Predicting Suicidality Using A Combined Genomic And Clinical Risk Assessment
Niculescu; Alexander Bogdan
U.S. patent application number 16/779229 was filed with the patent office on 2020-10-01 for predicting suicidality using a combined genomic and clinical risk assessment. The applicant listed for this patent is Indiana University Research and Technology Corporation, United States Government as Represented by the Department of Veterans Affairs. Invention is credited to Alexander Bogdan Niculescu.
| Application Number | 20200312425 16/779229 |
| Document ID | / |
| Family ID | 1000004896758 |
| Filed Date | 2020-10-01 |












View All Diagrams
| United States Patent Application | 20200312425 |
| Kind Code | A1 |
| Niculescu; Alexander Bogdan | October 1, 2020 |
PREDICTING SUICIDALITY USING A COMBINED GENOMIC AND CLINICAL RISK ASSESSMENT
Abstract
Biomarkers and methods for screening expression levels of the biomarkers for predicting suicidality (referred herein to suicidal ideation and actions, future hospitalizations and suicide completion) are disclosed. Also disclosed are quantitative questionnaires and mobile applications for assessing affective state and for assessing socio-demographic and psychological suicide risk factors, and their use to compute scores that can predict suicidality. Finally, an algorithm that combines biomarkers and computer apps for identifying subjects who are at risk for committing suicide is disclosed, as well as methods to mitigate and prevent suicidality based on the biomarkers and computer apps.
| Inventors: | Niculescu; Alexander Bogdan; (Indianapolis, IN) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 1000004896758 | ||||||||||
| Appl. No.: | 16/779229 | ||||||||||
| Filed: | January 31, 2020 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 15735304 | Dec 11, 2017 | |||
| PCT/US2016/036985 | Jun 10, 2016 | |||
| 16779229 | ||||
| 62278707 | Jan 14, 2016 | |||
| 62174880 | Jun 12, 2015 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C12Q 1/6883 20130101; Y02A 90/10 20180101; G01N 2800/304 20130101; G01N 2800/50 20130101; G16B 25/00 20190201; G16H 50/20 20180101; G16H 50/30 20180101; C12Q 1/6876 20130101; G16B 20/00 20190201; C12Q 2600/158 20130101; G01N 33/48 20130101; C12Q 1/68 20130101; G16H 10/20 20180101; G01N 33/6893 20130101; C12Q 2600/118 20130101 |
| International Class: | G16B 20/00 20060101 G16B020/00; G16B 25/00 20060101 G16B025/00; G01N 33/48 20060101 G01N033/48; C12Q 1/68 20060101 C12Q001/68; C12Q 1/6883 20060101 C12Q001/6883; G16H 50/20 20060101 G16H050/20; G16H 10/20 20060101 G16H010/20; G16H 50/30 20060101 G16H050/30; C12Q 1/6876 20060101 C12Q001/6876; G01N 33/68 20060101 G01N033/68 |
Goverment Interests
STATEMENT OF GOVERNMENT SUPPORT
[0002] This invention was made with government support under OD007363 awarded by National Institutes of Health and 2I01CX000139 merit award by the Veterans Administration. The Government has certain rights in the invention.
Claims
1-20. (canceled)
21. A method for assessing and mitigating suicidality in a subject in need thereof, comprising: determining an expression level of a panel of biomarkers in a biological sample from the subject, computing a score for the panel, based on the gene expression data for the biomarkers in the panel, which is z-scored for each of the biomarkers in the biomarker panel with a reference database, multiplying each biomarker z-scored value by a weight coefficient related to their functional evidence of involvement in suicidality to obtain a second score for each biomarker of the biomarker panel, with the resulting values for the increased in expression (risk) biomarkers being added, and the resulting values for the decreased in expression (protective) biomarkers being subtracted, wherein when the subject is male; the panel of biomarkers comprises: (i) solute carrier family 4 (sodium bicarbonate cotransporter), member 4 (SLC4A4), cell adhesion molecule 1 CADM1, dystrobrevin, alpha (DTNA), spermidine/spermine Nl-acetyl transferase 1 (SAT1), interleukin 6 (IL-6), RAS-like family 11 member B (RASL11B), glutamate receptor, Ionotropic, kainate 2 (GRIK2), histone cluster 1, H2bo (HIST1H2BO), jun proto--oncogene (JUN), and GRB2-associated binding protein 1 (GAB1), wherein the expression level of the biomarker(s) in the sample is increased relative to a reference expression level, denoting increased suicidality; or (ii) spindle and kinetochore associated complex subunit 2 (SKA2), CAP-GLY domain containing linker protein family, member 4 (CLIP4), kinesin family member 2C (KIF2C), kelch domain containing 3 (KLHDC3), chemokine (C-C motif) ligand 28 (CCL28), v-ets avian erythroblastosis virus E26 oncogene homolog (ERG), adenylate kinase 2 (AK2), myelin basic protein (MBP), and fatty acid desaturase 1 (FADS1), wherein the expression level of the biomarker(s) in the sample is decreased relative to a reference expression level, denoting increased suicidality; or wherein when the subject is female, and the panel of biomarkers comprises: (i) erythrocyte membrane protein band 4.1 like 5 (EPB41L5), HtrA serine peptidase 1 (HTRA1), deleted in primary ciliary dyskinesia homolog (DPCD), general transcription factor IIIC (GTF3C3), period circadian clock 1 (PERI), pyridoxal-dependent decarboxylase domain containing 1 (PDXDC1), kelch-like family member 28 (KLHL28), ubiquitin interaction motif containing 1 (UIMC1), sorting nexin family member 27 (SNX27), glutamate receptor ionotropic kainate 2 (GRIK2), wherein the expression level of the biomarker(s) in the sample is increased relative to a reference expression level, denoting increased suicidality; or (ii) phosphatidylinositol 3-kinase, catalytic subunit type 3 (PIK3C3), aldehyde dehydrogenase 3 family member A2 (ALDH3A2), ARP3 actin-related protein 3 homolog (yeast) (ACTR3), B-cell CLL (BCL2), MOB kinase activator 3B (MOB3B), casein kinase 1 alpha 1 (CSNK1A1), La ribonucleoprotein domain family member 4 (LARP4), zinc finger protein 548 (ZNF548), prolylcarboxypeptidase (angiotensinase C) (PRCP), and solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter) member B3 (SLC35B3), wherein the expression level of the biomarker(s) in the sample is decreased relative to a reference expression level, denoting increased suicidality; determining a reference score for the panel, obtained in a clinically relevant population identifying a difference between the score of the panel of biomarker(s) in the sample and the reference score of the panel of biomarker(s); and identifying the subject having suicidality based on the difference between the biomarker panel score of the subject relative to the biomarker panel score of reference; and administering to the subject identified as having suicidality a specific therapeutic drug(s) to treat suicidality, based on the specific biomarkers that are changed in the subject wherein the therapeutic drug (s) is selected from: (i) a group of psychiatric treatments: ketamine and other dissociants, lithium and other mood stabilizers, clozapine, chlorpromazine, prochlorperazine, and other antipsychotics, selegeline, fluoxetine, trimipramine, and other antidepressants, docosahexaenoic acid and other omega-3 fatty acids, and combinations thereof; or (ii) a group of new method of use/repurposed drugs consisting of: tocilizumab, tenoxicam, ramifenazone, and other anti-inflammatories; betulin, dl-alpha tocopherol, hesperidin, calcium folinate, harpagoside, rilmenidine, harman, homatropine, diphenhydramine, pirenperone, asiaticoside, adiphenine, metformin, chlorogenic acid, verapamil, metaraminol, yohimbine, trimethadione, and combinations thereof.
22. The method of claim 21, wherein before the step of generating the biomarker panel score, each biomarker is given a weighted coefficient, wherein the weighted coefficient is related to the importance of said each biomarker in assessing and predicting suicide risk.
23. The method of claim 21, wherein the biological sample is a peripheral tissue sample or a fluid.
24. The method of claim 21, wherein biomarker expression level measures RNA or protein of the biomarker in the biological sample.
25. The method of claim 21, wherein the subject is male, and the drug is selected based on the specific biomarkers that are changed in expression in the subject, and is selected from the group consisting of: thiamine, homatropine, vitexin, ergocalciferol, tropicamide, (-)-atenolol, haloperidol, spaglumic acid, and combinations thereof.
26. The method of claim 21, wherein the subject is female, and the drug is selected based on the specific biomarkers that are changed in expression in the subject, and is selected from the drug group consisting of: mifepristone, lansoprazole, nafcillin, betulin, and combinations thereof.
27. The method of claim 21, wherein the subject has a psychiatric disorder selected from the group consisting of: bipolar disorder, major depressive disorder, schizophrenia, schizoaffective disorder, anxiety disorders, post-traumatic stress disorder, and combinations thereof.
28. A method of assessing and mitigating suicidality in a subject in need thereof, comprising: calculating an Up-Suicide Scorebased on the equation: (Biomarker Panel Score)+(Suicidality Risk Score)+(Mood Score)+(Anxiety Score)=Up-Suicide Score; wherein the Biomarker Panel Score is obtained as per the method of claim 21; wherein the Suicidality Risk Score is calculated by (i) summing the binary results of the individual items in the CFI-S scale; wherein a yes/present answer generates a score of 1 and a no/absent answer generates a score of zero; and (ii) dividing the summed score by the number of items answered and multiplying by 100; wherein the individual items in the CFI-S scale are: lack of coping skills (cracks under pressure); dissatisfaction with present life; lack of hope for the future; current substance abuse; acute stresses: losses, grief; chronic stress: lack of positive relationships, social isolation; acute stress: rejection, history of excessive extroversion and impulsive behaviors (including rage, anger, physical fights, seeking revenge); acute/severe medical illness, pain; lack of children; Gender: Male; Personally knowing somebody who committed suicide; Psychiatric illness diagnosed and treated; past history of suicidal acts/gestures; Age: Older>60 or Younger<25; History of abuse: physical, sexual, emotional, neglect; History of command hallucinations of self-directed violence; Family history of suicide in blood relatives; With poor treatment compliance; Lack of religious beliefs; History of excessive introversion, conscientiousness; Chronic stress: perceived uselessness, not feeling needed, burden to extended kin; wherein the Mood Score is calculated by using a mood-rating scale; wherein the Anxiety Score is calculated by using an anxiety-rating scale; assessing the level of suicidality of the subject by comparing the subject's Up-Suicide Score to a reference Up-Suicide Score; administering a treatment for suicidality to the subject when the subject's Up-Suicide Score is greater than a reference Up-Suicide Score; and monitoring the subject's response to a treatment for suicidality by determining changes in the Up-Suicide Score after initiating a treatment.
29. The method of claim 28, wherein the method further comprises receiving, in a computer system, Biomarker Panel Score, Suicidality Risk Score, Mood Score, Anxiety Score, and/or Up-Suicide Score for the subject, the computer system comprising a database, wherein the database comprises a plurality of suicidality treatment profiles.
30. The method of claim 29, wherein the method further comprises a step of outputting from the computer system the identity of the suicidality treatment for administering to the subject.
31. The method of claim 28, wherein a user enters the Biomarker Panel Score, Suicidality Risk Score, Mood Score, Anxiety Score, and/or Up-Suicide Score of the subject in the computer system.
32. The method of claim 28, wherein the Biomarker Panel Score, Suicidality Risk Score, Mood Score, Anxiety Score, and/or Up-Suicide Score of the subject is received directly from equipment used in determining the subject's suicidality blood biomarker score.
33. The method of claim 28, wherein the Biomarker Panel Score, Suicidality Risk Score, Mood Score, and Anxiety Score of the subject are z-scored prior to the calculation of the Up-Suicide Score.
34. The method of claim 28, wherein the subject is male, the panel of biomarkers is (i) solute carrier family 4 (sodium bicarbonate cotransporter) member 4 (SLC4A4), cell adhesion molecule 1 CADM1, dystrobrevin alpha (DTNA), spermidine/spermine Nl-acetyl transferase 1 (SAT1), interleukin 6 (IL-6), RAS-like family 11 member B (RASL11B), glutamate receptor ionotropic kainate 2 (GRIK2), histone cluster 1 H2bo (HIST1H2BO), jun proto--oncogene (JUN), and GRB2-associated binding protein 1 (GAB1), wherein the expression level of the biomarker(s) in the sample is increased relative to a reference expression level, denoting increased suicidality; or (ii) spindle and kinetochore associated complex subunit 2 (SKA2), CAP-GLY domain containing linker protein family, member 4 (CLIP4), kinesin family member 2C (KIF2C), kelch domain containing 3 (KLHDC3), chemokine (C--C motif) ligand 28 (CCL28), v-ets avian erythroblastosis virus E26 oncogene homolog (ERG), adenylate kinase 2 (AK2), myelin basic protein (MBP), and fatty acid desaturase 1 (FADS1), wherein the expression level of the biomarker(s) in the sample is decreased relative to a reference expression level, denoting increased suicidality; or wherein when the subject is female, the panel of biomarkers is (i) erythrocyte membrane protein band 4.1 like 5 (EPB41L5), HtrA serine peptidase 1 (HTRA1), deleted in primary ciliary dyskinesia homolog (DPCD), general transcription factor IIIC polypeptide 3 (GTF3C3), period circadian clock 1 (PERI), pyridoxal-dependent decarboxylase domain containing 1 (PDXDC1), kelch-like family member 28 (KLHL28), ubiquitin interaction motif containing 1 (UIMC1), sorting nexin family member 27 (SNX27), glutamate receptor ionotropic kainate 2 (GRIK2), wherein the expression level of the biomarker(s) in the sample is increased relative to a reference expression level, denoting increased suicidality; or (ii) phosphatidylinositol 3-kinase, catalytic subunit type 3 (PIK3C3), aldehyde dehydrogenase 3 family member A2 (ALDH3A2), ARP3 actin-related protein 3 homolog (yeast) (ACTR3), B-cell CLL (BCL2), MOB kinase activator 3B (MOB3B), casein kinase 1 alpha 1 (CSNK1A1), La ribonucleoprotein domain family member 4 (LARP4), zinc finger protein 548 (ZNF548), prolylcarboxypeptidase (PRCP), solute carrier family 35 member B3 (SLC35B3), wherein the expression level of the biomarker(s) in the sample is decreased relative to a reference expression level, denoting increased suicidality.
35. A method of assessing and mitigating suicidality in a subject in need thereof, comprising: calculating a Suicidality Risk Score by adding the score of the individual items in the CFI-S scale, wherein a yes/present answer generates a score of 1 and a no/absent answer generates a score of zero, and dividing the summed score by the number of items answered and multiplying by 100, wherein the individual items in the CFI-S scale are: lack of coping skills (cracks under pressure); dissatisfaction with present life; lack of hope for the future; current substance abuse; acute stresses: losses, grief; chronic stress: lack of positive relationships, social isolation; acute stress: rejection, history of excessive extroversion and impulsive behaviors (including rage, anger, physical fights, seeking revenge); acute/severe medical illness, pain; lack of children; Gender: Male; Personally knowing somebody who committed suicide; Psychiatric illness diagnosed and treated; past history of suicidal acts/gestures; Age: Older>60 or Younger<25; History of abuse: physical, sexual, emotional, neglect; History of command hallucinations of self-directed violence; Family history of suicide in blood relatives; With poor treatment compliance; Lack of religious beliefs; History of excessive introversion, conscientiousness; Chronic stress: perceived uselessness, not feeling needed, burden to extended kin; assessing the level of suicidality of the subject by comparing the subject's Suicidality Risk Score to a reference Suicidality Risk Score; administering a treatment for suicidality to the subject when the subject's Suicidality Risk Score is greater than a reference Suicidality Risk Score; monitoring the response to a treatment of the subject by determining changes in the Suicidality Risk Score after initiation of a treatment; and decreasing the Suicidality Risk Score by targeting with psycho-social interventions and other treatments specific items of the CFI-S that are scored as yes/present in a particular subject.
36. The method of claim 35, wherein the method further comprises receiving, in a computer system, the scores of the individual items in the CFI-S scale or the subject's Suicidality Risk Score, the computer system comprising a database, wherein the database comprises a plurality of suicidality treatment profiles.
37. The method of claim 35, wherein the method further comprises a step of outputting from the computer system the identity of the targeted suicidality psycho-social intervention and other treatments for administering to the subject.
38. The method of claim 36, wherein a user enters the scores of the individual items in the CFI-S scale or subject's Suicidality Risk Score in the computer system.
39. The method of claim 36, wherein the scores of the individual items in the CFI-S scale or the subject's Suicidality Risk Score is received directly from equipment used in determining the subject's suicidality blood biomarker score.
40. The method of claim 35, wherein the subject's Suicidality Risk Score is z-scored along the Suicidality Risk Scores of subjects with similar Suicidality Risk Scores.
Description
CROSS REFERENCE TO RELATED APPLICATIONS
[0001] This application is a Continuation Application and claims priority to U.S. application Ser. No. 15/735,304 filed Dec. 11, 2017, which is the U.S. National Stage Application of International Patent Application PCT/US2016/036985, filed Jun. 10, 2016, which claims priority to and the benefit of U.S. Provisional Provisional Application No. 62/278,707, filed Jan. 14, 2016, and U.S. Provisional Application No. 62/174,880, filed on Jun. 12, 2015, the disclosures of which are hereby incorporated by reference in their entireties.
BACKGROUND OF THE DISCLOSURE
[0003] The present disclosure relates generally to biomarkers and their use for predicting a subject's risk of suicidality (e.g., suicide ideation and actions, future hospitalization due to suicidality, and suicide completion). More particularly, the present disclosure relates to gene expression biomarkers, and to methods of screening for biomarkers, for identifying subjects who are at risk of committing suicide, as well as for preventing and treating subjects for suicidality. The present disclosure further relates to quantitative clinical information assessments through questionnaires and mobile applications (referred to herein as "apps") for assessing affective state (mood and anxiety), for assessing socio-demographic and psychological suicide risk factors, and for identifying subjects who are at risk of committing suicide. Finally, the present disclosure relates to an algorithm for combining biomarkers and apps for identifying subjects who are at risk for committing suicide.
[0004] Suicide is a leading cause of death in psychiatric patients, and in society at large. Particularly, suicide accounts for one million deaths worldwide each year. Worldwide, one person dies every 40 seconds through suicide, a potentially preventable cause of death. Further, although women have a lower rate of suicide completion as compared to men, due in part to the less-violent methods used, women have a higher rate of suicide attempts. A limiting step in the ability to intervene is the lack of objective, reliable predictors. One cannot just ask individuals if they are suicidal, as the desire to not be stopped or future impulsive changes of mind may make their self-report of feelings, thoughts and plans unreliable.
[0005] There are currently no objective tools to assess and track changes in suicidal risk without asking the subjects directly. Such tools, however, could prove substantially advantageous as the subjects at risk often choose not to share their suicidal ideation or intent with others, for fear of stigma, hospitalization, or that their plans will be thwarted. The ability to assess and track changes in suicidal risk without asking a subject directly would further allow for intervening prior to suicide attempt and suicide completion by the subject.
[0006] Conventionally, a convergence of methods assessing the subject's internal subjective feelings and thoughts, along with external, more objective, ratings of actions and behaviors, are used de facto in clinical practice, albeit not in a formalized and systematic way. Accordingly, there exists a need to develop more quantitative and objective ways for predicting and tracking suicidal states. More particularly, it would be advantageous if objective tools and screening methods could be developed for determining expression levels of biomarkers to allow for determining suicidal risk and other psychotic depressed mood states, as well as monitoring a subject's response to treatments for lessening suicidal risk. The ability to assess and track changes in suicidal risk without asking a subject directly would further allow for intervening prior to suicide attempt and suicide completion by the subject.
BRIEF DESCRIPTION OF THE DISCLOSURE
[0007] The present disclosure is generally related to predicting state (suicidal ideation) and trait--future psychiatric hospitalizations for suicidality. The methods described herein increase the predictive accuracy for specifically identifying subjects who are at risk for committing suicide and for predicting future hospitalization due to suicidality. In one particular aspect, the methods described herein increase the predictive accuracy for specifically identifying subjects who are at risk for committing suicide and for predicting future hospitalization due to suicidality.
[0008] In one aspect, the present disclosure is directed to a method for predicting suicidality in a subject. The method comprises: obtaining an expression level of a blood biomarker in a sample obtained from the subject; obtaining a reference expression level of a blood biomarker; and identifying a difference between the expression level of the blood biomarker in a sample obtained from the subject and the reference expression level of a blood biomarker, wherein the difference in the expression level of the blood biomarker in the sample obtained from the subject and the reference expression level of the blood biomarker indicates a risk for suicide.
[0009] In another aspect, the present disclosure is directed to a method for mitigating suicidality in a subject in need thereof. The method comprises: obtaining an expression level of a blood biomarker in a sample obtained from the subject; obtaining a reference expression level of the blood biomarker; identifying a difference in the expression level of the blood biomarker in the sample and the reference expression level of the blood biomarker; and administering a treatment, wherein the treatment reduces the difference between the expression level of the blood biomarker in the sample and the reference expression level of the blood biomarker to mitigate suicidality in the subject.
[0010] In another aspect, the present disclosure is directed to a computer-implemented method for assessing mood, anxiety, and combinations thereof in the subject using a computer-implemented method for assessing mood, anxiety, and combinations thereof, the method implemented using a first computer device coupled to a memory device, the method comprising: receiving mood information, anxiety information, and combinations thereof into the first computer device; storing, by the first computer device, the mood information, anxiety information, and combinations thereof in the memory device; presenting, by the first computer device, in visual form the mood information, anxiety information, and combinations thereof to a second computer device; receiving a request from the second computer device for access to the mood information, anxiety information, and combinations thereof; and transmitting, by the first computer device, the mood information, anxiety information, and combinations thereof to the second computer device to assess mood, anxiety, and combinations thereof in the subject.
[0011] In another aspect, the present disclosure is directed to a computer-implemented method for assessing socio-demographic/psychological suicidal risk factors in the subject using a computer-implemented method for assessing socio-demographic/psychological suicidal risk factors in the subject, the method implemented using a first computer device coupled to a memory device, the method comprising: receiving socio-demographic/psychological suicidal risk factor information into the first computer device; storing, by the first computer device, the socio-demographic/psychological suicidal risk factor information in the memory device; presenting, by the first computer device, in visual form the socio-demographic/psychological suicidal risk factor information to a second computer device; receiving a request from the second computer device for access to socio-demographic/psychological suicidal risk factor information; and transmitting, by the first computer device, the socio-demographic/psychological suicidal risk factor information to the second computer device to assess the socio-demographic/psychological suicidal risk factors in the subject.
