Genomic Sequence Modification Method For Specifically Converting Nucleic Acid Bases Of Targeted Dna Sequence, And Molecular Comp
Kind Code
U.S. patent application number 16/838960 was filed with the patent office on 2020-08-06 for genomic sequence modification method for specifically converting nucleic acid bases of targeted dna sequence, and molecular comp. The applicant listed for this patent is NATIONAL UNIVERSITY CORPORATION KOBE UNIVERSITY. Invention is credited to Satomi Kojima, Akihiko Kondo, Keiji Nishida.
| Application Number | 20200248174 16/838960 |
| Document ID | 20200248174 / US20200248174 |
| Family ID | 1000004782499 |
| Filed Date | 2020-08-06 |
| Patent Application | download [pdf] |











View All Diagrams
| United States Patent Application | 20200248174 |
| Kind Code | A1 |
| Nishida; Keiji ; et al. | August 6, 2020 |
GENOMIC SEQUENCE MODIFICATION METHOD FOR SPECIFICALLY CONVERTING NUCLEIC ACID BASES OF TARGETED DNA SEQUENCE, AND MOLECULAR COMPLEX FOR USE IN SAME
Abstract
The invention provides a method of modifying a targeted site of a double stranded DNA, including a step of contacting a complex wherein a nucleic acid sequence-recognizing module that specifically binds to a target nucleotide sequence in a selected double stranded DNA and a nucleic acid base converting enzyme are linked, with the double stranded DNA, to convert one or more nucleotides in the targeted site to other one or more nucleotides or delete one or more nucleotides, or insert one or more nucleotides into the targeted site, without cleaving at least one strand of the double stranded DNA in the targeted site.
| Inventors: | Nishida; Keiji; (Kobe-shi, Hyogo, JP) ; Kondo; Akihiko; (Kobe-shi, Hyogo, JP) ; Kojima; Satomi; (Kobe-shi, Hyogo, JP) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 1000004782499 | ||||||||||
| Appl. No.: | 16/838960 | ||||||||||
| Filed: | April 2, 2020 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 15124021 | Nov 9, 2016 | 10655123 | ||
| PCT/JP2015/056436 | Mar 4, 2015 | |||
| 16838960 | ||||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C12N 15/1024 20130101; C07K 2319/00 20130101; C12N 9/22 20130101; C12N 15/11 20130101; C12Y 305/04005 20130101; C12N 9/78 20130101; C07K 2319/81 20130101; C12N 15/102 20130101; C12N 2310/3513 20130101 |
| International Class: | C12N 15/10 20060101 C12N015/10; C12N 9/22 20060101 C12N009/22; C12N 9/78 20060101 C12N009/78; C12N 15/11 20060101 C12N015/11 |
Foreign Application Data
| Date | Code | Application Number |
|---|---|---|
| Mar 5, 2014 | JP | 2014-043348 |
| Sep 30, 2014 | JP | 2014-201859 |
Claims
1. A method of modifying a targeted site of a double stranded DNA, comprising: contacting said double stranded DNA with at least one complex which comprises (i) a nucleic acid base converting enzyme linked to (ii) a nucleic acid sequence-recognizing module that specifically binds to a target nucleotide sequence in the targeted site of the double stranded DNA, thereby to convert one or more nucleotides in the targeted site to one or more different nucleotides or to delete one or more nucleotides in the targeted site or to insert one or more nucleotides into said targeted site, without introducing a double strand break (DSB) in said double stranded DNA in the targeted site, wherein the nucleic acid sequence-recognizing module is a CRISPR-Cas system, and wherein the CRISPR-Cas system comprises a nickase protein.
2. The method of claim 1 which comprises contacting the double stranded DNA with two or more complexes that each comprise a nucleic sequence-recognizing module that specifically binds to a different target nucleotide sequence.
3. The method of claim 2, wherein the different target nucleotide sequences are present in different genes.
4. The method of claim 1, wherein the nucleic acid base converting enzyme is a deaminase.
5. The method of claim 4, wherein the deaminase is a cytidine deaminase.
6. The method of claim 1, wherein the step of contacting comprises introducing a nucleic acid encoding the at least one complex into a cell which comprises the double stranded DNA.
7. The method of claim 6, wherein the cell is a prokaryotic cell, an eukaryotic cell, a microbial cell, a plant cell, an insect cell, an animal cell, a vertebrate cell, or a mammalian cell.
8. A method of modifying a targeted site in double stranded genomic DNA in each of two or more targeted alleles on homologous chromosomes in a polyploid cell, the method comprising: contacting said double stranded genomic DNA of the polyploid cell with at least one complex which comprises (i) a nucleic acid base converting enzyme linked to (ii) a nucleic acid sequence-recognizing module that specifically binds to a target nucleotide sequence in the targeted site in the double stranded genomic DNA in each of said two or more targeted alleles on homologous chromosomes in the polyploid cell, thereby to convert one or more nucleotides in said targeted site in the double stranded genomic DNA in each of said two or more targeted alleles on homologous chromosomes to one or more different nucleotides, or to delete one or more nucleotides in said targeted site in the double stranded genomic DNA in each of said two or more targeted alleles on homologous chromosomes, or to insert one or more nucleotides into said targeted site in the double stranded genomic DNA in each of said two or more targeted alleles on homologous chromosomes, without introducing a double strand break (DSB) in said double stranded genomic DNA, wherein the nucleic acid sequence-recognizing module is a CRISPR-Cas system, and wherein the CRISPR-Cas system comprises a nickase protein.
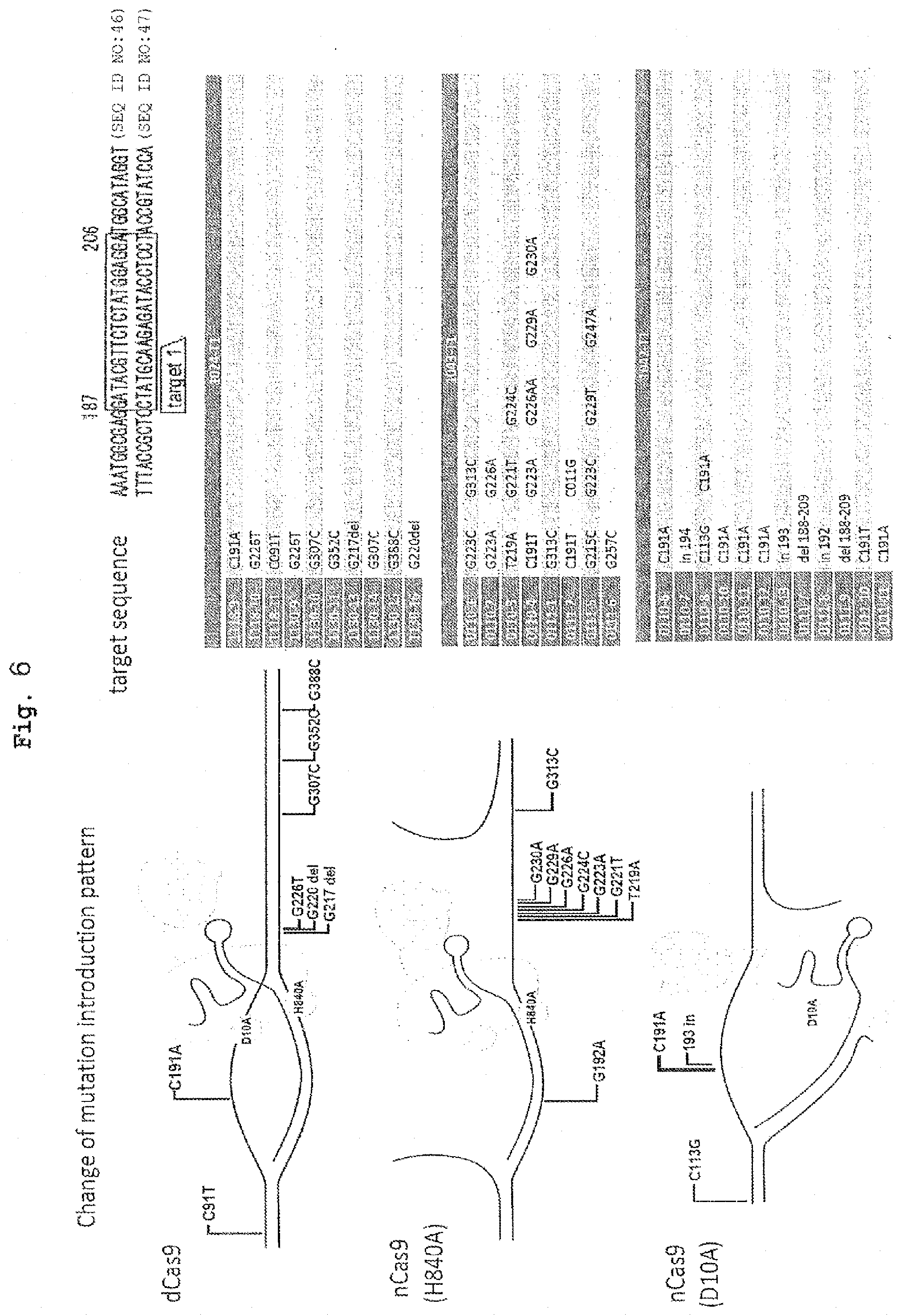
9. The method of claim 6, wherein the step of introducing the nucleic acid encoding the at least one complex into the cell comprises introducing an expression vector comprising the nucleic acid encoding the at least one complex into the cell, wherein the nucleic acid is under regulation of an inducible regulatory region, the method further comprising a step of inducing expression of the nudeic acid for an expression period to stabilize the conversion of one or more nucleotides in the targeted site to one or more different nucleotides, or the deletion of one or more nucleotides, or the insertion of one or more nucleotides into said targeted site in the double stranded DNA
10. The method of claim 9, wherein the target nucleotide sequence in the targeted site in the double stranded DNA is present in a gene essential for survival of the cell.
11. A nudeic acid-modifying enzyme complex, comprising: a nucleic acid base converting enzyme, linked to (ii) a nucleic acid sequence-recognizing module that specifically binds to a target nucleotide sequence in a targeted site of a double stranded DNA, wherein the nucleic acid sequence-recognizing module is a CRISPR-Cas system comprising either a Cas protein that is incapable of introducing a double strand break (DSB) in double stranded DNA or a Cas protein in which cleavage activity for only one strand of double stranded DNA has been inactivated, and wherein the complex is capable of converting one or more nucleotides in the targeted site to one or more other nudeotides, or is capable of deleting one or more nucleotides, or is capable of inserting one or more nucleotides into said targeted site, without introducing a double strand break (DSB) in double stranded DNA in the targeted site.
12. A nucleic add encoding the nucleic acid-modifying enzyme complex of claim 11.
13. The method of claim 1, wherein the nickase protein is a Cas9 D10A mutant nickase protein (nCas9(D10A)).
14. The method of claim 1, wherein the nickase protein is a Cas9 H840A mutant nickase protein (nCas9(H840A)).
15. The nucleic acid-modifying enzyme complex of claim 11, wherein only one of two DNA cleavage abilities of the Cas protein is inactivated.
16. A nucleic acid encoding the nucleic acid-modifying enzyme complex of claim 15.
Description
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] This patent application is a continuation of U.S. patent application Ser. No. 15/124,021, filed Nov. 9, 2016; which is the U.S. national phase of International Patent Application No. PCT/JP2015/056436, filed Mar. 4, 2015; which claims the benefit of Japanese Patent Application No. 2014-043348, filed on Mar. 5, 2014, and Japanese Patent Application No. 2014-201859, filed on Sep. 30, 2014, which are incorporated by reference in their entireties herein.
INCORPORATION-BY-REFERENCE OF MATERIAL ELECTRONICALLY SUBMITTED
[0002] Incorporated by reference in its entirety herein is a computer-readable nucleotide/amino acid sequence listing submitted concurrently herewith and identified as follows: 96.9 KB ASCII (Text) file named "150161_401C1_SEQ_LISTING.txt" created Mar. 30, 2020.
TECHNICAL FIELD
[0003] The present invention relates to a modification method of a genome sequence, which enables modification of a nucleic acid base in a particular region of a genome, without cleaving double-stranded DNA (with no cleavage or single strand cleavage), and without inserting a foreign DNA fragment, and a complex of a nucleic acid sequence-recognizing module and a nucleic acid base converting enzyme used therefor.
BACKGROUND ART
[0004] In recent years, genome editing is attracting attention as a technique for modifying the target gene and genome region of interest in various species. Conventionally, as a method of genome editing, a method utilizing an artificial nuclease comprising a combination of a molecule having a sequence-independent DNA cleavage ability and a molecule having a sequence recognition ability has been proposed (non-patent document 1).
[0005] For example, a method of performing recombination at a target gene locus in DNA in a plant cell or insect cell as a host, by using a zinc finger nuclease (ZFN) wherein a zinc finger DNA binding domain and a non-specific DNA cleavage domain are linked (patent document 1); a method of cleaving or modifying a target gene in a particular nucleotide sequence or a site adjacent thereto by using TALEN wherein a transcription activator-like (TAL) effector, which is a DNA binding module that the plant pathogenic bacteria Xanthomonas has, and a DNA endonuclease are linked (patent document 2); a method utilizing CRISPR-Cas9 system wherein DNA sequence CRISPR (Clustered Regularly interspaced short palindromic repeats), that functions in an acquired immune system possessed by eubacterium and archaebacterium, and nuclease Cas (CRISPR-associated) protein family having an important function along with CRISPR are combined (patent document 3) and the like have been reported. Furthermore, a method of cleaving a target gene in the vicinity of a particular sequence, by using artificial nuclease wherein a PPR protein configured to recognize a particular nucleotide sequence by a series of PPR motifs each consisting of 35 amino acids and recognizing one nucleic acid base, and nuclease are linked (patent document 4) has also been reported.
DOCUMENT LIST
Patent Documents
[0006] patent document 1: JP-B-4968498 [0007] patent document 2: National Publication of International Patent Application No. 2013-513389 [0008] patent document 3: National Publication of International Patent Application No. 2010-519929 [0009] patent document 4: JP-A-2013-128413 [0010] non-patent document [0011] non-patent document 1: Kelvin M Esvelt, Harris H Wang (2013) Genome-scale engineering for systems and synthetic biology, Molecular Systems Biology 9: 641
SUMMARY OF THE INVENTION
Problems to be Solved by the Invention
[0012] The genome editing techniques heretofore been proposed basically presuppose double-stranded DNA breaks (DSB). However, since they involve unexpected genome modifications, side effects such as strong cytotoxicity, chromosomal rearrangement and the like occur, and they have common problems of impaired reliability in gene therapy, extremely small number of surviving cells by nucleotide modification, and difficulty in genetic modification itself in primate ovum and unicellular microorganisms.
[0013] Therefore, an object of the present invention is to provide a novel method of genome editing for modifying a nucleic acid base of a particular sequence of a gene without DSB or insertion of foreign DNA fragment, i.e., by non-cleavage of a double stranded DNA or single strand cleavage, and a complex of a nucleic acid sequence-recognizing module and a nucleic acid base converting enzyme therefor.
Means of Solving the Problems
[0014] The present inventors have conducted intensive studies in an attempt to solve the above-mentioned problems and taken note of adopting base conversion by a conversion reaction of DNA base, without accompanying DSB. The base conversion reaction by a deamination reaction of DNA base is already known; however, targeting any site by recognizing a particular sequence of DNA, and specifically modifying the targeted DNA by base conversion of DNA bases has not been realized yet.
[0015] Therefore, deaminase, that catalyzes a deamination reaction, was used as an enzyme for such conversion of nucleic acid bases, and linked to a molecule having a DNA sequence recognition ability, thereby a genome sequence was modified by nucleic acid base conversion in a region containing a particular DNA sequence.
[0016] Specifically, CRISPR-Cas system (CRISPR-mutant Cas) was used. That is, a DNA encoding an RNA molecule, wherein genome specific CRISPR-RNA:crRNA (gRNA) containing a sequence complementary to a target sequence of a gene to be modified is linked to an RNA for recruiting Cas protein (trans-activating crRNA: tracrRNA) was produced. On the other hand, a DNA wherein a DNA encoding a mutant Cas protein (dCas), wherein cleavage ability of one or both strands of a double stranded DNA is inactivated and a deaminase gene are linked, was produced. These DNAs were introduced into a host yeast cell which comprises a gene to be modified. As a result, mutation could be introduced randomly within the range of several hundred nucleotides of the gene of interest including the target sequence. Compared to when a double mutant Cas protein, which do not cleave both of DNA strands in the double stranded DNA, was used, the mutation introduction efficiency increased when a mutant Cas protein which cleave of either one of the strands was used. In addition, it was clarified that the area of mutation region and variety of mutation vary depending on which of the DNA double strand is cleaved. Furthermore, mutation could be introduced extremely efficiently by targeting a plurality of regions in the target gene of interest. That is, a host cell introduced with DNA was seeded in a nonselective medium, and the sequence of the target gene of interest was examined in randomly selected colonies. As a result, introduction of mutation was confirmed in almost all colonies. It was also confirmed that genome editing can be simultaneously performed at a plurality of sites by targeting certain region in two or more target genes of interest. It was further demonstrated that the method can simultaneously introduce mutation into alleles of diploid or polyploid genomes, can introduce mutation into not only eukaryotic cells but also prokaryotic cells such as Escherichia coli, and is widely applicable irrespective of species. It was also found that editing of essential gene, which showed low efficiency heretofore, can be efficiently performed by transiently performing a nucleic acid base conversion reaction at a desired stage.
[0017] The present inventors have conducted further studies based on these findings and completed the present invention.
[0018] Accordingly, the present invention is as described below. [0019] [1] A method of modifying a targeted site of a double stranded DNA, comprising a step of contacting a complex wherein a nucleic acid sequence-recognizing module that specifically binds to a target nucleotide sequence in a selected double stranded DNA and a nucleic acid base converting enzyme are linked, with said double stranded DNA, to convert one or more nucleotides in the targeted site to other one or more nucleotides or delete one or more nucleotides, or insert one or more nucleotides into said targeted site, without cleaving at least one strand of said double stranded DNA in the targeted site. [0020] [2] The method of [1], wherein the nucleic acid sequence-recognizing module is selected from the group consisting of a CRISPR-Cas system wherein at least one DNA cleavage ability of Cas is inactivated, a zinc finger motif, a TAL effector and a PPR motif. [0021] [3] The method of [1], wherein the nucleic acid sequence-recognizing module is a CRISPR-Cas system wherein at least one DNA cleavage ability of Cas is inactivated. [0022] [4] The method of any of [1]-[3], which uses two or more kinds of nucleic acid sequence-recognizing modules each specifically binding to a different target nucleotide sequence. [0023] [5] The method of [4], wherein the different target nucleotide sequence is present in a different gene. [0024] [6] The method of any of [1]-[5], wherein the nucleic acid base converting enzyme is deaminase. [0025] [7] The method of the above-mentioned [6], wherein the deaminase is AID (AICDA). [0026] [8] The method of any of [1]- [7], wherein the double stranded DNA is contacted with the complex by introducing a nucleic acid encoding the complex into a cell having the double stranded DNA. [0027] [9] The method of [8], wherein the cell is a prokaryotic cell. [0028] [10] The method of [8], wherein the aforementioned cell is a eukaryotic cell. [0029] [11] The method of [8], wherein the cell is a cell of a microorganism. [0030] [12] The method of [8], wherein the cell is a plant cell. [0031] [13] The method of [8], wherein the cell is an insect cell. [0032] [14] The method of [8], wherein the cell is an animal cell. [0033] [15] The method of [8], wherein the aforementioned cell is a cell of a vertebrate. [0034] [16] The method of [8], wherein the cell is a mammalian cell. [0035] [17] The method of any of [9]- [16], wherein the cell is a polyploid cell, and a site in any targeted allele on a homologous chromosome is modified. [0036] [18] The method of any of [8]- [17], comprising a step of introducing an expression vector comprising a nucleic acid encoding the complex in a form permitting control of an expression period into the cell, and a step of inducing expression of the nucleic acid for a period necessary for stabilizing the modification of the targeted site in the double stranded DNA. [0037] [19] The method of the above-mentioned [18], wherein the target nucleotide sequence in the double stranded DNA is present in a gene essential for the cell. [0038] [20] A nucleic acid-modifying enzyme complex wherein a nucleic acid sequence-recognizing module that specifically binds to a target nucleotide sequence in a selected double stranded DNA and a nucleic acid base converting enzyme are linked, which converts one or more nucleotides in the targeted site to other one or more nucleotides or deletes one or more nucleotides, or inserts one or more nucleotides into said targeted site, without cleaving at least one strand of said double stranded DNA in the targeted site. [0039] [21] A nucleic acid encoding the nucleic acid-modifying enzyme complex of [20].
Effect of the Invention
[0040] According to the genome editing technique of the present invention, since it is not associated with insertion of a foreign DNA or double-stranded DNA breaks, the technique is superior in safety. The technique has some possibility of providing a solution in cases where conventional methods were considered as a gene recombination, and thus biologically or legally controversial. It is also theoretically possible to set a wide range of mutation introduction from a pin point of one base to several hundred bases, and the technique can also be applied to local evolution induction by introduction of random mutation into a particular limited region, which has been almost impossible heretofore.
BRIEF DESCRIPTION OF THE DRAWINGS
[0041] FIG. 1 is a schematic illustration showing a mechanism of the genetic modification method of the present invention using the CRISPR-Cas system.
[0042] FIG. 2 shows the results of verification, by using a budding yeast, of the effect of the genetic modification method of the present invention comprising a combination of a CRISPR-Cas system and PmCDA1 deaminase from Petromyzon marinus.
[0043] FIG. 3 shows changes in the number of surviving cells after expression induction when a CRISPR-Cas9 system using a D10A mutant of Cas9 having a nickase activity and a deaminase, PmCDA1, are used in combination (nCas9 D10A-PmCDA1), and when conventional Cas9 having a DNA double strand cleavage ability is used.
[0044] FIG. 4 shows the results when a plurality of expression constructs are constructed such that human AID deaminase and dCas9 are linked via SH3 domain and a binding ligand thereof, wherein the express constructs are introduced into a budding yeast together with two kinds of gRNA (targeting sequences of target 4 and target 5).
[0045] FIG. 5 shows that the mutation introduction efficiency is increased by the use of Cas9 that cleaves either DNA single strand.
[0046] FIG. 6 shows that in the case where a double stranded DNA is not cleaved, the area of mutation introduction region and frequency thereof change depending on which one of the single strands is cleaved.
[0047] FIG. 7 shows that extremely high mutation introduction efficiency can be realized by targeting two regions in proximity.
[0048] FIG. 8 shows that the genetic modification method of the present invention does not require selection by marker. It was found that mutation was introduced into all colonies sequenced.
[0049] FIG. 9 shows that a plurality of sites in a genome can be simultaneously edited by the genetic modification method of the present invention. The upper panel shows the nucleotide sequence and amino acid sequence of the target site of each gene, and an arrow on the nucleotide sequence shows the target nucleotide sequence. The number at the arrow end or arrow head indicates the position of the target nucleotide sequence terminus on ORF. The lower panel shows the results of sequencing of the target site in each 5 clones of red (R) and white (W) colonies. In the sequences, the nucleotides indicated with outline characters show occurrence of base conversion. As for responsiveness to canavanine (Can.sup.R), R shows resistance, and S shows sensitivity.
[0050] FIG. 10 shows that a mutation can be simultaneously introduced into both alleles on the homologous chromosome of diploid genome by the genetic modification method of the present invention. FIG. 10A shows homologous mutation introduction efficiency of Ade1 gene (upper panel) and can1 gene respectively. FIG. 10B shows that homologous mutation was actually introduced into red colony (lower panel). Also, occurrence of heterologous mutation in white colony was shown (upper panel).
[0051] FIG. 11 shows that genome editing of Escherichia coli, a prokaryotic cell, is possible by the genetic modification method of the present invention. FIG. 11A is a schematic illustration showing the plasmid used. FIG. 11B shows that a mutation (CAA.fwdarw.TAA) could be efficiently introduced by targeting a region in the galK gene. FIG. 11C shows the results of sequence analysis of each two clones of the respective colonies in a nonselective medium (none), a medium containing 25 .mu.g/ml rifampicin (Rif25) or a medium containing 50 .mu.g/ml rifampicin (Rif50). Introduction of a mutation imparting rifampicin resistance was confirmed (upper panel). The appearance frequency of rifampicin resistance strain was estimated to be about 10% (lower panel).
[0052] FIG. 12 shows control of the edited base sites by the length of guide RNA. FIG. 12A is a conceptual Figure of editing base site when the length of the target nucleotide sequence is 20 bases or 24 bases. FIG. 12B shows the results of editing by targeting gsiA gene and changing the length of the target nucleotide sequence. The mutated sites are shown with bold letters, "T" and "A" show introduction of complete mutation (C.fwdarw.T or G.fwdarw.A) into the clone, "t" shows that not less than 50% of mutation (C.fwdarw.T) is introduced into the clone (incomplete cloning), and "c" shows that the introduction efficiency of the mutation (C.fwdarw.T) into the clone is less than 50%.
[0053] FIG. 13 is a schematic illustration showing a temperature sensitive plasmid for mutation introduction, which was used in Example 11.
[0054] FIG. 14 shows the protocol of mutation introduction in Example 11.
[0055] FIG. 15 shows the results of introduction of mutation into the rpoB gene in Example 11.
[0056] FIG. 16 shows the results of introduction of mutation into the galK gene in Example 11.
DESCRIPTION OF EMBODIMENTS
[0057] The present invention provides a method of modifying a targeted site of a double stranded DNA by converting the target nucleotide sequence and nucleotides in the vicinity thereof in the double stranded DNA to other nucleotides, without cleaving at least one strand of the double stranded DNA to be modified. The method characteristically comprises a step of contacting a complex wherein a nucleic acid sequence-recognizing module that specifically binds to the target nucleotide sequence in the double stranded DNA and a nucleic acid base converting enzyme are linked, with the double stranded DNA to convert the targeted site, i.e., the target nucleotide sequence and nucleotides in the vicinity thereof, to other nucleotides.
