Trispecific Antibodies Specific For Her2 And A Blood Brain Barrier Receptor And Methods Of Use
Klein; Christian ; et al.
U.S. patent application number 16/724004 was filed with the patent office on 2020-09-24 for trispecific antibodies specific for her2 and a blood brain barrier receptor and methods of use. This patent application is currently assigned to Hoffmann-La Roche Inc.. The applicant listed for this patent is Hoffmann-La Roche Inc.. Invention is credited to Christian Klein, Julia Krueger, Ekkehard Moessner, Jens Niewoehner.
| Application Number | 20200299407 16/724004 |
| Document ID | / |
| Family ID | 1000004871987 |
| Filed Date | 2020-09-24 |








View All Diagrams
| United States Patent Application | 20200299407 |
| Kind Code | A1 |
| Klein; Christian ; et al. | September 24, 2020 |
TRISPECIFIC ANTIBODIES SPECIFIC FOR HER2 AND A BLOOD BRAIN BARRIER RECEPTOR AND METHODS OF USE
Abstract
The present invention relates to trispecific antibodies binding to HER2 and a blood-brain barrier receptor (BBB-R), methods for their production, pharmaceutical compositions containing said antibodies, and uses thereof.
| Inventors: | Klein; Christian; (Bonstetten, CH) ; Krueger; Julia; (Wuerzburg, DE) ; Moessner; Ekkehard; (Kreuzlingen, CH) ; Niewoehner; Jens; (Muenchen, DE) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Assignee: | Hoffmann-La Roche Inc. Little Falls NJ |
||||||||||
| Family ID: | 1000004871987 | ||||||||||
| Appl. No.: | 16/724004 | ||||||||||
| Filed: | December 20, 2019 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 15845970 | Dec 18, 2017 | |||
| 16724004 | ||||
| PCT/EP2016/064124 | Jun 20, 2016 | |||
| 15845970 | ||||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C07K 2317/732 20130101; C07K 2317/622 20130101; C07K 2317/94 20130101; C07K 16/18 20130101; C07K 16/2881 20130101; C07K 2317/60 20130101; C07K 2317/34 20130101; C07K 2317/33 20130101; A61K 2039/507 20130101; C07K 2317/515 20130101; C07K 2317/92 20130101; C07K 2317/41 20130101; C07K 2317/51 20130101; C07K 2317/73 20130101; C07K 16/32 20130101; A61K 2039/505 20130101; C07K 2317/734 20130101; C07K 2317/31 20130101; C07K 2317/24 20130101 |
| International Class: | C07K 16/32 20060101 C07K016/32; C07K 16/28 20060101 C07K016/28; C07K 16/18 20060101 C07K016/18 |
Foreign Application Data
| Date | Code | Application Number |
|---|---|---|
| Jun 24, 2015 | EP | 15173640.2 |
Claims
1. A trispecific antibody specifically binding to HER2 and a blood-brain barrier receptor (BBB-R), comprising a first monovalent antigen binding site specific for extracellular domain II of HER2 and a second monovalent antigen binding site specific for extracellular domain IV of HER2, and a third monovalent antigen binding site specific for a BBB-R.
2. The trispecific antibody of claim 1, wherein the BBB-R of the third antigen binding site is selected from the group consisting of transferrin receptor (TfR), insulin receptor, insulin-like growth factor receptor (IGF receptor), low density lipoprotein receptor-related protein 8 (LRP8), low density lipoprotein receptor-related protein 1 (LRP1), and heparin-binding epidermal growth factor-like growth factor (HB-EGF).
3. The trispecific antibody of claim 1 wherein the third antigen binding site is specific for the transferrin receptor
4. The trispecific antibody of claim 1 wherein the third antigen binding site specifically binds to an epitope in the transferrin receptor comprised within the amino acid sequence of SEQ ID NO: 202, 203 and/or 204.
5. The trispecific antibody claim 1, comprising a first Fab molecule capable of specific binding to extracellular domain II of HER2 and a second Fab molecule capable of specific binding to extracellular domain IV of HER2, wherein the sequence of the variable light chain of the first Fab molecule is identical to the sequence of the variable light chain of the second Fab molecule.
6. The trispecific antibody of claim 5, comprising (a) a first heavy chain comprising a heavy chain CDR1 comprising an amino acid sequence selected from the group consisting of SEQ ID NO: 55, SEQ ID NO: 58 and SEQ ID NO: 14; a heavy chain CDR 2 comprising an amino acid sequence selected from the group consisting of SEQ ID NO: 77; SEQ ID NO: 15 and SEQ ID NO: 60 and a heavy chain CDR 3 comprising an amino acid sequence selected from the group consisting of SEQ ID NO: 56 or SEQ ID NO: 59 and SEQ ID NO: 16, and (b) a second heavy chain comprising a heavy chain CDR1 comprising an amino acid sequence of SEQ ID NO: 20, a heavy chain CDR2 comprising an amino acid sequence of SEQ ID NO: 29 and a heavy chain CDR3 comprising an amino acid sequence selected from the group consisting of SEQ ID NO: 30 and SEQ ID NO: 79 (c) a first and a second light chain, wherein the variable light chains of the first and second light chain comprise the CDRs comprising an amino acid sequence of SEQ ID NO: 89, SEQ ID NO: 90 and SEQ ID NO: 19.
7. The trispecific antibody of claim 5, comprising two variable light chains comprising an amino acid sequence of SEQ ID NO: 54, a first heavy chain comprising a variable heavy chain comprising an amino acid sequence selected from the group consisting of SEQ ID NO: 64, SEQ ID NO: 70 and SEQ ID NO: 68, and a second heavy chain comprising a variable heavy chain comprising an amino acid sequence selected from the group consisting of SEQ ID NO: 92 and SEQ ID NO: 117.
8. The trispecific antibody of claim 1, comprising a first Fab molecule capable of specific binding to extracellular domain II of HER2 and a second Fab molecule capable of specific binding to extracellular domain IV of HER2, wherein either the variable regions or the constant regions of the heavy and light chain of at least one Fab fragment are exchanged.
9. The trispecific antibody of claim 8, wherein the first Fab molecule comprises a heavy chain CDR1 comprising an amino acid sequence of SEQ ID NO: 14, a heavy chain CDR2 comprising an amino acid sequence of SEQ ID NO: 15 and a heavy chain CDR3 comprising an amino acid sequence of SEQ ID NO: 16; and a light chain CDR1 comprising an amino acid sequence of SEQ ID NO: 11; a light chain CDR2 comprising an amino acid sequence of SEQ ID NO: 12 and a light chain CDR3 comprising an amino acid sequence of SEQ ID NO: 13, and wherein the second Fab molecule comprises a heavy chain CDR1 comprising an amino acid sequence of SEQ ID NO: 20; a heavy chain CDR2 comprising an amino acid sequence of SEQ ID NO: 108; a heavy chain CDR3 comprising an amino acid sequence of SEQ ID NO: 79; and a light chain CDR1 comprising an amino acid sequence of SEQ ID NO: 107, a light chain CDR2 comprising an amino acid sequence of SEQ ID NO: 18 and a light chain CDR3 comprising an amino acid sequence of SEQ ID NO: 19.
10. The trispecific antibody of claim 8, wherein the first Fab molecule comprises a heavy chain CDR1 comprising an amino acid sequence of SEQ ID NO: 14, a heavy chain CDR2 comprising an amino acid sequence of SEQ ID NO: 15 and a heavy chain CDR3 comprising an amino acid sequence of SEQ ID NO: 16; and a light chain CDR1 comprising an amino acid sequence of SEQ ID NO: 11, a light chain CDR2 comprising an amino acid sequence of SEQ ID NO: 12 and a light chain CDR3 comprising an amino acid sequence of SEQ ID NO: 13, and wherein the second Fab molecule comprises a heavy chain CDR1 comprising an amino acid sequence of SEQ ID NO: 20, a heavy chain CDR2 comprising an amino acid sequence of SEQ ID NO: 29, and a heavy chain CDR3 comprising an amino acid sequence selected from the group consisting of SEQ ID NO: 79, SEQ ID NO: 78, SEQ ID NO: 80, SEQ ID NO: 87, SEQ ID NO: 88; and a light chain CDR1 comprising an amino acid sequence selected from the group consisting of SEQ ID NO: 104, SEQ ID NO: 103 and SEQ ID NO: 158; a light chain CDR2 comprising an amino acid sequence of SEQ ID NO: 18 and a light chain CDR3 comprising an amino acid sequence of SEQ ID NO: 19.
11. The trispecific antibody of claim 8, wherein the first Fab molecule comprises a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 22 and a variable light chain comprising an amino acid sequence of SEQ ID NO: 24 and wherein the second Fab molecule comprises an amino acid sequence of SEQ ID NO: 105 and a light chain variable region comprising an amino acid sequence of SEQ ID NO: 106.
12. The trispecific antibody of claim 1, wherein the third antigen binding site specific for a BBB-R is a scFv or a scFab.
13. The trispecific antibody of claim 12, wherein the scFv or scFab is connected to the N-terminus of an IgG molecule comprsing the first and second antigen binding site.
14. The trispecific antibody of claim 12, wherein the scFv or scFab is connected to the C-terminus of the first or second subunit of the Fc domain.
15. The trispecific antibody of claim 1, wherein the third antigen binding site comprises a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 186, a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 187 and a heavy chain CDR3 comprising the amino acid sequence selected from the group of SEQ ID NO: 188, SEQ ID NO: 206 or SEQ ID NO: 174; and a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 189, a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 190 and a light chain CDR3 comprising the amino acid sequence of SEQ ID NO:191.
16. The trispecific antibody of claim 1, wherein the third antigen binding site comprises a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 172, a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 173 and a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 174; and a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 175, a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 176 and a light chain CDR3 comprising the amino acid sequence of SEQ ID NO:177.
17. The trispecific antibody of claim 1, wherein the third antigen binding site comprises a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 178 and a variable light chain comprising an amino acid sequence of SEQ ID NO: 179 or a humanized version thereof.
18. The trispecific antibody of claim 1, wherein the third antigen binding site comprises a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 192 or SEQ ID NO: 205 and a variable light chain comprising an amino acid sequence of SEQ ID NO: 193.
19. The trispecific antibody of claim 1, wherein the third antigen binding site comprises a heavy chain CDR1 of SEQ ID NO: 180, a heavy chain CDR2 of SEQ ID NO: 181 and a heavy chain CDR3 of SEQ ID NO: 182; and a light chain CDR1 of SEQ ID NO: 183, a light chain CDR2 of SEQ ID NO: 184 and a light chain CDR3 of SEQ ID NO: 185
20. The trispecific antibody of claim 1, wherein the third antigen binding site comprises a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 166 and a variable light chain comprising an amino acid sequence of SEQ ID NO: 167 or a humanized version thereof.
21. The trispecific antibody of claim 1, wherein the third antigen binding site comprises a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 92, SEQ ID NO: 195, SEQ ID NO: 196, SEQ ID NO: 197, SEQ ID NO: 198, SEQ ID NO: 199, or SEQ ID NO: 200, and a variable light chain comprising an amino acid sequence selected from the group of SEQ ID NO: 201, SEQ ID NO: 207, SEQ ID NO: 208, or SEQ ID NO:209.
22. A pharmaceutical composition comprising a bispecific antibody of claim 1.
23. The trispecific antibody of claim 1 for the treatment of cancer.
24. The trispecific antibody of claim 1 for use in the treatment of cancer.
25. The trispecific antibody of claim 1 for use as a medicament.
26. Use of the trispecific antibody of claim 1 in the manufacture of a medicament.
27. The use of claim 25, wherein the medicament is for treatment of cancer.
28. The treatment of claim 23, wherein the cancer is a HER2-positive cancer with brain metastases.
29. A nucleic acid sequence comprising a sequence encoding a heavy chain of a trispecific antibody of claim 1.
30. A nucleic acid sequence comprising a sequence encoding a light chain of a trispecific antibody of claim 1.
31. An expression vector comprising a nucleic acid sequence of claim 29.
32. A prokaryotic or eukaryotic host cell comprising a vector according to claim 31.
33. A method of producing an antibody comprising culturing the host cell of claim 32 so that the antibody is produced.
34. The invention as described herein, especially with reference to the foregoing examples.
Description
FIELD OF THE INVENTION
[0001] The present invention relates to trispecific antibodies binding to HER2 and a blood-brain barrier receptor (BBB-R), methods for their production, pharmaceutical compositions containing said antibodies, and uses thereof.
BACKGROUND
[0002] Antibodies specific for tumor-associated antigens are a valuable approach in cancer therapy because they mediate selective destruction of tumor cells, while leaving healthy cells and tissues undamaged. Treatment of cancers affecting the central nervous system (CNS) with large molecules such as antibodies remains challenging as brain penetration is severely limited by the largely impermeable blood-brain barrier (BBB). Past studies have shown that a very small percentage (approximately 0.1%) of an IgG circulating in the bloodstream crosses through the BBB into the CNS (Felgenhauer, Klin. Wschr. 52: 1158-1164 (1974)), where the CNS concentration of the antibody may be insufficient to permit a robust effect. Among the many strategies to overcome this obstacle is to utilize transcytosis trafficking pathways of endogenous receptors expressed at the brain capillary endothelium. Recombinant proteins such as monoclonal antibodies have been designed against these receptors to enable receptor-mediated delivery of large molecules to the brain. However, strategies to maximize brain uptake while minimizing reverse transcytosis back to the blood, and the extent of accumulation after therapeutic dosing, remain unexplored. Furthermore, whether antibodies that cross the BBB are pharmacodynamically functional is unknown.
[0003] Members of the ErbB family of receptor tyrosine kinases are important mediators of cell growth, differentiation and survival. The receptor family includes four distinct members, including epidermal growth factor receptor (EGFR or ErbB1), HER2 (ErbB2 or p185"e"), HER3 (ErbB3) and HER4 (ErbB4 or tyro2). HER2 is a transmembrane surface-bound receptor tyrosine kinase and is normally involved in the signal transduction pathways leading to cell growth and differentiation. HER2 is a promising target for treatment of breast cancer as it was found to be overexpressed in about one-quarter of breast cancer patients (Bange et al, 2001, Nature Medicine 7:548).
[0004] The murine monoclonal antibody 4D5 is targeting HER2 specifically in HER2 overexpressing cancer cells, while having no effect on cells expressing physiological levels of HER2. The humanized (4D5) monoclonal antibody (hu4D5) is commercially known as the drug Herceptin.RTM. (trastuzumab, rhuMAb HER2, U.S. Pat. No. 5,821,337), which gained FDA marketing approval in late 1998.
[0005] Herceptin was the first monoclonal antibody developed for the treatment of HER2-positive breast cancer and has increased survival times for patients so that they are now the same as for patients with HER2-negative breast cancer. Before Herceptin treatment, shorter survival outcomes were expected for patients diagnosed with HER2-positive breast cancer, compared to patients with HER2-negative disease. In the CLEOPATRA study, PERJETA in combination with Herceptin and chemotherapy has shown the extension of survival times for patients with this aggressive disease even further than Herceptin.
[0006] Pertuzumab (PERJETA.TM., rhuMab 2C4, U.S. Pat. No. 7,862,817) is a humanized monoclonal antibody, which is designed specifically to prevent the HER2 receptor from pairing (dimerising) with other HER receptors (EGFR/HER1, HER3 and HER4) on the surface of cells, a process that is believed to play a role in tumor growth and survival. The combination of PERJETA, Herceptin and chemotherapy is thought to provide a more comprehensive blockade of HER signaling pathways. PERJETA is approved in combination with Herceptin (trastuzumab) and docetaxel in adult patients with HER2-positive metastatic or locally recurrent unresectable breast cancer and gained FDA approval for neoadjuvant breast cancer treatment in September 2013. Pertuzumab binds to domain II of HER2, essential for dimerization, while Ttrastuzumab binds to extracellular domain IV of HER2.
[0007] Li et al (Cancer Research. 2013) describe bispecific, bivalent antibodies to ErbB2 that overcome trastuzumab resistance. The bispecific, bivalent antibodies described therein are based on the native Trastuzumab and Pertuzumab sequences.
[0008] Central nervous system (CNS) metastases are observed in up to half of patients with HER2-positive metastatic breast cancer (MBC), with incidence likely to continue to rise due to longer survival through improved systemic treatments. Drugs that specifically block HER2 to stop the growth of cancer cells are called HER2-targeted therapies. Examples of these drugs include trastuzumab (Herceptin), lapatinib (Tykerb), pertuzumab (Perjeta), and ado-trastuzumab emtansine (Kadcyla), commonly referred to as T-DM1. Some of these drugs may be used together with chemotherapy. Unfortunately, these drugs are not usually able to reach the brain as easily as they can reach the rest of the body, with lapatinib being a possible exception. Therefore, when cancer spreads to the brain it is usually treated with surgery and/or radiation therapy. While radiotherapy-based approaches can be effective, there are potential short- and long-term toxicities, and patients frequently progress. CNS response to existing systemic therapies has been generally poor, and there is a high unmet need with no approved treatment for CNS metastases in HER2-positive MBC.
[0009] Surprisingly the inventors of the present application found that optimizing the native Trastuzumab and Pertuzumab sequences and combining these optimized variants with a third binder targeting a blood brain barrier receptor (BBBR) results in a functional molecule with improved properties compared to the combination of the monospecific antibodies rhuMab 2C4 and hu 4D5. Hence the new trispecific format combines the superior characteristics of the bispecific HER2 antibodies known in the art with the advantages of a classical monospecific HER2 antibody and specific CNS targeting: The novel trispecific antibodies of the present invention are monovalent for the two different HER2 epitopes, resulting in the same avidity effect as the bivalent parental antibodies and specifically target the CNS via a BBBR binding moiety. In contrast, tetravalent antibodies may differ in their avidity for HER2 on cells. The avidity effect of the novel trispecific HER2 antibodies may result in a superior safety window on cell types with low HER2 expression such as in normal tissues or cardiac tissues where inhibition of HER2 and/or ADCC may not be desired.
[0010] In one aspect of the invention a trispecific antibody binding to HER2 and a blood-brain barrier receptor (BBB-R) is provided, comprising an IgG molecule wherein one of the Fab fragments is replaced by a crossover Fab fragment. Crossover Fab fragments are Fab fragments wherein either the variable regions or the constant regions of the heavy and light chain are exchanged. Bispecific antibody formats comprising crossover Fab fragments have been described, for example, in WO2009080252, WO2009080253, WO2009080251, WO2009080254, WO2010/136172, WO2010/145792 and WO2013/026831. The native Trastuzumab sequences have been optimized in their CDRs to improve the stability of the antibody CDRs against spontaneous chemical modification, the resulting sequences framework-grafted to avoid mispairing, resulting in highly potent trispecific antibodies that specifically bind to HER2; finally they can be produced with high yield and only low percentage of side products comparable to the conventional parental Her2 antibodies. Chain misparing of light chains resulting from the fact that both pertuzumab and trastuzumab are based on a comparable framework region has been overcome by grafting the CDRs on a completely novel antibody framework.
[0011] In another aspect of the invention trispecific antibodies binding to HER2 and a blood-brain barrier receptor (BBB-R) are provided wherein the two binding moieties for HER2 comprise identical light chains based on a consensus of the parental trastuzumab and pertuzumab light chains and the corresponding pertuzumab heavy chain has been remodeled. The use of this so-called `common light chain` principle, i.e. combining two binders that share one light chain but still have separate specificities, prevents light chain mispairing and in this particular case retains the epitope specificity of the parental antibodies. As a consequence, there are less side products during production, facilitating the homogenous preparation of the trispecific antigen binding molecules at high yields. Surprisingly the inventors of the present invention found that the bispecific HER2 antibodies in the monovalent common light chain format have an increased affinity to the pertuzumab epitope, and show superior inhibitory effects on cell proliferation and induction of cell dependent cytotoxicity (CDC) as compared to the combination of the parental antibodies. Complement dependent cytotoxicity (CDC) is very important for the optimal therapeutic monoclonal antibodies (mAb) function and is totally conserved even after a chemotherapy treatment. However, this activity is generated by some antibodies but not all of them.
SUMMARY
[0012] The present invention relates to a trispecific antibody specifically binding to HER2 and a blood-brain barrier receptor (BBB-R), comprising a first monovalent antigen binding site specific for extracellular domain II of HER2 and a second monovalent antigen binding site specific for extracellular domain IV of HER2, and a third monovalent antigen binding site specific for a BBB-R. In one aspect the BBB-R of the third antigen binding site is selected from the group consisting of transferrin receptor (TfR), insulin receptor, insulin-like growth factor receptor (IGF receptor), low density lipoprotein receptor-related protein 8 (LRP8), low density lipoprotein receptor-related protein 1 (LRP1), and heparin-binding epidermal growth factor-like growth factor (HB-EGF). In one aspect the third antigen binding site is specific for the transferrin receptor. In one aspect the third antigen binding site specifically binds to an epitope in the transferrin receptor comprised within the amino acid sequence of SEQ ID NO: 202, 203 and/or 204.
[0013] In one aspect the trispecific antibody specifically binding to HER2 and a blood-brain barrier receptor (BBB-R), comprises a first Fab molecule capable of specific binding to extracellular domain II of HER2 and a second Fab molecule capable of specific binding to extracellular domain IV of HER2, wherein the sequence of the variable light chain of the first Fab molecule is identical to the sequence of the variable light chain of the second Fab molecule. In one aspect the trispecific antibody specifically binding to HER2 and a blood-brain barrier receptor (BBB-R) comprises (a) a first heavy chain comprising a heavy chain CDR1 comprising an amino acid sequence selected from the group consisting of SEQ ID NO: 55, SEQ ID NO: 58 and SEQ ID NO: 14; a heavy chain CDR 2 comprising an amino acid sequence selected from the group consisting of SEQ ID NO: 77; SEQ ID NO: 15 and SEQ ID NO: 60 and a heavy chain CDR 3 comprising an amino acid sequence selected from the group consisting of SEQ ID NO: 56 or SEQ ID NO: 59 and SEQ ID NO: 16, and (b) a second heavy chain comprising a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 20, a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 29 and a heavy chain CDR3 comprising an amino acid sequence selected from the group consisting of SEQ ID NO: 30 and SEQ ID NO: 79; and (c) a first and a second light chain, wherein the variable light chains of the first and second light chain comprise the CDRs comprising the amino acid sequence of SEQ ID NO: 89, SEQ ID NO: 90 and SEQ ID NO: 19. In one aspect the trispecific antibody specifically binding to HER2 and a blood-brain barrier receptor (BBB-R) comprises two variable light chains comprising an amino acid sequence of SEQ ID NO: 54, a first heavy chain comprising a variable heavy chain comprising an amino acid sequence selected from the group consisting of SEQ ID NO: 64, SEQ ID NO: 70 and SEQ ID NO: 68, and a second heavy chain comprising a variable heavy chain comprising an amino acid sequence selected from the group consisting of SEQ ID NO: 92 and SEQ ID NO: 117. In one aspect the trispecific antibody specifically binding to HER2 and a blood-brain barrier receptor (BBB-R) comprises a first Fab molecule capable of specific binding to extracellular domain II of HER2 and a second Fab molecule capable of specific binding to extracellular domain IV of HER2, wherein either the variable regions or the constant regions of the heavy and light chain of at least one Fab fragment are exchanged. In one aspect the trispecific antibody specifically binding to HER2 and a blood-brain barrier receptor (BBB-R) comprises a first Fab molecule comprising a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 14, a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 15 and a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 16; and a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 11; a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 12 and a light chain CDR3 comprising the amino acid sequence of SEQ ID NO: 13, and a second Fab molecule comprising a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 20; a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 108; a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 79; and a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 107, a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 18 and a light chain CDR3 comprising the amino acid sequence of SEQ ID NO: 19. In one aspect the trispecific antibody specifically binding to HER2 and a blood-brain barrier receptor (BBB-R) comprises a first Fab molecule comprising a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 14, a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 15 and a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 16; and a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 11; a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 12 and a light chain CDR3 comprising the amino acid sequence of SEQ ID NO: 13, and a second Fab molecule comprising a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 20, a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 29, and a heavy chain CDR3 comprising an amino acid sequence selected from the group consisting of SEQ ID NO: 79, SEQ ID NO: 78, SEQ ID NO: 80, SEQ ID NO: 87, SEQ ID NO: 88; and a light chain CDR1 comprising an amino acid sequence selected from the group consisting of SEQ ID NO: 104, SEQ ID NO: 103 and SEQ ID NO: 158; a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 18 and a light chain CDR3 comprising the amino acid sequence of SEQ ID NO: 19. In one aspect the trispecific antibody specifically binding to HER2 and a blood-brain barrier receptor (BBB-R) comprises a first Fab molecule comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 22 and a variable light chain comprising an amino acid sequence of SEQ ID NO: 24 and wherein a second Fab molecule comprising an amino acid sequence of SEQ ID NO: 105 and a light chain variable region comprising an amino acid sequence of SEQ ID NO: 106. In one aspect the trispecific antibody specifically binding to HER2 and a blood-brain barrier receptor (BBB-R) comprises a third antigen binding site specific for a BBB-R selected from a scFv or a scFab. In one aspect the scFv or scFab is connected to the N-terminus of an IgG molecule comprsing the first and second antigen binding site. In one aspect the scFv or scFab is connected to the C-terminus of the first or second subunit of the Fc domain. In one aspect the trispecific antibody specifically binding to HER2 and a blood-brain barrier receptor (BBB-R) comprises a third antigen binding site comprising a 1. a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 186, a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 187 and a heavy chain CDR3 comprising the amino acid sequence selected from the group of SEQ ID NO: 188, SEQ ID NO: 206 or SEQ ID NO: 174; and a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 189, a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 190 and a light chain CDR3 comprising the amino acid sequence of SEQ ID NO:191. In one aspect the trispecific antibody specifically binding to HER2 and a blood-brain barrier receptor (BBB-R) comprises a third antigen binding site comprising a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 172, a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 173 and a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 174; and a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 175, a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 176 and a light chain CDR3 comprising the amino acid sequence of SEQ ID NO:177.
[0014] In one aspect the trispecific antibody specifically binding to HER2 and a blood-brain barrier receptor (BBB-R) comprises a third antigen binding site comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 178 and a variable light chain comprising an amino acid sequence of SEQ ID NO: 179 or a humanized version thereof. In one aspect the trispecific antibody specifically binding to HER2 and a blood-brain barrier receptor (BBB-R) comprises a third antigen binding site comprising 1. a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 192 or SEQ ID NO: 205 and a variable light chain comprising an amino acid sequence of SEQ ID NO: 193.
[0015] In one aspect the trispecific antibody specifically binding to HER2 and a blood-brain barrier receptor (BBB-R) comprises a third antigen binding site comprising a heavy chain CDR1 of SEQ ID NO: 180, a heavy chain CDR2 of SEQ ID NO: 181 and a heavy chain CDR3 of SEQ ID NO: 182; and a light chain CDR1 of SEQ ID NO: 183, a light chain CDR2 of SEQ ID NO: 184 and a light chain CDR3 of SEQ ID NO: 185. In one aspect the trispecific antibody specifically binding to HER2 and a blood-brain barrier receptor (BBB-R) comprises a third antigen binding site comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 166 and a variable light chain comprising an amino acid sequence of SEQ ID NO: 167 or a humanized version thereof. In one aspect the trispecific antibody specifically binding to HER2 and a blood-brain barrier receptor (BBB-R) comprises a third antigen binding site comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 194, SEQ ID NO: 195, SEQ ID NO: 196, SEQ ID NO: 197, SEQ ID NO: 198, SEQ ID NO: 199, or SEQ ID NO: 200, and a variable light chain comprising an amino acid sequence selected from the group of SEQ ID NO: 201, SEQ ID NO: 207, SEQ ID NO: 208, or SEQ ID NO:209.
[0016] In one aspect the trispecific antibody specifically binding to HER2 and a blood-brain barrier receptor (BBB-R) comprises a third antigen binding site comprising sequence alterations in the CDRs resulting in a reduced affinity.
[0017] In a second object the present invention relates to a pharmaceutical composition comprising a trispecific antibody of the present invention.
[0018] In a third object the present invention relates to a trispecific antibody of the present invention for the treatment of cancer. In another embodiment, use of the trispecific antibody as a medicament is provided. Preferably said use is for the treatment of cancer. In one aspect said cancer is a HER2-positive cancer with brain metastases.
[0019] In further objects the present invention relates to a nucleic acid sequence comprising a sequence encoding a heavy chain of a trispecific antibody of the present invention, a nucleic acid sequence comprising a sequence encoding a light chain of a trispecific antibody of the present invention, an expression vector comprising a nucleic acid sequence of the present invention and to a prokaryotic or eukaryotic host cell comprising a vector of the present invention. In addition a method of producing an antibody comprising culturing the host cell so that the antibody is produced is provided.
BRIEF DESCRIPTION OF THE FIGURES
[0020] FIGS. 1A-D: Schematic drawing of Trastuzumab and Pertuzumab bispecific antibodies in a 2+2 IgG-scFv format. The antibodies are bivalent for each antigen binding site and are able to bind two different paratopes in the ErbB2/HER2 receptor (antigen1=trastuzumab specificity, i.e. extracellular domain IV of HER2; antigen2=pertuzumab specificity extracellular domain II of HER2) (A): The single chain Fv (scFv) is fused C-terminally to the heavy chain in the order VH-VL (TvAB12, SEQ ID NOs 123 and 124). (B): The single chain Fv (scFv) is fused N-terminally to the light chain in the order VL-VH (TvAB13, SEQ ID NOs 125 and 126). (C) The single chain Fv (scFv) is fused C-terminally to the light chain in the order VL-VH (TvAB16: SEQ ID NOs 127 and 128, TvAB20: SEQ ID NOs 131 and 132). (D): The single chain Fv (scFv) is fused C-terminally to the heavy chain in the order VL-VH (TvAB17: SEQ ID NOs 129 and 130).
[0021] FIGS. 2A-B: Purification of Trastuzumab and Pertuzumab bispecific antibodies in a 2+2 IgG-scFv format. (A): Size-exclusion purification of TvAb12 (SEQ ID NOs 123 and 124) on a 26/60 Superdex 200 column. (B): SDS-Page analysis of main peak fraction originating from size-exclusion chromatography (NR=non-reducing, R=reducing conditions).
[0022] FIGS. 3A-B: Purification of Trastuzumab and Pertuzumab bispecific antibodies in a 2+2 IgG-scFv format. (A): Size-exclusion purification of TvAb16 (SEQ ID NOs 127 and 128) on a 26/60 Superdex 200 column. (B): SDS-Page analysis of main peak fraction originating from size-exclusion chromatography (NR=non-reducing, R=reducing conditions).
[0023] FIGS. 4A-B: Purification of Trastuzumab and Pertuzumab bispecific antibodies in a 2+2 IgG-scFv format. (A): Size-exclusion purification of TvAb20 (SEQ ID NOs 131 and 132) on a 26/60 Superdex 200 column. Main product peak marked with "1". (B) SDS-Page analysis of main peak fraction originating from size-exclusion chromatography (NR=non-reducing, R=reducing conditions).
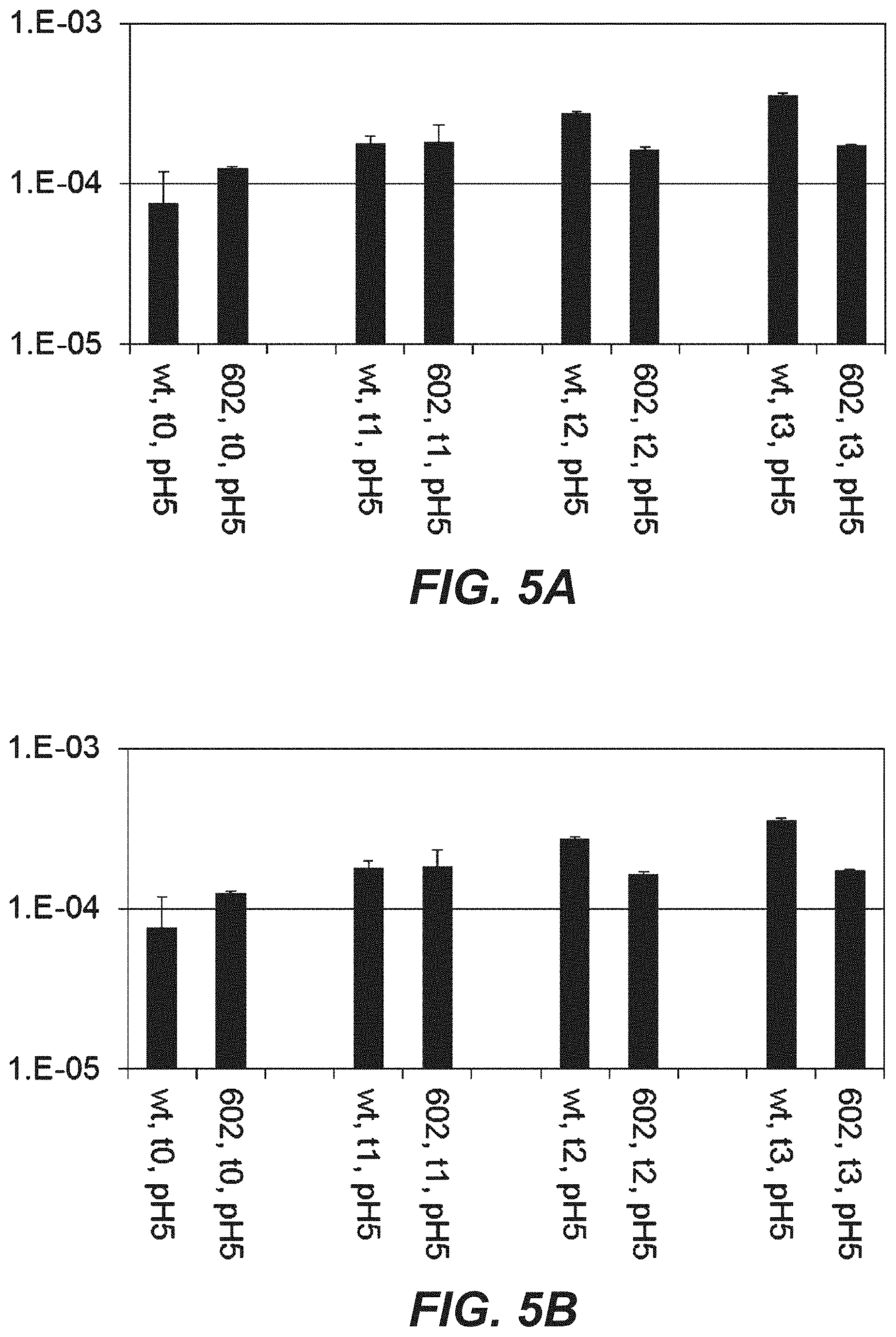
[0024] FIGS. 5A-B: Off-rates of Trastuzumab variants as determined by SPR method (ProteOn instrument) after incubating the samples for 1, 2, or 3 months at 40.degree. in buffer 40 mM Histidin, 150 mM NaCl, pH5.0. The off rates of the variant does not change over the investigated time period. "602": D98E mutation in heavy chain and T31V mutation in light chain.
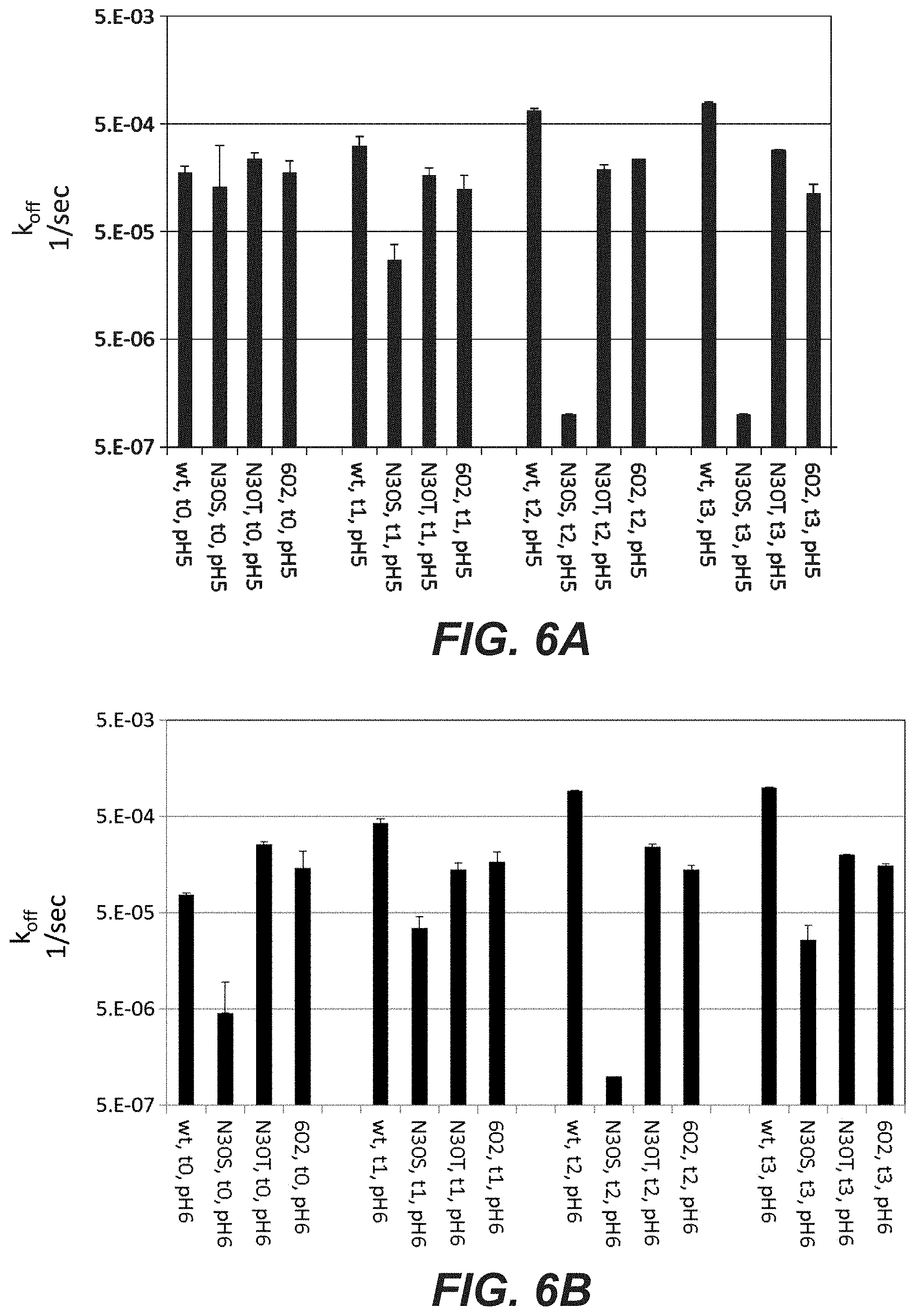
[0025] FIGS. 6A-C: Off-rates of Trastuzumab variants as determined by SPR method (ProteOn instrument) after incubating the samples for 1, 2, or 3 months at 40.degree. C. in 40 mM Histidin, 150 mM NaCl, at different pH. The off rates of the N30S variant were very slow, and therefore contain a high degree of uncertainty. "602": D98E mutation in heavy chain and T31V mutation in light chain, "N30T": D98E mutation in heavy chain and N30T mutation in light chain, "N30S": D98E mutation in heavy chain and N30S mutation in light chain. (A): pH5.0. (B): pH6.0, (C): pH7.4.
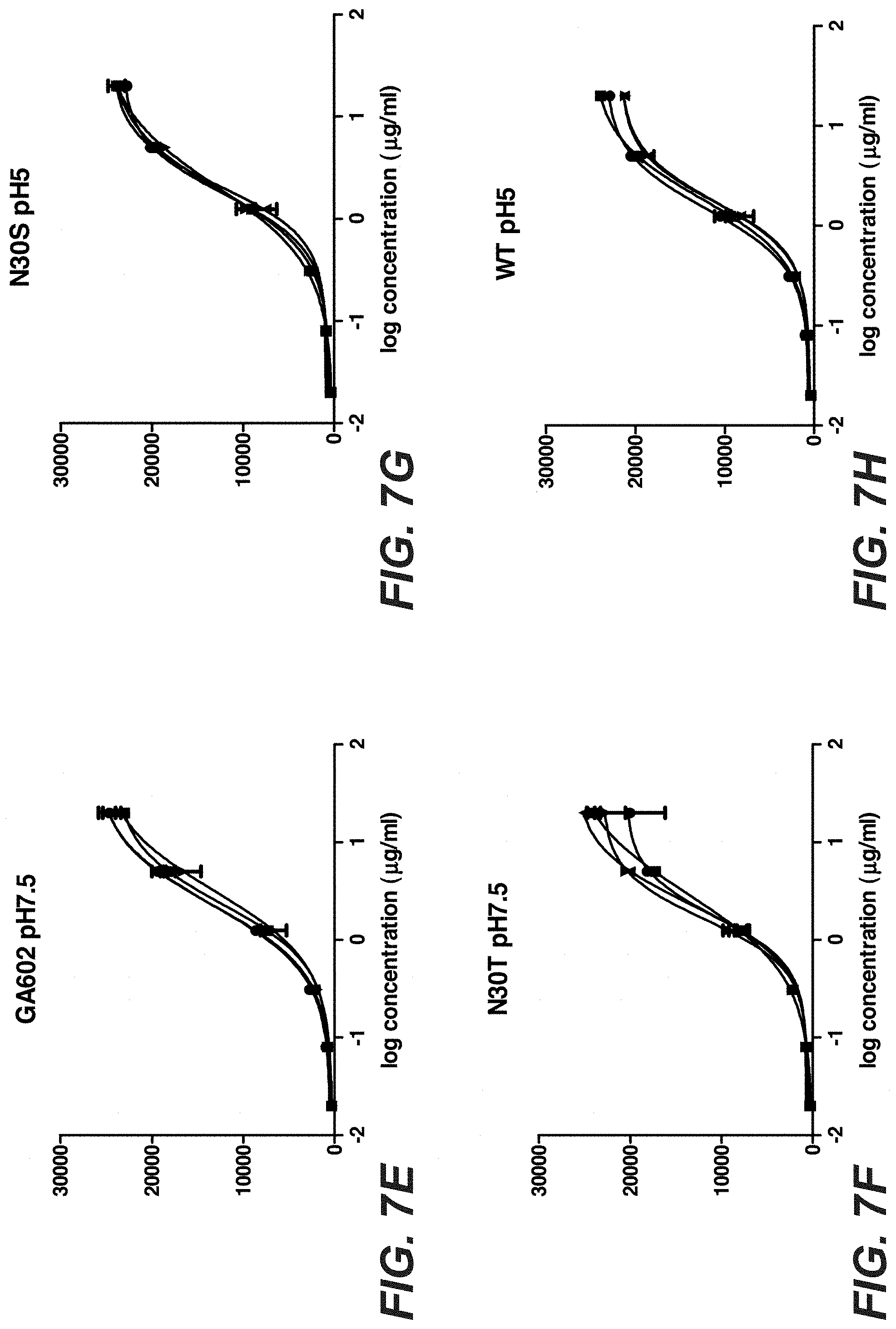
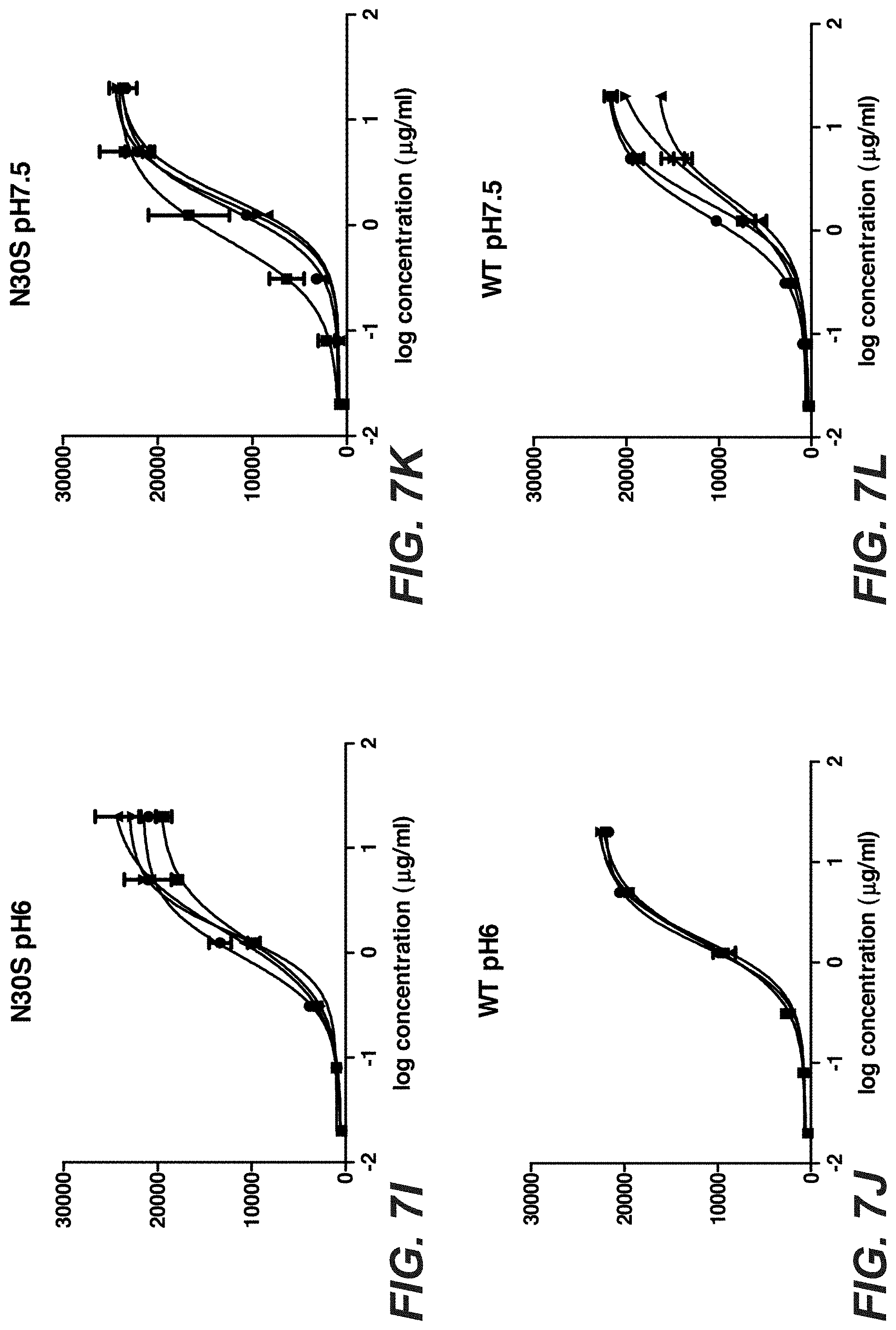
[0026] FIGS. 7A-L: Binding of Trastuzumab and Trastuzumab stabilization variants after stress to KPL-4 cells. Trastuzumab and 3 different stabilized Trastuzumab variants were incubated for one, two and three month in buffer with different pH values at 40.degree. C. The stressed antibodies were tested compared to the antibody at time point zero for binding to KPL-4 cells by flow cytometry. "602": D98E mutation in heavy chain and T31V mutation in light chain, "N30T": D98E mutation in heavy chain and N30T mutation in light chain, "N30S": D98E mutation in heavy chain and N30S mutation in light chain.
[0027] FIGS. 8A-F: Binding of Trastuzumab and Trastuzumab stabilization variants after stress to KPL-4 cells. Trastuzumab and the 2 stabilization variants GA602 (D98E mutation in heavy chain and T31V mutation in light chain) and GA603 (D98E mutation in heavy chain and T31V mutation in light chain and FcRN mutation T307Q und N434A) were incubated for one, two and three month in buffer 1 (40 mM Histidin 150 mM NaCl, pH5.0) or buffer 2 (2. 40 mM Histidin 150 mM NaCl, pH6.0) at 40.degree. C. The stressed antibodies were tested compared to the antibody at time point zero for binding to KPL-4 cells by flow cytometry.
[0028] FIGS. 9A-F: ADCC induction with Trastuzumab, GA602 and GA603 after stress on KPL-4 cells. Trastuzumab and the 2 stabilization variants GA602 (D98E mutation in heavy chain and T31V mutation in light chain) and GA603 (D98E mutation in heavy chain and T31V mutation in light chain and FcRN mutation T307Q und N434A) were incubated for one, two and three month in buffer 1 (40 mM Histidin 150 mM NaCl, pH 5.0) or buffer 2 (2. 40 mM Histidin 150 mM NaCl, pH6.0) at 40.degree. C. The stressed antibodies were tested compared to the antibody at time point zero for ADCC induction after 4 h on KPL-4 cells.
[0029] FIGS. 10A-B: Schematic drawing of Trastuzumab and Pertuzumab bispecific antibodies in a 1+1 format. (A): single chain Fab (scFab) based molecules (B): cross-over Fab (xFab) based molecules.
[0030] FIGS. 11A-B: Purification of CrossMab-XPer (SEQ ID NOs 109, 110, 96, 86). (A): SDS-PAGE showing the purified antibody molecule under reduced and non-reduced conditions. (B): HP-SEC analysis of purified CrossMab-XPer.
[0031] FIGS. 12A-B: Q-TOF mass spectrometry comparison of the spectra of CrossMab-XTra (top, SEQ ID NOs 119, 120, 121, 122) and CrossMab-CDRG (bottom, SEQ ID NOs 109, 110, 111, 112) estimating the integrity and purity of the antibody molecules.
[0032] FIGS. 13A-B: Proliferation inhibition by non-glycoengineered HER2 CrossMab (SEQ ID NOs 119, 120, 121, 122) after 5 days of incubation as measured in an AlamarBlue.RTM. assay. (A) BT474 cells (B) N87 cells.
[0033] FIGS. 14A-C: ADCC induced by different HER2 specific antibodies using (A) KPL-4, (B) T47D and (C) Calu-3 as target cells (E:T=25:1, effectors human PBMCs, incubation time 4 h). "HER2 crossmab wt": SEQ ID NOs 119, 120, 121, 122, non glycoengineered; "HER2 crossmab g2": SEQ ID NOs 119, 120, 121, 122, glycoengineered.
[0034] FIGS. 15A-C: Antitumor activity of different anti-Her2 antibodies in the Calu3 non-small cell lung cancer xenograft (Experiment: BispecHer2_PZ_Calu3_001). SCID beige mice with Calu3 xenograft tumors were treated i.p. once weekly at the indicated dosages for 7 weeks. Xolair a humanized IgG1 antibody targeting human IgE was used as a control. Statistical analysis based on medians at endpoint (day 85) reveals that compared to Xolair the bispecific HER2 antibodies suppressed tumor growth by 87.5% (s.); OmniE (SEQ ID NOs 145, 146) by 43.7% (n.s.); Crossmab_003 (SEQ ID NOs 119, 120, 121, 122, non glycoengineered) by 92.1% (s.); TvAb12 (SEQ ID NOs 123 and 124) by 59.8% (n.s.) and TvAb20 (SEQ ID NOs 131 and 132) by 12.6% (n.s.). Tumor growth curves are depicted as mean+/-SEM (n=8 in each group).
[0035] FIGS. 16A-C: Antitumor activity of different anti-Her2 antibodies in the KPL-4 breast cancer xenograft (Experiment: Bispec.Her2_PZ_KPL-4_002). SCID beige mice with KPL-4 xenograft tumors were treated i.p. once weekly at the indicated dosages for 5 weeks. Xolair a humanized IgG1 antibody targeting human IgE was used as a control. Statistical analysis based on medians at endpoint (day 59) reveals that compared to Xolair the bispecific HER2 antibodies suppressed tumor growth by 120.8% (s.); Crossmab_003 (SEQ ID NOs 119, 120, 121, 122, non glycoengineered) by 120.6% (s.); TvAb12 (SEQ ID NOs 123 and 124) by 70.1% (s.); TvAb20 (SEQ ID NOs 131 and 132) by 83.4% (s.). OmniE (SEQ ID NOs 145, 146) had no significant effect on tumor growth. Tumor growth curves are depicted as mean+/-SEM (n=9 in each group).
[0036] FIGS. 17A-C: Antitumor activity of anti-Her2_005 crossmab antibody (SEQ ID NOs 119, 120, 121, 122, non glycoengineered) in the KPL-4 breast cancer xenograft (Experiment: Bispec.Her2_PZ_KPL-4_003). SCID beige mice with KPL-4 xenograft tumors were treated i.p. once weekly with escalating dosages of the crossmab ranging from 1 to 20 mg/kg for 5 weeks. Xolair a humanized IgG1 antibody targeting human IgE was used as a control. Statistical analysis based on medians at endpoint (day 70) reveals that compared to Xolair the bispecific HER2 antibodies suppressed tumor growth by 121.8% (s.); The Her2 crossmab_005 suppressed tumor growth at a dosage of 1 mg/kg by 25.1% (n.s.); at 5 mg/kg by 112.3% (s.); at 10 mg/kg by 109.5% (s.) and by 20 mg/kg by 121.8% (s.). Tumor growth curves are depicted as mean+/-SEM (n=10 in each group).
[0037] FIGS. 18A-C: Antitumor activity of different anti-Her2 antibodies in the KPL-4 breast cancer xenograft (Experiment: Bispec.Her2_PZ_KPL-4_009). SCID beige mice with KPL-4 xenograft tumors were treated i.p. once weekly with the different compounds for 4 weeks. Xolair a humanized IgG1 antibody targeting human IgE was used as a control. Statistical analysis based on medians at endpoint (day 70) reveals that compared to Xolair the bispecific HER2 antibodies (each dosed at 5 mg/kg) suppressed tumor growth by 83.2% (s.) and both given at a dosage of 10 mg/kg each by 109.5% (s.). TvAb 16 (SEQ ID NOs 127 and 128) given at two different dosages (5 mg/kg and 10 mg/kg) had no significant anti-tumoral effect. TvAb20 (SEQ ID NOs 131 and 132), at a dosage of 5 mg/kg, suppressed tumor growth by 75.3% (s.) and at a dosage of 10 mg/kg by 59.8% (n.s.). Tumor growth curves are depicted as mean+/-SEM (n=10 in each group).
[0038] FIGS. 19A-F: SPR analysis of initial Pertuzumab/Trastuzumab hybrid light chains. SPR-based kinetic analyses of Pertuzumab, Trastuzumab, and sequence combinations with the initial Pertuzumab hybrid LCs harboring amino acid residues of the Trastuzumab LCDR3 region. Smooth lines represent a global fit of the data to a 1:1 interaction model. PertuzumabTrasL3: SEQ ID No: 26, PertuzumabTras Y91H: SEQ ID No: 28.
[0039] FIGS. 20A-B: SPR analysis of the Pertuzumab and Trastuzumab HCs in combination with the newly identified common light chain Pertuzumab (Tras.L3)(QM), SEQ ID No: 54. Shown is the binding of both antibodies to Her2 at different concentrations. Smooth lines represent a global fit of the data to a 1:1 interaction model.
[0040] FIGS. 21A-F: Characterization of the affinity-matured Pertuzumab clones identified by phage display. SPR analysis of the identified affinity-matured clones. Shown is the binding of bacterial Fabs to Her2 at different concentrations. Smooth lines represent a global fit of the data to a 1:1 interaction model. B2: SEQ ID No: 66, D1: SEQ ID No: 62, E1: SEQ ID No: 68, C8: SEQ ID No: 72, G2: SEQ ID No: 70, A1: SEQ ID No: 74.
[0041] FIG. 22: Schematic drawing of the bi-specific HER2 antibodies with a common light chain.
[0042] FIGS. 23A-F: Purification and analytical characterization of the bi-specific HER2 antibodies with a common light chain. The purification method involved an affinity step (protein A) followed by size exclusion chromatography (Superdex 200, GE Healthcare). The final product was analyzed and characterized by analytical size exclusion chromatography (Superdex 200 column). (A)(B): comprising D1der (SEQ ID NO: 64), (C)(D): comprising G2 (SEQ ID NO: 70), (E)(F): comprising E1 (SEQ ID NO: 68).
[0043] FIGS. 24A-D: SPR analysis of the Her2 knock-out variants. Shown are the sensograms of Trastuzumab and Pertuzumab binding to both knock-out variants. Smooth lines represent a global fit of the data to a 1:1 interaction model.
[0044] FIG. 25: Binding of bi-specific HER2 antibodies with a common light chain clone variants to KPL-4 cells. KPL-4 cells were stained with increasing concentrations of the indicated antibodies. The antibodies were detected with a FITC labeled anti-human secondary and the fluorescence was determined by flow cytometry. "Herceptarg CLC D1-der": SEQ ID NOs 64, 54, 92, "Herceptarg CLC G2/2": SEQ ID NOs 70, 54, 92, "Herceptarg CLC E1/1": SEQ ID NOs 68, 54, 92; "GA 604": SEQ ID NOs 109, 110, 111, 112.
[0045] FIGS. 26A-F: Proliferation inhibition of BT474, N87, and SkBr3 cells with bi-specific HER2 antibodies with common light chain clone variants. BT474 (A)(B), N87 (C)(D), and SkBr3 (E)(F) cells were treated with the three different Herceptarg variants. As controls Trastuzumab, Pertuzumab and the combination of both were included. After 5 days, proliferation inhibition was determined with CellTiter Glo. "Herceptarg CLC D1-der": SEQ ID NOs 64, 54, 92, "Herceptarg CLC G2/2": SEQ ID NOs 70, 54, 92, "Herceptarg CLC E1/1": SEQ ID NOs 68, 54, 92; "GA 604": SEQ ID NOs 109, 110, 111, 112.
[0046] FIGS. 27A-D: Killing of KPL-4 cells and MDA-MB 231 with bi-specific HER2 antibodies with common light chain variants. (A)(B) Antibody dependent killing of KPL-4 cells with PBMCs (E:T 25:1) or was determined by measuring LDH release after 4 h. (C)(D)Antibody dependent killing of MDA-MB 231 cells with PBMCs (E:T 5:1) was determined by measuring LDH release after 24 h. "Herceptarg CLC D1-der": SEQ ID NOs 64, 54, 92, "Herceptarg CLC G2/2": SEQ ID NOs 70, 54, 92, "Herceptarg CLC E1/1": SEQ ID NOs 68, 54, 92; "GA 604": SEQ ID NOs 109, 110, 111, 112.
[0047] FIG. 28: Proliferation inhibition of BT474 cells with bi-specific HER2 antibodies with common light chain clone variants. BT474 cells were treated with the different Herceptarg variants. As controls Trastuzumab, Pertuzumab and the combination of both were included. After 6 days, proliferation inhibition was determined with CellTiter Glo. "Herceptarg CLC D1-der wt": SEQ ID NOs 64, 54, 92, Herceptarg CLC D1-der G2'': SEQ ID NOs 64, 54, 92 (glycoengineered variant) "Herceptarg CrossMab": SEQ ID NOs 109, 110, 111, 112.
[0048] FIG. 29: C1q binding of Her2 antibodies on BT-474 cells. BT474 cells were incubated with the three Herceptarg variants. As controls Trastuzumab, Pertuzumab and the combination of both were included. "Herceptarg CLC D1-der wt": SEQ ID NOs 64, 54, 92, Herceptarg CLC D1-der G2'': SEQ ID NOs 64, 54, 92 (glycoengineered variant) "Herceptarg CrossMab": SEQ ID NOs 109, 110, 111, 112.
[0049] FIG. 30: CDC activation on BT-474 cells (LDH release). BT474 cells were incubated with the three Herceptarg variants. As controls Trastuzumab, Pertuzumab and the combination of both were included. "Herceptarg CLC D1-der wt": SEQ ID NOs 64, 54, 92, Herceptarg CLC D1-der G2'': SEQ ID NOs 64, 54, 92 (glycoengineered variant) "Herceptarg CrossMab": SEQ ID NOs 109, 110, 111, 112.
[0050] FIGS. 31A-B: CDC mediated killing of BT-474 cells (ACEA). BT474 cells were incubated with the three Herceptarg variants. As controls Trastuzumab, Pertuzumab and the combination of both were included. "Herceptarg CLC D1-der wt": SEQ ID NOs 64, 54, 92, Herceptarg CLC D1-der G2'': SEQ ID NOs 64, 54, 92 (glycoengineered variant) "Herceptarg CrossMab": SEQ ID NOs 109, 110, 111, 112.
[0051] FIG. 32: In vivo activity of bispecific antibodies. Tumor volume in mouse xenograft models after treatment with different Her2 bispecific molecules (10 mg/kg) was compared to treatment with Trastuzumab, Pertuzumab and the combination of both. "Herceptarg": SEQ ID NOs 64, 54, 92. "Control": Xolair, a non Her2 binding antibody.
[0052] FIG. 33: FACS binding of trispecific antibodies specific for HER2 and transferrin receptor to HER2+BT474.M1 cells. "Herceptarg-8D3" (SEQ ID NOs.: 165, 163, 161) and "Herceptarg (LALA)-8D3" (SEQ ID NOs.: 168, 169, 161) are trispecific, trivalent antibodies specific for extracellular domains II and IV of HER2 and the transferrin receptor. Positive control: "Herceptarg-LALA" (SEQ ID NOs: 170, 169, 161) and "Herceptarg" (SEQ ID NOs.: 171, 163, 161) are bispecific, bivalent antibodies specific for extracellular domains II and IV of HER2. Negative control "pTAU-8D3" is a bispecific antibody specific for the transferrin receptor and pTau (SEQ ID. NOs: 210, 211, 212).
[0053] FIG. 34: FACS binding of trispecific antibodies specific for HER2 and transferrin receptor to Transferrin receptor expressing (TfR+) BAF3 cells. "Herceptarg-8D3" (SEQ ID NOs.: 165, 163, 161) and "Herceptarg (LALA)-8D3" (SEQ ID NOs.: 168, 169, 161) are trispecific, trivalent antibodies specific for extracellular domains II and IV of HER2 and the transferrin receptor. Positive control: "Herceptarg-LALA" (SEQ ID NOs: 170, 169, 161) and "Herceptarg" (SEQ ID NOs.: 171, 163, 161) are bispecific, bivalent antibodies specific for extracellular domains II and IV of HER2. Positive control "pTAU-8D3" is a bispecific antibody specific for the transferrin receptor and pTau (SEQ ID. NOs: 210, 211, 212).
[0054] FIG. 35: Proliferation inhibition assay of trispecific antibodies specific for HER2 and transferrin receptor. Herceptarg-8D3'' (SEQ ID NOs.: 165, 163, 161) and "Herceptarg (LALA)-8D3" (SEQ ID NOs.: 168, 169, 161) are trispecific, trivalent antibodies specific for extracellular domains II and IV of HER2 and the transferrin receptor. Positive control: "Herceptarg-LALA" (SEQ ID NOs: 170, 169, 161) and "Herceptarg" (SEQ ID NOs.: 171, 163, 161) are bispecific, bivalent antibodies specific for extracellular domains II and IV of HER2. Negative control "pTAU-8D3" is a bispecific antibody specific for the transferrin receptor and pTau (SEQ ID. NOs: 210, 211, 212).
[0055] FIG. 36: Herceptarg(LALA)+/-scFab(8D3) penetration from vascular border into tumor tissue. Mean antibody signal in tumor tissue as a function of distance from the nearest tumor vessel shows that the brain shuttle (BS) increases penetration of Herceptarg into the brain tumor.
DETAILED DESCRIPTION OF EMBODIMENTS OF THE INVENTION
I. Definitions
[0056] Throughout the disclosure, the terms "ErbB2", "ErbB2 receptor", "c-Erb-B2", and "HER2" are used interchangeably, and, unless otherwise indicated, refer to a native sequence ErbB2 human polypeptide, or a functional derivative thereof "ber2", "erbB2" and "c-erb-B2" refer to the corresponding human gene.
[0057] Herein, "HER2 extracellular domain" or "HER2 ECD" refers to a domain of HER2 that is outside of a cell, either anchored to a cell membrane, or in circulation, including fragments thereof. The amino acid sequence of HER2 is shown in FIG. 1 of WO2013055874. In one embodiment, the extracellular domain of HER2 may comprise four domains: "Domain I" (amino acid residues from about 1-195; SEQ ID NO: 1 of WO2013055874), "Domain .PI." (amino acid residues from about 196-319; SEQ ID NO:2 of WO2013055874), "Domain III" (amino acid residues from about 320-488: SEQ ID NO:3 of WO2013055874), and "Domain IV" (amino acid residues from about 489-630; SEQ ID NO:4 of WO2013055874) (residue numbering without signal peptide). See Garrett et al. Mol. Cell. 11: 495-505 (2003), Cho et al. Nature All: 756-760 (2003), Franklin et al. Cancer Cell 5:317-328 (2004), and Plowman et al. Proc. Natl. Acad. Sci. 90:1746-1750 (1993), as well as FIG. 6 in WO2013055874.
[0058] "Antigen binding site specific for extracellular domain II of HER2" refers to the epitope of Pertuzumab (which is also known as recombinant humanized monoclonal antibody 2C4 (rhuMAb 2C4)), also depicted as "epitope 2C4". The "epitope 2C4" is the region in the extracellular domain of HER2 to which the murine antibody 2C4 and Pertuzumab bind (see e.g. WO2013055874). In order to screen for antibodies which bind essentially to the 2C4 epitope, a routine cross-blocking assay such as that described in Antibodies, A Laboratory Manual, Cold Spring Harbor Laboratory, Ed Harlow and David Lane (1988), can be performed. Preferably the antibody blocks 2C4's binding to HER2 by about 50% or more. Alternatively, epitope mapping can be performed to assess whether the antibody binds essentially to the 2C4 epitope of HER2. Epitope 2C4 comprises residues from Domain II (SEQ ID NO: 2 of WO2013055874) in the extracellular domain of HER2. 2C4 and Pertuzumab binds to the extracellular domain of HER2 at the junction of domains I, II and III (SEQ ID NOs: 1, 2, and 3 of WO2013055874, respectively). Franklin et al. Cancer Cell 5:317-328 (2004).
[0059] "Antigen binding site specific for extracellular domain IV of HER2" refers to the epitope of Trastuzumab, also depicted as "epitope 4D5". The "epitope 4D5" is the region in the extracellular domain of HER2 to which the antibody 4D5 (ATCC CRL 10463) and Trastuzumab bind. This epitope is close to the transmembrane domain of HER2, and within Domain IV of HER2 (SEQ ID NO: 4 of WO2013055874). To screen for antibodies which bind essentially to the 4D5 epitope, a routine cross-blocking assay such as that described in Antibodies, A Laboratory Manual, Cold Spring Harbor Laboratory, Ed Harlow and David Lane (1988), can be performed. Alternatively, epitope mapping can be performed to assess whether the antibody binds essentially to the 4D5 epitope of HER2 {e.g. any one or more residues in the region from about residue 529 to about residue 625, inclusive of the HER2 ECD, residue numbering including signal peptide).
[0060] The "blood-brain barrier" or "BBB" refers to the physiological barrier between the peripheral circulation and the brain and spinal cord which is formed by tight junctions within the brain capillary endothelial plasma membranes, creating a tight barrier that restricts the transport of molecules into the brain, even very small molecules such as urea (60 Daltons). The BBB within the brain, the blood-spinal cord barrier within the spinal cord, and the blood-retinal barrier within the retina are contiguous capillary barriers within the CNS, and are herein collectively referred to an the blood-brain barrier or BBB. The BBB also encompasses the blood-CSF barrier (choroid plexus) where the barrier is comprised of ependymal cells rather than capillary endothelial cells.
[0061] The term "blood brain barrier receptor" or "BBB-R" refers to an extracellular membrane-linked receptor protein expressed on brain endothelial cells which is capable of transporting molecules across the BBB and can be used to transport exogenous administrated molecules. Examples of BBB-R herein include: transferrin receptor (TfR), insulin receptor, insulin-like growth factor receptor (IGF-R), low density lipoprotein receptors including without limitation low density lipoprotein receptor-related protein 1 (LRP1) and low density lipoprotein receptor-related protein 8 (LRP8), and heparin-binding epidermal growth factor-like growth factor (HB-EGF). In one specific embodiment the BBB-R is a transferrin receptor, preferably a human transferrin receptor and/or a cynomolgous monkey transferrin receptor. The term "transferrin receptor" or "TfR" refers to a transmembrane glycoprotein (with a molecular weight of about 180,000) composed of two disulphide-bonded sub-units (each of apparent mo-lecular weight of about 90,000) involved in iron uptake in vertebrates. In one embodiment, the TfR herein is human TfR comprising the amino acid sequence as in Schneider et al. Nature 311:675-678 (1984). The TfR mediates receptor-mediated transcytosis (RMT)
[0062] An "acceptor human framework" for the purposes herein is a framework comprising the amino acid sequence of a light chain variable domain (VL) framework or a heavy chain variable domain (VH) framework derived from a human immunoglobulin framework or a human consensus framework, as defined below. An acceptor human framework "derived from" a human immunoglobulin framework or a human consensus framework may comprise the same amino acid sequence thereof, or it may contain amino acid sequence changes. In some embodiments, the number of amino acid changes are 10 or less, 9 or less, 8 or less, 7 or less, 6 or less, 5 or less, 4 or less, 3 or less, or 2 or less. In some embodiments, the VL acceptor human framework is identical in sequence to the VL human immunoglobulin framework sequence or human consensus framework sequence.
[0063] "Affinity" refers to the strength of the sum total of noncovalent interactions between a single binding site of a molecule (e.g., an antibody) and its binding partner (e.g., an antigen). Unless indicated otherwise, as used herein, "binding affinity" refers to intrinsic binding affinity which reflects a 1:1 interaction between members of a binding pair (e.g., antibody and antigen). The affinity of a molecule X for its partner Y can generally be represented by the dissociation constant (Kd). Affinity can be measured by common methods known in the art, including those described herein. Specific illustrative and exemplary embodiments for measuring binding affinity are described in the following.
[0064] An "affinity matured" antibody refers to an antibody with one or more alterations in one or more hypervariable regions (HVRs), compared to a parent antibody which does not possess such alterations, such alterations resulting in an improvement in the affinity of the antibody for antigen.
[0065] An "affinity reduced" antibody refers to an antibody with one or more alterations in one or more hypervariable regions (HVRs), compared to a parent antibody which does not possess such alterations, such alterations resulting in a reduction in the affinity of the antibody for antigen. Reduced affinity is advantageous for the exposure of the trispecific antibody specifically binding to HER2 and a blood-brain barrier receptor (BBB-R) in the brain.
[0066] The terms "a bispecific HER2 antibody" and "a bispecific antibody that specifically binds to HER2" are used interchangeably and refer to a bispecific antibody that is capable of binding HER2 on both extracellular domains II and IV, respectively, with sufficient affinity such that the antibody is useful as a diagnostic and/or therapeutic agent in targeting cells expressing HER2. In one embodiment, the extent of binding of a bispecific antibody that specifically binds to HER2 on both extracellular domains II and IV to an unrelated, non-HER2 protein is less than about 10% of the binding of the antibody to HER2 as measured, e.g., by a Enzyme-linked immunosorbent assay (ELISA), surface plasmon resonance (SPR) based assays (e.g. Biacore) or flow cytometry (FACS). In certain embodiments, a bispecific antibody that specifically binds to HER2 has a dissociation constant (Kd) of .ltoreq.1 .mu.M, .ltoreq.100 nM, .ltoreq.10 nM, .ltoreq.1 nM, .ltoreq.0.1 nM, .ltoreq.0.01 nM, or .ltoreq.0.001 nM (e.g. 10.sup.-8M or less, e.g. from 10.sup.-8M to 10.sup.-13M, e.g., from 10.sup.-9M to 10.sup.-13 M).
[0067] The terms "a trispecific antibody binding to HER2 and a blood-brain barrier receptor (BBB-R)" and "a trispecific antibody that specifically binds to HER2 and BBB-R" are used interchangeably and refer to a trispecific antibody that is capable of binding HER2 on both extracellular domains II and IV and a BBB-R, respectively, with sufficient affinity such that the antibody is useful as a diagnostic and/or therapeutic agent in targeting CNS cells expressing HER2. In one embodiment, the extent of binding of a trispecific antibody that specifically binds to HER2 on both extracellular domains II and IV and a BBB-R to an unrelated, non-HER2 or non BBB-R protein is less than about 10% of the binding of the antibody to HER2 or a BBB-R as measured, e.g., by a Enzyme-linked immunosorbent assay (ELISA), surface plasmon resonance (SPR) based assays (e.g. Biacore) or flow cytometry (FACS). In certain embodiments, the antigen binding site specifically binding to a BBB-R of the trispecific antibody has an off-rate of 0.07 to 0.005 l/s for the BBB-R, preferably as determined by SPR.
[0068] The term "antibody" herein is used in the broadest sense and encompasses various antibody structures, including but not limited to monoclonal antibodies, polyclonal antibodies, multispecific antibodies (e.g., trispecific antibodies), and antibody fragments so long as they exhibit the desired antigen-binding activity.
[0069] An "antibody fragment" refers to a molecule other than an intact antibody that comprises a portion of an intact antibody that binds the antigen to which the intact antibody binds. Examples of antibody fragments include but are not limited to Fv, Fab, Fab', Fab'-SH, F(ab')2; diabodies, cross-Fab fragments; linear antibodies; single-chain antibody molecules (e.g. scFv); and multispecific antibodies formed from antibody fragments. scFv antibodies are, e.g. described in Houston, J. S., Methods in Enzymol. 203 (1991) 46-96). In addition, antibody fragments comprise single chain polypeptides having the characteristics of a VH domain, namely being able to assemble together with a VL domain, or of a VL domain, namely being able to assemble together with a VH domain to a functional antigen binding site and thereby providing the antigen binding property of full length antibodies.
[0070] As used herein, "Fab fragment" refers to an antibody fragment comprising a light chain fragment comprising a VL domain and a constant domain of a light chain (CL), and a VH domain and a first constant domain (CH1) of a heavy chain. In one embodiment the trispecific antibodies of the invention comprise at least one Fab fragment, wherein either the variable regions or the constant regions of the heavy and light chain are exchanged. Due to the exchange of either the variable regions or the constant regions, said Fab fragment is also referred to as "cross-Fab fragment" or "xFab fragment" or "crossover Fab fragment". Two different chain compositions of a crossover Fab molecule are possible and comprised in the trispecific antibodies of the invention: On the one hand, the variable regions of the Fab heavy and light chain are exchanged, i.e. the crossover Fab molecule comprises a peptide chain composed of the light chain variable region (VL) and the heavy chain constant region (CH1), and a peptide chain composed of the heavy chain variable region (VH) and the light chain constant region (CL). This crossover Fab molecule is also referred to as CrossFab.sub.(VLVH). On the other hand, when the constant regions of the Fab heavy and light chain are exchanged, the crossover Fab molecule comprises a peptide chain composed of the heavy chain variable region (VH) and the light chain constant region (CL), and a peptide chain composed of the light chain variable region (VL) and the heavy chain constant region (CH1). This crossover Fab molecule is also referred to as CrossFab.sub.(CLCH1).
[0071] A "single chain Fab fragment" or "scFab" is a polypeptide consisting of an antibody heavy chain variable domain (VH), an antibody constant domain 1 (CH1), an antibody light chain variable domain (VL), an antibody light chain constant domain (CL) and a linker, wherein said antibody domains and said linker have one of the following orders in N-terminal to C-terminal direction: a) VH-CH1-linker-VL-CL, b) VL-CL-linker-VH-CH1, c) VH-CL-linker-VL-CH1 or d) VL-CH1-linker-VH-CL; and wherein said linker is a polypeptide of at least 30 amino acids, preferably between 32 and 50 amino acids. Said single chain Fab fragments a) VH-CH1-linker-VL-CL, b) VL-CL-linker-VH-CH1, c) VH-CL-linker-VL-CH1 and d) VL-CH1-linker-VH-CL, are stabilized via the natural disulfide bond between the CL domain and the CH1 domain. In addition, these single chain Fab molecules might be further stabilized by generation of interchain disulfide bonds via insertion of cysteine residues (e.g. position 44 in the variable heavy chain and position 100 in the variable light chain according to Kabat numbering). The term "N-terminus denotes the last amino acid of the N-terminus. The term "C-terminus denotes the last amino acid of the C-terminus.
[0072] By "fused" or "connected" is meant that the components (e.g. a Fab molecule and an Fc domain subunit) are linked by peptide bonds, either directly or via one or more peptide linkers.
[0073] The term "linker" as used herein refers to a peptide linker and is preferably a peptide with an amino acid sequence with a length of at least 5 amino acids, preferably with a length of 5 to 100, more preferably of 10 to 50 amino acids. In one embodiment said peptide linker is (GxS)n or (GxS)nGm with G=glycine, S=serine, and (x=3, n=3, 4, 5 or 6, and m=0, 1, 2 or 3) or (x=4, n=2, 3, 4 or 5 and m=0, 1, 2 or 3), preferably x=4 and n=2 or 3, more preferably with x=4, n=2. In one embodiment said peptide linker is (G4S).sub.2.
[0074] The term "immunoglobulin molecule" refers to a protein having the structure of a naturally occurring antibody. For example, immunoglobulins of the IgG class are heterotetrameric glycoproteins of about 150,000 daltons, composed of two light chains and two heavy chains that are disulfide-bonded. From N- to C-terminus, each heavy chain has a variable region (VH), also called a variable heavy domain or a heavy chain variable domain, followed by three constant domains (CH1, CH2, and CH3), also called a heavy chain constant region. Similarly, from N- to C-terminus, each light chain has a variable region (VL), also called a variable light domain or a light chain variable domain, followed by a constant light (CL) domain, also called a light chain constant region. The heavy chain of an immunoglobulin may be assigned to one of five types, called .alpha. (IgA), .delta. (IgD), .epsilon. (IgE), .gamma. (IgG), or .mu. (IgM), some of which may be further divided into subtypes, e.g. .gamma..sub.1 (IgG.sub.1), .gamma..sub.2 (IgG.sub.2), .gamma..sub.3 (IgG.sub.3), .gamma..sub.4 (IgG.sub.4), .alpha..sub.1 (IgA.sub.1) and .alpha..sub.2 (IgA.sub.2). The light chain of an immunoglobulin may be assigned to one of two types, called kappa (.kappa.) and lambda (.lamda.), based on the amino acid sequence of its constant domain. An immunoglobulin essentially consists of two Fab molecules and an Fc domain, linked via the immunoglobulin hinge region.
[0075] An "antibody that binds to the same epitope" as a reference antibody refers to an antibody that blocks binding of the reference antibody to its antigen in a competition assay by 50% or more, and conversely, the reference antibody blocks binding of the antibody to its antigen in a competition assay by 50% or more. An exemplary competition assay is provided herein.
[0076] The term "antigen binding domain" refers to the part of an antigen binding molecule that comprises the area which specifically binds to and is complementary to part or all of an antigen. Where an antigen is large, an antigen binding molecule may only bind to a particular part of the antigen, which part is termed an epitope. An antigen binding domain may be provided by, for example, one or more antibody variable domains (also called antibody variable regions). Preferably, an antigen binding domain comprises an antibody light chain variable region (VL) and an antibody heavy chain variable region (VH).
[0077] The term "chimeric" antibody refers to an antibody in which a portion of the heavy and/or light chain is derived from a particular source or species, while the remainder of the heavy and/or light chain is derived from a different source or species, usually prepared by recombinant DNA techniques. Chimeric antibodies comprising a rabbit variable region and a human constant region are preferred. Other preferred forms of "chimeric antibodies" encompassed by the present invention are those in which the constant region has been modified or changed from that of the original antibody to generate the properties according to the invention, especially in regard to C1q binding and/or Fc receptor (FcR) binding. Such chimeric antibodies are also referred to as "class-switched antibodies". Chimeric antibodies are the product of expressed immunoglobulin genes comprising DNA segments encoding immunoglobulin variable regions and DNA segments encoding immunoglobulin constant regions. Methods for producing chimeric antibodies involve conventional recombinant DNA and gene transfection techniques are well known in the art. See e.g. Morrison, S. L., et al., Proc. Natl. Acad. Sci. USA 81 (1984) 6851-6855; U.S. Pat. Nos. 5,202,238 and 5,204,244.
[0078] The term "cytotoxic agent" as used herein refers to a substance that inhibits or prevents a cellular function and/or causes cell death or destruction. Cytotoxic agents include, but are not limited to, radioactive isotopes (e.g., At.sup.211, I.sup.131, I.sup.125, Y.sup.90, Re.sup.186, Re.sup.188, Sm.sup.153, Bi.sup.212, P.sup.32, Pb.sup.212 and radioactive isotopes of Lu); chemotherapeutic agents or drugs (e.g., methotrexate, adriamicin, vinca alkaloids (vincristine, vinblastine, etoposide), doxorubicin, melphalan, mitomycin C, chlorambucil, daunorubicin or other intercalating agents); growth inhibitory agents; enzymes and fragments thereof such as nucleolytic enzymes; antibiotics; toxins such as small molecule toxins or enzymatically active toxins of bacterial, fungal, plant or animal origin, including fragments and/or variants thereof; and the various antitumor or anticancer agents disclosed below.
[0079] "Effector functions" refer to those biological activities attributable to the Fc region of an antibody, which vary with the antibody isotype. Examples of antibody effector functions include: C1q binding and complement dependent cytotoxicity (CDC); Fc receptor binding; antibody-dependent cell-mediated cytotoxicity (ADCC); antibody-dependent cellular phagocytosis (ADCP), cytokine secretion, immune complex-mediated antigen uptake by antigen presenting cells; down regulation of cell surface receptors (e.g. B cell receptor); and B cell activation.
[0080] As used herein, the terms "engineer, engineered, engineering", are considered to include any manipulation of the peptide backbone or the post-translational modifications of a naturally occurring or recombinant polypeptide or fragment thereof. Engineering includes modifications of the amino acid sequence, of the glycosylation pattern, or of the side chain group of individual amino acids, as well as combinations of these approaches.
[0081] The term "amino acid mutation" as used herein is meant to encompass amino acid substitutions, deletions, insertions, and modifications. Any combination of substitution, deletion, insertion, and modification can be made to arrive at the final construct, provided that the final construct possesses the desired characteristics, e.g., reduced binding to an Fc receptor, or increased association with another peptide. Amino acid sequence deletions and insertions include amino- and/or carboxy-terminal deletions and insertions of amino acids. Particular amino acid mutations are amino acid substitutions. For the purpose of altering e.g. the binding characteristics of an Fc region, non-conservative amino acid substitutions, i.e. replacing one amino acid with another amino acid having different structural and/or chemical properties, are particularly preferred. Amino acid substitutions include replacement by non-naturally occurring amino acids or by naturally occurring amino acid derivatives of the twenty standard amino acids (e.g. 4-hydroxyproline, 3-methylhistidine, ornithine, homoserine, 5-hydroxylysine). Amino acid mutations can be generated using genetic or chemical methods well known in the art. Genetic methods may include site-directed mutagenesis, PCR, gene synthesis and the like. It is contemplated that methods of altering the side chain group of an amino acid by methods other than genetic engineering, such as chemical modification, may also be useful. Various designations may be used herein to indicate the same amino acid mutation. For example, a substitution from proline at position 329 of the Fc domain to glycine can be indicated as 329G, G329, G329, P329G, or Pro329Gly.
[0082] An "effective amount" of an agent, e.g., a pharmaceutical formulation, refers to an amount effective, at dosages and for periods of time necessary, to achieve the desired therapeutic or prophylactic result.
[0083] The term "Fc domain" or "Fc region" herein is used to define a C-terminal region of an immunoglobulin heavy chain that contains at least a portion of the constant region. The term includes native sequence Fc regions and variant Fc regions. Although the boundaries of the Fc region of an IgG heavy chain might vary slightly, the human IgG heavy chain Fc region is usually defined to extend from Cys226, or from Pro230, to the carboxyl-terminus of the heavy chain. However, the C-terminal lysine (Lys447) of the Fc region may or may not be present. Unless otherwise specified herein, numbering of amino acid residues in the Fc region or constant region is according to the EU numbering system, also called the EU index, as described in Kabat et al., Sequences of Proteins of Immunological Interest, 5th Ed. Public Health Service, National Institutes of Health, Bethesda, Md., 1991. A "subunit" of an Fc domain as used herein refers to one of the two polypeptides forming the dimeric Fc domain, i.e. a polypeptide comprising C-terminal constant regions of an immunoglobulin heavy chain, capable of stable self-association. For example, a subunit of an IgG Fc domain comprises an IgG CH2 ("first subunit") and an IgG CH3 ("second subunit") constant domain.
[0084] A "modification promoting the association of the first and the second subunit of the Fc domain" is a manipulation of the peptide backbone or the post-translational modifications of an Fc domain subunit that reduces or prevents the association of a polypeptide comprising the Fc domain subunit with an identical polypeptide to form a homodimer. A modification promoting association as used herein particularly includes separate modifications made to each of the two Fc domain subunits desired to associate (i.e. the first and the second subunit of the Fc domain), wherein the modifications are complementary to each other so as to promote association of the two Fc domain subunits. For example, a modification promoting association may alter the structure or charge of one or both of the Fc domain subunits so as to make their association sterically or electrostatically favorable, respectively. Thus, (hetero)dimerization occurs between a polypeptide comprising the first Fc domain subunit and a polypeptide comprising the second Fc domain subunit, which might be non-identical in the sense that further components fused to each of the subunits (e.g. antigen binding moieties) are not the same. In some embodiments the modification promoting association comprises an amino acid mutation in the Fc domain, specifically an amino acid substitution. In a particular embodiment, the modification promoting association comprises a separate amino acid mutation, specifically an amino acid substitution, in each of the two subunits of the Fc domain.
[0085] "Framework" or "FR" refers to variable domain residues other than hypervariable region (HVR) residues. The FR of a variable domain generally consists of four FR domains: FR1, FR2, FR3, and FR4. Accordingly, the HVR and FR sequences generally appear in the following sequence in VH (or VL): FR1-H1(L1)-FR2-H2(L2)-FR3-H3(L3)-FR4.
[0086] The terms "full length antibody," "intact antibody," and "whole antibody" are used herein interchangeably to refer to an antibody having a structure substantially similar to a native antibody structure or having heavy chains that contain an Fc region as defined herein.
[0087] The terms "host cell," "host cell line," and "host cell culture" are used interchangeably and refer to cells into which exogenous nucleic acid has been introduced, including the progeny of such cells. Host cells include "transformants" and "transformed cells," which include the primary transformed cell and progeny derived therefrom without regard to the number of passages. Progeny may not be completely identical in nucleic acid content to a parent cell, but may contain mutations. Mutant progeny that have the same function or biological activity as screened or selected for in the originally transformed cell are included herein.
[0088] A "human antibody" is one which possesses an amino acid sequence which corresponds to that of an antibody produced by a human or a human cell or derived from a non-human source that utilizes human antibody repertoires or other human antibody-encoding sequences. This definition of a human antibody specifically excludes a humanized antibody comprising non-human antigen-binding residues. As also mentioned for chimeric and humanized antibodies according to the invention the term "human antibody" as used herein also comprises such antibodies which are modified in the constant region to generate the properties according to the invention, especially in regard to C1q binding and/or FcR binding, e.g. by "class switching" i.e. change or mutation of Fc parts (e.g. from IgG1 to IgG4 and/or IgG1/IgG4 mutation.)
[0089] The term "recombinant human antibody", as used herein, is intended to include all human antibodies that are prepared, expressed, created or isolated by recombinant means, such as antibodies isolated from a host cell such as a NS0 or CHO cell or from an animal (e.g. a mouse) that is transgenic for human immunoglobulin genes or antibodies expressed using a recombinant expression vector transfected into a host cell. Such recombinant human antibodies have variable and constant regions in a rearranged form. The recombinant human antibodies according to the invention have been subjected to in vivo somatic hypermutation. Thus, the amino acid sequences of the VH and VL regions of the recombinant antibodies are sequences that, while derived from and related to human germ line VH and VL sequences, may not naturally exist within the human antibody germ line repertoire in vivo.
[0090] A "human consensus framework" is a framework which represents the most commonly occurring amino acid residues in a selection of human immunoglobulin VL or VH framework sequences. Generally, the selection of human immunoglobulin VL or VH sequences is from a subgroup of variable domain sequences. Generally, the subgroup of sequences is a subgroup as in Kabat et al., Sequences of Proteins of Immunological Interest, Fifth Edition, NIH Publication 91-3242, Bethesda Md. (1991), vols. 1-3. In one embodiment, for the VL, the subgroup is subgroup kappa I as in Kabat et al., supra. In one embodiment, for the VH, the subgroup is subgroup III as in Kabat et al., supra.
[0091] A "humanized" antibody refers to a chimeric antibody comprising amino acid residues from non-human HVRs and amino acid residues from human FRs. In certain embodiments, a humanized antibody will comprise substantially all of at least one, and typically two, variable domains, in which all or substantially all of the HVRs (e.g., CDRs) correspond to those of a non-human antibody, and all or substantially all of the FRs correspond to those of a human antibody. A humanized antibody optionally may comprise at least a portion of an antibody constant region derived from a human antibody. A "humanized form" of an antibody, e.g., a non-human antibody, refers to an antibody that has undergone humanization. Other forms of "humanized antibodies" encompassed by the present invention are those in which the constant region has been additionally modified or changed from that of the original antibody to generate the properties according to the invention, especially in regard to C1q binding and/or Fc receptor (FcR) binding.
[0092] The term "hypervariable region" or "HVR," as used herein refers to each of the regions of an antibody variable domain which are hypervariable in sequence and/or form structurally defined loops ("hypervariable loops"). Generally, native four-chain antibodies comprise six HVRs; three in the VH (H1, H2, H3), and three in the VL (L1, L2, L3). HVRs generally comprise amino acid residues from the hypervariable loops and/or from the "complementarity determining regions" (CDRs), the latter being of highest sequence variability and/or involved in antigen recognition. Exemplary hypervariable loops occur at amino acid residues 26-32 (L1), 50-52 (L2), 91-96 (L3), 26-32 (H1), 53-55 (H2), and 96-101 (H3). (Chothia and Lesk, J. Mol. Biol. 196:901-917 (1987).) Exemplary CDRs (CDR-L1, CDR-L2, CDR-L3, CDR-H1, CDR-H2, and CDR-H3) occur at amino acid residues 24-34 of L1, 50-56 of L2, 89-97 of L3, 31-35B of H1, 50-65 of H2, and 95-102 of H3. (Kabat et al., Sequences of Proteins of Immunological Interest, 5th Ed. Public Health Service, National Institutes of Health, Bethesda, Md. (1991).) Hypervariable regions (HVRs) are also referred to as complementarity determining regions (CDRs), and these terms are used herein interchangeably in reference to portions of the variable region that form the antigen binding regions. This particular region has been described by Kabat et al., U.S. Dept. of Health and Human Services, "Sequences of Proteins of Immunological Interest" (1983) and by Chothia et al., J. Mol. Biol. 196:901-917 (1987), where the definitions include overlapping or subsets of amino acid residues when compared against each other. Nevertheless, application of either definition to refer to a CDR of an antibody or variants thereof is intended to be within the scope of the term as defined and used herein. The appropriate amino acid residues which encompass the CDRs as defined by each of the above cited references are set forth below in Table A as a comparison. The exact residue numbers which encompass a particular CDR will vary depending on the sequence and size of the CDR. Those skilled in the art can routinely determine which residues comprise a particular CDR given the variable region amino acid sequence of the antibody.
TABLE-US-00001 TABLE A CDR Definitions.sup.1 CDR Kabat Chothia AbM.sup.2 V.sub.H CDR1 31-35 26-32 26-35 V.sub.H CDR2 50-65 52-58 50-58 V.sub.H CDR3 95-102 95-102 95-102 V.sub.L CDR1 24-34 26-32 24-34 V.sub.L CDR2 50-56 50-52 50-56 V.sub.L CDR3 89-97 91-96 89-97 .sup.1Numbering of all CDR definitions in Table A is according to the numbering conventions set forth by Kabat et al. (see below). .sup.2"AbM" with a lowercase .sup."b" as used in Table A refers to the CDRs as defined by Oxford Molecular's .sup."AbM" antibody modeling software.
[0093] Kabat et al. also defined a numbering system for variable region sequences that is applicable to any antibody. One of ordinary skill in the art can unambiguously assign this system of "Kabat numbering" to any variable region sequence, without reliance on any experimental data beyond the sequence itself. As used herein, "Kabat numbering" refers to the numbering system set forth by Kabat et al., U.S. Dept. of Health and Human Services, "Sequence of Proteins of Immunological Interest" (1983). Unless otherwise specified, references to the numbering of specific amino acid residue positions in an antibody variable region are according to the Kabat numbering system.
[0094] With the exception of CDR1 in VH, CDRs generally comprise the amino acid residues that form the hypervariable loops. CDRs also comprise "specificity determining residues," or "SDRs," which are residues that contact antigen. SDRs are contained within regions of the CDRs called abbreviated-CDRs, or a-CDRs. Exemplary a-CDRs (a-CDR-L1, a-CDR-L2, a-CDR-L3, a-CDR-H1, a-CDR-H2, and a-CDR-H3) occur at amino acid residues 31-34 of L1, 50-55 of L2, 89-96 of L3, 31-35B of H1, 50-58 of H2, and 95-102 of H3. (See Almagro and Fransson, Front. Biosci. 13:1619-1633 (2008).) Unless otherwise indicated, HVR residues and other residues in the variable domain (e.g., FR residues) are numbered herein according to Kabat et al., supra.
[0095] An "immunoconjugate" is an antibody conjugated to one or more heterologous molecule(s), including but not limited to a cytotoxic agent.
[0096] An "individual" or "subject" is a mammal. Mammals include, but are not limited to, domesticated animals (e.g., cows, sheep, cats, dogs, and horses), primates (e.g., humans and non-human primates such as monkeys), rabbits, and rodents (e.g., mice and rats). In certain embodiments, the individual or subject is a human.
[0097] An "isolated" antibody is one which has been separated from a component of its natural environment. In some embodiments, an antibody is purified to greater than 95% or 99% purity as determined by, for example, electrophoretic (e.g., SDS-PAGE, isoelectric focusing (IEF), capillary electrophoresis) or chromatographic (e.g., ion exchange or reverse phase HPLC). For review of methods for assessment of antibody purity, see, e.g., Flatman et al., J. Chromatogr. B 848:79-87 (2007).
[0098] An "isolated" nucleic acid refers to a nucleic acid molecule that has been separated from a component of its natural environment. An isolated nucleic acid includes a nucleic acid molecule contained in cells that ordinarily contain the nucleic acid molecule, but the nucleic acid molecule is present extrachromosomally or at a chromosomal location that is different from its natural chromosomal location.
[0099] "Isolated nucleic acid encoding a trispecific antibody that specifically binds HER2 and a BBB-R" refers to one or more nucleic acid molecules encoding antibody heavy and light chains (or fragments thereof), including such nucleic acid molecule(s) in a single vector or separate vectors, and such nucleic acid molecule(s) present at one or more locations in a host cell.
[0100] The term "monoclonal antibody" as used herein refers to an antibody obtained from a population of substantially homogeneous antibodies, i.e., the individual antibodies comprising the population are identical and/or bind the same epitope, except for possible variant antibodies, e.g., containing naturally occurring mutations or arising during production of a monoclonal antibody preparation, such variants generally being present in minor amounts. In contrast to polyclonal antibody preparations, which typically include different antibodies directed against different determinants (epitopes), each monoclonal antibody of a monoclonal antibody preparation is directed against a single determinant on an antigen. Thus, the modifier "monoclonal" indicates the character of the antibody as being obtained from a substantially homogeneous population of antibodies, and is not to be construed as requiring production of the antibody by any particular method. For example, the monoclonal antibodies to be used in accordance with the present invention may be made by a variety of techniques, including but not limited to the hybridoma method, recombinant DNA methods, phage-display methods, and methods utilizing transgenic animals containing all or part of the human immunoglobulin loci, such methods and other exemplary methods for making monoclonal antibodies being described herein.
[0101] A "naked antibody" refers to an antibody that is not conjugated to a heterologous moiety (e.g., a cytotoxic moiety) or radiolabel. The naked antibody may be present in a pharmaceutical formulation.
[0102] "Native antibodies" refer to naturally occurring immunoglobulin molecules with varying structures. For example, native IgG antibodies are heterotetrameric glycoproteins of about 150,000 daltons, composed of two identical light chains and two identical heavy chains that are disulfide-bonded. From N- to C-terminus, each heavy chain has a variable region (VH), also called a variable heavy domain or a heavy chain variable domain, followed by three constant domains (CH1, CH2, and CH3). Similarly, from N- to C-terminus, each light chain has a variable region (VL), also called a variable light domain or a light chain variable domain, followed by a constant light (CL) domain. The light chain of an antibody may be assigned to one of two types, called kappa (.kappa.) and lambda (.lamda.), based on the amino acid sequence of its constant domain.
[0103] The term "package insert" is used to refer to instructions customarily included in commercial packages of therapeutic products, that contain information about the indications, usage, dosage, administration, combination therapy, contraindications and/or warnings concerning the use of such therapeutic products.
[0104] "No substantial cross-reactivity" means that a molecule (e.g., an antibody) does not recognize or specifically bind an antigen different from the actual target antigen of the molecule (e.g. an antigen closely related to the target antigen), particularly when compared to that target antigen. For example, an antibody may bind less than about 10% to less than about 5% to an antigen different from the actual target antigen, or may bind said antigen different from the actual target antigen at an amount consisting of less than about 10%, 9%, 8% 7%, 6%, 5%, 4%, 3%, 2%, 1%, 0.5%, 0.2%, or 0.1%, preferably less than about 2%, 1%, or 0.5%, and most preferably less than about 0.2% or 0.1% antigen different from the actual target antigen.
[0105] "Percent (%) amino acid sequence identity" with respect to a reference polypeptide sequence is defined as the percentage of amino acid residues in a candidate sequence that are identical with the amino acid residues in the reference polypeptide sequence, after aligning the sequences and introducing gaps, if necessary, to achieve the maximum percent sequence identity, and not considering any conservative substitutions as part of the sequence identity. Alignment for purposes of determining percent amino acid sequence identity can be achieved in various ways that are within the skill in the art, for instance, using publicly available computer software such as BLAST, BLAST-2, ALIGN or Megalign (DNASTAR) software. Those skilled in the art can determine appropriate parameters for aligning sequences, including any algorithms needed to achieve maximal alignment over the full length of the sequences being compared. For purposes herein, however, % amino acid sequence identity values are generated using the sequence comparison computer program ALIGN-2. The ALIGN-2 sequence comparison computer program was authored by Genentech, Inc., and the source code has been filed with user documentation in the U.S. Copyright Office, Washington D.C., 20559, where it is registered under U.S. Copyright Registration No. TXU510087. The ALIGN-2 program is publicly available from Genentech, Inc., South San Francisco, Calif., or may be compiled from the source code. The ALIGN-2 program should be compiled for use on a UNIX operating system, including digital UNIX V4.0D. All sequence comparison parameters are set by the ALIGN-2 program and do not vary.
[0106] In situations where ALIGN-2 is employed for amino acid sequence comparisons, the % amino acid sequence identity of a given amino acid sequence A to, with, or against a given amino acid sequence B (which can alternatively be phrased as a given amino acid sequence A that has or comprises a certain % amino acid sequence identity to, with, or against a given amino acid sequence B) is calculated as follows:
100 times the fraction X/Y
where X is the number of amino acid residues scored as identical matches by the sequence alignment program ALIGN-2 in that program's alignment of A and B, and where Y is the total number of amino acid residues in B. It will be appreciated that where the length of amino acid sequence A is not equal to the length of amino acid sequence B, the % amino acid sequence identity of A to B will not equal the % amino acid sequence identity of B to A. Unless specifically stated otherwise, all % amino acid sequence identity values used herein are obtained as described in the immediately preceding paragraph using the ALIGN-2 computer program.
[0107] The term "pharmaceutical formulation" refers to a preparation which is in such form as to permit the biological activity of an active ingredient contained therein to be effective, and which contains no additional components which are unacceptably toxic to a subject to which the formulation would be administered.
[0108] A "pharmaceutically acceptable carrier" refers to an ingredient in a pharmaceutical formulation, other than an active ingredient, which is nontoxic to a subject. A pharmaceutically acceptable carrier includes, but is not limited to, a buffer, excipient, stabilizer, or preservative.
[0109] As used herein, "treatment" (and grammatical variations thereof such as "treat" or "treating") refers to clinical intervention in an attempt to alter the natural course of the individual being treated, and can be performed either for prophylaxis or during the course of clinical pathology. Desirable effects of treatment include, but are not limited to, preventing occurrence or recurrence of disease, alleviation of symptoms, diminishment of any direct or indirect pathological consequences of the disease, preventing metastasis, decreasing the rate of disease progression, amelioration or palliation of the disease state, and remission or improved prognosis. In some embodiments, antibodies of the invention are used to delay development of a disease or to slow the progression of a disease.
[0110] The terms "cancer" and "cancerous" refer to or describe the physiological condition in mammals that is typically characterized by unregulated cell growth.
