Eribulin-based Antibody-drug Conjugates And Methods Of Use
Albone; Earl F. ; et al.
U.S. patent application number 16/702720 was filed with the patent office on 2020-09-24 for eribulin-based antibody-drug conjugates and methods of use. This patent application is currently assigned to EISAI R&D MANAGEMENT CO., LTD.. The applicant listed for this patent is EISAI R&D MANAGEMENT CO., LTD.. Invention is credited to Earl F. Albone, Xin Cheng, Daniel W. Custar, Keiji Furuuchi, Jing Li, Utpal Majumder, Toshimitsu Uenaka.
| Application Number | 20200297860 16/702720 |
| Document ID | / |
| Family ID | 1000004882066 |
| Filed Date | 2020-09-24 |












View All Diagrams
| United States Patent Application | 20200297860 |
| Kind Code | A1 |
| Albone; Earl F. ; et al. | September 24, 2020 |
ERIBULIN-BASED ANTIBODY-DRUG CONJUGATES AND METHODS OF USE
Abstract
Linker toxins and antibody-drug conjugates that bind to human oncology antigen targets such as folate receptor alpha and/or provide anti-tubulin drug activity are disclosed. The linker toxins and antibody-drug conjugates comprise an eribulin drug moiety and can be internalized into target antigen-expressing cells. The disclosure further relates to methods and compositions for use in the treatment of cancer by administering the antibody-drug conjugates provided herein.
| Inventors: | Albone; Earl F.; (Blue Bell, PA) ; Cheng; Xin; (Wallingford, PA) ; Custar; Daniel W.; (North Andover, MA) ; Furuuchi; Keiji; (Wynnewood, PA) ; Li; Jing; (Andover, MA) ; Majumder; Utpal; (Andover, MA) ; Uenaka; Toshimitsu; (West Chester, PA) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Assignee: | EISAI R&D MANAGEMENT CO.,
LTD. |
||||||||||
| Family ID: | 1000004882066 | ||||||||||
| Appl. No.: | 16/702720 | ||||||||||
| Filed: | December 4, 2019 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 15448497 | Mar 2, 2017 | 10548986 | ||
| 16702720 | ||||
| 62302562 | Mar 2, 2016 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C07K 2317/14 20130101; C07K 16/30 20130101; C07K 2317/40 20130101; C07K 2317/565 20130101; A61K 31/357 20130101; C07K 2317/77 20130101; A61K 47/6803 20170801; A61K 47/6849 20170801; A61K 47/6889 20170801; C07K 16/32 20130101; C07K 2317/92 20130101; C07K 16/28 20130101 |
| International Class: | A61K 47/68 20060101 A61K047/68; C07K 16/30 20060101 C07K016/30; A61K 31/357 20060101 A61K031/357; C07K 16/28 20060101 C07K016/28; C07K 16/32 20060101 C07K016/32 |
Claims
1-183. (canceled)
184. An antibody-drug conjugate of Formula (I): Ab-(L-D).sub.p (I) wherein Ab is an internalizing anti-folate receptor alpha antibody or internalizing antigen-binding fragment thereof comprising three heavy chain complementarity determining regions (HCDRs) comprising amino acid sequences of SEQ ID NO:2 (HCDR1), SEQ ID NO:3 (HCDR2), and SEQ ID NO:4 (HCDR3); and three light chain complementarity determining regions (LCDRs) comprising amino acid sequences of SEQ ID NO:7 (LCDR1), SEQ ID NO:8 (LCDR2), and SEQ ID NO:9 (LCDR3), as defined by the Kabat numbering system; or three heavy chain complementarity determining regions (HCDRs) comprising amino acid sequences of SEQ ID NO:13 (HCDR1), SEQ ID NO:14 (HCDR2), and SEQ ID NO:15 (HCDR3); and three light chain complementarity determining regions (LCDRs) comprising amino acid sequences of SEQ ID NO:16 (LCDR1), SEQ ID NO:17 (LCDR2), and SEQ ID NO:18 (LCDR3), as defined by the IMGT numbering system; D is eribulin; L is a cleavable linker comprising an antibody attachment group, a spacer unit comprising at least one polyethylene glycol (PEG) moiety, and a cleavable amino acid unit; and p is an integer from 1 to 8.
185. The antibody-drug conjugate of claim 184, wherein the spacer unit comprises -(PEG).sub.m- and m is an integer from 1 to 10.
186. The antibody-drug conjugate of claim 185, wherein m is an integer from 2 to 8.
187. The antibody-drug conjugate of claim 185, wherein m is 2.
188. The antibody-drug conjugate of claim 184, wherein the spacer unit is attached to the antibody or antigen-binding fragment through the antibody attachment group.
189. The antibody-drug conjugate of claim 184, wherein the antibody attachment group comprises a maleimide (Mal).
190. The antibody-drug conjugate of claim 184, wherein the antibody attachment group is attached to a cysteine residue on the antibody or antigen-binding fragment.
191. The antibody-drug conjugate of claim 184, wherein the spacer unit is attached to the cleavable amino acid unit.
192. The antibody-drug conjugate of claim 184, wherein the cleavable amino acid unit comprises valine (Val) attached to citrulline (Cit).
193. The antibody-drug conjugate of claim 184, wherein the cleavable linker comprises the antibody attachment group attached to the spacer unit attached to the cleavable amino acid unit.
194. The antibody-drug conjugate of claim 193, wherein the cleavable linker comprises Mal-spacer unit-Val-Cit.
195. The antibody-drug conjugate of claim 193, wherein the cleavable linker comprises Mal-(PEG).sub.2-Val-Cit.
196. The antibody-drug conjugate of claim 184, wherein the cleavable amino acid unit is covalently attached to eribulin directly or through an optional additional spacer unit.
197. The antibody-drug conjugate of claim 196, wherein the additional spacer unit is self-immolative.
198. The antibody-drug conjugate of claim 196, wherein the additional spacer unit comprises a p-aminobenzyloxycarbonyl (pAB).
199. The antibody-drug conjugate of claim 184, wherein the cleavable linker is covalently attached to eribulin via a C-35 amine.
200. The antibody-drug conjugate of claim 184, wherein the antibody or antigen-binding fragment comprises a heavy chain variable region comprising an amino acid sequence of SEQ ID NO:23, and a light chain variable region comprising an amino acid sequence of SEQ ID NO:24.
201. The antibody-drug conjugate of claim 184, wherein the antibody or antigen-binding fragment comprises a human IgG1 heavy chain constant domain.
202. The antibody-drug conjugate of claim 184, wherein the antibody or antigen-binding fragment comprises a human Ig kappa light chain constant domain.
203. The antibody-drug conjugate of claim 184, wherein p is 3 or 4.
204. A composition comprising multiple copies of the antibody-drug conjugate of claim 184, wherein the average p of the antibody-drug conjugates in the composition is from about 3.2 to about 4.4.
205. A pharmaceutical composition comprising the antibody-drug conjugate of claim 184, and a pharmaceutically acceptable carrier.
206. A method of treating a cancer in a patient, comprising administering to the patient a therapeutically effective amount of the antibody-drug conjugate of claim 184, wherein the cancer expresses folate receptor alpha.
207. The method of claim 206, wherein the cancer is a gastric cancer, an ovarian cancer, a lung cancer, a colorectal cancer, a breast cancer, an endometrial cancer, or an osteosarcoma.
208. A method of producing the antibody-drug conjugate of claim 184, comprising reacting the antibody or antigen-binding fragment with the cleavable linker and eribulin under conditions that allow conjugation.
Description
[0001] The present application is a continuation of U.S. patent application Ser. No. 15/448,497, filed Mar. 2, 2017, and claims the benefit of priority to U.S. Provisional Patent Application No. 62/302,562, filed Mar. 2, 2016, the entire contents of which are incorporated herein by reference.
[0002] The instant application contains a Sequence Listing which has been submitted electronically in ASCII format and is hereby incorporated by reference in its entirety. Said ASCII copy, created on Mar. 28, 2017, is named 08061 0024-00304 SL.txt and is 230,910 bytes in size.
[0003] The present disclosure relates to antibody drug conjugates (ADCs) that bind human oncology antigen targets such as folate receptor alpha and/or provide anti-tubulin drug activity. The disclosure further relates to methods and compositions useful in the treatment and diagnosis of cancers that express folate receptor alpha and/or are amenable to treatment by disrupting tubulin.
[0004] Cancer is among the leading causes of morbidity and mortality worldwide, with approximately 14 million new cases and 8.2 million cancer-related deaths in 2012. The most common causes of cancer death are cancers of: lung (1.59 million deaths); liver (745,000 deaths); stomach (723,000 deaths); colorectal (694,000 deaths); breast (521,000 deaths); and esophagus (400,000 deaths). The number of new cancer cases is expected to rise by about 70% over the next two decades, to approximately 22 million new cancer cases per year (World Cancer Report 2014).
[0005] Microtubules are dynamic filamentous cytoskeletal proteins that are involved in a variety of cellular functions, including intracellular migration and transport, cell signaling, and the maintenance of cell shape. Microtubules also play a critical role in mitotic cell division by forming the mitotic spindle required to segregate chromosomes into two daughter cells. The biological functions of microtubules in all cells are regulated in large part by their polymerization dynamics, which occurs by the reversible, non-covalent addition of a and 13 tubulin dimers at both ends of microtubules. This dynamic behavior and resulting control over microtubule length is vital to the proper functioning of the mitotic spindle. Even minor alteration of microtubule dynamics can engage the spindle checkpoint, arrest cell cycle progression at mitosis, and subsequently lead to cell death (Mukhtar et al. (2014) Mol. Cancer Ther. 13:275-84). Due to their rapid cell division, cancer cells are generally more sensitive to compounds that bind to tubulin and disrupt its normal function, as compared to normal cells. For this reason, tubulin inhibitors and other microtubule-targeted agents have become a promising class of drugs for the treatment of cancer (Dumontet and Jordan (2010) Nat. Rev. Drug Discov. 9:790-803).
[0006] Folate receptor alpha (FRA) is a glycophosphatidylinositol (GPI)-linked membrane protein that binds folate. While the role of FRA in the biology of normal and cancerous tissue is not fully understood, it is highly over-expressed on a high percentage of ovarian cancers of epithelial origin (O'Shannessy et al. (2013) Int. J. Gynecol. Pathol. 32(3):258-68), as well as in a percentage of non-small cell lung carcinomas (Christoph et al. (2014) Clin. Lung Cancer 15(5):320-30). FRA also has limited expression in normal tissues. These properties make FRA an attractive target for cancer immunotherapy.
[0007] The proto-oncogene human epidermal growth factor receptor 2 (HER2) encodes a transmembrane tyrosine kinase receptor that belongs to the human epidermal growth factor receptor (EGFR) family (King et al. (1985) Science 229:974-6). Overexpression of HER2 enables constitutive activation of growth factor signaling pathways, such as the PI3K-AKT-mTOR pathway, and thereby serves as an oncogenic driver in several types of cancers, including approximately 20% of invasive breast carcinomas (Slamon et al. (1989) Science 244:707-12; Gajria and Chandarlapaty (2011) Expert Rev. Anticancer Ther. 11:263-75). Given that HER2 amplification mediates the transformed phenotype, HER2 is another promising target for cancer treatment.
[0008] The present disclosure provides, in part, novel compounds with biological activity against tumor cells. The compounds may inhibit tumor growth in mammals, and may be useful for treating human cancer patients.
[0009] The present disclosure more specifically relates to antibody-drug conjugate compounds that are capable of binding, internalizing, and killing tumor cells (e.g., FRA-expressing tumor cells). Antibody-drug conjugate compounds comprising a linker that attaches a drug moiety to an antibody moiety are disclosed. Antibody-drug conjugate (ADC) compounds may be represented by Formula I:
Ab-(L-D).sub.p (I)
wherein Ab is an internalizing antibody or an internalizing antigen-binding fragment thereof which targets a tumor cell; D is eribulin; L is a cleavable linker that covalently attaches Ab to D; and p is an integer from 1 to 20.
[0010] In some embodiments, the linker is stable outside a cell, such that the ADC remains intact when present in extracellular conditions but is capable of being cleaved on internalization in a cell, e.g., a cancer cell. In some embodiments, the eribulin drug moiety is cleaved from the antibody moiety when the ADC enters a cell that expresses an antigen specific for the antibody moiety of the ADC, and cleavage releases an unmodified form of eribulin. In some embodiments, the linker comprises a cleavable moiety that is positioned such that no part of the linker or the antibody moiety remains bound to the eribulin drug moiety upon cleavage.
[0011] In some embodiments, the cleavable moiety in the linker is a cleavable peptide moiety. In some embodiments, an ADC that comprises a cleavable peptide moiety demonstrates lower aggregation levels, improved antibody:drug ratio, increased on-target killing of cancer cells, decreased off-target killing of non-cancer cells, and/or higher drug loading (p) relative to an ADC that comprises an alternate cleavable moiety. In some embodiments, adding a cleavable moiety increases cytotoxicity and/or potency relative to a non-cleavable linker. In some embodiments, the increased potency and/or cytotoxicity is in a cancer expressing moderate levels of the antigen targeted by the antibody moiety of the ADC (e.g., moderate FRA expression). In some embodiments, the cleavable peptide moiety is cleavable by an enzyme, and the linker is an enzyme-cleavable linker. In some embodiments, the enzyme is cathepsin, and the linker is a cathepsin-cleavable linker. In certain embodiments, the enzyme-cleavable linker (e.g., the cathepsin-cleavable linker) exhibits one or more of the improved properties mentioned above, as compared to an alternate cleavage mechanism.
[0012] In some embodiments, the cleavable peptide moiety in the linker comprises an amino acid unit. In some embodiments, the amino acid unit comprises valine-citrulline (Val-Cit). In some embodiments, an ADC that comprises Val-Cit demonstrates increased stability, decreased off-target cell killing, increased on-target cell killing, lower aggregation levels, and/or higher drug loading relative to an ADC that comprises an alternate amino acid unit or alternate cleavable moiety.
[0013] In some embodiments, the linker comprises at least one spacer unit joining the antibody moiety to the cleavable moiety. In some embodiments, the spacer unit in the linker may comprise at least one polyethylene glycol (PEG) moiety. The PEG moiety may, for example, comprise -(PEG).sub.m-, wherein m is an integer from 1 to 10. In some embodiments, the spacer unit in the linker comprises (PEG).sub.2. In some embodiments, an ADC that comprises a shorter spacer unit (e.g., (PEG).sub.2) demonstrates lower aggregation levels and/or higher drug loading relative to an ADC that comprises a longer spacer unit (e.g., (PEG).sub.8) despite the shorter linker length.
[0014] In some embodiments, the spacer unit in the linker attaches to the antibody moiety of the ADC via a maleimide moiety (Mal). In some embodiments, an ADC that comprises a linker attached to the antibody moiety via a Mal demonstrates higher drug loading relative to an ADC that comprises a linker attached to the antibody moiety via an alternate moiety. In some embodiments, the Mal in the linker is reactive with a cysteine residue on the antibody moiety. In some embodiments, the Mal in the linker is joined to the antibody moiety via a cysteine residue. In some embodiments, the Mal-spacer unit comprises a PEG moiety. In some embodiments, the linker comprises Mal-(PEG).sub.m, e.g., Mal-(PEG).sub.2. In some embodiments, the linker comprises Mal-(PEG).sub.2. In some embodiments, the Mal-spacer unit attaches the antibody moiety to the cleavable moiety in the linker. In some embodiments, the cleavable moiety in the linker is a cleavable peptide moiety, e.g., an amino acid unit. In some embodiments, the linker comprises Mal-(PEG).sub.2-Val-Cit.
[0015] In some embodiments, the cleavable moiety in the linker is directly joined to the eribulin drug moiety of the ADC, and the cleavable moiety is either directly connected to the antibody moiety or connected through a spacer unit. In some embodiments, a spacer unit also attaches the cleavable moiety in the linker to the eribulin drug moiety. In some embodiments, the spacer unit that attaches the cleavable moiety in the linker to the eribulin drug moiety is self-immolative. In some embodiments, the self-immolative spacer is capable of releasing unmodified eribulin in a target cell. In some embodiments, the self-immolative spacer unit comprises a p-aminobenzyl alcohol. In some embodiments, the self-immolative spacer unit comprises p-aminobenzyloxycarbonyl (pAB). The pAB in the linker, in some embodiments, attaches the cleavable moiety to the eribulin drug moiety. In some embodiments, the cleavable moiety is a cleavable peptide moiety, e.g., an amino acid unit. In some embodiments, the linker comprises Val-Cit-pAB. In some embodiments, the linker comprises Val-Cit-pAB and a PEG spacer unit joining the linker to the antibody moiety through a Mal.
[0016] In some embodiments, p is an integer from 1 to 6, from 2 to 5, or preferably, from 3 to 4. In the some embodiments, p is 4. In some embodiments, a pool of ADCs are provided, and the average p in the pool is about 4 (e.g., 3.5-4.5, such as about 3.8). In some embodiments, the linker comprises Mal-(PEG).sub.2-Val-Cit-pAB. In some embodiments, the linker comprises Mal-(PEG).sub.2-Val-Cit-pAB and p is 4. In some embodiments, a pool of ADCs are provided, wherein each ADC comprises a Mal-(PEG).sub.2-Val-Cit-pAB linker, and the average p in the pool is about 4 (e.g., 3.5-4.5, such as about 3.8).
[0017] In some embodiments, the internalizing antibody or internalizing antigen-binding fragment (Ab or Ab moiety) of the ADC is an anti-folate receptor alpha (FRA) antibody or internalizing antibody fragment, and can bind FRA-expressing tumor cells (i.e., the ADC targets FRA-expressing cells). In some embodiments, the ADC comprising an anti-FRA Ab moiety and a cleavable peptide moiety demonstrates lower aggregation levels, improved antibody:drug ratio, increased on-target killing of cancer cells, decreased off-target killing of non-cancer cells, higher drug loading (p), increased cytotoxicity, and/or potency relative to a non-cleavable linker or an alternate cleavage mechanism. In some embodiments, the increased potency and/or cytotoxicity is in a cancer expressing moderate levels of the antigen targeted by the antibody moiety of the ADC (e.g., moderate FRA expression). In some embodiments, the cleavable peptide moiety is cleavable by an enzyme, and the linker is an enzyme-cleavable linker. In some embodiments, the enzyme is cathepsin, and the linker is a cathepsin-cleavable linker. In certain embodiments, the enzyme-cleavable linker (e.g., the cathepsin-cleavable linker) exhibits one or more of the improved properties mentioned above, as compared to an alternate cleavage mechanism. In some embodiments, the linker is a Mal-(PEG).sub.m-Val-Cit-pAB.
[0018] In some embodiments, the internalizing antibody or internalizing antigen-binding fragment binds to folate receptor alpha (FRA) and targets FRA-expressing tumor cells. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment comprises three heavy chain complementarity determining regions (CDRs) and three light chain CDRs, wherein the heavy chain CDRs comprise heavy chain CDR1 consisting of SEQ ID NO:2, heavy chain CDR2 consisting of SEQ ID NO:3, and heavy chain CDR3 consisting of SEQ ID NO:4; and the three light chain CDRs comprise light chain CDR1 consisting of SEQ ID NO:7, light chain CDR2 consisting of SEQ ID NO:8, and light chain CDR3 consisting of SEQ ID NO:9, as defined by the Kabat numbering system; or wherein the heavy chain CDRs comprise heavy chain CDR1 consisting of SEQ ID NO:13, heavy chain CDR2 consisting of SEQ ID NO:14, and heavy chain CDR3 consisting of SEQ ID NO:15; and the light chain CDRs comprise light chain CDR1 consisting of SEQ ID NO:16, light chain CDR2 consisting of SEQ ID NO:17, and light chain CDR3 consisting of SEQ ID NO:18, as defined by the IMGT numbering system. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment comprises human framework sequences. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment comprises a heavy chain variable domain of SEQ ID NO:23 and a light chain variable domain of SEQ ID NO:24. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment comprises a human IgG1 heavy chain constant domain and an Ig kappa light chain constant domain. In some embodiments, the internalizing antibody or internalizing antigen-binding competes for binding and/or binds the same epitope as an antibody comprising a heavy chain variable domain of SEQ ID NO:23 and a light chain variable domain of SEQ ID NO:24. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment binds to an epitope comprising alanine-histadine-lysine-aspartic acid (AHKD) (SEQ ID NO:365) (O'Shannessy et al., (2011) Oncotarget 2:1227-43). In some embodiments, the internalizing antibody or internalizing antigen-binding fragment binds to an epitope comprising NTSQEAHKDVSYL (SEQ ID NO:366).
[0019] In some embodiments, the internalizing antibody or internalizing antigen-binding fragment is an internalizing anti-FRA antibody or internalizing antigen-binding fragment. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment comprises three heavy chain CDRs and three light chain CDRs, wherein the heavy chain CDRs comprise heavy chain CDR1 consisting of SEQ ID NO:2, heavy chain CDR2 consisting of SEQ ID NO:3, and heavy chain CDR3 consisting of SEQ ID NO:4; and the three light chain CDRs comprise light chain CDR1 consisting of SEQ ID NO:7, light chain CDR2 consisting of SEQ ID NO:8, and light chain CDR3 consisting of SEQ ID NO:9, as defined by the Kabat numbering system; or wherein the heavy chain CDRs comprise heavy chain CDR1 consisting of SEQ ID NO:13, heavy chain CDR2 consisting of SEQ ID NO:14, and heavy chain CDR3 consisting of SEQ ID NO:15; and the light chain CDRs comprise light chain CDR1 consisting of SEQ ID NO:16, light chain CDR2 consisting of SEQ ID NO:17, and light chain CDR3 consisting of SEQ ID NO:18, as defined by the IMGT numbering system; the linker comprises Mal-(PEG).sub.2-Val-Cit-pAB; and p is 4. In some embodiments, a pool of such ADCs are provided and p is about 4 (e.g., about 3.8). In some embodiments, the internalizing antibody or internalizing antigen-binding fragment comprises a heavy chain variable domain of SEQ ID NO:23 and a light chain variable domain of SEQ ID NO:24. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment comprises a human IgG1 heavy chain constant domain and an Ig kappa light chain constant domain. In some embodiments, the internalizing antibody or internalizing antigen-binding competes for binding and/or binds the same epitope as an antibody comprising a heavy chain variable domain of SEQ ID NO:23 and a light chain variable domain of SEQ ID NO:24. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment binds to an epitope comprising SEQ ID NO:365. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment binds to an epitope comprising SEQ ID NO:366.
[0020] In some embodiments, the internalizing antibody or internalizing antigen-binding fragment binds to human epidermal growth factor receptor 2 (her2) and targets her2-expressing tumor cells. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment comprises three heavy chain complementarity determining regions (CDRs) and three light chain CDRs, wherein the heavy chain CDRs comprise heavy chain CDR1 consisting of SEQ ID NO:71 heavy chain CDR2 consisting of SEQ ID NO:72, and heavy chain CDR3 consisting of SEQ ID NO:73; and the three light chain CDRs comprise light chain CDR1 consisting of SEQ ID NO:74, light chain CDR2 consisting of SEQ ID NO:75, and light chain CDR3 consisting of SEQ ID NO:76, as defined by the Kabat numbering system; or wherein the heavy chain CDRs comprise heavy chain CDR1 consisting of SEQ ID NO:191, heavy chain CDR2 consisting of SEQ ID NO:192, and heavy chain CDR3 consisting of SEQ ID NO:193; and the light chain CDRs comprise light chain CDR1 consisting of SEQ ID NO:194, light chain CDR2 consisting of SEQ ID NO:195, and light chain CDR3 consisting of SEQ ID NO:196, as defined by the IMGT numbering system. In some embodiments, the antibody or internalizing antigen-binding fragment comprises human framework sequences. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment comprises a heavy chain variable domain of SEQ ID NO:27 and a light chain variable domain of SEQ ID NO:28. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment comprises a human IgG1 heavy chain constant domain and an Ig kappa light chain constant domain. In some embodiments, the internalizing antibody or internalizing antigen-binding competes for binding and/or binds the same epitope as an antibody comprising a heavy chain variable domain of SEQ ID NO:27 and a light chain variable domain of SEQ ID NO:28.
[0021] In some embodiments, the internalizing antibody or internalizing antigen-binding fragment is an internalizing anti-her2 antibody or internalizing antigen-binding fragment. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment comprises three heavy chain CDRs and three light chain CDRs, wherein the heavy chain CDRs comprise heavy chain CDR1 consisting of SEQ ID NO:71 heavy chain CDR2 consisting of SEQ ID NO:72, and heavy chain CDR3 consisting of SEQ ID NO:73; and the three light chain CDRs comprise light chain CDR1 consisting of SEQ ID NO:74, light chain CDR2 consisting of SEQ ID NO:75, and light chain CDR3 consisting of SEQ ID NO:76, as defined by the Kabat numbering system; or wherein the heavy chain CDRs comprise heavy chain CDR1 consisting of SEQ ID NO:191, heavy chain CDR2 consisting of SEQ ID NO:192, and heavy chain CDR3 consisting of SEQ ID NO:193; and the light chain CDRs comprise light chain CDR1 consisting of SEQ ID NO:194, light chain CDR2 consisting of SEQ ID NO:195, and light chain CDR3 consisting of SEQ ID NO:196, as defined by the IMGT numbering system; the linker comprises Mal-(PEG).sub.2-Val-Cit-pAB; and p is 4. In some embodiments, a pool of such ADCs are provided and p is about 4 (e.g., about 3.8). In some embodiments, the internalizing antibody or internalizing antigen-binding fragment comprises a heavy chain variable domain of SEQ ID NO:27 and a light chain variable domain of SEQ ID NO:28. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment comprises a human IgG1 heavy chain constant domain and an Ig kappa light chain constant domain. In some embodiments, the internalizing antibody or internalizing antigen-binding competes for binding and/or binds the same epitope as an antibody comprising a heavy chain variable domain of SEQ ID NO:27 and a light chain variable domain of SEQ ID NO:28.
[0022] In some embodiments, the internalizing antibody or internalizing antigen-binding fragment binds to mesothelin (MSLN) and targets MSLN-expressing tumor cells. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment comprises three heavy chain complementarity determining regions (CDRs) and three light chain CDRs, wherein the heavy chain CDRs comprise heavy chain CDR1 consisting of SEQ ID NO:65 heavy chain CDR2 consisting of SEQ ID NO:66, and heavy chain CDR3 consisting of SEQ ID NO:67; and the three light chain CDRs comprise light chain CDR1 consisting of SEQ ID NO:68, light chain CDR2 consisting of SEQ ID NO:69, and light chain CDR3 consisting of SEQ ID NO:70, as defined by the Kabat numbering system; or wherein the heavy chain CDRs comprise heavy chain CDR1 consisting of SEQ ID NO:185, heavy chain CDR2 consisting of SEQ ID NO:186, and heavy chain CDR3 consisting of SEQ ID NO:187; and the light chain CDRs comprise light chain CDR1 consisting of SEQ ID NO:188, light chain CDR2 consisting of SEQ ID NO:189, and light chain CDR3 consisting of SEQ ID NO:190, as defined by the IMGT numbering system. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment comprises a heavy chain variable domain of SEQ ID NO:25 and a light chain variable domain of SEQ ID NO:26. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment comprises a human IgG1 heavy chain constant domain and an Ig kappa light chain constant domain. In some embodiments, the internalizing antibody or internalizing antigen-binding competes for binding and/or binds the same epitope as an antibody comprising a heavy chain variable domain of SEQ ID NO:25 and a light chain variable domain of SEQ ID NO:26.
[0023] In some embodiments, the internalizing antibody or internalizing antigen-binding fragment is an internalizing anti-MSLN antibody or internalizing antigen-binding fragment. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment comprises three heavy chain CDRs and three light chain CDRs, wherein the heavy chain CDRs comprise heavy chain CDR1 consisting of SEQ ID NO:65 heavy chain CDR2 consisting of SEQ ID NO:66, and heavy chain CDR3 consisting of SEQ ID NO:67; and the three light chain CDRs comprise light chain CDR1 consisting of SEQ ID NO:68, light chain CDR2 consisting of SEQ ID NO:69, and light chain CDR3 consisting of SEQ ID NO:70, as defined by the Kabat numbering system; or wherein the heavy chain CDRs comprise heavy chain CDR1 consisting of SEQ ID NO:185, heavy chain CDR2 consisting of SEQ ID NO:186, and heavy chain CDR3 consisting of SEQ ID NO:187; and the light chain CDRs comprise light chain CDR1 consisting of SEQ ID NO:188, light chain CDR2 consisting of SEQ ID NO:189, and light chain CDR3 consisting of SEQ ID NO:190, as defined by the IMGT numbering system; the linker comprises Mal-(PEG).sub.2-Val-Cit-pAB; and p is 4. In some embodiments, a pool of such ADCs are provided and p is about 4 (e.g., about 3.8). In some embodiments, the internalizing antibody or internalizing antigen-binding fragment comprises a heavy chain variable domain of SEQ ID NO:25 and a light chain variable domain of SEQ ID NO:26. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment comprises a human IgG1 heavy chain constant domain and an Ig kappa light chain constant domain. In some embodiments, the internalizing antibody or internalizing antigen-binding competes for binding and/or binds the same epitope as an antibody comprising a heavy chain variable domain of SEQ ID NO:25 and a light chain variable domain of SEQ ID NO:26.
[0024] Also provided herein are compositions comprising multiple copies of any of the described ADCs, wherein the average drug loading (average p) of the ADCs in the composition is between about 3 and 4, or about 3.5 to about 4.5, or about 4. In some embodiments, the average p is between about 3.2 and 3.8. In some embodiments, the average p is between about 3.6 and 4.4.
[0025] Also provided herein are compositions comprising -L-D, wherein D is eribulin; and L is a cleavable linker that covalently attaches to D. In some embodiments, the cleavable linker covalently attaches to the C-35 amine on eribulin. In some embodiments, the cleavable linker comprises Val-Cit. In some embodiments, the cleavable linker comprises a PEG spacer unit. In some embodiments, the cleavable linker comprises Mal-(PEG).sub.2-Val-Cit-pAB.
[0026] Further provided herein are pharmaceutical compositions comprising an ADC and a pharmaceutically acceptable diluent, carrier, and/or excipient.
[0027] Another aspect of the present disclosure includes therapeutic and diagnostic uses for the described ADC compounds and compositions, e.g., in treating cancer. Another aspect includes methods of treating a cancer that expresses an antigen targeted by the antibody moiety of the ADC, such as FRA. In various embodiments, methods are provided of killing or inhibiting the proliferation of tumor cells or cancer cells by administering a therapeutically effective amount and/or regimen of any one of the described ADCs. Another aspect includes methods for detecting tumor cells or cancer cells that express FRA using the disclosed ADCs, and methods of screening for cancer patients that will be responsive to treatment with the described ADCs. In some embodiments, the cancer is a gastric cancer, a serous ovarian cancer, a clear cell ovarian cancer, a non-small cell lung cancer, a colorectal cancer, a triple negative breast cancer, an endometrial cancer, a serous endometrial carcinoma, a lung carcinoid, or an osteosarcoma. Methods of producing the described ADCs are also disclosed.
BRIEF DESCRIPTION OF THE DRAWINGS
[0028] FIG. 1 shows one of the methodologies used to prepare MORAb-003 ADCs, as disclosed in certain embodiments. In this approach, unpaired cysteines are generated through partial reduction with limited molar equivalents of the non-thiol reducing agent TCEP. This approach preferentially reduces the interchain disulfide bonds that link the light chain and heavy chain (one pair per H-L pairing) and the two heavy chains in the hinge region (two pairs per H-H pairing in the case of human IgG1), while leaving the intrachain disulfide bonds intact.
[0029] FIG. 2 shows a method of synthesizing maleimide-(PEG).sub.2-Val-Cit-pAB-eribulin (mal-(PEG).sub.2-VCP-eribulin), as disclosed in certain embodiments.
[0030] FIG. 3 shows an SDS-PAGE analysis of reduction conditions for MORAb-003. Lanes are indicated to the right of the figure. Lane M corresponds to protein standard; lane 1 corresponds to untreated MORAb-003; lane 2 corresponds to 5.3 mg/mL reduced in 70.6 .mu.M TCEP; lane 3 corresponds to MORAb-003 5.3 mg/mL reduced in 141.2 .mu.M TCEP; lane 4 corresponds to MORAb-003 1.5 mg/mL reduced in 20 .mu.M TCEP; and lane 5 corresponds to MORAb-003 1.5 mg/mL reduced in 40 .mu.M TCEP. Identities of each band are indicated on the lower right gel. "H" indicates heavy chain. "L" indicates light chain.
[0031] FIG. 4 shows an SDS-PAGE analysis of reduction conditions for MORAb-003. Lane 1 corresponds to protein standard; lane 2 corresponds to untreated MORAb-003; lane 3 corresponds to MORAb-003 treated at a ratio of MORAb-003:TCEP of 1:1; lane 4 corresponds to MORAb-003 treated at a ratio of MORAb-003:TCEP of 1:2; lane 5 corresponds to MORAb-003 treated at a ratio of MORAb-003:TCEP of 1:3; and lane 6 corresponds to MORAb-003 treated at a ratio of MORAb-003:TCEP of 1:4.
[0032] FIG. 5 shows a non-reducing SDS-PAGE analysis of select MORAb-003 ADCs, including M-MMAE (lane 2), M-DM1 (lane 3), M-0026 (lane 4), M-0260 (lane 5), M-0267 (lane 6), M-0272 (lane 7), M-0285 (lane 8), M-0292 (lane 9), M-027-0381 (lane 10), and M-0284 (lane 11).
[0033] FIG. 6A shows the results of a bystander cytotoxicity assay of MORAb-003-maleimido-PEG2-Val-Cit-pAB-eribulin (M3-VCP-eribulin, or "MORAb-202"). FIG. 6B shows the results of a bystander cytotoxicity assay of MORAb-003-maleimido-(CH.sub.2).sub.5-Val-Cit-pAB-ER-001150828 (M3-ER-61318). FIG. 6C shows the results of a bystander cytotoxicity assay of MORAb-003-PEG-pAB-duostatin 3 (M3-027-0285). The information shown in the respective figure legends provides cell line:agent tested (cell line/cell lines cultured, seeding density of 1.sup.st/2.sup.nd cell line).
[0034] FIGS. 7A and 7B show drug-to-antibody ratio (DAR) distribution for ADCs MORAb-003-VCP-eribulin (FIG. 7A) and MORAb-003-0285 (FIG. 7B) relative to unconjugated MORAb-003, as disclosed in certain embodiments. Numbers over each peak indicate the DAR of the individual species.
[0035] FIG. 8 shows the results of a cytotoxicity analysis--competition of MORAb-003-VCP-eribulin with unconjugated MORAb-003 (2 .mu.M) in IGROV1 or SJSA-1 cells.
[0036] FIG. 9 shows body weight kinetics for each group of CD-1 mice (group average and SEM) treated with a single intravenous dose of vehicle (PBS), or MORAb-202 at 10, 20, 40, or 80 mg/kg.
[0037] FIG. 10 shows body weight kinetics for each group of CD-1 mice (group average and SEM) treated intravenously with PBS, or with eribulin at 0.4, 0.8, 1.6, or 3.2 mg/kg, according to a q4dx3 dosing regimen (doses administered once every four days for 3 doses total).
[0038] FIG. 11 shows tumor growth kinetics for each group of CB17-SCID mice implanted with hNSCLC NCI-H2110 cells (group average and SEM) and treated with a single intravenous dose of PBS, MORAb-003-VCP-eribulin (MORAb-202) at 1, 2.5, or 5 mg/kg, or MORAb-003-0285 at 5 mg/kg.
[0039] FIG. 12 shows tumor volumes of individual CB17-SCID mice implanted with hNSCLC NCI-H2110 cells, as well as group average and SEM, on day 17. Groups were treated with a single intravenous dose of PBS, MORAb-003-VCP-eribulin (MORAb-202) at 1, 2.5, or 5 mg/kg, or MORAb-003-0285 at 5 mg/kg.
[0040] FIG. 13 shows body weight kinetics for each group of NCI-H2110-implanted CB17-SCID mice (group average and SEM) treated with a single intravenous dose of PBS, MORAb-003-VCP-eribulin (MORAb-202) at 1, 2.5, or 5 mg/kg, or MORAb-003-0285 at 5 mg/kg.
[0041] FIG. 14 shows tumor growth kinetics for each group of NCI-H2110-implanted CB17-SCID mice (group average and SEM) treated intravenously with vehicle (PBS), or with eribulin at 0.5, 0.2, 0.8, or 1.6 mg/kg, according to a q4dx3 dosing regimen.
[0042] FIG. 15 shows tumor volumes of individual NCI-H2110-implanted CB17-SCID mice, as well as group average and SEM, on day 24. Groups were treated intravenously with vehicle (PBS), or with eribulin at 0.5, 0.2, 0.8, or 1.6 mg/kg, according to a q4dx3 dosing regimen.
[0043] FIG. 16 shows body weight change kinetics for each group of NCI-H2110-implanted CB17-SCID mice (group average and SEM) treated intravenously with vehicle (PBS), or with eribulin at 0.5, 0.2, 0.8, or 1.6 mg/kg, according to a q4dx3 dosing regimen.
[0044] FIG. 17 shows the potency of MORAb-003-VCP-eribulin (MORAb-202) on IGROV1, OVCAR3, NCI-H2110, A431-A3, and SJSA-1 cells, as measured by Crystal Violet cytotoxicity assay.
[0045] FIG. 18 shows tumor growth kinetics for each group of NCI-H2110-implanted CB17-SCID mice (group average and SEM) treated with a single intravenous dose of PBS, or MORAb-003-VCP-eribulin (MORAb-202) at 1, 2.5, or 5 mg/kg.
[0046] FIGS. 19A and 19B show tumor growth kinetics (FIG. 19A) and body weight change kinetics (FIG. 19B) for each group of NSCLC PDx (LXFA-737) tumor-bearing mice (group average and SEM) treated with a single intravenous dose of vehicle (PBS), MORAb-003 at 5 mg/kg, or MORAb-003-VCP-eribulin (MORAb-202) at 5 mg/kg.
[0047] FIGS. 20A and 20B show individual tumor volume ratios (FIG. 20A) and body weight change kinetics (FIG. 20B) for each group of endometrial cancer PDx (Endo-12961) tumor-bearing mice (group average and SEM) treated with a single intravenous dose of PBS, eribulin at 0.1 or 3.2 mg/kg, or MORAb-003-VCP-eribulin (MORAb-202) at 5 mg/kg. FIGS. 20C and 20D show tumor growth kinetics (FIG. 20C) and body weight change kinetics (FIG. 20D) for each group of endometrial cancer PDx (Endo-10590) tumor-bearing mice (group average and SEM) treated with a single intravenous dose of PBS, eribulin at 0.1 or 3.2 mg/kg, or MORAb-003-VCP-eribulin (MORAb-202) at 5 mg/kg.
[0048] FIG. 21A shows immunohistochemical (IHC) staining of tumor tissue in TNBC PDx (OD-BRE-0631) tumor-bearing mice with an anti-human IgG antibody. Tumor tissues from mice treated with a single intravenous dose of vehicle (right), or MORAb-003-VCP-eribulin (MORAb-202) at 5 mg/kg (left), were collected and stained 5 days post-treatment. FIG. 21B shows IHC staining of tumor tissue in TNBC PDx (OD-BRE-0631) tumor-bearing mice with an .alpha.-smooth muscle actin (SMA)-FITC antibody. Tumor tissues from untreated mice were collected 2 days prior to treatment (left), whereas tumor tissues from mice treated with a single intravenous dose of MORAb-003-VCP-eribulin (MORAb-202) at 5 mg/kg were collected 5 days post-treatment (right). FIG. 21C shows tumor growth kinetics for each group of TNBC PDx (OD-BRE-0631) tumor-bearing mice (group average and SEM) treated with a single intravenous dose of vehicle (PBS), or MORAb-003-VCP-eribulin (MORAb-202) at 5 mg/kg.
[0049] FIG. 22 shows the differentiation of human bone marrow-mesenchymal stem cells (BM-MSCs) in culture with MKN-74 cells following treatment with vehicle (PBS or ethanol), eribulin, MORAb-003, or MORAb-003-VCP-eribulin (MORAb-202), as measured by flow cytometry analysis. Stro-1.sup.+/CD105.sup.+, CD34.sup.+/CD31.sup.-, and NG2.sup.+ are markers of MSCs, adipocytes, and pericytes, respectively.
[0050] FIG. 23 shows the time course analysis of tumor tissues from NCI-H2110-implanted CB17-SCID mice treated with a single intravenous dose of vehicle (PBS), or MORAb-003-VCP-eribulin (MORAb-202) at 5 mg/kg, stained with an .alpha.-smooth muscle actin (SMA)-FITC antibody. Tumor tissues were collected and stained at day 0, and at days 3, 5, 7 and 9 post-treatment. Y-axis: %=[stained cells counted/total cells counted]*100. X-axis: day (total cells counted).
DETAILED DESCRIPTION OF ILLUSTRATIVE EMBODIMENTS
[0051] The disclosed compositions and methods may be understood more readily by reference to the following detailed description taken in connection with the accompanying figures, which form a part of this disclosure. It is to be understood that the disclosed compositions and methods are not limited to the specific compositions and methods described and/or shown herein, and that the terminology used herein is for the purpose of describing particular embodiments by way of example only and is not intended to be limiting of the claimed compositions and methods.
[0052] Throughout this text, the descriptions refer to compositions and methods of using said compositions. Where the disclosure describes or claims a feature or embodiment associated with a composition, such a feature or embodiment is equally applicable to the methods of using said composition. Likewise, where the disclosure describes or claims a feature or embodiment associated with a method of using a composition, such a feature or embodiment is equally applicable to the composition.
[0053] When a range of values is expressed, it includes embodiments using any particular value within the range. Further, reference to values stated in ranges includes each and every value within that range. All ranges are inclusive of their endpoints and combinable. When values are expressed as approximations, by use of the antecedent "about," it will be understood that the particular value forms another embodiment. Reference to a particular numerical value includes at least that particular value, unless the context clearly dictates otherwise. The use of "or" will mean "and/or" unless the specific context of its use dictates otherwise. All references cited herein are incorporated by reference for any purpose. Where a reference and the specification conflict, the specification will control.
[0054] It is to be appreciated that certain features of the disclosed compositions and methods, which are, for clarity, described herein in the context of separate embodiments, may also be provided in combination in a single embodiment. Conversely, various features of the disclosed compositions and methods that are, for brevity, described in the context of a single embodiment, may also be provided separately or in any subcombination.
Definitions
[0055] Various terms relating to aspects of the description are used throughout the specification and claims. Such terms are to be given their ordinary meaning in the art unless otherwise indicated. Other specifically defined terms are to be construed in a manner consistent with the definitions provided herein.
[0056] As used herein, the singular forms "a," "an," and "the" include plural forms unless the context clearly dictates otherwise.
[0057] The terms "about" or "approximately" in the context of numerical values and ranges refers to values or ranges that approximate or are close to the recited values or ranges such that the embodiment may perform as intended, such as having a desired amount of nucleic acids or polypeptides in a reaction mixture, as is apparent to the skilled person from the teachings contained herein. This is due, at least in part, to the varying properties of nucleic acid compositions, age, race, gender, anatomical and physiological variations and the inexactitude of biological systems. Thus, these terms encompass values beyond those resulting from systematic error.
[0058] The terms "antibody-drug conjugate," "antibody conjugate," "conjugate," "immunoconjugate," and "ADC" are used interchangeably, and refer to a compound or derivative thereof that is linked to an antibody (e.g., an anti-FRA antibody) and is defined by the generic formula: Ab-(L-D).sub.p (Formula I), wherein Ab=an antibody moiety (i.e., antibody or antigen-binding fragment), L=a linker moiety, D=a drug moiety, and p=the number of drug moieties per antibody moiety.
[0059] The term "antibody" is used in the broadest sense to refer to an immunoglobulin molecule that recognizes and specifically binds to a target, such as a protein, polypeptide, carbohydrate, polynucleotide, lipid, or combinations of the foregoing through at least one antigen recognition site within the variable region of the immunoglobulin molecule. The heavy chain of an antibody is composed of a heavy chain variable domain (V.sub.H) and a heavy chain constant region (C.sub.H). The light chain is composed of a light chain variable domain (V.sub.L) and a light chain constant domain (C.sub.L). For the purposes of this application, the mature heavy chain and light chain variable domains each comprise three complementarity determining regions (CDR1, CDR2 and CDR3) within four framework regions (FR1, FR2, FR3 and FR4) arranged from N-terminus to C-terminus: FR1, CDR1, FR2, CDR2, FR3, CDR3 and FR4. An "antibody" can be naturally occurring or man-made, such as monoclonal antibodies produced by conventional hybridoma technology. The term "antibody" includes full-length monoclonal antibodies and full-length polyclonal antibodies, as well as antibody fragments such as Fab, Fab', F(ab')2, Fv, and single chain antibodies. An antibody can be any one of the five major classes of immunoglobulins: IgA, IgD, IgE, IgG, and IgM, or subclasses thereof (e.g., isotypes IgG1, IgG2, IgG3, IgG4). The term further encompasses human antibodies, chimeric antibodies, humanized antibodies and any modified immunoglobulin molecule containing an antigen recognition site, so long as it demonstrates the desired biological activity.
[0060] The term "monoclonal antibody," as used herein, refers to an antibody obtained from a population of substantially homogeneous antibodies, i.e., the individual antibodies comprising the population are identical except for possible naturally occurring mutations that may be present in minor amounts. Monoclonal antibodies are highly specific, being directed against a single antigenic epitope. In contrast, conventional (polyclonal) antibody preparations typically include a multitude of antibodies directed against (or specific for) different epitopes. The modifier "monoclonal" indicates the character of the antibody as being obtained from a substantially homogeneous population of antibodies, and is not to be construed as requiring production of the antibody by any particular method. For example, the monoclonal antibodies to be used in accordance with the present disclosure may be made by the hybridoma method first described by Kohler et al. (1975) Nature 256:495, or may be made by recombinant DNA methods (see, e.g., U.S. Pat. No. 4,816,567). Monoclonal antibodies may also be isolated from phage antibody libraries using the techniques described in Clackson et al. (1991) Nature 352:624-8, and Marks et al. (1991) J. Mol. Biol. 222:581-97, for example.
[0061] The monoclonal antibodies described herein specifically include "chimeric" antibodies, in which a portion of the heavy and/or light chain is identical with or homologous to corresponding sequences in antibodies derived from a particular species or belonging to a particular antibody class or subclass, while the remainder of the chain(s) is identical with or homologous to corresponding sequences in antibodies derived from another species or belonging to another antibody class or subclass, as well as fragments of such antibodies, so long as they specifically bind the target antigen and/or exhibit the desired biological activity.
[0062] The term "human antibody," as used herein, refers an antibody produced by a human or an antibody having an amino acid sequence of an antibody produced by a human.
[0063] The term "chimeric antibody," as used herein, refers to antibodies wherein the amino acid sequence of the immunoglobulin molecule is derived from two or more species. In some instances, the variable regions of both heavy and light chains corresponds to the variable regions of antibodies derived from one species with the desired specificity, affinity, and activity while the constant regions are homologous to antibodies derived from another species (e.g., human) to minimize an immune response in the latter species.
[0064] As used herein, the term "humanized antibody" refers to forms of antibodies that contain sequences from non-human (e.g., murine) antibodies as well as human antibodies. Such antibodies are chimeric antibodies which contain minimal sequence derived from non-human immunoglobulin. In general, the humanized antibody will comprise substantially all of at least one, and typically two, variable domains, in which all or substantially all of the hypervariable loops correspond to those of a non-human immunoglobulin and all or substantially all of the framework (FR) regions are those of a human immunoglobulin sequence. The humanized antibody optionally also will comprise at least a portion of an immunoglobulin constant region (Fc), typically that of a human immunoglobulin. The humanized antibody can be further modified by the substitution of residues, either in the Fv framework region and/or within the replaced non-human residues to refine and optimize antibody specificity, affinity, and/or activity.
[0065] The term "antigen-binding fragment" or "antigen-binding portion" of an antibody, as used herein, refers to one or more fragments of an antibody that retain the ability to specifically bind to an antigen (e.g., FRA). Antigen-binding fragments preferably also retain the ability to internalize into an antigen-expressing cell. In some embodiments, antigen-binding fragments also retain immune effector activity. It has been shown that fragments of a full-length antibody can perform the antigen-binding function of a full-length antibody. Examples of binding fragments encompassed within the term "antigen-binding fragment" or "antigen-binding portion" of an antibody include (i) a Fab fragment, a monovalent fragment consisting of the V.sub.L, V.sub.H, C.sub.L, and C.sub.H1 domains; (ii) a F(ab').sub.2 fragment, a bivalent fragment comprising two Fab fragments linked by a disulfide bridge at the hinge region; (iii) a Fd fragment consisting of the V.sub.H and C.sub.H1 domains; (iv) a Fv fragment consisting of the V.sub.L and V.sub.H domains of a single arm of an antibody; (v) a dAb fragment, which comprises a single variable domain, e.g., a V.sub.H domain (see, e.g., Ward et al. (1989) Nature 341:544-6; and Winter et al., WO 90/05144); and (vi) an isolated complementarity determining region (CDR). Furthermore, although the two domains of the Fv fragment, V.sub.L and V.sub.H, are coded for by separate genes, they can be joined, using recombinant methods, by a synthetic linker that enables them to be made as a single protein chain in which the V.sub.L and V.sub.H regions pair to form monovalent molecules (known as single chain Fv (scFv)). See, e.g., Bird et al. (1988) Science 242:423-6; and Huston et al. (1988) Proc. Natl. Acad. Sci. USA 85:5879-83. Such single chain antibodies are also intended to be encompassed within the term "antigen-binding fragment" or "antigen-binding portion" of an antibody, and are known in the art as an exemplary type of binding fragment that can internalize into cells upon binding. See, e.g., Zhu et al. (2010) 9:2131-41; He et al. (2010) J. Nucl. Med. 51:427-32; and Fitting et al. (2015) MAbs 7:390-402. In certain embodiments, scFv molecules may be incorporated into a fusion protein. Other forms of single chain antibodies, such as diabodies are also encompassed. Diabodies are bivalent, bispecific antibodies in which V.sub.H and V.sub.L domains are expressed on a single polypeptide chain, but using a linker that is too short to allow for pairing between the two domains on the same chain, thereby forcing the domains to pair with complementary domains of another chain and creating two antigen binding sites (see e.g., Holliger et al. (1993) Proc. Natl. Acad. Sci. USA 90:6444-8; and Poljak et al. (1994) Structure 2:1121-3). Antigen-binding fragments are obtained using conventional techniques known to those of skill in the art, and the binding fragments are screened for utility (e.g., binding affinity, internalization) in the same manner as are intact antibodies. Antigen-binding fragments may be prepared by cleavage of the intact protein, e.g., by protease or chemical cleavage.
[0066] "Internalizing" as used herein in reference to an antibody or antigen-binding fragment refers to an antibody or antigen-binding fragment that is capable of being taken through the cell's lipid bilayer membrane to an internal compartment (i.e., "internalized") upon binding to the cell, preferably into a degradative compartment in the cell. For example, an internalizing anti-FRA antibody is one that is capable of being taken into the cell after binding to FRA on the cell membrane.
[0067] The term "folate receptor alpha" or "FRA," as used herein, refers to any native form of human FRA. The term encompasses full-length FRA (e.g., NCBI Reference Sequence: NP_000793; SEQ ID NO: 19), as well as any form of human FRA that results from cellular processing. The term also encompasses naturally occurring variants of FRA, including but not limited to splice variants, allelic variants, and isoforms. FRA can be isolated from a human, or may be produced recombinantly or by synthetic methods.
[0068] The term "anti-FRA antibody" or "antibody that specifically binds FRA" refers to any form of antibody or fragment thereof that specifically binds FRA, and encompasses monoclonal antibodies (including full length monoclonal antibodies), polyclonal antibodies, and biologically functional antibody fragments so long as they specifically bind FRA. Preferably the anti-FRA antibody used in the ADCs disclosed herein is an internalizing antibody or internalizing antibody fragment. MORAb-003 is an exemplary internalizing anti-human FRA antibody. As used herein, the terms "specific," "specifically binds," and "binds specifically" refer to the selective binding of the antibody to the target antigen epitope. Antibodies can be tested for specificity of binding by comparing binding to appropriate antigen to binding to irrelevant antigen or antigen mixture under a given set of conditions. If the antibody binds to the appropriate antigen with at least 2, 5, 7, and preferably 10 times more affinity than to irrelevant antigen or antigen mixture, then it is considered to be specific. In one embodiment, a specific antibody is one that only binds the FRA antigen, but does not bind (or exhibits minimal binding) to other antigens.
[0069] The term "human epidermal growth factor receptor 2," "her2," or "her2/neu," as used herein, refers to any native form of human her2. The term encompasses full-length her2 (e.g., NCBI Reference Sequence: NP_004439.2; SEQ ID NO: 21), as well as any form of human her2 that results from cellular processing. The term also encompasses naturally occurring variants of her2, including but not limited to splice variants, allelic variants, and isoforms. Her2 can be isolated from human, or may be produced recombinantly or by synthetic methods.
[0070] The term "anti-her2 antibody" or "antibody that specifically binds her2" refers to any form of antibody or fragment thereof that specifically binds her2, and encompasses monoclonal antibodies (including full length monoclonal antibodies), polyclonal antibodies, and biologically functional antibody fragments so long as they specifically bind her2. U.S. Pat. No. 5,821,337 (incorporated herein by reference) provides exemplary her2-binding sequences, including exemplary anti-her2 antibody sequences. Preferably the anti-her2 antibody used in the ADCs disclosed herein is an internalizing antibody or internalizing antibody fragment. Trastuzumab is an exemplary internalizing anti-human her2 antibody.
[0071] The term "epitope" refers to the portion of an antigen capable of being recognized and specifically bound by an antibody. When the antigen is a polypeptide, epitopes can be formed from contiguous amino acids or noncontiguous amino acids juxtaposed by tertiary folding of the polypeptide. The epitope bound by an antibody may be identified using any epitope mapping technique known in the art, including X-ray crystallography for epitope identification by direct visualization of the antigen-antibody complex, as well as monitoring the binding of the antibody to fragments or mutated variations of the antigen, or monitoring solvent accessibility of different parts of the antibody and the antigen. Exemplary strategies used to map antibody epitopes include, but are not limited to, array-based oligo-peptide scanning, limited proteolysis, site-directed mutagenesis, high-throughput mutagenesis mapping, hydrogen-deuterium exchange, and mass spectrometry (see, e.g., Gershoni et al. (2007) 21:145-56; and Hager-Braun and Tomer (2005) Expert Rev. Proteomics 2:745-56).
[0072] Competitive binding and epitope binning can also be used to determine antibodies sharing identical or overlapping epitopes. Competitive binding can be evaluated using a cross-blocking assay, such as the assay described in "Antibodies, A Laboratory Manual," Cold Spring Harbor Laboratory, Harlow and Lane (1.sup.st edition 1988, 2.sup.nd edition 2014). In some embodiments, competitive binding is identified when a test antibody or binding protein reduces binding of a reference antibody or binding protein to a target antigen such as FRA or her2 (e.g., a binding protein comprising CDRs and/or variable domains selected from those identified in Tables 2, 4, and 6), by at least about 50% in the cross-blocking assay (e.g., 50%, 60%, 70%, 80%, 90%, 95%, 99%, 99.5%, or more, or any percentage in between), and/or vice versa. In some embodiments, competitive binding can be due to shared or similar (e.g., partially overlapping) epitopes, or due to steric hindrance where antibodies or binding proteins bind at nearby epitopes. See, e.g., Tzartos, Methods in Molecular Biology (Morris, ed. (1998) vol. 66, pp. 55-66). In some embodiments, competitive binding can be used to sort groups of binding proteins that share similar epitopes, e.g., those that compete for binding can be "binned" as a group of binding proteins that have overlapping or nearby epitopes, while those that do not compete are placed in a separate group of binding proteins that do not have overlapping or nearby epitopes.
[0073] The term "k.sub.on" or "k.sub.a" refers to the on rate constant for association of an antibody to the antigen to form the antibody/antigen complex. The rate can be determined using standard assays, such as a Biacore or ELISA assay.
[0074] The term "k.sub.off" or "k.sub.a" refers to the off rate constant for dissociation of an antibody from the antibody/antigen complex. The rate can be determined using standard assays, such as a Biacore or ELISA assay.
[0075] The term "K.sub.D" refers to the equilibrium dissociation constant of a particular antibody-antigen interaction. K.sub.D is calculated by k.sub.a/k.sub.d. The rate can be determined using standard assays, such as a Biacore or ELISA assay.
[0076] The term "p" or "antibody:drug ratio" or "drug-to-antibody ratio" or "DAR" refers to the number of drug moieties per antibody moiety, i.e., drug loading, or the number of -L-D moieties per antibody or antigen-binding fragment (Ab) in ADCs of Formula I. In compositions comprising multiple copies of ADCs of Formula I, "p" refers to the average number of -L-D moieties per antibody or antigen-binding fragment, also referred to as average drug loading.
[0077] A "linker" or "linker moiety" is any chemical moiety that is capable of covalently joining a compound, usually a drug moiety such as a chemotherapeutic agent, to another moiety such as an antibody moiety. Linkers can be susceptible to or substantially resistant to acid-induced cleavage, peptidase-induced cleavage, light-based cleavage, esterase-induced cleavage, and/or disulfide bond cleavage, at conditions under which the compound or the antibody remains active.
[0078] The term "agent" is used herein to refer to a chemical compound, a mixture of chemical compounds, a biological macromolecule, or an extract made from biological materials. The term "therapeutic agent," "drug," or "drug moiety" refers to an agent that is capable of modulating a biological process and/or has biological activity.
[0079] The term "chemotherapeutic agent" or "anti-cancer agent" is used herein to refer to all chemical compounds that are effective in treating cancer regardless of mechanism of action. Inhibition of metastasis or angiogenesis is frequently a property of a chemotherapeutic agent. Non-limiting examples of chemotherapeutic agents include alkylating agents, for example, nitrogen mustards, ethyleneimine compounds, and alkyl sulphonates; antimetabolites, for example, folic acid, purine or pyrimidine antagonists; anti-mitotic agents, for example, anti-tubulin agents such as eribulin or eribulin mesylate (Halaven.TM.) or derivatives thereof, vinca alkaloids, and auristatins; cytotoxic antibiotics; compounds that damage or interfere with DNA expression or replication, for example, DNA minor groove binders; and growth factor receptor antagonists. In addition, chemotherapeutic agents include antibodies, biological molecules, and small molecules. A chemotherapeutic agent may be a cytotoxic or cytostatic agent. The term "cytostatic agent" refers to an agent that inhibits or suppresses cell growth and/or multiplication of cells.
[0080] The term "cytotoxic agent" refers to a substance that causes cell death primarily by interfering with a cell's expression activity and/or functioning. Examples of cytotoxic agents include, but are not limited to, anti-mitotic agents, such as eribulin, auristatins (e.g., monomethyl auristatin E (MMAE), monomethyl auristatin F (MMAF)), maytansinoids (e.g., maytansine), dolastatins, duostatins, cryptophycins, vinca alkaloids (e.g., vincristine, vinblastine), taxanes, taxols, and colchicines; anthracyclines (e.g., daunorubicin, doxorubicin, dihydroxyanthracindione); cytotoxic antibiotics (e.g., mitomycins, actinomycins, duocarmycins (e.g., CC-1065), auromycins, duomycins, calicheamicins, endomycins, phenomycins); alkylating agents (e.g., cisplatin); intercalating agents (e.g., ethidium bromide); topoisomerase inhibitors (e.g., etoposide, tenoposide); radioisotopes, such as At.sup.211, I.sup.131, I.sup.125, Y.sup.90, Re.sup.186, Re.sup.188, Sm.sup.153, Bi.sup.212 or .sup.213, P.sup.32, and radioactive isotopes of lutetium (e.g., Lu.sup.177); and toxins of bacterial, fungal, plant or animal origin (e.g., ricin (e.g., ricin A-chain), diphtheria toxin, Pseudomonas exotoxin A (e.g., PE40), endotoxin, mitogellin, combrestatin, restrictocin, gelonin, alpha-sarcin, abrin (e.g., abrin A-chain), modeccin (e.g., modeccin A-chain), curicin, crotin, Sapaonaria officinalis inhibitor, glucocorticoid).
[0081] The term "eribulin," as used herein, refers to a synthetic analog of halichondrin B, a macrocyclic compound that was originally isolated from the marine sponge Halichondria okadais. The term "eribulin drug moiety" refers to the component of an ADC that has the structure of eribulin, and is attached to the linker of the ADC via its C-35 amine. Eribulin is a microtubule dynamics inhibitor, which is thought to bind tubulin and induce cell cycle arrest at the G2/M phase by inhibiting mitotic spindle assembly. The term "eribulin mesylate" refers to the mesylate salt of eribulin, which is marketed under the trade name Halaven.TM..
[0082] The term "homolog" refers to a molecule which exhibits homology to another molecule, by for example, having sequences of chemical residues that are the same or similar at corresponding positions.
[0083] The term "inhibit" or "inhibition of," as used herein, means to reduce by a measurable amount, and can include but does not require complete prevention or inhibition.
[0084] The term "target-negative" or "target antigen-negative" refers to the absence of target antigen expression by a cell or tissue. The term "target-positive" or "target antigen-positive" refers to the presence of target antigen expression. For example, a cell or a cell line that does not express a target antigen may be described as target-negative, whereas a cell or cell line that expresses a target antigen may be described as target-positive.
[0085] The term "bystander killing" or "bystander effect" refers to the killing of target-negative cells in the presence of target-positive cells, wherein killing of target-negative cells is not observed in the absence of target-positive cells. Cell-to-cell contact, or at least proximity between target-positive and target-negative cells, enables bystander killing. This type of killing is distinguishable from "off-target killing," which refers to the indiscriminate killing of target-negative cells. "Off-target killing" may be observed in the absence of target-positive cells.
[0086] The term "cancer" refers to the physiological condition in mammals in which a population of cells is characterized by unregulated cell growth. Examples of cancers include, but are not limited to, carcinoma, lymphoma, blastoma, sarcoma, and leukemia. More particular examples of such cancers include squamous cell cancer, small cell lung cancer, nonsmall cell lung cancer, adenocarcinoma of the lung, squamous carcinoma of the lung, cancer of the peritoneum, hepatocellular cancer, gastrointestinal cancer, pancreatic cancer, glioblastoma, cervical cancer, ovarian cancer, liver cancer, bladder cancer, hepatoma, breast cancer (e.g., triple negative breast cancer), osteosarcoma, melanoma, colon cancer, colorectal cancer, endometrial (e.g., serous) or uterine cancer, salivary gland carcinoma, kidney cancer, liver cancer, prostate cancer, vulval cancer, thyroid cancer, hepatic carcinoma, and various types of head and neck cancers. Triple negative breast cancer refers to breast cancer that is negative for expression of the genes for estrogen receptor (ER), progesterone receptor (PR), or Her2/neu.
[0087] The terms "tumor" and "neoplasm" refer to any mass of tissue that results from excessive cell growth or proliferation, either benign or malignant, including precancerous lesions.
[0088] The terms "cancer cell" and "tumor cell" refer to individual cells or the total population of cells derived from a tumor, including both non-tumorigenic cells and cancer stem cells. As used herein, the term "tumor cell" will be modified by the term "non-tumorigenic" when referring solely to those tumor cells lacking the capacity to renew and differentiate to distinguish those tumor cells from cancer stem cells.
[0089] The terms "subject" and "patient" are used interchangeably herein to refer to any animal, such as any mammal, including but not limited to, humans, non-human primates, rodents, and the like. In some embodiments, the mammal is a mouse. In some embodiments, the mammal is a human.
[0090] The term "co-administration" or administration "in combination with" one or more therapeutic agents includes concurrent and consecutive administration in any order.
[0091] A "pharmaceutical composition" refers to a preparation which is in such form as to permit administration and subsequently provide the intended biological activity of the active ingredient(s) and/or to achieve a therapeutic effect, and which contains no additional components which are unacceptably toxic to a subject to which the formulation would be administered. The pharmaceutical composition may be sterile.
[0092] A "pharmaceutical excipient" comprises a material such as an adjuvant, a carrier, pH-adjusting and buffering agents, tonicity adjusting agents, wetting agents, preservative, and the like.
[0093] "Pharmaceutically acceptable" means approved or approvable by a regulatory agency of the Federal or a state government, or listed in the U.S. Pharmacopeia or other generally recognized pharmacopeia, for use in animals, and more particularly in humans.
[0094] An "effective amount" of an ADC as disclosed herein is an amount sufficient to perform a specifically stated purpose, for example to produce a therapeutic effect after administration, such as a reduction in tumor growth rate or tumor volume, a reduction in a symptom of cancer, or some other indicia of treatment efficacy. An effective amount can be determined in a routine manner in relation to the stated purpose. The term "therapeutically effective amount" refers to an amount of an ADC effective to treat a disease or disorder in a subject. In the case of cancer, a therapeutically effective amount of ADC can reduce the number of cancer cells, reduce tumor size, inhibit (e.g., slow or stop) tumor metastasis, inhibit (e.g., slow or stop) tumor growth, and/or relieve one or more symptoms. A "prophylactically effective amount" refers to an amount effective, at dosages and for periods of time necessary, to achieve the desired prophylactic result. Typically, since a prophylactic dose is used in subjects prior to or at an earlier stage of disease, the prophylactically effective amount will be less than the therapeutically effective amount.
[0095] As used herein, "to treat" or "therapeutic" and grammatically related terms, refer to any improvement of any consequence of disease, such as prolonged survival, less morbidity, and/or a lessening of side effects which are the byproducts of an alternative therapeutic modality. As is readily appreciated in the art, full eradication of disease is a preferred but albeit not a requirement for a treatment act. "Treatment" or "treat," as used herein, refers to the administration of a described ADC to a subject, e.g., a patient. The treatment can be to cure, heal, alleviate, relieve, alter, remedy, ameliorate, palliate, improve or affect the disorder, the symptoms of the disorder or the predisposition toward the disorder, e.g., a cancer.
[0096] In some embodiments, a labeled ADC is used. Suitable "labels" include radionuclides, enzymes, substrates, cofactors, inhibitors, fluorescent moieties, chemiluminescent moieties, magnetic particles, and the like.
[0097] By "protein," as used herein, is meant at least two covalently attached amino acids. The term encompasses polypeptides, oligopeptides, and peptides. In some embodiments, the two or more covalently attached amino acids are attached by a peptide bond. The protein may be made up of naturally occurring amino acids and peptide bonds, for example when the protein is made recombinantly using expression systems and host cells. Alternatively, the protein may include synthetic amino acids (e.g., homophenylalanine, citrulline, ornithine, and norleucine), or peptidomimetic structures, i.e., "peptide or protein analogs," such as peptoids. Peptoids are an exemplary class of peptidomimetics whose side chains are appended to the nitrogen atom of the peptide backbone, rather than to the .alpha.-carbons (as they are in amino acids), and have different hydrogen bonding and conformational characteristics in comparison to peptides (see, e.g., Simon et al. (1992) Proc. Natl. Acad. Sci. USA 89:9367). As such, peptoids can be resistant to proteolysis or other physiological or storage conditions, and effective at permeating cell membranes. Such synthetic amino acids may be incorporated in particular when the antibody is synthesized in vitro by conventional methods well known in the art. In addition, any combination of peptidomimetic, synthetic and naturally occurring residues/structures can be used. "Amino acid" also includes imino acid residues, such as proline and hydroxyproline. The amino acid "R group" or "side chain" may be in either the (L)- or the (S)-configuration. In a specific embodiment, the amino acids are in the (L)- or (5)-configuration.
[0098] A "recombinant protein" is a protein made using recombinant techniques using any techniques and methods known in the art, i.e., through the expression of a recombinant nucleic acid. Methods and techniques for the production of recombinant proteins are well known in the art.
[0099] An "isolated" protein is unaccompanied by at least some of the material with which it is normally associated in its natural state, for example constituting at least about 5%, or at least about 50% by weight of the total protein in a given sample. It is understood that the isolated protein may constitute from 5 to 99.9% by weight of the total protein content depending on the circumstances. For example, the protein may be made at a significantly higher concentration through the use of an inducible promoter or high expression promoter, such that the protein is made at increased concentration levels. The definition includes the production of an antibody in a wide variety of organisms and/or host cells that are known in the art.
[0100] For amino acid sequences, sequence identity and/or similarity may be determined using standard techniques known in the art, including, but not limited to, the local sequence identity algorithm of Smith and Waterman (1981) Adv. Appl. Math. 2:482, the sequence identity alignment algorithm of Needleman and Wunsch (1970) J. Mol. Biol. 48:443, the search for similarity method of Pearson and Lipman (1988) Proc. Nat. Acad. Sci. USA 85:2444, computerized implementations of these algorithms (GAP, BESTFIT, FASTA, and TFASTA in the Wisconsin Genetics Software Package, Genetics Computer Group, 575 Science Drive, Madison, Wis.), the Best Fit sequence program described by Devereux et al. (1984) Nucl. Acid Res. 12:387-95, preferably using the default settings, or by inspection. Preferably, percent identity is calculated by FastDB based upon the following parameters: mismatch penalty of 1; gap penalty of 1; gap size penalty of 0.33; and joining penalty of 30 ("Current Methods in Sequence Comparison and Analysis," Macromolecule Sequencing and Synthesis, Selected Methods and Applications, pp. 127-149 (1988), Alan R. Liss, Inc).
[0101] An example of a useful algorithm is PILEUP. PILEUP creates a multiple sequence alignment from a group of related sequences using progressive, pairwise alignments. It can also plot a tree showing the clustering relationships used to create the alignment. PILEUP uses a simplification of the progressive alignment method of Feng & Doolittle (1987) J. Mol. Evol. 35:351-60; the method is similar to that described by Higgins and Sharp (1989) CABIOS 5:151-3. Useful PILEUP parameters including a default gap weight of 3.00, a default gap length weight of 0.10, and weighted end gaps.
[0102] Another example of a useful algorithm is the BLAST algorithm, described in: Altschul et al. (1990) J. Mol. Biol. 215:403-10; Altschul et al. (1997) Nucleic Acids Res. 25:3389-402; and Karin et al. (1993) Proc. Natl. Acad. Sci. USA 90:5873-87. A particularly useful BLAST program is the WU-BLAST-2 program which was obtained from Altschul et al. (1996) Methods in Enzymology 266:460-80. WU-BLAST-2 uses several search parameters, most of which are set to the default values. The adjustable parameters are set with the following values: overlap span=1, overlap fraction=0.125, word threshold (T)=II. The HSP S and HSP S2 parameters are dynamic values and are established by the program itself depending upon the composition of the particular sequence and composition of the particular database against which the sequence of interest is being searched; however, the values may be adjusted to increase sensitivity.
[0103] An additional useful algorithm is gapped BLAST as reported by Altschul et al. (1993) Nucl. Acids Res. 25:3389-402. Gapped BLAST uses BLOSUM-62 substitution scores; threshold T parameter set to 9; the two-hit method to trigger ungapped extensions, charges gap lengths of k a cost of 10+k; Xu set to 16, and Xg set to 40 for database search stage and to 67 for the output stage of the algorithms. Gapped alignments are triggered by a score corresponding to about 22 bits.
[0104] Generally, the amino acid homology, similarity, or identity between proteins disclosed herein and variants thereof, including variants of FRA, variants of her2, variants of tubulin sequences, and variants of antibody variable domains (including individual variant CDRs), are at least 80% to the sequences depicted herein, and more typically with preferably increasing homologies or identities of at least 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, and almost 100% or 100%.
[0105] In a similar manner, "percent (%) nucleic acid sequence identity" with respect to the nucleic acid sequence of the antibodies and other proteins identified herein is defined as the percentage of nucleotide residues in a candidate sequence that are identical with the nucleotide residues in the coding sequence of the antigen binding protein. A specific method utilizes the BLASTN module of WU-BLAST-2 set to the default parameters, with overlap span and overlap fraction set to 1 and 0.125, respectively.
[0106] While the site or region for introducing an amino acid sequence variation is predetermined, the mutation per se need not be predetermined. For example, in order to optimize the performance of a mutation at a given site, random mutagenesis may be conducted at the target codon or region and the expressed antigen binding protein CDR variants screened for the optimal combination of desired activity. Techniques for making substitution mutations at predetermined sites in DNA having a known sequence are well known, for example, MI3 primer mutagenesis and PCR mutagenesis.
Antibody-Drug Conjugates
[0107] The compounds of the present disclosure include those with anti-cancer activity. In particular, the compounds include an antibody moiety (including an antigen-binding fragment thereof) conjugated (i.e., covalently attached by a linker) to a drug moiety, wherein the drug moiety when not conjugated to an antibody moiety has a cytotoxic or cytostatic effect. In various embodiments, the drug moiety exhibits reduced or no cytotoxicity when bound in a conjugate but resumes cytotoxicity after cleavage from the linker and antibody moiety. In various embodiments, the drug moiety exhibits reduced or no bystander killing when bound in a conjugate (e.g., using a non-cleavable linker) but exhibits increased bystander killing after cleavage from a conjugate (e.g., a conjugate having a cleavable Val-Cit cleavable moiety).
[0108] The development and production of an ADC for use as a human therapeutic agent, e.g., as an oncologic agent, may require more than the identification of an antibody capable of binding to a desired target or targets and attaching to a drug used on its own to treat cancer. Linking the antibody to the drug may have significant and unpredictable effects on the activity of one or both of the antibody and the drug, effects which will vary depending on the type of linker and/or drug chosen. In some embodiments, therefore, the components of the ADC are selected to (i) retain one or more therapeutic properties exhibited by the antibody and drug moieties in isolation, (ii) maintain the specific binding properties of the antibody moiety; (iii) optimize drug loading and drug-to-antibody ratios; (iv) allow delivery, e.g., intracellular delivery, of the drug moiety via stable attachment to the antibody moiety; (v) retain ADC stability as an intact conjugate until transport or delivery to a target site; (vi) minimize aggregation of the ADC prior to or after administration; (vii) allow for the therapeutic effect, e.g., cytotoxic effect, of the drug moiety after cleavage in the cellular environment; (viii) exhibit in vivo anti-cancer treatment efficacy comparable to or superior to that of the antibody and drug moieties in isolation; (ix) minimize off-target killing by the drug moiety; and/or (x) exhibit desirable pharmacokinetic and pharmacodynamics properties, formulatability, and toxicologic/immunologic profiles. Screening each of these properties may be needed to identify an improved ADC for therapeutic use (Ab et al. (2015) Mol. Cancer Ther. 14:1605-13).
[0109] In various embodiments, the ADCs disclosed herein exhibit unexpectedly favorable properties in some or each of the categories listed above. For instance, in some embodiments, ADC constructs comprising a Mal attachment to an antibody, a PEG spacer unit (preferably a short PEG spacer unit), and/or peptide cleavable linker (e.g., a Val-Cit linker) exhibit surprisingly favorable drug loading, aggregation, and/or stability profiles, and/or preserve antibody binding function, drug activity, and/or improved bystander killing, while reducing off-target killing, as compared to ADCs using other cleavable or non-cleavable linker structures.
[0110] In some embodiments, an ADC comprising a Mal-(PEG).sub.2-Val-Cit-pAB linker joining eribulin to an antibody (e.g., an anti-FRA antibody such as MORAb-003) exhibits particularly favorable properties across the listed categories, as compared to other cleavable or non-cleavable linkers joining eribulin to an antibody moiety. In some embodiments, an ADC comprising a Mal-(PEG).sub.2-Val-Cit-pAB linker joining eribulin to an antibody (e.g., an anti-FRA antibody such as MORAb-003) exhibits particularly favorable bystander killing properties as compared to an uncleavable ADC. In some embodiments, an ADC comprising a Mal-(PEG).sub.2-Val-Cit-pAB linker joining eribulin to an antibody (e.g., an anti-FRA antibody such as MORAb-003) exhibits particularly favorable bystander killing properties as compared to an ADC using alternate cleavable linker structures.
[0111] In some embodiments, an ADC comprising a Mal-(PEG).sub.2-Val-Cit-pAB linker joining eribulin to MORAb-003 exhibits a higher and more desirable drug:antibody ratio (i.e., a ratio of about 3-4) relative to an ADC, e.g., comprising a linker attached to the antibody via an alternate moiety (e.g., a succinimide moiety). In some embodiments, an ADC comprising a Mal-(PEG).sub.2-Val-Cit-pAB linker joining eribulin to MORAb-003 exhibits a higher and more desirable drug:antibody ratio, and/or lower aggregation levels, relative to an ADC, e.g., comprising a longer spacer unit (e.g., (PEG).sub.8). In some embodiments, an ADC comprising a Mal-(PEG).sub.2-Val-Cit-pAB linker joining eribulin to MORAb-003 demonstrates a higher and more desirable drug:antibody ratio, lower aggregation levels, increased on-target killing, and/or decreased off-target killing relative to an ADC, e.g., comprising an alternate cleavable moiety (i.e., a non-peptide cleavable moiety, such as a cleavable disulfide or sulfonamide). In some embodiments, an ADC comprising a Mal-(PEG).sub.2-Val-Cit-pAB linker joining eribulin to MORAb-003 demonstrates increased stability, increased on-target killing, decreased off-target killing, lower aggregation levels, and/or a higher and more desirable drug:antibody ratio relative to an ADC, e.g., comprising an alternate amino acid unit (e.g., Ala-Ala-Asn) or alternate cleavable moiety (e.g., a cleavable disulfide or sulfonamide).
[0112] In some embodiments, some or all of the desirable features described above for ADCs comprising a Mal-(PEG).sub.2-Val-Cit-pAB linker joining eribulin to MORAb-003 may be observed with ADCs comprising the Mal-(PEG).sub.2-Val-Cit-pAB-eribulin linker-toxin conjugated to an anti-her2 antibody such as trastuzumab, or an anti-mesothelin antibody.
[0113] The ADC compounds of the present disclosure may selectively deliver an effective dose of a cytotoxic or cytostatic agent to cancer cells or to tumor tissue. It has been discovered that the disclosed ADCs have potent cytotoxic and/or cytostatic activity against cells expressing the respective target antigen (e.g., FRA or her2). In some embodiments, the cytotoxic and/or cytostatic activity of the ADC is dependent on the target antigen expression level in a cell. In some embodiments, the disclosed ADCs are particularly effective at killing cancer cells expressing a high level of target antigen, as compared to cancer cells expressing the same antigen at a low level. In some embodiments, the disclosed ADCs are particularly effective at killing cancer cells expressing the target antigen at a moderate level, as compared to cancer cells expressing the same antigen at a low level. Exemplary high FRA-expressing cancers include but are not limited to ovarian cancer (e.g., serous ovarian cancer, clear cell ovarian cancer), lung carcinoid, triple negative breast cancer, endometrial cancer, and nonsmall cell lung cancer (e.g., adenocarcinoma). Exemplary moderate FRA-expressing cancers include but are not limited to gastric cancer and colorectal cancer. Exemplary low FRA-expressing cancers include but are not limited to melanoma and lymphoma. Exemplary high her2-expressing cancers include but are not limited to breast cancer, gastric cancer, esophageal cancer, ovarian cancer, and endometrial cancer. Exemplary moderate her2-expressing cancers include but are not limited to lung cancer and bladder cancer.
[0114] In some embodiments, cleavage of an ADC releases eribulin from the antibody moiety and linker. In some embodiments, cleavage and release of the eribulin improves cytotoxicity of the ADC. In some embodiments, an ADC comprising a cleavable linker is particularly effective at killing cancer cells, including bystander killing, as compared to comparable treatment with an ADC comprising a non-cleavable linker. In some embodiments, an ADC comprising a cleavable linker (e.g., a Val-Cit linker) demonstrates increased on-target cell killing and/or decreased off-target cell killing relative to an ADC comprising a non-cleavable linker (e.g., a non-cleavable (PEG).sub.2 or (PEG).sub.4 linker), particularly wherein the cells and/or cancer treated with the ADC do not express high levels of the target antigen.
[0115] In some embodiments, the disclosed ADCs also demonstrate bystander killing activity, but low off-target cytotoxicity. Without being bound by theory, the bystander killing activity of an ADC may be particularly beneficial where its penetration into a solid tumor is limited and/or target antigen expression among tumor cells is heterogeneous. In some embodiments, an ADC comprising a cleavable linker is particularly effective at bystander killing and/or demonstrates improved bystander killing activity, as compared to comparable treatment with an ADC comprising a non-cleavable linker.
[0116] Provided herein are ADC compounds comprising an antibody or antigen-binding fragment thereof (Ab) which targets a tumor cell, a drug moiety (D), and a linker moiety (L) that covalently attaches Ab to D. In certain aspects, the antibody or antigen-binding fragment is able to bind to a tumor-associated antigen (e.g., FRA or her2) with high specificity and high affinity. In certain embodiments, the antibody or antigen-binding fragment is internalized into a target cell upon binding, e.g., into a degradative compartment in the cell. Preferred ADCs are thus those that internalize upon binding to a target cell, undergo degradation, and release the drug moiety to kill cancer cells. The drug moiety may be released from the antibody and/or the linker moiety of the ADC by enzymatic action, hydrolysis, oxidation, or any other mechanism.
[0117] An exemplary ADC has Formula I:
Ab-(L-D).sub.p (I)
wherein Ab=antibody moiety (i.e., antibody or antigen-binding fragment), L=linker moiety, D=drug moiety, and p=the number of drug moieties per antibody moiety.
Antibodies
[0118] The antibody moiety (Ab) of Formula I includes within its scope any antibody or antigen-binding fragment that specifically binds to a target antigen on a cancer cell. The antibody or antigen-binding fragment may bind to a target antigen with a dissociation constant (K.sub.D) of .ltoreq.1 mM, .ltoreq.100 nM or .ltoreq.10 nM, or any amount in between, as measured by, e.g., BIAcore.RTM. analysis. In certain embodiments, the K.sub.D is 1 pM to 500 pM. In some embodiments, the K.sub.D is between 500 pM to 1 .mu.M, 1 .mu.M to 100 nM, or 100 mM to 10 nM.
[0119] In some embodiments, the antibody moiety is a four-chain antibody (also referred to as an immunoglobulin), comprising two heavy chains and two light chains. In some embodiments the antibody moiety is a two-chain half body (one light chain and one heavy chain), or an antigen-binding fragment of an immunoglobulin.
[0120] In some embodiments, the antibody moiety is an internalizing antibody or internalizing antigen-binding fragment thereof. In some embodiments, the internalizing antibody binds to a target cancer antigen expressed on the surface of a cell and enters the cell upon binding. In some embodiments, the drug moiety of the ADC is released from the antibody moiety of the ADC after the ADC enters and is present in a cell expressing the target cancer antigen (i.e., after the ADC has been internalized).
[0121] Amino acid and nucleic acid sequences of exemplary antibodies of the present disclosure are set forth in Tables 1-9.
TABLE-US-00001 TABLE 1 Antibodies niAb Class/Isotype Target MORAb-003 humanized human folate receptor alpha MORAb-009 mouse-human chimeric human mesothelin trastuzumab humanized human her2/neu 33011-xi rabbit-human chimeric human mesothelin 33011-zu humanized human mesothelin 111B10-xi rabbit-human chimeric human mesothelin 111B10-zu humanized human mesothelin 201C15-xi rabbit-human chimeric human mesothelin 201C15-zu humanized human mesothelin 346C6-xi rabbit-human chimeric human mesothelin 346C6-zu humanized human mesothelin Abbreviations: xi--chimeric; zu--humanized.
TABLE-US-00002 TABLE 2 Amino acid sequences of niAb variable regions mAb IgC chain SEQ ID NO Amino acid sequence 1 MORAb-003 Heavy chain 23 EVQLVESGGGVVQPGRSLRLSCSASGFT FSGYGLSWVRQAPGKGLEWVAMISSGGS YTYYADSVKGRFAISRDNAKNTLFLQMD SLRPEDTGVYFCARHGDDPAWFAYWGQG TPVTVSS 2 MORAb-003 Light chain 24 DIQLTQSPSSLSASVGDRVTITCSVSSS ISSNNLHWYQQKPGKAPKPWIYGTSNLA SGVPSRFSGSGSGTDYTFTISSLQPEDI ATYYCQQWSSYPYMYTFGQGTKVEIK 3 MORAb-009 Heavy chain 25 QVQLQQSGPELEKPGASVKISCKASGYS FTGYTMNWVKQSHGKS1EWIGLITPYNG ASSYNQKFRGKATLTVDKSSSTAYMDLL SLTSEDSAVYFCARGGYDGRGFDYWGSG TPVTVSS 4 MORAb-009 Light chain 26 DIELTQSPAIMSASPGEKVTMTCSASSS VSYMHWYQQKSGTSPKRWIYDTSKLASG VPGRFSGSGSGNSYSLTISSVEAEDDAT YYCQQWSKHPLTFGSGTKVEIK 5 trastuzumab Heavy chain 27 EVQLVESGGGLVQPGGSLRLSCAASGFN IKDTYIHWVRQAPGKGLEWVARIYPTNG YTRYADSVKGRFTISADTSKNTAYLQMN SLRAEDTAVYYCSRWGGDGFYAMDYWGQ GTLVTVSS 6 trastuzumab Light chain 28 DIQMTQSPSSLSASVGDRVTITCRASQD VNTAVAWYQQKPGKAPKLLIYSASFLYS GVPSRFSGSRSGTDFTLTISSLQPEDFA TYYCQQHYTTPPTFGOGTKVEIK 7 33011-xi Heavy chain 29 QSVEESGGRLVTPGTPLTLTCTVSGISL SSDAISWVRQAPGKGLEYIGIINGGGNT YYASWAKGRFTISKTSTTVDLKITSPTT EDTATYFCARGIQHGGGNSDYYYYGMDL WGPGTLVTVSS 8 33011-xi Light chain 30 EVLMTQTPSSVSAAVGDTVTIKCQASQS ISSVLSWYQQKPGQPPKLLIYLASTLAS GVPSRFSGSRSGTEFTLTISDLECDDAA TYYCQTNYGTSSSNYGFAFGGGTEVWK 9 33011-zu Heavy chain 31 EVQLVESGGGLVQPGGSLRLSCAASGIS ISSDAISWVRQAPGKGLEYIGIINGGGN TYYASWAKGRFTISRHNSKNTLYLQMNS LRAEDTAVYYCARGIQHGGGNSDYYYYG MDLWGQGTLVTVSS 10 33011-zu Light chain 32 DIQMTQSPSSLSASVGDRVTITCQASQS ISSVLSWYQQKPGKAPKLLIYLASTLAS GVPSRFSGSGSGTDFTLTISSLQCEDIA TYYCQTNYGTSSSNYGFAFGGGTKVEIK 11 111B10-xi Heavy chain 33 QSVEESGGRLVTPGTPLTLTCTVSGFSL NNYAMSWVRQAPGKGLEWIGSISTGGLA FYANWAKGRFTISRTSTTVDLKMTSLTT EDTATYFCGRNGGGSYIFYYFDLWGQGT LVTVSS 12 111B10-xi Light chain 34 AFELTQTPSSVEAAVGGTITIKCQASQS ISSYLSWYQQKPGQPPKLLIYSASTLAS GVSSRFKGSGSGTEYTLTISDLECADAA TYFCQSYYDIGTSTFGGGTEVVVK 13 111B10-zu Heavy chain 35 EVQLVESGGGLVQPGGSLRLSCAASGFS LNNYAMSWVRQAPGKGLEWIGSISTGGL AFYANWAKGRFTISRDNSKNTLYLQMNS LRAEDTAVYYCARNGGGSYIFYYFDLWG QGTLVTVSS 14 111B10-zu Light chain 36 DIQMTQSPSSLSASVGDRVTITCQASQS ISSYLSWYQQKPGKAPKLLIYSASTLAS GVPSRFSGSGSGTDFTLTISSLQCEDAA TYYCQSYYDIGTSTFGGGTKVEIK 15 201C15-xi Heavy chain 37 QSVKESGGRLVTPGTPLTLTCTVSGIDL SSYAMGWFRQAPGKGLEYIGTINIGGRV YYASWAKGRFTISRTSTTVDLKAPSLTA EDTATYFCARYYNGGSYDIWGPGTLVTV SL 16 201C15-xi Light chain 38 DVVMTQTPASASEPVGGTVTIKCQASES IYRVLAWYQQKPGQPPKLLIYDTSTLAS GAPSRFKGSGYGTEFTLTISGVQCEDAA TYYCQGGYYADSYGIAFGGGTEVVVK 17 201C15-zu Heavy chain 39 QVQLVESGGGLVQPGGSLRLSCSASGID ISSYAMGWVRQAPGKGLEYIGTINIGGR VYYASWAKGRFTISRDNSKNTLYLQMNS LRAEDTAVYYCARYYNGGSYDIWGQGTL VTVSS 18 201C15-zu Light chain 40 DIQMTQSPSTLSASVGDRVTITCQASES IYRVLAWYQQKPGKAPKLLIYDTSTLAS GVPSRFSGSGSGTEFTLTISSLQCDDAA TYYCQGGYYADSYGIAFGGGTKVEIK 19 346C6-xi Heavy chain 41 QSVEESGGRLVKPDESLTLTCTASGFSL SSYAMIWVRQAPGEGLEWIGTISTGGIT YYASWAKGRFTISKTSTTVDLKITSPTT EDTATYFCARGGYAASSAYYLPYYFDLW GQGTLVTVSS 20 346C6-xi Light chain 42 AAVLTQTPSPVSAAVGGTVTISCQSSQS VYNNNNLAWFQQKPGQPPKLLIYLASTL ASGVPSRFSGSGSGTQFTLTISGVQCDD AATYYCLGGCDDDADTFAFGGGTEVVVK 21 346C6-zu Heavy chain 43 EVQLVESGGGLVQPGGSLRLSCAASGFS LSSYAMIWVRQAPGKGLEWIGTISTGGI TYYASWAKGRFTISRDNSKNTLYLQMNS LRAEDTAVYYCARGGYAASSAYYLPYYF DLWGQGTLVTVSS 22 346C6-zu Light chain 44 DIQMTQSPSSLSASVGDRVTITCQSSQS VYNNNNLAWYQQKPGKVPKLLIYLASTL ASGVPSRFSGSGSGTDFTLTISSLQCED AATYYCLGGCDDDADTFAFGGGTKVEIK
TABLE-US-00003 TABLE 3 Nucleic acid sequences encoding mAb variable regions mAb IgG chain SEQ ID NO Nucleic acid sequence 1 MORAb-003 Heavy chain 45 GAGGTCCAACTGGTGGAGAGCGGTGGAG GTGTTGTGCAACCTGGCCGGTCCCTGCG CCTGTCCTGCTCCGCATCTGGCTTCACC TTCAGCGGCTATGGGTTGTCTTGGGTGA GACAGGCACCTGGAAAAGGTCTTGAGTG GGTTGCAATGATTAGTAGTGGTGGTAGT TATACCTACTATGCAGACAGTGTGAAGG GTAGATTTGCAATATCGCGAGACAACGC CAAGAACACATTGTTCCTGCAAATGGAC AGCCTGAGACCCGAAGACACCGGGGTCT ATTTTTGTGCAAGACATGGGGACGATCC CGCCTGGTTCGCTTATTGGGGCCAAGGG ACCCCGGTCACCGTCTCCTCA 2 MORAb-003 Light chain 46 GACATCCAGCTGACCCAGAGCCCAAGCA GCCTGAGCGCCAGCGTGGGTGACAGAGT GACCATCACCTGTAGTGTCAGCTCAAGT ATAAGTTCCAACAACTTGCACTGGTACC AGCAGAAGCCAGGTAAGGCTCCAAAGCC ATGGATCTACGGCACATCCAACCTGGCT TCTGGTGTGCCAAGCAGATTCAGCGGTA GCGGTAGCGGTACCGACTACACCTTCAC CATCAGCAGCCTCCAGCCAGAGGACATC GCCACCTACTACTGCCAACAGTGGAGTA GTTACCCGTACATGTACACGTTCGGCCA AGGGACCAAGGTGGAAATCAAA 3 MORAb-009 Heavy chain 47 CAGGTACAACTGCAGCAGTCTGGGCCTG AGCTGGAGAAGCCTGGCGCTTCAGTGAA GATATCCTGCAAGGCTTCTGGTTACTCA TTCACTGGCTACACCATGAACTGGGTGA AGCAGAGCCATGGAAAGAGCCTTGAGTG GATTGGACTTATTACTCCTTACAATGGT GCTTCTAGCTACAACCAGAAGTTCAGGG GCAAGGCCACATTAACTGTAGACAAGTC ATCCAGCACAGCCTACATGGACCTCCTC AGTCTGACATCTGAAGACTCTGCAGTCT ATTTCTGTGCAAGGGGGGGTTACGACGG GAGGGGTTTTGACTACTGGGGATCCGGG ACCCCGGTCACCGTCTCCTCA 4 MORAb-009 Light chain 48 GACATCGAGCTCACTCAGTCTCCAGCAA TCATGTCTGCATCTCCAGGGGAGAAGGT CACCATGACCTGCAGTGCCAGCTCAAGT GTAAGTTACATGCACTGGTACCAGCAGA AGTCAGGCACCTCCCCCAAAAGATGGAT TTATGACACATCCAAACTGGCTTCTGGA GTCCCAGGTCGCTTCAGTGGCAGTGGGT CTGGAAACTCTTACTCTCTCACAATCAG CAGCGTGGAGGCTGAAGATGATGCAACT TATTACTGCCAGCAGTGGAGTAAGCACC CTCTCACGTTCGGATCCGGGACCAAGGT GGAAATCAAA 5 33011-xi Heavy chain 49 CAGTCGGTGGAGGAGTCCGGGGGTCGCC TGGTCACGCCTGGGACACCCCTGACACT CACCTGCACCGTCTCTGGAATCTCCCTC AGTAGCGATGCAATAAGCTGGGTCCGCC AGGCTCCAGGGAAGGGGCTCGAATACAT CGGAATCATTAATGGTGGTGGTAACACA TACTACGCGAGCTGGGCGAAAGGCCGAT TCACCATCTCCAAAACCTCGACCACGGT GGATCTGAAAATCACCAGTCCGACAACC GAGGACACGGCCACCTATTTCTGTGCCA GAGGCATTCAACATGGTGGTGGTAATAG TGATTATTATTATTACGGCATGGACCTC TGGGGCCCAGGCACCCTGGTCACTGTCT CTTCA 6 33011-xi Light chain 50 GAAGTGTTGATGACCCAGACTCCATCCT CCGTGTCTGCAGCTGTGGGAGACACAGT CACCATCAAGTGCCAGGCCAGTCAGAGC ATTAGTAGTGTCTTGTCCTGGTATCAGC AGAAACCAGGGCAGCCTCCCAAGCTCCT GATCTATCTGGCATCCACTCTGGCATCT GGGGTCCCATCGCGGTTCAGCGGCAGTA GATCTGGGACAGAGTTCACTCTCACCAT CAGCGACCTGGAGTGTGACGATGCTGCC ACTTACTACTGTCAAACCAATTATGGTA CTAGTAGTAGTAATTATGGTTTTGCTTT CGGCGGAGGGACCGAGGTGGTCGTCAAA 7 33011-zu Heavy chain 51 GAAGTCCAACTGGTGGAAAGCGGGGGAG GACTGGTGCAGCCGGGCGGATCCCTCCG GCTGTCATGTGCTGCATCGGGAATTTCC CTCTCCTCCGACGCGATTAGCTGGGTCA GACAGGCCCCCGGAAAGGGGCTGGAGTA CATCGGTATCATCAACGGCGGCGGAAAC ACCTACTACGCCTCCTGGGCCAAGGGCC GCTTCACCATCTCGCGGCATAATTCCAA GAACACTCTGTACTTGCAAATGAACTCC CTGAGGGCCGAGGACACCGCCGTGTACT ACTGCGCGCGCGGCATCCAGCACGGTGG TGGAAACAGCGACTACTACTACTATGGG ATGGATCTGTGGGGCCAGGGAACTCTTG TGACCGTGTCGTCA 8 33011-zu Light chain 52 GACATTCAGATGACCCAGTCCCCAAGCT CGCTGTCCGCCTCCGTGGGCGACCGCGT GACCATCACGTGCCAGGCGTCCCAGTCA ATTAGCAGCGTGCTCTCCTGGTACCAAC AGAAGCCGGGGAAAGCACCCAAGCTGCT GATCTACTTGGCCTCCACTCTGGCCTCG GGAGTGCCTTCACGGTTCTCCGGATCGG GATCTGGTACTGATTTCACCCTCACCAT CTCGAGCCTTCAGTGCGAGGACATCGCT ACTTACTATTGTCAAACCAACTACGGAA CCTCCAGCTCCAACTACGGCTTTGCCTT CGGTGGCGGGACCAAGGTCGAAATCAAA 9 111B10-xi Heavy chain 53 CAGTCGGTGGAGGAGTCCGGGGGTCGCC TGGTCACGCCTGGGACACCCCTGACACT CACCTGCACAGTCTCTGGATTCTCCCTC AATAACTATGCAATGAGCTGGGTCCGCC AGGCTCCAGGGAAGGGGCTGGAATGGAT CGGATCCATTAGTACTGGTGGTCTCGCA TTCTACGCGAACTGGGCAAAAGGCCGAT TCACCATCTCCAGAACCTCGACCACGGT GGATCTGAAAATGACCAGTCTGACAACC GAGGACACGGCCACCTATTTCTGTGGCA GAAATGGTGGTGGTAGTTATATTTTCTA TTATTTTGACTTGTGGGGCCAAGGCACC CTCGTCACTGTCTCTTCA 10 111B10-xi Light chain 54 GCATTCGAATTGACCCAGACTCCATCCT CCGTGGAGGCAGCTGTGGGAGGCACAAT CACCATCAAGTGCCAGGCCAGTCAGAGC ATTAGTAGTTACTTATCCTGGTATCAGC AGAAACCAGGGCAGCCTCCCAAGCTCCT GATCTATTCTGCATCCACTCTGGCATCT GGGGTCTCATCGCGGTTCAAAGGCAGTG GATCTGGGACAGAGTACACTCTCACCAT CAGCGACCTGGAGTGTGCCGATGCTGCC ACTTACTTCTGTCAAAGCTATTATGATA TTGGTACTAGTACTTTCGGCGGAGGGAC CGAGGTGGTCGTCAAA 11 111B10-zu Heavy chain 55 GAAGTGCAGCTGGTGGAATCTGGCGGCG GACTGGTGCAGCCTGGCGGATCTCTGAG ACTGTCTTGTGCCGCCTCCGGCTTCTCC CTGAACAACTACGCCATGTCCTGGGTGC GACAGGCCCCTGGCAAAGGCCTGGAATG GATCGGCTCCATCAGCACAGGCGGCCTG GCCTTCTACGCCAATTGGGCCAAGGGCC GGTTCACCATCAGCCGGGACAACTCCAA GAACACCCTGTACCTCCAGATGAACTCC CTGCGGGCCGAGGACACCGCCGTGTACT ACTGTGCCAGAAACGGCGGAGGCTCCTA CATCTTCTACTACTTCGACCTGTGGGGC CAGGGCACCCTCGTGACAGTGTCATCT 12 111B10-zu Light chain 56 GATATTCAGATGACCCAGTCCCCCTCCA GCCTGTCCGCTTCTGTGGGCGACAGAGT GACCATCACCTGTCAGGCCTCCCAGTCC ATCTCCTCCTACCTGTCCTGGTATCAGC AGAAGCCCGGCAAGGCCCCCAAGCTGCT GATCTACTCTGCCTCCACACTGGCCTCC GGCGTGCCCTCTAGATTCTCCGGCTCTG GCTCTGGCACCGACTTTACCCTGACCAT CAGCTCCCTCCAGTGCGAGGATGCCGCC ACCTACTACTGCCAGTCCTACTACGACA TCGGCACCTCCACCTTCGGCGGAGGCAC CAAGGTGGAAATCAAA 13 201C15-xi Heavy chain 57 CAGTCAGTGAAGGAGTCCGGGGGTCGCC TGGTCACGCCTGGGACACCCCTGACACT CACCTGCACAGTCTCTGGAATCGACCTC AGTAGCTATGCAATGGGCTGGTTCCGCC AGGCTCCAGGGAAGGGGCTGGAATACAT CGGAACCATTAATATTGGTGGTCGCGTA TATTACGCGAGCTGGGCAAAAGGCCGAT TCACCATCTCCAGAACCTCGACCACGGT GGATCTGAAAGCGCCCAGTCTGACAGCC GAGGACACGGCCACCTATTTCTGTGCCA GATATTATAATGGTGGTAGTTATGACAT CTGGGGCCCAGGCACCCTGGTCACCGTC TCTTTA 14 201C15-xi Light chain 58 GATGTTGTGATGACCCAGACTCCAGCCT CCGCGTCTGAACCTGTGGGAGGCACAGT CACCATCAAGTGCCAGGCCAGTGAGAGC ATTTATCGCGTATTGGCCTGGTATCAGC AGAAACCAGGGCAGCCTCCCAAGCTCCT GATCTATGATACATCCACTCTGGCATCT GGGGCCCCATCGCGGTTCAAAGGCAGTG GATATGGGACAGAGTTCACTCTCACCAT CAGCGGCGTGCAGTGTGAAGATGCTGCC ACTTACTACTGTCAAGGCGGTTATTATG CTGATAGTTATGGTATTGCTTTCGGCGG AGGGACCGAGGTGGTGGTCAAA 15 201C15-zu Heavy chain 59 CAGGTGCAGCTGGTGGAATCTGGCGGAG GACTGGTGCAGCCTGGCGGCTCTCTGAG ACTGTCCTGTTCCGCCTCCGGAATCGAC CTGTCCTCCTACGCTATGGGCTGGGTGC GACAGGCTCCTGGCAAGGGCCTGGAGTA CATCGGCACCATCAACATCGGCGGCAGA GTGTACTACGCCTCCTGGGCCAAGGGCC GGTTCACCATCTCCAGAGACAACTCCAA GAACACCCTGTACCTCCAGATGAACTCC CTGCGGGCCGAGGACACCGCCGTGTACT ACTGCGCCCGGTACTACAACGGCGGCTC CTACGATATCTGGGGCCAGGGCACACTC GTGACCGTGTCCTCT 16 201C15-zu Light chain 60 GATATCCAGATGACCCAGTCCCCCTCCA CCCTGTCTGCCTCTGTGGGCGACAGAGT GACCATCACCTGTCAGGCCTCCGAGTCC ATCTACCGGGTGCTGGCCTGGTATCAGC AGAAGCCTGGCAAGGCCCCCAAGCTGCT GATCTACGACACCAGCACACTGGCCTCC GGCGTGCCCTCTAGATTCTCCGGCTCTG GCTCTGGCACCGAGTTTACCCTGACCAT CTCCAGCCTCCAGTGCGACGACGCCGCC ACCTACTATTGTCAGGGCGGCTACTACG CCGACTCCTACGGAATCGCTTTCGGCGG AGGCACCAAGGTGGAAATCAAA 17 346C6-xi Heavy chain 61 CAGTCGGTGGAGGAGTCCGGCGGTCGCC TGGTAAAGCCTGACGAATCCCTGACACT CACCTGCACAGCCTCTGGATTCTCCCTC AGTAGTTATGCAATGATCTGGGTCCGCC AGGCTCCAGGGGAGGGGCTGGAATGGAT CGGAACCATTAGTACTGGTGGTATCACA TACTACGCGAGCTGGGCGAAAGGCCGAT TCACCATCTCCAAAACCTCGACCACGGT GGATCTGAAAATCACCAGTCCGACAACC GAGGACACGGCCACCTATTTCTGTGCCA GAGGGGGATATGCTGCTAGTAGTGCTTA TTATCTCCCGTACTACTTTGACTTGTGG GGCCAAGGGACCCTGGTCACCGTCTCCT CA 18 346C6-xi Light chain 62 GCAGCCGTGCTGACCCAGACACCATCAC CCGTGTCTGCAGCTGTGGGAGGCACAGT CACCATCAGTTGCCAGTCCAGTCAGAGT GTTTATAATAATAACAACTTAGCCTGGT TTCAGCAGAAACCCGGGCAGCCTCCCAA GCTTCTGATCTATCTGGCATCCACTCTG GCATCTGGGGTCCCATCACGGTTCAGCG GCAGTGGATCTGGGACACAGTTCACTCT CACCATCAGCGGCGTGCAGTGTGACGAT GCTGCCACTTATTACTGTCTAGGTGGTT GTGATGATGATGCTGATACTTTTGCTTT CGGCGGAGGGACTGAGGTGGTGGTCAAA
19 346C6-zu Heavy chain 63 GAAGTGCAGCTGGTGGAATCTGGCGGCG GACTGGTGCAGCCTGGCGGATCTCTGAG ACTGTCTTGTGCCGCCTCCGGCTTCTCC CTGTCCTCCTACGCTATGATCTGGGTGC GACAGGCCCCTGGCAAGGGCCTGGAATG GATCGGCACCATCTCTACCGGCGGAATT ACCTACTACGCCTCCTGGGCCAAGGGCC GGTTCACCATCTCCAGAGACAACTCCAA GAACACCCTGTACCTCCAGATGAACTCC CTGCGGGCCGAGGACACCGCCGTGTACT ATTGTGCTAGAGGCGGCTACGCCGCCAG CTCCGCTTACTACCTGCCCTACTACTTC GACCTGTGGGGCCAGGGCACCCTCGTGA CAGTGTCATCT 20 346C6-zu Light chain 64 GATATTCAGATGACCCAGTCCCCCTCCA GCCTGTCCGCTTCTGTGGGCGACAGAGT GACCATCACCTGTCAGTCCTCCCAGTCC GTGTATAACAACAACAACCTGGCCTGGT ATCAGCAGAAACCCGGCAAGGTGCCCAA GCTGCTGATCTACCTGGCCTCCACACTG GCCTCTGGCGTGCCCTCTAGATTCTCCG GCTCTGGCTCTGGCACCGACTTTACCCT GACCATCAGCTCCCTCCAGTGCGAGGAT GCCGCCACCTACTATTGCCTGGGCGGCT GCGACGACGACGCCGATACCTTTGCTTT TGGCGGAGGCACCAAGGTGGAAATCAAA
TABLE-US-00004 TABLE 4 Amino acid sequences of mAb Kabat CDRs IgG SEQ ID Amino acid mAb chain NO sequence 1 MORAb-003 HC CDR1 2 GYGLS 2 MORAb-003 HC CDR2 3 MISSGGSYTYYADSVKG 3 MORAb-003 HC CDR3 4 HGDDPAWFAY 4 MORAb-003 LC CDR1 7 SVSSSISSNNLH 5 MORAb-003 LC CDR2 8 GTSNLAS 6 MORAb-003 LC CDR3 9 QQWSSYPYMYT 7 MORAb-009 HC CDR1 65 GYTMN 8 MORAb-009 HC CDR2 66 LITPYNGASSYNQKFRG 9 MORAb-009 HC CDR3 67 GGYDGRGFDY 10 MORAb-009 LC CDR1 68 SASSSVSYMH 11 MORAb-009 LC CDR2 69 DTSKLAS 12 MORAb-009 LC CDR3 70 QQWSKHPLT 13 trastuzumab HC CDR1 71 DTYIH 14 trastuzumab HC CDR2 72 RIYPTNGYTRYADSVKG 15 trastuzumab HC CDR3 73 WGGDGFYAMDY 16 trastuzumab LC CDR1 74 RASQDVNTAVA 17 trastuzumab LC CDR2 75 SASFLYS 18 trastuzumab LC CDR3 76 QQHYTTPPT 19 33011-xi HC CDR1 77 SDAIS 20 33011-xi HC CDR2 78 IINGGGNTYYASWAKG 21 33011-xi HC CDR3 79 GIQHGGGNSDYYYYGMDL 22 33011-xi LC CDR1 80 QASQSISSVLS 23 33011-xi LC CDR2 81 LASTLAS 24 33011-xi LC CDR3 82 QTNYGTSSSNYGFA 25 33011-zu HC CDR1 83 SDAIS 26 33011-zu HC CDR2 84 IINGGGNTYYASWAKG 27 33011-zu HC CDR3 85 GIQHGGGNSDYYYYGMDL 28 33011-zu LC CDR1 86 QASQSISSVLS 29 33011-zu LC CDR2 87 LASTLAS 30 33011-zu LC CDR3 88 QTNYGTSSSNYGFA 31 111B10-xi HC CDR1 89 NYAMS 32 111B10-xi HC CDR2 90 SISTGGLAFYANWAKG 33 111B10-xi HC CDR3 91 NGGGSYIFYYFDL 34 111B10-xi LC CDR1 92 QASQSISSYLS 35 111B10-xi LC CDR2 93 SASTLAS 36 111B10-xi LC CDR3 94 QSYYDIGTST 37 111B10-zu HC CDR1 95 NYAMS 38 111B10-zu HC CDR2 96 SISTGGLAFYANWAKG 39 111B10-zu HC CDR3 97 NGGGSYIFYYFDL 40 111B10-zu LC CDR1 98 QASQSISSYLS 41 111B10-zu LC CDR2 99 SASTLAS 42 111B10-zu LC CDR3 100 QSYYDIGTST 43 201C15-xi HC CDR1 101 SYAMG 44 201C15-xi HC CDR2 102 TINIGGRVYYASWAKG 45 201C15-xi HC CDR3 103 YYNGGSYDI 46 201C15-xi LC CDR1 104 QASESIYRVLA 47 201C15-xi LC CDR2 105 DTSTLAS 48 201C15-xi LC CDR3 106 QGGYYADSYGIA 49 201C15-zu HC CDR1 107 SYAMG 50 201C15-zu HC CDR2 108 TINIGGRVYYASWAKG 51 201C15-zu HC CDR3 109 YYNGGSYDI 52 201C15-zu LC CDR1 110 QASESIYRVLA 53 201C15-zu LC CDR2 111 DTSTLAS 54 201C15-zu LC CDR3 112 QGGYYADSYGIA 55 346C6-xi HC CDR1 113 SYAMI 56 346C6-xi HC CDR2 114 TISTGGITYYASWAKG 57 346C6-xi HC CDR3 115 GGYAASSAYYLPYYFDL 58 346C6-xi LC CDR1 116 QSSQSVYNNNNLA 59 346C6-xi LC CDR2 117 LASTLAS 60 346C6-xi LC CDR3 118 LGGCDDDADTFA 61 346C6-zu HC CDR1 119 SYAMI 62 346C6-zu HC CDR2 120 TISTGGITYYASWAKG 63 346C6-zu HC CDR3 121 GGYAASSAYYLPYYFDL 64 346C6-zu LC CDR1 122 QSSQSVYNNNNLA 65 346C6-zu LC CDR2 123 LASTLAS 66 346C6-zu LC CDR3 124 LGGCDDDADTFA