[0058] In the present invention, the "modification" of a double stranded DNA means that a nucleotide (e.g., dC) on a DNA strand is converted to another nucleotide (e.g., dT, dA or dG), or deleted, or a nucleotide or a nucleotide sequence is inserted between certain nucleotides on the DNA strand. While the double stranded DNA to be modified is not particularly limited, it is preferably a genomic DNA. The "targeted site" of a double stranded DNA means the entire or partial "target nucleotide sequence", which a nucleic acid sequence-recognizing module specifically recognizes and binds to, or the vicinity of the target nucleotide sequence (one or both of 5' upstream and 3' downstream), and the length thereof can be appropriately adjusted between 1 base and several hundred bases according to the object.
[0059] In the present invention, the "nucleic acid sequence-recognizing module" means a molecule or molecule complex having an ability to specifically recognize and bind to a particular nucleotide sequence (i.e., target nucleotide sequence) on a DNA strand. Binding of the nucleic acid sequence-recognizing module to a target nucleotide sequence enables a nucleic acid base converting enzyme linked to the module to specifically act on a targeted site of a double stranded DNA.
[0060] In the present invention, the "nucleic acid base converting enzyme" means an enzyme capable of converting a target nucleotide to another nucleotide by catalyzing a reaction for converting a substituent on a purine or pyrimidine ring on a DNA base to another group or atom, without cleaving the DNA strand.
[0061] In the present invention, the "nucleic acid-modifying enzyme complex" means a molecular complex comprising a complex of the above-mentioned nucleic acid sequence-recognizing module linked with a nucleic acid base converting enzyme, wherein the complex has nucleic acid base converting enzyme activity and is imparted with a particular nucleotide sequence recognition ability. The "complex" used herein encompasses not only one composed of a plurality of molecules, but also a single molecule having a nucleic acid sequence-recognizing module and a nucleic acid base converting enzyme such as a fusion protein.
[0062] The nucleic acid base converting enzyme used in the present invention is not particularly limited as long as it can catalyze the above-mentioned reaction, and examples thereof include deaminase belonging to the nucleic acid/nucleotide deaminase superfamily, which catalyzes a deamination reaction that converts an amino group to a carbonyl group. Preferable examples thereof include cytidine deaminase capable of converting cytosine or 5-methylcytosine to uracil or thymine, respectively, adenosine deaminase capable of converting adenine to hypoxanthine, guanosine deaminase capable of converting guanine to xanthine and the like. As cytidine deaminase, more preferred is activation-induced cytidine deaminase (hereinafter also referred to as AID), which is an enzyme that introduces a mutation into an immunoglobulin gene in the acquired immunity of vertebrate or the like.
[0063] While the origin of nucleic acid base converting enzyme is not particularly limited, for example, PmCDA1 (Petromyzon marinus cytosine deaminase 1) from Petromyzon marinus, or AID (Activation-induced cytidine deaminase; AICDA) from mammal (e.g., human, swine, bovine, horse, monkey etc) can be used. The base sequence and amino acid sequence of CDS of PmCDA1 are shown in SEQ ID NOs: 1 and 2, respectively, and the base sequence and amino acid sequence of CDS of human AID are shown in SEQ ID NOs: 3 and 4, respectively.
[0064] A target nucleotide sequence in a double stranded DNA to be recognized by the nucleic acid sequence-recognizing module in the nucleic acid-modifying enzyme complex of the present invention is not particularly limited as long as the module specifically binds to any sequence in the double stranded DNA. The length of the target nucleotide sequence only needs to be sufficient for specific binding of the nucleic acid sequence-recognizing module. For example, when mutation is introduced into a particular site in the genomic DNA of a mammal, it is not less than 12 nucleotides, preferably not less than 15 nucleotides, more preferably not less than 17 nucleotides, according to the genome size thereof. While the upper limit of the length is not particularly limited, it is preferably not more than 25 nucleotides, more preferably not more than 22 nucleotides.
[0065] As the nucleic acid sequence-recognizing module in the nucleic acid-modifying enzyme complex of the present invention, CRISPR-Cas system wherein at least one DNA cleavage ability of Cas is inactivated (CRISPR-mutant Cas), zinc finger motif, TAL effector and PPR motif and the like, as well as a fragment containing a DNA binding domain of a protein that specifically binds to DNA such as restriction enzyme, transcription factor, RNA polymerase or the like, and not having a DNA double strand cleavage ability and the like can be used, but the module is not limited thereto. Preferably, the modules include CRISPR-mutant Cas, zinc finger motif, TAL effector, PPR motif and the like.
[0066] A zinc finger motif is constructed by linking 3-6 different Cys2His2 type zinc finger units (1 finger recognizes about 3 bases), and can recognize a target nucleotide sequence of 9-18 bases. A zinc finger motif can be produced by a known method such as Modular assembly method (Nat Biotechnol (2002) 20: 135-141), OPEN method (Mol Cell (2008) 31: 294-301), CoDA method (Nat Methods (2011) 8: 67-69), Escherichia coli one-hybrid method (Nat Biotechnol (2008) 26:695-701) and the like. The above-mentioned patent document 1 can be referred to as for the detail of the zinc finger motif production.
[0067] A TAL effector has a module repeat structure with about 34 amino acids as a unit, and the 12th and 13th amino acid residues (called RVD) of one module determine the binding stability and base specificity. Since each module is highly independent, TAL effector specific to a target nucleotide sequence can be produced by simply linking the modules. For TAL effector, production methods utilizing an open resource (REAL method (Curr Protoc Mol Biol (2012) Chapter 12: Unit 12.15), FLASH method (Nat Biotechnol (2012) 30: 460-465), and Golden Gate method (Nucleic Acids Res (2011) 39: e82) etc) have been established, and a TAL effector for a target nucleotide sequence can be designed relatively easily. The above-mentioned patent document 2 can be referred to as for the detail of the production of TAL effector.
[0068] PPR motif is constructed such that a particular nucleotide sequence is recognized by a series of PPR motifs each consisting of 35 amino acids and recognizing one nucleic acid base, and recognizes a target base only by 1, 4 and ii(-2) amino acids of each motif. Motif configuration has no dependency, and is free of interference of motifs on both sides. Therefore, similar to TAL effector, a PPR protein specific to the target nucleotide sequence can be produced by simply linking PPR motifs. The above-mentioned patent document 4 can be referred to as for the detail of the production of PPR motif.
[0069] When a fragment of a restriction enzyme, transcription factor, RNA polymerase or the like is used, since the DNA binding domains of these proteins are well known, a fragment containing said domain and not having a DNA double strand cleavage ability can be easily designed and constructed.
[0070] Any of the above-mentioned nucleic acid sequence-recognizing module can be provided as a fusion protein with the above-mentioned nucleic acid base converting enzyme, or a protein binding domain such as SH3 domain, PDZ domain, GK domain, GB domain and the like and a binding partner thereof may be fused with a nucleic acid sequence-recognizing module and a nucleic acid base converting enzyme, respectively, and provided as a protein complex via an interaction of the domain and a binding partner thereof. Alternatively, a nucleic acid sequence-recognizing module and a nucleic acid base converting enzyme may be each fused with intein, and they can be linked by ligation after protein synthesis.
[0071] The nucleic acid-modifying enzyme complex of the present invention containing a complex (including fusion protein), wherein a nucleic acid sequence-recognizing module and a nucleic acid base converting enzyme are linked, may be contacted with a double stranded DNA as an enzyme reaction in a cell-free system. In view of the main object of the present invention, it is desirable to perform the contact by introducing a nucleic acid encoding the complex into a cell having the double stranded DNA of interest (e.g., genomic DNA).
[0072] Therefore, the nucleic acid sequence-recognizing module and the nucleic acid base converting enzyme are preferably prepared as a nucleic acid encoding a fusion protein thereof, or as nucleic acids encoding each of them in a form capable of forming a complex in a host cell after translation into a protein by utilizing a binding domain, intein or the like. The nucleic acid here may be a DNA or an RNA. When it is a DNA, it is preferably a double stranded DNA, and provided in the form of an expression vector disposed under regulation of a functional promoter in a host cell. When it is an RNA, it is preferably a single stranded RNA.
[0073] Since the complex of the present invention wherein a nucleic acid sequence-recognizing module and a nucleic acid base converting enzyme are linked, is not associated with double-stranded DNA breaks (DSB), genome editing with low toxicity is possible, and the genetic modification method of the present invention can be applied to a wide range of biological materials. Therefore, the cells into which nucleic acid encoding nucleic acid sequence-recognizing module and/or nucleic acid base converting enzyme is introduced can encompass cells of any species, from cells of microorganisms, such as bacterium,such as Escherichia coli and the like which are prokaryotes, such as yeast and the like which are lower eukaryotes, to cells of higher eukaryotes such as insect, plant and the like, and cells of vertebrates including mammals such as human and the like.
[0074] A DNA encoding a nucleic acid sequence-recognizing module such as zinc finger motif, TAL effector, PPR motif and the like can be obtained by any method mentioned above for each module. A DNA encoding a sequence-recognizing module of restriction enzyme, transcription factor, RNA polymerase and the like can be cloned by, for example, synthesizing an oligoDNA primer covering a region encoding a desired part of the protein (part containing DNA binding domain) based on the cDNA sequence information thereof, and amplifying by the RT-PCR method using, the total RNA or mRNA fraction prepared from the protein-producing cells as a template.
[0075] A DNA encoding a nucleic acid base converting enzyme can also be cloned similarly by synthesizing an oligoDNA primer based on the cDNA sequence information thereof, and amplifying by the RT-PCR method using, the total RNA or mRNA fraction prepared from the enzyme-producing cells as a template. For example, a DNA encoding PmCDA1 of Petromyzon marinus can be cloned by designing suitable primers for the upstream and downstream of CDS based on the cDNA sequence (accession No. EF094822) registered in the NCBI database, and cloning from mRNA Petromyzon marinus by the RT-PCR method. A DNA encoding human AID can be cloned by designing suitable primers for the upstream and downstream of CDS based on the cDNA sequence (accession No. AB040431) registered in the NCBI database, and cloning from, for example, mRNA from human lymph node by the RT-PCR method.
[0076] The cloned DNA may be directly, or after digestion with a restriction enzyme when desired, or after addition of a suitable linker and/or a nuclear localization signal (each organelle transfer signal when the target double stranded DNA of interest is mitochondria or chloroplast DNA), ligated with a DNA encoding a nucleic acid sequence-recognizing module to prepare a DNA encoding a fusion protein. Alternatively, a DNA encoding a nucleic acid sequence-recognizing module, and a DNA encoding a nucleic acid base converting enzyme may be each fused with a DNA encoding a binding domain or a binding partner thereof, or both DNAs may be fused with a DNA encoding a separation intein, whereby the nucleic acid sequence-recognizing conversion module and the nucleic acid base converting enzyme are translated in a host cell to form a complex. In these cases, a linker and/or a nuclear localization signal can be linked to a suitable position of one of or both DNAs when desired.
[0077] A DNA encoding a nucleic acid sequence-recognizing module and a DNA encoding a nucleic acid base converting enzyme can be obtained by chemically synthesizing the DNA strand, or by linking partly overlapping synthesized oligoDNA short strands by utilizing the PCR method and the Gibson Assembly method to construct a DNA encoding the full length thereof. The advantage of constructing a full-length DNA by chemical synthesis or a combination of PCR method or Gibson Assembly method is that the codon used can be designed in CDS full-length according to the host into which the DNA is introduced. In the expression of a heterologous DNA, the protein expression level is expected to increase by converting the DNA sequence thereof to a codon which is highly frequently used in the host organism. As the data of codon use frequency in host used, for example, the genetic code use frequency database (www.kazusa.or.jp/codon/index.html) disclosed in the home page of Kazusa DNA Research Institute can be used, or documents showing the codon use frequency in each host may be referred to. By reference to the obtained data and the DNA sequence to be introduced, codons showing low use frequency in the host from those used for the DNA sequence may be converted to a codon coding the same amino acid and showing high use frequency.
[0078] An expression vector containing a DNA encoding a nucleic acid sequence-recognizing module and/or a nucleic acid base converting enzyme can be produced, for example, by linking the DNA to the downstream of a promoter in a suitable expression vector.
[0079] As the expression vector, plasmids from Escherichia coli (e.g., pBR322, pBR325, pUC12, pUC13); plasmids from Bacillus subtilis (e.g., pUB110, pTP5, pC194); plasmids from yeast (e.g., pSH19, pSH15); insect cell expression plasmids (e.g., pFast-Bac); animal cell expression plasmids (e.g., pA1-11, pXT1, pRc/CMV, pRc/RSV, pcDNAI/Neo); bacteriophages such as .lamda. phage and the like; insect virus vectors such as baculovirus and the like (e.g., BmNPV, AcNPV); animal virus vectors such as retrovirus, vaccinia virus, adenovirus and the like, are used.
[0080] As the promoter, any promoter appropriate for a host used for gene expression can be used. In a conventional method involving DSB, since the survival rate of the host cell sometimes decreases markedly due to the toxicity, it is desirable to increase the number of cells by the start of the induction by using an inductive promoter. However, since sufficient cell proliferation can also be achieved by expressing the nucleic acid-modifying enzyme complex of the present invention, a constitutive promoter can also be used without limitation.
[0081] For example, when the host is an animal cell, SRa promoter, SV40 promoter, LTR promoter, CMV (cytomegalovirus) promoter, RSV (Rous sarcoma virus) promoter, MoMuLV (Moloney mouse leukemia virus) LTR, HSV-TK (simple herpes virus thymidine kinase) promoter and the like are used. Of these, CMV promoter, SRa promoter and the like are preferable.
[0082] When the host is Escherichia coli, trp promoter, lac promoter, recA promoter, .lamda.P.sub.L promoter, Ipp promoter, T7 promoter and the like are preferable.
[0083] When the host is genus Bacillus, SPO1 promoter, SPO2 promoter, penP promoter and the like are preferable.
[0084] When the host is a yeast, Ga1/10 promoter, PHO5 promoter, PGK promoter, GAP promoter, ADH promoter and the like are preferable.
[0085] When the host is an insect cell, polyhedrin promoter, P10 promoter and the like are preferable.
[0086] When the host is a plant cell, CaMV35S promoter, CaMV19S promoter, NOS promoter and the like are preferable.
[0087] As the expression vector, besides those mentioned above, one containing enhancer, splicing signal, terminator, polyA addition signal, a selection marker such as drug resistance gene, auxotrophic complementary gene and the like, replication origin and the like on demand can be used.
[0088] An RNA encoding a nucleic acid sequence-recognizing module and/or a nucleic acid base converting enzyme can be prepared by, for example, transcription to mRNA in an in vitro transcription system known per se by using a vector encoding DNA encoding the above-mentioned nucleic acid sequence-recognizing module and/or a nucleic acid base converting enzyme as a template.
[0089] A complex of a nucleic acid sequence-recognizing module and a nucleic acid base converting enzyme can be intracellularly expressed by introducing an expression vector containing a DNA encoding a nucleic acid sequence-recognizing module and/or a nucleic acid base converting enzyme into a host cell, and culturing the host cell.
[0090] As the host, genus Escherichia, genus Bacillus, yeast, insect cell, insect, animal cell and the like are used.
[0091] As the genus Escherichia, Escherichia coli K12- DH1 [Proc. Natl. Acad. Sci. USA, 60, 160 (1968)], Escherichia coli JM103 [Nucleic Acids Research, 9, 309 (1981)], Escherichia coli JA221 [Journal of Molecular Biology, 120, 517 (1978)], Escherichia coli HB101 [Journal of Molecular Biology, 41, 459 (1969)], Escherichia coli C600 [Genetics, 39, 440 (1954)] and the like are used.
[0092] As the genus Bacillus, Bacillus subtilis M1114 [Gene, 24, 255 (1983)], Bacillus subtilis 207-21 [Journal of Biochemistry, 95, 87 (1984)] and the like are used.
[0093] As the yeast, Saccharomyces cerevisiae AH22, AH22R.sup.-, NA87-11A, DKD-5D, 20B-12, Schizosaccharomyces pombe NCYC1913, NCYC2036, Pichia pastoris KM71 and the like are used.
[0094] As the insect cell when the virus is AcNPV, cells of established line from cabbage armyworm larva (Spodoptera frugiperda cell; Sf cell), MG1 cells from the mid-intestine of Trichoplusia ni, High Five.TM. cells from an egg of Trichoplusia ni, cells from Mamestra brassicae, cells from Estigmena acrea and the like are used. When the virus is BmNPV, cells of established line from Bombyx mori (Bombyx mori N cell; BmN cell) and the like are used as insect cells. As the Sf cell, for example, Sf9 cell (ATCC CRL1711), Sf21 cell [all above, In Vivo, 13, 213-217 (1977)] and the like are used.
[0095] As the insect, for example, larva of Bombyx mori, Drosophila, cricket and the like are used [Nature, 315, 592 (1985)].
[0096] As the animal cell, cell lines such as monkey COS-7 cell, monkey Vero cell, Chinese hamster ovary (CHO) cell, dhfr gene-deficient CHO cell, mouse L cell, mouse AtT-20 cell, mouse myeloma cell, rat GH3 cell, human FL cell and the like, pluripotent stem cells such as iPS cell, ES cell and the like of human and other mammals, and primary cultured cells prepared from various tissues are used. Furthermore, zebrafish embryo, Xenopus oocyte and the like can also be used.
[0097] As the plant cell, suspend cultured cells, callus, protoplast, leaf segment, root segment and the like prepared from various plants (e.g., grain such as rice, wheat, corn and the like, product crops such as tomato, cucumber, egg plant and the like, garden plants such as carnation, Eustoma russeffianum and the like, experiment plants such as tobacco, arabidopsis thaliana and the like) are used.
[0098] All the above-mentioned host cells may be haploid (monoploid), or polyploid (e.g., diploid, triploid, tetraploid and the like). In the conventional mutation introduction methods, mutation is, in principle, introduced into only one homologous chromosome to produce a heterologous geno-type. Therefore, the desired feature is not expressed unless it is a dominant mutation, and making it homologous inconveniently requires labor and time. In contrast, according to the present invention, since mutations can be introduced into all alleles on the homologous chromosome in the genome, desired feature can be expressed in a single generation even in the case of recessive mutation (FIG. 10), which is extremely useful since the problem of the conventional method can be solved.
[0099] An expression vector can be introduced by a known method (e.g., lysozyme method, competent method, PEG method, CaCl.sub.2 coprecipitation method, electroporation method, the microinjection method, the particle gun method, lipofection method, Agrobacterium method and the like) according to the kind of the host.
[0100] Escherichia coli can be transformed according to the methods described in, for example, Proc. Natl. Acad. Sci. USA, 69, 2110 (1972), Gene, 17, 107 (1982) and the like.
[0101] A vector can be introduced into the genus Bacillus according to the methods described in, for example, Molecular & General Genetics, 168, 111 (1979) and the like.
[0102] A vector can be introduced into a yeast according to the methods described in, for example, Methods in Enzymology, 194, 182-187 (1991), Proc. Natl. Acad. Sci. USA, 75, 1929 (1978) and the like.
[0103] A vector can be introduced into an insect cell and an insect according to the methods described in, for example, Bio/Technology, 6, 47-55 (1988) and the like.
[0104] A vector can be introduced into an animal cell according to the methods described in, for example, Cell Engineering additional volume 8, New Cell Engineering Experiment Protocol, 263-267 (1995) (published by Shujunsha), and Virology, 52, 456 (1973).
[0105] A cell introduced with a vector can be cultured according to a known method according to the kind of the host.
[0106] For example, when Escherichia coli or genus Bacillus is cultured, a liquid medium is preferable as a medium used for the culture. The medium preferably contains a carbon source, nitrogen source, inorganic substance and the like necessary for the growth of the transformant. Examples of the carbon source include glucose, dextrin, soluble starch, sucrose and the like; examples of the nitrogen source include inorganic or organic substances such as ammonium salts, nitrate salts, corn steep liquor, peptone, casein, meat extract, soybean cake, potato extract and the like; and examples of the inorganic substance include calcium chloride, sodium dihydrogen phosphate, magnesium chloride and the like. The medium may contain yeast extract, vitamins, growth promoting factor and the like. The pH of the medium is preferably about 5-about 8.
[0107] As a medium for culturing Escherichia coli, for example, M9 medium containing glucose, casamino acid [Journal of Experiments in Molecular Genetics, 431-433, Cold Spring Harbor Laboratory, New York 1972] is preferable. Where necessary, for example, agents such as 3.beta.-indolylacrylic acid may be added to the medium to ensure an efficient function of a promoter. Escherichia coli is cultured at generally about 15-about 43.degree. C. Where necessary, aeration and stirring may be performed.
[0108] The genus Bacillus is cultured at generally about 30-about 40.degree. C. Where necessary, aeration and stirring may be performed.
[0109] Examples of the medium for culturing yeast include Burkholder minimum medium [Proc. Natl. Acad. Sci. USA, 77, 4505 (1980)], SD medium containing 0.5% casamino acid [Proc. Natl. Acad. Sci. USA, 81, 5330 (1984)] and the like. The pH of the medium is preferably about 5-about 8. The culture is performed at generally about 20.degree. C.-about 35.degree. C. Where necessary, aeration and stirring may be performed.
[0110] As a medium for culturing an insect cell or insect, for example, Grace's Insect Medium [Nature, 195, 788 (1962)] containing an additive such as inactivated 10% bovine serum and the like as appropriate and the like are used. The pH of the medium is preferably about 6.2-about 6.4. The culture is performed at generally about 27.degree. C. Where necessary, aeration and stirring may be performed.
[0111] As a medium for culturing an animal cell, for example, minimum essential medium (MEM) containing about 5-about 20% of fetal bovine serum [Science, 122, 501 (1952)], Dulbecco's modified Eagle medium (DMEM) [Virology, 8, 396 (1959)], RPMI 1640 medium [The Journal of the American Medical Association, 199, 519 (1967)], 199 medium [Proceeding of the Society for the Biological Medicine, 73, 1 (1950)] and the like are used. The pH of the medium is preferably about 6-about 8. The culture is performed at generally about 30.degree. C.-about 40.degree. C. Where necessary, aeration and stirring may be performed.
[0112] As a medium for culturing a plant cell, for example, MS medium, LS medium, B5 medium and the like are used. The pH of the medium is preferably about 5-about 8. The culture is performed at generally about 20.degree. C.-about 30.degree. C. Where necessary, aeration and stirring may be performed.
[0113] As mentioned above, a complex of a nucleic acid sequence-recognizing module and a nucleic acid base converting enzyme, i.e., nucleic acid-modifying enzyme complex, can be expressed intracellularly.
[0114] An RNA encoding a nucleic acid sequence-recognizing module and/or a nucleic acid base converting enzyme can be introduced into a host cell by microinjection method, lipofection method and the like. RNA introduction can be performed once or multiple times (e.g., 2-5 times) at suitable intervals.
[0115] When a complex of a nucleic acid sequence-recognizing module and a nucleic acid base converting enzyme is expressed by an expression vector or RNA molecule introduced into the cell, the nucleic acid sequence-recognizing module specifically recognizes and binds to a target nucleotide sequence in the double stranded DNA (e.g., genomic DNA) of interest and, due to the action of the nucleic acid base converting enzyme linked to the nucleic acid sequence-recognizing module, base conversion occurs in the sense strand or antisense strand of the targeted site (whole or partial target nucleotide sequence or appropriately adjusted within several hundred bases including the vicinity thereof) and a mismatch occurs in the double stranded DNA (e.g., when cytidine deaminase such as PmCDA1, AID and the like is used as a nucleic acid base converting enzyme, cytosine on the sense strand or antisense strand at the targeted site is converted to uracil to cause U:G or G:U mismatch). When the mismatch is not correctly repaired, and when repaired such that a base of the opposite strand forms a pair with a base of the converted strand (T-A or A-T in the above-mentioned example), or when another nucleotide is further substituted (e.g., U.fwdarw.A, G) or when one to several dozen bases are deleted or inserted during repair, various mutations are introduced.
[0116] As for zinc finger motif, production of many actually functional zinc finger motifs is not easy, since production efficiency of a zinc finger that specifically binds to a target nucleotide sequence is not high and selection of a zinc finger having high binding specificity is not easy. While TAL effector and PPR motif have a high degree of freedom of target nucleic acid sequence recognition as compared to zinc finger motif, a problem remains in the efficiency since a large protein needs to be designed and constructed every time according to the target nucleotide sequence.
[0117] In contrast, since the CRISPR-Cas system recognizes the sequence of double stranded DNA of interest by a guide RNA complementary to the target nucleotide sequence, any sequence can be targeted by simply synthesizing an oligoDNA capable of specifically forming a hybrid with the target nucleotide sequence.
[0118] Therefore, in a more preferable embodiment of the present invention, a CRISPR-Cas system wherein at least one DNA cleavage ability of Cas is inactivated (CRISPR-mutant Cas) is used as a nucleic acid sequence-recognizing module.
[0119] FIG. 1 is a schematic illustration showing the double stranded DNA modification method of the present invention using CRISPR-mutant Cas as a nucleic acid sequence-recognizing module.
[0120] The nucleic acid sequence-recognizing module of the present invention using CRISPR-mutant Cas is provided as a complex of an RNA molecule consisting of a guide RNA complementary to the target nucleotide sequence and tracrRNA necessary for recruiting mutant Cas protein, and a mutant Cas protein.
