Radiolabeled Anti-pd-l1 Antibodies For Immuno-pet Imaging
Kelly; Marcus ; et al.
U.S. patent application number 16/915894 was filed with the patent office on 2020-11-05 for radiolabeled anti-pd-l1 antibodies for immuno-pet imaging. The applicant listed for this patent is REGENERON PHARMACEUTICALS, INC.. Invention is credited to Marcus Kelly, Dangshe Ma, William Olson, Gavin Thurston.
| Application Number | 20200345872 16/915894 |
| Document ID | / |
| Family ID | 1000004974790 |
| Filed Date | 2020-11-05 |

View All Diagrams
| United States Patent Application | 20200345872 |
| Kind Code | A1 |
| Kelly; Marcus ; et al. | November 5, 2020 |
RADIOLABELED ANTI-PD-L1 ANTIBODIES FOR IMMUNO-PET IMAGING
Abstract
Radiolabeled anti-PD-L1 antibodies and their use in immuno-PET imaging are provided herein. Included are methods of detecting the presence of PD-L1 proteins in a patient or sample.
| Inventors: | Kelly; Marcus; (New York, NY) ; Ma; Dangshe; (Millwood, NY) ; Olson; William; (Yorktown Heights, NY) ; Thurston; Gavin; (Briarcliff Manor, NY) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 1000004974790 | ||||||||||
| Appl. No.: | 16/915894 | ||||||||||
| Filed: | June 29, 2020 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 15829311 | Dec 1, 2017 | 10736976 | ||
| 16915894 | ||||
| 62428672 | Dec 1, 2016 | |||
| 62457267 | Feb 10, 2017 | |||
| 62569773 | Oct 9, 2017 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C07K 2317/565 20130101; A61K 51/1093 20130101; A61K 51/1045 20130101; C07K 16/2827 20130101; C07K 16/22 20130101; C07K 19/00 20130101; A61K 51/0474 20130101; A61K 2121/00 20130101; C07K 2317/515 20130101 |
| International Class: | A61K 51/10 20060101 A61K051/10; C07K 16/28 20060101 C07K016/28; A61K 51/04 20060101 A61K051/04; C07K 16/22 20060101 C07K016/22; C07K 19/00 20060101 C07K019/00 |
Claims
1.-10. (canceled)
11. A method for treating a tumor comprising: (a) selecting a subject with a solid tumor; (b) determining that the solid tumor is PD-L1-positive; and (c) administering one or more doses of an inhibitor of the PD-1/PD-L1 signaling axis to the subject in need thereof.
12. The method of claim 11, wherein step (b) comprises: (i) administering a radiolabeled antibody conjugate to the subject in need thereof, wherein the conjugate comprises: an antibody or antigen-binding fragment thereof that binds PD-L1, wherein said antibody or antigen-binding fragment thereof is covalently bonded to one or more moieties of formula (A): -L-M.sub.z (A) wherein L is a chelating moiety; M is a positron emitter; and z, independently at each occurrence, is 0 or 1; and wherein at least one of z is 1; and (ii) imaging localization of the radiolabeled antibody conjugate in the tumor by positron emission tomography (PET) imaging, wherein presence of the radiolabeled antibody conjugate in the tumor indicates that the tumor is PD-L1-positive.
13. The method of claim 12, wherein the subject is administered 0.1-10 mg/kg of the radiolabeled antibody conjugate.
14. The method of claim 12, wherein the radiolabeled antibody conjugate is administered sub-cutaneously or intravenously to the subject.
15. The method of claim 12, wherein PET imaging is done 2-7 days after administering the radiolabeled antibody conjugate.
16. The method of claim 12, wherein step (b) is carried out before treating the subject with an inhibitor of the PD-1/PD-L1 signaling axis.
17. The method of claim 12 further comprising: (a) administering the radiolabeled antibody conjugate after treating the subject with at least one dose of an inhibitor of the PD-1/PD-L1 signaling axis; and (b) imaging localization of the radiolabeled antibody conjugate in the tumor by PET imaging, wherein a decrease from the baseline in the area of localization of the radiolabeled antibody conjugate in the tumor indicates tumor regression.
18. The method of claim 17, wherein the subject is administered the radiolabeled antibody conjugate 1-20 weeks after administration of the inhibitor of the PD-1/PD-L1 signaling axis.
19. The method of claim 11, wherein the tumor is selected from the group consisting of blood cancer, brain cancer, renal cell cancer, ovarian cancer, bladder cancer, prostate cancer, breast cancer, hepatic cell carcinoma, bone cancer, colon cancer, non-small-cell lung cancer, squamous cell carcinoma of head and neck, colorectal cancer, mesothelioma, B cell lymphoma, and melanoma.
20. The method of claim 11, wherein the inhibitor of the PD-1/PD-L1 signaling axis is an anti-PD-1 antibody or antigen-binding fragment thereof.
21. The method of claim 20, wherein the anti-PD-1 antibody is selected from the group consisting of nivolumab, pembrolizumab and REGN2810.
22. The method of claim 11, wherein the inhibitor of the PD-1/PD-L1 signaling axis is an anti-PD-L1 antibody or antigen-binding fragment thereof.
23. The method of claim 22, wherein the anti-PD-L1 antibody is atezolizumab.
24. The method of claim 22, wherein the anti-PD-L1 antibody or antigen-binding fragment thereof comprises three heavy chain complementarity determining regions (HCDRs) in a heavy chain variable region (HCVR) of SEQ ID NO: 82; and three light chain complementarity determining regions (LCDRs) in a light chain variable region (LCVR) of SEQ ID NO: 90.
25. The method of claim 22, wherein the anti-PD-L1 antibody or antigen-binding fragment thereof comprises three HCDRs (HCDR1, HCDR2 and HCDR3) and three LCDRs (LCDR1, LCDR2 and LCDR3), wherein the HCDR1 comprises the amino acid sequence of SEQ ID NO: 84; the HCDR2 comprises the amino acid sequence of SEQ ID NO: 86; the HCDR3 comprises the amino acid sequence of SEQ ID NO: 88; the LCDR1 comprises the amino acid sequence of SEQ ID NO: 92; the LCDR2 comprises the amino acid sequence of SEQ ID NO: 94; and the LCDR3 comprises the amino acid sequence of SEQ ID NO: 96.
26. (canceled)
27. The method of claim 12, wherein the chelating moiety comprises desferrioxamine.
28. The method of claim 12, wherein the positron emitter is .sup.89Zr.
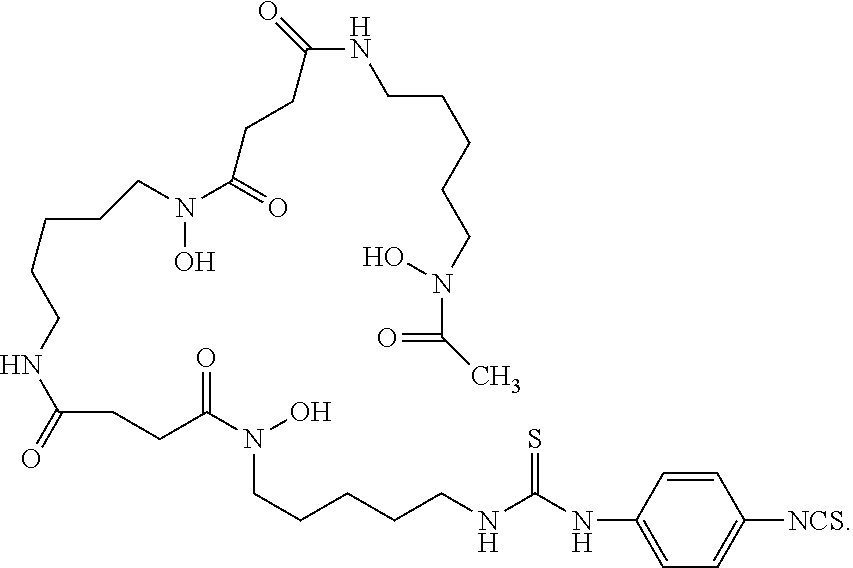
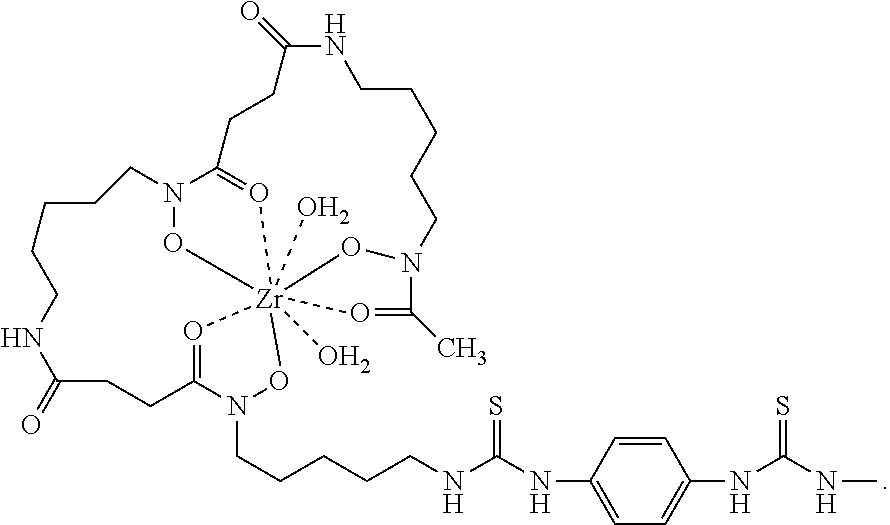
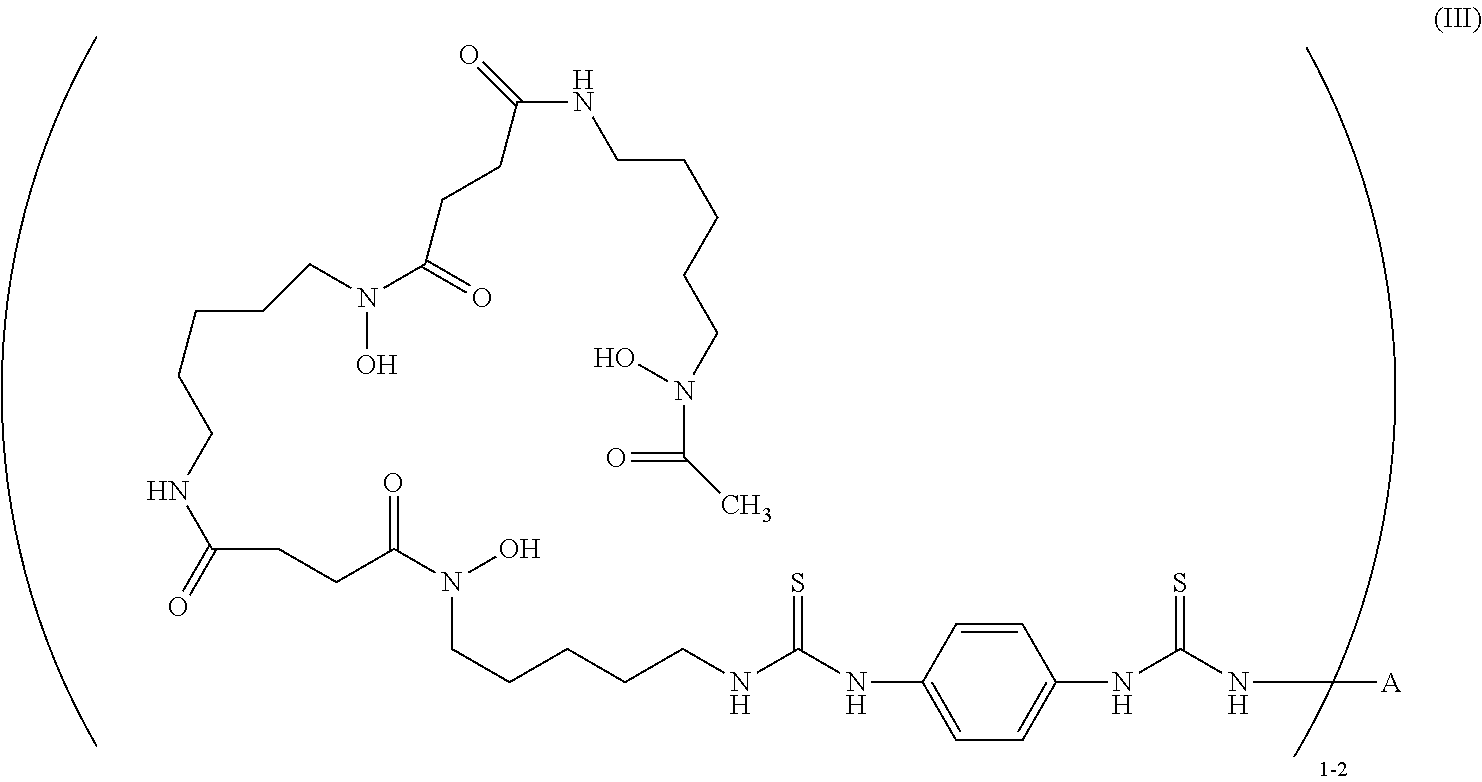
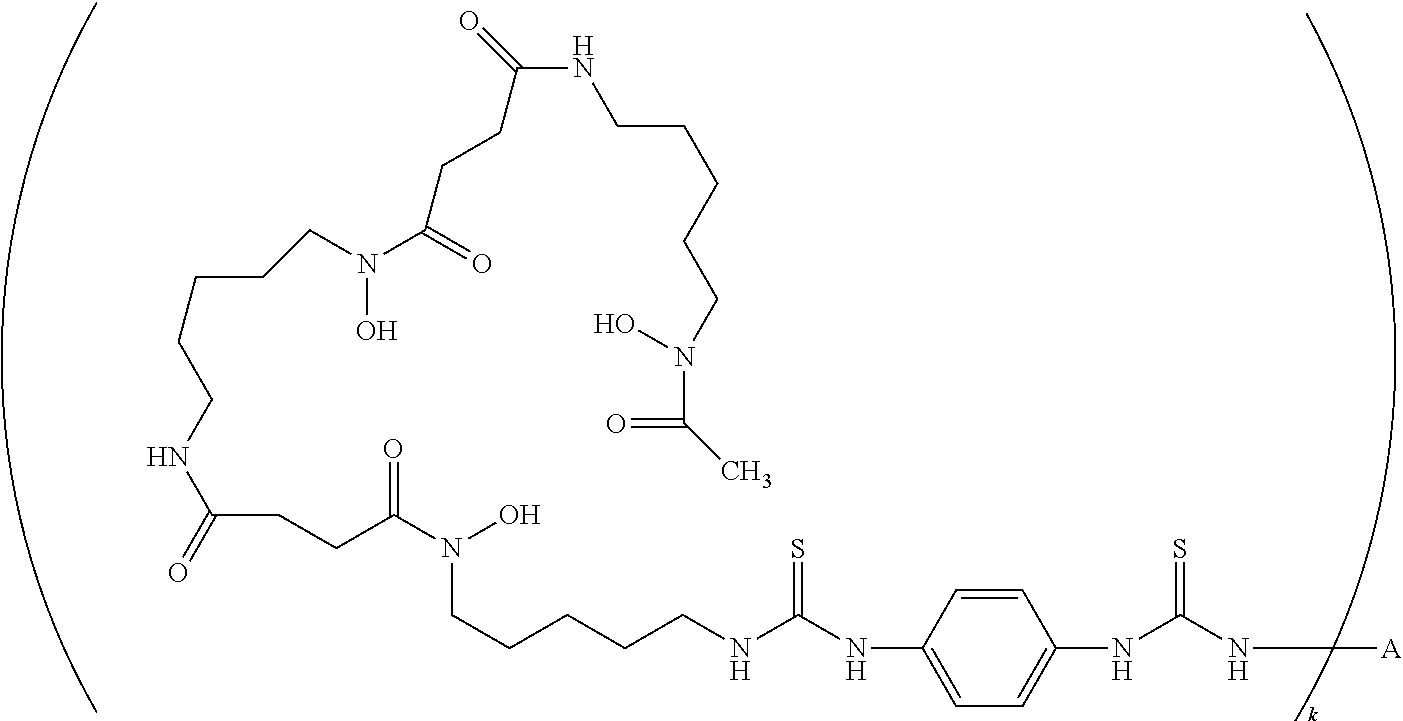
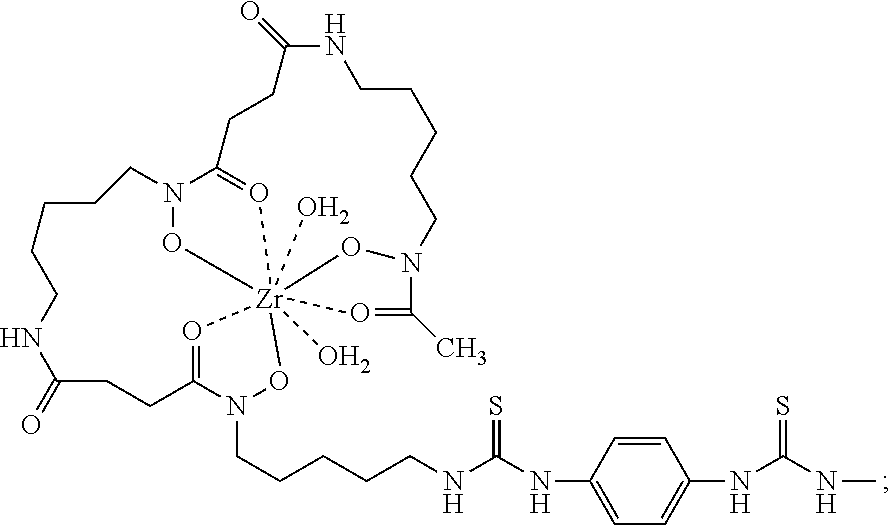
29. The method of claim 12, wherein -L-M is ##STR00006## wherein Zr is the positron emitter .sup.89Zr.
30. The method of claim 12, wherein antibody or antigen-binding fragment thereof is covalently bonded to one, two, or three moieties of Formula (A).
31. The method of claim 12, wherein the antibody has one or more properties selected from the group consisting of: (a) binds monomeric PD-L1 with a binding dissociation equilibrium constant (K.sub.D) of less than about 310 pM as measured in a surface plasmon resonance assay at 37.degree. C.; (b) binds monomeric human PD-L1 with a K.sub.D less than about 180 pM in a surface plasmon resonance assay at 25.degree. C.; (c) binds dimeric human PD-L1 with a K.sub.D of less than about 15 pM as measured in a surface plasmon resonance assay at 37.degree. C.; and (d) binds dimeric human PD-L1 with a K.sub.D less than about 8 pM in a surface plasmon resonance assay at 25.degree..
32. The method of claim 12, wherein the antibody comprises three heavy chain complementarity determining regions (HCDRs) in a heavy chain variable region (HCVR), wherein the HCVR has an amino acid sequence selected from the group consisting of SEQ ID NOs: 2, 34, 50, 82, 98, 146, 162, 178, 186, 234, 250, 266, 290, 306, 314, and 330; and three light chain complementarity determining regions (LCDRs) in a light chain variable region (LCVR), wherein the LCVR has an amino acid sequence selected from the group consisting of SEQ ID NOs: 10, 42, 58, 90, 106, 154, 170, 194, 242, 258, and 274.
Description
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] This application is a divisional application of U.S. patent application Ser. No. 15/829,311, filed Dec. 1, 2017, which claims the benefit under 34 U.S.C. .sctn. 119(e) of U.S. Provisional Application No. 62/428,672, filed Dec. 1, 2016, U.S. Provisional Application No. 62/457,267, filed Feb. 10, 2017, and U.S. Provisional Application No. 62/569,773, filed Oct. 9, 2017, all of which are herein specifically incorporated by reference in their entireties.
FIELD
[0002] This disclosure relates to radiolabeled anti-PD-L1 antibodies and their use in immuno-PET imaging.
SEQUENCE LISTING
[0003] An official copy of the sequence listing is submitted concurrently with the specification electronically via EFS-Web as an ASCII formatted sequence listing with a file name of "10305US01_Sequence_Listing_ST25.txt", a creation date of Dec. 1, 2017, and a size of about 117 KB. The sequence listing contained in this ASCII formatted document is part of the specification and is herein incorporated by reference in its entirety.
BACKGROUND
[0004] Programmed death-ligand 1 (PD-L1) (also called B7-H1 or CD274) is a 290 amino acid protein receptor ligand expressed widely on both lymphoid and non-lymphoid tissues such as CD4 and CD8 T-cells, macrophage lineage cells, peripheral tissues as well as on tumor cells, and virally-infected cells (Dong et al 1999, Nature Med.). PD-L1 binds to receptors PD-1 and B7-1 which belong to the CD28/CTLA-4 (cytotoxic T lymphocyte antigen)/ICOS (inducible co-stimulator) family of T-cell co-inhibitory receptors (Chen et al 2013, Nature Rev. Immunol. 13: 227-242) and attenuates the immune response by inhibiting T-cell activation. PD-L1 binding to PD-1 or B7-1 results in decreased T-cell proliferation and cytokine secretion, compromising humoral and cellular immune responses in diseases such as cancer, and viral infection. The expression of PD-L1 on tumor cells and virally-infected cells is exploited by tumors and chronic viral infections to evade immune response. PD-L1 is expressed on a wide variety of tumors and studies on animal models have shown that PD-L1 on tumors inhibits T-cell activation and lysis of tumor cells and may lead to increased death of tumor-specific T-cells. In chronic viral infections, PD-L1 expressed on virally-infected cells binds to PD-1 on virus-specific T-cells and these T-cells become "exhausted" with loss of effector functions and proliferative capacity (Freeman 2008, PNAS 105: 10275-10276). The PD-1: PD-L1 system also plays an important role in induced T-regulatory (Treg) cell development and in sustaining Treg function (Francisco et al 2010, Immunol. Rev. 236: 219-242). Blocking PD-L1 with antagonists, including monoclonal antibodies, has been studied in treatments of cancer and chronic viral infections (Ribas 2012, NEJM 366: 2517-2519; Freeman 2008, PNAS 105: 10275-10276; Sheridan 2012, Nature Biotechnology 30: 729-730).
[0005] Immuno-positron emission tomography (PET) is a diagnostic imaging tool that utilizes monoclonal antibodies labeled with positron emitters, combining the targeting properties of an antibody with the sensitivity of positron emission tomography cameras. See, e.g., The Oncologist, 12: 1379 (2007); Journal of Nuclear Medicine, 52(8): 1171 (2011). Immuno-PET enables the visualization and quantification of antigen and antibody accumulation in vivo and, as such, can serve as an important tool for diagnostics and complementing therapy. For example, immuno-PET can aid in the selection of potential patient candidates for a particular therapy, as well as in the monitoring of treatment.
[0006] As both PD1 and PD-L1 have emerged as targets for immunotherapy, there is need for diagnostic tools for anti-PD1 and/or anti-PD-L1 therapy, including, inter alia, diagnostic tools that enable the detection of suitable patient candidates for said therapy.
BRIEF SUMMARY
[0007] Included in this disclosure are radiolabeled anti-PD-L1 antibody conjugates for use in immuno-PET imaging.
[0008] In one aspect, the conjugate comprises an anti-PD-L1 antibody or antigen-binding fragment thereof, a chelating moiety, and a positron emitter.
[0009] Provided herein are also processes for synthesizing said conjugates and synthetic intermediates useful for the same.
[0010] Provided herein are also methods of imaging a tissue that expresses PD-L1, the methods comprising administering a radiolabeled anti-PD-L1 antibody conjugate described herein to the tissue; and visualizing the PD-L1 expression by positron emission tomography (PET) imaging.
[0011] Provided herein are also methods for detecting PD-L1 in a tissue, the methods comprising administering a radiolabeled anti-PD-L1 antibody conjugate described herein to the tissue; and visualizing the PD-L1 expression by PET imaging. In one embodiment, the tissue is present in a human subject. In certain embodiments, the subject is a non-human mammal. In certain embodiments, the subject has a disease or disorder such as cancer, an inflammatory disease, or an infection.
[0012] In some aspects, the subject is administered a dose of 5 mg, or 10 mg, or 20 mg, of a radiolabeled anti-PD-L1 antibody conjugate.
[0013] Provided herein are also methods for identifying a patient to be suitable for anti-tumor therapy comprising an inhibitor of the PD-1/PD-L1 signaling axis, the methods comprising selecting a patient with a solid tumor, administering a radiolabeled antibody conjugate described herein, and visualizing the administered radiolabeled antibody conjugate in the tumor by PET imaging wherein presence of the radiolabeled antibody conjugate in the tumor identifies the patient as suitable for anti-tumor therapy comprising an inhibitor of the PD-1/PD-L1 signaling axis.
[0014] Provided herein are also methods of treating a tumor, the methods comprising selecting a subject with a solid tumor; determining that the solid tumor is PD-L1-positive; and administering an anti-tumor therapy to the subject in need thereof. In certain embodiments, the anti-tumor therapy comprises an inhibitor of the PD-1/PD-L1 signaling axis (e.g., an anti-PD-1 antibody or an anti-PD-L1 antibody). In certain embodiments, the subject is administered a radiolabeled antibody conjugate described herein, and localization of the radiolabeled antibody conjugate is imaged via positron emission tomography (PET) imaging to determine if the tumor is PD-L1-positive.
[0015] Provided herein are also methods for monitoring the efficacy of an anti-tumor therapy in a subject, wherein the methods comprise selecting a subject with a solid tumor wherein the subject is being treated with an anti-tumor therapy; administering a radiolabeled conjugate described herein to the subject; imaging the localization of the administered radiolabeled conjugate in the tumor by PET imaging; and determining tumor growth, wherein a decrease from the baseline in uptake of the conjugate or radiolabeled signal indicates tumor regression and efficacy of the anti-tumor therapy. In certain embodiments, the anti-tumor therapy comprises an inhibitor of the PD-1/PD-L1 signaling axis (e.g., an anti-PD-1 antibody).
[0016] Provided herein are also methods for predicting response of a patient to an anti-tumor therapy comprising an inhibitor of the PD-1/PD-L1 signaling axis, the methods comprising selecting a patient with a solid tumor; and determining if the tumor is PD-L1-positive, wherein if the tumor is PD-L1-positive it indicates a positive response of the patient to an anti-tumor therapy comprising an inhibitor of the PD-1/PD-L1 signaling axis. In certain embodiments, the tumor is determined positive by administering a radiolabeled antibody conjugate of the present disclosure and localizing the radiolabeled antibody conjugate in the tumor by PET imaging wherein presence of the radiolabeled antibody conjugate in the tumor indicates that the tumor is PD-L1-positive.
BRIEF DESCRIPTION OF THE FIGURES
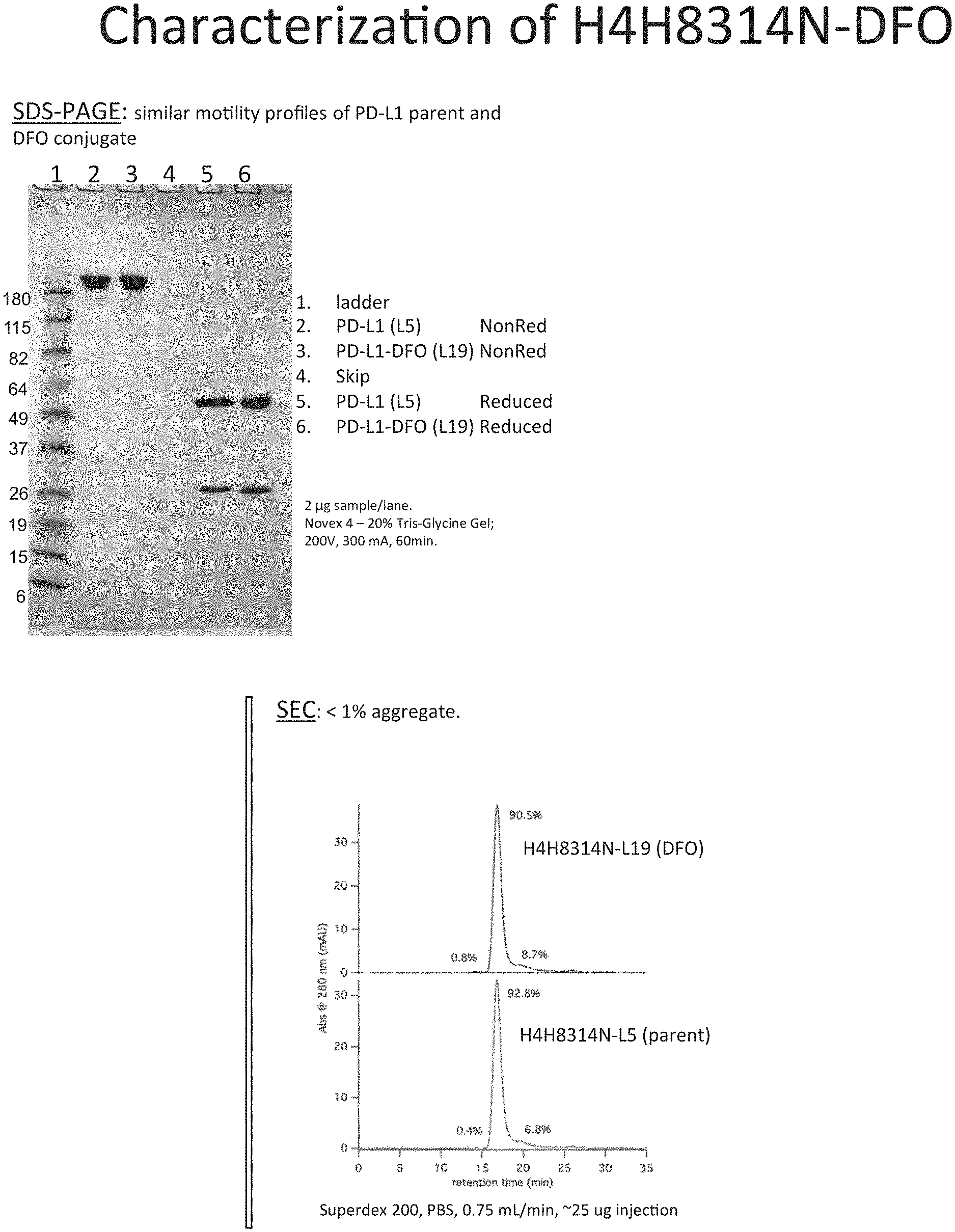
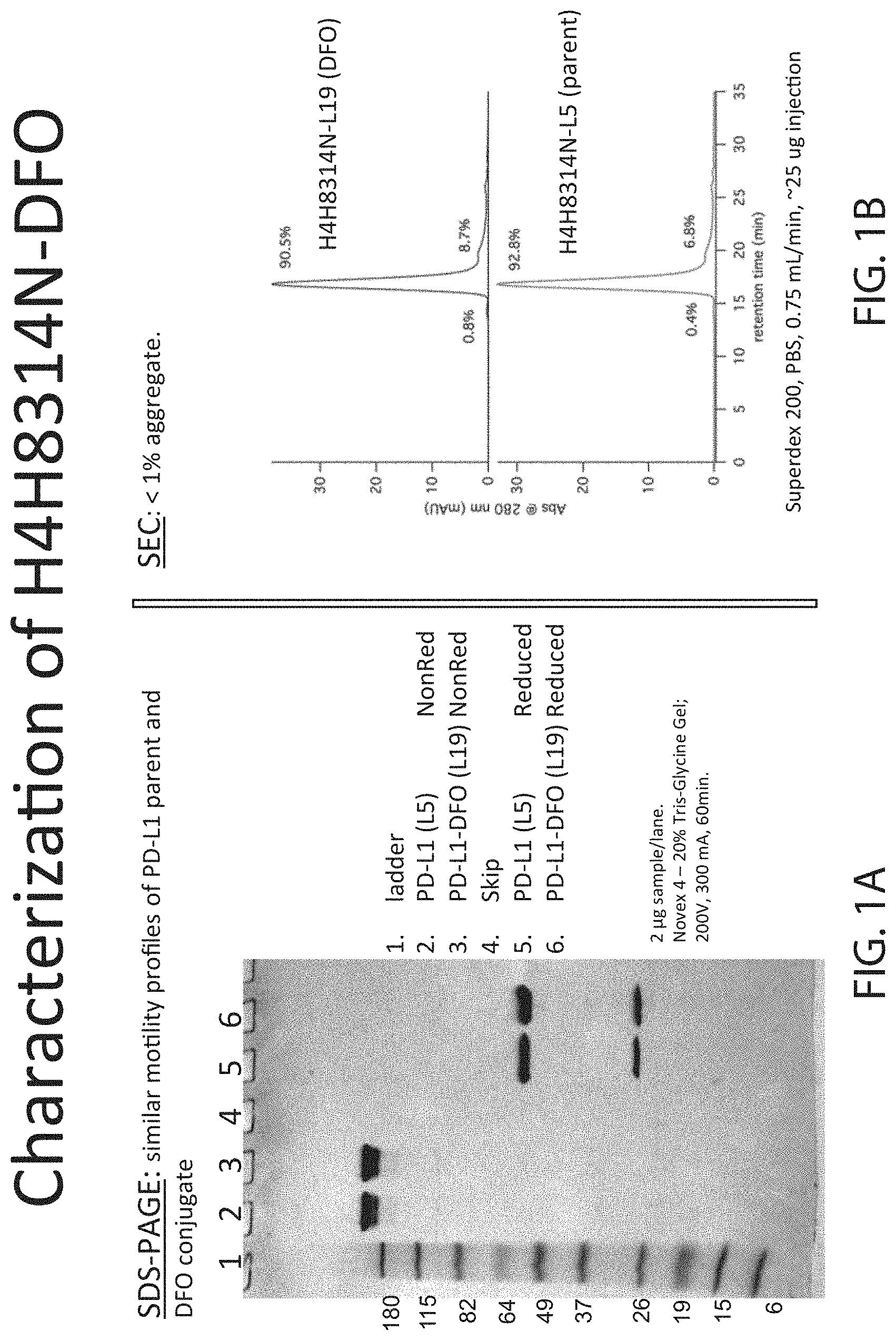
[0017] FIG. 1A depicts SDS-PAGE and FIG. 1B depicts SEC of un-modified anti-PD-L1 antibody and anti-PD-L1 DFO modified antibody.
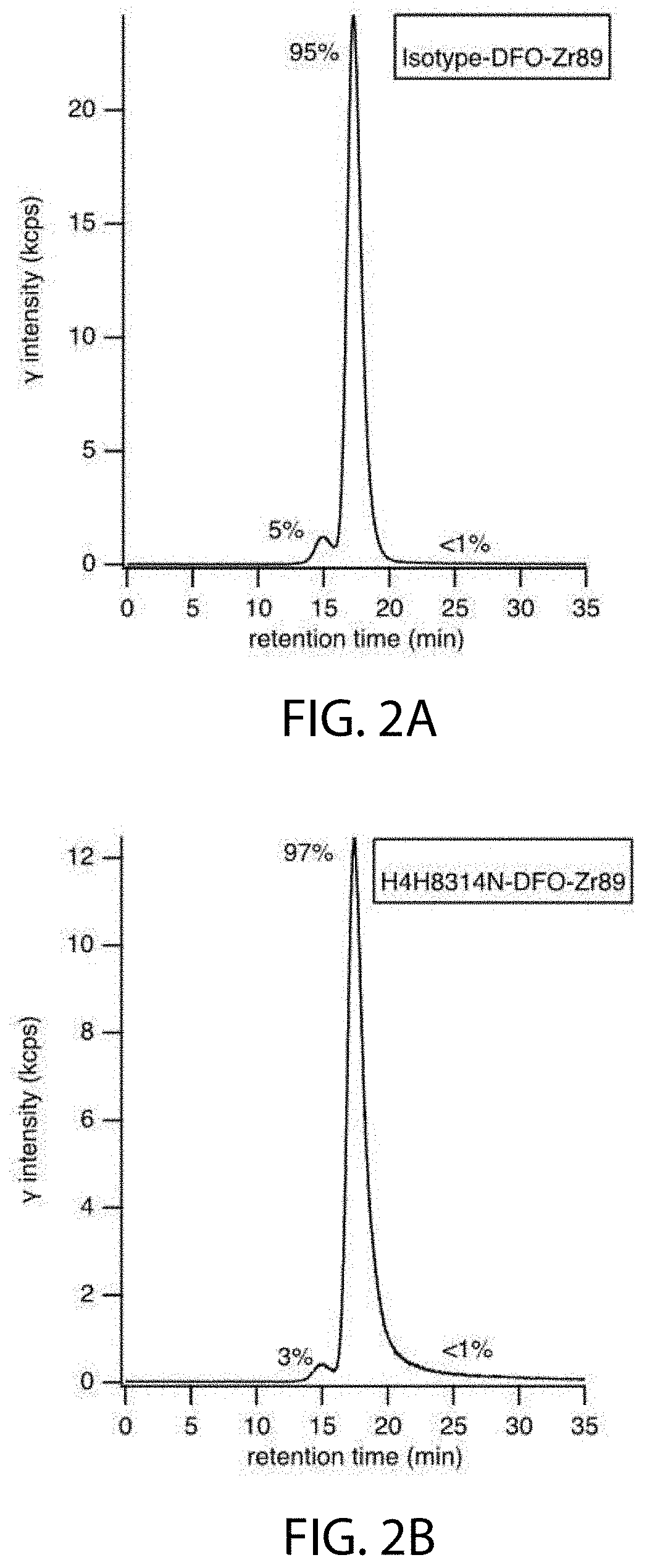
[0018] FIGS. 2A and 2B depict radio-SEC-HPLC after .sup.89Zr radiolabeling for Study 1.
