRKTE LLLVPSF >t16NtEAO [Nicotiani tabacum] (SEQ ID NO: 52) FLLRKWKNSNSQSKKLPPGPWKIPILGSMLHMIGGEPHHVLRDLAKKYGPLMHLQLGEISAVVVTS RDMAKEVLKTHDVVFASRPKIVAMDIICYNQSDIAFSPYGDHWRQMRKICVMELLNAKNVRSFSSI RRDEVVRLIDSIRSDSSSGELVNFTQRIIWFASSMTCRSAFGQVLKGQDIFAKKIREVIGLAEGFD VVDIFPTYKFLHVLSGMKRKLLNAHLKVDAIVEDVINEHKKNLAAGKSNGALGGEDLIDVLLRLMN DTSLQFPITNDNIKAVIVDMFAAGTETSSTTTVWAMAEMMKNPSVFTKAQAEVREAFRDKVSFDEN DVEELKYLKLVIKETLRLHPPSPLLVPRECREDTDINGYTIPAKTKVMVNVWALGRDPKYWDDAES FKPERFEQCSVDFFGNNFEFLPFGGGRRICPGMSFGLANLYLPLAQLLYHFDWKLPTGIMPRDLDL TELSGITIARKGGLYLNATPYQPSRE >t26CpVO [Citrus x paradisi] (SEQ ID NO: 53) WVWLRPKKLEKFLRQQGLKGNSYRLLFGDLKENSIELKEAKARPLSLDDDIAIRVNPFLHKLVNDY GKNSFMWFGPTPRVNIMNPDQIKAIFTKINDFQKVNSIPLARLLIVGLATLEGEKWAKHRKLINPA FHQEKLKLMLPAFYLSCIEIITKWEKQMSVEGSSELDVWPYLANLTSDVISRTAFGSSYEEGRRIF QLQAELAELTMQVFRSVHIPGWRFLETKRNRRMKEIDKEIRASLMGIIKNREKAMRAGEAANNDLL GILMETSFREIEEKGNNKNVGFSMNDVIEECKLFYFAGQETTSVLLNWTMVLLSKHQDWQERARQE VLQVFGNNKPDYDGLNHLKIVQMILYEVLRLYPPVTVLSRAVFKETKLGNLTLPAGVQIGLPMILV HQDPELWGDDAVEFKPERFAEGISKAAKNQVSYFPFALGPRICVGQNFALVEAKMATAMILQNYSF ELSPSYVHAPTAVPTLHPELGTQLILRKLWCKNN >t23AaAO [Artemesia annua] (SEQ ID NO: 54) FVYKFATRSKSTKKSLPEPWRLPIIGHMHKLIGTTPHRGVRDLARKYGSLMHLQLGEVPTIVVSSP KWAKEILTTYDITFANRPETLTGEIVLYHNTDVVLAPYGEYWRQLRKICTLELLSVKKVKSFQSLR EEECWNLVQEIKASGSGRPVNLSENVFKLIATILSRAAFGKGIKDQKELTEIVKEILRQTGGFDVA DIFPSKKFLHHLSGKRARLTSLRKKIDNLIDNLVAEHTVNTSSKTNETLLDVLLRLKDSAEFPLTS DNIKAIILDMFGAGTDTSSSTIEWAISELIKCPKAMEKVQAELRKALNGKEKIHEEDIQELSYLNM VIKETLRLHPPLPLVLPRECRQPVNLAGYNIPNKTKLIVNVFAINRDPEYWKDAEAFIPERFENSS ATVMGAEYEYLPFGAGRRMCPGAALGLANVQLPLANILYHFNWKLPNGVSYDQIDMTESSGATMQR KTELLLVPSF >t21AtKO [Arabidopsis thaliana] (SEQ ID NO: 55) FFFKKLLSFSRKNMSEVSTLPSVPVVPGFPVIGNLLQLKEKKPHKTFTRWSEIYGPIYSIKMGSSS LIVLNSTETAKEAMVTRFSSISTRKLSNALTVLTCDKSMVATSDYDDFHKLVKRCLLNGLLGANAQ KRKRHYRDALIENVSSKLHAHARDHPQEPVNFRAIFEHELFGVALKQAFGKDVESIYVKELGVTLS KDEIFKVLVHDMMEGAIDVDWRDFFPYLKWIPNKSFEARIQQKHKRRLAVMNALIQDRLKQNGSES DDDCYLNFLMSEAKTLTKEQIAILVWETIIETADTTLVTTEWAIYELAKHPSVQDRLCKEIQNVCG GEKFKEEQLSQVPYLNGVFHETLRKYSPAPLVPIRYAHEDTQIGGYHVPAGSEIAINIYGCNMDKK RWERPEDWWPERFLDDGKYETSDLHKTMAFGAGKRVCAGALQASLMAGIAIGRLVQEFEWKLRDGE EENVDTYGLTSQKLYPLMAIINPRRS >t30SrKO [Stevia rebaudiana] (SEQ ID NO: 56) WYLKSYTSARRSQSNHLPRVPEVPGVPLLGNLLQLKEKKPYMTFTRWAATYGPIYSIKTGATSMVV VSSNEIAKEALVTRFQSISTRNLSKALKVLTADKTMVAMSDYDDYHKTVKRHILTAVLGPNAQKKH RIHRDIMMDNISTQLHEFVKNNPEQEEVDLRKIFQSELFGLAMRQALGKDVESLYVEDLKITMNRD EIFQVLVVDPMMGAIDVDWRDFFPYLKWVPNKKFENTIQQMYIRREAVMKSLIKEHKKRIASGEKL NSYIDYLLSEAQTLTDQQLLMSLWEPIIESSDTTMVTTEWAMYELAKNPKLQDRLYRDIKSVCGSE KITEEHLSQLPYITAIFHETLRRHSPVPIIPLRHVHEDTVLGGYHVPAGTELAVNIYGCNMDKNVW ENPEEWMPERFMKENETIDFQKTMAFGGGKRVCAGSLQALLTASIGIGRMVQEFEWKLKDMTQEEV NTIGLTTQMLRPLRAIIKPRI >t52PpKO [Physcomitrella patens] (SEQ ID NO: 57) FVARTCLRNKKRLPPAIPGGLPVLGNLLQLTEKKPHRTFTAWSKEHGPIFTIKVGSVPQAVVNNSE IAKEVLVTKFASISKRQMPMALRVLTRDKTMVAMSDYGEEHRMLKKLVMTNLLGPTTQNKNRSLRD DALIGMIEGVLAELKASPTSPKVVNVRDYVQRSLFPFALQQVFGYIPDQVEVLELGTCVSTWDMFD ALVVAPLSAVINVDWRDFFPALRWIPNRSVEDLVRTVDFKRNSIMKALIRAQRMRLANLKEPPRCY ADIALTEATHLTEKQLEMSLWEPIIESADTTLVTSEWAMYEIAKNPDCQDRLYREIVSVAGTERMV TEDDLPNMPYLGAIIKETLRKYTPVPLIPSAFVEEDITLGGYDIPKGYQILVNLFAIANDPAVWSN PEKWDPERMLANKKVDMGFRDFSLMPFGAGKRMCAGITQAMFIIPMNVAALVQHCEWRLSPQEISN INNKIEDVVYLTTHKLSPLSCEATPRISHRLP >t15PsVO [Pleurotus sapidus] (SEQ ID NO: 58) MGKLHPLAIIPDYKGSMAASVTIFNKRTNPLDISVNQANDWPWRYAKTCVLSSDWALHEMIIHLNN THLVEEAVIVAAQRKLSPSHIVFRLLEPHWVVTLSLNALARSVLIPEVIVPIAGFSAPHIFQFIRE SFTNFDWKSLYVPADLESRFPVDQLNSPKFHNYAYARDINDMWTTLKKFVSSVLQDAQYYPDDAS VAGDTQIQAWCDEMRSGMGAGMTNFPESITTVDDLVNMVTMCIHIAAPQHTAVNYLQQYYQTFVSN KPSALFSPLPTSIAQLQKYTESDLMAALPLNAKRQWLLMAQIPYLLSMQVQEDENIVTYAANASTD KDPIIASAGRQLAADLKKLAAVFLVMSAQLDDQNTPYDVLAPEQLANAIVI >t20CiVO [Cichorium intvbus] (SEQ ID NO: 59) LLTRTTSKKNLLPEPWRLPIIGHMHHLIGTMPHRGVMELARKHGSLMHLQLGEVSTIVVSSPRWAK EVLTTYDITFANRPETLTGEIVAYHNTDIVLAPYGEYWRQLRKLCTLELLSNKKVKSFQSLREEEC WNLVKDIRSTGQGSPINLSENIFKMIATILSRAAFGKGIKDQMKFYELVKEILRLTGGFDVADIFP SKKLLHHLSGKRAKLTNIHNKLDNLINNIIAEHPGNRTSSSQETLLDVLLRLKESAEFPLTADNVK AVILDMFGAGTDTSSATIEWAISELIRCPRAMEKVQTELRQALNGKERIQEEDLQELNYLKLVIKE TLRLHPPLPLVMPRECREPCVLGGYDIPSKTKLIVNVFAINRDPEYWKDAETFMPERFENSPITVM GSEYEYLPFGAGRRMCPGAALGLANVELPLAHILYFNWKLPNGKTFEDLDMTESFGATVQRKTELL LVPTDFQTLTAST >t20HaGAO [Helianthus annuus] (SEQ ID NO: 60) LLTRPTSSKNRLPEPWRLPIIGHMHHLIGTMPHRGVMDLARKYGSLMHLQLGEVSAIVVSSPKWAK EILTTYDIPFANRPETLTGEIIAYHNTDIVLAPYGEYWRQLRKLCTLELLSVKKVKSFQSLREEEC WNLVQEIKASGSGTPFNLSEGIFKVIATVLSRAAFGKGIKDQKQFTEIVKEILRETGGFDVADIFP SKKFLHHLSGKRGRLTSIHNKLDSLINNLVAEHTVSKSSKVNETLLDVLLRLKNSEEFPLTADNVK AIILDMFGAGTDTSSATVEWAISELIRCPRAMEKVQAELRQALNGKERIKEEEIQDLPYLNLVIRE TLRLHPPLPLVMPRECRQAMNLAGYDVANKTKLIVNVFAINRDPEYWKDAESFNPERFENSNTTIM GADYEYLPFGAGRRMCPGSALGLANVQLPLANILYYFKWKLPNGASHDQLDMTESFGATVQRKTEL MLVPSF
Sequence CWU 1
1
641513PRTZingiber zerumbet 1Met Glu Ala Ile Ser Leu Phe Ser Pro Phe
Phe Phe Ile Thr Leu Phe1 5 10 15Leu Gly Phe Phe Ile Thr Leu Leu Ile
Lys Arg Ser Ser Arg Ser Ser 20 25 30Val His Lys Gln Gln Val Leu Leu
Ala Ser Leu Pro Pro Ser Pro Pro 35 40 45Arg Leu Pro Leu Ile Gly Asn
Ile His Gln Leu Val Gly Gly Asn Pro 50 55 60His Arg Ile Leu Leu Gln
Leu Ala Arg Thr His Gly Pro Leu Ile Cys65 70 75 80Leu Arg Leu Gly
Gln Val Asp Gln Val Val Ala Ser Ser Val Glu Ala 85 90 95Val Glu Glu
Ile Ile Lys Arg His Asp Leu Lys Phe Ala Asp Arg Pro 100 105 110Arg
Asp Leu Thr Phe Ser Arg Ile Phe Phe Tyr Asp Gly Asn Ala Val 115 120
125Val Met Thr Pro Tyr Gly Gly Glu Trp Lys Gln Met Arg Lys Ile Tyr
130 135 140Ala Met Glu Leu Leu Asn Ser Arg Arg Val Lys Ser Phe Ala
Ala Ile145 150 155 160Arg Glu Asp Val Ala Arg Lys Leu Thr Gly Glu
Ile Ala His Lys Ala 165 170 175Phe Ala Gln Thr Pro Val Ile Asn Leu
Ser Glu Met Val Met Ser Met 180 185 190Ile Asn Ala Ile Val Ile Arg
Val Ala Phe Gly Asp Lys Cys Lys Gln 195 200 205Gln Ala Tyr Phe Leu
His Leu Val Lys Glu Ala Met Ser Tyr Val Ser 210 215 220Ser Phe Ser
Val Ala Asp Met Tyr Pro Ser Leu Lys Phe Leu Asp Thr225 230 235
240Leu Thr Gly Leu Lys Ser Lys Leu Glu Gly Val His Gly Lys Leu Asp
245 250 255Lys Val Phe Asp Glu Ile Ile Ala Gln Arg Gln Ala Ala Leu
Ala Ala 260 265 270Glu Gln Ala Glu Glu Asp Leu Ile Ile Asp Val Leu
Leu Lys Leu Lys 275 280 285Asp Glu Gly Asn Gln Glu Phe Pro Ile Thr
Tyr Thr Ser Val Lys Ala 290 295 300Ile Val Met Glu Ile Phe Leu Ala
Gly Thr Glu Thr Ser Ser Ser Val305 310 315 320Ile Asp Trp Val Met
Ser Glu Leu Ile Lys Asn Pro Lys Ala Met Glu 325 330 335Lys Val Gln
Lys Glu Met Arg Glu Ala Met Gln Gly Lys Thr Lys Leu 340 345 350Glu
Glu Ser Asp Ile Pro Lys Phe Ser Tyr Leu Asn Leu Val Ile Lys 355 360
365Glu Thr Leu Arg Leu His Pro Pro Gly Pro Leu Leu Phe Pro Arg Glu
370 375 380Cys Arg Glu Thr Cys Glu Val Met Gly Tyr Arg Val Pro Ala
Gly Ala385 390 395 400Arg Leu Leu Ile Asn Ala Phe Ala Leu Ser Arg
Asp Glu Lys Tyr Trp 405 410 415Gly Ser Asp Ala Glu Ser Phe Lys Pro
Glu Arg Phe Glu Gly Ile Ser 420 425 430Val Asp Phe Lys Gly Ser Asn
Phe Glu Phe Met Pro Phe Gly Ala Gly 435 440 445Arg Arg Ile Cys Pro
Gly Met Thr Phe Gly Ile Ser Ser Val Glu Val 450 455 460Ala Leu Ala
His Leu Leu Phe His Phe Asp Trp Gln Leu Pro Gln Gly465 470 475
480Met Lys Ile Glu Asp Leu Asp Met Met Glu Val Ser Gly Met Ser Ala
485 490 495Thr Arg Arg Ser Pro Leu Leu Val Leu Ala Lys Leu Ile Ile
Pro Leu 500 505 510Pro2496PRTBarnadesia spinosa 2Met Glu Leu Thr
Leu Thr Thr Ser Leu Gly Leu Ala Val Phe Val Phe1 5 10 15Ile Leu Phe
Lys Leu Leu Thr Gly Ser Lys Ser Thr Lys Asn Ser Leu 20 25 30Pro Glu
Ala Trp Arg Leu Pro Ile Ile Gly His Met His His Leu Val 35 40 45Gly
Thr Leu Pro His Arg Gly Val Thr Asp Met Ala Arg Lys Tyr Gly 50 55
60Ser Leu Met His Leu Gln Leu Gly Glu Val Ser Thr Ile Val Val Ser65
70 75 80Ser Pro Arg Trp Ala Lys Glu Val Leu Thr Thr Tyr Asp Ile Thr
Phe 85 90 95Ala Asn Arg Pro Glu Thr Leu Thr Gly Glu Ile Val Ala Tyr
His Asn 100 105 110Thr Asp Ile Val Leu Ser Pro Tyr Gly Glu Tyr Trp
Arg Gln Leu Arg 115 120 125Lys Leu Cys Thr Leu Glu Leu Leu Ser Ala
Lys Lys Val Lys Ser Phe 130 135 140Gln Ser Leu Arg Glu Glu Glu Cys
Trp Asn Leu Val Lys Glu Val Arg145 150 155 160Ser Ser Gly Ser Gly
Ser Pro Val Asp Leu Ser Glu Ser Ile Phe Lys 165 170 175Leu Ile Ala
Thr Ile Leu Ser Arg Ala Ala Phe Gly Lys Gly Ile Lys 180 185 190Asp
Gln Arg Glu Phe Thr Glu Ile Val Lys Glu Ile Leu Arg Leu Thr 195 200
205Gly Gly Phe Asp Val Ala Asp Ile Phe Pro Ser Lys Lys Ile Leu His
210 215 220His Leu Ser Gly Lys Arg Ala Lys Leu Thr Asn Ile His Asn
Lys Leu225 230 235 240Asp Ser Leu Ile Asn Asn Ile Val Ser Glu His
Pro Gly Ser Arg Thr 245 250 255Ser Ser Ser Gln Glu Ser Leu Leu Asp
Val Leu Leu Arg Leu Lys Asp 260 265 270Ser Ala Glu Leu Pro Leu Thr
Ser Asp Asn Val Lys Ala Val Ile Leu 275 280 285Asp Met Phe Gly Ala
Gly Thr Asp Thr Ser Ser Ala Thr Ile Glu Trp 290 295 300Ala Ile Ser
Glu Leu Ile Arg Cys Pro Arg Ala Met Glu Lys Val Gln305 310 315
320Thr Glu Leu Arg Gln Ala Leu Asn Gly Lys Glu Arg Ile Gln Glu Glu
325 330 335Asp Ile Gln Glu Leu Ser Tyr Leu Lys Leu Val Ile Lys Glu
Thr Leu 340 345 350Arg Leu His Pro Pro Leu Pro Leu Val Met Pro Arg
Glu Cys Arg Glu 355 360 365Pro Cys Val Leu Ala Gly Tyr Glu Ile Pro
Thr Lys Thr Lys Leu Ile 370 375 380Val Asn Val Phe Ala Ile Asn Arg
Asp Pro Glu Tyr Trp Lys Asp Ala385 390 395 400Glu Thr Phe Met Pro
Glu Arg Phe Glu Asn Ser Pro Ile Asn Ile Met 405 410 415Gly Ser Glu
Tyr Glu Tyr Leu Pro Phe Gly Ala Gly Arg Arg Met Cys 420 425 430Pro
Gly Ala Ala Leu Gly Leu Ala Asn Val Glu Leu Pro Leu Ala His 435 440
445Ile Leu Tyr Tyr Phe Asn Trp Lys Leu Pro Asn Gly Ala Arg Leu Asp
450 455 460Glu Leu Asp Met Ser Glu Cys Phe Gly Ala Thr Val Gln Arg
Lys Ser465 470 475 480Glu Leu Leu Leu Val Pro Thr Ala Tyr Lys Thr
Ala Asn Asn Ser Ala 485 490 4953502PRTHyoscyamus muticus 3Met Gln
Phe Phe Ser Leu Val Ser Ile Phe Leu Phe Leu Ser Phe Leu1 5 10 15Phe
Leu Leu Arg Lys Trp Lys Asn Ser Asn Ser Gln Ser Lys Lys Leu 20 25
30Pro Pro Gly Pro Trp Lys Leu Pro Leu Leu Gly Ser Met Leu His Met
35 40 45Val Gly Gly Leu Pro His His Val Leu Arg Asp Leu Ala Lys Lys
Tyr 50 55 60Gly Pro Leu Met His Leu Gln Leu Gly Glu Val Ser Ala Val
Val Val65 70 75 80Thr Ser Pro Asp Met Ala Lys Glu Val Leu Lys Thr
His Asp Ile Ala 85 90 95Phe Ala Ser Arg Pro Lys Leu Leu Ala Pro Glu
Ile Val Cys Tyr Asn 100 105 110Arg Ser Asp Ile Ala Phe Cys Pro Tyr
Gly Asp Tyr Trp Arg Gln Met 115 120 125Arg Lys Ile Cys Val Leu Glu
Val Leu Ser Ala Lys Asn Val Arg Ser 130 135 140Phe Ser Ser Ile Arg
Arg Asp Glu Val Leu Arg Leu Val Asn Phe Val145 150 155 160Arg Ser
Ser Thr Ser Glu Pro Val Asn Phe Thr Glu Arg Leu Phe Leu 165 170
175Phe Thr Ser Ser Met Thr Cys Arg Ser Ala Phe Gly Lys Val Phe Lys
180 185 190Glu Gln Glu Thr Phe Ile Gln Leu Ile Lys Glu Val Ile Gly
Leu Ala 195 200 205Gly Gly Phe Asp Val Ala Asp Ile Phe Pro Ser Leu
Lys Phe Leu His 210 215 220Val Leu Thr Gly Met Glu Gly Lys Ile Met
Lys Ala His His Lys Val225 230 235 240Asp Ala Ile Val Glu Asp Val
Ile Asn Glu His Lys Lys Asn Leu Ala 245 250 255Met Gly Lys Thr Asn
Gly Ala Leu Gly Gly Glu Asp Leu Ile Asp Val 260 265 270Leu Leu Arg
Leu Met Asn Asp Gly Gly Leu Gln Phe Pro Ile Thr Asn 275 280 285Asp
Asn Ile Lys Ala Ile Ile Phe Asp Met Phe Ala Ala Gly Thr Glu 290 295
300Thr Ser Ser Ser Thr Leu Val Trp Ala Met Val Gln Met Met Arg
Asn305 310 315 320Pro Thr Ile Leu Ala Lys Ala Gln Ala Glu Val Arg
Glu Ala Phe Lys 325 330 335Gly Lys Glu Thr Phe Asp Glu Asn Asp Val
Glu Glu Leu Lys Tyr Leu 340 345 350Lys Leu Val Ile Lys Glu Thr Leu
Arg Leu His Pro Pro Val Pro Leu 355 360 365Leu Val Pro Arg Glu Cys
Arg Glu Glu Thr Glu Ile Asn Gly Tyr Thr 370 375 380Ile Pro Val Lys
Thr Lys Val Met Val Asn Val Trp Ala Leu Gly Arg385 390 395 400Asp
Pro Lys Tyr Trp Asp Asp Ala Asp Asn Phe Lys Pro Glu Arg Phe 405 410
415Glu Gln Cys Ser Val Asp Phe Ile Gly Asn Asn Phe Glu Tyr Leu Pro
420 425 430Phe Gly Gly Gly Arg Arg Ile Cys Pro Gly Ile Ser Phe Gly
Leu Ala 435 440 445Asn Val Tyr Leu Pro Leu Ala Gln Leu Leu Tyr His
Phe Asp Trp Lys 450 455 460Leu Pro Thr Gly Met Glu Pro Lys Asp Leu
Asp Leu Thr Glu Leu Val465 470 475 480Gly Val Thr Ala Ala Arg Lys
Ser Asp Leu Met Leu Val Ala Thr Pro 485 490 495Tyr Gln Pro Ser Arg
Glu 5004488PRTLactuca sativa 4Met Glu Leu Ser Ile Thr Thr Ser Ile
Ala Leu Ala Thr Ile Val Phe1 5 10 15Phe Leu Tyr Lys Leu Ala Thr Arg
Pro Lys Ser Thr Lys Lys Gln Leu 20 25 30Pro Glu Ala Ser Arg Leu Pro
Ile Ile Gly His Met His His Leu Ile 35 40 45Gly Thr Met Pro His Arg
Gly Val Met Asp Leu Ala Arg Lys His Gly 50 55 60Ser Leu Met His Leu
Gln Leu Gly Glu Val Ser Thr Ile Val Val Ser65 70 75 80Ser Pro Lys
Trp Ala Lys Glu Ile Leu Thr Thr Tyr Asp Ile Thr Phe 85 90 95Ala Asn
Arg Pro Glu Thr Leu Thr Gly Glu Ile Ile Ala Tyr His Asn 100 105
110Thr Asp Ile Val Leu Ala Pro Tyr Gly Glu Tyr Trp Arg Gln Leu Arg
115 120 125Lys Leu Cys Thr Leu Glu Leu Leu Ser Val Lys Lys Val Lys
Ser Phe 130 135 140Gln Ser Ile Arg Glu Glu Glu Cys Trp Asn Leu Val
Lys Glu Val Lys145 150 155 160Glu Ser Gly Ser Gly Lys Pro Ile Asn
Leu Ser Glu Ser Ile Phe Thr 165 170 175Met Ile Ala Thr Ile Leu Ser
Arg Ala Ala Phe Gly Lys Gly Ile Lys 180 185 190Asp Gln Arg Glu Phe
Thr Glu Ile Val Lys Glu Ile Leu Arg Gln Thr 195 200 205Gly Gly Phe
Asp Val Ala Asp Ile Phe Pro Ser Lys Lys Phe Leu His 210 215 220His
Leu Ser Gly Lys Arg Ala Arg Leu Thr Ser Ile His Lys Lys Leu225 230
235 240Asp Asn Leu Ile Asn Asn Ile Val Ala Glu His His Val Ser Thr
Ser 245 250 255Ser Lys Ala Asn Glu Thr Leu Leu Asp Val Leu Leu Arg
Leu Lys Asp 260 265 270Ser Ala Glu Phe Pro Leu Thr Ala Asp Asn Val
Lys Ala Ile Ile Leu 275 280 285Asp Met Phe Gly Ala Gly Thr Asp Thr
Ser Ser Ala Thr Val Glu Trp 290 295 300Ala Ile Ser Glu Leu Ile Arg
Cys Pro Arg Ala Met Glu Lys Val Gln305 310 315 320Ala Glu Leu Arg
Gln Ala Leu Asn Gly Lys Glu Lys Ile Gln Glu Glu 325 330 335Asp Ile
Gln Asp Leu Ala Tyr Leu Asn Leu Val Ile Arg Glu Thr Leu 340 345
350Arg Leu His Pro Pro Leu Pro Leu Val Met Pro Arg Glu Cys Arg Glu
355 360 365Pro Val Asn Leu Ala Gly Tyr Glu Ile Ala Asn Lys Thr Lys
Leu Ile 370 375 380Val Asn Val Phe Ala Ile Asn Arg Asp Pro Glu Tyr
Trp Lys Asp Ala385 390 395 400Glu Ala Phe Ile Pro Glu Arg Phe Glu
Asn Asn Pro Asn Asn Ile Met 405 410 415Gly Ala Asp Tyr Glu Tyr Leu
Pro Phe Gly Ala Gly Arg Arg Met Cys 420 425 430Pro Gly Ala Ala Leu
Gly Leu Ala Asn Val Gln Leu Pro Leu Ala Asn 435 440 445Ile Leu Tyr
His Phe Asn Trp Lys Leu Pro Asn Gly Ala Ser His Asp 450 455 460Gln
Leu Asp Met Thr Glu Ser Phe Gly Ala Thr Val Gln Arg Lys Thr465 470
475 480Glu Leu Leu Leu Val Pro Ser Phe 4855504PRTNicotiani tabacum
5Met Gln Phe Phe Ser Leu Val Ser Ile Phe Leu Phe Leu Ser Phe Leu1 5
10 15Phe Leu Leu Arg Lys Trp Lys Asn Ser Asn Ser Gln Ser Lys Lys
Leu 20 25 30Pro Pro Gly Pro Trp Lys Ile Pro Ile Leu Gly Ser Met Leu
His Met 35 40 45Ile Gly Gly Glu Pro His His Val Leu Arg Asp Leu Ala
Lys Lys Tyr 50 55 60Gly Pro Leu Met His Leu Gln Leu Gly Glu Ile Ser
Ala Val Val Val65 70 75 80Thr Ser Arg Asp Met Ala Lys Glu Val Leu
Lys Thr His Asp Val Val 85 90 95Phe Ala Ser Arg Pro Lys Ile Val Ala
Met Asp Ile Ile Cys Tyr Asn 100 105 110Gln Ser Asp Ile Ala Phe Ser
Pro Tyr Gly Asp His Trp Arg Gln Met 115 120 125Arg Lys Ile Cys Val
Met Glu Leu Leu Asn Ala Lys Asn Val Arg Ser 130 135 140Phe Ser Ser
Ile Arg Arg Asp Glu Val Val Arg Leu Ile Asp Ser Ile145 150 155
160Arg Ser Asp Ser Ser Ser Gly Glu Leu Val Asn Phe Thr Gln Arg Ile
165 170 175Ile Trp Phe Ala Ser Ser Met Thr Cys Arg Ser Ala Phe Gly
Gln Val 180 185 190Leu Lys Gly Gln Asp Ile Phe Ala Lys Lys Ile Arg
Glu Val Ile Gly 195 200 205Leu Ala Glu Gly Phe Asp Val Val Asp Ile
Phe Pro Thr Tyr Lys Phe 210 215 220Leu His Val Leu Ser Gly Met Lys
Arg Lys Leu Leu Asn Ala His Leu225 230 235 240Lys Val Asp Ala Ile
Val Glu Asp Val Ile Asn Glu His Lys Lys Asn 245 250 255Leu Ala Ala
Gly Lys Ser Asn Gly Ala Leu Gly Gly Glu Asp Leu Ile 260 265 270Asp
Val Leu Leu Arg Leu Met Asn Asp Thr Ser Leu Gln Phe Pro Ile 275 280
285Thr Asn Asp Asn Ile Lys Ala Val Ile Val Asp Met Phe Ala Ala Gly
290 295 300Thr Glu Thr Ser Ser Thr Thr Thr Val Trp Ala Met Ala Glu
Met Met305 310 315 320Lys Asn Pro Ser Val Phe Thr Lys Ala Gln Ala
Glu Val Arg Glu Ala 325 330 335Phe Arg Asp Lys Val Ser Phe Asp Glu
Asn Asp Val Glu Glu Leu Lys 340 345 350Tyr Leu Lys Leu Val Ile Lys
Glu Thr Leu Arg Leu His Pro Pro Ser 355 360 365Pro Leu Leu Val Pro
Arg Glu Cys Arg Glu Asp Thr Asp Ile Asn Gly 370 375 380Tyr Thr Ile
Pro Ala Lys Thr Lys Val Met Val Asn Val Trp Ala Leu385 390 395
400Gly Arg Asp Pro Lys Tyr Trp Asp Asp Ala Glu Ser Phe Lys Pro Glu
405 410 415Arg Phe Glu Gln Cys Ser Val Asp Phe Phe Gly Asn Asn Phe
Glu Phe 420 425 430Leu Pro Phe Gly Gly Gly Arg Arg Ile Cys Pro Gly
Met Ser Phe Gly 435 440 445Leu Ala Asn Leu Tyr Leu Pro Leu Ala Gln
Leu Leu Tyr His Phe Asp 450
455 460Trp Lys Leu Pro Thr Gly Ile Met Pro Arg Asp Leu Asp Leu Thr
Glu465 470 475 480Leu Ser Gly Ile Thr Ile Ala Arg Lys Gly Gly Leu
Tyr Leu Asn Ala 485 490 495Thr Pro Tyr Gln Pro Ser Arg Glu
5006522PRTCitrus x paradisi 6Met Glu Leu Pro Leu Lys Ser Ile Ala