[0121] The Cas protein used in the present invention is not particularly limited as long as it belongs to the CRISPR system, and is preferably Cas9. Examples of Cas9 include, but are not limited to, Cas9 (SpCas9 from Streptococcus pyogenes, Cas9 (StCas9) from Streptococcus thermophilus and the like, preferably SpCas9. As a mutant Cas used in the present invention, either a Cas having cleavage ability of both strands of the double stranded DNA is inactivated, or a Cas having nickase activity wherein only one of the cleavage ability of only one of the strands is inactivated, can be used. For example, in the case of SpCas9, a Dl OA mutant wherein the 10th Asp residue is converted to an Ala residue and lacking cleavage ability of a strand opposite to the strand forming a complementary strand with a guide RNA, or H840A mutant wherein the 840th His residue is converted to an Ala residue and lacking cleavage ability of strand complementary to guide RNA, or a double mutant thereof can be used, and another mutant Cas can be used similarly.
[0122] A nucleic acid base converting enzyme is provided as a complex with mutant Cas by a method similar to the linking scheme with the above-mentioned zinc finger and the like. Alternatively, a nucleic acid base converting enzyme and mutant Cas can also be linked by utilizing RNA scaffold with RNA aptamers MS2F6, PP7 and the like and binding proteins thereto. Guide RNA forms a complementary strand with the target nucleotide sequence, mutant Cas is recruited by the attached tracrRNA and mutant Cas recognizes DNA cleavage site recognition sequence PAM (protospacer adjacent motif) (when SpCas9 is used, PAM is 3 bases of NGG (N is any base), and, theoretically, can target any position on the genome). One or both DNAs cannot be cleaved, and, due to the action of the nucleic acid base converting enzyme linked to the mutant Cas, base conversion occurs in the targeted site (appropriately adjusted within several hundred bases including whole or partial target nucleotide sequence) and a mismatch occurs in the double stranded DNA. When the mismatch is not correctly repaired, and when repaired such that a base of the opposite strand forms a pair with a base of the converted strand, or when another nucleotide is further converted or when one to several dozen bases are deleted or inserted during repair, various mutations are introduced (see, e.g., FIG. 2).
[0123] Even when CRISPR-mutant Cas is used as a nucleic acid sequence-recognizing module, a nucleic acid sequence-recognizing module and a nucleic acid base converting enzyme are introduced, desirably in the form of a nucleic acid encoding same, into a cell having a double stranded DNA of interest, similar to when zinc finger and the like are used as a nucleic acid sequence-recognizing module.
[0124] A DNA encoding Cas can be cloned by a method similar to the above-mentioned method for a DNA encoding a nucleic acid base converting enzyme, from a cell producing the enzyme. A mutant Cas can be obtained by introducing a mutation to convert an amino acid residue of the part important for the DNA cleavage activity (e.g., 10th Asp residue and 840th His residue for Cas9, though not limited thereto) to another amino acid, into a DNA encoding cloned Cas, by a site specific mutation induction method known per se.
[0125] Alternatively, a DNA encoding mutant Cas can also be constructed as a DNA having codon usage suitable for expression in a host cell to be used, by a method similar to those mentioned above for a DNA encoding a nucleic acid sequence-recognizing module and a DNA encoding a nucleic acid base converting enzyme, and in a combination with chemical synthesis or PCR method or Gibson Assembly method. For example, CDS sequence and amino acid sequence optimized for the expression of SpCas9 in eukaryotic cells are shown in SEQ ID NOs: 5 and 6. In the sequence shown in SEQ ID NO: 5, when "A" is converted to "C" in base No. 29, a DNA encoding a Dl OA mutant can be obtained, and when "CA" is converted to "GC" in base Nos. 2518-2519, a DNA encoding an H840A mutant can be obtained.
[0126] A DNA encoding a mutant Cas and a DNA encoding a nucleic acid base converting enzyme may be linked to allow for expression as a fusion protein, or designed to be separately expressed using a binding domain, intein or the like, and form a complex in a host cell via protein-protein interaction or protein ligation.
[0127] The obtained DNA encoding a mutant Cas and/or a nucleic acid base converting enzyme can be inserted into the downstream of a promoter of an expression vector similar to the one mentioned above, according to the host.
[0128] On the other hand, a DNA encoding guide RNA and tracrRNA can be obtained by designing an oligoDNA sequence linking guide RNA sequence complementary to the target nucleotide sequence and known tracrRNA sequence (e.g., gttttagagctagaaatagcaagttaaaataaggctagtccgttatcaacttgaaaaagtggcaccgagtcgg- tggtgctttt; SEQ ID NO: 7) and chemically synthesizing using a DNA/RNA synthesizer.
[0129] While the length of the guide RNA sequence is not particularly limited as long as it can specifically bind to a target nucleotide sequence, for example, it is 15-30 nucleotides, preferably 18-24 nucleotides.
[0130] While a DNA encoding guide RNA and tracrRNA can also be inserted into an expression vector similar to the one mentioned above, according to the host. As the promoter, pol III promoter (e.g., SNR6, SNR52, SCR1, RPR1, U6, H1 promoter etc.) and terminator (e.g., T.sub.6 sequence) are preferably used.
[0131] An RNA encoding mutant Cas and/or a nucleic acid base converting enzyme can be prepared by, for example, transcription to mRNA in an in vitro transcription system known per se by using a vector encoding the above-mentioned mutant Cas and/or DNA encoding a nucleic acid base converting enzyme as a template.
[0132] Guide RNA-tracrRNA can be obtained by designing an oligoDNA sequence in which a sequence complementary to the target nucleotide sequence and known tracrRNA sequence are linked, and chemically synthesizing using a DNA/RNA synthesizer.
[0133] A DNA or RNA encoding mutant Cas and/or a nucleic acid base converting enzyme, guide RNA-tracrRNA or a DNA encoding same can be introduced into a host cell by a method similar to the above, according to the host.
[0134] Since conventional artificial nuclease accompanies Double-stranded DNA breaks (DSB), inhibition of growth and cell death assumedly caused by disordered cleavage (off-target cleavage) of chromosome may occur by targeting a sequence in the genome. The effect thereof is particularly fatal for many microorganisms and prokaryotes, and prevents applicability. In the present invention, mutation is introduced not by DNA cleavage but by a conversion reaction of the substituent on the DNA base (particularly deamination reaction), and therefore, drastic reduction of toxicity can be realized. In fact, as shown in the comparison tests using a budding yeast as a host in the below-mentioned Examples, when Cas9 having a conventional type of DSB activity is used, the number of surviving cells decreases by induction of expression, whereas it was confirmed that the cells continued to grow and the number of surviving cells increased by the technique of the present invention using a combination of mutant Cas and a nucleic acid base converting enzyme in combination (FIG. 3).
[0135] The modification of the double stranded DNA in the present invention does not preclude occurrence of cleavage of the double stranded DNA in a site other than the targeted site (appropriately adjusted within several hundred bases including whole or partial target nucleotide sequence). However, one of the greatest advantages of the present invention is avoidance of toxicity by off-target cleavage, which is generally applicable to any species. In one preferable embodiment, therefore, the modification of the double stranded DNA in the present invention is not associated with cleavage of DNA strand not only in a targeted site of a selected double stranded DNA but in other sites.
[0136] As shown in the below-mentioned Examples, when Cas having a nickase activity capable of cleaving only one of the strands of the double stranded DNA is used as a mutant Cas (FIG. 5), the mutation introduction efficiency increases as compared to when mutant Cas which is incapable of cleaving both strands is used. Therefore, for example, besides a nucleic acid sequence-recognizing module and a nucleic acid base converting enzyme, linking a protein having a nickase activity, thereby cleaving only a DNA single strand in the vicinity of the target nucleotide sequence, the mutation introduction efficiency can be improved while avoiding the strong toxicity of DSB.
[0137] Furthermore, a comparison of the effects of mutant Cas having two kinds of nickase activity of cleaving different strand reveals that using one of the mutant Cas results in mutated sites accumulating near the center of the target nucleotide sequence, and using another mutant Cas results in various mutations which are randomly introduced into region of several hundred bases from the target nucleotide sequence (FIG. 6). Therefore, by selecting a strand to be cleaved by the nickase, a mutation can be introduced into a particular nucleotide or nucleotide region at a pinpoint, or various mutations can be randomly introduced into a comparatively wide range, which can be properly adopted according to the object. For example, when the former technique is applied to genetically diseased iPS cell, a cell transplantation therapeutic agent with a lower risk of rejection can be produced by repairing mutation of the pathogenic gene in an iPS cell produced from the patients' own cell, and differentiating the cell into the somatic cell of interest.
[0138] Example 7 and the subsequent Examples mentioned below show that a mutation can be introduced into a particular nucleotide almost at a pinpoint. For pinpoint introduction of a mutation into a desired nucleotide, the target nucleotide sequence should be set to show certain regularity of the positional relationship between a nucleotide desired to be introduced with a mutation and the target nucleotide sequence. CRISPR-Cas system is used as a nucleic acid sequence-recognizing module and AID is used as a nucleic acid base converting enzyme, the target nucleotide sequence can be designed such that C (or G in the opposite strand) into which a mutation is desired to be introduced is at 2-5 nucleotides from the 5'-end of the target nucleotide sequence. As mentioned above, the length of the guide RNA sequence can be appropriately determined to fall between 15-30 nucleotides, preferably 18-24 nucleotides. Since the guide RNA sequence is a sequence complementary to the target nucleotide sequence, the length of the target nucleotide sequence changes when the length of the guide RNA sequence is changed; however, the regularity that a mutation is likely to be introduced into C or G at 2-5 nucleotides from the 5'-end irrespective of the length of the nucleotide, is maintained (FIG. 12). Therefore, by appropriately determining the length of the target nucleotide sequence (guide RNA as a complementary strand thereof), the site of a base into which a mutation can be introduced can be shifted. As a result, restriction by DNA cleavage site recognition sequence PAM (NGG) can also be removed, and the degree of freedom of mutation introduction becomes higher.
[0139] As shown in the below-mentioned Examples, when sequence-recognizing modules are produced corresponding to a plurality of target nucleotide sequences in proximity, and simultaneously used, the mutation introduction efficiency drastically increases relative to when a single nucleotide sequence is used as a target (FIG. 7). As the effect thereof, similar mutation induction is realized even when both target nucleotide sequences partly overlap or when the both are apart by about 600 bp. It can occur when both target nucleotide sequences are in the same direction (target nucleotide sequences are present on the same strand) (FIG. 7), and when they are opposed (target nucleotide sequences are present on each strand of double stranded DNA) (FIG. 4).
[0140] As shown in the below-mentioned Examples, the genome sequence modification method of the present invention can introduce mutation into almost all cells in which the nucleic acid-modifying enzyme complex of the present invention has been expressed, by selecting a suitable target nucleotide sequence (FIG. 8). Thus, insertion and selection of a selection marker gene, which are essential in the conventional genome editing, are not necessary. This dramatically facilitates and simplifies gene manipulation and extends the applicability to crop breeding and the like since a recombinant organism with foreign DNA is not produced.
[0141] Since the genome sequence modification method of the present invention shows extremely high mutation introduction efficiency, and does not require selection by markers, a plurality of DNA regions at completely different positions can be modified as targets (FIG. 9). Therefore, in one preferable embodiment of the present invention, two or more kinds of nucleic acid sequence-recognizing modules that specifically bind to different target nucleotide sequences (which may be present in one target gene of interest, or two or more different target genes of interest, which may be present on the same chromosome or different chromosomes) can be used. In this case, each one of these nucleic acid sequence-recognizing modules and nucleic acid base converting enzyme form a nucleic acid-modifying enzyme complex. Here, a common nucleic acid base converting enzyme can be used. For example, when CRISPR-Cas system is used as a nucleic acid sequence-recognizing module, a common complex of a Cas protein and a nucleic acid base converting enzyme (including fusion protein) is used, and two or more kinds of chimeric RNAs of tracrRNA and each of two or more guide RNAs that respectively form a complementary strand with a different target nucleotide sequences are produced and used as guide RNA-tracrRNAs. On the other hand, when zinc finger motif, TAL effector and the like are used as nucleic acid sequence-recognizing modules, for example, a nucleic acid base converting enzyme can be fused with a nucleic acid sequence-recognizing module that specifically binds to a different target nucleotide.
[0142] To express the nucleic acid-modifying enzyme complex of the present invention in a host cell, as mentioned above, an expression vector containing a DNA encoding the nucleic acid-modifying enzyme complex, or an RNA encoding the nucleic acid-modifying enzyme complex is introduced into a host cell. For efficient introduction of mutation, it is desirable to maintain an expression of nucleic acid-modifying enzyme complex at a given level or above for not less than a given period. From such aspect, introduction of an expression vector autonomously replicatable in a host cell (plasmid etc.) is reliable. However, since the plasmid etc. are foreign DNAs, they are preferably removed rapidly after successful introduction of mutation. Therefore, although it varies depending on the kind of host cell and the like, for example, the introduced plasmid is desirably removed from the host cell after a lapse of 6 hr-2 days from the introduction of an expression vector by using various plasmid removal methods which are well known in the art.
[0143] Alternatively, as long as sufficient expression of a nucleic acid-modifying enzyme complex for the introduction of mutation is achieved, it is also preferable to introduce mutation into the target double stranded DNA of interest by transient expression by using an expression vector without autonomous replicatability in a host cell (e.g., vector lacking replication origin that functions in a host cell and/or gene encoding protein necessary for replication etc.) or RNA.
[0144] Expression of target gene is suppressed while the nucleic acid-modifying enzyme complex of the present invention is expressed in a host cell to perform a nucleic acid base conversion reaction. Therefore, it was difficult to directly edit a gene essential for the survival of the host cell as a target gene (result in side effects such as growth inhibition of host, unstable mutation introduction efficiency, mutation of site different from target and the like). In the present invention, direct editing of an essential gene has been successfully and efficiently realized by causing a nucleic acid base conversion reaction at a desired stage, and transiently expressing the nucleic acid-modifying enzyme complex of the present invention in a host cell for a period necessary for stabilizing the modification of the targeted site. While the period necessary for a nucleic acid base conversion reaction and stabilizing the modification of the targeted site varies depending on the kind of the host cell, culture conditions and the like, host cells of 2-20 generations are generally considered to be necessary. For example, when the host cell is a yeast or bacterium (e.g., Escherichia coli), expression of a nucleic acid-modifying enzyme complex needs to be induced for 5-10 generations. Those of ordinary skill in the art can appropriately determine a preferable expression induction period based on the doubling time of the host cell under culture conditions used. For example, when a budding yeast is subjected to liquid culture in a 0.02% galactose inducer medium, the expression induction period is, for example, 20-40 hr. The expression induction period of the nucleic acid encoding the nucleic acid-modifying enzyme complex of the present invention may be extended beyond the above-mentioned "period necessary for establishing the modification of the targeted site" to the extent not causing side effects to the host cell.
[0145] As a means for transiently expressing the nucleic acid-modifying enzyme complex of the present invention at a desired stage for a desired period, a method comprising producing a construct (expression vector) containing a nucleic acid encoding the nucleic acid-modifying enzyme complex (a DNA encoding a guide RNA-tracrRNA and a DNA encoding a mutant Cas and nucleic acid base substitution enzyme in the case of CRISPR-Cas system), in a manner that the expression period can be controlled, and introducing the construct into a host cell can be used. The "manner that the expression period can be controlled" is specifically, for example, a nucleic acid encoding the nucleic acid-modifying enzyme complex of the present invention placed under regulation of an inducible regulatory region. While the "inducible regulatory region" is not particularly limited, it is, for example, an operon of a temperature sensitive (ts) mutation repressor and an operator regulated thereby in microorganism cells of bacterium (e.g., Escherichia coli), yeast and the like. Examples of the ts mutation repressor include, but are not limited to, ts mutation of cl repressor from .lamda. phage. In the case of .lamda. phage cl repressor (ts), it is linked to an operator to suppress expression of gene in the downstream at not more than 30.degree. C. (e.g., 28.degree. C.). At a high temperature of not less than 37.degree. C. (e.g., 42.degree. C.), it is dissociated from the operator to allow for induction of gene expression (FIGS. 13 and 14). Therefore, the period when the expression of the target gene is suppressed can be minimized by culturing a host cell introduced with a nucleic acid encoding nucleic acid-modifying enzyme complex generally at not more than 30.degree. C., raising the temperature to not less than 37.degree. C. at an appropriate stage, performing culture for a given period to carry out a nucleic acid base conversion reaction and, after introduction of mutation into the target gene, rapidly lowering the temperature to not more than 30.degree. C. Thus, even when an essential gene for the host cell is targeted, it can be efficiently edited while suppressing the side effects (FIG. 15).
[0146] When temperature sensitive mutation is utilized, for example, a temperature sensitive mutant of a protein necessary for autonomous replication of a vector is included in a vector containing a DNA encoding the nucleic acid-modifying enzyme complex of the present invention. As a result, autonomous replication becomes impossible rapidly after expression of the nucleic acid-modifying enzyme complex, and the vector naturally falls off during the cell division. Examples of the temperature sensitive mutant protein include, but are not limited to, a temperature sensitive mutant of Rep101 ori necessary for the replication of pSC101 ori. Rep101 ori (ts) acts on pSC101 ori to enable autonomous replication of plasmid at not more than 30.degree. C. (e.g., 28.degree. C.), but loses function at not less than 37.degree. C. (e.g., 42.degree. C.), and plasmid cannot replicate autonomously. Therefore, a combined use with cl repressor (ts) of the above-mentioned A phage simultaneously enables transient expression of the nucleic acid-modifying enzyme complex of the present invention, and removal of the plasmid.
[0147] On the other hand, when a higher eukaryotic cell such as animal cell, insect cell, plant cell and the like is used as a host cell, a DNA encoding the nucleic acid-modifying enzyme complex of the present invention is introduced into a host cell under regulation of an inducible promoter (e.g., metallothionein promoter (induced by heavy metal ion), heat shock protein promoter (induced by heat shock), Tet-ON/Tet-OFF system promoter (induced by addition or removal of tetracycline or a derivative thereof), steroid-responsive promoter (induced by steroid hormone or a derivative thereof) etc.), the induction substance is added to the medium (or removed from the medium) at an appropriate stage to induce expression of the nucleic acid-modifying enzyme complex, culture is performed for a given period to carry out a nucleic acid base conversion reaction and, introduction of mutation into the target gene, transient expression of the nucleic acid-modifying enzyme complex can be realized.
[0148] In Prokaryotic cells such as Escherichia coli and the like, inducible promoters can also be used. Examples of such inducible promoters include, but are not limited to, lac promoter (induced by IPTG), cspA promoter (induced by cold shock), araBAD promoter (induced by arabinose) and the like.
[0149] Alternatively, the above-mentioned inducible promoters can also be utilized as a vector removal mechanism when higher eukaryotic cells such as animal cell, insect cell, plant cell and the like are used as a host cell. That is, a vector is loaded with a replication origin that can function in a host cell, and a nucleic acid encoding a protein necessary for replication thereof (e.g., SV40 ori and large T antigen, oriP and EBNA-1 etc. for animal cells), and the expression of the nucleic acid encoding the protein is regulated by the above-mentioned inducible promoter. As a result, while the vector is autonomously replicatable in the presence of an induction substance, when the induction substance is removed, autonomous replication does not occur, and the vector naturally falls off during cell division (conversely, autonomous replication becomes impossible by the addition of tetracycline and doxycycline in the case of Tet-OFF system vector).
[0150] The present invention is explained in the following by referring to Examples, which are not to be construed as limitative.
EXAMPLE
[0151] In the below-mentioned Examples 1-6, experiments were performed as follows.
<Cell Line, Culture, Transformation, and Expression Induction>
[0152] Budding yeast Saccharomyces cerevisiae BY4741 strain (requiring leucine and uracil) was cultured in a standard YPDA medium or SD medium with a Dropout composition satisfying the auxotrophicity. The culture performed was standing culture in an agar plate or shaking culture in a liquid medium between 25.degree. C. and 30.degree. C. Transformation was performed by a lithium acetate method, and selection was made in SD medium matching appropriate auxotrophicity. For expression induction by galactose, after preculture overnight in an appropriate SD medium, culture in SR medium overnight with carbon source changed from 2% glucose to 2% raffinose, and further culture in SGal medium for 3 hr to about two nights with carbon source changed to 0.2-2% galactose were conducted for expression induction.
[0153] For the measurement of the number of surviving cells and Can1 mutation rate, a cell suspension was appropriately diluted, and applied on SD plate medium and SD-Arg+60 mg/I Canavanine plate medium or SD+300 mg/I Canavanine plate medium, and the number of colonies that emerge 3 days later was counted as the number of surviving cells. Using the number of surviving colonies in SD plate as the total number of cells, and the number of surviving colonies in Canavanine plate as the number of resistant mutant strain, the mutation rate was calculated and evaluated. The site of mutation introduction was identified by amplifying DNA fragments containing the target gene region of each strain by a colony PCR method, performing DNA sequencing, and performing an alignment analysis based on the sequence of Saccharomyces Genome Database (www.yeastgenome.org).
<Nucleic Acid Operation>
[0154] DNA was processed or constructed by any of PCR method, restriction enzyme treatment, ligation, Gibson Assembly method, and artificial chemical synthesis. For plasmid, as the yeast/ Escherichia coli shuttle vector, pRS315 for leucine selection and pRS426 for uracil selection were used as the backbone. Plasmid was amplified by Escherichia coli line XL-10 gold or DH5.alpha., and introduced into yeast by the lithium acetate method.
<Construct>
[0155] For inducible expression, budding yeast pGa1/10 (SEQ ID NO: 8), which is a bidirectional promoter inducible by galactose, was used. At the downstream of a promoter, a nuclear localization signal (ccc aag aag aag agg aag gtg; SEQ ID NO: 9(PKKKRV; encoding SEQ ID NO: 10)) was added to Cas9 gene ORF from Streptococcus pyogenes having a codon optimized for eukaryon expression (SEQ ID NO: 5) and ORF (SEQ ID NO: 1 or 3) of deaminase gene (PmCDA1 from Petromyzon marinus or hAID from human) was ligated via a linker sequence, and expressed as a fusion protein. As a linker sequence, GS linker (repeat of ggt gga gga ggt tct; SEQ ID NO: 11 (encoding GGGGS; SEQ ID NO: 12)), Flag.RTM. tag (gac tat aag gac cacgac gga gac tac aag gat cat gat att gat tac aaa gac gat gac gat aag; SEQ ID NO: 13 (encoding DYKDHDGDYKDHDIDYKDDDDK; SEQ ID NO: 14)), Strep-tag.RTM. (tgg agc cac ccg cag ttc gaa aaa; SEQ ID NO: 15 (encoding WSHPQFEK; SEQ ID NO: 16)), and other domains are selected and used in combination. Here, particularly, 2xGS, SH3 domain (SEQ ID NO: 17 and 18), and Flag.RTM. tag were ligated and used. As a terminator, ADH1 terminator from budding yeast (SEQ ID NO: 19) and Top2 terminator (SEQ ID NO: 20) were ligated. In the domain integration method, Cas9 gene ORF was ligated to SH3 domain via 2xGS linker to give one protein, deaminase was added with SH3 ligand sequence (SEQ ID NOs: 21 and 22) as another protein, and they were ligated to Ga1/10 promoter on both sides. And they were simultaneously expressed. These were incorporated into pRS315 plasmid.
[0156] In Cas9, mutation to convert the 10th aspartic acid to alanine (D10A, corresponding DNA sequence mutation a29c) and mutation to convert the 840th histidine to alanine (H840A, corresponding DNA sequence mutation ca2518gc) were introduced to remove cleavage ability of either side of DNA strand.
[0157] gRNA as a chimeric structure with tracrRNA (from Streptococcus pyogenes; SEQ ID NO: 7) was disposed between SNR52 promoter (SEQ ID NO: 23) and Sup4 terminator (SEQ ID NO: 24), and incorporated into pRS426 plasmid. As gRNA target base sequence, CAN1 gene ORF, 187-206 (gatacgttctctatggagga; SEQ ID NO: 25) (target 1), 786-805 (ttggagaaacccaggtgcct; SEQ ID NO: 26) (target 3), 793-812 (aacccaggtgcctggggtcc; SEQ ID NO: 27) (target 4), 563-582 (ttggccaagtcattcaattt; SEQ ID NO: 28) (target 2) and complementary strand sequence of 767-786 (ataacggaatccaactgggc; SEQ ID NO: 29) (target 5r) were used. For simultaneous expression of a plurality of targets, using a sequence from a promoter to a terminator as one set, and a plurality of the sets were incorporated into the same plasmid. They were introduced into cells along with Cas9-deaminase expression plasmid, intracellularly expressed, and a complex of gRNA-tracrRNA and Cas9-deaminase was formed.
Example 1: Modification of Genome Sequence by Linking DNA Sequence Recognition Ability of CRISPR-Cas to Deaminase PmCDA1
[0158] To test the effect of genome sequence modification technique of the present invention by utilizing deaminase and CRISPR-Cas nucleic acid sequence recognition ability, introduction of mutation into CAN1 gene encoding canavanine transporter, whose gene deficit results in canavanine-resistance, was attempted. As gRNA, a sequence complementary to 187-206 (target 1) of CAN1 gene ORF was used, a chimeric RNA expression vector obtained by linking thereto tracrRNA from Streptococcus pyogenes, and a vector expressing a protein obtained by fusing dCas9 with impaired nuclease activity by introducing mutations (D10A and H840A) into Cas9 (SpCas9) from Streptococcus pyogenes with PmCDA1 from Petromyzon marinus as deaminase were constructed, introduced into the budding yeast by the lithium acetate method, and coexpressed. The results are shown in FIG. 2. When cultured on a canavanine-containing SD plate, only the cells subjected to introduction and expression of gRNA-tracrRNA and dCas9-PmCDA1 formed colony. The resistant colony was picked up and the sequence of CAN1 gene region was determined. As a result, it was confirmed that a mutation was introduced into the target nucleotide sequence (target 1) and the vicinity thereof.
Example 2: Drastic Reduction of side Effects Toxicity
[0159] In conventional Cas9 and other artificial nucleases (ZFN, TALEN), inhibition of growth and cell death assumedly caused by disordered cleavage of chromosome occur by targeting a sequence in the genome. The effect thereof is particularly fatal for many microorganisms and prokaryotes, and prevents applicability.