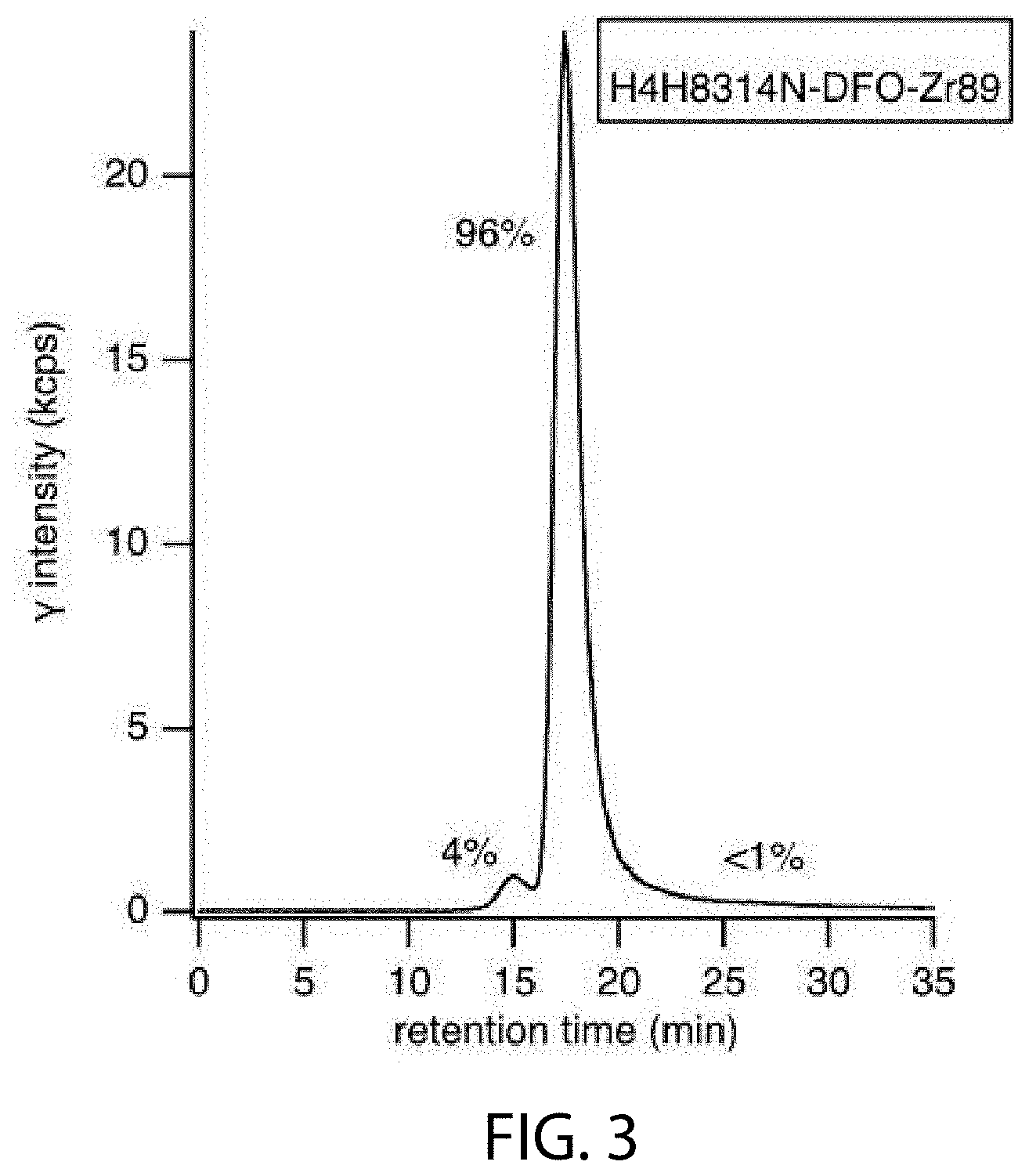
[0019] FIG. 3 depicts radio-SEC-HPLC of DFO-conjugate (anti-PD-L1) after .sup.89Zr radiolabeling for Study 2.
[0020] FIG. 4 depicts radio-SEC-HPLC SEC after .sup.89Zr radiolabeling Study 3.
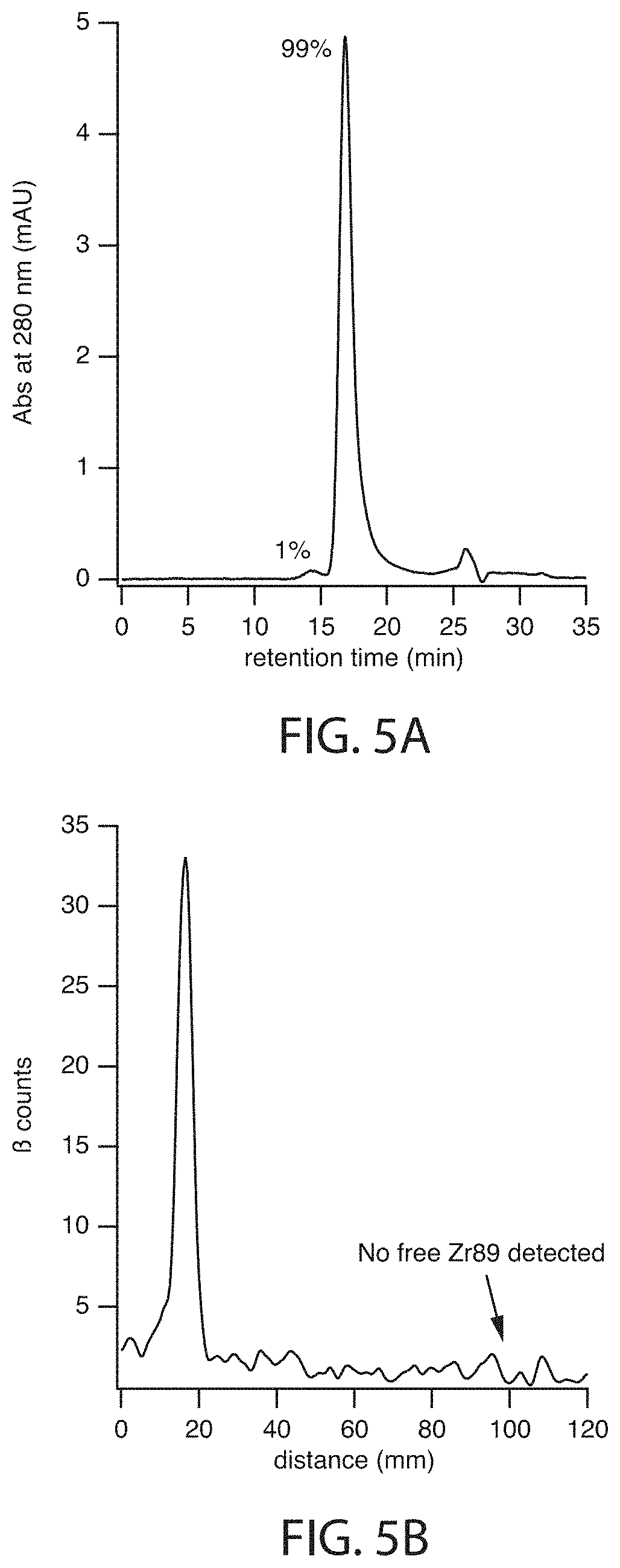
[0021] FIG. 5A depicts UV280-SEC-HPLC chromatogram and FIG. 5B depicts radio-iTLC trace after .sup.89Zr radiolabeling for Study 1.
[0022] FIGS. 6A, 6B, 6C, and 6D shows hPD-L1 expression by tumor cell lines MC38-cOVA/eGFP-mPD-L1-/-hPD-L1.sup.Tg (FIG. 6A), LOX-IMVI (FIG. 6B), MDA-MB-231 (FIG. 6C), and SK-Br-3 (FIG. 6D) in vitro, as described in Example 5 herein.
[0023] FIG. 7 shows hPD-L1 expression by MC38-cOVA/eGFP-mPD-L1-/-hPD-L1.sup.Tg and LOX-IMVI tumor cells with or without interferon-gamma treatment in vitro in a second experiment, as described in Example 5 herein.
[0024] FIGS. 8A, 8B, 8C, 8D, 8E, and 8F depict chromatograms generated by SEC-HPLC analysis using samples from radioimmunoconjugate preparations of .sup.89Zr-DFO-anti-PD-L1 antibody conjugate for studies shown in FIG. 8A, FIG. 8B, FIG. 8D, and FIG. 8E, and of isotype control radioimmunoconjugate .sup.89Zr-DFO-IgG4.sup.P for studies shown in FIG. 8C and FIG. 8F. Chromatograms for absorbance at 280 nm are shown in FIG. 8A-FIG. 8C and radio-chromatograms for intensity of .gamma.-emission are shown in FIG. 8D-FIG. 8F. In FIG. 8A-FIG. 8C, elution of buffer components was also detected. These peaks of salts in the sample buffer (retention time>25 min, asterisk "*") were excluded from the integration of peak areas. Peaks are labeled to indicate HMW (high molecular weight) immunoconjugate ("1"), monomeric immunoconjugate ("2"), unincorporated .sup.89Zr ("3"), and salts in the sample buffer ("*"). Abbreviations: mAU=milli absorbance units; cps=counts per second.
[0025] FIG. 9 provides ex vivo biodistribution data for .sup.89Zr-DFO-anti-PD-L1 antibody conjugate in PD-1hu/hu-PD-L1hu/hu mice. Sixteen mice (2 groups of 8 animals each) were administered a single IV dose of 50 .mu.Ci (1 mg/kg).sup.89Zr-DFO-anti-PD-L1 antibody conjugate on day 0 and were sacrificed on day 6 (black columns) or day 10 (gray columns) post dosing. Blood, collected via cardiac puncture, and the indicated harvested tissues were weighed and radioactivity was determined. The percent injected dose per gram (% ID/g) values for individual samples collected on day 6 or 10 were calculated relative to the radioactivity of a dose-standard from injected material (.sup.89Zr-DFO-anti-PD-L1 antibody conjugate) and the weight of the individual samples. Data are plotted as mean.+-.SD.
DETAILED DESCRIPTION
I. Definitions
[0026] Unless defined otherwise herein, all technical and scientific terms used herein have the same meaning as commonly understood by one of ordinary skill in the art to which the disclosed subject matter belongs.
[0027] The term "PD-L1" refers to programmed death-ligand 1, also known as CD274 and B7H1. The amino acid sequence of full-length PD-L1 is provided in GenBank as accession number NP_054862.1. The term "PD-L1" also includes protein variants of PD-L1. The term "PD-L1" includes recombinant PD-L1 or a fragment thereof. The term also encompasses PD-L1 or a fragment thereof coupled to, for example, histidine tag, mouse or human Fc, or a signal sequence such as ROR1. For example, the term includes sequences comprising a mouse Fc (mIgG2a) or human Fc (hIgG1) at the C-terminal, coupled to amino acid residues 19-239 of full-length PD-L1 (NP_054862.1). Protein variants comprise a histidine tag at the C-terminal, coupled to amino acid residues 19-239 of NP_054862.1. Unless specified as being from a non-human species, the term "PD-L1" means human PD-L1. PD-L1 is a 290 amino acid protein with extracellular IgV-like and IgC-like domains (amino acids 19-239 of full length PD-L1), a transmembrane domain and an intracellular domain of approximately 30 amino acids. PD-L1 is constitutively expressed on many cells such as antigen presenting cells (e.g., dendritic cells, macrophages, and B-cells) and on hematopoietic and non-hematopoietic cells (e.g., vascular endothelial cells, pancreatic islets, and sites of immune privilege). PD-L1 is also expressed on a wide variety of tumors, and virally-infected cells and is a component of the immunosuppressive milieu (Ribas 2012, NEJM 366: 2517-2519). PD-L1 binds to one of two T-cell co-inhibitors PD-1 and B7-1.
[0028] The term "PD-1" refers to the programmed death-1 protein, a T-cell co-inhibitor, also known as CD279. The amino acid sequence of full-length PD-1 is provided in GenBank as accession number NP_005009.2. The term also encompasses PD-1 or a fragment thereof coupled to, for example, histidine tag, mouse or human Fc, or a signal sequence such as ROR1. For example, the term includes sequences comprising a mouse Fc (mlgG2a) or human Fc (hIgG1) at the C-terminal, coupled to amino acid residues 25-170 of NP_005009.2 with a C93S change. PD-1 is a member of the CD28/CTLA-4/ICOS family of T-cell co-inhibitors. PD-1 is a 288-amino acid protein with an extracellular N-terminal domain which is IgV-like, a transmembrane domain and an intracellular domain containing an immunoreceptor tyrosine-based inhibitory (ITIM) motif and an immunoreceptor tyrosine-based switch (ITSM) motif (Chattopadhyay et al 2009, Immunol. Rev.). The PD-1 receptor has two ligands, PD-L1 and PD-L2.
[0029] The term "B7-1" refers to the T-lymphocyte activation antigen, also known as costimulatory factor CD80. B7-1 is a 288 amino acid membrane receptor with an extracellular N-terminal domain which comprises IgV-like (aa 37-138) and IgC-like (aa 154-232) regions, a transmembrane domain (aa 243-263) and a C-terminal intracellular region (aa 263-288). The amino acid sequence of full-length B7-1 is provided in GenBank as accession number NP_005182.1.
[0030] As used herein, the term "T-cell co-inhibitor" refers to a ligand and/or receptor which modulates the immune response via T-cell activation or suppression. The term "T-cell co-inhibitor", also known as T-cell co-signaling molecule, includes, but is not limited to, PD-1, lymphocyte activation gene 3 protein (LAG-3, also known as CD223), cytotoxic T-lymphocyte antigen-4 (CTLA-4), B and T lymphocyte attenuator (BTLA), CD-28, 2B4, LY108, T-cell immunoglobulin and mucin-3 (TIM3), T-cell immunoreceptor with immunoglobulin and ITIM (TIGIT; also known as VSIG9), leucocyte associated immunoglobulin-like receptor 1 (LAIR1; also known as CD305), inducible T-cell costimulator (ICOS; also known as CD278), B7-1 (CD80), and CD160.
[0031] The term "antibody", as used herein, is intended to refer to immunoglobulin molecules comprised of four polypeptide chains, two heavy (H) chains and two light (L) chains inter-connected by disulfide bonds (i.e., "full antibody molecules"), as well as multimers thereof (e.g. IgM) or antigen-binding fragments thereof. Each heavy chain is comprised of a heavy chain variable region ("HCVR" or "V.sub.H") and a heavy chain constant region (comprised of domains C.sub.H1, C.sub.H2 and C.sub.H3). Each light chain is comprised of a light chain variable region ("LCVR or "V.sub.L") and a light chain constant region (CL). The V.sub.H and V.sub.L regions can be further subdivided into regions of hypervariability, termed complementarity determining regions (CDR), interspersed with regions that are more conserved, termed framework regions (FR). Each V.sub.H and V.sub.L is composed of three CDRs and four FRs, arranged from amino-terminus to carboxy-terminus in the following order: FR1, CDR1, FR2, CDR2, FR3, CDR3, FR4. In certain embodiments, the FRs of the antibody (or antigen binding fragment thereof) may be identical to the human germline sequences, or may be naturally or artificially modified. An amino acid consensus sequence may be defined based on a side-by-side analysis of two or more CDRs.
[0032] Substitution of one or more CDR residues or omission of one or more CDRs is also possible. Antibodies have been described in the scientific literature in which one or two CDRs can be dispensed with for binding. Padlan et al. (1995 FASEB J. 9:133-139) analyzed the contact regions between antibodies and their antigens, based on published crystal structures, and concluded that only about one fifth to one third of CDR residues actually contact the antigen. Padlan also found many antibodies in which one or two CDRs had no amino acids in contact with an antigen (see also, Vajdos et al. 2002 J Mol Biol 320:415-428).
[0033] CDR residues not contacting antigen can be identified based on previous studies (for example residues H60-H65 in CDRH2 are often not required), from regions of Kabat CDRs lying outside Chothia CDRs, by molecular modeling and/or empirically. If a CDR or residue(s) thereof is omitted, it is usually substituted with an amino acid occupying the corresponding position in another human antibody sequence or a consensus of such sequences. Positions for substitution within CDRs and amino acids to substitute can also be selected empirically. Empirical substitutions can be conservative or non-conservative substitutions.
[0034] The fully human anti-PD-L1 monoclonal antibodies disclosed herein may comprise one or more amino acid substitutions, insertions and/or deletions in the framework and/or CDR regions of the heavy and light chain variable domains as compared to the corresponding germline sequences. Such mutations can be readily ascertained by comparing the amino acid sequences disclosed herein to germline sequences available from, for example, public antibody sequence databases. The present disclosure includes antibodies, and antigen-binding fragments thereof, which are derived from any of the amino acid sequences disclosed herein, wherein one or more amino acids within one or more framework and/or CDR regions are mutated to the corresponding residue(s) of the germline sequence from which the antibody was derived, or to the corresponding residue(s) of another human germline sequence, or to a conservative amino acid substitution of the corresponding germline residue(s) (such sequence changes are referred to herein collectively as "germline mutations"). A person of ordinary skill in the art, starting with the heavy and light chain variable region sequences disclosed herein, can easily produce numerous antibodies and antigen-binding fragments which comprise one or more individual germline mutations or combinations thereof. In certain embodiments, all of the framework and/or CDR residues within the V.sub.H and/or V.sub.L domains are mutated back to the residues found in the original germline sequence from which the antibody was derived. In other embodiments, only certain residues are mutated back to the original germline sequence, e.g., only the mutated residues found within the first 8 amino acids of FR1 or within the last 8 amino acids of FR4, or only the mutated residues found within CDR1, CDR2 or CDR3. In other embodiments, one or more of the framework and/or CDR residue(s) are mutated to the corresponding residue(s) of a different germline sequence (i.e., a germline sequence that is different from the germline sequence from which the antibody was originally derived). Furthermore, the antibodies of the present disclosure may contain any combination of two or more germline mutations within the framework and/or CDR regions, e.g., wherein certain individual residues are mutated to the corresponding residue of a particular germline sequence while certain other residues that differ from the original germline sequence are maintained or are mutated to the corresponding residue of a different germline sequence. Once obtained, antibodies and antigen-binding fragments that contain one or more germline mutations can be easily tested for one or more desired property such as, improved binding specificity, increased binding affinity, improved or enhanced antagonistic or agonistic biological properties (as the case may be), reduced immunogenicity, etc. Antibodies and antigen-binding fragments obtained in this general manner are encompassed within the present disclosure.
[0035] The present disclosure also includes fully human anti-PD-L1 monoclonal antibodies comprising variants of any of the HCVR, LCVR, and/or CDR amino acid sequences disclosed herein having one or more conservative substitutions. For example, the present disclosure includes anti-PD-L1 antibodies having HCVR, LCVR, and/or CDR amino acid sequences with, e.g., 10 or fewer, 8 or fewer, 6 or fewer, 4 or fewer, etc. conservative amino acid substitutions relative to any of the HCVR, LCVR, and/or CDR amino acid sequences disclosed herein.
[0036] The term "human antibody", as used herein, is intended to include antibodies having variable and constant regions derived from human germline immunoglobulin sequences. The human mAbs of the disclosure may include amino acid residues not encoded by human germline immunoglobulin sequences (e.g., mutations introduced by random or site-specific mutagenesis in vitro or by somatic mutation in vivo), for example in the CDRs and in particular CDR3. However, the term "human antibody", as used herein, is not intended to include mAbs in which CDR sequences derived from the germline of another mammalian species (e.g., mouse), have been grafted onto human FR sequences.
[0037] The term "multi-specific antigen-binding molecules", as used herein refers to bispecific, tri-specific or multi-specific antigen-binding molecules, and antigen-binding fragments thereof. Multi-specific antigen-binding molecules may be specific for different epitopes of one target polypeptide or may contain antigen-binding domains specific for epitopes of more than one target polypeptide. A multi-specific antigen-binding molecule can be a single multifunctional polypeptide, or it can be a multimeric complex of two or more polypeptides that are covalently or non-covalently associated with one another. The term "multi-specific antigen-binding molecules" includes antibodies of the present disclosure that may be linked to or co-expressed with another functional molecule, e.g., another peptide or protein. For example, an antibody or fragment thereof can be functionally linked (e.g., by chemical coupling, genetic fusion, non-covalent association or otherwise) to one or more other molecular entities, such as a protein or fragment thereof to produce a bi-specific or a multi-specific antigen-binding molecule with a second binding specificity. According to the present disclosure, the term "multi-specific antigen-binding molecules" also includes bi-specific, tri-specific or multi-specific antibodies or antigen-binding fragments thereof. In certain embodiments, an antibody of the present disclosure is functionally linked to another antibody or antigen-binding fragment thereof to produce a bispecific antibody with a second binding specificity. Bispecific and multi-specific antibodies of the present disclosure are described elsewhere herein.
[0038] The term "specifically binds," or "binds specifically to", or the like, means that an antibody or antigen-binding fragment thereof forms a complex with an antigen that is relatively stable under physiologic conditions. Specific binding can be characterized by an equilibrium dissociation constant of at least about 1.times.10.sup.-8 M or less (e.g., a smaller K.sub.D denotes a tighter binding). Methods for determining whether two molecules specifically bind are well known in the art and include, for example, equilibrium dialysis, surface plasmon resonance, and the like. As described herein, antibodies have been identified by surface plasmon resonance, e.g., BIACORE.TM., which bind specifically to PD-L1. Moreover, multi-specific antibodies that bind to one domain in PD-L1 and one or more additional antigens or a bi-specific that binds to two different regions of PD-L1 are nonetheless considered antibodies that "specifically bind", as used herein.
[0039] The terms "antigen-binding portion" of an antibody, "antigen-binding fragment" of an antibody, and the like, as used herein, include any naturally occurring, enzymatically obtainable, synthetic, or genetically engineered polypeptide or glycoprotein that specifically binds an antigen to form a complex. The terms "antigen-binding fragment" of an antibody, or "antibody fragment", as used herein, refers to one or more fragments of an antibody that retain the ability to bind to PD-L1.
[0040] An "isolated antibody", as used herein, is intended to refer to an antibody that is substantially free of other antibodies (Abs) having different antigenic specificities (e.g., an isolated antibody that specifically binds PD-L1, or a fragment thereof, is substantially free of Abs that specifically bind antigens other than PD-L1.
[0041] The term "surface plasmon resonance", as used herein, refers to an optical phenomenon that allows for the analysis of real-time biomolecular interactions by detection of alterations in protein concentrations within a biosensor matrix, for example using the BIACORE.TM. system (Pharmacia Biosensor AB, Uppsala, Sweden and Piscataway, N.J.).
[0042] The term "K.sub.D", as used herein, is intended to refer to the equilibrium dissociation constant of a particular antibody-antigen interaction.
[0043] The term "epitope" refers to an antigenic determinant that interacts with a specific antigen binding site in the variable region of an antibody molecule known as a paratope. A single antigen may have more than one epitope. Thus, different antibodies may bind to different areas on an antigen and may have different biological effects. The term "epitope" also refers to a site on an antigen to which B and/or T cells respond. It also refers to a region of an antigen that is bound by an antibody. Epitopes may be defined as structural or functional. Functional epitopes are generally a subset of the structural epitopes and have those residues that directly contribute to the affinity of the interaction. Epitopes may also be conformational, that is, composed of non-linear amino acids. In certain embodiments, epitopes may include determinants that are chemically active surface groupings of molecules such as amino acids, sugar side chains, phosphoryl groups, or sulfonyl groups, and, in certain embodiments, may have specific three-dimensional structural characteristics, and/or specific charge characteristics.
[0044] The term "substantial identity" or "substantially identical," when referring to a nucleic acid or fragment thereof, indicates that, when optimally aligned with appropriate nucleotide insertions or deletions with another nucleic acid (or its complementary strand), there is nucleotide sequence identity in at least about 90%, and more preferably at least about 95%, 96%, 97%, 98% or 99% of the nucleotide bases, as measured by any well-known algorithm of sequence identity, such as FASTA, BLAST or GAP.
[0045] As applied to polypeptides, the term "substantial similarity" or "substantially similar" means that two peptide sequences, when optimally aligned, such as by the programs GAP or BESTFIT using default gap weights, share at least 90% sequence identity, even more preferably at least 95%, 98% or 99% sequence identity. Preferably, residue positions, which are not identical, differ by conservative amino acid substitutions. A "conservative amino acid substitution" is one in which an amino acid residue is substituted by another amino acid residue having a side chain (R group) with similar chemical properties (e.g., charge or hydrophobicity). In general, a conservative amino acid substitution will not substantially change the functional properties of a protein. In cases where two or more amino acid sequences differ from each other by conservative substitutions, the percent or degree of similarity may be adjusted upwards to correct for the conservative nature of the substitution. Means for making this adjustment are well known to those of skill in the art. See, e.g., Pearson (1994) Methods Mol. Biol. 24: 307-331, which is herein incorporated by reference. Examples of groups of amino acids that have side chains with similar chemical properties include 1) aliphatic side chains: glycine, alanine, valine, leucine and isoleucine; 2) aliphatic-hydroxyl side chains: serine and threonine; 3) amide-containing side chains: asparagine and glutamine; 4) aromatic side chains: phenylalanine, tyrosine, and tryptophan; 5) basic side chains: lysine, arginine, and histidine; 6) acidic side chains: aspartate and glutamate, and 7) sulfur-containing side chains: cysteine and methionine. Preferred conservative amino acids substitution groups are: valine-leucine-isoleucine, phenylalanine-tyrosine, lysine-arginine, alanine-valine, glutamate-aspartate, and asparagine-glutamine. Alternatively, a conservative replacement is any change having a positive value in the PAM250 log-likelihood matrix disclosed in Gonnet et al. (1992) Science 256: 1443 45, herein incorporated by reference. A "moderately conservative" replacement is any change having a nonnegative value in the PAM250 log-likelihood matrix. Sequence similarity for polypeptides is typically measured using sequence analysis software. Protein analysis software matches similar sequences using measures of similarity assigned to various substitutions, deletions and other modifications, including conservative amino acid substitutions. For instance, GCG software contains programs such as GAP and BESTFIT which can be used with default parameters to determine sequence homology or sequence identity between closely related polypeptides, such as homologous polypeptides from different species of organisms or between a wild type protein and a mutein thereof. See, e.g., GCG Version 6.1. Polypeptide sequences also can be compared using FASTA with default or recommended parameters; a program in GCG Version 6.1. FASTA (e.g., FASTA2 and FASTA3) provides alignments and percent sequence identity of the regions of the best overlap between the query and search sequences (Pearson (2000) supra). Another preferred algorithm when comparing a sequence of the disclosure to a database containing a large number of sequences from different organisms is the computer program BLAST, especially BLASTP or TBLASTN, using default parameters. See, e.g., Altschul et al. (1990) J. Mol. Biol. 215: 403-410 and (1997) Nucleic Acids Res. 25:3389-3402, each of which is herein incorporated by reference.
[0046] By the phrase "therapeutically effective amount" is meant an amount that produces the desired effect for which it is administered. The exact amount will depend on the purpose of the treatment, and will be ascertainable by one skilled in the art using known techniques (see, for example, Lloyd (1999) The Art, Science and Technology of Pharmaceutical Compounding).
[0047] As used herein, the term "subject" refers to an animal, preferably a mammal, in need of amelioration, prevention and/or treatment of a disease or disorder such as chronic viral infection, cancer or autoimmune disease.
II. Radiolabeled Immunoconjugates of PD-L1 Antibodies for Immuno-PET Imaging
[0048] Provided herein are radiolabeled antigen-binding proteins that bind programmed death-ligand 1 (PD-L1). In some embodiments, the radiolabeled antigen-binding proteins comprise an antigen-binding protein covalently linked to one or more chelating moieties, which are chemical moieties that are capable of chelating a positron emitter.
[0049] In some embodiments, provided herein are antigen-binding proteins that bind PD-L1, e.g., antibodies, wherein said antigen-binding proteins that bind PD-L1 are covalently bonded to one or more moieties having the following structure:
-L-M.sub.z
wherein L is a chelating moiety; M is a positron emitter; and z, independently at each occurrence, is 0 or 1; and wherein at least one of z is 1.
[0050] In some embodiments, the radiolabeled antigen-binding protein is a compound of Formula (I):
M-L-A-[L-M.sub.z].sub.k (I)
A is a protein that binds PD-L1; L is a chelating moiety; M is a positron emitter; z is 0 or 1; and k is an integer from 0-30. In some embodiments, k is 1.
[0051] In certain embodiments, the radiolabeled antigen-binding protein is a compound of Formula (II):
A-[L-M].sub.k (II)
wherein A is a protein that binds PD-L1; L is a chelating moiety; M is a positron emitter; and k is an integer from 1-30.
[0052] In some embodiments, provided herein are compositions comprising a conjugate having the following structure:
A-L.sub.k
wherein A is a protein that binds PD-L1; L is a chelating moiety; and k is an integer from 1-30; wherein the conjugate is chelated with a positron emitter in an amount sufficient to provide a specific activity suitable for clinical PET imaging.
[0053] Suitable binding proteins, chelating moieties, and positron emitters are provided below.
[0054] A. PD-L1 Binding Proteins
[0055] Suitable PD-L1 binding protein are proteins that specifically bind to PD-L1, including those described in US Patent Publication No. US 2015-0203580 A1, incorporated herein by reference in its entirety. Exemplary anti-PD-L1 antibodies of the present disclosure are listed in Table 1 of US Patent Publication No. US 2015-0203580 A1, also presented below.
TABLE-US-00001 TABLE 1 Amino Acid Sequence Identifiers Antibody SEQ ID NOs: Designation HCVR HCDR1 HCDR2 HCDR3 LCVR LCDR1 LCDR2 LCDR3 H2M8306N 2 4 6 8 10 12 14 16 H2M8307N 18 20 22 24 26 28 30 32 H2M8309N 34 36 38 40 42 44 46 48 H2M8310N 50 52 54 56 58 60 62 64 H2M8312N 66 68 70 72 74 76 78 80 H2M8314N 82 84 86 88 90 92 94 96 H2M8316N 98 100 102 104 106 108 110 112 H2M8317N 114 116 118 120 122 124 126 128 H2M8321N 130 132 134 136 138 140 142 144 H2M8323N 146 148 150 152 154 156 158 160 H2M8718N 162 164 166 168 170 172 174 176 H2M8718N2 178 180 182 184 170 172 174 176 H2M8719N 186 188 190 192 194 196 198 200 H1H9323P 202 204 206 208 210 212 214 216 H1H9327P 218 220 222 224 226 228 230 232 H1H9329P 234 236 238 240 242 244 246 248 H1H9336P 250 252 254 256 258 260 262 264 H1H9344P2 266 268 270 272 274 276 278 280 H1H9345P2 282 284 286 288 274 276 278 280 H1H9351P2 290 292 294 296 274 276 278 280 H1H9354P2 298 300 302 304 274 276 278 280 H1H9364P2 306 308 310 312 274 276 278 280 H1H9373P2 314 316 318 320 274 276 278 280 H1H9382P2 322 324 326 328 274 276 278 280 H1H9387P2 330 332 334 336 274 276 278 280 H1H9396P2 338 340 342 344 274 276 278 280
Table 1 sets forth the amino acid sequence identifiers of the heavy chain variable regions (HCVRs), light chain variable regions (LCVRs), heavy chain complementarity determining regions (HCDR1, HCDR2 and HCDR3), and light chain complementarity determining regions (LCDR1, LCDR2 and LCDR3) of the exemplary anti-PD-L1 antibodies.
[0056] In some embodiments, the binding protein is an antibody or antigen binding fragment comprising an HCVR comprising an amino acid sequence selected from any of the HCVR amino acid sequences listed in Table 1, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity thereto.
[0057] In some embodiments, the binding protein is an antibody or antigen binding fragment comprising an LCVR comprising an amino acid sequence selected from any of the LCVR amino acid sequences listed in Table 1, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity thereto.
[0058] In some embodiments, the binding protein is an antibody or antigen binding fragment comprising an HCVR and an LCVR amino acid sequence pair (HCVR/LCVR) comprising any of the HCVR amino acid sequences listed in Table 1 paired with any of the LCVR amino acid sequences listed in Table 1. According to certain embodiments, the present disclosure provides antibodies, or antigen-binding fragments thereof, comprising an HCVR/LCVR amino acid sequence pair contained within any of the exemplary anti-PD-L1 antibodies listed in Table 1. In certain embodiments, the HCVR/LCVR amino acid sequence pair is selected from the group consisting of SEQ ID NOs: 2/10, 18/26, 34/42, 50/58, 66/74, 82/90, 98/106, 114/122, 130/138, 146/154, 162/170, 178/170, 186/194, 202/210, 218/226, 234/242, 250/258, 266/274, 282/274, 290/274, 298/274, 306/274, 314/274, 322/274, 330/274, and 338/274. In certain embodiments, the HCVR/LCVR amino acid sequence pair is selected from one of SEQ ID NOs: 82/90 (e.g., H2M8314N), 162/170 (e.g., H2M8718N), 306/274 (e.g., H1H9364P2), and 314/274 (e.g., H1H9373P2). In certain other embodiments, the HCVR/LCVR amino acid sequence pair is selected from one of SEQ ID NOs: 98/106 (e.g., H2M8316N), 146/154 (e.g., H2M8323N), 290/274 (e.g., H1H9351P2), and 330/274 (e.g., H1H9387P2).
[0059] In some embodiments, the binding protein is an antibody or antigen binding fragment comprising a heavy chain CDR1 (HCDR1) comprising an amino acid sequence selected from any of the HCDR1 amino acid sequences listed in Table 1 or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity.
[0060] In some embodiments, the binding protein is an antibody or antigen binding fragment comprising a heavy chain CDR2 (HCDR2) comprising an amino acid sequence selected from any of the HCDR2 amino acid sequences listed in Table 1 or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity.
[0061] In some embodiments, the binding protein is an antibody or antigen binding fragment comprising a heavy chain CDR3 (HCDR3) comprising an amino acid sequence selected from any of the HCDR3 amino acid sequences listed in Table 1 or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity.
[0062] In some embodiments, the binding protein is an antibody or antigen binding fragment comprising a light chain CDR1 (LCDR1) comprising an amino acid sequence selected from any of the LCDR1 amino acid sequences listed in Table 1 or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity.
[0063] In some embodiments, the binding protein is an antibody or antigen binding fragment comprising a light chain CDR2 (LCDR2) comprising an amino acid sequence selected from any of the LCDR2 amino acid sequences listed in Table 1 or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity.
[0064] In some embodiments, the binding protein is an antibody or antigen binding fragment comprising a light chain CDR3 (LCDR3) comprising an amino acid sequence selected from any of the LCDR3 amino acid sequences listed in Table 1 or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity.
[0065] In some embodiments, the binding protein is an antibody or antigen binding fragment comprising an HCDR3 and an LCDR3 amino acid sequence pair (HCDR3/LCDR3) comprising any of the HCDR3 amino acid sequences listed in Table 1 paired with any of the LCDR3 amino acid sequences listed in Table 1. According to certain embodiments, the present disclosure provides antibodies, or antigen-binding fragments thereof, comprising an HCDR3/LCDR3 amino acid sequence pair contained within any of the exemplary anti-PD-L1 antibodies listed in Table 1. In certain embodiments, the HCDR3/LCDR3 amino acid sequence pair is selected from the group consisting of SEQ ID NOs: 88/96 (e.g., H2M8314N), 168/176 (e.g., H2M8718N), 312/280 (e.g., H1H9364P2), and 320/280 (e.g., H1H9373P2). In certain other embodiments, the HCDR3/LCDR3 amino acid sequence pair is selected from the group consisting of SEQ ID NOs: 104/112 (e.g., H2M8316N), 152/160 (e.g., H2M8323N), 296/280 (e.g., H1H9351P2), and 336/280 (e.g., H1H9387P2).
[0066] In some embodiments, the binding protein is an antibody or antigen binding fragment comprising a set of six CDRs (i.e., HCDR1-HCDR2-HCDR3-LCDR1-LCDR2-LCDR3) contained within any of the exemplary anti-PD-L1 antibodies listed in Table 1. In certain embodiments, the HCDR1-HCDR2-HCDR3-LCDR1-LCDR2-LCDR3 amino acid sequence set is selected from the group consisting of SEQ ID NOs: 84-86-88-92-94-96 (e.g., H2M8314N); 164-166-168-172-174-176 (e.g., H2M8718N); 308-310-312-276-278-280 (e.g., H1H9364P2); and 316-318-320-276-278-280 (e.g., H1H9373P2). In certain other embodiments, the HCDR1-HCDR2-HCDR3-LCDR1-LCDR2-LCDR3 amino acid sequence set is selected from the group consisting of SEQ ID NOs: 100-102-104-108-110-112 (e.g., H2M8316N); 148-150-152-156-158-160 (e.g., H2M8323N); 292-294-296-276-278-280 (e.g., H1H9351P2); and 332-334-336-276-278-280 (e.g., H1H9387P2).
[0067] In some embodiments, the binding protein is an antibody or antigen binding fragment comprising a set of six CDRs (i.e., HCDR1-HCDR2-HCDR3-LCDR1-LCDR2-LCDR3) contained within an HCVR/LCVR amino acid sequence pair as defined by any of the exemplary anti-PD-L1 antibodies listed in Table 1. For example, in some embodiments, the binding protein is an antibody or antigen binding fragment comprising the HCDR1-HCDR2-HCDR3-LCDR1-LCDR2-LCDR3 amino acid sequences set contained within an HCVR/LCVR amino acid sequence pair selected from the group consisting of SEQ ID NOs: 82/90 (e.g., H2M8314N), 98/106 (e.g., H2M8316N), 146/154 (e.g., H2M8323N), 162/170 (e.g., H2M8718N), 290/274 (e.g., H1H9351P2), 306/274 (e.g., H1H9364P2), 314/274 (e.g., H1H9373P2) and 330/274 (e.g., H1H9387P2). Methods and techniques for identifying CDRs within HCVR and LCVR amino acid sequences are well known in the art and can be used to identify CDRs within the specified HCVR and/or LCVR amino acid sequences disclosed herein. Exemplary conventions that can be used to identify the boundaries of CDRs include, e.g., the Kabat definition, the Chothia definition, and the AbM definition. In general terms, the Kabat definition is based on sequence variability, the Chothia definition is based on the location of the structural loop regions, and the AbM definition is a compromise between the Kabat and Chothia approaches. See, e.g., Kabat, "Sequences of Proteins of Immunological Interest," National Institutes of Health, Bethesda, Md. (1991); Al-Lazikani et al., J. Mol. Biol. 273:927-948 (1997); and Martin et al., Proc. Natl. Acad. Sci. USA 86:9268-9272 (1989). Public databases are also available for identifying CDR sequences within an antibody.
[0068] In some embodiments, binding proteins are antibodies and antigen-binding fragments thereof that compete for specific binding to PD-L1 with an antibody or antigen-binding fragment thereof comprising the CDRs of a HCVR and the CDRs of a LCVR, wherein the HCVR and LCVR each has an amino acid sequence selected from the HCVR and LCVR sequences listed in Table 1.
[0069] In some embodiments, the binding proteins are isolated antibodies and antigen-binding fragments thereof that block PD-L1 binding to PD-1 or to B7-1. In some embodiments, the antibody or antigen-binding fragment thereof that blocks PD-L1 binding to PD-1 or to B7-1 may bind to the same epitope on PD-L1 as PD-1/B7-1 or may bind to a different epitope on PD-L1 as PD-1/B7-1. In certain embodiments, the antibodies of the disclosure that block PD-L1 binding to PD-1 or to B7-1 comprise the CDRs of an HCVR having an amino acid sequence selected from the group consisting of HCVR sequences listed in Table 1; and the CDRs of a LCVR having an amino acid sequence selected from the group consisting of LCVR sequences listed in Table 1.
[0070] In alternate embodiments, the present disclosure provides antibodies and antigen-binding fragments thereof that do not block PD-L1 binding to PD-1 or to B7-1. In certain embodiments, the present disclosure provides isolated antibodies or antigen-binding fragments thereof that bind PD-L1, wherein the antibodies or antigen-binding fragments thereof enhance PD-L1 binding to PD-1 or to B7-1. In some embodiments, the isolated antibodies or antigen-binding fragments thereof that enhance PD-L1 binding to PD-1/B7-1 comprise the CDRs of a HCVR, wherein the HCVR has an amino acid sequence selected from the group consisting of SEQ ID NOs: 18, 66, 114, 130, 202, 218, 266, 282, 298, 322 and 338; and the CDRs of a LCVR, wherein the LCVR has an amino acid sequence selected from the group consisting of SEQ ID NOs: 26, 74, 122, 138, 210, 226, and 274. In some embodiments, the isolated antibodies or antigen-binding fragments thereof comprise an HCVR/LCVR amino acid sequence pair selected from the group consisting of SEQ ID NOs: 18/26 (e.g., H2M8307N), 66/74 (e.g., H2M8312N), 114/122 (e.g., H2M8317N), 130/138 (e.g., H2M8321N), 202/210 (e.g., H1H9323P), 218/226 (e.g., H1H9327P), 266/274 (e.g., H1H9344P2), 282/274 (e.g., H1H9345P2), 298/274 (e.g., H1H9354P2), 322/274 (e.g., H1H9382P2), and 338/274 (e.g., H1H9396P2).