TABLE-US-00005 TABLE 5 Nucleic acid sequences encoding mAb Kabat CDRs mAb IgG chain SEQ ID NO Nucleic acid sequence 1 MORAb-003 HC CDR1 125 GGCTATGGGTTGTCT 2 MORAb-003 HC CDR2 126 ATGATTAGTAGTGGTGGTAGTTATACCTACTATG CAGACAGTGTGAAGGGT 3 MORAb-003 HC CDR3 127 CATGGGGACGATCCCGCCTGGTTCGCTTAT 4 MORAb-003 LC CDR1 128 AGTGTCAGCTCAAGTATAAGTTCCAACAACTTGC AC 5 MORAb-003 LC CDR2 129 GGCACATCCAACCTGGCTTCT 6 MORAb-003 LC CDR3 130 CAACAGTGGAGTAGTTACCCGTACATGTACACG 7 MORAb-009 HC CDR1 131 GGCTACACCATGAAC 8 MORAb-009 HC CDR2 132 CTTATTACTCCTTACAATGGTGCTTCTAGCTACA ACCAGAAGTTCAGGGGC 9 MORAb-009 HC CDR3 133 GGGGGTTACGACGGGAGGGGTTTTGACTAC 10 MORAb-009 LC CDR1 134 AGTGCCAGCTCAAGTGTAAGTTACATGCAC 11 MORAb-009 LC CDR2 135 GACACATCCAAACTGGCTTCT 12 MORAb-009 LC CDR3 136 CAGCAGTGGAGTAAGCACCCTCTCACG 13 33011-xi HC CDR1 137 AGCGATGCAATAAGC 14 33011-xi HC CDR2 138 ATCATTAATGGTGGTGGTAACACATACTACGCGA GCTGGGCGAAAGGC 15 33011-xi HC CDR3 139 GGCATTCAACATGGTGGTGGTAATAGTGATTATT ATTATTACGGCATGGACCTC 16 33011-xi LC CDR1 140 CAGGCCAGTCAGAGCATTAGTAGTGTCTTGTCC 17 33011-xi LC CDR2 141 CTGGCATCCACTCTGGCATCT 18 33011-xi LC CDR3 142 CAAACCAATTATGGTACTAGTAGTAGTAATTATG GTTTTGCT 19 33011-zu HC CDR1 143 TCCGACGCGATTAGC 20 33011-zu HC CDR2 144 ATCATCAACGGCGGCGGAAACACCTACTACGCCT CCTGGGCCAAGGGC 21 33011-zu HC CDR3 145 GGCATCCAGCACGGTGGTGGAAACAGCGACTACT ACTACTATGGGATGGATCTG 22 33011-zu LC CDR1 146 CAGGCGTCCCAGTCAATTAGCAGCGTGCTCTCC 23 33011-zu LC CDR2 147 TTGGCCTCCACTCTGGCCTCG 24 33011-zu LC CDR3 148 CAAACCAACTACGGAACCTCCAGCTCCAACTACG GCTTTGCC 25 111B10-xi HC CDR1 149 AACTATGCAATGAGC 26 111B10-xi HC CDR2 150 TCCATTAGTACTGGTGGTCTCGCATTCTACGCGA ACTGGGCAAAAGGC 27 111B10-xi HC CDR3 151 AATGGTGGTGGTAGTTATATTTTCTATTATTTTG ACTTG 28 111B10-xi LC CDR1 152 CAGGCCAGTCAGAGCATTAGTAGTTACTTATCC 29 111B10-xi LC CDR2 153 TCTGCATCCACTCTGGCATCT 30 111B10-xi LC CDR3 154 CAAAGCTATTATGATATTGGTACTAGTACT 31 111B10-zu HC CDR1 155 AACTACGCCATGTCC 32 111B10-zu HC CDR2 156 TCCATCAGCACAGGCGGCCTGGCCTTCTACGCCA ATTGGGCCAAGGGC 33 111B10-zu HC CDR3 157 AACGGCGGAGGCTCCTACATCTTCTACTACTTCG ACCTG 34 111B10-zu LC CDR1 158 CAGGCCTCCCAGTCCATCTCCTCCTACCTGTCC 35 111B10-zu LC CDR2 159 TCTGCCTCCACACTGGCCTCC 36 111B10-zu LC CDR3 160 CAGTCCTACTACGACATCGGCACCTCCACC 37 201C15-xi HC CDR1 161 AGCTATGCAATGGGC 38 201C15-xi HC CDR2 162 ACCATTAATATTGGTGGTCGCGTATATTACGCGA GCTGGGCAAAAGGC 39 201C15-xi HC CDR3 163 TATTATAATGGTGGTAGTTATGACATC 40 201C15-xi LC CDR1 164 CAGGCCAGTGAGAGCATTTATCGCGTATTGGCC 41 201C15-xi LC CDR2 165 GATACATCCACTCTGGCATCT 42 201C15-xi LC CDR3 166 CAAGGCGGTTATTATGCTGATAGTTATGGTATTG CT 43 201C15-zu HC CDR1 167 TCCTACGCTATGGGC 44 201C15-zu HC CDR2 168 ACCATCAACATCGGCGGCAGAGTGTACTACGCCT CCTGGGCCAAGGGC 45 201C15-zu HC CDR3 169 TACTACAACGGCGGCTCCTACGATATC 46 201C15-zu LC CDR1 170 CAGGCCTCCGAGTCCATCTACCGGGTGCTGGCC 47 201C15-zu LC CDR2 171 GACACCAGCACACTGGCCTCC 48 201C15-zu LC CDR3 172 CAGGGCGGCTACTACGCCGACTCCTACGGAATCG CT 49 346C6-xi HC CDR1 173 AGTTATGCAATGATC 50 346C6-xi HC CDR2 174 ACCATTAGTACTGGTGGTATCACATACTACGCGA GCTGGGCGAAAGGC 51 346C6-xi HC CDR3 175 GGGGGATATGCTGCTAGTAGTGCTTATTATCTCC CGTACTACTTTGACTTG 52 346C6-xi LC CDR1 176 CAGTCCTCCCAGTCCGTGTATAACAACAACAACC TGGCC 53 346C6-xi LC CDR2 177 CTGGCATCCACTCTGGCATCT 54 346C6-xi LC CDR3 178 CTAGGTGGTTGTGATGATGATGCTGATACTTTTG CT 55 346C6-zu HC CDR1 179 TCCTACGCTATGATC 56 346C6-zu HC CDR2 180 ACCATCTCTACCGGCGGAATTACCTACTACGCCT CCTGGGCCAAGGGC 57 346C6-zu HC CDR3 181 GGCGGCTACGCCGCCAGCTCCGCTTACTACCTGC CCTACTACTTCGACCTG 58 346C6-zu LC CDR1 182 CAGTCCTCCCAGTCCGTGTATAACAACAACAACC TGGCC 59 346C6-zu LC CDR2 183 CTGGCCTCCACACTGGCCTCT 60 346C6-zu LC CDR3 184 CTGGGCGGCTGCGACGACGACGCCGATACCTTTG CT
TABLE-US-00006 TABLE 6 Amino acid sequences of mAb IMGT CDRs IgG SEQ ID Amino acid mAb chain NO sequence 1 MORAb-003 HC CDR1 13 GFTFSGYG 2 MORAb-003 HC CDR2 14 ISSGGSYT 3 MORAb-003 HC CDR3 15 ARHGDDPAWFAY 4 MORAb-003 LC CDR1 16 SSISSNN 5 MORAb-003 LC CDR2 17 GTS 6 MORAb-003 LC CDR3 18 QQWSSYPYMYT 7 MORAb-009 HC CDR1 185 GYSFTGYT 8 MORAb-009 HC CDR2 186 ITPYNGAS 9 MORAb-009 HC CDR3 187 ARGGYDGRGFDY 10 MORAb-009 LC CDR1 188 SSVSY 11 MORAb-009 LC CDR2 189 DTS 12 MORAb-009 LC CDR3 190 QQWSKHPLT 13 trastuzumab HC CDR1 191 GFNIKDTY 14 trastuzumab HC CDR2 192 IYPTNGYT 15 trastuzumab HC CDR3 193 SRWGGDGFYAMDY 16 trastuzumab LC CDR1 194 QDVNTA 17 trastuzumab LC CDR2 195 SAS 18 trastuzumab LC CDR3 196 QQHYTTPPT 19 33011-xi HC CDR1 197 GISLSSDA 20 33011-xi HC CDR2 198 INGGGNT 21 33011-xi HC CDR3 199 ARGIQHGGGNSDYYYYGMDL 22 33011-xi LC CDR1 200 QSISSV 23 33011-xi LC CDR2 201 LAS 24 33011-xi LC CDR3 202 QTNYGTSSSNYGFA 25 33011-zu HC CDR1 203 GISLSSDA 26 33011-zu HC CDR2 204 INGGGNT 27 33011-zu HC CDR3 205 ARGIQHGGGNSDYYYYGMDL 28 33011-zu LC CDR1 206 QSISSV 29 33011-zu LC CDR2 207 LAS 30 33011-zu LC CDR3 208 QTNYGTSSSNYGFA 31 111B10-xi HC CDR1 209 GFSLNNYA 32 111B10-xi HC CDR2 210 ISTGGLA 33 111B10-xi HC CDR3 211 GRNGGGSYIFYYFDL 34 111B10-xi LC CDR1 212 QSISSY 35 111B10-xi LC CDR2 213 SAS 36 111B10-xi LC CDR3 214 QSYYDIGTST 37 111B10-zu HC CDR1 215 GFSLNNYA 38 111B10-zu HC CDR2 216 ISTGGLA 39 111B10-zu HC CDR3 217 ARNGGGSYIFYYFDL 40 111B10-zu LC CDR1 218 QSISSY 41 111B10-zu LC CDR2 219 SAS 42 111B10-zu LC CDR3 220 QSYYDIGTST 43 201C15-xi HC CDR1 221 GIDLSSYA 44 201C15-xi HC CDR2 222 INIGGRV 45 201C15-xi HC CDR3 223 ARYYNGGSYDI 46 201C15-xi LC CDR1 224 ESIYRV 47 201C15-xi LC CDR2 225 DTS 48 201C15-xi LC CDR3 226 QGGYYADSYGIA 49 201C15-zu HC CDR1 227 GIDLSSYA 50 201C15-zu HC CDR2 228 INIGGRV 51 201C15-zu HC CDR3 229 ARYYNGGSYDI 52 201C15-zu LC CDR1 230 ESIYRV 53 201C15-zu LC CDR2 231 DTS 54 201C15-zu LC CDR3 232 QGGYYADSYGIA 55 346C6-xi HC CDR1 233 GFSLSSYA 56 346C6-xi HC CDR2 234 ISTGGIT 57 346C6-xi HC CDR3 235 ARGGYAASSAYYLPYYFDL 58 346C6-xi LC CDR1 236 QSVYNNNN 59 346C6-xi LC CDR2 237 LAS 60 346C6-xi LC CDR3 238 LGGCDDDADTFA 61 346C6-zu HC CDR1 239 GFSLSSYA 62 346C6-zu HC CDR2 240 ISTGGIT 63 346C6-zu HC CDR3 241 ARGGYAASSAYYLPYYFDL 64 346C6-zu LC CDR1 242 QSVYNNNN 65 346C6-zu LC CDR2 243 LAS 66 346C6-zu LC CDR3 244 LGGCDDDADTFA
TABLE-US-00007 TABLE 7 Nucleic acid sequences encoding mAb IMGT CDRs IgG SEQ ID mAb chain NO Nucleic acid sequence 1 MORAb-003 HC CDR1 245 GGCTTCACCTTCAGCGGCTATGGG 2 MORAb-003 HC CDR2 246 ATTAGTAGTGGTGGTAGTTATACC 3 MORAb-003 HC CDR3 247 GCAAGACATGGGGACGATCCCGCCTGGTTCGCT TAT 4 MORAb-003 LC CDR1 248 TCAAGTATAAGTTCCAACAAC 5 MORAb-003 LC CDR2 249 GGCACATCC 6 MORAb-003 LC CDR3 250 CAACAGTGGAGTAGTTACCCGTACATGTACACG 7 MORAb-009 HC CDR1 251 GGTTACTCATTCACTGGCTACACC 8 MORAb-009 HC CDR2 252 ATTACTCCTTACAATGGTGCTTCT 9 MORAb-009 HC CDR3 253 GCAAGGGGGGGTTACGACGGGAGGGGTTTTGAC TAC 10 MORAb-009 LC CDR1 254 TCAAGTGTAAGTTAC 11 MORAb-009 LC CDR2 255 GACACATCC 12 MORAb-009 LC CDR3 256 CAGCAGTGGAGTAAGCACCCTCTCACG 13 33011-xi HC CDR1 257 GGAATCTCCCTCAGTAGCGATGCA 14 33011-xi HC CDR2 258 ATTAATGGTGGTGGTAACACA 15 33011-xi HC CDR3 259 GCCAGAGGCATTCAACATGGTGGTGGTAATAGT GATTATTATTATTACGGCATGGACCTC 16 33011-xi LC CDR1 260 CAGAGCATTAGTAGTGTC 17 33011-xi LC CDR2 261 CTGGCATCT 18 33011-xi LC CDR3 262 CAAACCAATTATGGTACTAGTAGTAGTAATTAT GGTTTTGCT 19 33011-zu HC CDR1 263 GGAATTTCCCTCTCCTCCGACGCG 20 33011-zu HC CDR2 264 ATCAACGGCGGCGGAAACACC 21 33011-zu HC CDR3 265 GCGCGCGGCATCCAGCACGGTGGTGGAAACAGC GACTACTACTACTATGGGATGGATCTG 22 33011-zu LC CDR1 266 CAGTCAATTAGCAGCGTG 23 33011-zu LC CDR2 267 TTGGCCTCC 24 33011-zu LC CDR3 268 CAAACCAACTACGGAACCTCCAGCTCCAACTAC GGCTTTGCC 25 111B10-xi HC CDR1 269 GGATTCTCCCTCAATAACTATGCA 26 111B10-xi HC CDR2 270 ATTAGTACTGGTGGTCTCGCA 27 111B10-xi HC CDR3 271 GGCAGAAATGGTGGTGGTAGTTATATTTTCTAT TATTTTGACTTG 28 111B10-xi LC CDR1 272 CAGAGCATTAGTAGTTAC 29 111B10-xi LC CDR2 273 TCTGCATCC 30 111B10-xi LC CDR3 274 CAAAGCTATTATGATATTGGTACTAGTACT 31 111B10-zu HC CDR1 275 GGCTTCTCCCTGAACAACTACGCC 32 111B10-zu HC CDR2 276 ATCAGCACAGGCGGCCTGGCC 33 111B10-zu HC CDR3 277 GCCAGAAACGGCGGAGGCTCCTACATCTTCTAC TACTTCGACCTG 34 111B10-zu LC CDR1 278 CAGTCCATCTCCTCCTAC 35 111B10-zu LC CDR2 279 TCTGCCTCC 36 111B10-zu LC CDR3 300 CAGTCCTACTACGACATCGGCACCTCCACC 37 201C15-xi HC CDR1 301 GGAATCGACCTCAGTAGCTATGCA 38 201C15-xi HC CDR2 302 ATTAATATTGGTGGTCGCGTA 39 201C15-xi HC CDR3 303 GCCAGATATTATAATGGTGGTAGTTATGACATC 40 201C15-xi LC CDR1 304 GAGAGCATTTATCGCGTA 41 201C15-xi LC CDR2 305 GATACATCC 42 201C15-xi LC CDR3 306 CAAGGCGGTTATTATGCTGATAGTTATGGTATT GCT 43 201C15-zu HC CDR1 307 GGAATCGACCTGTCCTCCTACGCT 44 201C15-zu HC CDR2 308 ATCAACATCGGCGGCAGAGTG 45 201C15-zu HC CDR3 309 GCCCGGTACTACAACGGCGGCTCCTACGATATC 46 201C15-zu LC CDR1 310 GAGTCCATCTACCGGGTG 47 201C15-zu LC CDR2 311 GACACCAGC 48 201C15-zu LC CDR3 312 CAGGGCGGCTACTACGCCGACTCCTACGGAATC GCT 49 346C6-xi HC CDR1 313 GGATTCTCCCTCAGTAGTTATGCA 50 346C6-xi HC CDR2 314 ATTAGTACTGGTGGTATCACA 51 346C6-xi HC CDR3 315 GCCAGAGGGGGATATGCTGCTAGTAGTGCTTAT TATCTCCCGTACTACTTTGACTTG 52 346C6-xi LC CDR1 316 CAGAGTGTTTATAATAATAACAAC 53 346C6-xi LC CDR2 317 CTGGCATCC 54 346C6-xi LC CDR3 318 CTAGGTGGTTGTGATGATGATGCTGATACTTTT GCT 55 346C6-zu HC CDR1 319 GGCTTCTCCCTGTCCTCCTACGCT 56 346C6-zu HC CDR2 320 ATCTCTACCGGCGGAATTACC 57 346C6-zu HC CDR3 321 GCTAGAGGCGGCTACGCCGCCAGCTCCGCTTAC TACCTGCCCTACTACTTCGACCTG 58 346C6-zu LC CDR1 322 CAGTCCGTGTATAACAACAACAAC 59 346C6-zu LC CDR2 323 CTGGCCTCC 60 346C6-zu LC CDR3 324 CTGGGCGGCTGCGACGACGACGCCGATACCTTT GCT
TABLE-US-00008 TABLE 8 Amino acid sequences of full-length mAb Ig chains SEQ ID mAb IgG chain NO Amino acid sequence 1 MORAb-003 Heavy chain 1 EVQLVESGGGVVQPGRSLRLSCSASGFTFSGY GLSWVRQAPGKGLEWVAMISSGGSYTYYADSV KGRFAISRDNAKNTLFLQMDSLRPEDTGVYFC ARHGDDPAWFAYWGQGTPVTVSSASTKGPSVF PLAPSSKSTSGGTAALGCLVKDYFPEPVTVSW NSGALTSGVHTFPAVLQSSGLYSISSVVTVPS SSLGTQTYICNVNHKPSNTKVDKKVEPKSCDK THTCPPCPAPELLGGPSVFLFPPKPKDTLMIS RTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHN AKTKPREEQYNSTYRVVSVLTVLHQDWLNGKE YKCKVSNKALPAPIEKTISKAKGQPREPQVYT LPPSRDELTKNQVSLTCLVKGFYPSDIAVEWE SNGQPENNYKTTPPVLDSDGSFFLYSKLTVDK SRWQQGNVFSCSVMHEALHNHYTQKSLSLSPG K 2 MORAb-003 Light chain 6 DIQLTQSPSSLSASVGDRVTITCSVSSSISSN NLHWYQQKPGKAPKPWIYGTSNLASGVPSRFS GSGSGTDYTFTISSLQPEDIATYYCQQWSSYP YMYTFGQGTKVEIKRTVAAPSVFIFPPSDEQL KSGTASVVCLLNNFYPREAKVQWKVDNALQSG NSQESVTEQDSKDSTYSLSSTLTLSKADYEKH KVYACEVTHQGLSSPVTKSFNRGEC 3 MORAb-009 Heavy chain 325 QVQLQQSGPELEKPGASVKISCKASGYSFTGY TMNWVKQSHGKSLEWIGLITPYNGASSYNQKF RGKATLTVDKSSSTAYMDLLSITSEDSAVYFC ARGGYDGRGFDYWGSGTPVTVSSASTKGPSVF PLAPSSKSTSGGTAALGCLVKDYFPEPVTVSW NSGALTSGVHTFPAVLQSSGLYSISSVVTVPS SSLGTQTYICNVNHKPSNTKVDKKVEPKSCDK THTCPPCPAPELLGGPSVFLFPPKPKDTLMIS RTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHN AKTKPREEQYNSTYRVVSVLTVLHQDWLNGKE YKCKVSNKALPAPIEKTISKAKGQPREPQVYT LPPSRDELTKNQVSLTCLVKGFYPSDIAVEWE SNGQPENNYKTTPPVLDSDGSFFLYSKLTVDK SRWQQGNVFSCSVMHEALHNHYTQKSLSLSPG K 4 MORAb-009 Light chain 326 DIELTQSPAIMSASPGEKVTMTCSASSSVSYM HWYQQKSGTSPKRWIYDTSKLASGVPGRFSGS GSGNSYSLTISSVEAEDDATYYCQQWSKHPLT FGSGTKVEIKRTVAAPSVFIFPPSDEQLKSGT ASVVCLLNNFYPREAKVQWKVDNALQSGNSQE SVTEQDSKDSTYSLSSTLTLSKADYEKHKVYA CEVTHQGLSSPVTKSFNRGEC 5 trastuzumab Heavy chain 327 EVQLVESGGGLVQPGGSLRLSCAASGFNIKDT YIHWVRQAPGKGLEWVARIYPTNGYTRYADSV KGRFTISADTSKNTAYLQMNSLRAEDTAVYYC SRWGGDGFYAMDYWGQGTLVTVSSASTKGPSV FPLAPSSKSTSGGTAALGCLVKDYFPEPVTVS WNSGALTSGVHTFPAVLQSSGLYSLSSVVTVP SSSLGTQTYICNVNHKPSNTKVDKKVEPPKSC DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLM ISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEV HNAKTKPREEQYNSTYRVVSVLTVLHQDWLNG KEYKCKVSNKALPAPIEKTISKAKGQPREPQV YTLPPSRDELTKNQVSLTCLVKGFYPSDIAVE WESNGQPENNYKTTPPVLDSDGSFFLYSKLTV DKSRWQQGNVFSCSVMHEALHNHYTQKSLSLS PGK 6 trastuzumab Light chain 328 DIQMTQSPSSLSASVGDRVTITCRASQDVNTA VAWYQQKPGKAPKLLIYSASFLYSGVPSRFSG SRSGTDFTLTISSLQPEDFATYYCQQHYTTPP TFGQGTKVEIKRTVAAPSVFIFPPSDEQLKSG TASVVCLLNNFYPREAKVQWKVDNALQSGNSQ ESVTEQDSKDSTYSLSSTLTLSKADYEKHKVY ACEVTHQGLSSPVTKSFNRGEC 7 33011-xi Heavy chain 329 QSVEESGGRLVTPGTPLTLTCTVSGISLSSDA ISWVRQAPGKGLEYIGIINGGGNTYYASWAKG RFTISKTSTTVDLKITSPTTEDTATYFCARGI QHGGGNSDYYYYGMDL WGPGTLVTVSSASTKGPSVFPLAPSSKSTSGG TAALGCLVKDYFPEPVTVSWNSGALTSGVHTF PAVLQSSGLYSISSVVTVPSSSLGTQTYICNV NHKPSNTKVDKKVEPKSCDKTHTCPPCPAPEL LGGPSVFLFPPKPKDTLMISRTPEVTCVVVDV SHEDPEVKFNWYVDGVEVHNAKTKPREEQYNS TYRVVSVLTVLHQDWLNGKEYKCKVSNKALPA PIEKTISKAKGQPREPQVYTLPPSRDELTKNQ VSLTCLVKGFYPSDIAVEWESNGQPENNYKTT PPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCS VMHEALHNHYTQKSLSLSPGK 8 33011-xi Light chain 330 EVLMTQTPSSVSAAVGDTVTIKCQASQSISSV LSWYQQKPGQPPKLLIYLASTLASGVPSRFSG SRSGTEFTLTISDLECDDAATYYCQTNYGTSS SNYGFAFGGGTEVVVKRTVAAPSVFIFPPSDE QLKSGTASVVCLLNNFYPREAKVQWKVDNALQ SGNSQESVTEQDSKDSTYSLSSTLTLSKADYE KHKVYACEVTHQGLSSPVTKSFNRGEC 9 33011-zu Heavy chain 331 EVQLVESGGGLVQPGGSLRLSCAASGISLSSD AISWVRQAPGKGLEYIGIINGGGNTYYASWAK GRFTISRHNSKNTLYLQMNSLRAEDTAVYYCA RGIQHGGGNSDYYYYGMDLWGQGTLVTVSSAS TKGPSVFPLAPSSKSTSGGTAALGCLVKDYFP EPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKV EPKSCDKTHTCPPCPAPELLGGPSVFLFPPKP KDTLMISRTPEVTCVVVDVSHEDPEVKFNWYV DGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQ DWLNGKEYKCKVSNKALPAPIEKTISKAKGQP REPQVYTLPPSRDELTKNQVSLTCLVKGFYPS DIAVEWESNGQPENNYKTTPPVLDSDGSFFLY SKLTVDKSRWQQGNVFSCSVMHEALHNHYTQK SLSLSPGK 10 33011-zu Light chain 332 DIQMTQSPSSLSASVGDRVTITCQASQSISSV LSWYQQKPGKAPKLLIYLASTLASGVPSRFSG SGSGTDFTLTISSLQCEDIATYYCQTNYGTSS SNYGFAFGGGTKVEIKRTVAAPSVFIFPPSDE QLKSGTASVVCLLNNFYPREAKVQWKVDNALQ SGNSQESVTEQDSKDSTYSLSSTLTLSKADYE KHKVYACEVTHQGLSSPVTKSFNRGEC 11 111B10-xi Heavy chain 333 QSVEESGGRLVTPGTPLTLTCTVSGFSLNNYA MSWVRQAPGKGLEWIGSISTGGLAFYANWAKG RFTISRTSTTVDLKMTSLTTEDTATYFCGRNG GGSYIFYYFDLWGQGTLVTVSSASTKGPSVFP LAPSSKSTSGGTAALGCLVKDYFPEPVTVSWN SGALTSGVHTFPAVLQSSGLYSLSSVVTVPSS SLGTQTYICNVNHKPSNTKVDKKVEPKSCDKT HTCPPCPAPELLGGPSVFLFPPKPKDTLMISR TPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNA KTKPREEQYNSTYRVVSVLTVLHQDWLNGKEY KCKVSNKALPAPIEKTISKAKGQPREPQVYTL PPSRDELTKNQVSLTCLVKGFYPSDIAVEWES NGQPENNYKTTPPVLDSDGSFFLYSKLTVDKS RWQQGNVFSCSVMHEALHNHYTQKSISISPGK 12 111B10-xi Light chain 334 AFELTQTPSSVEAAVGGTITIKCQASQSISSY LSWYQQKPGQPPKLLIYSASTLASGVSSRFKG SGSGTEYTLTISDLECADAATYFCQSYYDIGT STFGGGTEVVVKRTVAAPSVFIFPPSDEQLKS GTASVVCLLNNFYPREAKVQWKVDNALQSGNS QESVTEQDSKDSTYSLSSTLTLSKADYEKHKV YACEVTHQGLSSPVTKSFNRGEC 13 111B10-zu Heavy chain 335 EVQLVESGGGLVQPGGSLRLSCAASGFSLNNY AMSWVRQAPGKGLEWIGSISTGGLAFYANWAK GRFTISRDNSKNTLYLQMNSLRAEDTAVYYCA RNGGGSYIFYYFDLWGQGTLVTVSSASTKGPS VFPLAPSSKSTSGGTAALGCLVKDYFPEPVTV SWNSGALTSGVHTFPAVLQSSGLYSISSVVTV PSSSLGTQTYICNVNHKPSNTKVDKKVEPKSC DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLM ISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEV HNAKTKPREEQYNSTYRVVSVLTVLHQDWLNG KEYKCKVSNKALPAPIEKTISKAKGQPREPQV YTLPPSRDELTKNQVSLTCLVKGFYPSDIAVE WESNGQPENNYKTTPPVLDSDGSFFLYSKLTV DKSRWQQGNVFSCSVMHEALHNHYTQKSLSLS PGK 14 111B10-zu Light chain 336 DIQMTQSPSSLSASVGDRVTITCQASQSISSY LSWYQQKPGKAPKLLIYSASTLASGVPSRFSG SGSGTDFTLTISSLQCEDAATYYCQSYYDIGT STFGGGTKVEIKRTVAAPSVFIFPPSDEQLKS GTASVVCLLNNFYPREAKVQWKVDNALQSGNS QESVTEQDSKDSTYSLSSTLTLSKADYEKHKV YACEVTHQGLSSPVTKSFNRGEC 15 201C15-xi Heavy chain 337 QSVKESGGRLVTPGTPLTLTCTVSGIDLSSYA MGWFRQAPGKGLEYIGTINIGGRVYYASWAKG RFTISRTSTTVDLKAPSLTAEDTATYFCARYY NGGSYDIWGPGTLVTVSLASTKGPSVFPLAPS SKSTSGGTAALGCLVKDYFPEPVTVSWNSGAL TSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGT QTYICNVNHKPSNTKVDKKVEPKSCDKTHTCP PCPAPELLGGPSVFLFPPKPKDTLMISRTPEV TCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKP REEQYNSTYRVVSVLTVLHQDWLNGKEYKCKV SNKALPAPIEKTISKAKGQPREPQVYTLPPSR DELTKNQVSLTCLVKGFYPSDIAVEWESNGQP ENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQ GNVFSCSVMHEALHNHYTQKSLSLSPGK 16 201C15-xi Light chain 338 DVVMTQTPASASEPVGGTVTIKCQASESIYRV LAWYQQKPGQPPKLLIYDTSTLASGAPSRFKG SGYGTEFTLTISGVQCEDAATYYCQGGYYADS YGIAFGGGTEVVVKRTVAAPSVFIFPPSDEQL KSGTASVVCLLNNFYPREAKVQWKVDNALQSG NSQESVTEQDSKDSTYSLSSTLTLSKADYEKH KVYACEVTHQGLSSPVTKSFNRGEC 17 201C15-zu Heavy chain 339 QVQLVESGGGLVQPGGSLRLSCSASGIDLSSY AMGWVRQAPGKGLEYIGTINIGGRVYYASWAK GRFTISRDNSKNTLYLQMNSLRAEDTAVYYCA RYYNGGSYDIWGQGTLVTVSSASTKGPSVFPL APSSKSTSGGTAALGCLVKDYFPEPVTVSWNS GALTSGVHTFPAVLQSSGLYSISSVVTVPSSS LGTQTYICNVNHKPSNTKVDKKVEPKSCDKTH TCPPCPAPELLGGPSVFLFPPKPKDTLMISRT PEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAK TKPREEQYNSTYRVVSVLTVLHQDWLNGKEYK CKVSNKALPAPIEKTISKAKGQPREPQVYTLP PSRDELTKNQVSLTCLVKGFYPSDIAVEWESN GQPENNYKTTPPVLDSDGSFFLYSKLTVDKSR WQQGNVFSCSVMHEALHNHYTQKSLSLSPGK 18 201C15-zu Light chain 340 DIQMTQSPSTLSASVGDRVTITCQASESIYRV LAWYQQKPGKAPKLLIYDTSTLASGVPSRFSG SGSGTEFTLTISSLQCDDAATYYCQGGYYADS YGIAFGGGTKVEIKRTVAAPSVFIFPPSDEQL KSGTASVVCLLNNFYPREAKVQWKVDNALQSG NSQESVTEQDSKDSTYSLSSTLTLSKADYEKH KVYACEVTHQGLSSPVTKSFNRGEC 19 346C6-xi Heavy chain 341 QSVEESGGRLVKPDESLTLTCTASGFSLSSYA MIWVRQAPGEGLEWIGTISTGGITYYASWAKG RFTISKTSTTVDLKITSPTTEDTATYFCARGG YAASSAYYLPYYFDLWGQGTLVTVSSASTKGP SVFPLAPSSKSTSGGTAALGCLVKDYFPEPVT VSWNSGALTSGVHTFPAVLQSSGLYSLSSVVT VPSSSLGTQTYICNVNHKPSNTKVDKKVEPKS CDKTHTCPPCPAPELLGGPSVFLFPPKPKDTL MISRTPEVTCVVVDVSHEDPEVKFNWYVDGVE VHNAKTKPREEQYNSTYRVVSVLTVLHQDWLN GKEYKCKVSNKALPAPIEKTISKAKGQPREPQ VYTLPPSRDELTKNQVSLTCLVKGFYPSDIAV EWESNGQPENNYKTTPPVLDSDGSFFLYSKLT VDKSRWQQGNVFSCSVMHEALHNHYTQKSISL SPGK 20 346C6-xi Light chain 342 AAVLTQTPSPVSAAVGGTVTISCQSSQSVYNN NNLAWFQQKPGQPPKLLIYLASTLASGVPSRF SGSGSGTQFTLTISGVQCDDAATYYCLGGCDD DADTFAFGGGTEVVVKRTVAAPSVFIFPPSDE QLKSGTASVVCLLNNFYPREAKVQWKVDNALQ SGNSQESVTEQDSKDSTYSLSSTLTLSKADYE KHKVYACEVTHQGLSSPVTKSFNRGEC 21 346C6-zu Heavy chain 343 EVQLVESGGGLVQPGGSLRLSCAASGFSLSSY AMIWVRQAPGKGLEWIGTISTGGITYYASWAK GRFTISRDNSKNTLYLQMNSLRAEDTAVYYCA RGGYAASSAYYLPYYFDLWGQGTLVTVSSAST KGPSVFPLAPSSKSTSGGTAALGCLVKDYFPE PVTVSWNSGAITSGVHTFPAVLQSSGLYSISS VVTVPSSSLGTQTYICNVNHKPSNTKVDKKVE
PKSCDKTHTCPPCPAPELLGGPSVFLFPPKPK DTIMISRTPEVTCVVVDVSHEDPEVKFNWYVD GVEVHNAKTKPREEQYNSTYRVVSVLTVLHQD WLNGKEYKCKVSNKALPAPIEKTISKAKGQPR EPQVYTLPPSRDELTKNQVSLTCLVKGFYPSD IAVEWESNGQPENNYKTTPPVLDSDGSFFLYS KLTVDKSRWQQGNVFSCSVMHEALHNHYTQKS LSLSPGK 22 346C6-zu Light chain 344 DIQMTQSPSSLSASVGDRVTITCQSSQSVYNN NNLAWYQQKPGKVPKLLIYLASTLASGVPSRF SGSGSGTDFTLTISSLQCEDAATYYCLGGCDD DADTFAFGGGTKVEIKRTVAAPSVFIFPPSDE QLKSGTASVVCLLNNFYPREAKVQWKVDNALQ SGNSQESVTEQDSKDSTYSLSSTLTLSKADYE KHKVYACEVTHQGLSSPVTKSFNRGEC
TABLE-US-00009 TABLE 9 Nucleic acid sequences encoding full-length mAb Ig chains.sup.+ SEQ ID mAb IgG chain NO Nucleic acid sequence 1 MORAb-003 Heavy chain 345 GAGGTCCAACTGGTGGAGAGCGGTGGAGGTGTT GTGCAACCTGGCCGGTCCCTGCGCCTGTCCTGC TCCGCATCTGGCTTCACCTTCAGCGGCTATGGG TTGTCTTGGGTGAGACAGGCACCTGGAAAAGGT CTTGAGTGGGTTGCAATGATTAGTAGTGGTGGT AGTTATACCTACTATGCAGACAGTGTGAAGGGT AGATTTGCAATATCGCGAGACAACGCCAAGAAC ACATTGTTCCTGCAAATGGACAGCCTGAGACCC GAAGACACCGGGGTCTATTTTTGTGCAAGACAT GGGGACGATCCCGCCTGGTTCGCTTATTGGGGC CAAGGGACCCCGGTCACCGTCTCCTCAGCCTCC ACCAAGGGCCCATCGGTCTTCCCCCTGGCACCC TCCTCCAAGAGCACCTCTGGGGGCACAGCGGCC CTGGGCTGCCTGGTCAAGGACTACTTCCCCGAA CCGGTGACGGTGTCGTGGAACTCAGGCGCCCTG ACCAGCGGCGTGCACACCTTCCCGGCTGTCCTA CAGTCCTCAGGACTCTACTCCCTCAGCAGCGTG GTGACCGTGCCCTCCAGCAGCTTGGGCACCCAG ACCTACATCTGCAACGTGAATCACAAGCCCAGC AACACCAAGGTGGACAAGAAAGTTGAGCCCAAA TCTTGTGACAAAACTCACACATGCCCACCGTGC CCAGCACCTGAACTCCTGGGGGGACCGTCAGTC TTCCTCTTCCCCCCAAAACCCAAGGACACCCTC ATGATCTCCCGGACCCCTGAGGTCACATGCGTG GTGGTGGACGTGAGCCACGAAGACCCTGAGGTC AAGTTCAACTGGTACGTGGACGGCGTGGAGGTG CATAATGCCAAGACAAAGCCGCGGGAGGAGCAG TACAACAGCACGTACCGTGTGGTCAGCGTCCTC ACCGTCCTGCACCAGGACTGGCTGAATGGCAAG GAGTACAAGTGCAAGGTCTCCAACAAAGCCCTC CCAGCCCCCATCGAGAAAACCATCTCCAAAGCC AAAGGGCAGCCCCGAGAACCACAGGTGTACACC CTGCCCCCATCCCGGGATGAGCTGACCAAGAAC CAGGTCAGCCTGACCTGCCTGGTCAAAGGCTTC TATCCCAGCGACATCGCCGTGGAGTGGGAGAGC AATGGGCAGCCGGAGAACAACTACAAGACCACG CCTCCCGTGCTGGACTCCGACGGCTCCTTCTTC TTATATTCAAAGCTCACCGTGGACAAGAGCAGG TGGCAGCAGGGGAACGTCTTCTCATGCTCCGTG ATGCATGAGGCTCTGCACAACCACTACACGCAG AAGAGCCTCTCCCTGTCTCCCGGGAAATGA 2 MORAb-003 Light chain 346 GACATCCAGCTGACCCAGAGCCCAAGCAGCCTG AGCGCCAGCGTGGGTGACAGAGTGACCATCACC TGTAGTGTCAGCTCAAGTATAAGTTCCAACAAC TTGCACTGGTACCAGCAGAAGCCAGGTAAGGCT CCAAAGCCATGGATCTACGGCACATCCAACCTG GCTTCTGGTGTGCCAAGCAGATTCAGCGGTAGC GGTAGCGGTACCGACTACACCTTCACCATCAGC AGCCTCCAGCCAGAGGACATCGCCACCTACTAC TGCCAACAGTGGAGTAGTTACCCGTACATGTAC ACGTTCGGCCAAGGGACCAAGGTGGAAATCAAA CGAACTGTGGCTGCACCATCTGTCTTCATCTTC CCGCCATCTGATGAGCAGTTGAAATCTGGAACT GCCTCTGTTGTGTGCCTGCTGAATAACTTCTAT CCCAGAGAGGCCAAAGTACAGTGGAAGGTGGAT AACGCCCTCCAATCGGGTAACTCCCAGGAGAGT GTCACAGAGCAGGACAGCAAGGACAGCACCTAC AGCCTCAGCAGCACCCTGACGCTGAGCAAAGCA GACTACGAGAAACACAAAGTCTACGCCTGCGAA GTCACCCATCAGGGCCTGAGCTCGCCCGTCACA AAGAGCTTCAACAGGGGAGAGTGTTAA 3 MORAb-009 Heavy chain 347 CAGGTACAACTGCAGCAGTCTGGGCCTGAGCTG GAGAAGCCTGGCGCTTCAGTGAAGATATCCTGC AAGGCTTCTGGTTACTCATTCACTGGCTACACC ATGAACTGGGTGAAGCAGAGCCATGGAAAGAGC CTTGAGTGGATTGGACTTATTACTCCTTACAAT GGTGCTTCTAGCTACAACCAGAAGTTCAGGGGC AAGGCCACATTAACTGTAGACAAGTCATCCAGC ACAGCCTACATGGACCTCCTCAGTCTGACATCT GAAGACTCTGCAGTCTATTTCTGTGCAAGGGGG GGTTACGACGGGAGGGGTTTTGACTACTGGGGA TCCGGGACCCCGGTCACCGTCTCCTCAGCCTCC ACCAAGGGCCCATCGGTCTTCCCCCTGGCACCC TCCTCCAAGAGCACCTCTGGGGGCACAGCGGCC CTGGGCTGCCTGGTCAAGGACTACTTCCCCGAA CCGGTGACGGTGTCGTGGAACTCAGGCGCCCTG ACCAGCGGCGTGCACACCTTCCCGGCTGTCCTA CAGTCCTCAGGACTCTACTCCCTCAGCAGCGTG GTGACCGTGCCCTCCAGCAGCTTGGGCACCCAG ACCTACATCTGCAACGTGAATCACAAGCCCAGC AACACCAAGGTGGACAAGAAAGTTGAGCCCAAA TCTTGTGACAAAACTCACACATGCCCACCGTGC CCAGCACCTGAACTCCTGGGGGGACCGTCAGTC TTCCTCTTCCCCCCAAAACCCAAGGACACCCTC ATGATCTCCCGGACCCCTGAGGTCACATGCGTG GTGGTGGACGTGAGCCACGAAGACCCTGAGGTC AAGTTCAACTGGTACGTGGACGGCGTGGAGGTG CATAATGCCAAGACAAAGCCGCGGGAGGAGCAG TACAACAGCACGTACCGTGTGGTCAGCGTCCTC ACCGTCCTGCACCAGGACTGGCTGAATGGCAAG GAGTACAAGTGCAAGGTCTCCAACAAAGCCCTC CCAGCCCCCATCGAGAAAACCATCTCCAAAGCC AAAGGGCAGCCCCGAGAACCACAGGTGTACACC CTGCCCCCATCCCGGGATGAGCTGACCAAGAAC CAGGTCAGCCTGACCTGCCTGGTCAAAGGCTTC TATCCCAGCGACATCGCCGTGGAGTGGGAGAGC AATGGGCAGCCGGAGAACAACTACAAGACCACG CCTCCCGTGCTGGACTCCGACGGCTCCTTCTTC CTCTACAGCAAGCTCACCGTGGACAAGAGCAGG TGGCAGCAGGGGAACGTCTTCTCATGCTCCGTG ATGCATGAGGCTCTGCACAACCACTACACGCAG AAGAGCCTCTCCCTGTCTCCCGGGAAATGA 4 MORAb-009 Light chain 348 GACATCGAGCTCACTCAGTCTCCAGCAATCATG TCTGCATCTCCAGGGGAGAAGGTCACCATGACC TGCAGTGCCAGCTCAAGTGTAAGTTACATGCAC TGGTACCAGCAGAAGTCAGGCACCTCCCCCAAA AGATGGATTTATGACACATCCAAACTGGCTTCT GGAGTCCCAGGTCGCTTCAGTGGCAGTGGGTCT GGAAACTCTTACTCTCTCACAATCAGCAGCGTG GAGGCTGAAGATGATGCAACTTATTACTGCCAG CAGTGGAGTAAGCACCCTCTCACGTTCGGATCC GGGACCAAGGTGGAAATCAAACGAACTGTGGCT GCACCATCTGTCTTCATCTTCCCGCCATCTGAT GAGCAGTTGAAATCTGGAACTGCCTCTGTTGTG TGCCTGCTGAATAACTTCTATCCCAGAGAGGCC AAAGTACAGTGGAAGGTGGATAACGCCCTCCAA TCGGGTAACTCCCAGGAGAGTGTCACAGAGCAG GACAGCAAGGACAGCACCTACAGCCTCAGCAGC ACCCTGACGCTGAGCAAAGCAGACTACGAGAAA CACAAAGTCTACGCCTGCGAAGTCACCCATCAG GGCCTGAGCTCGCCCGTCACAAAGAGCTTCAAC AGGGGAGAGTGTTAA 5 33011-xi Heavy chain 349 CAGTCGGTGGAGGAGTCCGGGGGTCGCCTGGTC ACGCCTGGGACACCCCTGACACTCACCTGCACC GTCTCTGGAATCTCCCTCAGTAGCGATGCAATA AGCTGGGTCCGCCAGGCTCCAGGGAAGGGGCTC GAATACATCGGAATCATTAATGGTGGTGGTAAC ACATACTACGCGAGCTGGGCGAAAGGCCGATTC ACCATCTCCAAAACCTCGACCACGGTGGATCTG AAAATCACCAGTCCGACAACCGAGGACACGGCC ACCTATTTCTGTGCCAGAGGCATTCAACATGGT GGTGGTAATAGTGATTATTATTATTACGGCATG GACCTCTGGGGCCCAGGCACCCTGGTCACTGTC TCTTCAGCATCCACCAAGGGCCCATCGGTCTTC CCCCTGGCACCCTCCTCCAAGAGCACCTCTGGG GGCACAGCGGCCCTGGGCTGCCTGGTCAAGGAC TACTTCCCCGAACCGGTGACGGTGTCGTGGAAC TCAGGCGCCCTGACCAGCGGCGTGCACACCTTC CCGGCTGTCCTACAGTCCTCAGGACTCTACTCC CTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGC TTGGGCACCCAGACCTACATCTGCAACGTGAAT CACAAGCCCAGCAACACCAAGGTGGACAAGAAA GTTGAGCCCAAATCTTGTGACAAAACTCACACA TGCCCACCGTGCCCAGCACCTGAACTCCTGGGG GGACCGTCAGTCTTCCTCTTCCCCCCAAAACCC AAGGACACCCTCATGATCTCCCGGACCCCTGAG GTCACATGCGTGGTGGTGGACGTGAGCCACGAA GACCCTGAGGTCAAGTTCAACTGGTACGTGGAC GGCGTGGAGGTGCATAATGCCAAGACAAAGCCG CGGGAGGAGCAGTACAACAGCACGTACCGTGTG GTCAGCGTCCTCACCGTCCTGCACCAGGACTGG CTGAATGGCAAGGAGTACAAGTGCAAGGTCTCC AACAAAGCCCTCCCAGCCCCCATCGAGAAAACC ATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCA CAGGTGTACACCCTGCCCCCATCCCGGGATGAG CTGACCAAGAACCAGGTCAGCCTGACCTGCCTG GTCAAAGGCTTCTATCCCAGCGACATCGCCGTG GAGTGGGAGAGCAATGGGCAGCCGGAGAACAAC TACAAGACCACGCCTCCCGTGCTGGACTCCGAC GGCTCCTTCTTCTTATATTCAAAGCTCACCGTG GACAAGAGCAGGTGGCAGCAGGGGAACGTCTTC TCATGCTCCGTGATGCATGAGGCTCTGCACAAC CACTACACGCAGAAGAGCCTCTCCCTGTCTCCC GGGAAATGA 6 33011-xi Light chain 350 GAAGTGTTGATGACCCAGACTCCATCCTCCGTG TCTGCAGCTGTGGGAGACACAGTCACCATCAAG TGCCAGGCCAGTCAGAGCATTAGTAGTGTCTTG TCCTGGTATCAGCAGAAACCAGGGCAGCCTCCC AAGCTCCTGATCTATCTGGCATCCACTCTGGCA TCTGGGGTCCCATCGCGGTTCAGCGGCAGTAGA TCTGGGACAGAGTTCACTCTCACCATCAGCGAC CTGGAGTGTGACGATGCTGCCACTTACTACTGT CAAACCAATTATGGTACTAGTAGTAGTAATTAT GGTTTTGCTTTCGGCGGAGGGACCGAGGTGGTC GTCAAACGAACTGTGGCTGCACCATCTGTCTTC ATCTTCCCGCCATCTGATGAGCAGTTGAAATCT GGAACTGCCTCTGTTGTGTGCCTGCTGAATAAC TTCTATCCCAGAGAGGCCAAAGTACAGTGGAAG GTGGATAACGCCCTCCAATCGGGTAACTCCCAG GAGAGTGTCACAGAGCAGGACAGCAAGGACAGC ACCTACAGCCTCAGCAGCACCCTGACGCTGAGC AAAGCAGACTACGAGAAACACAAAGTCTACGCC TGCGAAGTCACCCATCAGGGCCTGAGCTCGCCC GTCACAAAGAGCTTCAACAGGGGAGAGTGTTGA 7 33011-zu Heavy chain 351 GAAGTCCAACTGGTGGAAAGCGGGGGAGGACTG GTGCAGCCGGGCGGATCCCTCCGGCTGTCATGT GCTGCATCGGGAATTTCCCTCTCCTCCGACGCG ATTAGCTGGGTCAGACAGGCCCCCGGAAAGGGG CTGGAGTACATCGGTATCATCAACGGCGGCGGA AACACCTACTACGCCTCCTGGGCCAAGGGCCGC TTCACCATCTCGCGGCATAATTCCAAGAACACT CTGTACTTGCAAATGAACTCCCTGAGGGCCGAG GACACCGCCGTGTACTACTGCGCGCGCGGCATC CAGCACGGTGGTGGAAACAGCGACTACTACTAC TATGGGATGGATCTGTGGGGCCAGGGAACTCTT GTGACCGTGTCGTCAGCATCCACCAAGGGCCCA TCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGC ACCTCTGGGGGCACAGCGGCCCTGGGCTGCCTG GTCAAGGACTACTTCCCCGAACCGGTGACGGTG TCGTGGAACTCAGGCGCCCTGACCAGCGGCGTG CACACCTTCCCGGCTGTCCTACAGTCCTCAGGA CTCTACTCCCTCAGCAGCGTGGTGACCGTGCCC TCCAGCAGCTTGGGCACCCAGACCTACATCTGC AACGTGAATCACAAGCCCAGCAACACCAAGGTG GACAAGAAAGTTGAGCCCAAATCTTGTGACAAA ACTCACACATGCCCACCGTGCCCAGCACCTGAA CTCCTGGGGGGACCGTCAGTCTTCCTCTTCCCC CCAAAACCCAAGGACACCCTCATGATCTCCCGG ACCCCTGAGGTCACATGCGTGGTGGTGGACGTG AGCCACGAAGACCCTGAGGTCAAGTTCAACTGG TACGTGGACGGCGTGGAGGTGCATAATGCCAAG ACAAAGCCGCGGGAGGAGCAGTACAACAGCACG TACCGTGTGGTCAGCGTCCTCACCGTCCTGCAC CAGGACTGGCTGAATGGCAAGGAGTACAAGTGC AAGGTCTCCAACAAAGCCCTCCCAGCCCCCATC GAGAAAACCATCTCCAAAGCCAAAGGGCAGCCC CGAGAACCACAGGTGTACACCCTGCCCCCATCC CGGGATGAGCTGACCAAGAACCAGGTCAGCCTG ACCTGCCTGGTCAAAGGCTTCTATCCCAGCGAC ATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCG GAGAACAACTACAAGACCACGCCTCCCGTGCTG GACTCCGACGGCTCCTTCTTCTTATATTCAAAG CTCACCGTGGACAAGAGCAGGTGGCAGCAGGGG AACGTCTTCTCATGCTCCGTGATGCATGAGGCT CTGCACAACCACTACACGCAGAAGAGCCTCTCC CTGTCTCCCGGGAAATGA 8 33011-zu Light chain 352 GACATTCAGATGACCCAGTCCCCAAGCTCGCTG TCCGCCTCCGTGGGCGACCGCGTGACCATCACG TGCCAGGCGTCCCAGTCAATTAGCAGCGTGCTC TCCTGGTACCAACAGAAGCCGGGGAAAGCACCC AAGCTGCTGATCTACTTGGCCTCCACTCTGGCC TCGGGAGTGCCTTCACGGTTCTCCGGATCGGGA TCTGGTACTGATTTCACCCTCACCATCTCGAGC CTTCAGTGCGAGGACATCGCTACTTACTATTGT CAAACCAACTACGGAACCTCCAGCTCCAACTAC GGCTTTGCCTTCGGTGGCGGGACCAAGGTCGAA ATCAAACGAACTGTGGCTGCACCATCTGTCTTC
ATCTTCCCGCCATCTGATGAGCAGTTGAAATCT GGAACTGCCTCTGTTGTGTGCCTGCTGAATAAC TTCTATCCCAGAGAGGCCAAAGTACAGTGGAAG GTGGATAACGCCCTCCAATCGGGTAACTCCCAG GAGAGTGTCACAGAGCAGGACAGCAAGGACAGC ACCTACAGCCTCAGCAGCACCCTGACGCTGAGC AAAGCAGACTACGAGAAACACAAAGTCTACGCC TGCGAAGTCACCCATCAGGGCCTGAGCTCGCCC GTCACAAAGAGCTTCAACAGGGGAGAGTGTTGA 9 111B10-xi Heavy chain 353 CAGTCGGTGGAGGAGTCCGGGGGTCGCCTGGTC ACGCCTGGGACACCCCTGACACTCACCTGCACA GTCTCTGGATTCTCCCTCAATAACTATGCAATG AGCTGGGTCCGCCAGGCTCCAGGGAAGGGGCTG GAATGGATCGGATCCATTAGTACTGGTGGTCTC GCATTCTACGCGAACTGGGCAAAAGGCCGATTC ACCATCTCCAGAACCTCGACCACGGTGGATCTG AAAATGACCAGTCTGACAACCGAGGACACGGCC ACCTATTTCTGTGGCAGAAATGGTGGTGGTAGT TATATTTTCTATTATTTTGACTTGTGGGGCCAA GGCACCCTCGTCACTGTCTCTTCAGCATCCACC AAGGGCCCATCGGTCTTCCCCCTGGCACCCTCC TCCAAGAGCACCTCTGGGGGCACAGCGGCCCTG GGCTGCCTGGTCAAGGACTACTTCCCCGAACCG GTGACGGTGTCGTGGAACTCAGGCGCCCTGACC AGCGGCGTGCACACCTTCCCGGCTGTCCTACAG TCCTCAGGACTCTACTCCCTCAGCAGCGTGGTG ACCGTGCCCTCCAGCAGCTTGGGCACCCAGACC TACATCTGCAACGTGAATCACAAGCCCAGCAAC ACCAAGGTGGACAAGAAAGTTGAGCCCAAATCT TGTGACAAAACTCACACATGCCCACCGTGCCCA GCACCTGAACTCCTGGGGGGACCGTCAGTCTTC CTCTTCCCCCCAAAACCCAAGGACACCCTCATG ATCTCCCGGACCCCTGAGGTCACATGCGTGGTG GTGGACGTGAGCCACGAAGACCCTGAGGTCAAG TTCAACTGGTACGTGGACGGCGTGGAGGTGCAT AATGCCAAGACAAAGCCGCGGGAGGAGCAGTAC AACAGCACGTACCGTGTGGTCAGCGTCCTCACC GTCCTGCACCAGGACTGGCTGAATGGCAAGGAG TACAAGTGCAAGGTCTCCAACAAAGCCCTCCCA GCCCCCATCGAGAAAACCATCTCCAAAGCCAAA GGGCAGCCCCGAGAACCACAGGTGTACACCCTG CCCCCATCCCGGGATGAGCTGACCAAGAACCAG GTCAGCCTGACCTGCCTGGTCAAAGGCTTCTAT CCCAGCGACATCGCCGTGGAGTGGGAGAGCAAT GGGCAGCCGGAGAACAACTACAAGACCACGCCT CCCGTGCTGGACTCCGACGGCTCCTTCTTCTTA TATTCAAAGCTCACCGTGGACAAGAGCAGGTGG CAGCAGGGGAACGTCTTCTCATGCTCCGTGATG CATGAGGCTCTGCACAACCACTACACGCAGAAG AGCCTCTCCCTGTCTCCCGGGAAATGA 10 111B10-xi Light chain 354 GCATTCGAATTGACCCAGACTCCATCCTCCGTG GAGGCAGCTGTGGGAGGCACAATCACCATCAAG TGCCAGGCCAGTCAGAGCATTAGTAGTTACTTA TCCTGGTATCAGCAGAAACCAGGGCAGCCTCCC AAGCTCCTGATCTATTCTGCATCCACTCTGGCA TCTGGGGTCTCATCGCGGTTCAAAGGCAGTGGA TCTGGGACAGAGTACACTCTCACCATCAGCGAC CTGGAGTGTGCCGATGCTGCCACTTACTTCTGT CAAAGCTATTATGATATTGGTACTAGTACTTTC GGCGGAGGGACCGAGGTGGTCGTCAAACGAACT GTGGCTGCACCATCTGTCTTCATCTTCCCGCCA TCTGATGAGCAGTTGAAATCTGGAACTGCCTCT GTTGTGTGCCTGCTGAATAACTTCTATCCCAGA GAGGCCAAAGTACAGTGGAAGGTGGATAACGCC CTCCAATCGGGTAACTCCCAGGAGAGTGTCACA GAGCAGGACAGCAAGGACAGCACCTACAGCCTC AGCAGCACCCTGACGCTGAGCAAAGCAGACTAC GAGAAACACAAAGTCTACGCCTGCGAAGTCACC CATCAGGGCCTGAGCTCGCCCGTCACAAAGAGC TTCAACAGGGGAGAGTGTTGA 11 111B10-zu Heavy chain 355 GAAGTGCAGCTGGTGGAATCTGGCGGCGGACTG GTGCAGCCTGGCGGATCTCTGAGACTGTCTTGT GCCGCCTCCGGCTTCTCCCTGAACAACTACGCC ATGTCCTGGGTGCGACAGGCCCCTGGCAAAGGC CTGGAATGGATCGGCTCCATCAGCACAGGCGGC CTGGCCTTCTACGCCAATTGGGCCAAGGGCCGG TTCACCATCAGCCGGGACAACTCCAAGAACACC CTGTACCTCCAGATGAACTCCCTGCGGGCCGAG GACACCGCCGTGTACTACTGTGCCAGAAACGGC GGAGGCTCCTACATCTTCTACTACTTCGACCTG TGGGGCCAGGGCACCCTCGTGACAGTGTCATCT GCATCCACCAAGGGCCCATCGGTCTTCCCCCTG GCACCCTCCTCCAAGAGCACCTCTGGGGGCACA GCGGCCCTGGGCTGCCTGGTCAAGGACTACTTC CCCGAACCGGTGACGGTGTCGTGGAACTCAGGC GCCCTGACCAGCGGCGTGCACACCTTCCCGGCT GTCCTACAGTCCTCAGGACTCTACTCCCTCAGC AGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGC ACCCAGACCTACATCTGCAACGTGAATCACAAG CCCAGCAACACCAAGGTGGACAAGAAAGTTGAG CCCAAATCTTGTGACAAAACTCACACATGCCCA CCGTGCCCAGCACCTGAACTCCTGGGGGGACCG TCAGTCTTCCTCTTCCCCCCAAAACCCAAGGAC ACCCTCATGATCTCCCGGACCCCTGAGGTCACA TGCGTGGTGGTGGACGTGAGCCACGAAGACCCT GAGGTCAAGTTCAACTGGTACGTGGACGGCGTG GAGGTGCATAATGCCAAGACAAAGCCGCGGGAG GAGCAGTACAACAGCACGTACCGTGTGGTCAGC GTCCTCACCGTCCTGCACCAGGACTGGCTGAAT GGCAAGGAGTACAAGTGCAAGGTCTCCAACAAA GCCCTCCCAGCCCCCATCGAGAAAACCATCTCC AAAGCCAAAGGGCAGCCCCGAGAACCACAGGTG TACACCCTGCCCCCATCCCGGGATGAGCTGACC AAGAACCAGGTCAGCCTGACCTGCCTGGTCAAA GGCTTCTATCCCAGCGACATCGCCGTGGAGTGG GAGAGCAATGGGCAGCCGGAGAACAACTACAAG ACCACGCCTCCCGTGCTGGACTCCGACGGCTCC TTCTTCTTATATTCAAAGCTCACCGTGGACAAG AGCAGGTGGCAGCAGGGGAACGTCTTCTCATGC TCCGTGATGCATGAGGCTCTGCACAACCACTAC ACGCAGAAGAGCCTCTCCCTGTCTCCCGGGAAA TGA 12 111B10-zu Light chain 356 GATATTCAGATGACCCAGTCCCCCTCCAGCCTG TCCGCTTCTGTGGGCGACAGAGTGACCATCACC TGTCAGGCCTCCCAGTCCATCTCCTCCTACCTG TCCTGGTATCAGCAGAAGCCCGGCAAGGCCCCC AAGCTGCTGATCTACTCTGCCTCCACACTGGCC TCCGGCGTGCCCTCTAGATTCTCCGGCTCTGGC TCTGGCACCGACTTTACCCTGACCATCAGCTCC CTCCAGTGCGAGGATGCCGCCACCTACTACTGC CAGTCCTACTACGACATCGGCACCTCCACCTTC GGCGGAGGCACCAAGGTGGAAATCAAACGAACT GTGGCTGCACCATCTGTCTTCATCTTCCCGCCA TCTGATGAGCAGTTGAAATCTGGAACTGCCTCT GTTGTGTGCCTGCTGAATAACTTCTATCCCAGA GAGGCCAAAGTACAGTGGAAGGTGGATAACGCC CTCCAATCGGGTAACTCCCAGGAGAGTGTCACA GAGCAGGACAGCAAGGACAGCACCTACAGCCTC AGCAGCACCCTGACGCTGAGCAAAGCAGACTAC GAGAAACACAAAGTCTACGCCTGCGAAGTCACC CATCAGGGCCTGAGCTCGCCCGTCACAAAGAGC TTCAACAGGGGAGAGTGTTGA 13 201C15-xi Heavy chain 357 CAGTCAGTGAAGGAGTCCGGGGGTCGCCTGGTC ACGCCTGGGACACCCCTGACACTCACCTGCACA GTCTCTGGAATCGACCTCAGTAGCTATGCAATG GGCTGGTTCCGCCAGGCTCCAGGGAAGGGGCTG GAATACATCGGAACCATTAATATTGGTGGTCGC GTATATTACGCGAGCTGGGCAAAAGGCCGATTC ACCATCTCCAGAACCTCGACCACGGTGGATCTG AAAGCGCCCAGTCTGACAGCCGAGGACACGGCC ACCTATTTCTGTGCCAGATATTATAATGGTGGT AGTTATGACATCTGGGGCCCAGGCACCCTGGTC ACCGTCTCTTTAGCATCCACCAAGGGCCCATCG GTCTTCCCCCTGGCACCCTCCTCCAAGAGCACC TCTGGGGGCACAGCGGCCCTGGGCTGCCTGGTC AAGGACTACTTCCCCGAACCGGTGACGGTGTCG TGGAACTCAGGCGCCCTGACCAGCGGCGTGCAC ACCTTCCCGGCTGTCCTACAGTCCTCAGGACTC TACTCCCTCAGCAGCGTGGTGACCGTGCCCTCC AGCAGCTTGGGCACCCAGACCTACATCTGCAAC GTGAATCACAAGCCCAGCAACACCAAGGTGGAC AAGAAAGTTGAGCCCAAATCTTGTGACAAAACT CACACATGCCCACCGTGCCCAGCACCTGAACTC CTGGGGGGACCGTCAGTCTTCCTCTTCCCCCCA AAACCCAAGGACACCCTCATGATCTCCCGGACC CCTGAGGTCACATGCGTGGTGGTGGACGTGAGC CACGAAGACCCTGAGGTCAAGTTCAACTGGTAC GTGGACGGCGTGGAGGTGCATAATGCCAAGACA AAGCCGCGGGAGGAGCAGTACAACAGCACGTAC CGTGTGGTCAGCGTCCTCACCGTCCTGCACCAG GACTGGCTGAATGGCAAGGAGTACAAGTGCAAG GTCTCCAACAAAGCCCTCCCAGCCCCCATCGAG AAAACCATCTCCAAAGCCAAAGGGCAGCCCCGA GAACCACAGGTGTACACCCTGCCCCCATCCCGG GATGAGCTGACCAAGAACCAGGTCAGCCTGACC TGCCTGGTCAAAGGCTTCTATCCCAGCGACATC GCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAG AACAACTACAAGACCACGCCTCCCGTGCTGGAC TCCGACGGCTCCTTCTTCTTATATTCAAAGCTC ACCGTGGACAAGAGCAGGTGGCAGCAGGGGAAC GTCTTCTCATGCTCCGTGATGCATGAGGCTCTG CACAACCACTACACGCAGAAGAGCCTCTCCCTG TCTCCCGGGAAATGA 14 201C15-xi Light chain 358 GATGTTGTGATGACCCAGACTCCAGCCTCCGCG TCTGAACCTGTGGGAGGCACAGTCACCATCAAG TGCCAGGCCAGTGAGAGCATTTATCGCGTATTG GCCTGGTATCAGCAGAAACCAGGGCAGCCTCCC AAGCTCCTGATCTATGATACATCCACTCTGGCA TCTGGGGCCCCATCGCGGTTCAAAGGCAGTGGA TATGGGACAGAGTTCACTCTCACCATCAGCGGC GTGCAGTGTGAAGATGCTGCCACTTACTACTGT CAAGGCGGTTATTATGCTGATAGTTATGGTATT GCTTTCGGCGGAGGGACCGAGGTGGTGGTCAAA CGAACTGTGGCTGCACCATCTGTCTTCATCTTC CCGCCATCTGATGAGCAGTTGAAATCTGGAACT GCCTCTGTTGTGTGCCTGCTGAATAACTTCTAT CCCAGAGAGGCCAAAGTACAGTGGAAGGTGGAT AACGCCCTCCAATCGGGTAACTCCCAGGAGAGT GTCACAGAGCAGGACAGCAAGGACAGCACCTAC AGCCTCAGCAGCACCCTGACGCTGAGCAAAGCA GACTACGAGAAACACAAAGTCTACGCCTGCGAA GTCACCCATCAGGGCCTGAGCTCGCCCGTCACA AAGAGCTTCAACAGGGGAGAGTGTTGA 15 201C15-zu Heavy chain 359 CAGGTGCAGCTGGTGGAATCTGGCGGAGGACTG GTGCAGCCTGGCGGCTCTCTGAGACTGTCCTGT TCCGCCTCCGGAATCGACCTGTCCTCCTACGCT ATGGGCTGGGTGCGACAGGCTCCTGGCAAGGGC CTGGAGTACATCGGCACCATCAACATCGGCGGC AGAGTGTACTACGCCTCCTGGGCCAAGGGCCGG TTCACCATCTCCAGAGACAACTCCAAGAACACC CTGTACCTCCAGATGAACTCCCTGCGGGCCGAG GACACCGCCGTGTACTACTGCGCCCGGTACTAC AACGGCGGCTCCTACGATATCTGGGGCCAGGGC ACACTCGTGACCGTGTCCTCTGCATCCACCAAG GGCCCATCGGTCTTCCCCCTGGCACCCTCCTCC AAGAGCACCTCTGGGGGCACAGCGGCCCTGGGC TGCCTGGTCAAGGACTACTTCCCCGAACCGGTG ACGGTGTCGTGGAACTCAGGCGCCCTGACCAGC GGCGTGCACACCTTCCCGGCTGTCCTACAGTCC TCAGGACTCTACTCCCTCAGCAGCGTGGTGACC GTGCCCTCCAGCAGCTTGGGCACCCAGACCTAC ATCTGCAACGTGAATCACAAGCCCAGCAACACC AAGGTGGACAAGAAAGTTGAGCCCAAATCTTGT GACAAAACTCACACATGCCCACCGTGCCCAGCA CCTGAACTCCTGGGGGGACCGTCAGTCTTCCTC TTCCCCCCAAAACCCAAGGACACCCTCATGATC TCCCGGACCCCTGAGGTCACATGCGTGGTGGTG GACGTGAGCCACGAAGACCCTGAGGTCAAGTTC AACTGGTACGTGGACGGCGTGGAGGTGCATAAT GCCAAGACAAAGCCGCGGGAGGAGCAGTACAAC AGCACGTACCGTGTGGTCAGCGTCCTCACCGTC CTGCACCAGGACTGGCTGAATGGCAAGGAGTAC AAGTGCAAGGTCTCCAACAAAGCCCTCCCAGCC CCCATCGAGAAAACCATCTCCAAAGCCAAAGGG CAGCCCCGAGAACCACAGGTGTACACCCTGCCC CCATCCCGGGATGAGCTGACCAAGAACCAGGTC AGCCTGACCTGCCTGGTCAAAGGCTTCTATCCC AGCGACATCGCCGTGGAGTGGGAGAGCAATGGG CAGCCGGAGAACAACTACAAGACCACGCCTCCC GTGCTGGACTCCGACGGCTCCTTCTTCTTATAT TCAAAGCTCACCGTGGACAAGAGCAGGTGGCAG CAGGGGAACGTCTTCTCATGCTCCGTGATGCAT GAGGCTCTGCACAACCACTACACGCAGAAGAGC CTCTCCCTGTCTCCCGGGAAATGA 16 201C15-zu Light chain 360 GATATCCAGATGACCCAGTCCCCCTCCACCCTG TCTGCCTCTGTGGGCGACAGAGTGACCATCACC TGTCAGGCCTCCGAGTCCATCTACCGGGTGCTG GCCTGGTATCAGCAGAAGCCTGGCAAGGCCCCC AAGCTGCTGATCTACGACACCAGCACACTGGCC TCCGGCGTGCCCTCTAGATTCTCCGGCTCTGGC TCTGGCACCGAGTTTACCCTGACCATCTCCAGC CTCCAGTGCGACGACGCCGCCACCTACTATTGT CAGGGCGGCTACTACGCCGACTCCTACGGAATC
GCTTTCGGCGGAGGCACCAAGGTGGAAATCAAA CGAACTGTGGCTGCACCATCTGTCTTCATCTTC CCGCCATCTGATGAGCAGTTGAAATCTGGAACT GCCTCTGTTGTGTGCCTGCTGAATAACTTCTAT CCCAGAGAGGCCAAAGTACAGTGGAAGGTGGAT AACGCCCTCCAATCGGGTAACTCCCAGGAGAGT GTCACAGAGCAGGACAGCAAGGACAGCACCTAC AGCCTCAGCAGCACCCTGACGCTGAGCAAAGCA GACTACGAGAAACACAAAGTCTACGCCTGCGAA GTCACCCATCAGGGCCTGAGCTCGCCCGTCACA AAGAGCTTCAACAGGGGAGAGTGTTGA 17 346C6-xi Heavy chain 361 CAGTCGGTGGAGGAGTCCGGCGGTCGCCTGGTA AAGCCTGACGAATCCCTGACACTCACCTGCACA GCCTCTGGATTCTCCCTCAGTAGTTATGCAATG ATCTGGGTCCGCCAGGCTCCAGGGGAGGGGCTG GAATGGATCGGAACCATTAGTACTGGTGGTATC ACATACTACGCGAGCTGGGCGAAAGGCCGATTC ACCATCTCCAAAACCTCGACCACGGTGGATCTG AAAATCACCAGTCCGACAACCGAGGACACGGCC ACCTATTTCTGTGCCAGAGGGGGATATGCTGCT AGTAGTGCTTATTATCTCCCGTACTACTTTGAC TTGTGGGGCCAAGGGACCCTGGTCACCGTCTCC TCAGCATCCACCAAGGGCCCATCGGTCTTCCCC CTGGCACCCTCCTCCAAGAGCACCTCTGGGGGC ACAGCGGCCCTGGGCTGCCTGGTCAAGGACTAC TTCCCCGAACCGGTGACGGTGTCGTGGAACTCA GGCGCCCTGACCAGCGGCGTGCACACCTTCCCG GCTGTCCTACAGTCCTCAGGACTCTACTCCCTC AGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTG GGCACCCAGACCTACATCTGCAACGTGAATCAC AAGCCCAGCAACACCAAGGTGGACAAGAAAGTT GAGCCCAAATCTTGTGACAAAACTCACACATGC CCACCGTGCCCAGCACCTGAACTCCTGGGGGGA CCGTCAGTCTTCCTCTTCCCCCCAAAACCCAAG GACACCCTCATGATCTCCCGGACCCCTGAGGTC ACATGCGTGGTGGTGGACGTGAGCCACGAAGAC CCTGAGGTCAAGTTCAACTGGTACGTGGACGGC GTGGAGGTGCATAATGCCAAGACAAAGCCGCGG GAGGAGCAGTACAACAGCACGTACCGTGTGGTC AGCGTCCTCACCGTCCTGCACCAGGACTGGCTG AATGGCAAGGAGTACAAGTGCAAGGTCTCCAAC AAAGCCCTCCCAGCCCCCATCGAGAAAACCATC TCCAAAGCCAAAGGGCAGCCCCGAGAACCACAG GTGTACACCCTGCCCCCATCCCGGGATGAGCTG ACCAAGAACCAGGTCAGCCTGACCTGCCTGGTC AAAGGCTTCTATCCCAGCGACATCGCCGTGGAG TGGGAGAGCAATGGGCAGCCGGAGAACAACTAC AAGACCACGCCTCCCGTGCTGGACTCCGACGGC TCCTTCTTCTTATATTCAAAGCTCACCGTGGAC AAGAGCAGGTGGCAGCAGGGGAACGTCTTCTCA TGCTCCGTGATGCATGAGGCTCTGCACAACCAC TACACGCAGAAGAGCCTCTCCCTGTCTCCCGGG AAATGA 18 346C6-xi Light chain 362 GCAGCCGTGCTGACCCAGACACCATCACCCGTG TCTGCAGCTGTGGGAGGCACAGTCACCATCAGT TGCCAGTCCAGTCAGAGTGTTTATAATAATAAC AACTTAGCCTGGTTTCAGCAGAAACCCGGGCAG CCTCCCAAGCTTCTGATCTATCTGGCATCCACT CTGGCATCTGGGGTCCCATCACGGTTCAGCGGC AGTGGATCTGGGACACAGTTCACTCTCACCATC AGCGGCGTGCAGTGTGACGATGCTGCCACTTAT TACTGTCTAGGTGGTTGTGATGATGATGCTGAT ACTTTTGCTTTCGGCGGAGGGACTGAGGTGGTG GTCAAACGAACTGTGGCTGCACCATCTGTCTTC ATCTTCCCGCCATCTGATGAGCAGTTGAAATCT GGAACTGCCTCTGTTGTGTGCCTGCTGAATAAC TTCTATCCCAGAGAGGCCAAAGTACAGTGGAAG GTGGATAACGCCCTCCAATCGGGTAACTCCCAG GAGAGTGTCACAGAGCAGGACAGCAAGGACAGC ACCTACAGCCTCAGCAGCACCCTGACGCTGAGC AAAGCAGACTACGAGAAACACAAAGTCTACGCC TGCGAAGTCACCCATCAGGGCCTGAGCTCGCCC GTCACAAAGAGCTTCAACAGGGGAGAGTGTTGA 19 346C6-zu Heavy chain 363 GAAGTGCAGCTGGTGGAATCTGGCGGCGGACTG GTGCAGCCTGGCGGATCTCTGAGACTGTCTTGT GCCGCCTCCGGCTTCTCCCTGTCCTCCTACGCT ATGATCTGGGTGCGACAGGCCCCTGGCAAGGGC CTGGAATGGATCGGCACCATCTCTACCGGCGGA ATTACCTACTACGCCTCCTGGGCCAAGGGCCGG TTCACCATCTCCAGAGACAACTCCAAGAACACC CTGTACCTCCAGATGAACTCCCTGCGGGCCGAG GACACCGCCGTGTACTATTGTGCTAGAGGCGGC TACGCCGCCAGCTCCGCTTACTACCTGCCCTAC TACTTCGACCTGTGGGGCCAGGGCACCCTCGTG ACAGTGTCATCTGCATCCACCAAGGGCCCATCG GTCTTCCCCCTGGCACCCTCCTCCAAGAGCACC TCTGGGGGCACAGCGGCCCTGGGCTGCCTGGTC AAGGACTACTTCCCCGAACCGGTGACGGTGTCG TGGAACTCAGGCGCCCTGACCAGCGGCGTGCAC ACCTTCCCGGCTGTCCTACAGTCCTCAGGACTC TACTCCCTCAGCAGCGTGGTGACCGTGCCCTCC AGCAGCTTGGGCACCCAGACCTACATCTGCAAC GTGAATCACAAGCCCAGCAACACCAAGGTGGAC AAGAAAGTTGAGCCCAAATCTTGTGACAAAACT CACACATGCCCACCGTGCCCAGCACCTGAACTC CTGGGGGGACCGTCAGTCTTCCTCTTCCCCCCA AAACCCAAGGACACCCTCATGATCTCCCGGACC CCTGAGGTCACATGCGTGGTGGTGGACGTGAGC CACGAAGACCCTGAGGTCAAGTTCAACTGGTAC GTGGACGGCGTGGAGGTGCATAATGCCAAGACA AAGCCGCGGGAGGAGCAGTACAACAGCACGTAC CGTGTGGTCAGCGTCCTCACCGTCCTGCACCAG GACTGGCTGAATGGCAAGGAGTACAAGTGCAAG GTCTCCAACAAAGCCCTCCCAGCCCCCATCGAG AAAACCATCTCCAAAGCCAAAGGGCAGCCCCGA GAACCACAGGTGTACACCCTGCCCCCATCCCGG GATGAGCTGACCAAGAACCAGGTCAGCCTGACC TGCCTGGTCAAAGGCTTCTATCCCAGCGACATC GCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAG AACAACTACAAGACCACGCCTCCCGTGCTGGAC TCCGACGGCTCCTTCTTCTTATATTCAAAGCTC ACCGTGGACAAGAGCAGGTGGCAGCAGGGGAAC GTCTTCTCATGCTCCGTGATGCATGAGGCTCTG CACAACCACTACACGCAGAAGAGCCTCTCCCTG TCTCCCGGGAAATGA 20 346C6-zu Light chain 364 GATATTCAGATGACCCAGTCCCCCTCCAGCCTG TCCGCTTCTGTGGGCGACAGAGTGACCATCACC TGTCAGTCCTCCCAGTCCGTGTATAACAACAAC AACCTGGCCTGGTATCAGCAGAAACCCGGCAAG GTGCCCAAGCTGCTGATCTACCTGGCCTCCACA CTGGCCTCTGGCGTGCCCTCTAGATTCTCCGGC TCTGGCTCTGGCACCGACTTTACCCTGACCATC AGCTCCCTCCAGTGCGAGGATGCCGCCACCTAC TATTGCCTGGGCGGCTGCGACGACGACGCCGAT ACCTTTGCTTTTGGCGGAGGCACCAAGGTGGAA ATCAAACGAACTGTGGCTGCACCATCTGTCTTC ATCTTCCCGCCATCTGATGAGCAGTTGAAATCT GGAACTGCCTCTGTTGTGTGCCTGCTGAATAAC TTCTATCCCAGAGAGGCCAAAGTACAGTGGAAG GTGGATAACGCCCTCCAATCGGGTAACTCCCAG GAGAGTGTCACAGAGCAGGACAGCAAGGACAGC ACCTACAGCCTCAGCAGCACCCTGACGCTGAGC AAAGCAGACTACGAGAAACACAAAGTCTACGCC TGCGAAGTCACCCATCAGGGCCTGAGCTCGCCC GTCACAAAGAGCTTCAACAGGGGAGAGTGTTGA .sup.+Nucleic acid sequences listed do not include leader sequences.
[0122] In various embodiments, an ADC disclosed herein may comprise any set of heavy and light chain variable domains listed in the tables above (e.g., MORAb-003 heavy and light chain variable domains, or trastuzumab heavy and light chain variable domains), or the set of six CDR sequences from the heavy and light chain set. In some embodiments, the ADC further comprises human heavy and light chain constant domains or fragments thereof. For instance, the ADC may comprise a human IgG heavy chain constant domain (such as an IgG1) and a human kappa or lambda light chain constant domain. In various embodiments, the antibody moiety of the described ADCs comprises a human immunoglobulin G subtype 1 (IgG1) heavy chain constant domain with a human Ig kappa light chain constant domain.
[0123] In various embodiments, the target cancer antigen for an ADC is folate receptor alpha ("FRA").
[0124] In various embodiments, the anti-FRA antibody or antigen-binding fragment thereof comprises three heavy chain CDRs and three light chain CDRs as follows: heavy chain CDR1 (HCDR1) consisting of SEQ ID NO:2, heavy chain CDR2 (HCDR2) consisting of SEQ ID NO:3, heavy chain CDR3 (HCDR3) consisting of SEQ ID NO:4; light chain CDR1 (LCDR1) consisting of SEQ ID NO:7, light chain CDR2 (LCDR2) consisting of SEQ ID NO:8, and light chain CDR3 (LCDR3) consisting of SEQ ID NO:9, as defined by the Kabat numbering system (Kabat, Sequences of Proteins of Immunological Interest (National Institutes of Health, Bethesda, Md. (1987 and 1991))).
[0125] In some embodiments, the anti-FRA antibody or antigen-binding fragment thereof comprises three heavy chain CDRs and three light chain CDRs as follows: heavy chain CDR1 consisting of SEQ ID NO:13, heavy chain CDR2 consisting of SEQ ID NO:14, heavy chain CDR3 consisting of SEQ ID NO:15; light chain CDR1 consisting of SEQ ID NO:16, light chain CDR2 consisting of SEQ ID NO:17, and light chain CDR3 consisting of SEQ ID NO:18, as defined by the IMGT numbering system (International ImMunoGeneTics Information System (IMGT.RTM.)).
[0126] In various embodiments, the anti-FRA antibody or antigen-binding fragment thereof comprises a heavy chain variable region comprising the amino acid sequence of SEQ ID NO:23, and a light chain variable region comprising the amino acid sequence of SEQ ID NO:24. In some embodiments, the anti-FRA antibody or antigen-binding fragment thereof comprises the heavy chain variable region amino acid sequence of SEQ ID NO:23 and the light chain variable region amino acid sequence of SEQ ID NO:24, or sequences that are at least 95% identical to the above-mentioned sequences. In some embodiments, the anti-FRA antibody or antigen-binding fragment thereof has a heavy chain variable region amino acid sequence that is at least 96%, at least 97%, at least 98%, or at least 99% identical to SEQ ID NO:23 and a light chain variable region amino acid sequence that is at least 96%, at least 97%, at least 98%, or at least 99% identical to SEQ ID NO:24.
[0127] In various embodiments, the anti-FRA antibody comprises a human IgG1 heavy chain constant domain with a human Ig kappa light chain constant domain.
[0128] In various embodiments, the anti-FRA antibody comprises the heavy chain amino acid sequence of SEQ ID NO:1 or a sequence that is at least 95% identical to SEQ ID NO:1, and the light chain amino acid sequence of SEQ ID NO:6 or a sequence that is at least 95% identical to SEQ ID NO:6. In particular embodiments, the antibody comprises the heavy chain amino acid sequence of SEQ ID NO:1 and the light chain amino acid sequence of SEQ ID NO:6, or sequences that are at least 95% identical to the above-mentioned sequences. In some embodiments, the anti-FRA antibody has a heavy chain amino acid sequence that is at least 96%, at least 97%, at least 98%, or at least 99% identical to SEQ ID NO:1 and/or a light chain amino acid sequence that is at least 96%, at least 97%, at least 98%, or at least 99% identical to SEQ ID NO:6. In some embodiments, the anti-FRA antibody comprises a heavy chain encoded by the nucleotide sequence of SEQ ID NO:11 (with the nucleotides encoding the leader sequence), or SEQ ID NO:345 (without the nucleotides encoding the leader sequence); and a light chain encoded by the nucleotide sequence of SEQ ID NO:12 (with the nucleotides encoding the leader sequence), or SEQ ID NO:346 (without the nucleotides encoding the leader sequence). In some embodiments, the heavy chain amino acid sequence lacks the C-terminal lysine. In various embodiments, the anti-FRA antibody has the amino acid sequence of the antibody produced by a cell line deposited under terms in accordance with the Budapest Treaty with the American Type Culture Collection (ATCC, 10801 University Blvd., Manassas, Va. 20110-2209) on Apr. 24, 2006, under the Accession No. PTA-7552, or such sequences lacking the heavy chain C-terminal lysine. In various embodiments, the anti-FRA antibody is MORAb-003 (USAN name: farletuzumab) (Ebel et al. (2007) Cancer Immunity 7:6), or an antigen-binding fragment thereof.
[0129] In various other embodiments, the target cancer antigen for an ADC is human epidermal growth factor receptor 2 ("her2").
[0130] In various embodiments, the anti-her2 antibody or antigen-binding fragment thereof comprises three heavy chain CDRs and three light chain CDRs as follows: heavy chain CDR1 (HCDR1) consisting of SEQ ID NO:71, heavy chain CDR2 (HCDR2) consisting of SEQ ID NO:72, heavy chain CDR3 (HCDR3) consisting of SEQ ID NO:73; light chain CDR1 (LCDR1) consisting of SEQ ID NO:74, light chain CDR2 (LCDR2) consisting of SEQ ID NO:75, and light chain CDR3 (LCDR3) consisting of SEQ ID NO:76, as defined by the Kabat numbering system.
[0131] In some embodiments, the anti-her2 antibody or antigen-binding fragment thereof comprises three heavy chain CDRs and three light chain CDRs as follows: heavy chain CDR1 consisting of SEQ ID NO:191, heavy chain CDR2 consisting of SEQ ID NO:192, heavy chain CDR3 consisting of SEQ ID NO:193; light chain CDR1 consisting of SEQ ID NO:194, light chain CDR2 consisting of SEQ ID NO:195, and light chain CDR3 consisting of SEQ ID NO:196, as defined by the IMGT numbering system.
[0132] In various embodiments, the anti-her2 antibody or antigen-binding fragment thereof comprises a heavy chain variable region comprising the amino acid sequence of SEQ ID NO:27, and a light chain variable region comprising the amino acid sequence of SEQ ID NO:28. In some embodiments, the anti-her2 antibody or antigen-binding fragment thereof comprises the heavy chain variable region amino acid sequence of SEQ ID NO:27 and the light chain variable region amino acid sequence of SEQ ID NO:28, or sequences that are at least 95% identical to the above-mentioned sequences. In some embodiments, the anti-her2 antibody or antigen-binding fragment thereof has a heavy chain variable region amino acid sequence that is at least 96%, at least 97%, at least 98%, or at least 99% identical to SEQ ID NO:27 and/or a light chain variable region amino acid sequence that is at least 96%, at least 97%, at least 98%, or at least 99% identical to SEQ ID NO:28.
[0133] In various embodiments, the anti-her2 antibody comprises a human IgG1 heavy chain constant domain and a human Ig kappa light chain constant domain.
[0134] In various embodiments, the anti-her2 antibody comprises the heavy chain amino acid sequence of SEQ ID NO:327 or a sequence that is at least 95% identical to SEQ ID NO:327, and the light chain amino acid sequence of SEQ ID NO:328 or a sequence that is at least 95% identical to SEQ ID NO:328. In particular embodiments, the antibody comprises the heavy chain amino acid sequence of SEQ ID NO:327 and the light chain amino acid sequence of SEQ ID NO:328, or sequences that are at least 95% identical to the above-mentioned sequences. In some embodiments, the anti-her2 antibody has a heavy chain amino acid sequence that is at least 96%, at least 97%, at least 98%, or at least 99% identical to SEQ ID NO:327 and a light chain amino acid sequence that is at least 96%, at least 97%, at least 98%, or at least 99% identical to SEQ ID NO:328. In various embodiments, the anti-her2 antibody is trastuzumab, or an antigen-binding fragment thereof.
[0135] In various embodiments, the anti-FRA antibody or antigen-binding fragment thereof comprises the three heavy chain CDRs and three light chain CDRs of MORAb-003 or wherein the CDRs include no more than one, two, three, four, five, or six amino acid additions, deletions or substitutions of HCDR1 (SEQ ID NO:2 according to Kabat, or SEQ ID NO:13 according to IMGT), HCDR2 (SEQ ID NO:3 according to Kabat, or SEQ ID NO:14 according to IMGT), HCDR3 (SEQ ID NO:4 according to Kabat, or SEQ ID NO:15 according to IMGT); LCDR1 (SEQ ID NO:7 according to Kabat, or SEQ ID NO:16 according to IMGT), LCDR2 (SEQ ID NO:8 according to Kabat, or SEQ ID NO:17 according to IMGT), and LCDR3 (SEQ ID NO:9 according to Kabat, or SEQ ID NO:18 according to IMGT).
[0136] In various other embodiments, the anti-her2 antibody or antigen-binding fragment thereof comprises the three heavy chain CDRs and three light chain CDRs of trastuzumab or wherein the CDRs include no more than one, two, three, four, five, or six amino acid additions, deletions or substitutions of HCDR1 (SEQ ID NO:71 according to Kabat, or SEQ ID NO:191 according to IMGT), HCDR2 (SEQ ID NO:72 according to Kabat, or SEQ ID NO:192 according to IMGT), HCDR3 (SEQ ID NO:73 according to Kabat, or SEQ ID NO:193 according to IMGT); LCDR1 (SEQ ID NO:74 according to Kabat, or SEQ ID NO:194 according to IMGT), LCDR2 (SEQ ID NO:75 according to Kabat, or SEQ ID NO:195 according to IMGT), and LCDR3 (SEQ ID NO:76 according to Kabat, or SEQ ID NO:196 according to IMGT).
[0137] In various embodiments, amino acid substitutions are of single residues. Insertions usually will be on the order of from about 1 to about 20 amino acid residues, although considerably larger insertions may be tolerated as long as biological function is retained (e.g., binding to FRA or her2). Deletions usually range from about 1 to about 20 amino acid residues, although in some cases deletions may be much larger. Substitutions, deletions, insertions, or any combination thereof may be used to arrive at a final derivative or variant. Generally these changes are done on a few amino acids to minimize the alteration of the molecule, particularly the immunogenicity and specificity of the antigen binding protein. However, larger changes may be tolerated in certain circumstances. Conservative substitutions are generally made in accordance with the following chart depicted as Table 10.
TABLE-US-00010 TABLE 10 Original Residue Exemplary Substitutions Ala Ser Arg Lys Asn Gln, His Asp Glu Cys Ser Gln Asn Glu Asp Gly Pro His Asn, Gln Ile Leu, Val Leu Ile, Val Lys Arg, Gln, Glu Met Leu, Ile Phe Met, Leu, Tyr Ser Thr Thr Ser Trp Tyr Tyr Trp, Phe Val Ile, Leu
[0138] Substantial changes in function or immunological identity are made by selecting substitutions that are less conservative than those shown in Table 10. For example, substitutions may be made which more significantly affect: the structure of the polypeptide backbone in the area of the alteration, for example the alpha-helical or beta-sheet structure; the charge or hydrophobicity of the molecule at the target site; or the bulk of the side chain. The substitutions which in general are expected to produce the greatest changes in the polypeptide's properties are those in which (a) a hydrophilic residue, e.g., seryl or threonyl, is substituted for (or by) a hydrophobic residue, e.g., leucyl, isoleucyl, phenylalanyl, valyl or alanyl; (b) a cysteine or proline is substituted for (or by) any other residue; (c) a residue having an electropositive side chain, e.g., lysyl, arginyl, or histidyl, is substituted for (or by) an electronegative residue, e.g., glutamyl or aspartyl; or (d) a residue having a bulky side chain, e.g., phenylalanine, is substituted for (or by) one not having a side chain, e.g., glycine.
[0139] In various embodiments where variant antibody sequences are used in an ADC, the variants typically exhibit the same qualitative biological activity and will elicit the same immune response, although variants may also be selected to modify the characteristics of the antigen binding proteins as needed. Alternatively, the variant may be designed such that the biological activity of the antigen binding protein is altered. For example, glycosylation sites may be altered or removed, as discussed herein.
[0140] Various antibodies may be used with the ADCs used herein to target cancer cells. As shown below, the linker-toxins in the ADCs disclosed herein are surprisingly effective with different tumor antigen-targeting antibodies. Suitable antigens expressed on tumor cells but not healthy cells, or expressed on tumor cells at a higher level than on healthy cells, are known in the art, as are antibodies directed against them. These antibodies may be used with the linkers and toxin (e.g., eribulin) disclosed herein. In some embodiments, the antibody moiety targets FRA. In some embodiments, the FRA-targeting antibody moiety is MORAb-003. In some embodiments, while the disclosed linkers and toxin (eribulin) are surprisingly effective with several different tumor-targeting antibodies, FRA-targeting antibody moieties such as MORAb-003 provided particularly improved drug:antibody ratio, tumor targeting, bystander killing, treatment efficacy, and reduced off-target killing. Improved treatment efficacy can be measured in vitro or in vivo, and may include reduced tumor growth rate and/or reduced tumor volume.
[0141] In certain embodiments, antibodies to other antigen targets are used and provide at least some of the favorable functional properties of an ADC comprising an FRA-targeting antibody moiety such as MORAb-003 (e.g., improved drug:antibody ratio, improved treatment efficacy, reduced off-target killing, etc.). In some embodiments, some or all of these favorable functional properties are observed when the disclosed linkers and toxin (eribulin) are conjugated to a her2-targeting antibody moiety such as trastuzumab. In some embodiments, the antibody moiety targets her2. In some embodiments, the her2-targeting antibody moiety is trastuzumab. In some embodiments, some or all of these favorable functional properties are observed when the disclosed linkers and toxin (eribulin) are conjugated to a MSLN-targeting antibody moiety such as MORAb-009. In some embodiments, the antibody moiety targets MSLN. In some embodiments, the MSLN-targeting antibody moiety is MORAb-009.
Linkers
[0142] In various embodiments, the linker in an ADC is stable extracellularly in a sufficient manner to be therapeutically effective. In some embodiments, the linker is stable outside a cell, such that the ADC remains intact when present in extracellular conditions (e.g., prior to transport or delivery into a cell). The term "intact," used in the context of an ADC, means that the antibody moiety remains attached to the drug moiety. As used herein, "stable," in the context of a linker or ADC comprising a linker, means that no more than 20%, no more than about 15%, no more than about 10%, no more than about 5%, no more than about 3%, or no more than about 1% of the linkers (or any percentage in between) in a sample of ADC are cleaved (or in the case of an overall ADC are otherwise not intact) when the ADC is present in extracellular conditions.
[0143] Whether a linker is stable extracellularly can be determined, for example, by including an ADC in plasma for a predetermined time period (e.g., 2, 4, 6, 8, 16, or 24 hours) and then quantifying the amount of free drug moiety present in the plasma. Stability may allow the ADC time to localize to target tumor cells and prevent the premature release of the drug, which could lower the therapeutic index of the ADC by indiscriminately damaging both normal and tumor tissues. In some embodiments, the linker is stable outside of a target cell and releases the drug moiety from the ADC once inside of the cell, such that the drug moiety can bind to its target (e.g., to microtubules). Thus, an effective linker will: (i) maintain the specific binding properties of the antibody moiety; (ii) allow delivery, e.g., intracellular delivery, of the drug moiety via stable attachment to the antibody moiety; (iii) remain stable and intact until the ADC has been transported or delivered to its target site; and (iv) allow for the therapeutic effect, e.g., cytotoxic effect, of the drug moiety after cleavage.
[0144] Linkers may impact the physico-chemical properties of an ADC. As many cytotoxic agents are hydrophobic in nature, linking them to the antibody with an additional hydrophobic moiety may lead to aggregation. ADC aggregates are insoluble and often limit achievable drug loading onto the antibody, which can negatively affect the potency of the ADC. Protein aggregates of biologics, in general, have also been linked to increased immunogenicity. As shown below, linkers disclosed herein result in ADCs with low aggregation levels and desirable levels of drug loading.
[0145] A linker may be "cleavable" or "non-cleavable" (Ducry and Stump, Bioconjugate Chem. (2010) 21:5-13). Cleavable linkers are designed to release the drug when subjected to certain environment factors, e.g., when internalized into the target cell, whereas non-cleavable linkers generally rely on the degradation of the antibody moiety itself.
[0146] In some embodiments, the linker is a non-cleavable linker. In some embodiments, the drug moiety of the ADC is released by degradation of the antibody moiety. Non-cleavable linkers tend to remain covalently associated with at least one amino acid of the antibody and the drug upon internalization by and degradation within the target cell. Non-cleavable linkers commonly include a thioether linkage, which is prepared by the conjugation of a thiol group on the drug or the antibody with a maleimide or haloacetamide group on the antibody or drug, respectively (Goldmacher et. al., In Cancer Drug Discovery and Development: Antibody-Drug Conjugates and Immunotoxins (G. L. Phillips ed., Springer, 2013)). An exemplary non-cleavable linker comprises thioether, cyclohexyl, N-succinimidyl 4-(N-maleimidomethyl) cyclohexane-1 carboxylate (SMCC), N-hydroxysuccinimide (NHS), or one or more polyethylene glycol (PEG) moieties, e.g., 1, 2, 3, 4, 5, or 6 PEG moieties. In some embodiments, the non-cleavable linker comprises (PEG).sub.2. In other embodiments, the non-cleavable linker comprises (PEG).sub.4.
[0147] In some embodiments, the linker is a cleavable linker. A cleavable linker refers to any linker that comprises a cleavable moiety. As used herein, the term "cleavable moiety" refers to any chemical bond that can be cleaved. Suitable cleavable chemical bonds are well known in the art and include, but are not limited to, acid labile bonds, protease/peptidase labile bonds, photolabile bonds, disulfide bonds, and esterase labile bonds. Linkers comprising a cleavable moiety can allow for the release of the drug moiety from the ADC via cleavage at a particular site in the linker. In various embodiments, cleavage of the antibody from the linked toxin activates or increases the activity of the toxin. In some embodiments, an ADC comprising a cleavable linker (e.g., a Val-Cit linker) demonstrates increased on-target cell killing and/or decreased off-target cell killing, as compared to an ADC comprising a non-cleavable linker (e.g., a non-cleavable (PEG).sub.2 or (PEG).sub.4 linker). In some embodiments, an ADC comprising a cleavable linker exhibits improved treatment efficacy relative to an ADC comprising a non-cleavable linker when the cells and/or the cancer treated with the ADC does not express high levels of the target antigen (e.g., FRA or her2). In some embodiments, cleavage of the antibody from the linked toxin is required to achieve improved treatment efficacy of an ADC, as measured in vitro and/or in vivo.
[0148] In some embodiments, the linker is cleavable under intracellular conditions, such that cleavage of the linker sufficiently releases the drug moiety from the antibody moiety in the intracellular environment to activate the drug and/or render the drug therapeutically effective. In some embodiments, the drug moiety is not cleaved from the antibody moiety until the ADC enters a cell that expresses an antigen specific for the antibody moiety of the ADC, and the drug moiety is cleaved from the antibody moiety upon entering the cell. In some embodiments, the linker comprises a cleavable moiety that is positioned such that no part of the linker or the antibody moiety remains bound to the drug moiety upon cleavage. Exemplary cleavable linkers include acid labile linkers, protease/peptidase-sensitive linkers, photolabile linkers, dimethyl-, disulfide-, or sulfonamide-containing linkers.
[0149] In some embodiments, the linker is a pH-sensitive linker, and is sensitive to hydrolysis at certain pH values. Typically, the pH-sensitive linker is cleavable under acidic conditions. This cleavage strategy generally takes advantage of the lower pH in the endosomal (pH.about.5-6) and lysosomal (pH.about.4.8) intracellular compartments, as compared to the cytosol (pH.about.7.4), to trigger hydrolysis of an acid labile group in the linker, such as a hydrazone (Jain et al. (2015) Pharm Res 32:3526-40). In some embodiments, the linker is an acid labile and/or hydrolyzable linker. For example, an acid labile linker that is hydrolyzable in the lysosome, and contains an acid labile group (e.g., a hydrazone, a semicarbazone, a thiosemicarbazone, a cis-aconitic amide, an orthoester, an acetal, a ketal, or the like) can be used. See, e.g., U.S. Pat. Nos. 5,122,368; 5,824,805; 5,622,929; Dubowchik and Walker (1999) Pharm. Therapeutics 83:67-123; Neville et al. (1989) Biol. Chem. 264:14653-61. Such linkers are relatively stable under neutral pH conditions, such as those in the blood, but are unstable at below pH 5.5 or 5.0, the approximate pH of the lysosome. In certain embodiments, the hydrolyzable linker is a thioether linker (such as, e.g., a thioether attached to the therapeutic agent via an acylhydrazone bond). See, e.g., U.S. Pat. No. 5,622,929.
[0150] In some embodiments, the linker is cleavable under reducing conditions. In some embodiments, the linker is cleavable in the presence of a reducing agent, such as glutathione or dithiothreitol. In some embodiments, the linker is a cleavable disulfide linker or a cleavable sulfonamide linker.
[0151] In some embodiments, the linker is a cleavable disulfide linker. A variety of disulfide linkers are known in the art, including, for example, those that can be formed using SATA (N-succinimidyl-5-acetylthioacetate), SPDP (N-succinimidyl-3-(2-pyridyldithio)propionate), SPDB (N-succinimidyl-3-(2-pyridyldithio)butyrate) and SMPT (N-succinimidyloxycarbonyl-alpha-methyl-alpha-(2-pyridyl-dithio)toluene), SPDB and SMPT. See, e.g., Thorpe et al. (1987) Cancer Res. 47:5924-31; Wawrzynczak et al., In Immunoconjugates: Antibody Conjugates in Radioimagery and Therapy of Cancer (C. W. Vogel ed., Oxford U. Press, 1987). See also U.S. Pat. No. 4,880,935. Disulfide linkers are typically used to exploit the abundance of intracellular thiols, which can facilitate the cleavage of their disulfide bonds. The intracellular concentrations of the most abundance intracellular thiol, reduced glutathione, are generally in the range of 1-10 nM, which is about 1,000-fold higher than that of the most abundant low-molecular thiol in the blood (i.e., cysteine) at about 5 .mu.M (Goldmacher et. al., In Cancer Drug Discovery and Development: Antibody-Drug Conjugates and Immunotoxins (G. L. Phillips ed., Springer, 2013)). The intracellular enzymes of the protein disulfide isomerase family may also contribute to the intracellular cleavage of a disulfide linker. As used herein, a cleavable disulfide linker refers to any linker that comprises a cleavable disulfide moiety. The term "cleavable disulfide moiety" refers to a disulfide bond that can be cleaved and/or reduced, e.g., by a thiol or enzyme. In some embodiments, the cleavable disulfide moiety is disulfidyl-dimethyl.
[0152] In some embodiments, the linker is a cleavable sulfonamide linker. As used herein, a cleavable sulfonamide linker refers to any linker that comprises a cleavable sulfonamide moiety. The term "cleavable sulfonamide moiety" refers to a sulfonamide group, i.e., sulfonyl group connected to an amine group, wherein the sulfur-nitrogen bond can be cleaved.
[0153] In some embodiments, the linker may be a dendritic type linker for covalent attachment of more than one drug moiety to an antibody moiety through a branching, multifunctional linker moiety. See, e.g., Sun et al. (2002) Bioorg. Med. Chem. Lett. 12:2213-5; Sun et al. (2003) Bioorg. Med. Chem. 11:1761-8. Dendritic linkers can increase the molar ratio of drug to antibody, i.e., drug loading, which is related to the potency of the ADC. Thus, where an antibody moiety bears only one reactive cysteine thiol group, for example, a multitude of drug moieties may be attached through a dendritic linker. In some embodiments, the linker moiety or linker-drug moiety may be attached to the antibody via reduced disulfide bridging chemistry or limited lysine utilization technology. See, e.g., Intl. Publ. Nos. WO2013173391 and WO2013173393.
[0154] In some embodiments, the linker is cleavable by a cleaving agent, e.g., an enzyme, that is present in the intracellular environment (e.g., within a lysosome or endosome or caveolea). The linker can be, e.g., a peptide linker that is cleaved by an intracellular peptidase or protease enzyme, including, but not limited to, a lysosomal or endosomal protease. In some embodiments, the linker is a cleavable peptide linker. As used herein, a cleavable peptide linker refers to any linker that comprises a cleavable peptide moiety. The term "cleavable peptide moiety" refers to any chemical bond linking amino acids (natural or synthetic amino acid derivatives) that can be cleaved by an agent that is present in the intracellular environment. For instance, a linker may comprise an alanine-alanine-asparagine (Ala-Ala-Asn) sequence or a valine-citrulline (Val-Cit) sequence that is cleavable by a peptidase such as cathepsin, e.g., cathepsin B.
[0155] In some embodiments, the linker is an enzyme-cleavable linker and a cleavable peptide moiety in the linker is cleavable by the enzyme. In some embodiments, the cleavable peptide moiety is cleavable by a lysosomal enzyme, e.g., cathepsin. In some embodiments, the linker is a cathepsin-cleavable linker. In some embodiments, the cleavable peptide moiety in the linker is cleavable by a lysosomal cysteine cathepsin, such as cathepsin B, C, F, H, K, L, O, S, V, X, or W. In some embodiments, the cleavable peptide moiety is cleavable by cathepsin B. An exemplary dipeptide that may be cleaved by cathepsin B is valine-citrulline (Val-Cit) (Dubowchik et al. (2002) Bioconjugate Chem. 13:855-69). In some embodiments, an ADC that comprises a cleavable peptide moiety demonstrates lower aggregation levels and/or higher drug loading (p) relative to an ADC that comprises an alternate cleavable moiety (e.g., a cleavable disulfide moiety or a cleavable sulfonamide moiety).
[0156] In some embodiments, the linker or the cleavable peptide moiety in the linker comprises an amino acid unit. In some embodiments, the amino acid unit allows for cleavage of the linker by a protease, thereby facilitating release of the drug moiety from the ADC upon exposure to one or more intracellular proteases, such as one or more lysosomal enzymes (Doronina et al. (2003) Nat. Biotechnol. 21:778-84; Dubowchik and Walker (1999) Pharm. Therapeutics 83:67-123). Exemplary amino acid units include, but are not limited to, dipeptides, tripeptides, tetrapeptides, and pentapeptides. Exemplary dipeptides include, but are not limited to, valine-citrulline (Val-Cit), alanine-asparagine (Ala-Asn), alanine-phenylalanine (Ala-Phe), phenylalanine-lysine (Phe-Lys), alanine-lysine (Ala-Lys), alanine-valine (Ala-Val), valine-alanine (Val-Ala), valine-lysine (Val-Lys), lysine-lysine (Lys-Lys), phenylalanine-citrulline (Phe-Cit), leucine-citrulline (Leu-Cit), isoleucine-citrulline (Ile-Cit), tryptophan-citrulline (Trp-Cit), and phenylalanine-alanine (Phe-Ala). Exemplary tripeptides include, but are not limited to, alanine-alanine-asparagine (Ala-Ala-Asn), glycine-valine-citrulline (Gly-Val-Cit), glycine-glycine-glycine (Gly-Gly-Gly), phenylalanine-phenylalanine-lysine (Phe-Phe-Lys), and glycine-phenylalanine-lysine (Gly-Phe-Lys). Other exemplary amino acid units include, but are not limited to, Gly-Phe-Leu-Gly (SEQ ID NO: 367), Ala-Leu-Ala-Leu (SEQ ID NO: 368), Phe-N.sup.9-tosyl-Arg, and Phe-N.sup.9-Nitro-Arg, as described in, e.g., U.S. Pat. No. 6,214,345. In some embodiments, the amino acid unit in the linker comprises Val-Cit. In some embodiments, the amino acid unit in the linker comprises Ala-Ala-Asn. In some embodiments, an ADC that comprises Val-Cit demonstrates decreased off-target cell killing, increased on-target cell killing, lower aggregation levels, and/or higher drug loading (p) relative to an ADC that comprises an alternate amino acid unit or an alternate cleavable moiety. An amino acid unit may comprise amino acid residues that occur naturally and/or minor amino acids and/or non-naturally occurring amino acid analogs, such as citrulline. Amino acid units can be designed and optimized for enzymatic cleavage by a particular enzyme, for example, a tumor-associated protease, a lysosomal protease such as cathepsin B, C, D, or S, or a plasmin protease.
[0157] In some embodiments, the linker in any of the ADCs disclosed herein may comprise at least one spacer unit joining the antibody moiety to the drug moiety. In some embodiments, the spacer unit joins a cleavage site (e.g., a cleavable peptide moiety) in the linker to the antibody moiety. In some embodiments, the linker, and/or spacer unit in the linker, is substantially hydrophilic. A hydrophilic linker may be used to reduce the extent to which the drug may be pumped out of resistant cancer cells through multiple drug resistance (MDR) or functionally similar transporters. In some aspects, the linker includes one or more polyethylene glycol (PEG) moieties, e.g., 1, 2, 3, 4, 5, or 6 PEG moieties. In some embodiments, the linker is a shorter PEG linker, and provides improved stability and reduced aggregation over longer PEG linkers.
[0158] In some embodiments, the spacer unit in the linker comprises one or more PEG moieties. In some embodiments, the spacer unit comprises -(PEG).sub.m-, and m is an integer from 1 to 10. In some embodiments, m ranges from 1 to 10; from 2 to 8; from 2 to 6; from 2 to 5; from 2 to 4; or from 2 to 3. In some embodiments, m is 8. In some embodiments, m is 4. In some embodiments, m is 3. In some embodiments, m is 2. In some embodiments, the spacer unit comprises (PEG).sub.2, (PEG).sub.4, (PEG)s, (PEG).sub.9, (PEG).sub.3-triazole-(PEG).sub.3, (PEG).sub.4-triazole-(PEG).sub.3, or dibenzylcyclooctene-triazole-(PEG).sub.3. In some preferred embodiments, the spacer unit comprises (PEG).sub.2. In some embodiments, an ADC that comprises a shorter spacer unit (e.g., (PEG).sub.2) demonstrates lower aggregation levels and/or higher drug loading (p) relative to an ADC that comprises a longer spacer unit (e.g., (PEG).sub.8).
[0159] In some embodiments, the spacer unit in the linker comprises an alkyl moiety. In some embodiments, the spacer unit comprises --(CH.sub.2).sub.n--, and n is an integer from 1 to 10 (i.e., n may be 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10). In some embodiments, n is 5. In some embodiments, an ADC that comprises a shorter spacer unit (e.g., (CH.sub.2).sub.5) demonstrates lower aggregation levels and/or higher drug loading (p) relative to an ADC that comprises a longer spacer unit (e.g., (PEG).sub.8).
[0160] A spacer unit may be used, for example, to link the antibody moiety to the drug moiety, either directly or indirectly. In some embodiments, the spacer unit links the antibody moiety to the drug moiety directly. In some embodiments, the antibody moiety and the drug moiety are attached via a spacer unit comprising one or more PEG moieties (e.g., (PEG).sub.2 or (PEG).sub.4). In some embodiments, the spacer unit links the antibody moiety to the drug moiety indirectly. In some embodiments, the spacer unit links the antibody moiety to the drug moiety indirectly through a cleavable moiety (e.g., a cleavable peptide, a cleavable disulfide, or a cleavable sulfonamide) and/or an attachment moiety to join the spacer unit to the antibody moiety, e.g., a maleimide moiety.
[0161] The spacer unit, in various embodiments, attaches to the antibody moiety (i.e., the antibody or antigen-binding fragment) via a maleimide moiety (Mal). In some embodiments, an ADC that comprises a linker attached to the antibody moiety via a maleimide moiety demonstrates higher drug loading (p) relative to an ADC that comprises a linker attached to the antibody moiety via an alternate attachment moiety such as a succinimide moiety.
[0162] A spacer unit that attaches to the antibody or antigen-binding fragment via a Mal is referred to herein as a "Mal-spacer unit." The term "maleimide moiety," as used herein, means a compound that contains a maleimide group and that is reactive with a sulfhydryl group, e.g., a sulfhydryl group of a cysteine residue on the antibody moiety. Other functional groups that are reactive with sulfhydryl groups (thiols) include, but are not limited to, iodoacetamide, bromoacetamide, vinyl pyridine, disulfide, pyridyl disulfide, isocyanate, and isothiocyanate. In some embodiments, the Mal-spacer unit is reactive with a cysteine residue on the antibody or antigen-binding fragment. In some embodiments, the Mal-spacer unit is joined to the antibody or antigen-binding fragment via the cysteine residue. In some embodiments, the Mal-spacer unit comprises a PEG moiety. In some embodiments, the Mal-spacer unit comprises an alkyl moiety.
[0163] In certain embodiments, the linker comprises the Mal-spacer unit and a cleavable peptide moiety. In some embodiments, the cleavable peptide moiety comprises an amino acid unit. In some embodiments, the amino acid unit comprises Val-Cit. In some embodiments, the amino acid unit comprises Ala-Ala-Asn. In some embodiments, the linker comprises the Mal-spacer unit and Val-Cit. In some embodiments, the linker comprises Mal-(PEG).sub.2 and Val-Cit. In some embodiments, the linker comprises Mal-(PEG). and Val-Cit, where m is 2 to 8 or 2 to 5, or 2, 3, 4, or 5. In some embodiments, the linker comprises Mal-(PEG).sub.8 and Val-Cit. In certain embodiments, the linker comprises Mal-(CH.sub.2).sub.5 and Val-Cit. In some embodiments, the linker comprises the Mal-spacer unit and Ala-Ala-Asn. In some embodiments, the linker comprises Mal-(PEG).sub.2 and Ala-Ala-Asn.
[0164] In some embodiments, the linker comprises the Mal-spacer unit and a cleavable disulfide moiety. In some embodiments, the cleavable disulfide moiety is disulfidyl-dimethyl. In some embodiments, the linker comprises the Mal-spacer unit and disulfidyl-dimethyl. In some embodiments, the linker comprises Mal-(PEG).sub.4-triazole-(PEG).sub.3 and disulfidyl-dimethyl.
[0165] In some embodiments, the linker comprises the Mal-spacer unit and a cleavable sulfonamide moiety. In some embodiments, the linker comprises Mal-(PEG).sub.4-triazole-(PEG).sub.3 and sulfonamide.
[0166] In various embodiments, the spacer unit attaches to the antibody or antigen-binding fragment via a succinimide moiety (OSu). A spacer unit that attaches to the antibody or antigen-binding fragment via an OSu is referred to herein as an "OSu-spacer unit." The term "succinimide moiety," as used herein, means a compound that contains a succinimide compound that is reactive with an amine group, e.g., an amine group of a lysine residue on the antibody moiety. An exemplary succinimide moiety is N-hydroxysuccinimide (NHS). In some embodiments, the OSu-spacer unit is reactive with a lysine residue on the antibody or antigen-binding fragment. In some embodiments, the OSu-spacer unit is joined to the antibody or antigen-binding fragment via the lysine residue. In some embodiments, the OSu-spacer unit comprises a PEG moiety. In some embodiments, the OSu-spacer unit comprises an alkyl moiety.
[0167] In certain embodiments, the linker comprises the OSu-spacer unit and a cleavable peptide moiety. In some embodiments, the cleavable peptide moiety comprises an amino acid unit. In some embodiments, the amino acid unit comprises Val-Cit. In some embodiments, the amino acid unit comprises Ala-Ala-Asn. In some embodiments, the linker comprises the OSu-spacer unit and Val-Cit. In some embodiments, the linker comprises OSu-(PEG).sub.2 and Val-Cit. In other embodiments, the linker comprises OSu-(PEG).sub.9 and Val-Cit. In other embodiments, the linker comprises OSu-(CH.sub.2).sub.5 and Val-Cit. In certain embodiments, the linker comprises OSu-(PEG).sub.3-triazole-(PEG).sub.3 and Val-Cit. In some embodiments, the linker comprises the OSu-spacer unit and Ala-Ala-Asn. In some embodiments, the linker comprises OSu-(PEG).sub.2 and Ala-Ala-Asn.
[0168] In some embodiments, the linker comprises the OSu-spacer unit and a cleavable disulfide moiety. In some embodiments, the cleavable disulfide moiety is disulfidyl-dimethyl. In some embodiments, the linker comprises the OSu-spacer unit and disulfidyl-dimethyl. In some embodiments, the linker comprises OSu-(PEG).sub.3-triazole-(PEG).sub.3 and disulfidyl-dimethyl. In other embodiments, the linker comprises OSu-dibenzylcyclooctene-triazole-(PEG).sub.3 and disulfidyl-dimethyl.
[0169] In some embodiments, the linker comprises the OSu-spacer unit and a cleavable sulfonamide moiety. In some embodiments, the linker comprises OSu-(PEG).sub.3-triazole-(PEG).sub.3 and sulfonamide. In other embodiments, the linker comprises OSu-dibenzylcyclooctene-triazole-(PEG).sub.3 and sulfonamide.
[0170] In some embodiments, the Mal-spacer unit or the OSu-spacer unit attaches the antibody moiety (i.e., the antibody or antigen-binding fragment) to the cleavable moiety in the linker. In some embodiments, the Mal-spacer unit or the OSu-spacer unit attaches the antibody or antigen-binding fragment to a cleavable peptide moiety. In some embodiments, the cleavable peptide moiety comprises an amino acid unit. In some embodiments, the linker comprises Mal-spacer unit-amino acid unit or OSu-spacer unit-amino acid unit. In some embodiments, the Mal-spacer unit or the OSu-spacer unit comprises a PEG moiety. In some embodiments, the Mal-spacer-unit or the OSu-spacer unit comprises an alkyl moiety. In some embodiments, the amino acid unit comprises Val-Cit. In other embodiments, the amino acid unit comprises Ala-Ala-Asn.
[0171] In some embodiments, the linker comprises the structure: Mal-spacer unit-Val-Cit. In some embodiments, the linker comprises the structure: Mal-(PEG).sub.2-Val-Cit. In some embodiments, the linker comprises the structure: Mal-(PEG).sub.2-Val-Cit-pAB. In some embodiments, the linker comprises Mal-(PEG).sub.8-Val-Cit. In certain embodiments, the linker comprises Mal-(CH.sub.2).sub.5-Val-Cit. In some embodiments, the linker comprises the Mal-spacer unit-Ala-Ala-Asn. In some embodiments, the linker comprises Mal-(PEG).sub.2-Ala-Ala-Asn.
[0172] In some embodiments, the linker comprises OSu-spacer unit-Val-Cit. In some embodiments, the linker comprises OSu-(PEG).sub.2-Val-Cit. In other embodiments, the linker comprises OSu-(PEG).sub.9-Val-Cit. In other embodiments, the linker comprises OSu-(CH.sub.2).sub.5-Val-Cit. In other embodiments, the linker comprises OSu-(PEG).sub.3-triazole-(PEG).sub.3-Val-Cit. In some embodiments, the linker comprises the OSu-spacer unit-Ala-Ala-Asn. In some embodiments, the linker comprises OSu-(PEG).sub.2-Ala-Ala-Asn.
[0173] In various embodiments, the Mal-spacer unit or the OSu-spacer unit attaches the antibody or antigen-binding fragment to a cleavable disulfide moiety. In some embodiments, the linker comprises Mal-spacer unit-disulfide or OSu-spacer unit-disulfide. In some embodiments, the disulfide is disulfidyl-dimethyl. In some embodiments, the linker comprises Mal-spacer unit-disulfidyl-dimethyl. In some embodiments, the linker comprises Mal-(PEG).sub.4-triazole-(PEG).sub.3-disulfidyl-dimethyl. In other embodiments, the linker comprises OSu-spacer unit-disulfidyl-dimethyl. In some embodiments, the linker comprises OSu-(PEG).sub.3-triazole-(PEG).sub.3-disulfidyl-dimethyl. In other embodiments, the linker comprises OSu-dibenzylcyclooctene-triazole-(PEG).sub.3-disulfidyl-dimethyl.
[0174] In certain embodiments, the Mal-spacer unit or the OSu-spacer unit attaches the antibody or antigen-binding fragment to a cleavable sulfonamide moiety. In some embodiments, the linker comprises Mal-spacer unit-sulfonamide or OSu-spacer unit-sulfonamide. In some embodiments, the linker comprises Mal-(PEG).sub.4-triazole-(PEG).sub.3-sulfonamide. In some embodiments, the linker comprises OSu-(PEG).sub.3-triazole-(PEG).sub.3-sulfonamide. In other embodiments, the linker comprises OSu-dibenzylcyclooctene-triazole-(PEG).sub.3-sulfonamide.
[0175] In various embodiments, the cleavable moiety in the linker is joined directly to the drug moiety. In other embodiments, another spacer unit is used to attach the cleavable moiety in the linker to the drug moiety. In various embodiments, the drug moiety is eribulin. In various embodiments, the eribulin is attached to the cleavable moiety in the linker by a spacer unit. In some embodiments, the eribulin is attached to the cleavable moiety in the linker by a self-immolative spacer unit. In certain embodiments, the eribulin is attached to the cleavable moiety in the linker by a self-immolative spacer unit, the cleavable moiety comprises Val-Cit, and a further spacer unit comprising PEG joins the cleavable moiety to the antibody moiety. In certain embodiments, the eribulin is joined to an anti-FRA antibody via a Mal-spacer unit in the linker joined to a Val-Cit cleavable moiety and a pAB self-immolative spacer unit. In certain other embodiments, the eribulin is joined to an anti-her2 antibody via a Mal-spacer unit in the linker joined to a Val-Cit cleavable moiety and a pAB self-immolative spacer unit.
[0176] A spacer unit may be "self-immolative" or "non-self-immolative." A "non-self-immolative" spacer unit is one in which part or all of the spacer unit remains bound to the drug moiety upon cleavage of the linker. Examples of non-self-immolative spacer units include, but are not limited to, a glycine spacer unit and a glycine-glycine spacer unit. Non-self-immolative spacer units may eventually degrade over time but do not readily release a linked native drug entirely under cellular conditions. A "self-immolative" spacer unit allows for release of the native drug moiety under intracellular conditions. A "native drug" is one where no part of the spacer unit or other chemical modification remains after cleavage/degradation of the spacer unit.
[0177] Self-immolation chemistry is known in the art and could be readily selected for the disclosed ADCs. In various embodiments, the spacer unit attaching the cleavable moiety in the linker to the drug moiety (e.g., eribulin) is self-immolative, and undergoes self-immolation concurrently with or shortly before/after cleavage of the cleavable moiety under intracellular conditions.
[0178] In certain embodiments, the self-immolative spacer unit in the linker comprises a p-aminobenzyl unit. In some embodiments, a p-aminobenzyl alcohol (pABOH) is attached to an amino acid unit or other cleavable moiety in the linker via an amide bond, and a carbamate, methylcarbamate, or carbonate is made between the pABOH and the drug moiety (Hamann et al. (2005) Expert Opin. Ther. Patents 15:1087-103). In some embodiments, the self-immolative spacer unit is or comprises p-aminobenzyloxycarbonyl (pAB). Without being bound by theory, it is thought that the self-immolation of pAB involves a spontaneous 1,6-elimination reaction (Jain et al. (2015) Pharm Res 32:3526-40).
[0179] In various embodiments, the structure of the p-aminobenzyloxycarbonyl (pAB) used in the disclosed ADCs is shown below:
##STR00001##
[0180] In various embodiments, the self-immolative spacer unit attaches the cleavable moiety in the linker to the C-35 amine on eribulin. In some embodiments, the self-immolative spacer unit is pAB. In some embodiments, the pAB attaches the cleavable moiety in the linker to the C-35 amine on eribulin. In some embodiments, the pAB undergoes self-immolation upon cleavage of the cleavable moiety, and eribulin is released from the ADC in its native, active form. In some embodiments, an anti-FRA antibody (e.g., MORAb-003) is joined to the C-35 amine of eribulin by a linker comprising Mal-(PEG).sub.2-Val-Cit-pAB. In other embodiments, an anti-her2 antibody (e.g., trastuzumab) is joined to the C-35 amine of eribulin by a linker comprising Mal-(PEG).sub.2-Val-Cit-pAB.
[0181] In some embodiments, the pAB undergoes self-immolation upon cleavage of a cleavable peptide moiety in the linker. In some embodiments, the cleavable peptide moiety comprises an amino acid unit. In some embodiments, the linker comprises amino acid unit-pAB. In some embodiments, the amino acid unit is Val-Cit. In some embodiments, the linker comprises Val-Cit-pAB (VCP). In certain embodiments, the amino acid unit is Ala-Ala-Asn. In some embodiments, the linker comprises Ala-Ala-Asn-pAB.
[0182] In some embodiments, the pAB undergoes self-immolation upon cleavage of a cleavable disulfide moiety in the linker. In some embodiments, the linker comprises disulfide-pAB. In some embodiments, the linker comprises disulfidyl-dimethyl-pAB.
[0183] In some embodiments, the pAB undergoes self-immolation upon cleavage of a cleavable sulfonamide moiety in the linker. In some embodiments, the linker comprises sulfonamide-pAB.
[0184] In various aspects, the antibody moiety of the ADC is conjugated to the drug moiety via a linker, wherein the linker comprises a Mal-spacer unit, a cleavable amino acid unit, and a pAB. In some embodiments, the spacer unit comprises a PEG moiety. In some embodiments, the spacer unit comprises an alkyl moiety. In some embodiments, the linker comprises Mal-(PEG).sub.2-amino acid unit-pAB. In some embodiments, the linker comprises Mal-(PEG).sub.2-Val-Cit-pAB. In other embodiments, the linker comprises Mal-(PEG).sub.2-Ala-Ala-Asn-pAB. In some embodiments, the linker comprises, Mal-(PEG).sub.8-amino acid unit-pAB. In some embodiments, the linker comprises Mal-(PEG).sub.8-Val-Cit-pAB. In some embodiments, the linker comprises Mal-(CH.sub.2).sub.5-amino acid unit-pAB. In some embodiments, the linker comprises Mal-(CH.sub.2).sub.5-Val-Cit-pAB.
[0185] In various embodiments, the antibody moiety of the ADC is conjugated to the drug moiety via a linker, wherein the linker comprises Mal-spacer unit-disulfide-pAB. In some embodiments, the spacer unit comprises a PEG moiety. In some embodiments, the linker comprises Mal-(PEG).sub.4-triazole-(PEG).sub.3-disulfide-pAB. In some embodiments, the linker comprises Mal-(PEG).sub.4-triazole-(PEG).sub.3-disulfidyl-dimethyl-pAB.
[0186] In some embodiments, the antibody moiety of the ADC is conjugated to the drug moiety via a linker, wherein the linker comprises Mal-spacer unit-sulfonamide-pAB. In some embodiments, the spacer unit comprises a PEG moiety. In some embodiments, the linker comprises Mal-(PEG).sub.4-triazole-(PEG).sub.3-sulfonamide-pAB.
[0187] In some aspects, the antibody moiety of the ADC is conjugated to the drug moiety via a linker, wherein the linker comprises OSu-spacer unit-amino acid unit-pAB. In some embodiments, the spacer unit comprises a PEG moiety. In some embodiments, the spacer unit comprises an alkyl moiety. In some embodiments, the linker comprises OSu-(PEG).sub.2-amino acid unit-pAB. In some embodiments, the linker comprises OSu-(PEG).sub.2-Val-Cit-pAB. In other embodiments, the linker comprises OSu-(PEG).sub.2-Ala-Ala-Asn-pAB. In some embodiments, the linker comprises, OSu-(PEG).sub.9-amino acid unit-pAB. In some embodiments, the linker comprises OSu-(PEG).sub.9-Val-Cit-pAB. In some embodiments, the linker comprises OSu-(CH.sub.2).sub.5-amino acid unit-pAB. In some embodiments, the linker comprises OSu-(CH.sub.2).sub.5-Val-Cit-pAB. In some embodiments, the linker comprises OSu-(PEG).sub.3-triazole-(PEG).sub.3-amino acid unit-pAB. In some embodiments, the linker comprises OSu-(PEG).sub.3-triazole-(PEG).sub.3-Val-Cit-pAB.
[0188] In some embodiments, the antibody moiety of the ADC is conjugated to the drug moiety via a linker, wherein the linker comprises OSu-spacer unit-disulfide-pAB. In some embodiments, the spacer unit comprises a PEG moiety. In some embodiments, the linker comprises OSu-(PEG).sub.3-triazole-(PEG).sub.3-disulfide-pAB. In some embodiments, the linker comprises OSu-(PEG).sub.3-triazole-(PEG).sub.3-disulfidyl-dimethyl-pAB. In some embodiments, the linker comprises OSu-dibenzylcyclooctene-triazole-(PEG).sub.3-disulfide-pAB. In some embodiments, the linker comprises OSu-dibenzylcyclooctene-triazole-(PEG).sub.3-disulfidyl-dimethyl-pAB.
[0189] In some embodiments, the antibody moiety of the ADC is conjugated to the drug moiety via a linker, wherein the linker comprises OSu-spacer unit-sulfonamide-pAB. In some embodiments, the spacer unit comprises a PEG moiety. In some embodiments, the linker comprises OSu-(PEG).sub.3-triazole-(PEG).sub.3-sulfonamide-pAB. In some embodiments, the linker comprises OSu-dibenzylcyclooctene-triazole-(PEG).sub.3-sulfonamide-pAB.
[0190] In various embodiments, the linker is designed to facilitate bystander killing (the killing of neighboring cells) through cleavage after cellular internalization and diffusion of the linker-drug moiety and/or the drug moiety alone to neighboring cells. In some embodiments, the linker promotes cellular internalization. In some embodiments, the linker is designed to minimize cleavage in the extracellular environment and thereby reduce toxicity to off-target tissue (e.g., non-cancerous tissue), while preserving ADC binding to target tissue and bystander killing of cancerous tissue that does not express an antigen targeted by the antibody moiety of an ADC, but surrounds target cancer tissue expressing that antigen. In some embodiments, a linker comprising a maleimide moiety (Mal), a polyethylene glycol (PEG) moiety, valine-citrulline (Val-Cit or "vc"), and a pAB provides these functional features. In some embodiments, a linker comprising Mal-(PEG).sub.2-Val-Cit-pAB is particularly effective in providing these functional features, e.g., when joining an anti-FRA antibody moiety such as MORAb-003 and a drug moiety such as eribulin. In some embodiments, at least some of these functional features may also be observed without an anti-FRA antibody moiety, and/or without MORAb-003. For instance, in some embodiments, a linker comprising Mal-(PEG).sub.2-Val-Cit-pAB is effective in providing some or all of these functional features, e.g., when joining an anti-her2 antibody moiety such as trastuzumab and a drug moiety such as eribulin.
[0191] In some embodiments, the antibody moiety is conjugated to the drug moiety via a linker comprising a maleimide moiety (Mal), a polyethylene glycol (PEG) moiety, valine citrulline (Val-Cit or "vc"), and a pAB. In these embodiments, the maleimide moiety covalently attaches the linker-drug moiety to the antibody moiety, and the pAB acts as a self-immolative spacer unit. Such linker may be referred to as the "m-vc-pAB" linker, the "Mal-VCP" linker, the "Mal-(PEG).sub.2-VCP" linker, or the "Mal-(PEG).sub.2-Val-Cit-pAB" linker. In some embodiments, the drug moiety is eribulin. The structure of Mal-(PEG).sub.2-Val-Cit-pAB-eribulin is provided in Table 46. The pAB of the Mal-(PEG).sub.2-Val-Cit-pAB linker is attached to the C-35 amine on eribulin.
[0192] It has been discovered that ADCs comprising Mal-(PEG).sub.2-Val-Cit-pAB-eribulin demonstrate a particular combination of desirable properties, particularly when paired with an anti-FRA antibody such as MORAb-003 or an antigen-binding fragment thereof. These properties include, but are not limited to, effective levels of drug loading (p.about.4), low aggregation levels, stability under storage conditions or when in circulation in the body (e.g., serum stability), retained affinity for target-expressing cells comparable to unconjugated antibody, potent cytotoxicity against target-expressing cells, low levels of off-target cell killing, high levels of bystander killing, and/or effective in vivo anti-cancer activity, all as compared to ADCs using other linker-toxin and/or antibody moieties. While numerous linker options and combinations of spacers and cleavage sites were known in the art and may provide certain benefits in one or more of these functional categories, the particular combination of a Mal-(PEG).sub.2-Val-Cit-pAB linker joining eribulin to an antibody moiety such as an anti-FRA antibody (e.g., MORAb-003) may provide good or superior properties across the spectrum of desirable functional properties for a therapeutic ADC. In some embodiments, the good or superior functional properties provided by the particular combination of a Mal-(PEG).sub.2-Val-Cit-pAB linker joining eribulin to an antibody moiety may be observed with this linker-toxin conjugated to, e.g., an anti-her 2 antibody such as trastuzumab.
[0193] In some embodiments, the ADC comprises Mal-(PEG).sub.2-Val-Cit-pAB-eribulin and an antibody moiety comprising an internalizing antibody or an antigen-binding fragment thereof that retains the ability to target and internalize in a tumor cell. In some embodiments, the ADC comprises Mal-(PEG).sub.2-Val-Cit-pAB-eribulin and an internalizing antibody or internalizing antigen-binding fragment thereof that targets an FRA-expressing tumor cell. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment thereof that targets an FRA-expressing tumor cell comprises three heavy chain complementarity determining regions (HCDRs) comprising amino acid sequences of SEQ ID NO:2 (HCDR1), SEQ ID NO:3 (HCDR2), and SEQ ID NO:4 (HCDR3); and three light chain complementarity determining regions (LCDRs) comprising amino acid sequences of SEQ ID NO:7 (LCDR1), SEQ ID NO:8 (LCDR2), and SEQ ID NO:9 (LCDR3), as defined by the Kabat numbering system; or three heavy chain complementarity determining regions (HCDRs) comprising amino acid sequences of SEQ ID NO:13 (HCDR1), SEQ ID NO:14 (HCDR2), and SEQ ID NO:15 (HCDR3); and three light chain complementarity determining regions (LCDRs) comprising amino acid sequences of SEQ ID NO:16 (LCDR1), SEQ ID NO:17 (LCDR2), and SEQ ID NO:18 (LCDR3), as defined by the IMGT numbering system. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment thereof that targets an FRA-expressing tumor cell comprises a heavy chain variable region comprising an amino acid sequence of SEQ ID NO:23, and a light chain variable region comprising an amino acid sequence of SEQ ID NO:24. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment thereof that targets an FRA-expressing tumor cell comprises a human IgG1 heavy chain constant domain and an Ig kappa light chain constant domain.
[0194] In some embodiments, the ADC has Formula I:
Ab-(L-D).sub.p (I)
wherein:
[0195] (i) Ab is an internalizing anti-folate receptor alpha (FRA) antibody or internalizing antigen-binding fragment thereof comprising three heavy chain complementarity determining regions (HCDRs) comprising amino acid sequences of SEQ ID NO:2 (HCDR1), SEQ ID NO:3 (HCDR2), and SEQ ID NO:4 (HCDR3); and three light chain complementarity determining regions (LCDRs) comprising amino acid sequences of SEQ ID NO:7 (LCDR1), SEQ ID NO:8 (LCDR2), and SEQ ID NO:9 (LCDR3), as defined by the Kabat numbering system; or three heavy chain complementarity determining regions (HCDRs) comprising amino acid sequences of SEQ ID NO:13 (HCDR1), SEQ ID NO:14 (HCDR2), and SEQ ID NO:15 (HCDR3); and three light chain complementarity determining regions (LCDRs) comprising amino acid sequences of SEQ ID NO:16 (LCDR1), SEQ ID NO:17 (LCDR2), and SEQ ID NO:18 (LCDR3), as defined by the IMGT numbering system;
[0196] (ii) D is eribulin;
[0197] (iii) L is a cleavable linker comprising Mal-(PEG).sub.2-Val-Cit-pAB; and
[0198] (iv) p is an integer from 1 to 20.
[0199] In some embodiments, the internalizing antibody or internalizing antigen-binding fragment thereof comprises a heavy chain variable region comprising an amino acid sequence of SEQ ID NO:23, and a light chain variable region comprising an amino acid sequence of SEQ ID NO:24. In some embodiments, the internalizing antibody is MORAb-003. In some embodiments, p is from 1 to 8, or 1 to 6. In some embodiments, p is from 2 to 8, or 2 to 5. In some embodiments, p is from 3 to 4. In some embodiments, p is 4.
[0200] In other embodiments, the ADC comprises Mal-(PEG).sub.2-Val-Cit-pAB-eribulin and an internalizing antibody or internalizing antigen-binding fragment thereof that targets a her2-expressing tumor cell. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment thereof that targets a her2-expressing tumor cell comprises three heavy chain complementarity determining regions (HCDRs) comprising amino acid sequences of SEQ ID NO:71 (HCDR1), SEQ ID NO:72 (HCDR2), and SEQ ID NO:73 (HCDR3); and three light chain complementarity determining regions (LCDRs) comprising amino acid sequences of SEQ ID NO:74 (LCDR1), SEQ ID NO:75 (LCDR2), and SEQ ID NO:76 (LCDR3), as defined by the Kabat numbering system; or three heavy chain complementarity determining regions (HCDRs) comprising amino acid sequences of SEQ ID NO:191 (HCDR1), SEQ ID NO:192 (HCDR2), and SEQ ID NO:193 (HCDR3); and three light chain complementarity determining regions (LCDRs) comprising amino acid sequences of SEQ ID NO:194 (LCDR1), SEQ ID NO:195 (LCDR2), and SEQ ID NO:196 (LCDR3), as defined by the IMGT numbering system. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment thereof that targets a her2-expressing tumor cell comprises a heavy chain variable region comprising an amino acid sequence of SEQ ID NO:27, and a light chain variable region comprising an amino acid sequence of SEQ ID NO:28. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment thereof that targets a her2-expressing tumor cell comprises a human IgG1 heavy chain constant domain and an Ig kappa light chain constant domain.
[0201] In some embodiments, the ADC has Formula I:
Ab-(L-D).sub.p (I)
wherein:
[0202] (i) Ab is an internalizing anti-human epidermal growth factor receptor 2 (her2) antibody or internalizing antigen-binding fragment thereof comprising three heavy chain complementarity determining regions (HCDRs) comprising amino acid sequences of SEQ ID NO:71 (HCDR1), SEQ ID NO:72 (HCDR2), and SEQ ID NO:73 (HCDR3); and three light chain complementarity determining regions (LCDRs) comprising amino acid sequences of SEQ ID NO:74 (LCDR1), SEQ ID NO:75 (LCDR2), and SEQ ID NO:76 (LCDR3), as defined by the Kabat numbering system; or three heavy chain complementarity determining regions (HCDRs) comprising amino acid sequences of SEQ ID NO:191 (HCDR1), SEQ ID NO:192 (HCDR2), and SEQ ID NO:193 (HCDR3); and three light chain complementarity determining regions (LCDRs) comprising amino acid sequences of SEQ ID NO:194 (LCDR1), SEQ ID NO:195 (LCDR2), and SEQ ID NO:196 (LCDR3), as defined by the IMGT numbering system;
[0203] (ii) D is eribulin;
[0204] (iii) L is a cleavable linker comprising Mal-(PEG).sub.2-Val-Cit-pAB; and
[0205] (iv) p is an integer from 1 to 20.
[0206] In some embodiments, the internalizing antibody or internalizing antigen-binding fragment thereof comprises a heavy chain variable region comprising an amino acid sequence of SEQ ID NO:27, and a light chain variable region comprising an amino acid sequence of SEQ ID NO:28. In some embodiments, the internalizing antibody is trastuzumab. In some embodiments, p is from 1 to 8, or 1 to 6. In some embodiments, p is from 2 to 8, or 2 to 5. In some embodiments, p is from 3 to 4. In some embodiments, p is 4.
[0207] In other embodiments, the ADC comprises Mal-(PEG).sub.2-Val-Cit-pAB-eribulin and an internalizing antibody or internalizing antigen-binding fragment thereof that targets a mesothelin (MSLN)-expressing tumor cell. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment thereof that targets a MSLN-expressing tumor cell comprises three heavy chain complementarity determining regions (HCDRs) comprising amino acid sequences of SEQ ID NO:65 (HCDR1), SEQ ID NO:66 (HCDR2), and SEQ ID NO:67 (HCDR3); and three light chain complementarity determining regions (LCDRs) comprising amino acid sequences of SEQ ID NO:68 (LCDR1), SEQ ID NO:69 (LCDR2), and SEQ ID NO:70 (LCDR3), as defined by the Kabat numbering system; or three heavy chain complementarity determining regions (HCDRs) comprising amino acid sequences of SEQ ID NO:185 (HCDR1), SEQ ID NO:186 (HCDR2), and SEQ ID NO:187 (HCDR3); and three light chain complementarity determining regions (LCDRs) comprising amino acid sequences of SEQ ID NO:188 (LCDR1), SEQ ID NO:189 (LCDR2), and SEQ ID NO:190 (LCDR3), as defined by the IMGT numbering system. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment thereof that targets a MSLN-expressing tumor cell comprises a heavy chain variable region comprising an amino acid sequence of SEQ ID NO:25, and a light chain variable region comprising an amino acid sequence of SEQ ID NO:26. In some embodiments, the internalizing antibody or internalizing antigen-binding fragment thereof that targets a MSLN-expressing tumor cell comprises a human IgG1 heavy chain constant domain and an Ig kappa light chain constant domain.
[0208] In some embodiments, the ADC has Formula I:
Ab-(L-D).sub.p (I)
wherein:
[0209] (i) Ab is an internalizing anti-mesothelin antibody or internalizing antigen-binding fragment thereof comprising three heavy chain complementarity determining regions (HCDRs) comprising amino acid sequences of SEQ ID NO:65 (HCDR1), SEQ ID NO:66 (HCDR2), and SEQ ID NO:67 (HCDR3); and three light chain complementarity determining regions (LCDRs) comprising amino acid sequences of SEQ ID NO:68 (LCDR1), SEQ ID NO:69 (LCDR2), and SEQ ID NO:70 (LCDR3), as defined by the Kabat numbering system; or three heavy chain complementarity determining regions (HCDRs) comprising amino acid sequences of SEQ ID NO:185 (HCDR1), SEQ ID NO:186 (HCDR2), and SEQ ID NO:187 (HCDR3); and three light chain complementarity determining regions (LCDRs) comprising amino acid sequences of SEQ ID NO:188 (LCDR1), SEQ ID NO:189 (LCDR2), and SEQ ID NO:190 (LCDR3), as defined by the IMGT numbering system;
[0210] (ii) D is eribulin;
[0211] (iii) L is a cleavable linker comprising Mal-(PEG).sub.2-Val-Cit-pAB; and
[0212] (iv) p is an integer from 1 to 20.
[0213] In some embodiments, the internalizing antibody or internalizing antigen-binding fragment thereof comprises a heavy chain variable region comprising an amino acid sequence of SEQ ID NO:25, and a light chain variable region comprising an amino acid sequence of SEQ ID NO:26. In some embodiments, the internalizing antibody is MORAb-003, MORAb-009, or trastuzumab. In some embodiments, p is from 1 to 8, or 1 to 6. In some embodiments, p is from 2 to 8, or 2 to 5. In some embodiments, p is from 3 to 4. In some embodiments, p is 4.
Drug Moieties
[0214] The drug moiety (D) of the ADCs described herein can be any chemotherapeutic agent. Useful classes of chemotherapeutic agents include, for example, anti-tubulin agents. In certain embodiments, the drug moiety is an anti-tubulin agent. Examples of anti-tubulin agents include cryptophycin and eribulin. The preferred drug moiety for use in the described ADCs is eribulin.