[0160] Therefore, to verify the safety and cell toxicity of the genome sequence modification technique of the present invention, a comparative test with conventional CRISPR-Cas9 was performed. Using sequences (targets 3, 4) in the CAN1 gene as gRNA target, the surviving cells were counted immediately after the start of expression induction by galactose and at 6 hr after the induction based on the colony forming ability on an SD plate. The results are shown in FIG. 3. In conventional Cas9, the growth was inhibited and cell death was induced, which decreased the number of surviving cells. In contrast, by the present technique (nCas9 D10A-PmCDA1), the cells could continue to grow, and the number of surviving cells drastically increased.
Example 3: Use of Different Linking Scheme
[0161] Whether mutation can be introduced into a targeted gene even when Cas9 and deaminase are not used as a fusion protein but when a nucleic acid-modifying enzyme complex is formed via a binding domain and a ligand thereof was examined. As Cas9, dCas9 used in Example 1 was used and human AID instead of PmCDA1 was used as deaminase. SH3 domain was fused with the former, and a binding ligand thereof was fused with the latter to produce various constructs shown in FIG. 4. In addition, sequences (target 4,5r) in the CAN1 gene were used as gRNA targets. These constructs were introduced into a budding yeast. As a result, even when dCas9 and deaminase were linked via the binding domain, mutation was efficiently introduced into the targeted site of the CAN1 gene (FIG. 4). The mutation introduction efficiency was remarkably improved by introducing a plurality of binding domains into dCas9. The main site of mutation introduction was 782nd (g782c) of ORF.
Example 4: High Efficiency and Changes in Mutation Pattern by Use of Nickase
[0162] In the same manner as in Example 1 except that D10A mutant nCas9 (D10A) that cleaves only a strand complementary to gRNA, or H840A mutant nCas9 (H840A) that cleaves only a strand opposite to a strand complementary to gRNA was used instead of dCas9, mutation was introduced into the CAN1 gene, and the sequence in the CAN1 gene region of the colony generated on a canavanine-containing SD plate was examined. It was found that the efficiency increases in the former (nCas9 (D10A)) as compared to dCas9 (FIG. 5), and the mutation gathers in the center of the target sequence (FIG. 6). Therefore, this method enables pinpoint introduction of mutation. On the other hand, it was found in the latter (nCas9 (H840A)) that a plurality of random mutations were introduced into a region of several hundred bases from the targeted nucleotide (FIG. 6) along with an increase in the efficiency as compared to dCas9 (FIG. 5).
[0163] Similar remarkable introduction of mutation could be confirmed even when the target nucleotide sequence was changed. In this genome editing system using CRISPR-Cas9 system and cytidine deaminase, it was confirmed as shown in Table 1 that cytosine present within the range of about 2-5 bp from the 5'-side of the target nucleotide sequence (20 bp) were preferentially deaminated. Therefore, by setting the target nucleotide sequence based on this regularity and further combining with nCas9 (D10A), precise genome editing of 1 nucleotide unit is possible. On the other hand, a plurality of mutations can be simultaneously inserted within the range of about several hundred bp in the vicinity of the target nucleotide sequence by using nCas9 (H840A). Furthermore, site specificity may possibly be further varied by changing the linking scheme of deaminase.
[0164] These results show that the kind of Cas9 protein can be changed properly according to the object.
TABLE-US-00001 TABLE 1 site of main position of CAN1 sequence mutation gene ORF (SEQ ID NO:) introduction 187-206 (target 1) Gatacgttctcta c191a, g226t tggagga (25) 563-582 (target 2) Ttggccaagtcat cc567at, tcaattt (28) c567del 786-805 (target 3) Ttggagaaaccca cc795tt, and ggtgcct (26) cc796tt 793-812 (target 4) Aacccaggtgcct ggggtcc (27) 767-786 (comple- Ataacggaatcca g782c mentary strand) actgggc (29) (target 5r)
Example 5: Efficiency Increases Synergistically by Targeting a Plurality of DNA Sequences in Proximity
[0165] Efficiency drastically increased by simultaneously using a plurality of targets in proximity rather than a single target (FIG. 7). In fact, 10-20% of cells had canavanine-resistant mutation (targets 3, 4). In the Figure, gRNA1 and gRNA2 target target 3 and target 4, respectively. As deaminase, PmCDA1 was used. The effect thereof was confirmed to occur not only when the sequences partly overlapped (targets 3, 4), but also when they were apart by about 600 bp (targets 1, 3). The effect was found both when the DNA sequences were in the same direction (targets 3, 4) and opposing (targets 4, 5) (FIG. 4).
Example 6: Genetic Modification not Requiring Selection Marker
[0166] As for the cells (Targets 3, 4) in which target 3 and target 4 were targeted in Example 5, 10 colonies were randomly selected from those grown on a non-selected (canavanine-free) plate (SD plate not containing Leu and Ura) and the sequences of the CAN1 gene region were determined. As a result, mutation was introduced into the targeted site of the CAN1 gene in all examined colonies (FIG. 8). That is, editing can be expected in almost all expressed cells by selecting a suitable target sequence according to the present invention. Therefore, insertion of a marker gene and selection, which are essential for the conventional gene manipulation, are not necessary. This dramatically facilitates and simplifies gene manipulation and extends the applicability to crop breeding and the like since a recombinant organism with foreign DNA is not produced.
[0167] In the following Examples, experiment techniques shared by Examples 1-6 were performed in the same manner as above.
Example 7: Simultaneous Editing of a Plurality of Sites (Different Gene)
[0168] In a general gene manipulation method, mutation of only one site is generally achieved by one operation due to various restrictions. Thus, whether a simultaneous mutation operation of a plurality of sites is possible using the method of the present invention was tested.
[0169] Using the ORF of positions 3 to 22 of Ade1 gene of budding yeast BY4741 strain as the first target nucleotide sequence (Ade1 target 5:GTCAATTACGAAGACTGAAC; SEQ ID NO: 30), and the ORF of positions 767-786 (complementary strand) of Can1 gene as the second target nucleotide sequence (Can1 target8 (786-767; ATAACGGAATCCAACTGGGC; SEQ ID NO: 29), both DNAs encoding chimeric RNAs of two kinds of gRNAs each containing a nucleotide sequence complementary thereto and tracrRNA (SEQ ID NO: 7) were placed on the same plasmid (pRS426), and introduced into BY4741 strain together with plasmid nCas9 D10A-PmCDA1 containing a nucleic acid encoding a fusion protein of mutant Cas9 and PmCDA1, and expressed, and introduction of mutation into the both genes was verified. The cells were cultured in an SD drop-out medium (uracil and leucine deficient; SD-UL) as a base, which maintains plasmid. The cells were appropriately diluted, applied on SD-UL and canavaine addition medium and allowed to form a colony. After 2 days of culture at 28.degree. C., colonies were observed, and the incidence of red colony due to Ade1 mutation, and the survival rate in a canavanine medium were respectively counted. The results are shown in Table 2.
TABLE-US-00002 TABLE 2 survival rate in incidence of red Canavanine red colony and medium colony medium (Can) Can survival rate SD-UL 0.54 .+-. 0.04 +canavanine 0.64 .+-. 0.14 0.51 .+-. 0.15 0.31 .+-. 0.04
[0170] As a phenotype, the proportion of introduction of mutation into both Ade1 gene and Can1 gene was high and about 31%.
[0171] Then, a colony on an SD-UL medium was subjected to PCR amplification followed by sequencing. Regions containing ORF of each of Ade1 and Can1 were amplified, and sequence information of about 500 b sequences surrounding the target sequence was obtained. To be specific, 5 red colonies and 5 white colonies were analyzed to find conversion of the 5th C of Ade1 gene ORF to G in all red colonies and the 5th C to T in all white colonies (FIG. 9). While the mutation rate of the target is 100%, as the mutation rate in light of the object of gene destruction, the desired mutation rate is 50% since the 5th C needs to be changed to G to be a stop codon. Similarly, as for the Can1 gene, mutation was confirmed in the 782nd G of ORF in all clones (FIG. 9); however, since only the mutation to C affords canavanine-resistance, the desired mutation rate is 70%. Desired mutations in both genes were simultaneously obtained in 40% clones (4 clones out of 10 clones) by investigation, and practically high efficiency was obtained.
Example 8: Editing of Polyploid Genome
[0172] Many organisms have diploid or polyploid genome. In the conventional mutation introduction methods, mutation is, in principle, introduced into only one homologous chromosome to produce a heterologous geno-type. Therefore, desired feature is not obtained unless it is a dominant mutation, and making it homologous requires labor and time. Thus, whether the technique of the present invention can introduce mutation into all target alleles on the homologous chromosome in the genome was tested.
[0173] That is, simultaneous editing of Ade1 and Can1 genes was performed in budding yeast YPH501 strain as a diploid strain. The phenotype of these gene mutations (red colony and canavanine-resistant) is a recessive phenotype, and therefore, these phenotypes do not appear unless both mutations of homologous gene (homologous mutation) are introduced.
[0174] Using the ORF of positions 1173-1154 (complementary strand) of Ade1 gene (Ade1 target 1: GTCAATAGGATCCCCTTTT; SEQ ID NO: 31) or of positions 3-22 (Ade1 target 5: GTCAATTACGAAGACTGAAC; SEQ ID NO: 30) as the first target nucleotide sequence, and the ORF of positions 767-786 (complementary strand) of Can1 gene as the second target nucleotide sequence (Can1 target8: ATAACGGAATCCAACTGGGC; SEQ ID NO: 29), both DNAs encoding chimeric RNAs of two kinds of gRNAs each containing a nucleotide sequence complementary thereto and tracrRNA (SEQ ID NO: 7) were placed on the same plasmid (pRS426), and introduced into BY4741 strain together with plasmid nCas9 D10A-PmCDA1 containing a nucleic acid encoding a fusion protein of mutant Cas9 and PmCDA1, and expressed, and introduction of mutation into each gene was verified.
[0175] As a result of colony count, it was found that each characteristic of phenotype could be obtained at a high probability (40% - 70%) (FIG. 10A).
[0176] To confirm mutation, Ade1 target region of each of white colony and red colony was sequenced to confirm overlapping of sequence signals indicating heterologous mutation in the target site of white colony (FIG. 10B, upper panel, G and T signals overlap at .dwnarw.). Phenotype was confirmed to be absent in colony with heterologous mutation. On the other hand, homologous mutation free of overlapping signal was confirmed in red colony (FIG. 10B, lower panel, T signal at .dwnarw.).
Example 9: Genome Editing in Escherichia coli
[0177] In this Example, it is demonstrated that this technique effectively functions in Escherichia coli, which is a representative bacterium model organism. Particularly, conventional nuclease type genome editing technique is fatal for bacteria, and the application is difficult. Thus, the superiority of this technique is emphasized. In combination with yeast, which is an eukaryote model cell, it is shown that this technique is widely applicable to any species irrespective of prokaryon and eukaryon.
[0178] Amino acid mutation of DlOA and H840A were introduced (dCas9) into Streptococcus pyogenes Cas9 gene containing bidirectional promoter region, and a construct to be expressed as a fusion protein with PmCDA1 via a linker sequence was constructed, and chimeric gRNAs encoding a sequence complementary to each of the target nucleotide sequences was simultaneously included in a plasmid (full-length nucleotide sequence is shown in SEQ ID NO: 32, in which sequence, a sequence complementary to each of the target sequences is introduced into the site of n.sub.20) (FIG. 11A).
[0179] First, the ORF of positions 426-445 (T CAA TGG GCT AAC TAC GTT C; SEQ ID NO: 33) of Escherichia coli galK gene was introduced as a target nucleotide sequence into a plasmid, various Escherichia coli strains (XL10-gold, DH5a, MG1655, BW25113) were transformed with the plasmid by calcium method or electroporation method, SOC medium was added, recovery culture was performed overnight, plasmid carrying cells were selected from ampicillin-containing LB medium, and colony was formed. Introduction of mutation was verified by direct-sequence from colony PCR. The results are shown in FIG. 11B.
[0180] Independent colony (1-3) was selected randomly, and sequence was analyzed. As a result, the 427-position C of ORF was converted to T (clones 2, 3) at a probability of not less than 60%, and the occurrence of gene destruction generating a stop codon (TAA) was confirmed.
[0181] Then, with a complementary sequence (5'-GGTCCATAAACTGAGACAGC-3'; SEQ ID NO: 34) of 1530-1549 base region of rpoB gene ORF, which is an essential gene, as a target, particular point mutation was introduced by a method similar to the above-mentioned method to try to impart rifampicin-resistant function to Escherichia coli. The sequences of colonies selected in a nonselective medium (none), a 25 .mu.g/ml rifampicin (Rif25) and 50 .mu.g/ml rifampicin (Rif50)-containing medium were analyzed. As a result, it was confirmed that conversion of the 1546-position G of ORF to A introduced amino acid mutation from Asp(GAC) to Asn(AAC), and rifampicin-resistance was imparted (FIG. 11C, upper panel). A 10-fold dilution series of the cell suspension after transformation treatment was spotted on a nonselective medium (none), a 25 .mu.g/ml rifampicin (Rif25) and 50 .mu.g/ml rifampicin (Rif50)-containing medium and cultured. As a result, it is estimated that rifampicin-resistant strain was obtained at about 10% frequency (FIG. 11C, lower panel).
[0182] As shown above, by this technique, a new function can be added by particular point mutation, rather than simple gene destruction. This technique is superior since essential gene is directly edited.
Example 10: Adjustment of editing base site by gRNA length
[0183] Conventionally, the gRNA length relative to a target nucleotide sequence was 20b as basic, and cytosine (or guanine in opposite strand) in a site of 2-5b from the 5'-terminus thereof (15-19b upstream of PAM sequence) is used as a mutation target. Whether expression of different gRNA length can shift the site of the base to be the target was examined (FIG. 12A).
[0184] Experimental Example performed on Escherichia coli is shown in FIG. 12B. A site containing many cytosines on Escherichia coli genome was searched for, and the experiment was performed using gsiA gene, which is a putative ABC-transporter. Substituted cytosine was examined while changing the length of the target to 24 bp, 22 bp, 20 bp, 18 bp to find that the 898th, 899th cytosine was substituted by thymine in the case of 20 bp (standard length). When the target site is longer than 20 bp, the 896th and 897th cytosines were also substituted, and when the target site was shorter, the 900th and 901st cytosines were also substituted. In fact, the target site could be shifted by changing the length of the gRNA.
Example 11: Development of Temperature Dependent Genome Editing Plasmid
[0185] A plasmid that induces expression of the nucleic acid-modifying enzyme complex of the present invention under high temperature conditions was designed. While optimizing efficiency by limitatively controlling the expression state, reduction of side effects (growth inhibition of host, unstable mutation introduction efficiency, mutation of site different from target and the like) was aimed. Simultaneously, a simultaneous and easy removal of plasmid after editing was intended by combining a mechanism for ceasing the replication of plasmid at a high temperature. The detail of the experiment is shown below.
[0186] With temperature sensitive plasmid pSC101-Rep101 system (sequence of pSC101 ori is shown in SEQ ID NO: 35, and sequence of temperature sensitive Rep101 is shown in SEQ ID NO: 36) as a backbone, temperature sensitive A repressor (c1857) system was used for expression induction. For genome editing, G113E mutation imparting RecA resistance was introduced into A repressor, to ensure normal function even under SOS response (SEQ ID NO: 37). dCas9-PmCDA1 (SEQ ID NO: 38) was ligated to Right Operator (SEQ ID NO: 39), and gRNA (SEQ ID NO: 40) was ligated to the downstream of Left Operator (SEQ ID NO: 41) to regulate the expression (full-length nucleotide sequence of the constructed expression vector is shown in SEQ ID NO: 42). During culture at not more than 30.degree. C., transcription of gRNA and expression of dCas9-PmCDA1 are suppressed, and the cells can grow normally. When cultured at not less than 37.degree. C., transcription of gRNA and expression of dCas9-PmCDA1 are induced, and replication of plasmid is suppressed simultaneously. Therefore, a nucleic acid-modifying enzyme complex necessary for genome editing is transiently supplied, and plasmid can be removed easily after editing (FIG. 13).
[0187] Specific protocol of the base substitution is shown in FIG. 14.
[0188] The culture temperature for plasmid construction is set at around 28.degree. C., and an Escherichia coli colony retaining the desired plasmid is first established. Then, the colony is directly used, or after plasmid extraction when the strain is changed, transformation with the target strain is performed again, and the obtained colony is used. Liquiud culture at 28.degree. C. is performed overnight. Thereafter, the colony is diluted with the medium, induction culture is performed at 42.degree. C. for 1 hr to overnight, the cell suspension is appropriately diluted and spread or spotted on a plate to acquire a single colony.
[0189] As a verification experiment, point mutation introduction into essential gene rpoB was performed. When rpoB, which is one of the RNA polymerase-constituting factors, is deleted or its function is lost, the Escherichia coli will not survive. On the other hand, it is known that resistance to antibiotic rifampicin (Rif) is acquired when point mutation is entered at a particular site. Therefore, aiming at such introduction of point mutation, a target site is selected and assay was performed.
[0190] The results are shown in FIG. 15. In the upper left panel, the left shows an LB (chloramphenicol addition) plate, and the right shows a rifampicin-added LB (chloramphenicol addition) plate, and samples with or without chloramphenicol were prepared and cultured at 28.degree. C. or 42.degree. C. When cultured at 28.degree. C., the rate of Rif resistance is low; however, when cultured at 42.degree. C., rifampicin resistance was obtained with extremely high efficiency. When the colonies (non-selection) obtained on LB were sequenced by 8 colonies, the 1546th guanine (G) was substituted by adenine (A) in not less than 60% of the strain cultured at 42.degree. C. (lower and upper left panels). It is clear that the base is also completely substituted in actual sequence spectrum (lower right panel).
[0191] Similarly, base substitution of galK, which is one of the factors involved in the galactose metabolism, was performed. Since metabolism of 2-deoxy-galactose (2DOG), which is an analogue of galactose, by galK is fatal to Escherichia coli, this was used as a selection method. Target site was set such that missense mutation is induced in target 8, and that stop codon is entered in target 12 (FIG. 16 lower right).
[0192] The results are shown in FIG. 16. In the upper left and lower left panels, the left shows an LB (chloramphenicol addition) plate, and the right shows a 2-DOG-added LB (chloramphenicol addition) plate, and samples with or without chloramphenicol were prepared and cultured at 28.degree. C. or 42.degree. C. In target 8, colony was produced only slightly on a 2-DOG addition plate (upper left panel), 3 colonies on LB (red frame) were sequenced to determine that the 61st cytosine (C) was substituted by thymine (T) in all colonies (upper right). This mutation is assumed to be insufficient to lose function of galK. On the other hand, in target 12, colony was obtained on 2-DOG addition plate by culture at 28.degree. C. and 42.degree. C. (lower left panel). 3 colonies on LB were sequenced to determine that the 271st cytosine was substituted by thymine in all colonies (lower right). It was shown that mutation can be also introduced stably and highly efficiently in such different targets.
[0193] The contents disclosed in any publication cited herein, including patents and patent applications, are hereby incorporated in their entireties by reference, to the extent that they have been disclosed herein.
[0194] This application is based on patent application Nos. 2014-43348 and 2014-201859 filed in Japan (filing dates: Mar. 5, 2014 and Sep. 30, 2014, respectively), the contents of which are incorporated in full herein.
INDUSTRIAL APPLICABILITY
[0195] The present invention makes it possible to safely introduce site specific mutation into any species without insertion of a foreign DNA or double-stranded DNA breaks. It is also possible to set a wide range of mutation introduction from a pin point of one base to several hundred bases, and the technique can also be applied to topical evolution induction by introduction of random mutation into a particular restricted region, which has been almost impossible heretofore, and is extremely useful.