Leu Thr Ile Val Ile Val Thr1 5 10 15Val Leu Thr Trp Ala Trp Arg Val
Leu Asn Trp Val Trp Leu Arg Pro 20 25 30Lys Lys Leu Glu Lys Phe Leu
Arg Gln Gln Gly Leu Lys Gly Asn Ser 35 40 45Tyr Arg Leu Leu Phe Gly
Asp Leu Lys Glu Asn Ser Ile Glu Leu Lys 50 55 60Glu Ala Lys Ala Arg
Pro Leu Ser Leu Asp Asp Asp Ile Ala Ile Arg65 70 75 80Val Asn Pro
Phe Leu His Lys Leu Val Asn Asp Tyr Gly Lys Asn Ser 85 90 95Phe Met
Trp Phe Gly Pro Thr Pro Arg Val Asn Ile Met Asn Pro Asp 100 105
110Gln Ile Lys Ala Ile Phe Thr Lys Ile Asn Asp Phe Gln Lys Val Asn
115 120 125Ser Ile Pro Leu Ala Arg Leu Leu Ile Val Gly Leu Ala Thr
Leu Glu 130 135 140Gly Glu Lys Trp Ala Lys His Arg Lys Leu Ile Asn
Pro Ala Phe His145 150 155 160Gln Glu Lys Leu Lys Leu Met Leu Pro
Ala Phe Tyr Leu Ser Cys Ile 165 170 175Glu Ile Ile Thr Lys Trp Glu
Lys Gln Met Ser Val Glu Gly Ser Ser 180 185 190Glu Leu Asp Val Trp
Pro Tyr Leu Ala Asn Leu Thr Ser Asp Val Ile 195 200 205Ser Arg Thr
Ala Phe Gly Ser Ser Tyr Glu Glu Gly Arg Arg Ile Phe 210 215 220Gln
Leu Gln Ala Glu Leu Ala Glu Leu Thr Met Gln Val Phe Arg Ser225 230
235 240Val His Ile Pro Gly Trp Arg Phe Leu Pro Thr Lys Arg Asn Arg
Arg 245 250 255Met Lys Glu Ile Asp Lys Glu Ile Arg Ala Ser Leu Met
Gly Ile Ile 260 265 270Lys Asn Arg Glu Lys Ala Met Arg Ala Gly Glu
Ala Ala Asn Asn Asp 275 280 285Leu Leu Gly Ile Leu Met Glu Thr Ser
Phe Arg Glu Ile Glu Glu His 290 295 300Gly Asn Asn Lys Asn Val Gly
Phe Ser Met Asn Asp Val Ile Glu Glu305 310 315 320Cys Lys Leu Phe
Tyr Phe Ala Gly Gln Glu Thr Thr Ser Val Leu Leu 325 330 335Asn Trp
Thr Met Val Leu Leu Ser Lys His Gln Asp Trp Gln Glu Arg 340 345
350Ala Arg Gln Glu Val Leu Gln Val Phe Gly Asn Asn Lys Pro Asp Tyr
355 360 365Asp Gly Leu Asn His Leu Lys Ile Val Gln Met Ile Leu Tyr
Glu Val 370 375 380Leu Arg Leu Tyr Pro Pro Val Thr Val Leu Ser Arg
Ala Val Phe Lys385 390 395 400Glu Thr Lys Leu Gly Asn Leu Thr Leu
Pro Ala Gly Val Gln Ile Gly 405 410 415Leu Pro Met Ile Leu Val His
Gln Asp Pro Glu Leu Trp Gly Asp Asp 420 425 430Ala Val Glu Phe Lys
Pro Glu Arg Phe Ala Glu Gly Ile Ser Lys Ala 435 440 445Ala Lys Asn
Gln Val Ser Tyr Phe Pro Phe Ala Leu Gly Pro Arg Ile 450 455 460Cys
Val Gly Gln Asn Phe Ala Leu Val Glu Ala Lys Met Ala Thr Ala465 470
475 480Met Ile Leu Gln Asn Tyr Ser Phe Glu Leu Ser Pro Ser Tyr Val
His 485 490 495Ala Pro Thr Ala Val Pro Thr Leu His Pro Glu Leu Gly
Thr Gln Leu 500 505 510Ile Leu Arg Lys Leu Trp Cys Lys Asn Asn 515
5207495PRTArtemesia annua 7Met Lys Ser Ile Leu Lys Ala Met Ala Leu
Ser Leu Thr Thr Ser Ile1 5 10 15Ala Leu Ala Thr Ile Leu Leu Phe Val
Tyr Lys Phe Ala Thr Arg Ser 20 25 30Lys Ser Thr Lys Lys Ser Leu Pro
Glu Pro Trp Arg Leu Pro Ile Ile 35 40 45Gly His Met His His Leu Ile
Gly Thr Thr Pro His Arg Gly Val Arg 50 55 60Asp Leu Ala Arg Lys Tyr
Gly Ser Leu Met His Leu Gln Leu Gly Glu65 70 75 80Val Pro Thr Ile
Val Val Ser Ser Pro Lys Trp Ala Lys Glu Ile Leu 85 90 95Thr Thr Tyr
Asp Ile Thr Phe Ala Asn Arg Pro Glu Thr Leu Thr Gly 100 105 110Glu
Ile Val Leu Tyr His Asn Thr Asp Val Val Leu Ala Pro Tyr Gly 115 120
125Glu Tyr Trp Arg Gln Leu Arg Lys Ile Cys Thr Leu Glu Leu Leu Ser
130 135 140Val Lys Lys Val Lys Ser Phe Gln Ser Leu Arg Glu Glu Glu
Cys Trp145 150 155 160Asn Leu Val Gln Glu Ile Lys Ala Ser Gly Ser
Gly Arg Pro Val Asn 165 170 175Leu Ser Glu Asn Val Phe Lys Leu Ile
Ala Thr Ile Leu Ser Arg Ala 180 185 190Ala Phe Gly Lys Gly Ile Lys
Asp Gln Lys Glu Leu Thr Glu Ile Val 195 200 205Lys Glu Ile Leu Arg
Gln Thr Gly Gly Phe Asp Val Ala Asp Ile Phe 210 215 220Pro Ser Lys
Lys Phe Leu His His Leu Ser Gly Lys Arg Ala Arg Leu225 230 235
240Thr Ser Leu Arg Lys Lys Ile Asp Asn Leu Ile Asp Asn Leu Val Ala
245 250 255Glu His Thr Val Asn Thr Ser Ser Lys Thr Asn Glu Thr Leu
Leu Asp 260 265 270Val Leu Leu Arg Leu Lys Asp Ser Ala Glu Phe Pro
Leu Thr Ser Asp 275 280 285Asn Ile Lys Ala Ile Ile Leu Asp Met Phe
Gly Ala Gly Thr Asp Thr 290 295 300Ser Ser Ser Thr Ile Glu Trp Ala
Ile Ser Glu Leu Ile Lys Cys Pro305 310 315 320Lys Ala Met Glu Lys
Val Gln Ala Glu Leu Arg Lys Ala Leu Asn Gly 325 330 335Lys Glu Lys
Ile His Glu Glu Asp Ile Gln Glu Leu Ser Tyr Leu Asn 340 345 350Met
Val Ile Lys Glu Thr Leu Arg Leu His Pro Pro Leu Pro Leu Val 355 360
365Leu Pro Arg Glu Cys Arg Gln Pro Val Asn Leu Ala Gly Tyr Asn Ile
370 375 380Pro Asn Lys Thr Lys Leu Ile Val Asn Val Phe Ala Ile Asn
Arg Asp385 390 395 400Pro Glu Tyr Trp Lys Asp Ala Glu Ala Phe Ile
Pro Glu Arg Phe Glu 405 410 415Asn Ser Ser Ala Thr Val Met Gly Ala
Glu Tyr Glu Tyr Leu Pro Phe 420 425 430Gly Ala Gly Arg Arg Met Cys
Pro Gly Ala Ala Leu Gly Leu Ala Asn 435 440 445Val Gln Leu Pro Leu
Ala Asn Ile Leu Tyr His Phe Asn Trp Lys Leu 450 455 460Pro Asn Gly
Val Ser Tyr Asp Gln Ile Asp Met Thr Glu Ser Ser Gly465 470 475
480Ala Thr Met Gln Arg Lys Thr Glu Leu Leu Leu Val Pro Ser Phe 485
490 4958509PRTArabidopsis thaliana 8Met Ala Phe Phe Ser Met Ile Ser
Ile Leu Leu Gly Phe Val Ile Ser1 5 10 15Ser Phe Ile Phe Ile Phe Phe
Phe Lys Lys Leu Leu Ser Phe Ser Arg 20 25 30Lys Asn Met Ser Glu Val
Ser Thr Leu Pro Ser Val Pro Val Val Pro 35 40 45Gly Phe Pro Val Ile
Gly Asn Leu Leu Gln Leu Lys Glu Lys Lys Pro 50 55 60His Lys Thr Phe
Thr Arg Trp Ser Glu Ile Tyr Gly Pro Ile Tyr Ser65 70 75 80Ile Lys
Met Gly Ser Ser Ser Leu Ile Val Leu Asn Ser Thr Glu Thr 85 90 95Ala
Lys Glu Ala Met Val Thr Arg Phe Ser Ser Ile Ser Thr Arg Lys 100 105
110Leu Ser Asn Ala Leu Thr Val Leu Thr Cys Asp Lys Ser Met Val Ala
115 120 125Thr Ser Asp Tyr Asp Asp Phe His Lys Leu Val Lys Arg Cys
Leu Leu 130 135 140Asn Gly Leu Leu Gly Ala Asn Ala Gln Lys Arg Lys
Arg His Tyr Arg145 150 155 160Asp Ala Leu Ile Glu Asn Val Ser Ser
Lys Leu His Ala His Ala Arg 165 170 175Asp His Pro Gln Glu Pro Val
Asn Phe Arg Ala Ile Phe Glu His Glu 180 185 190Leu Phe Gly Val Ala
Leu Lys Gln Ala Phe Gly Lys Asp Val Glu Ser 195 200 205Ile Tyr Val
Lys Glu Leu Gly Val Thr Leu Ser Lys Asp Glu Ile Phe 210 215 220Lys
Val Leu Val His Asp Met Met Glu Gly Ala Ile Asp Val Asp Trp225 230
235 240Arg Asp Phe Phe Pro Tyr Leu Lys Trp Ile Pro Asn Lys Ser Phe
Glu 245 250 255Ala Arg Ile Gln Gln Lys His Lys Arg Arg Leu Ala Val
Met Asn Ala 260 265 270Leu Ile Gln Asp Arg Leu Lys Gln Asn Gly Ser
Glu Ser Asp Asp Asp 275 280 285Cys Tyr Leu Asn Phe Leu Met Ser Glu
Ala Lys Thr Leu Thr Lys Glu 290 295 300Gln Ile Ala Ile Leu Val Trp
Glu Thr Ile Ile Glu Thr Ala Asp Thr305 310 315 320Thr Leu Val Thr
Thr Glu Trp Ala Ile Tyr Glu Leu Ala Lys His Pro 325 330 335Ser Val
Gln Asp Arg Leu Cys Lys Glu Ile Gln Asn Val Cys Gly Gly 340 345
350Glu Lys Phe Lys Glu Glu Gln Leu Ser Gln Val Pro Tyr Leu Asn Gly
355 360 365Val Phe His Glu Thr Leu Arg Lys Tyr Ser Pro Ala Pro Leu
Val Pro 370 375 380Ile Arg Tyr Ala His Glu Asp Thr Gln Ile Gly Gly
Tyr His Val Pro385 390 395 400Ala Gly Ser Glu Ile Ala Ile Asn Ile
Tyr Gly Cys Asn Met Asp Lys 405 410 415Lys Arg Trp Glu Arg Pro Glu
Asp Trp Trp Pro Glu Arg Phe Leu Asp 420 425 430Asp Gly Lys Tyr Glu
Thr Ser Asp Leu His Lys Thr Met Ala Phe Gly 435 440 445Ala Gly Lys
Arg Val Cys Ala Gly Ala Leu Gln Ala Ser Leu Met Ala 450 455 460Gly
Ile Ala Ile Gly Arg Leu Val Gln Glu Phe Glu Trp Lys Leu Arg465 470
475 480Asp Gly Glu Glu Glu Asn Val Asp Thr Tyr Gly Leu Thr Ser Gln
Lys 485 490 495Leu Tyr Pro Leu Met Ala Ile Ile Asn Pro Arg Arg Ser
500 5059513PRTStevia rebaudiana 9Met Asp Ala Val Thr Gly Leu Leu
Thr Val Pro Ala Thr Ala Ile Thr1 5 10 15Ile Gly Gly Thr Ala Val Ala
Leu Ala Val Ala Leu Ile Phe Trp Tyr 20 25 30Leu Lys Ser Tyr Thr Ser
Ala Arg Arg Ser Gln Ser Asn His Leu Pro 35 40 45Arg Val Pro Glu Val
Pro Gly Val Pro Leu Leu Gly Asn Leu Leu Gln 50 55 60Leu Lys Glu Lys
Lys Pro Tyr Met Thr Phe Thr Arg Trp Ala Ala Thr65 70 75 80Tyr Gly
Pro Ile Tyr Ser Ile Lys Thr Gly Ala Thr Ser Met Val Val 85 90 95Val
Ser Ser Asn Glu Ile Ala Lys Glu Ala Leu Val Thr Arg Phe Gln 100 105
110Ser Ile Ser Thr Arg Asn Leu Ser Lys Ala Leu Lys Val Leu Thr Ala
115 120 125Asp Lys Thr Met Val Ala Met Ser Asp Tyr Asp Asp Tyr His
Lys Thr 130 135 140Val Lys Arg His Ile Leu Thr Ala Val Leu Gly Pro
Asn Ala Gln Lys145 150 155 160Lys His Arg Ile His Arg Asp Ile Met
Met Asp Asn Ile Ser Thr Gln 165 170 175Leu His Glu Phe Val Lys Asn
Asn Pro Glu Gln Glu Glu Val Asp Leu 180 185 190Arg Lys Ile Phe Gln
Ser Glu Leu Phe Gly Leu Ala Met Arg Gln Ala 195 200 205Leu Gly Lys
Asp Val Glu Ser Leu Tyr Val Glu Asp Leu Lys Ile Thr 210 215 220Met
Asn Arg Asp Glu Ile Phe Gln Val Leu Val Val Asp Pro Met Met225 230
235 240Gly Ala Ile Asp Val Asp Trp Arg Asp Phe Phe Pro Tyr Leu Lys
Trp 245 250 255Val Pro Asn Lys Lys Phe Glu Asn Thr Ile Gln Gln Met
Tyr Ile Arg 260 265 270Arg Glu Ala Val Met Lys Ser Leu Ile Lys Glu
His Lys Lys Arg Ile 275 280 285Ala Ser Gly Glu Lys Leu Asn Ser Tyr
Ile Asp Tyr Leu Leu Ser Glu 290 295 300Ala Gln Thr Leu Thr Asp Gln
Gln Leu Leu Met Ser Leu Trp Glu Pro305 310 315 320Ile Ile Glu Ser
Ser Asp Thr Thr Met Val Thr Thr Glu Trp Ala Met 325 330 335Tyr Glu
Leu Ala Lys Asn Pro Lys Leu Gln Asp Arg Leu Tyr Arg Asp 340 345
350Ile Lys Ser Val Cys Gly Ser Glu Lys Ile Thr Glu Glu His Leu Ser
355 360 365Gln Leu Pro Tyr Ile Thr Ala Ile Phe His Glu Thr Leu Arg
Arg His 370 375 380Ser Pro Val Pro Ile Ile Pro Leu Arg His Val His
Glu Asp Thr Val385 390 395 400Leu Gly Gly Tyr His Val Pro Ala Gly
Thr Glu Leu Ala Val Asn Ile 405 410 415Tyr Gly Cys Asn Met Asp Lys
Asn Val Trp Glu Asn Pro Glu Glu Trp 420 425 430Asn Pro Glu Arg Phe
Met Lys Glu Asn Glu Thr Ile Asp Phe Gln Lys 435 440 445Thr Met Ala
Phe Gly Gly Gly Lys Arg Val Cys Ala Gly Ser Leu Gln 450 455 460Ala
Leu Leu Thr Ala Ser Ile Gly Ile Gly Arg Met Val Gln Glu Phe465 470
475 480Glu Trp Lys Leu Lys Asp Met Thr Gln Glu Glu Val Asn Thr Ile
Gly 485 490 495Leu Thr Thr Gln Met Leu Arg Pro Leu Arg Ala Ile Ile
Lys Pro Arg 500 505 510Ile10546PRTPhyscomitrella patens 10Met Ala
Lys His Leu Ala Thr Gln Leu Leu Gln Gln Trp Asn Glu Ala1 5 10 15Leu
Lys Thr Met Pro Pro Gly Phe Arg Thr Ala Gly Lys Ile Leu Val 20 25
30Trp Glu Glu Leu Ala Ser Asn Lys Val Leu Ile Thr Ile Ala Leu Ala
35 40 45Trp Val Leu Leu Phe Val Ala Arg Thr Cys Leu Arg Asn Lys Lys
Arg 50 55 60Leu Pro Pro Ala Ile Pro Gly Gly Leu Pro Val Leu Gly Asn
Leu Leu65 70 75 80Gln Leu Thr Glu Lys Lys Pro His Arg Thr Phe Thr
Ala Trp Ser Lys 85 90 95Glu His Gly Pro Ile Phe Thr Ile Lys Val Gly
Ser Val Pro Gln Ala 100 105 110Val Val Asn Asn Ser Glu Ile Ala Lys
Glu Val Leu Val Thr Lys Phe 115 120 125Ala Ser Ile Ser Lys Arg Gln
Met Pro Met Ala Leu Arg Val Leu Thr 130 135 140Arg Asp Lys Thr Met
Val Ala Met Ser Asp Tyr Gly Glu Glu His Arg145 150 155 160Met Leu
Lys Lys Leu Val Met Thr Asn Leu Leu Gly Pro Thr Thr Gln 165 170
175Asn Lys Asn Arg Ser Leu Arg Asp Asp Ala Leu Ile Gly Met Ile Glu
180 185 190Gly Val Leu Ala Glu Leu Lys Ala Ser Pro Thr Ser Pro Lys
Val Val 195 200 205Asn Val Arg Asp Tyr Val Gln Arg Ser Leu Phe Pro
Phe Ala Leu Gln 210 215 220Gln Val Phe Gly Tyr Ile Pro Asp Gln Val
Glu Val Leu Glu Leu Gly225 230 235 240Thr Cys Val Ser Thr Trp Asp
Met Phe Asp Ala Leu Val Val Ala Pro 245 250 255Leu Ser Ala Val Ile
Asn Val Asp Trp Arg Asp Phe Phe Pro Ala Leu 260 265 270Arg Trp Ile
Pro Asn Arg Ser Val Glu Asp Leu Val Arg Thr Val Asp 275 280 285Phe
Lys Arg Asn Ser Ile Met Lys Ala Leu Ile Arg Ala Gln Arg Met 290 295
300Arg Leu Ala Asn Leu Lys Glu Pro Pro Arg Cys Tyr Ala Asp Ile
Ala305 310 315 320Leu Thr Glu Ala Thr His Leu Thr Glu Lys Gln Leu
Glu Met Ser Leu 325 330 335Trp Glu Pro Ile Ile Glu Ser Ala Asp Thr
Thr Leu Val Thr Ser Glu 340 345 350Trp Ala Met Tyr Glu Ile Ala Lys
Asn Pro Asp Cys Gln Asp Arg Leu 355 360 365Tyr Arg Glu Ile Val Ser
Val Ala Gly Thr Glu Arg Met Val Thr Glu 370
375 380Asp Asp Leu Pro Asn Met Pro Tyr Leu Gly Ala Ile Ile Lys Glu
Thr385 390 395 400Leu Arg Lys Tyr Thr Pro Val Pro Leu Ile Pro Ser
Arg Phe Val Glu 405 410 415Glu Asp Ile Thr Leu Gly Gly Tyr Asp Ile
Pro Lys Gly Tyr Gln Ile 420 425 430Leu Val Asn Leu Phe Ala Ile Ala
Asn Asp Pro Ala Val Trp Ser Asn 435 440 445Pro Glu Lys Trp Asp Pro
Glu Arg Met Leu Ala Asn Lys Lys Val Asp 450 455 460Met Gly Phe Arg
Asp Phe Ser Leu Met Pro Phe Gly Ala Gly Lys Arg465 470 475 480Met
Cys Ala Gly Ile Thr Gln Ala Met Phe Ile Ile Pro Met Asn Val 485 490
495Ala Ala Leu Val Gln His Cys Glu Trp Arg Leu Ser Pro Gln Glu Ile
500 505 510Ser Asn Ile Asn Asn Lys Ile Glu Asp Val Val Tyr Leu Thr
Thr His 515 520 525Lys Leu Ser Pro Leu Ser Cys Glu Ala Thr Pro Arg
Ile Ser His Arg 530 535 540Leu Pro545111049PRTBacillus megaterium
11Met Thr Ile Lys Glu Met Pro Gln Pro Lys Thr Phe Gly Glu Leu Lys1
5 10 15Asn Leu Pro Leu Leu Asn Thr Asp Lys Pro Val Gln Ala Leu Met
Lys 20 25 30Ile Ala Asp Glu Leu Gly Glu Ile Phe Lys Phe Glu Ala Pro
Gly Arg 35 40 45Val Thr Arg Tyr Leu Ser Ser Gln Arg Leu Ile Lys Glu
Ala Cys Asp 50 55 60Glu Ser Arg Phe Asp Lys Asn Leu Ser Gln Ala Leu
Lys Phe Val Arg65 70 75 80Asp Phe Ala Gly Asp Gly Leu Ala Thr Ser
Trp Thr His Glu Lys Asn 85 90 95Trp Lys Lys Ala His Asn Ile Leu Leu
Pro Ser Phe Ser Gln Gln Ala 100 105 110Met Lys Gly Tyr His Ala Met
Met Val Asp Ile Ala Val Gln Leu Val 115 120 125Gln Lys Trp Glu Arg
Leu Asn Ala Asp Glu His Ile Glu Val Pro Glu 130 135 140Asp Met Thr
Arg Leu Thr Leu Asp Thr Ile Gly Leu Cys Gly Phe Asn145 150 155
160Tyr Arg Phe Asn Ser Phe Tyr Arg Asp Gln Pro His Pro Phe Ile Thr
165 170 175Ser Met Val Arg Ala Leu Asp Glu Ala Met Asn Lys Leu Gln
Arg Ala 180 185 190Asn Pro Asp Asp Pro Ala Tyr Asp Glu Asn Lys Arg
Gln Phe Gln Glu 195 200 205Asp Ile Lys Val Met Asn Asp Leu Val Asp
Lys Ile Ile Ala Asp Arg 210 215 220Lys Ala Ser Gly Glu Gln Ser Asp
Asp Leu Leu Thr His Met Leu Asn225 230 235 240Gly Lys Asp Pro Glu
Thr Gly Glu Pro Leu Asp Asp Glu Asn Ile Arg 245 250 255Tyr Gln Ile
Ile Thr Phe Leu Ile Ala Gly His Glu Thr Thr Ser Gly 260 265 270Leu
Leu Ser Phe Ala Leu Tyr Phe Leu Val Lys Asn Pro His Val Leu 275 280
285Gln Lys Ala Ala Glu Glu Ala Ala Arg Val Leu Val Asp Pro Val Pro
290 295 300Ser Tyr Lys Gln Val Lys Gln Leu Lys Tyr Val Gly Met Val
Leu Asn305 310 315 320Glu Ala Leu Arg Leu Trp Pro Thr Ile Pro Ala
Phe Ser Leu Tyr Ala 325 330 335Lys Glu Asp Thr Val Leu Gly Gly Glu
Tyr Pro Leu Glu Lys Gly Asp 340 345 350Glu Leu Met Val Leu Ile Pro
Gln Leu His Arg Asp Lys Thr Ile Trp 355 360 365Gly Asp Asp Val Glu
Glu Phe Arg Pro Glu Arg Phe Glu Asn Pro Ser 370 375 380Ala Ile Pro
Gln His Ala Phe Lys Pro Phe Gly Asn Gly Gln Arg Ala385 390 395
400Cys Ile Gly Gln Gln Phe Ala Leu His Glu Ala Thr Leu Val Leu Gly
405 410 415Met Met Leu Lys His Phe Asp Phe Glu Asp His Thr Asn Tyr
Glu Leu 420 425 430Asp Ile Lys Glu Thr Leu Thr Leu Lys Pro Glu Gly
Phe Val Val Lys 435 440 445Ala Lys Ser Lys Lys Ile Pro Leu Gly Gly
Ile Pro Ser Pro Ser Thr 450 455 460Glu Gln Ser Ala Lys Lys Val Arg
Lys Lys Ala Glu Asn Ala His Asn465 470 475 480Thr Pro Leu Leu Val
Leu Tyr Gly Ser Asn Met Gly Thr Ala Glu Gly 485 490 495Thr Ala Arg
Asp Leu Ala Asp Ile Ala Met Ser Lys Gly Phe Ala Pro 500 505 510Gln
Val Ala Thr Leu Asp Ser His Ala Gly Asn Leu Pro Arg Glu Gly 515 520
525Ala Val Leu Ile Val Thr Ala Ser Tyr Asn Gly His Pro Pro Asp Asn
530 535 540Ala Lys Gln Phe Val Asp Trp Leu Asp Gln Ala Ser Ala Asp
Glu Val545 550 555 560Lys Gly Val Arg Tyr Ser Val Phe Gly Cys Gly
Asp Lys Asn Trp Ala 565 570 575Thr Thr Tyr Gln Lys Val Pro Ala Phe
Ile Asp Glu Thr Leu Ala Ala 580 585 590Lys Gly Ala Glu Asn Ile Ala
Asp Arg Gly Glu Ala Asp Ala Ser Asp 595 600 605Asp Phe Glu Gly Thr
Tyr Glu Glu Trp Arg Glu His Met Trp Ser Asp 610 615 620Val Ala Ala
Tyr Phe Asn Leu Asp Ile Glu Asn Ser Glu Asp Asn Lys625 630 635
640Ser Thr Leu Ser Leu Gln Phe Val Asp Ser Ala Ala Asp Met Pro Leu
645 650 655Ala Lys Met His Gly Ala Phe Ser Thr Asn Val Val Ala Ser
Lys Glu 660 665 670Leu Gln Gln Pro Gly Ser Ala Arg Ser Thr Arg His
Leu Glu Ile Glu 675 680 685Leu Pro Lys Glu Ala Ser Tyr Gln Glu Gly
Asp His Leu Gly Val Ile 690 695 700Pro Arg Asn Tyr Glu Gly Ile Val
Asn Arg Val Thr Ala Arg Phe Gly705 710 715 720Leu Asp Ala Ser Gln
Gln Ile Arg Leu Glu Ala Glu Glu Glu Lys Leu 725 730 735Ala His Leu
Pro Leu Ala Lys Thr Val Ser Val Glu Glu Leu Leu Gln 740 745 750Tyr
Val Glu Leu Gln Asp Pro Val Thr Arg Thr Gln Leu Arg Ala Met 755 760
765Ala Ala Lys Thr Val Cys Pro Pro His Lys Val Glu Leu Glu Ala Leu
770 775 780Leu Glu Lys Gln Ala Tyr Lys Glu Gln Val Leu Ala Lys Arg
Leu Thr785 790 795 800Met Leu Glu Leu Leu Glu Lys Tyr Pro Ala Cys
Glu Met Lys Phe Ser 805 810 815Glu Phe Ile Ala Leu Leu Pro Ser Ile
Arg Pro Arg Tyr Tyr Ser Ile 820 825 830Ser Ser Ser Pro Arg Val Asp
Glu Lys Gln Ala Ser Ile Thr Val Ser 835 840 845Val Val Ser Gly Glu
Ala Trp Ser Gly Tyr Gly Glu Tyr Lys Gly Ile 850 855 860Ala Ser Asn
Tyr Leu Ala Glu Leu Gln Glu Gly Asp Thr Ile Thr Cys865 870 875
880Phe Ile Ser Thr Pro Gln Ser Glu Phe Thr Leu Pro Lys Asp Pro Glu
885 890 895Thr Pro Leu Ile Met Val Gly Pro Gly Thr Gly Val Ala Pro
Phe Arg 900 905 910Gly Phe Val Gln Ala Arg Lys Gln Leu Lys Glu Gln
Gly Gln Ser Leu 915 920 925Gly Glu Ala His Leu Tyr Phe Gly Cys Arg
Ser Pro His Glu Asp Tyr 930 935 940Leu Tyr Gln Glu Glu Leu Glu Asn