[0215] In various embodiments, the drug moiety is eribulin. In these embodiments, the linker of the ADC is attached via the C-35 amine on eribulin.
[0216] In various embodiments, the natural form of eribulin used for joining to the linker and antibody moiety is shown below:
##STR00002##
[0217] In certain embodiments, an intermediate, which is the precursor of the linker, is reacted with the drug moiety under appropriate conditions. In certain embodiments, reactive groups are used on the drug and/or the intermediate or linker. The product of the reaction between the drug and the intermediate, or the derivatized drug, is subsequently reacted with the antibody or antigen-binding fragment under appropriate conditions. Alternatively, the linker or intermediate may first be reacted with the antibody or a derivatized antibody, and then reacted with the drug or derivatized drug.
[0218] A number of different reactions are available for covalent attachment of drugs and/or linkers to the antibody moiety. This is often accomplished by reaction of one or more amino acid residues of the antibody molecule, including the amine groups of lysine, the free carboxylic acid groups of glutamic acid and aspartic acid, the sulfhydryl groups of cysteine, and the various moieties of the aromatic amino acids. For instance, non-specific covalent attachment may be undertaken using a carbodiimide reaction to link a carboxy (or amino) group on a compound to an amino (or carboxy) group on an antibody moiety. Additionally, bifunctional agents such as dialdehydes or imidoesters may also be used to link the amino group on a compound to an amino group on an antibody moiety. Also available for attachment of drugs to binding agents is the Schiff base reaction. This method involves the periodate oxidation of a drug that contains glycol or hydroxy groups, thus forming an aldehyde which is then reacted with the binding agent. Attachment occurs via formation of a Schiff base with amino groups of the binding agent. Isothiocyanates may also be used as coupling agents for covalently attaching drugs to binding agents. Other techniques are known to the skilled artisan and within the scope of the present disclosure.
Drug Loading
[0219] Drug loading is represented by p, and is also referred to herein as the drug-to-antibody ratio (DAR). Drug loading may range from 1 to 20 drug moieties per antibody moiety. In some embodiments, p is an integer from 1 to 20. In some embodiments, p is an integer from 1 to 10, 1 to 9, 1 to 8, 1 to 7, 1 to 6, 1 to 5, 1 to 4, 1 to 3, or 1 to 2. In some embodiments, p is an integer from 2 to 10, 2 to 9, 2 to 8, 2 to 7, 2 to 6, 2 to 5, 2 to 4, or 2 to 3. In some embodiments, p is an integer from 3 to 4. In other embodiments, p is 1, 2, 3, 4, 5, or 6, preferably 3 or 4.
[0220] Drug loading may be limited by the number of attachment sites on the antibody moiety. In some embodiments, the linker moiety (L) of the ADC attaches to the antibody moiety through a chemically active group on one or more amino acid residues on the antibody moiety. For example, the linker may be attached to the antibody moiety via a free amino, imino, hydroxyl, thiol, or carboxyl group (e.g., to the N- or C-terminus, to the epsilon amino group of one or more lysine residues, to the free carboxylic acid group of one or more glutamic acid or aspartic acid residues, or to the sulfhydryl group of one or more cysteine residues). The site to which the linker is attached can be a natural residue in the amino acid sequence of the antibody moiety, or it can be introduced into the antibody moiety, e.g., by DNA recombinant technology (e.g., by introducing a cysteine residue into the amino acid sequence) or by protein biochemistry (e.g., by reduction, pH adjustment, or hydrolysis).
[0221] In some embodiments, the number of drug moieties that can be conjugated to an antibody moiety is limited by the number of free cysteine residues. For example, where the attachment is a cysteine thiol group, an antibody may have only one or a few cysteine thiol groups, or may have only one or a few sufficiently reactive thiol groups through which a linker may be attached. Generally, antibodies do not contain many free and reactive cysteine thiol groups that may be linked to a drug moiety. Indeed, most cysteine thiol residues in antibodies exist as disulfide bridges. Over-attachment of linker-toxin to an antibody may destabilize the antibody by reducing the cysteine residues available to form disulfide bridges. Therefore, an optimal drug:antibody ratio should increase potency of the ADC (by increasing the number of attached drug moieties per antibody) without destabilizing the antibody moiety. In some embodiments, an optimal ratio may be about 3-4.
[0222] In some embodiments, a linker attached to an antibody moiety through a Mal moiety provides a ratio of about 3-4. In some embodiments, a linker attached to an antibody moiety through an alternate moiety (e.g., a OSu moiety) may provide a less optimal ratio (e.g., a lower ratio, such as about 0-3). In some embodiments, a linker comprising a short spacer unit (e.g., a short PEG spacer unit such as (PEG).sub.2 or (PEG).sub.4, or a short alkyl spacer unit such as (CH.sub.2).sub.5) provides a ratio of about 3-4. In some embodiments, a linker that comprises a longer spacer unit (e.g., (PEG).sub.8) may provide a less optimal ratio (e.g., a lower ratio, such as about 0-3). In some embodiments, a linker comprising a peptide cleavable moiety provides a ratio of about 3-4. In some embodiments, a linker that comprises an alternate cleavable moiety (e.g., a cleavable disulfide or a cleavable sulfonamide) may provide a less optimal ratio (e.g., a lower ratio, such as about 0-3). In some embodiments, an ADC comprising Mal-(PEG).sub.2-Val-Cit-pAB-eribulin joined to an antibody such as an anti-FRA antibody (e.g., MORAb-003) has a ratio of about 3-4. In some embodiments, a ratio of about 3-4 is observed with an ADC comprising Mal-(PEG).sub.2-Val-Cit-pAB-eribulin joined to a different antibody, such as an anti-her2 antibody (e.g., trastuzumab). In some embodiments, the optimal ratio observed with ADCs comprising the Mal-(PEG).sub.2-Val-Cit-pAB-eribulin linker-toxin is antibody-independent.
[0223] In some embodiments, an antibody moiety is exposed to reducing conditions prior to conjugation in order to generate one or more free cysteine residues. An antibody, in some embodiments, may be reduced with a reducing agent such as dithiothreitol (DTT) or tris(2-carboxyethyl)phosphine (TCEP), under partial or total reducing conditions, to generate reactive cysteine thiol groups. Unpaired cysteines may be generated through partial reduction with limited molar equivalents of TCEP, which preferentially reduces the interchain disulfide bonds which link the light chain and heavy chain (one pair per H-L pairing) and the two heavy chains in the hinge region (two pairs per H-H pairing in the case of human IgG1) while leaving the intrachain disulfide bonds intact (Stefano et al. (2013) Methods Mol. Biol. 1045:145-71). In embodiments, disulfide bonds within the antibodies are reduced electrochemically, e.g., by employing a working electrode that applies an alternating reducing and oxidizing voltage. This approach can allow for on-line coupling of disulfide bond reduction to an analytical device (e.g., an electrochemical detection device, an NMR spectrometer, or a mass spectrometer) or a chemical separation device (e.g., a liquid chromatograph (e.g., an HPLC) or an electrophoresis device (see, e.g., U.S. Publ. No. 20140069822)). In certain embodiments, an antibody is subjected to denaturing conditions to reveal reactive nucleophilic groups on amino acid residues, such as lysine or cysteine.
[0224] The drug loading of an ADC may be controlled in different ways, e.g., by: (i) limiting the molar excess of drug-linker intermediate or linker reagent relative to antibody; (ii) limiting the conjugation reaction time or temperature; (iii) partial or limiting reductive conditions for cysteine thiol modification; and/or (iv) engineering by recombinant techniques the amino acid sequence of the antibody such that the number and position of cysteine residues is modified for control of the number and/or position of linker-drug attachments.
[0225] In some embodiments, free cysteine residues are introduced into the amino acid sequence of the antibody moiety. For example, cysteine engineered antibodies can be prepared wherein one or more amino acids of a parent antibody are replaced with a cysteine amino acid. Any form of antibody may be so engineered, i.e. mutated. For example, a parent Fab antibody fragment may be engineered to form a cysteine engineered Fab referred to as a "ThioFab." Similarly, a parent monoclonal antibody may be engineered to form a "ThioMab." A single site mutation yields a single engineered cysteine residue in a ThioFab, whereas a single site mutation yields two engineered cysteine residues in a ThioMab, due to the dimeric nature of the IgG antibody. DNA encoding an amino acid sequence variant of the parent polypeptide can be prepared by a variety of methods known in the art (see, e.g., the methods described in WO2006/034488). These methods include, but are not limited to, preparation by site-directed (or oligonucleotide-mediated) mutagenesis, PCR mutagenesis, and cassette mutagenesis of an earlier prepared DNA encoding the polypeptide. Variants of recombinant antibodies may also be constructed also by restriction fragment manipulation or by overlap extension PCR with synthetic oligonucleotides. ADCs of Formula I include, but are not limited to, antibodies that have 1, 2, 3, or 4 engineered cysteine amino acids (Lyon et al. (2012) Methods Enzymol. 502:123-38). In some embodiments, one or more free cysteine residues are already present in an antibody moiety, without the use of engineering, in which case the existing free cysteine residues may be used to conjugate the antibody moiety to a drug moiety.
[0226] In some embodiments, higher drug loading (e.g., p>5) may cause aggregation, insolubility, toxicity, or loss of cellular permeability of certain antibody-drug conjugates. Higher drug loading may also negatively affect the pharmacokinetics (e.g., clearance) of certain ADCs. In some embodiments, lower drug loading (e.g., p<3) may reduce the potency of certain ADCs against target-expressing cells and/or bystander cells. In some embodiments, the drug loading for an ADC of the present disclosure ranges from 1 to about 8; from about 2 to about 6; from about 2 to about 5; from about 3 to about 5; or from about 3 to about 4.
[0227] Where more than one nucleophilic group reacts with a drug-linker intermediate or a linker moiety reagent followed by drug moiety reagent, in a reaction mixture comprising multiple copies of the antibody moiety and linker moiety, then the resulting product can be a mixture of ADC compounds with a distribution of one or more drug moieties attached to each copy of the antibody moiety in the mixture. In some embodiments, the drug loading in a mixture of ADCs resulting from a conjugation reaction ranges from 1 to 20 drug moieties attached per antibody moiety. The average number of drug moieties per antibody moiety (i.e., the average drug loading, or average p) may be calculated by any conventional method known in the art, e.g., by mass spectrometry (e.g., reverse-phase LC-MS), and/or high-performance liquid chromatography (e.g., HIC-HPLC). In some embodiments, the average number of drug moieties per antibody moiety is determined by hydrophobic interaction chromatography-high performance liquid chromatography (HIC-HPLC). In some embodiments, the average number of drug moieties per antibody moiety is determined by reverse-phase liquid chromatography-mass spectrometry (LC-MS). In some embodiments, the average number of drug moieties per antibody moiety is from about 3 to about 4; from about 3.1 to about 3.9; from about 3.2 to about 3.8; from about 3.2 to about 3.7; from about 3.2 to about 3.6; from about 3.3 to about 3.8; or from about 3.3 to about 3.7. In some embodiments, the average number of drug moieties per antibody moiety is from about 3.2 to about 3.8. In some embodiments, the average number of drug moieties per antibody moiety is about 3.8. In some embodiments, the average number of drug moieties per antibody moiety is from 3 to 4; from 3.1 to 3.9; from 3.2 to 3.8; from 3.2 to 3.7; from 3.2 to 3.6; from 3.3 to 3.8; or from 3.3 to 3.7. In some embodiments, the average number of drug moieties per antibody moiety is from 3.2 to 3.8. In some embodiments, the average number of drug moieties per antibody moiety is 3.8.
[0228] In some embodiments, the average number of drug moieties per antibody moiety is from about 3.5 to about 4.5; from about 3.6 to about 4.4; from about 3.7 to about 4.3; from about 3.7 to about 4.2; or from about 3.8 to about 4.2. In some embodiments, the average number of drug moieties per antibody moiety is from about 3.6 to about 4.4. In some embodiments, the average number of drug moieties per antibody moiety is about 4.0. In some embodiments, the average number of drug moieties per antibody moiety is from 3.5 to 4.5; from 3.6 to 4.4; from 3.7 to 4.3; from 3.7 to 4.2; or from 3.8 to 4.2. In some embodiments, the average number of drug moieties per antibody moiety is from 3.6 to 4.4. In some embodiments, the average number of drug moieties per antibody moiety is 4.0.
[0229] In various embodiments, the term "about" as used with respect to the average number of drug moieties per antibody moiety means+/-10%.
[0230] Individual ADC compounds, or "species," may be identified in the mixture by mass spectroscopy and separated by UPLC or HPLC, e.g. hydrophobic interaction chromatography (HIC-HPLC). In certain embodiments, a homogeneous or nearly homogenous ADC with a single loading value may be isolated from the conjugation mixture, e.g., by electrophoresis or chromatography.
[0231] In some embodiments, a drug loading and/or an average drug loading of about 4 provides beneficial properties. In some embodiments, a drug loading and/or an average drug loading of less than about 4 may result in an unacceptably high level of unconjugated antibody species, which can compete with the ADC for binding to a target antigen and/or provide for reduced treatment efficacy. In some embodiments, a drug loading and/or average drug loading of more than about 4 may result in an unacceptably high level of product heterogeneity and/or ADC aggregation. A drug loading and/or average drug loading of more than about 4 may also affect stability of the ADC, due to loss of one or more chemical bonds required to stabilize the antibody moiety.
[0232] In some embodiments, an ADC has Formula I:
Ab-(L-D).sub.p (I)
wherein:
[0233] (i) Ab is an internalizing anti-folate receptor alpha antibody or antigen-binding fragment thereof comprising a heavy chain variable region comprising an amino acid sequence of SEQ ID NO:23, and a light chain variable region comprising an amino acid sequence of SEQ ID NO:24;
[0234] (ii) D is eribulin;
[0235] (iii) L is a cleavable linker comprising Mal-(PEG).sub.2-Val-Cit-pAB; and
[0236] (iv) p is an integer from 3 to 4.
[0237] In other embodiments, an ADC has Formula I:
Ab-(L-D).sub.p (I)
wherein:
[0238] (i) Ab is an internalizing anti-human epidermal growth factor receptor 2 antibody or antigen-binding fragment thereof comprising a heavy chain variable region comprising an amino acid sequence of SEQ ID NO:27, and a light chain variable region comprising an amino acid sequence of SEQ ID NO:28;
[0239] (ii) D is eribulin;
[0240] (iii) L is a cleavable linker comprising Mal-(PEG).sub.2-Val-Cit-pAB; and
[0241] (iv) p is an integer from 3 to 4.
[0242] In some embodiments, p is 4.
[0243] The present disclosure includes methods of producing the described ADCs. Briefly, the ADCs comprise an antibody or antigen-binding fragment as the antibody moiety, a drug moiety, and a linker that joins the drug moiety and the antibody moiety. In some embodiments, the ADCs can be prepared using a linker having reactive functionalities for covalently attaching to the drug moiety and to the antibody moiety. For example, in some embodiments, a cysteine thiol of an antibody moiety can form a bond with a reactive functional group of a linker or a drug-linker intermediate (e.g., a maleimide moiety) to make an ADC. The generation of the ADCs can be accomplished by any technique known to the skilled artisan.
[0244] In some embodiments, an ADC is produced by contacting an antibody moiety with a linker and a drug moiety in a sequential manner, such that the antibody moiety is covalently linked to the linker first, and then the pre-formed antibody-linker intermediate reacts with the drug moiety. The antibody-linker intermediate may or may not be subjected to a purification step prior to contacting the drug moiety. In other embodiments, an ADC is produced by contacting an antibody moiety with a linker drug compound pre-formed by reacting a linker with a drug moiety. The pre-formed linker-drug compound may or may not be subjected to a purification step prior to contacting the antibody moiety. In other embodiments, the antibody moiety contacts the linker and the drug moiety in one reaction mixture, allowing simultaneous formation of the covalent bonds between the antibody moiety and the linker, and between the linker and the drug moiety. This method of producing ADCs may include a reaction, wherein the antibody moiety contacts the antibody moiety prior to the addition of the linker to the reaction mixture, and vice versa. In certain embodiments, an ADC is produced by reacting an antibody moiety with a linker joined to a drug moiety, such as Mal-(PEG).sub.2-Val-Cit-pAB-eribulin, under conditions that allow conjugation.
[0245] The ADCs prepared according to the methods described above may be subjected to a purification step. The purification step may involve any biochemical methods known in the art for purifying proteins, or any combination of methods thereof. These include, but are not limited to, tangential flow filtration (TFF), affinity chromatography, ion exchange chromatography, any charge or isoelectric point-based chromatography, mixed mode chromatography, e.g., CHT (ceramic hydroxyapatite), hydrophobic interaction chromatography, size exclusion chromatography, dialysis, filtration, selective precipitation, or any combination thereof
Therapeutic Uses and Compositions
[0246] Disclosed herein are methods of using the disclosed ADCs in treating a subject for a disorder, e.g., an oncologic disorder. ADCs may be administered alone or in combination with a second therapeutic agent, and may be administered in any pharmaceutically acceptable formulation, dosage, and dosing regimen. ADC treatment efficacy may be evaluated for toxicity as well as indicators of efficacy and adjusted accordingly. Efficacy measures include, but are not limited to, a cytostatic and/or cytotoxic effect observed in vitro or in vivo, reduced tumor volume, tumor growth inhibition, and/or prolonged survival.
[0247] Methods of determining whether an ADC exerts a cytostatic and/or cytotoxic effect on a cell are known. For example, the cytotoxic or cytostatic activity of an ADC can be measured by: exposing mammalian cells expressing a target protein of the ADC in a cell culture medium; culturing the cells for a period from about 6 hours to about 5 days; and measuring cell viability. Cell-based in vitro assays may also be used to measure viability (proliferation), cytotoxicity, and induction of apoptosis (caspase activation) of the ADC.
[0248] For determining whether an antibody-drug conjugate exerts a cytostatic effect, a thymidine incorporation assay may be used. For example, cancer cells expressing a target antigen at a density of 5,000 cells/well of a 96-well plated can be cultured for a 72-hour period and exposed to 0.5 .mu.Ci of .sup.3H-thymidine during the final 8 hours of the 72-hour period. The incorporation of .sup.3H-thymidine into cells of the culture is measured in the presence and absence of the ADC.
[0249] For determining cytotoxicity, necrosis or apoptosis (programmed cell death) may be measured. Necrosis is typically accompanied by increased permeability of the plasma membrane; swelling of the cell, and rupture of the plasma membrane. Apoptosis is typically characterized by membrane blebbing, condensation of cytoplasm, and the activation of endogenous endonucleases. Determination of any of these effects on cancer cells indicates that an ADC is useful in the treatment of cancers.
[0250] Cell viability may be measured, e.g., by determining in a cell the uptake of a dye such as neutral red, trypan blue, Crystal Violet, or ALAMAR.TM. blue (see, e.g., Page et al. (1993) Intl. J. Oncology 3:473-6). In such an assay, the cells are incubated in media containing the dye, the cells are washed, and the remaining dye, reflecting cellular uptake of the dye, is measured spectrophotometrically. In certain embodiments, in vitro potency of prepared ADCs is assessed using a Crystal Violet assay. Crystal Violet is a triarylmethane dye that accumulates in the nucleus of viable cells. In this assay, cells are exposed to the ADCs or control agents for a defined period of time, after which, cells are stained with crystal violet, washed copiously with water, then solubilized with 1% SDS and read spectrophotometrically. The protein-binding dye sulforhodamine B (SRB) can also be used to measure cytoxicity (Skehan et al. (1990) J. Natl. Cancer Inst. 82:1107-12).
[0251] Apoptosis can be quantitated, for example, by measuring DNA fragmentation. Commercial photometric methods for the quantitative in vitro determination of DNA fragmentation are available. Examples of such assays, including TUNEL (which detects incorporation of labeled nucleotides in fragmented DNA) and ELISA-based assays, are described in Biochemica (1999) No. 2, pp. 34-37 (Roche Molecular Biochemicals).
[0252] Apoptosis may also be determined by measuring morphological changes in a cell. For example, as with necrosis, loss of plasma membrane integrity can be determined by measuring uptake of certain dyes (e.g., a fluorescent dye such as, for example, acridine orange or ethidium bromide). A method for measuring apoptotic cell number has been described by Duke and Cohen, Current Protocols in Immunology (Coligan et al., eds. (1992) pp. 3.17.1-3.17.16). Cells also can be labeled with a DNA dye (e.g., acridine orange, ethidium bromide, or propidium iodide) and the cells observed for chromatin condensation and margination along the inner nuclear membrane. Other morphological changes that can be measured to determine apoptosis include, e.g., cytoplasmic condensation, increased membrane blebbing, and cellular shrinkage.
[0253] The disclosed ADCs may also be evaluated for bystander killing activity. Bystander killing activity may be determined, e.g., by an assay employing two cell lines, one positive for target antigen and one negative for target antigen. The cell lines are preferably labeled to differentiate them. For example, IGROV1 cells (FRA+) labeled with Nuclight.TM. Green (NLG) and HL-60 (FRA-) labeled with Nuclight.TM. Red (NLR) may be co-cultured, treated with an anti-FRA ADC followed by monitoring of cytotoxicity. Killing of the target antigen negative cells when mixed with target antigen positive cells is indicative of bystander killing, whereas killing of the target antigen negative cells in the absence of the target antigen positive cells is indicative of off-target killing.
[0254] In some aspects, the present disclosure features a method of killing, inhibiting or modulating the growth of, or interfering with the metabolism of, a cancer cell or tissue by disrupting tubulin. The method may be used with any subject where disruption of tubulin provides a therapeutic benefit. Subjects that may benefit from disrupting tubulin include, but are not limited to, those having or at risk of having a gastric cancer, ovarian cancer (e.g., serous ovarian cancer), lung cancer (e.g., non-small cell lung cancer), breast cancer (e.g., triple negative breast cancer), endometrial cancer (e.g., serous endometrial carcinoma), osteosarcoma, Kaposi's sarcoma, testicular germ cell cancer, leukemia, lymphoma (e.g., Hodgkin's disease, non-Hodgkin's lymphoma), myeloma, head and neck cancer, esophageal cancer, pancreatic cancer, prostate cancer, brain cancer (e.g., glioblastoma), thyroid cancer, colorectal cancer, and/or skin cancer (e.g., melanoma), or any metastases thereof (Dumontet and Jordan (2010) Nat. Rev. Drug Discov. 9:790-803). In various embodiments, the disclosed ADCs may be administered in any cell or tissue that expresses FRA, such as an FRA-expressing cancer cell or tissue. An exemplary embodiment includes a method of inhibiting FRA-mediated cell signaling or a method of killing a cell. The method may be used with any cell or tissue that expresses FRA, such as a cancerous cell or a metastatic lesion. Non-limiting examples of FRA-expressing cancers include gastric cancer, serous ovarian cancer, clear cell ovarian cancer, non-small cell lung cancer, colorectal cancer, triple negative breast cancer, endometrial cancer, serous endometrial carcinoma, lung carcinoid, and osteosarcoma. Non-limiting examples of FRA-expressing cells include IGROV1 and OVCAR3 human ovarian carcinoma cells, NCI-H2110 human non-small cell lung carcinoma cells, and cells comprising a recombinant nucleic acid encoding FRA or a portion thereof.
[0255] In various other embodiments, the disclosed ADCs may be administered in any cell or tissue that expresses her2, such as a her2-expressing cancer cell or tissue. An exemplary embodiment includes a method of inhibiting her2-mediated cell signaling or a method of killing a cell. The method may be used with any cell or tissue that expresses her2, such as a cancerous cell or a metastatic lesion. Non-limiting examples of her2-expressing cancers include breast cancer, gastric cancer, bladder cancer, urothelial cell carcinoma, esophageal cancer, lung cancer, cervical cancer, endometrial cancer, and ovarian cancer (English et al. (2013) Mol. Diagn. Ther. 17:85-99). Non-limiting examples of her2-expressing cells include NCI-N87-luc human gastric carcinoma cells, ZR75 and BT-474 human breast ductal carcinoma cells, and cells comprising a recombinant nucleic acid encoding her2 or a portion thereof.
[0256] In various other embodiments, the disclosed ADCs may be administered in any cell or tissue that expresses mesothelin (MSLN), such as a MSLN-expressing cancer cell or tissue. An exemplary embodiment includes a method of inhibiting MSLN-mediated cell signaling or a method of killing a cell. The method may be used with any cell or tissue that expresses MSLN, such as a cancerous cell or a metastatic lesion. Non-limiting examples of MSLN-expressing cancers include mesothelioma, pancreatic cancer (e.g., pancreatic adenocarcinoma), ovarian cancer, and lung cancer (e.g., lung adenocarcinoma) (Wang et al. (2012) PLoS ONE 7:e33214). Non-limiting examples of MSLN-expressing cells include OVCAR3 human ovarian carcinoma cells, HEC-251 human endometroid cells, H226 human lung squamous cell mesothelioma cells, and cells comprising a recombinant nucleic acid encoding MSLN or a portion thereof.
[0257] Exemplary methods include the steps of contacting the cell with an ADC, as described herein, in an effective amount, i.e., amount sufficient to kill the cell. The method can be used on cells in culture, e.g. in vitro, in vivo, ex vivo, or in situ. For example, cells that express FRA, her2, and/or MSLN (e.g., cells collected by biopsy of a tumor or metastatic lesion; cells from an established cancer cell line; or recombinant cells), can be cultured in vitro in culture medium and the contacting step can be effected by adding the ADC to the culture medium. The method will result in killing of cells expressing FRA, her2, and/or MSLN, including in particular tumor cells expressing FRA, her2, and/or MSLN. Alternatively, the ADC can be administered to a subject by any suitable administration route (e.g., intravenous, subcutaneous, or direct contact with a tumor tissue) to have an effect in vivo.
[0258] The in vivo effect of a disclosed ADC therapeutic composition can be evaluated in a suitable animal model. For example, xenogenic cancer models can be used, wherein cancer explants or passaged xenograft tissues are introduced into immune compromised animals, such as nude or SCID mice (Klein et al. (1997) Nature Med. 3:402-8). Efficacy may be predicted using assays that measure inhibition of tumor formation, tumor regression or metastasis, and the like.
[0259] In vivo assays that evaluate the promotion of apoptosis may also be used. In one embodiment, xenografts from tumor bearing mice treated with the therapeutic composition can be examined for the presence of apoptotic foci and compared to untreated control xenograft-bearing mice. The extent to which apoptotic foci are found in the tumors of the treated mice provides an indication of the therapeutic efficacy of the composition.
[0260] Further provided herein are methods of treating cancer. The ADCs disclosed herein can be administered to a non-human mammal or human subject for therapeutic purposes. The therapeutic methods entail administering to a mammal having a tumor a biologically effective amount of an ADC comprising a selected chemotherapeutic agent (e.g., eribulin) linked to a targeting antibody that binds to an antigen expressed, that is accessible to binding, or is localized on a cancer cell surface. An exemplary embodiment is a method of delivering a chemotherapeutic agent to a cell expressing FRA, comprising conjugating the chemotherapeutic agent to an antibody that immunospecifically binds to an FRA epitope and exposing the cell to the ADC. Exemplary tumor cells that express FRA for which the ADCs of the present disclosure are indicated include cells from a gastric cancer, a serous ovarian cancer, a nonsmall cell lung cancer, a colorectal cancer, a breast cancer (e.g., a triple negative breast cancer), a lung carcinoid, an osteosarcoma, an endometrial cancer, and an endometrial carcinoma with serous histology.
[0261] Another exemplary embodiment is a method of delivering a chemotherapeutic agent to a cell expressing her2, comprising conjugating the chemotherapeutic agent to an antibody that immunospecifically binds to a her2 epitope and exposing the cell to the ADC. Exemplary tumor cells that express her2 for which the ADCs of the present disclosure are indicated include cells from a breast cancer, a gastric cancer, a bladder cancer, an urothelial cell carcinoma, an esophageal cancer, a lung cancer, a cervical cancer, an endometrial cancer, and an ovarian cancer.
[0262] Another exemplary embodiment is a method of delivering a chemotherapeutic agent to a cell expressing MSLN, comprising conjugating the chemotherapeutic agent to an antibody that immunospecifically binds to a MSLN epitope and exposing the cell to the ADC. Exemplary tumor cells that express MSLN for which the ADCs of the present disclosure are indicated include cells from a mesothelioma, a pancreatic cancer (e.g., an pancreatic adenocarcinoma), an ovarian cancer, and a lung cancer (e.g., lung adenocarcinoma).
[0263] Another exemplary embodiment is a method of treating a patient having or at risk of having a cancer that expresses a target antigen for the antibody moiety of the ADC, such as FRA, her2, or MSLN, comprising administering to the patient a therapeutically effective amount of an ADC of the present disclosure. In some embodiments, the patient is non-responsive or poorly responsive to treatment with an anti-FRA antibody when administered alone, and/or treatment with a drug moiety (e.g., eribulin) when administered alone. In other embodiments, the patient is non-responsive or poorly responsive to treatment with an anti-her2 antibody when administered alone, and/or treatment with a drug moiety (e.g., eribulin) when administered alone. In other embodiments, the patient is non-responsive or poorly responsive to treatment with an anti-MSLN antibody when administered alone, and/or treatment with a drug moiety (e.g., eribulin) when administered alone. In other embodiments, the patient is intolerant to treatment with a drug moiety (e.g., eribulin) when administered alone. For instance, a patient may require doses of eribulin to treat a cancer that lead to systemic toxicity, which are overcome by targeted delivery to a cancer expressing a target antigen for the antibody moiety of the ADC such as FRA, her2, or MSLN, thereby reducing off-target killing.
[0264] Another exemplary embodiment is a method of reducing or inhibiting growth of an target antigen-expressing tumor (e.g., an FRA-expressing tumor, a her2-expressing tumor, or a MSLN-expressing tumor), comprising administering a therapeutically effective amount of an ADC. In some embodiments, the treatment is sufficient to reduce or inhibit the growth of the patient's tumor, reduce the number or size of metastatic lesions, reduce tumor load, reduce primary tumor load, reduce invasiveness, prolong survival time, and/or maintain or improve the quality of life. In some embodiments, the tumor is resistant or refractory to treatment with an anti-FRA antibody when administered alone, and/or treatment with a drug moiety (e.g., eribulin) when administered alone. In other embodiments, the tumor is resistant or refractory to treatment with an anti-her2 antibody when administered alone, and/or treatment with a drug moiety (e.g., eribulin) when administered alone. In some embodiments, the tumor is resistant or refractory to treatment with an anti-MSLN antibody when administered alone, and/or treatment with a drug moiety (e.g., eribulin) when administered alone.
[0265] Moreover, antibodies of the present disclosure may be administered to a non-human mammal expressing an antigen with which the ADC is capable of binding for veterinary purposes or as an animal model of human disease. Regarding the latter, such animal models may be useful for evaluating the therapeutic efficacy of the disclosed ADCs (e.g., testing of dosages and time courses of administration).
[0266] Further provided herein are therapeutic uses of the disclosed ADCs. An exemplary embodiment is the use of an ADC in the treatment of a target antigen-expressing cancer (e.g., an FRA-expressing cancer, a her2-expressing cancer, or a MSLN-expressing cancer). ADCs for use in the treatment of an target antigen-expressing cancer (e.g., an FRA-expressing cancer, a her2-expressing cancer, or a MSLN-expressing cancer) are also disclosed. Methods for identifying subjects having cancers that express FRA, her2, and/or MSLN are known in the art and may be used to identify suitable patients for treatment with a disclosed ADC.
[0267] Another exemplary embodiment is the use of an ADC in a method of manufacturing a medicament for the treatment of a target antigen-expressing cancer (e.g., an FRA-expressing cancer, a her2-expressing cancer, or a MSLN-expressing cancer).
[0268] The therapeutic compositions used in the practice of the foregoing methods may be formulated into pharmaceutical compositions comprising a pharmaceutically acceptable carrier suitable for the desired delivery method. An exemplary embodiment is a pharmaceutical composition comprising an ADC of the present disclosure and a pharmaceutically acceptable carrier. Suitable carriers include any material that, when combined with the therapeutic composition, retains the anti-tumor function of the therapeutic composition and is generally non-reactive with the patient's immune system. Pharmaceutically acceptable carriers include any and all solvents, dispersion media, coatings, antibacterial and antifungal agents, isotonic and absorption delaying agents, and the like that are physiologically compatible. Examples of pharmaceutically acceptable carriers include one or more of water, saline, phosphate buffered saline, dextrose, glycerol, ethanol, mesylate salt, and the like, as well as combinations thereof. In many cases, it will be preferable to include isotonic agents, for example, sugars, polyalcohols such as mannitol, sorbitol, or sodium chloride in the composition. Pharmaceutically acceptable carriers may further comprise minor amounts of auxiliary substances such as wetting or emulsifying agents, preservatives or buffers, which enhance the shelf life or effectiveness of the ADC.
[0269] Therapeutic formulations may be solubilized and administered via any route capable of delivering the therapeutic composition to the tumor site. Potentially effective routes of administration include, but are not limited to, intravenous, parenteral, intraperitoneal, intramuscular, intratumor, intradermal, intraorgan, orthotopic, and the like. Therapeutic protein preparations can be lyophilized and stored as sterile powders, preferably under vacuum, and then reconstituted in bacteriostatic water (containing for example, benzyl alcohol preservative) or in sterile water prior to injection. Therapeutic formulations may comprise an ADC or a pharmaceutically acceptable salt thereof, e.g., a mesylate salt.
[0270] The ADCs disclosed herein may be administered at a dosage ranging from about 0.2 mg/kg to about 10 mg/kg to a patient in need thereof. In some embodiments, the ADC is administered to the patient daily, bimonthly, or any time period in between. Dosages and administration protocols for the treatment of cancers using the foregoing methods will vary with the method and the target cancer, and will generally depend on a number of other factors appreciated in the art.
[0271] Various delivery systems are known and may be used to administer one or more ADCs of the present disclosure. Methods of administering the ADCs include, but are not limited to, parenteral administration (e.g., intradermal, intramuscular, intraperitoneal, intravenous and subcutaneous), epidural administration, intratumoral administration, and mucosal administration (e.g., intranasal and oral routes). In addition, pulmonary administration may be employed, e.g., by use of an inhaler or nebulizer, and formulation with an aerosolizing agent. See, e.g., the compositions and methods for pulmonary administration described in U.S. Pat. Nos. 6,019,968, 5,985,320, 5,985,309, 5,934,272, 5,874,064, 5,855,913, 5,290,540, and 4,880,078; and PCT Publ. Nos. WO 92/19244, WO 97/32572, WO 97/44013, WO 98/31346, and WO 99/66903. The ADCs may be administered by any convenient route, for example, by infusion or bolus injection, or by absorption through epithelial or mucocutaneous linings (e.g., oral mucosa, rectal and intestinal mucosa, etc.). Administration can be either systemic or local.
[0272] Therapeutic compositions disclosed herein may be sterile and stable under the conditions of manufacture and storage. In some embodiments, one or more of the ADCs, or pharmaceutical compositions, is supplied as a dry sterilized lyophilized powder or water free concentrate in a hermetically sealed container and can be reconstituted (e.g., with water or saline) to the appropriate concentration for administration to a subject. Preferably, one or more of the prophylactic or therapeutic agents or pharmaceutical compositions is supplied as a dry sterile lyophilized powder in a hermetically sealed container at a unit dosage of at least 5 mg, at least 10 mg, at least 15 mg, at least 25 mg, at least 35 mg, at least 45 mg, at least 50 mg, at least 75 mg, or at least 100 mg, or any amount in between. In some embodiments, the lyophilized ADCs or pharmaceutical compositions is stored at between 2.degree. C. and 8.degree. C. in the original container. In some embodiments, one or more of the ADCs or pharmaceutical compositions described herein is supplied in liquid form in a hermetically sealed container, e.g., a container indicating the quantity and concentration of the agent. In some embodiments, the liquid form of the administered composition is supplied in a hermetically sealed container of at least 0.25 mg/mL, at least 0.5 mg/mL, at least 1 mg/mL, at least 2.5 mg/mL, at least 5 mg/mL, at least 8 mg/mL, at least 10 mg/mL, at least 15 mg/mL, at least 25 mg/mL, at least 50 mg/mL, at least 75 mg/mL, or at least 100 mg/mL ADC. The liquid form may be stored at between 2.degree. C. and 8.degree. C. in the original container.
[0273] In some embodiments, the disclosed ADCs can be incorporated into a pharmaceutical composition suitable for parenteral administration. The injectable solution may be composed of either a liquid or lyophilized dosage form in a flint or amber vial, ampule, or pre-filled syringe, or other known delivery or storage device.
[0274] The compositions described herein may be in a variety of forms. These include, for example, liquid, semi-solid, and solid dosage forms, such as liquid solutions (e.g., injectable and infusible solutions), dispersions or suspensions, tablets, pills, powders, liposomes, and suppositories. The preferred form depends on the intended mode of administration and therapeutic application.
[0275] In various embodiments, treatment involves single bolus or repeated administration of the ADC preparation via an acceptable route of administration.
[0276] Patients may be evaluated for the levels of target antigen in a given sample (e.g. the levels of target antigen expressing cells) in order to assist in determining the most effective dosing regimen, etc. An exemplary embodiment is a method of determining whether a patient will be responsive to treatment with an ADC of the present disclosure, comprising providing a biological sample from the patient and contacting the biological sample with the ADC. Exemplary biological samples include tissue or body fluid, such as an inflammatory exudate, blood, serum, bowel fluid, stool sample, or tumor biopsy (e.g., a tumor biopsy derived from a patient having or at risk of a target antigen-expressing cancer, e.g., an FRA-expressing cancer, a her2-expressing cancer, or a MSLN-expressing cancer). In some embodiments, a sample (e.g., a tissue and/or body fluid) can be obtained from a subject, and a suitable immunological method can be used to detect and/or measure protein expression of the target antigen (e.g., FRA, her2, or MSLN). Such evaluations are also used for monitoring purposes throughout therapy, and are useful to gauge therapeutic success in combination with the evaluation of other parameters.
[0277] In some embodiments, the efficacy of an ADC may be evaluated by contacting a tumor sample from a subject with the ADC and evaluating tumor growth rate or volume. In some embodiments, when an ADC has been determined to be effective, it may be administered to the subject.
[0278] The above therapeutic approaches can be combined with any one of a wide variety of additional surgical, chemotherapy, or radiation therapy regimens.
[0279] Also disclosed herein are uses of one or more of the disclosed ADCs in the manufacture of a medicament for treating cancer, e.g., according to the methods described above. In some embodiments, the ADCs disclosed herein are used for treating cancer, e.g., according to the methods described above.
[0280] In various embodiments, kits for use in the laboratory and therapeutic applications described herein are within the scope of the present disclosure. Such kits may comprise a carrier, package, or container that is compartmentalized to receive one or more containers such as vials, tubes, and the like, each of the container(s) comprising one of the separate elements to be used in a method disclosed herein, along with a label or insert comprising instructions for use, such as a use described herein. Kits may comprise a container comprising a drug moiety. The present disclosure also provides one or more of the ADCs, or pharmaceutical compositions thereof, packaged in a hermetically sealed container, such as an ampoule or sachette, indicating the quantity of the agent.
[0281] Kits may comprise the container described above and one or more other containers associated therewith that comprise materials desirable from a commercial and user standpoint, including buffers, diluents, filters, needles, syringes; carrier, package, container, vial and/or tube labels listing contents and/or instructions for use, and package inserts with instructions for use.
[0282] A label may be present on or with the container to indicate that the composition is used for a specific therapy or non-therapeutic application, such as a prognostic, prophylactic, diagnostic, or laboratory application. A label may also indicate directions for either in vivo or in vitro use, such as those described herein. Directions and or other information may also be included on an insert(s) or label(s), which is included with or on the kit. The label may be on or associated with the container. A label may be on a container when letters, numbers, or other characters forming the label are molded or etched into the container itself. A label may be associated with a container when it is present within a receptacle or carrier that also holds the container, e.g., as a package insert. The label may indicate that the composition is used for diagnosing or treating a condition, such as a cancer a described herein.
[0283] It will be readily apparent to those skilled in the art that other suitable modifications and adaptations of the methods of the invention described herein are obvious and may be made using suitable equivalents without departing from the scope of the invention or the embodiments disclosed herein. Having now described the invention in detail, the same will be more clearly understood by reference to the following examples, which are included for purposes of illustration only and are not intended to be limiting.
Example 1
1. Materials and Methods
[0284] MORAb-003 used for the preparation of ADCs was from Lot # AA0312.
1.1 Cytotoxins
[0285] Structures of conjugatable cytotoxins are shown in Table 11.
TABLE-US-00011 TABLE 11 Conjugatable cytotoxins Compound Cleava- name Linker Cytotoxin bility Structure PEG3-Bz- disulfidyl- dimethyl- cryptophycin maleimido- PEG3- Benzyl- disulfidyl- dimethyl cryptophycin yes ##STR00003## LL2- cryptophycin LL2 cryptophycin yes ##STR00004## LL3- cryptophycin LL3 cryptophycin yes ##STR00005## VCP- cryptophycin maleimido- PEG2-Val- Cit-pAB cryptophycin yes ##STR00006## VCP- eribulin (ER- 001159569) maleimido- PEG2-Val- Cit-pAB eribulin yes ##STR00007## ER- 001161318 maleimido- (CH.sub.2).sub.5- Val-Cit- pAB ER- 001150828 (aziridino- maytanzine- P3) yes ##STR00008## ER- 001161319 maleimido- PEG2-Val- Cit-pAB ER- 001150828 (aziridino- maytanzine- P3) yes ##STR00009## ER- 001159200 maleimido- (CH.sub.2).sub.5 maytanzine DM1 No ##STR00010## M-MMAE maleimido- (CH.sub.2).sub.5- Val-Cit- pAB monomethyl auristatin E yes ##STR00011## NHS-PEG2- AuF NHS- PEG2 auristatin F no ##STR00012## M-DM1 SMCC maytansine DM1 no ##STR00013## M-0285 PEG-pAB duostatin 3 yes ##STR00014## M-0115 Asn-Ala duostatin-5 yes ##STR00015## M-172 cyclohexyl duostatin 3 no Reduced disulfide linking chemistry M-174 cyclohexyl duostatin 3 no Reduced disulfide linking chemistry M-158 PEG-pAB duostatin 10 yes Reduced disulfide linking chemistry M-0384 PEG- duostatin 14 no Reduced disulfide linking chemistry thioether M-0302 PEG-Asn duostatin 14 no Reduced disulfide linking chemistry M-292 PEG-Asn duostatin 14 yes Reduced disulfide linking chemistry M-0026 PEG duostatin 14 yes Reduced disulfide linking chemistry M-0267 PEG- duomycin 7 no Reduced disulfide linking chemistry thioether M-0272 Asn-Ala duomycin 7 yes Reduced disulfide linking chemistry M-0260 PEG-pAB duomycin 7 yes Reduced disulfide linking chemistry M-0276 Asn-Ala duomycin 7 yes Reduced disulfide linking chemistry M-015-0913 cyclohexyl duostatin 3 no Limited lysine utilization M-030-0132 PEG-pAB duostatin 6 yes Limited lysine utilization M-0161 cyclohexyl duostatin 10 no Limited lysine utilization M-0157 PEG-pAB duostatin 10 yes Limited lysine utilization M-027-0381 thioether duostatin 14 no Limited lysine utilization M-0025 PEG duostatin 14 no Limited lysine utilization M-0301 PEG-Asn duostatin 14 no Limited lysine utilization M-030-0011 PEG-pAB duostatin 14 yes Limited lysine utilization M-030-0291 PEG-Asn duostatin 14 yes Limited lysine utilization M-0114 PEG-pAB duostatin-5 yes Reduced disulfide bridging chemistry Abbreviations: Ala, alanine; Asn, asparagine; Cit, citrulline; NHS, N-hydroxystweinimide; pAB, p-aminobenzyloxycarbonyl; PEG, polyethylene glycol; SMCC, succinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate; Val, valine; VCP, Val-Cit-pAB.
1.2 Antibody-Drug Conjugation
1.2.1 Partial Reduction Using TCEP
[0286] Partial reduction conditions for MORAb-003 were established by varying concentration of the non-thiol reducing agent tris(2-carboxyethyl)phosphine (TCEP), antibody concentration, and time of reduction. MORAb-003 was buffer-exchanged into Dulbecco's Phosphate-Buffered Saline (DPBS) containing 1 mM ethylenediaminetetraacetic acid (EDTA), then concentrated to 10 mg/mL using centrifugal concentration with 10 kD molecular weight cut-off (MWCO) centrifugal filters. Antibodies were diluted to the appropriate concentration and TCEP was added at the indicated final concentration, and gently mixed for 1 hour at room temperature. TCEP was removed by desalting using 5 or 10 mL Zeba.TM. spin desalting columns with DPBS/1 mM EDTA as buffer (Thermo Fisher, 40 kD MWCO), according to the manufacturer's protocol. Samples were analyzed for free thiol content using the Thiol fluorometric quantification kit (Abcam), according to the manufacturer's protocol. SDS-PAGE analysis under non-reducing conditions was performed to determine extent and location of disulfide bond breakage, as described in section 1.3.3. In some cases, desalted MAbs were brought to 1-2 mg/mL by dilution in DPBS and subjected to biotinylation to determine conjugatability and drug-to-antibody (DAR) ratio. 10 mM maleimido-PEG2-biotin (Thermo Fisher) in dimethylsulfoxide (DMSO) was added to antibody (mAb) at a molar ratio of 10:1 and incubated at room temperature for 4 hours with gentle agitation. Following conjugation, unreacted compound was removed by desalting using Zeba.TM. spin desalting columns (Thermo Fisher). Samples were then analyzed by LC-MS for determination of DAR, as detailed in section 1.3.4.
1.2.2 Cytotoxin Conjugation
[0287] Partially-reduced antibody was brought to 2.5 mg/mL in 0.5.times.DPBS, 0.5 mM EDTA, and mixed thoroughly. Organic co-solvents, if used, were then added and mixed thoroughly. Co-solvents examined were propylene glycol (20% and 50% final concentration), dimethylsulfoxide (DMSO) (10%), N,N-dimethylformamide (20%), N,N-dimethylacetamide (20%), and N,N-dimethylpropionamide (20%). Maleimido-modified cytotoxin (6 mM stock in DMSO) was added to antibodies at a molar ratio of 1:6 (mAb:compound) and mixed thoroughly. Conjugation proceeded at room temperature for 3.5 hours, with gentle mixing. 50% propylene glycol at 50% was chosen as the final organic modifier and was used in all subsequent conjugation reactions.
1.2.3 Purification
[0288] Conjugated antibody was purified using 26/10 HiTrap.RTM. desalting column(s) (GE Healthcare) with chromatography performed on a fast protein liquid chromatography (FPLC) (GE Healthcare), in order to remove unreacted maleimido-cytotoxin and propylene glycol. MORAb-003 ADCs, including MORAb-003-mal-VCP-eribulin (MORAb-202), were formulated in DPBS (formulation buffer was used as running buffer during FPLC chromatography).
1.3 Biophysical Characterization
1.3.1 BCA Assay
[0289] Prepared bicinchoninic acid (BCA) reagent (200 .mu.L) was added to 25 .mu.L of serially-diluted ADCs or bovine gamma globin (Thermo Fisher) 2 mg/mL standard, and samples were mixed thoroughly. Samples were incubated at 37.degree. C. for 20 min. Plates were read at 595 nm on a SpectraMax.RTM. M5 plate reader (Molecular Devices). Data was analyzed using SoftMax.RTM. Pro (ver 3.2) with a 4-parameter fitting model.
1.3.2 SEC-HPLC Analysis
[0290] The antibody aggregation was analyzed by size-exclusion, high-performance liquid chromatography (SEC-HPLC) using an Agilent 1100. The mAb was diluted to 1 mg/mL in DPBS. The antibody (20 .mu.L) was injected onto a TSKgel.RTM. SuperSW guard column (4.6 mm.times.3.5 cm, 4 .mu.m pore size, Tosoh Bioscience), followed by a TSKgel.RTM. SuperSW3000 column (4.6 mm.times.30 cm, 4 .mu.m pore size), eluted from the column with 0.1 M sodium phosphate containing 0.15 M NaCl and 0.05% NaN.sub.3, at pH 7.4, at a flow rate of 0.3 mL/min for 20 min. All data were analyzed using Agilent ChemStation software. Percent aggregation was calculated as [PA.sub.aggregate/PA.sub.total]*100, where PA=integrated peak area.
1.3.3 SDS-PAGE Analysis
[0291] Protein samples (0.1-10 .mu.g) were brought to 1.times. with lithium dodecylsulfate (LDS) sample buffer. For non-reduced samples, incubation was performed at room temperature for 10 min prior to electrophoresis. For reduced samples, dithiothreitol (DTT) was added to a final concentration of 20 mM and samples were heated to 95.degree. C. for 10 min and placed on ice prior to electrophoresis. Samples were loaded on to 10-, 12-, or 15-well Bis-Tris SDS-PAGE gels (Thermo Fisher) with 1.times.MOPS or 1.times.MES as running buffer. Electrophoresis was performed at 185 V (constant voltage) for 1 hour. Gels were stained with InstantBlue staining solution (Expedeon) and destained in water. Documentation was performed on an UltraLum gel documentation system using 600 nm orange filters.
1.3.4 UPLC/ESI-MS Analysis of Drug-to-Antibody Ratio (DAR)
[0292] ADCs were deglycosylated using PNGase F (New England BioLabs). G7 buffer (10 .mu.L) and PNGase F (2 .mu.L) were added to the mAb (90 .mu.L, 1 mg/mL in DPBS). The reaction was incubated in a Discover microwave (CEM) for 2 cycles: (1) microwave power 10 W, 37.degree. C., 10 min, followed by a 5-min pause; (2) microwave power 2 W, 37.degree. C., 10 min. A portion of the sample was reduced by adding DTT to a final concentration of 20 mM, followed by incubation at 60.degree. C. for 3 min. Samples were then analyzed using a Waters Acquity Ultra Performance Liquid Chromatography (UPLC) and quadrupole time of flight (Q-Tof) Premier mass spectrometer. Samples (0.5-2 .mu.g each) were injected onto a MassPrep.TM. micro desalting column at 65.degree. C., eluted from the column with a 5 min equilibration in 95% of mobile phase A, a 10 min gradient (5-90% B), and a 10 min re-equilibration in 95% of mobile phase A, at 0.05 mL/min. Mobile phase A was 0.1% formic acid in water. Mobile phase B was 0.1% formic acid in acetonitrile. The Q-Tof mass spectrometer was run in positive ion, V-mode with detection in the range of 500-4000 m/z. The source parameters were as follows: capillary voltage, 2.25 kV (intact antibody)-2.50 kV (reduced antibody); sampling cone voltage, 65.0 V (intact antibody) or 50.0 V (reduced antibody); source temperature, 100.degree. C.; desolvation temperature, 250.degree. C.; desolvation gas flow, 550 L/hr. The protein peak was deconvoluted using the MassLynx.RTM. MaxEnt 1 function. Relative intensities of each unconjugated, singly-conjugated, and multiply-conjugated heavy and light chain masses were combined to calculate the overall DAR using the formula:
2[[I.sub.LC+1+2(I.sub.LC+2)+3(I.sub.LC+3)+ . . . n(I.sub.LC+n)]/.SIGMA.I.sub.LCtot]+2[[I.sub.HC+1+2(I.sub.HC+2)+3(I.sub.HC- +3)+ . . . n(I.sub.HC+n)]/.SIGMA.I.sub.HCtot]
where I.sub.LC+1 is mass intensity of light chain conjugated with one cytotoxin, I.sub.LC+2 is mass intensity of light chain conjugated with two cytotoxins, etc. I.sub.HC are the intensities from the corresponding conjugated heavy chains, and .SIGMA.I.sub.LCtot and .SIGMA.I.sub.HCtot are the combined intensities of all unconjugated and conjugated light chains and heavy chains, respectively.
1.3.5 HIC-HPLC DAR Analysis
[0293] In addition to DAR analysis by UPLC/electrospray ionization (ESI)-MS analysis, MORAb-003-vcp-eribulin DAR and MORAb-003-0285 DAR were also analyzed using hydrophobic interaction HPLC (HIC-HPLC). Samples were injected onto a TSKgel.RTM. Ether-5 PW, 7.5 mm ID.times.7.5 cm, 10 .mu.M pore size, and eluted from the column with a 3 min equilibration in 100% of mobile phase A, a 15 min gradient (0-100% B), a 5 min hold in 100% B, a 1 min change to 100% A, and a 5 min re-equilibration in 100% of mobile phase A, at 0.7 mL/min. Mobile phase A was 25 mM sodium phosphate, 1.5 M ammonium sulfate, pH 7.0. Mobile phase B was 25 mM sodium phosphate, 25% isopropanol, pH 7.0. Detection was done at 280 nm (reference 320 nm). DAR was determined by the formula:
[AUC.sub.+1+2(AUC.sub.+2)+3(AUC.sub.+3)+ . . . n(AUC.sub.+n).SIGMA.AUC.sub.tot]
where AUC.sub.+1 is the area under the curve for the mAb peak corresponding to ADC conjugated with one cytotoxin, AUC.sub.+2 is the area under the curve for the mAb peak corresponding to ADC conjugated with two cytotoxins, etc. .SIGMA.AUC.sub.tot is the combined area under the curve for all peaks.
1.4 Cytotoxicity Analyses
1.4.1 Crystal Violet Assay
[0294] IGROV1 (FR.sup.hi) and SJSA-1 (FR.sup.neg) cells were sub-cultured and seeded at 10,000 cells/well in complete growth medium in 96-well tissue culture plates, incubated at 37.degree. C., 5% CO.sub.2 overnight (16 hours). Typically, test reagents were serial diluted 1:4 in 2 mL deep-well dilution plates, starting at 1 .mu.M (10 dilutions total). 1004 of diluted samples were added to the cell plates (starting concentration of test samples at 500 nM). Plates were incubated at 37.degree. C., 5% CO.sub.2 for an additional 48 hours. Medium was discarded, plates were washed once with 2004 DPBS, stained with 50 .mu.L of 0.2% Crystal Violet solution at room temperature for 15 min, and then washed extensively with tap water. Plates were air-dried, and Crystal Violet was dissolved with 200 .mu.L of 1% SDS solution. Plates were read at 570 nm. Data was analyzed using GraphPad Prism 6. Assays were performed using a seeding density of 1,000 cells per well and compound exposure was for a total of 5 days. When shorter-term exposure was desired, medium containing cytotoxic agents was removed after 4 hours and replaced with fresh growth medium prior to 5-day incubation. For OVCAR3, CaOV3, and NCI-H2110, cells were seeded at 3,000 cells/well and incubated for 5 days with ADC. For competition experiments, titrated ADCs were pre-incubated with 2 .mu.M (final) unconjugated MORAb-003 prior to incubation with cells.
1.4.2 Bystander Killing Assay
[0295] The day before study commencement, Nuclight.TM. Green (NLG) IGROV1 cells were seeded at 5,000 cells/well into 96-well round bottom plates, followed by centrifugation at 1,000 rpm for 3 min at room temperature to ensure formation of a cell pellet. The plate was placed in the vessel of an Incucyte Zoom.RTM. (EssenBio science) and incubated at 37.degree. C./5% CO.sub.2 overnight. The program was set to collect images of cell growth, and to determine total numbers of nuclear green-stained and nuclear red-stained cells as well as phase-confluency of the cells every two hours. The day of the experiment, MORAb-003 ADC or free drug was diluted in complete RPMI medium and serially-diluted, starting at 400 nM. 504 of cytotoxin solution was added to the NLG-IGROV1 cells and incubated for 30 min. During the incubation period, Nuclight.TM. Red (NLR) HL-60 (FR.sup.neg) cells were diluted to 2.times.10.sup.5, 1.times.10.sup.5 or 5.times.10.sup.4 cell/mL with fresh media. 50 .mu.L of the NLR-HL60 cell suspension or medium alone was added to the NLG-IGROV1 wells, followed by centrifugation at 1,000 rpm for 3 min at room temperature to ensure re-formation of the cell pellet. The plate was placed back into the vessel of Incucyte Zoom (EssenBio science) and incubated at 37.degree. C./5% CO.sub.2 for up to 5 days. Relative cell growth of NLG-IGROV1 was determined by comparison to no ADC or free drug alone added samples using green cell counts. Relative cell growth of HL60 was done similarly, except that red cell count was determined. Determination of IC.sub.50 values for both NLG-IGROV1 and NLR-HL-60 was determined using Prism (GraphPad).
1.4.3 Serum Stability Assay
[0296] 20 .mu.L of MORAb-003 ADCs were thoroughly mixed with 80 .mu.L of DPBS, normal pooled human serum (Bioreclamation, Lot BRH552911), or normal pooled mouse serum (Bioreclamation, Lot MSE152591), and incubated at 37.degree. C. for 0, 4, 24, and 48 hours. Following incubation, samples were frozen and stored at -20.degree. C. until evaluation in cytotoxicity and binding assays. For cytotoxicity analyses, samples were evaluated on IGROV1 and SJSA-1 cells, as detailed in section 1.4.1. For binding assessment, samples were evaluated using a solution-based MSD ECL assay. Samples were incubated with biotinylated folate receptor alpha and sulfo-tag anti-MORAb-003 before capture on a streptavidin plate and detected using electrochemiluminescense with a MSD Sector Imager 2400.
2. Results
2.1 Preparation of MORAb-003 ADCs
[0297] In order to select the best combination of linker and cytotoxin to conjugate with MORAb-003, ADCs were prepared using three methodologies. According to the conjugation strategy shown in FIG. 1, unpaired cysteines are generated through partial reduction with limited molar equivalents of the non-thiol reducing agent TCEP. This strategy preferentially reduces the interchain disulfide bonds which link the light chain and heavy chain (one pair per H-L pairing) and the two heavy chains in the hinge region (two pairs per H-H pairing in the case of human IgG1), while leaving the intrachain disulfide bonds intact.
[0298] The second conjugation strategy for preparing MORAb-003 ADCs utilized reduced disulfide bridging chemistry. Reduced disulfide bridging chemistry rebridges the free thiols of the cysteine residues released during the partial reduction process, mimicking the role of the disulfide bond and thus retaining the stability and function of the ADC.
[0299] The third conjugation strategy for preparing MORAb-003 ADCs employed limited lysine utilization. Limited lysine utilization results in the conjugation of a very limited number of the estimated 70+ solvent-exposed lysines available on a typical human IgG molecule, and can potentially afford mixtures of ADC product with lower homogeneity relative to strategies involving cysteine modification.
2.1.1 Preparation of VCP-Eribulin for MORAb-003 ADCs
[0300] Eribulin (1) (10 mg, 14 .mu.mol) (FIG. 2) was dissolved in N,N-dimethylformamide (DMF) (1 mL), and mixed well. N,N-diisopropylethylamine (Hunig's Base or iPr.sub.2NEt) (3.6 .mu.L, 21 .mu.mol) and Fmoc-Val-Cit-para-aminobenzyl-para-nitrophenol (Fmoc-VCP-PNP) (2) (16 mg, 21 .mu.ma Concortis Biosystems, cat # VC1003) was added. The reaction mixture was stirred at room temperature for 4-16 hours, monitored using a ninhydrin test kit (Anaspec, cat #25241) until the reaction was completed. Diethylamine (Et.sub.2NH) (0.014 mL, 0.14 mmol) was then added to the reaction mixture, stirred for 2 hours at 18-25.degree. C. to remove the Fmoc protecting group. The reaction was monitored using a ninhydrin test kit. Upon completion, the solvent was evaporated under vacuum to afford crude VCP-eribulin (3) (16 mg), purified using a ZOBAX SB-C18 column (5 .mu.m pore size, 9.4.times.150 mm) on an Waters Alliance e2695 HPLC system in the mobile phase of H.sub.2O--CH.sub.3CN containing 0.1% formic acid, through a gradient of 15-70% B. VCP-eribulin (3) (16 mg) was dissolved in DMF (1 mL). Hunig's Base (7.2 .mu.L, 41 .mu.mol) and maleimido-PEG2-NHS (4) (9.7 mg, 27 .mu.mol) were added. The reaction mixture was stirred at 18-25.degree. C. for 3 hours. The reaction mixture was purified by HPLC (H.sub.2O--CH.sub.3CN) containing 0.1% formic acid) through a gradient of 15-70% B. Solvent was removed by lyophilization to yield mal-(PEG).sub.2-Val-Cit-p-aminobenzyloxycarbonyl (pAB)-eribulin (mal-(PEG).sub.2-VCP-eribulin) (5).
2.1.2 Optimization of Reduction Conditions
[0301] MORAb-003 ADCs were prepared by generating unpaired cysteines through partial reduction with limited molar equivalents of the non-thiol reducing agent tris(2-carboxyethyl)phosphine (TCEP). An initial investigation was performed on MORAb-003, whereby antibody concentration, TCEP concentration, and incubation time were varied, with the goal to generate an average of 4 conjugatable sites per antibody molecule. The number of free thiol sites was determined using a fluorometric thiol quantitation assay. The results of this analysis are shown in Table 12. The extent of H-H and H-L bond breakage following a 10 min, 30 min, 60 min, or 120 min incubation was also analyzed by SDS-PAGE (FIG. 3). For this analysis, non-reduced and reduced samples were loaded on an SDS-PAGE gel and electrophoresis was performed at 185 V for 1 hour. In FIG. 3, lane M corresponds to protein standard. Lane 1 corresponds to untreated, non-reduced MORAb-003. Lane 2 corresponds to MORAb-003 (5.3 mg/mL) reduced in 70.6 .mu.M TCEP. Lane 3 corresponds to MORAb-003 (5.3 mg/mL reduced) in 141.2 .mu.M TCEP. Lane 4 corresponds to MORAb-003 (1.5 mg/mL) reduced in 20 .mu.M TCEP. Lane 5 corresponds to MORAb-003 (1.5 mg/mL) reduced in 40 .mu.M TCEP. The identities of each band are indicated on the lower right gel. "H" indicates heavy chain, whereas "L" indicates light chain.
TABLE-US-00012 TABLE 12 Optimization of reduction conditions of MORAb-003 10 min 30 min 60 min 120 min Disulfide Disulfide Disulfide Disulfide MORAb-003 TCEP Free bonds Free bonds Free bonds Free bonds concentration concentration thiol reduced thiol reduced thiol reduced thiol reduced .mu.M (mg/ml) .mu.M .mu.M per MAb .mu.M per MAb .mu.M per MAb .mu.M per MAb 35.3 (5.3) 70.6 215 3.0 247.5 3.5 297.6 4.2 266.8 3.8 35.3 (5.3) 141.2 339 4.8 372.8 5.3 384.2 5.4 479.8 6.8 10 (1.5) 20 13.3 0.7 14.7 0.7 15.2 0.8 14.6 0.7 10 (1.5) 40 21.8 1.1 25.6 1.3 26.9 1.3 27.4 1.4
[0302] Analysis of the SDS-PAGE and thiol content suggested that 60 min incubation of 5.3 mg/mL mAb at 4-fold molar ratio of TCEP to mAb provided a reasonable starting point, as limited reduction of the intramolecular disulfides seemed to be present (as determined by the free thiol content), and very little unreduced mAb was remaining (unreduced mAb would act as a competitive inhibitor in in vitro and in vivo studies using prepared ADCs). Further studies were conducted with MORAb-003 at starting concentrations of 5.0 mg/mL to confirm this optimized molar ratio of TCEP to mAb using SDS-PAGE analysis (FIG. 4). In FIG. 4, lane 1 corresponds to protein standard. Lane 2 corresponds to untreated, non-reduced MORAb-003. Lane 3 corresponds to MORAb-003 treated at a ratio of MORAb-003:TCEP of 1:1. Lane 4 corresponds to MORAb-003 treated at a ratio of MORAb-003:TCEP of 1:2. Lane 5 corresponds to MORAb-003 treated at a ratio of MORAb-003:TCEP of 1:3. Lane 6 corresponds to MORAb-003 treated at a ratio of MORAb-003:TCEP of 1:4. Conjugation using maleimido-PEG2-biotin was also performed subsequent to reduction and TCEP removal, in order to simulate conjugation of cytotoxin for ADC preparation. DAR analysis was performed using LC-MS. The results of these studies are provided in Table 13.
TABLE-US-00013 TABLE 13 Optimization of reduction conditions of MORAb-003 - conjugation levels with maleimido-PEG2-biotin TCEP MORAb-003 TCEP:mAb TCEP (.mu.M) LC HC DAR 1 33.3 0.29 0.34 1.26 2 66.7 0.48 0.83 2.62 3 100 0.63 1.21 3.68 4 133.2 0.73 1.70 4.86 LC, light chain biotin level; HC, heavy chain biotin level; DAR, biotin per mAb [DAR = 2(LC) + 2(HC)].
[0303] Following biotin conjugation, free thiol analysis indicated that no free thiol was present in MORAb-003-biotin. This indicated that, following reduction of disulfide bonds, conjugation typically occurred at both thiols generated, and that any unconjugated, reduced disulfides underwent re-oxidation to reform disulfide bonds. The final conditions chosen for reduction for ADC generation were antibody concentration of 5.0 mg/mL, TCEP concentration of 110 .mu.M, and incubation time of 60 min. This leads to a mAb with a DAR of 4 following conjugation.
2.1.3 ADC Conjugation Optimization
[0304] As the first cytotoxin used for ADC preparation was cryptophycin, which is a hydrophobic compound, initial conjugation optimization experiments were performed with a "surrogate" anti-human mesothelin antibody having two unpaired cysteines available for conjugation (one per light chain) at specific locations. This greatly facilitates the analysis of conjugation efficiency by mass spectrometry, as only the light chain needs to be analyzed. Titration of propylene glycol during conjugation of maleimido-LL3-cryptophycin to the surrogate antibody was performed followed by analysis of conjugation efficiency of the light chain by LC-MS (Table 14).
TABLE-US-00014 TABLE 14 Optimization of propylene glycol concentration in conjugation reaction Propylene glycol (%) Conjugated Ab LC (%) 0 8% 20 48% 50 100% LC masses: unconjugated, 23536 Da; conjugated, 24367 Da.
[0305] 50% propylene glycol resulted in full occupation of the available sites, and was chosen as the final concentration to be used. No loss in binding of the mAb was observed following conjugation (data not shown), indicating that the propylene glycol did not have deleterious effects to the antibody. Thus, the final conjugation reaction conditions chosen were 2.5 mg/mL mAb final, 6:1 molar ratio of maleimido-linker-cytotoxin:mAb in 0.5.times.DPBS (final concentration after propylene glycol addition), 0.5 mM EDTA, 50% propylene glycol, pH 7.2 for 3.5-4 hours at room temperature. In these reactions, propylene glycol is added prior to addition of maleimido-linker-cytotoxin.
2.1.4 Preparation of ADCs and Biophysical Characterization
[0306] The established reduction and conjugation conditions, described in section 2.1.2, were used to prepare the first 10 MORAb-003 ADCs listed in Table 15. The remaining ADCs were prepared by either reduced disulfide bridging or limited lysine utilization, with the exceptions of M-MMAE and M-DM1. M-MMAE and M-DM1 were prepared by Concortis Biosystems, Inc., and were received in conjugated form.
[0307] Reduced disulfide bridging chemistry bridges across the free thiols produced during the partial reduction process, giving one cytotoxin per disulfide reduced. In theory, an antibody of DAR=4 would have both H-L and hinge disulfides re-bridged, providing an ADC with increased stability and homogeneity over traditional conjugation approaches. Limited lysine utilization results in the conjugation of a very limited number of the estimated 70+ solvent-exposed lysines available on a typical human IgG molecule. MORAb-003 conjugates prepared using this method resulted in a DAR of 2.0, suggesting that a single lysine was utilized per H-L pair.
[0308] All ADCs were purified by HiPrep 26/10 desalting chromatography and formulated into DPBS. DAR analysis was performed on all prepared ADCs by LC-MS and aggregation levels were determined by SEC-HPLC. The results of these DAR and aggregation analyses are listed in Table 15 next to the respective ADC.
TABLE-US-00015 TABLE 15 Biophysical analyses of MORAb-003 ADCs Compound name DAR Aggregation (%) 1 PEG3-Bz-disulfidyl-dimethyl- 3.7-3.9 29 cryptophycin 2 LL2-cryptophycin 3.2 18-36 3 LL3-cryptophycin 3.2-3.7 22-36 4 VCP-cryptophycin 3.4 50 5 VCP-eribulin 3.6 0-2.6 6 ER-001161318 3.5 3.2 7 ER-001161319 3.5 3.1 8 ER-001159200 2.8 9 M-MMAE 4.0 2 10 NHS-PEG2-AuF 5.0 11 M-DM1 3.6 1.8 12 M-0285 4.0 1.2 13 M-0115 4.0 0.4 14 M-172 3.1 3.6 15 M-174 2.8 4.4 16 M-158 4.5 3.8 17 M-0384 4.2 4.2 18 M-0302 4.3 3.3 19 M-292 4.0 4.5 20 M-0026 4.2 3.3 21 M-0267 4.0 2.9 22 M-0272 3.3 1.5 23 M-0260 3.2 1 24 M-0276 4.6 6.2 25 M-015-0913 2.0 <1 26 M-030-0132 2.0 <1 27 M-0161 2.1 2.4 28 M-0157 2.0 <1 29 M-027-0381 2.0 <1 30 M-0025 2.0 1.7 31 M-0301 2.0 1.4 32 M-030-0011 2.0 <1 33 M-030-0291 2.0 <1 34 M-0255 3.6 5.9 35 M-0114 4.0 3.9
[0309] DAR values for all ADCs were in the pre-determined range (DAR between 3 and 4). Aggregate levels for the cryptophycin-based ADCs were significantly higher than desired (>10%), whereas the eribulin-based (VCP-eribulin) and the maytansine-based maleimido-linker-cytotoxins (ER-001161318, ER-001161319, and M-MMAE) all demonstrated acceptable aggregate levels. An investigation into other organic co-solvents was performed on conjugation reactions to MORAb-003 using VCP-cryptophycin. Co-solvents tested were DMSO (10%), N,N-dimethylformamide (20%), N,N-dimethylacetamide (20%), and N,N-dimethylpropionamide (20%). Aggregate levels following conjugation using these co-solvents were all equal to, or higher than, 50% propylene glycol.
[0310] A non-reducing SDS-PAGE analysis was performed on a subset of the ADCs (FIG. 5). As DAR for all these ADCs was determined to be 4, it was thought that these ADCs should migrate as intact IgG of -160 kD, as both H-L and both hinge disulfides should be re-bridged. This subset of ADCs included M-MMAE (lane 2), M-DM1 (lane 3), M-0026 (lane 4), M-0260 (lane 5), M-0267 (lane 6), M-0272 (lane 7), M-0285 (lane 8), M-292 (lane 9), M-027-0381 (lane 10), and M-0384 (lane 11) (FIG. 5). In FIG. 5, lane 1 corresponds to protein standard.
[0311] It is clear from this analysis that, for the reduced disulfide bridging chemistry ADCs (lanes 4-9, 11), there is significant H-L monovalent species (80 kD), in addition to the intact ADC. This indicates that there is significant intra-chain hinge disulfide bridging, in addition to inter-chain hinge bridging. SEC-HPLC analysis indicates that the ADCs migrate as a single intact IgG, indicating that for those ADCs with intra-chain H-H bridging, the heavy chains are associated non-covalently in the final ADC.
2.2 In Vitro Potency Analyses of MORAb-003 ADCs
2.2.1 Cytotoxicity on IGROV1 and SJSA-1 Cells
[0312] In vitro potency of prepared ADCs was assessed using a Crystal Violet assay as detailed in section 1.4.1.
[0313] Initial screening of all MORAb-003 ADCs was performed on IGROV1 (FR.sup.hi(+++)) and SJSA-1 (FR.sup.neg(-)) cells. IGROV1 cells are of human ovarian epithelial carcinoma origin and express high levels of folate receptor alpha (FR), the target antigen of MORAb-003. SJSA-1 cells are a human osteosarcoma tumor cell line that are negative for folate receptor alpha. Screening of selected ADCs was also performed in CaOV3 (human ovarian carcinoma, FR.sup.med(++)), NCI-H2110 (human non-small cell lung carcinoma, FR.sup.med(++)) and/or OVCAR3 (human ovarian carcinoma, FR.sup.med(++)) cells. The results of this screening are provided in Table 16.
TABLE-US-00016 TABLE 16 Cytotoxicity (IC.sub.50) screening of MORAb-003 ADCs on various tumor cell lines Compound name IGROV1 SJSA-1 CaOV3 NCI-H2110 OVCAR3 PEG3-Bz-disulfidyl- 0.067 0.41 dimethyl-cryptophycin LL2-cryptophycin 0.023 4.7 0.33 LL3-cryptophycin 0.086 12.7 0.19 0.094 VCP-cryptophycin 0.03 ~100 0.02 VCP-eribulin 0.054 >100 3.7 0.73 0.16 ER-001161318 0.26 >100 3.1 ER-001161319 0.49 >100 11.3 ER-001159200 6.5 >100 9.2 M-MMAE 0.2 253 NHS-PEG2-AuF 0.2 >500 M-DM1 55 132 M-0285 0.3 >100 14 8.8 M-0115 0.54 >100 M-172 >500 >500 M-174 >500 >500 M-158 >500 >500 M-0384 2.25 2.45 M-0302 330 >500 M-292 1.7 >500 M-0026 1.38 540 M-0267 0.029 0.028 M-0272 0.252 1.02 M-0260 0.383 0.036 M-0276 0.43 30 M-015-0913 >500 >500 M-030-0132 >500 17.3 M-0161 >500 >500 M-0157 >500 >500 M-027-0381 14.5 28 M-0025 >500 >500 M-0301 >500 >500 M-030-0011 61.6 >500 M-030-0291 >500 105 M-0255 0.12 0.46 M-0114 144 >100 All values are IC.sub.50s in nM, and are mean values of replicate experiments, where performed.
[0314] VCP-eribulin ADC was potent (54 pM) on IGROV1 cells and had little killing on SJSA-1 cells. For these cell lines, the VCP-eribulin ADC demonstrated higher potency and specificity relative to ADCs with equivalent DAR values, such as M-MMAE and M-DM1. VCP-eribulin ADC also demonstrated potent cytotoxicity on additional FR-expressing tumor cell lines of ovarian (CaOV3 and OVCAR3) and non-small cell lung carcinoma (NC-H2110) origin.
[0315] ADCs VCP-eribulin, LL2-cryptophycin, LL3-cryptophycin, VCP-cryptophycin, ER-001161318, ER-001161319, and ER-001159200 displayed specific cytotoxicity (>2-logs of specificity) in CaOV3 (FR.sup.med(++)) cells. A number of these ADCs displayed sub-nanomolar potency. Cryptophycin conjugates also demonstrated high levels of potency (23 pM-86 pM) in IGROV1 cells, but, with the exception of the VCP-cryptophycin, also demonstrated measurable cytotoxicity on SJSA-1 cells. Cleavable maytansine conjugates ER-001161318 and ER-001161319 had intermediate potency on IGROV1 (0.26 nM and 0.49 nM), and little off-target killing of SJSA-1 cells.
[0316] All limited lysine utilization conjugates demonstrated no specificity and were not evaluated further. Cleavable conjugates using reduced disulfide bridging technology of duostatin-3 (M-0285), duostatin-5 (M-0115), and duostatin-14 (M-292 and M-0026) all demonstrated specific cytotoxicity on the IGROV1 cell line, with little cytotoxicity on the SJSA-1 cell line. Duostatin-3 and duostatin-5 conjugates, derivatives of auristatin, were slightly higher in potency then the duostatin-14 conjugates, which is a maytansine derivative. Potencies and specificities were comparable to the control M-MMAE conjugate, which uses a Val-Cit-pAB (VCP) linker attached to monomethyl E. Non-cleavable reduced disulfide chemistry conjugates all either lacked sufficient potency or specificity, and were not analyzed further.
2.2.2 Cytotoxicity on Human Folate Receptor-Expressing Ovarian Cancer Cell Line CaOV3
[0317] Potency of select MORAb-003 ADCs was also determined on human ovarian tumor cell lines OVCAR3 and CaOV3, as well as the human NSCLC cell line NCI-H2110 (Table 16). On the human ovarian cell line CaOV3, the cryptophycin conjugates demonstrated measurably higher potency than the VCP-eribulin conjugate, unlike that observed in IGROV1 cells. This may be due to the lower expression level of folate receptor alpha on CaOV3 cells compared with IGROV1, or the higher potency of cryptophycin on these cells, compared with eribulin. The maytansine-based conjugates ER-001161318, ER-001161319, and ER-001159200 all had potencies similar to, or lower than, VCP-eribulin.
2.3 Bystander Killing of VCP-Eribulin, ER-001161318, and M-0285
[0318] In order to assess bystander killing activity, an assay was set up using two labeled cell lines. In this assay, IGROV1 cells (FR.sup.hi) labeled with Nuclight.TM. Green and HL-60 (FR.sup.neg) labeled with Nuclight.TM. Red were co-cultured in different cell number ratios, and treated with titrations of MORAb-003 ADCs VCP-eribulin, ER-001161318, or M-0285. VCP-eribulin is an eribulin-based ADC comprising a maleimido-PEG2-Val-Cit-pAB cleavable linker, while ER-001161318 is maytansine-based ADC comprising a maleimido-(CH.sub.2).sub.5-Val-Cit-pAB cleavable linker and M-0285 is a duostatin-based ADC comprising a PEG-pAB cleavable linker. Cytotoxicity was monitored by an Incucyte Zoom.RTM. cell imager. The results of this bystander cytotoxicity assay are shown in Table 17 and FIGS. 6A-C.
TABLE-US-00017 TABLE 17 Bystander killing activity of VCP-eribulin on the co-culture of FR-positive and FR-negative cell lines EC.sub.50 (nM) HL-60 HL-60 IGROV-1 HL-60 (co-culture with IGROV-1) (eribulin) 0.0005972 39.74 0.2399 0.1702
[0319] When HL-60 (FR.sup.neg) cells were cultured at a 2:1 ratio to IGROV1 (FR.sup.hi) cells, treatment with MORAb003-VCP-eribulin resulted in a 2-log increase in killing of the HL-60 cells, compared with HL-60 cells alone (Table 17 and FIG. 6A). These data suggest that folate receptor alpha (FR) target-negative cells are killed more effectively by MORAb003-VCP-eribulin when co-cultured with FR target-positive cells, referred to herein as bystander killing. Bystander killing is distinguishable from off-target killing, which is defined as the killing of target-negative cells on their own, in the absence of and independent of co-culturing with target-positive cells. The observed increase in bystander killing was also almost identical to the increase observed following treatment of HL-60 cells with free eribulin, indicating a potential mechanism for the bystander effect. Without wishing to be bound by any theory, MORAb003-VCP-eribulin may be cleaved in or near FR-positive IGROV1 cells, which also undergo apoptosis and release free eribulin into culture. The released cytotoxin may kill FR-negative HL-60 cells.
[0320] In contrast, only a slight shift was observed for MORAb003-ER-001161318 (FIG. 6B), and no shift was observed with MORAb003-0285 (FIG. 6C). When the HL-60:IGROV1 ratio was lowered from 2:1 to 1:2, measurable killing of the HL-60 cells was observed, relative to HL-60 cells alone, for MORAb003-ER-001161318, while bystander effect still remained low, albeit detectable, for MORAb003-0285. These data suggest that, in terms of bystander killing, the MORAb-003 ADCs evaluated can be ranked as VCP-eribulin>ER-001161318>M-0285.
2.4 Serum Stability Analysis
[0321] Given the long circulating half-life in vivo of ADCs and the potential for toxicity if cytotoxins are released in circulation, ADCs should demonstrate stability in serum. MORAb-003 ADCs VCP-eribulin, ER-001161319, and M-0285 were preincubated in human or mouse serum at 37.degree. C. for up to 48 hours, then evaluated in a cytotoxicity assay with IGROV1 and SJSA-1 cells. ER-001161319 is maytansine-based ADC comprising the same cleavable linker as VCP-eribulin, maleimido-PEG2-Val-Cit-pAB. PBS and serum controls were included to correct for any serum effects on assay performance. The results of this study are shown in Table 18.
TABLE-US-00018 TABLE 18 Serum stability of selected MORAb-003 ADCs Cell-based cytotoxicity assay, EC.sub.50, nM MORAb003- MORAb003- VCP Eribulin ER001161319 MORAb003-0285 Human Mouse Human Mouse Human Mouse Time PBS Serum Serum PBS Serum Serum PBS Serum Serum IGROV1 0 hr-PBS 0.021 0.013 0.02 0.28 0.15 0.2 0.074 0.089 ND 0 hr-Serum 0.022 0.014 0.01 0.15 0.15 0.2 0.063 0.078 0.049 4 hr 0.03 0.018 0.019 0.14 0.17 0.25 0.065 0.075 0.049 24 hr 0.024 0.019 ND ND 0.27 0.9 0.059 0.074 0.044 48 hr 0.022 0.021 0.03 0.21 0.73 2.56 0.043 0.05 0.051 SJSA-1 0 hr-PBS >10 >10 >10 >10 >10 >10 >10 >10 >10 0 hr-Serum >10 >10 >10 >10 >10 >10 >10 >10 >10 4 hr >10 >10 >10 >10 >10 >10 >10 >10 >10 24 hr >10 >10 >10 >10 >10 >10 >10 >10 >10 48 hr >10 >10 >10 >10 >10 >10 >10 >10 >10 Bold text with an asterisk (*) indicates significant decrease in potency from T = 0 sample.
[0322] While VCP-eribulin and M-0285 were stable for at least 48 hours in either serum, ER-001161319 demonstrated a significant drop in potency after 48 hours. This may be due to the aziridino-carbamate linkage to the maytansine, which has not been described in the literature previously. The form of the compound released may not be highly potent, as no increase in cytotoxicity was seen on SJSA-1 cells.
2.5 In Vitro Studies with MORAb003-VCP-Eribulin
2.5.1 HIC-HPLC Analysis of DAR and Product Heterogeneity
[0323] MORAb003-VCP-eribulin and MORAb003-0285 were analyzed by HIC-HPLC in order to evaluate DAR by an alternate method and examine product heterogeneity and content of unconjugated antibody (competitor). MORAb003-VCP-eribulin was shown to have DAR species of 0, 2, 4, and 6, which is consistent with the method used for reduction and conjugation (FIG. 7A). Very low amounts of DAR=0 species were observed. Overall DAR, based on AUC calculations, was 3.80, consistent with values determined by LC-MS. MORAb003-0285 migrated as a single peak by HIC-HPLC, indicating a single DAR species (FIG. 7B). This was assigned as DAR 4.0.
2.5.2 Specificity by Competition Assay
[0324] Antigen specificity of MORAb-003-VCP-eribulin cytotoxicity was demonstrated for the VCP-eribulin conjugate using a competition assay format (FIG. 8). In this experiment, titrations of the MORAb-003-VCP-eribulin (starting concentration 100 nM) were co-incubated with 2 .mu.M unconjugated MORAb-003. Unconjugated MORAb-003 provided a 2-log shift in potency on IGROV1 cells, similar to results obtained with IMGN853, the anti-human folate receptor alpha-maytansine ADC from Immunogen now in Phase II clinical trials, on KB cells (Moore et al., 2015 American Society of Clinical Oncology (ASCO) Annual Meeting, Abstract 5518).
2.5.3 Cytotoxicity on NCI-H2110 NSCLC Cells
[0325] Cytotoxicity for both MORAb003-VCP-eribulin and MORAb003-0285 on the human NSCLC cell line NCI-H2110 was performed using a Crystal Violet assay. The results of this assay are shown in Table 16. MORAb003-VCP-eribulin had an IC.sub.50 of 0.73 nM, while MORAb003-0285 had an IC.sub.50 of 14 nM.
2.6 In Vivo Studies
2.6.1 Maximum Tolerated Dose (MTD) of MORAb-003-VCP-Eribulin (MORAb-202) in CD-1 Mouse Strain
[0326] Naive CD-1 mice were injected intravenously with 200 .mu.L of MORAb-202 according to the schedule in Table 19. Body weight was measured prior to dose on the dosing day, 24 hours post dose, and three times a week thereafter. The animals were observed for clinical well-being throughout the study duration. Two weeks after dosing, the terminal body weight was measured and recorded. Euthanized mice at the end of the study (and if any mouse euthanized or found dead during the study) were processed for necropsy. Organs were examined for signs of tissue damage.
TABLE-US-00019 TABLE 19 Study design Dose Group # Mice Treatment (mg/kg) Regimen Route 1 3 Vehicle* 0 single i.v. 2 MORAb-202 10 bolus 3 20 4 40 5 80
[0327] No significant body weight loss observed in any of the treatment groups compared with PBS-treated control group, or any clinical findings indicating toxicity during the treatment. Body weight of individual mice is shown in Table 20, and the group average and SEM is shown in Table 21. Body weight change kinetics for each group (group average and SEM) are shown in FIG. 9. MORAb-202 at doses up to 80 mg/kg via bolus intravenous administration produced no toxicity. Therefore, the MTD is above 80 mg/kg.
TABLE-US-00020 TABLE 20 Days Post Rx PBS control MORAb-202 10 mg/kg MORAb-202 20 mg/kg MORAb-202 40 mg/kg MORAb-202 80 mg/kg X A:Y1 A:Y2 A:Y3 B:Y1 B:Y2 B:Y3 C:Y1 C:Y2 C:Y3 D:Y1 D:Y2 D:Y3 E:Yl E:Y2 E:Y3 0 22.50 20.60 21.20 22.60 23.50 21.80 27.40 26.20 21.00 23.80 23.30 21.70 21.80 26.50 25.80 1 23.60 21.50 22.00 22.80 23.80 23.00 28.80 27.50 21.40 23.60 23.50 21.60 23.10 27.80 27.80 2 23.30 21.90 22.00 23.30 24.00 22.90 29.30 29.00 22.10 23.60 24.60 21.80 23.30 28.30 28.30 4 23.30 21.90 22.00 23.30 23.90 23.30 30.20 31.00 23.00 24.40 24.10 22.70 23.50 29.50 28.60 7 23.90 23.10 22.70 24.80 25.20 24.90 29.60 32.00 24.00 25.10 25.50 23.40 23.30 30.40 30.10 9 24.60 22.50 22.80 24.90 25.30 25.00 30.80 31.70 23.40 25.80 25.20 23.70 23.60 31.00 30.20 11 24.20 23.40 23.20 24.30 25.10 24.30 31.00 32.40 23.80 25.30 25.00 23.80 23.10 29.10 29.40 14 24.00 23.50 24.40 25.80 25.60 25.70 33.20 32.40 24.10 25.70 25.30 26.30 21.70 28.40 28.90
TABLE-US-00021 TABLE 21 PBS MORAb-202 10 mg/kg MORAb-202 20 mg/kg MORAb-202 40 mg/kg MORAB-202 80 mg/kg days post mean mean mean mean mean injections (g) sem n (g) sem n (g) sem n mean (g) n mean (g) n 0 21.4 0.6 3 22.6 0.5 3 24.9 2.0 3 22.9 0.6 3 24.7 1.5 3 1 22.4 0.6 3 23.2 0.3 3 25.9 2.3 3 22.9 0.7 3 26.2 1.6 3 2 22.4 0.5 3 25.4 0.3 3 26.8 2.4 3 23.3 0.8 3 26.6 1.7 3 4 22.4 0.5 3 23.5 0.2 3 28.1 2.5 3 23.7 0.5 3 27.2 1.9 3 7 23.2 0.4 5 25.0 0.1 3 28.5 2.4 3 24.7 0.6 3 27.9 2.3 3 9 23.3 0.7 3 25.1 0.1 3 28.6 2.6 3 24.9 0.6 3 28.3 2.3 3 11 23.6 0.3 3 24.6 0.3 3 29.1 2.7 5 24.7 0.5 3 27.2 2.1 3 14 24.0 0.3 3 25.7 0.1 3 29.9 2.9 3 25.8 0.3 3 26.3 2.5 3
2.6.2 Maximum Tolerated Dose of Eribulin in CD-1 Mice
[0328] Naive CD-1 mice were injected intravenously with 200 .mu.L of eribulin according to the schedule in Table 22. Body weight was measured three times a week including prior to dose on each dosing day and 24 hours following each dose. The animals were observed for clinical well-being throughout the study duration (two weeks after the last dose). The terminal body weight was measured and recorded. Euthanized mice at the end of the study (and if any mouse euthanized or found dead during the study) were processed for necropsy. Organs were examined for signs of tissue damage.
TABLE-US-00022 TABLE 22 Study design Dose Group # Mice Treatment (mg/kg) Regimen Route 1 3 PBS 0 q4dx3 i.v. 2 Eribulin 0.4 3 0.8 4 1.6 5 3.2
[0329] No significant body weight loss or clinical findings indicating toxicity observed in the animals administered eribulin at doses up to 1.6 mg/kg, using q4dx3 dosing regimen (once every four days for 3 doses total). Administration of 3.2 mg/kg with the same schedule induced piloerection in all three mice after the second dose. Severe weight loss (23% loss in one mouse, #552, after the second dose; 17% and 8% in the rest, #551 and #552, after the third dose) was observed, compared with PBS-treated control. No gross changes were observed in the organs of mice during necropsy. The body weight of individual mice is shown in Table 23, and the group average and SEM is shown in Table 24. Body weight change kinetics for each group (group average and SEM) are shown in FIG. 10.
[0330] Eribulin at doses up to 1.6 mg/kg, using q4dx3 dosing regimen, produced no toxicity, while 3.2 mg/kg induced severe weight loss. Therefore, the MTD of eribulin, in this study, is 1.6 mg/kg, q4dx3.
TABLE-US-00023 TABLE 23 Day PBS eribulin 0.4 mg/kg eribulin 0.8 mg/kg eribulin 1.6 mg/kg eribulin 3.2 mg/kg X A:Y1 A:Y2 A:Y3 B:Y1 B:Y2 B:Y3 C:Yl C:Y2 C:Y3 D:Y1 D:Y2 D:Y3 E:Y1 E:Y2 E:Y3 0 26.50 24.40 23.40 22.40 24.60 23.10 23.90 25.50 25.10 21.90 24.40 23.70 24.30 25.00 22.00 1 28.13 25.11 24.36 21.84 24.02 22.43 23.03 23.60 24.46 21.13 24.10 24.15 22.31 23.47 20.57 4 25.60 25.10 24.40 22.70 24.00 24.00 23.80 24.40 24.50 21.70 24.10 23.30 20.90 22.20 17.50 5 26.40 24.80 24.50 22.30 24.10 23.40 24.10 24.00 24.80 21.50 24.50 24.10 20.90 22.70 16.90 8 27.50 25.70 24.50 23.30 23.70 25.20 24.00 24.80 25.90 21.90 24.50 24.20 21.80 23.90 9003.00* 9 27.50 25.50 24.40 22.90 23.40 25.40 24.00 24.70 25.30 21.80 24.80 24.30 20.20 23.00 11 27.50 25.60 24.40 22.60 23.90 25.60 24.70 25.10 26.00 22.20 25.70 24.70 20.00 23.30 13 27.40 26.80 25.40 23.70 23.90 26.50 25.30 25.60 26.60 22.80 25.80 25.20 21.40 23.70 16 27.30 26.80 26.20 24.70 24.10 26.70 25.40 25.80 26.40 23.40 26.70 26.10 23.60 24.30 18 27.00 27.40 27.30 24.30 24.90 27.90 26.20 28.00 27.90 24.70 27.90 26.20 25.90 25.90 20 28.10 27.60 26.50 26.10 25.70 28.50 25.40 28.20 28.10 25.40 29.00 26.00 26.80 26.00 Each column represents an individual animal *9003: euthanized for weight loss >20%.
TABLE-US-00024 TABLE 24 PBS eribulin 0.4 mg/kg eribulin 0.8 mg/kg eribulin 1.6 mg/kg eribulin 3.2 mg/kg days post mean mean mean mean mean injections (g) sem n (g) sem n (g) sem n mean (g) n mean (g) n 0 24.8 0.9 3 23.4 0.6 3 24.8 0.5 3 23.3 0.7 3 23.8 0.9 3 1 25.9 1.2 3 22.8 0.7 3 23.7 0.4 3 23.1 1.0 3 22.1 0.8 3 4 25.0 0.3 3 23.6 0.4 3 24.2 0.2 3 23.0 0.7 3 20.2 1.4 3 5 25.2 0.6 3 23.3 0.5 3 24.3 0.3 3 23.4 0.9 3 20.2 1.7 3 8 25.9 0.9 3 24.1 0.6 3 24.9 0.6 3 23.5 0.8 3 22.9 0.9 2 9 25.8 0.9 3 23.9 0.8 3 24.7 0.4 3 23.6 0.9 3 21.6 1.1 2 11 25.8 0.9 3 24.0 0.9 3 25.3 0.4 3 24.2 1.0 3 21.7 1.3 2 13 26.5 0.6 3 24.7 0.9 3 25.8 0.4 3 24.6 0.9 3 22.6 0.9 2 16 26.8 0.3 3 25.2 0.8 3 25.9 0.3 3 25.4 1.0 3 24.0 0.3 2 18 27.2 0.1 3 25.7 1.1 3 27.4 0.6 3 26.3 0.9 3 25.9 0.0 2 20 27.4 0.5 3 26.8 0.9 3 27.2 0.9 3 26.8 1.1 3 26.4 0.3 2
2.6.3 Evaluation of Minimum Efficacious Dose of MORAb003-VCP-Eribulin (MORAb-202) in the hNSCLC NCI-H2110 Model in CB17-SCID Mice
[0331] Human NSCLC, NCI-H2110 cells, passage 47 were implanted subcutaneously in 30 CB17 SCID mice (female, 5 to 6 weeks old, weighing 20 grams). After 14 days post-implantation, mice were randomized into five groups. Average tumor volume in each group on the treatment day (Day 0) ranged between 154-175 mm.sup.3 (Table 27). The enrolled mice were treated with MORAb003-VCP-eribulin (MORAb-202) (Lot # NB2900-87E 10/07/15) at 1, 2.5, or 5 mg/kg, with MORAb-003-0285 (Lot #042-150-002) as control at 5 mg/kg, or with PBS, according to the study design (Table 25). Each group was removed from the study when tumor volume in any animal in the group was >2000 mm.sup.3. The last group was terminated on Day 61.
TABLE-US-00025 TABLE 25 Study design Dose Group # Mice Treatment (mg/kg) Regimen Route 1 5 PBS 0 single i.v. 2 5 MORAb-003-VCP- 1 bolus eribulin 3 5 MORAb-003-VCP- 2.5 eribulin 4 .sup. 4.sup.1 MORAb-003-VCP- 5 eribulin 5 5 MORAb003-0285 5 .sup.1Four mice in this group. One mouse was excluded from this group due to treatment injection error, which was verified by absence of compound in animal sera based on electrochemiluminenscent immunoassay (ECLIA) data.
[0332] The tumor volumes in individual mice are shown in Table 26, and the group average and SEM is shown in Table 27. Tumor growth kinetics for each group (group average and standard error of the mean, SEM) are shown in FIG. 11, and tumor volumes in individual mice, as well as group average and SEM, are shown in FIG. 12. Based on day 17 tumor volumes (when first tumor volume >2000 mm.sup.3 was observed), MORAb-202 caused tumor growth inhibition (TGI) of 47% at 1 mg/kg (p=0.002 vs. saline), TGI of 96% at 2.5 mg/kg (p<0.0001 vs. saline). However, the regressed tumors regrew one to two weeks after end of treatment. No tumor was detected in mice treated with 5 mg/kg of MORAb-202. These mice remained tumor free beyond 60 days after a single dose treatment. MORAb-003-0285 caused TGI of 89.7% at 5 mg/kg (p<0.0001 vs. saline).
[0333] Body weight of individual mice is shown in Table 28, and the group average and SEM is shown in Table 29. Body weight change kinetics for each group (group average and SEM) are shown in FIG. 13.
[0334] No significant body weight loss was observed in any of the treatment groups compared with control.
[0335] MORAb-202 showed significant effect on NCI-H2110 tumor growth. Tumor regression was achieved by a bolus treatment at 2.5 mg/kg with TGI of 94% (vs. PBS). Therefore, the minimum efficacious dose of MORAb-202 is 2.5 mg/kg, tested in this model. Complete tumor eradication was achieved by a single dose at 5 mg/kg. No tumor growth was observed for over 60 days.
TABLE-US-00026 TABLE 26 Tumor volumes days post ran- dom- MORab-202 ization PBS MORAb-202 1 mg/kg MORAb-202 2.5 mg/kg 5 mg/kg 003-0285 5 mg/kg 0 164 195 137 300 80 178 218 133 118 150 187 189 92 120 236 110 202 159 146 65 208 241 243 97 3 368 413 279 587 171 178 207 104 106 216 144 97 65 69 148 40 115 68 68 83 259 358 292 164 5 327 481 285 555 190 161 193 83 95 215 75 51 37 35 56 14 52 22 37 54 160 168 239 105 6 467 758 541 894 275 257 258 139 160 348 61 52 58 33 57 7 28 25 20 43 197 235 247 129 7 642 815 621 1055 395 317 306 182 167 476 64 54 53 36 57 8 48 16 20 52 192 255 266 128 10 891 1238 895 1328 662 506 494 230 285 708 24 37 35 15 71 0 0 0 0 39 155 240 181 86 12 993 1274 983 1519 1115 638 655 371 361 865 40 21 51 9 69 0 0 0 0 32 106 206 223 83 14 981 1410 1131 1695 971 848 812 402 418 901 41 30 37 0 89 0 0 0 0 31 115 235 157 79 17 1320 1723 1319 2089 1466 955 980 727 592 946 46 33 64 0 161 0 0 0 0 28 114 346 251 74 19 838 1030 856 602 953 56 37 90 0 282 0 0 0 0 27 144 438 359 94 24 102 37 197 0 702 0 0 0 0 46 391 1244 824 187 26 168 102 319 0 790 0 0 0 0 103 564 1470 1030 287 28 269 54 474 9 990 0 0 0 0 125 703 1898 1112 375 31 362 105 558 13 1187 0 0 0 0 225 1144 2427 1413 657 33 496 124 588 9 1461 0 0 0 0 35 573 212 669 16 1847 0 0 0 0 38 764 348 952 20 2367 0 0 0 0 40 0 0 0 0 42 0 0 0 0 45 0 0 0 0 47 0 0 0 0 52 0 0 0 0 54 0 0 0 0 59 0 0 0 0 61 0 0 0 0 Each column represents an individual animal.
TABLE-US-00027 TABLE 27 MORAb-202 MORAb-202 days post PBS 1 mg/kg 2.5 mg/kg randomization MEAN SEM N MEAN SEM N MEAN SEM N 0 175.2 36.41527 5 159.4 17.68781 5 164.8 25.8917 5 3 363.6 69.3831 5 162.2 24.14101 5 104.6 17.7581 5 5 367.6 66.21275 5 149.4 26.13343 5 50.8 7.242607 5 6 587 108.7468 5 232.4 37.74183 5 52.2 5.005179 5 7 705.6 109.7441 5 289.6 55.74694 5 52.8 4.611415 5 10 1002.8 122.532 5 444.6 85.61518 5 36.4 9.499597 5 12 1176.8 100.25 5 578 95.18355 5 38 10.62087 5 14 1237.6 138.8994 5 676.2 109.4307 5 39.4 14.30871 5 17 1583.4 146.0629 5 840 76.78507 5 60.8 27.09899 5 19 855.8 72.16584 5 93 49.35207 5 24 207.6 127.8177 5 26 275.8 138.3498 5 28 359.2 177.874 5 31 445 208.4929 5 33 535.6 255.2269 5 35 663.4 318.1881 5 38 890.2 402.5237 5 40 42 45 47 52 54 59 61 MORAb-202 MORAb-003-0285 days post 5 mg/kg 5 mg/kg randomization MEAN SEM N MEAN SEM N 0 154.25 16.95792 4 170.8 37.46065 5 3 72.75 13.88661 4 231.2 48.4055 5 5 31.25 7.500133 4 145.2 31.14683 5 6 20 4.140008 4 170.2 37.81015 5 7 23 7.76666 4 178.6 40.08123 5 10 0 0 4 140.2 35.30937 5 12 0 0 4 130 36.5513 5 14 0 0 4 123.4 34.69758 5 17 0 0 4 162.6 58.96373 5 19 0 0 4 212.4 79.06236 5 24 0 0 4 538.4 219.5123 5 26 0 0 4 690.8 249.2466 5 28 0 0 4 842.6 310.8641 5 31 0 0 4 1173.2 373.2365 5 33 0 0 4 35 0 0 4 38 0 0 4 40 0 0 4 42 0 0 4 45 0 0 4 47 0 0 4 52 0 0 4 54 0 0 4 59 0 0 4 61 0 0 4
TABLE-US-00028 TABLE 28 days post MORAb-202 MORAb-202 randomization PBS 1 mg/kg 2.5 mg/kg 0 19.1 18.2 18.4 18.9 18.8 19.1 18.6 19.3 20.6 18.4 17.8 18.1 18 19.8 16.3 3 19.6 18.2 18.9 18.9 19.3 19.3 18.4 20.2 20.9 18.6 18.4 19.1 18.6 19.9 16.4 6 19.7 18.4 18.4 19.1 19.1 19 18.3 20.3 21.3 19 18.5 19.4 18.7 20 16.4 7 19.7 18 18.9 18.8 18.9 18.9 18 20 21.2 18.9 18.7 18.7 18.7 19.7 16.5 10 19.7 18 19.2 18.5 19.1 18.4 18 20.1 20.9 19 19.3 19.7 18.8 19.8 16.6 12 19.8 17.7 19.1 18.4 19 18.3 17.8 20.5 20.9 19.5 18.9 20 19.7 20.2 17.2 14 18.8 17.4 18.4 18.2 17.5 17.9 17.7 20.3 21.2 19.9 18.8 19.6 19 19.3 17 17 18.8 17.2 18.3 17.5 17.2 17.4 17.7 20.4 20.7 19.2 18.8 19.8 19.7 19.2 17.3 19 16.7 17.2 19.9 20.7 18.9 18.3 19.8 18.7 19.5 16.8 24 18.8 20.2 19.2 19.9 16.9 26 18.9 19.6 18.9 19.5 16.5 28 18.8 19.6 19.5 19.6 16.6 31 18.9 20.1 19.6 20.7 17 33 18.9 19.8 19.4 21.2 17.6 35 19.2 19.7 19.5 20.7 17.4 38 19.6 20 19.7 20.6 18 40 42 45 47 52 54 59 61 days post MORab-202 MORAb-003-0285 randomization 5 mg/kg 5 mg/kg 0 17.6 18.7 16.1 19.7 20.5 17.4 18 17.4 18.8 3 17.5 18.8 15.9 19.9 20.8 17 18.1 16.3 18.5 6 17.5 19.3 16.3 19.6 20.8 17.7 18.2 16.8 18.7 7 17.4 19.4 16.5 19.2 20.6 17.7 18.5 16.8 19 10 17.6 19.4 16.7 20 20.5 18.2 18.6 17.5 20.3 12 17.9 19.6 16.9 20.2 20.4 18.3 18.8 18.1 20.4 14 17.5 19.3 17 19.2 20 18.2 18.9 18.4 19.7 17 17.9 20 17.3 19.7 19.8 17.9 18.9 18.6 19.5 19 18.1 20 17.1 20.2 19.7 18 19.3 18.4 19.6 24 18.5 20.7 17.5 20.2 20.1 18.5 20 19.1 18.4 26 18.3 20.7 17.6 19.7 20.6 18.4 19.9 18.6 19.1 28 18.6 21.3 17.6 20.1 20.8 18.5 19.9 18.8 19.5 31 18.6 20.4 17.9 20.9 20.7 18.2 20.9 19.6 19.5 33 18.8 19.6 18.2 21.3 35 18.7 20.2 18.1 19.6 38 18.8 20.4 19 17.2 40 19.4 20.4 18.7 19.3 42 19.9 20.4 18.8 20.6 45 19.8 21 18.3 21.7 47 19.7 20.7 18.3 21.1 52 20.2 21.1 18.5 21.9 54 20.3 21.4 18.6 22.6 59 20 21.6 18.8 21.7 61 20.5 22.1 19.2 21.3
TABLE-US-00029 TABLE 29 MORAb-202 MORAb-202 days post PBS 1 mg/kg 2.5 mg/kg randomization MEAN SEM N MEAN SEM N MEAN SEM N 0 18.68 0.165239 5 19.2 0.385328 5 18 0.554902 5 3 18.98 0.234959 5 19.48 0.47393 5 18.48 0.579842 5 6 18.94 0.245739 5 19.58 0.537015 5 18.6 0.609665 5 7 18.86 0.268971 5 19.4 0.549488 5 18.46 0.525953 5 10 18.9 0.29444 5 19.28 0.537015 5 18.84 0.585996 5 12 18.8 0.352933 5 19.4 0.600608 5 19.2 0.545849 5 14 18.06 0.267112 5 19.4 0.685817 5 18.74 0.454832 5 17 17.8 0.320373 5 19.08 0.673649 5 18.96 0.451533 5 19 18.68 0.764423 5 18.62 0.527655 5 24 19 0.579498 5 26 18.68 0.563279 5 23 18.82 0.573795 5 31 19.26 0.636533 5 33 19.38 0.585826 5 35 19.3 0.536644 5 38 19.58 0.430983 5 40 42 45 47 52 54 59 61 MORAb-202 MORAb-003-durostatin days post 5 mg/kg 5 mg/kg randomization MEAN SEM N MEAN SEM N 0 18.025 0.689078 4 18.42 0.578982 5 3 18.025 0.769253 4 18.14 0.76975 5 6 18.175 0.694839 4 18.44 0.667108 5 7 18.125 0.628577 4 18.52 0.638721 5 10 18.425 0.68618 4 19.02 0.590063 5 12 18.65 0.678513 4 19.2 0.502108 5 14 18.25 0.523801 4 19.04 0.352368 5 17 18.725 0.592675 4 18.94 0.33497 5 19 18.85 0.670634 4 19 0.338521 5 24 19.225 0.663539 4 19.32 0.313137 5 26 19.075 0.621135 4 19.32 0.41086 5 23 19.4 0.728103 4 19.5 0.407939 5 31 19.45 0.638148 4 19.78 0.484329 5 33 19.475 0.60047 4 35 19.15 0.416401 4 38 18.85 0.584918 4 40 19.45 0.314619 4 42 19.925 0.359691 4 45 20.2 0.665164 4 47 19.96 0.549137 4 52 20.425 0.651414 4 54 20.725 0.758819 4 59 20.525 0.620064 4 61 20.775 0.552051 4
2.6.4 Evaluation of Minimum Efficacious Dose of Eribulin in the hNSCLC NCI-H2110 Model in CB17-SCID Mice
[0336] Human NSCLC, H2110 cells, passage 46 were implanted subcutaneously in 30 CB17 SCID mice (female, 5 to 6 weeks old, weighing 20 grams). After 11 days post-implantation, mice were randomized into five groups. The five animals with the tumor volumes deviating the most from the average were excluded. Average tumor volume in each group on the treatment day (Day 0) ranged between 87.6-89.4 mm.sup.3 (Table 32). The enrolled mice were treated with eribulin (Lot # N1201193) at 0.05, 0.2, 0.8, or 1.6 mg/kg, or with PBS, according to the study design (Table 30). Each group was terminated, respectively, when tumor volume >2000 mm.sup.3 was first observed within the group. The study was terminated on Day 38 (30 days after the last dose).
TABLE-US-00030 TABLE 30 Study design Dose Group # Mice Treatment (mg/kg) Regimen Route 1 5 PBS 0 q4dx3 i.v. 2 Eribulin 0.05 3 0.2 4 4* 0.8 5 5 1.6
[0337] The tumor volumes in individual mice are shown in Table 31, and the group average and SEM is shown in Table 32. Tumor growth kinetics for each group (group average and SEM) are shown in FIG. 14, and tumor volumes in individual mice, as well as group average and SEM on Day 24 (when tumor volume >2000 mm.sup.3 were observed in PBS treated mice), are shown in FIG. 15. Eribulin caused TGI of 50.5% (with no tumor regression observed) at 0.05 mg/kg (p=0.0026 vs. saline); TGI of 99% at 0.2, 0.8, or 1.6 mg/kg (p values were <0.0001 for all 3 groups when compared to saline). The minimum efficacious dose that induced tumor regression is 0.2 mg/kg. However, majority of the regressed tumors in these mice (3/5 in 0.2 mg/kg group, 4/5 in 0.8 mg/kg group, and 2/5 in 1.6 mg/kg group) re-grew or remained measurable throughout the study duration (30 days after the last dose).
[0338] Body weight of individual mice is shown in Table 33, and the group average and SEM is shown in Table 34. Body weight change kinetics for each group (group average and SEM) are shown in FIG. 16.
[0339] No significant body weight loss in any of the treatment groups compared with saline-treated control group were observed. No clinical findings indicating toxicity during the treatment were observed.
[0340] Eribulin, at 0.2 mg/kg and higher, administered q4dX3 i.v., showed significant effect on H2110 tumor growth. Tumor regression was achieved. When a lower dose was administered (at 0.05 mg/kg), no tumor regression was achieved. Therefore, the minimum efficacious dose tested in this study is 0.2 mg/kg.