Sequence CWU 1
1
951624DNAPetromyzon marinusCDS(1)..(624) 1atg acc gac gct gag tac
gtg aga atc cat gag aag ttg gac atc tac 48Met Thr Asp Ala Glu Tyr
Val Arg Ile His Glu Lys Leu Asp Ile Tyr1 5 10 15acg ttt aag aaa cag
ttt ttc aac aac aaa aaa tcc gtg tcg cat aga 96Thr Phe Lys Lys Gln
Phe Phe Asn Asn Lys Lys Ser Val Ser His Arg 20 25 30tgc tac gtt ctc
ttt gaa tta aaa cga cgg ggt gaa cgt aga gcg tgt 144Cys Tyr Val Leu
Phe Glu Leu Lys Arg Arg Gly Glu Arg Arg Ala Cys 35 40 45ttt tgg ggc
tat gct gtg aat aaa cca cag agc ggg aca gaa cgt ggc 192Phe Trp Gly
Tyr Ala Val Asn Lys Pro Gln Ser Gly Thr Glu Arg Gly 50 55 60att cac
gcc gaa atc ttt agc att aga aaa gtc gaa gaa tac ctg cgc 240Ile His
Ala Glu Ile Phe Ser Ile Arg Lys Val Glu Glu Tyr Leu Arg65 70 75
80gac aac ccc gga caa ttc acg ata aat tgg tac tca tcc tgg agt cct
288Asp Asn Pro Gly Gln Phe Thr Ile Asn Trp Tyr Ser Ser Trp Ser Pro
85 90 95tgt gca gat tgc gct gaa aag atc tta gaa tgg tat aac cag gag
ctg 336Cys Ala Asp Cys Ala Glu Lys Ile Leu Glu Trp Tyr Asn Gln Glu
Leu 100 105 110cgg ggg aac ggc cac act ttg aaa atc tgg gct tgc aaa
ctc tat tac 384Arg Gly Asn Gly His Thr Leu Lys Ile Trp Ala Cys Lys
Leu Tyr Tyr 115 120 125gag aaa aat gcg agg aat caa att ggg ctg tgg
aac ctc aga gat aac 432Glu Lys Asn Ala Arg Asn Gln Ile Gly Leu Trp
Asn Leu Arg Asp Asn 130 135 140ggg gtt ggg ttg aat gta atg gta agt
gaa cac tac caa tgt tgc agg 480Gly Val Gly Leu Asn Val Met Val Ser
Glu His Tyr Gln Cys Cys Arg145 150 155 160aaa ata ttc atc caa tcg
tcg cac aat caa ttg aat gag aat aga tgg 528Lys Ile Phe Ile Gln Ser
Ser His Asn Gln Leu Asn Glu Asn Arg Trp 165 170 175ctt gag aag act
ttg aag cga gct gaa aaa cga cgg agc gag ttg tcc 576Leu Glu Lys Thr
Leu Lys Arg Ala Glu Lys Arg Arg Ser Glu Leu Ser 180 185 190att atg
att cag gta aaa ata ctc cac acc act aag agt cct gct gtt 624Ile Met
Ile Gln Val Lys Ile Leu His Thr Thr Lys Ser Pro Ala Val 195 200
2052208PRTPetromyzon marinus 2Met Thr Asp Ala Glu Tyr Val Arg Ile
His Glu Lys Leu Asp Ile Tyr1 5 10 15Thr Phe Lys Lys Gln Phe Phe Asn
Asn Lys Lys Ser Val Ser His Arg 20 25 30Cys Tyr Val Leu Phe Glu Leu
Lys Arg Arg Gly Glu Arg Arg Ala Cys 35 40 45Phe Trp Gly Tyr Ala Val
Asn Lys Pro Gln Ser Gly Thr Glu Arg Gly 50 55 60Ile His Ala Glu Ile
Phe Ser Ile Arg Lys Val Glu Glu Tyr Leu Arg65 70 75 80Asp Asn Pro
Gly Gln Phe Thr Ile Asn Trp Tyr Ser Ser Trp Ser Pro 85 90 95Cys Ala
Asp Cys Ala Glu Lys Ile Leu Glu Trp Tyr Asn Gln Glu Leu 100 105
110Arg Gly Asn Gly His Thr Leu Lys Ile Trp Ala Cys Lys Leu Tyr Tyr
115 120 125Glu Lys Asn Ala Arg Asn Gln Ile Gly Leu Trp Asn Leu Arg
Asp Asn 130 135 140Gly Val Gly Leu Asn Val Met Val Ser Glu His Tyr
Gln Cys Cys Arg145 150 155 160Lys Ile Phe Ile Gln Ser Ser His Asn
Gln Leu Asn Glu Asn Arg Trp 165 170 175Leu Glu Lys Thr Leu Lys Arg
Ala Glu Lys Arg Arg Ser Glu Leu Ser 180 185 190Ile Met Ile Gln Val
Lys Ile Leu His Thr Thr Lys Ser Pro Ala Val 195 200 2053600DNAHomo
sapiensCDS(1)..(600) 3atg gac agc ctc ttg atg aac cgg agg aag ttt
ctt tac caa ttc aaa 48Met Asp Ser Leu Leu Met Asn Arg Arg Lys Phe
Leu Tyr Gln Phe Lys1 5 10 15aat gtc cgc tgg gct aag ggt cgg cgt gag
acc tac ctg tgc tac gta 96Asn Val Arg Trp Ala Lys Gly Arg Arg Glu
Thr Tyr Leu Cys Tyr Val 20 25 30gtg aag agg cgt gac agt gct aca tcc
ttt tca ctg gac ttt ggt tat 144Val Lys Arg Arg Asp Ser Ala Thr Ser
Phe Ser Leu Asp Phe Gly Tyr 35 40 45ctt cgc aat aag aac ggc tgc cac
gtg gaa ttg ctc ttc ctc cgc tac 192Leu Arg Asn Lys Asn Gly Cys His
Val Glu Leu Leu Phe Leu Arg Tyr 50 55 60atc tcg gac tgg gac cta gac
cct ggc cgc tgc tac cgc gtc acc tgg 240Ile Ser Asp Trp Asp Leu Asp
Pro Gly Arg Cys Tyr Arg Val Thr Trp65 70 75 80ttc acc tcc tgg agc
ccc tgc tac gac tgt gcc cga cat gtg gcc gac 288Phe Thr Ser Trp Ser
Pro Cys Tyr Asp Cys Ala Arg His Val Ala Asp 85 90 95ttt ctg cga ggg
aac ccc tac ctc agt ctg agg atc ttc acc gcg cgc 336Phe Leu Arg Gly
Asn Pro Tyr Leu Ser Leu Arg Ile Phe Thr Ala Arg 100 105 110ctc tac
ttc tgt gag gac cgc aag gct gag ccc gag ggg ctg cgg cgg 384Leu Tyr
Phe Cys Glu Asp Arg Lys Ala Glu Pro Glu Gly Leu Arg Arg 115 120
125ctg cac cgc gcc ggg gtg caa ata gcc atc atg acc ttc aaa gat tat
432Leu His Arg Ala Gly Val Gln Ile Ala Ile Met Thr Phe Lys Asp Tyr
130 135 140ttt tac tgc tgg aat act ttt gta gaa aac cat gaa aga act
ttc aaa 480Phe Tyr Cys Trp Asn Thr Phe Val Glu Asn His Glu Arg Thr
Phe Lys145 150 155 160gcc tgg gaa ggg ctg cat gaa aat tca gtt cgt
ctc tcc aga cag ctt 528Ala Trp Glu Gly Leu His Glu Asn Ser Val Arg
Leu Ser Arg Gln Leu 165 170 175cgg cgc atc ctt ttg ccc ctg tat gag
gtt gat gac tta cga gac gca 576Arg Arg Ile Leu Leu Pro Leu Tyr Glu
Val Asp Asp Leu Arg Asp Ala 180 185 190ttt cgt act ttg gga ctt ctc
gac 600Phe Arg Thr Leu Gly Leu Leu Asp 195 2004200PRTHomo sapiens
4Met Asp Ser Leu Leu Met Asn Arg Arg Lys Phe Leu Tyr Gln Phe Lys1 5
10 15Asn Val Arg Trp Ala Lys Gly Arg Arg Glu Thr Tyr Leu Cys Tyr
Val 20 25 30Val Lys Arg Arg Asp Ser Ala Thr Ser Phe Ser Leu Asp Phe
Gly Tyr 35 40 45Leu Arg Asn Lys Asn Gly Cys His Val Glu Leu Leu Phe
Leu Arg Tyr 50 55 60Ile Ser Asp Trp Asp Leu Asp Pro Gly Arg Cys Tyr
Arg Val Thr Trp65 70 75 80Phe Thr Ser Trp Ser Pro Cys Tyr Asp Cys
Ala Arg His Val Ala Asp 85 90 95Phe Leu Arg Gly Asn Pro Tyr Leu Ser
Leu Arg Ile Phe Thr Ala Arg 100 105 110Leu Tyr Phe Cys Glu Asp Arg
Lys Ala Glu Pro Glu Gly Leu Arg Arg 115 120 125Leu His Arg Ala Gly
Val Gln Ile Ala Ile Met Thr Phe Lys Asp Tyr 130 135 140Phe Tyr Cys
Trp Asn Thr Phe Val Glu Asn His Glu Arg Thr Phe Lys145 150 155
160Ala Trp Glu Gly Leu His Glu Asn Ser Val Arg Leu Ser Arg Gln Leu
165 170 175Arg Arg Ile Leu Leu Pro Leu Tyr Glu Val Asp Asp Leu Arg
Asp Ala 180 185 190Phe Arg Thr Leu Gly Leu Leu Asp 195
20054116DNAArtificial SequenceStreptococcus pyogenes-derived Cas9
CDS optimized for eucaryotic expression.CDS(1)..(4116) 5atg gac aag
aag tac tcc att ggg ctc gat atc ggc aca aac agc gtc 48Met Asp Lys
Lys Tyr Ser Ile Gly Leu Asp Ile Gly Thr Asn Ser Val1 5 10 15ggt tgg
gcc gtc att acg gac gag tac aag gtg ccg agc aaa aaa ttc 96Gly Trp
Ala Val Ile Thr Asp Glu Tyr Lys Val Pro Ser Lys Lys Phe 20 25 30aaa
gtt ctg ggc aat acc gat cgc cac agc ata aag aag aac ctc att 144Lys
Val Leu Gly Asn Thr Asp Arg His Ser Ile Lys Lys Asn Leu Ile 35 40
45ggc gcc ctc ctg ttc gac tcc ggg gag acg gcc gaa gcc acg cgg ctc
192Gly Ala Leu Leu Phe Asp Ser Gly Glu Thr Ala Glu Ala Thr Arg Leu
50 55 60aaa aga aca gca cgg cgc aga tat acc cgc aga aag aat cgg atc
tgc 240Lys Arg Thr Ala Arg Arg Arg Tyr Thr Arg Arg Lys Asn Arg Ile
Cys65 70 75 80tac ctg cag gag atc ttt agt aat gag atg gct aag gtg
gat gac tct 288Tyr Leu Gln Glu Ile Phe Ser Asn Glu Met Ala Lys Val
Asp Asp Ser 85 90 95ttc ttc cat agg ctg gag gag tcc ttt ttg gtg gag
gag gat aaa aag 336Phe Phe His Arg Leu Glu Glu Ser Phe Leu Val Glu
Glu Asp Lys Lys 100 105 110cac gag cgc cac cca atc ttt ggc aat atc
gtg gac gag gtg gcg tac 384His Glu Arg His Pro Ile Phe Gly Asn Ile
Val Asp Glu Val Ala Tyr 115 120 125cat gaa aag tac cca acc ata tat
cat ctg agg aag aag ctt gta gac 432His Glu Lys Tyr Pro Thr Ile Tyr
His Leu Arg Lys Lys Leu Val Asp 130 135 140agt act gat aag gct gac
ttg cgg ttg atc tat ctc gcg ctg gcg cat 480Ser Thr Asp Lys Ala Asp
Leu Arg Leu Ile Tyr Leu Ala Leu Ala His145 150 155 160atg atc aaa
ttt cgg gga cac ttc ctc atc gag ggg gac ctg aac cca 528Met Ile Lys
Phe Arg Gly His Phe Leu Ile Glu Gly Asp Leu Asn Pro 165 170 175gac
aac agc gat gtc gac aaa ctc ttt atc caa ctg gtt cag act tac 576Asp
Asn Ser Asp Val Asp Lys Leu Phe Ile Gln Leu Val Gln Thr Tyr 180 185
190aat cag ctt ttc gaa gag aac ccg atc aac gca tcc gga gtt gac gcc
624Asn Gln Leu Phe Glu Glu Asn Pro Ile Asn Ala Ser Gly Val Asp Ala
195 200 205aaa gca atc ctg agc gct agg ctg tcc aaa tcc cgg cgg ctc
gaa aac 672Lys Ala Ile Leu Ser Ala Arg Leu Ser Lys Ser Arg Arg Leu
Glu Asn 210 215 220ctc atc gca cag ctc cct ggg gag aag aag aac ggc
ctg ttt ggt aat 720Leu Ile Ala Gln Leu Pro Gly Glu Lys Lys Asn Gly
Leu Phe Gly Asn225 230 235 240ctt atc gcc ctg tca ctc ggg ctg acc
ccc aac ttt aaa tct aac ttc 768Leu Ile Ala Leu Ser Leu Gly Leu Thr
Pro Asn Phe Lys Ser Asn Phe 245 250 255gac ctg gcc gaa gat gcc aag
ctt caa ctg agc aaa gac acc tac gat 816Asp Leu Ala Glu Asp Ala Lys
Leu Gln Leu Ser Lys Asp Thr Tyr Asp 260 265 270gat gat ctc gac aat
ctg ctg gcc cag atc ggc gac cag tac gca gac 864Asp Asp Leu Asp Asn
Leu Leu Ala Gln Ile Gly Asp Gln Tyr Ala Asp 275 280 285ctt ttt ttg
gcg gca aag aac ctg tca gac gcc att ctg ctg agt gat 912Leu Phe Leu
Ala Ala Lys Asn Leu Ser Asp Ala Ile Leu Leu Ser Asp 290 295 300att
ctg cga gtg aac acg gag atc acc aaa gct ccg ctg agc gct agt 960Ile
Leu Arg Val Asn Thr Glu Ile Thr Lys Ala Pro Leu Ser Ala Ser305 310
315 320atg atc aag cgc tat gat gag cac cac caa gac ttg act ttg ctg
aag 1008Met Ile Lys Arg Tyr Asp Glu His His Gln Asp Leu Thr Leu Leu
Lys 325 330 335gcc ctt gtc aga cag caa ctg cct gag aag tac aag gaa
att ttc ttc 1056Ala Leu Val Arg Gln Gln Leu Pro Glu Lys Tyr Lys Glu
Ile Phe Phe 340 345 350gat cag tct aaa aat ggc tac gcc gga tac att
gac ggc gga gca agc 1104Asp Gln Ser Lys Asn Gly Tyr Ala Gly Tyr Ile
Asp Gly Gly Ala Ser 355 360 365cag gag gaa ttt tac aaa ttt att aag
ccc atc ttg gaa aaa atg gac 1152Gln Glu Glu Phe Tyr Lys Phe Ile Lys
Pro Ile Leu Glu Lys Met Asp 370 375 380ggc acc gag gag ctg ctg gta
aag ctt aac aga gaa gat ctg ttg cgc 1200Gly Thr Glu Glu Leu Leu Val
Lys Leu Asn Arg Glu Asp Leu Leu Arg385 390 395 400aaa cag cgc act
ttc gac aat gga agc atc ccc cac cag att cac ctg 1248Lys Gln Arg Thr
Phe Asp Asn Gly Ser Ile Pro His Gln Ile His Leu 405 410 415ggc gaa
ctg cac gct atc ctc agg cgg caa gag gat ttc tac ccc ttt 1296Gly Glu
Leu His Ala Ile Leu Arg Arg Gln Glu Asp Phe Tyr Pro Phe 420 425
430ttg aaa gat aac agg gaa aag att gag aaa atc ctc aca ttt cgg ata
1344Leu Lys Asp Asn Arg Glu Lys Ile Glu Lys Ile Leu Thr Phe Arg Ile
435 440 445ccc tac tat gta ggc ccc ctc gcc cgg gga aat tcc aga ttc
gcg tgg 1392Pro Tyr Tyr Val Gly Pro Leu Ala Arg Gly Asn Ser Arg Phe
Ala Trp 450 455 460atg act cgc aaa tca gaa gag acc atc act ccc tgg
aac ttc gag gaa 1440Met Thr Arg Lys Ser Glu Glu Thr Ile Thr Pro Trp
Asn Phe Glu Glu465 470 475 480gtc gtg gat aag ggg gcc tct gcc cag
tcc ttc atc gaa agg atg act 1488Val Val Asp Lys Gly Ala Ser Ala Gln
Ser Phe Ile Glu Arg Met Thr 485 490 495aac ttt gat aaa aat ctg cct
aac gaa aag gtg ctt cct aaa cac tct 1536Asn Phe Asp Lys Asn Leu Pro
Asn Glu Lys Val Leu Pro Lys His Ser 500 505 510ctg ctg tac gag tac
ttc aca gtt tat aac gag ctc acc aag gtc aaa 1584Leu Leu Tyr Glu Tyr
Phe Thr Val Tyr Asn Glu Leu Thr Lys Val Lys 515 520 525tac gtc aca
gaa ggg atg aga aag cca gca ttc ctg tct gga gag cag 1632Tyr Val Thr
Glu Gly Met Arg Lys Pro Ala Phe Leu Ser Gly Glu Gln 530 535 540aag
aaa gct atc gtg gac ctc ctc ttc aag acg aac cgg aaa gtt acc 1680Lys
Lys Ala Ile Val Asp Leu Leu Phe Lys Thr Asn Arg Lys Val Thr545 550
555 560gtg aaa cag ctc aaa gaa gac tat ttc aaa aag att gaa tgt ttc
gac 1728Val Lys Gln Leu Lys Glu Asp Tyr Phe Lys Lys Ile Glu Cys Phe
Asp 565 570 575tct gtt gaa atc agc gga gtg gag gat cgc ttc aac gca
tcc ctg gga 1776Ser Val Glu Ile Ser Gly Val Glu Asp Arg Phe Asn Ala
Ser Leu Gly 580 585 590acg tat cac gat ctc ctg aaa atc att aaa gac
aag gac ttc ctg gac 1824Thr Tyr His Asp Leu Leu Lys Ile Ile Lys Asp
Lys Asp Phe Leu Asp 595 600 605aat gag gag aac gag gac att ctt gag
gac att gtc ctc acc ctt acg 1872Asn Glu Glu Asn Glu Asp Ile Leu Glu
Asp Ile Val Leu Thr Leu Thr 610 615 620ttg ttt gaa gat agg gag atg
att gaa gaa cgc ttg aaa act tac gct 1920Leu Phe Glu Asp Arg Glu Met
Ile Glu Glu Arg Leu Lys Thr Tyr Ala625 630 635 640cat ctc ttc gac
gac aaa gtc atg aaa cag ctc aag agg cgc cga tat 1968His Leu Phe Asp
Asp Lys Val Met Lys Gln Leu Lys Arg Arg Arg Tyr 645 650 655aca gga
tgg ggg cgg ctg tca aga aaa ctg atc aat ggg atc cga gac 2016Thr Gly
Trp Gly Arg Leu Ser Arg Lys Leu Ile Asn Gly Ile Arg Asp 660 665
670aag cag agt gga aag aca atc ctg gat ttt ctt aag tcc gat gga ttt
2064Lys Gln Ser Gly Lys Thr Ile Leu Asp Phe Leu Lys Ser Asp Gly Phe
675 680 685gcc aac cgg aac ttc atg cag ttg atc cat gat gac tct ctc
acc ttt 2112Ala Asn Arg Asn Phe Met Gln Leu Ile His Asp Asp Ser Leu
Thr Phe 690 695 700aag gag gac atc cag aaa gca caa gtt tct ggc cag
ggg gac agt ctt 2160Lys Glu Asp Ile Gln Lys Ala Gln Val Ser Gly Gln
Gly Asp Ser Leu705 710 715 720cac gag cac atc gct aat ctt gca ggt
agc cca gct atc aaa aag gga 2208His Glu His Ile Ala Asn Leu Ala Gly
Ser Pro Ala Ile Lys Lys Gly 725 730 735ata ctg cag acc gtt aag gtc
gtg gat gaa ctc gtc aaa gta atg gga 2256Ile Leu Gln Thr Val Lys Val
Val Asp Glu Leu Val Lys Val Met Gly 740 745 750agg cat aag ccc gag
aat atc gtt atc gag atg gcc cga gag aac caa 2304Arg His Lys Pro Glu
Asn Ile Val Ile Glu Met Ala Arg Glu Asn Gln 755 760 765act acc cag
aag gga cag aag aac agt agg gaa agg atg aag agg att 2352Thr Thr Gln
Lys Gly Gln Lys Asn Ser Arg Glu Arg Met Lys Arg Ile 770 775 780gaa
gag ggt ata aaa gaa ctg ggg tcc caa atc ctt aag gaa cac cca 2400Glu
Glu Gly Ile Lys Glu Leu Gly Ser Gln Ile Leu Lys Glu His Pro785 790
795 800gtt gaa aac acc cag ctt cag aat gag aag ctc tac ctg tac tac
ctg 2448Val Glu Asn Thr Gln Leu Gln Asn Glu Lys Leu Tyr Leu Tyr Tyr
Leu 805 810 815cag aac ggc agg gac atg tac gtg gat cag gaa ctg gac
atc aat cgg 2496Gln Asn Gly Arg Asp Met Tyr Val Asp Gln Glu Leu Asp
Ile Asn Arg 820 825 830ctc tcc gac tac gac gtg gat cat atc gtg ccc
cag tct ttt ctc aaa 2544Leu Ser Asp Tyr Asp Val Asp His Ile Val Pro
Gln Ser Phe Leu Lys 835 840 845gat gat tct att gat aat aaa
gtg ttg aca aga tcc gat aaa aat aga 2592Asp Asp Ser Ile Asp Asn Lys
Val Leu Thr Arg Ser Asp Lys Asn Arg 850 855 860ggg aag agt gat aac
gtc ccc tca gaa gaa gtt gtc aag aaa atg aaa 2640Gly Lys Ser Asp Asn
Val Pro Ser Glu Glu Val Val Lys Lys Met Lys865 870 875 880aat tat
tgg cgg cag ctg ctg aac gcc aaa ctg atc aca caa cgg aag 2688Asn Tyr
Trp Arg Gln Leu Leu Asn Ala Lys Leu Ile Thr Gln Arg Lys 885 890
895ttc gat aat ctg act aag gct gaa cga ggt ggc ctg tct gag ttg gat
2736Phe Asp Asn Leu Thr Lys Ala Glu Arg Gly Gly Leu Ser Glu Leu Asp
900 905 910aaa gcc ggc ttc atc aaa agg cag ctt gtt gag aca cgc cag
atc acc 2784Lys Ala Gly Phe Ile Lys Arg Gln Leu Val Glu Thr Arg Gln
Ile Thr 915 920 925aag cac gtg gcc caa att ctc gat tca cgc atg aac
acc aag tac gat 2832Lys His Val Ala Gln Ile Leu Asp Ser Arg Met Asn
Thr Lys Tyr Asp 930 935 940gaa aat gac aaa ctg att cga gag gtg aaa
gtt att act ctg aag tct 2880Glu Asn Asp Lys Leu Ile Arg Glu Val Lys
Val Ile Thr Leu Lys Ser945 950 955 960aag ctg gtc tca gat ttc aga
aag gac ttt cag ttt tat aag gtg aga 2928Lys Leu Val Ser Asp Phe Arg
Lys Asp Phe Gln Phe Tyr Lys Val Arg 965 970 975gag atc aac aat tac
cac cat gcg cat gat gcc tac ctg aat gca gtg 2976Glu Ile Asn Asn Tyr
His His Ala His Asp Ala Tyr Leu Asn Ala Val 980 985 990gta ggc act
gca ctt atc aaa aaa tat ccc aag ctt gaa tct gaa ttt 3024Val Gly Thr
Ala Leu Ile Lys Lys Tyr Pro Lys Leu Glu Ser Glu Phe 995 1000
1005gtt tac gga gac tat aaa gtg tac gat gtt agg aaa atg atc gca
3069Val Tyr Gly Asp Tyr Lys Val Tyr Asp Val Arg Lys Met Ile Ala
1010 1015 1020aag tct gag cag gaa ata ggc aag gcc acc gct aag tac
ttc ttt 3114Lys Ser Glu Gln Glu Ile Gly Lys Ala Thr Ala Lys Tyr Phe
Phe 1025 1030 1035tac agc aat att atg aat ttt ttc aag acc gag att
aca ctg gcc 3159Tyr Ser Asn Ile Met Asn Phe Phe Lys Thr Glu Ile Thr
Leu Ala 1040 1045 1050aat gga gag att cgg aag cga cca ctt atc gaa
aca aac gga gaa 3204Asn Gly Glu Ile Arg Lys Arg Pro Leu Ile Glu Thr
Asn Gly Glu 1055 1060 1065aca gga gaa atc gtg tgg gac aag ggt agg
gat ttc gcg aca gtc 3249Thr Gly Glu Ile Val Trp Asp Lys Gly Arg Asp
Phe Ala Thr Val 1070 1075 1080cgg aag gtc ctg tcc atg ccg cag gtg
aac atc gtt aaa aag acc 3294Arg Lys Val Leu Ser Met Pro Gln Val Asn
Ile Val Lys Lys Thr 1085 1090 1095gaa gta cag acc gga ggc ttc tcc
aag gaa agt atc ctc ccg aaa 3339Glu Val Gln Thr Gly Gly Phe Ser Lys
Glu Ser Ile Leu Pro Lys 1100 1105 1110agg aac agc gac aag ctg atc
gca cgc aaa aaa gat tgg gac ccc 3384Arg Asn Ser Asp Lys Leu Ile Ala
Arg Lys Lys Asp Trp Asp Pro 1115 1120 1125aag aaa tac ggc gga ttc
gat tct cct aca gtc gct tac agt gta 3429Lys Lys Tyr Gly Gly Phe Asp
Ser Pro Thr Val Ala Tyr Ser Val 1130 1135 1140ctg gtt gtg gcc aaa
gtg gag aaa ggg aag tct aaa aaa ctc aaa 3474Leu Val Val Ala Lys Val
Glu Lys Gly Lys Ser Lys Lys Leu Lys 1145 1150 1155agc gtc aag gaa
ctg ctg ggc atc aca atc atg gag cga tca agc 3519Ser Val Lys Glu Leu
Leu Gly Ile Thr Ile Met Glu Arg Ser Ser 1160 1165 1170ttc gaa aaa
aac ccc atc gac ttt ctc gag gcg aaa gga tat aaa 3564Phe Glu Lys Asn
Pro Ile Asp Phe Leu Glu Ala Lys Gly Tyr Lys 1175 1180 1185gag gtc
aaa aaa gac ctc atc att aag ctt ccc aag tac tct ctc 3609Glu Val Lys
Lys Asp Leu Ile Ile Lys Leu Pro Lys Tyr Ser Leu 1190 1195 1200ttt
gag ctt gaa aac ggc cgg aaa cga atg ctc gct agt gcg ggc 3654Phe Glu
Leu Glu Asn Gly Arg Lys Arg Met Leu Ala Ser Ala Gly 1205 1210
1215gag ctg cag aaa ggt aac gag ctg gca ctg ccc tct aaa tac gtt
3699Glu Leu Gln Lys Gly Asn Glu Leu Ala Leu Pro Ser Lys Tyr Val
1220 1225 1230aat ttc ttg tat ctg gcc agc cac tat gaa aag ctc aaa