[0012] In one aspect, the present disclosure is directed to a method for predicting suicidality in a subject. The method comprises: identifying a difference in the expression level of a blood biomarker in a sample obtained from a subject and a reference expression level of the blood biomarker by obtaining the expression level of the blood biomarker in a sample obtained from a subject; obtaining a reference expression level of a blood biomarker; analyzing the blood biomarker in the sample obtained from the subject and the reference expression level of the blood biomarker to detect the difference between the blood biomarker in the sample and the reference expression level of the blood biomarker; assessing mood, anxiety, and combinations thereof in the subject, using a first computer device coupled to a memory device, wherein the first computer device receives mood information, anxiety information, and combinations thereof into the first computer device; storing, by the first computer device, the mood information, anxiety information, and combinations thereof in the memory device; computing, by the first computer device, of the mood information, anxiety information, and combinations thereof, a score that can be used to predict suicidality; presenting, by the first computer device, in visual form the mood information, anxiety information, and combinations thereof to a second computer device; receiving a request from the second computer device for access to the mood information, anxiety information, and combinations thereof; and transmitting, by the first computer device, the mood information, anxiety information, and combinations thereof to the second computer device to assess mood, anxiety, and combinations thereof in the subject; assessing socio-demographic/psychological suicidal risk factors in the subject using the first computer device coupled to a memory device, wherein the first computer device receives socio-demographic/psychological suicidal risk factor information into the first computer device; storing, by the first computer device, the socio-demographic/psychological suicidal risk factor information in the memory device; computing, by the first computer device, of the socio-demographic/psychological suicidal risk factor information, a score that can be used to predict suicidality; presenting, by the first computer device, in visual form the socio-demographic/psychological suicidal risk factor information to the second computer device; receiving a request from the second computer device for access to socio-demographic/psychological suicidal risk factor information; and transmitting, by the first computer device, the socio-demographic/psychological suicidal risk factor information to the second computer device to assess the socio-demographic/psychological suicidal risk factors in the subject; and predicting suicidality in the subject by the combination of the difference between the expression level of the biomarker in the subject and the reference expression level of the blood biomarker; the assessment of mood, anxiety, and combinations thereof; and the assessment of socio-demographic/psychological suicidal risk factor information.
BRIEF DESCRIPTION OF THE DRAWINGS
[0013] The disclosure will be better understood, and features, aspects and advantages other than those set forth above will become apparent when consideration is given to the following detailed description thereof. Such detailed description makes reference to the following drawings, wherein:
[0014] FIGS. 1A-1C depict the Discovery cohort of Example 1: longitudinal within subject analysis. Phchp### is the study ID for each participant. V# denotes visit number (1, 2, 3, 4, 5, or 6). FIG. 1A depicts suicidal ideation (SI) scoring. FIG. 1B depicts subjects and visits. FIG. 1C depicts PhenoChipping: two-way unsupervised hierarchical clustering of all participant visits in the discovery cohort vs. 18 quantitative phenotypes measuring affective state and suicidality. SASS--Simplified Affective State Scale. A--Anxiety items (Anxiety, Uncertainty, Fear, Anger, Average). M--Mood items (Mood, Motivation, Movement, Thinking, Self-esteem, Interest, Appetite, Average). STAI-STATE is State Trait Anxiety Inventory, State Subscale. YMRS is Young Mania Rating Scale.
[0015] FIGS. 2A-2C depict the Biomarker Discovery, Prioritization and Validation of Example 1. FIG. 2A depicts Discovery--number of probe sets carried forward from the AP and DE analyses, with an internal score of 1 and above. Underline-increased in expression in High SI, bold-decreased in expression in High SI. FIG. 2B depicts Prioritization--CFG integration of multiple lines of evidence to prioritize suicide-relevant genes from the discovery step. FIG. 2C depicts Validation--Top CFG genes validated in the cohort of suicide completers, with a total score of 4 and above. All the genes shown were significantly changed in ANOVA from No SI to High SI to Suicide Completers. *survived Bonferroni correction. SAT1 (.times.3) had three different probe sets with the same total score of 8.
[0016] FIGS. 3A-3C depict the Convergent Functional Information for Suicide (CFI-S) Scale as analyzed in Example 1. FIG. 3A depicts Validation of scale. CFI-S levels in the Discovery Cohort and Suicide Completers. FIG. 3B depicts Validation of items. CFI-S was developed independently of any data from this Example by compiling known socio-demographic and clinical risk factors for suicide. It is composed of 22 items that assess the influence of mental health factors, as well as of life satisfaction, physical health, environmental stress, addictions, cultural factors known to influence suicidal behavior, and two demographic factors, age and gender. These 22 items are shown here validated in the discovery cohort and suicide completers in a manner similar to that for biomarkers. Additionally, a student's t-test was used to evaluate items that were increased in suicide completers when compared to living participants with high suicidal ideation. FIG. 3C depicts CFI-S predictions for suicidal ideation in the independent test cohort and predicting future hospitalizations due to suicidality.
[0017] FIGS. 4A & 4B depict the testing of Universal Predictor for Suicide (UP-Suicide). UP-Suicide is a combination of the best genomic data (top increased and decreased biomarkers from discovery and prioritization by CFG, and validation in suicide completers), and phenomic data (CFI-S and SASS). The graph in FIG. 4A depicts Area Under the Curve (AUC) for the UP-Suicide predicting suicidal ideation and hospitalizations within the first year in all participants, as well as separately in bipolar (BP), major depressive disorder (MDD), schizophrenia (SZ), and schizoaffective (SZA) participants. Two asterisks indicate the comparison survived Bonferroni correction for multiple comparisons. A single asterisk indicates nominal significance of p<0.05. Bold outline indicates that the UP-Suicide was synergistic to its components, i.e. performed better than the gene expression or phenomic markers individually. The table in FIG. 4B summarizes descriptive statistics for all participants together, as well as separately in BP, MDD, SZ, and SZA. Bold indicates the measure survived Bonferroni correction for 200 comparisons (20 genomic and phenomic markers/combinations.times.2 testing cohorts for SI and future hospitalizations in the first year.times.5 diagnostic categories--all, BP, MDD, SZA, SZ). Pearson correlation data in the suicidal ideation test cohort is shown for HAMD-SI vs. UP-Suicide, as well as Pearson correlation data in the hospitalization test cohort for frequency of hospitalizations for suicidality in the first year, and for frequency of hospitalizations for suicidality in all future available follow-up intervals (that varies among subjects, from 1 year to 8.5 years).
[0018] FIGS. 5A-5C depict prediction of Suicidal Ideation by UP-Suicide. The graph in FIG. 5A (top left) depicts Receiver operating curve identifying participants with suicidal ideation against participants with No SI or intermediate SI. The graph in FIG. 5A (top right) depicts suicidal ideation prediction. The Y axis contains the average UP-suicide scores with standard error for no SI, intermediate SI, and high SI. The graph in FIG. 5A (bottom right) is a Scatter plot depicting HAMD-SI score on the Y-axis and UP-Suicide score on the X axis with linear trendline. The table in FIG. 5B summarizes the descriptive statistics. ANOVA was performed between groups with no SI, intermediate SI, and high SI. FIG. 5C depicts the number of subjects correctly identified in the test cohort by categories based on thresholds in the discovery cohort. Category 1 means within 1 standard deviation above the average of High SI subjects in the discovery cohort, Category 2 means between 1 and 2 standard deviations above, and so on. Category -1 means within 1 standard deviation below the average of the No SI subjects in the discovery cohort, Category -2 means between 1 and 2 standard deviations below, and so on.
[0019] FIG. 6 depicts the Simplified Affective State Scale (SASS) questionnaire for measuring mood and anxiety.
[0020] FIGS. 7A & 7B depict a screen image of the SASS mobile app (FIG. 7A) and CFI-S mobile app (FIG. 7B).
[0021] FIGS. 8A & 8B summarize biological pathways and diseases as analyzed in Example 1.
[0022] FIG. 9 is a table summarizing the top biomarkers for all diagnoses, the top biomarkers for bipolar disorder, the top biomarkers for depression, the top biomarkers for schizoaffective disorder, and the top biomarkers for schizophrenia as analyzed in Example 1.
[0023] FIGS. 10A-10C depict biomarker discovery as analyzed in Example 2. Discovery cohort: longitudinal within-participant analysis. Phchp### is study ID for each participant. V# denotes visit number (1, 2, 3, 4, 5, or 6). FIG. 10A depicts suicidal ideation (SI) scoring. FIG. 10B depicts participants and visits. FIG. 10C depicts PhenoChipping: two-way unsupervised hierarchical clustering of all participant visits in the discovery cohort vs. 18 quantitative phenotypes measuring affective state and suicidality. SASS--Simplified Affective State Scale. A--Anxiety items (Anxiety, Uncertainty, Fear, Anger, Average). M--Mood items--Mood, Motivation, Movement, Thinking, Self-esteem, Interest, Appetite, Average). STAI-STATE is State Trait Anxiety Inventory, State Subscale. YMRS is Young Mania Rating Scale.
[0024] FIGS. 11A-11C depict biomarker prioritization and validation as analyzed in Example 2. FIG. 11A depicts Discovery--number of probesets carried forward from the AP and DE analyses, with an internal score of 1 and above. Underline-increased in expression in High SI, bold--decreased in expression in High SI. FIG. 11B depicts the Prioritization--CFG integration of multiple lines of evidence to prioritize suicide--relevant genes from the discovery step. FIG. 11C depicts Validation--Top CFG genes, with a total score of 4 and above, validated in the cohort of suicide completers. All the genes shown were significantly changed and survived Bonferroni correction in ANOVA from No SI to High SI to Suicide Completers. Some genes with (x n) after the symbol had multiple different probesets with the same total score.
[0025] FIGS. 12A & 12B depict Convergent Functional Information for Suicide (CFI-S) Scale as analyzed in Example 2. CFI-S was developed independently of any data from this Example, by compiling known socio-demographic and clinical risk factors for suicide. It is composed of 22 items that assess the influence of mental health factors, as well as of life satisfaction, physical health, environmental stress, addictions, cultural factors known to influence suicidal behavior, and two demographic factors, age and gender. FIG. 12A depicts testing of scale in females. Prediction of high suicidal ideation in females in a larger cohort that combines the discovery and test cohorts used for biomarker work. The table depicts individual items and their ability to differentiate between No SI and High SI. FIG. 12B depicts testing of the scale in males, in a larger cohort that combines the discovery and test cohorts used for the biomarker work in Example 1. The table depicts individual items and their ability to differentiate between No SI and High SI.
[0026] FIGS. 13A & 13B depict UP-Suicide predictions of suicidal ideation in the independent test cohort, and predicting future hospitalizations due to suicidality as analyzed in Example 2. FIG. 13A (Top left) depicts receiver operating curve identifying participants with suicidal ideation against participants with No SI or intermediate SI; (Top right): Y axis contains the average UP-Suicide scores with standard error of mean for no SI, intermediate SI, and high SI; (Bottom right): Scatter plot depicting HAMD-SI score on the Y-axis and UP-Suicide score on the X axis with linear trend line; and (Bottom Table) summarizes descriptive statistics. FIG. 13B (Top left) depicts receiver operating curve identifying participants with future hospitalizations due to suicidality against participants without future hospitalizations due to suicidality; (Top right): Y axis contains the average UP-Suicide scores with standard error of mean for no future hospitalizations due to suicidality and participants with future hospitalizations due to suicidality; (Bottom right): Scatter plot depicting frequency of future hospitalizations due to suicidality on the Y-axis and UP-Suicide score on the X axis with linear trend line; and (Bottom Table) summarizes descriptive statistics.
[0027] FIG. 14 is a table depicting the cohorts used in Example 2.
[0028] FIG. 15 is a table depicting biological pathways and diseases as analyzed in Example 2.
[0029] FIG. 16 is a table depicting UP-suicide predictions as analyzed in Example 2. UP-Suicide is composed of 50 validated biomarkers (18 increased in expression, 32 decreased in expression), along with clinical measures app scores (CFI-S, SASS). SASS is composed of Mood scale and Anxiety scale.
[0030] FIG. 17 depicts convergent functional information for suicide (CFI-S) App testing across genders. Prediction of high suicidal ideation in men and women in a larger cohort that combines the cohorts used in Examples 1 and 2 by gender. CFI-S was developed independently of any data from this disclosure, by compiling known socio-demographic and clinical risk factors for suicide. It is composed of 22 items that assess the influence of mental health factors, as well as of life satisfaction, physical health, environmental stress, addictions, cultural factors known to influence suicidal behavior, and two demographic factors, age and gender. The table depicts individual items and their ability to differentiate between No Suicidal Ideation and High Suicidal Ideation. These items provide clinical predictors and targets for psycho-therapeutic intervention.
[0031] FIG. 18 depicts convergent functional information for future hospitalization for suicide (CFI-S) App testing across genders. Particularly, prediction of future hospitalizations for suicidality in men and women in a larger cohort that combines the cohorts used in our studies by gender.
[0032] While the disclosure is susceptible to various modifications and alternative forms, specific embodiments thereof have been shown by way of example in the drawings and are herein described below in detail. It should be understood, however, that the description of specific embodiments is not intended to limit the disclosure to cover all modifications, equivalents and alternatives falling within the spirit and scope of the disclosure as defined by the appended claims.
DETAILED DESCRIPTION
[0033] Unless defined otherwise, all technical and scientific terms used herein have the same meaning as commonly understood by one of ordinary skill in the art to which the disclosure belongs. Although any methods and materials similar to or equivalent to those described herein can be used in the practice or testing of the present disclosure, the preferred methods and materials are described below.
[0034] New data for discovery, prioritization, validation and testing of next generation broader-spectrum blood biomarkers for suicidal ideation and behavior, across psychiatric diagnoses are disclosed. Also disclosed are two clinical information questionnaires in the form of apps, one for affective state (Simplified Affective Scale, SASS) and one for suicide risk factors (Convergent Functional Information for Suicide, CFI-S), that are useful in predicting suicidality. Both of these instruments do not directly ask about suicidal ideation. Also disclosed is a comprehensive universal predictor for suicide (UP-Suicide), composed of the combination of top biomarkers (from discovery, prioritization and validation), along with CFI-S, and SASS, which predicts in independent test cohorts suicidal ideation and future psychiatric hospitalizations for suicidality.
[0035] As disclosed herein, "patient psychiatric information" may include mood information, anxiety information, and other psychiatric symptom information and combinations thereof
[0036] As used herein, "predicting suicidality in a subject" is used herein to indicate in advance that a subject will attempt suicide and/or complete suicide.
[0037] As known by those skilled in the art, "suicidal ideation" refers to thoughts, feelings, intent, external actions and behaviors about completing suicide. Suicidal ideation can vary from fleeting thoughts to unsuccessful attempts. In some embodiments, the reference expression level of a biomarker can be obtained for a subject who has no suicidal ideation at the time the sample is obtained from the subject, but who later exhibits suicide ideation. As used herein, "suicidality" includes both suicide ideation and suicidal acts.
[0038] As used herein, "a reference expression level of a biomarker" refers to the expression level of a biomarker established for a subject with no suicidal ideation, expression level of a biomarker in a normal/healthy subject with no suicidal ideation as determined by one skilled in the art using established methods as described herein, and/or a known expression level of a biomarker obtained from literature. The reference expression level of the biomarker can further refer to the expression level of the biomarker established for a high suicide risk subject, including a population of high suicide risk subjects. The reference expression level of the biomarker can also refer to the expression level of the biomarker established for a low suicide risk subject, including a population of low suicide risk subjects. The reference expression level of the biomarker can also refer to the expression level of the biomarker established for any combination of subjects such as a subject with no suicidal ideation, expression level of the biomarker in a normal/healthy subject with no suicidal ideation, expression level of the biomarker for a subject who has no suicidal ideation at the time the sample is obtained from the subject, but who later exhibits suicide ideation, expression level of the biomarker as established for a high suicide risk subject, including a population of high suicide risk subjects, and expression level of the biomarker can also refer to the expression level of the biomarker established for a low suicide risk subject, including a population of low suicide risk subjects. The reference expression level of the biomarker can also refer to the expression level of the biomarker obtained from the subject to which the method is applied. As such, the change within a subject from visit to visit can indicate an increased or decreased risk for suicide. For example, a plurality of expression levels of a biomarker can be obtained from a plurality of samples obtained from the same subject and used to identify differences between the plurality of expression levels in each sample. Thus, in some embodiments, two or more samples obtained from the same subject can provide an expression level(s) of a blood biomarker and a reference expression level(s) of the blood biomarker.
[0039] As used herein, "expression level of a biomarker" refers to the process by which a gene product is synthesized from a gene encoding the biomarker as known by those skilled in the art. The gene product can be, for example, RNA (ribonucleic acid) and protein. Expression level can be quantitatively measured by methods known by those skilled in the art such as, for example, northern blotting, amplification, polymerase chain reaction, microarray analysis, tag-based technologies (e.g., serial analysis of gene expression and next generation sequencing such as whole transcriptome shotgun sequencing or RNA-Seq), Western blotting, enzyme linked immunosorbent assay (ELISA), and combinations thereof.
[0040] As used herein, a "difference" in the expression level of the biomarker refers to an increase or a decrease in the expression of a blood biomarker when analyzed against a reference expression level of the biomarker. In some embodiments, the "difference" refers to an increase or a decrease by about 1.2-fold or greater in the expression level of the biomarker as identified between a sample obtained from the subject and the reference expression level of the biomarker. In one embodiment, the difference in expression level is an increase or decrease by about 1.2 fold. As used herein "a risk for suicide" can refer to an increased (greater) risk that a subject will attempt to commit suicide and/or complete suicide For example, depending on the biomarker(s) selected, the difference in the expression level of the biomarker(s) can indicate an increased (greater) risk that a subject will attempt to commit suicide and/or complete suicide. Conversely, depending on the biomarker(s) selected, the difference in the expression level of the biomarker(s) can indicate a decreased (lower) risk that a subject will attempt to commit suicide and/or complete suicide.
[0041] In accordance with the present disclosure, biomarkers useful for objectively predicting, mitigating, and/or preventing suicidality in subjects have been discovered. In one aspect, the present disclosure is directed to a method for predicting suicidality in a subject. The method includes obtaining a reference expression level of a blood biomarker; and determining an expression level of the blood biomarker in a sample obtained from the subject. A change in the expression level of the blood biomarker in the sample obtained from the subject as compared to the reference expression level indicates suicidality. In some embodiments, the methods further include obtaining clinical risk factor information and clinical scale data such as for anxiety, mood and/or psychosis from the subject in addition to obtaining blood biomarker expression level in a sample obtained from the subject.
[0042] In one embodiment, the expression level of the blood biomarker in the sample obtained from the subject is increased as compared to the reference expression level of the biomarker. It has been found that an increase in the expression level of particular blood biomarkers in the sample obtained from the subject as compared to the reference expression level of the biomarker indicates a risk for suicide. Suitable biomarkers that indicate a risk for suicide when the expression level increases can be, for example, one or more biomarkers as listed in Table 1 and combinations thereof.
TABLE-US-00001 TABLE 1 Top Candidate Biomarker Genes - increase in expression Gene Gene Name Symbol interleukin 6 (interferon, beta 2) IL6 spermidine/spermine N1-acetyltransferase 1 SAT1 solute carrier family 4 (sodium bicarbonate cotrans- SLC4A4 porter), member 4 monoamine oxidase B MAOB Glutamate Receptor, Ionotropic, Kainate 2 GRIK2 Rho GTPase activating protein 26 ARHGAP26 B-cell CLL/lymphoma 2 BCL2 cadherin 4, type 1, R-cadherin (retinal) CDH4 chemokine (C-X-C motif) ligand 11 CXCL11 EMI domain containing 1 EMID1 family with sequence similarity 49, member B FAM49B GRB2-Associated Binding Protein 1 GAB1 GRINL1A complex locus 1 GCOM1 hippocalcin-like 1 HPCAL1 mitogen-activated protein kinase 9 MAPK9 nuclear paraspeckle assembly transcript 1 (non-protein NEAT1 coding) protein tyrosine kinase 2 PTK2 RAS-like, family 11, member B RASL11B small nucleolar RNA, H/ACA box 68 SNORA68 superoxide dismutase 2, mitochondrial SOD2 transcription factor 7-like 2 (T-cell specific, HMG- TCF7L2 box) v-raf murine sarcoma viral oncogene homolog B BRAF chromosome 1 open reading frame 61 C1orf61 Calreticulin CALR calcium/calmodulin-dependent protein kinase II beta CAMK2B caveolin 1, caveolae protein, 22 kDa CAV1 chromodomain helicase DNA binding protein 2 CHD2 clathrin, light chain A CLTA cAMP responsive element modulator CREM Cortactin CTTN dishevelled associated activator of morphogenesis 2 DAAM2 Dab, mitogen-responsive phosphoprotein, homolog 2 DAB2 (Drosophila) GABA(A) receptor-associated protein like 1 GABARAPL1 GABA(A) glutamate-ammonia ligase GLUL helicase with zinc finger HELZ immunoglobulin heavy constant gamma 1 (G1m IGHG1 marker) interleukin 1, beta IL1B jun proto-oncogene JUN jun B proto-oncogene JUNB lipoma HMGIC fusion partner LHFP myristoylated alanine-rich protein kinase C substrate MARCKS metallothionein 1E MT1E metallothionein 1H MT1H metallothionein 2A MT2A N-myc downstream regulated 1 NDRG1 nucleobindin 2 NUCB2 PHD finger protein 20-like 1 PHF20L1 phosphatase and tensin homolog PTEN reversion-inducing-cysteine-rich protein with kazal RECK motifs shisa family member 2 SHISA2 transmembrane 4 L six family member 1 TM4SF1 trophoblast glycoprotein TPBG tumor protein D52-like 1 TPD52L1 TSC22 domain family, member 3 TSC22D3 vacuole membrane protein 1 VMP1 ZFP36 ring finger protein ZFP36 zinc fingers and homeoboxes 2 ZHX2 UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, B4GALT1 polypeptide 1 BTB (POZ) domain containing 3 BTBD3 cell adhesion molecule 1 CADM1 chitobiase, di-N-acetyl- CTBS DEP domain containing 5 DEPDC5 dystrobrevin, alpha DTNA egf-like module containing, mucin-like, hormone EMR2 receptor-like 2 endogenous retrovirus group 3, member 2 ERV3-2 family with sequence similarity 183, FAM183CP member C, pseudogene histone cluster 1, H2bo HIST1H2BO potassium channel tetramerization domain containing KCTD21 21 Keratocan KERA laminin, beta 1 LAMB1 uncharacterized LOC100289061 LOC100129917 uncharacterized LOC285500 LOC285500 RAB36, member RAS oncogene family RAB36 uncharacterized LOC283352 RP11-66N7.2 transcription factor Dp-1 TFDP1 TMLHE antisense RNA 1 TMLHE-AS1 superoxide dismutase 2, mitochondrial SOD2 period circadian clock 1 PER1 Ras association (RalGDS) RAPH1 spondin 1, extracellular matrix protein SPON1 forkhead box P1 FOXP1 hepatitis A virus cellular receptor 2 HAVCR2 Rho GTPase activating protein 15 ARHGAP15 gap junction protein, alpha 1, 43 kDa GJA1 hes family bHLH transcription factor 1 HES1 HtrA serine peptidase 1 HTRA1 TIMP metallopeptidase inhibitor 1 TIMP1 erythrocyte membrane protein band 4.1 like 5 EPB41IL5 interleukin 1 receptor, type I IL1R1 intelectin 1 (galactofuranose binding) ITLN1 killer cell immunoglobulin-like receptor, two KIR2DL4 domains, long cytoplasmic tail, 4 nudix (nucleoside diphosphate linked moiety X)-type NUDT10 motif 10 pyridoxal-dependent decarboxylase domain containing PDXDC1 1 family with sequence similarity 214, member A FAM214A heat shock 60 kDa protein 1 (chaperonin) HSPD1 zinc finger, MYND-type containing 8 ZMYND8 adenylate kinase 2 AK2 AF4/FMR2 family, member 3 AFF3 mitochondrial ribosomal protein S5 MRPS5 v-akt murine thymoma viral oncogene homolog 3 AKT3 aspartate beta-hydroxylase ASPH ataxin 1 ATXN1 Brain and reproductive organ-expressed (TNFRSF1A BRE modulator) ClpB caseinolytic peptidase B homolog (E. coli) CLPB deleted in primary ciliary dyskinesia homolog (mouse) DPCD ECSIT signalling integrator ECSIT ectonucleoside triphosphate diphosphohydrolase 1 ENTPD1 EPH receptor B4 EPHB4 Fanconi anemia, complementation group I DANCI general transcription factor IIIC, polypeptide 3, 102 GTF3C3 kDa inter-alpha-trypsin inhibitor heavy chain family, ITIH5 member 5 kelch-like family member 28 KLHL28 major histocompatibility complex, class I-related MR1 protein inhibitor of activated STAT, 1 PIAS1 periphilin 1 PPHLN1 retinol dehydrogenase 13 (all-trans/9-cis) RDH13 strawberry notch homolog 1 (Drosophila) SBN01 sorting nexin family member 27 SNX27 single-stranded DNA binding protein 2 SSBP2 striatin, calmodulin binding protein STRN tetratricopeptide repeat domain 7A TTC7A ubiquitin interaction motif containing 1 UIMC1 Z-DNA binding protein 1 ZBP1 zinc finger protein 596 ZNF596 adaptor-related protein complex 3, sigma 2 subunit AP3S2