Ala Gln Ser Glu Gly Ile Ile Thr945 950 955 960Leu His Thr Ala Phe
Ser Arg Met Pro Asn Gln Pro Lys Thr Tyr Val 965 970 975Gln His Val
Met Glu Gln Asp Gly Lys Lys Leu Ile Glu Leu Leu Asp 980 985 990Gln
Gly Ala His Phe Tyr Ile Cys Gly Asp Gly Ser Gln Met Ala Pro 995
1000 1005Ala Val Glu Ala Thr Leu Met Lys Ser Tyr Ala Asp Val His
Gln 1010 1015 1020Val Ser Glu Ala Asp Ala Arg Leu Trp Leu Gln Gln
Leu Glu Glu 1025 1030 1035Lys Gly Arg Tyr Ala Lys Asp Val Trp Ala
Gly 1040 104512396PRTPleurotus sapidus 12Met Arg Tyr Gly Cys Ala
Ala Val Ala Leu Phe Tyr Leu Thr Ala Met1 5 10 15Gly Lys Leu His Pro
Leu Ala Ile Ile Pro Asp Tyr Lys Gly Ser Met 20 25 30Ala Ala Ser Val
Thr Ile Phe Asn Lys Arg Thr Asn Pro Leu Asp Ile 35 40 45Ser Val Asn
Gln Ala Asn Asp Trp Pro Trp Arg Tyr Ala Lys Thr Cys 50 55 60Val Leu
Ser Ser Asp Trp Ala Leu His Glu Met Ile Ile His Leu Asn65 70 75
80Asn Thr His Leu Val Glu Glu Ala Val Ile Val Ala Ala Gln Arg Lys
85 90 95Leu Ser Pro Ser His Ile Val Phe Arg Leu Leu Glu Pro His Trp
Val 100 105 110Val Thr Leu Ser Leu Asn Ala Leu Ala Arg Ser Val Leu
Ile Pro Glu 115 120 125Val Ile Val Pro Ile Ala Gly Phe Ser Ala Pro
His Ile Phe Gln Phe 130 135 140Ile Arg Glu Ser Phe Thr Asn Phe Asp
Trp Lys Ser Leu Tyr Val Pro145 150 155 160Ala Asp Leu Glu Ser Arg
Gly Phe Pro Val Asp Gln Leu Asn Ser Pro 165 170 175Lys Phe His Asn
Tyr Ala Tyr Ala Arg Asp Ile Asn Asp Met Trp Thr 180 185 190Thr Leu
Lys Lys Phe Val Ser Ser Val Leu Gln Asp Ala Gln Tyr Tyr 195 200
205Pro Asp Asp Ala Ser Val Ala Gly Asp Thr Gln Ile Gln Ala Trp Cys
210 215 220Asp Glu Met Arg Ser Gly Met Gly Ala Gly Met Thr Asn Phe
Pro Glu225 230 235 240Ser Ile Thr Thr Val Asp Asp Leu Val Asn Met
Val Thr Met Cys Ile 245 250 255His Ile Ala Ala Pro Gln His Thr Ala
Val Asn Tyr Leu Gln Gln Tyr 260 265 270Tyr Gln Thr Phe Val Ser Asn
Lys Pro Ser Ala Leu Phe Ser Pro Leu 275 280 285Pro Thr Ser Ile Ala
Gln Leu Gln Lys Tyr Thr Glu Ser Asp Leu Met 290 295 300Ala Ala Leu
Pro Leu Asn Ala Lys Arg Gln Trp Leu Leu Met Ala Gln305 310 315
320Ile Pro Tyr Leu Leu Ser Met Gln Val Gln Glu Asp Glu Asn Ile Val
325 330 335Thr Tyr Ala Ala Asn Ala Ser Thr Asp Lys Asp Pro Ile Ile
Ala Ser 340 345 350Ala Gly Arg Gln Leu Ala Ala Asp Leu Lys Lys Leu
Ala Ala Val Phe 355 360 365Leu Val Asn Ser Ala Gln Leu Asp Asp Gln
Asn Thr Pro Tyr Asp Val 370 375 380Leu Ala Pro Glu Gln Leu Ala Asn
Ala Ile Val Ile385 390 39513640PRTPleurotus ostreatus 13Met Ala Pro
Thr Met Ser Leu Ser Arg Ser Ala Leu Lys Asn Val His1 5 10 15Leu Pro
Tyr Met Val Gln His Pro Glu Pro Thr Asp Cys Ser Thr Ala 20 25 30Met
Lys His Ala Ala Glu Gly Tyr Asp Arg Ala Arg Gln Met Ile Ala 35 40
45Phe Leu Phe Asp Ile Leu Asp Tyr Glu Ser Ser Val Pro Gln Lys Phe
50 55 60Thr Pro Glu Glu Lys Lys Glu Lys Tyr Thr Trp Ser His Ser Asp
Lys65 70 75 80Phe Pro Pro His Leu Ala Ile Ile Pro Glu Asp Ile Asp
Val Pro Ala 85 90 95Tyr Ile Ile Phe Ser Ile Val Arg Leu Val Gln Thr
Leu Ser Ile Met 100 105 110Ser Gly Ile Gln Cys Asn Glu Arg Leu Ala
Pro Gly Pro Glu Gln Asn 115 120 125Thr Met Glu Lys Leu Thr Lys Trp
Asn Ala Glu Arg His Lys Asn Gln 130 135 140Gly Trp Val Lys Asp Met
Phe Asn Glu Pro Asn Ile Gly Leu Arg Asn145 150 155 160Asp Trp Tyr
Thr Asp Ala Val Phe Ala Gln Gln Phe Phe Thr Gly Pro 165 170 175Asn
Pro Thr Thr Ile Thr Leu Ala Ser Asp Thr Trp Met Lys Ala Phe 180 185
190Thr Glu Glu Ala Ala Ser Gln Gly Lys Arg Asp Leu Ile Ser Leu Phe
195 200 205Arg Ser Ala Pro Pro Asn Ser Phe Tyr Val Gln Asp Phe Ser
Asp Phe 210 215 220Arg Ala Arg Met Gly Ala Lys Pro Asp Glu Glu Leu
Cys Ala Thr Ser225 230 235 240Asp Gly Gly Val Thr Arg Tyr Gly Cys
Ala Ala Val Ala Leu Phe Tyr 245 250 255Leu Pro Pro Thr Gly Glu Leu
His Pro Leu Ala Ile Val Pro Asp Tyr 260 265 270Lys Gly Ser Met Ala
Ala Ser Ile Thr Leu Phe Asn Lys Arg Val Asp 275 280 285Pro Ser Asp
Ala Ser Val Asp Gln Ala Asn Asp Trp Pro Trp Arg Tyr 290 295 300Ala
Lys Thr Cys Val Leu Ser Ala Asp Trp Val Leu His Glu Met Ile305 310
315 320Ile His Leu Asn Asn Thr His Leu Val Gln Glu Ala Val Ile Val
Ala 325 330 335Val Gln Arg Thr Leu Pro Asp Ser His Ile Val Phe Arg
Leu Leu Lys 340 345 350Pro His Trp Val Val Thr Leu Ser Leu Asn Ala
Gln Ala Arg Ser Val 355 360 365Leu Ile Pro Glu Val Ile Val Pro Ile
Ala Gly Phe Ser Glu Leu Arg 370 375 380Ile Phe Gln Phe Val Gly His
Ala Phe Thr Asn Phe Asp Trp Lys Ala385 390 395 400Leu Tyr Val Pro
Thr Asp Leu Glu Phe Arg Gly Phe Pro Leu Asp Arg 405 410 415Leu Asp
Asp Asp Lys Phe His Asn Tyr Ala Tyr Ala Lys Asp Ile Lys 420 425
430Asp Met Trp Met Ala Leu Arg Lys Phe Val Ser Ser Val Leu Lys Asp
435 440 445Gly Lys Tyr Tyr Pro Asp Asp Ser Ala Val Ala Ala Asp Ala
Gln Ile 450 455 460Gln Asp Trp Cys Asp Glu Met Arg Ser Glu Lys Gly
Ala Gly Met Lys465 470 475 480Lys Phe Pro Glu Ser Ile Ser Thr Leu
Asp Asp Leu Ile Asp Met Val 485 490 495Thr Met Cys Ile His Ile Ala
Ala Pro Gln His Thr Ala Val Asn Tyr 500 505 510Leu Gln Gln Tyr Tyr
Gln Thr Phe Val Pro Asn Lys Pro Ser Ala Leu 515 520 525Phe Ser Pro
Leu Pro Thr Leu Leu Ser Gln Leu Glu Ser Tyr Thr Glu 530 535 540Ser
Asp Leu Met Ala Ala Leu Pro Leu Gly Ala Lys Gln Glu Trp Leu545 550
555 560Leu Met Ala Gln Val Pro Tyr Leu Leu Ser Lys Glu Val Glu Gln
Asp 565 570 575Gly Asn Ile Val Thr Tyr Ala Gly Thr Ala Ser Asn Asn
Glu Asp Pro 580 585 590Ile Ile Ala Ala Ala Gly Lys Glu Leu Ser Ala
Asp Leu Val Ile Leu 595 600 605Ala Gly Val Phe Leu Lys Asn Ser Glu
Lys Leu Asp Asp Gln Asn Thr 610 615 620Ala Tyr Asn Val Leu Ala Pro
Asp Gln Leu Ala Asn Ala Ile Val Ile625 630 635 64014495PRTCichorium
intybus 14Met Glu Ile Ser Ile Pro Thr Thr Leu Gly Leu Ala Val Ile
Ile Phe1 5 10 15Ile Ile Phe Lys Leu Leu Thr Arg Thr Thr Ser Lys Lys
Asn Leu Leu 20 25 30Pro Glu Pro Trp Arg Leu Pro Ile Ile Gly His Met
His His Leu Ile 35 40 45Gly Thr Met Pro His Arg Gly Val Met Glu Leu
Ala Arg Lys His Gly 50 55 60Ser Leu Met His Leu Gln Leu Gly Glu Val
Ser Thr Ile Val Val Ser65 70 75 80Ser Pro Arg Trp Ala Lys Glu Val
Leu Thr Thr Tyr Asp Ile Thr Phe 85 90 95Ala Asn Arg Pro Glu Thr Leu
Thr Gly Glu Ile Val Ala Tyr His Asn 100 105 110Thr Asp Ile Val Leu
Ala Pro Tyr Gly Glu Tyr Trp Arg Gln Leu Arg 115 120 125Lys Leu Cys
Thr Leu Glu Leu Leu Ser Asn Lys Lys Val Lys Ser Phe 130 135 140Gln
Ser Leu Arg Glu Glu Glu Cys Trp Asn Leu Val Lys Asp Ile Arg145 150
155 160Ser Thr Gly Gln Gly Ser Pro Ile Asn Leu Ser Glu Asn Ile Phe
Lys 165 170 175Met Ile Ala Thr Ile Leu Ser Arg Ala Ala Phe Gly Lys
Gly Ile Lys 180 185 190Asp Gln Met Lys Phe Thr Glu Leu Val Lys Glu
Ile Leu Arg Leu Thr 195
200 205Gly Gly Phe Asp Val Ala Asp Ile Phe Pro Ser Lys Lys Leu Leu
His 210 215 220His Leu Ser Gly Lys Arg Ala Lys Leu Thr Asn Ile His
Asn Lys Leu225 230 235 240Asp Asn Leu Ile Asn Asn Ile Ile Ala Glu
His Pro Gly Asn Arg Thr 245 250 255Ser Ser Ser Gln Glu Thr Leu Leu
Asp Val Leu Leu Arg Leu Lys Glu 260 265 270Ser Ala Glu Phe Pro Leu
Thr Ala Asp Asn Val Lys Ala Val Ile Leu 275 280 285Asp Met Phe Gly
Ala Gly Thr Asp Thr Ser Ser Ala Thr Ile Glu Trp 290 295 300Ala Ile
Ser Glu Leu Ile Arg Cys Pro Arg Ala Met Glu Lys Val Gln305 310 315
320Thr Glu Leu Arg Gln Ala Leu Asn Gly Lys Glu Arg Ile Gln Glu Glu
325 330 335Asp Leu Gln Glu Leu Asn Tyr Leu Lys Leu Val Ile Lys Glu
Thr Leu 340 345 350Arg Leu His Pro Pro Leu Pro Leu Val Met Pro Arg
Glu Cys Arg Glu 355 360 365Pro Cys Val Leu Gly Gly Tyr Asp Ile Pro
Ser Lys Thr Lys Leu Ile 370 375 380Val Asn Val Phe Ala Ile Asn Arg
Asp Pro Glu Tyr Trp Lys Asp Ala385 390 395 400Glu Thr Phe Met Pro
Glu Arg Phe Glu Asn Ser Pro Ile Thr Val Met 405 410 415Gly Ser Glu
Tyr Glu Tyr Leu Pro Phe Gly Ala Gly Arg Arg Met Cys 420 425 430Pro
Gly Ala Ala Leu Gly Leu Ala Asn Val Glu Leu Pro Leu Ala His 435 440
445Ile Leu Tyr Phe Asn Trp Lys Leu Pro Asn Gly Lys Thr Phe Glu Asp
450 455 460Leu Asp Met Thr Glu Ser Phe Gly Ala Thr Val Gln Arg Lys
Thr Glu465 470 475 480Leu Leu Leu Val Pro Thr Asp Phe Gln Thr Leu
Thr Ala Ser Thr 485 490 49515488PRTHelianthus annuus 15Met Glu Val
Ser Leu Thr Thr Ser Ile Ala Leu Ala Thr Ile Val Phe1 5 10 15Phe Leu
Tyr Lys Leu Leu Thr Arg Pro Thr Ser Ser Lys Asn Arg Leu 20 25 30Pro
Glu Pro Trp Arg Leu Pro Ile Ile Gly His Met His His Leu Ile 35 40
45Gly Thr Met Pro His Arg Gly Val Met Asp Leu Ala Arg Lys Tyr Gly
50 55 60Ser Leu Met His Leu Gln Leu Gly Glu Val Ser Ala Ile Val Val
Ser65 70 75 80Ser Pro Lys Trp Ala Lys Glu Ile Leu Thr Thr Tyr Asp
Ile Pro Phe 85 90 95Ala Asn Arg Pro Glu Thr Leu Thr Gly Glu Ile Ile
Ala Tyr His Asn 100 105 110Thr Asp Ile Val Leu Ala Pro Tyr Gly Glu
Tyr Trp Arg Gln Leu Arg 115 120 125Lys Leu Cys Thr Leu Glu Leu Leu
Ser Val Lys Lys Val Lys Ser Phe 130 135 140Gln Ser Leu Arg Glu Glu
Glu Cys Trp Asn Leu Val Gln Glu Ile Lys145 150 155 160Ala Ser Gly
Ser Gly Thr Pro Phe Asn Leu Ser Glu Gly Ile Phe Lys 165 170 175Val
Ile Ala Thr Val Leu Ser Arg Ala Ala Phe Gly Lys Gly Ile Lys 180 185
190Asp Gln Lys Gln Phe Thr Glu Ile Val Lys Glu Ile Leu Arg Glu Thr
195 200 205Gly Gly Phe Asp Val Ala Asp Ile Phe Pro Ser Lys Lys Phe
Leu His 210 215 220His Leu Ser Gly Lys Arg Gly Arg Leu Thr Ser Ile
His Asn Lys Leu225 230 235 240Asp Ser Leu Ile Asn Asn Leu Val Ala
Glu His Thr Val Ser Lys Ser 245 250 255Ser Lys Val Asn Glu Thr Leu
Leu Asp Val Leu Leu Arg Leu Lys Asn 260 265 270Ser Glu Glu Phe Pro
Leu Thr Ala Asp Asn Val Lys Ala Ile Ile Leu 275 280 285Asp Met Phe
Gly Ala Gly Thr Asp Thr Ser Ser Ala Thr Val Glu Trp 290 295 300Ala
Ile Ser Glu Leu Ile Arg Cys Pro Arg Ala Met Glu Lys Val Gln305 310
315 320Ala Glu Leu Arg Gln Ala Leu Asn Gly Lys Glu Arg Ile Lys Glu
Glu 325 330 335Glu Ile Gln Asp Leu Pro Tyr Leu Asn Leu Val Ile Arg
Glu Thr Leu 340 345 350Arg Leu His Pro Pro Leu Pro Leu Val Met Pro
Arg Glu Cys Arg Gln 355 360 365Ala Met Asn Leu Ala Gly Tyr Asp Val
Ala Asn Lys Thr Lys Leu Ile 370 375 380Val Asn Val Phe Ala Ile Asn
Arg Asp Pro Glu Tyr Trp Lys Asp Ala385 390 395 400Glu Ser Phe Asn
Pro Glu Arg Phe Glu Asn Ser Asn Thr Thr Ile Met 405 410 415Gly Ala
Asp Tyr Glu Tyr Leu Pro Phe Gly Ala Gly Arg Arg Met Cys 420 425
430Pro Gly Ser Ala Leu Gly Leu Ala Asn Val Gln Leu Pro Leu Ala Asn
435 440 445Ile Leu Tyr Tyr Phe Lys Trp Lys Leu Pro Asn Gly Ala Ser
His Asp 450 455 460Gln Leu Asp Met Thr Glu Ser Phe Gly Ala Thr Val
Gln Arg Lys Thr465 470 475 480Glu Leu Met Leu Val Pro Ser Phe
48516425PRTEscherichia coli 16Met Leu Glu Leu Leu Tyr Thr Ala Leu
Leu Tyr Leu Ile Gln Pro Leu1 5 10 15Ile Trp Ile Arg Leu Trp Val Arg
Gly Arg Lys Ala Pro Ala Tyr Arg 20 25 30Lys Arg Trp Gly Glu Arg Tyr
Gly Phe Tyr Arg His Pro Leu Lys Pro 35 40 45Gly Gly Ile Met Leu His
Ser Val Ser Val Gly Glu Thr Leu Ala Ala 50 55 60Ile Pro Leu Val Arg
Ala Leu Arg His Arg Tyr Pro Asp Leu Pro Ile65 70 75 80Thr Val Thr
Thr Met Thr Pro Thr Gly Ser Glu Arg Val Gln Ser Ala 85 90 95Phe Gly
Lys Asp Val Gln His Val Tyr Leu Pro Tyr Asp Leu Pro Asp 100 105
110Ala Leu Asn Arg Phe Leu Asn Lys Val Asp Pro Lys Leu Val Leu Ile
115 120 125Met Glu Thr Glu Leu Trp Pro Asn Leu Ile Ala Ala Leu His
Lys Arg 130 135 140Lys Ile Pro Leu Val Ile Ala Asn Ala Arg Leu Ser
Ala Arg Ser Ala145 150 155 160Ala Gly Tyr Ala Lys Leu Gly Lys Phe
Val Arg Arg Leu Leu Arg Arg 165 170 175Ile Thr Leu Ile Ala Ala Gln
Asn Glu Glu Asp Gly Ala Arg Phe Val 180 185 190Ala Leu Gly Ala Lys
Asn Asn Gln Val Thr Val Thr Gly Ser Leu Lys 195 200 205Phe Asp Ile
Ser Val Thr Pro Gln Leu Ala Ala Lys Ala Val Thr Leu 210 215 220Arg
Arg Gln Trp Ala Pro His Arg Pro Val Trp Ile Ala Thr Ser Thr225 230
235 240His Glu Gly Glu Glu Ser Val Val Ile Ala Ala His Gln Ala Leu
Leu 245 250 255Gln Gln Phe Pro Asn Leu Leu Leu Ile Leu Val Pro Arg
His Pro Glu 260 265 270Arg Phe Pro Asp Ala Ile Asn Leu Val Arg Gln
Ala Gly Leu Ser Tyr 275 280 285Ile Thr Arg Ser Ser Gly Glu Val Pro
Ser Thr Ser Thr Gln Val Val 290 295 300Val Gly Asp Thr Met Gly Glu
Leu Met Leu Leu Tyr Gly Ile Ala Asp305 310 315 320Leu Ala Phe Val
Gly Gly Ser Leu Val Glu Arg Gly Gly His Asn Pro 325 330 335Leu Glu
Ala Ala Ala His Ala Ile Pro Val Leu Met Gly Pro His Thr 340 345
350Phe Asn Phe Lys Asp Ile Cys Ala Arg Leu Glu Gln Ala Ser Gly Leu
355 360 365Ile Thr Val Thr Asp Ala Thr Thr Leu Ala Lys Glu Val Ser
Ser Leu 370 375 380Leu Thr Asp Ala Asp Tyr Arg Ser Phe Tyr Gly Arg
His Ala Val Glu385 390 395 400Val Leu Tyr Gln Asn Gln Gly Ala Leu
Gln Arg Leu Leu Gln Leu Leu 405 410 415Glu Pro Tyr Leu Pro Pro Lys
Thr His 420 4251766PRTEscherichia coli 17Met Asp Trp Leu Ala Lys
Tyr Trp Trp Ile Leu Val Ile Val Phe Leu1 5 10 15Val Gly Val Leu Leu
Asn Val Ile Lys Asp Leu Lys Arg Val Asp His 20 25 30Lys Lys Phe Leu
Ala Asn Lys Pro Glu Leu Pro Pro His Arg Asp Phe 35 40 45Asn Asp Lys
Trp Asp Asp Asp Asp Asp Trp Pro Lys Lys Asp Gln Pro 50 55 60Lys
Lys6518132PRTEscherichia coli 18Met Thr Trp Glu Tyr Ala Leu Ile Gly
Leu Val Val Gly Ile Ile Ile1 5 10 15Gly Ala Val Ala Met Arg Phe Gly
Asn Arg Lys Leu Arg Gln Gln Gln 20 25 30Ala Leu Gln Tyr Glu Leu Glu
Lys Asn Lys Ala Glu Leu Asp Glu Tyr 35 40 45Arg Glu Glu Leu Val Ser
His Phe Ala Arg Ser Ala Glu Leu Leu Asp 50 55 60Thr Met Ala His Asp
Tyr Arg Gln Leu Tyr Gln His Met Ala Lys Ser65 70 75 80Ser Ser Ser
Leu Leu Pro Glu Leu Ser Ala Glu Ala Asn Pro Phe Arg 85 90 95Asn Arg
Leu Ala Glu Ser Glu Ala Ser Asn Asp Gln Ala Pro Val Gln 100 105
110Met Pro Arg Asp Tyr Ser Glu Gly Ala Ser Gly Leu Leu Arg Thr Gly
115 120 125Ala Lys Arg Asp 13019294PRTEscherichia coli 19Met Lys
Pro Phe Leu Arg Trp Cys Phe Val Ala Thr Ala Leu Thr Leu1 5 10 15Ala
Gly Cys Ser Asn Thr Ser Trp Arg Lys Ser Glu Val Leu Ala Val 20 25
30Pro Leu Gln Pro Thr Leu Gln Gln Glu Val Ile Leu Ala Arg Met Glu
35 40 45Gln Ile Leu Ala Ser Arg Ala Leu Thr Asp Asp Glu Arg Ala Gln
Leu 50 55 60Leu Tyr Glu Arg Gly Val Leu Tyr Asp Ser Leu Gly Leu Arg
Ala Leu65 70 75 80Ala Arg Asn Asp Phe Ser Gln Ala Leu Ala Ile Arg
Pro Asp Met Pro 85 90 95Glu Val Phe Asn Tyr Leu Gly Ile Tyr Leu Thr
Gln Ala Gly Asn Phe 100 105 110Asp Ala Ala Tyr Glu Ala Phe Asp Ser
Val Leu Glu Leu Asp Pro Thr 115 120 125Tyr Asn Tyr Ala His Leu Asn
Arg Gly Ile Ala Leu Tyr Tyr Gly Gly 130 135 140Arg Asp Lys Leu Ala
Gln Asp Asp Leu Leu Ala Phe Tyr Gln Asp Asp145 150 155 160Pro Asn
Asp Pro Phe Arg Ser Leu Trp Leu Tyr Leu Ala Glu Gln Lys 165 170
175Leu Asp Glu Lys Gln Ala Lys Glu Val Leu Lys Gln His Phe Glu Lys
180 185 190Ser Asp Lys Glu Gln Trp Gly Trp Asn Ile Val Glu Phe Tyr
Leu Gly 195 200 205Asn Ile Ser Glu Gln Thr Leu Met Glu Arg Leu Lys
Ala Asp Ala Thr 210 215 220Asp Asn Thr Ser Leu Ala Glu His Leu Ser
Glu Thr Asn Phe Tyr Leu225 230 235 240Gly Lys Tyr Tyr Leu Ser Leu
Gly Asp Leu Asp Ser Ala Thr Ala Leu 245 250 255Phe Lys Leu Ala Val
Ala Asn Asn Val His Asn Phe Val Glu His Arg 260 265 270Tyr Ala Leu
Leu Glu Leu Ser Leu Leu Gly Gln Asp Gln Asp Asp Leu 275 280 285Ala
Glu Ser Asp Gln Gln 29020119PRTEscherichia coli 20Met Ser Lys Pro
Pro Leu Phe Phe Ile Val Ile Ile Gly Leu Ile Val1 5 10 15Val Ala Ala
Ser Phe Arg Phe Met Gln Gln Arg Arg Glu Lys Ala Asp 20 25 30Asn Asp
Met Ala Pro Leu Gln Gln Lys Leu Val Val Val Ser Asn Lys 35 40 45Arg
Glu Lys Pro Ile Asn Asp Arg Arg Ser Arg Gln Gln Glu Val Thr 50 55
60Pro Ala Gly Thr Ser Ile Arg Tyr Glu Ala Ser Phe Lys Pro Gln Ser65
70 75 80Gly Gly Met Glu Gln Thr Phe Arg Leu Asp Ala Gln Gln Tyr His
Ala 85 90 95Leu Thr Val Gly Asp Lys Gly Thr Leu Ser Tyr Lys Gly Thr
Arg Phe 100 105 110Val Ser Phe Val Gly Glu Gln
11521328PRTEscherichia coli 21Met Met Gln Asp Leu Arg Leu Ile Leu
Ile Ile Val Gly Ala Ile Ala1 5 10 15Ile Ile Ala Leu Leu Val His Gly
Phe Trp Thr Ser Arg Lys Glu Arg 20 25 30Ser Ser Met Phe Arg Asp Arg
Pro Leu Lys Arg Met Lys Ser Lys Arg 35 40 45Asp Asp Asp Ser Tyr Asp
Glu Asp Val Glu Asp Asp Glu Gly Val Gly 50 55 60Glu Val Arg Val His
Arg Val Asn His Ala Pro Ala Asn Ala Gln Glu65 70 75 80His Glu Ala
Ala Arg Pro Ser Pro Gln His Gln Tyr Gln Pro Pro Tyr 85 90 95Ala Ser
Ala Gln Pro Arg Gln Pro Val Gln Gln Pro Pro Glu Ala Gln 100 105
110Val Pro Pro Gln His Ala Pro His Pro Ala Gln Pro Val Gln Gln Pro
115 120 125Ala Tyr Gln Pro Gln Pro Glu Gln Pro Leu Gln Gln Pro Val
Ser Pro 130 135 140Gln Val Ala Pro Ala Pro Gln Pro Val His Ser Ala
Pro Gln Pro Ala145 150 155 160Gln Gln Ala Phe Gln Pro Ala Glu Pro
Val Ala Ala Pro Gln Pro Glu 165 170 175Pro Val Ala Glu Pro Ala Pro
Val Met Asp Lys Pro Lys Arg Lys Glu 180 185 190Ala Val Ile Ile Met
Asn Val Ala Ala His His Gly Ser Glu Leu Asn 195 200 205Gly Glu Leu
Leu Leu Asn Ser Ile Gln Gln Ala Gly Phe Ile Phe Gly 210 215 220Asp
Met Asn Ile Tyr His Arg His Leu Ser Pro Asp Gly Ser Gly Pro225 230
235 240Ala Leu Phe Ser Leu Ala Asn Met Val Lys Pro Gly Thr Phe Asp
Pro 245 250 255Glu Met Lys Asp Phe Thr Thr Pro Gly Val Thr Ile Phe
Met Gln Val 260 265 270Pro Ser Tyr Gly Asp Glu Leu Gln Asn Phe Lys
Leu Met Leu Gln Ser 275 280 285Ala Gln His Ile Ala Asp Glu Val Gly
Gly Val Val Leu Asp Asp Gln 290 295 300Arg Arg Met Met Thr Pro Gln
Lys Leu Arg Glu Tyr Gln Asp Ile Ile305 310 315 320Arg Glu Val Lys
Asp Ala Asn Ala 32522507PRTEscherichia coli 22Met Arg Asn Thr Leu
Ile Pro Ile Leu Val Ala Ile Cys Leu Phe Ile1 5 10 15Thr Gly Val Ala
Ile Leu Asn Ile Gln Leu Trp Tyr Ser Ala Lys Ala 20 25 30Glu Tyr Leu