TABLE-US-00031 TABLE 31 days post 1st dose vehicle eribulin 0.05 mg/kg eribulin 0.2 mg/kg eribulin 0.8 mg/kg eribulin 1.6 mg/kg 0 59 91 118 88 91 105 101 94 61 77 103 68 78 130 62 111 104 81 93 54 70 116 74 91 91 3 62 179 219 236 173 175 149 117 161 64 68 62 52 79 61 54 55 40 51 33 50 44 44 44 47 5 80 255 436 283 257 231 157 228 261 132 60 60 48 90 51 32 29 34 42 25 25 22 32 24 24 7 111 433 440 472 446 357 171 269 247 102 48 47 39 49 47 26 34 17 21 24 19 12 31 15 11 10 230 555 747 622 489 370 200 413 376 226 33 39 28 36 47 17 22 19 5 21 15 0 29 14 0 12 263 677 722 877 620 539 265 448 350 135 19 41 23 13 14 15 23 16 12 24 13 0 14 0 0 17 720 959 960 1158 885 725 514 751 620 531 0 0 0 0 13 17 38 0 0 26 0 0 0 0 0 19 862 1314 940 1097 941 869 437 908 776 837 27 39 29 29 16 0 20 18 0 27 0 0 19 0 0 24 1886 2308 1854 2760 1671 712 718 1489 1225 1040 0 15 19 23 11 14 0 20 0 33 18 0 19 0 0 26 0 24 0 11 14 0 8 14 14 15 8 0 17 0 0 28 0 7 0 14 83 0 16 20 14 17 0 0 16 0 0 31 0 16 0 10 31 0 10 15 26 29 11 0 17 0 0 33 0 27 0 13 22 0 13 8 18 44 8 0 28 0 0 35 0 19 0 16 42 0 13 0 22 50 14 0 17 0 0 38 0 19 0 14 45 0 11 13 13 54 11 0 20 0 0
TABLE-US-00032 TABLE 32 eribulin eribulin eribulin eribulin days post PBS 0.05 mg/kg 0.2 mg/kg 0.8 mg/kg 1.6 mg/kg 1st dose MEAN SEM N MEAN SEM N MEAN SEM N MEAN SEM N MEAN SEM N 0 89.4 9.34 5 87.6 8.18 5 88.2 12.56 5 88.6 10.02 5 88.4 8.11 5 3 173.8 30.31 5 133.2 19.74 5 64.4 4.45 5 46.6 4.31 5 45.8 1.20 5 5 262.2 56.43 5 201.8 24.37 5 61.8 7.43 5 32.4 2.83 5 25.4 1.72 5 7 380.4 67.55 5 229.2 43.40 5 46 1.79 5 24.4 2.83 5 17.6 3.62 5 10 528.6 85.83 5 317 43.21 5 36.6 3.17 5 16.8 3.07 5 11.6 5.42 5 12 631.8 101.42 5 347.4 70.14 5 22 5.07 5 18 2.34 5 5.4 3.30 5 17 936.4 70.46 5 628.2 48.40 5 2.6 2.60 5 16.2 7.39 5 0 0.00 5 10 1030.8 80.29 5 765.4 84.75 5 28 3.65 5 13 5.50 5 3.8 3.79 5 24 2095.8 195.76 5 1036.8 149.24 5 13.6 3.94 5 13.4 6.26 5 7.4 4.53 5 26 9.8 4.54 5 10.2 2.83 5 5 3.37 5 28 20.8 15.74 5 13.4 3.48 5 3.2 3.19 5 31 11.4 5.77 5 16 5.29 5 5.6 3.55 5 33 12.4 5.53 5 16.6 7.45 5 7.2 5.42 5 35 15.4 7.72 5 17 9.22 5 6.2 3.82 5 38 15.6 8.25 5 18.2 9.25 5 6.2 4.05 5
TABLE-US-00033 TABLE 33 days post 1st dose vehicle eribulin 0.05 mg/kg eribulin 0.2 mg/kg 0 18.5 16.7 19.1 20.4 19.6 19.1 16.4 18.6 20.1 17.9 18.2 18.5 16.7 19.8 18.9 3 18.8 16.6 19.6 20.9 20.0 19.4 17.1 18.5 20.4 18.7 18.6 18.5 16.9 19.6 19.8 5 18.8 16.8 19.3 21.2 20.0 19.4 16.5 18.4 20.4 19.4 18.5 19.1 16.9 20.2 20.1 7 18.6 16.5 19.3 21.2 19.8 19.4 16.3 18.7 20.3 19.1 18.5 18.5 17.1 19.6 20.7 8 18.3 16.7 18.8 21.1 19.6 19.3 16.3 18.4 20.4 19.4 18.6 19.1 17.0 19.6 19.9 9 18.4 16.4 18.5 21.0 19.5 18.8 16.1 18.6 19.9 19.1 18.3 20.0 16.8 19.5 19.5 10 19.0 17.2 19.2 21.6 20.3 19.7 16.3 19.4 20.5 20.0 18.7 19.4 17.1 20.0 20.1 12 19.0 15.9 18.5 21.3 19.2 18.8 15.9 18.6 19.6 19.7 18.3 19.3 16.9 19.7 19.9 17 18.8 15.5 17.8 20.4 18.3 17.5 16.0 18.5 18.0 19.7 19.0 19.3 17.4 20.5 20.0 19 18.9 15.6 17.2 20.6 18.1 17.8 16.3 18.4 18.0 19.4 19.1 19.0 17.1 21.1 21.0 24 18.2 15.8 17.4 20.3 18.2 18.8 16.5 18.8 17.4 18.0 19.3 20.2 18.8 21.7 22.0 26 19.8 20.9 18.6 22.0 20.4 28 20.1 20.6 18.1 21.5 21.0 31 18.7 20.1 18.7 22.0 21.2 33 20.0 20.3 18.4 22.2 21.8 35 19.5 20.1 18.7 22.1 21.4 38 19.8 20.8 18.3 21.6 21.6 days post 1st dose eribulin 0.8 mg/kg eribulin 1.6 mg/kg 0 18.6 17.8 18.4 17.8 18.1 18.2 18.3 19.4 16.3 19.0 3 18.8 18.0 18.4 18.1 19.0 17.8 18.3 20.1 15.8 19.6 5 18.9 18.1 18.5 17.6 18.2 18.1 18.1 19.5 16.0 19.3 7 19.1 18.1 19.0 17.8 19.0 18.1 18.2 19.5 16.3 19.4 8 19.2 18.0 19.0 17.7 18.8 18.0 18.3 20.1 16.2 19.4 9 19.2 17.7 18.5 17.5 18.5 17.4 18.0 19.6 15.8 19.4 10 19.0 17.7 18.9 17.8 19.1 18.1 18.5 21.0 16.1 20.0 12 18.9 17.8 19.0 17.8 19.0 18.0 18.8 20.5 16.3 20.1 17 19.2 18.0 18.8 18.1 19.1 19.0 19.2 21.0 16.4 19.4 19 19.1 17.7 19.4 18.7 19.1 19.1 19.5 21.1 16.3 19.8 24 20.1 18.5 20.3 19.1 20.3 19.4 20.6 21.7 18.1 20.7 26 20.3 18.1 19.9 19.3 20.9 19.5 20.7 21.6 18.3 20.6 28 20.3 17.8 20.2 19.6 20.6 19.6 20.3 21.2 17.6 21.1 31 20.1 18.2 20.3 19.5 20.7 19.8 20.4 21.9 18.1 21.9 33 20.2 18.3 21.0 19.2 20.2 20.1 19.9 21.7 17.9 20.7 35 20.1 17.8 21.0 19.3 20.3 20.3 18.2 21.8 18.2 20.9 38 20.4 18.1 21.4 19.4 21.2 20.0 21.0 21.9 18.4 20.3 Each column represents an individual animal.
TABLE-US-00034 TABLE 34 eribulin eribulin eribulin eribulin days post PBS 0.05 mg/kg 0.2 mg/kg 0.8 mg/kg 1.6 mg/kg 1st dose MEAN SEM N MEAN SEM N MEAN SEM N MEAN SEM N MEAN SEM N 0 18.9 0.62 5 18.4 0.62 5 18.4 0.51 5 18.1 0.16 5 18.2 0.53 5 3 19.2 0.73 5 18.8 0.54 5 18.7 0.51 5 18.5 0.19 5 18.3 0.75 5 5 19.2 0.73 5 18.8 0.66 5 19.0 0.60 5 18.3 0.22 5 18.2 0.62 5 7 19.1 0.77 5 18.8 0.67 5 18.9 0.60 5 18.6 0.27 5 18.3 0.58 5 10 18.9 0.72 5 18.8 0.69 5 18.8 0.51 5 18.5 0.29 5 18.4 0.67 5 12 18.8 0.76 5 18.5 0.65 5 18.8 0.59 5 18.3 0.30 5 18.0 0.69 5 17 19.5 0.73 5 19.2 0.74 5 19.1 0.55 5 18.5 0.32 5 18.7 0.84 5 19 18.8 0.86 5 18.5 0.68 5 18.8 0.56 5 18.5 0.28 5 18.7 0.77 5 24 18.2 0.79 5 17.9 0.60 5 19.3 0.53 5 18.6 0.24 5 19.0 0.73 5 26 19.5 0.74 5 18.8 0.30 5 19.2 0.79 5 28 20.4 0.63 5 19.7 0.36 5 20.1 0.62 5 31 20.3 0.56 5 19.7 0.48 5 20.1 0.57 5 33 20.3 0.59 5 19.7 0.50 5 20.0 0.66 5 35 20.1 0.66 5 19.8 0.43 5 20.4 0.71 5 38 20.5 0.68 5 19.8 0.47 5 20.1 0.62 5
Example 2
1. Materials and Methods
[0341] MORAb003-VCP-eribulin (MORAb-202) was synthesized by conjugating MORAb-003 (humanized anti-human folate receptor alpha) to the MAL-PEG2-Val-Cit-PAB-eribulin (ER-001159569) compound described in section 1.1 of Example 3. The conjugation method is described in section 1.4.1 of Example 4.
1.1 Tumor Models
[0342] Human tumor cell lines used in the additional in vitro evaluation of MORAb-202 include IGROV1 (human ovarian carcinoma, FR.sup.hi(+++), OVCAR3 (human ovarian carcinoma, FR.sup.med(++)), NCI-H2110 (human non-small cell lung carcinoma, FR.sup.med(++)), A431-A3 (A431 parental cell line stabily transfected with human mesothelin, FR.sup.lo(+/-)), SJSA-1 (human osteosarcoma, FR.sup.neg(-)), and HL-60 (human leukemia, FR.sup.neg(-)). All of these cell lines were obtained directly from the American Type Culture Collection (ATCC). For in vivo studies, non-small cell lung cancer, triple negative breast cancer, and endometrial cancer patient-derived xenograft mouse models were established and maintained at Oncotest GmbH (Freiburg, Germany), Oncodesign (Dijon, France), and EPO Berlin-Buch GmbH (Berlin, Germany), respectively.
1.2 In Vitro Cytotoxicity Analyses
1.2.1 Crystal Violet Assay
[0343] IGROV1 (FR.sup.hi(+++)), A431-A3 (FR.sup.lo(+/-)), and SJSA-1 (FR.sup.neg(-)) cells were sub-cultured and seeded at 10,000 cells/well in complete growth medium in 96-well tissue culture plates, incubated at 37.degree. C., 5% CO.sub.2 overnight (16 hours). Typically, test reagents were serially-diluted 1:4 in 2 mL deep-well dilution plates, starting at 1 .mu.M (10 dilutions total). 100 .mu.L of diluted samples were added to the cell plates (starting concentration of test samples at 100 nM). Plates were incubated at 37.degree. C., 5% CO.sub.2 for an additional 48 hours. Medium was discarded, plates were washed once with 200 .mu.L DPBS, stained with 50 .mu.L of 0.2% Crystal Violet solution at room temperature for 15 min, and then washed extensively with tap water. Plates were air-dried, and Crystal Violet was dissolved with 200 .mu.L of 1% SDS solution. Plates were read at 570 nm. Data was analyzed using GraphPad Prism 6. For OVCAR3 (FR.sup.med(++)) and NCI-H2110 (FR.sup.med(++)) cells were seeded at 3,000 cells/well and incubated for 5 days with MORAb-202.
1.3 In Vivo Studies
1.3.1 NCI-H2110 Xenograft Model
[0344] Animal Preparation:
[0345] CB17 SCID mice (female, 6 weeks old) were housed at 5 mice per ventilated cage. Sterilized food pellets and water bottle were available, ad lib, to the animals. Animals were acclimated for 5-7 days prior to tumor implantation.
[0346] Cell Culture:
[0347] Human NCI-H2110 cells were thawed from frozen stock (NB2813-65) and cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) in 5% CO.sub.2 at 37.degree. C. After two passages, upon reaching confluence at approximately 70%, the cells were harvested by using cell dissociation solution, washed twice with serum-free medium, and counted.
[0348] Tumor Implantation:
[0349] The cell suspension in serum-free medium was mixed with ice-cold matrigel at 1:1 (v:v) to a final concentration of 1.0.times.10.sup.8 cells/mL. Each mouse was injected subcutaneously with 100 .mu.L of the mixture at 1.0.times.10.sup.7 cells/mouse. A 27 G needle was used for all injections. Mice were monitored for clinical well-being and tumors were measured by digital caliper three times weekly, beginning on Day 3 post-implantation. Tumor volume (mm.sup.3) was calculated using the formula: W (mm).times.L (mm).times.D (mm).times..pi./6. When the tumors reached .about.100 mm.sup.3 (in an average of >70 to .about.130 mm.sup.3), the animals were randomized to 4-5 per group. The 5 animals with the tumor volumes deviating greatest from the average were excluded.
[0350] Study Design:
[0351] The enrolled experimental mice were injected intravenously with 200 .mu.L of vehicle or MORAb-202 at 1.0, 2.5, and 5 mg/kg, according to the study design (Table 35), on the day of randomization. Body weight was measured prior to dose, and two times per week during the study. At the end of the study, terminal body weight was measured and recorded. Animals were euthanized when the individual tumor volume exceeded 2000 mm.sup.3. Early termination criteria prior to reaching the maximum allowed tumor volume included: (1) tumor ulceration greater than 50% of the tumor (v:v); (2) paralysis; (3) body weight loss >20%; and (4) 50% of the animals within the group had met termination. Any mouse euthanized or found dead during the study was processed following the terminal procedure described above.
TABLE-US-00035 TABLE 35 Study design Dose Group # Mice Treatment (mg/kg) Regimen Route 1 5 Vehicle 0 single bolus i.v. 2 MORAb-202 1 3 2.5 4 5
1.3.2 Patient-Derived Xenograft (PDx) Models
1.3.2.1 Non-Small Cell Lung Cancer (NSCLC) PDx Model: LXFA-737 (Oncotest)
[0352] Tumor Implantation:
[0353] NSCLC tumor fragments were obtained from LXFA-737 tumor xenografts serially passaged in nude mice. After removal from donor mice, tumors were cut into fragments (3-4 mm edge length) and placed in phosphate-buffered saline (PBS) containing 10% penicillin/streptomycin. Recipient animals were anesthetized by inhalation of isoflurane and received unilateral or bilateral tumor implants subcutaneously in the flank. Tumor xenografts were implanted with one or two tumors per mouse at a take rate <65%. In the case of a bilateral take, one of these tumors was explanted prior to randomization. Animals and tumor implants were monitored daily until solid tumor growth was detectable in a sufficient number of animals. At randomization, the volume of growing tumors was determined. Animals fulfilling the randomization criteria (i.e. bearing tumors of 50-250 mm.sup.3, preferably 80-200 mm.sup.3) were distributed into experimental groups consisting of 5-6 animals per group, aiming at comparable median and mean group tumor volumes of approximately 100-120 mm.sup.3. Animals not used for experiments were euthanized. The day of randomization was designated as Day 0 of the experiment.
[0354] Study Design:
[0355] The enrolled experimental mice were injected intravenously with vehicle, MORAb-003 at 5 mg/kg, or MORAb-202 at 5 mg/kg, according to the study design (Table 36), on the day of randomization. Body weight was measured prior to dose on each dosing day, and two times per week during the study. At the end of the study, the terminal body weight was measured and recorded. Animals were euthanized when the individual tumor volume exceeded 2000 mm.sup.3.
TABLE-US-00036 TABLE 36 Study design Dose Group # Mice Treatment (mg/kg) Regimen Route 1 6 Vehicle 0 single bolus i.v. 2 6 MORAb-003 5 3 6 MORAb-202 5
1.3.2.2 Triple Negative Breast Cancer (TNBC) PDx Model: OD-BRE-0631 (Oncodesign)
[0356] Tumor Implantation:
[0357] Nine female SWISS nude mice were injected subcutaneously into the right flank with patient-derived TNBC tumor fragments. Tumor-bearing mice were euthanized when tumor volume reached 500-1000 mm.sup.3, and tumors were surgically excised. Tumor fragments (30-50 mg) were orthotopically implanted into the mammary fat pad region of 34 female SWISS nude mice 24 to 72 hours after a whole-body irradiation with a gamma-source (2 Gy, 60Co, BioMEP, France). When the tumors reached a mean volume of 200-300 mm.sup.3, 24 of the 34 total animals were randomized into two groups (n=12 animals) according to their individual tumor volume using Vivo Manager.RTM. software (Biosystemes, Couternon, France). A statistical test (analysis of variance) was performed to evaluate homogeneity between groups. The day of randomization was designated as Day 0 of the experiment.
[0358] Study Design:
[0359] On Day 1 (one day after randomization and two days prior to treatment), 3 mice from each of the two untreated groups were terminated. The remaining experimental mice were injected intravenously with vehicle or MORAb-202 at 5 mg/kg, according to the study design (Table 37), on Day 3. On Day 8 (five days after treatment), 3 mice from each of the two treated groups were terminated. Immediately following termination, tumor tissue was collected and fixed in 4% neutral buffered formalin for 24 to 48 hours, and then embedded in paraffin (Histosec.RTM., Merck, Darmstadt, Germany). The paraffin embedded sample was stored at room temperature for subsequent immunohistochemistry analysis.
TABLE-US-00037 TABLE 37 Study design Dose Group # Mice Treatment (mg/kg) Regimen Route 1 3 n/a n/a n/a n/a 9 Vehicle 0 single bolus i.v. 2 3 n/a n/a n/a n/a 9 MORAb-202 5 single bolus i.v.
[0360] Immunohistochemistry (IHC) Analysis:
[0361] IHC staining of formalin-fixed, paraffin-embedded tumor tissues were performed in order to evaluate both MORAb-202 occupation and cancer associated fibroblast expression. Prior to staining, slides were dewaxed and antigen was retrieved in a Lab Vision.TM. PT Module (Thermo Scientific), in citrate buffer (pH 6.0) pre-warmed to 85.degree. C., using the following program: warm to 97.degree. C.; incubate at 97.degree. C. for 30 min; and cool to 60.degree. C. Slides were then transferred to double distilled water at room temperature for 5 min. Staining was performed in a Lab Vision.TM. Autostainer 360 (Thermo Scientific). Briefly, slides were washed twice in 1.times. Tris-buffered saline/Tween-20 (TBST) for 6 min/wash. Tissue sections were then incubated in blocking buffer (300 .mu.L) (10% goat serum (Jackson Immunoresearch Laboratory Inc., Cat No. 005-000-121) diluted in 3% bovine serum albumin (BSA)-phosphate buffered saline (PBS)) for 1 hour, incubated in conjugated antibody (200 .mu.L) (Table 38) for 1 hour, and washed five times in 1.times.TBST for 6 min/wash. Slides were counterstained with DAPI in mounting media, and coverslipped slides were allowed to harden for 30 min. Slides were processed on a Panoramic MIDI scanner (3DHISTECH), and IHC images were analyzed using Halo software (Indica Labs). The antibodies used in this analysis targeted .alpha.-smooth muscle actin (SMA), which is a specific marker for cancer associated fibroblasts, and human IgG, which can detect the presence of MORAb-202.
TABLE-US-00038 TABLE 38 IHC antibodies Cat. Stock Working Antibody Conjugated Vendor No. Lot Solution Solution .alpha.-smooth FITC Sigma F3777 124M4775V 2.0 mg/mL 5.0 .mu.g/mL muscle actin (SMA)-FITC mouse IgGl, AF488 Biolegend 400129 B128493 0.2 mg/mL 1:1000 .kappa. isotype control goat anti- AF555 Mol. Probes A21433 1709318 n/a 1:200 human IgG
1.3.2.3 Endometrial Cancer PDx Models: Endo-12961 and Endo-10590 (EPO Berlin)
[0362] Tumor Implantation:
[0363] Endometrial cancer tumor fragments were obtained from serially passaged Endo-12961 and Endo-10590 tumor xenografts, and stored as stock in fluid nitrogen. Tumor fragments were implanted subcutaneously into the left flank of 40 NMRI nu/nu female mice, and tumor volume was monitored. Mice with a tumor volume of 100-160 mm.sup.3 were randomized into one of four groups (Groups A-D, Table 39). Satellite mice for randomization were included in a fifth group (Group E, Table 39). Each group consisted of 8 animals. The day of randomization was designated as Day 0 of the experiment.
[0364] Study Design:
[0365] The enrolled experimental mice were injected intravenously with PBS, eribulin at 3.2 mg/kg or 0.1 mg/kg, or MORAb-202 at 5 mg/kg, according to the study design (Table 39), on the day of randomization. Tumor growth was evaluated by the measurement of two perpendicular diameters twice weekly, and tumor volume (TV), relative tumor volume (RTV) and treated/control (T/C) values were calculated. Body weight was also evaluated twice weekly as a parameter for toxicity, with the calculation of the body weight per group and body weight changes (BWC) relative to the start of treatment. Animals were sacrificed when the individual tumor volume exceeded 1 cm.sup.3, or at the end of the study.
TABLE-US-00039 TABLE 39 Study design Dose Group # Mice Treatment (mg/kg) Regimen Route A 8 PBS 0 single bolus i.v. B Eribulin 3.2 C Eribulin 0.1 D MORAb-202 5 E n/a n/a n/a n/a
1.4 Mechanism of Action
[0366] 1.4.1 Three-Dimensional (3D) Co-Culture System in zPredicta
[0367] All mesenchymal stem cell (MSC)-containing 3D co-culture experiments were conducted in zPredicta, using organ-specific 3D extracellular matrix systems such as rStomach.TM.. Bone marrow mesenchymal stem cells (BM-MSCs) in rStomach.TM. were co-cultured with the Nuc Red Light MKN-74 gastric cancer cell line in quadruplicate in 48-well format for 12 days. MKN-74 cells had been previously shown to express enough folate receptor alpha (FR) for MORAb-202 treatment to induce cellular apoptosis. Prior to culture, BM-MSCs were evaluated for target antigen expression and for markers of MSC differentiation (Table 40) by flow cytometry.
TABLE-US-00040 TABLE 40 Markers of MSC differentiation Cell population Markers Mesenchymal stem cells (MSCs) Stro-1.sup.+/CD105.sup.+ Pre-adipocytes CD34.sup.+/CD31.sup.- Adipocytes Oil red Cancer associated fibroblasts Alpha-smooth muscle actin (aSMA), (CAFs) vimentin Pre-pericytes/pericytes NG2.sup.+, CD13.sup.+, CD146.sup.+ All FRA
[0368] rStomach.TM. cultures were treated with either MORAb-202, unconjugated MORAb-003 antibody, eribulin, or control, as described in Table 41. Controls included untreated and vehicle-treated (PBS and DMSO) cultures. MSC differentiation was monitored by light microscopy. Once visible differentiation was observed, samples were harvested for staining and flow cytometry analysis.
TABLE-US-00041 TABLE 41 Co-culture treatments Agent Working Concentration(s) MORAb-202 10 nM MORAb-003 (unconjugated antibody) 10 nM Eribulin 1.7 nM and 0.2 nM PBS DMSO 0.1% Untreated control
1.4.2 Time Course Analysis of Effect of MORAb-202 on Cancer Associated Fibroblasts
[0369] Subcutaneous H2110 xenograft tumor-bearing mice were prepared as described in section 1.3.1. Tumor samples were harvested at Days 0, 3, 5, 7, and 9 following administration of vehicle, or MORAb-202 at 5 mg/kg. Collected tumor samples were processed on slides, and the expression of cancer associated fibroblasts was analyzed by IHC as described in section 1.3.2.2.
2. Results
2.1 In Vitro Cytoxicity Analyses
2.1.1 Cytotoxicity of MORAb-202
[0370] In vitro potency of MORAb-202 was evaluated using a Crystal Violet assay, as detailed in section 1.2.1. Screening was performed on IGROV1 (FR.sup.hi(+++)), OVCAR3 (FR.sup.med(++)), NCI-H2110 (FR.sup.med(++)), A431-A3 (FR.sup.lo(+/-)), and SJSA-1 (FR.sup.neg(-)) cells. The results of this screening are provided in FIG. 17 and Table 42.
TABLE-US-00042 TABLE 42 Cytotoxicity (EC.sub.50) screening of MORAb-202 on various tumor cell lines EC.sub.50 (nM) IGROV I OVCAR3 NCI-H2110 A431-A3 SJSA-1 (FR+++) (FR++) (FR++) (FR+/-) (FR-) 0.01 0.16 0.74 23 >100
[0371] MORAb-202 exhibited folate receptor alpha expression-dependent cytotoxicity against tumor cell lines, and low levels of off-target killing. MORAb-202 demonstrated the highest level of potency (0.01 nM) on IGROV1 cells, with little cytotoxicity (>100 nM) on folate receptor alpha-negative SJSA-1 cells. Intermediate potency was observed in OVCAR3 and NCI-H2110 cells (0.16 nM and 0.74 nM).
2.2 In Vivo Studies
2.2.1 Efficacy of MORAb-202 in the NC1-H2110 Xenograft Model
[0372] Subcutaneous H2110 tumor-bearing mice were were injected intravenously with vehicle or MORAb-202 at 1, 2.5, and 5 mg/kg. Significant tumor regression was observed following a single dose of MORAb-202 at 5 mg/kg (FIG. 18 and Table 43). Using this xenograft model with high folate receptor alpha expression and single dose administrations, the therapeutic window for MORAb-202 was shown to be 1 mg/kg for tumor growth delay (with stable disease) and .gtoreq.2.5 mg/kg for tumor regression. In this study, MORAb-202 at a dose of 2.5 mg/kg resulted in a partial response, and MORAb-202 at a dose of 5 mg/kg resulted in a complete response.
TABLE-US-00043 TABLE 43 Anti-tumor activity of MORAb-202 in the NC1-H2110 xenograft model Tumor Volume, mm.sup.3 (Tumor Growth Inhibition, %) Day 17 Day 31 Vehicle (n = 5) 1583.4 .+-. 146.1 (100) n/a MORAb-202, 1 mg/kg, 840.0 .+-. 76.8 (53.1) n/a single dose (n = 5) MORAb-202, 2.5 mg/kg, 60.8 .+-. 27.1 (3.8) 1173.2 .+-. 373.2 single dose (n = 5) MORAb-202, 5 mg/kg, 0.0 (0.0) 0 (0.0) single dose (n = 4)
2.2.2 Efficacy of MORAb-202 in the NSCLC PDx Model: LXFA-737
[0373] Subcutaneous NSCLC PDx tumor-bearing mice were injected intravenously with vehicle, MORAb-003 at 5 mg/kg, or MORAb-202 at 5 mg/kg. A single dose of MORAb-202 (5 mg/kg) resulted in significant tumor regression in this model, in contrast to a single dose of unconjugated MORAb-003 antibody (5 mg/kg), which did not demonstrate significant anti-tumor activity (FIG. 19A). Five of the six total mice treated with MORAb-202 were considered to be tumor-free at Day 32 of the study (Table 44), and four remained tumor-free through Day 74 (termination of the study). In addition, no significant body weight loss was observed in the treatment group as compared to the vehicle-treated control group, indicating no toxicity during treatment (FIG. 19B).
TABLE-US-00044 TABLE 44 Anti-tumor activity of MORAb-202 in the NSCLC PDx model Tumor Volume, mm.sup.3 (Tumor Growth Inhibition, %) Day 21 Day 32 Day 74 Vehicle (n = 6) 1004.5 (100) 1561.3 (100) n/a MORAb-003, 860.7 (85.7) 1572.1 (100.7) n/a 5 mg/kg, single dose (n = 6) MORAb-202, 22.9 (2.3) 4.7 (0.3) 418.3 5 mg/kg, single (4/6 tumor-free) dose (n = 6)
2.2.3 Relative Efficacy of MORAb-202 and Eribulin in Endometrial Cancer PDx Models: Endo-12961 and Endo-10590
[0374] Endo-12961 and Endo-10590 xenografts express high levels of folate receptor alpha. Subcutaneous endometrial cancer PDx tumor-bearing mice were injected intravenously with PBS, eribulin at 3.2 mg/kg or 0.1 mg/kg, or MORAb-202 at 5 mg/kg. The maximum tolerated dose (MTD) of eribulin in this model is 3.2 mg/kg, whereas 0.1 mg/kg is equivalent to the dosage of eribulin provided by MORAb-202 administered at 5 mg/kg. Throughout the beginning of the study, significant anti-tumor activity was observed following treatment with MORAb-202 (5 mg/kg) and the MTD dose of eribulin (3.2 mg/kg) in both animal models, while no significant anti-tumor activity was observed following treatment with eribulin at 0.1 mg/kg (FIGS. 20A and 20C). However, regressed tumors in mice treated with eribulin at 3.2 mg/kg began to re-grow during the study duration, whereas no significant tumor re-growth was noted in mice treated with MORAb-202. In this study, MORAb-202 was found to be significantly more efficacious than eribulin. Eribulin treatment also temporarily affected body weight in the first week post-treatment (FIGS. 20B and 20D). In contrast, no body weight loss was observed in animals treated with MORAb-202.
2.3 Mechanism of Action of MORAb-202
2.3.1 IHC and Efficacy of MORAb-202 in the TNBC PDx Model: OD-BRE-0631
[0375] Subcutaneous TNBC PDx tumor-bearing mice were injected intravenously with vehicle or MORAb-202 at 5 mg/kg. Tumor tissue was collected from mice in each group prior to treatment (Day 1) and after treatment (Day 8). IHC analyses of the collected tumor tissues revealed that MORAb-202 occupies folate receptor alpha-expressing tumor cells five days post-treatment (Day 8), following administration on Day 3 as a single dose (5 mg/kg). Cell occupation was evaluated using an anti-human IgG antibody (FIG. 21A). MORAb-202 treatment was also shown to diminish the structure of cancer associated fibroblasts, as shown by IHC staining with an anti-.alpha.-smooth muscle actin (SMA)-FITC antibody (FIG. 21B). In terms of efficacy, MORAb-202 treatment resulted in maximum tumor regression at 11 days post-treatment, measured by a relative tumor volume (RTV) of 0.62 (FIG. 21C).
2.3.2 Effect of MORAb-202, MORAb-003, and Eribulin on 3D Co-Culture System
[0376] Bone marrow mesenchymal stem cells (BM-MSCs) in rStomach.TM. (zPredicta) were co-cultured with the MKN-74 gastric cancer cell line for 12 days. Prior to culture, BM-MSCs were evaluated for folate receptor alpha expression and for markers of MSC differentiation by flow cytometry. rStomach.TM. cultures were then treated with either MORAb-202, unconjugated MORAb-003 antibody, eribulin, or control. Once visible MSC differentiation was observed by light microscopy, samples were harvested for staining and flow cytometry analysis. The results of these analyses are shown in FIG. 22.
[0377] A total treatment duration of 7 days, with treatment replenishment during this period, was sufficient to produce a measureable effect on the differentiation of human BM-MSCs in culture with MKN-74 cells. Relative to vehicle control, treatment with MORAb-202 (10 nM) resulted in an increase in MSC and adipocyte populations, and a decrease in pericyte populations (Table 45). These data indicate that MORAb-202 may have a significant effect on the tumor microenvironment.
TABLE-US-00045 TABLE 45 Effect of MORAb-202, MORAb-003, and eribulin on 3D co-culture system Percentage of live cells Treatment MSCs Adipocytes Pericytes PBS 32.3% 0.72% 14.6% MORAb-202 43.7% 22.6% 11.4% MORAb-003 37.1% 0.69% 24.0% Eribulin 29.9% 2.68% 25.8%
2.3.3 Time Course Analysis of Effect of MORAb-202 on Cancer Associated Fibroblasts
[0378] Tumor samples were harvested from subcutaneous H2110 xenograft tumor-bearing mice at Days 0, 3, 5, 7, and 9 following administration of vehicle, or MORAb-202 at 5 mg/kg. Collected tumor samples were processed on slides, and cancer associated fibroblast (CAF) expression was analyzed by IHC. The CAF network structure, as evaluated and quantified by staining with an anti-.alpha.-smooth muscle actin (SMA)-FITC antibody, was prominent on Day 3 and Day 5, following administration of a single dose of MORAb-202 at 5 mg/kg (FIG. 23). However, by Day 7, the majority of this structure was significantly diminished.
Example 3
1. Materials and Methods
[0379] Conjugatable eribulin compounds having the structures shown in Table 46 were synthesized according to the following procedures, and used in the preparation of ADCs (Example 4).
[0380] All solvents used in the synthesis reactions were anhydrous grade (EMD Millipore). All solvents used for workup or purification were high performance liquid chromatography (HPLC) grade (EMD Millipore). Unless indicated otherwise, all chemicals were commercially available. Column chromatography was performed using a Biotage.RTM. SP4. Solvent removal was performed using either a rotary evaporator (BUchi Labortechik AG), or a centrifugal evaporator (Genevac, SP scientific). Preparative liquid chromatography-mass spectrometry (LC/MS) was conducted using a Waters AutoPurification System and an XTerra MS C18 column (5 .mu.m, 19 mm.times.100 mm) under acidic mobile phase conditions. Nuclear magnetic resonance (NMR) spectra were taken using deuterated chloroform (CDCl.sub.3) unless otherwise stated, and were recorded at 400 or 500 MHz using a Varian instrument (Agilent Technologies). Mass spectra were taken using a Waters Acquity Ultra Performance LC/MS. As used herein, the term "inerted" refers to replacement of the air in a reactor (e.g., a reaction vessel, a flask, a glass reactor) with an essentially moisture-free, inert gas, such as nitrogen or argon. Multiplicities are indicated using the following abbreviations: s=singlet, d=doublet, t=triplet, q=quartet, quint=quintet, sxt=sextet, m=multiplet, dd=doublet of doublets, ddd=doublet of doublets of doublets, dt=doublet of triplets, br s=a broad singlet.
TABLE-US-00046 TABLE 46 Conjugatable eribulin compounds ##STR00016## ##STR00017## ##STR00018## ##STR00019## ##STR00020## ##STR00021## ##STR00022## ##STR00023## ##STR00024## ##STR00025## ##STR00026## ##STR00027## ##STR00028## ##STR00029## ##STR00030## ##STR00031## ##STR00032## ##STR00033## ##STR00034## ##STR00035## ##STR00036##
1.1 Preparation of MAL-PEG2-Val-Cit-PAB-Eribulin (ER-001159569)
##STR00037##
[0382] Eribulin (ER-000086526) (61.5 mg, 0.074 mmol) was dissolved in N,N-dimethylformamide (DMF) (6.0 mL) and then mixed with Hunig Base (0.027 mL, 0.156 mmol) and Fmoc-Val-Cit-PAB-PNP (86 mg, 0.112 mmol). The reaction was stirred at room temperature for 18 hours until the coupling was complete, as determined by high performance liquid chromatography (HPLC) analysis. Diethylamine (0.078 mL, 0.745 mmol) was added to the mixture, and the mixture was stirred for an additional 2 hours until the reaction was complete. The solvent was removed by evaporation, and the residue was purified by flash chromatography to obtain Val-Cit-PAB-eribulin (ER-001228950) as a white solid (60 mg, 71% yield). .sup.1HNMR (400 MHz, CD.sub.3OD) .delta. ppm 7.56 (d, J=8.4 Hz, 2H), 7.32 (d, J=8.4 Hz, 2H), 5.14 (s, 1H), 5.06 (d, J=12.4 Hz, 1H), 5.03 (s, 1H), 5.01 (d, J=12.4 Hz, 1H), 4.87 (s, 1H), 4.83 (s, 1H), 4.71 (t, J=4.4 Hz, 1H), 4.62 (t, J=4.4 Hz, 1H), 4.57 (dd, J=4.8, 8.8 Hz, 1H), 4.47 (d, J=10.8 Hz, 1H), 4.32-4.27 (m, 2H), 4.18 (dd, J=4.8, 6.4 Hz, 1H), 4.13-4.07 (m, 2H), 3.98 (t, J=10.4 Hz, 1H), 3.88-3.82 (m, 3H), 3.76-3.70 (m, 4H), 3.60 (d, J=6.0 Hz, 1H), 3.38 (s, 3H), 3.26-3.10 (m, 3H), 2.93 (dd, J=2.0, 11.2 Hz, 1H), 2.91-2.84 (m, 1H), 2.75-2.64 (m, 2H), 2.44-2.29 (m, 5H), 2.21-1.97 (m, 8H), 1.93-1.83 (m, 3H), 1.79-1.72 (m, 5H), 1.68-1.29 (m, 8H), 1.11 (d, J=6.8 Hz, 3H), 1.07-1.01 (m, 1H), 1.06 (d, J=7.2 Hz, 3H), 1.02 (d, J=7.2 Hz, 3H). LCMS (M+H)=1135.7.
[0383] Val-Cit-PAB-eribulin (ER-001228950) (16 mg, 14 .mu.mol) was dissolved in DMF (1 mL). N,N-diisopropylethylamine (7.2 .mu.L, 41 .mu.mol) and Mal-PEG2-NHS (9.7 mg, 27 .mu.mol) were then added to this solution at room temperature, and the reaction mixture was stirred at room temperature for 1 hour. Upon completion of the reaction, the crude mixture was purified by reverse-phase HPLC using an acetonitrile-water mobile phase containing 0.1% formic acid. The collected fractions were concentrated under vacuum at room temperature in a non-heated water bath to yield Mal-PEG2-Val-Cit-PAB-eribulin (ER-001159569) (7.1 mg, 5.2 .mu.mol, 38% yield). .sup.1HNMR (400 MHz, CD.sub.3OD) .delta. ppm 7.59 (d, J=8.4 Hz, 2H), 7.31 (d, J=8.4 Hz, 2H), 6.81 (s, 2H), 5.13 (s, 1H), 5.06 (d, J=12.4 Hz, 1H), 5.02 (s, 1H), 5.01 (d, J=12.4 Hz, 1H), 4.87 (s, 1H), 4.82 (s, 1H), 4.71 (t, J=4.0 Hz, 1H), 4.61 (t, J=4.4 Hz, 1H), 4.50 (dd, J=5.2, 9.2 Hz, 1H), 4.47 (d, J=10.8 Hz, 1H), 4.32-4.27 (m, 2H), 4.19 (dd, J=6.8, 11.6 Hz, 1H), 4.13-4.07 (m, 2H), 3.98 (t, J=10.4 Hz, 1H), 3.88-3.82 (m, 3H), 3.76-3.64 (m, 6H), 3.62-3.51 (m, 6H), 3.38 (s, 3H), 3.22-3.08 (m, 4H), 2.93 (dd, J=2.4, 9.6 Hz, 1H), 2.92-2.84 (m, 1H), 2.76-2.63 (m, 2H), 2.52 (t, J=6.0 Hz, 2H), 2.44-2.29 (m, 5H), 2.21-1.97 (m, 8H), 1.93-1.83 (m, 3H), 1.80-1.66 (m, 5H), 1.66-1.28 (m, 10H), 1.11 (d, J=6.4 Hz, 3H), 1.07-1.01 (m, 1H), 0.99 (d, J=6.8 Hz, 3H), 0.97 (d, J=6.4 Hz, 3H). LCMS (M+H)=1374.9.
1.2 Preparation of NHS-PEG2-Val-Cit-PAB-Eribulin (ER-001236940)
##STR00038##
[0385] Val-Cit-PAB-eribulin (ER-001228950) (45 mg, 0.04 mmol) and bis(2,5-dioxopyrrolidin-1-yl) 3,3'-(ethane-1,2-diylbis(oxy))dipropanoate (79 mg, 0.198 mmol) were mixed in DMF (1.5 mL), and Et.sub.3N (44.2 .mu.l, 0.317 mmol) was then added. The mixture was stirred for 18 hours until the reaction was complete, as determined by HPLC analysis. The solvent was evaporated and the residue was purified by flash chromatography to obtain NHS-PEG2-Val-Cit-PAB-eribulin (ER-001236940) as a white solid (38 mg, 68% yield). .sup.1HNMR (400 MHz, CD.sub.3OD) .delta. ppm 7.58 (d, J=8.4 Hz, 2H), 7.33 (d, J=8.4 Hz, 2H), 5.14 (s, 1H), 5.05 (d, J=12.4 Hz, 1H), 5.03 (s, 1H), 5.01 (d, J=12.4 Hz, 1H), 4.87 (s, 1H), 4.83 (s, 1H), 4.71 (t, J=4.4 Hz, 1H), 4.62 (t, J=4.4 Hz, 1H), 4.51 (dd, J=4.8, 8.8 Hz, 1H), 4.50-4.47 (m, 1H), 4.32-4.27 (m, 2H), 4.21 (dd, J=4.8, 6.4 Hz, 1H), 4.14-4.08 (m, 2H), 3.99 (t, J=10.4 Hz, 1H), 3.88-3.82 (m, 3H), 3.78-3.70 (m, 4H), 3.62 (s, 2H), 3.62-3.58 (m, 1H), 3.50-3.46 (m, 2H), 3.39 (s, 4H), 3.36 (s, 3H), 3.22-3.08 (m, 3H), 2.93 (dd, J=2.0, 11.2 Hz, 1H), 2.91-2.87 (m, 1H), 2.84 (s, 2H), 2.80 (s, 2H), 2.75-2.64 (m, 2H), 2.59-2.52 (m, 2H), 2.44-2.29 (m, 5H), 2.21-1.97 (m, 10H), 1.93-1.83 (m, 3H), 1.79-1.72 (m, 5H), 1.68-1.29 (m, 8H), 1.11 (d, J=6.8 Hz, 3H), 1.08-0.98 (m, 1H), 1.00 (d, J=7.2 Hz, 3H), 0.98 (d, J=7.2 Hz, 3H). LCMS (M+H)=1421.0.
1.3 Preparation of NHS--(CH.sub.2).sub.5--Val-Cit-PAB-Eribulin (ER-001236941)
##STR00039##
[0386] Heptanedioic acid (1.6 g, 9.99 mmol) was dissolved in tetrahydrofuran (THF) (100 mL), and 1-hydroxypyrrolidine-2,5-dione (2.299 g, 19.98 mmol) was then added, followed by the addition of DCC (4.12 g, 19.98 mmol). The mixture was stirred at room temperature for 18 hours until HPLC analysis indicated the completion of the reaction. The solid was removed by filtration through a celite pad, and washed with THF (3.times.2 mL). The combined filtrate was concentrated and purified by flash chromatography to yield bis(2,5-dioxopyrrolidin-1-yl) heptanedioate (ER-001236140) as a white solid (2.5 g, 71% yield). .sup.1HNMR (400 MHz) .delta. ppm 2.83 (s, 8H), 2.64 (t, J=7.6 Hz, 4H), 1.80 (dt, J=7.6 Hz, 4H), 1.59-1.51 (m, 2H). LCMS (M+H)=355.2.
[0387] NHS--(CH.sub.2).sub.5-Val-Cit-PAB-eribulin (ER-001236941) was prepared (8.5 mg, 47% yield) from VCP-eribulin (ER-001228950) and bis(2,5-dioxopyrrolidin-1-yl) heptanedioate (ER-001236140) using the same procedure as described above for the preparation of NHS-PEG2-Val-Cit-PAB-eribulin (ER-001236940). .sup.1HNMR (400 MHz, CD.sub.3OD) .delta. ppm 7.56 (d, J=8.4 Hz, 2H), 7.30 (d, J=8.4 Hz, 2H), 5.13 (s, 1H), 5.04 (d, J=12.0 Hz, 1H), 5.01 (s, 1H), 5.00 (d, J=12.4 Hz, 1H), 4.86 (s, 1H), 4.82 (s, 1H), 4.70 (t, J=4.4 Hz, 1H), 4.60 (t, J=4.4 Hz, 1H), 4.50 (dd, J=4.8, 8.8 Hz, 1H), 4.46 (d, J=10.8 Hz, 1H), 4.36-4.25 (m, 2H), 4.17 (dd, J=4.8, 6.4 Hz, 1H), 4.13-4.06 (m, 2H), 3.97 (t, J=10.4 Hz, 1H), 3.87-3.80 (m, 3H), 3.74-3.68 (m, 2H), 3.37 (s, 3H), 3.20-3.06 (m, 4H), 2.94 (dd, J=2.0, 11.2 Hz, 1H), 2.90-2.82 (m, 1H), 2.82 (s, 4H), 2.74-2.65 (m, 2H), 2.61 (t, J=8.0 Hz, 2H), 2.46-2.26 (m, 7H), 2.24-1.81 (m, 13H), 1.78-1.28 (m, 19H), 1.10 (d, J=6.8 Hz, 3H), 1.06-0.96 (m, 1H), 0.97 (d, J=7.2 Hz, 3H), 0.95 (d, J=7.2 Hz, 3H). LCMS (M+H)=1375.1.
1.4 Preparation of Mal-(CH.sub.2).sub.5-Val-Cit-PAB-Eribulin (ER-001235638)
##STR00040##
[0388] Eribulin (ER-000086526) (10 mg, 0.012 mmol) was dissolved in DMF (1 mL), and mixed with MC-Val-Cit-PAB-PNP (9.02 mg, 0.012 mmol) and Hunig's Base (4.44 .mu.L, 0.025 mmol). The mixture was then stirred at room temperature for 12 hours until HPLC analysis indicated the completion of the reaction. The reaction mixture was concentrated and purified by flash chromatography to yield Mal-(CH.sub.2).sub.5-Val-Cit-PAB-eribulin (ER-001235638) as a white solid (11.3 mg, 63% yield). .sup.1HNMR (400 MHz, CD.sub.3OD) .delta. ppm 7.57 (d, J=8.4 Hz, 2H), 7.31 (d, J=8.4 Hz, 2H), 6.79 (s, 2H), 5.13 (s, 1H), 5.05 (d, J=12.4 Hz, 1H), 5.02 (s, 1H), 5.00 (d, J=12.4 Hz, 1H), 4.87 (s, 1H), 4.83 (s, 1H), 4.71 (t, J=4.4 Hz, 1H), 4.61 (t, J=4.4 Hz, 1H), 4.56-4.46 (m, 3H), 4.35-4.27 (m, 2H), 4.20-4.07 (m, 4H), 3.98 (t, J=10.8 Hz, 1H), 3.87-3.83 (m, 3H), 3.73-3.70 (m, 2H), 3.48 (t, J=7.6 Hz, 2H), 3.38 (s, 3H), 3.20-3.08 (m, 4H), 2.93 (dd, J=1.6, 9.6 Hz, 1H), 2.89-2.85 (m, 1H), 2.69 (dt, J=11.2, 16.8 Hz, 2H), 2.44-2.33 (m, 5H), 2.27-1.83 (m, 13H), 1.78-1.68 (m, 5H), 1.66-1.27 (m, 14H), 1.11 (d, J=7.2 Hz, 3H), 1.07-0.98 (m, 1H), 0.98 (d, J=7.2 Hz, 3H), 0.96 (d, J=7.2 Hz, 3H). LCMS (M+H)=1328.9.
1.5 Preparation of Mal-PEG8-Val-Cit-PAB-Eribulin (ER-001242287)
##STR00041##
[0390] VCP-eribulin (ER-001228950) (10 mg, 8.808 .mu.mol) and 2,5-dioxopyrrolidin-1-yl 1-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)-3-oxo-7,10,13,16,19,22,25,28-oct- aoxa-4-azahentriacontan-31-oate (6.07 mg, 8.808 .mu.mol) were mixed in DMF (1 mL), followed by the addition of Et.sub.3N (9.82 .mu.l, 0.07 mmol). The reaction mixture was stirred at room temperature for 18 hours until HPLC analysis indicated the completion of the reaction. The solvent was removed by evaporation, and the residue was purified by flash chromatography to yield Mal-PEG8-Val-Cit-PAB-eribulin (ER-001242287) as a white solid (3.0 mg, 20% yield). .sup.1HNMR (400 MHz, CD.sub.3OD) .delta. ppm 7.58 (d, J=8.4 Hz, 2H), 7.29 (d, J=8.4 Hz, 2H), 6.80 (s, 2H), 5.12 (s, 1H), 5.04 (d, J=12.4 Hz, 1H), 5.01 (s, 1H), 4.99 (d, J=12.4 Hz, 1H), 4.85 (s, 1H), 4.80 (s, 1H), 4.69 (t, J=4.4 Hz, 1H), 4.59 (t, J=4.4 Hz, 1H), 4.50-4.42 (m, 2H), 4.32-4.24 (m, 2H), 4.20-4.14 (m, 2H), 4.12-4.04 (m, 3H), 3.96 (t, J=10.4 Hz, 1H), 3.86-3.80 (m, 3H), 3.76-3.57 (m, 4H), 3.48 (t, J=6.0 Hz, 1H), 3.36 (s, 3H), 3.20-3.08 (m, 3H), 2.91 (dd, J=2.0, 11.2 Hz, 1H), 2.90-2.82 (m, 1H), 2.74-2.60 (m, 2H), 2.44-2.29 (m, 5H), 2.21-1.97 (m, 10H), 1.93-1.83 (m, 3H), 1.79-1.20 (m, 19H), 1.09 (d, J=6.8 Hz, 3H), 1.04-0.98 (m, 1H), 0.99 (d, J=7.2 Hz, 3H), 0.97 (d, J=7.2 Hz, 3H). LCMS (M+H)=1711.6.
1.6 Preparation of NHS-PEG9-Val-Cit-PAB-Eribulin (ER-001242288)
##STR00042##
[0392] NHS-PEG9-Val-Cit-PAB-eribulin (ER-001242288) was prepared (13 mg, 85% yield) from VCP-eribulin (ER-001228950) and BisNHS-PEG9 using the same procedure as described above for the preparation of NHS-PEG2-Val-Cit-PAB-eribulin (ER-001236940). .sup.1HNMR (400 MHz, CD.sub.3OD) .delta. ppm 7.61 (d, J=8.4 Hz, 2H), 7.32 (d, J=8.4 Hz, 2H), 5.16 (s, 1H), 5.06 (d, J=12.4 Hz, 1H), 5.01 (s, 1H), 5.00 (d, J=12.4 Hz, 1H), 4.87 (s, 1H), 4.82 (s, 1H), 4.71 (t, J=4.4 Hz, 1H), 4.61 (t, J=4.4 Hz, 1H), 4.52-4.45 (m, 2H), 4.34-4.26 (m, 2H), 4.20-4.19 (m, 1H), 4.14-4.06 (m, 2H), 3.98 (t, J=10.4 Hz, 1H), 3.88-3.80 (m, 3H), 3.76-3.70 (m, 4H), 3.66-3.58 (m, 37H), 3.38 (s, 3H), 3.24-3.10 (m, 3H), 2.93 (dd, J=2.0, 11.2 Hz, 1H), 2.91-2.84 (m, 1H), 2.84 (s, 4H), 2.76-2.64 (m, 2H), 2.58-2.50 (m, 4H), 2.46-2.28 (m, 5H), 2.22-1.96 (m, 8H), 1.91-1.82 (m, 3H), 1.79-1.68 (m, 5H), 1.64-1.24 (m, 8H), 1.11 (d, J=6.8 Hz, 3H), 1.08-0.96 (m, 1H), 0.99 (d, J=7.2 Hz, 3H), 0.97 (d, J=7.2 Hz, 3H). LCMS (M+H)=1729.7.
1.7 Preparation of NHS-PEG3-Triazole-PEG3-Val-Cit-PAB-Eribulin (ER-001243700)
##STR00043## ##STR00044##
[0394] VCP-eribulin (ER-001228950) (25 mg, 0.022 mmol) was dissolved in DMF (2.5 mL), and then mixed with Et.sub.3N (24.55 .mu.l, 0.176 mmol) and Azide-PEG3-NHS (8.34 mg, 0.024 mmol). The mixture was stirred at room temperature for 18 hours until HPLC analysis indicated the completion of the reaction. The mixture was concentrated under vacuum, and the residue was purified by prep-HPLC (MeCN and water with 0.1% formic acid). The fractions containing azide-PEG3-Val-Cit-PAB-eribulin were extracted with dichloromethane (CH.sub.2Cl.sub.2) (3.times.20 mL), and the CH.sub.2Cl.sub.2 was evaporated to obtain azide-PEG3-Val-Cit-PAB-eribulin (ER-001243116) as a white solid (18.9 mg, 63% yield). .sup.1HNMR (400 MHz, CD.sub.3OD) .delta. ppm 7.58 (d, J=8.4 Hz, 2H), 7.30 (d, J=8.4 Hz, 2H), 5.14 (s, 1H), 5.04 (d, J=12.4 Hz, 1H), 5.03 (s, 1H), 5.01 (d, J=12.4 Hz, 1H), 4.85 (s, 1H), 4.81 (s, 1H), 4.70 (t, J=4.4 Hz, 1H), 4.61 (t, J=4.4 Hz, 1H), 4.52-4.48 (m, 2H), 4.31-4.25 (m, 2H), 4.20-4.15 (m, 1H), 4.13-4.07 (m, 2H), 3.99 (t, J=10.4 Hz, 1H), 3.84-3.79 (m, 3H), 3.77-3.65 (m, 4H), 3.64-3.56 (m, 13H), 3.38 (s, 3H), 3.20-3.05 (m, 3H), 2.95-2.80 (m, 2H), 2.75-2.60 (m, 2H), 2.55-2.50 (m, 2H), 2.43-2.25 (m, 5H), 2.21-1.97 (m, 8H), 1.93-1.83 (m, 3H), 1.79-1.72 (m, 5H), 1.68-1.29 (m, 10H), 1.08 (d, J=6.8 Hz, 3H), 1.05-0.95 (m, 1H), 0.98 (d, J=7.2 Hz, 3H), 0.95 (d, J=7.2 Hz, 3H). LCMS (M+H)=1365.1.
[0395] Azide-PEG3-VCP-eribulin (ER-001243116) (9.6 mg, 7.035 .mu.mol) and 2,5-dioxopyrrolidin-1-yl 3-(2-(2-(prop-2-yn-1-yloxy)ethoxy)ethoxy)propanoate (6.61 mg, 0.021 mmol) were mixed in water (0.6 mL) and t-Butanol (1.8 mL). The mixture was bubbled with N2 was for 45 min. Copper iodide on amberlyst-21 (1.23 mmol/g, 10 mg) was added to the mixture and N2 was bubbled through the mixture for another 30 min. The reaction mixture was then stirred at room temperature for 72 hours until the complete consumption of the starting material. No desired NHS ester product was observed by LCMS analysis, only the hydrolyzed carboxylic acid. The mixture was filtered through a short celite pad to remove CuI resin. The filtrate was concentrated in vacuo, and the resulting residue was purified by preparative thin layer chromatography (prep-TLC) (20% MeOH/CH.sub.2Cl.sub.2) to obtain acid-PEG3-triazole-PEG3-Val-Cit-PAB-eribulin (ER-001243701) as a white solid (3.7 mg, 33% yield). LCMS (ES) (M+H)=1581.2.
[0396] Acid-PEG3-triazole-PEG3-Val-Cit-PAB-eribulin (ER-001243701) (3.0 mg, 1.898 .mu.mol) was dissolved in DMF (200 .mu.L) and 1-hydroxypyrrolidine-2,5-dione (0.437 mg, 3.796 .mu.mol) was added, followed by the addition of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) (0.728 mg, 3.796 .mu.mol). The reaction was approximately 50% complete after stirring at room temperature for 18 hours. EDC (1.46 mg, 7.8 .mu.mol) was added, and the mixture was stirred for another 18 hours until HPLC analysis indicated >95% conversion to NHS-PEG3-triazole-PEG3-Val-Cit-PAB-eribulin. The mixture was concentrated in vacuo, and the residue was purified by prep-TLC (15% MeOH/CH.sub.2Cl.sub.2) to yield NHS-PEG3-triazole-PEG3-Val-Cit-PAB-eribulin (ER-001243700) as a white solid (2.2 mg, 69% yield). .sup.1HNMR (400 MHz, CD.sub.3OD) .delta. ppm 8.00 (s, 1H), 7.59 (d, J=8.0 Hz, 2H), 7.31 (d, J=8.4 Hz, 2H), 5.13 (s, 1H), 5.04 (d, J=12.4 Hz, 1H), 5.02 (s, 1H), 5.00 (d, J=12.4 Hz, 1H), 4.87 (s, 1H), 4.83 (s, 1H), 4.71 (t, J=4.0 Hz, 1H), 4.63 (s, 2H), 4.61 (t, J=4.4 Hz, 1H), 4.57-4.55 (m, 2H), 4.51-4.45 (m, 1H), 4.32-4.28 (m, 2H), 4.21-4.17 (m, 2H), 4.13-4.10 (m, 2H), 3.98 (t, J=10.8 Hz, 1H), 3.88-3.80 (m, 5H), 3.75-3.70 (m, 4H), 3.68-3.55 (m, 18H), 3.45-3.40 (m, 2H), 3.38 (s, 3H), 3.20-3.08 (m, 4H), 2.93-2.80 (m, 2H), 2.75-2.50 (m, 2H), 2.68 (s, 4H), 2.48-2.30 (m, 7H), 2.28-1.92 (m, 10H), 1.90-1.68 (m, 8H), 1.65-1.27 (m, 8H), 1.11 (d, J=6.8 Hz, 3H), 1.05-0.95 (m, 1H), 0.99 (d, J=7.2 Hz, 3H), 0.97 (d, J=6.8 Hz, 3H). LCMS (M+H)=1678.3.
1.8 Preparation of Mal-PEG2-Ala-Ala-Asn-PAB-Eribulin (ER-001231679) and Mal-PEG2-(Ala-Ala-Asn-PAB)2-Eribulin (ER-001231690)
##STR00045##
[0398] Eribulin (ER-000086526) (10 mg, 0.014 mmol) was dissolved in DMF (0.5 mL), and mixed with Hunig's Base (3.59 .mu.L, 0.021 mmol). (9H-fluoren-9-yl)methyl ((S)-1-(((S)-1-(((S)-4-amino-1-((4-((((4-nitrophenoxy)carbonyl)oxy)methyl- )phenyl)amino)-1,4-dioxobutan-2-yl)amino)-1-oxopropan-2-yl)amino)-1-oxopro- pan-2-yl)carbamate (15.76 mg, 0.021 mmol) was then added, and the resulting yellow solution was stirred at room temperature for 3 days until HPLC analysis indicated the complete consumption of the starting material. Diethylamine (14.23 .mu.L, 0.137 mmol) was added to the reaction mixture, which was then stirred at room temperature for an additional 2 hours until there was 100% cleavage of Fmoc protection. The reaction mixture was concentrated to remove diethylamine, and the residue was re-dissolved in DMF (1.5 mL). Et.sub.3N (0.015 mL, 0.11 mmol) was added at room temperature, followed by the addition of 2,5-dioxopyrrolidin-1-yl 3-(2-(2-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)ethoxy)ethoxy)propanoate (9.71 mg, 0.027 mmol). The reaction mixture was stirred at room temperature for 16 hours until the reaction was complete, as determined by LCMS analysis. The mixture was concentrated under high vacuum, and purified by flash chromatography to obtain Mal-PEG2-Ala-Ala-Asn-PAB-eribulin (ER-001231679) (9.2 mg, 49% yield) and Mal-PEG2-(Ala-Ala-Asn-PAB)2-eribulin (ER-001231690) (6.0 mg, 18% yield) as colorless oils.
[0399] Mal-PEG2-Ala-Ala-Asn-PAB-eribulin (ER-001231679): .sup.1HNMR (400 MHz) .delta. ppm 9.23 (s, 1H), 8.00 (d, J=7.6 Hz, 1H), 7.61 (d, J=8.4 Hz, 2H), 7.38 (d, J=6.8 Hz, 1H), 7.24 (d, J=8.4 Hz, 2H), 7.13 (d, J=7.2 Hz, 1H), 6.68 (s, 2H), 6.30 (br s, 1H), 6.04-6.00 (m, 1H), 5.77 (br s, 1H), 5.42 (br s, 1H), 5.07 (s, 1H), 5.06-4.98 (m, 2H), 4.93 (s, 1H), 4.88 (s, 1H), 4.90-4.82 (m, 1H), 4.80 (s, 1H), 4.69 (t, J=4.0 Hz, 1H), 4.60 (t, J=4.0 Hz, 1H), 4.49-4.42 (m, 1H), 4.38-4.25 (m, 4H), 4.19 (t, J=4.8 Hz, 1H), 4.15-4.08 (m, 1H), 4.03 (t, J=4.8 Hz, 1H), 3.97-3.85 (m, 3H), 3.83-3.50 (m, 12H), 3.41 (s, 3H), 3.50-3.10 (m, 3H), 3.02-2.64 (m, 6H), 2.52-2.30 (m, 7H), 2.30-1.65 (m, 14H), 1.65-1.20 (m, 12H), 1.10 (d, J=6.8 Hz, 3H), 1.13-1.05 (m, 1H). LCMS (M+Na)=1396.6.
[0400] Mal-PEG2-(Ala-Ala-Asn-PAB)2-eribulin (ER-001231690): .sup.1HNMR (400 MHz, CD.sub.3OD) .delta. ppm 7.65 (d, J=8.4 Hz, 2H), 7.60 (d, J=8.4 Hz, 2H), 7.28 (d, J=8.8 Hz, 2H), 7.23 (d, J=8.4 Hz, 2H), 6.79 (s, 2H), 5.13 (s, 1H), 5.02 (s, 1H), 5.06-4.98 (m, 4H), 4.87 (s, 1H), 4.82 (s, 1H), 4.85-4.72 (m, 2H), 4.71 (t, J=4.8 Hz, 1H), 4.61 (t, J=4.4 Hz, 1H), 4.47 (d, J=11.2 Hz, 1H), 4.30-4.06 (m, 9H), 3.97 (t, J=4.8 Hz, 1H), 3.89-3.80 (m, 3H), 3.75-3.48 (m, 12H), 3.38 (s, 3H), 3.17 (d, J=6.8 Hz, 2H), 2.94-2.62 (m, 8H), 2.50-2.28 (m, 7H), 2.22-1.65 (m, 14H), 1.58-1.30 (m, 18H), 1.10 (d, J=6.8 Hz, 3H), 1.06-0.97 (m, 1H). LCMS (M+Na)=1802.8.
1.9 Preparation of NHS-PEG2-Ala-Ala-Asn-PAB-Eribulin (ER-001231691)
##STR00046##
[0402] Ala-Ala-Asn-PAB-eribulin (ER-001231678) was prepared (15 mg, quantitative yield) from eribulin (ER-000086526) and Fmoc-Ala-Ala-Asn-PAB-PNP using the same procedure as described above for the preparation of Val-Cit-PAB-eribulin (ER-001228950). LCMS (M+H)=1135.5.
[0403] NHS-PEG2-Ala-Ala-Asn-PAB-eribulin (ER-001231691) was prepared (12.4 mg, 64% yield) from Ala-Ala-Asn-PAB-eribulin (ER-001231678) and BisNHS-PEG2 using the same procedure as described above for the preparation of NHS-PEG2-Val-Cit-PAB-eribulin (ER-001236940). .sup.1HNMR (400 MHz) .delta. ppm 9.21 (s, 1H), 7.95 (d, J=8.0 Hz, 1H), 7.62 (d, J=8.8 Hz, 2H), 7.58-7.52 (m, 1H), 7.28 (br s, 1H), 7.24 (d, J=8.4 Hz, 2H), 7.10 (br s, 1H), 6.29 (d, J=12.4 Hz, 1H), 5.83 (br s, 1H), 5.38 (br s, 1H), 5.07 (s, 1H), 5.05-4.95 (m, 2H), 4.93 (s, 1H), 4.88 (s, 1H), 4.90-4.83 (m, 1H), 4.81 (s, 1H), 4.69 (t, J=4.4 Hz, 1H), 4.60 (t, J=4.4 Hz, 1H), 4.46-4.41 (m, 1H), 4.36-4.25 (m, 4H), 4.19 (dd, J=4.8, 6.0 Hz, 1H), 4.15-4.09 (m, 1H), 4.03 (dd, J=4.8, 6.0 Hz, 1H), 3.99-3.89 (m, 3H), 3.85-3.50 (m, 10H), 3.41 (s, 3H), 3.40-3.10 (m, 3H), 3.01-2.60 (m, 10H), 2.60-2.35 (m, 7H), 2.35-1.65 (m, 14H), 1.65-1.20 (m, 14H), 1.10 (d, J=6.8 Hz, 3H), 1.15-1.03 (m, 1H). LCMS (ES) (M+H)=1442.7.
1.10 Preparation of Azide-PEG3-Disulfide-PAB-Eribulin (ER-001237508)
##STR00047## ##STR00048##
[0405] 4-(((tert-butyldimethylsilyl)oxy)methyl)benzoic acid (1.0 g, 3.754 mmol) was dissolved in dichloromethane (DCM) (25 mL) cooled to 0.degree. C. Triethylamine (0.549 mL, 3.941 mmol) was then added, followed by diphenyl phosphorazidate (1.085 mg, 3.941 mmol). The reaction mixture was slowly warmed to room temperature and stirred for 14 hours. The crude mixture was diluted with ethyl acetate (EtOAc)/Hep (1:1, 100 mL), and passed through a short silica plug eluting with EtOAc/Hep (50%). The solvent was removed under vacuum to yield 1.10 g of 4-(((tert-butyldimethylsilyl)oxy)methyl)benzoyl azide (ER-001131970). .sup.1H NMR (400 MHz) .delta. ppm 7.98 (d, 2H, J=8.0 Hz), 7.40 (d, 2H, J=8.0 Hz), 4.79 (s, 2H), 0.94 (s, 9H), 0.10 (s, 6H).
[0406] 4-(((tert-butyldimethylsilyl)oxy)methyl) benzoyl azide (ER-001131970) (1.1 g, 3.775 mmol), dissolved in toluene (20 mL), was heated at 110.degree. C. for 3 hours. Although the product did not show as a single spot, thin layer chromatography (TLC) analysis indicated that the starting material was consumed. The reaction mixture was then cooled to room temperature, and transferred to a vial sealed under nitrogen and stored as a solution in toluene (1 mL=32.6 mg) at -20.degree. C.
[0407] Triethylamine (0.099 mL, 0.709 mmol) was added to a solution of tert-butyl((4-isocyanatobenzyl)oxy)dimethylsilane (165 mg, 0.626 mmol) in toluene (5 mL), followed by alcohol (90.0 mg, 0.591 mmol), and the reaction mixture was stirred for 6 hours at 36.degree. C. Progress of the reaction was monitored by UPLC/MS. A saturated solution of sodium hydrogen carbonate (NaHCO.sub.3) (10 mL) was then added, extracted with EtOAc/Hep (1:1, 60 mL), washed with brine, dried over sodium sulfate, and concentrated. The crude material was purified by flash chromatography (EtOAc/Hep 10% to 40%) to obtain 215 mg of 2-methyl-2-(methyldisulfanyl)propyl(4-(((tert-butyldimethylsilyl) oxy)methyl)phenyl)carbamate (ER-001131973). .sup.1H NMR (400 MHz) .delta. ppm 7.34 (d, 2H, J=8.4 Hz), 7.26 (d, 2H, J=7.6 Hz), 6.63 (br s, 1H), 4.69 (s, 2H), 4.17 (s, 2H), 2.42 (s, 3H), 1.35 (s, 6H), 0.93 (s, 9H), 0.08 (s, 6H).
[0408] 2-methyl-2-(methyldisulfanyl)propyl (4-(((tert-butyldimethylsilyl)oxy)methyl)phenyl)carbamate (ER-001131973) (198 mg, 0.476 mmol) and 2-(2-(2-(2-azidoethoxy)ethoxy)ethoxy)ethyl 4-methylbenzenesulfonate (325 mg, 0.87 mmol) were dissolved in DMF (6.6 mL). Cesium carbonate (621 mg, 1.905 mmol) was then added, followed by tetrabutylammoniumiodide (45 mg, 0.122 mmol), and the reaction mixture was stirred for 15 hours at 36.degree. C. Progress of the reaction was monitored by UPLC/MS. A saturated solution of NH.sub.4Cl (30 mL) was then added, extracted with EtOAc/Hep (2:1, 150 mL), washed with brine (10 mL), dried over sodium sulfate, and concentrated under vacuum. The crude material was purified by flash chromatography (EtOAc/Hep 20% to 50%) to obtain 248 mg of 2-methyl-2-(methyldisulfanyl)propyl (2-(2-(2-(2-azidoethoxy)ethoxy) ethoxy)ethyl)(4-(((tert-butyldimethylsilyl)oxy)methyl)phenyl)carbamate (ER-001140141). .sup.1HNMR (400 MHz) .delta. ppm 7.28 (d, 2H, J=8.4 Hz), 7.20 (d, 2H, J=8.0 Hz), 4.73 (s, 2H), 4.06 (br s, 2H), 3.83 (dd, 2H, J=6.4, 5.6 Hz), 3.68-3.56 (m, 12H), 3.37 (dd, 2H, J=5.6, 5.2 Hz), 2.33 (s, 3H), 1.14 (br s, 6H), 0.93 (s, 9H), 0.09 (s, 6H).
[0409] 2-methyl-2-(methyldisulfanyl)propyl (2-(2-(2-(2-azidoethoxy)ethoxy)ethoxy)ethyl)(4-(((tert-butyldimethylsilyl- )oxy)methyl)phenyl)carbamate (ER-001140141) (81 mg, 0.131 mmol) was dissolved in a mixture of methanol (5 mL) and water (0.5 mL). Acetic acid (0.5 mL, 8.734 mmol) was then added to the reaction mixture, and stirred for 14 hours at 38.degree. C. The reaction mixture was cooled to room temperature, and the solvent was removed under vacuum. The residue was diluted with EtOAc (30 mL), washed with water (2.times.5 mL), NaHCO.sub.3, and brine (3 mL), dried over sodium sulfate, and concentrated under vacuum. The crude material was purified by flash chromatography (EtOAc/Hep 30% to 90%) to obtain 61.0 mg of 2-methyl-2-(methyldisulfanyl)propyl (2-(2-(2-(2-azidoethoxy)ethoxy)ethoxy) ethyl)(4-(hydroxymethyl)phenyl)carbamate (ER-001140549). .sup.1HNMR (400 MHz) .delta. ppm 7.34 (d, 2H, J=8.8 Hz), 7.26 (d, 2H, J=8.0 Hz), 4.69 (d, 2H, J=4.4 Hz), 4.06 (br s, 2H), 3.84 (dd, 2H, J=6.2, 6.2 Hz), 3.66-3.56 (m, 12H), 3.37 (dd, 2H, J=5.2, 5.2 Hz), 2.33 (s, 3H), 1.74 (br s, 1H), 1.14 (br s, 6H).
[0410] 2-methyl-2-(methyldisulfanyl)propyl (2-(2-(2-(2-azidoethoxy)ethoxy)ethoxy)ethyl)(4-(hydroxymethyl)phenyl)carb- amate (ER-001140549) (60 mg, 0.119 mmol) was dissolved in DCM (2 mL) and Py (0.019 mL, 0.239 mmol) cooled to 0.degree. C. 4-nitrophenyl carbonochloridate (38.5 mg, 0.191 mmol) in DCM (2 mL) and dimethylaminopyridine (DMAP) (2.9 mg, 0.024 mmol) were then added, and the reaction mixture was stirred for 30 min at 0.degree. C. The reaction mixture was slowly warmed to room temperature, and stirred until the starting material was consumed (approximately 2.5 hours). The solvent was then removed under vacuum, and the residue was purified by flash chromatography (EtOAc/Hep 10% to 35%) to obtain 78 mg of 2-methyl-2-(methyldisulfanyl)propyl (2-(2-(2-(2-azidoethoxy)ethoxy)ethoxy) ethyl)(4-((((4-nitrophenoxy)carbonyl)oxy)methyl)phenyl)carbamate (ER-001140550). .sup.1H NMR (400 MHz) .delta. ppm 8.27 (dd, 2H, J=6.8, 2.4 Hz), 7.41 (d, 2H, J=8.8 Hz), 7.37 (dd, 2H, J=7.2, 2.4 Hz), 7.33 (d, 2H, J=8.8 Hz), 5.27 (s, 2H), 4.08 (br s, 2H), 3.85 (dd, 2H, J=5.8, 5.8 Hz), 3.66-3.57 (m, 12H), 3.36 (dd, 2H, J=5.2, 5.2 Hz), 2.33 (br s, 3H), 1.19 (br s, 6H).
[0411] 2-methyl-2-(methyldisulfanyl)propyl (2-(2-(2-(2-azidoethoxy)ethoxy)ethoxy)ethyl)(4-((((4-nitrophenoxy)carbony- l)oxy)methyl)phenyl)carbamate (ER-001140550) (30 mg, 0.045 mmol) in DCM (3 mL, 46.625 mmol) was placed in a 25-ml flask under nitrogen, and cooled to 0.degree. C. Amine (40.8 mg, 0.049 mmol) in DCM (2 mL) and Hunig's Base (0.024 mL, 0.135 mmol) were added, followed by DMAP (1.4 mg, 0.011 mmol). The reaction mixture was then slowly warmed to room temperature, stirred for 3 hours, concentrated under vacuum, and purified by flash chromatography (EtOAc/Hep 50% to 100%, followed by MeOH/EtOAc 3% to 8%) to obtain 45.0 mg of pure azide-PEG3-disulfide-PAB-eribulin (ER-001237508). .sup.1H NMR (400 MHz) .delta. ppm 7.32 (d, 2H, J=8.0 Hz), 7.25 (d, 2H, J=7.2 Hz), 5.28 (dd, 1H, J=5.6, 5.6 Hz), 5.11-5.04 (m, 3H), 4.93 (s, 1H), 4.88 (s, 1H), 4.81 (s, 1H), 4.69 (dd, 1H, J=4.4, 4.4 Hz), 4.60 (dd, 1H, J=4.2, 4.2 Hz), 4.36 (br s, 1H), 4.33 (dd, 1H, J=4.0, 2.0), 4.29 (ddd, 1H, J=9.6, 4.4, 4.4 Hz), 4.18 (dd, 1H, J=6.4, 4.4 Hz), 4.14-4.04 (m, 3H), 4.03 (dd, 1H, J=6.4, 4.4 Hz), 3.97-3.89 (m, 3H), 3.84-3.78 (m, 3H), 3.67-3.56 (m, 14H), 3.42 (s, 3H), 3.40-3.35 (m, 1H), 3.37 (dd, 2H, J=5.2, 5.2 Hz), 3.27 (d, 1H, J=3.2 Hz), 3.20 (ddd, 1H, J=12.8, 6.0, 6.0 Hz), 2.91-2.83 (m, 2H), 2.70 (dd, 1H, J=16.0, 10.0 Hz), 2.52-2.40 (m, 3H), 2.35-2.13 (m, 9H), 2.10-2.06 (m, 1H), 2.01-1.89 (m, 4H), 1.78-1.64 (m, 4H), 1.60-1.52 (m, 4H), 1.49-1.28 (m, 5H), 1.22-1.07 (m, 6H), 1.09 (d, 3H, J=6.0 Hz).
1.11 Preparation of Mal-PEG4-Triazole-PEG3-Disulfide-PAB-Eribulin (ER-001237504)
##STR00049##
[0413] A mixture of azide (9.0 mg, 7.151 .mu.mol) and 3-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)-N-(3,6,9,12-tetraoxapentadec-14-- yn-1-yl)propanamide (6.8 mg, 0.018 mmol) in tert-butanol (1.5 mL) and water (0.5 mL) was degassed for 45 min. Copper iodide on amberlyst-21 (1.23 mmol/g, 10 mg) was then added, and degassed for an additional 30 min. The reaction mixture was stirred at room temperature for 18 hours, and monitored by UPLC/MS. The majority of the starting material was consumed, and the desired product showed as a major peak. The mixture was then separated from resin, and purified on HPLC (acetonitril/water with 0.05% formic acid) to obtain 1.5 mg of Mal-PEG4-triazole-PEG3-disulfide-PAB-eribulin (ER-001237504). .sup.1H NMR (400 MHz) .delta. ppm 7.74 (s, 1H), 7.32 (d, 2H, J=8.4 Hz), 7.27-7.25 (m, 2H), 6.69 (br s, 2H), 5.43 (dd, 1H, J=5.6, 5.6 Hz), 5.14-5.06 (m, 3H), 4.95 (s, 1H), 4.89 (s, 1H), 4.82 (s, 1H), 4.70 (dd, 1H, J=4.4, 4.4 Hz), 4.66 (s, 2H), 4.62 (dd, 1H, J=4.4, 4.4 Hz), 4.52 (dd, 1H, J=5.2, 5.2 Hz), 4.38-4.31 (m, 2H), 4.30 (ddd, 1H, J=10.4, 4.0, 4.0 Hz), 4.20 (dd, 1H, J=6.4, 4.4 Hz), 4.16-4.05 (m, 3H), 4.04 (dd, 1H, J=6.4, 4.4 Hz), 3.99-3.91 (m, 3H), 3.87-3.80 (m, 6H), 3.70-3.59 (m, 22H), 3.53 (dd, 2H, J=5.2, 5.2 Hz), 3.44 (s, 3H), 3.43-3.36 (m, 3H), 3.29 (d, 1H, J=2.8 Hz), 3.18 (ddd, 1H, J=12.9, 6.2, 6.2 Hz), 2.92-2.84 (m, 2H), 2.72 (dd, 1H, J=16.0, 10.0 Hz), 2.54-2.42 (m, 5H), 2.37-1.90 (m, 19H), 178-1.52 (m, 3H), 1.50-1.14 (m, 16H), 1.10 (d, 3H, J=6.0 Hz). LCMS (M+H)=1642.1.
1.12 Preparation of NHS-PEG3-Triazole-PEG3-Disulfide-PAB-Eribulin (ER-001244129)
##STR00050##
[0415] A mixture of azide (9 mg, 7.151 .mu.mol) and 2,5-dioxopyrrolidin-1-yl 3-(2-(2-(prop-2-yn-1-yloxy)ethoxy)ethoxy)propanoate (4.5 mg, 14.30 .mu.mol) in tert-butanol (1 mL) and water (0.5 mL) was degassed for 45 min. Copper iodide on amberlyst-21 (1.23 mmol/g, 10 mg, 7.151 .mu.mol) was then added, and degassed for an additional 30 min. The reaction mixture was stirred room temperature for 18 hours, and monitored by UPLC/MS. The majority of the starting material was consumed, and the desired product showed as a major peak. The mixture was then separated from resin by filtration, extracted with DCM (15 mL), washed with brine (3.times.3 mL), dried over sodium sulfate, and concentrated under vacuum. The residue (5 mg, 3.39 .mu.mol) was azeotroped with toluene, dissolved in THF (1 mL), and cooled to 0.degree. C. DCC (4.2 mg, 0.02 mmol) was added, followed by 1-hydroxypyrrolidine-2,5-dione (2.2 mg, 0.019 mmol), and the reaction mixture was stirred at room temperature for 18 hours. The majority of the starting material was consumed, and the desired product showed as a major peak, as determined by UPLC/MS. The reaction mixture was then concentrated and purified on preparative TLC (DCM/i-propanol, 8%) to yield 2.5 mg of NHS-PEG3-triazole-PEG3-disulfide-PAB-eribulin (ER-001244129) as a colorless oil. .sup.1H NMR (400 MHz, CD.sub.2Cl.sub.2) .delta. ppm 7.72 (s, 1H), 7.32 (d, 2H, J=8.8 Hz), 7.25 (d, 2H, J=8.8 Hz), 5.08-5.04 (m, 3H), 4.93 (s, 1H), 4.85 (s, 1H), 4.78 (s, 1H), 4.64 (dd, 1H, J=4.4, 4.4 Hz), 4.58 (s, 2H), 4.55 (dd, 1H, J=4.4, 4.4 Hz), 4.48 (dd, 2H, J=5.0, 5.0 Hz), 4.32 (d, 1H, J=6.6 Hz), 4.27-4.22 (m, 2H), 4.14 (dd, 1H, J=6.6, 4.8 Hz), 4.10-4.01 (m, 3H), 4.00 (dd, 1H, J=6.8, 4.4 Hz), 3.92-3.78 (m, 9H), 3.65-3.53 (m, 19H), 3.44-3.39 (m, 4H), 3.37 (s, 3H), 3.26 (d, 1H, J=3.2 Hz), 3.13 (ddd, 1H, J=12.4, 6.0, 6.0 Hz), 2.91-2.73 (m, 11H), 2.70-2.64 (m, 2H), 2.54-2.41 (m, 3H), 2.38-1.80 (m, 16H), 1.74-1.52 (m, 3H), 1.41-1.13 (m, 10H), 1.07 (d, 3H, J=6.4 Hz). LCMS (M+H)=1572.3.
1.13 Preparation of Azide-PEG3-Sulfonamide-PAB-Eribulin (ER-001138856)
##STR00051##
[0417] 4-(((tert-butyldimethylsilyl)oxy)methyl)aniline (315 mg, 1.327 mmol) was dissolved in DCM (10 mL) cooled to 0.degree. C. Pyridine (0.268 mL, 3.317 mmol) was then added, followed by 5-cyanopyridine-2-sulfonyl chloride (365 mg, 1.801 mmol) in DCM (10 mL) over 15 min. The reaction mixture was slowly warmed to room temperature over 1 hour, and stirred for 2 hours. The reaction mixture was diluted with EtOAc (50 mL), washed with brine, dried over sodium sulfate, and concentrated under vacuum to obtain 610 mg (103%) of N-(4-(((tert-butyldimethylsilyl)oxy)methyl)phenyl)-5-cyanopyridine-2-sulf- onamide (ER-001137670). The crude product was reasonably pure, though colored. .sup.1H NMR (400 MHz) .delta. ppm 8.94 (dd, 1H, J=1.8, 0.6 Hz), 8.10 (dd, 1H, J=8.4, 2.0 Hz), 7.99 (dd, 1H, J=8.0, 0.8 Hz), 7.18 (d, 2H, J=8.2 Hz), 7.15 (br s, 1H), 7.11 (dd, 2H, J=6.8, 0.8 Hz), 4.64 (s, 2H), 0.90 (s, 9H), 0.05 (s, 6H).
[0418] N-(4-(((tert-butyldimethylsilyl)oxy)methyl)phenyl)-5-cyanopyridine-- 2-sulfonamide (ER-001137670) (105.0 mg, 0.26 mmol) and 2-(2-(2-(2-azidoethoxy)ethoxy)ethoxy)ethyl 4-methylbenzenesulfonate (143 mg, 0.383 mmol) were dissolved in DMF (4 mL). Potassium carbonate (K.sub.2CO.sub.3) (144 mg, 1.041 mmol) was then added, followed by tetrabutylammonium iodide (19.2 mg, 0.052 mmol), and the reaction mixture was stirred for 36 hours at 50.degree. C. Progress of the reaction was monitored by UPLC/MS. A saturated solution of NH.sub.4Cl (10 mL) was added, extracted with EtOAc/Hep (2:1, 30 mL), washed with brine, dried over sodium sulfate, and concentrated. The crude material was purified by flash chromatography (EtOAc/Hep 25% to 80%) to obtain 118.0 mg of N-(2-(2-(2-(2-azidoethoxy)ethoxy)ethoxy)ethyl)-N-(4-(((tert-butyldimethyl- silyl)oxy)methyl)phenyl)-5-cyanopyridine-2-sulfonamide (ER-001138452) (75%). .sup.1H NMR (400 MHz) .delta. ppm 8.99 (dd, 1H, J=1.8, 0.6 Hz), 8.08 (dd, 1H, J=8.2, 2.2 Hz), 7.86 (dd, 1H, J=8.0, 0.8 Hz), 7.24 (d, 2H, J=10 Hz), 7.09 (d, 2H, J=8.8 Hz), 4.69 (s, 2H), 4.06 (dd, 2H, J=6.0, 6.0 Hz), 3.67 (dd, 2H, J=5.2, 5.2 Hz), 3.65-3.62 (m, 4H), 3.58 (dd, 2H, J=6.2, 6.2 Hz), 3.56-3.53 (m, 4H), 3.38 (dd, 2H, J=5.2, 5.2 Hz), 0.93 (s, 9H), 0.08 (s, 6H).
[0419] N-(2-(2-(2-(2-azidoethoxy)ethoxy)ethoxy)ethyl)-N-(4-(((tert-butyldi- methylsilyl)oxy)methyl)phenyl)-5-cyanopyridine-2-sulfonamide (ER-001138452) (150 mg, 0.248 mmol) was dissolved in methanol (6 mL). Water (0.60 mL) was then added, followed by acetic acid (AcOH) (0.60 mL, 10.481 mmol). The reaction mixture was slowly warmed to 38.degree. C., and stirred for 14 hours. The majority of the solvent was removed under vacuum. The residue was diluted with EtOAc (30 mL), washed with water (2.times.5 mL), NaHCO.sub.3, and brine, dried over sodium sulfate, and concentrated under vacuum. The crude material was purified by flash chromatography (EtOAc/Hep 35% to 90%) to obtain 105.0 mg of N-(2-(2-(2-(2-azidoethoxy)ethoxy)ethoxy)ethyl)-5-cyano-N-(4-(hydroxymethy- l)phenyl)pyridine-2-sulfonamide (ER-001138455) (84%). .sup.1H NMR (400 MHz) .delta. ppm 8.99 (d, 1H, J=1.2 Hz), 8.09 (dd, 1H, J=8.4, 2.0 Hz), 7.88 (dd, 1H, J=8.4, 0.8 Hz), 7.30 (d, 2H, J=8.8 Hz), 7.15 (d, 2H, J=8.4 Hz), 4.67 (s, 2H), 4.06 (dd, 2H, J=6.2, 6.2 Hz), 3.66 (dd, 2H, J=5.0, 5.0 Hz), 3.65-3.58 (m, 6H), 3.55-3.51 (m, 4H), 3.38 (dd, 2H, J=5.2, 5.2 Hz.
[0420] N-(2-(2-(2-(2-azidoethoxy)ethoxy)ethoxy)ethyl)-5-cyano-N-(4-(hydrox- ymethyl)phenyl)pyridine-2-sulfonamide (ER-001138455) (45 mg, 0.092 mmol) was dissolved in DCM (3 mL), and cooled to 0.degree. C. following the addition of pyridine (0.015 mL, 0.183 mmol). 4-nitrophenyl carbonochloridate (20.3 mg, 0.101 mmol) in DCM (2 mL) and DMAP (2.3 mg, 0.018 mmol) was then added. The reaction mixture was slowly warmed to room temperature and stirred for 2 hours. UPLC/MS indicated that some starting material remained. The reaction mixture was then concentrated under vacuum, and purified by flash chromatography (EtOAc/Hep 12% to 40%) to obtain 35 mg of 4-4N-(2-(2-(2-(2-azidoethoxy)ethoxy)ethoxy)ethyl)-5-cyanopyridine)-2-sulf- onamido)benzyl (4-nitrophenyl) carbonate (ER-001235286) (58%), and 20 mg of starting material. .sup.1HNMR (400 MHz) .delta. ppm 8.99 (d, 1H, J=0.8 Hz), 8.27 (dd, 2H, J=9.2, 2.0 Hz), 8.12 (dd, 1H, J=7.6, 2.0 Hz), 7.92 (d, 1H, J=8.4 Hz), 7.38 (d, 4H, J=9.6 Hz), 7.26 (d, 2H, J=8.8 Hz), 5.45 (s, 2H), 4.06 (dd, 2H, J=5.8, 5.8 Hz), 3.67-3.58 (m, 8H), 3.58-3.50 (m, 4H), 3.38 (dd, 2H, J=6.1, 6.1 Hz).
[0421] 4-(N-(2-(2-(2-(2-azidoethoxy)ethoxy)ethoxy)ethyl)-5-cyanopyridine-2- -sulfonamido)benzyl (4-nitrophenyl) carbonate (ER-001235286) (35.0 mg, 0.053 mmol) was placed in a 25-mL flask under nitrogen, and cooled to 0.degree. C. Amine (48.5 mg, 0.059 mmol) in DCM (3 mL, 46.625 mmol) and Hunig's Base (0.037 mL, 0.214 mmol) was then added, followed by DMAP (2.61 mg, 0.021 mmol). The reaction mixture was stirred for 30 min at 0.degree. C., and then stirred for an additional 6 hours at room temperature. The reaction mixture was concentrated under vacuum, and purified by flash chromatography (EtOAc/Hep 50% to 100%, followed by MeOH/EtOAc 3% to 8%) to obtain 61.0 mg of pure azide-PEG3-sulfonamide-PAB-eribulin (ER-001138856). .sup.1H NMR (400 MHz) .delta. ppm 8.98 (d, 1H, J=1.2 Hz), 8.10 (dd, 1H, J=8.2, 1.8 Hz), 7.87 (d, 1H, J=8.0 Hz), 7.26 (d, 2H, J=6.8 Hz), 7.13 (d, 2H, J=8.4 Hz), 5.29 (dd, 1H, J=5.6, 5.6 Hz), 5.08-5.00 (m, 3H), 4.92 (s, 1H), 4.87 (s, 1H), 4.80 (s, 1H), 4.68 (dd, 1H, J=4.6, 4.6 Hz), 4.59 (dd, 1H, J=4.6, 4.6 Hz), 4.38-4.30 (m, 2H), 4.28 (ddd, 1H, J=10.4, 4.0, 4.0, Hz), 4.17 (dd, 1H, J=6.2, 4.6 Hz), 4.13-4.01 (m, 4H), 3.97-3.88 (m, 3H), 3.82-3.78 (m, 1H), 3.67-3.50 (m, 15H), 3.41 (s, 3H), 3.40-3.33 (m, 1H), 3.37 (dd, 2H, J=4.8, 4.8 Hz), 3.27 (d, 1H, J=3.2 Hz), 3.15 (ddd, 1H, J=12.8, 6.4, 6.4 Hz), 2.90-2.82 (m, 2H), 2.70 (dd, 1H, J=16.0, 10.0 Hz), 2.51-2.40 (m, 3H), 2.34-2.13 (m, 7H), 2.10-2.05 (m, 1H), 1.99-1.88 (m, 4H), 1.78-1.64 (m, 5H), 1.62-1.52 (m, 2H), 1.50-1.29 (m, 4H), 1.08 (d, 3H, J=6.8 Hz).
1.14 Preparation of Mal-PEG4-Triazole-PEG3-Sulfonamide-PAB-Eribulin (ER-001237505)
##STR00052##
[0423] A mixture of azide (10 mg, 8.023 .mu.mop and 3-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)-N-(3,6,9,12-tetraoxapentadec-14-- yn-1-yl)propanamide (9.20 mg, 0.024 mmol) in tert-butanol (2.1 mL) and water (0.7 mL) was degassed for 45 min. Copper iodide on amberlyst-21 (1.23 mmol/g, 15 mg) was then added, and degassed for an additional 30 min. The reaction mixture was stirred at room temperature for 18 hours, and was monitored by UPLC/MS. The majority of the starting material was consumed, and the desired product showed as a major peak. The reaction mixture was then separated from resin, and purified on preparative TLC (DCM/methanol, 7%) to yield 5.5 mg of Mal-PEG4-triazole-PEG3-sulfonamide-PAB-eribulin (ER-001237505). .sup.1H NMR (400 MHz, CD.sub.2Cl.sub.2) .delta. ppm 9.01 (s, 1H), 8.15 (dd, 1H, J=8.0, 1.8 Hz), 7.87 (d, 1H, J=8.0 Hz), 7.75 (s, 1H), 7.28 (d, 2H, J=8.0 Hz), 7.14 (d, 2H, J=8.4 Hz), 6.68 (s, 2H), 6.47 (br s, 1H), 5.44 (br s, 1H), 5.10-5.02 (m, 3H), 4.94 (s, 1H), 4.86 (s, 1H), 4.80 (s, 1H), 4.68 (dd, 1H, J=4.4, 4.4 Hz), 4.59 (s, 2H), 4.56 (dd, 1H, J=4.4, 4.4 Hz), 4.51 (dd, 2H, J=5.2, 5.2, Hz), 4.34 (d, 1H, J=7.6, Hz), 4.30-4.23 (m, 2H), 4.19-4.14 (m, 2H), 4.08 (dd, 1H, J=4.0, 4.0 Hz), 4.03-3.98 (m, 2H), 3.94-3.72 (m, 8H), 3.68-3.46 (m, 28H), 3.38 (s, 3H), 3.38-3.33 (m, 3H), 3.27 (d, 1H, J=3.2 Hz), 3.16-3.02 (m, 2H), 2.90-2.81 (m, 2H), 2.68 (dd, 1H, J=16.2, 9.8 Hz), 2.54-2.40 (m, 7H), 2.40-1.8 (m, 11H), 1.80-1.50 (m, 3H), 1.48-1.25 (m, 3H), 1.09 (d, 3H, J=6.4 Hz). LCMS (M+H)=1630.0.
1.15 Preparation of NHS-PEG3-Triazole-PEG3-Sulfonamide-PAB-Eribulin (ER-001244623)
##STR00053##
[0425] A mixture of azide (14 mg, 0.011 mmol) and 2,5-dioxopyrrolidin-1-yl 3-(2-(2-(prop-2-yn-1-yloxy)ethoxy)ethoxy)propanoate (8.80 mg, 0.028 mmol) in tert-butanol (2 mL) and water (1 mL) was degassed for 45 min. Copper iodide on amberlyst-21 (1.23 mmol/g, 20 mg) was then added, and degassed for an additional 30 min. The reaction mixture was stirred at room temperature for 18 hours, and was monitored by UPLC/MS. The majority of the starting material was consumed, and the desired product showed as a major peak. The reaction mixture was then separated from resin by extraction with DCM (2.times.10 mL). The DCM layer was washed with brine (4.times.5 mL), dried over sodium sulfate, and concentrated to the desired product (which was used in the next step without any further purification).
[0426] Crude acid (15.0 mg, 10.255 .mu.mol) was dissolved in THF (1.5 mL), and cooled to 0.degree. C. DCC (15.2 mg, 0.074 mmol) was then added, followed by 1-hydroxypyrrolidine-2,5-dione (8.3 mg, 0.072 mmol). The reaction mixture was stirred at room temperature for 18 hours. UPLC/MS indicated that the majority of the starting material was consumed, and the desired product showed as a major peak. The reaction mixture was concentrated, and purified on preparative TLC (DCM/i-propanol, 8%) to yield 2.5 mg of NHS-PEG3-triazole-PEG3-sulfonamide-PAB-eribulin (ER-001244623).
[0427] .sup.1H NMR (400 MHz, CD.sub.2Cl.sub.2) .delta. ppm 9.00 (s, 1H), 8.12 (d, 1H, J=8.4 Hz), 8.00 (d, 1H, J=8.0 Hz), 7.72 (s, 1H), 7.26 (d, 2H, J=8.0 Hz), 7.12 (d, 2H, J=8.0 Hz), 5.37 (br s, 1H), 5.08-5.02 (m, 3H), 4.93 (s, 1H), 4.85 (s, 1H), 4.78 (s, 1H), 4.66-4.62 (m, 1H), 4.58-4.56 (m, 4H), 4.33 (d, 1H, J=10.8 Hz), 4.29-4.21 (m, 2H), 4.10-3.96 (m, 4H), 3.93-3.76 (m, 6H), 3.74-3.44 (m, 27H), 3.36 (s, 3H), 3.34-3.24 (m, 2H), 3.15-3.06 (m, 1H), 2.97 (br s, 1H), 2.90-2.78 (m, 8H), 2.74-2.08 (m, 13H), 2.05-1.78 (m, 5H), 1.73-1.50 (m, 2H), 1.41-1.25 (m, 4H), 1.07 (d, 3H, J=6.0 Hz). LCMS (M+H)=1560.0.
1.16 Preparation of Mal-PEG2-Eribulin
[0428] Eribulin (5 mg, 7 .mu.mol) was dissolved in DMF (0.5 mL), and mixed with maleimido-PEG2-NHS (5 mg, 14 .mu.mol; Broadpharm, Cat No. BP-21680) and Hunig's base (2.4 .mu.L, 14 .mu.mol). The reaction mixture was stirred at room temperature for 2 hours. The reaction mixture was then purified by HPLC (water-acetonitrile gradient 30-70% containing 0.1% formic acid). Eluent was collect by mass, and lyophilized to dryness. Final yield was 3.7 mg (3.8 .mu.ma 54%). Predicted exact mass was 968.5 Da. Measured mass was 969.6 Da [M+H].
1.17 Preparation of Mal-PEG4-Eribulin
[0429] Eribulin (5 mg, 7 .mu.mol) was dissolved in DMF (0.5 mL), and mixed with maleimido-PEG4-NHS (6.2 mg, 14 .mu.mol; Broadpharm, Cat No. BP-20554) and Hunig's base (2.4 .mu.L, 14 .mu.mol). The reaction mixture was stirred at room temperature for 2 hours. The reaction mixture was then purified by HPLC (water-acetonitrile gradient 30-70% containing 0.1% formic acid). Eluent was collect by mass, and lyophilized to dryness. Final yield was 3.7 mg (3.5 .mu.mol, 50%). Predicted exact mass was 1056.5 Da. Measured mass was 1057.7 Da [M+H].
1.18 Preparation of Azido-PEG2-Eribulin
[0430] Eribulin (5 mg, 7 .mu.mol) was dissolved in DMF (0.5 mL), and mixed with azido-PEG2-NHS (4.2 mg, 14 .mu.mol; Broadpharm, Cat No. BP-20524) and Hunig's base (2.4 .mu.L, 14 .mu.mol). The reaction mixture was stirred at room temperature for 2 hours. The reaction mixture was then purified by HPLC (water-acetonitrile gradient 30-70% containing 0.1% formic acid). Eluent was collect by mass, and lyophilized to dryness. Final yield was 2.2 mg (2.4 .mu.mol, 34%). Predicted exact mass was 914.5 Da. Measured mass was 915.7 Da [M+H].
1.19 Preparation of Azido-PEG4-Eribulin
[0431] Eribulin (5 mg, 7 .mu.mol) was dissolved in DMF (0.5 mL), and mixed with azido-PEG4-NHS (5.5 mg, 14 .mu.mol; Broadpharm, Cat No. BP-20518) and Hunig's base (2.4 .mu.L, 14 .mu.mol). The reaction mixture was stirred at room temperature for 2 hours. The reaction mixture was then purified by HPLC (water-acetonitrile gradient 30-70% containing 0.1% formic acid). Eluent was collect by mass, and lyophilized to dryness. Final yield was 3.0 mg (3.0 .mu.mol, 43%). Predicted exact mass was 1002.5 Da. Measured mass was 1003.7 Da [M+H].
1.20 Preparation of azido-PEG4-Val-Cit-PAB-eribulin
[0432] Eribulin (15 mg, 21 .mu.mol) was dissolved in DMF (1.5 mL), and mixed well. Hunig's base (5.5 .mu.L, 32 .mu.mol) and Fmoc-VCP-PNP (24 mg, 22 .mu.mol; Levena Biopharma, Cat No. VC1003) were then added. The reaction mixture was stirred at room temperature overnight (16 hours). Upon completion of the reaction, diethylamine (20 .mu.L, 0.21 mmol) was added to the reaction mixture, and stirred for 2 hours at room temperature to remove the Fmoc protecting group. The deprotection reaction was monitored using a Waters SQD mass spectrometer. Upon completion of the reaction, the reaction mixture was transferred to a pre-weighed 1.5 mL microcentrifuge tube. The solvent was evaporated under vacuum using a refrigerated Centrivap concentrator with the temperature set at 30.degree. C. Yield was 16 mg (14 .mu.mol) of crude NH2-Val-Cit-pAB-eribulin (exact mass 1134.6 Da, 67% yield).
[0433] NH2-Val-Cit-pAB-eribulin (16 mg, 14.1 .mu.mol) was dissolved in DMF (1.5 mL). Hunig's Base (7.2 .mu.L, 41 .mu.mol) and azido-PEG4-NHS (11 mg, 28.2 .mu.mol) were then added. The reaction mixture was stirred at room temperature for 3 hours. The reaction mixture was then purified by HPLC (water--acetonitrile gradient 48-72% containing 0.1% formic acid). The eluent was collected at m/z 1409, and lyophilized to afford azido-PEG4-Val-Cit-PAB-eribulin (exact mass 1407.7 Da). 13 mg (9.2 .mu.mol) of azido-PEG4-Val-Cit-PAB-eribulin was obtained (65% step yield, 44% overall).
Example 4
1. Materials and Methods
[0434] All reagents used were obtained from commercial suppliers at research-grade or higher, unless otherwise indicated.
1.1 Antibodies
[0435] MORAb-003 (humanized anti-human folate receptor alpha, 25 mg/mL) and MORAb-009 (mouse-human chimeric anti-human mesothelin, 25 mg/mL) used in the following studies were from Lot # NB02962-19 and Lot #030A14, respectively. Trastuzumab was obtained commercially (Clingen), and was from Lot #503345.
[0436] Rabbit-human chimeric and humanized anti-human mesothelin antibodies having an unpaired cysteine at LCcys80 (Table 1) were expressed in 293F cells transiently or as stabily-selected pools. Conditioned medium was purified and decysteinylated as described in section 1.4.1.2.1.
1.2 Cytotoxins
[0437] Conjugatable eribulin compounds were synthesized as described in Example 3 (Table 46). Stocks (10 mM) were prepared in DMSO and stored at -20.degree. C. until use.
1.3 Tumor Cell Lines
[0438] Human tumor cell lines used in the analyses of MORAb-003, MORAb-009, and trastuzumab ADCs prepared with maleimido/succinimide (OSu)/azido-linker-eribulin compounds (Table 46) include IGROV1 (human ovarian carcinoma, FR.sup.hi, MSLN.sup.neg), NCI-H2110 (human non-small cell lung carcinoma, FR.sup.med, MSLN.sup.med), A431 (FR.sup.neg, MSLN.sup.neg), NCI-N87-luc (human gastric carcinoma, FR.sup.lo, MSLN.sup.med, her2.sup.hi), NUGC3 (human gastric adenocarcinoma, FR.sup.neg, MSLN.sup.neg, her2.sup.neg), ZR75 (human breast ductal carcinoma, FR.sup.neg, MSLN.sup.neg, her2.sup.med), and BT-474 (human breast ductal carcinoma, FR.sup.neg, MSLN.sup.neg, her2.sup.hi). Human tumor cell lines used in the analyses of rabbit-human chimeric and humanized anti-human mesothelin LCcys80 antibodies conjugated with MAL-PEG2-Val-Cit-PAB-eribulin (ER-001159569) were A3 (A431 stabily transfected with human mesothelin, MSLN.sup.hi), OVCAR3 (human ovarian carcinoma, MSLN.sup.hi), HEC-251 (human endometroid, MSLN.sup.med), H226 (human lung squamous cell mesothelioma, MSLN.sup.lo), and A431 parental (MSLN.sup.neg). All cell lines used were obtained directly from the American Type Culture Collection (ATCC), with the exceptions of IGROV1 (obtained from the National Cancer Institute, with permission) and A3 (generated at Morphotek from parental A431).
1.4 Antibody-Drug Conjugation
1.4.1 Cysteine-Based Conjugation Using Maleimides
1.4.1.1 Conjugation to Interchain Disulfides
1.4.1.1.1 Partial Reduction
[0439] MORAb-003 and MORAb-009 were buffer-exchanged into Dulbecco's phosphate-buffered saline (DPBS), and then concentrated to 20 mg/mL using centrifugal concentration. An equal volume of 270 .mu.M tris(2-carboxyethyl)phosphine (TCEP) in 1.times.DPBS with 2 mM EDTA was added, and the reduction was carried out by gentle mixing for 80 min at room temperature. Trastuzumab was partially-reduced in a similar manner, except the reduction was carried out by gentle mixing for 40 min at room temperature.
1.4.1.1.2 Conjugation
[0440] Maleimido-linker-eribulin compound (in DMSO) was conjugated to the partially reduced antibodies at a molar ratio of 1:6 (mAb:compound). The compound was added to 50% propylene glycol in DPBS and mixed well. An equal volume of partially-reduced antibody was then added, and mixed gently (final propylene glycol concentration of 25%). Conjugation proceeded for 3.5 to 4 hours at room temperature.
1.4.1.2 Conjugation to LCcys80
1.4.1.2.1 Decysteinylation
[0441] Using an AKTA Explorer (GE Healthcare), a protein A column (GE Healthcare) was equilibrated with 10 column volumes (CV) of 20 mM sodium phosphate, 10 mM EDTA, pH 7.2 (equilibration buffer). Conditioned medium was then loaded, followed by the washing of unbound material with 10 CV of equilibration buffer. The column was washed with 16 CV of 20 mM sodium phosphate, 10 mM EDTA, 5 mM cysteine, pH 7.2 at 0.5 mL/min for 16 hours to remove the capping group. The column was then washed with 60 CV of 20 mM Tris, pH 7.5 at 0.5 mL/min for 60 hours. The decysteinylated antibody was eluted using 5 CV of 0.1 M glycine, pH 2.9 and immediately neutralized using 5% volume of 2 M Tris, pH 9.0. The fractions containing the antibodies were pooled and dialyzed in DPBS using a MWCO 20K Slide-A-Lyzer (Thermo Fisher).
1.4.1.2.2 Conjugation
[0442] Decysteinylated antibody was brought to 5.0 mg/mL in DPBS, 1 mM EDTA, and 50% propylene glycol was prepared in DPBS, 1 mM EDTA. MAL-PEG2-Val-Cit-PAB-eribulin (ER-001159569) (12 mM in DMSO) was added to the 50% propylene glycol and mixed thoroughly. An equal volume of decysteinylated antibody was then added at a molar ratio of 1:4 (mAb:compound), and mixed gently. Conjugation proceeded for 3.5 to 4 hours at room temperature.
1.4.2 Amine-Based Conjugation Using Succinimides
1.4.2.1 Conjugation
[0443] Antibody (MORAb-003 or MORAb-009, non-reduced) was brought to 10.0 mg/mL in 0.1 M sodium bicarbonate, pH 8.3. 50% propylene glycol was prepared in 0.1 M sodium bicarbonate, pH 8.3. Succinimide (OSu)-linker-eribulin (in DMSO) was added to the 50% propylene glycol and mixed thoroughly. An equal volume of antibody was then added at a molar ratio of 1:4 (mAb:compound), and mixed thoroughly. Conjugation proceeded for 1 hour at room temperature. The conjugation reaction was quenched with the addition of 1:20 volume of 1 M Tris, pH 8.0, and the ADC was purified as described in section 1.4.4.
1.4.3 Two-Step Amine-Based Conjugation Using Strain-Promoted Alkyne-Azide Chemistry (SPAAC)
1.4.3.1 Dybenzylcyclooctyne (DBCO) Derivatization
[0444] Antibody (MORAb-003 or MORAb-009, non-reduced) was brought to 10.0 mg/mL in 0.1 M sodium bicarbonate, pH 8.3. 50% propylene glycol was prepared in 0.1 M sodium bicarbonate, pH 8.3. NHS-PEG4-DBCO (Click Chemistry Tools, 50 mM in DMSO) was added to the 50% propylene glycol and mixed thoroughly. An equal volume of antibody was then added at a molar ratio of 1:4 (mAb:compound), and mixed thoroughly. Conjugation proceeded for 1 hour at room temperature. Unreacted NHS-PEG4-DBCO was removed, as described in section 1.4.4.
1.4.3.2 Conjugation
[0445] 50% propylene glycol was prepared in DPBS. Azido-linker-eribulin compounds were added to the 50% propylene glycol and mixed thoroughly. An equal volume of the DBCO-modified MORAb-003 or MORAb-009 was then added to the mixture at a molar ratio of 1:4 (mAb:compound), and mixed thoroughly. SPAAC conjugation was allowed to proceed overnight at room temperature. Unreacted NHS-PEG4-DBCO was removed, as described in section 1.4.4.
1.4.4 Purification
[0446] Conjugated antibody was purified using HiTrap desalting column(s) (GE Healthcare). Chromatography was performed on a fast protein liquid chromatogaphy (FPLC) (GE Healthcare), using 1.times.DPBS as running buffer, in order to remove maleimido/OSu/azido-linker-eribulin and propylene glycol. Final protein content was determined by BCA assay, as described in section 1.3.1 of Example 1.
1.5 Biophysical Characterization
1.5.1 SEC-HPLC Analysis
[0447] The aggregation of ADCs was analyzed by size-exclusion, high-performance liquid chromatography (SEC-HPLC) using an Agilent 1260 HPLC. ADC was diluted to 1 mg/mL in DPBS. ADC (10 .mu.L) was then injected onto an Advanced SEC 300A guard column (4.6 mm.times.3.5 cm, 2.7 .mu.m pore size, Agilent), followed by a AdvancedBio 300A column (4.6 mm.times.30 cm, 2.7 .mu.m pore size). ADC was eluted from the column with 0.1 M sodium phosphate containing 0.15 M NaCl and 5% IPA, pH 7.4 at a flow rate of 0.25 mL/min for 28 min. All data were analyzed using Agilent ChemStation software. Percent aggregation was calculated as [PA.sub.aggregate/PA.sub.total]*100, where PA=integrated peak area.
1.5.2 HIC-HPLC Analysis of Drug-to-Antibody Ratio (DAR)
[0448] DAR was analyzed using hydrophobic interaction HPLC (HIC-HPLC). Samples were injected onto a TSKgel.RTM. Butyl-NPS, 4.6 mm ID.times.3.5 cm, 2.5 .mu.M nonporous size column (Tosoh Bioscience), and eluted with a 3 min equilibration in 100% of mobile phase A, a 15 min gradient (0-100% B), a 5 min hold in 100% B, a 1 min change to 100% A, and a 5 min re-equilibration in 100% of mobile phase A, at 0.7 mL/min. Mobile phase A was 25 mM sodium phosphate, 1.5 M ammonium sulfate, pH 7.0. Mobile phase B was 25 mM sodium phosphate, 25% isopropanol, pH 7.0. Detection was performed at 280 nm (reference 320 nm). DAR was determined by the formula:
[AUC.sub.+1+2(AUC.sub.+2)+3(AUC.sub.+3)+ . . . n(AUC.sub.+n)]/.SIGMA.AUC.sub.tot]
where AUC.sub.+1 is the area under the curve for the antibody peak corresponding to ADC conjugated with one cytotoxin, AUC.sub.+2 is the area under the curve for the antibody peak corresponding to ADC conjugated with two cytotoxins, etc. .SIGMA.AUC.sub.tot is the combined area under the curve for all peaks.
1.5.3 LC-MS DAR Analysis
[0449] DAR was also analyzed using an LC-MS method with a Waters Alliance HPLC with SQD/PDA detection. Samples were injected onto a Proteomix RP-1000 column (5 .mu.M, 1000 A, 4.6 mm.times.15 cm, Sepax) at 65.degree. C., and eluted with a 3 min equilibration in 25% B, a 27 min linear gradient from 25%-55% B, a 5 min hold at 55% B, a 1 min change to 90% B, a 5 min hold at 90% B, a 1 min change back to 25% B, and a 5 min reequilibration at 25% B. Mobile phase A was 0.1% TFA in water, and mobile phase B was 0.1% TFA in acetonitrile. The elute was then split (10:1) into PDA and SQD detectors. The SQD detector was set up as ES positive, capillary voltage at 3.2 kV, cone voltage at 40 V, extractor at 3 V, and RF lens at 0.2 V, source temperature at 150.degree. C., and desolvation temperature at 250.degree. C. Mass data was acquired at 200-2000 m/z for 40 min, continuum mode, scan time 1 second. Data was analyzed and deconvoluted offline using MassLynx and MaxEnt1. DAR was calculated using the formula:
2[[AUC.sub.LC+1+2(AUC.sub.LC+2)+3(AUC.sub.LC+3)+ . . . n(AUC.sub.LC+n)]/.SIGMA..sub.LCtot]+2[[AUC.sub.HC+1+2(AUC.sub.HC+2)+3(AUC- .sub.HC+3)+ . . . n(AUC.sub.HC+n)]/.SIGMA.AUC.sub.HCtot]
where AUC.sub.LC+1 is the area under the curve of the light chain peak conjugated with one cytotoxin, AUC.sub.LC+2 is the area under the curve of the light chain peak conjugated with two cytotoxins, etc. AUC.sub.HC is the area under the curve of the corresponding heavy chains, and .SIGMA.AUC.sub.LCtot and .SIGMA.AUC.sub.HCtot are the combined area under the curve of all unconjugated and conjugated light chains and heavy chains, respectively.
1.5.4 UPLC/ESI-MS DAR Analysis of LCcys80 ADCs
[0450] ADC (1 mg/mL) was reduced by adding DTT to a final concentration of 20 mM, followed by incubation at 60.degree. C. for 3 min. Samples were then analyzed using a Waters Acquity Ultra Performance Liquid Chromatography and Q-Tof Premier mass spectrometer. Samples (0.5-2 .mu.g each) were injected onto a MassPrep micro desalting column at 65.degree. C., eluted from the column with a 5 min equilibration in 95% of mobile phase A, a 10 min gradient (5-90% B), and a 10 min re-equilibration in 95% of mobile phase A, at 0.05 mL/min. Mobile phase A was 0.1% formic acid in water. Mobile phase B was 0.1% formic acid in acetonitrile. The Q-Tof mass spectrometer was run in positive ion, V-mode with detection in the range of 500-4000 m/z. The source parameters were as follows: capillary voltage, 2.25 kV (intact antibody)-2.50 kV (reduced antibody); sampling cone voltage, 65.0 V (intact antibody) or 50.0 V (reduced antibody); source temperature, 105.degree. C.; desolvation temperature, 250.degree. C.; desolvation gas flow, 550 L/hr. The light chain protein peak was deconvoluted using the MassLynx MaxEnt 1 function. Relative intensities of unconjugated and singly-conjugated light chain masses were used to calculate the overall DAR using the formula:
2[LC.sub.+1/.SIGMA.LC.sub.tot]
where LC.sub.+1 is mass intensity of light chain conjugated with one cytotoxin, and .SIGMA.LC.sub.tot is the combined intensities of unconjugated and conjugated light chain.
1.6 Binding Characterization
1.6.1 BIAcore
[0451] Antibody concentrations were adjusted to 2 .mu.g/mL in HBS-P+ buffer (GE Healthcare). Unmodified antibodies, or ADCs, were injected over an anti-human IgG sensor on a BIAcore T100 (GE Healthcare) for 1 min at a flow rate of 10 .mu.L/min. To record the antigen association to the captured antibody, a series of increasing concentrations of antigen was injected for 300 sec at a flow rate of 30 .mu.L/min. For anti-mesothelin antibodies, the range of concentrations was 10 nM-0.041 nM. For MORAb-003 and MORAb-009 ADCs, the range of concentrations was 100 nM-0.41 nM. The dissociation of antigen was monitored for 30 min at the same flow rate. The sensor surface was regenerated by injecting 3 M MgCl.sub.2 for 2.times.30 sec at a flow rate of 30 .mu.L/min. Sensograms were analyzed with Biacore T100 Evaluation Software using a 1:1 Langmuir binding model.
1.6.2 ELISA--Folate Receptor Alpha
[0452] Recombinant human folate receptor alpha was diluted to 115 ng/mL in coating buffer (50 mM carbonate-bicarbonate buffer, pH 9.6), and coated onto 96-well Maxisorp black plates (Thermo, Cat No. 43711, 100 .mu.L/well) at 4.degree. C., overnight. Coating solution was discarded and the plates were washed three times using 1.times.PBS with 0.05% Tween-20 (PBST) buffer. Plates were blocked in 300 .mu.L blocking buffer (1% BSA in PBST) at room temperature for 2 hours on an orbital shaker. MORAb-003 and MORAb-003 ADCs were diluted to 1000 ng/mL in blocking buffer, then serially-diluted 2-fold to obtain a range from 1000 ng/mL to 0.98 ng/mL. Blocking buffer was discarded and 100 .mu.L/well of diluted antibody was added to the plates. Plates were incubated at room temperature for 2 hours on an orbital shaker. Antibody solution was discarded and plates were washed three times using PBST. 100 .mu.L/well of goat-anti-human IgG (H+L)-HRP (1:10,000 dilution in blocking buffer) solution was added to the plates, and plates were incubated at room temperature for 1 hour on an orbital shaker. Secondary antibody solution was discarded and plates were washed three times using PBST. 100 .mu.L/well of QuantaBlu fluorogenic peroxidase substrate working solution (Thermo, Cat No. 15169) was added to the plates, and plates were incubated at room temperature for 30 min. Fluorescence was read at excitation 325 nm/emission 420 nm using a SpectraMax M5 (Molecular Devices). Data was analyzed using SoftMaxPro 5.4.2 software with 4-parameter fitting.
1.6.3 ELISA--Mesothelin
[0453] Recombinant human mesothelin was diluted to 1 .mu.g/mL in coating buffer (50 mM carbonate-bicarbonate buffer, pH 9.6), and coated onto 96-well Maxisorp black plates (Thermo, Cat No. 43711, 100 .mu.L/well) at 4.degree. C., overnight. Coating solution was discarded and the plates were washed three times using 1.times.PBS with 0.05% Tween-20 (PBST) buffer. Plates were blocked in 300 .mu.L blocking buffer (1% BSA in PBST) at room temperature for 2 hours on an orbital shaker. MORAb009 and MORAb-009 ADCs were diluted to 1000 ng/mL in blocking buffer, then serially-diluted 2.5-fold to obtain a range from 1000 ng/mL to 0.105 ng/mL. Blocking buffer was discarded and 1004/well of diluted antibody was added to the plates. Plates were incubated at room temperature for 2 hours on an orbital shaker. Antibody solution was discarded and plates were washed three times using PBST. 100 .mu.L/well of goat-anti-human IgG (H+L)-HRP (1:10,000 dilution in blocking buffer) solution was added to the plates, and plates were incubated at room temperature for 1 hour on an orbital shaker. Secondary antibody solution was discarded and plates were washed three times using PBST. 100 .mu.L/well of QuantaBlu fluorogenic peroxidase substrate working solution (Thermo, Cat No. 15169) was added to the plates, and plates were incubated at room temperature for 30 min. Fluorescence was read at excitation 325 nm/emission 420 nm using a SpectraMax M5 (Molecular Devices). Data was analyzed using SoftMaxPro 5.4.2 software with 4-parameter fitting.
1.7 Cytotoxicity Analyses
1.7.1 Crystal Violet Assay
[0454] IGROV1 (FR.sup.hi, MSLN.sup.neg), NCI-H2110 (FR.sup.med, MSLN.sup.med), and A431 (FR.sup.neg, MSLN.sup.neg) cells were sub-cultured and seeded at 5,000 cells/well in complete growth medium in 96 well tissue culture plates, incubated at 37.degree. C., 5% CO.sub.2 overnight (16 hours). Test reagents were serial diluted 1:3 in 2 mL deep-well dilution plates, starting at 200 nM (10 dilutions total). Diluted samples (100 .mu.L) were added to the cell plates (starting concentration of test samples at 100 nM). Plates were incubated at 37.degree. C., 5% CO.sub.2 for an additional 5 days. Medium was then discarded. The plates were washed once with 200 .mu.L DPBS, stained with 504 of 0.2% Crystal Violet solution at room temperature for 15 min, and then washed extensively with tap water. Plates were air-dried, and Crystal Violet was dissolved with 2004 of 1% SDS solution. Plates were read at 570 nm. Data was analyzed using GraphPad Prism 6.
2. Results
2.1 Biophysical Characterization of MORAb-003, MORAb-009, and Trastuzumab ADCs
[0455] MORAb-003 (humanized anti-human folate receptor alpha), MORAb-009 (mouse-human chimeric anti-human mesothelin), and trastuzumab (humanized anti-human her2) ADCs were prepared using the conjugatable eribulin compounds listed in Table 46 according to one of three conjugation methods, including: (1) partial reduction of antibody interchain disulfides using the non-thiol reductant TCEP, followed by conjugation using thiol-reactive maleimido-spacer-linker-eribulin constructs; (2) direct conjugation to antibody lysine residues using succinimide (OSu)-spacer-linker-eribulin constructs; and (3) conjugation to antibody lysine residues using a two-step approach, whereby OSu-PEG4-dibenzylcyclooctyne was first conjugated to lysine residues, then orthogonal conjugation of azido-spacer-linker-eribulin constructs was performed using SPAAC.
[0456] Following purification, aggregation levels for all MORAb-003, MORAb-009, and trastuzumab ADCs were determined by SEC-HPLC and the drug-to-antibody ratio (DAR) was analyzed using reverse-phase LC-MS and/or HIC-HPLC. The DAR for all maleimide-based ADCs was analyzed using both reverse-phase LC-MS and HIC-HPLC. A difference in DAR values of less than 0.3 was typically observed between the two methods. In contrast, the DAR for all ADCs prepared via conjugation through lysine residues was analyzed only by LC-MS, since the high degree of heterogeneity of these ADCs prevents the resolution of individual DAR species by HIC-HPLC. Binding to target antigen was also analyzed using ELISA, for MORAb-003 and MORAb-009 ADCs. The results of the DAR and aggregation analyses are shown in Table 47 next to the respective ADC.
TABLE-US-00047 TABLE 47 Biophysical analyses of MORAb-003, MORAb-009, and trastuzumab ADCs DAR Analysis Antigen Binding DAR DAR SEC-HPLC Analysis ELISA, ELISA, conjugation cleavage (LC- (HIC- % % % EC.sub.50, EC.sub.50, ADCs antibody chemistry spacer chemistry MS) HPLC) Aggr. Monomer Frag. ng/mL nM MORAb003 N/A N/A N/A 3.62 96.38 0 6.29 0.04 MORAb009 N/A N/A N/A 0 100 0 42.60 0.28 trastuzumab N/A N/A N/A 3.52 96.48 0 N/A N/A MORAb003- MORAb-003 maleimide PEG2 val-cit-pAB 3.58 3.91 3.12 96.88 0 22.60 0.15 ER1159569 (Lot NB3073-88L) MORAb009- MORAb-009 maleimide PEG2 val-cit-pAB 3.63 3.93 3.23 96.77 0 43.70 0.29 ER1159569 (Lot NB3073-88F) MORAb003- MORAb-003 maleimide PEG2 val-cit-pAB 4.80 4.88 3.21 96.79 0 18.20 0.12 ER1159569 (Lot NB3142-62A) MORAb009- MORAb-009 maleimide PEG2 val-cit-pAB 4.68 4.57 0.90 99.10 0 33.10 0.22 ER1159569 (Lot NB3142-62D) trastuzumab- trastuzumab maleimide PEG2 val-cit-pAB 3.10 3.11 1.26 98.74 0 N/A N/A ER1159569 MORAb003- MORAb-003 maleimide PEG8 val-cit-pAB 2.31 2.35 18.63 81.37 0 21.50 0.14 ER1242287 MORAb009- MORAb-009 maleimide PEG8 val-cit-pAB 1.13 2.00 11.24 88.76 0 58.60 0.39 ER1242287 MORAb003- MORAb-003 maleimide pentyl val-cit-pAB 3.65 3.89 3.95 96.05 0 15.30 0.10 ER1235638 MORAb009- MORAb-009 maleimide pentyl val-cit-pAB 3.99 4.10 4.5 95.5 0 65.60 0.44 ER1235638 MORAb003- MORAb-003 maleimide PEG2 ala-ala-asn- 3.60 3.83 3.09 96.91 0 18.30 0.12 ER1231679 pAB MORAb009- MORAb-009 maleimide PEG2 ala-ala-asn- 3.27 3.94 4.39 95.61 0 41.40 0.28 ER1231679 pAB MORAb003- MORAb-003 maleimide PEG2 ala-ala-asn- 3.02 3.23 4.44 95.56 0 8.92 0.06 ER1231690 pAB-ala-ala- asn-pAB MORAb009- MORAb-009 maleimide PEG2 ala-ala-asn- 2.36 3.17 6.22 93.78 0 58.70 0.39 ER1231690 pAB-ala-ala- asn-pAB MORAb003- MORAb-003 maleimide PEG4-triazole-PEG3 disylfidyl- 0.52 1.61 13.73 86.27 0 29.80 0.20 ER1237504 dimethyl- pAB MORAb009- MORAb-009 maleimide PEG4-triazole-PEG3 disylfidyl- 0.72 1.03 9.78 90.22 0 55.90 0.37 ER1237504 dimethyl- pAB MORAb003- MORAb-003 maleimide PEG4-triazole-PEG3 sulfonamide 1.85 3.88 5.72 94.28 0 18.30 0.12 ER1237505 MORAb009- MORAb-009 maleimide PEG4-triazole-PEG3 sulfonamide 2.33 3.91 5.44 94.56 0 61.00 0.41 ER1237505 MORAb003- MORAb-003 maleimide PEG2 non-cleavable 4.15 4.49 3.97 96.03 0 6.96 0.05 PEG2-eribulin MORAb009- MORAb-009 maleimide PEG2 non-cleavable 4.55 4.30 1.15 97.11 1.74 8.84 0.06 PEG2-eribulin MORAb003- MORAb-003 maleimide PEG4 non-cleavable 4.70 4.79 9.84 89.76 0 9.31 0.06 PEG4-eribulin MORAb009- MORAb-009 maleimide PEG4 non-cleavable 4.48 4.57 1.03 97.13 1.84 11.60 0.08 PEG4-eribulin MORAb003- MORAb-003 succinimide PEG2 val-cit-pAB 0.72 3.65 96.35 0 17.00 0.11 ER1236940 MORAb009- MORAb-009 succinimide PEG2 val-cit-pAB 0.89 2.75 97.25 0 66.30 0.44 ER1236940 MORAb003- MORAb-003 succinimide PEG9 val-cit-pAB 0.00 2.85 97.15 0 14.40 0.10 ER1242288 MORAb009- MORAb-009 succinimide PEG9 val-cit-pAB 0.21 1.69 98.31 0 15.30 0.10 ER1242288 MORAb003- MORAb-003 succinimide pentyl val-cit-pAB 0.77 3.13 96.87 0 13.00 0.09 ER1236941 MORAb009- MORAb-009 succinimide pentyl val-cit-pAB 0.93 3.04 96.96 0 44.60 0.30 ER1236941 MORAb003- MORAb-003 succinimide PEG3-triazole-PEG3 val-cit-pAB 0.00 3.92 96.08 0 6.22 0.04 ER1243700 MORAb009- MORAb-009 succinimide PEG3-triazole-PEG3 val-cit-pAB 0.06 1.97 98.03 0 46.70 0.31 ER1243700 MORAb003- MORAb-003 succinimide PEG2 ala-ala-asn- 0.37 3.46 96.54 0 11.50 0.08 ER1231691 pAB MORAb009- MORAb-009 succinimide PEG2 ala-ala-asn- 0.29 2.45 97.55 0 43.30 0.29 ER1231691 pAB MORAb003- MORAb-003 succinimide PEG3-triazole-PEG3 disylfidyl- 0.24 10.87 89.13 0 14.30 0.10 ER1244129 dimethyl- pAB MORAb009- MORAb-009 succinimide PEG3-triazole-PEG3 disylfidyl- 0.47 12.79 87.21 0 57.70 0.38 ER1244129 dimethyl- pAB MORAb003- MORAb-003 succinimide PEG3-triazole-PEG3 sulfonamide 0.55 5.21 94.79 0 4.54 0.03 ER1244623 MORAb009- MORAb-009 succinimide PEG3-triazole-PEG3 sulfonamide 1.14 0 100 0 39.00 0.26 ER1244623 MORAb003- MORAb-003 succinimide/ dibenzylcyclooctene- disylfidyl- 2.19 4.1 95.9 0 24.10 0.16 DBCO- click triazole-PEG3 dimethyl- ER1237508 pAB MORAb009- MORAb-009 succinimide/ dibenzylcyclooctene- disylfidyl- 2.33 0 100 0 53.80 0.36 DBCO- click triazole-PEG3 dimethyl- ER1237508 pAB MORAb003- MORAb-003 succinimide/ dibenzylcyclooctene- sulfonamide 1.82 3.49 96.51 0 15.00 0.10 DBCO- click triazole-PEG3 ER1138856 MORAb009- MORAb-009 succinimide/ dibenzylcyclooctene- sulfonamide 1.59 0 100 0 44.70 0.30 DBCO- click triazole-PEG3 ER1138856 MORAb003- MORAb-003 succinimide/ dibenzylcyclooctene- val-cit-pAB 3.09 2.87 97.13 0 16.00 0.11 DBCO-PEG4 click triazole-PEG4 VCP eribulin MORAb009- MORAb-009 succinimide/ dibenzylcyclooctene- val-cit-pAB 2.91 0.22 99.78 0 33.70 0.22 DBCO-PEG4 click triazole-PEG4 VCP eribulin MORAb003- MORAb-003 succinimide/ dibenzylcyclooctene- non-cleavable 3.43 3.88 96.12 0 19.10 0.13 DBCO-PEG2 click triazole-PEG2 eribulin MORAb009- MORAb-009 succinimide/ dibenzylcyclooctene- non-cleavable 3.07 1.15 98.85 0 23.30 0.16 DBCO-PEG2 click triazole-PEG2 eribulin MORAb003- MORAb-003 succinimide/ dibenzylcyclooctene- non-cleavable 2.96 3.64 96.36 0 13.30 0.09 DBCO-PEG4 click triazole-PEG4 eribulin MORAb009- MORAb-009 succinimide/ dibenzylcyclooctene- non-cleavable 2.8 1.12 98.88 0 45.20 0.30 DBCO-PEG4 click triazole-PEG4 eribulin Abbreviations: % Aggr., % aggregation; % Frag, % fragmentation.
2.1.1 MORAb-003, MORAb-009, and Trastuzumab ADCs
[0457] No significant differences between MORAb-003, MORAb-009, and trastuzumab were observed, in terms of both conjugation efficiency and biophysical parameters. All ADCs demonstrated similar DAR values and levels of aggregrate formation.
2.1.2 Maleimide-Based ADCs
[0458] For maleimide-based ADCs, both pentyl and PEG2 spacers paired with a val-cit-pAB cleavage site, and a PEG2 spacer paired with an ala-ala-asn-pAB cleavage site, provided DAR values between 3.5 and 4.0 by reverse-phase LC-MS and HIC-HPLC, in addition to low (<5%) aggregate levels. However, when the spacer was lengthened to PEG.sub.8 (paired with a val-cit-pAB cleavage site), aggregate levels increased (11-18%) and conjugation efficiency decreased, resulting in DAR values between 1.1 and 2.3. See, e.g., percent aggregation and DAR values of MORAb003/MORAb009-ER-001159569 (short PEG linker) and MORAb003/MORAb009-1242287 (long PEG linker) in Table 47.
[0459] For ADCs prepared with a disulfidyl-pAB cleavage site, low DAR values were observed (1.0-1.6), together with relatively high aggregate levels (10-14%). Significantly lower DAR values were observed when these ADCs were analyzed by LC-MS than by HIC-HPLC (see, e.g., LC-MS/HIC-HPLC DAR values for MORAb003/MORAb009-ER1237504 and MORAb003/MORAb009-ER1237505 in Table 47). This result suggests the linker cleavage site exhibits pH instability, as the mobile phase of LC-MS analysis is approximately 3.0, whereas the mobile phase of HIC-HPLC analysis is neutral.
[0460] For ADCs prepared with a sulfonamide cleavage site, low (<5%) aggregate levels were observed. Similar to the disulfidyl-pAB ADCs, lower DAR values were observed when analyzed by LC-MS (1.8-2.3) than by HIC-HPLC (3.9), which again indicates that the linker cleavage site exhibits pH instability.
[0461] For the PEG2 and PEG4 non-cleavable linkers, efficient conjugation was observed, resulting in DAR values between 4.0 and 4.7. MORAb-009 ADCs with these non-cleavable linkers also demonstrated low aggregation levels (<2%), while slightly higher aggregation levels were observed for the corresponding MORAb-003 ADCs (4% and 10% for PEG2 and PEG4, respectively).
2.1.3 Succinimide-Based ADCs
[0462] All ADCs prepared using succinimide coupled with spacer-linker-eribulin resulted in DAR values <1.0. To confirm that this lower conjugation efficiency (relative to maleimides) was not a consequence of the conjugation procedure itself, these ADCs were remade using a higher compound:antibody ratio and reanalyzed using the same DAR analysis methods. Similar results were obtained, which suggests, without being bound by theory, that lower DAR values are an inherent property of the combination of succinimide and eribulin, and that maleimides may be conjugated more efficiently. Efficiency of succinimide conjugation was increased through use of a two-step method, whereby DBCO was first added to the antibody using NHS-DBCO, followed by the addition of the azido compounds. This approach results in higher DAR values, as measured by reverse-phase HPLC analysis, as compared to conjugation directly to antibody lysine residues. For succinimide-based ADCs having sulfonamide (cleavable), val-cit-PAB (cleavable), or PEG2/PEG4 (non-cleavable) linkers, DAR values resulting from the two-step conjugation were similar to those determined for maleimide-based ADCs having a sulfonamide cleavage site. Without being bound by theory, this result again suggests that lower DAR values for succinimide-spacer-linker-eribulin conjugation reactions are an inherent property of the combination of succinimide and eribulin.
2.2 Binding Characterization of MORAb-003 and MORAb-009 ADCs
[0463] For MORAb-003 ADCs, no significant differences were observed between non-cleavable maleimide-based linker-eribulin ADCs and parental MORAb-003 in terms of target antigen binding. For other maleimide-based linker-eribulin MORAb-003 ADCs, a 2- to 3-fold loss in target antigen binding relative to parental MORAb-003 was typically observed by ELISA analysis. However, there was no apparent correlation between either linker length or linker composition and lower EC.sub.50 values. Similarly, for succinimide-based linker-eribulin MORAb-003 ADCs, a 0- to 3-fold loss in target antigen binding relative to unconjugated MORAb-003 was generally observed. Again, no correlation between either linker length or linker composition and lower EC.sub.50 values was apparent. For MORAb-009 ADCs, all ADCs had less than a 2-fold decrease in EC.sub.50 values, relative to parental MORAb-009.
2.3 In Vitro Cytoxicity Analyses of MORAb-003, MORAb-009, and Trastuzumab ADCs
[0464] In vitro potency of prepared MORAb-003, MORAb-009, and trastuzumab ADCs was evaluated using a Crystal Violet cell-based cytotoxicity assay. The cell lines selected for screening MORAb-003 and MORAb-009 ADCs were IGROV1, NCI-H2110, and A431. IGROV1 cells are of human ovarian epithelial carcinoma origin and express high levels of folate receptor alpha, but no mesothelin (i.e., MORAb-003-reactive). NCI-H2110 cells are of human non-small cell lung carcinoma origin and express moderate levels of both folate receptor alpha and mesothelin (i.e., MORAb-003- and MORAb-009-reactive). A431 control cells are of human epidermal carcinoma origin and do not express either target antigen. The results of this screening are shown in Table 48. MORAb-003, MORAb-009, and trastuzumab ADCs comprising the linker-toxin maleimido-PEG2-val-cit-pAB-eribulin (VCP-eribulin) were also evaluated in additional gastric and breast cancer cell lines, including NCI-N87 (FR.sup.lo, MSLN.sup.med, her2.sup.hi), BT-474 (FR.sup.neg, MSLN.sup.neg, her2.sup.hi), ZR-75 (FR.sup.neg, MSLN.sup.neg, her2.sup.med), and NUGC3 (FR.sup.neg, MSLN.sup.neg, her2.sup.neg). The results of this screening are shown in Table 49.
