Novel Metalloproteases
BABE; LILIA M. ; et al.
U.S. patent application number 16/881226 was filed with the patent office on 2020-09-17 for novel metalloproteases. The applicant listed for this patent is DANISCO US INC.. Invention is credited to LILIA M. BABE, ROOPA GHIRNIKAR, FRITS GOEDEGEBUUR, XIAOGANG GU, MARC KOLKMAN, JIAN YAO.
| Application Number | 20200291374 16/881226 |
| Document ID | / |
| Family ID | 1000004860225 |
| Filed Date | 2020-09-17 |











View All Diagrams
| United States Patent Application | 20200291374 |
| Kind Code | A1 |
| BABE; LILIA M. ; et al. | September 17, 2020 |
NOVEL METALLOPROTEASES
Abstract
Aspects of the present compositions and methods relate to novel metalloproteases, polynucleotides encoding the novel metalloproteases, and compositions and methods for use thereof.
| Inventors: | BABE; LILIA M.; (Emerald Hills, CA) ; GOEDEGEBUUR; FRITS; (VLAARDINGEN, NL) ; GHIRNIKAR; ROOPA; (SUNNYVALE, CA) ; GU; XIAOGANG; (SHANGHAI, CN) ; KOLKMAN; MARC; (OEGSTGEEST, NL) ; YAO; JIAN; (SUNNYVALE, CA) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 1000004860225 | ||||||||||
| Appl. No.: | 16/881226 | ||||||||||
| Filed: | May 22, 2020 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 15835551 | Dec 8, 2017 | 10696958 | ||
| 16881226 | ||||
| 14893473 | Nov 23, 2015 | |||
| PCT/US2014/039928 | May 29, 2014 | |||
| 15835551 | ||||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C12Y 304/24 20130101; C11D 3/38681 20130101; C12N 9/52 20130101; C11D 3/386 20130101; C12N 9/54 20130101; C12N 9/485 20130101 |
| International Class: | C12N 9/52 20060101 C12N009/52; C12N 9/48 20060101 C12N009/48; C12N 9/54 20060101 C12N009/54; C11D 3/386 20060101 C11D003/386 |
Foreign Application Data
| Date | Code | Application Number |
|---|---|---|
| May 29, 2013 | CN | 13/076384 |
| May 29, 2013 | CN | 13/076387 |
| May 29, 2013 | CN | 13/076398 |
| May 29, 2013 | CN | 13/076401 |
| May 29, 2013 | CN | 13/076406 |
| May 29, 2013 | CN | 13/076414 |
| May 29, 2013 | CN | 13/076415 |
| May 29, 2013 | CN | 13/076419 |
Claims
1. A polypeptide comprising an amino acid sequence having at least 60% sequence identity to the amino acid sequence selected from the group consisting of SEQ ID NOs: 3, 8, 13, 18, 23, 28, 33 and 38.
2. The polypeptide of claim 1, wherein said polypeptide has at least 80% sequence identity to the amino acid sequence selected from the group consisting of SEQ ID NOs: 3, 8, 13, 18, 23, 28, 33 and 38.
3. The polypeptide of any of claim 1 or 2, wherein said polypeptide has at least 95% sequence identity to the amino acid sequence selected from the group consisting of SEQ ID NOs: 3, 8, 13, 18, 23, 28, 33 and 38.
4. The polypeptide of any of the above claims, wherein said amino acid sequence is the amino acid sequence selected from the group consisting of SEQ ID NOs: 3, 8, 13, 18, 23, 28, 33 and 38.
5. The polypeptide of any of the above claims, wherein said polypeptide is derived from a member of the order Bacillales.
6. The polypeptide of any of the above claims, wherein said Bacillales member is a Paenibacillaceae family member.
7. The polypeptide of claim 6, wherein said Bacillales member is a Paenibacillus spp.
8. The polypeptide of any of claims 1-4, wherein said polypeptide is derived from a Planococcus species.
9. The polypeptide of any of the above claims, wherein said polypeptide has protease activity.
10. The polypeptide of claim 9, wherein said protease activity comprises casein hydrolysis, collagen hydrolysis, elastin hydrolysis, keratin hydrolysis, soy protein hydrolysis or corn meal protein hydrolysis.
11. The polypeptide of any of the above claims, wherein said polypeptide retains at least 50% of its maximal activity between pH 4.5 and 10.
12. The polypeptide of any of the above claims, wherein said polypeptide retains at least 50% of its maximal activity between 30.degree. C. and 70.degree. C.
13. The polypeptide of any of the above claims, wherein said polypeptide has cleaning activity in a detergent composition.
14. The polypeptide of claim 13, wherein said detergent composition is an ADW detergent composition.
15. The polypeptide of claim 13, wherein said detergent composition is a laundry detergent composition.
16. The polypeptide of claim 15, wherein said detergent composition is a liquid laundry detergent composition.
17. The polypeptide of claim 15, wherein said detergent composition is a powder laundry detergent composition.
18. The polypeptide of claim 13, wherein said detergent composition comprises a bleach component.
19. The polypeptide of any of the above claims, wherein said polypeptide is a recombinant polypeptide.
20. A composition comprising the polypeptide of any of the above claims.
21. The composition of claim 20, wherein said composition is a cleaning composition.
22. The composition of claim 21, wherein said composition is a detergent composition.
23. The composition of claim 22, wherein said detergent composition is selected from the group consisting of a laundry detergent, a fabric softening detergent, a dishwashing detergent, and a hard-surface cleaning detergent.
24. The composition of any of claims 20 to 22, wherein said composition further comprising a surfactant.
25. The composition of claim 24, wherein said surfactant is selected from the group consisting of an anionic surfactant, a cationic surfactant, a zwitterionic surfactant, a ampholytic surfactant, a semi-polar non-ionic surfactant, and a combination thereof.
26. The composition of claim 24, wherein said surfactant is an ionic surfactant.
27. The composition of claim 24, wherein said surfactant is a non-ionic surfactant.
28. The composition of any of claims 20-27, wherein said composition further comprises at least one calcium ion and/or zinc ion.
29. The composition of any of claims 20-28, wherein said composition further comprises at least one stabilizer.
30. The composition of any of claims 20-29, wherein said composition comprises from about 0.001 to about 0.1 weight % of said polypeptide.
31. The composition of any of claims 20-30, further comprising at least one bleaching agent.
32. The composition of any of claims 20-31, wherein said cleaning composition is phosphate-free.
33. The composition of any of claims 20-31, wherein said cleaning composition contains phosphate.
34. The composition of any of claims 20-33, further comprising at least one adjunct ingredient.
35. The composition of any of claims 20-34, wherein said composition is a granular, powder, solid, bar, liquid, tablet, gel, or paste composition.
36. The composition of any of claims 20-35, further comprising one or more additional enzymes or enzyme derivatives selected from the group consisting of acyl transferases, alpha-amylases, beta-amylases, alpha-galactosidases, arabinosidases, aryl esterases, beta-galactosidases, carrageenases, catalases, cellobiohydrolases, cellulases, chondroitinases, cutinases, endo-beta-1, 4-glucanases, endo-beta-mannanases, esterases, exo-mannanases, galactanases, glucoamylases, hemicellulases, hyaluronidases, keratinases, laccases, lactases, ligninases, lipases, lipoxygenases, mannanases, oxidases, pectate lyases, pectin acetyl esterases, pectinases, pentosanases, peroxidases, phenoloxidases, phosphatases, phospholipases, phytases, polygalacturonases, proteases, pullulanases, reductases, rhamnogalacturonases, beta-glucanases, tannases, transglutaminases, xylan acetyl-esterases, xylanases, xyloglucanases, and xylosidases, additional metallopotease enzymes and combinations thereof.
37. The composition of any of claims 20-36, wherein said composition is formulated at a pH of from about 5.5 to about 8.5.
38. A method for the pretreatment of animal feed comprising treating an animal feed pre-product with the polypeptide of any one of claims 1-19.
39. A method of cleaning, comprising contacting a surface or an item with a cleaning composition comprising the polypeptide of any one of claims 1-19.
40. A method of cleaning comprising contacting a surface or an item with the composition of any one of claims 20-37.
41. The method of claim 39 or 40, further comprising rinsing said surface or item after contacting said surface or item, respectively, with said composition.
42. The method of claims 39-41, wherein said item is dishware.
43. The method of any one of claims 39-41, wherein said item is fabric.
44. The method of any one of claims 39-43, further comprising the step of rinsing said surface or item after contacting said surface or item with said composition.
45. The method of claim 44, further comprising the step of drying said surface or item after said rinsing of said surface or item.
46. A method of cleaning a surface or item, comprising: providing the composition of any of claims 20-37 and a surface or item in need of cleaning; and contacting said composition with said surface or item in need of cleaning under conditions suitable for the cleansing of said surface of said surface or item, to produce a cleansed surface or item.
47. The method of claim 46, further comprising the step of rinsing said cleansed surface or item to produce a rinsed surface or item.
48. The method of any of claim 46 or 47, further comprising the step of drying said rinsed surface or item.
49. A method for producing the polypeptide of any of claims 1-19 comprising: a. stably transforming a host cell with an expression vector comprising a polynucleotide encoding the polypeptide of any of claims 1-19; b. cultivating said transformed host cell under conditions suitable for said host cell to produce said protease; and c. recovering said protease.
50. The method of claim 49, wherein said host cell is a filamentous fungus or bacterial cell.
51. The method of claim 49 or 50, wherein said host cell is selected from Bacillus spp., Streptomyces spp., Escherichia spp., Aspergillus spp., Trichoderma spp., Pseudomonas spp., Corynebacterium spp., Saccharomyces spp., or Pichia spp.
52. The method of any one of claims 49-51, wherein said expression vector comprises a polynucleotide sequence comprising: a. at least 70% sequence identity to the polynucleotide sequence selected from the group consisting of SEQ ID NOs: 4, 9, 14, 19, 24, 29, 34 and 39; or b. being capable of hybridizing to a probe derived from the polynucleotide sequence selected from the group consisting of SEQ ID NOs: 4, 9, 14, 19, 24, 29, 34 and 39 under conditions of intermediate to high stringency, or c. a polynucleotide sequence complementary to a polynucleotide sequence having at least 70% sequence identity to the polynucleotide sequence selected from the group consisting of SEQ ID NOs: 4, 9, 14, 19, 24, 29, 34 and 39.
53. The method of any one of claims 49-52, wherein said vector comprises a DNA sequence coding for a native or non-naturally occurring signal peptide.
54. The method of any one of claims 49-53, wherein said vector comprises a heterologous promoter and/or DNA sequence coding for a signal peptide.
55. The method of any one of claims 49-53, wherein said vector comprises a homologous promoter and/or DNA sequence coding for a signal peptide.
56. The method of any one of claims 49-65, wherein said host cell is cultivated in a culture media or a fermentation broth.
57. A nucleic acid sequence comprising a nucleic acid sequence: (i) having at least 70% identity to a sequence selected from the group consisting of SEQ ID NOs: 4, 9, 14, 19, 24, 29, 34 and 39, or (ii) being capable of hybridizing to a probe derived from the polynucleotide sequence selected from the group consisting of SEQ ID NOs: 4, 9, 14, 19, 24, 29, 34 and 39, under conditions of intermediate to high stringency, or (iii) being complementary to the polynucleotide sequence selected from the group consisting of SEQ ID NOs: 4, 9, 14, 19, 24, 29, 34 and 39.
58. A vector comprising the nucleic acid sequence of claim 57.
59. A host cell transformed with the vector of claim 58.
60. The host cell of claim 59 selected from Bacillus spp., Streptomyces spp., Escherichia spp., Aspergillus spp., Trichoderma spp., Pseudomonas spp., Corynebacterium spp., Saccharomyces spp., or Pichia spp.
61. The host cell of claim 59 or 60, wherein said Bacillus spp. is Bacillus subtilis.
62. A textile processing composition comprising the polypeptide of any one of claims 1-19.
63. An animal feed composition comprising the polypeptide of any one of claims 1-19.
64. A leather processing composition comprising the polypeptide of any one of claims 1-19.
65. A feather processing composition comprising the polypeptide or recombinant polypeptide of any one of claims 1-19.
66. A feather processing composition comprising the polypeptide or recombinant polypeptide of any one of claims 1-19.
67. A corn soy protein processing composition comprising the polypeptide or recombinant polypeptide of any one of claims 1-19.
Description
CROSS REFERENCE TO RELATED APPLICATIONS
[0001] This application is a Divisional of U.S. Ser. No. 15/835,551 filed Dec. 8, 2017, which is Continuation of U.S. Ser. No. 14/893,473, filed Nov. 23, 2015, which is a 371 of International Patent Application No. PCT/US2014/039928, filed May 29, 2014, which claims benefit of priority from International patent applications Serial No. PCT/CN2013/076419; Serial No. PCT/CN2013/076387; Serial No. PCT/CN2013/076401; Serial No. PCT/CN2013/076406; Serial No. PCT/CN2013/076414; Serial No. PCT/CN2013/076384; Serial No. PCT/CN2013/076398; and Serial No. PCT/CN2013/076415; all filed on 29 May 2013, the contents of which are incorporated herein by reference in their entirety.
SEQUENCE LISTING
[0002] The sequence listing submitted via EFS, in compliance with 37 C.F.R. .sctn. 1.52(e), is incorporated herein by reference. The sequence listing text file submitted via EFS contains the file "20200522_NB40167USPCD2_SeqLst" created on May 22, 2020, which is 207 kilo bytes in size.
FIELD OF THE INVENTION
[0003] The present disclosure relates to proteases and variants thereof. Compositions containing the proteases are suitable for use in cleaning, food and feed as well as in a variety of other industrial applications.
BACKGROUND
[0004] Metalloproteases (MPs) are among the hydrolases that mediate nucleophilic attack on peptide bonds using a water molecule coordinated in the active site. In their case, a divalent ion, such as zinc, activates the water molecule. This metal ion is held in place by amino acid ligands, usually 3 in number. The clan MA consists of zinc-dependent MPs in which two of the zinc ligands are the histidines in the motif: HisGluXXHis (SEQ ID NO: 41). This Glu is the catalytic residue. These are two domain proteases with the active site between the domains. In subclan MA(E), also known as Glu-zincins, the 3.sup.rd ligand is a Glu located C-terminal to the HDXXH (SEQ ID NO: 42) motif. Members of the families: M1, 3, 4, 13, 27 and 34 are all secreted proteases, almost exclusively from bacteria (Rawlings and Salvessen (2013) Handbook of Proteolytic Enzymes, Elsevier Press). They are generally active at elevated temperatures and this stability is attributed to calcium binding. Thermolysin-like proteases are found in the M4 family as defined by MEROPS (Rawlings et al., (2012) Nucleic Acids Res 40:D343-D350). Although proteases have long been known in the art of industrial enzymes, there remains a need for novel proteases that are suitable for particular conditions and uses.
SUMMARY
[0005] The present disclosure provides novel metalloprotease enzymes, nucleic acids encoding the same, and compositions and methods related to the production and use thereof.
[0006] In some embodiments, the invention is a polypeptide comprising an amino acid sequence having at least 60%, at least 80%, or at least 95% sequence identity to the amino acid sequence of SEQ ID NO: 3. In some embodiments, the invention is any of the above, wherein said polypeptide is derived from a member of the order Bacillales; family Bacillaceae, Paenibacillaceae, Alicyclobacillaceae, Lactobacillaceae, or a Bacillus, Alicyclobacillus, Geobacillus, Exiguobacterium, Lactobacillus, or Paenibacillus spp., such as Paenibacillus polymyxa. In some embodiments, the invention is any of the above, wherein said polypeptide is derived from a member of the Pseudococcidae, or a Planococcus spp., such as Planococcus donghaensis. In various embodiments of the invention, any of the above polypeptides has protease activity, such as azo-casein hydrolysis. In various embodiments of the invention, any of the above polypeptides retains at least 50% of its maximal activity between pH 5 and 9.5. In various embodiments of the invention, any of the above polypeptides retains at least 50% of its maximal activity between 30.degree. C. and 70.degree. C. In various embodiments of the invention, any of the above polypeptides has cleaning activity in a detergent composition, such as an ADW, laundry, liquid laundry, or powder laundry detergent composition.
[0007] In some embodiments, the invention is a polypeptide comprising an amino acid sequence having at least 60%, at least 80%, or at least 95% sequence identity to the amino acid sequence of SEQ ID NO: 8. In some embodiments, the invention is any of the above, wherein said polypeptide is derived from a member of the order Bacillales; family Bacillaceae, Paenibacillaceae, or Brevibacillaceae, or a Bacillus, Brevibacillus, or Paenibacillus spp., such as Paenibacillus sp. In some embodiments, the invention is any of the above, wherein said polypeptide is derived from Brevibacillus sp. In various embodiments of the invention, any of the above polypeptides has protease activity, such as azo-casein hydrolysis. In various embodiments of the invention, any of the above polypeptides retains at least 50% of its maximal activity between pH 5 and 10. In various embodiments of the invention, any of the above polypeptides retains at least 50% of its maximal activity between 35.degree. C. and 70.degree. C. In various embodiments of the invention, any of the above polypeptides has cleaning activity in a detergent composition, such as an ADW, laundry, liquid laundry, or powder laundry detergent composition.
[0008] In some embodiments, the invention is a polypeptide comprising an amino acid sequence having at least 60%, at least 80%, or at least 95% sequence identity to the amino acid sequence of SEQ ID NO: 13. In some embodiments, the invention is any of the above, wherein said polypeptide is derived from a member of the order Bacillales; family Bacillaceae, Paenibacillaceae, or Brevibacillaceae, or a Bacillus, Geobacillus, Brevibacillus, or Paenibacillus spp., such as Paenibacillus humicus. In some embodiments, the invention is any of the above, wherein said polypeptide is derived from Bacillus polymyxa. In various embodiments of the invention, any of the above polypeptides has protease activity, such as azo-casein hydrolysis. In various embodiments of the invention, any of the above polypeptides retains at least 50% of its maximal activity between pH 5 and 9.5. In various embodiments of the invention, any of the above polypeptides retains at least 50% of its maximal activity between 35.degree. C. and 70.degree. C. In various embodiments of the invention, any of the above polypeptides has cleaning activity in a detergent composition, such as an ADW, laundry, liquid laundry, or powder laundry detergent composition.
[0009] In some embodiments, the invention is a polypeptide comprising an amino acid sequence having at least 60%, at least 80%, or at least 95% sequence identity to the amino acid sequence of SEQ ID NO: 18. In some embodiments, the invention is any of the above, wherein said polypeptide is derived from a member of the order Bacillales; family Bacillaceae, Paenibacillaceae, or Brevibacillaceae, or a Bacillus, Geobacillus, Brevibacillus, or Paenibacillus spp., such as Paenibacillus ehimensis. In some embodiments, the invention is any of the above, wherein said polypeptide is derived from Brevibacillus sp. In various embodiments of the invention, any of the above polypeptides has protease activity, such as azo-casein hydrolysis. In various embodiments of the invention, any of the above polypeptides retains at least 50% of its maximal activity between pH 5 and 10.5. In various embodiments of the invention, any of the above polypeptides retains at least 50% of its maximal activity between 45.degree. C. and 75.degree. C. In various embodiments of the invention, any of the above polypeptides has cleaning activity in a detergent composition, such as an ADW, laundry, liquid laundry, or powder laundry detergent composition.
[0010] In some embodiments, the invention is a polypeptide comprising an amino acid sequence having at least 60%, at least 80%, or at least 95% sequence identity to the amino acid sequence of SEQ ID NO: 23. In some embodiments, the invention is any of the above, wherein said polypeptide is derived from a member of the order Bacillales; family Bacillaceae, Paenibacillaceae, Alicyclobacillaceae, Lactobacillaceae, or a Bacillus, Geobacillus, Alicyclobacillus, Brevibacillus, Paenibacillus, or Lactobacillus spp., such as Paenibacillus barcinonensis. In some embodiments, the invention is any of the above, wherein said polypeptide is derived from a member of the family Pseudococcidae, or a Planococcus spp., such as Planococcus donghaensis. In various embodiments of the invention, any of the above polypeptides has protease activity, such as azo-casein hydrolysis. In various embodiments of the invention, any of the above polypeptides retains at least 50% of its maximal activity between pH 5 and 10. In various embodiments of the invention, any of the above polypeptides retains at least 50% of its maximal activity between 35.degree. C. and 65.degree. C. In various embodiments of the invention, any of the above polypeptides has cleaning activity in a detergent composition, such as an ADW, laundry, liquid laundry, or powder laundry detergent composition.
[0011] In some embodiments, the invention is a polypeptide comprising an amino acid sequence having at least 60%, at least 80%, or at least 95% sequence identity to the amino acid sequence of SEQ ID NO: 28. In some embodiments, the invention is any of the above, wherein said polypeptide is derived from a member of the order Bacillales; family Bacillaceae, Paenibacillaceae, or a Bacillus, Brevibacillus, Paenibacillus, or Lactobacillus spp., such as Paenibacillus polymyxa. In some embodiments, the invention is any of the above, wherein said polypeptide is derived from a member of the family Pseudococcidae, or a Planococcus spp., such as Planococcus donghaensis. In various embodiments of the invention, any of the above polypeptides has protease activity, such as azo-casein hydrolysis. In various embodiments of the invention, any of the above polypeptides retains at least 50% of its maximal activity between pH 5 and 9.5. In various embodiments of the invention, any of the above polypeptides retains at least 50% of its maximal activity between 30.degree. C. and 65.degree. C. In various embodiments of the invention, any of the above polypeptides has cleaning activity in a detergent composition, such as an ADW, laundry, liquid laundry, or powder laundry detergent composition.
[0012] In some embodiments, the invention is a polypeptide comprising an amino acid sequence having at least 60%, at least 80%, or at least 95% sequence identity to the amino acid sequence of SEQ ID NO: 33. In some embodiments, the invention is any of the above, wherein said polypeptide is derived from a member of the order Bacillales; family Bacillaceae, Paenibacillaceae, or a Bacillus, Geobacillus, Brevibacillus, or Paenibacillus spp., such as Paenibacillus hunanensis. In some embodiments, the invention is any of the above, wherein said polypeptide is derived from Bacillus polymyxa. In various embodiments of the invention, any of the above polypeptides has protease activity, such as azo-casein hydrolysis. In various embodiments of the invention, any of the above polypeptides retains at least 50% of its maximal activity between pH 4.5 and 9.0. In various embodiments of the invention, any of the above polypeptides retains at least 50% of its maximal activity between 35.degree. C. and 70.degree. C. In various embodiments of the invention, any of the above polypeptides has cleaning activity in a detergent composition, such as an ADW, laundry, liquid laundry, or powder laundry detergent composition.
[0013] In some embodiments, the invention is a polypeptide comprising an amino acid sequence having at least 60%, at least 80%, or at least 95% sequence identity to the amino acid sequence of SEQ ID NO: 38. In some embodiments, the invention is any of the above, wherein said polypeptide is derived from a member of the order Bacillales; family Bacillaceae, Paenibacillaceae, Lactobacillaceae, or a Bacillus, Brevibacillus, Lactobacillus, Paenibacillus, or Geobacillus spp., such as Paenibacillus amylolyticus. In various embodiments of the invention, any of the above polypeptides has protease activity, such as azo-casein hydrolysis. In various embodiments of the invention, any of the above polypeptides retains at least 50% of its maximal activity between pH 5.5 and 10. In various embodiments of the invention, any of the above polypeptides retains at least 50% of its maximal activity between 35.degree. C. and 65.degree. C. In various embodiments of the invention, any of the above polypeptides has cleaning activity in a detergent composition, such as an ADW, laundry, liquid laundry, or powder laundry detergent composition.
[0014] In some embodiments, the invention is a composition comprising any of the above, such as a cleaning or detergent composition. In some embodiments, the composition further comprises a surfactant, at least one calcium ion and/or zinc ion, at least one stabilizer, at least one bleaching agent, and can contain phosphate, or be phosphate-free. In some embodiments, the composition further comprises one or more additional enzymes or enzyme derivatives selected from the group consisting of acyl transferases, alpha-amylases, beta-amylases, alpha-galactosidases, arabinosidases, aryl esterases, beta-galactosidases, carrageenases, catalases, cellobiohydrolases, cellulases, chondroitinases, cutinases, endo-beta-1, 4-glucanases, endo-beta-mannanases, esterases, exo-mannanases, galactanases, glucoamylases, hemicellulases, hyaluronidases, keratinases, laccases, lactases, ligninases, lipases, lipoxygenases, mannanases, oxidases, pectate lyases, pectin acetyl esterases, pectinases, pentosanases, peroxidases, phenoloxidases, phosphatases, phospholipases, phytases, polygalacturonases, proteases, pullulanases, reductases, rhamnogalacturonases, beta-glucanases, tannases, transglutaminases, xylan acetyl-esterases, xylanases, xyloglucanases, and xylosidases, and combinations thereof. In some embodiments, the composition is formulated at a pH of from about 5.5 to about 8.5. In some embodiments, the invention is a method of cleaning using any of the above polypeptides or compositions. In some embodiments, the invention is a textile processing composition, animal feed composition, leather processing composition, or feather processing composition.
BRIEF DESCRIPTION OF THE DRAWINGS
[0015] FIG. 1.1 provides a plasmid map of pGX085 (aprE-PspPro3), described in Example 1.2.
[0016] FIG. 1.2 provides a dose response curve of PspPro3 in the azo-casein assay.
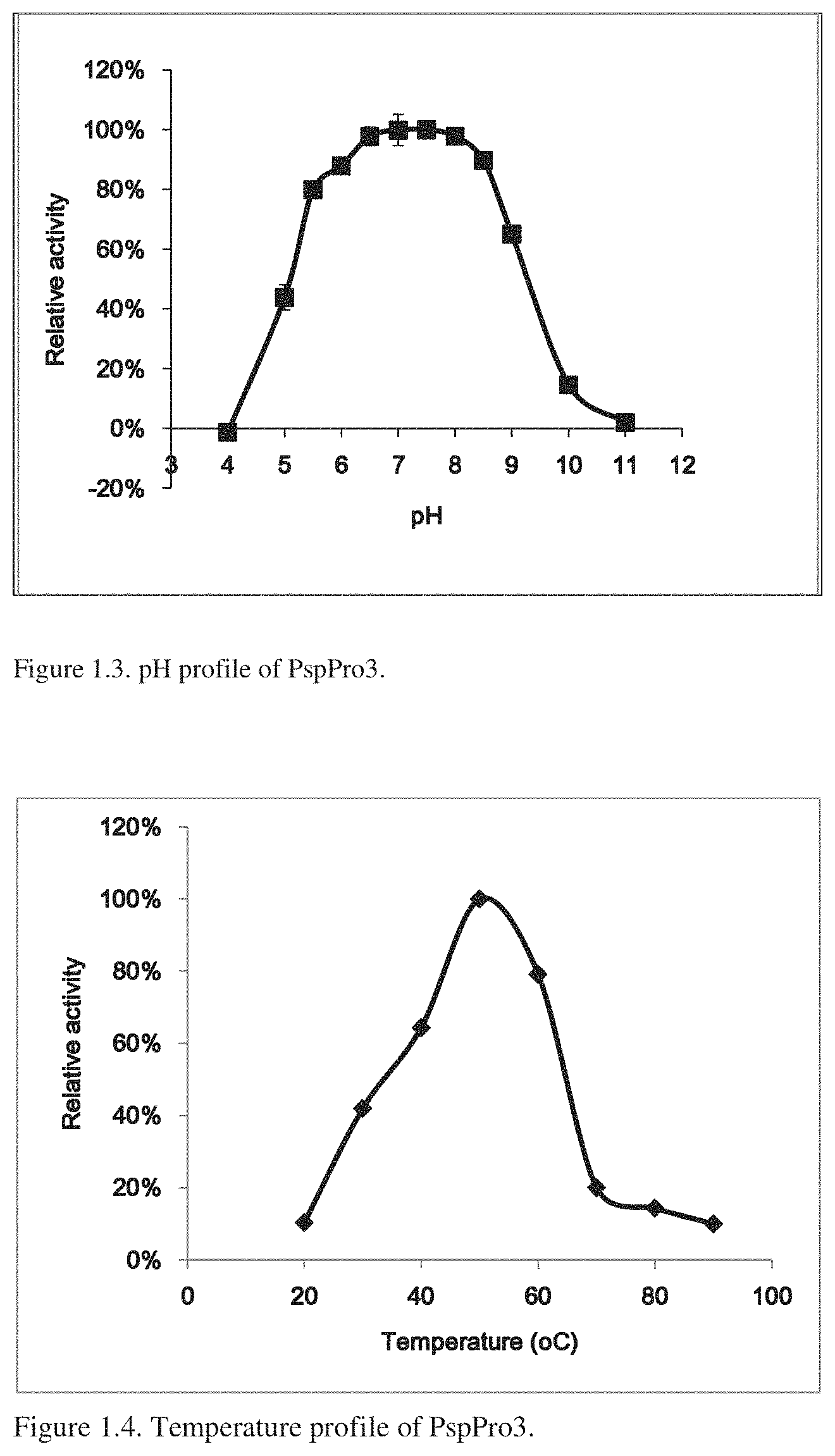
[0017] FIG. 1.3 provides the pH profile of PspPro3.
[0018] FIG. 1.4 provides the temperature profile of PspPro3.
[0019] FIG. 1.5A shows dose response for cleaning of PA-S-38 microswatches by PspPro3 protein in ADW detergent at pH 6 and 8.
[0020] FIG. 1.5B shows dose response for cleaning of PA-S-38 microswatches shows by PspPro3 protein in ADW detergent at pH 6 and 8 in the presence of bleach.
[0021] FIG. 1.6 shows cleaning performance of PspPro3 protein in liquid laundry detergent.
[0022] FIG. 1.7 (SEQ ID NOS: 3, 44, and 45, respectively) shows alignment of PspPro3 with other protein homologs.
[0023] FIG. 1.8 provides the phylogenetic tree for PspPro3 and its homologs.
[0024] FIG. 2.1 provides a plasmid map of pGX084 (aprE-PspPro2), described in Example 2.2.
[0025] FIG. 2.2 provides a dose response curve of PspPro2 in the azo-casein assay.
[0026] FIG. 2.3 provides the pH profile of purified PspPro2.
[0027] FIG. 2.4 provides the temperature profile of purified PspPro2.
[0028] FIG. 2.5A shows dose response for cleaning performance of PspPro2 at pH 6 in AT dish detergent with bleach.
[0029] FIG. 2.5B shows dose response for cleaning performance of purified PspPro2 at pH 8 in AT detergent with bleach.
[0030] FIG. 2.6A shows cleaning performance of PspPro2 protein in liquid laundry detergent.
[0031] FIG. 2.6B shows cleaning performance of PspPro2 protein in powder laundry detergent.
[0032] FIG. 2.7 (SEQ ID NOS: 8, 46, and 45, respectively) shows alignment of PspPro2 with other protein homologs.
[0033] FIG. 2.8 provides the phylogenetic tree for PspPro2 and its homologs.
[0034] FIG. 3.1 provides a plasmid map of pGX150 (aprE-PhuPro2), described in Example 3.2.
[0035] FIG. 3.2 provides a dose response curve of PhuPro2 in the azo-casein assay.
[0036] FIG. 3.3 provides the pH profile of purified PhuPro2.
[0037] FIG. 3.4 provides the temperature profile of purified PhuPro2.
[0038] FIG. 3.5A shows dose response for c leaning performance of PhuPro2 in AT dish detergent at pH 6.
[0039] FIG. 3.5B shows dose response for cleaning performance of PhuPro2 in AT dish detergent at pH 8.
[0040] FIG. 3.6 (SEQ ID NOS: 13, 47 and 45, respectively) shows alignment of PhuPro2 with other protein homologs.
[0041] FIG. 3.7 provides the phylogenetic tree for PhuPro2 and its homologs.
[0042] FIG. 4.1 provides a plasmid map of pGX148 (aprE-PehPro1), described in Example 4.2.
[0043] FIG. 4.2 provides a dose response curve of PehPro1 in the azo-casein assay.
[0044] FIG. 4.3 provides the pH profile of purified PehPro1.
[0045] FIG. 4.4 provides the temperature profile of purified PehPro1.
[0046] FIG. 4.5A shows dose response for cleaning performance of PehPro1 at pH 6 in AT dish detergent with bleach.
[0047] FIG. 4.5B shows dose response for cleaning performance of purified PehPro1 at pH 8 in AT detergent with bleach.
[0048] FIG. 4.6 (SEQ ID NOS: 18, 48, and 45, respectively) shows alignment of PehPro1 with other protein homologs.
[0049] FIG. 4.7 provides the phylogenetic tree for PehPro1 and its homologs.
[0050] FIG. 5.1 provides a plasmid map of pGX147 (aprE-PbaPro1), described in Example 5.2.
[0051] FIG. 5.2 provides a dose response curve of PbaPro1 in the azo-casein assay.
[0052] FIG. 5.3 provides the pH profile of purified PbaPro1.
[0053] FIG. 5.4 provides the temperature profile of purified PbaPro1.
[0054] FIG. 5.5A shows dose response for cleaning of PA-S-38 microswatches by PbaPro1protein in ADW detergent at pH 6.
[0055] FIG. 5.5B shows dose response for cleaning of PA-S-38 microswatches shows by PbaPro1protein in ADW detergent at pH 8.
[0056] FIG. 5.6 (SEQ ID NOS: 23, 49, and 45, respectively) shows the alignment of PbaPro1 with protease homologs.
[0057] FIG. 5.7 provides the phylogenetic tree for PbaPro1 and its homologs.
[0058] FIG. 6.1 provides a plasmid map of pGX138 (aprE-PpoPro1), described in Example 6.2.
[0059] FIG. 6.2 provides a dose response curve of PpoPro1 in the azo-casein assay.
[0060] FIG. 6.3 provides the pH profile of purified PpoPro1.
[0061] FIG. 6.4 provides the temperature profile of purified PpoPro1.
[0062] FIG. 6.5A shows dose response for cleaning of PA-S-38 microswatches by PpoPro1protein in ADW detergent at pH 6 in the presence of bleach.
[0063] FIG. 6.5B shows dose response for cleaning of PA-S-38 microswatches shows by PpoPro1protein in ADW detergent at pH 8 in the presence of bleach.
[0064] FIG. 6.6 (SEQ ID NOS: 28, 50, and 45, respectively) shows the alignment of PpoPro1 with protease homologs.
[0065] FIG. 6.7 provides the phylogenetic tree for PpoPro1 and its homologs.
[0066] FIG. 7.1 provides a plasmid map of pGX149 (aprE-PhuPro1), described in Example 7.2.
[0067] FIG. 7.2 provides a dose response curve of PhuPro1 in the azo-casein assay.
[0068] FIG. 7.3 provides the pH profile of purified PhuPro1.
[0069] FIG. 7.4 provides the temperature profile of purified PhuPro1.
[0070] FIG. 7.5A shows dose response for cleaning of PA-S-38 microswatches by PhuPro1 protein in ADW detergent at pH 6.
[0071] FIG. 7.5B shows dose response for cleaning of PA-S-38 microswatches shows by Phu Pro1protein in ADW detergent at pH 8.
[0072] FIG. 7.6 (SEQ ID NOS: 33, 51, and 45, respectively) shows alignment of PhuPro1 with other protein homologs.
[0073] FIG. 7.7 provides the phylogenetic tree for PhuPro1 and its homologs.
[0074] FIGS. 7.8A and 7.8B show cleaning performances of PhuPro1 and Purafect.RTM. Prime HA proteases.
[0075] FIG. 8.1 provides a plasmid map of pGX146 (aprE-PamPro1), described in Example 8.2.
[0076] FIG. 8.2 provides a dose response curve of PamPro1 in the azo-casein assay.
[0077] FIG. 8.3 provides the pH profile of purified PamPro1.
[0078] FIG. 8.4 provides the temperature profile of purified PamPro1.
[0079] FIG. 8.5A shows dose response for cleaning of PA-S-38 microswatches by PamPro1 protein in ADW detergent at pH 6.
[0080] FIG. 8.5B shows dose response for cleaning of PA-S-38 microswatches shows by PamPro1 protein in ADW detergent at pH 8.
[0081] FIG. 8.6 (SEQ ID NOS: 38, 52, and 45, respectively) shows the alignment of PamPro1 with protease homologs.
[0082] FIG. 8.7 provides the phylogenetic tree for PamPro1 and its homologs.
[0083] FIGS. 9.1A thru 9.1D (SEQ ID NOS: 53-62, 38, 23, 13, 63, 8, 28, 64, 3, 18, 33, 65-68, respectively) show the alignment of the various Paenibacillus metalloproteases with other bacterial metalloprotease homologs.
[0084] FIG. 9.2 provides the phylogenetic tree of the various Paenibacillus metalloproteases with other bacterial metalloprotease homologs.
DETAILED DESCRIPTION
[0085] The present invention provides novel metalloprotease enzymes, especially enzymes useful for detergent compositions cloned from various Paenibacillus sp. The compositions and methods are based, in part, on the observation that the novel metalloproteases of the present invention have proteolytic activity in the presence of detergent compositions. This feature makes metalloproteases of the present invention particularly well suited to and useful in a variety of cleaning applications where the enzyme can hydrolyze polypeptides in the presence of surfactants and other components found in detergent compositions. The invention includes compositions comprising at least one of the novel metalloprotease enzymes set forth herein. Some such compositions comprise detergent compositions. The metalloprotease enzymes of the present invention can be combined with other enzymes useful in detergent compositions. The invention also provides methods of cleaning using metalloprotease enzymes of the present invention.
Definitions and Abbreviations
[0086] Unless otherwise indicated, the practice of the present invention involves conventional techniques commonly used in molecular biology, protein engineering, microbiology, and recombinant DNA technology, which are within the skill of the art. Such techniques are known to those of skill in the art and are described in numerous texts and reference works well known to those of skill in the art. All patents, patent applications, articles and publications mentioned herein, both supra and infra, are hereby expressly incorporated herein by reference.
[0087] Unless defined otherwise herein, all technical and scientific terms used herein have the same meaning as commonly understood by one of ordinary skill in the art to which this invention pertains. Many technical dictionaries are known to those of skill in the art. Although any methods and materials similar or equivalent to those described herein find use in the practice of the present invention, some suitable methods and materials are described herein. Accordingly, the terms defined immediately below are more fully described by reference to the Specification as a whole. Also, as used herein, the singular "a", "an" and "the" includes the plural reference unless the context clearly indicates otherwise. Unless otherwise indicated, nucleic acids are written left to right in 5' to 3' orientation; amino acid sequences are written left to right in amino to carboxy orientation, respectively. It is to be understood that this invention is not limited to the particular methodology, protocols, and reagents described, as these may vary, depending upon the context they are used by those of skill in the art.
[0088] Furthermore, the headings provided herein are not limitations of the various aspects or embodiments of the invention.
[0089] It is intended that every maximum numerical limitation given throughout this specification includes every lower numerical limitation, as if such lower numerical limitations were expressly written herein. Every minimum numerical limitation given throughout this specification will include every higher numerical limitation, as if such higher numerical limitations were expressly written herein. Every numerical range given throughout this specification will include every narrower numerical range that falls within such broader numerical range, as if such narrower numerical ranges were all expressly written herein.
[0090] As used herein, the terms "protease" and "proteinase" refer to an enzyme that has the ability to break down proteins and peptides. A protease has the ability to conduct "proteolysis," by hydrolysis of peptide bonds that link amino acids together in a peptide or polypeptide chain forming the protein. This activity of a protease as a protein-digesting enzyme is referred to as "proteolytic activity." Many well known procedures exist for measuring proteolytic activity (See e.g., Kalisz, "Microbial Proteinases," In: Fiechter (ed.), Advances in Biochemical Engineering/Biotechnology, (1988)). For example, proteolytic activity may be ascertained by comparative assays which analyze the respective protease's ability to hydrolyze a suitable substrate. Exemplary substrates useful in the analysis of protease or proteolytic activity, include, but are not limited to, di-methyl casein (Sigma C-9801), bovine collagen (Sigma C-9879), bovine elastin (Sigma E-1625), and bovine keratin (ICN Biomedical 902111). Colorimetric assays utilizing these substrates are well known in the art (See e.g., WO 99/34011 and U.S. Pat. No. 6,376,450, both of which are incorporated herein by reference). The pNA peptidyl assay (See e.g., Del Mar et al., Anal. Biochem. 99:316-320 [1979]) also finds use in determining the active enzyme concentration. This assay measures the rate at which p-nitroaniline is released as the enzyme hydrolyzes a soluble synthetic substrate, such as succinyl-alanine-alanine-proline-phenylalanine-p-nitroanilide (suc-AAPF-pNA)(SEQ ID NO: 43). The rate of production of yellow color from the hydrolysis reaction is measured at 410 nm on a spectrophotometer and is proportional to the active enzyme concentration. In addition, absorbance measurements at 280 nanometers (nm) can be used to determine the total protein concentration in a sample of purified protein. The activity on substrate/protein concentration gives the enzyme specific activity.
[0091] As used herein, the term "variant polypeptide" refers to a polypeptide comprising an amino acid sequence that differs in at least one amino acid residue from the amino acid sequence of a parent or reference polypeptide (including but not limited to wild-type polypeptides).
[0092] As used herein, "the genus Bacillus" includes all species within the genus "Bacillus," as known to those of skill in the art, including but not limited to B. subtilis, B. licheniformis, B. lentus, B. brevis, B. stearothermophilus, B. alkalophilus, B. amyloliquefaciens, B. clausii, B. halodurans, B. megaterium, B. coagulans, B. circulans, and B. thuringiensis. It is recognized that the genus Bacillus continues to undergo taxonomical reorganization. Thus, it is intended that the genus include species that have been reclassified, including but not limited to such organisms as B. stearothermophilus, which is now named "Geobacillus stearothermophilus." The production of resistant endospores under stressful environmental conditions is considered the defining feature of the genus Bacillus, although this characteristic also applies to the recently named Alicyclobacillus, Amphibacillus, Aneurinibacillus, Paenibacillus, Brevibacillus, Filobacillus, Gracilibacillus, Halobacillus, Paenibacillus, Salibacillus, Thermobacillus, Ureibacillus, and Virgibacillus.
[0093] The terms "polynucleotide" and "nucleic acid," which are used interchangeably herein, refer to a polymer of any length of nucleotide monomers covalently bonded in a chain. DNA (deoxyribonucleic acid), a polynucleotide comprising deoxyribonucleotides, and RNA (ribonucleic acid), a polymer of ribonucleotides, are examples of polynucleotides or nucleic acids having distinct biological function. Polynucleotides or nucleic acids include, but are not limited to, a single-, double- or triple-stranded DNA, genomic DNA, cDNA, RNA, DNA-RNA hybrid, or a polymer comprising purine and pyrimidine bases, or other natural, chemically, biochemically modified, non-natural or derivatized nucleotide bases. The following are non-limiting examples of polynucleotides: genes, gene fragments, chromosomal fragments, expressed sequence tag(s) (EST(s)), exons, introns, messenger RNA (mRNA), transfer RNA (tRNA), ribosomal RNA (rRNA), ribozymes, complementary DNA (cDNA), recombinant polynucleotides, branched polynucleotides, plasmids, vectors, isolated DNA of any sequence, isolated RNA of any sequence, nucleic acid probes, and primers.
[0094] As used herein, the term "mutation" refers to changes made to a reference amino acid or nucleic acid sequence. It is intended that the term encompass substitutions, insertions and deletions.
[0095] As used herein, the term "vector" refers to a nucleic acid construct used to introduce or transfer nucleic acid(s) into a target cell or tissue. A vector is typically used to introduce foreign DNA into a cell or tissue. Vectors include plasmids, cloning vectors, bacteriophages, viruses (e.g., viral vector), cosmids, expression vectors, shuttle vectors, and the like. A vector typically includes an origin of replication, a multicloning site, and a selectable marker. The process of inserting a vector into a target cell is typically referred to as transformation. The present invention includes, in some embodiments, a vector that comprises a DNA sequence encoding a metalloprotease polypeptide (e.g., precursor or mature metalloprotease polypeptide) that is operably linked to a suitable prosequence (e.g., secretory, signal peptide sequence, etc.) capable of effecting the expression of the DNA sequence in a suitable host, and the folding and translocation of the recombinant polypeptide chain.
[0096] As used herein, the term "expression cassette," "expression plasmid" or "expression vector" refers to a nucleic acid construct or vector generated recombinantly or synthetically for the expression of a nucleic acid of interest in a target cell. An expression vector or expression cassette typically comprises a promoter nucleotide sequence that drives expression of the foreign nucleic acid. The expression vector or cassette also typically includes any other specified nucleic acid elements that permit transcription of a particular nucleic acid in a target cell. A recombinant expression cassette can be incorporated into a plasmid, chromosome, mitochondrial DNA, plastid DNA, virus, or nucleic acid fragment. Many prokaryotic and eukaryotic expression vectors are commercially available.
[0097] In some embodiments, the ends of the sequence are closed such that the DNA construct forms a closed circle. The nucleic acid sequence of interest, which is incorporated into the DNA construct, using techniques well known in the art, may be a wild-type, mutant, or modified nucleic acid. In some embodiments, the DNA construct comprises one or more nucleic acid sequences homologous to the host cell chromosome. In other embodiments, the DNA construct comprises one or more non-homologous nucleotide sequences. Once the DNA construct is assembled in vitro, it may be used, for example, to: 1) insert heterologous sequences into a desired target sequence of a host cell; and/or 2) mutagenize a region of the host cell chromosome (i.e., replace an endogenous sequence with a heterologous sequence); 3) delete target genes; and/or 4) introduce a replicating plasmid into the host. "DNA construct" is used interchangeably herein with "expression cassette."
[0098] As used herein, a "plasmid" refers to an extrachromosomal DNA molecule which is capable of replicating independently from the chromosomal DNA. A plasmid is double stranded (ds) and may be circular and is typically used as a cloning vector.
[0099] As used herein in the context of introducing a nucleic acid sequence into a cell, the term "introduced" refers to any method suitable for transferring the nucleic acid sequence into the cell. Such methods for introduction include but are not limited to protoplast fusion, transfection, transformation, electroporation, conjugation, and transduction (See e.g., Ferrari et al., "Genetics," in Hardwood et al. (eds.), Bacillus, Plenum Publishing Corp., pp. 57-72 [1989]).
[0100] Transformation refers to the genetic alteration of a cell which results from the uptake, optional genomic incorporation, and expression of genetic material (e.g., DNA).
[0101] As used herein, a nucleic acid is "operably linked" with another nucleic acid sequence when it is placed into a functional relationship with another nucleic acid sequence. For example, a promoter or enhancer is operably linked to a nucleotide coding sequence if the promoter affects the transcription of the coding sequence. A ribosome binding site may be operably linked to a coding sequence if it is positioned so as to facilitate translation of the coding sequence. Typically, "operably linked" DNA sequences are contiguous. However, enhancers do not have to be contiguous. Linking is accomplished by ligation at convenient restriction sites. If such sites do not exist, synthetic oligonucleotide adaptors or linkers may be used in accordance with conventional practice.
[0102] As used herein the term "gene" refers to a polynucleotide (e.g., a DNA segment), that encodes a polypeptide and includes regions preceding and following the coding regions as well as intervening sequences (introns) between individual coding segments (exons).
[0103] As used herein, "recombinant" when used with reference to a cell typically indicates that the cell has been modified by the introduction of a foreign nucleic acid sequence or that the cell is derived from a cell so modified. For example, a recombinant cell may comprise a gene not found in identical form within the native (non-recombinant) form of the cell, or a recombinant cell may comprise a native gene (found in the native form of the cell) but which has been modified and re-introduced into the cell. A recombinant cell may comprise a nucleic acid endogenous to the cell that has been modified without removing the nucleic acid from the cell; such modifications include those obtained by gene replacement, site-specific mutation, and related techniques known to those of ordinary skill in the art. Recombinant DNA technology includes techniques for the production of recombinant DNA in vitro and transfer of the recombinant DNA into cells where it may be expressed or propagated, thereby producing a recombinant polypeptide. "Recombination," "recombining," and "recombined" of polynucleotides or nucleic acids refer generally to the assembly or combining of two or more nucleic acid or polynucleotide strands or fragments to generate a new polynucleotide or nucleic acid. The recombinant polynucleotide or nucleic acid is sometimes referred to as a chimera. A nucleic acid or polypeptide is "recombinant" when it is artificial or engineered.
[0104] A nucleic acid or polynucleotide is said to "encode" a polypeptide if, in its native state or when manipulated by methods known to those of skill in the art, it can be transcribed and/or translated to produce the polypeptide or a fragment thereof. The anti-sense strand of such a nucleic acid is also said to encode the sequence.
[0105] "Host strain" or "host cell" refers to a suitable host for an expression vector comprising a DNA sequence of interest.
[0106] A "protein" or "polypeptide" comprises a polymeric sequence of amino acid residues. The terms "protein" and "polypeptide" are used interchangeably herein. The single and 3-letter code for amino acids as defined in conformity with the IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN) is used through out this disclosure. The single letter X refers to any of the twenty amino acids. It is also understood that a polypeptide may be coded for by more than one nucleotide sequence due to the degeneracy of the genetic code. Mutations can be named by the one letter code for the parent amino acid, followed by a position number and then the one letter code for the variant amino acid. For example, mutating glycine (G) at position 87 to serine (S) is represented as "G087S" or "G87S". Mutations can also be named by using the three letter code for an amino acid followed by its position in the polypeptide chain as counted from the N-terminus; for example, Ala10 for alanine at position 10. Multiple mutations are indicated by inserting a "-" between the mutations. Mutations at positions 87 and 90 are represented as either "G087S-A090Y" or "G87S-A90Y" or "G87S+A90Y" or "G087S+A090Y". For deletions, the one letter code "Z" is used. For an insertion relative to the parent sequence, the one letter code "Z" is on the left side of the position number. For a deletion, the one letter code "Z" is on the right side of the position number. For insertions, the position number is the position number before the inserted amino acid(s), plus 0.01 for each amino acid. For example, an insertion of three amino acids alanine (A), serine (S) and tyrosine (Y) between position 87 and 88 is shown as "Z087.01A-Z087.02S-Z087.03Y." Thus, combining all the mutations above plus a deletion at position 100 is: "G087S-Z087.01A-Z087.02S-Z087.03Y-A090Y-A100Z." When describing modifications, a position followed by amino acids listed in parentheses indicates a list of substitutions at that position by any of the listed amino acids. For example, 6(L,I) means position 6 can be substituted with a leucine or isoleucine.
[0107] A "prosequence" or "propetide sequence" refers to an amino acid sequence between the signal peptide sequence and mature protease sequence that is necessary for the proper folding and secretion of the protease; they are sometimes referred to as intramolecular chaperones. Cleavage of the prosequence or propeptide sequence results in a mature active protease. Bacterial metalloproteases are often expressed as pro-enzymes.
[0108] The term "signal sequence" or "signal peptide" refers to a sequence of amino acid residues that may participate in the secretion or direct transport of the mature or precursor form of a protein. The signal sequence is typically located N-terminal to the precursor or mature protein sequence. The signal sequence may be endogenous or exogenous. A signal sequence is normally absent from the mature protein. A signal sequence is typically cleaved from the protein by a signal peptidase after the protein is transported.
[0109] The term "mature" form of a protein, polypeptide, or peptide refers to the functional form of the protein, polypeptide, or peptide without the signal peptide sequence and propeptide sequence.
[0110] The term "precursor" form of a protein or peptide refers to a mature form of the protein having a prosequence operably linked to the amino or carbonyl terminus of the protein. The precursor may also have a "signal" sequence operably linked to the amino terminus of the prosequence. The precursor may also have additional polypeptides that are involved in post-translational activity (e.g., polypeptides cleaved therefrom to leave the mature form of a protein or peptide).
[0111] The term "wild-type" in reference to an amino acid sequence or nucleic acid sequence indicates that the amino acid sequence or nucleic acid sequence is native or naturally occurring sequence. As used herein, the term "naturally-occurring" refers to anything (e.g., proteins, amino acids, or nucleic acid sequences) that are found in nature.
[0112] As used herein, the term "non-naturally occurring" refers to anything that is not found in nature (e.g., recombinant nucleic acids and protein sequences produced in the laboratory), as modification of the wild-type sequence.
[0113] As used herein with regard to amino acid residue positions, "corresponding to" or "corresponds to" or "corresponds" refers to an amino acid residue at the enumerated position in a protein or peptide, or an amino acid residue that is analogous, homologous, or equivalent to an enumerated residue in a protein or peptide. As used herein, "corresponding region" generally refers to an analogous position in a related proteins or a reference protein.
[0114] The terms "derived from" and "obtained from" refer to not only a protein produced or producible by a strain of the organism in question, but also a protein encoded by a DNA sequence isolated from such strain and produced in a host organism containing such DNA sequence. Additionally, the term refers to a protein which is encoded by a DNA sequence of synthetic and/or cDNA origin and which has the identifying characteristics of the protein in question. To exemplify, "proteases derived from Bacillus" refers to those enzymes having proteolytic activity which are naturally produced by Bacillus, as well as to serine proteases like those produced by Bacillus sources but which through the use of genetic engineering techniques are produced by non-Bacillus organisms transformed with a nucleic acid encoding the serine proteases.
[0115] The term "identical" in the context of two nucleic acids or polypeptidesequences refers to the residues in the two sequences that are the same when aligned for maximum correspondence, as measured using one of the following sequence comparison or analysis algorithms.
[0116] As used herein, "homologous genes" refers to a pair of genes from different, but usually related species, which correspond to each other and which are identical or very similar to each other. The term encompasses genes that are separated by speciation (i.e., the development of new species) (e.g., orthologous genes), as well as genes that have been separated by genetic duplication (e.g., paralogous genes).
[0117] As used herein, "% identity or percent identity" refers to sequence similarity. Percent identity may be determined using standard techniques known in the art (See e.g., Smith and Waterman, Adv. Appl. Math. 2:482 [1981]; Needleman and Wunsch, J. Mol. Biol. 48:443 [1970]; Pearson and Lipman, Proc. Natl. Acad. Sci. USA 85:2444 [1988]; software programs such as GAP, BESTFIT, FASTA, and TFASTA in the Wisconsin Genetics Software Package (Genetics Computer Group, Madison, Wis.); and Devereux et al., Nucl. Acid Res. 12:387-395 [1984]). One example of a useful algorithm is PILEUP. PILEUP creates a multiple sequence alignment from a group of related sequences using progressive, pair-wise alignments. It can also plot a tree showing the clustering relationships used to create the alignment. PILEUP uses a simplification of the progressive alignment method of Feng and Doolittle (See, Feng and Doolittle, J. Mol. Evol. 35:351-360 [1987]). The method is similar to that described by Higgins and Sharp (See, Higgins and Sharp, CABIOS 5:151-153 [1989]). Useful PILEUP parameters include a default gap weight of 3.00, a default gap length weight of 0.10, and weighted end gaps. Other useful algorithm is the BLAST algorithms described by Altschul et al., (See, Altschul et al., J. Mol. Biol. 215:403-410 [1990]; and Karlin and Altschul, Proc. Natl. Acad. Sci. USA 90:5873-5787 [1993]). The BLAST program uses several search parameters, most of which are set to the default values.
[0118] The NCBI BLAST algorithm finds the most relevant sequences in terms of biological similarity but is not recommended for query sequences of less than 20 residues (Altschul, S F et al. (1997) Nucleic Acids Res. 25:3389-3402 and Schaffer, A A et al. (2001) Nucleic Acids Res. 29:2994-3005). Example default BLAST parameters for a nucleic acid sequence searches are: [0119] Neighboring words threshold: 11 [0120] E-value cutoff: 10 [0121] Scoring Matrix: NUC.3.1 (match=1, mismatch=-3) [0122] Gap Opening: 5 [0123] Gap Extension: 2 and the following parameters for amino acid sequence searches: [0124] Word size: 3 [0125] E-value cutoff: 10 [0126] Scoring Matrix: BLOSUM62 [0127] Gap Opening: 11 [0128] Gap extension: 1
[0129] A percent (%) amino acid sequence identity value is determined by the number of matching identical residues divided by the total number of residues of the "reference" sequence including any gaps created by the program for optimal/maximum alignment. If a sequence is 90% identical to SEQ ID NO: A, SEQ ID NO: A is is the "reference" sequence. BLAST algorithms refer the "reference" sequence as "query" sequence.
[0130] The CLUSTAL W algorithm is another example of a sequence alignment algorithm. See Thompson et al. (1994) Nucleic Acids Res. 22:4673-4680. Default parameters for the CLUSTAL W algorithm are: [0131] Gap opening penalty: 10.0 [0132] Gap extension penalty: 0.05 [0133] Protein weight matrix: BLOSUM series [0134] DNA weight matrix: IUB [0135] Delay divergent sequences %: 40 [0136] Gap separation distance: 8 [0137] DNA transitions weight: 0.50 [0138] List hydrophilic residues: GPSNDQEKR [0139] Use negative matrix: OFF [0140] Toggle Residue specific penalties: ON [0141] Toggle hydrophilic penalties: ON [0142] Toggle end gap separation penalty OFF.
[0143] In CLUSTAL algorithms, deletions occurring at either terminus are included. For example, a variant with five amino acid deletion at either terminus (or within the polypeptide) of a polypeptide of 500 amino acids would have a percent sequence identity of 99% (495/500 identical residues.times.100) relative to the "reference" polypeptide. Such a variant would be encompassed by a variant having "at least 99% sequence identity" to the polypeptide.
[0144] A polypeptide of interest may be said to be "substantially identical" to a reference polypeptide if the polypeptide of interest comprises an amino acid sequence having at least about 60%, least about 65%, least about 70%, at least about 75%, at least about 80%, at least about 85%, at least about 90%, at least about 91%, at least about 92%, at least about 93%, at least about 94%, at least about 95%, at least about 96%, at least about 97%, at least about 98%, at least about 99%, or at least about 99.5% sequence identity to the amino acid sequence of the reference polypeptide. The percent identity between two such polypeptides can be determined manually by inspection of the two optimally aligned polypeptide sequences or by using software programs or algorithms (e.g., BLAST, ALIGN, CLUSTAL) using standard parameters. One indication that two polypeptides are substantially identical is that the first polypeptide is immunologically cross-reactive with the second polypeptide. Typically, polypeptides that differ by conservative amino acid substitutions are immunologically cross-reactive. Thus, a polypeptide is substantially identical to a second polypeptide, for example, where the two peptides differ only by a conservative amino acid substitution or one or more conservative amino acid substitutions.
[0145] A nucleic acid of interest may be said to be "substantially identical" to a reference nucleic acid if the nucleic acid of interest comprises a nucleotide sequence having least about 60%, least about 65%, at least about 70%, at least about 75%, at least about 80%, at least about 85%, at least about 90%, at least about 91%, at least about 92%, at least about 93%, at least about 94%, at least about 95%, at least about 96%, at least about 97%, at least about 98%, at least about 99%, or at least about 99.5% sequence identity to the nucleotide sequence of the reference nucleic acid. The percent identity between two such nucleic acids can be determined manually by inspection of the two optimally aligned nucleic acid sequences or by using software programs or algorithms (e.g., BLAST, ALIGN, CLUSTAL) using standard parameters. One indication that two nucleic acid sequences are substantially identical is that the two nucleic acid molecules hybridize to each other under stringent conditions (e.g., within a range of medium to high stringency).
[0146] A nucleic acid or polynucleotide is "isolated" when it is at least partially or completely separated from other components, including but not limited to for example, other proteins, nucleic acids, cells, etc. Similarly, a polypeptide, protein or peptide is "isolated" when it is at least partially or completely separated from other components, including but not limited to for example, other proteins, nucleic acids, cells, etc. On a molar basis, an isolated species is more abundant than are other species in a composition. For example, an isolated species may comprise at least about 60%, about 65%, about 70%, about 75%, about 80%, about 85%, about 90%, about 91%, about 92%, about 93%, about 94%, about 95%, about 96%, about 97%, about 98%, about 99%, or about 100% (on a molar basis) of all macromolecular species present. Preferably, the species of interest is purified to essential homogeneity (i.e., contaminant species cannot be detected in the composition by conventional detection methods). Purity and homogeneity can be determined using a number of techniques well known in the art, such as agarose or polyacrylamide gel electrophoresis of a nucleic acid or a protein sample, respectively, followed by visualization upon staining. If desired, a high-resolution technique, such as high performance liquid chromatography (HPLC) or a similar means can be utilized for purification of the material.
[0147] "Hybridization" refers to the process by which one strand of nucleic acid forms a duplex with, i.e., base pairs with, a complementary strand. A nucleic acid sequence is considered to be "selectively hybridizable" to a reference nucleic acid sequence if the two sequences specifically hybridize to one another under moderate to high stringency hybridization and wash conditions. Hybridization conditions are based on the melting temperature (Tm) of the nucleic acid binding complex or probe. For example, "maximum stringency" typically occurs at about Tm-5.degree. C. (5.degree. below the Tm of the probe); "high stringency" at about 5-10.degree. C. below the Tm; "intermediate stringency" at about 10-20.degree. C. below the Tm of the probe; and "low stringency" at about 20-25.degree. C. below the Tm. Functionally, maximum stringency conditions can be used to identify sequences having strict identity or near-strict identity with the hybridization probe; while intermediate or low stringency hybridization can be used to identify or detect polynucleotide sequence homologs.
[0148] Moderate and high stringency hybridization conditions are well known in the art. Stringent hybridization conditions are exemplified by hybridization under the following conditions: 65.degree. C. and 0.1.times.SSC (where 1.times.SSC=0.15 M NaCl, 0.015 M Na3 citrate, pH 7.0). Hybridized, duplex nucleic acids are characterized by a melting temperature (T.sub.m), where one half of the hybridized nucleic acids are unpaired with the complementary strand. Mismatched nucleic acids within the duplex lower the T.sub.m. Very stringent hybridization conditions involve 68.degree. C. and 0.1.times.SSC. A nucleic acid encoding a variant metalloprotease can have a T.sub.m reduced by 1.degree. C.-3.degree. C. or more compared to a duplex formed between the nucleic acid and its identical complement.
[0149] Another example of high stringency conditions includes hybridization at about 42.degree. C. in 50% formamide, 5.times.SSC, 5.times.Denhardt's solution, 0.5% SDS and 100 .mu.g/ml denatured carrier DNA followed by washing two times in 2.times.SSC and 0.5% SDS at room temperature and two additional times in 0.1.times.SSC and 0.5% SDS at 42.degree. C. An example of moderate stringent conditions include an overnight incubation at 37.degree. C. in a solution comprising 20% formamide, 5.times.SSC (150 mM NaCl, 15 mM trisodium citrate), 50 mM sodium phosphate (pH 7.6), 5.times.Denhardt's solution, 10% dextran sulfate and 20 mg/ml denatured sheared salmon sperm DNA, followed by washing the filters in 1.times.SSC at about 37-50.degree. C. Those of skill in the art know how to adjust the temperature, ionic strength, etc. to accommodate factors such as probe length and the like.
[0150] The term "purified" as applied to nucleic acids or polypeptides generally denotes a nucleic acid or polypeptide that is essentially free from other components as determined by analytical techniques well known in the art (e.g., a purified polypeptide or polynucleotide forms a discrete band in an electrophoretic gel, chromatographic eluate, and/or a media subjected to density gradient centrifugation). For example, a nucleic acid or polypeptide that gives rise to essentially one band in an electrophoretic gel is "purified." A purified nucleic acid or polypeptide is at least about 50% pure, usually at least about 60%, about 65%, about 70%, about 75%, about 80%, about 85%, about 90%, about 91%, about 92%, about 93%, about 94%, about 95%, about 96%, about 9'7%, about 98%, about 99%, about 99.5%, about 99.6%, about 99.'7%, about 99.8% or more pure (e.g., percent by weight on a molar basis). In a related sense, the invention provides methods of enriching compositions for one or more molecules of the invention, such as one or more polypeptides or polynucleotides of the invention. A composition is enriched for a molecule when there is a substantial increase in the concentration of the molecule after application of a purification or enrichment technique. A substantially pure polypeptide or polynucleotide of the invention (e.g., substantially pure metalloprotease polypeptide or polynucleotide encoding a metalloprotease polypeptide of the invention, respectively) will typically comprise at least about 55%, about 60%, about 65%, about 70%, about 75%, about 80%, about 85%, about 90%, about 91%, about 92%, about 93%, about 94%, about 95%, about 96%, about 97%, about 98, about 99%, about 99.5% or more by weight (on a molar basis) of all macromolecular species in a particular composition.
[0151] The term "enriched" refers to a compound, polypeptide, cell, nucleic acid, amino acid, or other specified material or component that is present in a composition at a relative or absolute concentration that is higher than a starting composition.
[0152] In a related sense, the invention provides methods of enriching compositions for one or more molecules of the invention, such as one or more polypeptides of the invention (e.g., one or more metalloprotease polypeptides of the invention) or one or more nucleic acids of the invention (e.g., one or more nucleic acids encoding one or more metalloprotease polypeptides of the invention). A composition is enriched for a molecule when there is a substantial increase in the concentration of the molecule after application of a purification or enrichment technique. A substantially pure polypeptide or polynucleotide will typically comprise at least about 55%, about 60%, about 65%, about 70%, about 75%, about 80%, about 85%, about 90%, about 91%, about 92%, about 93%, about 94%, about 95%, about 96%, about 97%, about 98, about 99%, about 99.5% or more by weight (on a molar basis) of all macromolecular species in a particular composition.
[0153] As used herein, the term "functional assay" refers to an assay that provides an indication of a protein's activity. In some embodiments, the term refers to assay systems in which a protein is analyzed for its ability to function in its usual capacity. For example, in the case of a protease, a functional assay involves determining the effectiveness of the protease to hydrolyze a proteinaceous substrate.
[0154] The terms "modified nucleic acid sequence" and "modified gene" are used interchangeably herein to refer to a nucleic acid sequence that includes a deletion, insertion or interruption of naturally occurring (i.e., wild-type) nucleic acid sequence. In some embodiments, the expression product of the modified nucleic acid sequence is a truncated protein (e.g., if the modification is a deletion or interruption of the sequence). In some embodiments, the truncated protein retains biological activity. In alternative embodiments, the expression product of the modified nucleic acid sequence is an elongated protein (e.g., modifications comprising an insertion into the nucleic acid sequence). In some embodiments, a nucleotide insertion in the nucleic acid sequence leads to a truncated protein (e.g., when the insertion results in the formation of a stop codon). Thus, an insertion may result in either a truncated protein or an elongated protein as an expression product.
[0155] A "mutant" nucleic acid sequence typically refers to a nucleic acid sequence that has an alteration in at least one codon occurring in a host cell's wild-type sequence such that the expression product of the mutant nucleic acid sequence is a protein with an altered amino acid sequence relative to the wild-type protein. The expression product may have an altered functional capacity (e.g., enhanced enzymatic activity).
[0156] As used herein, the phrase "alteration in substrate specificity" refers to changes in the substrate specificity of an enzyme. In some embodiments, a change in substrate specificity is defined as a change in k.sub.cat and/or K.sub.m for a particular substrate, resulting from mutations of the enzyme or alteration of reaction conditions. The substrate specificity of an enzyme is determined by comparing the catalytic efficiencies it exhibits with different substrates. These determinations find particular use in assessing the efficiency of mutant enzymes, as it is generally desired to produce variant enzymes that exhibit greater ratios of k.sub.cat/K.sub.m for substrates of interest. However, it is not intended that the present invention be limited to any particular substrate composition or substrate specificity.
[0157] As used herein, "surface property" is used in reference to electrostatic charge, as well as properties such as the hydrophobicity and hydrophilicity exhibited by the surface of a protein. As used herein, the term "net charge" is defined as the sum of all charges present in a molecule. "Net charge changes" are made to a parent protein molecule to provide a variant that has a net charge that differs from that of the parent molecule (i.e., the variant has a net charge that is not the same as that of the parent molecule). For example, substitution of a neutral amino acid with a negatively charged amino acid or a positively charged amino acid with a neutral amino acid results in net charge of -1 with respect to the parent molecule. Substitution of a positively charged amino acid with a negatively charged amino acid results in a net charge of -2 with respect to the parent. Substitution of a neutral amino acid with a positively charged amino acid or a negatively charged amino acid with a neutral amino acid results in net charge of +1 with respect to the parent. Substitution of a negatively charged amino acid with a positively charged amino acid results in a net charge of +2 with respect to the parent. The net charge of a parent protein can also be altered by deletion and/or insertion of charged amino acids. A net change change applies to changes in charge of a variant versus a parent when measured at the same pH conditions.
[0158] The terms "thermally stable" and "thermostable" and "thermostability" refer to proteases that retain a specified amount of enzymatic activity after exposure to identified temperatures over a given period of time under conditions prevailing during the proteolytic, hydrolyzing, cleaning or other process of the invention, while being exposed to altered temperatures. "Altered temperatures" encompass increased or decreased temperatures. In some embodiments, the proteases retain at least about 50%, about 60%, about 70%, about 75%, about 80%, about 85%, about 90%, about 92%, about 95%, about 96%, about 97%, about 98%, or about 99% proteolytic activity after exposure to altered temperatures over a given time period, for example, at least about 60 minutes, about 120 minutes, about 180 minutes, about 240 minutes, about 300 minutes, etc.
[0159] The term "enhanced stability" in the context of an oxidation, chelator, thermal, chemical, autolytic and/or pH stable protease refers to a higher retained proteolytic activity over time as compared to other proteases (e.g., thermolysin proteases) and/or wild-type enzymes.
[0160] The term "diminished stability" in the context of an oxidation, chelator, thermal and/or pH stable protease refers to a lower retained proteolytic activity over time as compared to other proteases (e.g., thermolysin proteases) and/or wild-type enzymes.
