Tgf-b-receptor Ectodomain Fusion Molecules And Uses Thereof
Zwaagstra; John C. ; et al.
U.S. patent application number 15/755595 was filed with the patent office on 2020-07-23 for tgf-b-receptor ectodomain fusion molecules and uses thereof. This patent application is currently assigned to National Research Council of Canada. The applicant listed for this patent is National Research Council of Canada. Invention is credited to Maria Jaramillo, Anne E.G. Lenferink, Maureen D. O'Connor, Traian Sulea, John C. Zwaagstra.
| Application Number | 20200231652 15/755595 |
| Document ID | / |
| Family ID | 58186979 |
| Filed Date | 2020-07-23 |


View All Diagrams
| United States Patent Application | 20200231652 |
| Kind Code | A1 |
| Zwaagstra; John C. ; et al. | July 23, 2020 |
TGF-B-RECEPTOR ECTODOMAIN FUSION MOLECULES AND USES THEREOF
Abstract
The present invention relates, in general, to polypeptides capable of binding and neutralizing transforming growth factor beta (TGF-beta) ligands, and uses of these polypeptides for treating disorders related to TGF-beta expression or activation (e.g. cancer and fibrotic diseases), and methods of making such molecules.
| Inventors: | Zwaagstra; John C.; (Laval, CA) ; Sulea; Traian; (Kirkland, CA) ; Jaramillo; Maria; (Beaconsfield, CA) ; O'Connor; Maureen D.; (Beaconsfield, CA) ; Lenferink; Anne E.G.; (Lorraine, CA) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Assignee: | National Research Council of
Canada Ottawa ON |
||||||||||
| Family ID: | 58186979 | ||||||||||
| Appl. No.: | 15/755595 | ||||||||||
| Filed: | August 31, 2016 | ||||||||||
| PCT Filed: | August 31, 2016 | ||||||||||
| PCT NO: | PCT/IB2016/055204 | ||||||||||
| 371 Date: | February 27, 2018 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 62212058 | Aug 31, 2015 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C07K 14/71 20130101; C07K 16/30 20130101; C07K 2319/30 20130101; A61P 35/00 20180101; C07K 16/22 20130101; A61K 2039/505 20130101; C12N 15/62 20130101; C07K 19/00 20130101; C07K 2317/94 20130101 |
| International Class: | C07K 14/71 20060101 C07K014/71; C07K 16/22 20060101 C07K016/22; A61P 35/00 20060101 A61P035/00 |
Claims
1. A polypeptide construct comprising: a first portion comprising the second constant domain (C.sub.H2) and/or third constant domain (C.sub.H3) of an antibody heavy chain, and a second portion comprising at least two TGF-.beta. superfamily receptor ectodomains (T.beta.SR-ED) linked in tandem, wherein the N-terminus of the second portion is linked to the C-terminus of the first portion.
2. A polypeptide construct comprising: a first portion comprising the second constant domain (CH2) and/or third constant domain (CH3) of an antibody heavy chain, and a second portion comprising at least one TGF-.beta. superfamily receptor ectodomains (T.beta.SR-ED), wherein the N-terminus of the second portion is linked to the C-terminus of the first portion, and further wherein the first portion does not further comprise an antibody that binds to an antigen that is PD-L1, EGFR1, Her-2, CD4, CD6, CD20, CD25, MUC-1, IL-2, IL-6, or CTLA-4.
3. A polypeptide construct comprising: a first portion comprising the second constant domain (C.sub.H2) and/or third constant domain (C.sub.H3) of an antibody heavy chain, and a second portion comprising at least one TGF-.beta. superfamily receptor ectodomain (T.beta.SR-ED), wherein the N-terminus of the second portion is directly fused to the C-terminus of the first portion.
4. A polypeptide construct comprising a first portion comprising the second constant domain (C.sub.H2) and/or third constant domain (C.sub.H3) of an antibody heavy chain, and a second portion comprising at least one TGF-.beta. superfamily receptor ectodomain (T.beta.SR-ED), wherein the N-terminus of the second portion is linked to the C-terminus of the first portion, and wherein the polypeptide construct neutralizes TGF-.beta. with at least 100-fold more potency than the T.beta.SR-ED alone.
5. The polypeptide construct of claims 2-4, wherein the second portion comprises one T.beta.SR-ED.
6. The polypeptide construct of claim 5, wherein the second portion comprises two T.beta.SR-EDs.
7. The polypeptide construct according to claims 1-6, wherein the T.beta.SR-ED is a TGF-.beta. receptor type II ectodomain (T.beta.R-II-ED).
8. The polypeptide construct of claims 1-6, wherein the T.beta.SR-ED comprises a sequence selected from the group consisting of SEQ ID NO:35, SEQ ID NO:69, SEQ ID NO:75, SEQ ID NO:81, and a sequence substantially identical thereto.
9. The polypeptide construct of claims 1-8, wherein the second portion comprises a sequence selected from the group consisting of SEQ ID NO:43-SEQ ID NO:51, SEQ ID NO:61-SEQ ID NO:68, SEQ ID NO:73, SEQ ID NO:74, SEQ ID NO:79, SEQ ID NO:80, SEQ ID NO:85, SEQ ID NO:86, SEQ ID NO:88, and a sequence substantially identical thereto.
10. The polypeptide construct of any one of claims 1-8, wherein the first portion further comprises a C.sub.H1, a C.sub.H1 and V.sub.H, or C.sub.H1 and scFv.
11. The polypeptide construct of any one of claims 1-10, wherein the antibody heavy chain is of human origin.
12. The polypeptide construct of any one of claims 1-11, wherein the antibody heavy chain is selected from the group consisting of a human IgG1, IgG2, IgG3, or IgG4 heavy chain.
13. The polypeptide construct of any one of claims 1-12, wherein the antibody heavy chain is a human IgG1.
14. The polypeptide construct of claim 4, wherein the polypeptide construct shows longer in vivo half-life compared to the half-life of the second portion alone.
15. The polypeptide construct of any one of claims 1-14, wherein the polypeptide construct is a single chain polypeptide.
16. The polypeptide construct of any one of claims 1-15, wherein the polypeptide construct forms a dimeric polypeptide.
17. The polypeptide construct of claims 1-16, wherein the polypeptide construct is heterodimeric.
18. A polypeptide construct selected from the group consisting of any one of SEQ ID NO:91 to SEQ ID NO:120, and a sequence substantially identical thereto.
19. A polypeptide construct according to claims 1-16, wherein the construct comprises an antibody, antigen binding fragment thereof, or a targeting moiety.