Ala Gly Ala Arg Tyr Ala Ala Asn Asn Ile Asn His Ile 35 40 45Leu Glu
Glu Ala Ser Gln Ala Thr Gln Thr Ala Val Asn Ile Ala Gly 50 55 60Lys
Glu Cys Asn Leu Glu Glu Gln Tyr Gln Leu Gly Thr Glu Ala Ala65 70 75
80Leu Lys Pro His Leu Arg Thr Ile Ile Ile Leu Lys Gln Gly Ile Val
85 90 95Trp Cys Thr Ser Leu Pro Gly Asn Arg Val Leu Leu Ser Arg Ile
Pro 100 105 110Val Phe Pro Asp Ser Asn Leu Leu Leu Ala Pro Ala Ile
Asp Thr Val 115 120 125Asn Arg Leu Pro Ile Leu Leu Tyr Gln Asn Gln
Phe Ala Asp Thr Arg 130 135 140Ile Leu Val Thr Ile Ser Asp Gln His
Ile Arg Gly Ala Leu Asn Val145 150 155 160Pro Leu Lys Gly Val Arg
Tyr Val Leu Arg Val Ala Asp Asp Ile Ile 165 170 175Gly Pro Thr Gly
Asp Val Met Thr Leu Asn Gly His Tyr Pro Tyr Thr 180 185 190Glu Lys
Val His Ser Thr Lys Tyr His Phe Thr Ile Ile Phe Asn Pro 195 200
205Pro Pro Leu Phe Ser Phe Tyr Arg Leu Ile Asp Lys Gly Phe Gly Ile
210 215 220Leu Ile Phe Ile Leu Leu Ile Ala Cys Ala Ala Ala Phe Leu
Leu Asp225 230 235 240Arg Tyr Phe Asn Lys Ser Ala Thr Pro Glu Glu
Ile Leu Arg Arg Ala 245 250 255Ile Asn Asn Gly Glu Ile Val Pro Phe
Tyr Gln Pro Val Val Asn Gly 260 265 270Arg Glu Gly Thr Leu Arg Gly
Val Glu Val Leu Ala Arg Trp Lys Gln 275 280 285Pro His Gly Gly Tyr
Ile Ser Pro Ala Ala Phe Ile Pro Leu Ala Glu 290 295 300Lys Ser Gly
Leu Ile Val Pro Leu Thr Gln Ser Leu Met Asn Gln Val305
310 315 320Ala Arg Gln Met Asn Ala Ile Ala Ser Lys Leu Pro Glu Gly
Phe His 325 330 335Ile Gly Ile Asn Phe Ser Ala Ser His Ile Ile Ser
Pro Thr Phe Val 340 345 350Asp Glu Cys Leu Asn Phe Arg Asp Ser Phe
Thr Arg Arg Asp Leu Asn 355 360 365Leu Val Leu Glu Val Thr Glu Arg
Glu Pro Leu Asn Val Asp Glu Ser 370 375 380Leu Val Gln Arg Leu Asn
Ile Leu His Glu Asn Gly Phe Val Ile Ala385 390 395 400Leu Asp Asp
Phe Gly Thr Gly Tyr Ser Gly Leu Ser Tyr Leu His Asp 405 410 415Leu
His Ile Asp Tyr Ile Lys Ile Asp His Ser Phe Val Gly Arg Val 420 425
430Asn Ala Asp Pro Glu Ser Thr Arg Ile Leu Asp Cys Val Leu Asp Leu
435 440 445Ala Arg Lys Leu Ser Ile Ser Ile Val Ala Glu Gly Val Glu
Thr Lys 450 455 460Glu Gln Leu Asp Tyr Leu Asn Gln Asn Tyr Ile Thr
Phe Gln Gln Gly465 470 475 480Tyr Tyr Phe Tyr Lys Pro Val Thr Tyr
Ile Asp Leu Val Lys Ile Ile 485 490 495Leu Ser Lys Pro Lys Val Lys
Val Val Val Glu 500 50523271PRTEscherichia coli 23Met Gln Tyr Trp
Gly Lys Ile Ile Gly Val Ala Val Ala Leu Leu Met1 5 10 15Gly Gly Gly
Phe Trp Gly Val Val Leu Gly Leu Leu Ile Gly His Met 20 25 30Phe Asp
Lys Ala Arg Ser Arg Lys Met Ala Trp Phe Ala Asn Gln Arg 35 40 45Glu
Arg Gln Ala Leu Phe Phe Ala Thr Thr Phe Glu Val Met Gly His 50 55
60Leu Thr Lys Ser Lys Gly Arg Val Thr Glu Ala Asp Ile His Ile Ala65
70 75 80Ser Gln Leu Met Asp Arg Met Asn Leu His Gly Ala Ser Arg Thr
Ala 85 90 95Ala Gln Asn Ala Phe Arg Val Gly Lys Ser Asp Asn Tyr Pro
Leu Arg 100 105 110Glu Lys Met Arg Gln Phe Arg Ser Val Cys Phe Gly
Arg Phe Asp Leu 115 120 125Ile Arg Met Phe Leu Glu Ile Gln Ile Gln
Ala Ala Phe Ala Asp Gly 130 135 140Ser Leu His Pro Asn Glu Arg Ala
Val Leu Tyr Val Ile Ala Glu Glu145 150 155 160Leu Gly Ile Ser Arg
Ala Gln Phe Asp Gln Phe Leu Arg Met Met Gln 165 170 175Gly Gly Ala
Gln Phe Gly Gly Gly Tyr Gln Gln Gln Thr Gly Gly Gly 180 185 190Asn
Trp Gln Gln Ala Gln Arg Gly Pro Thr Leu Glu Asp Ala Cys Asn 195 200
205Val Leu Gly Val Lys Pro Thr Asp Asp Ala Thr Thr Ile Lys Arg Ala
210 215 220Tyr Arg Lys Leu Met Ser Glu His His Pro Asp Lys Leu Val
Ala Lys225 230 235 240Gly Leu Pro Pro Glu Met Met Glu Met Ala Lys
Gln Lys Ala Gln Glu 245 250 255Ile Gln Gln Ala Tyr Glu Leu Ile Lys
Gln Gln Lys Gly Phe Lys 260 265 27024349PRTEscherichia coli 24Met
Glu Leu Leu Ser Glu Tyr Gly Leu Phe Leu Ala Lys Ile Val Thr1 5 10
15Val Val Leu Ala Ile Ala Ala Ile Ala Ala Ile Ile Val Asn Val Ala
20 25 30Gln Arg Asn Lys Arg Gln Arg Gly Glu Leu Arg Val Asn Asn Leu
Ser 35 40 45Glu Gln Tyr Lys Glu Met Lys Glu Glu Leu Ala Ala Ala Leu
Met Asp 50 55 60Ser His Gln Gln Lys Gln Trp His Lys Ala Gln Lys Lys
Lys His Lys65 70 75 80Gln Glu Ala Lys Ala Ala Lys Ala Lys Ala Lys
Leu Gly Glu Val Ala 85 90 95Thr Asp Ser Lys Pro Arg Val Trp Val Leu
Asp Phe Lys Gly Ser Met 100 105 110Asp Ala His Glu Val Asn Ser Leu
Arg Glu Glu Ile Thr Ala Val Leu 115 120 125Ala Ala Phe Lys Pro Gln
Asp Gln Val Val Leu Arg Leu Glu Ser Pro 130 135 140Gly Gly Met Val
His Gly Tyr Gly Leu Ala Ala Ser Gln Leu Gln Arg145 150 155 160Leu
Arg Asp Lys Asn Ile Pro Leu Thr Val Thr Val Asp Lys Val Ala 165 170
175Ala Ser Gly Gly Tyr Met Met Ala Cys Val Ala Asp Lys Ile Val Ser
180 185 190Ala Pro Phe Ala Ile Val Gly Ser Ile Gly Val Val Ala Gln
Met Pro 195 200 205Asn Phe Asn Arg Phe Leu Lys Ser Lys Asp Ile Asp
Ile Glu Leu His 210 215 220Thr Ala Gly Gln Tyr Lys Arg Thr Leu Thr
Leu Leu Gly Glu Asn Thr225 230 235 240Glu Glu Gly Arg Glu Lys Phe
Arg Glu Glu Leu Asn Glu Thr His Gln 245 250 255Leu Phe Lys Asp Phe
Val Lys Arg Met Arg Pro Ser Leu Asp Ile Glu 260 265 270Gln Val Ala
Thr Gly Glu His Trp Tyr Gly Gln Gln Ala Val Glu Lys 275 280 285Gly
Leu Val Asp Glu Ile Asn Thr Ser Asp Glu Val Ile Leu Ser Leu 290 295
300Met Glu Gly Arg Glu Val Val Asn Val Arg Tyr Met Gln Arg Lys
Arg305 310 315 320Leu Ile Asp Arg Phe Thr Gly Ser Ala Ala Glu Ser
Ala Asp Arg Leu 325 330 335Leu Leu Arg Trp Trp Gln Arg Gly Gln Lys
Pro Leu Met 340 34525328PRTEscherichia coli 25Met Ile Glu Lys Ile
Trp Ser Gly Glu Ser Pro Leu Trp Arg Leu Leu1 5 10 15Leu Pro Leu Ser
Trp Leu Tyr Gly Leu Val Ser Gly Ala Ile Arg Leu 20 25 30Cys Tyr Lys
Leu Lys Leu Lys Arg Ala Trp Arg Ala Pro Val Pro Val 35 40 45Val Val
Val Gly Asn Leu Thr Ala Gly Gly Asn Gly Lys Thr Pro Val 50 55 60Val
Val Trp Leu Val Glu Gln Leu Gln Gln Arg Gly Ile Arg Val Gly65 70 75
80Val Val Ser Arg Gly Tyr Gly Gly Lys Ala Glu Ser Tyr Pro Leu Leu
85 90 95Leu Ser Ala Asp Thr Thr Thr Ala Gln Ala Gly Asp Glu Pro Val
Leu 100 105 110Ile Tyr Gln Arg Thr Asp Ala Pro Val Ala Val Ser Pro
Val Arg Ser 115 120 125Asp Ala Val Lys Ala Ile Leu Ala Gln His Pro
Asp Val Gln Ile Ile 130 135 140Val Thr Asp Asp Gly Leu Gln His Tyr
Arg Leu Ala Arg Asp Val Glu145 150 155 160Ile Val Val Ile Asp Gly
Val Arg Arg Phe Gly Asn Gly Trp Trp Leu 165 170 175Pro Ala Gly Pro
Met Arg Glu Arg Ala Gly Arg Leu Lys Ser Val Asp 180 185 190Ala Val
Ile Val Asn Gly Gly Val Pro Arg Ser Gly Glu Ile Pro Met 195 200
205His Leu Leu Pro Gly Gln Ala Val Asn Leu Arg Thr Gly Thr Arg Cys
210 215 220Asp Val Ala Gln Leu Glu His Val Val Ala Met Ala Gly Ile
Gly His225 230 235 240Pro Pro Arg Phe Phe Ala Thr Leu Lys Met Cys
Gly Val Gln Pro Glu 245 250 255Lys Cys Val Pro Leu Ala Asp His Gln
Ser Leu Asn His Ala Asp Val 260 265 270Ser Ala Leu Val Ser Ala Gly
Gln Thr Leu Val Met Thr Glu Lys Asp 275 280 285Ala Val Lys Cys Arg
Ala Phe Ala Glu Glu Asn Trp Trp Tyr Leu Pro 290 295 300Val Asp Ala
Gln Leu Ser Gly Asp Glu Pro Ala Lys Leu Leu Thr Gln305 310 315
320Leu Thr Leu Leu Ala Ser Gly Asn 32526121PRTEscherichia coli
26Met Asn Asn His Ala Thr Val Gln Ser Ser Ala Pro Val Ser Ala Ala1
5 10 15Pro Leu Leu Gln Val Ser Gly Ala Leu Ile Ala Ile Ile Ala Leu
Ile 20 25 30Leu Ala Ala Ala Trp Leu Val Lys Arg Leu Gly Phe Ala Pro
Lys Arg 35 40 45Thr Gly Val Asn Gly Leu Lys Ile Ser Ala Ser Ala Ser
Leu Gly Ala 50 55 60Arg Glu Arg Val Val Val Val Asp Val Glu Asp Ala
Arg Leu Val Leu65 70 75 80Gly Val Thr Ala Gly Gln Ile Asn Leu Leu
His Lys Leu Pro Pro Ser 85 90 95Ala Pro Thr Glu Glu Ile Pro Gln Thr
Asp Phe Gln Ser Val Met Lys 100 105 110Asn Leu Leu Lys Arg Ser Gly
Arg Ser 115 12027295PRTEscherichia coli 27Met Leu Ile Leu Leu Gly
Tyr Leu Val Val Leu Gly Thr Val Phe Gly1 5 10 15Gly Tyr Leu Met Thr
Gly Gly Ser Leu Gly Ala Leu Tyr Gln Pro Ala 20 25 30Glu Leu Val Ile
Ile Ala Gly Ala Gly Ile Gly Ser Phe Ile Val Gly 35 40 45Asn Asn Gly
Lys Ala Ile Lys Gly Thr Leu Lys Ala Leu Pro Leu Leu 50 55 60Phe Arg
Arg Ser Lys Tyr Thr Lys Ala Met Tyr Met Asp Leu Leu Ala65 70 75
80Leu Leu Tyr Arg Leu Met Ala Lys Ser Arg Gln Met Gly Met Phe Ser
85 90 95Leu Glu Arg Asp Ile Glu Asn Pro Arg Glu Ser Glu Ile Phe Ala
Ser 100 105 110Tyr Pro Arg Ile Leu Ala Asp Ser Val Met Leu Asp Phe
Ile Val Asp 115 120 125Tyr Leu Arg Leu Ile Ile Ser Gly His Met Asn
Thr Phe Glu Ile Glu 130 135 140Ala Leu Met Asp Glu Glu Ile Glu Thr
His Glu Ser Glu Ala Glu Val145 150 155 160Pro Ala Asn Ser Leu Ala
Leu Val Gly Asp Ser Leu Pro Ala Phe Gly 165 170 175Ile Val Ala Ala
Val Met Gly Val Val His Ala Leu Gly Ser Ala Asp 180 185 190Arg Pro
Ala Ala Glu Leu Gly Ala Leu Ile Ala His Ala Met Val Gly 195 200
205Thr Phe Leu Gly Ile Leu Leu Ala Tyr Gly Phe Ile Ser Pro Leu Ala
210 215 220Thr Val Leu Arg Gln Lys Ser Ala Glu Thr Ser Lys Met Met
Gln Cys225 230 235 240Val Lys Val Thr Leu Leu Ser Asn Leu Asn Gly
Tyr Ala Pro Pro Ile 245 250 255Ala Val Glu Phe Gly Arg Lys Thr Leu
Tyr Ser Ser Glu Arg Pro Ser 260 265 270Phe Ile Glu Leu Glu Glu His
Val Arg Ala Val Lys Asn Pro Gln Gln 275 280 285Gln Thr Thr Thr Glu
Glu Ala 290 29528293PRTEscherichia coli 28Met Met Arg Ile Ala Leu
Phe Leu Leu Thr Asn Leu Ala Val Met Val1 5 10 15Val Phe Gly Leu Val
Leu Ser Leu Thr Gly Ile Gln Ser Ser Ser Val 20 25 30Gln Gly Leu Met
Ile Met Ala Leu Leu Phe Gly Phe Gly Gly Ser Phe 35 40 45Val Ser Leu
Leu Met Ser Lys Trp Met Ala Leu Arg Ser Val Gly Gly 50 55 60Glu Val
Ile Glu Gln Pro Arg Asn Glu Arg Glu Arg Trp Leu Val Asn65 70 75
80Thr Val Ala Thr Gln Ala Arg Gln Ala Gly Ile Ala Met Pro Gln Val
85 90 95Ala Ile Tyr His Ala Pro Asp Ile Asn Ala Phe Ala Thr Gly Ala
Arg 100 105 110Arg Asp Ala Ser Leu Val Ala Val Ser Thr Gly Leu Leu
Gln Asn Met 115 120 125Ser Pro Asp Glu Ala Glu Ala Val Ile Ala His
Glu Ile Ser His Ile 130 135 140Ala Asn Gly Asp Met Val Thr Met Thr
Leu Ile Gln Gly Val Val Asn145 150 155 160Thr Phe Val Ile Phe Ile
Ser Arg Ile Leu Ala Gln Leu Ala Ala Gly 165 170 175Phe Met Gly Gly
Asn Arg Asp Glu Gly Glu Glu Ser Asn Gly Asn Pro 180 185 190Leu Ile
Tyr Phe Ala Val Ala Thr Val Leu Glu Leu Val Phe Gly Ile 195 200
205Leu Ala Ser Ile Ile Thr Met Trp Phe Ser Arg His Arg Glu Phe His
210 215 220Ala Asp Ala Gly Ser Ala Lys Leu Val Gly Arg Glu Lys Met
Ile Ala225 230 235 240Ala Leu Gln Arg Leu Lys Thr Ser Tyr Glu Pro
Gln Glu Ala Thr Ser 245 250 255Met Met Ala Leu Cys Ile Asn Gly Lys
Ser Lys Ser Leu Ser Glu Leu 260 265 270Phe Met Thr His Pro Pro Leu
Asp Lys Arg Ile Glu Ala Leu Arg Thr 275 280 285Gly Glu Tyr Leu Lys
29029441PRTEscherichia coli 29Met Ile Asn Arg Ile Val Ser Phe Phe
Ile Leu Cys Leu Val Leu Cys1 5 10 15Ile Pro Leu Cys Val Ala Tyr Phe
His Ser Gly Glu Leu Met Met Arg 20 25 30Phe Val Phe Phe Trp Pro Phe
Phe Met Ser Ile Met Trp Ile Val Gly 35 40 45Gly Val Tyr Phe Trp Val
Tyr Arg Glu Arg His Trp Pro Trp Gly Glu 50 55 60Asn Ala Pro Ala Pro
Gln Leu Lys Asp Asn Pro Ser Ile Ser Ile Ile65 70 75 80Ile Pro Cys
Phe Asn Glu Glu Lys Asn Val Glu Glu Thr Ile His Ala 85 90 95Ala Leu
Ala Gln Arg Tyr Glu Asn Ile Glu Val Ile Ala Val Asn Asp 100 105
110Gly Ser Thr Asp Lys Thr Arg Ala Ile Leu Asp Arg Met Ala Ala Gln
115 120 125Ile Pro His Leu Arg Val Ile His Leu Ala Gln Asn Gln Gly
Lys Ala 130 135 140Ile Ala Leu Lys Thr Gly Ala Ala Ala Ala Lys Ser
Glu Tyr Leu Val145 150 155 160Cys Ile Asp Gly Asp Ala Leu Leu Asp
Arg Asp Ala Ala Ala Tyr Ile 165 170 175Val Glu Pro Met Leu Tyr Asn
Pro Arg Val Gly Ala Val Thr Gly Asn 180 185 190Pro Arg Ile Arg Thr
Arg Ser Thr Leu Val Gly Lys Ile Gln Val Gly 195 200 205Glu Tyr Ser
Ser Ile Ile Gly Leu Ile Lys Arg Thr Gln Arg Ile Tyr 210 215 220Gly
Asn Val Phe Thr Val Ser Gly Val Ile Ala Ala Phe Arg Arg Ser225 230
235 240Ala Leu Ala Glu Val Gly Tyr Trp Ser Asp Asp Met Ile Thr Glu
Asp 245 250 255Ile Asp Ile Ser Trp Lys Leu Gln Leu Asn Gln Trp Thr
Ile Phe Tyr 260 265 270Glu Pro Arg Ala Leu Cys Trp Ile Leu Met Pro
Glu Thr Leu Lys Gly 275 280 285Leu Trp Lys Gln Arg Leu Arg Trp Ala
Gln Gly Gly Ala Glu Val Phe 290 295 300Leu Lys Asn Met Thr Arg Leu
Trp Arg Lys Glu Asn Phe Arg Met Trp305 310 315 320Pro Leu Phe Phe
Glu Tyr Cys Leu Thr Thr Ile Trp Ala Phe Thr Cys 325 330 335Leu Val
Gly Phe Ile Ile Tyr Ala Val Gln Leu Ala Gly Val Pro Leu 340 345
350Asn Ile Glu Leu Thr His Ile Ala Ala Thr His Thr Ala Gly Ile Leu
355 360 365Leu Cys Thr Leu Cys Leu Leu Gln Phe Ile Val Ser Leu Met
Ile Glu 370 375 380Asn Arg Tyr Glu His Asn Leu Thr Ser Ser Leu Phe
Trp Ile Ile Trp385 390 395 400Phe Pro Val Ile Phe Trp Met Leu Ser
Leu Ala Thr Thr Leu Val Ser 405 410 415Phe Thr Arg Val Met Leu Met
Pro Lys Lys Gln Arg Ala Arg Trp Val 420 425 430Ser Pro Asp Arg Gly
Ile Leu Arg Gly 435 44030131PRTEscherichia coli 30Met Thr Ser Arg
Phe Met Leu Ile Phe Ala Ala Ile Ser Gly Phe Ile1 5 10 15Phe Val Ala
Leu Gly Ala Phe Gly Ala His Val Leu Ser Lys Thr Met 20 25 30Gly Ala
Val Glu Met Gly Trp Ile Gln Thr Gly Leu Glu Tyr Gln Ala 35 40 45Phe
His Thr Leu Ala Ile Leu Gly Leu Ala Val Ala Met Gln Arg Arg 50 55
60Ile Ser Ile Trp Phe Tyr Trp Ser Ser Val Phe Leu Ala Leu Gly Thr65
70 75 80Val Leu Phe Ser Gly Ser Leu Tyr Cys Leu Ala Leu Ser His Leu
Arg 85 90 95Leu Trp Ala Phe Val Thr Pro Val Gly Gly Val Ser Phe Leu
Ala Gly 100 105 110Trp Ala Leu Met Leu Val Gly Ala Ile Arg Leu Lys
Arg Lys Gly Val 115 120 125Ser His Glu 13031655PRTEscherichia coli
31Met Asn Val Ile Lys Thr Ala Ile Cys Thr Leu Ile Thr Leu Pro Val1
5 10 15Gly Leu Gln Ala Ala Thr
Ser His Ser Ser Ser Met Thr Lys Asp Thr 20 25 30Ile Thr Val Val Ala
Thr Gly Asn Gln Asn Thr Val Phe Glu Thr Pro 35 40 45Ser Met Val Ser
Val Val Thr Asn Asp Thr Pro Trp Ser Lys Asn Ala 50 55 60Val Thr Ser
Ala Gly Met Leu Arg Gly Val Ala Gly Leu Ser Gln Thr65 70 75 80Gly
Ala Gly Arg Thr Asn Gly Gln Thr Phe Asn Leu Arg Gly Tyr Asp 85 90
95Lys Ser Gly Val Leu Val Leu Val Asp Gly Val Arg Gln Leu Ser Asp
100 105 110Met Ala Lys Ser Ser Gly Thr Tyr Leu Asp Pro Ala Leu Val
Lys Arg 115 120 125Ile Glu Val Val Arg Gly Pro Asn Ser Ser Leu Tyr
Gly Ser Gly Gly 130 135 140Leu Gly Gly Val Val Asp Phe Arg Thr Ala
Asp Ala Ala Asp Phe Leu145 150 155 160Pro Pro Gly Glu Thr Asn Gly
Val Ser Leu Trp Gly Asn Ile Ala Ser 165 170 175Gly Asp His Ser Thr
Gly Ser Gly Leu Thr Trp Phe Gly Lys Thr Glu 180 185 190Lys Thr Asp
Ala Leu Leu Ser Val Ile Met Arg Lys Arg Gly Ser Ile 195 200 205Tyr
Gln Ser Asp Gly Glu Arg Ala Pro Asn Lys Glu Lys Pro Ala Ala 210 215
220Leu Phe Ala Lys Gly Ser Val Ser Ile Thr Asp Ser Asn Lys Ala
Gly225 230 235 240Ala Ser Leu Arg Leu Tyr Arg Asn Ser Thr Thr Glu
Pro Gly Asn Pro 245 250 255Thr Leu Thr His Gly Asp Ser Gly Leu Arg
Asp Arg Lys Thr Ala Gln 260 265 270Asn Asp Met Gln Phe Trp Tyr Gln
Tyr Ala Pro Ala Asp Asn Ser Leu 275 280 285Ile Asn Val Lys Ser Thr
Leu Tyr Leu Ser Asp Ile Thr Val Lys Thr 290 295 300Asn Gly His Asn
Lys Thr Ala Glu Trp Arg Asn Asn Arg Thr Ser Gly305 310 315 320Val
Asn Val Val Asn Arg Ser His Ser Leu Ile Phe Pro Gly Ala His 325 330
335Gln Leu Ser Tyr Gly Ala Glu Tyr Tyr Arg Gln Gln Gln Lys Pro Glu
340 345 350Gly Thr Ala Thr Leu Tyr Pro Glu Gly His Ile Asp Phe Thr
Ser Leu 355 360 365Tyr Phe Gln Asp Glu Met Thr Met Glu Ser Tyr Pro
Val Asn Ile Ile 370 375 380Val Gly Ser Arg Tyr Asp Arg Tyr Asn Ser
Phe Asn Ala Arg Ala Gly385 390 395 400Glu Leu Asn Ala Glu Arg Leu
Ser Pro Arg Ala Ala Met Ser Val Ser 405 410 415Pro Thr Asp Trp Leu
Met Met Tyr Gly Ser Ile Ser Ser Ala Phe Arg 420 425 430Ala Pro Thr
Met Ala Glu Met Tyr Arg Asp Asp Val His Phe Tyr Arg 435 440 445Lys
Gly Lys Pro Asn Tyr Trp Val Pro Asn Leu Asn Leu Lys Pro Glu 450 455
460Asn Asn Thr Thr Arg Glu Ile Gly Ala Gly Ile Gln Leu Asp Ser
Leu465 470 475 480Leu Thr Asp Asn Asp Arg Leu Gln Leu Lys Gly Gly
Tyr Phe Gly Thr 485 490 495Asp Ala Arg Asn Tyr Ile Ala Thr Arg Val
Asp Met Lys Arg Met Arg 500 505 510Ser Tyr Ser Tyr Asn Val Ser Arg
Ala Arg Ile Trp Gly Trp Asp Ile 515 520 525Gln Gly Asn Tyr Gln Ser
Asp Tyr Val Asp Trp Met Leu Ser Tyr Asn 530 535 540Arg Thr Glu Ser
Met Asp Ala Ser Ser Arg Glu Trp Leu Gly Ser Gly545 550 555 560Asn
Pro Asp Thr Leu Ile Ser Asp Ile Ser Ile Pro Val Gly His Arg 565 570
575Gly Val Tyr Ala Gly Trp Arg Ala Glu Leu Ser Ala Pro Ala Thr His
580 585 590Val Lys Lys Gly Asp Pro Cys Gln Asp Gly Tyr Ala Ile His
Ser Phe 595 600 605Ser Leu Ser Tyr Lys Pro Val Ser Val Lys Gly Phe
Glu Ala Ser Val 610 615 620Thr Leu Asp Asn Ala Phe Asn Lys Leu Ala
Met Asn Gly Lys Gly Val625 630 635 640Pro Leu Ser Gly Arg Thr Val
Asn Leu Tyr Thr Arg Tyr Gln Trp 645 650 65532102PRTEscherichia coli
32Met Lys Tyr Leu Leu Ile Phe Leu Leu Val Leu Ala Ile Phe Val Ile1
5 10 15Ser Val Thr Leu Gly Ala Gln Asn Asp Gln Gln Val Thr Phe Asn
Tyr 20 25 30Leu Leu Ala Gln Gly Glu Tyr Arg Ile Ser Thr Leu Leu Ala
Val Leu 35 40 45Phe Ala Ala Gly Phe Ala Ile Gly Trp Leu Ile Cys Gly
Leu Phe Trp 50 55 60Leu Arg Val Arg Val Ser Leu Ala Arg Ala Glu Arg
Lys Ile Lys Arg65 70 75 80Leu Glu Asn Gln Leu Ser Pro Ala Thr Asp
Val Ala Val Val Pro His 85 90 95Ser Ser Ala Ala Lys Glu
10033710PRTStevia rebaudiana 33Met Gln Ser Asp Ser Val Lys Val Ser
Pro Phe Asp Leu Val Ser Ala1 5 10 15Ala Met Asn Gly Lys Ala Met Glu
Lys Leu Asn Ala Ser Glu Ser Glu 20 25 30Asp Pro Thr Thr Leu Pro Ala
Leu Lys Met Leu Val Glu Asn Arg Glu 35 40 45Leu Leu Thr Leu Phe Thr
Thr Ser Phe Ala Val Leu Ile Gly Cys Leu 50 55 60Val Phe Leu Met Trp