[0161] The term "cleaning activity" refers to a cleaning performance achieved by a metalloprotease polypeptide or reference protease under conditions prevailing during the proteolytic, hydrolyzing, cleaning, or other process of the invention. In some embodiments, cleaning performance of a metalloprotease polypeptide or reference protease may be determined by using various assays for cleaning one or more various enzyme sensitive stains on an item or surface (e.g., a stain resulting from food, grass, blood, ink, milk, oil, and/or egg protein). Cleaning performance of a variant or reference protease can be determined by subjecting the stain on the item or surface to standard wash condition(s) and assessing the degree to which the stain is removed by using various chromatographic, spectrophotometric, or other quantitative methodologies. Exemplary cleaning assays and methods are known in the art and include, but are not limited to those described in WO 99/34011 and U.S. Pat. No. 6,605,458, both of which are herein incorporated by reference, as well as those cleaning assays and methods included in the Examples provided below.
[0162] The term "cleaning effective amount" of a metalloprotease polypeptide or reference protease refers to the amount of protease that achieves a desired level of enzymatic activity in a specific cleaning composition. Such effective amounts are readily ascertained by one of ordinary skill in the art and are based on many factors, such as the particular protease used, the cleaning application, the specific composition of the cleaning composition, and whether a liquid or dry (e.g., granular, tablet, bar) composition is required, etc.
[0163] The term "cleaning adjunct material" refers to any liquid, solid, or gaseous material included in cleaning composition other than a metalloprotease polypeptide of the invention. In some embodiments, the cleaning compositions of the present invention include one or more cleaning adjunct materials. Each cleaning adjunct material is typically selected depending on the particular type and form of cleaning composition (e.g., liquid, granule, powder, bar, paste, spray, tablet, gel, foam, or other composition). Preferably, each cleaning adjunct material is compatible with the protease enzyme used in the composition.
[0164] The term "enhanced performance" in the context of cleaning activity refers to an increased or greater cleaning activity by an enzyme with respect to a parent or reference protein as measured on certain enzyme sensitive stains such as egg, milk, grass, ink, oil, and/or blood, as determined by usual evaluation after a standard wash cycle and/or multiple wash cycles.
[0165] The term "diminished performance" in the context of cleaning activity refers to a decreased or lesser cleaning activity by an enzyme on certain enzyme sensitive stains such as egg, milk, grass or blood, as determined by usual evaluation after a standard wash cycle and/or multiple wash cycles.
[0166] Cleaning compositions and cleaning formulations include any composition that is suited for cleaning, bleaching, disinfecting, and/or sterilizing any object, item, and/or surface. Such compositions and formulations include, but are not limited to for example, liquid and/or solid compositions, including cleaning or detergent compositions (e.g., liquid, tablet, gel, bar, granule, and/or solid laundry cleaning or detergent compositions and fine fabric detergent compositions; hard surface cleaning compositions and formulations, such as for glass, wood, ceramic and metal counter tops and windows; carpet cleaners; oven cleaners; fabric fresheners; fabric softeners; and textile, laundry booster cleaning or detergent compositions, laundry additive cleaning compositions, and laundry pre-spotter cleaning compositions; dishwashing compositions, including hand or manual dishwash compositions (e.g., "hand" or "manual" dishwashing detergents) and automatic dishwashing compositions (e.g., "automatic dishwashing detergents").
[0167] Cleaning composition or cleaning formulations, as used herein, include, unless otherwise indicated, granular or powder-form all-purpose or heavy-duty washing agents, especially cleaning detergents; liquid, granular, gel, solid, tablet, or paste-form all-purpose washing agents, especially the so-called heavy-duty liquid (HDL) detergent or heavy-duty powder detergent (HDD) types; liquid fine-fabric detergents; hand or manual dishwashing agents, including those of the high-foaming type; hand or manual dishwashing, automatic dishwashing, or dishware or tableware washing agents, including the various tablet, powder, solid, granular, liquid, gel, and rinse-aid types for household and institutional use; liquid cleaning and disinfecting agents, including antibacterial hand-wash types, cleaning bars, mouthwashes, denture cleaners, car shampoos, carpet shampoos, bathroom cleaners; hair shampoos and/or hair-rinses for humans and other animals; shower gels and foam baths and metal cleaners; as well as cleaning auxiliaries, such as bleach additives and "stain-stick" or pre-treat types. In some embodiments, granular compositions are in "compact" form; in some embodiments, liquid compositions are in a "concentrated" form.
[0168] As used herein, "fabric cleaning compositions" include hand and machine laundry detergent compositions including laundry additive compositions and compositions suitable for use in the soaking and/or pretreatment of stained fabrics (e.g., clothes, linens, and other textile materials).
[0169] As used herein, "non-fabric cleaning compositions" include non-textile (i.e., non-fabric) surface cleaning compositions, including, but not limited to for example, hand or manual or automatic dishwashing detergent compositions, oral cleaning compositions, denture cleaning compositions, and personal cleansing compositions.
[0170] As used herein, the term "fabric and/or hard surface cleaning and/or treatment composition" is a subset of cleaning and treatment compositions that includes, unless otherwise indicated, granular or powder-form all-purpose or "heavy-duty" washing agents, especially cleaning detergents; liquid, gel or paste-form all-purpose washing agents, especially the so-called heavy-duty liquid types; liquid fine-fabric detergents; hand dishwashing agents or light duty dishwashing agents, especially those of the high-foaming type; machine dishwashing agents, including the various tablet, granular, liquid and rinse-aid types for household and institutional use; liquid cleaning and disinfecting agents, car or carpet shampoos, bathroom cleaners including toilet bowl cleaners; fabric conditioning products including softening and/or freshening that may be in liquid, solid and/or dryer sheet form; as well as cleaning auxiliaries such as bleach additives and "stain-stick" or pre-treat types, substrate-laden products such as dryer added sheets. All of such products which are applicable may be in standard, concentrated or even highly concentrated form even to the extent that such products may in certain aspect be non-aqueous.
[0171] As used herein, the term "detergent composition" or "detergent formulation" is used in reference to a composition intended for use in a wash medium for the cleaning of soiled or dirty objects, including particular fabric and/or non-fabric objects or items. Such compositions of the present invention are not limited to any particular detergent composition or formulation. Indeed, in some embodiments, the detergents of the invention comprise at least one metalloprotease polypeptide of the invention and, in addition, one or more surfactants, transferase(s), hydrolytic enzymes, oxido reductases, builders (e.g., a builder salt), bleaching agents, bleach activators, bluing agents, fluorescent dyes, caking inhibitors, masking agents, enzyme activators, antioxidants, and/or solubilizers. In some instances, a builder salt is a mixture of a silicate salt and a phosphate salt, preferably with more silicate (e.g., sodium metasilicate) than phosphate (e.g., sodium tripolyphosphate). Some compositions of the invention, such as, but not limited to, cleaning compositions or detergent compositions, do not contain any phosphate (e.g., phosphate salt or phosphate builder).
[0172] As used herein, the term "bleaching" refers to the treatment of a material (e.g., fabric, laundry, pulp, etc.) or surface for a sufficient length of time and/or under appropriate pH and/or temperature conditions to effect a brightening (i.e., whitening) and/or cleaning of the material. Examples of chemicals suitable for bleaching include, but are not limited to, for example, ClO.sub.2, H.sub.2O.sub.2, peracids, NO.sub.2, etc.
[0173] As used herein, "wash performance" of a protease (e.g., a metalloprotease polypeptide of the invention) refers to the contribution of a metalloprotease polypeptide to washing that provides additional cleaning performance to the detergent as compared to the detergent without the addition of the metalloprotease polypeptide to the composition. Wash performance is compared under relevant washing conditions. In some test systems, other relevant factors, such as detergent composition, sud concentration, water hardness, washing mechanics, time, pH, and/or temperature, can be controlled in such a way that condition(s) typical for household application in a certain market segment (e.g., hand or manual dishwashing, automatic dishwashing, dishware cleaning, tableware cleaning, fabric cleaning, etc.) are imitated.
[0174] The term "relevant washing conditions" is used herein to indicate the conditions, particularly washing temperature, time, washing mechanics, sud concentration, type of detergent and water hardness, actually used in households in a hand dishwashing, automatic dishwashing, or laundry detergent market segment.
[0175] The term "improved wash performance" is used to indicate that a better end result is obtained in stain removal under relevant washing conditions, or that less metalloprotease polypeptide, on weight basis, is needed to obtain the same end result relative to the corresponding wild-type or starting parent protease.
[0176] As used herein, the term "disinfecting" refers to the removal of contaminants from the surfaces, as well as the inhibition or killing of microbes on the surfaces of items. It is not intended that the present invention be limited to any particular surface, item, or contaminant(s) or microbes to be removed.
[0177] The "compact" form of the cleaning compositions herein is best reflected by density and, in terms of composition, by the amount of inorganic filler salt. Inorganic filler salts are conventional ingredients of detergent compositions in powder form. In conventional detergent compositions, the filler salts are present in substantial amounts, typically about 17 to about 35% by weight of the total composition. In contrast, in compact compositions, the filler salt is present in amounts not exceeding about 15% of the total composition. In some embodiments, the filler salt is present in amounts that do not exceed about 10%, or more preferably, about 5%, by weight of the composition. In some embodiments, the inorganic filler salts are selected from the alkali and alkaline-earth-metal salts of sulfates and chlorides. In some embodiments, the filler salt is sodium sulfate.
[0178] As used herein in connection with a numerical value, the term "about" refers to a range of +/-0.5 of the numerical value, unless the term is otherwise specifically defined in context. For instance, the phrase a "pH value of about 6" refers to pH values of from 5.5 to 6.5, unless the pH value is specifically defined otherwise.
[0179] The position of an amino acid residue in a given amino acid sequence is typically numbered herein using the numbering of the position of the corresponding amino acid residue of the wild type Paenibacillus metalloprotease amino acid sequences shown in SEQ ID NOs: 3, 8, 13, 18, 23, 28, 33 or 38. The Paenibacillus sp. metalloprotease amino acid sequences, thus serves as a reference parent sequence. A given amino acid sequence, such as a metalloprotease enzyme amino acid sequence and variants thereof described herein, can be aligned with the wild type metalloprotease sequence (e.g., SEQ ID NO: 3) using an alignment algorithm as described herein, and an amino acid residue in the given amino acid sequence that aligns (preferably optimally aligns) with an amino acid residue in the wild type sequence can be conveniently numbered by reference to the corresponding amino acid residue in the metalloprotease sequence.
[0180] Oligonucleotide synthesis and purification steps are typically performed according to specifications. Techniques and procedures are generally performed according to conventional methods well known in the art and various general references that are provided throughout this document. Procedures therein are believed to be well known to those of ordinary skill in the art and are provided for the convenience of the reader.
Metalloprotease Polypeptides of the Present Invention
[0181] The present invention provides novel metalloprotease enzyme polypeptides, which may be collectively referred to as "enzymes of the invention" or "polypeptides of the invention." Polypeptides of the invention include isolated, recombinant, substantially pure, or non-naturally occurring polypeptides. In some embodiments, polypeptides of the invention are useful in cleaning applications and can be incorporated into cleaning compositions that are useful in methods of cleaning an item or a surface in need of cleaning.
[0182] In some embodiments, the enzyme of the present invention has 50, 60, 65, 70, 75, 80, 85, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99 or 100% identity to SEQ ID NOs: 3, 8, 13, 18, 23, 28, 33 or 38. In various embodiments, the enzyme of the present invention has 50, 60, 65, 70, 75, 80, 85, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99 or 100% identity to a metalloprotease enzyme from any genus in Tables 1.2, 2.2, 3.2, 4.2, 5.2, 6.2, 7.2 or 8.2.
[0183] In some embodiments, the enzyme of the present invention, including all embodiments supra, can be derived from a member of the order Bacillales or family Bacillaceae, Paenibacillaceae, Alicyclobacillaceae, or Lactobacillaceae. In some embodiments, the enzyme of the present invention, including all embodiments supra, can be derived from a Bacillus, Alicyclobacillus, Geobacillus, Exiguobacterium, Lactobacillus, or Paenibacillus species. In some embodiments, the enzyme of the present invention, including all embodiments supra, can be derived from a member of the Pseudococcidae family. In some embodiments, the enzyme of the present invention, including all embodiments supra, can be derived from a Planococcus species. Various enzyme metalloproteases have been found that have a high identity to each other and to the Paenibacillus enzymes as shown in SEQ ID NOs: 3, 8, 13, 18, 23, 28, 33 or 38.
[0184] In a particular embodiment, the invention is an enzyme derived from the genus Paenibacillus. In a particular embodiment, the invention is an enzyme derived from the genus Paenibacillus and from the species Paenibacillus sp., Paenibacillus ehimensis, Paenibacillus hunanensis, Paenibacillus barcinonensis, Paenibacillus amylolyticus, Paenibacillus humicus and Paenibacillus polymyxa.
[0185] Described are compositions and methods relating to enzymes cloned from Paenibacillus. The compositions and methods are based, in part, on the observation that cloned and expressed enzymes of the present invention have proteolytic activity in the presence of a detergent composition. Enzymes of the present invention also demonstrate excellent stability in detergent compositions. These features makes enzymes of the present invention well suited for use in a variety of cleaning applications, where the enzyme can hydrolyze proteins in the presence of surfactants and other components found in detergent compositions.
[0186] In some embodiments, the invention includes an isolated, recombinant, substantially pure, or non-naturally occurring enzyme having protease activity, which polypeptide comprises a polypeptide sequence having at least about 60%, 65%, 70%, 75%, 80%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% sequence identity to a parent enzyme as provided herein.
[0187] In some embodiments, the polypeptide of the present invention, is a polypeptide having a specified degree of amino acid sequence homology to the exemplified polypeptides, e.g., at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or even at least 99% sequence homology to the amino acid sequences of SEQ ID NOs: 3, 8, 13, 18, 23, 28, 33 or 38. Homology can be determined by amino acid sequence alignment, e.g., using a program such as BLAST, ALIGN, or CLUSTAL, as described herein.
[0188] Also provided are polypeptide enzymes of the present invention, having protease activity, said enzymes comprising an amino acid sequence which differs from the amino acid sequence of SEQ ID NOs: 3, 8, 13, 18, 23, 28, 33 or 38 by no more than 50, no more than 40, no more than 30, no more than 35, no more than 25, no more than 20, no more than 19, no more than 18, no more than 17, no more than 16, no more than 15, no more than 14, no more than 13, no more than 12, no more than 11, no more than 10, no more than 9, no more than 8, no more than 7, no more than 6, no more than 5, no more than 4, no more than 3, no more than 2, or no more than 1 amino acid residue(s), when aligned using any of the previously described alignment methods.
[0189] As noted above, the variant enzyme polypeptides of the invention have enzymatic activities (e.g., protease activities) and thus are useful in cleaning applications, including but not limited to, methods for cleaning dishware items, tableware items, fabrics, and items having hard surfaces (e.g., the hard surface of a table, table top, wall, furniture item, floor, ceiling, etc.). Exemplary cleaning compositions comprising one or more variant metalloprotease enzyme polypeptides of the invention are described infra. The enzymatic activity (e.g., protease enzyme activity) of an enzyme polypeptide of the invention can be determined readily using procedures well known to those of ordinary skill in the art. The Examples presented infra describe methods for evaluating the enzymatic activity and cleaning performance. The performance of polypeptide enzymes of the invention in removing stains (e.g., a protein stain such as blood/milk/ink or egg yolk), cleaning hard surfaces, or cleaning laundry, dishware or tableware item(s) can be readily determined using procedures well known in the art and/or by using procedures set forth in the Examples.
[0190] The metalloprotease polypeptides of the invention have protease activity such that they are useful in casein hydrolysis, collagen hydrolysis, elastin hydrolysis, keratin hydrolysis, soy protein hydrolysis or corn meal protein hydrolysis. Thus, the polypeptides of the invention find use in other applications such as pretreatments for food, feed, or protein degradation.
[0191] The polypeptides of the invention are also useful in pretreatment of animal feed products, such as soy protein, corn meal, and other protein rich components. Pretreatment of these animal feed products with a polypeptide of the invention may help in the breakdown of complex proteins into their hydrolysates which are easily digestible by animals.
[0192] In yet other embodiments, the disclosed metalloprotease polypeptides find use in hydrolysis of corn soy protein. The disclosed metalloprotease polypeptides may be used alone or in combination with other proteases, amylases or lipases to produce peptides and free amino acids from the corn or soy protein. In some embodiments, the recovered proteins, peptides or amino acids can be subsequently used in animal feed or human food products.
[0193] The polypeptides of the invention are also useful in treatment of wounds, particularly in wound debridement. Wound debridement is the removal of dead, damaged or infected tissue to improve the healing potential of the remaining healthy tissue. Debridement is an important part of the healing process for burns and other serious wounds. The wounds or burns may be treated with a composition comprising a polypeptide of the invention which would result in removal of unwanted damaged tissue and improving the healthy tissue.
[0194] The metalloprotease polypeptides of the present invention can have protease activity over a broad range of pH conditions. In some embodiments, the metalloprotease polypeptides have protease activity on azo-casein as a substrate, as demonstrated in Examples 3.1 to 3.8. In some embodiments, the metalloprotease polypeptides have protease activity at a pH of from about 3.0 to about 12.0. In some embodiments, the metalloprotease polypeptides have protease activity at a pH of from about 4.0 to about 10.5. In some embodiments, the metalloprotease polypeptides have at least 70% of maximal protease activity at a pH of from about 5.5 to about 9.0. In some embodiments, the metalloprotease polypeptides have at least 80% of maximal protease activity at a pH of from about 6.0 to about 8.5. In some embodiments, the metalloprotease polypeptides have maximal protease activity at a pH of about 7.5.
[0195] In some embodiments, the metalloprotease polypeptides of the present invention have protease activity at a temperature range of from about 10.degree. C. to about 100.degree. C. In some embodiments, the metalloprotease polypeptides of the present invention have protease activity at a temperature range of from about 20.degree. C. to about 90.degree. C. In some embodiments, the metalloprotease polypeptides have at least 70% of maximal protease activity at a temperature of from about 45.degree. C. to about 60.degree. C. In some embodiments, the metalloprotease polypeptides have maximal protease activity at a temperature of 50.degree. C.
[0196] In some embodiments, the metalloprotease polypeptides of the present invention demonstrate cleaning performance in a cleaning composition. Cleaning compositions often include ingredients harmful to the stability and performance of enzymes, making cleaning compositions a harsh environment for enzymes, e.g. metalloproteases, to retain function. Thus, it is not trivial for an enzyme to be put in a cleaning composition and expect enzymatic function (e.g. metalloprotease activity, such as demonstrated by cleaning performance). In some embodiments, the metalloprotease polypeptides of the present invention demonstrate cleaning performance in automatic dishwashing (ADW) detergent compositions. In some embodiments, the cleaning performance in automatic dishwashing (ADW) detergent compositions includes cleaning of egg yolk stains. In some embodiments, the metalloprotease polypeptides of the present invention demonstrate cleaning performance in laundry detergent compositions. In some embodiments, the cleaning performance in laundry detergent compositions includes cleaning of blood/milk/ink stains. In each of the cleaning compositions, the metalloprotease polypeptides of the present invention demonstrate cleaning performance with or without a bleach component.
[0197] The metalloprotease polypeptides of the invention have protease activity such that they are useful in casein hydrolysis, collagen hydrolysis, elastin hydrolysis, keratin hydrolysis, soy protein hydrolysis or corn meal protein hydrolysis. Thus, the polypeptides of the invention find use in other applications such as pretreatments for food, feed, or protein degradation.
[0198] A polypeptide of the invention can be subject to various changes, such as one or more amino acid insertions, deletions, and/or substitutions, either conservative or non-conservative, including where such changes do not substantially alter the enzymatic activity of the polypeptide. Similarly, a nucleic acid of the invention can also be subject to various changes, such as one or more substitutions of one or more nucleotides in one or more codons such that a particular codon encodes the same or a different amino acid, resulting in either a silent variation (e.g., when the encoded amino acid is not altered by the nucleotide mutation) or non-silent variation, one or more deletions of one or more nucleic acids (or codons) in the sequence, one or more additions or insertions of one or more nucleic acids (or codons) in the sequence, and/or cleavage of or one or more truncations of one or more nucleic acids (or codons) in the sequence. Many such changes in the nucleic acid sequence may not substantially alter the enzymatic activity of the resulting encoded polypeptide enzyme compared to the polypeptide enzyme encoded by the original nucleic acid sequence. A nucleic acid sequence of the invention can also be modified to include one or more codons that provide for optimum expression in an expression system (e.g., bacterial expression system), while, if desired, said one or more codons still encode the same amino acid(s).
[0199] In some embodiments, the present invention provides a genus of enzyme polypeptides having the desired enzymatic activity (e.g., protease enzyme activity or cleaning performance activity) which comprise sequences having the amino acid substitutions described herein and also which comprise one or more additional amino acid substitutions, such as conservative and non-conservative substitutions, wherein the polypeptide exhibits, maintains, or approximately maintains the desired enzymatic activity (e.g., proteolytic activity, as reflected in the cleaning activity or performance of the polypeptide enzymes of SEQ ID NOs: 3, 8, 13, 18, 23, 28, 33 and 38). Amino acid substitutions in accordance with the invention may include, but are not limited to, one or more non-conservative substitutions and/or one or more conservative amino acid substitutions. A conservative amino acid residue substitution typically involves exchanging a member within one functional class of amino acid residues for a residue that belongs to the same functional class (conservative amino acid residues are considered functionally homologous or conserved in calculating percent functional homology). A conservative amino acid substitution typically involves the substitution of an amino acid in an amino acid sequence with a functionally similar amino acid. For example, alanine, glycine, serine, and threonine are functionally similar and thus may serve as conservative amino acid substitutions for one another. Aspartic acid and glutamic acid may serve as conservative substitutions for one another.
[0200] Asparagine and glutamine may serve as conservative substitutions for one another. Arginine, lysine, and histidine may serve as conservative substitutions for one another. Isoleucine, leucine, methionine, and valine may serve as conservative substitutions for one another. Phenylalanine, tyrosine, and tryptophan may serve as conservative substitutions for one another. Other conservative amino acid substitution groups can be envisioned. For example, amino acids can be grouped by similar function or chemical structure or composition (e.g., acidic, basic, aliphatic, aromatic, sulfur-containing). For instance, an aliphatic grouping may comprise: Glycine (G), Alanine (A), Valine (V), Leucine (L), Isoleucine (I). Other groups containing amino acids that are considered conservative substitutions for one another include: aromatic: Phenylalanine (F), Tyrosine (Y), Tryptophan (W); sulfur-containing: Methionine (M), Cysteine (C); Basic: Arginine (R), Lysine (K), Histidine (H); Acidic: Aspartic acid (D), Glutamic acid (E); non-polar uncharged residues, Cysteine (C), Methionine (M), and Proline (P); hydrophilic uncharged residues: Serine (S), Threonine (T), Asparagine (N), and Glutamine (Q). Additional groupings of amino acids are well-known to those of skill in the art and described in various standard textbooks. Listing of a polypeptide sequence herein, in conjunction with the above substitution groups, provides an express listing of all conservatively substituted polypeptide sequences.
[0201] More conservative substitutions exist within the amino acid residue classes described above, which also or alternatively can be suitable. Conservation groups for substitutions that are more conservative include: valine-leucine-isoleucine, phenylalanine-tyrosine, lysine-arginine, alanine-valine, and asparagine-glutamine.
[0202] Conservatively substituted variations of a polypeptide sequence of the invention (e.g., variant metalloproteases of the invention) include substitutions of a small percentage, sometimes less than 25%, 20%, 15%, 14%, 13%, 12%, 11%, 10%, 9%, 8%, 7%, or 6% of the amino acids of the polypeptide sequence, or less than 5%, 4%, 3%, 2%, or 1%, or less than 10, 9, 8, 7, 6, 5, 4, 3, 2, or 1 amino acid substitution of the amino acids of the polypeptide sequence, with a conservatively selected amino acid of the same conservative substitution group.
[0203] As described elsewhere herein in greater detail and in the Examples provided herein, polypeptides of the invention may have cleaning abilities that may be compared to known proteases, including known metalloproteases.
Nucleic Acids of the Invention
[0204] The invention provides isolated, non-naturally occurring, or recombinant nucleic acids which may be collectively referred to as "nucleic acids of the invention" or "polynucleotides of the invention", which encode polypeptides of the invention. Nucleic acids of the invention, including all described below, are useful in recombinant production (e.g., expression) of polypeptides of the invention, typically through expression of a plasmid expression vector comprising a sequence encoding the polypeptide of interest or fragment thereof. As discussed above, polypeptides include metalloprotease polypeptides having enzymatic activity (e.g., proteolytic activity) which are useful in cleaning applications and cleaning compositions for cleaning an item or a surface (e.g., surface of an item) in need of cleaning.
[0205] In some embodiments, the invention provides an isolated, recombinant, substantially pure, or non-naturally occurring nucleic acid comprising a nucleotide sequence encoding any polypeptide (including any fusion protein, etc.) of the invention described above in the section entitled "Polypeptides of the Invention" and elsewhere herein. The invention also provides an isolated, recombinant, substantially pure, or non-naturally-occurring nucleic acid comprising a nucleotide sequence encoding a combination of two or more of any polypeptides of the invention described above and elsewhere herein. In some embodiments, the nucleic acids of the present invention has 50, 60, 65, 70, 75, 80, 85, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99 or 100% identity to SEQ ID NO: 4, 9, 14, 19, 24, 29, 34 and 39.
[0206] The present invention provides nucleic acids encoding a metalloprotease polypeptide of the present invention, wherein the metalloprotease polypeptide is a mature form having proteolytic activity, wherein the amino acid positions of the variant are numbered by correspondence with the amino acid sequence of Paenibacillus metalloprotease polypeptides set forth as SEQ ID NOs: 3, 8, 13, 18, 23, 28, 33 or 38.
[0207] Nucleic acids of the invention can be generated by using any suitable synthesis, manipulation, and/or isolation techniques, or combinations thereof. For example, a polynucleotide of the invention may be produced using standard nucleic acid synthesis techniques, such as solid-phase synthesis techniques that are well-known to those skilled in the art. In such techniques, fragments of up to 50 or more nucleotide bases are typically synthesized, then joined (e.g., by enzymatic or chemical ligation methods) to form essentially any desired continuous nucleic acid sequence. The synthesis of the nucleic acids of the invention can be also facilitated by any suitable method known in the art, including but not limited to chemical synthesis using the classical phosphoramidite method (See e.g., Beaucage et al. Tetrahedron Letters 22:1859-69 [1981]); or the method described by Matthes et al. (See, Matthes et al., EMBO J. 3:801-805 [1984], as is typically practiced in automated synthetic methods. Nucleic acids of the invention also can be produced by using an automatic DNA synthesizer. Customized nucleic acids can be ordered from a variety of commercial sources (e.g., The Midland Certified Reagent Company, the Great American Gene Company, Operon Technologies Inc., and DNA2.0). Other techniques for synthesizing nucleic acids and related principles are known in the art (See e.g., Itakura et al., Ann. Rev. Biochem. 53:323 [1984]; and Itakura et al., Science 198:1056 [1984]).
[0208] As indicated above, recombinant DNA techniques useful in modification of nucleic acids are well known in the art. For example, techniques such as restriction endonuclease digestion, ligation, reverse transcription and cDNA production, and polymerase chain reaction (e.g., PCR) are known and readily employed by those of skill in the art. Nucleotides of the invention may also be obtained by screening cDNA libraries using one or more oligonucleotide probes that can hybridize to or PCR-amplify polynucleotides which encode a metalloprotease polypeptide polypeptide(s) of the invention. Procedures for screening and isolating cDNA clones and PCR amplification procedures are well known to those of skill in the art and described in standard references known to those skilled in the art. Some nucleic acids of the invention can be obtained by altering a naturally occurring polynucleotide backbone (e.g., that encodes an enzyme or parent protease) by, for example, a known mutagenesis procedure (e.g., site-directed mutagenesis, site saturation mutagenesis, and in vitro recombination).
Methods for Making Modified Metalloprotease Polypeptides of the Invention
[0209] A variety of methods are known in the art that are suitable for generating modified polynucleotides of the invention that encode metalloprotease polypeptides of the invention, including, but not limited to, for example, site-saturation mutagenesis, scanning mutagenesis, insertional mutagenesis, deletion mutagenesis, random mutagenesis, site-directed mutagenesis, and directed-evolution, as well as various other recombinatorial approaches. Methods for making modified polynucleotides and proteins (e.g., metalloprotease polypeptides) include DNA shuffling methodologies, methods based on non-homologous recombination of genes, such as ITCHY (See, Ostermeier et al., 7:2139-44 [1999]), SCRACHY (See, Lutz et al. 98:11248-53 [2001]), SHIPREC (See, Sieber et al., 19:456-60 [2001]), and NRR (See, Bittker et al., 20:1024-9 [2001]; Bittker et al., 101:7011-6 [2004]), and methods that rely on the use of oligonucleotides to insert random and targeted mutations, deletions and/or insertions (See, Ness et al., 20:1251-5 [2002]; Coco et al., 20:1246-50 [2002]; Zha et al., 4:34-9 [2003]; Glaser et al., 149:3903-13 [1992]).
Vectors, Cells, and Methods for Producing Metalloprotease Polypeptides of the Invention
[0210] The present invention provides vectors comprising at least one metalloprotease polynucleotide of the invention described herein (e.g., a polynucleotide encoding a metalloprotease polypeptide of the invention described herein), expression vectors or expression cassettes comprising at least one nucleic acid or polynucleotide of the invention, isolated, substantially pure, or recombinant DNA constructs comprising at least one nucleic acid or polynucleotide of the invention, isolated or recombinant cells comprising at least one polynucleotide of the invention, and compositions comprising one or more such vectors, nucleic acids, expression vectors, expression cassettes, DNA constructs, cells, cell cultures, or any combination or mixtures thereof.
[0211] In some embodiments, the invention provides recombinant cells comprising at least one vector (e.g., expression vector or DNA construct) of the invention which comprises at least one nucleic acid or polynucleotide of the invention. Some such recombinant cells are transformed or transfected with such at least one vector. Such cells are typically referred to as host cells. Some such cells comprise bacterial cells, including, but are not limited to Bacillus sp. cells, such as B. subtilis cells. The invention also provides recombinant cells (e.g., recombinant host cells) comprising at least one metalloprotease polypeptide of the invention.
[0212] In some embodiments, the invention provides a vector comprising a nucleic acid or polynucleotide of the invention. In some embodiments, the vector is an expression vector or expression cassette in which a polynucleotide sequence of the invention which encodes a metalloprotease polypeptide of the invention is operably linked to one or additional nucleic acid segments required for efficient gene expression (e.g., a promoter operably linked to the polynucleotide of the invention which encodes a metalloprotease polypeptide of the invention). A vector may include a transcription terminator and/or a selection gene, such as an antibiotic resistance gene, that enables continuous cultural maintenance of plasmid-infected host cells by growth in antimicrobial-containing media.
[0213] An expression vector may be derived from plasmid or viral DNA, or in alternative embodiments, contains elements of both. Exemplary vectors include, but are not limited to pC194, pJH101, pE194, pHP13 (See, Harwood and Cutting [eds.], Chapter 3, Molecular Biological Methods for Bacillus, John Wiley & Sons [1990]; suitable replicating plasmids for B. subtilis include those listed on p. 92) See also, Perego, Integrational Vectors for Genetic Manipulations in Bacillus subtilis, in Sonenshein et al., [eds.] Bacillus subtilis and Other Gram-Positive Bacteria: Biochemistry, Physiology and Molecular Genetics, American Society for Microbiology, Washington, D.C. [1993], pp. 615-624), and p2JM103BBI.
[0214] For expression and production of a protein of interest (e.g., metalloprotease polypeptide) in a cell, at least one expression vector comprising at least one copy of a polynucleotide encoding the metalloprotease polypeptide, and in some instances comprising multiple copies, is transformed into the cell under conditions suitable for expression of the metalloprotease. In some embodiments of the present invention, a polynucleotide sequence encoding the metalloprotease polypeptide (as well as other sequences included in the vector) is integrated into the genome of the host cell, while in other embodiments, a plasmid vector comprising a polynucleotide sequence encoding the metalloprotease polypeptide remains as autonomous extra-chromosomal element within the cell. The invention provides both extrachromosomal nucleic acid elements as well as incoming nucleotide sequences that are integrated into the host cell genome. The vectors described herein are useful for production of the metalloprotease polypeptides of the invention. In some embodiments, a polynucleotide construct encoding the metalloprotease polypeptide is present on an integrating vector that enables the integration and optionally the amplification of the polynucleotide encoding the metalloprotease polypeptide into the host chromosome. Examples of sites for integration are well known to those skilled in the art. In some embodiments, transcription of a polynucleotide encoding a metalloprotease polypeptide of the invention is effectuated by a promoter that is the wild-type promoter for the selected precursor protease. In some other embodiments, the promoter is heterologous to the precursor protease, but is functional in the host cell. Specifically, examples of suitable promoters for use in bacterial host cells include, but are not limited to, for example, the amyE, amyQ, amyL, pstS, sacB, pSPAC, pAprE, pVeg, pHpaII promoters, the promoter of the B. stearothermophilus maltogenic amylase gene, the B. amyloliquefaciens (BAN) amylase gene, the B. subtilis alkaline protease gene, the B. clausii alkaline protease gene the B. pumilis xylosidase gene, the B. thuringiensis cryIIIA, and the B. licheniformis alpha-amylase gene. Additional promoters include, but are not limited to the A4 promoter, as well as phage Lambda PR or PL promoters, and the E. coli lac, trp or tac promoters.
[0215] Metalloprotease polypeptides of the present invention can be produced in host cells of any suitable microorganism, including bacteria and fungi. In some embodiments, metalloprotease polypeptides of the present invention can be produced in Gram-positive bacteria. In some embodiments, the host cells are Bacillus spp., Streptomyces spp., Escherichia spp., Aspergillus spp., Trichoderma spp., Pseudomonas spp., Corynebacterium spp., Saccharomyces spp., or Pichia spp. In some embodiments, the metalloprotease polypeptides are produced by Bacillus sp. host cells. Examples of Bacillus sp. host cells that find use in the production of the metalloprotease polypeptides of the invention include, but are not limited to B. licheniformis, B. lentus, B. subtilis, B. amyloliquefaciens, B. lentus, B. brevis, B. stearothermophilus, B. alkalophilus, B. coagulans, B. circulans, B. pumilis, B. thuringiensis, B. clausii, and B. megaterium, as well as other organisms within the genus Bacillus. In some embodiments, B. subtilis host cells are used for production of metalloprotease polypeptides. U.S. Pat. Nos. 5,264,366 and 4,760,025 (RE 34,606) describe various Bacillus host strains that can be used for producing metalloprotease polypeptide of the invention, although other suitable strains can be used.
[0216] Several bacterial strains that can be used to produce metalloprotease polypeptides of the invention include non-recombinant (i.e., wild-type) Bacillus sp. strains, as well as variants of naturally-occurring strains and/or recombinant strains. In some embodiments, the host strain is a recombinant strain, wherein a polynucleotide encoding a polypeptide of interest has been introduced into the host. In some embodiments, the host strain is a B. subtilis host strain and particularly a recombinant Bacillus subtilis host strain. Numerous B. subtilis strains are known, including, but not limited to for example, 1A6 (ATCC 39085), 168 (1A01), SB19, W23, Ts85, B637, PB1753 through PB1758, PB3360, JH642, 1A243 (ATCC 39,087), ATCC 21332, ATCC 6051, MI113, DE100 (ATCC 39,094), GX4931, PBT 110, and PEP 211strain (See e.g., Hoch et al., Genetics 73:215-228 [1973]; See also, U.S. Pat. Nos. 4,450,235 and 4,302,544, and EP 0134048, each of which is incorporated by reference in its entirety). The use of B. subtilis as an expression host cells is well known in the art (See e.g., Palva et al., Gene 19:81-87 [1982]; Fahnestock and Fischer, J. Bacteriol., 165:796-804 [1986]; and Wang et al., Gene 69:39-47 [1988]).
[0217] In some embodiments, the Bacillus host cell is a Bacillus sp. that includes a mutation or deletion in at least one of the following genes, degU, degS, degR and degQ. In some embodiments, the mutation is in a degU gene, and in some embodiments the mutation is degU(Hy)32 (See e.g., Msadek et al., J. Bacteriol. 172:824-834 [1990]; and Olmos et al., Mol. Gen. Genet. 253:562-567 [1997]). In some embodiments, the Bacillus host comprises a mutation or deletion in scoC4 (See e.g., Caldwell et al., J. Bacteriol. 183:7329-7340 [2001]); spollE (See e.g., Arigoni et al., Mol. Microbiol. 31:1407-1415 [1999]); and/or oppA or other genes of the opp operon (See e.g., Perego et al., Mol. Microbiol. 5:173-185 [1991]). Indeed, it is contemplated that any mutation in the opp operon that causes the same phenotype as a mutation in the oppA gene will find use in some embodiments of the altered Bacillus strain of the invention. In some embodiments, these mutations occur alone, while in other embodiments, combinations of mutations are present. In some embodiments, an altered Bacillus host cell strain that can be used to produce a metalloprotease polypeptide of the invention is a Bacillus host strain that already includes a mutation in one or more of the above-mentioned genes. In addition, Bacillus sp. host cells that comprise mutation(s) and/or deletions of endogenous protease genes find use. In some embodiments, the Bacillus host cell comprises a deletion of the aprE and the nprE genes. In other embodiments, the Bacillus sp. host cell comprises a deletion of 5 protease genes, while in other embodiments, the Bacillus sp. host cell comprises a deletion of 9 protease genes (See e.g., U.S. Pat. Appln. Pub. No. 2005/0202535, incorporated herein by reference).
[0218] Host cells are transformed with at least one nucleic acid encoding at least one metalloprotease polypeptide of the invention using any suitable method known in the art. Methods for introducing a nucleic acid (e.g., DNA) into Bacillus cells or E. coli cells utilizing plasmid DNA constructs or vectors and transforming such plasmid DNA constructs or vectors into such cells are well known. In some embodiments, the plasmids are subsequently isolated from E. coli cells and transformed into Bacillus cells. However, it is not essential to use intervening microorganisms such as E. coli, and in some embodiments, a DNA construct or vector is directly introduced into a Bacillus host.
[0219] Those of skill in the art are well aware of suitable methods for introducing nucleic acid sequences of the invention into Bacillus cells (See e.g., Ferrari et al., "Genetics," in Harwood et al. [eds.], Bacillus, Plenum Publishing Corp. [1989], pp. 57-72; Saunders et al., J. Bacteriol. 157:718-726 [1984]; Hoch et al., J. Bacteriol. 93:1925-1937 [1967]; Mann et al., Current Microbiol. 13:131-135 [1986]; Holubova, Folia Microbiol. 30:97 [1985]; Chang et al., Mol. Gen. Genet. 168:11-115 [1979]; Vorobjeva et al., FEMS Microbiol. Lett. 7:261-263 [1980]; Smith et al., Appl. Env. Microbiol. 51:634 [1986]; Fisher et al., Arch. Microbiol. 139:213-217 [1981]; and McDonald, J. Gen. Microbiol. 130:203 [1984]). Indeed, such methods as transformation, including protoplast transformation and transfection, transduction, and protoplast fusion are well known and suited for use in the present invention. Methods known in the art to transform Bacillus cells include such methods as plasmid marker rescue transformation, which involves the uptake of a donor plasmid by competent cells carrying a partially homologous resident plasmid (See, Contente et al., Plasmid 2:555-571 [1979]; Haima et al., Mol. Gen. Genet. 223:185-191 [1990]; Weinrauch et al., J. Bacteriol. 154:1077-1087 [1983]; and Weinrauch et al., J. Bacteriol. 169:1205-1211 [1987]). In this method, the incoming donor plasmid recombines with the homologous region of the resident "helper" plasmid in a process that mimics chromosomal transformation.
[0220] In addition to commonly used methods, in some embodiments, host cells are directly transformed with a DNA construct or vector comprising a nucleic acid encoding a metalloprotease polypeptide of the invention (i.e., an intermediate cell is not used to amplify, or otherwise process, the DNA construct or vector prior to introduction into the host cell). Introduction of the DNA construct or vector of the invention into the host cell includes those physical and chemical methods known in the art to introduce a nucleic acid sequence (e.g., DNA sequence) into a host cell without insertion into the host genome. Such methods include, but are not limited to calcium chloride precipitation, electroporation, naked DNA, liposomes and the like. In additional embodiments, DNA constructs or vector are co-transformed with a plasmid, without being inserted into the plasmid. In further embodiments, a selective marker is deleted from the altered Bacillus strain by methods known in the art (See, Stahl et al., J. Bacteriol. 158:411-418 [1984]; and Palmeros et al., Gene 247:255-264 [2000]).
[0221] In some embodiments, the transformed cells of the present invention are cultured in conventional nutrient media. The suitable specific culture conditions, such as temperature, pH and the like are known to those skilled in the art and are well described in the scientific literature. In some embodiments, the invention provides a culture (e.g., cell culture) comprising at least one metalloprotease polypeptide or at least one nucleic acid of the invention.
[0222] In some embodiments, host cells transformed with at least one polynucleotide sequence encoding at least one metalloprotease polypeptide of the invention are cultured in a suitable nutrient medium under conditions permitting the expression of the present protease, after which the resulting protease is recovered from the culture. In some embodiments, the protease produced by the cells is recovered from the culture medium by conventional procedures, including, but not limited to for example, separating the host cells from the medium by centrifugation or filtration, precipitating the proteinaceous components of the supernatant or filtrate by means of a salt (e.g., ammonium sulfate), chromatographic purification (e.g., ion exchange, gel filtration, affinity, etc.).
[0223] In some embodiments, a metalloprotease polypeptide produced by a recombinant host cell is secreted into the culture medium. A nucleic acid sequence that encodes a purification facilitating domain may be used to facilitate purification of proteins. A vector or DNA construct comprising a polynucleotide sequence encoding a metalloprotease polypeptide may further comprise a nucleic acid sequence encoding a purification facilitating domain to facilitate purification of the metalloprotease polypeptide (See e.g., Kroll et al., DNA Cell Biol. 12:441-53 [1993]). Such purification facilitating domains include, but are not limited to, for example, metal chelating peptides such as histidine-tryptophan modules that allow purification on immobilized metals (See, Porath, Protein Expr. Purif. 3:263-281 [1992]), protein A domains that allow purification on immobilized immunoglobulin, and the domain utilized in the FLAGS extension/affinity purification system. The inclusion of a cleavable linker sequence such as Factor XA or enterokinase (e.g., sequences available from Invitrogen, San Diego, Calif.) between the purification domain and the heterologous protein also find use to facilitate purification.
[0224] Assays for detecting and measuring the enzymatic activity of an enzyme, such as a metalloprotease polypeptide of the invention, are well known. Various assays for detecting and measuring activity of proteases (e.g., metalloprotease polypeptides of the invention), are also known to those of ordinary skill in the art. In particular, assays are available for measuring protease activity that are based on the release of acid-soluble peptides from casein or hemoglobin, measured as absorbance at 280 nm or colorimetrically using the Folin method. Other exemplary assays involve the solubilization of chromogenic substrates (See e.g., Ward, "Proteinases," in Fogarty (ed.)., Microbial Enzymes and Biotechnology, Applied Science, London, [1983], pp. 251-317). Other exemplary assays include, but are not limited to succinyl-Ala-Ala-Pro-Phe-para nitroanilide assay (suc-AAPF-pNA)(SEQ ID NO: 43) and the 2,4,6-trinitrobenzene sulfonate sodium salt assay (TNBS assay). Numerous additional references known to those in the art provide suitable methods (See e.g., Wells et al., Nucleic Acids Res. 11:7911-7925 [1983]; Christianson et al., Anal. Biochem. 223:119-129 [1994]; and Hsia et al., Anal Biochem. 242:221-227 [1999]).
[0225] A variety of methods can be used to determine the level of production of a mature protease (e.g., mature metalloprotease polypeptides of the present invention) in a host cell. Such methods include, but are not limited to, for example, methods that utilize either polyclonal or monoclonal antibodies specific for the protease. Exemplary methods include, but are not limited to enzyme-linked immunosorbent assays (ELISA), radioimmunoassays (MA), fluorescent immunoassays (FIA), and fluorescent activated cell sorting (FACS). These and other assays are well known in the art (See e.g., Maddox et al., J. Exp. Med. 158:1211 [1983]).
[0226] In some other embodiments, the invention provides methods for making or producing a mature metalloprotease polypeptide of the invention. A mature metalloprotease polypeptide does not include a signal peptide or a propeptide sequence. Some methods comprise making or producing a metalloprotease polypeptide of the invention in a recombinant bacterial host cell, such as for example, a Bacillus sp. cell (e.g., a B. subtilis cell). In some embodiments, the invention provides a method of producing a metalloprotease polypeptide of the invention, the method comprising cultivating a recombinant host cell comprising a recombinant expression vector comprising a nucleic acid encoding a metalloprotease polypeptide of the invention under conditions conducive to the production of the metalloprotease polypeptide. Some such methods further comprise recovering the metalloprotease polypeptide from the culture.
[0227] In some embodiments the invention provides methods of producing a metalloprotease polypeptide of the invention, the methods comprising: (a) introducing a recombinant expression vector comprising a nucleic acid encoding a metalloprotease polypeptide of the invention into a population of cells (e.g., bacterial cells, such as B. subtilis cells); and (b) culturing the cells in a culture medium under conditions conducive to produce the metalloprotease polypeptide encoded by the expression vector. Some such methods further comprise: (c) isolating the metalloprotease polypeptide from the cells or from the culture medium.
Fabric and Home Care Products
[0228] In some embodiments, the metalloprotease polypeptides of the present invention can be used in compositions comprising an adjunct material and a metalloprotease polypeptide, wherein the composition is a fabric and home care product.
[0229] In some embodiments, the fabric and home care product compositions comprising at least one metalloprotease polypeptide comprise one or more of the following ingredients (based on total composition weight): from about 0.0005 wt % to about 0.1 wt %, from about 0.001 wt % to about 0.05 wt %, or even from about 0.002 wt % to about 0.03 wt % of said metalloprotease polypeptide; and one or more of the following: from about 0.00003 wt % to about 0.1 wt % fabric hueing agent; from about 0.001 wt % to about 5 wt %, perfume capsules; from about 0.001 wt % to about 1 wt %, cold-water soluble brighteners; from about 0.00003 wt % to about 0.1 wt % bleach catalysts; from about 0.00003 wt % to about 0.1 wt % first wash lipases; from about 0.00003 wt % to about 0.1 wt % bacterial cleaning cellulases; and/or from about 0.05 wt % to about 20 wt % Guerbet nonionic surfactants.
[0230] In some embodiments, the fabric and home care product composition is a liquid laundry detergent or a dishwashing detergent, such as an automatic dishwashing (ADW) detergent or hand dishwashing detergent.
[0231] It is intended that the fabric and home care product is provided in any suitable form, including a fluid or solid, or granular, powder, solid, bar, liquid, tablet, gel, or paste form. The fabric and home care product may be in the form of a unit dose pouch, especially when in the form of a liquid, and typically the fabric and home care product is at least partially, or even completely, enclosed by a water-soluble pouch. In addition, in some embodiments of the fabric and home care products comprising at least one metalloprotease polypeptide, the fabric and home care product may have any combination of parameters and/or characteristics detailed above.
Compositions Having the Metalloprotease Polypeptide of the Present Invention
[0232] Unless otherwise noted, all component or composition levels provided herein are made in reference to the active level of that component or composition, and are exclusive of impurities, for example, residual solvents or by-products, which may be present in commercially available sources. Enzyme components weights are based on total active protein. All percentages and ratios are calculated by weight unless otherwise indicated. All percentages and ratios are calculated based on the total composition unless otherwise indicated. Compositions of the invention include cleaning compositions, such as detergent compositions. In the exemplified detergent compositions, the enzymes levels are expressed by pure enzyme by weight of the total composition and unless otherwise specified, the detergent ingredients are expressed by weight of the total compositions.
[0233] As indicated herein, in some embodiments, the cleaning compositions of the present invention further comprise adjunct materials including, but not limited to, surfactants, builders, bleaches, bleach activators, bleach catalysts, other enzymes, enzyme stabilizing systems, chelants, optical brighteners, soil release polymers, dye transfer agents, dispersants, suds suppressors, dyes, perfumes, colorants, filler salts, hydrotropes, photoactivators, fluorescers, fabric conditioners, hydrolyzable surfactants, preservatives, anti-oxidants, anti-shrinkage agents, anti-wrinkle agents, germicides, fungicides, color speckles, silvercare, anti-tarnish and/or anti-corrosion agents, alkalinity sources, solubilizing agents, carriers, processing aids, pigments, and pH control agents (See e.g., U.S. Pat. Nos. 6,610,642, 6,605,458, 5,705,464, 5,710,115, 5,698,504, 5,695,679, 5,686,014 and 5,646,101, all of which are incorporated herein by reference). Embodiments of specific cleaning composition materials are exemplified in detail below. In embodiments in which the cleaning adjunct materials are not compatible with the metalloprotease polypeptides of the present invention in the cleaning compositions, then suitable methods of keeping the cleaning adjunct materials and the protease(s) separated (i.e., not in contact with each other) until combination of the two components is appropriate are used. Such separation methods include any suitable method known in the art (e.g., gelcaps, encapsulation, tablets, physical separation, etc.).
[0234] The cleaning compositions of the present invention are advantageously employed for example, in laundry applications, hard surface cleaning, dishwashing applications, including automatic dishwashing and hand dishwashing, as well as cosmetic applications such as dentures, teeth, hair and skin. In addition, due to the unique advantages of increased effectiveness in lower temperature solutions, the enzymes of the present invention are ideally suited for laundry applications. Furthermore, the enzymes of the present invention find use in granular and liquid compositions.
[0235] The metalloprotease polypeptides of the present invention also find use in cleaning additive products. In some embodiments, low temperature solution cleaning applications find use. In some embodiments, the present invention provides cleaning additive products including at least one enzyme of the present invention is ideally suited for inclusion in a wash process when additional bleaching effectiveness is desired. Such instances include, but are not limited to low temperature solution cleaning applications. In some embodiments, the additive product is in its simplest form, one or more proteases. In some embodiments, the additive is packaged in dosage form for addition to a cleaning process. In some embodiments, the additive is packaged in dosage form for addition to a cleaning process where a source of peroxygen is employed and increased bleaching effectiveness is desired. Any suitable single dosage unit form finds use with the present invention, including but not limited to pills, tablets, gelcaps, or other single dosage units such as pre-measured powders or liquids. In some embodiments, filler(s) or carrier material(s) are included to increase the volume of such compositions. Suitable filler or carrier materials include, but are not limited to, various salts of sulfate, carbonate and silicate as well as talc, clay and the like. Suitable filler or carrier materials for liquid compositions include, but are not limited to water or low molecular weight primary and secondary alcohols including polyols and diols. Examples of such alcohols include, but are not limited to, methanol, ethanol, propanol and isopropanol. In some embodiments, the compositions contain from about 5% to about 90% of such materials. Acidic fillers find use to reduce pH. Alternatively, in some embodiments, the cleaning additive includes adjunct ingredients, as more fully described below.
[0236] The present cleaning compositions and cleaning additives require an effective amount of at least one of the metalloprotease polypeptides provided herein, alone or in combination with other proteases and/or additional enzymes. The required level of enzyme is achieved by the addition of one or more metalloprotease polypeptides of the present invention. Typically the present cleaning compositions comprise at least about 0.0001 weight percent, from about 0.0001 to about 10, from about 0.001 to about 1, or from about 0.01 to about 0.1 weight percent of at least one of the metalloprotease polypeptides of the present invention.
[0237] The cleaning compositions herein are typically formulated such that, during use in aqueous cleaning operations, the wash water will have a pH of from about 4.0 to about 11.5, or even from about 5.0 to about 11.5, or even from about 5.0 to about 8.0, or even from about 7.5 to about 10.5. Liquid product formulations are typically formulated to have a pH from about 3.0 to about 9.0 or even from about 3 to about 5. Granular laundry products are typically formulated to have a pH from about 9 to about 11. Techniques for controlling pH at recommended usage levels include the use of buffers, alkalis, acids, etc., and are well known to those skilled in the art.
[0238] Suitable "low pH cleaning compositions" typically have a pH of from about 3 to about 5, and are typically free of surfactants that hydrolyze in such a pH environment. Such surfactants include sodium alkyl sulfate surfactants that comprise at least one ethylene oxide moiety or even from about 1 to about 16 moles of ethylene oxide. Such cleaning compositions typically comprise a sufficient amount of a pH modifier, such as sodium hydroxide, monoethanolamine or hydrochloric acid, to provide such cleaning composition with a pH of from about 3 to about 5. Such compositions typically comprise at least one acid stable enzyme. In some embodiments, the compositions are liquids, while in other embodiments, they are solids. The pH of such liquid compositions is typically measured as a neat pH. The pH of such solid compositions is measured as a 10% solids solution of said composition wherein the solvent is distilled water. In these embodiments, all pH measurements are taken at 20.degree. C., unless otherwise indicated.
[0239] In some embodiments, when the metalloprotease polypeptide (s) is/are employed in a granular composition or liquid, it is desirable for the metalloprotease polypeptide to be in the form of an encapsulated particle to protect the metalloprotease polypeptide from other components of the granular composition during storage. In addition, encapsulation is also a means of controlling the availability of the metalloprotease polypeptide during the cleaning process. In some embodiments, encapsulation enhances the performance of the metalloprotease polypeptide (s) and/or additional enzymes. In this regard, the metalloprotease polypeptides of the present invention are encapsulated with any suitable encapsulating material known in the art. In some embodiments, the encapsulating material typically encapsulates at least part of the metalloprotease polypeptide (s) of the present invention. Typically, the encapsulating material is water-soluble and/or water-dispersible. In some embodiments, the encapsulating material has a glass transition temperature (Tg) of 0.degree. C. or higher. Glass transition temperature is described in more detail in WO 97/11151. The encapsulating material is typically selected from consisting of carbohydrates, natural or synthetic gums, chitin, chitosan, cellulose and cellulose derivatives, silicates, phosphates, borates, polyvinyl alcohol, polyethylene glycol, paraffin waxes, and combinations thereof. When the encapsulating material is a carbohydrate, it is typically selected from monosaccharides, oligosaccharides, polysaccharides, and combinations thereof. In some typical embodiments, the encapsulating material is a starch (See e.g., EP 0 922 499; U.S. Pat. Nos. 4,977,252; 5,354,559, and 5,935,826). In some embodiments, the encapsulating material is a microsphere made from plastic such as thermoplastics, acrylonitrile, methacrylonitrile, polyacrylonitrile, polymethacrylonitrile and mixtures thereof commercially available microspheres that find use include, but are not limited to those supplied by EXPANCEL.RTM. (Stockviksverken, Sweden), and PM 6545, PM 6550, PM 7220, PM 7228, EXTENDOSPHERES.RTM., LUXSIL.RTM., Q-CEL.RTM., and SPHERICEL.RTM. (PQ Corp., Valley Forge, Pa.).
[0240] As described herein, the metalloprotease polypeptides of the present invention find particular use in the cleaning industry, including, but not limited to laundry and dish detergents. These applications place enzymes under various environmental stresses. The metalloprotease polypeptides of the present invention provide advantages over many currently used enzymes, due to their stability under various conditions.
[0241] Indeed, there are a variety of wash conditions including varying detergent formulations, wash water volumes, wash water temperatures, and lengths of wash time, to which proteases involved in washing are exposed. In addition, detergent formulations used in different geographical areas have different concentrations of their relevant components present in the wash water. For example, European detergents typically have about 4500-5000 ppm of detergent components in the wash water, while Japanese detergents typically have approximately 667 ppm of detergent components in the wash water. In North America, particularly the United States, detergents typically have about 975 ppm of detergent components present in the wash water.
[0242] A low detergent concentration system includes detergents where less than about 800 ppm of the detergent components are present in the wash water. Japanese detergents are typically considered low detergent concentration system as they have approximately 667 ppm of detergent components present in the wash water.
[0243] A medium detergent concentration includes detergents where between about 800 ppm and about 2000 ppm of the detergent components are present in the wash water. North American detergents are generally considered to be medium detergent concentration systems as they have approximately 975 ppm of detergent components present in the wash water. Brazil typically has approximately 1500 ppm of detergent components present in the wash water.
[0244] A high detergent concentration system includes detergents where greater than about 2000 ppm of the detergent components are present in the wash water. European detergents are generally considered to be high detergent concentration systems as they have approximately 4500-5000 ppm of detergent components in the wash water.
[0245] Latin American detergents are generally high suds phosphate builder detergents and the range of detergents used in Latin America can fall in both the medium and high detergent concentrations as they range from 1500 ppm to 6000 ppm of detergent components in the wash water. As mentioned above, Brazil typically has approximately 1500 ppm of detergent components present in the wash water. However, other high suds phosphate builder detergent geographies, not limited to other Latin American countries, may have high detergent concentration systems up to about 6000 ppm of detergent components present in the wash water.
[0246] In light of the foregoing, it is evident that concentrations of detergent compositions in typical wash solutions throughout the world varies from less than about 800 ppm of detergent to about 6000 ppm in high suds phosphate builder geographies.
[0247] The concentrations of the typical wash solutions are determined empirically. For example, in the U.S., a typical washing machine holds a volume of about 64.4 L of wash solution. Accordingly, in order to obtain a concentration of about 975 ppm of detergent within the wash solution about 62.79 g of detergent composition must be added to the 64.4 L of wash solution. This amount is the typical amount measured into the wash water by the consumer using the measuring cup provided with the detergent.
[0248] As a further example, different geographies use different wash temperatures. The temperature of the wash water in Japan is typically less than that used in Europe. For example, the temperature of the wash water in North America and Japan is typically between about 10 and about 40.degree. C. (e.g., about 20.degree. C.), whereas the temperature of wash water in Europe is typically between about 30 and about 60.degree. C. (e.g., about 40.degree. C.). However, in the interest of saving energy, many consumers are switching to using cold water washing. In addition, in some further regions, cold water is typically used for laundry, as well as dish washing applications. In some embodiments, the "cold water washing" of the present invention utilizes "cold water detergent" suitable for washing at temperatures from about 10.degree. C. to about 40.degree. C., or from about 20.degree. C. to about 30.degree. C., or from about 15.degree. C. to about 25.degree. C., as well as all other combinations within the range of about 15.degree. C. to about 35.degree. C., and all ranges within 10.degree. C. to 40.degree. C.
[0249] As a further example, different geographies typically have different water hardness. Water hardness is usually described in terms of the grains per gallon mixed Ca.sup.2+/Mg.sup.2+. Hardness is a measure of the amount of calcium (Ca.sup.2+) and magnesium (Mg.sup.2+) in the water. Most water in the United States is hard, but the degree of hardness varies. Moderately hard (60-120 ppm) to hard (121-181 ppm) water has 60 to 181 parts per million (parts per million converted to grains per U.S. gallon is ppm # divided by 17.1 equals grains per gallon) of hardness minerals.
TABLE-US-00001 Water Grains per gallon Parts per million Soft less than 1.0 less than 17 Slightly hard 1.0 to 3.5 17 to 60 Moderately hard 3.5 to 7.0 60 to 120 Hard 7.0 to 10.5 120 to 180 Very hard greater than 10.5 greater than 180
[0250] European water hardness is typically greater than about 10.5 (for example about 10.5 to about 20.0) grains per gallon mixed Ca.sup.2+/Mg.sup.2+ (e.g., about 15 grains per gallon mixed Ca.sup.2+/Mg.sup.2+). North American water hardness is typically greater than Japanese water hardness, but less than European water hardness. For example, North American water hardness can be between about 3 to about 10 grains, about 3 to about 8 grains or about 6 grains. Japanese water hardness is typically lower than North American water hardness, usually less than about 4, for example about 3 grains per gallon mixed Ca.sup.2+/Mg.sup.2+.
[0251] Accordingly, in some embodiments, the present invention provides metalloprotease polypeptides that show surprising wash performance in at least one set of wash conditions (e.g., water temperature, water hardness, and/or detergent concentration). In some embodiments, the metalloprotease polypeptides of the present invention are comparable in wash performance to other metalloprotease polypeptide proteases. In some embodiments of the present invention, the metalloprotease polypeptides provided herein exhibit enhanced oxidative stability, enhanced thermal stability, enhanced cleaning capabilities under various conditions, and/or enhanced chelator stability. In addition, the metalloprotease polypeptides of the present invention find use in cleaning compositions that do not include detergents, again either alone or in combination with builders and stabilizers.
[0252] In some embodiments of the present invention, the cleaning compositions comprise at least one metalloprotease polypeptide of the present invention at a level from about 0.00001% to about 10% by weight of the composition and the balance (e.g., about 99.999% to about 90.0%) comprising cleaning adjunct materials by weight of composition. In some other embodiments of the present invention, the cleaning compositions of the present invention comprises at least one metalloprotease polypeptide at a level of about 0.0001% to about 10%, about 0.001% to about 5%, about 0.001% to about 2%, about 0.005% to about 0.5% by weight of the composition and the balance of the cleaning composition (e.g., about 99.9999% to about 90.0%, about 99.999% to about 98%, about 99.995% to about 99.5% by weight) comprising cleaning adjunct materials.
[0253] In some embodiments, the cleaning compositions of the present invention comprise one or more additional detergent enzymes, which provide cleaning performance and/or fabric care and/or dishwashing benefits. Examples of suitable enzymes include, but are not limited to, acyl transferases, alpha-amylases, beta-amylases, alpha-galactosidases, arabinosidases, aryl esterases, beta-galactosidases, carrageenases, catalases, cellobiohydrolases, cellulases, chondroitinases, cutinases, endo-beta-1, 4-glucanases, endo-beta-mannanases, esterases, exo-mannanases, galactanases, glucoamylases, hemicellulases, hyaluronidases, keratinases, laccases, lactases, ligninases, lipases, lipoxygenases, mannanases, oxidases, pectate lyases, pectin acetyl esterases, pectinases, pentosanases, peroxidases, phenoloxidases, phosphatases, phospholipases, phytases, polygalacturonases, proteases, pullulanases, reductases, rhamnogalacturonases, beta-glucanases, tannases, transglutaminases, xylan acetyl-esterases, xylanases, xyloglucanases, and xylosidases, or any combinations or mixtures thereof. In some embodiments, a combination of enzymes is used (i.e., a "cocktail") comprising conventional applicable enzymes like protease, lipase, cutinase and/or cellulase in conjunction with amylase is used.