[0111] A cancer cell with "HER receptor overexpression or amplification" is one which has significantly higher levels of a HER receptor protein or gene compared to a noncancerous cell of the same tissue type. Such overexpression may be caused by gene amplification or by increased transcription or translation. HER receptor overexpression or amplification may be determined in a diagnostic or prognostic assay by evaluating increased levels of the HER protein present on the surface of a cell {e.g. via an immunohistochemistry assay; IHC). Alternatively, or additionally, one may measure levels of HER-encoding nucleic acid in the cell, e.g. via in situ hybridization (ISH), including fluorescent in situ hybridization (FISH; see WO98/45479 published October, 1998) and chromogenic in situ hybridization (CISH; see, e.g. Tanner et al., Am. J. Pathol. 157(5): 1467-1472 (2000); Bella et al., J. Clin. Oncol. 26: (May 20 suppl; abstr 22147) (2008)), southern blotting, or polymerase chain reaction (PCR) techniques, such as quantitative real time PCR (qRT-PCR). One may also study HER receptor overexpression or amplification by measuring shed antigen {e.g., HER extracellular domain) in a biological fluid such as serum (see, e.g., U.S. Pat. No. 4,933,294 issued Jun. 12, 1990; WO91/05264 published Apr. 18, 1991; U.S. Pat. No. 5,401,638 issued Mar. 28, 1995; and Sias et al. J. Immunol. Methods 132: 73-80 (1990)). Aside from the above assays, various in vivo assays are available to the skilled practitioner. For example, one may expose cells within the body of the patient to an antibody which is optionally labeled with a detectable label, e.g. a radioactive isotope, and binding of the antibody to cells in the patient can be evaluated, e.g. by external scanning for radioactivity or by analyzing a biopsy taken from a patient previously exposed to the antibody.
[0112] A "HER2-positive" cancer comprises cancer cells which have higher than normal levels of HER2. Examples of HER2-positive cancer include HER2-positive breast cancer, HER2-positive gastric cancer and HER2-positive ovarian cancer. Optionally, HER2-positive cancer has an immunohistochemistry (IHC) score of 2+ or 3+ and/or an in situ hybridization (ISH) amplification ratio >2.0.
[0113] "Metastatic" cancer refers to cancer which has spread from one part of the body (e.g. the breast) to another part of the body. "HER2-positive cancer with brain metastases" refers to HER2-positive cancer which has spread from one part of the body (e.g. the breast) to the central nervous system (CNS). The term "variable region" or "variable domain" refers to the domain of an antibody heavy or light chain that is involved in binding the antibody to antigen. The variable domains of the heavy chain and light chain (VH and VL, respectively) of a native antibody generally have similar structures, with each domain comprising four conserved framework regions (FRs) and three hypervariable regions (HVRs). (See, e.g., Kindt et al. Kuby Immunology, 6.sup.th ed., W.H. Freeman and Co., page 91 (2007).) A single VH or VL domain may be sufficient to confer antigen-binding specificity. Furthermore, antibodies that bind a particular antigen may be isolated using a VH or VL domain from an antibody that binds the antigen to screen a library of complementary VL or VH domains, respectively. See, e.g., Portolano et al., J. Immunol. 150:880-887 (1993); Clarkson et al., Nature 352:624-628 (1991).
[0114] The term "antigen-binding site of an antibody" when used herein refer to the amino acid residues of an antibody which are responsible for antigen-binding. The antigen-binding portion of an antibody comprises amino acid residues from the "complementary determining regions" or "CDRs". "Framework" or "FR" regions are those variable domain regions other than the hypervariable region residues as herein defined. Therefore, the light and heavy chain variable domains of an antibody comprise from N- to C-terminus the domains FR1, CDR1, FR2, CDR2, FR3, CDR3, and FR4. Especially, CDR3 of the heavy chain is the region which contributes most to antigen binding and defines the antibody's properties. CDR and FR regions are determined according to the standard definition of Kabat et al., Sequences of Proteins of Immunological Interest, 5th ed., Public Health Service, National Institutes of Health, Bethesda, Md. (1991) and/or those residues from a "hypervariable loop".
[0115] Antibody specificity refers to selective recognition of the antibody for a particular epitope of an antigen. Natural antibodies, for example, are monospecific. The term "monospecific" antibody as used herein denotes an antibody that has one or more binding sites each of which bind to the same epitope of the same antigen.
[0116] "Bispecific antibodies" according to the disclosure are antibodies which have two different antigen-binding specificities. Antibodies of the present invention are specific for two different epitopes of HER2, i.e. the extracellular domains II and IV of HER2. The term "bispecific" antibody as used herein denotes an antibody that has at least two binding sites each of which bind to different epitopes of the same antigen.
[0117] "Trispecific antibodies" according to the disclosure are antibodies which have three different antigen-binding specificities. Antibodies of the present invention are specific for two different epitopes of HER2, i.e. the extracellular domains II and IV of HER2 and a BBB-R. The term "trispecific" antibody as used herein denotes an antibody that has at least three binding sites each of which bind to different epitopes.
[0118] Trispecific antibodies may also be used to localize cytotoxic agents to cells which express HER2 and/or a BBB-R. Trispecific antibodies can be prepared as full length antibodies or antibody fragments.
[0119] The term "valent" as used within the current application denotes the presence of a specified number of binding sites in an antibody molecule. As such, the terms "bivalent", "tetravalent", and "hexavalent" denote the presence of two binding sites, four binding sites, and six binding sites, respectively, in an antibody molecule. The trispecific antibodies according to the invention are at least "trivalent" and may be "multivalent" (e.g. "tetravalent" or "hexavalent").
[0120] Antibodies of the present invention have three binding sites and are trispecific. That is, the antibodies may be trispecific even in cases where there are more than three binding sites (i.e. that the antibody is multivalent). Trispecific antibodies of the invention include, for example, multivalent single chain antibodies, diabodies and triabodies, as well as antibodies having the constant domain structure of full length antibodies to which further antigen-binding sites (e.g., single chain Fv, a VH domain and/or a VL domain, Fab, or (Fab)2) are linked via one or more peptide-linkers. The antibodies can be full length from a single species, or be chimerized or humanized.
[0121] The term "vector," as used herein, refers to a nucleic acid molecule capable of propagating another nucleic acid to which it is linked. The term includes the vector as a self-replicating nucleic acid structure as well as the vector incorporated into the genome of a host cell into which it has been introduced. Certain vectors are capable of directing the expression of nucleic acids to which they are operatively linked. Such vectors are referred to herein as "expression vectors."
[0122] The term "amino acid" as used within this application denotes the group of naturally occurring carboxy .alpha.-amino acids comprising alanine (three letter code: ala, one letter code: A), arginine (arg, R), asparagine (asn, N), aspartic acid (asp, D), cysteine (cys, C), glutamine (gln, Q), glutamic acid (glu, E), glycine (gly, G), histidine (his, H), isoleucine (ile, I), leucine (leu, L), lysine (lys, K), methionine (met, M), phenylalanine (phe, F), proline (pro, P), serine (ser, S), threonine (thr, T), tryptophan (trp, W), tyrosine (tyr, Y), and valine (val, V).
[0123] As used herein, the expressions "cell", "cell line", and "cell culture" are used interchangeably and all such designations include progeny. Thus, the words "transfectants" and "transfected cells" include the primary subject cell and cultures derived there from without regard for the number of transfers. It is also understood that all progeny may not be precisely identical in DNA content, due to deliberate or inadvertent mutations. Variant progeny that have the same function or biological activity as screened for in the originally transformed cell are included.
[0124] "Affinity" refers to the strength of the sum total of noncovalent interactions between a single binding site of a molecule (e.g., an antibody) and its binding partner (e.g., an antigen). Unless indicated otherwise, as used herein, "binding affinity" refers to intrinsic binding affinity which reflects a 1:1 interaction between members of a binding pair (e.g., antibody and antigen). The affinity of a molecule X for its partner Y can generally be represented by the dissociation constant (Kd). Affinity can be measured by common methods known in the art, including those described herein. Specific illustrative and exemplary embodiments for measuring binding affinity are described in the following.
[0125] As used herein, the term "binding" or "specifically binding" refers to the binding of the antibody to an epitope of the antigen in an in-vitro assay, preferably in a surface plasmon resonance assay (SPR, BIAcore, GE-Healthcare Uppsala, Sweden). The affinity of the binding is defined by the terms ka (rate constant for the association of the antibody from the antibody/antigen complex), kD (dissociation constant), and KD (kD/ka). Binding or specifically binding means a binding affinity (KD) of 10-8 mol/1 or less, preferably 10-9 M to 10-13 mol/l.
[0126] Binding of the antibody to HER2 or a BBB-R can be investigated by a BIAcore assay (GE-Healthcare Uppsala, Sweden). The affinity of the binding is defined by the terms ka (rate constant for the association of the antibody from the antibody/antigen complex), kD (dissociation constant), and KD (kD/ka)
[0127] The term "epitope" includes any polypeptide determinant capable of specific binding to an antibody. In certain embodiments, epitope determinant include chemically active surface groupings of molecules such as amino acids, sugar side chains, phosphoryl, or sulfonyl, and, in certain embodiments, may have specific three dimensional structural characteristics, and or specific charge characteristics. An epitope is a region of an antigen that is bound by an antibody.
[0128] As used herein, the terms "engineer, engineered, engineering," particularly with the prefix "glyco-," as well as the term "glycosylation engineering" are considered to include any manipulation of the glycosylation pattern of a naturally occurring or recombinant polypeptide or fragment thereof. Glycosylation engineering includes metabolic engineering of the glycosylation machinery of a cell, including genetic manipulations of the oligosaccharide synthesis pathways to achieve altered glycosylation of glycoproteins expressed in cells. Furthermore, glycosylation engineering includes the effects of mutations and cell environment on glycosylation. In one embodiment, the glycosylation engineering is an alteration in glycosyltransferase activity. In a particular embodiment, the engineering results in altered glucosaminyltransferase activity and/or fucosyltransferase activity.
II. Compositions and Methods
[0129] In one aspect, the invention is based on trispecific antibodies specifically binding to HER2 and a BBB-R. Antibodies of the invention are useful, e.g., for the treatment or diagnosis of cancer, in particular a HER2-positive cancer with brain metastases. In one embodiment the trispecific antibody induces complement-dependent cytotoxicity (CDC) to a higher degree than the combination of Pertuzumab or Trastuzumab. In one such embodiment the complement dependent cytotoxicity of the trispecific antibody is determined by a LDH assay or a complement assay and compared to the complement dependent cytotoxicity of the combination of Pertuzumab and Trastuzumab as determined by the same assay. In one embodiment the complement dependent cytotoxicity is determined in vitro on cancer cells, preferably on breast cancer cells.
A. Exemplary BBB-R Binders Useful in the Trispecific Antibodies Specifically Binding to HER2 and a BBB-R
[0130] In one aspect of the invention, a trispecific antibody specifically binding to HER2 and a BBB-R is provided, wherein the antibody comprises a first monovalent antigen binding site that specifically binds the extracellular domain II of HER2, a second monovalent antigen binding site that specifically binds the extracellular domain IV of HER2 and a third monovalent antigen binding site specifically binding to a BBB-R.
[0131] In one embodiment the BBB-R of the third antigen binding site is selected from the group consisting of transferrin receptor (TfR), insulin receptor, insulin-like growth factor receptor (IGF receptor), low density lipoprotein receptor-related protein 8 (LRP8), low density lipoprotein receptor-related protein 1 (LRP1), and heparin-binding epidermal growth factor-like growth factor (HB-EGF). In one preferred embodiment the third antigen binding site is specific for the transferrin receptor, preferably the human transferrin receptor.
[0132] In one embodiment, the invention provides a trispecific antibody that specifically binds to extracellular domains II and IV of HER2 and to a BBB-R, comprising an antigen binding site that specifically binds to an epitope in the transferrin receptor comprised within the amino acid sequence of SEQ ID NO: 202, 203 or 204.
[0133] In one embodiment, the invention provides a trispecific antibody that specifically binds to extracellular domains II and IV of HER2 and to a BBB-R comprising a heavy chain comprising [0134] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 172; [0135] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 173; [0136] (c) a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 174; and a light chain comprising [0137] (a) a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 175; [0138] (b) a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 176; [0139] (c) a light chain CDR3 comprising the amino acid sequence of SEQ ID NO: 177.
[0140] In one embodiment, the invention provides a trispecific antibody that specifically binds to extracellular domains II and IV of HER2 and to a BBB-R comprising a variable light chain comprising an amino acid sequence of SEQ ID NO: 178, a heavy chain comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 179
[0141] In one embodiment, the invention provides a trispecific antibody that specifically binds to extracellular domains II and IV of HER2 and to a BBB-R comprising a humanized light chain variable domain derived from the variable light chain comprising an amino acid sequence of SEQ ID NO: 178, a humanized heavy chain variable domain derived from the heavy chain comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 179.
[0142] In one embodiment, the invention provides a trispecific antibody that specifically binds to extracellular domains II and IV of HER2 and to a BBB-R comprising a heavy chain comprising [0143] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 180; [0144] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 181; [0145] (c) a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 182; and a light chain comprising [0146] (a) a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 183; [0147] (b) a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 184; [0148] (c) a light chain CDR3 comprising the amino acid sequence of SEQ ID NO: 185.
[0149] In one embodiment, the invention provides a trispecific antibody that specifically binds to extracellular domains II and IV of HER2 and to a BBB-R comprising a variable light chain comprising an amino acid sequence of SEQ ID NO: 166, a heavy chain comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 167
[0150] In one embodiment, the invention provides a trispecific antibody that specifically binds to extracellular domains II and IV of HER2 and to a BBB-R comprising a humanized light chain variable domain derived from the variable light chain comprising an amino acid sequence of SEQ ID NO: 166, a humanized heavy chain variable domain derived from the heavy chain comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 167.
[0151] In one embodiment the monovalent antigen binding site specifically binding to the human Transferrin receptor is further optimized for improved brain exposure. Towards this end, the affinity of the antigen binding site specifically binding to the human Transferrin receptor is reduced by introducing certain mutations into its amino acid sequence.
[0152] The equilibrium dissociation constants (KD) are commonly used to describe molecular interactions. It is used as a measure of two molecules' interaction strength (e.g. affinity) with each other. Thus, the KD value is a measure for the strength of a bimolecular interaction.
[0153] However, the KD value as such does not describe the kinetics of the molecular interaction, i.e. from the KD value it cannot be deduced on the one hand how quickly the two molecules bind to each other (association rate constant or "on rate") and on the other hand how quickly the molecules dissociate (dissociation rate constant or "off-rate"). Characterizing bimolecular interactions only by their KD value neglects the fact that an identical KD value can be made up by extremely different (differing orders of magnitude) on and off-rates as the KD value is the ratio thereof. The on and off-rates are important for characterizing the binding behavior of molecules. The off-rate is especially important because it characterizes the binding duration of e.g. an antibody to its antigen. A long off-rate correlates to a slow dissociation of the formed complex whereas a short off-rate correlates to a quick dissociation.
[0154] In order to have a long-lasting (i.e. less frequent dosing requiring) or a tailor-made (e.g. depending on the surrounding conditions) interaction the off-rates have to be determined experimentally. This is even more important as it is next to impossible to predict the off-rate. Additionally the correlation between off-rate and binding affinity is poor as outlined above. For example, due to the fact that the KD value is the ratio of on and off-rate even weak binders can stay bound long to their target whereas tight binders can dissociate rapidly.
[0155] Herein are disclosed novel humanized and affinity reduced anti-transferrin receptor binders that have an off-rate for binding to the human transferrin receptor that is within a certain range in order to ensure proper BBB shuttling. It has been found that this range is defined at the one end by the off-rate of the murine anti-transferrin receptor antibody 128.1 (WO93/10819) determined by surface plasmon resonance for the cynomolgus transferrin receptor and at the other end by 5% of that off-rate (i.e. a 20-times slower dissociation). In one embodiment the monovalent antigen binding site specifically binding to the human Transferrin receptor (huTfR) and to the cynomolgus transferrin receptor (cyTfR) has an off-rate determined by surface plasmon resonance for the cynomolgus transferrin receptor between 0.1 l/s and 0.005 l/s.
[0156] In one embodiment the off-rate is determined at 500, 250, 125, 62.5, 31.25, 15.625 and 0 nM.
[0157] In one embodiment the off-rate is determined using a surface plasmon resonance chip with a biotin surface and a running buffer of 1.times.PBS supplemented with 250 mM sodium chloride at a flow rate of 10 .mu.L/min. In one embodiment the association is monitored for 180 seconds and the dissociation is monitored for 600 seconds. In one embodiment the off-rate is determined on a BIAcore T200. In one embodiment the off-rate is between 0.08 l/s and 0.008 l/s. In one embodiment of all aspects the off-rate is determined at 25.degree. C.
[0158] In one embodiment, the invention provides a trispecific antibody that specifically binds to extracellular domains II and IV of HER2 and to a BBB-R comprising a humanized, affinity reduced light chain variable domain derived from the variable light chain comprising an amino acid sequence of SEQ ID NO: 178, a humanized, affinity reduced heavy chain variable domain derived from the heavy chain comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 179.
[0159] In one embodiment, the invention provides a trispecific antibody that specifically binds to extracellular domains II and IV of HER2 and to a BBB-R comprising a humanized, affinity reduced light chain variable domain derived from the variable light chain comprising an amino acid sequence of SEQ ID NO: 166, a humanized, affinity reduced heavy chain variable domain derived from the heavy chain comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 167.
[0160] In one embodiment, the invention provides a trispecific antibody that specifically binds to extracellular domains II and IV of HER2 and to a BBB-R comprising
[0161] a heavy chain comprising [0162] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 186; [0163] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 187; [0164] (c) a heavy chain CDR3 comprising the amino acid sequence selected from the group of SEQ ID NO: 188, SEQ ID NO: 206 or SEQ ID NO: 174;
[0165] and a light chain comprising [0166] (a) a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 189; [0167] (b) a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 190; [0168] (c) a light chain CDR3 of SEQ ID NO: 191.
[0169] In one embodiment, the invention provides a trispecific antibody that specifically binds to extracellular domains II and IV of HER2 and to a BBB-R comprising a variable light chain comprising an amino acid sequence of SEQ ID NO: 192, a heavy chain comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 193 or SEQ ID NO: 205.
[0170] In one embodiment, the invention provides a trispecific antibody that specifically binds to extracellular domains II and IV of HER2 and to a BBB-R comprising a variable heavy chain comprising an amino acid sequence selected from the group of SEQ ID NO: 194, SEQ ID NO: 195, SEQ ID NO: 196, SEQ ID NO: 197, SEQ ID NO: 198, SEQ ID NO: 199, or SEQ ID NO: 200, and a variable light chain comprising an amino acid sequence selected from the group of SEQ ID NO: 201, SEQ ID NO: 207, SEQ ID NO: 208, or SEQ ID NO:209.
[0171] In one embodiment, the BBB-R antigen binding site comprises one single chain Fab (scFab) which is connected to a full length IgG antibody comprising the antigen binding sites specific for extracellular domains II and IV of HER2, wherein the scFab is connected either directly or by a linker to the C-terminal end of the Fc part of one of the heavy chains of the IgG antibody. In another embodiment the scFab is connected to the N-terminus of the IgG antibody, e.g. to the N-terminus of the variable light chain or heavy chain. In one embodiment the scFab is connected to the N-terminus of the IgG antibody with a linker.
[0172] In one embodiment, the BBB-R antigen binding site comprises one single chain Fv (scFv) which is connected to a full length IgG antibody comprising the antigen binding sites specific for extracellular domains II and IV of HER2, wherein the scFv is connected either directly or by a linker to the C-terminal end of the Fc part of one of the heavy chains of the IgG antibody. In another embodiment the scFv is connected to the N-terminus of the IgG antibody, e.g. to the N-terminus of the variable light chain or heavy chain. In one embodiment the scFv is connected to the N-terminus of the IgG antibody with a linker.
[0173] In one embodiment, the BBB-R antigen binding site comprises one crossover Fab fragment which is connected to a full length IgG antibody comprising the antigen binding sites specific for extracellular domains II and IV of HER2, wherein the crossover Fab fragment is connected either directly or by a linker to the C-terminal end of the Fc part of one of the heavy chains of the IgG antibody. In another embodiment the crossover Fab fragment is connected to the N-terminus of the IgG antibody, e.g. to the N-terminus of the variable light chain or heavy chain. In one embodiment the crossover Fab fragment is connected to the N-terminus of the IgG antibody with a linker. Crossover Fab fragments are Fab fragments wherein either the variable regions or the constant regions of the heavy and light chain are exchanged. Bispecific antibody formats comprising crossover Fab fragments have been described, for example, in WO2009080252, WO2009080253, WO2009080251, WO2009080254, WO2010/136172, WO2010/145792 and WO2013/026831.
[0174] In one embodiment, the BBB-R antigen binding site comprises one Fab fragment which is connected to a full length IgG antibody comprising the antigen binding sites specific for extracellular domains II and IV of HER2, wherein the Fab fragment is connected either directly or by a linker to the C-terminal end of the Fc part of one of the heavy chains of the IgG antibody. In another embodiment the Fab fragment is connected to the N-terminus of the IgG antibody, e.g. to the N-terminus of the variable light chain or heavy chain. In one embodiment the Fab fragment is connected to the N-terminus of the IgG antibody with a linker.
[0175] The term "linker" denotes a chemical linker or a peptidic linker that covalently connects BBB-R antigen binding site to the full length IgG antibody comprising the antigen binding sites specific for extracellular domains II and IV of HER2.
[0176] Single chain peptidic linkers, comprising of from one to twenty amino acid residues joined by peptide bonds, can be used. In certain embodiments, the amino acids are selected from the twenty naturally-occurring amino acids. In certain other embodiments, one or more of the amino acids are selected from glycine, alanine, proline, asparagine, glutamine and lysine. In other embodiments, the linker is a chemical linker. In certain embodiments, the linker is a single chain peptidic linker with an amino acid sequence with a length of at least 25 amino acid residues, in one preferred embodiment with a length of 32 to 50 amino acid residues. In one embodiment the peptidic linker is a (GxS)n linker with G=glycine, S=serine, (x=3, n=8, 9 or 10) or (x=4 and n=6, 7 or 8), in one embodiment with x=4, n=6 or 7, in one preferred embodiment with x=4, n=7. In one embodiment the linker is (G4S)4. In one embodiment the linker is (G4S)6G2.
[0177] Conjugation may be performed using a variety of chemical linkers. For example, the BBB-R antigen binding site may be connected to the full length IgG antibody comprising the antigen binding sites specific for extracellular domains II and IV of HER2 using a variety of bifunctional protein coupling agents such as N-succinimidyl-3-(2-pyridyldithio) propionate (SPDP), succinimidyl-4-(N-maleimidomethyl) cyclohexane-1-carboxylate (SMCC), iminothiolane (IT), bifunctional derivatives of imidoesters (such as dimethyl adipimidate HCl), active esters (such as disuccinimidyl suberate), aldehydes (such as glutaraldehyde), bis-azido compounds (such as bis (p-azidobenzoyl) hexanediamine), bis-diazonium derivatives (such as bis-(p-diazoniumbenzoyl)-ethylenediamine), diisocyanates (such as toluene 2,6-diisocyanate), and bis-active fluorine compounds (such as 1,5-difluoro-2,4-dinitrobenzene). The linker may be a "cleavable linker" facilitating release of the full length IgG antibody comprising the antigen binding sites specific for extracellular domains II and IV of HER2 upon delivery to the brain. For example, an acid-labile linker, peptidase-sensitive linker, photolabile linker, dimethyl linker or disulfide-containing linker (Chari et al, Cancer Res. 52 (1992) 127-131; U.S. Pat. No. 5,208,020) may be used.
[0178] Covalent conjugation can either be direct or via a linker. In certain embodiments, direct conjugation is by construction of a polypeptide fusion (i.e. by genetic fusion). In certain embodiments, direct conjugation is by formation of a covalent bond between a reactive group on one of the two portions of the BBB-R antigen binding site and a corresponding group or acceptor on the full length IgG antibody comprising the antigen binding sites specific for extracellular domains II and IV of HER2. In certain embodiments, direct conjugation is by modification (i.e. genetic modification) of one of the two molecules to be conjugated to include a reactive group (as non-limiting examples, a sulfhydryl group or a carboxyl group) that forms a covalent attachment to the other molecule to be conjugated under appropriate conditions. Conjugation may also be performed using a variety of linkers. For example, the BBB-R antigen binding site and the full length IgG antibody comprising the antigen binding sites specific for extracellular domains II and IV of HER2 may be conjugated using a variety of bifunctional protein coupling agents such as N-succinimidyl-3-(2-pyridyldithio) propionate (SPDP), succinimidyl-4-(N-maleimidomethyl) cyclohexane-1-carboxylate (SMCC), iminothiolane (IT), bifunctional derivatives of imidoesters (such as dimethyl adipimidate HCl), active esters (such as disuccinimidyl suberate), aldehydes (such as glutaraldehyde), bis-azido compounds (such as bis (p-azidobenzoyl) hexanediamine), bis-diazonium derivatives (such as bis-(p-diazoniumbenzoyl)-ethylenediamine), diisocyanates (such as toluene 2,6-diisocyanate), and bis-active fluorine compounds (such as 1,5-difluoro-2,4-dinitrobenzene). Peptidic linkers, comprised of from one to twenty amino acid residues joined by peptide bonds, may also be used. In certain such embodiments, the amino acid residues are selected from the twenty naturally-occurring amino acids. In certain other such embodiments, one or more of the amino acid residues are selected from glycine, alanine, proline, asparagine, glutamine and lysine. The linker may be a "cleavable linker" facilitating release of the the full length IgG antibody comprising the antigen binding sites specific for extracellular domains II and IV of HER2 upon delivery to the brain. For example, an acid-labile linker, peptidase-sensitive linker, photolabile linker, dimethyl linker or disulfide-containing linker (Chari et al, Cancer Res. 52 (1992) 127-131; U.S. Pat. No. 5,208,020) may be used. The above disclosed antigen binding site specifically binding to a BBB-R as outlined in this section are part of trispecific antibodies specifically binding to HER2 and a BBR-R. The antigen binding sites for HER2 as well as the antibody formats of the HER2 binders are described in sections II B and II C below.
B. Exemplary Trispecific Antibodies Specifically Binding to HER2 and a BBB-R with a Common Light Chain
[0179] In one aspect of the invention, a trispecific antibody specifically binding to HER2 and a BBB-R is provided, wherein the antibody comprises a first monovalent antigen binding site that specifically binds the extracellular domain II of HER2, a second monovalent antigen binding site that specifically binds the extracellular domain IV of HER2 and a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above. The inventors of the present invention surprisingly generated a trispecific antibody wherein both binding moieties (i.e. antigen binding sites) that specifically bind to HER2 share a common light chain that retains the efficacy of the parent monospecific antibodies in terms of inhibition of tumor cell proliferation and has an increased affinity to the extracellular domain II of HER2. The generation of a trispecific molecule with a common light chain that retains the binding properties of both of the parent antibodies is not straight-forward as the common CDRs of the hybrid light chain have to retain the binding specifity for both the extracellular domains II and IV of HER2. The use of this so-called `common light chain` principle, i.e. combining two binders that share one light chain but still have separate specificities, prevents light chain mispairing and in this particular case retains the epitope specificity of the parental antibodies. As a consequence, there are less side products during production, facilitating the homogenous preparation of HER2 trispecific antigen binding molecules at high yields. The heavy chain of Pertuzumab was further optimized, resulting in a more potent molecule in terms of affinity to the extracellular domain II of HER2. In addition, the Trastuzumab heavy chain has been stabilized by introducing certain mutations into the CDRs. The resulting molecule is superior to the parent Pertuzumab and Trastuzumab monospecific antibodies, plus targets the CNS through its BBB-R binding moiety. The trispecific HER2 antibodies in the monovalent common light chain format have an increased affinity to the pertuzumab epitope, and show superior inhibitory effects on cell proliferation as compared to the combination of the parental antibodies.
[0180] In one embodiment a trispecific antibody is provided, comprising a first Fab molecule capable of specific binding to extracellular domain II of HER2 and a second Fab molecule capable of specific binding to extracellular domain IV of HER2, wherein the sequence of the variable light chain of the first Fab molecule is identical to the sequence of the variable light chain of the second Fab molecule (i.e. the first and the second Fab molecule comprise a common light chain); and a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above.
[0181] In one embodiment, the invention provides a trispecific antibody that specifically binds to extracellular domains II and IV of HER2 and to a BBB-R comprising
[0182] a first heavy chain comprising [0183] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 55; [0184] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 77; [0185] (c) a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 56;
[0186] and a second heavy chain comprising [0187] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 20; [0188] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 29; [0189] (c) a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 30;
[0190] and a first and a second light chain comprising [0191] (a) a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 89; [0192] (b) a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 90; [0193] (c) a light chain CDR3 comprising the amino acid sequence of SEQ ID NO: 19.
[0194] In one embodiment, the invention provides a trispecific antibody that specifically binds to extracellular domains II and IV of HER2 and to a BBB-R comprising
[0195] a first heavy chain comprising [0196] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 14; [0197] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 60; [0198] (c) a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 16;
[0199] and a second heavy chain comprising [0200] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 20; [0201] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 29; [0202] (c) a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 30;
[0203] and a first and a second light chain comprising [0204] (a) a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 89; [0205] (b) a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 90; [0206] (c) a light chain CDR3 comprising the amino acid sequence of SEQ ID NO: 19.
[0207] In one embodiment, the invention provides a trispecific antibody that specifically binds to extracellular domains II and IV of HER2 and to a BBB-R comprising
[0208] a first heavy chain comprising [0209] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 58; [0210] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 15; [0211] (c) a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 59;
[0212] and a second heavy chain comprising [0213] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 20; [0214] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 29; [0215] (c) a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 30;
[0216] and a first and a second light chain comprising [0217] (a) a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 89; [0218] (b) a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 90; [0219] (c) a light chain CDR3 comprising the amino acid sequence of SEQ ID NO: 19.
[0220] In one embodiment the second heavy chain of any of the embodiments above bears at least one modification in the amino acid sequence that confers higher chemical stability to the CDRs, resulting in retained binding to HER2 under stress conditions. Modifications useful herein are e.g. D98E, D98N, D98T, G99A or G99S. Surprisingly the inventors found that some modifications of the CDRs did not only improve the stability of the molecule but also improved the binding affinity to HER2.
[0221] Hence in one embodiment, the invention provides a trispecific antibody that specifically binds to extracellular domains II and IV of HER2 and to a BBB-R comprising
[0222] a first heavy chain comprising [0223] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 55; [0224] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 77; [0225] (c) a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 56;
[0226] and a second heavy chain comprising [0227] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 20; [0228] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 29; [0229] (c) a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 79;
[0230] and a first and a second light chain comprising [0231] (a) a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 89; [0232] (b) a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 90; [0233] (c) a light chain CDR3 comprising the amino acid sequence of SEQ ID NO: 19.
[0234] In one embodiment, the invention provides a trispecific antibody that specifically binds to extracellular domains II and IV of HER2 and to a BBB-R comprising
[0235] a first heavy chain comprising [0236] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 14; [0237] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 60; [0238] (c) a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 16;
[0239] and a second heavy chain comprising [0240] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 20; [0241] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 29; [0242] (c) a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 79;
[0243] and a first and a second light chain comprising [0244] (a) a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 89; [0245] (b) a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 90; [0246] (c) a light chain CDR3 comprising the amino acid sequence of SEQ ID NO: 19.
[0247] In one embodiment, the invention provides a trispecific antibody that specifically binds to extracellular domains II and IV of HER2 and to a BBB-R comprising
[0248] a first heavy chain comprising [0249] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 58; [0250] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 15; [0251] (c) a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 59;
[0252] and a second heavy chain comprising [0253] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 20; [0254] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 29; [0255] (c) a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 79;
[0256] and a first and a second light chain comprising [0257] (a) a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 89; [0258] (b) a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 90; [0259] (c) a light chain CDR3 comprising the amino acid sequence of SEQ ID NO: 19.
[0260] In one embodiment, the trispecific antibody comprises two variable light chains comprising an amino acid sequence of SEQ ID NO 54 (i.e. a common light chain), a first heavy chain comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 64, and a second heavy chain comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO 92 and a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above.
[0261] In one embodiment, the trispecific antibody comprises two variable light chains comprising an amino acid sequence of SEQ ID NO 54 (i.e. a common light chain), a first heavy chain comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 70, and a second heavy chain comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO 92 and a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above.
[0262] In one embodiment, the trispecific antibody comprises two variable light chains comprising an amino acid sequence of SEQ ID NO 54 (i.e. a common light chain), a first heavy chain comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 68, and a second heavy chain comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 92 and a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above.
[0263] In one embodiment the second heavy chain of any of the embodiments above bears at least one modification in the amino acid sequence that confers stability to the CDRs and the binding to the target, e.g. D98E, D98N, D98T, G99A or G99S.
[0264] Hence in one embodiment, the trispecific antibody comprises two variable light chains comprising an amino acid sequence of SEQ ID NO: 54 (i.e. a common light chain), a first heavy chain comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 64, and a second heavy chain comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 117 and a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above.
[0265] In one embodiment, the trispecific antibody comprises two variable light chains comprising an amino acid sequence of SEQ ID NO:54 (i.e. a common light chain), a first heavy chain comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 70, and a second heavy chain comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 117 and a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above.
[0266] In one embodiment, the trispecific antibody comprises two variable light chains comprising an amino acid sequence of SEQ ID NO:54 (i.e. a common light chain), a first heavy chain comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 68, and a second heavy chain comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 117 and a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above.
[0267] In one embodiment, the trispecific antibody of the invention comprises a first heavy chain constant region comprising the amino acid sequence of SEQ ID NO: 114.
[0268] In one embodiment, the trispecific antibody of the invention comprises a second heavy chain constant region comprising the amino acid sequence of SEQ ID NO: 115.
[0269] In another embodiment the trispecific antibody of the invention comprises a first light chain constant region comprising the amino acid sequence of SEQ ID NO: 113.
[0270] In another embodiment the trispecific antibody of the invention comprises a second light chain constant region comprising the amino acid sequence of SEQ ID NO: 116.
[0271] In one embodiment a trispecific antibody is provided comprising SEQ ID NOs: 165, 163 and 161.
[0272] In one embodiment a trispecific antibody is provided comprising SEQ ID NOs: 168, 169 and 161.
[0273] In one embodiment the trispecific antibody of any of the above embodiments comprises a Fc domain modification that promotes heterodimerization as outlined in section D below.
C. Exemplary Trispecific Antibodies Specifically Binding to HER2 and a BBB-R Comprising a Crossover Fab Fragment
[0274] In one embodiment of the invention, a trispecific antibody specifically binding to HER2 and a BBB-R is provided, wherein the antibody comprises a first monovalent antigen binding site that specifically binds to the extracellular domain II of HER2, a second monovalent antigen binding site that specifically binds to the extracellular domain IV of HER2 and a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above. The inventors of the present invention generated a second trispecific antibody format wherein one of the binding moieties (i.e. antigen binding sites) for HER2 is a crossover Fab fragment. In one aspect of the invention a trispecific antibody is provided, comprising an IgG molecule, wherein one of the Fab fragments is replaced by a crossover Fab fragment. Crossover Fab fragments are Fab fragments wherein either the variable regions or the constant regions of the heavy and light chain are exchanged. Bispecific antibody formats comprising crossover Fab fragments have been described, for example, in WO2009080252, WO2009080253, WO2009080251, WO2009080254, WO2010/136172, WO2010/145792 and WO2013/026831. The native Trastuzumab sequence has been optimized by introducing modifications into the CDRs of both the variable heavy chain and the variable light chain to improve stability and affinity, the resulting sequences framework-grafted to avoid mispairing of the light chains in the trispecific molecule, resulting in highly potent trispecific antibodies that target HER2 and a BBB-R that can be produced with high yield and only low percentage of side products. In addition it shows superior inhibition of tumor cell proliferation as compared to the combination of the respective parental antibodies.
[0275] In one embodiment, the invention provides a trispecific antibody specifically binding to HER2 and a BBB-R comprising
[0276] a first antigen binding site specific for extracellular domain II of HER2, comprising [0277] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 14; [0278] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 15; [0279] (c) a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 16; [0280] (d) a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 11; [0281] (e) a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 12; [0282] (f) a light chain CDR3 comprising the amino acid sequence of SEQ ID NO: 13;
[0283] And a second antigen binding site specific for extracellular domain IV of HER2 comprising [0284] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 20; [0285] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 108; [0286] (c) a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 79; [0287] (d) a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 107; [0288] (e) a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 18; [0289] (f) a light chain CDR3 comprising the amino acid sequence of SEQ ID NO: 19;
[0290] In one embodiment, the trispecific antibody comprises a first antigen binding site specific for the extracellular domain II of HER2 comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 22 and a variable light chain comprising an amino acid sequence of SEQ ID NO: 24; and a second antigen binding site specific for the extracellular domain IV of HER2, comprising a heavy chain variable region comprising an amino acid sequence of SEQ ID NO: 105 and a light chain variable region comprising an amino acid sequence of SEQ ID NO: 106, and a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above.
[0291] In one embodiment, the invention provides a trispecific antibody that specifically binds to HER2 and a BBB-R comprising
[0292] a first antigen binding site specific for extracellular domain II of HER2 comprising [0293] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 14; [0294] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 15; [0295] (c) a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 16; [0296] (d) a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 11; [0297] (e) a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 12; [0298] (f) a light chain CDR3 comprising the amino acid sequence of SEQ ID NO: 13.
[0299] and a second antigen binding site specific for extracellular domain IV of HER2, comprising [0300] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 20; [0301] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 29; [0302] (c) a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 79; [0303] (d) a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 104; [0304] (e) a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 18; [0305] (f) a light chain CDR3 comprising the amino acid sequence of SEQ ID NO: 19.
[0306] In one embodiment, the trispecific antibody comprises a first antigen binding site specific for the extracellular domain II of HER2 comprising a variable heavy chain comprising an amino acid sequence of SEQ ID NO: 22 and a variable light chain comprising an amino acid sequence of SEQ ID NO: 24; and a second antigen binding site specific for the extracellular domain IV of HER2, comprising a heavy chain variable region comprising an amino acid sequence of SEQ ID NO: 117 and a light chain variable region comprising an amino acid sequence of SEQ ID NO: 118, and a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above.
[0307] In one embodiment, the invention provides a trispecific antibody that specifically binds to HER2 and a BBB-R comprising
[0308] a first antigen binding site specific for extracellular domain II of HER2 comprising [0309] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 14; [0310] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 15; [0311] (c) a heavy chain CDR3 comprising the amino acid sequence of SEQ ID NO: 16; [0312] (d) a light chain CDR1 comprising the amino acid sequence of SEQ ID NO: 11; [0313] (e) a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 12; [0314] (f) a light chain CDR3 comprising the amino acid sequence of SEQ ID NO: 13.
[0315] and a second antigen binding site specific for extracellular domain IV of HER2, comprising [0316] (a) a heavy chain CDR1 comprising the amino acid sequence of SEQ ID NO: 20; [0317] (b) a heavy chain CDR2 comprising the amino acid sequence of SEQ ID NO: 29; [0318] (c) a heavy chain CDR3 comprising the amino acid sequence selected from the group consisting of SEQ ID NO: 79, SEQ ID NO: 78, SEQ ID NO: 80, SEQ ID NO: 87, SEQ ID NO: 88; [0319] (d) a light chain CDR1 comprising the amino acid sequence selected from the group consisting of SEQ ID NO: 104, SEQ ID NO: 103 and SEQ ID NO: 158; [0320] (e) a light chain CDR2 comprising the amino acid sequence of SEQ ID NO: 18; [0321] (f) a light chain CDR3 comprising the amino acid sequence of SEQ ID NO: 19;
[0322] In one embodiment, the trispecific antibody of the invention comprises a first heavy chain constant region comprising the amino acid sequence of SEQ ID NO: 114.
[0323] In one embodiment, the trispecific antibody of the invention comprises a first heavy chain constant region comprising the amino acid sequence of SEQ ID NO: 114, wherein the C-terminal Lysine has been removed.
[0324] In one embodiment, the trispecific antibody of the invention comprises a second heavy chain constant region comprising the amino acid sequence of SEQ ID NO: 115.
[0325] In one embodiment, the trispecific antibody of the invention comprises a second heavy chain constant region comprising the amino acid sequence of SEQ ID NO: 115, wherein the C-terminal Lysine has been removed.
[0326] In another embodiment the trispecific antibody of the invention comprises a first light chain constant region comprising the amino acid sequence of SEQ ID NO: 113.
[0327] In another embodiment the trispecific antibody of the invention comprises a second light chain constant region comprising the amino acid sequence of SEQ ID NO: 116.
[0328] In one embodiment a trispecific antibody is provided comprising SEQ ID NOs: 109, 110, 111 and 112.
[0329] In one embodiment a trispecific antibody that specifically binds to HER2 and a BBB-R according to any of the above embodiments comprises
[0330] an Fc domain,
[0331] one Fab fragment comprising a first antigen binding site specific for the extracellular domain II of HER2,
[0332] and one Fab fragment comprising a second antigen binding site specific for the extracellular domain IV of HER2, [0333] wherein either the variable regions or the constant regions of the heavy and light chain of at least one Fab fragment are exchanged and [0334] a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above.
[0335] The triispecific antibodies described in this section are monovalent for the extracellular domain II of HER2 and monovalent for the extracellular domain IV of HER2. Due to the exchange of either the variable regions or the constant regions, the Fab fragment above is also referred to as "cross-Fab fragment" or "xFab fragment" or "crossover Fab fragment".
[0336] In one embodiment a trispecific antibody that specifically binds to HER2 and a BBB-R according to any of the above embodiments comprises
[0337] an Fc domain,
[0338] one Fab fragment comprising an antigen binding site specific for the extracellular domain II of HER2, wherein either the variable regions or the constant regions of the heavy and light chain are exchanged.
[0339] and one Fab fragment comprising an antigen binding site specific for the extracellular domain IV of HER2, and [0340] a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above.
[0341] In one embodiment a trispecific antibody that specifically binds to HER2 and a BBB-R according to any of the above embodiments comprises
[0342] an Fc domain,
[0343] one Fab fragment comprising an antigen binding site specific for the extracellular domain II of HER2,
[0344] and one Fab fragment comprising an antigen binding site specific for the extracellular domain IV of HER2, wherein either the variable regions or the constant regions of the heavy and light chain are exchanged, and [0345] a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above.
[0346] In one embodiment a trispecific antibody that specifically binds to HER2 and a BBB-R according to any of the above embodiments comprises
[0347] an Fc domain,
[0348] one Fab fragment comprising an antigen binding site specific for the extracellular domain II of HER2,
[0349] and one Fab fragment comprising an antigen binding site specific for the extracellular domain IV of HER2, wherein the variable regions of the heavy and light chain of the Fab fragment are exchanged; [0350] and a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above.
[0351] In one embodiment a trispecific antibody that specifically binds to HER2 and a BBB-R according to any of the above embodiments comprises
[0352] an Fc domain,
[0353] one Fab fragments comprising an antigen binding site specific for the extracellular domain II of HER2,
[0354] and one Fab fragment comprising an antigen binding site specific for the extracellular domain IV of HER2, wherein the constant regions of the heavy and light chain are exchanged, and a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above.
[0355] In one embodiment said trispecific antibody that specifically binds to HER2 and a BBB-R according to any of the above embodiments comprises an Fc domain to which two Fab fragments are fused to the N-terminus, wherein either the variable regions or the constant regions of the heavy and light chain of at least one Fab fragment are exchanged and a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above. In one embodiment the two Fab fragments are fused to the N-terminus of the Fc domain through an immunoglobulin hinge region. In one embodiment, the immunoglobulin hinge region is a human IgG1 hinge region. In one embodiment the Fab fragment comprising an antigen binding site specific for the extracellular domain II of HER2, the Fab fragment comprising an antigen binding site specific for the extracellular domain IV of HER2 and the Fc domain are part of an immunoglobulin molecule. In a particular embodiment the immunoglobulin molecule is an IgG class immunoglobulin. In an even more particular embodiment the immunoglobulin is an IgG1 subclass immunoglobulin. In another embodiment the immunoglobulin is an IgG4 subclass immunoglobulin. In a further particular embodiment the immunoglobulin is a human immunoglobulin. In other embodiments the immunoglobulin is a chimeric immunoglobulin or a humanized immunoglobulin.
[0356] In one embodiment a trispecific antibody that specifically binds to HER2 and a BBB-R according to any of the above embodiments comprises an Immunoglobulin G (IgG) molecule with one binding site specific for the extracellular domain II of HER2 and one binding site specific for the extracellular domain IV of HER2, wherein either the variable regions or the constant regions of the heavy and light chain of one arm (Fab fragment) of the IgG molecule are exchanged, and a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above.
[0357] In one embodiment a trispecific antibody that specifically binds to HER2 and a BBB-R according to any of the above embodiments comprises an Immunoglobulin G (IgG) molecule with one binding site specific for the extracellular domain II of HER2 and one binding site specific for the extracellular domain IV of HER2, wherein the variable regions of the heavy and light chain of one arm (Fab fragment) of the IgG molecule are exchanged, and a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above. In one embodiment the variable regions of the heavy and light chain of the one arm (Fab fragment) of the IgG molecule which comprises the binding site specific for the extracellular domain IV of HER2 are exchanged.
[0358] In one embodiment a trispecific antibody that specifically binds to HER2 and a BBB-R according to any of the above embodiments comprises an Immunoglobulin G (IgG) molecule with one binding site specific for the extracellular domain II of HER2 and one binding site specific for the extracellular domain IV of HER2, wherein the constant regions of the heavy and light chain of one arm (Fab fragment) of the IgG molecule are exchanged, and a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above. In one embodiment the constant regions of the heavy and light chain of the one arm (Fab fragment) of the IgG molecule which comprises the binding site specific for the extracellular domain IV of HER2 are exchanged.
[0359] In one embodiment a trispecific antibody that specifically binds to HER2 and a BBB-R according to any of the above embodiments comprises an Immunoglobulin G (IgG) molecule with one binding site specific for the extracellular domain II of HER2 and one binding site specific for the extracellular domain IV of HER2, wherein the complete VH-CH1 and VL-CL domains of one arm (Fab fragment) of the IgG molecule are exchanged; and a third monovalent antigen binding site specifically binding to a BBB-R as outlined in section II A above.
[0360] This means that at least one of the Fab fragments is fused to the N-terminus of the Fc domain via the light chain (VLCL). In one embodiment the other Fab fragment is fused to the N-terminus of the Fc domain via the heavy chain (VHCH1). In one embodiment both Fab fragments are fused to the N-terminus of the Fc domain through an immunoglobulin hinge region.
[0361] In one embodiment the trispecific antibody of any of the above embodiments comprises a Fc domain modification that promotes heterodimerization as outlined in section D below.
D. Fc Domain Modifications Promoting Heterodimerization
[0362] The trispecific antibodies binding to HER2 and a blood-brain barrier receptor (BBB-R) of the invention comprise different antigen binding moieties, fused to one or the other of the two subunits of the Fc domain, thus the two subunits of the Fc domain are typically comprised in two non-identical polypeptide chains. Recombinant co-expression of these polypeptides and subsequent dimerization leads to several possible combinations of the two polypeptides. To improve the yield and purity of the antibodies of the invention in recombinant production, it will thus be advantageous to introduce in the Fc domain of the trispecific antibodies of the invention a modification promoting the association of the desired polypeptides.
[0363] Accordingly, in particular embodiments the Fc domain of the trispecific antibodies of the invention comprises a modification promoting the association of the first and the second subunit of the Fc domain. The site of most extensive protein-protein interaction between the two subunits of a human IgG Fc domain is in the CH3 domain of the Fc domain. Thus, in one embodiment said modification is in the CH3 domain of the Fc domain.
[0364] In a specific embodiment said modification is a so-called "knob-into-hole" modification, comprising a "knob" modification in one of the two subunits of the Fc domain and a "hole" modification in the other one of the two subunits of the Fc domain.
[0365] The knob-into-hole technology is described e.g. in U.S. Pat. Nos. 5,731,168; 7,695,936; Ridgway et al., Prot Eng 9, 617-621 (1996) and Carter, J Immunol Meth 248, 7-15 (2001). Generally, the method involves introducing a protuberance ("knob") at the interface of a first polypeptide and a corresponding cavity ("hole") in the interface of a second polypeptide, such that the protuberance can be positioned in the cavity so as to promote heterodimer formation and hinder homodimer formation. Protuberances are constructed by replacing small amino acid side chains from the interface of the first polypeptide with larger side chains (e.g. tyrosine or tryptophan). Compensatory cavities of identical or similar size to the protuberances are created in the interface of the second polypeptide by replacing large amino acid side chains with smaller ones (e.g. alanine or threonine).
[0366] Accordingly, in a particular embodiment, in the CH3 domain of the first subunit of the Fc domain of the trispecific antibodies of the invention an amino acid residue is replaced with an amino acid residue having a larger side chain volume, thereby generating a protuberance within the CH3 domain of the first subunit which is positionable in a cavity within the CH3 domain of the second subunit, and in the CH3 domain of the second subunit of the Fc domain an amino acid residue is replaced with an amino acid residue having a smaller side chain volume, thereby generating a cavity within the CH3 domain of the second subunit within which the protuberance within the CH3 domain of the first subunit is positionable.
[0367] The protuberance and cavity can be made by altering the nucleic acid encoding the polypeptides, e.g. by site-specific mutagenesis, or by peptide synthesis.
[0368] In a specific embodiment, in the CH3 domain of the first subunit of the Fc domain the threonine residue at position 366 is replaced with a tryptophan residue (T366W), and in the CH3 domain of the second subunit of the Fc domain the tyrosine residue at position 407 is replaced with a valine residue (Y407V). In one embodiment, in the second subunit of the Fc domain additionally the threonine residue at position 366 is replaced with a serine residue (T366S) and the leucine residue at position 368 is replaced with an alanine residue (L368A).
[0369] In yet a further embodiment, in the first subunit of the Fc domain additionally the serine residue at position 354 is replaced with a cysteine residue (S354C), and in the second subunit of the Fc domain additionally the tyrosine residue at position 349 is replaced by a cysteine residue (Y349C). Introduction of these two cysteine residues results in formation of a disulfide bridge between the two subunits of the Fc domain, further stabilizing the dimer (Carter, J Immunol Methods 248, 7-15 (2001)).
[0370] In an alternative embodiment a modification promoting association of the first and the second subunit of the Fc domain comprises a modification mediating electrostatic steering effects, e.g. as described in PCT publication WO 2009/089004. Generally, this method involves replacement of one or more amino acid residues at the interface of the two Fc domain subunits by charged amino acid residues so that homodimer formation becomes electrostatically unfavorable but heterodimerization electrostatically favorable.
[0371] In one embodiment a trispecific antibodies binding to HER2 and a blood-brain barrier receptor (BBB-R) according to any of the above embodiments comprises an Immunoglobulin G (IgG) molecule comprising a first antigen binding site specific for extracellular domain II of HER2 and a second antigen binding site specific for extracellular domain IV of HER2 and a monovalent third antigen binding site specific for a BBB-R, wherein the Fc part of the first heavy chain comprises a first dimerization module and the Fc part of the second heavy chain comprises a second dimerization module allowing a heterodimerization of the two heavy chains of the IgG molecule.
[0372] In a further preferred embodiment, the first dimerization module comprises knobs and the second dimerization module comprises holes according to the knobs into holes strategy (see Carter P.; Ridgway J. B. B.; Presta L. G.: Immunotechnology, Volume 2, Number 1, February 1996, pp. 73-73(1)).
E. Nucleic Acid Sequences, Vectors and Methods of
[0373] The invention further provides isolated polynucleotides encoding trispecific antibodies binding to HER2 and a blood-brain barrier receptor (BBB-R) as described herein or a fragment thereof. The polynucleotides encoding trispecific antibodies of the invention may be expressed as a single polynucleotide that encodes the entire trispecific antigen binding molecule or as multiple (e.g., two or more) polynucleotides that are co-expressed. Polypeptides encoded by polynucleotides that are co-expressed may associate through, e.g., disulfide bonds or other means to form a functional trispecific antibody. For example, the light chain portion of a Fab fragment may be encoded by a separate polynucleotide from the portion of the trispecific antibody comprising the heavy chain portion of the Fab fragment, an Fc domain subunit and optionally (part of) another Fab fragment. When co-expressed, the heavy chain polypeptides will associate with the light chain polypeptides to form the Fab fragment. In another example, the portion of the trispecific antibody provided therein comprising one of the two Fc domain subunits and optionally (part of) one or more Fab fragments could be encoded by a separate polynucleotide from the portion of the trispecific antibody provided therein comprising the other of the two Fc domain subunits and optionally (part of) a Fab fragment. When co-expressed, the Fc domain subunits will associate to form the Fc domain.
[0374] In one embodiment, the present invention is directed to an isolated polynucleotide encoding a trispecific antibody of the invention or a fragment thereof, wherein the polynucleotide comprises a sequence that encodes a first variable heavy chain sequence as shown in SEQ ID NOs 63, 67 and 69. In one embodiment, the present invention is directed to an isolated polynucleotide encoding a trispecific antibody of the invention or a fragment thereof, wherein the polynucleotide comprises a sequence that encodes a second variable heavy chain sequence as shown in SEQ ID NOs 91 and 133.
[0375] In one embodiment, the present invention is directed to an isolated polynucleotide encoding a trispecific antibody of the invention or a fragment thereof, wherein the polynucleotide comprises a sequence that encodes a variable light chain sequence as shown in SEQ ID NO: 53.
[0376] In another embodiment, the present invention is directed to an isolated polynucleotide encoding a trispecific antibody or fragment thereof, wherein the polynucleotide comprises a sequence that encodes a polypeptide sequence as shown in SEQ ID NOs 83, 85, 91, 93, 95, 97, 99, 101, 63, 67, 69, 53, 21 and 23.
[0377] In another embodiment, the invention is directed to an isolated polynucleotide encoding a trispecific antibody of the invention or a fragment thereof, wherein the polynucleotide comprises a sequence that encodes a first variable heavy chain sequence that is at least about 80%, 85%, 90%, 95%, 96%, 97%, 98%, or 99% identical to an amino acid sequence in SEQ ID NOs 63, 67 and 69.
[0378] In another embodiment, the invention is directed to an isolated polynucleotide encoding a trispecific antibody of the invention or a fragment thereof, wherein the polynucleotide comprises a sequence that encodes a second variable heavy chain sequence that is at least about 80%, 85%, 90%, 95%, 96%, 97%, 98%, or 99% identical to an amino acid sequence in SEQ ID NOs 91 and 133.
[0379] In another embodiment, the invention is directed to an isolated polynucleotide encoding a trispecific antibody of the invention or a fragment thereof, wherein the polynucleotide comprises a sequence that encodes a variable light chain sequence that is at least about 80%, 85%, 90%, 95%, 96%, 97%, 98%, or 99% identical to an amino acid sequence in SEQ ID NO: 53.
[0380] In certain embodiments the polynucleotide or nucleic acid is DNA. In other embodiments, a polynucleotide of the present invention is RNA, for example, in the form of messenger RNA (mRNA). RNA of the present invention may be single stranded or double stranded.
[0381] In further objects the present invention relates to an expression vector comprising a nucleic acid sequence of the present invention and to a prokaryotic or eukaryotic host cell comprising a vector of the present invention. In addition a method of producing an antibody comprising culturing the host cell so that the antibody is produced is provided.
F. Fc Domain Modifications Reducing Fc Receptor Binding and/or Effector Function
[0382] In one aspect a trispecific antibodies binding to HER2 and a blood-brain barrier receptor (BBB-R) according to any of the above embodiments comprises an Immunoglobulin G (IgG) molecule wherein the Fc part is modified. The modified Fc part has a reduced binding affinity for the Fc.gamma. receptors compared to a wildtype Fc part.
[0383] The Fc domain of the trispecific antibodies of the invention consists of a pair of polypeptide chains comprising heavy chain domains of an immunoglobulin molecule. For example, the Fc domain of an immunoglobulin G (IgG) molecule is a dimer, each subunit of which comprises the CH2 and CH3 IgG heavy chain constant domains. The two subunits of the Fc domain are capable of stable association with each other.
[0384] In one embodiment according the invention the Fc domain of the trispecific antibodies of the invention is an IgG Fc domain. In a particular embodiment the Fc domain is an IgG.sub.1 Fc domain. In another embodiment the Fc domain is an IgG4 Fc domain. In a more specific embodiment, the Fc domain is an IgG4 Fc domain comprising an amino acid substitution at position S228 (Kabat numbering), particularly the amino acid substitution S228P. In a more specific embodiment, the Fc domain is an IgG4 Fc domain comprising amino acid substitutions L235E and S228P and P329G. This amino acid substitution reduces in vivo Fab arm exchange of IgG4 antibodies (see Stubenrauch et al., Drug Metabolism and Disposition 38, 84-91 (2010)). In a further particular embodiment the Fc domain is human.
[0385] The Fc domain confers favorable pharmacokinetic properties to the trispecific antibodies of the invention, including a long serum half-life which contributes to good accumulation in the target tissue and a favorable tissue-blood distribution ratio. At the same time it may, however, lead to undesirable targeting of the trispecific antibodies of the invention to cells expressing Fc receptors rather than to the preferred antigen-bearing cells. Accordingly, in particular embodiments the Fc domain of the the trispecific antibodies of the invention exhibits reduced binding affinity to an Fc receptor and/or reduced effector function, as compared to a native IgG.sub.1 Fc domain. In one such embodiment the Fc domain (or the trispecific antibodies of the invention comprising said Fc domain) exhibits less than 50%, preferably less than 20%, more preferably less than 10% and most preferably less than 5% of the binding affinity to an Fc receptor, as compared to a native IgG.sub.1 Fc domain (or a trispecific antibodies of the invention comprising a native IgG.sub.1 Fc domain), and/or less than 50%, preferably less than 20%, more preferably less than 10% and most preferably less than 5% of the effector function, as compared to a native IgG.sub.1 Fc domain domain (or a trispecific antibodies of the invention comprising a native IgG.sub.1 Fc domain). In one embodiment, the Fc domain (or the trispecific antibodies of the invention comprising said Fc domain) does not substantially bind to an Fc receptor and/or induce effector function. In a particular embodiment the Fc receptor is an Fc.gamma. receptor. In one embodiment the Fc receptor is a human Fc receptor. In one embodiment the Fc receptor is an activating Fc receptor. In a specific embodiment the Fc receptor is an activating human Fc.gamma. receptor, more specifically human Fc.gamma.RIIIa, Fc.gamma.RI or Fc.gamma.RIIa, most specifically human Fc.gamma.RIIIa. In one embodiment the Fc receptor is an inhibitory Fc receptor. In a specific embodiment the Fc receptor is an inhibitory human Fc.gamma. receptor, more specifically human FcgRIIB. In one embodiment the effector function is one or more of CDC, ADCC, ADCP, and cytokine secretion. In a particular embodiment the effector function is ADCC. In one embodiment the Fc domain domain exhibits substantially similar binding affinity to neonatal Fc receptor (FcRn), as compared to a native IgG.sub.1 Fc domain domain. Substantially similar binding to FcRn is achieved when the Fc domain (or the trispecific antibodies of the invention comprising said Fc domain) exhibits greater than about 70%, particularly greater than about 80%, more particularly greater than about 90% of the binding affinity of a native IgG.sub.1 Fc domain (or the trispecific antibodies of the invention comprising a native IgG.sub.1 Fc domain) to FcRn.
[0386] In certain embodiments the Fc domain is engineered to have reduced binding affinity to an Fc receptor and/or reduced effector function, as compared to a non-engineered Fc domain. In particular embodiments, the Fc domain of the trispecific antibodies of the invention comprises one or more amino acid mutation that reduces the binding affinity of the Fc domain to an Fc receptor and/or effector function. Typically, the same one or more amino acid mutation is present in each of the two subunits of the Fc domain. In one embodiment the amino acid mutation reduces the binding affinity of the Fc domain to an Fc receptor. In one embodiment the amino acid mutation reduces the binding affinity of the Fc domain to an Fc receptor by at least 2-fold, at least 5-fold, or at least 10-fold. In embodiments where there is more than one amino acid mutation that reduces the binding affinity of the Fc domain to the Fc receptor, the combination of these amino acid mutations may reduce the binding affinity of the Fc domain to an Fc receptor by at least 10-fold, at least 20-fold, or even at least 50-fold. In one embodiment the trispecific antibodies of the invention comprising an engineered Fc domain exhibits less than 20%, particularly less than 10%, more particularly less than 5% of the binding affinity to an Fc receptor as compared to a trispecific antibodies of the invention comprising a non-engineered Fc domain. In a particular embodiment the Fc receptor is an Fc.gamma. receptor. In some embodiments the Fc receptor is a human Fc receptor. In one embodiment the Fc receptor is an inhibitory Fc receptor. In a specific embodiment the Fc receptor is an inhibitory human Fc.gamma. receptor, more specifically human FcgRIIB. In some embodiments the Fc receptor is an activating Fc receptor. In a specific embodiment the Fc receptor is an activating human Fc.gamma. receptor, more specifically human Fc.gamma.RIIIa, Fc.gamma.RI or Fc.gamma.RIIa, most specifically human Fc.gamma.RIIIa. Preferably, binding to each of these receptors is reduced. In some embodiments binding affinity to a complement component, specifically binding affinity to C1q, is also reduced. In one embodiment binding affinity to neonatal Fc receptor (FcRn) is not reduced. Substantially similar binding to FcRn, i.e. preservation of the binding affinity of the Fc domain to said receptor, is achieved when the Fc domain (or the trispecific antibodies of the invention comprising said Fc domain) exhibits greater than about 70% of the binding affinity of a non-engineered form of the Fc domain (or the trispecific antibodies of the invention comprising said non-engineered form of the Fc domain) to FcRn. The Fc domain, or the trispecific antibodies of the invention of the invention comprising said Fc domain, may exhibit greater than about 80% and even greater than about 90% of such affinity. In certain embodiments the Fc domain of the trispecific antibodies of the invention is engineered to have reduced effector function, as compared to a non-engineered Fc domain. The reduced effector function can include, but is not limited to, one or more of the following: reduced complement dependent cytotoxicity (CDC), reduced antibody-dependent cell-mediated cytotoxicity (ADCC), reduced antibody-dependent cellular phagocytosis (ADCP), reduced cytokine secretion, reduced immune complex-mediated antigen uptake by antigen-presenting cells, reduced binding to NK cells, reduced binding to macrophages, reduced binding to monocytes, reduced binding to polymorphonuclear cells, reduced direct signaling inducing apoptosis, reduced dendritic cell maturation, or reduced T cell priming. In one embodiment the reduced effector function is one or more of reduced CDC, reduced ADCC, reduced ADCP, and reduced cytokine secretion. In a particular embodiment the reduced effector function is reduced ADCC. In one embodiment the reduced ADCC is less than 20% of the ADCC induced by a non-engineered Fc domain (or a trispecific antibody of the invention comprising a non-engineered Fc domain).
[0387] In one embodiment the amino acid mutation that reduces the binding affinity of the Fc domain to an Fc receptor and/or effector function is an amino acid substitution. In one embodiment the Fc domain comprises an amino acid substitution at a position of E233, L234, L235, N297, P331 and P329. In a more specific embodiment the Fc domain comprises an amino acid substitution at a position of L234, L235 and P329. In some embodiments the Fc domain comprises the amino acid substitutions L234A and L235A. In one such embodiment, the Fc domain is an IgG.sub.1 Fc domain, particularly a human IgG.sub.1 Fc domain. In one embodiment the Fc domain comprises an amino acid substitution at position P329. In a more specific embodiment the amino acid substitution is P329A or P329G, particularly P329G. In one embodiment the Fc domain comprises an amino acid substitution at position P329 and a further amino acid substitution at a position selected from E233, L234, L235, N297 and P331. In a more specific embodiment the further amino acid substitution is E233P, L234A, L235A, L235E, N297A, N297D or P331S. In particular embodiments the Fc domain comprises amino acid substitutions at positions P329, L234 and L235. In more particular embodiments the Fc domain comprises the amino acid mutations L234A, L235A and P329G ("P329G LALA"). In one such embodiment, the Fc domain is an IgG.sub.1 Fc domain, particularly a human IgG.sub.1 Fc domain. The "P329G LALA" combination of amino acid substitutions almost completely abolishes Fc.gamma. receptor binding of a human IgG.sub.1 Fc domain, as described in PCT patent application no. PCT/EP2012/055393, incorporated herein by reference in its entirety. PCT/EP2012/055393 also describes methods of preparing such mutant Fc domains and methods for determining its properties such as Fc receptor binding or effector functions.
[0388] IgG4 antibodies exhibit reduced binding affinity to Fc receptors and reduced effector functions as compared to IgG.sub.1 antibodies. Hence, in some embodiments the Fc domain of the trispecific antibodies of the invention is an IgG4 Fc domain, particularly a human IgG4 Fc domain. In one embodiment the IgG4 Fc domain comprises amino acid substitutions at position S228, specifically the amino acid substitution S228P. To further reduce its binding affinity to an Fc receptor and/or its effector function, in one embodiment the IgG4 Fc domain comprises an amino acid substitution at position L235, specifically the amino acid substitution L235E. In another embodiment, the IgG4 Fc domain comprises an amino acid substitution at position P329, specifically the amino acid substitution P329G. In a particular embodiment, the IgG4 Fc domain comprises amino acid substitutions at positions S228, L235 and P329, specifically amino acid substitutions S228P, L235E and P329G. Such IgG4 Fc domain mutants and their Fc.gamma. receptor binding properties are described in PCT patent application no. PCT/EP2012/055393, incorporated herein by reference in its entirety.
[0389] In a particular embodiment the Fc domain exhibiting reduced binding affinity to an Fc receptor and/or reduced effector function, as compared to a native IgG.sub.1 Fc domain, is a human IgG.sub.1 Fc domain comprising the amino acid substitutions L234A, L235A and optionally P329G, or a human IgG4 Fc domain comprising the amino acid substitutions S228P, L235E and optionally P329G.
[0390] In certain embodiments N-glycosylation of the Fc domain has been eliminated. In one such embodiment the Fc domain comprises an amino acid mutation at position N297, particularly an amino acid substitution replacing asparagine by alanine (N297A) or aspartic acid (N297D). In addition to the Fc domains described hereinabove and in PCT patent application no. PCT/EP2012/055393, Fc domains with reduced Fc receptor binding and/or effector function also include those with substitution of one or more of Fc domain residues 238, 265, 269, 270, 297, 327 and 329 (U.S. Pat. No. 6,737,056). Such Fc mutants include Fc mutants with substitutions at two or more of amino acid positions 265, 269, 270, 297 and 327, including the so-called "DANA" Fc mutant with substitution of residues 265 and 297 to alanine (U.S. Pat. No. 7,332,581).
[0391] Mutant Fc domains can be prepared by amino acid deletion, substitution, insertion or modification using genetic or chemical methods well known in the art. Genetic methods may include site-specific mutagenesis of the encoding DNA sequence, PCR, gene synthesis, and the like. The correct nucleotide changes can be verified for example by sequencing.
[0392] Binding to Fc receptors can be easily determined e.g. by ELISA, or by Surface Plasmon Resonance (SPR) using standard instrumentation such as a BIAcore instrument (GE Healthcare), and Fc receptors such as may be obtained by recombinant expression. A suitable such binding assay is described herein. Alternatively, binding affinity of Fc domains or cell activating trispecific antigen binding molecules comprising an Fc domain for Fc receptors may be evaluated using cell lines known to express particular Fc receptors, such as human NK cells expressing Fc.gamma.IIIa receptor.
[0393] Effector function of an Fc domain, or trispecific antibodies of the invention comprising an Fc domain, can be measured by methods known in the art. A suitable assay for measuring ADCC is described herein. Other examples of in vitro assays to assess ADCC activity of a molecule of interest are described in U.S. Pat. No. 5,500,362; Hellstrom et al. Proc Natl Acad Sci USA 83, 7059-7063 (1986) and Hellstrom et al., Proc Natl Acad Sci USA 82, 1499-1502 (1985); U.S. Pat. No. 5,821,337; Bruggemann et al., J Exp Med 166, 1351-1361 (1987). Alternatively, non-radioactive assays methods may be employed (see, for example, ACTI.TM. non-radioactive cytotoxicity assay for flow cytometry (CellTechnology, Inc. Mountain View, Calif.); and CytoTox 96.RTM. non-radioactive cytotoxicity assay (Promega, Madison, Wis.)). Useful effector cells for such assays include peripheral blood mononuclear cells (PBMC) and Natural Killer (NK) cells. Alternatively, or additionally, ADCC activity of the molecule of interest may be assessed in vivo, e.g. in a animal model such as that disclosed in Clynes et al., Proc Natl Acad Sci USA 95, 652-656 (1998).
[0394] In some embodiments, binding of the Fc domain to a complement component, specifically to C1q, is reduced. Accordingly, in some embodiments wherein the Fc domain is engineered to have reduced effector function, said reduced effector function includes reduced CDC. C1q binding assays may be carried out to determine whether the trispecific antibodies of the invention is able to bind C1q and hence has CDC activity. See e.g., C1q and C3c binding ELISA in WO 2006/029879 and WO 2005/100402. To assess complement activation, a CDC assay may be performed (see, for example, Gazzano-Santoro et al., J Immunol Methods 202, 163 (1996); Cragg et al., Blood 101, 1045-1052 (2003); and Cragg and Glennie, Blood 103, 2738-2743 (2004)).
[0395] The following section describes preferred embodiments of the trispecific antibodies of the invention comprising Fc domain modifications reducing Fc receptor binding and/or effector function.
G. Antibody Variants
[0396] In certain embodiments, amino acid sequence variants of the trispecific antibodies provided herein are contemplated, in addition to those described above. For example, it may be desirable to improve the binding affinity and/or other biological properties of the trispecific antibody. Amino acid sequence variants of a trispecific antibody may be prepared by introducing appropriate modifications into the nucleotide sequence encoding the trispecific antibody, or by peptide synthesis. Such modifications include, for example, deletions from, and/or insertions into and/or substitutions of residues within the amino acid sequences of the antibody. Any combination of deletion, insertion, and substitution can be made to arrive at the final construct, provided that the final construct possesses the desired characteristics, e.g., antigen-binding.
1. Substitution, Insertion, and Deletion Variants
[0397] In certain embodiments, antibody variants having one or more amino acid substitutions are provided. Sites of interest for substitutional mutagenesis include the HVRs and FRs. Conservative substitutions are shown in Table B under the heading of "conservative substitutions." More substantial changes are provided in Table B under the heading of "exemplary substitutions," and as further described below in reference to amino acid side chain classes. Amino acid substitutions may be introduced into an antibody of interest and the products screened for a desired activity, e.g., retained/improved antigen binding, decreased immunogenicity, or improved ADCC or CDC.
TABLE-US-00002 TABLE B Original Exemplary Preferred Residue Substitutions Substitutions Ala (A) Val; Leu; Ile Val Arg (R) Lys; Gln; Asn Lys Asn (N) Gln; His; Asp, Lys; Arg Gln Asp (D) Glu; Asn Glu Cys (C) Ser; Ala Ser Gln (Q) Asn; Glu Asn Glu (E) Asp; Gln Asp Gly (G) Ala Ala His (H) Asn; Gln; Lys; Arg Arg Ile (I) Leu; Val; Met; Ala; Phe; Norleucine Leu Leu (L) Norleucine; Ile; Val; Met; Ala; Phe Ile Lys (K) Arg; Gln; Asn Arg Met (M) Leu; Phe; Ile Leu Phe (F) Trp; Leu; Val; Ile; Ala; Tyr Tyr Pro (P) Ala Ala Ser (S) Thr Thr Thr (T) Val; Ser Ser Trp (W) Tyr; Phe Tyr Tyr (Y) Trp; Phe; Thr; Ser Phe Val (V) Ile; Leu; Met; Phe; Ala; Norleucine Leu
[0398] Amino acids may be grouped according to common side-chain properties: [0399] (1) hydrophobic: Norleucine, Met, Ala, Val, Leu, Ile; [0400] (2) neutral hydrophilic: Cys, Ser, Thr, Asn, Gln; [0401] (3) acidic: Asp, Glu; [0402] (4) basic: His, Lys, Arg; [0403] (5) residues that influence chain orientation: Gly, Pro; [0404] (6) aromatic: Trp, Tyr, Phe.
[0405] Non-conservative substitutions will entail exchanging a member of one of these classes for another class.
[0406] One type of substitutional variant involves substituting one or more hypervariable region residues of a parent antibody (e.g. a humanized or human antibody). Generally, the resulting variant(s) selected for further study will have modifications (e.g., improvements) in certain biological properties (e.g., increased affinity, reduced immunogenicity) relative to the parent antibody and/or will have substantially retained certain biological properties of the parent antibody. An exemplary substitutional variant is an affinity matured antibody, which may be conveniently generated, e.g., using phage display-based affinity maturation techniques such as those described herein. Briefly, one or more HVR residues are mutated and the variant antibodies displayed on phage and screened for a particular biological activity (e.g. binding affinity).
[0407] Alterations (e.g., substitutions) may be made in HVRs, e.g., to improve antibody affinity. Such alterations may be made in HVR "hotspots," i.e., residues encoded by codons that undergo mutation at high frequency during the somatic maturation process (see, e.g., Chowdhury, Methods Mol. Biol. 207:179-196 (2008)), and/or SDRs (a-CDRs), with the resulting variant VH or VL being tested for binding affinity. Affinity maturation by constructing and reselecting from secondary libraries has been described, e.g., in Hoogenboom et al. in Methods in Molecular Biology 178:1-37 (O'Brien et al., ed., Human Press, Totowa, N.J., (2001).) In some embodiments of affinity maturation, diversity is introduced into the variable genes chosen for maturation by any of a variety of methods (e.g., error-prone PCR, chain shuffling, or oligonucleotide-directed mutagenesis). A secondary library is then created. The library is then screened to identify any antibody variants with the desired affinity. Another method to introduce diversity involves HVR-directed approaches, in which several HVR residues (e.g., 4-6 residues at a time) are randomized. HVR residues involved in antigen binding may be specifically identified, e.g., using alanine scanning mutagenesis or modeling. CDR-H3 and CDR-L3 in particular are often targeted.
[0408] In certain embodiments, substitutions, insertions, or deletions may occur within one or more HVRs so long as such alterations do not substantially reduce the ability of the antibody to bind antigen. For example, conservative alterations (e.g., conservative substitutions as provided herein) that do not substantially reduce binding affinity may be made in HVRs. Such alterations may be outside of HVR "hotspots" or SDRs. In certain embodiments of the variant VH and VL sequences provided above, each HVR either is unaltered, or contains no more than one, two or three amino acid substitutions.
[0409] A useful method for identification of residues or regions of an antibody that may be targeted for mutagenesis is called "alanine scanning mutagenesis" as described by Cunningham and Wells (1989) Science, 244:1081-1085. In this method, a residue or group of target residues (e.g., charged residues such as arg, asp, his, lys, and glu) are identified and replaced by a neutral or negatively charged amino acid (e.g., alanine or polyalanine) to determine whether the interaction of the antibody with antigen is affected. Further substitutions may be introduced at the amino acid locations demonstrating functional sensitivity to the initial substitutions. Alternatively, or additionally, a crystal structure of an antigen-antibody complex to identify contact points between the antibody and antigen. Such contact residues and neighboring residues may be targeted or eliminated as candidates for substitution. Variants may be screened to determine whether they contain the desired properties.
[0410] Amino acid sequence insertions include amino- and/or carboxyl-terminal fusions ranging in length from one residue to polypeptides containing a hundred or more residues, as well as intrasequence insertions of single or multiple amino acid residues. Examples of terminal insertions include an antibody with an N-terminal methionyl residue. Other insertional variants of the antibody molecule include the fusion to the N- or C-terminus of the antibody to an enzyme (e.g. for ADEPT) or a polypeptide which increases the serum half-life of the antibody.
2. Glycosylation Variants
[0411] In certain embodiments, a trispecific antibody provided herein is altered to increase or decrease the extent to which the antibody is glycosylated. Addition or deletion of glycosylation sites to an antibody may be conveniently accomplished by altering the amino acid sequence such that one or more glycosylation sites is created or removed.
[0412] Where the trispecific antibody comprises an Fc region, the carbohydrate attached thereto may be altered. Native antibodies produced by mammalian cells typically comprise a branched, biantennary oligosaccharide that is generally attached by an N-linkage to Asn297 of the CH2 domain of the Fc region. See, e.g., Wright et al. TIBTECH 15:26-32 (1997). The oligosaccharide may include various carbohydrates, e.g., mannose, N-acetyl glucosamine (GlcNAc), galactose, and sialic acid, as well as a fucose attached to a GlcNAc in the "stem" of the biantennary oligosaccharide structure. In some embodiments, modifications of the oligosaccharide in a trispecific antibody of the invention may be made in order to create antibody variants with certain improved properties.