ggg tct 3744Asn Phe Leu Tyr Leu Ala Ser His Tyr Glu Lys Leu Lys Gly
Ser 1235 1240 1245ccc gaa gat aat gag cag aag cag ctg ttc gtg gaa
caa cac aaa 3789Pro Glu Asp Asn Glu Gln Lys Gln Leu Phe Val Glu Gln
His Lys 1250 1255 1260cac tac ctt gat gag atc atc gag caa ata agc
gaa ttc tcc aaa 3834His Tyr Leu Asp Glu Ile Ile Glu Gln Ile Ser Glu
Phe Ser Lys 1265 1270 1275aga gtg atc ctc gcc gac gct aac ctc gat
aag gtg ctt tct gct 3879Arg Val Ile Leu Ala Asp Ala Asn Leu Asp Lys
Val Leu Ser Ala 1280 1285 1290tac aat aag cac agg gat aag ccc atc
agg gag cag gca gaa aac 3924Tyr Asn Lys His Arg Asp Lys Pro Ile Arg
Glu Gln Ala Glu Asn 1295 1300 1305att atc cac ttg ttt act ctg acc
aac ttg ggc gcg cct gca gcc 3969Ile Ile His Leu Phe Thr Leu Thr Asn
Leu Gly Ala Pro Ala Ala 1310 1315 1320ttc aag tac ttc gac acc acc
ata gac aga aag cgg tac acc tct 4014Phe Lys Tyr Phe Asp Thr Thr Ile
Asp Arg Lys Arg Tyr Thr Ser 1325 1330 1335aca aag gag gtc ctg gac
gcc aca ctg att cat cag tca att acg 4059Thr Lys Glu Val Leu Asp Ala
Thr Leu Ile His Gln Ser Ile Thr 1340 1345 1350ggg ctc tat gaa aca
aga atc gac ctc tct cag ctc ggt gga gac 4104Gly Leu Tyr Glu Thr Arg
Ile Asp Leu Ser Gln Leu Gly Gly Asp 1355 1360 1365agc agg gct gac
4116Ser Arg Ala Asp 137061372PRTArtificial SequenceSynthetic
Construct 6Met Asp Lys Lys Tyr Ser Ile Gly Leu Asp Ile Gly Thr Asn
Ser Val1 5 10 15Gly Trp Ala Val Ile Thr Asp Glu Tyr Lys Val Pro Ser
Lys Lys Phe 20 25 30Lys Val Leu Gly Asn Thr Asp Arg His Ser Ile Lys
Lys Asn Leu Ile 35 40 45Gly Ala Leu Leu Phe Asp Ser Gly Glu Thr Ala
Glu Ala Thr Arg Leu 50 55 60Lys Arg Thr Ala Arg Arg Arg Tyr Thr Arg
Arg Lys Asn Arg Ile Cys65 70 75 80Tyr Leu Gln Glu Ile Phe Ser Asn
Glu Met Ala Lys Val Asp Asp Ser 85 90 95Phe Phe His Arg Leu Glu Glu
Ser Phe Leu Val Glu Glu Asp Lys Lys 100 105 110His Glu Arg His Pro
Ile Phe Gly Asn Ile Val Asp Glu Val Ala Tyr 115 120 125His Glu Lys
Tyr Pro Thr Ile Tyr His Leu Arg Lys Lys Leu Val Asp 130 135 140Ser
Thr Asp Lys Ala Asp Leu Arg Leu Ile Tyr Leu Ala Leu Ala His145 150
155 160Met Ile Lys Phe Arg Gly His Phe Leu Ile Glu Gly Asp Leu Asn
Pro 165 170 175Asp Asn Ser Asp Val Asp Lys Leu Phe Ile Gln Leu Val
Gln Thr Tyr 180 185 190Asn Gln Leu Phe Glu Glu Asn Pro Ile Asn Ala
Ser Gly Val Asp Ala 195 200 205Lys Ala Ile Leu Ser Ala Arg Leu Ser
Lys Ser Arg Arg Leu Glu Asn 210 215 220Leu Ile Ala Gln Leu Pro Gly
Glu Lys Lys Asn Gly Leu Phe Gly Asn225 230 235 240Leu Ile Ala Leu
Ser Leu Gly Leu Thr Pro Asn Phe Lys Ser Asn Phe 245 250 255Asp Leu
Ala Glu Asp Ala Lys Leu Gln Leu Ser Lys Asp Thr Tyr Asp 260 265
270Asp Asp Leu Asp Asn Leu Leu Ala Gln Ile Gly Asp Gln Tyr Ala Asp
275 280 285Leu Phe Leu Ala Ala Lys Asn Leu Ser Asp Ala Ile Leu Leu
Ser Asp 290 295 300Ile Leu Arg Val Asn Thr Glu Ile Thr Lys Ala Pro
Leu Ser Ala Ser305 310 315 320Met Ile Lys Arg Tyr Asp Glu His His
Gln Asp Leu Thr Leu Leu Lys 325 330 335Ala Leu Val Arg Gln Gln Leu
Pro Glu Lys Tyr Lys Glu Ile Phe Phe 340 345 350Asp Gln Ser Lys Asn
Gly Tyr Ala Gly Tyr Ile Asp Gly Gly Ala Ser 355 360 365Gln Glu Glu
Phe Tyr Lys Phe Ile Lys Pro Ile Leu Glu Lys Met Asp 370 375 380Gly
Thr Glu Glu Leu Leu Val Lys Leu Asn Arg Glu Asp Leu Leu Arg385 390
395 400Lys Gln Arg Thr Phe Asp Asn Gly Ser Ile Pro His Gln Ile His
Leu 405 410 415Gly Glu Leu His Ala Ile Leu Arg Arg Gln Glu Asp Phe
Tyr Pro Phe 420 425 430Leu Lys Asp Asn Arg Glu Lys Ile Glu Lys Ile
Leu Thr Phe Arg Ile 435 440 445Pro Tyr Tyr Val Gly Pro Leu Ala Arg
Gly Asn Ser Arg Phe Ala Trp 450 455 460Met Thr Arg Lys Ser Glu Glu
Thr Ile Thr Pro Trp Asn Phe Glu Glu465 470 475 480Val Val Asp Lys
Gly Ala Ser Ala Gln Ser Phe Ile Glu Arg Met Thr 485 490 495Asn Phe
Asp Lys Asn Leu Pro Asn Glu Lys Val Leu Pro Lys His Ser 500 505
510Leu Leu Tyr Glu Tyr Phe Thr Val Tyr Asn Glu Leu Thr Lys Val Lys
515 520 525Tyr Val Thr Glu Gly Met Arg Lys Pro Ala Phe Leu Ser Gly
Glu Gln 530 535 540Lys Lys Ala Ile Val Asp Leu Leu Phe Lys Thr Asn
Arg Lys Val Thr545 550 555 560Val Lys Gln Leu Lys Glu Asp Tyr Phe
Lys Lys Ile Glu Cys Phe Asp 565 570 575Ser Val Glu Ile Ser Gly Val
Glu Asp Arg Phe Asn Ala Ser Leu Gly 580 585 590Thr Tyr His Asp Leu
Leu Lys Ile Ile Lys Asp Lys Asp Phe Leu Asp 595 600 605Asn Glu Glu
Asn Glu Asp Ile Leu Glu Asp Ile Val Leu Thr Leu Thr 610 615 620Leu
Phe Glu Asp Arg Glu Met Ile Glu Glu Arg Leu Lys Thr Tyr Ala625 630
635 640His Leu Phe Asp Asp Lys Val Met Lys Gln Leu Lys Arg Arg Arg
Tyr 645 650 655Thr Gly Trp Gly Arg Leu Ser Arg Lys Leu Ile Asn Gly
Ile Arg Asp 660 665 670Lys Gln Ser Gly Lys Thr Ile Leu Asp Phe Leu
Lys Ser Asp Gly Phe 675 680 685Ala Asn Arg Asn Phe Met Gln Leu Ile
His Asp Asp Ser Leu Thr Phe 690 695 700Lys Glu Asp Ile Gln Lys Ala
Gln Val Ser Gly Gln Gly Asp Ser Leu705 710 715 720His Glu His Ile
Ala Asn Leu Ala Gly Ser Pro Ala Ile Lys Lys Gly 725 730 735Ile Leu
Gln Thr Val Lys Val Val Asp Glu Leu Val Lys Val Met Gly 740 745
750Arg His Lys Pro Glu Asn Ile Val Ile Glu Met Ala Arg Glu Asn Gln
755 760 765Thr Thr Gln Lys Gly Gln Lys Asn Ser Arg Glu Arg Met Lys
Arg Ile 770 775 780Glu Glu Gly Ile Lys Glu Leu Gly Ser Gln Ile Leu
Lys Glu His Pro785 790 795 800Val Glu Asn Thr Gln Leu Gln Asn Glu
Lys Leu Tyr Leu Tyr Tyr Leu 805 810 815Gln Asn Gly Arg Asp Met Tyr
Val Asp Gln Glu Leu Asp Ile Asn Arg 820 825 830Leu Ser Asp Tyr Asp
Val Asp His Ile Val Pro Gln Ser Phe Leu Lys 835 840 845Asp Asp Ser
Ile Asp Asn Lys Val Leu Thr Arg Ser Asp Lys Asn Arg 850 855 860Gly
Lys Ser Asp Asn Val Pro Ser Glu Glu Val Val Lys Lys Met Lys865 870
875 880Asn Tyr Trp Arg Gln Leu Leu Asn Ala Lys Leu Ile Thr Gln Arg
Lys 885 890 895Phe Asp Asn Leu Thr Lys Ala Glu Arg Gly Gly Leu Ser
Glu Leu Asp 900 905 910Lys Ala Gly Phe Ile Lys Arg Gln Leu Val Glu
Thr Arg Gln Ile Thr 915 920 925Lys His Val Ala Gln Ile Leu Asp Ser
Arg Met Asn Thr Lys Tyr Asp 930 935 940Glu Asn Asp Lys Leu Ile Arg
Glu Val Lys Val Ile Thr Leu Lys Ser945 950 955 960Lys Leu Val Ser
Asp Phe Arg Lys Asp Phe Gln Phe Tyr Lys Val Arg 965 970 975Glu Ile
Asn Asn Tyr His His Ala His Asp Ala Tyr Leu Asn Ala Val 980 985
990Val Gly Thr Ala Leu Ile Lys Lys Tyr Pro Lys Leu Glu Ser Glu Phe
995 1000 1005Val Tyr Gly Asp Tyr Lys Val Tyr Asp Val Arg Lys Met
Ile Ala 1010 1015 1020Lys Ser Glu Gln Glu Ile Gly Lys Ala Thr Ala
Lys Tyr Phe Phe 1025 1030 1035Tyr Ser Asn Ile Met Asn Phe Phe Lys
Thr Glu Ile Thr Leu Ala 1040 1045 1050Asn Gly Glu Ile Arg Lys Arg
Pro Leu Ile Glu Thr Asn Gly Glu 1055 1060 1065Thr Gly Glu Ile Val
Trp Asp Lys Gly Arg Asp Phe Ala Thr Val 1070 1075 1080Arg Lys Val
Leu Ser Met Pro Gln Val Asn Ile Val Lys Lys Thr 1085 1090 1095Glu
Val Gln Thr Gly Gly Phe Ser Lys Glu Ser Ile Leu Pro Lys 1100 1105
1110Arg Asn Ser Asp Lys Leu Ile Ala Arg Lys Lys Asp Trp Asp Pro
1115 1120 1125Lys Lys Tyr Gly Gly Phe Asp Ser Pro Thr Val Ala Tyr
Ser Val 1130 1135 1140Leu Val Val Ala Lys Val Glu Lys Gly Lys Ser
Lys Lys Leu Lys 1145 1150 1155Ser Val Lys Glu Leu Leu Gly Ile Thr
Ile Met Glu Arg Ser Ser 1160 1165 1170Phe Glu Lys Asn Pro Ile Asp
Phe Leu Glu Ala Lys Gly Tyr Lys 1175 1180 1185Glu Val Lys Lys Asp
Leu Ile Ile Lys Leu Pro Lys Tyr Ser Leu 1190 1195 1200Phe Glu Leu
Glu Asn Gly Arg Lys Arg Met Leu Ala Ser Ala Gly 1205 1210 1215Glu
Leu Gln Lys Gly Asn Glu Leu Ala Leu Pro Ser Lys Tyr Val 1220 1225
1230Asn Phe Leu Tyr Leu Ala Ser His Tyr Glu Lys Leu Lys Gly Ser
1235 1240 1245Pro Glu Asp Asn Glu Gln Lys Gln Leu Phe Val Glu Gln
His Lys 1250 1255 1260His Tyr Leu Asp Glu Ile Ile Glu Gln Ile Ser
Glu Phe Ser Lys 1265 1270 1275Arg Val Ile Leu Ala Asp Ala Asn Leu
Asp Lys Val Leu Ser Ala 1280 1285 1290Tyr Asn Lys His Arg Asp Lys
Pro Ile Arg Glu Gln Ala Glu Asn 1295 1300 1305Ile Ile His Leu Phe
Thr Leu Thr Asn Leu Gly Ala Pro Ala Ala 1310 1315 1320Phe Lys Tyr
Phe Asp Thr Thr Ile Asp Arg Lys Arg Tyr Thr Ser 1325 1330 1335Thr
Lys Glu Val Leu Asp Ala Thr Leu Ile His Gln Ser Ile Thr 1340 1345
1350Gly Leu Tyr Glu Thr Arg Ile Asp Leu Ser Gln Leu Gly Gly Asp
1355 1360 1365Ser Arg Ala Asp 1370783DNAStreptococcus pyogenes
7gttttagagc tagaaatagc aagttaaaat aaggctagtc cgttatcaac ttgaaaaagt
60ggcaccgagt cggtggtgct ttt 838665DNASaccharomyces cerevisiae
8tttcaaaaat tcttactttt tttttggatg gacgcaaaga agtttaataa tcatattaca
60tggcattacc accatataca tatccatata catatccata tctaatctta cttatatgtt
120gtggaaatgt aaagagcccc attatcttag cctaaaaaaa ccttctcttt
ggaactttca 180gtaatacgct taactgctca ttgctatatt gaagtacgga
ttagaagccg ccgagcgggt 240gacagccctc cgaaggaaga ctctcctccg
tgcgtcctcg tcttcaccgg tcgcgttcct 300gaaacgcaga tgtgcctcgc
gccgcactgc tccgaacaat aaagattcta caatactagc 360ttttatggtt
atgaagagga aaaattggca gtaacctggc cccacaaacc ttcaaatgaa
420cgaatcaaat taacaaccat aggatgataa tgcgattagt tttttagcct
tatttctggg 480gtaattaatc agcgaagcga tgatttttga tctattaaca
gatatataaa tgcaaaaact 540gcataaccac tttaactaat actttcaaca
ttttcggttt gtattacttc ttattcaaat 600gtaataaaag tatcaacaaa
aaattgttaa tatacctcta tactttaacg tcaaggagaa 660aaaac
665921DNAArtificial SequenceNuclear transition signal.CDS(1)..(21)
9ccc aag aag aag agg aag gtg 21Pro Lys Lys Lys Arg Lys Val1
5107PRTArtificial SequenceSynthetic Construct 10Pro Lys Lys Lys Arg
Lys Val1 51115DNAArtificial SequenceGS linkerCDS(1)..(15) 11ggt gga
gga ggt tct 15Gly Gly Gly Gly Ser1 5125PRTArtificial
SequenceSynthetic Construct 12Gly Gly Gly Gly Ser1
51366DNAArtificial
SequenceFlag tagCDS(1)..(66) 13gac tat aag gac cac gac gga gac tac
aag gat cat gat att gat tac 48Asp Tyr Lys Asp His Asp Gly Asp Tyr
Lys Asp His Asp Ile Asp Tyr1 5 10 15aaa gac gat gac gat aag 66Lys
Asp Asp Asp Asp Lys 201422PRTArtificial SequenceSynthetic Construct
14Asp Tyr Lys Asp His Asp Gly Asp Tyr Lys Asp His Asp Ile Asp Tyr1
5 10 15Lys Asp Asp Asp Asp Lys 201524DNAArtificial
SequenceStrep-tagCDS(1)..(24) 15tgg agc cac ccg cag ttc gaa aaa
24Trp Ser His Pro Gln Phe Glu Lys1 5168PRTArtificial
SequenceSynthetic Construct 16Trp Ser His Pro Gln Phe Glu Lys1
517171DNAArtificial SequenceSH3 domainCDS(1)..(171) 17gca gag tat
gtg cgg gcc ctc ttt gac ttt aat ggg aat gat gaa gaa 48Ala Glu Tyr
Val Arg Ala Leu Phe Asp Phe Asn Gly Asn Asp Glu Glu1 5 10 15gat ctt
ccc ttt aag aaa gga gac atc ctg aga atc cgg gat aag cct 96Asp Leu
Pro Phe Lys Lys Gly Asp Ile Leu Arg Ile Arg Asp Lys Pro 20 25 30gaa
gag cag tgg tgg aat gca gag gac agc gaa gga aag agg ggg atg 144Glu
Glu Gln Trp Trp Asn Ala Glu Asp Ser Glu Gly Lys Arg Gly Met 35 40
45att cct gtc cct tac gtg gag aag tat 171Ile Pro Val Pro Tyr Val
Glu Lys Tyr 50 551857PRTArtificial SequenceSynthetic Construct
18Ala Glu Tyr Val Arg Ala Leu Phe Asp Phe Asn Gly Asn Asp Glu Glu1
5 10 15Asp Leu Pro Phe Lys Lys Gly Asp Ile Leu Arg Ile Arg Asp Lys
Pro 20 25 30Glu Glu Gln Trp Trp Asn Ala Glu Asp Ser Glu Gly Lys Arg
Gly Met 35 40 45Ile Pro Val Pro Tyr Val Glu Lys Tyr 50
5519188DNASaccharomyces cerevisiae 19gcgaatttct tatgatttat
gatttttatt attaaataag ttataaaaaa aataagtgta 60tacaaatttt aaagtgactc
ttaggtttta aaacgaaaat tcttattctt gagtaactct 120ttcctgtagg
tcaggttgct ttctcaggta tagcatgagg tcgctcttat tgaccacacc 180tctaccgg
18820417DNASaccharomyces cerevisiae 20ataccaggca tggagcttat
ctggtccgtt cgagttttcg acgagtttgg agacattctt 60tatagatgtc cttttttttt
aatgatattc gttaaagaac aaaaagtcaa agcagtttaa 120cctaacacct
gttgttgatg ctacttgaaa caaggcttct aggcgaatac ttaaaaaggt
180aatttcaata gcggtttata tatctgtttg cttttcaaga tattatgtaa
acgcacgatg 240tttttcgccc aggctttatt ttttttgttg ttgttgtctt
ctcgaagaat tttctcgggc 300agatctttgt cggaatgtaa aaaagcgcgt
aattaaactt tctattatgc tgactaaaat 360ggaagtgatc accaaaggct
atttctgatt atataatcta gtcattactc gctcgag 4172133DNAArtificial
SequenceSH3-binding ligandCDS(1)..(33) 21cct cca cct gct ctg cca
cct aag aga agg aga 33Pro Pro Pro Ala Leu Pro Pro Lys Arg Arg Arg1
5 102211PRTArtificial SequenceSynthetic Construct 22Pro Pro Pro Ala
Leu Pro Pro Lys Arg Arg Arg1 5 1023269DNASaccharomyces cerevisiae
23tctttgaaaa gataatgtat gattatgctt tcactcatat ttatacagaa acttgatgtt
60ttctttcgag tatatacaag gtgattacat gtacgtttga agtacaactc tagattttgt
120agtgccctct tgggctagcg gtaaaggtgc gcattttttc acaccctaca
atgttctgtt 180caaaagattt tggtcaaacg ctgtagaagt gaaagttggt
gcgcatgttt cggcgttcga 240aacttctccg cagtgaaaga taaatgatc
2692414DNASaccharomyces cerevisiae 24tgttttttat gtct
142520DNASaccharomyces cerevisiae 25gatacgttct ctatggagga
202620DNASaccharomyces cerevisiae 26ttggagaaac ccaggtgcct
202720DNASaccharomyces cerevisiae 27aacccaggtg cctggggtcc
202820DNASaccharomyces cerevisiae 28ttggccaagt cattcaattt
202920DNASaccharomyces cerevisiae 29ataacggaat ccaactgggc
203020DNASaccharomyces cerevisiae 30gtcaattacg aagactgaac
203119DNASaccharomyces cerevisiae 31gtcaatagga tcccctttt
193210126DNAArtificial SequencePlasmid carrying dCas9-PmCDA1 fusion
protein and chimeric RNA targeting galK gene of
E.colimisc_feature(5561)..(5580)n is a, c, g, or t 32atcgccattc
gccattcagg ctgcgcaact gttgggaagg gcgatcggtg cgggcctctt 60cgctattacg
ccagctggcg aaagggggat gtgctgcaag gcgattaagt tgggtaacgc
120cagggttttc ccagtcacga cgttgtaaaa cgacggccag tgaattcgag
ctcggtaccc 180ggccgcaaac aacagataaa acgaaaggcc cagtctttcg
actgagcctt tcgttttatt 240tgatgcctgt caagtaacag caggactctt
agtggtgtgg agtattttta cctgaatcat 300aatggacaac tcgctccgtc
gtttttcagc tcgcttcaaa gtcttctcaa gccatctatt 360ctcattcaat
tgattgtgcg acgattggat gaatattttc ctgcaacatt ggtagtgttc
420acttaccatt acattcaacc caaccccgtt atctctgagg ttccacagcc
caatttgatt 480cctcgcattt ttctcgtaat agagtttgca agcccagatt
ttcaaagtgt ggccgttccc 540ccgcagctcc tggttatacc attctaagat
cttttcagcg caatctgcac aaggactcca 600ggatgagtac caatttatcg
tgaattgtcc ggggttgtcg cgcaggtatt cttcgacttt 660tctaatgcta
aagatttcgg cgtgaatgcc acgttctgtc ccgctctgtg gtttattcac
720agcatagccc caaaaacacg ctctacgttc accccgtcgt tttaattcaa
agagaacgta 780gcatctatgc gacacggatt ttttgttgtt gaaaaactgt
ttcttaaacg tgtagatgtc 840caacttctca tggattctca cgtactcagc
gtcggtcatc ctagacttat cgtcatcgtc 900tttgtaatca atatcatgat
ccttgtagtc tccgtcgtgg tccttatagt ctccggactc 960gagcctagac
ttatcgtcat cgtctttgta atcaatatca tgatccttgt agtctccgtc
1020gtggtcctta tagtctccgg aatacttctc cacgtaaggg acaggaatca
tccccctctt 1080tccttcgctg tcctctgcat tccaccactg ctcctcaggc
ttatcccgga ttctcaggat 1140gtctcctttc ttaaagggaa gatcctcttc
atcattccca ttaaagtcaa agagggctcg 1200cacatactca gcagaacctc
cacctccaga acctcctcca ccgtcacctc ctagctgact 1260caaatcaatg
cgtgtttcat aaagaccagt gatggattga tggataagag tggcatctaa
1320aacttctttt gtagacgtat atcgtttacg atcaattgtt gtatcaaaat
atttaaaagc 1380agcgggagct ccaagattcg tcaacgtaaa taaatgaata
atattttctg cttgttcacg 1440tattggtttg tctctatgtt tgttatatgc
actaagaact ttatctaaat tggcatctgc 1500taaaataaca cgcttagaaa
attcactgat ttgctcaata atctcatcta aataatgctt 1560atgctgctcc
acaaacaatt gtttttgttc gttatcttct ggactaccct tcaacttttc
1620ataatgacta gctaaatata aaaaattcac atatttgctt ggcagagcca
gctcatttcc 1680tttttgtaat tctccggcac tagccagcat ccgtttacga
ccgttttcta actcaaaaag 1740actatattta ggtagtttaa tgattaagtc
ttttttaact tccttatatc ctttagcttc 1800taaaaagtca atcggatttt
tttcaaagga acttctttcc ataattgtga tccctagtaa 1860ctctttaacg
gattttaact tcttcgattt ccctttttcc accttagcaa ccactaggac
1920tgaataagct accgttggac tatcaaaacc accatatttt tttggatccc
agtctttttt 1980acgagcaata agcttgtccg aatttctttt tggtaaaatt
gactccttgg agaatccgcc 2040tgtctgtact tctgttttct tgacaatatt
gacttggggc atggacaata ctttgcgcac 2100tgtggcaaaa tctcgccctt
tatcccagac aatttctcca gtttccccat tagtttcgat 2160tagagggcgt
ttgcgaatct ctccatttgc aagtgtaatt tctgttttga agaagttcat
2220gatattagag taaaagaaat attttgcggt tgctttgcct atttcttgct
cagacttagc 2280aatcatttta cgaacatcat aaactttata atcaccatag
acaaactccg attcaagttt 2340tggatatttc ttaatcaaag cagttccaac
gacggcattt agatacgcat catgggcatg 2400atggtaattg ttaatctcac
gtactttata gaattggaaa tcttttcgga agtcagaaac 2460taatttagat
tttaaggtaa tcactttaac ctctcgaata agtttatcat tttcatcgta
2520tttagtattc atgcgactat ccaaaatttg tgccacatgc ttagtgattt
ggcgagtttc 2580aaccaattgg cgtttgataa aaccagcttt atcaagttca
ctcaaacctc cacgttcagc 2640tttcgttaaa ttatcaaact tacgttgagt
gattaacttg gcgtttagaa gttgtctcca 2700atagtttttc atctttttga
ctacttcttc acttggaacg ttatccgatt taccacgatt 2760tttatcagaa
cgcgttaaga ccttattgtc tattgaatcg tctttaagga aactttgtgg
2820aacaatggca tcgacatcat aatcacttaa acgattaata tctaattctt
ggtccacata 2880catgtctctt ccattttgga gataatagag atagagcttt
tcattttgca attgagtatt 2940ttcaacagga tgctctttaa gaatctgact
tcctaattct ttgatacctt cttcgattcg 3000tttcatacgc tctcgcgaat
ttttctggcc cttttgagtt gtctgatttt cacgtgccat 3060ttcaataacg
atattttctg gcttatgccg ccccattact ttgaccaatt catcaacaac
3120ttttacagtc tgtaaaatac cttttttaat agcagggcta ccagctaaat
ttgcaatatg 3180ttcatgtaaa ctatcgcctt gtccagacac ttgtgctttt
tgaatgtctt ctttaaatgt 3240caaactatca tcatggatca gctgcataaa
attgcgattg gcaaaaccat ctgatttcaa 3300aaaatctaat attgttttgc
cagattgctt atccctaata ccattaatca attttcgaga 3360caaacgtccc
caaccagtat aacggcgacg tttaagctgt ttcatcacct tatcatcaaa
3420gaggtgagca tatgttttaa gtctttcctc aatcatctcc ctatcttcaa
ataaggtcaa 3480tgttaaaaca atatcctcta agatatcttc attttcttca
ttatccaaaa aatctttatc 3540tttaataatt tttagcaaat catggtaggt
acctaatgaa gcattaaatc tatcttcaac 3600tcctgaaatt tcaacactat
caaaacattc tatttttttg aaataatctt cttttaattg 3660cttaacggtt
acttttcgat ttgttttgaa gagtaaatca acaatggctt tcttctgttc
3720acctgaaaga aatgctggtt ttcgcattcc ttcagtaaca tatttgacct