[0043] In one particularly suitable embodiment, the subject is a male and the blood biomarker that increases in expression level as compared to the reference expression level is selected from solute carrier family 4 (sodium bicarbonate cotransporter), member 4 (SLC4A4), cell adhesion molecule 1 CADM1, dystrobrevin, alpha (DTNA), spermidine/spermine N1-acetyltransferase 1 (SAT1), interleukin 6 (interferon, beta 2) (IL6) and combinations thereof. In another embodiment, the subject is a female and the blood biomarker that increases in expression level as compared to the reference expression level is selected from erythrocyte membrane protein band 4.1 like 5 (EPB41L5), HtrA serine peptidase 1 (HTRA1), deleted in primary ciliary dyskinesia homolog (DPCD), general transcription factor IIIC, polypeptide 3, 102 kDa (GTF3C3), period circadian clock 1 (PER1), pyridoxal-dependent decarboxylase domain containing 1 (PDXDC1), kelch-like family member 28 (KLHL28), ubiquitin interaction motif containing 1 (UIMC1), sorting nexin family member 27 (SNX27) and combinations thereof.
[0044] In another embodiment, the expression level of the blood biomarker in the sample obtained from the subject is decreased as compared to the reference expression level of the biomarker. Suitable biomarkers that indicate a risk for suicide when the expression level decreases as compared to the reference expression level have been found to include, for example, one or more biomarkers as listed in Table 2 and combinations thereof.
TABLE-US-00002 TABLE 2 Top Candidate Biomarker Genes - decrease in expression Gene Name Gene Symbol spindle and kinetochore associated SKA2 complex subunit 2 coiled-coil domain containing 136 CCDC136 CD44 molecule (Indian blood group) CD44 fatty acid desaturase 1 FADS1 FK506 binding protein 5 FKBP5 forkhead box N3 FOXN3 hydroxyacyl-CoA dehydrogenase/3- HADHA ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit adenosylhomocysteinase-like 1 AHCYL1 AKT1 substrate 1 (proline-rich) AKT1S1 aldehyde dehydrogenase 3 family, ALDH3 A2 member A2 B-cell CLL/lymphoma 2 BCL2 C20orf27 calpain, small subunit 1 CAPNS1 CDC42 effector protein (Rho GTPase CDC42EP4 binding) 4 EH domain binding protein 1 EHBP1 eukaryotic translation initiation factor 5A EIF5A fumarate hydratase FH glycoprotein M6B GPM6B homeobox and leucine zipper encoding HOMEZ inhibitor of kappa light polypeptide gene IKBKB enhancer in B-cells, kinase beta integrin, beta 4 ITGB4 low density lipoprotein receptor adaptor LDLRAP1 protein 1 uncharacterized LOC728543 LOC728543 mitogen-activated protein kinase kinase 5 MAP2K5 neuromedin B NMB platelet-activating factor acetylhydrolase PAFAH1B2 1b, catalytic subunit 2 (30 kDa) pterin-4 alpha-carbinolamine PCBD2 dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 phosphatidylinositol-4-phosphate 3- PIK3C2A kinase, catalytic subunit type 2 alpha plakophilin 4 PKP4 solute carrier family 5 (sodium/ SLC5A3 myoinositol cotransporter), member 3 spectrin repeat containing, nuclear SYNE2 envelope 2 trans-golgi network protein 2 TGOLN2 trafficking protein, kinesin binding 2 TRAK2 adrenergic, beta, receptor kinase 1 ADRBK1 adenosylhomocysteinase-like 2 AHCYL2 aminoacyl tRNA synthetase complex- AIMP1 interacting multifunctional protein 1 ATPase, H+ transporting, lysosomal ATP6V0E1 9 kDa, V0 subunit e1 BRCA1/BRCA2-containing complex, BRCC3 subunit 3 2',3'-cyclic nucleotide 3' CNP phosphodiesterase collagen, type IX, alpha 2 COL9A2 cleavage and polyadenylation specific CPSF2 factor 2, 100 kDa cullin 4B CUL4B delta-like 1 (Drosophila) DLL1 dynein, axonemal, heavy chain 2 DNAH2 dipeptidyl-peptidase 4 DPP4 G2/M-phase specific E3 ubiquitin protein G2E3 ligase guanylate kinase 1 GUK1 Janus kinase 3 JAK3 lysosomal protein transmembrane 4 beta LAPTM4B lysophosphatidic acid receptor 1 LPAR1 membrane associated guanylate kinase, MAGI3 WW and PDZ domain containing 3 myelin basic protein MBP microspherule protein 1 MCRS1 myocyte enhancer factor 2C MEF2C opioid growth factor receptor OGFR protocadherin 9 PCDH9 pleckstrin homology domain containing, PLEKHB1 family B (evectins) member 1 polymerase (RNA) II (DNA directed) POLR2D polypeptide D protein kinase, cAMP-dependent, PRKACA catalytic, alpha protein kinase C, beta PRKCB proteasome (prosome, macropain) PSMB4 subunit, beta type, 4 RAB35, member RAS oncogene family RAB35 RNA binding motif protein, X-linked RBMX ribonuclease L (2',5'-oligoisoadenylate RNASEL synthetase-dependent) selenium binding protein 1 SELENBP1 solute carrier family 35, member E1 SLC35E1 synaptosomal-associated protein, 23 kDa SNAP23 transmembrane protein 254 TMEM254 transmembrane protein 259 TMEM259 tensin 1 TNS1 tripartite motif containing 23 TRIM23 tetraspanin 33 TSPAN33 pre-B lymphocyte 3 VPREB3 zinc finger, FYVE domain containing 21 ZFYVE21 zinc finger protein 519 ZNF519 cation channel, sperm associated 3 CATSPER3 chemokine (C-C motif) ligand 28 CCL28 CAP-GLY domain containing linker CLIP4 protein family, member 4 chromosome Y open reading frame 17 CYorf17 DDB1 and CUL4 associated factor 15 DCAF15 EPH receptor A10 EPHA10 v-ets avian erythroblastosis virus E26 ERG oncogene homolog heparan sulfate (glucosamine) 3-O- HS3ST3B1 sulfotransferase 3B1 IQ motif containing H IQCH kinesin family member 2C KIF2C kelch domain containing 3 KLHDC3 uncharacterized LOC100129917 LOC100129917 uncharacterized LOC100996345 LOC100996345 mediator complex subunit 21 MED21 PDX1 C-terminal inhibiting factor 1 PCIF1 plectin PLEC RAD23 homolog A (S. cerevisiae) RAD23A Rh-associated glycoprotein RHAG roundabout, axon guidance receptor, ROBO4 homolog 4 (Drosophila) ribosomal protein L6 pseudogene 17 RPL6P17 SET domain containing (lysine SETD8 methyltransferase) 8 SH3-domain GRB2-like endophilin B2 SH3GLB2 ST6 (alpha-N-acetyl-neuraminyl-2,3- ST6GALNAC4 beta-galactosyl-1,3)-N- acetylgalactosaminide alpha-2,6- sialyltransferase 4 testis expressed 10 TEX10 testis expressed 261 TEX261 thymosin beta 15B TMSB15B tubulin, gamma complex associated TUBGCP3 protein 3 thioredoxin reductase 2 TXNRD2 ubiquitin specific peptidase 12 USP12 vascular endothelial growth factor B VEGFB zinc finger and BTB domain containing ZBTB7A 7A glycogen synthase kinase 3 beta GSK3B adaptor-related protein complex 1, sigma AP1S2 2 subunit catalase CAT chromosome 18 open reading frame 54 C19orf54 long intergenic non-protein coding RNA LINC00342 342 MOB kinase activator 3B MOB3B phosphatidylinositol-4-phosphate 5- PIP5K1B kinase, type I, beta prolylcarboxypeptidase (angiotensinase PRCP C) CD200 receptor 1 CD200R1 CD84 molecule CD84 centrosomal protein 44 kDa CEP44 carnitine O-octanoyltransferase CROT DDB1 and CUL4 associated factor 5 DCAF5 DTW domain containing 2 DTWD2 endoplasmic reticulum protein 27 ERP27 family with sequence similarity 173, FAM173B member B glucosidase, alpha; neutral C GANC general transcription factor IIIC, GTF3C2 polypeptide 2, beta 110 kDa INO80 complex subunit D INO80D inositol polyphosphate-4-phosphatase, INPP4A type I, 107 kDa Jrk homolog (mouse) JRK potassium channel tetramerization KCTD5 domain containing 5 methyltransferase like 15 METTL15 phosphatidylinositol 3-kinase, catalytic PIK3C3 subunit type 3 RNA binding motif protein 48 RBM48 SWI/SNF Related, Matrix Associated, SMARCA2 Actin Dependent Regulator Of Chromatin, Subfamily A, Member 2 ubiquitin carboxyl-terminal hydrolase L5 UCHL5 vacuolar protein sorting 53 homolog VPS53 (S. cerevisiae) zinc finger protein 302 ZNF302 capping protein (actin filament) muscle CAPZA2 Z-line, alpha 2 leucine rich repeat containing 8 family, LRRC8B member B protein phosphatase, Mg2+ PPM1B ARP3 actin-related protein 3 homolog ACTR3 (yeast) SH2 domain containing 1A SH2D1A ALG13, UDP-N- ALG13 acetylglucosaminyltransferase subunit Rho GTPase activating protein 35 ARHGAP35 AT rich interactive domain 4B (RBP1- ARID4B like) charged multivesicular body protein 2B CHMP2B casein kinase 1, alpha 1 CSNK1A1 ethanolamine kinase 1 ETNK1 F-box and leucine-rich repeat protein 3 FBXL3 HECT and RLD domain containing E3 HERC4 ubiquitin protein ligase 4 jumonji domain containing 1C JMJD1C La ribonucleoprotein domain family, LARP4 member 4 muscleblind-like splicing regulator 1 MBNL1 mex-3 RNA binding family member C MEX3C nudix (nucleoside diphosphate linked NUDT6 moiety X)-type motif 6 polyhomeotic homolog 3 (Drosophila) PHC3 peroxiredoxin 3 PRDX3 Pvt1 oncogene (non-protein coding) PVT1 RAB22A, member RAS oncogene family RAB22A solute carrier family 35 (adenosine 3'- SLC35B3 phospho 5'-phosphosulfate transporter), member B3 small nuclear ribonucleoprotein 27 kDa SNRNP27 (U4 USP6 N-terminal like USP6NL WW domain containing adaptor with WAC coiled-coil wings apart-like homolog (Drosophila) WAPAL zinc finger, AN1-type domain 5 ZFAND5 zinc finger protein 117 ZNF117 zinc finger protein 141 ZNF141 zinc finger protein 548 ZNF548 signal sequence receptor, alpha SSR1
[0045] In one particularly suitable embodiment, the subject is a male and the blood biomarker that decreases in expression level as compared to the reference expression level is spindle and kinetochore associated complex subunit 2 (SKA2), CAP-GLY domain containing linker protein family, member 4 (CLIP4), kinesin family member 2C (KIF2C), kelch domain containing 3 (KLHDC3) and combinations thereof. In another embodiment, the subject is a female and the blood biomarker that decreases in expression level as compared to the reference expression level is selected from phosphatidylinositol 3-kinase, catalytic subunit type 3 (PIK3C3), aldehyde dehydrogenase 3 family, member A2 (ALDH3A2), ARP3 actin-related protein 3 homolog (yeast) (ACTR3), B-cell CLL (BCL2), MOB kinase activator 3B (MOB3B), casein kinase 1, alpha 1 (CSNK1A1), La ribonucleoprotein domain family, member 4 (LARP4), zinc finger protein 548 (ZNF548) and combinations thereof.
[0046] Table 3 further discloses the top biomarkers across gender having expression levels that increase or decrease (as indicated) as compared to the reference expression levels to predict suicidality.
TABLE-US-00003 TABLE 3 Top Universal Biomarkers for Suicide Across Genders Discovery in Significant Prediction Blood Validation of Suicidal Ideation (Direction of in Blood Across All and Gene Symbol Affymetrix Change)/ ANOVA p- Best In a Diagnostic Gene Name Probesets Score value/Score Group ROC AUC/p-value BCL2 203685_at (D)/1 5.98E-11/4 All B-cell 0.609/0.005 CLL/ Male SZ/SZA lymphoma 2 0.68/0.011 CD164 208654_s_at (D)/2 3.01E-08/4 All CD164 0.589/0.017 molecule, Male BP sialomucin 0.68/0.020 CD47 211075_s_at (D)/2 1.62E-17/4 All CD47 0.598/0.010 molecule Male SZ/SZA 0.67/0.016 DLG1 202514_at (D)/1 0.0000844 All discs, large 0.58/0.036 homolog 1 Male SZ/SZA (Drosophila) 0.65/0.030 DLG1 202516_s_at (D)/1 0.0000000000016/4 .sup. All discs, large 0.58/0.029 homolog 1 (Drosophila) DYRK2 202969_at (D)/1 0.00000000000017/4 All dual-specificity 0.58/0.034 tyrosine-(Y)- Male SZ/SZA phosphory- 0.68/0.010 lation regulated kinase 2 ITGB1BP1 203336_s_at (D)/1 0.000000025/4 .sup. All integrin beta 1 0.57/0.042 binding protein 1 APOE 203382_s_at (I)/1 3.44E-09/4 All apolipo- 0.59/0.021 protein E Male BP 0.71/0.0091 MRPS14 203800_s_at (D)/1 0.00000000039/4 .sup. Male SZ/SZA mitochondrial 0.69/0.0080 ribosomal protein S14 MRPS14 203801_at (D)/1 2.45E-17/4 All mitochondrial 0.60/0.0069 ribosomal protein Male SZ/SZA S14 0.68/0.011 IL6 205207_at (I)/1 1.82E-15/4 All interleukin 6 0.58/0.038 AKAP13 209534_x_at (I)/1 .sup. 0.000021/4 Male PTSD A kinase (PRKA) 0.78/0.0083 anchor protein 13 SECISBP2L 212450_at (D)/1 .sup. 0.000063/4 All SECIS binding 0.59/0.021 protein 2-like Male BP 0.71/0.0076 SOD2 215078_at (I)/2 2.27E-34/4 superoxide dismutase 2, mitochondrial LHFP 218656_s_at (I)/1 0.00000000040/4 .sup. All lipoma HMGIC 0.57/0.05 fusion partner Male MDD 0.69/0.034 SKA2 225686_at (D)/1 4.55E-03/2 All spindle and 0.62/0.003 kinetochore Male SZ/SZA associated 0.75/0.00063 complex subunit 2 GSK3B 226183_at (D)/1 2.19E-36/4 glycogen synthase kinase 3 beta ITPKB 232526_at AP 0.0000000045/4 All inositol- (I)/1 0.62/0.0019 trisphosphate 3- Male BP kinase B 0.76/0.0013 MTERF4 1557966_x_at (D)/2 6.72E-06/4 All mitochondrial 0.61/0.005 transcription Male SZ/SZA termination factor 0.72/0.0019 4 GDI2 200008_s_at (D)/2 1.52E-11/4 All GDP dissociation 0.59/0.013 inhibitor 2 Male BP 0.67/0.024 PRKAR1A 200605_s_at (D)/2 2.47E-06/4 Male BP protein kinase, 0.72/0.0059 cAMP- dependent, regulator, type I, alpha NR3C1 201866_s_at (D)/1 1.64E-03/2 Male BP nuclear receptor 0.67/0.029 subfamily 3, group C, member 1 (glucocorticoid receptor) ADK 204119_s_at DE 0.000000020/4 .sup. All adenosine kinase (D)/4 0.62/0.0026 Male SZ/SZA 0.66/0.019 PGK1 217383_at (D)/2 4.07E-07/4 Male SZ/SZA phosphoglycerate 0.63/0.046 kinase 1 ZFYVE21 219929_s_at (D)/2 5.96E-06/4 All zinc finger, 0.58/0.026 FYVE domain containing 21 RBM3 222026_at (D)/2 1.73E-05/4 RNA binding motif (RNP1, RRM) protein 3 FAM107B 223058_at (D)/2 2.36E-02/2 All family with 0.58/0.024 sequence Male BP similarity 107, 0.71/0.0079 member B ECHDC1 223087_at (D)/2 3.35E-09/4 All enoyl CoA 0.60/0.009 hydratase domain Male containing 1 SZ/SZA 0.66/0.019 TBL1XR1 235890_at AP 0.000000023/4 .sup. Male BP transducin (beta)- (D)/2 0.66/0.034 1 ike 1 X-linked receptor 1 LONRF2 235977_at (I)/1 1.48E-03/2 Male BP LON peptidase 0.73/0.0040 N-terminal domain and ring finger 2 QKI 211938_at (I)/2 1.88E-03/2 Male QKI, KH domain PTSD containing, RNA 0.77/0.011 binding YWHAH 242325_at (I)/2 6.65E-11/4 All tyrosine 3- 0.571/0.047 monooxygenase/ Male BP tryptophan 5- 0.66/0.033 monooxygenase activation protein, eta SLC4A4 210739_x_at (I)/1 7.74E-05/4 All solute carrier 0.64/0.00038 family 4 (sodium Male BP bicarbonate 0.77/0.00094 cotransporter), member 4 GDI2 200009_at (D)/1 .sup. 0.000015/4 All GDP dissociation 0.64/0.0006 inhibitor 2 Male SZ/SZA 0.72/0.0028 UQCRC2 200883_at (D)/1 .sup. 0.012/2 All ubiquinol- 0.61/0.0035 cytochrome c Male SZ/SZA reductase core 0.67/0.013 protein II CTNNB1 201533_at (D)/1 .sup. 0.0023/2 All catenin 0.59/0.018 (cadherin- Male BP associated 0.74/0.0037 protein), beta 1, 88 kDa PSMB4 202243_s_at (D)/1 6.55E-14/4 All proteasome 0.6/0.011 (prosome, Male SZ/SZA macropain) 0.68/0.010 subunit, beta type, 4 PRKACB 202742_s_at (D)/1 .sup. 0.00042/2 All protein kinase, 0.58/0.028 cAMP- dependent, catalytic, beta LPAR1 204036_at (D)/1 1.35003E-234 Male BP lysophosphatidic 0.68/0.022 acid receptor 1 HTR2C 207307_at (I)/1 4.30E-02/2 All 5-hydroxy- 0.583/0.025 tryptamine Male MDD (serotonin) 0.69/0.035 receptor 2C, G protein-coupled CTTN 214782_at DE 1.042E-19/4 Male BP cortactin (I)/1 0.76/0.0016 PDCL3 219043_s_at (D)/2 1.37E-02/2 All phosducin-like 3 0.6/0.009 Male SZ/SZA 0.65/0.030 SNX6 222410_s_at DE 0.0000068/4 All sorting nexin 6 (D)/1 0.62/0.0025 Male SZ/SZA 0.65/0.024 PIK3CA 231854_at DE 2.41E-37/4 All phosphatidyl- (D)/1 0.57/0.042 inositol-4,5- Male BP bisphosphate 3- 0.65/0.047 kinase, catalytic subunit alpha MBP 225408_at (D)/2 8.34E-07/4 myelin basic protein CCDC136 226972_s_at (D)/4 3.13E-03/2 coiled-coil domain containing 136 AIMP1 227605_at (D)/2 1.02E-05/4 All aminoacyl tRNA 0.60/0.007 synthetase Male SZ/SZA complex- 0.66/0.018 interacting multifunctional protein 1 PITHD1 229856_s_at (D)/4 0.000000067/4 .sup. Female BP PITH (C-terminal 0.83/0.031 proteasome- interacting domain of thioredoxin-like) domain containing 1 PCDH9 238919_at (D)/2 6.61E-05/4 protocadherin 9 CAPZA2 201238_s_at (D)/1 .sup. 0.00029/2 All capping protein 0.6/0.0086 (actin filament) Male BP muscle Z-line, 0.65/0.047 alpha 2 PSME4 237180_at (I)/1 2.64E-36/4 All Proteasome 0.6/0.011 Activator Subunit Male PTSD 4 0.79/0.0062 GABRB1 1557256_a_at (I)/1 .sup. 0.012/2 Male BP gamma- 0.74/0.0034 aminobutyric acid (GABA) A receptor, beta 1 CNP 1557943_at (D)/1 .sup. 0.019/2 2',3'-cyclic nucleotide 3'
phosphodiesterase RAP1A 202362_at (D)/1 .sup. 0.035/2 All RAP1A, member 0.6/0.011 of RAS oncogene Male BP family 0.71/0.0082 NGFR 205858_at (I)/1 2.24E-15/4 All nerve growth 0.59/0.018 factor receptor Male SZ/SZA 0.72/0.0020 CAMK2B 209956_s_at DE .sup. 0.00078/2 All calcium/calmodulin- (I)/1 0.62/0.0017 dependent Male BP protein kinase II 0.74/0.0029 beta CLN5 214252_s_at DE 1.79E-15/4 All ceroid- (D)/1 0.65/0.0002 lipofuscinosis, Male SZ/SZA neuronal 5 0.68/0.010 CLTA 216295_s_at DE 1.74E-15/4 All clathrin, light (D)/1 0.64/0.0006 chain A Male BP 0.73/0.0049 DOCK8 232843_s_at DE .sup. 0.0022/2 All dedicator of (D)/1 0.6/0.0079 cytokinesis 8 Male BP 0.78/0.00078 RARS2 232902_s_at DE .sup. 0.022/2 All arginyl-tRNA (D)/1 0.63/0.0014 synthetase 2, Male SZ/SZA mitochondrial 0.70/0.0043 PTK2 241453_at DE 2.87E-32/4 All protein tyrosine (I)/1 0.61/0.0045 kinase 2 Male MDD 0.69/0.033 PLCL1 241859_at (D)/1 .sup. 0.040/2 Male PTSD phospholipase 0.78/0.0083 C-like 1 LPAR1 204038_s_at (D)/2 1.66E-04/2 lysophosphatidic acid receptor 1 AK2 205996_s_at (D)/2 0.00000011/4.sup. All adenylate kinase 0.64/0.0005 2 Male SZ/SZA 0.74/0.0012 APLP2 208703_s_at (D)/2 3.65E-02/2 amyloid beta (A4) precursor- like protein 2 BACE1 224335_s_at (D/1 .sup. 0.00037/2 All beta-site APP- 0.58/0.032 cleaving enzyme Male BP 1 0.67/0.024 ELOVL5 214153_at (I)/1 .sup. 0.0028/2 Male PTSD ELOVL fatty 0.76/0.012 acid elongase 5 KIF2C 211519_s_at (D)/4 .sup. 0.014/2 kinesin family member 2C Significant Prediction of Future Hospitalizations Drugs that for Suicidality Convergent Modulate the Across All and Genetic and Brain Other Psychiatric Biomarker in Best in a Diagnostic Evidence For and Related Opposite Gene Symbol Group ROC AUC/ Involvement Disorders Direction Gene Name p-value in Suicide Evidence to Suicide BCL2 Male 5 Aging Omega-3 B-cell PTSD Alcoholism Lithium CLL/ 0.83/0.013 Anxiety BP lymphoma 2 Mood Disorders PTSD SZ CD164 Male 4 BP Clozapine CD164 PTSD Cocaine molecule, 0.96/0.0004 Dependence sialomucin Stress CD47 Male 4 MDD Clozapine CD47 PTSD Stress Omega-3 molecule 0.87/0.0048 SZ DLG1 Male 4 Alcoholism Omega-3 discs, large PTSD BP homolog 1 309/0.0023 MDD (Drosophila) SZ DLG1 Male 4 Alcoholism Omega-3 discs, large PTSD BP homolog 1 0.79/0.028 MDD (Drosophila) SZ DYRK2 Male 4 Aging Clozapine dual-specificity PTSD BP tyrosine-(Y)- 0.93/0.001 MDD phosphory- Sleep Disorders lation regulated kinase 2 ITGB1BP1 Male 4 Alzheimer's Disease Lithium integrin beta 1 PTSD BP binding 0.83/0.013 Mood Disorders protein 1 SZ APOE 6 Aggression Omega-3 apolipo- Aging protein E Alcoholism Alzheimer's Disease Autism Dementia Depression-related Longevity MDD Psychosis PTSD SZ MRPS14 Male 4 SZ Omega-3 mitochondrial PTSD ribosomal protein 0.84/0.0093 S14 MRPS14 Male 4 SZ Omega-3 mitochondrial PTSD ribosomal protein 0.77/0.035 S14 IL6 Female 6 Aggression interleukin 6 PTSD Anxiety 1/0.028 BP Cognition Dementia Depression Longevity MDD Mood Disorders Panic Psychosis PTSD Sleep Disorders Stress SZ AKAP13 All 4 Cocaine Clozapine A kinase (PRKA) 0.57/0.047 Dependence anchor protein 13 Male PTSD Panic 0.80/0.022 Stress SECISBP2L Male 4 Cocaine Clozapine SECIS binding PTSD Dependence protein 2-like 0.89/0.0034 MDD SZ SOD2 Male 5 Longevity Clozapine superoxide PTSD MDD dismutase 2, 0.85/0.010 Methamphetamine mitochondrial Abuse Mood Disorders SZ LHFP Male 4 SZ Omega-3 lipoma HMGIC MDD fusion partner 0.79/0.004 SKA2 Male 8 PTSD spindle and PTSD Stress kinetochore 0.84/0.0093 associated complex subunit 2 GSK3B Male 6 Aging Lithium glycogen PTSD Alcoholism synthase kinase 3 0.84/0.0093 BP beta Dementia Depression Mood Stabilizers response Lithium response MDD SZ ITPKB Male 4 Aging Omega-3 inositol- PTSD Alcoholism trisphosphate 3- 0.87/0.0048 Alzheimer's Disease kinase B Autism BP MDD Multiple Sclerosis Stress SZ SZA MTERF4 Male 4 Stress mitochondrial PTSD transcription 0.94/0.0006 termination factor 4 GDI2 4 BP Clozapine GDP dissociation MDD inhibitor 2 Mood Disorders SZ PRKAR1A Male 4 Alcoholism protein kinase, PTSD BP cAMP- 0.90/0.0023 Epilepsy dependent, Mood Disorders regulator, type I, Stress alpha SZ NR3C1 Male 5 Alcoholism Clozapine nuclear receptor PTSD Anxiety subfamily 3, 0.91/0.0015 BP group C, member Depression 1 (glucocorticoid Longevity receptor) MDD PTSD Response to escitalopram (SSRI) Response to Nortriptyline (TCA) Stress SZ ADK Male 0 Depression Omega-3 adenosine kinase PTSD 0.84/0.0093 PGK1 4 Alcoholism Clozapine phosphoglycerate BP kinase 1 MDD SZ SZA ZFYVE21 All 4 SZ zinc finger, 0.58/0.030 FYVE domain Male MDD containing 21 0.78/0.0044 RBM3 Female 4 Epilepsy Omega-3 RNA binding PTSD Response to Lithium Lithium motif (RNP1, 1/0.028 SZ RRM) protein 3 FAM107B Male 4 BP Lithium family with PTSD MDD sequence 0.93/0.001 Psychosis similarity 107, Response to Lithium member B Sleep Disorder SZ ECHDC1 Male 4 Addictions enoyl CoA PTSD BP hydratase domain 0.94/0.0006 PTSD containing 1 TBL1XR1 Female 2 Alcoholism Clozapine transducin (beta)- PTSD BP 1 ike 1 X-linked 1/0.028 Longevity receptor 1 LONRF2 Male 5 Stress Omega-3 LON peptidase PTSD BP N-terminal 0.77/0.039 domain and ring finger 2 QKI All 4 BP Omega-3 QKI, KH domain 0.58/0.031 Longevity containing, RNA MDD binding PTSD Stress SZ YWHAH 4 Alcoholism Omega-3 tyrosine 3- BP monooxygenase/ Longevity tryptophan 5- MDD monooxygenase SZ
activation protein, eta SLC4A4 6 Circadian solute carrier abnormalities family 4 (sodium Longevity bicarbonate MDD cotransporter), SZ member 4 GDI2 4 BP Clozapine GDP dissociation MDD inhibitor 2 Mood Disorders SZ UQCRC2 Male 4 ADHD Omega-3 ubiquinol- PTSD Alcohol cytochrome c 0.81/0.017 BP reductase core MDD protein II Multiple Sclerosis SZ CTNNB1 Male 4 MDD Clozapine catenin PTSD PTSD (cadherin- 0.80/0.022 Stress associated SZ protein), beta 1, 88 kDa PSMB4 Male 4 BP proteasome PTSD MDD (prosome, 0.80/0.022 SZ macropain) SZA subunit, beta type, 4 PRKACB Male 4 Alcohol Clozapine protein kinase, PTSD Alzheimer's Disease cAMP- 0.96/0.0004 BP dependent, Chronic Fatigue catalytic, beta Syndrome LPAR1 4 Aging Clozapine lysophosphatidic BP Omega-3 acid receptor 1 Longevity MDD Mood PTSD SZ HTR2C 6 Affective Disorder Clozapine 5-hydroxy- Alcohol tryptamine Antipsychotics (serotonin) BP receptor 2C, G MDD protein-coupled Mood Disorders Panic Disorder SZ CTTN 4 BP Clozapine cortactin Effect of valproate Omega-3 MDD Stress PDCL3 Male 5 Sleep Disorders phosducin-like 3 PTSD 0.80/0.022 SNX6 Male 4 Panic 0 sorting nexin 6 PTSD 0.86/0.0068 PIK3CA 4 Longevity Lithium phosphatidyl- MDD inositol-4,5- Stress bisphosphate 3- SZ kinase, catalytic subunit alpha MBP 4 Alcohol Clozapine myelin basic Alzheimer's Disease Omega-3 protein BP Lithium MDD Mood Disorders SZ CCDC136 4 Psychosis Clozapine coiled-coil domain containing 136 AIMP1 Male 4 aminoacyl tRNA PTSD synthetase 0.93/0.001 complex- interacting multifunctional protein 1 PITHD1 Male BP PITH (C-terminal PTSD Psychosis proteasome- 0.87/0.0048 SZ interacting domain of thioredoxin-like) domain containing 1 PCDH9 4 Aging Clozapine protocadherin 9 MDD Omega-3 Psychosis SZ CAPZA2 Male 4 BP capping protein PTSD MDD (actin filament) 0.93/0.001 PTSD muscle Z-line, SZ alpha 2 PSME4 4 Autism Proteasome Activator Subunit 4 GABRB1 4 Alcohol gamma- Autism aminobutyric Mood Stabilizers acid (GABA) A BP receptor, beta 1 MDD SZ SZA CNP Female 4 Alcohol Clozapine 2',3'-cyclic SZ/SZA Epilepsy Omega-3 nucleotide 3' 1/0.029 MDD phosphodiesterase Multiple Sclerosis Sleep Disorders SZ RAP1A Male 4 Longevity RAP1A, member PTSD SZ of RAS oncogene 0.83/0.013 SZA family NGFR 4 MDD nerve growth OCD factor receptor Panic Disorder SZ CAMK2B 4 Addictions Clozapine calcium/calmodulin- BP dependent SZ protein kinase II beta CLN5 Male 4 ceroid- PTSD lipofuscinosis, 0.84/0.0093 neuronal 5 CLTA 4 Alzheimer's Disease clathrin, light BP chain A MDD DOCK8 Male 4 ADHD dedicator of PTSD Longevity cytokinesis 8 0.76/0.044 RARS2 Male 4 PTSD arginyl-tRNA PTSD BP synthetase 2, 0.86/0.0068 mitochondrial PTK2 4 Alcohol 0 protein tyrosine Autism kinase 2 BP Circadian abnormalities MDD Psychosis Stress SZ PLCL1 4 Alcohol Clozapine phospholipase Psychosis C-like 1 SZ LPAR1 4 Aging Clozapine lysophosphatidic BP Omega-3 acid receptor 1 Longevity MDD Mood Disorders PTSD SZ AK2 2 BP adenylate kinase SZ 2 APLP2 4 BP Lithium amyloid beta Depression Omega-3 (A4) precursor- Effect of valproate like protein 2 Chronic Fatigue Syndrome BACE1 4 Alzheimer's Disease beta-site APP- Cocaine cleaving enzyme Dependence 1 MDD Psychosis ELOVL5 3 Alcohol ELOVL fatty Autism acid elongase 5 BP Circadian abnormalities Cocaine Dependence MDD Mood Disorders KIF2C kinesin family member 2C
[0047] Particularly suitable subjects are humans. Suitable subjects can also be experimental animals such as, for example, monkeys and rodents, that display a behavioral phenotype associated with suicide, for example, a mood disorder or psychosis. In one particular aspect, the subject is a female human. In another particular aspect, the subject is a male human.
[0048] In another aspect, the subject can further be diagnosed with a psychiatric disorder as known in the art. In particular aspects, the psychiatric disorder can be bipolar disorder, major depressive disorder, schizophrenia, and schizoaffective disorder, post-traumatic stress disorder, and combinations thereof.