[0254] In addition to the metalloprotease polypeptides provided herein, any other suitable protease finds use in the compositions of the present invention. Suitable proteases include those of animal, vegetable or microbial origin. In some embodiments, microbial proteases are used. In some embodiments, chemically or genetically modified mutants are included. In some embodiments, the protease is a serine protease, preferably an alkaline microbial protease or a trypsin-like protease. Examples of alkaline proteases include subtilisins, especially those derived from Bacillus (e.g., subtilisin, lentus, amyloliquefaciens, subtilisin Carlsberg, subtilisin 309, subtilisin 147 and subtilisin 168). Additional examples include those mutant proteases described in U.S. Pat. Nos. RE 34,606, 5,955,340, 5,700,676, 6,312,936, and 6,482,628, all of which are incorporated herein by reference. Additional protease examples include, but are not limited to trypsin (e.g., of porcine or bovine origin), and the Fusarium protease described in WO 89/06270. In some embodiments, commercially available protease enzymes that find use in the present invention include, but are not limited to MAXATASE.RTM., MAXACAL.TM. MAXAPEM.TM., OPTICLEAN.RTM., OPTIMASE.RTM., PROPERASE.RTM., PURAFECT.RTM., PURAFECT.RTM. OXP, PURAMAX.TM., EXCELLASE.TM., and PURAFAST.TM. (Genencor); ALCALASE.RTM., SAVINASE.RTM., PRIMASE.RTM., DURAZYM.TM., POLARZYME.RTM., OVOZYME.RTM., KANNASE.RTM., LIQUANASE.RTM., NEUTRASE.RTM., RELASE.RTM. and ESPERASE.RTM. (Novozymes); BLAP.TM. and BLAP.TM. variants (Henkel Kommanditgesellschaft auf Aktien, Duesseldorf, Germany), and KAP (B. alkalophilus subtilisin; Kao Corp., Tokyo, Japan). Various proteases are described in WO95/23221, WO 92/21760, WO 09/149200, WO 09/149144, WO 09/149145, WO 11/072099, WO 10/056640, WO 10/056653, WO 11/140364, WO 12/151534, U.S. Pat. Publ. No. 2008/0090747, and U.S. Pat. Nos. 5,801,039, 5,340,735, 5,500,364, 5,855,625, US RE 34,606, 5,955,340, 5,700,676, 6,312,936, and 6,482,628, and various other patents. In some further embodiments, metalloproteases find use in the present invention, including but not limited to the neutral metalloprotease described in WO 07/044993.
[0255] In addition, any suitable lipase finds use in the present invention. Suitable lipases include, but are not limited to those of bacterial or fungal origin. Chemically or genetically modified mutants are encompassed by the present invention. Examples of useful lipases include Humicola lanuginosa lipase (See e.g., EP 258 068, and EP 305 216), Rhizomucor miehei lipase (See e.g., EP 238 023), Candida lipase, such as C. antarctica lipase (e.g., the C. antarctica lipase A or B; See e.g., EP 214 761), Pseudomonas lipases such as P. alcaligenes lipase and P. pseudoalcaligenes lipase (See e.g., EP 218 272), P. cepacia lipase (See e.g., EP 331 376), P. stutzeri lipase (See e.g., GB 1,372,034), P. fluorescens lipase, Bacillus lipase (e.g., B. subtilis lipase [Dartois et al., Biochem. Biophys. Acta 1131:253-260 [1993]); B. stearothermophilus lipase [See e.g., JP 64/744992]; and B. pumilus lipase [See e.g., WO 91/16422]).
[0256] Furthermore, a number of cloned lipases find use in some embodiments of the present invention, including but not limited to Penicillium camembertii lipase (See, Yamaguchi et al., Gene 103:61-67 [1991]), Geotricum candidum lipase (See, Schimada et al., J. Biochem., 106:383-388 [1989]), and various Rhizopus lipases such as R. delemar lipase (See, Hass et al., Gene 109:117-113 [1991]), a R. niveus lipase (Kugimiya et al., Biosci. Biotech. Biochem. 56:716-719 [1992]) and R. oryzae lipase.
[0257] Other types of lipase polypeptide enzymes such as cutinases also find use in some embodiments of the present invention, including but not limited to the cutinase derived from Pseudomonas mendocina (See, WO 88/09367), and the cutinase derived from Fusarium solani pisi (See, WO 90/09446).
[0258] Additional suitable lipases include commercially available lipases such as M1 LIPASE.TM., LUMA FAST.TM., and LIPOMAX.TM. (Genencor); LIPEX.RTM., LIPOLASE.RTM. and LIPOLASE.RTM. ULTRA (Novozymes); and LIPASE P.TM. "Amano" (Amano Pharmaceutical Co. Ltd., Japan).
[0259] In some embodiments of the present invention, the cleaning compositions of the present invention further comprise lipases at a level from about 0.00001% to about 10% of additional lipase by weight of the composition and the balance of cleaning adjunct materials by weight of composition. In some other embodiments of the present invention, the cleaning compositions of the present invention also comprise lipases at a level of about 0.0001% to about 10%, about 0.001% to about 5%, about 0.001% to about 2%, about 0.005% to about 0.5% lipase by weight of the composition.
[0260] In some embodiments of the present invention, any suitable amylase finds use in the present invention. In some embodiments, any amylase (e.g., alpha and/or beta) suitable for use in alkaline solutions also find use. Suitable amylases include, but are not limited to those of bacterial or fungal origin. Chemically or genetically modified mutants are included in some embodiments. Amylases that find use in the present invention, include, but are not limited to a-amylases obtained from B. licheniformis (See e.g., GB 1,296,839). Additional suitable amylases include those found in WO9510603, WO9526397, WO9623874, WO9623873, WO9741213, WO9919467, WO0060060, WO0029560, WO9923211, WO9946399, WO0060058, WO0060059, WO9942567, WO0114532, WO02092797, WO0166712, WO0188107, WO0196537, WO0210355, WO9402597, WO0231124, WO9943793, WO9943794, WO2004113551, WO2005001064, WO2005003311, WO0164852, WO2006063594, WO2006066594, WO2006066596, WO2006012899, WO2008092919, WO2008000825, WO2005018336, WO2005066338, WO2009140504, WO2005019443, WO2010091221, WO2010088447, WO0134784, WO2006012902, WO2006031554, WO2006136161, WO2008101894, WO2010059413, WO2011098531, WO2011080352, WO2011080353, WO2011080354, WO2011082425, WO2011082429, WO2011076123, WO2011087836, WO2011076897, WO94183314, WO9535382, WO9909183, WO9826078, WO9902702, WO9743424, WO9929876, WO9100353, WO9605295, WO9630481, WO9710342, WO2008088493, WO2009149419, WO2009061381, WO2009100102, WO2010104675, WO2010117511, and WO2010115021. Commercially available amylases that find use in the present invention include, but are not limited to DURAMYL.RTM., TERMAMYL.RTM., FUNGAMYL.RTM., STAINZYME.RTM., STAINZYME PLUS.RTM., STAINZYME ULTRA.RTM., and BAN.TM. (Novozymes), as well as POWERASE.TM., RAPIDASE.RTM. and MAXAMYL.RTM. P (Genencor).
[0261] In some embodiments of the present invention, the cleaning compositions of the present invention further comprise amylases at a level from about 0.00001% to about 10% of additional amylase by weight of the composition and the balance of cleaning adjunct materials by weight of composition. In some other embodiments of the present invention, the cleaning compositions of the present invention also comprise amylases at a level of about 0.0001% to about 10%, about 0.001% to about 5%, about 0.001% to about 2%, about 0.005% to about 0.5% amylase by weight of the composition.
[0262] In some further embodiments, any suitable cellulase finds used in the cleaning compositions of the present invention. Suitable cellulases include, but are not limited to those of bacterial or fungal origin. Chemically or genetically modified mutants are included in some embodiments. Suitable cellulases include, but are not limited to Humicola insolens cellulases (See e.g., U.S. Pat. No. 4,435,307). Especially suitable cellulases are the cellulases having color care benefits (See e.g., EP 0 495 257). Commercially available cellulases that find use in the present include, but are not limited to CELLUZYME, CELLUCLEAN, CAREZYME (Novozymes), PURADEX AND REVITALENZ (Danisco US Inc.), and KAC-500(B) (Kao Corporation). In some embodiments, cellulases are incorporated as portions or fragments of mature wild-type or variant cellulases, wherein a portion of the N-terminus is deleted (See e.g., U.S. Pat. No. 5,874,276). Additional suitable cellulases include those found in WO2005054475, WO2005056787, U.S. Pat. Nos. 7,449,318, and 7,833,773. In some embodiments, the cleaning compositions of the present invention further comprise cellulases at a level from about 0.00001% to about 10% of additional cellulase by weight of the composition and the balance of cleaning adjunct materials by weight of composition. In some other embodiments of the present invention, the cleaning compositions of the present invention also comprise cellulases at a level of about 0.0001% to about 10%, about 0.001% to about 5%, about 0.001% to about 2%, about 0.005% to about 0.5% cellulase by weight of the composition.
[0263] Any mannanase suitable for use in detergent compositions also finds use in the present invention. Suitable mannanases include, but are not limited to those of bacterial or fungal origin. Chemically or genetically modified mutants are included in some embodiments. Various mannanases are known which find use in the present invention (See e.g., U.S. Pat. Nos. 6,566,114; 6,602,842; 5,476,775 and 6,440,991, and U.S. Prov. App. Ser. No. 61/739,267; all of which are incorporated herein by reference). Commercially available mannanases that find use in the present invention include, but are not limited to MANNASTAR, PURABRITE, and MANNAWAY. In some embodiments, the cleaning compositions of the present invention further comprise mannanases at a level from about 0.00001% to about 10% of additional mannanase by weight of the composition and the balance of cleaning adjunct materials by weight of composition. In some embodiments of the present invention, the cleaning compositions of the present invention also comprise mannanases at a level of about 0.0001% to about 10%, about 0.001% to about 5%, about 0.001% to about 2%, about 0.005% to about 0.5% mannanase by weight of the composition.
[0264] In some embodiments, peroxidases are used in combination with hydrogen peroxide or a source thereof (e.g., a percarbonate, perborate or persulfate) in the compositions of the present invention. In some alternative embodiments, oxidases are used in combination with oxygen. Both types of enzymes are used for "solution bleaching" (i.e., to prevent transfer of a textile dye from a dyed fabric to another fabric when the fabrics are washed together in a wash liquor), preferably together with an enhancing agent (See e.g., WO 94/12621 and WO 95/01426). Suitable peroxidases/oxidases include, but are not limited to those of plant, bacterial or fungal origin. Chemically or genetically modified mutants are included in some embodiments. In some embodiments, the cleaning compositions of the present invention further comprise peroxidase and/or oxidase enzymes at a level from about 0.00001% to about 10% of additional peroxidase and/or oxidase by weight of the composition and the balance of cleaning adjunct materials by weight of composition. In some other embodiments of the present invention, the cleaning compositions of the present invention also comprise, peroxidase and/or oxidase enzymes at a level of about 0.0001% to about 10%, about 0.001% to about 5%, about 0.001% to about 2%, about 0.005% to about 0.5% peroxidase and/or oxidase enzymes by weight of the composition.
[0265] In some embodiments, additional enzymes find use, including but not limited to perhydrolases (See e.g., WO 05/056782). In addition, in some embodiments, mixtures of the above mentioned enzymes are encompassed herein, in particular one or more additional protease, amylase, lipase, mannanase, and/or at least one cellulase. Indeed, it is contemplated that various mixtures of these enzymes will find use in the present invention. It is also contemplated that the varying levels of the metalloprotease polypeptide (s) and one or more additional enzymes may both independently range to about 10%, the balance of the cleaning composition being cleaning adjunct materials. The specific selection of cleaning adjunct materials are readily made by considering the surface, item, or fabric to be cleaned, and the desired form of the composition for the cleaning conditions during use (e.g., through the wash detergent use).
[0266] Examples of suitable cleaning adjunct materials include, but are not limited to, surfactants, builders, bleaches, bleach activators, bleach catalysts, other enzymes, enzyme stabilizing systems, chelants, optical brighteners, soil release polymers, dye transfer agents, dye transfer inhibiting agents, catalytic materials, hydrogen peroxide, sources of hydrogen peroxide, preformed peracids, polymeric dispersing agents, clay soil removal agents, structure elasticizing agents, dispersants, suds suppressors, dyes, perfumes, colorants, filler salts, hydrotropes, photoactivators, fluorescers, fabric conditioners, fabric softeners, carriers, hydrotropes, processing aids, solvents, pigments, hydrolyzable surfactants, preservatives, anti-oxidants, anti-shrinkage agents, anti-wrinkle agents, germicides, fungicides, color speckles, silvercare, anti-tarnish and/or anti-corrosion agents, alkalinity sources, solubilizing agents, carriers, processing aids, pigments, and pH control agents (See e.g., U.S. Pat. Nos. 6,610,642; 6,605,458; 5,705,464; 5,710,115; 5,698,504; 5,695,679; 5,686,014 and 5,646,101, all of which are incorporated herein by reference). Embodiments of specific cleaning composition materials are exemplified in detail below. In embodiments in which the cleaning adjunct materials are not compatible with the metalloprotease polypeptides of the present invention in the cleaning compositions, then suitable methods of keeping the cleaning adjunct materials and the protease(s) separated (i.e., not in contact with each other) until combination of the two components is appropriate are used. Such separation methods include any suitable method known in the art (e.g., gelcaps, encapsulation, tablets, physical separation, etc.).
[0267] In some embodiments, an effective amount of one or more metalloprotease polypeptide (s) provided herein is included in compositions useful for cleaning a variety of surfaces in need of proteinaceous stain removal. Such cleaning compositions include cleaning compositions for such applications as cleaning hard surfaces, fabrics, and dishes. Indeed, in some embodiments, the present invention provides fabric cleaning compositions, while in other embodiments, the present invention provides non-fabric cleaning compositions. Notably, the present invention also provides cleaning compositions suitable for personal care, including oral care (including dentrifices, toothpastes, mouthwashes, etc., as well as denture cleaning compositions), skin, and hair cleaning compositions. It is intended that the present invention encompass detergent compositions in any form (i.e., liquid, granular, bar, semi-solid, gels, emulsions, tablets, capsules, etc.).
[0268] By way of example, several cleaning compositions wherein the metalloprotease polypeptides of the present invention find use are described in greater detail below. In some embodiments in which the cleaning compositions of the present invention are formulated as compositions suitable for use in laundry machine washing method(s), the compositions of the present invention preferably contain at least one surfactant and at least one builder compound, as well as one or more cleaning adjunct materials preferably selected from organic polymeric compounds, bleaching agents, additional enzymes, suds suppressors, dispersants, lime-soap dispersants, soil suspension and anti-redeposition agents and corrosion inhibitors. In some embodiments, laundry compositions also contain softening agents (i.e., as additional cleaning adjunct materials). The compositions of the present invention also find use in detergent additive products in solid or liquid form. Such additive products are intended to supplement and/or boost the performance of conventional detergent compositions and can be added at any stage of the cleaning process. In some embodiments, the density of the laundry detergent compositions herein ranges from about 400 to about 1200 g/liter, while in other embodiments, it ranges from about 500 to about 950 g/liter of composition measured at 20.degree. C.
[0269] In embodiments formulated as compositions for use in manual dishwashing methods, the compositions of the invention preferably contain at least one surfactant and preferably at least one additional cleaning adjunct material selected from organic polymeric compounds, suds enhancing agents, group II metal ions, solvents, hydrotropes and additional enzymes.
[0270] In some embodiments, various cleaning compositions such as those provided in U.S. Pat. No. 6,605,458, find use with the metalloprotease polypeptides of the present invention. Thus, in some embodiments, the compositions comprising at least one metalloprotease polypeptide of the present invention is a compact granular fabric cleaning composition, while in other embodiments, the composition is a granular fabric cleaning composition useful in the laundering of colored fabrics, in further embodiments, the composition is a granular fabric cleaning composition which provides softening through the wash capacity, in additional embodiments, the composition is a heavy duty liquid fabric cleaning composition. In some embodiments, the compositions comprising at least one metalloprotease polypeptide of the present invention are fabric cleaning compositions such as those described in U.S. Pat. Nos. 6,610,642 and 6,376,450. In addition, the metalloprotease polypeptides of the present invention find use in granular laundry detergent compositions of particular utility under European or Japanese washing conditions (See e.g., U.S. Pat. No. 6,610,642).
[0271] In some alternative embodiments, the present invention provides hard surface cleaning compositions comprising at least one metalloprotease polypeptide provided herein. Thus, in some embodiments, the compositions comprising at least one metalloprotease polypeptide of the present invention is a hard surface cleaning composition such as those described in U.S. Pat. Nos. 6,610,642; 6,376,450, and 6,376,450.
[0272] In yet further embodiments, the present invention provides dishwashing compositions comprising at least one metalloprotease polypeptide provided herein. Thus, in some embodiments, the compositions comprising at least one metalloprotease polypeptide of the present invention is a hard surface cleaning composition such as those in U.S. Pat. Nos. 6,610,642 and 6,376,450. In some still further embodiments, the present invention provides dishwashing compositions comprising at least one metalloprotease polypeptide provided herein. In some further embodiments, the compositions comprising at least one metalloprotease polypeptide of the present invention comprise oral care compositions such as those in U.S. Pat. Nos. 6,376,450, and 6,376,450. The formulations and descriptions of the compounds and cleaning adjunct materials contained in the aforementioned U.S. Pat. Nos. 6,376,450; 6,605,458; 6,605,458, and 6,610,642, find use with the metalloprotease polypeptides provided herein.
[0273] The cleaning compositions of the present invention are formulated into any suitable form and prepared by any process chosen by the formulator, non-limiting examples of which are described in U.S. Pat. Nos. 5,879,584; 5,691,297; 5,574,005; 5,569,645; 5,565,422; 5,516,448; 5,489,392, and 5,486,303, all of which are incorporated herein by reference. When a low pH cleaning composition is desired, the pH of such composition is adjusted via the addition of a material such as monoethanolamine or an acidic material such as HCl.
[0274] In some embodiments, the cleaning compositions of the present invention can be formulated to have an alkaline pH under wash conditions, such as a pH of from about 8.0 to about 12.0, or from about 8.5 to about 11.0, or from about 9.0 to about 11.0. In some embodiments, the cleaning compositions of the present invention can be formulated to have a neutral pH under wash conditions, such as a pH of from about 5.0 to about 8.0, or from about 5.5 to about 8.0, or from about 6.0 to about 8.0, or from about 6.0 to about 7.5. In some embodiments, the neutral pH conditions can be measured when the cleaning composition is dissolved 1:100 (wt:wt) in de-ionised water at 20.degree. C., measured using a conventional pH meter.
[0275] While not essential for the purposes of the present invention, the non-limiting list of adjuncts illustrated hereinafter are suitable for use in the instant cleaning compositions. In some embodiments, these adjuncts are incorporated for example, to assist or enhance cleaning performance, for treatment of the substrate to be cleaned, or to modify the aesthetics of the cleaning composition as is the case with perfumes, colorants, dyes or the like. It is understood that such adjuncts are in addition to the metalloprotease polypeptides of the present invention. The precise nature of these additional components, and levels of incorporation thereof, will depend on the physical form of the composition and the nature of the cleaning operation for which it is to be used. Suitable adjunct materials include, but are not limited to, surfactants, builders, chelating agents, dye transfer inhibiting agents, deposition aids, dispersants, additional enzymes, and enzyme stabilizers, catalytic materials, bleach activators, bleach boosters, hydrogen peroxide, sources of hydrogen peroxide, preformed peracids, polymeric dispersing agents, clay soil removal/anti-redeposition agents, brighteners, suds suppressors, dyes, perfumes, structure elasticizing agents, fabric softeners, carriers, hydrotropes, processing aids and/or pigments. In addition to the disclosure below, suitable examples of such other adjuncts and levels of use are found in U.S. Pat. Nos. 5,576,282, 6,306,812, and 6,326,348, incorporated by reference. The aforementioned adjunct ingredients may constitute the balance of the cleaning compositions of the present invention.
[0276] In some embodiments, the cleaning compositions according to the present invention comprise an acidifying particle or an amino carboxylic builder. Examples of an amino carboxylic builder include aminocarboxylic acids, salts and derivatives thereof. In some embodiment, the amino carboxylic builder is an aminopolycarboxylic builder, such as glycine-N,N-diacetic acid or derivative of general formula MOOC--CHR-N(CH.sub.2COOM).sub.2 where R is C.sub.1-12 alkyl and M is alkali metal. In some embodiments, the amino carboxylic builder can be methylglycine diacetic acid (MGDA), GLDA (glutamic-N,N-diacetic acid), iminodisuccinic acid (IDS), carboxymethyl inulin and salts and derivatives thereof, aspartic acid-N-monoacetic acid (ASMA), aspartic acid-N,N-diacetic acid (ASDA), aspartic acid-N-monopropionic acid (ASMP), iminodisuccinic acid (IDA), N-(2-sulfomethyl) aspartic acid (SMAS), N-(2-sulfoethyl)aspartic acid (SEAS), N-(2-sulfomethyl)glutamic acid (SMGL), N-(2-sulfoethyl) glutamic acid (SEGL), IDS (iminodiacetic acid) and salts and derivatives thereof such as N-methyliminodiacetic acid (MIDA), alpha-alanine-N,N-diacetic acid (alpha-ALDA), serine-N,N-diacetic acid (SEDA), isoserine-N,Ndiacetic acid (ISDA), phenylalanine-N,N-diacetic acid (PHDA), anthranilic acid-N,N-diacetic acid (ANDA), sulfanilic acid-N,N-diacetic acid (SLDA), taurine-N,N-diacetic acid (TUDA) and sulfomethyl-N,N-diacetic acid (SMDA) and alkali metal salts and derivative thereof. In some embodiments, the acidifying particle has a weight geometric mean particle size of from about 400.mu. to about 1200.mu. and a bulk density of at least 550 g/L. In some embodiments, the acidifying particle comprises at least about 5% of the builder.
[0277] In some embodiments, the acidifying particle can comprise any acid, including organic acids and mineral acids. Organic acids can have one or two carboxyls and in some instances up to 15 carbons, especially up to 10 carbons, such as formic, acetic, propionic, capric, oxalic, succinic, adipic, maleic, fumaric, sebacic, malic, lactic, glycolic, tartaric and glyoxylic acids. In some embodiments, the acid is citric acid. Mineral acids include hydrochloric and sulphuric acid. In some instances, the acidifying particle of the invention is a highly active particle comprising a high level of amino carboxylic builder. Sulphuric acid has been found to further contribute to the stability of the final particle.
[0278] In some embodiments, the cleaning compositions according to the present invention comprise at least one surfactant and/or a surfactant system wherein the surfactant is selected from nonionic surfactants, anionic surfactants, cationic surfactants, ampholytic surfactants, zwitterionic surfactants, semi-polar nonionic surfactants and mixtures thereof. In some low pH cleaning composition embodiments (e.g., compositions having a neat pH of from about 3 to about 5), the composition typically does not contain alkyl ethoxylated sulfate, as it is believed that such surfactant may be hydrolyzed by such compositions the acidic contents. In some embodiments, the surfactant is present at a level of from about 0.1% to about 60%, while in alternative embodiments the level is from about 1% to about 50%, while in still further embodiments the level is from about 5% to about 40%, by weight of the cleaning composition.
[0279] In some embodiments, the cleaning compositions of the present invention comprise one or more detergent builders or builder systems. In some embodiments incorporating at least one builder, the cleaning compositions comprise at least about 1%, from about 3% to about 60% or even from about 5% to about 40% builder by weight of the cleaning composition. Builders include, but are not limited to, the alkali metal, ammonium and alkanolammonium salts of polyphosphates, alkali metal silicates, alkaline earth and alkali metal carbonates, aluminosilicates, polycarboxylate compounds, ether hydroxypolycarboxylates, copolymers of maleic anhydride with ethylene or vinyl methyl ether, 1, 3, 5-trihydroxy benzene-2, 4, 6-trisulphonic acid, and carboxymethyloxysuccinic acid, the various alkali metal, ammonium and substituted ammonium salts of polyacetic acids such as ethylenediamine tetraacetic acid and nitrilotriacetic acid, as well as polycarboxylates such as mellitic acid, succinic acid, citric acid, oxydisuccinic acid, polymaleic acid, benzene 1,3,5-tricarboxylic acid, carboxymethyloxysuccinic acid, and soluble salts thereof. Indeed, it is contemplated that any suitable builder will find use in various embodiments of the present invention.
[0280] In some embodiments, the builders form water-soluble hardness ion complexes (e.g., sequestering builders), such as citrates and polyphosphates (e.g., sodium tripolyphosphate and sodium tripolyphospate hexahydrate, potassium tripolyphosphate, and mixed sodium and potassium tripolyphosphate, etc.). It is contemplated that any suitable builder will find use in the present invention, including those known in the art (See e.g., EP 2 100 949).
[0281] In some embodiments, builders for use herein include phosphate builders and non-phosphate builders. In some embodiments, the builder is a phosphate builder. In some embodiments, the builder is a non-phosphate builder. If present, builders are used in a level of from 0.1% to 80%, or from 5 to 60%, or from 10 to 50% by weight of the composition. In some embodiments the product comprises a mixture of phosphate and non-phosphate builders.
[0282] Suitable phosphate builders include mono-phosphates, di-phosphates, tri-polyphosphates or oligomeric-poylphosphates, including the alkali metal salts of these compounds, including the sodium salts. In some embodiments, a builder can be sodium tripolyphosphate (STPP). Additionally, the composition can comprise carbonate and/or citrate, preferably citrate that helps to achieve a neutral pH composition of the invention. Other suitable non-phosphate builders include homopolymers and copolymers of polycarboxylic acids and their partially or completely neutralized salts, monomeric polycarboxylic acids and hydroxycarboxylic acids and their salts. In some embodiments, salts of the above mentioned compounds include the ammonium and/or alkali metal salts, i.e. the lithium, sodium, and potassium salts, including sodium salts. Suitable polycarboxylic acids include acyclic, alicyclic, hetero-cyclic and aromatic carboxylic acids, wherein in some embodiments, they can contain at least two carboxyl groups which are in each case separated from one another by, in some instances, no more than two carbon atoms.
[0283] In some embodiments, the cleaning compositions of the present invention contain at least one chelating agent. Suitable chelating agents include, but are not limited to copper, iron and/or manganese chelating agents and mixtures thereof. In embodiments in which at least one chelating agent is used, the cleaning compositions of the present invention comprise from about 0.1% to about 15% or even from about 3.0% to about 10% chelating agent by weight of the subject cleaning composition.
[0284] In some still further embodiments, the cleaning compositions provided herein contain at least one deposition aid. Suitable deposition aids include, but are not limited to, polyethylene glycol, polypropylene glycol, polycarboxylate, soil release polymers such as polytelephthalic acid, clays such as kaolinite, montmorillonite, atapulgite, illite, bentonite, halloysite, and mixtures thereof.
[0285] As indicated herein, in some embodiments, anti-redeposition agents find use in some embodiments of the present invention. In some embodiments, non-ionic surfactants find use. For example, in automatic dishwashing embodiments, non-ionic surfactants find use for surface modification purposes, in particular for sheeting, to avoid filming and spotting and to improve shine. These non-ionic surfactants also find use in preventing the re-deposition of soils. In some embodiments, the anti-redeposition agent is a non-ionic surfactant as known in the art (See e.g., EP 2 100 949). In some embodiments, the non-ionic surfactant can be ethoxylated nonionic surfactants, epoxy-capped poly(oxyalkylated) alcohols and amine oxides surfactants.
[0286] In some embodiments, the cleaning compositions of the present invention include one or more dye transfer inhibiting agents. Suitable polymeric dye transfer inhibiting agents include, but are not limited to, polyvinylpyrrolidone polymers, polyamine N-oxide polymers, copolymers of N-vinylpyrrolidone and N-vinylimidazole, polyvinyloxazolidones and polyvinylimidazoles or mixtures thereof. In embodiments in which at least one dye transfer inhibiting agent is used, the cleaning compositions of the present invention comprise from about 0.0001% to about 10%, from about 0.01% to about 5%, or even from about 0.1% to about 3% by weight of the cleaning composition.
[0287] In some embodiments, silicates are included within the compositions of the present invention. In some such embodiments, sodium silicates (e.g., sodium disilicate, sodium metasilicate, and crystalline phyllosilicates) find use. In some embodiments, silicates are present at a level of from about 1% to about 20%. In some embodiments, silicates are present at a level of from about 5% to about 15% by weight of the composition.
[0288] In some still additional embodiments, the cleaning compositions of the present invention also contain dispersants. Suitable water-soluble organic materials include, but are not limited to the homo- or co-polymeric acids or their salts, in which the polycarboxylic acid comprises at least two carboxyl radicals separated from each other by not more than two carbon atoms.
[0289] In some further embodiments, the enzymes used in the cleaning compositions are stabilized by any suitable technique. In some embodiments, the enzymes employed herein are stabilized by the presence of water-soluble sources of calcium and/or magnesium ions in the finished compositions that provide such ions to the enzymes. In some embodiments, the enzyme stabilizers include oligosaccharides, polysaccharides, and inorganic divalent metal salts, including alkaline earth metals, such as calcium salts, such as calcium formate. It is contemplated that various techniques for enzyme stabilization will find use in the present invention. For example, in some embodiments, the enzymes employed herein are stabilized by the presence of water-soluble sources of zinc (II), calcium (II) and/or magnesium (II) ions in the finished compositions that provide such ions to the enzymes, as well as other metal ions (e.g., barium (II), scandium (II), iron (II), manganese (II), aluminum (III), Tin (II), cobalt (II), copper (II), nickel (II), and oxovanadium (IV). Chlorides and sulfates also find use in some embodiments of the present invention. Examples of suitable oligosaccharides and polysaccharides (e.g., dextrins) are known in the art (See e.g., WO 07/145964). In some embodiments, reversible protease inhibitors also find use, such as boron-containing compounds (e.g., borate, 4-formyl phenyl boronic acid) and/or a tripeptide aldehyde find use to further improve stability, as desired.
[0290] In some embodiments, bleaches, bleach activators and/or bleach catalysts are present in the compositions of the present invention. In some embodiments, the cleaning compositions of the present invention comprise inorganic and/or organic bleaching compound(s). Inorganic bleaches include, but are not limited to perhydrate salts (e.g., perborate, percarbonate, perphosphate, persulfate, and persilicate salts). In some embodiments, inorganic perhydrate salts are alkali metal salts. In some embodiments, inorganic perhydrate salts are included as the crystalline solid, without additional protection, although in some other embodiments, the salt is coated. Any suitable salt known in the art finds use in the present invention (See e.g., EP 2 100 949).
[0291] In some embodiments, bleach activators are used in the compositions of the present invention. Bleach activators are typically organic peracid precursors that enhance the bleaching action in the course of cleaning at temperatures of 60.degree. C. and below. Bleach activators suitable for use herein include compounds which, under perhydrolysis conditions, give aliphatic peroxoycarboxylic acids having preferably from about 1 to about 10 carbon atoms, in particular from about 2 to about 4 carbon atoms, and/or optionally substituted perbenzoic acid. Additional bleach activators are known in the art and find use in the present invention (See e.g., EP 2 100 949).
[0292] In addition, in some embodiments and as further described herein, the cleaning compositions of the present invention further comprise at least one bleach catalyst. In some embodiments, the manganese triazacyclononane and related complexes find use, as well as cobalt, copper, manganese, and iron complexes. Additional bleach catalysts find use in the present invention (See e.g., U.S. Pat. Nos. 4,246,612, 5,227,084, 4,810410, WO 99/06521, and EP 2 100 949).
[0293] In some embodiments, the cleaning compositions of the present invention contain one or more catalytic metal complexes. In some embodiments, a metal-containing bleach catalyst finds use. In some embodiments, the metal bleach catalyst comprises a catalyst system comprising a transition metal cation of defined bleach catalytic activity, (e.g., copper, iron, titanium, ruthenium, tungsten, molybdenum, or manganese cations), an auxiliary metal cation having little or no bleach catalytic activity (e.g., zinc or aluminum cations), and a sequestrate having defined stability constants for the catalytic and auxiliary metal cations, particularly ethylenediaminetetraacetic acid, ethylenediaminetetra (methylenephosphonic acid) and water-soluble salts thereof are used (See e.g., U.S. Pat. No. 4,430,243). In some embodiments, the cleaning compositions of the present invention are catalyzed by means of a manganese compound. Such compounds and levels of use are well known in the art (See e.g., U.S. Pat. No. 5,576,282). In additional embodiments, cobalt bleach catalysts find use in the cleaning compositions of the present invention. Various cobalt bleach catalysts are known in the art (See e.g., U.S. Pat. Nos. 5,597,936 and 5,595,967) and are readily prepared by known procedures.
[0294] In some additional embodiments, the cleaning compositions of the present invention include a transition metal complex of a macropolycyclic rigid ligand (MRL). As a practical matter, and not by way of limitation, in some embodiments, the compositions and cleaning processes provided by the present invention are adjusted to provide on the order of at least one part per hundred million of the active MRL species in the aqueous washing medium, and in some embodiments, provide from about 0.005 ppm to about 25 ppm, more preferably from about 0.05 ppm to about 10 ppm, and most preferably from about 0.1 ppm to about 5 ppm, of the MRL in the wash liquor.
[0295] In some embodiments, transition-metals in the instant transition-metal bleach catalyst include, but are not limited to manganese, iron and chromium. MRLs also include, but are not limited to special ultra-rigid ligands that are cross-bridged (e.g., 5,12-diethyl-1,5,8,12-tetraazabicyclo[6.6.2]hexadecane). Suitable transition metal MRLs are readily prepared by known procedures (See e.g., WO 2000/32601, and U.S. Pat. No. 6,225,464).
[0296] In some embodiments, the cleaning compositions of the present invention comprise metal care agents. Metal care agents find use in preventing and/or reducing the tarnishing, corrosion, and/or oxidation of metals, including aluminum, stainless steel, and non-ferrous metals (e.g., silver and copper). Suitable metal care agents include those described in EP 2 100 949, WO 9426860 and WO 94/26859). In some embodiments, the metal care agent is a zinc salt. In some further embodiments, the cleaning compositions of the present invention comprise from about 0.1% to about 5% by weight of one or more metal care agent.
[0297] In some embodiments, the cleaning composition is a high density liquid (HDL) composition having a variant metalloprotease polypeptide protease. The HDL liquid laundry detergent can comprise a detersive surfactant (10%-40%) comprising anionic detersive surfactant (selected from a group of linear or branched or random chain, substituted or unsubstituted alkyl sulphates, alkyl sulphonates, alkyl alkoxylated sulphate, alkyl phosphates, alkyl phosphonates, alkyl carboxylates, and/or mixtures thereof); and optionally non-ionic surfactant (selected from a group of linear or branched or random chain, substituted or unsubstituted alkyl alkoxylated alcohol, for example a C.sub.8-C.sub.18 alkyl ethoxylated alcohol and/or C.sub.6-C.sub.12 alkyl phenol alkoxylates), optionally wherein the weight ratio of anionic detersive surfactant (with a hydrophilic index (HIc) of from 6.0 to 9) to non-ionic detersive surfactant is greater than 1:1.
[0298] The composition can comprise optionally, a surfactancy boosting polymer consisting of amphiphilic alkoxylated grease cleaning polymers (selected from a group of alkoxylated polymers having branched hydrophilic and hydrophobic properties, such as alkoxylated polyalkylenimines in the range of 0.05 wt %-10 wt %) and/or random graft polymers (typically comprising of hydrophilic backbone comprising monomers selected from the group consisting of: unsaturated C.sub.1-C.sub.6 carboxylic acids, ethers, alcohols, aldehydes, ketones, esters, sugar units, alkoxy units, maleic anhydride, saturated polyalcohols such as glycerol, and mixtures thereof; and hydrophobic side chain(s) selected from the group consisting of: C.sub.4-C.sub.25 alkyl group, polypropylene, polybutylene, vinyl ester of a saturated C-C.sub.6 mono-carboxylic acid, C.sub.1-C.sub.6 alkyl ester of acrylic or methacrylic acid, and mixtures thereof.
[0299] The composition can comprise additional polymers such as soil release polymers (include anionically end-capped polyesters, for example SRP1, polymers comprising at least one monomer unit selected from saccharide, dicarboxylic acid, polyol and combinations thereof, in random or block configuration, ethylene terephthalate-based polymers and co-polymers thereof in random or block configuration, for example Repel-o-tex SF, SF-2 and SRP6, Texcare SRA100, SRA300, SRN100, SRN170, SRN240, SRN300 and SRN325, Marloquest SL), anti-redeposition polymers (0.1 wt % to 10 wt %, include carboxylate polymers, such as polymers comprising at least one monomer selected from acrylic acid, maleic acid (or maleic anhydride), fumaric acid, itaconic acid, aconitic acid, mesaconic acid, citraconic acid, methylenemalonic acid, and any mixture thereof, vinylpyrrolidone homopolymer, and/or polyethylene glycol, molecular weight in the range of from 500 to 100,000 Da); cellulosic polymer (including those selected from alkyl cellulose, alkyl alkoxyalkyl cellulose, carboxyalkyl cellulose, alkyl carboxyalkyl cellulose examples of which include carboxymethyl cellulose, methyl cellulose, methyl hydroxyethyl cellulose, methyl carboxymethyl cellulose, and mixtures thereof) and polymeric carboxylate (such as maleate/acrylate random copolymer or polyacrylate homopolymer).
[0300] The composition can further comprise saturated or unsaturated fatty acid, preferably saturated or unsaturated C.sub.12-C.sub.24 fatty acid (0 wt % to 10 wt %); deposition aids (examples for which include polysaccharides, preferably cellulosic polymers, poly diallyl dimethyl ammonium halides (DADMAC), and co-polymers of DAD MAC with vinyl pyrrolidone, acrylamides, imidazoles, imidazolinium halides, and mixtures thereof, in random or block configuration, cationic guar gum, cationic cellulose such as cationic hydoxyethyl cellulose, cationic starch, cationic polyacylamides, and mixtures thereof.
[0301] The composition can further comprise dye transfer inhibiting agents examples of which include manganese phthalocyanine, peroxidases, polyvinylpyrrolidone polymers, polyamine N-oxide polymers, copolymers of N-vinylpyrrolidone and N-vinylimidazole, polyvinyloxazolidones and polyvinylimidazoles and/or mixtures thereof; chelating agents examples of which include ethylene-diamine-tetraacetic acid (EDTA); diethylene triamine penta methylene phosphonic acid (DTPMP); hydroxy-ethane diphosphonic acid (HEDP); ethylenediamine N,N'-disuccinic acid (EDDS); methyl glycine diacetic acid (MGDA); diethylene triamine penta acetic acid (DTPA); propylene diamine tetracetic acid (PDT A); 2-hydroxypyridine-N-oxide (HPNO); or methyl glycine diacetic acid (MGDA); glutamic acid N,N-diacetic acid (N,N-dicarboxymethyl glutamic acid tetrasodium salt (GLDA); nitrilotriacetic acid (NTA); 4,5-dihydroxy-m-benzenedisulfonic acid; citric acid and any salts thereof; N-hydroxyethylethylenediaminetri-acetic acid (HEDTA), triethylenetetraaminehexaacetic acid (TTHA), N-hydroxyethyliminodiacetic acid (HEIDA), dihydroxyethylglycine (DHEG), ethylenediaminetetrapropionic acid (EDTP) and derivatives thereof.
[0302] The composition can further comprise enzymes (0.01 wt % active enzyme to 0.03 wt % active enzyme) selected from a group of acyl transferases, alpha-amylases, beta-amylases, alpha-galactosidases, arabinosidases, aryl esterases, beta-galactosidases, carrageenases, catalases, cellobiohydrolases, cellulases, chondroitinases, cutinases, endo-beta-1, 4-glucanases, endo-beta-mannanases, esterases, exo-mannanases, galactanases, glucoamylases, hemicellulases, hyaluronidases, keratinases, laccases, lactases, ligninases, lipases, lipoxygenases, mannanases, oxidases, pectate lyases, pectin acetyl esterases, pectinases, pentosanases, peroxidases, phenoloxidases, phosphatases, phospholipases, phytases, polygalacturonases, proteases, pullulanases, reductases, rhamnogalacturonases, beta-glucanases, tannases, transglutaminases, xylan acetyl-esterases, xylanases, xyloglucanases, and xylosidases, and any mixture thereof. The composition may comprise an enzyme stabilizer (examples of which include polyols such as propylene glycol or glycerol, sugar or sugar alcohol, lactic acid, reversible protease inhibitor, boric acid, a boric acid derivative, e.g., an aromatic borate ester, or a phenyl boronic acid derivative such as 4-formylphenyl boronic acid, peptides or formate).
[0303] The composition can further comprise silicone or fatty-acid based suds suppressors; heuing dyes, calcium and magnesium cations, visual signaling ingredients, anti-foam (0.001 wt % to about 4.0 wt %), and/or structurant/thickener (0.01 wt % to 5 wt %, selected from the group consisting of diglycerides and triglycerides, ethylene glycol distearate, microcrystalline cellulose, cellulose based materials, microfiber cellulose, biopolymers, xanthan gum, gellan gum, and mixtures thereof).
[0304] Suitable detersive surfactants also include cationic detersive surfactants (selected from a group of alkyl pyridinium compounds, alkyl quarternary ammonium compounds, alkyl quarternary phosphonium compounds, alkyl ternary sulphonium compounds, and/or mixtures thereof); zwitterionic and/or amphoteric detersive surfactants (selected from a group of alkanolamine sulpho-betaines); ampholytic surfactants; semi-polar non-ionic surfactants and mixtures thereof.
[0305] The composition can be any liquid form, for example a liquid or gel form, or any combination thereof. The composition may be in any unit dose form, for example a pouch.
[0306] In some embodiments, the cleaning composition is a high density powder (HDD) composition having a variant metalloprotease polypeptide protease. The HDD powder laundry detergent can comprise a detersive surfactant including anionic detersive surfactants (selected from a group of linear or branched or random chain, substituted or unsubstituted alkyl sulphates, alkyl sulphonates, alkyl alkoxylated sulphate, alkyl phosphates, alkyl phosphonates, alkyl carboxylates and/or mixtures thereof), non-ionic detersive surfactant (selected from a group of linear or branched or random chain, substituted or unsubstituted C.sub.8-C.sub.18 alkyl ethoxylates, and/or C.sub.6-C.sub.12 alkyl phenol alkoxylates), cationic detersive surfactants (selected from a group of alkyl pyridinium compounds, alkyl quaternary ammonium compounds, alkyl quaternary phosphonium compounds, alkyl ternary sulphonium compounds, and mixtures thereof), zwitterionic and/or amphoteric detersive surfactants (selected from a group of alkanolamine sulpho-betaines); ampholytic surfactants; semi-polar non-ionic surfactants and mixtures thereof builders (phosphate free builders [for example zeolite builders examples of which include zeolite A, zeolite X, zeolite P and zeolite MAP in the range of 0 wt % to less than 10 wt %]; phosphate builders [examples of which include sodium tri-polyphosphate in the range of 0 wt % to less than 10 wt %]; citric acid, citrate salts and nitrilotriacetic acid or salt thereof in the range of less than 15 wt %); silicate salt (sodium or potassium silicate or sodium meta-silicate in the range of 0 wt % to less than 10 wt %, or layered silicate (SKS-6)); carbonate salt (sodium carbonate and/or sodium bicarbonate in the range of 0 wt % to less than 10 wt %); and bleaching agents (photobleaches, examples of which include sulfonated zinc phthalocyanines, sulfonated aluminum phthalocyanines, xanthenes dyes, and mixtures thereof; hydrophobic or hydrophilic bleach activators (examples of which include dodecanoyl oxybenzene sulfonate, decanoyl oxybenzene sulfonate, decanoyl oxybenzoic acid or salts thereof, 3,5,5-trimethy hexanoyl oxybenzene sulfonate, tetraacetyl ethylene diamine-TAED, and nonanoyloxybenzene sulfonate-NOB S, nitrile quats, and mixtures thereof; hydrogen peroxide; sources of hydrogen peroxide (inorganic perhydrate salts examples of which include mono or tetra hydrate sodium salt of perborate, percarbonate, persulfate, perphosphate, or persilicate); preformed hydrophilic and/or hydrophobic peracids (selected from a group consisting of percarboxylic acids and salts, percarbonic acids and salts, perimidic acids and salts, peroxymonosulfuric acids and salts) & mixtures thereof and/or bleach catalyst (such as imine bleach boosters examples of which include iminium cations and polyions; iminium zwitterions; modified amines; modified amine oxides; N-sulphonyl imines; N-phosphonyl imines; N-acyl imines; thiadiazole dioxides; perfluoroimines; cyclic sugar ketones and mixtures thereof; metal-containing bleach catalyst for example copper, iron, titanium, ruthenium, tungsten, molybdenum, or manganese cations along with an auxiliary metal cations such as zinc or aluminum and a sequestrate such as ethylenediaminetetraacetic acid, ethylenediaminetetra(methylenephos-phonic acid) and water-soluble salts thereof).
[0307] The composition can further comprise enzymes selected from a group of acyl transferases, alpha-amylases, beta-amylases, alpha-galactosidases, arabinosidases, aryl esterases, beta-galactosidases, carrageenases, catalases, cellobiohydrolases, cellulases, chondroitinases, cutinases, endo-beta-1, 4-glucanases, endo-beta-mannanases, esterases, exo-mannanases, galactanases, glucoamylases, hemicellulases, hyaluronidases, keratinases, laccases, lactases, ligninases, lipases, lipoxygenases, mannanases, oxidases, pectate lyases, pectin acetyl esterases, pectinases, pentosanases, peroxidases, phenoloxidases, phosphatases, phospholipases, phytases, polygalacturonases, proteases, pullulanases, reductases, rhamnogalacturonases, beta-glucanases, tannases, transglutaminases, xylan acetyl-esterases, xylanases, xyloglucanases, and xylosidases and any mixture thereof.
[0308] The composition can further comprise additional detergent ingredients including perfume microcapsules, starch encapsulated perfume accord, hueing agents, additional polymers including fabric integrity and cationic polymers, dye lock ingredients, fabric-softening agents, brighteners (for example C.I. Fluorescent brighteners), flocculating agents, chelating agents, alkoxylated polyamines, fabric deposition aids, and/or cyclodextrin.
[0309] In some embodiments, the cleaning composition is an automatic dishwashing (ADW) detergent composition having a metalloprotease of the present invention. The ADW detergent composition can comprise two or more non-ionic surfactants selected from a group of ethoxylated non-ionic surfactants, alcohol alkoxylated surfactants, epoxy-capped poly(oxyalkylated) alcohols, or amine oxide surfactants present in amounts from 0 to 10% by weight; builders in the range of 5-60% comprising either phosphate (mono-phosphates, di-phosphates, tri-polyphosphates or oligomeric-poylphosphates, preferred sodium tripolyphosphate-STPP or phosphate-free builders [amino acid based compounds, examples of which include MGDA (methyl-glycine-diacetic acid), and salts and derivatives thereof, GLDA (glutamic-N,Ndiacetic acid) and salts and derivatives thereof, IDS (iminodisuccinic acid) and salts and derivatives thereof, carboxy methyl inulin and salts and derivatives thereof and mixtures thereof, nitrilotriacetic acid (NTA), diethylene triamine penta acetic acid (DTPA), B-alaninediacetic acid (B-ADA) and their salts], homopolymers and copolymers of poly-carboxylic acids and their partially or completely neutralized salts, monomeric polycarboxylic acids and hydroxycarboxylic acids and their salts in the range of 0.5% to 50% by weight; sulfonated/carboxylated polymers (provide dimensional stability to the product) in the range of about 0.1% to about 50% by weight; drying aids in the range of about 0.1% to about 10% by weight (selected from polyesters, especially anionic polyesters optionally together with further monomers with 3 to 6 functionalities which are conducive to polycondensation, specifically acid, alcohol or ester functionalities, polycarbonate-, polyurethane- and/or polyurea-polyorganosiloxane compounds or precursor compounds thereof of the reactive cyclic carbonate and urea type); silicates in the range from about 1% to about 20% by weight (sodium or potassium silicates for example sodium disilicate, sodium meta-silicate and crystalline phyllosilicates); bleach-inorganic (for example perhydrate salts such as perborate, percarbonate, perphosphate, persulfate and persilicate salts) and organic (for example organic peroxyacids including diacyl and tetraacylperoxides, especially diperoxydodecanedioc acid, diperoxytetradecanedioc acid, and diperoxyhexadecanedioc acid); bleach activators-organic peracid precursors in the range from about 0.1% to about 10% by weight; bleach catalysts (selected from manganese triazacyclononane and related complexes, Co, Cu, Mn and Fe bispyridylamine and related complexes, and pentamine acetate cobalt(III) and related complexes); metal care agents in the range from about 0.1% to 5% by weight (selected from benzatriazoles, metal salts and complexes, and/or silicates); enzymes in the range from about 0.01 to 5.0 mg of active enzyme per gram of automatic dishwashing detergent composition (acyl transferases, alpha-amylases, beta-amylases, alpha-galactosidases, arabinosidases, aryl esterases, beta-galactosidases, carrageenases, catalases, cellobiohydrolases, cellulases, chondroitinases, cutinases, endo-beta-1, 4-glucanases, endo-beta-mannanases, esterases, exo-mannanases, galactanases, glucoamylases, hemicellulases, hyaluronidases, keratinases, laccases, lactases, ligninases, lipases, lipoxygenases, mannanases, oxidases, pectate lyases, pectin acetyl esterases, pectinases, pentosanases, peroxidases, phenoloxidases, phosphatases, phospholipases, phytases, polygalacturonases, proteases, pullulanases, reductases, rhamnogalacturonases, beta-glucanases, tannases, transglutaminases, xylan acetyl-esterases, xylanases, xyloglucanases, and xylosidases, and any mixture thereof); and enzyme stabilizer components (selected from oligosaccharides, polysaccharides and inorganic divalent metal salts).
[0310] The metalloproteases are normally incorporated into the detergent composition at a level of from 0.000001% to 5% of enzyme protein by weight of the composition, or from 0.00001% to 2%, or from 0.0001% to 1%, or from 0.001% to 0.75% of enzyme protein by weight of the composition.
Metalloprotease Polypeptides of the Present Invention for Use in Animal Feed
[0311] In a further aspect of the invention, the metalloprotease polypeptides of the present invention can be used as a component of an animal feed composition, animal feed additive and/or pet food comprising a metalloprotease and variants thereof. The present invention further relates to a method for preparing such an animal feed composition, animal feed additive composition and/or pet food comprising mixing the metalloprotease polypeptide with one or more animal feed ingredients and/or animal feed additive ingredients and/or pet food ingredients. Furthermore, the present invention relates to the use of the metalloprotease polypeptide in the preparation of an animal feed composition and/or animal feed additive composition and/or pet food.
[0312] The term "animal" includes all non-ruminant and ruminant animals. In a particular embodiment, the animal is a non-ruminant animal, such as a horse and a mono-gastric animal. Examples of mono-gastric animals include, but are not limited to, pigs and swine, such as piglets, growing pigs, sows; poultry such as turkeys, ducks, chicken, broiler chicks, layers; fish such as salmon, trout, tilapia, catfish and carps; and crustaceans such as shrimps and prawns. In a further embodiment the animal is a ruminant animal including, but not limited to, cattle, young calves, goats, sheep, giraffes, bison, moose, elk, yaks, water buffalo, deer, camels, alpacas, llamas, antelope, pronghorn and nilgai.
[0313] In the present context, it is intended that the term "pet food" is understood to mean a food for a household animal such as, but not limited to, dogs, cats, gerbils, hamsters, chinchillas, fancy rats, guinea pigs; avian pets, such as canaries, parakeets, and parrots; reptile pets, such as turtles, lizards and snakes; and aquatic pets, such as tropical fish and frogs.
[0314] The terms "animal feed composition," "feedstuff" and "fodder" are used interchangeably and can comprise one or more feed materials selected from the group comprising a) cereals, such as small grains (e.g., wheat, barley, rye, oats and combinations thereof) and/or large grains such as maize or sorghum; b) by products from cereals, such as corn gluten meal, Distillers Dried Grain Solubles (DDGS) (particularly corn based Distillers Dried Grain Solubles (cDDGS), wheat bran, wheat middlings, wheat shorts, rice bran, rice hulls, oat hulls, palm kernel, and citrus pulp; c) protein obtained from sources such as soya, sunflower, peanut, lupin, peas, fava beans, cotton, canola, fish meal, dried plasma protein, meat and bone meal, potato protein, whey, copra, sesame; d) oils and fats obtained from vegetable and animal sources; e) minerals and vitamins.
Metalloprotease Polypeptides of the Present Invention for Use in Textile Desizing
[0315] Also contemplated are compositions and methods of treating fabrics (e.g., to desize a textile) using a metalloprotease polypeptide of the present invention. Fabric-treating methods are well known in the art (see, e.g., U.S. Pat. No. 6,077,316). For example, the feel and appearance of a fabric can be improved by a method comprising contacting the fabric with a metalloprotease in a solution. The fabric can be treated with the solution under pressure.
[0316] A metalloprotease of the present invention can be applied during or after the weaving of a textile, or during the desizing stage, or one or more additional fabric processing steps. During the weaving of textiles, the threads are exposed to considerable mechanical strain. Prior to weaving on mechanical looms, warp yarns are often coated with sizing starch or starch derivatives to increase their tensile strength and to prevent breaking. A metalloprotease of the present invention can be applied during or after the weaving to remove these sizing starch or starch derivatives. After weaving, the metalloprotease can be used to remove the size coating before further processing the fabric to ensure a homogeneous and wash-proof result.
[0317] A metalloprotease of the present invention can be used alone or with other desizing chemical reagents and/or desizing enzymes to desize fabrics, including cotton-containing fabrics, as detergent additives, e.g., in aqueous compositions. An amylase also can be used in compositions and methods for producing a stonewashed look on indigo-dyed denim fabric and garments. For the manufacture of clothes, the fabric can be cut and sewn into clothes or garments, which are afterwards finished. In particular, for the manufacture of denim jeans, different enzymatic finishing methods have been developed. The finishing of denim garment normally is initiated with an enzymatic desizing step, during which garments are subjected to the action of proteolytic enzymes to provide softness to the fabric and make the cotton more accessible to the subsequent enzymatic finishing steps. The metalloprotease can be used in methods of finishing denim garments (e.g., a "bio-stoning process"), enzymatic desizing and providing softness to fabrics, and/or finishing process.
Metalloprotease Polypeptides of the Present Invention for Use in Paper Pulp Bleaching
[0318] The metalloprotease polypeptides described herein find further use in the enzyme aided bleaching of paper pulps such as chemical pulps, semi-chemical pulps, kraft pulps, mechanical pulps or pulps prepared by the sulfite method. In general terms, paper pulps are incubated with a metalloprotease polypeptide of the present invention under conditions suitable for bleaching the paper pulp.
[0319] In some embodiments, the pulps are chlorine free pulps bleached with oxygen, ozone, peroxide or peroxyacids. In some embodiments, the metalloprotease polypeptides are used in enzyme aided bleaching of pulps produced by modified or continuous pulping methods that exhibit low lignin contents. In some other embodiments, the metalloprotease polypeptides are applied alone or preferably in combination with xylanase and/or endoglucanase and/or alpha-galactosidase and/or cellobiohydrolase enzymes.
Metalloprotease Polypeptides of the Present Invention for Use in Protein Degradation
[0320] The metalloprotease polypeptides described herein find further use in the enzyme aided removal of proteins from animals and their subsequent degradation or disposal, such as feathers, skin, hair, hide, and the like. In some instances, immersion of the animal carcass in a solution comprising a metalloprotease polypeptide of the present invention can act to protect the skin from damage in comparison to the traditional immersion in scalding water or the defeathering process. In one embodiment, feathers can be sprayed with an isolated metalloprotase polypeptide of the present invention under conditions suitable for digesting or initiating degradation of the plumage. In some embodiments, a metalloprotease of the present invention can be used, as above, in combination with an oxidizing agent.
[0321] In some embodiments, removal of the oil or fat associated with raw feathers is assisted by using a metalloprotease polypeptide of the present invention. In some embodiments, the metalloprotease polypeptides are used in compositions for cleaning the feathers as well as to sanitize and partially dehydrate the fibers. In some other embodiments, the metalloprotease polypeptides are applied in a wash solution in combination with 95% ethanol or other polar organic solvent with or without a surfactant at about 0.5% (v/v).
[0322] In yet other embodiments, the disclosed metalloprotease polypeptides find use in recovering protein from plumage. The disclosed metalloprotease polypeptides may be used alone or in combination in suitable feather processing and proteolytic methods, such as those disclosed in PCT/EP2013/065362, PCT/EP2013/065363, and PCT/EP2013/065364, which are hereby incorporated by reference. In some embodiments, the recovered protein can be subsequently used in animal or fish feed.
EXPERIMENTAL
[0323] The claimed invention is described in further detail in the following examples which are not in any way intended to limit the scope of the invention as claimed.
Example 1.1
Cloning of Paenibacillus sp. Metalloprotease PspPro3
[0324] A strain of Paenibacillus sp. was selected as a potential source for enzymes which may be useful for various industrial applications. Genomic DNA for sequencing was obtained by first growing the strain on Heart Infusion agar plates (Difco) at 37.degree. C. for 24 hours. Cell material was scraped from the plates and used to prepare genomic DNA with the ZF Fungal/Bacterial DNA miniprep kit from Zymo (Cat No. D6005). The genomic DNA was used for genome sequencing. The entire genome of the Paenibacillus sp. strain was sequenced by BaseClear (Leiden, The Netherlands) using the Illumina's next generation sequencing technology. After assembly of the data, contigs were annotated by BioXpr (Namur, Belgium). One of the genes identified after annotation in Paenibacillus sp. encodes a metalloprotease and the sequence of this gene, called PspPro3, is provided in SEQ ID NO: 1. The corresponding protein encoded by the PspPro3 gene is shown in SEQ ID NO: 2. At the N-terminus, the protein has a signal peptide with a length of 26 amino acids as predicted by SignalP version 4.0 (Nordahl Petersen et al. (2011) Nature Methods, 8:785-786). The presence of a signal sequence suggests that PspPro3 is a secreted enzyme. The propeptide region was predicted based on protein sequence alignment with the Paenibacillus polymyxa Npr protein (Takekawa et al. (1991) Journal of Bacteriology, 173 (21): 6820-6825). The predicted mature region of PspPro3 protein is shown on SEQ ID NO: 3.
[0325] The nucleotide sequence of the PspPro3 gene isolated from Paenibacillus sp. is set forth as SEQ ID NO: 1. The sequence encoding the predicted native signal peptide is shown in italics:
TABLE-US-00002 ATGTTAATGAAAAAAGTATGGGTTTCGCTTCTTGGAGGAGCGATGTTAT TAGGGTCTGTAGCGTCTGGTGCATCAGCAGCGGAGAGTTCCGTTTCGGG GCCGGCTCAGCTTACGCCAACCTTCCATGCCGAACAATGGAAAGCACCT TCATCGGTATCGGGTGATGACATCGTATGGAGCTATTTAAATCGGCAAA AGAAAACGTTGCTGGGTACGGACAGCACCAGTGTCCGTGATCAATTCCG TATCGTAGATCGCACAAGCGACAAATCCGGCGTGAGCCATTATCGGCTG AAGCAATATGTAAACGGAATTCCCGTATATGGAGCTGAACAGACCATTC ATGTGGGCAAATCCGGTGAAGTGACCTCTTATCTGGGAGCCGTGATTAC TGAGGATCAGCAAGAAGAAGCTACGCAAGGTACAACTCCGAAAATCAGC GCTTCTGAAGCGGTCCATACCGCATATCAGGAGGCAGCTACACGGGTTC AAGCCCTCCCTACCTCCGATGATACGATTTCTAAAGATGCGGAGGAGCC AAGCAGTGTAAGCAAAGACACTTACTCCGAAGCAGCTAACAACGGAAAA ACGAGTTCTGTTGAAAAGGACAAGCTCAGCCTTGAGAAAGCGGCTGACC TGAAAGATAGCAAAATTGAAGCGGTGGAGGCAGAGCCAAACTCCATTGC CAAAATCGCCAACCTGCAGCCTGAGGTAGATCCTAAAGCCGAACTATAT TTCTATGCGAAGGGCGATGCATTGCAGCTGGTTTATGTGACTGAGGTTA ATATTTTGCAGCCTGCGCCGCTGCGTACACGCTACATCATTGACGCCAA TGATGGCAAAATCGTATCCCAGTATGACATCATTAATGAAGCGACAGGC ACAGGCAAAGGTGTACTCGGTGATACCAAAACATTCAACACTACTGCTT CCGGCAGCAGCTACCAGTTAAGAGATACGACTCGCGGGAATGGAATCGT GACTTACACGGCCTCCAACCGTCAAAGCATCCCAGGTACGATCCTGACC GATGCCGATAACGTATGGAATGATCCAGCCGGCGTGGATGCCCACGCTT ATGCAGCCAAAACCTATGATTATTATAAGGAAAAGTTCAATCGCAACAG CATTGACGGACGAGGCCTGCAGCTCCGTTCGACAGTTCATTACGGCAAT CGTTACAACAACGCCTTCTGGAACGGCTCCCAAATGACTTATGGAGACG GAGACGGCACCACATTTATCGCTTTTAGCGGTGATCCGGATGTAGTTGG TCATGAACTCACACACGGTGTTACGGAGTATACTTCCAATTTGGAATAT TACGGAGAATCCGGTGCGTTGAACGAGGCCTTCTCGGACATCATCGGCA ATGACATCCAGCGTAAAAACTGGCTTGTAGGCGATGATATTTACACGCC ACGCATTGCGGGTGATGCACTTCGTTCTATGTCCAATCCTACGCTGTAC GATCAACCGGATCACTATTCGAACTTGTACAGAGGCAGCTCCGATAACG GCGGCGTTCATACGAACAGCGGTATTATAAATAAAGCCTATTATCTGTT GGCACAAGGCGGCACCTTCCATGGTGTAACTGTCAATGGGATTGGCCGC GATGCAGCGGTTCAAATTTACTACAGCGCCTTTACGAACTACCTGACTT CTTCTTCTGACTTCTCCAATGCACGTGATGCCGTTGTACAAGCGGCAAA AGATCTCTACGGCGCGAGCTCGGCACAAGCTACCGCAGCAGCCAAATCT TTTGATGCTGTAGGCGTTAAC
[0326] The amino acid sequence of the PspPro3 precursor protein is set forth as SEQ ID NO: 2. The predicted signal peptide is shown in italics, and the predicted pro-peptide is shown in underlined text:
TABLE-US-00003 MLMKKVWVSLLGGAMLLGSVASGASAAESSVSGPAQLTPTFHAEQWKAP SSVSGDDIVWSYLNRQKKTLLGTDSTSVRDQFRIVDRTSDKSGVSHYRL KQYVNGIPVYGAEQTIHVGKSGEVTSYLGAVITEDQQEEATQGTTPKIS ASEAVHTAYQEAATRVQALPTSDDTISKDAEEPSSVSKDTYSEAANNGK TSSVEKDKLSLEKAADLKDSKIEAVEAEPNSIAKIANLQPEVDPKAELY FYAKGDALQLVYVTEVNILQPAPLRTRYIIDANDGKIVSQYDIINEATG TGKGVLGDTKTFNTTASGSSYQLRDTTRGNGIVTYTASNRQSIPGTILT DADNVWNDPAGVDAHAYAAKTYDYYKEKFNRNSIDGRGLQLRSTVHYGN RYNNAFWNGSQMTYGDGDGTTFIAFSGDPDVVGHELTHGVTEYTSNLEY YGESGALNEAFSDIIGNDIQRKNWLVGDDIYTPRIAGDALRSMSNPTLY DQPDHYSNLYRGSSDNGGVHTNSGIINKAYYLLAQGGTFHGVTVNGIGR DAAVQIYYSAFTNYLTSSSDFSNARDAVVQAAKDLYGASSAQATAAAKS FDAVGVN
[0327] The amino acid sequence of the predicted mature form of PspPro3 is set forth as SEQ ID NO: 3:
TABLE-US-00004 ATGTGKGVLGDTKTFNTTASGSSYQLRDTTRGNGIVTYTASNRQSIPGTI LTDADNVWNDPAGVDAHAYAAKTYDYYKEKFNRNSIDGRGLQLRSTVHYG NRYNNAFWNGSQMTYGDGDGTTFIAFSGDPDVVGHELTHGVTEYTSNLEY YGESGALNEAFSDIIGNDIQRKNWLVGDDIYTPRIAGDALRSMSNPTLYD QPDHYSNLYRGSSDNGGVHTNSGIINKAYYLLAQGGTFHGVTVNGIGRDA AVQIYYSAFTNYLTSSSDFSNARDAVVQAAKDLYGASSAQATAAAKSFDA VGVN
Example 1.2
Expression of Paenibacillus sp. Metalloprotease PspPro3
[0328] The DNA sequence of the propeptide-mature form of PspPro3 was synthesized and inserted into the Bacillus subtilis expression vector p2JM103BBI (Vogtentanz, Protein Expr Purif, 55:40-52, 2007) by Generay (Shanghai, China), resulting in plasmid pGX085 (AprE-PspPro3) (FIG. 1.1). Ligation of the gene encoding the PspPro3 protein into the digested vector resulted in the addition of three codons (Ala-Gly-Lys) between the 3' end of the Bacillus subtilis AprE signal sequence and the 5' end of the predicted PspPro3 native propeptide. The gene has an alternative start codon (GTG). As shown in FIG. 1.1, pGX085(AprE-PspPro3) contains an AprE promoter, an AprE signal sequence used to direct target protein secretion in B. subtilis, and the synthetic nucleotide sequence encoding the predicted propeptide and mature region of PspPro3 (SEQ ID NO: 4). The translation product of the synthetic AprE-PspPro3 gene is shown in SEQ ID NO: 5.
[0329] B. subtilis cells (degU.sup.Hy 32, .DELTA.scoC) were transformed with the pGX085(AprE-PspPro3) plasmid and the transformed cells were spread on Luria Agar plates supplemented with 5 ppm Chloramphenicol and 1.2% skim milk (Cat #232100, Difco). Colonies with the largest clear halos on the plates were selected and subjected to fermentation in a 250 ml shake flask with MBD medium (a MOPS based defined medium, supplemented with additional 5 mM CaCl.sub.2). The broth from the shake flasks was concentrated and buffer-exchanged into the loading buffer containing 20 mM Tris-HCl (pH 8.5), 1 mM CaCl.sub.2 and 10% propylene glycol using a VivaFlow 200 ultra filtration device (Sartorius Stedim). After filtering, this sample was applied to a 150 mL Q Sepharose High Performance column pre-equilibrated with the loading buffer above and PspPro3 was then eluted from the column via the loading buffer supplemented with a linear NaCl gradient from 0 to 0.7 M. The corresponding active purified protein fractions were further pooled and concentrated via 10K Amicon Ultra for further analyses.
[0330] The nucleotide sequence of the synthesized PspPro3 gene in plasmid pGX085(AprE-PspPro3) is depicted in SEQ ID NO: 4. The sequence encoding the predicted native signal peptide is shown in italics and the region encoding the three residue addition (AGK) is shown in bold:
TABLE-US-00005 GTGAGAAGCAAAAAATTGTGGATCAGCTTGTTGTTTGCGTTAACGTTAA TCTTTACGATGGCGTTCAGCAACATGAGCGCGCAGGCTGCTGGAAAAGC AGAATCATCAGTGTCAGGACCGGCTCAGCTTACGCCGACGTTTCATGCA GAGCAGTGGAAAGCACCGAGCAGCGTTAGCGGAGATGACATCGTGTGGA GCTACCTGAACAGACAGAAGAAAACGCTTCTTGGCACGGACAGCACGAG CGTCAGAGACCAGTTCAGAATCGTGGATAGAACAAGCGACAAAAGCGGC GTCAGCCATTATAGACTGAAGCAGTATGTGAACGGAATCCCGGTTTATG GCGCAGAACAAACAATCCATGTCGGAAAGAGCGGCGAAGTTACGAGCTA TCTGGGCGCGGTTATTACAGAGGACCAGCAAGAGGAGGCTACACAAGGC ACGACACCGAAAATTTCAGCATCAGAGGCAGTTCATACGGCCTACCAAG AAGCTGCAACGAGAGTTCAAGCCCTGCCTACGTCAGATGATACAATCAG CAAAGACGCTGAGGAACCTAGCTCAGTTAGCAAGGACACGTATAGCGAA GCCGCGAACAATGGCAAGACGTCAAGCGTGGAAAAAGACAAGCTTTCAC TGGAGAAGGCCGCTGATCTGAAAGACTCAAAGATCGAGGCTGTGGAAGC GGAACCGAATAGCATTGCAAAGATTGCCAACCTGCAACCGGAGGTGGAC CCGAAGGCGGAGCTGTATTTCTACGCTAAAGGCGATGCACTGCAACTGG TTTACGTCACGGAGGTTAACATCCTGCAGCCGGCACCGCTTAGAACGAG ATACATCATTGACGCGAACGACGGCAAGATCGTGAGCCAGTACGACATT ATCAACGAGGCCACGGGAACGGGCAAGGGAGTCCTTGGCGACACGAAGA CATTCAATACAACGGCCTCAGGCTCATCATACCAGCTGAGAGACACGAC GAGAGGCAACGGAATCGTCACGTACACGGCTAGCAATAGACAGAGCATT CCGGGCACAATCCTTACGGACGCAGACAATGTGTGGAATGACCCGGCAG GCGTGGACGCACATGCCTACGCAGCGAAGACGTACGACTACTACAAGGA GAAGTTCAACAGAAACAGCATCGACGGAAGAGGACTGCAACTTAGAAGC ACGGTGCATTACGGCAACAGATACAACAACGCTTTCTGGAACGGCAGCC AAATGACGTATGGAGACGGCGATGGAACAACGTTTATCGCATTCTCAGG CGACCCTGACGTTGTGGGACATGAACTGACGCATGGAGTCACAGAATAC ACGAGCAATCTGGAGTATTACGGAGAATCAGGCGCACTTAATGAGGCCT TCAGCGACATCATCGGAAACGACATCCAGAGAAAGAACTGGCTGGTTGG CGATGATATCTACACGCCGAGAATTGCGGGCGACGCGCTGAGATCAATG AGCAACCCTACGCTGTACGATCAGCCGGATCATTACAGCAACCTGTATA GAGGCTCAAGCGATAATGGCGGCGTGCATACAAACAGCGGCATCATCAA CAAAGCCTATTATCTGCTGGCGCAAGGCGGCACATTCCATGGCGTTACA GTTAATGGCATTGGCAGAGACGCAGCCGTGCAGATCTACTACAGCGCAT TCACGAATTACCTGACATCAAGCAGCGACTTTTCAAATGCAAGAGATGC AGTGGTGCAGGCGGCTAAAGACCTTTATGGAGCTTCAAGCGCTCAGGCC ACAGCTGCGGCAAAAAGCTTCGACGCGGTTGGAGTGAAT
[0331] The amino acid sequence of the PspPro3 precursor protein expressed from plasmid pGX085(AprE-PspPro3) is depicted in SEQ ID NO: 5. The predicted signal sequence is shown in italics, the three residue addition (AGK) shown in bold and the predicted pro-peptide is shown in underlined text.
TABLE-US-00006 MRSKKLWISLLFALTIJFTMAFSNMSAQAAGKAESSVSGPAOLTPTFHA EOWKAPSSVSGDDIVWSYLNROKKTLLGTDSTSVRDOFRIVDRTSDKSG VSHYREKOYVNGIPVYGAEOTIHVGKSGEVTSYLGAVITEDOOEEATOG TTPKISASEAVHTAYOEAATRVOALPTSDDTISKDAEEPSSVSKDTYSE AANNGKTSSVEKDKLSLEKAADLKDSKIEAVEAEPNSIAKIANLOPEVD PKAELYFYAKGDALOLVYVTEVNILOPAPLRTRYIIDANDGKIVSOYDI INEATGTGKGVLGDTKTFNTTASGSSYQLRDTTRGNGIVTYTASNRQSI PGTILTDADNVWNDPAGVDAHAYAAKTYDYYKEKFNRNSIDGRGLQLRS TVHYGNRYNNAFWNGSQMTYGDGDGTTFIAFSGDPDVVGHELTHGVTEY TSNLEYYGESGALNEAFSDIIGNDIQRKNWLVGDDIYTPRIAGDALRSM SNPTLYDQPDHYSNLYRGSSDNGGVHTNSGIINKAYYLLAQGTFHGVTV NGIGRDAAVQIYYSAFTNYLTSSSDFSNARDAVVQAAKDLYGASSAQAT AAAKSFDAVGVN
[0332] The amino acid sequence of the PspPro3 recombinant protein isolated from Bacillus subtilis culture was determined by tandem mass spectrometry, and shown below. It is the same as predicted and depicted in SEQ ID NO: 3.
TABLE-US-00007 ATGTGKGVLGDTKTFNTTASGSSYQLRDTTRGNGIVTYTASNRQSIPGTI LTDADNVWNDPAGVDAHAYAAKTYDYYKEKFNRNSIDGRGLQLRSTVHYG NRYNNAFWNGSQMTYGDGDGTTFIAFSGDPDVVGHELTHGVTEYTSNLEY YGESGALNEAFSDIIGNDIQRKNWLVGDDIYTPRIAGDALRSMSNPTLYD QPDHYSNLYRGSSDNGGVHTNSGIINKAYYLLAQGGTFHGVTVNGIGRDA AVQIYYSAFTNYLTSSSDFSNARDAVVQAAKDLYGASSAQATAAAKSFDA VGVN
Example 1.3
Proteolytic Activity of Metalloprotease PspPro3