[0071] In some embodiments, the binding proteins are antibodies and antigen-binding fragments thereof that bind specifically to PD-L1 from human or other species. In certain embodiments, the antibodies may bind to human PD-L1 and/or to cynomolgus PD-L1.
[0072] In some embodiments, the binding proteins are antibodies and antigen-binding fragments thereof that cross-compete for binding to PD-L1 with a reference antibody or antigen-binding fragment thereof comprising the CDRs of a HCVR and the CDRs of a LCVR, wherein the HCVR and LCVR each has an amino acid sequence selected from the HCVR and LCVR sequences listed in Table 1.
[0073] In one embodiment, the binding protein is an isolated antibody or antigen-binding fragment that has one or more of the following characteristics: (a) blocks the binding of PD-L1 to PD-1 or to B7-1; (b) binds specifically to human PD-L1 and/or cynomolgus PD-L1; (c) inhibits T-cell proliferation in a mixed lymphocyte reaction (MLR) assay; and (d) increases IL-2 and/or interferon-gamma secretion in a MLR assay.
[0074] In some embodiments, the binding protein is an antibody or antigen binding fragment thereof may bind specifically to PD-L1 in an agonist manner, i.e., it enhances or stimulates PD-L1 binding and/or activity; in other embodiments, the antibody can bind specifically to PD-L1 in an antagonist manner, i.e., it blocks PD-L1 from binding to its receptor.
[0075] In certain embodiments, the antibodies or antigen-binding fragments are bispecific comprising a first binding specificity to PD-L1 and a second binding specificity for a second target epitope. The second target epitope may be another epitope on PD-L1 or on a different protein such as a T-cell co-inhibitor. In certain embodiments, the target epitope may be on a different cell including e.g., a different T-cell, a B-cell, a tumor cell, an autoimmune tissue cell or a virally infected cell.
[0076] In some embodiments, the antibodies and antigen-binding fragments of antibodies bind monomeric PD-L1 (e.g., at 25.degree. C. or at 37.degree. C.) with a K.sub.D of less than about 318 pM as measured by surface plasmon resonance, e.g., using the assay format as defined in Example 3 of US Patent Publication No. US 2015-0203580 A1, or substantially similar assay. In certain embodiments, the antibodies or antigen-binding fragments thereof bind monomeric PD-L1 with a K.sub.D of less than about 300 pM, less than about 250 pM, less than about 150 pM, less than about 100 pM, or less than about 50 pM, as measured by surface plasmon resonance, e.g., using the assay format as defined in Example 3 of US Patent Publication No. US 2015-0203580 A1, or a substantially similar assay.
[0077] In some embodiments, the antibodies and antigen-binding fragments thereof bind dimeric PD-L1 (e.g., at 25.degree. C. or at 37.degree. C.) with a K.sub.D of less than about 15 pM as measured by surface plasmon resonance, e.g., using the assay format as defined in Example 3 of US Patent Publication No. US 2015-0203580 A1 or sustainably similar assay. In certain embodiments, the antibodies or antigen-binding fragments thereof bind dimeric PD-L1 with a K.sub.D of less than about 12 pM, less than about 10 pM, less than about 8 pM, or less than about 5 pM, as measured by surface plasmon resonance, e.g., using the assay format as defined in Example 3 of US Patent Publication No. US 2015-0203580 A1, or a substantially similar assay.
[0078] In some embodiments, the antibodies or antigen-binding fragments thereof bind cynomolgus (Macaca fascicularis) PD-L1 (e.g., at 25.degree. C. or at 37.degree. C.) with a K.sub.D of less than about 28 nM as measured by surface plasmon resonance, e.g., using the assay format as defined in Example 3 of US Patent Publication No. US 2015-0203580 A1. In certain embodiments, the antibodies or antigen-binding fragments thereof bind cynomolgus PD-L1 with a K.sub.D of less than about 25 nM, less than about 20 nM, less than about 15 nM, less than about 10 nM, or less than about 5 nM, as measured by surface plasmon resonance, e.g., using the assay format as defined in Example 3 of US Patent Publication No. US 2015-0203580 A1, or a substantially similar assay.
[0079] In some embodiments, the antibodies and antigen-binding fragments thereof bind PD-L1 with a dissociative half-life (t1/2) of greater than about 1 minute as measured by surface plasmon resonance at 25.degree. C. or 37.degree. C., e.g., using an assay format as defined in Example 3 of US Patent Publication No. US 2015-0203580 A1, or a substantially similar assay. In certain embodiments, the antibodies or antigen-binding fragments bind PD-L1 with a t1/2 of greater than about 5 minutes, greater than about 10 minutes, greater than about 30 minutes, greater than about 50 minutes, greater than about 60 minutes, greater than about 70 minutes, greater than about 80 minutes, greater than about 90 minutes, greater than about 100 minutes, greater than about 200 minutes, greater than about 300 minutes, greater than about 400 minutes, greater than about 500 minutes, greater than about 600 minutes, greater than about 700 minutes, or greater than about 800 minutes, as measured by surface plasmon resonance at 25.degree. C. or 37.degree. C., e.g., using an assay format as defined in Example 3 of US Patent Publication No. US 2015-0203580 A1 (e.g., mAb-capture or antigen-capture format), or a substantially similar assay.
[0080] In some embodiments, the antibodies or antigen-binding fragments thereof block PD-L1 binding to PD-1 with an IC.sub.50 of less than about 770 pM as determined using a ELISA-based immunoassay assay, e.g., as shown in Example 4 of US Patent Publication No. US 2015-0203580 A1, or a substantially similar assay. In some embodiments, the antibodies or antigen-binding fragments thereof block PD-L1 binding to B7-1 with an IC.sub.50 of less than about 10 nM as determined using a ELISA-based immunoassay assay, e.g., as shown in Example 4 of US Patent Publication No. US 2015-0203580 A1, or a substantially similar assay. In some embodiments, the antibodies and antigen-binding fragments thereof bind to PD-L1 and enhance the binding of PD-L1 to PD-1 or to B7-1.
[0081] In some embodiments, the antibodies bind to the extracellular domain of PD-L1 or to a fragment of the domain. In some embodiments, the antibodies bind to more than one domain (cross-reactive antibodies). In certain embodiments, the antibodies of the bind to an epitope located in the extracellular domain comprising amino acid residues 19-239 of NP_054862.1.
[0082] In certain embodiments, the antibodies function by blocking or inhibiting the PD-1-binding or the B7-1-binding activity associated with PD-L1 by binding to any other region or fragment of the full length protein. In certain embodiments, the antibodies attenuate or modulate the interaction between PD-L1 and PD-1/B7-1.
[0083] In certain embodiments, the antibodies are bi-specific antibodies. The bi-specific antibodies can bind one epitope in one domain and can also bind a second epitope in a different domain of PD-L1. In certain embodiments, the bi-specific antibodies bind two different epitopes in the same domain. In one embodiment, the multi-specific antigen-binding molecule comprises a first antigen-binding specificity wherein the first binding specificity comprises the extracellular domain or fragment thereof of PD-1; and a second antigen-binding specificity to another epitope of PD-L1. In another embodiment, the multi-specific antigen-binding molecule comprises a first antigen-binding specificity wherein the first binding specificity comprises the extracellular domain or fragment thereof of B7-1; and a second antigen-binding specificity to another epitope of PD-L1.
[0084] In one embodiment, the antibody or fragment thereof is a fully human monoclonal antibody or antigen-binding fragment thereof that binds to PD-L1, wherein the antibody or fragment thereof exhibits one or more of the following characteristics: (i) comprises a HCVR having an amino acid sequence selected from the group consisting of SEQ ID NO: 2, 18, 34, 50, 66, 82, 98, 114, 130, 146, 162, 178, 186, 202, 218, 234, 250, 258, 266, 274, 282, 290, 298, 306, 314, 322, 330 and 338, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity; (ii) comprises a LCVR having an amino acid sequence selected from the group consisting of SEQ ID NO: 10, 26, 42, 58, 74, 90, 106, 122, 138, 154, 170, 194, 210, 226, 242, 258, and 274, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity; (iii) comprises a HCDR3 domain having an amino acid sequence selected from the group consisting of SEQ ID NO: 8, 24, 40, 56, 72, 88, 104, 120, 136, 152, 168, 184, 192, 208, 224, 240, 256, 272, 280, 288, 296, 304, 312, 320, 328, 336 and 344, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity; and a LCDR3 domain having an amino acid sequence selected from the group consisting of SEQ ID NO: 16, 32, 48, 64, 80, 96, 112, 128, 144, 160, 176, 200, 216, 232, 248, 264, and 280, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity; (iv) comprises a HCDR1 domain having an amino acid sequence selected from the group consisting of SEQ ID NO: 4, 20, 36, 52, 68, 84, 100, 116, 132, 148, 164, 180, 188, 204, 220, 236, 252, 268, 284, 292, 300, 308, 316, 324, 332, and 340, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity; a HCDR2 domain having an amino acid sequence selected from the group consisting of SEQ ID NO: 6, 22, 38, 54, 70, 86, 102, 118, 134, 150, 166, 182, 190, 206, 222, 238, 254, 270, 286, 294, 302, 310, 318, 326, 334, and 342, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity; a LCDR1 domain having an amino acid sequence selected from the group consisting of SEQ ID NO: 12, 28, 44, 60, 76, 92, 108, 124, 140, 156, 172, 196, 212, 228, 244, 260, and 276, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity; and a LCDR2 domain having an amino acid sequence selected from the group consisting of SEQ ID NO: 14, 30, 46, 62, 78, 94, 110, 126, 142, 158, 174, 198, 214, 230, 246, 262, and 278, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity; (v) is a multi-specific antigen-binding molecule comprising a first binding specificity to PD-L1 and a second binding specificity to an antigen selected from the group consisting of PD-L1, a tumor specific antigen, a virally infected cell antigen, and a T-cell co-inhibitor; (vi) binds to human PD-L1 with a K.sub.D of about 4 pM to about 645 nM; (vii) binds to cynomolgus PD-L1 with a K.sub.D of about 70 pM to about 400 nM; (viii) blocks or enhances the binding of PD-L1 to PD-1 with an IC50.ltoreq.770 pM; (ix) blocks or enhances the binding of PD-L1 to B7-1 with an IC50.ltoreq.10 nM; (x) blocks PD-1-induced T-cell down-regulation and/or rescues T-cell signaling in a T-cell/APC luciferase reporter assay; (xi) stimulates T-cell proliferation and activity in a mixed lymphocyte reaction (MLR) assay; (xii) induces IL-2 and/or IFN.gamma. production in a MLR assay; and (xiii) suppresses tumor growth and increases survival in subjects with cancer.
[0085] In one embodiment, the antibody or fragment thereof is a fully human monoclonal antibody or antigen-binding fragment thereof that blocks PD-L1 binding to PD-1 or to B7-1, wherein the antibody or fragment thereof exhibits one or more of the following characteristics: (i) comprises a HCVR having an amino acid sequence selected from the group consisting of SEQ ID NO: 82, 98, 146, 162, 290, 306, 314, and 330, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity; (ii) comprises a LCVR having an amino acid sequence selected from the group consisting of SEQ ID NO: 90, 106, 154, 170, and 274, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity; (iii) comprises a HCDR3 domain having an amino acid sequence selected from the group consisting of SEQ ID NO: 88, 104, 152, 168, 296, 312, 320, and 336, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity; and a LCDR3 domain having an amino acid sequence selected from the group consisting of SEQ ID NO: 96, 112, 160, 176, and 280, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity; (iv) comprises a HCDR1 domain having an amino acid sequence selected from the group consisting of SEQ ID NO: 84, 100, 148, 164, 292, 308, 316, and 332, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity; a HCDR2 domain having an amino acid sequence selected from the group consisting of SEQ ID NO: 86, 102, 150, 166, 294, 310, 318, and 334, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity; a LCDR1 domain having an amino acid sequence selected from the group consisting of SEQ ID NO: 92, 108, 156, 172, and 276, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity; and a LCDR2 domain having an amino acid sequence selected from the group consisting of SEQ ID NO: 94, 110, 158, 174, and 278, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity; (v) is a multi-specific antigen-binding molecule comprising a first binding specificity to PD-L1 and a second binding specificity to an antigen selected from the group consisting of a different epitope of PD-L1, a tumor specific antigen, a virally-infected cell antigen, and a T-cell co-inhibitor; (vi) binds to human PD-L1 with a K.sub.D.ltoreq.10.sup.-10M; (vii) binds to cynomolgus PD-L1 with a K.sub.D.ltoreq.10.sup.-7M; (viii) blocks the binding of PD-L1 to PD-1 or to B7-1; (ix) blocks PD-1-induced T-cell down-regulation and/or rescues T-cell signaling in a T-cell/APC luciferase reporter assay; (xi) stimulates T-cell proliferation and activity in a mixed lymphocyte reaction (MLR) assay; (xii) induces IL-2 and/or IFN.gamma. production in a MLR assay; and (xiii) suppresses tumor growth and increases survival in subjects with cancer.
[0086] In certain embodiments, the anti-PD-L1 antibodies or antigen-binding fragments thereof bind an epitope within any one or more of the regions exemplified in PD-L1, either in natural form, or recombinantly produced, or to a fragment thereof. In some embodiments, the antibodies of the disclosure bind to an extracellular region comprising one or more amino acids selected from the group consisting of amino acid residues 19-239 of PD-L1. In some embodiments, the antibodies of the disclosure bind to a region comprising one or more amino acids selected from the group consisting of amino acid residues 1-221 of cynomolgus PD-L1.
[0087] In certain embodiments, the antibodies of the disclosure, as shown in Table 1, interact with at least one amino acid sequence selected from the group consisting of amino acid residues ranging from about position 19 to about position 130 of PD-L1; or amino acid residues ranging from about position 130 to about position 153 of PD-L1; or amino acid residues ranging from about position 153 to about position 210 of PD-L1; or to amino acid residues ranging from about position 210 to about position 239 of PD-L1.
[0088] In some embodiments, the anti-PD-L1 antibodies bind to the same epitope, or a portion of the epitope, as any of the specific exemplary antibodies described herein in Table 1, or an antibody having the CDR sequences of any of the exemplary antibodies described in Table 1. Likewise, suitable antibodies also include anti-PD-L1 antibodies that compete for binding to PD-L1 or a PD-L1 fragment with any of the specific exemplary antibodies described herein in Table 1, or an antibody having the CDR sequences of any of the exemplary antibodies described in Table 1. For example, suitable antibodies include anti-PD-L1 antibodies that cross-compete for binding to PD-L1 with one or more antibodies as defined in Example 6 of herein (e.g., H2aM8309N, H1H9329P, H1H9336P, H2aM8314N, H2aM8316N, H2AM8718N, H1H9387P2, H1H9351P2, H1H9364P2, H1H9373P2, and H2aM8306N). The present disclosure also includes anti-PD-L1 antibodies that cross-compete for binding to PD-L1 with one or more antibodies as defined in Example 6 of US Patent Publication No. US 2015-0203580 A1 (e.g., H1H9396P2, H2aM8317N, H2aM8321N, H1H9323P, H1H9382P2, H1H9344P2, H1H9345P2 and H1H9354P2).
[0089] The antibodies and antigen-binding fragments described herein specifically bind to PD-L1 and modulate the interaction of PD-L1 with PD-1 or with B7-1. The anti-PD-L1 antibodies may bind to PD-L1 with high affinity or with low affinity. In certain embodiments, the antibodies are blocking antibodies wherein the antibodies bind to PD-L1 and block the interaction of PD-L1 with PD-1 or with B7-1. In some embodiments, the blocking antibodies of the disclosure block the binding of PD-L1 to PD-1 or to B7-1 and/or stimulate or enhance T-cell activation. In some embodiments, the blocking antibodies are useful for stimulating or enhancing the immune response and/or for treating a subject suffering from cancer, or a chronic viral infection. The antibodies when administered to a subject in need thereof may reduce the chronic infection by a virus such as HIV, LCMV or HBV in the subject. They may be used to inhibit the growth of tumor cells in a subject. They may be used alone or as adjunct therapy with other therapeutic moieties or modalities known in the art for treating cancer, or viral infection. In certain embodiments, the anti-PD-L1 antibodies that bind to PD-L1 with a low affinity are used as multi-specific antigen-binding molecules wherein the first binding specificity binds to PD-L1 with a low affinity and the second binding specificity binds to an antigen selected from the group consisting of a different epitope of PD-L1, a T-cell co-inhibitor such as PD-1, a tumor specific antigen and an infected-cell-specific antigen.
[0090] In certain embodiments, the antibodies of the present disclosure are agonist antibodies, wherein the antibodies bind to PD-L1 and enhance the interaction of PD-L1 and PD-1/B7-1. In some embodiments, the activating antibodies enhance binding of PD-L1 to PD-1 or to B7-1 and/or inhibit or suppress T-cell activation. The activating antibodies of the present disclosure may be useful for inhibiting the immune response in a subject and/or for treating autoimmune disease.
[0091] In certain embodiments, the anti-PD-L1 antibodies are multi-specific antigen-binding molecules, wherein they comprise a first binding specificity to PD-L1 and a second binding specificity to an antigen selected from the group consisting of a different epitope of PD-L1, a T-cell co-inhibitor such as PD-1, a tumor specific antigen and an infected-cell-specific antigen. In certain embodiments, the first binding specificity binds to PD-L1 with low affinity, e.g., with a K.sub.D of 10.sup.-8 M, 10.sup.-7 M or more.
[0092] Certain anti-PD-L1 antibodies of the present disclosure are able to bind to and neutralize the activity of PD-L1, as determined by in vitro or in vivo assays. The ability of the antibodies of the disclosure to bind to and neutralize the activity of PD-L1 may be measured using any standard method known to those skilled in the art, including binding assays, or activity assays, as described herein.
[0093] Non-limiting, exemplary in vitro assays for measuring binding activity are illustrated in Example 3 of US Patent Publication No. US 2015-0203580 A1. In Example 3, the binding affinities and kinetic constants of human anti-PD-L1 antibodies for human PD-L1 and cynomolgus PD-L1 were determined by surface plasmon resonance and the measurements were conducted on a T200 Biacore instrument. In Examples 4 and 5 of US Patent Publication No. US 2015-0203580 A1, blocking assays were used to determine the ability of the anti-PD-L1 antibodies to block PD-L1-binding ability of PD-1 or to B7-1 in vitro. In Example 6 of US Patent Publication No. US 2015-0203580 A1, blocking assays were used to determine cross-competition between different anti-PD-L1 antibodies. Example 7 of US Patent Publication No. US 2015-0203580 A1 describes the binding of the antibodies to cells overexpressing PD-L1. In Example 8 of US 2015-0203580 A1, a luciferase assay was used to determine the ability of anti-PD-L1 antibodies to antagonize PD-1/PD-L1 signaling in T-cells.
[0094] Unless specifically indicated otherwise, the term "antibody," as used herein, shall be understood to encompass antibody molecules comprising two immunoglobulin heavy chains and two immunoglobulin light chains (i.e., "full antibody molecules") as well as antigen-binding fragments thereof. The terms "antigen-binding portion" of an antibody, "antigen-binding fragment" of an antibody, and the like, as used herein, include any naturally occurring, enzymatically obtainable, synthetic, or genetically engineered polypeptide or glycoprotein that specifically binds an antigen to form a complex. The terms "antigen-binding fragment" of an antibody, or "antibody fragment", as used herein, refers to one or more fragments of an antibody that retain the ability to specifically bind to PD-L1. An antibody fragment may include a Fab fragment, a F(ab').sub.2 fragment, a Fv fragment, a dAb fragment, a fragment containing a CDR, or an isolated CDR. In certain embodiments, the term "antigen-binding fragment" refers to a polypeptide or fragment thereof of a multi-specific antigen-binding molecule. In such embodiments, the term "antigen-binding fragment" includes, e.g., an extracellular domain of PD-1 which binds specifically to PD-L1. Antigen-binding fragments of an antibody may be derived, e.g., from full antibody molecules using any suitable standard techniques such as proteolytic digestion or recombinant genetic engineering techniques involving the manipulation and expression of DNA encoding antibody variable and (optionally) constant domains. Such DNA is known and/or is readily available from, e.g., commercial sources, DNA libraries (including, e.g., phage-antibody libraries), or can be synthesized. The DNA may be sequenced and manipulated chemically or by using molecular biology techniques, for example, to arrange one or more variable and/or constant domains into a suitable configuration, or to introduce codons, create cysteine residues, modify, add or delete amino acids, etc.
[0095] Non-limiting examples of antigen-binding fragments include: (i) Fab fragments; (ii) F(ab')2 fragments; (iii) Fd fragments; (iv) Fv fragments; (v) single-chain Fv (scFv) molecules; (vi) dAb fragments; and (vii) minimal recognition units consisting of the amino acid residues that mimic the hypervariable region of an antibody (e.g., an isolated complementarity determining region (CDR) such as a CDR3 peptide), or a constrained FR3-CDR3-FR4 peptide. Other engineered molecules, such as domain-specific antibodies, single domain antibodies, domain-deleted antibodies, chimeric antibodies, CDR-grafted antibodies, diabodies, triabodies, tetrabodies, minibodies, nanobodies (e.g. monovalent nanobodies, bivalent nanobodies, etc.), small modular immunopharmaceuticals (SMIPs), and shark variable IgNAR domains, are also encompassed within the expression "antigen-binding fragment," as used herein.
[0096] An antigen-binding fragment of an antibody will typically comprise at least one variable domain. The variable domain may be of any size or amino acid composition and will generally comprise at least one CDR, which is adjacent to or in frame with one or more framework sequences. In antigen-binding fragments having a V.sub.H domain associated with a V.sub.L domain, the V.sub.H and V.sub.L domains may be situated relative to one another in any suitable arrangement. For example, the variable region may be dimeric and contain V.sub.H-V.sub.H, V.sub.H-V.sub.L or V.sub.L-V.sub.L dimers. Alternatively, the antigen-binding fragment of an antibody may contain a monomeric V.sub.H or V.sub.L domain.
[0097] In certain embodiments, an antigen-binding fragment of an antibody may contain at least one variable domain covalently linked to at least one constant domain. Non-limiting, exemplary configurations of variable and constant domains that may be found within an antigen-binding fragment of an antibody of the present disclosure include: (i) V.sub.H-C.sub.H1; (ii) V.sub.H-C.sub.H2; (iii) V.sub.H-C.sub.H3; (iv) V.sub.H-C.sub.H1-C.sub.H2; (V) V.sub.H-C.sub.H1-C.sub.H2-C.sub.H3; (Vi) V.sub.H-C.sub.H2-C.sub.H3; V.sub.H--CL; V.sub.L--C.sub.H1; (ix) V.sub.L-C.sub.H2; (X) V.sub.L-C.sub.H3; (xi) V.sub.L-C.sub.H1-C.sub.H2; (XII) V.sub.L-C.sub.H1-C.sub.H2-C.sub.H3; (Xiii) V.sub.L-C.sub.H2-C.sub.H3; and (xiv) V.sub.L-CL. In any configuration of variable and constant domains, including any of the exemplary configurations listed above, the variable and constant domains may be either directly linked to one another or may be linked by a full or partial hinge or linker region. A hinge region may consist of at least 2 (e.g., 5, 10, 15, 20, 40, 60 or more) amino acids, which result in a flexible or semi-flexible linkage between adjacent variable and/or constant domains in a single polypeptide molecule. Moreover, an antigen-binding fragment of an antibody of the present disclosure may comprise a homo-dimer or hetero-dimer (or other multimer) of any of the variable and constant domain configurations listed above in non-covalent association with one another and/or with one or more monomeric V.sub.H or V.sub.L domain (e.g., by disulfide bond(s)).
[0098] As with full antibody molecules, antigen-binding fragments may be mono-specific or multi-specific (e.g., bi-specific). A multi-specific antigen-binding fragment of an antibody will typically comprise at least two different variable domains, wherein each variable domain is capable of specifically binding to a separate antigen or to a different epitope on the same antigen. Any multi-specific antibody format, including the exemplary bi-specific antibody formats disclosed herein, may be adapted for use in the context of an antigen-binding fragment of an antibody of the present disclosure using routine techniques available in the art.
[0099] The anti-PD-L1 antibodies and antibody fragments of the present disclosure encompass proteins having amino acid sequences that vary from those of the described antibodies, but that retain the ability to bind PD-L1. Such variant antibodies and antibody fragments comprise one or more additions, deletions, or substitutions of amino acids when compared to parent sequence, but exhibit biological activity that is essentially equivalent to that of the described antibodies. Likewise, the antibody-encoding DNA sequences of the present disclosure encompass sequences that comprise one or more additions, deletions, or substitutions of nucleotides when compared to the disclosed sequence, but that encode an antibody or antibody fragment that is essentially bioequivalent to an antibody or antibody fragment of the disclosure.
[0100] Two antigen-binding proteins, or antibodies, are considered bioequivalent if, for example, they are pharmaceutical equivalents or pharmaceutical alternatives whose rate and extent of absorption do not show a significant difference when administered at the same molar dose under similar experimental conditions, either single dose or multiple doses. Some antibodies will be considered equivalents or pharmaceutical alternatives if they are equivalent in the extent of their absorption but not in their rate of absorption and yet may be considered bioequivalent because such differences in the rate of absorption are intentional and are reflected in the labeling, are not essential to the attainment of effective body drug concentrations on, e.g., chronic use, and are considered medically insignificant for the particular drug product studied.
[0101] In one embodiment, two antigen-binding proteins are bioequivalent if there are no clinically meaningful differences in their safety, purity, or potency.
[0102] In one embodiment, two antigen-binding proteins are bioequivalent if a patient can be switched one or more times between the reference product and the biological product without an expected increase in the risk of adverse effects, including a clinically significant change in immunogenicity, or diminished effectiveness, as compared to continued therapy without such switching.
[0103] In one embodiment, two antigen-binding proteins are bioequivalent if they both act by a common mechanism or mechanisms of action for the condition or conditions of use, to the extent that such mechanisms are known.
[0104] Bioequivalence may be demonstrated by in vivo and/or in vitro methods. Bioequivalence measures include, e.g., (a) an in vivo test in humans or other mammals, in which the concentration of the antibody or its metabolites is measured in blood, plasma, serum, or other biological fluid as a function of time; (b) an in vitro test that has been correlated with and is reasonably predictive of human in vivo bioavailability data; (c) an in vivo test in humans or other mammals in which the appropriate acute pharmacological effect of the antibody (or its target) is measured as a function of time; and (d) in a well-controlled clinical trial that establishes safety, efficacy, or bioavailability or bioequivalence of an antibody.
[0105] Bioequivalent variants of the antibodies of the disclosure may be constructed by, for example, making various substitutions of residues or sequences or deleting terminal or internal residues or sequences not needed for biological activity. For example, cysteine residues not essential for biological activity can be deleted or replaced with other amino acids to prevent formation of unnecessary or incorrect intramolecular disulfide bridges upon renaturation. In other contexts, bioequivalent antibodies may include antibody variants comprising amino acid changes, which modify the glycosylation characteristics of the antibodies, e.g., mutations that eliminate or remove glycosylation.
[0106] According to certain embodiments of the present disclosure, anti-PD-L1 antibodies comprise an Fc domain comprising one or more mutations which enhance or diminish antibody binding to the FcRn receptor, e.g., at acidic pH as compared to neutral pH. For example, the present disclosure includes anti-PD-L1 antibodies comprising a mutation in the C.sub.H2 or a C.sub.H3 region of the Fc domain, wherein the mutation(s) increases the affinity of the Fc domain to FcRn in an acidic environment (e.g., in an endosome where pH ranges from about 5.5 to about 6.0). Such mutations may result in an increase in serum half-life of the antibody when administered to an animal. Non-limiting examples of such Fc modifications include, e.g., a modification at position 250 (e.g., E or Q); 250 and 428 (e.g., L or F); 252 (e.g., L/Y/F/W or T), 254 (e.g., S or T), and 256 (e.g., S/R/Q/E/D or T); or a modification at position 428 and/or 433 (e.g., H/L/R/S/P/Q or K) and/or 434 (e.g., A, W, H, F or Y [N434A, N434W, N434H, N434F or N434Y]); or a modification at position 250 and/or 428; or a modification at position 307 or 308 (e.g., 308F, V308F), and 434. In one embodiment, the modification comprises a 428L (e.g., M428L) and 434S (e.g., N434S) modification; a 428L, 2591 (e.g., V2591), and 308F (e.g., V308F) modification; a 433K (e.g., H433K) and a 434 (e.g., 434Y) modification; a 252, 254, and 256 (e.g., 252Y, 254T, and 256E) modification; a 250Q and 428L modification (e.g., T250Q and M428L); and a 307 and/or 308 modification (e.g., 308F or 308P). In yet another embodiment, the modification comprises a 265A (e.g., D265A) and/or a 297A (e.g., N297A) modification.
[0107] For example, the present disclosure includes anti-PD-L1 antibodies comprising an Fc domain comprising one or more pairs or groups of mutations selected from the group consisting of: 250Q and 248L (e.g., T250Q and M248L); 252Y, 254T and 256E (e.g., M252Y, S254T and T256E); 428L and 434S (e.g., M428L and N434S); 2571 and 3111 (e.g., P2571 and 03111); 2571 and 434H (e.g., P2571 and N434H); 376V and 434H (e.g., D376V and N434H); 307A, 380A and 434A (e.g., 1307A, E380A and N434A); and 433K and 434F (e.g., H433K and N434F). In one embodiment, the present disclosure includes anti-PD-L1 antibodies comprising an Fc domain comprising a S108P mutation in the hinge region of IgG4 to promote dimer stabilization. All possible combinations of the foregoing Fc domain mutations, and other mutations within the antibody variable domains disclosed herein, are contemplated within the scope of the present disclosure.
[0108] The present disclosure also includes anti-PD-L1 antibodies comprising a chimeric heavy chain constant (C.sub.H) region, wherein the chimeric C.sub.H region comprises segments derived from the C.sub.H regions of more than one immunoglobulin isotype. For example, the antibodies of the disclosure may comprise a chimeric C.sub.H region comprising part or all of a C.sub.H2 domain derived from a human IgG1, human IgG2 or human IgG4 molecule, combined with part or all of a C.sub.H3 domain derived from a human IgG1, human IgG2 or human IgG4 molecule. According to certain embodiments, the antibodies of the disclosure comprise a chimeric C.sub.H region having a chimeric hinge region. For example, a chimeric hinge may comprise an "upper hinge" amino acid sequence (amino acid residues from positions 216 to 227 according to EU numbering) derived from a human IgG1, a human IgG2 or a human IgG4 hinge region, combined with a "lower hinge" sequence (amino acid residues from positions 228 to 236 according to EU numbering) derived from a human IgG1, a human IgG2 or a human IgG4 hinge region. According to certain embodiments, the chimeric hinge region comprises amino acid residues derived from a human IgG1 or a human IgG4 upper hinge and amino acid residues derived from a human IgG2 lower hinge. An antibody comprising a chimeric C.sub.H region as described herein may, in certain embodiments, exhibit modified Fc effector functions without adversely affecting the therapeutic or pharmacokinetic properties of the antibody. (See, e.g., U.S. Ser. No. 14/170,166, filed Jan. 31, 2014, the disclosure of which is hereby incorporated by reference in its entirety).
[0109] B. Positron Emitters and Chelating Moieties
[0110] Suitable positron emitters include, but are not limited to, those that form stable complexes with the chelating moiety and have physical half-lives suitable for immuno-PET imaging purposes. Illustrative positron emitters include, but are not limited to, .sup.89Zr, .sup.68Ga, .sup.64Cu, .sup.44Sc, and .sup.86Y. Suitable positron emitters also include those that directly bond with the PD-L1 binding protein, including, but not limited to, .sup.76Br and .sup.124I, and those that are introduced via prosthetic group, e.g., 18F,
[0111] The chelating moieties described herein are chemical moieties that are covalently linked to the PD-L1 binding protein, e.g., anti-PD-L1 antibody and comprise a portion capable of chelating a positron emitter, i.e., capable of reacting with a positron emitter to form a coordinated chelate complex. Suitable moieties include those that allow efficient loading of the particular metal and form metal-chelator complexes that are sufficiently stable in vivo for diagnostic uses, e.g., immuno-PET imaging. Illustrative chelating moieties include those that minimize dissociation of the positron emitter and accumulation in mineral bone, plasma proteins, and/or bone marrow depositing to an extent suitable for diagnostic uses.
[0112] Examples of chelating moieties include, but are not limited to, those that form stable complexes with positron emitters .sup.89Zr, .sup.68Ga, .sup.64Cu, .sup.44Sc, and/or .sup.86Y. Illustrative chelating moieties include, but are not limited to, those described in Nature Protocols, 5(4): 739, 2010; Bioconjugate Chem., 26(12): 2579 (2015); Chem Commun (Camb), 51(12): 2301 (2015); Mol. Pharmaceutics, 12: 2142 (2015); Mol. Imaging Biol., 18: 344 (2015); Eur. J. Nucl. Med. Mol. Imaging, 37:250 (2010); Eur. J. Nucl. Med. Mol. Imaging (2016). doi:10.1007/s00259-016-3499-x; Bioconjugate Chem., 26(12): 2579 (2015); WO 2015/140212A1; and U.S. Pat. No. 5,639,879, incorporated by reference in their entireties.
[0113] Illustrative chelating moieties also include, but are not limited to, those that comprise desferrioxamine (DFO), 1,4,7,10-tetraacetic acid (DOTA), diethylenetriaminepentaacetic acid (DTPA), ethylenediaminetetraacetic acid (EDTA), (1,4,7,10-Tetraazacyclododecane-1,4,7,10-tetra(methylene phosphonic) acid (DOTP), 1R, 4R, 7R, 10R)-.quadrature.'.quadrature.''.quadrature.'''-Tetramethyl-1,4,7,10-tetr- aazacyclododecane-1,4,7,10-tetraacetic acid (DOTMA), 1,4,8,11-Tetraazacyclotetradecane-1,4,8, 11-tetraacetic acid (TETA), H.sub.4octapa, H.sub.6phospa, H.sub.2dedpa, H.sub.5decapa, H.sub.2azapa, HOPO, DO2A, 1,4,7,10-Tetrakis(carbamoylmethyl)-1,4,7,10-tetraazacyclododecane (DOTAM), 1,4,7-triazacyclononane-N,N',N''-triacetic acid (NOTA), 1,4,7,10-Tetrakis(carbamoylmethyl)-1,4,7,10-tetraazacyclododecane (DOTAM), 1,4,8,11-tetraazabicyclo[6.6.2]hexadecane-4, 11-dicetic acid (CB-TE2A), 1,4,7,10-Tetraazacyclododecane (Cyclen), 1,4,8,11-Tetraazacyclotetradecane (Cyclam), octadentate chelators, hexadentate chelators, phosphonate-based chelators, macrocyclic chelators, chelators comprising macrocyclic terephthalamide ligands, bifunctional chelators, fusarinine C and fusarinine C derivative chelators, triacetylfusarinine C (TAFC), ferrioxamine E (FOXE), ferrioxamine B (FOXB), ferrichrome A (FCHA), and the like.
[0114] In some embodiments, the chelating moieties are covalently bonded to the PD-L1 binding protein, e.g., antibody or antigen binding fragment thereof, via a linker moiety, which covalently attaches the chelating portion of the chelating moiety to the binding protein. In some embodiments, these linker moieties are formed from a reaction between a reactive moiety of the PD-L1 binding protein, e.g., cysteine or lysine of an antibody, and reactive moiety that is attached to a chelator, including, for example, a p-isothiocyanatobenyl group and the reactive moieties provided in the conjugation methods below. In addition, such linker moieties optionally comprise chemical groups used for purposes of adjusting polarity, solubility, steric interactions, rigidity, and/or the length between the chelating portion and PD-L1 binding protein.
[0115] C. Preparation of Radiolabeled Anti-PD-L1 Conjugates
[0116] The radiolabeled anti-PD-L1 protein conjugates can be prepared by (1) reacting a PD-L1 binding protein, e.g., antibody, with a molecule comprising a positron emitter chelator and a moiety reactive to the desirable conjugation site of the PD-L1 binding protein and (2) loading the desirable positron emitter.
[0117] Suitable conjugation sites include, but are not limited to, lysine and cysteine, both of which can be, for example, native or engineered, and can be, for example, present on the heavy or light chain of an antibody. Cysteine conjugation sites include, but are not limited to, those obtained from mutation, insertion, or reduction of antibody disulfide bonds. Methods for making cysteine engineered antibodies include, but are not limited to, those disclosed in WO2011/056983. Site-specific conjugation methods can also be used to direct the conjugation reaction to specific sites of an antibody, achieve desirable stoichiometry, and/or achieve desirable drug-to-antibody (DAR) ratios. Such conjugation methods are known to those of ordinary skill in the art and include, but are not limited to cysteine engineering and enzymatic and chemo-enzymatic methods, including, but not limited to, glutamine conjugation, Q295 conjugation, and transglutaminase-mediated conjugation, as well as those described in J. Clin. Immunol., 36: 100 (2016), incorporated herein by reference in its entirety. Suitable moieties reactive to the desirable conjugation site generally enable efficient and facile coupling of the PD-L1 binding protein, e.g., antibody and positron emitter chelator. Moieties reactive to lysine and cysteine sites include electrophilic groups, which are known to those of ordinary skill. In certain aspects, when the desired conjugation site is lysine, the reactive moiety is an isothiocyanate, e.g., p-isothiocyanatobenyl group or reactive ester. In certain aspects, when the desired conjugation site is cysteine, the reactive moiety is a maleimide.
[0118] When the chelator is desferrioxamine (DFO), suitable reactive moieties include, but are not limited to, an isothiocyantatobenzyl group, an n-hydroxysuccinimide ester,2,3,5,6 tetraflurorphenol ester, n-succinimidyl-S-acetylthioacetate, and those described in BioMed Research International, Vol 2014, Article ID 203601, incorporated herein by reference in its entirety. In certain embodiments, the PD-L1 binding protein is an antibody and the molecule comprising a positron emitter chelator and moiety reactive to the conjugation site is p-isothiocyantatobenzyl-desferrioxamine (p-SCN-Bn-DFO):
##STR00001##
[0119] Positron emitter loading is accomplished by incubating the PD-L1 binding protein chelator conjugate with the positron emitter for a time sufficient to allow coordination of said positron emitter to the chelator, e.g., by performing the methods described in the examples provided herein, or substantially similar method.
[0120] D. Illustrative Embodiments of Conjugates
[0121] Included in the instant disclosure are radiolabeled antibody conjugates comprising an antibody or antigen binding fragment thereof, that binds human program death ligand 1 (PD-L1), a chelating moiety, and a positron emitter.
[0122] In some embodiments, the chelating moiety comprises a chelator capable of forming a complex with .sup.89Zr. In certain embodiments, the chelating moiety comprises desferrioxamine. In certain embodiments, the chelating moiety is p-isothiocyanatobenzyl-desferrioxamine.
[0123] In some embodiments, the positron emitter is .sup.89Zr.
[0124] In some embodiments, the chelating moiety-to-antibody ratio of the conjugate is from 1 to 2.