Arg Arg Ser Ser Ser Lys Lys Leu Val Gln Asp65 70 75 80Pro Val Pro
Gln Val Ile Val Val Lys Lys Lys Glu Lys Glu Ser Glu 85 90 95Val Asp
Asp Gly Lys Lys Lys Val Ser Ile Phe Tyr Gly Thr Gln Thr 100 105
110Gly Thr Ala Glu Gly Phe Ala Lys Ala Leu Val Glu Glu Ala Lys Val
115 120 125Arg Tyr Glu Lys Thr Ser Phe Lys Val Ile Asp Leu Asp Asp
Tyr Ala 130 135 140Ala Asp Asp Asp Glu Tyr Glu Glu Lys Leu Lys Lys
Glu Ser Leu Ala145 150 155 160Phe Phe Phe Leu Ala Thr Tyr Gly Asp
Gly Glu Pro Thr Asp Asn Ala 165 170 175Ala Asn Phe Tyr Lys Trp Phe
Thr Glu Gly Asp Asp Lys Gly Glu Trp 180 185 190Leu Lys Lys Leu Gln
Tyr Gly Val Phe Gly Leu Gly Asn Arg Gln Tyr 195 200 205Glu His Phe
Asn Lys Ile Ala Ile Val Val Asp Asp Lys Leu Thr Glu 210 215 220Met
Gly Ala Lys Arg Leu Val Pro Val Gly Leu Gly Asp Asp Asp Gln225 230
235 240Cys Ile Glu Asp Asp Phe Thr Ala Trp Lys Glu Leu Val Trp Pro
Glu 245 250 255Leu Asp Gln Leu Leu Arg Asp Glu Asp Asp Thr Ser Val
Thr Thr Pro 260 265 270Tyr Thr Ala Ala Val Leu Glu Tyr Arg Val Val
Tyr His Asp Lys Pro 275 280 285Ala Asp Ser Tyr Ala Glu Asp Gln Thr
His Thr Asn Gly His Val Val 290 295 300His Asp Ala Gln His Pro Ser
Arg Ser Asn Val Ala Phe Lys Lys Glu305 310 315 320Leu His Thr Ser
Gln Ser Asp Arg Ser Cys Thr His Leu Glu Phe Asp 325 330 335Ile Ser
His Thr Gly Leu Ser Tyr Glu Thr Gly Asp His Val Gly Val 340 345
350Tyr Ser Glu Asn Leu Ser Glu Val Val Asp Glu Ala Leu Lys Leu Leu
355 360 365Gly Leu Ser Pro Asp Thr Tyr Phe Ser Val His Ala Asp Lys
Glu Asp 370 375 380Gly Thr Pro Ile Gly Gly Ala Ser Leu Pro Pro Pro
Phe Pro Pro Cys385 390 395 400Thr Leu Arg Asp Ala Leu Thr Arg Tyr
Ala Asp Val Leu Ser Ser Pro 405 410 415Lys Lys Val Ala Leu Leu Ala
Leu Ala Ala His Ala Ser Asp Pro Ser 420 425 430Glu Ala Asp Arg Leu
Lys Phe Leu Ala Ser Pro Ala Gly Lys Asp Glu 435 440 445Tyr Ala Gln
Trp Ile Val Ala Asn Gln Arg Ser Leu Leu Glu Val Met 450 455 460Gln
Ser Phe Pro Ser Ala Lys Pro Pro Leu Gly Val Phe Phe Ala Ala465 470
475 480Val Ala Pro Arg Leu Gln Pro Arg Tyr Tyr Ser Ile Ser Ser Ser
Pro 485 490 495Lys Met Ser Pro Asn Arg Ile His Val Thr Cys Ala Leu
Val Tyr Glu 500 505 510Thr Thr Pro Ala Gly Arg Ile His Arg Gly Leu
Cys Ser Thr Trp Met 515 520 525Lys Asn Ala Val Pro Leu Thr Glu Ser
Pro Asp Cys Ser Gln Ala Ser 530 535 540Ile Phe Val Arg Thr Ser Asn
Phe Arg Leu Pro Val Asp Pro Lys Val545 550 555 560Pro Val Ile Met
Ile Gly Pro Gly Thr Gly Leu Ala Pro Phe Arg Gly 565 570 575Phe Leu
Gln Glu Arg Leu Ala Leu Lys Glu Ser Gly Thr Glu Leu Gly 580 585
590Ser Ser Ile Phe Phe Phe Gly Cys Arg Asn Arg Lys Val Asp Phe Ile
595 600 605Tyr Glu Asp Glu Leu Asn Asn Phe Val Glu Thr Gly Ala Leu
Ser Glu 610 615 620Leu Ile Val Ala Phe Ser Arg Glu Gly Thr Ala Lys
Glu Tyr Val Gln625 630 635 640His Lys Met Ser Gln Lys Ala Ser Asp
Ile Trp Lys Leu Leu Ser Glu 645 650 655Gly Ala Tyr Leu Tyr Val Cys
Gly Asp Ala Lys Gly Met Ala Lys Asp 660 665 670Val His Arg Thr Leu
His Thr Ile Val Gln Glu Gln Gly Ser Leu Asp 675 680 685Ser Ser Lys
Ala Glu Leu Tyr Val Lys Asn Leu Gln Met Ser Gly Arg 690 695 700Tyr
Leu Arg Asp Val Trp705 71034692PRTArabidopsis thaliana 34Met Thr
Ser Ala Leu Tyr Ala Ser Asp Leu Phe Lys Gln Leu Lys Ser1 5 10 15Ile
Met Gly Thr Asp Ser Leu Ser Asp Asp Val Val Leu Val Ile Ala 20 25
30Thr Thr Ser Leu Ala Leu Val Ala Gly Phe Val Val Leu Leu Trp Lys
35 40 45Lys Thr Thr Ala Asp Arg Ser Gly Glu Leu Lys Pro Leu Met Ile
Pro 50 55 60Lys Ser Leu Met Ala Lys Asp Glu Asp Asp Asp Leu Asp Leu
Gly Ser65 70 75 80Gly Lys Thr Arg Val Ser Ile Phe Phe Gly Thr Gln
Thr Gly Thr Ala 85 90 95Glu Gly Phe Ala Lys Ala Leu Ser Glu Glu Ile
Lys Ala Arg Tyr Glu 100 105 110Lys Ala Ala Val Lys Val Ile Asp Leu
Asp Asp Tyr Ala Ala Asp Asp 115 120 125Asp Gln Tyr Glu Glu Lys Leu
Lys Lys Glu Thr Leu Ala Phe Phe Cys 130 135 140Val Ala Thr Tyr Gly
Asp Gly Glu Pro Thr Asp Asn Ala Ala Arg Phe145 150 155 160Ser Lys
Trp Phe Thr Glu Glu Asn Glu Arg Asp Ile Lys Leu Gln Gln 165 170
175Leu Ala Tyr Gly Val Phe Ala Leu Gly Asn Arg Gln Tyr Glu His Phe
180 185 190Asn Lys Ile Gly Ile Val Leu Asp Glu Glu Leu Cys Lys Lys
Gly Ala 195 200 205Lys Arg Leu Ile Glu Val Gly Leu Gly Asp Asp Asp
Gln Ser Ile Glu 210 215 220Asp Asp Phe Asn Ala Trp Lys Glu Ser Leu
Trp Ser Glu Leu Asp Lys225 230 235 240Leu Leu Lys Asp Glu Asp Asp
Lys Ser Val Ala Thr Pro Tyr Thr Ala 245 250 255Val Ile Pro Glu Tyr
Arg Val Val Thr His Asp Pro Arg Phe Thr Thr 260 265 270Gln Lys Ser
Met Glu Ser Asn Val Ala Asn Gly Asn Thr Thr Ile Asp 275 280 285Ile
His His Pro Cys Arg Val Asp Val Ala Val Gln Lys Glu Leu His 290 295
300Thr His Glu Ser Asp Arg Ser Cys Ile His Leu Glu Phe Asp Ile
Ser305 310 315 320Arg Thr Gly Ile Thr Tyr Glu Thr Gly Asp His Val
Gly Val Tyr Ala 325 330 335Glu Asn His Val Glu Ile Val Glu Glu Ala
Gly Lys Leu Leu Gly His 340 345 350Ser Leu Asp Leu Val Phe Ser Ile
His Ala Asp Lys Glu Asp Gly Ser 355 360 365Pro Leu Glu Ser Ala Val
Pro Pro Pro Phe Pro Gly Pro Cys Thr Leu 370 375 380Gly Thr Gly Leu
Ala Arg Tyr Ala Asp Leu Leu Asn Pro Pro Arg Lys385 390 395 400Ser
Ala Leu Val Ala Leu Ala Ala Tyr Ala Thr Glu Pro Ser Glu Ala 405 410
415Glu Lys Leu Lys His Leu Thr Ser Pro Asp Gly Lys Asp Glu Tyr Ser
420 425 430Gln Trp Ile Val Ala Ser Gln Arg Ser Leu Leu Glu Val Met
Ala Ala 435 440 445Phe Pro Ser Ala Lys Pro Pro Leu Gly Val Phe Phe
Ala Ala Ile Ala 450 455 460Pro Arg Leu Gln Pro Arg Tyr Tyr Ser Ile
Ser Ser Cys Gln Asp Trp465 470 475 480Ala Pro Ser Arg Val His Val
Thr Ser Ala Leu Val Tyr Gly Pro Thr 485 490 495Pro Thr Gly Arg Ile
His Lys Gly Val Cys Ser Thr Trp Met Lys Asn 500 505 510Ala Val Pro
Ala Glu Lys Ser His Glu Cys Ser Gly Ala Pro Ile Phe 515 520 525Ile
Arg Ala Ser Asn Phe Lys Leu Pro Ser Asn Pro Ser Thr Pro Ile 530 535
540Val Met Val Gly Pro Gly Thr Gly Leu Ala Pro Phe Arg Gly Phe
Leu545 550 555 560Gln Glu Arg Met Ala Leu Lys Glu Asp Gly Glu Glu
Leu Gly Ser Ser 565 570 575Leu Leu Phe Phe Gly Cys Arg Asn Arg Gln
Met Asp Phe Ile Tyr Glu 580 585 590Asp Glu Leu Asn Asn Phe Val Asp
Gln Gly Val Ile Ser Glu Leu Ile 595 600 605Met Ala Phe Ser Arg Glu
Gly Ala Gln Lys Glu Tyr Val Gln His Lys 610 615 620Met Met Glu Lys
Ala Ala Gln Val Trp Asp Leu Ile Lys Glu Glu Gly625 630 635 640Tyr
Leu Tyr Val Cys Gly Asp Ala Lys Gly Met Ala Arg Asp Val His 645 650
655Arg Thr Leu His Thr Ile Val Gln Glu Gln Glu Gly Val Ser Ser Ser
660 665 670Glu Ala Glu Ala Ile Val Lys Lys Leu Gln Thr Glu Gly Arg
Tyr Leu 675 680 685Arg Asp Val Trp 69035717PRTTaxus cuspidata 35Met
Gln Ala Asn Ser Asn Thr Val Glu Gly Ala Ser Gln Gly Lys Ser1 5 10
15Leu Leu Asp Ile Ser Arg Leu Asp His Ile Phe Ala Leu Leu Leu Asn
20 25 30Gly Lys Gly Gly Asp Leu Gly Ala Met Thr Gly Ser Ala Leu Ile
Leu 35 40 45Thr Glu Asn Ser Gln Asn Leu Met Ile Leu Thr Thr Ala Leu
Ala Val 50 55 60Leu Val Ala Cys Val Phe Phe Phe Val Trp Arg Arg Gly
Gly Ser Asp65 70 75 80Thr Gln Lys Pro Ala Val Arg Pro Thr Pro Leu
Val Lys Glu Glu Asp 85 90 95Glu Glu Glu Glu Asp Asp Ser Ala Lys Lys
Lys Val Thr Ile Phe Phe 100 105 110Gly Thr Gln Thr Gly Thr Ala Glu
Gly Phe Ala Lys Ala Leu Ala Glu 115 120 125Glu Ala Lys Ala Arg Tyr
Glu Lys Ala Val Phe Lys Val Val Asp Leu 130 135 140Asp Asn Tyr Ala
Ala Asp Asp Glu Gln Tyr Glu Glu Lys Leu Lys Lys145 150 155 160Glu
Lys Leu Ala Phe Phe Met Leu Ala Thr Tyr Gly Asp Gly Glu Pro 165 170
175Thr Asp Asn Ala Ala Arg Phe Tyr Lys Trp Phe Leu Glu Gly Lys Glu
180 185 190Arg Glu Pro Trp Leu Ser Asp Leu Thr Tyr Gly Val Phe Gly
Leu Gly 195 200 205Asn Arg Gln Tyr Glu His Phe Asn Lys Val Ala Lys
Ala Val Asp Glu 210 215 220Val Leu Ile Glu Gln Gly Ala Lys Arg Leu
Val Pro Val Gly Leu Gly225 230 235 240Asp Asp Asp Gln Cys Ile Glu
Asp Asp Phe Thr Ala Trp Arg Glu Gln 245 250 255Val Trp Pro Glu Leu
Asp Gln Leu Leu Arg Asp Glu Asp Asp Glu Pro 260 265 270Thr Ser Ala
Thr Pro Tyr Thr Ala Ala Ile Pro Glu Tyr Arg Val Glu 275 280 285Ile
Tyr Asp Ser Val Val Ser Val Tyr Glu Glu Thr His Ala Leu Lys 290 295
300Gln Asn Gly Gln Ala Val Tyr Asp Ile His His Pro Cys Arg Ser
Asn305 310 315 320Val Ala Val Arg Arg Glu Leu
His Thr Pro Leu Ser Asp Arg Ser Cys 325 330 335Ile His Leu Glu Phe
Asp Ile Ser Asp Thr Gly Leu Ile Tyr Glu Thr 340 345 350Gly Asp His
Val Gly Val His Thr Glu Asn Ser Ile Glu Thr Val Glu 355 360 365Glu
Ala Ala Lys Leu Leu Gly Tyr Gln Leu Asp Thr Ile Phe Ser Val 370 375
380His Gly Asp Lys Glu Asp Gly Thr Pro Leu Gly Gly Ser Ser Leu
Pro385 390 395 400Pro Pro Phe Pro Gly Pro Cys Thr Leu Arg Thr Ala
Leu Ala Arg Tyr 405 410 415Ala Asp Leu Leu Asn Pro Pro Arg Lys Ala
Ala Phe Leu Ala Leu Ala 420 425 430Ala His Ala Ser Asp Pro Ala Glu
Ala Glu Arg Leu Lys Phe Leu Ser 435 440 445Ser Pro Ala Gly Lys Asp
Glu Tyr Ser Gln Trp Val Thr Ala Ser Gln 450 455 460Arg Ser Leu Leu
Glu Ile Met Ala Glu Phe Pro Ser Ala Lys Pro Pro465 470 475 480Leu
Gly Val Phe Phe Ala Ala Ile Ala Pro Arg Leu Gln Pro Arg Tyr 485 490
495Tyr Ser Ile Ser Ser Ser Pro Arg Phe Ala Pro Ser Arg Ile His Val
500 505 510Thr Cys Ala Leu Val Tyr Gly Pro Ser Pro Thr Gly Arg Ile
His Lys 515 520 525Gly Val Cys Ser Asn Trp Met Lys Asn Ser Leu Pro
Ser Glu Glu Thr 530 535 540His Asp Cys Ser Trp Ala Pro Val Phe Val
Arg Gln Ser Asn Phe Lys545 550 555 560Leu Pro Ala Asp Ser Thr Thr
Pro Ile Val Met Val Gly Pro Gly Thr 565 570 575Gly Phe Ala Pro Phe
Arg Gly Phe Leu Gln Glu Arg Ala Lys Leu Gln 580 585 590Glu Ala Gly
Glu Lys Leu Gly Pro Ala Val Leu Phe Phe Gly Cys Arg 595 600 605Asn
Arg Gln Met Asp Tyr Ile Tyr Glu Asp Glu Leu Lys Gly Tyr Val 610 615
620Glu Lys Gly Ile Leu Thr Asn Leu Ile Val Ala Phe Ser Arg Glu
Gly625 630 635 640Ala Thr Lys Glu Tyr Val Gln His Lys Met Leu Glu
Lys Ala Ser Asp 645 650 655Thr Trp Ser Leu Ile Ala Gln Gly Gly Tyr
Leu Tyr Val Cys Gly Asp 660 665 670Ala Lys Gly Met Ala Arg Asp Val
His Arg Thr Leu His Thr Ile Val 675 680 685Gln Glu Gln Glu Ser Val
Asp Ser Ser Lys Ala Glu Phe Leu Val Lys 690 695 700Lys Leu Gln Met
Asp Gly Arg Tyr Leu Arg Asp Ile Trp705 710 71536705PRTArtemesia
annua 36Met Ala Gln Ser Thr Thr Ser Val Lys Leu Ser Pro Phe Asp Leu
Met1 5 10 15Thr Ala Leu Leu Asn Gly Lys Val Ser Phe Asp Thr Ser Asn
Thr Ser 20 25 30Asp Thr Asn Ile Pro Leu Ala Val Phe Met Glu Asn Arg
Glu Leu Leu 35 40 45Met Ile Leu Thr Thr Ser Val Ala Val Leu Ile Gly
Cys Val Val Val 50 55 60Leu Val Trp Arg Arg Ser Ser Ser Ala Ala Lys
Lys Ala Ala Glu Ser65 70 75 80Pro Val Ile Val Val Pro Lys Lys Val
Thr Glu Asp Glu Val Asp Asp 85 90 95Gly Arg Lys Lys Val Thr Val Phe
Phe Gly Thr Gln Thr Gly Thr Ala 100 105 110Glu Gly Phe Ala Lys Ala
Leu Val Glu Glu Ala Lys Ala Arg Tyr Glu 115 120 125Lys Ala Val Phe
Lys Val Ile Asp Leu Asp Asp Tyr Ala Ala Glu Asp 130 135 140Asp Glu
Tyr Glu Glu Lys Leu Lys Lys Glu Ser Leu Ala Phe Phe Phe145 150 155
160Leu Ala Thr Tyr Gly Asp Gly Glu Pro Thr Asp Asn Ala Ala Arg Phe
165 170 175Tyr Lys Trp Phe Thr Glu Gly Glu Glu Lys Gly Glu Trp Leu
Asp Lys 180 185 190Leu Gln Tyr Ala Val Phe Gly Leu Gly Asn Arg Gln
Tyr Glu His Phe 195 200 205Asn Lys Ile Ala Lys Val Val Asp Glu Lys
Leu Val Glu Gln Gly Ala 210 215 220Lys Arg Leu Val Pro Val Gly Met
Gly Asp Asp Asp Gln Cys Ile Glu225 230 235 240Asp Asp Phe Thr Ala
Trp Lys Glu Leu Val Trp Pro Glu Leu Asp Gln 245 250 255Leu Leu Arg
Asp Glu Asp Asp Thr Ser Val Ala Thr Pro Tyr Thr Ala 260 265 270Ala
Val Ala Glu Tyr Arg Val Val Phe His Asp Lys Pro Glu Thr Tyr 275 280
285Asp Gln Asp Gln Leu Thr Asn Gly His Ala Val His Asp Ala Gln His
290 295 300Pro Cys Arg Ser Asn Val Ala Val Lys Lys Glu Leu His Ser
Pro Leu305 310 315 320Ser Asp Arg Ser Cys Thr His Leu Glu Phe Asp
Ile Ser Asn Thr Gly 325 330 335Leu Ser Tyr Glu Thr Gly Asp His Val
Gly Val Tyr Val Glu Asn Leu 340 345 350Ser Glu Val Val Asp Glu Ala
Glu Lys Leu Ile Gly Leu Pro Pro His 355 360 365Thr Tyr Phe Ser Val
His Ala Asp Asn Glu Asp Gly Thr Pro Leu Gly 370 375 380Gly Ala Ser
Leu Pro Pro Pro Phe Pro Pro Cys Thr Leu Arg Lys Ala385 390 395
400Leu Ala Ser Tyr Ala Asp Val Leu Ser Ser Pro Lys Lys Ser Ala Leu
405 410 415Leu Ala Leu Ala Ala His Ala Thr Asp Ser Thr Glu Ala Asp
Arg Leu 420 425 430Lys Phe Leu Ala Ser Pro Ala Gly Lys Asp Glu Tyr
Ala Gln Trp Ile 435 440 445Val Ala Ser His Arg Ser Leu Leu Glu Val
Met Glu Ala Phe Pro Ser 450 455 460Ala Lys Pro Pro Leu Gly Val Phe
Phe Ala Ser Val Ala Pro Arg Leu465 470 475 480Gln Pro Arg Tyr Tyr
Ser Ile Ser Ser Ser Pro Arg Phe Ala Pro Asn 485 490 495Arg Ile His
Val Thr Cys Ala Leu Val Tyr Glu Gln Thr Pro Ser Gly 500 505 510Arg
Val His Lys Gly Val Cys Ser Thr Trp Met Lys Asn Ala Val Pro 515 520
525Met Thr Glu Ser Gln Asp Cys Ser Trp Ala Pro Ile Tyr Val Arg Thr
530 535 540Ser Asn Phe Arg Leu Pro Ser Asp Pro Lys Val Pro Val Ile
Met Ile545 550 555 560Gly Pro Gly Thr Gly Leu Ala Pro Phe Arg Gly
Phe Leu Gln Glu Arg 565 570 575Leu Ala Gln Lys Glu Ala Gly Thr Glu
Leu Gly Thr Ala Ile Leu Phe 580 585 590Phe Gly Cys Arg Asn Arg Lys
Val Asp Phe Ile Tyr Glu Asp Glu Leu 595 600 605Asn Asn Phe Val Glu
Thr Gly Ala Leu Ser Glu Leu Val Thr Ala Phe 610 615 620Ser Arg Glu
Gly Ala Thr Lys Glu Tyr Val Gln His Lys Met Thr Gln625 630 635
640Lys Ala Ser Asp Ile Trp Asn Leu Leu Ser Glu Gly Ala Tyr Leu Tyr
645 650 655Val Cys Gly Asp Ala Lys Gly Met Ala Lys Asp Val His Arg
Thr Leu 660 665 670His Thr Ile Val Gln Glu Gln Gly Ser Leu Asp Ser
Ser Lys Ala Glu 675 680 685Leu Tyr Val Lys Asn Leu Gln Met Ala Gly
Arg Tyr Leu Arg Asp Val 690 695 700Trp70537693PRTArabidopsis
thaliana 37Met Ala Thr Ser Ala Leu Tyr Ala Ser Asp Leu Phe Lys Gln
Leu Lys1 5 10 15Ser Ile Met Gly Thr Asp Ser Leu Ser Asp Asp Val Val
Leu Val Ile 20 25 30Ala Thr Thr Ser Leu Ala Leu Val Ala Gly Phe Val
Val Leu Leu Trp 35 40 45Lys Lys Thr Thr Ala Asp Arg Ser Gly Glu Leu
Lys Pro Leu Met Ile 50 55 60Pro Lys Ser Leu Met Ala Lys Asp Glu Asp
Asp Asp Leu Asp Leu Gly65 70 75 80Ser Gly Lys Thr Arg Val Ser Ile
Phe Phe Gly Thr Gln Thr Gly Thr 85 90 95Ala Glu Gly Phe Ala Lys Ala
Leu Ser Glu Glu Ile Lys Ala Arg Tyr 100 105 110Glu Lys Ala Ala Val
Lys Val Ile Asp Leu Asp Asp Tyr Ala Ala Asp 115 120 125Asp Asp Gln
Tyr Glu Glu Lys Leu Lys Lys Glu Thr Leu Ala Phe Phe 130 135 140Cys
Val Ala Thr Tyr Gly Asp Gly Glu Pro Thr Asp Asn Ala Ala Arg145 150
155 160Phe Tyr Lys Trp Phe Thr Glu Glu Asn Glu Arg Asp Ile Lys Leu
Gln 165 170 175Gln Leu Ala Tyr Gly Val Phe Ala Leu Gly Asn Arg Gln
Tyr Glu His 180 185 190Phe Asn Lys Ile Gly Ile Val Leu Asp Glu Glu
Leu Cys Lys Lys Gly 195 200 205Ala Lys Arg Leu Ile Glu Val Gly Leu
Gly Asp Asp Asp Gln Ser Ile 210 215 220Glu Asp Asp Phe Asn Ala Trp
Lys Glu Ser Leu Trp Ser Glu Leu Asp225 230 235 240Lys Leu Leu Lys
Asp Glu Asp Asp Lys Ser Val Ala Thr Pro Tyr Thr 245 250 255Ala Val
Ile Pro Glu Tyr Arg Val Val Thr His Asp Pro Arg Phe Thr 260 265
270Thr Gln Lys Ser Met Glu Ser Asn Val Ala Asn Gly Asn Thr Thr Ile
275 280 285Asp Ile His His Pro Cys Arg Val Asp Val Ala Val Gln Lys
Glu Leu 290 295 300His Thr His Glu Ser Asp Arg Ser Cys Ile His Leu
Glu Phe Asp Ile305 310 315 320Ser Arg Thr Gly Ile Thr Tyr Glu Thr
Gly Asp His Val Gly Val Tyr 325 330 335Ala Glu Asn His Val Glu Ile
Val Glu Glu Ala Gly Lys Leu Leu Gly 340 345 350His Ser Leu Asp Leu
Val Phe Ser Ile His Ala Asp Lys Glu Asp Gly 355 360 365Ser Pro Leu
Glu Ser Ala Val Pro Pro Pro Phe Pro Gly Pro Cys Thr 370 375 380Leu
Gly Thr Gly Leu Ala Arg Tyr Ala Asp Leu Leu Asn Pro Pro Arg385 390
395 400Lys Ser Ala Leu Val Ala Leu Ala Ala Tyr Ala Thr Glu Pro Ser
Glu 405 410 415Ala Glu Lys Leu Lys His Leu Thr Ser Pro Asp Gly Lys
Asp Glu Tyr 420 425 430Ser Gln Trp Ile Val Ala Ser Gln Arg Ser Leu
Leu Glu Val Met Ala 435 440 445Ala Phe Pro Ser Ala Lys Pro Pro Leu
Gly Val Phe Phe Ala Ala Ile 450 455 460Ala Pro Arg Leu Gln Pro Arg
Tyr Tyr Ser Ile Ser Ser Ser Pro Arg465 470 475 480Leu Ala Pro Ser
Arg Val His Val Thr Ser Ala Leu Val Tyr Gly Pro 485 490 495Thr Pro
Thr Gly Arg Ile His Lys Gly Val Cys Ser Thr Trp Met Lys 500 505
510Asn Ala Val Pro Ala Glu Lys Ser His Glu Cys Ser Gly Ala Pro Ile
515 520 525Phe Ile Arg Ala Ser Asn Phe Lys Leu Pro Ser Asn Pro Ser
Thr Pro 530 535 540Ile Val Met Val Gly Pro Gly Thr Gly Leu Ala Pro
Phe Arg Gly Phe545 550 555 560Leu Gln Glu Arg Met Ala Leu Lys Glu
Asp Gly Glu Glu Leu Gly Ser 565 570 575Ser Leu Leu Phe Phe Gly Cys
Arg Asn Arg Gln Met Asp Phe Ile Tyr 580 585 590Glu Asp Glu Leu Asn
Asn Phe Val Asp Gln Gly Val Ile Ser Glu Leu 595 600 605Ile Met Ala
Phe Ser Arg Glu Gly Ala Gln Lys Glu Tyr Val Gln His 610 615 620Lys
Met Met Glu Lys Ala Ala Gln Val Trp Asp Leu Ile Lys Glu Glu625 630
635 640Gly Tyr Leu Tyr Val Cys Gly Asp Ala Lys Gly Met Ala Arg Asp
Val 645 650 655His Arg Thr Leu His Thr Ile Val Gln Glu Gln Glu Gly
Val Ser Ser 660 665 670Ser Glu Ala Glu Ala Ile Val Lys Lys Leu Gln
Thr Glu Gly Arg Tyr 675 680 685Leu Arg Asp Val Trp
69038712PRTArabidopsis thaliana 38Met Ala Ser Ser Ser Ser Ser Ser
Ser Thr Ser Met Ile Asp Leu Met1 5 10 15Ala Ala Ile Ile Lys Gly Glu
Pro Val Ile Val Ser Asp Pro Ala Asn 20 25 30Ala Ser Ala Tyr Glu Ser
Val Ala Ala Glu Leu Ser Ser Met Leu Ile 35 40 45Glu Asn Arg Gln Phe
Ala Met Ile Val Thr Thr Ser Ile Ala Val Leu 50 55 60Ile Gly Cys Ile