[0049] In one embodiment, the subject can be diagnosed as having or as suspected of having bipolar disorder (BP) and the biomarker can be selected from DTNA; HS3ST3B1; CADM1; Unknown gene; KSR1; CD44; DAPP1; OPRM1; SPTBN1; AKT1S1; SAT1; C20orf27; and combinations thereof. As summarized in FIG. 17, the biomarker expression level can increase above a reference expression level of the biomarker or decrease below a reference expression level of the biomarker.
[0050] In another embodiment, the subject can be diagnosed as having or as suspected of having depression (MDD) and the biomarker can be selected from PHF20; EIF1B-AS1; TLN1; NUCKS1; DLK1; BBIP1; BDNF; SKA2; IL10; GATM; PRPF40A; and combinations thereof. As summarized in FIG. 17, the biomarker expression level can increase above a reference expression level of the biomarker or decrease below a reference expression level of the biomarker.
[0051] In another embodiment, the subject can be diagnosed as having or as suspected of having schizoaffective disorder (SZA) and the biomarker can be selected from USP48; NPRL3; TSPYL1; TMSB15B; IL6; TNS1; TNF; S100B; JUN; BATF2; ANXA11; and combinations thereof. As summarized in FIG. 17, the biomarker expression level can increase above a reference expression level of the biomarker or decrease below a reference expression level of the biomarker.
[0052] In another embodiment, the subject can be diagnosed as having or as suspected of having schizophrenia (SZ) and the biomarker can be selected from RP11-389C8.2; CYB561; LOC100128288; CDDC163P; C1orf61; SKA2; BDNF; HTR2A; SLC5A3; ATP6V0E1; JUN; LOC100131662; and combinations thereof. As summarized in FIG. 17, the biomarker expression level can increase above a reference expression level of the biomarker or decrease below a reference expression level of the biomarker.
[0053] A particularly suitable sample for which the expression level of a biomarker is determined can be, for example, blood, including whole blood, serum, plasma, leukocytes, and megakaryocytes.
[0054] The method can further include assessing mood, anxiety, and other like psychiatric symptoms, and combinations thereof in the subject using questionnaires and/or a computer-implemented method for assessing mood, anxiety, other like psychiatric symptoms, and combinations thereof. In one aspect, the method is implemented using a first computer device coupled to a memory device, the method comprising: receiving mood information, anxiety information, and combinations thereof into the first computer device; storing, by the first computer device, the mood information, anxiety information, and combinations thereof in the memory device; computing, by the first computer device, of the mood information, anxiety information, and combinations thereof, a score that can be used to predict suicidality; presenting, by the first computer device, in visual form the mood information, anxiety information, and combinations thereof to a second computer device; receiving a request from the second computer device for access to the mood information, anxiety information, and combinations thereof and transmitting, by the first computer device, the mood information, anxiety information, and combinations thereof to the second computer device to assess mood, anxiety, and combinations thereof in the subject. Suitable mood and anxiety information is described herein in more detail below.
[0055] The method can further include assessing socio-demographic/psychological suicidal risk factors in the subject using a computer-implemented method for assessing socio-demographic/psychological suicidal risk factors in the subject, the method implemented using a first computer device coupled to a memory device, the method comprising: receiving socio-demographic/psychological suicidal risk factor information into the first computer device; storing, by the first computer device, the socio-demographic/psychological suicidal risk factor information in the memory device; presenting, by the first computer device, in visual form the socio-demographic/psychological suicidal risk factor information to a second computer device; receiving a request from the second computer device for access to socio-demographic/psychological suicidal risk factor information; and transmitting, by the first computer device, the socio-demographic/psychological suicidal risk factor information to the second computer device to assess the socio-demographic/psychological suicidal risk factors in the subject. Suitable socio-demographic/psychological suicidal risk factors are described herein in more detail below.
[0056] In accordance with the present disclosure, biomarkers useful for objectively predicting future hospitalization due to suicidality in subjects have been discovered. In one aspect, the present disclosure is directed to a method for future hospitalization due to suicidality in a subject. The method includes obtaining a first expression level of a blood biomarker in an initial sample obtained from the subject; and determining a second expression level of the blood biomarker in a subsequent sample obtained from the subject, wherein an increase in the expression level of the blood biomarker in the subsequent sample obtained from the subject as compared to the expression level of the initial sample indicates a higher risk of future hospitalizations due to suicidality. In some embodiments, the methods further include obtaining clinical risk factor information and clinical scale data such as for anxiety, mood and/or psychosis from the subject in addition to obtaining blood biomarker expression level in a sample obtained from the subject.
[0057] Suitable biomarkers for predicting future hospitalization due to suicidality in a subject wherein an increase in the expression level of the blood biomarker occurs can be, for example, the blood biomarker(s) set forth in Table 1.
[0058] In another embodiment, the expression level of the blood biomarker in the sample obtained from the subject is increased as compared to the reference expression level of the biomarker. Suitable biomarkers that indicate a risk for future hospitalization due to suicidality when the expression level increases in males as compared to the reference expression level have been found to include, for example, solute carrier family 4 (sodium bicarbonate cotransporter), member 4 (SLC4A4), cell adhesion molecule 1 CADM1, dystrobrevin, alpha (DTNA), spermidine/spermine N1-acetyltransferase 1 (SAT1), interleukin 6 (interferon, beta 2) (IL6) and combinations thereof. Suitable biomarkers that indicate a risk for future hospitalization due to suicidality when the expression level increases in females as compared to the reference expression level have been found to include, for example, erythrocyte membrane protein band 4.1 like 5 (EPB41L5), HtrA serine peptidase 1 (HTRA1), deleted in primary ciliary dyskinesia homolog (DPCD), general transcription factor IIIC, polypeptide 3, 102 kDa (GTF3C3), period circadian clock 1 (PER1), pyridoxal-dependent decarboxylase domain containing 1 (PDXDC1), kelch-like family member 28 (KLHL28), ubiquitin interaction motif containing 1 (UIMC1), sorting nexin family member 27 (SNX27) and combinations thereof.
[0059] In another embodiment, the expression level of the blood biomarker in the sample obtained from the subject is decreased as compared to the reference expression level of the biomarker. Suitable biomarkers that indicate a risk for future hospitalization due to suicidality when the expression level decreases in males as compared to the reference expression level have been found to include, for example, spindle and kinetochore associated complex subunit 2 (SKA2), CAP-GLY domain containing linker protein family, member 4 (CLIP4), kinesin family member 2C (KIF2C), kelch domain containing 3 (KLHDC3) and combinations thereof. Suitable biomarkers that indicate a risk for future hospitalization due to suicidality when the expression level decreases in females as compared to the reference expression level have been found to include, for example, phosphatidylinositol 3-kinase, catalytic subunit type 3 (PIK3C3), aldehyde dehydrogenase 3 family, member A2 (ALDH3A2), ARP3 actin-related protein 3 homolog (yeast) (ACTR3), B-cell CLL (BCL2), MOB kinase activator 3B (MOB3B), casein kinase 1, alpha 1 (CSNK1A1), La ribonucleoprotein domain family, member 4 (LARP4), zinc finger protein 548 (ZNF548) and combinations thereof.
[0060] Particularly suitable subjects are humans. Suitable subjects can also be experimental animals such as, for example, monkeys and rodents, that display a behavioral phenotype associated with suicide, for example, a mood disorder or psychosis. In one particular embodiment, the subject is a female human. In another particular aspect, the subject is a male human.
[0061] In another aspect, the subject can further be diagnosed with a psychiatric disorder. The psychiatric disorder can be bipolar disorder, major depressive disorder, schizophrenia, and schizoaffective disorder, post-traumatic stress disorder and combinations thereof.
[0062] A particularly suitable sample for which the expression level of a biomarker is determined can be, for example, blood, including whole blood, serum, plasma, leukocytes, and megakaryocytes.
[0063] Suitable biomarkers found to have a difference in expression level include, for example, spermidine/spermine N1-acetyltransferase 1 (SAT1), interleukin 6 (interferon beta 2) (IL6), solute carrier family 4 (sodium bicarbonate cotransporter), member 4 (SLC4A4), spindle and kinetochore associated complex subunit 2 (SKA2), jun proto-oncogen (JUN), cell adhesion molecule 1 (CADM1), dystrobrevin alpha (DTNA), monoamine oxidase B (MAOB), myristoylated alanine-rich protein kinase C substrate (MARCKS), phosphatase and tensin homolog (PTEN), fatty acid desaturase 1 (FADS1), Rho GTPase activating protein 26 (ARHGAP26), B-cell CLL/lymphoma 2 (BCL2), cadherin 4 type 1 R cadherin (retinal) (CDH4), chemokine (C-X-C motif) ligand 11 (CXCL11), EMI domain containing 1 (EMID1), family with sequence similarity 49 member B (FAM49B), GRINUA complex locus (GCOM1), hippocalcin-like 1 (HPCAL1), mitogen-activated protein kinase 9 (MAPK9), nuclear paraspeckle assembly transcript 1 (NEAT1), protein tyrosine kinase 2 (PTK2), RAS-like family 11 member B (RASL11B), small nucleolar RNA H/ACA box 68 (SNORA68), superoxide dismutase 2 mitochondrial (SOD2), transcription factor 7-like 2 (T-cell specific HMG-box) (TCF7L2), v-raf murine sarcoma viral oncogene homolog (BRAF), Chromosome 1 Open Reading Frame 61 (C1orf61), calreticulin (CALR), calcium/calmodulin-dependent protein kinase II beta (CAMK2B), caveolin 1 caveolae proein 22 kDa (CAV1), chromodomain helicase DNA binding protein 2 (CHD2), cAMP responsive element modulators (CREM), cortactin (CTTN), disheveled associated activator of morphogenesis 2 (DAAM2), Dab mitogen responsive phosphoprotein homolog 2 (DAB2), GABA(A) receptor associated protein like 1 (GABARAPL1), glutamate-ammonia ligase (GLUL), helicase with zinc finger (HELZ), immunoglobulin heavy chain constant gamma 1 (IGHG1), interleukin 1 beta (IL1B), jun B proto-oncogen (JUNB), lipoma HMGIC fusion partner (LHFP), metallothionein 1 E (MT1E), metallothionein 1 H (MT1H), metallothionein 2 (MT2A), N-myc downstream regulated 1 (NDRG1), nucleobindin 2 (NUCB2), PHD finger protein 20-like 1 (PHF20L1), cysteine-rich protein with kazal motifs (RECK), shisa family member 2 (SHISA2), transmembrane 4 L six family member 1 (TM4SF1), trophoblast glycoprotein (TPBG), tumor protein D52-like 1 (TPD52L1), TSC22 domain family member 3 (TSC22D3), vacuole membrane protein 1 (VMP1), ZFP 36 ring finger protein (ZFP36), zink finger FYVE domain containing 21 (ZHX2), histone cluster 1 H2bo (HIST1H2BO), keratocan (KERA), transcription factor Dp-1 (TFDP1), Single-Stranded DNA Binding Protein 2 (SSBP2), Transcription Factor EC (TFEC), Diphosphoinositol Pentakisphosphate Kinase 1 (PPIP5K1), Fibroblast Growth Factor Receptor 1 Oncogene Partner 2 (FGFR1OP2), Zinc Finger MYND-Type Containing 8 (ZMYND8), Interferon Gamma (IFNG), Brain-Derived Neurotrophic Factor (BDNF), cAMP Responsive Element Binding Protein 1 (CREB1), Hes Family BHLH Transcription Factor 1 (HES1), Ankyrin Repeat And MYND Domain Containing 1 (ANKMY1), Aldehyde Dehydrogenase 3 Family Member A2 (ALDH3A2), Heparan Sulfate (Glucosamine) 3-O-Sulfotransferase 3B1 (HS3ST3B1), Kinase Suppressor Of Ras 1 (KSR1), Dual Adaptor Of Phosphotyrosine And 3-Phosphoinositides (DAPP1), Opioid Receptor Mu 1 (OPRM1), Spectrin Beta Non-Erythrocytic 1 (SPTBN1), PHD Finger Protein 20 (PHF20), EIF1B Antisense RNA 1 (EIF1B-AS1), Talin 1 (TLN1), Nuclear Casein Kinase And Cyclin-Dependent Kinase Substrate 1 (NUCKS1), Delta-Like 1 Homolog (DLK1), BBSome Interacting Protein 1 (BBIP1), Interleukin 10 (IL10), Glycine Amidinotransferase (GATM), PRP40 Pre-MRNA Processing Factor 40 Homolog A (PRPF40A), Ubiquitin Specific Peptidase 48 (USP48), Nitrogen Permease Regulator-Like 3 (NPRL3), Testis-Specific Y-Encoded-Like Protein-Like 1 (TSPYL1), thymosin beta 15B (TMSB15B), Minichromosome Maintenance Complex Component 8 (MCM8), tensin 1 (TNS1), Tumor Necrosis Factor (TNF), S100 Calcium Binding Protein B (S100B), Basic Leucine Zipper Transcription Factor ATF-Like 2 (BATF2), Annexin A11 (ANX11), RP11-389C8.2, Cytochrome B561 (CYB561), LOC100128288 (Uncharacterized LOC100128288), Coiled-Coil Domain Containing 163 Pseudogene (CCDC163P), 5-Hydroxytryptamine (Serotonin) Receptor 2A, G Protein-Coupled (HTR2A), Annexin A11 (ANXA11), Uncharacterized LOC100131662 (LOC100131662), Prolylcarboxypeptidase (Angiotensinase C; PRCP), and combinations thereof. See, FIG. 9 for a list of biomarkers identified as showing a difference in expression level.
[0064] In another aspect, the present disclosure is directed to a method for mitigating suicidality in a subject in need thereof. The method includes: obtaining an expression level of a blood biomarker in a sample obtained from the subject; obtaining a reference expression level of the blood biomarker; identifying a difference in the expression level of the blood biomarker in the sample as compared to the reference expression level of the blood biomarker; and administering a treatment, wherein the treatment reduces the difference between the expression level of the blood biomarker in the sample as compared to the reference expression level of the blood biomarker to mitigate suicidality in the subject. As used herein, "mitigate", "mitigating", and the like refer to making a condition less severe and/or preventing a condition. More particularly, the phrase "mitigate suicidality" refers to reducing suicide ideation in a subject and/or preventing suicide completion.
[0065] Suitable treatments can be a lifestyle modification, administering a therapy, and combinations thereof.
[0066] Suitable therapy can be a nutritional, a drug and psychotherapy.
[0067] Particularly suitable nutritionals can be omega-3 fatty acids, including, by way of example, docosahexaenoic acid (DHA).
[0068] Particularly suitable drugs include, for example, ketamine, lithium, clozapine, selegeline, tocilizumab, siltuximab, enkephalin, methionine, gevokizumab, gallium nitrate, vemurafenib, dabrafenib, oblimersen, rasagiline,(-)-gossypol, navitoclax, gemcitabine/paclitaxel, bortezomib/paclitaxel, ABT-199, paclitaxel/trastuzumab, paclitaxel/pertuzumab/trastuzumab, lapatinib/paclitaxel, doxorubicin/paclitaxel, epirubicin/paclitaxel, paclitaxel/topotecan, paclitaxel, canakinumab, tesevatinib, enzastaurin, fomepizole, miglitol, anakinra, and combinations thereof. Other suitable drugs, as well as biomarkers found to be changed in opposite direction in suicide versus in treatments with omega-3 fatty acids, lithium, clozapine, or antidepressants (MAOIs) as listed in Tables 4 & 5. These biomarkers could potentially be used to stratify patients to different treatment approaches, and monitor their responses.
TABLE-US-00004 TABLE 4 Top candidate biomarker genes - drugs that modulate expression of these markers in the opposite direction in male subjects Discovery (Change) Gene symbol/ Method/ Modulated by Modulated by Modulated by Other Gene Name Score Omega-3 Lithium Clozapine Drugs CCDC136 (D) (I) coiled-coil domain AP4 Mouse VT.sup.356 containing 136 CD44 (D) (I) CD44 molecule (Indian DE2 Mouse Blood.sup.356 blood group) IL6 (I) (D) tocilizumab interleukin 6 (interferon, AP2 Human Blood.sup.357 siltuximab beta 2) SAT1 (I) (D) spermidine/spermine N1- DE2 Mouse Blood.sup.358 acetyltransferase 1 DE1 MAOB (I) selegiline monoamine oxidase B DE1 ARHGAP26 (I) (D) Rho GTPase activating DE1 Mouse VT.sup.356 protein 26 BCL2 (D) (I) (I) B-cell CLL/lymphoma 2 DE1 Human Blood.sup.153 Rat Dentate gyrus Hippocampus.sup.359 EHBP1 (D) (I) VT.sup.356 EH domain binding protein DE 4 1 FAM49B (I) (D) family with sequence AP2 Mouse Blood.sup.358 similarity 49, member B HPCAL1 (I) (D) hippocalcin-like 1 DE2 Mouse VT.sup.356 MAPK9 (I) (D) mitogen-activated protein DE2 Mouse VT.sup.356 kinase 9 NEAT1 (I) (D) nuclear paraspeckle DE2 Mouse VT.sup.356 assembly transcript 1 (non- protein coding) RASL11B (I) (D) RAS-like, family 11, AP2 Mouse Caudate member B putamen.sup.356 TRAK2 (D) (I) (I) trafficking protein, kinesin DE2 Mouse Blood.sup.358 Mouse PFC.sup.360 binding 2 ADRBK1 adrenergic, beta, (D) (I) receptor kinase 1 DE1 Mouse PEC.sup.361 BRAE (I) Vemurafenib v-raf murine sarcoma viral DE1 Dabrafenib oncogene homolog B CAMK2B (I) (D) calcium/calmodulin- DE1 Mouse striatum.sup.362 dependent protein kinase II beta CNP (D) (I) (I) 2',3'-cyclic nucleotide 3' AP1 Mouse Hippocampus.sup.358 Mouse AMY.sup.356 phosphodiesterase CTTN cortactin (I) (D) (D) DE1 Mouse Blood.sup.358 Mouse VT.sup.356 G2E3 (D) (I) G2/M-phase specific E3 AP1 Mouse Hippocampus.sup.358 ubiquitin protein ligase GABARAPL1 GABA(A) (I) (D) receptor-associated protein DE1 Mouse Blood.sup.358 like 1 HELZ helicase with zinc (I) (D) finger DE1 Mouse Blood.sup.358 IL1B (I) (D) canakinumab interleukin 1, beta DE1 Mouse Blood.sup.358 gevokizumab gallium nitrate LHFP lipoma HMGIC (I) (D) fusion partner DE1 Mouse Blood.sup.358 LPAR1 lysophosphatidic (D) (I) (I) acid receptor 1 AP1 Mouse Hippocampus, Mouse AMY.sup.356 Blood.sup.358 MBP myelin basic protein (D) (I) (I) (I) AP1 Mouse Blood.sup.358 Oligodendrocy Mouse AMY and tes.sup.363 Blood.sup.356 Mouse Brain.sup.360 MEF2C myocyte enhancer (D) (I) factor 2C DE1 Mouse Hippocampus and VT.sup.356 NDRG1 (I) (D) N-myc downstream DE1 Mouse Blood.sup.358 regulated 1 OGFR (D) enkephalin opioid growth factor DE1 methionine receptor PCDH9 protocadherin 9 (D) (I) AP1 Mouse VT.sup.356 PHF20L1 (I) (D) (D) PHD finger protein 20-like 1 DE1 Mouse Blood.sup.358 Mouse Hippocampus.sup.356 PRKCB protein kinase C, (D) (I) beta DE1 Mouse PEC.sup.360 AP1 AMY.sup.364 RBMX RNA binding motif (D) (I) protein, X-linked DE1 Mouse NAC, Blood.sup.358 RNASEL ribonuclease L (D) (I) (2',5'-oligoisoadenylate AP1 Mouse Blood.sup.358 synthetase-dependent) SNAP23 synaptosomal- (D) (I) associated protein, 23 kDa AP1 Mouse Blood.sup.356 TM4SF1 transmembrane 4 (I) (D) L six family member 1 DE1 Mouse Blood.sup.358 TSPAN33 tetraspanin 33 (D) (I) (I) AP1 Mouse Blood.sup.358 Mouse VT.sup.356 VMP1 (I) (D) vacuole membrane protein 1 DE1 Mouse Blood.sup.358 ZFP36 (I) (D) (D) ZFP36 ring finger protein DE1 Mouse Blood.sup.358 Rat Brain.sup.365 BTBD3 (I) (D) BTB (POZ) domain DE 4 Mouse AMY.sup.358 containing 3 CADM1 (I) (D) cell adhesion molecule 1 DE4 Mouse VT.sup.356 CTBS (I) (D) chitobiase, di-N-acetyl- DE 4 VT.sup.356 LAMB1 (I) (D) laminin, beta 1 AP4 Mouse HIP.sup.358 PLEC (D) (I) plectin DE 4 Mouse VT.sup.356 RAD23A (D) (I) RAD23 homolog A DE 4 Mouse Blood.sup.358 (S. cerevisiae) SETD8 (D) (I) SET domain containing DE 4 Mouse Blood.sup.358 (lysine methyltransferase) 8 TXNRD2 (D) (I) thioredoxin reductase 2 AP4 Mouse Blood.sup.356 (I): increase in biomarker expression; (D): decrease in biomarker expression
TABLE-US-00005 TABLE 5 Top candidate biomarker genes - drugs that modulate expression of these markers in the opposite direction in female subjects Discovery (Change) Gene Symbol/ Method/ Modulated by Modulated by Modulated by Gene Name Score Omega-3 Lithium Clozapine Other Drugs Out of Validated Biomarkers (Bonferroni) (49 genes, 50 probesets) BCL2 (D) (I) (I) oblimersen, rasagiline, (-)- B-cell CLL DE/2 FC Hip gossypol, navitoclax, (Chen, Zeng et al. (Bai, Zhang et al. gemcitabine/paclitaxel, 1999) 2004) bortezomib/paclitaxel, ABT-199, (I) paclitaxel/trastuzumab, cerebellar granule paclitaxel/pertuzumab/trastuzumab, cells lapatinib/paclitaxel, (Chen and Chuang doxorubicin/paclitaxel, 1999) epirubicin/paclitaxel, (I) paclitaxel/topotecan, paclitaxel Human Blood (Lowthert, Leffert et al. 2012) (I) Astrocyte (Keshavarz, Emamghoreishi et al. 2013) (I) HIP (Chen, Rajkowska et al. 2000) (I) Dentate gyrus, HIP(Hammonds and Shim 2009) GSK3B (D) (I) enzastaurin glycogen synthase DE/1 FC (Fatemi, kinase 3 beta Reutiman et al. 2009) CAT (D) Oxidative Stress BP fomepizole catalase DE/2 (I) Plasma (de Sousa, Zarate et al. 2014) JUN (I) (D) (D) jun proto-oncogene DE/2 leukocytes FC DE/1 (Watanabe, Iga et al. (MacDonald, Eaton et 2014) al. 2005) MOB3B (D) (I) MOB kinase activator DE/1 PFC (females) (Le- 3B Niculescu, Case et al. 2011) NDRG1 (I) (D) N-myc downstream DE/1 Blood(Le-Niculescu, regulated 1 Case et al. 2011) SPON1 (D) (I) spondin 1, DE/1 VT extracellular matrix (Le-Niculescu, protein Balaraman et al. 2007) FOXP1 (I) (D) forkhead box P1 DE/4 Blood(Le-Niculescu, Case et al. 2011) HAVCR2 (I) (D) hepatitis A virus DE/4 PFC cellular receptor 2 (Jakovcevski, Bharadwaj et al. 2013) GJA1 (I) (D) (D) gap junction protein, DE/1 HIP (females) (Le- VT alpha 1, 43 kDa Niculescu, Case et al. (Le-Niculescu, 2011) Balaraman et al. 2007) CD84 (D) (I) CD84 molecule DE/2 Blood (Le-Niculescu, Balaraman et al. 2007) DCAF5 (D) (I) DDB1 and CUL4 DE/2 VT associated factor 5 (Le-Niculescu, Balaraman et al. 2007) GANC (D) miglitol glucosidase, alpha; DE/2 neutral C IL1R1 (I) anakinra interleukin 1 receptor, AP/1 type I INPP4A (D) (I) inositol polyphosphate- DE/1 VT 4-phosphatase, type I, (Le-Niculescu, 107 kDa Balaraman et al. 2007) JRK (D) (I) Jrk homolog (mouse) AP/2 Brain(Hammamieh, Chakraborty et al. 2014) PDXDC1 (I) (D) pyridoxal-dependent DE/2 VT decarboxylase domain (Le-Niculescu, containing 1 Balaraman et al. 2007) SMARCA2 (D) (I) SWI DE/1 HIP (males) (Le- Niculescu, Case et al. 2011) Out of Top Discovery and Prioritization Biomarkers(Non Bonferroni Validated, 65 genes) CLTA (I) (D) clathrin, light chain A DE/4 FC (MacDonald, Eaton et al. 2005) PPM1B (D) (I) protein phosphatase, DE/4 VT Mg2+ (Le-Niculescu, Balaraman et al. 2007) AFF3 (I) (D) AF4/FMR2 family, AP/4; Blood (Le- member 3 (I) Niculescu, Case et al. DE/1 2011) WAC (D) (I) WW domain DE/4 VT containing adaptor (Le-Niculescu, with coiled-coil Balaraman et al. 2007) AKT3 (I) enzastaurin v-akt murine thymoma AP/4 viral oncogene homolog 3 ARID4B (D) (I) AT rich interactive DE/4 HIP (males) (Le- domain 4B (RBP1- Niculescu, Case et al. like) 2011) ATXN1 (I) (D) ataxin 1 DE/4 Blood(Le-Niculescu, Case et al. 2011) BRE (I) (D) Brain and AP/4 VT reproductive organ- (Le-Niculescu, expressed (TNFRSF1A Balaraman et al. 2007) modulator) CSNK1A1 (D) (I) casein kinase 1, alpha DE/4 Blood(Le-Niculescu, 1 Case et al. 2011) ENTPD1 (I) (D) (D) ectonucleoside AP/4 Blood(Le-Niculescu, PFC triphosphate Case et al. 2011) (Jakovcevski, diphosphohydrolase 1 Bharadwaj et al. 2013) EPHB4 (I) tesevatinib EPH receptor B4 DE/4 ETNK1 (D) (I) ethanolamine kinase 1 AP/4 PFC (males)(Le- Niculescu, Case et al. 2011) ITIH5 (I) (D) (D) inter-alpha-trypsin AP/4 PFC inhibitor heavy chain Blood(Le-Niculescu, (Jakovcevski, family, member 5 Case et al. 2011) Bharadwaj et al. 2013) LARP4 (D) (I) La ribonucleoprotein DE/4 VT domain family, (Le-Niculescu, member 4 Balaraman et al. 2007) MBNL1 (D) (I) (I) muscleblind-like DE/4 HIP (males) (Le- Blood splicing regulator 1 Niculescu, Case et al. (Le-Niculescu, 2011) Balaraman et al. 2007) MR1 (I) Anti-Lymphocyte serum major DE/4 histocompatibility complex, class I- related PRDX3 (D) (I) peroxiredoxin 3 DE/4 Blood(Le-Niculescu, Case et al. 2011) RAB22A (D) (I) RAB22A, member DE/4 Blood RAS oncogene family (Le-Niculescu, Balaraman et al. 2007) SNX27 (I) (D) sorting nexin family AP/4 AMY member 27 (Le-Niculescu, Balaraman et al. 2007) SSBP2 (I) (D) (D) single-stranded DNA AP/4 Blood(Le-Niculescu, VT binding protein 2 Case et al. 2011) (Le-Niculescu, Balaraman et al. 2007) WAPAL (D) (I) (I) wings apart-like DE/4 SK-N-AS cells VT homolog (Drosophila) (ATCC derived from (Le-Niculescu, a human Balaraman et al. 2007) neuroblastoma cell (Seelan, Khalyfa et al. 2008) (I): increase in biomarker expression; (D): decrease in biomarker expression
[0069] More particularly, it has been found that BCL2, JUN, GHA1, ENTPD1, ITIH5, MBNL1, and SSBP2 are changed in expression by the above listed treatments, and in particular therapies such as nutritionals and drugs, suggesting these biomarkers may be core to the anti-suicidal mechanism of these drugs. Further, BCL2, CAT, and JUN may be useful blood pharmacogenomic markers of response to lithium. CD84, MBNL1, and RAB22A may be useful blood pharmacogenomic markers of response to clozapine. NDRG1, FOXP1, AFF3, ATXN1, CSNK1A1, ENTPD1, ITIH5, PRDX3, and SSBP2 may be useful blood pharmacogenomic markers of response to omega-3 fatty acids. Three existing drugs, used for other indications, have been identified as targeting the top suicide biomarkers identified in the present disclosure, and could potentially be re-purposed for testing in treatment of acute suicidality: anakinra (inhibiting ILR1), enzastaurin (inhibiting AKT3), and tesevatinib (inhibiting EPHB4). Additionally, Connectivity Map analyses (FIGS. 34A-34C) identified novel compounds that induce gene expression signatures that are the opposite of those present in suicide, and might generate leads and/or be tested for use to treat/prevent suicidality: betulin (an anti-cancer compound from the bark of birch trees), zalcitabine (an anti-HIV drug), and atractyloside (a toxic glycoside). Other common drugs identified by the Connectivity Map analyses are nafcillin, lansoprazole, mifepristone, LY294002, minoxidil, acetysalicilic acid, estradiol, buspirone, dicloxacillin, corticosterone, metformin, diphenhydramine, haloperidol, metaraminol, yohimbine, trimethadione and fluoxetine (see also Table 6, 7, and 8).