[0125] In a particular embodiment, chelating moiety is p-isothiocyanatobenzyl-desferrioxamine and the positron emitter is .sup.89Zr. In another particular embodiment, the chelating moiety is p-isothiocyanatobenzyl-desferrioxamine and the positron emitter is .sup.89Zr, and the chelating moiety-to-antibody ratio of the conjugate is from 1 to 2.
[0126] In some embodiments, provided herein are antigen-binding proteins that bind PD-L1, wherein said antigen-binding proteins that bind PD-L1 are covalently bonded to one or more moieties having the following structure:
-L-M.sub.z
wherein L is a chelating moiety; M is a positron emitter; and z, independently at each occurrence, is 0 or 1; and wherein at least one of z is 1. In certain embodiments, the radiolabeled antigen-binding protein is a compound of Formula (I):
M-L-A-[L-M.sub.z].sub.k (I)
A is a protein that binds PD-L1; L is a chelating moiety; M is a positron emitter; z is 0 or 1; and k is an integer from 0-30. In some embodiments, k is 1.
[0127] In some embodiments, L is:
##STR00002##
[0128] In some embodiments, M is .sup.89Zr.
[0129] In some embodiments, k is an integer from 1 to 2. In some embodiments, k is 1.
[0130] In some embodiments, -L-M is
##STR00003##
[0131] Included in the instant disclosure are also methods of synthesizing a radiolabeled antibody conjugates comprising contacting a compound of Formula (III):
##STR00004##
[0132] with .sup.89Zr, wherein A is an antibody or antigen-binding fragment thereof that binds PD-L1. In certain embodiments, the compound of Formula (III) is synthesized by contacting an antibody, or antigen binding fragment thereof, that binds PD-L1, with p-SCN-Bn-DFO.
[0133] Provided herein is also the product of the reaction between a compound of Formula (III) with .sup.89Zr.
[0134] Provided herein are compounds of Formula (III):
##STR00005##
wherein A is an antibody or antigen binding fragment thereof that binds PD-L1 and k is an integer from 1-30. In some embodiments, k is 1 or 2.
[0135] In some embodiments, provided herein are compositions comprising a conjugate having the following structure:
A-L.sub.k
wherein A is a protein that binds PD-L1; L is a chelating moiety; and k is an integer from 1-30; wherein the conjugate is chelated with a positron emitter in an amount sufficient to provide a specific activity suitable for clinical PET imaging. In some embodiments, the amount of chelated positron emitter is an amount sufficient to provide a specific activity of 1-3 mCi per 1-50 mg of the protein that binds PD-L1.
[0136] In some embodiments, the antibody or antigen-binding fragment thereof binds monomeric human programmed death-ligand 1 (PD-L1) with a binding dissociation equilibrium constant (K.sub.D) of less than about 310 pM as measured in a surface plasmon resonance assay at 37.degree. C.
[0137] In some embodiments, the antibody or antigen-binding fragment thereof binds monomeric human PD-L1 with a K.sub.D less than about 180 pM in a surface plasmon resonance assay at 25.degree. C.
[0138] In some embodiments, the antibody or antigen-binding fragment thereof binds dimeric human PD-L1 with a K.sub.D of less than about 15 pM as measured in a surface plasmon resonance assay at 37.degree. C.
[0139] In some embodiments, the antibody or antigen-binding fragment thereof that binds dimeric human PD-L1 with a K.sub.D less than about 8 pM in a surface plasmon resonance assay at 25.degree. C.
[0140] In some embodiments, the antibody or antigen-binding fragment thereof competes for binding to human PD-L1 with a reference antibody comprising the complementarity determining regions (CDRs) of a HCVR, wherein the HCVR has an amino acid sequence selected from the group consisting of HCVR sequences listed in Table 1; and the CDRs of a LCVR, wherein the LCVR has an amino acid sequence selected from the group consisting of LCVR sequences listed in Table 1. In some embodiments, the reference antibody or antigen-binding fragment thereof comprises an HCVR/LCVR amino acid sequence pair as set forth in Table 1. In some embodiments, the reference antibody comprises an HCVR/LCVR amino acid sequence pair selected from the group consisting of SEQ ID NOs: 82/90, 98/106, 146/154, 162/170, 290/274, 306/274, 314/274 and 330/274.
[0141] In some embodiments, the antibody or antigen-binding fragment thereof enhances PD-L1 binding to one of PD-1 or B7-1. In some embodiments, the antibody or antigen binding fragment thereof blocks PD-L1 binding to PD-1 and/or B7-1. In some embodiments, the antibody or antigen binding fragment thereof do not increase or decrease PD-L1 binding to its ligands.
[0142] In some embodiments, the antibody or antigen-binding fragment thereof comprises the complementarity determining regions (CDRs) of a HCVR, wherein the HCVR has an amino acid sequence selected from the group consisting of SEQ ID NOs: 18, 66, 114, 130, 202, 218, 266, 282, 298, 322, and 338; and the CDRs of a LCVR, wherein the LCVR has an amino acid sequence selected from the group consisting of SEQ ID NOs: 26, 74, 122, 138, 210, 226, and 274. In certain embodiments, the isolated antibody comprises an HCVR/LCVR amino acid sequence pair selected from the group consisting of SEQ ID NOs: 18/26, 66/74, 114/122, 130/138, 202/210, 218/226, 266/274, 282/274, 298/274, 322/274, and 338/274.
[0143] In some embodiments, the antibody is a human monoclonal antibody or antigen-binding fragment thereof that binds specifically to human PD-L1, wherein the antibody or antigen-binding fragment thereof comprises a heavy chain variable region (HCVR) having an amino acid sequence selected from the group consisting of HCVR sequences listed in Table 1.
[0144] In some embodiments, the antibody is a human monoclonal antibody or antigen-binding fragment thereof that binds specifically to human PD-L1, wherein the antibody or antigen-binding fragment thereof comprises a light chain variable region (LCVR) having an amino acid sequence selected from the group consisting of LCVR sequences listed in Table 1.
[0145] In some embodiments, the antibody a human monoclonal antibody or antigen-binding fragment thereof that binds specifically to human PD-L1, wherein the antibody or antigen-binding fragment thereof comprises (a) a HCVR having an amino acid sequence selected from the group consisting of HCVR sequences listed in Table 1; and (b) a LCVR having an amino acid sequence selected from the group consisting of LCVR sequences listed in Table 1.
[0146] In some embodiments, the antibody or antigen-binding fragment thereof comprises three heavy chain complementarity determining regions (CDRs) (HCDR1, HCDR2 and HCDR3) contained within any one of the heavy chain variable region (HCVR) sequences listed in Table 1; and three light chain CDRs (LCDR1, LCDR2 and LCDR3) contained within any one of the light chain variable region (LCVR) sequences listed in Table 1.
[0147] In some embodiments, the antibody or antigen-binding fragment thereof comprises: [0148] (a) a HCDR1 domain having an amino acid sequence selected from the group consisting of SEQ ID NOs: 4, 20, 36, 52, 68, 84, 100, 116, 132, 148, 164, 180, 188, 204, 220, 236, 252, 268, 284, 292, 300, 308, 316, 324, 332, and 340; [0149] (b) a HCDR2 domain having an amino acid sequence selected from the group consisting of SEQ ID NOs: 6, 22, 38, 54, 70, 86, 102, 118, 134, 150, 166, 182, 190, 206, 222, 238, 254, 270, 286, 294, 302, 310, 318, 326, 334, and 342; [0150] (c) a HCDR3 domain having an amino acid sequence selected from the group consisting of SEQ ID NOs: 8, 24, 40, 56, 72, 88, 104, 120, 136, 152, 168, 184, 192, 208, 224, 240, 256, 272, 288, 296, 304, 312, 320, 328, 336, and 344; [0151] (d) a LCDR1 domain having an amino acid sequence selected from the group consisting of SEQ ID NOs: 12, 28, 44, 60, 76, 92, 108, 124, 140, 156, 172, 196, 212, 228, 244, 260, and 276; [0152] (e) a LCDR2 domain having an amino acid sequence selected from the group consisting of SEQ ID NOs: 14, 30, 46, 62, 78, 94, 110, 126, 142, 158, 174, 198, 214, 230, 246, 262, and 278; and [0153] (f) a LCDR3 domain having an amino acid sequence selected from the group consisting of SEQ ID NOs: 16, 32, 48, 64, 80, 96, 112, 128, 144, 160, 176, 200, 216, 232, 248, 264, and 280.
[0154] In some embodiments, the antibody or antigen-binding fragment comprises a HCVR/LCVR amino acid sequence pair selected from the group consisting of SEQ ID NOs: 82/90, 98/106, 146/154, 162/170, 290/274, 306/274, 314/274 and 330/274
[0155] In some embodiments, the antibody or antigen-binding fragment thereof comprises the CDRs of a HCVR, wherein the HCVR has an amino acid sequence selected from the group consisting of SEQ ID NOs: 2, 34, 50, 82, 98, 146, 162, 178, 186, 234, 250, 290, 306, 314, and 330; and the CDRs of a LCVR, wherein the LCVR has an amino acid sequence selected from the group consisting of SEQ ID NOs: 10, 42, 58, 90, 106, 154, 170, 194, 242, 258, and 274.
[0156] E. Scaled Manufacturing for Production of Anti-PD-L1 Antibody-Chelator Conjugates
[0157] Included in the present disclosure are scaled-up manufacturing processes for producing anti-PD-L1 antibodies conjugated to a chelator. The anti-PD-L1 antibody-chelator conjugates are in a form suitable for radiolabeling.
[0158] Good manufacturing processes are adhered to in all aspects of production, including maintaining a sterile environment, practicing aseptic procedures, keeping records of all processes, and documenting product quality, purity, strength, and identity, and any deviations therefrom.
[0159] The scaled-up manufacturing process is, in some embodiments, much faster than the manufacturing process for research and development. In some embodiments, the scaled-up manufacturing process can take less than 12 hours, or less than 10 hours, or less than 8 hours, or less than 6 hours, or less than 4 hours, or less than or about 2 hours.
[0160] In some embodiments, a first step comprises ultrafiltration and diafiltration (UFDF), using a 30-50 kDa membrane, of the anti-PD-L1 antibody to remove excipients, conjugation interfering species, and salts that inhibit the conjugation process. Exemplary membrane polymers include polyethersulfone (PES), cellulose acetate (CA), and regenerated cellulose (RC). In this step, the antibody is buffer exchanged in a low ionic strength and non-interfering buffer solution. The buffer pH can be between about 4.5 to about 6, or about 5 to about 6, or about 5.3 to about 5.7, or about 5.5. Buffer systems contemplated as useful herein include any buffer system lacking a primary amine. Exemplary buffers include acetate, phosphate, or citrate buffers. The buffer provides protein stability during pre-conjugation processing. The process volume can be further reduced to concentrate the antibody, then sterile filtered.
[0161] Following the pre-conjugation UFDF, the concentrated and filtered antibody can be transferred into an amine free carbonate buffer system. The carbonate buffer system can have a pH in a range from about 8.5 to about 9.6, or from about 9.0 to about 9.6, or from about 9.2 to about 9.4, or from about 9.4 to about 9.6, or a pH of about 9.4.
[0162] A chelator, for example, DFO, in solvent is added to a target concentration into the buffer system containing the antibody, and additional solvent can be added to the solution to a desired percentage. The chelator can be added in molar excess of the antibody, for example, 3.5-5:1 chelator to antibody. The total reaction volume can be up to 5 L.
[0163] The reaction temperature and the reaction time are inversely related. For example, if the reaction temperature is higher, the reaction time is lower. If the reaction temperature is lower, the reaction time is higher. Illustratively, at a temperature above about 18.degree. C., the reaction may take less than 2 hours; at a temperature below 18.degree. C., the reaction may take more than 2 hours.
[0164] The conjugation reaction can be terminated by quenching, for example, by the addition of acetic acid.
[0165] In some embodiments, conjugation of the antibody with deferoxamine is performed to produce DFO-mAb conjugates. In some embodiments, conjugation of the antibody with p-SCN-Bn-deferoxamine is performed to produce DFO-mAb conjugates.
[0166] Exemplary solvents for the chelator include DMSO and DMA. Subsequent UFDF steps utilize membranes, and the membrane is chosen based on the solvent system used in the conjugation step. For example, DMA dissolves PES membranes, so the two could not be used in the same system.
[0167] Carbonate buffers are not preferred for stability of the conjugate during long term storage. Thus, once the antibody-chelator conjugates have been formed, they can be buffer exchanged into a buffer chosen specifically for long term storage and stability. Exemplary buffers include citrate, acetate, phosphate, arginine, and histidine buffers. A further UFDF step can be performed to remove residual salts and to provide a suitable concentration, excipient level, and pH of the conjugated monoclonal antibody. The resulting antibody-chelator conjugates can be sterile filtered and stored for subsequent formulation.
III. Methods of Using Radiolabeled Immunoconjugates
[0168] In certain aspects, the present disclosure provides diagnostic and therapeutic methods of use of the radiolabeled antibody conjugates of the present disclosure.
[0169] According to one aspect, the present disclosure provides methods of detecting PD-L1 in a tissue, the methods comprising administering a radiolabeled antibody conjugate of the provided herein to the tissue; and visualizing the PD-L1 expression by positron emission tomography (PET) imaging. In certain embodiments, the tissue comprises cells or cell lines. In certain embodiments, the tissue is present in a subject, wherein the subject is a mammal. In certain embodiments, the subject is a human subject. In certain embodiments, the subject has a disease or disorder selected from the group consisting of cancer, infectious disease and inflammatory disease. In one embodiment, the subject has cancer. In certain embodiments, the infectious disease is bacterial or viral infection caused by, for example, hepatitis B virus (HBV), hepatitis C virus (HCV), human immunodeficiency virus (HIV), and Mycobacterium tuberculosis.
[0170] According to one aspect, the present disclosure provides methods of imaging a tissue that expresses PD-L1 comprising administering a radiolabeled antibody conjugate of the present disclosure to the tissue; and visualizing the PD-L1 expression by positron emission tomography (PET) imaging. In one embodiment, the tissue is comprised in a tumor. In one embodiment, the tissue is comprised in a tumor cell culture or tumor cell line. In one embodiment, the tissue is comprised in a tumor lesion in a subject.
[0171] According to one aspect, the present disclosure provides methods for measuring response to a therapy, wherein the response to a therapy is measured by measuring inflammation. The methods, according to this aspect, comprise administering a radiolabeled antibody conjugate provided herein to a subject in need thereof and visualizing the PD-L1 expression by positron emission tomography (PET) imaging. In certain embodiments, the inflammation is present in a tumor in the subject. In certain embodiments, an increase in PD-L1 expression correlates to increase in inflammation in the tumor.
[0172] According to one aspect, the present disclosure provides methods for determining if a patient is suitable for anti-tumor therapy comprising an inhibitor of the PD-1/PD-L1 signaling axis, the methods comprising selecting a patient with a solid tumor, administering a radiolabeled antibody conjugate of the present disclosure, and localizing the administered radiolabeled antibody conjugate in the tumor by PET imaging wherein presence of the radiolabeled antibody conjugate in the tumor identifies the patient as suitable for anti-tumor therapy comprising an inhibitor of the PD-1/PD-L1 signaling axis.
[0173] According to one aspect, the present disclosure provides methods for identifying a candidate for anti-tumor therapy comprising an inhibitor of the PD-1/PD-L1 signaling axis, the methods comprising selecting a patient with a solid tumor, administering a radiolabeled antibody conjugate of the present disclosure, and localizing the administered radiolabeled antibody conjugate in the tumor by PET imaging wherein presence of the radiolabeled antibody conjugate in the tumor identifies the patient as suitable for anti-tumor therapy comprising an inhibitor of the PD-1/PD-L1 signaling axis.
[0174] According to one aspect, the present disclosure provides methods for predicting response of a patient to an anti-tumor therapy comprising an inhibitor of the PD-1/PD-L1 signaling axis, the methods comprising selecting a patient with a solid tumor, determining if the tumor is PD-L1-positive, wherein a positive response of the patient is predicted if the tumor is PD-L1-positive. In certain embodiments, the tumor is determined positive by administering a radiolabeled antibody conjugate of the present disclosure and localizing the radiolabeled antibody conjugate in the tumor by PET imaging wherein presence of the radiolabeled antibody conjugate in the tumor indicates that the tumor is PD-L1-positive.
[0175] According to one aspect, the present disclosure provides methods for detecting a PD-L1-positive tumor in a subject. The methods, according to this aspect, comprise selecting a subject with a solid tumor; administering a radiolabeled antibody conjugate of the present disclosure to the subject; and determining localization of the radiolabeled antibody conjugate by PET imaging, wherein presence of the radiolabeled antibody conjugate in a tumor indicates that the tumor is PD-L1-positive.
[0176] As used herein, the expression "a subject in need thereof" means a human or non-human mammal that exhibits one or more symptoms or indications of cancer, and/or who has been diagnosed with cancer, including a solid tumor and who needs treatment for the same. In many embodiments, the term "subject" may be interchangeably used with the term "patient". For example, a human subject may be diagnosed with a primary or a metastatic tumor and/or with one or more symptoms or indications including, but not limited to, unexplained weight loss, general weakness, persistent fatigue, loss of appetite, fever, night sweats, bone pain, shortness of breath, swollen abdomen, chest pain/pressure, enlargement of spleen, and elevation in the level of a cancer-related biomarker (e.g., CA125). The expression includes subjects with primary or established tumors. In specific embodiments, the expression includes human subjects that have and/or need treatment for a solid tumor, e.g., colon cancer, breast cancer, lung cancer, prostate cancer, skin cancer, liver cancer, bone cancer, ovarian cancer, cervical cancer, pancreatic cancer, head and neck cancer, and brain cancer. The term includes subjects with primary or metastatic tumors (advanced malignancies). In certain embodiments, the expression "a subject in need thereof" includes patients with a solid tumor that is resistant to or refractory to or is inadequately controlled by prior therapy (e.g., treatment with an anti-cancer agent). For example, the expression includes subjects who have been treated with one or more lines of prior therapy such as treatment with chemotherapy (e.g., carboplatin or docetaxel). In certain embodiments, the expression "a subject in need thereof" includes patients with a solid tumor which has been treated with one or more lines of prior therapy but which has subsequently relapsed or metastasized. In certain embodiments, the term includes subjects having an inflammatory disease or disorder including, but not limited to, cancer, rheumatoid arthritis, atherosclerosis, periodontitis, hay fever, heart disease, coronary artery disease, infectious disease, bronchitis, dermatitis, meningitis, asthma, tuberculosis, ulcerative colitis, Crohn's disease, inflammatory bowel disease, hepatitis, sinusitis, psoriasis, allergy, fibrosis, lupus, vasiculitis, ankylosing spondylitis, Graves' disease, Celiac disease, fibromyalgia, and transplant rejection.
[0177] In certain embodiments, the methods of the present disclosure are used in a subject with a solid tumor. The terms "tumor", "cancer" and "malignancy" are interchangeably used herein. As used herein, the term "solid tumor" refers to an abnormal mass of tissue that usually does not contain cysts or liquid areas. Solid tumors may be benign (not cancer) or malignant (cancer). For the purposes of the present disclosure, the term "solid tumor" means malignant solid tumors. The term includes different types of solid tumors named for the cell types that form them, viz. sarcomas, carcinomas and lymphomas. In certain embodiments, the term "solid tumor" includes cancers including, but not limited to, colorectal cancer, ovarian cancer, prostate cancer, breast cancer, brain cancer, cervical cancer, bladder cancer, anal cancer, uterine cancer, colon cancer, liver cancer, pancreatic cancer, lung cancer, endometrial cancer, bone cancer, testicular cancer, skin cancer, kidney cancer, stomach cancer, esophageal cancer, head and neck cancer, salivary gland cancer, and myeloma.
[0178] According to one aspect, the present disclosure provides methods of treating a tumor in a subject. The methods, according to this aspect, comprise selecting a subject with a solid tumor; determining that the tumor is PD-L1-positive; and administering one or more doses of an inhibitor of the PD-1/PD-L1 signaling axis. In certain embodiments, the tumor is determined to be PD-L1-positive by administering a radiolabeled antibody conjugate of the present disclosure to the subject; and visualizing the radiolabeled antibody conjugate in the tumor by PET imaging, wherein presence of the radiolabeled antibody conjugate in the tumor indicates that the tumor is PD-L1-positive.
[0179] As used herein, the terms "treat", "treating", or the like, mean to alleviate symptoms, eliminate the causation of symptoms either on a temporary or permanent basis, to delay or inhibit tumor growth, to reduce tumor cell load or tumor burden, to promote tumor regression, to cause tumor shrinkage, necrosis and/or disappearance, to prevent tumor recurrence, to prevent or inhibit metastasis, to inhibit metastatic tumor growth, and/or to increase duration of survival of the subject.
[0180] According to one aspect, the present disclosure provides methods for monitoring the efficacy of an anti-tumor therapy in a subject, wherein the methods comprise selecting a subject with a solid tumor wherein the subject is being treated with an anti-tumor therapy; administering a radiolabeled antibody conjugate of the present disclosure to the subject; imaging the localization of the administered radiolabeled conjugate in the tumor by PET imaging; and determining tumor growth, wherein a decrease from the baseline in radiolabeled signal indicates tumor regression and efficacy of the anti-tumor therapy. In certain embodiments, the anti-tumor therapy comprises an inhibitor of the PD-1/PD-L1 signaling axis (e.g., an anti-PD-1 antibody).
[0181] In certain embodiments, the present disclosure provides methods to assess changes in the inflammatory state of a tumor, the methods comprising selecting a subject with a solid tumor wherein the subject is being treated with an anti-tumor therapy; administering a radiolabeled antibody conjugate provided herein to the subject; and imaging the localization of the administered radiolabeled conjugate in the tumor by PET imaging, wherein an increase from the baseline in radiolabeled signal indicates increase in inflammation and efficacy of the anti-tumor therapy. In certain embodiments, the anti-tumor therapy comprises an inhibitor of the PD-1/PD-L1 signaling axis (e.g., an anti-PD-1 antibody).
[0182] As used herein, the term "baseline," with respect to the PD-L1 expression in the tumor, means the numerical value of uptake of the radiolabeled conjugate for a subject prior to or at the time of administration of a dose of anti-tumor therapy. The uptake of the radiolabeled conjugate is determined using methods known in the art (see, for example, Oosting et al 2015, J. Nucl. Med. 56: 63-69). In certain embodiments, the anti-tumor therapy comprises an inhibitor of the PD-1/PD-L1 signaling axis.
[0183] To determine whether there is tumor regression, the uptake of the radiolabeled conjugate is quantified at baseline and at one or more time points after administration of the inhibitor of the PD-1/PD-L1 signaling axis (e.g., an anti-PD-1 antibody). For example, the uptake of the administered radiolabeled antibody conjugate (e.g., radiolabeled anti-PD-L1 antibody conjugate) may be measured at day 2, day 3, day 4, day 5, day 6, day 7, day 8, day 9, day 10, day 11, day 12, day 14, day 15, day 22, day 25, day 29, day 36, day 43, day 50, day 57, day 64, day 71, day 85; or at the end of week 1, week 2, week 3, week 4, week 5, week 6, week 7, week 8, week 9, week 10, week 11, week 12, week 13, week 14, week 15, week 16, week 17, week 18, week 19, week 20, week 21, week 22, week 23, week 24, or longer, after the initial treatment with the inhibitor of the PD-1/PD-L1 signaling axis (e.g., an anti-PD-1 antibody). The difference between the value of the uptake at a particular time point following initiation of treatment and the value of the uptake at baseline is used to establish whether there has been a difference in amount of tumor tissue (tumor regression or progression). For example, a decrease from baseline in the uptake upon treatment with at least one dose of the inhibitor of the PD-1/PD-L1 signaling axis means tumor regression and indicates efficacy of the anti-tumor therapy.
[0184] In certain embodiments, the radiolabeled antibody conjugate is administered intravenously or subcutaneously to the subject. In certain embodiments, the radiolabeled antibody conjugate is administered intra-tumorally. Upon administration, the radiolabeled antibody conjugate is localized in the tumor. The localized radiolabeled antibody conjugate is imaged by PET imaging and the uptake of the radiolabeled antibody conjugate by the tumor is measured by methods known in the art. In certain embodiments, the imaging is carried out 1, 2, 3, 4, 5, 6 or 7 days after administration of the radiolabeled conjugate. In certain embodiments, the imaging is carried out on the same day upon administration of the radiolabeled antibody conjugate.
[0185] In certain embodiments, the antibody or antigen-binding fragment thereof that binds specifically to PD-L1. In certain embodiments, the anti-PD-L1 antibody comprises the CDRs of a HCVR, wherein the HCVR has an amino acid sequence selected from the group consisting of SEQ ID NOs: 2, 34, 50, 82, 98, 146, 162, 178, 186, 234, 250, 290, 306, 314, and 330; and the CDRs of a LCVR, wherein the LCVR has an amino acid sequence selected from the group consisting of SEQ ID NOs: 10, 42, 58, 90, 106, 154, 170, 194, 242, 258, and 274.
[0186] In certain embodiments, the inhibitor of the PD-1/PD-L1 signaling axis comprises an antibody or antigen-binding fragment thereof that binds specifically to PD-1. In certain embodiments, the anti-PD-1 antibody is selected from the group consisting of nivolumab, pembrolizumab and REGN2810. In certain other embodiments, the inhibitor of the PD-1/PD-L1 signaling axis comprises an antibody or antigen-binding fragment thereof that binds specifically to PD-L1. In one embodiment, the anti-PD-L1 antibody is atezolizumab. In one embodiment, the anti-PD-L1 antibody comprises an HCVR of SEQ ID NO: 82 and a LCVR of SEQ ID NO: 90.
IV. Examples
[0187] Certain embodiments of the disclosure are illustrated by the following non-limiting examples.
Example 1: Generation of Human Antibodies to PD-L1
[0188] Human anti PD-L1 antibodies, including those listed in Table 1, were prepared and characterized as described in US Patent Publication No. US 2015-0203580 A1, which is incorporated herein by reference in its entirety. In brief, human antibodies to PD-L1 were generated using a fragment of PD-L1 that ranges from about amino acids 19-239 of PD-L1 (Genbank Accession No. NP_054862.1). The immunogen was administered directly, with an adjuvant to stimulate the immune response, to a VELOCIMMUNE.RTM. mouse comprising DNA encoding human Immunoglobulin heavy and kappa light chain variable regions. The antibody immune response was monitored by a PD-L1-specific immunoassay. When a desired immune response was achieved splenocytes were harvested and fused with mouse myeloma cells to preserve their viability and form hybridoma cell lines. The hybridoma cell lines were screened and selected to identify cell lines that produce PD-L1-specific antibodies. Using this technique, and the immunogen described above, several anti-PD-L1 chimeric antibodies (i.e., antibodies possessing human variable domains and mouse constant domains) were obtained; exemplary antibodies generated in this manner were designated as H2M8306N, H2M8307N, H2M8309N, H2M8310N, H2M8312N, H2M8314N, H2M8316N, H2M8317N, H2M8321N, H2M8323N, H2M8718N, H2M8718N2, and H2M8719N.
[0189] Anti-PD-L1 antibodies were also isolated directly from antigen-positive B cells without fusion to myeloma cells, as described in U.S. 2007/0280945A1, herein specifically incorporated by reference in its entirety. Using this method, several fully human anti-PD-L1 antibodies (i.e., antibodies possessing human variable domains and human constant domains) were obtained; exemplary antibodies generated in this manner were designated as follows: H1H9323P, H1H9327P, H1H9329P, H1H9336P, H1H9344P2, H1H9345P2, H1H9351P2, H1H9354P2, H1H9364P2, H1H9373P2, H1H9382P2, H1H9387P2, and H1H9396P2.
Example 2: Conjugation of Anti-PD-L1 Antibody H4H8314N with p-SCN-Bn-DFO
[0190] In order to modify the parental anti-PD-L1 antibody, H4H8314N, and an isotype control antibody to be suitable for ImmunoPET studies with radiolabeling, a chelator, p-SCN-bn-Deferoxamine (DFO; Macrocylics, Cat #: B-705), was attached to the antibodies.
[0191] For the modification, H4H8314N was first buffer exchanged into PBS, pH 7.2 from histidine buffer by dialysis at 4.degree. C. overnight (Slide-A-Lyzer Dialysis Cassette G2 10 k MWCO; ThermoScientific) then buffer exchanged again using a PD-10 column (GE Healthcare, Cat. #: 17-0851-01) into a buffer composed of 50 mM carbonate buffer, 150 mM NaCl, pH 9.0 (conjugation buffer). To determine the concentration following the buffer exchanges, the samples were measured on a Nanodrop 2000 UV/VIS spectrometer (Thermo Scientific) using the MacVector sequence based extinction coefficient of 1.46 g/L (see Table 2). In 15 a mL polypropylene tube, 773.9 uL of H4H8314N (12.5 mg) was added to 1676.1 uL of conjugation buffer. In a separate vial, 29.3 uL of DMSO was added to 20.7 uL of DFO. In one-quarter increments, this DFO solution was added to the H1H8314N solution, each time gently being mixed by pipetting up-and-down. The final solution was 5 mg/mL H4H8314N in conjugation buffer, 2% DMSO with 6-fold mole-to-mole excess of DFO. This solution was allowed to incubate in a 3TC water bath with no additional stirring.
[0192] After 30 minutes at 3TC, the solution was promptly passed through a PD-10 desalting column (GE Healthcare, Cat. #: 17-0851-01), pre-equilibrated with a buffer containing 250 mM NaAcO at pH 5.4 (formulation buffer). The final solution was sterile-filtered via a syringe filter (Acrodisc 13 mm syringe filter, Pall Corporation, Cat #: 4602). The concentration and DFO-to-Antibody Ratio (DAR) was subsequently measured by UV/VIS spectroscopy. For the absorbance measurement, the DFO-conjugated antibody was measured against the formulation buffer at 252 nm (A252), 280 nm (A280) and 600 nm (A600). For the calculation, the background was corrected at each absorbance value using the equation:
A'.sub..lamda.=A.sub..lamda.-.lamda..sub.600
[0193] The antibody conjugate was tested for aggregation using SEC chromatography, with 25 ug of the sample injected onto a Superdex 200 column (GE Healthcare, Cat. No. 17-5175-01) monitored at 280 nm with a PBS mobile phase (0.75 mL/min). The antibody integrity was evaluated by SDS-PAGE 4-20% Tris/Gly pre-cast gel (Novex) with 2 ug of the sample loaded. The gel is shown in FIG. 1. The antibody concentration, conjugate concentration, and DAR were calculated using the equations below:
Antibody Concentration Calculation
[0194] Conc mAb ( mg / mL ) = A 280 ' 280 ##EQU00001##
Conjugate Concentration Calculation
[0195] Conc conjugate ( mg / mL ) = A 252 ' - 1.53 A 280 ' 252 - 1.53 280 ##EQU00002##
DAR Calculation
[0196] DAR = 252 A 280 ' - 280 A 252 ' 18800 A 252 ' - 28700 A 280 ' ##EQU00003##
TABLE-US-00002 TABLE 2 Molar extinction coefficients and molecular weight MW .epsilon.280 .epsilon.252 Antibody (g mol.sup.-1) (L g.sup.-1cm.sup.-1) (L g.sup.-1cm.sup.-1) H4H8314N 144984 1.46 0.553
TABLE-US-00003 TABLE 3 UV DAR, percent aggregate and concentration post DFO-attachment Concentration Antibody UV DAR (mg/mL) % aggregate H4H8314N 1.2 3.34 <1%
Example 3: .sup.89Zr Chelation of DFO Conjugated Monoclonal Antibodies
[0197] For use in ImmunoPET in vivo studies, the DFO-conjugated anti-PD-L1 antibody, H4H8314N, and a DFO-conjugated isotype control antibody were radiolabeled with .sup.89Zr.
[0198] DFO-conjugated antibody (250 or 750 ug) was first brought to 1.25 mg/mL in 1 M HEPES, pH 7.2. The recipe of DFO-Ab conjugate solution for each study is listed in Table 4. Separately, .sup.89Zr solution was prepared using the recipe for each corresponding study shown in Table 5. Stock .sup.89Zr-oxalic acid solution was obtained from Perkin Elmer or 3D Imaging. If the radioactivity concentration of the stock solution was low (see Table 5), a neutralization step was performed with 1 M borate, pH 9.0. The final radioactivity of the solution was first confirmed using a Capintec CRC-25R dose calibrator (Capintec #520), then immediately combined with the DFO-Ab conjugate solution, gently mixed (pipetting up-and-down) and subsequently incubated for 45 minutes at room temperature.
[0199] After the incubation, a small sample of each reaction mixture was taken for iTLC (instant thin layer liquid chromatography) to determine radiolabeling reaction yield and the remaining reaction mixtures were transferred to pre-equilibrated PD-10 columns (Vendor) with 250 mM sodium acetate at pH 5.4 for gravity fed desalting. Each PD-10 column took no more than 1.2 mL of reaction mixture (otherwise multiple columns were used). After the contents of the reaction entered the column bed, 1.6 mL of 250 mM sodium acetate at pH 5.4 (formulation buffer) was added; the flow through was discarded. An additional 1.8 mL of formulation buffer was added to the column, and the eluate was collected from each column. Next, approximately 500 uL of each solution was analyzed using a Nanodrop spectrophotometer (ThermoScientific). The final Ab concentration was calculated using the appropriate extinction coefficient and the absorption at 280 nm using the equation:
Concentration in mg/mL=Absorption at 280 nm/Extinction coefficient at 280 nm (found in Table 6)
[0200] The final mass measured in grams was recorded in Table 4. The radioactivity was then measured using the dose calibrator and reported in Table 5. The final material along with the material prior to the PD-10 column treatment, were then analyzed by iTLC. For this assay, 1 uL of each solution was added to the iTLC-SG-Glass microfiber chromatography paper impregnated with silica gel (Agilent Technologies, Cat # SG10001), developed in a TLC chamber with 20 mM citric acid buffer solution. The final material was also analyzed using a SEC-HPLC with UV 280 and radioisotope detector connected in series (Agilent 1260 with Lablogic Radio-TLC/HPLC Detector, SCAN-RAM) using a Superdex 200 column with PBS mobile phase at a flow rate of 0.75 mL/min. The radiotrace was used for the determining radiochemical purity by comparing the integration of the protein peak (.about.10 to 16 min) and free .sup.89Zr peak (.about.25 min). The monomeric purity was determined by comparing the integration of the oligomeric peak (10 min to .about.15 min) to the monomer (.about.16 min).
[0201] The specific activity and protein recovery (%) of each radiolabeled conjugate was determined using the following equations:
Mass of conjugate in mg=concentration in mg/mL.times.mass of solution in grams a.
Specific activity in mCi/mg=activity of vial in mCi/mass of conjugate in mg b.
Protein recovery=starting conjugate mass (mg)/Mass of conjugate in mg c.
[0202] Finally the appearance was noted and recorded in Table 7. Both UV280 and iTLC traces were performed on purified product.
[0203] The results are consolidated in Table 7. The radio-SEC-HPLC chromatograms are shown in FIGS. 2-4. An example of UV280 HPLC SEC chromatogram and radio-iTLC is shown in FIG. 5 for the .sup.89Zr radiolabeling, Study 1. The UV280-HPLC SEC chromatogram confirms the highly monomeric product (99%). The radio-iTLC trace was processed with a 7-point binomial smoothing function. The origin and solvent front was approximately 16 and 100 mm, respectively. No detectable .sup.89Zr was observed beyond 22 mm and corroborates the radiochemical purity determined by radio-SEC-HPLC SEC in FIG. 2B.
TABLE-US-00004 TABLE 4 DFO-antibody conjugate preparation for radiolabeling Radio- Conjugate Total Final labeling Study Radiolabeling Concentration mass volume Concentration # # Lots (mg/mL) DAR * (mg) (uL) (mg/mL) 1 1 Isotype-DFO- 3.7 1.6 250 200 1.25 .sup.89Zr 2 1 H4H8314N- 3.34 1.2 250 200 1.25 DFO-.sup.89Zr 3 2 H4H8314N- 3.34 1.2 750 600 1.25 DFO-.sup.89Zr 4 3 Isotype-DFO- 3.7 1.6 250 200 1.25 .sup.89Zr 5 3 H4H8314N- 3.34 1.2 250 200 1.25 DFO-.sup.89Zr * DAR is defined as the DFO to Antibody Ratio
TABLE-US-00005 TABLE 5 .sup.89Zr reaction solution preparation for radiolabeling Add'l 1M borate, Radio- .sup.89Zr- 1M oxalic pH 9.0 1M HEPES, Final Final Specific Radio - Study labeling oxalate acid added added pH 7.2 Vol Activity Activity labeling # Lots (uL) (uL) (uL) (uL) (uL) (uCi) (uCi/uL) 1 1 Isotype- 50 50 400 500 1000 1009 1.01 DFO-.sup.89Zr 2 1 H4H8314N- 50 50 400 500 1000 1000 1 DFO-.sup.89Zr 3 2 H4H8314N- 150 150 1200 1500 3000 3070 1.02 DFO-.sup.89Zr 4 3 Isotype- ~1 0 0 1000 1000 1680 1.68 DFO-.sup.89Zr 5 3 H4H8314N- ~1 0 0 1000 1000 1640 1.64 DFO-.sup.89Zr
TABLE-US-00006 TABLE 6 Extinction coefficients for conjugate lots Radiolabeling Lot .sub.280 (AU ml mg.sup.-1 cm.sup.-1) Isotype-DFO-.sup.89Zr 1.71 H4H8314N-DFO-.sup.89Zr 1.61
TABLE-US-00007 TABLE 7 Summary of .sup.89Zr labeled DFO-Ab conjugates for in vivo imaging and biodistribution studies Radio- Mono- chemical meric Protein Specific Radio- Study Conjugate Purity* Purity* Recovery Conc. Activity labeling # Lots Appearance (%) (%) (%) (mg/mL) (mCi/mg) 1 1 Isotype- Clear >99% >95% 60% 0.106 3.35 DFO-.sup.89Zr 2 1 H4H8341N- Clear >99% >95% 63% 0.121 2.75 DFO-.sup.89Zr 3 2 H4H8341N- Clear >99% >95% 62% 0.134 3.58 DFO-.sup.89Zr 4 3 Isotype- Clear >99% >95% 66% 0.074 5.38 DFO-.sup.89Zr 5 3 H4H8341N- Clear >99% >95% 74% 0.084 5.13 DFO-.sup.89Zr *by radio-SEC-HPLC
Example 4: Immunoreactivity
[0204] The immunoreactivity (IR) of the radiolabeled anti-PD-L1 antibody and isotype control antibody was measured as follows. For the initial studies, MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg cells were used and subsequently LOX-IMVI cells (see detailed description of cell lines in Example 5) were also used in the later study. In these assays, 20 ng of the respective .sup.89Zr labeled antibodies were added to 15.times.10.sup.6 MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg or 30.times.10.sup.6 LOX-IMVI cells in a final volume of 1 mL. Samples were incubated for 45 minutes with continuous mixing before undergoing 3 washes with media to remove any unbound antibody. The radioactivity of the test cell pellets was then counted in an automatic gamma counter (Wizard 2470, Perkin Elmer) against 2 reference standards containing the same 20 ng of .sup.89Zr labeled antibody. The percentage immunoreactivity was determined for the samples using the average of the standards as a measure of total activity.