[0333] The proteolytic activity of purified PspPro3 was measured in 50 mM Tris (pH 7), using azo-casein (Cat #74H7165, Megazyme) as a substrate. Prior to the reaction, the enzyme was diluted with Milli-Q water (Millipore) to specific concentrations. The azo-casein was dissolved in 100 mM Tris buffer (pH 7) to a final concentration of 1.5% (w/v). To initiate the reaction, 50 .mu.L of the diluted enzyme (or Milli-Q H.sub.2O alone as the blank control) was added to the non-binding 96-well microtiter Plate (96-MTP) (Corning Life Sciences, #3641) placed on ice, followed by the addition of 50 .mu.L of 1.5% azo-casein. After sealing the 96-MTP, the reaction was carried out in a Thermomixer (Eppendorf) at 40.degree. C. and 650 rpm for 10 min. The reaction was terminated by adding 100 .mu.L of 5% Trichloroacetic Acid (TCA). Following equilibration (5 min at the room temperature) and subsequent centrifugation (2000 g for 10 min at 4.degree. C.), 120 .mu.L supernatant was transferred to a new 96-MTP, and absorbance of the supernatant was measured at 440 nm (A.sub.440) using a SpectraMax 190. Net A.sub.440 was calculated by subtracting the A.sub.440 of the blank control from that of enzyme, and then plotted against different protein concentrations (from 1.25 ppm to 40 ppm). Each value was the mean of duplicate assays, and the value varies no more than 5%. The proteolytic activity is shown as Net A.sub.440. The proteolytic assay with azo-casein as the substrate (FIG. 1.2) indicates that PspPro3 is an active protease.
Example 1.4
pH Profile of Metalloprotease PspPro3
[0334] With azo-casein as the substrate, the pH profile of PspPro3 was studied in 12.5 mM acetate/Bis-Tris/HEPES/CHES buffer with different pH values (ranging from pH 4 to 11). To initiate the assay, 50 .mu.L of 25 mM acetate/Bis-Tris/HEPES/CHES buffer with a specific pH was first mixed with 2 .mu.L diluted enzyme (250 ppm in Milli-Q H.sub.2O) in a 96-MTP placed on ice, followed by the addition of 48 .mu.L of 1.5% (w/v) azo-casein prepared in H2O. The reaction was performed and analyzed as described in Example 1.3. Enzyme activity at each pH was reported as relative activity where the activity at the optimal pH was set to be 100%. The pH values tested were 4, 5, 5.5, 6, 6.5, 7, 7.5, 8, 8.5, 9, 10 and 11. Each value was the mean of triplicate assays. As shown in FIG. 1.3, the optimal pH of PspPro3 is 7.5, with greater than 70% of maximal activity retained between pH 5.5 and 9.
Example 1.5
Temperature Profile of Metalloprotease PspPro3
[0335] The temperature profiles of PspPro3 were analyzed in 50 mM Tris buffer (pH 7) using the azo-casein assay. The enzyme sample and azo-casein substrate were prepared as in Example 3. Prior to the reaction, 50 .mu.L of 1.5% azo-casein and 45 .mu.l Milli-Q H.sub.2O were mixed in a 2000_, PCR tube, which was then subsequently incubated in a Peltier Thermal Cycler (BioRad) at desired temperatures (i.e. 20-90.degree. C.) for 5 min. After the incubation, 5 .mu.L of diluted PspPro3 (100 ppm) or H.sub.2O (the blank control) was added to the substrate mixture, and the reaction was carried out in the Peltier Thermal Cycle for 10 min at different temperatures. To terminate the reaction, each assay mixture was transferred to a 96-MTP containing 100 .mu.L of 5% TCA per well. Subsequent centrifugation and absorbance measurement were performed as described in Example 1.3. The activity was reported as relative activity where the activity at the optimal temperature was set to be 100%. The tested temperatures were 20, 30, 40, 50, 60, 70, 80, and 90.degree. C. Each value was the mean of triplicate assays. The data in FIG. 1.4 suggest that PspPro3 showed an optimal temperature at 50.degree. C., and retained greater than 70% of its maximal activity between 45.degree. C. and 60.degree. C.
Example 1.6
Cleaning Performance of Metalloprotease PspPro3 in Automatic Dishwashing (ADW) Conditions
[0336] The cleaning performance of PspPro3 in automatic dishwashing (ADW) conditions was tested using PA-S-38 (egg yolk, with pigment, aged by heating) microswatches (CFT-Vlaardingen, The Netherlands) at pH 6 or 8 using a model automatic dishwashing (ADW) detergent. Prior to the reaction, purified PspPro3 were diluted with a dilution solution containing 10 mM NaCl, 0.1 mM CaCl.sub.2, 0.005% TWEEN.RTM. 80 and 10% propylene glycol to the desired concentrations. The reactions were performed in AT detergent (composition shown in Table 1.1) with 100 ppm water hardness (Ca.sup.2+:Mg.sup.2+=3:1), in the absence or presence of a bleach component (Peracid N,N-phthaloylaminoperoxycaproic acid-PAP). To initiate the reaction, 180 .mu.L of AT detergent buffered at pH 6 or 8 was added to a 96-MTP placed with PA-S-38 microswatches, followed by the addition of 20 .mu.L of diluted enzymes (or the dilution solution as the blank control). The 96-MTP was sealed and incubated in an incubator/shaker for 30 min at 50.degree. C. and 1150 rpm. After incubation, 1004, of wash liquid from each well was transferred to a new 96-MTP, and its absorbance was measured at 405 nm (A.sub.405) (referred here as the "Initial performance") using a spectrophotometer. The remaining wash liquid in the 96-MTP was discarded and the microswatches were rinsed once with 200 .mu.L water. Following the addition of 180 .mu.L of 0.1 M CAPS buffer (pH 10), the second incubation was carried out in the incubator/shaker at 50.degree. C. and 1150 rpm for 10 min. One hundred microliter of the resulting wash liquid was transferred to a new 96-MTP, and its absorbance measured at 405 nm (referred here as "Wash-off"). The sum of two absorbance measurements ("Initial performance" plus "Wash-off") gives the "Total performance", which measures the protease activity on the model stain. Dose response in cleaning the PA-S-38 microswatches at pH 6 and pH 8 for PspPro3 in AT detergent, in the absence or presence of bleach, is shown in FIGS. 5A and 5B, respectively.
TABLE-US-00008 TABLE 1.1 Composition of AT dish detergent formula with bleach Ingredient Concentration (mg/ml) MGDA (methylglycinediacetic acid) 0.143 Sodium citrate 1.86 Citric acid* varies PAP (peracid N, N-phthaloylaminoperoxycaproic acid) 0.057 Plurafac .RTM. LF 18B (a non-ionic surfactant) 0.029 Bismuthcitrate 0.006 Bayhibit .RTM. S (Phosphonobutantricarboxylic acid sodium salt) 0.006 Acusol .TM. 587 (a calcium polyphosphate inhibitor) 0.029 PEG 6000 0.043 PEG 1500 0.1 *The pH of the AT detergent is adjusted to the desired value (pH 6 or 8) by the addition of 0.9 M citric acid.
Example 1.7
Cleaning Performance of Metalloprotease PspPro3 in Laundry Conditions
[0337] The cleaning performance of PspPro3 protein in liquid laundry detergent was tested using EMPA-116 (cotton soiled with blood/milk/ink) microswatches (obtained from CFT Vlaardingen, The Netherlands) at pH 8.2 using a commercial detergent. Prior to the reaction, purified PspPro3 protein samples were diluted with a dilution solution (10 mM NaCl, 0.1 mM CaCl.sub.2, 0.005% TWEEN.RTM. 80 and 10% propylene glycol) to the desired concentrations; and the commercial detergent (Tide.RTM., Clean Breeze.RTM., Proctor & Gamble, USA, purchased September 2011) was incubated at 95.degree. C. for 1 hour to inactivate the enzymes present in the detergent. Proteolytic assays were subsequently performed to confirm the inactivation of proteases in the commercial detergent. The heat treated detergent was further diluted with 5 mM HEPES (pH 8.2) to a final concentration of 0.788 g/L. Meanwhile, the water hardness of the buffered liquid detergent was adjusted to 103 ppm (Ca.sup.2+:Mg.sup.2+=3:1). The specific conductivity of the buffered detergent was adjusted to either 0.62 mS/cm (low conductivity) or 3.5 mS/cm (high conductivity) by adjusting the NaCl concentration in the buffered detergent. To initiate the reaction, 190 .mu.l of either the high or low conductivity buffered detergent was added to a 96-MTP containing the EMPA-116 microswatches, followed by the addition of 10 .mu.l of diluted enzyme (or the dilution solution as blank control). The 96-MTP was sealed and incubated in an incubator/shaker for 20 min at 32.degree. C. and 1150 rpm. After incubation, 150 .mu.l of wash liquid from each well was transferred to a new 96-MTP, and its absorbance was measured at 600 nm using a spectrophotometer, which indicates the protease activity on the model stain; and Net A.sub.600 was subsequently calculated by subtracting the A.sub.600 of the blank control from that of the enzyme. Dose response for the cleaning of EMPA-116 microswatches in liquid laundry detergent at high or low conductivity is shown in FIG. 1.6.
Example 1.8
Comparison of PspPro3 to Other Metalloproteases
A. Identification of Homologous Proteases
[0338] Homologs were identified by a BLAST search (Altschul et al., Nucleic Acids Res, 25:3389-402, 1997) against the NCBI non-redundant protein database and the Genome Quest Patent database with search parameters set to default values. The mature protein amino acid sequence for PspPro3 (SEQ ID NO: 3) was used as the query sequence. Percent identity (PID) for both search sets is defined as the number of identical residues divided by the number of aligned residues in the pairwise alignment. Tables 1.2A and 1.2B provide a list of sequences with the percent identity to PspPro3. The length in Table 1.2 refers to the entire sequence length of the homologous proteases.
TABLE-US-00009 TABLE 1.2A List of sequences with percent identity to PspPro3 protein identified from the NCBI non-redundant protein database PID to Accession # PspPro3 Organism Length ZP_10321515.1 55 Bacillus macauensis ZFHKF-1 552 AAC43402.1 57 Alicyclobacillus acidocaldarius 546 P00800 57 Bacillus thermoproteolyticus 548 AAA22621.1 58 Geobacillus stearothermophilus 548 ZP_01862236.1 59 Bacillus sp. SG-1 560 YP_002884504.1 59 Exiguobacterium sp. AT1b 509 AEI46285.1 60 Paenibacillus mucilaginosus KNP414 525 ZP_08093424 60 Planococcus donghaensis MPA1U2 553 ZP_10324092.1 61 Bacillus macauensis ZFHKF-1 533 YP_006792441.1 61 Exiguobacterium antarcticum B7 498 AAK69076.1 63 Bacillus thuringiensis serovar finitimus 566 NP_976992.1 64 Bacillus cereus ATCC 10987 566 ZP_04321694 64 Bacillus cereus 566 BAA06144 64 Lactobacillus sp. 566 ZP_10241029.1 78 Paenibacillus peoriae KCTC 3763 599 YP_005073223 93 Paenibacillus terrae HPL-003 591 YP_003872179 94 Paenibacillus polymyxa E681 592 ZP_09775364 100 Paenibacillus sp. Aloe-11 593
TABLE-US-00010 TABLE 1.2B List of sequences with percent identity to PspPro3 protein identified from the Genome Quest Patent database PID to Patent # PspPro3 Organism Length US20120107907-0184 57.88 Bacillus caldoyticus 319 US20120107907-0177 57.88 Bacillus caldolyticus 544 WO2012110563-0002 58.2 Bacillus caldolyticus 319 EP2390321-0176 58.52 Bacillus stearothermophilis 548 US6518054-0002 59.22 Bacillus sp. 316 WO2004011619-0044 60.6 Empty 507 WO2004011619-0047 62.14 Empty 532 WO2004011619-0046 62.26 Empty 536 WO2012110563-0004 63.02 Bacillus megaterium 320 JP2002272453-0003 63.67 Empty 562 US8114656-0186 64.24 Bacillus brevis 304 WO2012110562-0005 64.52 Bacillus cereus 320 WO2007044993-0178 64.74 Bacillus thuringiensis 566 EP2178896-0184 65.38 Bacillus anthracis 566 WO2012110563-0005 65.48 Bacillus cereus 320 JP1995184649-0001 65.71 Lactobacillus sp. 566 US5962264-0004 65.81 Empty 566 US20120107907-0185 66.13 Bacillus cereus 317 US8114656-0187 93.36 Bacillus polymyxa 302 JP2005229807-0019 93.38 Paenibacillus polymyxa 566
B. Alignment of Homologous Protease Sequences
[0339] The amino acid sequence for mature PspPro3 (SEQ ID NO: 3) was aligned with thermolysin (P00800, Bacillus thermoproteolyticus) and protease from Paenibacillus sp. Aloe-11 (ZP_09775364) using CLUSTALW software (Thompson et al., Nucleic Acids Research, 22:4673-4680, 1994) with the default parameters. FIG. 1.7 shows the alignment of PspPro3 with these protease sequences.
C. Phylogenetic Tree
[0340] A phylogenetic tree for full length sequence of PspPro3 (SEQ ID NO: 2) was built using sequences of representative homologs from Tables 2A and the Neighbor Joining method (NJ) (Saitou, N.; and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing Guide Trees. MolBiol.Evol. 4, 406-425). The NJ method works on a matrix of distances between all pairs of sequences to be analyzed. These distances are related to the degree of divergence between the sequences. The phylodendron-phylogenetic tree printer software (http://iubio.bio.indiana.edu/treeapp/treeprint-form.html) was used to display the phylogenetic tree shown in FIG. 1.8.
Example 1.9
Terg-o-Tometer Performance Evaluation of PspPro3
[0341] The wash performance of PspPro3 was tested in a laundry detergent application using a Terg-o-Tometer (Instrument Marketing Services, Inc, Fairfield, N.J.). The performance evaluation was conducted at 32.degree. C. and 16.degree. C. The soil load consisted of two of each of the following stain swatches: EMPA116 Blood, Milk, Ink on cotton (Test materials AG, St. Gallen, Switzerland), EMPA117 Blood, Milk, Ink on polycotton (Test materials AG, St. Gallen, Switzerland), EMPA112 Cocoa on cotton (Test materials AG, St. Gallen, Switzerland), and CFT C-10 Pigment, Oil, and Milk content on cotton (Center for Testmaterials BV, Vlaardingen, Netherlands), plus extra white interlock knit fabric to bring the total fabric load to 40 g per beaker of the Terg-o-Tometer, which was filled with 1 L of deionized water. The water hardness was adjusted to 6 grains per gallon, and the pH in the beaker was buffered with 5 mM HEPES, pH 8.2. Heat inactivated Tide Regular HDL (Procter & Gamble), a commercial liquid detergent purchased in a local US supermarket, was used at 0.8 g/L. The detergent was inactivated before use by treatment at 92.degree. C. in a water bath for 2-3 hours followed by cooling to room temperature. Heat inactivation of commercial detergents serves to destroy the activity of enzymatic components while retaining the properties of the non-enzymatic components. Enzyme activity in the heat inactivated detergent was measured using the Suc-AAPF-pNA assay for measuring protease activity. The Purafect.RTM. Prime HA, (Genencor Int'l) and PspPro3 proteases were each added to final concentrations of 0, 0.2, 0.5, 1, and 1.5 ppm. The wash time was 12 minutes. After the wash treatment, all swatches were rinsed for 3 minutes and machine-dried at low heat.
[0342] Four of each types of swatch were measured before and after treatment by optical reflectance using a Tristimulus Minolta Meter CR-400. The difference in the L, a, b values was converted to total color difference (dE), as defined by the CIE-LAB color space. Cleaning of the stains is expressed as percent stain removal index (% SRI) by taking a ratio between the color difference before and after washing, and comparing it to the difference of unwashed soils (before wash) to unsoiled fabric, and averaging the eight values obtained by reading two different regions of each washed swatch and is reported in Tables 1.9A and 1.9B as Average % SRI (dE).+-.95CI. Table 1.9A summarizes the cleaning performance of PspPro3 at 32.degree. C. and Table 1.9B at 16.degree. C.
TABLE-US-00011 TABLE 1.9A Cleaning performance of PspPro3 at 32.degree. C. EMPA-116 EMPA-117 Purafect Prime Purafect Prime HA PspPro3 HA SprPro3 Average 95CI Average 95CI Average 95CI Average 95CI ppm % SRI [% SRI % SRI [% SRI % SRI [% SRI % SRI [% SRI enzyme (dE) (dE)] (dE) (dE)] (dE) (dE)] (dE) (dE)] 0 0.19 0.01 0.19 0.01 0.17 0.01 0.17 0.01 0.2 0.27 0.02 0.27 0.02 0.25 0.03 0.30 0.02 0.5 0.28 0.03 0.31 0.01 0.30 0.03 0.31 0.02 1 0.30 0.01 0.32 0.02 0.35 0.02 0.34 0.03 1.5 0.31 0.02 0.31 0.01 0.37 0.01 0.37 0.03 EMPA-112 CFT C-10 Purafect Prime Purafect Prime HA PspPro3 HA PspPro3 Average 95CI Average 95CI Average 95CI Average 95CI ppm % SRI [% SRI % SRI [% SRI % SRI [% SRI % SRI [% SRI enzyme (dE) (dE)] (dE) (dE)] (dE) (dE)] (dE) (dE)] 0 0.11 0.03 0.11 0.03 0.07 0.01 0.07 0.01 0.2 0.11 0.05 0.18 0.04 0.12 0.01 0.11 0.01 0.5 0.13 0.04 0.17 0.03 0.15 0.01 0.16 0.01 1 0.18 0.03 0.19 0.04 0.17 0.01 0.21 0.01 1.5 0.19 0.03 0.18 0.04 0.18 0.01 0.23 0.01
TABLE-US-00012 TABLE 1.9B Cleaning performance of PspPro3 at 16.degree. C. EMPA-116 EMPA-117 Purafect Prime Purafect Prime HA PspPro3 HA PspPro3 Average 95CI Average 95CI Average 95CI Average 95CI ppm % SRI [% SRI % SRI [% SRI % SRI [% SRI % SRI [% SRI enzyme (dE) (dE)] (dE) (dE)] (dE) (dE)] (dE) (dE)] 0 0.15 0.02 0.15 0.02 0 13 0.01 0 13 0.01 0.2 0.19 0.02 0.20 0.03 0.15 0.02 0.15 0.02 0.5 0.20 0.02 0.19 0.02 0.21 0.02 0.20 0.02 1 0.24 0.04 0.21 0.02 0.22 0.02 0.20 0.01 1.5 0.19 0.02 0.25 0.04 0.23 0.03 0.20 0.01 EMPA-112 CFT C-10 Purafect Prime Purafect Prime HA PspPro3 HA PspPro3 Average 95CI Average 95CI Average 95CI Average 95CI ppm % SRI [% SRI % SRI [% SRI % SRI [% SRI % SRI [% SRI enzyme (dE) (dE)] (dE) (dE)] (dE) (dE)] (dE) (dE)] 0 0.08 0.03 0.08 0.03 0.04 0.08 0.04 0.08 0.2 0.12 0.02 0.09 0.01 0.06 0.12 0.06 0.09 0.5 0.08 0.02 0.11 0.02 0.08 0.08 0.08 0.11 1 0.11 0.02 0.10 0.03 0.08 0.11 0.09 0.10 1.5 0.13 0.02 0.11 0.03 0.11 0.13 0.10 0.11
Example 2.1
Cloning of Metalloprotease PspPro2 from Paenibacillus sp.
[0343] A strain of Paenibacillus sp. was selected as a potential source for enzymes which may be useful for various industrial applications. Genomic DNA for sequencing was obtained by first growing the strain on Heart Infusion agar plates (Difco) at 37.degree. C. for 24 hr. Cell material was scraped from the plates and used to prepare genomic DNA with the ZF Fungal/Bacterial DNA miniprep kit from Zymo (Cat No. D6005). The genomic DNA was used for genome sequencing. The entire genome of the Paenibacillus sp. strain was sequenced by BaseClear (Leiden, The Netherlands) using the Illumina's next generation sequencing technology. After assembly of the data, contigs were annotated by BioXpr (Namur, Belgium). One of the genes identified after annotation in Paenibacillus sp. encodes a metalloprotease and the sequence of this gene, called PspPro2, is provided in SEQ ID NO: 6. The corresponding protein encoded by the PspPro2 gene is shown in SEQ ID NO: 7. At the N-terminus, the protein has a signal peptide with a length of 24 amino acids as predicted by SignalP version 4.0 (Nordahl Petersen et al. (2011) Nature Methods, 8:785-786). The presence of a signal sequence suggests that PspPro2 is a secreted enzyme. The propeptide region of PspPro2 was predicted based on protein sequence alignment with the Paenibacillus polymyxa Npr protein (Takekawa et al. (1991) Journal of Bacteriology, 173 (21): 6820-6825). The predicted mature region of PspPro2 is shown in SEQ ID NO: 8.
[0344] The nucleotide sequence of the PspPro2 gene isolated from Paenibacillus sp. is set forth as SEQ ID NO: 6. The sequence encoding the predicted native signal peptide is shown in italics:
TABLE-US-00013 ATGAAAAAAGTATGGGTTTCACTTCTTGGAGGAGCGATGTTATTAGGGG CTGTAGCACCAGGTGCATCAGCAGCAGAGCATTCTGTTCCTGATCCTAC TCAGCTAACACCGACCTTTCACGCCGAGCAATGGAAGGCTCCTTCCACG GTAACCGGCGACAATATTGTATGGAGCTATTTGAATCGACAAAAGAAAA CCTTATTGAATACAGACAGCACCAGTGTGCGTGATCAGTTCCGCATCAT TGATCGTACAAGCGACAAATCCGGTGCAAGCCATTATCGGCTCAAGCAA TATGTAAACGGGATCCCCGTATATGGGGCTGAACAGACCATTCATGTGA ACAACGCCGGTAAAGTAACCTCTTATTTGGGTGCTGTCATTTCAGAGGA TCAGCAGCAAGACGCGACCGAAGATACCACTCCAAAAATCAGCGCGACT GAAGCCGTTTATACCGCATATGCAGAAGCCGCTGCCCGGATTCAATCCT TCCCTTCCATCAATGATAGTCTTTCTGAGGCTAGTGAGGAACAAGGGAG TGAGAATCAAGGCAATGAGATTCAAAACATTGGGATTAAAAGCAGTGTA AGTAATGACACTTACGCAGAGGCGCATAACAACGTACTTTTAACCCCCG TTGACCAAGCAGAGCAAAGTTACATTGCCAAAATTGCTAATCTGGAGCC AAGTGTAGAGCCCAAAGCAGAATTATACATCTATCCAGATGGTGAGACT ACACGACTGGTTTATGTAACAGAGGTTAATATTCTTGAACCTGCGCCTC TGCGCACACGCTACTTCATTGATGCGAAAACCGGCAAAATCGTATTCCA GTATGACATCCTCAACCACGCAACAGGCACCGGCCGCGGCGTGGATGGC AAAACAAAATCATTTACGACTACAGCTTCAGGCAACCGGTATCAGTTGA AAGACACGACTCGCAGCAATGGAATCGTGACTTACACCGCTGGCAATCG CCAGACGACGCCAGGTACGATTTTGACCGATACAGATAATGTATGGGAG GACCCTGCGGCTGTTGATGCCCATGCCTACGCCATTAAAACCTATGACT ATTATAAGAATAAATTCGGTCGCGACAGTATTGATGGACGTGGCATGCA AATTCGTTCGACAGTCCATTACGGCAAAAAATATAACAATGCCTTCTGG AACGGCTCGCAAATGACCTACGGAGACGGAGACGGGTCCACATTTACCT TCTTCAGCGGCGATCCCGATGTCGTGGGGCATGAGCTCACCCACGGCGT CACCGAGTTCACCTCCAATTTGGAGTATTATGGTGAGTCCGGTGCATTG AACGAAGCCTTCTCGGATATTATCGGTAATGATATAGATGGCACCAGTT GGCTTCTTGGCGACGGCATTTATACGCCTAATATTCCAGGCGACGCTCT GCGTTCCCTGTCCGATCCTACACGATTCGGCCAGCCGGATCACTACTCC AATTTCTATCCGGACCCCAACAATGATGATGAAGGCGGAGTCCATACGA ACAGCGGTATTATCAACAAAGCCTATTATTTGCTGGCACAAGGCGGTAC GTCCCATGGTGTAACGGTAACTGGTATCGGACGCGAAGCGGCTGTATTC ATTTACTACAATGCCTTTACCAACTATTTGACCTCTACCTCCAACTTCT CTAACGCACGCGCTGCTGTTATACAGGCAGCCAAGGATTTTTATGGTGC TGATTCGCTGGCAGTAACCAGTGCTATTCAATCCTTTGATGCGGTAGGA ATCAAA
[0345] The amino acid sequence of the PspPro2 precursor protein is set forth as SEQ ID NO: 7. The predicted signal peptide is shown in italics, and the predicted pro-peptide is shown in underlined text:
TABLE-US-00014 MKKVWVSLLGGAMLLGAVAPGASAAEHSVPDPTQLTPTFHAEQWKAPSTV TGDNIVWSYLNRQKKTLLNTDSTSVRDQFRIIDRTSDKSGASHYRLKQYV NGIPVYGAEQTIHVNNAGKVTSYLGAVISEDQQQDATEDTTPKISATEAV YTAYAEAAARIQSFPSINDSLSEASEEQGSENQGNEIQNIGIKSSVSNDT YAEAHNNVLLTPVDQAEQSYIAKIANLEPSVEPKAELYIYPDGETTRLVY VTEVNILEPAPLRTRYFIDAKTGKIVFQYDILNHATGTGRGVDGKTKSFT TTASGNRYQLKDTTRSNGIVTYTAGNRQTTPGTILTDTDNVWEDPAAVDA HAYAIKTYDYYKNKFGRDSIDGRGMQIRSTVHYGKKYNNAFWNGSQMTYG DGDGSTFTFFSGDPDVVGHELTHGVTEFTSNLEYYGESGALNEAFSDIIG NDIDGTSWLLGDGIYTPNIPGDALRSLSDPTRFGQPDHYSNFYPDPNNDD EGGVHTNSGIINKAYYLLAQGGTSHGVTVTGIGREAAVFIYYNAFTNYLT STSNFSNARAAVIQAAKDFYGADSLAVTSAIQSFDAVGIK
[0346] The amino acid sequence of the predicted mature form of PspPro2 is set forth as SEQ ID NO: 8.
TABLE-US-00015 ATGTGRGVDGKTKSFTTTASGNRYQLKDTTRSNGIVTYTAGNRQTTPGTI LTDTDNVWEDPAAVDAHAYAIKTYDYYKNKFGRDSIDGRGMQIRSTVHYG KKYNNAFWNGSQMTYGDGDGSTFTFFSGDPDVVGHELTHGVTEFTSNLEY YGESGALNEAFSDIIGNDIDGTSWLLGDGIYTPNIPGDALRSLSDPTRFG QPDHYSNFYPDPNNDDEGGVHTNSGIINKAYYLLAQGGTSHGVTVTGIGR EAAVFIYYNAFTNYLTSTSNFSNARAAVIQAAKDFYGADSLAVTSAIQSF DAVGIK
Example 2.2
Expression of Paenibacillus sp. Metalloprotease PspPro2
[0347] The DNA sequence of the propeptide-mature form of PspPro2 was synthesized and inserted into the Bacillus subtilis expression vector p2JM103BBI (Vogtentanz, Protein Expr Purif, 55:40-52, 2007) by Generay (Shanghai, China), resulting in plasmid pGX084 (AprE-PspPro2) (FIG. 2.1). Ligation of this gene encoding the PspPro2 protein into the digested vector resulted in the addition of three codons (Ala-Gly-Lys) between the 3' end of the Bacillus subtilis AprE signal sequence and the 5' end of the predicted PspPro2 native propeptide. The gene has an alternative start codon (GTG). As shown in FIG. 2.1, pGX084(AprE-PspPro2) contains an AprE promoter, an AprE signal sequence used to direct target protein secretion in B. subtilis, and the synthetic nucleotide sequence encoding the predicted propeptide and mature regions of PspPro2, (SEQ ID NO: 9). The translation product of the synthetic AprE-PspPro2 gene is shown in SEQ ID NO: 10.
[0348] The pGX084(AprE-PspPro2) plasmid was transformed into B. subtilis cells (degU.sup.Hy 32, .DELTA.scoC) and the transformed cells were spread on Luria Agar plates supplemented with 5 ppm chloramphenicol and 1.2% skim milk (Cat #232100, Difco). Colonies with the largest clear halos on the plates were selected and subjected to fermentation in a 250 ml shake flask with MBD medium (a MOPS based defined medium, supplemented with additional 5 mM CaCl.sub.2).
[0349] The broth from the shake flasks was concentrated and buffer-exchanged into the loading buffer containing 20 mM Tris-HCl (pH 8.5) and 1 mM CaCl.sub.2 using a VivaFlow 200 ultra filtration device (Sartorius Stedim). After filtering, this sample was applied to a 150 ml Q Sepharose High Performance column pre-equilibrated with the loading buffer above, PspPro2 was eluted from the column with a linear salt gradient from 0 to 0.5 M NaCl in the loading buffer. The corresponding active fractions were collected, concentrated and buffer-exchanged again into the loading buffer described above. The sample was loaded onto a 20 ml DEAE Fast Flow column pre-equilibrated with the same loading buffer. PspPro2 was eluted from the column with a linear salt gradient from 0 to 0.3 M NaCl in the loading buffer. The corresponding active purified protein fractions were further pooled and concentrated via 10K Amicon Ultra for further analyses. The nucleotide sequence of the synthesized PspPro2 gene in plasmid pGX084 (AprE-PspPro2) is depicted in SEQ ID NO: 9. The sequence encoding the predicted native signal peptide is shown in italics and the oligo-nucleotide encoding the three residue addition (AGK) is shown in bold:
TABLE-US-00016 GTGAGAAGCAAAAAATTGTGGATCAGCTTGTTGTTTGCGTTAACGTTAA TCTTTACGATGGCGTTCAGCAACATGAGCGCGCAGGCTGCTGGAAAAGC AGAGCATTCAGTTCCTGACCCGACGCAACTTACACCGACATTTCATGCT GAGCAGTGGAAGGCACCGAGCACGGTCACGGGCGACAACATCGTGTGGA GCTACCTGAACAGACAGAAAAAGACGCTGCTGAACACGGACTCAACGAG CGTGAGAGACCAGTTCAGAATCATCGACAGAACGAGCGACAAGTCAGGC GCGTCACATTATAGACTGAAGCAGTACGTGAACGGCATCCCGGTCTACG GAGCCGAGCAAACGATCCATGTGAATAATGCGGGCAAAGTTACATCATA CCTGGGCGCCGTCATCTCAGAAGACCAGCAGCAAGATGCAACGGAGGAT ACAACACCGAAGATCAGCGCCACAGAAGCGGTCTATACGGCTTACGCCG AAGCGGCTGCAAGAATCCAGAGCTTCCCGTCAATTAATGACAGCCTGAG CGAAGCATCAGAGGAACAAGGCAGCGAGAACCAGGGCAATGAAATCCAA AACATCGGCATCAAGAGCAGCGTGTCAAACGACACGTATGCGGAGGCTC ATAACAACGTTCTGCTGACACCGGTCGATCAGGCCGAACAGAGCTATAT TGCAAAGATCGCGAATCTGGAGCCGTCAGTCGAGCCGAAGGCCGAGCTG TATATCTATCCGGACGGCGAGACGACGAGACTGGTGTACGTTACGGAGG TCAACATCCTTGAGCCTGCGCCGCTGAGAACAAGATACTTTATCGACGC CAAGACGGGCAAGATCGTGTTTCAGTACGATATCCTGAACCATGCGACG GGAACAGGCAGAGGCGTGGACGGCAAAACAAAATCATTCACGACAACGG CAAGCGGCAACAGATACCAGCTGAAGGACACAACAAGATCAAATGGCAT CGTCACATACACGGCCGGAAATAGACAGACGACGCCGGGAACGATTCTG ACGGATACAGATAACGTGTGGGAAGATCCGGCAGCAGTTGATGCACATG CATACGCGATCAAGACGTACGACTACTACAAGAACAAATTCGGAAGAGA TTCAATCGATGGAAGAGGCATGCAAATCAGATCAACGGTTCATTATGGC AAAAAGTACAACAATGCCTTCTGGAACGGCAGCCAAATGACATACGGCG ATGGAGACGGCTCAACGTTTACATTCTTTTCAGGCGACCCGGACGTCGT CGGCCATGAACTGACGCATGGCGTTACAGAGTTCACGAGCAACCTGGAG TATTACGGCGAATCAGGCGCACTGAATGAGGCTTTCAGCGACATCATTG GCAACGACATTGATGGCACATCATGGCTGCTTGGCGACGGCATTTACAC ACCTAACATTCCGGGCGATGCACTGAGAAGCCTGTCAGACCCTACGAGA TTCGGCCAACCTGACCATTACAGCAACTTCTACCCGGATCCTAATAACG ATGATGAGGGCGGAGTGCATACGAACAGCGGCATTATCAACAAAGCGTA CTATCTGCTGGCACAAGGCGGAACGTCACATGGAGTGACGGTGACAGGA ATCGGCAGAGAGGCGGCAGTGTTTATCTACTACAACGCCTTCACAAACT ACCTGACGAGCACGTCAAATTTCAGCAACGCTAGAGCGGCGGTCATCCA GGCAGCAAAGGACTTTTATGGAGCAGACTCACTGGCAGTTACGTCAGCA ATTCAGTCATTCGACGCAGTTGGAATTAAG
[0350] The amino acid sequence of the PspPro2 precursor protein expressed from plasmid pGX084(AprE-PspPro2) is depicted in SEQ ID NO: 10. The predicted signal sequence is shown in italics, the three residue addition (AGK) is shown in bold, and the predicted pro-peptide is shown in underlined text:
TABLE-US-00017 MRSKKLWISLLFALTLIFTMAFS1VMSAQAAGKAEHSVPDPTQLTPTFH AEQWKAPSTVTGDNIVWSYLNRQKKTLLNTDSTSVRDQFRIIDRTSDKS GASHYRLKQYVNGIPVYGAEQTIHVNNAGKVTSYLGAVISEDQQQDATE DTTPKISATEAVYTAYAEAAARIQSFPSINDSLSEASEEQGSENQGNEI QNIGIKSSVSNDTYAEAHNNVLLTPVDQAEQSYIAKIANLEPSVEPKAE LYIYPDGETTRLVYVTEVNILEPAPLRTRYFIDAKTGKIVFQYDILNHA TGTGRGVDGKTKSFTTTASGNRYQLKDTTRSNGIVTYTAGNRQTTPGTI LTDTDNVWEDPAAVDAHAYAIKTYDYYKNKFGRDSIDGRGMQIRSTVHY GKKYNNAFWNGSQMTYGDGDGSTFTFFSGDPDVVGHELTHGVTEFTSNL EYYGESGALNEAFSDIIGNDIDGTSWLLGDGIYTPNIPGDALRSLSDPT RFGQPDHYSNFYPDPNNDDEGGVHTNSGIINKAYYLLAQGGTSHGVTVT GIGREAAVFIYYNAFTNYLTSTSNFSNARAAVIQAAKDFYGADSLAVTS AIQSFDAVGIK
[0351] The amino acid sequence of the recombinant PspPro2 protein isolated from Bacillus subtilis culture was determined by tandem mass spectrometry, and shown below. It is the same as predicted and depicted in SEQ ID NO: 8.
TABLE-US-00018 ATGTGRGVDGKTKSFTTTASGNRYQLKDTTRSNGIVTYTAGNRQTTPGTI LTDTDNVWEDPAAVDAHAYAIKTYDYYKNKFGRDSIDGRGMQIRSTVHYG KKYNNAFWNGSQMTYGDGDGSTFTFFSGDPDVVGHELTHGVTEFTSNLEY YGESGALNEAFSDIIGNDIDGTSWLLGDGIYTPNIPGDALRSLSDPTRFG QPDHYSNFYPDPNNDDEGGVHTNSGIINKAYYLLAQGGTSHGVTVTGIGR EAAVFIYYNAFTNYLTSTSNFSNARAAVIQAAKDFYGADSLAVTSAIQSF DAVGIK
Example 2.3
Proteolytic Activity of Metalloprotease PspPro2
[0352] The proteolytic activity of purified PspPro2 was measured in 50 mM Tris (pH 7), using azo-casein (Cat #74H7165, Megazyme) as a substrate. Prior to the reaction, the enzyme was diluted with Milli-Q water (Millipore) to specific concentrations. The azo-casein was dissolved in 100 mM Tris buffer (pH 7) to a final concentration of 1.5% (w/v). To initiate the reaction, 50 .mu.l of the diluted enzyme (or Milli-Q H.sub.2O alone as the blank control) was added to the non-binding 96-well Microtiter Plate (96-MTP) (Corning Life Sciences, #3641) placed on ice, followed by the addition of 50 .mu.l of 1.5% azo-casein. After sealing the 96-MTP, the reaction was carried out in a Thermomixer (Eppendorf) at 40.degree. C. and 650 rpm for 10 min. The reaction was terminated by adding 100 .mu.l of 5% Trichloroacetic Acid (TCA). Following equilibration (5 min at the room temperature) and subsequent centrifugation (2000 g for 10 min at 4.degree. C.), 120 .mu.l supernatant was transferred to a new 96-MTP, and absorbance of the supernatant was measured at 440 nm (A.sub.440) using a SpectraMax 190. Net A.sub.440 was calculated by subtracting the A.sub.440 of the blank control from that of enzyme, and then plotted against different protein concentrations (from 1.25 ppm to 40 ppm). Each value was the mean of duplicate assays, and the value varies no more than 5%. The proteolytic activity is shown as Net A.sub.440. The proteolytic assays with azo-casein as the substrate (FIG. 2.2) indicate that PspPro2 is an active protease.
Example 2.4
pH Profile of Metalloprotease PspPro2
[0353] With azo-casein as the substrate, the pH profile of PspPro2 was studied in 12.5 mM acetate/Bis-Tris/HEPES/CHES buffer with different pH values (ranging from pH 4 to 11). To initiate the assay, 50 .mu.l of 25 mM acetate/Bis-Tris/HEPES/CHES buffer with a specific pH was first mixed with 2 .mu.l diluted enzyme (500 ppm in Milli-Q H.sub.2O) in a 96-MTP placed on ice, followed by the addition of 48 .mu.l of 1.5% (w/v) azo-casein prepared in H2O. The reaction was performed and analyzed as described in Example 2.3. Enzyme activity at each pH was reported as the relative activity, where the activity at the optimal pH was set to be 100%. The pH values tested were 4, 5, 5.5, 6, 6.5, 7, 7.5, 8, 8.5, 9, 10 and 11. Each result was the mean of triplicate assays. As shown in FIG. 2.3, the optimal pH of PspPro2 is 7.5 with greater than 70% of its maximal activity retained between pH 5.5 and 9.5.
Example 2.5
Temperature Profile of Metalloprotease PspPro2
[0354] The temperature profile of PspPro2 was analyzed in 50 mM Tris buffer (pH 7) using the azo-casein assay. The enzyme sample and azo-casein substrate were prepared as in Example 2.3.
[0355] Prior to the reaction, 50 .mu.l of 1.5% azo-casein and 45 .mu.l Milli-Q H.sub.2O were mixed in a 200 .mu.l PCR tube, which was then subsequently incubated in a Peltier Thermal Cycler (BioRad) at desired temperatures (i.e. 20-90.degree. C.) for 5 min. After the incubation, 5 .mu.l of diluted PspPro2 (200 ppm) or H.sub.2O (the blank control) was added to the substrate mixture, and the reaction was carried out in the Peltier Thermal Cycle for 10 min at different temperatures. To terminate the reaction, each assay mixture was transferred to a 96-MTP containing 100 .mu.l of 5% TCA per well. Subsequent centrifugation and absorbance measurement were performed as described in Example 3. The activity was reported as the relative activity, where the activity at the optimal temperature was set to be 100%. The tested temperatures are 20, 30, 40, 50, 60, 70, 80 and 90.degree. C. Each result was the mean of triplicate assays. The data in FIG. 2.4 suggest that PspPro2 showed an optimal temperature at 50.degree. C., and retained greater than 70% of its maximal activity between 40 and 65.degree. C.
Example 2.6
Cleaning Performance of Metalloprotease PspPro2 in Automatic Dishwashing (ADW) Conditions
[0356] The cleaning performance of PspPro2 protein in automatic dishwashing (ADW) conditions was tested using PA-S-38 (egg yolk, with pigment, aged by heating) microswatches (CFT-Vlaardingen, The Netherlands) at pH 6 and 8 using a model automatic dishwashing (ADW) detergent. Prior to the reaction, purified PspPro2 protein samples were diluted with the dilution solution containing 10 mM NaCl, 0.1 mM CaCl.sub.2, 0.005% TWEEN.RTM. 80 and 10% propylene glycol to the desired concentrations. The reactions were performed in AT detergent with 100 ppm water hardness (Ca.sup.2+:Mg.sup.2+=3:1), in the presence of a bleach component (Peracid N,N-phthaloylaminoperoxycaproic acid-PAP) (AT detergent composition shown in Table 1). To initiate the reaction, 180 .mu.l of the AT detergent solution at pH 6 or pH 8 was added to a 96-MTP placed with PA-S-38 microswatches, followed by the addition of 20 .mu.l of diluted enzymes (or the dilution solution as the blank control). The 96-MTP was sealed and incubated in an incubator/shaker for 30 min at 50.degree. C. and 1150 rpm. After incubation, 100 .mu.l of wash liquid from each well was transferred to a new 96-MTP, and its absorbance was measured at 405 nm (referred here as the "Initial performance") using a spectrophotometer. The remaining wash liquid in the 96-MTP was discarded and the microswatches were rinsed once with 200 .mu.l water. Following the addition of 180 .mu.l of 0.1 M CAPS buffer (pH 10), the second incubation was carried out in the incubator/shaker at 50.degree. C. and 1150 rpm for 10 min. One hundred microliters of the resulting wash liquid was transferred to a new 96-MTP, and its absorbance was measured at 405 nm (referred here as "Wash-off"). The sum of two absorbance measurements ("Initial performance" plus "Wash-off") gives the "Total performance", which measures the protease activity on the model stain. Dose response for cleaning of PA-S-38 microswatches at pH 6 and pH 8 for PspPro2 in AT detergent in the presence of bleach, is shown in FIGS. 2.5A and 2.5B, respectively.
TABLE-US-00019 TABLE 2.1 Composition of AT dish detergent with bleach Ingredient Concentration (mg/ml) MGDA (methylglycinediacetic acid) 0.143 Sodium citrate 1.86 Citric acid* varies PAP (peracid N, N-phthaloylaminoperoxycaproic acid) 0.057 Plurafac .RTM. LF 18B (a non-ionic surfactant) 0.029 Bismuthcitrate 0.006 Bayhibit .RTM. S (Phosphonobutantricarboxylic acid sodium salt) 0.006 Acusol .TM. 587 (a calcium polyphosphate inhibitor) 0.029 PEG 6000 0.043 PEG 1500 0.1 *The pH of the AT formula detergent is adjusted to the desired pH value (pH 6 or 8) by the addition of 0.9 M citric acid.
Example 2.7
Cleaning Performance of Metalloprotease PspPro2 in Laundry Conditions
A. Cleaning Performance in Liquid Laundry Detergent
[0357] The cleaning performance of PspPro2 protein in liquid laundry detergent was tested using EMPA-116 (cotton soiled with blood/milk/ink) microswatches (obtained from CFT Vlaardingen, The Netherlands) at pH 8.2 using a commercial detergent. Prior to the reaction, purified PspPro2 protein samples were diluted with a dilution solution (10 mM NaCl, 0.1 mM CaCl.sub.2), 0.005% TWEEN.RTM. 80 and 10% propylene glycol) to the desired concentrations; and the commercial detergent (Tide.RTM., Clean Breeze.RTM., Proctor & Gamble, USA, purchased September 2011) was incubated at 95.degree. C. for 1 hour to inactivate the enzymes present in the detergent. Proteolytic assays were subsequently performed to confirm the inactivation of proteases in the commercial detergent. The heat treated detergent was further diluted with 5 mM HEPES (pH 8.2) to a final concentration of 0.788 g/L. Meanwhile, the water hardness of the buffered liquid detergent was adjusted to 103 ppm (Ca.sup.2+:Mg.sup.2+=3:1). The specific conductivity of the buffered detergent was adjusted to either 0.62 mS/cm (low conductivity) or 3.5 mS/cm (high conductivity) by adjusting the NaCl concentration in the buffered detergent. To initiate the reaction, 190 .mu.l of either the high or low conductivity buffered detergent was added to a 96-MTP containing the EMPA-116 microswatches, followed by the addition of 10 .mu.l of diluted enzyme (or the dilution solution as blank control). The 96-MTP was sealed and incubated in an incubator/shaker for 20 min at 32.degree. C. and 1150 rpm. After incubation, 150 .mu.l of wash liquid from each well was transferred to a new 96-MTP, and its absorbance was measured at 600 nm using a spectrophotometer, which indicates the protease activity on the model stain; and Net A.sub.600 was subsequently calculated by subtracting the A.sub.600 of the blank control from that of the enzyme. Dose response for the cleaning of EMPA-116 microswatches in liquid laundry detergent at high or low conductivity is shown in FIG. 2.6A.
B. Cleaning Performance in Powder Laundry Detergent
[0358] The cleaning performance of PspPro2 protein in powder laundry detergent was tested using PA-S-38 (egg yolk, with pigment, aged by heating) microswatches (CFT-Vlaardingen, The Netherlands) using a commercial detergent. Prior to the reaction, purified PspPro2 protein samples were diluted with a dilution solution (10 mM NaCl, 0.1 mM CaCl.sub.2, 0.005% TWEEN.RTM. 80 and 10% propylene glycol) to the desired concentrations. The powder laundry detergent (Tide.RTM., Bleach Free, Proctor & Gamble, China, purchased in December 2011) was dissolved in water with 103 ppm water hardness (Ca.sup.2+:Mg.sup.2+=3:1) to a final concentration of 2 g/L (with conductivity of 2.3 mS/cm-low conductivity) or 5 g/L (with conductivity of 5.5 mS/cm-high conductivity). The detergents of different conductivities were subsequently heated in a microwave to near boiling in order to inactivate the enzymes present in the detergent. Proteolytic activity was measured following treatment to ensure that proteases in the commercial detergent had been inactivated. To initiate the reaction, 190 .mu.l of either the high or low conductivity heat-treated detergent was added to a 96-MTP containing the PA-S-38 microswatches, followed by the addition of 10 .mu.l of diluted enzyme (or the dilution solution as blank control). The 96-MTP was sealed and incubated in an incubator/shaker for 15 minutes at 32.degree. C. and 1150 rpm. After incubation, 150 .mu.l of wash liquid from each well was transferred to a new 96-MTP, and its absorbance was measured at 405 nm using a spectrophotometer, which indicates the protease activity on the model stain; and Net A.sub.405 was subsequently calculated by subtracting the A.sub.405 of the blank control from that of the enzyme. Dose response for the cleaning of PA-S-38 microswatches in powder laundry detergent at high or low conductivity is shown in FIG. 2.6B.
Example 2.8
Comparison of PspPro2 to Other Metalloproteases
Identification of Homologous Proteases
[0359] Homologs were identified by a BLAST search (Altschul et al., Nucleic Acids Res, 25:3389-402, 1997) against the NCBI non-redundant protein database and the Genome Quest Patent database with search parameters set to default values. The mature protein amino acid sequence for PspPro2 (SEQ ID NO: 8) is used as query sequence. Percent identity (PID) for both search sets is defined as the number of identical residues divided by the number of aligned residues in the pairwise alignment. Tables 2.2A and 2.2B provide a list of sequences with the percent identity to PspPro2. The length in Table 2.2 refers to the entire sequence length of the homologous proteases.
TABLE-US-00020 TABLE 2.2A List of sequences with percent identity to PspPro2 protein identified from the NCBI non-redundant protein database PID to Accession # PspPro2 Organism Length AAB02774.1 55 Geobacillus stearothermophilus 552 AAA22623.1 56 Bacillus caldolyticus 544 P00800 56 Bacillus thermoproteolyticus 548 YP_003670279.1 57 Geobacillus sp. C56-T3 546 BAD60997.1 57 Bacillus megaterium 562 ZP_02326503.1 58 Paenibacillus larvae subsp. larvae BRL-230010 520 ZP_08640523.1 58 Brevibacillus laterosporus LMG 15441 564 YP_003597483.1 58 Bacillus megaterium DSM 319 562 ZP_09069025.1 59 Paenibacillus larvae subsp. larvae B-3650 520 YP_001373863.1 59 Bacillus cytotoxicus NVH 391-98 565 ZP_04149724.1 59 Bacillus pseudomycoides DSM 12442 566 CAA43589.1 60 Brevibacillus brevis 527 ZP_10738945.1 60 Brevibacillus sp. CF112 528 ZP_04216147.1 60 Bacillus cereus Rock3-44 566 ZP_10575942.1 61 Brevibacillus sp. BC25 528 YP_002770810.1 62 Brevibacillus brevis NBRC 100599 528 ZP_08511445.1 63 Paenibacillus sp. HGF7 525 ZP_09077634.1 64 Paenibacillus elgii B69 524 ZP_09071078.1 67 Paenibacillus larvae subsp. larvae B-3650 529 YP_003872180.1 73 Paenibacillus polymyxa E681 587 YP_005073223.1 78 Paenibacillus terrae HPL-003 591 ZP_09775364.1 78 Paenibacillus sp. Aloe-11 593 YP_003948511.1 80 Paenibacillus polymyxa SC2 592 YP_005073224.1 94 Paenibacillus terrae HPL-003 595 ZP_10241029.1 96 Paenibacillus peoriae KCTC 3763 599 ZP_09775365.1 100 Paenibacillus sp. Aloe-11 580
TABLE-US-00021 TABLE 2.2B List of sequences with percent identity to PspPro2 protein identified from the Genome Quest database PID to Patent # PspPro2 Organism Length JP2002272453-0002 57.01 Bacillus megaterium 562 US6518054-0001 57.19 Bacillus sp. 319 EP2390321-0177 57.19 Bacillus caldolyticus 544 US20120107907-0176 57.19 Bacillus stearothermophilis 548 WO9520663-0003 57.51 empty 319 WO2012110562-0003 57.51 Geobacillus stearothermophilus 319 WO2012110563-0002 57.51 Bacillus caldolyticus 319 WO2004011619-0056 57.51 empty 546 WO2004011619-0003 57.51 empty 546 JP2002272453-0003 57.64 empty 562 US6518054-0002 57.88 Bacillus sp. 316 EP2178896-0184 58.15 Bacillus anthracis 566 WO2012110563-0004 58.28 Bacillus megaterium 320 JP1995184649-0001 58.79 Lactobacillus sp. 566 JP1994014788-0003 58.84 empty 317 US8114656-0178 59.42 Bacillus thuringiensis 566 WO2012110562-0005 59.49 Bacillus cereus 320 US5962264-0004 59.81 empty 566 US20120107907-0185 59.81 Bacillus cereus 317 US20120107907-0179 59.81 Bacillus cereus 566 WO2012110563-0005 60.13 Bacillus cereus 320 EP2390321-0186 60.33 Bacillus brevis 304 JP2005229807-0018 78.62 Paenibacillus polymyxa 566 EP2390321-0187 79.21 Bacillus polymyxa 302
B. Alignment of Homologous Protease Sequences
[0360] The amino acid sequence of mature PspPro2 (SEQ ID NO: 8) was aligned with thermolysin (P00800, Bacillus thermoproteolyticus) and protease from Paenibacillus sp. Aloe-11 (ZP_09775365.1) sequences using CLUSTALW software (Thompson et al., Nucleic Acids Research, 22:4673-4680, 1994) with the default parameters. FIG. 2.7 shows the alignment of PspPro2 with these protease sequences.
C. Phylogenetic Tree
[0361] A phylogenetic tree for precursor PspPro2 protein sequence (SEQ ID NO: 7) was built using sequences of representative homologs from Table 2A and the Neighbor Joining method (NJ) (Saitou, N.; and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing Guide Trees. MolBiol.Evol. 4, 406-425). The NJ method works on a matrix of distances between all pairs of sequences to be analyzed. These distances are related to the degree of divergence between the sequences. The phylodendron-phylogenetic tree printer software (http://iubio.bio.indiana.edu/treeapp/treeprint-form.html) was used to display the phylogenetic tree shown in FIG. 2.8.
Example 3.1
Cloning of Paenibacillus humicus Metalloprotease PhuPro2
[0362] A strain (DSM18784) of Paenibacillus humicus was selected as a potential source of enzymes which may be useful in various industrial applications. Genomic DNA for sequencing was obtained by first growing the strain on Heart Infusion agar plates (Difco) at 37.degree. C. for 24 hr. Cell material was scraped from the plates and used to prepare genomic DNA with the ZF Fungal/Bacterial DNA miniprep kit from Zymo (Cat No. D6005). The genomic DNA was used for genome sequencing. The entire genome of the Paenibacillus humicus strain was sequenced by BaseClear (Leiden, The Netherlands) using the Illumina's next generation sequencing technology. After assembly of the data, contigs were annotated by BioXpr (Namur, Belgium). One of the genes identified after annotation in Paenibacillus humicus encodes a metalloprotease and the sequence of this gene, called PhuPro2, is provided in SEQ ID NO: 11. The corresponding protein encoded by the PhuPro2 gene is shown in SEQ ID NO: 12. At the N-terminus, the protein has a signal peptide with a length of 23 amino acids as predicted by SignalP version 4.0 (Nordahl Petersen et al. (2011) Nature Methods, 8:785-786). The presence of a signal sequence suggests that PhuPro2 is a secreted enzyme. The propeptide region was predicted based on its protein sequence alignment with the Paenibacillus polymyxa Npr protein (Takekawa et al. (1991) Journal of Bacteriology, 173 (21): 6820-6825). The predicted mature region of PhuPro2 protein in shown in SEQ ID NO: 13. The nucleotide sequence of the PhuPro2 gene isolated from Paenibacillus humicus is set forth as SEQ ID NO: 11. The sequence encoding the predicted native signal peptide is shown in italics:
TABLE-US-00022 ATGAAAAAAATGATTCCTACTCTGCTCGGTACCGTATTGCTGCTTTC TTCCGCTTCCGCTGTCGCTGCTGAATCGCCAAGCCTCGGAGCGGCCG GAACTCCCGGGGTCAGCGTCGTGAACAATCAGCTCGTGACTCAATTC ATCGAGGCTTCCAAGGATGCCAAGATTGTCCCGGGCTCTTCCGAGGA TAAAATCTGGGCTTTCCTTGAAGGCCAGCAAGCAAAGCTGGGTGTAT CCGCAGCGGATGTAAAAACCTCGTTCCTGATCCAGAAGAAGGAAGTC GATCCGACTTCGGGCGTCGAGCATTTCCGCCTGCAGCAATATGTGAA TGGCATCCCGGTATATGGCGGTGACCAAACCATTCACATCGACAAGG CCGGCCAGGTTACGTCGTTCGTAGGAGCTGTTCTGCCGGCTCAAAAT CAAATCACGGCAAAATCCAGCGTACCAGCCATAAGCGCATCCGACGC TCTGGCTATCGCGGCGAAGGAAGCCAGTTCCCGCATCGGCGAGCTGG GAGCACAGGAGAAGACTCCGTCGGCTCAGCTGTACGTATATCCGGAA GGCAACGGGTCGCGTCTCGTCTACCAGACGGAAGTGAATGTGCTTGA GCCGCAGCCTCTGCGCACCCGCTATCTTATCGATGCGGCCGACGGCC ATATCGTGCAGCAGTACGATCTGATCGAGACGGCGACCGGTTCGGGC ACGGGCGTGCTGGGCGACAATAAGACGTTCCAGACGACTCTTTCCGG CAGCACGTACCAGCTGAAAGACACCACTCGCGGCAACGGCATCTACA CCTACACAGCCAGCAATCGGACCACGATTCCGGGCACGCTGCTGACG GACGCCGACAACGTATGGACGGATGGAGCCGCCGTCGATGCCCATAC TTATGCCGGAAAAGTATATGATTTCTACAAAACGAAGTTCGGACGCA ACAGCCTCGACGGCAACGGCCTGCTGATCCGTTCCTCGGTCCACTAC AGCAGCAGGTACAACAATGCCTTCTGGAACGGCACCCAGATTGTATT CGGCGACGGCGACGGCTCGACGTTCATTCCGCTGTCGGGCGATCTCG ACGTGGTCGGCCATGAGCTGTCCCACGGAGTCATCGAGTACACGTCC AACCTTCAATACCTCAATGAATCCGGCGCGCTGAACGAGTCCTATGC CGACGTCCTCGGCAACTCGATCCAGGCGAAAAACTGGCTTATCGGCG ACGATGTCTATACGCCTGGCATCTCCGGAGATGCTCTCCGTTCCATG TCCAACCCGACGCTTTACGGGCAGCCGGACAACTATGCCAACCGCTA TACGGGATCTTCCGACAACGGCGGCGTTCATACGAACAGCGGCATCA CGAACAAAGCGTTCTACCTGCTCGCCCAAGGCGGCACCCAGAACGGC GTTACCGTCGCCGGCATCGGGCGCGACGCAGCCGTGAACATTTTCTA CAACACAGTGGCCTATTACCTTACTTCCACTTCCAACTTCGCCGCGG CGAAGAACGCCTCGATCCAGGCAGCCAAAGACCTGTACGGAACGGGC TCCTCTTATGTCACCTCGGTGACCAATGCATTCAGAGCCGTAGGCCT G
[0363] The amino acid sequence of the PhuPro2 precursor protein is set forth as SEQ ID NO: 12. The predicted signal sequence is shown in italics, and the predicted propeptide is shown in underlined text:
TABLE-US-00023 MKKMIPTLLGTVLLLSSASAVAAESPSLGAAGTPGVSVVNNQLVTQF IEASKDAKIVPGSSEDKIWAFLEGQQAKLGVSAADVKTSFLIQKKEV DPTSGVEHFRLQQYVNGIPVYGGDQTIHIDKAGQVTSFVGAVLPAQN QITAKSSVPAISASDALAIAAKEASSRIGELGAQEKTPSAQLYVYPE GNGSRLVYQTEVNVLEPQPLRTRYLIDAADGHIVQQYDLIETATGSG TGVLGDNKTFQTTLSGSTYQLKDTTRGNGIYTYTASNRTTIPGTLLT DADNVWTDGAAVDAHTYAGKVYDFYKTKFGRNSLDGNGLLIRSSVHY SSRYNNAFWNGTQIVFGDGDGSTFIPLSGDLDVVGHELSHGVIEYTS NLQYLNESGALNESYADVLGNSIQAKNWLIGDDVYTPGISGDALRSM SNPTLYGQPDNYANRYTGSSDNGGVHTNSGITNKAFYLLAQGGTQNG VTVAGIGRDAAVNIFYNTVAYYLTSTSNFAAAKNASIQAAKDLYGTG SSYVTSVTNAFRAVGL
The amino acid sequence of the predicted mature form of PhuPro2 is set forth as SEQ ID NO: 13:
TABLE-US-00024 ATGSGTGVLGDNKTFQTTLSGSTYQLKDTTRGNGIYTYTASNRTTIP GTLLTDADNVWTDGAAVDAHTYAGKVYDFYKTKFGRNSLDGNGLLIR SSVHYSSRYNNAFWNGTQIVFGDGDGSTFIPLSGDLDVVGHELSHGV IEYTSNLQYLNESGALNESYADVLGNSIQAKNWLIGDDVYTPGISGD ALRSMSNPTLYGQPDNYANRYTGSSDNGGVHTNSGITNKAFYLLAQG GTQNGVTVAGIGRDAAVNIFYNTVAYYLTSTSNFAAAKNASIQAAKD LYGTGSSYVTSVTNAFRAVGL
Example 3.2
Expression of Paenibacillus humicus s Metalloprotease PhuPro2
[0364] The DNA sequence of the propeptide-mature form of PhuPro2 was synthesized and inserted into the Bacillus subtilis expression vector p2JM103BBI (Vogtentanz, Protein Expr Purif, 55:40-52, 2007) by Generay (Shanghai, China), resulting in plasmid pGX150(AprE-PhuPro2) (FIG. 1). Ligation of this gene encoding the PhuPro2 protein into the digested vector resulted in the addition of three codons (Ala-Gly-Lys) between the 3' end of the B. subtilis AprE signal sequence and the 5' end of the predicted PhuPro2 native propeptide. The gene has an alternative start codon (GTG). The resulting plasmid shown in FIG. 1 was labeled pGX150(AprE-PhuPro2). As shown in FIG. 3.1, pGX150(AprE-PhuPro2) contains an AprE promoter, an AprE signal sequence used to direct target protein secretion in B. subtilis, and the synthetic nucleotide sequence encoding the predicted propeptide and mature regions of PhuPro2 (SEQ ID NO: 14). The translation product of the synthetic AprE-PhuPro2 gene is shown in SEQ ID NO: 15.
[0365] The pGX150 (AprE-PhuPro2) plasmid was then transformed into B. subtilis cells (degU.sup.Hy 32, .DELTA.scoC) and the transformed cells were spread on Luria Agar plates supplemented with 5 ppm Chloramphenicol and 1.2% skim milk (Cat #232100, Difco). Colonies with the largest clear halos on the plates were selected and subjected to fermentation in a 250 ml shake flask with MBD medium (a MOPS based defined medium, supplemented with additional 5 mM CaCl.sub.2).
[0366] The broth from the shake flasks was concentrated and buffer-exchanged into the loading buffer containing 20 mM Tris-HCl (pH 8.5), 1 mM CaCl.sub.2 and 10% propylene glycol using a VivaFlow 200 ultra filtration device (Sartorius Stedim). After filtering, this sample was applied to an 80 ml Q Sepharose High Performance column pre-equilibrated with the loading buffer above; and the active flow-through fractions were collected and concentrated. The sample was loaded onto a 320 ml Superdex 75 gel filtration column pre-equilibrated with the loading buffer described above containing 0.15 M NaCl. The corresponding active purified protein fractions were further pooled and concentrated via 10K Amicon Ultra for further analyses.
[0367] The nucleotide sequence of the synthesized PhuPro2 gene in plasmid pGX150(AprE-PhuPro2) is depicted in SEQ ID NO: 14. The sequence encoding the three residue addition (AGK) is shown in bold:
TABLE-US-00025 GTGAGAAGCAAAAAATTGTGGATCAGCTTGTTGTTTGCGTTAACGTT AATCTTTACGATGGCGTTCAGCAACATGAGCGCGCAGGCTGCTGGAA AAGAATCACCGAGCCTTGGCGCTGCAGGAACACCGGGCGTTAGCGTT GTGAATAACCAACTGGTCACGCAGTTCATCGAAGCATCAAAAGACGC GAAAATTGTCCCTGGATCAAGCGAAGATAAGATTTGGGCATTTCTGG AAGGCCAGCAAGCAAAGCTTGGCGTCTCAGCTGCCGACGTGAAGACG AGCTTCCTGATCCAGAAGAAGGAGGTTGACCCGACATCAGGCGTTGA GCACTTTAGACTGCAACAGTACGTCAACGGCATCCCGGTTTATGGAG GCGATCAAACAATCCATATTGATAAGGCAGGCCAGGTCACATCATTC GTCGGAGCTGTCCTGCCGGCTCAGAACCAAATTACAGCAAAATCATC AGTTCCGGCAATTTCAGCCTCAGACGCTCTGGCAATCGCTGCCAAGG AGGCAAGCTCAAGAATTGGCGAACTGGGCGCACAAGAAAAGACACCG AGCGCCCAACTTTATGTCTATCCGGAGGGCAACGGAAGCAGACTGGT GTACCAGACAGAGGTCAATGTTCTGGAGCCGCAACCGCTGAGAACGA GATACCTTATCGATGCTGCGGATGGCCACATTGTTCAGCAATACGAC CTGATTGAGACAGCAACAGGAAGCGGAACGGGCGTGCTGGGCGACAA CAAGACGTTTCAGACAACACTTAGCGGCAGCACGTACCAACTTAAGG ACACGACGAGAGGCAATGGCATTTACACGTACACGGCCTCAAACAGA ACGACAATCCCAGGCACACTGCTGACGGATGCAGACAATGTTTGGAC GGACGGCGCAGCAGTTGACGCACACACGTACGCCGGCAAGGTGTACG ACTTTTACAAGACGAAGTTCGGCAGAAACAGCCTTGATGGAAATGGA CTGCTGATCAGAAGCAGCGTCCACTACAGCAGCAGATACAATAACGC CTTCTGGAACGGCACACAAATCGTCTTTGGCGATGGAGACGGATCAA CATTCATCCCGCTGTCAGGCGACCTGGACGTTGTGGGCCACGAGCTG AGCCACGGCGTCATCGAGTACACGAGCAACCTGCAGTACCTGAATGA AAGCGGCGCACTGAACGAGTCATATGCTGATGTGCTTGGCAATAGCA TCCAGGCCAAGAACTGGCTTATCGGAGACGACGTCTACACACCTGGC ATCAGCGGCGATGCTCTGAGAAGCATGAGCAATCCTACACTTTACGG CCAACCGGACAACTACGCGAATAGATATACGGGCAGCAGCGACAATG GCGGCGTTCATACAAACTCAGGCATCACGAACAAGGCGTTCTACCTG CTGGCACAGGGAGGCACGCAAAACGGCGTTACAGTTGCGGGCATTGG CAGAGATGCGGCCGTCAACATCTTCTACAACACAGTCGCCTACTACC TGACGAGCACGTCAAACTTCGCAGCGGCAAAGAACGCATCAATTCAA GCAGCAAAGGATCTGTACGGAACAGGCAGCTCATATGTCACGTCAGT TACGAATGCGTTTAGAGCCGTCGGCCTTTAA
[0368] The amino acid sequence of the PhuPro2 precursor protein expressed from plasmid pGX150(AprE-PhuPro2) is depicted in SEQ ID NO: 15. The predicted signal sequence is shown in italics, the three residue addition (AGK) is shown in bold, and the predicted propeptide is shown in underlined text.
TABLE-US-00026 (SEQ ID NO: 15) MRSKKLWISLLFALTLIFTMAFSNMSAQAAGKESPSLGAAGTPGVSV VNNQLVTQFIEASKDAKIVPGSSEDKIWAFLEGQQAKLGVSAADVKT SFLIQKKEVDPTSGVEHFRLQQYVNGIPVYGGDQTIHIDKAGQVTSF VGAVLPAQNQITAKSSVPAISASDALAIAAKEASSRIGELGAQEKTP SAQLYVYPEGNGSRLVYQTEVNVLEPQPLRTRYLIDAADGHIVQQYD LIETATGSGTGVLGDNKTFQTTLSGSTYQLKDTTRGNGIYTYTASNR TTIPGTLLTDADNVWTDGAAVDAHTYAGKVYDFYKTKFGRNSLDGNG LLIRSSVHYSSRYNNAFWNGTQIVFGDGDGSTFIPLSGDLDVVGHEL SHGVIEYTSNLQYLNESGALNESYADVLGNSIQAKNWLIGDDVYTPG ISGDALRSMSNPTLYGQPDNYANRYTGSSDNGGVHTNSGITNKAFYL LAQGGTQNGVTVAGIGRDAAVNIFYNTVAYYLTSTSNFAAAKNASIQ AAKDLYGTGSSYVTSVTNAFRAVGL.
Example 3.3
Proteolytic Activity of Metalloprotease PhuPro2
[0369] The proteolytic activity of purified metalloprotease PhuPro2 was measured in 50 mM Tris (pH 7), using azo-casein (Cat #74H7165, Megazyme) as a substrate. Prior to the reaction, the enzyme was diluted with Milli-Q water (Millipore) to specific concentrations. The azo-casein was dissolved in 100 mM Tris buffer (pH 7) to a final concentration of 1.5% (w/v). To initiate the reaction, 50 .mu.l of the diluted enzyme (or Milli-Q H.sub.2O alone as the blank control) was added to the non-binding 96-well Microtiter Plate (96-MTP) (Corning Life Sciences, #3641) placed on ice, followed by the addition of 50 .mu.l of 1.5% azo-casein. After sealing the 96-MTP, the reaction was carried out in a Thermomixer (Eppendorf) at 40.degree. C. and 650 rpm for 10 min. The reaction was terminated by adding 100 .mu.l of 5% Trichloroacetic Acid (TCA). Following equilibration (5 min at the room temperature) and subsequent centrifugation (2000 g for 10 min at 4.degree. C.), 120 .mu.l supernatant was transferred to a new 96-MTP, and absorbance of the supernatant was measured at 440 nm (A.sub.440) using a SpectraMax 190. Net A.sub.440 was calculated by subtracting the A.sub.440 of the blank control from that of enzyme, and then plotted against different protein concentrations (from 1.25 ppm to 40 ppm). Each value was the mean of triplicate assays.
[0370] The proteolytic activity is shown as Net A.sub.440. The proteolytic assay with azo-casein as the substrate (shown in FIG. 3.2) indicates that PhuPro2 is an active protease.
Example 3.4
pH Profile of Metalloprotease PhuPro2
[0371] With azo-casein as the substrate, the pH profile of metalloprotease PhuPro2 was studied in 12.5 mM acetate/Bis-Tris/HEPES/CHES buffer with different pH values (ranging from pH 4 to 11). To initiate the assay, 50 .mu.l of 25 mM acetate/Bis-Tris/HEPES/CHES buffer with a specific pH was first mixed with 2 .mu.l Milli-Q H.sub.2O diluted enzyme (125 ppm) in a 96-MTP placed on ice, followed by the addition of 48 .mu.l of 1.5% (w/v) azo-casein prepared in H2O. The reaction was performed and analyzed as described in Example 3.3. Enzyme activity at each pH was reported as the relative activity, where the activity at the optimal pH was set to be 100%. The pH values tested were 4, 5, 6, 7, 8, 9, 10 and 11. Each value was the mean of triplicate assays. As shown in FIG. 3.3, the optimal pH of PhuPro2 is 6, with greater than 70% of maximal activity retained between 5.5 and 8.5.
Example 3.5
Temperature Profile of Metalloprotease PhuPro2
[0372] The temperature profile of metalloprotease PhuPro2 was analyzed in 50 mM Tris buffer (pH 7) using the azo-casein assays. The enzyme sample and azo-casein substrate were prepared as in Example 3.3. Prior to the reaction, 50 .mu.l of 1.5% azo-casein and 45 .mu.l Milli-Q H.sub.2O were mixed in a 200 .mu.l PCR tube, which was then subsequently incubated in a Peltier Thermal Cycler (BioRad) at desired temperatures (i.e. 20-90.degree. C.) for 5 min. After the incubation, 5 .mu.l of diluted enzyme (50 ppm) or H.sub.2O (the blank control) was added to the substrate mixture, and the reaction was carried out in the Peltier Thermal Cycle for 10 min at different temperatures. To terminate the reaction, each assay mixture was transferred to a 96-MTP containing 100 .mu.l of 5% TCA per well. Subsequent centrifugation and absorbance measurement were performed as described in Example 3.3. The activity was reported as the relative activity, where the activity at the optimal temperature was set to be 100%. The tested temperatures are 20, 30, 40, 50, 60, 70, 80, and 90.degree. C. Each value was the mean of duplicate assays (the value varies no more than 5%). The data in FIG. 3.4 suggests that PhuPro2 showed an optimal temperature at 50.degree. C., and retained greater than 70% of its maximum activity between 45 and 65.degree. C.
Example 3.6
Cleaning Performance of Metalloprotease PhuPro2
[0373] The cleaning performance of PhuPro2 was tested using PA-S-38 (egg yolk, with pigment, aged by heating) microswatches (CFT-Vlaardingen, The Netherlands) at pH 6 and 8 using a model automatic dishwashing (ADW) detergent. Prior to the reaction, purified protease samples were diluted with a dilution solution containing 10 mM NaCl, 0.1 mM CaCl.sub.2, 0.005% TWEEN.RTM. 80 and 10% propylene glycol to the desired concentrations. The reactions were performed in AT detergent with 100 ppm water hardness (Ca.sup.2+:Mg.sup.2+=3:1) (detergent composition shown in Table 3.1). To initiate the reaction, 180 .mu.l of the AT detergent buffered at pH 6 or pH 8 was added to a 96-MTP placed with PA-S-38 microswatches, followed by the addition of 20 .mu.l of diluted enzymes (or the dilution solution as the blank control). The 96-MTP was sealed and incubated in an incubator/shaker for 30 min at 50.degree. C. and 1150 rpm. After incubation, 100 .mu.l of wash liquid from each well was transferred to a new 96-MTP, and its absorbance was measured at 405 nm (referred here as the "Initial performance") using a spectrophotometer. The remaining wash liquid in the 96-MTP was discarded and the microswatches were rinsed once with 200 .mu.l water. Following the addition of 180 .mu.l of 0.1 M CAPS buffer (pH 10), the second incubation was carried out in the incubator/shaker at 50.degree. C. and 1150 rpm for 10 min. One hundred microliters of the resulting wash liquid was transferred to a new 96-MTP, and its absorbance measured at 405 nm (referred here as the "Wash-off"). The sum of two absorbance measurements ("Initial performance" plus "Wash-off") gives the "Total performance", which measures the protease activity on the model stain; and Net A.sub.405 was subsequently calculated by subtracting the A.sub.405 of the "Total performance" of the blank control from that of the enzyme. Dose response in cleaning the PA-S-38 microswatches at pH 6 and pH 8 in AT dish detergent for PhuPro2 is shown in FIGS. 3.5A and 3.5B.
TABLE-US-00027 TABLE 3.1 Composition of AT dish detergent Ingredient Concentration (mg/ml) MGDA (methylglycinediacetic acid) 0.143 Sodium citrate 1.86 Citric acid* varies Plurafac .RTM. LF 18B (a non-ionic surfactant) 0.029 Bismuthcitrate 0.006 Bayhibit .RTM. S (Phosphonobutantricarboxylic acid sodium salt) 0.006 Acusol .TM. 587 (a calcium polyphosphate inhibitor) 0.029 PEG 6000 0.043 PEG 1500 0.1 *The pH of the AT formula detergent is adjusted to the desired value (pH 6 or 8) by the addition of 0.9 M citric acid.
Example 3.7
Comparison of PhuPro2 to Other Proteases
A. Identification of Homologous Proteases
[0374] Homologs were identified by a BLAST search (Altschul et al., Nucleic Acids Res, 25:3389-402, 1997) against the NCBI non-redundant protein database and the Genome Quest Patent database with search parameters set to default values. The predicted mature protein amino acid sequence for PhuPro2 (SEQ ID NO: 13) is used as the query sequences. Percent identity (PID) for both search sets is defined as the number of identical residues divided by the number of aligned residues in the pairwise alignment. Tables 3.2A and 3.2B provide a list of sequences with the percent identity to PhuPro2. The length in Table 3.2 refers to the entire sequence length of the homologous proteases.
TABLE-US-00028 TABLE 3.2A List of sequences with percent identity to PhuPro2 protein identified from the NCBI non-redundant protein database PID to Accession # PhuPro2 Organism Length P00800 59 Bacillus thermoproteolyticus 548 YP_003872180.1 59 Paenibacillus polymyxa E681 587 ZP_10575942.1 59 Brevibacillus sp. BC25 528 ZP_02326602.1 60 Paenibacillus larvae subsp. larvae 520 BRL-230010 ADM87306.1 61 Bacillus megaterium 562 ZP_09069025.1 61 Paenibacillus larvae subsp. 520 larvae B-3650 ZP_09069194.1 62 Paenibacillus larvae subsp. 502 Larvae B-3650 ZP_10738945.1 63 Brevibacillus sp. CF112 528 ZP_08511445.1 64 Paenibacillus sp. HGF7 525 ZP_09077634.1 65 Paenibacillus elgii B69 524 ZP_09775365.1 65 Paenibacillus sp. Aloe-11 580 ZP_09775364.1 70 Paenibacillus sp. Aloe-11 593 P29148 71 Paenibacillus polymyxa 590 ZP_10241030.1 71 Paenibacillus peoriae KCTC 3763 593 ZP_09071078.1 71 Paenibacillus larvae subsp. 529 larvae B-3650 YP_003872179.1 72 Paenibacillus polymyxa E681 592 YP_005073223.1 72 Paenibacillus terrae HPL-003 591
TABLE-US-00029 TABLE 3.2B List of sequences with percent identity to PhuPro2 protein identified from the Genome Quest Patent database PID to Patent ID # PhuPro2 Organism Length US20090208474-0030 59.22 Bacillus thermoproteolyticus 316 JP2002272453-0002 59.42 Bacillus megaterium 562 JP2006124323-0003 59.55 Bacillus thermoproteolyticus 316 US8114656-0183 59.87 Bacillus stearothermophilis 316 JP1989027475-0001 59.87 Bacillus subtilis 316 US20120009651-0002 59.87 Geobacillus caldoproteolyticus 548 JP2002272453-0003 60.45 empty 562 WO2012110563-0004 60.77 Bacillus megaterium 320 EP2390321-0186 62.25 Bacillus brevis 304 JP2005229807-0018 71.85 Paenibacillus polymyxa 566 US8114656-0187 72.09 Bacillus polymyxa 302
B. Alignment of Homologous Protease Sequences
[0375] The amino acid sequence of predicted mature PhuPro2 (SEQ ID NO: 13) protein was aligned with thermolysin (P00800, Bacillus thermoproteolyticus), and protease from Paenibacillus terrae HPL-003 (YP 005073223.1) sequences using CLUSTALW software (Thompson et al., Nucleic Acids Research, 22:4673-4680, 1994) with the default parameters. FIG. 3.6 shows the alignment of PhuPro2 with these protease sequences.
C. Phylogenetic Tree
[0376] A phylogenetic tree for full length sequence of PhuPro2 (SEQ ID NO: 12) was built using sequences of representative homologs from Table 2A and the Neighbor Joining method (NJ) (Saitou, N.; and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing Guide Trees. MolBiol.Evol. 4, 406-425). The NJ method works on a matrix of distances between all pairs of sequences to be analyzed. These distances are related to the degree of divergence between the sequences. The phylodendron-phylogenetic tree printer software (http://iubio.bio.indiana.edu/treeapp/treeprint-form.html) was used to display the phylogenetic tree shown in FIG. 3.7.
Example 4.1
Cloning of Paenibacillus ehimensis Metalloprotease PehPro1
[0377] A strain (DSM11029) of Paenibacillus ehimensis was selected as a potential source of enzymes which may be useful in various industrial applications. Genomic DNA for sequencing was obtained by first growing the strain on Heart Infusion agar plates (Difco) at 37.degree. C. for 24 hr. Cell material was scraped from the plates and used to prepare genomic DNA with the ZF Fungal/Bacterial DNA miniprep kit from Zymo (Cat No. D6005). The genomic DNA was used for genome sequencing. The entire genome of the Paenibacillus ehimensis strain was sequenced by BaseClear (Leiden, The Netherlands) using the Illumina's next generation sequencing technology. After assembly of the data, contigs were annotated by BioXpr (Namur, Belgium). One of the genes identified after annotation in Paenibacillus ehimensis encodes a metalloprotease and the sequence of this gene, called PehPro1, is provided in SEQ ID NO: 16. The corresponding protein encoded by the PehPro1 gene is shown in SEQ ID NO: 17. At the N-terminus, the protein has a signal peptide with a length of 23 amino acids as predicted by SignalP version 4.0 (Nordahl Petersen et al. (2011) Nature Methods, 8:785-786). The presence of a signal sequence suggests that PehPro1 is a secreted enzyme. The propeptide region was predicted based on protein sequence alignment with the Paenibacillus polymyxa Npr protein (Takekawa et al. (1991) Journal of Bacteriology, 173 (21): 6820-6825). The predicted mature region of PehPro1 protein is shown in SEQ ID NO: 18.
[0378] The nucleotide sequence of the PehPro1 gene isolated from Paenibacillus ehimensis is set forth as SEQ ID NO: 16. The sequence encoding the predicted native signal peptide is shown in italics:
TABLE-US-00030 ATGTTAAAAGTATGGGCATCGATTATTACAGGAGCATTTTTGCTCGG GAGCGTGCAAGGGGTGCAAGCTGCTCCACAAGATCAAGCTGCTCCCT TCGGAGGATTCACCCCTCAATTGATTACCGGGGAAAGCTGGAGTGCG CCGCAAGGAGTATCGGGAGAGGAAAAAATCTGGAAGTATCTCGAATC CAAGCAGGAAAGCTTCCAAATCGGCCAAACCGTTGATCTGAAAAAGC AATTGAAAATTATCGGCCAAACGACCGACGAGAAAACGGGAACCACG CATTACCGTCTACAGCAGTATGTGGGAGGCGTCCCCGTATACGGCGG CGTACAAACGATCCATGTCAACAAAGAAGGACAAGTTACCTCGCTGA TCGGCAGCCTGCTTCCCGACCAGCAGCAGCAAGTTTCGAAAAGCTTG AATTCGCAAATCAGCGAAGCGCAAGCCATCGCCGTGGCCCAGAAAGA TACCGAGGCCGCCGTCGGCAAGCTGGGTGAACCGCAAAAGACACCGG AAGCGGATCTGTACGTTTATTTACACAACGGACAACCGGTCCTCGCT TATGTGACCGAGGTTAACGTTCTCGAACCGGAGGCAATCCGGACGCG CTACTTCATCAGCGCCGAAGACGGCAGCATTTTATTCAAGTACGACA TCCTCGCTCACGCTACAGGTACCGGAAAAGGCGTGCTCGGAGATACG AAATCGTTCACGACCACGCAATCCGGCTCCACTTATCAATTGAAGGA TACGACGCGCGGGCAAGGTATCGTCACTTACAGCGCTGGCAACCGGT CCTCTCTGCCGGGAACGCTGCTCACCAGCTCCAGCAATATTTGGAAC GACGGCGCGGCGGTCGATGCGCATGCCTATACCGCCAAAGTGTACGA TTACTATAAAAACAAATTTGGCCGCAACAGCATTGACGGCAACGGCT TCCAGCTTAAATCGACCGTGCACTATTCCTCCAGATACAACAACGCC TTCTGGAACGGTGTGCAAATGGTGTACGGCGACGGCGACGGCGTAAC CTTCATTCCGTTCTCCGCCGATCCGGACGTCATCGGCCACGAATTGA CCCACGGCGTTACGGAACATACGGCCGGCCTGGAATACTACGGCGAA TCCGGAGCGCTGAACGAATCGATCTCCGATATTATCGGCAACGCGAT CGACGGCAAAAACTGGCTGATCGGCGACTTGATTTATACGCCGAATA CTCCCGGGGACGCCCTCCGCTCTATGGAGAACCCCAAGCTGTATAAC CAACCCGACCGCTATCAAGACCGCTATACGGGACCTTCCGATAACGG CGGCGTGCATATTAACAGCGGTATCAACAACAAAGCCTTCTACCTGA TCGCCCAAGGCGGCACGCACTATGGCGTCACCGTGAACGGGATCGGA CGCGATGCGGCTGTGCAAATTTTCTATGACGCCCTCATCAATTACCT GACTCCAACTTCGAACTTCTCGGCGATGCGCGCAGCAGCCATTCAAG CGGCAACCGACCTGTACGGAGCGAATTCTTCTCAAGTAAACGCTGTC AAAAAAGCGTATACTGCCGTCGGCGTGAAC
[0379] The amino acid sequence of the PehPro1 precursor protein is set forth as SEQ ID NO: 17. The predicted signal sequence is shown in italics, and the predicted propeptide is shown in underlined text:
TABLE-US-00031 MLKVWASIITGAFLLGSVQGVQAAPQDQAAPFGGFTPQLITGESWSA PQGVSGEEKIWKYLESKQESFQIGQTVDLKKQLKIIGQTTDEKTGTT HYRLQQYVGGVPVYGGVQTIHVNKEGQVTSLIGSLLPDQQQQVSKSL NSQISEAQAIAVAQKDTEAAVGKLGEPQKTPEADLYVYLHNGQPVLA YVTEVNVLEPEAIRTRYFISAEDGSILFKYDILAHATGTGKGVLGDT KSFTTTQSGSTYQLKDTTRGQGIVTYSAGNRSSLPGTLLTSSSNIWN DGAAVDAHAYTAKVYDYYKNKFGRNSIDGNGFQLKSTVHYSSRYNNA FWNGVQMVYGDGDGVTFIPFSADPDVIGHELTHGVTEHTAGLEYYGE SGALNESISDIIGNAIDGKNWLIGDLIYTPNTPGDALRSMENPKLYN QPDRYQDRYTGPSDNGGVHINSGINNKAFYLIAQGGTHYGVTVNGIG RDAAVQIFYDALINYLTPTSNFSAMRAAAIQAATDLYGANSSQVNAV KKAYTAVGVN
[0380] The amino acid sequence of the predicted mature form of PehPro1 is set forth as SEQ ID NO: 18:
TABLE-US-00032 ATGTGKGVLGDTKSFTTTQSGSTYQLKDTTRGQGIVTYSAGNRSSLP GTLLTSSSNIWNDGAAVDAHAYTAKVYDYYKNKFGRNSIDGNGFQLK STVHYSSRYNNAFWNGVQMVYGDGDGVTFIPFSADPDVIGHELTHGV TEHTAGLEYYGESGALNESISDIIGNAIDGKNWLIGDLIYTPNTPGD ALRSMENPKLYNQPDRYQDRYTGPSDNGGVHINSGINNKAFYLIAQG GTHYGVTVNGIGRDAAVQIFYDALINYLTPTSNFSAMRAAAIQAATD LYGANSSQVNAVKKAYTAVGVN
Example 4.2
Expression of Paenibacillus ehimensis Metalloprotease PehPro1
[0381] The DNA sequence of the propeptide-mature form of PehPro1 was synthesized and inserted into the Bacillus subtilis expression vector p2JM103BBI (Vogtentanz, Protein Expr Purif, 55:40-52, 2007) by Generay (Shanghai, China), resulting in plasmid pGX148(AprE-PehPro1) (FIG. 4.1). Ligation of this gene encoding the PehPro1 protein into the digested vector resulted in the addition of three codons (Ala-Gly-Lys) between the 3' end of the B. subtilis AprE signal sequence and the 5' end of the predicted PehPro1 native propeptide. The gene has an alternative start codon (GTG). The resulting plasmid shown in FIG. 1 was labeled pGX148(AprE-PehPro1). As shown in FIG. 1, pGX148(AprE-PehPro1) contains an AprE promoter, an AprE signal sequence used to direct target protein secretion in B. subtilis, and the synthetic nucleotide sequence encoding the predicted propeptide and mature regions of PehPro1 (SEQ ID NO: 19). The translation product of the synthetic AprE-PehPro1 gene is shown in SEQ ID NO: 20.
[0382] The pGX148(AprE-PehPro1) plasmid was then transformed into B. subtilis cells (degU.sup.Hy 32, .DELTA.scoC) and the transformed cells were spread on Luria Agar plates supplemented with 5 ppm Chloramphenicol and 1.2% skim milk (Cat #232100, Difco). Colonies with the largest clear halos on the plates were selected and subjected to fermentation in a 250 ml shake flask with MBD medium (a MOPS based defined medium. supplemented with additional 5 mM CaCl.sub.2)).
[0383] The broth from the shake flasks was concentrated and buffer-exchanged into the loading buffer containing 20 mM Tris-HCl (pH 8.5), 1 mM CaCl.sub.2) and 10% propylene glycol using a VivaFlow 200 ultra filtration device (Sartorius Stedim). After filtering, this sample was applied to an 80 ml Q Sepharose High Performance column pre-equilibrated with the loading buffer above, PehPro1 was eluted from the column with a linear salt gradient from 0 to 0.3 M NaCl in the loading buffer. The corresponding active fractions were collected, concentrated and buffer-exchanged again into the loading buffer described above. The sample was loaded onto a 40 ml DEAE Fast Flow column pre-equilibrated with the same loading buffer. PehPro1 was eluted from the column with a linear salt gradient from 0 to 0.15 M NaCl in the loading buffer. The corresponding active purified protein fractions were further pooled and concentrated via 10K Amicon Ultra for further analyses.
[0384] The nucleotide sequence of the synthesized PehPro1 gene in plasmid pGX148(AprE-PehPro1) is depicted in SEQ ID NO: 19. The sequence encoding the three residue addition (AGK) is shown in bold:
TABLE-US-00033 GTGAGAAGCAAAAAATTGTGGATCAGCTTGTTGTTTGCGTTAACGTT AATCTTTACGATGGCGTTCAGCAACATGAGCGCGCAGGCTGCTGGAA AAGCACCTCAAGATCAGGCAGCACCTTTTGGAGGCTTTACACCGCAA CTTATCACAGGCGAATCATGGTCAGCACCGCAGGGCGTTTCAGGCGA GGAAAAGATCTGGAAGTACCTTGAGAGCAAGCAGGAGTCATTTCAAA TCGGCCAGACAGTCGACCTGAAAAAGCAACTGAAGATCATCGGCCAA ACAACGGACGAAAAGACGGGCACGACGCATTATAGACTGCAACAATA TGTTGGCGGCGTGCCGGTTTATGGAGGCGTGCAAACAATCCACGTGA ACAAGGAAGGACAGGTCACGTCACTGATCGGCAGCCTGCTGCCGGAT CAGCAGCAACAAGTCTCAAAGAGCCTGAACTCACAAATTAGCGAGGC ACAAGCGATTGCAGTTGCACAAAAGGACACGGAAGCAGCTGTCGGCA AGCTGGGCGAACCGCAAAAAACACCTGAGGCTGACCTTTACGTCTAC CTGCATAACGGCCAGCCGGTCCTTGCGTACGTTACGGAAGTTAACGT GCTGGAGCCGGAGGCCATCAGAACGAGATACTTCATTAGCGCGGAGG ATGGAAGCATTCTGTTTAAGTACGATATTCTTGCTCACGCGACAGGC ACAGGCAAGGGCGTCCTTGGCGACACAAAAAGCTTCACGACAACGCA GAGCGGATCAACGTACCAGCTGAAAGATACAACAAGAGGACAAGGCA TCGTTACGTATTCAGCGGGCAATAGATCAAGCCTGCCGGGCACACTG CTGACATCAAGCTCAAACATTTGGAATGACGGCGCAGCAGTTGATGC CCATGCGTACACAGCCAAGGTGTACGACTACTATAAGAACAAGTTTG GCAGAAATAGCATCGACGGAAATGGATTTCAACTTAAATCAACGGTG CACTACTCATCAAGATATAACAATGCGTTTTGGAACGGAGTGCAGAT GGTCTACGGAGACGGCGACGGCGTGACATTTATTCCGTTTAGCGCCG ACCCGGACGTGATTGGACATGAACTGACACATGGAGTGACAGAGCAT ACGGCGGGACTGGAATATTACGGCGAAAGCGGCGCACTGAACGAAAG CATCTCAGACATTATTGGAAACGCAATCGATGGCAAAAACTGGCTGA TTGGCGATCTGATTTATACGCCGAATACACCGGGCGATGCACTGAGA TCAATGGAGAATCCGAAGCTGTACAACCAACCGGACAGATACCAAGA TAGATACACAGGACCGTCAGACAACGGCGGAGTCCATATCAACAGCG GAATCAATAACAAAGCCTTTTACCTGATCGCCCAAGGCGGAACGCAC TATGGCGTTACAGTCAATGGCATCGGAAGAGATGCCGCAGTTCAGAT TTTCTATGACGCGCTGATCAACTATCTGACGCCTACAAGCAATTTCT CAGCAATGAGAGCCGCAGCAATCCAAGCAGCCACGGATCTGTATGGA GCCAATTCATCACAAGTTAATGCTGTTAAGAAGGCTTATACGGCAGT GGGAGTTAACTAA
[0385] The amino acid sequence of the PehPro1 precursor protein expressed from plasmid pGX148(AprE-PehPro1) is depicted in SEQ ID NO: 20. The predicted signal sequence is shown in italics, the three residue addition (AGK) is shown in bold, and the predicted pro-peptide is shown in underlined text.
TABLE-US-00034 MRSKKLWISLLFALTLIFTMAFSNMSAQAAGKAPQDQAAPFGGFTPQ LITGESWSAPQGVSGEEKIWKYLESKQESFQIGQTVDLKKQLKIIGQ TTDEKTGTTHYRLQQYVGGVPVYGGVQTIHVNKEGQVTSLIGSLLPD QQQQVSKSLNSQISEAQAIAAQKDTEAAVGKLGEPQKTPEADLYVYL HNGQPVLAYVTEVNVLEPEAIRTRYFISAEDGSILFKYDILAHATGT GKGVLGDTKSFTTTQSGSTYQLKDTTRGQGIVTYSAGNRSSLPGTLL TSSSNIWNDGAAVDAHAYTAKVYDYYKNKFGRNSIDGNGFQLKSTVH YSSRYNNAFWNGVQMVYGDGDGVTFIPFSADPDVIGHELTHGVTEHT AGLEYYGESGALNESISDIIGNAIDGKNWLIGDLIYTPNTPGDALRS MENPKLYNQPDRYQDRYTGPSDNGGVHINSGINNKAFYLIAQGGTHY GVTVNGIGRDAAVQIFYDALINYLTPTSNFSAMRAAAIQAATDLYGA NSSQVNAVKKAYTAVGVN.
Example 4.3
Proteolytic Activity of Metalloprotease PehPro1
[0386] The proteolytic activity of purified metalloprotease PehPro1 was measured in 50 mM Tris (pH 7), using azo-casein (Cat #74H7165, Megazyme) as a substrate. Prior to the reaction, the enzyme was diluted with Milli-Q water (Millipore) to specific concentrations. The azo-casein was dissolved in 100 mM Tris buffer (pH 7) to a final concentration of 1.5% (w/v). To initiate the reaction, 50 .mu.l of the diluted enzyme (or Milli-Q H.sub.2O alone as the blank control) was added to the non-binding 96-well Microtiter Plate (96-MTP) (Corning Life Sciences, #3641) placed on ice, followed by the addition of 50 .mu.l of 1.5% azo-casein. After sealing the 96-MTP, the reaction was carried out in a Thermomixer (Eppendorf) at 40.degree. C. and 650 rpm for 10 min. The reaction was terminated by adding 100 .mu.l of 5% Trichloroacetic Acid (TCA). Following equilibration (5 min at the room temperature) and subsequent centrifugation (2000 g for 10 min at 4.degree. C.), 120 .mu.l supernatant was transferred to a new 96-MTP, and absorbance of the supernatant was measured at 440 nm (A.sub.440) using a SpectraMax 190. Net A.sub.440 was calculated by subtracting the A.sub.440 of the blank control from that of enzyme, and then plotted against different protein concentrations (from 1.25 ppm to 40 ppm). Each value was the mean of triplicate assays. The proteolytic activity is shown as Net A.sub.440. The proteolytic assay with azo-casein as the substrate (shown in FIG. 4.2) indicates that PehPro1 is an active protease.
Example 4.4
pH Profile of Metalloprotease PehPro1