20. A polypeptide construct according to claim 19, comprising the antibody, antigen binding fragment, or targeting moiety at the N-terminus of the first portion.
21. A polypeptide construct according to claim 19, wherein the antigen binding fragment may be selected from the group consisting of a Fv, scFv, Fab, or sdAb.
22. A polypeptide construct according to claim 19, wherein the antigen binding fragment binds to an antigen that is not PD-L1, EGFR1, Her-2, CD4, CD6, CD20, CD25, MUC-1, IL-2, IL-6, or CTLA-4.
23. A polypeptide construct according to claim 19, wherein the antibody is selected from the group consisting of Cetuximab, Avastin, Herceptin, Synagis, and FC5.
24. A polypeptide construct according to claim 23, wherein the antibody is Cetuximab.
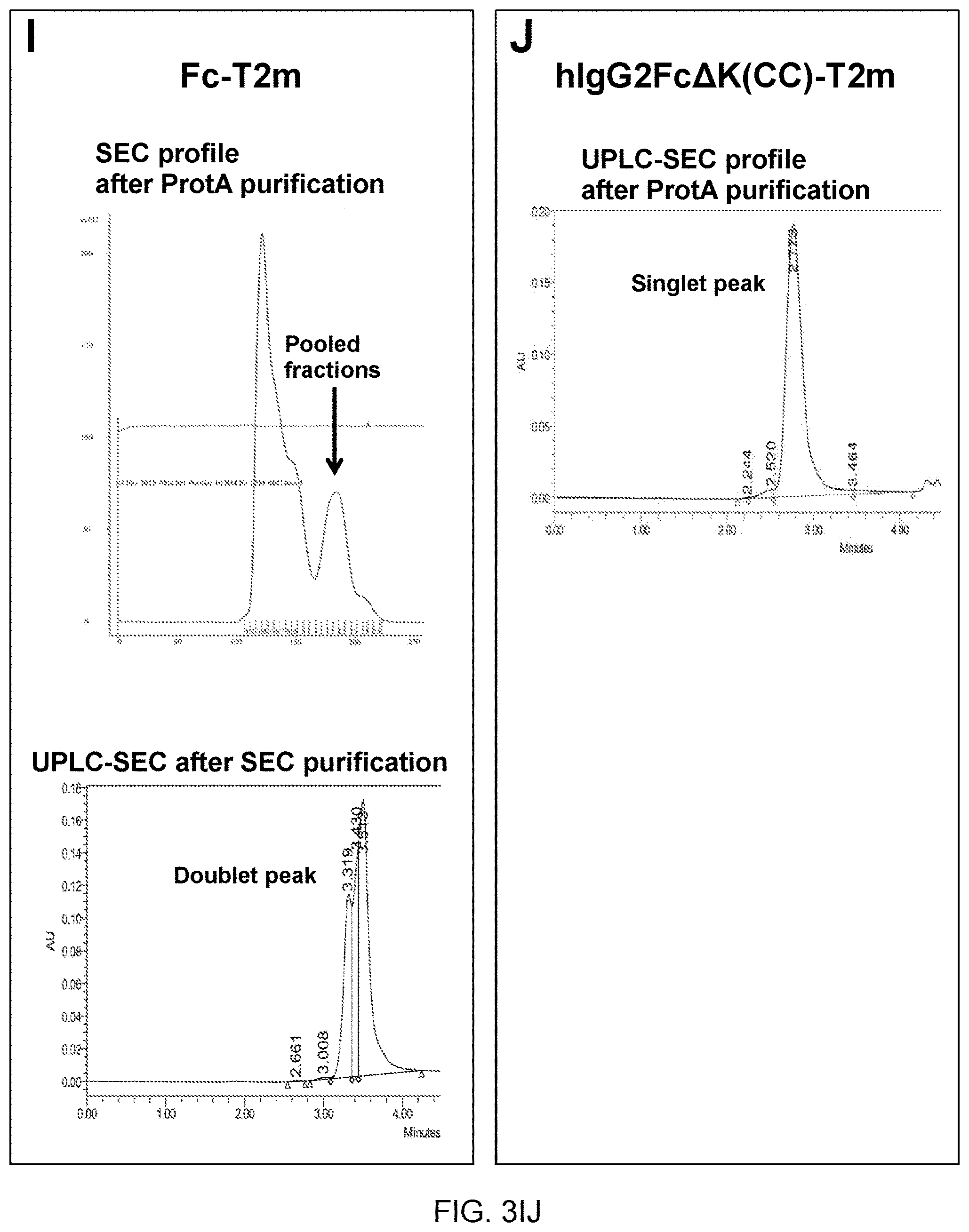
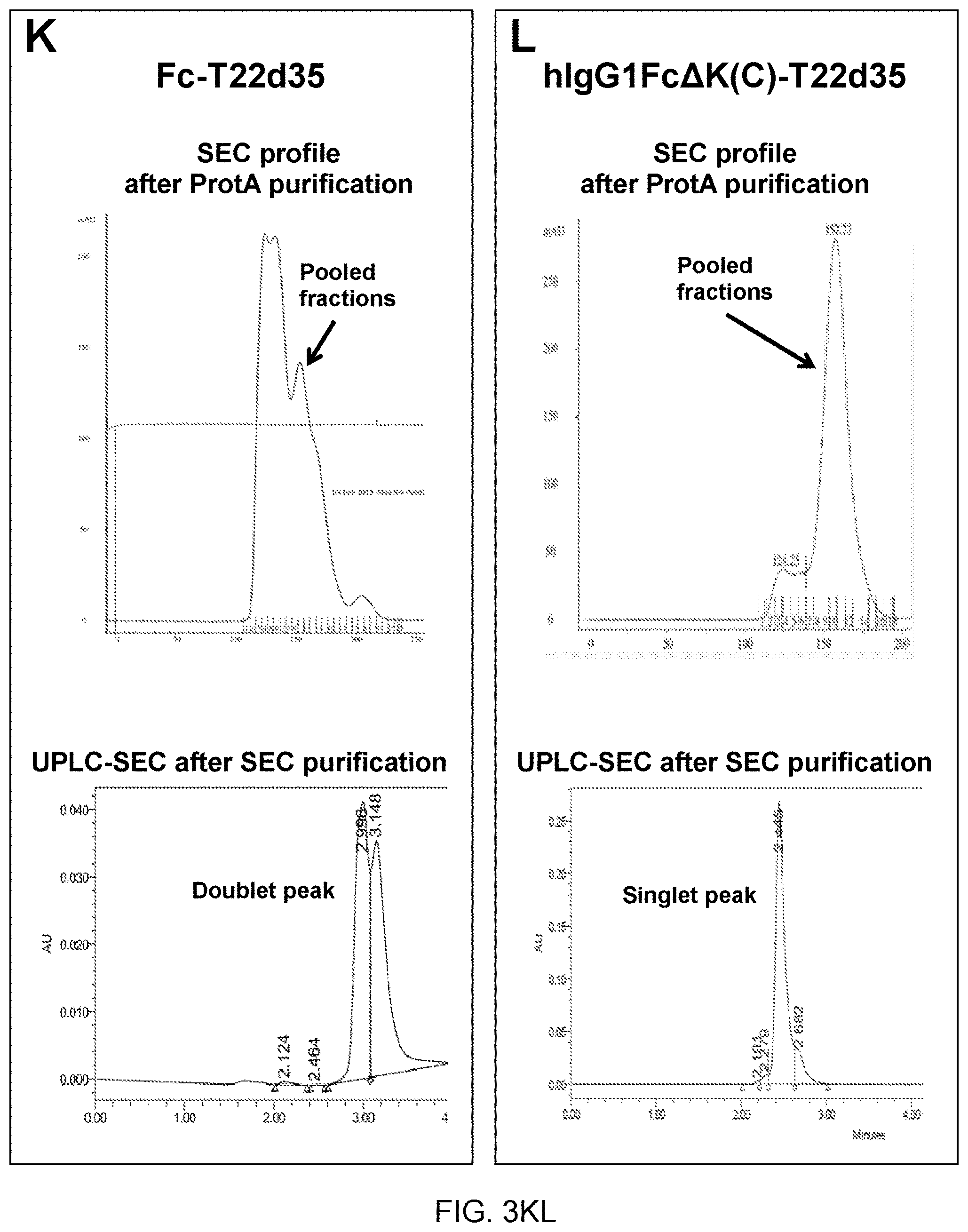
25. A polypeptide construct according to claim 19, wherein the targeting moiety comprises a poly-aspartate sequence motif for bone targeting.