[0205] As seen in Table 8, .sup.89Zr labeled anti-PD-L1 antibody retained immunoreactivity following conjugation and radiolabeling, with % IR ranging from 88 to 98% across the studies. The specificity of binding is apparent in the control antibodies having a background % IR of less than 1%.
TABLE-US-00008 TABLE 8 Immunoreactivity of .sup.89Zr chelated DFO-conjugates Study Study 1 Study 2 Study 3 Cell Line MC38-cOVA/eGFP- MC38-cOVA/eGFP- MC38-cOVA/eGFP- mPD-L1.sup.-/-hPD-L1.sup.Tg mPD-L1.sup.-/-hPD-L1.sup.Tg mPD-L1.sup.-/-hPD-L1.sup.Tg LOX-IMVI Antibody .sup.89Zr- .sup.89Zr- .sup.89Zr- .sup.89Zr- Anti-PD- .sup.89Zr- Anti-PD- .sup.89Zr- Anti-PD- .sup.89Zr- Anti-PD- .sup.89Zr- L1 Control L1 Control L1 Control L1 Control Cell 4048.4 29.6 8311.9 na 6262.4 68 5587.54 65.4 pellet activity Average 4536.5 6432.4 8567.2 na 6386.6 9544.8 6386.6 9544.8 Standard activity Percent 89.2 0.5 97.0 na 98.1 0.7 87.5 0.7 IR
Example 5: In Vitro and Ex Vivo Characterization of Human PD-L1 Expression on Tumor Cell Lines
[0206] Several tumor cell lines were studied to evaluate the expression level of human PD-L1, aiming at the detection of human PD-L1 expressed endogenously by tumors in vivo in either male NCr nude (Taconic, Hudson N.Y.) mice or in mice that were engineered to be homozygous for the expression of the extracellular domain of human PD-L1 in place of extracellular domain of mouse PD-L1 (PD-L1 Humin mice) on a 75% 057/B16/25% 129 strain background using VelociGene.RTM. technology (Valenzuela et al 2003, Nat. Biotechnol. 21: 652-659; US Patent Application Publication US2016/0157469).
[0207] Cell lines used in these studies include: 1) a murine colon carcinoma cell line MC38 (obtained from NCI at Frederick, Md., Laboratory of Tumor Immunology and Biology), which has been engineered in house to knock out murine PD-L1, but over-express full-length human PD-L1 and full-length chicken ovalbumin fused with eGFP, thus referred here as MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1Tg; 2) several human tumor cell lines: human melanoma cell line LOX-IMVI (endogenous PD-L1 positive line, obtained from NCI at Frederick, Md., Division of Cancer Treatment and Diagnosis; Tumor Repository), human breast cancer cell lines MDA-MB-231 (endogenous PD-L1 positive line) and SK-Br-3 (PD-L1 negative cell line) (both obtained from ATCC). In some cases, human PD-L1 was directly evaluated without any induction in vitro; in some cases, human PD-L1 expression was evaluated with overnight murine or human IFN.gamma. (100 ng/ml) treatment (obtained from Peprotech); in some cases, human PD-L1 was evaluated ex vivo on enzymatically dissociated tumor cells extracted from tumor bearing nude mice or humanized mice. All surface staining of human PD-L1 was performed following a standard protocol. Briefly, tumor cells were washed with PBS once, washed with ice cold staining buffer once, stained with commercial available fluorochrome directly conjugated anti-human PD-L1 antibody (eBioscience, clone MIH1) in staining buffer for 30 minutes on ice in the dark, and then washed with 2 mL of PBS once again. Fixable dye eFluor506 was also included following manufacturer's protocol (eBioscience, Cat #1T5983). Samples were acquired on BD FACSCanto II.TM. IVD10 equipped with DIVA v8. Data were further analyzed with FlowJo v10.0.6 or above.
[0208] PD-L1 expression by MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg cells prior to implantation and seven days post implantation in nude mice is shown in Table 9.
TABLE-US-00009 TABLE 9 Percentage of human PD-L1 positive MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg cells prior to implantation and 7 days post implantation in nude mice Isotype staining hPD-L1 staining Prior to implantation 0.6% 94.7% Post implantation 1.09% 74.0%
[0209] Prior to implantation, a vast majority of MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg cells were human PD-L1 positive, compared to isotype control staining. Seven days post implantation in nude mice and upon enzymatic and mechanical processing for tumor dissociation, .about.70% of MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg cells were still human PD-L1 positive.
[0210] PD-L1 expression by MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg cells prior to implantation and fourteen days post implantation in PD-L1 humanized mice is shown in Table 10.
TABLE-US-00010 TABLE 10 Percentage of human PD-L1 positive MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg cells prior to implantation and 14 days post implantation in PD-L1 humanized mice Isotype staining hPD-L1 staining Prior to implantation 0.2% 92.5% Post implantation 3.6 46.2%
[0211] Prior to implantation, a vast majority of MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg cells were human PD-L1 positive, compared to isotype control staining. Fourteen days post implantation in PD-1/PD-L1 double humanized mice and upon enzymatic and mechanical processing for tumor dissociation; .about.50% of MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg cells were still human PD-L1 positive.
[0212] PD-L1 expression by multiple tumor cell lines in vitro is shown in FIG. 6. To evaluate how comparable the expression level of PD-L1 by the engineered cell line (MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg) and other human tumor cell lines (LOX-IMVI melanoma cells, MDA-MB-231 breast cancer cells, and SK-Br-3 breast cancer cells) was, dose titration of anti-PD-L1 antibody staining was performed. FIG. 6 illustrates that MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg had the highest level of human PD-L1 expression (FIG. 6A) and SK-Br-3 had the lowest expression with no PD-L1 detectable (FIG. 6D), whereas PD-L1 expression by LOX-IMVI and MDA-MB-231 was moderate (about 5 times lower than MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg) (FIGS. 6B and 6C).
[0213] In a second experiment, further comparison between LOX-IMVI and MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1Tg was done with or without in vitro treatment by 100 ng/mL of hIFN.gamma./mIFN.gamma. overnight, respectively. FIG. 7 illustrated that median fluorescence intensity of PD-L1 reached the plateau at .about.150 nM of anti-PD-L1 antibody used for staining. At the baseline, PD-L1 expression by LOX-IMVI was moderate (about 6-7 times lower than MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1Tg). Upon treatment with mIFN.gamma., there was no change for PD-L1 staining on MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg, whereas 3-fold increase of human PD-L1 staining was seen in LOX-IMVI after treatment with hIFN.gamma..
[0214] Ex vivo PD-L1 expression by LOX-IMVI and MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg cells about three weeks post implantation in nude mice were shown in Tables 11 and 12.
TABLE-US-00011 TABLE 11 Percentage of PD-L1 positive LOX-IMVI and MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg cells ~3 weeks post implantation in nude mice Isotype hPD-L1 staining staining LOX-IMVI 0.2% 56.6% MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg 0.2% 96.2%
TABLE-US-00012 TABLE 12 Mean fluorescence intensity of PD-L1 by LOX-IMVI and MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg cells ~3 weeks post implantation in nude mice Tumor 1 Tumor 2 LOX-IMVI 8479.1 12121.5 MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg 49589.1 51445.0
[0215] Upon enzymatic and mechanical processing to allow for tumor dissociation, cells were stained with the anti-PD-L1 antibody (20 .mu.g/mL). The PD-L1 expression level on LOX-IMVI was about 5 times lower than that on MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg tumor cells.
Example 6: Selective Localization of Radiolabeled Anti-PD-L1 Antibody to hPD-L1 Positive Tumors in Nude Mice
[0216] To determine the in vivo localization of anti-PD-L1 antibody, Zirconium-89 labeled DFO-antibody conjugate was administered intravenously to nude mice bearing PD-L1 positive tumors.
[0217] The tumor line used for the study was a murine colon carcinoma cell-line referred to as MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1Tg, which has been engineered to knock out murine PD-L1 off the wild type MC38, but over-express full-length human PD-L1 and full-length chicken ovalbumin fused with eGFP. For the second study of tumors with endogenous expression of human PD-L1, the human melanoma cell line LOX-IMVI was used to establish tumors in vivo for subsequent anti-PD-L1 antibody localization studies.
[0218] The exemplary radiolabeled anti-PD-L1 antibody used for this study was H1H8314N, comprising HCVR/LCVR of SEQ ID NOs: 82/90.
[0219] For the first study, 1.times.10.sup.6 MC38-cOVA/eGFP-mPD-L1-/-hPD-L1.sup.Tg cells were implanted subcutaneously into the left flank of male 8-10 week old NCr nude mice (Taconic, Hudson N.Y.). For LOX-IMVI tumors, 1.times.10.sup.6 cells were implanted subcutaneously into the left flank of male 8-10 week old NCr nude mice. Once tumors had reached an average volume of 50-150 mm.sup.3 (.about.Day 7-10), mice were randomized into groups, and dosed with either .sup.89Zr labeled anti-PD-L1 DFO-antibody conjugate (H1H8314N) or a .sup.89Zr labeled non-binding isotype control DFO-antibody conjugate. The nude mice bearing MC38-cOVA/eGFP-mPD-L1-/-hPD-L1.sup.Tg tumors received 50.+-.1 uCi of .sup.89Zr labeled antibody with a protein dose .about.0.6 mg/kg. In the study using mice bearing LOX-IMVI tumors, mice received 35.+-.1 uCi of .sup.89Zr labeled antibody with a final antibody dose of 0.3 or 1 mg/kg.
[0220] PET imaging of antibody localization was assessed 6 days after administration of the antibodies. A Sofie Biosciences G8 PET/CT (Sofie Biosciences and Perkin Elmer) was used to acquire images). The instrument was pre-calibrated for detection of .sup.89Zr prior to image acquisition. The energy window ranged from 150 to 650 keV with a reconstructed resolution of 1.4 mm at the center of the field of view. Mice underwent induction anesthesia using isoflurane and were kept under continuous flow of isoflurane during imaging. Static 10-minute images were acquired using the G8 acquisition software and subsequently reconstructed using the pre-configured settings. Image data was corrected for decay and other parameters. CT images were acquired following PET acquisition and later co-registered with the PET images. Images were prepared using VivoQuant post-processing software (inviCRO Imaging Services).
[0221] For bio distribution, mice were euthanized at the final time-point (5-6 days post-dosing) and blood was collected via cardiac puncture. Tumors and normal tissues were then excised and placed in counting tubes. Weight for each sample were measured and recorded. Count data for .sup.89Zr in CPM was then collected by measuring samples on an automatic gamma counter (Wizard 2470, Perkin Elmer). The percent-injected dose per gram (% ID/g) was calculated for each sample using standards prepared from the injected material.
[0222] The average % ID/g for each antibody is presented in Table 13.
TABLE-US-00013 TABLE 13 Average % ID/g in analyzed tissues .sup.89Zr-Isotype .sup.89Zr-H1H8314N Control Antibody AVERAGE STDEV AVERAGE STDEV SAMPLE % ID/g % ID/g % ID/g % ID/g LIVER 3.1 0.4 0.9 0.9 SPLEEN 4.4 1.1 1.5 1.3 KIDNEY 4.0 0.7 1.4 1.6 BONE 5.1 2.6 1.7 1.6 LUNG 5.1 1.1 2.5 3.0 HEART 2.4 0.2 1.3 1.4 BLOOD 7.6 1.6 3.8 4.6 THYMUS 5.3 3.0 2.8 2.2 MC38-cOVA/eGFP- 55.3 12.2 3.0 3.3 mPD-L1.sup.-/-hPD-L1.sup.Tg S. BOWEL 1.5 0.3 0.6 0.6
[0223] From this, the clear high uptake in MC38-cOVA/eGFP-mPD-L1-/-hPD-L1.sup.Tg tumors was apparent over other normal tissues, with tumor uptake of 55.3% ID/g being significantly higher than the next highest uptake of 5.3% ID/g observed in the thymus. Tumor uptake was 7.3-fold and 17.8-fold higher than activity in blood and liver, respectively. The specificity of anti-PD-L1 uptake into tumor (55.3% ID/g) was apparent as compared to significantly reduced tumor uptake of 3% observed for the non-binding isotype control antibody. Pilot PET imaging performed here demonstrated a clear localization of the .sup.89Zr labeled anti-PD-L1 DFO-antibody conjugate to the MC38-cOVA/eGFP-mPD-L1-/-hPD-L1.sup.Tg tumors. Little background signal was observed in the animals at this Day 6 post-dosing time-point. In contrast to the clear tumor localization that was apparent using anti-PD-L1 antibody, only faint background activity was apparent in imaging of the control antibody in this model. Imaging clearly indicated high, specific uptake of anti-PD-L1 antibody in human PD-L1 positive tumor, showing the localization of .sup.89Zr radiolabeled anti-PD-L1 antibody to a MC38-cOVA/eGFP-mPD-L1-/-hPD-L1.sup.Tg tumor in an NCr nude mouse.
[0224] In a second study, the ability of anti-PD-L1 antibody to selectively target tumors expressing endogenous levels of human PD-L1 antigen was assessed. Here, mice bearing human LOX-IMVI melanoma tumors received .sup.89Zr labeled antibody at doses of 0.3 and 1 mg/kg. Again, blood, tumor and tissues were taken at Day 6 post-injection and the % ID/g for the samples was calculated. The average % ID/g for each antibody is presented in Table 14.
TABLE-US-00014 TABLE 14 Average % ID/G in analyzed tissues from second study (LOX-IMVI tumors) .sup.89Zr-Isotype control .sup.89Zr-DFO-H1H8314N .sup.89Zr-DFO-H1H8314N antibody 0.3 mg/kg 1 mg/kg 1 mg/kg AVERAGE STDEV AVERAGE STDEV AVERAGE STDEV SAMPLE % ID/g % ID/g % ID/g % ID/g % ID/g % ID/g LIVER 2.9 0.3 3.3 0.2 3.9 0.3 SPLEEN 4.2 0.2 4.3 0.9 4.2 0.7 KIDNEY 4.3 0.4 4.3 0.8 3.4 0.4 BONE 3.2 0.6 2.7 0.5 3.6 0.4 LUNG 5.7 1.0 6.6 1.6 5.9 1.2 HEART 3.2 0.8 3.2 0.4 2.9 0.6 BLOOD 8.1 1.4 9.5 1.0 11.1 6.2 THYMUS 5.3 2.3 5.6 0.7 4.9 1.4 LOX-IMVI 20.6 2.7 10.6 2.6 12.0 1.8 TUMOR S.BOWEL 1.5 0.2 1.8 0.4 2.0 0.3
[0225] At the lower 0.3 mg/kg dose, clear targeting to tumor over normal tissues was observed, with a 20.6% ID/g observed in the LOX-IMVI tumors. When mice received the higher 1 mg/kg dose, reduced tumor uptake 10.6% ID/g of was observed relative to the 0.3 mg/kg level. This suggests that the higher protein dose and possibly the subsequent higher fraction of unlabeled antibody led to blocking of tumor uptake by the .sup.89Zr labeled anti-PD-L1 antibody. In accordance with this, PET imaging conducted immediately prior to the biodistribution study also showed that uptake of anti-PD-L1 antibody at the 1 mg/kg dose was roughly equivalent to that of the control antibody. At the lower dose of 0.3 mg/kg, a clear increase in tumor localization of the anti-PD-L1 antibody was apparent relative to control antibody. Overall, the PET images and the biodistribution data demonstrate specific targeting of the LOX-IMVI tumors at the 0.3 mg/kg dose of anti-PD-L1 antibody.
Example 7: Selective Localization of Radiolabeled Anti-PD-L1 Antibody to hPD-L1 Positive Tumors in Mice
[0226] This Example describes the in vivo localization of a Zirconium-89 labeled DFO-anti-PD-L1 antibody conjugate in mice humanized for PD-L1. The exemplary antibody used in this Example was H1H8314N, comprising HCVR/LCVR of SEQ ID NOs: 82/90.
[0227] Mice humanized for PD-L1 were engineered using VelociGene.RTM. technology (Valenzuela et al 2003, Nat. Biotechnol. 21: 652-659; US Patent Application Publication US2016/0157469).
[0228] The tumor line used was a murine colon carcinoma cell-line referred to as MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg, engineered in-house to express full-length chicken ovalbumin fused with eGFP and to knock out murine PD-L1 off the wild type MC38, but over-express full-length human PD-L1.
[0229] 1.times.10.sup.6 cells of MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg were implanted subcutaneously into the left flank of male humanized PD-L1 mice. Once tumors had reached an average volume of 50-150 mm.sup.3 (.about.Day 7), mice were randomized into groups, and dosed with either .sup.89Zr labeled anti-PD-L1 DFO-antibody conjugate or a .sup.89Zr labeled non-binding isotype control DFO-antibody conjugate. The mice received 50.+-.1 uCi of .sup.89Zr labeled antibody with a final protein dose of 1 or 3 mg/kg.
[0230] PET imaging of antibody localization was assessed 6 days after administration of the antibodies. A Sofie Biosciences G8 PET/CT (Sofie Biosciences and Perkin Elmer) was used to acquire images). The instrument was pre-calibrated for detection of .sup.89Zr prior to image acquisition. The energy window ranged from 150 to 650 keV with a reconstructed resolution of 1.4 mm at the center of the field of view. Mice underwent induction anesthesia using isoflurane and were kept under continuous flow of isoflurane during imaging. Static 10-minute images were acquired using the G8 acquisition software and subsequently reconstructed using the pre-configured settings. Image data was corrected for decay and other parameters. CT images were acquired following PET acquisition and later co-registered with the PET images. Images were prepared using VivoQuant post-processing software (inviCRO Imaging Services).
[0231] For biodistribution, mice were euthanized at the final time-point (5-6 days post-dosing) and blood was collected via cardiac puncture. Tumors and normal tissues were then excised and placed in counting tubes. Weight for each sample were measured and recorded. Count data for .sup.89Zr in CPM was then collected by measuring samples on an automatic gamma counter (Wizard 2470, Perkin Elmer). The percent-injected dose per gram (% ID/g) was calculated for each sample using standards prepared from the injected material.
Results
[0232] Humanized PD-L1 mice bearing MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg tumors received .sup.89Zr labeled anti-PD-L1 DFO-antibody conjugate at a final antibody dose of 1 or 3 mg/kg. Blood, tumor and tissues were taken and weighed at Day 6 post-injection and the % ID/g for the samples was calculated based on the counts from each sample. The average % ID/g for dose at 1 and 3 mg/kg is presented in Table 15 and Table 16 respectively.
TABLE-US-00015 TABLE 15 Average % ID/g in analyzed tissues of anti-PD-L1 antibody at 1mg/kg AVERAGE STDEV SAMPLE % ID/g % ID/g LIVER 8.6 1.5 SPLEEN 14.1 1.1 KIDNEY 7.8 1.0 BONE 4.5 1.4 LUNG 7.9 3.0 HEART 4.3 1.1 BLOOD 9.1 4.6 THYMUS 9.7 3.5 MC38-cOVA/eGFP- 34.1 18.0 mPD-L1.sup.-/-hPD-L1.sup.Tg S. BOWEL 2.4 0.9
[0233] At the 1 mg/kg dose level, clear tumor targeting of the MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg tumors is apparent with a % ID/g of 34.1% despite the expression of PD-L1 in normal tissues in these humanized mice. At this dose, some localization of the .sup.89Zr labeled anti-PD-L1 antibody was apparent in the spleen, where antibody uptake of 14.1% ID/g was observed. Such uptake is expected because of the normal expression of human PD-L1 in place of mouse PD-L1 expression of human PD-L1 in the spleen. At the 3 mg/kg antibody dose, localization of .sup.89Zr-DFO-anti-PD-L1 antibody conjugate to the spleen was reduced, as uptake now averaged 9.7% ID/g in mice that received this antibody dose (Table 16).
TABLE-US-00016 TABLE 16 Average % ID/g in analyzed tissues of anti-PD-L1 antibody at 3 mg/kg AVERAGE STDEV SAMPLE % ID/g % ID/g LIVER 6.7 1.4 SPLEEN 9.7 1.3 KIDNEY 7.0 1.1 BONE 3.6 0.6 LUNG 11.0 1.0 HEART 4.7 0.7 BLOOD 12.4 2.1 THYMUS 7.6 0.5 MC38-cOVA/eGFP- 28.7 13.1 mPD-L1.sup.-/-hPD-L1.sup.Tg S. BOWEL 0.4 0.2
[0234] Clear tumor targeting was still observed at the 3 mg/kg dose, with an average of 28.7% ID/g taken up by the MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg tumors. Therefore although reduced normal tissue localization was apparent in imaging the 3 mg/kg dose, clear localization of anti-PD-L1 labeled antibody to the MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg tumors remained clear at this dose. Overall, these results indicate that clear targeting of the MC38-cOVA/eGFP-mPD-L1.sup.-/-hPD-L1.sup.Tg tumors was possible in mice expressing PD-L1 on regular sites of normal tissue expression.
[0235] The results from the studies performed here clearly demonstrate that anti-PD-L1 antibody labeled with .sup.89Zr can significantly and specifically localize to tumors. One may envisage a scenario where the anti-PD-L1 antibody is used in the selection of patients with PD-L1 positive tumors for subsequent treatment with inhibitors of the PD-1/PD-L1 signaling axis.
Example 8: Scaled-Up Manufacturing Process for Producing DFO-Anti-PD-L1 Antibody Conjugates
[0236] This example details the scaled-up manufacturing process for preparing the anti-PD-L1 antibody to be suitable for radiolabeling by attaching p-SCN-bn-Deferoxamine (DFO) to the anti-PD-L1 antibody (mAb, H4H8314N) described herein: (1) ultrafiltration and diafiltration (UFDF) processes prior to mAb conjugation removes excipients that inhibit the conjugation process; (2) following the pre-conjugation UFDF, conjugation of the mAb with p-SCN-Bn-deferoxamine is performed to produce DFO-mAb conjugates; and (3) a post-conjugation UFDF to remove residual salts provides a suitable concentration, excipient level, and pH of the conjugated monoclonal antibody. The resulting DFO-mAb conjugates are then provided in a buffered state with improved stability for subsequent formulation.
(1) Pre-Conjugation Ultrafiltration and Diafiltration (UFDF)
[0237] 100 g anti-PD-L1 antibody was buffer exchanged into a 5 mM acetate buffer solution having a pH of 5.50 using a Sius Prostream (TangenX Technology Corporation) membrane (membrane capacity of .ltoreq.500 g/m.sup.2) to remove residual salts prior to conjugation. The process volume was reduced to further concentrate the antibody, then the antibody was sterile filtered using a Sartopore 2 (Sartorius) membrane having a 0.45/0.2 .mu.m (heterogeneous PES double layer) or equivalent pore size. The acetate buffer temperature was kept at a target temperature of 20.+-.5.degree. C. The solutions were well mixed.
(2) Conjugation
[0238] The concentrated and filtered antibody (20 g) was transferred into a conjugation vessel containing an amine free carbonate buffer system (56 mM Carbonate, 167 mM Sodium Chloride, pH 9.40) resulting in negligible levels of residual acetate. DFO (25 mM p-SCN-Bn-Deferoxamine) was solubilized in DMSO and added to the conjugation vessel, along with additional DMSO such that the DMSO was present in a final amount of 5%. DFO was added in molar excess at a ratio of 4.5:1 DFO to mAb. The total reaction volume equaled 2.0 L. The buffer system was mixed throughout the addition of the reaction ingredients and throughout the reaction time.
[0239] The reaction temperature was controlled for specific time by using an equation which relates temperature to reaction time. In this instance, the reaction temperature was held at 18.degree. C. for 120 minutes. The reaction was quenched by the addition of 2M acetic acid (23 mL/L), resulting in the solution having a pH of 6.
(3) Post-Conjugation UFDF
[0240] After the conjugation step, the quenched DFO-mAb conjugation solution was buffer exchanged into histidine buffer (10 mM Histidine, pH 5.50 with 0.0005% (w/v) super refined polysorbate 80 added as a shear protectant) to remove residual process salts, DMSO, and unreacted DFO. Once diafiltered, the solution was then concentrated and subsequently formulated. The histidine buffer was selected for long term storage of protein at -80.degree. C. The same Sius Prostream membrane mentioned in step (1) was used in the final UFDF step. The resulting concentrated DFO-mAb conjugate solution was sterile filtered using the Sartopore 2 filter mentioned above.
[0241] UV-DAR (target of 1.5) and protein concentration determination was performed as described in Example 2.
TABLE-US-00017 TABLE 17 Molar Extinction Coefficients and Molecular Weight MW 280 252 Antibody (g mol.sup.-1) (L g.sup.-1cm.sup.-1) (L g.sup.-1cm.sup.-1) H4H8314N 144984 211480 80172
Example 9: Predicted Whole Body and Tissue Exposure of Radioactivity in Human Subjects to be Given an IV Dose of .sup.89Zr-DFO-Anti-PD-L1 Antibody Conjugate
[0242] The purpose of the following experiment was to estimate the predicted whole body and tissue exposures to radioactivity in human subjects due to an intravenous (IV) dose of .sup.89Zr-DFO-anti-PD-L1 antibody conjugate. The exemplary anti-PD-L1 antibody used in the radiolabeled conjugate was H4H8314N.
Characterization of Radioimmunoconjugates
[0243] Anti-PD-L1 immunoconjugate (DFO-Ab) and isotype control immunoconjugate (DFO-IgG4.sup.P Control) were radiolabeled and purified for use in in vivo imaging and biodistribution studies. SEC-HPLC analysis and a MC38/mPD-L1.sup.-/-/hPD-L1 (murine MC38 colon adenocarcinoma cells engineered to knock out mouse PD-L1 and stably express human PD-L1) cell-based in vitro assay were performed to characterize the resultant radioimmunoconjugates.
Monomeric and Radiochemical Purity
[0244] SEC-HPLC using UV- and .gamma.-emission detectors was performed to assess monomeric and radiochemical purity. Results for radioimmunoconjugate preparations of .sup.89Zr-DFO-anti-PD-L1 antibody conjugate and of isotype control radioimmunoconjugate .sup.89Zr-DFO-IgG4.sup.P are shown in FIG. 8.
[0245] Analysis of chromatograms for absorption at 280 nm was performed to evaluate the relative amounts of high molecular weight (HMW) and monomeric protein in the radioimmunoconjugate preparations. As summarized in Table 18, the monomeric peaks (a readout of monomeric purity) constitute 99.6, 99.2, and 98.6%, respectively, of the total protein peak area for preparations of .sup.89Zr-DFO-anti-PD-L1 antibody conjugate and isotype control .sup.89Zr-DFO-IgG4.sup.P; low levels of HMW species (0.4, 0.8, and 1.4%, respectively) were also detected. Low molecular weight (LMW) species were not observed for any of the tested samples.
[0246] Analysis of radio-chromatograms for .gamma.-emission was performed to evaluate the relative amounts of .sup.89Zr incorporated into radioimmunoconjugates compared with unincorporated .sup.89Zr (such as free .sup.89Zr or .sup.89Zr chelated with free DFO-derivatives). As summarized in Table 18, the peaks for unincorporated .sup.89Zr constitute.ltoreq.1.1% of the total .gamma.-emission peak area, while the combined peaks for radiolabeled monomeric and HMW species (a readout of radiochemical purity) constitute 98.9, 99.5, and 99.5%, respectively, of the total .gamma.-emission peak area for preparations of .sup.89Zr-DFO-anti-PD-L1 antibody conjugate and isotype control .sup.89Zr-DFO-IgG4.sup.P.
TABLE-US-00018 TABLE 18 Summary of SEC-HPLC Data Approximate Peak Area (%) Retention UV- Radio- Peak Time Chroma- Chroma- Number Species (min) togram togram .sup.89Zr-DFO-H4H8314N Study 1 1 HMW 13 0.4 1.1 2 Monomer 16 99.6 97.8 3 Unincorporated .sup.89Zr 26 n/a 1.1 .sup.89Zr-DFO-H4H8314N Study 2 1 HMW 14 0.8 1.3 2 Monomer 16 99.2 98.2 3 Unincorporated .sup.89Zr 26 n/a 0.5 .sup.89Zr-DFO-IgG4.sup.P Control 1 HMW 13 1.4 1.5 2 Monomer 16 98.6 98.0 3 Unincorporated .sup.89Zr 26 n/a 0.5
Numerical values for SEC-HPLC analysis graphically represented in FIG. 8. UV-chromatogram indicated the chromatogram for absorption at 280 nm and radio-chromatogram indicates the chromatogram for intensity of .gamma.-emission. HMW: high molecular weight; n/a: not applicable.
Immunoreactivity
[0247] The immunoreactivity, a measure of the percent of radiolabeled, conjugated antibody that is capable of binding its antigen, was determined by incubating .sup.89Zr-DFO-anti-PD-L1 antibody conjugate with MC38/mPD-L1.sup.-/-/hPD-L1 cells. The 2 tested lots of .sup.89Zr-DFO-anti-PD-L1 antibody conjugate demonstrated 84.5 and 88.8% immunoreactivity on MC38/mPD-L1.sup.-/-/hPD-L1 cells (Table 19). Background, nonspecific immunoreactivity of 8.8% was observed for the isotype control radioimmunoconjugate.
TABLE-US-00019 TABLE 19 Immunoreactivity of .sup.89Zr labeled anti-PD-L1 DFO-antibody conjugate and isotype control .sup.89Zr-DFO-IgG4.sup.P Radioimmunoconjugate Immunoreactivity .sup.89Zr-DFO-anti-PD-L1 antibody 84.5% conjugate (lot 1) .sup.89Zr-DFO-anti-PD-L1 antibody 88.8% conjugate (lot 2) isotype control .sup.89Zr-DFO-IgG4.sup.P 8.8%
[0248] In conclusion, two separate lots of .sup.89Zr-DFO-anti-PD-L1 antibody conjugate showed high immunoreactivity, percentage of monomer, and radiochemical purity.
.sup.89Zr-DFO-Anti-PD-L1 Biodistribution in Mice
[0249] This experiment evaluated the biodistribution of the anti-human PD-L1 radioimmunoconjugate, .sup.89Zr-DFO-anti-PD-L1 antibody conjugate, over time following administration of a single 50 .mu.Ci (1 mg/kg) intravenous (IV) dose to PD-L1/PD-1-humanized mice (PD-1hu/huPD-L1hu/hu). Since H4H8314N does not bind mouse PD-L1, the portion of the mouse PD-L1 gene encoding the PD-L1 ectodomain was replaced by the corresponding human sequence in PD-1hu/hu-PD-L1hu/hu mice. In this strain, the ectodomain of mouse PD-1 was similarly humanized. These mice were not subjected to immune/inflammatory challenge, and are therefore expected to have unstimulated levels of PD-L1 expression on immune cells. Two groups of 8 animals each were sacrificed 6 days (144 hours) or 10 days (240 hours) post dosing, blood was collected and the following tissues were harvested: heart, lungs, liver, spleen, kidneys, stomach, small intestine, caecum, large intestine, bone (femur), thymus, muscle, bladder, and brain. The percentage of radioactivity of the total injected dose (% ID) localized to specific tissues or blood was determined and reported as average % ID per gram (% ID/g) of tissue. In advance of sacrifice, immuno-PET/computed tomography (CT) images were acquired 1, 24, 48, 72, 144, 192 (10-day group only), and 240 (10-day group only) hours post dosing from the same animals.
[0250] Relative to .sup.89Zr levels in blood, uptake of .sup.89Zr-DFO-anti-PD-L1 antibody conjugate into specific tissues was negligible throughout the 10-day study period, as evaluated by ex vivo tissue analysis (Table 20 and FIG. 9) and in vivo imaging. Compared with blood (9.4.+-.2.2% ID/g), all harvested tissues, with the exception of spleen, demonstrated lower .sup.89Zr levels (.ltoreq.6.7% ID/g) on day 6 post dosing. A small degree of target-mediated .sup.89Zr-DFO-anti-PD-L1 antibody conjugate uptake (10.2.+-.1.9% ID/g) was observed in the spleen, in agreement with PD-L1 expression on splenocytes, as demonstrated by flow cytometry. At 10 days post-dosing, .sup.89Zr levels in blood had decreased 7.8-fold relative to day 6 post dosing, suggesting a mouse-anti-human antibody (MAHA) response affecting .sup.89Zr-DFO-anti-PD-L1 antibody conjugate levels. This observed MAHA response is likely due to the fact that the target, PD-L1, is expressed on antigen-presenting cells (Francisco, 2010), leading to the presentation of the human antibody to the mouse immune system and subsequent MAHA formation. In parallel, .sup.89Zr levels in the liver were 4.1-fold increased on day 10 compared with day 6 post dosing, possibly as a result of MAHA/.sup.89Zr-DFO-anti-PD-L1 antibody conjugate immune complex (IC) formation and subsequent liver-mediated IC clearance (Rojko, 2014). Whole animal in vivo PET imaging did not uncover marked tissue-specific uptake of .sup.89Zr-DFO-anti-PD-L1 antibody conjugate beyond a low signal for spleen and the MAHA-mediated accumulation in the liver described above.
[0251] In summary, marked target-mediated uptake of .sup.89Zr-DFO-anti-PD-L1 antibody conjugate into specific tissues above .sup.89Zr levels in blood was not observed over a 6-day period in PD-L1/PD-1-humanized mice administered a single IV dose of 1 mg/kg (50 .mu.Ci) of .sup.89Zr-DFO-anti-PD-L1 antibody conjugate with the exception of the spleen, where a small degree of target-mediated uptake was observed in agreement with the demonstrated expression of PD-L1 on splenocytes. Data collected beyond day 6 until the end of the study on day 10 post dosing were affected by a MAHA response.
TABLE-US-00020 TABLE 20 Average Ex Vivo Biodistribution Data .sup.89Zr Levels on Day 6 .sup.89Zr Levels on Day 10 post Dosing (% ID/g) post Dosing (% ID/g) Tissue Average SD Average SD Blood 9.4 2.2 1.2 1.4 Heart 3.1 0.6 1.2 0.4 Lungs 5.9 0.7 2.6 0.7 Liver 4.9 1.9 20.2 7.8 Spleen 10.2 1.9 12.1 3.0 Kidneys 5.3 1.1 3.9 1.3 Stomach 0.9 0.3 0.4 0.1 Small Intestine 1.5 0.3 0.9 0.1 Caecum 1.0 0.2 0.6 0.2 Large Intestine 1.4 0.3 0.7 0.2 Bone (Femur) 6.3 2.1 6.9 1.4 Thymus 6.7 1.6 5.3 1.1 Muscle 0.9 0.1 0.5 0.1 Bladder 4.3 2.1 1.7 0.9 Brain 0.4 0.1 0.2 0.1 Abbreviation: % ID/g = Percent injected dose per gram (of tissue)
Estimates of Whole Body and Tissue Exposures to Radioactivity in Humans
[0252] This experiment used PET/CT image data for four PD-1/PD-L1-humanized male mice and four PD-1/PD-L1-humanized female mice imaged at 1, 24, 48, 72, 144, 192, and 240 hours following single IV administration of 50 .mu.Ci (1 mg/kg) of .sup.89Zr-DFO-anti-PD-L1 antibody conjugate. The data generated by administration of this clinically relevant dose was used in calculating estimates of human exposure to radioactivity. Tissue concentration data was determined using volume of interest (V01) analysis.
[0253] For radiation dosimetry estimation, the mean residence time was determined for the following regions: brain, stomach contents, heart contents, kidneys, liver, lungs, muscle, red marrow, spleen, bladder contents, and remainder of body. These mean residence time values were used as an input into the OLINDA/EXM 1.1 software program to estimate the mean absorbed tissue doses and effective dose in humans.
[0254] The effective human dose for .sup.89Zr-DFO-anti-PD-L1 antibody conjugate was estimated to be 0.513 mSv/MBq (millisievert/megabecquerel) in the adult male and 0.622 mSv/MBq in the adult female. The organs predicted to have the highest absorbed dose in humans were the spleen and liver. The estimated absorbed dose in the spleen was 0.856 mSv/MBq in the adult male and 1.12 mSv/MBq in the adult female. The estimated absorbed dose in the liver was 0.764 mSv/MBq in the adult male and 0.974 mSv/MBq in the adult female.
[0255] Average decay-corrected percent of the injected dose per mL (DC % ID/mL) values for male and female mice (n=4 male, n=4 female) for each VOI are summarized in Table 21.
TABLE-US-00021 TABLE 21 Biodistribution Data Average Decay-corrected Percent Injected Dose Per mL (DC % ID/mL) .+-. SD Time (h) 1 24 48 Sex Female Male Female Male Female Brain 1.365 .+-. 0.115 1.190 .+-. 0.050 0.903 .+-. 0.115 0.538 .+-. 0.071 0.640 .+-. 0.079 Lungs 12.503 .+-. 1.146 12.498 .+-. 0.414 8.293 .+-. 0.635 7.155 .+-. 1.175 6.715 .+-. 0.370 Liver 12.298 .+-. 0.664 12.078 .+-. 0.372 9.058 .+-. 0.793 7.200 .+-. 0.499 8.113 .+-. 0.969 Heart 27.688 .+-. 1.942 25.695 .+-. 0.934 15.685 .+-. 1.223 13.323 .+-. 1.133 12.088 .+-. 0.883 Kidneys 11.430 .+-. 0.387 12.100 .+-. 0.872 7.345 .+-. 0.322 6.783 .+-. 0.811 6.418 .+-. 0.761 Spleen 15.263 .+-. 2.166 15.860 .+-. 0.974 14.135 .+-. 2.010 11.265 .+-. 1.706 13.675 .+-. 2.195 Bladder 6.045 .+-. 3.910 9.688 .+-. 4.991 1.653 .+-. 0.107 1.820 .+-. 0.283 1.443 .+-. 0.205 Muscle 1.608 .+-. 0.182 1.435 .+-. 0.198 2.608 .+-. 0.196 1.780 .+-. 0.137 2.368 .+-. 0.259 Stomach 3.238 .+-. 1.063 3.978 .+-. 0.632 2.875 .+-. 0.921 3.073 .+-. 0.566 2.478 .+-. 0.296 Bone 3.683 .+-. 1.418 3.023 .+-. 0.244 3.310 .+-. 0.330 2.738 .+-. 0.171 4.600 .+-. 0.511 Average Decay-corrected Percent Injected Dose Per mL (DC % ID/mL) .+-. SD Time (h) 48 72 144 Sex Male Female Male Female Male Brain 0.548 .+-. 0.218 0.685 .+-. 0.096 0.623 .+-. 0.224 0.465 .+-. 0.231 0.398 .+-. 0.073 Lungs 5.888 .+-. 0.990 6.060 .+-. 0.708 5.558 .+-. 0.385 4.863 .+-. 0.316 4.585 .+-. 0.339 Liver 6.125 .+-. 0.858 7.838 .+-. 0.932 6.203 .+-. 0.483 9.423 .+-. 1.885 6.208 .+-. 1.428 Heart 10.25 .+-. 1.335 11.740 .+-. 1.553 9.915 .+-. 0.171 8.140 .+-. 0.598 7.463 .+-. 0.768 Kidneys 5.565 .+-. 0.680 6.475 .+-. 0.493 5.568 .+-. 0.550 5.643 .+-. 0.222 4.815 .+-. 0.450 Spleen 9.388 .+-. 1.389 13.655 .+-. 3.606 9.920 .+-. 1.414 15.105 .+-. 2.959 10.303 .+-. 1.102 Bladder 1.403 .+-. 0.160 1.318 .+-. 0.108 1.710 .+-. 0.346 1.115 .+-. 0.224 1.293 .+-. 0.430 Muscle 1.955 .+-. 0.339 2.408 .+-. 0.181 2.148 .+-. 0.176 2.095 .+-. 0.168 1.918 .+-. 0.144 Stomach 2.238 .+-. 0.487 2.260 .+-. 0.306 2.233 .+-. 0.491 2.380 .+-. 0.405 1.665 .+-. 0.148 Bone 3.493 .+-. 0.716 4.850 .+-. 1.292 4.658 .+-. 1.399 8.993 .+-. 1.057 7.635 .+-. 0.872
[0256] Estimated human mean residence time (MRT) values are provided in Table 22 for each of the source organs. MRT in the remainder of the body was obtained by subtracting the sum of all source organ residence times from the reciprocal of the .sup.89Zr decay constant (Huang et al., Biodistribution, toxicity and radiation dosimetry studies of the serotonin transporter radioligand 4-[18F]-ADAM in rats and monkeys. Eur J Nucl Med Mol Imaging, 2010; 37: 545-555). This represents a conservative estimation of the cumulative tissue radioactivity.