TABLE-US-00048 TABLE 48 Cytotoxicity (IC.sub.50) screening of MORAb-003 and MORAb-009 ADCs on IGROV1, NCI-H2110, and A431 cells Cytotoxicity Analysis IGROV1 NCI-H2110 A431 conjugation cleavage (FR.sup.hi, MSLN.sup.neg) (FR.sup.med, MSLN.sup.med) (FR.sup.neg, MSLN.sup.neg) ADCs antibody chemistry spacer chemistry IC.sub.50 (nM) SD IC.sub.50 (nM) SD IC.sub.50 (nM) SD MORAb003 N/A N/A N/A N/A N/A N/A N/A N/A N/A MORAb009 N/A N/A N/A N/A N/A N/A N/A N/A N/A trastuzumab N/A N/A N/A eribulin N/A N/A N/A N/A 0.320 0.212 0.199 0.034 0.653 0.159 MORAb003- MORAb-003 maleimide PEG2 val-cit-pAB 0.155 0.064 3.685 0.417 >100 ER1159569 (Lot NB3073-88L) MORAb009- MORAb-009 maleimide PEG2 val-cit-pAB 9.450 2.093 14.945 1.747 >100 ER1159569 (Lot NB3073-88F) MORAb003- MORAb-003 maleimide PEG2 val-cit-pAB 0.020 1.550 >100 ER1159569 (Lot NB3142-62A) MORAb009- MORAb-009 maleimide PEG2 val-cit-pAB 5.687 6.784 >100 ER1159569 (Lot NB3142-62D) trastuzumab- trastuzumab maleimide PEG2 val-cit-pAB ER1159569 MORAb003- MORAb-003 maleimide PEG8 val-cit-pAB 0.115 0.035 7.065 0.417 85.960 ER1242287 MORAb009- MORAb-009 maleimide PEG8 val-cit-pAB 25.765 8.478 34.455 3.033 >100 ER1242287 MORAb003- MORAb-003 maleimide pentyl val-cit-pAB 0.105 0.092 3.920 1.032 >100 ER1235638 MORAb009- MORAb-009 maleimide pentyl val-cit-pAB 6.830 0.962 13.965 6.611 >100 ER1235638 MORAb003- MORAb-003 maleimide PEG2 ala-ala-asn- 0.080 0.028 3.800 0.566 31.630 1.202 ER1231679 pAB MORAb009- MORAb-009 maleimide PEG2 ala-ala-asn- 8.890 0.976 7.080 1.867 34.390 3.536 ER1231679 pAB MORAb003- MORAb-003 maleimide PEG2 ala-ala-asn- 0.125 0.021 4.745 2.114 38.555 0.403 ER1231690 pAB-ala-ala- asn-pAB MORAb009- MORAb-009 maleimide PEG2 ala-ala-asn- 16.980 5.176 12.310 3.422 54.960 5.360 ER1231690 pAB-ala-ala- asn-pAB MORAb003- MORAb-003 maleimide PEG4-triazole- disylfidyl- 0.265 0.092 0.845 0.177 7.005 0.290 ER1237504 PEG3 dimethyl- pAB MORAb009- MORAb-009 maleimide PEG4-triazole- disylfidyl- 6.375 2.751 1.220 0.325 8.130 0.608 ER1237504 PEG3 dimethyl- pAB MORAb003- MORAb-003 maleimide PEG4-triazole- sulfonamide 0.370 0.269 0.690 0.283 6.800 0.834 ER1237505 PEG3 MORAb009- MORAb-009 maleimide PEG4-triazole- sulfonamide 6.370 3.012 0.990 0.453 9.030 1.527 ER1237505 PEG3 MORAb003- MORAb-003 maleimide PEG2 non-cleavable 0.330 38.300 >100 PEG2-eribulin MORAb009- MORAb-009 maleimide PEG2 non-cleavable 42.770 50.040 >100 PEG2-eribulin MORAb003- MORAb -003 maleimide PEG4 non-cleavable 0.277 21.630 >100 PEG4-eribulin MORAb009- MORAb-009 maleimide PEG4 non-cleavable 76.320 31.600 >100 PEG4-eribulin MORAb003- MORAb-003 succinimide PEG2 val-cit-pAB 0.325 0.106 30.545 3.132 >100 ER1236940 MORAb009- MORAb-009 succinimide PEG2 val-cit-pAB 31.915 2.510 36.500 11.031 90.060 ER1236940 MORAb003- MORAb-003 succinimide PEG9 val-cit-pAB 38.105 45.601 64.010 8.075 >100 ER1242288 MORAb009- MORAb-009 succinimide PEG9 val-cit-pAB >100 >100 >100 ER1242288 MORAb003- MORAb-003 succinimide pentyl val-cit-pAB 0.330 0.071 42.105 12.594 >100 ER1236941 MORAb009- MORAb-009 succinimide pentyl val-cit-pAB >100 49.485 13.569 >100 ER1236941 MORAb003- MORAb-003 succinimide PEG3-triazole- val-cit-pAB 1.150 >100 >100 ER1243700 PEG3 MORAb009- MORAb-009 succinimide PEG3-triazole- val-cit-pAB >100 >100 >100 ER1243700 PEG3 MORAb003- MORAb-003 succinimide PEG2 ala-ala-asn- 12.320 31.795 4.448 >100 ER1231691 pAB MORAb009- MORAb-009 succinimide PEG2 ala-ala-asn- >100 20.000 5.954 >100 ER1231691 pAB MORAb003- MORAb-003 succinimide PEG3-triazole- disylfidyl- 0.370 0.184 0.750 0.071 12.005 1.534 ER1244129 PEG3 dimethyl-pAB MORAb009- MORAb-009 succinimide PEG3-triazole- disylfidyl- 6.595 4.052 0.840 0.057 9.230 0.014 ER1244129 PEG3 dimethyl-pAB MORAb003- MORAb-003 succinimide PEG3-triazole- sulfonamide 0.980 0.396 1.820 0.410 37.235 15.733 ER1244623 PEG3 MORAb009- MORAb-009 succinimide PEG3-triazole- sulfonamide 24.505 4.702 2.235 0.629 36.665 14.206 ER1244623 PEG3 MORAb003- MORAb-003 succinimide/ dibenzyl- disylfidyl- 0.545 0.389 0.900 0.071 9.670 0.382 DBCO- click cyclooctene- dimethyl-pAB ER1237508 triazole-PEG3 MORAb009- MORAb-009 succinimide/ dibenzyl- disylfidyl- 10.245 3.486 1.040 0.297 11.280 2.277 DBCO- click cyclooctene- dimethyl-pAB ER1237508 triazole-PEG3 MORAb003- MORAb-003 succinimide/ dibenzyl- sulfonamide 1.775 1.421 1.655 0.007 24.990 2.022 DBCO- click cyclooctene- ER1138856 triazole-PEG3 MORAb009- MORAb-009 succinimide/ dibenzyl- sulfonamide 19.155 5.438 1.960 0.113 28.070 0.636 DBCO- click cyclooctene- ER1138856 triazole-PEG3 MORAb003- MORAb-003 succinimide/ dibenzyl- val-cit-pAB 0.038 4.281 >100 DBCO-PEG4 click cyclooctene- VCP eribulin triazole-PEG4 MORAb009- MORAb-009 succinimide/ dibenzyl- val-cit-pAB 12.960 31.400 >100 DBCO-PEG4 click cyclooctene- VCP eribulin triazole-PEG4 MORAb003- MORAb-003 succinimide/ dibenzyl- non-cleavable 4.250 38.070 >100 DBCO-PEG2 click cyclooctene- eribulin triazole-PEG2 MORAb009- MORAb-009 succinimide/ dibenzyl- non-cleavable 75.680 85.680 >100 DBCO-PEG2 click cyclooctene- eribulin triazole-PEG2 MORAb003- MORAb-003 succinimide/ dibenzyl- non-cleavable 1.323 46.280 >100 DBCO-PEG4 click cyclooctene- eribulin triazole-PEG4 MORAb009- MORAb-009 succinimide/ dibenzyl- non-cleavable 61.490 39.330 >100 DBCO-PEG4 click cyclooctene- eribulin triazole-PEG4 All IC.sub.50 values are in nM, and represent mean values of replicate experiments. SD--standard deviation.
TABLE-US-00049 TABLE 49 Cytotoxicity (IC.sub.50) screening of MORAb-003, MORAb-009, and trastuzumab ADCs on NCI-N87, BT-474, ZR-75, and NUGC3 cells Cytotoxicity Analysis NCI-N87-Luc BT-474 ZR-75-1 NUGC3-Luc (FR.sup.lo, MSLN.sup.med, (FRn.sup.eg, MSLN.sup.neg, (FR.sup.neg, MSLN.sup.neg, (FR.sup.neg, MSLN.sup.neg, conjugation cleavage her2.sup.hi) her2.sup.hi) her2.sup.med) her2.sup.neg) ADCs antibody chemistry spacer chemistry IC.sub.50 (nM) IC.sub.50 (nM) IC.sub.50 (nM) IC.sub.50 (nM) MORAb003 N/A N/A N/A MORAb009 N/A N/A N/A trastuzumab N/A N/A N/A 0.78 0.641 >100 >100 eribulin N/A N/A N/A N/A 0.257 0.151 0.236 0.445 MORAb003- MORAb-003 maleimide PEG2 val-cit-pAB ER1159569 (Lot NB3073-88L) MORAb009- MORAb-009 maleimide PEG2 val-cit-pAB ER1159569 (Lot NB3073-88F) MORAb003- MORAb-003 maleimide PEG2 val-cit-pAB 4.528 11.46 14.74 20.45 ER1159569 (Lot NB3142-62A) MORAb009- MORAb-009 maleimide PEG2 val-cit-pAB 0.013 10.21 12.8 29.93 ER1159569 (Lot NB3142-62D) trastuzumab- trastuzumab maleimide PEG2 val-cit-pAB 0.006 0.003 0.023 20.06 ER1159569 All IC50 values are in nM, and represent mean values of replicate experiments. SD--standard deviation.
2.3.1 Cytotoxicity of Maleimide-Based ADCs
[0465] All maleimide-based MORAb-003 and MORAb-009 ADCs displayed specific cytotoxicity on IGROV1 cells, with a 2-3 orders of magnitude difference in potency observed between antibodies. The val-cit-pAB-eribulin MORAb-003 ADCs demonstrated higher potency on the IGROV1 cell line than either the PEG2 or PEG4 non-cleavable MORAb-003 ADCs, but fold-specificity was unchanged. Similar trends were observed for MORAb-009 ADCs, with the non-cleavable MORAb-009 ADCs demonstrating lower cytotoxicity on IGROV1 cells than val-cit-pAB-eribulin MORAb-009 ADCs.
[0466] Maleimide-based MORAb-009 ADCs with disulfidyl- and sulfonamide-based linkers demonstrated higher potency on the NCI-H2110 cell line than the IGROV1 cell line. This may be due to the potential instability of the linkers in culture, as described below. Potent cytotoxicity was also observed with the corresponding MORAb-003 ADCs. In contrast, maleimide-based MORAb-003 and MORAb-009 ADCs with non-cleavable linkers demonstrated relatively low potency on NCI-H2110 cells. Without being bound by theory, this result suggests that with lower target expression, efficient cleavage and release of the payload may improve cytotoxicity.
[0467] ADCs with a val-cit-pAB enzyme-cleavable linker or a non-cleavable linker demonstrated low levels of off-target killing on A431 control cells (IC.sub.50>100 nM), whereas ADCs with an ala-ala-asn-pAB enzyme-cleavable linker displayed weak but detectable killing of these control cells. This indicates that val-cit-pAB enzyme-cleavable linkers may be more stable in culture ala-ala-asn-pAB enzyme-cleavable linkers. In addition, MORAb-009 ADCs with a shorter PEG2 spacer demonstrated higher cytoxicity in IGROV1 cells than corresponding ADCs with a longer PEG.sub.8 spacer. This same trend was observed in NCI-H2110 cells for both MORAb-003 and MORAb-009 ADCs, with shorter spacer lengths resulting in higher cytotoxicity.
[0468] ADCs with sulfonamide-based linkers generally demonstrated higher DAR values and lower aggregate levels than the corresponding ADCs with disulfidyl-based linkers. However, nM-level killing of A431 control cells was observed in both of these categories of ADCs, suggesting that the disulfidyl- and sulfonamide-based linkers were less stable in culture than the enzyme-cleavable linkers under the assay conditions examined.
[0469] The specific linker-toxin maleimido-PEG2-val-cit-pAB-eribulin (VCP-eribulin) was further examined for specificity and potency on different gastric and breast cancer cell lines. VCP-eribulin was conjugated to MORAb-003 and MORAb-009, in addition to the anti-human her2 antibody trastuzumab. MORAb-003-VCP-eribulin demonstrated weak but specific killing on NCI-N87 cells, which express low levels of folate receptor alpha (FR), and little killing on the remaining three FR-negative cell lines. MORAb-009-VCP-eribulin also demonstrated potent cytotoxicity on NCI-N87 cells, which express moderate levels of mesothelin. Trastuzumab-VCP-eribulin was very potent (3-6 pM, IC.sub.50) on NCI-N87 and BT-474 cells, the two cell lines that express high levels of her2, and also potent on ZR-75 breast cancer cells, which only moderately express her2. MORAb-003, MORAb-009, and trastuzumab VCP-eribulin ADCs all demonstrated low cytotoxicity on NUGC3 cells, with do not express FR, mesothelin, or her2, the respective target antigens.
2.3.2 Cytoxicity of Succinimide-Based ADCs
[0470] Trends in cytotoxicity of the succinimide-based ADCs were similar to the maleimide-based ADCs for IGROV1 cells, with PEG.sub.8 spacer ADCs demonstrating low cytotoxicity in addition to low DAR values. Lower cytotoxicity on both IGROV1 and NCI-H2110 cells was generally observed for succinimide-based ADCs with enzyme-cleavable linkers compared with the corresponding maleimide-based ADCs, which was most likely due to their lower DAR values. Off-target killing of A431 cells was also observed with the disulfidyl- and sulfonamide-based linkers, similar to the corresponding maleimide-based ADCs. This points to increased instability potentially arising from the cleavage site, rather than the conjugation chemistry.
[0471] When a two-step conjugation was performed, higher DAR values were observed relative to those obtained with the direct succinimide conjugation approach. These higher DAR values correlated with higher potency. For the VCP-eribulin MORAb-003 ADC, potent cytotoxicity on both IGROV1 and NCI-H2110 cells was observed. While non-cleavable MORAb-003 ADCs demonstrated potency on IGROV1 cells (1-4 nM), they were still less potent than the VCP-eribulin MORAb-003 ADC prepared with this method (38 pM), even though DAR values were comparable. In addition, non-cleavable MORAb-003 ADCs prepared using the two-step method were slightly less potent than the corresponding maleimide-based ADCs on the IGROV1 cell line, which may be due to their lower DAR values. Similar to their maleimide-based counterparts, non-cleavable ADCs prepared using the two-step method also lost nearly all cytotoxicity on NCI-2110 cells.
2.4 Biophysical Characterization of Anti-Human Mesothelin (LCcys80) ADCs
[0472] MAL-PEG2-Val-Cit-PAB-eribulin (ER-001159569) was conjugated to eight different anti-human mesothelin antibodies (Table 1). Binding affinities of the parental antibodies were determined by BIAcore analysis, as described above in section 1.6.1. Aggregation levels for all anti-human mesothelin ADCs were determined by SEC-HPLC and the DAR was analyzed using HIC-HPLC. In vitro potency was evaluated using a Crystal Violet cell-based cytotoxicity assay in A3 (A431 stabily transfected with human mesothelin (MSLN), MSLN.sup.hi), OVCAR3 (human ovarian, MSLN.sup.hi), HEC-251 (human endometroid, MSLN.sup.med), H226 (human lung squamous cell mesothelioma,) MSLN.sup.lo, and A431 parental (MSLN.sup.neg) cells. The results of the DAR, aggregation, and cytotoxicity analyses are shown in Table 50.
TABLE-US-00050 TABLE 50 Biophysical characterization of anti-human mesothelin (LCcys80) ADCs Parental MAb Affinity ADC k.sub.a k.sub.d SEC-HPLC Cell based Cytotoxicity assay, (10.sup.5M.sup.-l (10.sup.-3 K.sub.D Payload HIC % % EC50, nM sec.sup.-1) sec.sup.-1) (10.sup.-9M) Drug-linker DAR aggregate monomer A431 OVCAR3 HEC-251 H226 A3 33O11 xi 1.92 8.97 91.03 40.67 0.008 3.950 >100 0.14 zu 2.2 0.65 3.4 ER-001159569-000 1.69 1.42 98.58 ~100 0.064 26.500 >100 0.28 111B10 xi 6.5 3.9 6.3 ER-001159569-000 1.90 4.25 95.75 38.10 0.004 13.960 ~100 0.05 zu 5.1 3 6.5 ER-001159569-000 1.81 3.64 96.36 68.92 0.014 27.42 >100 0.12 201C15 xi 2.4 0.26 1.1 ER-001159569-000 1.85 1.62 98.38 48.50 0.004 14.82 ~100 0.27 zu 3.1 1.1 4.2 ER-001159569-000 1.80 5.84 94.16 68.88 0.290 20.42 >100 0.41 346C6 xi 3.8 0.49 1.4 ER-001159569-000 1.56 5.28 94.72 34.49 0.087 5.73 ~100 0.11 zu 133 93 8.9 ER-001159569-000 1.63 4.48 95.52 72.86 1.180 32.54 >100 0.55 Abbreviations: xi--chimeric; zu--humanized
[0473] All anti-human mesothelin ADCs retained low aggregation levels (<10% aggregate) and demonstrated high potency on target cell lines. High potency was observed on A3 and OVCAR3, whereas HEC-251 and H226 cells were relatively resistant to ADC cytotoxicity.
Selected Sequences:
TABLE-US-00051 [0474] SEQ ID NO: 1 (MORAb-003 Heavy chain (HC)) 1 EVQLVESGGG VVQPGRSLRL SCSASGFTFS GYGLSWVRQA PGKGLEWVAM 51 ISSGGSYTYY ADSVKGRFAI SRDNAKNTLF LQMDSLRPED TGVYFCARHG 101 DDPAWFAYWG QGTPVTVSSA STKGPSVFPL APSSKSTSGG TAALGCLVKD 151 YFPEPVTVSW NSGALTSGVH TFPAVLQSSG LYSLSSVVTV PSSSLGTQTY 201 ICNVNHKPSN TKVDKKVEPK SCDKTHTCPP CPAPELLGGP SVFLFPPKPK 251 DTLMISRTPE VTCVVVDVSH EDPEVKFNWY VDGVEVHNAK TKPREEQYNS 301 TYRVVSVLTV LHQDWLNGKE YKCKVSNKAL PAPIEKTISK AKGQPREPQV 351 YTLPPSRDEL TKNQVSLTCL VKGFYPSDIA VEWESNGQPE NNYKTTPPVL 101 DSDGSFFLYS KLTVDKSRWQ QGNVFSCSVM HEALHNHYTQ KSLSLSPGK SEQ ID NO: 2 (MORAb-003 HC CDR1; Kabat): GYGLS SEQ ID NO: 3 (MORAb-003 HC CDR2; Kabat): MISSGGSYTYYADSVKG SEQ ID NO: 4 (MORAb-003 HC CDR3; Kabat): HGDDPAWFAY SEQ ID NO: 5 (MORAb-003 Heavy Chain full length pre-protein amino acid sequence; leader sequence underlined) 1 MGWSCIILFL VATATGVHSE VQLVESGGGV VQPGRSLRLS CSASGFTFSG 51 YGLSWVRQAP GKGLEWVAMI SSGGSYTYYA DSVKGRFAIS RDNAKNTLFL 101 QMDSLRPEDT GVYFCARHGD DPAWFAYWGQ GTPVTVSSAS TKGPSVFPLA 151 PSSKSTSGGT AALGCLVKDY FPEPVTVSWN SGALTSGVHT FPAVLQSSGL 201 YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KVDKKVEPKS CDKTHTCPPC 251 PAPELLGGPS VFLFPPKPKD TLMISRTPEV TCVVVDVSHE DPEVKFNWYV 301 DGVEVHNAKT KPREEQYNST YRVVSVLTVL HQDWLNGKEY KCKVSNKALP 351 APIEKTISKA KGQPREPQVY TLPPSRDELT KNQVSLTCLV KGFYPSDIAV 401 EWESNGQPEN NYKTTPPVLD SDGSFFLYSK LTVDKSRWQQ GNVFSCSVMH 451 EALHNHYTQK SLSLSPGK SEQ ID NO: 6 (MORAb-003 Light chain (LC)) 1 DIQLTQSPSS LSASVGDRVT ITCSVSSSIS SNNLHWYQQK PGKAPKPWIY 51 GTSNLASGVP SRFSGSGSGT DYTFTISSLQ PEDIATYYCQ QWSSYPYMYT 101 FGQGTKVEIK RTVAAPSVFI FPPSDEQLKS GTASVVCLLN NFYPREAKVQ 151 WKVDNALQSG NSQESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT 201 HQGLSSPVTK SFNRGEC SEQ ID NO: 7 (MORAb-003 LC CDR1; Kabat): SVSSSISSNNLH SEQ ID NO: 8 (MORAb-003 LC CDR2: Kabat): GTSNLAS SEQ ID NO: 9 (MORAb-003 LC CDR3; Kabat): QQWSSYPYMYT SEQ ID NO: 10 MORAb-003 Light Chain full length pre-protein amino acid sequence (leader sequence underlined) 1 MGWSCIILFL VATATGVHSD IQLTQSPSSL SASVGDRVTI TCSVSSSISS 51 NNLHWYQQKP GKAPKPWIYG TSNLASGVPS RFSGSGSGTD YTFTISSLQP 101 EDIATYYCQQ WSSYPYMYTF GQGTKVEIKR TVAAPSVFIF PPSDEQLKSG 151 TASVVCLLNN FYPREAKVQW KVDNALQSGN SQESVTEQDS KDSTYSLSST 201 LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC SEQ ID NO: 11 (MORAb-003 HC nt) 1 ATGGGATGGA GCTGTATCAT CCTCTTCTTG GTAGCAACAG CTACAGGTGT 51 CCACTCCGAG GTCCAACTGG TGGAGAGCGG TGGAGGTGTT GTGCAACCTG 101 GCCGGTCCCT GCGCCTGTCC TGCTCCGCAT CTGGCTTCAC CTTCAGCGGC 151 TATGGGTTGT CTTGGGTGAG ACAGGCACCT GGAAAAGGTC TTGAGTGGGT 201 TGCAATGATT AGTAGTGGTG GTAGTTATAC CTACTATGCA GACAGTGTGA 251 AGGGTAGATT TGCAATATCG CGAGACAACG CCAAGAACAC ATTGTTCCTG 301 CAAATGGACA GCCTGAGACC CGAAGACACC GGGGTCTATT TTTGTGCAAG 351 ACATGGGGAC GATCCCGCCT GGTTCGCTTA TTGGGGCCAA GGGACCCCGG 401 TCACCGTCTC CTCAGCCTCC ACCAAGGGCC CATCGGTCTT CCCCCTGGCA 451 CCCTCCTCCA AGAGCACCTC TGGGGGCACA GCGGCCCTGG GCTGCCTGGT 501 CAAGGACTAC TTCCCCGAAC CGGTGACGGT GTCGTGGAAC TCAGGCGCCC 551 TGACCAGCGG CGTGCACACC TTCCCGGCTG TCCTACAGTC CTCAGGACTC 601 TACTCCCTCA GCAGCGTGGT GACCGTGCCC TCCAGCAGCT TGGGCACCCA 651 GACCTACATC TGCAACGTGA ATCACAAGCC CAGCAACACC AAGGTGGACA 701 AGAAAGTTGA GCCCAAATCT TGTGACAAAA CTCACACATG CCCACCGTGC 751 CCAGCACCTG AACTCCTGGG GGGACCGTCA GTCTTCCTCT TCCCCCCAAA 801 ACCCAAGGAC ACCCTCATGA TCTCCCGGAC CCCTGAGGTC ACATGCGTGG 851 TGGTGGACGT GAGCCACGAA GACCCTGAGG TCAAGTTCAA CTGGTACGTG 901 GACGGCGTGG AGGTGCATAA TGCCAAGACA AAGCCGCGGG AGGAGCAGTA 951 CAACAGCACG TACCGTGTGG TCAGCGTCCT CACCGTCCTG CACCAGGACT 1001 GGCTGAATGG CAAGGAGTAC AAGTGCAAGG TCTCCAACAA AGCCCTCCCA 1051 GCCCCCATCG AGAAAACCAT CTCCAAAGCC AAAGGGCAGC CCCGAGAACC 1101 ACAGGTGTAC ACCCTGCCCC CATCCCGGGA TGAGCTGACC AAGAACCAGG 1151 TCAGCCTGAC CTGCCTGGTC AAAGGCTTCT ATCCCAGCGA CATCGCCGTG 1201 GAGTGGGAGA GCAATGGGCA GCCGGAGAAC AACTACAAGA CCACGCCTCC 1251 CGTGCTGGAC TCCGACGGCT CCTTCTTCTT ATATTCAAAG CTCACCGTGG 1301 ACAAGAGCAG GTGGCAGCAG GGGAACGTCT TCTCATGCTC CGTGATGCAT 1351 GAGGCTCTGC ACAACCACTA CACGCAGAAG AGCCTCTCCC TGTCTCCCGG 1401 GAAATGA SEQ ID NO: 12 (MORAb-003 LC nt) 1 ATGGGATGGA GCTGTATCAT CCTCTTCTTG GTAGCAACAG CTACAGGTGT 51 CCACTCCGAC ATCCAGCTGA CCCAGAGCCC AAGCAGCCTG AGCGCCAGCG 101 TGGGTGACAG AGTGACCATC ACCTGTAGTG TCAGCTCAAG TATAAGTTCC 151 AACAACTTGC ACTGGTACCA GCAGAAGCCA GGTAAGGCTC CAAAGCCATG 201 GATCTACGGC ACATCCAACC TGGCTTCTGG TGTGCCAAGC AGATTCAGCG 251 GTAGCGGTAG CGGTACCGAC TACACCTTCA CCATCAGCAG CCTCCAGCCA 301 GAGGACATCG CCACCTACTA CTGCCAACAG TGGAGTAGTT ACCCGTACAT 351 GTACACGTTC GGCCAAGGGA CCAAGGTGGA AATCAAACGA ACTGTGGCTG 401 CACCATCTGT CTTCATCTTC CCGCCATCTG ATGAGCAGTT GAAATCTGGA 451 ACTGCCTCTG TTGTGTGCCT GCTGAATAAC TTCTATCCCA GAGAGGCCAA 501 AGTACAGTGG AAGGTGGATA ACGCCCTCCA ATCGGGTAAC TCCCAGGAGA 551 GTGTCACAGA GCAGGACAGC AAGGACAGCA CCTACAGCCT CAGCAGCACC 601 CTGACGCTGA GCAAAGCAGA CTACGAGAAA CACAAAGTCT ACGCCTGCGA 651 AGTCACCCAT CAGGGCCTGA GCTCGCCCGT CACAAAGAGC TTCAACAGGG 101 GAGAGTGTTA A SEQ ID NO: 13 (MORAb-003 HC CDR1; IMGT): GFTFSGYG SEQ ID NO: 14 (MORAb-003 HC CDR2; IMGT): ISSGGSYT SEQ ID NO: 15 (MORAb-003 HC CDR3; IMGT): ARHGDDPAWFAY SEQ ID NO: 16 (MORAb-003 LC CDR1; IMGT): SSISSNN SEQ ID NO: 17 (MORAb-003 LC CDR2; IMGT): GTS SEQ ID NO: 18 (MORAb-003 LC CDR3; IMGT): QQWSSYPYMYT SEQ ID NO: 19 (human FRA) 1 maqrmttqll lllvwvavvg eaqtriawar tellnvcmna khhkekpgpe dklheqcrpw 61 rknaccstnt sqeahkdvsy lyrfnwnhcg emapackrhf iqdtclyecs pnlgpwiqqv 121 dqswrkervl nvplckedce qwwedcrtsy tcksnwhkgw nwtsgfnkca vgaacqpfhf 181 yfptptvlcn eiwthsykvs nysrgsgrci qmwfdpaqgn pneevarfya aamsgagpwa 241 awpfllslal mllwlls SEQ ID NO: 20 (human FRA nucleotide) 1 cattccttgg tgccactgac cacagctctt tcttcaggga cagacatggc tcagcggatg 61 acaacacagc tgctgctcct tctagtgtgg gtggctgtag taggggaggc tcagacaagg 121 attgcatggg ccaggactga gcttctcaat gtctgcatga acgccaagca ccacaaggaa 181 aagccaggcc ccgaggacaa gttgcatgag cagtgtcgac cctggaggaa gaatgcctgc 241 tgttctacca acaccagcca ggaagcccat aaggatgttt cctacctata tagattcaac 301 tggaaccact gtggagagat ggcacctgcc tgcaaacggc atttcatcca ggacacctgc 361 ctctacgagt gctcccccaa cttggggccc tggatccagc aggtggatca gagctggcgc 421 aaagagcggg tactgaacgt gcccctgtgc aaagaggact gtgagcaatg gtgggaagat 481 tgtcgcacct cctacacctg caagagcaac tggcacaagg gctggaactg gacttcaggg 541 tttaacaagt gcgcagtggg agctgcctgc caacctttcc atttctactt ccccacaccc 601 actgttctgt gcaatgaaat ctggactcac tcctacaagg tcagcaacta cagccgaggg 661 agtggccgct gcatccagat gtggttcgac ccagcccagg gcaaccccaa tgaggaggtg 721 gcgaggttct atgctgcagc catgagtggg gctgggccct gggcagcctg gcctttcctg 781 cttagcctgg ccctaatgct gctgtggctg ctcagctgac ctccttttac cttctgatac 841 ctggaaatcc ctgccctgtt cagccccaca gctcccaact atttggttcc tgctccatgg 901 tcgggcctct gacagccact ttgaataaac cagacaccgc acatgtgtct tgagaattat 961 ttggaaaaaa aaaaaaaaaa aa SEQ ID NO: 21 (human her2) 1 melaalcrwg lllallppga astqvctgtd mklrlpaspe thldmlrhly qgcqvvqgnl 61 eltylptnas lsflqdiqev qgyvliahnq vrqvplqrlr ivrgtqlfed nyalavldng 121 dpinnttpvt gaspgglrel qlrslteilk ggvliqrnpq lcyqdtilwk difhknnqla 181 ltlidtnrsr achpcspmck gsrcwgesse dcqsltrtvc aggcarckgp lptdccheqc 241 aagctgpkhs dclaclhfnh sgicelhcpa lvtyntdtfe smpnpegryt fgascvtacp 301 ynylstdvgs ctivcplhnq evtaedgtqr cekcskpcar vcyglgmehl revravtsan 361 igefagckki fgslaflpes fdgdpasnta plqpeqlqvf etleeitgyl yisawpdslp 421 dlsvfqnlqv irgrilhnga ysltlqglgi swlglrslre lgsglalihh nthlcfvhtv 481 pwdqlfrnph qallhtanrp edecvgegla chqlcarghc wgpgptqcvn csqflrgqec 541 veecrvlqgl preyvnarhc lpchpecqpq ngsvtcfgpe adqcvacahy kdppfcvarc 601 psgvkpdlsy mpiwkfpdee gacqpcpinc thscvdlddk gcpaeqrasp ltsiisavvg 661 illvvvlgvv fgilikrrqq kirkytmrrl lqetelvepl tpsgampnqa qmrilketel
721 rkvkvlgsga fgtvykgiwi pdgenvkipv aikvlrents pkankeilde ayvmagvgsp 781 yvsrllgicl tstvqlvtql mpygclldhv renrgrlgsq dllnwcmclia kgmsyledvr 841 lvhrdlaarn vlvkspnhvk itdfglarll dideteyhad ggkvpikwma lesilrrrft 901 hqsdvwsygv tvwelmtfga kpydgipare ipdllekger lpqppictid vymimvkcwm 961 idsecrprfr elvsefsrma rdpqrfvviq nedlgpaspl dstfyrslle dddmgdlvda 1021 eeylvpqqgf fcpdpapgag gmvhhrhrss strsgggdlt lglepseeea prsplapseg 1081 agsdvfdgdl gmgaakglqs lpthdpsplq rysedptvpl psetdgyvap ltcspqpeyv 1141 nqpdvrpqpp spregplpaa rpagatlerp ktlspgkngv vkdvfafgga venpeyltpq 1201 ggaapqphpp pafspafdnl yywdqdpper gappstfkgt ptaenpeylg ldvpv SEQ ID NO: 22 (human her2 nucleotide) 1 ATGGAGCTGG CGGCCTTGTG CCGCTGGGGG CTCCTCCTCG CCCTCTTGCC CCCCGGAGCC 61 GCGAGCACCC AAGTGTGCAC CGGCACAGAC ATGAAGCTGC GGCTCCCTGC CAGTCCCGAG 121 ACCCACCTGG ACATGCTCCG CCACCTCTAC CAGGGCTGCC AGGTGGTGCA GGGAAACCTG 181 GAACTCACCT ACCTGCCCAC CAATGCCAGC CTGTCCTTCC TGCAGGATAT CCAGGAGGTG 241 CAGGGCTACG TGCTCATCGC TCACAACCAA GTGAGGCAGG TCCCACTGCA GAGGCTGCGG 301 ATTGTGCGAG GCACCCAGCT CTTTGAGGAC AACTATGCCC TGGCCGTGCT AGACAATGGA 361 GACCCGCTGA ACAATACCAC CCCTGTCACA GGGGCCTCCC CAGGAGGCCT GCGGGAGCTG 421 CAGCTTCGAA GCCTCACAGA GATCTTGAAA GGAGGGGTCT TGATCCAGCG GAACCCCCAG 481 CTCTGCTACC AGGACACGAT TTTGTGGAAG GACATCTTCC ACAAGAACAA CCAGCTGGCT 541 CTCACACTGA TAGACACCAA CCGCTCTCGG GCCTGCCACC CCTGTTCTCC GATGTGTAAG 601 GGCTCCCGCT GCTGGGGAGA GAGTTCTGAG GATTGTCAGA GCCTGACGCG CACTGTCTGT 661 GCCGGTGGCT GTGCCCGCTG CAAGGGGCCA CTGCCCACTG ACTGCTGCCA TGAGCAGTGT 721 GCTGCCGGCT GCACGGGCCC CAAGCACTCT GACTGCCTGG CCTGCCTCCA CTTCAACCAC 781 AGTGGCATCT GTGAGCTGCA CTGCCCAGCC CTGGTCACCT ACAACACAGA CACGTTTGAG 841 TCCATGCCCA ATCCCGAGGG CCGGTATACA TTCGGCGCCA GCTGTGTGAC TGCCTGTCCC 901 TACAACTACC TTTCTACGGA CGTGGGATCC TGCACCCTCG TCTGCCCCCT GCACAACCAA 961 GAGGTGACAG CAGAGGATGG AACACAGCGG TGTGAGAAGT GCAGCAAGCC CTGTGCCCGA 1021 GTGTGCTATG GTCTGGGCAT GGAGCACTTG CGAGAGGTGA GGGCAGTTAC CAGTGCCAAT 1081 ATCCAGGAGT TTGCTGGCTG CAAGAAGATC TTTGGGAGCC TGGCATTTCT GCCGGAGAGC 1141 TTTGATGGGG ACCCAGCCTC CAACACTGCC CCGCTCCAGC CAGAGCAGCT CCAAGTGTTT 1201 GAGACTCTGG AAGAGATCAC AGGTTACCTA TACATCTCAG CATGGCCGGA CAGCCTGCCT 1261 GACCTCAGCG TCTTCCAGAA CCTGCAAGTA ATCCGGGGAC GAATTCTGCA CAATGGCGCC 1321 TACTCGCTGA CCCTGCAAGG GCTGGGCATC AGCTGGCTGG GGCTGCGCTC ACTGAGGGAA 1381 CTGGGCAGTG GACTGGCCCT CATCCACCAT AACACCCACC TCTGCTTCGT GCACACGGTG 1441 CCCTGGGACC AGCTCTTTCG GAACCCGCAC CAAGCTCTGC TCCACACTGC CAACCGGCCA 1501 GAGGACGAGT GTGTGGGCGA GGGCCTGGCC TGCCACCAGC TGTGCGCCCG AGGGCACTGC 1561 TGGGGTCCAG GGCCCACCCA GTGTGTCAAC TGCAGCCAGT TCCTTCGGGG CCAGGAGTGC 1621 GTGGAGGAAT GCCGAGTACT GCAGGGGCTC CCCAGGGAGT ATGTGAATGC CAGGCACTGT 1681 TTGCCGTGCC ACCCTGAGTG TCAGCCCCAG AATGGCTCAG TGACCTGTTT TGGACCGGAG 1741 GCTGACCAGT GTGTGGCCTG TGCCCACTAT AAGGACCCTC CCTTCTGCGT GGCCCGCTGC 1801 CCCAGCGGTG TGAAACCTGA CCTCTCCTAC ATGCCCATCT GGAAGTTTCC AGATGAGGAG 1861 GGCGCATGCC AGCCTTGCCC CATCAACTGC ACCCACTCCT GTGTGGACCT GGATGACAAG 1921 GGCTGCCCCG CCGAGCAGAG AGCCAGCCCT CTGACGTCCA TCATCTCTGC GGTGGTTGGC 1981 ATTCTGCTGG TCGTGGTCTT GGGGGTGGTC TTTGGGATCC TCATCAAGCG ACGGCAGCAG 2041 AAGATCCGGA AGTACACGAT GCGGAGACTG CTGCAGGAAA CGGAGCTGGT GGAGCCGCTG 2101 ACACCTAGCG GAGCGATGCC CAACCAGGCG CAGATGCGGA TCCTGAAAGA GACGGAGCTG 2161 AGGAAGGTGA AGGTGCTTGG ATCTGGCGCT TTTGGCACAG TCTACAAGGG CATCTGGATC 2221 CCTGATGGGG AGAATGTGAA AATTCCAGTG GCCATCAAAG TGTTGAGGGA AAACACATCC 2281 CCCAAAGCCA ACAAAGAAAT CTTAGACGAA GCATACGTGA TGGCTGGTGT GGGCTCCCCA 2341 TATGTCTCCC GCCTTCTGGG CATCTGCCTG ACATCCACGG TGCAGCTGGT GACACAGCTT 2401 ATGCCCTATG GCTGCCTCTT AGACCATGTC CGGGAAAACC GCGGACGCCT GGGCTCCCAG 2461 GACCTGCTGA ACTGGTGTAT GCAGATTGCC AAGGGGATGA GCTACCTGGA GGATGTGCGG 2521 CTCGTACACA GGGACTTGGC CGCTCGGAAC GTGCTGGTCA AGAGTCCCAA CCATGTCAAA 2581 ATTACAGACT TCGGGCTGGC TCGGCTGCTG GACATTGACG AGACAGAGTA CCATGCAGAT 2641 GGGGGCAAGG TGCCCATCAA GTGGATGGCG CTGGAGTCCA TTCTCCGCCG GCGGTTCACC 2701 CACCAGAGTG ATGTGTGGAG TTATGGTGTG ACTGTGTGGG AGCTGATGAC TTTTGGGGCC 2761 AAACCTTACG ATGGGATCCC AGCCCGGGAG ATCCCTGACC TGCTGGAAAA GGGGGAGCGG 2821 CTGCCCCAGC CCCCCATCTG CACCATTGAT GTCTACATGA TCATGGTCAA ATGTTGGATG 2881 ATTGACTCTG AATGTCGGCC AAGATTCCGG GAGTTGGTGT CTGAATTCTC CCGCATGGCC 2941 AGGGACCCCC AGCGCTTTGT GGTCATCCAG AATGAGGACT TGGGCCCAGC CAGTCCCTTG 3001 GACAGCACCT TCTACCGCTC ACTGCTGGAG GACGATGACA TGGGGGACCT GGTGGATGCT 3061 GAGGAGTATC TGGTACCCCA GCAGGGCTTC TTCTGTCCAG ACCCTGCCCC GGGCGCTGGG 3121 GGCATGGTCC ACCACAGGCA CCGCAGCTCA TCTACCAGGA GTGGCGGTGG GGACCTGACA 3181 CTAGGGCTGG AGCCCTCTGA AGAGGAGGCC CCCAGGTCTC CACTGGCACC CTCCGAAGGG 3241 GCTGGCTCCG ATGTATTTGA TGGTGACCTG GGAATGGGGG CAGCCAAGGG GCTGCAAAGC 3301 CTCCCCACAC ATGACCCCAG CCCTCTACAG CGGTACAGTG AGGACCCCAC AGTACCCCTG 3361 CCCTCTGAGA CTGATGGCTA CGTTGCCCCC CTGACCTGCA GCCCCCAGCC TGAATATGTG 3421 AACCAGCCAG ATGTTCGGCC CCAGCCCCCT TCGCCCCGAG AGGGCCCTCT GCCTGCTGCC 3481 CGACCTGCTG GTGCCACTCT GGAAAGGCCC AAGACTCTCT CCCCAGGGAA GAATGGGGTC 3541 GTCAAAGACG TTTTTGCCTT TGGGGGTGCC GTGGAGAACC CCGAGTACTT GACACCCCAG 3601 GGAGGAGCTG CCCCTCAGCC CCACCCTCCT CCTGCCTTCA GCCCAGCCTT CGACAACCTC 3661 TATTACTGGG ACCAGGACCC ACCAGAGCGG GGGGCTCCAC CCAGCACCTT CAAAGGGACA 3721 CCTACGGCAG AGAACCCAGA GTACCTGGGT CTGGACGTGC CAGTGTGA
Sequence CWU 1 SEQUENCE LISTING <160> NUMBER OF SEQ ID
NOS: 368 <210> SEQ ID NO 1 <211> LENGTH: 449
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE: 1
Glu Val Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg 1 5
10 15 Ser Leu Arg Leu Ser Cys Ser Ala Ser Gly Phe Thr Phe Ser Gly
Tyr 20 25 30 Gly Leu Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu
Glu Trp Val 35 40 45 Ala Met Ile Ser Ser Gly Gly Ser Tyr Thr Tyr
Tyr Ala Asp Ser Val 50 55 60 Lys Gly Arg Phe Ala Ile Ser Arg Asp
Asn Ala Lys Asn Thr Leu Phe 65 70 75 80 Leu Gln Met Asp Ser Leu Arg
Pro Glu Asp Thr Gly Val Tyr Phe Cys 85 90 95 Ala Arg His Gly Asp
Asp Pro Ala Trp Phe Ala Tyr Trp Gly Gln Gly 100 105 110 Thr Pro Val
Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser Val Phe 115 120 125 Pro
Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala Leu 130 135
140 Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp
145 150 155 160 Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro
Ala Val Leu 165 170 175 Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val
Val Thr Val Pro Ser 180 185 190 Ser Ser Leu Gly Thr Gln Thr Tyr Ile
Cys Asn Val Asn His Lys Pro 195 200 205 Ser Asn Thr Lys Val Asp Lys
Lys Val Glu Pro Lys Ser Cys Asp Lys 210 215 220 Thr His Thr Cys Pro
Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro 225 230 235 240 Ser Val
Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser 245 250 255
Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp 260
265 270 Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His
Asn 275 280 285 Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr
Tyr Arg Val 290 295 300 Val Ser Val Leu Thr Val Leu His Gln Asp Trp
Leu Asn Gly Lys Glu 305 310 315 320 Tyr Lys Cys Lys Val Ser Asn Lys
Ala Leu Pro Ala Pro Ile Glu Lys 325 330 335 Thr Ile Ser Lys Ala Lys
Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr 340 345 350 Leu Pro Pro Ser
Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr 355 360 365 Cys Leu
Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu 370 375 380
Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu 385
390 395 400 Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val
Asp Lys 405 410 415 Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser
Val Met His Glu 420 425 430 Ala Leu His Asn His Tyr Thr Gln Lys Ser
Leu Ser Leu Ser Pro Gly 435 440 445 Lys <210> SEQ ID NO 2
<211> LENGTH: 5 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 2 Gly Tyr Gly Leu Ser 1 5 <210> SEQ ID
NO 3 <211> LENGTH: 17 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 3 Met Ile Ser Ser Gly Gly Ser Tyr Thr
Tyr Tyr Ala Asp Ser Val Lys 1 5 10 15 Gly <210> SEQ ID NO 4
<211> LENGTH: 10 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 4 His Gly Asp Asp Pro Ala Trp Phe Ala Tyr 1 5
10 <210> SEQ ID NO 5 <211> LENGTH: 468 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polypeptide <400> SEQUENCE: 5 Met Gly Trp
Ser Cys Ile Ile Leu Phe Leu Val Ala Thr Ala Thr Gly 1 5 10 15 Val
His Ser Glu Val Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln 20 25
30 Pro Gly Arg Ser Leu Arg Leu Ser Cys Ser Ala Ser Gly Phe Thr Phe
35 40 45 Ser Gly Tyr Gly Leu Ser Trp Val Arg Gln Ala Pro Gly Lys
Gly Leu 50 55 60 Glu Trp Val Ala Met Ile Ser Ser Gly Gly Ser Tyr
Thr Tyr Tyr Ala 65 70 75 80 Asp Ser Val Lys Gly Arg Phe Ala Ile Ser
Arg Asp Asn Ala Lys Asn 85 90 95 Thr Leu Phe Leu Gln Met Asp Ser
Leu Arg Pro Glu Asp Thr Gly Val 100 105 110 Tyr Phe Cys Ala Arg His
Gly Asp Asp Pro Ala Trp Phe Ala Tyr Trp 115 120 125 Gly Gln Gly Thr
Pro Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro 130 135 140 Ser Val
Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr 145 150 155
160 Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr
165 170 175 Val Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr
Phe Pro 180 185 190 Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser
Ser Val Val Thr 195 200 205 Val Pro Ser Ser Ser Leu Gly Thr Gln Thr
Tyr Ile Cys Asn Val Asn 210 215 220 His Lys Pro Ser Asn Thr Lys Val
Asp Lys Lys Val Glu Pro Lys Ser 225 230 235 240 Cys Asp Lys Thr His
Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu 245 250 255 Gly Gly Pro
Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu 260 265 270 Met
Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser 275 280
285 His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu
290 295 300 Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn
Ser Thr 305 310 315 320 Tyr Arg Val Val Ser Val Leu Thr Val Leu His
Gln Asp Trp Leu Asn 325 330 335 Gly Lys Glu Tyr Lys Cys Lys Val Ser
Asn Lys Ala Leu Pro Ala Pro 340 345 350 Ile Glu Lys Thr Ile Ser Lys
Ala Lys Gly Gln Pro Arg Glu Pro Gln 355 360 365 Val Tyr Thr Leu Pro
Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val 370 375 380 Ser Leu Thr
Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val 385 390 395 400
Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro 405
410 415 Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu
Thr 420 425 430 Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser
Cys Ser Val 435 440 445 Met His Glu Ala Leu His Asn His Tyr Thr Gln
Lys Ser Leu Ser Leu 450 455 460 Ser Pro Gly Lys 465 <210> SEQ
ID NO 6 <211> LENGTH: 217 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 6 Asp Ile Gln Leu Thr Gln Ser Pro
Ser Ser Leu Ser Ala Ser Val Gly 1 5 10 15 Asp Arg Val Thr Ile Thr
Cys Ser Val Ser Ser Ser Ile Ser Ser Asn 20 25 30 Asn Leu His Trp
Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Pro Trp 35 40 45 Ile Tyr
Gly Thr Ser Asn Leu Ala Ser Gly Val Pro Ser Arg Phe Ser 50 55 60
Gly Ser Gly Ser Gly Thr Asp Tyr Thr Phe Thr Ile Ser Ser Leu Gln 65
70 75 80 Pro Glu Asp Ile Ala Thr Tyr Tyr Cys Gln Gln Trp Ser Ser
Tyr Pro 85 90 95 Tyr Met Tyr Thr Phe Gly Gln Gly Thr Lys Val Glu
Ile Lys Arg Thr 100 105 110 Val Ala Ala Pro Ser Val Phe Ile Phe Pro
Pro Ser Asp Glu Gln Leu 115 120 125 Lys Ser Gly Thr Ala Ser Val Val
Cys Leu Leu Asn Asn Phe Tyr Pro 130 135 140 Arg Glu Ala Lys Val Gln
Trp Lys Val Asp Asn Ala Leu Gln Ser Gly 145 150 155 160 Asn Ser Gln
Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr 165 170 175 Ser
Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His 180 185
190 Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val
195 200 205 Thr Lys Ser Phe Asn Arg Gly Glu Cys 210 215 <210>
SEQ ID NO 7 <211> LENGTH: 12 <212> TYPE: PRT
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 7 Ser Val Ser Ser Ser Ile
Ser Ser Asn Asn Leu His 1 5 10 <210> SEQ ID NO 8 <211>
LENGTH: 7 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 8 Gly Thr Ser Asn Leu Ala Ser 1 5 <210> SEQ ID NO 9
<211> LENGTH: 11 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 9 Gln Gln Trp Ser Ser Tyr Pro Tyr Met Tyr Thr
1 5 10 <210> SEQ ID NO 10 <211> LENGTH: 236 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polypeptide <400> SEQUENCE: 10 Met Gly
Trp Ser Cys Ile Ile Leu Phe Leu Val Ala Thr Ala Thr Gly 1 5 10 15
Val His Ser Asp Ile Gln Leu Thr Gln Ser Pro Ser Ser Leu Ser Ala 20
25 30 Ser Val Gly Asp Arg Val Thr Ile Thr Cys Ser Val Ser Ser Ser
Ile 35 40 45 Ser Ser Asn Asn Leu His Trp Tyr Gln Gln Lys Pro Gly
Lys Ala Pro 50 55 60 Lys Pro Trp Ile Tyr Gly Thr Ser Asn Leu Ala
Ser Gly Val Pro Ser 65 70 75 80 Arg Phe Ser Gly Ser Gly Ser Gly Thr
Asp Tyr Thr Phe Thr Ile Ser 85 90 95 Ser Leu Gln Pro Glu Asp Ile
Ala Thr Tyr Tyr Cys Gln Gln Trp Ser 100 105 110 Ser Tyr Pro Tyr Met
Tyr Thr Phe Gly Gln Gly Thr Lys Val Glu Ile 115 120 125 Lys Arg Thr
Val Ala Ala Pro Ser Val Phe Ile Phe Pro Pro Ser Asp 130 135 140 Glu
Gln Leu Lys Ser Gly Thr Ala Ser Val Val Cys Leu Leu Asn Asn 145 150
155 160 Phe Tyr Pro Arg Glu Ala Lys Val Gln Trp Lys Val Asp Asn Ala
Leu 165 170 175 Gln Ser Gly Asn Ser Gln Glu Ser Val Thr Glu Gln Asp
Ser Lys Asp 180 185 190 Ser Thr Tyr Ser Leu Ser Ser Thr Leu Thr Leu
Ser Lys Ala Asp Tyr 195 200 205 Glu Lys His Lys Val Tyr Ala Cys Glu
Val Thr His Gln Gly Leu Ser 210 215 220 Ser Pro Val Thr Lys Ser Phe
Asn Arg Gly Glu Cys 225 230 235 <210> SEQ ID NO 11
<211> LENGTH: 1407 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 11 atgggatgga gctgtatcat
cctcttcttg gtagcaacag ctacaggtgt ccactccgag 60 gtccaactgg
tggagagcgg tggaggtgtt gtgcaacctg gccggtccct gcgcctgtcc 120
tgctccgcat ctggcttcac cttcagcggc tatgggttgt cttgggtgag acaggcacct
180 ggaaaaggtc ttgagtgggt tgcaatgatt agtagtggtg gtagttatac
ctactatgca 240 gacagtgtga agggtagatt tgcaatatcg cgagacaacg
ccaagaacac attgttcctg 300 caaatggaca gcctgagacc cgaagacacc
ggggtctatt tttgtgcaag acatggggac 360 gatcccgcct ggttcgctta
ttggggccaa gggaccccgg tcaccgtctc ctcagcctcc 420 accaagggcc
catcggtctt ccccctggca ccctcctcca agagcacctc tgggggcaca 480
gcggccctgg gctgcctggt caaggactac ttccccgaac cggtgacggt gtcgtggaac
540 tcaggcgccc tgaccagcgg cgtgcacacc ttcccggctg tcctacagtc
ctcaggactc 600 tactccctca gcagcgtggt gaccgtgccc tccagcagct
tgggcaccca gacctacatc 660 tgcaacgtga atcacaagcc cagcaacacc
aaggtggaca agaaagttga gcccaaatct 720 tgtgacaaaa ctcacacatg
cccaccgtgc ccagcacctg aactcctggg gggaccgtca 780 gtcttcctct
tccccccaaa acccaaggac accctcatga tctcccggac ccctgaggtc 840
acatgcgtgg tggtggacgt gagccacgaa gaccctgagg tcaagttcaa ctggtacgtg
900 gacggcgtgg aggtgcataa tgccaagaca aagccgcggg aggagcagta
caacagcacg 960 taccgtgtgg tcagcgtcct caccgtcctg caccaggact
ggctgaatgg caaggagtac 1020 aagtgcaagg tctccaacaa agccctccca
gcccccatcg agaaaaccat ctccaaagcc 1080 aaagggcagc cccgagaacc
acaggtgtac accctgcccc catcccggga tgagctgacc 1140 aagaaccagg
tcagcctgac ctgcctggtc aaaggcttct atcccagcga catcgccgtg 1200
gagtgggaga gcaatgggca gccggagaac aactacaaga ccacgcctcc cgtgctggac
1260 tccgacggct ccttcttctt atattcaaag ctcaccgtgg acaagagcag
gtggcagcag 1320 gggaacgtct tctcatgctc cgtgatgcat gaggctctgc
acaaccacta cacgcagaag 1380 agcctctccc tgtctcccgg gaaatga 1407
<210> SEQ ID NO 12 <211> LENGTH: 711 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 12 atgggatgga
gctgtatcat cctcttcttg gtagcaacag ctacaggtgt ccactccgac 60
atccagctga cccagagccc aagcagcctg agcgccagcg tgggtgacag agtgaccatc
120 acctgtagtg tcagctcaag tataagttcc aacaacttgc actggtacca
gcagaagcca 180 ggtaaggctc caaagccatg gatctacggc acatccaacc
tggcttctgg tgtgccaagc 240 agattcagcg gtagcggtag cggtaccgac
tacaccttca ccatcagcag cctccagcca 300 gaggacatcg ccacctacta
ctgccaacag tggagtagtt acccgtacat gtacacgttc 360 ggccaaggga
ccaaggtgga aatcaaacga actgtggctg caccatctgt cttcatcttc 420
ccgccatctg atgagcagtt gaaatctgga actgcctctg ttgtgtgcct gctgaataac
480 ttctatccca gagaggccaa agtacagtgg aaggtggata acgccctcca
atcgggtaac 540 tcccaggaga gtgtcacaga gcaggacagc aaggacagca
cctacagcct cagcagcacc 600 ctgacgctga gcaaagcaga ctacgagaaa
cacaaagtct acgcctgcga agtcacccat 660 cagggcctga gctcgcccgt
cacaaagagc ttcaacaggg gagagtgtta a 711 <210> SEQ ID NO 13
<211> LENGTH: 8 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 13 Gly Phe Thr Phe Ser Gly Tyr Gly 1 5
<210> SEQ ID NO 14 <211> LENGTH: 8 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 14 Ile Ser Ser Gly Gly Ser
Tyr Thr 1 5 <210> SEQ ID NO 15 <211> LENGTH: 12
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 15 Ala
Arg His Gly Asp Asp Pro Ala Trp Phe Ala Tyr 1 5 10 <210> SEQ
ID NO 16 <211> LENGTH: 7 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 16 Ser Ser Ile Ser Ser Asn Asn 1 5
<210> SEQ ID NO 17 <211> LENGTH: 3 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 17 Gly Thr Ser 1
<210> SEQ ID NO 18 <211> LENGTH: 11 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 18 Gln Gln Trp Ser Ser Tyr
Pro Tyr Met Tyr Thr 1 5 10 <210> SEQ ID NO 19 <211>
LENGTH: 257 <212> TYPE: PRT <213> ORGANISM: Homo
sapiens <400> SEQUENCE: 19 Met Ala Gln Arg Met Thr Thr Gln
Leu Leu Leu Leu Leu Val Trp Val 1 5 10 15 Ala Val Val Gly Glu Ala
Gln Thr Arg Ile Ala Trp Ala Arg Thr Glu 20 25 30 Leu Leu Asn Val
Cys Met Asn Ala Lys His His Lys Glu Lys Pro Gly 35 40 45 Pro Glu
Asp Lys Leu His Glu Gln Cys Arg Pro Trp Arg Lys Asn Ala 50 55 60
Cys Cys Ser Thr Asn Thr Ser Gln Glu Ala His Lys Asp Val Ser Tyr 65
70 75 80 Leu Tyr Arg Phe Asn Trp Asn His Cys Gly Glu Met Ala Pro
Ala Cys 85 90 95 Lys Arg His Phe Ile Gln Asp Thr Cys Leu Tyr Glu
Cys Ser Pro Asn 100 105 110 Leu Gly Pro Trp Ile Gln Gln Val Asp Gln
Ser Trp Arg Lys Glu Arg 115 120 125 Val Leu Asn Val Pro Leu Cys Lys
Glu Asp Cys Glu Gln Trp Trp Glu 130 135 140 Asp Cys Arg Thr Ser Tyr
Thr Cys Lys Ser Asn Trp His Lys Gly Trp 145 150 155 160 Asn Trp Thr
Ser Gly Phe Asn Lys Cys Ala Val Gly Ala Ala Cys Gln 165 170 175 Pro
Phe His Phe Tyr Phe Pro Thr Pro Thr Val Leu Cys Asn Glu Ile 180 185
190 Trp Thr His Ser Tyr Lys Val Ser Asn Tyr Ser Arg Gly Ser Gly Arg
195 200 205 Cys Ile Gln Met Trp Phe Asp Pro Ala Gln Gly Asn Pro Asn
Glu Glu 210 215 220 Val Ala Arg Phe Tyr Ala Ala Ala Met Ser Gly Ala
Gly Pro Trp Ala 225 230 235 240 Ala Trp Pro Phe Leu Leu Ser Leu Ala
Leu Met Leu Leu Trp Leu Leu 245 250 255 Ser <210> SEQ ID NO
20 <211> LENGTH: 982 <212> TYPE: DNA <213>
ORGANISM: Homo sapiens <400> SEQUENCE: 20 cattccttgg
tgccactgac cacagctctt tcttcaggga cagacatggc tcagcggatg 60
acaacacagc tgctgctcct tctagtgtgg gtggctgtag taggggaggc tcagacaagg
120 attgcatggg ccaggactga gcttctcaat gtctgcatga acgccaagca
ccacaaggaa 180 aagccaggcc ccgaggacaa gttgcatgag cagtgtcgac
cctggaggaa gaatgcctgc 240 tgttctacca acaccagcca ggaagcccat
aaggatgttt cctacctata tagattcaac 300 tggaaccact gtggagagat
ggcacctgcc tgcaaacggc atttcatcca ggacacctgc 360 ctctacgagt
gctcccccaa cttggggccc tggatccagc aggtggatca gagctggcgc 420
aaagagcggg tactgaacgt gcccctgtgc aaagaggact gtgagcaatg gtgggaagat
480 tgtcgcacct cctacacctg caagagcaac tggcacaagg gctggaactg
gacttcaggg 540 tttaacaagt gcgcagtggg agctgcctgc caacctttcc
atttctactt ccccacaccc 600 actgttctgt gcaatgaaat ctggactcac
tcctacaagg tcagcaacta cagccgaggg 660 agtggccgct gcatccagat
gtggttcgac ccagcccagg gcaaccccaa tgaggaggtg 720 gcgaggttct
atgctgcagc catgagtggg gctgggccct gggcagcctg gcctttcctg 780
cttagcctgg ccctaatgct gctgtggctg ctcagctgac ctccttttac cttctgatac
840 ctggaaatcc ctgccctgtt cagccccaca gctcccaact atttggttcc
tgctccatgg 900 tcgggcctct gacagccact ttgaataaac cagacaccgc
acatgtgtct tgagaattat 960 ttggaaaaaa aaaaaaaaaa aa 982 <210>
SEQ ID NO 21 <211> LENGTH: 1255 <212> TYPE: PRT
<213> ORGANISM: Homo sapiens <400> SEQUENCE: 21 Met Glu
Leu Ala Ala Leu Cys Arg Trp Gly Leu Leu Leu Ala Leu Leu 1 5 10 15
Pro Pro Gly Ala Ala Ser Thr Gln Val Cys Thr Gly Thr Asp Met Lys 20
25 30 Leu Arg Leu Pro Ala Ser Pro Glu Thr His Leu Asp Met Leu Arg
His 35 40 45 Leu Tyr Gln Gly Cys Gln Val Val Gln Gly Asn Leu Glu
Leu Thr Tyr 50 55 60 Leu Pro Thr Asn Ala Ser Leu Ser Phe Leu Gln
Asp Ile Gln Glu Val 65 70 75 80 Gln Gly Tyr Val Leu Ile Ala His Asn
Gln Val Arg Gln Val Pro Leu 85 90 95 Gln Arg Leu Arg Ile Val Arg
Gly Thr Gln Leu Phe Glu Asp Asn Tyr 100 105 110 Ala Leu Ala Val Leu
Asp Asn Gly Asp Pro Leu Asn Asn Thr Thr Pro 115 120 125 Val Thr Gly
Ala Ser Pro Gly Gly Leu Arg Glu Leu Gln Leu Arg Ser 130 135 140 Leu
Thr Glu Ile Leu Lys Gly Gly Val Leu Ile Gln Arg Asn Pro Gln 145 150
155 160 Leu Cys Tyr Gln Asp Thr Ile Leu Trp Lys Asp Ile Phe His Lys
Asn 165 170 175 Asn Gln Leu Ala Leu Thr Leu Ile Asp Thr Asn Arg Ser
Arg Ala Cys 180 185 190 His Pro Cys Ser Pro Met Cys Lys Gly Ser Arg
Cys Trp Gly Glu Ser 195 200 205 Ser Glu Asp Cys Gln Ser Leu Thr Arg
Thr Val Cys Ala Gly Gly Cys 210 215 220 Ala Arg Cys Lys Gly Pro Leu
Pro Thr Asp Cys Cys His Glu Gln Cys 225 230 235 240 Ala Ala Gly Cys
Thr Gly Pro Lys His Ser Asp Cys Leu Ala Cys Leu 245 250 255 His Phe
Asn His Ser Gly Ile Cys Glu Leu His Cys Pro Ala Leu Val 260 265 270
Thr Tyr Asn Thr Asp Thr Phe Glu Ser Met Pro Asn Pro Glu Gly Arg 275
280 285 Tyr Thr Phe Gly Ala Ser Cys Val Thr Ala Cys Pro Tyr Asn Tyr
Leu 290 295 300 Ser Thr Asp Val Gly Ser Cys Thr Leu Val Cys Pro Leu
His Asn Gln 305 310 315 320 Glu Val Thr Ala Glu Asp Gly Thr Gln Arg
Cys Glu Lys Cys Ser Lys 325 330 335 Pro Cys Ala Arg Val Cys Tyr Gly
Leu Gly Met Glu His Leu Arg Glu 340 345 350 Val Arg Ala Val Thr Ser
Ala Asn Ile Gln Glu Phe Ala Gly Cys Lys 355 360 365 Lys Ile Phe Gly
Ser Leu Ala Phe Leu Pro Glu Ser Phe Asp Gly Asp 370 375 380 Pro Ala
Ser Asn Thr Ala Pro Leu Gln Pro Glu Gln Leu Gln Val Phe 385 390 395
400 Glu Thr Leu Glu Glu Ile Thr Gly Tyr Leu Tyr Ile Ser Ala Trp Pro
405 410 415 Asp Ser Leu Pro Asp Leu Ser Val Phe Gln Asn Leu Gln Val
Ile Arg 420 425 430 Gly Arg Ile Leu His Asn Gly Ala Tyr Ser Leu Thr
Leu Gln Gly Leu 435 440 445 Gly Ile Ser Trp Leu Gly Leu Arg Ser Leu
Arg Glu Leu Gly Ser Gly 450 455 460 Leu Ala Leu Ile His His Asn Thr
His Leu Cys Phe Val His Thr Val 465 470 475 480 Pro Trp Asp Gln Leu
Phe Arg Asn Pro His Gln Ala Leu Leu His Thr 485 490 495 Ala Asn Arg
Pro Glu Asp Glu Cys Val Gly Glu Gly Leu Ala Cys His 500 505 510 Gln
Leu Cys Ala Arg Gly His Cys Trp Gly Pro Gly Pro Thr Gln Cys 515 520
525 Val Asn Cys Ser Gln Phe Leu Arg Gly Gln Glu Cys Val Glu Glu Cys
530 535 540 Arg Val Leu Gln Gly Leu Pro Arg Glu Tyr Val Asn Ala Arg
His Cys 545 550 555 560 Leu Pro Cys His Pro Glu Cys Gln Pro Gln Asn
Gly Ser Val Thr Cys 565 570 575 Phe Gly Pro Glu Ala Asp Gln Cys Val
Ala Cys Ala His Tyr Lys Asp 580 585 590 Pro Pro Phe Cys Val Ala Arg
Cys Pro Ser Gly Val Lys Pro Asp Leu 595 600 605 Ser Tyr Met Pro Ile
Trp Lys Phe Pro Asp Glu Glu Gly Ala Cys Gln 610 615 620 Pro Cys Pro
Ile Asn Cys Thr His Ser Cys Val Asp Leu Asp Asp Lys 625 630 635 640
Gly Cys Pro Ala Glu Gln Arg Ala Ser Pro Leu Thr Ser Ile Ile Ser 645
650 655 Ala Val Val Gly Ile Leu Leu Val Val Val Leu Gly Val Val Phe
Gly 660 665 670 Ile Leu Ile Lys Arg Arg Gln Gln Lys Ile Arg Lys Tyr
Thr Met Arg 675 680 685 Arg Leu Leu Gln Glu Thr Glu Leu Val Glu Pro
Leu Thr Pro Ser Gly 690 695 700 Ala Met Pro Asn Gln Ala Gln Met Arg
Ile Leu Lys Glu Thr Glu Leu 705 710 715 720 Arg Lys Val Lys Val Leu
Gly Ser Gly Ala Phe Gly Thr Val Tyr Lys 725 730 735 Gly Ile Trp Ile
Pro Asp Gly Glu Asn Val Lys Ile Pro Val Ala Ile 740 745 750 Lys Val
Leu Arg Glu Asn Thr Ser Pro Lys Ala Asn Lys Glu Ile Leu 755 760 765
Asp Glu Ala Tyr Val Met Ala Gly Val Gly Ser Pro Tyr Val Ser Arg 770
775 780 Leu Leu Gly Ile Cys Leu Thr Ser Thr Val Gln Leu Val Thr Gln
Leu 785 790 795 800 Met Pro Tyr Gly Cys Leu Leu Asp His Val Arg Glu
Asn Arg Gly Arg 805 810 815 Leu Gly Ser Gln Asp Leu Leu Asn Trp Cys
Met Gln Ile Ala Lys Gly 820 825 830 Met Ser Tyr Leu Glu Asp Val Arg
Leu Val His Arg Asp Leu Ala Ala 835 840 845 Arg Asn Val Leu Val Lys
Ser Pro Asn His Val Lys Ile Thr Asp Phe 850 855 860 Gly Leu Ala Arg
Leu Leu Asp Ile Asp Glu Thr Glu Tyr His Ala Asp 865 870 875 880 Gly
Gly Lys Val Pro Ile Lys Trp Met Ala Leu Glu Ser Ile Leu Arg 885 890
895 Arg Arg Phe Thr His Gln Ser Asp Val Trp Ser Tyr Gly Val Thr Val
900 905 910 Trp Glu Leu Met Thr Phe Gly Ala Lys Pro Tyr Asp Gly Ile
Pro Ala 915 920 925 Arg Glu Ile Pro Asp Leu Leu Glu Lys Gly Glu Arg
Leu Pro Gln Pro 930 935 940 Pro Ile Cys Thr Ile Asp Val Tyr Met Ile
Met Val Lys Cys Trp Met 945 950 955 960 Ile Asp Ser Glu Cys Arg Pro
Arg Phe Arg Glu Leu Val Ser Glu Phe 965 970 975 Ser Arg Met Ala Arg
Asp Pro Gln Arg Phe Val Val Ile Gln Asn Glu 980 985 990 Asp Leu Gly
Pro Ala Ser Pro Leu Asp Ser Thr Phe Tyr Arg Ser Leu 995 1000 1005
Leu Glu Asp Asp Asp Met Gly Asp Leu Val Asp Ala Glu Glu Tyr 1010
1015 1020 Leu Val Pro Gln Gln Gly Phe Phe Cys Pro Asp Pro Ala Pro
Gly 1025 1030 1035 Ala Gly Gly Met Val His His Arg His Arg Ser Ser
Ser Thr Arg 1040 1045 1050 Ser Gly Gly Gly Asp Leu Thr Leu Gly Leu
Glu Pro Ser Glu Glu 1055 1060 1065 Glu Ala Pro Arg Ser Pro Leu Ala
Pro Ser Glu Gly Ala Gly Ser 1070 1075 1080 Asp Val Phe Asp Gly Asp
Leu Gly Met Gly Ala Ala Lys Gly Leu 1085 1090 1095 Gln Ser Leu Pro
Thr His Asp Pro Ser Pro Leu Gln Arg Tyr Ser 1100 1105 1110 Glu Asp
Pro Thr Val Pro Leu Pro Ser Glu Thr Asp Gly Tyr Val 1115 1120 1125
Ala Pro Leu Thr Cys Ser Pro Gln Pro Glu Tyr Val Asn Gln Pro 1130
1135 1140 Asp Val Arg Pro Gln Pro Pro Ser Pro Arg Glu Gly Pro Leu
Pro 1145 1150 1155 Ala Ala Arg Pro Ala Gly Ala Thr Leu Glu Arg Pro
Lys Thr Leu 1160 1165 1170 Ser Pro Gly Lys Asn Gly Val Val Lys Asp
Val Phe Ala Phe Gly 1175 1180 1185 Gly Ala Val Glu Asn Pro Glu Tyr
Leu Thr Pro Gln Gly Gly Ala 1190 1195 1200 Ala Pro Gln Pro His Pro
Pro Pro Ala Phe Ser Pro Ala Phe Asp 1205 1210 1215 Asn Leu Tyr Tyr
Trp Asp Gln Asp Pro Pro Glu Arg Gly Ala Pro 1220 1225 1230 Pro Ser
Thr Phe Lys Gly Thr Pro Thr Ala Glu Asn Pro Glu Tyr 1235 1240 1245
Leu Gly Leu Asp Val Pro Val 1250 1255 <210> SEQ ID NO 22
<211> LENGTH: 3768 <212> TYPE: DNA <213>
ORGANISM: Homo sapiens <400> SEQUENCE: 22 atggagctgg
cggccttgtg ccgctggggg ctcctcctcg ccctcttgcc ccccggagcc 60
gcgagcaccc aagtgtgcac cggcacagac atgaagctgc ggctccctgc cagtcccgag
120 acccacctgg acatgctccg ccacctctac cagggctgcc aggtggtgca
gggaaacctg 180 gaactcacct acctgcccac caatgccagc ctgtccttcc
tgcaggatat ccaggaggtg 240 cagggctacg tgctcatcgc tcacaaccaa
gtgaggcagg tcccactgca gaggctgcgg 300 attgtgcgag gcacccagct
ctttgaggac aactatgccc tggccgtgct agacaatgga 360 gacccgctga
acaataccac ccctgtcaca ggggcctccc caggaggcct gcgggagctg 420
cagcttcgaa gcctcacaga gatcttgaaa ggaggggtct tgatccagcg gaacccccag
480 ctctgctacc aggacacgat tttgtggaag gacatcttcc acaagaacaa
ccagctggct 540 ctcacactga tagacaccaa ccgctctcgg gcctgccacc
cctgttctcc gatgtgtaag 600 ggctcccgct gctggggaga gagttctgag
gattgtcaga gcctgacgcg cactgtctgt 660 gccggtggct gtgcccgctg
caaggggcca ctgcccactg actgctgcca tgagcagtgt 720 gctgccggct
gcacgggccc caagcactct gactgcctgg cctgcctcca cttcaaccac 780
agtggcatct gtgagctgca ctgcccagcc ctggtcacct acaacacaga cacgtttgag
840 tccatgccca atcccgaggg ccggtataca ttcggcgcca gctgtgtgac
tgcctgtccc 900 tacaactacc tttctacgga cgtgggatcc tgcaccctcg
tctgccccct gcacaaccaa 960 gaggtgacag cagaggatgg aacacagcgg
tgtgagaagt gcagcaagcc ctgtgcccga 1020 gtgtgctatg gtctgggcat
ggagcacttg cgagaggtga gggcagttac cagtgccaat 1080 atccaggagt
ttgctggctg caagaagatc tttgggagcc tggcatttct gccggagagc 1140
tttgatgggg acccagcctc caacactgcc ccgctccagc cagagcagct ccaagtgttt
1200 gagactctgg aagagatcac aggttaccta tacatctcag catggccgga
cagcctgcct 1260 gacctcagcg tcttccagaa cctgcaagta atccggggac
gaattctgca caatggcgcc 1320 tactcgctga ccctgcaagg gctgggcatc
agctggctgg ggctgcgctc actgagggaa 1380 ctgggcagtg gactggccct
catccaccat aacacccacc tctgcttcgt gcacacggtg 1440 ccctgggacc
agctctttcg gaacccgcac caagctctgc tccacactgc caaccggcca 1500
gaggacgagt gtgtgggcga gggcctggcc tgccaccagc tgtgcgcccg agggcactgc
1560 tggggtccag ggcccaccca gtgtgtcaac tgcagccagt tccttcgggg
ccaggagtgc 1620 gtggaggaat gccgagtact gcaggggctc cccagggagt
atgtgaatgc caggcactgt 1680 ttgccgtgcc accctgagtg tcagccccag
aatggctcag tgacctgttt tggaccggag 1740 gctgaccagt gtgtggcctg
tgcccactat aaggaccctc ccttctgcgt ggcccgctgc 1800 cccagcggtg
tgaaacctga cctctcctac atgcccatct ggaagtttcc agatgaggag 1860
ggcgcatgcc agccttgccc catcaactgc acccactcct gtgtggacct ggatgacaag
1920 ggctgccccg ccgagcagag agccagccct ctgacgtcca tcatctctgc
ggtggttggc 1980 attctgctgg tcgtggtctt gggggtggtc tttgggatcc
tcatcaagcg acggcagcag 2040 aagatccgga agtacacgat gcggagactg
ctgcaggaaa cggagctggt ggagccgctg 2100 acacctagcg gagcgatgcc
caaccaggcg cagatgcgga tcctgaaaga gacggagctg 2160 aggaaggtga
aggtgcttgg atctggcgct tttggcacag tctacaaggg catctggatc 2220
cctgatgggg agaatgtgaa aattccagtg gccatcaaag tgttgaggga aaacacatcc
2280 cccaaagcca acaaagaaat cttagacgaa gcatacgtga tggctggtgt
gggctcccca 2340 tatgtctccc gccttctggg catctgcctg acatccacgg
tgcagctggt gacacagctt 2400 atgccctatg gctgcctctt agaccatgtc
cgggaaaacc gcggacgcct gggctcccag 2460 gacctgctga actggtgtat
gcagattgcc aaggggatga gctacctgga ggatgtgcgg 2520 ctcgtacaca
gggacttggc cgctcggaac gtgctggtca agagtcccaa ccatgtcaaa 2580
attacagact tcgggctggc tcggctgctg gacattgacg agacagagta ccatgcagat
2640 gggggcaagg tgcccatcaa gtggatggcg ctggagtcca ttctccgccg
gcggttcacc 2700 caccagagtg atgtgtggag ttatggtgtg actgtgtggg
agctgatgac ttttggggcc 2760 aaaccttacg atgggatccc agcccgggag
atccctgacc tgctggaaaa gggggagcgg 2820 ctgccccagc cccccatctg
caccattgat gtctacatga tcatggtcaa atgttggatg 2880 attgactctg
aatgtcggcc aagattccgg gagttggtgt ctgaattctc ccgcatggcc 2940
agggaccccc agcgctttgt ggtcatccag aatgaggact tgggcccagc cagtcccttg
3000 gacagcacct tctaccgctc actgctggag gacgatgaca tgggggacct
ggtggatgct 3060 gaggagtatc tggtacccca gcagggcttc ttctgtccag
accctgcccc gggcgctggg 3120 ggcatggtcc accacaggca ccgcagctca
tctaccagga gtggcggtgg ggacctgaca 3180 ctagggctgg agccctctga
agaggaggcc cccaggtctc cactggcacc ctccgaaggg 3240 gctggctccg
atgtatttga tggtgacctg ggaatggggg cagccaaggg gctgcaaagc 3300
ctccccacac atgaccccag ccctctacag cggtacagtg aggaccccac agtacccctg
3360 ccctctgaga ctgatggcta cgttgccccc ctgacctgca gcccccagcc
tgaatatgtg 3420 aaccagccag atgttcggcc ccagccccct tcgccccgag
agggccctct gcctgctgcc 3480 cgacctgctg gtgccactct ggaaaggccc
aagactctct ccccagggaa gaatggggtc 3540 gtcaaagacg tttttgcctt
tgggggtgcc gtggagaacc ccgagtactt gacaccccag 3600 ggaggagctg
cccctcagcc ccaccctcct cctgccttca gcccagcctt cgacaacctc 3660
tattactggg accaggaccc accagagcgg ggggctccac ccagcacctt caaagggaca
3720 cctacggcag agaacccaga gtacctgggt ctggacgtgc cagtgtga 3768
<210> SEQ ID NO 23 <211> LENGTH: 119 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 23 Glu Val Gln Leu Val
Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg 1 5 10 15 Ser Leu Arg
Leu Ser Cys Ser Ala Ser Gly Phe Thr Phe Ser Gly Tyr 20 25 30 Gly
Leu Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45 Ala Met Ile Ser Ser Gly Gly Ser Tyr Thr Tyr Tyr Ala Asp Ser Val
50 55 60 Lys Gly Arg Phe Ala Ile Ser Arg Asp Asn Ala Lys Asn Thr
Leu Phe 65 70 75 80 Leu Gln Met Asp Ser Leu Arg Pro Glu Asp Thr Gly
Val Tyr Phe Cys 85 90 95 Ala Arg His Gly Asp Asp Pro Ala Trp Phe
Ala Tyr Trp Gly Gln Gly 100 105 110 Thr Pro Val Thr Val Ser Ser 115
<210> SEQ ID NO 24 <211> LENGTH: 110 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 24 Asp Ile Gln Leu Thr
Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly 1 5 10 15 Asp Arg Val
Thr Ile Thr Cys Ser Val Ser Ser Ser Ile Ser Ser Asn 20 25 30 Asn
Leu His Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Pro Trp 35 40
45 Ile Tyr Gly Thr Ser Asn Leu Ala Ser Gly Val Pro Ser Arg Phe Ser
50 55 60 Gly Ser Gly Ser Gly Thr Asp Tyr Thr Phe Thr Ile Ser Ser
Leu Gln 65 70 75 80 Pro Glu Asp Ile Ala Thr Tyr Tyr Cys Gln Gln Trp
Ser Ser Tyr Pro 85 90 95 Tyr Met Tyr Thr Phe Gly Gln Gly Thr Lys
Val Glu Ile Lys 100 105 110 <210> SEQ ID NO 25 <211>
LENGTH: 119 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polypeptide
<400> SEQUENCE: 25 Gln Val Gln Leu Gln Gln Ser Gly Pro Glu
Leu Glu Lys Pro Gly Ala 1 5 10 15 Ser Val Lys Ile Ser Cys Lys Ala
Ser Gly Tyr Ser Phe Thr Gly Tyr 20 25 30 Thr Met Asn Trp Val Lys
Gln Ser His Gly Lys Ser Leu Glu Trp Ile 35 40 45 Gly Leu Ile Thr
Pro Tyr Asn Gly Ala Ser Ser Tyr Asn Gln Lys Phe 50 55 60 Arg Gly
Lys Ala Thr Leu Thr Val Asp Lys Ser Ser Ser Thr Ala Tyr 65 70 75 80
Met Asp Leu Leu Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Phe Cys 85
90 95 Ala Arg Gly Gly Tyr Asp Gly Arg Gly Phe Asp Tyr Trp Gly Ser
Gly 100 105 110 Thr Pro Val Thr Val Ser Ser 115 <210> SEQ ID
NO 26 <211> LENGTH: 106 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 26 Asp Ile Glu Leu Thr Gln Ser
Pro Ala Ile Met Ser Ala Ser Pro Gly 1 5 10 15 Glu Lys Val Thr Met
Thr Cys Ser Ala Ser Ser Ser Val Ser Tyr Met 20 25 30 His Trp Tyr
Gln Gln Lys Ser Gly Thr Ser Pro Lys Arg Trp Ile Tyr 35 40 45 Asp
Thr Ser Lys Leu Ala Ser Gly Val Pro Gly Arg Phe Ser Gly Ser 50 55
60 Gly Ser Gly Asn Ser Tyr Ser Leu Thr Ile Ser Ser Val Glu Ala Glu
65 70 75 80 Asp Asp Ala Thr Tyr Tyr Cys Gln Gln Trp Ser Lys His Pro
Leu Thr 85 90 95 Phe Gly Ser Gly Thr Lys Val Glu Ile Lys 100 105
<210> SEQ ID NO 27 <211> LENGTH: 120 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 27 Glu Val Gln Leu Val
Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg
Leu Ser Cys Ala Ala Ser Gly Phe Asn Ile Lys Asp Thr 20 25 30 Tyr
Ile His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45 Ala Arg Ile Tyr Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Asp Ser Val
50 55 60 Lys Gly Arg Phe Thr Ile Ser Ala Asp Thr Ser Lys Asn Thr
Ala Tyr 65 70 75 80 Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala
Val Tyr Tyr Cys 85 90 95 Ser Arg Trp Gly Gly Asp Gly Phe Tyr Ala
Met Asp Tyr Trp Gly Gln 100 105 110 Gly Thr Leu Val Thr Val Ser Ser
115 120 <210> SEQ ID NO 28 <211> LENGTH: 107
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE: 28
Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly 1 5
10 15 Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Asp Val Asn Thr
Ala 20 25 30 Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys
Leu Leu Ile 35 40 45 Tyr Ser Ala Ser Phe Leu Tyr Ser Gly Val Pro
Ser Arg Phe Ser Gly 50 55 60 Ser Arg Ser Gly Thr Asp Phe Thr Leu
Thr Ile Ser Ser Leu Gln Pro 65 70 75 80 Glu Asp Phe Ala Thr Tyr Tyr
Cys Gln Gln His Tyr Thr Thr Pro Pro 85 90 95 Thr Phe Gly Gln Gly
Thr Lys Val Glu Ile Lys 100 105 <210> SEQ ID NO 29
<211> LENGTH: 123 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 29 Gln Ser Val Glu Glu Ser Gly
Gly Arg Leu Val Thr Pro Gly Thr Pro 1 5 10 15 Leu Thr Leu Thr Cys
Thr Val Ser Gly Ile Ser Leu Ser Ser Asp Ala 20 25 30 Ile Ser Trp
Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Tyr Ile Gly 35 40 45 Ile
Ile Asn Gly Gly Gly Asn Thr Tyr Tyr Ala Ser Trp Ala Lys Gly 50 55
60 Arg Phe Thr Ile Ser Lys Thr Ser Thr Thr Val Asp Leu Lys Ile Thr
65 70 75 80 Ser Pro Thr Thr Glu Asp Thr Ala Thr Tyr Phe Cys Ala Arg
Gly Ile 85 90 95 Gln His Gly Gly Gly Asn Ser Asp Tyr Tyr Tyr Tyr
Gly Met Asp Leu 100 105 110 Trp Gly Pro Gly Thr Leu Val Thr Val Ser
Ser 115 120 <210> SEQ ID NO 30 <211> LENGTH: 112
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE: 30
Glu Val Leu Met Thr Gln Thr Pro Ser Ser Val Ser Ala Ala Val Gly 1 5
10 15 Asp Thr Val Thr Ile Lys Cys Gln Ala Ser Gln Ser Ile Ser Ser
Val 20 25 30 Leu Ser Trp Tyr Gln Gln Lys Pro Gly Gln Pro Pro Lys
Leu Leu Ile 35 40 45 Tyr Leu Ala Ser Thr Leu Ala Ser Gly Val Pro
Ser Arg Phe Ser Gly 50 55 60 Ser Arg Ser Gly Thr Glu Phe Thr Leu
Thr Ile Ser Asp Leu Glu Cys 65 70 75 80 Asp Asp Ala Ala Thr Tyr Tyr
Cys Gln Thr Asn Tyr Gly Thr Ser Ser 85 90 95 Ser Asn Tyr Gly Phe
Ala Phe Gly Gly Gly Thr Glu Val Val Val Lys 100 105 110 <210>
SEQ ID NO 31 <211> LENGTH: 126 <212> TYPE: PRT
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 31 Glu Val Gln Leu Val
Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg
Leu Ser Cys Ala Ala Ser Gly Ile Ser Leu Ser Ser Asp 20 25 30 Ala
Ile Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Tyr Ile 35 40
45 Gly Ile Ile Asn Gly Gly Gly Asn Thr Tyr Tyr Ala Ser Trp Ala Lys
50 55 60 Gly Arg Phe Thr Ile Ser Arg His Asn Ser Lys Asn Thr Leu
Tyr Leu 65 70 75 80 Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val
Tyr Tyr Cys Ala 85 90 95 Arg Gly Ile Gln His Gly Gly Gly Asn Ser
Asp Tyr Tyr Tyr Tyr Gly 100 105 110 Met Asp Leu Trp Gly Gln Gly Thr
Leu Val Thr Val Ser Ser 115 120 125 <210> SEQ ID NO 32
<211> LENGTH: 112 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 32 Asp Ile Gln Met Thr Gln Ser
Pro Ser Ser Leu Ser Ala Ser Val Gly 1 5 10 15 Asp Arg Val Thr Ile
Thr Cys Gln Ala Ser Gln Ser Ile Ser Ser Val 20 25 30 Leu Ser Trp
Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45 Tyr
Leu Ala Ser Thr Leu Ala Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55
60 Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Cys
65 70 75 80 Glu Asp Ile Ala Thr Tyr Tyr Cys Gln Thr Asn Tyr Gly Thr
Ser Ser 85 90 95 Ser Asn Tyr Gly Phe Ala Phe Gly Gly Gly Thr Lys
Val Glu Ile Lys 100 105 110 <210> SEQ ID NO 33 <211>
LENGTH: 118 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polypeptide
<400> SEQUENCE: 33 Gln Ser Val Glu Glu Ser Gly Gly Arg Leu
Val Thr Pro Gly Thr Pro 1 5 10 15 Leu Thr Leu Thr Cys Thr Val Ser
Gly Phe Ser Leu Asn Asn Tyr Ala 20 25 30 Met Ser Trp Val Arg Gln
Ala Pro Gly Lys Gly Leu Glu Trp Ile Gly 35 40 45 Ser Ile Ser Thr
Gly Gly Leu Ala Phe Tyr Ala Asn Trp Ala Lys Gly 50 55 60 Arg Phe
Thr Ile Ser Arg Thr Ser Thr Thr Val Asp Leu Lys Met Thr 65 70 75 80
Ser Leu Thr Thr Glu Asp Thr Ala Thr Tyr Phe Cys Gly Arg Asn Gly 85
90 95 Gly Gly Ser Tyr Ile Phe Tyr Tyr Phe Asp Leu Trp Gly Gln Gly
Thr 100 105 110 Leu Val Thr Val Ser Ser 115 <210> SEQ ID NO
34 <211> LENGTH: 108 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 34 Ala Phe Glu Leu Thr Gln Thr
Pro Ser Ser Val Glu Ala Ala Val Gly 1 5 10 15 Gly Thr Ile Thr Ile
Lys Cys Gln Ala Ser Gln Ser Ile Ser Ser Tyr 20 25 30 Leu Ser Trp
Tyr Gln Gln Lys Pro Gly Gln Pro Pro Lys Leu Leu Ile 35 40 45 Tyr
Ser Ala Ser Thr Leu Ala Ser Gly Val Ser Ser Arg Phe Lys Gly 50 55
60 Ser Gly Ser Gly Thr Glu Tyr Thr Leu Thr Ile Ser Asp Leu Glu Cys
65 70 75 80 Ala Asp Ala Ala Thr Tyr Phe Cys Gln Ser Tyr Tyr Asp Ile
Gly Thr 85 90 95 Ser Thr Phe Gly Gly Gly Thr Glu Val Val Val Lys
100 105 <210> SEQ ID NO 35 <211> LENGTH: 121
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE: 35
Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5
10 15 Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Ser Leu Asn Asn
Tyr 20 25 30 Ala Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu
Glu Trp Ile 35 40 45 Gly Ser Ile Ser Thr Gly Gly Leu Ala Phe Tyr
Ala Asn Trp Ala Lys 50 55 60 Gly Arg Phe Thr Ile Ser Arg Asp Asn
Ser Lys Asn Thr Leu Tyr Leu 65 70 75 80 Gln Met Asn Ser Leu Arg Ala
Glu Asp Thr Ala Val Tyr Tyr Cys Ala 85 90 95 Arg Asn Gly Gly Gly
Ser Tyr Ile Phe Tyr Tyr Phe Asp Leu Trp Gly 100 105 110 Gln Gly Thr
Leu Val Thr Val Ser Ser 115 120 <210> SEQ ID NO 36
<211> LENGTH: 108 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 36 Asp Ile Gln Met Thr Gln Ser
Pro Ser Ser Leu Ser Ala Ser Val Gly 1 5 10 15 Asp Arg Val Thr Ile
Thr Cys Gln Ala Ser Gln Ser Ile Ser Ser Tyr 20 25 30 Leu Ser Trp
Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45 Tyr
Ser Ala Ser Thr Leu Ala Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55
60 Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Cys
65 70 75 80 Glu Asp Ala Ala Thr Tyr Tyr Cys Gln Ser Tyr Tyr Asp Ile
Gly Thr 85 90 95 Ser Thr Phe Gly Gly Gly Thr Lys Val Glu Ile Lys
100 105 <210> SEQ ID NO 37 <211> LENGTH: 114
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE: 37
Gln Ser Val Lys Glu Ser Gly Gly Arg Leu Val Thr Pro Gly Thr Pro 1 5
10 15 Leu Thr Leu Thr Cys Thr Val Ser Gly Ile Asp Leu Ser Ser Tyr
Ala 20 25 30 Met Gly Trp Phe Arg Gln Ala Pro Gly Lys Gly Leu Glu
Tyr Ile Gly 35 40 45 Thr Ile Asn Ile Gly Gly Arg Val Tyr Tyr Ala
Ser Trp Ala Lys Gly 50 55 60 Arg Phe Thr Ile Ser Arg Thr Ser Thr
Thr Val Asp Leu Lys Ala Pro 65 70 75 80 Ser Leu Thr Ala Glu Asp Thr
Ala Thr Tyr Phe Cys Ala Arg Tyr Tyr 85 90 95 Asn Gly Gly Ser Tyr
Asp Ile Trp Gly Pro Gly Thr Leu Val Thr Val 100 105 110 Ser Leu
<210> SEQ ID NO 38 <211> LENGTH: 110 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 38 Asp Val Val Met Thr
Gln Thr Pro Ala Ser Ala Ser Glu Pro Val Gly 1 5 10 15 Gly Thr Val
Thr Ile Lys Cys Gln Ala Ser Glu Ser Ile Tyr Arg Val 20 25 30 Leu
Ala Trp Tyr Gln Gln Lys Pro Gly Gln Pro Pro Lys Leu Leu Ile 35 40
45 Tyr Asp Thr Ser Thr Leu Ala Ser Gly Ala Pro Ser Arg Phe Lys Gly
50 55 60 Ser Gly Tyr Gly Thr Glu Phe Thr Leu Thr Ile Ser Gly Val
Gln Cys 65 70 75 80 Glu Asp Ala Ala Thr Tyr Tyr Cys Gln Gly Gly Tyr
Tyr Ala Asp Ser 85 90 95 Tyr Gly Ile Ala Phe Gly Gly Gly Thr Glu
Val Val Val Lys 100 105 110 <210> SEQ ID NO 39 <211>
LENGTH: 117 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polypeptide
<400> SEQUENCE: 39 Gln Val Gln Leu Val Glu Ser Gly Gly Gly
Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg Leu Ser Cys Ser Ala
Ser Gly Ile Asp Leu Ser Ser Tyr 20 25 30 Ala Met Gly Trp Val Arg
Gln Ala Pro Gly Lys Gly Leu Glu Tyr Ile 35 40 45 Gly Thr Ile Asn
Ile Gly Gly Arg Val Tyr Tyr Ala Ser Trp Ala Lys 50 55 60 Gly Arg
Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr Leu 65 70 75 80
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys Ala 85
90 95 Arg Tyr Tyr Asn Gly Gly Ser Tyr Asp Ile Trp Gly Gln Gly Thr
Leu 100 105 110 Val Thr Val Ser Ser 115 <210> SEQ ID NO 40
<211> LENGTH: 110 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 40 Asp Ile Gln Met Thr Gln Ser
Pro Ser Thr Leu Ser Ala Ser Val Gly 1 5 10 15 Asp Arg Val Thr Ile
Thr Cys Gln Ala Ser Glu Ser Ile Tyr Arg Val 20 25 30 Leu Ala Trp
Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45 Tyr
Asp Thr Ser Thr Leu Ala Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55
60 Ser Gly Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Cys
65 70 75 80 Asp Asp Ala Ala Thr Tyr Tyr Cys Gln Gly Gly Tyr Tyr Ala
Asp Ser 85 90 95 Tyr Gly Ile Ala Phe Gly Gly Gly Thr Lys Val Glu
Ile Lys 100 105 110 <210> SEQ ID NO 41 <211> LENGTH:
122 <212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE: 41
Gln Ser Val Glu Glu Ser Gly Gly Arg Leu Val Lys Pro Asp Glu Ser 1 5
10 15 Leu Thr Leu Thr Cys Thr Ala Ser Gly Phe Ser Leu Ser Ser Tyr
Ala 20 25 30 Met Ile Trp Val Arg Gln Ala Pro Gly Glu Gly Leu Glu
Trp Ile Gly 35 40 45 Thr Ile Ser Thr Gly Gly Ile Thr Tyr Tyr Ala
Ser Trp Ala Lys Gly 50 55 60 Arg Phe Thr Ile Ser Lys Thr Ser Thr
Thr Val Asp Leu Lys Ile Thr 65 70 75 80 Ser Pro Thr Thr Glu Asp Thr
Ala Thr Tyr Phe Cys Ala Arg Gly Gly 85 90 95 Tyr Ala Ala Ser Ser
Ala Tyr Tyr Leu Pro Tyr Tyr Phe Asp Leu Trp 100 105 110 Gly Gln Gly
Thr Leu Val Thr Val Ser Ser 115 120 <210> SEQ ID NO 42
<211> LENGTH: 112 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 42 Ala Ala Val Leu Thr Gln Thr
Pro Ser Pro Val Ser Ala Ala Val Gly 1 5 10 15 Gly Thr Val Thr Ile
Ser Cys Gln Ser Ser Gln Ser Val Tyr Asn Asn 20 25 30 Asn Asn Leu
Ala Trp Phe Gln Gln Lys Pro Gly Gln Pro Pro Lys Leu 35 40 45 Leu
Ile Tyr Leu Ala Ser Thr Leu Ala Ser Gly Val Pro Ser Arg Phe 50 55
60 Ser Gly Ser Gly Ser Gly Thr Gln Phe Thr Leu Thr Ile Ser Gly Val
65 70 75 80 Gln Cys Asp Asp Ala Ala Thr Tyr Tyr Cys Leu Gly Gly Cys
Asp Asp 85 90 95 Asp Ala Asp Thr Phe Ala Phe Gly Gly Gly Thr Glu
Val Val Val Lys 100 105 110 <210> SEQ ID NO 43 <211>
LENGTH: 125 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polypeptide
<400> SEQUENCE: 43 Glu Val Gln Leu Val Glu Ser Gly Gly Gly
Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg Leu Ser Cys Ala Ala
Ser Gly Phe Ser Leu Ser Ser Tyr 20 25 30 Ala Met Ile Trp Val Arg
Gln Ala Pro Gly Lys Gly Leu Glu Trp Ile 35 40 45 Gly Thr Ile Ser
Thr Gly Gly Ile Thr Tyr Tyr Ala Ser Trp Ala Lys 50 55 60 Gly Arg
Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr Leu 65 70 75 80
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys Ala 85
90 95 Arg Gly Gly Tyr Ala Ala Ser Ser Ala Tyr Tyr Leu Pro Tyr Tyr
Phe 100 105 110 Asp Leu Trp Gly Gln Gly Thr Leu Val Thr Val Ser Ser
115 120 125 <210> SEQ ID NO 44 <211> LENGTH: 112
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE: 44
Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly 1 5
10 15 Asp Arg Val Thr Ile Thr Cys Gln Ser Ser Gln Ser Val Tyr Asn
Asn 20 25 30 Asn Asn Leu Ala Trp Tyr Gln Gln Lys Pro Gly Lys Val
Pro Lys Leu 35 40 45 Leu Ile Tyr Leu Ala Ser Thr Leu Ala Ser Gly
Val Pro Ser Arg Phe 50 55 60 Ser Gly Ser Gly Ser Gly Thr Asp Phe
Thr Leu Thr Ile Ser Ser Leu 65 70 75 80 Gln Cys Glu Asp Ala Ala Thr
Tyr Tyr Cys Leu Gly Gly Cys Asp Asp 85 90 95 Asp Ala Asp Thr Phe
Ala Phe Gly Gly Gly Thr Lys Val Glu Ile Lys 100 105 110 <210>
SEQ ID NO 45 <211> LENGTH: 357 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 45 gaggtccaac
tggtggagag cggtggaggt gttgtgcaac ctggccggtc cctgcgcctg 60
tcctgctccg catctggctt caccttcagc ggctatgggt tgtcttgggt gagacaggca
120 cctggaaaag gtcttgagtg ggttgcaatg attagtagtg gtggtagtta
tacctactat 180 gcagacagtg tgaagggtag atttgcaata tcgcgagaca
acgccaagaa cacattgttc 240 ctgcaaatgg acagcctgag acccgaagac
accggggtct atttttgtgc aagacatggg 300 gacgatcccg cctggttcgc
ttattggggc caagggaccc cggtcaccgt ctcctca 357 <210> SEQ ID NO
46 <211> LENGTH: 330 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 46 gacatccagc tgacccagag
cccaagcagc ctgagcgcca gcgtgggtga cagagtgacc 60 atcacctgta
gtgtcagctc aagtataagt tccaacaact tgcactggta ccagcagaag 120
ccaggtaagg ctccaaagcc atggatctac ggcacatcca acctggcttc tggtgtgcca
180 agcagattca gcggtagcgg tagcggtacc gactacacct tcaccatcag
cagcctccag 240 ccagaggaca tcgccaccta ctactgccaa cagtggagta
gttacccgta catgtacacg 300 ttcggccaag ggaccaaggt ggaaatcaaa 330
<210> SEQ ID NO 47 <211> LENGTH: 357 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 47 caggtacaac
tgcagcagtc tgggcctgag ctggagaagc ctggcgcttc agtgaagata 60
tcctgcaagg cttctggtta ctcattcact ggctacacca tgaactgggt gaagcagagc
120 catggaaaga gccttgagtg gattggactt attactcctt acaatggtgc
ttctagctac 180 aaccagaagt tcaggggcaa ggccacatta actgtagaca
agtcatccag cacagcctac 240 atggacctcc tcagtctgac atctgaagac
tctgcagtct atttctgtgc aagggggggt 300 tacgacggga ggggttttga
ctactgggga tccgggaccc cggtcaccgt ctcctca 357 <210> SEQ ID NO
48 <211> LENGTH: 318 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 48 gacatcgagc tcactcagtc
tccagcaatc atgtctgcat ctccagggga gaaggtcacc 60 atgacctgca
gtgccagctc aagtgtaagt tacatgcact ggtaccagca gaagtcaggc 120
acctccccca aaagatggat ttatgacaca tccaaactgg cttctggagt cccaggtcgc
180 ttcagtggca gtgggtctgg aaactcttac tctctcacaa tcagcagcgt
ggaggctgaa 240 gatgatgcaa cttattactg ccagcagtgg agtaagcacc
ctctcacgtt cggatccggg 300 accaaggtgg aaatcaaa 318 <210> SEQ
ID NO 49 <211> LENGTH: 369 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 49 cagtcggtgg aggagtccgg
gggtcgcctg gtcacgcctg ggacacccct gacactcacc 60 tgcaccgtct
ctggaatctc cctcagtagc gatgcaataa gctgggtccg ccaggctcca 120
gggaaggggc tcgaatacat cggaatcatt aatggtggtg gtaacacata ctacgcgagc
180 tgggcgaaag gccgattcac catctccaaa acctcgacca cggtggatct
gaaaatcacc 240 agtccgacaa ccgaggacac ggccacctat ttctgtgcca
gaggcattca acatggtggt 300 ggtaatagtg attattatta ttacggcatg
gacctctggg gcccaggcac cctggtcact 360 gtctcttca 369 <210> SEQ
ID NO 50 <211> LENGTH: 336 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 50 gaagtgttga tgacccagac
tccatcctcc gtgtctgcag ctgtgggaga cacagtcacc 60 atcaagtgcc
aggccagtca gagcattagt agtgtcttgt cctggtatca gcagaaacca 120
gggcagcctc ccaagctcct gatctatctg gcatccactc tggcatctgg ggtcccatcg
180 cggttcagcg gcagtagatc tgggacagag ttcactctca ccatcagcga
cctggagtgt 240 gacgatgctg ccacttacta ctgtcaaacc aattatggta
ctagtagtag taattatggt 300 tttgctttcg gcggagggac cgaggtggtc gtcaaa
336 <210> SEQ ID NO 51 <211> LENGTH: 378 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polynucleotide <400> SEQUENCE: 51
gaagtccaac tggtggaaag cgggggagga ctggtgcagc cgggcggatc cctccggctg
60 tcatgtgctg catcgggaat ttccctctcc tccgacgcga ttagctgggt
cagacaggcc 120 cccggaaagg ggctggagta catcggtatc atcaacggcg
gcggaaacac ctactacgcc 180 tcctgggcca agggccgctt caccatctcg
cggcataatt ccaagaacac tctgtacttg 240 caaatgaact ccctgagggc
cgaggacacc gccgtgtact actgcgcgcg cggcatccag 300 cacggtggtg
gaaacagcga ctactactac tatgggatgg atctgtgggg ccagggaact 360
cttgtgaccg tgtcgtca 378 <210> SEQ ID NO 52 <211>
LENGTH: 336 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polynucleotide
<400> SEQUENCE: 52 gacattcaga tgacccagtc cccaagctcg
ctgtccgcct ccgtgggcga ccgcgtgacc 60 atcacgtgcc aggcgtccca
gtcaattagc agcgtgctct cctggtacca acagaagccg 120 gggaaagcac
ccaagctgct gatctacttg gcctccactc tggcctcggg agtgccttca 180
cggttctccg gatcgggatc tggtactgat ttcaccctca ccatctcgag ccttcagtgc
240 gaggacatcg ctacttacta ttgtcaaacc aactacggaa cctccagctc
caactacggc 300 tttgccttcg gtggcgggac caaggtcgaa atcaaa 336
<210> SEQ ID NO 53 <211> LENGTH: 354 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 53 cagtcggtgg
aggagtccgg gggtcgcctg gtcacgcctg ggacacccct gacactcacc 60
tgcacagtct ctggattctc cctcaataac tatgcaatga gctgggtccg ccaggctcca
120 gggaaggggc tggaatggat cggatccatt agtactggtg gtctcgcatt
ctacgcgaac 180 tgggcaaaag gccgattcac catctccaga acctcgacca
cggtggatct gaaaatgacc 240 agtctgacaa ccgaggacac ggccacctat
ttctgtggca gaaatggtgg tggtagttat 300 attttctatt attttgactt
gtggggccaa ggcaccctcg tcactgtctc ttca 354 <210> SEQ ID NO 54
<211> LENGTH: 324 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 54 gcattcgaat tgacccagac
tccatcctcc gtggaggcag ctgtgggagg cacaatcacc 60 atcaagtgcc
aggccagtca gagcattagt agttacttat cctggtatca gcagaaacca 120
gggcagcctc ccaagctcct gatctattct gcatccactc tggcatctgg ggtctcatcg
180 cggttcaaag gcagtggatc tgggacagag tacactctca ccatcagcga
cctggagtgt 240 gccgatgctg ccacttactt ctgtcaaagc tattatgata
ttggtactag tactttcggc 300 ggagggaccg aggtggtcgt caaa 324
<210> SEQ ID NO 55 <211> LENGTH: 363 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 55 gaagtgcagc
tggtggaatc tggcggcgga ctggtgcagc ctggcggatc tctgagactg 60
tcttgtgccg cctccggctt ctccctgaac aactacgcca tgtcctgggt gcgacaggcc
120 cctggcaaag gcctggaatg gatcggctcc atcagcacag gcggcctggc
cttctacgcc 180 aattgggcca agggccggtt caccatcagc cgggacaact
ccaagaacac cctgtacctc 240 cagatgaact ccctgcgggc cgaggacacc
gccgtgtact actgtgccag aaacggcgga 300 ggctcctaca tcttctacta
cttcgacctg tggggccagg gcaccctcgt gacagtgtca 360 tct 363 <210>
SEQ ID NO 56 <211> LENGTH: 324 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 56 gatattcaga
tgacccagtc cccctccagc ctgtccgctt ctgtgggcga cagagtgacc 60
atcacctgtc aggcctccca gtccatctcc tcctacctgt cctggtatca gcagaagccc
120 ggcaaggccc ccaagctgct gatctactct gcctccacac tggcctccgg
cgtgccctct 180 agattctccg gctctggctc tggcaccgac tttaccctga
ccatcagctc cctccagtgc 240 gaggatgccg ccacctacta ctgccagtcc
tactacgaca tcggcacctc caccttcggc 300 ggaggcacca aggtggaaat caaa 324
<210> SEQ ID NO 57 <211> LENGTH: 342 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 57 cagtcagtga
aggagtccgg gggtcgcctg gtcacgcctg ggacacccct gacactcacc 60
tgcacagtct ctggaatcga cctcagtagc tatgcaatgg gctggttccg ccaggctcca
120 gggaaggggc tggaatacat cggaaccatt aatattggtg gtcgcgtata
ttacgcgagc 180 tgggcaaaag gccgattcac catctccaga acctcgacca
cggtggatct gaaagcgccc 240 agtctgacag ccgaggacac ggccacctat
ttctgtgcca gatattataa tggtggtagt 300 tatgacatct ggggcccagg
caccctggtc accgtctctt ta 342 <210> SEQ ID NO 58 <211>
LENGTH: 330 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polynucleotide
<400> SEQUENCE: 58 gatgttgtga tgacccagac tccagcctcc
gcgtctgaac ctgtgggagg cacagtcacc 60 atcaagtgcc aggccagtga
gagcatttat cgcgtattgg cctggtatca gcagaaacca 120 gggcagcctc
ccaagctcct gatctatgat acatccactc tggcatctgg ggccccatcg 180
cggttcaaag gcagtggata tgggacagag ttcactctca ccatcagcgg cgtgcagtgt
240 gaagatgctg ccacttacta ctgtcaaggc ggttattatg ctgatagtta
tggtattgct 300 ttcggcggag ggaccgaggt ggtggtcaaa 330 <210> SEQ
ID NO 59 <211> LENGTH: 351 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 59 caggtgcagc tggtggaatc
tggcggagga ctggtgcagc ctggcggctc tctgagactg 60 tcctgttccg
cctccggaat cgacctgtcc tcctacgcta tgggctgggt gcgacaggct 120
cctggcaagg gcctggagta catcggcacc atcaacatcg gcggcagagt gtactacgcc
180 tcctgggcca agggccggtt caccatctcc agagacaact ccaagaacac
cctgtacctc 240 cagatgaact ccctgcgggc cgaggacacc gccgtgtact
actgcgcccg gtactacaac 300 ggcggctcct acgatatctg gggccagggc
acactcgtga ccgtgtcctc t 351 <210> SEQ ID NO 60 <211>
LENGTH: 330 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polynucleotide
<400> SEQUENCE: 60 gatatccaga tgacccagtc cccctccacc
ctgtctgcct ctgtgggcga cagagtgacc 60 atcacctgtc aggcctccga
gtccatctac cgggtgctgg cctggtatca gcagaagcct 120 ggcaaggccc
ccaagctgct gatctacgac accagcacac tggcctccgg cgtgccctct 180
agattctccg gctctggctc tggcaccgag tttaccctga ccatctccag cctccagtgc
240 gacgacgccg ccacctacta ttgtcagggc ggctactacg ccgactccta
cggaatcgct 300 ttcggcggag gcaccaaggt ggaaatcaaa 330 <210> SEQ
ID NO 61 <211> LENGTH: 366 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 61 cagtcggtgg aggagtccgg
cggtcgcctg gtaaagcctg acgaatccct gacactcacc 60 tgcacagcct
ctggattctc cctcagtagt tatgcaatga tctgggtccg ccaggctcca 120
ggggaggggc tggaatggat cggaaccatt agtactggtg gtatcacata ctacgcgagc
180 tgggcgaaag gccgattcac catctccaaa acctcgacca cggtggatct
gaaaatcacc 240 agtccgacaa ccgaggacac ggccacctat ttctgtgcca
gagggggata tgctgctagt 300 agtgcttatt atctcccgta ctactttgac
ttgtggggcc aagggaccct ggtcaccgtc 360 tcctca 366 <210> SEQ ID
NO 62 <211> LENGTH: 336 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 62 gcagccgtgc tgacccagac
accatcaccc gtgtctgcag ctgtgggagg cacagtcacc 60 atcagttgcc
agtccagtca gagtgtttat aataataaca acttagcctg gtttcagcag 120
aaacccgggc agcctcccaa gcttctgatc tatctggcat ccactctggc atctggggtc
180 ccatcacggt tcagcggcag tggatctggg acacagttca ctctcaccat
cagcggcgtg 240 cagtgtgacg atgctgccac ttattactgt ctaggtggtt
gtgatgatga tgctgatact 300 tttgctttcg gcggagggac tgaggtggtg gtcaaa
336 <210> SEQ ID NO 63 <211> LENGTH: 375 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polynucleotide <400> SEQUENCE: 63
gaagtgcagc tggtggaatc tggcggcgga ctggtgcagc ctggcggatc tctgagactg
60 tcttgtgccg cctccggctt ctccctgtcc tcctacgcta tgatctgggt
gcgacaggcc 120 cctggcaagg gcctggaatg gatcggcacc atctctaccg
gcggaattac ctactacgcc 180 tcctgggcca agggccggtt caccatctcc
agagacaact ccaagaacac cctgtacctc 240 cagatgaact ccctgcgggc
cgaggacacc gccgtgtact attgtgctag aggcggctac 300 gccgccagct
ccgcttacta cctgccctac tacttcgacc tgtggggcca gggcaccctc 360
gtgacagtgt catct 375 <210> SEQ ID NO 64 <211> LENGTH:
336 <212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polynucleotide <400> SEQUENCE:
64 gatattcaga tgacccagtc cccctccagc ctgtccgctt ctgtgggcga
cagagtgacc 60 atcacctgtc agtcctccca gtccgtgtat aacaacaaca
acctggcctg gtatcagcag 120 aaacccggca aggtgcccaa gctgctgatc
tacctggcct ccacactggc ctctggcgtg 180 ccctctagat tctccggctc
tggctctggc accgacttta ccctgaccat cagctccctc 240 cagtgcgagg
atgccgccac ctactattgc ctgggcggct gcgacgacga cgccgatacc 300
tttgcttttg gcggaggcac caaggtggaa atcaaa 336 <210> SEQ ID NO
65 <211> LENGTH: 5 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 65 Gly Tyr Thr Met Asn 1 5
<210> SEQ ID NO 66 <211> LENGTH: 17 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 66 Leu Ile Thr Pro Tyr Asn
Gly Ala Ser Ser Tyr Asn Gln Lys Phe Arg 1 5 10 15 Gly <210>
SEQ ID NO 67 <211> LENGTH: 10 <212> TYPE: PRT
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 67 Gly Gly Tyr Asp Gly Arg
Gly Phe Asp Tyr 1 5 10 <210> SEQ ID NO 68 <211> LENGTH:
10 <212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 68 Ser
Ala Ser Ser Ser Val Ser Tyr Met His 1 5 10 <210> SEQ ID NO 69
<211> LENGTH: 7 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 69 Asp Thr Ser Lys Leu Ala Ser 1 5
<210> SEQ ID NO 70 <211> LENGTH: 9 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 70 Gln Gln Trp Ser Lys His
Pro Leu Thr 1 5 <210> SEQ ID NO 71 <211> LENGTH: 5
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 71 Asp
Thr Tyr Ile His 1 5 <210> SEQ ID NO 72 <211> LENGTH: 17
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 72 Arg
Ile Tyr Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Asp Ser Val Lys 1 5 10
15 Gly <210> SEQ ID NO 73 <211> LENGTH: 11 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic peptide <400> SEQUENCE: 73 Trp Gly Gly
Asp Gly Phe Tyr Ala Met Asp Tyr 1 5 10 <210> SEQ ID NO 74
<211> LENGTH: 11 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 74 Arg Ala Ser Gln Asp Val Asn Thr Ala Val
Ala 1 5 10 <210> SEQ ID NO 75 <211> LENGTH: 7
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 75 Ser
Ala Ser Phe Leu Tyr Ser 1 5 <210> SEQ ID NO 76 <211>
LENGTH: 9 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 76 Gln Gln His Tyr Thr Thr Pro Pro Thr 1 5 <210>
SEQ ID NO 77 <211> LENGTH: 5 <212> TYPE: PRT
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 77 Ser Asp Ala Ile Ser 1 5
<210> SEQ ID NO 78 <211> LENGTH: 16 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 78 Ile Ile Asn Gly Gly Gly
Asn Thr Tyr Tyr Ala Ser Trp Ala Lys Gly 1 5 10 15 <210> SEQ
ID NO 79 <211> LENGTH: 18 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 79 Gly Ile Gln His Gly Gly Gly Asn
Ser Asp Tyr Tyr Tyr Tyr Gly Met 1 5 10 15 Asp Leu <210> SEQ
ID NO 80 <211> LENGTH: 11 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 80 Gln Ala Ser Gln Ser Ile Ser Ser
Val Leu Ser 1 5 10 <210> SEQ ID NO 81 <211> LENGTH: 7
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 81 Leu
Ala Ser Thr Leu Ala Ser 1 5 <210> SEQ ID NO 82 <211>
LENGTH: 14 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 82 Gln Thr Asn Tyr Gly Thr Ser Ser Ser Asn Tyr Gly Phe
Ala 1 5 10 <210> SEQ ID NO 83 <211> LENGTH: 5
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 83 Ser
Asp Ala Ile Ser 1 5 <210> SEQ ID NO 84 <211> LENGTH: 16
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 84 Ile
Ile Asn Gly Gly Gly Asn Thr Tyr Tyr Ala Ser Trp Ala Lys Gly 1 5 10
15 <210> SEQ ID NO 85 <211> LENGTH: 18 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic peptide <400> SEQUENCE: 85 Gly Ile Gln
His Gly Gly Gly Asn Ser Asp Tyr Tyr Tyr Tyr Gly Met 1 5 10 15 Asp
Leu <210> SEQ ID NO 86 <211> LENGTH: 11 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic peptide <400> SEQUENCE: 86 Gln Ala Ser
Gln Ser Ile Ser Ser Val Leu Ser 1 5 10 <210> SEQ ID NO 87
<211> LENGTH: 7 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 87 Leu Ala Ser Thr Leu Ala Ser 1 5
<210> SEQ ID NO 88 <211> LENGTH: 14 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 88 Gln Thr Asn Tyr Gly Thr
Ser Ser Ser Asn Tyr Gly Phe Ala 1 5 10 <210> SEQ ID NO 89
<211> LENGTH: 5 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 89 Asn Tyr Ala Met Ser 1 5 <210> SEQ ID
NO 90 <211> LENGTH: 16 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 90 Ser Ile Ser Thr Gly Gly Leu Ala
Phe Tyr Ala Asn Trp Ala Lys Gly 1 5 10 15 <210> SEQ ID NO 91
<211> LENGTH: 13 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 91 Asn Gly Gly Gly Ser Tyr Ile Phe Tyr Tyr
Phe Asp Leu 1 5 10 <210> SEQ ID NO 92 <211> LENGTH: 11
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 92 Gln
Ala Ser Gln Ser Ile Ser Ser Tyr Leu Ser 1 5 10 <210> SEQ ID
NO 93 <211> LENGTH: 7 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 93 Ser Ala Ser Thr Leu Ala Ser 1 5
<210> SEQ ID NO 94 <211> LENGTH: 10 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 94 Gln Ser Tyr Tyr Asp Ile
Gly Thr Ser Thr 1 5 10 <210> SEQ ID NO 95 <211> LENGTH:
5 <212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 95 Asn
Tyr Ala Met Ser 1 5 <210> SEQ ID NO 96 <211> LENGTH: 16
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 96 Ser
Ile Ser Thr Gly Gly Leu Ala Phe Tyr Ala Asn Trp Ala Lys Gly 1 5 10
15 <210> SEQ ID NO 97 <211> LENGTH: 13 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic peptide <400> SEQUENCE: 97 Asn Gly Gly
Gly Ser Tyr Ile Phe Tyr Tyr Phe Asp Leu 1 5 10 <210> SEQ ID
NO 98 <211> LENGTH: 11 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 98 Gln Ala Ser Gln Ser Ile Ser Ser
Tyr Leu Ser 1 5 10 <210> SEQ ID NO 99 <211> LENGTH: 7
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 99 Ser
Ala Ser Thr Leu Ala Ser 1 5 <210> SEQ ID NO 100 <211>
LENGTH: 10 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 100 Gln Ser Tyr Tyr Asp Ile Gly Thr Ser Thr 1 5 10
<210> SEQ ID NO 101 <211> LENGTH: 5 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 101 Ser Tyr Ala Met Gly 1 5
<210> SEQ ID NO 102 <211> LENGTH: 16 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 102 Thr Ile Asn Ile Gly Gly
Arg Val Tyr Tyr Ala Ser Trp Ala Lys Gly 1 5 10 15 <210> SEQ
ID NO 103 <211> LENGTH: 9 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 103 Tyr Tyr Asn Gly Gly Ser Tyr Asp
Ile 1 5 <210> SEQ ID NO 104 <211> LENGTH: 11
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 104
Gln Ala Ser Glu Ser Ile Tyr Arg Val Leu Ala 1 5 10 <210> SEQ
ID NO 105 <211> LENGTH: 7 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 105 Asp Thr Ser Thr Leu Ala Ser 1 5
<210> SEQ ID NO 106 <211> LENGTH: 12 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 106 Gln Gly Gly Tyr Tyr Ala
Asp Ser Tyr Gly Ile Ala 1 5 10 <210> SEQ ID NO 107
<211> LENGTH: 5 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 107 Ser Tyr Ala Met Gly 1 5 <210> SEQ
ID NO 108 <211> LENGTH: 16 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 108 Thr Ile Asn Ile Gly Gly Arg Val
Tyr Tyr Ala Ser Trp Ala Lys Gly 1 5 10 15 <210> SEQ ID NO 109
<211> LENGTH: 9 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 109 Tyr Tyr Asn Gly Gly Ser Tyr Asp Ile 1 5
<210> SEQ ID NO 110 <211> LENGTH: 11 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 110 Gln Ala Ser Glu Ser Ile
Tyr Arg Val Leu Ala 1 5 10 <210> SEQ ID NO 111 <211>
LENGTH: 7 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 111 Asp Thr Ser Thr Leu Ala Ser 1 5 <210> SEQ ID NO
112 <211> LENGTH: 12 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 112 Gln Gly Gly Tyr Tyr Ala Asp Ser
Tyr Gly Ile Ala 1 5 10 <210> SEQ ID NO 113 <211>
LENGTH: 5 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 113 Ser Tyr Ala Met Ile 1 5 <210> SEQ ID NO 114
<211> LENGTH: 16 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 114 Thr Ile Ser Thr Gly Gly Ile Thr Tyr Tyr
Ala Ser Trp Ala Lys Gly 1 5 10 15 <210> SEQ ID NO 115
<211> LENGTH: 17 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 115 Gly Gly Tyr Ala Ala Ser Ser Ala Tyr Tyr
Leu Pro Tyr Tyr Phe Asp 1 5 10 15 Leu <210> SEQ ID NO 116
<211> LENGTH: 13 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 116 Gln Ser Ser Gln Ser Val Tyr Asn Asn Asn
Asn Leu Ala 1 5 10 <210> SEQ ID NO 117 <211> LENGTH: 7
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 117
Leu Ala Ser Thr Leu Ala Ser 1 5 <210> SEQ ID NO 118
<211> LENGTH: 12 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 118 Leu Gly Gly Cys Asp Asp Asp Ala Asp Thr
Phe Ala 1 5 10 <210> SEQ ID NO 119 <211> LENGTH: 5
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 119
Ser Tyr Ala Met Ile 1 5 <210> SEQ ID NO 120 <211>
LENGTH: 16 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 120 Thr Ile Ser Thr Gly Gly Ile Thr Tyr Tyr Ala Ser Trp
Ala Lys Gly 1 5 10 15 <210> SEQ ID NO 121 <211> LENGTH:
17 <212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 121
Gly Gly Tyr Ala Ala Ser Ser Ala Tyr Tyr Leu Pro Tyr Tyr Phe Asp 1 5
10 15 Leu <210> SEQ ID NO 122 <211> LENGTH: 13
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 122
Gln Ser Ser Gln Ser Val Tyr Asn Asn Asn Asn Leu Ala 1 5 10
<210> SEQ ID NO 123 <211> LENGTH: 7 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 123 Leu Ala Ser Thr Leu Ala
Ser 1 5 <210> SEQ ID NO 124 <211> LENGTH: 12
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 124
Leu Gly Gly Cys Asp Asp Asp Ala Asp Thr Phe Ala 1 5 10 <210>
SEQ ID NO 125 <211> LENGTH: 15 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 125 ggctatgggt
tgtct 15 <210> SEQ ID NO 126 <211> LENGTH: 51
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 126 atgattagta gtggtggtag ttatacctac tatgcagaca
gtgtgaaggg t 51 <210> SEQ ID NO 127 <211> LENGTH: 30
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 127 catggggacg atcccgcctg gttcgcttat 30 <210> SEQ
ID NO 128 <211> LENGTH: 36 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 128 agtgtcagct caagtataag
ttccaacaac ttgcac 36 <210> SEQ ID NO 129 <211> LENGTH:
21 <212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 129 ggcacatcca acctggcttc t 21 <210> SEQ ID NO 130
<211> LENGTH: 33 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 130 caacagtgga gtagttaccc
gtacatgtac acg 33 <210> SEQ ID NO 131 <211> LENGTH: 15
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 131 ggctacacca tgaac 15 <210> SEQ ID NO 132
<211> LENGTH: 51 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 132 cttattactc cttacaatgg
tgcttctagc tacaaccaga agttcagggg c 51 <210> SEQ ID NO 133
<211> LENGTH: 30 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 133 gggggttacg acgggagggg
ttttgactac 30 <210> SEQ ID NO 134 <211> LENGTH: 30
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 134 agtgccagct caagtgtaag ttacatgcac 30 <210> SEQ
ID NO 135 <211> LENGTH: 21 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 135 gacacatcca aactggcttc t
21 <210> SEQ ID NO 136 <211> LENGTH: 27 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic oligonucleotide <400> SEQUENCE: 136
cagcagtgga gtaagcaccc tctcacg 27 <210> SEQ ID NO 137
<211> LENGTH: 15 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 137 agcgatgcaa taagc 15
<210> SEQ ID NO 138 <211> LENGTH: 48 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 138 atcattaatg
gtggtggtaa cacatactac gcgagctggg cgaaaggc 48 <210> SEQ ID NO
139 <211> LENGTH: 54 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 139 ggcattcaac atggtggtgg
taatagtgat tattattatt acggcatgga cctc 54 <210> SEQ ID NO 140
<211> LENGTH: 33 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 140 caggccagtc agagcattag
tagtgtcttg tcc 33 <210> SEQ ID NO 141 <211> LENGTH: 21
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 141 ctggcatcca ctctggcatc t 21 <210> SEQ ID NO 142
<211> LENGTH: 42 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 142 caaaccaatt atggtactag
tagtagtaat tatggttttg ct 42 <210> SEQ ID NO 143 <211>
LENGTH: 15 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 143 tccgacgcga ttagc 15 <210> SEQ ID NO
144 <211> LENGTH: 48 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 144 atcatcaacg gcggcggaaa
cacctactac gcctcctggg ccaagggc 48 <210> SEQ ID NO 145
<211> LENGTH: 54 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 145 ggcatccagc acggtggtgg
aaacagcgac tactactact atgggatgga tctg 54 <210> SEQ ID NO 146
<211> LENGTH: 33 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 146 caggcgtccc agtcaattag
cagcgtgctc tcc 33 <210> SEQ ID NO 147 <211> LENGTH: 21
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 147 ttggcctcca ctctggcctc g 21 <210> SEQ ID NO 148
<211> LENGTH: 42 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 148 caaaccaact acggaacctc
cagctccaac tacggctttg cc 42 <210> SEQ ID NO 149 <211>
LENGTH: 15 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 149 aactatgcaa tgagc 15 <210> SEQ ID NO
150 <211> LENGTH: 48 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 150 tccattagta ctggtggtct
cgcattctac gcgaactggg caaaaggc 48 <210> SEQ ID NO 151
<211> LENGTH: 39 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 151 aatggtggtg gtagttatat
tttctattat tttgacttg 39 <210> SEQ ID NO 152 <211>
LENGTH: 33 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 152 caggccagtc agagcattag tagttactta tcc 33
<210> SEQ ID NO 153 <211> LENGTH: 21 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 153 tctgcatcca
ctctggcatc t 21 <210> SEQ ID NO 154 <211> LENGTH: 30
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 154 caaagctatt atgatattgg tactagtact 30 <210> SEQ
ID NO 155 <211> LENGTH: 15 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 155 aactacgcca tgtcc 15
<210> SEQ ID NO 156 <211> LENGTH: 48 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 156 tccatcagca
caggcggcct ggccttctac gccaattggg ccaagggc 48 <210> SEQ ID NO
157 <211> LENGTH: 39 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 157 aacggcggag gctcctacat
cttctactac ttcgacctg 39 <210> SEQ ID NO 158 <211>
LENGTH: 33 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 158 caggcctccc agtccatctc ctcctacctg tcc 33
<210> SEQ ID NO 159 <211> LENGTH: 21 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 159 tctgcctcca
cactggcctc c 21 <210> SEQ ID NO 160 <211> LENGTH: 30
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 160 cagtcctact acgacatcgg cacctccacc 30 <210> SEQ
ID NO 161 <211> LENGTH: 15 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 161 agctatgcaa tgggc 15
<210> SEQ ID NO 162 <211> LENGTH: 48 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 162 accattaata
ttggtggtcg cgtatattac gcgagctggg caaaaggc 48 <210> SEQ ID NO
163 <211> LENGTH: 27 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 163 tattataatg gtggtagtta
tgacatc 27 <210> SEQ ID NO 164 <211> LENGTH: 33
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 164 caggccagtg agagcattta tcgcgtattg gcc 33 <210>
SEQ ID NO 165 <211> LENGTH: 21 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 165 gatacatcca
ctctggcatc t 21 <210> SEQ ID NO 166 <211> LENGTH: 36
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 166 caaggcggtt attatgctga tagttatggt attgct 36
<210> SEQ ID NO 167 <211> LENGTH: 15 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 167 tcctacgcta
tgggc 15 <210> SEQ ID NO 168 <211> LENGTH: 48
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 168 accatcaaca tcggcggcag agtgtactac gcctcctggg ccaagggc
48 <210> SEQ ID NO 169 <211> LENGTH: 27 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic oligonucleotide <400> SEQUENCE: 169
tactacaacg gcggctccta cgatatc 27 <210> SEQ ID NO 170
<211> LENGTH: 33 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 170 caggcctccg agtccatcta
ccgggtgctg gcc 33 <210> SEQ ID NO 171 <211> LENGTH: 21
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 171 gacaccagca cactggcctc c 21 <210> SEQ ID NO 172
<211> LENGTH: 36 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 172 cagggcggct actacgccga
ctcctacgga atcgct 36 <210> SEQ ID NO 173 <211> LENGTH:
15 <212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 173 agttatgcaa tgatc 15 <210> SEQ ID NO 174
<211> LENGTH: 48 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 174 accattagta ctggtggtat
cacatactac gcgagctggg cgaaaggc 48 <210> SEQ ID NO 175
<211> LENGTH: 51 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 175 gggggatatg ctgctagtag
tgcttattat ctcccgtact actttgactt g 51 <210> SEQ ID NO 176
<211> LENGTH: 39 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 176 cagtcctccc agtccgtgta
taacaacaac aacctggcc 39 <210> SEQ ID NO 177 <211>
LENGTH: 21 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 177 ctggcatcca ctctggcatc t 21 <210>
SEQ ID NO 178 <211> LENGTH: 36 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 178 ctaggtggtt
gtgatgatga tgctgatact tttgct 36 <210> SEQ ID NO 179
<211> LENGTH: 15 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 179 tcctacgcta tgatc 15
<210> SEQ ID NO 180 <211> LENGTH: 48 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 180 accatctcta
ccggcggaat tacctactac gcctcctggg ccaagggc 48 <210> SEQ ID NO
181 <211> LENGTH: 51 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 181 ggcggctacg ccgccagctc
cgcttactac ctgccctact acttcgacct g 51 <210> SEQ ID NO 182
<211> LENGTH: 39 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 182 cagtcctccc agtccgtgta
taacaacaac aacctggcc 39 <210> SEQ ID NO 183 <211>
LENGTH: 21 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 183 ctggcctcca cactggcctc t 21 <210>
SEQ ID NO 184 <211> LENGTH: 36 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 184 ctgggcggct
gcgacgacga cgccgatacc tttgct 36 <210> SEQ ID NO 185
<211> LENGTH: 8 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 185 Gly Tyr Ser Phe Thr Gly Tyr Thr 1 5
<210> SEQ ID NO 186 <211> LENGTH: 8 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 186 Ile Thr Pro Tyr Asn Gly
Ala Ser 1 5 <210> SEQ ID NO 187 <211> LENGTH: 12
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 187
Ala Arg Gly Gly Tyr Asp Gly Arg Gly Phe Asp Tyr 1 5 10 <210>
SEQ ID NO 188 <211> LENGTH: 5 <212> TYPE: PRT
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 188 Ser Ser Val Ser Tyr 1 5
<210> SEQ ID NO 189 <211> LENGTH: 3 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 189 Asp Thr Ser 1
<210> SEQ ID NO 190 <211> LENGTH: 9 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 190 Gln Gln Trp Ser Lys His
Pro Leu Thr 1 5 <210> SEQ ID NO 191 <211> LENGTH: 8
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 191
Gly Phe Asn Ile Lys Asp Thr Tyr 1 5 <210> SEQ ID NO 192
<211> LENGTH: 8 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 192 Ile Tyr Pro Thr Asn Gly Tyr Thr 1 5
<210> SEQ ID NO 193 <211> LENGTH: 13 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 193 Ser Arg Trp Gly Gly Asp
Gly Phe Tyr Ala Met Asp Tyr 1 5 10 <210> SEQ ID NO 194
<211> LENGTH: 6 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 194 Gln Asp Val Asn Thr Ala 1 5 <210>
SEQ ID NO 195 <211> LENGTH: 3 <212> TYPE: PRT
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 195 Ser Ala Ser 1
<210> SEQ ID NO 196 <211> LENGTH: 9 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 196 Gln Gln His Tyr Thr Thr
Pro Pro Thr 1 5 <210> SEQ ID NO 197 <211> LENGTH: 8
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 197
Gly Ile Ser Leu Ser Ser Asp Ala 1 5 <210> SEQ ID NO 198
<211> LENGTH: 7 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 198 Ile Asn Gly Gly Gly Asn Thr 1 5
<210> SEQ ID NO 199 <211> LENGTH: 20 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 199 Ala Arg Gly Ile Gln His
Gly Gly Gly Asn Ser Asp Tyr Tyr Tyr Tyr 1 5 10 15 Gly Met Asp Leu
20 <210> SEQ ID NO 200 <211> LENGTH: 6 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic peptide <400> SEQUENCE: 200 Gln Ser Ile
Ser Ser Val 1 5 <210> SEQ ID NO 201 <211> LENGTH: 3
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 201
Leu Ala Ser 1 <210> SEQ ID NO 202 <211> LENGTH: 14
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 202
Gln Thr Asn Tyr Gly Thr Ser Ser Ser Asn Tyr Gly Phe Ala 1 5 10
<210> SEQ ID NO 203 <211> LENGTH: 8 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 203 Gly Ile Ser Leu Ser Ser
Asp Ala 1 5 <210> SEQ ID NO 204 <211> LENGTH: 7
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 204
Ile Asn Gly Gly Gly Asn Thr 1 5 <210> SEQ ID NO 205
<211> LENGTH: 20 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 205 Ala Arg Gly Ile Gln His Gly Gly Gly Asn
Ser Asp Tyr Tyr Tyr Tyr 1 5 10 15 Gly Met Asp Leu 20 <210>
SEQ ID NO 206 <211> LENGTH: 6 <212> TYPE: PRT
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 206 Gln Ser Ile Ser Ser Val
1 5 <210> SEQ ID NO 207 <211> LENGTH: 3 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic peptide <400> SEQUENCE: 207 Leu Ala Ser 1
<210> SEQ ID NO 208 <211> LENGTH: 14 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 208 Gln Thr Asn Tyr Gly Thr
Ser Ser Ser Asn Tyr Gly Phe Ala 1 5 10 <210> SEQ ID NO 209
<211> LENGTH: 8 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 209 Gly Phe Ser Leu Asn Asn Tyr Ala 1 5
<210> SEQ ID NO 210 <211> LENGTH: 7 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 210 Ile Ser Thr Gly Gly Leu
Ala 1 5 <210> SEQ ID NO 211 <211> LENGTH: 15
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 211
Gly Arg Asn Gly Gly Gly Ser Tyr Ile Phe Tyr Tyr Phe Asp Leu 1 5 10
15 <210> SEQ ID NO 212 <211> LENGTH: 6 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic peptide <400> SEQUENCE: 212 Gln Ser Ile
Ser Ser Tyr 1 5 <210> SEQ ID NO 213 <211> LENGTH: 3
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 213
Ser Ala Ser 1 <210> SEQ ID NO 214 <211> LENGTH: 10
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 214
Gln Ser Tyr Tyr Asp Ile Gly Thr Ser Thr 1 5 10 <210> SEQ ID
NO 215 <211> LENGTH: 8 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 215 Gly Phe Ser Leu Asn Asn Tyr Ala 1
5 <210> SEQ ID NO 216 <211> LENGTH: 7 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 216 Ile Ser Thr Gly Gly Leu
Ala 1 5 <210> SEQ ID NO 217 <211> LENGTH: 15
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 217
Ala Arg Asn Gly Gly Gly Ser Tyr Ile Phe Tyr Tyr Phe Asp Leu 1 5 10
15 <210> SEQ ID NO 218 <211> LENGTH: 6 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic peptide <400> SEQUENCE: 218 Gln Ser Ile
Ser Ser Tyr 1 5 <210> SEQ ID NO 219 <211> LENGTH: 3
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 219
Ser Ala Ser 1 <210> SEQ ID NO 220 <211> LENGTH: 10
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 220
Gln Ser Tyr Tyr Asp Ile Gly Thr Ser Thr 1 5 10 <210> SEQ ID
NO 221 <211> LENGTH: 8 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 221 Gly Ile Asp Leu Ser Ser Tyr Ala 1
5 <210> SEQ ID NO 222 <211> LENGTH: 7 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 222 Ile Asn Ile Gly Gly Arg
Val 1 5 <210> SEQ ID NO 223 <211> LENGTH: 11
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 223
Ala Arg Tyr Tyr Asn Gly Gly Ser Tyr Asp Ile 1 5 10 <210> SEQ
ID NO 224 <211> LENGTH: 6 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 224 Glu Ser Ile Tyr Arg Val 1 5
<210> SEQ ID NO 225 <211> LENGTH: 3 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 225 Asp Thr Ser 1
<210> SEQ ID NO 226 <211> LENGTH: 12 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 226 Gln Gly Gly Tyr Tyr Ala
Asp Ser Tyr Gly Ile Ala 1 5 10 <210> SEQ ID NO 227
<211> LENGTH: 8 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 227 Gly Ile Asp Leu Ser Ser Tyr Ala 1 5
<210> SEQ ID NO 228 <211> LENGTH: 7 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 228 Ile Asn Ile Gly Gly Arg
Val 1 5 <210> SEQ ID NO 229 <211> LENGTH: 11
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 229
Ala Arg Tyr Tyr Asn Gly Gly Ser Tyr Asp Ile 1 5 10 <210> SEQ
ID NO 230 <211> LENGTH: 6 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 230 Glu Ser Ile Tyr Arg Val 1 5
<210> SEQ ID NO 231 <211> LENGTH: 3 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 231 Asp Thr Ser 1
<210> SEQ ID NO 232 <211> LENGTH: 12 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 232 Gln Gly Gly Tyr Tyr Ala
Asp Ser Tyr Gly Ile Ala 1 5 10 <210> SEQ ID NO 233
<211> LENGTH: 8 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 233 Gly Phe Ser Leu Ser Ser Tyr Ala 1 5
<210> SEQ ID NO 234 <211> LENGTH: 7 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 234 Ile Ser Thr Gly Gly Ile
Thr 1 5 <210> SEQ ID NO 235 <211> LENGTH: 19
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 235
Ala Arg Gly Gly Tyr Ala Ala Ser Ser Ala Tyr Tyr Leu Pro Tyr Tyr 1 5
10 15 Phe Asp Leu <210> SEQ ID NO 236 <211> LENGTH: 8
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 236
Gln Ser Val Tyr Asn Asn Asn Asn 1 5 <210> SEQ ID NO 237
<211> LENGTH: 3 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 237 Leu Ala Ser 1 <210> SEQ ID NO 238
<211> LENGTH: 12 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 238 Leu Gly Gly Cys Asp Asp Asp Ala Asp Thr
Phe Ala 1 5 10 <210> SEQ ID NO 239 <211> LENGTH: 8
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 239
Gly Phe Ser Leu Ser Ser Tyr Ala 1 5 <210> SEQ ID NO 240
<211> LENGTH: 7 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 240 Ile Ser Thr Gly Gly Ile Thr 1 5
<210> SEQ ID NO 241 <211> LENGTH: 19 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 241 Ala Arg Gly Gly Tyr Ala
Ala Ser Ser Ala Tyr Tyr Leu Pro Tyr Tyr 1 5 10 15 Phe Asp Leu
<210> SEQ ID NO 242 <211> LENGTH: 8 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 242 Gln Ser Val Tyr Asn Asn
Asn Asn 1 5 <210> SEQ ID NO 243 <211> LENGTH: 3
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 243
Leu Ala Ser 1 <210> SEQ ID NO 244 <211> LENGTH: 12
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 244
Leu Gly Gly Cys Asp Asp Asp Ala Asp Thr Phe Ala 1 5 10 <210>
SEQ ID NO 245 <211> LENGTH: 24 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 245 ggcttcacct
tcagcggcta tggg 24 <210> SEQ ID NO 246 <211> LENGTH: 24
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 246 attagtagtg gtggtagtta tacc 24 <210> SEQ ID NO
247 <211> LENGTH: 36 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 247 gcaagacatg gggacgatcc
cgcctggttc gcttat 36 <210> SEQ ID NO 248 <211> LENGTH:
21 <212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 248 tcaagtataa gttccaacaa c 21 <210> SEQ ID NO 249
<211> LENGTH: 9 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 249 ggcacatcc 9 <210>
SEQ ID NO 250 <211> LENGTH: 33 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 250 caacagtgga
gtagttaccc gtacatgtac acg 33 <210> SEQ ID NO 251 <211>
LENGTH: 24 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 251 ggttactcat tcactggcta cacc 24 <210>
SEQ ID NO 252 <211> LENGTH: 24 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 252 attactcctt
acaatggtgc ttct 24 <210> SEQ ID NO 253 <211> LENGTH: 36
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 253 gcaagggggg gttacgacgg gaggggtttt gactac 36
<210> SEQ ID NO 254 <211> LENGTH: 15 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 254 tcaagtgtaa
gttac 15 <210> SEQ ID NO 255 <211> LENGTH: 9
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 255 gacacatcc 9 <210> SEQ ID NO 256 <211>
LENGTH: 27 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 256 cagcagtgga gtaagcaccc tctcacg 27
<210> SEQ ID NO 257 <211> LENGTH: 24 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 257 ggaatctccc
tcagtagcga tgca 24 <210> SEQ ID NO 258 <211> LENGTH: 21
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 258 attaatggtg gtggtaacac a 21 <210> SEQ ID NO 259
<211> LENGTH: 60 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 259 gccagaggca ttcaacatgg
tggtggtaat agtgattatt attattacgg catggacctc 60 <210> SEQ ID
NO 260 <211> LENGTH: 18 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 260 cagagcatta gtagtgtc 18
<210> SEQ ID NO 261 <211> LENGTH: 9 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 261 ctggcatct 9
<210> SEQ ID NO 262 <211> LENGTH: 42 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 262 caaaccaatt
atggtactag tagtagtaat tatggttttg ct 42 <210> SEQ ID NO 263
<211> LENGTH: 24 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 263 ggaatttccc tctcctccga
cgcg 24 <210> SEQ ID NO 264 <211> LENGTH: 21
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 264 atcaacggcg gcggaaacac c 21 <210> SEQ ID NO 265
<211> LENGTH: 60 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 265 gcgcgcggca tccagcacgg
tggtggaaac agcgactact actactatgg gatggatctg 60 <210> SEQ ID
NO 266 <211> LENGTH: 18 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 266 cagtcaatta gcagcgtg 18
<210> SEQ ID NO 267 <211> LENGTH: 9 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 267 ttggcctcc 9
<210> SEQ ID NO 268 <211> LENGTH: 42 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 268 caaaccaact
acggaacctc cagctccaac tacggctttg cc 42 <210> SEQ ID NO 269
<211> LENGTH: 24 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 269 ggattctccc tcaataacta
tgca 24 <210> SEQ ID NO 270 <211> LENGTH: 21
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 270 attagtactg gtggtctcgc a 21 <210> SEQ ID NO 271
<211> LENGTH: 45 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 271 ggcagaaatg gtggtggtag
ttatattttc tattattttg acttg 45 <210> SEQ ID NO 272
<211> LENGTH: 18 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 272 cagagcatta gtagttac 18
<210> SEQ ID NO 273 <211> LENGTH: 9 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 273 tctgcatcc 9
<210> SEQ ID NO 274 <211> LENGTH: 30 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 274 caaagctatt
atgatattgg tactagtact 30 <210> SEQ ID NO 275 <211>
LENGTH: 24 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 275 ggcttctccc tgaacaacta cgcc 24 <210>
SEQ ID NO 276 <211> LENGTH: 21 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 276 atcagcacag
gcggcctggc c 21 <210> SEQ ID NO 277 <211> LENGTH: 45
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 277 gccagaaacg gcggaggctc ctacatcttc tactacttcg acctg 45
<210> SEQ ID NO 278 <211> LENGTH: 18 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 278 cagtccatct
cctcctac 18 <210> SEQ ID NO 279 <211> LENGTH: 9
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 279 tctgcctcc 9 <210> SEQ ID NO 280 <400>
SEQUENCE: 280 000 <210> SEQ ID NO 281 <400> SEQUENCE:
281 000 <210> SEQ ID NO 282 <400> SEQUENCE: 282 000
<210> SEQ ID NO 283 <400> SEQUENCE: 283 000 <210>
SEQ ID NO 284 <400> SEQUENCE: 284 000 <210> SEQ ID NO
285 <400> SEQUENCE: 285 000 <210> SEQ ID NO 286
<400> SEQUENCE: 286 000 <210> SEQ ID NO 287 <400>
SEQUENCE: 287 000 <210> SEQ ID NO 288 <400> SEQUENCE:
288 000 <210> SEQ ID NO 289 <400> SEQUENCE: 289 000
<210> SEQ ID NO 290 <400> SEQUENCE: 290 000 <210>
SEQ ID NO 291 <400> SEQUENCE: 291 000 <210> SEQ ID NO
292 <400> SEQUENCE: 292 000 <210> SEQ ID NO 293
<400> SEQUENCE: 293 000 <210> SEQ ID NO 294 <400>
SEQUENCE: 294 000 <210> SEQ ID NO 295 <400> SEQUENCE:
295 000 <210> SEQ ID NO 296 <400> SEQUENCE: 296 000
<210> SEQ ID NO 297 <400> SEQUENCE: 297 000 <210>
SEQ ID NO 298 <400> SEQUENCE: 298 000 <210> SEQ ID NO
299 <400> SEQUENCE: 299 000 <210> SEQ ID NO 300
<211> LENGTH: 30 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 300 cagtcctact acgacatcgg
cacctccacc 30 <210> SEQ ID NO 301 <211> LENGTH: 24
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 301 ggaatcgacc tcagtagcta tgca 24 <210> SEQ ID NO
302 <211> LENGTH: 21 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 302 attaatattg gtggtcgcgt a
21 <210> SEQ ID NO 303 <211> LENGTH: 33 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic oligonucleotide <400> SEQUENCE: 303
gccagatatt ataatggtgg tagttatgac atc 33 <210> SEQ ID NO 304
<211> LENGTH: 18 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 304 gagagcattt atcgcgta 18
<210> SEQ ID NO 305 <211> LENGTH: 9 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 305 gatacatcc 9