[0387] With azo-casein as the substrate, the pH profile of metalloprotease PehPro1 was studied in 12.5 mM acetate/Bis-Tris/HEPES/CHES buffer with different pH values (ranging from pH 4 to 11). To initiate the assay, 50 .mu.l of 25 mM acetate/Bis-Tris/HEPES/CHES buffer with a specific pH was first mixed with 2 .mu.l Milli-Q H.sub.2O diluted enzyme (250 ppm) in a 96-MTP placed on ice, followed by the addition of 48 .mu.l of 1.5% (w/v) azo-casein prepared in H2O. The reaction was performed and analyzed as described in Example 4.3. Enzyme activity at each pH was reported as the relative activity, where the activity at the optimal pH was set to be 100%. The pH values tested were 4, 5, 6, 7, 8, 9, 10 and 11. Each value was the mean of triplicate assays. As shown in FIG. 4.3, the optimal pH of PehPro1 is 7, with greater than 70% of maximal activity retained between 5.5 and 9.5.
Example 4.5
Temperature Profile of Metalloprotease PehPro1
[0388] The temperature profile of metalloprotease PehPro1 was analyzed in 50 mM Tris buffer (pH 7) using the azo-casein assays. The enzyme sample and azo-casein substrate were prepared as in Example 4.3. Prior to the reaction, 50 .mu.l of 1.5% azo-casein and 45 .mu.l Milli-Q H.sub.2O were mixed in a 200 .mu.l PCR tube, which was then subsequently incubated in a Peltier Thermal Cycler (BioRad) at desired temperatures (i.e. 20-90.degree. C.) for 5 min. After the incubation, 5 .mu.l of diluted enzyme (100 ppm) or H.sub.2O (the blank control) was added to the substrate mixture, and the reaction was carried out in the Peltier Thermal Cycle for 10 min at different temperatures. To terminate the reaction, each assay mixture was transferred to a 96-MTP containing 100 .mu.l of 5% TCA per well. Subsequent centrifugation and absorbance measurement were performed as described in Example 4.3. The activity was reported as the relative activity, where the activity at the optimal temperature was set to be 100%. The tested temperatures are 20, 30, 40, 50, 60, 70, 80, and 90.degree. C. Each value was the mean of duplicate assays (the value varies no more than 5%). The data in FIG. 4.4 suggest that PehPro1 showed an optimal temperature at 70.degree. C., and retained greater than 70% of its maximum activity between 60 and 75.degree. C.
Example 4.6
Cleaning Performance of Metalloprotease PehPro1
[0389] The cleaning performance of PehPro1 was tested using PA-S-38 (egg yolk, with pigment, aged by heating) microswatches (CFT-Vlaardingen, The Netherlands) at pH 6 and 8 using a model automatic dishwashing (ADW) detergent. Prior to the reaction, purified protease samples were diluted with a dilution solution containing 10 mM NaCl, 0.1 mM CaCl.sub.2, 0.005% TWEEN.RTM. 80 and 10% propylene glycol to the desired concentrations. The reactions were performed in AT detergent with 100 ppm water hardness (Ca.sup.2+:Mg.sup.2+=3:1) (detergent composition shown in Table 4.1). To initiate the reaction, 180 .mu.l of the AT detergent buffered at pH 6 or pH 8 was added to a 96-MTP placed with PA-S-38 microswatches, followed by the addition of 20 .mu.l of diluted enzymes (or the dilution solution as the blank control). The 96-MTP was sealed and incubated in an incubator/shaker for 30 min at 50.degree. C. and 1150 rpm. After incubation, 100 .mu.l of wash liquid from each well was transferred to a new 96-MTP, and its absorbance was measured at 405 nm (referred here as the "Initial performance") using a spectrophotometer. The remaining wash liquid in the 96-MTP was discarded and the microswatches were rinsed once with 200 .mu.l water. Following the addition of 180 .mu.l of 0.1 M CAPS buffer (pH 10), the second incubation was carried out in the incubator/shaker at 50.degree. C. and 1150 rpm for 10 min. One hundred microliters of the resulting wash liquid was transferred to a new 96-MTP, and its absorbance measured at 405 nm (referred here as the "Wash-off"). The sum of two absorbance measurements ("Initial performance" plus "Wash-off") gives the "Total performance", which measures the protease activity on the model stain; and Net A.sub.405 was subsequently calculated by subtracting the A.sub.405 of the "Total performance" of the blank control from that of the enzyme. Dose response in cleaning the PA-S-38 microswatches at pH 6 and pH 8 for PehPro1 in AT detergent is shown in FIGS. 4.5A and 4.5B.
TABLE-US-00035 TABLE 4.1 Composition of AT dish detergent Ingredient Concentration (mg/ml) MGDA (methylglycinediacetic acid) 0.143 Sodium citrate 1.86 Citric acid* varies Plurafac .RTM. LF 18B (a non-ionic surfactant) 0.029 Bismuthcitrate 0.006 Bayhibit .RTM. S 0.006 (Phosphonobutantricarboxylic acid sodium salt) Acusol .TM. 587 (a calcium polyphosphate 0.029 inhibitor) PEG 6000 0.043 PEG 1500 0.1 *The pH of the AT formula detergent is adjusted to the desired value (pH 6 or 8) by the addition of 0.9M citric acid.
Example 4.7
Comparison of PehPro1 to Other Proteases
A. Identification of Homologous Proteases
[0390] Homologs were identified by a BLAST search (Altschul et al., Nucleic Acids Res, 25:3389-402, 1997) against the NCBI non-redundant protein database and the Genome Quest Patent database with search parameters set to default values. The mature protein amino acid sequence for PehPro1 (SEQ ID NO: 18) was used as the query sequence. Percent identity (PID) for both search sets is defined as the number of identical residues divided by the number of aligned residues in the pairwise alignment. Tables 4.2A and 4.2B provide a list of sequences with the percent identity to PehPro1. The length in Table 4.2 refers to the entire sequence length of the homologous proteases.
TABLE-US-00036 TABLE 4.2A List of sequences with percent identity to PehPro1 protein identified from the NCBI non-redundant protein database Accession # PID to PehPro1 Organism Length ZP_09077634.1 88 Paenibacillus elgii B69 524 ZP_09071078.1 74 Paenibacillus larvae subsp. larvae B-3650 529 YP_003872179.1 74 Paenibacillus polymyxa E681 592 P29148 73 Paenibacillus polymyxa 590 P43263 68 Brevibacillus brevis 527 ZP_09775365.1 68 Paenibacillus sp. Aloe-11 580 ZP_10241029.1 67 Paenibacillus peoriae KCTC 3763 599 ZP_10575942.1 66 Brevibacillus sp. BC25 528 YP_002770810.1 67 Brevibacillus brevis NBRC 100599 528 ZP_08640523.1 64 Brevibacillus laterosporus LMG 15441 564 YP_004646155.1 63 Paenibacillus mucilaginosus KNP414 525 ZP_08093424.1 60 Planococcus donghaensis MPA1U2 553 YP_003670279.1 59 Geobacillus sp. C56-T3 546 P00800 59 Bacillus thermoproteolyticus 548
TABLE-US-00037 TABLE 4.2B List of sequences with percent identity to PehPro1 protein identified from the Genome Quest Patent database PID to Patent ID # PehPro1 Organism Length JP2005229807-0019 74.5 Paenibacillus polymyxa 566 US20120107907-0187 74.09 Bacillus polymyxa 302 US8114656-0186 68.21 Bacillus brevis 304 WO2004011619-0044 63.25 empty 507 EP2390321-0185 62.9 Bacillus cereus 317 WO2012110563-0004 62.7 Bacillus megaterium 320 WO2012110563-0005 62.58 Bacillus cereus 320 JP1995184649-0001 62.5 Lactobacillus sp. 566 JP2005333991-0002 62.38 empty 562 EP2178896-0184 62.18 Bacillus anthracis 566 JP1994014788-0003 61.94 empty 317 EP2390321-0178 61.86 Bacillus thuringiensis 566 US6518054-0002 60.84 Bacillus sp. 316 US8114656-0176 60.13 Bacillus stearothermophilus 548 US6103512-0003 59.81 empty 319 US20120107907-0184 59.49 Bacillus caldoyticus 319
B. Alignment of Homologous Protease Sequences
[0391] The amino acid sequence of predicted mature PehPro1 (SEQ ID NO: 18) was aligned with thermolysin (P00800, Bacillus thermoproteolyticus) and protease from Paenibacillus elgii B69 (ZP_09077634.1) using CLUSTALW software (Thompson et al., Nucleic Acids Research, 22:4673-4680, 1994) with the default parameters. FIG. 4.6 shows the alignment of PehPro1 with these protease sequences.
C. Phylogenetic Tree
[0392] A phylogenetic tree for precursor protein PehPro1 (SEQ ID NO: 17) was built using sequences of representative homologs from Table 2A and the Neighbor Joining method (NJ) (Saitou, N.; and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing Guide Trees. MolBiol.Evol. 4, 406-425). The NJ method works on a matrix of distances between all pairs of sequences to be analyzed. These distances are related to the degree of divergence between the sequences. The phylodendron-phylogenetic tree printer software (http://iubio.bio.indiana.edu/treeapp/treeprint-form.html) was used to display the phylogenetic tree shown in FIG. 4.7.
Example 5.1
Cloning of Paenibacillus barcinonensis Metalloprotease PbaPro1
[0393] A strain (DSM15478) of Paenibacillus barcinonensis was selected as a potential source of enzymes which may be useful in various industrial applications. Genomic DNA for sequencing was obtained by first growing the strain on Heart Infusion agar plates (Difco) at 37.degree. C. for 24 hr. Cell material was scraped from the plates and used to prepare genomic DNA with the ZF Fungal/Bacterial DNA miniprep kit from Zymo (Cat No. D6005). The genomic DNA was used for genome sequencing. The entire genome of the Paenibacillus barcinonensis strain was sequenced by BaseClear (Leiden, The Netherlands) using the Illumina's next generation sequencing technology. After assembly of the data, contigs were annotated by BioXpr (Namur, Belgium). One of the genes identified after annotation in Paenibacillus barcinonensis encodes a metalloprotease and the sequence of this gene, called PbaPro1, is provided in SEQ ID NO: 21. The corresponding protein encoded by the PbaPro1 gene is shown in SEQ ID NO: 22. At the N-terminus, the protein has a signal peptide with a length of 25 amino acids as predicted by SignalP version 4.0 (Nordahl Petersen et al. (2011) Nature Methods, 8:785-786). The presence of a signal sequence suggests that PbaPro1 is a secreted enzyme. The propeptide region was predicted based on protein sequence alignment with the Paenibacillus polymyxa Npr protein (Takekawa et al. (1991) Journal of Bacteriology, 173 (21): 6820-6825). The predicted mature region of PbaPro1 protein is shown in SEQ ID NO: 23.
[0394] The nucleotide sequence of the PbaPro1 gene isolated from Paenibacillus barcinonensis is set forth as SEQ ID NO: 21. The sequence encoding the predicted native signal peptide is shown in italics:
TABLE-US-00038 ATGAAATTGACCAAAATTATGCCAACAATTCTTGCAGGAGCTCTTTT GCTCACATCCCTGTCCTCTGCAGCAGCAATGCCGTTATCTGACTCAT CCATTCCATTTGAGGGCCCCTACACCTCCGAGGAGAGTATTCTGTTG AACAACAACCCGGACGAAATGATTTATAATTTTCTTGCACAACAAGA GCAATTTCTGAATGCCGACGTCAAAGGACAGCTCAAAATCATTAAAC GCAACACAGACACTTCCGGCATCAGACACTTTCGTCTGAAGCAATAC ATCAAAGGTGTTCCGGTTTACGGCGCAGAACAAACGATCCATCTGGA CAAGAACGGAGCTGTAACTTCCGCACTCGGCGATCTTCCGCCAATTG AAGAACAGGCTGTTCCGAATGATGGCGTTCCCGCAATCAGTGCAGAC GATGCCATCCGTGCCGCCGAGAATGAAGCCACCTCCCGTCTTGGAGA GCTTGGCGCACCAGAGCTTGAGCCAAAGGCCGAATTAAACATTTATC ATCATGAAGATGACGGACAAACCTACCTCGTTTACATTACGGAAGTT AACGTGCTTGAGCCTTCCCCGCTACGGACCAAATATTTTATTAACGC CCTTGATGGAAGCATCGTATCTCAATACGATATTATCAACTTTGCCA CAGGCACCGGTACAGGCGTGCATGGTGATACCAAAACACTGACGACA ACTCAATCCGGCAGCACCTATCAGCTGAAAGATACAACTCGTGGAAA AGGCATTCAAACCTATACTGCGAACAATCGCTCCTCGCTTCCAGGCA GCTTGTCTACCAGTTCCAATAACGTATGGACAGACCGTGCAGCTGTA GATGCGCACGCCTATGCTGCCGCCACATATGACTTCTACAAAAACAA ATTCAATCGCAACGGCATTGACGGAAACGGGCTGTTGATTCGCTCTA CAGTGCATTATGGCTCCAACTATAAAAACGCCTTCTGGAACGGAGCA CAGATTGTCTATGGAGATGGCGATGGCATCGAGTTCGGTCCCTTCTC CGGTGATCTCGATGTTGTCGGACATGAATTGACACACGGGGTGATTG AATATACAGCCAATCTCGAATATCGCAATGAGCCGGGTGCTTTAAAC GAAGCTTTTGCCGACATTATGGGGAACACCATCGAAAGCAAAAACTG GCTGCTTGGCGACGGAATCTATACTCCAAACATTCCAGGTGATGCCC TGCGCTCGTTATCCGACCCTACGCTGTATAACCAGCCTGACAAATAC AGTGATCGCTACACTGGCTCTCAGGATAATGGCGGTGTGCATATCAA CAGCGGGATCATTAACAAAGCATATTATCTTGCAGCCCAAGGCGGTA CTCATAACGGGGTAACCGTTAGCGGCATCGGCCGGGATAAAGCAGTA CGTATTTTCTATAGCACGCTGGTGAACTACCTGACGCCAACCTCCAA ATTTGCAGCAGCCAAAACAGCGACAATTCAGGCAGCCAAGGACCTGT ACGGTGCCAATTCCGCTGAAGCTACGGCAATCACCAAAGCTTATCAA GCGGTAGGTTTG
[0395] The amino acid sequence of the PbaPro1 precursor protein is set forth as SEQ ID NO: 22. The predicted signal sequence is shown in italics, and the predicted pro-peptide is shown in underlined text:
TABLE-US-00039 MKLTKIMPTILAGALLLTSLSSAAAMPLSDSSIPFEGPYTSEESILL NNNPDEMIYNFLAQQEQFLNADVKGQLKIIKRNTDTSGIRHFRLKQY IKGVPVYGAEQTIHLDKNGAVTSALGDLPPIEEQAVPNDGVPAISAD DAIRAAENEATSRLGELGAPELEPKAELNIYHHEDDGQTYLVYITEV NVLEPSPLRTKYFINALDGSIVSQYDIINFATGTGTGVHGDTKTLTT TQSGSTYQLKDTTRGKGIQTYTANNRSSLPGSLSTSSNNVWTDRAAV DAHAYAAATYDFYKNKFNRNGIDGNGLLIRSTVHYGSNYKNAFWNGA QIVYGDGDGIEFGPFSGDLDVVGHELTHGVIEYTANLEYRNEPGALN EAFADIMGNTIESKNWLLGDGIYTPNIPGDALRSLSDPTLYNQPDKY SDRYTGSQDNGGVHINSGIINKAYYLAAQGGTHNGVTVSGIGRDKAV RIFYSTLVNYLTPTSKFAAAKTATIQAAKDLYGANSAEATAITKAYQ AVGL
[0396] The amino acid sequence of the predicted mature form of PbaPro1 is set forth as SEQ ID NO: 23:
TABLE-US-00040 ATGTGTGVHGDTKTLTTTQSGSTYQLKDTTRGKGIQTYTANNRSSLP GSLSTSSNNVWTDRAAVDAHAYAAATYDFYKNKFNRNGIDGNGLLIR STVHYGSNYKNAFWNGAQIVYGDGDGIEFGPFSGDLDVVGHELTHGV IEYTANLEYRNEPGALNEAFADIMGNTIESKNWLLGDGIYTPNIPGD ALRSLSDPTLYNQPDKYSDRYTGSQDNGGVHINSGIINKAYYLAAQG GTHNGVTVSGIGRDKAVRIFYSTLVNYLTPTSKFAAAKTATIQAAKD LYGANSAEATAITKAYQAVGL
Example 5.2
Expression of Paenibacillus barcinonensis Metalloprotease PbaPro1
[0397] The DNA sequence of the propeptide-mature form of PbaPro1 was synthesized and inserted into the Bacillus subtilis expression vector p2JM103BBI (Vogtentanz, Protein Expr Purif, 55:40-52, 2007) by Generay (Shanghai, China), resulting in plasmid pGX147(AprE-PbaPro1) (FIG. 5.1). Ligation of this gene encoding the PbaPro1 protein into the digested vector resulted in the addition of three codons (Ala-Gly-Lys) between the 3' end of the B. subtilis AprE signal sequence and the 5' end of the predicted PbaPro1 native propeptide. The gene has an alternative start codon (GTG). The resulting plasmid shown in FIG. 1 was labeled pGX147(AprE-PbaPro1). As shown in FIG. 5.1, pGX147(AprE-PbaPro1) contains an AprE promoter, an AprE signal sequence used to direct target protein secretion in B. subtilis, and the synthetic nucleotide sequence encoding the predicted propeptide and mature regions of PbaPro1 (SEQ ID NO: 24). The translation product of the synthetic AprE-PbaPro1 gene is shown in SEQ ID NO: 25.
[0398] The pGX147(AprE-PbaPro1) plasmid was then transformed into B. subtilis cells (degU.sup.Hy 32, .DELTA.scoC) and the transformed cells were spread on Luria Agar plates supplemented with 5 ppm Chloramphenicol and 1.2% skim milk (Cat #232100, Difco). Colonies with the largest clear halos on the plates were selected and subjected to fermentation in a 250 ml shake flask with MBD medium (a MOPS based defined medium. supplemented with additional 5 mM CaCl.sub.2).
[0399] The broth from the shake flasks was concentrated and buffer-exchanged into the loading buffer containing 20 mM Tris-HCl (pH 8.5), 1 mM CaCl.sub.2 and 10% propylene glycol using a VivaFlow 200 ultra filtration device (Sartorius Stedim). After filtering, this sample was applied to an 80 ml Q Sepharose High Performance column pre-equilibrated with the loading buffer above; and the active flow-through fractions were collected and concentrated. The sample was loaded onto a 320 ml Superdex 75 gel filtration column pre-equilibrated with the loading buffer described above containing 0.15 M NaCl. The corresponding active purified protein fractions were further pooled and concentrated via 10K Amicon Ultra for further analyses.
[0400] The nucleotide sequence of the synthesized PbaPro1 gene in plasmid pGX147(AprE-PbaPro1) is depicted in SEQ ID NO: 24. The sequence encoding the three residue addition (AGK) is shown in bold:
TABLE-US-00041 GTGAGAAGCAAAAAATTGTGGATCAGCTTGTTGTTTGCGTTAACGTT AATCTTTACGATGGCGTTCAGCAACATGAGCGCGCAGGCTGCTGGAA AAATGCCTCTGTCAGACAGCAGCATTCCGTTTGAGGGCCCGTACACA TCAGAAGAAAGCATCCTGCTGAACAACAACCCGGACGAGATGATCTA CAATTTCCTGGCACAGCAGGAGCAGTTCCTGAACGCAGACGTGAAGG GCCAGCTGAAAATCATCAAAAGAAACACAGACACGAGCGGCATCAGA CACTTCAGACTGAAGCAGTACATCAAGGGCGTCCCGGTTTACGGCGC TGAGCAGACAATCCACCTGGACAAAAATGGCGCAGTGACGAGCGCAC TTGGAGATCTGCCGCCGATTGAAGAGCAAGCAGTCCCGAACGATGGC GTTCCGGCGATTAGCGCTGATGACGCTATCAGAGCCGCGGAAAACGA AGCGACGTCAAGACTGGGAGAACTTGGCGCACCGGAACTTGAACCGA AGGCGGAACTGAACATCTATCACCACGAAGACGATGGACAGACGTAC CTGGTGTACATCACGGAGGTGAATGTGCTGGAGCCGTCACCGCTGAG AACAAAATACTTCATCAATGCGCTGGATGGCAGCATCGTTAGCCAAT ACGACATCATTAACTTCGCCACAGGCACGGGCACAGGCGTTCATGGC GACACAAAAACGCTTACGACAACACAGTCAGGCTCAACGTACCAGCT GAAAGACACAACAAGAGGCAAGGGCATCCAGACGTATACAGCCAATA ACAGAAGCTCACTTCCGGGCTCACTGTCAACAAGCAGCAATAATGTC TGGACGGACAGAGCTGCAGTGGACGCGCACGCGTATGCTGCGGCCAC GTACGACTTCTACAAGAACAAGTTCAACAGAAACGGCATTGATGGCA ACGGCCTGCTTATTAGAAGCACGGTCCACTACGGCTCAAACTACAAG AATGCGTTTTGGAACGGCGCCCAAATTGTTTATGGCGATGGAGACGG CATCGAGTTCGGACCTTTTAGCGGCGACCTGGATGTGGTCGGACATG AACTGACGCACGGCGTTATCGAGTATACGGCGAATCTGGAATACAGA AATGAACCGGGCGCTCTGAATGAGGCCTTCGCGGATATCATGGGCAA CACAATTGAGAGCAAAAACTGGCTTCTGGGCGACGGAATCTACACGC CGAACATTCCGGGAGATGCACTGAGATCACTGAGCGACCCTACGCTG TACAACCAGCCGGACAAATACAGCGACAGATACACGGGATCACAGGA CAATGGCGGCGTCCATATTAACTCAGGCATCATCAACAAAGCGTATT ATCTGGCAGCTCAAGGCGGCACGCATAATGGCGTCACAGTTAGCGGA ATCGGCAGAGACAAGGCCGTCAGAATTTTCTACTCAACGCTGGTGAA CTACCTGACACCGACAAGCAAGTTTGCAGCCGCCAAAACAGCCACGA TTCAGGCAGCAAAGGACCTGTACGGAGCGAACTCAGCAGAGGCCACA GCGATTACGAAGGCTTATCAAGCCGTGGGACTGTAA
[0401] The amino acid sequence of the PbaPro1 precursor protein expressed from plasmid pGX147(AprE-PbaPro1) is depicted in SEQ ID NO: 25. The predicted signal sequence is shown in italics, the three residue addition (AGK) is shown in bold, and the predicted pro-peptide is shown in underlined text.
TABLE-US-00042 MRSKKLWISLLFALTLIFTMAFSNMSAQAAGKMPLSDSSIPFEGPYT SEESILLNNNPDEMIYNFLAQQEQFLNADVKGQLKIIKRNTDTSGIR HFRLKQYIKGVPVYGAEQTIHLDKNGAVTSALGDLPPIEEQAVPNDG VPAISADDAIRAAENEATSRLGELGAPELEPKAELNIYHHEDDGQTY LVYITEVNVLEPSPLRTKYFINALDGSIVSQYDIINFATGTGTGVHG DTKTLTTTQSGSTYQLKDTTRGKGIQTYTANNRSSLPGSLSTSSNNV WTDRAAVDAHAYAAATYDFYKNKFNRNGIDGNGLLIRSTVHYGSNYK NAFWNGAQIVYGDGDGIEFGPFSGDLDVVGHELTHGVIEYTANLEYR NEPGALNEAFADIMGNTIESKNWLLGDGIYTPNIPGDALRSLSDPTL YNQPDKYSDRYTGSQDNGGVHINSGIINKAYYLAAQGGTHNGVTVSG IGRDKAVRIFYSTLVNYLTPTSKFAAAKTATIQAAKDLYGANSAEAT AITKAYQAVGL
Example 5.3
Proteolytic Activity of Metalloprotease PbaPro1
[0402] The proteolytic activity of purified metalloprotease PbaPro1 was measured in 50 mM Tris (pH 7), using azo-casein (Cat #74H7165, Megazyme) as a substrate. Prior to the reaction, the enzyme was diluted with Milli-Q water (Millipore) to specific concentrations. The azo-casein was dissolved in 100 mM Tris buffer (pH 7) to a final concentration of 1.5% (w/v). To initiate the reaction, 50 .mu.l of the diluted enzyme (or Milli-Q H.sub.2O alone as the blank control) was added to the non-binding 96-well Microtiter Plate (96-MTP) (Corning Life Sciences, #3641) placed on ice, followed by the addition of 50 .mu.l of 1.5% azo-casein. After sealing the 96-MTP, the reaction was carried out in a Thermomixer (Eppendorf) at 40.degree. C. and 650 rpm for 10 min. The reaction was terminated by adding 100 .mu.l of 5% Trichloroacetic Acid (TCA). Following equilibration (5 min at the room temperature) and subsequent centrifugation (2000 g for 10 min at 4.degree. C.), 120 .mu.l supernatant was transferred to a new 96-MTP, and absorbance of the supernatant was measured at 440 nm (A.sub.440) using a SpectraMax 190. Net A.sub.440 was calculated by subtracting the A.sub.440 of the blank control from that of enzyme, and then plotted against different protein concentrations (from 1.25 ppm to 40 ppm). Each value was the mean of triplicate assays.
[0403] The proteolytic activities are shown as Net A.sub.44o. The proteolytic assay with azo-casein as the substrate (shown in FIG. 5.2) indicates that PbaPro1 is an active protease.
Example 5.4
pH Profile of Metalloprotease PbaPro1
[0404] With azo-casein as the substrate, the pH profile of metalloprotease PbaPro1 was studied in 12.5 mM acetate/Bis-Tris/HEPES/CHES buffer with different pH values (ranging from pH 5 to 11). To initiate the assay, 50 .mu.l of 25 mM acetate/Bis-Tris/HEPES/CHES buffer with a specific pH was first mixed with 2 .mu.l Milli-Q H.sub.2O diluted enzyme (125 ppm) in a 96-MTP placed on ice, followed by the addition of 48 .mu.l of 1.5% (w/v) azo-casein prepared in H2O. The reaction was performed and analyzed as described in Example 5.3. Enzyme activity at each pH was reported as the relative activity, where the activity at the optimal pH was set to be 100%. The pH values tested were 5, 6, 7, 8, 9, 10 and 11. Each value was the mean of triplicate assays. As shown in FIG. 5.3, the optimal pH of PbaPro1 is 8, with greater than 70% of maximal activity retained between 7 and 9.
Example 5.5
Temperature Profile of Metalloprotease PbaPro1
[0405] The temperature profiles of metalloprotease PbaPro1 was analyzed in 50 mM Tris buffer (pH 7) using the azo-casein assays. The enzyme sample and azo-casein substrate were prepared as in Example 5.3. Prior to the reaction, 50 .mu.l of 1.5% azo-casein and 45 .mu.l Milli-Q H.sub.2O were mixed in a 200 .mu.l PCR tube, which was then subsequently incubated in a Peltier Thermal Cycler (BioRad) at desired temperatures (i.e. 20-90.degree. C.) for 5 min. After the incubation, 5 .mu.l of diluted enzyme (50 ppm) or H.sub.2O (the blank control) was added to the substrate mixture, and the reaction was carried out in the Peltier Thermal Cycle for 10 min at different temperatures. To terminate the reaction, each assay mixture was transferred to a 96-MTP containing 100 .mu.l of 5% TCA per well. Subsequent centrifugation and absorbance measurement were performed as described in Example 5.3. The activity was reported as the relative activity, where the activity at the optimal temperature was set to be 100%. The tested temperatures are 20, 30, 40, 50, 60, 70, 80, and 90.degree. C. Each value was the mean of duplicate assays (the value varies no more than 5%). The data in FIG. 5.4 suggest that PbaPro1 showed an optimal temperature at 50.degree. C., and retained greater than 70% of its maximum activity between 45 and 55.degree. C.
Example 5.6
Cleaning Performance of Metalloprotease PbaPro1
[0406] The cleaning performance of PbaPro1 was tested using PA-S-38 (egg yolk, with pigment, aged by heating) microswatches (CFT-Vlaardingen, The Netherlands) at pH 6 and 8 using a model automatic dishwashing (ADW) detergent. Prior to the reaction, purified protease samples were diluted with a dilution solution containing 10 mM NaCl, 0.1 mM CaCl.sub.2, 0.005% TWEEN.RTM. 80 and 10% propylene glycol to the desired concentrations. The reactions were performed in AT detergent with 100 ppm water hardness (Ca.sup.2+:Mg.sup.2+=3:1) (detergent composition shown in Table 5.1). To initiate the reaction, 180 .mu.l of the AT detergent buffered at pH 6 or pH 8 was added to a 96-MTP placed with PA-S-38 microswatches, followed by the addition of 20 .mu.l of diluted enzymes (or the dilution solution as the blank control). The 96-MTP was sealed and incubated in an incubator/shaker for 30 min at 50.degree. C. and 1150 rpm. After incubation, 100 .mu.l of wash liquid from each well was transferred to a new 96-MTP, and its absorbance was measured at 405 nm (referred here as the "Initial performance") using a spectrophotometer. The remaining wash liquid in the 96-MTP was discarded and the microswatches were rinsed once with 200 .mu.l water. Following the addition of 180 .mu.l of 0.1 M CAPS buffer (pH 10), the second incubation was carried out in the incubator/shaker at 50.degree. C. and 1150 rpm for 10 min. One hundred microliters of the resulting wash liquid was transferred to a new 96-MTP, and its absorbance measured at 405 nm (referred here as the "Wash-off"). The sum of two absorbance measurements ("Initial performance" plus "Wash-off") gives the "Total performance", which measures the protease activity on the model stain; and Net A.sub.405 was subsequently calculated by subtracting the A.sub.405 of the "Total performance" of the blank control from that of the enzyme. Dose response in cleaning the PA-S-38 microswatches at pH 6 and pH 8 in AT detergent for PbaPro1 is shown in FIGS. 5.5A and 5.5B.
TABLE-US-00043 TABLE 5.1 Composition of AT dish detergent formula with bleach Concentration Ingredient (mg/ml) MGDA (methylglycinediacetic acid) 0.143 Sodium citrate 1.86 Citric acid* varies PAP (peracid N,N-phthaloylaminoperoxycaproic acid) 0.057 Plurafac .RTM. LF 18B (a non-ionic surfactant) 0.029 Bismuthcitrate 0.006 Bayhibit .RTM. S (Phosphonobutantricarboxylic 0.006 acid sodium salt) Acusol .TM. 587 (a calcium polyphosphate inhibitor) 0.029 PEG 6000 0.043 PEG 1500 0.1 *The pH of the AT formula detergent is adjusted to the desired value (pH 6 or 8) by the addition of 0.9M citric acid.
Example 5.7
Comparison of PbaPro1 to Other Proteases
A. Identification of Homologous Proteases
[0407] Homologs were identified by a BLAST search (Altschul et al., Nucleic Acids Res, 25:3389-402, 1997) against the NCBI non-redundant protein database and the Genome Quest Patent database with search parameters set to default values. The predicted mature protein amino acid sequence for PbaPro1 (SEQ ID NO: 23) was used as the query sequence. Percent identity (PID) for both search sets is defined as the number of identical residues divided by the number of aligned residues in the pairwise alignment. Tables 5.2A and 5.2B provide a list of sequences with the percent identity to PbaPro1. The length in Table 5.2 refers to the entire sequence length of the homologous proteases.
TABLE-US-00044 TABLE 5.2A List of sequences with percent identity to PbaPro1 protein identified from the NCBI non-redundant protein database PID to Accession # PbaPro1 Organism Length AAB02774.1 56 Geobacillus stearothermophilus 552 P00800 56 Bacillus stermoproteolyticus 548 AAA22623.1 57 Bacillus caldolyticus 544 YP_003670279.1 57 Geobacillus sp. C56-T3 546 AAC43402.1 57 Alicyclobacillus acidocaldarius 546 YP_003597483.1 57 Bacillus megaterium DSM 319 562 ZP_08093424.1 57 Planococcus donghaensis MPA1U2 553 ZP_08640523.1 59 Brevibacillus laterosporus 564 LMG 15441 ZP_04216147.1 59 Bacillus cereus Rock3-44 566 YP_001373863.1 60 Bacillus cytotoxicus NVH 391-98 565 YP_004646155.1 60 Paenibacillus mucilaginosus 525 KNP414 ZP_10738945.1 61 Brevibacillus sp. CF112 528 CAA43589.1 63 Brevibacillus brevis 527 ZP_02326602.1 64 Paenibacillus larvae subsp. 520 larvae BRL-230010 ZP_02326503.1 65 Paenibacillus larvae subsp. larvae 520 B-3650 ZP_09077634.1 66 Paenibacillus elgii B69 524 ZP_08511445.1 68 Paenibacillus sp. HGF7 525 ZP_09775364.1 70 Paenibacillus sp. Aloe-11 593 YP_005073223.1 70 Paenibacillus terrae HPL-003 591 ZP_10241030.1 70 Paenibacillus peoriae KCTC 3763 593 YP_003948511.11 71 Paenibacillus polymyxa SC2 592
TABLE-US-00045 TABLE 5.2B List of sequences with percent identity to PbaPro1 protein identified from the Genome Quest Patent database PID to Patent # PbaPro1 Organism Length JP2005333991-0002 56.91 562 WO2012110562-0007 56.96 Bacillus cereus 320 WO2012110562-0006 57.23 Bacillus megaterium 320 EP2390321-0178 57.23 Bacillus thuringiensis 566 EP2390321-0184 57.56 Bacillus caldoyticus 319 WO2007044993-0184 57.56 Bacillus sp. 319 US20120107907-0177 57.56 Bacillus caldolyticus 544 CN102168095-0002 57.88 319 WO2012110562-0004 57.88 Bacillus caldolyticus 319 WO2012110562-0003 57.88 Geobacillus stearothermophilus 319 WO2004011619-0056 57.88 546 JP1995184649-0001 57.88 Lactobacillus sp. 566 JP2010535248-0240 57.88 Bacillus anthracis 566 US6518054-0001 58.2 Bacillus sp. 319 US6103512-0003 58.2 319 WO2011163237-0001 58.2 Geobacillus stearothermophilus 548 JP1994014788-0003 58.25 317 US8114656-0185 58.9 Bacillus cereus 317 US20120107907-0179 58.9 Bacillus cereus 566 WO2012110563-0005 59.22 Bacillus cereus 320 WO2004011619-0044 59.6 507 US20120107907-0186 63.25 Bacillus brevis 304 JP2005229807-0018 70.86 Paenibacillus polymyxa 566 EP2390321-0187 71.1 Bacillus polymyxa 302 JP2009511072-0203 71.1 Paenibacillus polymyxa 302
B. Alignment of Homologous Protease Sequences
[0408] The amino acid sequence of the predicted mature PbaPro1 (SEQ ID NO: 23) was aligned with Thermolysin (P00800, Bacillus thermoproteolyticus), and protease from Paenibacillus polymyxa SC2 (YP 003948511.1) using CLUSTALW software (Thompson et al., Nucleic Acids Research, 22:4673-4680, 1994) with the default parameters. FIG. 5.6 shows the alignment of PbaPro1 with these protease sequences.
C. Phylogenetic Tree
[0409] A phylogenetic tree for full length sequence of PbaPro1 (SEQ ID NO: 22) was built using sequences of representative homologs from Table 2A and the Neighbor Joining method (NJ) (Saitou, N.; and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing Guide Trees. MolBiol.Evol. 4, 406-425). The NJ method works on a matrix of distances between all pairs of sequences to be analyzed. These distances are related to the degree of divergence between the sequences. The phylodendron-phylogenetic tree printer software (http://iubio.bio.indiana.edu/treeapp/treeprint-form.html) was used to display the phylogenetic tree shown in FIG. 5.7.
Example 6.1
Cloning of Paenibacillus polymyxa SC2 Metalloprotease PpoPro1
[0410] The nucleic acid sequence for the PpoPro1 gene was identified in the NCBI database (NCBI Reference Sequence: NC 014622.1 from 4536397-4538175) and is provided in SEQ ID NO: 26. The corresponding protein encoded by the PpoPro1 gene is shown in SEQ ID NO: 27. At the N-terminus, the protein has a signal peptide with a length of 24 amino acids as predicted by SignalP version 4.0 (Nordahl Petersen et al. (2011) Nature Methods, 8:785-786). The presence of a signal sequence suggests that PpoPro1 is a secreted enzyme. The propeptide region was predicted based on protein sequence alignment with the Paenibacillus polymyxa Npr protein (Takekawa et al. (1991) Journal of Bacteriology, 173 (21): 6820-6825). The predicted mature region of PpoPro1 protein is shown in SEQ ID NO: 28.
[0411] The nucleotide sequence of the PpoPro1 gene identified from NCBI database is set forth as SEQ ID NO: 26. The sequence encoding the predicted native signal peptide is shown in italics:
TABLE-US-00046 ATGAAAAAAGTATGGGTTTCGCTTCTTGGAGGAGCTATGTTATTAGGGTCT GTCGCGTCTGGTGCATCTGCGGAGAGTTCCGTTTCGGGGCCAGCTCAGCTT ACACCGACCTTCCACGCCGAGCAATGGAAAGCACCTACCTCGGTATCGGGG GATGACATTGTATGGAGCTATTTAAATCGACAAAAGAAATCGTTGCTGGGT GTGGATAGCTCCAGTGTACGTGAACAATTCCGAATCGTTGATCGCACAAGC GACAAATCCGGTGTAAGCCATTATCGACTGAAGCAGTATGTAAACGGAATT CCCGTGTATGGAGCTGAACAAACTATTCATGTGGGCAAATCTGGTGAGGTC ACCTCTTACTTAGGAGCGGTGGTTAATGAGGATCAGCAGGCAGAAGCTACG CAAGGTACAACTCCAAAAATCAGCGCTTCTGAAGCGGTCTACACCGCATAT AAAGAAGCAGCTGCACGGATTGAAGCCCTCCCTACCTCCGACGATACTATT TCTAAAGACGCTGAGGAGCCAAGCAGTGTAAGTAAAGATACTTACGCCGAA GCAGCTAACAACGAAAAAACGCTTTCTGTTGATAAGGACGAGCTGAGTCTT GATCAGGCATCTGTCCTGAAAGATAGCAAAATTGAAGCAGTGGAACCAGAA AAAAGTTCCATTGCCAAAATCGCTAATCTGCAGCCTGAAGTAGATCCTAAA GCAGAACTCTACTACTACCCTAAGGGGGATGACCTGCTGCTGGTTTATGTA ACAGAAGTTAATGTTTTAGAACCTGCCCCACTGCGTACCCGCTACATTATT GATGCCAATGACGGCAGCATCGTATTCCAGTATGACATCATTAATGAAGCG ACAGGCACAGGTAAAGGTGTGCTTGGTGATTCCAAATCGTTCACTACTACC GCTTCCGGCAGTAGCTACCAGTTAAAAGATACAACACGCGGTAACGGAATC GTGACTTACACGGCCTCCAACCGTCAAAGCATCCCAGGTACCATTTTGACA GATGCCGATAATGTATGGAATGATCCAGCTGGTGTGGACGCCCATGCGTAT GCTGCTAAAACCTATGATTACTATAAAGCCAAATTTGGACGCAACAGCATT GACGGACGCGGTCTGCAACTTCGTTCGACGGTCCATTACGGTAGTCGCTAC AACAATGCCTTCTGGAACGGCTCCCAAATGACTTATGGAGATGGAGATGGT AGCACATTTATCGCCTTCAGCGGGGACCCCGATGTAGTAGGACATGAACTT ACGCATGGTGTCACAGAGTATACTTCGAATTTGGAATATTACGGAGAGTCC GGCGCATTGAATGAAGCTTTCTCAGACGTTATCGGGAATGACATTCAGCGC AAAAACTGGCTTGTAGGCGATGATATTTACACGCCAAACATTGCAGGCGAT GCCCTTCGCTCAATGTCCAATCCAACCCTGTACGATCAACCAGATCACTAT TCCAACCTGTACAGAGGCAGCTCCGATAACGGCGGTGTTCACACCAACAGC GGTATTATCAATAAAGCTTACTACTTGTTAGCACAAGGTGGTAATTTCCAT GGCGTAACTGTAAATGGAATTGGCCGTGATGCAGCGGTGCAAATTTACTAC AGTGCCTTTACGAACTACCTGACTTCTTCTTCCGACTTCTCCAACGCACGT GCTGCTGTGATCCAAGCCGCAAAAGATCTGTACGGGGCGAACTCAGCAGAA GCAACTGCAGCTGCCAAGTCTTTTGACGCTGTAGGCGTAAACTAA
[0412] The amino acid sequence of the PpoPro1 precursor protein is set forth as SEQ ID NO: 27. The predicted signal sequence is shown in italics, and the predicted propeptide is shown in underlined text:
TABLE-US-00047 MKKVWVSLLGGAMLLGSVASGASAESSVSGPAQLTPTFHAEQWKAPTSVSG DDIVWSYLNRQKKSLLGVDSSSVREQFRIVDRTSDKSGVSHYRLKQYVNGI PVYGAEQTIHVGKSGEVTSYLGAVVNEDQQAEATQGTTPKISASEAVYTAY KEAAARIEALPTSDDTISKDAEEPSSVSKDTYAEAANNEKTLSVDKDELSL DQASVLKDSKIEAVEPEKSSIAKIANLQPEVDPKAELYYYPKGDDLLLVYV TEVNVLEPAPLRTRYIIDANDGSIVFQYDIINEATGTGKGVLGDSKSFTTT ASGSSYQLKDTTRGNGIVTYTASNRQSIPGTILTDADNVWNDPAGVDAHAY AAKTYDYYKAKFGRNSIDGRGLQLRSTVHYGSRYNNAFWNGSQMTYGDGDG STFIAFSGDPDVVGHELTHGVTEYTSNLEYYGESGALNEAFSDVIGNDIQR KNWLVGDDIYTPNIAGDALRSMSNPTLYDQPDHYSNLYRGSSDNGGVHTNS GIINKAYYLLAQGGNFHGVTVNGIGRDAAVQIYYSAFTNYLTSSSDFSNAR AAVIQAAKDLYGANSAEATAAAKSFDAVGVN
[0413] The amino acid sequence of the predicted mature form of PpoPro1 is set forth as SEQ ID NO: 28:
TABLE-US-00048 ATGTGKGVLGDSKSFTTTASGSSYQLKDTTRGNGIVTYTASNRQSIPGTIL TDADNVWNDPAGVDAHAYAAKTYDYYKAKFGRNSIDGRGLQLRSTVHYGSR YNNAFWNGSQMTYGDGDGSTFIAFSGDPDVVGHELTHGVTEYTSNLEYYGE SGALNEAFSDVIGNDIQRKNWLVGDDIYTPNIAGDALRSMSNPTLYDQPDH YSNLYRGSSDNGGVHTNSGIINKAYYLLAQGGNFHGVTVNGIGRDAAVQIY YSAFTNYLTSSSDFSNARAAVIQAAKDLYGANSAEATAAAKSFDAVGVN
Example 6.2
Expression of Paenibacillus polymyxa SC2 Metalloprotease PpoPro1
[0414] The DNA sequence of the propeptide-mature form of PpoPro1 was synthesized and inserted into the Bacillus subtilis expression vector p2JM103BBI (Vogtentanz, Protein Expr Purif, 55:40-52, 2007) by Generay (Shanghai, China), resulting in plasmid pGX138(AprE-PpoPro1) (FIG. 1). Ligation of this gene encoding the PpoPro1 protein into the digested vector resulted in the addition of three codons (Ala-Gly-Lys) between the 3' end of the B. subtilis AprE signal sequence and the 5' end of the predicted PpoPro1 native propeptide. The gene has an alternative start codon (GTG). The resulting plasmid shown in FIG. 6.1, labeled pGX138(AprE-PpoPro1 contains an AprE promoter, an AprE signal sequence used to direct target protein secretion in B. subtilis, and the synthetic nucleotide sequence encoding the predicted propeptide and mature regions of PpoPro1 (SEQ ID NO: 29). The translation product of the synthetic AprE-PpoPro1 gene is shown in SEQ ID NO: 30.
[0415] The pGX138(AprE-PpoPro1) plasmid was then transformed into B. subtilis cells (degU.sup.Hy 32, .DELTA.scoC) and the transformed cells were spread on Luria Agar plates supplemented with 5 ppm Chloramphenicol and 1.2% skim milk (Cat #232100, Difco). Colonies with the largest clear halos on the plates were selected and subjected to fermentation in a 250 ml shake flask with MBD medium (a MOPS based defined medium, supplemented with additional 5 mM CaCl.sub.2)).
[0416] The broth from the shake flasks was concentrated and buffer-exchanged into the loading buffer containing 20 mM Tris-HCl (pH 8.5), 1 mM CaCl.sub.2) and 10% propylene glycol using a VivaFlow 200 ultra filtration device (Sartorius Stedim). After filtering, this sample was applied to an 80 ml Q Sepharose High Performance column pre-equilibrated with the loading buffer above, PpoPro1 was eluted from the column with a linear salt gradient from 0 to 0.25 M NaCl in the loading buffer. The corresponding active fractions were collected and concentrated. The sample was loaded onto a 320 ml Superdex 75 gel filtration column pre-equilibrated with the loading buffer described above containing 0.15 M NaCl. The corresponding active purified protein fractions were further pooled and concentrated via 10K Amicon Ultra for further analyses.
[0417] The nucleotide sequence of the synthesized PpoPro1 gene in plasmid pGX138(AprE-PpoPro1) is depicted in SEQ ID NO: 29. The sequence encoding the three residue addition (AGK) is shown in bold:
TABLE-US-00049 GTGAGAAGCAAAAAATTGTGGATCAGCTTGTTGTTTGCGTTAACGTTAATC TTTACGATGGCGTTCAGCAACATGAGCGCGCAGGCTGCTGGAAAAGAATCA TCAGTGTCAGGACCGGCTCAGCTTACACCGACATTTCACGCAGAACAATGG AAGGCTCCGACGTCAGTTTCAGGAGACGACATCGTGTGGAGCTACCTGAAT AGACAGAAGAAAAGCCTGCTGGGAGTGGATAGCAGCAGCGTCAGAGAGCAG TTCAGAATCGTTGACAGAACGAGCGACAAAAGCGGAGTCAGCCATTATAGA CTGAAGCAGTACGTGAATGGCATCCCGGTTTATGGCGCAGAGCAGACAATT CATGTTGGCAAGAGCGGAGAAGTCACAAGCTATCTGGGCGCTGTGGTCAAT GAAGATCAACAAGCCGAGGCTACACAGGGAACAACGCCGAAAATTAGCGCC TCAGAGGCAGTCTACACGGCGTACAAAGAAGCGGCTGCAAGAATCGAAGCC CTGCCGACATCAGACGATACAATTTCAAAAGATGCGGAGGAGCCGAGCTCA GTTAGCAAGGATACATACGCGGAAGCCGCAAACAATGAGAAAACACTGAGC GTGGACAAGGACGAGCTGTCACTTGATCAGGCTAGCGTCCTTAAAGACAGC AAGATCGAGGCCGTTGAGCCTGAAAAGTCATCAATTGCGAAAATCGCCAAT CTGCAACCTGAAGTCGACCCGAAGGCGGAACTGTACTACTACCCGAAAGGC GATGACCTGCTTCTGGTGTACGTCACGGAAGTGAACGTCCTGGAACCGGCA CCGCTGAGAACAAGATACATCATCGACGCGAACGACGGAAGCATCGTCTTC CAGTATGACATTATCAACGAAGCAACGGGAACGGGCAAAGGCGTTCTTGGA GACTCAAAGAGCTTCACGACAACGGCTTCAGGAAGCAGCTACCAGCTGAAA GACACGACGAGAGGAAACGGAATCGTCACATATACGGCGTCAAACAGACAA AGCATCCCTGGCACAATCCTGACGGATGCTGACAACGTTTGGAATGATCCG GCTGGCGTGGATGCCCATGCTTATGCGGCAAAAACGTATGACTATTACAAG GCGAAGTTCGGCAGAAATTCAATCGATGGCAGAGGACTGCAGCTTAGAAGC ACGGTGCACTACGGATCAAGATATAACAATGCCTTCTGGAACGGCAGCCAG ATGACATACGGAGACGGAGATGGAAGCACATTTATTGCATTCAGCGGCGAC CCTGATGTGGTTGGCCATGAGCTGACGCATGGCGTTACAGAATATACGAGC AATCTTGAATACTACGGCGAGTCAGGCGCTCTGAACGAGGCATTTAGCGAT GTTATCGGCAATGACATCCAGAGAAAAAACTGGCTGGTGGGCGACGATATT TACACGCCTAATATCGCTGGCGATGCCCTTAGATCAATGTCAAACCCGACG CTGTATGATCAGCCTGACCACTACTCAAACCTGTATAGAGGCTCATCAGAT AACGGAGGCGTCCATACGAATAGCGGCATCATTAACAAGGCATATTATCTT CTGGCCCAGGGCGGCAATTTTCATGGAGTGACGGTTAATGGAATTGGAAGA GACGCAGCCGTCCAAATCTACTACAGCGCTTTCACGAACTACCTTACATCA AGCTCAGACTTTAGCAATGCCAGAGCTGCTGTTATCCAGGCAGCGAAGGAT CTTTACGGCGCCAACTCAGCCGAAGCTACGGCCGCAGCTAAATCATTTGAT GCAGTGGGCGTTAAT
[0418] The amino acid sequence of the PpoPro1 precursor protein expressed from plasmid pGX138(AprE-PpoPro1) is depicted in SEQ ID NO: 30. The predicted signal sequence is shown in italics, the three residue addition (AGK) is shown in bold, and the predicted pro-peptide is shown in underlined text.
TABLE-US-00050 MRSKKLWISLLFALTLIFTMAFSNMSAQAAGKESSVSGPAQLTPTFHAEQW KAPTSVSGDDIVWSYLNRQKKSLLGVDSSSVREQFRIVDRTSDKSGVSHYR LKQYVNGIPVYGAEQTIHVGKSGEVTSYLGAVVNEDQQAEATQGTTPKISA SEAVYTAYKEAAARIEALPTSDDTISKDAEEPSSVSKDTYAEAANNEKTLS VDKDELSLDQASVLKDSKIEAVEPEKSSIAKIANLQPEVDPKAELYYYPKG DDLLLVYVTEVNVLEPAPLRTRYIIDANDGSIVFQYDIINEATGTGKGVLG DSKSFTTTASGSSYQLKDTTRGNGIVTYTASNRQSIPGTILTDADNVWNDP AGVDAHAYAAKTYDYYKAKFGRNSIDGRGLQLRSTVHYGSRYNNAFWNGSQ MTYGDGDGSTFIAFSGDPDVVGHELTHGVTEYTSNLEYYGESGALNEAFSD VIGNDIQRKNWLVGDDIYTPNIAGDALRSMSNPTLYDQPDHYSNLYRGSSD NGGVHTNSGIINKAYYLLAQGGNFHGVTVNGIGRDAAVQIYYSAFTNYLTS SSDFSNARAAVIQAAKDLYGANSAEATAAAKSFDAVGVN
Example 6.3
Proteolytic Activity of Metalloprotease PpoPro1
[0419] The proteolytic activity of purified PpoPro1 was measured in 50 mM Tris (pH 7), using azo-casein (Cat #74H7165, Megazyme) as a substrate. Prior to the reaction, the enzyme was diluted with Milli-Q water (Millipore) to specific concentrations. The azo-casein was dissolved in 100 mM Tris buffer (pH 7) to a final concentration of 1.5% (w/v). To initiate the reaction, 50 .mu.L of the diluted enzyme (or Milli-Q H.sub.2O alone as the blank control) was added to the non-binding 96-well microtiter Plate (96-MTP) (Corning Life Sciences, #3641) placed on ice, followed by the addition of 50 .mu.L of 1.5% azo-casein. After sealing the 96-MTP, the reaction was carried out in a Thermomixer (Eppendorf) at 40.degree. C. and 650 rpm for 10 min. The reaction was terminated by adding 100 .mu.L of 5% Trichloroacetic Acid (TCA). Following equilibration (5 min at the room temperature) and subsequent centrifugation (2000 g for 10 min at 4.degree. C.), 120 .mu.L supernatant was transferred to a new 96-MTP, and absorbance of the supernatant was measured at 440 nm (A.sub.440) using a SpectraMax 190. Net A.sub.440 was calculated by subtracting the A.sub.440 of the blank control from that of enzyme, and then plotted against different protein concentrations (from 1.25 ppm to 40 ppm). Each value was the mean of duplicate assays, and the value varies no more than 5%. The proteolytic activity is shown as Net A.sub.44o. The proteolytic assay with azo-casein as the substrate (FIG. 6.2) indicates PpoPro1 is an active protease.
Example 4
pH Profile of Metalloprotease PpoPro1
[0420] With azo-casein as the substrate, the pH profile of PpoPro1 was studied in 12.5 mM acetate/Bis-Tris/HEPES/CHES buffer with different pH values (ranging from pH 4 to 11). To initiate the assay, 50 .mu.L of 25 mM acetate/Bis-Tris/HEPES/CHES buffer with a specific pH was first mixed with 2 .mu.L diluted enzyme (250 ppm in Milli-Q H.sub.2O) in a 96-MTP placed on ice, followed by the addition of 48 .mu.L of 1.5% (w/v) azo-casein prepared in H2O. The reaction was performed and analyzed as described in Example 6.3. Enzyme activity at each pH was reported as relative activity where the activity at the optimal pH was set to be 100%. The pH values tested were 4, 5, 6, 7, 8, 9, 10 and 11. Each value was the mean of triplicate assays. As shown in FIG. 6.3, the optimal pH of PpoPro1 is about 7, with greater than 70% of maximal activity retained between 5.5 and 8.5.
Example 6.5
Temperature Profile of Metalloprotease PpoPro1
[0421] The temperature profile of PpoPro1 was analyzed in 50 mM Tris buffer (pH 7) using the azo-casein assay. The enzyme sample and azo-casein substrate were prepared as in Example 6.3. Prior to the reaction, 50 .mu.L of 1.5% azo-casein and 45 .mu.l Milli-Q H.sub.2O were mixed in a 2000_, PCR tube, which was then subsequently incubated in a Peltier Thermal Cycler (BioRad) at desired temperatures (i.e. 20-90.degree. C.) for 5 min. After the incubation, 5 .mu.L of diluted PpoPro1 (100 ppm) or H.sub.2O (the blank control) was added to the substrate mixture, and the reaction was carried out in the Peltier Thermal Cycle for 10 min at different temperatures. To terminate the reaction, each assay mixture was transferred to a 96-MTP containing 100 .mu.L of 5% TCA per well. Subsequent centrifugation and absorbance measurement were performed as described in Example 6.3. The activity was reported as relative activity where the activity at the optimal temperature was set to be 100%. The tested temperatures were 20, 30, 40, 50, 60, 70, 80, and 90.degree. C. Each value was the mean of duplicate assays (the value varies no more than 5%). The data in FIG. 6.4 suggests that PpoPro1 showed an optimal temperature at 50.degree. C., and retained greater than 70% of its maximum activity between 40 and 55.degree. C.
Example 6.6
Cleaning Performance of Metalloprotease PpoPro1
[0422] The cleaning performance of PpoPro1 was tested using PA-S-38 (egg yolk, with pigment, aged by heating) microswatches (CFT-Vlaardingen, The Netherlands) at pH 6 or 8 using a model automatic dishwashing (ADW) detergent (AT detergent). Prior to the reaction, purified PpoPro1 was diluted with a dilution solution containing 10 mM NaCl, 0.1 mM CaCl.sub.2, 0.005% TWEEN.RTM. 80 and 10% propylene glycol to the desired concentrations. The reactions were performed in AT detergent (composition shown in Table 6.1) with 100 ppm water hardness (Ca.sup.2+:Mg.sup.2+=3:1), in the presence of a bleach component ((Peracid N,N-phthaloylaminoperoxycaproic acid-PAP). To initiate the reaction, 180 .mu.L of AT detergent buffered at pH 6 or 8 was added to a 96-MTP placed with PA-S-38 microswatches, followed by the addition of 20 .mu.L of diluted enzymes (or the dilution solution as the blank control). The 96-MTP was sealed and incubated in an incubator/shaker for 30 min at 50.degree. C. and 1150 rpm. After incubation, 100 .mu.L of wash liquid from each well was transferred to a new 96-MTP, and its absorbance was measured at 405 nm (referred here as the "Initial performance") using a spectrophotometer. The remaining wash liquid in the 96-MTP was discarded and the microswatches were rinsed once with 200 .mu.L water. Following the addition of 180 .mu.L of 0.1 M CAPS buffer (pH 10), the second incubation was carried out in the incubator/shaker at 50.degree. C. and 1150 rpm for 10 min. One hundred microliter of the resulting wash liquid was transferred to a new 96-MTP, and its absorbance measured at 405 nm (referred here as "Wash-off"). The sum of two absorbance measurements ("Initial performance" plus "Wash-off") gives the "Total performance", which measures the protease activity on the model stain; and Net A.sub.405 was subsequently calculated by subtracting the A.sub.405 of the "Total performance" of the blank control from that of the enzyme. Dose response in cleaning the PA-S-38 microswatches at pH 6 and pH 8 for PpoPro1 in AT dish detergent, in the presence of bleach, is shown in FIGS. 6.5A and 6.5B.
TABLE-US-00051 TABLE 6.1 Composition of AT dish detergent formula with bleach Concentration Ingredient (mg/ml) MGDA (methylglycinediacetic acid) 0.143 Sodium citrate 1.86 Citric acid* varies PAP (peracid N,N-phthaloylaminoperoxycaproic 0.057 acid) Plurafac .RTM. LF 18B (a non-ionic surfactant) 0.029 Bismuthcitrate 0.006 Bayhibit .RTM. S (Phosphonobutantricarboxylic 0.006 acid sodium salt) Acusol .TM. 587 (a calcium polyphosphate 0.029 inhibitor) PEG 6000 0.043 PEG 1500 0.1 *The pH of the AT formula detergent is adjusted to the desired value (pH 6 or 8) by the addition of 0.9M citric acid.
Example 6.7
Comparison of PpoPro1 to Other Metalloproteases
Identification of Homologous Proteases
[0423] Homologs were identified by a BLAST search (Altschul et al., Nucleic Acids Res, 25:3389-402, 1997) against the NCBI non-redundant protein database and the Genome Quest Patent database with search parameters set to default values. The predicted mature protein amino acid sequence for PpoPro1 (SEQ ID NO: 28) was used as the query sequence. Percent identity (PID) for both search sets is defined as the number of identical residues divided by the number of aligned residues in the pairwise alignment. Tables 6.2A and 6.2B provide a list of sequences with the percent identity to PpoPro1. The length in Table 6.2 refers to the entire sequence length of the homologous proteases.
TABLE-US-00052 TABLE 6.2A List of sequences with percent identity to PpoPro1 protein identified from the NCBI non-redundant protein database PID to Accession # PpoPro1 Organism Length P00800 56 Bacillus thermoproteolyticus 548 ZP_08640523.1 57 Brevibacillus laterosporus LMG 564 15441 AAA22623.1 57 Bacillus caldolyticus 544 ZP_08093424.1 59 Planococcus donghaensis MPA1U2 553 ZP_10738945.1 60 Brevibacillus sp. CF112 528 CAA43589.1 62 Brevibacillus brevis 527 ZP_02326503.1 62 Paenibacillus larvae subsp. 520 larvae BRL-230010 YP_005495105.1 63 Bacillus megaterium WSH-002 562 YP_001373863.1 64 Bacillus cytotoxicus NVH 391-98 565 ZP_04310163.1 64 Bacillus cereus BGSC 6E1 581 BAA06144.1 64 Lactobacillus sp.] 566 ZP_08511445.1 65 Paenibacillus sp. HGF7 525 ZP_04216147.1 65 Bacillus cereus Rock3-44 566 ZP_09071078.1 68 Paenibacillus larvae subsp. larvae B- 3650 ZP_09077634.1 69 Paenibacillus elgii B69 524 YP_005073224.1 79 Paenibacillus terrae HPL-003 595 ZP_10241029.1 80 Paenibacillus peoriae KCTC 3763 599 YP_005073223.1 93 Paenibacillus terrae HPL-003 591 ZP_10241030.1 95 Paenibacillus peoriae KCTC 3763 593 ZP_09775364.1 95 Paenibacillus sp. Aloe-11 593 YP_003872179.1 97 Paenibacillus polymyxa E681 592 YP_003948511.1 100 Paenibacillus polymyxa SC2 592
TABLE-US-00053 TABLE 6.2B List of sequences with percent identity to PpoPro1 protein identified from the Genome Quest Patent database PID to Patent # PpoPro1 Organism Length US20120107907-0187 97.34 Bacillus polymyxa 302 US5962264-0004 65.48 empty 566 WO2012110563-0005 65.16 Bacillus cereus 320 JP1994070791-0002 64.52 empty 317 WO2012110562-0005 64.19 Bacillus cereus 320 WO2012110563-0004 63.34 Bacillus megaterium 320 JP2002272453-0002 61.98 Bacillus megaterium 562 WO2004011619-0047 61.49 empty 532 EP2390321-0186 62.58 Bacillus brevis 304 US6518054-0002 59.22 Bacillus sp. 316 US6518054-0001 58.52 Bacillus sp. 319 US20120107907-0176 58.52 Bacillus stearothermophilis 548 JP2005229807-0019 93.05 Paenibacillus polymyxa 566 WO2012110562-0003 58.2 Geobacillus 319 stearothermophilus WO2004011619-0044 59.27 empty 507 EP2390321-0185 66.13 Bacillus cereus 317 JP1995184649-0001 65.71 Lactobacillus sp. 566 EP2178896-0184 65.38 Bacillus anthracis 566
Alignment of Homologous Protease Sequences
[0424] The amino acid sequence of predicted mature PpoPro1 (SEQ ID NO: 28) was aligned with thermolysin (P00800, Bacillus thermoproteolyticus) and protease from Paenibacillus polymyxa SC2 (YP 003948511.1) using CLUSTALW software (Thompson et al., Nucleic Acids Research, 22:4673-4680, 1994) with the default parameters. FIG. 6.6 shows the alignment of PpoPro1 with these protease sequences.
Phylogenetic Tree
[0425] A phylogenetic tree for precursor PpoPro1 (SEQ ID NO: 27) was built using sequences of representative homologs from Tables 6.2A and the Neighbor Joining method (NJ) (Saitou, N.; and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing Guide Trees. MolBiol.Evol. 4, 406-425). The NJ method works on a matrix of distances between all pairs of sequences to be analyzed. These distances are related to the degree of divergence between the sequences. The phylodendron-phylogenetic tree printer software (http://iubio.bio.indiana.edu/treeapp/treeprint-form.html) was used to display the phylogenetic tree shown in FIG. 6.7.
Example 7.1
Cloning of Paenibacillus hunanensis Metalloprotease PhuPro1
[0426] A strain (DSM22170) of Paenibacillus hunanensis was selected as a potential source of enzymes which may be useful in various industrial applications. Genomic DNA for sequencing was obtained by first growing the strain on Heart Infusion agar plates (Difco) at 37.degree. C. for 24 hr. Cell material was scraped from the plates and used to prepare genomic DNA with the ZF Fungal/Bacterial DNA miniprep kit from Zymo (Cat No. D6005). The genomic DNA was used for genome sequencing. The entire genome of the Paenibacillus hunanensis strain was sequenced by BaseClear (Leiden, The Netherlands) using the Illumina's next generation sequencing technology. After assembly of the data, contigs were annotated by BioXpr (Namur, Belgium). One of the genes identified after annotation in Paenibacillus hunanensis encodes a metalloprotease and the sequence of this gene, called PhuPro1, is provided in SEQ ID NO: 31. This gene has an alternative start codon (TTG). The corresponding protein encoded by the PhuPro1 gene is shown in SEQ ID NO: 32. At the N-terminus, the protein has a signal peptide with a length of 23 amino acids as predicted by SignalP version 4.0 (Nordahl Petersen et al. (2011) Nature Methods, 8:785-786). The presence of a signal sequence suggests that PhuPro1 is a secreted enzyme. The propeptide region was predicted based on protein sequence alignment with the Paenibacillus polymyxa Npr protein (Takekawa et al. (1991) Journal of Bacteriology, 173 (21): 6820-6825). The predicted mature region of PhuPro1 protein is shown in SEQ ID NO: 33.
[0427] The nucleotide sequence of the PhuPro1 gene isolated from Paenibacillus hunanensis is set forth as SEQ ID NO: 31. The sequence encoding the predicted native signal peptide is shown in italics:
TABLE-US-00054 TTGAAAAAAACAGTTGGTCTTTTACTTGCAGGTAGCTTGCTCGTTGGTGCT ACAACGTCCGCTTTCGCAGCAGAAGCAAATGATCTGGCACCACTCGGTGAT TACACGCCAAAATTGATTACGCAAGCAACAGGCATCACTGGCGCTAGTGGC GATGCTAAAGTATGGAAGTTCCTGGAGAAGCAAAAACGTACCATCGTAACC GATGATGCAGCTTCTGCTGATGTGAAGGAATTGTTTGAGATCACAAAACGT CAATCCGATTCTCAAACCGGTACAGAGCACTATCGCCTGAACCAAACCTTT AAAGGCATCCCAGTCTATGGCGCAGAGCAAACACTGCACTTTGACAAATCC GGCAATGTATCTCTGTACATGGGTCAGGTTGTTGAGGATGTGTCCGCTAAA CTGGAAGCTTCCGATTCCAAAAAAGGCGTAACTGAGGATGTATACGCTTCG GATACGAAAAATGATCTGGTAACACCAGAAATCAGCGCTTCTCAAGCCATC TCGATTGCTGAAAAGGATGCAGCTTCCAAAATCGGCTCCCTCGGCGAAGCA CAAAAAACGCCAGAAGCGAAGCTGTATATCTACGCTCCTGAGGATCAAGCA GCACGTCTGGCTTATGTGACAGAAGTAAACGTACTGGAGCCATCTCCGCTG CGTACTCGCTATTTTGTAGATGCAAAAACAGGTTCGATCCTGTTCCAATAT GATCTGATTGAGCATGCAACAGGTACAGGTAAAGGGGTACTGGGTGATACC AAGTCCTTCACTGTAGGTACTTCCGGTTCTTCCTATGTGATGACTGATAGC ACGCGTGGAAAAGGTATCCAAACCTACACGGCGTCTAACCGCACATCACTG CCAGGTAGCACTGTAACGAGCAGCAGCAGCACATTTAACGATCCAGCATCT GTCGATGCCCATGCGTATGCACAAAAAGTATATGATTTCTACAAATCCAAC TTTAACCGCAACAGCATCGACGGTAATGGTCTGGCTATCCGCTCCACTACG CACTATTCCACACGTTATAACAATGCGTTCTGGAATGGTTCCCAAATGGTA TACGGTGATGGCGATGGTTCGCAATTCATCGCATTCTCCGGCGACCTTGAC GTAGTAGGTCACGAGCTGACACACGGTGTAACCGAGTACACAGCGAACCTG GAATACTATGGTCAATCCGGTGCACTGAACGAATCCATTTCGGATATCTTT GGTAACACAATCGAAGGTAAAAACTGGATGGTAGGCGATGCGATCTACACA CCAGGCGTATCCGGCGATGCTCTTCGCTACATGGATGATCCAACAAAAGGT GGACAACCAGCGCGTATGGCAGATTACAACAACACAAGCGCTGATAATGGC GGTGTACACACAAACAGTGGTATCCCGAATAAAGCATACTACTTGCTGGCA CAGGGTGGCACATTTGGCGGTGTAAATGTAACAGGTATCGGTCGCTCGCAA GCGATCCAGATCGTTTACCGTGCACTAACATACTACCTGACATCCACATCT AACTTCTCGAACTACCGTTCTGCAATGGTGCAAGCATCTACAGACCTGTAC GGTGCAAACTCTACACAAACAACAGCGGTGAAAAACTCGCTGAGCGCAGTA GGCATTAAC
[0428] The amino acid sequence of the PhuPro1 precursor protein is set forth as SEQ ID NO: 32. The predicted signal sequence is shown in italics, and the predicted pro-peptide is shown in underlined text:
TABLE-US-00055 MKKTVGLLLAGSLLVGATTSAFAAEANDLAPLGDYTPKLITQATGITGASG DAKVWKFLEKQKRTIVTDDAASADVKELFEITKRQSDSQTGTEHYRLNQTF KGIPVYGAEQTLHFDKSGNVSLYMGQVVEDVSAKLEASDSKKGVTEDVYAS DTKNDLVTPEISASQAISIAEKDAASKIGSLGEAQKTPEAKLYIYAPEDQA ARLAYVTEVNVLEPSPLRTRYFVDAKTGSILFQYDLIEHATGTGKGVLGDT KSFTVGTSGSSYVMTDSTRGKGIQTYTASNRTSLPGSTVTSSSSTFNDPAS VDAHAYAQKVYDFYKSNFNRNSIDGNGLAIRSTTHYSTRYNNAFWNGSQMV YGDGDGSQFIAFSGDLDVVGHELTHGVTEYTANLEYYGQSGALNESISDIE GNTIEGKNAVMVGDAIYTPGVSGDALRYMDDPTKGGQPARMADYNNTSADN GGVHTNSGIPNKAYYLLAQGGTFGGVNVTGIGRSQAIQIVYRALTYYLTST SNFSNYRSAMVQASTDLYGANSTQTTAVKNSLSAVGIN
[0429] The amino acid sequence of the predicted mature form of PhuPro1 is set forth as SEQ ID NO: 33:
TABLE-US-00056 ATGTGKGVLGDTKSFTVGTSGSSYVMTDSTRGKGIQTYTASNRTSLPGSTV TSSSSTFNDPASVDAHAYAQKVYDFYKSNFNRNSIDGNGLAIRSTTHYSTR YNNAFWNGSQMVYGDGDGSQFIAFSGDLDVVGHELTHGVTEYTANLEYYGQ SGALNESISDIFGNTIEGKNWMVGDAIYTPGVSGDALRYMDDPTKGGQPAR MADYNNTSADNGGVHTNSGIPNKAYYLLAQGGTFGGVNVTGIGRSQAIQIV YRALTYYLTSTSNFSNYRSAMVQASTDLYGANSTQTTAVKNSLSAVGIN
Example 7.2
Expression of Paenibacillus hunanensis Metalloprotease PhuPro1
[0430] The DNA sequence of the propeptide-mature form of PhuPro1 was synthesized and inserted into the Bacillus subtilis expression vector p2JM103BBI (Vogtentanz, Protein Expr Purif, 55:40-52, 2007) by Generay (Shanghai, China), resulting in plasmid pGX149(AprE-PhuPro1) (FIG. 7.1). Ligation of this gene encoding the PhuPro1 protein into the digested vector resulted in the addition of three codons (Ala-Gly-Lys) between the 3' end of the B. subtilis AprE signal sequence and the 5' end of the predicted PhuPro1 native propeptide. The gene has an alternative start codon (GTG). The resulting plasmid shown in FIG. 1, labeled pGX149(AprE-PhuPro1) contains an AprE promoter, an AprE signal sequence used to direct target protein secretion in B. subtilis, and the synthetic nucleotide sequence encoding the predicted propeptide and mature regions of PhuPro1 (SEQ ID NO: 34). The translation product of the synthetic AprE-PhuPro1 gene is shown in SEQ ID NO: 35.
[0431] The pGX149(AprE-PhuPro1) plasmid was then transformed into B. subtilis cells (degU.sup.Hy 32, .DELTA.scoC) and the transformed cells were spread on Luria Agar plates supplemented with 5 ppm Chloramphenicol and 1.2% skim milk (Cat #232100, Difco). Colonies with the largest clear halos on the plates were selected and subjected to fermentation in a 250 ml shake flask with MBD medium (a MOPS based defined medium, supplemented with additional 5 mM CaCl.sub.2).
[0432] The broth from the shake flasks was concentrated and buffer-exchanged into the loading buffer containing 20 mM Tris-HCl (pH 8.5), 1 mM CaCl.sub.2 and 10% propylene glycol using a VivaFlow 200 ultra filtration device (Sartorius Stedim). After filtering, this sample was applied to a 80 ml Q Sepharose High Performance column pre-equilibrated with the loading buffer above; and the active flow-through fractions were collected and concentrated via 10K Amicon Ultra for further analyses.
[0433] The nucleotide sequence of the synthesized PhuPro1 gene in plasmid pGX149(AprE-PhuPro1) is depicted in SEQ ID NO: 34. The sequence encoding the three residue addition (AGK) is shown in bold:
TABLE-US-00057 GTGAGAAGCAAAAAATTGTGGATCAGCTTGTTGTTTGCGTTAACGTTAATC TTTACGATGGCGTTCAGCAACATGAGCGCGCAGGCTGCTGGAAAAGCAGAA GCTAATGATCTTGCCCCGCTTGGCGATTATACACCGAAGCTTATTACACAG GCAACGGGAATTACAGGCGCATCAGGCGATGCGAAGGTGTGGAAGTTCCTG GAGAAGCAGAAGAGAACGATTGTCACGGACGACGCCGCAAGCGCGGATGTC AAGGAGCTGTTCGAGATCACGAAGAGACAGAGCGATAGCCAGACGGGAACG GAGCATTACAGACTGAACCAGACGTTCAAGGGCATTCCGGTCTACGGAGCT GAACAAACGCTGCATTTTGATAAAAGCGGCAACGTCTCACTGTACATGGGC CAAGTCGTTGAGGACGTTAGCGCCAAACTTGAGGCTAGCGACAGCAAGAAA GGCGTCACAGAAGATGTCTACGCGTCAGACACGAAAAACGACCTGGTTACA CCGGAAATCTCAGCTTCACAGGCCATCTCAATTGCAGAGAAAGACGCAGCG TCAAAAATCGGCTCACTGGGCGAGGCTCAGAAAACGCCGGAGGCGAAACTT TACATCTACGCCCCTGAGGACCAGGCTGCGAGACTGGCTTACGTGACAGAA GTTAATGTGCTGGAGCCGTCACCGCTTAGAACGAGATATTTCGTGGACGCA AAGACGGGCAGCATTCTGTTTCAGTACGATCTTATCGAACACGCGACAGGC ACAGGAAAGGGAGTTCTGGGAGACACAAAAAGCTTCACGGTTGGCACGTCA GGCAGCAGCTACGTGATGACAGACAGCACGAGAGGCAAGGGCATTCAAACG TATACAGCGAGCAACAGAACAAGCCTGCCGGGAAGCACAGTCACGAGCTCA TCATCAACGTTTAATGACCCGGCCTCAGTGGATGCTCACGCATACGCGCAG AAAGTGTACGACTTCTACAAAAGCAACTTCAATAGAAACAGCATCGACGGA AACGGCCTTGCGATCAGAAGCACGACGCACTACAGCACAAGATACAACAAC GCCTTCTGGAACGGCAGCCAAATGGTTTACGGCGATGGCGACGGATCACAG TTTATCGCATTTAGCGGAGACCTGGACGTCGTTGGCCATGAGCTGACACAT GGCGTTACGGAGTACACAGCAAACCTGGAATACTATGGCCAGTCAGGCGCC CTTAACGAGAGCATCAGCGACATTTTTGGCAATACGATCGAAGGAAAGAAC TGGATGGTCGGCGACGCAATCTACACACCGGGCGTTTCAGGCGATGCACTG AGATATATGGACGACCCGACAAAGGGCGGACAGCCGGCCAGAATGGCGGAT TACAATAATACGTCAGCAGATAACGGCGGCGTGCATACAAATAGCGGCATC CCTAACAAAGCATATTACCTGCTTGCGCAAGGAGGAACATTTGGCGGCGTG AATGTTACGGGCATTGGCAGATCACAAGCGATTCAGATCGTTTACAGAGCG CTGACGTACTACCTTACGAGCACGAGCAATTTTAGCAACTACAGAAGCGCA ATGGTGCAGGCAAGCACGGATCTGTATGGCGCAAATTCAACACAAACGACG GCGGTCAAGAATAGCCTTTCAGCAGTGGGCATTAACTAA
[0434] The amino acid sequence of the PhuPro1 precursor protein expressed from plasmid pGX149(AprE-PhuPro1) is depicted in SEQ ID NO: 35. The predicted signal sequence is shown in italics, the three residue addition (AGK) is shown in bold, and the predicted pro-peptide is shown in underlined text.
TABLE-US-00058 MRSKKLWISLLFALTLIFTMAFSNMSAQAAGKAEANDLAPLGDYTPKLITQ ATGITGASGDAKVWKFLEKQKRTIVTDDAASADVKELFEITKRQSDSQTGT EHYRLNQTFKGIPVYGAEQTLHFDKSGNVSLYMGQVVEDVSAKLEASDSKK GVTEDVYASDTKNDLVTPEISASQAISIAEKDAASKIGSLGEAQKTPEAKL YIYAPEDQAARLAYVTEVNVLEPSPLRTRYFVDAKTGSILFQYDLIEHATG TGKGVLGDTKSFTVGTSGSSYVMTDSTRGKGIQTYTASNRTSLPGSTVTSS SSTFNDPASVDAHAYAQKVYDFYKSNFNRNSIDGNGLAIRSTTHYSTRYNN AFWNGSQMVYGDGDGSQFIAFSGDLDVVGHELTHGVTEYTANLEYYGQSGA LNESISDIFGNTIEGKNWMVGDAIYTPGVSGDALRYMDDPTKGGQPARMAD YNNTSADNGGVHTNSGIPNKAYYLLAQGGTFGGVNVTGIGRSQAIQIVYRA LTYYLTSTSNFSNYRSAMVQASTDLYGANSTQTTAVKNSLSAVGIN
Example 7.3
Proteolytic Activity of Metalloprotease PhuPro1
[0435] The proteolytic activity of purified metalloprotease PhuPro1 was measured in 50 mM Tris (pH 7), using azo-casein (Cat #74H7165, Megazyme) as a substrate. Prior to the reaction, the enzyme was diluted with Milli-Q water (Millipore) to specific concentrations. The azo-casein was dissolved in 100 mM Tris buffer (pH 7) to a final concentration of 1.5% (w/v). To initiate the reaction, 50 .mu.l of the diluted enzyme (or Milli-Q H.sub.2O alone as the blank control) was added to the non-binding 96-well Microtiter Plate (96-MTP) (Corning Life Sciences, #3641) placed on ice, followed by the addition of 50 .mu.l of 1.5% azo-casein. After sealing the 96-MTP, the reaction was carried out in a Thermomixer (Eppendorf) at 40.degree. C. and 650 rpm for 10 min. The reaction was terminated by adding 100 .mu.l of 5% Trichloroacetic Acid (TCA). Following equilibration (5 min at the room temperature) and subsequent centrifugation (2000 g for 10 min at 4.degree. C.), 120 .mu.l supernatant was transferred to a new 96-MTP, and absorbance of the supernatant was measured at 440 nm (A.sub.440) using a SpectraMax 190. Net A.sub.440 was calculated by subtracting the A.sub.440 of the blank control from that of enzyme, and then plotted against different protein concentrations (from 1.25 ppm to 40 ppm). Each value was the mean of triplicate assays. The proteolytic activity is shown as Net A.sub.440. The proteolytic assay with azo-casein as the substrate (shown in FIG. 7.2) indicates that PhuPro1 is an active protease.
Example 7.4
pH Profile of Metalloprotease PhuPro1
[0436] With azo-casein as the substrate, the pH profile of metalloprotease PhuPro1 was studied in 12.5 mM acetate/Bis-Tris/HEPES/CHES buffer with different pH values (ranging from pH 4 to 11). To initiate the assay, 50 .mu.l of 25 mM acetate/Bis-Tris/HEPES/CHES buffer with a specific pH was first mixed with 2 .mu.l Milli-Q H.sub.2O diluted enzyme (125 ppm) in a 96-MTP placed on ice, followed by the addition of 48 .mu.l of 1.5% (w/v) azo-casein prepared in H2O. The reaction was performed and analyzed as described in Example 3. Enzyme activity at each pH was reported as the relative activity, where the activity at the optimal pH was set to be 100%. The pH values tested were 4, 5, 6, 7, 8, 9, 10 and 11. Each value was the mean of triplicate assays. As shown in FIG. 7.3, the optimal pH of PhuPro1 is about 6, with greater than 70% of maximal activity retained between 5 and 8.
Example 7.5
Temperature Profile of Metalloprotease PhuPro1
[0437] The temperature profile of metalloprotease PhuPro1 was analyzed in 50 mM Tris buffer (pH 7) using the azo-casein assays. The enzyme sample and azo-casein substrate were prepared as in Example 7.3. Prior to the reaction, 50 .mu.l of 1.5% azo-casein and 45 .mu.l Milli-Q H.sub.2O were mixed in a 200 .mu.l PCR tube, which was then subsequently incubated in a Peltier Thermal Cycler (BioRad) at desired temperatures (i.e. 20-90.degree. C.) for 5 min. After the incubation, 5 .mu.l of diluted enzyme (50 ppm) or H.sub.2O (the blank control) was added to the substrate mixture, and the reaction was carried out in the Peltier Thermal Cycle for 10 min at different temperatures. To terminate the reaction, each assay mixture was transferred to a 96-MTP containing 100 .mu.l of 5% TCA per well. Subsequent centrifugation and absorbance measurement were performed as described in Example 7.3. The activity was reported as the relative activity, where the activity at the optimal temperature was set to be 100%. The tested temperatures are 20, 30, 40, 50, 60, 70, 80, and 90.degree. C. Each value was the mean of duplicate assays (the value varies no more than 5%). The data in FIG. 7.4 suggests that PhuPro1 showed an optimal temperature at 60.degree. C., and retained greater than 70% of its maximum activity between 45 and 65.degree. C.
Example 7.6
Cleaning Performance of Metalloprotease PhuPro1
[0438] The cleaning performance of PhuPro1 was tested using PA-S-38 (egg yolk, with pigment, aged by heating) microswatches (CFT-Vlaardingen, The Netherlands) at pH 6 and 8 using a model automatic dishwashing (ADW) detergent. Prior to the reaction, purified protease samples were diluted with a dilution solution containing 10 mM NaCl, 0.1 mM CaCl.sub.2, 0.005% TWEEN.RTM. 80 and 10% propylene glycol to the desired concentrations. The reactions were performed in AT detergent with 100 ppm water hardness (Ca.sup.2+:Mg.sup.2+=3:1) (detergent composition shown in Table 7.1). To initiate the reaction, 180 .mu.l of the AT detergent buffered at pH 6 or pH 8 was added to a 96-MTP placed with PA-S-38 microswatches, followed by the addition of 20 .mu.l of diluted enzymes (or the dilution solution as the blank control). The 96-MTP was sealed and incubated in an incubator/shaker for 30 min at 50.degree. C. and 1150 rpm. After incubation, 100 .mu.l of wash liquid from each well was transferred to a new 96-MTP, and its absorbance was measured at 405 nm (referred here as the "Initial performance") using a spectrophotometer. The remaining wash liquid in the 96-MTP was discarded and the microswatches were rinsed once with 200 .mu.l water. Following the addition of 180 .mu.l of 0.1 M CAPS buffer (pH 10), the second incubation was carried out in the incubator/shaker at 50.degree. C. and 1150 rpm for 10 min. One hundred microliters of the resulting wash liquid was transferred to a new 96-MTP, and its absorbance measured at 405 nm (referred here as the "Wash-off"). The sum of two absorbance measurements ("Initial performance" plus "Wash-off") gives the "Total performance", which measures the protease activity on the model stain; and Net A.sub.405 was subsequently calculated by subtracting the A.sub.405 of the "Total performance" of the blank control from that of the enzyme. Dose response in cleaning the PA-S-38 microswatches at pH 6 and pH 8 in AT detergent for PhuPro1 is shown in FIGS. 7.5A and 7.5B.
TABLE-US-00059 TABLE 7.1 Composition of AT detergent Concentration Ingredient (mg/ml) MGDA (methylglycinediacetic acid) 0.143 Sodium citrate 1.86 Citric acid* varies Plurafac .RTM. LF 18B (a non-ionic surfactant) 0.029 Bismuthcitrate 0.006 Bayhibit .RTM. S (Phosphonobutantricarboxylic 0.006 acid sodium salt) Acusol .TM. 587 (a calcium polyphosphate 0.029 inhibitor) PEG 6000 0.043 PEG 1500 0.1 *The pH of the AT formula detergent is adjusted to the desired value (pH 6 or 8) by the addition of 0.9M citric acid.
Example 7.7
Comparison of PhuPro1 to Other Proteases
A. Identification of Homologous Proteases
[0439] Homologs were identified by a BLAST search (Altschul et al., Nucleic Acids Res, 25:3389-402, 1997) against the NCBI non-redundant protein database and the Genome Quest Patent database with search parameters set to default values. The predicted mature protein amino acid sequence for PhuPro1 (SEQ ID NO: 33) was used as the query sequence. Percent identity (PID) for both search sets is defined as the number of identical residues divided by the number of aligned residues in the pairwise alignment. Tables 7.2A and 7.2B provide a list of sequences with the percent identity to PhuPro1. The length in Table 7.2 refers to the entire sequence length of the homologous proteases.
TABLE-US-00060 TABLE 7.2A List of sequences with percent identity to PhuPro1 protein identified from the NCBI non-redundant protein database Accession # PID to PhuPro1 Organism Length P00800 55 Bacillus thermoproteolyticus 548 AAB02774.1 55 Geobacillus stearothermophilus 552 EJS73098.1 56 Bacillus cereus BAG2X1-3 566 BAD60997.1 56 Bacillus megaterium 562 ZP_04216147.1 57 Bacillus cereus Rock3-44 566 YP_893436.1 56 Bacillus thuringiensis str. Al Hakam 566 ZP_08640523.1 58 Brevibacillus laterosporus 564 ZP_09069194.1 59 Paenibacillus larvae subsp. larvae B-3650 502 YP_002770810.1 60 Brevibacillus brevis 528 ZP_08511445.1 61 Paenibacillus sp. HGF7 525 P43263 61 Brevibacillus brevis 527 ZP_09775365.1 62 Paenibacillus sp. Aloe-11 580 ZP_09077634.1 66 Paenibacillus elgii B69 524 P29148 68 NPRE_PAEPO 590 ZP_09775364.1 69 Paenibacillus sp. Aloe-11 593 ZP_10241030.1 69 Paenibacillus peoriae KCTC 3763 593 YP_005073223.1 69 Paenibacillus terrae HPL-003 591
TABLE-US-00061 TABLE 7.2B List of sequences with percent identity to PhuPro1 protein identified from the Genome Quest Patent database PID to Patent ID # PhuPro1 Organism Length WO2012110562-0003 56.23 Geobacillus stearothermophilus 319 US6518054-0001 56.55 Bacillus sp. 319 JP2002272453-0002 56.69 Bacillus megaterium 562 US20090123467-0184 56.73 Bacillus anthracis 566 US6103512-0003 56.87 319 EP0867512-0002 56.96 316 WO2012110562-0005 57.1 Bacillus cereus 320 WO2012110563-0005 58.06 Bacillus cereus 320 US20120107907-0187 68.44 Bacillus polymyxa 302
B. Alignment of Homologous Protease Sequences
[0440] The amino acid sequence of predicted mature PhuPro1 (SEQ ID NO: 33) protein was aligned with Proteinase T (P00800, Bacillus thermoproteolyticus), and protease from Paenibacillus terrae HPL-003 (YP 005073223.1) using CLUSTALW software (Thompson et al., Nucleic Acids Research, 22:4673-4680, 1994) with the default parameters. FIG. 7.6 shows the alignment of PhuPro1 with these protease sequences.
C. Phylogenetic Tree
[0441] A phylogenetic tree for full length sequence of PhuPro1 (SEQ ID NO: 2) was built using sequences of representative homologs from Table 2A and the Neighbor Joining method (NJ) (Saitou, N.; and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing Guide Trees. MolBiol.Evol. 4, 406-425). The NJ method works on a matrix of distances between all pairs of sequences to be analyzed. These distances are related to the degree of divergence between the sequences. The phylodendron-phylogenetic tree printer software (http://iubio.bio.indiana.edu/treeapp/treeprint-form.html) was used to display the phylogenetic tree shown in FIG. 7.7.
Example 7.8
Terg-o-Tometer Performance Evaluation of PhuPro1
[0442] The wash performance of PhuPro1 was tested in a laundry detergent application using a Terg-o-Tometer (Instrument Marketing Services, Inc, Fairfield, N.J.). The performance evaluation was conducted at 32.degree. C. and 16.degree. C. The soil load consisted of two of each of the following stain swatches: EMPA116 Blood, Milk, Ink on cotton (Test materials AG, St. Gallen, Switzerland), EMPA117 Blood, Milk, Ink on polycotton (Test materials AG, St. Gallen, Switzerland), EMPA112 Cocoa on cotton (Test materials AG, St. Gallen, Switzerland), and CFT C-10 Pigment, Oil, and Milk content on cotton (Center for Testmaterials BV, Vlaardingen, Netherlands), plus extra white interlock knit fabric to bring the total fabric load to 40 g per beaker of the Terg-o-Tometer, which was filled with 1 L of deionized water. The water hardness was adjusted to 6 grains per gallon, and the pH in the beaker was buffered with 5 mM HEPES, pH 8.2. Heat inactivated Tide Regular HDL (Proctor & Gamble), a commercial liquid detergent purchased in a local US supermarket, was used at 0.8 g/L. The detergent was inactivated before use by treatment at 92.degree. C. in a water bath for 2-3 hours followed by cooling to room temperature. Heat inactivation of commercial detergents serves to destroy the activity of enzymatic components while retaining the properties of the non-enzymatic components. Enzyme activity in the heat inactivated detergent was measured using the Suc-AAPF-pNA assay for measuring protease activity. The Purafect.RTM. Prime HA, (Genencor Int'l) and PhuPro1 proteases were each added to final concentrations of 1 ppm. A control sample with no enzyme was included. The wash time was 12 minutes. After the wash treatment, all swatches were rinsed for 3 minutes and machine-dried at low heat.
[0443] Four of each type of swatch were measured before and after treatment by optical reflectance using a Tristimulus Minolta Meter CR-400. The difference in the L, a, b values was converted to total color difference (dE), as defined by the CIE-LAB color space. Cleaning of the stains is expressed as percent stain removal index (% SRI) by taking a ratio between the color difference before and after washing, and comparing it to the difference of unwashed soils (before wash) to unsoiled fabric, and averaging the eight values obtained by reading two different regions of each washed swatch. Cleaning performances of PhuPro1 and Purafect.RTM. Prime HA proteases at 32.degree. C. are shown in Tables 7.8A and FIG. 7.8A and at 16.degree. C. are shown in Table 7.8B and FIG. 7.8B.
TABLE-US-00062 TABLE 7.8A Cleaning performance of PhuPro1 at 32.degree. C. EMPA-116 EMPA-117 Purafect Prime Purafect Prime HA PhuPro1 HA PhuPro1 Average 95CI Average 95CI Average 95CI Average 95CI ppm % SRI [% SRI % SRI [% SRI % SRI [% SRI % SRI [% SRI enzyme (dE) (dE)] (dE) (dE)] (dE) (dE)] (dE) (dE)] 0 0.25 0.02 0.25 0.02 0.19 0.02 0.19 0.02 0.2 0.31 0.02 0.31 0.01 0.31 0.03 0.32 0.04 0.5 0.34 0.02 0.33 0.03 0.34 0.02 0.37 0.02 1 0.35 0.03 0.36 0.02 0.38 0.03 0.42 0.03 1.5 0.36 0.02 0.37 0.03 0.35 0.03 0.43 0.03 EMPA-112 CFT C-10 Purafect Prime Purafect Prime HA PhuPro1 HA PhuPro1 Average 95CI Average 95CI Average 95CI Average 95CI ppm % SRI [% SRI % SRI [% SRI % SRI [% SRI % SRI [% SRI enzyme (dE) (dE)] (dE) (dE)] (dE) (dE)] (dE) (dE)] 0 0.15 0.03 0.15 0.03 0.07 0.01 0.07 0.01 0.2 0.17 0.04 0.14 0.02 0.11 0.01 0.15 0.01 0.5 0.19 0.02 0.19 0.04 0.13 0.01 0.16 0.03 1 0.20 0.03 0.22 0.03 0.17 0.01 0.17 0.01 1.5 0.24 0.03 0.25 0.04 0.17 0.02 0.20 0.02
TABLE-US-00063 TABLE 7.8B Cleaning performance of PhuPro1 at 16.degree. C. EMPA-116 EMPA-117 Purafect Prime Purafect Prime HA PhuPro1 HA PhuPro1 Average 95CI Average 95CI Average 95CI Average 95CI ppm % SRI [% SRI % SRI [% SRI % SRI [% SRI % SRI [% SRI enzyme (dE) (dE)] (dE) (dE)] (dE) (dE)] (dE) (dE)] 0 0.14 0.02 0.14 0.02 0 12 0.01 0 12 0.01 0.2 0.19 0.02 0.17 0.03 0.17 0.02 0.14 0.03 0.5 0.22 0.03 0.28 0.04 0.20 0.03 0.22 0.01 1 0.24 0.02 0.26 0.02 0.20 0.01 0.24 0.04 1.5 0.23 0.03 0.26 0.03 0.23 0.02 0.25 0.02 EMPA-112 CFT C-10 Purafect Prime Purafect Prime HA PhuPro1 HA PhuPro1 Average 95CI Average 95CI Average 95CI Average 95I ppm % SRI [% SRI % SRI [% SRI % SRI [% SRI % SRI [% SRI enzyme (dE) (dE)] (dE) (dE)] (dE) (dE)] (dE) (dE)] 0 0.09 0.03 0.09 0.03 0.07 0.01 0.07 0.01 0.2 0.07 0.01 0.09 0.02 0.08 0.02 0.06 0.01 0.5 0.11 0.02 0.12 0.03 0.10 0.01 0.09 0.01 1 0.11 0.02 0.12 0.02 0.13 0.01 0.15 0.01 1.5 0.13 0.03 0.19 0.03 0.13 0.01 0.11 0.01
Example 8.1
Cloning of Paenibacillus amylolyticus Metalloprotease PamPro1
[0444] A strain (DSM11747) of Paenibacillus amylolyticus was selected as a potential source of enzymes which may be useful in various industrial applications. Genomic DNA for sequencing was obtained by first growing the strain on Heart Infusion agar plates (Difco) at 37.degree. C. for 24 hr. Cell material was scraped from the plates and used to prepare genomic DNA with the ZF Fungal/Bacterial DNA miniprep kit from Zymo (Cat No. D6005). The genomic DNA was used for genome sequencing. The entire genome of the Paenibacillus amylolyticus strain was sequenced by BaseClear (Leiden, The Netherlands) using the Illumina's next generation sequencing technology. After assembly of the data, contigs were annotated by BioXpr (Namur, Belgium). One of the genes identified after annotation in Paenibacillus amylolyticus encodes a metalloprotease and the sequence of this gene, called PamPro1, is provided in SEQ ID NO: 36. The corresponding protein encoded by the PamPro1 gene is shown in SEQ ID NO: 37. At the N-terminus, the protein has a signal peptide with a length of 25 amino acids as predicted by SignalP version 4.0 (Nordahl Petersen et al. (2011) Nature Methods, 8:785-786). The presence of a signal sequence suggests that PamPro1 is a secreted enzyme. The propeptide region was predicted based on protein sequence alignment with the Paenibacillus polymyxa Npr protein (Takekawa et al. (1991) Journal of Bacteriology, 173 (21): 6820-6825). The predicted mature region of PamPro1 protein is shown in SEQ ID NO: 3.