TABLE-US-00006 TABLE 6 Therapeutic Compounds for Suicidality across Gender Therapeutic compound/Drug Score* fluoxetine -0.812 betulin -0.812 dl-alpha tocopherol -0.821 haloperidol -0.823 hesperidin -0.824 calcium folinate -0.825 harpagoside -0.826 trimipramine -0.836 rilmenidine -0.845 tenoxicam -0.851 chlorpromazine -0.852 harman -0.858 homatropine -0.863 ramifenazone -0.864 clozapine -0.866 diphenhydramine -0.873 prochlorperazine -0.874 pirenperone -0.876 asiaticoside -0.886 adiphenine -0.923 verapamil -0.922 metaraminol -0.936 vohimbine -0.958 metformin -0.983 trimethadione -1 chlorogenic acid -1 *Score of -1 means maximum opposite effect.
TABLE-US-00007 TABLE 7 Therapeutic Compounds for Suicidality in Men Therapeutic compound/drug Score* thiamine -0.778 homatropine -0.789 vitexin -0.794 ergocalciferol -0.801 tropicamide -0.801 (-)-atenolol -0.817 betulin -0.905 spaglumic acid -1 *Score of -1 means maximum opposite effect.
TABLE-US-00008 TABLE 8 Therapeutic Compounds for Suicidality in Women Therapeutic compound/drug Score* mifepristone -0.797 lansoprazole -0.888 nafcillin -0.895 betulin -1 *Score of -1 means maximum opposite effect.
[0070] In another aspect, the subject can further be diagnosed with a psychiatric disorder. The psychiatric disorder can be any psychiatric disorder known in the art, including, for example, bipolar disorder, major depressive disorder, schizophrenia, and schizoaffective disorder, post-traumatic stress disorder, and combinations thereof.
[0071] In another aspect, the present disclosure is directed to a questionnaire and/or a computer-implemented method for assessing mood, anxiety, and combinations thereof in the subject using a computer-implemented method for assessing mood, anxiety, and the like, and combinations thereof. In one aspect, the method is implemented using a computer device coupled to a memory device. The method implemented using a first computer device coupled to a memory device includes receiving mood information, anxiety information, and combinations thereof into the first computer device; storing, by the first computer device, the mood information, anxiety information, and combinations thereof in the memory device; presenting, by the first computer device, in visual form the mood information, anxiety information, and combinations thereof to a second computer device; receiving a request from the second computer device for access to the mood information, anxiety information, and combinations thereof and transmitting, by the first computer device, the mood information, anxiety information, and combinations thereof to the second computer device to assess mood, anxiety, and combinations thereof in the subject.
[0072] Mood information includes information relating to a subject's mood, motivation, movement, thinking, self-esteem, interest, appetite, and combinations thereof Anxiety information includes information relating to a subjects anxiety, uncertainty, fear, anger, and combinations thereof. Particular mood and anxiety information assessed can include: determining how good is the subject's mood; determining the subject's motivation, drive, determination to do things right now; determining how high is the subject's physical energy and the amount of moving about that the subject feels like doing right now; determining how high is the subject's mental energy and thinking activity going on in the subject's mind right now; determining how good the subject feels about himself/herself and his/her accomplishments right now; determining how high the subject's interest to do things that are fun and enjoyable right now; determining how high the subjects appetite and desire for food is right now; determining how anxious the subject is right now; determining how uncertain about things the subject is right now; determining how frightened about things the subject feels right now; determining how angry about things the subject feels right now; determining events or actions the subject thinks are influencing how the subject feels right now; determining additional feelings the subject has right now; and combinations thereof. As illustrated in FIG. 6, the mood and anxiety information can be assessed by having the subject rate each piece of information on a scale of lowest to highest.
[0073] The subject of the method can further be diagnosed as having a psychiatric disorder selected from bipolar disorder, major depressive disorder, schizophrenia, and schizoaffective disorder, post-traumatic stress disorder, and combinations thereof.
[0074] In another aspect, the present disclosure is directed to a computer-implemented method for assessing socio-demographic/psychological suicidal risk factors in the subject using a computer-implemented method for assessing socio-demographic/psychological suicidal risk factors in the subject, the method implemented using a computer device coupled to a memory device. The method includes: receiving socio-demographic/psychological suicidal risk factor information into the first computer device; storing, by the first computer device, the socio-demographic/psychological suicidal risk factor information in the memory device; presenting, by the first computer device, in visual form the socio-demographic/psychological suicidal risk factor information to a second computer device; receiving a request from the second computer device for access to socio-demographic/psychological suicidal risk factor information; and transmitting, by the first computer device, the socio-demographic/psychological suicidal risk factor information to the second computer device to assess the socio-demographic/psychological suicidal risk factors in the subject.
[0075] Socio-demographic and clinical risk factors for suicide includes items for assessing the influence of mental health factors, as well as of life satisfaction, physical health, environmental stress, addictions, cultural factors known to influence suicidal behavior, and two demographic factors, age and gender. Socio-demographic/psychological suicidal risk factors assessed can include: lack of coping skills when faced with stress; dissatisfaction with current life; lack of hope for the future; current substance abuse; acute loss/grief; psychiatric illness diagnosed and treated; poor treatment compliance; family history of suicide in blood relatives; personally knowing somebody who committed suicide; history of abuse (such as physical abuse, sexual abuse, emotional abuse, and neglect); acute/severe medical illness (including acute pain); chronic stress (including perceived uselessness, not feeling needed, and burden to extended kin); history of excessive introversion/conscientiousness (including planned suicide attempts); past history of suicidal acts/gestures; lack of religious beliefs; rejection; lack of positive relationships/social isolation; history of excessive extroversion and impulsive behavior (including rage, anger, physical fights and seeking revenge); lack of children/not in touch with children/not helping care for children; history of command hallucinations of self-directed violence; age (older than 60 years or younger than 25 years); gender; and combinations thereof.
[0076] The socio-demographic/psychological suicidal risk factors can be assessed by having the subject provide an answer to the above factors such as a yes answer, a no answer and a not applicable answer.
[0077] The subject of the method can further be diagnosed as having a psychiatric disorder selected from bipolar disorder, major depressive disorder, schizophrenia, and schizoaffective disorder, post-traumatic stress disorder, and combinations thereof.
[0078] In another aspect, the present disclosure is directed to a method for predicting suicidality in a subject. The method includes: identifying a difference in the expression level of a blood biomarker in a sample obtained from a subject and a reference expression level of the blood biomarker by obtaining the expression level of the blood biomarker in a sample obtained from a subject; obtaining a reference expression level of a blood biomarker; analyzing the blood biomarker in the sample obtained from the subject and the reference expression level of the blood biomarker to detect the difference between the blood biomarker in the sample and the reference expression level of the blood biomarker; assessing mood, anxiety, and combinations thereof, using a first computer device coupled to a memory device, wherein the first computer device receives mood information, anxiety information, and combinations thereof into the first computer device; storing, by the first computer device, the mood information, anxiety information, and combinations thereof in the memory device; presenting, by the first computer device, in visual form the mood information, anxiety information, and combinations thereof to a second computer device; receiving a request from the second computer device for access to the mood information, anxiety information, and combinations thereof; and transmitting, by the first computer device, the mood information, anxiety information, and combinations thereof to the second computer device to assess mood, anxiety, and combinations thereof in the subject; assessing socio-demographic/psychological suicidal risk factors in the subject using the first computer device coupled to a memory device, wherein the first computer device receives socio-demographic/psychological suicidal risk factor information into the first computer device; storing, by the first computer device, the socio-demographic/psychological suicidal risk factor information in the memory device; presenting, by the first computer device, in visual form the socio-demographic/psychological suicidal risk factor information to the second computer device; receiving a request from the second computer device for access to socio-demographic/psychological suicidal risk factor information; and transmitting, by the first computer device, the socio-demographic/psychological suicidal risk factor information to the second computer device to assess the socio-demographic/psychological suicidal risk factors in the subject; and predicting suicidality in the subject by the combination of the difference between the expression level of the biomarker in the subject and the reference expression level of the blood biomarker; the assessment of mood, anxiety, and combinations thereof; and the assessment of socio-demographic/psychological suicidal risk factor information.
[0079] As used herein, while the methods are described as using a first and second computer device, it should be understood that more or less than two computer devices may be used to perform the methods of the present disclosure. Particularly, three computer devices, or four computer devices or even five or more computer devices can be used to perform the methods without departing from the scope of the present disclosure.
[0080] In one aspect, the present disclosure is directed to a method for predicting future hospitalization of a subject due to suicidality. The method includes: identifying a difference in the expression level of a blood biomarker in a sample obtained from a subject and a reference expression level of the blood biomarker by obtaining the expression level of the blood biomarker in a sample obtained from a subject; obtaining a reference expression level of a blood biomarker; analyzing the blood biomarker in the sample obtained from the subject and the reference expression level of the blood biomarker to detect the difference between the blood biomarker in the sample and the reference expression level of the blood biomarker; assessing mood, anxiety, and combinations thereof, using a first computer device coupled to a memory device, wherein the first computer device receives mood information, anxiety information, and combinations thereof into the first computer device; storing, by the first computer device, the mood information, anxiety information, and combinations thereof in the memory device; presenting, by the first computer device, in visual form the mood information, anxiety information, and combinations thereof to a second computer device; receiving a request from the second computer device for access to the mood information, anxiety information, and combinations thereof; and transmitting, by the first computer device, the mood information, anxiety information, and combinations thereof to the second computer device to assess mood, anxiety, and combinations thereof in the subject; assessing socio-demographic/psychological suicidal risk factors in the subject using the first computer device coupled to a memory device, wherein the first computer device receives socio-demographic/psychological suicidal risk factor information into the first computer device; storing, by the first computer device, the socio-demographic/psychological suicidal risk factor information in the memory device; presenting, by the first computer device, in visual form the socio-demographic/psychological suicidal risk factor information to a second computer device; receiving a request from the second computer device for access to socio-demographic/psychological suicidal risk factor information; and transmitting, by the first computer device, the socio-demographic/psychological suicidal risk factor information to the second computer device to assess the socio-demographic/psychological suicidal risk factors in the subject; and predicting future hospitalization of the subject due to suicidality by the combination of the difference between the expression level of the biomarker in the subject and the reference expression level of the blood biomarker; the assessment of mood, anxiety, and combinations thereof; and the assessment of socio-demographic/psychological suicidal risk factor information.
[0081] Suitable biomarkers for use in the method for predicting suicide ideation in a subject and the method for predicting future hospitalization a subject due to suicidality include those described herein.
[0082] Mood information for use in the method for predicting suicide ideation in a subject and the method for predicting future hospitalization of a subject due to suicidality includes information relating to a subject's mood, motivation, movement, thinking, self-esteem, interest, appetite, and combinations thereof as described herein. Anxiety information includes information relating to a subjects anxiety, uncertainty, fear, anger, and combinations thereof as described herein.
[0083] Socio-demographic and clinical risk factors for suicide for use in the method for predicting suicide ideation in a subject and the method for predicting future hospitalization of a subject due to suicidality include items for assessing the influence of mental health factors, as well as of life satisfaction, physical health, environmental stress, addictions, cultural factors known to influence suicidal behavior, and two demographic factors, age and gender as described herein.
EXAMPLES
[0084] Methods
[0085] Human Blood Gene Expression Experiments and Analyses
[0086] RNA extraction. Whole blood (2.5-5 ml) was collected into each PaxGene tube by routine venipuncture. PaxGene tubes contain proprietary reagents for the stabilization of RNA. RNA was extracted and processed.
[0087] Microarrays. Biotin-labeled aRNAs were hybridized to Affymetrix HG-U133 Plus 2.0 GeneChips (Affymetrix; with over 40 000 genes and expressed sequence tags), according to the manufacturer's protocols. Arrays were stained using standard Affymetrix protocols for antibody signal amplification and scanned on an Affymetrix GeneArray 2500 scanner with a target intensity set at 250. Quality-control measures, including 30/50 ratios for glyceraldehyde 3-phosphate dehydrogenase and .beta.-actin, scale factors, background and Q-values, were within acceptable limits.
[0088] Analysis. The participant's SI scores at the time of blood collection (0--no suicidal ideation (SI) compared with 2 and above--high SI) were used. Gene expression differences between the no SI and the high SI visits were analyzed using a within-participant design, then an across-participants summation (FIGS. 1C and 10C).
[0089] Gene Expression Analysis in the Discovery Cohort
[0090] Data was analyzed in two ways: an Absent-Present (AP) approach and a differential expression (DE) approach. The AP approach may capture turning on and off of genes, and the DE approach may capture gradual changes in expression. For the AP approach, Affymetrix Microarray Suite Version 5.0 (MASS) was used to generate Absent (A), Marginal (M), or Present (P) calls for each probe set on the chip (Affymetrix U133 Plus 2.0 GeneChips) for all participants in the discovery cohort. For the DE approach, all Affymetrix microarray data was imported as Cel. files into Partek Genomic Suites 6.6 software package (Partek Incorporated, St Louis, Mo., USA). Using only the perfect match values, a robust multi-array analysis (RMA) was run, background corrected with quantile normalization and a median polish probe set summarization, to obtain the normalized expression levels of all probe sets for each chip. RMA was performed independently for each of the 6 diagnoses used in the study, to avoid potential artefacts due to different ranges of gene expression in different diagnoses (Niculescu et al. MP 2015). Then the participants' normalized data was extracted from these RMA and assembled for the different cohorts used in the Example.
[0091] A/P analysis. For the longitudinal within participant AP analysis, comparisons were made within participant between sequential visits to identify changes in gene expression from Absent to Present that track changes in phene expression (suicidal ideation, "SI") from No SI to High SI. For a comparison, if there was a change from A to P tracking a change from No SI to High SI, or a change from P to A tracking a change from High SI to No SI, that was given a score of +1 (increased biomarker in High SI). If the change was in opposite direction in the gene vs the phene (SI), that was given a score of -1 (decreased biomarker in High SI). If there was no change in gene expression between visits, despite a change of phene expression (suicidal ideation), or a change in gene expression between visits, despite no change in phene expression (suicidal ideation), that was given a score of 0 (not tracking as a biomarker). If there was no change in gene expression and no change in suicidal ideation between visits, that was given a score of +1 if there was concordance (P-P with High SI-High SI, or A-A with No SI-No SI), or a score of -1 if there was the opposite (A-A with High SI-High SI, or P-P with No SI-No SI). If the changes were to M (moderate) instead of P, the values used were 0.5 or -0.5. These values were then summed up across the comparisons in each participant, resulting in a participant score for each gene/probeset in each participant. A perfection bonus was also used. If the gene expression perfectly tracked the suicidal ideation in a participant that had at least two comparisons (3 visits), that probe set was rewarded by a doubling of its participant score. Additionally, a non-tracking correction was used. If there was no change in gene expression in any of the comparisons for a particular participant, that overall participant score for that probe set in that participant was zero.
[0092] DE analysis. For the longitudinal within participant DE analysis, fold changes (FC) in gene expression were calculated between sequential visits within each participant. Scoring methodology was similar to that used above for AP. Probe sets that had a FC.gtoreq.1.2 were scored+1 (increased in High SI) or -1 (decreased in High SI). FC.gtoreq.1.1 were scored+0.5 or -0.5. FC lower than 1.1 were considered no change. The only difference between the DE and the AP analyses was when scoring comparisons where there was no phene expression (SI) change between visits and no change in gene expression between visits (FC lower than 1.1). In that case, the comparison received the same score as the nearest preceding comparison where there was a change in SI from visit to visit. If no preceding comparison with a change in SI was available, then it was given the same score as the nearest subsequent comparison where there was a change in SI. Also for DE, a perfection bonus and a non-tracking correction was used. If the gene expression perfectly tracked the suicidal ideation in a participant who had at least two comparisons (3 visits), that probe set was rewarded by a doubling of its score. If there was no change in gene expression in any of the comparisons for a particular participant, that overall participant score for that probe set in that participant was zero.
[0093] Internal score. Once scores within each participant were calculated, an algebraic sum across all participants was obtained for each probe set. Probe sets were then given internal CFG points based upon these algebraic sum scores. Probe sets with scores above the 33% of the distribution (for increased probe sets and decreased probe sets) received 1 point, those above 50% of the distribution received 2 points, and those above 80% of the distribution received 4 points.
[0094] In Example 1, for AP analyses, 23 probe sets received 4 points, 581 probe sets received 2 points, and 2077 probe sets received 1 point, for a total of 2681 probe sets. For DE analyses, 31 probe sets received 4 points, 1294 probe sets received 2 points, and 5839 probe sets received 1 point, for a total of 7164 probe sets. The overlap between the two discovery methods is shown in FIG. 2A. For Example 2, for AP analyses, 30 probesets received 4 points, 647 probesets with 2 points, and 2596 probesets with 1 point, for a total of 3273 probesets. For DE analyses, 95 probesets received 4 points, 2215 probesets with 2 points, and 7520 probesets with 1 point, for a total of 9829 probesets. The overlap between the two discovery methods for probesets with an internal score of 1 is shown in FIG. 11A.
[0095] Different probe sets may be found by the two methods due to differences in scope (DE capturing genes that were present in both visits of a comparison (i.e. PP, but are changed in expression), thresholds (what makes the 33% change cutoff across participants varies between methods), and technical detection levels (what is considered in the noise range varies between the methods).
[0096] In total, 9413 probe sets were identified with an internal CFG score of 1. Gene names for the probe sets were identified using NetAffyx (Affymetrix) and Partek for Affymetrix HG-U133 Plus 2.0 GeneChips, followed by GeneCards to confirm the primary gene symbol. In addition, for those probe sets that were not assigned a gene name by NetAffyx or Partek, the UCSC Genome Browser was used to directly map them to known genes, with the following limitations. In case the probe set fell in an intron, that particular gene was assumed to be implicated. Only one gene was assigned to each probe set. Genes were then scored using manually curated CFG databases as described below (FIGS. 2C and 11C).
[0097] Convergent Functional Genomics
[0098] Databases. Manually curated databases of all the human gene expression (postmortem brain, blood and cell cultures), human genetics (association, copy number variations and linkage), and animal model gene expression and genetic studies published to date on psychiatric disorders was established (Laboratory of Neurophenomics, Indiana University School of Medicine, www.neurophenomics.info). The databases include only primary literature data and do not include review papers or other secondary data integration analyses to avoid redundancy and circularity. These large and constantly updated databases have been used for CFG cross validation and prioritization (FIGS. 2B, 2C, 11B and 11C). For Example 2, data from 442 papers on suicide were present in the databases at the time of the CFG analyses (genetic studies-164, brain studies-192, peripheral fluids-86).
[0099] Human postmortem brain gene expression evidence. Converging evidence was scored for a gene if there were published reports of human postmortem data showing changes in expression of that gene or changes in protein levels in brains from participants who died from suicide.
[0100] Human blood and other peripheral tissue gene expression data. Converging evidence was scored for a gene if there were published reports of human blood, lymphoblastoid cell lines, CSF, or other peripheral tissue data showing changes in expression of that gene or changes in protein levels in participants who had a history of suicidality or who died from suicide.
[0101] Human genetic evidence (association and linkage). To designate convergence for a particular gene, the gene had to have independent published evidence of association or linkage for suicide. For linkage, the location of each gene was obtained through GeneCards (http://www.genecards.org), and the sex averaged cM location of the start of the gene was then obtained through http://compgen.rutgers.edu/mapinterpolator. For linkage convergence, the start of the gene had to map within 5 cM of the location of a marker linked to the disorder.
[0102] CFG scoring. For CFG analysis (FIGS. 2C and 11C), the external cross-validating lines of evidence were weighted such that findings in human postmortem brain tissue, the target organ, were prioritized over peripheral tissue findings and genetic findings, by giving them twice as many points. Human brain expression evidence was given 4 points, whereas human peripheral evidence was given 2 points, and human genetic evidence was given a maximum of 2 points for association and 1 point for linkage. Each line of evidence was capped in such a way that any positive findings within that line of evidence resulted in maximum points, regardless of how many different studies support that single line of evidence, to avoid potential popularity biases. In addition to the external CFG score, genes were also prioritized based upon the initial gene expression analyses used to identify them. Probe sets identified by gene expression analyses could receive a maximum of 4 points. Thus, the maximum possible total CFG score for each gene was 12 points (4 points for the internal CFG score and 8 points for the external CFG score). The scoring system was decided upon before the analyses were carried out. Twice as much weight was given to external CFG than to internal CFG in order to increase generalizability and avoid fit to cohort of the prioritized genes. It is recognized that other ways of scoring the lines of evidence may give slightly different results in terms of prioritization, if not in terms of the list of genes per se. Nevertheless, it is believed that this simple scoring system provides a good separation of genes based on gene expression evidence and on independent cross-validating evidence in the field (FIGS. 2B and 11B).