ttgtcaattc 3780gttataaacc gtaaaatact cataaagcaa actatgtttt
ggtagtactt tttcatttgg 3840aagattttta tcaaagtttg tcatgcgttc
aataaatgat tgagctgaag cacctttatc 3900gacaacttct tcaaaattcc
atggggtaat tgtttcttca gacttccgag tcatccatgc 3960aaaacgacta
ttgccacgcg ccaatggacc aacataataa ggaattcgaa aagtcaagat
4020tttttcaatc ttctcacgat tgtcttttaa aaatggataa aagtcttctt
gtcttctcaa 4080aatagcatgc agctcaccca agtgaatttg atggggaata
gagccgttgt caaaggtccg 4140ttgcttgcgc agcaaatctt cacgatttag
tttcaccaat aattcctcag taccatccat 4200tttttctaaa attggtttga
taaatttata aaattcttct tggctagctc ccccatcaat 4260ataacctgca
tatccgtttt ttgattgatc aaaaaagatt tctttatact tttctggaag
4320ttgttgtcga actaaagctt ttaaaagagt caagtcttga tgatgttcat
cgtagcgttt 4380aatcattgaa gctgataggg gagccttagt tatttcagta
tttactctta ggatatctga 4440aagtaaaata gcatctgata aattcttagc
tgccaaaaac aaatcagcat attgatctcc 4500aatttgcgcc aataaattat
ctaaatcatc atcgtaagta tcttttgaaa gctgtaattt 4560agcatcttct
gccaaatcaa aatttgattt aaaattaggg gtcaaaccca atgacaaagc
4620aatgagattc ccaaataagc catttttctt ctcaccgggg agctgagcaa
tgagattttc 4680taatcgtctt gatttactca atcgtgcaga aagaatcgct
ttagcatcta ctccacttgc 4740gttaataggg ttttcttcaa ataattgatt
gtaggtttgt accaactgga taaatagttt 4800gtccacatca ctattatcag
gatttaaatc tccctcaatc aaaaaatgac cacgaaactt 4860aatcatatgc
gctaaggcca aatagattaa gcgcaaatcc gctttatcag tagaatctac
4920caattttttt cgcagatgat agatagttgg atatttctca tgataagcaa
cttcatctac 4980tatatttcca aaaataggat gacgttcatg cttcttgtct
tcttccacca aaaaagactc 5040ttcaagtcga tgaaagaaac tatcatctac
tttcgccatc tcatttgaaa aaatctcctg 5100tagataacaa atacgattct
tccgacgtgt ataccttcta cgagctgtcc gtttgagacg 5160agtcgcttcc
gctgtctctc cactgtcaaa taaaagagcc cctataagat tttttttgat
5220actgtggcgg tctgtatttc ccagaacctt gaacttttta gacggaacct
tatattcatc 5280agtgatcacc gcccatccga cgctatttgt gccgatagct
aagcctattg agtatttctt 5340atccattttt gcctcctaaa atgggccctt
taaattaaat ccataatgag tttgatgatt 5400tcaataatag ttttaatgac
ctccgaaatt agtttaatat gctttaattt ttctttttca 5460aaatatctct
tcaaaaaata ttacccaata cttaataata aatagattat aacacaaaat
5520tcttttgaca agtagtttat tttgttataa ttctatagta nnnnnnnnnn
nnnnnnnnnn 5580gttttagagc tagaaatagc aagttaaaat aaggctagtc
cgttatcaac ttgaaaaagt 5640ggcaccgagt cggtgctttt tttgatactt
ctattctact ctgactgcaa accaaaaaaa 5700caagcgcttt caaaacgctt
gttttatcat ttttagggaa attaatctct taatcctttt 5760atcattctac
atttaggcgc tgccatcttg ctaaacctac taagctccac aggatgattt
5820cgtaatcccg caagaggccc ggcagtaccg gcataaccaa gcctatgcct
acagcatcca 5880gggtgacggt gccgaggatg acgatgagcg cattgttaga
tttcatacac ggtgcctgac 5940tgcgttagca atttaactgt gataaactac
cgcattaaag cttatcgatg ataagctgtc 6000aaacatgaga attacaactt
atatcgtatg gggctgactt caggtgctac atttgaagag 6060ataaattgca
ctgaaatcta gtcggatcct cgctcactga ctcgctgcgc tcggtcgttc
6120ggctgcggcg agcggtatca gctcactcaa aggcggtaat acggttatcc
acagaatcag 6180gggataacgc aggaaagaac atgtgagcaa aaggccagca
aaaggccagg aaccgtaaaa 6240aggccgcgtt gctggcgttt ttccataggc
tccgcccccc tgacgagcat cacaaaaatc 6300gacgctcaag tcagaggtgg
cgaaacccga caggactata aagataccag gcgtttcccc 6360ctggaagctc
cctcgtgcgc tctcctgttc cgaccctgcc gcttaccgga tacctgtccg
6420cctttctccc ttcgggaagc gtggcgcttt ctcatagctc acgctgtagg
tatctcagtt 6480cggtgtaggt cgttcgctcc aagctgggct gtgtgcacga
accccccgtt cagcccgacc 6540gctgcgcctt atccggtaac tatcgtcttg
agtccaaccc ggtaagacac gacttatcgc 6600cactggcagc agccactggt
aacaggatta gcagagcgag gtatgtaggc ggtgctacag 6660agttcttgaa
gtggtggcct aactacggct acactagaag gacagtattt ggtatctgcg
6720ctctgctgaa gccagttacc ttcggaaaaa gagttggtag ctcttgatcc
ggcaaacaaa 6780ccaccgctgg tagcggtggt ttttttgttt gcaagcagca
gattacgcgc agaaaaaaag 6840gatctcaaga agatcctttg atcttttcta
cggggtctga cgctcagtgg aacgaaaact 6900cacgttaagg gattttggtc
atgagattat caaaaaggat cttcacctag atccttttaa 6960attaaaaatg
aagttttaaa tcaatctaaa gtatatatga gtaaacttgg tctgacagtt
7020accaatgctt aatcagtgag gcacctatct cagcgatctg tctatttcgt
tcatccatag 7080ttgcctgact ccccgtcgtg tagataacta cgatacggga
gggcttacca tctggcccca 7140gtgctgcaat gataccgcga gaaccacgct
caccggctcc agatttatca gcaataaacc 7200agccagccgg aagggccgag
cgcagaagtg gtcctgcaac tttatccgcc tccatccagt 7260ctattaattg
ttgccgggaa gctagagtaa gtagttcgcc agttaatagt ttgcgcaacg
7320ttgttgccat tgctgcaggc atcgtggtgt cacgctcgtc gtttggtatg
gcttcattca 7380gctccggttc ccaacgatca aggcgagtta catgatcccc
catgttgtgc aaaaaagcgg 7440ttagctcctt cggtcctccg atcgttgtca
gaagtaagtt ggccgcagtg ttatcactca 7500tggttatggc agcactgcat
aattctctta ctgtcatgcc atccgtaaga tgcttttctg 7560tgactggtga
gtactcaacc aagtcattct gagaatagtg tatgcggcga ccgagttgct
7620cttgcccggc gtcaacacgg gataataccg cgccacatag cagaacttta
aaagtgctca 7680tcattggaaa acgttcttcg gggcgaaaac tctcaaggat
cttaccgctg ttgagatcca 7740gttcgatgta acccactcgt gcacccaact
gatcttcagc atcttttact ttcaccagcg 7800tttctgggtg agcaaaaaca
ggaaggcaaa atgccgcaaa aaagggaata agggcgacac 7860ggaaatgttg
aatactcata ctcttccttt ttcaatatta ttgaagcatt tatcagggtt
7920attgtctcat gagcggatac atatttgaat gtatttagaa aaataaacaa
ataggggttc 7980cgcgcacatt tccccgaaaa gtgccacctg acgtcaatgc
cgagcgaaag cgagccgaag 8040ggtagcattt acgttagata accccctgat
atgctccgac gctttatata gaaaagaaga 8100ttcaactagg taaaatctta
atataggttg agatgataag gtttataagg aatttgtttg 8160ttctaatttt
tcactcattt tgttctaatt tcttttaaca aatgttcttt tttttttaga
8220acagttatga tatagttaga atagtttaaa ataaggagtg agaaaaagat
gaaagaaaga 8280tatggaacag tctataaagg ctctcagagg ctcatagacg
aagaaagtgg agaagtcata 8340gaggtagaca agttataccg taaacaaacg
tctggtaact tcgtaaaggc atatatagtg 8400caattaataa gtatgttaga
tatgattggc ggaaaaaaac ttaaaatcgt taactatatc 8460ctagataatg
tccacttaag taacaataca atgatagcta caacaagaga aatagcaaaa
8520gctacaggaa caagtctaca aacagtaata acaacactta aaatcttaga
agaaggaaat 8580attataaaaa gaaaaactgg agtattaatg ttaaaccctg
aactactaat gagaggcgac 8640gaccaaaaac aaaaatacct cttactcgaa
tttgggaact ttgagcaaga ggcaaatgaa 8700atagattgac ctcccaataa
caccacgtag ttattgggag gtcaatctat gaaatgcgat 8760taagcttttt
ctaattcaca taagcgtgca ggtttaaagt acataaaaaa tataatgaaa
8820aaaagcatca ttatactaac gttataccaa cattatactc tcattatact
aattgcttat 8880tccaatttcc tattggttgg aaccaacagg cgttagtgtg
ttgttgagtt ggtactttca 8940tgggattaat cccatgaaac ccccaaccaa
ctcgccaaag ctttggctaa cacacacgcc 9000attccaacca atagttttct
cggcataaag ccatgctctg acgcttaaat gcactaatgc 9060cttaaaaaaa
cattaaagtc taacacacta gacttattta cttcgtaatt aagtcgttaa
9120accgtgtgct ctacgaccaa aagtataaaa cctttaagaa ctttcttttt
tcttgtaaaa 9180aaagaaacta gataaatctc tcatatcttt tattcaataa
tcgcatcaga ttgcagtata 9240aatttaacga tcactcatca tgttcatatt
tatcagagct cgtgctataa ttatactaat 9300tttataagga ggaaaaaata
aagagggtta taatgaacga gaaaaatata aaacacagtc 9360aaaactttat
tacttcaaaa cataatatag ataaaataat gacaaatata agattaaatg
9420aacatgataa tatctttgaa atcggctcag gaaaagggca ttttaccctt
gaattagtac 9480agaggtgtaa tttcgtaact gccattgaaa tagaccataa
attatgcaaa actacagaaa 9540ataaacttgt tgatcacgat aatttccaag
ttttaaacaa ggatatattg cagtttaaat 9600ttcctaaaaa ccaatcctat
aaaatatttg gtaatatacc ttataacata agtacggata 9660taatacgcaa
aattgttttt gatagtatag ctgatgagat ttatttaatc gtggaatacg
9720ggtttgctaa aagattatta aatacaaaac gctcattggc attattttta
atggcagaag 9780ttgatatttc tatattaagt atggttccaa gagaatattt
tcatcctaaa cctaaagtga 9840atagctcact tatcagatta aatagaaaaa
aatcaagaat atcacacaaa gataaacaga 9900agtataatta tttcgttatg
aaatgggtta acaaagaata caagaaaata tttacaaaaa 9960atcaatttaa
caattcctta aaacatgcag gaattgacga tttaaacaat attagctttg
10020aacaattctt atctcttttc aatagctata aattatttaa taagtaagtt
aagggatgca 10080taaactgcat cccttaactt gtttttcgtg tacctatttt ttgtga
101263320DNAEscherichia coli 33tcaatgggct aactacgttc
203420DNAEscherichia coli 34ggtccataaa ctgagacagc
2035223DNAEscherichia coli 35gagttataca cagggctggg atctattctt
tttatctttt tttattcttt ctttattcta 60taaattataa ccacttgaat ataaacaaaa
aaaacacaca aaggtctagc ggaatttaca 120gagggtctag cagaatttac
aagttttcca gcaaaggtct agcagaattt acagataccc 180acaactcaaa
ggaaaaggac tagtaattat cattgactag ccc 22336951DNAEscherichia coli
36atgtctgaat tagttgtttt caaagcaaat gaactagcga ttagtcgcta tgacttaacg
60gagcatgaaa ccaagctaat tttatgctgt gtggcactac tcaaccccac gattgaaaac
120cctacaatga aagaacggac ggtatcgttc acttataacc aatacgttca
gatgatgaac 180atcagtaggg aaaatgctta tggtgtatta gctaaagcaa
ccagagagct gatgacgaga 240actgtggaaa tcaggaatcc tttggttaaa
ggctttgaga ttttccagtg gacaaactat 300gccaagttct caagcgaaaa
attagaatta gtttttagtg aagagatatt gccttatctt 360ttccagttaa
aaaaattcat aaaatataat ctggaacatg ttaagtcttt tgaaaacaaa
420tactctatga ggatttatga gtggttatta aaagaactaa cacaaaagaa
aactcacaag 480gcaaatatag
agattagcct tgatgaattt aagttcatgt taatgcttga aaataactac
540catgagttta aaaggcttaa ccaatgggtt ttgaaaccaa taagtaaaga
tttaaacact 600tacagcaata tgaaattggt ggttgataag cgaggccgcc
cgactgatac gttgattttc 660caagttgaac tagatagaca aatggatctc
gtaaccgaac ttgagaacaa ccagataaaa 720atgaatggtg acaaaatacc
aacaaccatt acatcagatt cctacctaca taacggacta 780agaaaaacac
tacacgatgc tttaactgca aaaattcagc tcaccagttt tgaggcaaaa
840tttttgagtg acatgcaaag taagcatgat ctcaatggtt cgttctcatg
gctcacgcaa 900aaacaacgaa ccacactaga gaacatactg gctaaatacg
gaaggatctg a 95137714DNABacteriophage lambda 37tcagccaaac
gtctcttcag gccactgact agcgataact ttccccacaa cggaacaact 60ctcattgcat
gggatcattg ggtactgtgg gtttagtggt tgtaaaaaca cctgaccgct
120atccctgatc agtttcttga aggtaaactc atcaccccca agtctggcta
tgcagaaatc 180acctggctca acagcctgct cagggtcaac gagaattaac
attccgtcag gaaagcttgg 240cttggagcct gttggtgcgg tcatggaatt
accttcaacc tcaagccaga atgcagaatc 300actggctttt ttggttgtgc
ttacccatct ctccgcatca cctttggtaa aggttctaag 360cttaggtgag
aacatccctg cctgaacatg agaaaaaaca gggtactcat actcacttct
420aagtgacggc tgcatactaa ccgcttcata catctcgtag atttctctgg
cgattgaagg 480gctaaattct tcaacgctaa ctttgagaat ttttgtaagc
aatgcggcgt tataagcatt 540taatgcattg atgccattaa ataaagcacc
aacgcctgac tgccccatcc ccatcttgtc 600tgcgacagat tcctgggata
agccaagttc atttttcttt ttttcataaa ttgctttaag 660gcgacgtgcg
tcctcaagct gctcttgtgt taatggtttc ttttttgtgc tcat
714385097DNAArtificial SequencedCas9-PmCDA1 38atggataaga aatactcaat
aggcttagct atcggcacaa atagcgtcgg atgggcggtg 60atcactgatg aatataaggt
tccgtctaaa aagttcaagg ttctgggaaa tacagaccgc 120cacagtatca
aaaaaaatct tataggggct cttttatttg acagtggaga gacagcggaa
180gcgactcgtc tcaaacggac agctcgtaga aggtatacac gtcggaagaa
tcgtatttgt 240tatctacagg agattttttc aaatgagatg gcgaaagtag
atgatagttt ctttcatcga 300cttgaagagt cttttttggt ggaagaagac
aagaagcatg aacgtcatcc tatttttgga 360aatatagtag atgaagttgc
ttatcatgag aaatatccaa ctatctatca tctgcgaaaa 420aaattggtag
attctactga taaagcggat ttgcgcttaa tctatttggc cttagcgcat
480atgattaagt ttcgtggtca ttttttgatt gagggagatt taaatcctga
taatagtgat 540gtggacaaac tatttatcca gttggtacaa acctacaatc
aattatttga agaaaaccct 600attaacgcaa gtggagtaga tgctaaagcg
attctttctg cacgattgag taaatcaaga 660cgattagaaa atctcattgc
tcagctcccc ggtgagaaga aaaatggctt atttgggaat 720ctcattgctt
tgtcattggg tttgacccct aattttaaat caaattttga tttggcagaa
780gatgctaaat tacagctttc aaaagatact tacgatgatg atttagataa
tttattggcg 840caaattggag atcaatatgc tgatttgttt ttggcagcta
agaatttatc agatgctatt 900ttactttcag atatcctaag agtaaatact
gaaataacta aggctcccct atcagcttca 960atgattaaac gctacgatga
acatcatcaa gacttgactc ttttaaaagc tttagttcga 1020caacaacttc
cagaaaagta taaagaaatc ttttttgatc aatcaaaaaa cggatatgca
1080ggttatattg atgggggagc tagccaagaa gaattttata aatttatcaa
accaatttta 1140gaaaaaatgg atggtactga ggaattattg gtgaaactaa
atcgtgaaga tttgctgcgc 1200aagcaacgga cctttgacaa cggctctatt
ccccatcaaa ttcacttggg tgagctgcat 1260gctattttga gaagacaaga
agacttttat ccatttttaa aagacaatcg tgagaagatt 1320gaaaaaatct
tgacttttcg aattccttat tatgttggtc cattggcgcg tggcaatagt
1380cgttttgcat ggatgactcg gaagtctgaa gaaacaatta ccccatggaa
ttttgaagaa 1440gttgtcgata aaggtgcttc agctcaatca tttattgaac
gcatgacaaa ctttgataaa 1500aatcttccaa atgaaaaagt actaccaaaa
catagtttgc tttatgagta ttttacggtt 1560tataacgaat tgacaaaggt
caaatatgtt actgaaggaa tgcgaaaacc agcatttctt 1620tcaggtgaac
agaagaaagc cattgttgat ttactcttca aaacaaatcg aaaagtaacc
1680gttaagcaat taaaagaaga ttatttcaaa aaaatagaat gttttgatag
tgttgaaatt 1740tcaggagttg aagatagatt taatgcttca ttaggtacct
accatgattt gctaaaaatt 1800attaaagata aagatttttt ggataatgaa
gaaaatgaag atatcttaga ggatattgtt 1860ttaacattga ccttatttga
agatagggag atgattgagg aaagacttaa aacatatgct 1920cacctctttg
atgataaggt gatgaaacag cttaaacgtc gccgttatac tggttgggga
1980cgtttgtctc gaaaattgat taatggtatt agggataagc aatctggcaa
aacaatatta 2040gattttttga aatcagatgg ttttgccaat cgcaatttta
tgcagctgat ccatgatgat 2100agtttgacat ttaaagaaga cattcaaaaa
gcacaagtgt ctggacaagg cgatagttta 2160catgaacata ttgcaaattt
agctggtagc cctgctatta aaaaaggtat tttacagact 2220gtaaaagttg
ttgatgaatt ggtcaaagta atggggcggc ataagccaga aaatatcgtt
2280attgaaatgg cacgtgaaaa tcagacaact caaaagggcc agaaaaattc
gcgagagcgt 2340atgaaacgaa tcgaagaagg tatcaaagaa ttaggaagtc
agattcttaa agagcatcct 2400gttgaaaata ctcaattgca aaatgaaaag
ctctatctct attatctcca aaatggaaga 2460gacatgtatg tggaccaaga
attagatatt aatcgtttaa gtgattatga tgtcgatgcc 2520attgttccac
aaagtttcct taaagacgat tcaatagaca ataaggtctt aacgcgttct
2580gataaaaatc gtggtaaatc ggataacgtt ccaagtgaag aagtagtcaa
aaagatgaaa 2640aactattgga gacaacttct aaacgccaag ttaatcactc
aacgtaagtt tgataattta 2700acgaaagctg aacgtggagg tttgagtgaa
cttgataaag ctggttttat caaacgccaa 2760ttggttgaaa ctcgccaaat
cactaagcat gtggcacaaa ttttggatag tcgcatgaat 2820actaaatacg
atgaaaatga taaacttatt cgagaggtta aagtgattac cttaaaatct
2880aaattagttt ctgacttccg aaaagatttc caattctata aagtacgtga
gattaacaat 2940taccatcatg cccatgatgc gtatctaaat gccgtcgttg
gaactgcttt gattaagaaa 3000tatccaaaac ttgaatcgga gtttgtctat
ggtgattata aagtttatga tgttcgtaaa 3060atgattgcta agtctgagca
agaaataggc aaagcaaccg caaaatattt cttttactct 3120aatatcatga
acttcttcaa aacagaaatt acacttgcaa atggagagat tcgcaaacgc
3180cctctaatcg aaactaatgg ggaaactgga gaaattgtct gggataaagg
gcgagatttt 3240gccacagtgc gcaaagtatt gtccatgccc caagtcaata
ttgtcaagaa aacagaagta 3300cagacaggcg gattctccaa ggagtcaatt
ttaccaaaaa gaaattcgga caagcttatt 3360gctcgtaaaa aagactggga
tccaaaaaaa tatggtggtt ttgatagtcc aacggtagct 3420tattcagtcc
tagtggttgc taaggtggaa aaagggaaat cgaagaagtt aaaatccgtt
3480aaagagttac tagggatcac aattatggaa agaagttcct ttgaaaaaaa
tccgattgac 3540tttttagaag ctaaaggata taaggaagtt aaaaaagact
taatcattaa actacctaaa 3600tatagtcttt ttgagttaga aaacggtcgt
aaacggatgc tggctagtgc cggagaatta 3660caaaaaggaa atgagctggc
tctgccaagc aaatatgtga attttttata tttagctagt 3720cattatgaaa
agttgaaggg tagtccagaa gataacgaac aaaaacaatt gtttgtggag
3780cagcataagc attatttaga tgagattatt gagcaaatca gtgaattttc
taagcgtgtt 3840attttagcag atgccaattt agataaagtt cttagtgcat
ataacaaaca tagagacaaa 3900ccaatacgtg aacaagcaga aaatattatt
catttattta cgttgacgaa tcttggagct 3960cccgctgctt ttaaatattt
tgatacaaca attgatcgta aacgatatac gtctacaaaa 4020gaagttttag
atgccactct tatccatcaa tccatcactg gtctttatga aacacgcatt
4080gatttgagtc agctaggagg tgacggtgga ggaggttctg gaggtggagg
ttctgctgag 4140tatgtgcgag ccctctttga ctttaatggg aatgatgaag
aggatcttcc ctttaagaaa 4200ggagacatcc tgagaatccg ggataagcct
gaggagcagt ggtggaatgc agaggacagc 4260gaaggaaaga gggggatgat
tcctgtccct tacgtggaga agtattccgg agactataag 4320gaccacgacg
gagactacaa ggatcatgat attgattaca aagacgatga cgataagtct
4380aggctcgagt ccggagacta taaggaccac gacggagact acaaggatca
tgatattgat 4440tacaaagacg atgacgataa gtctaggatg accgacgctg
agtacgtgag aatccatgag 4500aagttggaca tctacacgtt taagaaacag
tttttcaaca acaaaaaatc cgtgtcgcat 4560agatgctacg ttctctttga
attaaaacga cggggtgaac gtagagcgtg tttttggggc 4620tatgctgtga
ataaaccaca gagcgggaca gaacgtggca ttcacgccga aatctttagc
4680attagaaaag tcgaagaata cctgcgcgac aaccccggac aattcacgat
aaattggtac 4740tcatcctgga gtccttgtgc agattgcgct gaaaagatct
tagaatggta taaccaggag 4800ctgcggggga acggccacac tttgaaaatc
tgggcttgca aactctatta cgagaaaaat 4860gcgaggaatc aaattgggct
gtggaacctc agagataacg gggttgggtt gaatgtaatg 4920gtaagtgaac
actaccaatg ttgcaggaaa atattcatcc aatcgtcgca caatcaattg
4980aatgagaata gatggcttga gaagactttg aagcgagctg aaaaacgacg
gagcgagttg 5040tccattatga ttcaggtaaa aatactccac accactaaga
gtcctgctgt tacttga 509739105DNAEscherichia coli 39acgttaaatc
tatcaccgca agggataaat atctaacacc gtgcgtgttg actattttac 60ctctggcggt
gataatggtt gcagggccca ttttaggagg caaaa 10540247DNAArtificial
SequencegRNA 40ggtttagcaa gatggcagcg cctaaatgta gaatgataaa
aggattaaga gattaatttc 60cctaaaaatg ataaaacaag cgttttgaaa gcgcttgttt
ttttggtttg cagtcagagt 120agaatagaag tatcaaaaaa agcaccgact
cggtgccact ttttcaagtt gataacggac 180tagccttatt ttaacttgct
atttctagct ctaaaactga gaccatcccg ggtctctact 240gcagaat
2474164DNAEscherichia coli 41tatcaccgcc agtggtattt atgtcaacac
cgccagagat aatttatcac cgcagatggt 60tatc 644210867DNAArtificial
SequencePlasmid 42gtcggaactg actaaagtag tgagttatac acagggctgg
gatctattct ttttatcttt 60ttttattctt tctttattct ataaattata accacttgaa
tataaacaaa aaaaacacac 120aaaggtctag cggaatttac agagggtcta
gcagaattta caagttttcc agcaaaggtc 180tagcagaatt tacagatacc
cacaactcaa aggaaaagga ctagtaatta tcattgacta 240gcccatctca
attggtatag tgattaaaat cacctagacc aattgagatg tatgtctgaa
300ttagttgttt tcaaagcaaa tgaactagcg attagtcgct atgacttaac
ggagcatgaa 360accaagctaa ttttatgctg tgtggcacta ctcaacccca
cgattgaaaa ccctacaatg 420aaagaacgga cggtatcgtt cacttataac