[0445] The nucleotide sequence of the PamPro1 gene isolated from Paenibacillus amylolyticus is set forth as SEQ ID NO: 36. The sequence encoding the predicted native signal peptide is shown in italics:
TABLE-US-00064 ATGAAATTCGCCAAAGTTATGCCAACAATTCTTGGAGGAGCTCTTTTGCTC GCTTCCGTATCCTCTGCTACTGCAGCTCCAGTGTCTGATCAATCCATTCCA CTTCAGGCCCCTTATGCCTCTGAGGGGGGTATTCCATTGAACAGTGGAACA GATGACACTATCTTTAATTATCTTGGACAGCAGGAACAATTTCTGAATTCC GATGTGAAATCCCAGCTCAAAATTGTCAAAAGAAACACAGATACATCTGGC GTAAGACACTTCCGCCTGAAACAGTATATTAAAGGTATCCCGGTTTATGGT GCAGAACAGACGGTCCACCTGGACAAAACCGGAGCCGTGAGCTCCGCACTT GGCGATCTTCCACCGATTGAAGAGCAGGCCATTCCGAATGATGGTGTAGCC GAGATCAGCGGAGAAGACGCGATCCAGATTGCAACCGAAGAAGCAACCTCC CGGATTGGAGAGCTTGGTGCCGCGGAAATCACGCCTCAAGCTGAATTGAAC ATCTATCATCATGAAGAAGATGGTCAGACATATCTGGTTTACATTACGGAA GTAAACGTACTGGAACCTGCCCCTCTACGGACCAAATATTTCATTAACGCA GTGGATGGCAGTATCGTATCCCAGTTTGACCTCATTAACTTCGCTACTGGA ACAGGTACAGGTGTACTCGGTGATACCAAAACCCTGACAACCACCCAATCC GGCAGCACCTTCCAACTGAAAGACACCACTCGTGGCAATGGCATCCAAACG TATACGGCAAACAATGGCTCCTCACTGCCTGGTAGCTTGCTTACAGATTCG GATAATGTATGGACCGATCGTGCAGGTGTAGATGCTCATGCTCATGCCGCT GCTACGTATGATTTCTACAAAAACAAATTCAACCGTAACGGTATTAATGGT AACGGATTGTTGATCAGATCAACCGTGCACTACGGCTCCAATTACAATAAC GCCTTCTGGAACGGGGCACAGATTGTCTTTGGTGACGGAGATGGAACGATG TTCCGATCCCTGTCTGGTGATCTGGATGTTGTGGGTCATGAATTGACGCAT GGTGTTATTGAATATACAGCCAATCTGGAATATCGCAATGAACCAGGTGCA CTCAATGAAGCCTTTGCCGATATTTTCGGTAATACGATCCAAAGCAAAAAC TGGCTGCTCGGTGATGATATCTACACACCTAACACTCCAGGAGATGCGCTG CGCTCCCTCTCCAACCCTACATTGTATGGTCAACCTGACAAATACAGCGAT CGCTACACAGGCTCACAGGACAACGGCGGTGTCCATATCAACAGTGGTATC ATCAATAAAGCCTATTTCCTTGCTGCTCAAGGCGGAACACATAATGGTGTG ACTGTTACCGGAATCGGCCGGGATAAAGCGATCCAGATTTTCTACAGCACA CTGGTGAACTACCTGACACCAACGTCCAAATTTGCCGCTGCCAAAACAGCT ACCATTCAAGCAGCCAAAGATCTGTACGGAGCAACTTCCGCTGAAGCTACT GCTATTACCAAAGCATATCAAGCTGTAGGCCTG
[0446] The amino acid sequence of the PamPro1 precursor protein is set forth as SEQ ID NO: 37. The predicted signal sequence is shown in italics, and the predicted propeptide is shown in underlined text:
TABLE-US-00065 MKFAKVMPTILGGALLLASVSSATAAPVSDQSIPLQAPYASEGGIPLNSGT DDTIFNYLGQQEQFLNSDVKSQLKIVKRNTDTSGVRHFRLKQYIKGIPVYG AEQTVHLDKTGAVSSALGDLPPIEEQAIPNDGVAEISGEDAIQIATEEATS RIGELGAAEITPQAELNIYHHEEDGQTYLVYITEVNVLEPAPLRTKYFINA VDGSIVSQFDLINFATGTGTGVLGDTKTLTTTQSGSTFQLKDTTRGNGIQT YTANNGSSLPGSLLTDSDNVWTDRAGVDAHAHAAATYDFYKNKFNRNGING NGLLIRSTVHYGSNYNNAFWNGAQIVFGDGDGTMFRSLSGDLDVVGHELTH GVIEYTANLEYRNEPGALNEAFADIFGNTIQSKNWLLGDDIYTPNTPGDAL RSLSNPTLYGQPDKYSDRYTGSQDNGGVHINSGIINKAYFLAAQGGTHNGV TVTGIGRDKAIQIFYSTLVNYLTPTSKFAAAKTATIQAAKDLYGATSAEAT AITKAYQAVGL
[0447] The amino acid sequence of the predicted mature form of PamPro1 is set forth as SEQ ID NO: 38:
TABLE-US-00066 ATGTGTGVLGDTKTLTTTQSGSTFQLKDTTRGNGIQTYTANNGSSLPGSLL TDSDNVWTDRAGVDAHAHAAATYDFYKNKFNRNGINGNGLLIRSTVHYGSN YNNAFWNGAQIVFGDGDGTMFRSLSGDLDVVGHELTHGVIEYTANLEYRNE PGALNEAFADIFGNTIQSKNWLLGDDIYTPNTPGDALRSLSNPTLYGQPDK YSDRYTGSQDNGGVHINSGIINKAYFLAAQGGTHNGVTVTGIGRDKAIQIF YSTLVNYLTPTSKFAAAKTATIQAAKDLYGATSAEATAITKAYQAVGL
Example 8.2
Expression of Paenibacillus amylolyticus Metalloprotease PamPro1
[0448] The DNA sequence of the propeptide-mature form of PamPro1 was synthesized and inserted into the Bacillus subtilis expression vector p2JM103BBI (Vogtentanz, Protein Expr Purif, 55:40-52, 2007) by Generay (Shanghai, China), resulting in plasmid pGX146(AprE-PamPro1) (FIG. 1). Ligation of this gene encoding the PamPro1 protein into the digested vector resulted in the addition of three codons (Ala-Gly-Lys) between the 3' end of the B. subtilis AprE signal sequence and the 5' end of the predicted PamPro1 native propeptide. The gene has an alternative start codon (GTG). The resulting plasmid shown in FIG. 8.1, labeled pGX146(AprE-PamPro1) contains an AprE promoter, an AprE signal sequence used to direct target protein secretion in B. subtilis, and the synthetic nucleotide sequence encoding the predicted propeptide and mature regions of PamPro1 (SEQ ID NO: 39). The translation product of the synthetic AprE-PamPro1 gene is shown in SEQ ID NO: 40.
[0449] The pGX146(AprE-PamPro1) plasmid was then transformed into B. subtilis cells (degU.sup.Hy 32, .DELTA.scoC) and the transformed cells were spread on Luria Agar plates supplemented with 5 ppm Chloramphenicol and 1.2% skim milk (Cat #232100, Difco). Colonies with the largest clear halos on the plates were selected and subjected to fermentation in a 250 ml shake flask with MBD medium (a MOPS based defined medium, supplemented with additional 5 mM CaCl.sub.2).
[0450] The broth from the shake flasks was concentrated and buffer-exchanged into the loading buffer containing 20 mM Tris-HCl (pH 8.5), 1 mM CaCl.sub.2 and 10% propylene glycol using a VivaFlow 200 ultra filtration device (Sartorius Stedim). After filtering, this sample was applied to an 80 ml Q Sepharose High Performance column pre-equilibrated with the loading buffer above; and the active flow-through fractions were collected and concentrated. The sample was loaded onto a 320 ml Superdex 75 gel filtration column pre-equilibrated with the loading buffer described above containing 0.15 M NaCl. The corresponding active purified protein fractions were further pooled and concentrated via 10K Amicon Ultra for further analyses.
[0451] The nucleotide sequence of the synthesized PamPro1 gene in plasmid pGX146(AprE-PamPro1) is depicted in SEQ ID NO: 39. The sequence encoding the three residue addition (AGK) is shown in bold:
TABLE-US-00067 GTGAGAAGCAAAAAATTGTGGATCAGCTTGTTGTTTGCGTTAACGTTAATC TTTACGATGGCGTTCAGCAACATGAGCGCGCAGGCTGCTGGAAAAGCTCCG GTTAGCGACCAGTCAATCCCTCTTCAAGCACCGTATGCCAGCGAAGGAGGC ATTCCGCTTAACAGCGGCACGGACGACACGATTTTCAATTACCTGGGCCAA CAGGAGCAGTTCCTGAACAGCGACGTCAAGAGCCAGCTGAAGATCGTCAAA AGAAACACAGACACATCAGGCGTGAGACACTTCAGACTGAAGCAATACATC AAGGGCATCCCGGTTTATGGCGCTGAACAAACGGTTCACCTGGACAAAACA GGCGCAGTTTCATCAGCACTGGGAGATCTGCCGCCGATTGAAGAGCAAGCA ATCCCGAATGATGGAGTTGCGGAAATTAGCGGCGAGGATGCAATCCAAATC GCGACGGAGGAGGCTACATCAAGAATTGGAGAACTTGGCGCAGCGGAGATT ACACCGCAGGCTGAACTGAACATCTATCACCATGAGGAAGACGGCCAGACG TACCTGGTTTACATTACGGAAGTGAACGTGCTGGAACCGGCACCTCTGAGA ACAAAGTACTTTATCAACGCGGTTGACGGCAGCATCGTCTCACAGTTCGAC CTGATTAACTTCGCCACGGGAACAGGAACGGGCGTTCTTGGAGACACAAAG ACGCTGACGACGACGCAGTCAGGCAGCACATTCCAGCTGAAGGACACAACA AGAGGCAACGGCATCCAAACGTACACGGCGAACAATGGATCATCACTGCCG GGCTCACTGCTGACGGATTCAGATAACGTGTGGACGGATAGAGCTGGCGTT GACGCGCATGCTCACGCTGCTGCGACGTACGACTTCTACAAGAACAAGTTC AACAGAAACGGCATTAACGGAAATGGCCTGCTGATCAGAAGCACGGTGCAT TATGGCTCAAACTACAACAACGCTTTTTGGAACGGCGCACAGATCGTGTTT GGCGACGGCGATGGCACAATGTTTAGAAGCCTGTCAGGAGACCTGGATGTG GTGGGCCACGAACTGACGCACGGCGTGATCGAGTATACGGCGAACCTTGAA TATAGAAACGAGCCGGGAGCACTGAATGAGGCGTTCGCGGACATTTTCGGC AACACAATCCAGAGCAAAAACTGGCTGCTGGGCGACGATATCTATACACCG AACACACCGGGCGATGCACTGAGATCACTGTCAAATCCGACGCTGTATGGC CAACCGGATAAGTACTCAGACAGATATACGGGCAGCCAAGACAATGGCGGC GTTCACATCAACTCAGGCATCATCAACAAGGCTTACTTCCTTGCGGCCCAA GGAGGAACACATAACGGCGTTACAGTTACAGGCATTGGCAGAGACAAGGCG ATCCAGATCTTTTACAGCACGCTGGTGAACTACCTGACACCTACGTCAAAG TTTGCCGCAGCGAAAACAGCAACAATTCAGGCGGCTAAAGACCTGTACGGA GCGACATCAGCCGAGGCCACAGCAATTACAAAAGCATATCAAGCAGTTGGC CTTTAA
[0452] The amino acid sequence of the PamPro1 precursor protein expressed from plasmid pGX146(AprE-PamPro1) is depicted in SEQ ID NO: 40. The predicted signal sequence is shown in italics, the three residue addition (AGK) is shown in bold, and the predicted pro-peptide is shown in underlined text.
TABLE-US-00068 MRSKKLWISLLFALTLIFTMAFSNMSAQAAGKAPVSDQSIPLQAPYASEGG IPLNSGTDDTIFNYLGQQEQFLNSDVKSQLKIVKRNTDTSGVRHFRLKQYI KGIPVYGAEQTVHLDKTGAVSSALGDLPPIEEQAIPNDGVAEISGEDAIQI ATEEATSRIGELGAAEITPQAELNIYHHEEDGQTYLVYITEVNVLEPAPLR TKYFINAVDGSIVSQFDLINFATGTGTGVLGDTKTLTTTQSGSTFQLKDTT RGNGIQTYTANNGSSLPGSLLTDSDNVWTDRAGVDAHAHAAATYDFYKNKE NRNGINGNGLLIRSTVHYGSNYNNAFWNGAQIVFGDGDGTMFRSLSGDLDV VGHELTHGVIEYTANLEYRNEPGALNEAFADIFGNTIQSKNWLLGDDIYTP NTPGDALRSLSNPTLYGQPDKYSDRYTGSQDNGGVHINSGIINKAYFLAAQ GGTHNGVTVTGIGRDKAIQIFYSTLVNYLTPTSKFAAAKTATIQAAKDLYG ATSAEATAITKAYQAVGL
Example 8.3
Proteolytic Activity of Metalloprotease PamPro1
[0453] The proteolytic activity of purified metalloprotease PamPro1 was measured in 50 mM Tris (pH 7), using azo-casein (Cat #74H7165, Megazyme) as a substrate. Prior to the reaction, the enzyme was diluted with Milli-Q water (Millipore) to specific concentrations. The azo-casein was dissolved in 100 mM Tris buffer (pH 7) to a final concentration of 1.5% (w/v). To initiate the reaction, 50 .mu.l of the diluted enzyme (or Milli-Q H.sub.2O alone as the blank control) was added to the non-binding 96-well Microtiter Plate (96-MTP) (Corning Life Sciences, #3641) placed on ice, followed by the addition of 50 .mu.l of 1.5% azo-casein. After sealing the 96-MTP, the reaction was carried out in a Thermomixer (Eppendorf) at 40.degree. C. and 650 rpm for 10 min. The reaction was terminated by adding 100 .mu.l of 5% Trichloroacetic Acid (TCA). Following equilibration (5 min at the room temperature) and subsequent centrifugation (2000 g for 10 min at 4.degree. C.), 120 .mu.l supernatant was transferred to a new 96-MTP, and absorbance of the supernatant was measured at 440 nm (A.sub.440) using a SpectraMax 190. Net A.sub.440 was calculated by subtracting the A.sub.440 of the blank control from that of enzyme, and then plotted against different protein concentrations (from 1.25 ppm to 40 ppm). Each value was the mean of triplicate assays. The proteolytic activity is shown as Net A.sub.44o. The proteolytic assay with azo-casein as the substrate (shown in FIG. 8.2) indicates that PamPro1 is an active protease.
Example 8.4
pH Profiles of Metalloprotease PamPro1
[0454] With azo-casein as the substrate, the pH profiles of metalloprotease PamPro1 were studied in 12.5 mM acetate/Bis-Tris/HEPES/CHES buffer with different pH values (ranging from pH 4 to 11). To initiate the assay, 50 .mu.l of 25 mM acetate/Bis-Tris/HEPES/CHES buffer with a specific pH was first mixed with 2 .mu.l Milli-Q H.sub.2O diluted enzyme (125 ppm) in a 96-MTP placed on ice, followed by the addition of 48 .mu.l of 1.5% (w/v) azo-casein prepared in H2O. The reaction was performed and analyzed as described in Example 8.3. Enzyme activity at each pH was reported as the relative activity, where the activity at the optimal pH was set to be 100%. The pH values tested were 4, 5, 6, 7, 8, 9, 10 and 11. Each value was the mean of triplicate assays. As shown in FIG. 8.3, the optimal pH of PamPro1 is about 8, with greater than 70% of maximal activity retained between 7 and 9.5.
Example 8.5
Temperature Profile of Metalloprotease PamPro1
[0455] The temperature profile of metalloprotease PamPro1 was analyzed in 50 mM Tris buffer (pH 7) using the azo-casein assays. The enzyme sample and azo-casein substrate were prepared as in Example 8.3. Prior to the reaction, 50 .mu.l of 1.5% azo-casein and 45 .mu.l Milli-Q H.sub.2O were mixed in a 200 .mu.l PCR tube, which was then subsequently incubated in a Peltier Thermal Cycler (BioRad) at desired temperatures (i.e. 20-90.degree. C.) for 5 min. After the incubation, 5 .mu.l of diluted enzyme (50 ppm) or H.sub.2O (the blank control) was added to the substrate mixture, and the reaction was carried out in the Peltier Thermal Cycle for 10 min at different temperatures. To terminate the reaction, each assay mixture was transferred to a 96-MTP containing 100 .mu.l of 5% TCA per well. Subsequent centrifugation and absorbance measurement were performed as described in Example 8.3. The activity was reported as the relative activity, where the activity at the optimal temperature was set to be 100%. The tested temperatures are 20, 30, 40, 50, 60, 70, 80, and 90.degree. C. Each value was the mean of duplicate assays (the value varies no more than 5%). The data in FIG. 8.4 suggest that PamPro1 showed an optimal temperature at about 50.degree. C., and retained greater than 70% of its maximum activity between 45 and 55.degree. C.
Example 8.6
Cleaning Performance of Metalloprotease PamPro1
[0456] The cleaning performance of PamPro1 was tested using PA-S-38 (egg yolk, with pigment, aged by heating) microswatches (CFT-Vlaardingen, The Netherlands) at pH 6 and 8 using a model automatic dishwashing (ADW) detergent. Prior to the reaction, purified protease samples were diluted with a dilution solution containing 10 mM NaCl, 0.1 mM CaCl.sub.2, 0.005% TWEEN.RTM. 80 and 10% propylene glycol to the desired concentrations. The reactions were performed in AT detergent with 100 ppm water hardness (Ca.sup.2+:Mg.sup.2+=3:1) (detergent composition shown in Table 8.1). To initiate the reaction, 180 .mu.l of the AT detergent buffered at pH 6 or pH 8 was added to a 96-MTP placed with PA-S-38 microswatches, followed by the addition of 20 .mu.l of diluted enzymes (or the dilution solution as the blank control). The 96-MTP was sealed and incubated in an incubator/shaker for 30 min at 50.degree. C. and 1150 rpm. After incubation, 100 .mu.l of wash liquid from each well was transferred to a new 96-MTP, and its absorbance was measured at 405 nm (referred here as the "Initial performance") using a spectrophotometer. The remaining wash liquid in the 96-MTP was discarded and the microswatches were rinsed once with 200 .mu.l water. Following the addition of 180 .mu.l of 0.1 M CAPS buffer (pH 10), the second incubation was carried out in the incubator/shaker at 50.degree. C. and 1150 rpm for 10 min. One hundred microliters of the resulting wash liquid was transferred to a new 96-MTP, and its absorbance measured at 405 nm (referred here as the "Wash-off"). The sum of two absorbance measurements ("Initial performance" plus "Wash-off") gives the "Total performance", which measures the protease activity on the model stain; and Net A.sub.405 was subsequently calculated by subtracting the A.sub.405 of the "Total performance" of the blank control from that of the enzyme. Dose response in cleaning the PA-S-38 microswatches at pH 6 and pH 8 in AT dish detergent for PamPro1 is shown in FIGS. 5A and 5B.
TABLE-US-00069 TABLE 8.1 Composition of AT dish detergent formula with bleach Ingredient Concentration (mg/ml) MGDA (methylglycinediacetic acid) 0.143 Sodium citrate 1.86 Citric acid* varies PAP (peracid N,N- 0.057 phthaloylaminoperoxycaproic acid) Plurafac .RTM. LF 18B (a non-ionic surfactant) 0.029 Bismuthcitrate 0.006 Bayhibit .RTM. S (Phosphonobutantricarboxylic 0.006 acid sodium salt) Acusol .TM. 587 (a calcium polyphosphate 0.029 inhibitor) PEG 6000 0.043 PEG 1500 0.1 *The pH of the AT formula detergent is adjusted to the desired value (pH 6 or 8) by the addition of 0.9M citric acid.
Example 8.7
Comparison of PamPro1 to Other Proteases
A. Identification of Homologous Proteases
[0457] Homologs were identified by a BLAST search (Altschul et al., Nucleic Acids Res, 25:3389-402, 1997) against the NCBI non-redundant protein database and the Genome Quest Patent database with search parameters set to default values. The predicted mature protein amino acid sequence for PamPro1 (SEQ ID NO: 38) was used as the query sequence. Percent identity (PID) for both search sets is defined as the number of identical residues divided by the number of aligned residues in the pairwise alignment. Tables 8.2A and 8.2B provide a list of sequences with the percent identity to PamPro1. The length in Table 8.2 refers to the entire sequence length of the homologous proteases.
TABLE-US-00070 TABLE 8.2A List of sequences with percent identity to PamPro1 protein identified from the NCBI non-redundant protein database PID to Accession # PamPro1 Organism Length P23384 56 Bacillus caldolyticus 544 P00800 56 Bacillus thermoproteolyticus 548 ZP_08640523.1 57 Brevibacillus laterosporus 564 LMG 15441 BAA06144.1 57 Lactobacillus sp. 566 YP_003872180.1 58 Paenibacillus polymyxa E681 587 ZP_04149724.1 59 Bacillus pseudomycoides 566 DSM 12442 EJR46541.1 60 Bacillus cereus VD107 566 YP_001373863.1 60 Bacillus cytotoxicus NVH 391-98 565 ZP_10738945.1 61 Brevibacillus sp. CF112 528 YP_004646155.1 61 Paenibacillus mucilaginosus 525 KNP414 ZP_02326602.1 62 Paenibacillus larvae subsp. 520 larvae BRL-230010 P43263 63 Brevibacillus brevis 527 ZP_09775365.1 64 Paenibacillus sp. Aloe-11 580 ZP_09077634.1 65 Paenibacillus elgii B69 529 ZP_09071078.1 68 Paenibacillus larvae subsp. 529 larvae B-3650 ZP_08511445.1 69 Paenibacillus sp. HGF7 525 YP_005073223.1 70 Paenibacillus terrae HPL-003 591 YP_003948511.1 71 Paenibacillus polymyxa SC2 592 ZP_10241030.1 71 Paenibacillus peoriae KCTC 3763 593
TABLE-US-00071 TABLE 8.2B List of sequences with percent identity to PamPro1 protein identified from the Genome Quest Patent database PID to Patent # PamPro1 Organism Length US7335504-0030 56.63 Bacillus thermoproteolyticus 316 US20120107907-0184 56.91 Bacillus caldoyticus 319 JP2006124323-0003 56.96 Bacillus thermoproteolyticus 316 JP1993199872-0001 56.96 Bacillus sp. 316 JP1997000255-0001 56.96 empty 548 US6518054-0001 57.23 Bacillus sp. 319 US20120107907-0176 57.23 Bacillus stearothermophilis 548 US8114656-0183 57.28 Bacillus stearothermophilis 316 US20120009651-0002 57.28 Geobacillus 548 caldoproteolyticus JP2011103791-0020 57.28 Geobacillus 552 stearothermophilus WO2012110562-0006 57.88 Bacillus megaterium 320 EP2390321-0178 57.88 Bacillus thuringiensis 566 US6518054-0002 57.93 Bacillus sp. 316 WO2012110562-0007 58.25 Bacillus cereus 320 JP1995184649-0001 58.52 Lactobacillus sp. 566 EP2178896-0184 58.52 Bacillus anthracis 566 EP2390321-0195 59.55 Bacillus cereus 317 WO2012110563-0005 59.87 Bacillus cereus 320 US20080293610-0186 63.25 Bacillus brevis 304 JP2005229807-0018 71.19 Paenibacillus polymyxa 566 US8114656-0187 71.43 Bacillus polymyxa 302
B. Alignment of Homologous Protease Sequences
[0458] The amino acid sequence of the predicted mature PamPro1 (SEQ ID NO: 38) was aligned with thermolysin (P00800, Bacillus thermoproteolyticus), and protease from Paenibacillus peoriae KCTC 3763 (YP 005073223.1) using CLUSTALW software (Thompson et al., Nucleic Acids Research, 22:4673-4680, 1994) with the default parameters. FIG. 8.6 shows the alignment of PamPro1 with these protease sequences.
C. Phylogenetic Tree
[0459] A phylogenetic tree for full length sequences of PamPro1 (SEQ ID NO: 37) was built using sequences of representative homologs from Table 8.2A and the Neighbor Joining method (NJ) (Saitou, N.; and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing Guide Trees. MolBiol.Evol. 4, 406-425). The NJ method works on a matrix of distances between all pairs of sequences to be analyzed. These distances are related to the degree of divergence between the sequences. The phylodendron-phylogenetic tree printer software (http://iubio.bio.indiana.edu/treeapp/treeprint-form.html) was used to display the phylogenetic tree shown in FIG. 8.7.
Example 9
Comparison of the Various Paenibacillus Metalloproteases with Other Bacterial Metalloprotease Homologs
A. Alignment of Homologous Protease Sequences
[0460] The amino acid sequence of the predicted mature sequences for the Paenibacillus proteases described in Examples 1.1 to 8.7 were aligned with related bacterial metalloproteases using CLUSTALW software (Thompson et al., Nucleic Acids Research, 22:4673-4680, 1994) with the default parameters. FIG. 9.1 shows the alignment of the various Paenibacillus metalloproteases with other bacterial metalloprotease homologs.
B. Phylogenetic Tree
[0461] A phylogenetic tree for full length sequences of the metalloproteases aligned in FIG. 9.1 was created using the Neighbor Joining method (NJ) (Saitou, N.; and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing Guide Trees. MolBiol.Evol. 4, 406-425). The NJ method works on a matrix of distances between all pairs of sequences to be analyzed. These distances are related to the degree of divergence between the sequences. The phylodendron-phylogenetic tree printer software (http://iubio.bio.indiana.edu/treeapp/treeprint-form.html) was used to display the phylogenetic tree shown in FIG. 9.2, where one can observe the clustering of the sequences from Paenibacillus genus.
[0462] While preferred embodiments of the present invention have been shown and described herein, it will be obvious to those skilled in the art that such embodiments are provided by way of example only. Numerous variations, changes, and substitutions will now occur to those skilled in the art without departing from the invention. It should be understood that various alternatives to the embodiments of the invention described herein can be employed in practicing the invention. It is intended that the following claims define the scope of the invention and that methods and structures within the scope of these claims and their equivalents be covered thereby.
Sequence CWU 1
1
6811785DNAPaenibacillus sp.misc_feature(1)..(1785)nucleotide
sequence of the PspPro3 gene isolated from Paenibacillus sp.
1atgttaatga aaaaagtatg ggtttcgctt cttggaggag cgatgttatt agggtctgta
60gcgtctggtg catcagcagc ggagagttcc gtttcggggc cggctcagct tacgccaacc
120ttccatgccg aacaatggaa agcaccttca tcggtatcgg gtgatgacat
cgtatggagc 180tatttaaatc ggcaaaagaa aacgttgctg ggtacggaca
gcaccagtgt ccgtgatcaa 240ttccgtatcg tagatcgcac aagcgacaaa
tccggcgtga gccattatcg gctgaagcaa 300tatgtaaacg gaattcccgt
atatggagct gaacagacca ttcatgtggg caaatccggt 360gaagtgacct
cttatctggg agccgtgatt actgaggatc agcaagaaga agctacgcaa
420ggtacaactc cgaaaatcag cgcttctgaa gcggtccata ccgcatatca
ggaggcagct 480acacgggttc aagccctccc tacctccgat gatacgattt
ctaaagatgc ggaggagcca 540agcagtgtaa gcaaagacac ttactccgaa
gcagctaaca acggaaaaac gagttctgtt 600gaaaaggaca agctcagcct
tgagaaagcg gctgacctga aagatagcaa aattgaagcg 660gtggaggcag
agccaaactc cattgccaaa atcgccaacc tgcagcctga ggtagatcct
720aaagccgaac tatatttcta tgcgaagggc gatgcattgc agctggttta
tgtgactgag 780gttaatattt tgcagcctgc gccgctgcgt acacgctaca
tcattgacgc caatgatggc 840aaaatcgtat cccagtatga catcattaat
gaagcgacag gcacaggcaa aggtgtactc 900ggtgatacca aaacattcaa
cactactgct tccggcagca gctaccagtt aagagatacg 960actcgcggga
atggaatcgt gacttacacg gcctccaacc gtcaaagcat cccaggtacg
1020atcctgaccg atgccgataa cgtatggaat gatccagccg gcgtggatgc
ccacgcttat 1080gcagccaaaa cctatgatta ttataaggaa aagttcaatc
gcaacagcat tgacggacga 1140ggcctgcagc tccgttcgac agttcattac
ggcaatcgtt acaacaacgc cttctggaac 1200ggctcccaaa tgacttatgg
agacggagac ggcaccacat ttatcgcttt tagcggtgat 1260ccggatgtag
ttggtcatga actcacacac ggtgttacgg agtatacttc caatttggaa
1320tattacggag aatccggtgc gttgaacgag gccttctcgg acatcatcgg
caatgacatc 1380cagcgtaaaa actggcttgt aggcgatgat atttacacgc
cacgcattgc gggtgatgca 1440cttcgttcta tgtccaatcc tacgctgtac
gatcaaccgg atcactattc gaacttgtac 1500agaggcagct ccgataacgg
cggcgttcat acgaacagcg gtattataaa taaagcctat 1560tatctgttgg
cacaaggcgg caccttccat ggtgtaactg tcaatgggat tggccgcgat
1620gcagcggttc aaatttacta cagcgccttt acgaactacc tgacttcttc
ttctgacttc 1680tccaatgcac gtgatgccgt tgtacaagcg gcaaaagatc
tctacggcgc gagctcggca 1740caagctaccg cagcagccaa atcttttgat
gctgtaggcg ttaac 17852595PRTPaenibacillus
sp.misc_feature(1)..(595)amino acid sequence of the PspPro3
precursor protein 2Met Leu Met Lys Lys Val Trp Val Ser Leu Leu Gly
Gly Ala Met Leu1 5 10 15Leu Gly Ser Val Ala Ser Gly Ala Ser Ala Ala
Glu Ser Ser Val Ser 20 25 30Gly Pro Ala Gln Leu Thr Pro Thr Phe His
Ala Glu Gln Trp Lys Ala 35 40 45Pro Ser Ser Val Ser Gly Asp Asp Ile
Val Trp Ser Tyr Leu Asn Arg 50 55 60Gln Lys Lys Thr Leu Leu Gly Thr
Asp Ser Thr Ser Val Arg Asp Gln65 70 75 80Phe Arg Ile Val Asp Arg
Thr Ser Asp Lys Ser Gly Val Ser His Tyr 85 90 95Arg Leu Lys Gln Tyr
Val Asn Gly Ile Pro Val Tyr Gly Ala Glu Gln 100 105 110Thr Ile His
Val Gly Lys Ser Gly Glu Val Thr Ser Tyr Leu Gly Ala 115 120 125Val
Ile Thr Glu Asp Gln Gln Glu Glu Ala Thr Gln Gly Thr Thr Pro 130 135
140Lys Ile Ser Ala Ser Glu Ala Val His Thr Ala Tyr Gln Glu Ala
Ala145 150 155 160Thr Arg Val Gln Ala Leu Pro Thr Ser Asp Asp Thr
Ile Ser Lys Asp 165 170 175Ala Glu Glu Pro Ser Ser Val Ser Lys Asp
Thr Tyr Ser Glu Ala Ala 180 185 190Asn Asn Gly Lys Thr Ser Ser Val
Glu Lys Asp Lys Leu Ser Leu Glu 195 200 205Lys Ala Ala Asp Leu Lys
Asp Ser Lys Ile Glu Ala Val Glu Ala Glu 210 215 220Pro Asn Ser Ile
Ala Lys Ile Ala Asn Leu Gln Pro Glu Val Asp Pro225 230 235 240Lys
Ala Glu Leu Tyr Phe Tyr Ala Lys Gly Asp Ala Leu Gln Leu Val 245 250
255Tyr Val Thr Glu Val Asn Ile Leu Gln Pro Ala Pro Leu Arg Thr Arg
260 265 270Tyr Ile Ile Asp Ala Asn Asp Gly Lys Ile Val Ser Gln Tyr
Asp Ile 275 280 285Ile Asn Glu Ala Thr Gly Thr Gly Lys Gly Val Leu
Gly Asp Thr Lys 290 295 300Thr Phe Asn Thr Thr Ala Ser Gly Ser Ser
Tyr Gln Leu Arg Asp Thr305 310 315 320Thr Arg Gly Asn Gly Ile Val
Thr Tyr Thr Ala Ser Asn Arg Gln Ser 325 330 335Ile Pro Gly Thr Ile
Leu Thr Asp Ala Asp Asn Val Trp Asn Asp Pro 340 345 350Ala Gly Val
Asp Ala His Ala Tyr Ala Ala Lys Thr Tyr Asp Tyr Tyr 355 360 365Lys
Glu Lys Phe Asn Arg Asn Ser Ile Asp Gly Arg Gly Leu Gln Leu 370 375
380Arg Ser Thr Val His Tyr Gly Asn Arg Tyr Asn Asn Ala Phe Trp
Asn385 390 395 400Gly Ser Gln Met Thr Tyr Gly Asp Gly Asp Gly Thr
Thr Phe Ile Ala 405 410 415Phe Ser Gly Asp Pro Asp Val Val Gly His
Glu Leu Thr His Gly Val 420 425 430Thr Glu Tyr Thr Ser Asn Leu Glu
Tyr Tyr Gly Glu Ser Gly Ala Leu 435 440 445Asn Glu Ala Phe Ser Asp
Ile Ile Gly Asn Asp Ile Gln Arg Lys Asn 450 455 460Trp Leu Val Gly
Asp Asp Ile Tyr Thr Pro Arg Ile Ala Gly Asp Ala465 470 475 480Leu
Arg Ser Met Ser Asn Pro Thr Leu Tyr Asp Gln Pro Asp His Tyr 485 490
495Ser Asn Leu Tyr Arg Gly Ser Ser Asp Asn Gly Gly Val His Thr Asn
500 505 510Ser Gly Ile Ile Asn Lys Ala Tyr Tyr Leu Leu Ala Gln Gly
Gly Thr 515 520 525Phe His Gly Val Thr Val Asn Gly Ile Gly Arg Asp
Ala Ala Val Gln 530 535 540Ile Tyr Tyr Ser Ala Phe Thr Asn Tyr Leu
Thr Ser Ser Ser Asp Phe545 550 555 560Ser Asn Ala Arg Asp Ala Val
Val Gln Ala Ala Lys Asp Leu Tyr Gly 565 570 575Ala Ser Ser Ala Gln
Ala Thr Ala Ala Ala Lys Ser Phe Asp Ala Val 580 585 590Gly Val Asn
5953304PRTPaenibacillus sp.misc_feature(1)..(304)amino acid
sequence of the predicted mature form of PspPro3 3Ala Thr Gly Thr
Gly Lys Gly Val Leu Gly Asp Thr Lys Thr Phe Asn1 5 10 15Thr Thr Ala
Ser Gly Ser Ser Tyr Gln Leu Arg Asp Thr Thr Arg Gly 20 25 30Asn Gly
Ile Val Thr Tyr Thr Ala Ser Asn Arg Gln Ser Ile Pro Gly 35 40 45Thr
Ile Leu Thr Asp Ala Asp Asn Val Trp Asn Asp Pro Ala Gly Val 50 55
60Asp Ala His Ala Tyr Ala Ala Lys Thr Tyr Asp Tyr Tyr Lys Glu Lys65
70 75 80Phe Asn Arg Asn Ser Ile Asp Gly Arg Gly Leu Gln Leu Arg Ser
Thr 85 90 95Val His Tyr Gly Asn Arg Tyr Asn Asn Ala Phe Trp Asn Gly
Ser Gln 100 105 110Met Thr Tyr Gly Asp Gly Asp Gly Thr Thr Phe Ile
Ala Phe Ser Gly 115 120 125Asp Pro Asp Val Val Gly His Glu Leu Thr
His Gly Val Thr Glu Tyr 130 135 140Thr Ser Asn Leu Glu Tyr Tyr Gly
Glu Ser Gly Ala Leu Asn Glu Ala145 150 155 160Phe Ser Asp Ile Ile
Gly Asn Asp Ile Gln Arg Lys Asn Trp Leu Val 165 170 175Gly Asp Asp
Ile Tyr Thr Pro Arg Ile Ala Gly Asp Ala Leu Arg Ser 180 185 190Met
Ser Asn Pro Thr Leu Tyr Asp Gln Pro Asp His Tyr Ser Asn Leu 195 200
205Tyr Arg Gly Ser Ser Asp Asn Gly Gly Val His Thr Asn Ser Gly Ile
210 215 220Ile Asn Lys Ala Tyr Tyr Leu Leu Ala Gln Gly Gly Thr Phe
His Gly225 230 235 240Val Thr Val Asn Gly Ile Gly Arg Asp Ala Ala
Val Gln Ile Tyr Tyr 245 250 255Ser Ala Phe Thr Asn Tyr Leu Thr Ser
Ser Ser Asp Phe Ser Asn Ala 260 265 270Arg Asp Ala Val Val Gln Ala
Ala Lys Asp Leu Tyr Gly Ala Ser Ser 275 280 285Ala Gln Ala Thr Ala
Ala Ala Lys Ser Phe Asp Ala Val Gly Val Asn 290 295
30041803DNAArtificial SequenceSynthetic nucleotide sequence of the
synthesized PspPro3 gene in plasmid pGX085(AprE- PspPro3)
4gtgagaagca aaaaattgtg gatcagcttg ttgtttgcgt taacgttaat ctttacgatg
60gcgttcagca acatgagcgc gcaggctgct ggaaaagcag aatcatcagt gtcaggaccg
120gctcagctta cgccgacgtt tcatgcagag cagtggaaag caccgagcag
cgttagcgga 180gatgacatcg tgtggagcta cctgaacaga cagaagaaaa
cgcttcttgg cacggacagc 240acgagcgtca gagaccagtt cagaatcgtg
gatagaacaa gcgacaaaag cggcgtcagc 300cattatagac tgaagcagta
tgtgaacgga atcccggttt atggcgcaga acaaacaatc 360catgtcggaa
agagcggcga agttacgagc tatctgggcg cggttattac agaggaccag
420caagaggagg ctacacaagg cacgacaccg aaaatttcag catcagaggc
agttcatacg 480gcctaccaag aagctgcaac gagagttcaa gccctgccta
cgtcagatga tacaatcagc 540aaagacgctg aggaacctag ctcagttagc
aaggacacgt atagcgaagc cgcgaacaat 600ggcaagacgt caagcgtgga
aaaagacaag ctttcactgg agaaggccgc tgatctgaaa 660gactcaaaga
tcgaggctgt ggaagcggaa ccgaatagca ttgcaaagat tgccaacctg
720caaccggagg tggacccgaa ggcggagctg tatttctacg ctaaaggcga
tgcactgcaa 780ctggtttacg tcacggaggt taacatcctg cagccggcac
cgcttagaac gagatacatc 840attgacgcga acgacggcaa gatcgtgagc
cagtacgaca ttatcaacga ggccacggga 900acgggcaagg gagtccttgg
cgacacgaag acattcaata caacggcctc aggctcatca 960taccagctga
gagacacgac gagaggcaac ggaatcgtca cgtacacggc tagcaataga
1020cagagcattc cgggcacaat ccttacggac gcagacaatg tgtggaatga
cccggcaggc 1080gtggacgcac atgcctacgc agcgaagacg tacgactact
acaaggagaa gttcaacaga 1140aacagcatcg acggaagagg actgcaactt
agaagcacgg tgcattacgg caacagatac 1200aacaacgctt tctggaacgg
cagccaaatg acgtatggag acggcgatgg aacaacgttt 1260atcgcattct
caggcgaccc tgacgttgtg ggacatgaac tgacgcatgg agtcacagaa
1320tacacgagca atctggagta ttacggagaa tcaggcgcac ttaatgaggc
cttcagcgac 1380atcatcggaa acgacatcca gagaaagaac tggctggttg
gcgatgatat ctacacgccg 1440agaattgcgg gcgacgcgct gagatcaatg
agcaacccta cgctgtacga tcagccggat 1500cattacagca acctgtatag
aggctcaagc gataatggcg gcgtgcatac aaacagcggc 1560atcatcaaca
aagcctatta tctgctggcg caaggcggca cattccatgg cgttacagtt
1620aatggcattg gcagagacgc agccgtgcag atctactaca gcgcattcac
gaattacctg 1680acatcaagca gcgacttttc aaatgcaaga gatgcagtgg
tgcaggcggc taaagacctt 1740tatggagctt caagcgctca ggccacagct
gcggcaaaaa gcttcgacgc ggttggagtg 1800aat 18035601PRTArtificial
SequenceSynthetic amino acid sequence of the PspPro3 precursor
protein expressed from plasmid pGX085(AprE- PspPro3) 5Met Arg Ser
Lys Lys Leu Trp Ile Ser Leu Leu Phe Ala Leu Thr Leu1 5 10 15Ile Phe
Thr Met Ala Phe Ser Asn Met Ser Ala Gln Ala Ala Gly Lys 20 25 30Ala
Glu Ser Ser Val Ser Gly Pro Ala Gln Leu Thr Pro Thr Phe His 35 40
45Ala Glu Gln Trp Lys Ala Pro Ser Ser Val Ser Gly Asp Asp Ile Val
50 55 60Trp Ser Tyr Leu Asn Arg Gln Lys Lys Thr Leu Leu Gly Thr Asp
Ser65 70 75 80Thr Ser Val Arg Asp Gln Phe Arg Ile Val Asp Arg Thr
Ser Asp Lys 85 90 95Ser Gly Val Ser His Tyr Arg Leu Lys Gln Tyr Val
Asn Gly Ile Pro 100 105 110Val Tyr Gly Ala Glu Gln Thr Ile His Val
Gly Lys Ser Gly Glu Val 115 120 125Thr Ser Tyr Leu Gly Ala Val Ile
Thr Glu Asp Gln Gln Glu Glu Ala 130 135 140Thr Gln Gly Thr Thr Pro
Lys Ile Ser Ala Ser Glu Ala Val His Thr145 150 155 160Ala Tyr Gln
Glu Ala Ala Thr Arg Val Gln Ala Leu Pro Thr Ser Asp 165 170 175Asp
Thr Ile Ser Lys Asp Ala Glu Glu Pro Ser Ser Val Ser Lys Asp 180 185
190Thr Tyr Ser Glu Ala Ala Asn Asn Gly Lys Thr Ser Ser Val Glu Lys
195 200 205Asp Lys Leu Ser Leu Glu Lys Ala Ala Asp Leu Lys Asp Ser
Lys Ile 210 215 220Glu Ala Val Glu Ala Glu Pro Asn Ser Ile Ala Lys
Ile Ala Asn Leu225 230 235 240Gln Pro Glu Val Asp Pro Lys Ala Glu
Leu Tyr Phe Tyr Ala Lys Gly 245 250 255Asp Ala Leu Gln Leu Val Tyr
Val Thr Glu Val Asn Ile Leu Gln Pro 260 265 270Ala Pro Leu Arg Thr
Arg Tyr Ile Ile Asp Ala Asn Asp Gly Lys Ile 275 280 285Val Ser Gln
Tyr Asp Ile Ile Asn Glu Ala Thr Gly Thr Gly Lys Gly 290 295 300Val
Leu Gly Asp Thr Lys Thr Phe Asn Thr Thr Ala Ser Gly Ser Ser305 310
315 320Tyr Gln Leu Arg Asp Thr Thr Arg Gly Asn Gly Ile Val Thr Tyr
Thr 325 330 335Ala Ser Asn Arg Gln Ser Ile Pro Gly Thr Ile Leu Thr
Asp Ala Asp 340 345 350Asn Val Trp Asn Asp Pro Ala Gly Val Asp Ala
His Ala Tyr Ala Ala 355 360 365Lys Thr Tyr Asp Tyr Tyr Lys Glu Lys
Phe Asn Arg Asn Ser Ile Asp 370 375 380Gly Arg Gly Leu Gln Leu Arg
Ser Thr Val His Tyr Gly Asn Arg Tyr385 390 395 400Asn Asn Ala Phe
Trp Asn Gly Ser Gln Met Thr Tyr Gly Asp Gly Asp 405 410 415Gly Thr
Thr Phe Ile Ala Phe Ser Gly Asp Pro Asp Val Val Gly His 420 425
430Glu Leu Thr His Gly Val Thr Glu Tyr Thr Ser Asn Leu Glu Tyr Tyr
435 440 445Gly Glu Ser Gly Ala Leu Asn Glu Ala Phe Ser Asp Ile Ile
Gly Asn 450 455 460Asp Ile Gln Arg Lys Asn Trp Leu Val Gly Asp Asp
Ile Tyr Thr Pro465 470 475 480Arg Ile Ala Gly Asp Ala Leu Arg Ser
Met Ser Asn Pro Thr Leu Tyr 485 490 495Asp Gln Pro Asp His Tyr Ser
Asn Leu Tyr Arg Gly Ser Ser Asp Asn 500 505 510Gly Gly Val His Thr
Asn Ser Gly Ile Ile Asn Lys Ala Tyr Tyr Leu 515 520 525Leu Ala Gln
Gly Gly Thr Phe His Gly Val Thr Val Asn Gly Ile Gly 530 535 540Arg
Asp Ala Ala Val Gln Ile Tyr Tyr Ser Ala Phe Thr Asn Tyr Leu545 550
555 560Thr Ser Ser Ser Asp Phe Ser Asn Ala Arg Asp Ala Val Val Gln
Ala 565 570 575Ala Lys Asp Leu Tyr Gly Ala Ser Ser Ala Gln Ala Thr
Ala Ala Ala 580 585 590Lys Ser Phe Asp Ala Val Gly Val Asn 595
60061770DNAPaenibacillus sp.misc_feature(1)..(1770)nucleotide
sequence of the PspPro2 gene isolated from Paenibacillus sp.
6atgaaaaaag tatgggtttc acttcttgga ggagcgatgt tattaggggc tgtagcacca
60ggtgcatcag cagcagagca ttctgttcct gatcctactc agctaacacc gacctttcac
120gccgagcaat ggaaggctcc ttccacggta accggcgaca atattgtatg
gagctatttg 180aatcgacaaa agaaaacctt attgaataca gacagcacca
gtgtgcgtga tcagttccgc 240atcattgatc gtacaagcga caaatccggt
gcaagccatt atcggctcaa gcaatatgta 300aacgggatcc ccgtatatgg
ggctgaacag accattcatg tgaacaacgc cggtaaagta 360acctcttatt
tgggtgctgt catttcagag gatcagcagc aagacgcgac cgaagatacc
420actccaaaaa tcagcgcgac tgaagccgtt tataccgcat atgcagaagc
cgctgcccgg 480attcaatcct tcccttccat caatgatagt ctttctgagg
ctagtgagga acaagggagt 540gagaatcaag gcaatgagat tcaaaacatt
gggattaaaa gcagtgtaag taatgacact 600tacgcagagg cgcataacaa
cgtactttta acccccgttg accaagcaga gcaaagttac 660attgccaaaa
ttgctaatct ggagccaagt gtagagccca aagcagaatt atacatctat
720ccagatggtg agactacacg actggtttat gtaacagagg ttaatattct
tgaacctgcg 780cctctgcgca cacgctactt cattgatgcg aaaaccggca
aaatcgtatt ccagtatgac 840atcctcaacc acgcaacagg caccggccgc
ggcgtggatg gcaaaacaaa atcatttacg 900actacagctt caggcaaccg
gtatcagttg aaagacacga ctcgcagcaa tggaatcgtg 960acttacaccg
ctggcaatcg ccagacgacg ccaggtacga ttttgaccga tacagataat
1020gtatgggagg accctgcggc tgttgatgcc catgcctacg ccattaaaac
ctatgactat 1080tataagaata aattcggtcg cgacagtatt gatggacgtg
gcatgcaaat tcgttcgaca 1140gtccattacg gcaaaaaata taacaatgcc
ttctggaacg gctcgcaaat gacctacgga 1200gacggagacg ggtccacatt
taccttcttc agcggcgatc ccgatgtcgt ggggcatgag 1260ctcacccacg
gcgtcaccga gttcacctcc aatttggagt attatggtga gtccggtgca
1320ttgaacgaag ccttctcgga tattatcggt aatgatatag atggcaccag
ttggcttctt 1380ggcgacggca tttatacgcc taatattcca ggcgacgctc
tgcgttccct gtccgatcct 1440acacgattcg gccagccgga tcactactcc
aatttctatc cggaccccaa caatgatgat 1500gaaggcggag tccatacgaa
cagcggtatt atcaacaaag cctattattt gctggcacaa 1560ggcggtacgt
cccatggtgt aacggtaact
ggtatcggac gcgaagcggc tgtattcatt 1620tactacaatg cctttaccaa
ctatttgacc tctacctcca acttctctaa cgcacgcgct 1680gctgttatac
aggcagccaa ggatttttat ggtgctgatt cgctggcagt aaccagtgct
1740attcaatcct ttgatgcggt aggaatcaaa 17707590PRTPaenibacillus
sp.misc_feature(1)..(590)amino acid sequence of the PspPro2
precursor protein 7Met Lys Lys Val Trp Val Ser Leu Leu Gly Gly Ala
Met Leu Leu Gly1 5 10 15Ala Val Ala Pro Gly Ala Ser Ala Ala Glu His
Ser Val Pro Asp Pro 20 25 30Thr Gln Leu Thr Pro Thr Phe His Ala Glu
Gln Trp Lys Ala Pro Ser 35 40 45Thr Val Thr Gly Asp Asn Ile Val Trp
Ser Tyr Leu Asn Arg Gln Lys 50 55 60Lys Thr Leu Leu Asn Thr Asp Ser
Thr Ser Val Arg Asp Gln Phe Arg65 70 75 80Ile Ile Asp Arg Thr Ser
Asp Lys Ser Gly Ala Ser His Tyr Arg Leu 85 90 95Lys Gln Tyr Val Asn
Gly Ile Pro Val Tyr Gly Ala Glu Gln Thr Ile 100 105 110His Val Asn
Asn Ala Gly Lys Val Thr Ser Tyr Leu Gly Ala Val Ile 115 120 125Ser
Glu Asp Gln Gln Gln Asp Ala Thr Glu Asp Thr Thr Pro Lys Ile 130 135
140Ser Ala Thr Glu Ala Val Tyr Thr Ala Tyr Ala Glu Ala Ala Ala
Arg145 150 155 160Ile Gln Ser Phe Pro Ser Ile Asn Asp Ser Leu Ser
Glu Ala Ser Glu 165 170 175Glu Gln Gly Ser Glu Asn Gln Gly Asn Glu
Ile Gln Asn Ile Gly Ile 180 185 190Lys Ser Ser Val Ser Asn Asp Thr
Tyr Ala Glu Ala His Asn Asn Val 195 200 205Leu Leu Thr Pro Val Asp
Gln Ala Glu Gln Ser Tyr Ile Ala Lys Ile 210 215 220Ala Asn Leu Glu
Pro Ser Val Glu Pro Lys Ala Glu Leu Tyr Ile Tyr225 230 235 240Pro
Asp Gly Glu Thr Thr Arg Leu Val Tyr Val Thr Glu Val Asn Ile 245 250
255Leu Glu Pro Ala Pro Leu Arg Thr Arg Tyr Phe Ile Asp Ala Lys Thr
260 265 270Gly Lys Ile Val Phe Gln Tyr Asp Ile Leu Asn His Ala Thr
Gly Thr 275 280 285Gly Arg Gly Val Asp Gly Lys Thr Lys Ser Phe Thr
Thr Thr Ala Ser 290 295 300Gly Asn Arg Tyr Gln Leu Lys Asp Thr Thr
Arg Ser Asn Gly Ile Val305 310 315 320Thr Tyr Thr Ala Gly Asn Arg
Gln Thr Thr Pro Gly Thr Ile Leu Thr 325 330 335Asp Thr Asp Asn Val
Trp Glu Asp Pro Ala Ala Val Asp Ala His Ala 340 345 350Tyr Ala Ile
Lys Thr Tyr Asp Tyr Tyr Lys Asn Lys Phe Gly Arg Asp 355 360 365Ser
Ile Asp Gly Arg Gly Met Gln Ile Arg Ser Thr Val His Tyr Gly 370 375
380Lys Lys Tyr Asn Asn Ala Phe Trp Asn Gly Ser Gln Met Thr Tyr
Gly385 390 395 400Asp Gly Asp Gly Ser Thr Phe Thr Phe Phe Ser Gly
Asp Pro Asp Val 405 410 415Val Gly His Glu Leu Thr His Gly Val Thr
Glu Phe Thr Ser Asn Leu 420 425 430Glu Tyr Tyr Gly Glu Ser Gly Ala
Leu Asn Glu Ala Phe Ser Asp Ile 435 440 445Ile Gly Asn Asp Ile Asp
Gly Thr Ser Trp Leu Leu Gly Asp Gly Ile 450 455 460Tyr Thr Pro Asn
Ile Pro Gly Asp Ala Leu Arg Ser Leu Ser Asp Pro465 470 475 480Thr
Arg Phe Gly Gln Pro Asp His Tyr Ser Asn Phe Tyr Pro Asp Pro 485 490
495Asn Asn Asp Asp Glu Gly Gly Val His Thr Asn Ser Gly Ile Ile Asn
500 505 510Lys Ala Tyr Tyr Leu Leu Ala Gln Gly Gly Thr Ser His Gly
Val Thr 515 520 525Val Thr Gly Ile Gly Arg Glu Ala Ala Val Phe Ile
Tyr Tyr Asn Ala 530 535 540Phe Thr Asn Tyr Leu Thr Ser Thr Ser Asn
Phe Ser Asn Ala Arg Ala545 550 555 560Ala Val Ile Gln Ala Ala Lys
Asp Phe Tyr Gly Ala Asp Ser Leu Ala 565 570 575Val Thr Ser Ala Ile
Gln Ser Phe Asp Ala Val Gly Ile Lys 580 585 5908306PRTPaenibacillus
sp.misc_feature(1)..(306)amino acid sequence of the predicted
mature form of PspPro2 8Ala Thr Gly Thr Gly Arg Gly Val Asp Gly Lys
Thr Lys Ser Phe Thr1 5 10 15Thr Thr Ala Ser Gly Asn Arg Tyr Gln Leu
Lys Asp Thr Thr Arg Ser 20 25 30Asn Gly Ile Val Thr Tyr Thr Ala Gly
Asn Arg Gln Thr Thr Pro Gly 35 40 45Thr Ile Leu Thr Asp Thr Asp Asn
Val Trp Glu Asp Pro Ala Ala Val 50 55 60Asp Ala His Ala Tyr Ala Ile
Lys Thr Tyr Asp Tyr Tyr Lys Asn Lys65 70 75 80Phe Gly Arg Asp Ser
Ile Asp Gly Arg Gly Met Gln Ile Arg Ser Thr 85 90 95Val His Tyr Gly
Lys Lys Tyr Asn Asn Ala Phe Trp Asn Gly Ser Gln 100 105 110Met Thr
Tyr Gly Asp Gly Asp Gly Ser Thr Phe Thr Phe Phe Ser Gly 115 120
125Asp Pro Asp Val Val Gly His Glu Leu Thr His Gly Val Thr Glu Phe
130 135 140Thr Ser Asn Leu Glu Tyr Tyr Gly Glu Ser Gly Ala Leu Asn
Glu Ala145 150 155 160Phe Ser Asp Ile Ile Gly Asn Asp Ile Asp Gly
Thr Ser Trp Leu Leu 165 170 175Gly Asp Gly Ile Tyr Thr Pro Asn Ile
Pro Gly Asp Ala Leu Arg Ser 180 185 190Leu Ser Asp Pro Thr Arg Phe
Gly Gln Pro Asp His Tyr Ser Asn Phe 195 200 205Tyr Pro Asp Pro Asn
Asn Asp Asp Glu Gly Gly Val His Thr Asn Ser 210 215 220Gly Ile Ile
Asn Lys Ala Tyr Tyr Leu Leu Ala Gln Gly Gly Thr Ser225 230 235
240His Gly Val Thr Val Thr Gly Ile Gly Arg Glu Ala Ala Val Phe Ile
245 250 255Tyr Tyr Asn Ala Phe Thr Asn Tyr Leu Thr Ser Thr Ser Asn
Phe Ser 260 265 270Asn Ala Arg Ala Ala Val Ile Gln Ala Ala Lys Asp
Phe Tyr Gly Ala 275 280 285Asp Ser Leu Ala Val Thr Ser Ala Ile Gln
Ser Phe Asp Ala Val Gly 290 295 300Ile Lys30591794DNAArtificial
SequenceSynthetic nucleotide sequence of the synthesized PspPro2
gene in plasmid pGX084 (AprE-PspPro2) 9gtgagaagca aaaaattgtg
gatcagcttg ttgtttgcgt taacgttaat ctttacgatg 60gcgttcagca acatgagcgc
gcaggctgct ggaaaagcag agcattcagt tcctgacccg 120acgcaactta
caccgacatt tcatgctgag cagtggaagg caccgagcac ggtcacgggc
180gacaacatcg tgtggagcta cctgaacaga cagaaaaaga cgctgctgaa
cacggactca 240acgagcgtga gagaccagtt cagaatcatc gacagaacga
gcgacaagtc aggcgcgtca 300cattatagac tgaagcagta cgtgaacggc
atcccggtct acggagccga gcaaacgatc 360catgtgaata atgcgggcaa
agttacatca tacctgggcg ccgtcatctc agaagaccag 420cagcaagatg
caacggagga tacaacaccg aagatcagcg ccacagaagc ggtctatacg
480gcttacgccg aagcggctgc aagaatccag agcttcccgt caattaatga
cagcctgagc 540gaagcatcag aggaacaagg cagcgagaac cagggcaatg
aaatccaaaa catcggcatc 600aagagcagcg tgtcaaacga cacgtatgcg
gaggctcata acaacgttct gctgacaccg 660gtcgatcagg ccgaacagag
ctatattgca aagatcgcga atctggagcc gtcagtcgag 720ccgaaggccg
agctgtatat ctatccggac ggcgagacga cgagactggt gtacgttacg
780gaggtcaaca tccttgagcc tgcgccgctg agaacaagat actttatcga
cgccaagacg 840ggcaagatcg tgtttcagta cgatatcctg aaccatgcga
cgggaacagg cagaggcgtg 900gacggcaaaa caaaatcatt cacgacaacg
gcaagcggca acagatacca gctgaaggac 960acaacaagat caaatggcat
cgtcacatac acggccggaa atagacagac gacgccggga 1020acgattctga
cggatacaga taacgtgtgg gaagatccgg cagcagttga tgcacatgca
1080tacgcgatca agacgtacga ctactacaag aacaaattcg gaagagattc
aatcgatgga 1140agaggcatgc aaatcagatc aacggttcat tatggcaaaa
agtacaacaa tgccttctgg 1200aacggcagcc aaatgacata cggcgatgga
gacggctcaa cgtttacatt cttttcaggc 1260gacccggacg tcgtcggcca
tgaactgacg catggcgtta cagagttcac gagcaacctg 1320gagtattacg
gcgaatcagg cgcactgaat gaggctttca gcgacatcat tggcaacgac
1380attgatggca catcatggct gcttggcgac ggcatttaca cacctaacat
tccgggcgat 1440gcactgagaa gcctgtcaga ccctacgaga ttcggccaac
ctgaccatta cagcaacttc 1500tacccggatc ctaataacga tgatgagggc
ggagtgcata cgaacagcgg cattatcaac 1560aaagcgtact atctgctggc
acaaggcgga acgtcacatg gagtgacggt gacaggaatc 1620ggcagagagg
cggcagtgtt tatctactac aacgccttca caaactacct gacgagcacg
1680tcaaatttca gcaacgctag agcggcggtc atccaggcag caaaggactt
ttatggagca 1740gactcactgg cagttacgtc agcaattcag tcattcgacg
cagttggaat taag 179410598PRTArtificial SequenceSynthetic amino acid
sequence of the PspPro2 precursor protein expressed from plasmid
pGX084(AprE-PspPro2) 10Met Arg Ser Lys Lys Leu Trp Ile Ser Leu Leu
Phe Ala Leu Thr Leu1 5 10 15Ile Phe Thr Met Ala Phe Ser Asn Met Ser
Ala Gln Ala Ala Gly Lys 20 25 30Ala Glu His Ser Val Pro Asp Pro Thr
Gln Leu Thr Pro Thr Phe His 35 40 45Ala Glu Gln Trp Lys Ala Pro Ser
Thr Val Thr Gly Asp Asn Ile Val 50 55 60Trp Ser Tyr Leu Asn Arg Gln
Lys Lys Thr Leu Leu Asn Thr Asp Ser65 70 75 80Thr Ser Val Arg Asp
Gln Phe Arg Ile Ile Asp Arg Thr Ser Asp Lys 85 90 95Ser Gly Ala Ser
His Tyr Arg Leu Lys Gln Tyr Val Asn Gly Ile Pro 100 105 110Val Tyr
Gly Ala Glu Gln Thr Ile His Val Asn Asn Ala Gly Lys Val 115 120
125Thr Ser Tyr Leu Gly Ala Val Ile Ser Glu Asp Gln Gln Gln Asp Ala
130 135 140Thr Glu Asp Thr Thr Pro Lys Ile Ser Ala Thr Glu Ala Val
Tyr Thr145 150 155 160Ala Tyr Ala Glu Ala Ala Ala Arg Ile Gln Ser
Phe Pro Ser Ile Asn 165 170 175Asp Ser Leu Ser Glu Ala Ser Glu Glu
Gln Gly Ser Glu Asn Gln Gly 180 185 190Asn Glu Ile Gln Asn Ile Gly
Ile Lys Ser Ser Val Ser Asn Asp Thr 195 200 205Tyr Ala Glu Ala His
Asn Asn Val Leu Leu Thr Pro Val Asp Gln Ala 210 215 220Glu Gln Ser
Tyr Ile Ala Lys Ile Ala Asn Leu Glu Pro Ser Val Glu225 230 235
240Pro Lys Ala Glu Leu Tyr Ile Tyr Pro Asp Gly Glu Thr Thr Arg Leu
245 250 255Val Tyr Val Thr Glu Val Asn Ile Leu Glu Pro Ala Pro Leu
Arg Thr 260 265 270Arg Tyr Phe Ile Asp Ala Lys Thr Gly Lys Ile Val
Phe Gln Tyr Asp 275 280 285Ile Leu Asn His Ala Thr Gly Thr Gly Arg
Gly Val Asp Gly Lys Thr 290 295 300Lys Ser Phe Thr Thr Thr Ala Ser
Gly Asn Arg Tyr Gln Leu Lys Asp305 310 315 320Thr Thr Arg Ser Asn
Gly Ile Val Thr Tyr Thr Ala Gly Asn Arg Gln 325 330 335Thr Thr Pro
Gly Thr Ile Leu Thr Asp Thr Asp Asn Val Trp Glu Asp 340 345 350Pro
Ala Ala Val Asp Ala His Ala Tyr Ala Ile Lys Thr Tyr Asp Tyr 355 360
365Tyr Lys Asn Lys Phe Gly Arg Asp Ser Ile Asp Gly Arg Gly Met Gln
370 375 380Ile Arg Ser Thr Val His Tyr Gly Lys Lys Tyr Asn Asn Ala
Phe Trp385 390 395 400Asn Gly Ser Gln Met Thr Tyr Gly Asp Gly Asp
Gly Ser Thr Phe Thr 405 410 415Phe Phe Ser Gly Asp Pro Asp Val Val
Gly His Glu Leu Thr His Gly 420 425 430Val Thr Glu Phe Thr Ser Asn
Leu Glu Tyr Tyr Gly Glu Ser Gly Ala 435 440 445Leu Asn Glu Ala Phe
Ser Asp Ile Ile Gly Asn Asp Ile Asp Gly Thr 450 455 460Ser Trp Leu
Leu Gly Asp Gly Ile Tyr Thr Pro Asn Ile Pro Gly Asp465 470 475
480Ala Leu Arg Ser Leu Ser Asp Pro Thr Arg Phe Gly Gln Pro Asp His
485 490 495Tyr Ser Asn Phe Tyr Pro Asp Pro Asn Asn Asp Asp Glu Gly
Gly Val 500 505 510His Thr Asn Ser Gly Ile Ile Asn Lys Ala Tyr Tyr
Leu Leu Ala Gln 515 520 525Gly Gly Thr Ser His Gly Val Thr Val Thr
Gly Ile Gly Arg Glu Ala 530 535 540Ala Val Phe Ile Tyr Tyr Asn Ala
Phe Thr Asn Tyr Leu Thr Ser Thr545 550 555 560Ser Asn Phe Ser Asn
Ala Arg Ala Ala Val Ile Gln Ala Ala Lys Asp 565 570 575Phe Tyr Gly
Ala Asp Ser Leu Ala Val Thr Ser Ala Ile Gln Ser Phe 580 585 590Asp
Ala Val Gly Ile Lys 595111599DNAPaenibacillus
humicusmisc_feature(1)..(1599)nucleotide sequence of the PhuPro2
gene isolated from Paenibacillus humicus 11atgaaaaaaa tgattcctac
tctgctcggt accgtattgc tgctttcttc cgcttccgct 60gtcgctgctg aatcgccaag
cctcggagcg gccggaactc ccggggtcag cgtcgtgaac 120aatcagctcg
tgactcaatt catcgaggct tccaaggatg ccaagattgt cccgggctct
180tccgaggata aaatctgggc tttccttgaa ggccagcaag caaagctggg
tgtatccgca 240gcggatgtaa aaacctcgtt cctgatccag aagaaggaag
tcgatccgac ttcgggcgtc 300gagcatttcc gcctgcagca atatgtgaat
ggcatcccgg tatatggcgg tgaccaaacc 360attcacatcg acaaggccgg
ccaggttacg tcgttcgtag gagctgttct gccggctcaa 420aatcaaatca
cggcaaaatc cagcgtacca gccataagcg catccgacgc tctggctatc
480gcggcgaagg aagccagttc ccgcatcggc gagctgggag cacaggagaa
gactccgtcg 540gctcagctgt acgtatatcc ggaaggcaac gggtcgcgtc
tcgtctacca gacggaagtg 600aatgtgcttg agccgcagcc tctgcgcacc
cgctatctta tcgatgcggc cgacggccat 660atcgtgcagc agtacgatct
gatcgagacg gcgaccggtt cgggcacggg cgtgctgggc 720gacaataaga
cgttccagac gactctttcc ggcagcacgt accagctgaa agacaccact
780cgcggcaacg gcatctacac ctacacagcc agcaatcgga ccacgattcc
gggcacgctg 840ctgacggacg ccgacaacgt atggacggat ggagccgccg
tcgatgccca tacttatgcc 900ggaaaagtat atgatttcta caaaacgaag
ttcggacgca acagcctcga cggcaacggc 960ctgctgatcc gttcctcggt
ccactacagc agcaggtaca acaatgcctt ctggaacggc 1020acccagattg
tattcggcga cggcgacggc tcgacgttca ttccgctgtc gggcgatctc
1080gacgtggtcg gccatgagct gtcccacgga gtcatcgagt acacgtccaa
ccttcaatac 1140ctcaatgaat ccggcgcgct gaacgagtcc tatgccgacg
tcctcggcaa ctcgatccag 1200gcgaaaaact ggcttatcgg cgacgatgtc
tatacgcctg gcatctccgg agatgctctc 1260cgttccatgt ccaacccgac
gctttacggg cagccggaca actatgccaa ccgctatacg 1320ggatcttccg
acaacggcgg cgttcatacg aacagcggca tcacgaacaa agcgttctac
1380ctgctcgccc aaggcggcac ccagaacggc gttaccgtcg ccggcatcgg
gcgcgacgca 1440gccgtgaaca ttttctacaa cacagtggcc tattacctta
cttccacttc caacttcgcc 1500gcggcgaaga acgcctcgat ccaggcagcc
aaagacctgt acggaacggg ctcctcttat 1560gtcacctcgg tgaccaatgc
attcagagcc gtaggcctg 159912533PRTPaenibacillus
humicusmisc_feature(1)..(533)amino acid sequence of the PhuPro2
precursor protein 12Met Lys Lys Met Ile Pro Thr Leu Leu Gly Thr Val
Leu Leu Leu Ser1 5 10 15Ser Ala Ser Ala Val Ala Ala Glu Ser Pro Ser
Leu Gly Ala Ala Gly 20 25 30Thr Pro Gly Val Ser Val Val Asn Asn Gln
Leu Val Thr Gln Phe Ile 35 40 45Glu Ala Ser Lys Asp Ala Lys Ile Val
Pro Gly Ser Ser Glu Asp Lys 50 55 60Ile Trp Ala Phe Leu Glu Gly Gln
Gln Ala Lys Leu Gly Val Ser Ala65 70 75 80Ala Asp Val Lys Thr Ser
Phe Leu Ile Gln Lys Lys Glu Val Asp Pro 85 90 95Thr Ser Gly Val Glu
His Phe Arg Leu Gln Gln Tyr Val Asn Gly Ile 100 105 110Pro Val Tyr
Gly Gly Asp Gln Thr Ile His Ile Asp Lys Ala Gly Gln 115 120 125Val
Thr Ser Phe Val Gly Ala Val Leu Pro Ala Gln Asn Gln Ile Thr 130 135
140Ala Lys Ser Ser Val Pro Ala Ile Ser Ala Ser Asp Ala Leu Ala
Ile145 150 155 160Ala Ala Lys Glu Ala Ser Ser Arg Ile Gly Glu Leu
Gly Ala Gln Glu 165 170 175Lys Thr Pro Ser Ala Gln Leu Tyr Val Tyr
Pro Glu Gly Asn Gly Ser 180 185 190Arg Leu Val Tyr Gln Thr Glu Val
Asn Val Leu Glu Pro Gln Pro Leu 195 200 205Arg Thr Arg Tyr Leu Ile
Asp Ala Ala Asp Gly His Ile Val Gln Gln 210 215 220Tyr Asp Leu Ile
Glu Thr Ala Thr Gly Ser Gly Thr Gly Val Leu Gly225 230 235 240Asp
Asn Lys Thr Phe Gln Thr Thr Leu Ser Gly Ser Thr Tyr Gln Leu 245 250
255Lys Asp Thr Thr Arg Gly Asn Gly Ile Tyr Thr Tyr Thr Ala Ser Asn
260 265 270Arg Thr Thr Ile Pro
Gly Thr Leu Leu Thr Asp Ala Asp Asn Val Trp 275 280 285Thr Asp Gly
Ala Ala Val Asp Ala His Thr Tyr Ala Gly Lys Val Tyr 290 295 300Asp
Phe Tyr Lys Thr Lys Phe Gly Arg Asn Ser Leu Asp Gly Asn Gly305 310
315 320Leu Leu Ile Arg Ser Ser Val His Tyr Ser Ser Arg Tyr Asn Asn
Ala 325 330 335Phe Trp Asn Gly Thr Gln Ile Val Phe Gly Asp Gly Asp
Gly Ser Thr 340 345 350Phe Ile Pro Leu Ser Gly Asp Leu Asp Val Val
Gly His Glu Leu Ser 355 360 365His Gly Val Ile Glu Tyr Thr Ser Asn
Leu Gln Tyr Leu Asn Glu Ser 370 375 380Gly Ala Leu Asn Glu Ser Tyr
Ala Asp Val Leu Gly Asn Ser Ile Gln385 390 395 400Ala Lys Asn Trp
Leu Ile Gly Asp Asp Val Tyr Thr Pro Gly Ile Ser 405 410 415Gly Asp
Ala Leu Arg Ser Met Ser Asn Pro Thr Leu Tyr Gly Gln Pro 420 425
430Asp Asn Tyr Ala Asn Arg Tyr Thr Gly Ser Ser Asp Asn Gly Gly Val
435 440 445His Thr Asn Ser Gly Ile Thr Asn Lys Ala Phe Tyr Leu Leu
Ala Gln 450 455 460Gly Gly Thr Gln Asn Gly Val Thr Val Ala Gly Ile
Gly Arg Asp Ala465 470 475 480Ala Val Asn Ile Phe Tyr Asn Thr Val
Ala Tyr Tyr Leu Thr Ser Thr 485 490 495Ser Asn Phe Ala Ala Ala Lys
Asn Ala Ser Ile Gln Ala Ala Lys Asp 500 505 510Leu Tyr Gly Thr Gly
Ser Ser Tyr Val Thr Ser Val Thr Asn Ala Phe 515 520 525Arg Ala Val
Gly Leu 53013303PRTPaenibacillus humicusmisc_feature(1)..(303)amino
acid sequence of the predicted mature form of PhuPro2 13Ala Thr Gly
Ser Gly Thr Gly Val Leu Gly Asp Asn Lys Thr Phe Gln1 5 10 15Thr Thr
Leu Ser Gly Ser Thr Tyr Gln Leu Lys Asp Thr Thr Arg Gly 20 25 30Asn
Gly Ile Tyr Thr Tyr Thr Ala Ser Asn Arg Thr Thr Ile Pro Gly 35 40
45Thr Leu Leu Thr Asp Ala Asp Asn Val Trp Thr Asp Gly Ala Ala Val
50 55 60Asp Ala His Thr Tyr Ala Gly Lys Val Tyr Asp Phe Tyr Lys Thr
Lys65 70 75 80Phe Gly Arg Asn Ser Leu Asp Gly Asn Gly Leu Leu Ile
Arg Ser Ser 85 90 95Val His Tyr Ser Ser Arg Tyr Asn Asn Ala Phe Trp
Asn Gly Thr Gln 100 105 110Ile Val Phe Gly Asp Gly Asp Gly Ser Thr
Phe Ile Pro Leu Ser Gly 115 120 125Asp Leu Asp Val Val Gly His Glu
Leu Ser His Gly Val Ile Glu Tyr 130 135 140Thr Ser Asn Leu Gln Tyr
Leu Asn Glu Ser Gly Ala Leu Asn Glu Ser145 150 155 160Tyr Ala Asp
Val Leu Gly Asn Ser Ile Gln Ala Lys Asn Trp Leu Ile 165 170 175Gly
Asp Asp Val Tyr Thr Pro Gly Ile Ser Gly Asp Ala Leu Arg Ser 180 185
190Met Ser Asn Pro Thr Leu Tyr Gly Gln Pro Asp Asn Tyr Ala Asn Arg
195 200 205Tyr Thr Gly Ser Ser Asp Asn Gly Gly Val His Thr Asn Ser
Gly Ile 210 215 220Thr Asn Lys Ala Phe Tyr Leu Leu Ala Gln Gly Gly
Thr Gln Asn Gly225 230 235 240Val Thr Val Ala Gly Ile Gly Arg Asp
Ala Ala Val Asn Ile Phe Tyr 245 250 255Asn Thr Val Ala Tyr Tyr Leu
Thr Ser Thr Ser Asn Phe Ala Ala Ala 260 265 270Lys Asn Ala Ser Ile
Gln Ala Ala Lys Asp Leu Tyr Gly Thr Gly Ser 275 280 285Ser Tyr Val
Thr Ser Val Thr Asn Ala Phe Arg Ala Val Gly Leu 290 295
300141629DNAArtificial SequenceSynthetic nucleotide sequence of the
synthesized PhuPro2 gene in plasmid pGX150(AprE- PhuPro2)
14gtgagaagca aaaaattgtg gatcagcttg ttgtttgcgt taacgttaat ctttacgatg
60gcgttcagca acatgagcgc gcaggctgct ggaaaagaat caccgagcct tggcgctgca
120ggaacaccgg gcgttagcgt tgtgaataac caactggtca cgcagttcat
cgaagcatca 180aaagacgcga aaattgtccc tggatcaagc gaagataaga
tttgggcatt tctggaaggc 240cagcaagcaa agcttggcgt ctcagctgcc
gacgtgaaga cgagcttcct gatccagaag 300aaggaggttg acccgacatc
aggcgttgag cactttagac tgcaacagta cgtcaacggc 360atcccggttt
atggaggcga tcaaacaatc catattgata aggcaggcca ggtcacatca
420ttcgtcggag ctgtcctgcc ggctcagaac caaattacag caaaatcatc
agttccggca 480atttcagcct cagacgctct ggcaatcgct gccaaggagg
caagctcaag aattggcgaa 540ctgggcgcac aagaaaagac accgagcgcc
caactttatg tctatccgga gggcaacgga 600agcagactgg tgtaccagac
agaggtcaat gttctggagc cgcaaccgct gagaacgaga 660taccttatcg
atgctgcgga tggccacatt gttcagcaat acgacctgat tgagacagca
720acaggaagcg gaacgggcgt gctgggcgac aacaagacgt ttcagacaac
acttagcggc 780agcacgtacc aacttaagga cacgacgaga ggcaatggca
tttacacgta cacggcctca 840aacagaacga caatcccagg cacactgctg
acggatgcag acaatgtttg gacggacggc 900gcagcagttg acgcacacac
gtacgccggc aaggtgtacg acttttacaa gacgaagttc 960ggcagaaaca
gccttgatgg aaatggactg ctgatcagaa gcagcgtcca ctacagcagc
1020agatacaata acgccttctg gaacggcaca caaatcgtct ttggcgatgg
agacggatca 1080acattcatcc cgctgtcagg cgacctggac gttgtgggcc
acgagctgag ccacggcgtc 1140atcgagtaca cgagcaacct gcagtacctg
aatgaaagcg gcgcactgaa cgagtcatat 1200gctgatgtgc ttggcaatag
catccaggcc aagaactggc ttatcggaga cgacgtctac 1260acacctggca
tcagcggcga tgctctgaga agcatgagca atcctacact ttacggccaa
1320ccggacaact acgcgaatag atatacgggc agcagcgaca atggcggcgt
tcatacaaac 1380tcaggcatca cgaacaaggc gttctacctg ctggcacagg
gaggcacgca aaacggcgtt 1440acagttgcgg gcattggcag agatgcggcc
gtcaacatct tctacaacac agtcgcctac 1500tacctgacga gcacgtcaaa
cttcgcagcg gcaaagaacg catcaattca agcagcaaag 1560gatctgtacg
gaacaggcag ctcatatgtc acgtcagtta cgaatgcgtt tagagccgtc
1620ggcctttaa 162915542PRTArtificial SequenceSynthetic amino acid
sequence of the PhuPro2 precursor protein expressed from plasmid
pGX150(AprE- PhuPro2) 15Met Arg Ser Lys Lys Leu Trp Ile Ser Leu Leu
Phe Ala Leu Thr Leu1 5 10 15Ile Phe Thr Met Ala Phe Ser Asn Met Ser
Ala Gln Ala Ala Gly Lys 20 25 30Glu Ser Pro Ser Leu Gly Ala Ala Gly
Thr Pro Gly Val Ser Val Val 35 40 45Asn Asn Gln Leu Val Thr Gln Phe
Ile Glu Ala Ser Lys Asp Ala Lys 50 55 60Ile Val Pro Gly Ser Ser Glu
Asp Lys Ile Trp Ala Phe Leu Glu Gly65 70 75 80Gln Gln Ala Lys Leu
Gly Val Ser Ala Ala Asp Val Lys Thr Ser Phe 85 90 95Leu Ile Gln Lys
Lys Glu Val Asp Pro Thr Ser Gly Val Glu His Phe 100 105 110Arg Leu
Gln Gln Tyr Val Asn Gly Ile Pro Val Tyr Gly Gly Asp Gln 115 120
125Thr Ile His Ile Asp Lys Ala Gly Gln Val Thr Ser Phe Val Gly Ala
130 135 140Val Leu Pro Ala Gln Asn Gln Ile Thr Ala Lys Ser Ser Val
Pro Ala145 150 155 160Ile Ser Ala Ser Asp Ala Leu Ala Ile Ala Ala
Lys Glu Ala Ser Ser 165 170 175Arg Ile Gly Glu Leu Gly Ala Gln Glu
Lys Thr Pro Ser Ala Gln Leu 180 185 190Tyr Val Tyr Pro Glu Gly Asn
Gly Ser Arg Leu Val Tyr Gln Thr Glu 195 200 205Val Asn Val Leu Glu
Pro Gln Pro Leu Arg Thr Arg Tyr Leu Ile Asp 210 215 220Ala Ala Asp
Gly His Ile Val Gln Gln Tyr Asp Leu Ile Glu Thr Ala225 230 235
240Thr Gly Ser Gly Thr Gly Val Leu Gly Asp Asn Lys Thr Phe Gln Thr
245 250 255Thr Leu Ser Gly Ser Thr Tyr Gln Leu Lys Asp Thr Thr Arg
Gly Asn 260 265 270Gly Ile Tyr Thr Tyr Thr Ala Ser Asn Arg Thr Thr
Ile Pro Gly Thr 275 280 285Leu Leu Thr Asp Ala Asp Asn Val Trp Thr
Asp Gly Ala Ala Val Asp 290 295 300Ala His Thr Tyr Ala Gly Lys Val
Tyr Asp Phe Tyr Lys Thr Lys Phe305 310 315 320Gly Arg Asn Ser Leu
Asp Gly Asn Gly Leu Leu Ile Arg Ser Ser Val 325 330 335His Tyr Ser
Ser Arg Tyr Asn Asn Ala Phe Trp Asn Gly Thr Gln Ile 340 345 350Val
Phe Gly Asp Gly Asp Gly Ser Thr Phe Ile Pro Leu Ser Gly Asp 355 360
365Leu Asp Val Val Gly His Glu Leu Ser His Gly Val Ile Glu Tyr Thr
370 375 380Ser Asn Leu Gln Tyr Leu Asn Glu Ser Gly Ala Leu Asn Glu
Ser Tyr385 390 395 400Ala Asp Val Leu Gly Asn Ser Ile Gln Ala Lys
Asn Trp Leu Ile Gly 405 410 415Asp Asp Val Tyr Thr Pro Gly Ile Ser
Gly Asp Ala Leu Arg Ser Met 420 425 430Ser Asn Pro Thr Leu Tyr Gly
Gln Pro Asp Asn Tyr Ala Asn Arg Tyr 435 440 445Thr Gly Ser Ser Asp
Asn Gly Gly Val His Thr Asn Ser Gly Ile Thr 450 455 460Asn Lys Ala
Phe Tyr Leu Leu Ala Gln Gly Gly Thr Gln Asn Gly Val465 470 475
480Thr Val Ala Gly Ile Gly Arg Asp Ala Ala Val Asn Ile Phe Tyr Asn
485 490 495Thr Val Ala Tyr Tyr Leu Thr Ser Thr Ser Asn Phe Ala Ala
Ala Lys 500 505 510Asn Ala Ser Ile Gln Ala Ala Lys Asp Leu Tyr Gly
Thr Gly Ser Ser 515 520 525Tyr Val Thr Ser Val Thr Asn Ala Phe Arg
Ala Val Gly Leu 530 535 540161581DNAPaenibacillus
ehimensismisc_feature(1)..(1581)nucleotide sequence of the PehPro1
gene isolated from Paenibacillus ehimensis 16atgttaaaag tatgggcatc
gattattaca ggagcatttt tgctcgggag cgtgcaaggg 60gtgcaagctg ctccacaaga
tcaagctgct cccttcggag gattcacccc tcaattgatt 120accggggaaa
gctggagtgc gccgcaagga gtatcgggag aggaaaaaat ctggaagtat
180ctcgaatcca agcaggaaag cttccaaatc ggccaaaccg ttgatctgaa
aaagcaattg 240aaaattatcg gccaaacgac cgacgagaaa acgggaacca
cgcattaccg tctacagcag 300tatgtgggag gcgtccccgt atacggcggc
gtacaaacga tccatgtcaa caaagaagga 360caagttacct cgctgatcgg
cagcctgctt cccgaccagc agcagcaagt ttcgaaaagc 420ttgaattcgc
aaatcagcga agcgcaagcc atcgccgtgg cccagaaaga taccgaggcc
480gccgtcggca agctgggtga accgcaaaag acaccggaag cggatctgta
cgtttattta 540cacaacggac aaccggtcct cgcttatgtg accgaggtta
acgttctcga accggaggca 600atccggacgc gctacttcat cagcgccgaa
gacggcagca ttttattcaa gtacgacatc 660ctcgctcacg ctacaggtac
cggaaaaggc gtgctcggag atacgaaatc gttcacgacc 720acgcaatccg
gctccactta tcaattgaag gatacgacgc gcgggcaagg tatcgtcact
780tacagcgctg gcaaccggtc ctctctgccg ggaacgctgc tcaccagctc
cagcaatatt 840tggaacgacg gcgcggcggt cgatgcgcat gcctataccg
ccaaagtgta cgattactat 900aaaaacaaat ttggccgcaa cagcattgac
ggcaacggct tccagcttaa atcgaccgtg 960cactattcct ccagatacaa
caacgccttc tggaacggtg tgcaaatggt gtacggcgac 1020ggcgacggcg
taaccttcat tccgttctcc gccgatccgg acgtcatcgg ccacgaattg
1080acccacggcg ttacggaaca tacggccggc ctggaatact acggcgaatc
cggagcgctg 1140aacgaatcga tctccgatat tatcggcaac gcgatcgacg
gcaaaaactg gctgatcggc 1200gacttgattt atacgccgaa tactcccggg
gacgccctcc gctctatgga gaaccccaag 1260ctgtataacc aacccgaccg
ctatcaagac cgctatacgg gaccttccga taacggcggc 1320gtgcatatta
acagcggtat caacaacaaa gccttctacc tgatcgccca aggcggcacg
1380cactatggcg tcaccgtgaa cgggatcgga cgcgatgcgg ctgtgcaaat
tttctatgac 1440gccctcatca attacctgac tccaacttcg aacttctcgg
cgatgcgcgc agcagccatt 1500caagcggcaa ccgacctgta cggagcgaat
tcttctcaag taaacgctgt caaaaaagcg 1560tatactgccg tcggcgtgaa c
158117527PRTPaenibacillus ehimensismisc_feature(1)..(527)amino acid
sequence of the PehPro1 precursor protein 17Met Leu Lys Val Trp Ala
Ser Ile Ile Thr Gly Ala Phe Leu Leu Gly1 5 10 15Ser Val Gln Gly Val
Gln Ala Ala Pro Gln Asp Gln Ala Ala Pro Phe 20 25 30Gly Gly Phe Thr
Pro Gln Leu Ile Thr Gly Glu Ser Trp Ser Ala Pro 35 40 45Gln Gly Val
Ser Gly Glu Glu Lys Ile Trp Lys Tyr Leu Glu Ser Lys 50 55 60Gln Glu
Ser Phe Gln Ile Gly Gln Thr Val Asp Leu Lys Lys Gln Leu65 70 75
80Lys Ile Ile Gly Gln Thr Thr Asp Glu Lys Thr Gly Thr Thr His Tyr
85 90 95Arg Leu Gln Gln Tyr Val Gly Gly Val Pro Val Tyr Gly Gly Val
Gln 100 105 110Thr Ile His Val Asn Lys Glu Gly Gln Val Thr Ser Leu
Ile Gly Ser 115 120 125Leu Leu Pro Asp Gln Gln Gln Gln Val Ser Lys
Ser Leu Asn Ser Gln 130 135 140Ile Ser Glu Ala Gln Ala Ile Ala Val
Ala Gln Lys Asp Thr Glu Ala145 150 155 160Ala Val Gly Lys Leu Gly
Glu Pro Gln Lys Thr Pro Glu Ala Asp Leu 165 170 175Tyr Val Tyr Leu
His Asn Gly Gln Pro Val Leu Ala Tyr Val Thr Glu 180 185 190Val Asn
Val Leu Glu Pro Glu Ala Ile Arg Thr Arg Tyr Phe Ile Ser 195 200
205Ala Glu Asp Gly Ser Ile Leu Phe Lys Tyr Asp Ile Leu Ala His Ala
210 215 220Thr Gly Thr Gly Lys Gly Val Leu Gly Asp Thr Lys Ser Phe
Thr Thr225 230 235 240Thr Gln Ser Gly Ser Thr Tyr Gln Leu Lys Asp
Thr Thr Arg Gly Gln 245 250 255Gly Ile Val Thr Tyr Ser Ala Gly Asn
Arg Ser Ser Leu Pro Gly Thr 260 265 270Leu Leu Thr Ser Ser Ser Asn
Ile Trp Asn Asp Gly Ala Ala Val Asp 275 280 285Ala His Ala Tyr Thr
Ala Lys Val Tyr Asp Tyr Tyr Lys Asn Lys Phe 290 295 300Gly Arg Asn
Ser Ile Asp Gly Asn Gly Phe Gln Leu Lys Ser Thr Val305 310 315
320His Tyr Ser Ser Arg Tyr Asn Asn Ala Phe Trp Asn Gly Val Gln Met
325 330 335Val Tyr Gly Asp Gly Asp Gly Val Thr Phe Ile Pro Phe Ser
Ala Asp 340 345 350Pro Asp Val Ile Gly His Glu Leu Thr His Gly Val
Thr Glu His Thr 355 360 365Ala Gly Leu Glu Tyr Tyr Gly Glu Ser Gly
Ala Leu Asn Glu Ser Ile 370 375 380Ser Asp Ile Ile Gly Asn Ala Ile
Asp Gly Lys Asn Trp Leu Ile Gly385 390 395 400Asp Leu Ile Tyr Thr
Pro Asn Thr Pro Gly Asp Ala Leu Arg Ser Met 405 410 415Glu Asn Pro
Lys Leu Tyr Asn Gln Pro Asp Arg Tyr Gln Asp Arg Tyr 420 425 430Thr
Gly Pro Ser Asp Asn Gly Gly Val His Ile Asn Ser Gly Ile Asn 435 440
445Asn Lys Ala Phe Tyr Leu Ile Ala Gln Gly Gly Thr His Tyr Gly Val
450 455 460Thr Val Asn Gly Ile Gly Arg Asp Ala Ala Val Gln Ile Phe
Tyr Asp465 470 475 480Ala Leu Ile Asn Tyr Leu Thr Pro Thr Ser Asn
Phe Ser Ala Met Arg 485 490 495Ala Ala Ala Ile Gln Ala Ala Thr Asp
Leu Tyr Gly Ala Asn Ser Ser 500 505 510Gln Val Asn Ala Val Lys Lys
Ala Tyr Thr Ala Val Gly Val Asn 515 520 52518304PRTPaenibacillus
ehimensismisc_feature(1)..(304)amino acid sequence of the predicted
mature form of PehPro1 18Ala Thr Gly Thr Gly Lys Gly Val Leu Gly
Asp Thr Lys Ser Phe Thr1 5 10 15Thr Thr Gln Ser Gly Ser Thr Tyr Gln
Leu Lys Asp Thr Thr Arg Gly 20 25 30Gln Gly Ile Val Thr Tyr Ser Ala
Gly Asn Arg Ser Ser Leu Pro Gly 35 40 45Thr Leu Leu Thr Ser Ser Ser
Asn Ile Trp Asn Asp Gly Ala Ala Val 50 55 60Asp Ala His Ala Tyr Thr
Ala Lys Val Tyr Asp Tyr Tyr Lys Asn Lys65 70 75 80Phe Gly Arg Asn
Ser Ile Asp Gly Asn Gly Phe Gln Leu Lys Ser Thr 85 90 95Val His Tyr
Ser Ser Arg Tyr Asn Asn Ala Phe Trp Asn Gly Val Gln 100 105 110Met
Val Tyr Gly Asp Gly Asp Gly Val Thr Phe Ile Pro Phe Ser Ala 115 120
125Asp Pro Asp Val Ile Gly His Glu Leu Thr His Gly Val Thr Glu His
130 135 140Thr Ala Gly Leu Glu Tyr Tyr Gly Glu Ser Gly Ala Leu Asn
Glu Ser145 150 155 160Ile Ser Asp Ile Ile Gly Asn Ala Ile Asp Gly
Lys Asn Trp Leu Ile 165 170 175Gly Asp Leu Ile Tyr Thr Pro Asn Thr
Pro Gly Asp Ala Leu Arg Ser 180 185
190Met Glu Asn Pro Lys Leu Tyr Asn Gln Pro Asp Arg Tyr Gln Asp Arg
195 200 205Tyr Thr Gly Pro Ser Asp Asn Gly Gly Val His Ile Asn Ser
Gly Ile 210 215 220Asn Asn Lys Ala Phe Tyr Leu Ile Ala Gln Gly Gly
Thr His Tyr Gly225 230 235 240Val Thr Val Asn Gly Ile Gly Arg Asp
Ala Ala Val Gln Ile Phe Tyr 245 250 255Asp Ala Leu Ile Asn Tyr Leu
Thr Pro Thr Ser Asn Phe Ser Ala Met 260 265 270Arg Ala Ala Ala Ile
Gln Ala Ala Thr Asp Leu Tyr Gly Ala Asn Ser 275 280 285Ser Gln Val
Asn Ala Val Lys Lys Ala Tyr Thr Ala Val Gly Val Asn 290 295
300191611DNAArtificial SequenceSynthetic nucleotide sequence of the
synthesized PehPro1 gene in plasmid pGX148(AprE- PehPro1)
19gtgagaagca aaaaattgtg gatcagcttg ttgtttgcgt taacgttaat ctttacgatg
60gcgttcagca acatgagcgc gcaggctgct ggaaaagcac ctcaagatca ggcagcacct
120tttggaggct ttacaccgca acttatcaca ggcgaatcat ggtcagcacc
gcagggcgtt 180tcaggcgagg aaaagatctg gaagtacctt gagagcaagc
aggagtcatt tcaaatcggc 240cagacagtcg acctgaaaaa gcaactgaag
atcatcggcc aaacaacgga cgaaaagacg 300ggcacgacgc attatagact
gcaacaatat gttggcggcg tgccggttta tggaggcgtg 360caaacaatcc
acgtgaacaa ggaaggacag gtcacgtcac tgatcggcag cctgctgccg
420gatcagcagc aacaagtctc aaagagcctg aactcacaaa ttagcgaggc
acaagcgatt 480gcagttgcac aaaaggacac ggaagcagct gtcggcaagc
tgggcgaacc gcaaaaaaca 540cctgaggctg acctttacgt ctacctgcat
aacggccagc cggtccttgc gtacgttacg 600gaagttaacg tgctggagcc
ggaggccatc agaacgagat acttcattag cgcggaggat 660ggaagcattc
tgtttaagta cgatattctt gctcacgcga caggcacagg caagggcgtc
720cttggcgaca caaaaagctt cacgacaacg cagagcggat caacgtacca
gctgaaagat 780acaacaagag gacaaggcat cgttacgtat tcagcgggca
atagatcaag cctgccgggc 840acactgctga catcaagctc aaacatttgg
aatgacggcg cagcagttga tgcccatgcg 900tacacagcca aggtgtacga
ctactataag aacaagtttg gcagaaatag catcgacgga 960aatggatttc
aacttaaatc aacggtgcac tactcatcaa gatataacaa tgcgttttgg
1020aacggagtgc agatggtcta cggagacggc gacggcgtga catttattcc
gtttagcgcc 1080gacccggacg tgattggaca tgaactgaca catggagtga
cagagcatac ggcgggactg 1140gaatattacg gcgaaagcgg cgcactgaac
gaaagcatct cagacattat tggaaacgca 1200atcgatggca aaaactggct
gattggcgat ctgatttata cgccgaatac accgggcgat 1260gcactgagat
caatggagaa tccgaagctg tacaaccaac cggacagata ccaagataga
1320tacacaggac cgtcagacaa cggcggagtc catatcaaca gcggaatcaa
taacaaagcc 1380ttttacctga tcgcccaagg cggaacgcac tatggcgtta
cagtcaatgg catcggaaga 1440gatgccgcag ttcagatttt ctatgacgcg
ctgatcaact atctgacgcc tacaagcaat 1500ttctcagcaa tgagagccgc
agcaatccaa gcagccacgg atctgtatgg agccaattca 1560tcacaagtta
atgctgttaa gaaggcttat acggcagtgg gagttaacta a
161120536PRTArtificial SequenceSynthetic amino acid sequence of the
PehPro1 precursor protein expressed from plasmid pGX148(AprE-
PehPro1) 20Met Arg Ser Lys Lys Leu Trp Ile Ser Leu Leu Phe Ala Leu
Thr Leu1 5 10 15Ile Phe Thr Met Ala Phe Ser Asn Met Ser Ala Gln Ala
Ala Gly Lys 20 25 30Ala Pro Gln Asp Gln Ala Ala Pro Phe Gly Gly Phe
Thr Pro Gln Leu 35 40 45Ile Thr Gly Glu Ser Trp Ser Ala Pro Gln Gly
Val Ser Gly Glu Glu 50 55 60Lys Ile Trp Lys Tyr Leu Glu Ser Lys Gln
Glu Ser Phe Gln Ile Gly65 70 75 80Gln Thr Val Asp Leu Lys Lys Gln
Leu Lys Ile Ile Gly Gln Thr Thr 85 90 95Asp Glu Lys Thr Gly Thr Thr
His Tyr Arg Leu Gln Gln Tyr Val Gly 100 105 110Gly Val Pro Val Tyr
Gly Gly Val Gln Thr Ile His Val Asn Lys Glu 115 120 125Gly Gln Val
Thr Ser Leu Ile Gly Ser Leu Leu Pro Asp Gln Gln Gln 130 135 140Gln
Val Ser Lys Ser Leu Asn Ser Gln Ile Ser Glu Ala Gln Ala Ile145 150
155 160Ala Val Ala Gln Lys Asp Thr Glu Ala Ala Val Gly Lys Leu Gly
Glu 165 170 175Pro Gln Lys Thr Pro Glu Ala Asp Leu Tyr Val Tyr Leu
His Asn Gly 180 185 190Gln Pro Val Leu Ala Tyr Val Thr Glu Val Asn
Val Leu Glu Pro Glu 195 200 205Ala Ile Arg Thr Arg Tyr Phe Ile Ser
Ala Glu Asp Gly Ser Ile Leu 210 215 220Phe Lys Tyr Asp Ile Leu Ala
His Ala Thr Gly Thr Gly Lys Gly Val225 230 235 240Leu Gly Asp Thr
Lys Ser Phe Thr Thr Thr Gln Ser Gly Ser Thr Tyr 245 250 255Gln Leu
Lys Asp Thr Thr Arg Gly Gln Gly Ile Val Thr Tyr Ser Ala 260 265
270Gly Asn Arg Ser Ser Leu Pro Gly Thr Leu Leu Thr Ser Ser Ser Asn
275 280 285Ile Trp Asn Asp Gly Ala Ala Val Asp Ala His Ala Tyr Thr
Ala Lys 290 295 300Val Tyr Asp Tyr Tyr Lys Asn Lys Phe Gly Arg Asn
Ser Ile Asp Gly305 310 315 320Asn Gly Phe Gln Leu Lys Ser Thr Val
His Tyr Ser Ser Arg Tyr Asn 325 330 335Asn Ala Phe Trp Asn Gly Val
Gln Met Val Tyr Gly Asp Gly Asp Gly 340 345 350Val Thr Phe Ile Pro
Phe Ser Ala Asp Pro Asp Val Ile Gly His Glu 355 360 365Leu Thr His
Gly Val Thr Glu His Thr Ala Gly Leu Glu Tyr Tyr Gly 370 375 380Glu
Ser Gly Ala Leu Asn Glu Ser Ile Ser Asp Ile Ile Gly Asn Ala385 390
395 400Ile Asp Gly Lys Asn Trp Leu Ile Gly Asp Leu Ile Tyr Thr Pro
Asn 405 410 415Thr Pro Gly Asp Ala Leu Arg Ser Met Glu Asn Pro Lys
Leu Tyr Asn 420 425 430Gln Pro Asp Arg Tyr Gln Asp Arg Tyr Thr Gly
Pro Ser Asp Asn Gly 435 440 445Gly Val His Ile Asn Ser Gly Ile Asn
Asn Lys Ala Phe Tyr Leu Ile 450 455 460Ala Gln Gly Gly Thr His Tyr
Gly Val Thr Val Asn Gly Ile Gly Arg465 470 475 480Asp Ala Ala Val
Gln Ile Phe Tyr Asp Ala Leu Ile Asn Tyr Leu Thr 485 490 495Pro Thr
Ser Asn Phe Ser Ala Met Arg Ala Ala Ala Ile Gln Ala Ala 500 505
510Thr Asp Leu Tyr Gly Ala Asn Ser Ser Gln Val Asn Ala Val Lys Lys
515 520 525Ala Tyr Thr Ala Val Gly Val Asn 530
535211563DNAPaenibacillus
barcinonensismisc_feature(1)..(1563)nucleotide sequence of the
PbaPro1 gene isolated from Paenibacillus barcinonensis 21atgaaattga
ccaaaattat gccaacaatt cttgcaggag ctcttttgct cacatccctg 60tcctctgcag
cagcaatgcc gttatctgac tcatccattc catttgaggg cccctacacc
120tccgaggaga gtattctgtt gaacaacaac ccggacgaaa tgatttataa
ttttcttgca 180caacaagagc aatttctgaa tgccgacgtc aaaggacagc
tcaaaatcat taaacgcaac 240acagacactt ccggcatcag acactttcgt
ctgaagcaat acatcaaagg tgttccggtt 300tacggcgcag aacaaacgat
ccatctggac aagaacggag ctgtaacttc cgcactcggc 360gatcttccgc
caattgaaga acaggctgtt ccgaatgatg gcgttcccgc aatcagtgca
420gacgatgcca tccgtgccgc cgagaatgaa gccacctccc gtcttggaga
gcttggcgca 480ccagagcttg agccaaaggc cgaattaaac atttatcatc
atgaagatga cggacaaacc 540tacctcgttt acattacgga agttaacgtg
cttgagcctt ccccgctacg gaccaaatat 600tttattaacg cccttgatgg
aagcatcgta tctcaatacg atattatcaa ctttgccaca 660ggcaccggta
caggcgtgca tggtgatacc aaaacactga cgacaactca atccggcagc
720acctatcagc tgaaagatac aactcgtgga aaaggcattc aaacctatac
tgcgaacaat 780cgctcctcgc ttccaggcag cttgtctacc agttccaata
acgtatggac agaccgtgca 840gctgtagatg cgcacgccta tgctgccgcc
acatatgact tctacaaaaa caaattcaat 900cgcaacggca ttgacggaaa
cgggctgttg attcgctcta cagtgcatta tggctccaac 960tataaaaacg
ccttctggaa cggagcacag attgtctatg gagatggcga tggcatcgag
1020ttcggtccct tctccggtga tctcgatgtt gtcggacatg aattgacaca
cggggtgatt 1080gaatatacag ccaatctcga atatcgcaat gagccgggtg
ctttaaacga agcttttgcc 1140gacattatgg ggaacaccat cgaaagcaaa
aactggctgc ttggcgacgg aatctatact 1200ccaaacattc caggtgatgc
cctgcgctcg ttatccgacc ctacgctgta taaccagcct 1260gacaaataca
gtgatcgcta cactggctct caggataatg gcggtgtgca tatcaacagc
1320gggatcatta acaaagcata ttatcttgca gcccaaggcg gtactcataa
cggggtaacc 1380gttagcggca tcggccggga taaagcagta cgtattttct
atagcacgct ggtgaactac 1440ctgacgccaa cctccaaatt tgcagcagcc
aaaacagcga caattcaggc agccaaggac 1500ctgtacggtg ccaattccgc
tgaagctacg gcaatcacca aagcttatca agcggtaggt 1560ttg
156322521PRTPaenibacillus barcinonensismisc_feature(1)..(521)amino
acid sequence of the PbaPro1 precursor protein 22Met Lys Leu Thr
Lys Ile Met Pro Thr Ile Leu Ala Gly Ala Leu Leu1 5 10 15Leu Thr Ser
Leu Ser Ser Ala Ala Ala Met Pro Leu Ser Asp Ser Ser 20 25 30Ile Pro
Phe Glu Gly Pro Tyr Thr Ser Glu Glu Ser Ile Leu Leu Asn 35 40 45Asn
Asn Pro Asp Glu Met Ile Tyr Asn Phe Leu Ala Gln Gln Glu Gln 50 55
60Phe Leu Asn Ala Asp Val Lys Gly Gln Leu Lys Ile Ile Lys Arg Asn65
70 75 80Thr Asp Thr Ser Gly Ile Arg His Phe Arg Leu Lys Gln Tyr Ile
Lys 85 90 95Gly Val Pro Val Tyr Gly Ala Glu Gln Thr Ile His Leu Asp
Lys Asn 100 105 110Gly Ala Val Thr Ser Ala Leu Gly Asp Leu Pro Pro
Ile Glu Glu Gln 115 120 125Ala Val Pro Asn Asp Gly Val Pro Ala Ile
Ser Ala Asp Asp Ala Ile 130 135 140Arg Ala Ala Glu Asn Glu Ala Thr
Ser Arg Leu Gly Glu Leu Gly Ala145 150 155 160Pro Glu Leu Glu Pro
Lys Ala Glu Leu Asn Ile Tyr His His Glu Asp 165 170 175Asp Gly Gln
Thr Tyr Leu Val Tyr Ile Thr Glu Val Asn Val Leu Glu 180 185 190Pro
Ser Pro Leu Arg Thr Lys Tyr Phe Ile Asn Ala Leu Asp Gly Ser 195 200
205Ile Val Ser Gln Tyr Asp Ile Ile Asn Phe Ala Thr Gly Thr Gly Thr
210 215 220Gly Val His Gly Asp Thr Lys Thr Leu Thr Thr Thr Gln Ser
Gly Ser225 230 235 240Thr Tyr Gln Leu Lys Asp Thr Thr Arg Gly Lys
Gly Ile Gln Thr Tyr 245 250 255Thr Ala Asn Asn Arg Ser Ser Leu Pro
Gly Ser Leu Ser Thr Ser Ser 260 265 270Asn Asn Val Trp Thr Asp Arg
Ala Ala Val Asp Ala His Ala Tyr Ala 275 280 285Ala Ala Thr Tyr Asp
Phe Tyr Lys Asn Lys Phe Asn Arg Asn Gly Ile 290 295 300Asp Gly Asn
Gly Leu Leu Ile Arg Ser Thr Val His Tyr Gly Ser Asn305 310 315
320Tyr Lys Asn Ala Phe Trp Asn Gly Ala Gln Ile Val Tyr Gly Asp Gly
325 330 335Asp Gly Ile Glu Phe Gly Pro Phe Ser Gly Asp Leu Asp Val
Val Gly 340 345 350His Glu Leu Thr His Gly Val Ile Glu Tyr Thr Ala
Asn Leu Glu Tyr 355 360 365Arg Asn Glu Pro Gly Ala Leu Asn Glu Ala
Phe Ala Asp Ile Met Gly 370 375 380Asn Thr Ile Glu Ser Lys Asn Trp
Leu Leu Gly Asp Gly Ile Tyr Thr385 390 395 400Pro Asn Ile Pro Gly
Asp Ala Leu Arg Ser Leu Ser Asp Pro Thr Leu 405 410 415Tyr Asn Gln
Pro Asp Lys Tyr Ser Asp Arg Tyr Thr Gly Ser Gln Asp 420 425 430Asn
Gly Gly Val His Ile Asn Ser Gly Ile Ile Asn Lys Ala Tyr Tyr 435 440
445Leu Ala Ala Gln Gly Gly Thr His Asn Gly Val Thr Val Ser Gly Ile
450 455 460Gly Arg Asp Lys Ala Val Arg Ile Phe Tyr Ser Thr Leu Val
Asn Tyr465 470 475 480Leu Thr Pro Thr Ser Lys Phe Ala Ala Ala Lys
Thr Ala Thr Ile Gln 485 490 495Ala Ala Lys Asp Leu Tyr Gly Ala Asn
Ser Ala Glu Ala Thr Ala Ile 500 505 510Thr Lys Ala Tyr Gln Ala Val
Gly Leu 515 52023303PRTPaenibacillus
barcinonensismisc_feature(1)..(303)amino acid sequence of the
predicted mature form of PbaPro1 23Ala Thr Gly Thr Gly Thr Gly Val
His Gly Asp Thr Lys Thr Leu Thr1 5 10 15Thr Thr Gln Ser Gly Ser Thr
Tyr Gln Leu Lys Asp Thr Thr Arg Gly 20 25 30Lys Gly Ile Gln Thr Tyr
Thr Ala Asn Asn Arg Ser Ser Leu Pro Gly 35 40 45Ser Leu Ser Thr Ser
Ser Asn Asn Val Trp Thr Asp Arg Ala Ala Val 50 55 60Asp Ala His Ala
Tyr Ala Ala Ala Thr Tyr Asp Phe Tyr Lys Asn Lys65 70 75 80Phe Asn
Arg Asn Gly Ile Asp Gly Asn Gly Leu Leu Ile Arg Ser Thr 85 90 95Val
His Tyr Gly Ser Asn Tyr Lys Asn Ala Phe Trp Asn Gly Ala Gln 100 105
110Ile Val Tyr Gly Asp Gly Asp Gly Ile Glu Phe Gly Pro Phe Ser Gly
115 120 125Asp Leu Asp Val Val Gly His Glu Leu Thr His Gly Val Ile
Glu Tyr 130 135 140Thr Ala Asn Leu Glu Tyr Arg Asn Glu Pro Gly Ala
Leu Asn Glu Ala145 150 155 160Phe Ala Asp Ile Met Gly Asn Thr Ile
Glu Ser Lys Asn Trp Leu Leu 165 170 175Gly Asp Gly Ile Tyr Thr Pro
Asn Ile Pro Gly Asp Ala Leu Arg Ser 180 185 190Leu Ser Asp Pro Thr
Leu Tyr Asn Gln Pro Asp Lys Tyr Ser Asp Arg 195 200 205Tyr Thr Gly
Ser Gln Asp Asn Gly Gly Val His Ile Asn Ser Gly Ile 210 215 220Ile
Asn Lys Ala Tyr Tyr Leu Ala Ala Gln Gly Gly Thr His Asn Gly225 230
235 240Val Thr Val Ser Gly Ile Gly Arg Asp Lys Ala Val Arg Ile Phe
Tyr 245 250 255Ser Thr Leu Val Asn Tyr Leu Thr Pro Thr Ser Lys Phe
Ala Ala Ala 260 265 270Lys Thr Ala Thr Ile Gln Ala Ala Lys Asp Leu
Tyr Gly Ala Asn Ser 275 280 285Ala Glu Ala Thr Ala Ile Thr Lys Ala
Tyr Gln Ala Val Gly Leu 290 295 300241587DNAArtificial
SequenceSynthetic nucleotide sequence of the synthesized PbaPro1
gene in plasmid pGX147(AprE- PbaPro1) 24gtgagaagca aaaaattgtg
gatcagcttg ttgtttgcgt taacgttaat ctttacgatg 60gcgttcagca acatgagcgc
gcaggctgct ggaaaaatgc ctctgtcaga cagcagcatt 120ccgtttgagg
gcccgtacac atcagaagaa agcatcctgc tgaacaacaa cccggacgag
180atgatctaca atttcctggc acagcaggag cagttcctga acgcagacgt
gaagggccag 240ctgaaaatca tcaaaagaaa cacagacacg agcggcatca
gacacttcag actgaagcag 300tacatcaagg gcgtcccggt ttacggcgct
gagcagacaa tccacctgga caaaaatggc 360gcagtgacga gcgcacttgg
agatctgccg ccgattgaag agcaagcagt cccgaacgat 420ggcgttccgg
cgattagcgc tgatgacgct atcagagccg cggaaaacga agcgacgtca
480agactgggag aacttggcgc accggaactt gaaccgaagg cggaactgaa
catctatcac 540cacgaagacg atggacagac gtacctggtg tacatcacgg
aggtgaatgt gctggagccg 600tcaccgctga gaacaaaata cttcatcaat
gcgctggatg gcagcatcgt tagccaatac 660gacatcatta acttcgccac
aggcacgggc acaggcgttc atggcgacac aaaaacgctt 720acgacaacac
agtcaggctc aacgtaccag ctgaaagaca caacaagagg caagggcatc
780cagacgtata cagccaataa cagaagctca cttccgggct cactgtcaac
aagcagcaat 840aatgtctgga cggacagagc tgcagtggac gcgcacgcgt
atgctgcggc cacgtacgac 900ttctacaaga acaagttcaa cagaaacggc
attgatggca acggcctgct tattagaagc 960acggtccact acggctcaaa
ctacaagaat gcgttttgga acggcgccca aattgtttat 1020ggcgatggag
acggcatcga gttcggacct tttagcggcg acctggatgt ggtcggacat
1080gaactgacgc acggcgttat cgagtatacg gcgaatctgg aatacagaaa
tgaaccgggc 1140gctctgaatg aggccttcgc ggatatcatg ggcaacacaa
ttgagagcaa aaactggctt 1200ctgggcgacg gaatctacac gccgaacatt
ccgggagatg cactgagatc actgagcgac 1260cctacgctgt acaaccagcc
ggacaaatac agcgacagat acacgggatc acaggacaat 1320ggcggcgtcc
atattaactc aggcatcatc aacaaagcgt attatctggc agctcaaggc
1380ggcacgcata atggcgtcac agttagcgga atcggcagag acaaggccgt
cagaattttc 1440tactcaacgc tggtgaacta cctgacaccg acaagcaagt
ttgcagccgc caaaacagcc 1500acgattcagg cagcaaagga cctgtacgga
gcgaactcag cagaggccac agcgattacg 1560aaggcttatc aagccgtggg actgtaa
158725528PRTArtificial SequenceSynthetic amino acid sequence of the