Val Met Leu Val Trp Arg Arg Ser Gly Ser Gly Asn65 70 75 80Ser Lys
Arg Val Glu Pro Leu Lys Pro Leu Val Ile Lys Pro Arg Glu 85 90 95Glu
Glu Ile Asp Asp Gly Arg Lys Lys Val Thr Ile Phe Phe Gly Thr 100 105
110Gln Thr Gly Thr Ala Glu Gly Phe Ala Lys Ala Leu Gly Glu Glu Ala
115 120 125Lys Ala Arg Tyr Glu Lys Thr Arg Phe Lys Ile Val Asp Leu
Asp Asp 130 135 140Tyr Ala Ala Asp Asp Asp Glu Tyr Glu Glu Lys Leu
Lys Lys Glu Asp145 150 155 160Val Ala Phe Phe Phe Leu Ala Thr Tyr
Gly Asp Gly Glu Pro Thr Asp 165 170 175Asn Ala Ala Arg Phe Tyr Lys
Trp Phe Thr Glu Gly Asn Asp Arg Gly 180 185 190Glu Trp Leu Lys Asn
Leu Lys Tyr Gly Val Phe Gly Leu Gly Asn Arg 195 200 205Gln Tyr Glu
His Phe Asn Lys Val Ala Lys Val Val Asp Asp Ile Leu 210 215 220Val
Glu Gln Gly Ala Gln Arg Leu Val Gln Val Gly Leu Gly Asp Asp225 230
235 240Asp Gln Cys Ile Glu Asp Asp Phe Thr Ala Trp Arg Glu Ala Leu
Trp 245 250 255Pro Glu Leu Asp Thr Ile Leu Arg Glu Glu Gly Asp Thr
Ala Val Ala 260 265 270Thr Pro Tyr Thr Ala Ala Val Leu Glu Tyr Arg
Val Ser Ile His Asp 275 280 285Ser Glu Asp Ala Lys Phe Asn Asp Ile
Asn Met Ala Asn Gly Asn Gly 290 295 300Tyr Thr Val Phe Asp Ala Gln
His Pro Tyr Lys Ala Asn Val Ala Val305 310 315 320Lys Arg Glu Leu
His Thr Pro Glu Ser Asp Arg Ser Cys Ile His Leu 325 330 335Glu Phe
Asp Ile Ala Gly Ser Gly Leu Thr Tyr Glu Thr Gly Asp His 340 345
350Val Gly Val Leu Cys Asp Asn Leu Ser Glu Thr Val Asp Glu Ala Leu
355 360 365Arg Leu Leu Asp Met Ser Pro Asp Thr Tyr Phe Ser Leu His
Ala Glu 370 375 380Lys Glu Asp Gly Thr Pro Ile Ser Ser Ser Leu Pro
Pro Pro Phe Pro385 390 395 400Pro Cys Asn Leu Arg Thr Ala Leu Thr
Arg Tyr Ala Cys Leu Leu Ser 405 410 415Ser Pro Lys Lys Ser Ala Leu
Val Ala Leu Ala Ala His Ala Ser Asp 420 425 430Pro Thr Glu Ala Glu
Arg Leu Lys His Leu Ala Ser Pro Ala Gly Lys 435 440 445Asp Glu Tyr
Ser Lys Trp Val Val Glu Ser Gln Arg Ser Leu Leu Glu 450 455 460Val
Met Ala Glu Phe Pro Ser Ala Lys Pro Pro Leu Gly Val Phe Phe465 470
475 480Ala Gly Val Ala Pro Arg Leu Gln Pro Arg Phe Tyr Ser Ile Ser
Ser 485 490 495Ser Pro Lys Ile Ala Glu Thr Arg Ile His Val Thr Cys
Ala Leu Val 500 505 510Tyr Glu Lys Met Pro Thr Gly Arg Ile His Lys
Gly Val Cys Ser Thr 515 520 525Trp Met Lys Asn Ala Val Pro Tyr Glu
Lys Ser Glu Asn Cys Ser Ser 530 535 540Ala Pro Ile Phe Val Arg Gln
Ser Asn Phe Lys Leu Pro Ser Asp Ser545 550 555 560Lys Val Pro Ile
Ile Met Ile Gly Pro Gly Thr Gly Leu Ala Pro Phe 565 570 575Arg Gly
Phe Leu Gln Glu Arg Leu Ala Leu Val Glu Ser Gly Val Glu 580 585
590Leu Gly Pro Ser Val Leu Phe Phe Gly Cys Arg Asn Arg Arg Met Asp
595 600 605Phe Ile Tyr Glu Glu Glu Leu Gln Arg Phe Val Glu Ser Gly
Ala Leu 610 615 620Ala Glu Leu Ser Val Ala Phe Ser Arg Glu Gly Pro
Thr Lys Glu Tyr625 630 635 640Val Gln His Lys Met Met Asp Lys Ala
Ser Asp Ile Trp Asn Met Ile 645 650 655Ser Gln Gly Ala Tyr Leu Tyr
Val Cys Gly Asp Ala Lys Gly Met Ala 660 665 670Arg Asp Val His Arg
Ser
Leu His Thr Ile Ala Gln Glu Gln Gly Ser 675 680 685Met Asp Ser Thr
Lys Ala Glu Gly Phe Val Lys Asn Leu Gln Thr Ser 690 695 700Gly Arg
Tyr Leu Arg Asp Val Trp705 71039713PRTArabidopsis thaliana 39Met
Ala Ser Ser Ser Ser Ser Ser Ser Thr Ser Met Ile Asp Leu Met1 5 10
15Ala Ala Ile Ile Lys Gly Glu Pro Val Ile Val Ser Asp Pro Ala Asn
20 25 30Ala Ser Ala Tyr Glu Ser Val Ala Ala Glu Leu Ser Ser Met Leu
Ile 35 40 45Glu Asn Arg Gln Phe Ala Met Ile Val Thr Thr Ser Ile Ala
Val Leu 50 55 60Ile Gly Cys Ile Val Met Leu Val Trp Arg Arg Ser Gly
Ser Gly Asn65 70 75 80Ser Lys Arg Val Glu Pro Leu Lys Pro Leu Val
Ile Lys Pro Arg Glu 85 90 95Glu Glu Ile Asp Asp Gly Arg Lys Lys Val
Thr Ile Phe Phe Gly Thr 100 105 110Gln Thr Gly Thr Ala Glu Gly Phe
Ala Lys Ala Leu Gly Glu Glu Ala 115 120 125Lys Ala Arg Tyr Glu Lys
Thr Arg Phe Lys Ile Val Asp Leu Asp Asp 130 135 140Tyr Ala Ala Asp
Asp Asp Glu Tyr Glu Glu Lys Leu Lys Lys Glu Asp145 150 155 160Val
Ala Phe Phe Phe Leu Ala Thr Tyr Gly Asp Gly Glu Pro Thr Asp 165 170
175Asn Ala Ala Arg Phe Tyr Lys Trp Phe Thr Glu Gly Asn Asp Arg Gly
180 185 190Glu Trp Leu Lys Asn Leu Lys Tyr Gly Val Phe Gly Leu Gly
Asn Arg 195 200 205Gln Tyr Glu His Phe Asn Lys Val Ala Lys Val Val
Asp Asp Ile Leu 210 215 220Val Glu Gln Gly Ala Gln Arg Leu Val Gln
Val Gly Leu Gly Asp Asp225 230 235 240Asp Gln Cys Ile Glu Asp Asp
Phe Thr Ala Trp Arg Glu Ala Leu Trp 245 250 255Pro Glu Leu Asp Thr
Ile Leu Arg Glu Glu Gly Asp Thr Ala Val Ala 260 265 270Thr Pro Tyr
Thr Ala Ala Val Leu Glu Tyr Arg Val Ser Ile His Asp 275 280 285Ser
Glu Asp Ala Lys Phe Asn Asp Ile Thr Leu Ala Asn Gly Asn Gly 290 295
300Tyr Thr Val Phe Asp Ala Gln His Pro Tyr Lys Ala Asn Val Ala
Val305 310 315 320Lys Arg Glu Leu His Thr Pro Glu Ser Asp Arg Ser
Cys Ile His Leu 325 330 335Glu Phe Asp Ile Ala Gly Ser Gly Leu Thr
Met Lys Leu Gly Asp His 340 345 350Val Gly Val Leu Cys Asp Asn Leu
Ser Glu Thr Val Asp Glu Ala Leu 355 360 365Arg Leu Leu Asp Met Ser
Pro Asp Thr Tyr Phe Ser Leu His Ala Glu 370 375 380Lys Glu Asp Gly
Thr Pro Ile Ser Ser Ser Leu Pro Pro Pro Phe Pro385 390 395 400Pro
Cys Asn Leu Arg Thr Ala Leu Thr Arg Tyr Ala Cys Leu Leu Ser 405 410
415Ser Pro Lys Lys Ser Ala Leu Val Ala Leu Ala Ala His Ala Ser Asp
420 425 430Pro Thr Glu Ala Glu Arg Leu Lys His Leu Ala Ser Pro Ala
Gly Lys 435 440 445Asp Glu Tyr Ser Lys Trp Val Val Glu Ser Gln Arg
Ser Leu Leu Glu 450 455 460Val Met Ala Glu Phe Pro Ser Ala Lys Pro
Pro Leu Gly Val Phe Phe465 470 475 480Ala Gly Val Ala Pro Arg Leu
Gln Pro Arg Phe Tyr Ser Ile Ser Ser 485 490 495Ser Pro Lys Ile Ala
Glu Thr Arg Ile His Val Thr Cys Ala Leu Val 500 505 510Tyr Glu Lys
Met Pro Thr Gly Arg Ile His Lys Gly Val Cys Ser Thr 515 520 525Trp
Met Lys Asn Ala Val Pro Tyr Glu Lys Ser Glu Lys Leu Phe Leu 530 535
540Gly Arg Pro Ile Phe Val Arg Gln Ser Asn Phe Lys Leu Pro Ser
Asp545 550 555 560Ser Lys Val Pro Ile Ile Met Ile Gly Pro Gly Thr
Gly Leu Ala Pro 565 570 575Phe Arg Gly Phe Leu Gln Glu Arg Leu Ala
Leu Val Glu Ser Gly Val 580 585 590Glu Leu Gly Pro Ser Val Leu Phe
Phe Gly Cys Arg Asn Arg Arg Met 595 600 605Asp Phe Ile Tyr Glu Glu
Glu Leu Gln Arg Phe Val Glu Ser Gly Ala 610 615 620Leu Ala Glu Leu
Ser Val Ala Phe Ser Arg Glu Gly Pro Thr Lys Glu625 630 635 640Tyr
Val Gln His Lys Met Met Asp Lys Ala Ser Asp Ile Trp Asn Met 645 650
655Ile Ser Gln Gly Ala Tyr Leu Tyr Val Cys Gly Asp Ala Lys Gly Met
660 665 670Ala Arg Asp Val His Arg Ser Leu His Thr Ile Ala Gln Glu
Gln Gly 675 680 685Ser Met Asp Ser Thr Lys Ala Glu Gly Phe Val Lys
Asn Leu Gln Thr 690 695 700Ser Gly Arg Tyr Leu Arg Asp Val Trp705
71040711PRTStevia rebaudiana 40Met Ala Gln Ser Asp Ser Val Lys Val
Ser Pro Phe Asp Leu Val Ser1 5 10 15Ala Ala Met Asn Gly Lys Ala Met
Glu Lys Leu Asn Ala Ser Glu Ser 20 25 30Glu Asp Pro Thr Thr Leu Pro
Ala Leu Lys Met Leu Val Glu Asn Arg 35 40 45Glu Leu Leu Thr Leu Phe
Thr Thr Ser Phe Ala Val Leu Ile Gly Cys 50 55 60Leu Val Phe Leu Met
Trp Arg Arg Ser Ser Ser Lys Lys Leu Val Gln65 70 75 80Asp Pro Val
Pro Gln Val Ile Val Val Lys Lys Lys Glu Lys Glu Ser 85 90 95Glu Val
Asp Asp Gly Lys Lys Lys Val Ser Ile Phe Tyr Gly Thr Gln 100 105
110Thr Gly Thr Ala Glu Gly Phe Ala Lys Ala Leu Val Glu Glu Ala Lys
115 120 125Val Arg Tyr Glu Lys Thr Ser Phe Lys Val Ile Asp Leu Asp
Asp Tyr 130 135 140Ala Ala Asp Asp Asp Glu Tyr Glu Glu Lys Leu Lys
Lys Glu Ser Leu145 150 155 160Ala Phe Phe Phe Leu Ala Thr Tyr Gly
Asp Gly Glu Pro Thr Asp Asn 165 170 175Ala Ala Asn Phe Tyr Lys Trp
Phe Thr Glu Gly Asp Asp Lys Gly Glu 180 185 190Leu Leu Lys Lys Leu
Gln Tyr Gly Val Phe Gly Leu Gly Asn Arg Gln 195 200 205Tyr Glu His
Phe Asn Lys Ile Ala Ile Val Val Asp Asp Lys Leu Thr 210 215 220Glu
Met Gly Ala Lys Arg Leu Val Pro Val Gly Leu Gly Asp Asp Asp225 230
235 240Gln Cys Ile Glu Asp Asp Phe Thr Ala Trp Lys Glu Leu Val Trp
Pro 245 250 255Glu Leu Asp Gln Leu Leu Arg Asp Glu Asp Asp Thr Ser
Val Thr Thr 260 265 270Pro Tyr Thr Ala Ala Val Leu Glu Tyr Arg Val
Val Tyr His Asp Lys 275 280 285Pro Ala Asp Ser Tyr Ala Glu Asp Gln
Thr His Thr Asn Gly His Val 290 295 300Val His Asp Ala Gln His Pro
Ser Arg Ser Asn Val Ala Phe Lys Lys305 310 315 320Glu Leu His Thr
Ser Gln Ser Asp Arg Ser Cys Thr His Leu Glu Phe 325 330 335Asp Ile
Ser His Thr Gly Leu Ser Tyr Glu Thr Gly Asp His Val Gly 340 345
350Val Tyr Ser Glu Asn Leu Ser Glu Val Val Asp Glu Ala Leu Lys Leu
355 360 365Leu Gly Leu Ser Pro Asp Thr Tyr Phe Ser Val His Ala Asp
Lys Glu 370 375 380Asp Gly Thr Pro Ile Gly Gly Ala Ser Leu Pro Pro
Pro Phe Pro Pro385 390 395 400Cys Thr Leu Arg Asp Ala Leu Thr Arg
Tyr Ala Asp Val Leu Ser Ser 405 410 415Pro Lys Lys Val Ala Leu Leu
Ala Leu Ala Ala His Ala Ser Asp Pro 420 425 430Ser Glu Ala Asp Arg
Leu Lys Phe Leu Ala Ser Pro Ala Gly Lys Asp 435 440 445Glu Tyr Ala
Gln Trp Ile Val Ala Asn Gln Arg Ser Leu Leu Glu Val 450 455 460Met
Gln Ser Phe Pro Ser Ala Lys Pro Pro Leu Gly Val Phe Phe Ala465 470
475 480Ala Val Ala Pro Arg Leu Gln Pro Arg Tyr Tyr Ser Ile Ser Ser
Ser 485 490 495Pro Lys Met Ser Pro Asn Arg Ile His Val Thr Cys Ala
Leu Val Tyr 500 505 510Glu Thr Thr Pro Ala Gly Arg Ile His Arg Gly
Leu Cys Ser Thr Trp 515 520 525Met Lys Asn Ala Val Pro Leu Thr Glu
Ser Pro Asp Cys Ser Gln Ala 530 535 540Ser Ile Phe Val Arg Thr Ser
Asn Phe Arg Leu Pro Val Asp Pro Lys545 550 555 560Val Pro Val Ile
Met Ile Gly Pro Gly Thr Gly Leu Ala Pro Phe Arg 565 570 575Gly Phe
Leu Gln Glu Arg Leu Ala Leu Lys Glu Ser Gly Thr Glu Leu 580 585
590Gly Ser Ser Ile Phe Phe Phe Gly Cys Arg Asn Arg Lys Val Asp Phe
595 600 605Ile Tyr Glu Asp Glu Leu Asn Asn Phe Val Glu Thr Gly Ala
Leu Ser 610 615 620Glu Leu Ile Val Ala Phe Ser Arg Glu Gly Thr Ala
Lys Glu Tyr Val625 630 635 640Gln His Lys Met Ser Gln Lys Ala Ser
Asp Ile Trp Lys Leu Leu Ser 645 650 655Glu Gly Ala Tyr Leu Tyr Val
Cys Gly Asp Ala Lys Gly Met Ala Lys 660 665 670Asp Val His Arg Thr
Leu His Thr Ile Val Gln Glu Gln Gly Ser Leu 675 680 685Asp Ser Ser
Lys Ala Glu Leu Tyr Val Lys Asn Leu Gln Met Ser Gly 690 695 700Arg
Tyr Leu Arg Asp Val Trp705 71041710PRTStevia rebaudiana 41Met Ala
Gln Ser Glu Ser Val Glu Ala Ser Thr Ile Asp Leu Met Thr1 5 10 15Ala
Val Leu Lys Asp Thr Val Ile Asp Thr Ala Asn Ala Ser Asp Asn 20 25
30Gly Asp Ser Lys Met Pro Pro Ala Leu Ala Met Met Phe Glu Ile Arg
35 40 45Asp Leu Leu Leu Ile Leu Thr Thr Ser Val Ala Val Leu Val Gly
Cys 50 55 60Phe Val Val Leu Val Trp Lys Arg Ser Ser Gly Lys Lys Ser
Gly Lys65 70 75 80Glu Leu Glu Pro Pro Lys Ile Val Val Pro Lys Arg
Arg Leu Glu Gln 85 90 95Glu Val Asp Asp Gly Lys Lys Lys Val Thr Ile
Phe Phe Gly Thr Gln 100 105 110Thr Gly Thr Ala Glu Gly Phe Ala Lys
Ala Leu Phe Glu Glu Ala Lys 115 120 125Ala Arg Tyr Glu Lys Ala Ala
Phe Lys Val Ile Asp Leu Asp Asp Tyr 130 135 140Ala Ala Asp Leu Asp
Glu Tyr Ala Glu Lys Leu Lys Lys Glu Thr Tyr145 150 155 160Ala Phe
Phe Phe Leu Ala Thr Tyr Gly Asp Gly Glu Pro Thr Asp Asn 165 170
175Ala Ala Lys Phe Tyr Lys Trp Phe Thr Glu Gly Asp Glu Lys Gly Val
180 185 190Trp Leu Gln Lys Leu Gln Tyr Gly Val Phe Gly Leu Gly Asn
Arg Gln 195 200 205Tyr Glu His Phe Asn Lys Ile Gly Ile Val Val Asp
Asp Gly Leu Thr 210 215 220Glu Gln Gly Ala Lys Arg Ile Val Pro Val
Gly Leu Gly Asp Asp Asp225 230 235 240Gln Ser Ile Glu Asp Asp Phe
Ser Ala Trp Lys Glu Leu Val Trp Pro 245 250 255Glu Leu Asp Leu Leu
Leu Arg Asp Glu Asp Asp Lys Ala Ala Ala Thr 260 265 270Pro Tyr Thr
Ala Ala Ile Pro Glu Tyr Arg Val Val Phe His Asp Lys 275 280 285Pro
Asp Ala Phe Ser Asp Asp His Thr Gln Thr Asn Gly His Ala Val 290 295
300His Asp Ala Gln His Pro Cys Arg Ser Asn Val Ala Val Lys Lys
Glu305 310 315 320Leu His Thr Pro Glu Ser Asp Arg Ser Cys Thr His
Leu Glu Phe Asp 325 330 335Ile Ser His Thr Gly Leu Ser Tyr Glu Thr
Gly Asp His Val Gly Val 340 345 350Tyr Cys Glu Asn Leu Ile Glu Val
Val Glu Glu Ala Gly Lys Leu Leu 355 360 365Gly Leu Ser Thr Asp Thr
Tyr Phe Ser Leu His Ile Asp Asn Glu Asp 370 375 380Gly Ser Pro Leu
Gly Gly Pro Ser Leu Gln Pro Pro Phe Pro Pro Cys385 390 395 400Thr
Leu Arg Lys Ala Leu Thr Asn Tyr Ala Asp Leu Leu Ser Ser Pro 405 410
415Lys Lys Ser Thr Leu Leu Ala Leu Ala Ala His Ala Ser Asp Pro Thr
420 425 430Glu Ala Asp Arg Leu Arg Phe Leu Ala Ser Arg Glu Gly Lys
Asp Glu 435 440 445Tyr Ala Glu Trp Val Val Ala Asn Gln Arg Ser Leu
Leu Glu Val Met 450 455 460Glu Ala Phe Pro Ser Ala Arg Pro Pro Leu
Gly Val Phe Phe Ala Ala465 470 475 480Val Ala Pro Arg Leu Gln Pro
Arg Tyr Tyr Ser Ile Ser Ser Ser Pro 485 490 495Lys Met Glu Pro Asn
Arg Ile His Val Thr Cys Ala Leu Val Tyr Glu 500 505 510Lys Thr Pro
Ala Gly Arg Ile His Lys Gly Ile Cys Ser Thr Trp Met 515 520 525Lys
Asn Ala Val Pro Leu Thr Glu Ser Gln Asp Cys Ser Trp Ala Pro 530 535
540Ile Phe Val Arg Thr Ser Asn Phe Arg Leu Pro Ile Asp Pro Lys
Val545 550 555 560Pro Val Ile Met Ile Gly Pro Gly Thr Gly Leu Ala
Pro Phe Arg Gly 565 570 575Phe Leu Gln Glu Arg Leu Ala Leu Lys Glu
Ser Gly Thr Glu Leu Gly 580 585 590Ser Ser Ile Leu Phe Phe Gly Cys
Arg Asn Arg Lys Val Asp Tyr Ile 595 600 605Tyr Glu Asn Glu Leu Asn
Asn Phe Val Glu Asn Gly Ala Leu Ser Glu 610 615 620Leu Asp Val Ala
Phe Ser Arg Asp Gly Pro Thr Lys Glu Tyr Val Gln625 630 635 640His
Lys Met Thr Gln Lys Ala Ser Glu Ile Trp Asn Met Leu Ser Glu 645 650
655Gly Ala Tyr Leu Tyr Val Cys Gly Asp Ala Lys Gly Met Ala Lys Asp
660 665 670Val His Arg Thr Leu His Thr Ile Val Gln Glu Gln Gly Ser
Leu Asp 675 680 685Ser Ser Lys Ala Glu Leu Tyr Val Lys Asn Leu Gln
Met Ser Gly Arg 690 695 700Tyr Leu Arg Asp Val Trp705
71042708PRTStevia rebaudiana 42Met Ala Gln Ser Asn Ser Val Lys Ile
Ser Pro Leu Asp Leu Val Thr1 5 10 15Ala Leu Phe Ser Gly Lys Val Leu
Asp Thr Ser Asn Ala Ser Glu Ser 20 25 30Gly Glu Ser Ala Met Leu Pro
Thr Ile Ala Met Ile Met Glu Asn Arg 35 40 45Glu Leu Leu Met Ile Leu
Thr Thr Ser Val Ala Val Leu Ile Gly Cys 50 55 60Val Val Val Leu Val
Trp Arg Arg Ser Ser Thr Lys Lys Ser Ala Leu65 70 75 80Glu Pro Pro
Val Ile Val Val Pro Lys Arg Val Gln Glu Glu Glu Val 85 90 95Asp Asp
Gly Lys Lys Lys Val Thr Val Phe Phe Gly Thr Gln Thr Gly 100 105
110Thr Ala Glu Gly Phe Ala Lys Ala Leu Val Glu Glu Ala Lys Ala Arg
115 120 125Tyr Glu Lys Ala Val Phe Lys Val Ile Asp Leu Asp Asp Tyr
Ala Ala 130 135 140Asp Asp Asp Glu Tyr Glu Glu Lys Leu Lys Lys Glu
Ser Leu Ala Phe145 150 155 160Phe Phe Leu Ala Thr Tyr Gly Asp Gly
Glu Pro Thr Asp Asn Ala Ala 165 170 175Arg Phe Tyr Lys Trp Phe Thr
Glu Gly Asp Ala Lys Gly Glu Trp Leu 180 185 190Asn Lys Leu Gln Tyr
Gly Val Phe Gly Leu Gly Asn Arg Gln Tyr Glu 195 200 205His Phe Asn
Lys Ile Ala Lys Val Val Asp Asp Gly Leu Val Glu Gln 210 215 220Gly
Ala Lys Arg Leu Val Pro Val Gly Leu Gly Asp Asp Asp Gln Cys225 230
235 240Ile Glu Asp Asp Phe Thr Ala Trp Lys Glu Leu Val Trp Pro Glu
Leu 245 250 255Asp Gln Leu Leu Arg Asp Glu Asp Asp Thr Thr Val Ala
Thr Pro Tyr 260 265 270Thr Ala Ala Val Ala Glu Tyr Arg Val Val Phe
His Glu Lys Pro Asp 275 280 285Ala Leu Ser Glu Asp Tyr Ser Tyr Thr
Asn Gly His
Ala Val His Asp 290 295 300Ala Gln His Pro Cys Arg Ser Asn Val Ala
Val Lys Lys Glu Leu His305 310 315 320Ser Pro Glu Ser Asp Arg Ser
Cys Thr His Leu Glu Phe Asp Ile Ser 325 330 335Asn Thr Gly Leu Ser
Tyr Glu Thr Gly Asp His Val Gly Val Tyr Cys 340 345 350Glu Asn Leu
Ser Glu Val Val Asn Asp Ala Glu Arg Leu Val Gly Leu 355 360 365Pro
Pro Asp Thr Tyr Phe Ser Ile His Thr Asp Ser Glu Asp Gly Ser 370 375
380Pro Leu Gly Gly Ala Ser Leu Pro Pro Pro Phe Pro Pro Cys Thr
Leu385 390 395 400Arg Lys Ala Leu Thr Cys Tyr Ala Asp Val Leu Ser
Ser Pro Lys Lys 405 410 415Ser Ala Leu Leu Ala Leu Ala Ala His Ala
Thr Asp Pro Ser Glu Ala 420 425 430Asp Arg Leu Lys Phe Leu Ala Ser
Pro Ala Gly Lys Asp Glu Tyr Ser 435 440 445Gln Trp Ile Val Ala Ser
Gln Arg Ser Leu Leu Glu Val Met Glu Ala 450 455 460Phe Pro Ser Ala
Lys Pro Ser Leu Gly Val Phe Phe Ala Ser Val Ala465 470 475 480Pro
Arg Leu Gln Pro Arg Tyr Tyr Ser Ile Ser Ser Ser Pro Lys Met 485 490
495Ala Pro Asp Arg Ile His Val Thr Cys Ala Leu Val Tyr Glu Lys Thr
500 505 510Pro Ala Gly Arg Ile His Lys Gly Val Cys Ser Thr Trp Met
Lys Asn 515 520 525Ala Val Pro Met Thr Glu Ser Gln Asp Cys Ser Trp
Ala Pro Ile Tyr 530 535 540Val Arg Thr Ser Asn Phe Arg Leu Pro Ser
Asp Pro Lys Val Pro Val545 550 555 560Ile Met Ile Gly Pro Gly Thr
Gly Leu Ala Pro Phe Arg Gly Phe Leu 565 570 575Gln Glu Arg Leu Ala
Leu Lys Glu Ala Gly Thr Asp Leu Gly Leu Ser 580 585 590Ile Leu Phe
Phe Gly Cys Arg Asn Arg Lys Val Asp Phe Ile Tyr Glu 595 600 605Asn
Glu Leu Asn Asn Phe Val Glu Thr Gly Ala Leu Ser Glu Leu Ile 610 615
620Val Ala Phe Ser Arg Glu Gly Pro Thr Lys Glu Tyr Val Gln His
Lys625 630 635 640Met Ser Glu Lys Ala Ser Asp Ile Trp Asn Leu Leu
Ser Glu Gly Ala 645 650 655Tyr Leu Tyr Val Cys Gly Asp Ala Lys Gly
Met Ala Lys Asp Val His 660 665 670Arg Thr Leu His Thr Ile Val Gln
Glu Gln Gly Ser Leu Asp Ser Ser 675 680 685Lys Ala Glu Leu Tyr Val
Lys Asn Leu Gln Met Ser Gly Arg Tyr Leu 690 695 700Arg Asp Val
Trp70543706PRTPelargonium graveolens 43Met Ala Gln Ser Ser Ser Gly
Ser Met Ser Pro Phe Asp Phe Met Thr1 5 10 15Ala Ile Ile Lys Gly Lys
Met Glu Pro Ser Asn Ala Ser Leu Gly Ala 20 25 30Ala Gly Glu Val Thr