TABLE-US-00022 TABLE 22 Human Mean Residence Times (h) Physical Decay.sup.1 Biexponential Fit.sup.2 Organ/Tissue Female Male Female Male Brain 0.398 0.364 0.372 0.344 Stomach Contents 0.511 0.476 0.492 0.480 Heart Contents 2.433 2.279 2.290 2.154 Kidneys 0.868 0.818 0.832 0.794 Liver 5.902 5.919 8.240 5.938 Lungs 2.508 2.772 2.411 2.642 Muscle 17.635 23.677 13.348 17.182 Red Marrow 2.777 2.024 2.613 1.913 Spleen 0.996 0.871 1.053 0.910 Bladder Contents 0.299 0.491 0.315 0.405 Remainder of Body 78.794 73.430 81.157 80.361 .sup.1Mean residence time calculated assuming only physical decay following day 6 time point .sup.2Mean residence time calculated from a biexponential fit of the data
[0257] The estimated absorbed tissue doses for all target organs for the OLINDA/EXM 1.1 adult male and adult female phantoms are provided in Table 23. The effective dose, defined by the International Commission on Radiological Protection (ICRP) (International Commission on Radiological Protection. 1990 Recommendations of the International Commission on Radiological Protection. ICRP Publication 60, Pergamon Press, New York, 1991) is a quantity that is calculated by multiplying the absorbed dose for a given organ by a stochastic risk weighting factor and adding the weighted doses together. Estimated effective doses are provided at the end of Table 23. These values represent a conservative estimation of radioactive absorbed doses.
TABLE-US-00023 TABLE 23 Estimated Human Tissue Absorbed Doses and Effective Dose Physical Decay.sup.1 Biexponential Fit.sup.2 Adult Adult Adult Adult Male Female Male Female Organ/Tissue (mSv/MBq) (mSv/MBq) (mSv/MBq) (mSv/MBq) Adrenals 0.561 0.702 0.567 0.726 Brain 0.179 0.237 0.182 0.234 Breasts 0.366 0.459 0.379 0.466 Gallbladder Wall 0.601 0.692 0.610 0.751 LLI Wall 0.519 0.652 0.530 0.651 Small Intestine 0.563 0.600 0.582 0.605 Stomach Wall 0.575 0.714 0.584 0.718 ULI Wall 0.553 0.685 0.571 0.700 Heart Wall 0.789 0.973 0.781 0.964 Kidney 0.650 0.773 0.641 0.774 Liver 0.764 0.974 0.764 1.220 Lungs 0.575 0.705 0.561 0.700 Muscle 0.396 0.481 0.381 0.464 Ovaries 0.533 0.645 0.542 0.642 Pancreas 0.597 0.743 0.606 0.765 Red Marrow 0.480 0.591 0.483 0.587 Osteogenic Cells 0.604 0.777 0.625 0.779 Skin 0.291 0.373 0.297 0.374 Spleen 0.856 1.120 0.876 1.160 Testes 0.399 NA 0.407 NA Thymus 0.481 0.605 0.484 0.601 Thyroid 0.417 0.484 0.423 0.480 Urinary Bladder 0.580 0.496 0.559 0.494 Wall Uterus 0.545 0.638 0.554 0.636 Total Body 0.440 0.550 0.440 0.554 Effective Dose 0.513 0.622 0.516 0.625 .sup.1Absorbed doses calculated from MRT assuming only physical decay following day 6 time point .sup.2Absorbed doses calculated from MRT with a biexponential fit of the data Abbreviations: LLI = lower large intestine, ULI = upper large intestine, NA = not applicable
[0258] The estimated human tissue absorbed doses and effective human dose (Table 23) from the physical decay and the biexponential fit methods were similar. The physical decay method was selected to produce the final set of estimated human tissue absorbed doses and effective dose due to the apparent MAHA response in this murine model. Therefore, the effective human dose for .sup.89Zr-DFO-anti-PD-L1 antibody conjugate was estimated to be 0.513 mSv/MBq in the adult male and 0.622 mSv/MBq in the adult female. The organs predicted to have the highest absorbed dose in humans are the spleen and liver. The estimated absorbed dose in the spleen was 0.856 mSv/MBq in the adult male and 1.12 mSv/MBq in the adult female. The estimated absorbed dose in the liver was 0.764 mSv/MBq in the adult male and 0.974 mSv/MBq in the adult female.
Example 10: ImmunoPET Imaging of PD-L1 in Tumors Using an .sup.89Zr-DFO-Anti-PD-L1 Antibody Conjugate in Patients with Advanced Thoracic Malignancies
[0259] The primary objective of this study is to determine the safety and tolerability of .sup.89Zr-DFO-anti-PD-L1 antibody conjugate, in which the anti-PD-L1 antibody used in the radiolabeled conjugate is H4H8314N. The secondary objectives of the study are: [0260] Study part A only: To establish adequate mass dose of .sup.89Zr-DFO-anti-PD-L1 antibody conjugate and optimal post-infusion imaging time, as assessed by imaging and blood draw after tracer infusion. [0261] Study part B only: To establish test/re-test reliability of PET measures as assessed on two separate tracer infusions at optimal mass dose and imaging time point as determined in Part A. [0262] To characterize the pharmacokinetic (PK) profile of .sup.89Zr-DFO-anti-PD-L1 antibody conjugate based on tracer plasma activity concentration.
[0263] This is an open label, 2-part study designed to evaluate the safety and tolerability of .sup.89Zr-DFO-anti-PD-L1. Study Part A will establish an adequate mass dose and activity dose of .sup.89Zr-DFO-anti-PD-L1 and an optimal post-infusion imaging time. Test/re-test variability of .sup.89Zr-DFO-anti-PD-L1 will be evaluated in Part B.
[0264] All patients will undergo screening procedures. Patients who meet the eligibility criteria will undergo .sup.18F-fluorodeoxyglucose (.sup.18F-FDG) PET/computed tomography (CT) and diagnostic CT scans to assess lesion viability, location, and dimensions. These scans will not be required if adequate quality images are available that were acquired within 28 days of the expected first dose of .sup.89Zr-DFO-anti-PD-L1.
Part A
[0265] Three sequential dose cohorts are planned to be treated open-label with .sup.89Zr-DFO-anti-PD-L1 at 5 mg, 10 mg, or 20 mg.
[0266] After infusion with .sup.89Zr-DFO-anti-PD-L1, patients will undergo .sup.89Zr-DFO-anti-PD-L1 PET/CT scans on day 1, day 4.+-.1 and day 7.+-.1. Additional imaging may be performed up to day 10. Patients will undergo safety assessments and provide samples for hematology, chemistry, immune safety assays, pharmacokinetics, anti-drug antibody analysis, and biomarker analysis.
[0267] Patients will continue to undergo safety evaluations, including physical examination, vital signs, and documentation of Adverse Events (AEs), up to day 21 after the infusion of the .sup.89Zr-DFO-anti-PD-L1 tracer.
[0268] Dose escalation decisions to identify an adequate dose will be informed by safety and tolerability data and by evaluation of immune-positron emission tomography (iPET) positivity and tracer plasma activity concentration, as described below.
Dose Cohorts in Part A
[0269] Up to 3 ascending mass dose cohorts are planned. For each mass dose cohort, an initial 2 patients will be dosed, with at a minimum 48-hour interval between the dosing of each patient. Upon completion of the day 7.+-.1 day PET/CT scan for the second patient at a given mass dose, all available imaging, tracer plasma activity concentration, clinical dosimetry, and safety data will be reviewed. Based upon this review, a decision will be made to: [0270] Expand the cohort 6 patients, if there is tumor uptake positivity/tumor localization in at least 1 patient, as defined by a tumor-to-blood ratio>1 [0271] Ascend to the next mass dose cohort if there is inadequate tumor uptake and plasma tracer activity concentration, with adequate defined by blood standardized uptake value (SUV) range of 1 to 5 at the optimum imaging time point [0272] Proceed with the next mass dose cohort at a lower mass dose, based on inadequate tumor uptake and adequate plasma tracer activity concentration.
[0273] If tumor localization is inadequate in at least 2 patients at all three proposed mass dose levels, and this is determined to be due to low image signal-to-noise, the activity dose will be increased up to a maximum of 185 MBq for further expansion of previously tested mass dose cohorts.
Part B
[0274] Study Part B will begin once an adequate .sup.89Zr-DFO-anti-PD-L1 dose and an optimal imaging time have been determined in Part A. On day 1 of Part B, patients will receive the tracer mass dose. Subsequent to receiving the tracer, patients will undergo a scan at the optimal time as identified in Part A. Patients in Part B will receive a second tracer dose and scan after an inter-dose interval of 14 to 28 days. The actual timing of the second tracer dose after the interval will be determined based on results from Part A.
[0275] Patients will undergo safety assessments, including physical examination, vital signs, and documentation of adverse events (AEs) during and after visits where .sup.89Zr-DFO-anti-PD-L1 tracer is administered. During these visits, patients will provide samples for PK, hematology, chemistry, and immune safety assays.
[0276] For both Part A and Part B, patients will continue to undergo safety evaluations, including physical examination, vital signs, and documentation of AEs, up to 21 days after the last infusion of the .sup.89Zr-DFO-anti-PD-L1 tracer.
Study Duration
[0277] For Part A, patients will have a screening period of up to 28 days (4 weeks) and a follow-up period of up to 21 days (approximately 3 weeks) after infusion of the tracer dose. The duration of study Part A is approximately 7 weeks, including the screening period.
[0278] For Part B, patients will have a screening period of up to 28 days (4 weeks), an inter-infusion interval of up to 28 days (4 weeks), and a 21-day (3 week) safety follow-up period that includes the second scan period. The total duration of the study for each patient will be up to 11 weeks, including the screening period.
[0279] The end of study for this study is defined as the last visit of the last patient.
[0280] For study Part A, 3 sequential dose levels of up to 6 patients each are planned per cohort, with 3 cohorts planned, for a total of up to 18 patients. For study Part B, up to 10 patients will be enrolled. Enrollment of a maximum of 28 patients in a single study site is planned for the entire study.
Patient Target Population
[0281] The target population will consist of patients 18 years of age or older with advanced thoracic malignancies and PD-L 1 IHC score on a diagnostic or subsequent biopsy of 1% (positive PD-L1 IHC score by 22C3 PharmDx assay, Dako North America Inc.). [0282] For Part A, the thoracic malignancies will be limited to NSCLC, gastro-esophageal junction adenocarcinoma, and gastric cancer, with PD-L1 score of .gtoreq.1% by IHC. [0283] For Part B, all patients with advanced thoracic malignancies and a PD-L1 score of .gtoreq.1% by IHC will be eligible. Patients must also have stable disease as per RECIST 1.1 between the two most recent imaging studies.
[0284] All patients requiring therapy should be on standard of care therapy.
Treatment
[0285] .sup.89Zr-DFO-anti-PD-L1, a radioimmunoconjugate formed by covalently conjugating bifunctional chelator (p-SCN-Bn-DFO) to H4H8314N (anti-PD-L1 monoclonal antibody) and radiolabeling this compound with .sup.89Zr. .sup.89Zr-DFO-anti-PD-L1 is supplied in an aqueous buffered vehicle.
[0286] For Part A, .sup.89Zr-DFO-anti-PD-L1 will be administered IV on day 1 (baseline). For Part B, .sup.89Zr-DFO-anti-PD-L1 will be administered IV on day 1 and day 7.+-.3. Actual timing of the second dose in Part B will be determined from results in Part A.
[0287] The .sup.89Zr-DFO-anti-PD-L1 tracer will be administered at a dose level well below the estimated cumulative exposure levels in humans based on PK models and lower than the levels at which currently available anti-PD-1 agents are used for anti-cancer treatment. This study will exclude patients who are currently treated with anti-PD-L1 to avoid competition for target.
Endpoints
[0288] The primary endpoint in the study is the incidence and severity of Treatment-emergent adverse events (TEAEs) through day 21 of the last dose of tracer infusion in patients with thoracic malignancies dosed with .sup.89Zr-DFO-anti-PD-L1.
[0289] For Part A only, the study will establish an adequate mass dose and activity dose of .sup.89Zr-DFO-anti-PD-L1 and optimal post-infusion imaging time, and the following will be determined via blood drawing and imaging at day 1, 4, and 7 after tracer infusion: [0290] Standardized uptake value of .sup.89Zr-DFO-anti-PD-L1 in the blood pool, with subsequent calculation of tumor-to-blood ratios at the time of imaging [0291] Clinical dosimetry based on the absorbed dose and effective tissue radiation, as calculated from PET image acquisition data and tracer activity concentration in blood [0292] Standardized Uptake Values (SUVs) across the tumor regions of interest (ROIs) [0293] Maximal SUVs (SUVmax) within tumor ROIs [0294] Plasma tracer activity concentration, expressed as SUV, with calculation of area under the curve through day 7 (AUC.sub.0-7)
[0295] For Part B only, the study will establish the test/re-test reliability of .sup.89Zr-DFO-anti-PD-L1 PET measures, and the following will be determined from measures of 2 separate tracer infusions at an adequate mass dose and optimal imaging time points, as determined from Part A: [0296] Blood pool SUV with subsequent calculation of tumor-to-blood ratio [0297] SUVs across the tumor ROIs [0298] SUVmax within the tumor ROIs [0299] Biodistribution of .sup.89Zr-DFO-anti-PD-L1
[0300] The resulting data will be indicative of the safety and tolerability of .sup.89Zr-DFO-anti-PD-L1 in humans.
[0301] The embodiments and examples described above are intended to be merely illustrative and non-limiting. Those skilled in the art will recognize or will be able to ascertain using no more than routine experimentation, numerous equivalents of specific compounds, materials and procedures. All such equivalents are considered to be within the scope and are encompassed by the appended claims.
Sequence CWU 1
1
3441363DNAArtificial Sequencesynthetic 1gaggtgcagc tggtggagtc
tgggggaggc ttggtccagc ctggggggtc cctgagactc 60tcctgtgcag cctctggatt
cacctttagt aggttttgga tgagctgggt ccgccaggct 120ccagggaagg
ggctggagtg ggtggccaac ataaaccaag atggaactga gaaatactat
180gtggactctg tgaagggccg attcaccatc tccagagaca acgccaagaa
ctcactgtat 240ctgcaaatga acagcctgag agccggggac acggctgtgt
attactgtgc gaatacgtat 300tacgattttt ggagtggtca ctttgactac
tggggccagg gaaccctggt caccgtctcc 360tca 3632121PRTArtificial
Sequencesynthetic 2Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val
Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe
Thr Phe Ser Arg Phe 20 25 30Trp Met Ser Trp Val Arg Gln Ala Pro Gly
Lys Gly Leu Glu Trp Val 35 40 45Ala Asn Ile Asn Gln Asp Gly Thr Glu
Lys Tyr Tyr Val Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg
Asp Asn Ala Lys Asn Ser Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu
Arg Ala Gly Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Asn Thr Tyr Tyr
Asp Phe Trp Ser Gly His Phe Asp Tyr Trp Gly 100 105 110Gln Gly Thr
Leu Val Thr Val Ser Ser 115 120324DNAArtificial Sequencesynthetic
3ggattcacct ttagtaggtt ttgg 2448PRTArtificial Sequencesynthetic
4Gly Phe Thr Phe Ser Arg Phe Trp1 5524DNAArtificial
Sequencesynthetic 5ataaaccaag atggaactga gaaa 2468PRTArtificial
Sequencesynthetic 6Ile Asn Gln Asp Gly Thr Glu Lys1
5742DNAArtificial Sequencesynthetic 7gcgaatacgt attacgattt
ttggagtggt cactttgact ac 42814PRTArtificial Sequencesynthetic 8Ala
Asn Thr Tyr Tyr Asp Phe Trp Ser Gly His Phe Asp Tyr1 5
109321DNAArtificial Sequencesynthetic 9gacatccaga tgacccagtc
tccttccacc ctgtctgcat ctgtaggaga cagagtcacc 60atcacttgtc gggccagtca
gagtattagt aattggttgg cctggtatca gcagaaacca 120gggaaagccc
ctaagctcct gatctataag gcgtctagtt tagaaagtgg ggtcccatca
180aggttcagcg gcagtggatc tgggacagaa ttcactctca ccatcagcag
cctgcagcct 240gatgattttg caacttatta ctgccaacag tatcatagtt
attcgtacac ttttggccag 300gggaccaagc tggagatcaa a
32110107PRTArtificial Sequencesynthetic 10Asp Ile Gln Met Thr Gln
Ser Pro Ser Thr Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile
Thr Cys Arg Ala Ser Gln Ser Ile Ser Asn Trp 20 25 30Leu Ala Trp Tyr
Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Lys Ala
Ser Ser Leu Glu Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly
Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75
80Asp Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Tyr His Ser Tyr Ser Tyr
85 90 95Thr Phe Gly Gln Gly Thr Lys Leu Glu Ile Lys 100
1051118DNAArtificial Sequencesynthetic 11cagagtatta gtaattgg
18126PRTArtificial Sequencesynthetic 12Gln Ser Ile Ser Asn Trp1
5139DNAArtificial Sequencesynthetic 13aaggcgtct 9143PRTArtificial
Sequencesynthetic 14Lys Ala Ser11527DNAArtificial Sequencesynthetic
15caacagtatc atagttattc gtacact 27169PRTArtificial
Sequencesynthetic 16Gln Gln Tyr His Ser Tyr Ser Tyr Thr1
517363DNAArtificial Sequencesynthetic 17caggagcacc tggtggagtc
tgggggaggc gtggtccagc ctgggaggtc cctgagactc 60tcctgtgaag cgtctggatt
caccttcagt aactttggca tgcactgggt ccgccaggct 120ccaggcaagg
ggctggagtg ggtggcagct ttatggtctg atggaagtaa taaatactat
180gcagactccg tgaagggtcg agtcaccatc tccagagaca attccaagaa
cacactgtat 240ctgcaaatga acagcctgag agccgaggac acggctgtct
attactgtgc gagagggaga 300ggagcccccg gtattccgat ttttgggtac
tggggccagg gaaccctggt caccgtctcc 360tca 36318121PRTArtificial
Sequencesynthetic 18Gln Glu His Leu Val Glu Ser Gly Gly Gly Val Val
Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Glu Ala Ser Gly Phe
Thr Phe Ser Asn Phe 20 25 30Gly Met His Trp Val Arg Gln Ala Pro Gly
Lys Gly Leu Glu Trp Val 35 40 45Ala Ala Leu Trp Ser Asp Gly Ser Asn
Lys Tyr Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Val Thr Ile Ser Arg
Asp Asn Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu
Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Gly Arg Gly
Ala Pro Gly Ile Pro Ile Phe Gly Tyr Trp Gly 100 105 110Gln Gly Thr
Leu Val Thr Val Ser Ser 115 1201924DNAArtificial Sequencesynthetic
19ggattcacct tcagtaactt tggc 24208PRTArtificial Sequencesynthetic
20Gly Phe Thr Phe Ser Asn Phe Gly1 52124DNAArtificial
Sequencesynthetic 21ttatggtctg atggaagtaa taaa 24228PRTArtificial
Sequencesynthetic 22Leu Trp Ser Asp Gly Ser Asn Lys1
52342DNAArtificial Sequencesynthetic 23gcgagaggga gaggagcccc
cggtattccg atttttgggt ac 422414PRTArtificial Sequencesynthetic
24Ala Arg Gly Arg Gly Ala Pro Gly Ile Pro Ile Phe Gly Tyr1 5
1025321DNAArtificial Sequencesynthetic 25gacatccaga tgacccagtc
tccatcctcc ctgtctgcat ctgttggaga cagagtcacc 60atcacttgcc gggcaagtca
gggcattaga aatgatttag gctggtatca gcagaaacca 120gggaaagccc
ctaagcgcct gatctatact gcatccagtt tgcaaagtgg ggtcccatca
180agattcagcg gcagtggatc tgggacagaa ttcactctca caatcagcag
cctacagcct 240gaagattttg caacttatta ctgtctacaa cataatagtt
accctctcac attcggcgga 300gggaccaagg tggcgatcaa a
32126107PRTArtificial Sequencesynthetic 26Asp Ile Gln Met Thr Gln
Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile
Thr Cys Arg Ala Ser Gln Gly Ile Arg Asn Asp 20 25 30Leu Gly Trp Tyr
Gln Gln Lys Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr Thr Ala
Ser Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly
Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75
80Glu Asp Phe Ala Thr Tyr Tyr Cys Leu Gln His Asn Ser Tyr Pro Leu
85 90 95Thr Phe Gly Gly Gly Thr Lys Val Ala Ile Lys 100
1052718DNAArtificial Sequencesynthetic 27cagggcatta gaaatgat
18286PRTArtificial Sequencesynthetic 28Gln Gly Ile Arg Asn Asp1
5299DNAArtificial Sequencesynthetic 29actgcatcc 9303PRTArtificial
Sequencesynthetic 30Thr Ala Ser13127DNAArtificial Sequencesynthetic
31ctacaacata atagttaccc tctcaca 27329PRTArtificial
Sequencesynthetic 32Leu Gln His Asn Ser Tyr Pro Leu Thr1
533390DNAArtificial Sequencesynthetic 33gaggtgcagc tggtggagtc
tgggggaggc ttggtaaagc ctggggggtc ccttagactc 60tcctgtgcag cctctggatt
cactttcagt aacgcctgga tgagctgggt ccgccaggct 120ccagggaagg
ggctggagtg ggttggccgt attaaaagga aaactgatgg tgggacaaca
180gactacgctg cacccgtgaa aggcagattc accatctcaa gagatgattc
aaaaaatacg 240ctgcatctgc aaatgaacag cctgaaaacc gaggacacag
ccgtgtatta ctgtaccaca 300gatgatattg tagttgtacc agctgttatg
agggaatact acttcggtat ggacgtctgg 360ggccaaggga ccacggtcac
cgtctcctca 39034130PRTArtificial Sequencesynthetic 34Glu Val Gln
Leu Val Glu Ser Gly Gly Gly Leu Val Lys Pro Gly Gly1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Asn Ala 20 25 30Trp
Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Gly Arg Ile Lys Arg Lys Thr Asp Gly Gly Thr Thr Asp Tyr Ala Ala
50 55 60Pro Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp Ser Lys Asn
Thr65 70 75 80Leu His Leu Gln Met Asn Ser Leu Lys Thr Glu Asp Thr
Ala Val Tyr 85 90 95Tyr Cys Thr Thr Asp Asp Ile Val Val Val Pro Ala
Val Met Arg Glu 100 105 110Tyr Tyr Phe Gly Met Asp Val Trp Gly Gln
Gly Thr Thr Val Thr Val 115 120 125Ser Ser 1303524DNAArtificial
Sequencesynthetic 35ggattcactt tcagtaacgc ctgg 24368PRTArtificial
Sequencesynthetic 36Gly Phe Thr Phe Ser Asn Ala Trp1
53730DNAArtificial Sequencesynthetic 37attaaaagga aaactgatgg
tgggacaaca 303810PRTArtificial Sequencesynthetic 38Ile Lys Arg Lys
Thr Asp Gly Gly Thr Thr1 5 103963DNAArtificial Sequencesynthetic
39accacagatg atattgtagt tgtaccagct gttatgaggg aatactactt cggtatggac
60gtc 634021PRTArtificial Sequencesynthetic 40Thr Thr Asp Asp Ile
Val Val Val Pro Ala Val Met Arg Glu Tyr Tyr1 5 10 15Phe Gly Met Asp
Val 2041321DNAArtificial Sequencesynthetic 41gacatccaga tgacccagtc
tccatcctcc ctgtctgcat ctgtaggaga cagagtcacc 60atcacttgcc ggacaagtca
gggcattaga aatgatttag gctggtatca gcagaaacca 120gggaaagccc
ctaagcgcct gatctatgct gcatccagtt tgcaaagtgg ggtcccatca
180aggttcagcg gcagtggatc tgggacagaa ttcactctca caatcagcag
cctgcagcct 240gaagattttg caacttatta ctgtctacag cataataatt
acccgtacac ttttggccag 300gggaccaagc tggagatcaa a
32142107PRTArtificial Sequencesynthetic 42Asp Ile Gln Met Thr Gln
Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile
Thr Cys Arg Thr Ser Gln Gly Ile Arg Asn Asp 20 25 30Leu Gly Trp Tyr
Gln Gln Lys Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr Ala Ala
Ser Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly
Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75
80Glu Asp Phe Ala Thr Tyr Tyr Cys Leu Gln His Asn Asn Tyr Pro Tyr
85 90 95Thr Phe Gly Gln Gly Thr Lys Leu Glu Ile Lys 100
1054318DNAArtificial Sequencesynthetic 43cagggcatta gaaatgat
18446PRTArtificial Sequencesynthetic 44Gln Gly Ile Arg Asn Asp1
5459DNAArtificial Sequencesynthetic 45gctgcatcc 9463PRTArtificial
Sequencesynthetic 46Ala Ala Ser14727DNAArtificial Sequencesynthetic
47ctacagcata ataattaccc gtacact 27489PRTArtificial
Sequencesynthetic 48Leu Gln His Asn Asn Tyr Pro Tyr Thr1
549363DNAArtificial Sequencesynthetic 49caggtgcaat tggtgcagtc
tggggcggag gtgaagaagc ctggggcctc agtgcaggtc 60tcctgcaagg cttctggata
ctccttcacc ggctactata tacactgggt gcgacaggcc 120cctggacaag
gacttgagtg gatgggatgg atcaacccta acagtggcac caaaaagtat
180gcacacaagt ttcagggcag ggtcaccatg accagggaca cgtccatcga
cacagcctac 240atgattttga gcagtctgat atccgacgac acggccgtgt
attactgtgc gagagatgag 300gactggaact ttgggagctg gttcgactcc
tggggccagg gaaccctggt caccgtctcc 360tca 36350121PRTArtificial
Sequencesynthetic 50Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Gln Val Ser Cys Lys Ala Ser Gly Tyr
Ser Phe Thr Gly Tyr 20 25 30Tyr Ile His Trp Val Arg Gln Ala Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Trp Ile Asn Pro Asn Ser Gly Thr
Lys Lys Tyr Ala His Lys Phe 50 55 60Gln Gly Arg Val Thr Met Thr Arg
Asp Thr Ser Ile Asp Thr Ala Tyr65 70 75 80Met Ile Leu Ser Ser Leu
Ile Ser Asp Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Glu Asp
Trp Asn Phe Gly Ser Trp Phe Asp Ser Trp Gly 100 105 110Gln Gly Thr
Leu Val Thr Val Ser Ser 115 1205124DNAArtificial Sequencesynthetic
51ggatactcct tcaccggcta ctat 24528PRTArtificial Sequencesynthetic
52Gly Tyr Ser Phe Thr Gly Tyr Tyr1 55324DNAArtificial
Sequencesynthetic 53atcaacccta acagtggcac caaa 24548PRTArtificial
Sequencesynthetic 54Ile Asn Pro Asn Ser Gly Thr Lys1
55542DNAArtificial Sequencesynthetic 55gcgagagatg aggactggaa
ctttgggagc tggttcgact cc 425614PRTArtificial Sequencesynthetic
56Ala Arg Asp Glu Asp Trp Asn Phe Gly Ser Trp Phe Asp Ser1 5
1057336DNAArtificial Sequencesynthetic 57gatattgtga tgacccagac
tccactctcc tcacctgtca cccttggaca gccggcctcc 60atctcctgca ggtctagtca
aaccctcgta cacggtgatg gaaacacgta cttgagttgg 120attcagcaga
ggccaggcca gcctccgaga ctcctcattt ataaggtttc taatcagttc
180tctggggtcc cagacagatt cagtggcagt ggggcaggga cagatttcac
actgaaaatc 240agcagggtgg aagctgagga tgtcgggctt tatttctgca
tgcaagctac acattttccg 300atcaccttcg gccaagggac acgactggag attaaa
33658112PRTArtificial Sequencesynthetic 58Asp Ile Val Met Thr Gln
Thr Pro Leu Ser Ser Pro Val Thr Leu Gly1 5 10 15Gln Pro Ala Ser Ile
Ser Cys Arg Ser Ser Gln Thr Leu Val His Gly 20 25 30Asp Gly Asn Thr
Tyr Leu Ser Trp Ile Gln Gln Arg Pro Gly Gln Pro 35 40 45Pro Arg Leu
Leu Ile Tyr Lys Val Ser Asn Gln Phe Ser Gly Val Pro 50 55 60Asp Arg
Phe Ser Gly Ser Gly Ala Gly Thr Asp Phe Thr Leu Lys Ile65 70 75
80Ser Arg Val Glu Ala Glu Asp Val Gly Leu Tyr Phe Cys Met Gln Ala
85 90 95Thr His Phe Pro Ile Thr Phe Gly Gln Gly Thr Arg Leu Glu Ile
Lys 100 105 1105933DNAArtificial Sequencesynthetic 59caaaccctcg
tacacggtga tggaaacacg tac 336011PRTArtificial Sequencesynthetic
60Gln Thr Leu Val His Gly Asp Gly Asn Thr Tyr1 5 10619DNAArtificial
Sequencesynthetic 61aaggtttct 9623PRTArtificial Sequencesynthetic
62Lys Val Ser16327DNAArtificial Sequencesynthetic 63atgcaagcta
cacattttcc gatcacc 27649PRTArtificial Sequencesynthetic 64Met Gln
Ala Thr His Phe Pro Ile Thr1 565363DNAArtificial Sequencesynthetic
65caggtacacc tggtgcagtc tggggctgag gtgaagaagc ctggggcctc agtgaaggtc