26. A polypeptide construct according to claim 25, wherein the targeting moiety comprises D10.
27. A polypeptide construct according to any preceding claim wherein the construct is a dimeric polypeptide.
28. A polypeptide construct according to claim 27, wherein the dimeric polypeptide comprises: a first single chain polypeptide comprising a first portion comprising the second constant domain (C.sub.H2) and third constant domain (C.sub.H3) of an antibody heavy chain, and a heavy chain variable region of a given antibody; a second portion comprising one or more TGF-.beta. superfamily receptor ectodomains (T.beta.SR-ED), wherein the N-terminus of the second portion is linked to the C-terminus of the first portion, and a second single chain polypeptide comprising a first portion comprising the second constant domain (C.sub.H2) and third constant domain (C.sub.H3) of an antibody heavy chain, and a light chain variable region of said given antibody; a second portion comprising one or more TGF-.beta. superfamily receptor ectodomain (T.beta.SR-ED) which is the same or different from the ectodomain(s) in the first polypeptide, wherein the N-terminus of the second portion is linked to the C-terminus of the first portion.
29. A nucleic acid molecule encoding the polypeptide construct of any preceding claim.
30. A vector comprising the nucleic acid molecule of claim 29.
31. A composition comprising one or more than one independently selected polypeptide construct of any one of claims 1 to 30 and a pharmaceutically-acceptable carrier, diluent, or excipient.
32. A transgenic cellular host comprising the nucleic acid molecule of claim 29 or a vector of claim 30.
33. The transgenic cellular host of claim 32, further comprising a second nucleic acid molecule or a second vector encoding a second polypeptide construct different from the first polypeptide construct.
34. The use of a polypeptide construct according to any one of claims 1-28, for treatment of a medical condition, disease or disorder.
35. The use according to claim 34, wherein the medical condition, disease or disorder comprises cancer, ocular diseases, fibrotic diseases, or genetic disorders of connective tissue.
Description
FIELD OF THE INVENTION
[0001] The present invention relates to TGF-.beta. superfamily receptor ectodomain fusion molecules and uses thereof. More specifically, the present invention relates to TGF-.beta. superfamily receptor ectodomain fusion molecules and their use in TGF-.beta. superfamily ligand neutralization.
BACKGROUND OF THE INVENTION
[0002] TGF-.beta. is part of a superfamily of over 30 ligands that regulate several physiological processes, including cell proliferation, migration and differentiation. Perturbation of their levels and/or signaling gives rise to significant pathological effects. For instance, TGF-.beta. and activin ligands play critical pathogenic roles in many diseases including cancer (Hawinkels & Ten Dijke, 2011; Massague et al, 2000; Rodgarkia-Dara et al, 2006). TGF-.beta., in particular, is considered as a critical regulator of tumor progression and is overexpressed by most tumor types. It favors tumorigenesis in part by inducing an epithelial-mesenchymal transition (EMT) in the epithelial tumor cells, leading to aggressive metastasis (Thiery et al, 2009). TGF-.beta. also promotes tumorigenesis by acting as a powerful suppressor of the immune response in the tumor microenvironment (Li et al, 2006). In fact, TGF-.beta. is recognized as one of the most potent immunosuppressive factors present in the tumor microenvironment. TGF-.beta. interferes with the differentiation, proliferation and survival of many immune cell types, including dendritic cells, macrophages, NK cells, neutrophils, B-cells and T-cells; thus, it modulates both innate and adaptive immunity (Santarpia et al, 2015; Yang et al, 2010). The importance of TGF-beta in the tumor microenvironment is highlighted by evidence showing that, in several tumor types (including melanoma, lung, pancreatic, colorectal, hepatic and breast), elevated levels of TGF-.beta. ligand are correlated with disease progression and recurrence, metastasis, and mortality. Hence, significant effort has been invested in devising anti-tumor therapeutic approaches that involve TGF-.beta. inhibition (Arteaga, 2006; Mourskaia et al, 2007; Wojtowicz-Praga, 2003).
[0003] One approach to developing therapeutic agents that inhibit TGF-.beta. function has been to use antibodies or soluble decoy receptors (also termed receptor ectodomain (ED)-based ligand traps) to bind and sequester ligand, thereby blocking access of ligand to its normal cell surface receptors (Zwaagstra et al, 2012). In general, receptor ED-based traps are a class of therapeutic agents that are able to sequester a wide range of ligands and that can be optimized using protein engineering approaches (Economides et al, 2003; Holash et al, 2002; Jin et al, 2009).