Ala Met Ile Leu Asp Asn Arg Glu Leu Val Met 35 40 45Ile Leu Thr Thr
Ser Ile Ala Val Leu Ile Gly Cys Val Val Val Phe 50 55 60Ile Trp Arg
Arg Ser Ser Ser Gln Thr Pro Thr Ala Val Gln Pro Leu65 70 75 80Lys
Pro Leu Leu Ala Lys Glu Thr Glu Ser Glu Val Asp Asp Gly Lys 85 90
95Gln Lys Val Thr Ile Phe Phe Gly Thr Gln Thr Gly Thr Ala Glu Gly
100 105 110Phe Ala Lys Ala Leu Ala Asp Glu Ala Lys Ala Arg Tyr Asp
Lys Val 115 120 125Thr Phe Lys Val Val Asp Leu Asp Asp Tyr Ala Ala
Asp Asp Glu Glu 130 135 140Tyr Glu Glu Lys Leu Lys Lys Glu Thr Leu
Ala Phe Phe Phe Leu Ala145 150 155 160Thr Tyr Gly Asp Gly Glu Pro
Thr Asp Asn Ala Ala Arg Phe Tyr Lys 165 170 175Trp Phe Leu Glu Gly
Lys Glu Arg Gly Glu Trp Leu Gln Asn Leu Lys 180 185 190Phe Gly Val
Phe Gly Leu Gly Asn Arg Gln Tyr Glu His Phe Asn Lys 195 200 205Ile
Ala Ile Val Val Asp Glu Ile Leu Ala Glu Gln Gly Gly Lys Arg 210 215
220Leu Ile Ser Val Gly Leu Gly Asp Asp Asp Gln Cys Ile Glu Asp
Asp225 230 235 240Phe Thr Ala Trp Arg Glu Ser Leu Trp Pro Glu Leu
Asp Gln Leu Leu 245 250 255Arg Asp Glu Asp Asp Thr Thr Val Ser Thr
Pro Tyr Thr Ala Ala Val 260 265 270Leu Glu Tyr Arg Val Val Phe His
Asp Pro Ala Asp Ala Pro Thr Leu 275 280 285Glu Lys Ser Tyr Ser Asn
Ala Asn Gly His Ser Val Val Asp Ala Gln 290 295 300His Pro Leu Arg
Ala Asn Val Ala Val Arg Arg Glu Leu His Thr Pro305 310 315 320Ala
Ser Asp Arg Ser Cys Thr His Leu Glu Phe Asp Ile Ser Gly Thr 325 330
335Gly Ile Ala Tyr Glu Thr Gly Asp His Val Gly Val Tyr Cys Glu Asn
340 345 350Leu Ala Glu Thr Val Glu Glu Ala Leu Glu Leu Leu Gly Leu
Ser Pro 355 360 365Asp Thr Tyr Phe Ser Val His Ala Asp Lys Glu Asp
Gly Thr Pro Leu 370 375 380Ser Gly Ser Ser Leu Pro Pro Pro Phe Pro
Pro Cys Thr Leu Arg Thr385 390 395 400Ala Leu Thr Leu His Ala Asp
Leu Leu Ser Ser Pro Lys Lys Ser Ala 405 410 415Leu Leu Ala Leu Ala
Ala His Ala Ser Asp Pro Thr Glu Ala Asp Arg 420 425 430Leu Arg His
Leu Ala Ser Pro Ala Gly Lys Asp Glu Tyr Ala Gln Trp 435 440 445Ile
Val Ala Ser Gln Arg Ser Leu Leu Glu Val Met Ala Glu Phe Pro 450 455
460Ser Ala Lys Pro Pro Leu Gly Val Phe Phe Ala Ser Val Ala Pro
Arg465 470 475 480Leu Gln Pro Arg Tyr Tyr Ser Ile Ser Ser Ser Pro
Arg Ile Ala Pro 485 490 495Ser Arg Ile His Val Thr Cys Ala Leu Val
Tyr Glu Lys Thr Pro Thr 500 505 510Gly Arg Val His Lys Gly Val Cys
Ser Thr Trp Met Lys Asn Ser Val 515 520 525Pro Ser Glu Lys Ser Asp
Glu Cys Ser Trp Ala Pro Ile Phe Val Arg 530 535 540Gln Ser Asn Phe
Lys Leu Pro Ala Asp Ala Lys Val Pro Ile Ile Met545 550 555 560Ile
Gly Pro Gly Thr Gly Leu Ala Pro Phe Arg Gly Phe Leu Gln Glu 565 570
575Arg Leu Ala Leu Lys Glu Ala Gly Thr Glu Leu Gly Pro Ser Ile Leu
580 585 590Phe Phe Gly Cys Arg Asn Ser Lys Met Asp Tyr Ile Tyr Glu
Asp Glu 595 600 605Leu Asp Asn Phe Val Gln Asn Gly Ala Leu Ser Glu
Leu Val Leu Ala 610 615 620Phe Ser Arg Glu Gly Pro Thr Lys Glu Tyr
Val Gln His Lys Met Met625 630 635 640Glu Lys Ala Ser Asp Ile Trp
Asn Leu Ile Ser Gln Gly Ala Tyr Leu 645 650 655Tyr Val Cys Gly Asp
Ala Lys Gly Met Ala Arg Asp Val His Arg Thr 660 665 670Leu His Thr
Ile Ala Gln Glu Gln Gly Ser Leu Asp Ser Ser Lys Ala 675 680 685Glu
Ser Met Val Lys Asn Leu Gln Met Ser Gly Arg Tyr Leu Arg Asp 690 695
700Val Trp705447PRTArtificial sequenceSynthetic sequence 44Gly Ser
Gly Gly Gly Gly Ser1 5458PRTArtificial sequenceSynthetic sequence
45Gly Ser Gly Glu Ala Ala Ala Lys1 54613PRTArtificial
sequenceSynthetic sequence 46Gly Ser Gly Glu Ala Ala Ala Lys Glu
Ala Ala Ala Lys1 5 10479PRTArtificial sequenceSynthetic sequence
47Gly Ser Gly Met Gly Ser Ser Ser Asn1 548491PRTZingiber zerumbet
48Leu Leu Ile Lys Arg Ser Ser Arg Ser Ser Val His Lys Gln Gln Val1
5 10 15Leu Leu Ala Ser Leu Pro Pro Ser Pro Pro Arg Leu Pro Leu Ile
Gly 20 25 30Asn Ile His Gln Leu Val Gly Gly Asn Pro His Arg Ile Leu
Leu Gln 35 40 45Leu Ala Arg Thr His Gly Pro Leu Ile Cys Leu Arg Leu
Gly Gln Val 50 55 60Asp Gln Val Val Ala Ser Ser Val Glu Ala Val Glu
Glu Ile Ile Lys65 70 75 80Arg His Asp Leu Lys Phe Ala Asp Arg Pro
Arg Asp Leu Thr Phe Ser 85 90 95Arg Ile Phe Phe Tyr Asp Gly Asn Ala
Val Val Met Thr Pro Tyr Gly 100 105 110Gly Glu Trp Lys Gln Met Arg
Lys Ile Tyr Ala Met Glu Leu Leu Asn 115 120 125Ser Arg Arg Val Lys
Ser Phe Ala Ala Ile Arg Glu Asp Val Ala Arg 130 135 140Lys Leu Thr
Gly Glu Ile Ala His Lys Ala Phe Ala Gln Thr Pro Val145 150 155
160Ile Asn Leu Ser Glu Met Val Met Ser Met Ile Asn Ala Ile Val Ile
165 170 175Arg Val Ala Phe Gly Asp Lys Cys Lys Gln Gln Ala Tyr Phe
Leu His 180 185 190Leu Val Lys Glu Ala Met Ser Tyr Val Ser Ser Phe
Ser Val Ala Asp 195 200 205Met Tyr Pro Ser Leu Lys Phe Leu Asp Thr
Leu Thr Gly Leu Lys Ser 210 215 220Lys Leu Glu Gly Val His Gly Lys
Leu Asp Lys Val Phe Asp Glu Ile225 230 235 240Ile Ala Gln Arg Gln
Ala Ala Leu Ala Ala Glu Gln Ala Glu Glu Asp 245 250 255Leu Ile Ile
Asp Val Leu Leu Lys Leu Lys Asp Glu Gly Asn Gln Glu 260 265 270Phe
Pro Ile Thr Tyr Thr Ser Val Lys Ala Ile Val Met Glu Ile Phe 275 280
285Leu Ala Gly Thr Glu Thr Ser Ser Ser Val Ile Asp Trp Val Met Ser
290 295 300Glu Leu Ile Lys Asn Pro Lys Ala Met Glu Lys Val Gln Lys
Glu Met305 310 315 320Arg Glu Ala Met Gln Gly Lys Thr Lys Leu Glu
Glu Ser Asp Ile Pro 325 330 335Lys Phe Ser Tyr Leu Asn Leu Val Ile
Lys Glu Thr Leu Arg Leu His 340 345 350Pro Pro Gly Pro Leu Leu Phe
Pro Arg Glu Cys Arg Glu Thr Cys Glu 355 360 365Val Met Gly Tyr Arg
Val Pro Ala Gly Ala Arg Leu Leu Ile Asn Ala 370 375 380Phe Ala Leu
Ser Arg Asp Glu Lys Tyr Trp Gly Ser Asp Ala Glu Ser385 390 395
400Phe Lys Pro Glu Arg Phe Glu Gly Ile Ser Val Asp Phe Lys Gly Ser
405 410 415Asn Phe Glu Phe Met Pro Phe Gly Ala Gly Arg Arg Ile Cys
Pro Gly 420 425 430Met Thr Phe Gly Ile Ser Ser Val Glu Val Ala Leu
Ala His Leu Leu 435 440 445Phe His Phe Asp Trp Gln Leu Pro Gln Gly
Met Lys Ile Glu Asp Leu 450 455 460Asp Met Met Glu Val Ser Gly Met
Ser Ala Thr Arg Arg Ser Pro Leu465 470 475 480Leu Val Leu Ala Lys
Leu Ile Ile Pro Leu Pro 485 49049476PRTBarnadesia spinosa 49Leu Leu
Thr Gly Ser Lys Ser Thr Lys Asn Ser Leu Pro Glu Ala Trp1 5 10 15Arg
Leu Pro Ile Ile Gly His Met His His Leu Val Gly Thr Leu Pro 20 25
30His Arg Gly Val Thr Asp Met Ala Arg Lys Tyr Gly Ser Leu Met His
35 40 45Leu Gln Leu Gly Glu Val Ser Thr Ile Val Val Ser Ser Pro Arg
Trp 50 55 60Ala Lys Glu Val Leu Thr Thr Tyr Asp Ile Thr Phe Ala Asn
Arg Pro65 70 75 80Glu Thr Leu Thr Gly Glu Ile Val Ala Tyr His Asn
Thr Asp Ile Val 85 90 95Leu Ser Pro Tyr Gly Glu Tyr Trp Arg Gln Leu
Arg Lys Leu Cys Thr 100 105 110Leu Glu Leu Leu Ser Ala Lys Lys Val
Lys Ser Phe Gln Ser Leu Arg 115 120 125Glu Glu Glu Cys Trp Asn Leu
Val Lys Glu Val Arg Ser Ser Gly Ser 130 135 140Gly Ser Pro Val Asp
Leu Ser Glu Ser Ile Phe Lys Leu Ile Ala Thr145 150 155 160Ile Leu
Ser Arg Ala Ala Phe Gly Lys Gly Ile Lys Asp Gln Arg Glu 165 170
175Phe Thr Glu Ile Val Lys Glu Ile Leu Arg Leu Thr Gly Gly Phe Asp
180 185 190Val Ala Asp Ile Phe Pro Ser Lys Lys Ile Leu His His Leu
Ser Gly 195 200 205Lys Arg Ala Lys Leu Thr Asn Ile His Asn Lys Leu
Asp Ser Leu Ile 210 215 220Asn Asn Ile Val Ser Glu His Pro Gly Ser
Arg Thr Ser Ser Ser Gln225 230 235 240Glu Ser Leu Leu Asp Val Leu
Leu Arg Leu Lys Asp Ser Ala Glu Leu 245 250 255Pro Leu Thr Ser Asp
Asn Val Lys Ala Val Ile Leu Asp Met Phe Gly 260 265 270Ala Gly Thr
Asp Thr Ser Ser Ala Thr Ile Glu Trp Ala Ile Ser Glu 275 280 285Leu
Ile Arg Cys Pro Arg Ala Met Glu Lys Val Gln Thr Glu Leu Arg 290 295
300Gln Ala Leu Asn Gly Lys Glu Arg Ile Gln Glu Glu Asp Ile Gln
Glu305 310 315 320Leu Ser Tyr Leu Lys Leu Val Ile Lys Glu Thr Leu
Arg Leu His Pro 325 330 335Pro Leu Pro Leu Val Met Pro Arg Glu Cys
Arg Glu Pro Cys Val Leu 340 345 350Ala Gly Tyr Glu Ile Pro Thr Lys
Thr Lys Leu Ile Val Asn Val Phe 355 360 365Ala Ile Asn Arg Asp Pro
Glu Tyr Trp Lys Asp Ala Glu Thr Phe Met 370 375 380Pro Glu Arg Phe
Glu Asn Ser Pro Ile Asn Ile Met Gly Ser Glu Tyr385 390 395 400Glu
Tyr Leu Pro Phe Gly Ala Gly Arg Arg Met Cys Pro Gly Ala Ala 405 410
415Leu Gly Leu Ala Asn Val Glu Leu Pro Leu Ala His Ile Leu Tyr Tyr
420 425 430Phe Asn Trp Lys Leu Pro Asn Gly Ala Arg Leu Asp Glu Leu
Asp Met 435 440 445Ser Glu Cys Phe Gly Ala Thr Val Gln Arg Lys Ser
Glu Leu Leu Leu 450 455 460Val Pro Thr Ala Tyr Lys Thr Ala Asn Asn
Ser Ala465 470 47550486PRTHyoscyamus muticus 50Phe Leu Leu Arg Lys
Trp Lys Asn Ser Asn Ser Gln Ser Lys Lys Leu1 5 10 15Pro Pro Gly Pro
Trp Lys Leu Pro Leu Leu Gly Ser Met Leu His Met 20 25 30Val Gly Gly
Leu Pro His His Val Leu Arg Asp Leu Ala Lys Lys Tyr 35 40 45Gly Pro
Leu Met His Leu Gln Leu Gly Glu Val Ser Ala Val Val Val 50 55 60Thr
Ser Pro Asp Met Ala Lys Glu Val Leu Lys Thr His Asp Ile Ala65 70 75
80Phe Ala Ser Arg Pro Lys Leu Leu Ala Pro Glu Ile Val Cys Tyr Asn
85 90 95Arg Ser Asp Ile Ala Phe Cys Pro Tyr Gly Asp Tyr Trp Arg Gln
Met 100 105 110Arg Lys Ile Cys Val Leu Glu Val Leu Ser Ala Lys Asn
Val Arg Ser 115 120 125Phe Ser Ser Ile Arg Arg Asp Glu Val Leu Arg
Leu Val Asn Phe Val 130 135 140Arg Ser Ser Thr Ser Glu Pro Val Asn
Phe Thr Glu Arg Leu Phe Leu145 150 155 160Phe Thr Ser Ser Met Thr
Cys Arg Ser Ala Phe Gly Lys Val Phe Lys 165 170 175Glu Gln Glu Thr
Phe Ile Gln Leu Ile Lys Glu Val Ile Gly Leu Ala 180 185 190Gly Gly
Phe Asp Val Ala Asp Ile Phe Pro Ser Leu Lys Phe Leu His 195 200
205Val Leu Thr Gly Met Glu Gly Lys Ile Met Lys Ala His His Lys Val
210 215 220Asp Ala Ile Val Glu Asp Val Ile Asn Glu His Lys Lys Asn
Leu Ala225 230 235 240Met Gly Lys Thr Asn Gly Ala Leu Gly Gly Glu
Asp Leu Ile Asp Val 245 250 255Leu Leu Arg Leu Met Asn Asp Gly Gly
Leu Gln Phe Pro Ile Thr Asn 260 265 270Asp Asn Ile Lys Ala Ile Ile
Phe Asp Met Phe Ala Ala Gly Thr Glu 275 280 285Thr Ser Ser Ser Thr
Leu Val Trp Ala Met Val Gln Met Met Arg Asn 290 295 300Pro Thr Ile
Leu Ala Lys Ala Gln Ala Glu Val Arg Glu Ala Phe Lys305 310 315
320Gly Lys Glu Thr Phe Asp Glu Asn Asp Val Glu Glu Leu Lys Tyr Leu
325 330 335Lys Leu Val Ile Lys Glu Thr Leu Arg Leu His Pro Pro Val
Pro Leu 340 345 350Leu Val Pro Arg Glu Cys Arg Glu Glu Thr Glu Ile
Asn Gly Tyr Thr 355 360 365Ile Pro Val Lys Thr Lys Val Met Val Asn
Val Trp Ala Leu Gly Arg 370 375 380Asp Pro Lys Tyr Trp Asp Asp Ala
Asp Asn Phe Lys Pro Glu Arg Phe385 390 395 400Glu Gln Cys Ser Val
Asp Phe Ile Gly Asn Asn Phe Glu Tyr Leu Pro 405 410 415Phe Gly Gly
Gly Arg Arg Ile Cys Pro Gly Ile Ser Phe Gly Leu Ala 420 425 430Asn
Val Tyr Leu Pro Leu Ala Gln Leu Leu Tyr His Phe Asp Trp Lys 435 440
445Leu Pro Thr Gly Met Glu Pro Lys Asp Leu Asp Leu Thr Glu Leu Val
450 455 460Gly Val Thr Ala Ala Arg Lys Ser Asp Leu Met Leu Val Ala
Thr Pro465 470 475 480Tyr Gln Pro Ser Arg Glu 48551469PRTLactuca
sativa 51Lys Leu Ala Thr Arg Pro Lys Ser Thr Lys Lys Gln Leu Pro
Glu Ala1 5 10 15Ser Arg Leu Pro Ile Ile Gly His Met His His Leu Ile
Gly Thr Met 20 25 30Pro His Arg Gly Val Met Asp Leu Ala Arg Lys His
Gly Ser Leu Met 35 40 45His Leu Gln Leu Gly Glu Val Ser Thr Ile Val
Val Ser Ser Pro Lys 50 55 60Trp Ala Lys Glu Ile Leu Thr Thr Tyr Asp
Ile Thr Phe Ala Asn Arg65 70 75 80Pro Glu Thr Leu Thr Gly Glu Ile
Ile Ala Tyr His Asn Thr Asp Ile 85 90 95Val Leu Ala Pro Tyr Gly Glu
Tyr Trp Arg Gln Leu Arg Lys Leu Cys 100 105 110Thr Leu Glu Leu Leu
Ser Val Lys Lys Val Lys Ser Phe Gln Ser Ile 115 120 125Arg Glu Glu
Glu Cys Trp Asn Leu Val Lys Glu Val Lys Glu Ser Gly 130 135 140Ser
Gly Lys Pro Ile Asn Leu Ser Glu Ser Ile Phe Thr Met Ile Ala145 150
155 160Thr Ile Leu Ser Arg Ala Ala Phe Gly Lys Gly Ile Lys Asp Gln
Arg 165 170 175Glu Phe Thr Glu Ile Val Lys Glu Ile Leu Arg Gln Thr
Gly Gly Phe 180 185 190Asp Val Ala Asp Ile Phe Pro Ser Lys Lys Phe
Leu His His Leu Ser 195 200 205Gly Lys Arg Ala Arg Leu Thr Ser Ile
His Lys Lys Leu Asp Asn Leu 210 215 220Ile Asn Asn Ile Val Ala Glu
His His Val Ser Thr Ser Ser Lys Ala225 230 235 240Asn Glu Thr Leu
Leu Asp Val Leu Leu Arg Leu Lys Asp Ser Ala Glu 245 250 255Phe Pro
Leu Thr Ala Asp Asn Val Lys Ala Ile Ile Leu Asp Met Phe 260 265
270Gly Ala Gly Thr Asp Thr Ser Ser Ala Thr Val Glu Trp Ala Ile Ser
275 280 285Glu Leu Ile Arg Cys Pro Arg Ala Met Glu Lys Val Gln Ala
Glu Leu 290 295 300Arg Gln Ala Leu Asn Gly Lys Glu Lys Ile Gln Glu
Glu Asp Ile Gln305 310 315 320Asp Leu Ala Tyr Leu Asn Leu Val Ile
Arg Glu Thr Leu Arg Leu His 325 330 335Pro Pro Leu Pro Leu Val Met
Pro Arg Glu Cys Arg Glu Pro Val Asn 340 345 350Leu Ala Gly Tyr Glu
Ile Ala Asn Lys Thr Lys Leu Ile Val Asn Val 355 360 365Phe Ala Ile
Asn Arg Asp Pro Glu Tyr Trp Lys Asp Ala Glu Ala Phe 370 375 380Ile
Pro Glu Arg Phe Glu Asn Asn Pro Asn Asn Ile Met Gly Ala Asp385 390
395 400Tyr Glu Tyr Leu Pro Phe Gly Ala Gly Arg Arg Met Cys Pro Gly
Ala 405 410 415Ala Leu Gly Leu Ala Asn Val Gln Leu Pro Leu Ala Asn
Ile Leu Tyr 420 425 430His Phe Asn Trp Lys Leu Pro Asn Gly Ala Ser
His Asp Gln Leu Asp 435 440 445Met Thr Glu Ser Phe Gly Ala Thr Val
Gln Arg Lys Thr Glu Leu Leu 450 455 460Leu Val Pro Ser
Phe46552488PRTNicotiani tabacum 52Phe Leu Leu Arg Lys Trp Lys Asn
Ser Asn Ser Gln Ser Lys Lys Leu1 5 10 15Pro Pro Gly Pro Trp Lys Ile
Pro Ile Leu Gly Ser Met Leu His Met 20 25 30Ile Gly Gly Glu Pro His
His Val Leu Arg Asp Leu Ala Lys Lys Tyr 35 40 45Gly Pro Leu Met His
Leu Gln Leu Gly Glu Ile Ser Ala Val Val Val 50 55 60Thr Ser Arg Asp
Met Ala Lys Glu Val Leu Lys Thr His Asp Val Val65 70 75 80Phe Ala
Ser Arg Pro Lys Ile Val Ala Met Asp Ile Ile Cys Tyr Asn 85 90 95Gln
Ser Asp Ile Ala Phe Ser Pro Tyr Gly Asp His Trp Arg Gln Met 100 105
110Arg Lys Ile Cys Val Met Glu Leu Leu Asn Ala Lys Asn Val Arg Ser
115 120 125Phe Ser Ser Ile Arg Arg Asp Glu Val Val Arg Leu Ile Asp
Ser Ile 130 135 140Arg Ser Asp Ser Ser Ser Gly Glu Leu Val Asn Phe
Thr Gln Arg Ile145 150 155 160Ile Trp Phe Ala Ser Ser Met Thr Cys
Arg Ser Ala Phe Gly Gln Val 165 170 175Leu Lys Gly Gln Asp Ile Phe
Ala Lys Lys Ile Arg Glu Val Ile Gly 180 185 190Leu Ala Glu Gly Phe
Asp Val Val Asp Ile Phe Pro Thr Tyr Lys Phe 195 200 205Leu His Val
Leu Ser Gly Met Lys Arg Lys Leu Leu Asn Ala His Leu 210 215 220Lys
Val Asp Ala Ile Val Glu Asp Val Ile Asn Glu His Lys Lys Asn225 230
235 240Leu Ala Ala Gly Lys Ser Asn Gly Ala Leu Gly Gly Glu Asp Leu
Ile 245 250 255Asp Val Leu Leu Arg Leu Met Asn Asp Thr Ser Leu Gln
Phe Pro Ile 260 265 270Thr Asn Asp Asn Ile Lys Ala Val Ile Val Asp
Met Phe Ala Ala Gly 275 280 285Thr Glu Thr Ser Ser Thr Thr Thr Val
Trp Ala Met Ala Glu Met Met 290 295 300Lys Asn Pro Ser Val Phe Thr
Lys Ala Gln Ala Glu Val Arg Glu Ala305 310 315 320Phe Arg Asp Lys
Val Ser Phe Asp Glu Asn Asp Val Glu Glu Leu Lys 325 330 335Tyr Leu
Lys Leu Val Ile Lys Glu Thr Leu Arg Leu His Pro Pro Ser 340 345
350Pro Leu Leu Val Pro Arg Glu Cys Arg Glu Asp Thr Asp Ile Asn Gly
355 360 365Tyr Thr Ile Pro Ala Lys Thr Lys Val Met Val Asn Val Trp
Ala Leu 370 375 380Gly Arg Asp Pro Lys Tyr Trp Asp Asp Ala Glu Ser
Phe Lys Pro Glu385 390 395 400Arg Phe Glu Gln Cys Ser Val Asp Phe
Phe Gly Asn Asn Phe Glu Phe 405 410 415Leu Pro Phe Gly Gly Gly Arg
Arg Ile Cys Pro Gly Met Ser Phe Gly 420 425 430Leu Ala Asn Leu Tyr
Leu Pro Leu Ala Gln Leu Leu Tyr His Phe Asp 435 440 445Trp Lys Leu
Pro Thr Gly Ile Met Pro Arg Asp Leu Asp Leu Thr Glu 450 455 460Leu
Ser Gly Ile Thr Ile Ala Arg Lys Gly Gly Leu Tyr Leu Asn Ala465 470
475 480Thr Pro Tyr Gln Pro Ser Arg Glu 48553496PRTCitrus x paradisi
53Trp Val Trp Leu Arg Pro Lys Lys Leu Glu Lys Phe Leu Arg Gln Gln1
5 10 15Gly Leu Lys Gly Asn Ser Tyr Arg Leu Leu Phe Gly Asp Leu Lys
Glu 20 25 30Asn Ser Ile Glu Leu Lys Glu Ala Lys Ala Arg Pro Leu Ser
Leu Asp 35 40 45Asp Asp Ile Ala Ile Arg Val Asn Pro Phe Leu His Lys
Leu Val Asn 50 55 60Asp Tyr Gly Lys Asn Ser Phe Met Trp Phe Gly Pro
Thr Pro Arg Val65 70 75 80Asn Ile Met Asn Pro Asp Gln Ile Lys Ala
Ile Phe Thr Lys Ile Asn 85 90 95Asp Phe Gln Lys Val Asn Ser Ile Pro
Leu Ala Arg Leu Leu Ile Val 100 105 110Gly Leu Ala Thr Leu Glu Gly
Glu Lys Trp Ala Lys His Arg Lys Leu 115 120 125Ile Asn Pro Ala Phe
His Gln Glu Lys Leu Lys Leu Met Leu Pro Ala 130 135 140Phe Tyr Leu
Ser Cys Ile Glu Ile Ile Thr Lys Trp Glu Lys Gln Met145 150 155
160Ser Val Glu Gly Ser Ser Glu Leu Asp Val Trp Pro Tyr Leu Ala Asn
165 170 175Leu Thr Ser Asp Val Ile Ser Arg Thr Ala Phe Gly Ser Ser
Tyr Glu 180 185 190Glu Gly Arg Arg Ile Phe Gln Leu Gln Ala Glu Leu
Ala Glu Leu Thr 195 200 205Met Gln Val Phe Arg Ser Val His Ile Pro
Gly Trp Arg Phe Leu Pro 210 215 220Thr Lys Arg Asn Arg Arg Met Lys
Glu Ile Asp Lys Glu Ile Arg Ala225 230 235 240Ser Leu Met Gly Ile
Ile Lys Asn Arg Glu Lys Ala Met Arg Ala Gly 245 250 255Glu Ala Ala
Asn Asn Asp Leu Leu Gly Ile Leu Met Glu Thr Ser Phe 260 265 270Arg
Glu Ile Glu Glu His Gly Asn Asn Lys Asn Val Gly Phe Ser Met 275 280
285Asn Asp Val Ile Glu Glu Cys Lys Leu Phe Tyr Phe Ala Gly Gln Glu
290 295 300Thr Thr Ser Val Leu Leu Asn Trp Thr Met Val Leu Leu Ser
Lys His305 310 315 320Gln Asp Trp Gln Glu Arg Ala Arg Gln Glu Val
Leu Gln Val Phe Gly 325 330 335Asn Asn Lys Pro Asp Tyr Asp Gly Leu
Asn His Leu Lys Ile Val Gln 340 345 350Met Ile Leu Tyr Glu Val Leu
Arg Leu Tyr Pro Pro Val Thr Val Leu 355 360 365Ser Arg Ala Val Phe