[0413] In one embodiment, trispecific antibody variants are provided having a carbohydrate structure that lacks fucose attached (directly or indirectly) to an Fc region. For example, the amount of fucose in such antibody may be from 1% to 80%, from 1% to 65%, from 5% to 65% or from 20% to 40%. The amount of fucose is determined by calculating the average amount of fucose within the sugar chain at Asn297, relative to the sum of all glycostructures attached to Asn 297 (e. g. complex, hybrid and high mannose structures) as measured by MALDI-TOF mass spectrometry, as described in WO 2008/077546, for example. Asn297 refers to the asparagine residue located at about position 297 in the Fc region (Eu numbering of Fc region residues); however, Asn297 may also be located about .+-.3 amino acids upstream or downstream of position 297, i.e., between positions 294 and 300, due to minor sequence variations in antibodies. Such fucosylation variants may have improved ADCC function. See, e.g., US Patent Publication Nos. US 2003/0157108 (Presta, L.); US 2004/0093621 (Kyowa Hakko Kogyo Co., Ltd). Examples of publications related to "defucosylated" or "fucose-deficient" antibody variants include: US 2003/0157108; WO 2000/61739; WO 2001/29246; US 2003/0115614; US 2002/0164328; US 2004/0093621; US 2004/0132140; US 2004/0110704; US 2004/0110282; US 2004/0109865; WO 2003/085119; WO 2003/084570; WO 2005/035586; WO 2005/035778; WO2005/053742; WO2002/031140; Okazaki et al. J. Mol. Biol. 336:1239-1249 (2004); Yamane-Ohnuki et al. Biotech. Bioeng. 87: 614 (2004). Examples of cell lines capable of producing defucosylated antibodies include Lec13 CHO cells deficient in protein fucosylation (Ripka et al. Arch. Biochem. Biophys. 249:533-545 (1986); US Pat Appl No US 2003/0157108 A1, Presta, L; and WO 2004/056312 A1, Adams et al., especially at Example 11), and knockout cell lines, such as alpha-1,6-fucosyltransferase gene, FUT8, knockout CHO cells (see, e.g., Yamane-Ohnuki et al. Biotech. Bioeng. 87: 614 (2004); Kanda, Y. et al., Biotechnol. Bioeng., 94(4):680-688 (2006); and WO2003/085107).
[0414] Trispecific antibodies variants are further provided with bisected oligosaccharides, e.g., in which a biantennary oligosaccharide attached to the Fc region of the trispecific antibody is bisected by GlcNAc. Such trispecific antibody variants may have reduced fucosylation and/or improved ADCC function. Examples of such antibody variants are described, e.g., in WO 2003/011878 (Jean-Mairet et al.); U.S. Pat. No. 6,602,684 (Umana et al.); and US 2005/0123546 (Umana et al.). Antibody variants with at least one galactose residue in the oligosaccharide attached to the Fc region are also provided. Such antibody variants may have improved CDC function. Such antibody variants are described, e.g., in WO 1997/30087 (Patel et al.); WO 1998/58964 (Raju, S.); and WO 1999/22764 (Raju, S.).
3. Cysteine Engineered Antibody Variants
[0415] In certain embodiments, it may be desirable to create cysteine engineered trispecific antibodies, e.g., "thioMAbs," in which one or more residues of a trispecific antibody are substituted with cysteine residues. In particular embodiments, the substituted residues occur at accessible sites of the trispecific antibody. By substituting those residues with cysteine, reactive thiol groups are thereby positioned at accessible sites of the antibody and may be used to conjugate the antibody to other moieties, such as drug moieties or linker-drug moieties, to create an immunoconjugate. In certain embodiments, any one or more of the following residues may be substituted with cysteine: V205 (Kabat numbering) of the light chain; A118 (EU numbering) of the heavy chain; and S400 (EU numbering) of the heavy chain Fc region. Cysteine engineered antibodies may be generated as described, e.g., in U.S. Pat. No. 7,521,541.
H. Recombinant Methods and Compositions
[0416] Trispecific antibodies of the invention may be obtained, for example, by solid-state peptide synthesis (e.g. Merrifield solid phase synthesis) or recombinant production. For recombinant production one or more polynucleotide encoding the trispecific antibodies (or fragments), e.g., as described above, is isolated and inserted into one or more vectors for further cloning and/or expression in a host cell. Such polynucleotide may be readily isolated and sequenced using conventional procedures. In one embodiment a vector, preferably an expression vector, comprising one or more of the polynucleotides of the invention is provided. Methods which are well known to those skilled in the art can be used to construct expression vectors containing the coding sequence of a trispecific antibody (fragment) along with appropriate transcriptional/translational control signals. These methods include in vitro recombinant DNA techniques, synthetic techniques and in vivo recombination/genetic recombination. See, for example, the techniques described in Maniatis et al., MOLECULAR CLONING: A LABORATORY MANUAL, Cold Spring Harbor Laboratory, N.Y. (1989); and Ausubel et al., CURRENT PROTOCOLS IN MOLECULAR BIOLOGY, Greene Publishing Associates and Wiley Interscience, N.Y (1989). The expression vector can be part of a plasmid, virus, or may be a nucleic acid fragment. The expression vector includes an expression cassette into which the polynucleotide encoding the trispecific antibody (fragment) (i.e. the coding region) is cloned in operable association with a promoter and/or other transcription or translation control elements. As used herein, a "coding region" is a portion of nucleic acid which consists of codons translated into amino acids. Although a "stop codon" (TAG, TGA, or TAA) is not translated into an amino acid, it may be considered to be part of a coding region, if present, but any flanking sequences, for example promoters, ribosome binding sites, transcriptional terminators, introns, 5' and 3' untranslated regions, and the like, are not part of a coding region. Two or more coding regions can be present in a single polynucleotide construct, e.g. on a single vector, or in separate polynucleotide constructs, e.g. on separate (different) vectors. Furthermore, any vector may contain a single coding region, or may comprise two or more coding regions, e.g. a vector of the present invention may encode one or more polypeptides, which are post- or co-translationally separated into the final proteins via proteolytic cleavage. In addition, a vector, polynucleotide, or nucleic acid of the invention may encode heterologous coding regions, either fused or unfused to a polynucleotide encoding the trispecific antibody (fragment) of the invention, or variant or derivative thereof. Heterologous coding regions include without limitation specialized elements or motifs, such as a secretory signal peptide or a heterologous functional domain. An operable association is when a coding region for a gene product, e.g. a polypeptide, is associated with one or more regulatory sequences in such a way as to place expression of the gene product under the influence or control of the regulatory sequence(s). Two DNA fragments (such as a polypeptide coding region and a promoter associated therewith) are "operably associated" if induction of promoter function results in the transcription of mRNA encoding the desired gene product and if the nature of the linkage between the two DNA fragments does not interfere with the ability of the expression regulatory sequences to direct the expression of the gene product or interfere with the ability of the DNA template to be transcribed. Thus, a promoter region would be operably associated with a nucleic acid encoding a polypeptide if the promoter was capable of effecting transcription of that nucleic acid. The promoter may be a cell-specific promoter that directs substantial transcription of the DNA only in predetermined cells. Other transcription control elements, besides a promoter, for example enhancers, operators, repressors, and transcription termination signals, can be operably associated with the polynucleotide to direct cell-specific transcription. Suitable promoters and other transcription control regions are disclosed herein. A variety of transcription control regions are known to those skilled in the art. These include, without limitation, transcription control regions, which function in vertebrate cells, such as, but not limited to, promoter and enhancer segments from cytomegaloviruses (e.g. the immediate early promoter, in conjunction with intron-A), simian virus 40 (e.g. the early promoter), and retroviruses (such as, e.g. Rous sarcoma virus). Other transcription control regions include those derived from vertebrate genes such as actin, heat shock protein, bovine growth hormone and rabbit a-globin, as well as other sequences capable of controlling gene expression in eukaryotic cells. Additional suitable transcription control regions include tissue-specific promoters and enhancers as well as inducible promoters (e.g. promoters inducible tetracyclins). Similarly, a variety of translation control elements are known to those of ordinary skill in the art. These include, but are not limited to ribosome binding sites, translation initiation and termination codons, and elements derived from viral systems (particularly an internal ribosome entry site, or IRES, also referred to as a CITE sequence). The expression cassette may also include other features such as an origin of replication, and/or chromosome integration elements such as retroviral long terminal repeats (LTRs), or adeno-associated viral (AAV) inverted terminal repeats (ITRs).
[0417] Polynucleotide and nucleic acid coding regions of the present invention may be associated with additional coding regions which encode secretory or signal peptides, which direct the secretion of a polypeptide encoded by a polynucleotide of the present invention. For example, if secretion of the trispecific antibody is desired, DNA encoding a signal sequence may be placed upstream of the nucleic acid encoding a trispecific antibody of the invention or a fragment thereof. According to the signal hypothesis, proteins secreted by mammalian cells have a signal peptide or secretory leader sequence which is cleaved from the mature protein once export of the growing protein chain across the rough endoplasmic reticulum has been initiated. Those of ordinary skill in the art are aware that polypeptides secreted by vertebrate cells generally have a signal peptide fused to the N-terminus of the polypeptide, which is cleaved from the translated polypeptide to produce a secreted or "mature" form of the polypeptide. In certain embodiments, the native signal peptide, e.g. an immunoglobulin heavy chain or light chain signal peptide is used, or a functional derivative of that sequence that retains the ability to direct the secretion of the polypeptide that is operably associated with it. Alternatively, a heterologous mammalian signal peptide, or a functional derivative thereof, may be used. For example, the wild-type leader sequence may be substituted with the leader sequence of human tissue plasminogen activator (TPA) or mouse .beta.-glucuronidase.
[0418] DNA encoding a short protein sequence that could be used to facilitate later purification (e.g. a histidine tag) or assist in labeling the trispecific antibody may be included within or at the ends of the trispecific antibody (fragment) encoding polynucleotide.
[0419] In a further embodiment, a host cell comprising one or more polynucleotides of the invention is provided. In certain embodiments a host cell comprising one or more vectors of the invention is provided. The polynucleotides and vectors may incorporate any of the features, singly or in combination, described herein in relation to polynucleotides and vectors, respectively. In one such embodiment a host cell comprises (e.g. has been transformed or transfected with) a vector comprising a polynucleotide that encodes (part of) a trispecific antibody of the invention. As used herein, the term "host cell" refers to any kind of cellular system which can be engineered to generate the trispecific antibodies of the invention or fragments thereof. Host cells suitable for replicating and for supporting expression of trispecific antibodies are well known in the art. Such cells may be transfected or transduced as appropriate with the particular expression vector and large quantities of vector containing cells can be grown for seeding large scale fermenters to obtain sufficient quantities of the trispecific antibody for clinical applications. Suitable host cells include prokaryotic microorganisms, such as E. coli, or various eukaryotic cells, such as Chinese hamster ovary cells (CHO), insect cells, or the like. For example, polypeptides may be produced in bacteria in particular when glycosylation is not needed. After expression, the polypeptide may be isolated from the bacterial cell paste in a soluble fraction and can be further purified. In addition to prokaryotes, eukaryotic microbes such as filamentous fungi or yeast are suitable cloning or expression hosts for polypeptide-encoding vectors, including fungi and yeast strains whose glycosylation pathways have been "humanized", resulting in the production of a polypeptide with a partially or fully human glycosylation pattern. See Gerngross, Nat Biotech 22, 1409-1414 (2004), and Li et al., Nat Biotech 24, 210-215 (2006). Suitable host cells for the expression of (glycosylated) polypeptides are also derived from multicellular organisms (invertebrates and vertebrates). Examples of invertebrate cells include plant and insect cells. Numerous baculoviral strains have been identified which may be used in conjunction with insect cells, particularly for transfection of Spodoptera frugiperda cells. Plant cell cultures can also be utilized as hosts. See e.g. U.S. Pat. Nos. 5,959,177, 6,040,498, 6,420,548, 7,125,978, and 6,417,429 (describing PLANTIBODIES.TM. technology for producing antibodies in transgenic plants). Vertebrate cells may also be used as hosts. For example, mammalian cell lines that are adapted to grow in suspension may be useful. Other examples of useful mammalian host cell lines are monkey kidney CV1 line transformed by SV40 (COS-7); human embryonic kidney line (293 or 293T cells as described, e.g., in Graham et al., J Gen Virol 36, 59 (1977)), baby hamster kidney cells (BHK), mouse sertoli cells (TM4 cells as described, e.g., in Mather, Biol Reprod 23, 243-251 (1980)), monkey kidney cells (CV1), African green monkey kidney cells (VERO-76), human cervical carcinoma cells (HELA), canine kidney cells (MDCK), buffalo rat liver cells (BRL 3A), human lung cells (W138), human liver cells (Hep G2), mouse mammary tumor cells (MMT 060562), TRI cells (as described, e.g., in Mather et al., Annals N.Y. Acad Sci 383, 44-68 (1982)), MRC 5 cells, and FS4 cells. Other useful mammalian host cell lines include Chinese hamster ovary (CHO) cells, including dhfr.sup.- CHO cells (Urlaub et al., Proc Natl Acad Sci USA 77, 4216 (1980)); and myeloma cell lines such as YO, NS0, P3X63 and Sp2/0. For a review of certain mammalian host cell lines suitable for protein production, see, e.g., Yazaki and Wu, Methods in Molecular Biology, Vol. 248 (B.K.C. Lo, ed., Humana Press, Totowa, N.J.), pp. 255-268 (2003). Host cells include cultured cells, e.g., mammalian cultured cells, yeast cells, insect cells, bacterial cells and plant cells, to name only a few, but also cells comprised within a transgenic animal, transgenic plant or cultured plant or animal tissue. In one embodiment, the host cell is a eukaryotic cell, preferably a mammalian cell, such as a Chinese Hamster Ovary (CHO) cell, a human embryonic kidney (HEK) cell or a lymphoid cell (e.g., Y0, NS0, Sp20 cell).
[0420] Standard technologies are known in the art to express foreign genes in these systems. Cells expressing a polypeptide comprising either the heavy or the light chain of an antigen binding domain such as an antibody, may be engineered so as to also express the other of the antibody chains such that the expressed product is an antibody that has both a heavy and a light chain.
[0421] In one embodiment, a method of producing a trispecific antibody according to the invention is provided, wherein the method comprises culturing a host cell comprising a polynucleotide encoding the trispecific antibody, as provided herein, under conditions suitable for expression of the trispecific antibody, and recovering the trispecific antibody from the host cell (or host cell culture medium).
[0422] The components of the trispecific antibody are genetically fused to each other. Trispecific antibodies can be designed such that its components are fused directly to each other or indirectly through a linker sequence. The composition and length of the linker may be determined in accordance with methods well known in the art and may be tested for efficacy. Examples of linker sequences between different components of trispecific antibodies are found in the sequences provided herein. Additional sequences may also be included to incorporate a cleavage site to separate the individual components of the fusion if desired, for example an endopeptidase recognition sequence.
[0423] In certain embodiments the Fab fragments forming part of the trispecific antibody comprise at least an antibody variable region capable of binding an antigenic determinant. Variable regions can form part of and be derived from naturally or non-naturally occurring antibodies and fragments thereof. Methods to produce polyclonal antibodies and monoclonal antibodies are well known in the art (see e.g. Harlow and Lane, "Antibodies, a laboratory manual", Cold Spring Harbor Laboratory, 1988). Non-naturally occurring antibodies can be constructed using solid phase-peptide synthesis, can be produced recombinantly (e.g. as described in U.S. Pat. No. 4,186,567) or can be obtained, for example, by screening combinatorial libraries comprising variable heavy chains and variable light chains (see e.g. U.S. Pat. No. 5,969,108 to McCafferty).
[0424] Any animal species of antibody, antibody fragment, antigen binding domain or variable region can be used in the trispecific antibodies of the invention. Non-limiting antibodies, antibody fragments, antigen binding domains or variable regions useful in the present invention can be of murine, primate, or human origin. If the trispecific antibody is intended for human use, a chimeric form of antibody may be used wherein the constant regions of the antibody are from a human. A humanized or fully human form of the antibody can also be prepared in accordance with methods well known in the art (see e. g. U.S. Pat. No. 5,565,332 to Winter). Humanization may be achieved by various methods including, but not limited to (a) grafting the non-human (e.g., donor antibody) CDRs onto human (e.g. recipient antibody) framework and constant regions with or without retention of critical framework residues (e.g. those that are important for retaining good antigen binding affinity or antibody functions), (b) grafting only the non-human specificity-determining regions (SDRs or a-CDRs; the residues critical for the antibody-antigen interaction) onto human framework and constant regions, or (c) transplanting the entire non-human variable domains, but "cloaking" them with a human-like section by replacement of surface residues. Humanized antibodies and methods of making them are reviewed, e.g., in Almagro and Fransson, Front Biosci 13, 1619-1633 (2008), and are further described, e.g., in Riechmann et al., Nature 332, 323-329 (1988); Queen et al., Proc Natl Acad Sci USA 86, 10029-10033 (1989); U.S. Pat. Nos. 5,821,337, 7,527,791, 6,982,321, and 7,087,409; Jones et al., Nature 321, 522-525 (1986); Morrison et al., Proc Natl Acad Sci 81, 6851-6855 (1984); Morrison and Oi, Adv Immunol 44, 65-92 (1988); Verhoeyen et al., Science 239, 1534-1536 (1988); Padlan, Molec Immun 31(3), 169-217 (1994); Kashmiri et al., Methods 36, 25-34 (2005) (describing SDR (a-CDR) grafting); Padlan, Mol Immunol 28, 489-498 (1991) (describing "resurfacing"); Dall'Acqua et al., Methods 36, 43-60 (2005) (describing "FR shuffling"); and Osbourn et al., Methods 36, 61-68 (2005) and Klimka et al., Br J Cancer 83, 252-260 (2000) (describing the "guided selection" approach to FR shuffling). Human antibodies and human variable regions can be produced using various techniques known in the art. Human antibodies are described generally in van Dijk and van de Winkel, Curr Opin Pharmacol 5, 368-74 (2001) and Lonberg, Curr Opin Immunol 20, 450-459 (2008). Human variable regions can form part of and be derived from human monoclonal antibodies made by the hybridoma method (see e.g. Monoclonal Antibody Production Techniques and Applications, pp. 51-63 (Marcel Dekker, Inc., New York, 1987)). Human antibodies and human variable regions may also be prepared by administering an immunogen to a transgenic animal that has been modified to produce intact human antibodies or intact antibodies with human variable regions in response to antigenic challenge (see e.g. Lonberg, Nat Biotech 23, 1117-1125 (2005). Human antibodies and human variable regions may also be generated by isolating Fv clone variable region sequences selected from human-derived phage display libraries (see e.g., Hoogenboom et al. in Methods in Molecular Biology 178, 1-37 (O'Brien et al., ed., Human Press, Totowa, N.J., 2001); and McCafferty et al., Nature 348, 552-554; Clackson et al., Nature 352, 624-628 (1991)). Phage typically display antibody fragments, either as single-chain Fv (scFv) fragments or as Fab fragments.
[0425] In certain embodiments, the Fab fragments useful in the present invention are engineered to have enhanced binding affinity according to, for example, the methods disclosed in U.S. Pat. Appl. Publ. No. 2004/0132066, the entire contents of which are hereby incorporated by reference. The ability of the trispecific antibody of the invention to bind to a specific antigenic determinant can be measured either through an enzyme-linked immunosorbent assay (ELISA) or other techniques familiar to one of skill in the art, e.g. surface plasmon resonance technique (analyzed on a BIACORE T100 system) (Liljeblad, et al., Glyco J 17, 323-329 (2000)), and traditional binding assays (Heeley, Endocr Res 28, 217-229 (2002)). Competition assays may be used to identify an antibody, antibody fragment, antigen binding domain or variable domain that competes with a reference antibody for binding to a particular antigen. In certain embodiments, such a competing antibody binds to the same epitope (e.g. a linear or a conformational epitope) that is bound by the reference antibody. Detailed exemplary methods for mapping an epitope to which an antibody binds are provided in Morris (1996) "Epitope Mapping Protocols," in Methods in Molecular Biology vol. 66 (Humana Press, Totowa, N.J.). In an exemplary competition assay, immobilized antigen is incubated in a solution comprising a first labeled antibody that binds to the antigen and a second unlabeled antibody that is being tested for its ability to compete with the first antibody for binding to the antigen. The second antibody may be present in a hybridoma supernatant. As a control, immobilized antigen is incubated in a solution comprising the first labeled antibody but not the second unlabeled antibody. After incubation under conditions permissive for binding of the first antibody to the antigen, excess unbound antibody is removed, and the amount of label associated with immobilized antigen is measured.
[0426] If the amount of label associated with immobilized antigen is substantially reduced in the test sample relative to the control sample, then that indicates that the second antibody is competing with the first antibody for binding to the antigen. See Harlow and Lane (1988) Antibodies: A Laboratory Manual ch. 14 (Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.). Trispecific antibodies prepared as described herein may be purified by art-known techniques such as high performance liquid chromatography, ion exchange chromatography, gel electrophoresis, affinity chromatography, size exclusion chromatography, and the like. The actual conditions used to purify a particular protein will depend, in part, on factors such as net charge, hydrophobicity, hydrophilicity etc., and will be apparent to those having skill in the art. For affinity chromatography purification an antibody, ligand, receptor or antigen can be used to which the trispecific antibody binds. For example, for affinity chromatography purification of trispecific antibodies of the invention, a matrix with protein A or protein G may be used. Sequential Protein A or G affinity chromatography and size exclusion chromatography can be used to isolate a trispecific antibody essentially as described in the Examples. The purity of the trispecific antibody can be determined by any of a variety of well known analytical methods including gel electrophoresis, high pressure liquid chromatography, and the like.
I. Assays
[0427] Trispecific antibodies binding to HER2 and a blood-brain barrier receptor (BBB-R) provided herein may be identified, screened for, or characterized for their physical/chemical properties and/or biological activities by various assays known in the art.
1. Affinity Assays
[0428] The affinity of the trispecific antibodies binding to HER2 and a blood-brain barrier receptor (BBB-R) can be determined in accordance with the methods set forth in the Examples by surface plasmon resonance (SPR), using standard instrumentation such as a BIAcore instrument (GE Healthcare), and receptors or target proteins such as may be obtained by recombinant expression. Alternatively, binding of trispecific antibody provided therein to HER2 and/or a BBB-R may be evaluated using cell lines expressing the particular receptor or target antigen, for example by flow cytometry (FACS). A specific illustrative and exemplary embodiment for measuring binding affinity is described in the following and in the Examples below.
[0429] According to one embodiment, K.sub.D is measured by surface plasmon resonance using a BIACORE.RTM. T100 machine (GE Healthcare) at 25.degree. C.
[0430] To analyze the interaction between the Fc-portion and Fc receptors, His-tagged recombinant Fc-receptor is captured by an anti-Penta His antibody (Qiagen) immobilized on CMS chips and the trispecific constructs are used as analytes. Briefly, carboxymethylated dextran biosensor chips (CMS, GE Healthcare) are activated with N-ethyl-N'-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) according to the supplier's instructions. Anti Penta-His antibody is diluted with 10 mM sodium acetate, pH 5.0, to 40 .mu.g/ml before injection at a flow rate of 5 .mu.l/min to achieve approximately 6500 response units (RU) of coupled protein. Following the injection of the ligand, 1 M ethanolamine is injected to block unreacted groups. Subsequently the Fc-receptor is captured for 60 s at 4 or 10 nM. For kinetic measurements, four-fold serial dilutions of the trispecific construct (range between 500 nM and 4000 nM) are injected in HBS-EP (GE Healthcare, 10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.05% Surfactant P20, pH 7.4) at 25.degree. C. at a flow rate of 30 .mu.l/min for 120 s.
[0431] To determine the affinity to the target antigen, trispecific constructs are captured by an anti human Fab specific antibody (GE Healthcare) that is immobilized on an activated CM5-sensor chip surface as described for the anti Penta-His antibody. The final amount of coupled protein is approximately 12000 RU. The trispecific constructs are captured for 90 s at 300 nM. The target antigens are passed through the flow cells for 180 s at a concentration range from 250 to 1000 nM with a flowrate of 30 .mu.l/min. The dissociation is monitored for 180 s.
[0432] Bulk refractive index differences are corrected for by subtracting the response obtained on reference flow cell. The steady state response was used to derive the dissociation constant K.sub.D by non-linear curve fitting of the Langmuir binding isotherm. Association rates (k.sub.on) and dissociation rates (k.sub.off) are calculated using a simple one-to-one Langmuir binding model (BIACORE.RTM. T100 Evaluation Software version 1.1.1) by simultaneously fitting the association and dissociation sensorgrams. The equilibrium dissociation constant (K.sub.D) is calculated as the ratio k.sub.off/k.sub.on. See, e.g., Chen et al., J Mol Biol 293, 865-881 (1999).
2. Binding Assays and Other Assays
[0433] In one aspect, a trispecific antibody of the invention is tested for its antigen binding activity, e.g., by known methods such as ELISA, Western blot, etc.
[0434] In another aspect, competition assays may be used to identify an antibody that competes with a specific anti-HER2 for binding to HER2. In certain embodiments, such a competing antibody binds to the same epitope (e.g., a linear or a conformational epitope) that is bound by a specific anti-HER2 antibody. In another aspect, competition assays may be used to identify an antibody that competes with a specific anti-BBB-R for binding to BBB-R. In certain embodiments, such a competing antibody binds to the same epitope (e.g., a linear or a conformational epitope) that is bound by a specific anti-BBB-R antibody. Detailed exemplary methods for mapping an epitope to which an antibody binds are provided in Morris (1996) "Epitope Mapping Protocols," in Methods in Molecular Biology vol. 66 (Humana Press, Totowa, N.J.). Further methods are described in the example section.
3. Activity Assays
[0435] In one aspect, assays are provided for identifying trispecific antibodies specific for HER2 and a BBB-R thereof having biological activity. Biological activity may include, e.g., DNA fragmentation, induction of apoptosis and lysis of targeted cells. Antibodies having such biological activity in vivo and/or in vitro are also provided. In one embodiment said activity is induction of complement-dependent cytotoxicity (CDC). In one embodiment said trispecific antibodies induce CDC to a higher degree than pertuzumab or trastuzumab alone. In one embodiment said trispecific antibodies induce CDC to at least an about 10 times higher degree, an about 20 times higher degree or an about 30 times higher degree than pertuzumab or trastuzumab alone. In another embodiment said trispecific antibodies induce CDC to a higher degree than the combination of pertuzumab or trastuzumab. In another embodiment said trispecific antibodies induce CDC to about a 30%, 40%, 50% or 60%, or at least a 30% to 70%, or at least a 40% to 60% higher degree than the combination of pertuzumab or trastuzumab. The complement-dependent cytotoxicity (CDC) assay can be performed with non heat-treated serum or commercially available complement fractions (see e.g. Lazar, G. A. et al. Engineered antibody Fc variants with enhanced effector function. Proc. Natl Acad. Sci. USA 103, 4005-4010 (2006)). Target cell killing can be assessed by several cell viability reagents such as Alamar Blue (Lazar, G. A. et al. Engineered antibody Fc variants with enhanced effector function. Proc. Natl Acad. Sci. USA 103, 4005-4010 (2006), Idusogie, E. E. et al. Engineered antibodies with increased activity to recruit complement. J. Immunol. 166, 2571-2575 (2001)), CellTiter-Glo (see e.g. Zhao, X. et al. Targeting C-type lectin-like molecule-1 for antibody-mediated immunotherapy in acute myeloid leukemia. Haematologica 95, 71-78 (2009)), LDH release (see e.g. Konishi, E., Kitai, Y. & Kondo, T. Utilization of complement-dependent cytotoxicity to measure low levels of antibodies: application to nonstructural protein 1 in a model of Japanese encephalitis virus. Clin. Vaccine Immunol. 15, 88-94 (2008) and the examples disclosed herein) or calcein-AM release. In some embodiments said degree of induction of CDC is determined by a LDH release assay or a complement assay measuring binding of complement protein C1q to the antibodies of the invention bound to a cellular antigen. The CDC induction of the trispecific antibody is then compared to the CDC induction of either Pertuzumab or Trastuzumab alone, or the combination of Pertuzumab and Trastuzumab, with all values for CDC induction being assayed in the same assay, with the same cell line and the same respective antibody concentration. If performed in a microtiterplate, the capability of the trispecific antibodies and the controls (either Pertuzumab or Trastuzumab alone, or the combination of Pertuzumab and Trastuzumab) to induce CDC are preferably measured in the same microtiterplate using the same assay. Exemplary assays are disclosed, e.g. in example 18 or 19. In one embodiment said induction of CDC is determined on cancer cells, e.g. breast cancer cells.
[0436] In certain embodiments, a trispecific antibody of the invention is tested for such biological activity. Assays for detecting cell lysis (e.g. by measurement of LDH release) or apoptosis (e.g. using the TUNEL assay) are well known in the art. Assays for measuring ADCC or CDC are also described in WO 2004/065540 (see Example 1 therein), the entire content of which is incorporated herein by reference.
J. Pharmaceutical Formulations
[0437] Pharmaceutical formulations of a trispecific antibody specific for HER2 and a BBB-R as described herein are prepared by mixing such trispecific antibody having the desired degree of purity with one or more optional pharmaceutically acceptable carriers (Remington's Pharmaceutical Sciences 16th edition, Osol, A. Ed. (1980)), in the form of lyophilized formulations or aqueous solutions. Pharmaceutically acceptable carriers are generally nontoxic to recipients at the dosages and concentrations employed, and include, but are not limited to: buffers such as phosphate, citrate, and other organic acids; antioxidants including ascorbic acid and methionine; preservatives (such as octadecyldimethylbenzyl ammonium chloride; hexamethonium chloride; benzalkonium chloride; benzethonium chloride; phenol, butyl or benzyl alcohol; alkyl parabens such as methyl or propyl paraben; catechol; resorcinol; cyclohexanol; 3-pentanol; and m-cresol); low molecular weight (less than about 10 residues) polypeptides; proteins, such as serum albumin, gelatin, or immunoglobulins; hydrophilic polymers such as polyvinylpyrrolidone; amino acids such as glycine, glutamine, asparagine, histidine, arginine, or lysine; monosaccharides, disaccharides, and other carbohydrates including glucose, mannose, or dextrins; chelating agents such as EDTA; sugars such as sucrose, mannitol, trehalose or sorbitol; salt-forming counter-ions such as sodium; metal complexes (e.g. Zn-protein complexes); and/or non-ionic surfactants such as polyethylene glycol (PEG). Exemplary pharmaceutically acceptable carriers herein further include insterstitial drug dispersion agents such as soluble neutral-active hyaluronidase glycoproteins (sHASEGP), for example, human soluble PH-20 hyaluronidase glycoproteins, such as rHuPH20 (HYLENEX.RTM., Baxter International, Inc.). Certain exemplary sHASEGPs and methods of use, including rHuPH20, are described in US Patent Publication Nos. 2005/0260186 and 2006/0104968. In one aspect, a sHASEGP is combined with one or more additional glycosaminoglycanases such as chondroitinases.
[0438] Exemplary lyophilized antibody formulations are described in U.S. Pat. No. 6,267,958. Aqueous antibody formulations include those described in U.S. Pat. No. 6,171,586 and WO2006/044908, the latter formulations including a histidine-acetate buffer.
[0439] The formulation herein may also contain more than one active ingredients as necessary for the particular indication being treated, preferably those with complementary activities that do not adversely affect each other. Such active ingredients are suitably present in combination in amounts that are effective for the purpose intended.
[0440] Active ingredients may be entrapped in microcapsules prepared, for example, by coacervation techniques or by interfacial polymerization, for example, hydroxymethylcellulose or gelatin-microcapsules and poly-(methylmethacylate) microcapsules, respectively, in colloidal drug delivery systems (for example, liposomes, albumin microspheres, microemulsions, nano-particles and nanocapsules) or in macroemulsions. Such techniques are disclosed in Remington's Pharmaceutical Sciences 16th edition, Osol, A. Ed. (1980).
[0441] Sustained-release preparations may be prepared. Suitable examples of sustained-release preparations include semipermeable matrices of solid hydrophobic polymers containing the antibody, which matrices are in the form of shaped articles, e.g. films, or microcapsules.
[0442] The formulations to be used for in vivo administration are generally sterile. Sterility may be readily accomplished, e.g., by filtration through sterile filtration membranes.
K. Therapeutic Methods and Compositions
[0443] Any of the trispecific antibodies specific for HER2 and a BBB-R provided herein may be used in therapeutic methods.
[0444] In one aspect, a trispecific antibody specific for HER2 and a BBB-R for use as a medicament is provided. In further aspects, a trispecific antibody specific for HER2 and a BBB-R for use in treating cancer is provided. In certain embodiments, a trispecific antibody specific for HER2 and a BBB-R for use in a method of treatment is provided. In certain embodiments, the invention provides a trispecific antibody specific for HER2 and a BBB-R for use in a method of treating an individual having cancer comprising administering to the individual an effective amount of the trispecific antibody specific for HER2 and a BBB-R. In one such embodiment, the method further comprises administering to the individual an effective amount of at least one additional therapeutic agent, e.g., as described below. An "individual" according to any of the above embodiments is preferably a human.
[0445] In a further aspect, the invention provides for the use of a trispecific antibody specific for HER2 and a BBB-R in the manufacture or preparation of a medicament. In one embodiment, the medicament is for treatment of cancer. In one embodiment said cancer is HER2-positive cancer with brain metastases. In a further embodiment, the medicament is for use in a method of treating cancer comprising administering to an individual having cancer an effective amount of the medicament. In one such embodiment, the method further comprises administering to the individual an effective amount of at least one additional therapeutic agent, e.g., as described below. An "individual" according to any of the above embodiments may be a human.
[0446] In a further aspect, the invention provides a method for treating cancer. In one embodiment, the method comprises administering to an individual having cancer an effective amount of a trispecific antibody specific for HER2 and a BBB-R. In one such embodiment, the method further comprises administering to the individual an effective amount of at least one additional therapeutic agent, as described below. An "individual" according to any of the above embodiments may be a human.
[0447] In a further aspect, the invention provides pharmaceutical formulations comprising any of the trispecific antibodies specific for HER2 and a BBB-R provided herein, e.g., for use in any of the above therapeutic methods. In one embodiment, a pharmaceutical formulation comprises any of the trispecific antibodies specific for HER2 and a BBB-R provided herein and a pharmaceutically acceptable carrier. In another embodiment, a pharmaceutical formulation comprises any of the trispecific antibodies specific for HER2 and a BBB-R provided herein and at least one additional therapeutic agent, e.g., as described below.
[0448] A trispecific antibody of the invention can be administered by any suitable means, including parenteral, intrapulmonary, and intranasal, and, if desired for local treatment, intralesional administration. Parenteral infusions include intramuscular, intravenous, intraarterial, intraperitoneal, or subcutaneous administration. Dosing can be by any suitable route, e.g. by injections, such as intravenous or subcutaneous injections, depending in part on whether the administration is brief or chronic. Various dosing schedules including but not limited to single or multiple administrations over various time-points, bolus administration, and pulse infusion are contemplated herein.
[0449] Trispecific antibodies of the invention would be formulated, dosed, and administered in a fashion consistent with good medical practice. Factors for consideration in this context include the particular disorder being treated, the particular mammal being treated, the clinical condition of the individual patient, the cause of the disorder, the site of delivery of the agent, the method of administration, the scheduling of administration, and other factors known to medical practitioners. The trispecific antibody need not be, but is optionally formulated with one or more agents currently used to prevent or treat the disorder in question. The effective amount of such other agents depends on the amount of antibody present in the formulation, the type of disorder or treatment, and other factors discussed above. These are generally used in the same dosages and with administration routes as described herein, or about from 1 to 99% of the dosages described herein, or in any dosage and by any route that is empirically/clinically determined to be appropriate.
[0450] For the prevention or treatment of disease, the appropriate dosage of a trispecific antibody of the invention will depend on the type of disease to be treated, the type of antibody, the severity and course of the disease, whether the trispecific antibody is administered for preventive or therapeutic purposes, previous therapy, the patient's clinical history and response to the trispecific antibody, and the discretion of the attending physician. The trispecific antibody is suitably administered to the patient at one time or over a series of treatments. Depending on the type and severity of the disease, about 1 .mu.g/kg to 15 mg/kg (e.g. 0.1 mg/kg-10 mg/kg) of the trispecific antibody can be an initial candidate dosage for administration to the patient, whether, for example, by one or more separate administrations, or by continuous infusion. One typical daily dosage might range from about 1 .mu.g/kg to 100 mg/kg or more, depending on the factors mentioned above. For repeated administrations over several days or longer, depending on the condition, the treatment would generally be sustained until a desired suppression of disease symptoms occurs. One exemplary dosage of the trispecific antibody would be in the range from about 0.05 mg/kg to about 10 mg/kg. Thus, one or more doses of about 0.5 mg/kg, 2.0 mg/kg, 4.0 mg/kg or 10 mg/kg may be administered to the patient. Such doses may be administered intermittently, e.g. every week or every three weeks (e.g. such that the patient receives from about two to about twenty, or e.g. about six doses of the trispecific antibody). An initial higher loading dose, followed by one or more lower doses may be administered. However, other dosage regimens may be useful. The progress of this therapy is easily monitored by conventional techniques and assays.
[0451] It is understood that any of the above formulations or therapeutic methods may be carried out using an immunoconjugate of the invention in place of or in addition to a trispecific antibody specific for HER2 and a BBB-R of the invention.
I. Articles of Manufacture
[0452] In another aspect of the invention, an article of manufacture containing materials useful for the treatment, prevention and/or diagnosis of the disorders described above is provided. The article of manufacture comprises a container and a label or package insert on or associated with the container. Suitable containers include, for example, bottles, vials, syringes, IV solution bags, etc. The containers may be formed from a variety of materials such as glass or plastic. The container holds a composition which is by itself or combined with another composition effective for treating, preventing and/or diagnosing the condition and may have a sterile access port (for example the container may be an intravenous solution bag or a vial having a stopper pierceable by a hypodermic injection needle). At least one active agent in the composition is a trispecific antibody of the invention. The label or package insert indicates that the composition is used for treating the condition of choice. Moreover, the article of manufacture may comprise (a) a first container with a composition contained therein, wherein the composition comprises a trispecific antibody of the invention; and (b) a second container with a composition contained therein, wherein the composition comprises a further cytotoxic or otherwise therapeutic agent. The article of manufacture in this embodiment of the invention may further comprise a package insert indicating that the compositions can be used to treat a particular condition. Alternatively, or additionally, the article of manufacture may further comprise a second (or third) container comprising a pharmaceutically-acceptable buffer, such as bacteriostatic water for injection (BWFI), phosphate-buffered saline, Ringer's solution and dextrose solution. It may further include other materials desirable from a commercial and user standpoint, including other buffers, diluents, filters, needles, and syringes.
[0453] It is understood that any of the above articles of manufacture may include an immunoconjugate of the invention in place of or in addition to a trispecific antibody specific for HER2 and a BBB-R of the invention.
M. Immunoconjugates
[0454] The invention also provides immunoconjugates comprising an trispecific antibody specific for HER2 and a BBB-R herein conjugated to one or more cytotoxic agents, such as chemotherapeutic agents or drugs, growth inhibitory agents, toxins (e.g., protein toxins, enzymatically active toxins of bacterial, fungal, plant, or animal origin, or fragments thereof), or radioactive isotopes.
[0455] In one embodiment, an immunoconjugate is an antibody-drug conjugate (ADC) in which an antibody is conjugated to one or more drugs, including but not limited to a maytansinoid (see U.S. Pat. Nos. 5,208,020, 5,416,064 and European Patent EP 0 425 235 B1); an auristatin such as monomethylauristatin drug moieties DE and DF (MMAE and MMAF) (see U.S. Pat. Nos. 5,635,483 and 5,780,588, and 7,498,298); a dolastatin; a calicheamicin or derivative thereof (see U.S. Pat. Nos. 5,712,374, 5,714,586, 5,739,116, 5,767,285, 5,770,701, 5,770,710, 5,773,001, and 5,877,296; Hinman et al., Cancer Res. 53:3336-3342 (1993); and Lode et al., Cancer Res. 58:2925-2928 (1998)); an anthracycline such as daunomycin or doxorubicin (see Kratz et al., Current Med. Chem. 13:477-523 (2006); Jeffrey et al., Bioorganic & Med. Chem. Letters 16:358-362 (2006); Torgov et al., Bioconj. Chem. 16:717-721 (2005); Nagy et al., Proc. Natl. Acad. Sci. USA 97:829-834 (2000); Dubowchik et al., Bioorg. & Med. Chem. Letters 12:1529-1532 (2002); King et al., J. Med. Chem. 45:4336-4343 (2002); and U.S. Pat. No. 6,630,579); methotrexate; vindesine; a taxane such as docetaxel, paclitaxel, larotaxel, tesetaxel, and ortataxel; a trichothecene; and CC1065.
[0456] In another embodiment, an immunoconjugate comprises an antibody as described herein conjugated to an enzymatically active toxin or fragment thereof, including but not limited to diphtheria A chain, nonbinding active fragments of diphtheria toxin, exotoxin A chain (from Pseudomonas aeruginosa), ricin A chain, abrin A chain, modeccin A chain, alpha-sarcin, Aleurites fordii proteins, dianthin proteins, Phytolaca americana proteins (PAPI, PAPII, and PAP-S), Momordica charantia inhibitor, curcin, crotin, Sapaonaria officinalis inhibitor, gelonin, mitogellin, restrictocin, phenomycin, enomycin, and the tricothecenes.
[0457] In another embodiment, an immunoconjugate comprises an antibody as described herein conjugated to a radioactive atom to form a radioconjugate. A variety of radioactive isotopes are available for the production of radioconjugates. Examples include At.sup.211, I.sup.131, I.sup.125, Y.sup.90, Re.sup.186, Re.sup.188, Sm.sup.153, Bi.sup.212, P.sup.32, Pb.sup.212 and radioactive isotopes of Lu. When the radioconjugate is used for detection, it may comprise a radioactive atom for scintigraphic studies, for example tc99m or 1123, or a spin label for nuclear magnetic resonance (NMR) imaging (also known as magnetic resonance imaging, mri), such as iodine-123 again, iodine-131, indium-111, fluorine-19, carbon-13, nitrogen-15, oxygen-17, gadolinium, manganese or iron.
[0458] Conjugates of an antibody and cytotoxic agent may be made using a variety of bifunctional protein coupling agents such as N-succinimidyl-3-(2-pyridyldithio) propionate (SPDP), succinimidyl-4-(N-maleimidomethyl) cyclohexane-1-carboxylate (SMCC), iminothiolane (IT), bifunctional derivatives of imidoesters (such as dimethyl adipimidate HCl), active esters (such as disuccinimidyl suberate), aldehydes (such as glutaraldehyde), bis-azido compounds (such as bis (p-azidobenzoyl) hexanediamine), bis-diazonium derivatives (such as bis-(p-diazoniumbenzoyl)-ethylenediamine), diisocyanates (such as toluene 2,6-diisocyanate), and bis-active fluorine compounds (such as 1,5-difluoro-2,4-dinitrobenzene). For example, a ricin immunotoxin can be prepared as described in Vitetta et al., Science 238:1098 (1987). Carbon-14-labeled 1-isothiocyanatobenzyl-3-methyldiethylene triaminepentaacetic acid (MX-DTPA) is an exemplary chelating agent for conjugation of radionucleotide to the antibody. See WO94/11026. The linker may be a "cleavable linker" facilitating release of a cytotoxic drug in the cell. For example, an acid-labile linker, peptidase-sensitive linker, photolabile linker, dimethyl linker or disulfide-containing linker (Chari et al., Cancer Res. 52:127-131 (1992); U.S. Pat. No. 5,208,020) may be used.
[0459] The immunuoconjugates or ADCs herein expressly contemplate, but are not limited to such conjugates prepared with cross-linker reagents including, but not limited to, BMPS, EMCS, GMBS, HBVS, LC-SMCC, MBS, MPBH, SBAP, SIA, SIAB, SMCC, SMPB, SMPH, sulfo-EMCS, sulfo-GMBS, sulfo-KMUS, sulfo-MBS, sulfo-SIAB, sulfo-SMCC, and sulfo-SMPB, and SVSB (succinimidyl-(4-vinylsulfone)benzoate) which are commercially available (e.g., from Pierce Biotechnology, Inc., Rockford, Ill., U.S.A).
III. EXAMPLES
[0460] The following are examples of methods and compositions of the invention. It is understood that various other embodiments may be practiced, given the general description provided above.
[0461] Although the foregoing invention has been described in some detail by way of illustration and example for purposes of clarity of understanding, the descriptions and examples should not be construed as limiting the scope of the invention. The disclosures of all patent and scientific literature cited herein are expressly incorporated in their entirety by reference.
Example 1: Materials and Methods
[0462] Unless stated otherwise the following general methods have been applied:
Recombinant DNA Techniques
[0463] Standard methods were used to manipulate DNA as described in Sambrook, J. et al., Molecular cloning: A laboratory manual; Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., 1989. The molecular biological reagents were used according to the manufacturer's instructions.
DNA and Protein Sequence Analysis and Sequence Data Management
[0464] General information regarding the nucleotide sequences of human immunoglobulins light and heavy chains is given in: Kabat, E., A., et al., (1991) Sequences of Proteins of Immunological Interest, Fifth Ed., NIH Publication No 91-3242. Amino acids of antibody chains are numbered according to EU numbering (Edelman, G. M., et al., PNAS 63 (1969) 78-85; Kabat, E. A., et al., (1991) Sequences of Proteins of Immunological Interest, Fifth Ed., NIH Publication No 91-3242). The GCG's (Genetics Computer Group, Madison, Wis.) software package version 10.2 and Infomax's Vector NTI Advance suite version 8.0 was used for sequence creation, mapping, analysis, annotation and illustration.
DNA Sequencing
[0465] DNA sequences were determined by double strand sequencing performed at SequiServe (Vaterstetten, Germany) and Geneart AG (Regensburg, Germany).
Example 2: Generation of Trastuzumab and Pertuzumab Bispecific Antibodies in a 2+2 IgG-scFv Format
Gene Synthesis
[0466] Desired gene segments were prepared by Geneart AG (Regensburg, Germany) from synthetic oligonucleotides and PCR products by automated gene synthesis. The gene segments which are flanked by singular restriction endonuclease cleavage sites were cloned into pGA18 (ampR) plasmids. The plasmid DNA was purified from transformed bacteria and concentration determined by UV spectroscopy. The DNA sequence of the subcloned gene fragments was confirmed by DNA sequencing.
Construction of the Expression Plasmids
[0467] The following expression vector was used for the construction of all heavy and light chain encoding expression plasmids. The vector is composed of the following elements: [0468] a hygromycin resistance gene as a selection marker, [0469] an origin of replication, oriP, of Epstein-Barr virus (EBV), [0470] an origin of replication from the vector pUC18 which allows replication of this plasmid in E. coli [0471] a beta-lactamase gene which confers ampicillin resistance in E. coli, [0472] the immediate early enhancer and promoter from the human cytomegalovirus (HCMV), [0473] the human 1-immunoglobulin polyadenylation ("poly A") signal sequence, and
[0474] The immunoglobulin genes comprising the heavy or light chain were prepared by gene synthesis and cloned into pGA18 (ampR) plasmids as described above. Variable heavy chain constructs were constructed by directional cloning using unique restriction sites. Variable light chain constructs were ordered as gene synthesis comprising VL and CL and constructed by directional cloning using unique restriction sites. The final expression vectors were transformed into E. coli cells, expression plasmid DNA was isolated (Miniprep) and subjected to restriction enzyme analysis and DNA sequencing. Correct clones were grown in 150 ml LB-Amp medium, again plasmid DNA was isolated (Maxiprep) and sequence integrity confirmed by DNA sequencing.
Transient Expression of Immunoglobulin Variants in HEK293 Cells
[0475] Recombinant immunoglobulin variants were expressed by transient transfection of human embryonic kidney 293-F cells using the FreeStyle.TM. 293 Expression System according to the manufacturer's instruction (Invitrogen, USA). For small scale test expressions 30 ml of 0.5.times.10.sup.6 HEK293F cells/ml were seeded one day prior to transfection. The next day, plasmid DNA (1 .mu.g DNA per ml culture volume) was mixed with 1.2 ml Opti-MEM.RTM. I Reduced Serum Medium (Invitrogen, Carlsbad, Calif., USA) followed by addition of 40 .mu.l of 293Fectin.TM. Transfection Reagent (Invitrogen, Carlsbad, Calif., USA). The mixture was incubated for 15 min at room temperature and added drop wise to the cells. One day post-transfection each flask was fed with 300 .mu.l L-Glutamine (200 mM, Sigma-Aldrich, Steinheim, Germany) and 600 .mu.l feed7 containing L-asparagine, amino acids, trace elements, ammonium-Fe(III) citrate, ethanolamine, trace elements, D-glucose, FreeStyle medium without RPMI. Three days post-transfection cell concentration, viability and glucose concentration in the medium were determined using an automated cell viability analyzer (Vi-CELL.TM. XR, Beckman Coulter, Fullerton, Calif., USA) and a glucose meter (Accu-CHEKO Sensor comfort, Roche Diagnostics GmbH, Mannheim, Germany). In addition each flask was fed with 300 .mu.l of L-glutamine, 300 .mu.l non-essential amino acids solution (PAN.TM. Biotech, Aidenbach, Germany), 300 .mu.l sodium pyruvate (100 mM, Gibco, Invitrogen), 1.2 ml feed7 and ad 5 g/L glucose (D-(+)-Glucose solution 45%, Sigma). Finally, six days post-transfection antibodies were harvested by centrifugation at 3500 rpm in a X3R Multifuge (Heraeus, Buckinghamshire, England) for 15 min at ambient temperature, the supernatant was sterile filtered through a Steriflip filter unit (0.22 mm Millipore Express PLUS PES membrane, Millipore, Bedford, Mass.) and stored at -20.degree. C. until further use. Large scale transfections up to 5 L were scaled linearly.
Purification of Bispecific and Control Antibodies
[0476] Bispecific antibodies were purified from cell culture supernatants by affinity chromatography using Protein A-Sepharose.TM. (GE Healthcare, Sweden) and Superdex200 size exclusion chromatography. Briefly, sterile filtered cell culture supernatants were applied on a HiTrap ProteinA HP (5 ml) column equilibrated with PBS buffer (10 mM Na.sub.2HPO.sub.4, 1 mM KH.sub.2PO.sub.4, 137 mM NaCl and 2.7 mM KCl, pH 7.4). Unbound proteins were washed out with equilibration buffer. Antibody and antibody variants were eluted with 0.1 M citrate buffer, pH 2.8, and the protein containing fractions were neutralized with 0.1 ml 1 M Tris, pH 8.5. Eluted protein fractions were pooled, concentrated with an Amicon Ultra centrifugal filter device (MWCO: 30 K, Millipore) to a volume of 3 ml and loaded on a Superdex200 HiLoad 120 ml 16/60 gel filtration column (GE Healthcare, Sweden) equilibrated with 20 mM Histidin, 140 mM NaCl, pH 6.0. Fractions containing purified bispecific and control antibodies with less than 5% high molecular weight aggregates were pooled and stored as 1.0 mg/ml aliquots at -80.degree. C.
Protein Quantification
[0477] Proteins were quantified by affinity chromatography using the automated Ultimate 3000 system (Dionex, Idstein, Germany) with a pre-packed Poros.RTM. A protein A column (Applied Biosystems, Foster City, Calif., USA). All samples were loaded in buffer A (0.2 M Na.sub.2HPO.sub.4.[2H.sub.2O], pH 7.4) and eluted in buffer B (0.1 M citric acid, 0.2 M NaCl, pH 2.5). In order to determine the protein concentration an extinction coefficient of 1.62 was used for all samples.
Analysis of Purified Proteins
[0478] The protein concentration of purified protein samples was determined by measuring the optical density (OD) at 280 nm, using the molar extinction coefficient calculated on the basis of the amino acid sequence. Purity and molecular weight of bispecific and control antibodies were analyzed by SDS-PAGE in the presence and absence of a reducing agent (5 mM 1,4-dithiotreitol) and staining with Coomassie brilliant blue. The NuPAGE.RTM. Pre-Cast gel system (Invitrogen, USA) was used according to the manufacturer's instruction (4-20% Tris-Glycine gels). The aggregate content of bispecific and control antibody samples was analyzed by high-performance SEC using a Superdex 200 analytical size-exclusion column (GE Healthcare, Sweden) in 200 mM KH.sub.2PO.sub.4, 250 mM KCl, pH 7.0 running buffer at 25.degree. C. 25 .mu.g protein were injected on the column at a flow rate of 0.5 ml/min and eluted isocratic over 50 minutes. Integrity of the amino acid backbone of reduced bispecific antibody light and heavy chains was verified by NanoElectrospray Q-TOF mass spectrometry after removal of N-glycans by enzymatic treatment with Peptide-N-Glycosidase F (Roche Molecular Biochemicals).
Analytical HPLC
[0479] Antibodies were analyzed using a Agilent HPLC 1100 (Agilent Technologies, Palo Alto, Calif., USA) with a TSK-GEL G3000SW gel filtration column (7.5 mm ID.times.30 cm, TosoHaas Corp., Montgomeryville, Pa., USA). 18 .mu.l of the eluted proteins were loaded onto the column in Buffer A (0.05 M K.sub.2HPO.sub.4/KH.sub.2PO.sub.4 in 300 mM NaCl, pH 7.5) and separated based on size.
Reducing and Non-Reducing SDS-PAGE
[0480] 7 .mu.l of the eluted proteins were mixed with 2.times. sample buffer (NuPAGE.RTM. LDS Sample buffer, Invitrogen, Carlsbad, Calif., USA) and another 7 .mu.l were mixed with 2.times. sample buffer containing 10% reducing agent (NuPAGE.RTM. Sample Reducing Agent, Invitrogen, Carlsbad, Calif., USA). Samples were heated to 70.degree. for 10 min and loaded onto a pre-cast NuPAGE.RTM. 4-12% BisTris Gel (Invitrogen, Carlsbad, Calif., USA). The gel was run for 45 min at 200V and 125 mA. Afterwards the gel was washed three times with Millipore water and stained with SimplyBlue.TM. SafeStain (Invitrogen, Carlsbad, Calif., USA). The gel was destained overnight in Millipore water. FIGS. 1A-D depict schematically the different variants of Trastuzumab and Pertuzumab bispecific antibodies in a 2+2 IgG-scFv format. All bispecific antibodies are bivalent for each antigen binding site and bind two different paratopes in the ErbB2/HER2 receptor (antigen1=trastuzumab specificity; antigen2=pertuzumab specificity). All bispecific antibodies in a 2+2 IgG-scFv format described herein are non frame-work grafted, non-CDR optimized, not glycoengineered and do not bear any mutation in the Fc part.
[0481] FIGS. 2A-B, 3A-B and 4A-B show exemplary size-exclusion purification graphs, SDS-PAGE analysis and analytical HPLC of variants of Trastuzumab and Pertuzumab bispecific antibodies in a 2+2 IgG-scFv format. No data shown for TvAB17, TvAB13 and variants Herceptin-scFv A to E; all variants of Trastuzumab and Pertuzumab bispecific antibodies in a 2+2 IgG-scFv format were produced with the same quality.
TABLE-US-00003 TABLE 1a Sequences of Trastuzumab and Pertuzumab bispecific antibodies in a 2 + 2 IgG-scFv format SEQ ID NO Name Sequence TvAB_12_2431_TrastuzumabHCscFvOmnitarg(HC_LC) 123 Light chain diqmtqspsslsasvgdrvtitcrasqdvntavawyqqkpgkapklliysasflysgvpsrfsgs (kappa) rsgtdftltisslqpedfatyycqqhyttpptfgqgtkveikrtvaapsvfifppsdeqlksgta- svv [Trastuzumab, cllnnfypreakvqwkvdnalqsgnsqesvteqdskdstyslsstltlskadyekhkvyacevt 1016] hqglsspvtksfnrgec 124 Heavy evqlvesggglvqpggslrlscaasgfnikdtyihwvrqapgkglewvariyptngytryadsv chain kgrftisadtskntaylqmnslraedtavyycsrwggdgfyamdywgqgtlvtvssastkgps [Trastuzumab, vfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvtvps + scFv sslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsvflfppkpkdtlmis Omnitarg, rtpevtcvvvdvshedpevkfnwyvdgvevhnaktkpreeqynstyrvvsyltvlhqdwlng RB40] keykckvsnkalpapiektiskakgqprepqvytlppsreemtknqvsltclvkgfypsdiave wesngqpennykttppvldsdgsfflyskltvdksrwqqgnvfscsvmhealhnhytqkslsl spgkggggsggggsggggsevqlvesggglvqpggslrlscaasgftftdytmdwvrqapgk clewvadvnpnsggsiynqrfkgrftlsvdrskntlylqmnslraedtavyycarnlgpsfyfdy wgqgtlvtvssggggsggggsggggsggggsdiqmtqspsslsasvgdrvtitckasqdvsig vawyqqkpgkapklliysasyrytgvpsrfsgsgsgtdftltisslqpedfatyycqqyyiypytf gcgtkveik TvAB13 [TvAb13_1330scFvTrastuzumab(LC_HC)OmnitargLC_IntronA_cDNA] 125 Light chain diqmtqspsslsasvgdrvtitcrasqdvntavawyqqkpgkapklliysasflysgvpsrfsgs [scFv rsgtdftltisslqpedfatyycqqhyttpptfgqgtkveikggggsggggsggggsevqlvesg Trastuzumab + gglvqpggslrlscaasgfnikdtyihwvrqapgkglewvariyptngytryadsvkgrftisad Omnitarg, tskntaylqmnslraedtavyycsrwggdgfyamdywgqgtlvtvssggggsggggsgggg RB34] sdiqmtqspsslsasvgdrvtitckasqdvsigvawyqqkpgkapklliysasyrytgvpsrfsg sgsgtdftltisslqpedfatyycqqyyiypytfgqgtkveikrtvaapsvfifppsdeqlksgtas vvcllnnfypreakvqwkvdnalqsgnsqesvteqdskdstyslsstltlskadyekhkvyace vthqglsspvtksfnrgec 126 Heavy evqlvesggglvqpggslrlscaasgftftdytmdwvrqapgkglewvadvnpnsggsiynqr chain fkgrftlsvdrskntlylqmnshaedtavyycarnlgpsfyfdywgqgtlvtvssastkgpsvfp (Omnitarg, lapsskstsggtaalgclvkdyfpepvtvswnsgaitsgvhtfpavlqssglyslssvvtvpssslg RB33) tqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsvflfppkpkdtlmisrtpe vtcvvvdvshedpevkfnwyvdgvevhnaktkpreeqynstyrvvsvltvlhqdwlngkey kckvsnkalpapiektiskakgqprepqvytlppsrdeltknqvsltclvkgfypsdiavewesn gqpennykttppvldsdgsfflyskltvdksrwqqgnvfscsvmhealhnhytqkslslspgk TvAB16 [TVAb16_2330TrastuzumabLCseFvOmnitarg(LC_HC)] 127 Light chaindiqmtqspsslsasvgdrvtitcrasqdvntavawyqqkpgkapklliysasflysgvp- srfsgs [Trastuzumab + rsgtdifitisslqpedfatyycqqhyttpptfgqgtkveikrtvaapsvfifppsdeqlksgtasvv scFvOmnitarg, cllnnfypreakvqwkvdnalqsgnsqesvteqdskdstyslsstltlskadyekhkvyacevt RB35] hqglsspvtksfnrgecggggsggggsggggsdiqmtqspsslsasvgdrvtitckasqdvsig vawyqqkpgkapklliysasyrytgvpsrfsgsgsgtdftltisslqpedfatyycqqyyiypytf gqgtkveikggggsggggsggggsevqlvesggglvqpggslrlscaasgftftdytmdwvrq apgkglewvadvnpnsggsiynqrfkgrfftsvdrskntlylqmnslraedtavyycarrilgps fyfdywgqgtlvtvss 128 Heavy evqlvesggglvqpggslrlscaasgfnikdtyihwvrqapgkglewvariyptngytryadsv chain kgrftisadtskntaylqmnslraedtavyycsrwggdgfyamdywgqgtlvtvssastkgps [Trastuzumab, vfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvtvps 1036] sslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsvflfppkpkdtlmis rtpevtcvvvdvshedpevkfnwyvdgvevhnaktkpreeqynstyrvvsyltvlhqdwing keykckvsnkalpapiektiskakgqprepqvytlppsrdeltknqvsltclvkgfypsdiave wesngqpennykttppvldsdgsfflyskltvdksrwqqgnvfscsvmhealhnhytqkslsl spgk TvAB17 [TvAb17_2431_TrastuzumabHCseFvOmnitarg(LC_HC)] 129 Light chain diqmtqspsslsasvgdrvtitcrasqdvntavawyqqkpgkapklliysasflysgvpsrfsgs (kappa) rsgtdftltisslqpedfatyycqqhyttpptfgqgtkveikrtvaapsvfifppsdeqlksgta- svv [Trastuzumab, cllnnfypreakvqwkvdnalqsgnsqesvteqdskdstyslsstltlskadyekhkvyacevt 1016] hqglsspvtksfnrgec 130 Heavy evqlvesggglvqpggslrlscaasgfnikdtyihwvrqapgkglewvariyptngytryadsv chain kgrftisadtskntaylqmnslraedtavyycsrwggdgfyamdywgqgtlvtvssastkgps [Trastuzumab, vfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvtvps + sslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsvflfppkpkdtlmis scFvOmnitarg, rtpevtcvvvdvshedpevkfnwyvdgvevhnaktkpreeqynstyrvvsyltvlhqdwing RB43] keykckvsnkalpapiektiskakgqprepqvytlppsreemtknqvsltclvkgfypsdiave wesngqpennykttppvldsdgsfflyskltvdksrwqqgnvfscsvmhealhnhytqkslsl spgkggggsggggsggggsdiqmtqspsslsasvgdrvtitckasqdvsigvawyqqkpgka pklliysasyrytgvpsrfsgsgsgtdftltisslqpedfatyycqqyyiypytfgcgtkveikggg gsggggsggggsggggsevqlvesggglvqpggslrlscaasgftftdytmdwvrqapgkcle wvadvnpnsggsiynqrfkgrftlsvdrskntlylqmnslraedtavyycarnlgpsfyfdywg qgtlvtvss TvAB20 [TVAb20_4441TrastuzumabLCseFvOmnitarg(LC_HC)] 131 Light chain diqmtqspsslsasvgdrvtitcrasqdvntavawyqqkpgkapklliysasflysgvpsrfsgs [Trastuzumab, rsgtdftltisslqpedfatyycqqhyttpptfgqgtkveikrtvaapsvfifppsdeqlksgtasvv + scFv cllnnfypreakvqwkvdnalqsgnsqesvteqdskdstyslsstltlskadyekhkvyacevt Omnitarg, hqglsspvtksfnrgecggggsggggsggggsggggsdiqmtqspsslsasvgdrvtitckas- q RB61] dvsigvawyqqkpgkapklliysasyrytgvpsrfsgsgsgtdftltisslqpedfatyycqqyyi ypytfgcgtkveikggggsggggsggggsggggsevqlvesggglvqpggslrlscaasgftft dytmdwvrqapgkclewvadvnpnsggsiynqrfkgrftlsvdrskntlylqmnslraedtav yycarnlgpsfyfdywgqgtlvtvss 132 Heavy evqlvesggglvqpggslrlscaasgfnikdtyihwvrqapgkglewvariyptngytryadsv chain kgrftisadtskntaylqmnslraedtavyycsrwggdgfyamdywgqgtlvtvssastkgps [Trastuzumab, vfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvtvps 1036] sslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsvflfppkpkdtlmis rtpevtcvvvdvshedpevkfnwyvdgvevhnaktkpreeqynstyrvvsyltvlhqdwing keykckvsnkalpapiektiskakgqprepqvytlppsrdeltknqvsltclvkgfypsdiave wesngqpennykttppvldsdgsfflyskltvdksrwqqgnvfscsvmhealhnhytqkslsl spgk Herceptarg 2 + 2 OmniE 145 Heavy chainevqlvesggglvqpggslrlscaasgftftdytmdwvrqapgkglewvadvnpnsggsi- ynqrfkgrftls with scFv vdrskntlylqmnslraedtavyycarnlgpsfyfdywgqgtlvtvssastkgpsvfplapss- kstsggtaal Trastuzumab gclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvytypssslgtqtyicnvnhkpsntkvdkkv stabilized epkscdlcthtcppcpapellggpsvflfppkpkdtlmisrtpevtcvvvdvshedpevkfnwyvdgvevh with naktkpreeqynstyrvvsvltvlhqdwlngkeykckvsnkalpapiektiskakgqprepqvytlpp- srde disulphide ltknqvsltclvkgfypsdiavewesngqpennykttppvldsdgsfflyskltydksrwqqgnyfscsvmh bonding ealhnhytqkslslspgkggggsggggsevqlvesggglvqpggslrlscaasgfnikdtyihwv- rqapgk glewvariyptngytryadsvkgrftisadtskntaylqmnslraedtavyycsrwggegfyamdywgcg tlytvssggggsggggsggggsdiqmtqspsslsasvgdrytitcrasqdvnvavawyqqkpgkcpklliy sasflysgvpsrfsgsrsgtdftltisslqpedfatyycqqhyltpptfgqgtkveik 146 Pertuzumab diqmtqspsslsasvgdrvtitckasqdvsigvawyqqkpgkapklliysasyrytgvpsrfsgsgsgtdftl- t light chain isslqpedfatyycqqyyiypylfgqgtkveikrtvaapsvfifppsdeqlksgtasvvcllnnfyprealwq- w kvdnalqsgnsqesvteqdskdstyslsstltlskadyekhkvyacevthqglsspvtksfnrgec
TABLE-US-00004 TABLE 1 B Variants of TvAB12_2431_TrastuzumabHCscFvOnmitarg(HC_LC) with a stabilizing disulphide bond in the scFv part to reduce aggregation levels Name of molecule Disulphide position (VH-VL) Trastuzumab_scFv_WT WT Trastuzumab_scFv_A 44-100 Trastuzumab_scFv_B 45-98 Trastuzumab_scFv_C 101-46 Trastuzumab_scFv_D 103-44 Trastuzumab_scFv_E 105-43
Example 3: Proliferation Inhibition Assay with Trastuzumab and Pertuzumab Bispecific Antibodies in a 2+2 IgG-scFv Format
Cell Lines
[0482] MDA-MB 175 VII cells were maintained in DMEM/F12 medium (Gibco) supplemented with 10% fetal calf serum and 2 mL L-glutamine. Propagation of cell lines followed standard cell culture protocols.
[0483] The ability of the bispecific antibodies to inhibit proliferation was assessed in the cell line MDA-MB-175 VII. MDA-MB-175 VII were cultured in DMEM/F12 medium (Gibco) supplemented with 10% fetal calf serum, 2 mM L-glutamine. Cells in the logarithmic growth phase were detached, counted and 2.times.10e4 cells were seeded in 100 .mu.L medium per well of a 96-well cell culture plate. Cells were maintained overnight in the incubator and the following day 100 .mu.L of the respective antibodies diluted in medium were added in form of a dilution series to the cells. After a total incubation time of 6 days cell growth was assessed in an Alamar Blue (Invitrogen) assay. The assay was performed as recommended by the manufacturer.
TABLE-US-00005 TABLE 2 shows the potency of selected bispecific antibodies in the proliferation assay. Antibody SEQ ID NO: EC50 [nM] Herceptarg WT 1,84 Herceptarg A 1,89 Herceptarg B 2,66 Herceptarg C 2,56 Herceptarg D 1,63 Herceptarg E 1,75 TvAb 12 123/124 4,90 CrossMab 119/120/121/122 4,75 Pertuzumab 2,11 Pertuzumab + Herceptin 1,70 Herceptin 2,92
Example 4: Herceptarg Stabilisation
[0484] The aspartate isomerization site at position 98 of the heavy chain and the asparagine deamidation site at position 30 of the light chain are stability hotspots of trastuzumab. Those two positions affect the stability and integrity of the antigen binding capacity of the antibody. This problem was overcome by introducing the lyophilized formulations using either sodium succinate or histidine buffer. In order to increase stability and storage half-life we intended to replace those known sources of instability by amino acids, or amino acid stretches that should have a higher intrinsic stability. We tested herefore the replacement of Asp98 by Glu in the heavy chain and the replacement of Asn30 by Ser, as well Thr31 by Val in the light chain. Those mutations were abbreviated D98E, N30S, and T31V. T31V does not directly influence the deamidation of N30, but it was assumed that the residue adjacent on the C-terminal side of an Asparagine would influence the stability properties of a polypeptide chain.
[0485] The antibody samples were incubated over a period of either 1, 2, or 3 months at 40.degree. C. in one of the three buffers: 40 mM Histidin, 150 mM NaCl, pH5.0, 40 mM Histidin, 150 mM NaCl, pH6.0, or 40 mM Histidin, 150 mM NaCL, pH7.4. The protein concentration during this period was always 1 mg/ml. After the indicated time points, samples were taken and shock frozen in liquid nitrogen, and then kept at -80.degree. C. until further analysis. This analysis was carried out on a ProteOn XPR36 instrument (BioRad). Approximately 700 RU of Her2, respectively, were immobilized on 2 channels of a GLM chip using amine coupling (vertical orientation). Trastuzumab variants were measured in duplicates at 6 different analyte concentrations (100, 50, 25, 12.5, 6.25, 0 nM) by injections in horizontal orientation at 100 .mu.l/min. Association rate were recorded for 180s, the dissociation rate for 600s. Regeneration was performed by two pulses of 10 mM glycine pH 1.5 and 50 mM NaOH for 60s at 150 .mu.l/min (horizontal orientation). In total four different Trastuzumab variants were measured. In the first experiment (Tables 3 and 4), only the unmodified Trastuzumab and variant 602 (D98E of heavy chain and T31V of light chain) were tested. In the second experiment the unmodified Trastuzumab, variant 602 (D98E of heavy chain and T31V of light chain) and variants VH:D98E/VL: N30S VH:D98E/VL: N30T were tested (Table 5, 6, 7).
Results:
[0486] All variants show the same affinity towards recombinant Her2 antigen as the parental Trastuzumab molecule. After exposure to pH5 or pH6 at 40.degree. C., Trastuzumab lost affinity by a factor .about.5, mainly driven by increasing the off-rate and keeping the on-rate unchanged. Variant 602 showed almost undistinguishable affinities before and after pH stress. Variant N30S had a higher affinity from the beginning compared to the parental Trastuzumab, which stayed approximately constant during the stress conditions. The variants D98E (VH) together with either N30S, N30T, or T31V (VL) were used in the further experiments. Results are shown in FIGS. 5A-B and 6A-C and the tables below.
TABLE-US-00006 TABLE 3 Kinetic affinity parameters of Trastuzumab variants as determined by SPR method (ProteOn instrument) after incubating the samples for 1, 2, or 3 months at 40.degree. in 40 mM Histidin, 150 mM NaCl, pH 5.0 buffer. Each measurement was done in duplicate and both experimental values are shown. Tested were the resynthesized Trastuzumab (wt) and compared to the D98E T31V variant in the heavy and light chain, respectively (named clone 602). t0 t1 pH 5 ka kd KD ka kd KD wt 1 2.4E+05 1.1E-04 4.3E-10 3.3E+05 1.6E-04 5.0E-10 2 2.4E+05 4.4E-05 1.8E-10 2.8E+05 1.9E-04 7.0E-10 602 1 2.6E+05 1.2E-04 4.6E-10 2.7E+05 2.2E-04 8.0E-10 2 2.3E+05 1.3E-04 5.4E-10 2.8E+05 1.4E-04 5.2E-10 t2 t3 pH 5 ka kd KD ka kd KD wt 1 2.9E+05 2.6E-04 9.2E-10 2.8E+05 3.4E-04 1.2E-09 2 2.8E+05 2.8E-04 9.9E-10 2.9E+05 3.6E-04 1.3E-09 602 1 2.9E+05 1.7E-04 5.8E-10 3.0E+05 1.7E-04 5.8E-10 2 2.9E+05 1.6E-04 5.3E-10 3.1E+05 1.7E-04 5.5E-10
TABLE-US-00007 TABLE 4 Kinetic affinity parameters of Trastuzumab variants similar to table 3. Here the samples are incubated at 40.degree. in 40 mM Histidin, 150 mM NaCl, pH 6.0 for the same time intervals as before. t0 t1 pH 6 ka kd KD ka kd KD wt 1 2.8E+05 9.5E-05 3.4E-10 3.0E+05 1.5E-04 5.1E-10 2 2.8E+05 8.6E-05 3.1E-10 2.8E+05 2.1E-04 7.7E-10 602 1 2.9E+05 1.4E-04 4.8E-10 3.0E+05 1.6E-04 5.2E-10 2 2.8E+05 1.4E-04 5.0E-10 2.9E+05 1.5E-04 5.1E-10 t2 t3 pH 6 ka kd KD ka kd KD wt 1 2.5E+05 3.1E-04 1.2E-09 2.3E+05 4.0E-04 1.7E-09 2 2.6E+05 3.3E-04 1.3E-09 2.4E+05 4.1E-04 1.8E-09 602 1 3.0E+05 1.6E-04 5.3E-10 3.0E+05 1.8E-04 5.9E-10 2 2.9E+05 1.6E-04 5.5E-10 2.9E+05 1.7E-04 5.7E-10
TABLE-US-00008 TABLE 5 Kinetic affinity parameters of Trastuzumab variants similar to table 3. Here the samples are incubated at 40.degree. in 40 mM Histidin, 150 mM NaCl, pH 5.0 for the same time intervals as before. Samples included are the resynthesized Trastuzumab (wt), the D98E T31V variant in the heavy and light chain, respectively (named clone 602. This was produced in CHO instead of HEK). Also the light chain variants N30S, and N30T (both have the D98E heavy chain variant) t0 t1 pH 5 ka kd KD ka kd KD 602 1 2.20E+05 1.40E-04 6.34E-10 2.32E+05 9.14E-05 3.95E-10 CHO 2 2.32E+05 2.13E-04 9.21E-10 2.10E+05 1.54E-04 7.35E-10 N30T 1 2.19E+05 2.11E-04 9.64E-10 2.21E+05 1.46E-04 6.61E-10 2 2.18E+05 2.61E-04 1.20E-09 2.05E+05 1.85E-04 9.02E-10 N30S * 1 2.59E+05 2.59E-19 1.00E-24 2.54E+05 1.93E-05 7.58E-11 2 2.44E+05 2.36E-17 9.68E-23 2.42E+05 3.47E-05 1.43E-10 wt 1 2.51E+05 1.60E-04 6.39E-10 2.30E+05 3.62E-04 1.58E-09 2 2.29E+05 1.93E-04 8.43E-10 2.13E+05 2.66E-04 1.25E-09 t2 t3 pH 5 ka kd KD ka kd KD 602 1 2.32E+05 2.36E-04 1.02E-09 2.01E+05 9.67E-05 4.81E-10 CHO 2 2.37E+05 2.35E-04 9.89E-10 2.08E+05 1.30E-04 6.25E-10 N30T 1 2.20E+05 1.77E-04 8.05E-10 2.31E+05 2.87E-04 1.24E-09 2 2.01E+05 2.02E-04 1.00E-09 2.27E+05 2.82E-04 1.24E-09 N30S * 1 2.49E+05 1.16E-16 4.64E-22 2.54E+05 4.11E-18 1.62E-23 2 2.37E+05 3.71E-16 1.57E-21 2.37E+05 6.06E-17 2.55E-22 wt 1 2.38E+05 6.47E-04 2.72E-09 2.33E+05 7.72E-04 3.32E-09 2 2.26E+05 6.88E-04 3.05E-09 2.18E+05 7.91E-04 3.62E-09 * Due to slow dissociation rates, the off-rates and the dissociation constant contain a high degree of uncertainty.
TABLE-US-00009 TABLE 6 Kinetic affinity parameters of Trastuzumab variants similar to table 3. Here the samples are incubated at 40.degree. in 40 mM Histidin, 150 mM NaCl, pH 6.0 for the same time intervals as before. Samples included are the resynthesized Trastuzumab (wt), the D98E T31V variant in the heavy and light chain, respectively (named clone 602. This was produced in CHO instead of HEK). Also the light chain variants N30S, and N30T (both have the D98E heavy chain variant) t0 t1 pH 6 ka kd KD ka kd KD 602 1 2.07E+05 9.33E-05 4.51E-10 2.30E+05 1.37E-04 5.94E-10 CHO 2 2.16E+05 1.97E-04 9.12E-10 2.16E+05 2.02E-04 9.32E-10 N30T 1 2.24E+05 2.45E-04 1.09E-09 2.03E+05 1.24E-04 6.10E-10 2 2.31E+05 2.71E-04 1.17E-09 1.93E+05 1.57E-04 8.13E-10 N30S * 1 2.56E+05 1.18E-17 4.62E-23 2.65E+05 2.77E-05 1.04E-10 2 2.48E+05 8.14E-06 3.28E-11 2.51E+05 4.23E-05 1.68E-10 wt 1 2.23E+05 7.43E-05 3.33E-10 2.22E+05 4.60E-04 2.07E-09 2 2.23E+05 7.98E-05 3.59E-10 2.21E+05 4.03E-04 1.82E-09 t2 t3 pH 6 ka kd KD ka kd KD 602 1 2.19E+05 1.29E-04 5.87E-10 2.14E+05 1.50E-04 7.01E-10 CHO 2 2.07E+05 1.52E-04 7.31E-10 2.13E+05 1.60E-04 7.50E-10 N30T 1 2.11E+05 2.27E-04 1.08E-09 2.08E+05 2.01E-04 9.67E-10 2 2.05E+05 2.55E-04 1.24E-09 2.03E+05 1.98E-04 9.75E-10 N30S * 1 2.38E+05 8.97E-19 3.77E-24 2.36E+05 1.84E-05 7.80E-11 2 2.25E+05 4.30E-17 1.91E-22 2.33E+05 3.41E-05 1.46E-10 wt 1 2.31E+05 9.24E-04 4.00E-09 1.89E+05 9.91E-04 5.24E-09 2 2.20E+05 9.31E-04 4.24E-09 1.78E+05 9.95E-04 5.58E-09 * Due to slow dissociation rates, the off-rates and the dissociation constant contain a high degree of uncertainty.
TABLE-US-00010 TABLE 7 Kinetic affinity parameters of Trastuzumab variants similar to table 3. Here the samples are incubated at 40.degree. in 40 mM Histidin, 150 mM NaCl, pH 7.4 for the same time intervals as before. Samples included are the resynthesized Trastuzumab (wt), the D98E T31V variant in the heavy and light chain, respectively (named clone 602. This was produced in CHO instead of HEK). Also the light chain variants N30S, and N30T (both have the D98E heavy chain variant) t0 t1 pH 7.4 ka kd KD ka kd KD 602 1 2.15E+05 1.23E-04 5.71E-10 2.32E+05 1.80E-04 7.78E-10 CHO 2 2.17E+05 2.08E-04 9.55E-10 2.19E+05 2.31E-04 1.06E-09 N30T 1 2.46E+05 2.19E-04 8.87E-10 2.15E+05 1.86E-04 8.65E-10 2 2.20E+05 2.30E-04 1.04E-09 2.00E+05 2.03E-04 1.02E-09 N30S 1 2.58E+05 1.35E-06 5.25E-12 2.60E+05 2.55E-05 9.82E-11 2 2.36E+05 2.24E-05 9.46E-11 2.49E+05 6.31E-18 2.53E-23 wt 1 2.25E+05 7.49E-05 3.33E-10 1.99E+05 8.80E-04 4.43E-09 2 2.12E+05 9.85E-05 4.65E-10 2.00E+05 8.78E-04 4.40E-09 t2 t3 pH 7.4 ka kd KD ka kd KD 602 1 1.97E+05 1.05E-04 5.34E-10 2.02E+05 2.24E-04 1.11E-09 CHO 2 1.82E+05 1.33E-04 7.30E-10 2.00E+05 2.14E-04 1.07E-09 N30T 1 2.12E+05 2.64E-04 1.25E-09 1.94E+05 1.41E-04 7.26E-10 2 1.98E+05 2.84E-04 1.44E-09 1.73E+05 1.54E-04 8.87E-10 N30S 1 2.43E+05 7.28E-21 2.99E-26 2.37E+05 6.30E-05 2.66E-10 2 2.27E+05 1.24E-05 5.46E-11 2.21E+05 6.55E-05 2.97E-10 wt 1 1.64E+05 1.35E-03 8.27E-09 1.55E+05 1.79E-03 1.15E-08 2 1.67E+05 1.29E-03 7.72E-09 1.45E+05 1.63E-03 1.12E-08 * Due to slow dissociation rates, the off-rates and the dissociation constant contain a high degree of uncertainty.
Example 5: Binding of Trastuzumab and Trastuzumab Stabilization Variants after Stress to KPL-4 Cells
Binding
[0487] KPL-4 cells were harvested and resuspended in FACS buffer. 0.2 Mio cells were seeded into a 96 well round bottom plate. The plate was centrifuged at 400 g for 3 min to pellet the cells. The supernatant was removed and the cells were resuspended in 40 .mu.l of the diluted antibodies. The plate was incubated for 30 min at 4.degree. C. to allow binding of the antibodies. To remove unbound antibodies the cells were centrifuged again and washed twice with FACS buffer. To detect the antibodies the cells were resuspended in 12 .mu.l diluted secondary goat anti-human Fc specific FITC-labeled secondary antibody (Jackson ImmunoResearch #109-096-098) and incubated again for 30 min at 4.degree. C. Afterwards the cells were washed twice with FACS buffer, resuspended in 200 .mu.l FACS buffer and the fluorescence was measured with BD CantoII.
ADCC
[0488] Target cells were harvested, washed, stained with calcein (Invitrogen), resuspended in AIM V.RTM. medium (Life Technologies), and plated at a concentration of 3.times.10.sup.4 cells/well. The respective antibody dilutions were added in triplicates to the cells and incubated for 10 min before addition of the effector cells (peripheral blood mononuclear effector cells [PBMCs]). Effector (E) and target (T) cells were then incubated for the indicated time at 37.degree. C. at the indicated E:T ratio (triplicates for all samples). After incubation the cells were washed once with PBS and then lysed with borate buffer. Calcein retention was measured in a Wallac Victor3 1420 Multilabel Counter. ADCC was calculated using the following formula:
Percentage ADCC = ( [ sample release - spontaneous release maximal release - spontaneous release ] ) .times. 1 0 0 . ##EQU00001##
[0489] Spontaneous release, corresponding to target cells incubated with effector cells without antibody, was defined as 0% cytotoxicity, and maximal release (target cells lysed with 1% Triton X-100) was defined as 100% cytotoxicity. The average percentage of ADCC and standard deviations of the triplicates of each experiment were calculated.
[0490] Results are shown in FIGS. 7A-L, 8A-F and 9A-F.
Example 6: Generation of Herceptarg CrossMab and Framework Grafting on Novel LC06 Based Framework to Achieve Less Mispairing
Gene Synthesis
[0491] Desired gene segments were prepared by Geneart AG (Regensburg, Germany) from synthetic oligonucleotides and PCR products by automated gene synthesis. The gene segments encoding heavy or light chains with C-terminal attachment of scFv antibody fragments, "knobs-into-hole" antibody heavy chains carrying S354C and T366W mutations and "knobs-into-hole" heavy chains carrying Y349C, T366S, L368A and Y407V mutations in the CH3 domain in combination with unmodified VH domains, crossed C kappa domains or scFab antibody fragments as well as unmodified antibody light chains or CH1 domain exchanged light chains are flanked by singular restriction endonuclease cleavage sites (BamHI-XbaI, BamHI-XmnI or BamHI-KpnI) and were cloned into pGA18 (ampR) plasmids. The plasmid DNA was purified from transformed bacteria and concentration determined by UV spectroscopy. The DNA sequence of the subcloned gene fragments was confirmed by DNA sequencing. All constructs were designed with a 5'-end DNA sequence coding for a leader peptide (MGWSCIILFLVATATGVHS), which targets proteins for secretion in eukaryotic cells.
Construction of the Expression Plasmids
[0492] The expression vector that was used for the construction of all "knobs-into-hole" heavy chain as well as antibody light chain encoding expression plasmids comprises the following elements: [0493] a hygromycin resistance gene as a selection marker, [0494] an origin of replication, oriP, of Epstein-Barr virus (EBV), [0495] an origin of replication from the vector pUC18 which allows replication of this plasmid in E. coli [0496] a beta-lactamase gene which confers ampicillin resistance in E. coli, [0497] the immediate early enhancer and promoter from the human cytomegalovirus (HCMV), [0498] the human 1-immunoglobulin polyadenylation ("poly A") signal sequence, and [0499] unique BamHI and XbaI restriction sites.
[0500] The immunoglobulin genes comprising heavy or light chains with C-terminal attachment of scFv antibody fragments, "knobs-into-hole" heavy chains with unmodified VH domains, crossed C kappa domains or scFab fragments as well as unmodified light chains or CH1 domain exchanged light chains were prepared by gene synthesis and cloned into pGA18 (ampR) plasmids as described. The pG18 (ampR) plasmids carrying the synthesized DNA segments and the expression vector were digested with BamHI and XbaI, BamHI and XmnI or BamHI and KpnI restriction enzymes (Roche Molecular Biochemicals) and subjected to agarose gel electrophoresis. Purified heavy or light chains with C-terminal attachment of scFv antibody fragments, "knobs-into-hole" heavy and unmodified or domain exchanged light chain encoding DNA segments were then ligated to the isolated expression vector BamHI/XbaI, BamHI/XmnI or BamHI/KpnI fragment resulting in the final expression vectors. The final expression vectors were transformed into E. coli cells, expression plasmid DNA was isolated (Miniprep) and subjected to restriction enzyme analysis and DNA sequencing. Correct clones were grown in 150 ml LB-Amp medium, again plasmid DNA was isolated (Maxiprep) and sequence integrity confirmed by DNA sequencing.
Transient Expression of Bispecific Antibodies in HEK293 Cells
[0501] Recombinant bispecific antibodies were expressed by transient transfection of human embryonic kidney 293-F cells using the FreeStyle.TM. 293 Expression System according to the manufacturer's instruction (Invitrogen, USA). Briefly, suspension FreeStyle.TM. 293-F cells were cultivated in FreeStyle.TM. 293 Expression medium at 37.degree. C./8% CO.sub.2 and the cells were seeded in fresh medium at a density of 1.times.10.sup.6 viable cells/ml one day before transfection. For transfection, DNA was prepared in 10 ml Dulbecco's PBS (PAA, Austria) using 162.5 .mu.l of 293-Free.TM. Transfection Reagent (Merck, USA) and 125 .mu.g of heavy with N-terminal attachment of scFab encoding DNA in a plasmid ratio of 1:1 with "Knobs-into-hole" heavy chain 1 and 2 and light chain plasmid DNA in a 1:1:1 molar ratio in 250 ml final transfection volume. For transfection of Cross Mabs, a plasmid ratio of 1:1:1:1, 1:1:1:2, 1:1:1:4, 1:1:1:8 of "Knobs-into-hole" heavy chain 1:unmodified light chain:C kappa domain exchanged "Knobs-into-hole" heavy chain 2:CH1 domain exchanged light chain was prepared. The CH1-VL light chain plasmid ration was used at 1.times., 2.times., 4.times. and 8.times. molar ratios to assess the optimization of chain pairing using a combination of CE-SDS and Q-TOF spectrometry. Antibody containing cell culture supernatants were harvested 7 days after transfection by centrifugation at 14000 g for 30 minutes and filtered through a sterile filter (0.22 .mu.m). Supernatants were stored at -20.degree. C. until purification. The sequences of the resulting antibodies are shown below in table 8.
Preparation of the Glycoengineered Derivatives of Bispecific <Her2GlyMab> Antibodies
[0502] Glycoengineered derivatives of bispecific <Her2GlyMab> antibodies were produced by co-transfecting HEK293-EBNA cells with the mammalian antibody heavy and light chain expression vectors using a calcium phosphate-transfection approach. Exponentially growing HEK293-EBNA cells were transfected by the calcium phosphate method. For the production of the glycoengineered antibody, the cells were co-transfected with a plasmid for a fusion GnTIII polypeptide expression and a second plasmid for mannosidase II expression, respectively. Plasmid ratios of bispecific antibodies were added as described in the material and methods section above. Cells were grown as adherent monolayer cultures in T flasks using DMEM culture medium supplemented with 10% FCS, and were transfected when they were between 50 and 80% confluent. For the transfection of a T75 flask, 7.5 (to 8) million cells were seeded 24 hours before transfection in ca 14 ml DMEM culture medium supplemented with FCS (at 10% V/V final), (eventually 250 .mu.g/ml neomycin,) and cells were placed at 37.degree. C. in an incubator with a 5% CO2 atmosphere overnight. For each T75 flask to be transfected, a solution of DNA, CaCl2 and water was prepared by mixing 47 .mu.g total plasmid vector DNA, 235 .mu.l of a 1M CaCl2 solution, and adding water to a final volume of 469 .mu.l. To this solution, 469 .mu.l of a 50 mM HEPES, 280 mM NaCl, 1.5 mM Na.sub.2HPO.sub.4 solution at pH 7.05 were added, mixed immediately for 10 sec and left to stand at room temperature for 20 sec. The suspension was diluted with ca. 12 ml of DMEM supplemented with 2% FCS, and added to the T75 in place of the existing medium. The cells were incubated at 37.degree. C., 5% CO2 for about 17 to 20 hours, then medium was replaced with ca. 12 ml DMEM, 10% FCS. The conditioned culture medium was harvested 5 to 7 days post-transfection centrifuged for 5 min at 210-300 .mu.g, sterile filtered through a 0.22 .mu.m filter (or alternatively centrifuged for 5 min at 1200 rpm, followed by a second centrifugation for 10 min at 4000 rpm) and kept at 4.degree. C.
[0503] Glycoengineered antibodies were purified and formulated as described above for the non-glycoengineered antibodies. The oligosaccharides attached to the Fc region of the antibodies were analysed as described below to determine the amount of fucose.
Framework Grafting
[0504] Rationale: Similar framework of trastuzumab and pertuzumab allows mis-pairing of light chains: Both Trastuzumab and Pertuzumab have a V.sub.HIII V.sub.LkI framework and therefore the light chain interface affinity to both heavy chains is identical.
[0505] In order to avoid mispairing of light chains in the crossMab Herceptarg bispecific antibody, the Trastuzumab framework of "CrossMabXPer Her2GlyMab" was exchanged with the framework of the non-related antibody LC06. Trastuzumab is related to germlines hVH3_66 and hVK1D_39 whereas the antibody LC06 corresponds to germlines hVbase_VH1_1 germline and hVL_3, respectively. Pertuzumab is related to hVH3_23 and hVK1D_13 showing a very similar framework system compared to Trastuzumab.
[0506] Both germline acceptor frameworks of LC06 are different from the Trastuzumab framework, especially, the LC06 lambda light chain, compared to the Trastzumab kappa light chain. The antibody Fab crystal structure of Trastuzumab has been superimposed and the compatibility of the framewords have been structrally evaluated. CDRI, II and III of the Trastuzumab light chain were grafted onto the new Lambda framework of LC06. Mutations included were D98E and N30S, the N30S was used in favour of T31V because of the increase in affinity to the Her2 extracellular domain with the N30S modification. It was thought that any reduction of the Kd caused by the CDR grafting could be compensated by the use of the N30S mutation. Some accommodations in the acceptor frameworks were required in order to get the CDRs in their biological active conformation; for example A(LC06 lambda) at the Kabat position 71 of the light chain has been backmutated to F(Trastuzumab kappa). The original VHIII-VLkI (Kappa I family) framework of Pertuzumab was maintained. The resulting bispecific antibody is depicted as "CrossMab-CDRG Her2GlyMab", with sequences as shown in Table 8 below.