60tcctgcaagg cttctggata caccttcacc ggctactata tacattgggt gcgacaggcc
120cctggacacg ggcttgagtg gatgggatgg ctcaacccta atactggtac
cacaaagtat 180atacagaact ttcagggcag ggtcaccatg accagggaca
cgtccagcag cacagcctac 240atggagctga ccaggctgag atctgacgac
acggccgtgt attactgtgc gagagatgag 300gactggaatt atgggagctg
gttcgacacc tggggccagg gaaccctggt cacagtctcc 360tca
36366121PRTArtificial Sequencesynthetic 66Gln Val His Leu Val Gln
Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser
Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Tyr Ile His Trp
Val Arg Gln Ala Pro Gly His Gly Leu Glu Trp Met 35 40 45Gly Trp Leu
Asn Pro Asn Thr Gly Thr Thr Lys Tyr Ile Gln Asn Phe 50 55 60Gln Gly
Arg Val Thr Met Thr Arg Asp Thr Ser Ser Ser Thr Ala Tyr65 70 75
80Met Glu Leu Thr Arg Leu Arg Ser Asp Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Arg Asp Glu Asp Trp Asn Tyr Gly Ser Trp Phe Asp Thr Trp
Gly 100 105 110Gln Gly Thr Leu Val Thr Val Ser Ser 115
1206724DNAArtificial Sequencesynthetic 67ggatacacct tcaccggcta ctat
24688PRTArtificial Sequencesynthetic 68Gly Tyr Thr Phe Thr Gly Tyr
Tyr1 56924DNAArtificial Sequencesynthetic 69ctcaacccta atactggtac
caca 24708PRTArtificial Sequencesynthetic 70Leu Asn Pro Asn Thr Gly
Thr Thr1 57142DNAArtificial Sequencesynthetic 71gcgagagatg
aggactggaa ttatgggagc
tggttcgaca cc 427214PRTArtificial Sequencesynthetic 72Ala Arg Asp
Glu Asp Trp Asn Tyr Gly Ser Trp Phe Asp Thr1 5 1073336DNAArtificial
Sequencesynthetic 73gatattgtaa tgacccagac tccactctcc tcacctgtca
cccttggaca gccggcctcc 60atctcctgca ggtctagtcc aagcctcgta cacagtgatg
gaaacaccta cttgagttgg 120cttcagcaga ggccaggcca gcctccaaga
ctcctaattt ataagatttc taaccgattc 180tctggggtcc cagacagatt
cagtggcagt ggggcaggga cagatttcac gctgaaaatc 240agcagggtgg
aagctgagga tgtcggggtt tattactgca tgcaagctac acattttccg
300atcaccttcg gccaagggac acgactggag attaga 33674112PRTArtificial
Sequencesynthetic 74Asp Ile Val Met Thr Gln Thr Pro Leu Ser Ser Pro
Val Thr Leu Gly1 5 10 15Gln Pro Ala Ser Ile Ser Cys Arg Ser Ser Pro
Ser Leu Val His Ser 20 25 30Asp Gly Asn Thr Tyr Leu Ser Trp Leu Gln
Gln Arg Pro Gly Gln Pro 35 40 45Pro Arg Leu Leu Ile Tyr Lys Ile Ser
Asn Arg Phe Ser Gly Val Pro 50 55 60Asp Arg Phe Ser Gly Ser Gly Ala
Gly Thr Asp Phe Thr Leu Lys Ile65 70 75 80Ser Arg Val Glu Ala Glu
Asp Val Gly Val Tyr Tyr Cys Met Gln Ala 85 90 95Thr His Phe Pro Ile
Thr Phe Gly Gln Gly Thr Arg Leu Glu Ile Arg 100 105
1107533DNAArtificial Sequencesynthetic 75ccaagcctcg tacacagtga
tggaaacacc tac 337611PRTArtificial Sequencesynthetic 76Pro Ser Leu
Val His Ser Asp Gly Asn Thr Tyr1 5 10779DNAArtificial
Sequencesynthetic 77aagatttct 9783PRTArtificial Sequencesynthetic
78Lys Ile Ser17927DNAArtificial Sequencesynthetic 79atgcaagcta
cacattttcc gatcacc 27809PRTArtificial Sequencesynthetic 80Met Gln
Ala Thr His Phe Pro Ile Thr1 581360DNAArtificial Sequencesynthetic
81gaggtgcagc tggtggaatc tgggggaggt gtggtgcggc ctggggggtc cctgagactc
60tcctgtgcag cctctggatt cacttttgat gattatggca tgacctgggt ccgccaagct
120ccagggaggg gcctggaatg ggtctctggt attcattggc atggtaaacg
cacaggttat 180gcagactctg tgaagggccg attcaccata tccagagaca
acgccaagaa atccctgtat 240ctgcaaatga acagtctgaa aggcgaggac
acggccttgt atcattgtgt gaggggggga 300atgagtacag gggactggtt
cgacccctgg ggccagggaa ccctggtcat cgtctcctca 36082120PRTArtificial
Sequencesynthetic 82Glu Val Gln Leu Val Glu Ser Gly Gly Gly Val Val
Arg Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe
Thr Phe Asp Asp Tyr 20 25 30Gly Met Thr Trp Val Arg Gln Ala Pro Gly
Arg Gly Leu Glu Trp Val 35 40 45Ser Gly Ile His Trp His Gly Lys Arg
Thr Gly Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg
Asp Asn Ala Lys Lys Ser Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu
Lys Gly Glu Asp Thr Ala Leu Tyr His Cys 85 90 95Val Arg Gly Gly Met
Ser Thr Gly Asp Trp Phe Asp Pro Trp Gly Gln 100 105 110Gly Thr Leu
Val Ile Val Ser Ser 115 1208324DNAArtificial Sequencesynthetic
83ggattcactt ttgatgatta tggc 24848PRTArtificial Sequencesynthetic
84Gly Phe Thr Phe Asp Asp Tyr Gly1 58524DNAArtificial
Sequencesynthetic 85attcattggc atggtaaacg caca 24868PRTArtificial
Sequencesynthetic 86Ile His Trp His Gly Lys Arg Thr1
58739DNAArtificial Sequencesynthetic 87gtgagggggg gaatgagtac
aggggactgg ttcgacccc 398813PRTArtificial Sequencesynthetic 88Val
Arg Gly Gly Met Ser Thr Gly Asp Trp Phe Asp Pro1 5
1089324DNAArtificial Sequencesynthetic 89gacatccaga tgacccagtc
tccatcctcc ctgtctgcat ctctaggaga cagagtcacc 60atcacttgcc gggcaagtca
gagcattaac agttatttaa attggtatca gcagaaacca 120gggaaagccc
ctaaactcct gatctatgtt gcatccagtt tgcaaagtgg ggtcccatca
180aggttcagtg gcagtggatc tgggacagaa ttcactctca ccatcagcaa
tctgcaacct 240gaagattttg caacttacta ctgtcaacag agttacagta
cccctccgat caccttcggc 300caagggacac gactggagat taaa
32490108PRTArtificial Sequencesynthetic 90Asp Ile Gln Met Thr Gln
Ser Pro Ser Ser Leu Ser Ala Ser Leu Gly1 5 10 15Asp Arg Val Thr Ile
Thr Cys Arg Ala Ser Gln Ser Ile Asn Ser Tyr 20 25 30Leu Asn Trp Tyr
Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Val Ala
Ser Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly
Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser Asn Leu Gln Pro65 70 75
80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Ser Tyr Ser Thr Pro Pro
85 90 95Ile Thr Phe Gly Gln Gly Thr Arg Leu Glu Ile Lys 100
1059118DNAArtificial Sequencesynthetic 91cagagcatta acagttat
18926PRTArtificial Sequencesynthetic 92Gln Ser Ile Asn Ser Tyr1
5939DNAArtificial Sequencesynthetic 93gttgcatcc 9943PRTArtificial
Sequencesynthetic 94Val Ala Ser19530DNAArtificial Sequencesynthetic
95caacagagtt acagtacccc tccgatcacc 309610PRTArtificial
Sequencesynthetic 96Gln Gln Ser Tyr Ser Thr Pro Pro Ile Thr1 5
1097360DNAArtificial Sequencesynthetic 97gaggtgcagc tggtggagtc
tgggggaggt gtggtacggc cgggggggtc cctgagactc 60tcctgtgcag cctctggatt
cacctttgat gattatggca tgacctgggt ccgccaagtt 120ccagggaagg
ggctggagtg ggtctctggt attcattgga gtggtagaag cacaggttat
180gcagactctg tgaagggccg attcaccatc tccagagaca acgccaagaa
ctccctgtat 240ctgcaaatga acagtctgag agccgaggac acggccttgt
attactgtgc gaggggggga 300atgagtacgg gggactggtt cgacccctgg
ggccagggaa ccctggtcac cgtctcctca 36098120PRTArtificial
Sequencesynthetic 98Glu Val Gln Leu Val Glu Ser Gly Gly Gly Val Val
Arg Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe
Thr Phe Asp Asp Tyr 20 25 30Gly Met Thr Trp Val Arg Gln Val Pro Gly
Lys Gly Leu Glu Trp Val 35 40 45Ser Gly Ile His Trp Ser Gly Arg Ser
Thr Gly Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg
Asp Asn Ala Lys Asn Ser Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu
Arg Ala Glu Asp Thr Ala Leu Tyr Tyr Cys 85 90 95Ala Arg Gly Gly Met
Ser Thr Gly Asp Trp Phe Asp Pro Trp Gly Gln 100 105 110Gly Thr Leu
Val Thr Val Ser Ser 115 1209924DNAArtificial Sequencesynthetic
99ggattcacct ttgatgatta tggc 241008PRTArtificial Sequencesynthetic
100Gly Phe Thr Phe Asp Asp Tyr Gly1 510124DNAArtificial
Sequencesynthetic 101attcattgga gtggtagaag caca 241028PRTArtificial
Sequencesynthetic 102Ile His Trp Ser Gly Arg Ser Thr1
510339DNAArtificial Sequencesynthetic 103gcgagggggg gaatgagtac
gggggactgg ttcgacccc 3910413PRTArtificial Sequencesynthetic 104Ala
Arg Gly Gly Met Ser Thr Gly Asp Trp Phe Asp Pro1 5
10105324DNAArtificial Sequencesynthetic 105gacatccaga tgacccagtc
tccatcctcc ctgtctgcat ctgtaggaga cagagtcacc 60atcacttgcc gggcaagtca
gagcattagc agctatttaa attggtatca gcagaaacca 120gggaaagccc
ctaagctcct gatctatgtt gcatccagtt tgcaaagtgg ggtcccatca
180aggttcagtg gcagtggatc tgggacagat ttcactctca ccatcagcag
tctgcaacct 240gaagattttg caacttacta ctgtcaacag agttacagta
cccctccgat caccttcggc 300caagggacac gactggagat taaa
324106108PRTArtificial Sequencesynthetic 106Asp Ile Gln Met Thr Gln
Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile
Thr Cys Arg Ala Ser Gln Ser Ile Ser Ser Tyr 20 25 30Leu Asn Trp Tyr
Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Val Ala
Ser Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly
Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75
80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Ser Tyr Ser Thr Pro Pro
85 90 95Ile Thr Phe Gly Gln Gly Thr Arg Leu Glu Ile Lys 100
10510718DNAArtificial Sequencesynthetic 107cagagcatta gcagctat
181086PRTArtificial Sequencesynthetic 108Gln Ser Ile Ser Ser Tyr1
51099DNAArtificial Sequencesynthetic 109gttgcatcc
91103PRTArtificial Sequencesynthetic 110Val Ala
Ser111130DNAArtificial Sequencesynthetic 111caacagagtt acagtacccc
tccgatcacc 3011210PRTArtificial Sequencesynthetic 112Gln Gln Ser
Tyr Ser Thr Pro Pro Ile Thr1 5 10113345DNAArtificial
Sequencesynthetic 113gaggtgcagt tggtggagtc tggaggaggc ttggtccagc
ctggggggtc cctaagactc 60tcctgtgcag cctctgggtt caccgtcggt agtaactaca
tgaactgggt ccgtcaggct 120ccagggaagg gactggagtg ggtctcagtt
atttatagtg gtggtagtac atactacgca 180gattccgtga agggccgatt
caccatctcc agactcactt ccaagaacac actgtatctt 240caaatgagca
gcctgagacc tgaggacacg gccgtgtatt attgtgcgag agggattagg
300ggtctggacg tctggggcca agggaccacg gtcaccgtct cttca
345114115PRTArtificial Sequencesynthetic 114Glu Val Gln Leu Val Glu
Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser
Cys Ala Ala Ser Gly Phe Thr Val Gly Ser Asn 20 25 30Tyr Met Asn Trp
Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ser Val Ile
Tyr Ser Gly Gly Ser Thr Tyr Tyr Ala Asp Ser Val Lys 50 55 60Gly Arg
Phe Thr Ile Ser Arg Leu Thr Ser Lys Asn Thr Leu Tyr Leu65 70 75
80Gln Met Ser Ser Leu Arg Pro Glu Asp Thr Ala Val Tyr Tyr Cys Ala
85 90 95Arg Gly Ile Arg Gly Leu Asp Val Trp Gly Gln Gly Thr Thr Val
Thr 100 105 110Val Ser Ser 11511524DNAArtificial Sequencesynthetic
115gggttcaccg tcggtagtaa ctac 241168PRTArtificial Sequencesynthetic
116Gly Phe Thr Val Gly Ser Asn Tyr1 511721DNAArtificial
Sequencesynthetic 117atttatagtg gtggtagtac a 211187PRTArtificial
Sequencesynthetic 118Ile Tyr Ser Gly Gly Ser Thr1
511927DNAArtificial Sequencesynthetic 119gcgagaggga ttaggggtct
ggacgtc 271209PRTArtificial Sequencesynthetic 120Ala Arg Gly Ile
Arg Gly Leu Asp Val1 5121324DNAArtificial Sequencesynthetic
121gacatccaga tgacccagtc tccatcctcc ctgtctgcat ctgtaggaga
cagagtcacc 60atcacttgcc gggcaagtca gaccattaac atctatttaa attggtatca
gcagaaacca 120gggagagccc ctaggctcct gatctatgct gcatccagtt
tgcaaagtgg ggtcccatca 180aggttcagtg gcagtggatc tgggacagat
ttcactctca ccatcagcag tctgcaacct 240gaagattttg caacttacta
ctgtcaccag agttacagta cccctccgat caccttcggc 300caagggacac
gactggagat taaa 324122108PRTArtificial Sequencesynthetic 122Asp Ile
Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp
Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Thr Ile Asn Ile Tyr 20 25
30Leu Asn Trp Tyr Gln Gln Lys Pro Gly Arg Ala Pro Arg Leu Leu Ile
35 40 45Tyr Ala Ala Ser Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser
Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu
Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys His Gln Ser Tyr
Ser Thr Pro Pro 85 90 95Ile Thr Phe Gly Gln Gly Thr Arg Leu Glu Ile
Lys 100 10512318DNAArtificial Sequencesynthetic 123cagaccatta
acatctat 181246PRTArtificial Sequencesynthetic 124Gln Thr Ile Asn
Ile Tyr1 51259DNAArtificial Sequencesynthetic 125gctgcatcc
91263PRTArtificial Sequencesynthetic 126Ala Ala
Ser112730DNAArtificial Sequencesynthetic 127caccagagtt acagtacccc
tccgatcacc 3012810PRTArtificial Sequencesynthetic 128His Gln Ser
Tyr Ser Thr Pro Pro Ile Thr1 5 10129345DNAArtificial
Sequencesynthetic 129gaggaacggt tggtggagtc tggaggagac ttggtccagc
ctggggggtc cctgagactc 60tcctgtgcag cctctggcat caccgtcggt actaattata
tgaactgggt ccgccaggct 120ccagggaagg gactggagtg ggtctcagtt
atttctagcg gtggtaatac acactacgca 180gactccgtga agggccgatt
cattatgtcc agacaaactt ccaaaaacac gctgtatctt 240cagatgaata
gcctggaaac tgaggacacg gccgtatatt attgtgcgag ggggatcaga
300ggtttggacg tctggggcca agggaccatg gtcaccgtct cctca
345130115PRTArtificial Sequencesynthetic 130Glu Glu Arg Leu Val Glu
Ser Gly Gly Asp Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser
Cys Ala Ala Ser Gly Ile Thr Val Gly Thr Asn 20 25 30Tyr Met Asn Trp
Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ser Val Ile
Ser Ser Gly Gly Asn Thr His Tyr Ala Asp Ser Val Lys 50 55 60Gly Arg
Phe Ile Met Ser Arg Gln Thr Ser Lys Asn Thr Leu Tyr Leu65 70 75
80Gln Met Asn Ser Leu Glu Thr Glu Asp Thr Ala Val Tyr Tyr Cys Ala
85 90 95Arg Gly Ile Arg Gly Leu Asp Val Trp Gly Gln Gly Thr Met Val
Thr 100 105 110Val Ser Ser 11513124DNAArtificial Sequencesynthetic
131ggcatcaccg tcggtactaa ttat 241328PRTArtificial Sequencesynthetic
132Gly Ile Thr Val Gly Thr Asn Tyr1 513321DNAArtificial
Sequencesynthetic 133atttctagcg gtggtaatac a 211347PRTArtificial
Sequencesynthetic 134Ile Ser Ser Gly Gly Asn Thr1
513527DNAArtificial Sequencesynthetic 135gcgaggggga tcagaggttt
ggacgtc 271369PRTArtificial Sequencesynthetic 136Ala Arg Gly Ile
Arg Gly Leu Asp Val1 5137324DNAArtificial Sequencesynthetic
137gacatccaga tgacccagtc tccatcctcc ctgtctgcat ctgtaggaga
cagagtcacc 60atcacttgcc gggcaagtca gagcatgagc agctatttaa attggtatca
gcagaaacca 120gggagagccc ctaagctcct gatctttgct gcatccagtt
tgcaaagtgg ggtcccatca 180aggttcagtg gcagtggatc tgggacagat
ttcactctca ccatcagcag tctgcaacct 240gaagattttg caacttacta
ctgtcaacag agttacagta cccctccgat caccttcggc 300caagggacac
gactggagat taaa 324138108PRTArtificial Sequencesynthetic 138Asp Ile
Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp
Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Ser Met Ser Ser Tyr 20 25
30Leu Asn Trp Tyr Gln Gln Lys Pro Gly Arg Ala Pro Lys Leu Leu Ile
35 40 45Phe Ala Ala Ser Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser
Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu
Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Ser Tyr
Ser Thr Pro Pro 85 90 95Ile Thr Phe Gly Gln Gly Thr Arg Leu Glu Ile
Lys 100 10513918DNAArtificial Sequencesynthetic 139cagagcatga
gcagctat 181406PRTArtificial Sequencesynthetic 140Gln Ser Met Ser
Ser Tyr1 51419DNAArtificial Sequencesynthetic 141gctgcatcc
91423PRTArtificial Sequencesynthetic 142Ala Ala
Ser114330DNAArtificial Sequencesynthetic 143caacagagtt acagtacccc
tccgatcacc 3014410PRTArtificial Sequencesynthetic 144Gln Gln Ser
Tyr Ser Thr Pro Pro Ile Thr1 5 10145354DNAArtificial
Sequencesynthetic 145caggtccagc tggtgcagtc tggggctgag gtgaagatgc
ctgggtcctc ggtgagggtc 60tcctgcaagg cttctggagg catcttcagc agttctacta
tcagttgggt gcgacaggcc 120cctggacaag ggcttgaatg gatgggagag
atcatccctg
tctttggtac agtaaactac 180gcacagaagt tccaggacag agtcatattt
accgcggacg aatctacgac tacagcctac 240atggagctga gcagcctgaa
atctggggac acggccgtat atttctgtgc gcgaaattgg 300ggattaggct
ctttttatat ctggggccaa gggacaatgg tcaccgtctc ttca
354146118PRTArtificial Sequencesynthetic 146Gln Val Gln Leu Val Gln
Ser Gly Ala Glu Val Lys Met Pro Gly Ser1 5 10 15Ser Val Arg Val Ser
Cys Lys Ala Ser Gly Gly Ile Phe Ser Ser Ser 20 25 30Thr Ile Ser Trp
Val Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Glu Ile
Ile Pro Val Phe Gly Thr Val Asn Tyr Ala Gln Lys Phe 50 55 60Gln Asp
Arg Val Ile Phe Thr Ala Asp Glu Ser Thr Thr Thr Ala Tyr65 70 75
80Met Glu Leu Ser Ser Leu Lys Ser Gly Asp Thr Ala Val Tyr Phe Cys
85 90 95Ala Arg Asn Trp Gly Leu Gly Ser Phe Tyr Ile Trp Gly Gln Gly
Thr 100 105 110Met Val Thr Val Ser Ser 11514724DNAArtificial
Sequencesynthetic 147ggaggcatct tcagcagttc tact 241488PRTArtificial
Sequencesynthetic 148Gly Gly Ile Phe Ser Ser Ser Thr1
514924DNAArtificial Sequencesynthetic 149atcatccctg tctttggtac agta
241508PRTArtificial Sequencesynthetic 150Ile Ile Pro Val Phe Gly
Thr Val1 515133DNAArtificial Sequencesynthetic 151gcgcgaaatt
ggggattagg ctctttttat atc 3315211PRTArtificial Sequencesynthetic
152Ala Arg Asn Trp Gly Leu Gly Ser Phe Tyr Ile1 5
10153324DNAArtificial Sequencesynthetic 153gaaattgtgt tgacgcagtc
tccaggcacc ctgtctttgt ctccagggga aagagccacc 60ctctcctgca gggccagtca
gagttttaac ttcaactact tagcctggta ccagcagaaa 120cctggccagg
ctcccagact cctcatctat ggtgcatcca gcagggccac tggcatccca
180gacaggttca gtggcagtgg gtctgggaca gacttcactc tcaccatcaa
caggctggag 240cctgaagatt ttggagtgtt ttattgtcag cagtatgaaa
gcgcaccttg gacgttcggc 300caagggacca aggtggaaat caaa
324154108PRTArtificial Sequencesynthetic 154Glu Ile Val Leu Thr Gln
Ser Pro Gly Thr Leu Ser Leu Ser Pro Gly1 5 10 15Glu Arg Ala Thr Leu
Ser Cys Arg Ala Ser Gln Ser Phe Asn Phe Asn 20 25 30Tyr Leu Ala Trp
Tyr Gln Gln Lys Pro Gly Gln Ala Pro Arg Leu Leu 35 40 45Ile Tyr Gly
Ala Ser Ser Arg Ala Thr Gly Ile Pro Asp Arg Phe Ser 50 55 60Gly Ser
Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Asn Arg Leu Glu65 70 75
80Pro Glu Asp Phe Gly Val Phe Tyr Cys Gln Gln Tyr Glu Ser Ala Pro
85 90 95Trp Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys 100
10515521DNAArtificial Sequencesynthetic 155cagagtttta acttcaacta c
211567PRTArtificial Sequencesynthetic 156Gln Ser Phe Asn Phe Asn
Tyr1 51579DNAArtificial Sequencesynthetic 157ggtgcatcc
91583PRTArtificial Sequencesynthetic 158Gly Ala
Ser115927DNAArtificial Sequencesynthetic 159cagcagtatg aaagcgcacc
ttggacg 271609PRTArtificial Sequencesynthetic 160Gln Gln Tyr Glu
Ser Ala Pro Trp Thr1 5161345DNAArtificial Sequencesynthetic
161gaggtgcagc ttgtagagtc tgggggagac ttggtacatc ctggcaggtc
cctgagactc 60tcctgtgcag cctctggttt cccctttgat gagtatgcca tgcactgggt
ccggcaagtt 120ccagggaagg gcctggagtg ggtctcaggt attagttgga
gtaataataa cataggctat 180gcggactctg tgaagggccg attcaccatc
tccagagaca acgccaaaaa ctccctgtat 240ctacaaatga acagtctgag
acctgaggac acggcctttt attactgtgc aaaatctgga 300atctttgact
cctggggcca gggaaccctg gtcaccgtct cctca 345162115PRTArtificial
Sequencesynthetic 162Glu Val Gln Leu Val Glu Ser Gly Gly Asp Leu
Val His Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Pro Phe Asp Glu Tyr 20 25 30Ala Met His Trp Val Arg Gln Val Pro
Gly Lys Gly Leu Glu Trp Val 35 40 45Ser Gly Ile Ser Trp Ser Asn Asn
Asn Ile Gly Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser
Arg Asp Asn Ala Lys Asn Ser Leu Tyr65 70 75 80Leu Gln Met Asn Ser
Leu Arg Pro Glu Asp Thr Ala Phe Tyr Tyr Cys 85 90 95Ala Lys Ser Gly
Ile Phe Asp Ser Trp Gly Gln Gly Thr Leu Val Thr 100 105 110Val Ser
Ser 11516324DNAArtificial Sequencesynthetic 163ggtttcccct
ttgatgagta tgcc 241648PRTArtificial Sequencesynthetic 164Gly Phe
Pro Phe Asp Glu Tyr Ala1 516524DNAArtificial Sequencesynthetic
165attagttgga gtaataataa cata 241668PRTArtificial Sequencesynthetic
166Ile Ser Trp Ser Asn Asn Asn Ile1 516724DNAArtificial
Sequencesynthetic 167gcaaaatctg gaatctttga ctcc 241688PRTArtificial
Sequencesynthetic 168Ala Lys Ser Gly Ile Phe Asp Ser1
5169315DNAArtificial Sequencesynthetic 169gacatccaga tgacccagtc
tccatcctcc ctgtctgcat ctgtaggaga cagagtcacc 60atcacttgcc gggcaagtca
gagcattagc agctatttaa attggtatca gcagaaacca 120gggaagctcc
tgatctatgc tgcatccagt ttgcaaagtg gggtcccatc acggttcagt
180ggcggtggat ctgggacaga tttcactctc accatcagca gtctgcgacc
tgaagatttt 240gcaacttact actgtcaaca gagttactgt acccctccga
tcaccttcgg ccaagggaca 300cgactggaga ttaaa 315170105PRTArtificial
Sequencesynthetic 170Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu
Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser
Gln Ser Ile Ser Ser Tyr 20 25 30Leu Asn Trp Tyr Gln Gln Lys Pro Gly
Lys Leu Leu Ile Tyr Ala Ala 35 40 45Ser Ser Leu Gln Ser Gly Val Pro
Ser Arg Phe Ser Gly Gly Gly Ser 50 55 60Gly Thr Asp Phe Thr Leu Thr
Ile Ser Ser Leu Arg Pro Glu Asp Phe65 70 75 80Ala Thr Tyr Tyr Cys
Gln Gln Ser Tyr Cys Thr Pro Pro Ile Thr Phe 85 90 95Gly Gln Gly Thr
Arg Leu Glu Ile Lys 100 10517118DNAArtificial Sequencesynthetic
171cagagcatta gcagctat 181726PRTArtificial Sequencesynthetic 172Gln
Ser Ile Ser Ser Tyr1 51739DNAArtificial Sequencesynthetic
173gctgcatcc 91743PRTArtificial Sequencesynthetic 174Ala Ala
Ser117530DNAArtificial Sequencesynthetic 175caacagagtt actgtacccc
tccgatcacc 3017610PRTArtificial Sequencesynthetic 176Gln Gln Ser
Tyr Cys Thr Pro Pro Ile Thr1 5 10177354DNAArtificial
Sequencesynthetic 177gaggtgcagc tggtggagtc cgggggaggc gtggtccagc
ctgggaggtc cctgagactc 60tcctgtgcag cctctggatt caccttcagt agttatggca
tgcactgggt ccgccaggct 120ccaggcaagg gactggagtg ggtgacactt
atatcatatg agggaaggaa taaatactat 180gcagactccg tgaagggccg
attcaccatt tccagagaca attccaagaa cacgctgtat 240ctgcaaatga
acagcctgag agctgaggac acggctgtat attactgtgc gaaagatagg
300accctttacg gtatggacgt ctggggccaa ggaaccacgg tcaccgtctc ctca
354178118PRTArtificial Sequencesynthetic 178Glu Val Gln Leu Val Glu
Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser
Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30Gly Met His Trp
Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Thr Leu Ile
Ser Tyr Glu Gly Arg Asn Lys Tyr Tyr Ala Asp Ser Val 50 55 60Lys Gly
Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr65 70 75
80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Lys Asp Arg Thr Leu Tyr Gly Met Asp Val Trp Gly Gln Gly
Thr 100 105 110Thr Val Thr Val Ser Ser 11517924DNAArtificial
Sequencesynthetic 179ggattcacct tcagtagtta tggc 241808PRTArtificial
Sequencesynthetic 180Gly Phe Thr Phe Ser Ser Tyr Gly1
518124DNAArtificial Sequencesynthetic 181atatcatatg agggaaggaa taaa
241828PRTArtificial Sequencesynthetic 182Ile Ser Tyr Glu Gly Arg
Asn Lys1 518333DNAArtificial Sequencesynthetic 183gcgaaagata
ggacccttta cggtatggac gtc 3318411PRTArtificial Sequencesynthetic
184Ala Lys Asp Arg Thr Leu Tyr Gly Met Asp Val1 5
10185363DNAArtificial Sequencesynthetic 185caggtcacct tgagggagtc
tggtcctgcg ctggtgaaaa ccacacagac cctcacactg 60acctgcacct tctctgggtt
ctcactcagc actaatagaa tgtgtgtgac ctggatccgt 120cagcccccag
ggaaggccct ggagtggctt gcgcgcattg attgggatgg tgttaaatac
180tacaacacat ctctgaagac caggctcacc atctccaagg acacctccaa
aaaccaggtg 240gtccttacaa tgaccaacat ggaccctgtg gacacagcca
ctttttactg tgcacggtcg 300acttcgttga ctttttacta ctttgactac
tggggccagg gaaccctggt caccgtctcc 360tca 363186121PRTArtificial
Sequencesynthetic 186Gln Val Thr Leu Arg Glu Ser Gly Pro Ala Leu
Val Lys Thr Thr Gln1 5 10 15Thr Leu Thr Leu Thr Cys Thr Phe Ser Gly
Phe Ser Leu Ser Thr Asn 20 25 30Arg Met Cys Val Thr Trp Ile Arg Gln
Pro Pro Gly Lys Ala Leu Glu 35 40 45Trp Leu Ala Arg Ile Asp Trp Asp
Gly Val Lys Tyr Tyr Asn Thr Ser 50 55 60Leu Lys Thr Arg Leu Thr Ile
Ser Lys Asp Thr Ser Lys Asn Gln Val65 70 75 80Val Leu Thr Met Thr
Asn Met Asp Pro Val Asp Thr Ala Thr Phe Tyr 85 90 95Cys Ala Arg Ser
Thr Ser Leu Thr Phe Tyr Tyr Phe Asp Tyr Trp Gly 100 105 110Gln Gly
Thr Leu Val Thr Val Ser Ser 115 12018730DNAArtificial
Sequencesynthetic 187gggttctcac tcagcactaa tagaatgtgt
3018810PRTArtificial Sequencesynthetic 188Gly Phe Ser Leu Ser Thr
Asn Arg Met Cys1 5 1018921DNAArtificial Sequencesynthetic
189attgattggg atggtgttaa a 211907PRTArtificial Sequencesynthetic
190Ile Asp Trp Asp Gly Val Lys1 519139DNAArtificial
Sequencesynthetic 191gcacggtcga cttcgttgac tttttactac tttgactac
3919213PRTArtificial Sequencesynthetic 192Ala Arg Ser Thr Ser Leu
Thr Phe Tyr Tyr Phe Asp Tyr1 5 10193324DNAArtificial
Sequencesynthetic 193gacatccaga tgacccagtc tccatcctcc ctgtctgcat
ctgtaggaga cagagtcacc 60atcacttgcc gggcaagtca gagcattagc agctatttaa
attggtatca gcagaaacca 120gggaaagccc ctaagctcct gatctatgct
gcatccagtt tgcaaagtgg ggtcccatca 180aggttcagtg gcagtggatc
tgggacagat ttcactctca ccatcagcag tctgcaacct 240gaagattttg
caacttacta ctgtcaacag agttacagta cccctccgat caccttcggc
300caagggacac gactggagat taaa 324194108PRTArtificial
Sequencesynthetic 194Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu
Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser
Gln Ser Ile Ser Ser Tyr 20 25 30Leu Asn Trp Tyr Gln Gln Lys Pro Gly
Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ala Ala Ser Ser Leu Gln Ser
Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe
Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr
Tyr Tyr Cys Gln Gln Ser Tyr Ser Thr Pro Pro 85 90 95Ile Thr Phe Gly
Gln Gly Thr Arg Leu Glu Ile Lys 100 10519518DNAArtificial
Sequencesynthetic 195cagagcatta gcagctat 181966PRTArtificial
Sequencesynthetic 196Gln Ser Ile Ser Ser Tyr1 51979DNAArtificial
Sequencesynthetic 197gctgcatcc 91983PRTArtificial Sequencesynthetic
198Ala Ala Ser119930DNAArtificial Sequencesynthetic 199caacagagtt
acagtacccc tccgatcacc 3020010PRTArtificial Sequencesynthetic 200Gln
Gln Ser Tyr Ser Thr Pro Pro Ile Thr1 5 10201345DNAArtificial
Sequencesynthetic 201gaggtgcagc tggtggagtc tggaggaggc ttggtccagc
cgggggggtc cctgagactc 60tcctgtgcag cctctgagtt caccgtcggt accaaccaca
tgaactgggt ccgccaggct 120ccagggaagg gactggagtg ggtctcagtt
atttatagcg gtggtaacac attctacgca 180gactccgtga agggccgatt
caccatctcc agacacactt ccaagaacac gctgtatctt 240caaatgaaca
gcctgacagc agaggacacg gccgtatatt actgtgcgcg aggattgggg
300ggtatggacg tctggggcca agggaccacg gtcaccgtct cctca
345202115PRTArtificial Sequencesynthetic 202Glu Val Gln Leu Val Glu
Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser
Cys Ala Ala Ser Glu Phe Thr Val Gly Thr Asn 20 25 30His Met Asn Trp
Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ser Val Ile
Tyr Ser Gly Gly Asn Thr Phe Tyr Ala Asp Ser Val Lys 50 55 60Gly Arg
Phe Thr Ile Ser Arg His Thr Ser Lys Asn Thr Leu Tyr Leu65 70 75
80Gln Met Asn Ser Leu Thr Ala Glu Asp Thr Ala Val Tyr Tyr Cys Ala
85 90 95Arg Gly Leu Gly Gly Met Asp Val Trp Gly Gln Gly Thr Thr Val
Thr 100 105 110Val Ser Ser 11520324DNAArtificial Sequencesynthetic
203gagttcaccg tcggtaccaa ccac 242048PRTArtificial Sequencesynthetic
204Glu Phe Thr Val Gly Thr Asn His1 520521DNAArtificial
Sequencesynthetic 205atttatagcg gtggtaacac a 212067PRTArtificial
Sequencesynthetic 206Ile Tyr Ser Gly Gly Asn Thr1
520727DNAArtificial Sequencesynthetic 207gcgcgaggat tggggggtat
ggacgtc 272089PRTArtificial Sequencesynthetic 208Ala Arg Gly Leu