[0103] Pathway Analyses
[0104] IPA 9.0 (Ingenuity Systems, Redwood City, Calif., USA), GeneGO MetaCore (Encinitas, Calif.), and Kyoto Encyclopedia of Genes and Genomes (through the Partek Genomics Suite 6.6 software package) were used to analyze the biological roles, including top canonical pathways, and diseases, of the candidate genes resulting from this work, as well as to identify genes in the dataset that are the target of existing drugs (FIGS. 8, 15 and 17). The analyses was run together for all the AP and DE probe sets with a total CFG score.gtoreq.4, then for those of them that showed stepwise change in the suicide completers validation cohort, then for those of them that were nominally significant, and finally for those of them that survived Bonferroni correction.
[0105] Validation Analyses
[0106] For validation of the candidate biomarker genes, which of the top candidate genes (CFG score of 4 or above) that were stepwise changed in expression from the No SI group to the High SI group to the suicide completers group, were examined. The empirical cutoff of 33% of the maximum possible CFG score of 12 was used, which also permits the inclusion of potentially novel genes with maximal internal CFG score, but no external CFG score. Statistical analyses were performed in SPSS using one-way ANOVA and Bonferonni corrections.
[0107] For the AP analyses, the Affymetrix microarray data files were imported from the participants in the validation cohort of suicide completers into MAS5, alongside the data files from the participants in the discovery cohort.
[0108] For the DE analyses, Cel. files were imported into Partek Genomic Suites. A RMA was then run, background corrected with quantile normalization, and a median polish probe set summarization of all the chips from the validation cohort to obtain the normalized expression levels of all probe sets for each chip. Partek normalizes expression data into a log base of 2 for visualization purposes. Expression data was non-log-transformed by taking 2 to the power of the transformed expression value. The non-log-transformed expression data was then used to compare expression levels of biomarkers in the different groups.
[0109] Testing Analyses
[0110] The test cohort for suicidal ideation and the test cohort for future hospitalizations analyses were assembled out of data that was RMA normalized by diagnosis. Phenomic (clinical) and gene expression markers used for predictions were z-scored by diagnosis, to be able to combine different markers into panels and to avoid potential artefacts due to different ranges of phene expression and gene expression in different diagnoses. Markers were combined by computing the average of the increased risk markers minus the average of the decreased risk markers. Predictions were performed using R-studio.
[0111] Predicting Suicidal Ideation. Receiver-operating characteristic (ROC) analyses between marker levels and suicidal ideation (SI) were performed by assigning participants with a HAMD SI score of 0-1 into the no SI category, and participants with a HAMD-SI score of 2 and greater into the SI category. Additionally, ANOVA was performed between no (HAMD-SI 0), moderate (HAMD-SI 1), and high SI participants (HAMD-SI 2 and above) and Pearson R (one-tail) was calculated between HAMD-SI scores and marker levels.
[0112] Predicting Future Hospitalizations for Suicidality. Analyses for hospitalizations in the first year following testing were conducted on data for all the participants for which data was collected. For each participant in the test cohort for future hospitalizations, the Example visit with highest levels for the marker or combination of markers was selected as index visit (or with the lowest levels, in the case of decreased markers). ROC analyses between marker levels and future hospitalizations were performed based on assigning if participants had been hospitalized for suicidality (suicide ideation, suicide attempts) or not following the index testing visit. Additionally, a one tailed t-test with unequal variance was performed between groups of participants with and without hospitalizations for suicidality. Pearson R (one-tail) correlation was performed between hospitalization frequency (number of hospitalizations for suicidality divided by duration of follow-up) and biomarker score. The correlation analysis for hospitalizations frequency was also conducted for all future hospitalizations due to suicide beyond one year, as this calculation, unlike the ROC and t-test, accounts for the actual length of follow-up, which varied beyond one year from participant to participant.
Example 1
[0113] In this Example, male subjects were analyzed for predicting suicidal ideation and future hospitalizations for suicidality.
[0114] Human Participants
[0115] Data was obtained from four cohorts: one live psychiatric participants discovery cohort (within-participant changes in suicidal ideation; n=37 out of 217); one postmortem coroner's office validation cohort (suicide completers; n=26); and two live psychiatric participants test cohorts--one for predicting suicidal ideation (n=108) and one for predicting future hospitalizations for suicidality (n=157).
[0116] Live psychiatric participants were recruited from the patient population at the Indianapolis VA Medical Center. All participants understood and signed informed consent forms detailing the research goals, procedure, caveats and safeguards. Participants completed diagnostic assessments by an extensive structured clinical interview--Diagnostic Interview for Genetic Studies--at a baseline visit, followed by up to six testing visits, 3-6 months apart or whenever a hospitalization occurred. At each testing visit, they received a series of psychiatric rating scales, including the Hamilton Rating Scale for Depression-17, which includes a suicidal ideation (SI) rating item (FIGS. 1A-1C), and blood was drawn. Whole blood (10 ml) was collected in two RNA-stabilizing PAXgene tubes, labeled with an anonymized ID number, and stored at -80 degrees C. in a locked freezer until the time of future processing. Whole-blood (predominantly lymphocyte) RNA was extracted for microarray gene expression studies from the PAXgene tubes, as detailed below. This Example focused on a male population because of the demographics of the catchment area (primarily male in a VA Medical Center), and to minimize any potential gender-related effects on gene expression, which would have decreased the discriminative power of the analysis given the relatively small sample size.
[0117] The within participant discovery cohort, from which the biomarker data were derived, consisted of 37 male participants with psychiatric disorders, with multiple visits, who each had at least one diametric change in SI scores from no SI to high SI from one testing visit to another testing visit. There was 1 participant with 6 visits, 1 participant with 5 visits, 1 participant with 4 visits, 23 participants with 3 visits each, and 11 participants with 2 visits each, resulting in a total of 106 blood samples for subsequent microarray studies (FIG. 1B).
[0118] The postmortem cohort, in which the top biomarker findings were validated, consisted of a demographically matched cohort of 24 male violent suicide completers obtained through the Marion County coroner's office (FIG. 9). A last observed alive postmortem interval of 24 hours or less was required, and the cases selected had completed suicide by means other than overdose, which could affect gene expression. 14 participants completed suicide by gunshot to head or chest, 8 by hanging, 1 by electrocution and 1 by slit wrist. Next of kin signed informed consent at the coroner's office for donation of tissues and fluids for research. The samples were collected as part of the INBRAIN initiative (Indiana Center for Biomarker Research in Neuropsychiatry).
[0119] The independent test cohort for predicting suicidal ideation consisted of 108 male participants with psychiatric disorders, demographically matched with the discovery cohort with one or multiple testing visits in the lab, with either no SI, intermediate SI, or high SI, resulting in a total of 223 blood samples in whom whole-genome blood gene expression data were obtained.
[0120] The test cohort for predicting future hospitalizations consisted of male participants in whom whole-genome blood gene expression data were obtained at testing visits over the years as part of a longitudinal study. If the participants had multiple testing visits, the visit with the highest marker (or combination of markers) levels was selected for the analyses. The participants' subsequent number of psychiatric hospitalizations, with or without suicidality, was tabulated from electronic medical records. All participants had at least one year of follow-up or more at the VA Medical Center since the time of the testing visits in the lab. Participants were evaluated for the presence of future hospitalizations for suicidality, and for the frequency of such hospitalizations. A hospitalization was deemed to be without suicidality if suicidality was not listed as a reason for admission, and no SI was described in the admission and discharge medical notes. Conversely, a hospitalization was deemed to be because of suicidality if suicidal acts or intent was listed as a reason for admission, and SI was described in the admission and discharge medical notes.
[0121] Medications
[0122] The participants in the discovery cohort were all diagnosed with various psychiatric disorders (e.g., BP, MDD, SZA, SZ, PTSD). The participants were on a variety of different psychiatric medications: mood stabilizer, antidepressants, antipsychotics, benzodiazepines and others. Medications can have a strong influence on gene expression. However, the identification of differentially expressed genes was based on within-participant analyses, which factor out not only genetic background effects but also medication effects, as the participants had no major medication changes between visits. Moreover, there was no consistent pattern in any particular type of medication, or between any change in medications and SI, in the rare instances where there were changes in medications between visits.
[0123] Results
[0124] The top increased and decreased biomarkers after the discovery for ideation (CADM1, CLIP4, DTNA, KIF2C), prioritization with CFG for prior evidence (SAT1, SKA2, SLC4A4), and validation for behavior in suicide completers (IL6, MBP, JUN, KLHDC3) steps were tested in a completely independent test cohort of psychiatric participants for prediction of suicidal ideation (n=108), and in a future follow-up cohort of psychiatric participants (n=157) for prediction of psychiatric hospitalizations due to suicidality. The best individual biomarker across psychiatric diagnoses for predicting suicidal ideation was SLC4A4, with 72% accuracy. For bipolar disorder in particular, SLC4A4 predicted suicidal ideation with 93% accuracy, and future hospitalizations with 70% accuracy. Two new clinical information apps, one for affective state (Simplified Affective Scale, SASS) and one for suicide risk factors (Convergent Functional Information for Suicide, CFI-S) are disclosed, and how well they predict suicidal ideation across psychiatric diagnoses (85% accuracy for SASS, 89% accuracy for CFI-S). Also disclosed is that the integration of the top biomarkers and the clinical information into a universal predictive measure (UP-Suicide) was able to predict suicidal ideation across psychiatric diagnoses with 92% accuracy. For bipolar disorder, it was able to predict suicidal ideation with 98% accuracy and future hospitalizations with 94% accuracy.
[0125] For discovery, two differential expression methodologies were used: Absent/Present (AP) (reflecting on/off of transcription) and Differential Expression (DE) (reflecting more subtle gradual changes in expression levels). Genes that tracked suicidal ideation in each participant were identified. Three thresholds were used for increased in expression genes and for decreased in expression genes: .gtoreq.33% (low), .gtoreq.50% (medium), and .gtoreq.80% (high) of the maximum scoring increased and decreased gene across participants. These differentially expressed genes were then prioritized using a Bayesian-like Convergent Functional Genomics (CFG) approach (FIGS. 2A-2C), integrating all the previously published human genetic evidence, postmortem brain gene expression evidence, and peripheral fluids evidence for suicide available in the field as of September 2014 to identify and prioritize disease relevant genomic biomarkers, extracting generalizable signal out of potential cohort-specific noise and genetic heterogeneity. For validation, genes whose levels of expression were changed stepwise significantly from no suicidal ideation to high suicidal ideation to suicide completion, and who survived Bonferroni correction for multiple comparisons, were carried forward. The overall best biomarkers for suicidal ideation across diagnostic groups was identified. The top genes after discovery were DTNA and KIF2C from AP, CADM1 and CLIP4 from DE. The top genes after prioritization with CFG were SLC4A4 and SKA2 from AP; and SAT1 and SKA2 from DE. The top genes after validation in suicide completers were IL6 and MBP from AP; and JUN and KLHDC3 from DE (FIG. 2C). Notably, the SAT1 finding is a replication and expansion of previously reported results identifying SAT1 as a biomarker for suicidality (Le-Niculescu et al. 2013), and the SKA2 finding is an independent replication of a previous report identifying SKA2 as a biomarker for suicidality (Kaminsky et al. 2014). A number of other genes identified are completely novel in terms of their involvement in suicidality.
[0126] To understand the biology represented by the biomarkers identified, and derive some mechanistic and practical insights, unbiased biological pathway analyses and hypothesis driven mechanistic queries, overall disease involvement and specific neuropsychiatric disorders queries, and overall drug modulation along with targeted queries for omega-3, lithium and clozapine were conducted. Administration of omega-3s in particular may be a mass-deployable therapeutic and preventive strategy.
[0127] The sets of biomarkers identified have biological roles in immune and inflammatory response, growth factor regulation, mTOR signaling, stress, and perhaps overall the switch between cell survival and proliferation vs. apoptosis (FIG. 8). An extrapolation can be made and model proposed whereas suicide is a whole body apoptosis (or "self-poptosis") in response to perceived stressful life events.
[0128] Evidence for the involvement of the biomarkers for suicidality was also examined for involvement in other psychiatric disorders, allowing for analysis of context and specificity FIGS. 8 and 9). SKA2, HADHA, SNORA68, RASL11B, CXCL11, HOMEZ, LOC728543, AHCYL1, LDLRAP1, NEAT1 and PAFAH1B2 appeared to be relatively specific for suicide, based on the evidence to date. SAT1, IL6, FOXN3 and FKBP5 were less specific for suicide, having equally high evidence for involvement in suicide and in other psychiatric disorders, possibly mediating stress response as a common denominator. CADM1, discovered in this Example as a top biomarker for suicide, had previous evidence for involvement in other psychiatric disorders, such as ASD and BP. Interestingly, it was identified in a previous study as a blood biomarker increased in expression in low mood states in bipolar participants, and it is increased in expression in the current Example in high suicidal ideation states. Increased expression of CADM1 is associated with decreased cellular proliferation and with apoptosis, and this gene is decreased in expression or silenced in certain types of cancers.
[0129] A 22-item scale and app for suicide risk, Convergent Functional Information for Suicidality (CFI-S), was also developed, which integrates information about known life events, mental health, physical health, stress, addictions, and cultural factors that can influence suicide risk. Clinical risk predictors and scales are of high interest in the military and in the general population at large. The scale disclosed herein builds on those excellent prior achievements, while aiming for comprehensiveness, simplicity and quantification similar to a polygenic risk score. CFI-S is able to distinguish between individuals who committed suicide (coroner's cases, information obtained from the next of kin, n=35) and those high risk participants who did not, but had experienced changes in suicidal ideation (e.g., the discovery cohort of psychiatric participants described herein). Items of the CFI-S scale that were the most significantly different were analyzed. Seven (7) items that were significantly different were identified, 5 of which survived Bonferroni correction: lack of coping skills when faced with stress (p=3.35E-11), dissatisfaction with current life (p=2.77E-06), lack of hope for the future (4.58E-05), current substance abuse (p=1.25E-04), and acute loss/grief (p=9.45E-4). The top item was inability to cope with stress, which was independently consistent with the biological mechanistic results discussed above.
[0130] CFI-S provided good accuracy (ROC AUC 0.70, p-value 0.006) at predicting future hospitalizations for suicidality in the first year, across diagnostic groups. CFI-Suicide had very good accuracy (AUC 0.89, p-value 3.53E-13) at predicting suicidal ideation in psychiatric participants across diagnostic groups. Within diagnostic groups, in affective disorders, the accuracy was even higher. CFI-S had excellent accuracy at predicting high suicidal ideation in bipolar participants (AUC 0.97, p-value 1.75E-06) and in depression participants (AUC 0.95, p-value 7.98E-06).
[0131] Previously, the TASS (Total Affective State Scale) was developed and described for measuring mood and anxiety. The wording used in TASS was simplified and a new app was developed for an 11 item scale for measuring mood and anxiety, the Simplified Affective State Scale (SASS). The SASS is a set of 11 visual analog scales (7 for mood, 4 for anxiety) that provides a number ranging from 0 to 100 for mood state and for anxiety state.
[0132] SASS had very good accuracy (AUC 0.85, 9.96E-11) at predicting suicidal ideation in psychiatric participants across diagnostic groups. Within diagnostic groups, in affective disorders, the accuracy was even higher (AUC 0.87) in both bipolar disorder and depression. SASS also had good accuracy (AUC 0.71, p-value 0.008) at predicting future hospitalizations for suicidality in the first year following testing.
[0133] The best single biomarker predictor for suicidal ideation state across all diagnostic groups was SLC4A4, the top increased biomarker from AP CFG prioritization (AUC 0.72, p-value 2.41E-05). Within diagnostic groups, the accuracy was even higher. SLC4A4 had very good accuracy at predicting future high suicidal ideation in bipolar participants (AUC 0.93, p-value 9.45E-06) and good accuracy in schizophrenia participants (AUC 0.76, p-value 0.030). SLC4A4 is a sodium-bicarbonate co-transporter that regulates intracellular pH, and possibly apoptosis. Very little is known to date about its roles in the brain, thus representing a completely novel finding.
[0134] SKA2, the top decreased biomarker from AP and DE CFG, had good accuracy at predicting suicidal ideation across all diagnostic groups (AUC 0.69), and even better accuracy in bipolar participants (AUC 0.76, p-value 0.011) and schizophrenia participants (AUC 0.82).
[0135] The best single top biomarker predictor for future hospitalizations for suicidal behavior in the first year across all diagnostic groups was SAT1, the top increased biomarker from the DE CFG (AUC 0.55). Within diagnostic groups, in affective disorders, the SAT1 prediction accuracies were higher (depression AUC 0.62, bipolar AUC 0.63).
[0136] The a priori primary endpoint was a combined universal predictor for suicide (UP-Suicide), composed of the top biomarkers from discovery, prioritization and validation (n=11), along with CFI-Suicide, and SASS. UP-Suicide is an excellent predictor of suicidal ideation across all disorders in the independent cohort of psychiatric participants (AUC 0.92). UP-Suicide also has good predictive ability for future psychiatric hospitalizations for suicidality in the first year of follow-up (AUC 0.70). The predictive ability of UP-Suicide is notably higher in affective disorder participants (bipolar, depression) (FIGS. 4A & 4B).
Example 2
[0137] In this Example, female subjects were analyzed for predicting suicidal ideation and future hospitalizations for suicidality.
[0138] Human Participants
[0139] Four cohorts were used: one live psychiatric participants discovery cohort (within-participant changes in suicidal ideation; n=12 out of 51); one postmortem coroner's office validation cohort (suicide completers; n=6); and two live psychiatric participants test cohorts--one for predicting suicidal ideation (n=33), and one for predicting future hospitalizations for suicidality (n=24).
[0140] The live psychiatric participants were part of a larger longitudinal cohort that was continuously being collected. Participants were recruited from the patient population at the Indianapolis VA Medical Center and Indiana University School of Medicine through referrals from care providers, the use of brochures left in plain sight in public places and mental health clinics, and through word of mouth. All participants understood and signed informed consent forms detailing the research goals, procedure, caveats and safeguards. Participants completed diagnostic assessments by an extensive structured clinical interview--Diagnostic Interview for Genetic Studies--at a baseline visit, followed by up to six testing visits, 3-6 months apart or whenever a new psychiatric hospitalization occurred. At each testing visit, they received a series of psychiatric rating scales, including the Hamilton Rating Scale for Depression-17, which includes a suicidal ideation (SI) rating item (FIG. 10A), and the blood was drawn. Whole blood (10 ml) was collected in two RNA-stabilizing PAXgene tubes, labeled with an anonymized ID number, and stored at -80 degrees C. in a locked freezer until the time of future processing. Whole-blood (predominantly lymphocyte) RNA was extracted for microarray gene expression studies from the PAXgene tubes, as detailed below. This Exampled focused on a female population.
[0141] The within participant discovery cohort, from which the biomarker data were derived, consisted of 12 female participants with psychiatric disorders and multiple visits in the lab, who each had at least one diametric change in SI scores from no SI to high SI from one testing visit to another. There were 7 participants with 3 visits each, and 5 participants with 2 visits each, resulting in a total of 31 blood samples for subsequent microarray studies (FIGS. 10B and 10C).
[0142] The postmortem cohort, in which the top biomarker findings were validated for behavior, consisted of a demographically matched cohort of 6 female violent suicide completers obtained through the Marion County coroner's office (FIG. 14). A last observed alive postmortem interval of 24 hours or less was required, and the cases selected had completed suicide by means other than overdose, which could affect gene expression. 5 participants completed suicide by gunshot to head or chest, and 1 by asphyxiation. Next of kin signed informed consent at the coroner's office for donation of blood for research. The samples were collected as part of the INBRAIN initiative (Indiana Center for Biomarker Research in Neuropsychiatry).
[0143] The independent test cohort for predicting suicidal ideation (FIG. 14) consisted of 33 female participants with psychiatric disorders, demographically matched with the discovery cohort, with one or multiple testing visits in the lab, with either no SI, intermediate SI, or high SI, resulting in a total of 74 blood samples in whom whole-genome blood gene expression data were obtained (FIG. 14).
[0144] The test cohort for predicting future hospitalizations (FIG. 14) consisted of 24 female participants in whom whole-genome blood gene expression data were obtained at testing visits over the years as part of a longitudinal study. If the participants had multiple testing visits, the visit with the highest marker (or combination of markers) levels was selected for the analyses (so called "high watermark" or index visit). The participants' subsequent number of psychiatric hospitalizations, with or without suicidality (ideation or attempt), was tabulated from electronic medical records. Participants were evaluated for the presence of future hospitalizations for suicidality, and for the frequency of such hospitalizations. A hospitalization was deemed to be without suicidality if suicidality was not listed as a reason for admission, and no SI was described in the admission and discharge medical notes. Conversely, a hospitalization was deemed to be because of suicidality if suicidal acts or intent was listed as a reason for admission, and/or SI was described in the admission and discharge medical notes.
[0145] Medications
[0146] The participants in the discovery cohort were all diagnosed with various psychiatric disorders (FIG. 14). Their psychiatric medications were listed in their electronic medical records, and documented at the time of each testing visit. The participants were on a variety of different psychiatric medications: mood stabilizers, antidepressants, antipsychotics, benzodiazepines and others (data not shown). Medications can have a strong influence on gene expression. However, discovery of differentially expressed genes was based on within--participant analyses, which factor out not only genetic background effects but also medication effects, as the participants had no major medication changes between visits. Moreover, there was no consistent pattern in any particular type of medication, or between any change in medications and SI, in the rare instances where there were changes in medications between visits.
[0147] Clock Gene Database