[0004] Previously, a novel protein engineering design strategy was used to generate single-chain, bivalent traps that are able to potently neutralize members of the TGF-.beta. superfamily of ligands due to avidity effects (Zwaagstra et al, 2012) [WO 2008/113185; WO 2010/031168]. In this case, bivalency was achieved via covalent linkage of two T.beta.RII ectodomains using portions of the intrinsically disordered regions (IDR) that flank the structured, ligand-binding domain of T.beta.RII-ED. One example of these single-chain bivalent traps, T22d35, exhibited TGF-.beta. neutralization potencies .about.100-fold higher than the monovalent non-engineered T.beta.RII ectodomain, though it did not neutralize the TGF-.beta.2 isoform and had a relatively short circulating half-life.
[0005] While research to date indicates that single-chain TGF-.beta. traps have promising therapeutic potential, their circulating half-lives and manufacturability present challenges to the commercial application.
SUMMARY OF THE INVENTION
[0006] The present invention relates to TGF-.beta. superfamily receptor ectodomain fusion molecules and uses thereof. More specifically, the present invention relates to TGF-.beta. superfamily receptor ectodomain fusion molecules and their use in TGF-.beta. superfamily ligand neutralization.
[0007] In some aspects, the invention relates to TGF-.beta. superfamily receptor ectodomain-based polypeptides that are similar to typical Fc fusions in design, in that the ectodomain is fused to a dimeric antibody constant domain. In particular, with respect to the present polypeptides, the Fc portion occupies the N-terminal position. Fc fusions in the prior art typically provide the Fc portion at the C-terminal end of the fusion. As will be evident from the results presented herein, this difference in orientation provides a number of significant advantages.
[0008] In other aspects, the present polypeptides incorporate at least two TGF-.beta. superfamily receptor ectodomains that are linked in tandem to the C-terminus of an antibody constant domain.
[0009] Thus, there is provided a polypeptide construct comprising: a first portion comprising the second constant domain (C.sub.H2) and/or third constant domain (C.sub.H3) of an antibody heavy chain, and a second portion comprising at least two TGF-.beta. superfamily receptor ectodomains (T.beta.SR-ED) linked in tandem; wherein the N-terminus of the second portion is linked to the C-terminus of the first portion.
[0010] There is also provided a polypeptide construct comprising: a first portion comprising the second constant domain (CH2) and/or third constant domain (CH3) of an antibody heavy chain, and a second portion comprising at least one TGF-.beta. superfamily receptor ectodomains (T.beta.SR-ED), wherein the N-terminus of the second portion is linked to the C-terminus of the first portion, and further wherein the first portion does not further comprise an antibody that binds to an antigen that is PD-L1, EGFR1, Her-2, CD4, CD6, CD20, CD25, MUC-1, IL-2, IL-6, or CTLA-4.
[0011] There is provided a polypeptide construct comprising: a first portion comprising the second constant domain (C.sub.H2) and/or third constant domain (C.sub.H3) of an antibody heavy chain, and a second portion comprising at least one TGF-.beta. superfamily receptor ectodomain (T.beta.SR-ED), wherein the N-terminus of the second portion is directly fused to the C-terminus of the first portion.
[0012] In an embodiment, there is provided a polypeptide construct comprising a first portion comprising the second constant domain (C.sub.H2) and/or third constant domain (C.sub.H3) of an antibody heavy chain, and a second portion comprising at least one TGF-.beta. superfamily receptor ectodomain (T.beta.SR-ED), wherein the N-terminus of the second portion is linked to the C-terminus of the first portion, and wherein the polypeptide construct neutralizes TGF-.beta. with at least 100-fold more potency than the T.beta.SR-ED alone.
[0013] In a preferred embodiment, the second portion comprises one, two, or multiple TGF-.beta. superfamily receptor ectodomain (T.beta.SR-ED). In a preferred embodiment, the T.beta.SR-ED is a TGF-.beta. receptor type II ectodomain (TI.beta.R-II-ED). In a preferred embodiment, the T.beta.SR-ED comprises a sequence selected from the group consisting of SEQ ID NO:35, SEQ ID NO:69, SEQ ID NO:75, SEQ ID NO:81, and a sequence substantially identical thereto.
[0014] The second portion may comprise a sequence selected from the group consisting of SEQ ID NO:43-SEQ ID NO:51, SEQ ID NO:61-SEQ ID NO:68, SEQ ID NO:73, SEQ ID NO:74, SEQ ID NO:79, SEQ ID NO:80, SEQ ID NO:85, SEQ ID NO:86, SEQ ID NO:88, and a sequence substantially identical thereto.
[0015] In a preferred embodiment, the first portion of a polypeptide construct of the present invention further comprises a C.sub.H1, a C.sub.H1 and V.sub.H, or C.sub.H1 and scFv.
[0016] There is provided a polypeptide construct of the present invention wherein the antibody heavy chain is of human origin. In a preferred embodiment, the antibody heavy chain is selected from the group consisting of a human IgG1, IgG2, IgG3, or IgG4 heavy chain. More preferably, the antibody heavy chain is a human IgG1.
[0017] In accordance with the present invention, the polypeptide construct shows longer in vivo half-life compared to the half-life of the second portion alone.
[0018] There is provided a polypeptide construct of the present invention, wherein the polypeptide construct is a single chain polypeptide.
[0019] In an embodiment, the polypeptide construct forms a dimeric polypeptide. In another embodiment, the polypeptide construct is heterodimeric.
[0020] There is provided a polypeptide construct selected from the group consisting of any one of SEQ ID NO:91 to SEQ ID NO:120, and a sequence substantially identical thereto.
[0021] There is provided a polypeptide construct according to the present invention, wherein the construct comprises an antibody, antigen binding fragment thereof, or a targeting moiety. In a preferred embodiment, the antibody, the antigen binding fragment, or the targeting moiety is at the N-terminus of the first portion.
[0022] In a preferred embodiment, the antigen binding fragment may be selected from the group consisting of a Fv, scFv, Fab, or sdAb. In a preferred embodiment, the antigen binding fragment binds to any antigen, provided that it is not PD-L1, EGFR1, Her-2, CD4, CD6, CD20, CD25, MUC-1, IL-2, IL-6, or CTLA-4.
[0023] In a preferred embodiment, a polypeptide construct of the present invention comprises an antibody selected from the group consisting of Cetuximab, Avastin, Herceptin, Synagis, and FC5. In a preferred embodiment, the antibody is Cetuximab.
[0024] In a preferred embodiment, a polypeptide construct of the present invention comprises a targeting moiety, wherein the targeting moiety comprises a poly-aspartate sequence motif for bone targeting. In a preferred embodiment, the targeting moiety comprises D10.