TABLE-US-00011 TABLE 8 Sequences of Herceptarg CrossMab bispecific antibodies. Construct SEQ ID NO Sequence OAscFab1 Her2GlyMab scFab 133 diqmtqspsslsasvgdrvtitcrasqdvnvavawyqqkpgkapklliysasflysgvpsrfsg Trastuzumab srsgtdftltisslqpedfatyycqqhyttpptfgqgtkveikrtvaapsvfifppsdeqlksgtas heavy chain 1 vvcllnnfypreakvqwkvdnalqsgnsqesvteqdskdstyslsstltlskadyekhkvyac evthqglsspvtksfnrgecggggsggggsggggsggggsggggsggggsggevqlvesgg glvqpggslrlscaasgfnikdtyihwvrqapgkglewvariyptngytryadsvkgrftisadt skntaylqmnslraedtavyycsrwggegfyamdywgqgtlvtvssastkgpsvfplapssk stsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvtvpssslgtqtyi cnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsvflfppkpkdtlmisrtpevtcv vvdvshedpevkfnwyvdgvevhnaktkpreeqynstyrvvsyltvlhqdwlngkeykck vsnkalpapiektiskakgqprepqvytlppcrdeltknqvslwclvkgfypsdiavewesng qpennykttppvldsdgsfflyskltvdksrwqqgnvfscsvmhealhnhytqkslslspgk Pertuzumab 134 evqlvesggglvqpggslrlscaasgftftdytmdwvrqapgkglewvadvnpnsggsiyn heavy chain 2 qrfkgrftlsvdrskntlylqmnslraedtavyycarnlgpsfyfdywgqgtlvtvssastkgps vfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvtvp ssslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsvflfppkpkdtlm isrtpevtcvvvdvshedpevkfnwyydgvevhnaktkpreeqynstyrvvsvltvlhqdwl ngkeykckvsnkalpapiektiskakgqprepqvctlppsrdeltknqvslscavkgfypsdi avewesngqpennykttppyldsdgsfflyskltvdksrwqqgnyfscsvmhealhnhytq kslslspgk Pertuzumab 135 diqmtqspsslsasvgdrytitckasqdysigvawyqqkpgkapklliysasyrytgvpsrfsg light chain 1 sgsgtdftltisslqpedfatyycqqyyiypytfgqgtkveikrtvaapsvfifppsdeqlksgtas vycllnnfypreakvqwkvdnalqsgnsqesvteqdskdstyslsstltlskadyekhkvyac evthqglsspvtksfnrgec OAscFab2 Her2GlyMab Pertuzumab 136 evqlvesggglvqpggslrlscaasgftftdytmdwyrqapgkglewvadynpnsggsiyn heavy chain 1 qrfkgrftlsvdrskntlylqmnslraedtavyycarnlgpsfyfdywgqgtlvtvssastkgps vfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvtvp ssslgtqtyicnynhkpsntkvdkkvepkscdkthtcppcpapellggpsvflfppkpkdtlm isrtpevtcvvvdvshedpevkfnwyydgvevhnaktkpreeqynstyrvvsvltvlhqdwl ngkeykckvsnkalpapiektiskakgqprepqvctlppsrdeltknqvslscavkgfypsdi avewesngqpennykttppvldsdgsfflvskltvdksrwqqgnvfscsvmhealhnhytq kslslspgk Pertuzumab 137 diqmtqspsslsasvgdrvtitckasqdvsigvawyqqkpgkapklliysasyrytgvpsrfsg light chain 1 sgsgtdftltisslqpedfatyycqqyyiypytfgqgtkveikrtvaapsyfifppsdeqlksgtas vvcllnnfypreakvqwkvdnalqsgnsqesvteqdskdstyslsstltlskadyekhkvyac evthqglsspytksfnrgec scFab 138 evqlvesggglvqpggslrlscaasgfnikdtyihwyrqapgkglewvariyptngytryads Trastuzumab vkgrftisadtskntaylqmnslraedtavyycsrwggegfyamdywgqgtlvtvssastkg heavy chain 2 psvfplapsskstsggtaalgclvkdvfpepvtvswnsgaltsgvhtfpavlqssglyslssvvt vpssslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsvflfppkpkdt lmisrtpevtcvvvdvshedpevkfnwyydgvevhnaktkpreeqynstyrvvsvltvlhq dwlngkeykckvsnkalpapiektiskakgqprepqvytlpperdeltknqvslwclykgfy psdiavewesngqpennykttppvldsdgsfflyskltvdksrwqqgnyfscsvmhealhn hytqkslslspgkggggsggggsggggsggggsggggsggggsggdiqmtqspsslsasvg drvtitcrasqdynvavawyqqkpgkapklliysasflysgypsrfsgsrsgtdftltisslqped fatyycqqhyttpptfgqgtkveikrtvaapsvfifppsdeqlksgtasvvcllnnfypreakyq wkydnalqsgnsqesvteqdskdstyslsstltlskadyekhkvyacevthqglsspvtksfnr gec OAscFabPer1 Her2GlyMab scFab 139 evqlvesggglvqpggslrlscaasgftftdytmdwvrqapgkglewvadvnpnsggsiyn Pertuzumab qrfkgrftlsvdrskntlylqmnslraedtavyycarnlgpsfyfdywgqgtlvtvssastkgps heavy chain 1 vfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvtvp ssslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsvflfppkpkdtlm isrtpevtcvvvdvshedpevkfnwyvdgvevhnaktkpreeqynstyrvvsvltvlhqdwl ngkeykckvsnkalpapiektiskakgqprepqvctlppsrdeltknqvslscavkgfypsdi avewesngqpennykttppyldsdgsfflyskltvdksrwqqgnvfscsvmhealhnhytq kslslspgkggggsggggsggggsggggsggggsggggsggdiqmtqspsslsasvgdrvt itckasqdvsigvawyqqkpgkapklliysasyrytgvpsrfsgsgsgtdftltisslqpedfaty ycqqyyiypytfgqgtkveikrtvaapsvfifppsdeqlksgtasvvcllnnfypreakvqwk vdnalqsgnsqesvteqdskdstyslsstltlskadyekhkvyacevthqglsspytksfnrgec Trastuzumab 140 evqlvesggglvqpggslrlscaasgfnikdtyihwvrqapgkglewvariyptngytryads heavy chain 2 vkgrftisadtskntaylqmnslraedtavyycsrwggegfyamdywgqgtlytyssastkg psvfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvt vpssslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsvflfppkpkdt lmisrtpevtcvvvdvshedpevkfnwyydgvevhnaktkpreeqynstyrvvsvltvlhq dwlngkeykckvsnkalpapiektiskakgqprepqvytlppcrdeltknqvslwclkgfy psdiavewesngqpennykttppyldsdgsfflyskltvdksrwqqgnvfscsvmhealhn hytqkslslspgk Trastuzumab 141 diqmtqspsslsasvgdrvtitcrasqdvnvavawyqqkpgkapklliysasflysgypsrfsg light chain 2 srsgtdftltisslqpedfatyycqqhyttpptfgqgtkveikrtvaapsyfifppsdeqlksgtas vvcllnnfypreakvqwkydnalqsgnsqesvteqdskdstyslsstltlskadyekhkvyac evthqglsspvtksfnrge OAscFabPer2 Her2GlyMab Trastuzumab 142 evqlvesggglvqpggslrlscaasgfnikdtyihwvrqapgkglewvariyptngytryads heavy chain 1 vkgrftisadtskntaylqmnslraedtavyycsrwggegfyamdywgqgtlvtvssastkg psvfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvt vpssslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsvflfppkpkdt lmisrtpevtcvvvdvshedpevkfnwyvdgvevhnaktkpreeqynstyrvvsvltvlhq dwlngkeykckvsnkalpapiektiskakgqprepqvytlppcrdeltknqvslwclvkgfy psdiavewesngqpennykttppyldsdgsfflyskltvdksrwqqgnvfscsvmhealhn hytqkslslspgk Trastuzumab 143 diqmtqspsslsasvgdrvtitcrasqdvnvavawyqqkpgkapklliysasflysgypsrfsg light chain 1 srsgtdftltisslqpedfatyycqqhyttpptfgqgtkveikrtvaapsyfifppsdeqlksgtas vvcllnnfypreakvqwkydnalqsgnsqesvteqdskdstyslsstltlskadyekhkvyac evthqglsspvtksfnrgec scFab 144 evqlvesggglvqpggslrlscaasgftftdytmdwvrqapgkglewvadvnpnsggsiyn Pertuzumab qrfkgrftlsvdrskntlylqmnslraedtavyycarnlgpsfyfdywgqgtlvtvssastkgps heavy chain 2 vfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvtvp ssslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsvflfppkpkdtlm isrtpevtcvvvdvshedpevkfnwyydgvevhnaktkpreeqynstyrvvsvltvlhqdwl ngkeykckvsnkalpapiektiskakgqprepqvctlppsrdeltknqvslscavkgfypsdi avewesngqpennykttppvldsdgsfflyskltvdksrwqqgnvfscsvmhealhnhytq kslslspgkggggsggggsggggsggggsggggsggggsggdiqmtqspsslsasvgdrvt itckasqdysigvawyqqkpgkapklliysasyrytgvpsrfsgsgsgtdftltisslqpedfaty ycqqyyiypytfgqgtkveikrtvaapsyfifppsdeqlksgtasvvcllnnfypreakvqwk vdnalqsgnsqesvteqdskdstyslsstltlskadyekhkvyacevthqglsspvtksfnrgec CrossMab-XPer Her2G1yMab XPertuzumab 109 evqlvesggglvqpggslrlscaasgftftdytmdwvrqapgkglewvadvnpnsggsiyn heavy chain qrfkgrftlsydrskntlylqmnslraedtavyycarnlgpsfyfdywgqgtlvtvssasvaaps vfifppsdeqlksgtasvvcllnnfypreakvqwkvdnalqsgnsqesvteqdskdstyslsst ltlskadyekhkvyacevthqglsspvtksfnrgecdkthtcppcpapellggpsvflfppkpk dtlmisrtpevtcvvvdvshedpevkfnwyvdgvevhnaktkpreeqynstyrvvsvltvlh qdwlngkeykckvsnkalpapiektiskakgqprepqvytlpperdeltknqvslwclvkgf ypsdiavewesngqpennykttppvldsdgsfflyskltvdksrwqqgnvfscsvmhealh nhytqkslslspgk XPertuzumab 110 diqmtqspsslsasvgdrvtitckasqdvsigvawyqqkpgkapklliysasyrytgvpsrfsg light chain sgsgtdftltisslqpedfatyycqqyyiypytfgqgtkveikssastkgpsvfplapsskstsggt aalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvtypssslgtqtyicnynh kpsntkvdkkvepksc Trastuzumab 96 evqlvesggglvqpggslrlscaasgfnikdtyihwvrqapgkglewvariyptngytryads heavy chain vkgrftisadtskntaylqmnslraedtavyycsrwggegfyamdywgqgtlvtvssastkg (VHD98E CH1) psvfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvt vpssslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsvflfppkpkdt lmisrtpevtcvvvdvshedpevkfnwyydgvevhnaktkpreeqynstyrvvsvltvlhq dwlngkeykckvsnkalpapiektiskakgqprepqvctlppsrdeltknqvslscavkgfyp sdiavewesngqpennykttppvldsdgsfflyskltvdksrwqqgnvfscsvmhealhnh ytqkslslspgk Trastuzumab 86 diqmtqspsslsasvgdrvtitcrasqdvnvavawyqqkpgkapklliysasflysgvpsrfsg light chain srsgtdftltisslqpedfatyycqqhyttpptfgqgtkveikrtvaapsyfifppsdeqlksgtas (VL T31V CL) vvcllnnfypreakvqwkvdnalqsgnsqesvteqdskdstyslsstltlskadyekhkvyac evthqglsspvtksfnrgec CrossMab-XTra Her2GlyMab Pertuzumab 119 evqlvesggglvqpggslrlscaasgftftdytmdwvrqapgkglewvadvnpnsggsiyn heavy chain qrfkgrftlsydrskntlylqmnslraedtavyycarnlgpsfyfdywgqgtlvtvssasvaaps vfifppsdeqlksgtasvvcllnnfypreakvqwkvdnalqsgnsqesvteqdskdstyslsst ltlskadyekhkvyacevthqglsspvtksfnrgecdkthtcppcpapellggpsvflfppkpk dtlmisrtpevtcvvvdvshedpevkfnwyydgvevhnaktkpreeqynstyrvvsvltvlh qdwlngkeykckvsnkalpapiektiskakgqprepqvctlppsrdeltknqvslscavkgfy psdiavewesngqpennykttppvldsdgsfflvskltvdksrwqqgnvfscsvmhealhn hytqkslslspgk Pertuzumab 120 diqmtqspsslsasvgdrvtitckasqdvsigvawyqqkpgkapklliysasyrytgvpsrfsg light chain sgsgtdftltisslqpedfatyycqqyyiypytfgqgtkveikrtvaapsvfifppsdeqlksgtas vvcllnnfypreakvqwkydnalqsgnsqesvteqdskdstyslsstltlskadyekhkvyac evthqglsspytksfnrgec XTrastuzumab 121 evqlvesggglvqpggslrlscaasgfnikdtyihwvrqapgkglewvariyptngytryads heavy chain vkgrftisadtskntaylqmnslraedtavyycsrwggegfyamdywgqgtlvtvssastkg psvfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvt vpssslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsvflfppkpkdt lmisrtpevtcvvvdvshedpevkfnwyydgvevhnaktkpreeqynstyrvvsvltvlhq dwlngkeykckvsnkalpapiektiskakgqprepqvytlpperdeltknqvslwclvkgfy psdiavewesngqpennykttppvldsdgsfflyskltvdksrwqqgnvfscsvmhealhn hytqkslslspgk XTrastuzumab 122 diqmtqspsslsasvgdrvtitcrasqdvnvavawyqqkpgkapklliysasflysgvpsrfsg light chain srsgtdftltisslqpedfatyycqqhyttpptfgqgtkveikssastkgpsvfplapsskstsggta algclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvtvpssslgtqtyicnvnhk psntkvdkkvepksc Optimized Trastuzumab sequences: CrossMab-XTra Her2GlyMab and CrossMab-XPer Her2GlyMab Trastuzumab 117 evqlvesggglvqpggslrlscaasgfnikdtyihwyrqapgkglewvariyptngytryads VH (D98E) vkgrftisadtskntaylqmnslraedtavyycsrwggegfyamdywgqgtlvtvss Trastuzumab 115 astkgpsvfplapsskstsggtaalgclykdyfpepvtvswnsgaltsgvhtfpavlqssglysl CH1 ssvvtvpssslgtqtyicnvnhkpsntkvdkkvepkscdkth Trastuzumab 118 diqmtqspsslsasvgdrvtitcrasqdvnvavawyqqkpgkapklliysasflysgvpsrfsg VL (T31V) srsgtdftltisslqpedfatyycqqhyttpptfgqgtkveikr Trastuzumab 116 tvaapsvfifppsdeqlksgtasvvcllnnfypreakvqwkvdnalqsgnsqesvteqdskds CL tyslsstltlskadyekhkvyacevthqglsspvtksfnrgec Trastuzumab 20 gfnikdtyih VH CDR1 Trastuzumab 29 riyptngytryadsvkg VH CDR2 Trastuzumab 79 wggegfyamdy VH CDR3 Trastuzumab 104 rasqdvnvava VL CDR1 Trastuzumab 18 sasflys VL CDR2 Trastuzumab 19 qqhyttppt VL CDR3
CrossMab-CDRG Her2GlyMab XPertuzumab 109 evqlvesggglvqpggslrlscaasgftftdytmdwvrqapgkglewvadvnpnsggsiyn heavy chain qrfkgrftlsvdrskntlylqmnslraedtavyycarnlgpsfyfdywgqgtlvtvssasvaaps (VHCL) vfifppsdeqlksgtasvvcllnnfypreakvqwkvdnalqsgnsqesvteqdskdstyslsst ltlskadyekhkvyacevthqglsspvtksfnrgecdkthtcppcpapellggpsvflfppkpk dtlmisrtpevtcvvvdvshedpevkfnwyvdgvevhnaktkpreeqynstyrvvsvltvlh qdwlngkeykckvsnkalpapiektiskakgqprepqvytlpperdeltknqvslwclvkgf ypsdiavewesngqpennykttppvldsdgsfflyskltvdksrwqqgnvfscsvmhealh nhytqkslslspgk Pertuzumab 110 diqmtqspsslsasvgdrvtitckasqdvsigvawyqqkpgkapklliysasyrytgvpsrfsg light chain sgsgtdftltisslqpedfatyycqqyyiypytfgqgtkveikssastkgpsvfplapsskstsggt (VLCH1) aalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvtvpssslgtqtyicnvnh kpsntkvdkkvepksc Trastuzumab 111 qvqlvqsgaevkkpgasvkvsckasgfnikdtyihwvrqapgqglewmgriyptngytrya CDRG heavy qkfqgrvtmtrdtsistaymelsrlrsddtavyycsrwggegfyamdywgqgtmvtvssast chain kgpsvfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssv (VHCH1) vtvpssslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsvflfppkpk dtlmisrtpevtcvvvdvshedpevkfnwyvdgvevhnaktkpreeqynstyrvvsvltvlh qdwlngkeykckvsnkalpapiektiskakgqprepqvctlppsrdeltknqvslscavkgfy psdiavewesngqpennykttppvldsdgsfflvskltvdksrwqqgnvfscsvmhealhn hytqkslslspgk Trastuzumab 112 diqltqppsvsvapgqtaritcgasqdvstavawyqqkpgqapvlvvysasflysgipsrfsgs CDRG light rsgtdftltisrveagdeadyycqqhyttpptfgtgtkvtvlrtvaapsvfifppsdeqlksgtasv chain (VLCL) vcllnnfypreakvqwkvdnalqsgnsqesvteqdskdstyslssfttlskadyekhkvyace vthqglsspvtksfnrgec Trastuzumab 105 evqlvqsgaevkkpgasvkvsekasgfnikdtyihwvrqapgqglewmgriyptngytrya CDRG VH qkfqgrvtmtrdtsistaymelsrlrsddtavyycsrwggegfyamdywgqgtmvtvss (D98E, CDRG) Trastuzumab 115 astkgpsvfplapsskstsggtaalgelvkdyfpepvtvswnsgaltsgvhtfpavlqssglysl CH1 ssvvtvpssslgtqtyicnvnhkpsntkvdkkvepksedkth Trastuzumab 106 diqltqppsvsvapgqtaritcgasqdvstavawyqqkpgqapvlvvysasflysgipsrfsgs CDRG VL rsgtdftltisrveagdeadyycqqhyttpptfgtgtkvtvlr (N30T, CDRG) Trastuzumab 116 tvaapsvfifppsdeqlksgtasvvcllnnfypreakvqwkvdnalqsgnsqesvteqdskds CL tyslsstltlskadyekhkvyacevthqglsspvtksfnrgec Trastuzumab 20 gfnikdtyih CDRG VH CDR1 Trastuzumab 108 riyptngytryaqkfqg CDRG VH CDR2 Trastuzumab 79 wggegfyamdy CDRG VH CDR3 Trastuzumab 107 gasqdvstava CDRG VL CDR1 Trastuzumab 18 sasflys CDRG VL CDR2 Trastuzumab 19 qqhyttppt CDRG VL CDR3 Pertuzumab sequences in CrossMab-CDRG Her2GlyMab, CrossMab-XTra Her2GlyMab and CrossMab-XPer Her2GlyMab: see Pertuzumab wt (parent) sequences Table 32
Purification of Bispecific Antibodies
[0507] Bispecific antibodies were purified from cell culture supernatants by affinity chromatography using MabSelectSure-Sepharose.TM. (GE Healthcare, Sweden) and Superdex 200 size exclusion (GE Healthcare, Sweden) chromatography. Briefly, sterile filtered cell culture supernatants were captured on a MabSelect SuRe resin equilibrated with PBS buffer (10 mM Na.sub.2HPO.sub.4, 1 mM KH.sub.2PO.sub.4, 137 mM NaCl and 2.7 mM KCl, pH 7.4), washed with equilibration buffer and eluted with 25 mM sodium citrate at pH 3.0. The eluted protein fractions were pooled, neutralized with 2M Tris, pH 9.0 and further purified by size exclusion chromatography using a Superdex 200 26/60 GL (GE Healthcare, Sweden) column equilibrated with 20 mM histidine, 140 mM NaCl, pH 6.0. Size exclusion chromatography fractions were analysed by CE-SDS (Caliper Life Science, USA) and bispecific antibody containing fractions were pooled and stored at -80.degree. C.
Analysis of Purified Proteins
[0508] The protein concentration of purified protein samples was determined by measuring the optical density (OD) at 280 nm, using the molar extinction coefficient calculated on the basis of the amino acid sequence. Purity, antibody integrity and molecular weight of bispecific and control antibodies were analyzed by CE-SDS using microfluidic Labchip technology (Caliper Life Science, USA). 5 .mu.l of protein solution was prepared for CE-SDS analysis using the HT Protein Express Reagent Kit according manufacturer's instructions and analysed on LabChip GXII system using a HT Protein Express Chip. Data were analyzed using LabChip GX Software version 3.0.618.0. The aggregate content of bispecific and control antibody samples was analyzed by high-performance SEC using a Superdex 200 analytical size-exclusion column (GE Healthcare, Sweden) in 200 mM KH.sub.2PO.sub.4, 250 mM KCl, pH 7.0 running buffer at 25.degree. C. 25 .mu.g protein were injected on the column at a flow rate of 0.5 ml/min and eluted isocratic over 50 minutes.
Mass Spectrometry
[0509] The integrity of the amino acid backbone of reduced bispecific antibody light and heavy chains was verified by NanoElectrospray Q-TOF mass spectrometry after removal of N-glycans by enzymatic treatment with Peptide-N-Glycosidase F (Roche Molecular Biochemicals). The amount of heavy and light chain mis-pairing was also quantified.
Surface Plasmon Resonance
Instrument: BIAacore T100 (GE Healthcare)
[0510] Software: Biacore T100 Control, Version 2.02/2.03 [0511] Biacore T100 Evaluation, Version 2.02/2.03 [0512] Biacore B3000 (Biacore) [0513] Software: Biacore B3000 Control, Version 4.1.2 [0514] BIAEvaluation, Version 4.1.1
Assayformat Chip: CMS-Chip
[0515] Kinetic constants and resulting affinities of <Her2GlyMab>-molecules were measured for both "Trastuzumab"- and "Pertuzumab"--functionalities respecitively. These two functionalities were distinguished via pre-complexation of Her2 with either amine coupled parental MAb "Trastuzumab" (FC1/2) or "Pertuzumab" FC3/4). Complex formation of parental MAb and Her2 commensed after injection of Her2 ECD.
[0516] As a consequence of pre-complexed parental MAb "Trastuzumab"/Her2 all "Trastuzumab"-binding sites are saturated, but all "Pertuzumab"-binding sites are available and vice versa. Finally the binding of the "<Her2GlyMab>"-molecules to be analyzed was measured via injections using increasing concentrations with each cycle. Association and dissociation observed were calculated with a Langmuir 1:1 binding model. To minimize dissociation of Her2 during the measurements, the kinetic constants were measured at T=25.degree. C.
Amine Coupling of Capture Molecules
[0517] Standard amine coupling on flow cells 1 to 4 according to the manufacturer's instructions: CMS Chip, T=25.degree. C., running buffer: HBS-N buffer, activation by mixture of EDC/NHS, aimed at 800 RU; the parental Abs "Trastuzumab" or "Pertuzumab" were diluted in coupling buffer sodium acetate, pH 4.5, c=2-3 .mu.g/mL; finally remaining activated carboxyl groups were blocked by injection of 1 M Ethanolamine.
[0518] Chip surface on flow cell 1 and 3 (with either amine coupled parental mAb "Trastuzumab" or "Pertuzumab") were used as reference control surface for correction of possible buffer-effects or non-specific binding.
Kinetic Characterization of <Her2GlyMab> Molecules at 25.degree. C.
[0519] Running buffer: PBS
[0520] All samples were diluted with running buffer+1 mg/mL BSA.
[0521] Capturing of HER2 ECD on flow cells 2 and 4: c=100 nM, flow 5 .mu.l/min, time 120 sec.
[0522] Analyte samples: A classical concentration series of the <Her2GlyMab>-molecules were analyzed at five concentrations (c=300, 100, 33.33, 11.11 and 3.7 nM.) at a flow rate of 50 .mu.l/min was injected. Singles for each concentration, one as a duplicate; association time: 180 sec., dissociation time: 900 sec.
[0523] Final regeneration was performed after each cycle using 10 mM Glycin pH 2.5 for amine coupled Mab "Trastuzumab" and 25 mM NaOH for amine coupled Mab "Pertuzumab", contact time each 60 sec, flow rate 30 .mu.l/min.
[0524] Kinetic parameters were calculated by using double referencing (control reference: binding of analyte to Mabs Trastuzumab and Pertuzumab respectively; Flow Cell: 1 respectively 3), concentration "0" used as the blank. Calculations were performed with model `Langmuir binding 1:1, RI (refractive index)=0.
Results: Expression & Purification Bispecific, Bivalent <Her2GlyMab> Antibody Molecules
[0525] According the procedures described in the materials and methods above, the bispecific, bivalent <Her2GlyMab> antibody molecules OAscFab1, OAscFab2, OAscFabPer1, OAscFabPer2, CrossMab-XPer, CrossMab-XTra and CrossMab-CDRG were expressed and purified. In each molecule the VH and VL of part are based on optimized Trastuzumab sequences with mutations T31V or N30T in the variable light chain and mutation D98E in the heavy chain and the Pertuzumab parent sequence respectively. The schematic structure of the antibodies is shown in FIGS. 10A-B. The sequences are shown in Table 8 above.
[0526] Expression of <Her2GlyMab> antibody molecules OAscFab1 Her2 GlyMab, OAscFab2 Her2 GlyMab, OAscFabPer1 Her2 GlyMab, OAscFabPer2 Her2 GlyMab (all glycoengineered, Knobs-into-holes, with the Trastuzumab scFab bearing D98E and T31V mutations), CrossMab-XPer Her2 GlyMab, CrossMab-XTra Her2 GlyMab (both glycoengineered, Knobs-into-holes, with the Trastuzumab cross-Fab bearing D98E and T31V mutations), and CrossMab-CDRG Her2 GlyMab (glycoengineered, Knobs-into-holes, CDR grafted, with the Trastuzumab cross-Fab bearing D98E and N30S mutations) was confirmed by western blotting and HP-SEC (FIGS. 11A-B). Purification of OAscFab1, OAscFab2, OAscFabPer1, OAscFabPer2, CrossMab-XPer, CrossMab XTra and CrossMab-CDRG led to the yields shown in Table 9. All OAscFab constructs showed less than 90% monomer post purification, the reduced purity of these molecules could not be increased by optimization of the plasmid ratios in expression (data not shown). However, optimization of the plasmid chain ratios in expression proved to increase the monomeric fraction present post purification of the CrossMab antibodies as described below.
TABLE-US-00012 TABLE 9 Her2GlyMab Purification -- Analysis of the percentage aggregation post protein A and SEC purification using HP-SEC and the respective protein yields calculated by UV spectroscopy A280 of the glycoengineered constructs. Purity Expression Purity Expression post yield post post yield post protein A protein A SEC SEC Name (%) (mg/L) (%) (mg/L) OAscFab1 48.4 144 90 31.6 (SEQ ID NOs 133, 134, 135) OAscFab2 41 42.9 93.6 12.6 (SEQ ID NOs 136, 137, 138) OAscFabPerl 41.6 43.4 88.8 4.6 (SEQ ID NOs 139, 140, 141) OAscFabPer2 38.2 34.2 86.7 6.6 (SEQ ID NOs 142, 143, 144) CrossMab-XTra 65.1 28.4 73 10.6 (SEQ ID NOs 119, 120, 121, 122) CrossMab-XPer 76.4 30.9 85 17.9 (SEQ ID NOs 109, 110, 96, 86) CrossMab-CDRG 73 31.5 95 11.9 (SEQ ID NOs 109, 110, 111, 112)
Example 7: Expression & Purification Bispecific, Bivalent <Her2GlyMab> Antibody Molecules Optimization of the Plasmid Ratios Used in Expression
CrossMab-XTra
[0527] According the procedures described in the materials and methods above, the bispecific, bivalent <Her2GlyMab> antibody molecule CrossMab-XTra, was expressed with molar plasmid ratios of 1:1:1:1, 1:1:1:2, 1:1:1:4 and 1:1:1:8 and purified. Expression of CrossMab-XTra was confirmed by Western blot. After Protein A purification of cell culture supernatants the construct showed at a 1:1:1:1 equimolar plasmid ratio approximately 73% of bispecific antibody with the expected molecular weight of approximately 148 kDa as detected by Q-TOF MS; 11% containing 2.times. Pertuzumab light chains paired with Pertuzumab and XTrastuzumab heavy chains; 9% intact XTrastuzumab antibodies (both heavy and light chains originating from XTrastuzumab) with the formation of the heavy chain hole-hole association; 4% XTrastuzumab heavy chain hole-hole association combined with Pertuzumab light chains only; and 3% XTrastuzumab heavy chain hole-hole association with 1.times. XTrastuzumab light chain and 1.times. Pertuzumab light chain.
[0528] The 1:1:1:2 plasmid ratio where the molar ratio of the crossed Trastuzumab light chain (XHerLC) was expressed 2-fold, showed approximately 81% of bispecific antibody with the expected molecular weight of approximately 148 kDa as detected by Q-TOF MS; 1% containing 2.times. Pertuzumab light chains paired with Pertuzumab and XTrastuzumab heavy chains; 16% intact XTrastuzumab antibodies (both heavy and light chains originating from XTrastuzumab) with the formation of the heavy chain hole-hole association; XTrastuzumab heavy chain hole-hole association combined with Pertuzumab light chains only were not detected; and 1% XTrastuzumab heavy chain hole-hole association with 1.times. XTrastuzumab light chain and 1.times. Pertuzumab light chain.
[0529] The 1:1:1:4 plasmid ratio where the molar ratio of the crossed Trastuzumab light chain was expressed 4-fold, showed approximately 64% of bispecific antibody with the expected molecular weight of approximately 148 kDa as detected by Q-TOF MS; 2.times. Pertuzumab light chains paired with Pertuzumab and XTrastuzumab heavy chains was not detected, however, 12% 2.times. XTrastuzumab light chains paired with Pertuzumab and XTrastuzumab heavy chains was detected for the first time at this ratio; 24% intact XTrastuzumab antibodies (both heavy and light chains originating from XTrastuzumab) with the formation of the heavy chain hole-hole association; XTrastuzumab heavy chain hole-hole association combined with Pertuzumab light chains only were not detected; XTrastuzumab heavy chain hole-hole association with 1.times. XTrastuzumab light chain and 1.times. Pertuzumab light chain were not detected.
[0530] The 1:1:1:8 plasmid ratio where the molar ratio of the crossed Trastuzumab light chain was expressed 8-fold, showed approximately 45% of bispecific antibody with the expected molecular weight of approximately 148 kDa as detected by Q-TOF MS; 2.times. Pertuzumab light chains paired with Pertuzumab and XTrastuzumab heavy chains was not detected, however, 28% 2.times. XTrastuzumab light chains paired with Pertuzumab and XTrastuzumab heavy chains was detected for the first time at this ratio; 27% intact XTrastuzumab antibodies (both heavy and light chains originating from XTrastuzumab) with the formation of the heavy chain hole-hole association; XTrastuzumab heavy chain hole-hole association combined with Pertuzumab light chains only were not detected; XTrastuzumab heavy chain hole-hole association with 1.times. XTrastuzumab light chain and 1.times. Pertuzumab light chain were not detected.
TABLE-US-00013 TABLE 10 CrossMab-XTra Her2GlyMab-Q-TOF analysis and quantification of the by-product profile of the Her2GlyMab antibody constructs comparing the optimization plasmid titration of the crossed Trastuzumab light chain (XHer LC) N.D. -- Not Detected CrossMab-XTra CrossMab-XTra CrossMab-XTra CrossMab-XTra Detected protein Her2GlyMab Her2GlyMab Her2GlyMab Her2GlyMab species in MS 1:1:1:1 1:1:1:2 1:1:1:4 1:1:1:8 1 .times. XHer HC; 1 .times. N.D. N.D. ~12% ~28% Per HC; 2 .times. XHer LC 2 .times. XHer HC; 2 .times. ~9% ~16% ~24% ~27% XHer LC CrossMab-XTra ~73% ~81% ~64% ~45% Her2GlyMab (100%) 2 .times. XHer HC; 1 .times. ~3% ~1% N.D. N.D. XHer LC; 1 .times. Per HC 1 .times. XHer HC; 1 .times. ~11% ~1% N.D. N.D. Per HC; 2 .times. Per LC 2 .times. XHer HC; 2 .times. ~4% N.D. N.D. N.D. Per LC
CrossMab-XPer
[0531] After Protein A purification of cell culture supernatants the construct showed at a 1:1:1:1 equimolar plasmid ratio approximately 85% of bispecific antibody with the expected molecular weight of approximately 148 kDa as detected by Q-TOF MS; 2% containing 2.times. XPertuzumab light chains paired with XPertuzumab and Trastuzumab heavy chains; 1% intact Trastuzumab antibodies (both heavy and light chains originating from Trastuzumab) with the formation of the heavy chain hole-hole association; 12% 2.times. Trastuzumab light chains paired with XPertuzumab and Trastuzumab heavy chains. No further species were detected.
[0532] The 1:1:1:2 plasmid ratio where the molar ratio of the crossed Pertuzumab light chain (XPerLC) was expressed 2-fold, showed approximately 89% of bispecific antibody with the expected molecular weight of approximately 148 kDa as detected by Q-TOF MS; 7% containing 2.times. XPertuzumab light chains paired with XPertuzumab and Trastuzumab heavy chains; intact Trastuzumab antibodies (both heavy and light chains originating from Trastuzumab) with the formation of the heavy chain hole-hole association were not detected; 4% 2.times. Trastuzumab light chains paired with XPertuzumab and Trastuzumab heavy chains.
[0533] The 1:1:1:4 plasmid ratio where the molar ratio of the crossed Pertuzumab light chain (XPerLC) was expressed 4-fold, showed approximately 74% of bispecific antibody with the expected molecular weight of approximately 148 kDa as detected by Q-TOF MS; 25% containing 2.times. XPertuzumab light chains paired with XPertuzumab and Trastuzumab heavy chains; intact Trastuzumab antibodies (both heavy and light chains originating from Trastuzumab) with the formation of the heavy chain hole-hole association were not detected; 1% 2.times. Trastuzumab light chains paired with XPertuzumab and Trastuzumab heavy chains.
[0534] The 1:1:1:8 plasmid ratio where the molar ratio of the crossed Pertuzumab light chain (XPerLC) was expressed 8-fold, showed approximately 52% of bispecific antibody with the expected molecular weight of approximately 148 kDa as detected by Q-TOF MS; 48% containing 2.times. XPertuzumab light chains paired with XPertuzumab and Trastuzumab heavy chains; intact Trastuzumab antibodies (both heavy and light chains originating from Trastuzumab) with the formation of the heavy chain hole-hole association were not detected; 2.times. Trastuzumab light chains paired with XPertuzumab and Trastuzumab heavy chains were not detected.
CrossMab-CDRG
[0535] After Protein A purification of cell culture supernatants the construct showed at a 1:1:1:1 equimolar plasmid ratio approximately 95% of bispecific antibody with the expected molecular weight of approximately 148 kDa as detected by Q-TOF MS and 5% containing 2.times. XPertuzumab light chains paired with XPertuzumab and Trastuzumab (HerCDRG) heavy chains. No further species were detected, therefore, further optimization of the plamid ratios was not performed. The MS spectra performed on the antibodies CrossMab-XHer and CrossMab-CDRG to compare the byproduct profile can be seen in FIGS. 12A-B.
TABLE-US-00014 TABLE 11 CrossMab-XPer Her2GlyMab-Q-TOF analysis and quantification of the by- product profile of the Her2GlyMab antibody constructs comparing the optimization plasmid titration of the crossed Trastuzumab light chain (XHer LC) N.D. -- Not Detected CrossMab-XPer CrossMab-XPer CrossMab-XPer CrossMab-XPer Detected protein Her2GlyMab Her2GlyMab Her2GlyMab Her2GlyMab species in MS 1:1:1:1 1:1:1:2 1:1:1:4 1:1:1:8 1 .times. Her HC; 1 .times. ~2% ~7% ~25% ~48% XPer HC; 2 .times. XPer LC 2 .times. Her HC; 2 .times. ~1% n.d. n.d. n.d. Her LC CrossMab-XPer ~85% ~89% ~74% ~52% Her2GlyMab (100%) 1 .times. Her HC; 1 .times. ~12% ~4% ~1% n.d. XPer HC; 2 .times. Her LC
TABLE-US-00015 TABLE 12 CrossMab-CDRG Her2GlyMab-Q-TOF analysis and quantificaion of the by-product profile of the Her2GlyMab antibody construct detecting the presence of mispaired antibody heavy and light chains. N.D. -- Not Detected CrossMab- CDRG Her2GlyMab Detected protein species in MS 1:1:1:1 2 .times. XPer HC; 2 .times. XPer LC N.D. 2 .times. HerCDRG HC; 2 .times. HerCDRG LC N.D. CrossMab-CDRG Her2GlyMab (100%) ~95% 1 .times. HerCDRG HC; 1 .times. XPer HC; 2 .times. XPer LC ~5%
Example 8: Simultaneous Binding of Bispecific Antibodies to Both Antigens
[0536] The binding of the bispecific antibody was analyzed via BIAcore as described above.
[0537] In separate assay format samples (CrossMabXPer as well as for OAscFab1 and OAscFab2) were proven to be functional for Trastuzumab as well as Pertuzumab specificity. CrossMabXPer showed kinetic constants and resulting affinities in the same order of magnitude as the positive controls. Except for a slightly reduced k.sub.a-rate constant for the Trastuzumab mediated binding OAscFab1 and OAscFab2 showed kinetic constants and resulting affinities in the same order of magnitude as the positive controls i.e the parental Mabs.
[0538] Partly bivalent binding of the positive controls-depending on the ligand density on the CM5-Chip may cause the variation of the dissociation rate constants in the two experiments depicted in this example.
TABLE-US-00016 TABLE 13 SPR analysis of the Her2GlyMab affinities - The association and dissociation rates of the antibodies were measure using a BIAcoreT100 with a CM5-Chip at 25.degree. C. T = 25.degree. C. analyzed -- k.sub.a k.sub.d t(1/2) K.sub.D Experiment function analyte [M.sup.-1 s.sup.-1] [s.sup.-1] [min] [M] UJ2530 "Trastuzumab" Pertuzumab no binding as expected UJ2530 "Trastuzumab" CrossMabXPer 1.0E+05 7.6E-05 151.9 7.4E-10 UJ2530 "Pertuzumab" Trastuzumab no binding as expected UJ2530 "Pertuzumab" Pertuzumab 4.7E+05 9.8E-05 118.1 2.1E-10 UJ2530 "Pertuzumab" CrossMabXPer 2.0E+05 1.8E-04 66.0 8.7E-10 UJ2530_b "Trastuzumab" Trastuzumab 7.0E+05 3.0E-05 382.9 4.3E-11 UJ2530_b "Trastuzumab" Pertuzumab no binding as expected UJ2530_b "Trastuzumab" OAscFab1 1.6E+04 8.7E-05 133.5 5.4E-09 UJ2530_b "Trastuzumab" OAscFab2 1.8E+04 1.1E-04 100.6 6.2E-09 UJ2530_b "Pertuzumab" Trastuzumab no binding as expected UJ2530_b "Pertuzumab" Pertuzumab 2.4E+05 2.6E-04 44.8 1.1E-09 UJ2530_b "Pertuzumab" OAscFab1 1.2E+05 2.6E-04 44.3 2.2E-09 UJ2530_b "Pertuzumab" OAscFab2 1.7E+05 2.6E-04 44.8 1.5E-09
Example 9: In Vitro Evaluation of 1+1 Herceptarg CrossMAb and Glyocoengineered Herceptarg Crossmab
Proliferation Inhibition Assay
[0539] AlamarBlue.RTM. (Invitrogen) was used for the measurement of the metabolic activity and proliferation of (A) BT474 and (B) N87 cells after a 5 day incubation in presence of HER2 CrossMab (CrossMab-XTra Her2GlyMab, SEQ ID NOs 119, 120, 121, 122), Trastuzumab, Pertuzumab or the combination of Trastuzumab/Pertuzumab. The bioreduction of the dye reduces the amount of the oxidized form (blue) and concomitantly increases the fluorescent intermediate (red).
[0540] Target cells were harvested, washed, resuspended in RPMI 1640 (Gibco)+10% FCS+1% GlutaMAX.TM. (Gibco) and plated at a concentration of 1.times.10.sup.4 cells/well. Cells were incubated for 3 hours in the cell incubator before respective antibody dilutions were added. Plates were gently shaked and incubated for 5 days in the cell incubator.
[0541] 25 .mu.l/well of Alamar Blue were added to the plate and incubated for 7 h in the incubator.
[0542] Absorbance was monitored at 584 nm and 612 nm in a Wallac Victor3 1420 Multilabel Counter.
[0543] For calculation of the percentage of proliferation inhibition, non-treated controls samples were included in the assay and defined as 100% proliferation. The average percentage of proliferation inhibition of the triplicates of each experiment was calculated.
[0544] Results are shown in FIGS. 13A-B.
ADCC Assay
[0545] ADCC mediated by HER2 CrossMab (CrossMab-XTra Her2GlyMab, SEQ ID NOs 119, 120, 121, 122), Trastuzumab, Pertuzumab or the combo of Trastuzumab/Pertuzumab was assessed on KPL-4 (A), T47D (B) and Calu-3 (C) cells.
[0546] Target cells were harvested, washed, resuspended in AIM V.RTM. medium (Life Technologies), and plated at a concentration of 3.times.10.sup.4 cells/well. The respective antibody dilutions were added in triplicates to the cells and incubated for 10 min before addition of the effector cells (peripheral blood mononuclear effector cells [PBMCs]). Effector (E) and target (T) cells were then incubated for 4 h at 37.degree. C. at an E:T ratio of 25:1 (triplicates for all samples). Lactate dehydrogenase (LDH) release was measured using the LDH Cytotoxicity Detection Kit (Roche Applied Science). ADCC was calculated using the following formula:
Percentage ADCC = ( [ sample release - spontaneous release maximal release - spontaneous release ] ) .times. 1 00. ##EQU00002##
[0547] Spontaneous release, corresponding to target cells incubated with effector cells without antibody, was defined as 0% cytotoxicity, with maximal release (target cells lysed with 1% Triton X-100) defined as 100% cytotoxicity. The average percentage of ADCC and standard deviations of the triplicates of each experiment were calculated. Results are shown in FIGS. 14A-C.
Example 10: In Vivo Characterization of HER2 CrossMab: Effect of Bispecific Antibodies Targeting HER2 on Tumor Growth in Calu3 Lung Cancer and KPL4 Breast Cancer Xenograft
In Vitro Cultured Cells--Calu3
[0548] This human lung adenocarcinoma cancer cell line has been established from a human caucasian male with lung cancer. Cells were obtained from Chugai Pharmaceuticals Co., Ltd. and passaged in house for working cell bank. Tumor cells are routinely cultured in RPMI medium (PAN Biotech, Germany) supplemented with 10% fetal bovine serum glutamine (PAN Biotech, Germany) at 37.degree. C. in a water-saturated atmosphere at 5% CO2. Culture passage is performed with trypsin/EDTA 1.times. (PAN) splitting twice/week. Cell passage P6 is used for in vivo study.
In Vitro Cultured Cells--KPL-4
[0549] This human breast cancer cell line has been established from the malignant pleural effusion of a breast cancer patient with an inflammatory skin metastasis. Cells have been provided by Professor J. Kurebayashi (Kawasaki Medical School, Kurashiki, Japan).Tumor cells are routinely cultured in DMEM medium (PAN Biotech, Germany) supplemented with 10% fetal bovine serum (PAN Biotech, Germany) and 2 mM L-glutamine (PAN Biotech, Germany) at 37.degree. C. in a water-saturated atmosphere at 5% CO2. Culture passage is performed with trypsin/EDTA 1.times. (PAN) splitting twice/week. Cell passage P6 is used for in vivo study.
Animals
[0550] Female SCID beige (C.B.-17) mice; age 10-12 weeks; body weight 18-20 g (Charles River Germany, Sulzfeld) or female BALB/C nu/nu mice; age 8-10 weeks; body weight >20 g (Bomholtgard, Denmark) are maintained under specific-pathogen-free condition with daily cycles of 12 h light/12 h darkness according to international guidelines (GV-Solas; Felasa; TierschG). After arrival animals are housed in the quarantine part of the animal facility for one week to get accustomed to new environment and for observation. Continuous health monitoring is carried out on regular basis. Diet food (Alltromin) and water (acidified pH 2.5-3) are provided ad libitum. The experimental study was reviewed and approved by local government; registration no. 55.2-1-54-2531.2-3-08 Scid-beige orthotop rodent breast cancer model and 211-2531.2-16/00. 1.2.2 subcutan tumor model.
Tumor Cell Injection
[0551] At the day of injection tumor cells are harvested (trypsin-EDTA) from culture flasks (Greiner TriFlask) and transferred into 50 ml culture medium, washed once and resuspended in PBS. After an additional washing step with PBS and filtration (cell strainer; Falcon O100 .mu.m) the final cell titer is adjusted to 1.5.times.108/ml. Tumor cell suspension is carefully mixed with transfer pipette to avoid cell aggregation. Anesthesia is performed using a Stephens inhalation unit for small animals with preincubation chamber (plexiglas), individual mouse nose-mask (silicon) and not flammable or explosive anesthesia compound Isoflurane (Pharmacia-Upjohn, Germany) in a closed circulation system. Two days before injection, coat of the SCID beige mice are shaved. For subcutaneous injection of Calu3 cells, skin of anaesthetized animals is carefully lifted up with an anatomic forceps and 100 .mu.l cell suspension (=5.0.times.10e6 cells) is injected subcutaneously in the right flank of the animals. Cell suspension is filled into a 1.0 ml tuberculin syringe (Braun, Melsungen) using a wide injection needle (0.45.times.25 mm). KPL-4 cells (3.times.10e6 cells) are injected orthotopically in a volume of 20 .mu.l into the right penultimate inguinal mammary fat pad of each anesthetized mouse. For the orthotopic implantation, the cell suspension is injected through the skin under the nipple using a using a Hamilton microliter syringe and a 30Gx1/2'' needle.
Monitoring
[0552] Animals are controlled daily for detection of clinical symptoms of adverse effects. For monitoring throughout the experiment the body weight of the animals is documented two times weekly and the tumor volume is measured by caliper twice weekly. Tumor volume was calculated according to NCI protocol (Tumor weight=1/2ab.sup.2, where "a" and "b" are the long and the short diameters of the tumor, respectively).Termination criteria were the critical tumor mass (up to 1.7 g or O>1.5 cm), body weight loss more than 20% from baseline, tumor ulceration or poor general condition of the animals. Study exclusion criteria for the animals are described and approved in the corresponding "Tierversuchsanzeige".
Treatment of Animals
[0553] Mice were randomized for tumor volume, for KPL-4 a mean of 80 mm.sup.3, for Calu3 a mean of 100 mm.sup.3. Mice were treated once weekly with a volume of 10 ml/kg intra peritoneal. For combination treatment Trastuzumab was given first and Pertuzumab was given 24 hrs thereafter. Results are shown in Tables 14 to 17 and FIGS. 15A-C, 16A-C, 17A-C and 18A-C.
TABLE-US-00017 TABLE 14 BispecHer2_Pz_Calu3_001 (FIGS. 15A-C): CrossMAb_003 non-ge: non glycoengineered CrossMab-XTra Her2GlyMab (SEQ ID NOs 119, 120, 121, 122), negative control: anti-IgE antibody (Omalizumab), Herceptarg 2 + 2 OmniE: Pertuzumab antibody with the Trastuzumab scFV added onto the c-terminus of the heavy chains. The trastuzumab scFv contains a stabilizing disulphide bond between VH 105-VL 43 (SEQ ID NOs: 145, 146), TvAb12 and TvAb20: scFv 2 + 2 HER2 bispecific antibodies, see example 2. Cumulative No of Dose Route/Mode of No of dose Group animals Compound (mg/kg) administration treatments (mg/kg) 1 8 negative control 10 i.p. once weekly 7 70 2 8 Trastuzumab + 10 i.p. once weekly 7 70 Pertuzumab 10 i.p. once weekly 7 70 3 8 Herceptarg 2 + 2 13.5 i.p. once weekly 7 94.5 OmniE 4 8 CrossMAb_003 20 i.p. once weekly 7 140 non-ge 5 8 TvAb12 11.3 i.p. once weekly 7 79.1 6 8 TvAb20 11.4 i.p. once weekly 7 79.8
TABLE-US-00018 TABLE 15 BispecHER2_PZ_KPL-4_002 (FIGS. 16A-C): CrossMAb_003 non-ge: non glycoengineered CrossMab-XTra Her2GlyMab (SEQ ID NOs 119, 120, 121, 122), negative control: anti-IgE antibody (Omalizumab), Herceptarg 2 + 2 OmniE: Pertuzumab antibody with the Trastuzumab scFV added onto the c-terminal of the heavy chains. The trastuzumab scFv contains a stabilizing disulphide bond between VH 105-VL 43 (SEQ ID NOs: 145, 146), TvAb12 and TvAb20: scFv 2 + 2 HER2 bispecific antibodies, see example 2. Cumulative No of Dose Route/Mode of No of dose Group animals Compound (mg/kg) administration treatments (mg/kg) 1 9 Negative control 10 i.p. once weekly 5 50 2 9 Trastuzumab + 10 i.p. once weekly 5 50 Pertuzumab 10 i.p. once weekly 50 3 9 Herceptarg 2 + 2 13.5 i.p. once weekly 5 67.5 OmniE 4 9 CrossMAb_003 20 i.p. once weekly 5 100 non-ge 5 9 TvAb12 11.3 i.p. once weekly 5 56.5 6 9 TvAb20 11.42 i.p. once weekly 5 57.1
TABLE-US-00019 TABLE 16 Bispec.HER2_PZ_KPL-4_003 (FIGS. 17A-C): CrossMAb_005 non glycoengineered CrossMab-XTra Her2GlyMab (SEQ ID NOs 119, 120, 121, 122), negative control: anti-IgE antibody (Omalizumab). Cumulative No of Dose Route/Mode of No of dose Group animals Compound (mg/kg) administration treatments (mg/kg) 1 10 Negative control 10 i.p. once weekly 5 50 2 10 Trastuzumab + 10 i.p. once weekly 5 50 Pertuzumab 10 i.p. once weekly 50 3 10 Her2_Crossmab_005 20 i.p. once weekly 5 100 4 10 Her2_Crossmab_005 10 i.p. once weekly 5 50 5 10 Her2_Crossmab_005 5 i.p. once weekly 5 25 6 10 Her2_Crossmab_005 1 i.p. once weekly 5 5
TABLE-US-00020 TABLE 17 Exploratory_PZ_KPL-4_009 (FIGS. 18A-C): negative control: anti-IgE antibody (Omalizumab), TvAb16 and TvAb20: scFv 2 + 2 HER2 bispecific antibodies, see example 2. Cumulative No of Dose Route/Mode of No of dose Group animals Compound (mg/kg) administration treatments (mg/kg) 1 10 Negative 10 i.p. once weekly 4 40 control 2 10 Trastuzumab + 10 i.p. once weekly 4 40 Pertuzumab 10 i.p. once weekly 4 40 3 10 Trastuzumab + 5 i.p. once weekly 4 20 Pertuzumab 5 i.p. once weekly 4 20 4 10 TvAb16 10 i.p. once weekly 4 40 5 10 TvAb16 5 i.p. once weekly 4 20 6 10 TvAb20 10 i.p. once weekly 4 40 7 10 TvAb20 5 i.p. once weekly 4 20
Example 11: Generation of a Common Light Chain for Trastuzumab and Pertuzumab
Gene Synthesis
[0554] Desired gene segments, where required, were either generated by PCR using appropriate templates or were synthesized at Geneart AG (Regensburg, Germany) from synthetic oligonucleotides and PCR products by automated gene synthesis. In cases where no exact gene sequence was available, oligonucleotide primers were designed based on sequences from closest homologues and the genes were isolated by RT-PCR from RNA originating from the appropriate tissue. The gene segments flanked by singular restriction endonuclease cleavage sites were cloned into standard cloning/sequencing vectors. The plasmid DNA was purified from transformed bacteria and concentration determined by UV spectroscopy. The DNA sequence of the subcloned gene fragments was confirmed by DNA sequencing. Gene segments were designed with suitable restriction sites to allow subcloning into the respective expression vectors. All constructs were designed with a 5'-end DNA sequence coding for a leader peptide which targets proteins for secretion in eukaryotic cells. SEQ ID NOs: 155, 156, and 157 give exemplary leader peptides.
Cloning of Antigen Expression Vectors
[0555] A DNA fragment encoding amino acids 1 to 629 of matured Tyrosine kinase-type cell surface receptor HER2 (Her2, Uniprot: P04626) was cloned in frame into a mammalian recipient vector containing an N-terminal leader sequence. In addition, the construct contains a C-terminal avi-tag allowing specific biotinylation during co-expression with Bir A biotin ligase and a His-tag used for purification by immobilized-metal affinity chromatography (IMAC) (SEQ ID NOs 1 and 2).
[0556] The antigen expression is generally driven by an MPSV promoter and transcription is terminated by a synthetic polyA signal sequence located downstream of the CDS. In addition to the expression cassette, each vector contains an EBV oriP sequence for autonomous replication in EBV-EBNA expressing cell lines.
Production and Purification of Antigens and Antibodies
[0557] Both antigens and antibodies were transiently transfected into HEK 293 cells, stably expressing the EBV-derived protein EBNA. A simultaneously co-transfected plasmid encoding biotin ligase Bir A allowed avi tag-specific biotinlylation in vivo. The proteins were then purified using a protein A column followed by gel filtration.
Design of Trastuzumab/Pertuzumab Common Light Chains
[0558] For the generation of a common light chain (CLC) for Trastuzumab and Pertuzumab, the individual light chains (LC) were analyzed and compared. Sequence analysis revealed that both variable domains originate from the same germline sequence. Given that the Trastuzumab LCDR3 in general and residue H91 in particular interact specifically with the Trastuzumab-specific epitope on Her2, the first attempt to create a Trastuzumab/Pertuzumab CLC was done as follows: Either the complete LCDR3 region or only residue H91 of Trastuzumab substituted the corresponding positions in the Pertuzumab LC. As a result, a hybrid LC construct was created encoding Pertuzumab-derived LCDR1 and 2 but harboring Trastuzumab-derived LCDR3 amino acid residues. The resulting CLCs, named either "Pertuzumab (Tras.L3) LC" (DNA sequence of variable domains listed as SEQ ID NO: 25) or "Pertuzumab (Tras.Y91H) LC" (SEQ ID NO: 27), were co-expressed with either the Trastuzumab or the Pertuzumab HC. The resulting four antibodies (protein sequence of variable domains listed as SEQ ID NOs: 22 and 26, "Pertuzumab HC".times."Pertuzumab (Tras.L3) LC"; SEQ ID NO: 92 and 26, "Trastuzumab HC".times."Pertuzumab (Tras.L3) LC"; SEQ ID NOs: 22 and 28, "Pertuzumab HC".times."Pertuzumab (Trast.Y91H) LC"; SEQ ID NO: 92 and 28, "Trastuzumab HC".times."Pertuzumab (Tras.Y91H) LC") were purified from mammalian-derived cell culture supernatant and binding to Her2 was measured and compared with the respective parental antibodies (SEQ ID NOs: 22 and 24, "Pertuzumab HC".times."Pertuzumab LC"; SEQ ID NO: 92 and 82, "Trastuzumab HC".times."Trastuzumab LC") by SPR.
Affinity-Determination by SPR Using BioRad's ProteOn XPR36 Biosensor
[0559] The Affinity (K.sub.D) of the new antibody chain combinations was measured by surface plasmon resonance using a ProteOn XPR36 instrument (Biorad) at 25.degree. C. In a first step, 6500 RU of anti-human IgG (Sigma 12136, polyclonal goat antibody) recognizing hu IgG (Fc-specific) was immobilized on all 6 channels of a GLM chip by Amine coupling(NaAcetate pH4, 30 .mu.l/min, 300s) (vertical orientation).
[0560] Each antibody was diluted with PBST (10 mM phosphate, 150 mM sodium chloride pH 7.4, 0.005% Tween 20) to 2 .mu.g/ml, and then injected for 60s at 30 .mu.l/minute to achieve immobilization levels of about 400 response units (RU) in vertical orientation. Injection of Her2: For one-shot kinetics measurements, injection direction was changed to horizontal orientation, three-fold dilution series of purified Her2 (varying concentration ranges between 300 and 3.7 nM) were injected simultaneously at 100 .mu.l/min along separate channels 1-5, with association times of 180s, and dissociation times of 600s. Buffer (PBST) was injected along the sixth channel to provide an "in-line" blank for referencing. Regeneration was performed by two pulses of 10 mM glycine pH 1.5 and 50 mM NaOH for 30s at 1000/min (horizontal orientation). Association rate constants (k.sub.on) and dissociation rate constants (k.sub.off) were calculated using a simple one-to-one Langmuir binding model in ProteOn Manager v3.1 software by simultaneously fitting the association and dissociation sensorgrams. The equilibrium dissociation constant (K.sub.D) was calculated as the ratio k.sub.on/k.sub.on.
[0561] As expected, both control antibodies Trastuzumab and Pertuzumab recognized Her2 with their known affinities in the low nanomolar or sub-nanomolar range. However, while the affinity of Pertuzumab HC in combination with the newly designed "Pertuzumab (Tras.Y91H) LC" was slightly reduced, the same light chain in combination with Trastuzumab HC did not yield any detectable binding. This indicates that a single Trastuzumab-derived point mutation (Y91H) in the Pertuzumab LC is not sufficient translate binding to Her2 in this chain combination. This is in contrast to the second CLC variant "Pertuzumab LC (Trast. L3)" which resulted in weak binding when combined with Trastuzumab HC, while binding was reduced but clearly visible when co-expressed with the Pertuzumab HC. A summary of the kinetic and thermodynamic measurements is given in FIGS. 19A-F and Table 18. Based on this finding, the CLC "Pertuzumab LC (Trast. L3)" was further modified in order to restore Her2 binding in combination with Trastuzumab HC while affecting the affinity as little as possible when co-expressed with Pertuzumab HC.
TABLE-US-00021 TABLE 18 Kinetic and thermodynamic parameters of Trastuzumab, Pertuzumab, and hybrid molecules thereof Clone ka [1/Ms] kd [1/s] KD [M] Trastuzumab 1.01E+05 2.24E-04 2.21E-09 Pertuzumab 8.08E+04 6.69E-05 1.47E-10 Pertuzumab HC 6.47E+04 1.73E-03 2.67E-08 Pertuzumab (Tras.L3) LC Pertuzumab HC 8.99E+04 1.98E-04 2.21E-09 Pertuzumab(Tras.Y91H) LC Trastuzumab HC 4.19E+04 0.05 1.09E-06 Pertuzumab(Tras.L3) LC Trastuzumab HC N/A N/A N/A Pertuzumab(Tras.Y91H) LC
Example 12: Affinity-Improvement of the Common Light Chain
[0562] Introduction and Characterization of Additional Trastuzumab-Specific LCDR Residues into the Pertuzumab (Tras.L3) LC
[0563] Based on the results of the previous SPR measurement, binding of the designed CLC "Pertuzumab (Trast. L3) LC" in combination with the Pertuzumab HC led to a reduced but still well detectable binding. In contrast, co-expression of the CLC with Trastuzumab HC resulted in a very weak affinity in the micromolar range and was significantly reduced compared to the parental Trastuzumab antibody.
[0564] In order to further evolve the generation of a CLC and improve the binding in combination with the Trastuzumab HC, additional Trastuzumab-specific amino acid of the LCDR1, 2, and 3 as well in framework 3 region that are specific for Trastuzumab were individually introduced into the sequence of the previously designed "Pertuzumab (Trast.L3) LC" (DNA sequence of variable domains listed as SEQ ID NO: 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, and 51). The resulting constructs (protein sequence of variable domains listed as SEQ ID NO: 32, 34, 36, 38, 40, 42, 44, 46, 48, 50, and 52) were co-expressed with the Trastuzumab HC (SEQ ID NO: 92) or the Pertuzumab HC (SEQ ID NO: 22) and purified from mammalian-derived cell culture supernatant. Binding to Her2 was measured and compared with the respective parental antibodies.
Affinity-Determination of the "Pertuzumab (Trast.L3) LC" Variants by SPR
[0565] The Affinity (K.sub.D) of the new antibody chain combinations was measured by surface plasmon resonance using a ProteOn XPR36 instrument (Biorad) and was performed as described before. A summary of the kinetic and thermodynamic measurements is given in table 19.
[0566] Interestingly, none of the additional single mutations in the "Pertuzumab (Tras.L3) LC" variants significantly affected the affinity when combined with the Pertuzumab and the affinity was comparable to "Pertuzumab HC".times."Pertuzumab (Tras.L3) LC". However, combination of the "Pertuzumab (Tras.L3) LC" variants with the Trastuzumab HC resulted in significant differences. While back mutations to the Pertuzumab sequence in the LCDR3 region (P94Y and T96Y) (protein sequence of variable domains listed as SEQ ID NOs: 50 and 52) completely abolished binding to Her2, introduction of Pertuzumab-specific mutations (SEQ ID NOs: 32, 34, 36, 38, 40, 42, 44, 46, and 48) yielded an equal or improved affinity compared to the initial combination "Trastuzumab HC".times."Pertuzumab (Tras.L3) LC". In particular, introduction of the mutations I31T, G32A, Y53F, and G66R led to the most significant improvement in binding.
Simultaneous Introduction of Most Significant Trastuzumab-Specific Residues into the "Pertuzumab (Tras.L3) LC"
[0567] Based on the binding measurement described above, introduction of the 4 most relevant Trastuzumab-specific mutations into the "Pertuzumab (Tras.L3) LC" was combined. The protein sequences of the variable domains containing either the individual mutations are listed as SEQ ID NOs: 36, 40, 42, and 48.) The chain containing the "quadruple mutation" was named "Pertuzumab (Tras.L3) (QM) LC" (SEQ ID NOs: 54).
Affinity-Determination of the "Pertuzumab (Trast.L3)(QM) LC" by SPR
[0568] In order to characterize the new "Pertuzumab (Tras.L3) (QM) LC" and to determine the binding affinity when combined with either Pertuzumab HC or Trastuzumab HC, a further SPR experiment was performed. The Affinity (K.sub.D) of the new antibody chain combinations was measured using a ProteOn XPR36 instrument (Biorad) and was performed as described before. A summary of the kinetic and thermodynamic measurements is given in FIGS. 20A-B and Table 20.
[0569] Similar to the individual mutations that were introduced and characterized before, the affinity of the antibody comprising the Pertuzumab HC and the "Pertuzumab (Tras.L3) (QM) LC" did not significantly decrease compared to the initial chain combination "Pertuzumab HC".times."Pertuzumab (Tras.L3) LC". For both chain combinations, the affinity was in the range of 26 to 34 nM. Interestingly, the combination of the "Trastuzumab HC" with the "Pertuzumab (Tras.L3) (QM) LC" leads to a very strong improvement of the affinity to Her2. Furthermore, its affinity of 200 pM is even better than the measured affinity of the parental antibody Trastuzumab (2.2 nM). Given that the combination of "Pertuzumab (Tras.L3) (QM) LC" with Trastuzumab HC fully restores (or even exceeds) binding to Her2 and considering that the combination with the Pertuzumab HC diminishes but not abrogates binding to Her2, it was apparent to use the "Pertuzumab (Tras.L3) (QM) LC" as a CLC for both Her2 specificities and restore binding to the Pertuzumab-specific epitope by affinity-maturation of the Pertuzumab HC.
TABLE-US-00022 TABLE 20 Kinetic and thermodynamic parameters of antibodies comprising either a Trastuzumab or Pertuzumab HC and the final CLC Clone ka [1/Ms] kd [1/s] KD [M] Pertuzumab HC 9.46E+04 3.24E-03 3.42E-08 Pertuzumab (Tras.L3)(QM) LC Trastuzumab HC 2.69E+05 5.42E-05 2.02E-10 Pertuzumab (Tras.L3)(QM) LC
Example 13: Affinity Maturation of the Pertuzumab Heavy Chain
Generation of Pertuzumab-Based H1/H3 and H2 Affinity Maturation Libraries
[0570] Generation of affinity-matured Pertutzumab-derived heavy chains was carried out by phage display using standard protocols (Silacci et al, 2005).
[0571] For the generation of Pertuzumab-derived HCs with improved affinity when jointly expressed with "Pertuzumab (Tras.L3) (QM) LC" a maturation library randomized in CDR1 and 3 or in CDR2 was generated. The sequence of the Pertuzumab HC (SEQ ID NO: 22) and of the "Pertuzumab (Tras.L3) (QM) LC" (SEQ ID NO: 54) was cloned into a phagemid and used as a template for the randomization. For the generation of the Pertuzumab HC affinity maturation library randomized in CDR1 and 3, three fragments were assembled by "splicing by overlapping extension" (SOE) PCR and cloned into the phage vector. The following primer combinations were used to generate the library fragments: fragment 1 (LMB3 (SEQ ID NO: 147) and AM_omni_H1_TN-ba (SEQ ID NO: 148), fragment 2 (RJH108 (omni_3'H1_fo) (SEQ ID NO: 149) and RJH109 (omni_5'H3_re) (SEQ ID NO: 150), and fragment 3 (AM_omni_H3_TN_fo (SEQ ID NO: 151) and RJH99 (SEQ ID NO: 152). After assembly of sufficient amounts of full length randomized fragment, it was digested with MunI/NheI alongside with identically treated acceptor phagemid vector. 6 ug of Fab library insert were ligated with 24 ug of phagemid vector. Purified ligations were used for 60 transformations resulting in 6.times.10 exp9 transformants. Phagemid particles displaying the Pertuzumab affinity maturation library were rescued and purified by PEG/NaCl purification to be used for selections.
[0572] The generation of the CDR2-randomized Pertuzumab HC affinity maturation library was done similarly, but only 2 fragments were generated, assembled and cloned into the phagemid using the same restriction enzymes as before. The following primer combinations were used to generate the library fragments: fragment 1 (LMB3 (SEQ ID NO: 147) and RJH110 (omni_5'H2_ba) (SEQ ID NO: 153) and fragment 2 (AM_omni_h2_TN_fo (SEQ ID NO: 154) and RJH99 (SEQ ID NO:152). Purified ligations were used for 60 transformations resulting in 4.times.10 exp9 transformants. Phagemid particles displaying the Pertuzumab affinity maturation library were rescued and purified by PEG/NaCl purification to be used for selections.
TABLE-US-00023 TABLE 21 a) Primer combinations for the generation of the CDR1 and 3-randomized Pertuzumab affinity-maturation library Pertuzumab HC affinity maturation (CDR1 and 3) fragment 5'Primer 3'Primer PCR1 LMB3 AM_omni_H1_TN-ba PCR2 RJH108 (omni_3'H1_fo) RJH109 (omni_5'H3_re) PCR3 AM_omni_H3_TN_fo RJH99
TABLE-US-00024 TABLE 21 b Primer sequences for the generation of the CDR1 and 3-randomized Pertuzumab affinity-maturation library Pertuzumab HC affinity maturation (CDR1 and 3) SEQ ID Name Sequence 147 LMB3 CAGGAAACAGCTATGACCATGATTAC 148 AM_omni_ CCGGTGCCTGACGAACCCAATCCAT 4 3 2 1 H1_TN-ba AAAGGTAAAACCGCTTGCTGCACAGCTC 1 T = 60%, S/G/R/N/D = 20% (4% each), rest = 20% (1.7% each) 2 D = 60%, S/N/T/A/R/E/Q/G = 30% (3.8% each), rest = 10% (1.1% each) 3 Y = 60%, F/S/H/N/D/T = 30% (5.0% each), rest = 10% (0.9%) 4 T = 60%, A/G/V/S/P/D/N = 30% (4.3% each), rest = 10% (1.0%) 149 RJH108 ATGGATTGGGTTCGTCAGGCACCGGGTAAAGG (omni_3' H1_fo) 150 RJH109 ATTACGTGCACAATAATACACTGCGGTATCCTC (omni_5' H3_re) 151 AM_omni_ TACCGCAGTGTATTATTGTGCACGT 4a 5 5a 6 7 TTC H3_TN_fo 8 TTTGATTATTGGGGTCAGGGCACCCTGGTTAC 4a N = 60%, G/D/E/QN/S/A/P/R/L/T/Y = 40% (3.3% each) 5 L = 60%, G/Y/S/A/D/T/R/P/V/N/W/F/I/E = 40% (2.9% each) 5a G = 60%, Y/S/A/D/T/R/P/LN/N/W/F/I/E = 40% (2.9% each) 6 P = 60%, G/Y/S/A/D/T/R/L/V/N/W/F/I/E = 40% (2.9% each) 7 S = 60%, G/Y/P/A/D/T/R/L/V/N/W/F/I/E = 40% (2.9% each) 8 Y = 60%, G/A/P/W/S/D/T/F/R/K/H = 40% (3.6% each) 152 RJH99 GGCTGAGACTCCTCAAGAGAAGGATTAG
TABLE-US-00025 TABLE 22 a) Primer combinations for the generation of the CDR2- randomized Pertuzumab affinity-maturation library Pertuzumab HC affinity maturation (CDR2) fragment 5'Primer 3'Primer PCR1 LMB3 RJH110(omni_5'H2_ba)
TABLE-US-00026 TABLE 22 b Primer combinations for the generation of the CDR2-randomized Pertuzumab affinity-maturation library Pertuzumab HC affinity maturation (CDR2) SEQ ID Name Sequence 153 RJH110_ ATTAACATCTGCAACCCATTCCAGACCTTTAC (omni_5' H2_ba) 154 AM_omni_ GGTCTGGAATGGGTTGCAGATGTTAAT 9 10 11 12 h2_TN_fo GGT 13 ATT 14 AAC 15 CGTTTTAAAGGTCGTTTTACCCTGAG 9 P = 60%, G/A/S/T/D/N/F/Y = 30% (3.8% each), rest = 10% (1.0% each) 10 N = 60%, S/D/G/T/R/A = 30% (5.0% each), rest = 10% (0.8%) 11 S = 60%, G = 10%, rest = 30% (1.8% each)
Selection of Affinity Matured Pertuzumab HC-Derived Clones
[0573] Selections against the extracellular domain (ECD) of human Her2 were carried out using HEK293-expressed protein. The antigen was enzymatically biotinylated by co-expression of the biotin ligase Bir A via an N-terminal avi-tag. Panning rounds were performed in solution according to the following pattern: 1. binding of .about.10.sup.12 phagemid particles to 10 nM biotinylated Her2 ECD for 0.5 h in a total volume of 1 ml, 2. capture of biotinylated Her2 ECD and specifically bound phage particles by addition of 5.4.times.10.sup.7 streptavidin-coated magnetic beads for 10 min, 3. washing of beads using 5.times.1 ml PBS/Tween20 and 5.times.1 ml PBS, 4. elution of phage particles by addition of 1 ml 100 mM TEA for 10 min and neutralization by adding 500 .mu.l 1M Tris/HCl pH 7.4, 5. re-infection of exponentially growing E. coli TG1 bacteria, and 6.infection with helperphage VCSM13 and subsequent PEG/NaC1 precipitation of phagemid particles to be used in subsequent selection rounds. Selections were carried out over 3 rounds using decreasing (from 20.times.10.sup.-9M to 1.times.10.sup.-9M) antigen concentrations. In round 2 and 3, capture of antigen:phage complexes was performed using neutravidin plates instead of streptavidin beads. In addition, neutravidin plates were washed for 3h in 2 l PBS. Specific binders were identified by ELISA as follows: 100 .mu.l of 30 nM biotinylated Her2 ECD per well were coated on neutravidin plates. Fab-containing bacterial supernatants were added and binding Fabs were detected via their Flag-tags by using an anti-Flag/HRP secondary antibody. ELISA-positive clones were bacterially expressed as soluble Fab fragments in 96-well format and supernatants were subjected to a kinetic screening experiment by SPR-analysis using Proteon XPR36.
Affinity-Determination of Affinity-Matured Pertuzumab HC Variants by SPR
[0574] The Affinity (K.sub.D) of the new Pertuzumab HC variants was measured by surface plasmon resonance. In a first step, 7000 RU of polyclonal anti-human Fab antibody were immobilized on all 6 channels of a GLM chip by Amine coupling (NaAcetate pH4.5, 300/min, 300s) (vertical orientation).
[0575] Each antibody-containing bacterial supernatant was filtered and 3-fold diluted with PBS, and then injected for 180s at 30 .mu.l/minute to achieve immobilization levels of between 100 and 400 response units (RU) in vertical orientation. Injection of Her2: For one-shot kinetics measurements, injection direction was changed to horizontal orientation, two-fold dilution series of purified Her2 (varying concentration ranges between 100 and 6.25 nM) were injected simultaneously at 1000/min along separate channels 1-5, with association times of 180s, and dissociation times of 1000s. Buffer (PBST) was injected along the sixth channel to provide an "in-line" blank for referencing. Regeneration was performed by two pulses of 10 mM glycine pH 1.5 and 50 mM NaOH for 30s at 100 .mu.l/min (horizontal orientation). Association rate constants (k.sub.on) and dissociation rate constants (k.sub.off) were calculated using a simple one-to-one Langmuir binding model in ProteOn Manager v3.1 software by simultaneously fitting the association and dissociation sensorgrams. The equilibrium dissociation constant (K.sub.D) was calculated as the ratio k.sub.off/k.sub.on. Clones expressing Fabs with the highest affinity constants were identified and the heavy chains of the corresponding phagemids were sequenced. The thermodynamic measurement of the most affine Pertuzumab HC variants (protein sequence of variable domains listed as SEQ ID NOs: 62, 66, 68, 70, 72, and 74) is summarized in Table 23 and FIGS. 21A-F.
TABLE-US-00027 TABLE 23 Affinity of selected affinity matured Pertuzumab clone variants in combination with the common light chain (CLC) Pertuzumab heavy chain affinity (nM) Pertuzumab 34 Aff.mat. clone D1 1 Aff.mat. clone B2 1 Aff.mat. clone E1 0.5 Aff.mat. clone G2 1 Aff.mat. clone C8 3 Aff.mat. clone A1 1
Example 14: Characterization of the Trastuzumab/Pertuzumab Bispecific Antibodies
[0576] Generation of theTrastuzumab/Pertuzumab Bi-Specific Anti-Her Antibodies with a CLC
[0577] In the following step, the affinity-matured Pertuzumab HC as well as the Trastuzumab HC were expressed in combination with the CLC named "Pertuzumab (Tras.L3) (QM) LC" in a bispecific antibody format. For the generation of such bispecific antibodies with a CLC, heterodimerization of the 2 different HCs was achieved by application of the knob-into-hole technology. Variable domains of the Pertuzumab affinity-matured clones E1 and G2 (protein sequence of variable domains listed as SEQ ID NOs: 68 and 70) as well as a clone "D1-derived" (D1-der) sequence (SEQ ID NOs: 64) were cloned into a human IgG1 HC containing the "hole" mutations in domain CH3. A schematic overview of the bispecific antibody with a CLC is shown in FIG. 22. Affinity-maturation clone "D1-der" combines the CDR1 and 3 mutations of clone D1 (SEQ ID NOs: 62) with additional CDR2 mutations found in other selected clones. The variable domain of the Tratuzumab HC (SEQ ID NO: 92) was cloned into a human IgG1 HC harboring the "knob" mutations in domain CH3. The resulting constructs, called "Herceptarg" constructs, were co-expressed and purified from mammalian-derived cell culture supernatant. A summary of the analytical data for all three bi-specific antibodies is shown clones in FIGS. 23A-F and Table 24.
TABLE-US-00028 TABLE 24 Production of HER2 antibody with common light chain yield/liter Antibody (mg/l) % Monomer Herceptarg CLC G2 41.3 100 Herceptarg CLC E1 72.1 96 Herceptarg CLC D1- 20.1 100 der.
Generation of Her2 Knock-Out Antigen Variants
[0578] In order to analyze and characterize binding of the Herceptarg bi-specific antibodies to each of both epitopes on Her2, Her2 knock-out variants were designed. In these variants, either of the two specific epitopes was deleted by mutation of the amino acids that interact with the respective antibody chains.
[0579] For expression and purification, the respective DNA fragments were fused in frame to an N-terminal leader sequence and a C-terminal human IgG1 Fc coding fragment serving as solubility- and purification tag. An avi-tag at the C-terminal end of the Fc fragment allowed in vivo biotinylation. In order to express the antigen in a monomeric form, these Fc chains contained the "knob" mutations (SEQ ID NOs: 5 and 6, Her2 ECD-Fc(knob); SEQ ID NOs: 7 and 8, Her2 ECD (Pertuzumab KO)-Fc(knob); SEQ ID NOs: 9 and 10, Her2 ECD (Trastuzumab KO)-Fc(knob)) and was co-expressed in combination with an "Fc-hole" counterpart (SEQ ID NOs: 3 and 4).
Affinity-Determination of the Herceptarg Bi-Specific Antibodies by SPR
[0580] The Affinity (K.sub.D) of the new bi-specific antibodies to each of their epitopes in her2 was measured by SPR using a ProteOn XPR36 instrument (Biorad) at 25.degree. C. In a first step, 11000 RU of a polyclonal goat anti-human IgG (Sigma 12136) recognizing human IgG (Fc-specific) was immobilized on all 6 channels of a GLM chip by Amine coupling (NaAcetate pH4, 30 .mu.l/min, 300s) (vertical orientation).
[0581] Each antibody was diluted with PBST (10 mM phosphate, 150 mM sodium chloride pH 7.4, 0.005% Tween 20) to 2 .mu.g/ml, and then injected for 60s at 30 .mu.l/minute to achieve immobilization levels of about 400 response units (RU) in vertical orientation. Injection of Her2: For one-shot kinetics measurements, injection direction was changed to horizontal orientation, two-fold dilution series of purified monovalent Her2-Fc protein constructs (varying concentration ranges between 100 and 6.25 nM) were injected simultaneously at 100 .mu.l/min along separate channels 1-5, with association times of 180s, and dissociation times of 600s. Buffer (PBST) was injected along the sixth channel to provide an "in-line" blank for referencing. Regeneration was performed by two pulses of 10 mM glycine pH 1.5 and 50 mM NaOH for 30s at 100 .mu.l/min (horizontal orientation). Association rate constants (k.sub.on) and dissociation rate constants (k.sub.off) were calculated using a simple one-to-one Langmuir binding model in ProteOn Manager v3.1 software by simultaneously fitting the association and dissociation sensorgrams. The equilibrium dissociation constant (K.sub.D) was calculated as the ratio k.sub.off/k.sub.on.
[0582] All antibodies including Trastuzumab, Pertuzumab as well as three bi-specific Herceptarg constructs comprising the affinity-matured Pertuzumab HCs, the Trastuzumab HC, and the CLC "Pertuzumab (Trast.L3) (QM)" were tested for binding to both Her2 knock-out variants. As expected, both Trastuzumab and Pertuzumab bind to their respective Her2 epitope with the excepted affinity. Binding to their corresponding knock-out variant was abolished (FIGS. 24A-D). These results demonstrate that the use of Her2 knock-out epitope variants allows dissecting the binding of the bispecific antibodies and analyzing the individual affinity of both specificities. Determination of the individual K.sub.D values of each Herceptarg clone variant revealed a constant binding affinity to the Trastuzumab epitope in the expected range of 0.6 to 1.8 nM. In contrast, binding to the Pertuzumab epitope depends on the affinity-matured Pertuzumab HC. Among the three Herceptarg clone variants, clone "D1-der" was found to have the highest affinity (0.16 nM). A summary of the Thermodynamic data is shown in table 25. In summary, this experiment confirms that we were able to generate a bi-specific antibody with a CLC and specific for the epitopes Trastuzumab and Pertuzumab.
Binding Analysis of the Herceptarg Clone Variants to KPL-4 Cells
[0583] KPL-4 cells were harvested and resuspended in FACS buffer. 0.2 Mio cells were seeded into a 96 well round bottom plate. The plate was centrifuged at 400 g for 3 min to pellet the cells. The supernatant was removed and the cells were resuspended in 40 .mu.l of the diluted antibodies. The plate was incubated for 30 min at 4.degree. C. to allow binding of the antibodies. To remove unbound antibodies the cells were centrifuged again and washed twice with FACS buffer. To detect the antibodies the cells were resuspended in 12 .mu.l diluted secondary goat anti-human Fc specific FITC-labeled secondary antibody (Jackson ImmunoResearch #109-096-098) and incubated again for 30 min at 4.degree. C. Afterwards the cells were washed twice with FACS buffer, resuspended in 200 .mu.l FACS buffer and the fluorescence was measured with BD Cantoll. Results are shown in FIG. 25.
TABLE-US-00029 TABLE 25 Affinity of selected bi-specific antibodies affinity matured Pertuzumab clone variants in combination with the CLC KD (nM) Trastuzumab KD (nM) Pertuzumab clone Epitope Epitope Trastuzumab 2.17 N/A Pertuzumab N/A 0.62 Herceptarg E1 0.72 1.32 Herceptarg G2 1.82 9.65 Herceptarg D1-derived 0.6 0.16 Herceptarg crossmab 0.8 0.84
TABLE-US-00030 TABLE 26 Proliferation inhibition of various cell types (IC50 values with confidence interval) cell Trastuzumab + Herceptarg Herceptarg Herceptarg line Trastuzumab Pertuzumab Pertuzumab CLC E1/1 CLC D1-der CLC G2/2 GA604 BT474 110.8 205.4 222.3 114.1 78.38 89.99 125.4 (96.7-127.0) (156-270.7) (176.5-280.1) (106-122.7) (69.03- 89) (82.3-98.4) (113.6-138.4) N87 83.13 205.8 312.9 136.5 92.7 119.1 109.6 (59.7-115.7) (126.3-335) (156.5-625.6) (126.6-147) (80.7-106.5) (107.6-132) (98.3-122.2) SkBr3 73.73 nd 55.63 69.01 47.54 92.59 59.84 (43.54-124.8) (25.97-119.2) (55.78-85.4) (26.61-84.9) (70.1-122.3) (50.2-71.4)
Proliferation Inhibition Mediated by the Herceptarg Binding Variants
[0584] Target cells were harvested, washed, resuspended in RPMI 1640 (Gibco)+10% FCS+1% GlutaMAX.TM. (Gibco) and plated at a concentration of 5.times.10.sup.3 cells/well. Cells were incubated for 4 hours in the cell incubator before respective antibody dilutions were added. Plates were gently shaked and incubated for 5 days in the cell incubator. The plates were equilibrated to room temperature and 100 .mu.l/well of the freshly prepared CellTiter Glo (Promega) substrate were added to each well. Luminescence was measured in a Wallac Victor3 1420 Multilabel Counter. Results are shown in Table 26 and FIGS. 26A-F.
Herceptarg Clones-Mediated Antibody-Dependent Cell-Mediated Cytotoxicity (ADCC)
[0585] Target cells were harvested, washed, resuspended in AIM V.RTM. medium (Life Technologies), and plated at a concentration of 3.times.10.sup.4 cells/well. The respective antibody dilutions were added in triplicates to the cells and incubated for 10 min before addition of the effector cells (peripheral blood mononuclear effector cells [PBMCs]). Effector (E) and target (T) cells were then incubated for the indicated time at 37.degree. C. at the indicated E:T ratio (triplicates for all samples). Lactate dehydrogenase (LDH) release was measured using the LDH Cytotoxicity Detection Kit (Roche Applied Science). ADCC was calculated using the following formula:
Percentage ADCC = ( [ sample release - spontaneous release maximal release - spontaneous release ] ) .times. 1 00. ##EQU00003##
[0586] Spontaneous release, corresponding to target cells incubated with effector cells without antibody, was defined as 0% cytotoxicity, and maximal release (target cells lysed with 1% Triton X-100) was defined as 100% cytotoxicity. The average percentage of ADCC and standard deviations of the triplicates of each experiment were calculated. Results are shown in FIGS. 27A-D.
TABLE-US-00031 TABLE 27 Leader Peptides SEQ ID Protein sequence 155 MDWTWRILFLVAAATGAHS 156 MDMRVPAQLLGLLLLWFPGARC 157 MGWSCIILFLVATATGVHS
Example 15: Proliferation Assay
[0587] 1.times.10.sup.4 BT-474 cells/well were cultured in RPMI/10% FCS in a 96-well flat bottom plate. After 24 hrs growth medium was removed and titrated amounts of indicated antibodies were added (premixed in culture medium) to a final volume of 100 .mu.l. To determine the number of viable cells in culture, the CellTiter-Glo Luminescent Cell Viability Assay was performed by quantifying the present ATP levels as an indicator of metabolically active cells. Thus, after six days of culture, 100 .mu.l CellTiter-Glo Reaction Mix (Promega, cat.no. # G7571) was added to the cells, shook for 2 min before 75 .mu.l of the lysate was transferred to a separate 96-well flat bottom titer plate (Costar, cat.no. #3917). After additional mixing, luminescence was assessed according to the manufacturer's instructions using a Tecan Infinite Reader.
[0588] Results are shown in Table 28 and FIG. 28.
[0589] In the proliferation assay it was shown that the bispecific Her2 antibodies inhibited proliferation of BT-474 cells more potently than Pertuzumab or Trastuzumab alone or in combination. The following bispecific Her2 antibodies were tested: Herceptarg CLC D1-der wt": SEQ ID NOs 64, 54, 92, Herceptarg CLC D1-der G2": SEQ ID NOs 64, 54, 92 (glycoengineered variant) "Herceptarg CrossMab": SEQ ID NOs 109, 110, 111, 112.
TABLE-US-00032 TABLE 28 IC50 BT474 proliferation assay Antibody treatment IC50 Proliferation [nM] Trastuzumab n.d. Pertuzumab n.d. Trastuzumab + Pertuzumab 6.20 Herceptarg CLC D1-der. wt 3.31 Herceptarg CLC D1-der. G2 3.93 Herceptarg CrossMab 4.75
Example 16: C1q FACS Binding Assay
[0590] 3.times.10.sup.5 BT-474 cells were incubated with 10 .mu.g/ml of indicated antibody on ice. The following bispecific Her2 antibodies were tested: Herceptarg CLC D1-der wt": SEQ ID NOs 64, 54, 92, Herceptarg CLC D1-der G2": SEQ ID NOs 64, 54, 92 (glycoengineered variant) "Herceptarg CrossMab": SEQ ID NOs 109, 110, 111, 112.
[0591] After 30 min, 10 .mu.g/ml C1q (Sigma, C1740) was added and additionally incubated for 20 min. After washing, cells were counterstained with commercial PE-labeled anti-C1q antibody (Cedarlane, CL7611PE-SP). After further incubation (30 min, ice), cells were washed twice and analyzed on a FACS Canto II.
[0592] Results are shown in FIG. 29 and Table 29. This C1q assay illustrates the binding of recombinant complement factor C1q to different Her antibodies on BT-474 cells. It was shown that the highest C1q binding resulted upon treatment with the combination of Trastuzumab and Pertuzumab, followed by the two CLC bispecific Her2 antibodies. Treatment with the Crossmab resulted only in a slightly elevated C1q binding.
TABLE-US-00033 TABLE 29 C1q binding assay PE-signal antibody/antibodies (geomean) trastuzumab 282 pertuzumab 344 combination of trastuzumab and pertuzumab 2157 bispecific anti-HER2 antibody, common light chain 1439 bispecific anti-HER2 antibody, common light chain, 1036 glycoengineered bispecific anti-HER2 antibody, CrossMab format 489
Example 17: LDH Assay with Baby Rabbit Complement (BRC)
[0593] CHO-K1 Nxre19 cells (IL15R transfected CHO-K1) were seeded at 10,000 cells/well on 96-well flat bottom cell culture plates (NUNC, 100 .mu.L/well) and cultivated overnight. IL15-Fc fusion polypeptide was added (25 .mu.L/well in 5-fold end-concentration) and incubated for one hour. Thereafter, one vial of Baby Rabbit complement (Cedarlane, Cat. No. CL3441) was reconstituted with 1 mL of Aqua bidest. The complement solution was diluted with medium and 25 .mu.L added to the wells. After four hours the plates were centrifuged at 200 g and 100 .mu.L/well were transferred to another 96-well flat bottom plate. Thereafter 100 .mu.L of LDH reaction mix (Cytotoxicity Detection Kit, Roche Diagnostic GmbH, Mannheim, Germany) was added. After an incubation time of 20 min. at 37.degree. C. optical density (OD) was measured at 492/690 nm on a Tecan Sunrise reader.
TABLE-US-00034 TABLE 30 LDH assay with BRC signal [OD] sample BRC 1/40 BRC 1/30 9000 ng/ml IL15-Fc-fusion with HUC 11.3 12.3 3000 ng/ml IL15-Fc-fusion with HUC 12.3 17.0 1000 ng/ml IL15-Fc-fusion with HUC 10.2 13.6 333.3 ng/ml IL15-Fc-fusion with HUC 7.8 12.2 111.1 ng/ml IL15-Fc-fusion with HUC 8.3 13.0 37.04 ng/ml IL15-Fc-fusion with HUC 14.9 19.7 12.35 ng/ml IL15-Fc-fusion with HUC 43.2 53.0 4.12 ng/ml IL15-Fc-fusion with HUC 41.5 63.8 0 ng/ml IL15-Fc-fusion with HUC 42.4 48.4
[0594] It can be seen that BRC has a low background toxicity and shows dose dependent complement toxicity.
Example 18: CDC (Complement Dependent Cytotoxicity) Activation on BT-474 Cells (LDH Release)
[0595] 1.times.10.sup.4 cells/well were incubated with 10 .mu.g/ml of the indicated antibodies for 30 min at 37.degree. C. in 150 .mu.l. The following bispecific Her2 antibodies were tested: Herceptarg CLC D1-der wt": SEQ ID NOs 64, 54, 92, Herceptarg CLC D1-der G2": SEQ ID NOs 64, 54, 92 (glycoengineered variant) "Herceptarg CrossMab": SEQ ID NOs 109, 110, 111, 112.
[0596] Then 50 .mu.l Baby Rabbit Complement (Cedarlane, cat. no. CL3441, batch no. 6312) was added and incubated for further 2 hrs. Then, the s/n was transferred and mixed with 50 .mu.l LDH Reaction Mix (Roche) and, after a further incubation of 15 min, extinktion (Ex.) at 490/620 nm was analyzed on a Tecan Sunrise Reader. The specific antibody dependent toxicity (mean+/-SD of n=4) on BT-474 cell was calculated as follows:
(Ex. sample-Ex. spontanous lysis/Ex. maximal lysis-spontanous lysis).times.100.
[0597] Results are shown in FIG. 30.
[0598] This CDC assay shows the release of LDH as a marker for dying/dead cells upon treatment of different anti-Her2 antibodies (formats, combination) in the presence of baby rabbit complement. Here, the combination of Trastuzumab and Pertuzumab resulted in a significant induction of CDC, whereas the parental antibodies alone did not. Surprisingly, both CLC Herceptarg variants provoked even superior CDC effects, whereas the Herceptarg Crossmab treatment results in a CDC reaction less effective than the combination of the parental antibodies.
Example 19: CDC (Complement Dependent Cytotoxicity)-Mediated Killing of BT-474 Cells (ACEA)
[0599] 1.times.10.sup.4 BT-474 cells/well were seeded on 96-well E-Plates (ACEA Biosciences Inc.) and grown overnight in an Xcelligence device. Growth medium was removed and cells were washed once with serum-free AIM-V medium (Gibco). 50 .mu.l/well AIM-V medium and 50 .mu.l antibody in AIM-V (3-fold end concentration) were added and incubated for 20 min. 50 .mu.l Baby Rabbit Complement (Cedarlane) was added and CellIndex (CI; as representative for the viability of the cells) was measured every 5 minutes (see curve). The following bispecific Her2 antibodies were tested: Herceptarg CLC D1-der wt": SEQ ID NOs 64, 54, 92, Herceptarg CLC D1-der G2": SEQ ID NOs 64, 54, 92 (glycoengineered variant) "Herceptarg CrossMab": SEQ ID NOs 109, 110, 111, 112.
[0600] Specific CDC was calculated according following formula, whereas CI is the normalized cell index:
% CDC = CI Complement control - CI sample CI Complement control .times. 100 ##EQU00004##
[0601] At two representative time points (1 hr and 2 hrs after starting the reaction, specific lysis (=CDC-induced cell death) was calculated and shown in the diagram (mean+/SEM of n=4).
[0602] Results are shown in FIGS. 31A-B and Table 31. This CDC assay illustrates a change in the cell index as a marker for dying/dead cells upon treatment with different anti-Her2 antibodies (formats, combination) in the presence of baby rabbit complement: Here, the combination of Trastuzumab and Pertuzumab resulted in a significant induction of CDC, whereas the parental antibodies alone did not. Surprisingly, both CLC Herceptarg variants provoked even superior CDC effects, whereas the Herceptarg Crossmab treatment results in a CDC reaction less effective than the combination of the parental antibodies.
[0603] One possible reason for the superiority of Herceptarg CLC D1-der may be the slightly higher affinity to the Trastuzumab epitope as well as the significantly higher affinity to the Pertuzumab epitope (see also table 25).
TABLE-US-00035 TABLE 31 CDC (complement dependent cytotoxicity)- mediated killing of BT-474 cells (ACEA) specific lysis [% cell index ACEA] antibody/antibodies 1 hour 2 hours trastuzumab -3.5 .+-. 0.6 -6.5 .+-. 0.8 pertuzumab -5.3 .+-. 1.0 -8.3 .+-. 2.1 combination of trastuzumab and 20.9 .+-. 6.7 26.3 .+-. 7.0 pertuzumab bispecific anti-HER2 antibody, common 31.8 .+-. 3.4 38.9 .+-. 3.7 light chain (D1 der) bispecific anti-HER2 antibody, common 28.8 .+-. 2.6 35.8 .+-. 2.6 light chain, glycoengineered (D1 der) bispecific anti-HER2 antibody, 12.9 .+-. 1.4 22.7 .+-. 1.6 CrossMab format
Example 20: Mouse Xenograft Studies
Cell Line KPL4
[0604] This human breast cancer cell line has been established from the malignant pleural effusion of a breast cancer patient with an inflammatory skin metastasis. Cells have been provided by Professor J. Kurebayashi (Kawasaki Medical School, Kurashiki, Japan).Tumor cells were routinely cultured in DMEM medium (PAN Biotech, Germany) supplemented with 10% fetal bovine serum (PAN Biotech, Germany) and 2 mM L-glutamine (PAN Biotech, Germany) at 37.degree. C. in a water-saturated atmosphere at 5% CO2. Culture passage was performed with trypsin/EDTA 1.times. (PAN) splitting twice/week. Cell passage P6 was used for in vivo study.
Mice
[0605] Female SCID beige (C.B.-17) mice; age 10-12 weeks; body weight 18-20 g (Charles River Germany, Sulzfeld); body weight >20 g are maintained under specific-pathogen-free condition with daily cycles of 12 h light/12 h darkness according to international guidelines (GV-Solas; Felasa; TierschG). After arrival animals were housed in the quarantine part of the animal facility for one week to get accustomed to new environment and for observation. Continuous health monitoring was carried out on regular basis. Diet food (Alltromin) and water were provided ad libitum. The experimental study was reviewed and approved by local government.
Tumor Cell Injection
[0606] At the day of injection tumor cells were harvested (trypsin-EDTA) from culture flasks (Greiner TriFlask) and transferred into 50 ml culture medium, washed once and resuspended in PBS. After an additional washing step with PBS and filtration (cell strainer; Falcon O100 .mu.m) the final cell titer was adjusted to 1.5.times.10e8/ml. Tumor cell suspension was carefully mixed with transfer pipette to avoid cell aggregation. Anesthesia was performed using a Stephens inhalation unit for small animals with preincubation chamber (plexiglas), individual mouse nose-mask (silicon) and not flammable or explosive anesthesia compound Isoflurane (Pharmacia-Upjohn, Germany) in a closed circulation system. Two days before injection, coat of the SCID beige mice were shaved and KPL-4 cells (3.times.10e6 cells) were injected orthotopically in a volume of 20 .mu.l (using a Hamilton microliter syringe and a 30Gx1/2'' needle) into the right penultimate inguinal mammary fat pad of each anesthetized mouse. The cell suspension was injected through the skin under the nipple.
Monitoring
[0607] Animals were controlled daily for detection of clinical symptoms of adverse effects. For monitoring throughout the experiment the body weight of the animals was documented two times weekly and the tumor volume was measured by caliper twice weekly. Tumor volume was calculated according to NCI protocol (Tumor weight=1/2ab2, where "a" and "b" are the long and the short diameters of the tumor, respectively). Termination criteria were the critical tumor mass (up to 1.7 g or O>1.5 cm), body weight loss more than 20% from baseline, tumor ulceration or poor general condition of the animals. Study exclusion criteria for the animals are described and approved in the corresponding "Tierversuchsanzeige".
Treatment
[0608] Mice were randomized for tumor volume of 80 mm.sup.3 and subsequently treated once weekly with a volume of 10 ml/kg intra peritoneal. For combination treatment Herceptin was given first and Perjeta was given 24 hrs thereafter.
[0609] The following bispecific Her2 antibodies was tested: Herceptarg CLC-D1-der: SEQ ID NOs 64, 54, 92. Results are shown in FIG. 32.
TABLE-US-00036 No of Route/Mode of Group animals Compound Dose (mg/kg) administration 1 9 Control (Xolair) 10 i.p. once weekly 2 9 Herceptin 10 i.p. once weekly 3 9 Perjeta 10 i.p. once weekly 4 9 Herceptin plus 10 plus 10 i.p. once weekly Perjeta 5 8 Herceptarg CLC- 10 i.p. once weekly D1-der
TABLE-US-00037 TABLE 32 Parent sequences of Pertuzumab and Trastuzumab SEQ ID NO Name Sequence Pertuzumab wt (parent) sequences 22 Pertuzumab EVQLVESGGGLVQPGGSLRLSCAASGFTFTDYTMDWVRQAPGKGLEW wt VH VADVNPNSGGSIYNQRFKGRFTLSVDRSKNTLYLQMNSLRAEDTAVY (10289) YCARNLGPSFYFDYWGQGTLVTVSS 21 Pertuzumab GAGGTGCAGCTGGTCGAGTCTGGCGGCGGACTGGTGCAGCCTGGCGG wt VH CAGCCTGAGACTGAGCTGCGCCGCCAGCGGCTTCACCTTCACCGACT (10289) DNA ACACCATGGACTGGGTGCGGCAGGCCCCTGGCAAGGGCCTGGAATGG GTGGCCGACGTGAACCCCAACAGCGGCGGCAGCATCTACAACCAGCG GTTCAAGGGCCGGTTCACCCTGAGCGTGGACAGAAGCAAGAACACCC TGTACCTCCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTAC TACTGCGCCCGGAACCTGGGCCCCAGCTTCTACTTCGACTACTGGGG CCAGGGCACCCTGGTGACCGTGAGCAGCGCTAGCACCAAGGGCCCAT CGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACA GCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGAC GGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCC CGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGC 14 Pertuzumab GFTFTDYTMD wt VH CDR1 15 Pertuzumab DVNPNSGGSIYNQRFKG wt VH CDR2 16 Pertuzumab NLGPSFYFDY wt VH CDR3 114 Pertuzumab ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALT wt CH1 SGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKV DKKV 24 Pertuzumab DIQMTQSPSSLSASVGDRVTITCKASQDVSIGVAWYQQKPGKAPKLL wt VL IYSASYRYTGVPSRFSGSGSGTDFILTISSLQPEDFATYYCQQYYIY (10290) PYTFGQGTKVEIK 23 Pertuzumab GACATCCAGATGACCCAGAGCCCCAGCAGCCTGAGCGCCAGCGTGGG wt VL CGACAGAGTGACCATCACCTGCAAGGCCAGCCAGGACGTGTCCATCG (10290) DNA GCGTGGCCTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTG ATCTACAGCGCCAGCTACCGGTACACAGGCGTGCCCAGCCGGTTCAG CGGCAGCGGCTCCGGCACCGACTTCACCCTGACCATCAGCAGCCTGC AGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGTACTACATCTAC CCCTACACCTTCGGCCAGGGCACCAAGGTGGAGATCAAG 11 Pertuzumab KASQDVSIGVA wt VL CDR1 12 Pertuzumab SASYRYT wt VL CDR2 13 Pertuzumab QQYYIYPYT wt VL CDR3 113 Pertuzumab RTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNAL wt CL QSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGL SSPVTKSFNRGEC Trastuzumab wt (parent) sequences 82 Trastuzumab DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKLL VL (4245) IYSASFLYSGVPSRFSGSRSGTDFTLTISSLQPEDFATYYCQQHYTT PPTFGQGTKVEIK 81 Trastuzumab GACATCCAGATGACCCAGAGCCCAAGCTCTCTGTCTGCCTCTGTGGG VL (4245) DNA CGACAGAGTGACCATCACCTGCAGAGCCAGCCAGGACGTGAACACAG CCGTGGCCTGGTATCAGCAGAAGCCAGGCAAGGCCCCAAAGCTGCTG ATCTACAGCGCCAGCTTCCTGTACAGCGGCGTGCCAAGCAGATTCAG CGGCAGCAGAAGCGGCACAGACTTCACCCTGACCATCAGCAGCCTGC AGCCAGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACC CCACCAACCTTCGGACAGGGCACCAAGGTGGAGATCAAG 17 Trastuzumab RASQDVNTAVA wt VL CDR1 18 Trastuzumab SASFLYS wt VL CDR2 19 Trastuzumab QQHYTTPPT wt VL CDR3 116 Trastuzumab RTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNAL wt CL QSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGL SSPVTKSFNRGEC 92 Trastuzumab EVQLVESGGGLVQPGGSLRLSCAASGFNIKDTYIHWVRQAPGKGLEW (11345) VARIYPTNGYTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVY YCSRWGGDGFYAMDYWGQGTLVTVSS 91 Trastuzumab GAAGTGCAATTGGTGGAAAGCGGCGGCGGCCTGGTGCAACCGGGCGG (11345) DNA CAGCCTGCGTCTGAGCTGCGCGGCCTCCGGATTTAACATAAAGGACA CATACATCCACTGGGTGCGCCAAGCACCTGGGAAGGGTCTCGAGTGG GTGGCTCGGATTTACCCAACAAATGGCTACACCAGGTATGCGGATAG CGTGAAAGGCCGTTTTACCATTTCAGCTGATACTTCGAAGAACACCG CCTATCTGCAAATGAACAGCCTGCGTGCGGAAGATACGGCCGTGTAT TATTGCTCGCGTTGGGGAGGAGACGGGTTCTATGCTATGGATTACTG GGGCCAAGGCACCCTGGTGACGGTTAGCTCA 20 Trastuzumab GFNIKDTYIH wt VH CDR1 29 Trastuzumab RIYPTNGYTRYADSVKG wt VH CDR2 30 Trastuzumab WGGDGFYAMDY wt VH CDR3 115 Trastuzumab ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALT wt CH1 SGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKV DKKV
TABLE-US-00038 TABLE 33 Sequences of antibodies with common light chain SEQ ID NO Name Sequence Pertuzumab/Trastuzumab hybrid light chains 26 Pertuzumab DIQMTQSPSSLSASVGDRVTITCKASQDVSIGVAWYQQKPGKAPKLL VL (Trast. IYSASYRYTGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQHYTT L3) PPTFGQGTKVEIK (10403) 25 Pertuzumab GACATCCAGATGACCCAGAGCCCCAGCAGCCTGAGCGCCAGCGTGGG VL (Trast. CGACAGAGTGACCATCACATGCAAGGCCAGCCAGGACGTGTCCATCG L3) GCGTGGCCTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTG (10403)DNA ATCTACAGCGCCAGCTACCGGTACACCGGCGTGCCCAGCAGATTCAG CGGCAGCGGCTCCGGCACCGACTTCACCCTGACCATCAGCAGCCTGC AGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACC CCCCCCACCTTCGGCCAGGGCACCAAGGTGGAAATCAAG 28 Pertuzumab DIQMTQSPSSLSASVGDRVTITCKASQDVSIGVAWYQQKPGKAPKLL VL (Trast. IYSASYRYTGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQHYIY (10404) PYTFGQGTKVEIK 27 Pertuzumab GACATCCAGATGACCCAGAGCCCCAGCAGCCTGAGCGCCAGCGTGGG VL (Trast. CGACAGAGTGACCATCACCTGCAAGGCCAGCCAGGACGTGTCCATCG H91) GCGTGGCCTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTG (10404)DNA ATCTACAGCGCCAGCTACCGGTACACAGGCGTGCCCAGCCGGTTCAG CGGCAGCGGCTCCGGCACCGACTTCACCCTGACCATCAGCAGCCTGC AGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACATCTAC CCCTACACCTTCGGCCAGGGCACCAAGGTGGAGATCAAG 32 Pertuzumab DIQMTQSPSSLSASVGDRVTITCRASQDVSIGVAWYQQKPGKAPKLL VL IYSASYRYTGVPSRFSGSGSGTDTTLTISSLQPEDFATYYCQQHYTT (Tras.L3) PPTFGQGTKVEIK K24R (10949) 31 Pertuzumab GACATCCAGATGACCCAGAGCCCCAGCAGCCTGAGCGCCAGCGTGGG VL CGACAGAGTGACCATCACATGCCGGGCCAGCCAGGACGTGTCCATCG (Tras.L3) GCGTGGCCTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTG K24R ATCTACAGCGCCAGCTACCGGTACACCGGCGTGCCCAGCAGATTCAG (10949)DNA CGGCAGCGGCTCCGGCACCGACTTCACCCTGACCATCAGCAGCCTGC AGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACC CCCCCCACCTTCGGCCAGGGCACCAAGGTGGAAATCAAG 34 Pertuzumab DIQMTQSPSSLSASVGDRVTITCKASQDVNIGVAWYQQKPGKAPKLL VL IYSASYRYTGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQHYTT (Tras.L3) PPTFGQGTKVEIK S30N (10950) 33 Pertuzumab GACATCCAGATGACCCAGAGCCCCAGCAGCCTGAGCGCCAGCGTGGG VL(Tras.L3) CGACAGAGTGACCATCACATGCAAGGCCAGCCAGGACGTGAACATCG S30N GCGTGGCCTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTG (10950)DNA ATCTACAGCGCCAGCTACCGGTACACCGGCGTGCCCAGCAGATTCAG CGGCAGCGGCTCCGGCACCGACTTCACCCTGACCATCAGCAGCCTGC AGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACC CCCCCCACCTTCGGCCAGGGCACCAAGGTGGAAATCAAG 36 Pertuzumab DIQMTQSPSSLSASVGDRVTITCKASQDVSTGVAWYQQKPGKAPKLL VL IYSASYRYTGVPSRFSGSGSGTDFTLTISSTQPEDFATYYCQQHYTT (Tras.L3) PPTFGQGTKVEIK I31T (10951) 35 Pertuzumab GACATCCAGATGACCCAGAGCCCCAGCAGCCTGAGCGCCAGCGTGGG VL CGACAGAGTGACCATCACATGCAAGGCCAGCCAGGACGTGTCCACCG (Tras.L3) GCGTGGCCTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTG I31T ATCTACAGCGCCAGCTACCGGTACACCGGCGTGCCCAGCAGATTCAG (10951)DNA CGGCAGCGGCTCCGGCACCGACTTCACCCTGACCATCAGCAGCCTGC AGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACC CCCCCCACCTTCGGCCAGGGCACCAAGGTGGAAATCAAG 38 Pertuzumab DIQMTQSPSSLSASVGDRVTITCKASQDVSVGVAWYQQKPGKAPKLL VL IYSASYRYTGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQHYTT (Tras.L3) PPTFGQGTKVEIK I31V (10952) 37 Pertuzumab GACATCCAGATGACCCAGAGCCCCAGCAGCCTGAGCGCCAGCGTGGG VL CGACAGAGTGACCATCACATGCAAGGCCAGCCAGGACGTGTCCGTCG (Tras.L3) GCGTGGCCTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTG I31V ATCTACAGCGCCAGCTACCGGTACACCGGCGTGCCCAGCAGATTCAG (10952)DNA CGGCAGCGGCTCCGGCACCGACTTCACCCTGACCATCAGCAGCCTGC AGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACC CCCCCCACCTTCGGCCAGGGCACCAAGGTGGAAATCAAG 40 Pertuzumab DIQMTQSPSSLSASVGDRVTITCKASQDVSIAVAWYQQKPGKAPKLL VL IYSASYRYTGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQHYTT (Tras.L3) PPTFGQGTKVEIK G32A (10953) 39 Pertuzumab GACATCCAGATGACCCAGAGCCCCAGCAGCCTGAGCGCCAGCGTGGG VL CGACAGAGTGACCATCACATGCAAGGCCAGCCAGGACGTGTCCATCG (Tras.L3) CCGTGGCCTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTG G32A ATCTACAGCGCCAGCTACCGGTACACCGGCGTGCCCAGCAGATTCAG (10953)DNA CGGCAGCGGCTCCGGCACCGACTTCACCCTGACCATCAGCAGCCTGC AGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACC CCCCCCACCTTCGGCCAGGGCACCAAGGTGGAAATCAAG 42 Pertuzumab DIQMTQSPSSLSASVGDRVTITCKASQDVSIGVAWYQQKPGKAPKLL VL IYSASFRYTGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQHYTT (Tras.L3) PPTFGQGTKVEIK Y53F (10954) 41 Pertuzumab GACATCCAGATGACCCAGAGCCCCAGCAGCCTGAGCGCCAGCGTGGG VL(Tras.L3) CGACAGAGTGACCATCACATGCAAGGCCAGCCAGGACGTGTCCATCG Y53F GCGTGGCCTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTG (10954) DNA ATCTACAGCGCCAGCTTCCGGTACACCGGCGTGCCCAGCAGATTCAG CGGCAGCGGCTCCGGCACCGACTTCACCCTGACCATCAGCAGCCTGC AGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACC CCCCCCACCTTCGGCCAGGGCACCAAGGTGGAAATCAAG 44 Pertuzumab DIQMTQSPSSLSASVGDRVTITCKASQDVSIGVAWYQQKPGKAPKLL VL IYSASYLYTGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQHYTT (Tras.L3) PPTFGQGTKVEIK R54L (10955) 43 Pertuzumab GACATCCAGATGACCCAGAGCCCCAGCAGCCTGAGCGCCAGCGTGGG (Tras.L3) CGACAGAGTGACCATCACATGCAAGGCCAGCCAGGACGTGTCCATCG R54L GCGTGGCCTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTG (10955)DNA ATCTACAGCGCCAGCTACCTGTACACCGGCGTGCCCAGCAGATTCAG CGGCAGCGGCTCCGGCACCGACTTCACCCTGACCATCAGCAGCCTGC AGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACC CCCCCCACCTTCGGCCAGGGCACCAAGGTGGAAATCAAG 46 Pertuzumab DIQMTQSPSSLSASVGDRVTITCKASQDVSIGVAWYQQKPGKAPKLL (Tras.L3) IYSASYRYSGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQHYTT T56S PPTFGQGTKVEIK (10956) 45 Pertuzumab GACATCCAGATGACCCAGAGCCCCAGCAGCCTGAGCGCCAGCGTGGG (Tras.L3) CGACAGAGTGACCATCACATGCAAGGCCAGCCAGGACGTGTCCATCG T56S GCGTGGCCTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTG (10956)DNA ATCTACAGCGCCAGCTACCGGTACAGCGGCGTGCCCAGCAGATTCAG CGGCAGCGGCTCCGGCACCGACTTCACCCTGACCATCAGCAGCCTGC AGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACC CCCCCCACCTTCGGCCAGGGCACCAAGGTGGAAATCAAG 48 Pertuzumab DIQMTQSPSSLSASVGDRVTITCKASQDVSIGVAWYQQKPGKAPKLL (Tras.L3) IYSASYRYTGVPSRFSGSRSGTDFTLTISSLQPEDFATYYCQQHYTT G66R PPTFGQGTKVEIK (10957) 47 Pertuzumab GACATCCAGATGACCCAGAGCCCCAGCAGCCTGAGCGCCAGCGTGGG (Tras.L3) CGACAGAGTGACCATCACATGCAAGGCCAGCCAGGACGTGTCCATCG G66R GCGTGGCCTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTG (10957) DNA ATCTACAGCGCCAGCTACCGGTACACCGGCGTGCCCAGCAGATTCAG CGGCAGCCGCTCCGGCACCGACTTCACCCTGACCATCAGCAGCCTGC AGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACC CCCCCCACCTTCGGCCAGGGCACCAAGGTGGAAATCAAG 50 Pertuzumab DIQMTQSPSSLSASVGDRVTITCKASQDVSIGVAWYQQKPGKAPKLL (Tras.L3) IYSASYRYTGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQHYTY T94Y PPTFGQGTKVEIK (10958) 49 Pertuzumab ACATCCAGATGACCCAGAGCCCCAGCAGCCTGAGCGCCAGCGTGGGC (Tras.L3) GACAGAGTGACCATCACATGCAAGGCCAGCCAGGACGTGTCCATCGG T94Y CGTGGCCTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGA (10958)DNA TCTACAGCGCCAGCTACCGGTACACCGGCGTGCCCAGCAGATTCAGC GGCAGCGGCTCCGGCACCGACTTCACCCTGACCATCAGCAGCCTGCA GCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCTACC CCCCCACCTTCGGCCAGGGCACCAAGGTGGAAATCAAG 52 Pertuzumab DIQMTQSPSSLSASVGDRVTITCKASQDVSIGVAWYQQKPGKAPKLL (Tras.L3) IYSASYRYTGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQHYTT P96Y PYTFGQGTKVEIK (10959) 51 Pertuzumab GACATCCAGATGACCCAGAGCCCCAGCAGCCTGAGCGCCAGCGTGGG (Tras.L3) CGACAGAGTGACCATCACATGCAAGGCCAGCCAGGACGTGTCCATCG P96Y GCGTGGCCTGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTG (10959)DNA ATCTACAGCGCCAGCTACCGGTACACCGGCGTGCCCAGCAGATTCAG CGGCAGCGGCTCCGGCACCGACTTCACCCTGACCATCAGCAGCCTGC AGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACC CCCTACACCTTCGGCCAGGGCACCAAGGTGGAAATCAAG 54 Pertuzumab DIQMTQSPSSLSASVGDRVTITCKASQDVSTAVAWYQQKPGKAPKLL (Tras.L3) IYSASFRYTGVPSRFSGSRSGTDFTLTISSLQPEDFATYYCQQHYTT (Qm) PPTFGQGTKVEIK (11055) 53 Pertuzumab GACATCCAGATGACCCAGAGCCCCAGCAGCCTGTCTGCCAGCGTGGG (Tras.L3) CGACAGAGTGACCATCACATGCAAGGCCAGCCAGGACGTGTCCACAG (Qm) CCGTGGCCTGGTATCAGCAGAAGCCTGGCAAGGCCCCCAAGCTGCTG (11055)DNA ATCTACAGCGCCAGCTTCCGGTACACCGGCGTGCCCAGCAGATTCAG CGGCAGCAGATCCGGCACCGACTTCACCCTGACCATCAGCTCCCTGC AGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACC CCCCCCACATTTGGCCAGGGCACCAAGGTGGAAATCAAG 89 Pertuzumab KASQDVSTAVA (Tras.L3) (QM)-CDR1 90 Pertuzumab SASFRYT (Tras.L3) (QM)-CDR2 19 Pertuzumab QQHYTTPPT (Tras.L3) (QM)-CDR3 Pertuzumab/Trastuzumab hybrid light chain affinity matured VH clones 62 Pertuzumab EVQLVESGGGLVQPGGSLRLSCAASGFTFNDYTMDWVRQAPGKGLEW aff.mat. VADVNPNSGGSIYNQRFKGRFTLSVDRSKNTLYLQMNSLRAEDTAVY clone D1 YCARNLGPFFYFDYWGQGTLVTVSS 61 Pertuzumab GAGGTGCAATTGGTTGAAAGCGGTGGTGGTCTGGTTCAGCCTGGTGG aff.mat. TAGCCTGCGTCTGAGCTGTGCAGCAAGCGGTTTTACCTTTAACGATT clone D1 ATACCATGGATTGGGTTCGTCAGGCACCGGGTAAAGGTCTGGAATGG DNA GTTGCAGATGTTAATCCGAATAGCGGTGGTAGCATTTATAACCAGCG TTTTAAAGGTCGTTTTACCCTGAGCGTTGATCGTAGCAAAAATACCC TGTATCTGCAAATGAATAGTCTGCGTGCAGAGGATACCGCAGTGTAT TATTGTGCACGTAACCTGGGTCCGTTCTTCTACTTTGATTATTGGGG TCAGGGCACCCTGGTTACCGTTAGCAGC 55 Pertuzumab GFTFNDYTMD aff.mat. clone D1- VH CDR1 15 Pertuzumab DVNPNSGGSIYNQRFKG aff.mat. clone D1- VH CDR2 56 Pertuzumab NLGPFFYFDY aff.mat. clone D1- VH CDR3 64 Pertuzumab EVQLVESGGGLVQPGGSLRLSCAASGFTFNDYTMDWVRQAPGKGLEW aff.mat. VADVNPNSGGSIVNRRFKGRFTLSVDRSKNTLYLQMNSLRAEDTAVY clone D1- YCARNLGPFFYFDYWGQGTLVTVSS derived 63 Pertuzumab GAGGTGCAATTGGTTGAAAGCGGTGGTGGTCTGGTTCAGCCTGGTGG aff.mat. TAGCCTGCGTCTGAGCTGTGCAGCAAGCGGTTTTACCTTTAACGATT clone D1- ATACCATGGATTGGGTTCGTCAGGCACCGGGTAAAGGTCTGGAATGG derived, GTTGCAGATGTTAATCCGAATAGCGGTGGTAGCATTGTTAACCGTCG DNA TTTTAAAGGTCGTTTTACCCTGAGCGTTGATCGTAGCAAAAATACCC TGTATCTGCAAATGAATAGTCTGCGTGCAGAGGATACCGCAGTGTAT TATTGTGCACGTAACCTGGGTCCGTTCTTCTACTTTGATTATTGGGG TCAGGGCACCCTGGTTACCGTTAGCAGC