PbaPro1 precursor protein expressed from plasmid
pGX147(AprE-PbaPro1) 25Met Arg Ser Lys Lys Leu Trp Ile Ser Leu Leu
Phe Ala Leu Thr Leu1 5 10 15Ile Phe Thr Met Ala Phe Ser Asn Met Ser
Ala Gln Ala Ala Gly Lys 20 25 30Met Pro Leu Ser Asp Ser Ser Ile Pro
Phe Glu Gly Pro Tyr Thr Ser 35 40 45Glu Glu Ser Ile Leu Leu Asn Asn
Asn Pro Asp Glu Met Ile Tyr
Asn 50 55 60Phe Leu Ala Gln Gln Glu Gln Phe Leu Asn Ala Asp Val Lys
Gly Gln65 70 75 80Leu Lys Ile Ile Lys Arg Asn Thr Asp Thr Ser Gly
Ile Arg His Phe 85 90 95Arg Leu Lys Gln Tyr Ile Lys Gly Val Pro Val
Tyr Gly Ala Glu Gln 100 105 110Thr Ile His Leu Asp Lys Asn Gly Ala
Val Thr Ser Ala Leu Gly Asp 115 120 125Leu Pro Pro Ile Glu Glu Gln
Ala Val Pro Asn Asp Gly Val Pro Ala 130 135 140Ile Ser Ala Asp Asp
Ala Ile Arg Ala Ala Glu Asn Glu Ala Thr Ser145 150 155 160Arg Leu
Gly Glu Leu Gly Ala Pro Glu Leu Glu Pro Lys Ala Glu Leu 165 170
175Asn Ile Tyr His His Glu Asp Asp Gly Gln Thr Tyr Leu Val Tyr Ile
180 185 190Thr Glu Val Asn Val Leu Glu Pro Ser Pro Leu Arg Thr Lys
Tyr Phe 195 200 205Ile Asn Ala Leu Asp Gly Ser Ile Val Ser Gln Tyr
Asp Ile Ile Asn 210 215 220Phe Ala Thr Gly Thr Gly Thr Gly Val His
Gly Asp Thr Lys Thr Leu225 230 235 240Thr Thr Thr Gln Ser Gly Ser
Thr Tyr Gln Leu Lys Asp Thr Thr Arg 245 250 255Gly Lys Gly Ile Gln
Thr Tyr Thr Ala Asn Asn Arg Ser Ser Leu Pro 260 265 270Gly Ser Leu
Ser Thr Ser Ser Asn Asn Val Trp Thr Asp Arg Ala Ala 275 280 285Val
Asp Ala His Ala Tyr Ala Ala Ala Thr Tyr Asp Phe Tyr Lys Asn 290 295
300Lys Phe Asn Arg Asn Gly Ile Asp Gly Asn Gly Leu Leu Ile Arg
Ser305 310 315 320Thr Val His Tyr Gly Ser Asn Tyr Lys Asn Ala Phe
Trp Asn Gly Ala 325 330 335Gln Ile Val Tyr Gly Asp Gly Asp Gly Ile
Glu Phe Gly Pro Phe Ser 340 345 350Gly Asp Leu Asp Val Val Gly His
Glu Leu Thr His Gly Val Ile Glu 355 360 365Tyr Thr Ala Asn Leu Glu
Tyr Arg Asn Glu Pro Gly Ala Leu Asn Glu 370 375 380Ala Phe Ala Asp
Ile Met Gly Asn Thr Ile Glu Ser Lys Asn Trp Leu385 390 395 400Leu
Gly Asp Gly Ile Tyr Thr Pro Asn Ile Pro Gly Asp Ala Leu Arg 405 410
415Ser Leu Ser Asp Pro Thr Leu Tyr Asn Gln Pro Asp Lys Tyr Ser Asp
420 425 430Arg Tyr Thr Gly Ser Gln Asp Asn Gly Gly Val His Ile Asn
Ser Gly 435 440 445Ile Ile Asn Lys Ala Tyr Tyr Leu Ala Ala Gln Gly
Gly Thr His Asn 450 455 460Gly Val Thr Val Ser Gly Ile Gly Arg Asp
Lys Ala Val Arg Ile Phe465 470 475 480Tyr Ser Thr Leu Val Asn Tyr
Leu Thr Pro Thr Ser Lys Phe Ala Ala 485 490 495Ala Lys Thr Ala Thr
Ile Gln Ala Ala Lys Asp Leu Tyr Gly Ala Asn 500 505 510Ser Ala Glu
Ala Thr Ala Ile Thr Lys Ala Tyr Gln Ala Val Gly Leu 515 520
525261779DNAPaenibacillus polymyxa
SC2misc_feature(1)..(1779)nucleotide sequence of the PpoPro1 gene
identified from NCBI database 26atgaaaaaag tatgggtttc gcttcttgga
ggagctatgt tattagggtc tgtcgcgtct 60ggtgcatctg cggagagttc cgtttcgggg
ccagctcagc ttacaccgac cttccacgcc 120gagcaatgga aagcacctac
ctcggtatcg ggggatgaca ttgtatggag ctatttaaat 180cgacaaaaga
aatcgttgct gggtgtggat agctccagtg tacgtgaaca attccgaatc
240gttgatcgca caagcgacaa atccggtgta agccattatc gactgaagca
gtatgtaaac 300ggaattcccg tgtatggagc tgaacaaact attcatgtgg
gcaaatctgg tgaggtcacc 360tcttacttag gagcggtggt taatgaggat
cagcaggcag aagctacgca aggtacaact 420ccaaaaatca gcgcttctga
agcggtctac accgcatata aagaagcagc tgcacggatt 480gaagccctcc
ctacctccga cgatactatt tctaaagacg ctgaggagcc aagcagtgta
540agtaaagata cttacgccga agcagctaac aacgaaaaaa cgctttctgt
tgataaggac 600gagctgagtc ttgatcaggc atctgtcctg aaagatagca
aaattgaagc agtggaacca 660gaaaaaagtt ccattgccaa aatcgctaat
ctgcagcctg aagtagatcc taaagcagaa 720ctctactact accctaaggg
ggatgacctg ctgctggttt atgtaacaga agttaatgtt 780ttagaacctg
ccccactgcg tacccgctac attattgatg ccaatgacgg cagcatcgta
840ttccagtatg acatcattaa tgaagcgaca ggcacaggta aaggtgtgct
tggtgattcc 900aaatcgttca ctactaccgc ttccggcagt agctaccagt
taaaagatac aacacgcggt 960aacggaatcg tgacttacac ggcctccaac
cgtcaaagca tcccaggtac cattttgaca 1020gatgccgata atgtatggaa
tgatccagct ggtgtggacg cccatgcgta tgctgctaaa 1080acctatgatt
actataaagc caaatttgga cgcaacagca ttgacggacg cggtctgcaa
1140cttcgttcga cggtccatta cggtagtcgc tacaacaatg ccttctggaa
cggctcccaa 1200atgacttatg gagatggaga tggtagcaca tttatcgcct
tcagcgggga ccccgatgta 1260gtaggacatg aacttacgca tggtgtcaca
gagtatactt cgaatttgga atattacgga 1320gagtccggcg cattgaatga
agctttctca gacgttatcg ggaatgacat tcagcgcaaa 1380aactggcttg
taggcgatga tatttacacg ccaaacattg caggcgatgc ccttcgctca
1440atgtccaatc caaccctgta cgatcaacca gatcactatt ccaacctgta
cagaggcagc 1500tccgataacg gcggtgttca caccaacagc ggtattatca
ataaagctta ctacttgtta 1560gcacaaggtg gtaatttcca tggcgtaact
gtaaatggaa ttggccgtga tgcagcggtg 1620caaatttact acagtgcctt
tacgaactac ctgacttctt cttccgactt ctccaacgca 1680cgtgctgctg
tgatccaagc cgcaaaagat ctgtacgggg cgaactcagc agaagcaact
1740gcagctgcca agtcttttga cgctgtaggc gtaaactaa
177927592PRTPaenibacillus polymyxa SC2misc_feature(1)..(592)amino
acid sequence of the PpoPro1 precursor protein 27Met Lys Lys Val
Trp Val Ser Leu Leu Gly Gly Ala Met Leu Leu Gly1 5 10 15Ser Val Ala
Ser Gly Ala Ser Ala Glu Ser Ser Val Ser Gly Pro Ala 20 25 30Gln Leu
Thr Pro Thr Phe His Ala Glu Gln Trp Lys Ala Pro Thr Ser 35 40 45Val
Ser Gly Asp Asp Ile Val Trp Ser Tyr Leu Asn Arg Gln Lys Lys 50 55
60Ser Leu Leu Gly Val Asp Ser Ser Ser Val Arg Glu Gln Phe Arg Ile65
70 75 80Val Asp Arg Thr Ser Asp Lys Ser Gly Val Ser His Tyr Arg Leu
Lys 85 90 95Gln Tyr Val Asn Gly Ile Pro Val Tyr Gly Ala Glu Gln Thr
Ile His 100 105 110Val Gly Lys Ser Gly Glu Val Thr Ser Tyr Leu Gly
Ala Val Val Asn 115 120 125Glu Asp Gln Gln Ala Glu Ala Thr Gln Gly
Thr Thr Pro Lys Ile Ser 130 135 140Ala Ser Glu Ala Val Tyr Thr Ala
Tyr Lys Glu Ala Ala Ala Arg Ile145 150 155 160Glu Ala Leu Pro Thr
Ser Asp Asp Thr Ile Ser Lys Asp Ala Glu Glu 165 170 175Pro Ser Ser
Val Ser Lys Asp Thr Tyr Ala Glu Ala Ala Asn Asn Glu 180 185 190Lys
Thr Leu Ser Val Asp Lys Asp Glu Leu Ser Leu Asp Gln Ala Ser 195 200
205Val Leu Lys Asp Ser Lys Ile Glu Ala Val Glu Pro Glu Lys Ser Ser
210 215 220Ile Ala Lys Ile Ala Asn Leu Gln Pro Glu Val Asp Pro Lys
Ala Glu225 230 235 240Leu Tyr Tyr Tyr Pro Lys Gly Asp Asp Leu Leu
Leu Val Tyr Val Thr 245 250 255Glu Val Asn Val Leu Glu Pro Ala Pro
Leu Arg Thr Arg Tyr Ile Ile 260 265 270Asp Ala Asn Asp Gly Ser Ile
Val Phe Gln Tyr Asp Ile Ile Asn Glu 275 280 285Ala Thr Gly Thr Gly
Lys Gly Val Leu Gly Asp Ser Lys Ser Phe Thr 290 295 300Thr Thr Ala
Ser Gly Ser Ser Tyr Gln Leu Lys Asp Thr Thr Arg Gly305 310 315
320Asn Gly Ile Val Thr Tyr Thr Ala Ser Asn Arg Gln Ser Ile Pro Gly
325 330 335Thr Ile Leu Thr Asp Ala Asp Asn Val Trp Asn Asp Pro Ala
Gly Val 340 345 350Asp Ala His Ala Tyr Ala Ala Lys Thr Tyr Asp Tyr
Tyr Lys Ala Lys 355 360 365Phe Gly Arg Asn Ser Ile Asp Gly Arg Gly
Leu Gln Leu Arg Ser Thr 370 375 380Val His Tyr Gly Ser Arg Tyr Asn
Asn Ala Phe Trp Asn Gly Ser Gln385 390 395 400Met Thr Tyr Gly Asp
Gly Asp Gly Ser Thr Phe Ile Ala Phe Ser Gly 405 410 415Asp Pro Asp
Val Val Gly His Glu Leu Thr His Gly Val Thr Glu Tyr 420 425 430Thr
Ser Asn Leu Glu Tyr Tyr Gly Glu Ser Gly Ala Leu Asn Glu Ala 435 440
445Phe Ser Asp Val Ile Gly Asn Asp Ile Gln Arg Lys Asn Trp Leu Val
450 455 460Gly Asp Asp Ile Tyr Thr Pro Asn Ile Ala Gly Asp Ala Leu
Arg Ser465 470 475 480Met Ser Asn Pro Thr Leu Tyr Asp Gln Pro Asp
His Tyr Ser Asn Leu 485 490 495Tyr Arg Gly Ser Ser Asp Asn Gly Gly
Val His Thr Asn Ser Gly Ile 500 505 510Ile Asn Lys Ala Tyr Tyr Leu
Leu Ala Gln Gly Gly Asn Phe His Gly 515 520 525Val Thr Val Asn Gly
Ile Gly Arg Asp Ala Ala Val Gln Ile Tyr Tyr 530 535 540Ser Ala Phe
Thr Asn Tyr Leu Thr Ser Ser Ser Asp Phe Ser Asn Ala545 550 555
560Arg Ala Ala Val Ile Gln Ala Ala Lys Asp Leu Tyr Gly Ala Asn Ser
565 570 575Ala Glu Ala Thr Ala Ala Ala Lys Ser Phe Asp Ala Val Gly
Val Asn 580 585 59028304PRTPaenibacillus polymyxa
SC2misc_feature(1)..(304)amino acid sequence of the predicted
mature form of PpoPro1 28Ala Thr Gly Thr Gly Lys Gly Val Leu Gly
Asp Ser Lys Ser Phe Thr1 5 10 15Thr Thr Ala Ser Gly Ser Ser Tyr Gln
Leu Lys Asp Thr Thr Arg Gly 20 25 30Asn Gly Ile Val Thr Tyr Thr Ala
Ser Asn Arg Gln Ser Ile Pro Gly 35 40 45Thr Ile Leu Thr Asp Ala Asp
Asn Val Trp Asn Asp Pro Ala Gly Val 50 55 60Asp Ala His Ala Tyr Ala
Ala Lys Thr Tyr Asp Tyr Tyr Lys Ala Lys65 70 75 80Phe Gly Arg Asn
Ser Ile Asp Gly Arg Gly Leu Gln Leu Arg Ser Thr 85 90 95Val His Tyr
Gly Ser Arg Tyr Asn Asn Ala Phe Trp Asn Gly Ser Gln 100 105 110Met
Thr Tyr Gly Asp Gly Asp Gly Ser Thr Phe Ile Ala Phe Ser Gly 115 120
125Asp Pro Asp Val Val Gly His Glu Leu Thr His Gly Val Thr Glu Tyr
130 135 140Thr Ser Asn Leu Glu Tyr Tyr Gly Glu Ser Gly Ala Leu Asn
Glu Ala145 150 155 160Phe Ser Asp Val Ile Gly Asn Asp Ile Gln Arg
Lys Asn Trp Leu Val 165 170 175Gly Asp Asp Ile Tyr Thr Pro Asn Ile
Ala Gly Asp Ala Leu Arg Ser 180 185 190Met Ser Asn Pro Thr Leu Tyr
Asp Gln Pro Asp His Tyr Ser Asn Leu 195 200 205Tyr Arg Gly Ser Ser
Asp Asn Gly Gly Val His Thr Asn Ser Gly Ile 210 215 220Ile Asn Lys
Ala Tyr Tyr Leu Leu Ala Gln Gly Gly Asn Phe His Gly225 230 235
240Val Thr Val Asn Gly Ile Gly Arg Asp Ala Ala Val Gln Ile Tyr Tyr
245 250 255Ser Ala Phe Thr Asn Tyr Leu Thr Ser Ser Ser Asp Phe Ser
Asn Ala 260 265 270Arg Ala Ala Val Ile Gln Ala Ala Lys Asp Leu Tyr
Gly Ala Asn Ser 275 280 285Ala Glu Ala Thr Ala Ala Ala Lys Ser Phe
Asp Ala Val Gly Val Asn 290 295 300291800DNAArtificial
SequenceSynthetic nucleotide sequence of the synthesized PpoPro1
gene in plasmid pGX138(AprE-PpoPro1) 29gtgagaagca aaaaattgtg
gatcagcttg ttgtttgcgt taacgttaat ctttacgatg 60gcgttcagca acatgagcgc
gcaggctgct ggaaaagaat catcagtgtc aggaccggct 120cagcttacac
cgacatttca cgcagaacaa tggaaggctc cgacgtcagt ttcaggagac
180gacatcgtgt ggagctacct gaatagacag aagaaaagcc tgctgggagt
ggatagcagc 240agcgtcagag agcagttcag aatcgttgac agaacgagcg
acaaaagcgg agtcagccat 300tatagactga agcagtacgt gaatggcatc
ccggtttatg gcgcagagca gacaattcat 360gttggcaaga gcggagaagt
cacaagctat ctgggcgctg tggtcaatga agatcaacaa 420gccgaggcta
cacagggaac aacgccgaaa attagcgcct cagaggcagt ctacacggcg
480tacaaagaag cggctgcaag aatcgaagcc ctgccgacat cagacgatac
aatttcaaaa 540gatgcggagg agccgagctc agttagcaag gatacatacg
cggaagccgc aaacaatgag 600aaaacactga gcgtggacaa ggacgagctg
tcacttgatc aggctagcgt ccttaaagac 660agcaagatcg aggccgttga
gcctgaaaag tcatcaattg cgaaaatcgc caatctgcaa 720cctgaagtcg
acccgaaggc ggaactgtac tactacccga aaggcgatga cctgcttctg
780gtgtacgtca cggaagtgaa cgtcctggaa ccggcaccgc tgagaacaag
atacatcatc 840gacgcgaacg acggaagcat cgtcttccag tatgacatta
tcaacgaagc aacgggaacg 900ggcaaaggcg ttcttggaga ctcaaagagc
ttcacgacaa cggcttcagg aagcagctac 960cagctgaaag acacgacgag
aggaaacgga atcgtcacat atacggcgtc aaacagacaa 1020agcatccctg
gcacaatcct gacggatgct gacaacgttt ggaatgatcc ggctggcgtg
1080gatgcccatg cttatgcggc aaaaacgtat gactattaca aggcgaagtt
cggcagaaat 1140tcaatcgatg gcagaggact gcagcttaga agcacggtgc
actacggatc aagatataac 1200aatgccttct ggaacggcag ccagatgaca
tacggagacg gagatggaag cacatttatt 1260gcattcagcg gcgaccctga
tgtggttggc catgagctga cgcatggcgt tacagaatat 1320acgagcaatc
ttgaatacta cggcgagtca ggcgctctga acgaggcatt tagcgatgtt
1380atcggcaatg acatccagag aaaaaactgg ctggtgggcg acgatattta
cacgcctaat 1440atcgctggcg atgcccttag atcaatgtca aacccgacgc
tgtatgatca gcctgaccac 1500tactcaaacc tgtatagagg ctcatcagat
aacggaggcg tccatacgaa tagcggcatc 1560attaacaagg catattatct
tctggcccag ggcggcaatt ttcatggagt gacggttaat 1620ggaattggaa
gagacgcagc cgtccaaatc tactacagcg ctttcacgaa ctaccttaca
1680tcaagctcag actttagcaa tgccagagct gctgttatcc aggcagcgaa
ggatctttac 1740ggcgccaact cagccgaagc tacggccgca gctaaatcat
ttgatgcagt gggcgttaat 180030600PRTArtificial SequenceSynthetic
amino acid sequence of the PpoPro1 precursor protein expressed from
plasmid pGX138(AprE-PpoPro1) 30Met Arg Ser Lys Lys Leu Trp Ile Ser
Leu Leu Phe Ala Leu Thr Leu1 5 10 15Ile Phe Thr Met Ala Phe Ser Asn
Met Ser Ala Gln Ala Ala Gly Lys 20 25 30Glu Ser Ser Val Ser Gly Pro
Ala Gln Leu Thr Pro Thr Phe His Ala 35 40 45Glu Gln Trp Lys Ala Pro
Thr Ser Val Ser Gly Asp Asp Ile Val Trp 50 55 60Ser Tyr Leu Asn Arg
Gln Lys Lys Ser Leu Leu Gly Val Asp Ser Ser65 70 75 80Ser Val Arg
Glu Gln Phe Arg Ile Val Asp Arg Thr Ser Asp Lys Ser 85 90 95Gly Val
Ser His Tyr Arg Leu Lys Gln Tyr Val Asn Gly Ile Pro Val 100 105
110Tyr Gly Ala Glu Gln Thr Ile His Val Gly Lys Ser Gly Glu Val Thr
115 120 125Ser Tyr Leu Gly Ala Val Val Asn Glu Asp Gln Gln Ala Glu
Ala Thr 130 135 140Gln Gly Thr Thr Pro Lys Ile Ser Ala Ser Glu Ala
Val Tyr Thr Ala145 150 155 160Tyr Lys Glu Ala Ala Ala Arg Ile Glu
Ala Leu Pro Thr Ser Asp Asp 165 170 175Thr Ile Ser Lys Asp Ala Glu
Glu Pro Ser Ser Val Ser Lys Asp Thr 180 185 190Tyr Ala Glu Ala Ala
Asn Asn Glu Lys Thr Leu Ser Val Asp Lys Asp 195 200 205Glu Leu Ser
Leu Asp Gln Ala Ser Val Leu Lys Asp Ser Lys Ile Glu 210 215 220Ala
Val Glu Pro Glu Lys Ser Ser Ile Ala Lys Ile Ala Asn Leu Gln225 230
235 240Pro Glu Val Asp Pro Lys Ala Glu Leu Tyr Tyr Tyr Pro Lys Gly
Asp 245 250 255Asp Leu Leu Leu Val Tyr Val Thr Glu Val Asn Val Leu
Glu Pro Ala 260 265 270Pro Leu Arg Thr Arg Tyr Ile Ile Asp Ala Asn
Asp Gly Ser Ile Val 275 280 285Phe Gln Tyr Asp Ile Ile Asn Glu Ala
Thr Gly Thr Gly Lys Gly Val 290 295 300Leu Gly Asp Ser Lys Ser Phe
Thr Thr Thr Ala Ser Gly Ser Ser Tyr305 310 315 320Gln Leu Lys Asp
Thr Thr Arg Gly Asn Gly Ile Val Thr Tyr Thr Ala 325 330 335Ser Asn
Arg Gln Ser Ile Pro Gly Thr Ile Leu Thr Asp Ala Asp Asn 340 345
350Val Trp Asn Asp Pro Ala Gly Val Asp Ala His Ala Tyr Ala Ala Lys
355 360 365Thr Tyr Asp Tyr Tyr Lys Ala Lys Phe Gly Arg Asn Ser Ile
Asp Gly 370 375 380Arg Gly Leu Gln Leu Arg Ser Thr Val His Tyr Gly
Ser Arg Tyr Asn385 390 395 400Asn Ala Phe Trp Asn Gly Ser Gln Met
Thr Tyr Gly Asp Gly Asp Gly 405 410 415Ser Thr Phe Ile Ala Phe
Ser
Gly Asp Pro Asp Val Val Gly His Glu 420 425 430Leu Thr His Gly Val
Thr Glu Tyr Thr Ser Asn Leu Glu Tyr Tyr Gly 435 440 445Glu Ser Gly
Ala Leu Asn Glu Ala Phe Ser Asp Val Ile Gly Asn Asp 450 455 460Ile
Gln Arg Lys Asn Trp Leu Val Gly Asp Asp Ile Tyr Thr Pro Asn465 470
475 480Ile Ala Gly Asp Ala Leu Arg Ser Met Ser Asn Pro Thr Leu Tyr
Asp 485 490 495Gln Pro Asp His Tyr Ser Asn Leu Tyr Arg Gly Ser Ser
Asp Asn Gly 500 505 510Gly Val His Thr Asn Ser Gly Ile Ile Asn Lys
Ala Tyr Tyr Leu Leu 515 520 525Ala Gln Gly Gly Asn Phe His Gly Val
Thr Val Asn Gly Ile Gly Arg 530 535 540Asp Ala Ala Val Gln Ile Tyr
Tyr Ser Ala Phe Thr Asn Tyr Leu Thr545 550 555 560Ser Ser Ser Asp
Phe Ser Asn Ala Arg Ala Ala Val Ile Gln Ala Ala 565 570 575Lys Asp
Leu Tyr Gly Ala Asn Ser Ala Glu Ala Thr Ala Ala Ala Lys 580 585
590Ser Phe Asp Ala Val Gly Val Asn 595 600311641DNAPaenibacillus
hunanensismisc_feature(1)..(1641)nucleotide sequence of the PhuPro1
gene isolated from Paenibacillus hunanensis 31ttgaaaaaaa cagttggtct
tttacttgca ggtagcttgc tcgttggtgc tacaacgtcc 60gctttcgcag cagaagcaaa
tgatctggca ccactcggtg attacacgcc aaaattgatt 120acgcaagcaa
caggcatcac tggcgctagt ggcgatgcta aagtatggaa gttcctggag
180aagcaaaaac gtaccatcgt aaccgatgat gcagcttctg ctgatgtgaa
ggaattgttt 240gagatcacaa aacgtcaatc cgattctcaa accggtacag
agcactatcg cctgaaccaa 300acctttaaag gcatcccagt ctatggcgca
gagcaaacac tgcactttga caaatccggc 360aatgtatctc tgtacatggg
tcaggttgtt gaggatgtgt ccgctaaact ggaagcttcc 420gattccaaaa
aaggcgtaac tgaggatgta tacgcttcgg atacgaaaaa tgatctggta
480acaccagaaa tcagcgcttc tcaagccatc tcgattgctg aaaaggatgc
agcttccaaa 540atcggctccc tcggcgaagc acaaaaaacg ccagaagcga
agctgtatat ctacgctcct 600gaggatcaag cagcacgtct ggcttatgtg
acagaagtaa acgtactgga gccatctccg 660ctgcgtactc gctattttgt
agatgcaaaa acaggttcga tcctgttcca atatgatctg 720attgagcatg
caacaggtac aggtaaaggg gtactgggtg ataccaagtc cttcactgta
780ggtacttccg gttcttccta tgtgatgact gatagcacgc gtggaaaagg
tatccaaacc 840tacacggcgt ctaaccgcac atcactgcca ggtagcactg
taacgagcag cagcagcaca 900tttaacgatc cagcatctgt cgatgcccat
gcgtatgcac aaaaagtata tgatttctac 960aaatccaact ttaaccgcaa
cagcatcgac ggtaatggtc tggctatccg ctccactacg 1020cactattcca
cacgttataa caatgcgttc tggaatggtt cccaaatggt atacggtgat
1080ggcgatggtt cgcaattcat cgcattctcc ggcgaccttg acgtagtagg
tcacgagctg 1140acacacggtg taaccgagta cacagcgaac ctggaatact
atggtcaatc cggtgcactg 1200aacgaatcca tttcggatat ctttggtaac
acaatcgaag gtaaaaactg gatggtaggc 1260gatgcgatct acacaccagg
cgtatccggc gatgctcttc gctacatgga tgatccaaca 1320aaaggtggac
aaccagcgcg tatggcagat tacaacaaca caagcgctga taatggcggt
1380gtacacacaa acagtggtat cccgaataaa gcatactact tgctggcaca
gggtggcaca 1440tttggcggtg taaatgtaac aggtatcggt cgctcgcaag
cgatccagat cgtttaccgt 1500gcactaacat actacctgac atccacatct
aacttctcga actaccgttc tgcaatggtg 1560caagcatcta cagacctgta
cggtgcaaac tctacacaaa caacagcggt gaaaaactcg 1620ctgagcgcag
taggcattaa c 164132547PRTPaenibacillus
hunanensismisc_feature(1)..(547)amino acid sequence of the PhuPro1
precursor protein 32Met Lys Lys Thr Val Gly Leu Leu Leu Ala Gly Ser
Leu Leu Val Gly1 5 10 15Ala Thr Thr Ser Ala Phe Ala Ala Glu Ala Asn
Asp Leu Ala Pro Leu 20 25 30Gly Asp Tyr Thr Pro Lys Leu Ile Thr Gln
Ala Thr Gly Ile Thr Gly 35 40 45Ala Ser Gly Asp Ala Lys Val Trp Lys
Phe Leu Glu Lys Gln Lys Arg 50 55 60Thr Ile Val Thr Asp Asp Ala Ala
Ser Ala Asp Val Lys Glu Leu Phe65 70 75 80Glu Ile Thr Lys Arg Gln
Ser Asp Ser Gln Thr Gly Thr Glu His Tyr 85 90 95Arg Leu Asn Gln Thr
Phe Lys Gly Ile Pro Val Tyr Gly Ala Glu Gln 100 105 110Thr Leu His
Phe Asp Lys Ser Gly Asn Val Ser Leu Tyr Met Gly Gln 115 120 125Val
Val Glu Asp Val Ser Ala Lys Leu Glu Ala Ser Asp Ser Lys Lys 130 135
140Gly Val Thr Glu Asp Val Tyr Ala Ser Asp Thr Lys Asn Asp Leu
Val145 150 155 160Thr Pro Glu Ile Ser Ala Ser Gln Ala Ile Ser Ile
Ala Glu Lys Asp 165 170 175Ala Ala Ser Lys Ile Gly Ser Leu Gly Glu
Ala Gln Lys Thr Pro Glu 180 185 190Ala Lys Leu Tyr Ile Tyr Ala Pro
Glu Asp Gln Ala Ala Arg Leu Ala 195 200 205Tyr Val Thr Glu Val Asn
Val Leu Glu Pro Ser Pro Leu Arg Thr Arg 210 215 220Tyr Phe Val Asp
Ala Lys Thr Gly Ser Ile Leu Phe Gln Tyr Asp Leu225 230 235 240Ile
Glu His Ala Thr Gly Thr Gly Lys Gly Val Leu Gly Asp Thr Lys 245 250
255Ser Phe Thr Val Gly Thr Ser Gly Ser Ser Tyr Val Met Thr Asp Ser
260 265 270Thr Arg Gly Lys Gly Ile Gln Thr Tyr Thr Ala Ser Asn Arg
Thr Ser 275 280 285Leu Pro Gly Ser Thr Val Thr Ser Ser Ser Ser Thr
Phe Asn Asp Pro 290 295 300Ala Ser Val Asp Ala His Ala Tyr Ala Gln
Lys Val Tyr Asp Phe Tyr305 310 315 320Lys Ser Asn Phe Asn Arg Asn
Ser Ile Asp Gly Asn Gly Leu Ala Ile 325 330 335Arg Ser Thr Thr His
Tyr Ser Thr Arg Tyr Asn Asn Ala Phe Trp Asn 340 345 350Gly Ser Gln
Met Val Tyr Gly Asp Gly Asp Gly Ser Gln Phe Ile Ala 355 360 365Phe
Ser Gly Asp Leu Asp Val Val Gly His Glu Leu Thr His Gly Val 370 375
380Thr Glu Tyr Thr Ala Asn Leu Glu Tyr Tyr Gly Gln Ser Gly Ala
Leu385 390 395 400Asn Glu Ser Ile Ser Asp Ile Phe Gly Asn Thr Ile
Glu Gly Lys Asn 405 410 415Trp Met Val Gly Asp Ala Ile Tyr Thr Pro
Gly Val Ser Gly Asp Ala 420 425 430Leu Arg Tyr Met Asp Asp Pro Thr
Lys Gly Gly Gln Pro Ala Arg Met 435 440 445Ala Asp Tyr Asn Asn Thr
Ser Ala Asp Asn Gly Gly Val His Thr Asn 450 455 460Ser Gly Ile Pro
Asn Lys Ala Tyr Tyr Leu Leu Ala Gln Gly Gly Thr465 470 475 480Phe
Gly Gly Val Asn Val Thr Gly Ile Gly Arg Ser Gln Ala Ile Gln 485 490
495Ile Val Tyr Arg Ala Leu Thr Tyr Tyr Leu Thr Ser Thr Ser Asn Phe
500 505 510Ser Asn Tyr Arg Ser Ala Met Val Gln Ala Ser Thr Asp Leu
Tyr Gly 515 520 525Ala Asn Ser Thr Gln Thr Thr Ala Val Lys Asn Ser
Leu Ser Ala Val 530 535 540Gly Ile Asn54533304PRTPaenibacillus
hunanensismisc_feature(1)..(304)amino acid sequence of the
predicted mature form of PhuPro1 33Ala Thr Gly Thr Gly Lys Gly Val
Leu Gly Asp Thr Lys Ser Phe Thr1 5 10 15Val Gly Thr Ser Gly Ser Ser
Tyr Val Met Thr Asp Ser Thr Arg Gly 20 25 30Lys Gly Ile Gln Thr Tyr
Thr Ala Ser Asn Arg Thr Ser Leu Pro Gly 35 40 45Ser Thr Val Thr Ser
Ser Ser Ser Thr Phe Asn Asp Pro Ala Ser Val 50 55 60Asp Ala His Ala
Tyr Ala Gln Lys Val Tyr Asp Phe Tyr Lys Ser Asn65 70 75 80Phe Asn
Arg Asn Ser Ile Asp Gly Asn Gly Leu Ala Ile Arg Ser Thr 85 90 95Thr
His Tyr Ser Thr Arg Tyr Asn Asn Ala Phe Trp Asn Gly Ser Gln 100 105
110Met Val Tyr Gly Asp Gly Asp Gly Ser Gln Phe Ile Ala Phe Ser Gly
115 120 125Asp Leu Asp Val Val Gly His Glu Leu Thr His Gly Val Thr
Glu Tyr 130 135 140Thr Ala Asn Leu Glu Tyr Tyr Gly Gln Ser Gly Ala
Leu Asn Glu Ser145 150 155 160Ile Ser Asp Ile Phe Gly Asn Thr Ile
Glu Gly Lys Asn Trp Met Val 165 170 175Gly Asp Ala Ile Tyr Thr Pro
Gly Val Ser Gly Asp Ala Leu Arg Tyr 180 185 190Met Asp Asp Pro Thr
Lys Gly Gly Gln Pro Ala Arg Met Ala Asp Tyr 195 200 205Asn Asn Thr
Ser Ala Asp Asn Gly Gly Val His Thr Asn Ser Gly Ile 210 215 220Pro
Asn Lys Ala Tyr Tyr Leu Leu Ala Gln Gly Gly Thr Phe Gly Gly225 230
235 240Val Asn Val Thr Gly Ile Gly Arg Ser Gln Ala Ile Gln Ile Val
Tyr 245 250 255Arg Ala Leu Thr Tyr Tyr Leu Thr Ser Thr Ser Asn Phe
Ser Asn Tyr 260 265 270Arg Ser Ala Met Val Gln Ala Ser Thr Asp Leu
Tyr Gly Ala Asn Ser 275 280 285Thr Gln Thr Thr Ala Val Lys Asn Ser
Leu Ser Ala Val Gly Ile Asn 290 295 300341671DNAArtificial
SequenceSynthetic nucleotide sequence of the synthesized PhuPro1
gene in plasmid pGX149(AprE- PhuPro1) 34gtgagaagca aaaaattgtg
gatcagcttg ttgtttgcgt taacgttaat ctttacgatg 60gcgttcagca acatgagcgc
gcaggctgct ggaaaagcag aagctaatga tcttgccccg 120cttggcgatt
atacaccgaa gcttattaca caggcaacgg gaattacagg cgcatcaggc
180gatgcgaagg tgtggaagtt cctggagaag cagaagagaa cgattgtcac
ggacgacgcc 240gcaagcgcgg atgtcaagga gctgttcgag atcacgaaga
gacagagcga tagccagacg 300ggaacggagc attacagact gaaccagacg
ttcaagggca ttccggtcta cggagctgaa 360caaacgctgc attttgataa
aagcggcaac gtctcactgt acatgggcca agtcgttgag 420gacgttagcg
ccaaacttga ggctagcgac agcaagaaag gcgtcacaga agatgtctac
480gcgtcagaca cgaaaaacga cctggttaca ccggaaatct cagcttcaca
ggccatctca 540attgcagaga aagacgcagc gtcaaaaatc ggctcactgg
gcgaggctca gaaaacgccg 600gaggcgaaac tttacatcta cgcccctgag
gaccaggctg cgagactggc ttacgtgaca 660gaagttaatg tgctggagcc
gtcaccgctt agaacgagat atttcgtgga cgcaaagacg 720ggcagcattc
tgtttcagta cgatcttatc gaacacgcga caggcacagg aaagggagtt
780ctgggagaca caaaaagctt cacggttggc acgtcaggca gcagctacgt
gatgacagac 840agcacgagag gcaagggcat tcaaacgtat acagcgagca
acagaacaag cctgccggga 900agcacagtca cgagctcatc atcaacgttt
aatgacccgg cctcagtgga tgctcacgca 960tacgcgcaga aagtgtacga
cttctacaaa agcaacttca atagaaacag catcgacgga 1020aacggccttg
cgatcagaag cacgacgcac tacagcacaa gatacaacaa cgccttctgg
1080aacggcagcc aaatggttta cggcgatggc gacggatcac agtttatcgc
atttagcgga 1140gacctggacg tcgttggcca tgagctgaca catggcgtta
cggagtacac agcaaacctg 1200gaatactatg gccagtcagg cgcccttaac
gagagcatca gcgacatttt tggcaatacg 1260atcgaaggaa agaactggat
ggtcggcgac gcaatctaca caccgggcgt ttcaggcgat 1320gcactgagat
atatggacga cccgacaaag ggcggacagc cggccagaat ggcggattac
1380aataatacgt cagcagataa cggcggcgtg catacaaata gcggcatccc
taacaaagca 1440tattacctgc ttgcgcaagg aggaacattt ggcggcgtga
atgttacggg cattggcaga 1500tcacaagcga ttcagatcgt ttacagagcg
ctgacgtact accttacgag cacgagcaat 1560tttagcaact acagaagcgc
aatggtgcag gcaagcacgg atctgtatgg cgcaaattca 1620acacaaacga
cggcggtcaa gaatagcctt tcagcagtgg gcattaacta a
167135556PRTArtificial SequenceSynthetic amino acid sequence of the
PhuPro1 precursor protein expressed from plasmid pGX149(AprE-
PhuPro1) 35Met Arg Ser Lys Lys Leu Trp Ile Ser Leu Leu Phe Ala Leu
Thr Leu1 5 10 15Ile Phe Thr Met Ala Phe Ser Asn Met Ser Ala Gln Ala
Ala Gly Lys 20 25 30Ala Glu Ala Asn Asp Leu Ala Pro Leu Gly Asp Tyr
Thr Pro Lys Leu 35 40 45Ile Thr Gln Ala Thr Gly Ile Thr Gly Ala Ser
Gly Asp Ala Lys Val 50 55 60Trp Lys Phe Leu Glu Lys Gln Lys Arg Thr
Ile Val Thr Asp Asp Ala65 70 75 80Ala Ser Ala Asp Val Lys Glu Leu
Phe Glu Ile Thr Lys Arg Gln Ser 85 90 95Asp Ser Gln Thr Gly Thr Glu
His Tyr Arg Leu Asn Gln Thr Phe Lys 100 105 110Gly Ile Pro Val Tyr
Gly Ala Glu Gln Thr Leu His Phe Asp Lys Ser 115 120 125Gly Asn Val
Ser Leu Tyr Met Gly Gln Val Val Glu Asp Val Ser Ala 130 135 140Lys
Leu Glu Ala Ser Asp Ser Lys Lys Gly Val Thr Glu Asp Val Tyr145 150
155 160Ala Ser Asp Thr Lys Asn Asp Leu Val Thr Pro Glu Ile Ser Ala
Ser 165 170 175Gln Ala Ile Ser Ile Ala Glu Lys Asp Ala Ala Ser Lys
Ile Gly Ser 180 185 190Leu Gly Glu Ala Gln Lys Thr Pro Glu Ala Lys
Leu Tyr Ile Tyr Ala 195 200 205Pro Glu Asp Gln Ala Ala Arg Leu Ala
Tyr Val Thr Glu Val Asn Val 210 215 220Leu Glu Pro Ser Pro Leu Arg
Thr Arg Tyr Phe Val Asp Ala Lys Thr225 230 235 240Gly Ser Ile Leu
Phe Gln Tyr Asp Leu Ile Glu His Ala Thr Gly Thr 245 250 255Gly Lys
Gly Val Leu Gly Asp Thr Lys Ser Phe Thr Val Gly Thr Ser 260 265
270Gly Ser Ser Tyr Val Met Thr Asp Ser Thr Arg Gly Lys Gly Ile Gln
275 280 285Thr Tyr Thr Ala Ser Asn Arg Thr Ser Leu Pro Gly Ser Thr
Val Thr 290 295 300Ser Ser Ser Ser Thr Phe Asn Asp Pro Ala Ser Val
Asp Ala His Ala305 310 315 320Tyr Ala Gln Lys Val Tyr Asp Phe Tyr
Lys Ser Asn Phe Asn Arg Asn 325 330 335Ser Ile Asp Gly Asn Gly Leu
Ala Ile Arg Ser Thr Thr His Tyr Ser 340 345 350Thr Arg Tyr Asn Asn
Ala Phe Trp Asn Gly Ser Gln Met Val Tyr Gly 355 360 365Asp Gly Asp
Gly Ser Gln Phe Ile Ala Phe Ser Gly Asp Leu Asp Val 370 375 380Val
Gly His Glu Leu Thr His Gly Val Thr Glu Tyr Thr Ala Asn Leu385 390
395 400Glu Tyr Tyr Gly Gln Ser Gly Ala Leu Asn Glu Ser Ile Ser Asp
Ile 405 410 415Phe Gly Asn Thr Ile Glu Gly Lys Asn Trp Met Val Gly
Asp Ala Ile 420 425 430Tyr Thr Pro Gly Val Ser Gly Asp Ala Leu Arg
Tyr Met Asp Asp Pro 435 440 445Thr Lys Gly Gly Gln Pro Ala Arg Met
Ala Asp Tyr Asn Asn Thr Ser 450 455 460Ala Asp Asn Gly Gly Val His
Thr Asn Ser Gly Ile Pro Asn Lys Ala465 470 475 480Tyr Tyr Leu Leu
Ala Gln Gly Gly Thr Phe Gly Gly Val Asn Val Thr 485 490 495Gly Ile
Gly Arg Ser Gln Ala Ile Gln Ile Val Tyr Arg Ala Leu Thr 500 505
510Tyr Tyr Leu Thr Ser Thr Ser Asn Phe Ser Asn Tyr Arg Ser Ala Met
515 520 525Val Gln Ala Ser Thr Asp Leu Tyr Gly Ala Asn Ser Thr Gln
Thr Thr 530 535 540Ala Val Lys Asn Ser Leu Ser Ala Val Gly Ile
Asn545 550 555361563DNAPaenibacillus
amylolyticusmisc_feature(1)..(1563)nucleotide sequence of the
PamPro1 gene isolated from Paenibacillus amylolyticus 36atgaaattcg
ccaaagttat gccaacaatt cttggaggag ctcttttgct cgcttccgta 60tcctctgcta
ctgcagctcc agtgtctgat caatccattc cacttcaggc cccttatgcc
120tctgaggggg gtattccatt gaacagtgga acagatgaca ctatctttaa
ttatcttgga 180cagcaggaac aatttctgaa ttccgatgtg aaatcccagc
tcaaaattgt caaaagaaac 240acagatacat ctggcgtaag acacttccgc
ctgaaacagt atattaaagg tatcccggtt 300tatggtgcag aacagacggt
ccacctggac aaaaccggag ccgtgagctc cgcacttggc 360gatcttccac
cgattgaaga gcaggccatt ccgaatgatg gtgtagccga gatcagcgga
420gaagacgcga tccagattgc aaccgaagaa gcaacctccc ggattggaga
gcttggtgcc 480gcggaaatca cgcctcaagc tgaattgaac atctatcatc
atgaagaaga tggtcagaca 540tatctggttt acattacgga agtaaacgta
ctggaacctg cccctctacg gaccaaatat 600ttcattaacg cagtggatgg
cagtatcgta tcccagtttg acctcattaa cttcgctact 660ggaacaggta
caggtgtact cggtgatacc aaaaccctga caaccaccca atccggcagc
720accttccaac tgaaagacac cactcgtggc aatggcatcc aaacgtatac
ggcaaacaat 780ggctcctcac tgcctggtag cttgcttaca gattcggata
atgtatggac cgatcgtgca 840ggtgtagatg ctcatgctca tgccgctgct
acgtatgatt tctacaaaaa caaattcaac 900cgtaacggta ttaatggtaa
cggattgttg atcagatcaa ccgtgcacta cggctccaat 960tacaataacg
ccttctggaa cggggcacag attgtctttg gtgacggaga tggaacgatg
1020ttccgatccc tgtctggtga tctggatgtt gtgggtcatg aattgacgca
tggtgttatt 1080gaatatacag ccaatctgga atatcgcaat gaaccaggtg
cactcaatga agcctttgcc 1140gatattttcg gtaatacgat ccaaagcaaa
aactggctgc tcggtgatga tatctacaca 1200cctaacactc caggagatgc
gctgcgctcc ctctccaacc ctacattgta tggtcaacct 1260gacaaataca
gcgatcgcta cacaggctca caggacaacg gcggtgtcca tatcaacagt
1320ggtatcatca
ataaagccta tttccttgct gctcaaggcg gaacacataa tggtgtgact
1380gttaccggaa tcggccggga taaagcgatc cagattttct acagcacact
ggtgaactac 1440ctgacaccaa cgtccaaatt tgccgctgcc aaaacagcta
ccattcaagc agccaaagat 1500ctgtacggag caacttccgc tgaagctact
gctattacca aagcatatca agctgtaggc 1560ctg 156337521PRTPaenibacillus
amylolyticusmisc_feature(1)..(521)amino acid sequence of the
PamPro1 precursor protein 37Met Lys Phe Ala Lys Val Met Pro Thr Ile
Leu Gly Gly Ala Leu Leu1 5 10 15Leu Ala Ser Val Ser Ser Ala Thr Ala
Ala Pro Val Ser Asp Gln Ser 20 25 30Ile Pro Leu Gln Ala Pro Tyr Ala
Ser Glu Gly Gly Ile Pro Leu Asn 35 40 45Ser Gly Thr Asp Asp Thr Ile
Phe Asn Tyr Leu Gly Gln Gln Glu Gln 50 55 60Phe Leu Asn Ser Asp Val
Lys Ser Gln Leu Lys Ile Val Lys Arg Asn65 70 75 80Thr Asp Thr Ser
Gly Val Arg His Phe Arg Leu Lys Gln Tyr Ile Lys 85 90 95Gly Ile Pro
Val Tyr Gly Ala Glu Gln Thr Val His Leu Asp Lys Thr 100 105 110Gly
Ala Val Ser Ser Ala Leu Gly Asp Leu Pro Pro Ile Glu Glu Gln 115 120
125Ala Ile Pro Asn Asp Gly Val Ala Glu Ile Ser Gly Glu Asp Ala Ile
130 135 140Gln Ile Ala Thr Glu Glu Ala Thr Ser Arg Ile Gly Glu Leu
Gly Ala145 150 155 160Ala Glu Ile Thr Pro Gln Ala Glu Leu Asn Ile
Tyr His His Glu Glu 165 170 175Asp Gly Gln Thr Tyr Leu Val Tyr Ile
Thr Glu Val Asn Val Leu Glu 180 185 190Pro Ala Pro Leu Arg Thr Lys
Tyr Phe Ile Asn Ala Val Asp Gly Ser 195 200 205Ile Val Ser Gln Phe
Asp Leu Ile Asn Phe Ala Thr Gly Thr Gly Thr 210 215 220Gly Val Leu
Gly Asp Thr Lys Thr Leu Thr Thr Thr Gln Ser Gly Ser225 230 235
240Thr Phe Gln Leu Lys Asp Thr Thr Arg Gly Asn Gly Ile Gln Thr Tyr
245 250 255Thr Ala Asn Asn Gly Ser Ser Leu Pro Gly Ser Leu Leu Thr
Asp Ser 260 265 270Asp Asn Val Trp Thr Asp Arg Ala Gly Val Asp Ala
His Ala His Ala 275 280 285Ala Ala Thr Tyr Asp Phe Tyr Lys Asn Lys
Phe Asn Arg Asn Gly Ile 290 295 300Asn Gly Asn Gly Leu Leu Ile Arg
Ser Thr Val His Tyr Gly Ser Asn305 310 315 320Tyr Asn Asn Ala Phe
Trp Asn Gly Ala Gln Ile Val Phe Gly Asp Gly 325 330 335Asp Gly Thr
Met Phe Arg Ser Leu Ser Gly Asp Leu Asp Val Val Gly 340 345 350His
Glu Leu Thr His Gly Val Ile Glu Tyr Thr Ala Asn Leu Glu Tyr 355 360
365Arg Asn Glu Pro Gly Ala Leu Asn Glu Ala Phe Ala Asp Ile Phe Gly
370 375 380Asn Thr Ile Gln Ser Lys Asn Trp Leu Leu Gly Asp Asp Ile
Tyr Thr385 390 395 400Pro Asn Thr Pro Gly Asp Ala Leu Arg Ser Leu
Ser Asn Pro Thr Leu 405 410 415Tyr Gly Gln Pro Asp Lys Tyr Ser Asp
Arg Tyr Thr Gly Ser Gln Asp 420 425 430Asn Gly Gly Val His Ile Asn
Ser Gly Ile Ile Asn Lys Ala Tyr Phe 435 440 445Leu Ala Ala Gln Gly
Gly Thr His Asn Gly Val Thr Val Thr Gly Ile 450 455 460Gly Arg Asp
Lys Ala Ile Gln Ile Phe Tyr Ser Thr Leu Val Asn Tyr465 470 475
480Leu Thr Pro Thr Ser Lys Phe Ala Ala Ala Lys Thr Ala Thr Ile Gln
485 490 495Ala Ala Lys Asp Leu Tyr Gly Ala Thr Ser Ala Glu Ala Thr
Ala Ile 500 505 510Thr Lys Ala Tyr Gln Ala Val Gly Leu 515
52038303PRTPaenibacillus amylolyticusmisc_feature(1)..(303)amino
acid sequence of the predicted mature form of PamPro1 38Ala Thr Gly
Thr Gly Thr Gly Val Leu Gly Asp Thr Lys Thr Leu Thr1 5 10 15Thr Thr
Gln Ser Gly Ser Thr Phe Gln Leu Lys Asp Thr Thr Arg Gly 20 25 30Asn
Gly Ile Gln Thr Tyr Thr Ala Asn Asn Gly Ser Ser Leu Pro Gly 35 40
45Ser Leu Leu Thr Asp Ser Asp Asn Val Trp Thr Asp Arg Ala Gly Val
50 55 60Asp Ala His Ala His Ala Ala Ala Thr Tyr Asp Phe Tyr Lys Asn
Lys65 70 75 80Phe Asn Arg Asn Gly Ile Asn Gly Asn Gly Leu Leu Ile
Arg Ser Thr 85 90 95Val His Tyr Gly Ser Asn Tyr Asn Asn Ala Phe Trp
Asn Gly Ala Gln 100 105 110Ile Val Phe Gly Asp Gly Asp Gly Thr Met
Phe Arg Ser Leu Ser Gly 115 120 125Asp Leu Asp Val Val Gly His Glu
Leu Thr His Gly Val Ile Glu Tyr 130 135 140Thr Ala Asn Leu Glu Tyr
Arg Asn Glu Pro Gly Ala Leu Asn Glu Ala145 150 155 160Phe Ala Asp
Ile Phe Gly Asn Thr Ile Gln Ser Lys Asn Trp Leu Leu 165 170 175Gly
Asp Asp Ile Tyr Thr Pro Asn Thr Pro Gly Asp Ala Leu Arg Ser 180 185
190Leu Ser Asn Pro Thr Leu Tyr Gly Gln Pro Asp Lys Tyr Ser Asp Arg
195 200 205Tyr Thr Gly Ser Gln Asp Asn Gly Gly Val His Ile Asn Ser
Gly Ile 210 215 220Ile Asn Lys Ala Tyr Phe Leu Ala Ala Gln Gly Gly
Thr His Asn Gly225 230 235 240Val Thr Val Thr Gly Ile Gly Arg Asp
Lys Ala Ile Gln Ile Phe Tyr 245 250 255Ser Thr Leu Val Asn Tyr Leu
Thr Pro Thr Ser Lys Phe Ala Ala Ala 260 265 270Lys Thr Ala Thr Ile
Gln Ala Ala Lys Asp Leu Tyr Gly Ala Thr Ser 275 280 285Ala Glu Ala
Thr Ala Ile Thr Lys Ala Tyr Gln Ala Val Gly Leu 290 295
300391587DNAArtificial SequenceSynthetic nucleotide sequence of the
synthesized PamPro1 gene in plasmid pGX146(AprE- PamPro1)
39gtgagaagca aaaaattgtg gatcagcttg ttgtttgcgt taacgttaat ctttacgatg
60gcgttcagca acatgagcgc gcaggctgct ggaaaagctc cggttagcga ccagtcaatc
120cctcttcaag caccgtatgc cagcgaagga ggcattccgc ttaacagcgg
cacggacgac 180acgattttca attacctggg ccaacaggag cagttcctga
acagcgacgt caagagccag 240ctgaagatcg tcaaaagaaa cacagacaca
tcaggcgtga gacacttcag actgaagcaa 300tacatcaagg gcatcccggt
ttatggcgct gaacaaacgg ttcacctgga caaaacaggc 360gcagtttcat
cagcactggg agatctgccg ccgattgaag agcaagcaat cccgaatgat
420ggagttgcgg aaattagcgg cgaggatgca atccaaatcg cgacggagga
ggctacatca 480agaattggag aacttggcgc agcggagatt acaccgcagg
ctgaactgaa catctatcac 540catgaggaag acggccagac gtacctggtt
tacattacgg aagtgaacgt gctggaaccg 600gcacctctga gaacaaagta
ctttatcaac gcggttgacg gcagcatcgt ctcacagttc 660gacctgatta
acttcgccac gggaacagga acgggcgttc ttggagacac aaagacgctg
720acgacgacgc agtcaggcag cacattccag ctgaaggaca caacaagagg
caacggcatc 780caaacgtaca cggcgaacaa tggatcatca ctgccgggct
cactgctgac ggattcagat 840aacgtgtgga cggatagagc tggcgttgac
gcgcatgctc acgctgctgc gacgtacgac 900ttctacaaga acaagttcaa
cagaaacggc attaacggaa atggcctgct gatcagaagc 960acggtgcatt
atggctcaaa ctacaacaac gctttttgga acggcgcaca gatcgtgttt
1020ggcgacggcg atggcacaat gtttagaagc ctgtcaggag acctggatgt
ggtgggccac 1080gaactgacgc acggcgtgat cgagtatacg gcgaaccttg
aatatagaaa cgagccggga 1140gcactgaatg aggcgttcgc ggacattttc
ggcaacacaa tccagagcaa aaactggctg 1200ctgggcgacg atatctatac
accgaacaca ccgggcgatg cactgagatc actgtcaaat 1260ccgacgctgt
atggccaacc ggataagtac tcagacagat atacgggcag ccaagacaat
1320ggcggcgttc acatcaactc aggcatcatc aacaaggctt acttccttgc
ggcccaagga 1380ggaacacata acggcgttac agttacaggc attggcagag
acaaggcgat ccagatcttt 1440tacagcacgc tggtgaacta cctgacacct
acgtcaaagt ttgccgcagc gaaaacagca 1500acaattcagg cggctaaaga
cctgtacgga gcgacatcag ccgaggccac agcaattaca 1560aaagcatatc
aagcagttgg cctttaa 158740528PRTArtificial SequenceSynthetic amino
acid sequence of the PamPro1 precursor protein expressed from
plasmid pGX146(AprE- PamPro1) 40Met Arg Ser Lys Lys Leu Trp Ile Ser
Leu Leu Phe Ala Leu Thr Leu1 5 10 15Ile Phe Thr Met Ala Phe Ser Asn
Met Ser Ala Gln Ala Ala Gly Lys 20 25 30Ala Pro Val Ser Asp Gln Ser
Ile Pro Leu Gln Ala Pro Tyr Ala Ser 35 40 45Glu Gly Gly Ile Pro Leu
Asn Ser Gly Thr Asp Asp Thr Ile Phe Asn 50 55 60Tyr Leu Gly Gln Gln
Glu Gln Phe Leu Asn Ser Asp Val Lys Ser Gln65 70 75 80Leu Lys Ile
Val Lys Arg Asn Thr Asp Thr Ser Gly Val Arg His Phe 85 90 95Arg Leu
Lys Gln Tyr Ile Lys Gly Ile Pro Val Tyr Gly Ala Glu Gln 100 105
110Thr Val His Leu Asp Lys Thr Gly Ala Val Ser Ser Ala Leu Gly Asp
115 120 125Leu Pro Pro Ile Glu Glu Gln Ala Ile Pro Asn Asp Gly Val
Ala Glu 130 135 140Ile Ser Gly Glu Asp Ala Ile Gln Ile Ala Thr Glu
Glu Ala Thr Ser145 150 155 160Arg Ile Gly Glu Leu Gly Ala Ala Glu
Ile Thr Pro Gln Ala Glu Leu 165 170 175Asn Ile Tyr His His Glu Glu
Asp Gly Gln Thr Tyr Leu Val Tyr Ile 180 185 190Thr Glu Val Asn Val
Leu Glu Pro Ala Pro Leu Arg Thr Lys Tyr Phe 195 200 205Ile Asn Ala
Val Asp Gly Ser Ile Val Ser Gln Phe Asp Leu Ile Asn 210 215 220Phe
Ala Thr Gly Thr Gly Thr Gly Val Leu Gly Asp Thr Lys Thr Leu225 230
235 240Thr Thr Thr Gln Ser Gly Ser Thr Phe Gln Leu Lys Asp Thr Thr
Arg 245 250 255Gly Asn Gly Ile Gln Thr Tyr Thr Ala Asn Asn Gly Ser
Ser Leu Pro 260 265 270Gly Ser Leu Leu Thr Asp Ser Asp Asn Val Trp
Thr Asp Arg Ala Gly 275 280 285Val Asp Ala His Ala His Ala Ala Ala
Thr Tyr Asp Phe Tyr Lys Asn 290 295 300Lys Phe Asn Arg Asn Gly Ile
Asn Gly Asn Gly Leu Leu Ile Arg Ser305 310 315 320Thr Val His Tyr
Gly Ser Asn Tyr Asn Asn Ala Phe Trp Asn Gly Ala 325 330 335Gln Ile
Val Phe Gly Asp Gly Asp Gly Thr Met Phe Arg Ser Leu Ser 340 345
350Gly Asp Leu Asp Val Val Gly His Glu Leu Thr His Gly Val Ile Glu
355 360 365Tyr Thr Ala Asn Leu Glu Tyr Arg Asn Glu Pro Gly Ala Leu
Asn Glu 370 375 380Ala Phe Ala Asp Ile Phe Gly Asn Thr Ile Gln Ser
Lys Asn Trp Leu385 390 395 400Leu Gly Asp Asp Ile Tyr Thr Pro Asn
Thr Pro Gly Asp Ala Leu Arg 405 410 415Ser Leu Ser Asn Pro Thr Leu
Tyr Gly Gln Pro Asp Lys Tyr Ser Asp 420 425 430Arg Tyr Thr Gly Ser
Gln Asp Asn Gly Gly Val His Ile Asn Ser Gly 435 440 445Ile Ile Asn
Lys Ala Tyr Phe Leu Ala Ala Gln Gly Gly Thr His Asn 450 455 460Gly
Val Thr Val Thr Gly Ile Gly Arg Asp Lys Ala Ile Gln Ile Phe465 470
475 480Tyr Ser Thr Leu Val Asn Tyr Leu Thr Pro Thr Ser Lys Phe Ala
Ala 485 490 495Ala Lys Thr Ala Thr Ile Gln Ala Ala Lys Asp Leu Tyr
Gly Ala Thr 500 505 510Ser Ala Glu Ala Thr Ala Ile Thr Lys Ala Tyr
Gln Ala Val Gly Leu 515 520 525415PRTArtificial SequenceSynthetic
peptidemisc_feature(3)..(4)Xaa can be any naturally occurring amino
acid 41His Glu Xaa Xaa His1 5425PRTArtificial SequenceSynthetic
peptidemisc_feature(3)..(4)Xaa can be any naturally occurring amino
acid 42His Asp Xaa Xaa His1 5434PRTArtificial SequenceSynthetic
peptideMOD_RES(1)..(1)Succinyl at 5'-endMOD_RES(4)..(4)Para
nitroanilide (pNA) at 3'-end 43Ala Ala Pro
Phe144306PRTPaenibacillus
sp.misc_feature(1)..(306)Paenibacillus_sp_Aloe-11 44Asn Glu Ala Thr
Gly Thr Gly Lys Gly Val Leu Gly Asp Thr Lys Thr1 5 10 15Phe Asn Thr
Thr Ala Ser Gly Ser Ser Tyr Gln Leu Arg Asp Thr Thr 20 25 30Arg Gly
Asn Gly Ile Val Thr Tyr Thr Ala Ser Asn Arg Gln Ser Ile 35 40 45Pro
Gly Thr Ile Leu Thr Asp Ala Asp Asn Val Trp Asn Asp Pro Ala 50 55
60Gly Val Asp Ala His Ala Tyr Ala Ala Lys Thr Tyr Asp Tyr Tyr Lys65
70 75 80Glu Lys Phe Asn Arg Asn Ser Ile Asp Gly Arg Gly Leu Gln Leu
Arg 85 90 95Ser Thr Val His Tyr Gly Asn Arg Tyr Asn Asn Ala Phe Trp
Asn Gly 100 105 110Ser Gln Met Thr Tyr Gly Asp Gly Asp Gly Thr Thr
Phe Ile Ala Phe 115 120 125Ser Gly Asp Pro Asp Val Val Gly His Glu
Leu Thr His Gly Val Thr 130 135 140Glu Tyr Thr Ser Asn Leu Glu Tyr
Tyr Gly Glu Ser Gly Ala Leu Asn145 150 155 160Glu Ala Phe Ser Asp
Ile Ile Gly Asn Asp Ile Gln Arg Lys Asn Trp 165 170 175Leu Val Gly
Asp Asp Ile Tyr Thr Pro Arg Ile Ala Gly Asp Ala Leu 180 185 190Arg
Ser Met Ser Asn Pro Thr Leu Tyr Asp Gln Pro Asp His Tyr Ser 195 200
205Asn Leu Tyr Arg Gly Ser Ser Asp Asn Gly Gly Val His Thr Asn Ser
210 215 220Gly Ile Ile Asn Lys Ala Tyr Tyr Leu Leu Ala Gln Gly Gly
Thr Phe225 230 235 240His Gly Val Thr Val Asn Gly Ile Gly Arg Asp
Ala Ala Val Gln Ile 245 250 255Tyr Tyr Ser Ala Phe Thr Asn Tyr Leu
Thr Ser Ser Ser Asp Phe Ser 260 265 270Asn Ala Arg Asp Ala Val Val
Gln Ala Ala Lys Asp Leu Tyr Gly Ala 275 280 285Ser Ser Ala Gln Ala
Thr Ala Ala Ala Lys Ser Phe Asp Ala Val Gly 290 295 300Val
Asn30545316PRTB.
thermoproteolyticusmisc_feature(1)..(316)B_thermoproteolyticus_P00800
45Ile Thr Gly Thr Ser Thr Val Gly Val Gly Arg Gly Val Leu Gly Asp1
5 10 15Gln Lys Asn Ile Asn Thr Thr Tyr Ser Thr Tyr Tyr Tyr Leu Gln
Asp 20 25 30Asn Thr Arg Gly Asn Gly Ile Phe Thr Tyr Asp Ala Lys Tyr
Arg Thr 35 40 45Thr Leu Pro Gly Ser Leu Trp Ala Asp Ala Asp Asn Gln
Phe Phe Ala 50 55 60Ser Tyr Asp Ala Pro Ala Val Asp Ala His Tyr Tyr
Ala Gly Val Thr65 70 75 80Tyr Asp Tyr Tyr Lys Asn Val His Asn Arg
Leu Ser Tyr Asp Gly Asn 85 90 95Asn Ala Ala Ile Arg Ser Ser Val His
Tyr Ser Gln Gly Tyr Asn Asn 100 105 110Ala Phe Trp Asn Gly Ser Gln
Met Val Tyr Gly Asp Gly Asp Gly Gln 115 120 125Thr Phe Ile Pro Leu
Ser Gly Gly Ile Asp Val Val Ala His Glu Leu 130 135 140Thr His Ala
Val Thr Asp Tyr Thr Ala Gly Leu Ile Tyr Gln Asn Glu145 150 155
160Ser Gly Ala Ile Asn Glu Ala Ile Ser Asp Ile Phe Gly Thr Leu Val
165 170 175Glu Phe Tyr Ala Asn Lys Asn Pro Asp Trp Glu Ile Gly Glu
Asp Val 180 185 190Tyr Thr Pro Gly Ile Ser Gly Asp Ser Leu Arg Ser
Met Ser Asp Pro 195 200 205Ala Lys Tyr Gly Asp Pro Asp His Tyr Ser
Lys Arg Tyr Thr Gly Thr 210 215 220Gln Asp Asn Gly Gly Val His Ile
Asn Ser Gly Ile Ile Asn Lys Ala225 230 235 240Ala Tyr Leu Ile Ser
Gln Gly Gly Thr His Tyr Gly Val Ser Val Val 245 250 255Gly Ile Gly
Arg Asp Lys Leu Gly Lys Ile Phe Tyr Arg Ala Leu Thr 260 265 270Gln
Tyr Leu Thr Pro Thr Ser Asn Phe Ser Gln Leu Arg Ala Ala Ala 275 280
285Val Gln Ser Ala Thr Asp Leu Tyr Gly Ser Thr Ser Gln Glu Val Ala
290 295 300Ser Val Lys Gln Ala Phe Asp Ala Val Gly Val Lys305 310
31546306PRTPaenibacillus sp.
Aloe-11misc_feature(1)..(306)ZP_09775365.1_P_sp_Aloe-11 46Ala Thr
Gly Thr
Gly Arg Gly Val Asp Gly Lys Thr Lys Ser Phe Thr1 5 10 15Thr Thr Ala
Ser Gly Asn Arg Tyr Gln Leu Lys Asp Thr Thr Arg Ser 20 25 30Asn Gly
Ile Val Thr Tyr Thr Ala Gly Asn Arg Gln Thr Thr Pro Gly 35 40 45Thr
Ile Leu Thr Asp Thr Asp Asn Val Trp Glu Asp Pro Ala Ala Val 50 55
60Asp Ala His Ala Tyr Ala Ile Lys Thr Tyr Asp Tyr Tyr Lys Asn Lys65
70 75 80Phe Gly Arg Asp Ser Ile Asp Gly Arg Gly Met Gln Ile Arg Ser
Thr 85 90 95Val His Tyr Gly Lys Lys Tyr Asn Asn Ala Phe Trp Asn Gly
Ser Gln 100 105 110Met Thr Tyr Gly Asp Gly Asp Gly Ser Thr Phe Thr
Phe Phe Ser Gly 115 120 125Asp Pro Asp Val Val Gly His Glu Leu Thr
His Gly Val Thr Glu Phe 130 135 140Thr Ser Asn Leu Glu Tyr Tyr Gly
Glu Ser Gly Ala Leu Asn Glu Ala145 150 155 160Phe Ser Asp Ile Ile
Gly Asn Asp Ile Asp Gly Thr Ser Trp Leu Leu 165 170 175Gly Asp Gly
Ile Tyr Thr Pro Asn Ile Pro Gly Asp Ala Leu Arg Ser 180 185 190Leu
Ser Asp Pro Thr Arg Phe Gly Gln Pro Asp His Tyr Ser Asn Phe 195 200
205Tyr Pro Asp Pro Asn Asn Asp Asp Glu Gly Gly Val His Thr Asn Ser
210 215 220Gly Ile Ile Asn Lys Ala Tyr Tyr Leu Leu Ala Gln Gly Gly
Thr Ser225 230 235 240His Gly Val Thr Val Thr Gly Ile Gly Arg Glu
Ala Ala Val Phe Ile 245 250 255Tyr Tyr Asn Ala Phe Thr Asn Tyr Leu
Thr Ser Thr Ser Asn Phe Ser 260 265 270Asn Ala Arg Ala Ala Val Ile
Gln Ala Ala Lys Asp Phe Tyr Gly Ala 275 280 285Asp Ser Leu Ala Val
Thr Ser Ala Ile Gln Ser Phe Asp Ala Val Gly 290 295 300Ile
Lys30547304PRTP.
terraemisc_feature(1)..(304)P_terrae_HPL-003_YP_005073223. 47Ala
Thr Gly Thr Gly Lys Gly Val Leu Gly Asp Thr Lys Ser Phe Asn1 5 10
15Thr Thr Gln Ser Gly Ser Ser Tyr Gln Leu Lys Asp Thr Thr Arg Gly
20 25 30Asn Gly Ile Val Thr Tyr Thr Ala Ser Asn Arg Gln Thr Ile Pro
Gly 35 40 45Thr Leu Leu Thr Asp Ala Asp Asn Val Trp Asn Asp Pro Ala
Gly Val 50 55 60Asp Ala His Ala Tyr Ala Ala Lys Thr Tyr Asp Tyr Tyr
Lys Asp Lys65 70 75 80Phe Gly Arg Asn Ser Ile Asp Gly Arg Gly Leu
Gln Leu Arg Ser Thr 85 90 95Val His Tyr Gly Ser Arg Tyr Asn Asn Ala
Phe Trp Asn Gly Ser Gln 100 105 110Met Thr Tyr Gly Asp Gly Asp Gly
Thr Thr Phe Ile Ala Phe Ser Gly 115 120 125Asp Pro Asp Val Val Gly
His Glu Leu Thr His Gly Val Thr Glu Tyr 130 135 140Thr Ser Asn Leu
Asp Tyr Tyr Gly Glu Ser Gly Ala Leu Asn Glu Ser145 150 155 160Phe
Ser Asp Ile Ile Gly Asn Asp Ile Gln Arg Lys Asn Trp Leu Val 165 170
175Gly Asp Asp Ile Tyr Thr Pro Ser Ile Ala Gly Asp Ala Leu Arg Ser
180 185 190Met Ser Asn Pro Thr Leu Tyr Asp Gln Pro Asp His Tyr Ser
Asn Leu 195 200 205Tyr Lys Gly Ser Ser Asp Asn Gly Gly Val His Thr
Asn Ser Gly Ile 210 215 220Ile Asn Lys Ala Tyr Tyr Leu Leu Ala Gln
Gly Gly Thr Phe His Asn225 230 235 240Val Thr Val Ser Gly Ile Gly
Arg Asp Ala Ala Val Gln Ile Tyr Tyr 245 250 255Ser Ala Phe Thr Asn
Tyr Leu Thr Ser Thr Ser Asn Phe Ser Asn Thr 260 265 270Arg Ala Ala
Val Val Gln Ala Ala Lys Asp Leu Tyr Gly Ala Asn Ser 275 280 285Ala
Gln Ala Thr Ala Ala Ala Lys Ser Phe Asp Ala Val Gly Val Asn 290 295
30048301PRTPaenibacillus
elgiimisc_feature(1)..(301)Paenibacillus_elgii_B69_ZP_090 48Ala Thr
Gly Thr Gly Lys Gly Val Leu Gly Asp Thr Lys Ser Phe Thr1 5 10 15Thr
Thr Gln Ser Gly Ser Ser Tyr Gln Leu Lys Asp Thr Thr Arg Gly 20 25
30Gln Gly Ile Val Thr Tyr Ser Ala Gly Asn Arg Thr Ser Leu Pro Gly
35 40 45Ser Leu Leu Thr Ser Thr Asn Asn Ile Trp Asn Asp Gly Ser Ala
Val 50 55 60Asp Ala His Ala Tyr Thr Gly Lys Val Tyr Asp Tyr Tyr Lys
Asn Lys65 70 75 80Phe Gly Arg Asn Ser Ile Asp Gly Asn Gly Leu Gln
Leu Lys Ser Thr 85 90 95Val His Tyr Ser Thr Arg Tyr Asn Asn Ala Phe
Trp Asn Gly Val Gln 100 105 110Met Val Tyr Gly Asp Gly Asp Gly Val
Thr Phe Arg Ser Phe Pro Ala 115 120 125Asp Pro Asp Val Ile Gly His
Glu Leu Thr His Gly Val Thr Glu Ser 130 135 140Thr Ala Gly Leu Glu
Tyr Tyr Gly Glu Ser Gly Ala Leu Asn Glu Ser145 150 155 160Ile Ser
Asp Ile Phe Gly Asn Ala Ile Glu Gly Lys Asn Trp Leu Ile 165 170
175Gly Asp Leu Ile Thr Leu Asn Ala Gly Ala Leu Arg Ser Met Glu Asn
180 185 190Pro Lys Leu Tyr Arg Gln Pro Asp Arg Tyr Gln Asp Arg Tyr
Thr Gly 195 200 205Pro Ser Asp Asn Gly Gly Val His Thr Asn Ser Gly
Ile Asn Asn Lys 210 215 220Ala Phe His Leu Ile Ala Gln Gly Gly Thr
His Tyr Gly Val Thr Val225 230 235 240Asn Gly Ile Gly Arg Ser Ala
Ala Glu Gln Ile Phe Tyr Asp Ala Leu 245 250 255Thr His Tyr Leu Thr
Pro Thr Ser Asn Phe Ser Ala Ile Arg Ala Ala 260 265 270Ala Ile Gln
Ala Ala Thr Asp Ser Phe Gly Ala Asn Ser Ser Gln Val 275 280 285Asp
Ala Val Lys Lys Ala Tyr Asn Ala Val Gly Val Asn 290 295
30049306PRTPaenibacillus polymyxa
SC2misc_feature(1)..(306)P_polymyxa_SC2 49Asn Glu Ala Thr Gly Thr
Gly Lys Gly Val Leu Gly Asp Ser Lys Ser1 5 10 15Phe Thr Thr Thr Ala
Ser Gly Ser Ser Tyr Gln Leu Lys Asp Thr Thr 20 25 30Arg Gly Asn Gly
Ile Val Thr Tyr Thr Ala Ser Asn Arg Gln Ser Ile 35 40 45Pro Gly Thr
Ile Leu Thr Asp Ala Asp Asn Val Trp Asn Asp Pro Ala 50 55 60Gly Val
Asp Ala His Ala Tyr Ala Ala Lys Thr Tyr Asp Tyr Tyr Lys65 70 75
80Ala Lys Phe Gly Arg Asn Ser Ile Asp Gly Arg Gly Leu Gln Leu Arg
85 90 95Ser Thr Val His Tyr Gly Ser Arg Tyr Asn Asn Ala Phe Trp Asn
Gly 100 105 110Ser Gln Met Thr Tyr Gly Asp Gly Asp Gly Ser Thr Phe
Ile Ala Phe 115 120 125Ser Gly Asp Pro Asp Val Val Gly His Glu Leu
Thr His Gly Val Thr 130 135 140Glu Tyr Thr Ser Asn Leu Glu Tyr Tyr
Gly Glu Ser Gly Ala Leu Asn145 150 155 160Glu Ala Phe Ser Asp Val
Ile Gly Asn Asp Ile Gln Arg Lys Asn Trp 165 170 175Leu Val Gly Asp
Asp Ile Tyr Thr Pro Asn Ile Ala Gly Asp Ala Leu 180 185 190Arg Ser
Met Ser Asn Pro Thr Leu Tyr Asp Gln Pro Asp His Tyr Ser 195 200
205Asn Leu Tyr Arg Gly Ser Ser Asp Asn Gly Gly Val His Thr Asn Ser
210 215 220Gly Ile Ile Asn Lys Ala Tyr Tyr Leu Leu Ala Gln Gly Gly
Asn Phe225 230 235 240His Gly Val Thr Val Asn Gly Ile Gly Arg Asp
Ala Ala Val Gln Ile 245 250 255Tyr Tyr Ser Ala Phe Thr Asn Tyr Leu
Thr Ser Ser Ser Asp Phe Ser 260 265 270Asn Ala Arg Ala Ala Val Ile
Gln Ala Ala Lys Asp Leu Tyr Gly Ala 275 280 285Asn Ser Ala Glu Ala
Thr Ala Ala Ala Lys Ser Phe Asp Ala Val Gly 290 295 300Val
Asn30550306PRTPaenibacillus polymyxa
SC2misc_feature(1)..(306)P_polymyxa_SC2_YP_003948511.1 50Asn Glu
Ala Thr Gly Thr Gly Lys Gly Val Leu Gly Asp Ser Lys Ser1 5 10 15Phe
Thr Thr Thr Ala Ser Gly Ser Ser Tyr Gln Leu Lys Asp Thr Thr 20 25
30Arg Gly Asn Gly Ile Val Thr Tyr Thr Ala Ser Asn Arg Gln Ser Ile
35 40 45Pro Gly Thr Ile Leu Thr Asp Ala Asp Asn Val Trp Asn Asp Pro
Ala 50 55 60Gly Val Asp Ala His Ala Tyr Ala Ala Lys Thr Tyr Asp Tyr
Tyr Lys65 70 75 80Ala Lys Phe Gly Arg Asn Ser Ile Asp Gly Arg Gly
Leu Gln Leu Arg 85 90 95Ser Thr Val His Tyr Gly Ser Arg Tyr Asn Asn
Ala Phe Trp Asn Gly 100 105 110Ser Gln Met Thr Tyr Gly Asp Gly Asp
Gly Ser Thr Phe Ile Ala Phe 115 120 125Ser Gly Asp Pro Asp Val Val
Gly His Glu Leu Thr His Gly Val Thr 130 135 140Glu Tyr Thr Ser Asn
Leu Glu Tyr Tyr Gly Glu Ser Gly Ala Leu Asn145 150 155 160Glu Ala
Phe Ser Asp Val Ile Gly Asn Asp Ile Gln Arg Lys Asn Trp 165 170
175Leu Val Gly Asp Asp Ile Tyr Thr Pro Asn Ile Ala Gly Asp Ala Leu
180 185 190Arg Ser Met Ser Asn Pro Thr Leu Tyr Asp Gln Pro Asp His
Tyr Ser 195 200 205Asn Leu Tyr Arg Gly Ser Ser Asp Asn Gly Gly Val
His Thr Asn Ser 210 215 220Gly Ile Ile Asn Lys Ala Tyr Tyr Leu Leu
Ala Gln Gly Gly Asn Phe225 230 235 240His Gly Val Thr Val Asn Gly
Ile Gly Arg Asp Ala Ala Val Gln Ile 245 250 255Tyr Tyr Ser Ala Phe
Thr Asn Tyr Leu Thr Ser Ser Ser Asp Phe Ser 260 265 270Asn Ala Arg
Ala Ala Val Ile Gln Ala Ala Lys Asp Leu Tyr Gly Ala 275 280 285Asn
Ser Ala Glu Ala Thr Ala Ala Ala Lys Ser Phe Asp Ala Val Gly 290 295
300Val Asn30551304PRTP.
terraemisc_feature(1)..(304)P_terrae_HPL-003_YP_005073223 51Ala Thr
Gly Thr Gly Lys Gly Val Leu Gly Asp Thr Lys Ser Phe Asn1 5 10 15Thr
Thr Gln Ser Gly Ser Ser Tyr Gln Leu Lys Asp Thr Thr Arg Gly 20 25
30Asn Gly Ile Val Thr Tyr Thr Ala Ser Asn Arg Gln Thr Ile Pro Gly
35 40 45Thr Leu Leu Thr Asp Ala Asp Asn Val Trp Asn Asp Pro Ala Gly
Val 50 55 60Asp Ala His Ala Tyr Ala Ala Lys Thr Tyr Asp Tyr Tyr Lys
Asp Lys65 70 75 80Phe Gly Arg Asn Ser Ile Asp Gly Arg Gly Leu Gln
Leu Arg Ser Thr 85 90 95Val His Tyr Gly Ser Arg Tyr Asn Asn Ala Phe
Trp Asn Gly Ser Gln 100 105 110Met Thr Tyr Gly Asp Gly Asp Gly Thr
Thr Phe Ile Ala Phe Ser Gly 115 120 125Asp Pro Asp Val Val Gly His
Glu Leu Thr His Gly Val Thr Glu Tyr 130 135 140Thr Ser Asn Leu Asp
Tyr Tyr Gly Glu Ser Gly Ala Leu Asn Glu Ser145 150 155 160Phe Ser
Asp Ile Ile Gly Asn Asp Ile Gln Arg Lys Asn Trp Leu Val 165 170
175Gly Asp Asp Ile Tyr Thr Pro Ser Ile Ala Gly Asp Ala Leu Arg Ser
180 185 190Met Ser Asn Pro Thr Leu Tyr Asp Gln Pro Asp His Tyr Ser
Asn Leu 195 200 205Tyr Lys Gly Ser Ser Asp Asn Gly Gly Val His Thr
Asn Ser Gly Ile 210 215 220Ile Asn Lys Ala Tyr Tyr Leu Leu Ala Gln
Gly Gly Thr Phe His Asn225 230 235 240Val Thr Val Ser Gly Ile Gly
Arg Asp Ala Ala Val Gln Ile Tyr Tyr 245 250 255Ser Ala Phe Thr Asn
Tyr Leu Thr Ser Thr Ser Asn Phe Ser Asn Thr 260 265 270Arg Ala Ala
Val Val Gln Ala Ala Lys Asp Leu Tyr Gly Ala Asn Ser 275 280 285Ala
Gln Ala Thr Ala Ala Ala Lys Ser Phe Asp Ala Val Gly Val Asn 290 295
30052309PRTP. peoriaemisc_feature(1)..(309)P_peoriae_KCTC 52Asp Ile
Ile Asn Glu Ala Thr Gly Thr Gly Lys Gly Val Leu Gly Asp1 5 10 15Thr
Lys Ser Phe Thr Thr Thr Ala Ser Gly Ser Ser Tyr Gln Leu Arg 20 25
30Asp Thr Thr Arg Gly Asn Gly Ile Val Thr Tyr Thr Ala Ser Asn Arg
35 40 45Gln Ser Ile Pro Gly Thr Ile Leu Thr Asp Ala Asp Asn Val Trp
Asn 50 55 60Asp Pro Ala Gly Val Asp Ala His Ala Tyr Ala Ala Lys Thr
Tyr Asp65 70 75 80Tyr Tyr Lys Glu Lys Phe Asn Arg Asn Ser Ile Asp
Gly Arg Gly Leu 85 90 95Gln Leu Arg Ser Thr Val His Tyr Gly Asn Arg
Tyr Asn Asn Ala Phe 100 105 110Trp Asn Gly Ser Gln Met Thr Tyr Gly
Asp Gly Asp Gly Thr Thr Phe 115 120 125Ile Ala Phe Ser Gly Asp Pro
Asp Val Val Gly His Glu Leu Thr His 130 135 140Gly Val Thr Glu Tyr
Thr Ser Asn Leu Glu Tyr Tyr Gly Glu Ser Gly145 150 155 160Ala Leu
Asn Glu Ser Phe Ser Asp Ile Ile Gly Asn Asp Ile Gln Arg 165 170
175Lys Asn Trp Leu Val Gly Asp Asp Ile Tyr Thr Pro Arg Ile Ala Gly
180 185 190Asp Ala Leu Arg Ser Met Ser Asn Pro Thr Leu Tyr Asp Gln
Pro Asp 195 200 205His Tyr Ser Asn Leu Tyr Arg Gly Ser Ser Asp Asn
Gly Gly Val His 210 215 220Thr Asn Ser Gly Ile Ile Asn Lys Ala Tyr
Tyr Leu Leu Ala Gln Gly225 230 235 240Gly Thr Phe His Gly Val Thr
Val Asn Gly Ile Gly Arg Asp Ala Ala 245 250 255Val Gln Ile Tyr Tyr
Ser Ala Phe Thr Asn Tyr Leu Thr Ser Ser Ser 260 265 270Asp Phe Ser
Asn Ala Arg Asp Ala Val Val Gln Ala Ala Lys Asp Leu 275 280 285Tyr
Gly Ala Ser Ser Ala Gln Ala Thr Ala Ala Ala Lys Ala Phe Asp 290 295
300Ala Val Gly Val Asn30553316PRTBacillus
thermoproteolyticusmisc_feature(1)..(316)1KEI.A 53Ile Thr Gly Thr
Ser Thr Val Gly Val Gly Arg Gly Val Leu Gly Asp1 5 10 15Gln Lys Asn
Ile Asn Thr Thr Tyr Ser Thr Tyr Tyr Tyr Leu Gln Asp 20 25 30Asn Thr
Arg Gly Asn Gly Ile Phe Thr Tyr Asp Ala Lys Tyr Arg Thr 35 40 45Thr
Leu Pro Gly Ser Leu Trp Ala Asp Ala Asp Asn Gln Phe Phe Ala 50 55
60Ser Tyr Asp Ala Pro Ala Val Asp Ala His Tyr Tyr Ala Gly Val Thr65
70 75 80Tyr Asp Tyr Tyr Lys Asn Val His Asn Arg Leu Ser Tyr Asp Gly
Asn 85 90 95Asn Ala Ala Ile Arg Ser Ser Val His Tyr Ser Gln Gly Tyr
Asn Asn 100 105 110Ala Phe Trp Asn Gly Ser Gln Met Val Tyr Gly Asp
Gly Asp Gly Gln 115 120 125Thr Phe Ile Pro Leu Ser Gly Gly Ile Asp
Val Val Ala His Glu Leu 130 135 140Thr His Ala Val Thr Asp Tyr Thr
Ala Gly Leu Ile Tyr Gln Asn Glu145 150 155 160Ser Gly Ala Ile Asn
Glu Ala Ile Ser Asp Ile Phe Gly Thr Leu Val 165 170 175Glu Phe Tyr
Ala Asn Lys Asn Pro Asp Trp Glu Ile Gly Glu Asp Val 180 185 190Tyr
Thr Pro Gly Ile Ser Gly Asp Ser Leu Arg Ser Met Ser Asp Pro 195 200
205Ala Lys Tyr Gly Asp Pro Asp His Tyr Ser Lys Arg Tyr Thr Gly Thr
210 215 220Gln Asp Asn Gly Gly Val His Ile Asn Ser Gly Ile Ile Asn
Lys Ala225 230 235 240Ala Tyr Leu Ile Ser Gln Gly Gly Thr His Tyr
Gly Val Ser Val Val 245 250 255Gly Ile Gly Arg Asp Lys Leu Gly Lys
Ile Phe Tyr Arg Ala Leu Thr 260
265 270Gln Tyr Leu Thr Pro Thr Ser Asn Phe Ser Gln Leu Arg Ala Ala
Ala 275 280 285Val Gln Ser Ala Thr Asp Leu Tyr Gly Ser Thr Ser Gln
Glu Val Ala 290 295 300Ser Val Lys Gln Ala Phe Asp Ala Val Gly Val
Lys305 310 31554316PRTB.
caldolyticusmisc_feature(1)..(316)B_caldolyticus_AAA22623.1 54Thr
Ser Thr Val Gly Val Gly Arg Gly Val Leu Gly Asp Gln Lys Tyr1 5 10
15Ile Asn Thr Thr Tyr Ser Ser Tyr Tyr Gly Tyr Tyr Tyr Leu Gln Asp
20 25 30Asn Thr Arg Gly Ser Gly Ile Phe Thr Tyr Asp Gly Arg Asn Arg
Thr 35 40 45Val Leu Pro Gly Ser Leu Trp Ala Asp Gly Asp Asn Gln Phe
Phe Ala 50 55 60Ser Tyr Asp Ala Ala Ala Val Asp Ala His Tyr Tyr Ala
Gly Val Val65 70 75 80Tyr Asp Tyr Tyr Lys Asn Val His Gly Arg Leu
Ser Tyr Asp Gly Ser 85 90 95Asn Ala Ala Ile Arg Ser Thr Val His Tyr
Gly Arg Gly Tyr Asn Asn 100 105 110Ala Phe Trp Asn Gly Ser Gln Met
Val Tyr Gly Asp Gly Asp Gly Gln 115 120 125Thr Phe Leu Pro Phe Ser
Gly Gly Ile Asp Val Val Gly His Glu Leu 130 135 140Thr His Ala Val
Thr Asp Tyr Thr Ala Gly Leu Val Tyr Gln Asn Glu145 150 155 160Ser
Gly Ala Ile Asn Glu Ala Met Ser Asp Ile Phe Gly Thr Leu Val 165 170
175Glu Phe Tyr Ala Asn Arg Asn Pro Asp Trp Glu Ile Gly Glu Asp Ile
180 185 190Tyr Thr Pro Gly Val Ala Gly Asp Ala Leu Arg Ser Met Ser
Asp Pro 195 200 205Ala Lys Tyr Gly Asp Pro Asp His Tyr Ser Lys Arg
Tyr Thr Gly Thr 210 215 220Gln Asp Asn Gly Gly Val His Thr Asn Ser
Gly Ile Ile Asn Lys Ala225 230 235 240Ala Tyr Leu Leu Ser Gln Gly
Gly Val His Tyr Gly Val Ser Val Thr 245 250 255Gly Ile Gly Arg Asp
Lys Met Gly Lys Ile Phe Tyr Arg Ala Leu Val 260 265 270Tyr Tyr Leu
Thr Pro Thr Ser Asn Phe Ser Gln Leu Arg Ala Ala Cys 275 280 285Val
Gln Ala Ala Ala Asp Leu Tyr Gly Ser Thr Ser Gln Glu Val Asn 290 295
300Ser Val Lys Gln Ala Phe Asn Ala Val Gly Val Tyr305 310
31555292PRTB. anthracismisc_feature(1)..(292)B_anthracis_NP843132.1
55Val Thr Gly Thr Asn Ala Val Gly Thr Gly Lys Gly Val Leu Gly Asp1
5 10 15Thr Lys Ser Leu Asn Thr Thr Leu Ser Ala Ser Ser Tyr Tyr Leu
Gln 20 25 30Asp Asn Thr Arg Gly Ala Thr Ile Phe Thr Tyr Asp Ala Lys
Asn Arg 35 40 45Ser Thr Leu Pro Gly Thr Leu Trp Val Asp Ala Asp Asn
Val Phe Asn 50 55 60Ala Ala Tyr Asp Ala Ala Ala Val Asp Ala His Tyr
Tyr Ala Gly Arg65 70 75 80Thr Tyr Asp Tyr Tyr Lys Ala Thr Phe Asn
Arg Asn Ser Ile Asn Asp 85 90 95Ala Gly Ala Pro Leu Lys Ser Thr Val
His Tyr Gly Ser Arg Tyr Asn 100 105 110Asn Ala Phe Trp Asn Gly Ser
Gln Met Val Tyr Gly Asp Gly Asp Gly 115 120 125Val Thr Phe Thr Ser
Leu Ser Gly Gly Ile Asp Val Ile Gly His Glu 130 135 140Leu Thr His
Ala Val Thr Glu Tyr Ser Ser Asp Leu Ile Tyr Gln Asn145 150 155
160Glu Ser Gly Ala Leu Asn Glu Ala Ile Ser Asp Val Phe Gly Thr Leu
165 170 175Val Glu Tyr Tyr Asp Asn Arg Asn Pro Asp Trp Glu Ile Gly
Glu Asp 180 185 190Ile Tyr Thr Pro Gly Lys Ala Gly Asp Ala Leu Arg
Ser Met Ser Asp 195 200 205Pro Thr Lys Tyr Gly Asp Pro Asp His Tyr
Ser Lys Arg Tyr Thr Gly 210 215 220Thr Gly Asp Asn Gly Gly Val His
Thr Asn Ser Gly Ile Ile Asn Lys225 230 235 240Ala Ala Tyr Leu Leu
Ala Asn Gly Gly Thr His Tyr Gly Val Thr Val 245 250 255Asn Gly Ile
Gly Lys Asp Lys Val Gly Ala Ile Tyr Tyr Arg Ala Asn 260 265 270Thr
Gln Tyr Phe Thr Gln Ser Thr Thr Phe Ser Gln Ala Arg Ala Gly 275 280
285Leu Val Gln Ala 29056317PRTB.
thuringiensismisc_feature(1)..(317)B_thuringiensis_YP893436.1 56Val
Thr Gly Thr Asn Ala Val Gly Thr Gly Lys Gly Val Leu Gly Asp1 5 10
15Thr Lys Ser Leu Asn Thr Thr Leu Ser Ala Ser Ser Tyr Tyr Leu Gln
20 25 30Asp Asn Thr Arg Gly Ala Thr Ile Phe Thr Tyr Asp Ala Lys Asn
Arg 35 40 45Ser Thr Leu Pro Gly Thr Leu Trp Val Asp Ala Asp Asn Val
Phe Asn 50 55 60Ala Ala Tyr Asp Ala Ala Ala Val Asp Ala His Tyr Tyr
Ala Gly Lys65 70 75 80Thr Tyr Asp Tyr Tyr Lys Ala Thr Phe Asn Arg
Asn Ser Ile Asn Asp 85 90 95Ala Gly Ala Pro Leu Lys Ser Thr Val His
Tyr Gly Ser Arg Tyr Asn 100 105 110Asn Ala Phe Trp Asn Gly Ser Gln
Met Val Tyr Gly Asp Gly Asp Gly 115 120 125Val Thr Phe Thr Ser Leu
Ser Gly Gly Ile Asp Val Ile Gly His Glu 130 135 140Leu Thr His Ala
Val Thr Glu Tyr Ser Ser Asp Leu Ile Tyr Gln Asn145 150 155 160Glu
Ser Gly Ala Leu Asn Glu Ala Ile Ser Asp Val Phe Gly Thr Leu 165 170
175Val Glu Phe Tyr Asp Asn Arg Asn Pro Asp Trp Glu Ile Gly Glu Asp
180 185 190Ile Tyr Thr Pro Gly Lys Ala Gly Asp Ala Leu Arg Ser Met
Ser Asp 195 200 205Pro Thr Lys Tyr Gly Asp Pro Asp His Tyr Ser Lys
Arg Tyr Thr Gly 210 215 220Thr Gly Asp Asn Gly Gly Val His Thr Asn
Ser Gly Ile Ile Asn Lys225 230 235 240Ala Ala Tyr Leu Leu Ala Asn
Gly Gly Thr His Tyr Gly Val Thr Val 245 250 255Asn Gly Ile Gly Lys
Asp Lys Val Gly Ala Ile Tyr Tyr Arg Ala Asn 260 265 270Thr Gln Tyr
Phe Thr Gln Ser Thr Thr Phe Ser Gln Ala Arg Ala Gly 275 280 285Leu
Val Gln Ala Ala Thr Asp Leu Tyr Gly Ala Ser Ser Ala Glu Val 290 295
300Ala Ala Val Lys Gln Ser Tyr Ser Ala Val Gly Val Asn305 310
31557314PRTB. cereusmisc_feature(1)..(314)B_cereus_ZP04310163.1
57Thr Asn Ala Val Gly Thr Gly Lys Gly Val Leu Gly Asp Thr Lys Ser1
5 10 15Leu Asn Thr Thr Leu Ser Ala Ser Ser Tyr Tyr Leu Gln Asp Asn
Thr 20 25 30Arg Gly Ala Thr Ile Phe Thr Tyr Asp Ala Lys Asn Arg Ser
Thr Leu 35 40 45Pro Gly Thr Leu Trp Val Asp Ala Asp Asn Val Phe Asn
Ala Ala Tyr 50 55 60Asp Ala Ala Ala Val Asp Ala His Tyr Tyr Ala Gly
Lys Thr Tyr Asp65 70 75 80Tyr Tyr Lys Ala Thr Phe Asn Arg Asn Ser
Ile Asn Asp Ala Gly Ala 85 90 95Pro Leu Lys Ser Thr Val His Tyr Gly
Ser Arg Tyr Asn Asn Ala Phe 100 105 110Trp Asn Gly Ser Gln Met Val
Tyr Gly Asp Gly Asp Gly Val Thr Phe 115 120 125Thr Ser Leu Ser Gly
Gly Ile Asp Val Ile Gly His Glu Leu Thr His 130 135 140Ala Val Thr
Glu Tyr Ser Ser Asp Leu Ile Tyr Gln Asn Glu Ser Gly145 150 155
160Ala Leu Asn Glu Ala Ile Ser Asp Val Phe Gly Thr Leu Val Glu Phe
165 170 175Tyr Asp Asn Arg Asn Pro Asp Trp Glu Ile Gly Glu Asp Ile
Tyr Thr 180 185 190Pro Gly Lys Ala Gly Asp Ala Leu Arg Ser Met Ser
Asp Pro Thr Lys 195 200 205Tyr Gly Asp Pro Asp His Tyr Ser Lys Arg
Tyr Thr Gly Thr Gly Asp 210 215 220Asn Gly Gly Val His Thr Asn Ser
Gly Ile Ile Asn Lys Ala Ala Tyr225 230 235 240Leu Leu Ala Asn Gly
Gly Thr His Tyr Gly Val Thr Val Asn Gly Ile 245 250 255Gly Lys Asp
Lys Val Gly Ala Ile Tyr Tyr Arg Ala Asn Thr Gln Tyr 260 265 270Phe
Thr Gln Ser Thr Thr Phe Ser Gln Ala Arg Ala Gly Leu Val Gln 275 280
285Ala Ala Ala Asp Leu Tyr Gly Ala Ser Ser Ala Glu Val Ala Ala Val
290 295 300Lys Gln Ser Tyr Ser Ala Val Gly Val Asn305
31058317PRTLactobacillus
sp.misc_feature(1)..(317)Lactobacillus_sp_BAA06144.1 58Val Thr Gly
Thr Asn Ala Val Gly Thr Gly Lys Gly Val Leu Gly Asp1 5 10 15Thr Lys
Ser Leu Asn Thr Thr Leu Ser Ala Ser Ser Tyr Tyr Leu Gln 20 25 30Asp
Asn Thr Arg Gly Ala Thr Ile Phe Thr Tyr Asp Ala Lys Asn Arg 35 40
45Ser Thr Leu Pro Gly Thr Leu Trp Val Asp Ala Asp Asn Val Phe Asn
50 55 60Ala Ala Tyr Asp Ala Ala Ala Val Asp Ala His Tyr Tyr Ala Gly
Lys65 70 75 80Thr Tyr Asp Tyr Tyr Lys Ala Thr Phe Asn Arg Asn Ser
Ile Asn Asp 85 90 95Ala Gly Ala Pro Leu Lys Ser Thr Val His Tyr Gly
Ser Lys Tyr Asn 100 105 110Asn Ala Phe Trp Asn Gly Ser Gln Met Val
Tyr Gly Asp Gly Asp Gly 115 120 125Val Thr Phe Thr Ser Leu Ser Gly
Gly Ile Asp Val Ile Gly His Glu 130 135 140Leu Thr His Ala Val Thr
Glu Tyr Ser Ser Asp Leu Ile Tyr Gln Asn145 150 155 160Glu Ser Gly
Ala Leu Asn Glu Ala Ile Ser Asp Val Phe Gly Thr Leu 165 170 175Val
Glu Tyr Tyr Asp Asn Arg Asn Pro Asp Trp Glu Ile Gly Glu Asp 180 185
190Ile Tyr Thr Pro Gly Lys Ala Gly Asp Ala Leu Arg Ser Met Ser Asp
195 200 205Pro Thr Lys Tyr Gly Asp Pro Asp His Tyr Ser Lys Arg Tyr
Thr Gly 210 215 220Thr Ser Asp Asn Gly Gly Val His Thr Asn Ser Gly
Ile Ile Asn Lys225 230 235 240Ala Ala Tyr Leu Leu Ala Asn Gly Gly
Thr His Tyr Gly Val Thr Val 245 250 255Asn Gly Ile Gly Lys Asp Lys
Val Gly Ala Ile Tyr Tyr Arg Ala Asn 260 265 270Thr Gln Tyr Phe Thr
Gln Ser Thr Thr Phe Ser Gln Ala Arg Ala Gly 275 280 285Leu Val Gln
Ala Ala Ala Asp Leu Tyr Gly Ala Ser Ser Ala Glu Val 290 295 300Ala
Ala Val Lys Gln Ser Tyr Ser Ala Val Gly Val Asn305 310
31559317PRTBacillus thermoproteolyticusmisc_feature(1)..(317)1NPC.A
59Val Thr Gly Thr Asn Lys Val Gly Thr Gly Lys Gly Val Leu Gly Asp1
5 10 15Thr Lys Ser Leu Asn Thr Thr Leu Ser Gly Ser Ser Tyr Tyr Leu
Gln 20 25 30Asp Asn Thr Arg Gly Ala Thr Ile Phe Thr Tyr Asp Ala Lys
Asn Arg 35 40 45Ser Thr Leu Pro Gly Thr Leu Trp Ala Asp Ala Asp Asn
Val Phe Asn 50 55 60Ala Ala Tyr Asp Ala Ala Ala Val Asp Ala His Tyr
Tyr Ala Gly Lys65 70 75 80Thr Tyr Asp Tyr Tyr Lys Ala Thr Phe Asn
Arg Asn Ser Ile Asn Asp 85 90 95Ala Gly Ala Pro Leu Lys Ser Thr Val
His Tyr Gly Ser Asn Tyr Asn 100 105 110Asn Ala Phe Trp Asn Gly Ser
Gln Met Val Tyr Gly Asp Gly Asp Gly 115 120 125Val Thr Phe Thr Ser
Leu Ser Gly Gly Ile Asp Val Ile Gly His Glu 130 135 140Leu Thr His
Ala Val Thr Glu Asn Ser Ser Asn Leu Ile Tyr Gln Asn145 150 155
160Glu Ser Gly Ala Leu Asn Glu Ala Ile Ser Asp Ile Phe Gly Thr Leu
165 170 175Val Glu Phe Tyr Asp Asn Arg Asn Pro Asp Trp Glu Ile Gly
Glu Asp 180 185 190Ile Tyr Thr Pro Gly Lys Ala Gly Asp Ala Leu Arg
Ser Met Ser Asp 195 200 205Pro Thr Lys Tyr Gly Asp Pro Asp His Tyr
Ser Lys Arg Tyr Thr Gly 210 215 220Ser Ser Asp Asn Gly Gly Val His
Thr Asn Ser Gly Ile Ile Asn Lys225 230 235 240Gln Ala Tyr Leu Leu
Ala Asn Gly Gly Thr His Tyr Gly Val Thr Val 245 250 255Thr Gly Ile
Gly Lys Asp Lys Leu Gly Ala Ile Tyr Tyr Arg Ala Asn 260 265 270Thr
Gln Tyr Phe Thr Gln Ser Thr Thr Phe Ser Gln Ala Arg Ala Gly 275 280
285Ala Val Gln Ala Ala Ala Asp Leu Tyr Gly Ala Asn Ser Ala Glu Val
290 295 300Ala Ala Val Lys Gln Ser Phe Ser Ala Val Gly Val Asn305
310 31560317PRTB.
cytotoxicusmisc_feature(1)..(317)B_cytotoxicus_YP001373863.1 60Val
Thr Gly Thr Asn Ala Val Gly Thr Gly Thr Gly Val Leu Gly Asp1 5 10
15Lys Lys Ser Ile Asn Thr Thr Leu Ser Gly Ser Thr Tyr Tyr Leu Gln
20 25 30Asp Asn Thr Arg Gly Ala Gln Ile Phe Thr Tyr Asp Ala Lys Asn
Arg 35 40 45Ser Ser Leu Pro Gly Thr Leu Trp Ala Asp Val Asp Asn Ala
Phe His 50 55 60Ala Lys Tyr Asp Ala Ala Ala Val Asp Ala His Tyr Tyr
Ala Gly Val65 70 75 80Thr Tyr Asp Tyr Tyr Lys Asn Thr Phe Asn Arg
Asn Ser Ile Asn Asp 85 90 95Ala Gly Ala Ala Leu Lys Ser Thr Val His
Tyr Gly Ser Asn Tyr Asn 100 105 110Asn Ala Phe Trp Asn Gly Ser Gln
Met Val Tyr Gly Asp Gly Asp Gly 115 120 125Val Thr Phe Thr Ser Leu
Ser Gly Gly Ile Asp Val Ile Gly His Glu 130 135 140Leu Thr His Ala
Val Thr Glu Tyr Ser Ser Asn Leu Ile Tyr Gln Tyr145 150 155 160Glu
Ser Gly Ala Leu Asn Glu Ala Ile Ser Asp Ile Phe Gly Thr Leu 165 170
175Val Glu Tyr Tyr Asp Asn Arg Asn Pro Asp Trp Glu Ile Gly Glu Asp
180 185 190Ile Tyr Thr Pro Gly Lys Ala Gly Asp Ala Leu Arg Ser Met
Ser Asp 195 200 205Pro Thr Lys Tyr Gly Asp Pro Asp His Tyr Ser Lys
Arg Tyr Thr Gly 210 215 220Ser Gly Asp Asn Gly Gly Val His Thr Asn
Ser Gly Ile Ile Asn Lys225 230 235 240Ala Ala Tyr Leu Leu Ala Asn
Gly Gly Thr His Tyr Gly Val Thr Val 245 250 255Asn Gly Ile Gly Lys
Asp Lys Val Gly Ala Ile Tyr Tyr Arg Ala Asn 260 265 270Thr Gln Tyr
Phe Thr Gln Ser Thr Thr Phe Ser Gln Ala Arg Ala Gly 275 280 285Leu
Val Gln Ala Ala Ala Asp Leu Tyr Gly Ala Asn Ser Ala Glu Val 290 295
300Thr Ala Val Lys Gln Ser Tyr Asp Ala Val Gly Val Lys305 310
31561314PRTB.
megateriummisc_feature(1)..(314)B_megaterium_YP005495105.1 61Thr
Asn Ala Ile Gly Ser Gly Lys Gly Val Leu Gly Asp Thr Lys Ser1 5 10
15Leu Lys Thr Thr Leu Ser Gly Ser Ala Tyr Tyr Leu Gln Asp Asn Thr
20 25 30Arg Gly Ala Thr Ile Tyr Thr Tyr Asp Ala Lys Asn Arg Thr Ser
Leu 35 40 45Pro Gly Thr Leu Trp Ala Asp Thr Asp Asn Thr Tyr Asn Ala
Thr Arg 50 55 60Asp Ala Ala Ala Val Asp Ala His Tyr Tyr Ala Gly Val
Thr Tyr Asp65 70 75 80Tyr Tyr Lys Asn Lys Phe Asn Arg Asn Ser Tyr
Asp Asn Ala Gly Ala 85 90 95Pro Leu Lys Ser Thr Val His Tyr Ser Ser
Gly Tyr Asn Asn Ala Phe 100 105 110Trp Asn Gly Ser Gln Met Val Tyr
Gly Asp Gly Asp Gly Thr Thr Phe 115 120 125Val Pro Leu Ser Gly Gly
Leu Asp Val Ile Gly His Glu Leu Thr His 130 135 140Ala Val Thr Glu
Arg Ser Ser Asn Leu Ile Tyr Gln Tyr Glu Ser Gly145 150 155 160Ala
Leu
Asn Glu Ala Ile Ser Asp Ile Phe Gly Thr Leu Val Glu Tyr 165 170
175Tyr Asp Asn Arg Asn Pro Asp Trp Glu Ile Gly Glu Asp Ile Tyr Thr
180 185 190Pro Gly Thr Ser Gly Asp Ala Leu Arg Ser Met Ser Asn Pro
Ala Lys 195 200 205Tyr Gly Asp Pro Asp His Tyr Ser Lys Arg Tyr Thr
Gly Ser Ser Asp 210 215 220Asn Gly Gly Val His Thr Asn Ser Gly Ile
Ile Asn Lys Ala Ala Tyr225 230 235 240Leu Leu Ala Asn Gly Gly Thr
His Tyr Gly Val Thr Val Thr Gly Ile 245 250 255Gly Gly Asp Lys Leu
Gly Lys Ile Tyr Tyr Arg Ala Asn Thr Leu Tyr 260 265 270Phe Thr Gln
Ser Thr Thr Phe Ser Gln Ala Arg Ala Gly Leu Val Gln 275 280 285Ala
Ala Ala Asp Leu Tyr Gly Ser Gly Ser Gln Glu Val Ile Ser Val 290 295
300Gly Lys Ser Phe Asp Ala Val Gly Val Gln305 31062322PRTBacillus
sp. SG-1misc_feature(1)..(322)B_sp_SG-1_ZP01858398.1 62Val Ser Gly
Thr Asp Gln Val Gly Thr Gly Lys Gly Val Leu Gly Asp1 5 10 15Thr Lys
Ser Leu Asn Thr Thr Leu Ser Asn Gly Thr Tyr Tyr Leu Gln 20 25 30Asp
Asn Thr Arg Gly Gly Gly Ile Met Thr Tyr Asp Met Lys Asn Arg 35 40
45Thr Phe Phe Pro Gln Phe Tyr Leu Pro Gly Ser Leu Trp Ser Asp Ala
50 55 60Asp Asn Val Tyr Asn Gln Ala Tyr Asp Ala Ala Ala Val Asp Ala
His65 70 75 80Tyr Phe Ala Gly Ala Thr Phe Asp Tyr Tyr Lys Asp Val
Phe Gly Arg 85 90 95Asn Ser Tyr Asp Asn Lys Gly Thr Thr Ile Gln Ser
Ser Val His Tyr 100 105 110Ser Lys Asn Tyr Asn Asn Ala Phe Trp Asn
Gly Ser Gln Met Val Tyr 115 120 125Gly Asp Gly Asp Gly Thr Thr Phe
Ile Pro Leu Ser Gly Gly Leu Asp 130 135 140Val Val Ala His Glu Leu
Thr His Ala Val Thr Asp Thr Ser Ser Asp145 150 155 160Leu Val Tyr
Gln Asn Glu Ser Gly Ala Leu Asn Glu Ala Ile Ser Asp 165 170 175Ile
Phe Gly Thr Leu Val Glu Tyr His Glu Asn His Asn Pro Asp Phe 180 185
190Glu Ile Gly Glu Asp Ile Tyr Thr Pro Asn Thr Pro Asn Asp Ala Leu
195 200 205Arg Ser Met Ser Asp Pro Ala Lys Tyr Gly Asp Pro Asp His
Tyr Ser 210 215 220Val Arg Tyr Thr Gly Thr Gln Asp Asn Gly Gly Val
His Ile Asn Ser225 230 235 240Gly Ile Ile Asn Lys Gln Ala Tyr Leu
Leu Ser Glu Gly Gly Thr His 245 250 255Tyr Gly Val Asn Val Thr Gly
Ile Gly Arg Glu Lys Leu Gly Glu Ile 260 265 270Tyr Tyr Arg Met Asn
Thr Val Tyr Leu Thr Ala Ser Ser Thr Phe Ser 275 280 285Gln Ala Arg
Ser Ala Ala Val Gln Ala Ala Ser Asp Leu Tyr Gly Ser 290 295 300Asn
Ser Pro Glu Val Gln Ser Val Asn Gln Ser Phe Asp Ala Val Gly305 310
315 320Ile Asn63306PRTPaenibacillus
peoriaemisc_feature(1)..(306)PpePro1 63Ala Thr Gly Thr Gly Arg Gly
Val Asp Gly Val Thr Lys Ser Phe Thr1 5 10 15Thr Thr Ala Ser Gly Asn
Gly Tyr Gln Leu Lys Asp Thr Thr Arg Ser 20 25 30Asn Gly Ile Val Thr
Tyr Thr Ala Asn Asn Arg Gln Thr Thr Pro Gly 35 40 45Thr Ile Met Thr
Asp Ala Asp Asn Val Trp Asn Asp Pro Ala Ala Val 50 55 60Asp Ala His
Ala Tyr Ala Ile Lys Thr Tyr Asp Tyr Tyr Lys Asn Lys65 70 75 80Phe
Gly Arg Asp Ser Ile Asp Gly Arg Gly Met Gln Ile Arg Ser Thr 85 90
95Val His Tyr Gly Lys Lys Tyr Val Asn Ala Phe Trp Asn Gly Ser Gln
100 105 110Met Thr Tyr Gly Asp Gly Asp Gly Ser Thr Phe Thr Phe Phe
Ser Gly 115 120 125Asp Pro Asp Val Val Gly His Glu Leu Thr His Gly
Val Thr Glu Phe 130 135 140Thr Ser Asn Leu Glu Tyr Tyr Gly Glu Ser
Gly Ala Leu Asn Glu Ala145 150 155 160Phe Ser Asp Ile Ile Gly Asn
Asp Ile Asp Gly Ala Asn Trp Leu Leu 165 170 175Gly Asp Gly Ile Tyr
Thr Pro Gly Ile Pro Gly Asp Ala Leu Arg Ser 180 185 190Leu Ser Asp
Pro Thr Arg Phe Gly Gln Pro Asp His Tyr Ser Asn Phe 195 200 205Tyr
Pro Asp Pro Asn Asn Asp Asp Glu Gly Gly Val His Thr Asn Ser 210 215
220Gly Ile Ile Asn Lys Ala Tyr Tyr Leu Leu Ala Gln Gly Gly Thr
Ser225 230 235 240His Gly Val Lys Val Thr Gly Ile Gly Arg Glu Ala
Ala Val Phe Ile 245 250 255Tyr Tyr Asn Ala Phe Thr Asn Tyr Leu Thr
Ser Thr Ser Asn Phe Ser 260 265 270Asn Ala Arg Ala Ala Val Ile Gln
Ala Ala Lys Asp Phe Tyr Gly Ala 275 280 285Asp Ser Leu Ala Val Thr
Ser Ala Ile Lys Ser Phe Asp Ala Val Gly 290 295 300Ile
Lys30564304PRTPaenibacillus polymyxamisc_feature(1)..(304)PpoPro2
64Ala Thr Gly Thr Gly Lys Gly Val Leu Gly Asp Thr Lys Ser Phe Thr1
5 10 15Thr Thr Ala Ser Gly Ser Ser Tyr Gln Leu Lys Asp Thr Thr Arg
Gly 20 25 30Asn Gly Ile Val Thr Tyr Thr Ala Ser Asn Arg Gln Ser Ile
Pro Gly 35 40 45Thr Leu Leu Thr Asp Ala Asp Asn Val Trp Asn Asp Pro
Ala Gly Val 50 55 60Asp Ala His Ala Tyr Ala Ala Lys Thr Tyr Asp Tyr
Tyr Lys Ser Lys65 70 75 80Phe Gly Arg Asp Ser Val Asp Gly Arg Gly
Leu Gln Leu Arg Ser Thr 85 90 95Val His Tyr Gly Ser Arg Tyr Asn Asn
Ala Phe Trp Asn Gly Ser Gln 100 105 110Met Thr Tyr Gly Asp Gly Asp
Gly Ser Thr Phe Ile Ala Phe Ser Gly 115 120 125Asp Pro Asp Val Val
Gly His Glu Leu Thr His Gly Val Thr Glu Tyr 130 135 140Thr Ser Asn
Leu Glu Tyr Tyr Gly Glu Ser Gly Ala Leu Asn Glu Ala145 150 155
160Phe Ser Asp Val Ile Gly Asn Asp Ile Gln Arg Lys Asn Trp Leu Val
165 170 175Gly Asp Asp Ile Tyr Thr Pro Asn Ile Ala Gly Asp Ala Leu
Arg Ser 180 185 190Met Ser Asn Pro Thr Leu Tyr Asp Gln Pro Asp His
Tyr Ser Asn Leu 195 200 205Tyr Lys Gly Ser Ser Asp Asn Gly Gly Val
His Thr Asn Ser Gly Ile 210 215 220Ile Asn Lys Ala Tyr Tyr Leu Leu
Ala Gln Gly Gly Thr Phe His Gly225 230 235 240Val Ala Val Asn Gly
Ile Gly Arg Asp Ala Ala Val Gln Ile Tyr Tyr 245 250 255Ser Ala Phe
Thr Asn Tyr Leu Thr Ser Ser Ser Asp Phe Ser Asn Ala 260 265 270Arg
Ala Ala Val Ile Gln Ala Ala Lys Asp Leu Tyr Gly Ala Asn Ser 275 280
285Ala Glu Ala Thr Ala Ala Ala Lys Ser Phe Asp Ala Val Gly Val Asn
290 295 30065303PRTPaenibacillus
terraemisc_feature(1)..(303)PtePro1 65Ala Thr Gly Thr Gly Val Gly
Val Leu Gly Asp Thr Lys Thr Phe Thr1 5 10 15Thr Thr Gln Ser Gly Thr
Gln Tyr Val Met Gln Asp Thr Thr Arg Gly 20 25 30Gly Gly Ile Val Thr
Tyr Ser Ala Gly Asn Thr Gln Ser Leu Pro Gly 35 40 45Thr Leu Met Arg
Asp Thr Asp Asn Val Trp Thr Asp Pro Ala Ala Val 50 55 60Asp Ala His
Ala Tyr Ala Ala Val Val Tyr Asp Tyr Phe Lys Asn Asn65 70 75 80Phe
Asn Arg Asp Ser Leu Asp Gly Arg Gly Met Ala Ile Lys Ser Thr 85 90
95Val His Tyr Gly Ser Arg Tyr Asn Asn Ala Phe Trp Asn Gly Thr Gln
100 105 110Ile Ala Tyr Gly Asp Gly Asp Gly Thr Thr Phe Arg Ala Phe
Ser Gly 115 120 125Asp Leu Asp Val Ile Gly His Glu Leu Thr His Gly
Ile Thr Glu Lys 130 135 140Thr Ala Gly Leu Ile Tyr Gln Gly Glu Ser
Gly Ala Leu Asn Glu Ser145 150 155 160Ile Ser Asp Val Phe Gly Asn
Thr Ile Gln Gly Lys Asn Trp Leu Ile 165 170 175Gly Asp Asp Ile Tyr
Thr Pro Ser Ile Pro Gly Asp Ala Leu Arg Ser 180 185 190Met Glu Asn
Pro Thr Leu Phe Asn Gln Pro Asp His Tyr Ser Asn Ile 195 200 205Tyr
Arg Gly Ser Asp Asp Asn Gly Gly Val His Thr Asn Ser Gly Ile 210 215
220Pro Asn Lys Ala Phe Tyr Leu Leu Ala Gln Gly Gly Thr His Arg
Gly225 230 235 240Val Ser Val Thr Gly Ile Gly Arg Gly Asp Ala Ala
Lys Ile Val Tyr 245 250 255Lys Ala Leu Thr Tyr Tyr Leu Thr Ser Thr
Ser Asn Phe Ala Ala Met 260 265 270Arg Gln Ala Ala Ile Ser Ser Ala
Thr Asp Leu Phe Gly Ala Asn Ser 275 280 285Ala Gln Val Asn Ser Val
Lys Ala Ala Tyr Ala Ala Val Gly Ile 290 295
30066304PRTBrevibacillus brevismisc_feature(1)..(304)BbrPro1 66Val
Thr Ala Thr Gly Lys Gly Val Leu Gly Asp Thr Lys Gln Phe Glu1 5 10
15Thr Thr Lys Gln Gly Ser Thr Tyr Met Leu Lys Asp Thr Thr Arg Gly
20 25 30Lys Gly Ile Glu Thr Tyr Thr Ala Asn Asn Arg Thr Ser Leu Pro
Gly 35 40 45Thr Leu Met Thr Asp Ser Asp Asn Tyr Trp Thr Asp Gly Ala
Ala Val 50 55 60Asp Ala His Ala His Ala Gln Lys Thr Tyr Asp Tyr Phe
Arg Asn Val65 70 75 80His Asn Arg Asn Ser Tyr Asp Gly Asn Gly Ala
Val Ile Arg Ser Thr 85 90 95Val His Tyr Ser Thr Arg Tyr Asn Asn Ala
Phe Trp Asn Gly Ser Gln 100 105 110Met Val Tyr Gly Asp Gly Asp Gly
Thr Thr Phe Leu Pro Leu Ser Gly 115 120 125Gly Leu Asp Val Val Ala
His Glu Leu Thr His Ala Val Thr Glu Arg 130 135 140Thr Ala Gly Leu
Val Tyr Gln Asn Glu Ser Gly Ala Leu Asn Glu Ser145 150 155 160Met
Ser Asp Ile Phe Gly Ala Met Val Asp Asn Asp Asp Trp Leu Met 165 170
175Gly Glu Asp Ile Tyr Thr Pro Gly Arg Ser Gly Asp Ala Leu Arg Ser
180 185 190Leu Gln Asp Pro Ala Ala Tyr Gly Asp Pro Asp His Tyr Ser
Lys Arg 195 200 205Tyr Thr Gly Ser Gln Asp Asn Gly Gly Val His Thr
Asn Ser Gly Ile 210 215 220Asn Asn Lys Ala Ala Tyr Leu Leu Ala Glu
Gly Gly Thr His Tyr Gly225 230 235 240Val Arg Val Asn Gly Ile Gly
Arg Thr Asp Thr Ala Lys Ile Tyr Tyr 245 250 255His Ala Leu Thr His
Tyr Leu Thr Pro Tyr Ser Asn Phe Ser Ala Met 260 265 270Arg Arg Ala
Ala Val Leu Ser Ala Thr Asp Leu Phe Gly Ala Asn Ser 275 280 285Arg
Gln Val Gln Ala Val Asn Ala Ala Tyr Asp Ala Val Gly Val Lys 290 295
30067300PRTBacillus subtilismisc_feature(1)..(300)NprE 67Ala Ala
Thr Thr Gly Thr Gly Thr Thr Leu Lys Gly Lys Thr Val Ser1 5 10 15Leu
Asn Ile Ser Ser Glu Ser Gly Lys Tyr Val Leu Arg Asp Leu Ser 20 25
30Lys Pro Thr Gly Thr Gln Ile Ile Thr Tyr Asp Leu Gln Asn Arg Glu
35 40 45Tyr Asn Leu Pro Gly Thr Leu Val Ser Ser Thr Thr Asn Gln Phe
Thr 50 55 60Thr Ser Ser Gln Arg Ala Ala Val Asp Ala His Tyr Asn Leu
Gly Lys65 70 75 80Val Tyr Asp Tyr Phe Tyr Gln Lys Phe Asn Arg Asn
Ser Tyr Asp Asn 85 90 95Lys Gly Gly Lys Ile Val Ser Ser Val His Tyr
Gly Ser Arg Tyr Asn 100 105 110Asn Ala Ala Trp Ile Gly Asp Gln Met
Ile Tyr Gly Asp Gly Asp Gly 115 120 125Ser Phe Phe Ser Pro Leu Ser
Gly Ser Met Asp Val Thr Ala His Glu 130 135 140Met Thr His Gly Val
Thr Gln Glu Thr Ala Asn Leu Asn Tyr Glu Asn145 150 155 160Gln Pro
Gly Ala Leu Asn Glu Ser Phe Ser Asp Val Phe Gly Tyr Phe 165 170
175Asn Asp Thr Glu Asp Trp Asp Ile Gly Glu Asp Ile Thr Val Ser Gln
180 185 190Pro Ala Leu Arg Ser Leu Ser Asn Pro Thr Lys Tyr Gly Gln
Pro Asp 195 200 205Asn Phe Lys Asn Tyr Lys Asn Leu Pro Asn Thr Asp
Ala Gly Asp Tyr 210 215 220Gly Gly Val His Thr Asn Ser Gly Ile Pro
Asn Lys Ala Ala Tyr Asn225 230 235 240Thr Ile Thr Lys Ile Gly Val
Asn Lys Ala Glu Gln Ile Tyr Tyr Arg 245 250 255Ala Leu Thr Val Tyr
Leu Thr Pro Ser Ser Thr Phe Lys Asp Ala Lys 260 265 270Ala Ala Leu
Ile Gln Ser Ala Arg Asp Leu Tyr Gly Ser Gln Asp Ala 275 280 285Ala
Ser Val Glu Ala Ala Trp Asn Ala Val Gly Leu 290 295
30068300PRTBacillus subtilismisc_feature(1)..(300)NprE_variant
68Ala Ala Thr Thr Gly Thr Gly Thr Thr Leu Lys Gly Lys Thr Val Ser1
5 10 15Leu Asn Ile Ser Ser Glu Ser Gly Lys Tyr Val Leu Arg Asp Leu
Ser 20 25 30Lys Pro Thr Gly Thr Gln Ile Ile Thr Tyr Asp Leu Gln Asn
Arg Glu 35 40 45Tyr Asn Leu Pro Gly Thr Leu Val Ser Ser Thr Thr Asn
Gln Phe Thr 50 55 60Thr Ser Ser Gln Arg Ala Ala Val Asp Ala His Tyr
Asn Leu Gly Lys65 70 75 80Val Tyr Asp Tyr Phe Tyr Gln Lys Phe Asn
Arg Asn Ser Tyr Asp Asn 85 90 95Lys Gly Gly Lys Ile Val Ser Ser Val
His Tyr Gly Ser Arg Tyr Asn 100 105 110Asn Ala Ala Trp Ile Gly Asp
Gln Met Ile Tyr Gly Asp Gly Asp Gly 115 120 125Ile Leu Phe Ser Pro
Leu Ser Gly Ser Leu Asp Val Thr Ala His Glu 130 135 140Met Thr His
Gly Val Thr Gln Glu Thr Ala Asn Leu Asn Tyr Glu Asn145 150 155
160Gln Pro Gly Ala Leu Asn Glu Ser Phe Ser Asp Val Phe Gly Tyr Phe
165 170 175Asn Asp Thr Glu Asp Trp Asp Ile Gly Glu Asp Ile Thr Ile
Ser Gln 180 185 190Pro Ala Leu Arg Ser Leu Ser Asn Pro Thr Lys Tyr
Gly Gln Pro Asp 195 200 205Asn Phe Lys Asn Tyr Lys Asn Leu Pro Asn
Thr Pro Ala Gly Asp Tyr 210 215 220Gly Gly Val His Thr Asn Ser Gly
Ile Pro Asn Lys Ala Ala Tyr Asn225 230 235 240Thr Ile Thr Lys Ile
Gly Val Asn Lys Ala Glu Gln Ile Tyr Tyr Arg 245 250 255Ala Leu Thr
Val Tyr Leu Thr Pro Ser Ser Thr Phe Lys Asp Ala Lys 260 265 270Ala
Ala Leu Ile Gln Ser Ala Arg Asp Leu Tyr Gly Ser Gln Asp Ala 275 280
285Ala Ser Val Glu Ala Ala Trp Asn Ala Val Gly Leu 290 295 300
References
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014