[0025] There is provided a polypeptide construct according to the present invention wherein the construct is a dimeric polypeptide; wherein the dimeric polypeptide comprises: a first single chain polypeptide comprising a first portion comprising the second constant domain (C.sub.H2) and third constant domain (C.sub.H3) of an antibody heavy chain, and a heavy chain variable region of a given antibody; a second portion comprising one or more TGF-.beta. superfamily receptor ectodomains (T.beta.SR-ED), wherein the N-terminus of the second portion is linked to the C-terminus of the first portion, and a second single chain polypeptide comprising a first portion comprising the second constant domain (C.sub.H2) and third constant domain (C.sub.H3) of an antibody heavy chain, and a light chain variable region of said given antibody; a second portion comprising one or more TGF-.beta. superfamily receptor ectodomain (T.beta.SR-ED) which is the same or different from the ectodomain(s) in the first polypeptide, wherein the N-terminus of the second portion is linked to the C-terminus of the first portion.
[0026] There is also provided a nucleic acid molecule encoding the polypeptide construct of the present invention. There is also provided a vector comprising the nucleic acid molecule of claim the present invention.
[0027] There is also provided a composition comprising one or more than one independently selected polypeptide construct of the present invention and a pharmaceutically-acceptable carrier, diluent, or excipient.
[0028] There is also provided a transgenic cellular host comprising the nucleic acid molecule or a vector of the present invention. The transgenic cellular host further comprising a second nucleic acid molecule or a second vector encoding a second polypeptide construct different from the first polypeptide construct.
[0029] There is also provided the use of a polypeptide construct according to the present invention for treatment of a medical condition, disease or disorder; wherein the medi medical condition, disease or disorder comprises, but is not limited to, cancer, ocular diseases, fibrotic diseases, or genetic disorders of connective tissue.
[0030] In a preferred embodiment, there therefore provided a polypeptide construct comprising: [0031] a first portion comprising the second constant domain (CH2) and/or third constant domain (CH3) of an antibody heavy chain, and [0032] a second portion comprising at least two TGF-.beta. superfamily receptor ectodomains (T.beta.SR-ED), [0033] wherein the N-terminus of the second portion is linked to the C-terminus of the first portion.
[0034] The antibody constant domain can further comprise, either linked thereto or formed integrally therewith, a binding agent such as a full size antibody, a ligand or any other protein of interest. In the alternative, the antibody constant domain comprises only the CH2 and/or CH3 regions, and not a full size antibody. In these and other types of constructs, the CH2 and/or CH3 region can be altered by deleting or substituting amino acids including one or more of the cysteines that provide cross-linking when the present constructs are provided as dimeric constructs.
[0035] In other aspects of the present invention, there is provided a polypeptide construct that incorporates one or more such ectodomains. When the constructs comprise only one ectodomain linked to the antibody constant domain, then the construct is further characterized by at least one of the following: (1) when the constant domain further comprises a full sized antibody, that antibody does not bind effectively to PD-L1 or to an immunoregulatory antigen selected, (2) the constant domain comprises only the CH2 and/or CH3 regions, (3) the constant domain comprises an amino acid alteration relative to a wild type counterpart, such as a cysteine residue alteration; and (4) the first portion is linked to the second portion directly and without intervening amino acids.
[0036] In another of its aspects, the present invention provides a polypeptide construct comprising [0037] a first portion comprising the second constant domain (C.sub.H2) and/or third constant domain (C.sub.H3) of an antibody heavy chain, and [0038] a second portion comprising at least one TGF-.beta. superfamily receptor ectodomain (T.beta.SR-ED), wherein
[0039] the N-terminus of the second portion is linked to the C-terminus of the first portion. These polypeptide constructs can neutralize TGF-.beta., and with at least 100-fold more potency than the T.beta.SR-ED alone.
[0040] The second portion of the polypeptide construct of the present invention may comprise one or two or more T.beta.SR-ED. In a preferred embodiment the construct comprises two or more independently selected ectodomains linked in tandem and to the C-terminus of the constant domain. The T.beta.SR-ED may be selected from the group consisting of a TGF-.beta. receptor type II ectodomain (T.beta.RII-ED), a bone morphogenetic protein receptor type la ectodomain (BMPR-ED), an activin receptor type IIa ectodomain (ActRIIA-ED), and an activin receptor type IIb ectodomain (ActRIIb-ED). In another preferred embodiment, the ectodomain is a TI.beta.R-II ectodomain.
[0041] In the polypeptide construct described herein, the first portion further may comprise a C.sub.H1, a C.sub.H1 and V.sub.H, or a C.sub.H1 and scFv. It may constitute an Fc region, an antibody, or any ligand binding agent or moiety.
[0042] The polypeptide construct of the present invention may comprise a C.sub.H2 and C.sub.H3 from an antibody heavy chain that is of human or mouse origin. For example, and without wishing to be limiting, the antibody heavy chain may be selected from the group consisting of a human IgG1, IgG2, IgG3, or IgG4 heavy chain. In embodiments, the constant domain in the constructs is CH2 per se, or CH3 per se or CH2-CH3.
[0043] The polypeptide construct described herein may show longer in vivo half-life compared to the half-life of T.beta.SR-ED alone.
[0044] In one example, the polypeptide construct of the present invention may be a single chain polypeptide. The polypeptide construct as described herein may also form a dimeric polypeptide. This dimeric polypeptide may be heterodimeric.
[0045] The present invention further provides a polypeptide construct comprising [0046] a first portion comprising the second constant domain (C.sub.H2) and/or third constant domain (C.sub.H3) of an antibody heavy chain, and [0047] a second portion comprising at least one TGF-.beta. superfamily receptor ectodomain (T.beta.SR-ED),
[0048] wherein the N-terminus of the second portion is linked to the C-terminus of the first portion; additionally, in the construct as just described, the first portion is not derived from certain antibodies discussed infra.
[0049] The present invention also provides a polypeptide construct, comprising: [0050] a first single chain polypeptide comprising a first portion comprising the second constant domain (C.sub.H2) and/or third constant domain (C.sub.H3) of an antibody heavy chain, and a heavy chain variable region of a given antibody; and a second portion comprising one or more TGF-.beta. superfamily receptor ectodomains (T.beta.SR-ED),
[0051] wherein the N-terminus of the second portion is linked to the C-terminus of the first portion, and [0052] a second single chain polypeptide comprising a first portion comprising the second constant domain (C.sub.H2) and/or third constant domain (C.sub.H3) of an antibody heavy chain, and a light chain variable region of said given antibody; and a second portion comprising one or more TGF-.beta. superfamily receptor ectodomain (T.beta.SR-ED) which is the same or different from the ectodomain(s) in the first polypeptide, wherein the N-terminus of the second portion is linked to the C-terminus of the first portion.