55 Pertuzumab GFTFNDYTMD aff.mat. clone D1- derived VH CDR1 77 Pertuzumab DVNPNSGGSIVNRRFKG aff.mat. clone D1- derived VH CDR2 56 Pertuzumab NLGPFFYFDY aff.mat. clone D1- derived VH CDR3 66 Pertuzumab EVQLVESGGGLVQPGGSLRLSCAASGFTFNDYTMDWFRQAPGKGLEW aff.mat. VADVNPNSGGSIYNQRFKGRFTLSVDRSKNTLYLQMNSLRAEDTAVY clone B2 YCARNLGPNFYFDYWGQGTLVTVSS 65 Pertuzumab GAGGTGCAATTGGTTGAAAGCGGTGGTGGTCTGGTTCAGCCTGGTGG aff.mat. TAGCCTGCGTCTGAGCTGTGCAGCAAGCGGTTTTACCTTTAACGATT clone B2, ATACCATGGATTGGTTTCGTCAGGCACCGGGTAAAGGTCTGGAATGG DNA GTTGCAGATGTTAATCCGAATAGCGGTGGTAGCATTTATAACCAGCG TTTTAAAGGTCGTTTTACCCTGAGCGTTGATCGTAGCAAAAATACCC TGTATCTGCAAATGAATAGTCTGCGTGCAGAGGATACCGCAGTGTAT TATTGTGCACGTAATCTGGGTCCGAACTTCTACTTTGATTATTGGGG TCAGGGCACCCTGGTTACCGTTAGCAGC 55 Pertuzumab GFTFNDYTMD aff.mat. clone B2- VH CDR1 15 Pertuzumab DVNPNSGGSIYNQRFKG aff.mat. clone B2- VH CDR2 57 Pertuzumab NLGPNFYFDY aff.mat. clone B2- VH CDR3 68 Pertuzumab EVQLVESGGGLVQPGGSLRLSCAASGFTFADYTMDWVRQAPGKGLEW aff.mat. VADVNPNSGGSIYNQRFKGRFTLSVDRSKNTLYLQMNSLRAEDTAVY clone E1 YCARNLGPWFYFDYWGQGTLVTVSS 67 Pertuzumab GAGGTGCAATTGGTTGAAAGCGGTGGTGGTCTGGTTCAGCCTGGTGG aff.mat. TAGCCTGCGTCTGAGCTGTGCAGCAAGCGGTTTTACCTTTGCAGATT clone E1, ATACCATGGATTGGGTTCGTCAGGCACCGGGTAAAGGTCTGGAATGG DNA GTTGCAGATGTTAATCCGAATAGCGGTGGTAGCATTTATAACCAGCG TTTTAAAGGTCGTTTTACCCTGAGCGTTGATCGTAGCAAAAATACCC TGTATCTGCAAATGAATAGTCTGCGTGCAGAGGATACCGCAGTGTAT TATTGTGCACGTAATCTGGGTCCGTGGTTCTACTTTGATTATTGGGG TCAGGGCACCCTGGTTACCGTTAGCAGC 58 Pertuzumab GFTFADYTMD aff.mat. clone E1- VH CDR1 15 Pertuzumab DVNPNSGGSIYNQRFKG aff.mat. clone E1- VH CDR2 59 Pertuzumab NLGPWFYFDY aff.mat. clone E1- VH CDR3 70 Pertuzumab EVQLVESGGGLVQPGGSLRLSCAASGFTFTDYTMDWVRQAPGKGLEW aff.mat. VADVNPNSGGYIVNRRFKGRFTLSVDRSKNTLYLQMNSLRAEDTAVY clone G2 YCARNLGPSFYFDYWGQGTLVTVSS 69 Pertuzumab GAGGTGCAATTGGTTGAAAGCGGTGGTGGTCTGGTTCAGCCTGGTGG aff.mat. TAGCCTGCGTCTGAGCTGTGCAGCAAGCGGTTTTACCTTTACCGATT clone G2, ACACAATGGATTGGGTTCGTCAGGCACCGGGTAAAGGTCTGGAATGG DNA GTTGCAGATGTTAATCCGAACTCTGGTGGTTACATTGTTAACCGTCG TTTTAAAGGTCGTTTTACCCTGAGCGTTGATCGTAGCAAAAATACCC TGTATCTGCAAATGAATAGTCTGCGTGCAGAGGATACCGCAGTGTAT TATTGTGCACGTAATCTGGGTCCGAGCTTCTATTTTGATTATTGGGG TCAGGGCACCCTGGTTACCGTTAGCAGC 14 Pertuzumab GFTFTDYTMD aff.mat. clone G2- VH CDR1 60 Pertuzumab DVNPNSGGYIVNRRFKG aff.mat. clone G2- VH CDR2 16 Pertuzumab NLGPSFYFDY aff.mat. clone G2- VH CDR3 72 Pertuzumab EVQLVESGGGLVQPGGSLRLSCAASGFTFTDYTMDWVRQAPGKGLEW aff.mat. VADVNPNSGGSIMNRRFKGRFTLSVDRSKNTLYLQMNSLRAEDTAVY clone C8 YCARNLGPSFYFDYWGQGTLVTVSS 71 Pertuzumab GAGGTGCAATTGGTTGAAAGCGGTGGTGGTCTGGTTCAGCCTGGTGG aff.mat. TAGCCTGCGTCTGAGCTGTGCAGCAAGCGGTTTTACCTTTACCGATT clone C8, ACACAATGGATTGGGTTCGTCAGGCACCGGGTAAAGGTCTGGAATGG DNA GTTGCAGATGTTAATCCGAACTCTGGTGGTTCTATTATGAACCGTCG TTTTAAAGGTCGTTTTACCCTGAGCGTTGATCGTAGCAAAAATACCC TGTATCTGCAAATGAATAGTCTGCGTGCAGAGGATACCGCAGTGTAT TATTGTGCACGTAATCTGGGTCCGAGCTTCTATTTTGATTATTGGGG TCAGGGCACCCTGGTTACCGTTAGCAGC 14 Pertuzumab GFTFTDYTMD aff.mat. clone C8- VH CDR1 75 Pertuzumab DVNPNSGGSIMNRRFKG aff.mat. clone C8- VH CDR2 16 Pertuzumab NLGPSFYFDY aff.mat. clone C8- VH CDR3 74 Pertuzumab EVQLVESGGGLVQPGGSLRLSCAASGFTFTDYTMDWVRQAPGKGLEW aff.mat. VADVNPNSGGSIVNQRFKGRFTLSVDRSKNTLYLQMNSLRAEDTAVY clone A1 YCARNLGPWFYFDYWGQGTLVTVSS 73 Pertuzumab GAGGTGCAATTGGTTGAAAGCGGTGGTGGTCTGGTTCAGCCTGGTGG aff.mat. TAGCCTGCGTCTGAGCTGTGCAGCAAGCGGTTTTACCTTTACCGATT clone A1, ACACAATGGATTGGGTTCGTCAGGCACCGGGTAAAGGTCTGGAATGG DNA GTTGCAGATGTTAATCCGAACTCTGGTGGTTCTATTGTTAACCAGCG TTTTAAAGGTCGTTTTACCCTGAGCGTTGATCGTAGCAAAAATACCC TGTATCTGCAAATGAATAGTCTGCGTGCAGAGGATACCGCAGTGTAT TATTGTGCACGTAATCTGGGTCCGTGGTTCTACTTTGATTATTGGGG TCAGGGCACCCTGGTTACCGTTAGCAGC 14 Pertuzumab GFTFTDYTMD aff.mat. clone A1- VH CDR1 76 Pertuzumab DVNPNSGGSIVNQRFKG aff.mat. clone A1- VH CDR2 59 Pertuzumab NLGPWFYFDY aff.mat. clone A1- VH CDR3 Trastuzumab Stabilization Variants 84 Trastuzumab DIQMTQSPSSLSASVGDRVTITCRASQDVNAAVAWYQQKPGKAPKLL VL T31A IYSASFLYSGVPSRFSGSRSGTDFTLTISSTQPEDFATYYCQQHYTT (6641) PPTFGQGTKVEIK 83 Trastuzumab GATATCCAGATGACCCAGTCTCCATCCTCCCTGTCTGCATCTGTAGG VL T31A AGACAGAGTCACCATCACTTGCCGGGCAAGTCAGGACGTGAACGCCG (6641) CTGTAGCGTGGTACCAGCAGAAACCAGGTAAGGCACCGAAGCTATTA ATTTATAGTGCGAGCTTCCTGTACAGTGGGGTCCCGTCGCGTTTTAG CGGCTCTCGATCCGGCACGGATTTTACCCTGACCATTAGCAGCCTGC AGCCTGAAGACTTTGCGACATATTATTGCCAACAGCACTACACAACT CCTCCCACCTTTGGCCAGGGTACGAAAGTTGAAATTAAA 103 Trastuzumab RASQDVNAAVA VL T31A (6641)CDR1 18 Trastuzumab SASFLYS VL T31A (6641)CDR2 19 Trastuzumab QQHYTTPPT VL T31A (6641)CDR3 86 Trastuzumab DIQMTQSPSSLSASVGDRVTITCRASQDVNVAVAWYQQKPGKAPKLL VL T31 IYSASFLYSGVPSRFSGSRSGTDFTLTISSTQPEDFATYYCQQHYTT (6642)V PPTFGQGTKVEIK 85 Trastuzumab GATATCCAGATGACCCAGTCTCCATCCTCCCTGTCTGCATCTGTAGG VL T31V AGACAGAGTCACCATCACTTGCCGGGCAAGTCAGGACGTGAACGTGG (6642)DNA CTGTAGCGTGGTACCAGCAGAAACCAGGTAAGGCACCGAAGCTATTA ATTTATAGTGCGAGCTTCCTGTACAGTGGGGTCCCGTCGCGTTTTAG CGGCTCTCGATCCGGCACGGATTTTACCCTGACCATTAGCAGCCTGC AGCCTGAAGACTTTGCGACATATTATTGCCAACAGCACTACACAACT CCTCCCACCTTTGGCCAGGGTACGAAAGTTGAAATTAAAG 104 Trastuzumab RASQDVNVAVA VL T31V (6642)CDR1 18 Trastuzumab SASFLYS VL T31V (6642)CDR2 19 Trastuzumab QQHYTTPPT VL T31V (6642)CDR3 158 Trastuzumab RASQDVSTAVA VL N30S CDR1 94 Trastuzumab EVQLVESGGGLVQPGGSLRLSCAASGFNIKDTYIHWVRQAPGKGLEW VH (D98N) VARTYPINGYTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVY (6636) YCSRWGGNGFYAMDYWGQGTLVTVSS 93 Trastuzumab GAAGTGCAATTGGTGGAAAGCGGCGGCGGCCTGGTGCAACCGGGCGG VH (D98N) CAGCCTGCGTCTGAGCTGCGCGGCCTCCGGATTTAACATAAAGGACA (6636)DNA CATACATCCACTGGGTGCGCCAAGCACCTGGGAAGGGTCTCGAGTGG GTGGCTCGGATTTACCCAACAAATGGCTACACCAGGTATGCGGATAG CGTGAAAGGCCGTTTTACCATTTCAGCTGATACTTCGAAGAACACCG CCTATCTGCAAATGAACAGCCTGCGTGCGGAAGATACGGCCGTGTAT TATTGCTCGCGTTGGGGAGGAAACGGGTTCTATGCTATGGATTACTG GGGCCAAGGCACCCTGGTGACGGTTAGCTCA 20 Trastuzumab GFNIKDTYIH VH (D98N) (6636)CDR1 29 Trastuzumab RIYPTNGYTRYADSVKG VH (D98N) (6636)CDR2 78 Trastuzumab WGGNGFYAMDY VH (D98N) (6636)CDR3 96 Trastuzumab EVQLVESGGGLVQPGGSLRLSCAASGFNIKDTYIHWVRQAPGKGLEW VH (D98E) VARTYPINGYTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVY (6637) YCSRWGGEGFYAMDYWGQGTLVTVSS 95 Trastuzumab GAAGTGCAATTGGTGGAAAGCGGCGGCGGCCTGGTGCAACCGGGCGG VH (D98E) CAGCCTGCGTCTGAGCTGCGCGGCCTCCGGATTTAACATAAAGGACA (6637)DNA CATACATCCACTGGGTGCGCCAAGCACCTGGGAAGGGTCTCGAGTGG GTGGCTCGGATTTACCCAACAAATGGCTACACCAGGTATGCGGATAG CGTGAAAGGCCGTTTTACCATTTCAGCTGATACTTCGAAGAACACCG CCTATCTGCAAATGAACAGCCTGCGTGCGGAAGATACGGCCGTGTAT TATTGCTCGCGTTGGGGAGGAGAGGGGTTCTATGCTATGGATTACTG GGGCCAAGGCACCCTGGTGACGGTTAGCTCA 20 Trastuzumab GFNIKDTYIH
VH (D98E) (6637)CDR1 29 Trastuzumab RIYPTNGYTRYADSVKG VH (D98E) (6637)CDR2 79 Trastuzumab WGGEGFYAMDY VH (D98E) (6637)CDR3 98 Trastuzumab EVQLVESGGGLVQPGGSLRLSCAASGFNIKDTYIHWVRQAPGKGLEW VH (D98T) VARTYPINGYTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVY (6638) YCSRWGGTGFYAMDYWGQGTLVTVSS 97 Trastuzumab GAAGTGCAATTGGTGGAAAGCGGCGGCGGCCTGGTGCAACCGGGCGG VH (D98T) CAGCCTGCGTCTGAGCTGCGCGGCCTCCGGATTTAACATAAAGGACA (6638)DNA CATACATCCACTGGGTGCGCCAAGCACCTGGGAAGGGTCTCGAGTGG GTGGCTCGGATTTACCCAACAAATGGCTACACCAGGTATGCGGATAG CGTGAAAGGCCGTTTTACCATTTCAGCTGATACTTCGAAGAACACCG CCTATCTGCAAATGAACAGCCTGCGTGCGGAAGATACGGCCGTGTAT TATTGCTCGCGTTGGGGAGGAACCGGGTTCTATGCTATGGATTACTG GGGCCAAGGCACCCTGGTGACGGTTAGCTCA 20 Trastuzumab GFNIKDTYIH VH (D98T) (6638)CDR1 29 Trastuzumab RIYPTNGYTRYADSVKG VH (D98T) (6638)CDR2 80 Trastuzumab WGGTGFYAMDY VH (D98T) (6638)CDR3 100 Trastuzumab EVQLVESGGGLVQPGGSLRLSCAASGFNIKDTYIHWVRQAPGKGLEW VH (G99A) VARIYPTNGYTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVY (6639) YCSRWGGDAFYAMDYWGQGTLVTVSS 99 Trastuzumab GAAGTGCAATTGGTGGAAAGCGGCGGCGGCCTGGTGCAACCGGGCGG VH (G99A) CAGCCTGCGTCTGAGCTGCGCGGCCTCCGGATTTAACATAAAGGACA (6639)DNA CATACATCCACTGGGTGCGCCAAGCACCTGGGAAGGGTCTCGAGTGG GTGGCTCGGATTTACCCAACAAATGGCTACACCAGGTATGCGGATAG CGTGAAAGGCCGTTTTACCATTTCAGCTGATACTTCGAAGAACACCG CCTATCTGCAAATGAACAGCCTGCGTGCGGAAGATACGGCCGTGTAT TATTGCTCGCGTTGGGGAGGAGACGCCTTCTATGCTATGGATTACTG GGGCCAAGGCACCCTGGTGACGGTTAGCTCA 20 Trastuzumab GFNIKDTYIH VH (G99A) (6639)CDR1 29 Trastuzumab RIYPTNGYTRYADSVKG VH (G99A) (6639)CDR2 87 Trastuzumab WGGDAFYAMDY VH (G99A) (6639)CDR3 102 Trastuzumab EVQLVESGGGLVQPGGSLRLSCAASGFNIKDTYIHWVRQAPGKGLEW VH (G99S) VARIYPTNGYTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVY (6640) YCSRWGGDSFYAMDYWGQGTLVTVSS 101 Trastuzumab GAAGTGCAATTGGTGGAAAGCGGCGGCGGCCTGGTGCAACCGGGCGG VH (G99S) CAGCCTGCGTCTGAGCTGCGCGGCCTCCGGATTTAACATAAAGGACA (6640)DNA CATACATCCACTGGGTGCGCCAAGCACCTGGGAAGGGTCTCGAGTGG GTGGCTCGGATTTACCCAACAAATGGCTACACCAGGTATGCGGATAG CGTGAAAGGCCGTTTTACCATTTCAGCTGATACTTCGAAGAACACCG CCTATCTGCAAATGAACAGCCTGCGTGCGGAAGATACGGCCGTGTAT TATTGCTCGCGTTGGGGAGGAGACAGCTTCTATGCTATGGATTACTG GGGCCAAGGCACCCTGGTGACGGTTAGCTCA 20 Trastuzumab GFNIKDTYIH VH (G99S) (6640)CDR1 29 Trastuzumab RIYPTNGYTRYADSVKG VH (G99S) (6640)CDR2 88 Trastuzumab WGGDSFYAMDY VH (G99S) (6640)CDR3
TABLE-US-00039 TABLE 34 Antigens SEQ ID NO Name Sequence 2 Her2 ECD MGWSCIILFLVATATGVHSTQVCTGTDMKLRLPASPETHLDMLRHLY QGCQVVQGNLELTYLPTNASLSFLQDIQEVQGYVLIAHNQVRQVPLQ RLRIVRGTQLFEDNYALAVLDNGDPLNNTTPVTGASPGGLRELQLRS LTEILKGGVLIQRNPQLCYQDTILWKDIFHKNNQLALTLIDTNRSRA CHPCSPMCKGSRCWGESSEDCQSLTRTVCAGGCARCKGPLPTDCCHE QCAAGCTGPKHSDCLACLHFNHSGICELHCPALVTYNTDTFESMPNP EGRYTFGASCVTACPYNYLSTDVGSCTLVCPLHNQEVTAEDGTQRCE KCSKPCARVCYGLGMEHLREVRAVISANIQEFAGCKKIFGSLAFLPE SFDGDPASNTAPLQPEQLQVFETLEEITGYLYISAWPDSLPDLSVFQ NLQVIRGRILHNGAYSLTLQGLGISWLGLRSLRELGSGLALIHHNTH LCFVHTVPWDQLFRNPHQALLHTANRPEDECVGEGLACHQLCARGHC WGPGPTQCVNCSQFLRGQECVEECRVLQGLPREYVNARHCLPCHPEC QPQNGSVICFGLEADQCVACAHYKDPPFCVARCPSGVKPDLSYMPIW KFPDEEGACQPCPINCTHSCVDLDDKGCPAEQRASPLTVDEQLYFQG GSGLNDIFEAQKIEWHEARAHHHHHH 1 Her2 ECD ACCCAAGTGTGCACCGGCACAGACATGAAGCTGCGGCTCCCTGCCAG DNA TCCCGAGACCCACCTGGACATGCTCCGCCACCTCTACCAGGGCTGCC AGGTGGTGCAGGGAAACCTGGAACTCACCTACCTGCCCACCAATGCC AGCCTGTCCTTCCTGCAGGATATCCAGGAGGTGCAGGGCTACGTGCT CATCGCTCACAACCAAGTGAGGCAGGTCCCACTGCAGAGGCTGCGGA TTGTGCGAGGCACCCAGCTCTTTGAGGACAACTATGCCCTGGCCGTG CTAGACAATGGAGACCCGCTGAACAATACCACCCCTGTCACAGGGGC CTCCCCAGGAGGCCTGCGGGAGCTGCAGCTTCGAAGCCTCACAGAGA TCTTGAAAGGAGGGGTCTTGATCCAGCGGAACCCCCAGCTCTGCTAC CAGGACACGATTTTGTGGAAGGACATCTTCCACAAGAACAACCAGCT GGCTCTCACACTGATAGACACCAACCGCTCTCGGGCCTGCCACCCCT GTTCTCCGATGTGTAAGGGCTCCCGCTGCTGGGGAGAGAGTTCTGAG GATTGTCAGAGCCTGACGCGCACTGTCTGTGCCGGTGGCTGTGCCCG CTGCAAGGGGCCACTGCCCACTGACTGCTGCCATGAGCAGTGTGCTG CCGGCTGCACGGGCCCCAAGCACTCTGACTGCCTGGCCTGCCTCCAC TTCAACCACAGTGGCATCTGTGAGCTGCACTGCCCAGCCCTGGTCAC CTACAACACAGACACGTTTGAGTCCATGCCCAATCCCGAGGGCCGGT ATACATTCGGCGCCAGCTGTGTGACTGCCTGTCCCTACAACTACCTT TCTACGGACGTGGGATCCTGCACCCTCGTCTGCCCCCTGCACAACCA AGAGGTGACAGCAGAGGATGGAACACAGCGGTGTGAGAAGTGCAGCA AGCCCTGTGCCCGAGTGTGCTATGGTCTGGGCATGGAGCACTTGCGA GAGGTGAGGGCAGTTACCAGTGCCAATATCCAGGAGTTTGCTGGCTG CAAGAAGATCTTTGGGAGCCTGGCATTTCTGCCGGAGAGCTTTGATG GGGACCCAGCCTCCAACACTGCCCCGCTCCAGCCAGAGCAGCTCCAA GTGTTTGAGACTCTGGAAGAGATCACAGGTTACCTATACATCTCAGC ATGGCCGGACAGCCTGCCTGACCTCAGCGTCTTCCAGAACCTGCAAG TAATCCGGGGACGAATTCTGCACAATGGCGCCTACTCGCTGACCCTG CAAGGGCTGGGCATCAGCTGGCTGGGGCTGCGCTCACTGAGGGAACT GGGCAGTGGACTGGCCCTCATCCACCATAACACCCACCTCTGCTTCG TGCACACGGTGCCCTGGGACCAGCTCTTTCGGAACCCGCACCAAGCT CTGCTCCACACTGCCAACCGGCCAGAGGACGAGTGTGTGGGCGAGGG CCTGGCCTGCCACCAGCTGTGCGCCCGAGGGCACTGCTGGGGTCCAG GGCCCACCCAGTGTGTCAACTGCAGCCAGTTCCTTCGGGGCCAGGAG TGCGTGGAGGAATGCCGAGTACTGCAGGGGCTCCCCAGGGAGTATGT GAATGCCAGGCACTGTTTGCCGTGCCACCCTGAGTGTCAGCCCCAGA ATGGCTCAGTGACCTGTTTTGGACTGGAGGCTGACCAGTGTGTGGCC TGTGCCCACTATAAGGACCCTCCCTTCTGCGTGGCCCGCTGCCCCAG CGGTGTGAAACCTGACCTCTCCTACATGCCCATCTGGAAGTTTCCAG ATGAGGAGGGCGCATGCCAGCCTTGCCCCATCAACTGCACCCACTCC TGTGTGGACCTGGATGACAAGGGCTGCCCCGCCGAGCAGAGAGCCAG CCCTCTGACGGTCGACGAACAGTTATATTTTCAGGGCGGCTCAGGCC TGAACGACATCTTCGAGGCCCAGAAGATCGAGTGGCACGAGGCTCGA GCTCACCACCATCACCATCAC 4 Fc (hole) DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVS HEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWL NGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKN QVSLSCAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVS KLTVDKSRWQQGNVFSCSVMHEALHNRFTQKSLSLSPGK 3 Fc (hole) DNA GACAAAACTCACACATGCCCACCGTGCCCAGCACCTGAACTCCTGGG GGGACCGTCAGTCTTCCTCTTCCCCCCAAAACCCAAGGACACCCTCA TGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTGGACGTGAGC CACGAAGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGGA GGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTACAACAGCA CGTACCGTGTGGTCAGCGTCCTCACCGTCCTGCACCAGGACTGGCTG AATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAGCCCTCCCAGC CCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAAC CACAGGTGTGCACCCTGCCCCCATCCCGGGATGAGCTGACCAAGAAC CAGGTCAGCCTCTCGTGCGCAGTCAAAGGCTTCTATCCCAGCGACAT CGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGA CCACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCGTGAGC AAGCTCACCGTGGACAAGAGCAGGTGGCAGCAGGGGAACGTCTTCTC ATGCTCCGTGATGCATGAGGCTCTGCACAACCGCTTCACGCAGAAGA GCCTCTCCCTGTCTCCGGGTAAA 6 Her2 ECD- TQVCTGTDMKLRLPASPETHLDMLRHLYQGCQVVQGNLELTYLPTNA Fc (knob) SLSFLQDIQEVQGYVLIAHNQVRQVPLQRLRIVRGTQLFEDNYALAV LDNGDPLNNTTPVTGASPGGLRELQLRSLTEILKGGVLIQRNPQLCY QDTILWKDIFHKNNQLALTLIDTNRSRACHPCSPMCKGSRCWGESSE DCQSLTRTVCAGGCARCKGPLPTDCCHEQCAAGCTGPKHSDCLACLH FNHSGICELHCPALVTYNTDTFESMPNPEGRYTFGASCVTACPYNYL STDVGSCTLVCPLHNQEVTAEDGTQRCEKCSKPCARVCYGLGMEHLR EVRAVTSANIQEFAGCKKIFGSLAFLPESFDGDPASNTAPLQPEQLQ VFETLEEITGYLYISAWPDSLPDLSVFQNLQVIRGRILHNGAYSLTL QGLGISWLGLRSLRELGSGLALIHHNTHLCFVHTVPWDQLFRNPHQA LLHTANRPEDECVGEGLACHQLCARGHCWGPGPTQCVNCSQFLRGQE CVEECRVLQGLPREYVNARHCLPCHPECQPQNGSVTCFGLEADQCVA CAHYKDPPFCVARCPSGVKPDLSYMPIWKFPDEEGACQPCPINCTHS CVDLDDKGCPAEQRASPLTVDGGSPTPPTPGGGSADKTHTCPPCPAP ELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYV DGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNK ALPAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFY PSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQG NVFSCSVMHEALHNHYTQKSLSLSPGKSGGLNDIFEAQKIEWHE 5 Her2 ECD- ACCCAAGTGTGCACCGGCACAGACATGAAGCTGCGGCTCCCTGCCAG Fc (knob) DNA TCCCGAGACCCACCTGGACATGCTCCGCCACCTCTACCAGGGCTGCC AGGTGGTGCAGGGAAACCTGGAACTCACCTACCTGCCCACCAATGCC AGCCTGTCCTTCCTGCAGGATATCCAGGAGGTGCAGGGCTACGTGCT CATCGCTCACAACCAAGTGAGGCAGGTCCCACTGCAGAGGCTGCGGA TTGTGCGAGGCACCCAGCTCTTTGAGGACAACTATGCCCTGGCCGTG CTAGACAATGGAGACCCGCTGAACAATACCACCCCTGTCACAGGGGC CTCCCCAGGAGGCCTGCGGGAGCTGCAGCTTCGAAGCCTCACAGAGA TCTTGAAAGGAGGGGTCTTGATCCAGCGGAACCCCCAGCTCTGCTAC CAGGACACGATTTTGTGGAAGGACATCTTCCACAAGAACAACCAGCT GGCTCTCACACTGATAGACACCAACCGCTCTCGGGCCTGCCACCCCT GTTCTCCGATGTGTAAGGGCTCCCGCTGCTGGGGAGAGAGTTCTGAG GATTGTCAGAGCCTGACGCGCACTGTCTGTGCCGGTGGCTGTGCCCG CTGCAAGGGGCCACTGCCCACTGACTGCTGCCATGAGCAGTGTGCTG CCGGCTGCACGGGCCCCAAGCACTCTGACTGCCTGGCCTGCCTCCAC TTCAACCACAGTGGCATCTGTGAGCTGCACTGCCCAGCCCTGGTCAC CTACAACACAGACACGTTTGAGTCCATGCCCAATCCCGAGGGCCGGT ATACATTCGGCGCCAGCTGTGTGACTGCCTGTCCCTACAACTACCTT TCTACGGACGTGGGATCCTGCACCCTCGTCTGCCCCCTGCACAACCA AGAGGTGACAGCAGAGGATGGAACACAGCGGTGTGAGAAGTGCAGCA AGCCCTGTGCCCGAGTGTGCTATGGTCTGGGCATGGAGCACTTGCGA GAGGTGAGGGCAGTTACCAGTGCCAATATCCAGGAGTTTGCTGGCTG CAAGAAGATCTTTGGGAGCCTGGCATTTCTGCCGGAGAGCTTTGATG GGGACCCAGCCTCCAACACTGCCCCGCTCCAGCCAGAGCAGCTCCAA GTGTTTGAGACTCTGGAAGAGATCACAGGTTACCTATACATCTCAGC ATGGCCGGACAGCCTGCCTGACCTCAGCGTCTTCCAGAACCTGCAAG TAATCCGGGGACGAATTCTGCACAATGGCGCCTACTCGCTGACCCTG CAAGGGCTGGGCATCAGCTGGCTGGGGCTGCGCTCACTGAGGGAACT GGGCAGTGGACTGGCCCTCATCCACCATAACACCCACCTCTGCTTCG TGCACACGGTGCCCTGGGACCAGCTCTTTCGGAACCCGCACCAAGCT CTGCTCCACACTGCCAACCGGCCAGAGGACGAGTGTGTGGGCGAGGG CCTGGCCTGCCACCAGCTGTGCGCCCGAGGGCACTGCTGGGGTCCAG GGCCCACCCAGTGTGTCAACTGCAGCCAGTTCCTTCGGGGCCAGGAG TGCGTGGAGGAATGCCGAGTACTGCAGGGGCTCCCCAGGGAGTATGT GAATGCCAGGCACTGTTTGCCGTGCCACCCTGAGTGTCAGCCCCAGA ATGGCTCAGTGACCTGTTTTGGACTGGAGGCTGACCAGTGTGTGGCC TGTGCCCACTATAAGGACCCTCCCTTCTGCGTGGCCCGCTGCCCCAG CGGTGTGAAACCTGACCICTCCTACATGCCCATCTGGAAGTTTCCAG ATGAGGAGGGCGCATGCCAGCCTTGCCCCATCAACTGCACCCACTCC TGTGTGGACCTGGATGACAAGGGCTGCCCCGCCGAGCAGAGAGCCAG CCCTCTGACGGTCGACGGTGGTAGTCCGACACCTCCGACACCCGGGG GTGGTTCTGCAGACAAAACTCACACATGCCCACCGTGCCCAGCACCT GAACTCCTGGGGGGACCGTCAGTCTTCCTCTTCCCCCCAAAACCCAA GGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGG TGGACGTGAGCCACGAAGACCCTGAGGTCAAGTTCAACTGGTACGTG GACGGCGTGGAGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCA GTACAACAGCACGTACCGTGTGGTCAGCGTCCTCACCGTCCTGCACC AGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAA GCCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCA GCCCCGAGAACCACAGGTGTACACCCTGCCCCCATGCCGGGATGAGC TGACCAAGAACCAGGTCAGCCTGTGGTGCCTGGTCAAAGGCTTCTAT CCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAA CAACTACAAGACCACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCT TCCTCTACAGCAAGCTCACCGTGGACAAGAGCAGGTGGCAGCAGGGG AACGTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAACCACTA CACGCAGAAGAGCCTCTCCCTGTCTCCGGGTAAATCCGGAGGCCTGA ACGACATCTTCGAGGCCCAGAAGATTGAATGGCACGAG 8 Her2 TQVCTGTDMKLRLPASPETHLDMLRHLYQGCQVVQGNLELTYLPTNA ECD (pertuzumab SLSFLQDIQEVQGYVLIAHNQVRQVPLQRLRIVRGTQLFEDNYALAV KO- LDNGDPLNNTTPVTGASPGGLRELQLRSLTEILKGGVLIQRNPQLCY Fc (knob) QDTILWKDIFHKNNQLALTLIDTNRSRACHPCSPMCKGSRCWGESSE DCQSLTRTVCAGGCARCKGPLPTDCCHEQCAAGCTGPKHSDCLACLH FNHSGICELHCPALVTYNTDTRESMPNPEGRYRFGASCVTACPYNYL STDRGSCTLVCPLANQEVTAEDGTQRCEKCSKPCARVCYGLGMEHLR EVRAVTSANIQEFAGCKKIFGSLAFLPESFDGDPASNTAPLQPEQLQ VFETLEEITGYLYISAWPDSLPDLSVFQNLQVIRGRILHNGAYSLTL QGLGISWLGLRSLRELGSGLALIHHNTHLCFVHTVPWDQLFRNPHQA LLHTANRPEDECVGEGLACHQLCARGHCWGPGPTQCVNCSQFLRGQE CVEECRVLQGLPREYVNARHCLPCHPECQPQNGSVTCFGLEADQCVA CAHYKDPPFCVARCPSGVKPDLSYMPIWKFPDEEGACQPCPINCTHS CVDLDDKGCPAEQRASPLTVDGGSPTPPTPGGGSADKTHTCPPCPAP ELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYV DGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNK ALPAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFY PSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQG NVFSCSVMHEALHNHYTQKSLSLSPGKSGGLNDIFEAQKIEWHE 7 Her2 ACCCAAGTGTGCACCGGCACAGACATGAAGCTGCGGCTCCCTGCCAG ECD (pertuzumab TCCCGAGACCCACCTGGACATGCTCCGCCACCTCTACCAGGGCTGCC KO)- AGGTGGTGCAGGGAAACCTGGAACTCACCTACCTGCCCACCAATGCC Fc (knob) DNA AGCCTGTCCTTCCTGCAGGATATCCAGGAGGTGCAGGGCTACGTGCT CATCGCTCACAACCAAGTGAGGCAGGTCCCACTGCAGAGGCTGCGGA TTGTGCGAGGCACCCAGCTCTTTGAGGACAACTATGCCCTGGCCGTG CTAGACAATGGAGACCCGCTGAACAATACCACCCCTGTCACAGGGGC CTCCCCAGGAGGCCTGCGGGAGCTGCAGCTTCGAAGCCTCACAGAGA TCTTGAAAGGAGGGGTCTTGATCCAGCGGAACCCCCAGCTCTGCTAC CAGGACACGATTTTGTGGAAGGACATCTTCCACAAGAACAACCAGCT GGCTCTCACACTGATAGACACCAACCGCTCTCGGGCCTGCCACCCCT GTTCTCCGATGTGTAAGGGCTCCCGCTGCTGGGGAGAGAGTTCTGAG GATTGTCAGAGCCTGACGCGCACTGTCTGTGCCGGTGGCTGTGCCCG CTGCAAGGGGCCACTGCCCACTGACTGCTGCCATGAGCAGTGTGCTG CCGGCTGCACGGGCCCCAAGCACTCTGACTGCCTGGCCTGCCTCCAC TTCAACCACAGTGGCATCTGTGAGCTGCACTGCCCAGCCCTGGTCAC CTACAACACAGACACGCGGGAGTCCATGCCCAATCCCGAGGGCCGGT ATAGATTCGGCGCCAGCTGTGTGACTGCCTGTCCCTACAACTACCTT TCTACGGACCGGGGATCCTGCACCCTCGTCTGCCCCCTGGCCAACCA AGAGGTGACAGCAGAGGATGGAACACAGCGGTGTGAGAAGTGCAGCA AGCCCTGTGCCCGAGTGTGCTATGGTCTGGGCATGGAGCACTTGCGA GAGGTGAGGGCAGTTACCAGTGCCAATATCCAGGAGTTTGCTGGCTG CAAGAAGATCTTTGGGAGCCTGGCATTTCTGCCGGAGAGCTTTGATG GGGACCCAGCCTCCAACACTGCCCCGCTCCAGCCAGAGCAGCTCCAA GTGTTTGAGACTCTGGAAGAGATCACAGGTTACCTATACATCTCAGC ATGGCCGGACAGCCTGCCTGACCTCAGCGTCTTCCAGAACCTGCAAG TAATCCGGGGACGAATTCTGCACAATGGCGCCTACTCGCTGACCCTG CAAGGGCTGGGCATCAGCTGGCTGGGGCTGCGCTCACTGAGGGAACT GGGCAGTGGACTGGCCCTCATCCACCATAACACCCACCTCTGCTTCG TGCACACGGTGCCCTGGGACCAGCTCTTTCGGAACCCGCACCAAGCT CTGCTCCACACTGCCAACCGGCCAGAGGACGAGTGTGTGGGCGAGGG CCTGGCCTGCCACCAGCTGTGCGCCCGAGGGCACTGCTGGGGTCCAG GGCCCACCCAGTGTGTCAACTGCAGCCAGTTCCTTCGGGGCCAGGAG TGCGTGGAGGAATGCCGAGTACTGCAGGGGCTCCCCAGGGAGTATGT GAATGCCAGGCACTGTTTGCCGTGCCACCCTGAGTGTCAGCCCCAGA ATGGCTCAGTGACCTGTTTTGGACTGGAGGCTGACCAGTGTGTGGCC TGTGCCCACTATAAGGACCCTCCCTTCTGCGTGGCCCGCTGCCCCAG CGGTGTGAAACCTGACCTCTCCTACATGCCCATCTGGAAGTTTCCAG ATGAGGAGGGCGCATGCCAGCCTTGCCCCATCAACTGCACCCACTCC TGTGTGGACCTGGATGACAAGGGCTGCCCCGCCGAGCAGAGAGCCAG CCCTCTGACGGTCGACGGTGGTAGTCCGACACCTCCGACACCCGGGG GTGGTTCTGCAGACAAAACTCACACATGCCCACCGTGCCCAGCACCT GAACTCCTGGGGGGACCGTCAGTCTTCCTCTTCCCCCCAAAACCCAA GGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGG TGGACGTGAGCCACGAAGACCCTGAGGTCAAGTTCAACTGGTACGTG GACGGCGTGGAGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCA GTACAACAGCACGTACCGTGTGGTCAGCGTCCTCACCGTCCTGCACC AGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAA GCCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCA GCCCCGAGAACCACAGGTGTACACCCTGCCCCCATGCCGGGATGAGC TGACCAAGAACCAGGTCAGCCTGTGGTGCCTGGTCAAAGGCTTCTAT CCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAA CAACTACAAGACCACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCT TCCTCTACAGCAAGCTCACCGTGGACAAGAGCAGGTGGCAGCAGGGG AACGTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAACCACTA CACGCAGAAGAGCCTCTCCCTGTCTCCGGGTAAATCCGGAGGCCTGA ACGACATCTTCGAGGCCCAGAAGATTGAATGGCACGAG 10 Her2 TQVCTGTDMKLRLPASPETHLDMLRHLYQGCQVVQGNLELTYLPTNA ECD (trastuzumab SLSFLQDIQEVQGYVLIAHNQVRQVPLQRLRIVRGTQLFEDNYALAV KO)- LDNGDPLNNTTPVTGASPGGLRELQLRSLTEILKGGVLIQRNPQLCY Fc (knob) QDTILWKDIFHKNNQLALTLIDTNRSRACHPCSPMCKGSRCWGESSE DCQSLTRTVCAGGCARCKGPLPTDCCHEQCAAGCTGPKHSDCLACLH FNHSGICELHCPALVTYNTDTFESMPNPEGRYTFGASCVTACPYNYL
STDVGSCTLVCPLHNQEVTAEDGTQRCEKCSKPCARVCYGLGMEHLR EVRAVTSANIQEFAGCKKIFGSLAFLPESFDGDPASNTAPLQPEQLQ VFETLEEITGYLYISAWPDSLPDLSVFQNLQVIRGRILHNGAYSLTL QGLGISWLGLRSLRELGSGLALIHHNTHLCFVHTVPWDQLFRNPHQA LLHTANRPEDECVGEGLACHQLCARGHCWGPGPTQCVNCSQFLRGQE CVEECRVLQGLPREYVNARHCLPCHPECQPQNGSVTCFGLEARQCVA CAHYKDRRCVARCPSGVKPDLSYMPIWKFPDEEGACQPCPINCTHSC VDLDDKGCPAEQRASPLTVDGGSPTPPTPGGGSADKTHTCPPCPAPE LLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVD GVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKA LPAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYP SDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGN VFSCSVMHEALHNHYTQKSLSLSPGKSGGLNDIFEAQKIEWHE 9 Her2 ACCCAAGTGTGCACCGGCACAGACATGAAGCTGCGGCTCCCTGCCAG ECD (trastuzumab TCCCGAGACCCACCTGGACATGCTCCGCCACCTCTACCAGGGCTGCC KO)- AGGTGGTGCAGGGAAACCTGGAACTCACCTACCTGCCCACCAATGCC Fc (knob) DNA AGCCTGTCCTTCCTGCAGGATATCCAGGAGGTGCAGGGCTACGTGCT CATCGCTCACAACCAAGTGAGGCAGGTCCCACTGCAGAGGCTGCGGA TTGTGCGAGGCACCCAGCTCTTTGAGGACAACTATGCCCTGGCCGTG CTAGACAATGGAGACCCGCTGAACAATACCACCCCTGTCACAGGGGC CTCCCCAGGAGGCCTGCGGGAGCTGCAGCTTCGAAGCCTCACAGAGA TCTTGAAAGGAGGGGTCTTGATCCAGCGGAACCCCCAGCTCTGCTAC CAGGACACGATTTTGTGGAAGGACATCTTCCACAAGAACAACCAGCT GGCTCTCACACTGATAGACACCAACCGCTCTCGGGCCTGCCACCCCT GTTCTCCGATGTGTAAGGGCTCCCGCTGCTGGGGAGAGAGTTCTGAG GATTGTCAGAGCCTGACGCGCACTGTCTGTGCCGGTGGCTGTGCCCG CTGCAAGGGGCCACTGCCCACTGACTGCTGCCATGAGCAGTGTGCTG CCGGCTGCACGGGCCCCAAGCACTCTGACTGCCTGGCCTGCCTCCAC TTCAACCACAGTGGCATCTGTGAGCTGCACTGCCCAGCCCTGGTCAC CTACAACACAGACACGTTTGAGTCCATGCCCAATCCCGAGGGCCGGT ATACATTCGGCGCCAGCTGTGTGACTGCCTGTCCCTACAACTACCTT TCTACGGACGTGGGATCCTGCACCCTCGTCTGCCCCCTGCACAACCA AGAGGTGACAGCAGAGGATGGAACACAGCGGTGTGAGAAGTGCAGCA AGCCCTGTGCCCGAGTGTGCTATGGTCTGGGCATGGAGCACTTGCGA GAGGTGAGGGCAGTTACCAGTGCCAATATCCAGGAGTTTGCTGGCTG CAAGAAGATCTTTGGGAGCCTGGCATTTCTGCCGGAGAGCTTTGATG GGGACCCAGCCTCCAACACTGCCCCGCTCCAGCCAGAGCAGCTCCAA GTGTTTGAGACTCTGGAAGAGATCACAGGTTACCTATACATCTCAGC ATGGCCGGACAGCCTGCCTGACCTCAGCGTCTTCCAGAACCTGCAAG TAATCCGGGGACGAATTCTGCACAATGGCGCCTACTCGCTGACCCTG CAAGGGCTGGGCATCAGCTGGCTGGGGCTGCGCTCACTGAGGGAACT GGGCAGTGGACTGGCCCTCATCCACCATAACACCCACCTCTGCTTCG TGCACACGGTGCCCTGGGACCAGCTCTTTCGGAACCCGCACCAAGCT CTGCTCCACACTGCCAACCGGCCAGAGGACGAGTGTGTGGGCGAGGG CCTGGCCTGCCACCAGCTGTGCGCCCGAGGGCACTGCTGGGGTCCAG GGCCCACCCAGTGTGTCAACTGCAGCCAGTTCCTTCGGGGCCAGGAG TGCGTGGAGGAATGCCGAGTACTGCAGGGGCTCCCCAGGGAGTATGT GAATGCCAGGCACTGTTTGCCGTGCCACCCTGAGTGTCAGCCCCAGA ATGGCTCAGTGACCTGTTTTGGACTGGAGGCTCGGCAGTGTGTGGCC TGTGCCCACTATAAGGACAGACGGTGCGTGGCCCGCTGCCCCAGCGG TGTGAAACCTGACCTCTCCTACATGCCCATCTGGAAGTTTCCAGATG AGGAGGGCGCATGCCAGCCTTGCCCCATCAACTGCACCCACTCCTGT GTGGACCTGGATGACAAGGGCTGCCCCGCCGAGCAGAGAGCCAGCCC TCTGACGGTCGACGGTGGTAGTCCGACACCTCCGACACCCGGGGGTG GTTCTGCAGACAAAACTCACACATGCCCACCGTGCCCAGCACCTGAA CTCCTGGGGGGACCGTCAGTCTTCCTCTTCCCCCCAAAACCCAAGGA CACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTGG ACGTGAGCCACGAAGACCCTGAGGTCAAGTTCAACTGGTACGTGGAC GGCGTGGAGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTA CAACAGCACGTACCGTGTGGTCAGCGTCCTCACCGTCCTGCACCAGG ACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAGCC CTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCC CCGAGAACCACAGGTGTACACCCTGCCCCCATGCCGGGATGAGCTGA CCAAGAACCAGGTCAGCCTGTGGTGCCTGGTCAAAGGCTTCTATCCC AGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAA CTACAAGACCACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCC TCTACAGCAAGCTCACCGTGGACAAGAGCAGGTGGCAGCAGGGGAAC GTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAACCACTACAC GCAGAAGAGCCTCTCCCTGTCTCCGGGTAAATCCGGAGGCCTGAACG ACATCTTCGAGGCCCAGAAGATTGAATGGCACGAG
TABLE-US-00040 TABLE 35 Full-length antibody sequences of common light chain antibody ''D1 der'' SEQ ID NO Name Sequence 159 Trastuzumab EVQLVESGGGLVQPGGSLRLSCAASGFNIKDTYIHWVRQAPGKGLEW VHCH1-Fc VARIYPTNGYTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVY KNOB YCSRWGGDGFYAMDYWGQGTLVTVSSASTKGPSVFPLAPSSKSTSGG TAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSV VTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAP ELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYV DGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNK ALPAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFY PSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQG NVFSCSVMHEALHNHYTQKSLSLSPGK 160 Trastuzumab GAAGTGCAATTGGTGGAAAGCGGCGGCGGCCTGGTGCAACCGGGCGGCAGCC VHCH1-Fc TGCGTCTGAGCTGCGCGGCCTCCGGATTTAACATAAAGGACACATACATCCA KNOB DNA CTGGGTGCGCCAAGCACCTGGGAAGGGTCTCGAGTGGGTGGCTCGGATTTAC CCAACAAATGGCTACACCAGGTATGCGGATAGCGTGAAAGGCCGTTTTACCA TTTCAGCTGATACTTCGAAGAACACCGCCTATCTGCAAATGAACAGCCTGCG TGCGGAAGATACGGCCGTGTATTATTGCTCGCGTTGGGGAGGAGACGGGTTC TATGCTATGGATTACTGGGGCCAAGGCACCCTGGTGACGGTTAGCTCAGCTA GCACCAAGGGCCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTC TGGGGGCACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCG GTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCC CGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGT GCCCTCCAGCAGCTTGGGCACCCAGACCTACATCTGCAACGTGAATCACAAG CCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTGACAAAA CTCACACATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGT CTTCCTCTTCCCCCCAAAACCCAAGGACACCCTCATGATCTCCCGGACCCCT GAGGTCACATGCGTGGTGGTGGACGTGAGCCACGAAGACCCTGAGGTCAAGT TCAACTGGTACGTGGACGGCGTGGAGGTGCATAATGCCAAGACAAAGCCGCG GGAGGAGCAGTACAACAGCACGTACCGTGTGGTCAGCGTCCTCACCGTCCTG CACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAG CCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCG AGAACCACAGGTGTACACCCTGCCCCCATGCCGGGATGAGCTGACCAAGAAC CAGGTCAGCCTGTGGTGCCTGGTCAAAGGCTTCTATCCCAGCGACATCGCCG TGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGACCACGCCTCC CGTGCTGGACTCCGACGGCTCCTTCTTCCTCTACAGCAAGCTCACCGTGGAC AAGAGCAGGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGTGATGCATGAGG CTCTGCACAACCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGTAAA 161 Common DIQMTQSPSSLSASVGDRVTITCKASQDVSTAVAWYQQKPGKAPKLLIYSAS light chain FRYTGVPSRFSGSRSGTDFTLTISSLQPEDFATYYCQQHYTTPPTFGQGTKV VLCL EIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQS GNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKS FNRGEC 162 Common GACATCCAGATGACCCAGAGCCCCAGCAGCCTGTCTGCCAGCGTGGGCGACAGAGTG light chain ACCATCACATGCAAGGCCAGCCAGGACGTGTCCACAGCCGTGGCCTGGTATCAGCAG VLCL-DNA AAGCCTGGCAAGGCCCCCAAGCTGCTGATCTACAGCGCCAGCTTCCGGTACACCGGC GTGCCCAGCAGATTCAGCGGCAGCAGATCCGGCACCGACTTCACCCTGACCATCAGC TCCCTGCAGCCCGAGGACTTCGCCACCTACTACTGCCAGCAGCACTACACCACCCCC CCCACATTTGGCCAGGGCACCAAGGTGGAAATCAAGCGTACGGTGGCTGCACCATCT GTCTTCATCTTCCCGCCATCTGATGAGCAGTTGAAATCTGGAACTGCCTCTGTTGTG TGCCTGCTGAATAACTTCTATCCCAGAGAGGCCAAAGTACAGTGGAAGGTGGATAAC GCCCTCCAATCGGGTAACTCCCAGGAGAGTGTCACAGAGCAGGACAGCAAGGACAGC ACCTACAGCCTCAGCAGCACCCTGACGCTGAGCAAAGCAGACTACGAGAAACACAAA GTCTACGCCTGCGAAGTCACCCATCAGGGCCTGAGCTCGCCCGTCACAAAGAGCTTC AACAGGGGAGAGTGT 163 Pertuzumab EVQLVESGGGLVQPGGSLRLSCAASGFTFNDYTMDWVRQAPGKGLEWVADVN VHCH1 Fc PNSGGSIVNRRFKGRFTLSVDRSKNTLYLQMNSLRAEDTAVYYCARNLGPFF hole YFDYWGQGTLVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPV TVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKP SNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPE VTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLH QDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKNQ VSLSCAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDK SRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK 164 Pertuzumab GAAGTTCAGCTGGTTGAAAGCGGTGGTGGTCTGGTTCAGCCTGGTGGTAGCC VHCH1 Fc TGCGTCTGAGCTGTGCAGCAAGCGGTTTTACCTTTAACGATTATACCATGGA hole DNA TTGGGTTCGTCAGGCACCGGGTAAAGGTCTGGAATGGGTTGCAGATGTTAAT CCGAATAGCGGTGGTAGCATTGTTAACCGTCGTTTTAAAGGTCGTTTTACCC TGAGCGTTGATCGTAGCAAAAATACCCTGTATCTGCAAATGAATAGTCTGCG TGCAGAGGATACCGCAGTGTATTATTGTGCACGTAACCTGGGTCCGTTCTTC TACTTTGATTATTGGGGTCAGGGCACCCTGGTTACCGTTAGCAGCGCTAGCA CCAAGGGCCCAAGCGTGTTCCCTCTGGCCCCCAGCAGCAAGAGCACAAGCGG CGGAACAGCCGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAGCCCGTG ACAGTGTCCTGGAACAGCGGAGCCCTGACCAGCGGCGTGCACACCTTTCCAG CCGTGCTGCAGAGCAGCGGCCTGTACAGCCTGAGCAGCGTGGTCACAGTGCC TAGCAGCAGCCTGGGCACCCAGACCTACATCTGCAACGTGAACCACAAGCCC AGCAACACCAAGGTGGACAAGAAGGTGGAGCCCAAGAGCTGCGACAAGACCC ACACCTGTCCCCCTTGTCCTGCCCCTGAGCTGCTGGGCGGACCCAGCGTGTT CCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGCCGGACCCCCGAA GTGACCTGCGTGGTGGTGGACGTGTCCCACGAGGACCCTGAAGTGAAGTTCA ATTGGTACGTGGACGGCGTGGAGGTGCACAATGCCAAGACCAAGCCCCGGGA GGAACAGTACAACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCAC CAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTCTCCAACAAGGCCC TGCCTGCCCCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCAGAGA ACCCCAGGTGTGCACCCTGCCCCCCAGCAGAGATGAGCTGACCAAGAACCAG GTGTCCCTGAGCTGTGCCGTCAAGGGCTTCTACCCCAGCGATATCGCCGTGG AGTGGGAGAGCAACGGCCAGCCTGAGAACAACTACAAGACCACCCCCCCTGT GCTGGACAGCGACGGCAGCTTCTTCCTGGTGTCCAAACTGACCGTGGACAAG AGCCGGTGGCAGCAGGGCAACGTGTTCAGCTGCAGCGTGATGCACGAGGCCC TGCACAACCACTACACCCAGAAGTCCCTGAGCCTGAGCCCCGGCAAG
Example 21: Generation of Trivalent HER2 (Extracellular Domain IV)--HER2 (Extracellular Domain II)--Transferrin Specific Antibodies
Generation of the Expression Plasmids
[0610] Desired proteins were expressed by transient transfection of human embryonic kidney cells (HEK 293). For the expression of a desired gene/protein (e.g. antibody-Fab multimeric protein) a transcription unit comprising the following functional elements was used: [0611] The immediate early enhancer and promoter from the human cytomegalovirus (P-CMV) including intron A, [0612] a murine immunoglobulin heavy chain signal sequence (SS), [0613] a gene/protein to be expressed (e.g. full length antibody heavy chain), and [0614] the bovine growth hormone polyadenylation sequence (BGH pA).
[0615] Besides the expression unit/cassette including the desired gene to be expressed the basic/standard mammalian expression plasmid contains: [0616] An origin of replication from the vector pUC18 which allows replication of this plasmid in E. coli, and [0617] a beta-lactamase gene which confers ampicillin resistance in E. coli Expression Plasmids Coding for the Following Antibody+/-scFab Proteins were Constructed:
1. BBB-0580 Trivalent Herceptarg-scFab(8D3)
[0618] Heavy chain knob (Herceptin(D98E)(IgG1) knob SS-(G4S)4-VL-Ck-(G4S)6-GG-VH-CH1) (Seq. 21832/SEQ ID NO.: 165)
[0619] Composition of the knob Herceptin(D98E)-scFab(8D3) heavy chain fusion protein: [0620] Herceptin(D89E) human IgG1 heavy chain without C-terminal Lys containing CH3 knob mutation T366W and the S354C mutation for the formation of an additional disulfide bridge [0621] Glycine-Serine-linker [0622] Variable light chain domain (VL) variant of the mouse 8D3 anti-transferrin antibody (Boado et al., 2009) [0623] Human C-kappa light chain [0624] Glycine-Serine-linker [0625] Variable heavy chain domain (VL) variant of the mouse 8D3 anti-transferrin antibody (Boado et al., 2009) [0626] Human IgG1 CH1 heavy chain domain
[0627] Heavy chain hole (Pertuzumab(aff.mat.)(IgG1) hole SS (Seq. ID NO.: 163) Composition of the hole Pertuzumab(aff.mat.) heavy chain protein: [0628] Pertuzumab(aff.mat.) human IgG1 heavy chain containing the CH3 hole mutations T366S, Y407V and L368A and the Y349C mutation for the formation of an additional disulfide bridge
[0629] Light chain (CLC-Herc combo) (clone D1-derSEQ ID NO.: 161) [0630] See example 14
2. BBB-0581 Trivalent Herceptarg(LALA)-scFab(8D3)
[0631] Heavy chain knob (Herceptin(D98E)(IgG1) LALA-PG knob SS-(G4S)4-VL-Ck-(G4S)6-GG-VH-CH1) (Seq. 21834, SEQ ID. NO.: 168)
[0632] Composition of the knob Herceptin(D98E)-scFab(8D3) heavy chain fusion protein: [0633] Herceptin(D89E) human IgG1 heavy chain without C-terminal Lys containing: [0634] CH2 mutations L234A, L235A and P329G to impair Fc gamma receptor-mediated interaction with immune effector cells as well as complement activation [0635] CH3 knob mutation T366W and the S354C mutation for the formation of an additional disulfide bridge [0636] Glycine-Serine-linker [0637] Variable light chain domain (VL) variant of the mouse 8D3 anti-transferrin antibody (Boado et al., 2009), SEQ ID NO.: 178 [0638] Human C-kappa light chain [0639] Glycine-Serine-linker [0640] Variable heavy chain domain (VL) variant of the mouse 8D3 anti-transferrin antibody (Boado et al., 2009), SEQ ID NO.: 179 [0641] Human IgG1 CH1 heavy chain domain
[0642] Heavy chain hole (Pertuzumab(aff.mat.)(IgG1) LALA-PG hole SS (Seq. 21836, SEQ ID NO.: 169)
[0643] Composition of the hole Pertuzumab(aff.mat.) heavy chain protein: [0644] Pertuzumab(aff.mat.) human IgG1 heavy chain containing: [0645] CH2 mutations L234A, L235A and P329G to impair Fc gamma receptor-mediated interaction with immune effector cells as well as complement activation [0646] CH3 hole mutations T366S, L368A and Y407V and the Y349C mutation for the formation of an additional disulfide bridge [0647] CH3 hole mutations H435R and Y436F to avoid protein purification of heavy chain hole-hole dimers
[0648] Light chain (CLC-Herc combo) (clone D1-derSEQ ID NO.: 161) [0649] See example 14 3. BBB-0582 Bivalent Herceptarg(LALA) (Control Molecule without BBB-R Binder)
[0650] Heavy chain knob (Herceptin(D98E)(IgG1) LALA-PG (Seq. 21835, SEQ ID. NO.: 170) Composition of the knob Herceptin(D98E) heavy chain protein: [0651] Herceptin(D89E) human IgG1 heavy chain containing: [0652] CH2 mutations L234A, L235A and P329G to impair Fc gamma receptor-mediated interaction with immune effector cells as well as complement activation [0653] CH3 knob mutation T366W and the S354C mutation for the formation of an additional disulfide bridge
[0654] Heavy chain hole (Pertuzumab(aff.mat.)(IgG1) LALA-PG hole SS (Seq. 21836 SEQ ID. NO.: 163)
[0655] Composition of the hole Pertuzumab(aff.mat.) heavy chain protein: [0656] Pertuzumab(aff.mat.) human IgG1 heavy chain containing: [0657] CH2 mutations L234A, L235A and P329G to impair Fc gamma receptor-mediated interaction with immune effector cells as well as complement activation [0658] CH3 hole mutations T366S, L368A and Y407V and the Y349C mutation for the formation of an additional disulfide bridge [0659] CH3 hole mutations H435R and Y436F to avoid protein purification of hole-hole dimers
[0660] Light chain (CLC-Herc combo) (clone D1-derSEQ ID NO.: 161) [0661] See example 14 4. Herceptarg (Control Molecule without BBB-R Binder)
[0662] Heavy chain knob (Herceptin(D98E)(IgG1) (SEQ ID NO.: 171)
[0663] Composition of the knob Herceptin(D98E) heavy chain protein: [0664] Herceptin(D89E) human IgG1 heavy chain containing CH3 knob mutation T366W and the S354C mutation for the formation of an additional disulfide bridge
[0665] Heavy chain hole (Pertuzumab(aff.mat.)(IgG1) hole SS (Seq. 21836 SEQ ID. NO.: 163) Composition of the hole Pertuzumab(aff.mat.) heavy chain protein: [0666] Pertuzumab(aff.mat.) human IgG1 heavy chain containing CH3 hole mutations T366S, L368A and Y407V and the Y349C mutation for the formation of an additional disulfide bridge
[0667] Light chain (CLC-Herc combo) (clone D1-derSEQ ID NO.: 161) [0668] See example 14
[0669] Sequences are shown in tables 36 and 37.
[0670] Ref Boado R J, Zhang Y, Wang Y, Pardridge W M. Engineering and expression of a chimeric transferrin receptor monoclonal antibody for blood-brain barrier delivery in the mouse. Biotechnol Bioeng. 2009; 102(4):1251-8.
Example 22: Purification of Trivalent HER2 (Extracellular Domain IV)-HER2 (Extracellular Domain II)-Transferrin Specific Antibodies and Control Antibodies without BBB-R Binder
[0671] The antibody chains were generated by transient transfection of HEK293 cells (human embryonic kidney cell line 293-derived) cultivated in F17 Medium (Invitrogen Corp.). For transfection 293-Free Transfection Reagent (Merck, Millipore) was used. The antibody chains were expressed from three different plasmids, coding for the trivalent Herceptarg-scFab(8D3) knob and hole heavy chains and the Herceptarg light chain or the bivalent Herceptarg knob and hole heavy chains and the Herceptarg light chain, respectively. For transfection the three plasmids were used at plasmid ratios of 2:1:1 (LC: HC knob: HC hole) for trivalent Herceptarg-scFab(8D3), of 1:1:2 (LC: HC knob: HC hole) for trivalent Herceptarg(LALA)-scFab(8D3) and bivalent Herceptarg(LALA) and of 2:1:1 (LC: HC knob: HC hole) for bivalent Herceptarg. Transfections were performed as specified in the manufacturer's instructions. Antibody-containing cell culture supernatants were harvested seven days after transfection. Supernatants were stored frozen until purification.
[0672] Proteins were purified from filtered cell culture supernatants. Supernatants were applied to a protein A Sepharose column (GE Healthcare) and washed with PBS pH 7.4. Elution of antibodies was achieved with 100 mM Citarate buffer at pH 3.0 followed by immediate neutralization of the sample to pH 6.5. Aggregated protein and other byproducts were separated from monomeric antibodies by size exclusion chromatography (SEC; Superdex 200; GE Healthcare) in 20 mM histidine, 140 mM NaCl, pH 6.0. Every single fraction was analyzed on analytical SEC (MabPac SEC-1, Thermo Scientific) and on a chip-based capillary electrophoresis system (CE-SDS, LabChipGX, Caliper) for the quantification of incompletely assembled molecules and other byproducts. Monomeric antibody fractions without byproducts were pooled. After concentration using a MILLIPOREAmicon Ultra (30 molecular weight cut off) centrifugal concentrator the protein was stored at -80.degree. C. Analytical characterization of the endproduct was done by UV protein determination, CE-SDS, size exclusion chromatography, mass spectrometry and also by endotoxin determination.
Example 23: Binding Analysis of Herceptarg-scFab(8D3) and Control Antibodies to BT474-M1 and BA/F3 Cells
[0673] HER2+ BT474-M1 and TfR+ BA/F3 cells were used as target cells for binding analysis of Herceptarg-scFab(8D3) and Herceptarg control molecules. BT474-M1 cells were maintained in RPMI 1640 supplemented with 10% fetal calf serum and 2 mL L-glutamine and BA/F3 cells were maintained in RPMI 1640, 10% FCS, 2 mM L-glutamine and 10 ng/ml rmIL-3. Propagation of cell lines followed standard cell culture protocols.
[0674] BT474-M1 and BA/F3 cells were harvested and resuspended in FACS buffer. 0.2 Mio cells were seeded into a 96 well round bottom plate. The plate was centrifuged at 350 g for 3 min to pellet the cells. The supernatant was removed and the cells were resuspended in 120 .mu.l of the diluted antibodies. The plate was incubated for 1 h at 4.degree. C. to allow binding of the antibodies. To remove unbound antibodies the cells were centrifuged again and washed twice with FACS buffer. To detect the antibodies the cells were resuspended in 100 .mu.l diluted goat anti-human Fc.gamma. specific PE-labeled secondary antibody (Jackson ImmunoResearch #109-116-170) and incubated again for 30 min at 4.degree. C. Afterwards the cells were washed twice with FACS buffer, resuspended in 50 .mu.l FACS buffer and the fluorescence was measured with BD Canto II. Results are shown in FIGS. 33 and 34. All Herceptarg antibodies specifically bind to HER2+ BT474-M1 cells and the Herceptarg-scFab(8D3) fusion proteins bind to TfR+ BA/F3 cells.
Example 24: Proliferation Inhibition Assay with Trivalent HER2 (Extracellular Domain IV)-HER2 (Extracellular Domain II)-Transferrin Specific Antibodies and Control Antibodies without BBB-R Binder on HER2+BT474-M1 Cells
[0675] The ability of the Herceptarg molecules to inhibit proliferation was assessed in the cell line BT474-M1. Cells in the logarithmic growth phase were detached, counted and 4.times.10.sup.3 cells were seeded in 60 .mu.L medium per well of a 96-well cell culture plate. Cells were maintained overnight in the incubator and the following day 60 .mu.L of the respective antibodies diluted in medium were added in form of a dilution series to the cells. After a total incubation time of 5 days cell growth was assessed by CellTiter-Glo.RTM. Luminescent Cell Viability Assay. The assay was performed as recommended by the manufacturer.
[0676] FIG. 35 shows inhibition of BT474-M1 cell proliferation after incubation with Herceptarg+/-scFab(8D3) molecules.
TABLE-US-00041 TABLE 36 BBB-R binders SEQ ID NO Description Sequence 172 2/99 CDR-H1 FSLSSY 173 2/99 CDR-H2 YIWSGGSTDYASWA 174 2/99 CDR-H3 RYGTSYPDYGDANGFDP 175 2/99 CDR-L1 QASQSISSYLS 176 2/99 CDR-L2 RAS 177 2/99 CDR-L3 CYSSSNVDN 178 2/99 VH QSMEESGGRLVTPGTPLTLTCTVSGFSLSSYAMSWVRQAPGKGLEWIGYIWSG GSTDYASWAKGRFTISKTSTTVDLKITSPTTEDTATYFCARRYGTSYPDYGDA NGFDPWGPGTLVTVSS 179 2/99 VL AYDMTQTPASVEVAVGGTVTIKCQASQSISSYLSWYQQKPGQRPKLLIYRAST LASGVSSRFKGSGSGTQFTLTISGVECADAATYYCQQCYSSSNVDNTFGGGTE VVVKR 180 4/94 CDR-H1 GFNIKDT 181 4/94 CDR-H2 RIDPANGDTKCDPKFQ 182 4/94 CDR-H3 YLYPYYFD 183 4/94 CDR-L1 SESVDTY 184 4/94 CDR-L2 GAS 185 4/94 CDR-L3 TYNYPL 166 4/94 VH EVQLQQSGAVLVKPGASVKLSCPASGFNIKDTY1HWVIQRPEQGLEWIGRIDP ANGDTKCDPKFQVKATITADTSSNTAYLQLSSLTSEDTAVYFCVRDYLYPYYF DFWGQGTTLTVSS 167 4/94 VL KIVMTQSPKSMSMSVGERVTLNCRASESVDTYVSWYQQKPEQSPELLIYGAS NRYTGVPDRFTGSGSATDFTLTISSVQAEDLADYYCGQTYNYPLTFGAGTKLE LKR 186 Humanized SYAMS 2/99 CDR-H1 187 Humanized YIWSGGSTDY ASWAKS 2/99 CDR-H2 188 Humanized RYGTSYPDYGDASGFDP 2/99CDR-H3 206 Humanized RYGTSYPDYGDAQGFDP 2/99CDR-H3 variant 189 Humanized RASQSISSYL A 2/99 CDR-L1 190 Humanized RASTLAS 2/99 CDR-L2 191 Humanized QQNYASSNVD NT 2/99 CDR-L3 192 Humanized QSMQESGPGL VKPSQTLSLT CTVSGFSLSS YAMSWIRQHP 2/99 VH GKGLEWIGYIWSGGSTDYASWAKSRVTISK TSTTVSLKLS SVTAADTAVY YCARRYGTSYPDYGDASGFDPWGQGTLVTV SS 205 Humanized QSMQESGPGL VKPSQTLSLT CTVSGFSLSS YAMSWIRQHP GKGLEWIGYI 2/99 VH WSGGSTDYAS WAKSRVTISK TSTTVSLKLS SVTAADTAVY variant YCARRYGTSYPDYGDAQGFDPWGQGTLVTVSS 193 Humanized AIQLTQSPSS LSASVGDRVT ITCRASQSIS SYLAWYQQKP GKAPKLLIYR 2/99 VL ASTLASGVPS RFSGSGSGTD FTLTISSLQP EDFATYYCQQ NYASSNVDNT FGGGTKVEI 194 Humanized QVQLVQSGAE VKKPGASVKV SCKASGFN1K DTYIHWVIQA PGQGLEWMGR 2/99 VH IDPANGDTKS DPKFQVRVTI TADTSTSTVY MELSSLRSED TAVYYCVRDY variant 1 LYPYYFDFWG QGTTVTVSS 195 Humanized QVQLVQSGAE VKKPGASVKV SCKASGFNIK DTYIHWVIQA PGQGLEWMGR 2/99 VH IDPANGDTKS APKFQVRVTI TADTSTSTVY MELSSLRSED TAVYYCVRDY variant 2 LYPYYFDFWG QGTTVTVSS 196 Humanized QVQLVQSGAE VKKPGASVKV SCKASGFNIK DTYIHWVIQA PGQGLEWMGR 2/99 VH IDPANGDTKS variant 3 DPKFQGRVTI TADTSTSTVY MELSSLRSED TAVYYCVRDY LYPYYFDFWG QGTTVTVSS 197 Humanized QVQLVQSGAE VKKPGSSVKV SCKASGFNIK DTY1HWVIQA PGQGLEWMGR 2/99 VH IDPANGDTKS DPKFQGRVTI TADESTSTAY MELSSLRSED variant 4 TAVYYCVRDY LYPYYFDFWG QGTTVTVSS 198 Humanized QVQLVQSGAE VKKPGSSVKV SCKASGFNIK DTYIHWVIQR PGQGLEWMGR 2/99 VH IDPANGDTKS variant 5 DPKFQGRVTI TADESTSTAY MELSSLRSED TAVYYCVRDY LYPYYFDFWG QGTTVTVSS 199 Humanized QVQLQESGPG LVKPSETLSL TCTVSGFN1K DTYIHWVIQR PGKGLEWIGR 2/99 VH IDPANGDTKS DPSLQSRVTI SADTSKNQAS LKLSSVTAAD variant 6 TAVYYCVRDY LYPYYFDFWG QGTTVTVSS 200 Humanized EVQLVESGGG LVQPGGSLRL SCAASGFNIK DTYIHWVIQA PGKGLEWVGR 2/99 VH IDPANGDTKS variant 7 DPSVQVRATI SADTSKNTAY LQMNSLRAED TAVYYCVRDY LYPYYFDFWG QGTTVTVSS 201 Humanized KIQMTQSPSS LSASVGDRVT ITCRASESVD TYVSWYQQKP GKAPKLLIYG 2/99 VL ASNRYTGVPS RFSGSGSGTD FTLTISSLQP EDFADYYCGQ TYNYPLTFGQ variant 1 GTKLEI 207 Humanized KIVLTQSPGT LSLSPGERAT LSCRASESVD TYVSWYQQKP GQAPRLLIYG 2/99 VL ASNRYTGVPD variant 2 RFSGSGSGTD FTLTISRLEP EDFADYYCGQ TYNYPLTFGQ GTKLEI 208 Humanized EIVLTQSPGT LSLSPGERAT LSCRASESVD TYVSWYQQKP GQAPRLLIYG 2/99 VL ASNRYTGVPD variant 3 RFSGSGSGTD FTLTISRLEP EDFADYYCGQ TYNYPLTFGQ GTKLEI 209 Humanized KIVLTQSPGT LSLSPGERAT LSCRASESVD TYVSWYQQKP GQAPRLLIYG 2/99 VL ASNRYTGIPD variant 4 RFSGSGSGTD FTLTISRLEP EDFADYYCGQ TYNYPLTFGQ GTKLEI
TABLE-US-00042 TABLE 37 Trivalent HER2 (extracellular domain IV)-HER2 (extracellular domain II)- Transferrin specific antibodies and control antibodies without BBB-R binder SEQ ID NO Description Sequence BBB-0580/Herceptarg-scFab(8D3) 165 Her-knob- evqlvesggglvqpggslrlscaasgfnikdtyihwvrqapgkglewvariyptngytry scFab(8D3) adsvkgrftisadtskntaylqmnslraedtavyycsrwggegfyamdywgqgtlvtvss Heavy astkgpsvfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssgl chain yslssvvtvpssslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsv 21832 flfppkpkdtlmisrtpevtcvvvdvshedpevkfnwyvdgvevhnaktkpreeqynst yrvvsvltvlhqdwilgkeykckvsnkalpapiektiskakgqprepqvytlpperdelt knqvslwclvkgfypsdiavewesngqpennykttppvldsdgsfflyskltvdksrwq qgnvfscsvmhealhnhytqkslslspgggsggggsggggsggggsdiqmtqspasls asleeivtitcqasqdignwlawyqqkpgkspqlliygatsladgvpsrfsgsrsgtqfslki srvqvedigiyyclqayntpwtfgggtkveikrtvaapsvfifppsdeqlksgtasvvclln nfypreakvqwkvdnalqsgnsqesvteqdskdstyslsstltlskadyekhkvyacevt hqglsspvtksfnrgecggggsggggsggggsggggsggggsggggsggevqlvesgg glvqpgnsltlscvasgftfsnygmhwirqapkkglewiamiyydsskmnyadtvkgr ftisrdnskntlylemnslrsedtamyycavptshyvvdvwgqgvsvtvssastkgpsvf plapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvtv pssslgtqtyicnvnhkpsntkvdkkvepksc 163 Per-hole evqlvesggglvqpggslrlscaasgftfndytmdwvrqapgkglewvadvnpnsggsi Heavy vnrrfkgrftlsvdrskntlylqmnslraedtavyycarnlgpffyfdywgqgtlvtvssast chain kgpsvfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglysl 21833 ssvvtvpssslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsvflf ppkpkdtlmisrtpevtcvvvdvshedpevkfnwyvdgvevhnaktkpreeqynstyr vvsvltvlhqdwlngkeykekvsnkalpapiektiskakgqprepqvctlppsrdeltkn qvslscavkgfypsdiavewesngqpennykttppvldsdgsfflvskltvdksrwqqg nvfscsvmhealhnhytqkslslspgk 161 Her/Per diqmtqspsslsasvgdrvtitckasqdvstavawyqqkpgkapklliysasfrytgvpsr CLC fsgsrsgtdftltisslqpedfatyycqqhyttpptfgqgtkveik 21831 rtvaapsvfifppsdeqlksgtasvvcllnnfypreakvqwkvdnalqsgnsqesvteqds kdstyslsstltlskadyekhkvyacevthqglsspvtksfnrgec BBB-0581/Herceptarg(LALA-PG)-scFab(8D3) 168 Her-knob- evqlvesggglvqpggslrlscaasgfnikdtyihwvrqapgkglewvariyptngytry LALA-PG- adsvkgrftisadtskntaylqmnslraedtavyycsrwggegfyamdywgqgtlvtvss scFab(8D3) astkgpsvfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssgl Heavy yslssvvtvpssslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapeaaggps chain vflfppkpkdtlmisrtpevtcvvvdvshedpevkfnwyvdgvevhnaktkpreeqyn 21834 styrvvsvltvlhqdwlngkeykekvsnkalgapiektiskakgqprepqvytlpperdel tknqvslwelvkgfypsdiavewesngqpennykttppvldsdgsfflyskltvdksrw qqgnvfsesvmhealhnhytqkslslspgggsggggsggggsggggsdiqmtqspasl sasleeivtitegasqdignwlawyqqkpgkspqlliygatsladgvpsrfsgsrsgtqfslk isrvqvedigiyyclqayntpwtfgggtkveikrtvaapsvfifppsdeqlksgtasvvcll nnfypreakvqwkvdnalqsgnsqesvteqdskdstyslsstltlskadyekhkvyacev thqglsspvtksfnrgecggggsggggsggggsggggsggggsggggsggevqlvesg gglvqpgnsltlscvasgftfsnygmhwirqapkkglewiamiyydsskmnyadtvkg rftisrdnskntlylemnslrsedtamyycavptshyvvdvwgqgvsvtvssastkgpsv fplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvtv pssslgtqtyicnvnhkpsntkvdkkvepksc 169 Per-hole Evqlvesggglvqpggslrlscaasgftfndytmdwvrqapgkglewvadvnpnsggs LALA-PG ivnrrfkgrftlsvdrskntlylqmnslraedtavyycarnlgpffyfdywgqgtlvtvssas Heavy tkgpsvfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglys chain lssvvtvpssslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapeaaggpsvf 21836 lfppkpkdtlmisrtpevtcvvvdvshedpevkfnwyvdgvevhnaktkpreeqynst yrvvsvltvlhqdwlngkeykekvsnkalgapiektiskakgqprepqvctlppsrdeltk nqvslscavkgfypsdiavewesngqpennykttppvldsdgsfflvskltvdksrwqq gnvfscsvmhealhnrftqkslslspgk 161 Her/Per diqmtqspsslsasvgdrvtitckasqdvstavawyqqkpgkapklliysasfrytgvpsr CLC fsgsrsgtdftltisslqpedfatyycqqhyttpptfgqgtkveik 21831 rtvaapsvfifppsdeqlksgtasvvcllnnfypreakvqwkvdnalqsgnsqesvteqds kdstyslsstltlskadyekhkvyacevthqglsspvtksfnrgec BBB-0582/Herceptarg-LALA-PG (control molecule without BBB-R binder) 170 Her-knob- evqlvesggglvqpggslrlscaasgfnikdtyihwvrqapgkglewvariyptngytry LALA-PG adsvkgrftisadtskntaylqmnslraedtavyycsrwggegfyamdywgqgtlvtvss Heavy astkgpsvfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssgl Chain yslssvvtvpssslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapeaaggps 21835 vflfppkpkdtlmisrtpevtcvvvdvshedpevkfnwyvdgvevhnaktkpreeqyn styrvvsvltvlhqdwlngkeykekvsnkalgapiektiskakgqprepqvytlpperdel tknqvslwelvkgfypsdiavewesngqpennykttppvldsdgsfflyskltvdksrw qqgnvfscsvmhealhnhytqkslslspgk 169 Per-hole Evqlvesggglvqpggslrlscaasgftfndytmdwvrqapgkglewvadvnpnsggs LALA-PG ivnrrfkgrftlsvdrskntlylqmnslraedtavyycarnlgpffyfdywgqgtlvtvssas Heavy tkgpsvfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglys chain lssvvtvpssslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapeaaggpsvf 21836 lfppkpkdtlmisrtpevtcvvvdvshedpevkfnwyvdgvevhnaktkpreeqynst yrvvsyltvlhqdwlngkeykekvsnkalgapiektiskakgqprepqvctlppsrdeltk nqvslscavkgfypsdiavewesngqpennykttppvldsdgsfflvskltvdksrwqq gnvfscsvmhealhnrftqkslslspgk 161 Her/Per diqmtqspsslsasvgdrvtitckasqdvstavawyqqkpgkapklliysasfrytgvpsr CLC fsgsrsgtdftltisslqpedfatyycqqhyttpptfgqgtkveik 21831 rtvaapsvfifppsdeqlksgtasvvcllnnfypreakvqwkvdnalqsgnsqesvteqds kdstyslsstltlskadyekhkvyacevthqglsspvtksfnrgec Herceptarg (control molecule without BBB-R binder) 171 pETR13573 evqlvesggglvqpggslrlscaasgfnikdtyihwvrqapgkglewvariyptngytry Her-knob adsvkgrftisadtskntaylqmnslraedtavyycsrwggegfyamdywgqgtlvtvss heavy chain astkgpsvfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssgl yslssvvtvpssslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsv flfppkpkdtlmisrtpevtcvvvdvshedpevkfnwyvdgvevhnaktkpreeqynst yrvvsvltvlhqdwlngkeykekvsnkalpapiektiskakgqprepqvytlpperdelt knqvslwclvkgfypsdiavewesngqpennykttppvldsdgsfflyskltvdksrwq qgnvfscsvmhealhnhytqkslslspgk 163 Per-hole evqlvesggglvqpggslrlscaasgftfndytmdwvrqapgkglewvadvnpnsggsi Heavy vnrrfkgrftlsvdrskntlylqmnslraedtavyycarnlgpffyfdywgqgtlvtvssast chain kgpsvfplapsskstsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglysl 21833 ssvvtvpssslgtqtyicnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsvflf ppkpkdtlmisrtpevtcvvvdvshedpevkfnwyvdgvevhnaktkpreeqynstyr vvsvltvlhqdwlngkeykekvsnkalpapiektiskakgqprepqvctlppsrdeltkn qvslscavkgfypsdiavewesngqpennykttppvldsdgsfflvskltvdksrwqqg nvfscsvmhealhnhytqkslslspgk 161 Her/Per diqmtqspsslsasvgdrvtitckasqdvstavawyqqkpgkapklliysasfrytgvpsr CLC fsgsrsgtdftltisslqpedfatyycqqhyttpptfgqgtkveik 21831 rtvaapsvfifppsdeqlksgtasvvcllnnfypreakvqwkvdnalqsgnsqesvteqds kdstyslsstltlskadyekhkvyacevthqglsspvtksfnrgec
TABLE-US-00043 TABLE 38 Sequence of pTAU-BBB-R control molecule SEQ ID NO Description Sequence 210 8936_pTau_ Aqvltqttspvsaavgstvtiscqssqsvrtnklawfqqkpgqppkrliysastldfgvpsrf RbMab86_VL_ sasgsgtqftltisdvqeddaatyyclgyfdesiadevafgggtevvykrtvaapsvfifpps huIgkappa deqlksgtasvvcllnnfypreakvqwkvdnalqsgnsqesvteqdskdstyslsstltlsk adyekhkvyacevthqglsspvtksfnrgec 211 8975-pTau- Qsveesggrlvtpgtpltltetvsgfslssnainwvrqapgkglewigyiavsgntyyasw Rb86- akgrftiskasttvdlkmtsptaedtgtyfcgksniwgpgtlvtvslastkgpsvfplapssk hugamma1-SS- stsggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvtvpssslgtq hole tyicnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsvflfppkpkdtlmisrt pevtcvvvdvshedpevkfnwyvdgvevhnaktkpreeqynstyrvvsyltvlhqdwl ngkeykckvsnkalpapiektiskakgqprepqvctlppsrdeltknqvslscavkgfyp sdiavewesngqpennykttppyldsdgsfflyskltvdksrwqqgnyfscsvmhealh nhytqkslslspgk 212 8976-pTau- qsveesggrlvtpgtpltltctvsgfslssnainwvrqapgkglewigyiaysgntyyaswa Rb86- kgrftiskasttvdlkmtsptaedtgtyfcgksniwgpgtlvtvslastkgpsvfplapsskst hugamma1-SS- sggtaalgclvkdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvtvpssslgtqty knob-mTfR- icnvnhkpsntkvdkkvepkscdkthtcppcpapellggpsvflfppkpkdtlmisrtpe 8D3-scFab vtcvvvdvshedpevkfnwyydgvevhnaktkpreeqynstyrvvsvltvlhqdwlng keykckvsnkalpapiektiskakgqprepqvytlppcrdeltknqvslwclvkgfypsd iavewesngqpennykttppyldsdgsfflyskltvdksrwqqgnyfscsvmhealhnh ytqkslslspgggsggggsggggsggggsdiqmtqspaslsasleeivtitcqasqdignw lawyqqkpgkspqlliygatsladgvpsrfsgsrsgtqfslkisrvqvedigiyyclqaynt pwtfgggtkveikrtvaapsyfifppsdeqlksgtasyycllnnfypreakvqwkydnal qsgnsqesvteqdskdstyslsstltlskadyekhkvyacevthqglsspvtksfnrgecgg ggsggggsggggsggggsggggsggggsggevqlvesggglyqpgnsltlscvasgftf snygmhwirqapkkglewiamiyydsskmnyadtvkgrftisrdnskntlylemnslr sedtamyycayptshyvvdvwgqgvsvtvssastkgpsvfplapsskstsggtaalgclv kdyfpepvtvswnsgaltsgvhtfpavlqssglyslssvvtvpssslgtqtyicnynhkpsn tkvdkkvepksc
Example 25: Characterization of Monovalent Anti TfR Antibodies
[0677] Identification of human and cynomolgus TfR-binding antibodies by cell ELISA To screen rabbit B-cell or mouse hybridoma supernatants for antibodies recognizing human and cynomolgus TfR, a cell ELISA using stably transfected CHO-K1 cells way employed. Stable transfectants were obtained by transfecting CHO-K1 cells with expression plasmids containing expression cassettes for the human or cynomolgus TfR as well as for neomycin-phosphotransferase. After transfection, cells were diluted in growth medium containing 500 .mu.g/mL G418 (Life Technologies). After appearance of growing clones, cells were detached, stained with MEM-75 (Abcam) or 13E4 (Life Technologies) and PE-labeled secondary antibodies for human or cynomolgus TfR, and highly fluorescent cells sorted as single cells into 96-well-plate wells (FACS Aria). After 7 days of growth, clones were again checked for TfR expression and best expressing clones selected for cell ELISA experiments.
[0678] Briefly, 15,000 cells were seeded per well of a 384-well plate and incubated for 18 h at 37.degree. C., 5% CO2. Supernatant was removed using an automated washer (BIOTEK), and 30 .mu.L of antibody-containing supernatant added to each well, followed by 24 .mu.L of growth medium. After 2 hours of incubation, wells were emptied and 30 .mu.L of 0.05% glutaraldehyde in PBS added for 45 min. at RT. After 3 washes with PBS/0.025% Tween20 (PBST), 30 .mu.L of anti-rabbit-HRP or anti-mouse-HRP (Southern Biotech) diluted 1:5000 in Blocking buffer was added and plates incubated for 1 hour at RT. Wells were washed 6 times with PBST and signal was generated using 30 .mu.L of TMB per well and absorbance measured at 450 nm.
Cloning and Expression of Anti-TfR Antibodies
[0679] Recombinant DNA techniques
[0680] Standard methods were used to manipulate DNA as described in Sambrook, J. et al., Molecular cloning: A laboratory manual; Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., 1989. The molecular biological reagents were used according to the manufacturer's instructions. [0681] Gene and oligonucleotide synthesis
[0682] Desired gene segments were prepared by chemical synthesis at Geneart GmbH (Regensburg, Germany). The synthesized gene fragments were cloned into an E. coli plasmid for propagation/amplification. The DNA sequences of subcloned gene fragments were verified by DNA sequencing. Alternatively, short synthetic DNA fragments were assembled by annealing chemically synthesized oligonucleotides or via PCR. The respective oligonucleotides were prepared by metabion GmbH (Planegg-Martinsried, Germany).
PCR Amplification of V-Domains
[0683] Total RNA was prepared from B-cells lysate (resuspended in RLT buffer--Qiagen--Cat. N.degree. 79216) using the NucleoSpin 8/96 RNA kit (Macherey&Nagel; 740709.4, 740698) according to manufacturer's protocol. RNA was eluted with 60 .mu.L RNAse free water. 6 .mu.L of RNA was used to generate cDNA by reverse transcriptase reaction using the Superscript III First-Strand Synthesis SuperMix (Invitrogen 18080-400) and an oligo-dT-primer according to the manufacturer's instructions. All steps were performed on a Hamilton ML Star System. 4 .mu.L of cDNA were used to amplify the immunoglobulin heavy and light chain variable regions (VH and VL) with the AccuPrime SuperMix (Invitrogen 12344-040) in a final volume of 50 .mu.L using the primers rbHC.up and rbHC.do for the heavy chain, rbLC.up and rbLC.do for the light chain of Wild Type Rabbit B cells and BcPCR FHLC leader.fw and BcPCR huCkappa.rev for the light chain of transgenic rabbit B-cells (see Table below). All forward primers were specific for the signal peptide (of respectively VH and VL) whereas the reverse primers were specific for the constant regions (of respectively VH and VL). The PCR conditions for the RbVH+RbVL were as follows: Hot start at 94.degree. C. for 5 min.; 35 cycles of 20 sec. at 94.degree. C., 20 sec. at 70.degree. C., 45 sec. at 68.degree. C., and a final extension at 68.degree. C. for 7 min. The PCR conditions for the HuVL were as follows: Hot start at 94.degree. C. for 5 min.; 40 cycles of 20 sec. at 94.degree. C., 20 sec. at 52.degree. C., 45 sec. at 68.degree. C., and a final extension at 68.degree. C. for 7 min.
TABLE-US-00044 TABLE 39 rbHC.up AAGCTTGCCACCATGGAGACTGGGCTGCG (SEQ ID NO: 213) CTGGCTTC rbHCf.do CCATTGGTGAGGGTGCCCGAG (SEQ ID NO: 214) rbLC.up AAGCTTGCCACCATGGACAYGAGGGCCCC (SEQ ID NO: 215) CACTC rbLC.do CAGAGTRCTGCTGAGGTTGTAGGTAC (SEQ ID NO: 216) BcPCR_FHLC_leader. ATGGACATGAGGGTCCCCGC fw (SEQ ID NO: 217) BcPCR_huCkappa.rev GATTTCAACTGCTCATCAGATGGC (SEQ ID NO: 218)
[0684] 8 .mu.L of 50 .mu.L PCR solution were loaded on a 48 E-Gel 2% (Invitrogen G8008-02). Positive PCR reactions were cleaned using the NucleoSpin Extract II kit (Macherey&Nagel; 740609250) according to manufacturer's protocol and eluted in 50 .mu.L elution buffer. All cleaning steps were performed on a Hamilton ML Starlet System.
Recombinant Expression of Rabbit Monoclonal Bivalent Antibodies
[0685] For recombinant expression of rabbit monoclonal bivalent antibodies, PCR-products coding for VH or VL were cloned as cDNA into expression vectors by the overhang cloning method (RS Haun et al., BioTechniques (1992) 13, 515-518; M Z Li et al., Nature Methods (2007) 4, 251-256). The expression vectors contained an expression cassette consisting of a 5' CMV promoter including intron A, and a 3' BGH poly adenylation sequence. In addition to the expression cassette, the plasmids contained a pUC18-derived origin of replication and a beta-lactamase gene conferring ampicillin resistance for plasmid amplification in E. coli. Three variants of the basic plasmid were used: one plasmid containing the rabbit IgG constant region designed to accept the VH regions while two additional plasmids containing rabbit or human kappa LC constant region to accept the VL regions.
[0686] Linearized expression plasmids coding for the kappa or gamma constant region and VL/VH inserts were amplified by PCR using overlapping primers.
[0687] Purified PCR products were incubated with T4 DNA-polymerase which generated single-strand overhangs. The reaction was stopped by dCTP addition.
[0688] In the next step, plasmid and insert were combined and incubated with recA which induced site specific recombination. The recombined plasmids were transformed into E. coli. The next day the grown colonies were picked and tested for correct recombined plasmid by plasmid preparation, restriction analysis and DNA-sequencing.
[0689] For antibody expression, the isolated HC and LC plasmids were transiently co-transfected into HEK293 cells and the supernatants were harvested after 1 week.
[0690] Generation of vectors for the expression of rabbit monoclonal monovalent antibodies
[0691] For recombinant expression of selected candidates as monoclonal monovalent antibodies rabbit constant regions of all VH chains were converted into human constant regions enclosing the knob-mutation in the CH3 segment. For VL chains derived from rabbit wild-type B-cells, rabbit C kappa constant regions were converted into human. 4 .mu.L of cDNA of the selected candidates were used to amplify the immunoglobulin heavy and light chain variable regions with the AccuPrime SuperMix (Invitrogen 12344-040) in a final volume of 50 .mu.L with forward primers specific for the signal peptide and reverse primers specific for the CDR3-J region with (at the 3' end) overlap sequence (20 bp) homologous to the human constant regions (respectively of VH and VL). The PCR conditions for the VH and VL chain amplification were as follows: Hot start at 94.degree. C. for 5 min; 35 cycles of 20 sec. at 94.degree. C., 20 sec. at 68.degree. C., 45 sec. at 68.degree. C., and a final extension at 68.degree. C. for 7 min.
[0692] PCR-products coding for VH or VL were cloned as cDNA into expression vectors by the overhang cloning method (R S Haun et al., BioTechniques (1992) 13, 515-518; M Z Li et al., Nature Methods (2007) 4, 251-256). The expression vectors contained an expression cassette consisting of a 5' CMV promoter including intron A, and a 3' BGH poly adenylation sequence. In addition to the expression cassette, the plasmids contained a pUC18-derived origin of replication and a beta-lactamase gene conferring ampicillin resistance for plasmid amplification in E. coli. Two variants of the basic plasmid were used: one plasmid containing the human IgG constant region designed to accept the new amplified VH chain and a second plasmid containing the human kappa LC constant region to accept the VL chain.
[0693] Linearized expression plasmids coding for the kappa or gamma constant region and VL/VH inserts were amplified by PCR using overlapping primers.
[0694] Purified PCR products were incubated with T4 DNA-polymerase which generated single-strand overhangs. The reaction was stopped by dCTP addition.
[0695] In the next step, plasmid and insert were combined and incubated with recA which induced site specific recombination. The recombined plasmids were transformed into E. coli. The next day the grown colonies were picked and tested for correct recombined plasmid by plasmid preparation, restriction analysis and DNA-sequencing.
Transient Expression of the Monovalent Anti-TfR Antibodies
[0696] The antibodies were generated in vivo in transiently transfected HEK293 cells (human embryonic kidney cell line 293-derived) cultivated in F17 Medium (Invitrogen Corp.). For transfection "293-Free" Transfection Reagent (Novagen) was used. Antibodies and antibody-based modified molecules as described above were expressed from individual expression plasmids. Transfections were performed as specified in the manufacturer's instructions. Recombinant protein-containing cell culture supernatants were harvested three to seven days after transfection. Supernatants were stored at reduced temperature (e.g. -80.degree. C.) until purification. General information regarding the recombinant expression of human immunoglobulins in e.g. HEK293 cells is given in: Meissner, P. et al., Biotechnol. Bioeng. 75 (2001) 197-203.
hCMEC/D3 Cell Culture for Transcytosis Assays
[0697] Medium and supplements for hCMEC/D3 (Weksler, B. B. et al., FASEB J. 19 (2005), 1872-1874) were obtained from Lonza. hCMEC/D3 cells (passages 26-29) were cultured to confluence on collagen-coated coverslips (microscopy) or flasks in EBM2 medium containing 2.5% FBS, a quarter of the supplied growth factors and fully complemented with supplied hydrocortisone, gentamycin and ascorbic acid.
[0698] For all transcytosis assays, high density pore (1.times.108 pores/cm2) PET membrane filter inserts (0.4 .mu.m, 12 mm diameter) were used in 12-well cell culture plates. Optimum media volumes were calculated to be 400 .mu.L and 1600 .mu.L for apical and basolateral chambers, respectively. Apical chambers of filter inserts were coated with rat tail collagen I (7.5 .mu.g/cm2) followed by fibronectin (5 .mu.g/mL), each incubation lasting for 1 hour at RT. hCMEC/D3 cells were grown to confluent monolayers (approx. 2.times.105 cells/cm2) for 10-12 days in EMB2 medium.
Transcytosis Assay of Monovalent Transferrin Receptor Specific Antibodies
[0699] The entire assay was performed in serum-free EBM2 medium. Filter inserts with cells were incubated apically with monovalent antibodies (concentration: 2.67 .mu.g/mL) for 1 hour at 37.degree. C. following which the entire apical and basolateral media were collected. From these values, paracellular flux was calculated. The monolayers were washed at RT in serum-free medium apically (400 .mu.L) and basolaterally (1600 .mu.L) 3.times.3-5 min. each. All the washes were collected to monitor efficiency of removal of the unbound antibody. Pre-warmed medium was added to the apical chamber and the filters transferred to a fresh 12 well plate (blocked overnight with PBS containing 1% BSA) containing 1600 .mu.L pre-warmed medium. At this point, cells on filters were lysed in 500 .mu.L RIPA buffer in order to determine specific antibody uptake. The remaining filters were incubated at 37.degree. C. and samples collected at various time points to determine apical and/or basolateral release of antibody. The content of antibody in the samples was quantified using a highly sensitive IgG ELISA. For each time point, data were generated from three filter cell cultures.
Sensitive IgG ELISA after Transcytosis Assay
[0700] The entire procedure was performed at RT using an automated washer for the wash steps. A 384-well plate was coated with 30 .mu.L/well of 1 .mu.g/mL anti-human/mouse-IgG, Fc .andgate.-specific in PBS for 2 hours followed by 1 hour incubation in blocking buffer PBS containing 1% BSA or 1% CroteinC for human and mouse IgG assays, respectively). Serially diluted samples from the transcytosis assay and standard concentrations of the antibody used in the transcytosis assay were added to the plate and incubated for 2 hours. After four washes, 30 .mu.L/well of 50 ng/mL anti-human/mouse-F(ab)2-Biotin in blocking buffer was added and incubated for a further 2 hours. Following 6 washes, 30 .mu.L/well of 50 ng/mL (hulgG assay) or 100 ng/mL (mIgG assay) Poly-HRP40-Streptavidin (Fitzgerald; in PBS containing 1% BSA and 0.05% Tween-20) was added and incubated for 30 min. After 4 washes, immune complexes were detected by addition of 30 .mu.L/well of BM Chemiluminescence Substrate (Roche). The luminescence signal was measured using a luminescence plate reader and concentration calculated using the fitted standard curve. The sensitivity of the assay ranged from 10 pg/mL to 10 ng/mL.
[0701] Rabbit anti-transferrin antibody clone 2/99 showed properties comparable to that of the anti-transferrin receptor antibody 128.1. This can be seen from the following Table.
TABLE-US-00045 TABLE 40 Characterization of antibody 2/99 (SEQ ID NO. 178/179) loading total total transcytosis transcytosis total total sum % of basolateral apical % loading % basolateral apical transported mAb % of mAb of mAb sum transported % origin [pg] basolateral [pg] [pg] [pg] 128.1 128.1 128.1 of mAb 128.1 mAb 2226 34 757 1229 1986 100 100 100 100 128.1 clone- 2773 36 998 1346 2344 125 132 110 118 2/99 max. EC50 max. geo. BIAcore [ng/mL] geo. EC50 mean ratio ratio BIAcore BIAcore BIAcore t1/2 FACS mean [ng/mL] Cyno EC50 max off-rate t1/2 off-rate Cyno ratio hTfR- hTfR- FACS TfR- Cyno/ Cyno/ huTfR huTfR cyTfR TfR t1/2 origin CHO CHO cyTfR CHO human human [1/s] [min] [1/s] [min] human/Cyno mAb 96 78200 314 52100 3.3 0.6 6.06E-04 19 5.47E-02 0 90.2 128.1 clone- 275 55600 241 52000 0.9 1.0 6.16E-04 19 2.77E-04 42 0.4 2/99
[0702] The mouse anti-human transferrin-receptor antibody 128.1 (WO 93/10819) was taken as reference. The transferrin receptor binder 2/99 shows a transcytosis loading of 705 pg, whereof after 4 hours 170 pg (=24% of loading) can be found in the basolateral compartment and 294 pg (=42% of loading) can be found in the apical compartment.
[0703] The transferrin receptor binder 4/94 shows a transcytosis loading of 3510 pg, whereof after 4 hours 748 pg (=21% of loading) can be found in the basolateral compartment and 1503 pg (=43% of loading) can be found in the apical compartment.
Example 26: Epitope Mapping by Cell ELISA of CHO Cells Transfected with hTfR Mutants
[0704] In order to be able determine the epitope regions on human transferrin receptor (hTfR), mutations were introduced into the hTfR sequence at positions, where a cluster of surface-exposed amino acids had different amino acids in the aligned mouse TfR sequence (see Table below), following the rationale that in spite of the significant homology between human and mouse TfR (77% identity), no antibodies directed to the extracellular part are known which show good cross-reactivity between both orthologous. Cloning of plasmids with the corresponding mutations is described above. To map binding of human TfR binders to those epitopes, CHO-K1 cells were transiently transfected with the described plasmids and antibody binding measured in a cell ELISA. Briefly, 104 cells were plated per well of a 96-well plate the day before experiment in normal growth medium (RPMI/10% FCS). The other day, medium was changed to OPTI-MEM Serum-Reduced Medium (Gibco), and 10 .mu.L of a mixture of 1200 OPTI-MEM, 12 .mu.g plasmid DNA and 12 .mu.L XtremeGENE transfection reagent (Roche) were added to the wells after 30 minutes of pre-incubation. Cells were incubated for 2 days at 37.degree. C./7.5 CO2, then medium was removed and TfR antibodies added at concentrations between 1 nM and 100 nM in growth medium, followed by 2 h incubation at 4.degree. C. Afterwards, antibody solutions were replaced by 0.05% glutaraldehyde in PBS and cells fixed for 15 min. at RT, then washed twice with PBS and incubated with HRP-conjugated anti-human-Fc secondary antibody (BioRad; 1:2000 in ELISA Blocking Reagent (Roche)) for 1.5 hours at RT. Signal was generated after 3 washes with PBS using 50 .mu.L of TMB per well and absorbance measured at 450 nm.
TABLE-US-00046 TABLE 41 Plasmid # mutations in hTfR 10188 -- 18909 Thr518Asp/Gln520Lys/Phe521Ser/Gln524Arg 18910 Arg325Gln 18911 Ser355Ala/Asp356Arg/Lys358Asn/Thr359Ile 18912 Asp204Gln/Lys205Ser/Arg208Asn 18913 Lys574Gly/Glu575Ala/Ile577Thr/Glu578Gln 18914 Ala196Ile/Gln197Gly/Asn198Gln/Ser199Asn/Val200Met/ Ile201Val/Ile202Thr/Val203Ile/Asp204Val/Lys205Gln/ Asn206Ser/Gly207Asn/Arg208Gly/Leu209Asn/Val210Leu/ Tyr211Asp/Leu212Pro 18974 Asp245Glu/Tyr247Ser/Thr248Tyr/Pro249Ser
Example 27:Surface Plasmon Resonance-Based Binding Assay for Human TfR-Antibody Interaction
[0705] The binding experiment were carried out on a BIAcore B 4000 (GE Healthcare) equipped with C1 sensorchip (GE Healthcare, cat.no. BR1005-35) pre-treated with anti-human Fab antibody (GE Healthcare, cat.no 28-9583-25) using a standard amine coupling chemistry procedure accordingly to the vendor's manual.
[0706] For kinetic measurements the sample antibody was immobilized applying a contact time of 60 seconds and a flow rate of 10 .mu.L/min in phosphate buffer saline pH 7.4, 0.05 Tween 20 at 25.degree. C. Recombinant His6-tagged human transferrin receptor (R&D systems, cat.no 2474-TR-050) was applied in increasing concentrations and the signal monitored over the time. An average time span of 150 seconds of association time and 600 seconds of dissociation time at 304/min flow rate was recorded. Data were fit using a 1:1 binding model (Langmuir isotherm).
TABLE-US-00047 TABLE 42 Sequences of the epitopes of human transferrin receptor (hTfR) SEQ ID NO Sequence 202 IGQNMVTIVQSNGNL 203 NMVTIVQSNGNLDPV 204 QSNGNLDPVESPEGY
Example 28: Surface Plasmon Resonance-Based Binding Assay for Human TfR-Antibody Interaction
[0707] The binding experiment were carried out on a BIAcore B 4000 (GE Healthcare) equipped with C1 sensor chip (GE Healthcare, cat.no. BR1005-35) pre-treated with anti-human Fab antibody (GE Healthcare, cat.no 28-9583-25) using a standard amine coupling chemistry procedure accordingly to the vendor's manual.
[0708] For kinetic measurements the sample antibody was immobilized applying a contact time of 60 seconds and a flow rate of 10 .mu.L/min in phosphate buffer saline pH 7.4, 0.05 Tween 20 at 25.degree. C. Recombinant His6-tagged human transferrin receptor (R&D systems, cat.no 2474-TR-050) was applied in increasing concentrations and the signal monitored over the time. An average time span of 150 seconds of association time and 600 seconds of dissociation time at 304/min flow rate was recorded. Data were fit using a 1:1 binding model (Langmuir isotherm).
Example 29: Humanization of the VH and VL Domains of Murine and Rabbit Anti-Transferrin Receptor Antibody
[0709] The non-human anti-transferrin receptor antibodies were humanized as follows: Based on the characterization of encoding sequences and amino acid sequences that comprise the VH and VL domains of a the non-human anti-transferrin receptor antibodies of the IgG1 class with kappa light chain, a corresponding humanized anti-transferrin receptor antibody was generated by CDR grafting an backward/forward mutations based on the human germline framework VH4_3 and VK1_10 combination for clone 299.
[0710] The humanized antibodies of clone 2/99 as reported herein were not available by applying standard humanization techniques. It was required to introduce non-standard mutations in the amino acid sequence in order to obtain a humanized antibody with transferrin receptor binding off-rates within the intended range. This is especially important as the antibodies as reported herein are being developed for crossing the human blood-brain-barrier to shuttle the HER bispecific antibody as the therapeutic payload into the brain.
[0711] It has been found that in order to obtain a suitable and developable humanized antibody two cysteine amino acid residues in the light chain of the parental rabbit antibody had to be replaced by a proline and an asparagine amino acid residue, respectively. In addition to be within the given off-rate range a serine residue present in the middle of the rabbit CDRL3 had to be replaced by an alanine residue.
[0712] Is has further been found that it is advantageous to change three amino acid residues in the heavy chain at positions 65, 100 g and 105 (numbering according to Kabat).
[0713] All numbering as used herein is based on the Kabat variable domain numbering scheme.
[0714] Anti-transferrin receptor antibodies that specifically bind to human transferrin receptor (huTfR) and cynomolgus transferrin receptor (cyTfR) were obtained.
Example 30: Method to Determine Human/Cynomolgus TfR Receptor Affinity
[0715] In this example the method for the determination of the human transferrin receptor affinities for the comparison of the dissociation behavior is outlined.
[0716] For all analysis the Biotin CAPture Kit from GE Healthcare (Instruction 28-9242-34 AB) was used. First the chip was rehydrated by docking it in the BIAcore T200 instrument. After that, the chip was left on standby with running buffer overnight. For surface preparation the Biotin CAPture Reagent was diluted 1:100 in running buffer (1.times.PBS, supplemented with 0.25 M NaCl). This solution was injected on Flow Cells 1 to 4 for 360 sec with a flow rate of 2 .mu.L/min. Next the sensor surface was conditioned with three one-minute injections of regeneration solution provided in the Biotin CAPture Kit. This has to be done for the docking procedure or for the first time or after storage. A 100 nM human or cynomolgus, respectively, mono-biotinylated transferrin receptor solution should be injected on Flow Cell 2 for 30 sec at a flow rate of 10 .mu.L/min. For affinity determination injections with six concentrations (500, 250, 125, 62.5, 31.25, 15.625 and 0 nM) were applied. They were injected on the "hu-TfR-flow cell" (e.g. Flow Cell 2 as prepared as outlined above) with an injection time of 180 sec (association) and a flow rate of 10 .mu.L/min. After dissociation phase of 600 sec the surface was regenerated according to the manufacturer's instructions with the regeneration solution provided in the Biotin CAPture Kit and the next cycle was carried out.
[0717] The kinetic data was evaluated using the BIAcore T200 evaluation software. Especially the dissociation rate constants of the different human transferrin receptor binders were taken into account after the application of the 1:1 Langmuir binding model.
Example 31: B4000 Relative Ranking of Human/Cynomolgus Transferrin Receptor Dissociation
[0718] A CAP sensor chip (provided in the Biotin CAPture Kit, series S #28-9202-34 GE) was mounted into a BIAcore B4000 system, normalized and addressed in a hydrodynamic manner, according to the manufacturer's instructions. In the first cycle the CAP reagent (as provided in the Kit) was addressed to spot 1, 2, 4 and 5 with a flow rate of 10 .mu.L/min for 300 sec. The human transferrin receptor capture took place in spot 1 (human transferrin receptor-biotinylated) and spot 5 (cynomolgus transferrin receptor-biotinylated) with a flow rate of 10 .mu.L/min and a contact time of 30 sec. The receptors were diluted to a concentration of 50 nM with running buffer (1.times.PBS #28995084,GE Healthcare, supplemented with 0.25 M NaCl). The antibodies were injected into all flow cells in a concentration series of 100 nM, 50 nM, 25 nM and 0 nM for 180 sec at a flow speed of 30 .mu.L/min. Dissociation time was set to 300 sec. Regeneration of the whole complex from the CAP chip has been performed utilizing the regeneration solution provided in the Biotin CAPture Kit (120 sec with a flow rate of 10 .mu.L/min). To control the active protein concentration a second cycle was performed in spot 5 utilizing biotinylated protein A (# P2165-2MG, Sigma) with a flow rate of 10 .mu.L/min and a contact time of 30 sec. Spot 1 was kept empty in this control cycle. The antibodies and the regeneration were handled like in cycle 1. Relevant kinetic data was calculated using the BIAcore B4000 evaluation software. The dissociation from the human transferrin receptor has been determined applying the 1:1 dissociation fit.
Results
[0719] The mouse anti-human transferrin-receptor antibody 128.1 (WO 93/10819) was taken as reference. The new humanized anti-transferrin receptor antibodies that specifically bind to human transferrin receptor (huTfR) and cynomolgus transferrin receptor (cyTfR) had an off-rate for the human transferrin receptor that was equal to or less than (i.e. at most) that of the anti-transferrin receptor antibody 128.1 for the cynomolgus transferrin receptor, whereby the off-rates are determined by surface plasmon resonance and bound with an off-rate for the human transferrin receptor that was between and including 0.1 l/s and 0.005 l/s.
[0720] In the following Table the off-rates of humanization variants of the rabbit light chain variable domain of clone 2/99 in combination with humanization variants of the rabbit heavy chain variable domain of clone 2/99 are shown. Binding partner was human transferrin receptor (determined at 25.degree. C.).
TABLE-US-00048 TABLE 43 VH VL 0 (rb) 1 2 5 6 7 8 9 11 12 0 (rb) 4.34E-04 9.08E-04 8.06E-04 7.72E-04 6.63E-04 5.15E-04 4.06E-04 9.01E-04 9.05E-04 9.21E-04 1 5.69E-03 1.00E-03 1.00E-03 1.00E-03 1.00E-03 7.52E-03 3.19E-03 6.94E-03 1.00E-03 1.00E-03 2 1.25E-03 2.86E-03 2.75E-03 2.41E-03 1.87E-03 1.31E-03 1.01E-03 3.99E-03 3.85E-03 6.35E-03 3 1.32E-03 4.31E-03 3.84E-03 3.16E-03 2.82E-03 1.45E-03 1.00E-03 4.17E-03 5.65E-03 5.86E-03 4 1.36E-03 2.56E-03 2.63E-03 2.38E-03 1.87E-03 1.25E-03 7.88E-04 2.70E-03 3.88E-03 3.11E-03 5 1.94E-03 2.71E-03 2.62E-03 2.53E-03 1.66E-03 1.35E-03 1.07E-03 3.50E-03 4.56E-03 5.82E-03 6 1.90E-03 5.38E-03 5.55E-03 4.64E-03 3.06E-03 1.97E-03 1.40E-03 6.83E-03 6.71E-03 7.05E-03 7 4.63E-03 7.33E-03 7.50E-03 6.97E-03 5.63E-03 3.66E-03 2.31E-03 7.61E-03 7.81E-03 7.71E-03 8 1.39E-03 4.85E-03 3.94E-03 3.78E-03 3.01E-03 1.72E-03 1.16E-03 5.23E-03 5.52E-03 5.31E-03 9- 1.41E-03 2.46E-03 2.21E-03 2.03E-03 1.41E-03 1.21E-03 1.01E-03 2.52E-03 2.42E-03 2.19E-03 NYA (SEQ ID NO 193) 10 1.88E-03 6.77E-03 6.49E-03 6.53E-03 4.55E-03 2.64E-03 1.73E-03 7.19E-03 7.16E-03 7.79E-03 12 5.41E-03 7.05E-03 8.14E-03 1.00E-03 7.78E-03 7.75E-03 6.72E-03 1.00E-03 7.87E-03 1.00E-03 14 1.78E-03 2.99E-03 2.44E-03 2.33E-03 2.20E-03 1.53E-03 1.04E-03 3.32E-03 3.51E-03 5.46E-03 15 6.63E-03 6.69E-03 6.38E-03 6.37E-03 4.21E-03 2.73E-03 1.81E-03 7.39E-03 7.09E-03 7.76E-03 17 1.49E-03 7.56E-03 7.12E-03 7.45E-03 7.17E-03 1.87E-03 1.12E-03 4.25E-03 7.55E-03 7.27E-03 VH 23- VL 13 15 16 17 18 19 20 21 22 DANG 0 (rb) 4.82E-04 7.63E-04 6.53E-04 4.13E-04 1.09E-03 1.00E-03 1.11E-03 5.65E-04 5.06E-04 3.38E-04 1 1.00E-03 7.72E-03 7.71E-03 4.33E-03 1.00E-03 1.00E-03 1.00E-03 7.71E-03 5.78E-03 2.80E-03 2 2.46E-03 2.16E-03 1.97E-03 1.05E-03 4.89E-03 7.69E-03 5.25E-03 1.38E-03 1.26E-03 7.15E-04 3 2.77E-03 2.07E-03 1.73E-03 8.43E-04 6.65E-03 1.00E-03 7.15E-03 1.94E-03 1.43E-03 7.83E-04 4 1.30E-03 1.34E-03 1.27E-03 7.23E-04 3.35E-03 1.00E-03 4.36E-03 1.46E-03 1.18E-03 7.61E-04 5 2.18E-03 2.14E-03 2.23E-03 1.23E-03 3.49E-03 1.00E-03 3.52E-03 1.37E-03 1.41E-03 8.80E-04 6 3.65E-03 3.50E-03 3.39E-03 1.71E-03 6.74E-03 1.00E-03 6.06E-03 2.07E-03 2.14E-03 1.16E-03 7 6.68E-03 5.43E-03 5.25E-03 2.33E-03 7.66E-03 1.00E-03 7.14E-03 2.38E-03 3.37E-03 1.55E-03 8 2.47E-03 2.09E-03 1.97E-03 1.11E-03 6.77E-03 6.71E-03 6.58E-03 1.74E-03 1.65E-03 9.41E-04 9- 1.39E-03 1.42E-03 1.36E-03 9.34E-04 2.21E-03 6.17E-03 1.89E-03 1.13E-03 1.26E-03 7.69E-04 NYA (SEQ ID NO 193) 10 5.89E-03 3.99E-03 4.24E-03 1.88E-03 7.46E-03 1.00E-03 7.05E-03 2.11E-03 2.09E-03 1.20E-03 12 7.85E-03 7.64E-03 7.54E-03 2.84E-03 1.00E-03 1.00E-03 1.00E-03 7.54E-03 6.44E-03 1.87E-03 14 2.22E-03 1.94E-03 1.75E-03 1.05E-03 3.00E-03 7.96E-03 2.39E-03 1.03E-03 1.12E-03 7.33E-04 15 7.11E-03 6.03E-03 4.77E-03 1.56E-03 7.58E-03 1.00E-03 7.85E-03 1.92E-03 1.79E-03 9.86E-04 17 3.39E-03 1.69E-03 1.66E-03 9.88E-04 5.30E-03 1.00E-03 4.57E-03 9.97E-04 9.38E-04 6.94E-04
[0721] The combination of VH23-DANG with VL09-NYA (SEQ ID NO 193) was chosen as starting point for further engineering to develop a binding site that is reflecting the properties of the antibody 128.1 with respect to the binding to human transferrin receptor more closely. In the following Table the off-rates of different exemplary variants of VH23-DANG and VL9-NYA (SEQ ID NO 193) as well as different other variable domain humanization variants for the human transferrin receptor are shown in comparison (determined according to Example 31 at 25.degree. C.).
TABLE-US-00049 TABLE 44 VK9- NYA (SEQ ID NO VK9- VK9- VK9- VH567- VK12- VK17- VK15- VK2- VK6- 193) SYA GYS CYS P . . . NYA HYS TYS AYS SYS SYS VH23- 8.83E-03 3.96E-03 4.62E-03 1.53E-02 2.29E-03 6.97E-03 5.22E-03 1.32E-02 DANG VH23- 3.95E-02 4.84E-04 3.73E-03 2.33E-03 DASG (SEQ ID NO 192) VH23- 1.49E-02 7.55E-04 3.75E-03 1.51E-03 DAQG (SEQ ID NO 205) VH9- 1.82E-03 6.52E-03 4.02E-02 DANG VH9- 1.25E-03 DAQG VH7- 3.08E-02 DANG reference: 128.1 = 7.78E-02 (determined for the cynomolgus transferrin receptor).
[0722] In the following Table the kinetic data of different exemplary variants of VH23 and VL9 are shown in comparison (determined according to Example 30).
TABLE-US-00050 TABLE 45 BIAcore Assay @ 25.degree. C. TfR kd [s.sup.-1] ka [s.sup.-1 M.sup.-1] kD [M] mAh 128.1 cynomolgus 7.33E-02 5.41E+05 1.36E-07 VH23-DASG/ human 3.95E-02 8.83E+04 4.47E-07 VL09-NYA (SEQ ID NO 192/193) VH23-DAQG/ human 1.37E-02 1.21E+05 1.13E-07 VL09-NYA (SEQ ID NO 205/193) VH23-DANG/ human 8.83E-03 1.55E+05 5.72E-08 VL09-NYA
[0723] In the following Table the off-rates of humanization variants of the murine light chain variable domain of clone 4/94 in combination with humanization variants of the murine heavy chain variable domain of clone 4/94 are shown. Binding partner was human transferrin receptor.
TABLE-US-00051 TABLE 46 VH 1 2 3 4 (SEQ ID (SEQ ID (SEQ ID (SEQ ID VL 0 (mu) NO 201) NO 207) NO 208) NO 209) 0 (mu) 1.36E-04 1.37E-04 1.56E-04 1.48E-04 1.77E-04 1 3.70E-04 4.10E-04 4.54E-04 4.76E-04 4.48E-04 (SEQ ID NO 194) 2 3.64E-04 3.99E-04 4.26E-04 4.15E-04 4.38E-04 (SEQ ID NO 195) 3 3.39E-04 3.86E-04 4.30E-04 4.34E-04 4.52E-04 (SEQ ID NO 196) 4 5.42E-04 6.57E-04 7.03E-04 6.83E-04 7.05E-04 (SEQ ID NO 197) 5 5.44E-04 6.84E-04 7.17E-04 7.17E-04 7.28E-04 (SEQ ID NO 198) 6 3.81E-04 4.86E-04 5.27E-04 5.50E-04 5.52E-04 (SEQ ID NO 199) 7 2.32E-04 2.74E-04 2.99E-04 3.06E-04 3.26E-04 (SEQ ID NO 200)
Example 32: In Vivo Characterization of Herceptarg-scFab(8D3)
[0724] A Her2-expressing intracranial tumor xenograft model was used to compare the brain tumor penetration of Herceptarg-brain shuttle to Herceptarg alone by 3D fluorescence ultramicroscopy.
Cell Lines
[0725] The human breast cancer cell line BT474 M1 was maintained in RPMI 1640 supplemented with 10% fetal calf serum and 2 mL L-glutamine. Propagation of cell lines followed standard cell culture protocols.
Intracranial Implantation of Tumor Cells in Mice
[0726] Female severe combined immunodeficient hairless outbred (SHO) mice (weight 20 to 27 g) were obtained from Charles River and were 7 to 9 weeks of age at initiation of experiments. Mice were anesthetized with i.p. injection of 100 mg/kg ketamine and 10 mg/kg xylazine. Scalp was disinfected and removed over the whole right hemisphere of the brain to expose the cranium. The periosteum was removed by a bone scraper. Next, a 0.7-mm burr hole in diameter was drilled 2 mm right of midline and 2 mm anterior to bregma with a dental drill. Mice were then placed in a small animal stereotactic frame and BT-474 M1 cell suspension (5.times.10.sup.5 cells in 3 .mu.l) was injected using a Hamilton syringe with a 26s-gauge needle to a depth of 3 mm into the brain tissue. After injection, needle was slowly withdrawn and the hole was sealed with Cyano veneer tissue glue. All animal studies were approved by the local government (file number 55.2-1-54-2532.0-76-14, Government of Upper Bavaria, Germany).
Application of Substances
[0727] 30 days after tumor cell implantation mice received an i.v. injection of Herceptarg(LALA)-scFab(8D3)-Alexa Fluor 750 (5 mg/kg, n=7) or Herceptarg(LALA)-Alexa Fluor 750 (3.75 mg/kg; n=5) 6 h before dissection. Five minutes before necropsy, mice were injected i.v. with 100 .mu.g lectin-Alexa Fluor 750 solution to allow visualization of blood vessels ex vivo. Substances were systemically given via the tail vein in a volume of 150 .mu.l.
Necropsy, Fixation and Optical Clearance of Brains
[0728] Mice were sacrificed by cervical dislocation. Brains were explanted and transferred to 10% buffered formalin for about 12 hours at room temperature in the dark and thereafter were dehydrated in a graded tetrahydrofuran series (50%, 70%, 80%, 100% for 90 minutes each, 100% over night and 100% for 90 minutes). Afterwards brains were cleared in dibenzyl ether for 2 days until the specimen became optically transparent.
3D Imaging of Brains by Ultramicroscopy
[0729] The optical transparent samples were placed in a commercial ultramicroscope, equipped with an Imager 3QE camera (both LaVision Biotec), a 2 My PLAPO 2VC objective lens (Olympus) and a supercontinuum white light laser (SuperK EXTREME 80 mHz VIS; NKT Photonics). The samples were scanned with a standard magnification of 0.63 in 5.1 .mu.m thick virtual sections with filter settings appropriate for Alexa Fluor 647 (excitation range 655/15 nm, emission range 680/30 nm) and Alexa Fluor 750 (excitation range 747/33 nm, emission range 786/22 nm) signal detection. Tiff raw data were converted to dicom files and visualized using the OsiriX software (Pixmeo). A 3D brain tumor region of interest (ROI) was defined according to the dimensions of chaotic vessel structures seen by lectin-Alexa Fluor 750 staining. The determined 3D ROI was imported to 3D-reconstructed mab-Alexa Fluor 750 data files to read out mean antibody fluorescent signals within the brain tumors.
[0730] The distinct fluorescent signal intensities of mab-Alexa Fluor 750 at different distances from nearest lectin-Alexa Fluor 647-stained blood vessels were quantified. This was done with a set of custom developed image analysis algorithms implemented as a "Definiens Cognition Network Technology" rule set (Definiens AG).
[0731] Statistical analysis was performed by GraphPad Prism software (GraphPad Software, Inc.) using 2-way ANOVA plus Sidak's multiple comparison test (FIG. 36). The significance levels are indicated by asterisks (**p<0.01 and ****P<0.0001). All data are presented as means.+-.standard error of the mean.
Sequence CWU 1
1
21811995DNAArtificial sequenceHer2 ECD DNA 1acccaagtgt gcaccggcac
agacatgaag ctgcggctcc ctgccagtcc cgagacccac 60ctggacatgc tccgccacct
ctaccagggc tgccaggtgg tgcagggaaa cctggaactc 120acctacctgc
ccaccaatgc cagcctgtcc ttcctgcagg atatccagga ggtgcagggc
180tacgtgctca tcgctcacaa ccaagtgagg caggtcccac tgcagaggct
gcggattgtg 240cgaggcaccc agctctttga ggacaactat gccctggccg
tgctagacaa tggagacccg 300ctgaacaata ccacccctgt cacaggggcc
tccccaggag gcctgcggga gctgcagctt 360cgaagcctca cagagatctt
gaaaggaggg gtcttgatcc agcggaaccc ccagctctgc 420taccaggaca
cgattttgtg gaaggacatc ttccacaaga acaaccagct ggctctcaca
480ctgatagaca ccaaccgctc tcgggcctgc cacccctgtt ctccgatgtg
taagggctcc 540cgctgctggg gagagagttc tgaggattgt cagagcctga
cgcgcactgt ctgtgccggt 600ggctgtgccc gctgcaaggg gccactgccc
actgactgct gccatgagca gtgtgctgcc 660ggctgcacgg gccccaagca
ctctgactgc ctggcctgcc tccacttcaa ccacagtggc 720atctgtgagc
tgcactgccc agccctggtc acctacaaca cagacacgtt tgagtccatg
780cccaatcccg agggccggta tacattcggc gccagctgtg tgactgcctg
tccctacaac 840tacctttcta cggacgtggg atcctgcacc ctcgtctgcc
ccctgcacaa ccaagaggtg 900acagcagagg atggaacaca gcggtgtgag
aagtgcagca agccctgtgc ccgagtgtgc 960tatggtctgg gcatggagca
cttgcgagag gtgagggcag ttaccagtgc caatatccag 1020gagtttgctg
gctgcaagaa gatctttggg agcctggcat ttctgccgga gagctttgat
1080ggggacccag cctccaacac tgccccgctc cagccagagc agctccaagt
gtttgagact 1140ctggaagaga tcacaggtta cctatacatc tcagcatggc
cggacagcct gcctgacctc 1200agcgtcttcc agaacctgca agtaatccgg
ggacgaattc tgcacaatgg cgcctactcg 1260ctgaccctgc aagggctggg
catcagctgg ctggggctgc gctcactgag ggaactgggc 1320agtggactgg
ccctcatcca ccataacacc cacctctgct tcgtgcacac ggtgccctgg
1380gaccagctct ttcggaaccc gcaccaagct ctgctccaca ctgccaaccg
gccagaggac 1440gagtgtgtgg gcgagggcct ggcctgccac cagctgtgcg
cccgagggca ctgctggggt 1500ccagggccca cccagtgtgt caactgcagc
cagttccttc ggggccagga gtgcgtggag 1560gaatgccgag tactgcaggg
gctccccagg gagtatgtga atgccaggca ctgtttgccg 1620tgccaccctg
agtgtcagcc ccagaatggc tcagtgacct gttttggact ggaggctgac
1680cagtgtgtgg cctgtgccca ctataaggac cctcccttct gcgtggcccg
ctgccccagc 1740ggtgtgaaac ctgacctctc ctacatgccc atctggaagt
ttccagatga ggagggcgca 1800tgccagcctt gccccatcaa ctgcacccac
tcctgtgtgg acctggatga caagggctgc 1860cccgccgagc agagagccag
ccctctgacg gtcgacgaac agttatattt tcagggcggc 1920tcaggcctga
acgacatctt cgaggcccag aagatcgagt ggcacgaggc tcgagctcac
1980caccatcacc atcac 19952684PRTArtificial sequenceHer2 ECD 2Met
Gly Trp Ser Cys Ile Ile Leu Phe Leu Val Ala Thr Ala Thr Gly1 5 10
15Val His Ser Thr Gln Val Cys Thr Gly Thr Asp Met Lys Leu Arg Leu
20 25 30Pro Ala Ser Pro Glu Thr His Leu Asp Met Leu Arg His Leu Tyr
Gln 35 40 45Gly Cys Gln Val Val Gln Gly Asn Leu Glu Leu Thr Tyr Leu
Pro Thr 50 55 60Asn Ala Ser Leu Ser Phe Leu Gln Asp Ile Gln Glu Val
Gln Gly Tyr65 70 75 80Val Leu Ile Ala His Asn Gln Val Arg Gln Val
Pro Leu Gln Arg Leu 85 90 95Arg Ile Val Arg Gly Thr Gln Leu Phe Glu
Asp Asn Tyr Ala Leu Ala 100 105 110Val Leu Asp Asn Gly Asp Pro Leu
Asn Asn Thr Thr Pro Val Thr Gly 115 120 125Ala Ser Pro Gly Gly Leu
Arg Glu Leu Gln Leu Arg Ser Leu Thr Glu 130 135 140Ile Leu Lys Gly
Gly Val Leu Ile Gln Arg Asn Pro Gln Leu Cys Tyr145 150 155 160Gln
Asp Thr Ile Leu Trp Lys Asp Ile Phe His Lys Asn Asn Gln Leu 165 170
175Ala Leu Thr Leu Ile Asp Thr Asn Arg Ser Arg Ala Cys His Pro Cys
180 185 190Ser Pro Met Cys Lys Gly Ser Arg Cys Trp Gly Glu Ser Ser
Glu Asp 195 200 205Cys Gln Ser Leu Thr Arg Thr Val Cys Ala Gly Gly
Cys Ala Arg Cys 210 215 220Lys Gly Pro Leu Pro Thr Asp Cys Cys His
Glu Gln Cys Ala Ala Gly225 230 235 240Cys Thr Gly Pro Lys His Ser
Asp Cys Leu Ala Cys Leu His Phe Asn 245 250 255His Ser Gly Ile Cys
Glu Leu His Cys Pro Ala Leu Val Thr Tyr Asn 260 265 270Thr Asp Thr
Phe Glu Ser Met Pro Asn Pro Glu Gly Arg Tyr Thr Phe 275 280 285Gly
Ala Ser Cys Val Thr Ala Cys Pro Tyr Asn Tyr Leu Ser Thr Asp 290 295
300Val Gly Ser Cys Thr Leu Val Cys Pro Leu His Asn Gln Glu Val
Thr305 310 315 320Ala Glu Asp Gly Thr Gln Arg Cys Glu Lys Cys Ser
Lys Pro Cys Ala 325 330 335Arg Val Cys Tyr Gly Leu Gly Met Glu His