D00015

D00016

D00017

D00018

D00019

D00020

D00021

D00022

D00023

D00024

D00025

D00026

D00027

D00028

D00029

D00030
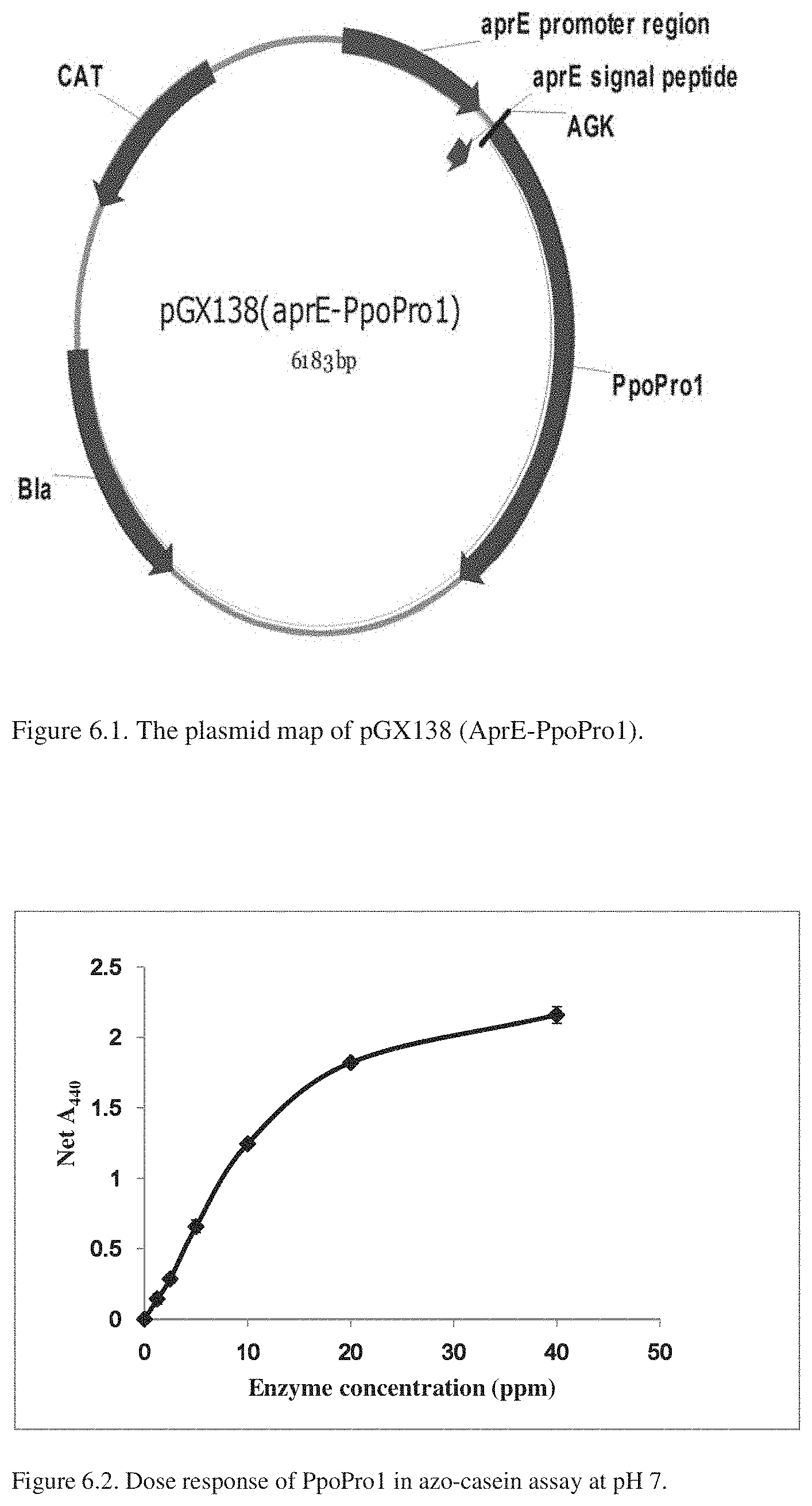
D00031

D00032

D00033

D00034

D00035

D00036

D00037

D00038

D00039

D00040

D00041

D00042

D00043

D00044

D00045

D00046

D00047

D00048

D00049

D00050

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.