[0053] In alternative constructs of the present invention, the polypeptide construct comprises an antibody Fc fragment linked at the C-terminus of each heavy chain to at least one TGF-.beta. superfamily receptor ectodomain (T.beta.SR-ED), as described above. In embodiments the receptor ectodomain portion comprises two independently selected ectodomains that are linked in tandem, i.e., in a linear manner. In some embodiments, the ectodomains are the same in sequence, or least the same with respect to their target ligand. The construct may further comprise a binding fragment or moiety at the N-terminus of the Fc; the binding fragment may be selected from the group consisting of a Fv, scFv, Fab, or sdAb, or any other binding moiety such as a motif for bone targeting, also as described above. In the polypeptide constructs as described above, the TGF-.beta. receptor ectodomain does not interfere in the native function or specificity of the binding fragment.
[0054] The present invention also provides a nucleic acid molecule encoding the polypeptide constructs as described herein. A vector comprising the nucleic acid molecule just described is also encompassed by the invention. The invention also includes a transgenic cellular host comprising the nucleic acid molecule or a vector as described herein; the cellular host may further include a second nucleic acid molecule or a second vector encoding a second polypeptide construct different from the first polypeptide construct. Systems used to produce the present polypeptides can be secretion systems, particularly in the case where dimerization through disulfide bridges is required, and the expression polynucleotides thus encode secretion signals that are cleaved by the host upon secretion into the culturing medium.