[0148] In this Example, a database was compiled of genes associated with circadian function, by using a combination of review papers (Zhang et al. Cell 2009; 139(1):19-210, McCarthy and Welsh Journal of biological rhythms 2012; 27(5):339-352) and searches of existing databases CircaDB (circadb.hogeneschlab.org), GeneCards (www.genecards.org), and GenAtlas (genatlas.medecine.univ-paris5.fr). Using the data, a total of 1280 genes were identified that show circadian functioning. The genes were classified into "core" clock genes, i.e. those genes that are the main engine driving circadian function (n=18), "immediate" clock genes, i.e. the genes that directly input or output to the core clock (n=331), and "distant" clock genes, i.e. genes that directly input or output to the immediate clock genes (n=1,119).
[0149] Clinical Measures
[0150] The Simplified Affective State Scale (SASS) is an 11-item scale for measuring mood and anxiety. The SASS has a set of 11 visual analog scales (7 for mood, 4 for anxiety) that ends up providing a number ranging from 0 to 100 for mood state, and the same for anxiety state. Also developed is an Android app version.
[0151] In some embodiments, the systems and methods described utilize a computer implemented method for assessing suicidal risk factors based upon patient psychiatric information further including mood information, anxiety information, and other psychiatric symptom information. Any and all such patient psychiatric information may be represented as a quantitative rating on a defined analog scale, such as the ratings and scales described above. Further, as used herein, such patient psychiatric information may be processed using an associated processing algorithm. The associated processing algorithm may include calculating mean values for each component of patient psychiatric information and then assigning a suitable weighting to each calculated mean value. The processing algorithm may thus use the quantitative ratings of the patient psychiatric information as inputs to calculate a diagnostic output score. The diagnostic output score may be used to compare to reference scores (from a diagnostic database) associated with patients having psychiatric symptom information (e.g., psychiatric disorder diagnosis or lack thereof) similar to the patient. By such comparison, the diagnostic output score may be assigned a percentile. The diagnostic output score may also be compared to the reference scores in the diagnostic database associated with individuals with no suicidality and high suicidality. Thus, if the diagnostic output score meets or exceeds a high suicidality reference score, a patient may be marked as at risk for suicide. Conversely, if the diagnostic output score meets or falls below a low suicidality reference score, a patient may be marked as not at risk for suicide.
[0152] Convergent Functional Information for Suicidality (CFI-S) is a 22-item scale and Android app for suicide risk, which integrates, in a simple binary fashion (Yes-1, No-0), similar to a polygenic risk score, information about known life events, mental health, physical health, stress, addictions, and cultural factors that can influence suicide risk. The scale was administered at participant testing visits (n=39), or scored based on retrospective electronic medical record information and Diagnostic Interview for Genetic Testing (DIGS) information (n=48). When information was not available for an item, it was not scored (NA).
[0153] In other embodiments, the systems and methods described utilize a computer implemented method for assessing suicidal risk factors based upon socio-demographic/psychological suicidal risk factors. Any and all such socio-demographic/psychological suicidal risk factors may be represented as a quantitative rating on a defined analog scale, such as the ratings and scales described above. Further, as used herein, such socio-demographic/psychological suicidal risk factors may be processed using an associated processing algorithm. The associated processing algorithm may include calculating mean values for each component socio-demographic/psychological suicidal risk factor and then assigning a suitable weighting to each calculated mean value. The processing algorithm may thus use the quantitative ratings of the socio-demographic/psychological suicidal risk factors as inputs to calculate a diagnostic output score. The diagnostic output score may be used to compare to reference scores (from a diagnostic database) associated with patients having socio-demographic/psychological suicidal risk factors similar to the patient. By such comparison, the diagnostic output score may be assigned a percentile. The diagnostic output score may also be compared to the reference scores in the diagnostic database associated with individuals with no suicidality and high suicidality. Thus, if the diagnostic output score meets or exceeds a high suicidality reference score, a patient may be marked as at risk for suicide. Conversely, if the diagnostic output score meets or falls below a low suicidality reference score, a patient may be marked as not at risk for suicide.
[0154] In some computer-implemented methods described above and herein, multiple computing devices may interact with one another (e.g., first and second computer devices). To protect data and privacy, such requests and transmissions are made using data encryption.
[0155] Combining Gene Expression and Clinical Measures
[0156] The Universal Predictor for Suicide (UP-Suicide) construct, the primary endpoint, was decided upon as part of a apriori study design to be broad-spectrum, and combine the top Bonferroni validated biomarkers with the phenomic (clinical) markers (SASS and CFI-S).
[0157] Results
[0158] Discovery of Biomarkers for Suicidal Ideation
[0159] A whole-genome gene expression profiling was conducted in the blood samples from a longitudinally followed cohort of female participants with psychiatric disorders that predispose to suicidality. The samples were collected at repeated visits, 3-6 months apart. State information about suicidal ideation (SI) was collected from a questionnaire (HAMD) administered at the time of each blood draw. Out of 51 female psychiatric participants (with a total of 123 visits) followed longitudinally in this Example, with a diagnosis of BP, MDD, SZ and SZA, there were 12 participants that switched from a no SI (SI score of 0) to a high SI state (SI score of 2 and above) at different visits, which was the intended discovery group (FIG. 10B). A within-participant design was used to analyze data from these 12 participants and their 31 visits. A within-participant design factors out genetic variability, as well as some medications, lifestyle, and demographic effects on gene expression, permitting identification of relevant signal with Ns as small as 1. Another benefit of a within-participant design may be accuracy/consistency of self-report of psychiatric symptoms (`phene expression`), similar in rationale to the signal detection benefits it provides in gene expression.
[0160] For discovery, two differential expression methodologies were used: Absent/Present (AP) (reflecting on/off of transcription), and Differential Expression (DE) (reflecting more subtle gradual changes in expression levels). The genes that tracked suicidal ideation in each participant were identified in the analyses. Three thresholds were used for increased in expression genes and for decreased in expression genes: .gtoreq.33.3% (low), .gtoreq.50% (medium), and .gtoreq.80% (high) of the maximum scoring increased and decreased gene across participants. Such a restrictive approach was used as a way of minimizing false positives, even at the risk of having false negatives. For example, there were genes on each of the two lists, from AP and DE analyses, that had clear prior evidence for involvement in suicidality, such as AKAP10 (31.7%) and MED28 (31.8%) from AP, and S 100B (31.7%) and SKA2 (31.4%) for DE, but were not included in subsequent analyses because they did not meet the apriori set 33.3% threshold. Notably, SKA2 reproduces the results in males (Example 1).
[0161] Prioritization of Biomarkers Based on Prior Evidence in the Field
[0162] These differentially expressed genes were then prioritized using a Bayesian-like Convergent Functional Genomics (CFG) approach (FIGS. 11B and 11C) integrating all the previously published human genetic evidence, postmortem brain gene expression evidence, and peripheral fluids evidence for suicide in the field available at the time of this analyses (i.e., September 2015). This is a way of identifying and prioritizing disease relevant genomic biomarkers, extracting generalizable signal out of potential cohort-specific noise and genetic heterogeneity. The manually curated databases of the psychiatric genomic and proteomic literature to date were used in CFG analyses. The CFG approach is thus a de facto field-wide collaboration.
[0163] Validation of Biomarkers for Behavior in Suicide Completers
[0164] For validation in suicide completers, 1471 genes were used that had a CFG score of 4 and above, from AP and DE, reflecting either maximum internal score from discovery or additional external literature cross-validating evidence. Out of these, 882 did not show any stepwise change in suicide completers (NC--non-concordant). As such, they may be involved primarily in ideation and not in behavior. The remaining 589 genes (40.0%) had levels of expression that were changed stepwise from no suicidal ideation to high suicidal ideation to suicide completion. 396 of these genes (26.9%) were nominally significant, and 49 genes (50 probesets--two for JUN) (3.33%) survived Bonferroni correction for multiple comparisons (FIG. 11C). These genes are likely involved in suicidal ideation and suicidal behavior. (A person can have suicidal ideation without suicidal behavior, but cannot have suicidal behavior without suicidal ideation).
[0165] Selection of Biomarkers for Testing of Predictive Ability
[0166] For testing, Bonferroni validated biomarkers (49 genes, 50 probesets) were focused on. A secondary analysis of the top scoring biomarkers from both discovery and prioritization (65 genes) was conducted so as to avoid potential false negatives in the validation step due to possible postmortem artefacts or extreme stringency of statistical cutoff. The top CFG scoring genes after the Bonferroni validation step were BCL2 and GSK3B. The top CFG scoring genes from the discovery and prioritization steps were FAM214A, CLTA, HSPD1, and ZMYND8. Notably, all have co-directional gene expression changes evidence in brains of suicide completers in studies form other groups.
[0167] Biological Understanding
[0168] Unbiased biological pathway analyses and hypothesis driven mechanistic queries, overall disease involvement and specific neuropsychiatric disorders queries, and overall drug modulation along with targeted queries for omega-3, lithium and clozapine were studied (FIGS. 15 and 17). Administration of omega-3s in particular may be a mass-deployable therapeutic and preventive strategy.
[0169] The sets of biomarkers identified have biological roles in inflammation, neurotrophins, inositol signaling, stress response, and perhaps overall the switch between cell survival and proliferation vs. apoptosis (FIG. 15).
[0170] The involvement of these biomarkers for suicidality in other psychiatric disorders were also analyzed. FAM214A, MOB3B, ZNF548, and ARHGAP35 were relatively specific for suicide, based on the evidence to date in the field, and were also identified co-directionally in the previous male work (Example 1). BCL2, GSK3B, HSPD1, and PER1 were less specific for suicide, having equally high evidence for involvement in suicide and in other psychiatric disorders. BCL2 was also identified co-directionally in Example 1.
[0171] HSPD1, found to be a top biomarker in this Example, increased in expression in suicidality, and was also increased in expression in the blood following anti-depressant treatment. Thus, this may be a useful biomarker for treatment-emergent suicidal ideation (TESI).
[0172] Further, a number of the genes changed in expression in opposite direction in suicide in this Example vs. high mood in Example 1--SSBP2, ZNF596, suggesting that suicidal participants are in a low mood state. Also, some of the top suicide biomarkers are changed in expression in the same direction as in high psychosis participants in a previous psychosis biomarker study--HERC4, PIP5K1B, SLC35B3, SNX27, KIR2DL4, NUDT10, suggesting that suicidal participants may be in a psychosis-like state. Taken together, the data indicates that suicidality could be viewed as a psychotic dysphoric state. This molecularly informed view is consistent with the emerging clinical evidence in the field.
[0173] A number of top biomarkers identified have biological roles that are related to the core circadian clock (such as PER1), or modulate the circadian clock (such as CSNK1A1), or show at least some circadian pattern (such as HTRA1). To be able to ascertain all the genes in the dataset that were circadian and do estimates for enrichment, a database from literature was compiled of all the known genes that fall into these three categories, numbering a total of 1468 genes. Using an estimate of about 21,000 genes in the human genome, that gives about 7% of genes having some circadian pattern. Out of the 49 Bonferroni validated biomarker genes, 7 had circadian evidence (14.3%), suggesting a 2-fold enrichment for circadian genes.
[0174] Additionally, biological pathway analyses were conducted on the genes that, after discovery and prioritization, were stepwise changed in suicide completers (n=882) and may be involved in ideation and behavior vs. those that were not stepwise changed (n=589), and that may only be involved in ideation. The genes involved in ideation map to pathways related to PI3K signaling. The genes involved in behavior map to pathways related to glucocorticoid receptor signaling. This is consistent with ideation being related to neurotrophic factors, and behavior being related to stress.
[0175] Clinical Information
[0176] A 22-item scale and app were used for suicide risk, Convergent Functional Information for Suicidality (CFI-S), which scores in a simple binary fashion and integrates information about known life events, mental health, physical health, stress, addictions, and cultural factors that can influence suicide risk. Determining which items of the CFI-S scale were the most significantly different between no and high suicidal ideation live participants was analyzed (FIG. 12A). Seven items were identified that were significantly different: lack of positive relationships/social isolation (p=0.004), substance abuse (p=0.0071), history of impulsive behaviors (p=0.015), lack of religious beliefs (p=0.018), past history of suicidal acts/gestures (p=0.025), rejection (p=0.029), and history of command auditory hallucinations (p=0.045) (FIG. 12B). It is noted that lack of positive relationships/social isolation was the second top item in males as well. Social isolation increases vulnerability to stress, which is independently consistent with the biological marker results.
[0177] Testing for Predictive Ability
[0178] The best single increased (risk) biomarker predictor for suicidal ideation state was EPB41L5 (ROC AUC 0.68, p-value 0.06; Pearson Correlation 0.22, p-value 0.03), an increased in expression, Bonferroni validated biomarker (FIG. 16). This biomarker was also identified co-directionally in Example 1, and has no evidence for involvement in other psychiatric disorders. The best single decreased (protective) biomarker predictor for suicidal ideation is PIK3C3 (ROC AUC 0.65, p-value 0.1; Pearson Correlation -0.21, p-value 0.037), a decreased in expression, Bonferroni validated biomarker (FIG. 16). PIK3C3 is also decreased in expression in postmortem brains in depression.
[0179] The best single increased (risk) biomarker predictor for future hospitalizations for suicidality was HTRA1 (ROC AUC 0.84, p-value 0.01; Cox Regression Hazard Ratio 4.55, p-value 0.01), an increased in expression, Bonferroni validated biomarker (FIG. 16). HTRA1 is also increased in expression in the blood of schizophrenics. The best single decreased (protective) biomarker predictor for future hospitalizations for suicidality was CSNK1A1 (ROC AUC 0.96, p-value 0.0007; Cox Regression Hazard Ratio 620.5, p-value 0.02), a top discovery and prioritization, non-Bonferroni validated biomarker (FIG. 16). This biomarker was also identified co-directionally in Example 1. CSNK1A1 (casein kinase 1, alpha 1) is a circadian clock gene, part of the input into the core clock. It decreased in expression in suicidality, and decreased in postmortem brains of alcoholics. It has further been found to be increased in expression by mood stabilizers and by omega-3 fatty acids. PIK3C3 was also found to be a good predictor for future hospitalizations for suicidality (ROC AUC 0.9, p-value 0.011) (FIG. 16).
[0180] BCL2, the top CFG scoring biomarker from validation, had good accuracy at predicting future hospitalizations for suicidality (ROC AUC 0.89, p-value 0.007; Cox Regression Hazard Ratio 3.08, p-value 0.01) (FIG. 16). In the panel of 50 validated biomarkers, BioM-50, had even better accuracy at predicting future hospitalizations for suicidality (ROC AUC 0.94, p-value 0.002; Cox Regression Hazard Ratio 89.46, p-value 0.02) (FIG. 16). Overall, in women, blood biomarkers seemed to perform better for predicting future hospitalizations for suicidality (trait) than for predicting suicidal ideation (state). This is different than the trend seen in Example 1, where blood biomarkers were somewhat better predictors of state than of trait.
[0181] CFI-S had very good accuracy (ROC AUC 0.84, p-value 0.002; Pearson Correlation 0.39, p-value 0.001) at predicting suicidal ideation in psychiatric participants across diagnostic groups. The other app, SASS, also had very good accuracy (ROC AUC 0.81, p-value 0.003; Pearson Correlation 0.38, p-value 0.0005) at predicting suicidal ideation in women psychiatric participants. The combination of the apps was synergistic (ROC AUC 0.87, p-value 0.0009; Pearson Correlation 0.48, p-value 0.0001). Thus, even without the benefit of potentially more costly and labor intensive blood biomarker testing, clinically useful predictions could be made with the apps.
[0182] The apriori primary endpoint was a combined universal predictor for suicide (UP-Suicide), composed of CFI-S and SASS, along with the Bonferroni validated biomarkers (n=50) resulting from the sequential discovery for ideation, prioritization with CFG, and validation for behavior in suicide completers steps. UP-Suicide was a good predictor of suicidal ideation (ROC AUC 0.82, p-value 0.003; Pearson Correlation 0.43, p-value 0.0003) (FIGS. 13A, 13B and 16). UP-Suicide also had good predictive ability for future psychiatric hospitalizations for suicidality (ROC AUC 0.78, p-value 0.032; Cox Regression Hazard Ratio 9.61, p-value 0.01).
[0183] Discussion
[0184] The present Example identified markers involved in suicidal ideation and suicidal behavior, including suicide completion, in females. Markers involved in behavior may be on a continuum with some of the markers involved in ideation, varying in the degree of expression changes from less severe (ideation) to more severe (behavior). One cannot have suicidal behavior without suicidal ideation, but it may be possible to have suicidal ideation without suicidal behavior.
[0185] 50 biomarkers were found to have survived Bonferroni correction (49 genes; one gene, JUN, had two different probesets that validated). Additionally, 65 other biomarkers that were non Bonferroni, but had maximum internal score of 4 in discovery and a CFG score of 6 and above, meaning that in addition to strong evidence in this Example, they also had prior independent evidence of involvement in suicide from other studies, were also studied. These additional biomarkers are likely involved in suicide, but did not make the Bonferroni validation cutoff due to its stringency or potential technical/postmortem artefact reasons (FIGS. 26 and 30).
[0186] Data validating the CFI-S in women in the combined discovery and test cohort of live psychiatric participants was analyzed (FIGS. 12A and 12B) and compared with similar analyses in men (Example 1). The chronic stress of lack of positive relationships/social isolation was identified as the top differential item in women, which is consistent with biological data from the biomarker side of this Example.
[0187] In assessing anxiety and mood, it was shown that anxiety measures cluster with suicidal ideation and CFI-S, and mood measures are in the opposite cluster, suggesting that the participants have high suicidal ideation when they have high anxiety and low mood (FIG. 10C).
[0188] The rationale for identifying blood biomarkers as opposed to brain biomarkers is a pragmatic one--the brain cannot be readily accessed in live individuals. Other peripheral fluids, such as CSF, require more invasive and painful procedures. Nevertheless, it is likely that many of the peripheral blood transcriptomic changes are not necessarily mirroring what is happening in the brain, and vice-versa. The keys to finding peripheral biomarkers are, first, to have a powerful discovery approach, such as the within-participant design, that closely tracks the phenotype you are trying to measure and reduces noise. Second, cross-validating and prioritizing the results with other lines of evidence, such as brain gene expression and genetic data, are important in order to establish relevance and generalizability of findings. Third, it is important to validate for behavior in an independent cohort with a robust and relevant phenotype, in this case suicide completers. Fourth, testing for predictive ability in independent/prospective cohorts is a must.
[0189] Biomarkers that survive such a rigorous step-wise discovery, prioritization, validation and testing process are likely directly relevant to the disorder studied. As such, whether they are involved in other psychiatric disorders or are relatively specific for suicide, and whether they are the modulated by existing drugs in general, and drugs known to treat suicidality in particular were evaluated.
[0190] A series of biomarkers have been identified that seem to be changed in opposite direction in suicide vs. in treatments with omega-3 fatty acids, lithium, clozapine. These biomarkers could potentially be used to stratify patients to different treatment approaches, and monitor their response.
[0191] BCL2, JUN, GHA1, ENTPD1, ITIH5, MBNL1, and SSBP2 were changed in expression by two of these three treatments, suggesting they may be core to the anti-suicidal mechanism of these drugs. BCL2, CAT, and JUN may be useful blood pharmacogenomic markers of response to lithium. CD84, MBNL1, and RAB22A may be useful blood pharmacogenomic markers of response to clozapine. NDRG1, FOXP1, AFF3, ATXN1, CSNK1A1, ENTPD1, ITIH5, PRDX3, and SSBP2 may be useful blood pharmacogenomic markers of response to omega-3 fatty acids. Three existing drugs used for other indications have been identified as targeting the top suicide biomarkers identified, and could potentially be re-purposed for testing in treatment of acute suicidality: anakinra (inhibiting ILR1), enzastaurin (inhibiting AKT3), and tesevatinib (inhibiting EPHB4). Additionally, Connectivity Map (ref) analyses identified compounds that induced gene expression signatures that were the opposite of those present in suicide, and might generate leads and/or be tested for use to treat/prevent suicidality: betulin (an anti-cancer compound from the bark of birch trees), zalcitabine (an anti-HIV drug), and atractyloside (a toxic glycoside). Other common drugs identified by the Connectivity Map analyses were nafcillin, lansoprazole, mifepristone, LY294002, minoxidil, acetysalicilic acid, estradiol, buspirone, dicloxacillin, corticosterone, metformin, diphenhydramine, haloperidol, and fluoxetine.
[0192] Of note, a number of biomarkers from the current Example in women reproduced and were co-directional with previous findings in Example 1 (BCL2, ALDH3A2, FAM214A, CLTA, ZMYND8, JUN), whereas others had changes in opposite directions (GSK3B, HSPD1, AK2, CAT), underlying the issue of biological context and differences in suicidality between the two genders.
[0193] Disclosed herein are instruments (biomarkers and applications) for predicting suicidality, that do not require asking the person assessed if they have suicidal thoughts, as individuals who are truly suicidal often do not share that information with people close to them or with clinicians. The widespread use of such risk prediction tests as part of routine or targeted healthcare assessments will lead to early disease interception followed by preventive lifestyle modifications or treatment. Biomarkers identified herein for suicidal ideation are enriched for genes involved in neuronal connectivity and schizophrenia. Biomarkers identified herein also validated for suicide behavior are enriched for genes involved in neuronal activity and mood.
[0194] Worldwide, one person dies every 40 seconds through suicide, a potentially preventable tragedy. A limiting step in the ability to intervene is the lack of objective, reliable predictors. A powerful within-participant discovery approach is disclosed herein in which genes that change in expression between no suicidal ideation and high suicidal ideation states were identified. The methods disclosed herein do not require asking the person assessed if they have thoughts of suicide, as individuals who are truly suicidal often do not share that information with clinicians. The widespread use of such risk prediction tests as part of routine or targeted healthcare assessments will lead to early disease interception followed by preventive lifestyle modifications and proactive treatment.
[0195] In view of the above, it will be seen that the several advantages of the disclosure are achieved and other advantageous results attained. As various changes could be made in the above methods without departing from the scope of the disclosure, it is intended that all matter contained in the above description and shown in the accompanying drawings shall be interpreted as illustrative and not in a limiting sense.
[0196] When introducing elements of the present disclosure or the various versions, embodiment(s) or aspects thereof, the articles "a", "an", "the" and "said" are intended to mean that there are one or more of the elements. The terms "comprising", "including" and "having" are intended to be inclusive and mean that there may be additional elements other than the listed elements.
* * * * *
References
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014