Lys Glu Thr Lys Leu Gly Asn Leu Thr Leu Pro 370 375 380Ala Gly Val
Gln Ile Gly Leu Pro Met Ile Leu Val His Gln Asp Pro385 390 395
400Glu Leu Trp Gly Asp Asp Ala Val Glu Phe Lys Pro Glu Arg Phe Ala
405 410 415Glu Gly Ile Ser Lys Ala Ala Lys Asn Gln Val Ser Tyr Phe
Pro Phe 420 425 430Ala Leu Gly Pro Arg Ile Cys Val Gly Gln Asn Phe
Ala Leu Val Glu 435 440 445Ala Lys Met Ala Thr Ala Met Ile Leu Gln
Asn Tyr Ser Phe Glu Leu 450 455 460Ser Pro Ser Tyr Val His Ala Pro
Thr Ala Val Pro Thr Leu His Pro465 470 475 480Glu Leu Gly Thr Gln
Leu Ile Leu Arg Lys Leu Trp Cys Lys Asn Asn 485 490
49554472PRTArtemesia annua 54Phe Val Tyr Lys Phe Ala Thr Arg Ser
Lys Ser Thr Lys Lys Ser Leu1 5 10 15Pro Glu Pro Trp Arg Leu Pro Ile
Ile Gly His Met His His Leu Ile 20 25 30Gly Thr Thr Pro His Arg Gly
Val Arg Asp Leu Ala Arg Lys Tyr Gly 35 40 45Ser Leu Met His Leu Gln
Leu Gly Glu Val Pro Thr Ile Val Val Ser 50 55 60Ser Pro Lys Trp Ala
Lys Glu Ile Leu Thr Thr Tyr Asp Ile Thr Phe65 70 75 80Ala Asn Arg
Pro Glu Thr Leu Thr Gly Glu Ile Val Leu Tyr His Asn 85 90 95Thr Asp
Val Val Leu Ala Pro Tyr Gly Glu Tyr Trp Arg Gln Leu Arg 100 105
110Lys Ile Cys Thr Leu Glu Leu Leu Ser Val Lys Lys Val Lys Ser Phe
115 120 125Gln Ser Leu Arg Glu Glu Glu Cys Trp Asn Leu Val Gln Glu
Ile Lys 130 135 140Ala Ser Gly Ser Gly Arg Pro Val Asn Leu Ser Glu
Asn Val Phe Lys145 150 155 160Leu Ile Ala Thr Ile Leu Ser Arg Ala
Ala Phe Gly Lys Gly Ile Lys 165 170 175Asp Gln Lys Glu Leu Thr Glu
Ile Val Lys Glu Ile Leu Arg Gln Thr 180 185 190Gly Gly Phe Asp Val
Ala Asp Ile Phe Pro Ser Lys Lys Phe Leu His 195 200 205His Leu Ser
Gly Lys Arg Ala Arg Leu Thr Ser Leu Arg Lys Lys Ile 210 215 220Asp
Asn Leu Ile Asp Asn Leu Val Ala Glu His Thr Val Asn Thr Ser225 230
235 240Ser Lys Thr Asn Glu Thr Leu Leu Asp Val Leu Leu Arg Leu Lys
Asp 245 250 255Ser Ala Glu Phe Pro Leu Thr Ser Asp Asn Ile Lys Ala
Ile Ile Leu 260 265 270Asp Met Phe Gly Ala Gly Thr Asp Thr Ser Ser
Ser Thr Ile Glu Trp 275 280 285Ala Ile Ser Glu Leu Ile Lys Cys Pro
Lys Ala Met Glu Lys Val Gln 290 295 300Ala Glu Leu Arg Lys Ala Leu
Asn Gly Lys Glu Lys Ile His Glu Glu305 310 315 320Asp Ile Gln Glu
Leu Ser Tyr Leu Asn Met Val Ile Lys Glu Thr Leu 325 330 335Arg Leu
His Pro Pro Leu Pro Leu Val Leu Pro Arg Glu Cys Arg Gln 340 345
350Pro Val Asn Leu Ala Gly Tyr Asn Ile Pro Asn Lys Thr Lys Leu Ile
355 360 365Val Asn Val Phe Ala Ile Asn Arg Asp Pro Glu Tyr Trp Lys
Asp Ala 370 375 380Glu Ala Phe Ile Pro Glu Arg Phe Glu Asn Ser Ser
Ala Thr Val Met385 390 395 400Gly Ala Glu Tyr Glu Tyr Leu Pro Phe
Gly Ala Gly Arg Arg Met Cys 405 410 415Pro Gly Ala Ala Leu Gly Leu
Ala Asn Val Gln Leu Pro Leu Ala Asn 420 425 430Ile Leu Tyr His Phe
Asn Trp Lys Leu Pro Asn Gly Val Ser Tyr Asp 435 440 445Gln Ile Asp
Met Thr Glu Ser Ser Gly Ala Thr Met Gln Arg Lys Thr 450 455 460Glu
Leu Leu Leu Val Pro Ser Phe465 47055488PRTArabidopsis thaliana
55Phe Phe Phe Lys Lys Leu Leu Ser Phe Ser Arg Lys Asn Met Ser Glu1
5 10 15Val Ser Thr Leu Pro Ser Val Pro Val Val Pro Gly Phe Pro Val
Ile 20 25 30Gly Asn Leu Leu Gln Leu Lys Glu Lys Lys Pro His Lys Thr
Phe Thr 35 40 45Arg Trp Ser Glu Ile Tyr Gly Pro Ile Tyr Ser Ile Lys
Met Gly Ser 50 55 60Ser Ser Leu Ile Val Leu Asn Ser Thr Glu Thr Ala
Lys Glu Ala Met65 70 75 80Val Thr Arg Phe Ser Ser Ile Ser Thr Arg
Lys Leu Ser Asn Ala Leu 85 90 95Thr Val Leu Thr Cys Asp Lys Ser Met
Val Ala Thr Ser Asp Tyr Asp 100 105 110Asp Phe His Lys Leu Val Lys
Arg Cys Leu Leu Asn Gly Leu Leu Gly 115 120 125Ala Asn Ala Gln Lys
Arg Lys Arg His Tyr Arg Asp Ala Leu Ile Glu 130 135 140Asn Val Ser
Ser Lys Leu His Ala His Ala Arg Asp His Pro Gln Glu145 150 155
160Pro Val Asn Phe Arg Ala Ile Phe Glu His Glu Leu Phe Gly Val Ala
165 170 175Leu Lys Gln Ala Phe Gly Lys Asp Val Glu Ser Ile Tyr Val
Lys Glu 180 185 190Leu Gly Val Thr Leu Ser Lys Asp Glu Ile Phe Lys
Val Leu Val His 195 200 205Asp Met Met Glu Gly Ala Ile Asp Val Asp
Trp Arg Asp Phe Phe Pro 210 215 220Tyr Leu Lys Trp Ile Pro Asn Lys
Ser Phe Glu Ala Arg Ile Gln Gln225 230 235 240Lys His Lys Arg Arg
Leu Ala Val Met Asn Ala Leu Ile Gln Asp Arg 245 250 255Leu Lys Gln
Asn Gly Ser Glu Ser Asp Asp Asp Cys Tyr Leu Asn Phe 260 265 270Leu
Met Ser Glu Ala Lys Thr Leu Thr Lys Glu Gln Ile Ala Ile Leu 275 280
285Val Trp Glu Thr Ile Ile Glu Thr Ala Asp Thr Thr Leu Val Thr Thr
290 295 300Glu Trp Ala Ile Tyr Glu Leu Ala Lys His Pro Ser Val Gln
Asp Arg305 310 315 320Leu Cys Lys Glu Ile Gln Asn Val Cys Gly Gly
Glu Lys Phe Lys Glu 325 330 335Glu Gln Leu Ser Gln Val Pro Tyr Leu
Asn Gly Val Phe His Glu Thr 340 345 350Leu Arg Lys Tyr Ser Pro Ala
Pro Leu Val Pro Ile Arg Tyr Ala His 355 360 365Glu Asp Thr Gln
Ile Gly Gly Tyr His Val Pro Ala Gly Ser Glu Ile 370 375 380Ala Ile
Asn Ile Tyr Gly Cys Asn Met Asp Lys Lys Arg Trp Glu Arg385 390 395
400Pro Glu Asp Trp Trp Pro Glu Arg Phe Leu Asp Asp Gly Lys Tyr Glu
405 410 415Thr Ser Asp Leu His Lys Thr Met Ala Phe Gly Ala Gly Lys
Arg Val 420 425 430Cys Ala Gly Ala Leu Gln Ala Ser Leu Met Ala Gly
Ile Ala Ile Gly 435 440 445Arg Leu Val Gln Glu Phe Glu Trp Lys Leu
Arg Asp Gly Glu Glu Glu 450 455 460Asn Val Asp Thr Tyr Gly Leu Thr
Ser Gln Lys Leu Tyr Pro Leu Met465 470 475 480Ala Ile Ile Asn Pro
Arg Arg Ser 48556483PRTStevia rebaudiana 56Trp Tyr Leu Lys Ser Tyr
Thr Ser Ala Arg Arg Ser Gln Ser Asn His1 5 10 15Leu Pro Arg Val Pro
Glu Val Pro Gly Val Pro Leu Leu Gly Asn Leu 20 25 30Leu Gln Leu Lys
Glu Lys Lys Pro Tyr Met Thr Phe Thr Arg Trp Ala 35 40 45Ala Thr Tyr
Gly Pro Ile Tyr Ser Ile Lys Thr Gly Ala Thr Ser Met 50 55 60Val Val
Val Ser Ser Asn Glu Ile Ala Lys Glu Ala Leu Val Thr Arg65 70 75
80Phe Gln Ser Ile Ser Thr Arg Asn Leu Ser Lys Ala Leu Lys Val Leu
85 90 95Thr Ala Asp Lys Thr Met Val Ala Met Ser Asp Tyr Asp Asp Tyr
His 100 105 110Lys Thr Val Lys Arg His Ile Leu Thr Ala Val Leu Gly
Pro Asn Ala 115 120 125Gln Lys Lys His Arg Ile His Arg Asp Ile Met
Met Asp Asn Ile Ser 130 135 140Thr Gln Leu His Glu Phe Val Lys Asn
Asn Pro Glu Gln Glu Glu Val145 150 155 160Asp Leu Arg Lys Ile Phe
Gln Ser Glu Leu Phe Gly Leu Ala Met Arg 165 170 175Gln Ala Leu Gly
Lys Asp Val Glu Ser Leu Tyr Val Glu Asp Leu Lys 180 185 190Ile Thr
Met Asn Arg Asp Glu Ile Phe Gln Val Leu Val Val Asp Pro 195 200
205Met Met Gly Ala Ile Asp Val Asp Trp Arg Asp Phe Phe Pro Tyr Leu
210 215 220Lys Trp Val Pro Asn Lys Lys Phe Glu Asn Thr Ile Gln Gln
Met Tyr225 230 235 240Ile Arg Arg Glu Ala Val Met Lys Ser Leu Ile
Lys Glu His Lys Lys 245 250 255Arg Ile Ala Ser Gly Glu Lys Leu Asn
Ser Tyr Ile Asp Tyr Leu Leu 260 265 270Ser Glu Ala Gln Thr Leu Thr
Asp Gln Gln Leu Leu Met Ser Leu Trp 275 280 285Glu Pro Ile Ile Glu
Ser Ser Asp Thr Thr Met Val Thr Thr Glu Trp 290 295 300Ala Met Tyr
Glu Leu Ala Lys Asn Pro Lys Leu Gln Asp Arg Leu Tyr305 310 315
320Arg Asp Ile Lys Ser Val Cys Gly Ser Glu Lys Ile Thr Glu Glu His
325 330 335Leu Ser Gln Leu Pro Tyr Ile Thr Ala Ile Phe His Glu Thr
Leu Arg 340 345 350Arg His Ser Pro Val Pro Ile Ile Pro Leu Arg His
Val His Glu Asp 355 360 365Thr Val Leu Gly Gly Tyr His Val Pro Ala
Gly Thr Glu Leu Ala Val 370 375 380Asn Ile Tyr Gly Cys Asn Met Asp
Lys Asn Val Trp Glu Asn Pro Glu385 390 395 400Glu Trp Asn Pro Glu
Arg Phe Met Lys Glu Asn Glu Thr Ile Asp Phe 405 410 415Gln Lys Thr
Met Ala Phe Gly Gly Gly Lys Arg Val Cys Ala Gly Ser 420 425 430Leu
Gln Ala Leu Leu Thr Ala Ser Ile Gly Ile Gly Arg Met Val Gln 435 440
445Glu Phe Glu Trp Lys Leu Lys Asp Met Thr Gln Glu Glu Val Asn Thr
450 455 460Ile Gly Leu Thr Thr Gln Met Leu Arg Pro Leu Arg Ala Ile
Ile Lys465 470 475 480Pro Arg Ile57494PRTPhyscomitrella patens
57Phe Val Ala Arg Thr Cys Leu Arg Asn Lys Lys Arg Leu Pro Pro Ala1
5 10 15Ile Pro Gly Gly Leu Pro Val Leu Gly Asn Leu Leu Gln Leu Thr
Glu 20 25 30Lys Lys Pro His Arg Thr Phe Thr Ala Trp Ser Lys Glu His
Gly Pro 35 40 45Ile Phe Thr Ile Lys Val Gly Ser Val Pro Gln Ala Val
Val Asn Asn 50 55 60Ser Glu Ile Ala Lys Glu Val Leu Val Thr Lys Phe
Ala Ser Ile Ser65 70 75 80Lys Arg Gln Met Pro Met Ala Leu Arg Val
Leu Thr Arg Asp Lys Thr 85 90 95Met Val Ala Met Ser Asp Tyr Gly Glu
Glu His Arg Met Leu Lys Lys 100 105 110Leu Val Met Thr Asn Leu Leu
Gly Pro Thr Thr Gln Asn Lys Asn Arg 115 120 125Ser Leu Arg Asp Asp
Ala Leu Ile Gly Met Ile Glu Gly Val Leu Ala 130 135 140Glu Leu Lys
Ala Ser Pro Thr Ser Pro Lys Val Val Asn Val Arg Asp145 150 155
160Tyr Val Gln Arg Ser Leu Phe Pro Phe Ala Leu Gln Gln Val Phe Gly
165 170 175Tyr Ile Pro Asp Gln Val Glu Val Leu Glu Leu Gly Thr Cys
Val Ser 180 185 190Thr Trp Asp Met Phe Asp Ala Leu Val Val Ala Pro
Leu Ser Ala Val 195 200 205Ile Asn Val Asp Trp Arg Asp Phe Phe Pro
Ala Leu Arg Trp Ile Pro 210 215 220Asn Arg Ser Val Glu Asp Leu Val
Arg Thr Val Asp Phe Lys Arg Asn225 230 235 240Ser Ile Met Lys Ala
Leu Ile Arg Ala Gln Arg Met Arg Leu Ala Asn 245 250 255Leu Lys Glu
Pro Pro Arg Cys Tyr Ala Asp Ile Ala Leu Thr Glu Ala 260 265 270Thr
His Leu Thr Glu Lys Gln Leu Glu Met Ser Leu Trp Glu Pro Ile 275 280
285Ile Glu Ser Ala Asp Thr Thr Leu Val Thr Ser Glu Trp Ala Met Tyr
290 295 300Glu Ile Ala Lys Asn Pro Asp Cys Gln Asp Arg Leu Tyr Arg
Glu Ile305 310 315 320Val Ser Val Ala Gly Thr Glu Arg Met Val Thr
Glu Asp Asp Leu Pro 325 330 335Asn Met Pro Tyr Leu Gly Ala Ile Ile
Lys Glu Thr Leu Arg Lys Tyr 340 345 350Thr Pro Val Pro Leu Ile Pro
Ser Arg Phe Val Glu Glu Asp Ile Thr 355 360 365Leu Gly Gly Tyr Asp
Ile Pro Lys Gly Tyr Gln Ile Leu Val Asn Leu 370 375 380Phe Ala Ile
Ala Asn Asp Pro Ala Val Trp Ser Asn Pro Glu Lys Trp385 390 395
400Asp Pro Glu Arg Met Leu Ala Asn Lys Lys Val Asp Met Gly Phe Arg
405 410 415Asp Phe Ser Leu Met Pro Phe Gly Ala Gly Lys Arg Met Cys
Ala Gly 420 425 430Ile Thr Gln Ala Met Phe Ile Ile Pro Met Asn Val
Ala Ala Leu Val 435 440 445Gln His Cys Glu Trp Arg Leu Ser Pro Gln
Glu Ile Ser Asn Ile Asn 450 455 460Asn Lys Ile Glu Asp Val Val Tyr
Leu Thr Thr His Lys Leu Ser Pro465 470 475 480Leu Ser Cys Glu Ala
Thr Pro Arg Ile Ser His Arg Leu Pro 485 49058381PRTPleurotus
sapidus 58Met Gly Lys Leu His Pro Leu Ala Ile Ile Pro Asp Tyr Lys
Gly Ser1 5 10 15Met Ala Ala Ser Val Thr Ile Phe Asn Lys Arg Thr Asn
Pro Leu Asp 20 25 30Ile Ser Val Asn Gln Ala Asn Asp Trp Pro Trp Arg
Tyr Ala Lys Thr 35 40 45Cys Val Leu Ser Ser Asp Trp Ala Leu His Glu
Met Ile Ile His Leu 50 55 60Asn Asn Thr His Leu Val Glu Glu Ala Val
Ile Val Ala Ala Gln Arg65 70 75 80Lys Leu Ser Pro Ser His Ile Val
Phe Arg Leu Leu Glu Pro His Trp 85 90 95Val Val Thr Leu Ser Leu Asn
Ala Leu Ala Arg Ser Val Leu Ile Pro 100 105 110Glu Val Ile Val Pro
Ile Ala Gly Phe Ser Ala Pro His Ile Phe Gln 115 120 125Phe Ile Arg
Glu Ser Phe Thr Asn Phe Asp Trp Lys Ser Leu Tyr Val 130 135 140Pro
Ala Asp Leu Glu Ser Arg Gly Phe Pro Val Asp Gln Leu Asn Ser145 150
155 160Pro Lys Phe His Asn Tyr Ala Tyr Ala Arg Asp Ile Asn Asp Met
Trp 165 170 175Thr Thr Leu Lys Lys Phe Val Ser Ser Val Leu Gln Asp
Ala Gln Tyr 180 185 190Tyr Pro Asp Asp Ala Ser Val Ala Gly Asp Thr
Gln Ile Gln Ala Trp 195 200 205Cys Asp Glu Met Arg Ser Gly Met Gly
Ala Gly Met Thr Asn Phe Pro 210 215 220Glu Ser Ile Thr Thr Val Asp
Asp Leu Val Asn Met Val Thr Met Cys225 230 235 240Ile His Ile Ala
Ala Pro Gln His Thr Ala Val Asn Tyr Leu Gln Gln 245 250 255Tyr Tyr
Gln Thr Phe Val Ser Asn Lys Pro Ser Ala Leu Phe Ser Pro 260 265
270Leu Pro Thr Ser Ile Ala Gln Leu Gln Lys Tyr Thr Glu Ser Asp Leu
275 280 285Met Ala Ala Leu Pro Leu Asn Ala Lys Arg Gln Trp Leu Leu
Met Ala 290 295 300Gln Ile Pro Tyr Leu Leu Ser Met Gln Val Gln Glu
Asp Glu Asn Ile305 310 315 320Val Thr Tyr Ala Ala Asn Ala Ser Thr
Asp Lys Asp Pro Ile Ile Ala 325 330 335Ser Ala Gly Arg Gln Leu Ala
Ala Asp Leu Lys Lys Leu Ala Ala Val 340 345 350Phe Leu Val Asn Ser
Ala Gln Leu Asp Asp Gln Asn Thr Pro Tyr Asp 355 360 365Val Leu Ala
Pro Glu Gln Leu Ala Asn Ala Ile Val Ile 370 375
38059475PRTCichorium intybus 59Leu Leu Thr Arg Thr Thr Ser Lys Lys
Asn Leu Leu Pro Glu Pro Trp1 5 10 15Arg Leu Pro Ile Ile Gly His Met
His His Leu Ile Gly Thr Met Pro 20 25 30His Arg Gly Val Met Glu Leu
Ala Arg Lys His Gly Ser Leu Met His 35 40 45Leu Gln Leu Gly Glu Val
Ser Thr Ile Val Val Ser Ser Pro Arg Trp 50 55 60Ala Lys Glu Val Leu
Thr Thr Tyr Asp Ile Thr Phe Ala Asn Arg Pro65 70 75 80Glu Thr Leu
Thr Gly Glu Ile Val Ala Tyr His Asn Thr Asp Ile Val 85 90 95Leu Ala
Pro Tyr Gly Glu Tyr Trp Arg Gln Leu Arg Lys Leu Cys Thr 100 105
110Leu Glu Leu Leu Ser Asn Lys Lys Val Lys Ser Phe Gln Ser Leu Arg
115 120 125Glu Glu Glu Cys Trp Asn Leu Val Lys Asp Ile Arg Ser Thr
Gly Gln 130 135 140Gly Ser Pro Ile Asn Leu Ser Glu Asn Ile Phe Lys
Met Ile Ala Thr145 150 155 160Ile Leu Ser Arg Ala Ala Phe Gly Lys
Gly Ile Lys Asp Gln Met Lys 165 170 175Phe Thr Glu Leu Val Lys Glu
Ile Leu Arg Leu Thr Gly Gly Phe Asp 180 185 190Val Ala Asp Ile Phe
Pro Ser Lys Lys Leu Leu His His Leu Ser Gly 195 200 205Lys Arg Ala
Lys Leu Thr Asn Ile His Asn Lys Leu Asp Asn Leu Ile 210 215 220Asn
Asn Ile Ile Ala Glu His Pro Gly Asn Arg Thr Ser Ser Ser Gln225 230
235 240Glu Thr Leu Leu Asp Val Leu Leu Arg Leu Lys Glu Ser Ala Glu
Phe 245 250 255Pro Leu Thr Ala Asp Asn Val Lys Ala Val Ile Leu Asp
Met Phe Gly 260 265 270Ala Gly Thr Asp Thr Ser Ser Ala Thr Ile Glu
Trp Ala Ile Ser Glu 275 280 285Leu Ile Arg Cys Pro Arg Ala Met Glu
Lys Val Gln Thr Glu Leu Arg 290 295 300Gln Ala Leu Asn Gly Lys Glu
Arg Ile Gln Glu Glu Asp Leu Gln Glu305 310 315 320Leu Asn Tyr Leu
Lys Leu Val Ile Lys Glu Thr Leu Arg Leu His Pro 325 330 335Pro Leu
Pro Leu Val Met Pro Arg Glu Cys Arg Glu Pro Cys Val Leu 340 345
350Gly Gly Tyr Asp Ile Pro Ser Lys Thr Lys Leu Ile Val Asn Val Phe
355 360 365Ala Ile Asn Arg Asp Pro Glu Tyr Trp Lys Asp Ala Glu Thr
Phe Met 370 375 380Pro Glu Arg Phe Glu Asn Ser Pro Ile Thr Val Met
Gly Ser Glu Tyr385 390 395 400Glu Tyr Leu Pro Phe Gly Ala Gly Arg
Arg Met Cys Pro Gly Ala Ala 405 410 415Leu Gly Leu Ala Asn Val Glu
Leu Pro Leu Ala His Ile Leu Tyr Phe 420 425 430Asn Trp Lys Leu Pro
Asn Gly Lys Thr Phe Glu Asp Leu Asp Met Thr 435 440 445Glu Ser Phe
Gly Ala Thr Val Gln Arg Lys Thr Glu Leu Leu Leu Val 450 455 460Pro
Thr Asp Phe Gln Thr Leu Thr Ala Ser Thr465 470
47560468PRTHelianthus annuus 60Leu Leu Thr Arg Pro Thr Ser Ser Lys
Asn Arg Leu Pro Glu Pro Trp1 5 10 15Arg Leu Pro Ile Ile Gly His Met
His His Leu Ile Gly Thr Met Pro 20 25 30His Arg Gly Val Met Asp Leu
Ala Arg Lys Tyr Gly Ser Leu Met His 35 40 45Leu Gln Leu Gly Glu Val
Ser Ala Ile Val Val Ser Ser Pro Lys Trp 50 55 60Ala Lys Glu Ile Leu
Thr Thr Tyr Asp Ile Pro Phe Ala Asn Arg Pro65 70 75 80Glu Thr Leu
Thr Gly Glu Ile Ile Ala Tyr His Asn Thr Asp Ile Val 85 90 95Leu Ala
Pro Tyr Gly Glu Tyr Trp Arg Gln Leu Arg Lys Leu Cys Thr 100 105
110Leu Glu Leu Leu Ser Val Lys Lys Val Lys Ser Phe Gln Ser Leu Arg
115 120 125Glu Glu Glu Cys Trp Asn Leu Val Gln Glu Ile Lys Ala Ser
Gly Ser 130 135 140Gly Thr Pro Phe Asn Leu Ser Glu Gly Ile Phe Lys
Val Ile Ala Thr145 150 155 160Val Leu Ser Arg Ala Ala Phe Gly Lys
Gly Ile Lys Asp Gln Lys Gln 165 170 175Phe Thr Glu Ile Val Lys Glu
Ile Leu Arg Glu Thr Gly Gly Phe Asp 180 185 190Val Ala Asp Ile Phe
Pro Ser Lys Lys Phe Leu His His Leu Ser Gly 195 200 205Lys Arg Gly
Arg Leu Thr Ser Ile His Asn Lys Leu Asp Ser Leu Ile 210 215 220Asn
Asn Leu Val Ala Glu His Thr Val Ser Lys Ser Ser Lys Val Asn225 230
235 240Glu Thr Leu Leu Asp Val Leu Leu Arg Leu Lys Asn Ser Glu Glu
Phe 245 250 255Pro Leu Thr Ala Asp Asn Val Lys Ala Ile Ile Leu Asp
Met Phe Gly 260 265 270Ala Gly Thr Asp Thr Ser Ser Ala Thr Val Glu
Trp Ala Ile Ser Glu 275 280 285Leu Ile Arg Cys Pro Arg Ala Met Glu
Lys Val Gln Ala Glu Leu Arg 290 295 300Gln Ala Leu Asn Gly Lys Glu
Arg Ile Lys Glu Glu Glu Ile Gln Asp305 310 315 320Leu Pro Tyr Leu
Asn Leu Val Ile Arg Glu Thr Leu Arg Leu His Pro 325 330 335Pro Leu
Pro Leu Val Met Pro Arg Glu Cys Arg Gln Ala Met Asn Leu 340 345
350Ala Gly Tyr Asp Val Ala Asn Lys Thr Lys Leu Ile Val Asn Val Phe
355 360 365Ala Ile Asn Arg Asp Pro Glu Tyr Trp Lys Asp Ala Glu Ser
Phe Asn 370 375 380Pro Glu Arg Phe Glu Asn Ser Asn Thr Thr Ile Met
Gly Ala Asp Tyr385 390 395 400Glu Tyr Leu Pro Phe Gly Ala Gly Arg
Arg Met Cys Pro Gly Ser Ala 405 410 415Leu Gly Leu Ala Asn Val Gln
Leu Pro Leu Ala Asn Ile Leu Tyr Tyr 420 425 430Phe Lys Trp Lys Leu
Pro Asn Gly Ala Ser His Asp Gln Leu Asp Met 435 440 445Thr Glu Ser
Phe Gly Ala Thr Val Gln Arg Lys Thr Glu Leu Met Leu 450 455 460Val
Pro Ser Phe4656112PRTArtificial SequenceSynthetic sequence 61His
Val Tyr Gly Arg Ala Val Val Glu Gln Trp Arg1 5 106211PRTArtificial
SequenceSynthetic sequence 62Val Tyr Gly Arg Ala Val Val Glu Gln
Trp Arg1 5 106312PRTArtificial SequenceSynthetic Sequence 63Phe Trp
Tyr Leu Lys Ser
Tyr Thr Ser Ala Arg Arg1 5 106411PRTArtificial SequenceSynthetic
Sequence 64Trp Tyr Leu Lys Ser Tyr Thr Ser Ala Arg Arg1 5 10
References
D00000

D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012
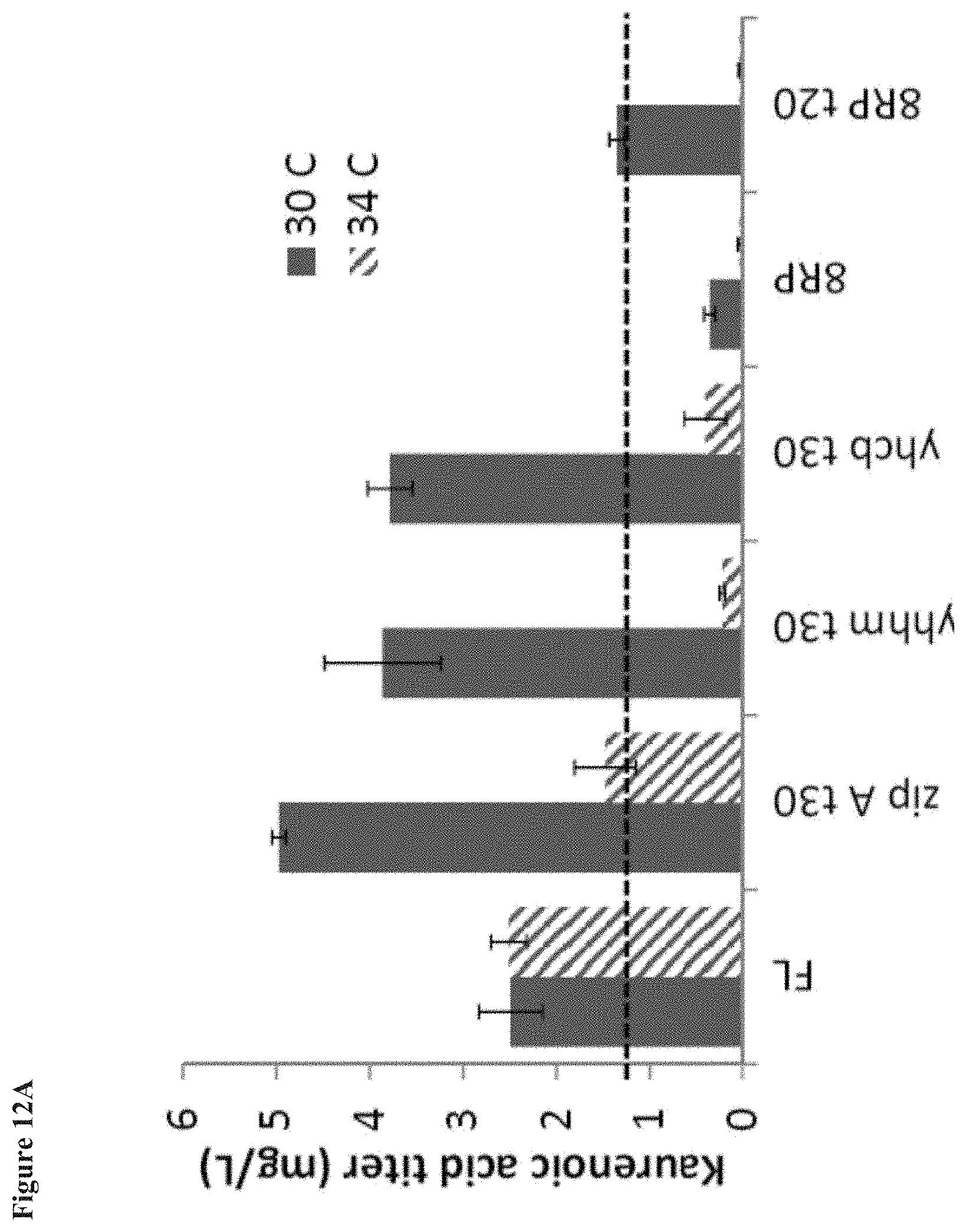
D00013
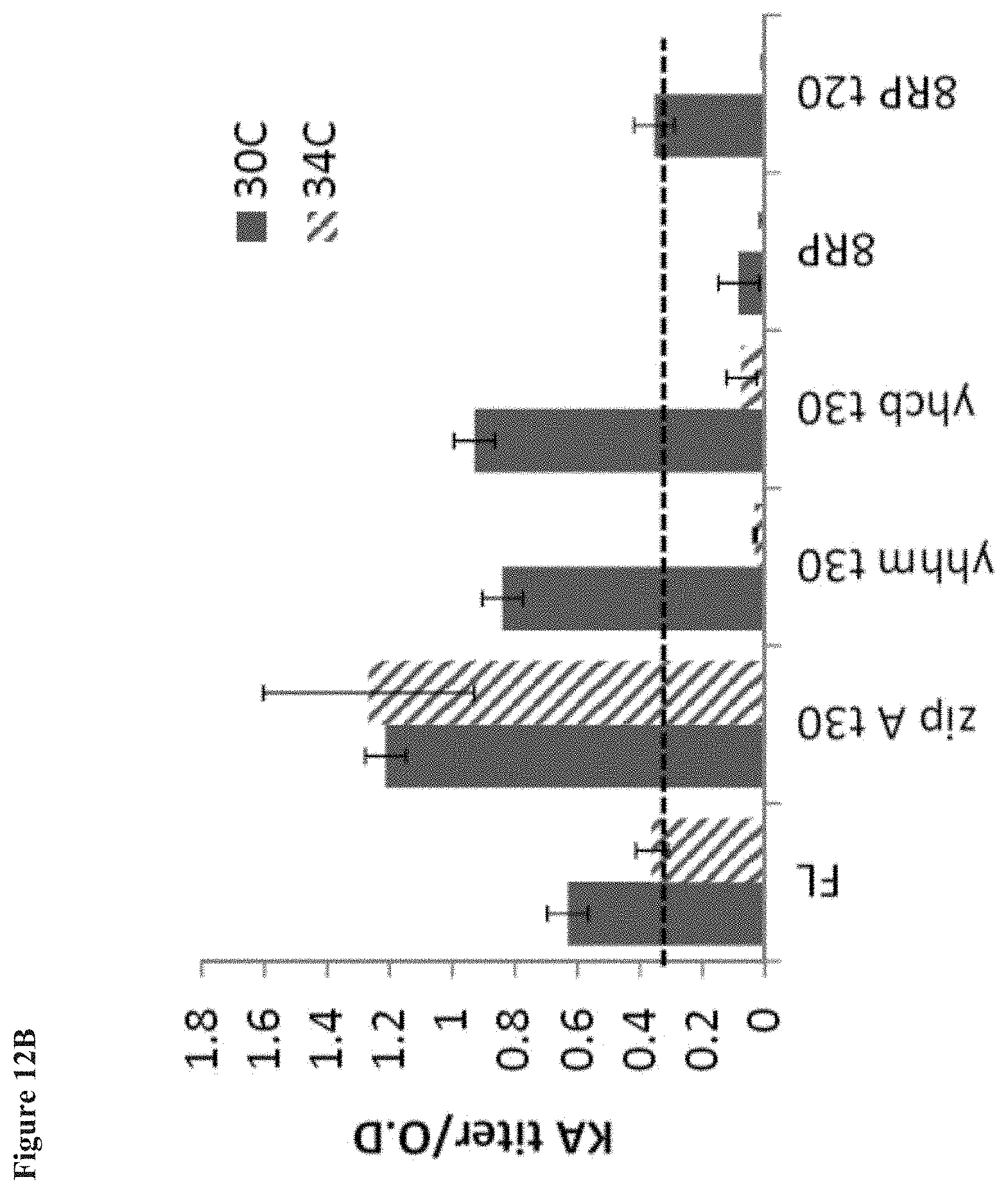
D00014
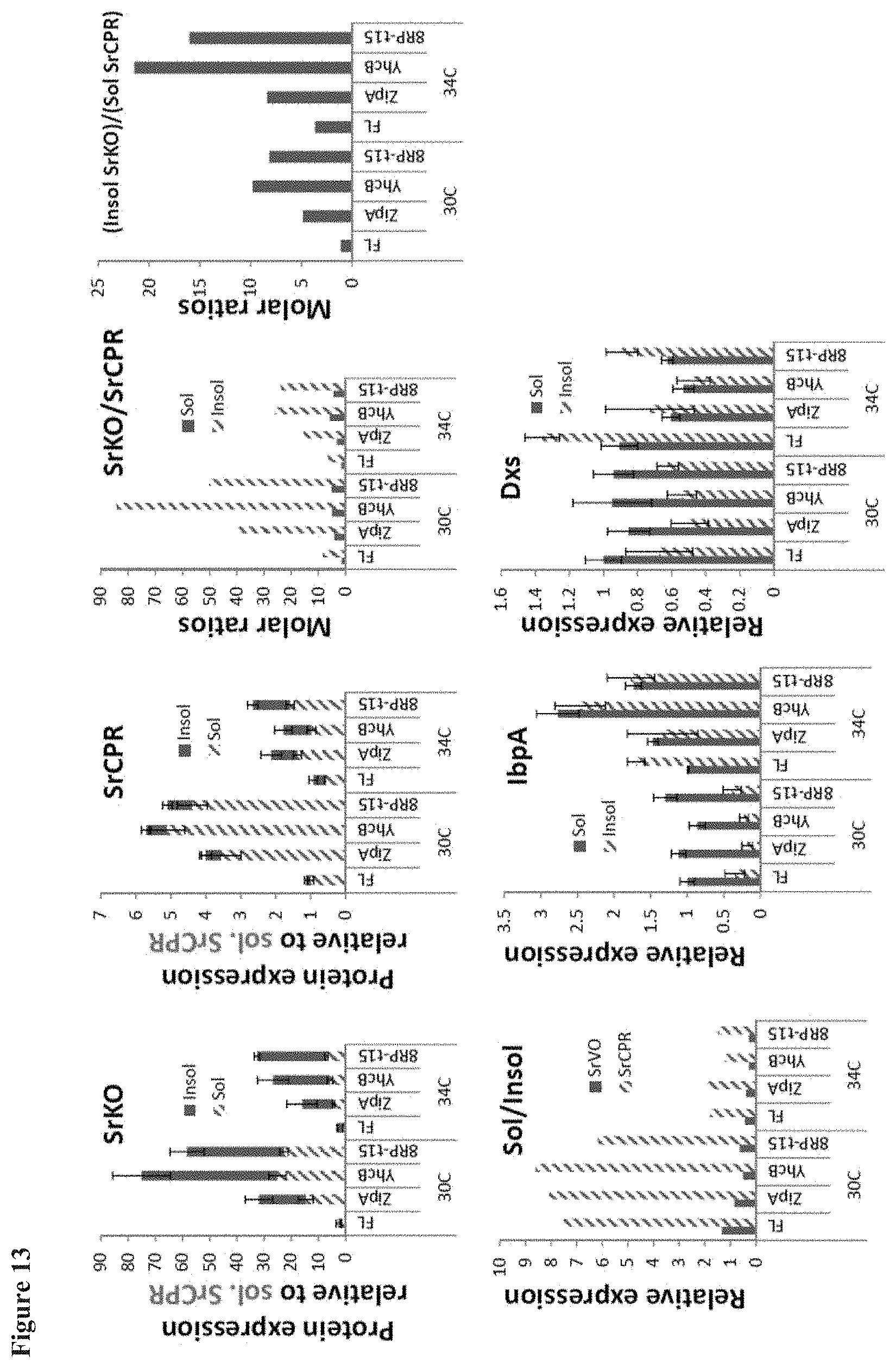
S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.