Leu Arg Glu Val Arg Ala 340 345 350Val Thr Ser Ala Asn Ile Gln Glu
Phe Ala Gly Cys Lys Lys Ile Phe 355 360 365Gly Ser Leu Ala Phe Leu
Pro Glu Ser Phe Asp Gly Asp Pro Ala Ser 370 375 380Asn Thr Ala Pro
Leu Gln Pro Glu Gln Leu Gln Val Phe Glu Thr Leu385 390 395 400Glu
Glu Ile Thr Gly Tyr Leu Tyr Ile Ser Ala Trp Pro Asp Ser Leu 405 410
415Pro Asp Leu Ser Val Phe Gln Asn Leu Gln Val Ile Arg Gly Arg Ile
420 425 430Leu His Asn Gly Ala Tyr Ser Leu Thr Leu Gln Gly Leu Gly
Ile Ser 435 440 445Trp Leu Gly Leu Arg Ser Leu Arg Glu Leu Gly Ser
Gly Leu Ala Leu 450 455 460Ile His His Asn Thr His Leu Cys Phe Val
His Thr Val Pro Trp Asp465 470 475 480Gln Leu Phe Arg Asn Pro His
Gln Ala Leu Leu His Thr Ala Asn Arg 485 490 495Pro Glu Asp Glu Cys
Val Gly Glu Gly Leu Ala Cys His Gln Leu Cys 500 505 510Ala Arg Gly
His Cys Trp Gly Pro Gly Pro Thr Gln Cys Val Asn Cys 515 520 525Ser
Gln Phe Leu Arg Gly Gln Glu Cys Val Glu Glu Cys Arg Val Leu 530 535
540Gln Gly Leu Pro Arg Glu Tyr Val Asn Ala Arg His Cys Leu Pro
Cys545 550 555 560His Pro Glu Cys Gln Pro Gln Asn Gly Ser Val Thr
Cys Phe Gly Leu 565 570 575Glu Ala Asp Gln Cys Val Ala Cys Ala His
Tyr Lys Asp Pro Pro Phe 580 585 590Cys Val Ala Arg Cys Pro Ser Gly
Val Lys Pro Asp Leu Ser Tyr Met 595 600 605Pro Ile Trp Lys Phe Pro
Asp Glu Glu Gly Ala Cys Gln Pro Cys Pro 610 615 620Ile Asn Cys Thr
His Ser Cys Val Asp Leu Asp Asp Lys Gly Cys Pro625 630 635 640Ala
Glu Gln Arg Ala Ser Pro Leu Thr Val Asp Glu Gln Leu Tyr Phe 645 650
655Gln Gly Gly Ser Gly Leu Asn Asp Ile Phe Glu Ala Gln Lys Ile Glu
660 665 670Trp His Glu Ala Arg Ala His His His His His His 675
6803681DNAArtificial sequenceFc (hole) DNA 3gacaaaactc acacatgccc
accgtgccca gcacctgaac tcctgggggg accgtcagtc 60ttcctcttcc ccccaaaacc
caaggacacc ctcatgatct cccggacccc tgaggtcaca 120tgcgtggtgg
tggacgtgag ccacgaagac cctgaggtca agttcaactg gtacgtggac
180ggcgtggagg tgcataatgc caagacaaag ccgcgggagg agcagtacaa
cagcacgtac 240cgtgtggtca gcgtcctcac cgtcctgcac caggactggc
tgaatggcaa ggagtacaag 300tgcaaggtct ccaacaaagc cctcccagcc
cccatcgaga aaaccatctc caaagccaaa 360gggcagcccc gagaaccaca
ggtgtgcacc ctgcccccat cccgggatga gctgaccaag 420aaccaggtca
gcctctcgtg cgcagtcaaa ggcttctatc ccagcgacat cgccgtggag
480tgggagagca atgggcagcc ggagaacaac tacaagacca cgcctcccgt
gctggactcc 540gacggctcct tcttcctcgt gagcaagctc accgtggaca
agagcaggtg gcagcagggg 600aacgtcttct catgctccgt gatgcatgag
gctctgcaca accgcttcac gcagaagagc 660ctctccctgt ctccgggtaa a
6814227PRTArtificial sequenceFc (hole) 4Asp Lys Thr His Thr Cys Pro
Pro Cys Pro Ala Pro Glu Leu Leu Gly1 5 10 15Gly Pro Ser Val Phe Leu
Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 20 25 30Ile Ser Arg Thr Pro
Glu Val Thr Cys Val Val Val Asp Val Ser His 35 40 45Glu Asp Pro Glu
Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val 50 55 60His Asn Ala
Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr65 70 75 80Arg
Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly 85 90
95Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile
100 105 110Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro
Gln Val 115 120 125Cys Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys
Asn Gln Val Ser 130 135 140Leu Ser Cys Ala Val Lys Gly Phe Tyr Pro
Ser Asp Ile Ala Val Glu145 150 155 160Trp Glu Ser Asn Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro 165 170 175Val Leu Asp Ser Asp
Gly Ser Phe Phe Leu Val Ser Lys Leu Thr Val 180 185 190Asp Lys Ser
Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met 195 200 205His
Glu Ala Leu His Asn Arg Phe Thr Gln Lys Ser Leu Ser Leu Ser 210 215
220Pro Gly Lys22552670DNAArtificial sequenceHer2 ECD-Fc(knob) DNA
5acccaagtgt gcaccggcac agacatgaag ctgcggctcc ctgccagtcc cgagacccac
60ctggacatgc tccgccacct ctaccagggc tgccaggtgg tgcagggaaa cctggaactc
120acctacctgc ccaccaatgc cagcctgtcc ttcctgcagg atatccagga
ggtgcagggc 180tacgtgctca tcgctcacaa ccaagtgagg caggtcccac
tgcagaggct gcggattgtg 240cgaggcaccc agctctttga ggacaactat
gccctggccg tgctagacaa tggagacccg 300ctgaacaata ccacccctgt
cacaggggcc tccccaggag gcctgcggga gctgcagctt 360cgaagcctca
cagagatctt gaaaggaggg gtcttgatcc agcggaaccc ccagctctgc
420taccaggaca cgattttgtg gaaggacatc ttccacaaga acaaccagct
ggctctcaca 480ctgatagaca ccaaccgctc tcgggcctgc cacccctgtt
ctccgatgtg taagggctcc 540cgctgctggg gagagagttc tgaggattgt
cagagcctga cgcgcactgt ctgtgccggt 600ggctgtgccc gctgcaaggg
gccactgccc actgactgct gccatgagca gtgtgctgcc 660ggctgcacgg
gccccaagca ctctgactgc ctggcctgcc tccacttcaa ccacagtggc
720atctgtgagc tgcactgccc agccctggtc acctacaaca cagacacgtt
tgagtccatg 780cccaatcccg agggccggta tacattcggc gccagctgtg
tgactgcctg tccctacaac 840tacctttcta cggacgtggg atcctgcacc
ctcgtctgcc ccctgcacaa ccaagaggtg 900acagcagagg atggaacaca
gcggtgtgag aagtgcagca agccctgtgc ccgagtgtgc 960tatggtctgg
gcatggagca cttgcgagag gtgagggcag ttaccagtgc caatatccag
1020gagtttgctg gctgcaagaa gatctttggg agcctggcat ttctgccgga
gagctttgat 1080ggggacccag cctccaacac tgccccgctc cagccagagc
agctccaagt gtttgagact 1140ctggaagaga tcacaggtta cctatacatc
tcagcatggc cggacagcct gcctgacctc 1200agcgtcttcc agaacctgca
agtaatccgg ggacgaattc tgcacaatgg cgcctactcg 1260ctgaccctgc
aagggctggg catcagctgg ctggggctgc gctcactgag ggaactgggc
1320agtggactgg ccctcatcca ccataacacc cacctctgct tcgtgcacac
ggtgccctgg 1380gaccagctct ttcggaaccc gcaccaagct ctgctccaca
ctgccaaccg gccagaggac 1440gagtgtgtgg gcgagggcct ggcctgccac
cagctgtgcg cccgagggca ctgctggggt 1500ccagggccca cccagtgtgt
caactgcagc cagttccttc ggggccagga gtgcgtggag 1560gaatgccgag
tactgcaggg gctccccagg gagtatgtga atgccaggca ctgtttgccg
1620tgccaccctg agtgtcagcc ccagaatggc tcagtgacct gttttggact
ggaggctgac 1680cagtgtgtgg cctgtgccca ctataaggac cctcccttct
gcgtggcccg ctgccccagc 1740ggtgtgaaac ctgacctctc ctacatgccc
atctggaagt ttccagatga ggagggcgca 1800tgccagcctt gccccatcaa
ctgcacccac tcctgtgtgg acctggatga caagggctgc 1860cccgccgagc
agagagccag ccctctgacg gtcgacggtg gtagtccgac acctccgaca
1920cccgggggtg gttctgcaga caaaactcac acatgcccac cgtgcccagc
acctgaactc 1980ctggggggac cgtcagtctt cctcttcccc ccaaaaccca
aggacaccct catgatctcc 2040cggacccctg aggtcacatg cgtggtggtg
gacgtgagcc acgaagaccc tgaggtcaag 2100ttcaactggt acgtggacgg
cgtggaggtg cataatgcca agacaaagcc gcgggaggag 2160cagtacaaca
gcacgtaccg tgtggtcagc gtcctcaccg tcctgcacca ggactggctg
2220aatggcaagg agtacaagtg caaggtctcc aacaaagccc tcccagcccc
catcgagaaa 2280accatctcca aagccaaagg gcagccccga gaaccacagg
tgtacaccct gcccccatgc 2340cgggatgagc tgaccaagaa ccaggtcagc
ctgtggtgcc tggtcaaagg cttctatccc 2400agcgacatcg ccgtggagtg
ggagagcaat gggcagccgg agaacaacta caagaccacg 2460cctcccgtgc
tggactccga cggctccttc ttcctctaca gcaagctcac cgtggacaag
2520agcaggtggc agcaggggaa cgtcttctca tgctccgtga tgcatgaggc
tctgcacaac 2580cactacacgc agaagagcct ctccctgtct ccgggtaaat
ccggaggcct gaacgacatc 2640ttcgaggccc agaagattga atggcacgag
26706890PRTArtificial sequenceHer2 ECD-Fc(knob) 6Thr Gln Val Cys
Thr Gly Thr Asp Met Lys Leu Arg Leu Pro Ala Ser1 5 10 15Pro Glu Thr
His Leu Asp Met Leu Arg His Leu Tyr Gln Gly Cys Gln 20 25 30Val Val
Gln Gly Asn Leu Glu Leu Thr Tyr Leu Pro Thr Asn Ala Ser 35 40 45Leu
Ser Phe Leu Gln Asp Ile Gln Glu Val Gln Gly Tyr Val Leu Ile 50 55
60Ala His Asn Gln Val Arg Gln Val Pro Leu Gln Arg Leu Arg Ile Val65
70 75 80Arg Gly Thr Gln Leu Phe Glu Asp Asn Tyr Ala Leu Ala Val Leu
Asp 85 90 95Asn Gly Asp Pro Leu Asn Asn Thr Thr Pro Val Thr Gly Ala
Ser Pro 100 105 110Gly Gly Leu Arg Glu Leu Gln Leu Arg Ser Leu Thr
Glu Ile Leu Lys 115 120 125Gly Gly Val Leu Ile Gln Arg Asn Pro Gln
Leu Cys Tyr Gln Asp Thr 130 135 140Ile Leu Trp Lys Asp Ile Phe His
Lys Asn Asn Gln Leu Ala Leu Thr145 150 155 160Leu Ile Asp Thr Asn
Arg Ser Arg Ala Cys His Pro Cys Ser Pro Met 165 170 175Cys Lys Gly
Ser Arg Cys Trp Gly Glu Ser Ser Glu Asp Cys Gln Ser 180 185 190Leu
Thr Arg Thr Val Cys Ala Gly Gly Cys Ala Arg Cys Lys Gly Pro 195 200
205Leu Pro Thr Asp Cys Cys His Glu Gln Cys Ala Ala Gly Cys Thr Gly
210 215 220Pro Lys His Ser Asp Cys Leu Ala Cys Leu His Phe Asn His
Ser Gly225 230 235 240Ile Cys Glu Leu His Cys Pro Ala Leu Val Thr
Tyr Asn Thr Asp Thr 245 250 255Phe Glu Ser Met Pro Asn Pro Glu Gly
Arg Tyr Thr Phe Gly Ala Ser 260 265 270Cys Val Thr Ala Cys Pro Tyr
Asn Tyr Leu Ser Thr Asp Val Gly Ser 275 280 285Cys Thr Leu Val Cys
Pro Leu His Asn Gln Glu Val Thr Ala Glu Asp 290 295 300Gly Thr Gln
Arg Cys Glu Lys Cys Ser Lys Pro Cys Ala Arg Val Cys305 310 315
320Tyr Gly Leu Gly Met Glu His Leu Arg Glu Val Arg Ala Val Thr Ser
325 330 335Ala Asn Ile Gln Glu Phe Ala Gly Cys Lys Lys Ile Phe Gly
Ser Leu 340 345 350Ala Phe Leu Pro Glu Ser Phe Asp Gly Asp Pro Ala
Ser Asn Thr Ala 355 360 365Pro Leu Gln Pro Glu Gln Leu Gln Val Phe
Glu Thr Leu Glu Glu Ile 370 375 380Thr Gly Tyr Leu Tyr Ile Ser Ala
Trp Pro Asp Ser Leu Pro Asp Leu385 390 395 400Ser Val Phe Gln Asn
Leu Gln Val Ile Arg Gly Arg Ile Leu His Asn 405 410 415Gly Ala Tyr
Ser Leu Thr Leu Gln Gly Leu Gly Ile Ser Trp Leu Gly 420 425 430Leu
Arg Ser Leu Arg Glu Leu Gly Ser Gly Leu Ala Leu Ile His His 435 440
445Asn Thr His Leu Cys Phe Val His Thr Val Pro Trp Asp Gln Leu Phe
450 455 460Arg Asn Pro His Gln Ala Leu Leu His Thr Ala Asn Arg Pro
Glu Asp465 470 475 480Glu Cys Val Gly Glu Gly Leu Ala Cys His Gln
Leu Cys Ala Arg Gly 485 490 495His Cys Trp Gly Pro Gly Pro Thr Gln
Cys Val Asn Cys Ser Gln Phe 500 505 510Leu Arg Gly Gln Glu Cys Val
Glu Glu Cys Arg Val Leu Gln Gly Leu 515 520 525Pro Arg Glu Tyr Val
Asn Ala Arg His Cys Leu Pro Cys His Pro Glu 530 535 540Cys Gln Pro
Gln Asn Gly Ser Val Thr Cys Phe Gly Leu Glu Ala Asp545 550 555
560Gln Cys Val Ala Cys Ala His Tyr Lys Asp Pro Pro Phe Cys Val Ala
565 570 575Arg Cys Pro Ser Gly Val Lys Pro Asp Leu Ser Tyr Met Pro
Ile Trp 580 585 590Lys Phe Pro Asp Glu Glu Gly Ala Cys Gln Pro Cys
Pro Ile Asn Cys 595 600 605Thr His Ser Cys Val Asp Leu Asp Asp Lys
Gly Cys Pro Ala Glu Gln 610 615 620Arg Ala Ser Pro Leu Thr Val Asp
Gly Gly Ser Pro Thr Pro Pro Thr625
630 635 640Pro Gly Gly Gly Ser Ala Asp Lys Thr His Thr Cys Pro Pro
Cys Pro 645 650 655Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe Leu
Phe Pro Pro Lys 660 665 670Pro Lys Asp Thr Leu Met Ile Ser Arg Thr
Pro Glu Val Thr Cys Val 675 680 685Val Val Asp Val Ser His Glu Asp
Pro Glu Val Lys Phe Asn Trp Tyr 690 695 700Val Asp Gly Val Glu Val
His Asn Ala Lys Thr Lys Pro Arg Glu Glu705 710 715 720Gln Tyr Asn
Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His 725 730 735Gln
Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys 740 745
750Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln
755 760 765Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Cys Arg Asp
Glu Leu 770 775 780Thr Lys Asn Gln Val Ser Leu Trp Cys Leu Val Lys
Gly Phe Tyr Pro785 790 795 800Ser Asp Ile Ala Val Glu Trp Glu Ser
Asn Gly Gln Pro Glu Asn Asn 805 810 815Tyr Lys Thr Thr Pro Pro Val
Leu Asp Ser Asp Gly Ser Phe Phe Leu 820 825 830Tyr Ser Lys Leu Thr
Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val 835 840 845Phe Ser Cys
Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln 850 855 860Lys
Ser Leu Ser Leu Ser Pro Gly Lys Ser Gly Gly Leu Asn Asp Ile865 870
875 880Phe Glu Ala Gln Lys Ile Glu Trp His Glu 885
89072670DNAArtificial sequenceHer2 ECD(pertuzumab KO)-Fc(knob)DNA
7acccaagtgt gcaccggcac agacatgaag ctgcggctcc ctgccagtcc cgagacccac
60ctggacatgc tccgccacct ctaccagggc tgccaggtgg tgcagggaaa cctggaactc
120acctacctgc ccaccaatgc cagcctgtcc ttcctgcagg atatccagga
ggtgcagggc 180tacgtgctca tcgctcacaa ccaagtgagg caggtcccac
tgcagaggct gcggattgtg 240cgaggcaccc agctctttga ggacaactat
gccctggccg tgctagacaa tggagacccg 300ctgaacaata ccacccctgt
cacaggggcc tccccaggag gcctgcggga gctgcagctt 360cgaagcctca
cagagatctt gaaaggaggg gtcttgatcc agcggaaccc ccagctctgc
420taccaggaca cgattttgtg gaaggacatc ttccacaaga acaaccagct
ggctctcaca 480ctgatagaca ccaaccgctc tcgggcctgc cacccctgtt
ctccgatgtg taagggctcc 540cgctgctggg gagagagttc tgaggattgt
cagagcctga cgcgcactgt ctgtgccggt 600ggctgtgccc gctgcaaggg
gccactgccc actgactgct gccatgagca gtgtgctgcc 660ggctgcacgg
gccccaagca ctctgactgc ctggcctgcc tccacttcaa ccacagtggc
720atctgtgagc tgcactgccc agccctggtc acctacaaca cagacacgcg
ggagtccatg 780cccaatcccg agggccggta tagattcggc gccagctgtg
tgactgcctg tccctacaac 840tacctttcta cggaccgggg atcctgcacc
ctcgtctgcc ccctggccaa ccaagaggtg 900acagcagagg atggaacaca
gcggtgtgag aagtgcagca agccctgtgc ccgagtgtgc 960tatggtctgg
gcatggagca cttgcgagag gtgagggcag ttaccagtgc caatatccag
1020gagtttgctg gctgcaagaa gatctttggg agcctggcat ttctgccgga
gagctttgat 1080ggggacccag cctccaacac tgccccgctc cagccagagc
agctccaagt gtttgagact 1140ctggaagaga tcacaggtta cctatacatc
tcagcatggc cggacagcct gcctgacctc 1200agcgtcttcc agaacctgca
agtaatccgg ggacgaattc tgcacaatgg cgcctactcg 1260ctgaccctgc
aagggctggg catcagctgg ctggggctgc gctcactgag ggaactgggc
1320agtggactgg ccctcatcca ccataacacc cacctctgct tcgtgcacac
ggtgccctgg 1380gaccagctct ttcggaaccc gcaccaagct ctgctccaca
ctgccaaccg gccagaggac 1440gagtgtgtgg gcgagggcct ggcctgccac
cagctgtgcg cccgagggca ctgctggggt 1500ccagggccca cccagtgtgt
caactgcagc cagttccttc ggggccagga gtgcgtggag 1560gaatgccgag
tactgcaggg gctccccagg gagtatgtga atgccaggca ctgtttgccg
1620tgccaccctg agtgtcagcc ccagaatggc tcagtgacct gttttggact
ggaggctgac 1680cagtgtgtgg cctgtgccca ctataaggac cctcccttct
gcgtggcccg ctgccccagc 1740ggtgtgaaac ctgacctctc ctacatgccc
atctggaagt ttccagatga ggagggcgca 1800tgccagcctt gccccatcaa
ctgcacccac tcctgtgtgg acctggatga caagggctgc 1860cccgccgagc
agagagccag ccctctgacg gtcgacggtg gtagtccgac acctccgaca
1920cccgggggtg gttctgcaga caaaactcac acatgcccac cgtgcccagc
acctgaactc 1980ctggggggac cgtcagtctt cctcttcccc ccaaaaccca
aggacaccct catgatctcc 2040cggacccctg aggtcacatg cgtggtggtg
gacgtgagcc acgaagaccc tgaggtcaag 2100ttcaactggt acgtggacgg
cgtggaggtg cataatgcca agacaaagcc gcgggaggag 2160cagtacaaca
gcacgtaccg tgtggtcagc gtcctcaccg tcctgcacca ggactggctg
2220aatggcaagg agtacaagtg caaggtctcc aacaaagccc tcccagcccc
catcgagaaa 2280accatctcca aagccaaagg gcagccccga gaaccacagg
tgtacaccct gcccccatgc 2340cgggatgagc tgaccaagaa ccaggtcagc
ctgtggtgcc tggtcaaagg cttctatccc 2400agcgacatcg ccgtggagtg
ggagagcaat gggcagccgg agaacaacta caagaccacg 2460cctcccgtgc
tggactccga cggctccttc ttcctctaca gcaagctcac cgtggacaag
2520agcaggtggc agcaggggaa cgtcttctca tgctccgtga tgcatgaggc
tctgcacaac 2580cactacacgc agaagagcct ctccctgtct ccgggtaaat
ccggaggcct gaacgacatc 2640ttcgaggccc agaagattga atggcacgag
26708890PRTArtificial sequenceHer2 ECD(pertuzumab KO)-Fc(knob) 8Thr
Gln Val Cys Thr Gly Thr Asp Met Lys Leu Arg Leu Pro Ala Ser1 5 10
15Pro Glu Thr His Leu Asp Met Leu Arg His Leu Tyr Gln Gly Cys Gln
20 25 30Val Val Gln Gly Asn Leu Glu Leu Thr Tyr Leu Pro Thr Asn Ala
Ser 35 40 45Leu Ser Phe Leu Gln Asp Ile Gln Glu Val Gln Gly Tyr Val
Leu Ile 50 55 60Ala His Asn Gln Val Arg Gln Val Pro Leu Gln Arg Leu
Arg Ile Val65 70 75 80Arg Gly Thr Gln Leu Phe Glu Asp Asn Tyr Ala
Leu Ala Val Leu Asp 85 90 95Asn Gly Asp Pro Leu Asn Asn Thr Thr Pro
Val Thr Gly Ala Ser Pro 100 105 110Gly Gly Leu Arg Glu Leu Gln Leu
Arg Ser Leu Thr Glu Ile Leu Lys 115 120 125Gly Gly Val Leu Ile Gln
Arg Asn Pro Gln Leu Cys Tyr Gln Asp Thr 130 135 140Ile Leu Trp Lys
Asp Ile Phe His Lys Asn Asn Gln Leu Ala Leu Thr145 150 155 160Leu
Ile Asp Thr Asn Arg Ser Arg Ala Cys His Pro Cys Ser Pro Met 165 170
175Cys Lys Gly Ser Arg Cys Trp Gly Glu Ser Ser Glu Asp Cys Gln Ser
180 185 190Leu Thr Arg Thr Val Cys Ala Gly Gly Cys Ala Arg Cys Lys
Gly Pro 195 200 205Leu Pro Thr Asp Cys Cys His Glu Gln Cys Ala Ala
Gly Cys Thr Gly 210 215 220Pro Lys His Ser Asp Cys Leu Ala Cys Leu
His Phe Asn His Ser Gly225 230 235 240Ile Cys Glu Leu His Cys Pro
Ala Leu Val Thr Tyr Asn Thr Asp Thr 245 250 255Arg Glu Ser Met Pro
Asn Pro Glu Gly Arg Tyr Arg Phe Gly Ala Ser 260 265 270Cys Val Thr
Ala Cys Pro Tyr Asn Tyr Leu Ser Thr Asp Arg Gly Ser 275 280 285Cys
Thr Leu Val Cys Pro Leu Ala Asn Gln Glu Val Thr Ala Glu Asp 290 295
300Gly Thr Gln Arg Cys Glu Lys Cys Ser Lys Pro Cys Ala Arg Val
Cys305 310 315 320Tyr Gly Leu Gly Met Glu His Leu Arg Glu Val Arg
Ala Val Thr Ser 325 330 335Ala Asn Ile Gln Glu Phe Ala Gly Cys Lys
Lys Ile Phe Gly Ser Leu 340 345 350Ala Phe Leu Pro Glu Ser Phe Asp
Gly Asp Pro Ala Ser Asn Thr Ala 355 360 365Pro Leu Gln Pro Glu Gln
Leu Gln Val Phe Glu Thr Leu Glu Glu Ile 370 375 380Thr Gly Tyr Leu
Tyr Ile Ser Ala Trp Pro Asp Ser Leu Pro Asp Leu385 390 395 400Ser
Val Phe Gln Asn Leu Gln Val Ile Arg Gly Arg Ile Leu His Asn 405 410
415Gly Ala Tyr Ser Leu Thr Leu Gln Gly Leu Gly Ile Ser Trp Leu Gly
420 425 430Leu Arg Ser Leu Arg Glu Leu Gly Ser Gly Leu Ala Leu Ile
His His 435 440 445Asn Thr His Leu Cys Phe Val His Thr Val Pro Trp
Asp Gln Leu Phe 450 455 460Arg Asn Pro His Gln Ala Leu Leu His Thr
Ala Asn Arg Pro Glu Asp465 470 475 480Glu Cys Val Gly Glu Gly Leu
Ala Cys His Gln Leu Cys Ala Arg Gly 485 490 495His Cys Trp Gly Pro
Gly Pro Thr Gln Cys Val Asn Cys Ser Gln Phe 500 505 510Leu Arg Gly
Gln Glu Cys Val Glu Glu Cys Arg Val Leu Gln Gly Leu 515 520 525Pro
Arg Glu Tyr Val Asn Ala Arg His Cys Leu Pro Cys His Pro Glu 530 535
540Cys Gln Pro Gln Asn Gly Ser Val Thr Cys Phe Gly Leu Glu Ala
Asp545 550 555 560Gln Cys Val Ala Cys Ala His Tyr Lys Asp Pro Pro
Phe Cys Val Ala 565 570 575Arg Cys Pro Ser Gly Val Lys Pro Asp Leu
Ser Tyr Met Pro Ile Trp 580 585 590Lys Phe Pro Asp Glu Glu Gly Ala
Cys Gln Pro Cys Pro Ile Asn Cys 595 600 605Thr His Ser Cys Val Asp
Leu Asp Asp Lys Gly Cys Pro Ala Glu Gln 610 615 620Arg Ala Ser Pro
Leu Thr Val Asp Gly Gly Ser Pro Thr Pro Pro Thr625 630 635 640Pro
Gly Gly Gly Ser Ala Asp Lys Thr His Thr Cys Pro Pro Cys Pro 645 650
655Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys
660 665 670Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr
Cys Val 675 680 685Val Val Asp Val Ser His Glu Asp Pro Glu Val Lys
Phe Asn Trp Tyr 690 695 700Val Asp Gly Val Glu Val His Asn Ala Lys
Thr Lys Pro Arg Glu Glu705 710 715 720Gln Tyr Asn Ser Thr Tyr Arg
Val Val Ser Val Leu Thr Val Leu His 725 730 735Gln Asp Trp Leu Asn
Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys 740 745 750Ala Leu Pro
Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln 755 760 765Pro
Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Cys Arg Asp Glu Leu 770 775
780Thr Lys Asn Gln Val Ser Leu Trp Cys Leu Val Lys Gly Phe Tyr
Pro785 790 795 800Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln
Pro Glu Asn Asn 805 810 815Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser
Asp Gly Ser Phe Phe Leu 820 825 830Tyr Ser Lys Leu Thr Val Asp Lys
Ser Arg Trp Gln Gln Gly Asn Val 835 840 845Phe Ser Cys Ser Val Met
His Glu Ala Leu His Asn His Tyr Thr Gln 850 855 860Lys Ser Leu Ser
Leu Ser Pro Gly Lys Ser Gly Gly Leu Asn Asp Ile865 870 875 880Phe
Glu Ala Gln Lys Ile Glu Trp His Glu 885 89092667DNAArtificial
sequenceHer2 ECD(trastuzumab KO)-Fc(knob)DNA 9acccaagtgt gcaccggcac
agacatgaag ctgcggctcc ctgccagtcc cgagacccac 60ctggacatgc tccgccacct
ctaccagggc tgccaggtgg tgcagggaaa cctggaactc 120acctacctgc
ccaccaatgc cagcctgtcc ttcctgcagg atatccagga ggtgcagggc
180tacgtgctca tcgctcacaa ccaagtgagg caggtcccac tgcagaggct
gcggattgtg 240cgaggcaccc agctctttga ggacaactat gccctggccg
tgctagacaa tggagacccg 300ctgaacaata ccacccctgt cacaggggcc
tccccaggag gcctgcggga gctgcagctt 360cgaagcctca cagagatctt
gaaaggaggg gtcttgatcc agcggaaccc ccagctctgc 420taccaggaca
cgattttgtg gaaggacatc ttccacaaga acaaccagct ggctctcaca
480ctgatagaca ccaaccgctc tcgggcctgc cacccctgtt ctccgatgtg
taagggctcc 540cgctgctggg gagagagttc tgaggattgt cagagcctga
cgcgcactgt ctgtgccggt 600ggctgtgccc gctgcaaggg gccactgccc
actgactgct gccatgagca gtgtgctgcc 660ggctgcacgg gccccaagca
ctctgactgc ctggcctgcc tccacttcaa ccacagtggc 720atctgtgagc
tgcactgccc agccctggtc acctacaaca cagacacgtt tgagtccatg
780cccaatcccg agggccggta tacattcggc gccagctgtg tgactgcctg
tccctacaac 840tacctttcta cggacgtggg atcctgcacc ctcgtctgcc
ccctgcacaa ccaagaggtg 900acagcagagg atggaacaca gcggtgtgag
aagtgcagca agccctgtgc ccgagtgtgc 960tatggtctgg gcatggagca
cttgcgagag gtgagggcag ttaccagtgc caatatccag 1020gagtttgctg
gctgcaagaa gatctttggg agcctggcat ttctgccgga gagctttgat
1080ggggacccag cctccaacac tgccccgctc cagccagagc agctccaagt
gtttgagact 1140ctggaagaga tcacaggtta cctatacatc tcagcatggc
cggacagcct gcctgacctc 1200agcgtcttcc agaacctgca agtaatccgg
ggacgaattc tgcacaatgg cgcctactcg 1260ctgaccctgc aagggctggg
catcagctgg ctggggctgc gctcactgag ggaactgggc 1320agtggactgg
ccctcatcca ccataacacc cacctctgct tcgtgcacac ggtgccctgg
1380gaccagctct ttcggaaccc gcaccaagct ctgctccaca ctgccaaccg
gccagaggac 1440gagtgtgtgg gcgagggcct ggcctgccac cagctgtgcg
cccgagggca ctgctggggt 1500ccagggccca cccagtgtgt caactgcagc
cagttccttc ggggccagga gtgcgtggag 1560gaatgccgag tactgcaggg
gctccccagg gagtatgtga atgccaggca ctgtttgccg 1620tgccaccctg
agtgtcagcc ccagaatggc tcagtgacct gttttggact ggaggctcgg
1680cagtgtgtgg cctgtgccca ctataaggac agacggtgcg tggcccgctg
ccccagcggt 1740gtgaaacctg acctctccta catgcccatc tggaagtttc
cagatgagga gggcgcatgc 1800cagccttgcc ccatcaactg cacccactcc
tgtgtggacc tggatgacaa gggctgcccc 1860gccgagcaga gagccagccc
tctgacggtc gacggtggta gtccgacacc tccgacaccc 1920gggggtggtt
ctgcagacaa aactcacaca tgcccaccgt gcccagcacc tgaactcctg
1980gggggaccgt cagtcttcct cttcccccca aaacccaagg acaccctcat
gatctcccgg 2040acccctgagg tcacatgcgt ggtggtggac gtgagccacg
aagaccctga ggtcaagttc 2100aactggtacg tggacggcgt ggaggtgcat
aatgccaaga caaagccgcg ggaggagcag 2160tacaacagca cgtaccgtgt
ggtcagcgtc ctcaccgtcc tgcaccagga ctggctgaat 2220ggcaaggagt
acaagtgcaa ggtctccaac aaagccctcc cagcccccat cgagaaaacc
2280atctccaaag ccaaagggca gccccgagaa ccacaggtgt acaccctgcc
cccatgccgg 2340gatgagctga ccaagaacca ggtcagcctg tggtgcctgg
tcaaaggctt ctatcccagc 2400gacatcgccg tggagtggga gagcaatggg
cagccggaga acaactacaa gaccacgcct 2460cccgtgctgg actccgacgg
ctccttcttc ctctacagca agctcaccgt ggacaagagc 2520aggtggcagc
aggggaacgt cttctcatgc tccgtgatgc atgaggctct gcacaaccac
2580tacacgcaga agagcctctc cctgtctccg ggtaaatccg gaggcctgaa
cgacatcttc 2640gaggcccaga agattgaatg gcacgag 266710889PRTArtificial
sequenceHer2 ECD(trastuzumab KO)-Fc(knob) 10Thr Gln Val Cys Thr Gly
Thr Asp Met Lys Leu Arg Leu Pro Ala Ser1 5 10 15Pro Glu Thr His Leu
Asp Met Leu Arg His Leu Tyr Gln Gly Cys Gln 20 25 30Val Val Gln Gly
Asn Leu Glu Leu Thr Tyr Leu Pro Thr Asn Ala Ser 35 40 45Leu Ser Phe
Leu Gln Asp Ile Gln Glu Val Gln Gly Tyr Val Leu Ile 50 55 60Ala His
Asn Gln Val Arg Gln Val Pro Leu Gln Arg Leu Arg Ile Val65 70 75
80Arg Gly Thr Gln Leu Phe Glu Asp Asn Tyr Ala Leu Ala Val Leu Asp
85 90 95Asn Gly Asp Pro Leu Asn Asn Thr Thr Pro Val Thr Gly Ala Ser
Pro 100 105 110Gly Gly Leu Arg Glu Leu Gln Leu Arg Ser Leu Thr Glu
Ile Leu Lys 115 120 125Gly Gly Val Leu Ile Gln Arg Asn Pro Gln Leu
Cys Tyr Gln Asp Thr 130 135 140Ile Leu Trp Lys Asp Ile Phe His Lys
Asn Asn Gln Leu Ala Leu Thr145 150 155 160Leu Ile Asp Thr Asn Arg
Ser Arg Ala Cys His Pro Cys Ser Pro Met 165 170 175Cys Lys Gly Ser
Arg Cys Trp Gly Glu Ser Ser Glu Asp Cys Gln Ser 180 185 190Leu Thr
Arg Thr Val Cys Ala Gly Gly Cys Ala Arg Cys Lys Gly Pro 195 200
205Leu Pro Thr Asp Cys Cys His Glu Gln Cys Ala Ala Gly Cys Thr Gly
210 215 220Pro Lys His Ser Asp Cys Leu Ala Cys Leu His Phe Asn His
Ser Gly225 230 235 240Ile Cys Glu Leu His Cys Pro Ala Leu Val Thr
Tyr Asn Thr Asp Thr 245 250 255Phe Glu Ser Met Pro Asn Pro Glu Gly
Arg Tyr Thr Phe Gly Ala Ser 260 265 270Cys Val Thr Ala Cys Pro Tyr
Asn Tyr Leu Ser Thr Asp Val Gly Ser 275 280 285Cys Thr Leu Val Cys
Pro Leu His Asn Gln Glu Val Thr Ala Glu Asp 290 295 300Gly Thr Gln
Arg Cys Glu Lys Cys Ser Lys Pro Cys Ala Arg Val Cys305 310 315
320Tyr Gly Leu Gly Met Glu His Leu Arg Glu Val Arg Ala Val Thr Ser
325 330 335Ala Asn Ile Gln Glu Phe Ala Gly Cys Lys Lys Ile Phe Gly
Ser Leu 340 345 350Ala Phe Leu Pro Glu Ser Phe Asp Gly Asp Pro Ala
Ser Asn Thr Ala 355 360 365Pro Leu Gln Pro Glu Gln Leu Gln Val Phe
Glu Thr Leu Glu Glu Ile 370 375 380Thr Gly Tyr Leu Tyr Ile Ser Ala
Trp Pro Asp Ser Leu Pro Asp Leu385 390 395 400Ser Val Phe Gln
Asn Leu Gln Val Ile Arg Gly Arg Ile Leu His Asn 405 410 415Gly Ala
Tyr Ser Leu Thr Leu Gln Gly Leu Gly Ile Ser Trp Leu Gly 420 425
430Leu Arg Ser Leu Arg Glu Leu Gly Ser Gly Leu Ala Leu Ile His His
435 440 445Asn Thr His Leu Cys Phe Val His Thr Val Pro Trp Asp Gln
Leu Phe 450 455 460Arg Asn Pro His Gln Ala Leu Leu His Thr Ala Asn
Arg Pro Glu Asp465 470 475 480Glu Cys Val Gly Glu Gly Leu Ala Cys
His Gln Leu Cys Ala Arg Gly 485 490 495His Cys Trp Gly Pro Gly Pro
Thr Gln Cys Val Asn Cys Ser Gln Phe 500 505 510Leu Arg Gly Gln Glu
Cys Val Glu Glu Cys Arg Val Leu Gln Gly Leu 515 520 525Pro Arg Glu
Tyr Val Asn Ala Arg His Cys Leu Pro Cys His Pro Glu 530 535 540Cys
Gln Pro Gln Asn Gly Ser Val Thr Cys Phe Gly Leu Glu Ala Arg545 550
555 560Gln Cys Val Ala Cys Ala His Tyr Lys Asp Arg Arg Cys Val Ala
Arg 565 570 575Cys Pro Ser Gly Val Lys Pro Asp Leu Ser Tyr Met Pro
Ile Trp Lys 580 585 590Phe Pro Asp Glu Glu Gly Ala Cys Gln Pro Cys
Pro Ile Asn Cys Thr 595 600 605His Ser Cys Val Asp Leu Asp Asp Lys
Gly Cys Pro Ala Glu Gln Arg 610 615 620Ala Ser Pro Leu Thr Val Asp
Gly Gly Ser Pro Thr Pro Pro Thr Pro625 630 635 640Gly Gly Gly Ser
Ala Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala 645 650 655Pro Glu
Leu Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro 660 665
670Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val
675 680 685Val Asp Val Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp
Tyr Val 690 695 700Asp Gly Val Glu Val His Asn Ala Lys Thr Lys Pro
Arg Glu Glu Gln705 710 715 720Tyr Asn Ser Thr Tyr Arg Val Val Ser
Val Leu Thr Val Leu His Gln 725 730 735Asp Trp Leu Asn Gly Lys Glu
Tyr Lys Cys Lys Val Ser Asn Lys Ala 740 745 750Leu Pro Ala Pro Ile
Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro 755 760 765Arg Glu Pro
Gln Val Tyr Thr Leu Pro Pro Cys Arg Asp Glu Leu Thr 770 775 780Lys
Asn Gln Val Ser Leu Trp Cys Leu Val Lys Gly Phe Tyr Pro Ser785 790
795 800Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn
Tyr 805 810 815Lys Thr Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe
Phe Leu Tyr 820 825 830Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln
Gln Gly Asn Val Phe 835 840 845Ser Cys Ser Val Met His Glu Ala Leu
His Asn His Tyr Thr Gln Lys 850 855 860Ser Leu Ser Leu Ser Pro Gly
Lys Ser Gly Gly Leu Asn Asp Ile Phe865 870 875 880Glu Ala Gln Lys
Ile Glu Trp His Glu 8851111PRTArtificial sequencePertuzumab wt VL
CDR1 11Lys Ala Ser Gln Asp Val Ser Ile Gly Val Ala1 5
10127PRTArtificial sequencePertuzumab wt VL CDR2 12Ser Ala Ser Tyr
Arg Tyr Thr1 5139PRTArtificial sequencePertuzumab wt VL CDR3 13Gln
Gln Tyr Tyr Ile Tyr Pro Tyr Thr1 51410PRTArtificial
sequencePertuzumab wt VH CDR1 14Gly Phe Thr Phe Thr Asp Tyr Thr Met
Asp1 5 101517PRTArtificial sequencePertuzumab wt VH CDR2 15Asp Val
Asn Pro Asn Ser Gly Gly Ser Ile Tyr Asn Gln Arg Phe Lys1 5 10
15Gly1610PRTArtificial sequencePertuzumab wt VH CDR3 16Asn Leu Gly
Pro Ser Phe Tyr Phe Asp Tyr1 5 101711PRTArtificial
sequenceTrastuzumab wt VL CDR1 17Arg Ala Ser Gln Asp Val Asn Thr
Ala Val Ala1 5 10187PRTArtificial sequenceTrastuzumab VL CDR2 18Ser
Ala Ser Phe Leu Tyr Ser1 5199PRTArtificial sequenceTrastuzumab VL
CDR3 19Gln Gln His Tyr Thr Thr Pro Pro Thr1 52010PRTArtificial
sequenceTrastuzumab VH CDR1 20Gly Phe Asn Ile Lys Asp Thr Tyr Ile
His1 5 1021558DNAArtificial sequencePertuzumab wt VH (10289) DNA
21gaggtgcagc tggtcgagtc tggcggcgga ctggtgcagc ctggcggcag cctgagactg
60agctgcgccg ccagcggctt caccttcacc gactacacca tggactgggt gcggcaggcc
120cctggcaagg gcctggaatg ggtggccgac gtgaacccca acagcggcgg
cagcatctac 180aaccagcggt tcaagggccg gttcaccctg agcgtggaca
gaagcaagaa caccctgtac 240ctccagatga acagcctgcg ggccgaggac
accgccgtgt actactgcgc ccggaacctg 300ggccccagct tctacttcga
ctactggggc cagggcaccc tggtgaccgt gagcagcgct 360agcaccaagg
gcccatcggt cttccccctg gcaccctcct ccaagagcac ctctgggggc
420acagcggccc tgggctgcct ggtcaaggac tacttccccg aaccggtgac
ggtgtcgtgg 480aactcaggcg ccctgaccag cggcgtgcac accttcccgg
ctgtcctaca gtcctcagga 540ctctactccc tcagcagc 55822119PRTArtificial
sequencePertuzumab wt VH (10289) 22Glu Val Gln Leu Val Glu Ser Gly
Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala
Ala Ser Gly Phe Thr Phe Thr Asp Tyr 20 25 30Thr Met Asp Trp Val Arg
Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Asp Val Asn Pro
Asn Ser Gly Gly Ser Ile Tyr Asn Gln Arg Phe 50 55 60Lys Gly Arg Phe
Thr Leu Ser Val Asp Arg Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln
Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala
Arg Asn Leu Gly Pro Ser Phe Tyr Phe Asp Tyr Trp Gly Gln Gly 100 105
110Thr Leu Val Thr Val Ser Ser 11523321DNAArtificial
sequencePertuzumab wt VL (10290) DNA 23gacatccaga tgacccagag
ccccagcagc ctgagcgcca gcgtgggcga cagagtgacc 60atcacctgca aggccagcca
ggacgtgtcc atcggcgtgg cctggtatca gcagaagccc 120ggcaaggccc
ccaagctgct gatctacagc gccagctacc ggtacacagg cgtgcccagc
180cggttcagcg gcagcggctc cggcaccgac ttcaccctga ccatcagcag
cctgcagccc 240gaggacttcg ccacctacta ctgccagcag tactacatct
acccctacac cttcggccag 300ggcaccaagg tggagatcaa g
32124107PRTArtificial sequencePertuzumab wt VL (10290) 24Asp Ile
Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp
Arg Val Thr Ile Thr Cys Lys Ala Ser Gln Asp Val Ser Ile Gly 20 25
30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile
35 40 45Tyr Ser Ala Ser Tyr Arg Tyr Thr Gly Val Pro Ser Arg Phe Ser
Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu
Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Tyr Tyr
Ile Tyr Pro Tyr 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys
100 10525321DNAArtificial sequencePertuzumab VL (Trast. L3) (10403)
DNA 25gacatccaga tgacccagag ccccagcagc ctgagcgcca gcgtgggcga
cagagtgacc 60atcacatgca aggccagcca ggacgtgtcc atcggcgtgg cctggtatca
gcagaagccc 120ggcaaggccc ccaagctgct gatctacagc gccagctacc
ggtacaccgg cgtgcccagc 180agattcagcg gcagcggctc cggcaccgac
ttcaccctga ccatcagcag cctgcagccc 240gaggacttcg ccacctacta
ctgccagcag cactacacca ccccccccac cttcggccag 300ggcaccaagg
tggaaatcaa g 32126107PRTArtificial sequencePertuzumab VL (Trast.
L3) (10403) 26Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala
Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Lys Ala Ser Gln Asp
Val Ser Ile Gly 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala
Pro Lys Leu Leu Ile 35 40 45Tyr Ser Ala Ser Tyr Arg Tyr Thr Gly Val
Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu
Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr
Cys Gln Gln His Tyr Thr Thr Pro Pro 85 90 95Thr Phe Gly Gln Gly Thr
Lys Val Glu Ile Lys 100 10527321DNAArtificial sequencePertuzumab VL
(Trast. H91) (10404) DNA 27gacatccaga tgacccagag ccccagcagc
ctgagcgcca gcgtgggcga cagagtgacc 60atcacctgca aggccagcca ggacgtgtcc
atcggcgtgg cctggtatca gcagaagccc 120ggcaaggccc ccaagctgct
gatctacagc gccagctacc ggtacacagg cgtgcccagc 180cggttcagcg
gcagcggctc cggcaccgac ttcaccctga ccatcagcag cctgcagccc
240gaggacttcg ccacctacta ctgccagcag cactacatct acccctacac
cttcggccag 300ggcaccaagg tggagatcaa g 32128107PRTArtificial
sequencePertuzumab VL (Trast. H91) (10404) 28Asp Ile Gln Met Thr
Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr
Ile Thr Cys Lys Ala Ser Gln Asp Val Ser Ile Gly 20 25 30Val Ala Trp
Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ser
Ala Ser Tyr Arg Tyr Thr Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser
Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75
80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln His Tyr Ile Tyr Pro Tyr
85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys 100
1052917PRTArtificial sequenceTrastuzumab VH CDR2 29Arg Ile Tyr Pro
Thr Asn Gly Tyr Thr Arg Tyr Ala Asp Ser Val Lys1 5 10
15Gly3011PRTArtificial sequenceTrastuzumab wt VH CDR3 30Trp Gly Gly
Asp Gly Phe Tyr Ala Met Asp Tyr1 5 1031321DNAArtificial
sequencePertuzumab VL (Tras.L3) K24R (10949) DNA 31gacatccaga
tgacccagag ccccagcagc ctgagcgcca gcgtgggcga cagagtgacc 60atcacatgcc
gggccagcca ggacgtgtcc atcggcgtgg cctggtatca gcagaagccc
120ggcaaggccc ccaagctgct gatctacagc gccagctacc ggtacaccgg
cgtgcccagc 180agattcagcg gcagcggctc cggcaccgac ttcaccctga
ccatcagcag cctgcagccc 240gaggacttcg ccacctacta ctgccagcag
cactacacca ccccccccac cttcggccag 300ggcaccaagg tggaaatcaa g
32132107PRTArtificial sequencePertuzumab VL (Tras.L3) K24R (10949)
32Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1
5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Asp Val Ser Ile
Gly 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu
Leu Ile 35 40 45Tyr Ser Ala Ser Tyr Arg Tyr Thr Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln
His Tyr Thr Thr Pro Pro 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu
Ile Lys 100 10533321DNAArtificial sequencePertuzumab VL(Tras.L3)
S30N (10950) DNA 33gacatccaga tgacccagag ccccagcagc ctgagcgcca
gcgtgggcga cagagtgacc 60atcacatgca aggccagcca ggacgtgaac atcggcgtgg
cctggtatca gcagaagccc 120ggcaaggccc ccaagctgct gatctacagc
gccagctacc ggtacaccgg cgtgcccagc 180agattcagcg gcagcggctc
cggcaccgac ttcaccctga ccatcagcag cctgcagccc 240gaggacttcg
ccacctacta ctgccagcag cactacacca ccccccccac cttcggccag
300ggcaccaagg tggaaatcaa g 32134107PRTArtificial sequencePertuzumab
VL (Tras.L3) S30N (10950) 34Asp Ile Gln Met Thr Gln Ser Pro Ser Ser
Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Lys Ala
Ser Gln Asp Val Asn Ile Gly 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro
Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ser Ala Ser Tyr Arg Tyr
Thr Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp
Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala
Thr Tyr Tyr Cys Gln Gln His Tyr Thr Thr Pro Pro 85 90 95Thr Phe Gly
Gln Gly Thr Lys Val Glu Ile Lys 100 10535321DNAArtificial
sequencePertuzumab VL (Tras.L3) I31T (10951) DNA 35gacatccaga
tgacccagag ccccagcagc ctgagcgcca gcgtgggcga cagagtgacc 60atcacatgca
aggccagcca ggacgtgtcc accggcgtgg cctggtatca gcagaagccc
120ggcaaggccc ccaagctgct gatctacagc gccagctacc ggtacaccgg
cgtgcccagc 180agattcagcg gcagcggctc cggcaccgac ttcaccctga
ccatcagcag cctgcagccc 240gaggacttcg ccacctacta ctgccagcag
cactacacca ccccccccac cttcggccag 300ggcaccaagg tggaaatcaa g
32136107PRTArtificial sequencePertuzumab VL (Tras.L3) I31T (10951)
36Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1
5 10 15Asp Arg Val Thr Ile Thr Cys Lys Ala Ser Gln Asp Val Ser Thr
Gly 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu
Leu Ile 35 40 45Tyr Ser Ala Ser Tyr Arg Tyr Thr Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln
His Tyr Thr Thr Pro Pro 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu
Ile Lys 100 10537321DNAArtificial sequencePertuzumab VL (Tras.L3)
I31V (10952) DNA 37gacatccaga tgacccagag ccccagcagc ctgagcgcca
gcgtgggcga cagagtgacc 60atcacatgca aggccagcca ggacgtgtcc gtcggcgtgg
cctggtatca gcagaagccc 120ggcaaggccc ccaagctgct gatctacagc
gccagctacc ggtacaccgg cgtgcccagc 180agattcagcg gcagcggctc
cggcaccgac ttcaccctga ccatcagcag cctgcagccc 240gaggacttcg
ccacctacta ctgccagcag cactacacca ccccccccac cttcggccag
300ggcaccaagg tggaaatcaa g 32138107PRTArtificial sequencePertuzumab
VL (Tras.L3) I31V (10952) 38Asp Ile Gln Met Thr Gln Ser Pro Ser Ser
Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Lys Ala
Ser Gln Asp Val Ser Val Gly 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro
Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ser Ala Ser Tyr Arg Tyr
Thr Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp
Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala
Thr Tyr Tyr Cys Gln Gln His Tyr Thr Thr Pro Pro 85 90 95Thr Phe Gly
Gln Gly Thr Lys Val Glu Ile Lys 100 10539321DNAArtificial
sequencePertuzumab VL (Tras.L3) G32A (10953) DNA 39gacatccaga
tgacccagag ccccagcagc ctgagcgcca gcgtgggcga cagagtgacc 60atcacatgca
aggccagcca ggacgtgtcc atcgccgtgg cctggtatca gcagaagccc
120ggcaaggccc ccaagctgct gatctacagc gccagctacc ggtacaccgg
cgtgcccagc 180agattcagcg gcagcggctc cggcaccgac ttcaccctga
ccatcagcag cctgcagccc 240gaggacttcg ccacctacta ctgccagcag
cactacacca ccccccccac cttcggccag 300ggcaccaagg tggaaatcaa g
32140107PRTArtificial sequencePertuzumab VL (Tras.L3) G32A (10953)
40Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1
5 10 15Asp Arg Val Thr Ile Thr Cys Lys Ala Ser Gln Asp Val Ser Ile
Ala 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu
Leu Ile 35 40 45Tyr Ser Ala Ser Tyr Arg Tyr Thr Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln
His Tyr Thr Thr Pro Pro 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu
Ile Lys 100 10541321DNAArtificial sequencePertuzumab VL (Tras.L3)
Y53F (10954) DNA 41gacatccaga tgacccagag ccccagcagc ctgagcgcca
gcgtgggcga cagagtgacc 60atcacatgca aggccagcca ggacgtgtcc atcggcgtgg
cctggtatca gcagaagccc 120ggcaaggccc ccaagctgct gatctacagc
gccagcttcc ggtacaccgg cgtgcccagc 180agattcagcg gcagcggctc
cggcaccgac ttcaccctga ccatcagcag cctgcagccc 240gaggacttcg
ccacctacta ctgccagcag cactacacca ccccccccac cttcggccag
300ggcaccaagg tggaaatcaa g 32142107PRTArtificial sequencePertuzumab
VL
(Tras.L3) Y53F (10954) 42Asp Ile Gln Met Thr Gln Ser Pro Ser Ser
Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Lys Ala
Ser Gln Asp Val Ser Ile Gly 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro
Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ser Ala Ser Phe Arg Tyr
Thr Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp
Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala
Thr Tyr Tyr Cys Gln Gln His Tyr Thr Thr Pro Pro 85 90 95Thr Phe Gly
Gln Gly Thr Lys Val Glu Ile Lys 100 10543321DNAArtificial
sequencePertuzumab (Tras.L3) R54L (10955) DNA 43gacatccaga
tgacccagag ccccagcagc ctgagcgcca gcgtgggcga cagagtgacc 60atcacatgca
aggccagcca ggacgtgtcc atcggcgtgg cctggtatca gcagaagccc
120ggcaaggccc ccaagctgct gatctacagc gccagctacc tgtacaccgg
cgtgcccagc 180agattcagcg gcagcggctc cggcaccgac ttcaccctga
ccatcagcag cctgcagccc 240gaggacttcg ccacctacta ctgccagcag
cactacacca ccccccccac cttcggccag 300ggcaccaagg tggaaatcaa g
32144107PRTArtificial sequencePertuzumab VL (Tras.L3) R54L (10955)
44Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1
5 10 15Asp Arg Val Thr Ile Thr Cys Lys Ala Ser Gln Asp Val Ser Ile
Gly 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu
Leu Ile 35 40 45Tyr Ser Ala Ser Tyr Leu Tyr Thr Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln
His Tyr Thr Thr Pro Pro 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu
Ile Lys 100 10545321DNAArtificial sequencePertuzumab (Tras.L3) T56S
(10956) DNA 45gacatccaga tgacccagag ccccagcagc ctgagcgcca
gcgtgggcga cagagtgacc 60atcacatgca aggccagcca ggacgtgtcc atcggcgtgg
cctggtatca gcagaagccc 120ggcaaggccc ccaagctgct gatctacagc
gccagctacc ggtacagcgg cgtgcccagc 180agattcagcg gcagcggctc
cggcaccgac ttcaccctga ccatcagcag cctgcagccc 240gaggacttcg
ccacctacta ctgccagcag cactacacca ccccccccac cttcggccag
300ggcaccaagg tggaaatcaa g 32146107PRTArtificial sequencePertuzumab
(Tras.L3) T56S (10956) 46Asp Ile Gln Met Thr Gln Ser Pro Ser Ser
Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Lys Ala
Ser Gln Asp Val Ser Ile Gly 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro
Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ser Ala Ser Tyr Arg Tyr
Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp
Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala
Thr Tyr Tyr Cys Gln Gln His Tyr Thr Thr Pro Pro 85 90 95Thr Phe Gly
Gln Gly Thr Lys Val Glu Ile Lys 100 10547321DNAArtificial
sequencePertuzumab (Tras.L3) G66R (10957) DNA 47gacatccaga
tgacccagag ccccagcagc ctgagcgcca gcgtgggcga cagagtgacc 60atcacatgca
aggccagcca ggacgtgtcc atcggcgtgg cctggtatca gcagaagccc
120ggcaaggccc ccaagctgct gatctacagc gccagctacc ggtacaccgg
cgtgcccagc 180agattcagcg gcagccgctc cggcaccgac ttcaccctga
ccatcagcag cctgcagccc 240gaggacttcg ccacctacta ctgccagcag
cactacacca ccccccccac cttcggccag 300ggcaccaagg tggaaatcaa g
32148107PRTArtificial sequencePertuzumab (Tras.L3) G66R (10957)
48Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1
5 10 15Asp Arg Val Thr Ile Thr Cys Lys Ala Ser Gln Asp Val Ser Ile
Gly 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu
Leu Ile 35 40 45Tyr Ser Ala Ser Tyr Arg Tyr Thr Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Arg Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln
His Tyr Thr Thr Pro Pro 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu
Ile Lys 100 10549320DNAArtificial sequencePertuzumab (Tras.L3) T94Y
(10958) DNA 49acatccagat gacccagagc cccagcagcc tgagcgccag
cgtgggcgac agagtgacca 60tcacatgcaa ggccagccag gacgtgtcca tcggcgtggc
ctggtatcag cagaagcccg 120gcaaggcccc caagctgctg atctacagcg
ccagctaccg gtacaccggc gtgcccagca 180gattcagcgg cagcggctcc
ggcaccgact tcaccctgac catcagcagc ctgcagcccg 240aggacttcgc
cacctactac tgccagcagc actacaccta cccccccacc ttcggccagg
300gcaccaaggt ggaaatcaag 32050107PRTArtificial sequencePertuzumab
(Tras.L3) T94Y (10958) 50Asp Ile Gln Met Thr Gln Ser Pro Ser Ser
Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Lys Ala
Ser Gln Asp Val Ser Ile Gly 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro
Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ser Ala Ser Tyr Arg Tyr
Thr Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp
Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala
Thr Tyr Tyr Cys Gln Gln His Tyr Thr Tyr Pro Pro 85 90 95Thr Phe Gly
Gln Gly Thr Lys Val Glu Ile Lys 100 10551321DNAArtificial
sequencePertuzumab (Tras.L3) P96Y (10959) DNA 51gacatccaga
tgacccagag ccccagcagc ctgagcgcca gcgtgggcga cagagtgacc 60atcacatgca
aggccagcca ggacgtgtcc atcggcgtgg cctggtatca gcagaagccc
120ggcaaggccc ccaagctgct gatctacagc gccagctacc ggtacaccgg
cgtgcccagc 180agattcagcg gcagcggctc cggcaccgac ttcaccctga
ccatcagcag cctgcagccc 240gaggacttcg ccacctacta ctgccagcag
cactacacca ccccctacac cttcggccag 300ggcaccaagg tggaaatcaa g
32152107PRTArtificial sequencePertuzumab (Tras.L3) P96Y (10959)
52Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1
5 10 15Asp Arg Val Thr Ile Thr Cys Lys Ala Ser Gln Asp Val Ser Ile
Gly 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu
Leu Ile 35 40 45Tyr Ser Ala Ser Tyr Arg Tyr Thr Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln
His Tyr Thr Thr Pro Tyr 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu
Ile Lys 100 10553321DNAArtificial sequencePertuzumab (Tras.L3) (QM)
(11055) DNA 53gacatccaga tgacccagag ccccagcagc ctgtctgcca
gcgtgggcga cagagtgacc 60atcacatgca aggccagcca ggacgtgtcc acagccgtgg
cctggtatca gcagaagcct 120ggcaaggccc ccaagctgct gatctacagc
gccagcttcc ggtacaccgg cgtgcccagc 180agattcagcg gcagcagatc
cggcaccgac ttcaccctga ccatcagctc cctgcagccc 240gaggacttcg
ccacctacta ctgccagcag cactacacca ccccccccac atttggccag
300ggcaccaagg tggaaatcaa g 32154107PRTArtificial sequencePertuzumab
(Tras.L3) (QM) (11055) 54Asp Ile Gln Met Thr Gln Ser Pro Ser Ser
Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Lys Ala
Ser Gln Asp Val Ser Thr Ala 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro
Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ser Ala Ser Phe Arg Tyr
Thr Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Arg Ser Gly Thr Asp
Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala
Thr Tyr Tyr Cys Gln Gln His Tyr Thr Thr Pro Pro 85 90 95Thr Phe Gly
Gln Gly Thr Lys Val Glu Ile Lys 100 1055510PRTArtificial
sequencePertuzumab aff.mat. clone D1- VH CDR1 55Gly Phe Thr Phe Asn
Asp Tyr Thr Met Asp1 5 105610PRTArtificial sequencePertuzumab
aff.mat. clone D1- VH CDR3 56Asn Leu Gly Pro Phe Phe Tyr Phe Asp
Tyr1 5 105710PRTArtificial sequencePertuzumab aff.mat. clone B2- VH
CDR3 57Asn Leu Gly Pro Asn Phe Tyr Phe Asp Tyr1 5
105810PRTArtificial sequencePertuzumab aff.mat. clone E1- VH CDR1
58Gly Phe Thr Phe Ala Asp Tyr Thr Met Asp1 5 105910PRTArtificial
sequencePertuzumab aff.mat. clone E1- VH CDR3 59Asn Leu Gly Pro Trp
Phe Tyr Phe Asp Tyr1 5 106017PRTArtificial sequencePertuzumab
aff.mat. clone G2- VH CDR2 60Asp Val Asn Pro Asn Ser Gly Gly Tyr
Ile Val Asn Arg Arg Phe Lys1 5 10 15Gly61357DNAArtificial
sequencePertuzumab aff.mat. clone D1 DNA 61gaggtgcaat tggttgaaag
cggtggtggt ctggttcagc ctggtggtag cctgcgtctg 60agctgtgcag caagcggttt
tacctttaac gattatacca tggattgggt tcgtcaggca 120ccgggtaaag
gtctggaatg ggttgcagat gttaatccga atagcggtgg tagcatttat
180aaccagcgtt ttaaaggtcg ttttaccctg agcgttgatc gtagcaaaaa
taccctgtat 240ctgcaaatga atagtctgcg tgcagaggat accgcagtgt
attattgtgc acgtaacctg 300ggtccgttct tctactttga ttattggggt
cagggcaccc tggttaccgt tagcagc 35762119PRTArtificial
sequencePertuzumab aff.mat. clone D1 62Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys
Ala Ala Ser Gly Phe Thr Phe Asn Asp Tyr 20 25 30Thr Met Asp Trp Val
Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Asp Val Asn
Pro Asn Ser Gly Gly Ser Ile Tyr Asn Gln Arg Phe 50 55 60Lys Gly Arg
Phe Thr Leu Ser Val Asp Arg Ser Lys Asn Thr Leu Tyr65 70 75 80Leu
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90
95Ala Arg Asn Leu Gly Pro Phe Phe Tyr Phe Asp Tyr Trp Gly Gln Gly
100 105 110Thr Leu Val Thr Val Ser Ser 11563357DNAArtificial
sequencePertuzumab aff.mat. clone D1-derived, DNA 63gaggtgcaat
tggttgaaag cggtggtggt ctggttcagc ctggtggtag cctgcgtctg 60agctgtgcag
caagcggttt tacctttaac gattatacca tggattgggt tcgtcaggca
120ccgggtaaag gtctggaatg ggttgcagat gttaatccga atagcggtgg
tagcattgtt 180aaccgtcgtt ttaaaggtcg ttttaccctg agcgttgatc
gtagcaaaaa taccctgtat 240ctgcaaatga atagtctgcg tgcagaggat
accgcagtgt attattgtgc acgtaacctg 300ggtccgttct tctactttga
ttattggggt cagggcaccc tggttaccgt tagcagc 35764119PRTArtificial
sequencePertuzumab aff.mat. clone D1-derived 64Glu Val Gln Leu Val
Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu
Ser Cys Ala Ala Ser Gly Phe Thr Phe Asn Asp Tyr 20 25 30Thr Met Asp
Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Asp
Val Asn Pro Asn Ser Gly Gly Ser Ile Val Asn Arg Arg Phe 50 55 60Lys
Gly Arg Phe Thr Leu Ser Val Asp Arg Ser Lys Asn Thr Leu Tyr65 70 75
80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Arg Asn Leu Gly Pro Phe Phe Tyr Phe Asp Tyr Trp Gly Gln
Gly 100 105 110Thr Leu Val Thr Val Ser Ser 11565357DNAArtificial
sequencePertuzumab aff.mat. clone B2, DNA 65gaggtgcaat tggttgaaag
cggtggtggt ctggttcagc ctggtggtag cctgcgtctg 60agctgtgcag caagcggttt
tacctttaac gattatacca tggattggtt tcgtcaggca 120ccgggtaaag
gtctggaatg ggttgcagat gttaatccga atagcggtgg tagcatttat
180aaccagcgtt ttaaaggtcg ttttaccctg agcgttgatc gtagcaaaaa
taccctgtat 240ctgcaaatga atagtctgcg tgcagaggat accgcagtgt
attattgtgc acgtaatctg 300ggtccgaact tctactttga ttattggggt
cagggcaccc tggttaccgt tagcagc 35766119PRTArtificial
sequencePertuzumab aff.mat. clone B2 66Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys
Ala Ala Ser Gly Phe Thr Phe Asn Asp Tyr 20 25 30Thr Met Asp Trp Phe
Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Asp Val Asn
Pro Asn Ser Gly Gly Ser Ile Tyr Asn Gln Arg Phe 50 55 60Lys Gly Arg
Phe Thr Leu Ser Val Asp Arg Ser Lys Asn Thr Leu Tyr65 70 75 80Leu
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90
95Ala Arg Asn Leu Gly Pro Asn Phe Tyr Phe Asp Tyr Trp Gly Gln Gly
100 105 110Thr Leu Val Thr Val Ser Ser 11567357DNAArtificial
sequencePertuzumab aff.mat. clone E1, DNA 67gaggtgcaat tggttgaaag
cggtggtggt ctggttcagc ctggtggtag cctgcgtctg 60agctgtgcag caagcggttt
tacctttgca gattatacca tggattgggt tcgtcaggca 120ccgggtaaag
gtctggaatg ggttgcagat gttaatccga atagcggtgg tagcatttat
180aaccagcgtt ttaaaggtcg ttttaccctg agcgttgatc gtagcaaaaa
taccctgtat 240ctgcaaatga atagtctgcg tgcagaggat accgcagtgt
attattgtgc acgtaatctg 300ggtccgtggt tctactttga ttattggggt
cagggcaccc tggttaccgt tagcagc 35768119PRTArtificial
sequencePertuzumab aff.mat. clone E1 68Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys
Ala Ala Ser Gly Phe Thr Phe Ala Asp Tyr 20 25 30Thr Met Asp Trp Val
Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Asp Val Asn
Pro Asn Ser Gly Gly Ser Ile Tyr Asn Gln Arg Phe 50 55 60Lys Gly Arg
Phe Thr Leu Ser Val Asp Arg Ser Lys Asn Thr Leu Tyr65 70 75 80Leu
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90
95Ala Arg Asn Leu Gly Pro Trp Phe Tyr Phe Asp Tyr Trp Gly Gln Gly
100 105 110Thr Leu Val Thr Val Ser Ser 11569357DNAArtificial
sequencePertuzumab aff.mat. clone G2, DNA 69gaggtgcaat tggttgaaag
cggtggtggt ctggttcagc ctggtggtag cctgcgtctg 60agctgtgcag caagcggttt
tacctttacc gattacacaa tggattgggt tcgtcaggca 120ccgggtaaag
gtctggaatg ggttgcagat gttaatccga actctggtgg ttacattgtt
180aaccgtcgtt ttaaaggtcg ttttaccctg agcgttgatc gtagcaaaaa
taccctgtat 240ctgcaaatga atagtctgcg tgcagaggat accgcagtgt
attattgtgc acgtaatctg 300ggtccgagct tctattttga ttattggggt
cagggcaccc tggttaccgt tagcagc 35770119PRTArtificial
sequencePertuzumab aff.mat. clone G2 70Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys
Ala Ala Ser Gly Phe Thr Phe Thr Asp Tyr 20 25 30Thr Met Asp Trp Val
Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Asp Val Asn
Pro Asn Ser Gly Gly Tyr Ile Val Asn Arg Arg Phe 50 55 60Lys Gly Arg
Phe Thr Leu Ser Val Asp Arg Ser Lys Asn Thr Leu Tyr65 70 75 80Leu
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90
95Ala Arg Asn Leu Gly Pro Ser Phe Tyr Phe Asp Tyr Trp Gly Gln Gly
100 105 110Thr Leu Val Thr Val Ser Ser 11571357DNAArtificial
sequencePertuzumab aff.mat. clone C8, DNA 71gaggtgcaat tggttgaaag
cggtggtggt ctggttcagc ctggtggtag cctgcgtctg 60agctgtgcag caagcggttt
tacctttacc gattacacaa tggattgggt tcgtcaggca 120ccgggtaaag
gtctggaatg ggttgcagat gttaatccga actctggtgg ttctattatg
180aaccgtcgtt ttaaaggtcg ttttaccctg agcgttgatc gtagcaaaaa
taccctgtat 240ctgcaaatga atagtctgcg tgcagaggat accgcagtgt
attattgtgc acgtaatctg 300ggtccgagct tctattttga ttattggggt
cagggcaccc tggttaccgt tagcagc 35772119PRTArtificial
sequencePertuzumab aff.mat. clone C8 72Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys
Ala Ala Ser Gly Phe Thr Phe Thr Asp Tyr 20 25 30Thr Met Asp Trp Val
Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Asp Val Asn
Pro Asn Ser Gly Gly Ser Ile Met Asn Arg Arg Phe 50 55 60Lys Gly Arg
Phe Thr Leu Ser Val Asp Arg Ser Lys Asn Thr Leu Tyr65 70 75 80Leu
Gln Met Asn Ser Leu Arg
Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asn Leu Gly Pro
Ser Phe Tyr Phe Asp Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val Thr
Val Ser Ser 11573357DNAArtificial sequencePertuzumab aff.mat. clone
A1 DNA 73gaggtgcaat tggttgaaag cggtggtggt ctggttcagc ctggtggtag
cctgcgtctg 60agctgtgcag caagcggttt tacctttacc gattacacaa tggattgggt
tcgtcaggca 120ccgggtaaag gtctggaatg ggttgcagat gttaatccga
actctggtgg ttctattgtt 180aaccagcgtt ttaaaggtcg ttttaccctg
agcgttgatc gtagcaaaaa taccctgtat 240ctgcaaatga atagtctgcg
tgcagaggat accgcagtgt attattgtgc acgtaatctg 300ggtccgtggt
tctactttga ttattggggt cagggcaccc tggttaccgt tagcagc
35774119PRTArtificial sequencePertuzumab aff.mat. clone A1 74Glu
Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10
15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Thr Asp Tyr
20 25 30Thr Met Asp Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Val 35 40 45Ala Asp Val Asn Pro Asn Ser Gly Gly Ser Ile Val Asn Gln
Arg Phe 50 55 60Lys Gly Arg Phe Thr Leu Ser Val Asp Arg Ser Lys Asn
Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr
Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asn Leu Gly Pro Trp Phe Tyr Phe
Asp Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val Thr Val Ser Ser
1157517PRTArtificial sequencePertuzumab aff.mat. clone C8- VH CDR2
75Asp Val Asn Pro Asn Ser Gly Gly Ser Ile Met Asn Arg Arg Phe Lys1
5 10 15Gly7617PRTArtificial sequencePertuzumab aff.mat. clone A1-
VH CDR2 76Asp Val Asn Pro Asn Ser Gly Gly Ser Ile Val Asn Gln Arg
Phe Lys1 5 10 15Gly7717PRTArtificial sequencePertuzumab aff.mat.
clone D1-derived VH CDR2 77Asp Val Asn Pro Asn Ser Gly Gly Ser Ile
Val Asn Arg Arg Phe Lys1 5 10 15Gly7811PRTArtificial
sequenceTrastuzumab VH (D98N) (6636) CDR3 78Trp Gly Gly Asn Gly Phe
Tyr Ala Met Asp Tyr1 5 107911PRTArtificial sequenceTrastuzumab VH
CDR3 79Trp Gly Gly Glu Gly Phe Tyr Ala Met Asp Tyr1 5
108011PRTArtificial sequenceTrastuzumab VH (D98T) (6638) CDR3 80Trp
Gly Gly Thr Gly Phe Tyr Ala Met Asp Tyr1 5 1081321DNAArtificial
sequenceTrastuzumab VL (4245) DNA 81gacatccaga tgacccagag
cccaagctct ctgtctgcct ctgtgggcga cagagtgacc 60atcacctgca gagccagcca
ggacgtgaac acagccgtgg cctggtatca gcagaagcca 120ggcaaggccc
caaagctgct gatctacagc gccagcttcc tgtacagcgg cgtgccaagc
180agattcagcg gcagcagaag cggcacagac ttcaccctga ccatcagcag
cctgcagcca 240gaggacttcg ccacctacta ctgccagcag cactacacca
ccccaccaac cttcggacag 300ggcaccaagg tggagatcaa g
32182107PRTArtificial sequenceTrastuzumab VL (4245) 82Asp Ile Gln
Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg
Val Thr Ile Thr Cys Arg Ala Ser Gln Asp Val Asn Thr Ala 20 25 30Val
Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40
45Tyr Ser Ala Ser Phe Leu Tyr Ser Gly Val Pro Ser Arg Phe Ser Gly
50 55 60Ser Arg Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln
Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln His Tyr Thr
Thr Pro Pro 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys 100
10583321DNAArtificial sequenceTrastuzumab VL T31A (6641) DNA
83gatatccaga tgacccagtc tccatcctcc ctgtctgcat ctgtaggaga cagagtcacc
60atcacttgcc gggcaagtca ggacgtgaac gccgctgtag cgtggtacca gcagaaacca
120ggtaaggcac cgaagctatt aatttatagt gcgagcttcc tgtacagtgg
ggtcccgtcg 180cgttttagcg gctctcgatc cggcacggat tttaccctga
ccattagcag cctgcagcct 240gaagactttg cgacatatta ttgccaacag
cactacacaa ctcctcccac ctttggccag 300ggtacgaaag ttgaaattaa a
32184107PRTArtificial sequenceTrastuzumab VL T31A (6641) 84Asp Ile
Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp
Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Asp Val Asn Ala Ala 20 25
30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile
35 40 45Tyr Ser Ala Ser Phe Leu Tyr Ser Gly Val Pro Ser Arg Phe Ser
Gly 50 55 60Ser Arg Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu
Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln His Tyr
Thr Thr Pro Pro 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys
100 10585322DNAArtificial sequenceTrastuzumab VL T31V (6642) DNA
85gatatccaga tgacccagtc tccatcctcc ctgtctgcat ctgtaggaga cagagtcacc
60atcacttgcc gggcaagtca ggacgtgaac gtggctgtag cgtggtacca gcagaaacca
120ggtaaggcac cgaagctatt aatttatagt gcgagcttcc tgtacagtgg
ggtcccgtcg 180cgttttagcg gctctcgatc cggcacggat tttaccctga
ccattagcag cctgcagcct 240gaagactttg cgacatatta ttgccaacag
cactacacaa ctcctcccac ctttggccag 300ggtacgaaag ttgaaattaa ag
32286107PRTArtificial sequenceTrastuzumab VL T31V (6642) 86Asp Ile
Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp
Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Asp Val Asn Val Ala 20 25
30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile
35 40 45Tyr Ser Ala Ser Phe Leu Tyr Ser Gly Val Pro Ser Arg Phe Ser
Gly 50 55 60Ser Arg Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu
Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln His Tyr
Thr Thr Pro Pro 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys
100 1058711PRTArtificial sequenceTrastuzumab VH (G99A) (6639) CDR3
87Trp Gly Gly Asp Ala Phe Tyr Ala Met Asp Tyr1 5
108811PRTArtificial sequenceTrastuzumab VH (G99S) (6640) CDR3 88Trp
Gly Gly Asp Ser Phe Tyr Ala Met Asp Tyr1 5 108911PRTArtificial
sequencePertuzumab (Tras.L3) (QM)-CDR1 89Lys Ala Ser Gln Asp Val
Ser Thr Ala Val Ala1 5 10907PRTArtificial sequencePertuzumab
(Tras.L3) (QM)-CDR2 90Ser Ala Ser Phe Arg Tyr Thr1
591360DNAArtificial sequenceTrastuzumab VH (11345) DNA 91gaagtgcaat
tggtggaaag cggcggcggc ctggtgcaac cgggcggcag cctgcgtctg 60agctgcgcgg
cctccggatt taacataaag gacacataca tccactgggt gcgccaagca
120cctgggaagg gtctcgagtg ggtggctcgg atttacccaa caaatggcta
caccaggtat 180gcggatagcg tgaaaggccg ttttaccatt tcagctgata
cttcgaagaa caccgcctat 240ctgcaaatga acagcctgcg tgcggaagat
acggccgtgt attattgctc gcgttgggga 300ggagacgggt tctatgctat
ggattactgg ggccaaggca ccctggtgac ggttagctca 36092120PRTArtificial
sequenceTrastuzumab VH (11345) 92Glu Val Gln Leu Val Glu Ser Gly
Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala
Ala Ser Gly Phe Asn Ile Lys Asp Thr 20 25 30Tyr Ile His Trp Val Arg
Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Arg Ile Tyr Pro
Thr Asn Gly Tyr Thr Arg Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe
Thr Ile Ser Ala Asp Thr Ser Lys Asn Thr Ala Tyr65 70 75 80Leu Gln
Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ser
Arg Trp Gly Gly Asp Gly Phe Tyr Ala Met Asp Tyr Trp Gly Gln 100 105
110Gly Thr Leu Val Thr Val Ser Ser 115 12093360DNAArtificial
sequenceTrastuzumab VH (D98N) (6636) DNA 93gaagtgcaat tggtggaaag
cggcggcggc ctggtgcaac cgggcggcag cctgcgtctg 60agctgcgcgg cctccggatt
taacataaag gacacataca tccactgggt gcgccaagca 120cctgggaagg
gtctcgagtg ggtggctcgg atttacccaa caaatggcta caccaggtat
180gcggatagcg tgaaaggccg ttttaccatt tcagctgata cttcgaagaa
caccgcctat 240ctgcaaatga acagcctgcg tgcggaagat acggccgtgt
attattgctc gcgttgggga 300ggaaacgggt tctatgctat ggattactgg
ggccaaggca ccctggtgac ggttagctca 36094120PRTArtificial
sequenceTrastuzumab VH (D98N) (6636) 94Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys
Ala Ala Ser Gly Phe Asn Ile Lys Asp Thr 20 25 30Tyr Ile His Trp Val
Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Arg Ile Tyr
Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg
Phe Thr Ile Ser Ala Asp Thr Ser Lys Asn Thr Ala Tyr65 70 75 80Leu
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90
95Ser Arg Trp Gly Gly Asn Gly Phe Tyr Ala Met Asp Tyr Trp Gly Gln
100 105 110Gly Thr Leu Val Thr Val Ser Ser 115
12095360DNAArtificial sequenceTrastuzumab VH (D98E) (6637) DNA
95gaagtgcaat tggtggaaag cggcggcggc ctggtgcaac cgggcggcag cctgcgtctg
60agctgcgcgg cctccggatt taacataaag gacacataca tccactgggt gcgccaagca
120cctgggaagg gtctcgagtg ggtggctcgg atttacccaa caaatggcta
caccaggtat 180gcggatagcg tgaaaggccg ttttaccatt tcagctgata
cttcgaagaa caccgcctat 240ctgcaaatga acagcctgcg tgcggaagat
acggccgtgt attattgctc gcgttgggga 300ggagaggggt tctatgctat
ggattactgg ggccaaggca ccctggtgac ggttagctca 36096120PRTArtificial
sequenceTrastuzumab VH (D98E) (6637) 96Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys
Ala Ala Ser Gly Phe Asn Ile Lys Asp Thr 20 25 30Tyr Ile His Trp Val
Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Arg Ile Tyr
Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg
Phe Thr Ile Ser Ala Asp Thr Ser Lys Asn Thr Ala Tyr65 70 75 80Leu
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90
95Ser Arg Trp Gly Gly Glu Gly Phe Tyr Ala Met Asp Tyr Trp Gly Gln
100 105 110Gly Thr Leu Val Thr Val Ser Ser 115
12097360DNAArtificial sequenceTrastuzumab VH (D98T) (6638) DNA
97gaagtgcaat tggtggaaag cggcggcggc ctggtgcaac cgggcggcag cctgcgtctg
60agctgcgcgg cctccggatt taacataaag gacacataca tccactgggt gcgccaagca
120cctgggaagg gtctcgagtg ggtggctcgg atttacccaa caaatggcta
caccaggtat 180gcggatagcg tgaaaggccg ttttaccatt tcagctgata
cttcgaagaa caccgcctat 240ctgcaaatga acagcctgcg tgcggaagat
acggccgtgt attattgctc gcgttgggga 300ggaaccgggt tctatgctat
ggattactgg ggccaaggca ccctggtgac ggttagctca 36098120PRTArtificial
sequenceTrastuzumab VH (D98T) (6638) 98Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys
Ala Ala Ser Gly Phe Asn Ile Lys Asp Thr 20 25 30Tyr Ile His Trp Val
Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Arg Ile Tyr
Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg
Phe Thr Ile Ser Ala Asp Thr Ser Lys Asn Thr Ala Tyr65 70 75 80Leu
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90
95Ser Arg Trp Gly Gly Thr Gly Phe Tyr Ala Met Asp Tyr Trp Gly Gln
100 105 110Gly Thr Leu Val Thr Val Ser Ser 115
12099360DNAArtificial sequenceTrastuzumab VH (G99A) (6639) DNA
99gaagtgcaat tggtggaaag cggcggcggc ctggtgcaac cgggcggcag cctgcgtctg
60agctgcgcgg cctccggatt taacataaag gacacataca tccactgggt gcgccaagca
120cctgggaagg gtctcgagtg ggtggctcgg atttacccaa caaatggcta
caccaggtat 180gcggatagcg tgaaaggccg ttttaccatt tcagctgata
cttcgaagaa caccgcctat 240ctgcaaatga acagcctgcg tgcggaagat
acggccgtgt attattgctc gcgttgggga 300ggagacgcct tctatgctat
ggattactgg ggccaaggca ccctggtgac ggttagctca 360100120PRTArtificial
sequenceTrastuzumab VH (G99A) (6639) 100Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys
Ala Ala Ser Gly Phe Asn Ile Lys Asp Thr 20 25 30Tyr Ile His Trp Val
Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Arg Ile Tyr
Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg
Phe Thr Ile Ser Ala Asp Thr Ser Lys Asn Thr Ala Tyr65 70 75 80Leu
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90
95Ser Arg Trp Gly Gly Asp Ala Phe Tyr Ala Met Asp Tyr Trp Gly Gln
100 105 110Gly Thr Leu Val Thr Val Ser Ser 115
120101360DNAArtificial sequenceTrastuzumab VH (G99S) (6640) DNA
101gaagtgcaat tggtggaaag cggcggcggc ctggtgcaac cgggcggcag
cctgcgtctg 60agctgcgcgg cctccggatt taacataaag gacacataca tccactgggt
gcgccaagca 120cctgggaagg gtctcgagtg ggtggctcgg atttacccaa
caaatggcta caccaggtat 180gcggatagcg tgaaaggccg ttttaccatt
tcagctgata cttcgaagaa caccgcctat 240ctgcaaatga acagcctgcg
tgcggaagat acggccgtgt attattgctc gcgttgggga 300ggagacagct
tctatgctat ggattactgg ggccaaggca ccctggtgac ggttagctca
360102120PRTArtificial sequenceTrastuzumab VH (G99S) (6640) 102Glu
Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10
15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Asn Ile Lys Asp Thr
20 25 30Tyr Ile His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Val 35 40 45Ala Arg Ile Tyr Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Asp
Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Ala Asp Thr Ser Lys Asn
Thr Ala Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr
Ala Val Tyr Tyr Cys 85 90 95Ser Arg Trp Gly Gly Asp Ser Phe Tyr Ala
Met Asp Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser
115 12010311PRTArtificial sequenceTrastuzumab VL T31A (6641) CDR1
103Arg Ala Ser Gln Asp Val Asn Ala Ala Val Ala1 5
1010411PRTArtificial sequenceTrastuzumab VL CDR1 104Arg Ala Ser Gln
Asp Val Asn Val Ala Val Ala1 5 10105120PRTArtificial
sequenceTrastuzumab CDRG VH (D98E, CDRG) 105Glu Val Gln Leu Val Gln
Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser
Cys Lys Ala Ser Gly Phe Asn Ile Lys Asp Thr 20 25 30Tyr Ile His Trp
Val Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Arg Ile
Tyr Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Gln Lys Phe 50 55 60Gln Gly
Arg Val Thr Met Thr Arg Asp Thr Ser Ile Ser Thr Ala Tyr65 70 75
80Met Glu Leu Ser Arg Leu Arg Ser Asp Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ser Arg Trp Gly Gly Glu Gly Phe Tyr Ala Met Asp Tyr Trp Gly
Gln 100 105 110Gly Thr Met Val Thr Val Ser Ser 115
120106107PRTArtificial sequenceTrastuzumab CDRG VL (N30T, CDRG)
106Asp Ile Gln Leu Thr Gln Pro Pro Ser Val Ser Val Ala Pro Gly Gln1
5 10 15Thr Ala Arg Ile Thr Cys Gly Ala Ser Gln Asp Val Ser Thr Ala
Val 20 25 30Ala Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro Val Leu Val
Val Tyr 35 40 45Ser Ala Ser Phe Leu Tyr Ser Gly Ile Pro Ser Arg Phe
Ser Gly Ser 50 55 60Arg Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Arg
Val Glu Ala Gly65 70 75 80Asp Glu Ala Asp Tyr Tyr Cys Gln Gln His
Tyr Thr Thr Pro Pro Thr 85 90 95Phe Gly Thr Gly Thr Lys Val Thr Val
Leu Arg 100 10510711PRTArtificial sequenceTrastuzumab CDRG VL CDR1
107Gly Ala Ser Gln Asp Val Ser Thr Ala Val Ala1 5
1010817PRTArtificial sequenceTrastuzumab CDRG VH CDR2 108Arg Ile
Tyr Pro Thr Asn Gly Tyr Thr
Arg Tyr Ala Gln Lys Phe Gln1 5 10 15Gly109453PRTArtificial
sequenceXPertuzumab heavy chain 109Glu Val Gln Leu Val Glu Ser Gly
Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala
Ala Ser Gly Phe Thr Phe Thr Asp Tyr 20 25 30Thr Met Asp Trp Val Arg
Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Asp Val Asn Pro
Asn Ser Gly Gly Ser Ile Tyr Asn Gln Arg Phe 50 55 60Lys Gly Arg Phe
Thr Leu Ser Val Asp Arg Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln
Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala
Arg Asn Leu Gly Pro Ser Phe Tyr Phe Asp Tyr Trp Gly Gln Gly 100 105
110Thr Leu Val Thr Val Ser Ser Ala Ser Val Ala Ala Pro Ser Val Phe
115 120 125Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly Thr Ala
Ser Val 130 135 140Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala
Lys Val Gln Trp145 150 155 160Lys Val Asp Asn Ala Leu Gln Ser Gly
Asn Ser Gln Glu Ser Val Thr 165 170 175Glu Gln Asp Ser Lys Asp Ser
Thr Tyr Ser Leu Ser Ser Thr Leu Thr 180 185 190Leu Ser Lys Ala Asp
Tyr Glu Lys His Lys Val Tyr Ala Cys Glu Val 195 200 205Thr His Gln
Gly Leu Ser Ser Pro Val Thr Lys Ser Phe Asn Arg Gly 210 215 220Glu
Cys Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu225 230
235 240Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp
Thr 245 250 255Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val
Val Asp Val 260 265 270Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp
Tyr Val Asp Gly Val 275 280 285Glu Val His Asn Ala Lys Thr Lys Pro
Arg Glu Glu Gln Tyr Asn Ser 290 295 300Thr Tyr Arg Val Val Ser Val
Leu Thr Val Leu His Gln Asp Trp Leu305 310 315 320Asn Gly Lys Glu
Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala 325 330 335Pro Ile
Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro 340 345
350Gln Val Tyr Thr Leu Pro Pro Cys Arg Asp Glu Leu Thr Lys Asn Gln
355 360 365Val Ser Leu Trp Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp
Ile Ala 370 375 380Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn
Tyr Lys Thr Thr385 390 395 400Pro Pro Val Leu Asp Ser Asp Gly Ser
Phe Phe Leu Tyr Ser Lys Leu 405 410 415Thr Val Asp Lys Ser Arg Trp
Gln Gln Gly Asn Val Phe Ser Cys Ser 420 425 430Val Met His Glu Ala
Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser 435 440 445Leu Ser Pro
Gly Lys 450110212PRTArtificial sequenceXPertuzumab light chain
110Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1
5 10 15Asp Arg Val Thr Ile Thr Cys Lys Ala Ser Gln Asp Val Ser Ile
Gly 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu
Leu Ile 35 40 45Tyr Ser Ala Ser Tyr Arg Tyr Thr Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln
Tyr Tyr Ile Tyr Pro Tyr 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu
Ile Lys Ser Ser Ala Ser Thr 100 105 110Lys Gly Pro Ser Val Phe Pro
Leu Ala Pro Ser Ser Lys Ser Thr Ser 115 120 125Gly Gly Thr Ala Ala
Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu 130 135 140Pro Val Thr
Val Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His145 150 155
160Thr Phe Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser
165 170 175Val Val Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr
Ile Cys 180 185 190Asn Val Asn His Lys Pro Ser Asn Thr Lys Val Asp
Lys Lys Val Glu 195 200 205Pro Lys Ser Cys 210111450PRTArtificial
sequenceTrastuzumab CDRG heavy chain (VHCH1) 111Gln Val Gln Leu Val
Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val
Ser Cys Lys Ala Ser Gly Phe Asn Ile Lys Asp Thr 20 25 30Tyr Ile His
Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Arg
Ile Tyr Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Gln Lys Phe 50 55 60Gln
Gly Arg Val Thr Met Thr Arg Asp Thr Ser Ile Ser Thr Ala Tyr65 70 75
80Met Glu Leu Ser Arg Leu Arg Ser Asp Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ser Arg Trp Gly Gly Glu Gly Phe Tyr Ala Met Asp Tyr Trp Gly
Gln 100 105 110Gly Thr Met Val Thr Val Ser Ser Ala Ser Thr Lys Gly
Pro Ser Val 115 120 125Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser
Gly Gly Thr Ala Ala 130 135 140Leu Gly Cys Leu Val Lys Asp Tyr Phe
Pro Glu Pro Val Thr Val Ser145 150 155 160Trp Asn Ser Gly Ala Leu
Thr Ser Gly Val His Thr Phe Pro Ala Val 165 170 175Leu Gln Ser Ser
Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro 180 185 190Ser Ser
Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys 195 200
205Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp
210 215 220Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu
Gly Gly225 230 235 240Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys
Asp Thr Leu Met Ile 245 250 255Ser Arg Thr Pro Glu Val Thr Cys Val
Val Val Asp Val Ser His Glu 260 265 270Asp Pro Glu Val Lys Phe Asn
Trp Tyr Val Asp Gly Val Glu Val His 275 280 285Asn Ala Lys Thr Lys
Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg 290 295 300Val Val Ser
Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys305 310 315
320Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu
325 330 335Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln
Val Cys 340 345 350Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn
Gln Val Ser Leu 355 360 365Ser Cys Ala Val Lys Gly Phe Tyr Pro Ser
Asp Ile Ala Val Glu Trp 370 375 380Glu Ser Asn Gly Gln Pro Glu Asn
Asn Tyr Lys Thr Thr Pro Pro Val385 390 395 400Leu Asp Ser Asp Gly
Ser Phe Phe Leu Val Ser Lys Leu Thr Val Asp 405 410 415Lys Ser Arg
Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His 420 425 430Glu
Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro 435 440
445Gly Lys 450112213PRTArtificial sequenceTrastuzumab CDRG light
chain (VLCL) 112Asp Ile Gln Leu Thr Gln Pro Pro Ser Val Ser Val Ala
Pro Gly Gln1 5 10 15Thr Ala Arg Ile Thr Cys Gly Ala Ser Gln Asp Val
Ser Thr Ala Val 20 25 30Ala Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro
Val Leu Val Val Tyr 35 40 45Ser Ala Ser Phe Leu Tyr Ser Gly Ile Pro
Ser Arg Phe Ser Gly Ser 50 55 60Arg Ser Gly Thr Asp Phe Thr Leu Thr
Ile Ser Arg Val Glu Ala Gly65 70 75 80Asp Glu Ala Asp Tyr Tyr Cys
Gln Gln His Tyr Thr Thr Pro Pro Thr 85 90 95Phe Gly Thr Gly Thr Lys
Val Thr Val Leu Arg Thr Val Ala Ala Pro 100 105 110Ser Val Phe Ile
Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly Thr 115 120 125Ala Ser
Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala Lys 130 135
140Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln
Glu145 150 155 160Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr
Ser Leu Ser Ser 165 170 175Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu
Lys His Lys Val Tyr Ala 180 185 190Cys Glu Val Thr His Gln Gly Leu
Ser Ser Pro Val Thr Lys Ser Phe 195 200 205Asn Arg Gly Glu Cys
210113107PRTArtificial sequencePertuzumab wt CL 113Arg Thr Val Ala
Ala Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu1 5 10 15Gln Leu Lys
Ser Gly Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe 20 25 30Tyr Pro
Arg Glu Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln 35 40 45Ser
Gly Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser 50 55
60Thr Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu65
70 75 80Lys His Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser
Ser 85 90 95Pro Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 100
10511498PRTArtificial sequencePertuzumab wt CH1 114Ala Ser Thr Lys
Gly Pro Ser Val Phe Pro Leu Ala Pro Ser Ser Lys1 5 10 15Ser Thr Ser
Gly Gly Thr Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr 20 25 30Phe Pro
Glu Pro Val Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser 35 40 45Gly
Val His Thr Phe Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser 50 55
60Leu Ser Ser Val Val Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr65
70 75 80Tyr Ile Cys Asn Val Asn His Lys Pro Ser Asn Thr Lys Val Asp
Lys 85 90 95Lys Val115107PRTArtificial sequenceTrastuzumab CH1
115Ala Ser Thr Lys Gly Pro Ser Val Phe Pro Leu Ala Pro Ser Ser Lys1
5 10 15Ser Thr Ser Gly Gly Thr Ala Ala Leu Gly Cys Leu Val Lys Asp
Tyr 20 25 30Phe Pro Glu Pro Val Thr Val Ser Trp Asn Ser Gly Ala Leu
Thr Ser 35 40 45Gly Val His Thr Phe Pro Ala Val Leu Gln Ser Ser Gly
Leu Tyr Ser 50 55 60Leu Ser Ser Val Val Thr Val Pro Ser Ser Ser Leu
Gly Thr Gln Thr65 70 75 80Tyr Ile Cys Asn Val Asn His Lys Pro Ser
Asn Thr Lys Val Asp Lys 85 90 95Lys Val Glu Pro Lys Ser Cys Asp Lys
Thr His 100 105116106PRTArtificial sequenceTrastuzumab CL 116Thr
Val Ala Ala Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln1 5 10
15Leu Lys Ser Gly Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr
20 25 30Pro Arg Glu Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln
Ser 35 40 45Gly Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp
Ser Thr 50 55 60Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp
Tyr Glu Lys65 70 75 80His Lys Val Tyr Ala Cys Glu Val Thr His Gln
Gly Leu Ser Ser Pro 85 90 95Val Thr Lys Ser Phe Asn Arg Gly Glu Cys
100 105117120PRTArtificial sequenceTrastuzumab VH (D98E) 117Glu Val
Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser
Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Asn Ile Lys Asp Thr 20 25
30Tyr Ile His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val
35 40 45Ala Arg Ile Tyr Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Asp Ser
Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Ala Asp Thr Ser Lys Asn Thr
Ala Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala
Val Tyr Tyr Cys 85 90 95Ser Arg Trp Gly Gly Glu Gly Phe Tyr Ala Met
Asp Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser 115
120118108PRTArtificial sequenceTrastuzumab VL (T31V) 118Asp Ile Gln
Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg
Val Thr Ile Thr Cys Arg Ala Ser Gln Asp Val Asn Val Ala 20 25 30Val
Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40
45Tyr Ser Ala Ser Phe Leu Tyr Ser Gly Val Pro Ser Arg Phe Ser Gly
50 55 60Ser Arg Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln
Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln His Tyr Thr
Thr Pro Pro 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg
100 105119453PRTArtificial sequencePertuzumab heavy chain 119Glu
Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10
15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Thr Asp Tyr
20 25 30Thr Met Asp Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Val 35 40 45Ala Asp Val Asn Pro Asn Ser Gly Gly Ser Ile Tyr Asn Gln
Arg Phe 50 55 60Lys Gly Arg Phe Thr Leu Ser Val Asp Arg Ser Lys Asn
Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr
Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asn Leu Gly Pro Ser Phe Tyr Phe
Asp Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val Thr Val Ser Ser Ala
Ser Val Ala Ala Pro Ser Val Phe 115 120 125Ile Phe Pro Pro Ser Asp
Glu Gln Leu Lys Ser Gly Thr Ala Ser Val 130 135 140Val Cys Leu Leu
Asn Asn Phe Tyr Pro Arg Glu Ala Lys Val Gln Trp145 150 155 160Lys
Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln Glu Ser Val Thr 165 170
175Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser Ser Thr Leu Thr
180 185 190Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr Ala Cys
Glu Val 195 200 205Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser
Phe Asn Arg Gly 210 215 220Glu Cys Asp Lys Thr His Thr Cys Pro Pro
Cys Pro Ala Pro Glu Leu225 230 235 240Leu Gly Gly Pro Ser Val Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr 245 250 255Leu Met Ile Ser Arg
Thr Pro Glu Val Thr Cys Val Val Val Asp Val 260 265 270Ser His Glu
Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val 275 280 285Glu
Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser 290 295
300Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp
Leu305 310 315 320Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys
Ala Leu Pro Ala 325 330 335Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys
Gly Gln Pro Arg Glu Pro 340 345 350Gln Val Cys Thr Leu Pro Pro Ser
Arg Asp Glu Leu Thr Lys Asn Gln 355 360 365Val Ser Leu Ser Cys Ala
Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala 370 375 380Val Glu Trp Glu
Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr385 390 395 400Pro
Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Val Ser Lys Leu 405
410
415Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser
420 425 430Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser
Leu Ser 435 440 445Leu Ser Pro Gly Lys 450120214PRTArtificial
sequencePertuzumab light chain 120Asp Ile Gln Met Thr Gln Ser Pro
Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys
Lys Ala Ser Gln Asp Val Ser Ile Gly 20 25 30Val Ala Trp Tyr Gln Gln
Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ser Ala Ser Tyr
Arg Tyr Thr Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly
Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp
Phe Ala Thr Tyr Tyr Cys Gln Gln Tyr Tyr Ile Tyr Pro Tyr 85 90 95Thr
Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala Ala 100 105
110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly
115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg
Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser
Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu Gln Asp Ser Lys
Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys
Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala Cys Glu Val Thr
His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200 205Phe Asn Arg
Gly Glu Cys 210121450PRTArtificial sequenceXTrastuzumab heavy chain
121Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1
5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Asn Ile Lys Asp
Thr 20 25 30Tyr Ile His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu
Trp Val 35 40 45Ala Arg Ile Tyr Pro Thr Asn Gly Tyr Thr Arg Tyr Ala
Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Ala Asp Thr Ser Lys
Asn Thr Ala Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ser Arg Trp Gly Gly Glu Gly Phe Tyr
Ala Met Asp Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser
Ser Ala Ser Thr Lys Gly Pro Ser Val 115 120 125Phe Pro Leu Ala Pro
Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala 130 135 140Leu Gly Cys
Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser145 150 155
160Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val
165 170 175Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr
Val Pro 180 185 190Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn
Val Asn His Lys 195 200 205Pro Ser Asn Thr Lys Val Asp Lys Lys Val
Glu Pro Lys Ser Cys Asp 210 215 220Lys Thr His Thr Cys Pro Pro Cys
Pro Ala Pro Glu Leu Leu Gly Gly225 230 235 240Pro Ser Val Phe Leu
Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile 245 250 255Ser Arg Thr
Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu 260 265 270Asp
Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His 275 280
285Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg
290 295 300Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn
Gly Lys305 310 315 320Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu
Pro Ala Pro Ile Glu 325 330 335Lys Thr Ile Ser Lys Ala Lys Gly Gln
Pro Arg Glu Pro Gln Val Tyr 340 345 350Thr Leu Pro Pro Cys Arg Asp
Glu Leu Thr Lys Asn Gln Val Ser Leu 355 360 365Trp Cys Leu Val Lys
Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp 370 375 380Glu Ser Asn
Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val385 390 395
400Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp
405 410 415Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val
Met His 420 425 430Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu
Ser Leu Ser Pro 435 440 445Gly Lys 450122212PRTArtificial
sequenceXTrastuzumab light chain 122Asp Ile Gln Met Thr Gln Ser Pro
Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys
Arg Ala Ser Gln Asp Val Asn Val Ala 20 25 30Val Ala Trp Tyr Gln Gln
Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ser Ala Ser Phe
Leu Tyr Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Arg Ser Gly
Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp
Phe Ala Thr Tyr Tyr Cys Gln Gln His Tyr Thr Thr Pro Pro 85 90 95Thr
Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Ser Ser Ala Ser Thr 100 105
110Lys Gly Pro Ser Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser
115 120 125Gly Gly Thr Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe
Pro Glu 130 135 140Pro Val Thr Val Ser Trp Asn Ser Gly Ala Leu Thr
Ser Gly Val His145 150 155 160Thr Phe Pro Ala Val Leu Gln Ser Ser
Gly Leu Tyr Ser Leu Ser Ser 165 170 175Val Val Thr Val Pro Ser Ser
Ser Leu Gly Thr Gln Thr Tyr Ile Cys 180 185 190Asn Val Asn His Lys
Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu 195 200 205Pro Lys Ser
Cys 210123214PRTArtificial sequenceLight chain (kappa)
[Trastuzumab, 1016] 123Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu
Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser
Gln Asp Val Asn Thr Ala 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly
Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ser Ala Ser Phe Leu Tyr Ser
Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Arg Ser Gly Thr Asp Phe
Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr
Tyr Tyr Cys Gln Gln His Tyr Thr Thr Pro Pro 85 90 95Thr Phe Gly Gln
Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser
Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly 115 120
125Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala
130 135 140Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn
Ser Gln145 150 155 160Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser
Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys Ala Asp
Tyr Glu Lys His Lys Val Tyr 180 185 190Ala Cys Glu Val Thr His Gln
Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200 205Phe Asn Arg Gly Glu
Cys 210124711PRTArtificial sequenceHeavy chain [Trastuzumab + scFv
Omnitarg, RB40] 124Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val
Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe
Asn Ile Lys Asp Thr 20 25 30Tyr Ile His Trp Val Arg Gln Ala Pro Gly
Lys Gly Leu Glu Trp Val 35 40 45Ala Arg Ile Tyr Pro Thr Asn Gly Tyr
Thr Arg Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Ala
Asp Thr Ser Lys Asn Thr Ala Tyr65 70 75 80Leu Gln Met Asn Ser Leu
Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ser Arg Trp Gly Gly
Asp Gly Phe Tyr Ala Met Asp Tyr Trp Gly Gln 100 105 110Gly Thr Leu
Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser Val 115 120 125Phe
Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala 130 135
140Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val
Ser145 150 155 160Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr
Phe Pro Ala Val 165 170 175Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser
Ser Val Val Thr Val Pro 180 185 190Ser Ser Ser Leu Gly Thr Gln Thr
Tyr Ile Cys Asn Val Asn His Lys 195 200 205Pro Ser Asn Thr Lys Val
Asp Lys Lys Val Glu Pro Lys Ser Cys Asp 210 215 220Lys Thr His Thr
Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly225 230 235 240Pro
Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile 245 250
255Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu
260 265 270Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu
Val His 275 280 285Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn
Ser Thr Tyr Arg 290 295 300Val Val Ser Val Leu Thr Val Leu His Gln
Asp Trp Leu Asn Gly Lys305 310 315 320Glu Tyr Lys Cys Lys Val Ser
Asn Lys Ala Leu Pro Ala Pro Ile Glu 325 330 335Lys Thr Ile Ser Lys
Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr 340 345 350Thr Leu Pro
Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser Leu 355 360 365Thr
Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp 370 375
380Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro
Val385 390 395 400Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys
Leu Thr Val Asp 405 410 415Lys Ser Arg Trp Gln Gln Gly Asn Val Phe
Ser Cys Ser Val Met His 420 425 430Glu Ala Leu His Asn His Tyr Thr
Gln Lys Ser Leu Ser Leu Ser Pro 435 440 445Gly Lys Gly Gly Gly Gly
Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly 450 455 460Ser Glu Val Gln
Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly465 470 475 480Gly
Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Thr Asp 485 490
495Tyr Thr Met Asp Trp Val Arg Gln Ala Pro Gly Lys Cys Leu Glu Trp
500 505 510Val Ala Asp Val Asn Pro Asn Ser Gly Gly Ser Ile Tyr Asn
Gln Arg 515 520 525Phe Lys Gly Arg Phe Thr Leu Ser Val Asp Arg Ser
Lys Asn Thr Leu 530 535 540Tyr Leu Gln Met Asn Ser Leu Arg Ala Glu
Asp Thr Ala Val Tyr Tyr545 550 555 560Cys Ala Arg Asn Leu Gly Pro
Ser Phe Tyr Phe Asp Tyr Trp Gly Gln 565 570 575Gly Thr Leu Val Thr
Val Ser Ser Gly Gly Gly Gly Ser Gly Gly Gly 580 585 590Gly Ser Gly
Gly Gly Gly Ser Gly Gly Gly Gly Ser Asp Ile Gln Met 595 600 605Thr
Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly Asp Arg Val Thr 610 615
620Ile Thr Cys Lys Ala Ser Gln Asp Val Ser Ile Gly Val Ala Trp
Tyr625 630 635 640Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile
Tyr Ser Ala Ser 645 650 655Tyr Arg Tyr Thr Gly Val Pro Ser Arg Phe
Ser Gly Ser Gly Ser Gly 660 665 670Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro Glu Asp Phe Ala 675 680 685Thr Tyr Tyr Cys Gln Gln
Tyr Tyr Ile Tyr Pro Tyr Thr Phe Gly Cys 690 695 700Gly Thr Lys Val
Glu Ile Lys705 710125471PRTArtificial sequenceLight chain [scFv
Trastuzumab + Omnitarg, RB34] 125Asp Ile Gln Met Thr Gln Ser Pro
Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys
Arg Ala Ser Gln Asp Val Asn Thr Ala 20 25 30Val Ala Trp Tyr Gln Gln
Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ser Ala Ser Phe
Leu Tyr Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Arg Ser Gly
Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp
Phe Ala Thr Tyr Tyr Cys Gln Gln His Tyr Thr Thr Pro Pro 85 90 95Thr
Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Gly Gly Gly Gly Ser 100 105
110Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Glu Val Gln Leu Val Glu
115 120 125Ser Gly Gly Gly Leu Val Gln Pro Gly Gly Ser Leu Arg Leu
Ser Cys 130 135 140Ala Ala Ser Gly Phe Asn Ile Lys Asp Thr Tyr Ile
His Trp Val Arg145 150 155 160Gln Ala Pro Gly Lys Gly Leu Glu Trp
Val Ala Arg Ile Tyr Pro Thr 165 170 175Asn Gly Tyr Thr Arg Tyr Ala
Asp Ser Val Lys Gly Arg Phe Thr Ile 180 185 190Ser Ala Asp Thr Ser
Lys Asn Thr Ala Tyr Leu Gln Met Asn Ser Leu 195 200 205Arg Ala Glu
Asp Thr Ala Val Tyr Tyr Cys Ser Arg Trp Gly Gly Asp 210 215 220Gly
Phe Tyr Ala Met Asp Tyr Trp Gly Gln Gly Thr Leu Val Thr Val225 230
235 240Ser Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly
Gly 245 250 255Ser Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser
Ala Ser Val 260 265 270Gly Asp Arg Val Thr Ile Thr Cys Lys Ala Ser
Gln Asp Val Ser Ile 275 280 285Gly Val Ala Trp Tyr Gln Gln Lys Pro
Gly Lys Ala Pro Lys Leu Leu 290 295 300Ile Tyr Ser Ala Ser Tyr Arg
Tyr Thr Gly Val Pro Ser Arg Phe Ser305 310 315 320Gly Ser Gly Ser
Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln 325 330 335Pro Glu
Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Tyr Tyr Ile Tyr Pro 340 345
350Tyr Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala
355 360 365Ala Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu
Lys Ser 370 375 380Gly Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe
Tyr Pro Arg Glu385 390 395 400Ala Lys Val Gln Trp Lys Val Asp Asn
Ala Leu Gln Ser Gly Asn Ser 405 410 415Gln Glu Ser Val Thr Glu Gln
Asp Ser Lys Asp Ser Thr Tyr Ser Leu 420 425 430Ser Ser Thr Leu Thr
Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val 435 440 445Tyr Ala Cys
Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys 450 455 460Ser
Phe Asn Arg Gly Glu Cys465 470126449PRTArtificial sequenceHeavy
chain (Omnitarg, RB33) 126Glu Val Gln Leu Val Glu Ser Gly Gly Gly
Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser
Gly Phe Thr Phe Thr Asp Tyr 20 25 30Thr Met Asp Trp Val Arg Gln Ala
Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Asp Val Asn Pro Asn Ser
Gly Gly Ser Ile Tyr Asn Gln Arg Phe 50 55 60Lys Gly Arg Phe Thr Leu
Ser Val Asp Arg Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn
Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asn
Leu Gly Pro Ser Phe Tyr Phe Asp Tyr Trp Gly Gln Gly 100 105 110Thr
Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly
Pro Ser Val Phe 115 120 125Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser
Gly Gly Thr Ala Ala Leu 130 135 140Gly Cys Leu Val Lys Asp Tyr Phe
Pro Glu Pro Val Thr Val Ser Trp145 150 155 160Asn Ser Gly Ala Leu
Thr Ser Gly Val His Thr Phe Pro Ala Val Leu 165 170 175Gln Ser Ser
Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser 180 185 190Ser
Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro 195 200
205Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp Lys
210 215 220Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly
Gly Pro225 230 235 240Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp
Thr Leu Met Ile Ser 245 250 255Arg Thr Pro Glu Val Thr Cys Val Val
Val Asp Val Ser His Glu Asp 260 265 270Pro Glu Val Lys Phe Asn Trp
Tyr Val Asp Gly Val Glu Val His Asn 275 280 285Ala Lys Thr Lys Pro
Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val 290 295 300Val Ser Val
Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu305 310 315
320Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys
325 330 335Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val
Tyr Thr 340 345 350Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln
Val Ser Leu Thr 355 360 365Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp
Ile Ala Val Glu Trp Glu 370 375 380Ser Asn Gly Gln Pro Glu Asn Asn
Tyr Lys Thr Thr Pro Pro Val Leu385 390 395 400Asp Ser Asp Gly Ser
Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys 405 410 415Ser Arg Trp
Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu 420 425 430Ala
Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly 435 440
445Lys127470PRTArtificial sequenceLight chain [Trastuzumab +
scFvOmnitarg, RB35] 127Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu
Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser
Gln Asp Val Asn Thr Ala 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly
Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ser Ala Ser Phe Leu Tyr Ser
Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Arg Ser Gly Thr Asp Phe
Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr
Tyr Tyr Cys Gln Gln His Tyr Thr Thr Pro Pro 85 90 95Thr Phe Gly Gln
Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser
Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly 115 120
125Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala
130 135 140Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn
Ser Gln145 150 155 160Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser
Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys Ala Asp
Tyr Glu Lys His Lys Val Tyr 180 185 190Ala Cys Glu Val Thr His Gln
Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200 205Phe Asn Arg Gly Glu
Cys Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser 210 215 220Gly Gly Gly
Gly Ser Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu225 230 235
240Ser Ala Ser Val Gly Asp Arg Val Thr Ile Thr Cys Lys Ala Ser Gln
245 250 255Asp Val Ser Ile Gly Val Ala Trp Tyr Gln Gln Lys Pro Gly
Lys Ala 260 265 270Pro Lys Leu Leu Ile Tyr Ser Ala Ser Tyr Arg Tyr
Thr Gly Val Pro 275 280 285Ser Arg Phe Ser Gly Ser Gly Ser Gly Thr
Asp Phe Thr Leu Thr Ile 290 295 300Ser Ser Leu Gln Pro Glu Asp Phe
Ala Thr Tyr Tyr Cys Gln Gln Tyr305 310 315 320Tyr Ile Tyr Pro Tyr
Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys 325 330 335Gly Gly Gly
Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Glu 340 345 350Val
Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly Ser 355 360
365Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Thr Asp Tyr Thr
370 375 380Met Asp Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Val Ala385 390 395 400Asp Val Asn Pro Asn Ser Gly Gly Ser Ile Tyr
Asn Gln Arg Phe Lys 405 410 415Gly Arg Phe Thr Leu Ser Val Asp Arg
Ser Lys Asn Thr Leu Tyr Leu 420 425 430Gln Met Asn Ser Leu Arg Ala
Glu Asp Thr Ala Val Tyr Tyr Cys Ala 435 440 445Arg Asn Leu Gly Pro
Ser Phe Tyr Phe Asp Tyr Trp Gly Gln Gly Thr 450 455 460Leu Val Thr
Val Ser Ser465 470128450PRTArtificial sequenceHeavy chain
[Trastuzumab, 1036] 128Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu
Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Asn Ile Lys Asp Thr 20 25 30Tyr Ile His Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Arg Ile Tyr Pro Thr Asn Gly
Tyr Thr Arg Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser
Ala Asp Thr Ser Lys Asn Thr Ala Tyr65 70 75 80Leu Gln Met Asn Ser
Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ser Arg Trp Gly
Gly Asp Gly Phe Tyr Ala Met Asp Tyr Trp Gly Gln 100 105 110Gly Thr
Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser Val 115 120
125Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala
130 135 140Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr
Val Ser145 150 155 160Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His
Thr Phe Pro Ala Val 165 170 175Leu Gln Ser Ser Gly Leu Tyr Ser Leu
Ser Ser Val Val Thr Val Pro 180 185 190Ser Ser Ser Leu Gly Thr Gln
Thr Tyr Ile Cys Asn Val Asn His Lys 195 200 205Pro Ser Asn Thr Lys
Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp 210 215 220Lys Thr His
Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly225 230 235
240Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile
245 250 255Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser
His Glu 260 265 270Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly
Val Glu Val His 275 280 285Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln
Tyr Asn Ser Thr Tyr Arg 290 295 300Val Val Ser Val Leu Thr Val Leu
His Gln Asp Trp Leu Asn Gly Lys305 310 315 320Glu Tyr Lys Cys Lys
Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu 325 330 335Lys Thr Ile
Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr 340 345 350Thr
Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu 355 360
365Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp
370 375 380Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro
Pro Val385 390 395 400Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser
Lys Leu Thr Val Asp 405 410 415Lys Ser Arg Trp Gln Gln Gly Asn Val
Phe Ser Cys Ser Val Met His 420 425 430Glu Ala Leu His Asn His Tyr
Thr Gln Lys Ser Leu Ser Leu Ser Pro 435 440 445Gly Lys
450129214PRTArtificial sequenceLight chain (kappa) [Trastuzumab,
1016] 129Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser
Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Asp Val
Asn Thr Ala 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro
Lys Leu Leu Ile 35 40 45Tyr Ser Ala Ser Phe Leu Tyr Ser Gly Val Pro
Ser Arg Phe Ser Gly 50 55 60Ser Arg Ser Gly Thr Asp Phe Thr Leu Thr
Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys
Gln Gln His Tyr Thr Thr Pro Pro 85 90 95Thr Phe Gly Gln Gly Thr Lys
Val Glu Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile
Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser
Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys
Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150
155 160Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu
Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His
Lys Val Tyr 180 185 190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser
Pro Val Thr Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys
210130711PRTArtificial sequenceHeavy chain [Trastuzumab +
scFvOmnitarg, RB43] 130Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu
Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Asn Ile Lys Asp Thr 20 25 30Tyr Ile His Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Arg Ile Tyr Pro Thr Asn Gly