<210> SEQ ID NO 306 <211> LENGTH: 36 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 306 caaggcggtt
attatgctga tagttatggt attgct 36 <210> SEQ ID NO 307
<211> LENGTH: 24 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 307 ggaatcgacc tgtcctccta
cgct 24 <210> SEQ ID NO 308 <211> LENGTH: 21
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 308 atcaacatcg gcggcagagt g 21 <210> SEQ ID NO 309
<211> LENGTH: 33 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 309 gcccggtact acaacggcgg
ctcctacgat atc 33 <210> SEQ ID NO 310 <211> LENGTH: 18
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 310 gagtccatct accgggtg 18 <210> SEQ ID NO 311
<211> LENGTH: 9 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 311 gacaccagc 9 <210>
SEQ ID NO 312 <211> LENGTH: 36 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 312 cagggcggct
actacgccga ctcctacgga atcgct 36 <210> SEQ ID NO 313
<211> LENGTH: 24 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 313 ggattctccc tcagtagtta
tgca 24 <210> SEQ ID NO 314 <211> LENGTH: 21
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 314 attagtactg gtggtatcac a 21 <210> SEQ ID NO 315
<211> LENGTH: 57 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 315 gccagagggg gatatgctgc
tagtagtgct tattatctcc cgtactactt tgacttg 57 <210> SEQ ID NO
316 <211> LENGTH: 24 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 316 cagagtgttt ataataataa
caac 24 <210> SEQ ID NO 317 <211> LENGTH: 9 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic oligonucleotide <400> SEQUENCE: 317
ctggcatcc 9 <210> SEQ ID NO 318 <211> LENGTH: 36
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 318 ctaggtggtt gtgatgatga tgctgatact tttgct 36
<210> SEQ ID NO 319 <211> LENGTH: 24 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 319 ggcttctccc
tgtcctccta cgct 24 <210> SEQ ID NO 320 <211> LENGTH: 21
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 320 atctctaccg gcggaattac c 21 <210> SEQ ID NO 321
<211> LENGTH: 57 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 321 gctagaggcg gctacgccgc
cagctccgct tactacctgc cctactactt cgacctg 57 <210> SEQ ID NO
322 <211> LENGTH: 24 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 322 cagtccgtgt ataacaacaa
caac 24 <210> SEQ ID NO 323 <211> LENGTH: 9 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic oligonucleotide <400> SEQUENCE: 323
ctggcctcc 9 <210> SEQ ID NO 324 <211> LENGTH: 36
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 324 ctgggcggct gcgacgacga cgccgatacc tttgct 36
<210> SEQ ID NO 325 <211> LENGTH: 449 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 325 Gln Val Gln Leu Gln
Gln Ser Gly Pro Glu Leu Glu Lys Pro Gly Ala 1 5 10 15 Ser Val Lys
Ile Ser Cys Lys Ala Ser Gly Tyr Ser Phe Thr Gly Tyr 20 25 30 Thr
Met Asn Trp Val Lys Gln Ser His Gly Lys Ser Leu Glu Trp Ile 35 40
45 Gly Leu Ile Thr Pro Tyr Asn Gly Ala Ser Ser Tyr Asn Gln Lys Phe
50 55 60 Arg Gly Lys Ala Thr Leu Thr Val Asp Lys Ser Ser Ser Thr
Ala Tyr 65 70 75 80 Met Asp Leu Leu Ser Leu Thr Ser Glu Asp Ser Ala
Val Tyr Phe Cys 85 90 95 Ala Arg Gly Gly Tyr Asp Gly Arg Gly Phe
Asp Tyr Trp Gly Ser Gly 100 105 110 Thr Pro Val Thr Val Ser Ser Ala
Ser Thr Lys Gly Pro Ser Val Phe 115 120 125 Pro Leu Ala Pro Ser Ser
Lys Ser Thr Ser Gly Gly Thr Ala Ala Leu 130 135 140 Gly Cys Leu Val
Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp 145 150 155 160 Asn
Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val Leu 165 170
175 Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser
180 185 190 Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His
Lys Pro 195 200 205 Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys
Ser Cys Asp Lys 210 215 220 Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Glu Leu Leu Gly Gly Pro 225 230 235 240 Ser Val Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser 245 250 255 Arg Thr Pro Glu Val
Thr Cys Val Val Val Asp Val Ser His Glu Asp 260 265 270 Pro Glu Val
Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn 275 280 285 Ala
Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val 290 295
300 Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu
305 310 315 320 Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro
Ile Glu Lys 325 330 335 Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr 340 345 350 Leu Pro Pro Ser Arg Asp Glu Leu Thr
Lys Asn Gln Val Ser Leu Thr 355 360 365 Cys Leu Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu Trp Glu 370 375 380 Ser Asn Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu 385 390 395 400 Asp Ser
Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys 405 410 415
Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu 420
425 430 Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro
Gly 435 440 445 Lys <210> SEQ ID NO 326 <211> LENGTH:
213 <212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE:
326 Asp Ile Glu Leu Thr Gln Ser Pro Ala Ile Met Ser Ala Ser Pro Gly
1 5 10 15 Glu Lys Val Thr Met Thr Cys Ser Ala Ser Ser Ser Val Ser
Tyr Met 20 25 30 His Trp Tyr Gln Gln Lys Ser Gly Thr Ser Pro Lys
Arg Trp Ile Tyr 35 40 45 Asp Thr Ser Lys Leu Ala Ser Gly Val Pro
Gly Arg Phe Ser Gly Ser 50 55 60 Gly Ser Gly Asn Ser Tyr Ser Leu
Thr Ile Ser Ser Val Glu Ala Glu 65 70 75 80 Asp Asp Ala Thr Tyr Tyr
Cys Gln Gln Trp Ser Lys His Pro Leu Thr 85 90 95 Phe Gly Ser Gly
Thr Lys Val Glu Ile Lys Arg Thr Val Ala Ala Pro 100 105 110 Ser Val
Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly Thr 115 120 125
Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala Lys 130
135 140 Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln
Glu 145 150 155 160 Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr
Ser Leu Ser Ser 165 170 175 Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu
Lys His Lys Val Tyr Ala 180 185 190 Cys Glu Val Thr His Gln Gly Leu
Ser Ser Pro Val Thr Lys Ser Phe 195 200 205 Asn Arg Gly Glu Cys 210
<210> SEQ ID NO 327 <211> LENGTH: 451 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 327 Glu Val Gln Leu Val
Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg
Leu Ser Cys Ala Ala Ser Gly Phe Asn Ile Lys Asp Thr 20 25 30 Tyr
Ile His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45 Ala Arg Ile Tyr Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Asp Ser Val
50 55 60 Lys Gly Arg Phe Thr Ile Ser Ala Asp Thr Ser Lys Asn Thr
Ala Tyr 65 70 75 80 Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala
Val Tyr Tyr Cys 85 90 95 Ser Arg Trp Gly Gly Asp Gly Phe Tyr Ala
Met Asp Tyr Trp Gly Gln 100 105 110 Gly Thr Leu Val Thr Val Ser Ser
Ala Ser Thr Lys Gly Pro Ser Val 115 120 125 Phe Pro Leu Ala Pro Ser
Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala 130 135 140 Leu Gly Cys Leu
Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser 145 150 155 160 Trp
Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val 165 170
175 Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro
180 185 190 Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn
His Lys 195 200 205 Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro
Pro Lys Ser Cys 210 215 220 Asp Lys Thr His Thr Cys Pro Pro Cys Pro
Ala Pro Glu Leu Leu Gly 225 230 235 240 Gly Pro Ser Val Phe Leu Phe
Pro Pro Lys Pro Lys Asp Thr Leu Met 245 250 255 Ile Ser Arg Thr Pro
Glu Val Thr Cys Val Val Val Asp Val Ser His 260 265 270 Glu Asp Pro
Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val 275 280 285 His
Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr 290 295
300 Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly
305 310 315 320 Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro
Ala Pro Ile 325 330 335 Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro
Arg Glu Pro Gln Val 340 345 350 Tyr Thr Leu Pro Pro Ser Arg Asp Glu
Leu Thr Lys Asn Gln Val Ser 355 360 365 Leu Thr Cys Leu Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala Val Glu 370 375 380 Trp Glu Ser Asn Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro 385 390 395 400 Val Leu
Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val 405 410 415
Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met 420
425 430 His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu
Ser 435 440 445 Pro Gly Lys 450 <210> SEQ ID NO 328
<211> LENGTH: 214 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 328 Asp Ile Gln Met Thr Gln Ser
Pro Ser Ser Leu Ser Ala Ser Val Gly 1 5 10 15 Asp Arg Val Thr Ile
Thr Cys Arg Ala Ser Gln Asp Val Asn Thr Ala 20 25 30 Val Ala Trp
Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45 Tyr
Ser Ala Ser Phe Leu Tyr Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55
60 Ser Arg Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro
65 70 75 80 Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln His Tyr Thr Thr
Pro Pro 85 90 95 Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg
Thr Val Ala Ala 100 105 110 Pro Ser Val Phe Ile Phe Pro Pro Ser Asp
Glu Gln Leu Lys Ser Gly 115 120 125 Thr Ala Ser Val Val Cys Leu Leu
Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140 Lys Val Gln Trp Lys Val
Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln 145 150 155 160 Glu Ser Val
Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175 Ser
Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185
190 Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser
195 200 205 Phe Asn Arg Gly Glu Cys 210 <210> SEQ ID NO 329
<211> LENGTH: 453 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 329 Gln Ser Val Glu Glu Ser Gly
Gly Arg Leu Val Thr Pro Gly Thr Pro 1 5 10 15 Leu Thr Leu Thr Cys
Thr Val Ser Gly Ile Ser Leu Ser Ser Asp Ala 20 25 30 Ile Ser Trp
Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Tyr Ile Gly 35 40 45 Ile
Ile Asn Gly Gly Gly Asn Thr Tyr Tyr Ala Ser Trp Ala Lys Gly 50 55
60 Arg Phe Thr Ile Ser Lys Thr Ser Thr Thr Val Asp Leu Lys Ile Thr
65 70 75 80 Ser Pro Thr Thr Glu Asp Thr Ala Thr Tyr Phe Cys Ala Arg
Gly Ile 85 90 95 Gln His Gly Gly Gly Asn Ser Asp Tyr Tyr Tyr Tyr
Gly Met Asp Leu 100 105 110 Trp Gly Pro Gly Thr Leu Val Thr Val Ser
Ser Ala Ser Thr Lys Gly 115 120 125 Pro Ser Val Phe Pro Leu Ala Pro
Ser Ser Lys Ser Thr Ser Gly Gly 130 135 140 Thr Ala Ala Leu Gly Cys
Leu Val Lys Asp Tyr Phe Pro Glu Pro Val 145 150 155 160 Thr Val Ser
Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe 165 170 175 Pro
Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val 180 185
190 Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val
195 200 205 Asn His Lys Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu
Pro Lys 210 215 220 Ser Cys Asp Lys Thr His Thr Cys Pro Pro Cys Pro
Ala Pro Glu Leu 225 230 235 240 Leu Gly Gly Pro Ser Val Phe Leu Phe
Pro Pro Lys Pro Lys Asp Thr 245 250 255 Leu Met Ile Ser Arg Thr Pro
Glu Val Thr Cys Val Val Val Asp Val 260 265 270 Ser His Glu Asp Pro
Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val 275 280 285 Glu Val His
Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser 290 295 300 Thr
Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu 305 310
315 320 Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro
Ala 325 330 335 Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro
Arg Glu Pro 340 345 350 Gln Val Tyr Thr Leu Pro Pro Ser Arg Asp Glu
Leu Thr Lys Asn Gln 355 360 365 Val Ser Leu Thr Cys Leu Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala 370 375 380 Val Glu Trp Glu Ser Asn Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr 385 390 395 400 Pro Pro Val Leu
Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu 405 410 415 Thr Val
Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser 420 425 430
Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser 435
440 445 Leu Ser Pro Gly Lys 450 <210> SEQ ID NO 330
<211> LENGTH: 219 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 330 Glu Val Leu Met Thr Gln Thr
Pro Ser Ser Val Ser Ala Ala Val Gly 1 5 10 15 Asp Thr Val Thr Ile
Lys Cys Gln Ala Ser Gln Ser Ile Ser Ser Val 20 25 30 Leu Ser Trp
Tyr Gln Gln Lys Pro Gly Gln Pro Pro Lys Leu Leu Ile 35 40 45 Tyr
Leu Ala Ser Thr Leu Ala Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55
60 Ser Arg Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser Asp Leu Glu Cys
65 70 75 80 Asp Asp Ala Ala Thr Tyr Tyr Cys Gln Thr Asn Tyr Gly Thr
Ser Ser 85 90 95 Ser Asn Tyr Gly Phe Ala Phe Gly Gly Gly Thr Glu
Val Val Val Lys 100 105 110 Arg Thr Val Ala Ala Pro Ser Val Phe Ile
Phe Pro Pro Ser Asp Glu 115 120 125 Gln Leu Lys Ser Gly Thr Ala Ser
Val Val Cys Leu Leu Asn Asn Phe 130 135 140 Tyr Pro Arg Glu Ala Lys
Val Gln Trp Lys Val Asp Asn Ala Leu Gln 145 150 155 160 Ser Gly Asn
Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser 165 170 175 Thr
Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu 180 185
190 Lys His Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser
195 200 205 Pro Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 210 215
<210> SEQ ID NO 331 <211> LENGTH: 456 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 331 Glu Val Gln Leu Val
Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg
Leu Ser Cys Ala Ala Ser Gly Ile Ser Leu Ser Ser Asp 20 25 30 Ala
Ile Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Tyr Ile 35 40
45 Gly Ile Ile Asn Gly Gly Gly Asn Thr Tyr Tyr Ala Ser Trp Ala Lys
50 55 60 Gly Arg Phe Thr Ile Ser Arg His Asn Ser Lys Asn Thr Leu
Tyr Leu 65 70 75 80 Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val
Tyr Tyr Cys Ala 85 90 95 Arg Gly Ile Gln His Gly Gly Gly Asn Ser
Asp Tyr Tyr Tyr Tyr Gly 100 105 110 Met Asp Leu Trp Gly Gln Gly Thr
Leu Val Thr Val Ser Ser Ala Ser 115 120 125 Thr Lys Gly Pro Ser Val
Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr 130 135 140 Ser Gly Gly Thr
Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro 145 150 155 160 Glu
Pro Val Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val 165 170
175 His Thr Phe Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser
180 185 190 Ser Val Val Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr
Tyr Ile 195 200 205 Cys Asn Val Asn His Lys Pro Ser Asn Thr Lys Val
Asp Lys Lys Val 210 215 220 Glu Pro Lys Ser Cys Asp Lys Thr His Thr
Cys Pro Pro Cys Pro Ala 225 230 235 240 Pro Glu Leu Leu Gly Gly Pro
Ser Val Phe Leu Phe Pro Pro Lys Pro 245 250 255 Lys Asp Thr Leu Met
Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val 260 265 270 Val Asp Val
Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val 275 280 285 Asp
Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln 290 295
300 Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln
305 310 315 320 Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser
Asn Lys Ala 325 330 335 Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys
Ala Lys Gly Gln Pro 340 345 350 Arg Glu Pro Gln Val Tyr Thr Leu Pro
Pro Ser Arg Asp Glu Leu Thr 355 360 365 Lys Asn Gln Val Ser Leu Thr
Cys Leu Val Lys Gly Phe Tyr Pro Ser 370 375 380 Asp Ile Ala Val Glu
Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr 385 390 395 400 Lys Thr
Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr 405 410 415
Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe 420
425 430 Ser Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln
Lys 435 440 445 Ser Leu Ser Leu Ser Pro Gly Lys 450 455 <210>
SEQ ID NO 332 <211> LENGTH: 219 <212> TYPE: PRT
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 332 Asp Ile Gln Met Thr
Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly 1 5 10 15 Asp Arg Val
Thr Ile Thr Cys Gln Ala Ser Gln Ser Ile Ser Ser Val 20 25 30 Leu
Ser Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40
45 Tyr Leu Ala Ser Thr Leu Ala Ser Gly Val Pro Ser Arg Phe Ser Gly
50 55 60 Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu
Gln Cys 65 70 75 80 Glu Asp Ile Ala Thr Tyr Tyr Cys Gln Thr Asn Tyr
Gly Thr Ser Ser 85 90 95 Ser Asn Tyr Gly Phe Ala Phe Gly Gly Gly
Thr Lys Val Glu Ile Lys 100 105 110 Arg Thr Val Ala Ala Pro Ser Val
Phe Ile Phe Pro Pro Ser Asp Glu 115 120 125 Gln Leu Lys Ser Gly Thr
Ala Ser Val Val Cys Leu Leu Asn Asn Phe 130 135 140 Tyr Pro Arg Glu
Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln 145 150 155 160 Ser
Gly Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser 165 170
175 Thr Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu
180 185 190 Lys His Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu
Ser Ser 195 200 205 Pro Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 210
215 <210> SEQ ID NO 333 <211> LENGTH: 448 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polypeptide <400> SEQUENCE: 333 Gln Ser
Val Glu Glu Ser Gly Gly Arg Leu Val Thr Pro Gly Thr Pro 1 5 10 15
Leu Thr Leu Thr Cys Thr Val Ser Gly Phe Ser Leu Asn Asn Tyr Ala 20
25 30 Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Ile
Gly 35 40 45 Ser Ile Ser Thr Gly Gly Leu Ala Phe Tyr Ala Asn Trp
Ala Lys Gly 50 55 60 Arg Phe Thr Ile Ser Arg Thr Ser Thr Thr Val
Asp Leu Lys Met Thr 65 70 75 80 Ser Leu Thr Thr Glu Asp Thr Ala Thr
Tyr Phe Cys Gly Arg Asn Gly 85 90 95 Gly Gly Ser Tyr Ile Phe Tyr
Tyr Phe Asp Leu Trp Gly Gln Gly Thr 100 105 110 Leu Val Thr Val Ser
Ser Ala Ser Thr Lys Gly Pro Ser Val Phe Pro 115 120 125 Leu Ala Pro
Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala Leu Gly 130 135 140 Cys
Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp Asn 145 150
155 160 Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val Leu
Gln 165 170 175 Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val
Pro Ser Ser 180 185 190 Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val
Asn His Lys Pro Ser 195 200 205 Asn Thr Lys Val Asp Lys Lys Val Glu
Pro Lys Ser Cys Asp Lys Thr 210 215 220 His Thr Cys Pro Pro Cys Pro
Ala Pro Glu Leu Leu Gly Gly Pro Ser 225 230 235 240 Val Phe Leu Phe
Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg 245 250 255 Thr Pro
Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro 260 265 270
Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala 275
280 285 Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val
Val 290 295 300 Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly
Lys Glu Tyr 305 310 315 320 Lys Cys Lys Val Ser Asn Lys Ala Leu Pro
Ala Pro Ile Glu Lys Thr 325 330 335 Ile Ser Lys Ala Lys Gly Gln Pro
Arg Glu Pro Gln Val Tyr Thr Leu 340 345 350 Pro Pro Ser Arg Asp Glu
Leu Thr Lys Asn Gln Val Ser Leu Thr Cys 355 360 365 Leu Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser 370 375 380 Asn Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp 385 390 395
400 Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser
405 410 415 Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His
Glu Ala 420 425 430 Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu
Ser Pro Gly Lys 435 440 445 <210> SEQ ID NO 334 <211>
LENGTH: 215 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polypeptide
<400> SEQUENCE: 334 Ala Phe Glu Leu Thr Gln Thr Pro Ser Ser
Val Glu Ala Ala Val Gly 1 5 10 15 Gly Thr Ile Thr Ile Lys Cys Gln
Ala Ser Gln Ser Ile Ser Ser Tyr 20 25 30 Leu Ser Trp Tyr Gln Gln
Lys Pro Gly Gln Pro Pro Lys Leu Leu Ile 35 40 45 Tyr Ser Ala Ser
Thr Leu Ala Ser Gly Val Ser Ser Arg Phe Lys Gly 50 55 60 Ser Gly
Ser Gly Thr Glu Tyr Thr Leu Thr Ile Ser Asp Leu Glu Cys 65 70 75 80
Ala Asp Ala Ala Thr Tyr Phe Cys Gln Ser Tyr Tyr Asp Ile Gly Thr 85
90 95 Ser Thr Phe Gly Gly Gly Thr Glu Val Val Val Lys Arg Thr Val
Ala 100 105 110 Ala Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln
Leu Lys Ser 115 120 125 Gly Thr Ala Ser Val Val Cys Leu Leu Asn Asn
Phe Tyr Pro Arg Glu 130 135 140 Ala Lys Val Gln Trp Lys Val Asp Asn
Ala Leu Gln Ser Gly Asn Ser 145 150 155 160 Gln Glu Ser Val Thr Glu
Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu 165 170 175 Ser Ser Thr Leu
Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val 180 185 190 Tyr Ala
Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys 195 200 205
Ser Phe Asn Arg Gly Glu Cys 210 215 <210> SEQ ID NO 335
<211> LENGTH: 451 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 335 Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg Leu Ser
Cys Ala Ala Ser Gly Phe Ser Leu Asn Asn Tyr 20 25 30 Ala Met Ser
Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Ile 35 40 45 Gly
Ser Ile Ser Thr Gly Gly Leu Ala Phe Tyr Ala Asn Trp Ala Lys 50 55
60 Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr Leu
65 70 75 80 Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr
Cys Ala 85 90 95 Arg Asn Gly Gly Gly Ser Tyr Ile Phe Tyr Tyr Phe
Asp Leu Trp Gly 100 105 110 Gln Gly Thr Leu Val Thr Val Ser Ser Ala
Ser Thr Lys Gly Pro Ser 115 120 125 Val Phe Pro Leu Ala Pro Ser Ser
Lys Ser Thr Ser Gly Gly Thr Ala 130 135 140 Ala Leu Gly Cys Leu Val
Lys Asp Tyr Phe Pro Glu Pro Val Thr Val 145 150 155 160 Ser Trp Asn
Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala 165 170 175 Val
Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val 180 185
190 Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His
195 200 205 Lys Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys
Ser Cys 210 215 220 Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Glu Leu Leu Gly 225 230 235 240 Gly Pro Ser Val Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met 245 250 255 Ile Ser Arg Thr Pro Glu Val
Thr Cys Val Val Val Asp Val Ser His 260 265 270 Glu Asp Pro Glu Val
Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val 275 280 285 His Asn Ala
Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr 290 295 300 Arg
Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly 305 310
315 320 Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro
Ile 325 330 335 Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu
Pro Gln Val 340 345 350 Tyr Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr
Lys Asn Gln Val Ser 355 360 365 Leu Thr Cys Leu Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu 370 375 380 Trp Glu Ser Asn Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro 385 390 395 400 Val Leu Asp Ser
Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val 405 410 415 Asp Lys
Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met 420 425 430
His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser 435
440 445 Pro Gly Lys 450 <210> SEQ ID NO 336 <211>
LENGTH: 215 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polypeptide
<400> SEQUENCE: 336 Asp Ile Gln Met Thr Gln Ser Pro Ser Ser
Leu Ser Ala Ser Val Gly 1 5 10 15 Asp Arg Val Thr Ile Thr Cys Gln
Ala Ser Gln Ser Ile Ser Ser Tyr 20 25 30 Leu Ser Trp Tyr Gln Gln
Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45 Tyr Ser Ala Ser
Thr Leu Ala Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60 Ser Gly
Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Cys 65 70 75 80
Glu Asp Ala Ala Thr Tyr Tyr Cys Gln Ser Tyr Tyr Asp Ile Gly Thr 85
90 95 Ser Thr Phe Gly Gly Gly Thr Lys Val Glu Ile Lys Arg Thr Val
Ala 100 105 110 Ala Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln
Leu Lys Ser 115 120 125 Gly Thr Ala Ser Val Val Cys Leu Leu Asn Asn
Phe Tyr Pro Arg Glu 130 135 140 Ala Lys Val Gln Trp Lys Val Asp Asn
Ala Leu Gln Ser Gly Asn Ser 145 150 155 160 Gln Glu Ser Val Thr Glu
Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu 165 170 175 Ser Ser Thr Leu
Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val 180 185 190 Tyr Ala
Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys 195 200 205
Ser Phe Asn Arg Gly Glu Cys 210 215 <210> SEQ ID NO 337
<211> LENGTH: 444 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 337 Gln Ser Val Lys Glu Ser Gly
Gly Arg Leu Val Thr Pro Gly Thr Pro 1 5 10 15 Leu Thr Leu Thr Cys
Thr Val Ser Gly Ile Asp Leu Ser Ser Tyr Ala 20 25 30 Met Gly Trp
Phe Arg Gln Ala Pro Gly Lys Gly Leu Glu Tyr Ile Gly 35 40 45 Thr
Ile Asn Ile Gly Gly Arg Val Tyr Tyr Ala Ser Trp Ala Lys Gly 50 55
60 Arg Phe Thr Ile Ser Arg Thr Ser Thr Thr Val Asp Leu Lys Ala Pro
65 70 75 80 Ser Leu Thr Ala Glu Asp Thr Ala Thr Tyr Phe Cys Ala Arg
Tyr Tyr 85 90 95 Asn Gly Gly Ser Tyr Asp Ile Trp Gly Pro Gly Thr
Leu Val Thr Val 100 105 110 Ser Leu Ala Ser Thr Lys Gly Pro Ser Val
Phe Pro Leu Ala Pro Ser 115 120 125 Ser Lys Ser Thr Ser Gly Gly Thr
Ala Ala Leu Gly Cys Leu Val Lys 130 135 140 Asp Tyr Phe Pro Glu Pro
Val Thr Val Ser Trp Asn Ser Gly Ala Leu 145 150 155 160 Thr Ser Gly
Val His Thr Phe Pro Ala Val Leu Gln Ser Ser Gly Leu 165 170 175 Tyr
Ser Leu Ser Ser Val Val Thr Val Pro Ser Ser Ser Leu Gly Thr 180 185
190 Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro Ser Asn Thr Lys Val
195 200 205 Asp Lys Lys Val Glu Pro Lys Ser Cys Asp Lys Thr His Thr
Cys Pro 210 215 220 Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser
Val Phe Leu Phe 225 230 235 240 Pro Pro Lys Pro Lys Asp Thr Leu Met
Ile Ser Arg Thr Pro Glu Val 245 250 255 Thr Cys Val Val Val Asp Val
Ser His Glu Asp Pro Glu Val Lys Phe 260 265 270 Asn Trp Tyr Val Asp
Gly Val Glu Val His Asn Ala Lys Thr Lys Pro 275 280 285 Arg Glu Glu
Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr 290 295 300 Val
Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val 305 310
315 320 Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys
Ala 325 330 335 Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro
Pro Ser Arg 340 345 350 Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr
Cys Leu Val Lys Gly 355 360 365 Phe Tyr Pro Ser Asp Ile Ala Val Glu
Trp Glu Ser Asn Gly Gln Pro 370 375 380 Glu Asn Asn Tyr Lys Thr Thr
Pro Pro Val Leu Asp Ser Asp Gly Ser 385 390 395 400 Phe Phe Leu Tyr
Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln 405 410 415 Gly Asn
Val Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn His 420 425 430
Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 435 440 <210>
SEQ ID NO 338 <211> LENGTH: 217 <212> TYPE: PRT
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 338 Asp Val Val Met Thr
Gln Thr Pro Ala Ser Ala Ser Glu Pro Val Gly 1 5 10 15 Gly Thr Val
Thr Ile Lys Cys Gln Ala Ser Glu Ser Ile Tyr Arg Val 20 25 30 Leu
Ala Trp Tyr Gln Gln Lys Pro Gly Gln Pro Pro Lys Leu Leu Ile 35 40
45 Tyr Asp Thr Ser Thr Leu Ala Ser Gly Ala Pro Ser Arg Phe Lys Gly
50 55 60 Ser Gly Tyr Gly Thr Glu Phe Thr Leu Thr Ile Ser Gly Val
Gln Cys 65 70 75 80 Glu Asp Ala Ala Thr Tyr Tyr Cys Gln Gly Gly Tyr
Tyr Ala Asp Ser 85 90 95 Tyr Gly Ile Ala Phe Gly Gly Gly Thr Glu
Val Val Val Lys Arg Thr 100 105 110 Val Ala Ala Pro Ser Val Phe Ile
Phe Pro Pro Ser Asp Glu Gln Leu 115 120 125 Lys Ser Gly Thr Ala Ser
Val Val Cys Leu Leu Asn Asn Phe Tyr Pro 130 135 140 Arg Glu Ala Lys
Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly 145 150 155 160 Asn
Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr 165 170
175 Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His
180 185 190 Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser
Pro Val 195 200 205 Thr Lys Ser Phe Asn Arg Gly Glu Cys 210 215
<210> SEQ ID NO 339 <211> LENGTH: 447 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 339 Gln Val Gln Leu Val
Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg
Leu Ser Cys Ser Ala Ser Gly Ile Asp Leu Ser Ser Tyr 20 25 30 Ala
Met Gly Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Tyr Ile 35 40
45 Gly Thr Ile Asn Ile Gly Gly Arg Val Tyr Tyr Ala Ser Trp Ala Lys
50 55 60 Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu
Tyr Leu 65 70 75 80 Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val
Tyr Tyr Cys Ala 85 90 95 Arg Tyr Tyr Asn Gly Gly Ser Tyr Asp Ile
Trp Gly Gln Gly Thr Leu 100 105 110 Val Thr Val Ser Ser Ala Ser Thr
Lys Gly Pro Ser Val Phe Pro Leu 115 120 125 Ala Pro Ser Ser Lys Ser
Thr Ser Gly Gly Thr Ala Ala Leu Gly Cys 130 135 140 Leu Val Lys Asp
Tyr Phe Pro Glu Pro Val Thr Val Ser Trp Asn Ser 145 150 155 160 Gly
Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val Leu Gln Ser 165 170
175 Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser Ser Ser
180 185 190 Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro
Ser Asn 195 200 205 Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys
Asp Lys Thr His 210 215 220 Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu
Leu Gly Gly Pro Ser Val 225 230 235 240 Phe Leu Phe Pro Pro Lys Pro
Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250 255 Pro Glu Val Thr Cys
Val Val Val Asp Val Ser His Glu Asp Pro Glu 260 265 270 Val Lys Phe
Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 275 280 285 Thr
Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser 290 295
300 Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys
305 310 315 320 Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu
Lys Thr Ile 325 330 335 Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln
Val Tyr Thr Leu Pro 340 345 350 Pro Ser Arg Asp Glu Leu Thr Lys Asn
Gln Val Ser Leu Thr Cys Leu 355 360 365 Val Lys Gly Phe Tyr Pro Ser
Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375 380 Gly Gln Pro Glu Asn
Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser 385 390 395 400 Asp Gly
Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg 405 410 415
Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu 420
425 430 His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys
435 440 445 <210> SEQ ID NO 340 <211> LENGTH: 217
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE:
340 Asp Ile Gln Met Thr Gln Ser Pro Ser Thr Leu Ser Ala Ser Val Gly
1 5 10 15 Asp Arg Val Thr Ile Thr Cys Gln Ala Ser Glu Ser Ile Tyr
Arg Val 20 25 30 Leu Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro
Lys Leu Leu Ile 35 40 45 Tyr Asp Thr Ser Thr Leu Ala Ser Gly Val
Pro Ser Arg Phe Ser Gly 50 55 60 Ser Gly Ser Gly Thr Glu Phe Thr
Leu Thr Ile Ser Ser Leu Gln Cys 65 70 75 80 Asp Asp Ala Ala Thr Tyr
Tyr Cys Gln Gly Gly Tyr Tyr Ala Asp Ser 85 90 95 Tyr Gly Ile Ala
Phe Gly Gly Gly Thr Lys Val Glu Ile Lys Arg Thr 100 105 110 Val Ala
Ala Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu 115 120 125
Lys Ser Gly Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro 130
135 140 Arg Glu Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser
Gly 145 150 155 160 Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys
Asp Ser Thr Tyr 165 170 175 Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys
Ala Asp Tyr Glu Lys His 180 185 190 Lys Val Tyr Ala Cys Glu Val Thr
His Gln Gly Leu Ser Ser Pro Val 195 200 205 Thr Lys Ser Phe Asn Arg
Gly Glu Cys 210 215 <210> SEQ ID NO 341 <211> LENGTH:
452 <212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE:
341 Gln Ser Val Glu Glu Ser Gly Gly Arg Leu Val Lys Pro Asp Glu Ser
1 5 10 15 Leu Thr Leu Thr Cys Thr Ala Ser Gly Phe Ser Leu Ser Ser
Tyr Ala 20 25 30 Met Ile Trp Val Arg Gln Ala Pro Gly Glu Gly Leu
Glu Trp Ile Gly 35 40 45 Thr Ile Ser Thr Gly Gly Ile Thr Tyr Tyr
Ala Ser Trp Ala Lys Gly 50 55 60 Arg Phe Thr Ile Ser Lys Thr Ser
Thr Thr Val Asp Leu Lys Ile Thr 65 70 75 80 Ser Pro Thr Thr Glu Asp
Thr Ala Thr Tyr Phe Cys Ala Arg Gly Gly 85 90 95 Tyr Ala Ala Ser
Ser Ala Tyr Tyr Leu Pro Tyr Tyr Phe Asp Leu Trp 100 105 110 Gly Gln
Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro 115 120 125
Ser Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr 130
135 140 Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val
Thr 145 150 155 160 Val Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val
His Thr Phe Pro 165 170 175 Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser
Leu Ser Ser Val Val Thr 180 185 190 Val Pro Ser Ser Ser Leu Gly Thr
Gln Thr Tyr Ile Cys Asn Val Asn 195 200 205 His Lys Pro Ser Asn Thr
Lys Val Asp Lys Lys Val Glu Pro Lys Ser 210 215 220 Cys Asp Lys Thr
His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu 225 230 235 240 Gly
Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu 245 250
255 Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser
260 265 270 His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly
Val Glu 275 280 285 Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln
Tyr Asn Ser Thr 290 295 300 Tyr Arg Val Val Ser Val Leu Thr Val Leu
His Gln Asp Trp Leu Asn 305 310 315 320 Gly Lys Glu Tyr Lys Cys Lys
Val Ser Asn Lys Ala Leu Pro Ala Pro 325 330 335 Ile Glu Lys Thr Ile
Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln 340 345 350 Val Tyr Thr
Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val 355 360 365 Ser
Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val 370 375
380 Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro
385 390 395 400 Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser
Lys Leu Thr 405 410 415 Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val
Phe Ser Cys Ser Val 420 425 430 Met His Glu Ala Leu His Asn His Tyr
Thr Gln Lys Ser Leu Ser Leu 435 440 445 Ser Pro Gly Lys 450
<210> SEQ ID NO 342 <211> LENGTH: 219 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 342 Ala Ala Val Leu Thr
Gln Thr Pro Ser Pro Val Ser Ala Ala Val Gly 1 5 10 15 Gly Thr Val
Thr Ile Ser Cys Gln Ser Ser Gln Ser Val Tyr Asn Asn 20 25 30 Asn
Asn Leu Ala Trp Phe Gln Gln Lys Pro Gly Gln Pro Pro Lys Leu 35 40
45 Leu Ile Tyr Leu Ala Ser Thr Leu Ala Ser Gly Val Pro Ser Arg Phe
50 55 60 Ser Gly Ser Gly Ser Gly Thr Gln Phe Thr Leu Thr Ile Ser
Gly Val 65 70 75 80 Gln Cys Asp Asp Ala Ala Thr Tyr Tyr Cys Leu Gly
Gly Cys Asp Asp 85 90 95 Asp Ala Asp Thr Phe Ala Phe Gly Gly Gly
Thr Glu Val Val Val Lys 100 105 110 Arg Thr Val Ala Ala Pro Ser Val
Phe Ile Phe Pro Pro Ser Asp Glu 115 120 125 Gln Leu Lys Ser Gly Thr
Ala Ser Val Val Cys Leu Leu Asn Asn Phe 130 135 140 Tyr Pro Arg Glu
Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln 145 150 155 160 Ser
Gly Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser 165 170
175 Thr Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu
180 185 190 Lys His Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu
Ser Ser 195 200 205 Pro Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 210
215 <210> SEQ ID NO 343 <211> LENGTH: 455 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polypeptide <400> SEQUENCE: 343 Glu Val
Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15
Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Ser Leu Ser Ser Tyr 20
25 30 Ala Met Ile Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Ile 35 40 45 Gly Thr Ile Ser Thr Gly Gly Ile Thr Tyr Tyr Ala Ser
Trp Ala Lys 50 55 60 Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys
Asn Thr Leu Tyr Leu 65 70 75 80 Gln Met Asn Ser Leu Arg Ala Glu Asp
Thr Ala Val Tyr Tyr Cys Ala 85 90 95 Arg Gly Gly Tyr Ala Ala Ser
Ser Ala Tyr Tyr Leu Pro Tyr Tyr Phe 100 105 110 Asp Leu Trp Gly Gln
Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr 115 120 125 Lys Gly Pro
Ser Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser 130 135 140 Gly
Gly Thr Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu 145 150
155 160 Pro Val Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val
His 165 170 175 Thr Phe Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser
Leu Ser Ser 180 185 190 Val Val Thr Val Pro Ser Ser Ser Leu Gly Thr
Gln Thr Tyr Ile Cys 195 200 205 Asn Val Asn His Lys Pro Ser Asn Thr
Lys Val Asp Lys Lys Val Glu 210 215 220 Pro Lys Ser Cys Asp Lys Thr
His Thr Cys Pro Pro Cys Pro Ala Pro 225 230 235 240 Glu Leu Leu Gly
Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys 245 250 255 Asp Thr
Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val 260 265 270
Asp Val Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp 275
280 285 Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln
Tyr 290 295 300 Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu
His Gln Asp 305 310 315 320 Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys
Val Ser Asn Lys Ala Leu 325 330 335 Pro Ala Pro Ile Glu Lys Thr Ile
Ser Lys Ala Lys Gly Gln Pro Arg 340 345 350 Glu Pro Gln Val Tyr Thr
Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys 355 360 365 Asn Gln Val Ser
Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp 370 375 380 Ile Ala
Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys 385 390 395
400 Thr Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser
405 410 415 Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val
Phe Ser 420 425 430 Cys Ser Val Met His Glu Ala Leu His Asn His Tyr
Thr Gln Lys Ser 435 440 445 Leu Ser Leu Ser Pro Gly Lys 450 455
<210> SEQ ID NO 344 <211> LENGTH: 219 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 344 Asp Ile Gln Met Thr
Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly 1 5 10 15 Asp Arg Val
Thr Ile Thr Cys Gln Ser Ser Gln Ser Val Tyr Asn Asn 20 25 30 Asn
Asn Leu Ala Trp Tyr Gln Gln Lys Pro Gly Lys Val Pro Lys Leu 35 40
45 Leu Ile Tyr Leu Ala Ser Thr Leu Ala Ser Gly Val Pro Ser Arg Phe
50 55 60 Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu 65 70 75 80 Gln Cys Glu Asp Ala Ala Thr Tyr Tyr Cys Leu Gly
Gly Cys Asp Asp 85 90 95 Asp Ala Asp Thr Phe Ala Phe Gly Gly Gly
Thr Lys Val Glu Ile Lys 100 105 110 Arg Thr Val Ala Ala Pro Ser Val
Phe Ile Phe Pro Pro Ser Asp Glu 115 120 125 Gln Leu Lys Ser Gly Thr
Ala Ser Val Val Cys Leu Leu Asn Asn Phe 130 135 140 Tyr Pro Arg Glu
Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln 145 150 155 160 Ser
Gly Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser 165 170
175 Thr Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu
180 185 190 Lys His Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu
Ser Ser 195 200 205 Pro Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 210
215 <210> SEQ ID NO 345 <211> LENGTH: 1350 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polynucleotide <400> SEQUENCE: 345
gaggtccaac tggtggagag cggtggaggt gttgtgcaac ctggccggtc cctgcgcctg
60 tcctgctccg catctggctt caccttcagc ggctatgggt tgtcttgggt
gagacaggca 120 cctggaaaag gtcttgagtg ggttgcaatg attagtagtg
gtggtagtta tacctactat 180 gcagacagtg tgaagggtag atttgcaata
tcgcgagaca acgccaagaa cacattgttc 240 ctgcaaatgg acagcctgag
acccgaagac accggggtct atttttgtgc aagacatggg 300 gacgatcccg
cctggttcgc ttattggggc caagggaccc cggtcaccgt ctcctcagcc 360
tccaccaagg gcccatcggt cttccccctg gcaccctcct ccaagagcac ctctgggggc
420 acagcggccc tgggctgcct ggtcaaggac tacttccccg aaccggtgac
ggtgtcgtgg 480 aactcaggcg ccctgaccag cggcgtgcac accttcccgg
ctgtcctaca gtcctcagga 540 ctctactccc tcagcagcgt ggtgaccgtg
ccctccagca gcttgggcac ccagacctac 600 atctgcaacg tgaatcacaa
gcccagcaac accaaggtgg acaagaaagt tgagcccaaa 660 tcttgtgaca
aaactcacac atgcccaccg tgcccagcac ctgaactcct ggggggaccg 720
tcagtcttcc tcttcccccc aaaacccaag gacaccctca tgatctcccg gacccctgag
780 gtcacatgcg tggtggtgga cgtgagccac gaagaccctg aggtcaagtt
caactggtac 840 gtggacggcg tggaggtgca taatgccaag acaaagccgc
gggaggagca gtacaacagc 900 acgtaccgtg tggtcagcgt cctcaccgtc
ctgcaccagg actggctgaa tggcaaggag 960 tacaagtgca aggtctccaa
caaagccctc ccagccccca tcgagaaaac catctccaaa 1020 gccaaagggc
agccccgaga accacaggtg tacaccctgc ccccatcccg ggatgagctg 1080
accaagaacc aggtcagcct gacctgcctg gtcaaaggct tctatcccag cgacatcgcc
1140 gtggagtggg agagcaatgg gcagccggag aacaactaca agaccacgcc
tcccgtgctg 1200 gactccgacg gctccttctt cttatattca aagctcaccg
tggacaagag caggtggcag 1260 caggggaacg tcttctcatg ctccgtgatg
catgaggctc tgcacaacca ctacacgcag 1320 aagagcctct ccctgtctcc
cgggaaatga 1350 <210> SEQ ID NO 346 <211> LENGTH: 654
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polynucleotide <400> SEQUENCE:
346 gacatccagc tgacccagag cccaagcagc ctgagcgcca gcgtgggtga
cagagtgacc 60 atcacctgta gtgtcagctc aagtataagt tccaacaact
tgcactggta ccagcagaag 120 ccaggtaagg ctccaaagcc atggatctac
ggcacatcca acctggcttc tggtgtgcca 180 agcagattca gcggtagcgg
tagcggtacc gactacacct tcaccatcag cagcctccag 240 ccagaggaca
tcgccaccta ctactgccaa cagtggagta gttacccgta catgtacacg 300
ttcggccaag ggaccaaggt ggaaatcaaa cgaactgtgg ctgcaccatc tgtcttcatc
360 ttcccgccat ctgatgagca gttgaaatct ggaactgcct ctgttgtgtg
cctgctgaat 420 aacttctatc ccagagaggc caaagtacag tggaaggtgg
ataacgccct ccaatcgggt 480 aactcccagg agagtgtcac agagcaggac
agcaaggaca gcacctacag cctcagcagc 540 accctgacgc tgagcaaagc
agactacgag aaacacaaag tctacgcctg cgaagtcacc 600 catcagggcc
tgagctcgcc cgtcacaaag agcttcaaca ggggagagtg ttaa 654 <210>
SEQ ID NO 347 <211> LENGTH: 1350 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 347 caggtacaac
tgcagcagtc tgggcctgag ctggagaagc ctggcgcttc agtgaagata 60
tcctgcaagg cttctggtta ctcattcact ggctacacca tgaactgggt gaagcagagc
120 catggaaaga gccttgagtg gattggactt attactcctt acaatggtgc
ttctagctac 180 aaccagaagt tcaggggcaa ggccacatta actgtagaca
agtcatccag cacagcctac 240 atggacctcc tcagtctgac atctgaagac
tctgcagtct atttctgtgc aagggggggt 300 tacgacggga ggggttttga
ctactgggga tccgggaccc cggtcaccgt ctcctcagcc 360 tccaccaagg
gcccatcggt cttccccctg gcaccctcct ccaagagcac ctctgggggc 420
acagcggccc tgggctgcct ggtcaaggac tacttccccg aaccggtgac ggtgtcgtgg
480 aactcaggcg ccctgaccag cggcgtgcac accttcccgg ctgtcctaca
gtcctcagga 540 ctctactccc tcagcagcgt ggtgaccgtg ccctccagca
gcttgggcac ccagacctac 600 atctgcaacg tgaatcacaa gcccagcaac
accaaggtgg acaagaaagt tgagcccaaa 660 tcttgtgaca aaactcacac
atgcccaccg tgcccagcac ctgaactcct ggggggaccg 720 tcagtcttcc
tcttcccccc aaaacccaag gacaccctca tgatctcccg gacccctgag 780
gtcacatgcg tggtggtgga cgtgagccac gaagaccctg aggtcaagtt caactggtac
840 gtggacggcg tggaggtgca taatgccaag acaaagccgc gggaggagca
gtacaacagc 900 acgtaccgtg tggtcagcgt cctcaccgtc ctgcaccagg
actggctgaa tggcaaggag 960 tacaagtgca aggtctccaa caaagccctc
ccagccccca tcgagaaaac catctccaaa 1020 gccaaagggc agccccgaga
accacaggtg tacaccctgc ccccatcccg ggatgagctg 1080 accaagaacc
aggtcagcct gacctgcctg gtcaaaggct tctatcccag cgacatcgcc 1140
gtggagtggg agagcaatgg gcagccggag aacaactaca agaccacgcc tcccgtgctg
1200 gactccgacg gctccttctt cctctacagc aagctcaccg tggacaagag
caggtggcag 1260 caggggaacg tcttctcatg ctccgtgatg catgaggctc
tgcacaacca ctacacgcag 1320 aagagcctct ccctgtctcc cgggaaatga 1350
<210> SEQ ID NO 348 <211> LENGTH: 642 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 348 gacatcgagc
tcactcagtc tccagcaatc atgtctgcat ctccagggga gaaggtcacc 60
atgacctgca gtgccagctc aagtgtaagt tacatgcact ggtaccagca gaagtcaggc
120 acctccccca aaagatggat ttatgacaca tccaaactgg cttctggagt
cccaggtcgc 180 ttcagtggca gtgggtctgg aaactcttac tctctcacaa
tcagcagcgt ggaggctgaa 240 gatgatgcaa cttattactg ccagcagtgg
agtaagcacc ctctcacgtt cggatccggg 300 accaaggtgg aaatcaaacg
aactgtggct gcaccatctg tcttcatctt cccgccatct 360 gatgagcagt
tgaaatctgg aactgcctct gttgtgtgcc tgctgaataa cttctatccc 420
agagaggcca aagtacagtg gaaggtggat aacgccctcc aatcgggtaa ctcccaggag
480 agtgtcacag agcaggacag caaggacagc acctacagcc tcagcagcac
cctgacgctg 540 agcaaagcag actacgagaa acacaaagtc tacgcctgcg
aagtcaccca tcagggcctg 600 agctcgcccg tcacaaagag cttcaacagg
ggagagtgtt aa 642 <210> SEQ ID NO 349 <211> LENGTH:
1362 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polynucleotide
<400> SEQUENCE: 349 cagtcggtgg aggagtccgg gggtcgcctg
gtcacgcctg ggacacccct gacactcacc 60 tgcaccgtct ctggaatctc
cctcagtagc gatgcaataa gctgggtccg ccaggctcca 120 gggaaggggc
tcgaatacat cggaatcatt aatggtggtg gtaacacata ctacgcgagc 180
tgggcgaaag gccgattcac catctccaaa acctcgacca cggtggatct gaaaatcacc
240 agtccgacaa ccgaggacac ggccacctat ttctgtgcca gaggcattca
acatggtggt 300 ggtaatagtg attattatta ttacggcatg gacctctggg
gcccaggcac cctggtcact 360 gtctcttcag catccaccaa gggcccatcg
gtcttccccc tggcaccctc ctccaagagc 420 acctctgggg gcacagcggc
cctgggctgc ctggtcaagg actacttccc cgaaccggtg 480 acggtgtcgt
ggaactcagg cgccctgacc agcggcgtgc acaccttccc ggctgtccta 540
cagtcctcag gactctactc cctcagcagc gtggtgaccg tgccctccag cagcttgggc
600 acccagacct acatctgcaa cgtgaatcac aagcccagca acaccaaggt
ggacaagaaa 660 gttgagccca aatcttgtga caaaactcac acatgcccac
cgtgcccagc acctgaactc 720 ctggggggac cgtcagtctt cctcttcccc
ccaaaaccca aggacaccct catgatctcc 780 cggacccctg aggtcacatg
cgtggtggtg gacgtgagcc acgaagaccc tgaggtcaag 840 ttcaactggt
acgtggacgg cgtggaggtg cataatgcca agacaaagcc gcgggaggag 900
cagtacaaca gcacgtaccg tgtggtcagc gtcctcaccg tcctgcacca ggactggctg
960 aatggcaagg agtacaagtg caaggtctcc aacaaagccc tcccagcccc
catcgagaaa 1020 accatctcca aagccaaagg gcagccccga gaaccacagg
tgtacaccct gcccccatcc 1080 cgggatgagc tgaccaagaa ccaggtcagc
ctgacctgcc tggtcaaagg cttctatccc 1140 agcgacatcg ccgtggagtg
ggagagcaat gggcagccgg agaacaacta caagaccacg 1200 cctcccgtgc
tggactccga cggctccttc ttcttatatt caaagctcac cgtggacaag 1260
agcaggtggc agcaggggaa cgtcttctca tgctccgtga tgcatgaggc tctgcacaac
1320 cactacacgc agaagagcct ctccctgtct cccgggaaat ga 1362
<210> SEQ ID NO 350 <211> LENGTH: 660 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 350 gaagtgttga
tgacccagac tccatcctcc gtgtctgcag ctgtgggaga cacagtcacc 60
atcaagtgcc aggccagtca gagcattagt agtgtcttgt cctggtatca gcagaaacca
120 gggcagcctc ccaagctcct gatctatctg gcatccactc tggcatctgg
ggtcccatcg 180 cggttcagcg gcagtagatc tgggacagag ttcactctca
ccatcagcga cctggagtgt 240 gacgatgctg ccacttacta ctgtcaaacc
aattatggta ctagtagtag taattatggt 300 tttgctttcg gcggagggac
cgaggtggtc gtcaaacgaa ctgtggctgc accatctgtc 360 ttcatcttcc
cgccatctga tgagcagttg aaatctggaa ctgcctctgt tgtgtgcctg 420
ctgaataact tctatcccag agaggccaaa gtacagtgga aggtggataa cgccctccaa
480 tcgggtaact cccaggagag tgtcacagag caggacagca aggacagcac
ctacagcctc 540 agcagcaccc tgacgctgag caaagcagac tacgagaaac
acaaagtcta cgcctgcgaa 600 gtcacccatc agggcctgag ctcgcccgtc
acaaagagct tcaacagggg agagtgttga 660 <210> SEQ ID NO 351
<211> LENGTH: 1371 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 351 gaagtccaac tggtggaaag
cgggggagga ctggtgcagc cgggcggatc cctccggctg 60 tcatgtgctg
catcgggaat ttccctctcc tccgacgcga ttagctgggt cagacaggcc 120
cccggaaagg ggctggagta catcggtatc atcaacggcg gcggaaacac ctactacgcc
180 tcctgggcca agggccgctt caccatctcg cggcataatt ccaagaacac
tctgtacttg 240 caaatgaact ccctgagggc cgaggacacc gccgtgtact
actgcgcgcg cggcatccag 300 cacggtggtg gaaacagcga ctactactac
tatgggatgg atctgtgggg ccagggaact 360 cttgtgaccg tgtcgtcagc
atccaccaag ggcccatcgg tcttccccct ggcaccctcc 420 tccaagagca
cctctggggg cacagcggcc ctgggctgcc tggtcaagga ctacttcccc 480
gaaccggtga cggtgtcgtg gaactcaggc gccctgacca gcggcgtgca caccttcccg
540 gctgtcctac agtcctcagg actctactcc ctcagcagcg tggtgaccgt
gccctccagc 600 agcttgggca cccagaccta catctgcaac gtgaatcaca
agcccagcaa caccaaggtg 660 gacaagaaag ttgagcccaa atcttgtgac
aaaactcaca catgcccacc gtgcccagca 720 cctgaactcc tggggggacc
gtcagtcttc ctcttccccc caaaacccaa ggacaccctc 780 atgatctccc
ggacccctga ggtcacatgc gtggtggtgg acgtgagcca cgaagaccct 840
gaggtcaagt tcaactggta cgtggacggc gtggaggtgc ataatgccaa gacaaagccg
900 cgggaggagc agtacaacag cacgtaccgt gtggtcagcg tcctcaccgt
cctgcaccag 960 gactggctga atggcaagga gtacaagtgc aaggtctcca
acaaagccct cccagccccc 1020 atcgagaaaa ccatctccaa agccaaaggg
cagccccgag aaccacaggt gtacaccctg 1080 cccccatccc gggatgagct
gaccaagaac caggtcagcc tgacctgcct ggtcaaaggc 1140 ttctatccca
gcgacatcgc cgtggagtgg gagagcaatg ggcagccgga gaacaactac 1200
aagaccacgc ctcccgtgct ggactccgac ggctccttct tcttatattc aaagctcacc
1260 gtggacaaga gcaggtggca gcaggggaac gtcttctcat gctccgtgat
gcatgaggct 1320 ctgcacaacc actacacgca gaagagcctc tccctgtctc
ccgggaaatg a 1371 <210> SEQ ID NO 352 <211> LENGTH: 660
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polynucleotide <400> SEQUENCE:
352 gacattcaga tgacccagtc cccaagctcg ctgtccgcct ccgtgggcga
ccgcgtgacc 60 atcacgtgcc aggcgtccca gtcaattagc agcgtgctct
cctggtacca acagaagccg 120 gggaaagcac ccaagctgct gatctacttg
gcctccactc tggcctcggg agtgccttca 180 cggttctccg gatcgggatc
tggtactgat ttcaccctca ccatctcgag ccttcagtgc 240 gaggacatcg
ctacttacta ttgtcaaacc aactacggaa cctccagctc caactacggc 300
tttgccttcg gtggcgggac caaggtcgaa atcaaacgaa ctgtggctgc accatctgtc
360 ttcatcttcc cgccatctga tgagcagttg aaatctggaa ctgcctctgt
tgtgtgcctg 420 ctgaataact tctatcccag agaggccaaa gtacagtgga
aggtggataa cgccctccaa 480 tcgggtaact cccaggagag tgtcacagag
caggacagca aggacagcac ctacagcctc 540 agcagcaccc tgacgctgag
caaagcagac tacgagaaac acaaagtcta cgcctgcgaa 600 gtcacccatc
agggcctgag ctcgcccgtc acaaagagct tcaacagggg agagtgttga 660
<210> SEQ ID NO 353 <211> LENGTH: 1347 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polynucleotide <400> SEQUENCE: 353
cagtcggtgg aggagtccgg gggtcgcctg gtcacgcctg ggacacccct gacactcacc
60 tgcacagtct ctggattctc cctcaataac tatgcaatga gctgggtccg
ccaggctcca 120 gggaaggggc tggaatggat cggatccatt agtactggtg
gtctcgcatt ctacgcgaac 180 tgggcaaaag gccgattcac catctccaga
acctcgacca cggtggatct gaaaatgacc 240 agtctgacaa ccgaggacac
ggccacctat ttctgtggca gaaatggtgg tggtagttat 300 attttctatt
attttgactt gtggggccaa ggcaccctcg tcactgtctc ttcagcatcc 360
accaagggcc catcggtctt ccccctggca ccctcctcca agagcacctc tgggggcaca
420 gcggccctgg gctgcctggt caaggactac ttccccgaac cggtgacggt
gtcgtggaac 480 tcaggcgccc tgaccagcgg cgtgcacacc ttcccggctg
tcctacagtc ctcaggactc 540 tactccctca gcagcgtggt gaccgtgccc
tccagcagct tgggcaccca gacctacatc 600 tgcaacgtga atcacaagcc
cagcaacacc aaggtggaca agaaagttga gcccaaatct 660 tgtgacaaaa
ctcacacatg cccaccgtgc ccagcacctg aactcctggg gggaccgtca 720
gtcttcctct tccccccaaa acccaaggac accctcatga tctcccggac ccctgaggtc
780 acatgcgtgg tggtggacgt gagccacgaa gaccctgagg tcaagttcaa
ctggtacgtg 840 gacggcgtgg aggtgcataa tgccaagaca aagccgcggg
aggagcagta caacagcacg 900 taccgtgtgg tcagcgtcct caccgtcctg
caccaggact ggctgaatgg caaggagtac 960 aagtgcaagg tctccaacaa
agccctccca gcccccatcg agaaaaccat ctccaaagcc 1020 aaagggcagc
cccgagaacc acaggtgtac accctgcccc catcccggga tgagctgacc 1080
aagaaccagg tcagcctgac ctgcctggtc aaaggcttct atcccagcga catcgccgtg
1140 gagtgggaga gcaatgggca gccggagaac aactacaaga ccacgcctcc
cgtgctggac 1200 tccgacggct ccttcttctt atattcaaag ctcaccgtgg
acaagagcag gtggcagcag 1260 gggaacgtct tctcatgctc cgtgatgcat
gaggctctgc acaaccacta cacgcagaag 1320 agcctctccc tgtctcccgg gaaatga
1347 <210> SEQ ID NO 354 <211> LENGTH: 648 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polynucleotide <400> SEQUENCE: 354
gcattcgaat tgacccagac tccatcctcc gtggaggcag ctgtgggagg cacaatcacc
60 atcaagtgcc aggccagtca gagcattagt agttacttat cctggtatca
gcagaaacca 120 gggcagcctc ccaagctcct gatctattct gcatccactc
tggcatctgg ggtctcatcg 180 cggttcaaag gcagtggatc tgggacagag
tacactctca ccatcagcga cctggagtgt 240 gccgatgctg ccacttactt
ctgtcaaagc tattatgata ttggtactag tactttcggc 300 ggagggaccg
aggtggtcgt caaacgaact gtggctgcac catctgtctt catcttcccg 360
ccatctgatg agcagttgaa atctggaact gcctctgttg tgtgcctgct gaataacttc
420 tatcccagag aggccaaagt acagtggaag gtggataacg ccctccaatc
gggtaactcc 480 caggagagtg tcacagagca ggacagcaag gacagcacct
acagcctcag cagcaccctg 540 acgctgagca aagcagacta cgagaaacac
aaagtctacg cctgcgaagt cacccatcag 600 ggcctgagct cgcccgtcac
aaagagcttc aacaggggag agtgttga 648 <210> SEQ ID NO 355
<211> LENGTH: 1356 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 355 gaagtgcagc tggtggaatc
tggcggcgga ctggtgcagc ctggcggatc tctgagactg 60 tcttgtgccg
cctccggctt ctccctgaac aactacgcca tgtcctgggt gcgacaggcc 120
cctggcaaag gcctggaatg gatcggctcc atcagcacag gcggcctggc cttctacgcc
180 aattgggcca agggccggtt caccatcagc cgggacaact ccaagaacac
cctgtacctc 240 cagatgaact ccctgcgggc cgaggacacc gccgtgtact
actgtgccag aaacggcgga 300 ggctcctaca tcttctacta cttcgacctg
tggggccagg gcaccctcgt gacagtgtca 360 tctgcatcca ccaagggccc
atcggtcttc cccctggcac cctcctccaa gagcacctct 420 gggggcacag
cggccctggg ctgcctggtc aaggactact tccccgaacc ggtgacggtg 480
tcgtggaact caggcgccct gaccagcggc gtgcacacct tcccggctgt cctacagtcc
540 tcaggactct actccctcag cagcgtggtg accgtgccct ccagcagctt
gggcacccag 600 acctacatct gcaacgtgaa tcacaagccc agcaacacca
aggtggacaa gaaagttgag 660 cccaaatctt gtgacaaaac tcacacatgc
ccaccgtgcc cagcacctga actcctgggg 720 ggaccgtcag tcttcctctt
ccccccaaaa cccaaggaca ccctcatgat ctcccggacc 780 cctgaggtca
catgcgtggt ggtggacgtg agccacgaag accctgaggt caagttcaac 840
tggtacgtgg acggcgtgga ggtgcataat gccaagacaa agccgcggga ggagcagtac
900 aacagcacgt accgtgtggt cagcgtcctc accgtcctgc accaggactg
gctgaatggc 960 aaggagtaca agtgcaaggt ctccaacaaa gccctcccag
cccccatcga gaaaaccatc 1020 tccaaagcca aagggcagcc ccgagaacca
caggtgtaca ccctgccccc atcccgggat 1080 gagctgacca agaaccaggt
cagcctgacc tgcctggtca aaggcttcta tcccagcgac 1140 atcgccgtgg
agtgggagag caatgggcag ccggagaaca actacaagac cacgcctccc 1200
gtgctggact ccgacggctc cttcttctta tattcaaagc tcaccgtgga caagagcagg
1260 tggcagcagg ggaacgtctt ctcatgctcc gtgatgcatg aggctctgca
caaccactac 1320 acgcagaaga gcctctccct gtctcccggg aaatga 1356
<210> SEQ ID NO 356 <211> LENGTH: 648 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 356 gatattcaga
tgacccagtc cccctccagc ctgtccgctt ctgtgggcga cagagtgacc 60
atcacctgtc aggcctccca gtccatctcc tcctacctgt cctggtatca gcagaagccc
120 ggcaaggccc ccaagctgct gatctactct gcctccacac tggcctccgg
cgtgccctct 180 agattctccg gctctggctc tggcaccgac tttaccctga
ccatcagctc cctccagtgc 240 gaggatgccg ccacctacta ctgccagtcc
tactacgaca tcggcacctc caccttcggc 300 ggaggcacca aggtggaaat
caaacgaact gtggctgcac catctgtctt catcttcccg 360 ccatctgatg
agcagttgaa atctggaact gcctctgttg tgtgcctgct gaataacttc 420
tatcccagag aggccaaagt acagtggaag gtggataacg ccctccaatc gggtaactcc
480 caggagagtg tcacagagca ggacagcaag gacagcacct acagcctcag
cagcaccctg 540 acgctgagca aagcagacta cgagaaacac aaagtctacg
cctgcgaagt cacccatcag 600 ggcctgagct cgcccgtcac aaagagcttc
aacaggggag agtgttga 648 <210> SEQ ID NO 357 <211>
LENGTH: 1335 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polynucleotide
<400> SEQUENCE: 357 cagtcagtga aggagtccgg gggtcgcctg
gtcacgcctg ggacacccct gacactcacc 60 tgcacagtct ctggaatcga
cctcagtagc tatgcaatgg gctggttccg ccaggctcca 120 gggaaggggc
tggaatacat cggaaccatt aatattggtg gtcgcgtata ttacgcgagc 180
tgggcaaaag gccgattcac catctccaga acctcgacca cggtggatct gaaagcgccc
240 agtctgacag ccgaggacac ggccacctat ttctgtgcca gatattataa
tggtggtagt 300 tatgacatct ggggcccagg caccctggtc accgtctctt
tagcatccac caagggccca 360 tcggtcttcc ccctggcacc ctcctccaag
agcacctctg ggggcacagc ggccctgggc 420 tgcctggtca aggactactt
ccccgaaccg gtgacggtgt cgtggaactc aggcgccctg 480 accagcggcg
tgcacacctt cccggctgtc ctacagtcct caggactcta ctccctcagc 540
agcgtggtga ccgtgccctc cagcagcttg ggcacccaga cctacatctg caacgtgaat
600 cacaagccca gcaacaccaa ggtggacaag aaagttgagc ccaaatcttg
tgacaaaact 660 cacacatgcc caccgtgccc agcacctgaa ctcctggggg
gaccgtcagt cttcctcttc 720 cccccaaaac ccaaggacac cctcatgatc
tcccggaccc ctgaggtcac atgcgtggtg 780 gtggacgtga gccacgaaga
ccctgaggtc aagttcaact ggtacgtgga cggcgtggag 840 gtgcataatg
ccaagacaaa gccgcgggag gagcagtaca acagcacgta ccgtgtggtc 900
agcgtcctca ccgtcctgca ccaggactgg ctgaatggca aggagtacaa gtgcaaggtc
960 tccaacaaag ccctcccagc ccccatcgag aaaaccatct ccaaagccaa
agggcagccc 1020 cgagaaccac aggtgtacac cctgccccca tcccgggatg
agctgaccaa gaaccaggtc 1080 agcctgacct gcctggtcaa aggcttctat
cccagcgaca tcgccgtgga gtgggagagc 1140 aatgggcagc cggagaacaa
ctacaagacc acgcctcccg tgctggactc cgacggctcc 1200 ttcttcttat
attcaaagct caccgtggac aagagcaggt ggcagcaggg gaacgtcttc 1260
tcatgctccg tgatgcatga ggctctgcac aaccactaca cgcagaagag cctctccctg
1320 tctcccggga aatga 1335 <210> SEQ ID NO 358 <211>
LENGTH: 654 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polynucleotide
<400> SEQUENCE: 358 gatgttgtga tgacccagac tccagcctcc
gcgtctgaac ctgtgggagg cacagtcacc 60 atcaagtgcc aggccagtga
gagcatttat cgcgtattgg cctggtatca gcagaaacca 120 gggcagcctc
ccaagctcct gatctatgat acatccactc tggcatctgg ggccccatcg 180
cggttcaaag gcagtggata tgggacagag ttcactctca ccatcagcgg cgtgcagtgt
240 gaagatgctg ccacttacta ctgtcaaggc ggttattatg ctgatagtta
tggtattgct 300 ttcggcggag ggaccgaggt ggtggtcaaa cgaactgtgg
ctgcaccatc tgtcttcatc 360 ttcccgccat ctgatgagca gttgaaatct
ggaactgcct ctgttgtgtg cctgctgaat 420 aacttctatc ccagagaggc
caaagtacag tggaaggtgg ataacgccct ccaatcgggt 480 aactcccagg
agagtgtcac agagcaggac agcaaggaca gcacctacag cctcagcagc 540
accctgacgc tgagcaaagc agactacgag aaacacaaag tctacgcctg cgaagtcacc
600 catcagggcc tgagctcgcc cgtcacaaag agcttcaaca ggggagagtg ttga 654
<210> SEQ ID NO 359 <211> LENGTH: 1344 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polynucleotide <400> SEQUENCE: 359
caggtgcagc tggtggaatc tggcggagga ctggtgcagc ctggcggctc tctgagactg
60 tcctgttccg cctccggaat cgacctgtcc tcctacgcta tgggctgggt
gcgacaggct 120 cctggcaagg gcctggagta catcggcacc atcaacatcg
gcggcagagt gtactacgcc 180 tcctgggcca agggccggtt caccatctcc
agagacaact ccaagaacac cctgtacctc 240 cagatgaact ccctgcgggc
cgaggacacc gccgtgtact actgcgcccg gtactacaac 300 ggcggctcct
acgatatctg gggccagggc acactcgtga ccgtgtcctc tgcatccacc 360
aagggcccat cggtcttccc cctggcaccc tcctccaaga gcacctctgg gggcacagcg
420 gccctgggct gcctggtcaa ggactacttc cccgaaccgg tgacggtgtc
gtggaactca 480 ggcgccctga ccagcggcgt gcacaccttc ccggctgtcc
tacagtcctc aggactctac 540 tccctcagca gcgtggtgac cgtgccctcc
agcagcttgg gcacccagac ctacatctgc 600 aacgtgaatc acaagcccag
caacaccaag gtggacaaga aagttgagcc caaatcttgt 660 gacaaaactc
acacatgccc accgtgccca gcacctgaac tcctgggggg accgtcagtc 720
ttcctcttcc ccccaaaacc caaggacacc ctcatgatct cccggacccc tgaggtcaca
780 tgcgtggtgg tggacgtgag ccacgaagac cctgaggtca agttcaactg
gtacgtggac 840 ggcgtggagg tgcataatgc caagacaaag ccgcgggagg
agcagtacaa cagcacgtac 900 cgtgtggtca gcgtcctcac cgtcctgcac
caggactggc tgaatggcaa ggagtacaag 960 tgcaaggtct ccaacaaagc
cctcccagcc cccatcgaga aaaccatctc caaagccaaa 1020 gggcagcccc
gagaaccaca ggtgtacacc ctgcccccat cccgggatga gctgaccaag 1080
aaccaggtca gcctgacctg cctggtcaaa ggcttctatc ccagcgacat cgccgtggag
1140 tgggagagca atgggcagcc ggagaacaac tacaagacca cgcctcccgt
gctggactcc 1200 gacggctcct tcttcttata ttcaaagctc accgtggaca
agagcaggtg gcagcagggg 1260 aacgtcttct catgctccgt gatgcatgag
gctctgcaca accactacac gcagaagagc 1320 ctctccctgt ctcccgggaa atga
1344 <210> SEQ ID NO 360 <211> LENGTH: 654 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polynucleotide <400> SEQUENCE: 360
gatatccaga tgacccagtc cccctccacc ctgtctgcct ctgtgggcga cagagtgacc
60 atcacctgtc aggcctccga gtccatctac cgggtgctgg cctggtatca
gcagaagcct 120 ggcaaggccc ccaagctgct gatctacgac accagcacac
tggcctccgg cgtgccctct 180 agattctccg gctctggctc tggcaccgag
tttaccctga ccatctccag cctccagtgc 240 gacgacgccg ccacctacta
ttgtcagggc ggctactacg ccgactccta cggaatcgct 300 ttcggcggag
gcaccaaggt ggaaatcaaa cgaactgtgg ctgcaccatc tgtcttcatc 360
ttcccgccat ctgatgagca gttgaaatct ggaactgcct ctgttgtgtg cctgctgaat
420 aacttctatc ccagagaggc caaagtacag tggaaggtgg ataacgccct
ccaatcgggt 480 aactcccagg agagtgtcac agagcaggac agcaaggaca
gcacctacag cctcagcagc 540 accctgacgc tgagcaaagc agactacgag
aaacacaaag tctacgcctg cgaagtcacc 600 catcagggcc tgagctcgcc
cgtcacaaag agcttcaaca ggggagagtg ttga 654 <210> SEQ ID NO 361
<211> LENGTH: 1359 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 361 cagtcggtgg aggagtccgg
cggtcgcctg gtaaagcctg acgaatccct gacactcacc 60 tgcacagcct
ctggattctc cctcagtagt tatgcaatga tctgggtccg ccaggctcca 120
ggggaggggc tggaatggat cggaaccatt agtactggtg gtatcacata ctacgcgagc
180 tgggcgaaag gccgattcac catctccaaa acctcgacca cggtggatct
gaaaatcacc 240 agtccgacaa ccgaggacac ggccacctat ttctgtgcca
gagggggata tgctgctagt 300 agtgcttatt atctcccgta ctactttgac
ttgtggggcc aagggaccct ggtcaccgtc 360 tcctcagcat ccaccaaggg
cccatcggtc ttccccctgg caccctcctc caagagcacc 420 tctgggggca
cagcggccct gggctgcctg gtcaaggact acttccccga accggtgacg 480
gtgtcgtgga actcaggcgc cctgaccagc ggcgtgcaca ccttcccggc tgtcctacag
540 tcctcaggac tctactccct cagcagcgtg gtgaccgtgc cctccagcag
cttgggcacc 600 cagacctaca tctgcaacgt gaatcacaag cccagcaaca
ccaaggtgga caagaaagtt 660 gagcccaaat cttgtgacaa aactcacaca
tgcccaccgt gcccagcacc tgaactcctg 720 gggggaccgt cagtcttcct
cttcccccca aaacccaagg acaccctcat gatctcccgg 780 acccctgagg
tcacatgcgt ggtggtggac gtgagccacg aagaccctga ggtcaagttc 840
aactggtacg tggacggcgt ggaggtgcat aatgccaaga caaagccgcg ggaggagcag
900 tacaacagca cgtaccgtgt ggtcagcgtc ctcaccgtcc tgcaccagga
ctggctgaat 960 ggcaaggagt acaagtgcaa ggtctccaac aaagccctcc
cagcccccat cgagaaaacc 1020 atctccaaag ccaaagggca gccccgagaa
ccacaggtgt acaccctgcc cccatcccgg 1080 gatgagctga ccaagaacca
ggtcagcctg acctgcctgg tcaaaggctt ctatcccagc 1140 gacatcgccg
tggagtggga gagcaatggg cagccggaga acaactacaa gaccacgcct 1200
cccgtgctgg actccgacgg ctccttcttc ttatattcaa agctcaccgt ggacaagagc
1260 aggtggcagc aggggaacgt cttctcatgc tccgtgatgc atgaggctct
gcacaaccac 1320 tacacgcaga agagcctctc cctgtctccc gggaaatga 1359
<210> SEQ ID NO 362 <211> LENGTH: 660 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 362 gcagccgtgc
tgacccagac accatcaccc gtgtctgcag ctgtgggagg cacagtcacc 60
atcagttgcc agtccagtca gagtgtttat aataataaca acttagcctg gtttcagcag
120 aaacccgggc agcctcccaa gcttctgatc tatctggcat ccactctggc
atctggggtc 180 ccatcacggt tcagcggcag tggatctggg acacagttca
ctctcaccat cagcggcgtg 240 cagtgtgacg atgctgccac ttattactgt
ctaggtggtt gtgatgatga tgctgatact 300 tttgctttcg gcggagggac
tgaggtggtg gtcaaacgaa ctgtggctgc accatctgtc 360 ttcatcttcc
cgccatctga tgagcagttg aaatctggaa ctgcctctgt tgtgtgcctg 420
ctgaataact tctatcccag agaggccaaa gtacagtgga aggtggataa cgccctccaa
480 tcgggtaact cccaggagag tgtcacagag caggacagca aggacagcac
ctacagcctc 540 agcagcaccc tgacgctgag caaagcagac tacgagaaac
acaaagtcta cgcctgcgaa 600 gtcacccatc agggcctgag ctcgcccgtc
acaaagagct tcaacagggg agagtgttga 660 <210> SEQ ID NO 363
<211> LENGTH: 1368 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 363 gaagtgcagc tggtggaatc
tggcggcgga ctggtgcagc ctggcggatc tctgagactg 60 tcttgtgccg
cctccggctt ctccctgtcc tcctacgcta tgatctgggt gcgacaggcc 120
cctggcaagg gcctggaatg gatcggcacc atctctaccg gcggaattac ctactacgcc
180 tcctgggcca agggccggtt caccatctcc agagacaact ccaagaacac
cctgtacctc 240 cagatgaact ccctgcgggc cgaggacacc gccgtgtact
attgtgctag aggcggctac 300 gccgccagct ccgcttacta cctgccctac
tacttcgacc tgtggggcca gggcaccctc 360 gtgacagtgt catctgcatc
caccaagggc ccatcggtct tccccctggc accctcctcc 420 aagagcacct
ctgggggcac agcggccctg ggctgcctgg tcaaggacta cttccccgaa 480
ccggtgacgg tgtcgtggaa ctcaggcgcc ctgaccagcg gcgtgcacac cttcccggct
540 gtcctacagt cctcaggact ctactccctc agcagcgtgg tgaccgtgcc
ctccagcagc 600 ttgggcaccc agacctacat ctgcaacgtg aatcacaagc
ccagcaacac caaggtggac 660 aagaaagttg agcccaaatc ttgtgacaaa
actcacacat gcccaccgtg cccagcacct 720 gaactcctgg ggggaccgtc
agtcttcctc ttccccccaa aacccaagga caccctcatg 780 atctcccgga
cccctgaggt cacatgcgtg gtggtggacg tgagccacga agaccctgag 840
gtcaagttca actggtacgt ggacggcgtg gaggtgcata atgccaagac aaagccgcgg
900 gaggagcagt acaacagcac gtaccgtgtg gtcagcgtcc tcaccgtcct
gcaccaggac 960 tggctgaatg gcaaggagta caagtgcaag gtctccaaca
aagccctccc agcccccatc 1020 gagaaaacca tctccaaagc caaagggcag
ccccgagaac cacaggtgta caccctgccc 1080 ccatcccggg atgagctgac
caagaaccag gtcagcctga cctgcctggt caaaggcttc 1140 tatcccagcg
acatcgccgt ggagtgggag agcaatgggc agccggagaa caactacaag 1200
accacgcctc ccgtgctgga ctccgacggc tccttcttct tatattcaaa gctcaccgtg
1260 gacaagagca ggtggcagca ggggaacgtc ttctcatgct ccgtgatgca
tgaggctctg 1320 cacaaccact acacgcagaa gagcctctcc ctgtctcccg
ggaaatga 1368 <210> SEQ ID NO 364 <211> LENGTH: 660
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polynucleotide <400> SEQUENCE:
364 gatattcaga tgacccagtc cccctccagc ctgtccgctt ctgtgggcga
cagagtgacc 60 atcacctgtc agtcctccca gtccgtgtat aacaacaaca
acctggcctg gtatcagcag 120 aaacccggca aggtgcccaa gctgctgatc
tacctggcct ccacactggc ctctggcgtg 180 ccctctagat tctccggctc
tggctctggc accgacttta ccctgaccat cagctccctc 240 cagtgcgagg
atgccgccac ctactattgc ctgggcggct gcgacgacga cgccgatacc 300
tttgcttttg gcggaggcac caaggtggaa atcaaacgaa ctgtggctgc accatctgtc
360 ttcatcttcc cgccatctga tgagcagttg aaatctggaa ctgcctctgt
tgtgtgcctg 420 ctgaataact tctatcccag agaggccaaa gtacagtgga
aggtggataa cgccctccaa 480 tcgggtaact cccaggagag tgtcacagag
caggacagca aggacagcac ctacagcctc 540 agcagcaccc tgacgctgag
caaagcagac tacgagaaac acaaagtcta cgcctgcgaa 600 gtcacccatc
agggcctgag ctcgcccgtc acaaagagct tcaacagggg agagtgttga 660
<210> SEQ ID NO 365 <211> LENGTH: 4 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 365 Ala His Lys Asp 1
<210> SEQ ID NO 366 <211> LENGTH: 13 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 366 Asn Thr Ser Gln Glu Ala
His Lys Asp Val Ser Tyr Leu 1 5 10 <210> SEQ ID NO 367
<211> LENGTH: 4 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 367 Gly Phe Leu Gly 1 <210> SEQ ID NO
368 <211> LENGTH: 4 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 368 Ala Leu Ala Leu 1
1 SEQUENCE LISTING <160> NUMBER OF SEQ ID NOS: 368
<210> SEQ ID NO 1 <211> LENGTH: 449 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 1 Glu Val Gln Leu Val
Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg 1 5 10 15 Ser Leu Arg
Leu Ser Cys Ser Ala Ser Gly Phe Thr Phe Ser Gly Tyr 20 25 30 Gly
Leu Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45 Ala Met Ile Ser Ser Gly Gly Ser Tyr Thr Tyr Tyr Ala Asp Ser Val
50 55 60 Lys Gly Arg Phe Ala Ile Ser Arg Asp Asn Ala Lys Asn Thr
Leu Phe 65 70 75 80 Leu Gln Met Asp Ser Leu Arg Pro Glu Asp Thr Gly
Val Tyr Phe Cys 85 90 95 Ala Arg His Gly Asp Asp Pro Ala Trp Phe
Ala Tyr Trp Gly Gln Gly 100 105 110 Thr Pro Val Thr Val Ser Ser Ala
Ser Thr Lys Gly Pro Ser Val Phe 115 120 125 Pro Leu Ala Pro Ser Ser
Lys Ser Thr Ser Gly Gly Thr Ala Ala Leu 130 135 140 Gly Cys Leu Val
Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp 145 150 155 160 Asn
Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val Leu 165 170
175 Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser
180 185 190 Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His
Lys Pro 195 200 205 Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys
Ser Cys Asp Lys 210 215 220 Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Glu Leu Leu Gly Gly Pro 225 230 235 240 Ser Val Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser 245 250 255 Arg Thr Pro Glu Val
Thr Cys Val Val Val Asp Val Ser His Glu Asp 260 265 270 Pro Glu Val
Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn 275 280 285 Ala
Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val 290 295
300 Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu
305 310 315 320 Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro
Ile Glu Lys 325 330 335 Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr 340 345 350 Leu Pro Pro Ser Arg Asp Glu Leu Thr
Lys Asn Gln Val Ser Leu Thr 355 360 365 Cys Leu Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu Trp Glu 370 375 380 Ser Asn Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu 385 390 395 400 Asp Ser
Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys 405 410 415
Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu 420
425 430 Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro
Gly 435 440 445 Lys <210> SEQ ID NO 2 <211> LENGTH: 5
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 2 Gly
Tyr Gly Leu Ser 1 5 <210> SEQ ID NO 3 <211> LENGTH: 17
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 3 Met
Ile Ser Ser Gly Gly Ser Tyr Thr Tyr Tyr Ala Asp Ser Val Lys 1 5 10
15 Gly <210> SEQ ID NO 4 <211> LENGTH: 10 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic peptide <400> SEQUENCE: 4 His Gly Asp Asp
Pro Ala Trp Phe Ala Tyr 1 5 10 <210> SEQ ID NO 5 <211>
LENGTH: 468 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polypeptide
<400> SEQUENCE: 5 Met Gly Trp Ser Cys Ile Ile Leu Phe Leu Val
Ala Thr Ala Thr Gly 1 5 10 15 Val His Ser Glu Val Gln Leu Val Glu
Ser Gly Gly Gly Val Val Gln 20 25 30 Pro Gly Arg Ser Leu Arg Leu
Ser Cys Ser Ala Ser Gly Phe Thr Phe 35 40 45 Ser Gly Tyr Gly Leu
Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu 50 55 60 Glu Trp Val
Ala Met Ile Ser Ser Gly Gly Ser Tyr Thr Tyr Tyr Ala 65 70 75 80 Asp
Ser Val Lys Gly Arg Phe Ala Ile Ser Arg Asp Asn Ala Lys Asn 85 90
95 Thr Leu Phe Leu Gln Met Asp Ser Leu Arg Pro Glu Asp Thr Gly Val
100 105 110 Tyr Phe Cys Ala Arg His Gly Asp Asp Pro Ala Trp Phe Ala
Tyr Trp 115 120 125 Gly Gln Gly Thr Pro Val Thr Val Ser Ser Ala Ser
Thr Lys Gly Pro 130 135 140 Ser Val Phe Pro Leu Ala Pro Ser Ser Lys
Ser Thr Ser Gly Gly Thr 145 150 155 160 Ala Ala Leu Gly Cys Leu Val
Lys Asp Tyr Phe Pro Glu Pro Val Thr 165 170 175 Val Ser Trp Asn Ser
Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro 180 185 190 Ala Val Leu
Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr 195 200 205 Val
Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn 210 215
220 His Lys Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser
225 230 235 240 Cys Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Glu Leu Leu 245 250 255 Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys
Pro Lys Asp Thr Leu 260 265 270 Met Ile Ser Arg Thr Pro Glu Val Thr
Cys Val Val Val Asp Val Ser 275 280 285 His Glu Asp Pro Glu Val Lys
Phe Asn Trp Tyr Val Asp Gly Val Glu 290 295 300 Val His Asn Ala Lys
Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr 305 310 315 320 Tyr Arg
Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn 325 330 335
Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro 340
345 350 Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro
Gln 355 360 365 Val Tyr Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys
Asn Gln Val 370 375 380 Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro
Ser Asp Ile Ala Val 385 390 395 400 Glu Trp Glu Ser Asn Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro 405 410 415 Pro Val Leu Asp Ser Asp
Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr 420 425 430 Val Asp Lys Ser
Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val 435 440 445 Met His
Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu 450 455 460
Ser Pro Gly Lys
465 <210> SEQ ID NO 6 <211> LENGTH: 217 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polypeptide <400> SEQUENCE: 6 Asp Ile Gln
Leu Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly 1 5 10 15 Asp
Arg Val Thr Ile Thr Cys Ser Val Ser Ser Ser Ile Ser Ser Asn 20 25
30 Asn Leu His Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Pro Trp
35 40 45 Ile Tyr Gly Thr Ser Asn Leu Ala Ser Gly Val Pro Ser Arg
Phe Ser 50 55 60 Gly Ser Gly Ser Gly Thr Asp Tyr Thr Phe Thr Ile
Ser Ser Leu Gln 65 70 75 80 Pro Glu Asp Ile Ala Thr Tyr Tyr Cys Gln
Gln Trp Ser Ser Tyr Pro 85 90 95 Tyr Met Tyr Thr Phe Gly Gln Gly
Thr Lys Val Glu Ile Lys Arg Thr 100 105 110 Val Ala Ala Pro Ser Val
Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu 115 120 125 Lys Ser Gly Thr
Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro 130 135 140 Arg Glu
Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly 145 150 155
160 Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr
165 170 175 Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu
Lys His 180 185 190 Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu
Ser Ser Pro Val 195 200 205 Thr Lys Ser Phe Asn Arg Gly Glu Cys 210
215 <210> SEQ ID NO 7 <211> LENGTH: 12 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic peptide <400> SEQUENCE: 7 Ser Val Ser Ser
Ser Ile Ser Ser Asn Asn Leu His 1 5 10 <210> SEQ ID NO 8
<211> LENGTH: 7 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 8 Gly Thr Ser Asn Leu Ala Ser 1 5 <210>
SEQ ID NO 9 <211> LENGTH: 11 <212> TYPE: PRT
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 9 Gln Gln Trp Ser Ser Tyr
Pro Tyr Met Tyr Thr 1 5 10 <210> SEQ ID NO 10 <211>
LENGTH: 236 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polypeptide
<400> SEQUENCE: 10 Met Gly Trp Ser Cys Ile Ile Leu Phe Leu
Val Ala Thr Ala Thr Gly 1 5 10 15 Val His Ser Asp Ile Gln Leu Thr
Gln Ser Pro Ser Ser Leu Ser Ala 20 25 30 Ser Val Gly Asp Arg Val
Thr Ile Thr Cys Ser Val Ser Ser Ser Ile 35 40 45 Ser Ser Asn Asn
Leu His Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro 50 55 60 Lys Pro
Trp Ile Tyr Gly Thr Ser Asn Leu Ala Ser Gly Val Pro Ser 65 70 75 80
Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Tyr Thr Phe Thr Ile Ser 85
90 95 Ser Leu Gln Pro Glu Asp Ile Ala Thr Tyr Tyr Cys Gln Gln Trp
Ser 100 105 110 Ser Tyr Pro Tyr Met Tyr Thr Phe Gly Gln Gly Thr Lys
Val Glu Ile 115 120 125 Lys Arg Thr Val Ala Ala Pro Ser Val Phe Ile
Phe Pro Pro Ser Asp 130 135 140 Glu Gln Leu Lys Ser Gly Thr Ala Ser
Val Val Cys Leu Leu Asn Asn 145 150 155 160 Phe Tyr Pro Arg Glu Ala
Lys Val Gln Trp Lys Val Asp Asn Ala Leu 165 170 175 Gln Ser Gly Asn
Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp 180 185 190 Ser Thr
Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr 195 200 205
Glu Lys His Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser 210
215 220 Ser Pro Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 225 230 235
<210> SEQ ID NO 11 <211> LENGTH: 1407 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 11 atgggatgga
gctgtatcat cctcttcttg gtagcaacag ctacaggtgt ccactccgag 60
gtccaactgg tggagagcgg tggaggtgtt gtgcaacctg gccggtccct gcgcctgtcc
120 tgctccgcat ctggcttcac cttcagcggc tatgggttgt cttgggtgag
acaggcacct 180 ggaaaaggtc ttgagtgggt tgcaatgatt agtagtggtg
gtagttatac ctactatgca 240 gacagtgtga agggtagatt tgcaatatcg
cgagacaacg ccaagaacac attgttcctg 300 caaatggaca gcctgagacc
cgaagacacc ggggtctatt tttgtgcaag acatggggac 360 gatcccgcct
ggttcgctta ttggggccaa gggaccccgg tcaccgtctc ctcagcctcc 420
accaagggcc catcggtctt ccccctggca ccctcctcca agagcacctc tgggggcaca
480 gcggccctgg gctgcctggt caaggactac ttccccgaac cggtgacggt
gtcgtggaac 540 tcaggcgccc tgaccagcgg cgtgcacacc ttcccggctg
tcctacagtc ctcaggactc 600 tactccctca gcagcgtggt gaccgtgccc
tccagcagct tgggcaccca gacctacatc 660 tgcaacgtga atcacaagcc
cagcaacacc aaggtggaca agaaagttga gcccaaatct 720 tgtgacaaaa
ctcacacatg cccaccgtgc ccagcacctg aactcctggg gggaccgtca 780
gtcttcctct tccccccaaa acccaaggac accctcatga tctcccggac ccctgaggtc
840 acatgcgtgg tggtggacgt gagccacgaa gaccctgagg tcaagttcaa
ctggtacgtg 900 gacggcgtgg aggtgcataa tgccaagaca aagccgcggg
aggagcagta caacagcacg 960 taccgtgtgg tcagcgtcct caccgtcctg
caccaggact ggctgaatgg caaggagtac 1020 aagtgcaagg tctccaacaa
agccctccca gcccccatcg agaaaaccat ctccaaagcc 1080 aaagggcagc
cccgagaacc acaggtgtac accctgcccc catcccggga tgagctgacc 1140
aagaaccagg tcagcctgac ctgcctggtc aaaggcttct atcccagcga catcgccgtg
1200 gagtgggaga gcaatgggca gccggagaac aactacaaga ccacgcctcc
cgtgctggac 1260 tccgacggct ccttcttctt atattcaaag ctcaccgtgg
acaagagcag gtggcagcag 1320 gggaacgtct tctcatgctc cgtgatgcat
gaggctctgc acaaccacta cacgcagaag 1380 agcctctccc tgtctcccgg gaaatga
1407 <210> SEQ ID NO 12 <211> LENGTH: 711 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polynucleotide <400> SEQUENCE: 12
atgggatgga gctgtatcat cctcttcttg gtagcaacag ctacaggtgt ccactccgac
60 atccagctga cccagagccc aagcagcctg agcgccagcg tgggtgacag
agtgaccatc 120 acctgtagtg tcagctcaag tataagttcc aacaacttgc
actggtacca gcagaagcca 180 ggtaaggctc caaagccatg gatctacggc
acatccaacc tggcttctgg tgtgccaagc 240 agattcagcg gtagcggtag
cggtaccgac tacaccttca ccatcagcag cctccagcca 300 gaggacatcg
ccacctacta ctgccaacag tggagtagtt acccgtacat gtacacgttc 360
ggccaaggga ccaaggtgga aatcaaacga actgtggctg caccatctgt cttcatcttc
420 ccgccatctg atgagcagtt gaaatctgga actgcctctg ttgtgtgcct
gctgaataac 480 ttctatccca gagaggccaa agtacagtgg aaggtggata
acgccctcca atcgggtaac 540 tcccaggaga gtgtcacaga gcaggacagc
aaggacagca cctacagcct cagcagcacc 600 ctgacgctga gcaaagcaga
ctacgagaaa cacaaagtct acgcctgcga agtcacccat 660
cagggcctga gctcgcccgt cacaaagagc ttcaacaggg gagagtgtta a 711
<210> SEQ ID NO 13 <211> LENGTH: 8 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 13 Gly Phe Thr Phe Ser Gly
Tyr Gly 1 5 <210> SEQ ID NO 14 <211> LENGTH: 8
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 14 Ile
Ser Ser Gly Gly Ser Tyr Thr 1 5 <210> SEQ ID NO 15
<211> LENGTH: 12 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 15 Ala Arg His Gly Asp Asp Pro Ala Trp Phe
Ala Tyr 1 5 10 <210> SEQ ID NO 16 <211> LENGTH: 7
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 16 Ser
Ser Ile Ser Ser Asn Asn 1 5 <210> SEQ ID NO 17 <211>
LENGTH: 3 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 17 Gly Thr Ser 1 <210> SEQ ID NO 18 <211>
LENGTH: 11 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 18 Gln Gln Trp Ser Ser Tyr Pro Tyr Met Tyr Thr 1 5 10
<210> SEQ ID NO 19 <211> LENGTH: 257 <212> TYPE:
PRT <213> ORGANISM: Homo sapiens <400> SEQUENCE: 19 Met
Ala Gln Arg Met Thr Thr Gln Leu Leu Leu Leu Leu Val Trp Val 1 5 10
15 Ala Val Val Gly Glu Ala Gln Thr Arg Ile Ala Trp Ala Arg Thr Glu
20 25 30 Leu Leu Asn Val Cys Met Asn Ala Lys His His Lys Glu Lys
Pro Gly 35 40 45 Pro Glu Asp Lys Leu His Glu Gln Cys Arg Pro Trp
Arg Lys Asn Ala 50 55 60 Cys Cys Ser Thr Asn Thr Ser Gln Glu Ala
His Lys Asp Val Ser Tyr 65 70 75 80 Leu Tyr Arg Phe Asn Trp Asn His
Cys Gly Glu Met Ala Pro Ala Cys 85 90 95 Lys Arg His Phe Ile Gln
Asp Thr Cys Leu Tyr Glu Cys Ser Pro Asn 100 105 110 Leu Gly Pro Trp
Ile Gln Gln Val Asp Gln Ser Trp Arg Lys Glu Arg 115 120 125 Val Leu
Asn Val Pro Leu Cys Lys Glu Asp Cys Glu Gln Trp Trp Glu 130 135 140
Asp Cys Arg Thr Ser Tyr Thr Cys Lys Ser Asn Trp His Lys Gly Trp 145
150 155 160 Asn Trp Thr Ser Gly Phe Asn Lys Cys Ala Val Gly Ala Ala
Cys Gln 165 170 175 Pro Phe His Phe Tyr Phe Pro Thr Pro Thr Val Leu
Cys Asn Glu Ile 180 185 190 Trp Thr His Ser Tyr Lys Val Ser Asn Tyr
Ser Arg Gly Ser Gly Arg 195 200 205 Cys Ile Gln Met Trp Phe Asp Pro
Ala Gln Gly Asn Pro Asn Glu Glu 210 215 220 Val Ala Arg Phe Tyr Ala
Ala Ala Met Ser Gly Ala Gly Pro Trp Ala 225 230 235 240 Ala Trp Pro
Phe Leu Leu Ser Leu Ala Leu Met Leu Leu Trp Leu Leu 245 250 255 Ser
<210> SEQ ID NO 20 <211> LENGTH: 982 <212> TYPE:
DNA <213> ORGANISM: Homo sapiens <400> SEQUENCE: 20
cattccttgg tgccactgac cacagctctt tcttcaggga cagacatggc tcagcggatg
60 acaacacagc tgctgctcct tctagtgtgg gtggctgtag taggggaggc
tcagacaagg 120 attgcatggg ccaggactga gcttctcaat gtctgcatga
acgccaagca ccacaaggaa 180 aagccaggcc ccgaggacaa gttgcatgag
cagtgtcgac cctggaggaa gaatgcctgc 240 tgttctacca acaccagcca
ggaagcccat aaggatgttt cctacctata tagattcaac 300 tggaaccact
gtggagagat ggcacctgcc tgcaaacggc atttcatcca ggacacctgc 360
ctctacgagt gctcccccaa cttggggccc tggatccagc aggtggatca gagctggcgc
420 aaagagcggg tactgaacgt gcccctgtgc aaagaggact gtgagcaatg
gtgggaagat 480 tgtcgcacct cctacacctg caagagcaac tggcacaagg
gctggaactg gacttcaggg 540 tttaacaagt gcgcagtggg agctgcctgc
caacctttcc atttctactt ccccacaccc 600 actgttctgt gcaatgaaat
ctggactcac tcctacaagg tcagcaacta cagccgaggg 660 agtggccgct
gcatccagat gtggttcgac ccagcccagg gcaaccccaa tgaggaggtg 720
gcgaggttct atgctgcagc catgagtggg gctgggccct gggcagcctg gcctttcctg
780 cttagcctgg ccctaatgct gctgtggctg ctcagctgac ctccttttac
cttctgatac 840 ctggaaatcc ctgccctgtt cagccccaca gctcccaact
atttggttcc tgctccatgg 900 tcgggcctct gacagccact ttgaataaac
cagacaccgc acatgtgtct tgagaattat 960 ttggaaaaaa aaaaaaaaaa aa 982
<210> SEQ ID NO 21 <211> LENGTH: 1255 <212> TYPE:
PRT <213> ORGANISM: Homo sapiens <400> SEQUENCE: 21 Met
Glu Leu Ala Ala Leu Cys Arg Trp Gly Leu Leu Leu Ala Leu Leu 1 5 10
15 Pro Pro Gly Ala Ala Ser Thr Gln Val Cys Thr Gly Thr Asp Met Lys
20 25 30 Leu Arg Leu Pro Ala Ser Pro Glu Thr His Leu Asp Met Leu
Arg His 35 40 45 Leu Tyr Gln Gly Cys Gln Val Val Gln Gly Asn Leu
Glu Leu Thr Tyr 50 55 60 Leu Pro Thr Asn Ala Ser Leu Ser Phe Leu
Gln Asp Ile Gln Glu Val 65 70 75 80 Gln Gly Tyr Val Leu Ile Ala His
Asn Gln Val Arg Gln Val Pro Leu 85 90 95 Gln Arg Leu Arg Ile Val
Arg Gly Thr Gln Leu Phe Glu Asp Asn Tyr 100 105 110 Ala Leu Ala Val
Leu Asp Asn Gly Asp Pro Leu Asn Asn Thr Thr Pro 115 120 125 Val Thr
Gly Ala Ser Pro Gly Gly Leu Arg Glu Leu Gln Leu Arg Ser 130 135 140
Leu Thr Glu Ile Leu Lys Gly Gly Val Leu Ile Gln Arg Asn Pro Gln 145
150 155 160 Leu Cys Tyr Gln Asp Thr Ile Leu Trp Lys Asp Ile Phe His
Lys Asn 165 170 175 Asn Gln Leu Ala Leu Thr Leu Ile Asp Thr Asn Arg
Ser Arg Ala Cys 180 185 190 His Pro Cys Ser Pro Met Cys Lys Gly Ser
Arg Cys Trp Gly Glu Ser 195 200 205 Ser Glu Asp Cys Gln Ser Leu Thr
Arg Thr Val Cys Ala Gly Gly Cys 210 215 220 Ala Arg Cys Lys Gly Pro
Leu Pro Thr Asp Cys Cys His Glu Gln Cys 225 230 235 240 Ala Ala Gly
Cys Thr Gly Pro Lys His Ser Asp Cys Leu Ala Cys Leu 245 250 255 His
Phe Asn His Ser Gly Ile Cys Glu Leu His Cys Pro Ala Leu Val 260 265
270
Thr Tyr Asn Thr Asp Thr Phe Glu Ser Met Pro Asn Pro Glu Gly Arg 275
280 285 Tyr Thr Phe Gly Ala Ser Cys Val Thr Ala Cys Pro Tyr Asn Tyr
Leu 290 295 300 Ser Thr Asp Val Gly Ser Cys Thr Leu Val Cys Pro Leu
His Asn Gln 305 310 315 320 Glu Val Thr Ala Glu Asp Gly Thr Gln Arg
Cys Glu Lys Cys Ser Lys 325 330 335 Pro Cys Ala Arg Val Cys Tyr Gly
Leu Gly Met Glu His Leu Arg Glu 340 345 350 Val Arg Ala Val Thr Ser
Ala Asn Ile Gln Glu Phe Ala Gly Cys Lys 355 360 365 Lys Ile Phe Gly
Ser Leu Ala Phe Leu Pro Glu Ser Phe Asp Gly Asp 370 375 380 Pro Ala
Ser Asn Thr Ala Pro Leu Gln Pro Glu Gln Leu Gln Val Phe 385 390 395
400 Glu Thr Leu Glu Glu Ile Thr Gly Tyr Leu Tyr Ile Ser Ala Trp Pro
405 410 415 Asp Ser Leu Pro Asp Leu Ser Val Phe Gln Asn Leu Gln Val
Ile Arg 420 425 430 Gly Arg Ile Leu His Asn Gly Ala Tyr Ser Leu Thr
Leu Gln Gly Leu 435 440 445 Gly Ile Ser Trp Leu Gly Leu Arg Ser Leu
Arg Glu Leu Gly Ser Gly 450 455 460 Leu Ala Leu Ile His His Asn Thr
His Leu Cys Phe Val His Thr Val 465 470 475 480 Pro Trp Asp Gln Leu
Phe Arg Asn Pro His Gln Ala Leu Leu His Thr 485 490 495 Ala Asn Arg
Pro Glu Asp Glu Cys Val Gly Glu Gly Leu Ala Cys His 500 505 510 Gln
Leu Cys Ala Arg Gly His Cys Trp Gly Pro Gly Pro Thr Gln Cys 515 520
525 Val Asn Cys Ser Gln Phe Leu Arg Gly Gln Glu Cys Val Glu Glu Cys
530 535 540 Arg Val Leu Gln Gly Leu Pro Arg Glu Tyr Val Asn Ala Arg
His Cys 545 550 555 560 Leu Pro Cys His Pro Glu Cys Gln Pro Gln Asn
Gly Ser Val Thr Cys 565 570 575 Phe Gly Pro Glu Ala Asp Gln Cys Val
Ala Cys Ala His Tyr Lys Asp 580 585 590 Pro Pro Phe Cys Val Ala Arg
Cys Pro Ser Gly Val Lys Pro Asp Leu 595 600 605 Ser Tyr Met Pro Ile
Trp Lys Phe Pro Asp Glu Glu Gly Ala Cys Gln 610 615 620 Pro Cys Pro
Ile Asn Cys Thr His Ser Cys Val Asp Leu Asp Asp Lys 625 630 635 640
Gly Cys Pro Ala Glu Gln Arg Ala Ser Pro Leu Thr Ser Ile Ile Ser 645
650 655 Ala Val Val Gly Ile Leu Leu Val Val Val Leu Gly Val Val Phe
Gly 660 665 670 Ile Leu Ile Lys Arg Arg Gln Gln Lys Ile Arg Lys Tyr
Thr Met Arg 675 680 685 Arg Leu Leu Gln Glu Thr Glu Leu Val Glu Pro
Leu Thr Pro Ser Gly 690 695 700 Ala Met Pro Asn Gln Ala Gln Met Arg
Ile Leu Lys Glu Thr Glu Leu 705 710 715 720 Arg Lys Val Lys Val Leu
Gly Ser Gly Ala Phe Gly Thr Val Tyr Lys 725 730 735 Gly Ile Trp Ile
Pro Asp Gly Glu Asn Val Lys Ile Pro Val Ala Ile 740 745 750 Lys Val
Leu Arg Glu Asn Thr Ser Pro Lys Ala Asn Lys Glu Ile Leu 755 760 765
Asp Glu Ala Tyr Val Met Ala Gly Val Gly Ser Pro Tyr Val Ser Arg 770
775 780 Leu Leu Gly Ile Cys Leu Thr Ser Thr Val Gln Leu Val Thr Gln
Leu 785 790 795 800 Met Pro Tyr Gly Cys Leu Leu Asp His Val Arg Glu
Asn Arg Gly Arg 805 810 815 Leu Gly Ser Gln Asp Leu Leu Asn Trp Cys
Met Gln Ile Ala Lys Gly 820 825 830 Met Ser Tyr Leu Glu Asp Val Arg
Leu Val His Arg Asp Leu Ala Ala 835 840 845 Arg Asn Val Leu Val Lys
Ser Pro Asn His Val Lys Ile Thr Asp Phe 850 855 860 Gly Leu Ala Arg
Leu Leu Asp Ile Asp Glu Thr Glu Tyr His Ala Asp 865 870 875 880 Gly
Gly Lys Val Pro Ile Lys Trp Met Ala Leu Glu Ser Ile Leu Arg 885 890
895 Arg Arg Phe Thr His Gln Ser Asp Val Trp Ser Tyr Gly Val Thr Val
900 905 910 Trp Glu Leu Met Thr Phe Gly Ala Lys Pro Tyr Asp Gly Ile
Pro Ala 915 920 925 Arg Glu Ile Pro Asp Leu Leu Glu Lys Gly Glu Arg
Leu Pro Gln Pro 930 935 940 Pro Ile Cys Thr Ile Asp Val Tyr Met Ile
Met Val Lys Cys Trp Met 945 950 955 960 Ile Asp Ser Glu Cys Arg Pro
Arg Phe Arg Glu Leu Val Ser Glu Phe 965 970 975 Ser Arg Met Ala Arg
Asp Pro Gln Arg Phe Val Val Ile Gln Asn Glu 980 985 990 Asp Leu Gly
Pro Ala Ser Pro Leu Asp Ser Thr Phe Tyr Arg Ser Leu 995 1000 1005
Leu Glu Asp Asp Asp Met Gly Asp Leu Val Asp Ala Glu Glu Tyr 1010
1015 1020 Leu Val Pro Gln Gln Gly Phe Phe Cys Pro Asp Pro Ala Pro
Gly 1025 1030 1035 Ala Gly Gly Met Val His His Arg His Arg Ser Ser
Ser Thr Arg 1040 1045 1050 Ser Gly Gly Gly Asp Leu Thr Leu Gly Leu
Glu Pro Ser Glu Glu 1055 1060 1065 Glu Ala Pro Arg Ser Pro Leu Ala
Pro Ser Glu Gly Ala Gly Ser 1070 1075 1080 Asp Val Phe Asp Gly Asp
Leu Gly Met Gly Ala Ala Lys Gly Leu 1085 1090 1095 Gln Ser Leu Pro
Thr His Asp Pro Ser Pro Leu Gln Arg Tyr Ser 1100 1105 1110 Glu Asp
Pro Thr Val Pro Leu Pro Ser Glu Thr Asp Gly Tyr Val 1115 1120 1125
Ala Pro Leu Thr Cys Ser Pro Gln Pro Glu Tyr Val Asn Gln Pro 1130
1135 1140 Asp Val Arg Pro Gln Pro Pro Ser Pro Arg Glu Gly Pro Leu
Pro 1145 1150 1155 Ala Ala Arg Pro Ala Gly Ala Thr Leu Glu Arg Pro
Lys Thr Leu 1160 1165 1170 Ser Pro Gly Lys Asn Gly Val Val Lys Asp
Val Phe Ala Phe Gly 1175 1180 1185 Gly Ala Val Glu Asn Pro Glu Tyr
Leu Thr Pro Gln Gly Gly Ala 1190 1195 1200 Ala Pro Gln Pro His Pro
Pro Pro Ala Phe Ser Pro Ala Phe Asp 1205 1210 1215 Asn Leu Tyr Tyr
Trp Asp Gln Asp Pro Pro Glu Arg Gly Ala Pro 1220 1225 1230 Pro Ser
Thr Phe Lys Gly Thr Pro Thr Ala Glu Asn Pro Glu Tyr 1235 1240 1245
Leu Gly Leu Asp Val Pro Val 1250 1255 <210> SEQ ID NO 22
<211> LENGTH: 3768 <212> TYPE: DNA <213>
ORGANISM: Homo sapiens <400> SEQUENCE: 22 atggagctgg
cggccttgtg ccgctggggg ctcctcctcg ccctcttgcc ccccggagcc 60
gcgagcaccc aagtgtgcac cggcacagac atgaagctgc ggctccctgc cagtcccgag
120 acccacctgg acatgctccg ccacctctac cagggctgcc aggtggtgca
gggaaacctg 180 gaactcacct acctgcccac caatgccagc ctgtccttcc
tgcaggatat ccaggaggtg 240 cagggctacg tgctcatcgc tcacaaccaa
gtgaggcagg tcccactgca gaggctgcgg 300 attgtgcgag gcacccagct
ctttgaggac aactatgccc tggccgtgct agacaatgga 360 gacccgctga
acaataccac ccctgtcaca ggggcctccc caggaggcct gcgggagctg 420
cagcttcgaa gcctcacaga gatcttgaaa ggaggggtct tgatccagcg gaacccccag
480 ctctgctacc aggacacgat tttgtggaag gacatcttcc acaagaacaa
ccagctggct 540 ctcacactga tagacaccaa ccgctctcgg gcctgccacc
cctgttctcc gatgtgtaag 600 ggctcccgct gctggggaga gagttctgag
gattgtcaga gcctgacgcg cactgtctgt 660 gccggtggct gtgcccgctg
caaggggcca ctgcccactg actgctgcca tgagcagtgt 720 gctgccggct
gcacgggccc caagcactct gactgcctgg cctgcctcca cttcaaccac 780
agtggcatct gtgagctgca ctgcccagcc ctggtcacct acaacacaga cacgtttgag
840 tccatgccca atcccgaggg ccggtataca ttcggcgcca gctgtgtgac
tgcctgtccc 900 tacaactacc tttctacgga cgtgggatcc tgcaccctcg
tctgccccct gcacaaccaa 960 gaggtgacag cagaggatgg aacacagcgg
tgtgagaagt gcagcaagcc ctgtgcccga 1020 gtgtgctatg gtctgggcat
ggagcacttg cgagaggtga gggcagttac cagtgccaat 1080 atccaggagt
ttgctggctg caagaagatc tttgggagcc tggcatttct gccggagagc 1140
tttgatgggg acccagcctc caacactgcc ccgctccagc cagagcagct ccaagtgttt
1200 gagactctgg aagagatcac aggttaccta tacatctcag catggccgga
cagcctgcct 1260 gacctcagcg tcttccagaa cctgcaagta atccggggac
gaattctgca caatggcgcc 1320 tactcgctga ccctgcaagg gctgggcatc
agctggctgg ggctgcgctc actgagggaa 1380 ctgggcagtg gactggccct
catccaccat aacacccacc tctgcttcgt gcacacggtg 1440 ccctgggacc
agctctttcg gaacccgcac caagctctgc tccacactgc caaccggcca 1500
gaggacgagt gtgtgggcga gggcctggcc tgccaccagc tgtgcgcccg agggcactgc
1560 tggggtccag ggcccaccca gtgtgtcaac tgcagccagt tccttcgggg
ccaggagtgc 1620
gtggaggaat gccgagtact gcaggggctc cccagggagt atgtgaatgc caggcactgt
1680 ttgccgtgcc accctgagtg tcagccccag aatggctcag tgacctgttt
tggaccggag 1740 gctgaccagt gtgtggcctg tgcccactat aaggaccctc
ccttctgcgt ggcccgctgc 1800 cccagcggtg tgaaacctga cctctcctac
atgcccatct ggaagtttcc agatgaggag 1860 ggcgcatgcc agccttgccc
catcaactgc acccactcct gtgtggacct ggatgacaag 1920 ggctgccccg
ccgagcagag agccagccct ctgacgtcca tcatctctgc ggtggttggc 1980
attctgctgg tcgtggtctt gggggtggtc tttgggatcc tcatcaagcg acggcagcag
2040 aagatccgga agtacacgat gcggagactg ctgcaggaaa cggagctggt
ggagccgctg 2100 acacctagcg gagcgatgcc caaccaggcg cagatgcgga
tcctgaaaga gacggagctg 2160 aggaaggtga aggtgcttgg atctggcgct
tttggcacag tctacaaggg catctggatc 2220 cctgatgggg agaatgtgaa
aattccagtg gccatcaaag tgttgaggga aaacacatcc 2280 cccaaagcca
acaaagaaat cttagacgaa gcatacgtga tggctggtgt gggctcccca 2340
tatgtctccc gccttctggg catctgcctg acatccacgg tgcagctggt gacacagctt
2400 atgccctatg gctgcctctt agaccatgtc cgggaaaacc gcggacgcct
gggctcccag 2460 gacctgctga actggtgtat gcagattgcc aaggggatga
gctacctgga ggatgtgcgg 2520 ctcgtacaca gggacttggc cgctcggaac
gtgctggtca agagtcccaa ccatgtcaaa 2580 attacagact tcgggctggc
tcggctgctg gacattgacg agacagagta ccatgcagat 2640 gggggcaagg
tgcccatcaa gtggatggcg ctggagtcca ttctccgccg gcggttcacc 2700
caccagagtg atgtgtggag ttatggtgtg actgtgtggg agctgatgac ttttggggcc
2760 aaaccttacg atgggatccc agcccgggag atccctgacc tgctggaaaa
gggggagcgg 2820 ctgccccagc cccccatctg caccattgat gtctacatga
tcatggtcaa atgttggatg 2880 attgactctg aatgtcggcc aagattccgg
gagttggtgt ctgaattctc ccgcatggcc 2940 agggaccccc agcgctttgt
ggtcatccag aatgaggact tgggcccagc cagtcccttg 3000 gacagcacct
tctaccgctc actgctggag gacgatgaca tgggggacct ggtggatgct 3060
gaggagtatc tggtacccca gcagggcttc ttctgtccag accctgcccc gggcgctggg
3120 ggcatggtcc accacaggca ccgcagctca tctaccagga gtggcggtgg
ggacctgaca 3180 ctagggctgg agccctctga agaggaggcc cccaggtctc
cactggcacc ctccgaaggg 3240 gctggctccg atgtatttga tggtgacctg
ggaatggggg cagccaaggg gctgcaaagc 3300 ctccccacac atgaccccag
ccctctacag cggtacagtg aggaccccac agtacccctg 3360 ccctctgaga
ctgatggcta cgttgccccc ctgacctgca gcccccagcc tgaatatgtg 3420
aaccagccag atgttcggcc ccagccccct tcgccccgag agggccctct gcctgctgcc
3480 cgacctgctg gtgccactct ggaaaggccc aagactctct ccccagggaa
gaatggggtc 3540 gtcaaagacg tttttgcctt tgggggtgcc gtggagaacc
ccgagtactt gacaccccag 3600 ggaggagctg cccctcagcc ccaccctcct
cctgccttca gcccagcctt cgacaacctc 3660 tattactggg accaggaccc
accagagcgg ggggctccac ccagcacctt caaagggaca 3720 cctacggcag
agaacccaga gtacctgggt ctggacgtgc cagtgtga 3768 <210> SEQ ID
NO 23 <211> LENGTH: 119 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 23 Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Val Val Gln Pro Gly Arg 1 5 10 15 Ser Leu Arg Leu Ser
Cys Ser Ala Ser Gly Phe Thr Phe Ser Gly Tyr 20 25 30 Gly Leu Ser
Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45 Ala
Met Ile Ser Ser Gly Gly Ser Tyr Thr Tyr Tyr Ala Asp Ser Val 50 55
60 Lys Gly Arg Phe Ala Ile Ser Arg Asp Asn Ala Lys Asn Thr Leu Phe
65 70 75 80 Leu Gln Met Asp Ser Leu Arg Pro Glu Asp Thr Gly Val Tyr
Phe Cys 85 90 95 Ala Arg His Gly Asp Asp Pro Ala Trp Phe Ala Tyr
Trp Gly Gln Gly 100 105 110 Thr Pro Val Thr Val Ser Ser 115
<210> SEQ ID NO 24 <211> LENGTH: 110 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 24 Asp Ile Gln Leu Thr
Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly 1 5 10 15 Asp Arg Val
Thr Ile Thr Cys Ser Val Ser Ser Ser Ile Ser Ser Asn 20 25 30 Asn
Leu His Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Pro Trp 35 40
45 Ile Tyr Gly Thr Ser Asn Leu Ala Ser Gly Val Pro Ser Arg Phe Ser
50 55 60 Gly Ser Gly Ser Gly Thr Asp Tyr Thr Phe Thr Ile Ser Ser
Leu Gln 65 70 75 80 Pro Glu Asp Ile Ala Thr Tyr Tyr Cys Gln Gln Trp
Ser Ser Tyr Pro 85 90 95 Tyr Met Tyr Thr Phe Gly Gln Gly Thr Lys
Val Glu Ile Lys 100 105 110 <210> SEQ ID NO 25 <211>
LENGTH: 119 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polypeptide
<400> SEQUENCE: 25 Gln Val Gln Leu Gln Gln Ser Gly Pro Glu
Leu Glu Lys Pro Gly Ala 1 5 10 15 Ser Val Lys Ile Ser Cys Lys Ala
Ser Gly Tyr Ser Phe Thr Gly Tyr 20 25 30 Thr Met Asn Trp Val Lys
Gln Ser His Gly Lys Ser Leu Glu Trp Ile 35 40 45 Gly Leu Ile Thr
Pro Tyr Asn Gly Ala Ser Ser Tyr Asn Gln Lys Phe 50 55 60 Arg Gly
Lys Ala Thr Leu Thr Val Asp Lys Ser Ser Ser Thr Ala Tyr 65 70 75 80
Met Asp Leu Leu Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Phe Cys 85
90 95 Ala Arg Gly Gly Tyr Asp Gly Arg Gly Phe Asp Tyr Trp Gly Ser
Gly 100 105 110 Thr Pro Val Thr Val Ser Ser 115 <210> SEQ ID
NO 26 <211> LENGTH: 106 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 26 Asp Ile Glu Leu Thr Gln Ser
Pro Ala Ile Met Ser Ala Ser Pro Gly 1 5 10 15 Glu Lys Val Thr Met
Thr Cys Ser Ala Ser Ser Ser Val Ser Tyr Met 20 25 30 His Trp Tyr
Gln Gln Lys Ser Gly Thr Ser Pro Lys Arg Trp Ile Tyr 35 40 45 Asp
Thr Ser Lys Leu Ala Ser Gly Val Pro Gly Arg Phe Ser Gly Ser 50 55
60 Gly Ser Gly Asn Ser Tyr Ser Leu Thr Ile Ser Ser Val Glu Ala Glu
65 70 75 80 Asp Asp Ala Thr Tyr Tyr Cys Gln Gln Trp Ser Lys His Pro
Leu Thr 85 90 95 Phe Gly Ser Gly Thr Lys Val Glu Ile Lys 100 105
<210> SEQ ID NO 27 <211> LENGTH: 120 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 27 Glu Val Gln Leu Val
Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg
Leu Ser Cys Ala Ala Ser Gly Phe Asn Ile Lys Asp Thr 20 25 30 Tyr
Ile His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45 Ala Arg Ile Tyr Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Asp Ser Val
50 55 60 Lys Gly Arg Phe Thr Ile Ser Ala Asp Thr Ser Lys Asn Thr
Ala Tyr 65 70 75 80 Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala
Val Tyr Tyr Cys 85 90 95 Ser Arg Trp Gly Gly Asp Gly Phe Tyr Ala
Met Asp Tyr Trp Gly Gln 100 105 110 Gly Thr Leu Val Thr Val Ser Ser
115 120 <210> SEQ ID NO 28 <211> LENGTH: 107
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 28 Asp Ile Gln Met Thr
Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly 1 5 10 15 Asp Arg Val
Thr Ile Thr Cys Arg Ala Ser Gln Asp Val Asn Thr Ala 20 25 30 Val
Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40
45 Tyr Ser Ala Ser Phe Leu Tyr Ser Gly Val Pro Ser Arg Phe Ser Gly
50 55 60 Ser Arg Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu
Gln Pro 65 70 75 80 Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln His Tyr
Thr Thr Pro Pro 85 90 95 Thr Phe Gly Gln Gly Thr Lys Val Glu Ile
Lys 100 105 <210> SEQ ID NO 29 <211> LENGTH: 123
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE: 29
Gln Ser Val Glu Glu Ser Gly Gly Arg Leu Val Thr Pro Gly Thr Pro 1 5
10 15 Leu Thr Leu Thr Cys Thr Val Ser Gly Ile Ser Leu Ser Ser Asp
Ala 20 25 30 Ile Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu
Tyr Ile Gly 35 40 45 Ile Ile Asn Gly Gly Gly Asn Thr Tyr Tyr Ala
Ser Trp Ala Lys Gly 50 55 60 Arg Phe Thr Ile Ser Lys Thr Ser Thr
Thr Val Asp Leu Lys Ile Thr 65 70 75 80 Ser Pro Thr Thr Glu Asp Thr
Ala Thr Tyr Phe Cys Ala Arg Gly Ile 85 90 95 Gln His Gly Gly Gly
Asn Ser Asp Tyr Tyr Tyr Tyr Gly Met Asp Leu 100 105 110 Trp Gly Pro
Gly Thr Leu Val Thr Val Ser Ser 115 120 <210> SEQ ID NO 30
<211> LENGTH: 112 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 30 Glu Val Leu Met Thr Gln Thr
Pro Ser Ser Val Ser Ala Ala Val Gly 1 5 10 15 Asp Thr Val Thr Ile
Lys Cys Gln Ala Ser Gln Ser Ile Ser Ser Val 20 25 30 Leu Ser Trp
Tyr Gln Gln Lys Pro Gly Gln Pro Pro Lys Leu Leu Ile 35 40 45 Tyr
Leu Ala Ser Thr Leu Ala Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55
60 Ser Arg Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser Asp Leu Glu Cys
65 70 75 80 Asp Asp Ala Ala Thr Tyr Tyr Cys Gln Thr Asn Tyr Gly Thr
Ser Ser 85 90 95 Ser Asn Tyr Gly Phe Ala Phe Gly Gly Gly Thr Glu
Val Val Val Lys 100 105 110 <210> SEQ ID NO 31 <211>
LENGTH: 126 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polypeptide
<400> SEQUENCE: 31 Glu Val Gln Leu Val Glu Ser Gly Gly Gly
Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg Leu Ser Cys Ala Ala
Ser Gly Ile Ser Leu Ser Ser Asp 20 25 30 Ala Ile Ser Trp Val Arg
Gln Ala Pro Gly Lys Gly Leu Glu Tyr Ile 35 40 45 Gly Ile Ile Asn
Gly Gly Gly Asn Thr Tyr Tyr Ala Ser Trp Ala Lys 50 55 60 Gly Arg
Phe Thr Ile Ser Arg His Asn Ser Lys Asn Thr Leu Tyr Leu 65 70 75 80
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys Ala 85
90 95 Arg Gly Ile Gln His Gly Gly Gly Asn Ser Asp Tyr Tyr Tyr Tyr
Gly 100 105 110 Met Asp Leu Trp Gly Gln Gly Thr Leu Val Thr Val Ser
Ser 115 120 125 <210> SEQ ID NO 32 <211> LENGTH: 112
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE: 32
Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly 1 5
10 15 Asp Arg Val Thr Ile Thr Cys Gln Ala Ser Gln Ser Ile Ser Ser
Val 20 25 30 Leu Ser Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys
Leu Leu Ile 35 40 45 Tyr Leu Ala Ser Thr Leu Ala Ser Gly Val Pro
Ser Arg Phe Ser Gly 50 55 60 Ser Gly Ser Gly Thr Asp Phe Thr Leu
Thr Ile Ser Ser Leu Gln Cys 65 70 75 80 Glu Asp Ile Ala Thr Tyr Tyr
Cys Gln Thr Asn Tyr Gly Thr Ser Ser 85 90 95 Ser Asn Tyr Gly Phe
Ala Phe Gly Gly Gly Thr Lys Val Glu Ile Lys 100 105 110 <210>
SEQ ID NO 33 <211> LENGTH: 118 <212> TYPE: PRT
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 33 Gln Ser Val Glu Glu
Ser Gly Gly Arg Leu Val Thr Pro Gly Thr Pro 1 5 10 15 Leu Thr Leu
Thr Cys Thr Val Ser Gly Phe Ser Leu Asn Asn Tyr Ala 20 25 30 Met
Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Ile Gly 35 40
45 Ser Ile Ser Thr Gly Gly Leu Ala Phe Tyr Ala Asn Trp Ala Lys Gly
50 55 60 Arg Phe Thr Ile Ser Arg Thr Ser Thr Thr Val Asp Leu Lys
Met Thr 65 70 75 80 Ser Leu Thr Thr Glu Asp Thr Ala Thr Tyr Phe Cys
Gly Arg Asn Gly 85 90 95 Gly Gly Ser Tyr Ile Phe Tyr Tyr Phe Asp
Leu Trp Gly Gln Gly Thr 100 105 110 Leu Val Thr Val Ser Ser 115
<210> SEQ ID NO 34 <211> LENGTH: 108 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 34 Ala Phe Glu Leu Thr
Gln Thr Pro Ser Ser Val Glu Ala Ala Val Gly 1 5 10 15 Gly Thr Ile
Thr Ile Lys Cys Gln Ala Ser Gln Ser Ile Ser Ser Tyr 20 25 30 Leu
Ser Trp Tyr Gln Gln Lys Pro Gly Gln Pro Pro Lys Leu Leu Ile 35 40
45 Tyr Ser Ala Ser Thr Leu Ala Ser Gly Val Ser Ser Arg Phe Lys Gly
50 55 60 Ser Gly Ser Gly Thr Glu Tyr Thr Leu Thr Ile Ser Asp Leu
Glu Cys 65 70 75 80 Ala Asp Ala Ala Thr Tyr Phe Cys Gln Ser Tyr Tyr
Asp Ile Gly Thr 85 90 95 Ser Thr Phe Gly Gly Gly Thr Glu Val Val
Val Lys 100 105 <210> SEQ ID NO 35 <211> LENGTH: 121
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE: 35
Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5
10 15
Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Ser Leu Asn Asn Tyr 20
25 30 Ala Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Ile 35 40 45 Gly Ser Ile Ser Thr Gly Gly Leu Ala Phe Tyr Ala Asn
Trp Ala Lys 50 55 60 Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys
Asn Thr Leu Tyr Leu 65 70 75 80 Gln Met Asn Ser Leu Arg Ala Glu Asp
Thr Ala Val Tyr Tyr Cys Ala 85 90 95 Arg Asn Gly Gly Gly Ser Tyr
Ile Phe Tyr Tyr Phe Asp Leu Trp Gly 100 105 110 Gln Gly Thr Leu Val
Thr Val Ser Ser 115 120 <210> SEQ ID NO 36 <211>
LENGTH: 108 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polypeptide
<400> SEQUENCE: 36 Asp Ile Gln Met Thr Gln Ser Pro Ser Ser
Leu Ser Ala Ser Val Gly 1 5 10 15 Asp Arg Val Thr Ile Thr Cys Gln
Ala Ser Gln Ser Ile Ser Ser Tyr 20 25 30 Leu Ser Trp Tyr Gln Gln
Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45 Tyr Ser Ala Ser
Thr Leu Ala Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60 Ser Gly
Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Cys 65 70 75 80
Glu Asp Ala Ala Thr Tyr Tyr Cys Gln Ser Tyr Tyr Asp Ile Gly Thr 85
90 95 Ser Thr Phe Gly Gly Gly Thr Lys Val Glu Ile Lys 100 105
<210> SEQ ID NO 37 <211> LENGTH: 114 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 37 Gln Ser Val Lys Glu
Ser Gly Gly Arg Leu Val Thr Pro Gly Thr Pro 1 5 10 15 Leu Thr Leu
Thr Cys Thr Val Ser Gly Ile Asp Leu Ser Ser Tyr Ala 20 25 30 Met
Gly Trp Phe Arg Gln Ala Pro Gly Lys Gly Leu Glu Tyr Ile Gly 35 40
45 Thr Ile Asn Ile Gly Gly Arg Val Tyr Tyr Ala Ser Trp Ala Lys Gly
50 55 60 Arg Phe Thr Ile Ser Arg Thr Ser Thr Thr Val Asp Leu Lys
Ala Pro 65 70 75 80 Ser Leu Thr Ala Glu Asp Thr Ala Thr Tyr Phe Cys
Ala Arg Tyr Tyr 85 90 95 Asn Gly Gly Ser Tyr Asp Ile Trp Gly Pro
Gly Thr Leu Val Thr Val 100 105 110 Ser Leu <210> SEQ ID NO
38 <211> LENGTH: 110 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 38 Asp Val Val Met Thr Gln Thr
Pro Ala Ser Ala Ser Glu Pro Val Gly 1 5 10 15 Gly Thr Val Thr Ile
Lys Cys Gln Ala Ser Glu Ser Ile Tyr Arg Val 20 25 30 Leu Ala Trp
Tyr Gln Gln Lys Pro Gly Gln Pro Pro Lys Leu Leu Ile 35 40 45 Tyr
Asp Thr Ser Thr Leu Ala Ser Gly Ala Pro Ser Arg Phe Lys Gly 50 55
60 Ser Gly Tyr Gly Thr Glu Phe Thr Leu Thr Ile Ser Gly Val Gln Cys
65 70 75 80 Glu Asp Ala Ala Thr Tyr Tyr Cys Gln Gly Gly Tyr Tyr Ala
Asp Ser 85 90 95 Tyr Gly Ile Ala Phe Gly Gly Gly Thr Glu Val Val
Val Lys 100 105 110 <210> SEQ ID NO 39 <211> LENGTH:
117 <212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE: 39
Gln Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5
10 15 Ser Leu Arg Leu Ser Cys Ser Ala Ser Gly Ile Asp Leu Ser Ser
Tyr 20 25 30 Ala Met Gly Trp Val Arg Gln Ala Pro Gly Lys Gly Leu
Glu Tyr Ile 35 40 45 Gly Thr Ile Asn Ile Gly Gly Arg Val Tyr Tyr
Ala Ser Trp Ala Lys 50 55 60 Gly Arg Phe Thr Ile Ser Arg Asp Asn
Ser Lys Asn Thr Leu Tyr Leu 65 70 75 80 Gln Met Asn Ser Leu Arg Ala
Glu Asp Thr Ala Val Tyr Tyr Cys Ala 85 90 95 Arg Tyr Tyr Asn Gly
Gly Ser Tyr Asp Ile Trp Gly Gln Gly Thr Leu 100 105 110 Val Thr Val
Ser Ser 115 <210> SEQ ID NO 40 <211> LENGTH: 110
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE: 40
Asp Ile Gln Met Thr Gln Ser Pro Ser Thr Leu Ser Ala Ser Val Gly 1 5
10 15 Asp Arg Val Thr Ile Thr Cys Gln Ala Ser Glu Ser Ile Tyr Arg
Val 20 25 30 Leu Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys
Leu Leu Ile 35 40 45 Tyr Asp Thr Ser Thr Leu Ala Ser Gly Val Pro
Ser Arg Phe Ser Gly 50 55 60 Ser Gly Ser Gly Thr Glu Phe Thr Leu
Thr Ile Ser Ser Leu Gln Cys 65 70 75 80 Asp Asp Ala Ala Thr Tyr Tyr
Cys Gln Gly Gly Tyr Tyr Ala Asp Ser 85 90 95 Tyr Gly Ile Ala Phe
Gly Gly Gly Thr Lys Val Glu Ile Lys 100 105 110 <210> SEQ ID
NO 41 <211> LENGTH: 122 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 41 Gln Ser Val Glu Glu Ser Gly
Gly Arg Leu Val Lys Pro Asp Glu Ser 1 5 10 15 Leu Thr Leu Thr Cys
Thr Ala Ser Gly Phe Ser Leu Ser Ser Tyr Ala 20 25 30 Met Ile Trp
Val Arg Gln Ala Pro Gly Glu Gly Leu Glu Trp Ile Gly 35 40 45 Thr
Ile Ser Thr Gly Gly Ile Thr Tyr Tyr Ala Ser Trp Ala Lys Gly 50 55
60 Arg Phe Thr Ile Ser Lys Thr Ser Thr Thr Val Asp Leu Lys Ile Thr
65 70 75 80 Ser Pro Thr Thr Glu Asp Thr Ala Thr Tyr Phe Cys Ala Arg
Gly Gly 85 90 95 Tyr Ala Ala Ser Ser Ala Tyr Tyr Leu Pro Tyr Tyr
Phe Asp Leu Trp 100 105 110 Gly Gln Gly Thr Leu Val Thr Val Ser Ser
115 120 <210> SEQ ID NO 42 <211> LENGTH: 112
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE: 42
Ala Ala Val Leu Thr Gln Thr Pro Ser Pro Val Ser Ala Ala Val Gly 1 5
10 15 Gly Thr Val Thr Ile Ser Cys Gln Ser Ser Gln Ser Val Tyr Asn
Asn 20 25 30 Asn Asn Leu Ala Trp Phe Gln Gln Lys Pro Gly Gln Pro
Pro Lys Leu 35 40 45 Leu Ile Tyr Leu Ala Ser Thr Leu Ala Ser Gly
Val Pro Ser Arg Phe 50 55 60
Ser Gly Ser Gly Ser Gly Thr Gln Phe Thr Leu Thr Ile Ser Gly Val 65
70 75 80 Gln Cys Asp Asp Ala Ala Thr Tyr Tyr Cys Leu Gly Gly Cys
Asp Asp 85 90 95 Asp Ala Asp Thr Phe Ala Phe Gly Gly Gly Thr Glu
Val Val Val Lys 100 105 110 <210> SEQ ID NO 43 <211>
LENGTH: 125 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polypeptide
<400> SEQUENCE: 43 Glu Val Gln Leu Val Glu Ser Gly Gly Gly
Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg Leu Ser Cys Ala Ala
Ser Gly Phe Ser Leu Ser Ser Tyr 20 25 30 Ala Met Ile Trp Val Arg
Gln Ala Pro Gly Lys Gly Leu Glu Trp Ile 35 40 45 Gly Thr Ile Ser
Thr Gly Gly Ile Thr Tyr Tyr Ala Ser Trp Ala Lys 50 55 60 Gly Arg
Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr Leu 65 70 75 80
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys Ala 85
90 95 Arg Gly Gly Tyr Ala Ala Ser Ser Ala Tyr Tyr Leu Pro Tyr Tyr
Phe 100 105 110 Asp Leu Trp Gly Gln Gly Thr Leu Val Thr Val Ser Ser
115 120 125 <210> SEQ ID NO 44 <211> LENGTH: 112
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE: 44
Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly 1 5
10 15 Asp Arg Val Thr Ile Thr Cys Gln Ser Ser Gln Ser Val Tyr Asn
Asn 20 25 30 Asn Asn Leu Ala Trp Tyr Gln Gln Lys Pro Gly Lys Val
Pro Lys Leu 35 40 45 Leu Ile Tyr Leu Ala Ser Thr Leu Ala Ser Gly
Val Pro Ser Arg Phe 50 55 60 Ser Gly Ser Gly Ser Gly Thr Asp Phe
Thr Leu Thr Ile Ser Ser Leu 65 70 75 80 Gln Cys Glu Asp Ala Ala Thr
Tyr Tyr Cys Leu Gly Gly Cys Asp Asp 85 90 95 Asp Ala Asp Thr Phe
Ala Phe Gly Gly Gly Thr Lys Val Glu Ile Lys 100 105 110 <210>
SEQ ID NO 45 <211> LENGTH: 357 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 45 gaggtccaac
tggtggagag cggtggaggt gttgtgcaac ctggccggtc cctgcgcctg 60
tcctgctccg catctggctt caccttcagc ggctatgggt tgtcttgggt gagacaggca
120 cctggaaaag gtcttgagtg ggttgcaatg attagtagtg gtggtagtta
tacctactat 180 gcagacagtg tgaagggtag atttgcaata tcgcgagaca
acgccaagaa cacattgttc 240 ctgcaaatgg acagcctgag acccgaagac
accggggtct atttttgtgc aagacatggg 300 gacgatcccg cctggttcgc
ttattggggc caagggaccc cggtcaccgt ctcctca 357 <210> SEQ ID NO
46 <211> LENGTH: 330 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 46 gacatccagc tgacccagag
cccaagcagc ctgagcgcca gcgtgggtga cagagtgacc 60 atcacctgta
gtgtcagctc aagtataagt tccaacaact tgcactggta ccagcagaag 120
ccaggtaagg ctccaaagcc atggatctac ggcacatcca acctggcttc tggtgtgcca
180 agcagattca gcggtagcgg tagcggtacc gactacacct tcaccatcag
cagcctccag 240 ccagaggaca tcgccaccta ctactgccaa cagtggagta
gttacccgta catgtacacg 300 ttcggccaag ggaccaaggt ggaaatcaaa 330
<210> SEQ ID NO 47 <211> LENGTH: 357 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 47 caggtacaac
tgcagcagtc tgggcctgag ctggagaagc ctggcgcttc agtgaagata 60
tcctgcaagg cttctggtta ctcattcact ggctacacca tgaactgggt gaagcagagc
120 catggaaaga gccttgagtg gattggactt attactcctt acaatggtgc
ttctagctac 180 aaccagaagt tcaggggcaa ggccacatta actgtagaca
agtcatccag cacagcctac 240 atggacctcc tcagtctgac atctgaagac
tctgcagtct atttctgtgc aagggggggt 300 tacgacggga ggggttttga
ctactgggga tccgggaccc cggtcaccgt ctcctca 357 <210> SEQ ID NO
48 <211> LENGTH: 318 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 48 gacatcgagc tcactcagtc
tccagcaatc atgtctgcat ctccagggga gaaggtcacc 60 atgacctgca
gtgccagctc aagtgtaagt tacatgcact ggtaccagca gaagtcaggc 120
acctccccca aaagatggat ttatgacaca tccaaactgg cttctggagt cccaggtcgc