[0055] Compositions comprising one or more than one independently selected polypeptide construct described herein and a pharmaceutically-acceptable carrier, diluent, or excipient are also encompassed by the present invention.
[0056] Additional aspects and advantages of the present invention will be apparent in view of the following description. The detailed descriptions and examples, while indicating preferred embodiments of the invention, are given by way of illustration only, as various changes and modifications within the scope of the invention will become apparent to those skilled in the art in light of the teachings of this invention.
BRIEF DESCRIPTION OF THE DRAWINGS
[0057] These and other features of the invention will now be described by way of example, with reference to the appended drawings, wherein:
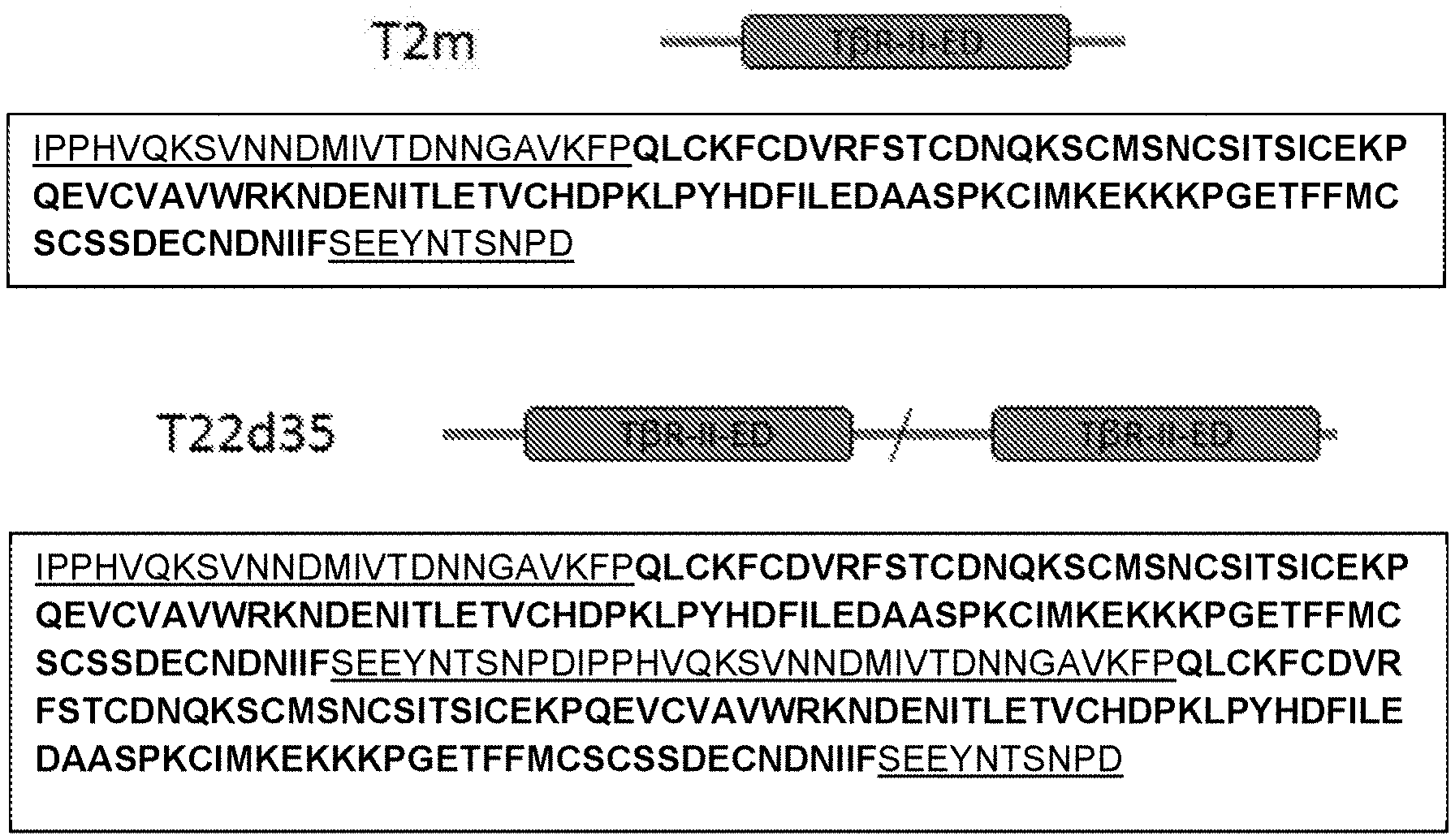
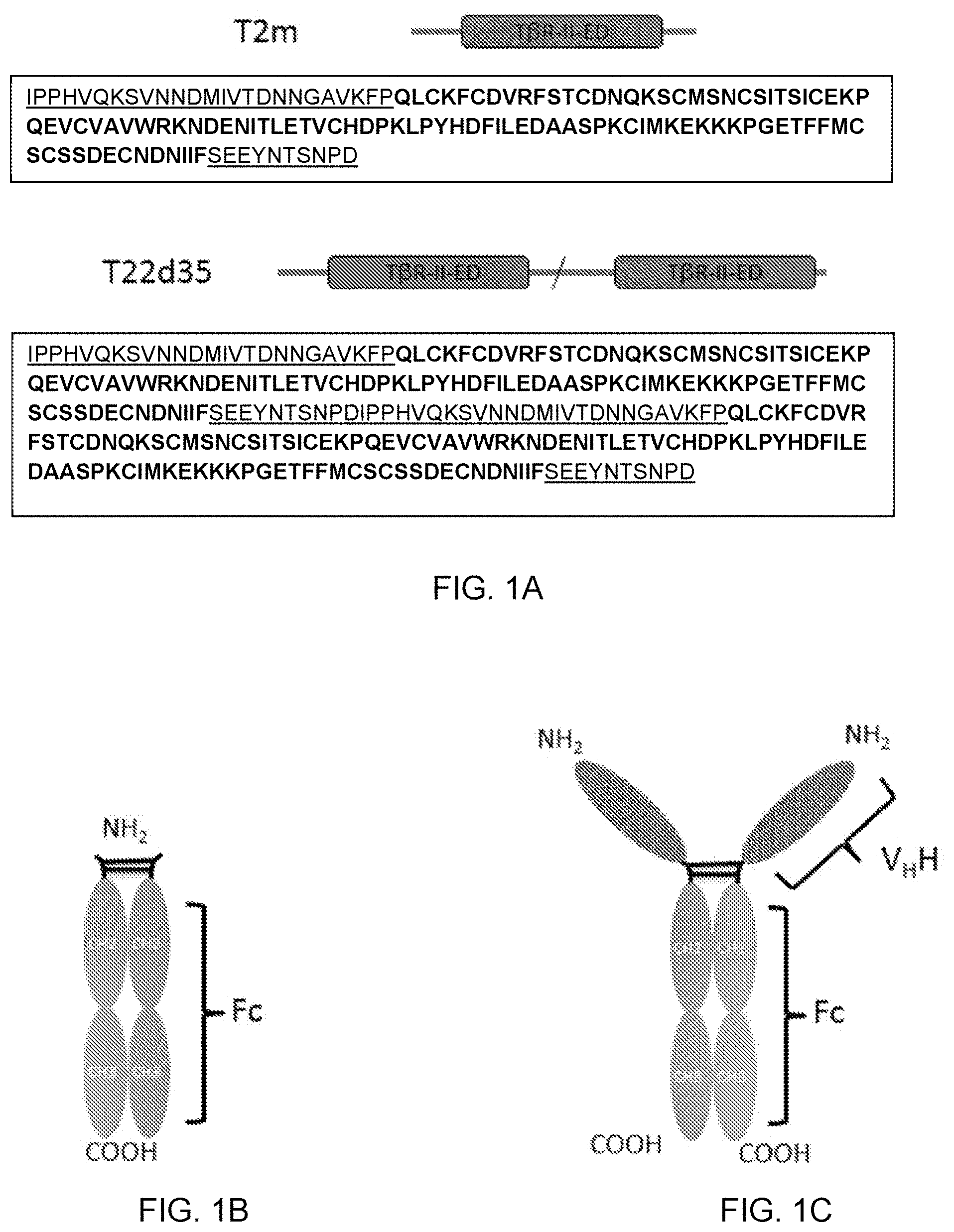
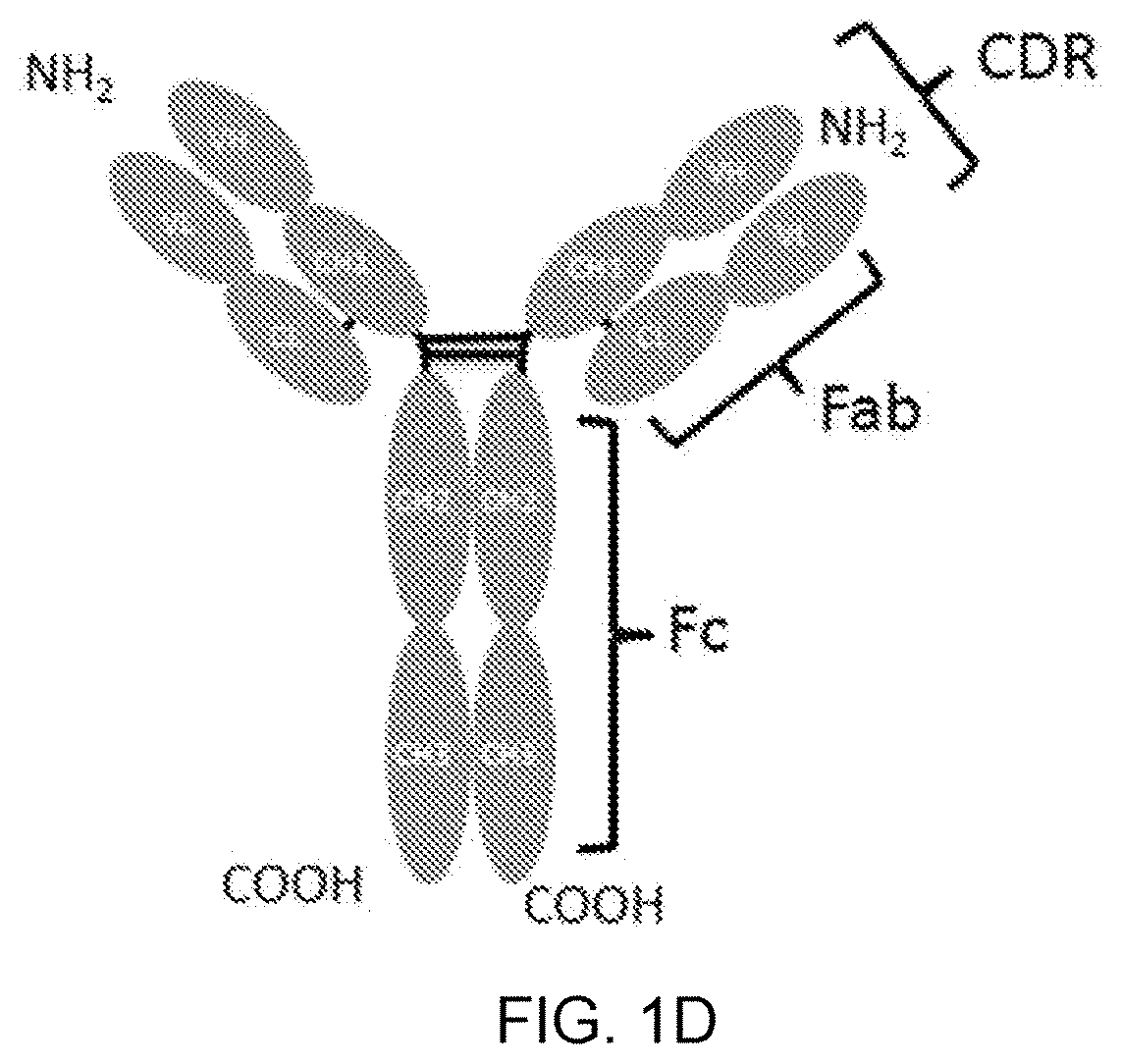
[0058] FIG. 1A is a schematic diagram showing TGF-.beta. Type II receptor ectodomain (T.beta.RII-ED)-based molecules T2m and T22d35 along with their sequences (SEQ ID NO:43 and 46, respectively). Natural linker sequences (SEQ ID NO:36, 39 and 40) are underlined and depicted as dark grey lines; the sequence of the TI.beta.R-II-ED structured domain (SEQ ID NO:35) is shown in bold, and the domain labeled and depicted in dark grey; the site of the fusion of natural linkers is depicted by a slash. FIGS. 1B-D are schematic diagrams of IgG Fc-based scaffolds: an IgG Fc region (FIG. 1B), a V.sub.HH-IgG Fc (comprising a V.sub.HH single domain antibody fused to the N-terminus of an Fc region; FIG. 1C), and a full-size antibody (FIG. 1D).