Tyr Thr Arg Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser
Ala Asp Thr Ser Lys Asn Thr Ala Tyr65 70 75 80Leu Gln Met Asn Ser
Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ser Arg Trp Gly
Gly Asp Gly Phe Tyr Ala Met Asp Tyr Trp Gly Gln 100 105 110Gly Thr
Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser Val 115 120
125Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala
130 135 140Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr
Val Ser145 150 155 160Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His
Thr Phe Pro Ala Val 165 170 175Leu Gln Ser Ser Gly Leu Tyr Ser Leu
Ser Ser Val Val Thr Val Pro 180 185 190Ser Ser Ser Leu Gly Thr Gln
Thr Tyr Ile Cys Asn Val Asn His Lys 195 200 205Pro Ser Asn Thr Lys
Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp 210 215 220Lys Thr His
Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly225 230 235
240Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile
245 250 255Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser
His Glu 260 265 270Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly
Val Glu Val His 275 280 285Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln
Tyr Asn Ser Thr Tyr Arg 290 295 300Val Val Ser Val Leu Thr Val Leu
His Gln Asp Trp Leu Asn Gly Lys305 310 315 320Glu Tyr Lys Cys Lys
Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu 325 330 335Lys Thr Ile
Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr 340 345 350Thr
Leu Pro Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser Leu 355 360
365Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp
370 375 380Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro
Pro Val385 390 395 400Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser
Lys Leu Thr Val Asp 405 410 415Lys Ser Arg Trp Gln Gln Gly Asn Val
Phe Ser Cys Ser Val Met His 420 425 430Glu Ala Leu His Asn His Tyr
Thr Gln Lys Ser Leu Ser Leu Ser Pro 435 440 445Gly Lys Gly Gly Gly
Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly 450 455 460Ser Asp Ile
Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val465 470 475
480Gly Asp Arg Val Thr Ile Thr Cys Lys Ala Ser Gln Asp Val Ser Ile
485 490 495Gly Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys
Leu Leu 500 505 510Ile Tyr Ser Ala Ser Tyr Arg Tyr Thr Gly Val Pro
Ser Arg Phe Ser 515 520 525Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu
Thr Ile Ser Ser Leu Gln 530 535 540Pro Glu Asp Phe Ala Thr Tyr Tyr
Cys Gln Gln Tyr Tyr Ile Tyr Pro545 550 555 560Tyr Thr Phe Gly Cys
Gly Thr Lys Val Glu Ile Lys Gly Gly Gly Gly 565 570 575Ser Gly Gly
Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser 580 585 590Glu
Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 595 600
605Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Thr Asp Tyr
610 615 620Thr Met Asp Trp Val Arg Gln Ala Pro Gly Lys Cys Leu Glu
Trp Val625 630 635 640Ala Asp Val Asn Pro Asn Ser Gly Gly Ser Ile
Tyr Asn Gln Arg Phe 645 650 655Lys Gly Arg Phe Thr Leu Ser Val Asp
Arg Ser Lys Asn Thr Leu Tyr 660 665 670Leu Gln Met Asn Ser Leu Arg
Ala Glu Asp Thr Ala Val Tyr Tyr Cys 675 680 685Ala Arg Asn Leu Gly
Pro Ser Phe Tyr Phe Asp Tyr Trp Gly Gln Gly 690 695 700Thr Leu Val
Thr Val Ser Ser705 710131480PRTArtificial sequenceLight chain
[Trastuzumab + scFv Omnitarg, RB61] 131Asp Ile Gln Met Thr Gln Ser
Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr
Cys Arg Ala Ser Gln Asp Val Asn Thr Ala 20 25 30Val Ala Trp Tyr Gln
Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ser Ala Ser
Phe Leu Tyr Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Arg Ser
Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu
Asp Phe Ala Thr Tyr Tyr Cys Gln Gln His Tyr Thr Thr Pro Pro 85 90
95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala Ala
100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys
Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr
Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn Ala Leu
Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu Gln Asp
Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr Leu
Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala Cys Glu
Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200 205Phe
Asn Arg Gly Glu Cys Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser 210 215
220Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Asp Ile Gln Met Thr
Gln225 230 235 240Ser Pro Ser Ser Leu Ser Ala Ser Val Gly Asp Arg
Val Thr Ile Thr 245 250
255Cys Lys Ala Ser Gln Asp Val Ser Ile Gly Val Ala Trp Tyr Gln Gln
260 265 270Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile Tyr Ser Ala Ser
Tyr Arg 275 280 285Tyr Thr Gly Val Pro Ser Arg Phe Ser Gly Ser Gly
Ser Gly Thr Asp 290 295 300Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro
Glu Asp Phe Ala Thr Tyr305 310 315 320Tyr Cys Gln Gln Tyr Tyr Ile
Tyr Pro Tyr Thr Phe Gly Cys Gly Thr 325 330 335Lys Val Glu Ile Lys
Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly 340 345 350Gly Gly Gly
Ser Gly Gly Gly Gly Ser Glu Val Gln Leu Val Glu Ser 355 360 365Gly
Gly Gly Leu Val Gln Pro Gly Gly Ser Leu Arg Leu Ser Cys Ala 370 375
380Ala Ser Gly Phe Thr Phe Thr Asp Tyr Thr Met Asp Trp Val Arg
Gln385 390 395 400Ala Pro Gly Lys Cys Leu Glu Trp Val Ala Asp Val
Asn Pro Asn Ser 405 410 415Gly Gly Ser Ile Tyr Asn Gln Arg Phe Lys
Gly Arg Phe Thr Leu Ser 420 425 430Val Asp Arg Ser Lys Asn Thr Leu
Tyr Leu Gln Met Asn Ser Leu Arg 435 440 445Ala Glu Asp Thr Ala Val
Tyr Tyr Cys Ala Arg Asn Leu Gly Pro Ser 450 455 460Phe Tyr Phe Asp
Tyr Trp Gly Gln Gly Thr Leu Val Thr Val Ser Ser465 470 475
480132450PRTArtificial sequenceHeavy chain [Trastuzumab, 1036]
132Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1
5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Asn Ile Lys Asp
Thr 20 25 30Tyr Ile His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu
Trp Val 35 40 45Ala Arg Ile Tyr Pro Thr Asn Gly Tyr Thr Arg Tyr Ala
Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Ala Asp Thr Ser Lys
Asn Thr Ala Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ser Arg Trp Gly Gly Asp Gly Phe Tyr
Ala Met Asp Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser
Ser Ala Ser Thr Lys Gly Pro Ser Val 115 120 125Phe Pro Leu Ala Pro
Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala 130 135 140Leu Gly Cys
Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser145 150 155
160Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val
165 170 175Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr
Val Pro 180 185 190Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn
Val Asn His Lys 195 200 205Pro Ser Asn Thr Lys Val Asp Lys Lys Val
Glu Pro Lys Ser Cys Asp 210 215 220Lys Thr His Thr Cys Pro Pro Cys
Pro Ala Pro Glu Leu Leu Gly Gly225 230 235 240Pro Ser Val Phe Leu
Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile 245 250 255Ser Arg Thr
Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu 260 265 270Asp
Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His 275 280
285Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg
290 295 300Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn
Gly Lys305 310 315 320Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu
Pro Ala Pro Ile Glu 325 330 335Lys Thr Ile Ser Lys Ala Lys Gly Gln
Pro Arg Glu Pro Gln Val Tyr 340 345 350Thr Leu Pro Pro Ser Arg Asp
Glu Leu Thr Lys Asn Gln Val Ser Leu 355 360 365Thr Cys Leu Val Lys
Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp 370 375 380Glu Ser Asn
Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val385 390 395
400Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp
405 410 415Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val
Met His 420 425 430Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu
Ser Leu Ser Pro 435 440 445Gly Lys 450133696PRTArtificial
sequencescFab Trastuzumab heavy chain 1 133Asp Ile Gln Met Thr Gln
Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile
Thr Cys Arg Ala Ser Gln Asp Val Asn Val Ala 20 25 30Val Ala Trp Tyr
Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ser Ala
Ser Phe Leu Tyr Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Arg
Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75
80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln His Tyr Thr Thr Pro Pro
85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala
Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu
Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe
Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn Ala
Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu Gln
Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr
Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala Cys
Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200
205Phe Asn Arg Gly Glu Cys Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser
210 215 220Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly
Ser Gly225 230 235 240Gly Gly Gly Ser Gly Gly Glu Val Gln Leu Val
Glu Ser Gly Gly Gly 245 250 255Leu Val Gln Pro Gly Gly Ser Leu Arg
Leu Ser Cys Ala Ala Ser Gly 260 265 270Phe Asn Ile Lys Asp Thr Tyr
Ile His Trp Val Arg Gln Ala Pro Gly 275 280 285Lys Gly Leu Glu Trp
Val Ala Arg Ile Tyr Pro Thr Asn Gly Tyr Thr 290 295 300Arg Tyr Ala
Asp Ser Val Lys Gly Arg Phe Thr Ile Ser Ala Asp Thr305 310 315
320Ser Lys Asn Thr Ala Tyr Leu Gln Met Asn Ser Leu Arg Ala Glu Asp
325 330 335Thr Ala Val Tyr Tyr Cys Ser Arg Trp Gly Gly Glu Gly Phe
Tyr Ala 340 345 350Met Asp Tyr Trp Gly Gln Gly Thr Leu Val Thr Val
Ser Ser Ala Ser 355 360 365Thr Lys Gly Pro Ser Val Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr 370 375 380Ser Gly Gly Thr Ala Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro385 390 395 400Glu Pro Val Thr Val
Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val 405 410 415His Thr Phe
Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser 420 425 430Ser
Val Val Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile 435 440
445Cys Asn Val Asn His Lys Pro Ser Asn Thr Lys Val Asp Lys Lys Val
450 455 460Glu Pro Lys Ser Cys Asp Lys Thr His Thr Cys Pro Pro Cys
Pro Ala465 470 475 480Pro Glu Leu Leu Gly Gly Pro Ser Val Phe Leu
Phe Pro Pro Lys Pro 485 490 495Lys Asp Thr Leu Met Ile Ser Arg Thr
Pro Glu Val Thr Cys Val Val 500 505 510Val Asp Val Ser His Glu Asp
Pro Glu Val Lys Phe Asn Trp Tyr Val 515 520 525Asp Gly Val Glu Val
His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln 530 535 540Tyr Asn Ser
Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln545 550 555
560Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala
565 570 575Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly
Gln Pro 580 585 590Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Cys Arg
Asp Glu Leu Thr 595 600 605Lys Asn Gln Val Ser Leu Trp Cys Leu Val
Lys Gly Phe Tyr Pro Ser 610 615 620Asp Ile Ala Val Glu Trp Glu Ser
Asn Gly Gln Pro Glu Asn Asn Tyr625 630 635 640Lys Thr Thr Pro Pro
Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr 645 650 655Ser Lys Leu
Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe 660 665 670Ser
Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys 675 680
685Ser Leu Ser Leu Ser Pro Gly Lys 690 695134449PRTArtificial
sequencePertuzumab heavy chain 2 134Glu Val Gln Leu Val Glu Ser Gly
Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala
Ala Ser Gly Phe Thr Phe Thr Asp Tyr 20 25 30Thr Met Asp Trp Val Arg
Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Asp Val Asn Pro
Asn Ser Gly Gly Ser Ile Tyr Asn Gln Arg Phe 50 55 60Lys Gly Arg Phe
Thr Leu Ser Val Asp Arg Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln
Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala
Arg Asn Leu Gly Pro Ser Phe Tyr Phe Asp Tyr Trp Gly Gln Gly 100 105
110Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser Val Phe
115 120 125Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala
Ala Leu 130 135 140Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val
Thr Val Ser Trp145 150 155 160Asn Ser Gly Ala Leu Thr Ser Gly Val
His Thr Phe Pro Ala Val Leu 165 170 175Gln Ser Ser Gly Leu Tyr Ser
Leu Ser Ser Val Val Thr Val Pro Ser 180 185 190Ser Ser Leu Gly Thr
Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro 195 200 205Ser Asn Thr
Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp Lys 210 215 220Thr
His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro225 230
235 240Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile
Ser 245 250 255Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser
His Glu Asp 260 265 270Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly
Val Glu Val His Asn 275 280 285Ala Lys Thr Lys Pro Arg Glu Glu Gln
Tyr Asn Ser Thr Tyr Arg Val 290 295 300Val Ser Val Leu Thr Val Leu
His Gln Asp Trp Leu Asn Gly Lys Glu305 310 315 320Tyr Lys Cys Lys
Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys 325 330 335Thr Ile
Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Cys Thr 340 345
350Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Ser
355 360 365Cys Ala Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu
Trp Glu 370 375 380Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr
Pro Pro Val Leu385 390 395 400Asp Ser Asp Gly Ser Phe Phe Leu Val
Ser Lys Leu Thr Val Asp Lys 405 410 415Ser Arg Trp Gln Gln Gly Asn
Val Phe Ser Cys Ser Val Met His Glu 420 425 430Ala Leu His Asn His
Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly 435 440
445Lys135214PRTArtificial sequencePertuzumab light chain 1 135Asp
Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10
15Asp Arg Val Thr Ile Thr Cys Lys Ala Ser Gln Asp Val Ser Ile Gly
20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu
Ile 35 40 45Tyr Ser Ala Ser Tyr Arg Tyr Thr Gly Val Pro Ser Arg Phe
Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser
Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Tyr
Tyr Ile Tyr Pro Tyr 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile
Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro
Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys
Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp
Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu
Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170
175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr
180 185 190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr
Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys 210136449PRTArtificial
sequencePertuzumab heavy chain 1 136Glu Val Gln Leu Val Glu Ser Gly
Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala
Ala Ser Gly Phe Thr Phe Thr Asp Tyr 20 25 30Thr Met Asp Trp Val Arg
Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Asp Val Asn Pro
Asn Ser Gly Gly Ser Ile Tyr Asn Gln Arg Phe 50 55 60Lys Gly Arg Phe
Thr Leu Ser Val Asp Arg Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln
Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala
Arg Asn Leu Gly Pro Ser Phe Tyr Phe Asp Tyr Trp Gly Gln Gly 100 105
110Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser Val Phe
115 120 125Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala
Ala Leu 130 135 140Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val
Thr Val Ser Trp145 150 155 160Asn Ser Gly Ala Leu Thr Ser Gly Val
His Thr Phe Pro Ala Val Leu 165 170 175Gln Ser Ser Gly Leu Tyr Ser
Leu Ser Ser Val Val Thr Val Pro Ser 180 185 190Ser Ser Leu Gly Thr
Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro 195 200 205Ser Asn Thr
Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp Lys 210 215 220Thr
His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro225 230
235 240Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile
Ser 245 250 255Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser
His Glu Asp 260 265 270Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly
Val Glu Val His Asn 275 280 285Ala Lys Thr Lys Pro Arg Glu Glu Gln
Tyr Asn Ser Thr Tyr Arg Val 290 295 300Val Ser Val Leu Thr Val Leu
His Gln Asp Trp Leu Asn Gly Lys Glu305 310 315 320Tyr Lys Cys Lys
Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys 325 330 335Thr Ile
Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Cys Thr 340 345
350Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Ser
355 360 365Cys Ala Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu
Trp Glu 370 375 380Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr
Pro Pro Val Leu385 390 395 400Asp Ser Asp Gly Ser Phe Phe Leu Val
Ser
Lys Leu Thr Val Asp Lys 405 410 415Ser Arg Trp Gln Gln Gly Asn Val
Phe Ser Cys Ser Val Met His Glu 420 425 430Ala Leu His Asn His Tyr
Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly 435 440
445Lys137214PRTArtificial sequencePertuzumab light chain 1 137Asp
Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10
15Asp Arg Val Thr Ile Thr Cys Lys Ala Ser Gln Asp Val Ser Ile Gly
20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu
Ile 35 40 45Tyr Ser Ala Ser Tyr Arg Tyr Thr Gly Val Pro Ser Arg Phe
Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser
Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Tyr
Tyr Ile Tyr Pro Tyr 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile
Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro
Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys
Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp
Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu
Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170
175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr
180 185 190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr
Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys 210138696PRTArtificial
sequencescFab Trastuzumab heavy chain 2 138Glu Val Gln Leu Val Glu
Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser
Cys Ala Ala Ser Gly Phe Asn Ile Lys Asp Thr 20 25 30Tyr Ile His Trp
Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Arg Ile
Tyr Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Asp Ser Val 50 55 60Lys Gly
Arg Phe Thr Ile Ser Ala Asp Thr Ser Lys Asn Thr Ala Tyr65 70 75
80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ser Arg Trp Gly Gly Glu Gly Phe Tyr Ala Met Asp Tyr Trp Gly
Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly
Pro Ser Val 115 120 125Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser
Gly Gly Thr Ala Ala 130 135 140Leu Gly Cys Leu Val Lys Asp Tyr Phe
Pro Glu Pro Val Thr Val Ser145 150 155 160Trp Asn Ser Gly Ala Leu
Thr Ser Gly Val His Thr Phe Pro Ala Val 165 170 175Leu Gln Ser Ser
Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro 180 185 190Ser Ser
Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys 195 200
205Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp
210 215 220Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu
Gly Gly225 230 235 240Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys
Asp Thr Leu Met Ile 245 250 255Ser Arg Thr Pro Glu Val Thr Cys Val
Val Val Asp Val Ser His Glu 260 265 270Asp Pro Glu Val Lys Phe Asn
Trp Tyr Val Asp Gly Val Glu Val His 275 280 285Asn Ala Lys Thr Lys
Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg 290 295 300Val Val Ser
Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys305 310 315
320Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu
325 330 335Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln
Val Tyr 340 345 350Thr Leu Pro Pro Cys Arg Asp Glu Leu Thr Lys Asn
Gln Val Ser Leu 355 360 365Trp Cys Leu Val Lys Gly Phe Tyr Pro Ser
Asp Ile Ala Val Glu Trp 370 375 380Glu Ser Asn Gly Gln Pro Glu Asn
Asn Tyr Lys Thr Thr Pro Pro Val385 390 395 400Leu Asp Ser Asp Gly
Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp 405 410 415Lys Ser Arg
Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His 420 425 430Glu
Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro 435 440
445Gly Lys Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly
450 455 460Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly
Gly Ser465 470 475 480Gly Gly Asp Ile Gln Met Thr Gln Ser Pro Ser
Ser Leu Ser Ala Ser 485 490 495Val Gly Asp Arg Val Thr Ile Thr Cys
Arg Ala Ser Gln Asp Val Asn 500 505 510Val Ala Val Ala Trp Tyr Gln
Gln Lys Pro Gly Lys Ala Pro Lys Leu 515 520 525Leu Ile Tyr Ser Ala
Ser Phe Leu Tyr Ser Gly Val Pro Ser Arg Phe 530 535 540Ser Gly Ser
Arg Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu545 550 555
560Gln Pro Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln His Tyr Thr Thr
565 570 575Pro Pro Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg
Thr Val 580 585 590Ala Ala Pro Ser Val Phe Ile Phe Pro Pro Ser Asp
Glu Gln Leu Lys 595 600 605Ser Gly Thr Ala Ser Val Val Cys Leu Leu
Asn Asn Phe Tyr Pro Arg 610 615 620Glu Ala Lys Val Gln Trp Lys Val
Asp Asn Ala Leu Gln Ser Gly Asn625 630 635 640Ser Gln Glu Ser Val
Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser 645 650 655Leu Ser Ser
Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys 660 665 670Val
Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr 675 680
685Lys Ser Phe Asn Arg Gly Glu Cys 690 695139695PRTArtificial
sequencescFab Pertuzumab heavy chain 1 139Glu Val Gln Leu Val Glu
Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser
Cys Ala Ala Ser Gly Phe Thr Phe Thr Asp Tyr 20 25 30Thr Met Asp Trp
Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Asp Val
Asn Pro Asn Ser Gly Gly Ser Ile Tyr Asn Gln Arg Phe 50 55 60Lys Gly
Arg Phe Thr Leu Ser Val Asp Arg Ser Lys Asn Thr Leu Tyr65 70 75
80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Arg Asn Leu Gly Pro Ser Phe Tyr Phe Asp Tyr Trp Gly Gln
Gly 100 105 110Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro
Ser Val Phe 115 120 125Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly
Gly Thr Ala Ala Leu 130 135 140Gly Cys Leu Val Lys Asp Tyr Phe Pro
Glu Pro Val Thr Val Ser Trp145 150 155 160Asn Ser Gly Ala Leu Thr
Ser Gly Val His Thr Phe Pro Ala Val Leu 165 170 175Gln Ser Ser Gly
Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser 180 185 190Ser Ser
Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro 195 200
205Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp Lys
210 215 220Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly
Gly Pro225 230 235 240Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp
Thr Leu Met Ile Ser 245 250 255Arg Thr Pro Glu Val Thr Cys Val Val
Val Asp Val Ser His Glu Asp 260 265 270Pro Glu Val Lys Phe Asn Trp
Tyr Val Asp Gly Val Glu Val His Asn 275 280 285Ala Lys Thr Lys Pro
Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val 290 295 300Val Ser Val
Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu305 310 315
320Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys
325 330 335Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val
Cys Thr 340 345 350Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln
Val Ser Leu Ser 355 360 365Cys Ala Val Lys Gly Phe Tyr Pro Ser Asp
Ile Ala Val Glu Trp Glu 370 375 380Ser Asn Gly Gln Pro Glu Asn Asn
Tyr Lys Thr Thr Pro Pro Val Leu385 390 395 400Asp Ser Asp Gly Ser
Phe Phe Leu Val Ser Lys Leu Thr Val Asp Lys 405 410 415Ser Arg Trp
Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu 420 425 430Ala
Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly 435 440
445Lys Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser
450 455 460Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly
Ser Gly465 470 475 480Gly Asp Ile Gln Met Thr Gln Ser Pro Ser Ser
Leu Ser Ala Ser Val 485 490 495Gly Asp Arg Val Thr Ile Thr Cys Lys
Ala Ser Gln Asp Val Ser Ile 500 505 510Gly Val Ala Trp Tyr Gln Gln
Lys Pro Gly Lys Ala Pro Lys Leu Leu 515 520 525Ile Tyr Ser Ala Ser
Tyr Arg Tyr Thr Gly Val Pro Ser Arg Phe Ser 530 535 540Gly Ser Gly
Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln545 550 555
560Pro Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Tyr Tyr Ile Tyr Pro
565 570 575Tyr Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg Thr
Val Ala 580 585 590Ala Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu
Gln Leu Lys Ser 595 600 605Gly Thr Ala Ser Val Val Cys Leu Leu Asn
Asn Phe Tyr Pro Arg Glu 610 615 620Ala Lys Val Gln Trp Lys Val Asp
Asn Ala Leu Gln Ser Gly Asn Ser625 630 635 640Gln Glu Ser Val Thr
Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu 645 650 655Ser Ser Thr
Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val 660 665 670Tyr
Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys 675 680
685Ser Phe Asn Arg Gly Glu Cys 690 695140450PRTArtificial
sequenceTrastuzumab heavy chain 2 140Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys
Ala Ala Ser Gly Phe Asn Ile Lys Asp Thr 20 25 30Tyr Ile His Trp Val
Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Arg Ile Tyr
Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg
Phe Thr Ile Ser Ala Asp Thr Ser Lys Asn Thr Ala Tyr65 70 75 80Leu
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90
95Ser Arg Trp Gly Gly Glu Gly Phe Tyr Ala Met Asp Tyr Trp Gly Gln
100 105 110Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro
Ser Val 115 120 125Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly
Gly Thr Ala Ala 130 135 140Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro
Glu Pro Val Thr Val Ser145 150 155 160Trp Asn Ser Gly Ala Leu Thr
Ser Gly Val His Thr Phe Pro Ala Val 165 170 175Leu Gln Ser Ser Gly
Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro 180 185 190Ser Ser Ser
Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys 195 200 205Pro
Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp 210 215
220Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly
Gly225 230 235 240Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp
Thr Leu Met Ile 245 250 255Ser Arg Thr Pro Glu Val Thr Cys Val Val
Val Asp Val Ser His Glu 260 265 270Asp Pro Glu Val Lys Phe Asn Trp
Tyr Val Asp Gly Val Glu Val His 275 280 285Asn Ala Lys Thr Lys Pro
Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg 290 295 300Val Val Ser Val
Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys305 310 315 320Glu
Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu 325 330
335Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr
340 345 350Thr Leu Pro Pro Cys Arg Asp Glu Leu Thr Lys Asn Gln Val
Ser Leu 355 360 365Trp Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile
Ala Val Glu Trp 370 375 380Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr
Lys Thr Thr Pro Pro Val385 390 395 400Leu Asp Ser Asp Gly Ser Phe
Phe Leu Tyr Ser Lys Leu Thr Val Asp 405 410 415Lys Ser Arg Trp Gln
Gln Gly Asn Val Phe Ser Cys Ser Val Met His 420 425 430Glu Ala Leu
His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro 435 440 445Gly
Lys 450141213PRTArtificial sequenceTrastuzumab light chain 2 141Asp
Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10
15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Asp Val Asn Val Ala
20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu
Ile 35 40 45Tyr Ser Ala Ser Phe Leu Tyr Ser Gly Val Pro Ser Arg Phe
Ser Gly 50 55 60Ser Arg Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser
Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln His
Tyr Thr Thr Pro Pro 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile
Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro
Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys
Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp
Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu
Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170
175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr
180 185 190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr
Lys Ser 195 200 205Phe Asn Arg Gly Glu 210142450PRTArtificial
sequenceTrastuzumab heavy chain 1 142Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys
Ala Ala Ser Gly Phe Asn Ile Lys Asp Thr 20 25 30Tyr Ile His Trp Val
Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Arg Ile Tyr
Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg
Phe Thr Ile Ser Ala Asp Thr Ser Lys Asn Thr Ala Tyr65 70 75 80Leu
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90
95Ser Arg Trp Gly Gly Glu Gly Phe Tyr Ala Met Asp Tyr Trp Gly Gln
100 105 110Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys
Gly Pro Ser Val 115 120 125Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr
Ser Gly Gly Thr Ala Ala 130 135 140Leu Gly Cys Leu Val Lys Asp Tyr
Phe Pro Glu Pro Val Thr Val Ser145 150 155 160Trp Asn Ser Gly Ala
Leu Thr Ser Gly Val His Thr Phe Pro Ala Val 165 170 175Leu Gln Ser
Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro 180 185 190Ser
Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys 195 200
205Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp
210 215 220Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu
Gly Gly225 230 235 240Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys
Asp Thr Leu Met Ile 245 250 255Ser Arg Thr Pro Glu Val Thr Cys Val
Val Val Asp Val Ser His Glu 260 265 270Asp Pro Glu Val Lys Phe Asn
Trp Tyr Val Asp Gly Val Glu Val His 275 280 285Asn Ala Lys Thr Lys
Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg 290 295 300Val Val Ser
Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys305 310 315
320Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu
325 330 335Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln
Val Tyr 340 345 350Thr Leu Pro Pro Cys Arg Asp Glu Leu Thr Lys Asn
Gln Val Ser Leu 355 360 365Trp Cys Leu Val Lys Gly Phe Tyr Pro Ser
Asp Ile Ala Val Glu Trp 370 375 380Glu Ser Asn Gly Gln Pro Glu Asn
Asn Tyr Lys Thr Thr Pro Pro Val385 390 395 400Leu Asp Ser Asp Gly
Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp 405 410 415Lys Ser Arg
Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His 420 425 430Glu
Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro 435 440
445Gly Lys 450143214PRTArtificial sequenceTrastuzumab light chain 1
143Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1
5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Asp Val Asn Val
Ala 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu
Leu Ile 35 40 45Tyr Ser Ala Ser Phe Leu Tyr Ser Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Arg Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln
His Tyr Thr Thr Pro Pro 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu
Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro
Pro Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val
Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln
Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150 155
160Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser
165 170 175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys
Val Tyr 180 185 190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro
Val Thr Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys
210144695PRTArtificial sequencescFab Pertuzumab heavy chain 2
144Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1
5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Thr Asp
Tyr 20 25 30Thr Met Asp Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu
Trp Val 35 40 45Ala Asp Val Asn Pro Asn Ser Gly Gly Ser Ile Tyr Asn
Gln Arg Phe 50 55 60Lys Gly Arg Phe Thr Leu Ser Val Asp Arg Ser Lys
Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asn Leu Gly Pro Ser Phe Tyr
Phe Asp Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val Thr Val Ser Ser
Ala Ser Thr Lys Gly Pro Ser Val Phe 115 120 125Pro Leu Ala Pro Ser
Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala Leu 130 135 140Gly Cys Leu
Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp145 150 155
160Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val Leu
165 170 175Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val
Pro Ser 180 185 190Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val
Asn His Lys Pro 195 200 205Ser Asn Thr Lys Val Asp Lys Lys Val Glu
Pro Lys Ser Cys Asp Lys 210 215 220Thr His Thr Cys Pro Pro Cys Pro
Ala Pro Glu Leu Leu Gly Gly Pro225 230 235 240Ser Val Phe Leu Phe
Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser 245 250 255Arg Thr Pro
Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp 260 265 270Pro
Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn 275 280
285Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val
290 295 300Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly
Lys Glu305 310 315 320Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro
Ala Pro Ile Glu Lys 325 330 335Thr Ile Ser Lys Ala Lys Gly Gln Pro
Arg Glu Pro Gln Val Cys Thr 340 345 350Leu Pro Pro Ser Arg Asp Glu
Leu Thr Lys Asn Gln Val Ser Leu Ser 355 360 365Cys Ala Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu 370 375 380Ser Asn Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu385 390 395
400Asp Ser Asp Gly Ser Phe Phe Leu Val Ser Lys Leu Thr Val Asp Lys
405 410 415Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met
His Glu 420 425 430Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser
Leu Ser Pro Gly 435 440 445Lys Gly Gly Gly Gly Ser Gly Gly Gly Gly
Ser Gly Gly Gly Gly Ser 450 455 460Gly Gly Gly Gly Ser Gly Gly Gly
Gly Ser Gly Gly Gly Gly Ser Gly465 470 475 480Gly Asp Ile Gln Met
Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val 485 490 495Gly Asp Arg
Val Thr Ile Thr Cys Lys Ala Ser Gln Asp Val Ser Ile 500 505 510Gly
Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu 515 520
525Ile Tyr Ser Ala Ser Tyr Arg Tyr Thr Gly Val Pro Ser Arg Phe Ser
530 535 540Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser
Leu Gln545 550 555 560Pro Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln
Tyr Tyr Ile Tyr Pro 565 570 575Tyr Thr Phe Gly Gln Gly Thr Lys Val
Glu Ile Lys Arg Thr Val Ala 580 585 590Ala Pro Ser Val Phe Ile Phe
Pro Pro Ser Asp Glu Gln Leu Lys Ser 595 600 605Gly Thr Ala Ser Val
Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu 610 615 620Ala Lys Val
Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser625 630 635
640Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu
645 650 655Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His
Lys Val 660 665 670Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser
Pro Val Thr Lys 675 680 685Ser Phe Asn Arg Gly Glu Cys 690
695145701PRTArtificial sequenceHeavy chain with scFv Trastuzumab
stabilized with disulphide bonding 145Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys
Ala Ala Ser Gly Phe Thr Phe Thr Asp Tyr 20 25 30Thr Met Asp Trp Val
Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Asp Val Asn
Pro Asn Ser Gly Gly Ser Ile Tyr Asn Gln Arg Phe 50 55 60Lys Gly Arg
Phe Thr Leu Ser Val Asp Arg Ser Lys Asn Thr Leu Tyr65 70 75 80Leu
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90
95Ala Arg Asn Leu Gly Pro Ser Phe Tyr Phe Asp Tyr Trp Gly Gln Gly
100 105 110Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser
Val Phe 115 120 125Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly
Thr Ala Ala Leu 130 135 140Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu
Pro Val Thr Val Ser Trp145 150 155 160Asn Ser Gly Ala Leu Thr Ser
Gly Val His Thr Phe Pro Ala Val Leu 165 170 175Gln Ser Ser Gly Leu
Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser 180 185 190Ser Ser Leu
Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro 195 200 205Ser
Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp Lys 210 215
220Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly
Pro225 230 235 240Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr
Leu Met Ile Ser 245 250 255Arg Thr Pro Glu Val Thr Cys Val Val Val
Asp Val Ser His Glu Asp 260 265 270Pro Glu Val Lys Phe Asn Trp Tyr
Val Asp Gly Val Glu Val His Asn 275 280 285Ala Lys Thr Lys Pro Arg
Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val 290 295 300Val Ser Val Leu
Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu305 310 315 320Tyr
Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys 325 330
335Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr
340 345 350Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser
Leu Thr 355 360 365Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala
Val Glu Trp Glu 370 375 380Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys
Thr Thr Pro Pro Val Leu385 390 395 400Asp Ser Asp Gly Ser Phe Phe
Leu Tyr Ser Lys Leu Thr Val Asp Lys 405 410 415Ser Arg Trp Gln Gln
Gly Asn Val Phe Ser Cys Ser Val Met His Glu 420 425 430Ala Leu His
Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly 435 440 445Lys
Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Glu Val Gln Leu Val 450 455
460Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly Ser Leu Arg Leu
Ser465 470 475 480Cys Ala Ala Ser Gly Phe Asn Ile Lys Asp Thr Tyr
Ile His Trp Val 485 490 495Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Val Ala Arg Ile Tyr Pro 500 505 510Thr Asn Gly Tyr Thr Arg Tyr Ala
Asp Ser Val Lys Gly Arg Phe Thr 515 520 525Ile Ser Ala Asp Thr Ser
Lys Asn Thr Ala Tyr Leu Gln Met Asn Ser 530 535 540Leu Arg Ala Glu
Asp Thr Ala Val Tyr Tyr Cys Ser Arg Trp Gly Gly545 550 555 560Glu
Gly Phe Tyr Ala Met Asp Tyr Trp Gly Cys Gly Thr Leu Val Thr 565 570
575Val Ser Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly
580 585 590Gly Ser Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser
Ala Ser 595 600 605Val Gly Asp Arg Val Thr Ile Thr Cys Arg Ala Ser
Gln Asp Val Asn 610 615 620Val Ala Val Ala Trp Tyr Gln Gln Lys Pro
Gly Lys Cys Pro Lys Leu625 630 635 640Leu Ile Tyr Ser Ala Ser Phe
Leu Tyr Ser Gly Val Pro Ser Arg Phe 645 650 655Ser Gly Ser Arg Ser
Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu 660 665 670Gln Pro Glu
Asp Phe Ala Thr Tyr Tyr Cys Gln Gln His Tyr Thr Thr 675 680 685Pro
Pro Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys 690 695
700146214PRTArtificial sequencePertuzumab light chain 146Asp Ile
Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp
Arg Val Thr Ile Thr Cys Lys Ala Ser Gln Asp Val Ser Ile Gly 20 25
30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile
35 40 45Tyr Ser Ala Ser Tyr Arg Tyr Thr Gly Val Pro Ser Arg Phe Ser
Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu
Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Tyr Tyr
Ile Tyr Pro Tyr 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys
Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser
Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu
Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys
Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser
Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170
175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr
180 185 190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr
Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys 21014726DNAArtificial
sequenceLMB3 147caggaaacag ctatgaccat gattac 2614857DNAArtificial
sequenceAM_omni_H1_TN-bamisc_feature(26)..(26)T=60%, S/G/R/N/D=20%
(4% each), rest=20% (1.7% each)misc_feature(27)..(27)D=60%,
S/N/T/A/R/E/Q/G=30% (3.8% each), rest=10% (1.1%
each)misc_feature(28)..(28)Y=60%, F/S/H/N/D/T=30% (5.0% each),
rest=10% (0.9%)misc_feature(29)..(29)T=60%, A/G/V/S/P/D/N=30% (4.3%
each), rest=10% (1.0%) 148ccggtgcctg acgaacccaa tccatnnnna
aaggtaaaac cgcttgctgc acagctc 5714932DNAArtificial sequenceRJH108
(omni_3'H1_fo) 149atggattggg ttcgtcaggc accgggtaaa gg
3215033DNAArtificial sequenceRJH109 (omni_5'H3_re) 150attacgtgca
caataataca ctgcggtatc ctc 3315166DNAArtificial
sequenceAM_omni_H3_TN_fomisc_feature(26)..(26)N=60%,
G/D/E/Q/V/S/A/P/R/L/T/Y=40% (3.3% each)misc_feature(27)..(27)L=60%,
G/Y/S/A/D/T/R/P/V/N/W/F/I/E=40% (2.9%
each)misc_feature(28)..(28)G=60%, Y/S/A/D/T/R/P/L/V/N/W/F/I/E=40%
(2.9% each)misc_feature(29)..(29)P=60%,
G/Y/S/A/D/T/R/L/V/N/W/F/I/E=40% (2.9%
each)misc_feature(30)..(30)S=60%, G/Y/P/A/D/T/R/L/V/N/W/F/I/E=40%
(2.9% each)misc_feature(34)..(34)Y=60%, G/A/P/W/S/D/T/F/R/K/H=40%
(3.6% each) 151taccgcagtg tattattgtg cacgtnnnnn ttcntttgat
tattggggtc agggcaccct 60ggttac 6615228DNAArtificial sequenceRJH99
152ggctgagact cctcaagaga aggattag 2815332DNAArtificial
sequenceRJH110(omni_5'H2_ba) 153attaacatct gcaacccatt ccagaccttt ac
3215469DNAArtificial
sequenceAM_omni_h2_TN_fomisc_feature(28)..(28)P=60%,
G/A/S/T/D/N/F/Y=30% (3.8% each), rest=10% (1.0%
each)misc_feature(29)..(29)N=60%, S/D/G/T/R/A=30% (5.0% each),
rest=10%
(0.8%)misc_feature(30)..(30)S=60%, G=10%, rest=30% (1.8%
each)misc_feature(31)..(31)misc_feature(31)..(31)n is a, c, g, or
tmisc_feature(35)..(35)misc_feature(35)..(35)n is a, c, g, or
tmisc_feature(39)..(39)misc_feature(39)..(39)misc_feature(39)..(39)n
is a, c, g, or tmisc_feature(43)..(43)n is a, c, g, or t
154ggtctggaat gggttgcaga tgttaatnnn nggtnattna acncgtttta
aaggtcgttt 60taccctgag 6915519PRTArtificial sequenceLeader Peptide
1 155Met Asp Trp Thr Trp Arg Ile Leu Phe Leu Val Ala Ala Ala Thr
Gly1 5 10 15Ala His Ser15622PRTArtificial sequenceLeader Peptide 2
156Met Asp Met Arg Val Pro Ala Gln Leu Leu Gly Leu Leu Leu Leu Trp1
5 10 15Phe Pro Gly Ala Arg Cys 2015719PRTArtificial sequenceLeader
Peptide 3 157Met Gly Trp Ser Cys Ile Ile Leu Phe Leu Val Ala Thr
Ala Thr Gly1 5 10 15Val His Ser15811PRTArtificial
sequenceTrastuzumab VL N30S CDR1 158Arg Ala Ser Gln Asp Val Ser Thr
Ala Val Ala1 5 10159450PRTArtificial sequenceTrastuzumab VHCH1- Fc
KNOB 159Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly
Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Asn Ile Lys
Asp Thr 20 25 30Tyr Ile His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu
Glu Trp Val 35 40 45Ala Arg Ile Tyr Pro Thr Asn Gly Tyr Thr Arg Tyr
Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Ala Asp Thr Ser
Lys Asn Thr Ala Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu
Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ser Arg Trp Gly Gly Asp Gly Phe
Tyr Ala Met Asp Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser Val 115 120 125Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala 130 135 140Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser145 150 155
160Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val
165 170 175Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr
Val Pro 180 185 190Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn
Val Asn His Lys 195 200 205Pro Ser Asn Thr Lys Val Asp Lys Lys Val
Glu Pro Lys Ser Cys Asp 210 215 220Lys Thr His Thr Cys Pro Pro Cys
Pro Ala Pro Glu Leu Leu Gly Gly225 230 235 240Pro Ser Val Phe Leu
Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile 245 250 255Ser Arg Thr
Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu 260 265 270Asp
Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His 275 280
285Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg
290 295 300Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn
Gly Lys305 310 315 320Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu
Pro Ala Pro Ile Glu 325 330 335Lys Thr Ile Ser Lys Ala Lys Gly Gln
Pro Arg Glu Pro Gln Val Tyr 340 345 350Thr Leu Pro Pro Cys Arg Asp
Glu Leu Thr Lys Asn Gln Val Ser Leu 355 360 365Trp Cys Leu Val Lys
Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp 370 375 380Glu Ser Asn
Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val385 390 395
400Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp
405 410 415Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val
Met His 420 425 430Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu
Ser Leu Ser Pro 435 440 445Gly Lys 4501601350PRTArtificial
sequenceTrastuzumab VHCH1- Fc KNOB DNA 160Gly Ala Ala Gly Thr Gly
Cys Ala Ala Thr Thr Gly Gly Thr Gly Gly1 5 10 15Ala Ala Ala Gly Cys
Gly Gly Cys Gly Gly Cys Gly Gly Cys Cys Thr 20 25 30Gly Gly Thr Gly
Cys Ala Ala Cys Cys Gly Gly Gly Cys Gly Gly Cys 35 40 45Ala Gly Cys
Cys Thr Gly Cys Gly Thr Cys Thr Gly Ala Gly Cys Thr 50 55 60Gly Cys
Gly Cys Gly Gly Cys Cys Thr Cys Cys Gly Gly Ala Thr Thr65 70 75
80Thr Ala Ala Cys Ala Thr Ala Ala Ala Gly Gly Ala Cys Ala Cys Ala
85 90 95Thr Ala Cys Ala Thr Cys Cys Ala Cys Thr Gly Gly Gly Thr Gly
Cys 100 105 110Gly Cys Cys Ala Ala Gly Cys Ala Cys Cys Thr Gly Gly
Gly Ala Ala 115 120 125Gly Gly Gly Thr Cys Thr Cys Gly Ala Gly Thr
Gly Gly Gly Thr Gly 130 135 140Gly Cys Thr Cys Gly Gly Ala Thr Thr
Thr Ala Cys Cys Cys Ala Ala145 150 155 160Cys Ala Ala Ala Thr Gly
Gly Cys Thr Ala Cys Ala Cys Cys Ala Gly 165 170 175Gly Thr Ala Thr
Gly Cys Gly Gly Ala Thr Ala Gly Cys Gly Thr Gly 180 185 190Ala Ala
Ala Gly Gly Cys Cys Gly Thr Thr Thr Thr Ala Cys Cys Ala 195 200
205Thr Thr Thr Cys Ala Gly Cys Thr Gly Ala Thr Ala Cys Thr Thr Cys
210 215 220Gly Ala Ala Gly Ala Ala Cys Ala Cys Cys Gly Cys Cys Thr
Ala Thr225 230 235 240Cys Thr Gly Cys Ala Ala Ala Thr Gly Ala Ala
Cys Ala Gly Cys Cys 245 250 255Thr Gly Cys Gly Thr Gly Cys Gly Gly
Ala Ala Gly Ala Thr Ala Cys 260 265 270Gly Gly Cys Cys Gly Thr Gly
Thr Ala Thr Thr Ala Thr Thr Gly Cys 275 280 285Thr Cys Gly Cys Gly
Thr Thr Gly Gly Gly Gly Ala Gly Gly Ala Gly 290 295 300Ala Cys Gly
Gly Gly Thr Thr Cys Thr Ala Thr Gly Cys Thr Ala Thr305 310 315
320Gly Gly Ala Thr Thr Ala Cys Thr Gly Gly Gly Gly Cys Cys Ala Ala
325 330 335Gly Gly Cys Ala Cys Cys Cys Thr Gly Gly Thr Gly Ala Cys
Gly Gly 340 345 350Thr Thr Ala Gly Cys Thr Cys Ala Gly Cys Thr Ala
Gly Cys Ala Cys 355 360 365Cys Ala Ala Gly Gly Gly Cys Cys Cys Ala
Thr Cys Gly Gly Thr Cys 370 375 380Thr Thr Cys Cys Cys Cys Cys Thr
Gly Gly Cys Ala Cys Cys Cys Thr385 390 395 400Cys Cys Thr Cys Cys
Ala Ala Gly Ala Gly Cys Ala Cys Cys Thr Cys 405 410 415Thr Gly Gly
Gly Gly Gly Cys Ala Cys Ala Gly Cys Gly Gly Cys Cys 420 425 430Cys
Thr Gly Gly Gly Cys Thr Gly Cys Cys Thr Gly Gly Thr Cys Ala 435 440
445Ala Gly Gly Ala Cys Thr Ala Cys Thr Thr Cys Cys Cys Cys Gly Ala
450 455 460Ala Cys Cys Gly Gly Thr Gly Ala Cys Gly Gly Thr Gly Thr
Cys Gly465 470 475 480Thr Gly Gly Ala Ala Cys Thr Cys Ala Gly Gly
Cys Gly Cys Cys Cys 485 490 495Thr Gly Ala Cys Cys Ala Gly Cys Gly
Gly Cys Gly Thr Gly Cys Ala 500 505 510Cys Ala Cys Cys Thr Thr Cys
Cys Cys Gly Gly Cys Thr Gly Thr Cys 515 520 525Cys Thr Ala Cys Ala
Gly Thr Cys Cys Thr Cys Ala Gly Gly Ala Cys 530 535 540Thr Cys Thr
Ala Cys Thr Cys Cys Cys Thr Cys Ala Gly Cys Ala Gly545 550 555
560Cys Gly Thr Gly Gly Thr Gly Ala Cys Cys Gly Thr Gly Cys Cys Cys
565 570 575Thr Cys Cys Ala Gly Cys Ala Gly Cys Thr Thr Gly Gly Gly
Cys Ala 580 585 590Cys Cys Cys Ala Gly Ala Cys Cys Thr Ala Cys Ala
Thr Cys Thr Gly 595 600 605Cys Ala Ala Cys Gly Thr Gly Ala Ala Thr
Cys Ala Cys Ala Ala Gly 610 615 620Cys Cys Cys Ala Gly Cys Ala Ala
Cys Ala Cys Cys Ala Ala Gly Gly625 630 635 640Thr Gly Gly Ala Cys
Ala Ala Gly Ala Ala Ala Gly Thr Thr Gly Ala 645 650 655Gly Cys Cys
Cys Ala Ala Ala Thr Cys Thr Thr Gly Thr Gly Ala Cys 660 665 670Ala
Ala Ala Ala Cys Thr Cys Ala Cys Ala Cys Ala Thr Gly Cys Cys 675 680
685Cys Ala Cys Cys Gly Thr Gly Cys Cys Cys Ala Gly Cys Ala Cys Cys
690 695 700Thr Gly Ala Ala Cys Thr Cys Cys Thr Gly Gly Gly Gly Gly
Gly Ala705 710 715 720Cys Cys Gly Thr Cys Ala Gly Thr Cys Thr Thr
Cys Cys Thr Cys Thr 725 730 735Thr Cys Cys Cys Cys Cys Cys Ala Ala
Ala Ala Cys Cys Cys Ala Ala 740 745 750Gly Gly Ala Cys Ala Cys Cys
Cys Thr Cys Ala Thr Gly Ala Thr Cys 755 760 765Thr Cys Cys Cys Gly
Gly Ala Cys Cys Cys Cys Thr Gly Ala Gly Gly 770 775 780Thr Cys Ala
Cys Ala Thr Gly Cys Gly Thr Gly Gly Thr Gly Gly Thr785 790 795
800Gly Gly Ala Cys Gly Thr Gly Ala Gly Cys Cys Ala Cys Gly Ala Ala
805 810 815Gly Ala Cys Cys Cys Thr Gly Ala Gly Gly Thr Cys Ala Ala
Gly Thr 820 825 830Thr Cys Ala Ala Cys Thr Gly Gly Thr Ala Cys Gly
Thr Gly Gly Ala 835 840 845Cys Gly Gly Cys Gly Thr Gly Gly Ala Gly
Gly Thr Gly Cys Ala Thr 850 855 860Ala Ala Thr Gly Cys Cys Ala Ala
Gly Ala Cys Ala Ala Ala Gly Cys865 870 875 880Cys Gly Cys Gly Gly
Gly Ala Gly Gly Ala Gly Cys Ala Gly Thr Ala 885 890 895Cys Ala Ala
Cys Ala Gly Cys Ala Cys Gly Thr Ala Cys Cys Gly Thr 900 905 910Gly
Thr Gly Gly Thr Cys Ala Gly Cys Gly Thr Cys Cys Thr Cys Ala 915 920
925Cys Cys Gly Thr Cys Cys Thr Gly Cys Ala Cys Cys Ala Gly Gly Ala
930 935 940Cys Thr Gly Gly Cys Thr Gly Ala Ala Thr Gly Gly Cys Ala
Ala Gly945 950 955 960Gly Ala Gly Thr Ala Cys Ala Ala Gly Thr Gly
Cys Ala Ala Gly Gly 965 970 975Thr Cys Thr Cys Cys Ala Ala Cys Ala
Ala Ala Gly Cys Cys Cys Thr 980 985 990Cys Cys Cys Ala Gly Cys Cys
Cys Cys Cys Ala Thr Cys Gly Ala Gly 995 1000 1005Ala Ala Ala Ala
Cys Cys Ala Thr Cys Thr Cys Cys Ala Ala Ala 1010 1015 1020Gly Cys
Cys Ala Ala Ala Gly Gly Gly Cys Ala Gly Cys Cys Cys 1025 1030
1035Cys Gly Ala Gly Ala Ala Cys Cys Ala Cys Ala Gly Gly Thr Gly
1040 1045 1050Thr Ala Cys Ala Cys Cys Cys Thr Gly Cys Cys Cys Cys
Cys Ala 1055 1060 1065Thr Gly Cys Cys Gly Gly Gly Ala Thr Gly Ala
Gly Cys Thr Gly 1070 1075 1080Ala Cys Cys Ala Ala Gly Ala Ala Cys
Cys Ala Gly Gly Thr Cys 1085 1090 1095Ala Gly Cys Cys Thr Gly Thr
Gly Gly Thr Gly Cys Cys Thr Gly 1100 1105 1110Gly Thr Cys Ala Ala
Ala Gly Gly Cys Thr Thr Cys Thr Ala Thr 1115 1120 1125Cys Cys Cys
Ala Gly Cys Gly Ala Cys Ala Thr Cys Gly Cys Cys 1130 1135 1140Gly
Thr Gly Gly Ala Gly Thr Gly Gly Gly Ala Gly Ala Gly Cys 1145 1150
1155Ala Ala Thr Gly Gly Gly Cys Ala Gly Cys Cys Gly Gly Ala Gly
1160 1165 1170Ala Ala Cys Ala Ala Cys Thr Ala Cys Ala Ala Gly Ala
Cys Cys 1175 1180 1185Ala Cys Gly Cys Cys Thr Cys Cys Cys Gly Thr
Gly Cys Thr Gly 1190 1195 1200Gly Ala Cys Thr Cys Cys Gly Ala Cys
Gly Gly Cys Thr Cys Cys 1205 1210 1215Thr Thr Cys Thr Thr Cys Cys
Thr Cys Thr Ala Cys Ala Gly Cys 1220 1225 1230Ala Ala Gly Cys Thr
Cys Ala Cys Cys Gly Thr Gly Gly Ala Cys 1235 1240 1245Ala Ala Gly
Ala Gly Cys Ala Gly Gly Thr Gly Gly Cys Ala Gly 1250 1255 1260Cys
Ala Gly Gly Gly Gly Ala Ala Cys Gly Thr Cys Thr Thr Cys 1265 1270
1275Thr Cys Ala Thr Gly Cys Thr Cys Cys Gly Thr Gly Ala Thr Gly
1280 1285 1290Cys Ala Thr Gly Ala Gly Gly Cys Thr Cys Thr Gly Cys
Ala Cys 1295 1300 1305Ala Ala Cys Cys Ala Cys Thr Ala Cys Ala Cys
Gly Cys Ala Gly 1310 1315 1320Ala Ala Gly Ala Gly Cys Cys Thr Cys
Thr Cys Cys Cys Thr Gly 1325 1330 1335Thr Cys Thr Cys Cys Gly Gly
Gly Thr Ala Ala Ala 1340 1345 1350161214PRTArtificial
sequenceCommon light chain VLCL 161Asp Ile Gln Met Thr Gln Ser Pro
Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys
Lys Ala Ser Gln Asp Val Ser Thr Ala 20 25 30Val Ala Trp Tyr Gln Gln
Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ser Ala Ser Phe
Arg Tyr Thr Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Arg Ser Gly
Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp
Phe Ala Thr Tyr Tyr Cys Gln Gln His Tyr Thr Thr Pro Pro 85 90 95Thr
Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala Ala 100 105
110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly
115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg
Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser
Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu Gln Asp Ser Lys
Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys
Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala Cys Glu Val Thr
His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200 205Phe Asn Arg
Gly Glu Cys 210162642PRTArtificial sequenceCommon light chain VLCL
- DNA 162Gly Ala Cys Ala Thr Cys Cys Ala Gly Ala Thr Gly Ala Cys
Cys Cys1 5 10 15Ala Gly Ala Gly Cys Cys Cys Cys Ala Gly Cys Ala Gly
Cys Cys Thr 20 25 30Gly Thr Cys Thr Gly Cys Cys Ala Gly Cys Gly Thr
Gly Gly Gly Cys 35 40 45Gly Ala Cys Ala Gly Ala Gly Thr Gly Ala Cys
Cys Ala Thr Cys Ala 50 55 60Cys Ala Thr Gly Cys Ala Ala Gly Gly Cys
Cys Ala Gly Cys Cys Ala65 70 75 80Gly Gly Ala Cys Gly Thr Gly Thr
Cys Cys Ala Cys Ala Gly Cys Cys 85 90 95Gly Thr Gly Gly Cys Cys Thr
Gly Gly Thr Ala Thr Cys Ala Gly Cys 100 105 110Ala Gly Ala Ala Gly
Cys Cys Thr Gly Gly Cys Ala Ala Gly Gly Cys 115 120 125Cys Cys Cys
Cys Ala Ala Gly Cys Thr Gly Cys Thr Gly Ala Thr Cys 130 135 140Thr
Ala Cys Ala Gly Cys Gly Cys Cys Ala Gly Cys Thr Thr Cys Cys145 150
155 160Gly Gly Thr Ala Cys Ala Cys Cys Gly Gly Cys Gly Thr Gly Cys
Cys 165 170 175Cys Ala Gly Cys Ala Gly Ala Thr Thr Cys Ala Gly Cys
Gly Gly Cys 180 185 190Ala Gly Cys Ala Gly Ala Thr Cys Cys Gly Gly
Cys Ala Cys Cys Gly 195 200 205Ala Cys Thr Thr Cys Ala Cys Cys Cys
Thr Gly Ala Cys Cys Ala Thr 210 215 220Cys Ala Gly Cys Thr Cys Cys
Cys Thr Gly Cys Ala Gly Cys Cys Cys225 230 235 240Gly Ala Gly Gly
Ala Cys Thr Thr Cys Gly Cys Cys Ala Cys Cys Thr 245 250 255Ala Cys
Thr Ala
Cys Thr Gly Cys Cys Ala Gly Cys Ala Gly Cys Ala 260 265 270Cys Thr
Ala Cys Ala Cys Cys Ala Cys Cys Cys Cys Cys Cys Cys Cys 275 280
285Ala Cys Ala Thr Thr Thr Gly Gly Cys Cys Ala Gly Gly Gly Cys Ala
290 295 300Cys Cys Ala Ala Gly Gly Thr Gly Gly Ala Ala Ala Thr Cys
Ala Ala305 310 315 320Gly Cys Gly Thr Ala Cys Gly Gly Thr Gly Gly
Cys Thr Gly Cys Ala 325 330 335Cys Cys Ala Thr Cys Thr Gly Thr Cys
Thr Thr Cys Ala Thr Cys Thr 340 345 350Thr Cys Cys Cys Gly Cys Cys
Ala Thr Cys Thr Gly Ala Thr Gly Ala 355 360 365Gly Cys Ala Gly Thr
Thr Gly Ala Ala Ala Thr Cys Thr Gly Gly Ala 370 375 380Ala Cys Thr
Gly Cys Cys Thr Cys Thr Gly Thr Thr Gly Thr Gly Thr385 390 395
400Gly Cys Cys Thr Gly Cys Thr Gly Ala Ala Thr Ala Ala Cys Thr Thr
405 410 415Cys Thr Ala Thr Cys Cys Cys Ala Gly Ala Gly Ala Gly Gly
Cys Cys 420 425 430Ala Ala Ala Gly Thr Ala Cys Ala Gly Thr Gly Gly
Ala Ala Gly Gly 435 440 445Thr Gly Gly Ala Thr Ala Ala Cys Gly Cys
Cys Cys Thr Cys Cys Ala 450 455 460Ala Thr Cys Gly Gly Gly Thr Ala
Ala Cys Thr Cys Cys Cys Ala Gly465 470 475 480Gly Ala Gly Ala Gly
Thr Gly Thr Cys Ala Cys Ala Gly Ala Gly Cys 485 490 495Ala Gly Gly
Ala Cys Ala Gly Cys Ala Ala Gly Gly Ala Cys Ala Gly 500 505 510Cys
Ala Cys Cys Thr Ala Cys Ala Gly Cys Cys Thr Cys Ala Gly Cys 515 520
525Ala Gly Cys Ala Cys Cys Cys Thr Gly Ala Cys Gly Cys Thr Gly Ala
530 535 540Gly Cys Ala Ala Ala Gly Cys Ala Gly Ala Cys Thr Ala Cys
Gly Ala545 550 555 560Gly Ala Ala Ala Cys Ala Cys Ala Ala Ala Gly
Thr Cys Thr Ala Cys 565 570 575Gly Cys Cys Thr Gly Cys Gly Ala Ala
Gly Thr Cys Ala Cys Cys Cys 580 585 590Ala Thr Cys Ala Gly Gly Gly
Cys Cys Thr Gly Ala Gly Cys Thr Cys 595 600 605Gly Cys Cys Cys Gly
Thr Cys Ala Cys Ala Ala Ala Gly Ala Gly Cys 610 615 620Thr Thr Cys
Ala Ala Cys Ala Gly Gly Gly Gly Ala Gly Ala Gly Thr625 630 635
640Gly Thr163449PRTArtificial sequencePertuzumab VHCH1 Fc hole
163Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1
5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Asn Asp
Tyr 20 25 30Thr Met Asp Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu
Trp Val 35 40 45Ala Asp Val Asn Pro Asn Ser Gly Gly Ser Ile Val Asn
Arg Arg Phe 50 55 60Lys Gly Arg Phe Thr Leu Ser Val Asp Arg Ser Lys
Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asn Leu Gly Pro Phe Phe Tyr
Phe Asp Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val Thr Val Ser Ser
Ala Ser Thr Lys Gly Pro Ser Val Phe 115 120 125Pro Leu Ala Pro Ser
Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala Leu 130 135 140Gly Cys Leu
Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp145 150 155
160Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val Leu
165 170 175Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val
Pro Ser 180 185 190Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val
Asn His Lys Pro 195 200 205Ser Asn Thr Lys Val Asp Lys Lys Val Glu
Pro Lys Ser Cys Asp Lys 210 215 220Thr His Thr Cys Pro Pro Cys Pro
Ala Pro Glu Leu Leu Gly Gly Pro225 230 235 240Ser Val Phe Leu Phe
Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser 245 250 255Arg Thr Pro
Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp 260 265 270Pro
Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn 275 280
285Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val
290 295 300Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly
Lys Glu305 310 315 320Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro
Ala Pro Ile Glu Lys 325 330 335Thr Ile Ser Lys Ala Lys Gly Gln Pro
Arg Glu Pro Gln Val Cys Thr 340 345 350Leu Pro Pro Ser Arg Asp Glu
Leu Thr Lys Asn Gln Val Ser Leu Ser 355 360 365Cys Ala Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu 370 375 380Ser Asn Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu385 390 395
400Asp Ser Asp Gly Ser Phe Phe Leu Val Ser Lys Leu Thr Val Asp Lys
405 410 415Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met
His Glu 420 425 430Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser
Leu Ser Pro Gly 435 440 445Lys1641347DNAArtificial
sequencePertuzumab VHCH1 Fc hole DNA 164gaagttcagc tggttgaaag
cggtggtggt ctggttcagc ctggtggtag cctgcgtctg 60agctgtgcag caagcggttt
tacctttaac gattatacca tggattgggt tcgtcaggca 120ccgggtaaag
gtctggaatg ggttgcagat gttaatccga atagcggtgg tagcattgtt
180aaccgtcgtt ttaaaggtcg ttttaccctg agcgttgatc gtagcaaaaa
taccctgtat 240ctgcaaatga atagtctgcg tgcagaggat accgcagtgt
attattgtgc acgtaacctg 300ggtccgttct tctactttga ttattggggt
cagggcaccc tggttaccgt tagcagcgct 360agcaccaagg gcccaagcgt
gttccctctg gcccccagca gcaagagcac aagcggcgga 420acagccgccc
tgggctgcct ggtcaaggac tacttccccg agcccgtgac agtgtcctgg
480aacagcggag ccctgaccag cggcgtgcac acctttccag ccgtgctgca
gagcagcggc 540ctgtacagcc tgagcagcgt ggtcacagtg cctagcagca
gcctgggcac ccagacctac 600atctgcaacg tgaaccacaa gcccagcaac
accaaggtgg acaagaaggt ggagcccaag 660agctgcgaca agacccacac
ctgtccccct tgtcctgccc ctgagctgct gggcggaccc 720agcgtgttcc
tgttcccccc aaagcccaag gacaccctga tgatcagccg gacccccgaa
780gtgacctgcg tggtggtgga cgtgtcccac gaggaccctg aagtgaagtt
caattggtac 840gtggacggcg tggaggtgca caatgccaag accaagcccc
gggaggaaca gtacaacagc 900acctaccggg tggtgtccgt gctgaccgtg
ctgcaccagg actggctgaa cggcaaagag 960tacaagtgca aggtctccaa
caaggccctg cctgccccca tcgagaaaac catcagcaag 1020gccaagggcc
agcccagaga accccaggtg tgcaccctgc cccccagcag agatgagctg
1080accaagaacc aggtgtccct gagctgtgcc gtcaagggct tctaccccag
cgatatcgcc 1140gtggagtggg agagcaacgg ccagcctgag aacaactaca
agaccacccc ccctgtgctg 1200gacagcgacg gcagcttctt cctggtgtcc
aaactgaccg tggacaagag ccggtggcag 1260cagggcaacg tgttcagctg
cagcgtgatg cacgaggccc tgcacaacca ctacacccag 1320aagtccctga
gcctgagccc cggcaag 1347165934PRTArtificial
sequenceHer-knob-scFab(8D3) Heavy chain (21832) 165Glu Val Gln Leu
Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg
Leu Ser Cys Ala Ala Ser Gly Phe Asn Ile Lys Asp Thr 20 25 30Tyr Ile
His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala
Arg Ile Tyr Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Asp Ser Val 50 55
60Lys Gly Arg Phe Thr Ile Ser Ala Asp Thr Ser Lys Asn Thr Ala Tyr65
70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr
Cys 85 90 95Ser Arg Trp Gly Gly Glu Gly Phe Tyr Ala Met Asp Tyr Trp
Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys
Gly Pro Ser Val 115 120 125Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr
Ser Gly Gly Thr Ala Ala 130 135 140Leu Gly Cys Leu Val Lys Asp Tyr
Phe Pro Glu Pro Val Thr Val Ser145 150 155 160Trp Asn Ser Gly Ala
Leu Thr Ser Gly Val His Thr Phe Pro Ala Val 165 170 175Leu Gln Ser
Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro 180 185 190Ser
Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys 195 200
205Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp
210 215 220Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu
Gly Gly225 230 235 240Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys
Asp Thr Leu Met Ile 245 250 255Ser Arg Thr Pro Glu Val Thr Cys Val
Val Val Asp Val Ser His Glu 260 265 270Asp Pro Glu Val Lys Phe Asn
Trp Tyr Val Asp Gly Val Glu Val His 275 280 285Asn Ala Lys Thr Lys
Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg 290 295 300Val Val Ser
Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys305 310 315
320Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu
325 330 335Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln
Val Tyr 340 345 350Thr Leu Pro Pro Cys Arg Asp Glu Leu Thr Lys Asn
Gln Val Ser Leu 355 360 365Trp Cys Leu Val Lys Gly Phe Tyr Pro Ser
Asp Ile Ala Val Glu Trp 370 375 380Glu Ser Asn Gly Gln Pro Glu Asn
Asn Tyr Lys Thr Thr Pro Pro Val385 390 395 400Leu Asp Ser Asp Gly
Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp 405 410 415Lys Ser Arg
Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His 420 425 430Glu
Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro 435 440
445Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly
450 455 460Gly Gly Ser Asp Ile Gln Met Thr Gln Ser Pro Ala Ser Leu
Ser Ala465 470 475 480Ser Leu Glu Glu Ile Val Thr Ile Thr Cys Gln
Ala Ser Gln Asp Ile 485 490 495Gly Asn Trp Leu Ala Trp Tyr Gln Gln
Lys Pro Gly Lys Ser Pro Gln 500 505 510Leu Leu Ile Tyr Gly Ala Thr
Ser Leu Ala Asp Gly Val Pro Ser Arg 515 520 525Phe Ser Gly Ser Arg
Ser Gly Thr Gln Phe Ser Leu Lys Ile Ser Arg 530 535 540Val Gln Val
Glu Asp Ile Gly Ile Tyr Tyr Cys Leu Gln Ala Tyr Asn545 550 555
560Thr Pro Trp Thr Phe Gly Gly Gly Thr Lys Val Glu Ile Lys Arg Thr
565 570 575Val Ala Ala Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu
Gln Leu 580 585 590Lys Ser Gly Thr Ala Ser Val Val Cys Leu Leu Asn
Asn Phe Tyr Pro 595 600 605Arg Glu Ala Lys Val Gln Trp Lys Val Asp
Asn Ala Leu Gln Ser Gly 610 615 620Asn Ser Gln Glu Ser Val Thr Glu
Gln Asp Ser Lys Asp Ser Thr Tyr625 630 635 640Ser Leu Ser Ser Thr
Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His 645 650 655Lys Val Tyr
Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val 660 665 670Thr
Lys Ser Phe Asn Arg Gly Glu Cys Gly Gly Gly Gly Ser Gly Gly 675 680
685Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly
690 695 700Gly Ser Gly Gly Gly Gly Ser Gly Gly Glu Val Gln Leu Val
Glu Ser705 710 715 720Gly Gly Gly Leu Val Gln Pro Gly Asn Ser Leu
Thr Leu Ser Cys Val 725 730 735Ala Ser Gly Phe Thr Phe Ser Asn Tyr
Gly Met His Trp Ile Arg Gln 740 745 750Ala Pro Lys Lys Gly Leu Glu
Trp Ile Ala Met Ile Tyr Tyr Asp Ser 755 760 765Ser Lys Met Asn Tyr
Ala Asp Thr Val Lys Gly Arg Phe Thr Ile Ser 770 775 780Arg Asp Asn
Ser Lys Asn Thr Leu Tyr Leu Glu Met Asn Ser Leu Arg785 790 795
800Ser Glu Asp Thr Ala Met Tyr Tyr Cys Ala Val Pro Thr Ser His Tyr
805 810 815Val Val Asp Val Trp Gly Gln Gly Val Ser Val Thr Val Ser
Ser Ala 820 825 830Ser Thr Lys Gly Pro Ser Val Phe Pro Leu Ala Pro
Ser Ser Lys Ser 835 840 845Thr Ser Gly Gly Thr Ala Ala Leu Gly Cys
Leu Val Lys Asp Tyr Phe 850 855 860Pro Glu Pro Val Thr Val Ser Trp
Asn Ser Gly Ala Leu Thr Ser Gly865 870 875 880Val His Thr Phe Pro
Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu 885 890 895Ser Ser Val
Val Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr 900 905 910Ile
Cys Asn Val Asn His Lys Pro Ser Asn Thr Lys Val Asp Lys Lys 915 920
925Val Glu Pro Lys Ser Cys 930166119PRTArtificial sequence4/94 VH
166Glu Val Gln Leu Gln Gln Ser Gly Ala Val Leu Val Lys Pro Gly Ala1
5 10 15Ser Val Lys Leu Ser Cys Pro Ala Ser Gly Phe Asn Ile Lys Asp
Thr 20 25 30Tyr Ile His Trp Val Ile Gln Arg Pro Glu Gln Gly Leu Glu
Trp Ile 35 40 45Gly Arg Ile Asp Pro Ala Asn Gly Asp Thr Lys Cys Asp
Pro Lys Phe 50 55 60Gln Val Lys Ala Thr Ile Thr Ala Asp Thr Ser Ser
Asn Thr Ala Tyr65 70 75 80Leu Gln Leu Ser Ser Leu Thr Ser Glu Asp
Thr Ala Val Tyr Phe Cys 85 90 95Val Arg Asp Tyr Leu Tyr Pro Tyr Tyr
Phe Asp Phe Trp Gly Gln Gly 100 105 110Thr Thr Leu Thr Val Ser Ser
115167108PRTArtificial sequence4/94 VL 167Lys Ile Val Met Thr Gln
Ser Pro Lys Ser Met Ser Met Ser Val Gly1 5 10 15Glu Arg Val Thr Leu
Asn Cys Arg Ala Ser Glu Ser Val Asp Thr Tyr 20 25 30Val Ser Trp Tyr
Gln Gln Lys Pro Glu Gln Ser Pro Glu Leu Leu Ile 35 40 45Tyr Gly Ala
Ser Asn Arg Tyr Thr Gly Val Pro Asp Arg Phe Thr Gly 50 55 60Ser Gly
Ser Ala Thr Asp Phe Thr Leu Thr Ile Ser Ser Val Gln Ala65 70 75
80Glu Asp Leu Ala Asp Tyr Tyr Cys Gly Gln Thr Tyr Asn Tyr Pro Leu
85 90 95Thr Phe Gly Ala Gly Thr Lys Leu Glu Leu Lys Arg 100
105168934PRTArtificial sequenceHer-knob-LALA-PG-scFab(8D3) Heavy
chain (21834) 168Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val
Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe
Asn Ile Lys Asp Thr 20 25 30Tyr Ile His Trp Val Arg Gln Ala Pro Gly
Lys Gly Leu Glu Trp Val 35 40 45Ala Arg Ile Tyr Pro Thr Asn Gly Tyr
Thr Arg Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Ala
Asp Thr Ser Lys Asn Thr Ala Tyr65 70 75 80Leu Gln Met Asn Ser Leu
Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ser Arg Trp Gly Gly
Glu Gly Phe Tyr Ala Met Asp Tyr Trp Gly Gln 100 105 110Gly Thr Leu
Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser Val 115 120 125Phe
Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala 130 135
140Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val
Ser145 150 155 160Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr
Phe Pro Ala Val 165 170 175Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser
Ser Val Val Thr Val Pro 180 185 190Ser Ser Ser Leu Gly Thr Gln Thr
Tyr Ile Cys Asn Val Asn His Lys 195 200 205Pro Ser Asn Thr
Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp 210 215 220Lys Thr
His Thr Cys Pro Pro Cys Pro Ala Pro Glu Ala Ala Gly Gly225 230 235
240Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile
245 250 255Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser
His Glu 260 265 270Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly
Val Glu Val His 275 280 285Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln
Tyr Asn Ser Thr Tyr Arg 290 295 300Val Val Ser Val Leu Thr Val Leu
His Gln Asp Trp Leu Asn Gly Lys305 310 315 320Glu Tyr Lys Cys Lys
Val Ser Asn Lys Ala Leu Gly Ala Pro Ile Glu 325 330 335Lys Thr Ile
Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr 340 345 350Thr
Leu Pro Pro Cys Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu 355 360
365Trp Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp
370 375 380Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro
Pro Val385 390 395 400Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser
Lys Leu Thr Val Asp 405 410 415Lys Ser Arg Trp Gln Gln Gly Asn Val
Phe Ser Cys Ser Val Met His 420 425 430Glu Ala Leu His Asn His Tyr
Thr Gln Lys Ser Leu Ser Leu Ser Pro 435 440 445Gly Gly Gly Ser Gly
Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly 450 455 460Gly Gly Ser
Asp Ile Gln Met Thr Gln Ser Pro Ala Ser Leu Ser Ala465 470 475
480Ser Leu Glu Glu Ile Val Thr Ile Thr Cys Gln Ala Ser Gln Asp Ile
485 490 495Gly Asn Trp Leu Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ser
Pro Gln 500 505 510Leu Leu Ile Tyr Gly Ala Thr Ser Leu Ala Asp Gly
Val Pro Ser Arg 515 520 525Phe Ser Gly Ser Arg Ser Gly Thr Gln Phe
Ser Leu Lys Ile Ser Arg 530 535 540Val Gln Val Glu Asp Ile Gly Ile
Tyr Tyr Cys Leu Gln Ala Tyr Asn545 550 555 560Thr Pro Trp Thr Phe
Gly Gly Gly Thr Lys Val Glu Ile Lys Arg Thr 565 570 575Val Ala Ala
Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu 580 585 590Lys
Ser Gly Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro 595 600
605Arg Glu Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly
610 615 620Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser
Thr Tyr625 630 635 640Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala
Asp Tyr Glu Lys His 645 650 655Lys Val Tyr Ala Cys Glu Val Thr His
Gln Gly Leu Ser Ser Pro Val 660 665 670Thr Lys Ser Phe Asn Arg Gly
Glu Cys Gly Gly Gly Gly Ser Gly Gly 675 680 685Gly Gly Ser Gly Gly
Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly 690 695 700Gly Ser Gly
Gly Gly Gly Ser Gly Gly Glu Val Gln Leu Val Glu Ser705 710 715
720Gly Gly Gly Leu Val Gln Pro Gly Asn Ser Leu Thr Leu Ser Cys Val
725 730 735Ala Ser Gly Phe Thr Phe Ser Asn Tyr Gly Met His Trp Ile
Arg Gln 740 745 750Ala Pro Lys Lys Gly Leu Glu Trp Ile Ala Met Ile
Tyr Tyr Asp Ser 755 760 765Ser Lys Met Asn Tyr Ala Asp Thr Val Lys
Gly Arg Phe Thr Ile Ser 770 775 780Arg Asp Asn Ser Lys Asn Thr Leu
Tyr Leu Glu Met Asn Ser Leu Arg785 790 795 800Ser Glu Asp Thr Ala
Met Tyr Tyr Cys Ala Val Pro Thr Ser His Tyr 805 810 815Val Val Asp
Val Trp Gly Gln Gly Val Ser Val Thr Val Ser Ser Ala 820 825 830Ser
Thr Lys Gly Pro Ser Val Phe Pro Leu Ala Pro Ser Ser Lys Ser 835 840
845Thr Ser Gly Gly Thr Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe
850 855 860Pro Glu Pro Val Thr Val Ser Trp Asn Ser Gly Ala Leu Thr
Ser Gly865 870 875 880Val His Thr Phe Pro Ala Val Leu Gln Ser Ser
Gly Leu Tyr Ser Leu 885 890 895Ser Ser Val Val Thr Val Pro Ser Ser
Ser Leu Gly Thr Gln Thr Tyr 900 905 910Ile Cys Asn Val Asn His Lys
Pro Ser Asn Thr Lys Val Asp Lys Lys 915 920 925Val Glu Pro Lys Ser
Cys 930169449PRTArtificial sequencePer-hole LALA-PG Heavy chain
(21836) 169Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro
Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe
Asn Asp Tyr 20 25 30Thr Met Asp Trp Val Arg Gln Ala Pro Gly Lys Gly
Leu Glu Trp Val 35 40 45Ala Asp Val Asn Pro Asn Ser Gly Gly Ser Ile
Val Asn Arg Arg Phe 50 55 60Lys Gly Arg Phe Thr Leu Ser Val Asp Arg
Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala
Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asn Leu Gly Pro Phe
Phe Tyr Phe Asp Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser Val Phe 115 120 125Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala Leu 130 135 140Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp145 150
155 160Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val
Leu 165 170 175Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr
Val Pro Ser 180 185 190Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn
Val Asn His Lys Pro 195 200 205Ser Asn Thr Lys Val Asp Lys Lys Val
Glu Pro Lys Ser Cys Asp Lys 210 215 220Thr His Thr Cys Pro Pro Cys
Pro Ala Pro Glu Ala Ala Gly Gly Pro225 230 235 240Ser Val Phe Leu
Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser 245 250 255Arg Thr
Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp 260 265
270Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn
275 280 285Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr
Arg Val 290 295 300Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu
Asn Gly Lys Glu305 310 315 320Tyr Lys Cys Lys Val Ser Asn Lys Ala
Leu Gly Ala Pro Ile Glu Lys 325 330 335Thr Ile Ser Lys Ala Lys Gly
Gln Pro Arg Glu Pro Gln Val Cys Thr 340 345 350Leu Pro Pro Ser Arg
Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Ser 355 360 365Cys Ala Val
Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu 370 375 380Ser
Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu385 390
395 400Asp Ser Asp Gly Ser Phe Phe Leu Val Ser Lys Leu Thr Val Asp
Lys 405 410 415Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val
Met His Glu 420 425 430Ala Leu His Asn Arg Phe Thr Gln Lys Ser Leu
Ser Leu Ser Pro Gly 435 440 445Lys170450PRTArtificial
sequenceHer-knob-LALA-PG Heavy Chain (21835) 170Glu Val Gln Leu Val
Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu
Ser Cys Ala Ala Ser Gly Phe Asn Ile Lys Asp Thr 20 25 30Tyr Ile His
Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Arg
Ile Tyr Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Asp Ser Val 50 55 60Lys
Gly Arg Phe Thr Ile Ser Ala Asp Thr Ser Lys Asn Thr Ala Tyr65 70 75
80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ser Arg Trp Gly Gly Glu Gly Phe Tyr Ala Met Asp Tyr Trp Gly
Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly
Pro Ser Val 115 120 125Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser
Gly Gly Thr Ala Ala 130 135 140Leu Gly Cys Leu Val Lys Asp Tyr Phe
Pro Glu Pro Val Thr Val Ser145 150 155 160Trp Asn Ser Gly Ala Leu
Thr Ser Gly Val His Thr Phe Pro Ala Val 165 170 175Leu Gln Ser Ser
Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro 180 185 190Ser Ser
Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys 195 200
205Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp
210 215 220Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Ala Ala
Gly Gly225 230 235 240Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys
Asp Thr Leu Met Ile 245 250 255Ser Arg Thr Pro Glu Val Thr Cys Val
Val Val Asp Val Ser His Glu 260 265 270Asp Pro Glu Val Lys Phe Asn
Trp Tyr Val Asp Gly Val Glu Val His 275 280 285Asn Ala Lys Thr Lys
Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg 290 295 300Val Val Ser
Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys305 310 315
320Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Gly Ala Pro Ile Glu
325 330 335Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln
Val Tyr 340 345 350Thr Leu Pro Pro Cys Arg Asp Glu Leu Thr Lys Asn
Gln Val Ser Leu 355 360 365Trp Cys Leu Val Lys Gly Phe Tyr Pro Ser
Asp Ile Ala Val Glu Trp 370 375 380Glu Ser Asn Gly Gln Pro Glu Asn
Asn Tyr Lys Thr Thr Pro Pro Val385 390 395 400Leu Asp Ser Asp Gly
Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp 405 410 415Lys Ser Arg
Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His 420 425 430Glu
Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro 435 440
445Gly Lys 450171450PRTArtificial sequencepETR13573Her-knob heavy
chain 171Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro
Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Asn Ile
Lys Asp Thr 20 25 30Tyr Ile His Trp Val Arg Gln Ala Pro Gly Lys Gly
Leu Glu Trp Val 35 40 45Ala Arg Ile Tyr Pro Thr Asn Gly Tyr Thr Arg
Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Ala Asp Thr
Ser Lys Asn Thr Ala Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala
Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ser Arg Trp Gly Gly Glu Gly
Phe Tyr Ala Met Asp Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr
Val Ser Ser Ala Ser Thr Lys Gly Pro Ser Val 115 120 125Phe Pro Leu
Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala 130 135 140Leu
Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser145 150
155 160Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala
Val 165 170 175Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
Thr Val Pro 180 185 190Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys
Asn Val Asn His Lys 195 200 205Pro Ser Asn Thr Lys Val Asp Lys Lys
Val Glu Pro Lys Ser Cys Asp 210 215 220Lys Thr His Thr Cys Pro Pro
Cys Pro Ala Pro Glu Leu Leu Gly Gly225 230 235 240Pro Ser Val Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile 245 250 255Ser Arg
Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu 260 265
270Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His
275 280 285Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr
Tyr Arg 290 295 300Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp
Leu Asn Gly Lys305 310 315 320Glu Tyr Lys Cys Lys Val Ser Asn Lys
Ala Leu Pro Ala Pro Ile Glu 325 330 335Lys Thr Ile Ser Lys Ala Lys
Gly Gln Pro Arg Glu Pro Gln Val Tyr 340 345 350Thr Leu Pro Pro Cys
Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu 355 360 365Trp Cys Leu
Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp 370 375 380Glu
Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val385 390
395 400Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val
Asp 405 410 415Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser
Val Met His 420 425 430Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser
Leu Ser Leu Ser Pro 435 440 445Gly Lys 4501726PRTArtificial
sequence2/99 CDR-H1 172Phe Ser Leu Ser Ser Tyr1 517314PRTArtificial
sequence2/99 CDR-H2 173Tyr Ile Trp Ser Gly Gly Ser Thr Asp Tyr Ala
Ser Trp Ala1 5 1017417PRTArtificial sequence2/99 CDR-H3 174Arg Tyr
Gly Thr Ser Tyr Pro Asp Tyr Gly Asp Ala Asn Gly Phe Asp1 5 10
15Pro17511PRTArtificial sequence2/99 CDR-L1 175Gln Ala Ser Gln Ser
Ile Ser Ser Tyr Leu Ser1 5 101763PRTArtificial sequence2/99 CDR-L2
176Arg Ala Ser11779PRTArtificial sequence2/99 CDR-L3 177Cys Tyr Ser
Ser Ser Asn Val Asp Asn1 5178122PRTArtificial sequence2/99 VH
178Gln Ser Met Glu Glu Ser Gly Gly Arg Leu Val Thr Pro Gly Thr Pro1
5 10 15Leu Thr Leu Thr Cys Thr Val Ser Gly Phe Ser Leu Ser Ser Tyr
Ala 20 25 30Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Ile Gly 35 40 45Tyr Ile Trp Ser Gly Gly Ser Thr Asp Tyr Ala Ser Trp
Ala Lys Gly 50 55 60Arg Phe Thr Ile Ser Lys Thr Ser Thr Thr Val Asp
Leu Lys Ile Thr65 70 75 80Ser Pro Thr Thr Glu Asp Thr Ala Thr Tyr
Phe Cys Ala Arg Arg Tyr 85 90 95Gly Thr Ser Tyr Pro Asp Tyr Gly Asp
Ala Asn Gly Phe Asp Pro Trp 100 105 110Gly Pro Gly Thr Leu Val Thr
Val Ser Ser 115 120179111PRTArtificial sequence2/99 VL 179Ala Tyr
Asp Met Thr Gln Thr Pro Ala Ser Val Glu Val Ala Val Gly1 5 10 15Gly
Thr Val Thr Ile Lys Cys Gln Ala Ser Gln Ser Ile Ser Ser Tyr 20 25
30Leu Ser Trp Tyr Gln Gln Lys Pro Gly Gln Arg Pro Lys Leu Leu Ile
35 40 45Tyr Arg Ala Ser Thr Leu Ala Ser Gly Val Ser Ser Arg Phe Lys
Gly 50 55 60Ser Gly Ser Gly Thr Gln Phe Thr Leu Thr Ile Ser Gly Val
Glu Cys65 70 75 80Ala Asp Ala Ala Thr Tyr Tyr Cys Gln Gln Cys Tyr
Ser Ser Ser Asn 85 90 95Val Asp Asn Thr Phe Gly Gly Gly Thr Glu Val
Val Val Lys Arg 100 105 1101807PRTArtificial sequence4/94 CDR-H1
180Gly Phe Asn Ile Lys Asp Thr1 518116PRTArtificial sequence4/94
CDR-H2 181Arg Ile Asp Pro Ala Asn Gly Asp Thr Lys Cys Asp Pro Lys
Phe Gln1 5 10 151828PRTArtificial sequence4/94 CDR-H3
182Tyr Leu Tyr Pro Tyr Tyr Phe Asp1 51837PRTArtificial sequence4/94
CDR-L1 183Ser Glu Ser Val Asp Thr Tyr1 51843PRTArtificial
sequence4/94 CDR-L2 184Gly Ala Ser11856PRTArtificial sequence4/94
CDR-L3 185Thr Tyr Asn Tyr Pro Leu1 51865PRTArtificial
sequenceHumanized 2/99 CDR-H1 186Ser Tyr Ala Met Ser1
518716PRTArtificial sequenceHumanized 2/99 CDR-H2 187Tyr Ile Trp
Ser Gly Gly Ser Thr Asp Tyr Ala Ser Trp Ala Lys Ser1 5 10
1518817PRTArtificial sequenceHumanized 2/99 CDR-H3 188Arg Tyr Gly
Thr Ser Tyr Pro Asp Tyr Gly Asp Ala Ser Gly Phe Asp1 5 10
15Pro18911PRTArtificial sequenceHumanized 2/99 CDR-L1 189Arg Ala
Ser Gln Ser Ile Ser Ser Tyr Leu Ala1 5 101907PRTArtificial
sequenceHumanized 2/99 CDR-L2 190Arg Ala Ser Thr Leu Ala Ser1
519112PRTArtificial sequenceHumanized 2/99 CDR-L3 191Gln Gln Asn
Tyr Ala Ser Ser Asn Val Asp Asn Thr1 5 10192122PRTArtificial
sequenceHumanized 2/99 VH 192Gln Ser Met Gln Glu Ser Gly Pro Gly
Leu Val Lys Pro Ser Gln Thr1 5 10 15Leu Ser Leu Thr Cys Thr Val Ser
Gly Phe Ser Leu Ser Ser Tyr Ala 20 25 30Met Ser Trp Ile Arg Gln His
Pro Gly Lys Gly Leu Glu Trp Ile Gly 35 40 45Tyr Ile Trp Ser Gly Gly
Ser Thr Asp Tyr Ala Ser Trp Ala Lys Ser 50 55 60Arg Val Thr Ile Ser
Lys Thr Ser Thr Thr Val Ser Leu Lys Leu Ser65 70 75 80Ser Val Thr
Ala Ala Asp Thr Ala Val Tyr Tyr Cys Ala Arg Arg Tyr 85 90 95Gly Thr
Ser Tyr Pro Asp Tyr Gly Asp Ala Ser Gly Phe Asp Pro Trp 100 105
110Gly Gln Gly Thr Leu Val Thr Val Ser Ser 115
120193109PRTArtificial sequenceHumanized 2/99 VL 193Ala Ile Gln Leu
Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val
Thr Ile Thr Cys Arg Ala Ser Gln Ser Ile Ser Ser Tyr 20 25 30Leu Ala
Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr
Arg Ala Ser Thr Leu Ala Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55
60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65
70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Asn Tyr Ala Ser Ser
Asn 85 90 95Val Asp Asn Thr Phe Gly Gly Gly Thr Lys Val Glu Ile 100
105194119PRTArtificial sequenceHumanized 2/99 VH variant 1 194Gln
Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10
15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Phe Asn Ile Lys Asp Thr
20 25 30Tyr Ile His Trp Val Ile Gln Ala Pro Gly Gln Gly Leu Glu Trp
Met 35 40 45Gly Arg Ile Asp Pro Ala Asn Gly Asp Thr Lys Ser Asp Pro
Lys Phe 50 55 60Gln Val Arg Val Thr Ile Thr Ala Asp Thr Ser Thr Ser
Thr Val Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr
Ala Val Tyr Tyr Cys 85 90 95Val Arg Asp Tyr Leu Tyr Pro Tyr Tyr Phe
Asp Phe Trp Gly Gln Gly 100 105 110Thr Thr Val Thr Val Ser Ser
115195119PRTArtificial sequenceHumanized 2/99 VH variant 2 195Gln
Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10
15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Phe Asn Ile Lys Asp Thr
20 25 30Tyr Ile His Trp Val Ile Gln Ala Pro Gly Gln Gly Leu Glu Trp
Met 35 40 45Gly Arg Ile Asp Pro Ala Asn Gly Asp Thr Lys Ser Ala Pro
Lys Phe 50 55 60Gln Val Arg Val Thr Ile Thr Ala Asp Thr Ser Thr Ser
Thr Val Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr
Ala Val Tyr Tyr Cys 85 90 95Val Arg Asp Tyr Leu Tyr Pro Tyr Tyr Phe
Asp Phe Trp Gly Gln Gly 100 105 110Thr Thr Val Thr Val Ser Ser
115196119PRTArtificial sequenceHumanized 2/99 VH variant 3 196Gln
Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10
15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Phe Asn Ile Lys Asp Thr
20 25 30Tyr Ile His Trp Val Ile Gln Ala Pro Gly Gln Gly Leu Glu Trp
Met 35 40 45Gly Arg Ile Asp Pro Ala Asn Gly Asp Thr Lys Ser Asp Pro
Lys Phe 50 55 60Gln Gly Arg Val Thr Ile Thr Ala Asp Thr Ser Thr Ser
Thr Val Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr
Ala Val Tyr Tyr Cys 85 90 95Val Arg Asp Tyr Leu Tyr Pro Tyr Tyr Phe
Asp Phe Trp Gly Gln Gly 100 105 110Thr Thr Val Thr Val Ser Ser
115197119PRTArtificial sequenceHumanized 2/99 VH variant 4 197Gln
Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ser1 5 10
15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Phe Asn Ile Lys Asp Thr
20 25 30Tyr Ile His Trp Val Ile Gln Ala Pro Gly Gln Gly Leu Glu Trp
Met 35 40 45Gly Arg Ile Asp Pro Ala Asn Gly Asp Thr Lys Ser Asp Pro
Lys Phe 50 55 60Gln Gly Arg Val Thr Ile Thr Ala Asp Glu Ser Thr Ser
Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr
Ala Val Tyr Tyr Cys 85 90 95Val Arg Asp Tyr Leu Tyr Pro Tyr Tyr Phe
Asp Phe Trp Gly Gln Gly 100 105 110Thr Thr Val Thr Val Ser Ser
115198119PRTArtificial sequenceHumanized 2/99 VH variant 5 198Gln
Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ser1 5 10
15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Phe Asn Ile Lys Asp Thr
20 25 30Tyr Ile His Trp Val Ile Gln Arg Pro Gly Gln Gly Leu Glu Trp
Met 35 40 45Gly Arg Ile Asp Pro Ala Asn Gly Asp Thr Lys Ser Asp Pro
Lys Phe 50 55 60Gln Gly Arg Val Thr Ile Thr Ala Asp Glu Ser Thr Ser
Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr
Ala Val Tyr Tyr Cys 85 90 95Val Arg Asp Tyr Leu Tyr Pro Tyr Tyr Phe
Asp Phe Trp Gly Gln Gly 100 105 110Thr Thr Val Thr Val Ser Ser
115199119PRTArtificial sequenceHumanized 2/99 VH variant 6 199Gln
Val Gln Leu Gln Glu Ser Gly Pro Gly Leu Val Lys Pro Ser Glu1 5 10
15Thr Leu Ser Leu Thr Cys Thr Val Ser Gly Phe Asn Ile Lys Asp Thr
20 25 30Tyr Ile His Trp Val Ile Gln Arg Pro Gly Lys Gly Leu Glu Trp
Ile 35 40 45Gly Arg Ile Asp Pro Ala Asn Gly Asp Thr Lys Ser Asp Pro
Ser Leu 50 55 60Gln Ser Arg Val Thr Ile Ser Ala Asp Thr Ser Lys Asn
Gln Ala Ser65 70 75 80Leu Lys Leu Ser Ser Val Thr Ala Ala Asp Thr
Ala Val Tyr Tyr Cys 85 90 95Val Arg Asp Tyr Leu Tyr Pro Tyr Tyr Phe
Asp Phe Trp Gly Gln Gly 100 105 110Thr Thr Val Thr Val Ser Ser
115200119PRTArtificial sequenceHumanized 2/99 VH variant 7 200Glu
Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10
15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Asn Ile Lys Asp Thr
20 25 30Tyr Ile His Trp Val Ile Gln Ala Pro Gly Lys Gly Leu Glu Trp
Val 35 40 45Gly Arg Ile Asp Pro Ala Asn Gly Asp Thr Lys Ser Asp Pro
Ser Val 50 55 60Gln Val Arg Ala Thr Ile Ser Ala Asp Thr Ser Lys Asn
Thr Ala Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr
Ala Val Tyr Tyr Cys 85 90 95Val Arg Asp Tyr Leu Tyr Pro Tyr Tyr Phe
Asp Phe Trp Gly Gln Gly 100 105 110Thr Thr Val Thr Val Ser Ser
115201106PRTArtificial sequenceHumanized 2/99 VL variant 1 201Lys
Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10
15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Glu Ser Val Asp Thr Tyr
20 25 30Val Ser Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu
Ile 35 40 45Tyr Gly Ala Ser Asn Arg Tyr Thr Gly Val Pro Ser Arg Phe
Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser
Leu Gln Pro65 70 75 80Glu Asp Phe Ala Asp Tyr Tyr Cys Gly Gln Thr
Tyr Asn Tyr Pro Leu 85 90 95Thr Phe Gly Gln Gly Thr Lys Leu Glu Ile
100 10520215PRTArtificial sequenceHuman transferring receptor
(hTFR) epitope 1 202Ile Gly Gln Asn Met Val Thr Ile Val Gln Ser Asn
Gly Asn Leu1 5 10 1520315PRTArtificial sequenceHuman transferring
receptor (hTFR) epitope 2 203Asn Met Val Thr Ile Val Gln Ser Asn
Gly Asn Leu Asp Pro Val1 5 10 1520415PRTArtificial sequenceHuman
transferring receptor (hTFR) epitope 3 204Gln Ser Asn Gly Asn Leu
Asp Pro Val Glu Ser Pro Glu Gly Tyr1 5 10 15205122PRTArtificial
sequenceHumanized 2/99 VH variant 205Gln Ser Met Gln Glu Ser Gly
Pro Gly Leu Val Lys Pro Ser Gln Thr1 5 10 15Leu Ser Leu Thr Cys Thr
Val Ser Gly Phe Ser Leu Ser Ser Tyr Ala 20 25 30Met Ser Trp Ile Arg
Gln His Pro Gly Lys Gly Leu Glu Trp Ile Gly 35 40 45Tyr Ile Trp Ser
Gly Gly Ser Thr Asp Tyr Ala Ser Trp Ala Lys Ser 50 55 60Arg Val Thr
Ile Ser Lys Thr Ser Thr Thr Val Ser Leu Lys Leu Ser65 70 75 80Ser
Val Thr Ala Ala Asp Thr Ala Val Tyr Tyr Cys Ala Arg Arg Tyr 85 90
95Gly Thr Ser Tyr Pro Asp Tyr Gly Asp Ala Gln Gly Phe Asp Pro Trp
100 105 110Gly Gln Gly Thr Leu Val Thr Val Ser Ser 115
12020617PRTArtificial sequenceHumanized 2/99 CDR-H3 variant 206Arg
Tyr Gly Thr Ser Tyr Pro Asp Tyr Gly Asp Ala Gln Gly Phe Asp1 5 10
15Pro207106PRTArtificial sequenceHumanized 2/99 VL variant 2 207Lys
Ile Val Leu Thr Gln Ser Pro Gly Thr Leu Ser Leu Ser Pro Gly1 5 10
15Glu Arg Ala Thr Leu Ser Cys Arg Ala Ser Glu Ser Val Asp Thr Tyr
20 25 30Val Ser Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro Arg Leu Leu
Ile 35 40 45Tyr Gly Ala Ser Asn Arg Tyr Thr Gly Val Pro Asp Arg Phe
Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Arg
Leu Glu Pro65 70 75 80Glu Asp Phe Ala Asp Tyr Tyr Cys Gly Gln Thr
Tyr Asn Tyr Pro Leu 85 90 95Thr Phe Gly Gln Gly Thr Lys Leu Glu Ile
100 105208106PRTArtificial sequenceHumanized 2/99 VL variant 3
208Glu Ile Val Leu Thr Gln Ser Pro Gly Thr Leu Ser Leu Ser Pro Gly1
5 10 15Glu Arg Ala Thr Leu Ser Cys Arg Ala Ser Glu Ser Val Asp Thr
Tyr 20 25 30Val Ser Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro Arg Leu
Leu Ile 35 40 45Tyr Gly Ala Ser Asn Arg Tyr Thr Gly Val Pro Asp Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Arg Leu Glu Pro65 70 75 80Glu Asp Phe Ala Asp Tyr Tyr Cys Gly Gln
Thr Tyr Asn Tyr Pro Leu 85 90 95Thr Phe Gly Gln Gly Thr Lys Leu Glu
Ile 100 105209106PRTArtificial sequenceHumanized 2/99 VL variant 4
209Lys Ile Val Leu Thr Gln Ser Pro Gly Thr Leu Ser Leu Ser Pro Gly1
5 10 15Glu Arg Ala Thr Leu Ser Cys Arg Ala Ser Glu Ser Val Asp Thr
Tyr 20 25 30Val Ser Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro Arg Leu
Leu Ile 35 40 45Tyr Gly Ala Ser Asn Arg Tyr Thr Gly Ile Pro Asp Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Arg Leu Glu Pro65 70 75 80Glu Asp Phe Ala Asp Tyr Tyr Cys Gly Gln
Thr Tyr Asn Tyr Pro Leu 85 90 95Thr Phe Gly Gln Gly Thr Lys Leu Glu
Ile 100 105210219PRTArtificial
sequence8936_pTau_RbMab86_VL_huIgkappa 210Ala Gln Val Leu Thr Gln
Thr Thr Ser Pro Val Ser Ala Ala Val Gly1 5 10 15Ser Thr Val Thr Ile
Ser Cys Gln Ser Ser Gln Ser Val Arg Thr Asn 20 25 30Lys Leu Ala Trp
Phe Gln Gln Lys Pro Gly Gln Pro Pro Lys Arg Leu 35 40 45Ile Tyr Ser
Ala Ser Thr Leu Asp Phe Gly Val Pro Ser Arg Phe Ser 50 55 60Ala Ser
Gly Ser Gly Thr Gln Phe Thr Leu Thr Ile Ser Asp Val Gln65 70 75
80Cys Asp Asp Ala Ala Thr Tyr Tyr Cys Leu Gly Tyr Phe Asp Cys Ser
85 90 95Ile Ala Asp Cys Val Ala Phe Gly Gly Gly Thr Glu Val Val Val
Lys 100 105 110Arg Thr Val Ala Ala Pro Ser Val Phe Ile Phe Pro Pro
Ser Asp Glu 115 120 125Gln Leu Lys Ser Gly Thr Ala Ser Val Val Cys
Leu Leu Asn Asn Phe 130 135 140Tyr Pro Arg Glu Ala Lys Val Gln Trp
Lys Val Asp Asn Ala Leu Gln145 150 155 160Ser Gly Asn Ser Gln Glu
Ser Val Thr Glu Gln Asp Ser Lys Asp Ser 165 170 175Thr Tyr Ser Leu
Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu 180 185 190Lys His
Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser 195 200
205Pro Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 210
215211438PRTArtificial sequence8975-pTau-Rb86-hugamma1-SS-hole
211Gln Ser Val Glu Glu Ser Gly Gly Arg Leu Val Thr Pro Gly Thr Pro1
5 10 15Leu Thr Leu Thr Cys Thr Val Ser Gly Phe Ser Leu Ser Ser Asn
Ala 20 25 30Ile Asn Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Ile Gly 35 40 45Tyr Ile Ala Val Ser Gly Asn Thr Tyr Tyr Ala Ser Trp
Ala Lys Gly 50 55 60Arg Phe Thr Ile Ser Lys Ala Ser Thr Thr Val Asp
Leu Lys Met Thr65 70 75 80Ser Pro Thr Ala Glu Asp Thr Gly Thr Tyr
Phe Cys Gly Lys Ser Asn 85 90 95Ile Trp Gly Pro Gly Thr Leu Val Thr
Val Ser Leu Ala Ser Thr Lys 100 105 110Gly Pro Ser Val Phe Pro Leu
Ala Pro Ser Ser Lys Ser Thr Ser Gly 115 120 125Gly Thr Ala Ala Leu
Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro 130 135 140Val Thr Val
Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr145 150 155
160Phe Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val
165 170 175Val Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile
Cys Asn 180 185 190Val Asn His Lys Pro Ser Asn Thr Lys Val Asp Lys
Lys Val Glu Pro 195 200 205Lys Ser Cys Asp Lys Thr His Thr Cys Pro
Pro Cys Pro Ala Pro Glu 210 215 220Leu Leu Gly Gly Pro Ser Val Phe
Leu Phe Pro Pro Lys Pro Lys Asp225 230 235 240Thr Leu Met Ile Ser
Arg Thr Pro Glu Val Thr Cys Val Val Val Asp 245 250 255Val Ser His
Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly 260 265 270Val
Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln
Tyr Asn 275 280 285Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu
His Gln Asp Trp 290 295 300Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val
Ser Asn Lys Ala Leu Pro305 310 315 320Ala Pro Ile Glu Lys Thr Ile
Ser Lys Ala Lys Gly Gln Pro Arg Glu 325 330 335Pro Gln Val Cys Thr
Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn 340 345 350Gln Val Ser
Leu Ser Cys Ala Val Lys Gly Phe Tyr Pro Ser Asp Ile 355 360 365Ala
Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr 370 375
380Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Val Ser
Lys385 390 395 400Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn
Val Phe Ser Cys 405 410 415Ser Val Met His Glu Ala Leu His Asn His
Tyr Thr Gln Lys Ser Leu 420 425 430Ser Leu Ser Pro Gly Lys
435212922PRTArtificial
sequence8976-pTau-Rb86-hugamma1-SS-knob-mTfR-8D3-scFab 212Gln Ser
Val Glu Glu Ser Gly Gly Arg Leu Val Thr Pro Gly Thr Pro1 5 10 15Leu
Thr Leu Thr Cys Thr Val Ser Gly Phe Ser Leu Ser Ser Asn Ala 20 25
30Ile Asn Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Ile Gly
35 40 45Tyr Ile Ala Val Ser Gly Asn Thr Tyr Tyr Ala Ser Trp Ala Lys
Gly 50 55 60Arg Phe Thr Ile Ser Lys Ala Ser Thr Thr Val Asp Leu Lys
Met Thr65 70 75 80Ser Pro Thr Ala Glu Asp Thr Gly Thr Tyr Phe Cys
Gly Lys Ser Asn 85 90 95Ile Trp Gly Pro Gly Thr Leu Val Thr Val Ser
Leu Ala Ser Thr Lys 100 105 110Gly Pro Ser Val Phe Pro Leu Ala Pro
Ser Ser Lys Ser Thr Ser Gly 115 120 125Gly Thr Ala Ala Leu Gly Cys
Leu Val Lys Asp Tyr Phe Pro Glu Pro 130 135 140Val Thr Val Ser Trp
Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr145 150 155 160Phe Pro
Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val 165 170
175Val Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn
180 185 190Val Asn His Lys Pro Ser Asn Thr Lys Val Asp Lys Lys Val
Glu Pro 195 200 205Lys Ser Cys Asp Lys Thr His Thr Cys Pro Pro Cys
Pro Ala Pro Glu 210 215 220Leu Leu Gly Gly Pro Ser Val Phe Leu Phe
Pro Pro Lys Pro Lys Asp225 230 235 240Thr Leu Met Ile Ser Arg Thr
Pro Glu Val Thr Cys Val Val Val Asp 245 250 255Val Ser His Glu Asp
Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly 260 265 270Val Glu Val
His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn 275 280 285Ser
Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp 290 295
300Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu
Pro305 310 315 320Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly
Gln Pro Arg Glu 325 330 335Pro Gln Val Tyr Thr Leu Pro Pro Cys Arg
Asp Glu Leu Thr Lys Asn 340 345 350Gln Val Ser Leu Trp Cys Leu Val
Lys Gly Phe Tyr Pro Ser Asp Ile 355 360 365Ala Val Glu Trp Glu Ser
Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr 370 375 380Thr Pro Pro Val
Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys385 390 395 400Leu
Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys 405 410
415Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu
420 425 430Ser Leu Ser Pro Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly
Gly Gly 435 440 445Gly Ser Gly Gly Gly Gly Ser Asp Ile Gln Met Thr
Gln Ser Pro Ala 450 455 460Ser Leu Ser Ala Ser Leu Glu Glu Ile Val
Thr Ile Thr Cys Gln Ala465 470 475 480Ser Gln Asp Ile Gly Asn Trp
Leu Ala Trp Tyr Gln Gln Lys Pro Gly 485 490 495Lys Ser Pro Gln Leu
Leu Ile Tyr Gly Ala Thr Ser Leu Ala Asp Gly 500 505 510Val Pro Ser
Arg Phe Ser Gly Ser Arg Ser Gly Thr Gln Phe Ser Leu 515 520 525Lys
Ile Ser Arg Val Gln Val Glu Asp Ile Gly Ile Tyr Tyr Cys Leu 530 535
540Gln Ala Tyr Asn Thr Pro Trp Thr Phe Gly Gly Gly Thr Lys Val
Glu545 550 555 560Ile Lys Arg Thr Val Ala Ala Pro Ser Val Phe Ile
Phe Pro Pro Ser 565 570 575Asp Glu Gln Leu Lys Ser Gly Thr Ala Ser
Val Val Cys Leu Leu Asn 580 585 590Asn Phe Tyr Pro Arg Glu Ala Lys
Val Gln Trp Lys Val Asp Asn Ala 595 600 605Leu Gln Ser Gly Asn Ser
Gln Glu Ser Val Thr Glu Gln Asp Ser Lys 610 615 620Asp Ser Thr Tyr
Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp625 630 635 640Tyr
Glu Lys His Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu 645 650
655Ser Ser Pro Val Thr Lys Ser Phe Asn Arg Gly Glu Cys Gly Gly Gly
660 665 670Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly
Gly Gly 675 680 685Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly
Gly Glu Val Gln 690 695 700Leu Val Glu Ser Gly Gly Gly Leu Val Gln
Pro Gly Asn Ser Leu Thr705 710 715 720Leu Ser Cys Val Ala Ser Gly
Phe Thr Phe Ser Asn Tyr Gly Met His 725 730 735Trp Ile Arg Gln Ala
Pro Lys Lys Gly Leu Glu Trp Ile Ala Met Ile 740 745 750Tyr Tyr Asp
Ser Ser Lys Met Asn Tyr Ala Asp Thr Val Lys Gly Arg 755 760 765Phe
Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr Leu Glu Met 770 775
780Asn Ser Leu Arg Ser Glu Asp Thr Ala Met Tyr Tyr Cys Ala Val
Pro785 790 795 800Thr Ser His Tyr Val Val Asp Val Trp Gly Gln Gly
Val Ser Val Thr 805 810 815Val Ser Ser Ala Ser Thr Lys Gly Pro Ser
Val Phe Pro Leu Ala Pro 820 825 830Ser Ser Lys Ser Thr Ser Gly Gly
Thr Ala Ala Leu Gly Cys Leu Val 835 840 845Lys Asp Tyr Phe Pro Glu
Pro Val Thr Val Ser Trp Asn Ser Gly Ala 850 855 860Leu Thr Ser Gly
Val His Thr Phe Pro Ala Val Leu Gln Ser Ser Gly865 870 875 880Leu
Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser Ser Ser Leu Gly 885 890
895Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro Ser Asn Thr Lys
900 905 910Val Asp Lys Lys Val Glu Pro Lys Ser Cys 915
92021337DNAArtificial sequencerbHC.up 213aagcttgcca ccatggagac
tgggctgcgc tggcttc 3721421DNAArtificial sequencerbHCf.do
214ccattggtga gggtgcccga g 2121534DNAArtificial sequencerbLC.up
215aagcttgcca ccatggacay gagggccccc actc 3421626DNAArtificial
sequencerbLC.do 216cagagtrctg ctgaggttgt aggtac
2621720DNAArtificial sequenceBcPCR_FHLC_leader.fw 217atggacatga
gggtccccgc 2021824DNAArtificial sequenceBcPCR_huCkappa.rev
218gatttcaact gctcatcaga tggc 24
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013
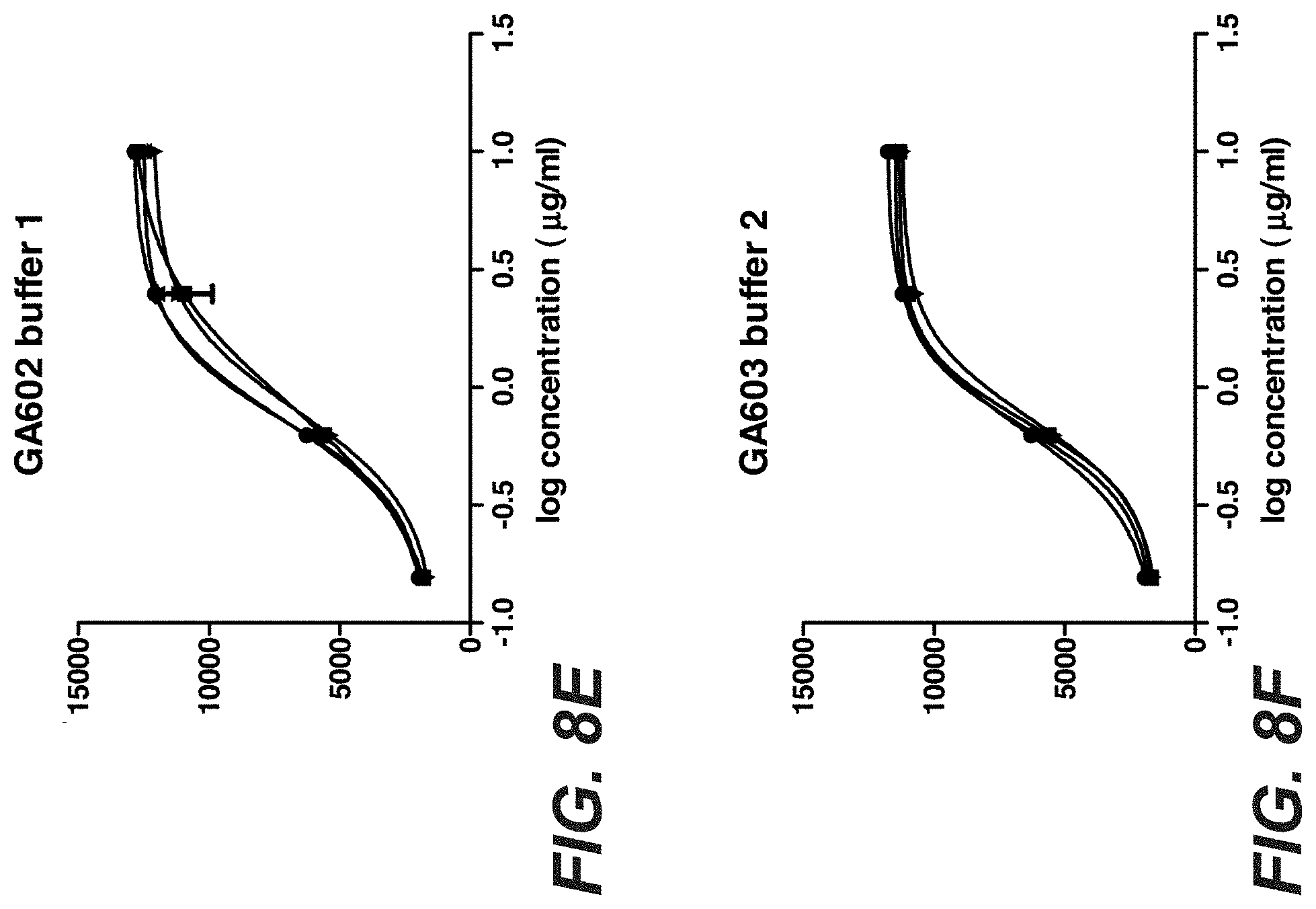
D00014