caatacgttc agatgatgaa catcagtagg 480gaaaatgctt atggtgtatt
agctaaagca accagagagc tgatgacgag aactgtggaa 540atcaggaatc
ctttggttaa aggctttgag attttccagt ggacaaacta tgccaagttc
600tcaagcgaaa aattagaatt agtttttagt gaagagatat tgccttatct
tttccagtta 660aaaaaattca taaaatataa tctggaacat gttaagtctt
ttgaaaacaa atactctatg 720aggatttatg agtggttatt aaaagaacta
acacaaaaga aaactcacaa ggcaaatata 780gagattagcc ttgatgaatt
taagttcatg ttaatgcttg aaaataacta ccatgagttt 840aaaaggctta
accaatgggt tttgaaacca ataagtaaag atttaaacac ttacagcaat
900atgaaattgg tggttgataa gcgaggccgc ccgactgata cgttgatttt
ccaagttgaa 960ctagatagac aaatggatct cgtaaccgaa cttgagaaca
accagataaa aatgaatggt 1020gacaaaatac caacaaccat tacatcagat
tcctacctac ataacggact aagaaaaaca 1080ctacacgatg ctttaactgc
aaaaattcag ctcaccagtt ttgaggcaaa atttttgagt 1140gacatgcaaa
gtaagcatga tctcaatggt tcgttctcat ggctcacgca aaaacaacga
1200accacactag agaacatact ggctaaatac ggaaggatct gaggttctta
tggctcttgt 1260atctatcagt gaagcatcaa gactaacaaa caaaagtaga
acaactgttc accgttacat 1320atcaaaggga aaactgtcca tatgcacaga
gataatctca tgaccaaaac cggtagctag 1380aggggccgca ttaggcaccc
caggctttac actttatgct tccggctcgt ataatgtgtg 1440gattttgagt
taggatccgg cgagattttc aggagctaag gaagctaaaa tggagaaaaa
1500aatcactgga tataccaccg ttgatatatc ccaatggcat cgtaaagaac
attttgaggc 1560atttcagtca gttgctcaat gtacctataa ccagaccgtt
cagctggata ttacggcctt 1620tttaaagacc gtaaagaaaa ataagcacaa
gttttatccg gcctttattc acattcttgc 1680ccgcctgatg aatgctcatc
cggaattccg tatggcaatg aaagacggtg agctggtgat 1740atgggatagt
gttcaccctt gttacaccgt tttccatgag caaactgaaa cgttttcatc
1800gctctggagt gaataccacg acgatttccg gcagtttcta cacatatatt
cgcaagatgt 1860ggcgtgttac ggtgaaaacc tggcctattt ccctaaaggg
tttattgaga atatgttttt 1920cgtctcagcc aatccctggg tgagtttcac
cagttttgat ttaaacgtgg ccaatatgga 1980caacttcttc gcccccgttt
tcaccatggg caaatattat acgcaaggcg acaaggtgct 2040gatgccgctg
gcgattcagg ttcatcatgc cgtctgtgat ggcttccatg tcggcagaat
2100gcttaatgaa ttacaacagt actgcgatga gtggcagggc ggggcgtaaa
cgcgtggatc 2160cggcttacta aaagccagat aacagtatgc gtatttgcgc
gctgattttt gcggtctaga 2220ggtttagcaa gatggcagcg cctaaatgta
gaatgataaa aggattaaga gattaatttc 2280cctaaaaatg ataaaacaag
cgttttgaaa gcgcttgttt ttttggtttg cagtcagagt 2340agaatagaag
tatcaaaaaa agcaccgact cggtgccact ttttcaagtt gataacggac
2400tagccttatt ttaacttgct atttctagct ctaaaactga gaccatcccg
ggtctctact 2460gcagaattat caccgccagt ggtatttatg tcaacaccgc
cagagataat ttatcaccgc 2520agatggttat cgatgaagat tcttgctcaa
ttgttatcag ctatgcgccg accagaacac 2580cttgccgatc agccaaacgt
ctcttcaggc cactgactag cgataacttt ccccacaacg 2640gaacaactct
cattgcatgg gatcattggg tactgtgggt ttagtggttg taaaaacacc
2700tgaccgctat ccctgatcag tttcttgaag gtaaactcat cacccccaag
tctggctatg 2760cagaaatcac ctggctcaac agcctgctca gggtcaacga
gaattaacat tccgtcagga 2820aagcttggct tggagcctgt tggtgcggtc
atggaattac cttcaacctc aagccagaat 2880gcagaatcac tggctttttt
ggttgtgctt acccatctct ccgcatcacc tttggtaaag 2940gttctaagct
taggtgagaa catccctgcc tgaacatgag aaaaaacagg gtactcatac
3000tcacttctaa gtgacggctg catactaacc gcttcataca tctcgtagat
ttctctggcg 3060attgaagggc taaattcttc aacgctaact ttgagaattt
ttgtaagcaa tgcggcgtta 3120taagcattta atgcattgat gccattaaat
aaagcaccaa cgcctgactg ccccatcccc 3180atcttgtctg cgacagattc
ctgggataag ccaagttcat ttttcttttt ttcataaatt 3240gctttaaggc
gacgtgcgtc ctcaagctgc tcttgtgtta atggtttctt ttttgtgctc
3300atacgttaaa tctatcaccg caagggataa atatctaaca ccgtgcgtgt
tgactatttt 3360acctctggcg gtgataatgg ttgcagggcc cattttagga
ggcaaaaatg gataagaaat 3420actcaatagg cttagctatc ggcacaaata
gcgtcggatg ggcggtgatc actgatgaat 3480ataaggttcc gtctaaaaag
ttcaaggttc tgggaaatac agaccgccac agtatcaaaa 3540aaaatcttat
aggggctctt ttatttgaca gtggagagac agcggaagcg actcgtctca
3600aacggacagc tcgtagaagg tatacacgtc ggaagaatcg tatttgttat
ctacaggaga 3660ttttttcaaa tgagatggcg aaagtagatg atagtttctt
tcatcgactt gaagagtctt 3720ttttggtgga agaagacaag aagcatgaac
gtcatcctat ttttggaaat atagtagatg 3780aagttgctta tcatgagaaa
tatccaacta tctatcatct gcgaaaaaaa ttggtagatt 3840ctactgataa
agcggatttg cgcttaatct atttggcctt agcgcatatg attaagtttc
3900gtggtcattt tttgattgag ggagatttaa atcctgataa tagtgatgtg
gacaaactat 3960ttatccagtt ggtacaaacc tacaatcaat tatttgaaga
aaaccctatt aacgcaagtg 4020gagtagatgc taaagcgatt ctttctgcac
gattgagtaa atcaagacga ttagaaaatc 4080tcattgctca gctccccggt
gagaagaaaa atggcttatt tgggaatctc attgctttgt 4140cattgggttt
gacccctaat tttaaatcaa attttgattt ggcagaagat gctaaattac
4200agctttcaaa agatacttac gatgatgatt tagataattt attggcgcaa
attggagatc 4260aatatgctga tttgtttttg gcagctaaga atttatcaga
tgctatttta ctttcagata 4320tcctaagagt aaatactgaa ataactaagg
ctcccctatc agcttcaatg attaaacgct 4380acgatgaaca tcatcaagac
ttgactcttt taaaagcttt agttcgacaa caacttccag 4440aaaagtataa
agaaatcttt tttgatcaat caaaaaacgg atatgcaggt tatattgatg
4500ggggagctag ccaagaagaa ttttataaat ttatcaaacc aattttagaa
aaaatggatg 4560gtactgagga attattggtg aaactaaatc gtgaagattt
gctgcgcaag caacggacct 4620ttgacaacgg ctctattccc catcaaattc
acttgggtga gctgcatgct attttgagaa 4680gacaagaaga cttttatcca
tttttaaaag acaatcgtga gaagattgaa aaaatcttga 4740cttttcgaat
tccttattat gttggtccat tggcgcgtgg caatagtcgt tttgcatgga
4800tgactcggaa gtctgaagaa acaattaccc catggaattt tgaagaagtt
gtcgataaag 4860gtgcttcagc tcaatcattt attgaacgca tgacaaactt
tgataaaaat cttccaaatg 4920aaaaagtact accaaaacat agtttgcttt
atgagtattt tacggtttat aacgaattga 4980caaaggtcaa atatgttact
gaaggaatgc gaaaaccagc atttctttca ggtgaacaga 5040agaaagccat
tgttgattta ctcttcaaaa caaatcgaaa agtaaccgtt aagcaattaa
5100aagaagatta tttcaaaaaa atagaatgtt ttgatagtgt tgaaatttca
ggagttgaag 5160atagatttaa tgcttcatta ggtacctacc atgatttgct
aaaaattatt aaagataaag 5220attttttgga taatgaagaa aatgaagata
tcttagagga tattgtttta acattgacct 5280tatttgaaga tagggagatg
attgaggaaa gacttaaaac atatgctcac ctctttgatg 5340ataaggtgat
gaaacagctt aaacgtcgcc gttatactgg ttggggacgt ttgtctcgaa
5400aattgattaa tggtattagg gataagcaat ctggcaaaac aatattagat
tttttgaaat 5460cagatggttt tgccaatcgc aattttatgc agctgatcca
tgatgatagt ttgacattta 5520aagaagacat tcaaaaagca caagtgtctg
gacaaggcga tagtttacat gaacatattg 5580caaatttagc tggtagccct
gctattaaaa aaggtatttt acagactgta aaagttgttg 5640atgaattggt
caaagtaatg gggcggcata agccagaaaa tatcgttatt gaaatggcac
5700gtgaaaatca gacaactcaa aagggccaga aaaattcgcg agagcgtatg
aaacgaatcg 5760aagaaggtat caaagaatta ggaagtcaga ttcttaaaga
gcatcctgtt gaaaatactc 5820aattgcaaaa tgaaaagctc tatctctatt
atctccaaaa tggaagagac atgtatgtgg 5880accaagaatt agatattaat
cgtttaagtg attatgatgt cgatgccatt gttccacaaa 5940gtttccttaa
agacgattca atagacaata aggtcttaac gcgttctgat aaaaatcgtg
6000gtaaatcgga taacgttcca agtgaagaag tagtcaaaaa gatgaaaaac
tattggagac 6060aacttctaaa cgccaagtta atcactcaac gtaagtttga
taatttaacg aaagctgaac 6120gtggaggttt gagtgaactt gataaagctg
gttttatcaa acgccaattg gttgaaactc 6180gccaaatcac taagcatgtg
gcacaaattt tggatagtcg catgaatact aaatacgatg 6240aaaatgataa
acttattcga gaggttaaag tgattacctt aaaatctaaa ttagtttctg
6300acttccgaaa agatttccaa ttctataaag tacgtgagat taacaattac
catcatgccc 6360atgatgcgta tctaaatgcc gtcgttggaa ctgctttgat
taagaaatat ccaaaacttg 6420aatcggagtt tgtctatggt gattataaag
tttatgatgt tcgtaaaatg attgctaagt 6480ctgagcaaga aataggcaaa
gcaaccgcaa aatatttctt ttactctaat atcatgaact 6540tcttcaaaac
agaaattaca cttgcaaatg gagagattcg caaacgccct ctaatcgaaa
6600ctaatgggga aactggagaa attgtctggg ataaagggcg agattttgcc
acagtgcgca 6660aagtattgtc catgccccaa gtcaatattg tcaagaaaac
agaagtacag acaggcggat 6720tctccaagga gtcaatttta ccaaaaagaa
attcggacaa gcttattgct cgtaaaaaag 6780actgggatcc aaaaaaatat
ggtggttttg atagtccaac ggtagcttat tcagtcctag 6840tggttgctaa
ggtggaaaaa gggaaatcga agaagttaaa atccgttaaa gagttactag
6900ggatcacaat tatggaaaga agttcctttg aaaaaaatcc gattgacttt
ttagaagcta 6960aaggatataa ggaagttaaa aaagacttaa tcattaaact
acctaaatat agtctttttg 7020agttagaaaa cggtcgtaaa cggatgctgg
ctagtgccgg agaattacaa aaaggaaatg 7080agctggctct gccaagcaaa
tatgtgaatt ttttatattt agctagtcat tatgaaaagt 7140tgaagggtag
tccagaagat aacgaacaaa aacaattgtt tgtggagcag cataagcatt
7200atttagatga gattattgag caaatcagtg aattttctaa gcgtgttatt
ttagcagatg 7260ccaatttaga taaagttctt agtgcatata acaaacatag
agacaaacca atacgtgaac 7320aagcagaaaa tattattcat ttatttacgt
tgacgaatct tggagctccc gctgctttta 7380aatattttga tacaacaatt
gatcgtaaac gatatacgtc tacaaaagaa gttttagatg 7440ccactcttat
ccatcaatcc atcactggtc tttatgaaac acgcattgat ttgagtcagc
7500taggaggtga cggtggagga ggttctggag gtggaggttc tgctgagtat
gtgcgagccc 7560tctttgactt taatgggaat gatgaagagg atcttccctt
taagaaagga gacatcctga 7620gaatccggga taagcctgag gagcagtggt
ggaatgcaga ggacagcgaa ggaaagaggg 7680ggatgattcc tgtcccttac
gtggagaagt attccggaga ctataaggac cacgacggag 7740actacaagga
tcatgatatt gattacaaag acgatgacga taagtctagg ctcgagtccg
7800gagactataa ggaccacgac ggagactaca aggatcatga tattgattac
aaagacgatg 7860acgataagtc taggatgacc gacgctgagt acgtgagaat
ccatgagaag ttggacatct 7920acacgtttaa gaaacagttt ttcaacaaca
aaaaatccgt gtcgcataga tgctacgttc 7980tctttgaatt aaaacgacgg
ggtgaacgta
gagcgtgttt ttggggctat gctgtgaata 8040aaccacagag cgggacagaa
cgtggcattc acgccgaaat ctttagcatt agaaaagtcg 8100aagaatacct
gcgcgacaac cccggacaat tcacgataaa ttggtactca tcctggagtc
8160cttgtgcaga ttgcgctgaa aagatcttag aatggtataa ccaggagctg
cgggggaacg 8220gccacacttt gaaaatctgg gcttgcaaac tctattacga
gaaaaatgcg aggaatcaaa 8280ttgggctgtg gaacctcaga gataacgggg
ttgggttgaa tgtaatggta agtgaacact 8340accaatgttg caggaaaata
ttcatccaat cgtcgcacaa tcaattgaat gagaatagat 8400ggcttgagaa
gactttgaag cgagctgaaa aacgacggag cgagttgtcc attatgattc
8460aggtaaaaat actccacacc actaagagtc ctgctgttac ttgacaggca
tcaaataaaa 8520cgaaaggctc agtcgaaaga ctgggccttt cgttttatct
gttgtttgcg gccgggtacc 8580gagctcgaat tcactggccg tcgttttaca
acgtcgtgac tgggaaaacc ctggcgttac 8640ccaacttaat cgccttgcag
cacatccccc tttcgccagc tggcgtaata gcgaagaggc 8700ccgcaccgat
cgcccttccc aacagttgcg cagcctgaat ggcgaatggc gattcacaaa
8760aaataggtac acgaaaaaca agttaaggga tgcagtttat gcatccctta
acttacttat 8820taaataattt atagctattg aaaagagata agaattgttc
aaagctaata ttgtttaaat 8880cgtcaattcc tgcatgtttt aaggaattgt
taaattgatt ttttgtaaat attttcttgt 8940attctttgtt aacccatttc
ataacgaaat aattatactt ctgtttatct ttgtgtgata 9000ttcttgattt
ttttctattt aatctgataa gtgagctatt cactttaggt ttaggatgaa
9060aatattctct tggaaccata cttaatatag aaatatcaac ttctgccatt
aaaaataatg 9120ccaatgagcg ttttgtattt aataatcttt tagcaaaccc
gtattccacg attaaataaa 9180tctcatcagc tatactatca aaaacaattt
tgcgtattat atccgtactt atgttataag 9240gtatattacc aaatatttta
taggattggt ttttaggaaa tttaaactgc aatatatcct 9300tgtttaaaac
ttggaaatta tcgtgatcaa caagtttatt ttctgtagtt ttgcataatt
9360tatggtctat ttcaatggca gttacgaaat tacacctctg tactaattca
agggtaaaat 9420gcccttttcc tgagccgatt tcaaagatat tatcatgttc
atttaatctt atatttgtca 9480ttattttatc tatattatgt tttgaagtaa
taaagttttg actgtgtttt atatttttct 9540cgttcattat aaccctcttt
attttttcct ccttataaaa ttagtataat tatagcacga 9600gctctgataa
atatgaacat gatgagtgat cgttaaattt atactgcaat ctgatgcgat
9660tattgaataa aagatatgag agatttatct agtttctttt tttacaagaa
aaaagaaagt 9720tcttaaaggt tttatacttt tggtcgtaga gcacacggtt
taacgactta attacgaagt 9780aaataagtct agtgtgttag actttaatgt
ttttttaagg cattagtgca tttaagcgtc 9840agagcatggc tttatgccga
gaaaactatt ggttggaatg gcgtgtgtgt tagccaaagc 9900tttggcgagt
tggttggggg tttcatggga ttaatcccat gaaagtacca actcaacaac
9960acactaacgc ctgttggttc caaccaatag gaaattggaa taagcaatta
gtataatgag 10020agtataatgt tggtataacg ttagtataat gatgcttttt
ttcattatat tttttatgta 10080ctttaaacct gcacgcttat gtgaattaga
aaaagcttaa tcgcatttca tagattgacc 10140tcccaataac tacgtggtgt
tattgggagg tcaatctatt tcatttgcct cttgctcaaa 10200gttcccaaat
tcgagtaaga ggtatttttg tttttggtcg tcgcctctca ttagtagttc
10260agggtttaac attaatactc cagtttttct ttttataata tttccttctt
ctaagatttt 10320aagtgttgtt attactgttt gtagacttgt tcctgtagct
tttgctattt ctcttgttgt 10380agctatcatt gtattgttac ttaagtggac
attatctagg atatagttaa cgattttaag 10440tttttttccg ccaatcatat
ctaacatact tattaattgc actatatatg cctttacgaa 10500gttaccagac
gtttgtttac ggtataactt gtctacctct atgacttctc cactttcttc
10560gtctatgagc ctctgagagc ctttatagac tgttccatat ctttctttca
tctttttctc 10620actccttatt ttaaactatt ctaactatat cataactgtt
ctaaaaaaaa aagaacattt 10680gttaaaagaa attagaacaa aatgagtgaa
aaattagaac aaacaaattc cttataaacc 10740ttatcatctc aacctatatt
aagattttac ctagttgaat cttcttttct atataaagcg 10800tcggagcata
tcagggggtt atctaacgta aatgctaccc ttcggctcgc tttcgctcgg 10860cattgac
108674366DNAartificial sequenceCAN1 ORF 43ggggttaccg gcccagttgg
attccgttat tggagaaacc caggtgcctg gggtccaggt 60ataata
664466DNAartificial sequenceCAN1 ORF 44ccccaatggc cgggtcaacc
taaggcaata acctctttgg gtccacggac cccaggtcca 60tattat
664566DNAartificial sequencemutant sequence 45ggggttaccg gcccagttgg
attcccttat tggagaaacc caggtgcctg gggtccaggt 60ataata
664640DNAartificial sequencetarget sequence 46aaatggcgag gatacgttct
ctatggagga tggcataggt 404740DNAartificial sequencetarget sequence
47tttaccgctc ctatgcaaga gatacctcct accgtatcca 404847DNAartificial
sequenceCAN1 ORF 48gattccgtta ttggagaaac ccaggtgcct ggggtccagg
tataata 474947DNAartificial sequenceCAN1 ORF 49ctaaggcaat
aacctctttg ggtccacgga ccccaggtcc atattat 475040DNAartificial
sequenceAde1 50taacgataat gtcaattacg aagactgaac tggacggtat
405111PRTartificial sequenceAde1 51Met Ser Ile Thr Lys Thr Glu Leu
Asp Gly Ile1 5 105240DNAartificial sequenceAde1 52taacgataat
gtgaattacg aagactgaac tggacggtat 405340DNAartificial sequenceAde1
53taacgataat gttaattacg aagactgaac tggacggtat 405440DNAartificial
sequenceCan1 54ggggttaccg gcccagttgg attccgttat tggagaaacc
405513PRTartificial sequenceCan1 55Gly Val Thr Gly Pro Val Gly Phe
Arg Tyr Trp Arg Asn1 5 105640DNAartificial sequenceCan1
56ggggttaccg gcccagttgg attcccttat tggagaaacc 405740DNAartificial
sequenceCan1 57ggggttaccg gcccagttgg attccattat tggagaaacc
405840DNAartificial sequenceCan1 58ggggttaccg gcccagttga attcccttat
tggagaaacc 405940DNAartificial sequenceCan1 59ggggttaccg gcccagttgg
attcctttat tggagaaacc 406021DNAartificial sequenceWT-Ade1
60acgataatgt caattacgaa g 216120DNAartificial sequencegalK
61tcaatgggct aactacgttc 206220DNAartificial sequencegalK
62ttaatgggct aactacgttc 206320DNAartificial sequencerpoB
63gctgtctcag tttatggacc 206420DNAartificial sequencerpoB
64cgacagagtc aaatacctgg 206520DNAartificial sequencerpoB
65gctgtctcag tttatggacc 206620DNAartificial sequencerpoB
66gctgtctcag tttatgaacc 206720DNAartificial sequencerpoB
67gctgtctcag tttataaacc 206820DNAartificial sequencerpoB
68cgacagagtc aaatacttgg 206960DNAartificial sequencetarget
69tcgcttgaac atccagcgaa acaggccccc cccatcgagc agaaaacggt ggtggatggc
607060DNAartificial sequencetarget 70tcgcttgaac atccagcgaa
acaggttccc cccatcgagc agaaaacggt ggtggatggc 607160DNAartificial
sequencetarget 71tcgcttgaac atccagcgaa acaggtcccc cccatcgagc
agaaaacggt ggtggatggc 607260DNAartificial sequencetarget
72tcgcttgaac atccagcgaa acaggccccc cccatcgagc agaaaacggt ggtggatggc
607360DNAartificial sequencetarget 73tcgcttgaac atccagcgaa
acaggccccc cccatcgagc agaaaacggt ggtggatggc 607460DNAartificial
sequencetarget 74tcgcttgaac atccagcgaa acaggccccc cccatcgagc
agaaaacggt ggtggatggc 607560DNAartificial sequencetarget
75tcgcttgaac atccagcgaa acaggtcccc cccatcgagc agaaaacggt ggtggatggc
607660DNAartificial sequencetarget 76tcgcttgaac atccagcgaa
acaggccccc cccatcgagc agaaaacggt ggtggatggc 607760DNAartificial
sequencetarget 77tcgcttgaac atccagcgaa acaggccccc cccatcgagc
agaaaacggt ggtggatggc 607860DNAartificial sequencetarget
78tcgcttgaac atccagcgaa acaggcctcc cccatcgagc agaaaacggt ggtggatggc
607960DNAartificial sequencetarget 79tcgcttgaac atccagcgaa
acaggccttc cccatcgagc agaaaacggt ggtggatggc 608060DNAartificial
sequencetarget 80tcgcttgaac atccagcgaa acaggccttc cccatcgagc
agaaaacggt ggtggatggc 608160DNAartificial sequencetarget
81tcgcttgaac atccagcgaa acaggccttc cccatcgagc agaaaacggt ggtggatggc
608260DNAartificial sequencetarget 82tcgcttgaac atccagcgaa
acaggccttc cccatcgagc agaaaacggt ggtggatggc 608360DNAartificial
sequencetarget 83tcgcttgaac atccagcgaa acaggccccc cccatcgagc
agaaaacggt ggtggatggc 608460DNAartificial sequencetarget
84tcgcttgaac atccagcgaa acaggccttt cccatcgagc agaaaacggt ggtggatggc
608560DNAartificial sequencetarget 85tcgcttgaac atccagcgaa
acaggccctt cccatcgagc agaaaacggt ggtggatggc 608660DNAartificial
sequencetarget 86tcgcttgaac atccagcgaa acaggcccct cccatcgagc
agaaaacggt ggtggatggc 608760DNAartificial sequencetarget
87tcgcttgaac atccagcgaa acaggcccca cccatcgagc agaaaacggt ggtggatggc
608860DNAartificial sequencetarget 88tcgcttgaac atccagcgaa
acaggcccct tccatcgagc agaaaacggt ggtggatggc 608960DNAartificial
sequencetarget 89tcgcttgaac atccagcgaa acaggccccc cccatcgagc
agaaaacggt ggtggatggc 609020DNAartificial sequencerpoB1r
90gctgtctcag tttatggacc 209120DNAartificial sequencerpoB1r
91gctgtctcag tttatgaacc 209220DNAartificial sequencegalK 8
92actcacacca ttcaggcgcc 209320DNAartificial sequencegalK 8
93acttacacca ttcaggcgcc 209421DNAartificial sequencegalK 12
94tcaatgggct aactacgttc g 219521DNAartificial sequencegalK 12
95ttaatgggct aactacgttc g 21
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014

D00015
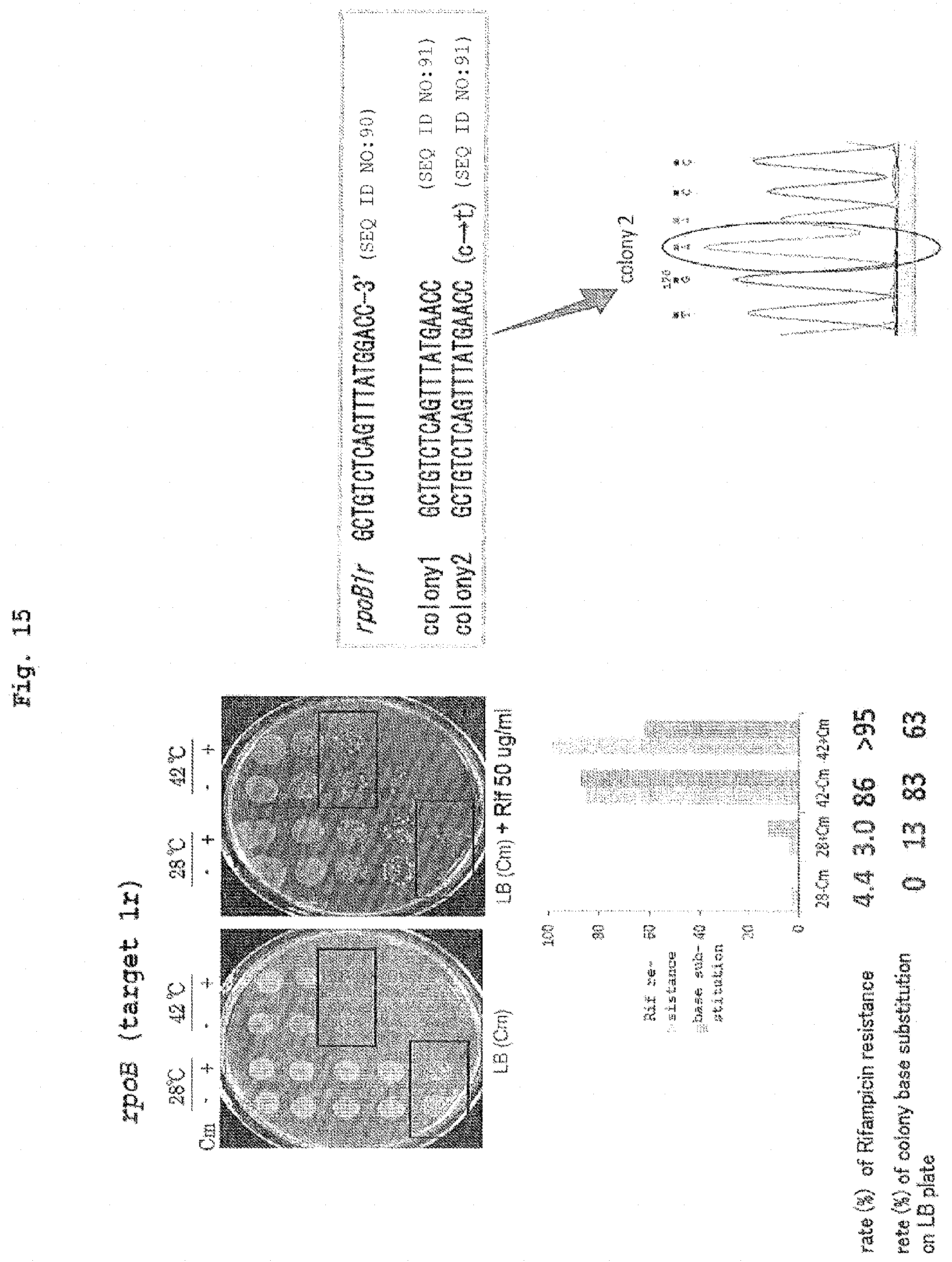
D00016

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.