180 ttcagtggca gtgggtctgg aaactcttac tctctcacaa tcagcagcgt
ggaggctgaa 240 gatgatgcaa cttattactg ccagcagtgg agtaagcacc
ctctcacgtt cggatccggg 300 accaaggtgg aaatcaaa 318 <210> SEQ
ID NO 49 <211> LENGTH: 369 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 49 cagtcggtgg aggagtccgg
gggtcgcctg gtcacgcctg ggacacccct gacactcacc 60 tgcaccgtct
ctggaatctc cctcagtagc gatgcaataa gctgggtccg ccaggctcca 120
gggaaggggc tcgaatacat cggaatcatt aatggtggtg gtaacacata ctacgcgagc
180 tgggcgaaag gccgattcac catctccaaa acctcgacca cggtggatct
gaaaatcacc 240 agtccgacaa ccgaggacac ggccacctat ttctgtgcca
gaggcattca acatggtggt 300 ggtaatagtg attattatta ttacggcatg
gacctctggg gcccaggcac cctggtcact 360 gtctcttca 369 <210> SEQ
ID NO 50 <211> LENGTH: 336 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 50 gaagtgttga tgacccagac
tccatcctcc gtgtctgcag ctgtgggaga cacagtcacc 60 atcaagtgcc
aggccagtca gagcattagt agtgtcttgt cctggtatca gcagaaacca 120
gggcagcctc ccaagctcct gatctatctg gcatccactc tggcatctgg ggtcccatcg
180 cggttcagcg gcagtagatc tgggacagag ttcactctca ccatcagcga
cctggagtgt 240 gacgatgctg ccacttacta ctgtcaaacc aattatggta
ctagtagtag taattatggt 300 tttgctttcg gcggagggac cgaggtggtc gtcaaa
336 <210> SEQ ID NO 51 <211> LENGTH: 378 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polynucleotide <400> SEQUENCE: 51
gaagtccaac tggtggaaag cgggggagga ctggtgcagc cgggcggatc cctccggctg
60 tcatgtgctg catcgggaat ttccctctcc tccgacgcga ttagctgggt
cagacaggcc 120 cccggaaagg ggctggagta catcggtatc atcaacggcg
gcggaaacac ctactacgcc 180 tcctgggcca agggccgctt caccatctcg
cggcataatt ccaagaacac tctgtacttg 240 caaatgaact ccctgagggc
cgaggacacc gccgtgtact actgcgcgcg cggcatccag 300 cacggtggtg
gaaacagcga ctactactac tatgggatgg atctgtgggg ccagggaact 360
cttgtgaccg tgtcgtca 378
<210> SEQ ID NO 52 <211> LENGTH: 336 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 52 gacattcaga
tgacccagtc cccaagctcg ctgtccgcct ccgtgggcga ccgcgtgacc 60
atcacgtgcc aggcgtccca gtcaattagc agcgtgctct cctggtacca acagaagccg
120 gggaaagcac ccaagctgct gatctacttg gcctccactc tggcctcggg
agtgccttca 180 cggttctccg gatcgggatc tggtactgat ttcaccctca
ccatctcgag ccttcagtgc 240 gaggacatcg ctacttacta ttgtcaaacc
aactacggaa cctccagctc caactacggc 300 tttgccttcg gtggcgggac
caaggtcgaa atcaaa 336 <210> SEQ ID NO 53 <211> LENGTH:
354 <212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polynucleotide <400> SEQUENCE:
53 cagtcggtgg aggagtccgg gggtcgcctg gtcacgcctg ggacacccct
gacactcacc 60 tgcacagtct ctggattctc cctcaataac tatgcaatga
gctgggtccg ccaggctcca 120 gggaaggggc tggaatggat cggatccatt
agtactggtg gtctcgcatt ctacgcgaac 180 tgggcaaaag gccgattcac
catctccaga acctcgacca cggtggatct gaaaatgacc 240 agtctgacaa
ccgaggacac ggccacctat ttctgtggca gaaatggtgg tggtagttat 300
attttctatt attttgactt gtggggccaa ggcaccctcg tcactgtctc ttca 354
<210> SEQ ID NO 54 <211> LENGTH: 324 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 54 gcattcgaat
tgacccagac tccatcctcc gtggaggcag ctgtgggagg cacaatcacc 60
atcaagtgcc aggccagtca gagcattagt agttacttat cctggtatca gcagaaacca
120 gggcagcctc ccaagctcct gatctattct gcatccactc tggcatctgg
ggtctcatcg 180 cggttcaaag gcagtggatc tgggacagag tacactctca
ccatcagcga cctggagtgt 240 gccgatgctg ccacttactt ctgtcaaagc
tattatgata ttggtactag tactttcggc 300 ggagggaccg aggtggtcgt caaa 324
<210> SEQ ID NO 55 <211> LENGTH: 363 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 55 gaagtgcagc
tggtggaatc tggcggcgga ctggtgcagc ctggcggatc tctgagactg 60
tcttgtgccg cctccggctt ctccctgaac aactacgcca tgtcctgggt gcgacaggcc
120 cctggcaaag gcctggaatg gatcggctcc atcagcacag gcggcctggc
cttctacgcc 180 aattgggcca agggccggtt caccatcagc cgggacaact
ccaagaacac cctgtacctc 240 cagatgaact ccctgcgggc cgaggacacc
gccgtgtact actgtgccag aaacggcgga 300 ggctcctaca tcttctacta
cttcgacctg tggggccagg gcaccctcgt gacagtgtca 360 tct 363 <210>
SEQ ID NO 56 <211> LENGTH: 324 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 56 gatattcaga
tgacccagtc cccctccagc ctgtccgctt ctgtgggcga cagagtgacc 60
atcacctgtc aggcctccca gtccatctcc tcctacctgt cctggtatca gcagaagccc
120 ggcaaggccc ccaagctgct gatctactct gcctccacac tggcctccgg
cgtgccctct 180 agattctccg gctctggctc tggcaccgac tttaccctga
ccatcagctc cctccagtgc 240 gaggatgccg ccacctacta ctgccagtcc
tactacgaca tcggcacctc caccttcggc 300 ggaggcacca aggtggaaat caaa 324
<210> SEQ ID NO 57 <211> LENGTH: 342 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 57 cagtcagtga
aggagtccgg gggtcgcctg gtcacgcctg ggacacccct gacactcacc 60
tgcacagtct ctggaatcga cctcagtagc tatgcaatgg gctggttccg ccaggctcca
120 gggaaggggc tggaatacat cggaaccatt aatattggtg gtcgcgtata
ttacgcgagc 180 tgggcaaaag gccgattcac catctccaga acctcgacca
cggtggatct gaaagcgccc 240 agtctgacag ccgaggacac ggccacctat
ttctgtgcca gatattataa tggtggtagt 300 tatgacatct ggggcccagg
caccctggtc accgtctctt ta 342 <210> SEQ ID NO 58 <211>
LENGTH: 330 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polynucleotide
<400> SEQUENCE: 58 gatgttgtga tgacccagac tccagcctcc
gcgtctgaac ctgtgggagg cacagtcacc 60 atcaagtgcc aggccagtga
gagcatttat cgcgtattgg cctggtatca gcagaaacca 120 gggcagcctc
ccaagctcct gatctatgat acatccactc tggcatctgg ggccccatcg 180
cggttcaaag gcagtggata tgggacagag ttcactctca ccatcagcgg cgtgcagtgt
240 gaagatgctg ccacttacta ctgtcaaggc ggttattatg ctgatagtta
tggtattgct 300 ttcggcggag ggaccgaggt ggtggtcaaa 330 <210> SEQ
ID NO 59 <211> LENGTH: 351 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 59 caggtgcagc tggtggaatc
tggcggagga ctggtgcagc ctggcggctc tctgagactg 60 tcctgttccg
cctccggaat cgacctgtcc tcctacgcta tgggctgggt gcgacaggct 120
cctggcaagg gcctggagta catcggcacc atcaacatcg gcggcagagt gtactacgcc
180 tcctgggcca agggccggtt caccatctcc agagacaact ccaagaacac
cctgtacctc 240 cagatgaact ccctgcgggc cgaggacacc gccgtgtact
actgcgcccg gtactacaac 300 ggcggctcct acgatatctg gggccagggc
acactcgtga ccgtgtcctc t 351 <210> SEQ ID NO 60 <211>
LENGTH: 330 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polynucleotide
<400> SEQUENCE: 60 gatatccaga tgacccagtc cccctccacc
ctgtctgcct ctgtgggcga cagagtgacc 60 atcacctgtc aggcctccga
gtccatctac cgggtgctgg cctggtatca gcagaagcct 120 ggcaaggccc
ccaagctgct gatctacgac accagcacac tggcctccgg cgtgccctct 180
agattctccg gctctggctc tggcaccgag tttaccctga ccatctccag cctccagtgc
240 gacgacgccg ccacctacta ttgtcagggc ggctactacg ccgactccta
cggaatcgct 300 ttcggcggag gcaccaaggt ggaaatcaaa 330 <210> SEQ
ID NO 61 <211> LENGTH: 366 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 61 cagtcggtgg aggagtccgg
cggtcgcctg gtaaagcctg acgaatccct gacactcacc 60 tgcacagcct
ctggattctc cctcagtagt tatgcaatga tctgggtccg ccaggctcca 120
ggggaggggc tggaatggat cggaaccatt agtactggtg gtatcacata ctacgcgagc
180 tgggcgaaag gccgattcac catctccaaa acctcgacca cggtggatct
gaaaatcacc 240 agtccgacaa ccgaggacac ggccacctat ttctgtgcca
gagggggata tgctgctagt 300 agtgcttatt atctcccgta ctactttgac
ttgtggggcc aagggaccct ggtcaccgtc 360 tcctca 366 <210> SEQ ID
NO 62 <211> LENGTH: 336 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence:
Synthetic
polynucleotide <400> SEQUENCE: 62 gcagccgtgc tgacccagac
accatcaccc gtgtctgcag ctgtgggagg cacagtcacc 60 atcagttgcc
agtccagtca gagtgtttat aataataaca acttagcctg gtttcagcag 120
aaacccgggc agcctcccaa gcttctgatc tatctggcat ccactctggc atctggggtc
180 ccatcacggt tcagcggcag tggatctggg acacagttca ctctcaccat
cagcggcgtg 240 cagtgtgacg atgctgccac ttattactgt ctaggtggtt
gtgatgatga tgctgatact 300 tttgctttcg gcggagggac tgaggtggtg gtcaaa
336 <210> SEQ ID NO 63 <211> LENGTH: 375 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polynucleotide <400> SEQUENCE: 63
gaagtgcagc tggtggaatc tggcggcgga ctggtgcagc ctggcggatc tctgagactg
60 tcttgtgccg cctccggctt ctccctgtcc tcctacgcta tgatctgggt
gcgacaggcc 120 cctggcaagg gcctggaatg gatcggcacc atctctaccg
gcggaattac ctactacgcc 180 tcctgggcca agggccggtt caccatctcc
agagacaact ccaagaacac cctgtacctc 240 cagatgaact ccctgcgggc
cgaggacacc gccgtgtact attgtgctag aggcggctac 300 gccgccagct
ccgcttacta cctgccctac tacttcgacc tgtggggcca gggcaccctc 360
gtgacagtgt catct 375 <210> SEQ ID NO 64 <211> LENGTH:
336 <212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polynucleotide <400> SEQUENCE:
64 gatattcaga tgacccagtc cccctccagc ctgtccgctt ctgtgggcga
cagagtgacc 60 atcacctgtc agtcctccca gtccgtgtat aacaacaaca
acctggcctg gtatcagcag 120 aaacccggca aggtgcccaa gctgctgatc
tacctggcct ccacactggc ctctggcgtg 180 ccctctagat tctccggctc
tggctctggc accgacttta ccctgaccat cagctccctc 240 cagtgcgagg
atgccgccac ctactattgc ctgggcggct gcgacgacga cgccgatacc 300
tttgcttttg gcggaggcac caaggtggaa atcaaa 336 <210> SEQ ID NO
65 <211> LENGTH: 5 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 65 Gly Tyr Thr Met Asn 1 5
<210> SEQ ID NO 66 <211> LENGTH: 17 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 66 Leu Ile Thr Pro Tyr Asn
Gly Ala Ser Ser Tyr Asn Gln Lys Phe Arg 1 5 10 15 Gly <210>
SEQ ID NO 67 <211> LENGTH: 10 <212> TYPE: PRT
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 67 Gly Gly Tyr Asp Gly Arg
Gly Phe Asp Tyr 1 5 10 <210> SEQ ID NO 68 <211> LENGTH:
10 <212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 68 Ser
Ala Ser Ser Ser Val Ser Tyr Met His 1 5 10 <210> SEQ ID NO 69
<211> LENGTH: 7 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 69 Asp Thr Ser Lys Leu Ala Ser 1 5
<210> SEQ ID NO 70 <211> LENGTH: 9 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 70 Gln Gln Trp Ser Lys His
Pro Leu Thr 1 5 <210> SEQ ID NO 71 <211> LENGTH: 5
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 71 Asp
Thr Tyr Ile His 1 5 <210> SEQ ID NO 72 <211> LENGTH: 17
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 72 Arg
Ile Tyr Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Asp Ser Val Lys 1 5 10
15 Gly <210> SEQ ID NO 73 <211> LENGTH: 11 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic peptide <400> SEQUENCE: 73 Trp Gly Gly
Asp Gly Phe Tyr Ala Met Asp Tyr 1 5 10 <210> SEQ ID NO 74
<211> LENGTH: 11 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 74 Arg Ala Ser Gln Asp Val Asn Thr Ala Val
Ala 1 5 10 <210> SEQ ID NO 75 <211> LENGTH: 7
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 75 Ser
Ala Ser Phe Leu Tyr Ser 1 5 <210> SEQ ID NO 76 <211>
LENGTH: 9 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 76 Gln Gln His Tyr Thr Thr Pro Pro Thr 1 5
<210> SEQ ID NO 77 <211> LENGTH: 5 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 77 Ser Asp Ala Ile Ser 1 5
<210> SEQ ID NO 78 <211> LENGTH: 16 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 78 Ile Ile Asn Gly Gly Gly
Asn Thr Tyr Tyr Ala Ser Trp Ala Lys Gly 1 5 10 15 <210> SEQ
ID NO 79 <211> LENGTH: 18 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 79 Gly Ile Gln His Gly Gly Gly Asn
Ser Asp Tyr Tyr Tyr Tyr Gly Met 1 5 10 15 Asp Leu <210> SEQ
ID NO 80 <211> LENGTH: 11 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 80 Gln Ala Ser Gln Ser Ile Ser Ser
Val Leu Ser 1 5 10 <210> SEQ ID NO 81 <211> LENGTH: 7
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 81 Leu
Ala Ser Thr Leu Ala Ser 1 5 <210> SEQ ID NO 82 <211>
LENGTH: 14 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 82 Gln Thr Asn Tyr Gly Thr Ser Ser Ser Asn Tyr Gly Phe
Ala 1 5 10 <210> SEQ ID NO 83 <211> LENGTH: 5
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 83 Ser
Asp Ala Ile Ser 1 5 <210> SEQ ID NO 84 <211> LENGTH: 16
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 84 Ile
Ile Asn Gly Gly Gly Asn Thr Tyr Tyr Ala Ser Trp Ala Lys Gly 1 5 10
15 <210> SEQ ID NO 85 <211> LENGTH: 18 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic peptide <400> SEQUENCE: 85 Gly Ile Gln
His Gly Gly Gly Asn Ser Asp Tyr Tyr Tyr Tyr Gly Met 1 5 10 15 Asp
Leu <210> SEQ ID NO 86 <211> LENGTH: 11 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic peptide <400> SEQUENCE: 86 Gln Ala Ser
Gln Ser Ile Ser Ser Val Leu Ser 1 5 10 <210> SEQ ID NO 87
<211> LENGTH: 7 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 87 Leu Ala Ser Thr Leu Ala Ser 1 5
<210> SEQ ID NO 88 <211> LENGTH: 14 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 88 Gln Thr Asn Tyr Gly Thr
Ser Ser Ser Asn Tyr Gly Phe Ala 1 5 10 <210> SEQ ID NO 89
<211> LENGTH: 5 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 89 Asn Tyr Ala Met Ser 1 5 <210> SEQ ID
NO 90 <211> LENGTH: 16 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 90 Ser Ile Ser Thr Gly Gly Leu Ala
Phe Tyr Ala Asn Trp Ala Lys Gly 1 5 10 15 <210> SEQ ID NO 91
<211> LENGTH: 13 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 91 Asn Gly Gly Gly Ser Tyr Ile Phe Tyr Tyr
Phe Asp Leu 1 5 10 <210> SEQ ID NO 92 <211> LENGTH: 11
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 92 Gln
Ala Ser Gln Ser Ile Ser Ser Tyr Leu Ser 1 5 10 <210> SEQ ID
NO 93 <211> LENGTH: 7 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence:
Synthetic
peptide <400> SEQUENCE: 93 Ser Ala Ser Thr Leu Ala Ser 1 5
<210> SEQ ID NO 94 <211> LENGTH: 10 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 94 Gln Ser Tyr Tyr Asp Ile
Gly Thr Ser Thr 1 5 10 <210> SEQ ID NO 95 <211> LENGTH:
5 <212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 95 Asn
Tyr Ala Met Ser 1 5 <210> SEQ ID NO 96 <211> LENGTH: 16
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 96 Ser
Ile Ser Thr Gly Gly Leu Ala Phe Tyr Ala Asn Trp Ala Lys Gly 1 5 10
15 <210> SEQ ID NO 97 <211> LENGTH: 13 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic peptide <400> SEQUENCE: 97 Asn Gly Gly
Gly Ser Tyr Ile Phe Tyr Tyr Phe Asp Leu 1 5 10 <210> SEQ ID
NO 98 <211> LENGTH: 11 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 98 Gln Ala Ser Gln Ser Ile Ser Ser
Tyr Leu Ser 1 5 10 <210> SEQ ID NO 99 <211> LENGTH: 7
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 99 Ser
Ala Ser Thr Leu Ala Ser 1 5 <210> SEQ ID NO 100 <211>
LENGTH: 10 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 100 Gln Ser Tyr Tyr Asp Ile Gly Thr Ser Thr 1 5 10
<210> SEQ ID NO 101 <211> LENGTH: 5 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 101 Ser Tyr Ala Met Gly 1 5
<210> SEQ ID NO 102 <211> LENGTH: 16 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 102 Thr Ile Asn Ile Gly Gly
Arg Val Tyr Tyr Ala Ser Trp Ala Lys Gly 1 5 10 15 <210> SEQ
ID NO 103 <211> LENGTH: 9 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 103 Tyr Tyr Asn Gly Gly Ser Tyr Asp
Ile 1 5 <210> SEQ ID NO 104 <211> LENGTH: 11
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 104
Gln Ala Ser Glu Ser Ile Tyr Arg Val Leu Ala 1 5 10 <210> SEQ
ID NO 105 <211> LENGTH: 7 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 105 Asp Thr Ser Thr Leu Ala Ser 1 5
<210> SEQ ID NO 106 <211> LENGTH: 12 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 106 Gln Gly Gly Tyr Tyr Ala
Asp Ser Tyr Gly Ile Ala 1 5 10 <210> SEQ ID NO 107
<211> LENGTH: 5 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 107 Ser Tyr Ala Met Gly 1 5 <210> SEQ
ID NO 108 <211> LENGTH: 16 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 108 Thr Ile Asn Ile Gly Gly Arg Val
Tyr Tyr Ala Ser Trp Ala Lys Gly 1 5 10 15 <210> SEQ ID NO 109
<211> LENGTH: 9 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 109 Tyr Tyr Asn Gly Gly Ser Tyr Asp Ile 1 5
<210> SEQ ID NO 110 <211> LENGTH: 11 <212> TYPE:
PRT
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 110 Gln Ala Ser Glu Ser Ile
Tyr Arg Val Leu Ala 1 5 10 <210> SEQ ID NO 111 <211>
LENGTH: 7 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 111 Asp Thr Ser Thr Leu Ala Ser 1 5 <210> SEQ ID NO
112 <211> LENGTH: 12 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 112 Gln Gly Gly Tyr Tyr Ala Asp Ser
Tyr Gly Ile Ala 1 5 10 <210> SEQ ID NO 113 <211>
LENGTH: 5 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 113 Ser Tyr Ala Met Ile 1 5 <210> SEQ ID NO 114
<211> LENGTH: 16 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 114 Thr Ile Ser Thr Gly Gly Ile Thr Tyr Tyr
Ala Ser Trp Ala Lys Gly 1 5 10 15 <210> SEQ ID NO 115
<211> LENGTH: 17 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 115 Gly Gly Tyr Ala Ala Ser Ser Ala Tyr Tyr
Leu Pro Tyr Tyr Phe Asp 1 5 10 15 Leu <210> SEQ ID NO 116
<211> LENGTH: 13 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 116 Gln Ser Ser Gln Ser Val Tyr Asn Asn Asn
Asn Leu Ala 1 5 10 <210> SEQ ID NO 117 <211> LENGTH: 7
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 117
Leu Ala Ser Thr Leu Ala Ser 1 5 <210> SEQ ID NO 118
<211> LENGTH: 12 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 118 Leu Gly Gly Cys Asp Asp Asp Ala Asp Thr
Phe Ala 1 5 10 <210> SEQ ID NO 119 <211> LENGTH: 5
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 119
Ser Tyr Ala Met Ile 1 5 <210> SEQ ID NO 120 <211>
LENGTH: 16 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 120 Thr Ile Ser Thr Gly Gly Ile Thr Tyr Tyr Ala Ser Trp
Ala Lys Gly 1 5 10 15 <210> SEQ ID NO 121 <211> LENGTH:
17 <212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 121
Gly Gly Tyr Ala Ala Ser Ser Ala Tyr Tyr Leu Pro Tyr Tyr Phe Asp 1 5
10 15 Leu <210> SEQ ID NO 122 <211> LENGTH: 13
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 122
Gln Ser Ser Gln Ser Val Tyr Asn Asn Asn Asn Leu Ala 1 5 10
<210> SEQ ID NO 123 <211> LENGTH: 7 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 123 Leu Ala Ser Thr Leu Ala
Ser 1 5 <210> SEQ ID NO 124 <211> LENGTH: 12
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 124
Leu Gly Gly Cys Asp Asp Asp Ala Asp Thr Phe Ala 1 5 10 <210>
SEQ ID NO 125 <211> LENGTH: 15 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 125 ggctatgggt
tgtct 15 <210> SEQ ID NO 126 <211> LENGTH: 51
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 126
atgattagta gtggtggtag ttatacctac tatgcagaca gtgtgaaggg t 51
<210> SEQ ID NO 127 <211> LENGTH: 30 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 127 catggggacg
atcccgcctg gttcgcttat 30 <210> SEQ ID NO 128 <211>
LENGTH: 36 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 128 agtgtcagct caagtataag ttccaacaac ttgcac
36 <210> SEQ ID NO 129 <211> LENGTH: 21 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic oligonucleotide <400> SEQUENCE: 129
ggcacatcca acctggcttc t 21 <210> SEQ ID NO 130 <211>
LENGTH: 33 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 130 caacagtgga gtagttaccc gtacatgtac acg 33
<210> SEQ ID NO 131 <211> LENGTH: 15 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 131 ggctacacca
tgaac 15 <210> SEQ ID NO 132 <211> LENGTH: 51
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 132 cttattactc cttacaatgg tgcttctagc tacaaccaga
agttcagggg c 51 <210> SEQ ID NO 133 <211> LENGTH: 30
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 133 gggggttacg acgggagggg ttttgactac 30 <210> SEQ
ID NO 134 <211> LENGTH: 30 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 134 agtgccagct caagtgtaag
ttacatgcac 30 <210> SEQ ID NO 135 <211> LENGTH: 21
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 135 gacacatcca aactggcttc t 21 <210> SEQ ID NO 136
<211> LENGTH: 27 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 136 cagcagtgga gtaagcaccc
tctcacg 27 <210> SEQ ID NO 137 <211> LENGTH: 15
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 137 agcgatgcaa taagc 15 <210> SEQ ID NO 138
<211> LENGTH: 48 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 138 atcattaatg gtggtggtaa
cacatactac gcgagctggg cgaaaggc 48 <210> SEQ ID NO 139
<211> LENGTH: 54 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 139 ggcattcaac atggtggtgg
taatagtgat tattattatt acggcatgga cctc 54 <210> SEQ ID NO 140
<211> LENGTH: 33 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 140 caggccagtc agagcattag
tagtgtcttg tcc 33 <210> SEQ ID NO 141 <211> LENGTH: 21
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 141 ctggcatcca ctctggcatc t 21 <210> SEQ ID NO 142
<211> LENGTH: 42 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 142 caaaccaatt atggtactag
tagtagtaat tatggttttg ct 42 <210> SEQ ID NO 143 <211>
LENGTH: 15 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 143 tccgacgcga ttagc 15 <210> SEQ ID NO
144 <211> LENGTH: 48 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 144
atcatcaacg gcggcggaaa cacctactac gcctcctggg ccaagggc 48 <210>
SEQ ID NO 145 <211> LENGTH: 54 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 145 ggcatccagc
acggtggtgg aaacagcgac tactactact atgggatgga tctg 54 <210> SEQ
ID NO 146 <211> LENGTH: 33 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 146 caggcgtccc agtcaattag
cagcgtgctc tcc 33 <210> SEQ ID NO 147 <211> LENGTH: 21
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 147 ttggcctcca ctctggcctc g 21 <210> SEQ ID NO 148
<211> LENGTH: 42 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 148 caaaccaact acggaacctc
cagctccaac tacggctttg cc 42 <210> SEQ ID NO 149 <211>
LENGTH: 15 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 149 aactatgcaa tgagc 15 <210> SEQ ID NO
150 <211> LENGTH: 48 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 150 tccattagta ctggtggtct
cgcattctac gcgaactggg caaaaggc 48 <210> SEQ ID NO 151
<211> LENGTH: 39 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 151 aatggtggtg gtagttatat
tttctattat tttgacttg 39 <210> SEQ ID NO 152 <211>
LENGTH: 33 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 152 caggccagtc agagcattag tagttactta tcc 33
<210> SEQ ID NO 153 <211> LENGTH: 21 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 153 tctgcatcca
ctctggcatc t 21 <210> SEQ ID NO 154 <211> LENGTH: 30
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 154 caaagctatt atgatattgg tactagtact 30 <210> SEQ
ID NO 155 <211> LENGTH: 15 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 155 aactacgcca tgtcc 15
<210> SEQ ID NO 156 <211> LENGTH: 48 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 156 tccatcagca
caggcggcct ggccttctac gccaattggg ccaagggc 48 <210> SEQ ID NO
157 <211> LENGTH: 39 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 157 aacggcggag gctcctacat
cttctactac ttcgacctg 39 <210> SEQ ID NO 158 <211>
LENGTH: 33 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 158 caggcctccc agtccatctc ctcctacctg tcc 33
<210> SEQ ID NO 159 <211> LENGTH: 21 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 159 tctgcctcca
cactggcctc c 21 <210> SEQ ID NO 160 <211> LENGTH: 30
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 160 cagtcctact acgacatcgg cacctccacc 30 <210> SEQ
ID NO 161 <211> LENGTH: 15 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 161 agctatgcaa tgggc 15
<210> SEQ ID NO 162 <211> LENGTH: 48 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide
<400> SEQUENCE: 162 accattaata ttggtggtcg cgtatattac
gcgagctggg caaaaggc 48 <210> SEQ ID NO 163 <211>
LENGTH: 27 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 163 tattataatg gtggtagtta tgacatc 27
<210> SEQ ID NO 164 <211> LENGTH: 33 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 164 caggccagtg
agagcattta tcgcgtattg gcc 33 <210> SEQ ID NO 165 <211>
LENGTH: 21 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 165 gatacatcca ctctggcatc t 21 <210>
SEQ ID NO 166 <211> LENGTH: 36 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 166 caaggcggtt
attatgctga tagttatggt attgct 36 <210> SEQ ID NO 167
<211> LENGTH: 15 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 167 tcctacgcta tgggc 15
<210> SEQ ID NO 168 <211> LENGTH: 48 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 168 accatcaaca
tcggcggcag agtgtactac gcctcctggg ccaagggc 48 <210> SEQ ID NO
169 <211> LENGTH: 27 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 169 tactacaacg gcggctccta
cgatatc 27 <210> SEQ ID NO 170 <211> LENGTH: 33
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 170 caggcctccg agtccatcta ccgggtgctg gcc 33 <210>
SEQ ID NO 171 <211> LENGTH: 21 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 171 gacaccagca
cactggcctc c 21 <210> SEQ ID NO 172 <211> LENGTH: 36
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 172 cagggcggct actacgccga ctcctacgga atcgct 36
<210> SEQ ID NO 173 <211> LENGTH: 15 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 173 agttatgcaa
tgatc 15 <210> SEQ ID NO 174 <211> LENGTH: 48
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 174 accattagta ctggtggtat cacatactac gcgagctggg cgaaaggc
48 <210> SEQ ID NO 175 <211> LENGTH: 51 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic oligonucleotide <400> SEQUENCE: 175
gggggatatg ctgctagtag tgcttattat ctcccgtact actttgactt g 51
<210> SEQ ID NO 176 <211> LENGTH: 39 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 176 cagtcctccc
agtccgtgta taacaacaac aacctggcc 39 <210> SEQ ID NO 177
<211> LENGTH: 21 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 177 ctggcatcca ctctggcatc t
21 <210> SEQ ID NO 178 <211> LENGTH: 36 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic oligonucleotide <400> SEQUENCE: 178
ctaggtggtt gtgatgatga tgctgatact tttgct 36 <210> SEQ ID NO
179 <211> LENGTH: 15 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 179 tcctacgcta tgatc 15
<210> SEQ ID NO 180 <211> LENGTH: 48 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide
<400> SEQUENCE: 180 accatctcta ccggcggaat tacctactac
gcctcctggg ccaagggc 48 <210> SEQ ID NO 181 <211>
LENGTH: 51 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 181 ggcggctacg ccgccagctc cgcttactac
ctgccctact acttcgacct g 51 <210> SEQ ID NO 182 <211>
LENGTH: 39 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 182 cagtcctccc agtccgtgta taacaacaac
aacctggcc 39 <210> SEQ ID NO 183 <211> LENGTH: 21
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 183 ctggcctcca cactggcctc t 21 <210> SEQ ID NO 184
<211> LENGTH: 36 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 184 ctgggcggct gcgacgacga
cgccgatacc tttgct 36 <210> SEQ ID NO 185 <211> LENGTH:
8 <212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 185
Gly Tyr Ser Phe Thr Gly Tyr Thr 1 5 <210> SEQ ID NO 186
<211> LENGTH: 8 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 186 Ile Thr Pro Tyr Asn Gly Ala Ser 1 5
<210> SEQ ID NO 187 <211> LENGTH: 12 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 187 Ala Arg Gly Gly Tyr Asp
Gly Arg Gly Phe Asp Tyr 1 5 10 <210> SEQ ID NO 188
<211> LENGTH: 5 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 188 Ser Ser Val Ser Tyr 1 5 <210> SEQ
ID NO 189 <211> LENGTH: 3 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 189 Asp Thr Ser 1 <210> SEQ ID
NO 190 <211> LENGTH: 9 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 190 Gln Gln Trp Ser Lys His Pro Leu
Thr 1 5 <210> SEQ ID NO 191 <211> LENGTH: 8 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic peptide <400> SEQUENCE: 191 Gly Phe Asn
Ile Lys Asp Thr Tyr 1 5 <210> SEQ ID NO 192 <211>
LENGTH: 8 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 192 Ile Tyr Pro Thr Asn Gly Tyr Thr 1 5 <210> SEQ
ID NO 193 <211> LENGTH: 13 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 193 Ser Arg Trp Gly Gly Asp Gly Phe
Tyr Ala Met Asp Tyr 1 5 10 <210> SEQ ID NO 194 <211>
LENGTH: 6 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 194 Gln Asp Val Asn Thr Ala 1 5 <210> SEQ ID NO 195
<211> LENGTH: 3 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 195 Ser Ala Ser 1 <210> SEQ ID NO 196
<211> LENGTH: 9 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 196 Gln Gln His Tyr Thr Thr Pro Pro Thr 1 5
<210> SEQ ID NO 197 <211> LENGTH: 8 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide
<400> SEQUENCE: 197 Gly Ile Ser Leu Ser Ser Asp Ala 1 5
<210> SEQ ID NO 198 <211> LENGTH: 7 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 198 Ile Asn Gly Gly Gly Asn
Thr 1 5 <210> SEQ ID NO 199 <211> LENGTH: 20
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 199
Ala Arg Gly Ile Gln His Gly Gly Gly Asn Ser Asp Tyr Tyr Tyr Tyr 1 5
10 15 Gly Met Asp Leu 20 <210> SEQ ID NO 200 <211>
LENGTH: 6 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 200 Gln Ser Ile Ser Ser Val 1 5 <210> SEQ ID NO 201
<211> LENGTH: 3 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 201 Leu Ala Ser 1 <210> SEQ ID NO 202
<211> LENGTH: 14 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 202 Gln Thr Asn Tyr Gly Thr Ser Ser Ser Asn
Tyr Gly Phe Ala 1 5 10 <210> SEQ ID NO 203 <211>
LENGTH: 8 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 203 Gly Ile Ser Leu Ser Ser Asp Ala 1 5 <210> SEQ
ID NO 204 <211> LENGTH: 7 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 204 Ile Asn Gly Gly Gly Asn Thr 1 5
<210> SEQ ID NO 205 <211> LENGTH: 20 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 205 Ala Arg Gly Ile Gln His
Gly Gly Gly Asn Ser Asp Tyr Tyr Tyr Tyr 1 5 10 15 Gly Met Asp Leu
20 <210> SEQ ID NO 206 <211> LENGTH: 6 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic peptide <400> SEQUENCE: 206 Gln Ser Ile
Ser Ser Val 1 5 <210> SEQ ID NO 207 <211> LENGTH: 3
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 207
Leu Ala Ser 1 <210> SEQ ID NO 208 <211> LENGTH: 14
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 208
Gln Thr Asn Tyr Gly Thr Ser Ser Ser Asn Tyr Gly Phe Ala 1 5 10
<210> SEQ ID NO 209 <211> LENGTH: 8 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 209 Gly Phe Ser Leu Asn Asn
Tyr Ala 1 5 <210> SEQ ID NO 210 <211> LENGTH: 7
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 210
Ile Ser Thr Gly Gly Leu Ala 1 5 <210> SEQ ID NO 211
<211> LENGTH: 15 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 211 Gly Arg Asn Gly Gly Gly Ser Tyr Ile Phe
Tyr Tyr Phe Asp Leu 1 5 10 15 <210> SEQ ID NO 212 <211>
LENGTH: 6 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 212 Gln Ser Ile Ser Ser Tyr 1 5 <210> SEQ ID NO 213
<211> LENGTH: 3 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 213 Ser Ala Ser 1
<210> SEQ ID NO 214 <211> LENGTH: 10 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 214 Gln Ser Tyr Tyr Asp Ile
Gly Thr Ser Thr 1 5 10 <210> SEQ ID NO 215 <211>
LENGTH: 8 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 215 Gly Phe Ser Leu Asn Asn Tyr Ala 1 5 <210> SEQ
ID NO 216 <211> LENGTH: 7 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 216 Ile Ser Thr Gly Gly Leu Ala 1 5
<210> SEQ ID NO 217 <211> LENGTH: 15 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 217 Ala Arg Asn Gly Gly Gly
Ser Tyr Ile Phe Tyr Tyr Phe Asp Leu 1 5 10 15 <210> SEQ ID NO
218 <211> LENGTH: 6 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 218 Gln Ser Ile Ser Ser Tyr 1 5
<210> SEQ ID NO 219 <211> LENGTH: 3 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 219 Ser Ala Ser 1
<210> SEQ ID NO 220 <211> LENGTH: 10 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 220 Gln Ser Tyr Tyr Asp Ile
Gly Thr Ser Thr 1 5 10 <210> SEQ ID NO 221 <211>
LENGTH: 8 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 221 Gly Ile Asp Leu Ser Ser Tyr Ala 1 5 <210> SEQ
ID NO 222 <211> LENGTH: 7 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 222 Ile Asn Ile Gly Gly Arg Val 1 5
<210> SEQ ID NO 223 <211> LENGTH: 11 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 223 Ala Arg Tyr Tyr Asn Gly
Gly Ser Tyr Asp Ile 1 5 10 <210> SEQ ID NO 224 <211>
LENGTH: 6 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 224 Glu Ser Ile Tyr Arg Val 1 5 <210> SEQ ID NO 225
<211> LENGTH: 3 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 225 Asp Thr Ser 1 <210> SEQ ID NO 226
<211> LENGTH: 12 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 226 Gln Gly Gly Tyr Tyr Ala Asp Ser Tyr Gly
Ile Ala 1 5 10 <210> SEQ ID NO 227 <211> LENGTH: 8
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 227
Gly Ile Asp Leu Ser Ser Tyr Ala 1 5 <210> SEQ ID NO 228
<211> LENGTH: 7 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 228 Ile Asn Ile Gly Gly Arg Val 1 5
<210> SEQ ID NO 229 <211> LENGTH: 11 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 229 Ala Arg Tyr Tyr Asn Gly
Gly Ser Tyr Asp Ile 1 5 10 <210> SEQ ID NO 230 <211>
LENGTH: 6 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 230
Glu Ser Ile Tyr Arg Val 1 5 <210> SEQ ID NO 231 <211>
LENGTH: 3 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 231 Asp Thr Ser 1 <210> SEQ ID NO 232 <211>
LENGTH: 12 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 232 Gln Gly Gly Tyr Tyr Ala Asp Ser Tyr Gly Ile Ala 1 5
10 <210> SEQ ID NO 233 <211> LENGTH: 8 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic peptide <400> SEQUENCE: 233 Gly Phe Ser
Leu Ser Ser Tyr Ala 1 5 <210> SEQ ID NO 234 <211>
LENGTH: 7 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 234 Ile Ser Thr Gly Gly Ile Thr 1 5 <210> SEQ ID NO
235 <211> LENGTH: 19 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 235 Ala Arg Gly Gly Tyr Ala Ala Ser
Ser Ala Tyr Tyr Leu Pro Tyr Tyr 1 5 10 15 Phe Asp Leu <210>
SEQ ID NO 236 <211> LENGTH: 8 <212> TYPE: PRT
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 236 Gln Ser Val Tyr Asn Asn
Asn Asn 1 5 <210> SEQ ID NO 237 <211> LENGTH: 3
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 237
Leu Ala Ser 1 <210> SEQ ID NO 238 <211> LENGTH: 12
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 238
Leu Gly Gly Cys Asp Asp Asp Ala Asp Thr Phe Ala 1 5 10 <210>
SEQ ID NO 239 <211> LENGTH: 8 <212> TYPE: PRT
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 239 Gly Phe Ser Leu Ser Ser
Tyr Ala 1 5 <210> SEQ ID NO 240 <211> LENGTH: 7
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic peptide <400> SEQUENCE: 240
Ile Ser Thr Gly Gly Ile Thr 1 5 <210> SEQ ID NO 241
<211> LENGTH: 19 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 241 Ala Arg Gly Gly Tyr Ala Ala Ser Ser Ala
Tyr Tyr Leu Pro Tyr Tyr 1 5 10 15 Phe Asp Leu <210> SEQ ID NO
242 <211> LENGTH: 8 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 242 Gln Ser Val Tyr Asn Asn Asn Asn 1
5 <210> SEQ ID NO 243 <211> LENGTH: 3 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 243 Leu Ala Ser 1
<210> SEQ ID NO 244 <211> LENGTH: 12 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic peptide <400> SEQUENCE: 244 Leu Gly Gly Cys Asp Asp
Asp Ala Asp Thr Phe Ala 1 5 10 <210> SEQ ID NO 245
<211> LENGTH: 24 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 245 ggcttcacct tcagcggcta
tggg 24 <210> SEQ ID NO 246 <211> LENGTH: 24
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 246 attagtagtg gtggtagtta tacc 24 <210> SEQ ID NO
247 <211> LENGTH: 36 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 247 gcaagacatg gggacgatcc cgcctggttc gcttat 36
<210> SEQ ID NO 248 <211> LENGTH: 21 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 248 tcaagtataa
gttccaacaa c 21 <210> SEQ ID NO 249 <211> LENGTH: 9
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 249 ggcacatcc 9 <210> SEQ ID NO 250 <211>
LENGTH: 33 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 250 caacagtgga gtagttaccc gtacatgtac acg 33
<210> SEQ ID NO 251 <211> LENGTH: 24 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 251 ggttactcat
tcactggcta cacc 24 <210> SEQ ID NO 252 <211> LENGTH: 24
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 252 attactcctt acaatggtgc ttct 24 <210> SEQ ID NO
253 <211> LENGTH: 36 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 253 gcaagggggg gttacgacgg
gaggggtttt gactac 36 <210> SEQ ID NO 254 <211> LENGTH:
15 <212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 254 tcaagtgtaa gttac 15 <210> SEQ ID NO 255
<211> LENGTH: 9 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 255 gacacatcc 9 <210>
SEQ ID NO 256 <211> LENGTH: 27 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 256 cagcagtgga
gtaagcaccc tctcacg 27 <210> SEQ ID NO 257 <211> LENGTH:
24 <212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 257 ggaatctccc tcagtagcga tgca 24 <210> SEQ ID NO
258 <211> LENGTH: 21 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 258 attaatggtg gtggtaacac a
21 <210> SEQ ID NO 259 <211> LENGTH: 60 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic oligonucleotide <400> SEQUENCE: 259
gccagaggca ttcaacatgg tggtggtaat agtgattatt attattacgg catggacctc
60 <210> SEQ ID NO 260 <211> LENGTH: 18 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic oligonucleotide <400> SEQUENCE: 260
cagagcatta gtagtgtc 18 <210> SEQ ID NO 261 <211>
LENGTH: 9 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 261 ctggcatct 9 <210> SEQ ID NO 262
<211> LENGTH: 42 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 262 caaaccaatt atggtactag
tagtagtaat tatggttttg ct 42 <210> SEQ ID NO 263 <211>
LENGTH: 24 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 263 ggaatttccc tctcctccga cgcg 24 <210>
SEQ ID NO 264 <211> LENGTH: 21 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 264 atcaacggcg
gcggaaacac c 21 <210> SEQ ID NO 265 <211> LENGTH: 60
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 265 gcgcgcggca
tccagcacgg tggtggaaac agcgactact actactatgg gatggatctg 60
<210> SEQ ID NO 266 <211> LENGTH: 18 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 266 cagtcaatta
gcagcgtg 18 <210> SEQ ID NO 267 <211> LENGTH: 9
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 267 ttggcctcc 9 <210> SEQ ID NO 268 <211>
LENGTH: 42 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 268 caaaccaact acggaacctc cagctccaac
tacggctttg cc 42 <210> SEQ ID NO 269 <211> LENGTH: 24
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 269 ggattctccc tcaataacta tgca 24 <210> SEQ ID NO
270 <211> LENGTH: 21 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 270 attagtactg gtggtctcgc a
21 <210> SEQ ID NO 271 <211> LENGTH: 45 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic oligonucleotide <400> SEQUENCE: 271
ggcagaaatg gtggtggtag ttatattttc tattattttg acttg 45 <210>
SEQ ID NO 272 <211> LENGTH: 18 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 272 cagagcatta
gtagttac 18 <210> SEQ ID NO 273 <211> LENGTH: 9
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 273 tctgcatcc 9 <210> SEQ ID NO 274 <211>
LENGTH: 30 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic oligonucleotide
<400> SEQUENCE: 274 caaagctatt atgatattgg tactagtact 30
<210> SEQ ID NO 275 <211> LENGTH: 24 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 275 ggcttctccc
tgaacaacta cgcc 24 <210> SEQ ID NO 276 <211> LENGTH: 21
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 276 atcagcacag gcggcctggc c 21 <210> SEQ ID NO 277
<211> LENGTH: 45 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 277 gccagaaacg gcggaggctc
ctacatcttc tactacttcg acctg 45 <210> SEQ ID NO 278
<211> LENGTH: 18 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 278 cagtccatct cctcctac 18
<210> SEQ ID NO 279 <211> LENGTH: 9 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 279 tctgcctcc 9
<210> SEQ ID NO 280 <400> SEQUENCE: 280 000 <210>
SEQ ID NO 281 <400> SEQUENCE: 281 000 <210> SEQ ID NO
282 <400> SEQUENCE: 282 000 <210> SEQ ID NO 283
<400> SEQUENCE: 283 000 <210> SEQ ID NO 284 <400>
SEQUENCE: 284 000 <210> SEQ ID NO 285 <400> SEQUENCE:
285 000 <210> SEQ ID NO 286
<400> SEQUENCE: 286 000 <210> SEQ ID NO 287 <400>
SEQUENCE: 287 000 <210> SEQ ID NO 288 <400> SEQUENCE:
288 000 <210> SEQ ID NO 289 <400> SEQUENCE: 289 000
<210> SEQ ID NO 290 <400> SEQUENCE: 290 000 <210>
SEQ ID NO 291 <400> SEQUENCE: 291 000 <210> SEQ ID NO
292 <400> SEQUENCE: 292 000 <210> SEQ ID NO 293
<400> SEQUENCE: 293 000 <210> SEQ ID NO 294 <400>
SEQUENCE: 294 000 <210> SEQ ID NO 295 <400> SEQUENCE:
295 000 <210> SEQ ID NO 296 <400> SEQUENCE: 296 000
<210> SEQ ID NO 297 <400> SEQUENCE: 297 000 <210>
SEQ ID NO 298 <400> SEQUENCE: 298 000 <210> SEQ ID NO
299 <400> SEQUENCE: 299 000 <210> SEQ ID NO 300
<211> LENGTH: 30 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 300 cagtcctact acgacatcgg
cacctccacc 30 <210> SEQ ID NO 301 <211> LENGTH: 24
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 301 ggaatcgacc tcagtagcta tgca 24 <210> SEQ ID NO
302 <211> LENGTH: 21 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 302 attaatattg gtggtcgcgt a
21 <210> SEQ ID NO 303 <211> LENGTH: 33 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic oligonucleotide <400> SEQUENCE: 303
gccagatatt ataatggtgg tagttatgac atc 33 <210> SEQ ID NO 304
<211> LENGTH: 18 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 304 gagagcattt atcgcgta 18
<210> SEQ ID NO 305 <211> LENGTH: 9 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 305 gatacatcc 9
<210> SEQ ID NO 306 <211> LENGTH: 36 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 306 caaggcggtt
attatgctga tagttatggt attgct 36 <210> SEQ ID NO 307
<211> LENGTH: 24 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 307 ggaatcgacc tgtcctccta
cgct 24 <210> SEQ ID NO 308 <211> LENGTH: 21
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 308 atcaacatcg gcggcagagt g 21 <210> SEQ ID NO 309
<211> LENGTH: 33 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 309 gcccggtact acaacggcgg
ctcctacgat atc 33 <210> SEQ ID NO 310 <211> LENGTH: 18
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 310 gagtccatct accgggtg 18 <210> SEQ ID NO 311
<211> LENGTH: 9 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 311 gacaccagc 9 <210>
SEQ ID NO 312 <211> LENGTH: 36 <212> TYPE: DNA
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 312 cagggcggct
actacgccga ctcctacgga atcgct 36 <210> SEQ ID NO 313
<211> LENGTH: 24 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 313 ggattctccc tcagtagtta
tgca 24 <210> SEQ ID NO 314 <211> LENGTH: 21
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 314 attagtactg gtggtatcac a 21 <210> SEQ ID NO 315
<211> LENGTH: 57 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 315 gccagagggg gatatgctgc
tagtagtgct tattatctcc cgtactactt tgacttg 57 <210> SEQ ID NO
316 <211> LENGTH: 24 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 316 cagagtgttt ataataataa
caac 24 <210> SEQ ID NO 317 <211> LENGTH: 9 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic oligonucleotide <400> SEQUENCE: 317
ctggcatcc 9 <210> SEQ ID NO 318 <211> LENGTH: 36
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 318 ctaggtggtt gtgatgatga tgctgatact tttgct 36
<210> SEQ ID NO 319 <211> LENGTH: 24 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic oligonucleotide <400> SEQUENCE: 319 ggcttctccc
tgtcctccta cgct 24 <210> SEQ ID NO 320 <211> LENGTH: 21
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 320 atctctaccg gcggaattac c 21 <210> SEQ ID NO 321
<211> LENGTH: 57 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 321 gctagaggcg gctacgccgc
cagctccgct tactacctgc cctactactt cgacctg 57 <210> SEQ ID NO
322 <211> LENGTH: 24 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
oligonucleotide <400> SEQUENCE: 322 cagtccgtgt ataacaacaa
caac 24 <210> SEQ ID NO 323 <211> LENGTH: 9 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic oligonucleotide <400> SEQUENCE: 323
ctggcctcc 9 <210> SEQ ID NO 324 <211> LENGTH: 36
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic oligonucleotide <400>
SEQUENCE: 324 ctgggcggct gcgacgacga cgccgatacc tttgct 36
<210> SEQ ID NO 325 <211> LENGTH: 449 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 325 Gln Val Gln Leu Gln
Gln Ser Gly Pro Glu Leu Glu Lys Pro Gly Ala 1 5 10 15 Ser Val Lys
Ile Ser Cys Lys Ala Ser Gly Tyr Ser Phe Thr Gly Tyr 20 25 30 Thr
Met Asn Trp Val Lys Gln Ser His Gly Lys Ser Leu Glu Trp Ile 35 40
45 Gly Leu Ile Thr Pro Tyr Asn Gly Ala Ser Ser Tyr Asn Gln Lys Phe
50 55 60 Arg Gly Lys Ala Thr Leu Thr Val Asp Lys Ser Ser Ser Thr
Ala Tyr 65 70 75 80 Met Asp Leu Leu Ser Leu Thr Ser Glu Asp Ser Ala
Val Tyr Phe Cys 85 90 95 Ala Arg Gly Gly Tyr Asp Gly Arg Gly Phe
Asp Tyr Trp Gly Ser Gly 100 105 110 Thr Pro Val Thr Val Ser Ser Ala
Ser Thr Lys Gly Pro Ser Val Phe 115 120 125 Pro Leu Ala Pro Ser Ser
Lys Ser Thr Ser Gly Gly Thr Ala Ala Leu 130 135 140 Gly Cys Leu Val
Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp 145 150 155 160 Asn
Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val Leu 165 170
175 Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser
180 185 190 Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His
Lys Pro 195 200 205 Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys
Ser Cys Asp Lys 210 215 220 Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Glu Leu Leu Gly Gly Pro 225 230 235 240
Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser 245
250 255 Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu
Asp 260 265 270 Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu
Val His Asn 275 280 285 Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn
Ser Thr Tyr Arg Val 290 295 300 Val Ser Val Leu Thr Val Leu His Gln
Asp Trp Leu Asn Gly Lys Glu 305 310 315 320 Tyr Lys Cys Lys Val Ser
Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys 325 330 335 Thr Ile Ser Lys
Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr 340 345 350 Leu Pro
Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr 355 360 365
Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu 370
375 380 Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val
Leu 385 390 395 400 Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu
Thr Val Asp Lys 405 410 415 Ser Arg Trp Gln Gln Gly Asn Val Phe Ser
Cys Ser Val Met His Glu 420 425 430 Ala Leu His Asn His Tyr Thr Gln
Lys Ser Leu Ser Leu Ser Pro Gly 435 440 445 Lys <210> SEQ ID
NO 326 <211> LENGTH: 213 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 326 Asp Ile Glu Leu Thr Gln Ser
Pro Ala Ile Met Ser Ala Ser Pro Gly 1 5 10 15 Glu Lys Val Thr Met
Thr Cys Ser Ala Ser Ser Ser Val Ser Tyr Met 20 25 30 His Trp Tyr
Gln Gln Lys Ser Gly Thr Ser Pro Lys Arg Trp Ile Tyr 35 40 45 Asp
Thr Ser Lys Leu Ala Ser Gly Val Pro Gly Arg Phe Ser Gly Ser 50 55
60 Gly Ser Gly Asn Ser Tyr Ser Leu Thr Ile Ser Ser Val Glu Ala Glu
65 70 75 80 Asp Asp Ala Thr Tyr Tyr Cys Gln Gln Trp Ser Lys His Pro
Leu Thr 85 90 95 Phe Gly Ser Gly Thr Lys Val Glu Ile Lys Arg Thr
Val Ala Ala Pro 100 105 110 Ser Val Phe Ile Phe Pro Pro Ser Asp Glu
Gln Leu Lys Ser Gly Thr 115 120 125 Ala Ser Val Val Cys Leu Leu Asn
Asn Phe Tyr Pro Arg Glu Ala Lys 130 135 140 Val Gln Trp Lys Val Asp
Asn Ala Leu Gln Ser Gly Asn Ser Gln Glu 145 150 155 160 Ser Val Thr
Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser Ser 165 170 175 Thr
Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr Ala 180 185
190 Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser Phe
195 200 205 Asn Arg Gly Glu Cys 210 <210> SEQ ID NO 327
<211> LENGTH: 451 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 327 Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg Leu Ser
Cys Ala Ala Ser Gly Phe Asn Ile Lys Asp Thr 20 25 30 Tyr Ile His
Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45 Ala
Arg Ile Tyr Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Asp Ser Val 50 55
60 Lys Gly Arg Phe Thr Ile Ser Ala Asp Thr Ser Lys Asn Thr Ala Tyr
65 70 75 80 Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr
Tyr Cys 85 90 95 Ser Arg Trp Gly Gly Asp Gly Phe Tyr Ala Met Asp
Tyr Trp Gly Gln 100 105 110 Gly Thr Leu Val Thr Val Ser Ser Ala Ser
Thr Lys Gly Pro Ser Val 115 120 125 Phe Pro Leu Ala Pro Ser Ser Lys
Ser Thr Ser Gly Gly Thr Ala Ala 130 135 140 Leu Gly Cys Leu Val Lys
Asp Tyr Phe Pro Glu Pro Val Thr Val Ser 145 150 155 160 Trp Asn Ser
Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val 165 170 175 Leu
Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro 180 185
190 Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys
195 200 205 Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Pro Lys
Ser Cys 210 215 220 Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Glu Leu Leu Gly 225 230 235 240 Gly Pro Ser Val Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met 245 250 255 Ile Ser Arg Thr Pro Glu Val
Thr Cys Val Val Val Asp Val Ser His 260 265 270 Glu Asp Pro Glu Val
Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val 275 280 285 His Asn Ala
Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr 290 295 300 Arg
Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly 305 310
315 320 Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro
Ile 325 330 335 Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu
Pro Gln Val 340 345 350 Tyr Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr
Lys Asn Gln Val Ser 355 360 365 Leu Thr Cys Leu Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu 370 375 380 Trp Glu Ser Asn Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro 385 390 395 400 Val Leu Asp Ser
Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val 405 410 415 Asp Lys
Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met 420 425 430
His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser 435
440 445 Pro Gly Lys 450 <210> SEQ ID NO 328 <211>
LENGTH: 214 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polypeptide
<400> SEQUENCE: 328 Asp Ile Gln Met Thr Gln Ser Pro Ser Ser
Leu Ser Ala Ser Val Gly 1 5 10 15 Asp Arg Val Thr Ile Thr Cys Arg
Ala Ser Gln Asp Val Asn Thr Ala 20 25 30 Val Ala Trp Tyr Gln Gln
Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45 Tyr Ser Ala Ser
Phe Leu Tyr Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60 Ser Arg
Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro 65 70 75 80
Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln His Tyr Thr Thr Pro Pro 85
90 95 Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala
Ala 100 105 110 Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu
Lys Ser Gly 115 120 125 Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe
Tyr Pro Arg Glu Ala 130 135 140 Lys Val Gln Trp Lys Val Asp Asn Ala
Leu Gln Ser Gly Asn Ser Gln 145 150 155 160 Glu Ser Val Thr Glu Gln
Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175 Ser Thr Leu Thr
Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190 Ala Cys
Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200 205
Phe Asn Arg Gly Glu Cys 210 <210> SEQ ID NO 329 <211>
LENGTH: 453
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE:
329 Gln Ser Val Glu Glu Ser Gly Gly Arg Leu Val Thr Pro Gly Thr Pro
1 5 10 15 Leu Thr Leu Thr Cys Thr Val Ser Gly Ile Ser Leu Ser Ser
Asp Ala 20 25 30 Ile Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu
Glu Tyr Ile Gly 35 40 45 Ile Ile Asn Gly Gly Gly Asn Thr Tyr Tyr
Ala Ser Trp Ala Lys Gly 50 55 60 Arg Phe Thr Ile Ser Lys Thr Ser
Thr Thr Val Asp Leu Lys Ile Thr 65 70 75 80 Ser Pro Thr Thr Glu Asp
Thr Ala Thr Tyr Phe Cys Ala Arg Gly Ile 85 90 95 Gln His Gly Gly
Gly Asn Ser Asp Tyr Tyr Tyr Tyr Gly Met Asp Leu 100 105 110 Trp Gly
Pro Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly 115 120 125
Pro Ser Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly 130
135 140 Thr Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro
Val 145 150 155 160 Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly
Val His Thr Phe 165 170 175 Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr
Ser Leu Ser Ser Val Val 180 185 190 Thr Val Pro Ser Ser Ser Leu Gly
Thr Gln Thr Tyr Ile Cys Asn Val 195 200 205 Asn His Lys Pro Ser Asn
Thr Lys Val Asp Lys Lys Val Glu Pro Lys 210 215 220 Ser Cys Asp Lys
Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu 225 230 235 240 Leu
Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr 245 250
255 Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val
260 265 270 Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp
Gly Val 275 280 285 Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu
Gln Tyr Asn Ser 290 295 300 Thr Tyr Arg Val Val Ser Val Leu Thr Val
Leu His Gln Asp Trp Leu 305 310 315 320 Asn Gly Lys Glu Tyr Lys Cys
Lys Val Ser Asn Lys Ala Leu Pro Ala 325 330 335 Pro Ile Glu Lys Thr
Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro 340 345 350 Gln Val Tyr
Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln 355 360 365 Val
Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala 370 375
380 Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr
385 390 395 400 Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr
Ser Lys Leu 405 410 415 Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn
Val Phe Ser Cys Ser 420 425 430 Val Met His Glu Ala Leu His Asn His
Tyr Thr Gln Lys Ser Leu Ser 435 440 445 Leu Ser Pro Gly Lys 450
<210> SEQ ID NO 330 <211> LENGTH: 219 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 330 Glu Val Leu Met Thr
Gln Thr Pro Ser Ser Val Ser Ala Ala Val Gly 1 5 10 15 Asp Thr Val
Thr Ile Lys Cys Gln Ala Ser Gln Ser Ile Ser Ser Val 20 25 30 Leu
Ser Trp Tyr Gln Gln Lys Pro Gly Gln Pro Pro Lys Leu Leu Ile 35 40
45 Tyr Leu Ala Ser Thr Leu Ala Ser Gly Val Pro Ser Arg Phe Ser Gly
50 55 60 Ser Arg Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser Asp Leu
Glu Cys 65 70 75 80 Asp Asp Ala Ala Thr Tyr Tyr Cys Gln Thr Asn Tyr
Gly Thr Ser Ser 85 90 95 Ser Asn Tyr Gly Phe Ala Phe Gly Gly Gly
Thr Glu Val Val Val Lys 100 105 110 Arg Thr Val Ala Ala Pro Ser Val
Phe Ile Phe Pro Pro Ser Asp Glu 115 120 125 Gln Leu Lys Ser Gly Thr
Ala Ser Val Val Cys Leu Leu Asn Asn Phe 130 135 140 Tyr Pro Arg Glu
Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln 145 150 155 160 Ser
Gly Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser 165 170
175 Thr Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu
180 185 190 Lys His Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu
Ser Ser 195 200 205 Pro Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 210
215 <210> SEQ ID NO 331 <211> LENGTH: 456 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polypeptide <400> SEQUENCE: 331 Glu Val
Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15
Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Ile Ser Leu Ser Ser Asp 20
25 30 Ala Ile Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Tyr
Ile 35 40 45 Gly Ile Ile Asn Gly Gly Gly Asn Thr Tyr Tyr Ala Ser
Trp Ala Lys 50 55 60 Gly Arg Phe Thr Ile Ser Arg His Asn Ser Lys
Asn Thr Leu Tyr Leu 65 70 75 80 Gln Met Asn Ser Leu Arg Ala Glu Asp
Thr Ala Val Tyr Tyr Cys Ala 85 90 95 Arg Gly Ile Gln His Gly Gly
Gly Asn Ser Asp Tyr Tyr Tyr Tyr Gly 100 105 110 Met Asp Leu Trp Gly
Gln Gly Thr Leu Val Thr Val Ser Ser Ala Ser 115 120 125 Thr Lys Gly
Pro Ser Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr 130 135 140 Ser
Gly Gly Thr Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro 145 150
155 160 Glu Pro Val Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly
Val 165 170 175 His Thr Phe Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr
Ser Leu Ser 180 185 190 Ser Val Val Thr Val Pro Ser Ser Ser Leu Gly
Thr Gln Thr Tyr Ile 195 200 205 Cys Asn Val Asn His Lys Pro Ser Asn
Thr Lys Val Asp Lys Lys Val 210 215 220 Glu Pro Lys Ser Cys Asp Lys
Thr His Thr Cys Pro Pro Cys Pro Ala 225 230 235 240 Pro Glu Leu Leu
Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro 245 250 255 Lys Asp
Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val 260 265 270
Val Asp Val Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val 275
280 285 Asp Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu
Gln 290 295 300 Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val
Leu His Gln 305 310 315 320 Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys
Lys Val Ser Asn Lys Ala 325 330 335 Leu Pro Ala Pro Ile Glu Lys Thr
Ile Ser Lys Ala Lys Gly Gln Pro 340 345 350 Arg Glu Pro Gln Val Tyr
Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr 355 360 365 Lys Asn Gln Val
Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser 370 375 380 Asp Ile
Ala Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr 385 390 395
400 Lys Thr Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr
405 410 415 Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn
Val Phe 420 425 430 Ser Cys Ser Val Met His Glu Ala Leu His Asn His
Tyr Thr Gln Lys 435 440 445 Ser Leu Ser Leu Ser Pro Gly Lys 450 455
<210> SEQ ID NO 332
<211> LENGTH: 219 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 332 Asp Ile Gln Met Thr Gln Ser
Pro Ser Ser Leu Ser Ala Ser Val Gly 1 5 10 15 Asp Arg Val Thr Ile
Thr Cys Gln Ala Ser Gln Ser Ile Ser Ser Val 20 25 30 Leu Ser Trp
Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45 Tyr
Leu Ala Ser Thr Leu Ala Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55
60 Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Cys
65 70 75 80 Glu Asp Ile Ala Thr Tyr Tyr Cys Gln Thr Asn Tyr Gly Thr
Ser Ser 85 90 95 Ser Asn Tyr Gly Phe Ala Phe Gly Gly Gly Thr Lys
Val Glu Ile Lys 100 105 110 Arg Thr Val Ala Ala Pro Ser Val Phe Ile
Phe Pro Pro Ser Asp Glu 115 120 125 Gln Leu Lys Ser Gly Thr Ala Ser
Val Val Cys Leu Leu Asn Asn Phe 130 135 140 Tyr Pro Arg Glu Ala Lys
Val Gln Trp Lys Val Asp Asn Ala Leu Gln 145 150 155 160 Ser Gly Asn
Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser 165 170 175 Thr
Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu 180 185
190 Lys His Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser
195 200 205 Pro Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 210 215
<210> SEQ ID NO 333 <211> LENGTH: 448 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 333 Gln Ser Val Glu Glu
Ser Gly Gly Arg Leu Val Thr Pro Gly Thr Pro 1 5 10 15 Leu Thr Leu
Thr Cys Thr Val Ser Gly Phe Ser Leu Asn Asn Tyr Ala 20 25 30 Met
Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Ile Gly 35 40
45 Ser Ile Ser Thr Gly Gly Leu Ala Phe Tyr Ala Asn Trp Ala Lys Gly
50 55 60 Arg Phe Thr Ile Ser Arg Thr Ser Thr Thr Val Asp Leu Lys
Met Thr 65 70 75 80 Ser Leu Thr Thr Glu Asp Thr Ala Thr Tyr Phe Cys
Gly Arg Asn Gly 85 90 95 Gly Gly Ser Tyr Ile Phe Tyr Tyr Phe Asp
Leu Trp Gly Gln Gly Thr 100 105 110 Leu Val Thr Val Ser Ser Ala Ser
Thr Lys Gly Pro Ser Val Phe Pro 115 120 125 Leu Ala Pro Ser Ser Lys
Ser Thr Ser Gly Gly Thr Ala Ala Leu Gly 130 135 140 Cys Leu Val Lys
Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp Asn 145 150 155 160 Ser
Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val Leu Gln 165 170
175 Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser Ser
180 185 190 Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys
Pro Ser 195 200 205 Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser
Cys Asp Lys Thr 210 215 220 His Thr Cys Pro Pro Cys Pro Ala Pro Glu
Leu Leu Gly Gly Pro Ser 225 230 235 240 Val Phe Leu Phe Pro Pro Lys
Pro Lys Asp Thr Leu Met Ile Ser Arg 245 250 255 Thr Pro Glu Val Thr
Cys Val Val Val Asp Val Ser His Glu Asp Pro 260 265 270 Glu Val Lys
Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala 275 280 285 Lys
Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val 290 295
300 Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr
305 310 315 320 Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile
Glu Lys Thr 325 330 335 Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro
Gln Val Tyr Thr Leu 340 345 350 Pro Pro Ser Arg Asp Glu Leu Thr Lys
Asn Gln Val Ser Leu Thr Cys 355 360 365 Leu Val Lys Gly Phe Tyr Pro
Ser Asp Ile Ala Val Glu Trp Glu Ser 370 375 380 Asn Gly Gln Pro Glu
Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp 385 390 395 400 Ser Asp
Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser 405 410 415
Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala 420
425 430 Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly
Lys 435 440 445 <210> SEQ ID NO 334 <211> LENGTH: 215
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE:
334 Ala Phe Glu Leu Thr Gln Thr Pro Ser Ser Val Glu Ala Ala Val Gly
1 5 10 15 Gly Thr Ile Thr Ile Lys Cys Gln Ala Ser Gln Ser Ile Ser
Ser Tyr 20 25 30 Leu Ser Trp Tyr Gln Gln Lys Pro Gly Gln Pro Pro
Lys Leu Leu Ile 35 40 45 Tyr Ser Ala Ser Thr Leu Ala Ser Gly Val
Ser Ser Arg Phe Lys Gly 50 55 60 Ser Gly Ser Gly Thr Glu Tyr Thr
Leu Thr Ile Ser Asp Leu Glu Cys 65 70 75 80 Ala Asp Ala Ala Thr Tyr
Phe Cys Gln Ser Tyr Tyr Asp Ile Gly Thr 85 90 95 Ser Thr Phe Gly
Gly Gly Thr Glu Val Val Val Lys Arg Thr Val Ala 100 105 110 Ala Pro
Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser 115 120 125
Gly Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu 130
135 140 Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn
Ser 145 150 155 160 Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser
Thr Tyr Ser Leu 165 170 175 Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp
Tyr Glu Lys His Lys Val 180 185 190 Tyr Ala Cys Glu Val Thr His Gln
Gly Leu Ser Ser Pro Val Thr Lys 195 200 205 Ser Phe Asn Arg Gly Glu
Cys 210 215 <210> SEQ ID NO 335 <211> LENGTH: 451
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE:
335 Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly
1 5 10 15 Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Ser Leu Asn
Asn Tyr 20 25 30 Ala Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly
Leu Glu Trp Ile 35 40 45 Gly Ser Ile Ser Thr Gly Gly Leu Ala Phe
Tyr Ala Asn Trp Ala Lys 50 55 60 Gly Arg Phe Thr Ile Ser Arg Asp
Asn Ser Lys Asn Thr Leu Tyr Leu 65 70 75 80 Gln Met Asn Ser Leu Arg
Ala Glu Asp Thr Ala Val Tyr Tyr Cys Ala 85 90 95 Arg Asn Gly Gly
Gly Ser Tyr Ile Phe Tyr Tyr Phe Asp Leu Trp Gly 100 105 110 Gln Gly
Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125
Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130
135 140 Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr
Val 145 150 155 160 Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His
Thr Phe Pro Ala 165 170 175 Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu
Ser Ser Val Val Thr Val 180 185 190 Pro Ser Ser Ser Leu Gly Thr Gln
Thr Tyr Ile Cys Asn Val Asn His
195 200 205 Lys Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys
Ser Cys 210 215 220 Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Glu Leu Leu Gly 225 230 235 240 Gly Pro Ser Val Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met 245 250 255 Ile Ser Arg Thr Pro Glu Val
Thr Cys Val Val Val Asp Val Ser His 260 265 270 Glu Asp Pro Glu Val
Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val 275 280 285 His Asn Ala
Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr 290 295 300 Arg
Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly 305 310
315 320 Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro
Ile 325 330 335 Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu
Pro Gln Val 340 345 350 Tyr Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr
Lys Asn Gln Val Ser 355 360 365 Leu Thr Cys Leu Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu 370 375 380 Trp Glu Ser Asn Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro 385 390 395 400 Val Leu Asp Ser
Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val 405 410 415 Asp Lys
Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met 420 425 430
His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser 435
440 445 Pro Gly Lys 450 <210> SEQ ID NO 336 <211>
LENGTH: 215 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polypeptide
<400> SEQUENCE: 336 Asp Ile Gln Met Thr Gln Ser Pro Ser Ser
Leu Ser Ala Ser Val Gly 1 5 10 15 Asp Arg Val Thr Ile Thr Cys Gln
Ala Ser Gln Ser Ile Ser Ser Tyr 20 25 30 Leu Ser Trp Tyr Gln Gln
Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45 Tyr Ser Ala Ser
Thr Leu Ala Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60 Ser Gly
Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Cys 65 70 75 80
Glu Asp Ala Ala Thr Tyr Tyr Cys Gln Ser Tyr Tyr Asp Ile Gly Thr 85
90 95 Ser Thr Phe Gly Gly Gly Thr Lys Val Glu Ile Lys Arg Thr Val
Ala 100 105 110 Ala Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln
Leu Lys Ser 115 120 125 Gly Thr Ala Ser Val Val Cys Leu Leu Asn Asn
Phe Tyr Pro Arg Glu 130 135 140 Ala Lys Val Gln Trp Lys Val Asp Asn
Ala Leu Gln Ser Gly Asn Ser 145 150 155 160 Gln Glu Ser Val Thr Glu
Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu 165 170 175 Ser Ser Thr Leu
Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val 180 185 190 Tyr Ala
Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys 195 200 205
Ser Phe Asn Arg Gly Glu Cys 210 215 <210> SEQ ID NO 337
<211> LENGTH: 444 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 337 Gln Ser Val Lys Glu Ser Gly
Gly Arg Leu Val Thr Pro Gly Thr Pro 1 5 10 15 Leu Thr Leu Thr Cys
Thr Val Ser Gly Ile Asp Leu Ser Ser Tyr Ala 20 25 30 Met Gly Trp
Phe Arg Gln Ala Pro Gly Lys Gly Leu Glu Tyr Ile Gly 35 40 45 Thr
Ile Asn Ile Gly Gly Arg Val Tyr Tyr Ala Ser Trp Ala Lys Gly 50 55
60 Arg Phe Thr Ile Ser Arg Thr Ser Thr Thr Val Asp Leu Lys Ala Pro
65 70 75 80 Ser Leu Thr Ala Glu Asp Thr Ala Thr Tyr Phe Cys Ala Arg
Tyr Tyr 85 90 95 Asn Gly Gly Ser Tyr Asp Ile Trp Gly Pro Gly Thr
Leu Val Thr Val 100 105 110 Ser Leu Ala Ser Thr Lys Gly Pro Ser Val
Phe Pro Leu Ala Pro Ser 115 120 125 Ser Lys Ser Thr Ser Gly Gly Thr
Ala Ala Leu Gly Cys Leu Val Lys 130 135 140 Asp Tyr Phe Pro Glu Pro
Val Thr Val Ser Trp Asn Ser Gly Ala Leu 145 150 155 160 Thr Ser Gly
Val His Thr Phe Pro Ala Val Leu Gln Ser Ser Gly Leu 165 170 175 Tyr
Ser Leu Ser Ser Val Val Thr Val Pro Ser Ser Ser Leu Gly Thr 180 185
190 Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro Ser Asn Thr Lys Val
195 200 205 Asp Lys Lys Val Glu Pro Lys Ser Cys Asp Lys Thr His Thr
Cys Pro 210 215 220 Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser
Val Phe Leu Phe 225 230 235 240 Pro Pro Lys Pro Lys Asp Thr Leu Met
Ile Ser Arg Thr Pro Glu Val 245 250 255 Thr Cys Val Val Val Asp Val
Ser His Glu Asp Pro Glu Val Lys Phe 260 265 270 Asn Trp Tyr Val Asp
Gly Val Glu Val His Asn Ala Lys Thr Lys Pro 275 280 285 Arg Glu Glu
Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr 290 295 300 Val
Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val 305 310
315 320 Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys
Ala 325 330 335 Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro
Pro Ser Arg 340 345 350 Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr
Cys Leu Val Lys Gly 355 360 365 Phe Tyr Pro Ser Asp Ile Ala Val Glu
Trp Glu Ser Asn Gly Gln Pro 370 375 380 Glu Asn Asn Tyr Lys Thr Thr
Pro Pro Val Leu Asp Ser Asp Gly Ser 385 390 395 400 Phe Phe Leu Tyr
Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln 405 410 415 Gly Asn
Val Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn His 420 425 430
Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 435 440 <210>
SEQ ID NO 338 <211> LENGTH: 217 <212> TYPE: PRT
<213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 338 Asp Val Val Met Thr
Gln Thr Pro Ala Ser Ala Ser Glu Pro Val Gly 1 5 10 15 Gly Thr Val
Thr Ile Lys Cys Gln Ala Ser Glu Ser Ile Tyr Arg Val 20 25 30 Leu
Ala Trp Tyr Gln Gln Lys Pro Gly Gln Pro Pro Lys Leu Leu Ile 35 40
45 Tyr Asp Thr Ser Thr Leu Ala Ser Gly Ala Pro Ser Arg Phe Lys Gly
50 55 60 Ser Gly Tyr Gly Thr Glu Phe Thr Leu Thr Ile Ser Gly Val
Gln Cys 65 70 75 80 Glu Asp Ala Ala Thr Tyr Tyr Cys Gln Gly Gly Tyr
Tyr Ala Asp Ser 85 90 95 Tyr Gly Ile Ala Phe Gly Gly Gly Thr Glu
Val Val Val Lys Arg Thr 100 105 110 Val Ala Ala Pro Ser Val Phe Ile
Phe Pro Pro Ser Asp Glu Gln Leu 115 120 125 Lys Ser Gly Thr Ala Ser
Val Val Cys Leu Leu Asn Asn Phe Tyr Pro 130 135 140 Arg Glu Ala Lys
Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly 145 150 155 160 Asn
Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr 165 170
175 Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His
180 185 190 Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser
Pro Val 195 200 205
Thr Lys Ser Phe Asn Arg Gly Glu Cys 210 215 <210> SEQ ID NO
339 <211> LENGTH: 447 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polypeptide <400> SEQUENCE: 339 Gln Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg Leu Ser
Cys Ser Ala Ser Gly Ile Asp Leu Ser Ser Tyr 20 25 30 Ala Met Gly
Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Tyr Ile 35 40 45 Gly
Thr Ile Asn Ile Gly Gly Arg Val Tyr Tyr Ala Ser Trp Ala Lys 50 55
60 Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr Leu
65 70 75 80 Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr
Cys Ala 85 90 95 Arg Tyr Tyr Asn Gly Gly Ser Tyr Asp Ile Trp Gly
Gln Gly Thr Leu 100 105 110 Val Thr Val Ser Ser Ala Ser Thr Lys Gly
Pro Ser Val Phe Pro Leu 115 120 125 Ala Pro Ser Ser Lys Ser Thr Ser
Gly Gly Thr Ala Ala Leu Gly Cys 130 135 140 Leu Val Lys Asp Tyr Phe
Pro Glu Pro Val Thr Val Ser Trp Asn Ser 145 150 155 160 Gly Ala Leu
Thr Ser Gly Val His Thr Phe Pro Ala Val Leu Gln Ser 165 170 175 Ser
Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser Ser Ser 180 185
190 Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro Ser Asn
195 200 205 Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp Lys
Thr His 210 215 220 Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly
Gly Pro Ser Val 225 230 235 240 Phe Leu Phe Pro Pro Lys Pro Lys Asp
Thr Leu Met Ile Ser Arg Thr 245 250 255 Pro Glu Val Thr Cys Val Val
Val Asp Val Ser His Glu Asp Pro Glu 260 265 270 Val Lys Phe Asn Trp
Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 275 280 285 Thr Lys Pro
Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser 290 295 300 Val
Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys 305 310
315 320 Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr
Ile 325 330 335 Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr
Thr Leu Pro 340 345 350 Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val
Ser Leu Thr Cys Leu 355 360 365 Val Lys Gly Phe Tyr Pro Ser Asp Ile
Ala Val Glu Trp Glu Ser Asn 370 375 380 Gly Gln Pro Glu Asn Asn Tyr
Lys Thr Thr Pro Pro Val Leu Asp Ser 385 390 395 400 Asp Gly Ser Phe
Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg 405 410 415 Trp Gln
Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu 420 425 430
His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 435 440
445 <210> SEQ ID NO 340 <211> LENGTH: 217 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polypeptide <400> SEQUENCE: 340 Asp Ile
Gln Met Thr Gln Ser Pro Ser Thr Leu Ser Ala Ser Val Gly 1 5 10 15
Asp Arg Val Thr Ile Thr Cys Gln Ala Ser Glu Ser Ile Tyr Arg Val 20
25 30 Leu Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu
Ile 35 40 45 Tyr Asp Thr Ser Thr Leu Ala Ser Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60 Ser Gly Ser Gly Thr Glu Phe Thr Leu Thr Ile
Ser Ser Leu Gln Cys 65 70 75 80 Asp Asp Ala Ala Thr Tyr Tyr Cys Gln
Gly Gly Tyr Tyr Ala Asp Ser 85 90 95 Tyr Gly Ile Ala Phe Gly Gly
Gly Thr Lys Val Glu Ile Lys Arg Thr 100 105 110 Val Ala Ala Pro Ser
Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu 115 120 125 Lys Ser Gly
Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro 130 135 140 Arg
Glu Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly 145 150
155 160 Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr
Tyr 165 170 175 Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr
Glu Lys His 180 185 190 Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly
Leu Ser Ser Pro Val 195 200 205 Thr Lys Ser Phe Asn Arg Gly Glu Cys
210 215 <210> SEQ ID NO 341 <211> LENGTH: 452
<212> TYPE: PRT <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polypeptide <400> SEQUENCE:
341 Gln Ser Val Glu Glu Ser Gly Gly Arg Leu Val Lys Pro Asp Glu Ser
1 5 10 15 Leu Thr Leu Thr Cys Thr Ala Ser Gly Phe Ser Leu Ser Ser
Tyr Ala 20 25 30 Met Ile Trp Val Arg Gln Ala Pro Gly Glu Gly Leu
Glu Trp Ile Gly 35 40 45 Thr Ile Ser Thr Gly Gly Ile Thr Tyr Tyr
Ala Ser Trp Ala Lys Gly 50 55 60 Arg Phe Thr Ile Ser Lys Thr Ser
Thr Thr Val Asp Leu Lys Ile Thr 65 70 75 80 Ser Pro Thr Thr Glu Asp
Thr Ala Thr Tyr Phe Cys Ala Arg Gly Gly 85 90 95 Tyr Ala Ala Ser
Ser Ala Tyr Tyr Leu Pro Tyr Tyr Phe Asp Leu Trp 100 105 110 Gly Gln
Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro 115 120 125
Ser Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr 130
135 140 Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val
Thr 145 150 155 160 Val Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val
His Thr Phe Pro 165 170 175 Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser
Leu Ser Ser Val Val Thr 180 185 190 Val Pro Ser Ser Ser Leu Gly Thr
Gln Thr Tyr Ile Cys Asn Val Asn 195 200 205 His Lys Pro Ser Asn Thr
Lys Val Asp Lys Lys Val Glu Pro Lys Ser 210 215 220 Cys Asp Lys Thr
His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu 225 230 235 240 Gly
Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu 245 250
255 Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser
260 265 270 His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly
Val Glu 275 280 285 Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln
Tyr Asn Ser Thr 290 295 300 Tyr Arg Val Val Ser Val Leu Thr Val Leu
His Gln Asp Trp Leu Asn 305 310 315 320 Gly Lys Glu Tyr Lys Cys Lys
Val Ser Asn Lys Ala Leu Pro Ala Pro 325 330 335 Ile Glu Lys Thr Ile
Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln 340 345 350 Val Tyr Thr
Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val 355 360 365 Ser
Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val 370 375
380 Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro
385 390 395 400 Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser
Lys Leu Thr 405 410 415 Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val
Phe Ser Cys Ser Val 420 425 430 Met His Glu Ala Leu His Asn His Tyr
Thr Gln Lys Ser Leu Ser Leu 435 440 445 Ser Pro Gly Lys 450
<210> SEQ ID NO 342 <211> LENGTH: 219 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 342 Ala Ala Val Leu Thr
Gln Thr Pro Ser Pro Val Ser Ala Ala Val Gly 1 5 10 15 Gly Thr Val
Thr Ile Ser Cys Gln Ser Ser Gln Ser Val Tyr Asn Asn 20 25 30 Asn
Asn Leu Ala Trp Phe Gln Gln Lys Pro Gly Gln Pro Pro Lys Leu 35 40
45 Leu Ile Tyr Leu Ala Ser Thr Leu Ala Ser Gly Val Pro Ser Arg Phe
50 55 60 Ser Gly Ser Gly Ser Gly Thr Gln Phe Thr Leu Thr Ile Ser
Gly Val 65 70 75 80 Gln Cys Asp Asp Ala Ala Thr Tyr Tyr Cys Leu Gly
Gly Cys Asp Asp 85 90 95 Asp Ala Asp Thr Phe Ala Phe Gly Gly Gly
Thr Glu Val Val Val Lys 100 105 110 Arg Thr Val Ala Ala Pro Ser Val
Phe Ile Phe Pro Pro Ser Asp Glu 115 120 125 Gln Leu Lys Ser Gly Thr
Ala Ser Val Val Cys Leu Leu Asn Asn Phe 130 135 140 Tyr Pro Arg Glu
Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln 145 150 155 160 Ser
Gly Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser 165 170
175 Thr Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu
180 185 190 Lys His Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu
Ser Ser 195 200 205 Pro Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 210
215 <210> SEQ ID NO 343 <211> LENGTH: 455 <212>
TYPE: PRT <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polypeptide <400> SEQUENCE: 343 Glu Val
Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15
Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Ser Leu Ser Ser Tyr 20
25 30 Ala Met Ile Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Ile 35 40 45 Gly Thr Ile Ser Thr Gly Gly Ile Thr Tyr Tyr Ala Ser
Trp Ala Lys 50 55 60 Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys
Asn Thr Leu Tyr Leu 65 70 75 80 Gln Met Asn Ser Leu Arg Ala Glu Asp
Thr Ala Val Tyr Tyr Cys Ala 85 90 95 Arg Gly Gly Tyr Ala Ala Ser
Ser Ala Tyr Tyr Leu Pro Tyr Tyr Phe 100 105 110 Asp Leu Trp Gly Gln
Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr 115 120 125 Lys Gly Pro
Ser Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser 130 135 140 Gly
Gly Thr Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu 145 150
155 160 Pro Val Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val
His 165 170 175 Thr Phe Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser
Leu Ser Ser 180 185 190 Val Val Thr Val Pro Ser Ser Ser Leu Gly Thr
Gln Thr Tyr Ile Cys 195 200 205 Asn Val Asn His Lys Pro Ser Asn Thr
Lys Val Asp Lys Lys Val Glu 210 215 220 Pro Lys Ser Cys Asp Lys Thr
His Thr Cys Pro Pro Cys Pro Ala Pro 225 230 235 240 Glu Leu Leu Gly
Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys 245 250 255 Asp Thr
Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val 260 265 270
Asp Val Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp 275
280 285 Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln
Tyr 290 295 300 Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu
His Gln Asp 305 310 315 320 Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys
Val Ser Asn Lys Ala Leu 325 330 335 Pro Ala Pro Ile Glu Lys Thr Ile
Ser Lys Ala Lys Gly Gln Pro Arg 340 345 350 Glu Pro Gln Val Tyr Thr
Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys 355 360 365 Asn Gln Val Ser
Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp 370 375 380 Ile Ala
Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys 385 390 395
400 Thr Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser
405 410 415 Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val
Phe Ser 420 425 430 Cys Ser Val Met His Glu Ala Leu His Asn His Tyr
Thr Gln Lys Ser 435 440 445 Leu Ser Leu Ser Pro Gly Lys 450 455
<210> SEQ ID NO 344 <211> LENGTH: 219 <212> TYPE:
PRT <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polypeptide <400> SEQUENCE: 344 Asp Ile Gln Met Thr
Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly 1 5 10 15 Asp Arg Val
Thr Ile Thr Cys Gln Ser Ser Gln Ser Val Tyr Asn Asn 20 25 30 Asn
Asn Leu Ala Trp Tyr Gln Gln Lys Pro Gly Lys Val Pro Lys Leu 35 40
45 Leu Ile Tyr Leu Ala Ser Thr Leu Ala Ser Gly Val Pro Ser Arg Phe
50 55 60 Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu 65 70 75 80 Gln Cys Glu Asp Ala Ala Thr Tyr Tyr Cys Leu Gly
Gly Cys Asp Asp 85 90 95 Asp Ala Asp Thr Phe Ala Phe Gly Gly Gly
Thr Lys Val Glu Ile Lys 100 105 110 Arg Thr Val Ala Ala Pro Ser Val
Phe Ile Phe Pro Pro Ser Asp Glu 115 120 125 Gln Leu Lys Ser Gly Thr
Ala Ser Val Val Cys Leu Leu Asn Asn Phe 130 135 140 Tyr Pro Arg Glu
Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln 145 150 155 160 Ser
Gly Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser 165 170
175 Thr Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu
180 185 190 Lys His Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu
Ser Ser 195 200 205 Pro Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 210
215 <210> SEQ ID NO 345 <211> LENGTH: 1350 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polynucleotide <400> SEQUENCE: 345
gaggtccaac tggtggagag cggtggaggt gttgtgcaac ctggccggtc cctgcgcctg
60 tcctgctccg catctggctt caccttcagc ggctatgggt tgtcttgggt
gagacaggca 120 cctggaaaag gtcttgagtg ggttgcaatg attagtagtg
gtggtagtta tacctactat 180 gcagacagtg tgaagggtag atttgcaata
tcgcgagaca acgccaagaa cacattgttc 240 ctgcaaatgg acagcctgag
acccgaagac accggggtct atttttgtgc aagacatggg 300 gacgatcccg
cctggttcgc ttattggggc caagggaccc cggtcaccgt ctcctcagcc 360
tccaccaagg gcccatcggt cttccccctg gcaccctcct ccaagagcac ctctgggggc
420 acagcggccc tgggctgcct ggtcaaggac tacttccccg aaccggtgac
ggtgtcgtgg 480 aactcaggcg ccctgaccag cggcgtgcac accttcccgg
ctgtcctaca gtcctcagga 540 ctctactccc tcagcagcgt ggtgaccgtg
ccctccagca gcttgggcac ccagacctac 600 atctgcaacg tgaatcacaa
gcccagcaac accaaggtgg acaagaaagt tgagcccaaa 660 tcttgtgaca
aaactcacac atgcccaccg tgcccagcac ctgaactcct ggggggaccg 720
tcagtcttcc tcttcccccc aaaacccaag gacaccctca tgatctcccg gacccctgag
780 gtcacatgcg tggtggtgga cgtgagccac gaagaccctg aggtcaagtt
caactggtac 840 gtggacggcg tggaggtgca taatgccaag acaaagccgc
gggaggagca gtacaacagc 900 acgtaccgtg tggtcagcgt cctcaccgtc
ctgcaccagg actggctgaa tggcaaggag 960
tacaagtgca aggtctccaa caaagccctc ccagccccca tcgagaaaac catctccaaa
1020 gccaaagggc agccccgaga accacaggtg tacaccctgc ccccatcccg
ggatgagctg 1080 accaagaacc aggtcagcct gacctgcctg gtcaaaggct
tctatcccag cgacatcgcc 1140 gtggagtggg agagcaatgg gcagccggag
aacaactaca agaccacgcc tcccgtgctg 1200 gactccgacg gctccttctt
cttatattca aagctcaccg tggacaagag caggtggcag 1260 caggggaacg
tcttctcatg ctccgtgatg catgaggctc tgcacaacca ctacacgcag 1320
aagagcctct ccctgtctcc cgggaaatga 1350 <210> SEQ ID NO 346
<211> LENGTH: 654 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 346 gacatccagc tgacccagag
cccaagcagc ctgagcgcca gcgtgggtga cagagtgacc 60 atcacctgta
gtgtcagctc aagtataagt tccaacaact tgcactggta ccagcagaag 120
ccaggtaagg ctccaaagcc atggatctac ggcacatcca acctggcttc tggtgtgcca
180 agcagattca gcggtagcgg tagcggtacc gactacacct tcaccatcag
cagcctccag 240 ccagaggaca tcgccaccta ctactgccaa cagtggagta
gttacccgta catgtacacg 300 ttcggccaag ggaccaaggt ggaaatcaaa
cgaactgtgg ctgcaccatc tgtcttcatc 360 ttcccgccat ctgatgagca
gttgaaatct ggaactgcct ctgttgtgtg cctgctgaat 420 aacttctatc
ccagagaggc caaagtacag tggaaggtgg ataacgccct ccaatcgggt 480
aactcccagg agagtgtcac agagcaggac agcaaggaca gcacctacag cctcagcagc
540 accctgacgc tgagcaaagc agactacgag aaacacaaag tctacgcctg
cgaagtcacc 600 catcagggcc tgagctcgcc cgtcacaaag agcttcaaca
ggggagagtg ttaa 654 <210> SEQ ID NO 347 <211> LENGTH:
1350 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polynucleotide
<400> SEQUENCE: 347 caggtacaac tgcagcagtc tgggcctgag
ctggagaagc ctggcgcttc agtgaagata 60 tcctgcaagg cttctggtta
ctcattcact ggctacacca tgaactgggt gaagcagagc 120 catggaaaga
gccttgagtg gattggactt attactcctt acaatggtgc ttctagctac 180
aaccagaagt tcaggggcaa ggccacatta actgtagaca agtcatccag cacagcctac
240 atggacctcc tcagtctgac atctgaagac tctgcagtct atttctgtgc
aagggggggt 300 tacgacggga ggggttttga ctactgggga tccgggaccc
cggtcaccgt ctcctcagcc 360 tccaccaagg gcccatcggt cttccccctg
gcaccctcct ccaagagcac ctctgggggc 420 acagcggccc tgggctgcct
ggtcaaggac tacttccccg aaccggtgac ggtgtcgtgg 480 aactcaggcg
ccctgaccag cggcgtgcac accttcccgg ctgtcctaca gtcctcagga 540
ctctactccc tcagcagcgt ggtgaccgtg ccctccagca gcttgggcac ccagacctac
600 atctgcaacg tgaatcacaa gcccagcaac accaaggtgg acaagaaagt
tgagcccaaa 660 tcttgtgaca aaactcacac atgcccaccg tgcccagcac
ctgaactcct ggggggaccg 720 tcagtcttcc tcttcccccc aaaacccaag
gacaccctca tgatctcccg gacccctgag 780 gtcacatgcg tggtggtgga
cgtgagccac gaagaccctg aggtcaagtt caactggtac 840 gtggacggcg
tggaggtgca taatgccaag acaaagccgc gggaggagca gtacaacagc 900
acgtaccgtg tggtcagcgt cctcaccgtc ctgcaccagg actggctgaa tggcaaggag
960 tacaagtgca aggtctccaa caaagccctc ccagccccca tcgagaaaac
catctccaaa 1020 gccaaagggc agccccgaga accacaggtg tacaccctgc
ccccatcccg ggatgagctg 1080 accaagaacc aggtcagcct gacctgcctg
gtcaaaggct tctatcccag cgacatcgcc 1140 gtggagtggg agagcaatgg
gcagccggag aacaactaca agaccacgcc tcccgtgctg 1200 gactccgacg
gctccttctt cctctacagc aagctcaccg tggacaagag caggtggcag 1260
caggggaacg tcttctcatg ctccgtgatg catgaggctc tgcacaacca ctacacgcag
1320 aagagcctct ccctgtctcc cgggaaatga 1350 <210> SEQ ID NO
348 <211> LENGTH: 642 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 348 gacatcgagc tcactcagtc
tccagcaatc atgtctgcat ctccagggga gaaggtcacc 60 atgacctgca
gtgccagctc aagtgtaagt tacatgcact ggtaccagca gaagtcaggc 120
acctccccca aaagatggat ttatgacaca tccaaactgg cttctggagt cccaggtcgc
180 ttcagtggca gtgggtctgg aaactcttac tctctcacaa tcagcagcgt
ggaggctgaa 240 gatgatgcaa cttattactg ccagcagtgg agtaagcacc
ctctcacgtt cggatccggg 300 accaaggtgg aaatcaaacg aactgtggct
gcaccatctg tcttcatctt cccgccatct 360 gatgagcagt tgaaatctgg
aactgcctct gttgtgtgcc tgctgaataa cttctatccc 420 agagaggcca
aagtacagtg gaaggtggat aacgccctcc aatcgggtaa ctcccaggag 480
agtgtcacag agcaggacag caaggacagc acctacagcc tcagcagcac cctgacgctg
540 agcaaagcag actacgagaa acacaaagtc tacgcctgcg aagtcaccca
tcagggcctg 600 agctcgcccg tcacaaagag cttcaacagg ggagagtgtt aa 642
<210> SEQ ID NO 349 <211> LENGTH: 1362 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polynucleotide <400> SEQUENCE: 349
cagtcggtgg aggagtccgg gggtcgcctg gtcacgcctg ggacacccct gacactcacc
60 tgcaccgtct ctggaatctc cctcagtagc gatgcaataa gctgggtccg
ccaggctcca 120 gggaaggggc tcgaatacat cggaatcatt aatggtggtg
gtaacacata ctacgcgagc 180 tgggcgaaag gccgattcac catctccaaa
acctcgacca cggtggatct gaaaatcacc 240 agtccgacaa ccgaggacac
ggccacctat ttctgtgcca gaggcattca acatggtggt 300 ggtaatagtg
attattatta ttacggcatg gacctctggg gcccaggcac cctggtcact 360
gtctcttcag catccaccaa gggcccatcg gtcttccccc tggcaccctc ctccaagagc
420 acctctgggg gcacagcggc cctgggctgc ctggtcaagg actacttccc
cgaaccggtg 480 acggtgtcgt ggaactcagg cgccctgacc agcggcgtgc
acaccttccc ggctgtccta 540 cagtcctcag gactctactc cctcagcagc
gtggtgaccg tgccctccag cagcttgggc 600 acccagacct acatctgcaa
cgtgaatcac aagcccagca acaccaaggt ggacaagaaa 660 gttgagccca
aatcttgtga caaaactcac acatgcccac cgtgcccagc acctgaactc 720
ctggggggac cgtcagtctt cctcttcccc ccaaaaccca aggacaccct catgatctcc
780 cggacccctg aggtcacatg cgtggtggtg gacgtgagcc acgaagaccc
tgaggtcaag 840 ttcaactggt acgtggacgg cgtggaggtg cataatgcca
agacaaagcc gcgggaggag 900 cagtacaaca gcacgtaccg tgtggtcagc
gtcctcaccg tcctgcacca ggactggctg 960 aatggcaagg agtacaagtg
caaggtctcc aacaaagccc tcccagcccc catcgagaaa 1020 accatctcca
aagccaaagg gcagccccga gaaccacagg tgtacaccct gcccccatcc 1080
cgggatgagc tgaccaagaa ccaggtcagc ctgacctgcc tggtcaaagg cttctatccc
1140 agcgacatcg ccgtggagtg ggagagcaat gggcagccgg agaacaacta
caagaccacg 1200 cctcccgtgc tggactccga cggctccttc ttcttatatt
caaagctcac cgtggacaag 1260 agcaggtggc agcaggggaa cgtcttctca
tgctccgtga tgcatgaggc tctgcacaac 1320 cactacacgc agaagagcct
ctccctgtct cccgggaaat ga 1362 <210> SEQ ID NO 350 <211>
LENGTH: 660 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polynucleotide
<400> SEQUENCE: 350 gaagtgttga tgacccagac tccatcctcc
gtgtctgcag ctgtgggaga cacagtcacc 60 atcaagtgcc aggccagtca
gagcattagt agtgtcttgt cctggtatca gcagaaacca 120 gggcagcctc
ccaagctcct gatctatctg gcatccactc tggcatctgg ggtcccatcg 180
cggttcagcg gcagtagatc tgggacagag ttcactctca ccatcagcga cctggagtgt
240 gacgatgctg ccacttacta ctgtcaaacc aattatggta ctagtagtag
taattatggt 300 tttgctttcg gcggagggac cgaggtggtc gtcaaacgaa
ctgtggctgc accatctgtc 360 ttcatcttcc cgccatctga tgagcagttg
aaatctggaa ctgcctctgt tgtgtgcctg 420 ctgaataact tctatcccag
agaggccaaa gtacagtgga aggtggataa cgccctccaa 480 tcgggtaact
cccaggagag tgtcacagag caggacagca aggacagcac ctacagcctc 540
agcagcaccc tgacgctgag caaagcagac tacgagaaac acaaagtcta cgcctgcgaa
600 gtcacccatc agggcctgag ctcgcccgtc acaaagagct tcaacagggg
agagtgttga 660 <210> SEQ ID NO 351 <211> LENGTH: 1371
<212> TYPE: DNA <213> ORGANISM: Artificial Sequence
<220> FEATURE: <223> OTHER INFORMATION: Description of
Artificial Sequence: Synthetic polynucleotide <400> SEQUENCE:
351 gaagtccaac tggtggaaag cgggggagga ctggtgcagc cgggcggatc
cctccggctg 60 tcatgtgctg catcgggaat ttccctctcc tccgacgcga
ttagctgggt cagacaggcc 120 cccggaaagg ggctggagta catcggtatc
atcaacggcg gcggaaacac ctactacgcc 180
tcctgggcca agggccgctt caccatctcg cggcataatt ccaagaacac tctgtacttg
240 caaatgaact ccctgagggc cgaggacacc gccgtgtact actgcgcgcg
cggcatccag 300 cacggtggtg gaaacagcga ctactactac tatgggatgg
atctgtgggg ccagggaact 360 cttgtgaccg tgtcgtcagc atccaccaag
ggcccatcgg tcttccccct ggcaccctcc 420 tccaagagca cctctggggg
cacagcggcc ctgggctgcc tggtcaagga ctacttcccc 480 gaaccggtga
cggtgtcgtg gaactcaggc gccctgacca gcggcgtgca caccttcccg 540
gctgtcctac agtcctcagg actctactcc ctcagcagcg tggtgaccgt gccctccagc
600 agcttgggca cccagaccta catctgcaac gtgaatcaca agcccagcaa
caccaaggtg 660 gacaagaaag ttgagcccaa atcttgtgac aaaactcaca
catgcccacc gtgcccagca 720 cctgaactcc tggggggacc gtcagtcttc
ctcttccccc caaaacccaa ggacaccctc 780 atgatctccc ggacccctga
ggtcacatgc gtggtggtgg acgtgagcca cgaagaccct 840 gaggtcaagt
tcaactggta cgtggacggc gtggaggtgc ataatgccaa gacaaagccg 900
cgggaggagc agtacaacag cacgtaccgt gtggtcagcg tcctcaccgt cctgcaccag
960 gactggctga atggcaagga gtacaagtgc aaggtctcca acaaagccct
cccagccccc 1020 atcgagaaaa ccatctccaa agccaaaggg cagccccgag
aaccacaggt gtacaccctg 1080 cccccatccc gggatgagct gaccaagaac
caggtcagcc tgacctgcct ggtcaaaggc 1140 ttctatccca gcgacatcgc
cgtggagtgg gagagcaatg ggcagccgga gaacaactac 1200 aagaccacgc
ctcccgtgct ggactccgac ggctccttct tcttatattc aaagctcacc 1260
gtggacaaga gcaggtggca gcaggggaac gtcttctcat gctccgtgat gcatgaggct
1320 ctgcacaacc actacacgca gaagagcctc tccctgtctc ccgggaaatg a 1371
<210> SEQ ID NO 352 <211> LENGTH: 660 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 352 gacattcaga
tgacccagtc cccaagctcg ctgtccgcct ccgtgggcga ccgcgtgacc 60
atcacgtgcc aggcgtccca gtcaattagc agcgtgctct cctggtacca acagaagccg
120 gggaaagcac ccaagctgct gatctacttg gcctccactc tggcctcggg
agtgccttca 180 cggttctccg gatcgggatc tggtactgat ttcaccctca
ccatctcgag ccttcagtgc 240 gaggacatcg ctacttacta ttgtcaaacc
aactacggaa cctccagctc caactacggc 300 tttgccttcg gtggcgggac
caaggtcgaa atcaaacgaa ctgtggctgc accatctgtc 360 ttcatcttcc
cgccatctga tgagcagttg aaatctggaa ctgcctctgt tgtgtgcctg 420
ctgaataact tctatcccag agaggccaaa gtacagtgga aggtggataa cgccctccaa
480 tcgggtaact cccaggagag tgtcacagag caggacagca aggacagcac
ctacagcctc 540 agcagcaccc tgacgctgag caaagcagac tacgagaaac
acaaagtcta cgcctgcgaa 600 gtcacccatc agggcctgag ctcgcccgtc
acaaagagct tcaacagggg agagtgttga 660 <210> SEQ ID NO 353
<211> LENGTH: 1347 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 353 cagtcggtgg aggagtccgg
gggtcgcctg gtcacgcctg ggacacccct gacactcacc 60 tgcacagtct
ctggattctc cctcaataac tatgcaatga gctgggtccg ccaggctcca 120
gggaaggggc tggaatggat cggatccatt agtactggtg gtctcgcatt ctacgcgaac
180 tgggcaaaag gccgattcac catctccaga acctcgacca cggtggatct
gaaaatgacc 240 agtctgacaa ccgaggacac ggccacctat ttctgtggca
gaaatggtgg tggtagttat 300 attttctatt attttgactt gtggggccaa
ggcaccctcg tcactgtctc ttcagcatcc 360 accaagggcc catcggtctt
ccccctggca ccctcctcca agagcacctc tgggggcaca 420 gcggccctgg
gctgcctggt caaggactac ttccccgaac cggtgacggt gtcgtggaac 480
tcaggcgccc tgaccagcgg cgtgcacacc ttcccggctg tcctacagtc ctcaggactc
540 tactccctca gcagcgtggt gaccgtgccc tccagcagct tgggcaccca
gacctacatc 600 tgcaacgtga atcacaagcc cagcaacacc aaggtggaca
agaaagttga gcccaaatct 660 tgtgacaaaa ctcacacatg cccaccgtgc
ccagcacctg aactcctggg gggaccgtca 720 gtcttcctct tccccccaaa
acccaaggac accctcatga tctcccggac ccctgaggtc 780 acatgcgtgg
tggtggacgt gagccacgaa gaccctgagg tcaagttcaa ctggtacgtg 840
gacggcgtgg aggtgcataa tgccaagaca aagccgcggg aggagcagta caacagcacg
900 taccgtgtgg tcagcgtcct caccgtcctg caccaggact ggctgaatgg
caaggagtac 960 aagtgcaagg tctccaacaa agccctccca gcccccatcg
agaaaaccat ctccaaagcc 1020 aaagggcagc cccgagaacc acaggtgtac
accctgcccc catcccggga tgagctgacc 1080 aagaaccagg tcagcctgac
ctgcctggtc aaaggcttct atcccagcga catcgccgtg 1140 gagtgggaga
gcaatgggca gccggagaac aactacaaga ccacgcctcc cgtgctggac 1200
tccgacggct ccttcttctt atattcaaag ctcaccgtgg acaagagcag gtggcagcag
1260 gggaacgtct tctcatgctc cgtgatgcat gaggctctgc acaaccacta
cacgcagaag 1320 agcctctccc tgtctcccgg gaaatga 1347 <210> SEQ
ID NO 354 <211> LENGTH: 648 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 354 gcattcgaat tgacccagac
tccatcctcc gtggaggcag ctgtgggagg cacaatcacc 60 atcaagtgcc
aggccagtca gagcattagt agttacttat cctggtatca gcagaaacca 120
gggcagcctc ccaagctcct gatctattct gcatccactc tggcatctgg ggtctcatcg
180 cggttcaaag gcagtggatc tgggacagag tacactctca ccatcagcga
cctggagtgt 240 gccgatgctg ccacttactt ctgtcaaagc tattatgata
ttggtactag tactttcggc 300 ggagggaccg aggtggtcgt caaacgaact
gtggctgcac catctgtctt catcttcccg 360 ccatctgatg agcagttgaa
atctggaact gcctctgttg tgtgcctgct gaataacttc 420 tatcccagag
aggccaaagt acagtggaag gtggataacg ccctccaatc gggtaactcc 480
caggagagtg tcacagagca ggacagcaag gacagcacct acagcctcag cagcaccctg
540 acgctgagca aagcagacta cgagaaacac aaagtctacg cctgcgaagt
cacccatcag 600 ggcctgagct cgcccgtcac aaagagcttc aacaggggag agtgttga
648 <210> SEQ ID NO 355 <211> LENGTH: 1356 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polynucleotide <400> SEQUENCE: 355
gaagtgcagc tggtggaatc tggcggcgga ctggtgcagc ctggcggatc tctgagactg
60 tcttgtgccg cctccggctt ctccctgaac aactacgcca tgtcctgggt
gcgacaggcc 120 cctggcaaag gcctggaatg gatcggctcc atcagcacag
gcggcctggc cttctacgcc 180 aattgggcca agggccggtt caccatcagc
cgggacaact ccaagaacac cctgtacctc 240 cagatgaact ccctgcgggc
cgaggacacc gccgtgtact actgtgccag aaacggcgga 300 ggctcctaca
tcttctacta cttcgacctg tggggccagg gcaccctcgt gacagtgtca 360
tctgcatcca ccaagggccc atcggtcttc cccctggcac cctcctccaa gagcacctct
420 gggggcacag cggccctggg ctgcctggtc aaggactact tccccgaacc
ggtgacggtg 480 tcgtggaact caggcgccct gaccagcggc gtgcacacct
tcccggctgt cctacagtcc 540 tcaggactct actccctcag cagcgtggtg
accgtgccct ccagcagctt gggcacccag 600 acctacatct gcaacgtgaa
tcacaagccc agcaacacca aggtggacaa gaaagttgag 660 cccaaatctt
gtgacaaaac tcacacatgc ccaccgtgcc cagcacctga actcctgggg 720
ggaccgtcag tcttcctctt ccccccaaaa cccaaggaca ccctcatgat ctcccggacc
780 cctgaggtca catgcgtggt ggtggacgtg agccacgaag accctgaggt
caagttcaac 840 tggtacgtgg acggcgtgga ggtgcataat gccaagacaa
agccgcggga ggagcagtac 900 aacagcacgt accgtgtggt cagcgtcctc
accgtcctgc accaggactg gctgaatggc 960 aaggagtaca agtgcaaggt
ctccaacaaa gccctcccag cccccatcga gaaaaccatc 1020 tccaaagcca
aagggcagcc ccgagaacca caggtgtaca ccctgccccc atcccgggat 1080
gagctgacca agaaccaggt cagcctgacc tgcctggtca aaggcttcta tcccagcgac
1140 atcgccgtgg agtgggagag caatgggcag ccggagaaca actacaagac
cacgcctccc 1200 gtgctggact ccgacggctc cttcttctta tattcaaagc
tcaccgtgga caagagcagg 1260 tggcagcagg ggaacgtctt ctcatgctcc
gtgatgcatg aggctctgca caaccactac 1320 acgcagaaga gcctctccct
gtctcccggg aaatga 1356 <210> SEQ ID NO 356 <211>
LENGTH: 648 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polynucleotide
<400> SEQUENCE: 356 gatattcaga tgacccagtc cccctccagc
ctgtccgctt ctgtgggcga cagagtgacc 60 atcacctgtc aggcctccca
gtccatctcc tcctacctgt cctggtatca gcagaagccc 120 ggcaaggccc
ccaagctgct gatctactct gcctccacac tggcctccgg cgtgccctct 180
agattctccg gctctggctc tggcaccgac tttaccctga ccatcagctc cctccagtgc
240 gaggatgccg ccacctacta ctgccagtcc tactacgaca tcggcacctc
caccttcggc 300 ggaggcacca aggtggaaat caaacgaact gtggctgcac
catctgtctt catcttcccg 360 ccatctgatg agcagttgaa atctggaact
gcctctgttg tgtgcctgct gaataacttc 420 tatcccagag aggccaaagt
acagtggaag gtggataacg ccctccaatc gggtaactcc 480
caggagagtg tcacagagca ggacagcaag gacagcacct acagcctcag cagcaccctg
540 acgctgagca aagcagacta cgagaaacac aaagtctacg cctgcgaagt
cacccatcag 600 ggcctgagct cgcccgtcac aaagagcttc aacaggggag agtgttga
648 <210> SEQ ID NO 357 <211> LENGTH: 1335 <212>
TYPE: DNA <213> ORGANISM: Artificial Sequence <220>
FEATURE: <223> OTHER INFORMATION: Description of Artificial
Sequence: Synthetic polynucleotide <400> SEQUENCE: 357
cagtcagtga aggagtccgg gggtcgcctg gtcacgcctg ggacacccct gacactcacc
60 tgcacagtct ctggaatcga cctcagtagc tatgcaatgg gctggttccg
ccaggctcca 120 gggaaggggc tggaatacat cggaaccatt aatattggtg
gtcgcgtata ttacgcgagc 180 tgggcaaaag gccgattcac catctccaga
acctcgacca cggtggatct gaaagcgccc 240 agtctgacag ccgaggacac
ggccacctat ttctgtgcca gatattataa tggtggtagt 300 tatgacatct
ggggcccagg caccctggtc accgtctctt tagcatccac caagggccca 360
tcggtcttcc ccctggcacc ctcctccaag agcacctctg ggggcacagc ggccctgggc
420 tgcctggtca aggactactt ccccgaaccg gtgacggtgt cgtggaactc
aggcgccctg 480 accagcggcg tgcacacctt cccggctgtc ctacagtcct
caggactcta ctccctcagc 540 agcgtggtga ccgtgccctc cagcagcttg
ggcacccaga cctacatctg caacgtgaat 600 cacaagccca gcaacaccaa
ggtggacaag aaagttgagc ccaaatcttg tgacaaaact 660 cacacatgcc
caccgtgccc agcacctgaa ctcctggggg gaccgtcagt cttcctcttc 720
cccccaaaac ccaaggacac cctcatgatc tcccggaccc ctgaggtcac atgcgtggtg
780 gtggacgtga gccacgaaga ccctgaggtc aagttcaact ggtacgtgga
cggcgtggag 840 gtgcataatg ccaagacaaa gccgcgggag gagcagtaca
acagcacgta ccgtgtggtc 900 agcgtcctca ccgtcctgca ccaggactgg
ctgaatggca aggagtacaa gtgcaaggtc 960 tccaacaaag ccctcccagc
ccccatcgag aaaaccatct ccaaagccaa agggcagccc 1020 cgagaaccac
aggtgtacac cctgccccca tcccgggatg agctgaccaa gaaccaggtc 1080
agcctgacct gcctggtcaa aggcttctat cccagcgaca tcgccgtgga gtgggagagc
1140 aatgggcagc cggagaacaa ctacaagacc acgcctcccg tgctggactc
cgacggctcc 1200 ttcttcttat attcaaagct caccgtggac aagagcaggt
ggcagcaggg gaacgtcttc 1260 tcatgctccg tgatgcatga ggctctgcac
aaccactaca cgcagaagag cctctccctg 1320 tctcccggga aatga 1335
<210> SEQ ID NO 358 <211> LENGTH: 654 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 358 gatgttgtga
tgacccagac tccagcctcc gcgtctgaac ctgtgggagg cacagtcacc 60
atcaagtgcc aggccagtga gagcatttat cgcgtattgg cctggtatca gcagaaacca
120 gggcagcctc ccaagctcct gatctatgat acatccactc tggcatctgg
ggccccatcg 180 cggttcaaag gcagtggata tgggacagag ttcactctca
ccatcagcgg cgtgcagtgt 240 gaagatgctg ccacttacta ctgtcaaggc
ggttattatg ctgatagtta tggtattgct 300 ttcggcggag ggaccgaggt
ggtggtcaaa cgaactgtgg ctgcaccatc tgtcttcatc 360 ttcccgccat
ctgatgagca gttgaaatct ggaactgcct ctgttgtgtg cctgctgaat 420
aacttctatc ccagagaggc caaagtacag tggaaggtgg ataacgccct ccaatcgggt
480 aactcccagg agagtgtcac agagcaggac agcaaggaca gcacctacag
cctcagcagc 540 accctgacgc tgagcaaagc agactacgag aaacacaaag
tctacgcctg cgaagtcacc 600 catcagggcc tgagctcgcc cgtcacaaag
agcttcaaca ggggagagtg ttga 654 <210> SEQ ID NO 359
<211> LENGTH: 1344 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 359 caggtgcagc tggtggaatc
tggcggagga ctggtgcagc ctggcggctc tctgagactg 60 tcctgttccg
cctccggaat cgacctgtcc tcctacgcta tgggctgggt gcgacaggct 120
cctggcaagg gcctggagta catcggcacc atcaacatcg gcggcagagt gtactacgcc
180 tcctgggcca agggccggtt caccatctcc agagacaact ccaagaacac
cctgtacctc 240 cagatgaact ccctgcgggc cgaggacacc gccgtgtact
actgcgcccg gtactacaac 300 ggcggctcct acgatatctg gggccagggc
acactcgtga ccgtgtcctc tgcatccacc 360 aagggcccat cggtcttccc
cctggcaccc tcctccaaga gcacctctgg gggcacagcg 420 gccctgggct
gcctggtcaa ggactacttc cccgaaccgg tgacggtgtc gtggaactca 480
ggcgccctga ccagcggcgt gcacaccttc ccggctgtcc tacagtcctc aggactctac
540 tccctcagca gcgtggtgac cgtgccctcc agcagcttgg gcacccagac
ctacatctgc 600 aacgtgaatc acaagcccag caacaccaag gtggacaaga
aagttgagcc caaatcttgt 660 gacaaaactc acacatgccc accgtgccca
gcacctgaac tcctgggggg accgtcagtc 720 ttcctcttcc ccccaaaacc
caaggacacc ctcatgatct cccggacccc tgaggtcaca 780 tgcgtggtgg
tggacgtgag ccacgaagac cctgaggtca agttcaactg gtacgtggac 840
ggcgtggagg tgcataatgc caagacaaag ccgcgggagg agcagtacaa cagcacgtac
900 cgtgtggtca gcgtcctcac cgtcctgcac caggactggc tgaatggcaa
ggagtacaag 960 tgcaaggtct ccaacaaagc cctcccagcc cccatcgaga
aaaccatctc caaagccaaa 1020 gggcagcccc gagaaccaca ggtgtacacc
ctgcccccat cccgggatga gctgaccaag 1080 aaccaggtca gcctgacctg
cctggtcaaa ggcttctatc ccagcgacat cgccgtggag 1140 tgggagagca
atgggcagcc ggagaacaac tacaagacca cgcctcccgt gctggactcc 1200
gacggctcct tcttcttata ttcaaagctc accgtggaca agagcaggtg gcagcagggg
1260 aacgtcttct catgctccgt gatgcatgag gctctgcaca accactacac
gcagaagagc 1320 ctctccctgt ctcccgggaa atga 1344 <210> SEQ ID
NO 360 <211> LENGTH: 654 <212> TYPE: DNA <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 360 gatatccaga tgacccagtc
cccctccacc ctgtctgcct ctgtgggcga cagagtgacc 60 atcacctgtc
aggcctccga gtccatctac cgggtgctgg cctggtatca gcagaagcct 120
ggcaaggccc ccaagctgct gatctacgac accagcacac tggcctccgg cgtgccctct
180 agattctccg gctctggctc tggcaccgag tttaccctga ccatctccag
cctccagtgc 240 gacgacgccg ccacctacta ttgtcagggc ggctactacg
ccgactccta cggaatcgct 300 ttcggcggag gcaccaaggt ggaaatcaaa
cgaactgtgg ctgcaccatc tgtcttcatc 360 ttcccgccat ctgatgagca
gttgaaatct ggaactgcct ctgttgtgtg cctgctgaat 420 aacttctatc
ccagagaggc caaagtacag tggaaggtgg ataacgccct ccaatcgggt 480
aactcccagg agagtgtcac agagcaggac agcaaggaca gcacctacag cctcagcagc
540 accctgacgc tgagcaaagc agactacgag aaacacaaag tctacgcctg
cgaagtcacc 600 catcagggcc tgagctcgcc cgtcacaaag agcttcaaca
ggggagagtg ttga 654 <210> SEQ ID NO 361 <211> LENGTH:
1359 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polynucleotide
<400> SEQUENCE: 361 cagtcggtgg aggagtccgg cggtcgcctg
gtaaagcctg acgaatccct gacactcacc 60 tgcacagcct ctggattctc
cctcagtagt tatgcaatga tctgggtccg ccaggctcca 120 ggggaggggc
tggaatggat cggaaccatt agtactggtg gtatcacata ctacgcgagc 180
tgggcgaaag gccgattcac catctccaaa acctcgacca cggtggatct gaaaatcacc
240 agtccgacaa ccgaggacac ggccacctat ttctgtgcca gagggggata
tgctgctagt 300 agtgcttatt atctcccgta ctactttgac ttgtggggcc
aagggaccct ggtcaccgtc 360 tcctcagcat ccaccaaggg cccatcggtc
ttccccctgg caccctcctc caagagcacc 420 tctgggggca cagcggccct
gggctgcctg gtcaaggact acttccccga accggtgacg 480 gtgtcgtgga
actcaggcgc cctgaccagc ggcgtgcaca ccttcccggc tgtcctacag 540
tcctcaggac tctactccct cagcagcgtg gtgaccgtgc cctccagcag cttgggcacc
600 cagacctaca tctgcaacgt gaatcacaag cccagcaaca ccaaggtgga
caagaaagtt 660 gagcccaaat cttgtgacaa aactcacaca tgcccaccgt
gcccagcacc tgaactcctg 720 gggggaccgt cagtcttcct cttcccccca
aaacccaagg acaccctcat gatctcccgg 780 acccctgagg tcacatgcgt
ggtggtggac gtgagccacg aagaccctga ggtcaagttc 840 aactggtacg
tggacggcgt ggaggtgcat aatgccaaga caaagccgcg ggaggagcag 900
tacaacagca cgtaccgtgt ggtcagcgtc ctcaccgtcc tgcaccagga ctggctgaat
960 ggcaaggagt acaagtgcaa ggtctccaac aaagccctcc cagcccccat
cgagaaaacc 1020 atctccaaag ccaaagggca gccccgagaa ccacaggtgt
acaccctgcc cccatcccgg 1080 gatgagctga ccaagaacca ggtcagcctg
acctgcctgg tcaaaggctt ctatcccagc 1140 gacatcgccg tggagtggga
gagcaatggg cagccggaga acaactacaa gaccacgcct 1200 cccgtgctgg
actccgacgg ctccttcttc ttatattcaa agctcaccgt ggacaagagc 1260
aggtggcagc aggggaacgt cttctcatgc tccgtgatgc atgaggctct gcacaaccac
1320 tacacgcaga agagcctctc cctgtctccc gggaaatga 1359 <210>
SEQ ID NO 362
<211> LENGTH: 660 <212> TYPE: DNA <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic
polynucleotide <400> SEQUENCE: 362 gcagccgtgc tgacccagac
accatcaccc gtgtctgcag ctgtgggagg cacagtcacc 60 atcagttgcc
agtccagtca gagtgtttat aataataaca acttagcctg gtttcagcag 120
aaacccgggc agcctcccaa gcttctgatc tatctggcat ccactctggc atctggggtc
180 ccatcacggt tcagcggcag tggatctggg acacagttca ctctcaccat
cagcggcgtg 240 cagtgtgacg atgctgccac ttattactgt ctaggtggtt
gtgatgatga tgctgatact 300 tttgctttcg gcggagggac tgaggtggtg
gtcaaacgaa ctgtggctgc accatctgtc 360 ttcatcttcc cgccatctga
tgagcagttg aaatctggaa ctgcctctgt tgtgtgcctg 420 ctgaataact
tctatcccag agaggccaaa gtacagtgga aggtggataa cgccctccaa 480
tcgggtaact cccaggagag tgtcacagag caggacagca aggacagcac ctacagcctc
540 agcagcaccc tgacgctgag caaagcagac tacgagaaac acaaagtcta
cgcctgcgaa 600 gtcacccatc agggcctgag ctcgcccgtc acaaagagct
tcaacagggg agagtgttga 660 <210> SEQ ID NO 363 <211>
LENGTH: 1368 <212> TYPE: DNA <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic polynucleotide
<400> SEQUENCE: 363 gaagtgcagc tggtggaatc tggcggcgga
ctggtgcagc ctggcggatc tctgagactg 60 tcttgtgccg cctccggctt
ctccctgtcc tcctacgcta tgatctgggt gcgacaggcc 120 cctggcaagg
gcctggaatg gatcggcacc atctctaccg gcggaattac ctactacgcc 180
tcctgggcca agggccggtt caccatctcc agagacaact ccaagaacac cctgtacctc
240 cagatgaact ccctgcgggc cgaggacacc gccgtgtact attgtgctag
aggcggctac 300 gccgccagct ccgcttacta cctgccctac tacttcgacc
tgtggggcca gggcaccctc 360 gtgacagtgt catctgcatc caccaagggc
ccatcggtct tccccctggc accctcctcc 420 aagagcacct ctgggggcac
agcggccctg ggctgcctgg tcaaggacta cttccccgaa 480 ccggtgacgg
tgtcgtggaa ctcaggcgcc ctgaccagcg gcgtgcacac cttcccggct 540
gtcctacagt cctcaggact ctactccctc agcagcgtgg tgaccgtgcc ctccagcagc
600 ttgggcaccc agacctacat ctgcaacgtg aatcacaagc ccagcaacac
caaggtggac 660 aagaaagttg agcccaaatc ttgtgacaaa actcacacat
gcccaccgtg cccagcacct 720 gaactcctgg ggggaccgtc agtcttcctc
ttccccccaa aacccaagga caccctcatg 780 atctcccgga cccctgaggt
cacatgcgtg gtggtggacg tgagccacga agaccctgag 840 gtcaagttca
actggtacgt ggacggcgtg gaggtgcata atgccaagac aaagccgcgg 900
gaggagcagt acaacagcac gtaccgtgtg gtcagcgtcc tcaccgtcct gcaccaggac
960 tggctgaatg gcaaggagta caagtgcaag gtctccaaca aagccctccc
agcccccatc 1020 gagaaaacca tctccaaagc caaagggcag ccccgagaac
cacaggtgta caccctgccc 1080 ccatcccggg atgagctgac caagaaccag
gtcagcctga cctgcctggt caaaggcttc 1140 tatcccagcg acatcgccgt
ggagtgggag agcaatgggc agccggagaa caactacaag 1200 accacgcctc
ccgtgctgga ctccgacggc tccttcttct tatattcaaa gctcaccgtg 1260
gacaagagca ggtggcagca ggggaacgtc ttctcatgct ccgtgatgca tgaggctctg
1320 cacaaccact acacgcagaa gagcctctcc ctgtctcccg ggaaatga 1368
<210> SEQ ID NO 364 <211> LENGTH: 660 <212> TYPE:
DNA <213> ORGANISM: Artificial Sequence <220> FEATURE:
<223> OTHER INFORMATION: Description of Artificial Sequence:
Synthetic polynucleotide <400> SEQUENCE: 364 gatattcaga
tgacccagtc cccctccagc ctgtccgctt ctgtgggcga cagagtgacc 60
atcacctgtc agtcctccca gtccgtgtat aacaacaaca acctggcctg gtatcagcag
120 aaacccggca aggtgcccaa gctgctgatc tacctggcct ccacactggc
ctctggcgtg 180 ccctctagat tctccggctc tggctctggc accgacttta
ccctgaccat cagctccctc 240 cagtgcgagg atgccgccac ctactattgc
ctgggcggct gcgacgacga cgccgatacc 300 tttgcttttg gcggaggcac
caaggtggaa atcaaacgaa ctgtggctgc accatctgtc 360 ttcatcttcc
cgccatctga tgagcagttg aaatctggaa ctgcctctgt tgtgtgcctg 420
ctgaataact tctatcccag agaggccaaa gtacagtgga aggtggataa cgccctccaa
480 tcgggtaact cccaggagag tgtcacagag caggacagca aggacagcac
ctacagcctc 540 agcagcaccc tgacgctgag caaagcagac tacgagaaac
acaaagtcta cgcctgcgaa 600 gtcacccatc agggcctgag ctcgcccgtc
acaaagagct tcaacagggg agagtgttga 660 <210> SEQ ID NO 365
<211> LENGTH: 4 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 365 Ala His Lys Asp 1 <210> SEQ ID NO
366 <211> LENGTH: 13 <212> TYPE: PRT <213>
ORGANISM: Artificial Sequence <220> FEATURE: <223>
OTHER INFORMATION: Description of Artificial Sequence: Synthetic
peptide <400> SEQUENCE: 366 Asn Thr Ser Gln Glu Ala His Lys
Asp Val Ser Tyr Leu 1 5 10 <210> SEQ ID NO 367 <211>
LENGTH: 4 <212> TYPE: PRT <213> ORGANISM: Artificial
Sequence <220> FEATURE: <223> OTHER INFORMATION:
Description of Artificial Sequence: Synthetic peptide <400>
SEQUENCE: 367 Gly Phe Leu Gly 1 <210> SEQ ID NO 368
<211> LENGTH: 4 <212> TYPE: PRT <213> ORGANISM:
Artificial Sequence <220> FEATURE: <223> OTHER
INFORMATION: Description of Artificial Sequence: Synthetic peptide
<400> SEQUENCE: 368 Ala Leu Ala Leu 1










