D00015

D00016

D00017

D00018

D00019
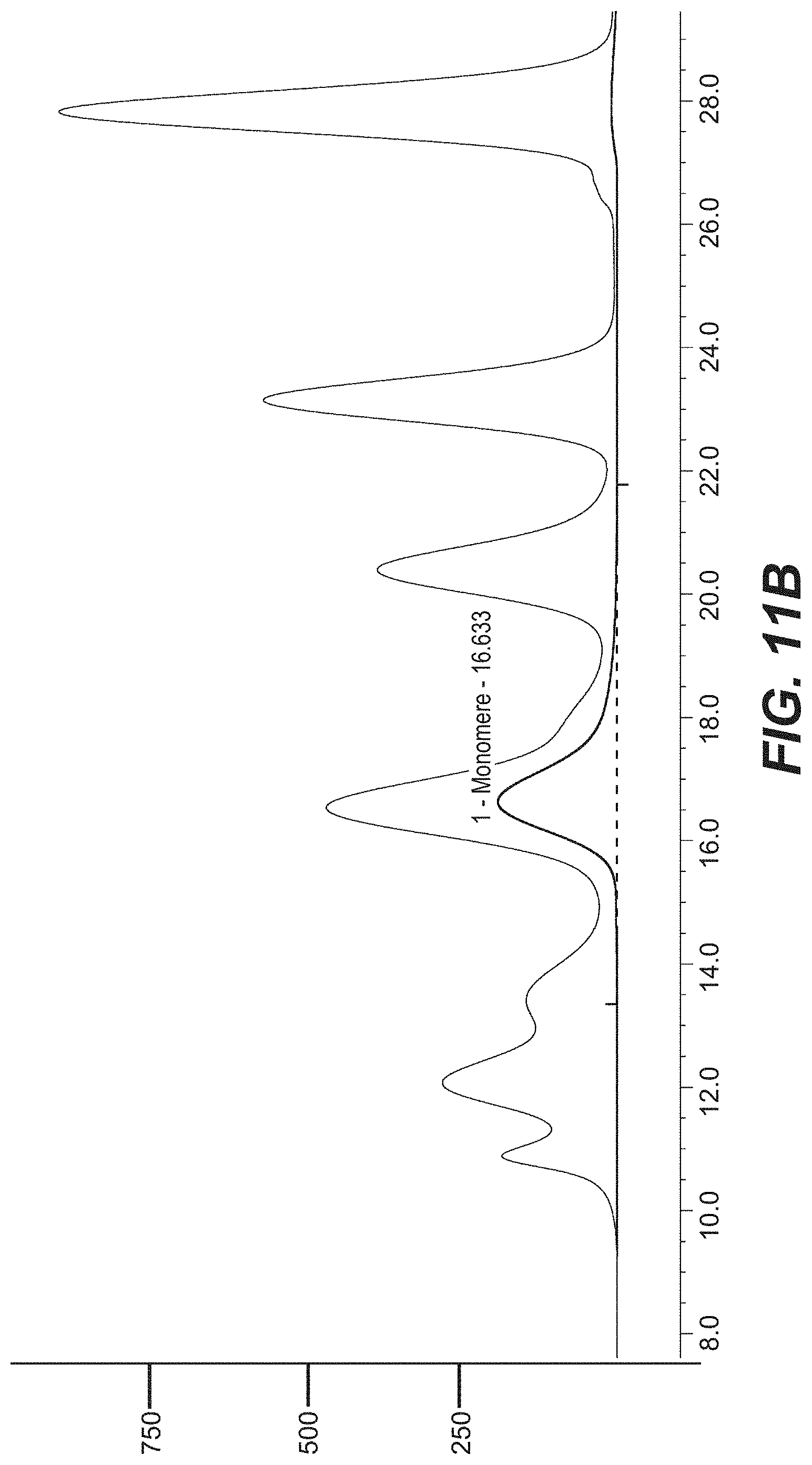
D00020

D00021

D00022

D00023

D00024

D00025
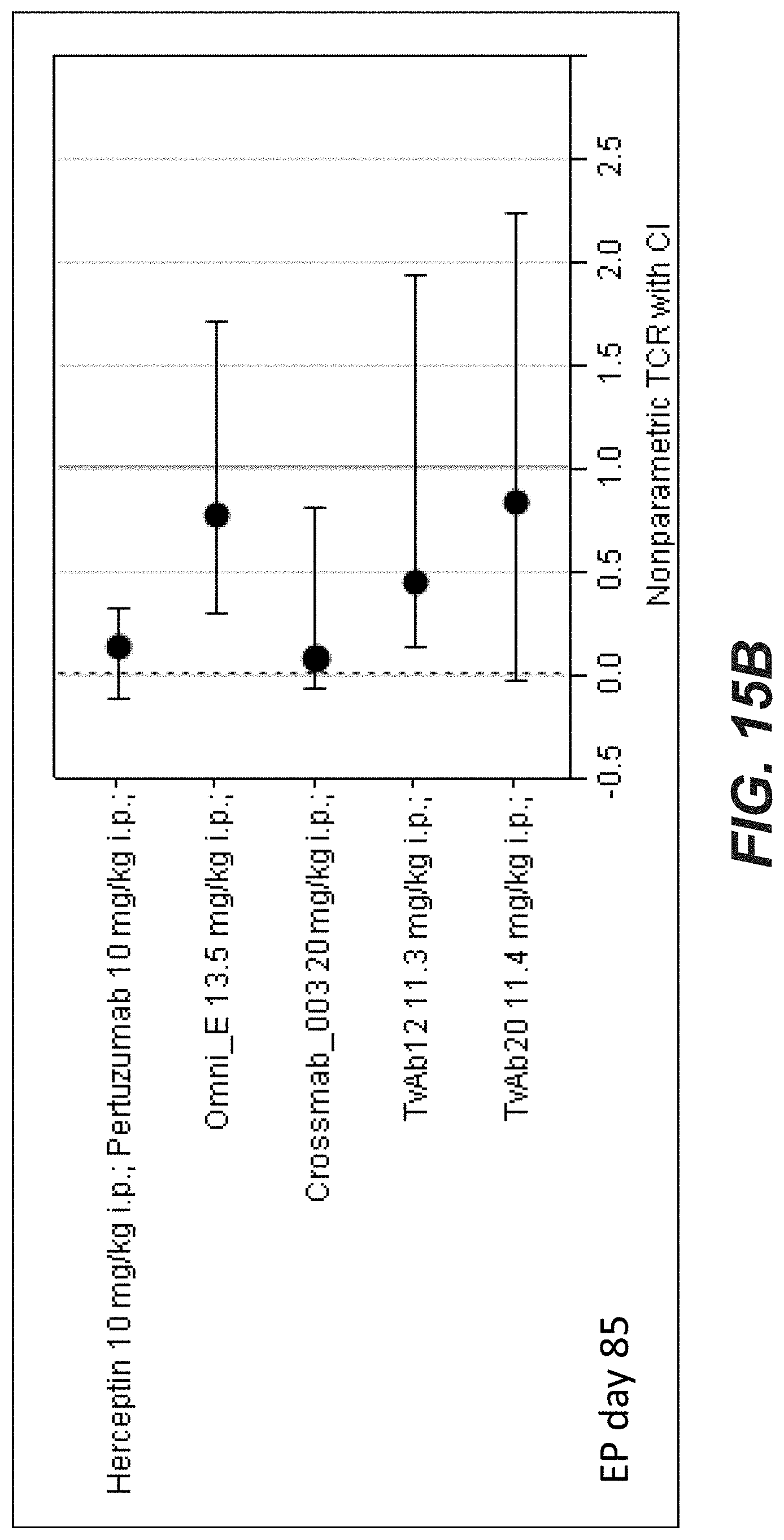
D00026

D00027
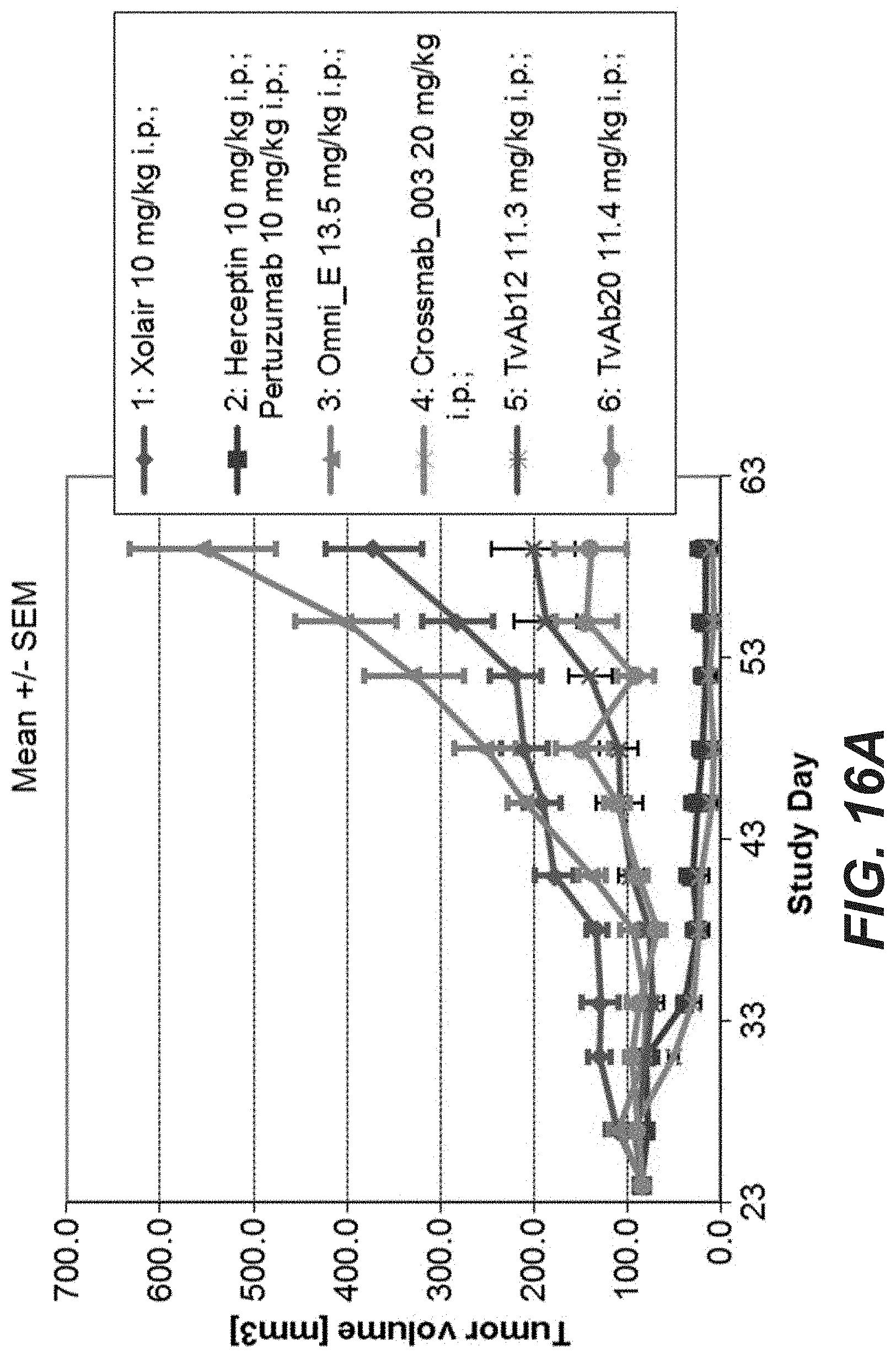
D00028

D00029

D00030

D00031

D00032
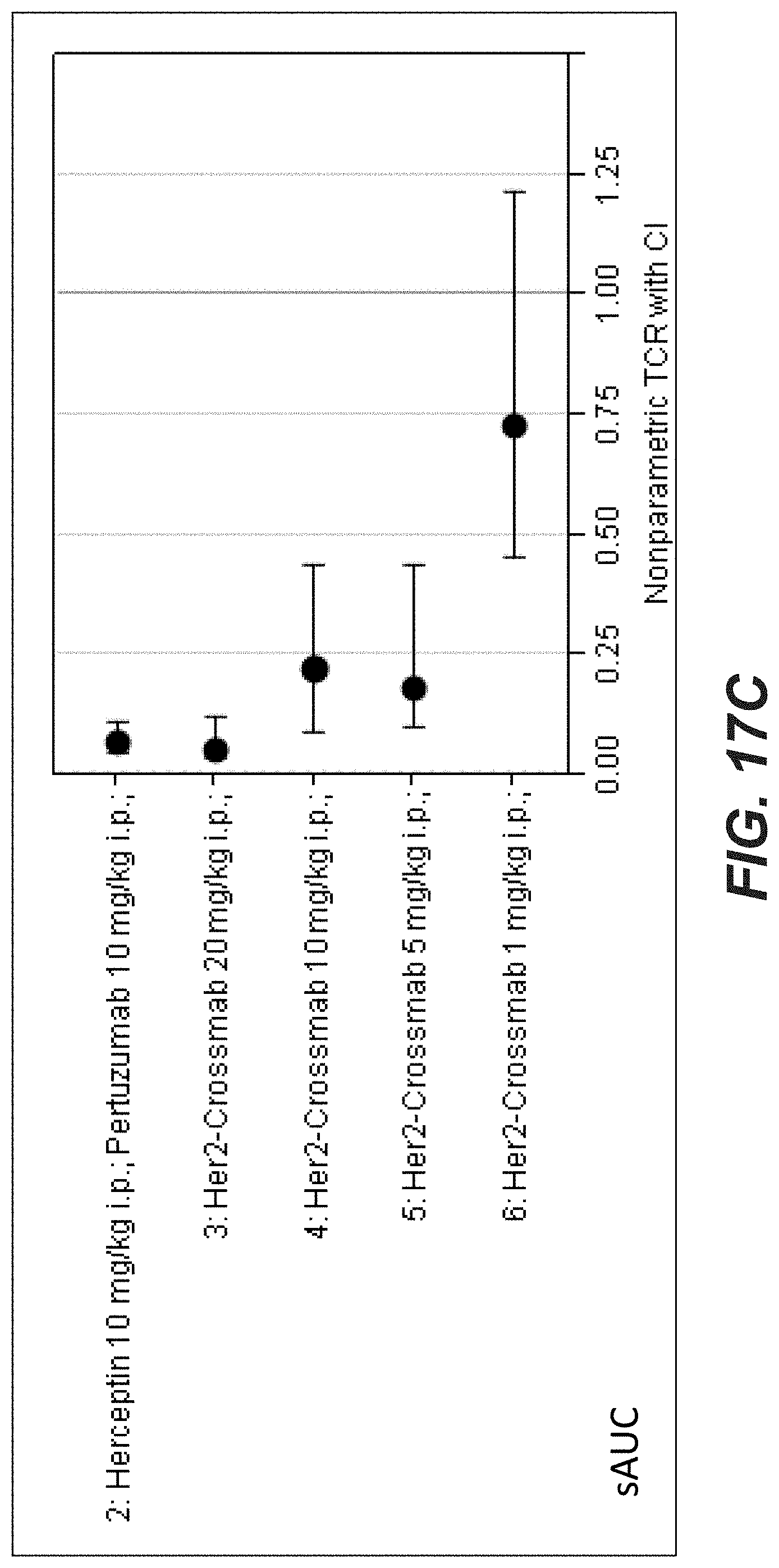
D00033

D00034

D00035

D00036

D00037

D00038

D00039

D00040
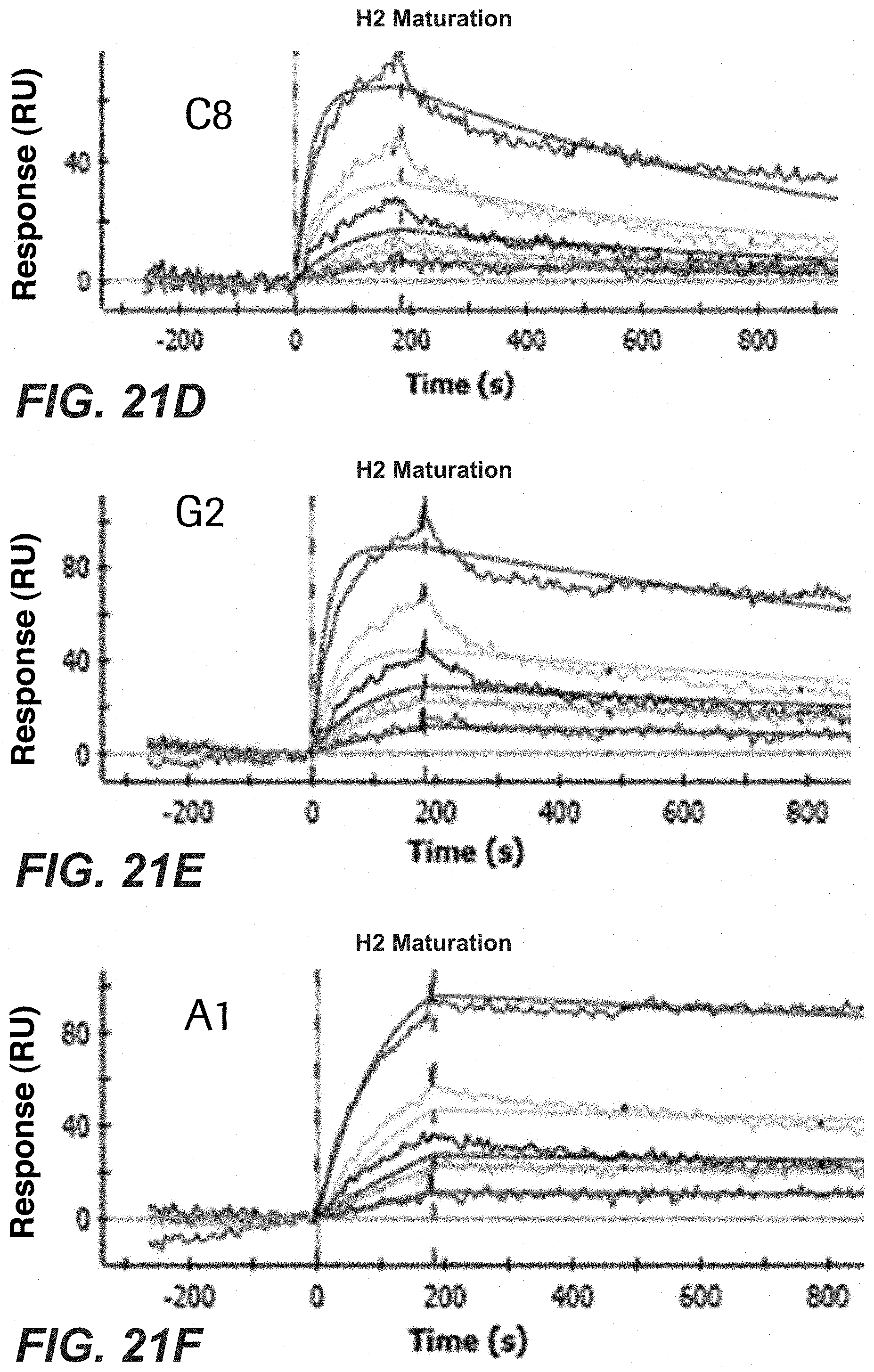
D00041
D00042

D00043

D00044
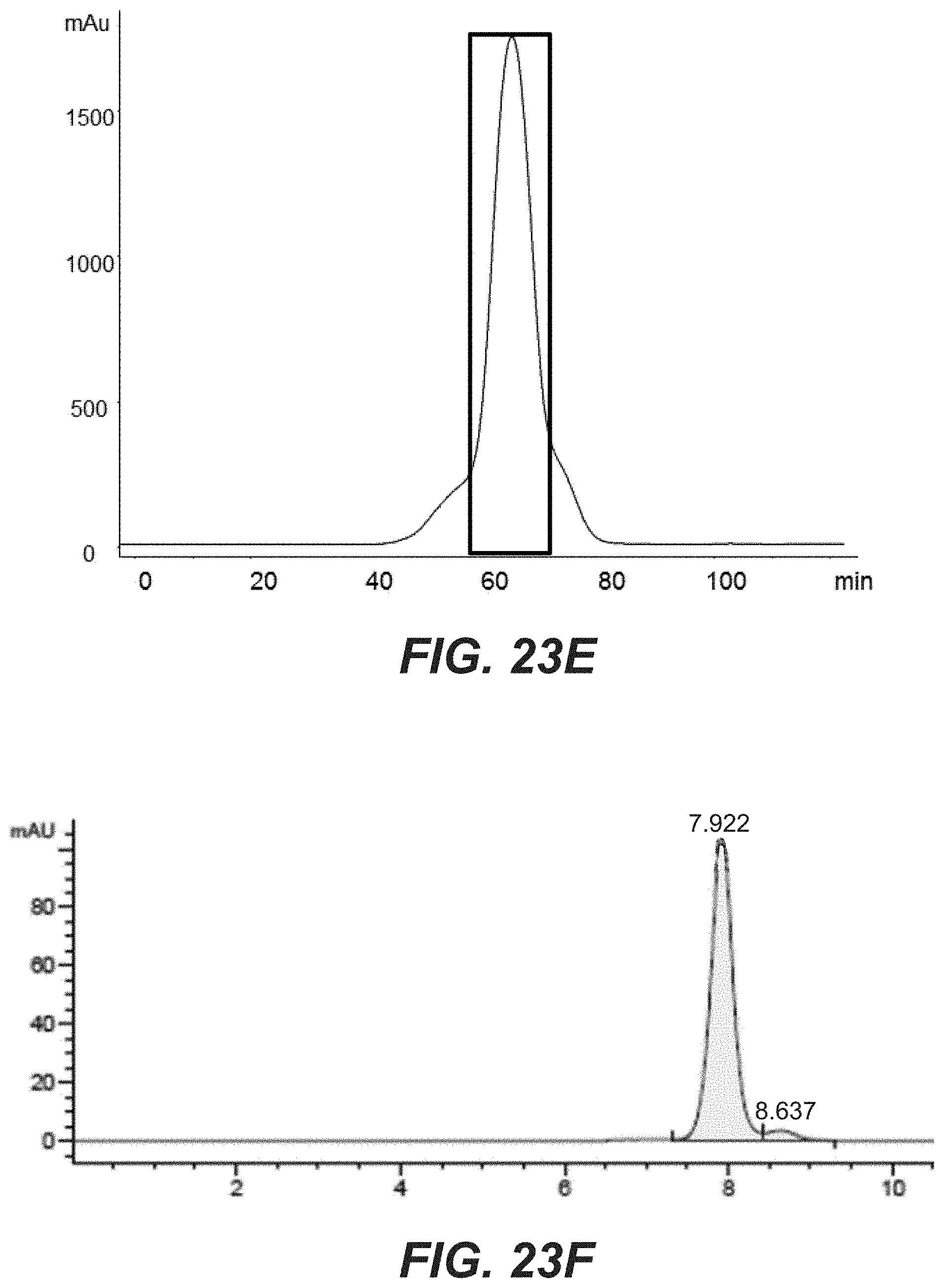
D00045

D00046

D00047
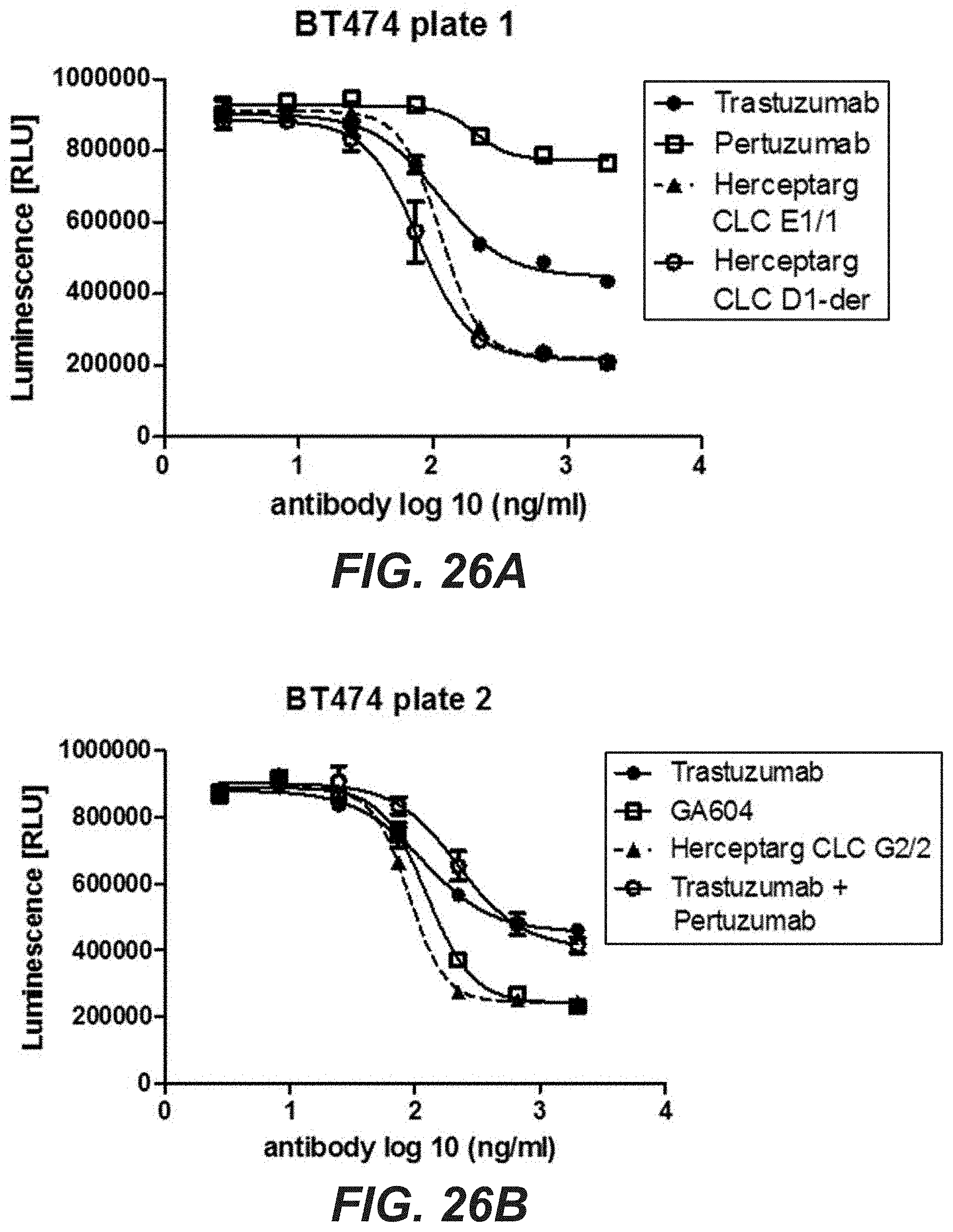
D00048

D00049

D00050
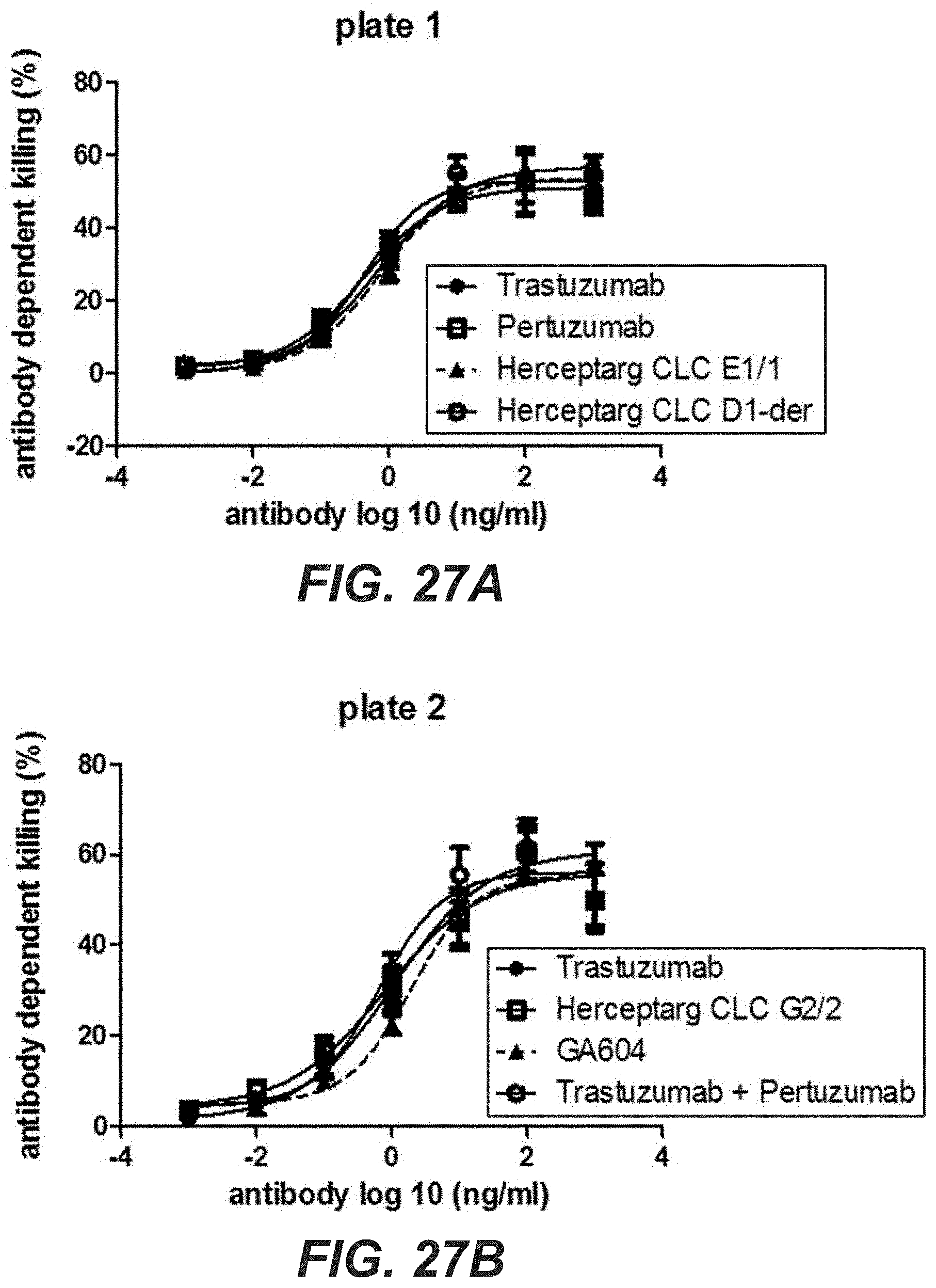
D00051

D00052

D00053

D00054
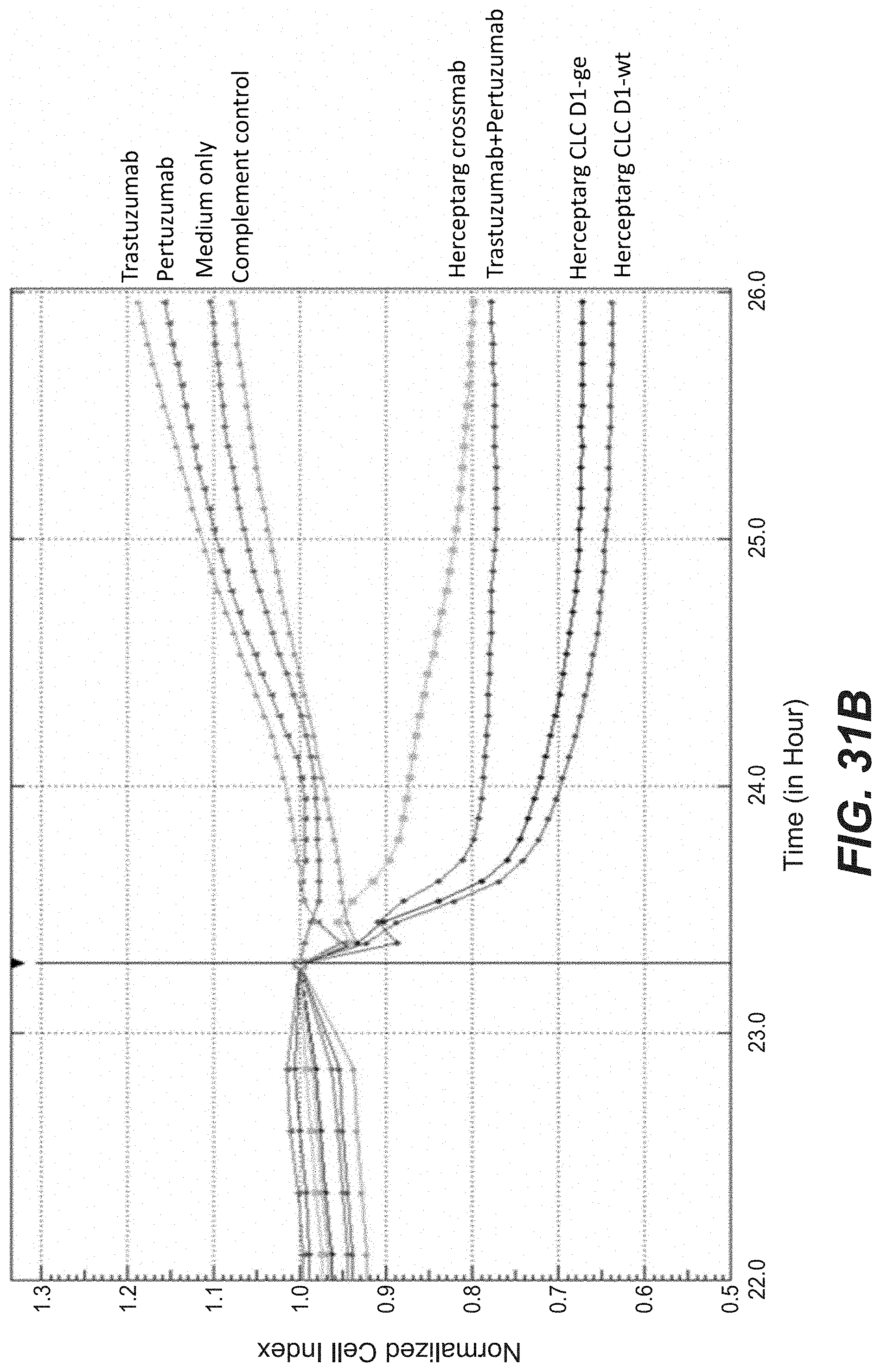
D00055
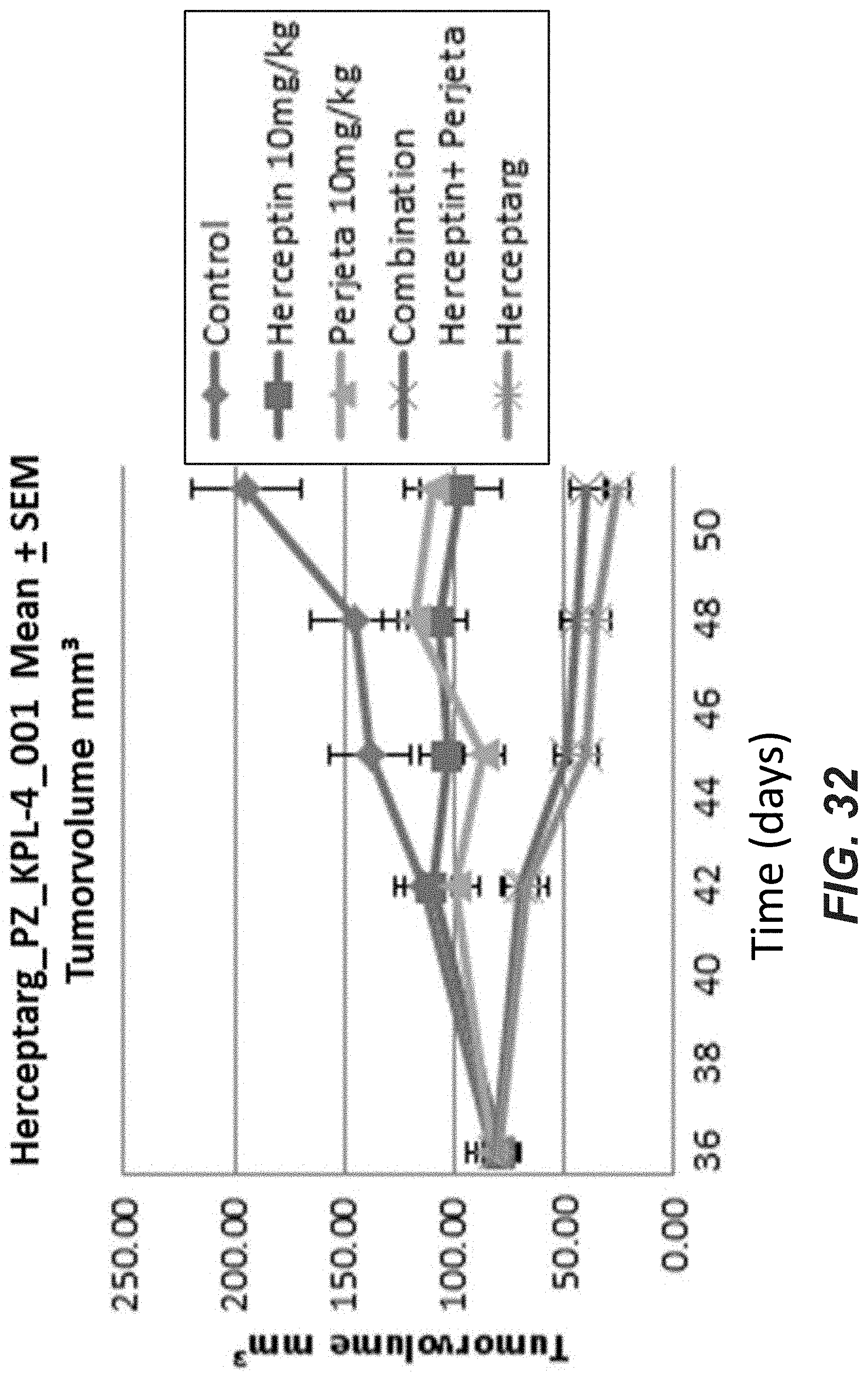
D00056

D00057
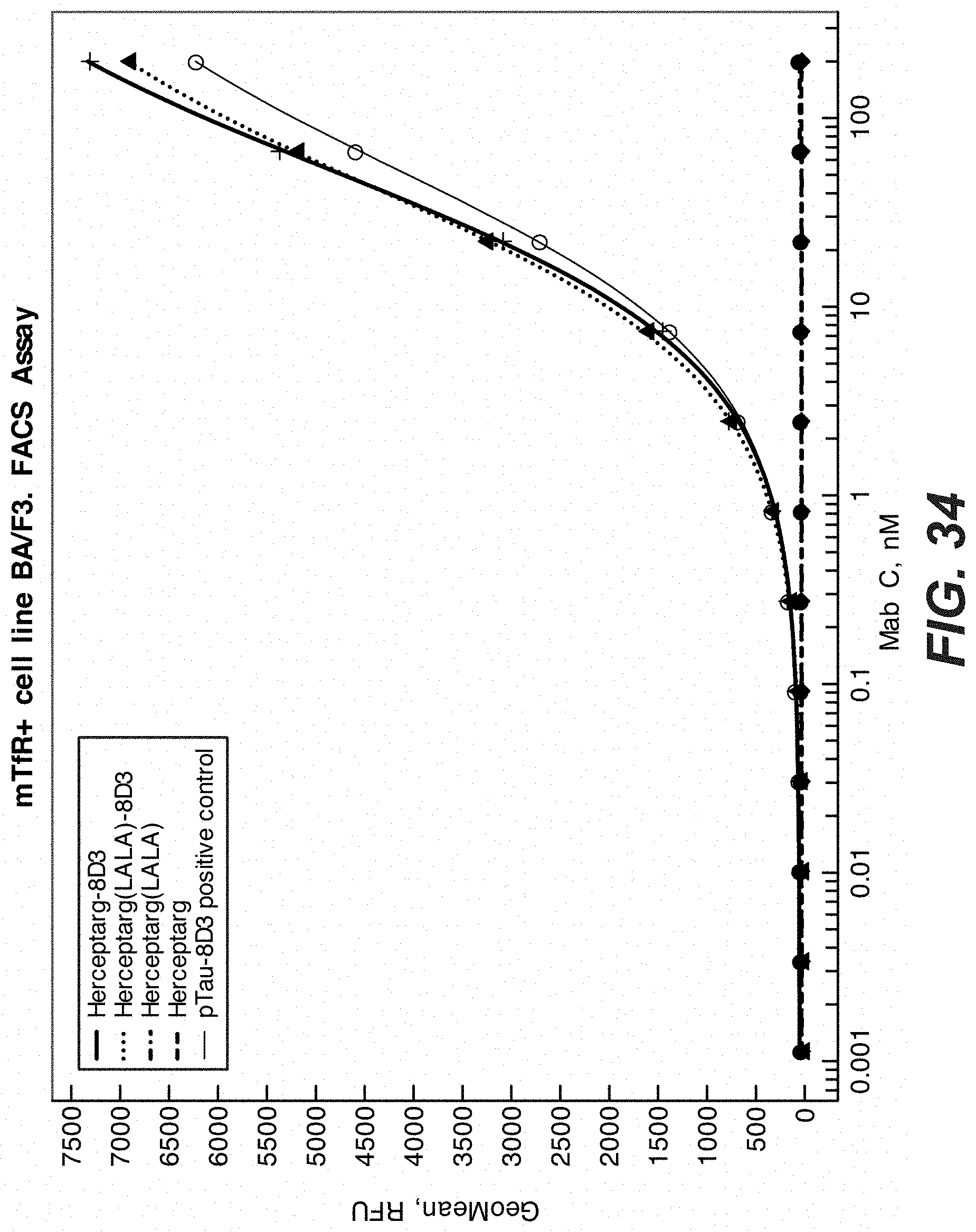
D00058

D00059

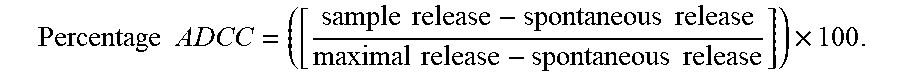



P00001
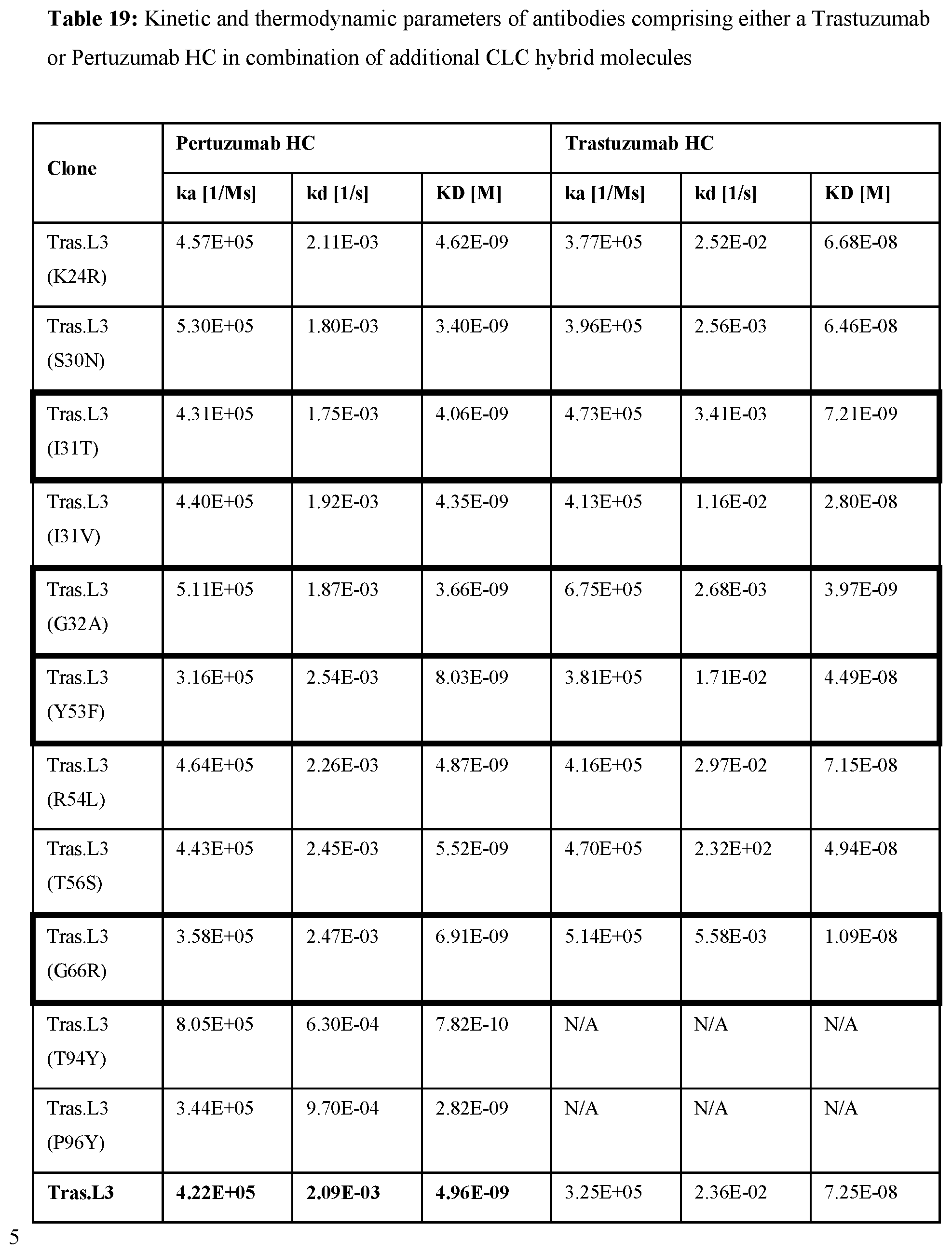
S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.