D00015

D00016

D00017

D00018

D00019

D00020

D00021

D00022

D00023

D00024

D00025

D00026

D00027

D00028

D00029

D00030
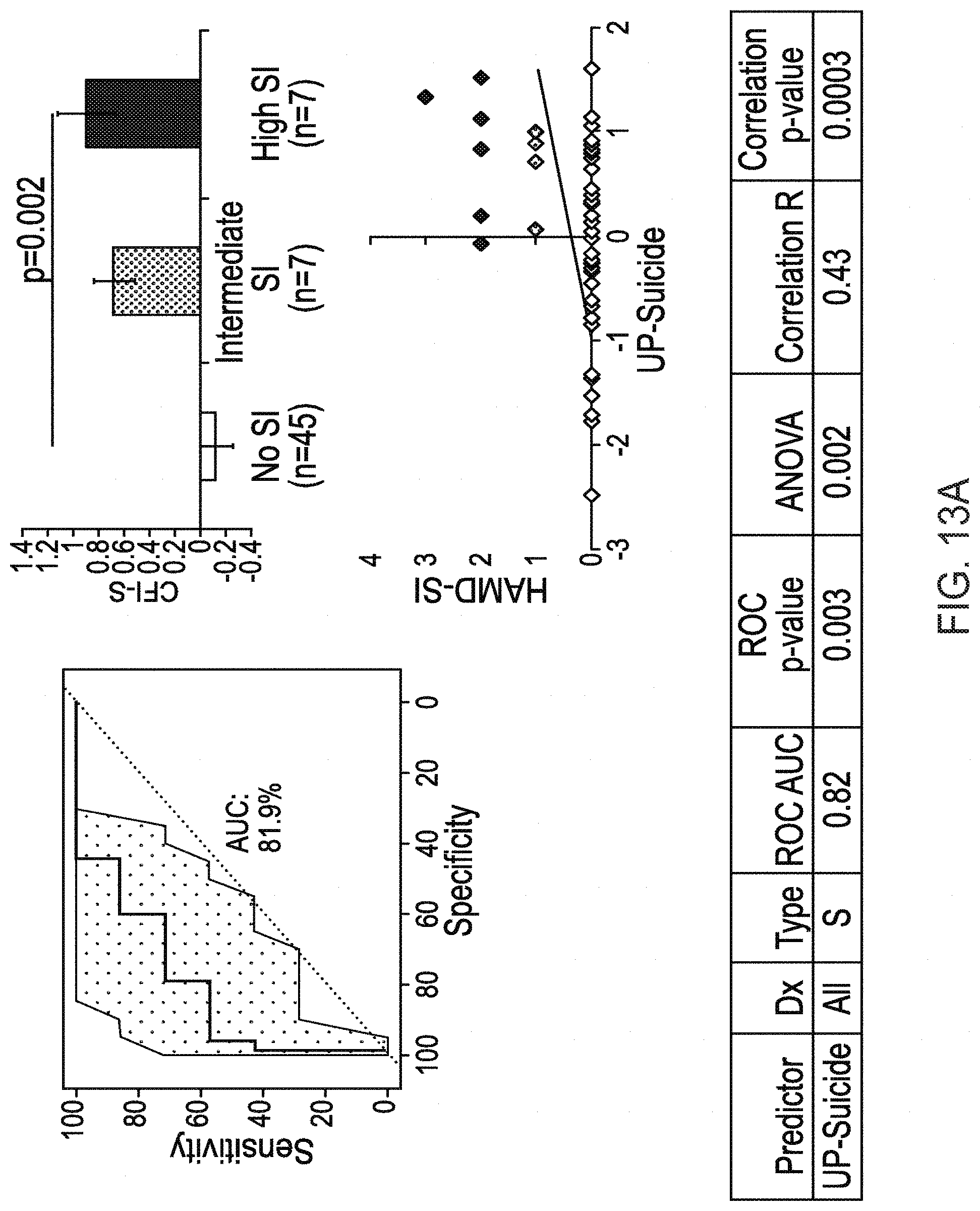
D00031

D00032

D00033

D00034

D00035

D00036

D00037

D00038

D00039

D00040

D00041

XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.