Gly Gly Met Asp Val1 5209321DNAArtificial Sequencesynthetic
209gacatccaga tgacccagtc tccatcctcc ctgtctgcat ctgtaggaga
cagagtcacc 60atcacttgcc gggcgagtca ggtcattagc aattatttag cctggtatca
gcagaaacca 120gggaaagttc ctaggctcct gatctatgct gcatccactt
tgcaatcagg ggtcccatct 180cgtttcagtg gcagtggatc tgggacagat
ttcactctca ccatcagcag cctgcagcct 240gaagatgttg caacttatta
ctgtcaaaag tataacagtg cccctcggac gttcggccaa 300gggaccaagg
tggaaatcaa a 321210107PRTArtificial Sequencesynthetic 210Asp Ile
Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp
Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Val Ile Ser Asn Tyr 20 25
30Leu Ala Trp Tyr Gln Gln Lys Pro Gly Lys Val Pro Arg Leu Leu Ile
35 40 45Tyr Ala Ala Ser Thr Leu Gln Ser Gly Val Pro Ser Arg Phe Ser
Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu
Gln Pro65 70 75 80Glu Asp Val Ala Thr Tyr Tyr Cys Gln Lys Tyr Asn
Ser Ala Pro Arg 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys
100 10521118DNAArtificial Sequencesynthetic 211caggtcatta gcaattat
182126PRTArtificial Sequencesynthetic 212Gln Val Ile Ser Asn Tyr1
52139DNAArtificial Sequencesynthetic 213gctgcatcc
92143PRTArtificial Sequencesynthetic 214Ala Ala
Ser121527DNAArtificial Sequencesynthetic 215caaaagtata acagtgcccc
tcggacg 272169PRTArtificial Sequencesynthetic 216Gln Lys Tyr Asn
Ser Ala Pro Arg Thr1 5217360DNAArtificial Sequencesynthetic
217gaggtgcagc tggtggagtc tgggggaggc ttggtccagc ggggggagtc
cctgagactt 60tactgtgcag cctctggatt cacctttagt aaatattgga tgaactgggt
ccgccaggct 120ccagggaagg ggctggagtg ggtggccaac ataaagggag
atggaagtga gaaatactat 180gtggactctg tgaagggccg gttcaccatc
tccagagaca acgccaagaa ctcactatat 240ctacaaatga acagcctgag
agccgaggac acggctgttt attactgtgc gagagattat 300tggggatcag
gctactactt tgacttctgg ggccagggaa ccctggtcac cgtctcctca
360218120PRTArtificial Sequencesynthetic 218Glu Val Gln Leu Val Glu
Ser Gly Gly Gly Leu Val Gln Arg Gly Glu1 5 10 15Ser Leu Arg Leu Tyr
Cys Ala Ala Ser Gly Phe Thr Phe Ser Lys Tyr 20 25 30Trp Met Asn Trp
Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Asn Ile
Lys Gly Asp Gly Ser Glu Lys Tyr Tyr Val Asp Ser Val 50 55 60Lys Gly
Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn Ser Leu Tyr65 70 75
80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Arg Asp Tyr Trp Gly Ser Gly Tyr Tyr Phe Asp Phe Trp Gly
Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser 115
12021924DNAArtificial Sequencesynthetic 219ggattcacct ttagtaaata
ttgg 242208PRTArtificial Sequencesynthetic 220Gly Phe Thr Phe Ser
Lys Tyr Trp1 522124DNAArtificial Sequencesynthetic 221ataaagggag
atggaagtga gaaa 242228PRTArtificial Sequencesynthetic 222Ile Lys
Gly Asp Gly Ser Glu Lys1 522339DNAArtificial Sequencesynthetic
223gcgagagatt attggggatc aggctactac tttgacttc 3922413PRTArtificial
Sequencesynthetic 224Ala Arg Asp Tyr Trp Gly Ser Gly Tyr Tyr Phe
Asp Phe1 5 10225321DNAArtificial Sequencesynthetic 225gacatccaga
tgacccagtc tccatcctcc ctgtctgcat ctgtaggaga cagagtcacc 60atcacttgcc
gggcaagtca gaacattaac aactatttaa attggtatca gcagaaacca
120gggaaagccc ctaaactcct gatctatgct gcatccagtt tccaaaatgc
ggtcccatca 180aggttcagtg gcagtggatc tgggacagat ttcactctca
ccatcagcag tctgcaacct 240gaagattttg caacttacta ctgtcaacag
agttacaata ccccgctcac tttcggcggg 300gggaccaagg tggagatcaa a
321226107PRTArtificial Sequencesynthetic 226Asp Ile Gln Met Thr Gln
Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile
Thr Cys Arg Ala Ser Gln Asn Ile Asn Asn Tyr 20 25 30Leu Asn Trp Tyr
Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ala Ala
Ser Ser Phe Gln Asn Ala Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly
Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75
80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Ser Tyr Asn Thr Pro Leu
85 90 95Thr Phe Gly Gly Gly Thr Lys Val Glu Ile Lys 100
10522718DNAArtificial Sequencesynthetic 227cagaacatta acaactat
182286PRTArtificial Sequencesynthetic 228Gln Asn Ile Asn Asn Tyr1
52299DNAArtificial Sequencesynthetic 229gctgcatcc
92303PRTArtificial Sequencesynthetic 230Ala Ala
Ser123127DNAArtificial Sequencesynthetic 231caacagagtt acaatacccc
gctcact 272329PRTArtificial Sequencesynthetic 232Gln Gln Ser Tyr
Asn Thr Pro Leu Thr1 5233390DNAArtificial Sequencesynthetic
233gaggtgcagc tggtggagtc tgggggaggc ttggtccagt ctggggggtc
cctgagactc 60tcctgtgcag cctctggatt cacctttagt agctattgga tgagctgggt
ccgccaggct 120ccagggaagg ggctggagtg ggtggccaac ataaagcaag
atggaagtga gaaatactat 180gtggactctg tgaagggccg attcaccatc
tccagagaca acgccaagaa ctcactgtat 240ctgcaaatga acagcctgag
agccgatgac acggctgtgt attactgtgc gagagatgat 300attgtagtag
taccagctcc tatgggatat tactactact acttcggtat ggacgtctgg
360ggccaaggga ccacggtcac cgtctcctca 390234130PRTArtificial
Sequencesynthetic 234Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu
Val Gln Ser Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Ser Ser Tyr 20 25 30Trp Met Ser Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Asn Ile Lys Gln Asp Gly Ser
Glu Lys Tyr Tyr Val Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser
Arg Asp Asn Ala Lys Asn Ser Leu Tyr65 70 75 80Leu Gln Met Asn Ser
Leu Arg Ala Asp Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Asp
Ile Val Val Val Pro Ala Pro Met Gly Tyr Tyr Tyr 100 105 110Tyr Tyr
Phe Gly Met Asp Val Trp Gly Gln Gly Thr Thr Val Thr Val 115 120
125Ser Ser 13023524DNAArtificial Sequencesynthetic 235ggattcacct
ttagtagcta ttgg 242368PRTArtificial Sequencesynthetic 236Gly Phe
Thr Phe Ser Ser Tyr Trp1 523724DNAArtificial Sequencesynthetic
237ataaagcaag atggaagtga gaaa 242388PRTArtificial Sequencesynthetic
238Ile Lys Gln Asp Gly Ser Glu Lys1 523969DNAArtificial
Sequencesynthetic 239gcgagagatg atattgtagt agtaccagct cctatgggat
attactacta ctacttcggt 60atggacgtc 6924023PRTArtificial
Sequencesynthetic 240Ala Arg Asp Asp Ile Val Val Val Pro Ala Pro
Met Gly Tyr Tyr Tyr1 5 10 15Tyr Tyr Phe Gly Met Asp Val
20241321DNAArtificial Sequencesynthetic 241gacatccaga tgacccagtc
tccatcctcc ctgtctgcat ctgtaggaga cagagtcacc 60atcacttgcc gggcaagtca
gggcattaga aatgatttag gctggtatca gcagaaacca 120gggaaagccc
ctaagcgcct gatctatgct gcatccagtt tgcaaagtgg ggtcccatca
180aggttcagcg gcagtggatc tgggacagaa ttcactctca caatcagcag
cctgcagcct 240gaagattttg caacttatta ctgtctacag cataatagtt
acccgtacac ttttggccag 300gggaccaagc tggagatcaa a
321242107PRTArtificial Sequencesynthetic 242Asp Ile Gln Met Thr Gln
Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile
Thr Cys Arg Ala Ser Gln Gly Ile Arg Asn Asp 20 25 30Leu Gly Trp Tyr
Gln Gln Lys Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr Ala Ala
Ser Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly
Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75
80Glu Asp Phe Ala Thr Tyr Tyr Cys Leu Gln His Asn Ser Tyr Pro Tyr
85 90 95Thr Phe Gly Gln Gly Thr Lys Leu Glu Ile Lys 100
10524318DNAArtificial Sequencesynthetic 243cagggcatta gaaatgat
182446PRTArtificial Sequencesynthetic 244Gln Gly Ile Arg Asn Asp1
52459DNAArtificial Sequencesynthetic 245gctgcatcc
92463PRTArtificial Sequencesynthetic 246Ala Ala
Ser124727DNAArtificial Sequencesynthetic 247ctacagcata atagttaccc
gtacact 272489PRTArtificial Sequencesynthetic 248Leu Gln His Asn
Ser Tyr Pro Tyr Thr1 5249369DNAArtificial Sequencesynthetic
249gaagtgcagc tggtggagtc tgggggaggc ttggttcagc ctggcaggtc
cctgagactc 60tcctgtgcag cctctggatt cacctttgat gattttgcca tgcactgggt
ccgacaagct 120ccagggaagg gcctggagtg ggtctcaggt attagttgga
ctggtggtaa catggactat 180gcgaactctg tgaagggccg attcaccatc
tccagagagg acgccaagaa ttccctgtat 240ctgcaaatga acagtctgag
agctgcggac acggccttgt attactgtgt aaaagatata 300agggggatag
tggctacggg gggggctttt gatatctggg gccgagggac aatggtcacc 360gtctcttca
369250123PRTArtificial Sequencesynthetic 250Glu Val Gln Leu Val Glu
Ser Gly Gly Gly Leu Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser
Cys Ala Ala Ser Gly Phe Thr Phe Asp Asp Phe 20 25 30Ala Met His Trp
Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ser Gly Ile
Ser Trp Thr Gly Gly Asn Met Asp Tyr Ala Asn Ser Val 50 55 60Lys Gly
Arg Phe Thr Ile Ser Arg Glu Asp Ala Lys Asn Ser Leu Tyr65 70 75
80Leu Gln Met Asn Ser Leu Arg Ala Ala Asp Thr Ala Leu Tyr Tyr Cys
85 90 95Val Lys Asp Ile Arg Gly Ile Val Ala Thr Gly Gly Ala Phe Asp
Ile 100 105 110Trp Gly Arg Gly Thr Met Val Thr Val Ser Ser 115
12025124DNAArtificial Sequencesynthetic 251ggattcacct ttgatgattt
tgcc 242528PRTArtificial Sequencesynthetic 252Gly Phe Thr Phe Asp
Asp Phe Ala1 525324DNAArtificial Sequencesynthetic 253attagttgga
ctggtggtaa catg 242548PRTArtificial Sequencesynthetic 254Ile Ser
Trp Thr Gly Gly Asn Met1 525548DNAArtificial Sequencesynthetic
255gtaaaagata taagggggat agtggctacg gggggggctt ttgatatc
4825616PRTArtificial Sequencesynthetic 256Val Lys Asp Ile Arg Gly
Ile Val Ala Thr Gly Gly Ala Phe Asp Ile1 5 10 15257321DNAArtificial
Sequencesynthetic 257gacatccaga tgacccagtc tccatcctcc ctgtctgcat
ctgtaggaga cagagtcacc 60atctcttgcc gggcaagtca gaccattagc acttatttaa
attggtttca gcagaaacca 120gggaaagccc ctaagctcct gatctatgtt
gtgtccagtt tgcaaagtgg ggtcccatca 180aggttcagtg gcagtggatc
tgggacagat ttcactctca ccatcagcag tctgcaacct 240gaagattttg
caacttatta ctgtcaacag agttacagta ccccattcac tttcggccct
300gggaccaaag tggatatcaa a 321258107PRTArtificial Sequencesynthetic
258Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1
5 10 15Asp Arg Val Thr Ile Ser Cys Arg Ala Ser Gln Thr Ile Ser Thr
Tyr 20 25 30Leu Asn Trp Phe Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu
Leu Ile 35 40 45Tyr Val Val Ser Ser Leu Gln Ser Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln
Ser Tyr Ser Thr Pro Phe 85 90 95Thr Phe Gly Pro Gly Thr Lys Val Asp
Ile Lys 100 10525918DNAArtificial Sequencesynthetic 259cagaccatta
gcacttat 182606PRTArtificial Sequencesynthetic 260Gln Thr Ile Ser
Thr Tyr1 52619DNAArtificial Sequencesynthetic 261gttgtgtcc
92623PRTArtificial Sequencesynthetic 262Val Val
Ser126327DNAArtificial Sequencesynthetic 263caacagagtt acagtacccc
attcact 272649PRTArtificial Sequencesynthetic 264Gln Gln Ser Tyr
Ser Thr Pro Phe Thr1 5265345DNAArtificial Sequencesynthetic
265gaggtgcagc tggtggagtc tggaggaggc ttggtccagc cgggggggtc
cctgagactc 60tcctgtgcag cctctggatt caccgtcggt accaactaca tgaactgggt
ccgccaggct 120ccagggaagg gactggagtg gatctcagtt atttatagcg
gtggtagcac attctacgca 180gactccgtga agggccgatt caccatctcc
agacagactt cccagaacac gctgtatctt 240caaatgaaca gcctgagacc
tgaggacacg gccgtatatt actgtgcgag aggtatacgt 300ggttttgata
tctggggcca agggacaatg gtcaccgtct cttca 345266115PRTArtificial
Sequencesynthetic 266Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu
Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Thr Val Gly Thr Asn 20 25 30Tyr Met Asn Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Trp Ile 35 40 45Ser Val Ile Tyr Ser Gly Gly Ser
Thr Phe Tyr Ala Asp Ser Val Lys 50 55 60Gly Arg Phe Thr Ile Ser Arg
Gln Thr Ser Gln Asn Thr Leu Tyr Leu65 70 75 80Gln Met Asn Ser Leu
Arg Pro Glu Asp Thr Ala Val Tyr Tyr Cys Ala 85 90 95Arg Gly Ile Arg
Gly Phe Asp Ile Trp Gly Gln Gly Thr Met Val Thr 100 105 110Val Ser
Ser 11526724DNAArtificial Sequencesynthetic 267ggattcaccg
tcggtaccaa ctac 242688PRTArtificial Sequencesynthetic 268Gly Phe
Thr Val Gly Thr Asn Tyr1 526921DNAArtificial Sequencesynthetic
269atttatagcg gtggtagcac a 212707PRTArtificial Sequencesynthetic
270Ile Tyr Ser Gly Gly Ser Thr1 527127DNAArtificial
Sequencesynthetic 271gcgagaggta tacgtggttt tgatatc
272729PRTArtificial Sequencesynthetic 272Ala Arg Gly Ile Arg Gly
Phe Asp Ile1 5273324DNAArtificial Sequencesynthetic 273gacatccaga
tgacccagtc tccatcctcc ctgtctgcat ctgtaggaga cagagtcacc 60atcacttgcc
gggcaagtca gagcattagc agctatttaa attggtatca gcagaaacca
120gggaaagccc ctaagctcct gatctatgct gcatccagtt tgcaaagtgg
ggtcccgtca 180aggttcagtg gcagtggatc tgggacagat ttcactctca
ccatcagcag tctgcaacct 240gaagattttg caacttacta ctgtcaacag
agttacagta cccctccgat caccttcggc 300caagggacac gactggagat taaa
324274108PRTArtificial Sequencesynthetic 274Asp Ile Gln Met Thr Gln
Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile
Thr Cys Arg Ala Ser Gln Ser Ile Ser Ser Tyr 20 25 30Leu Asn Trp Tyr
Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ala Ala
Ser Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly
Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75
80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Ser Tyr Ser Thr Pro Pro
85 90 95Ile Thr Phe Gly Gln Gly Thr Arg Leu Glu Ile Lys 100
10527518DNAArtificial Sequencesynthetic 275cagagcatta gcagctat
182766PRTArtificial Sequencesynthetic 276Gln Ser Ile Ser Ser Tyr1
52779DNAArtificial Sequencesynthetic 277gctgcatcc
92783PRTArtificial Sequencesynthetic 278Ala Ala
Ser127930DNAArtificial Sequencesynthetic 279caacagagtt acagtacccc
tccgatcacc 3028010PRTArtificial Sequencesynthetic 280Gln Gln Ser
Tyr Ser Thr Pro Pro Ile Thr1 5 10281345DNAArtificial
Sequencesynthetic 281gaggtgcagc tggtggagtc tggaggaggc ttggtccagc
cgggggggtc cctgagactc 60tcctgtgcag cctctgggtt taccatcagt accaactaca
tgaactgggt ccgccaggct 120ccagggaagg ggctggagtg ggtcgcagtt
atttatagca gtggttccac atactatatc 180gactccgtga agggccgatt
caccatctcc agactcactt ccaagaacac ggtgtatctt 240caaatgagca
gcctgaattc tgaagacacg gccgtgtatt actgtgcgag ggggatcagg
300ggttttgata tttggggcca agggacaatg gtcaccgtct cttca
345282115PRTArtificial Sequencesynthetic 282Glu Val Gln Leu Val Glu
Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser
Cys Ala Ala Ser Gly Phe Thr Ile Ser Thr Asn 20 25 30Tyr Met Asn Trp
Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Val Ile
Tyr Ser Ser Gly Ser Thr Tyr Tyr Ile Asp Ser Val Lys 50 55 60Gly Arg
Phe Thr Ile Ser Arg Leu Thr Ser Lys Asn Thr Val Tyr Leu65 70 75
80Gln Met Ser Ser Leu Asn Ser Glu Asp Thr Ala Val Tyr Tyr Cys Ala
85 90 95Arg Gly Ile Arg Gly Phe Asp Ile Trp Gly Gln Gly Thr Met Val
Thr 100 105 110Val Ser Ser 11528324DNAArtificial Sequencesynthetic
283gggtttacca tcagtaccaa ctac 242848PRTArtificial Sequencesynthetic
284Gly Phe Thr Ile Ser Thr Asn Tyr1 528521DNAArtificial
Sequencesynthetic 285atttatagca gtggttccac a 212867PRTArtificial
Sequencesynthetic 286Ile Tyr Ser Ser Gly Ser Thr1
528727DNAArtificial Sequencesynthetic 287gcgaggggga tcaggggttt
tgatatt 272889PRTArtificial Sequencesynthetic 288Ala Arg Gly Ile
Arg Gly Phe Asp Ile1 5289372DNAArtificial Sequencesynthetic
289gaagtgcagc tggtggagtc ggggggaggc ttggtacagc ctggcaggtc
cctgagactc 60tcctgtgcag cctctggatt caccattgat gatagtgcca tgcactgggt
ccggcaaact 120ccagggaagg gcctggagtg ggtctcaggt attagttgga
aaagtggtag cataggttat 180gcggactctg tgaggggccg attcaccatc
tccagagaca acgccaagaa ttccctctat 240ctgcaaatga acagtctgag
agttgaggac acggccttgt attactgtgt aaaagatata 300aggggcaact
ggaactacgg gggaaactgg ttcgacccct ggggccaggg aaccctggtc
360actgtctcct ca 372290124PRTArtificial Sequencesynthetic 290Glu
Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Arg1 5 10
15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Ile Asp Asp Ser
20 25 30Ala Met His Trp Val Arg Gln Thr Pro Gly Lys Gly Leu Glu Trp
Val 35 40 45Ser Gly Ile Ser Trp Lys Ser Gly Ser Ile Gly Tyr Ala Asp
Ser Val 50 55 60Arg Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn
Ser Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Val Glu Asp Thr
Ala Leu Tyr Tyr Cys 85 90 95Val Lys Asp Ile Arg Gly Asn Trp Asn Tyr
Gly Gly Asn Trp Phe Asp 100 105 110Pro Trp Gly Gln Gly Thr Leu Val
Thr Val Ser Ser 115 12029124DNAArtificial Sequencesynthetic
291ggattcacca ttgatgatag tgcc 242928PRTArtificial Sequencesynthetic
292Gly Phe Thr Ile Asp Asp Ser Ala1 529324DNAArtificial
Sequencesynthetic 293attagttgga aaagtggtag cata 242948PRTArtificial
Sequencesynthetic 294Ile Ser Trp Lys Ser Gly Ser Ile1
529551DNAArtificial Sequencesynthetic 295gtaaaagata taaggggcaa
ctggaactac gggggaaact ggttcgaccc c 5129617PRTArtificial
Sequencesynthetic 296Val Lys Asp Ile Arg Gly Asn Trp Asn Tyr Gly
Gly Asn Trp Phe Asp1 5 10 15Pro297345DNAArtificial
Sequencesynthetic 297gaggtgcagc tggtggagtc tggaggaggc ttggtccagc
ctggggggtc cctgagactc 60tcatgtgaag cctctgggtt caccgtcggt gtcaaccaca
tgaactgggt ccgccaggct 120ccagggaagg gtctggagtg ggtctcagtt
attttcagta gtggtaggac attctacgga 180gactacgtga aggggcgatt
aaccatcttc agacaaacct cccagaacac ggtgtatctt 240caaatgaata
gcctgagaag tgaggacacg gccatatatt actgtgcgag agggattggc
300ggtttggaca tctggggccg agggacaatg gtcaccgtct cttca
345298115PRTArtificial Sequencesynthetic 298Glu Val Gln Leu Val Glu
Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser
Cys Glu Ala Ser Gly Phe Thr Val Gly Val Asn 20 25 30His Met Asn Trp
Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ser Val Ile
Phe Ser Ser Gly Arg Thr Phe Tyr Gly Asp Tyr Val Lys 50 55 60Gly Arg
Leu Thr Ile Phe Arg Gln Thr Ser Gln Asn Thr Val Tyr Leu65 70 75
80Gln Met Asn Ser Leu Arg Ser Glu Asp Thr Ala Ile Tyr Tyr Cys Ala
85 90 95Arg Gly Ile Gly Gly Leu Asp Ile Trp Gly Arg Gly Thr Met Val
Thr 100 105 110Val Ser Ser 11529924DNAArtificial Sequencesynthetic
299gggttcaccg tcggtgtcaa ccac 243008PRTArtificial Sequencesynthetic
300Gly Phe Thr Val Gly Val Asn His1 530121DNAArtificial
Sequencesynthetic 301attttcagta gtggtaggac a 213027PRTArtificial
Sequencesynthetic 302Ile Phe Ser Ser Gly Arg Thr1
530327DNAArtificial Sequencesynthetic 303gcgagaggga ttggcggttt
ggacatc 273049PRTArtificial Sequencesynthetic 304Ala Arg Gly Ile
Gly Gly Leu Asp Ile1 5305369DNAArtificial Sequencesynthetic
305gaagtgcagc tggtggagtc tgggggaggc ttggttcagc ctggcaggtc
cctaagactc 60tcctgtgcag cctctggatt cacctttgat gattatgcct tgcactgggt
ccggcaagct 120ccagggaagg gcctggagtg ggtctcaggt attagttgga
ctggtggtac tatagactat 180gcggactctg tgaagggccg attcaccatc
tccagagaca acgccaagaa ctccctgtat 240ctgcaaatga gcagtctgag
aactgaggac acggccatat attactgtac aagagatatc 300cgggggaact
ggaagtacgg aggctggttc gacccctggg gccagggaac cctggtcacc 360gtctcctca
369306123PRTArtificial Sequencesynthetic 306Glu Val Gln Leu Val Glu
Ser Gly Gly Gly Leu Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser
Cys Ala Ala Ser Gly Phe Thr Phe Asp Asp Tyr 20 25 30Ala Leu His Trp
Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ser Gly Ile
Ser Trp Thr Gly Gly Thr Ile Asp Tyr Ala Asp Ser Val 50 55 60Lys Gly
Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn Ser Leu Tyr65 70 75
80Leu Gln Met Ser Ser Leu Arg Thr Glu Asp Thr Ala Ile Tyr Tyr Cys
85 90 95Thr Arg Asp Ile Arg Gly Asn Trp Lys Tyr Gly Gly Trp Phe Asp
Pro 100 105 110Trp Gly Gln Gly Thr Leu Val Thr Val Ser Ser 115
12030724DNAArtificial Sequencesynthetic 307ggattcacct ttgatgatta
tgcc 243088PRTArtificial Sequencesynthetic 308Gly Phe Thr Phe Asp
Asp Tyr Ala1 530924DNAArtificial Sequencesynthetic 309attagttgga
ctggtggtac tata 243108PRTArtificial Sequencesynthetic 310Ile Ser
Trp Thr Gly Gly Thr Ile1 531148DNAArtificial Sequencesynthetic
311acaagagata tccgggggaa ctggaagtac ggaggctggt tcgacccc
4831216PRTArtificial Sequencesynthetic 312Thr Arg Asp Ile Arg Gly
Asn Trp Lys Tyr Gly Gly Trp Phe Asp Pro1 5 10 15313360DNAArtificial
Sequencesynthetic 313caggtgcagc tggtgcagtc tgggactgag gtgaagaagc
ctggggcctc agtgaaggtc 60tcctgcaagg cttctggata caccttcacc gcctactata
tgcactgggt gcgacaggcc 120cctggtcaag gacttgactg gatgggatgg
atcagcccta acagtggttt cacaaactat 180gcacagaagt ttcagggcag
ggtcaccatg accagggaca cgtccatcaa cacattttat 240atggagctga
gtggactgag atctgacgac acggccgtat attactgtgc gcgagagggt
300tctactcacc acaattcttt cgacccctgg ggccagggaa ccctggtcac
cgtctcctca 360314120PRTArtificial Sequencesynthetic 314Gln Val Gln
Leu Val Gln Ser Gly Thr Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val
Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Ala Tyr 20 25 30Tyr
Met His Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Asp Trp Met 35 40
45Gly Trp Ile Ser Pro Asn Ser Gly Phe Thr Asn Tyr Ala Gln Lys Phe
50 55 60Gln Gly Arg Val Thr Met Thr Arg Asp Thr Ser Ile Asn Thr Phe
Tyr65 70 75 80Met Glu Leu Ser Gly Leu Arg Ser Asp Asp Thr Ala Val
Tyr Tyr Cys 85 90 95Ala Arg Glu Gly Ser Thr His His Asn Ser Phe Asp
Pro Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser 115
12031524DNAArtificial Sequencesynthetic 315ggatacacct tcaccgccta
ctat 243168PRTArtificial Sequencesynthetic 316Gly Tyr Thr Phe Thr
Ala Tyr Tyr1 531724DNAArtificial Sequencesynthetic 317atcagcccta
acagtggttt caca 243188PRTArtificial Sequencesynthetic 318Ile Ser
Pro Asn Ser Gly Phe Thr1 531939DNAArtificial Sequencesynthetic
319gcgcgagagg gttctactca ccacaattct ttcgacccc 3932013PRTArtificial
Sequencesynthetic 320Ala Arg Glu Gly Ser Thr His His Asn Ser Phe
Asp Pro1 5 10321342DNAArtificial Sequencesynthetic 321gaggtgcagc
tggtggagtc tggaggaggc ttggtccaac cgggggggtc cctgaggctc 60tcctgtgcag
cctctgggtt caccgtcggt actaacttca tgaattgggt ccgccaggct
120ccagggaagg ggctggagtg ggtctcagcg atttatagcg gtggtaccgc
taactacgca 180gactccgtga agggccgatt caccatttcc agagacactt
ccaggaacac gctgtatctt 240caaatgaaca gcctgagaac tgaggacacg
gccgtttatt attgtgcgag aggggggggt 300atggacgtct ggggccaagg
gaccacggtc accgtctcct ca 342322114PRTArtificial Sequencesynthetic
322Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1
5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Val Gly Thr
Asn 20 25 30Phe Met Asn Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu
Trp Val 35 40 45Ser Ala Ile Tyr Ser Gly Gly Thr Ala Asn Tyr Ala Asp
Ser Val Lys 50 55 60Gly Arg Phe Thr Ile Ser Arg Asp Thr Ser Arg Asn
Thr Leu Tyr Leu65 70 75 80Gln Met Asn Ser Leu Arg Thr Glu Asp Thr
Ala Val Tyr Tyr Cys Ala 85 90 95Arg Gly Gly Gly Met Asp Val Trp Gly
Gln Gly Thr Thr Val Thr Val 100 105 110Ser Ser32324DNAArtificial
Sequencesynthetic 323gggttcaccg tcggtactaa cttc 243248PRTArtificial
Sequencesynthetic 324Gly Phe Thr Val Gly Thr Asn Phe1
532521DNAArtificial Sequencesynthetic 325atttatagcg gtggtaccgc t
213267PRTArtificial Sequencesynthetic 326Ile Tyr Ser Gly Gly Thr
Ala1 532724DNAArtificial Sequencesynthetic 327gcgagagggg ggggtatgga
cgtc 243288PRTArtificial Sequencesynthetic 328Ala Arg Gly Gly Gly
Met Asp Val1 5329354DNAArtificial Sequencesynthetic 329caggtccagc
tggtgcagtc tggggctgag gtgaagaagc ctgggtcctc ggtgaaggtc 60tcctgcaagg
cttctggagg caccttcaac acctatgttc tcagctgggt gcgacaggcc
120cctggacaag ggcttgagtg gatgggagag atcatcccta tcttaggtgc
agcaaactac 180gcacagaact tccagggcag agtcactttt accacggacg
aatccacgaa tacagcctac 240atggacctga gcagcctaag atctgaggac
acggccgtgt attactgtgc gagagatcgg 300acctccgggg ggttcgaccc
ctggggccag ggaaccctgg tcactgtctc ctca 354330118PRTArtificial
Sequencesynthetic 330Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val
Lys Lys Pro Gly Ser1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly
Gly Thr Phe Asn Thr Tyr 20 25 30Val Leu Ser Trp Val Arg Gln Ala Pro
Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Glu Ile Ile Pro Ile Leu Gly
Ala Ala Asn Tyr Ala Gln Asn Phe 50 55 60Gln Gly Arg Val Thr Phe Thr
Thr Asp Glu Ser Thr Asn Thr Ala Tyr65 70 75 80Met Asp Leu Ser Ser
Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Arg
Thr Ser Gly Gly Phe Asp Pro Trp Gly Gln Gly Thr 100 105 110Leu Val
Thr Val Ser Ser 11533124DNAArtificial Sequencesynthetic
331ggaggcacct tcaacaccta tgtt 243328PRTArtificial Sequencesynthetic
332Gly Gly Thr Phe Asn Thr Tyr Val1 533324DNAArtificial
Sequencesynthetic 333atcatcccta tcttaggtgc agca 243348PRTArtificial
Sequencesynthetic 334Ile Ile Pro Ile Leu Gly Ala Ala1
533533DNAArtificial Sequencesynthetic 335gcgagagatc ggacctccgg
ggggttcgac ccc 3333611PRTArtificial Sequencesynthetic 336Ala Arg
Asp Arg Thr Ser Gly Gly Phe Asp Pro1 5 10337357DNAArtificial
Sequencesynthetic 337caggttcagc tggtgcagtc tggagctgag gtggagaagc
ctggggcctc agtgaaggtc 60tcctgcaagg cttctggtta catctttacc cactatggta
tcagctgggt gcgacaggcc 120cctggacaag gacttgagtg ggtgggctgg
atcagccctt acaatggtta cacagactat 180gcacagaaac tccagggcag
agtcaccttg accacagaca catccacgac cacagcctac 240atggagctga
ggaacctgag atctgacgac acggccatgt attactgttc gagagggagg
300ggcccttact ggtccttcga tctctggggc cgtggcaccc tggtcaccgt ctcctca
357338119PRTArtificial Sequencesynthetic 338Gln Val Gln Leu Val Gln
Ser Gly Ala Glu Val Glu Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser
Cys Lys Ala Ser Gly Tyr Ile Phe Thr His Tyr 20 25 30Gly Ile Ser Trp
Val Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Val 35 40 45Gly Trp Ile
Ser Pro Tyr Asn Gly Tyr Thr Asp Tyr Ala Gln Lys Leu 50 55 60Gln Gly
Arg Val Thr Leu Thr Thr Asp Thr Ser Thr Thr Thr Ala Tyr65 70 75
80Met Glu Leu Arg Asn Leu Arg Ser Asp Asp Thr Ala Met Tyr Tyr Cys
85 90 95Ser Arg Gly Arg Gly Pro Tyr Trp Ser Phe Asp Leu Trp Gly Arg
Gly 100 105 110Thr Leu Val Thr Val Ser Ser 11533924DNAArtificial
Sequencesynthetic 339ggttacatct ttacccacta tggt 243408PRTArtificial
Sequencesynthetic 340Gly Tyr Ile Phe Thr His Tyr Gly1
534124DNAArtificial Sequencesynthetic 341atcagccctt acaatggtta caca
243428PRTArtificial Sequencesynthetic 342Ile Ser Pro Tyr Asn Gly
Tyr Thr1 534336DNAArtificial Sequencesynthetic 343tcgagaggga
ggggccctta ctggtccttc gatctc 3634412PRTArtificial Sequencesynthetic
344Ser Arg Gly Arg Gly Pro Tyr Trp Ser Phe Asp Leu1 5 10






D00000

D00001

D00002

D00003

D00004

D00005

D00006
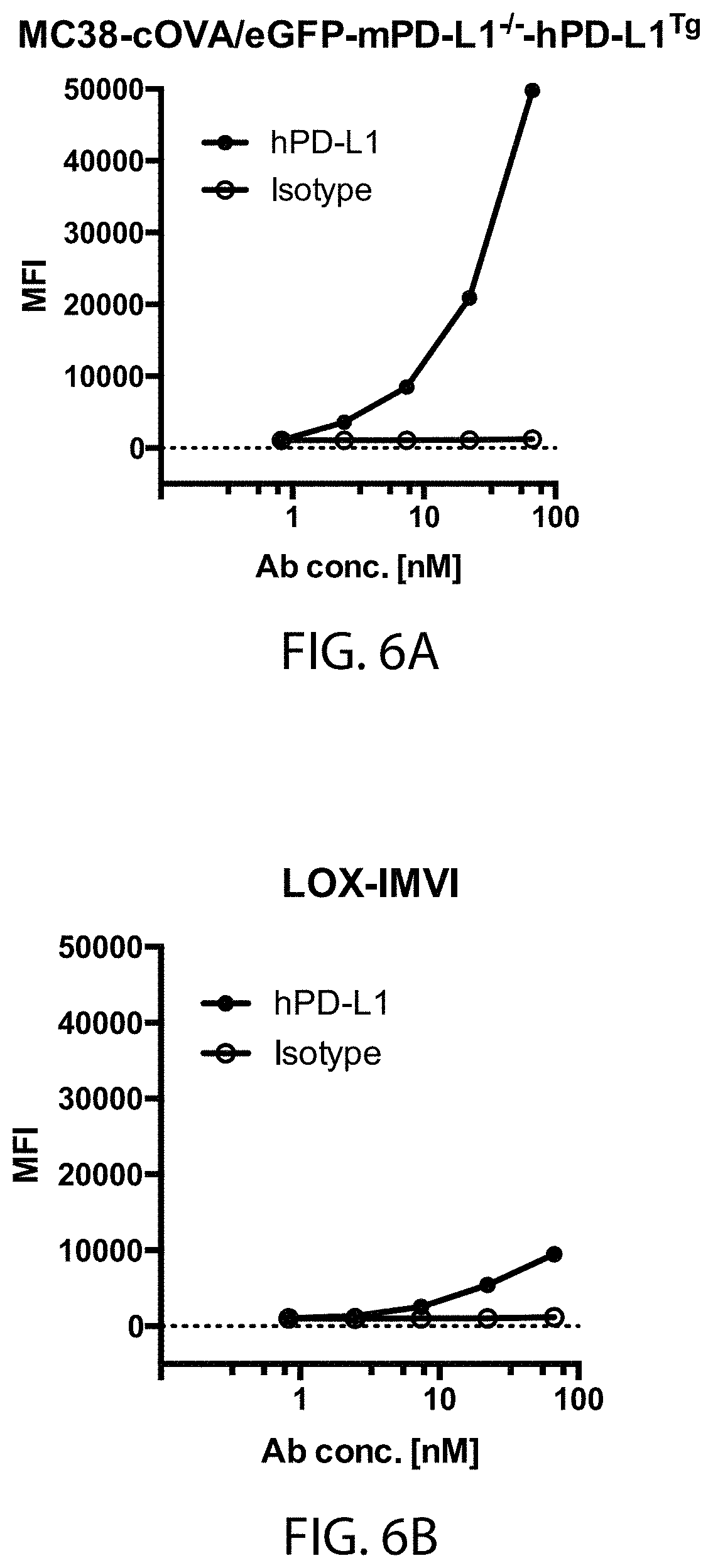
D00007

D00008
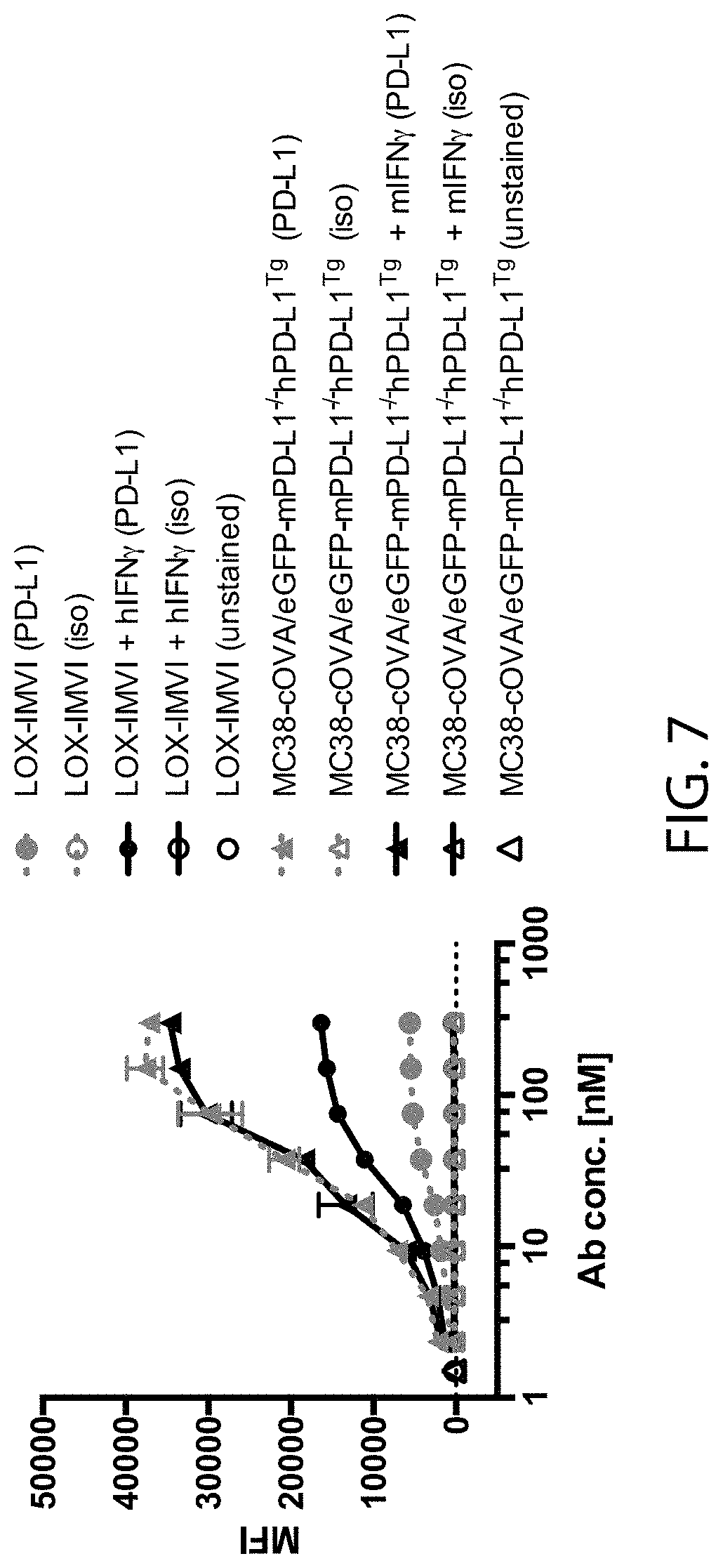
D00009
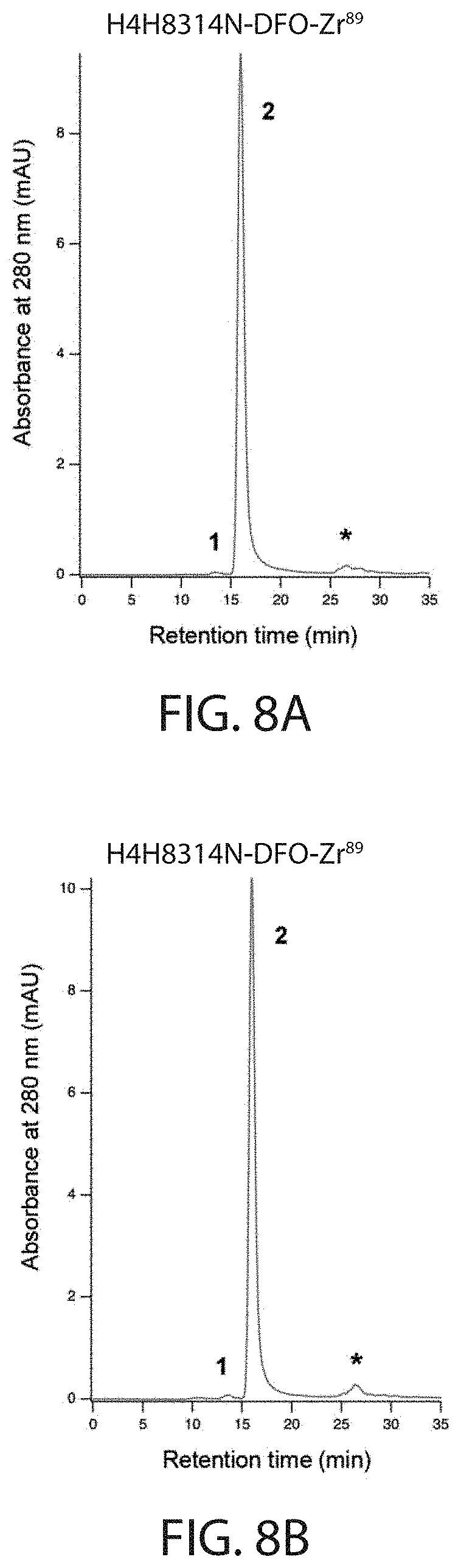
D00010
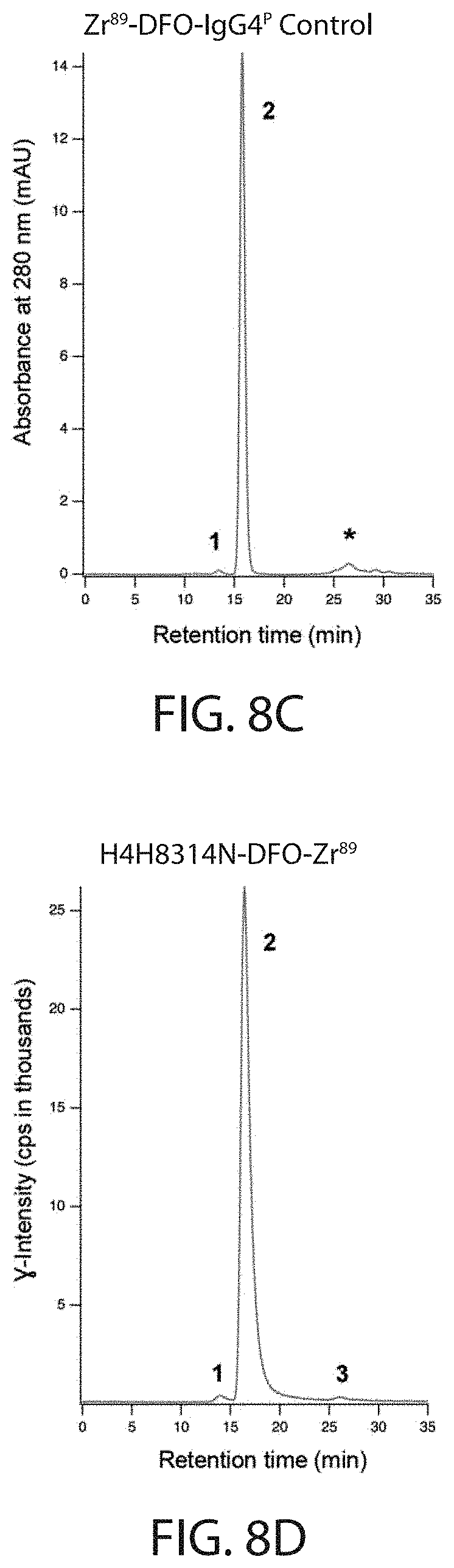
D00011
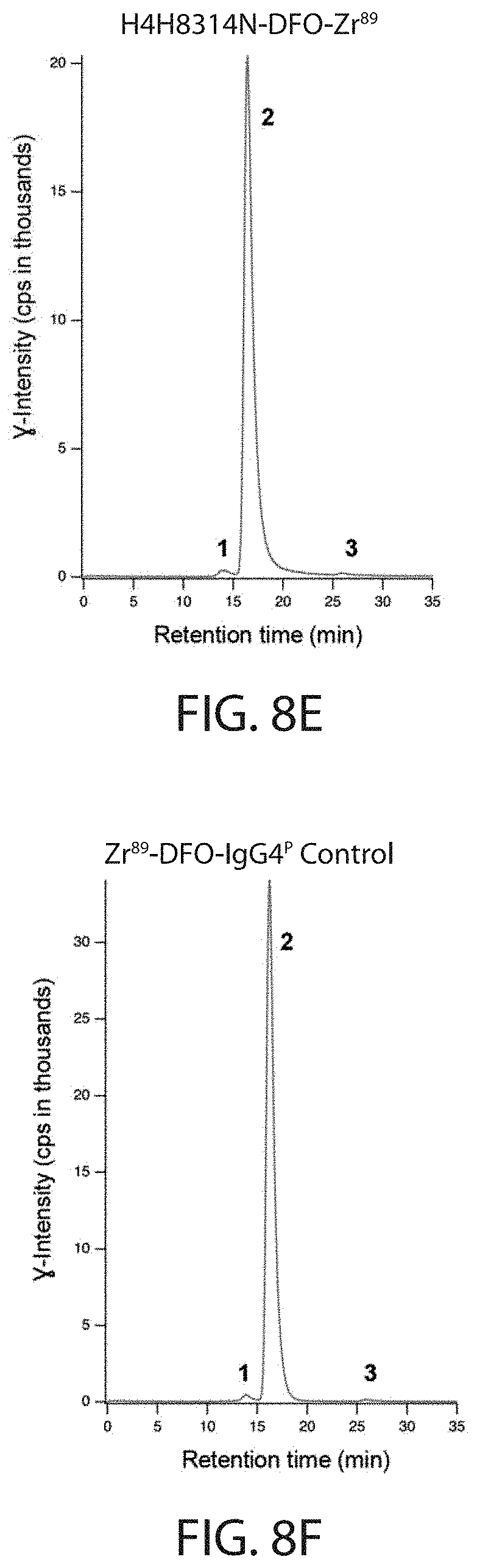
D00012
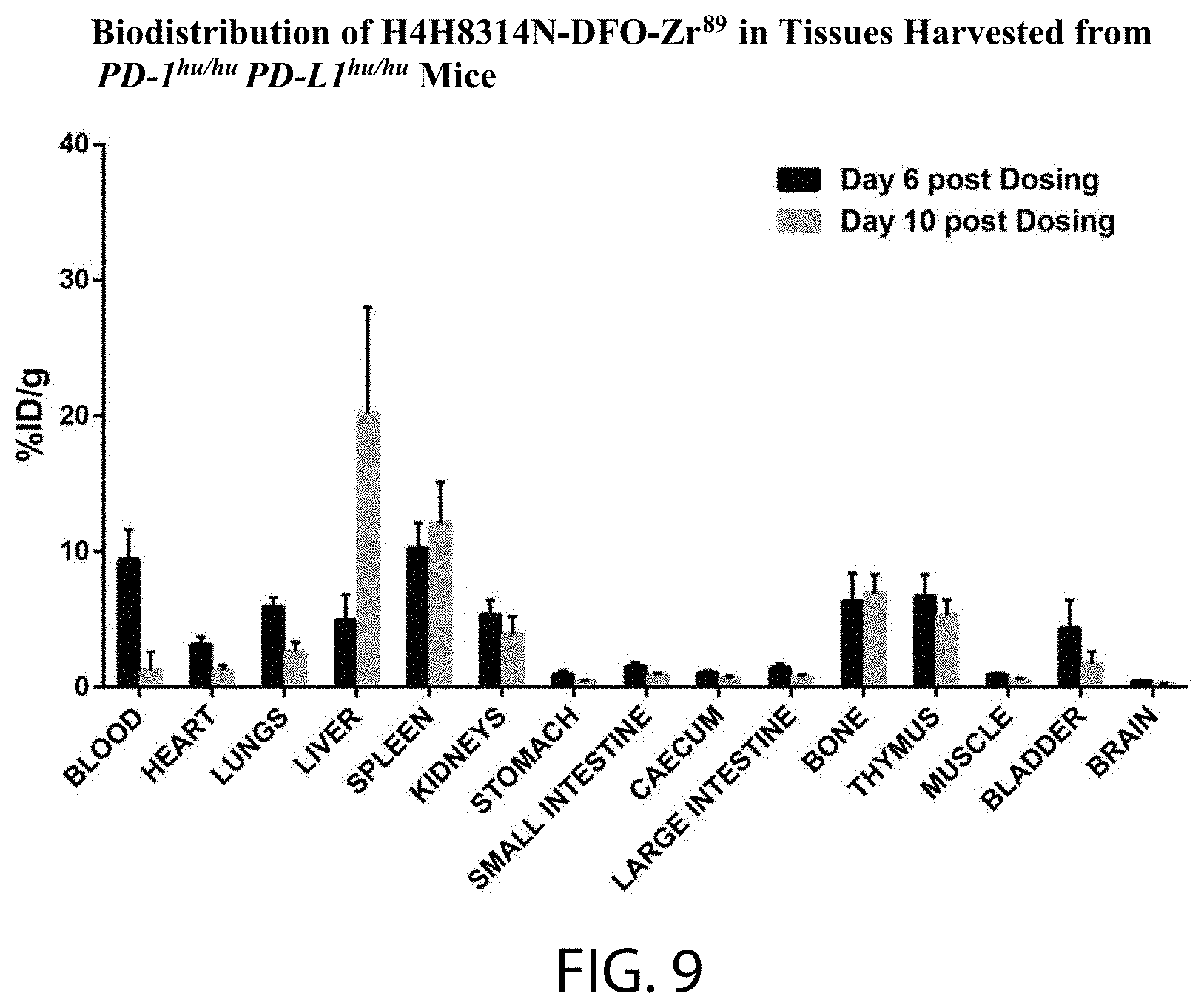
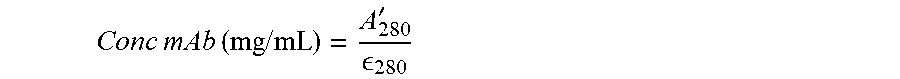
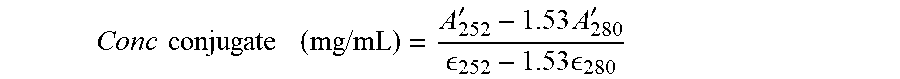
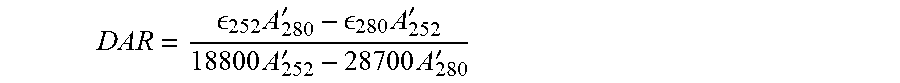
S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.