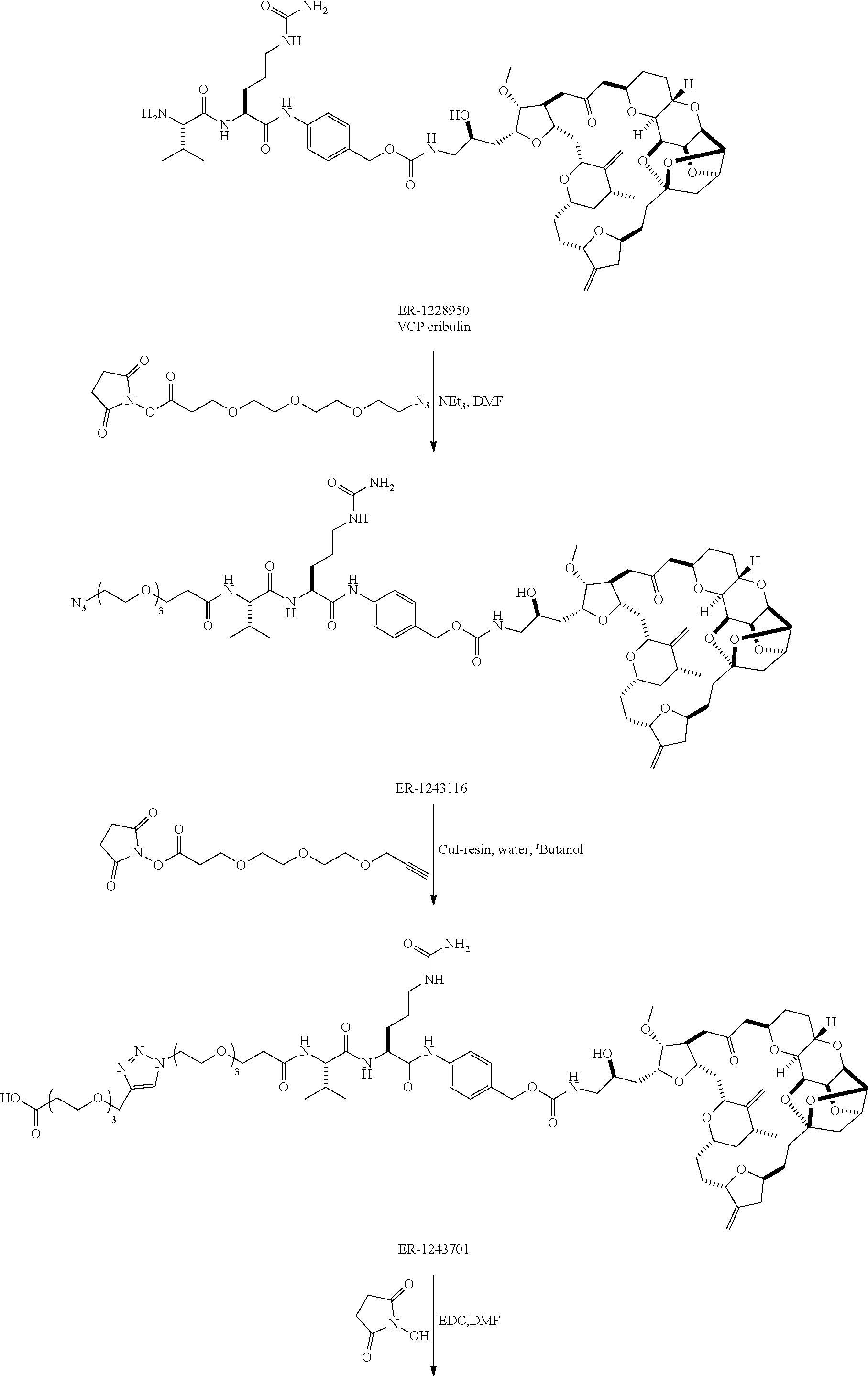


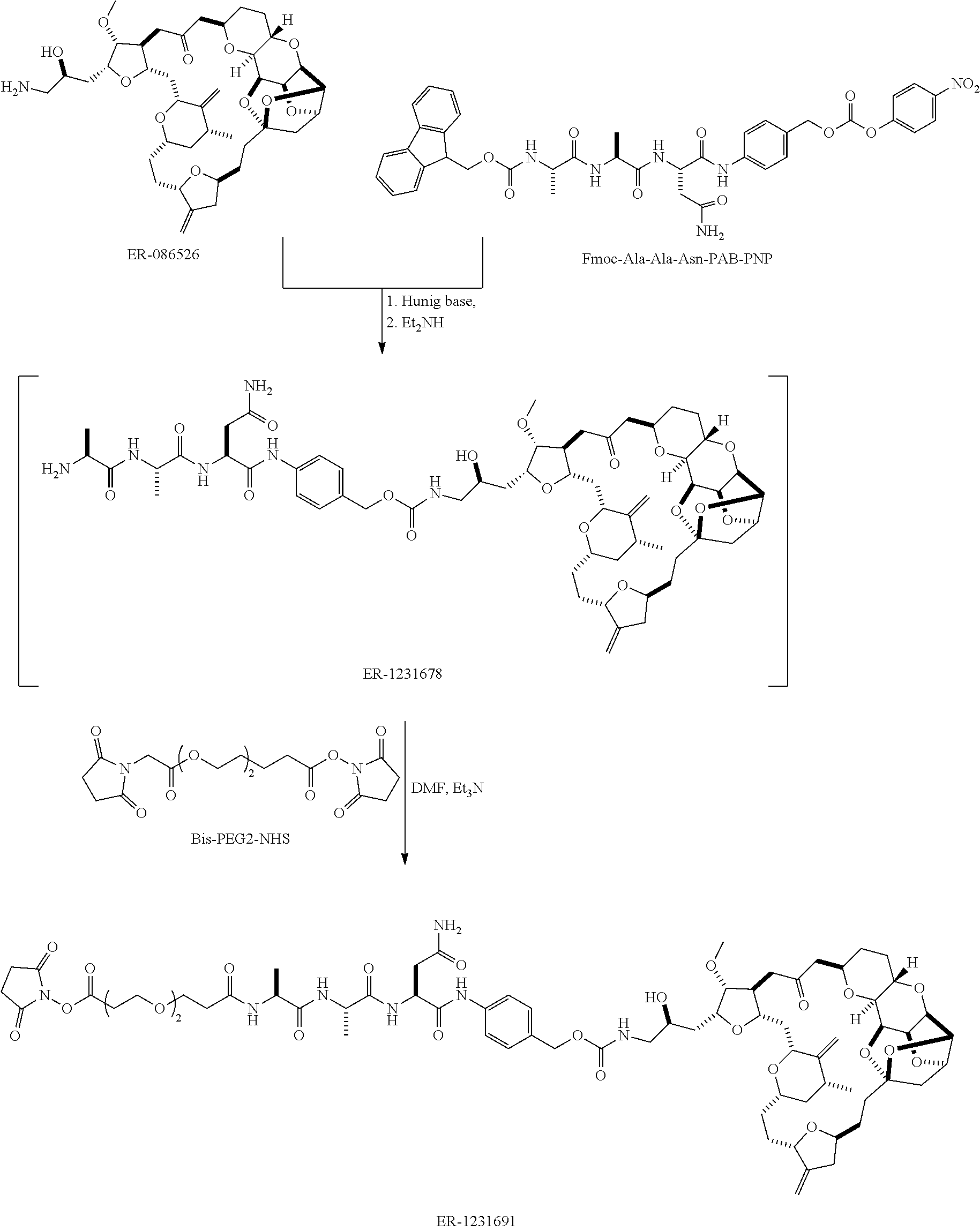







D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014

D00015

D00016

D00017

D00018

D00019

D00020

D00021

D00022

D00023

D00024
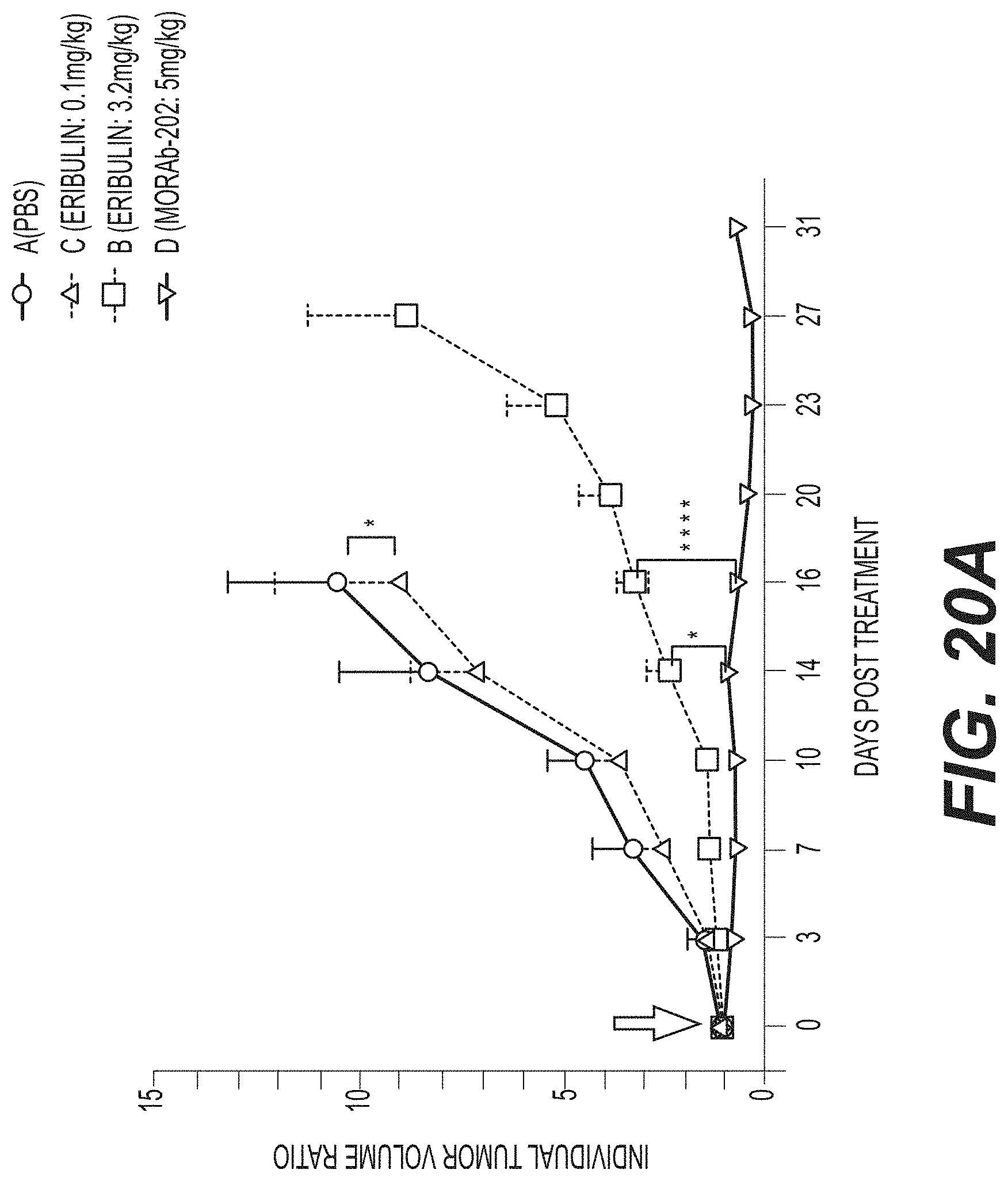
D00025

D00026

D00027

D00028

D00029

D00030

D00031

D00032

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.