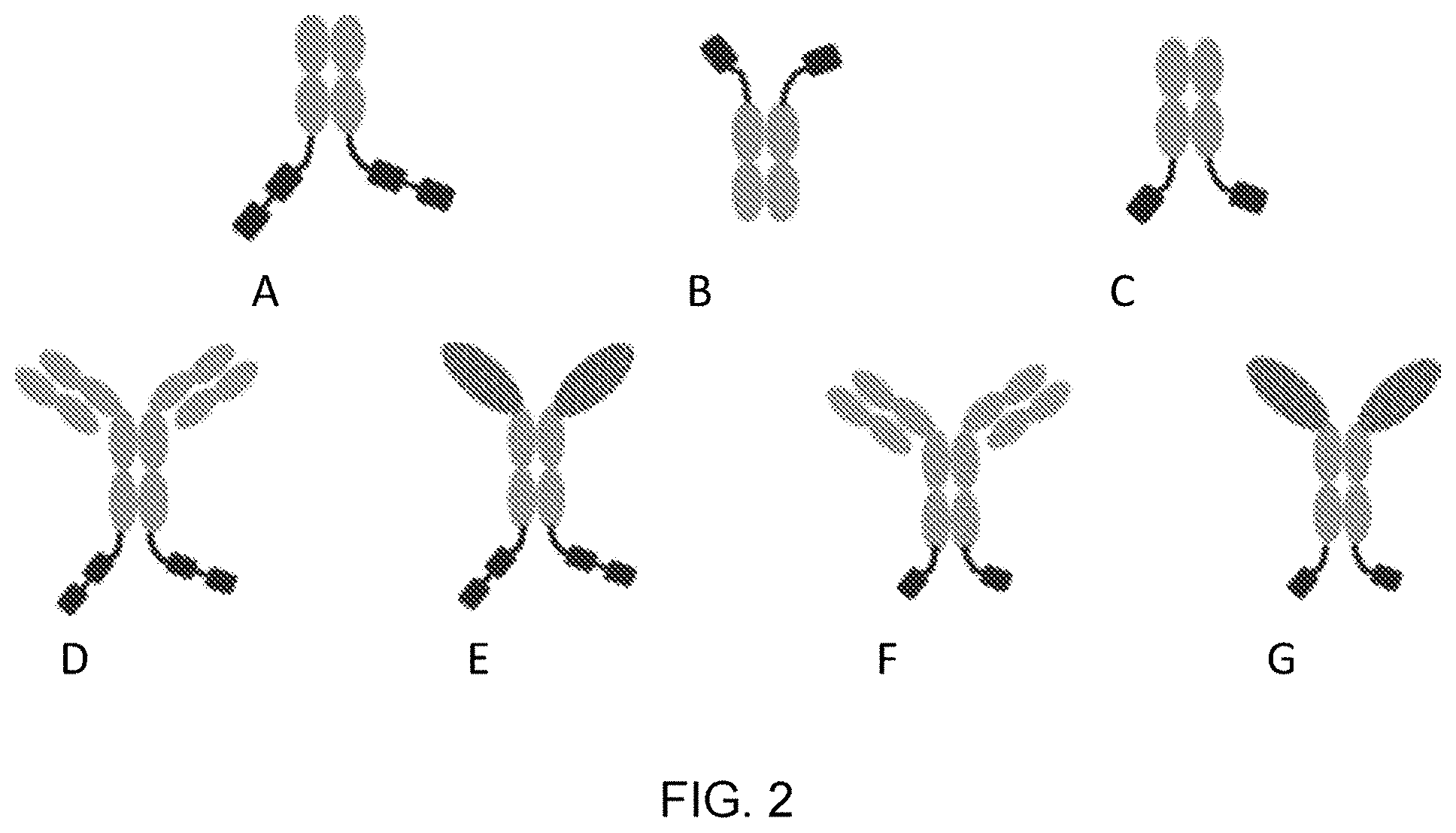
[0059] FIG. 2 is a schematic representation of TGF-.beta. superfamily receptor-ectodomain-based fusion constructs of the present invention. (A) represents constructs in which T22d35 (dark grey) is fused to the C-terminus of IgG Fc regions (IgG isoforms 1, 2, 3 or 4) with no Fab or other functional binding moiety at the N-terminus (Fc-T22d35, A), (D) represents constructs in which T22d35 (dark grey) is fused to the C-terminus of the IgG Fc region of full-size antibodies with heavy and light chain Fabs (FSA-T22d35, D), (E) represents constructs in which T22d35 (dark grey) is fused to the C-terminus of IgG Fc regions that have a non-Fab binding/localization moiety at the N-terminus, such as the variable region of a camelid V.sub.HH antibody (V.sub.HH-Fc-T22d35) or a deca-aspartate motif for bone targeting (D10-Fc-T22d35). Similarly, (B), (C) and (F) and (G) represent constructs in which T2m (the TGF-.beta. Type II receptor ectodomain, T.beta.RII-ED--dark grey) is fused at the N-terminus of an IgG Fc (T2m-Fc, B) or the C-terminus of IgG Fc regions with no Fab or other functional binding moiety at the N-terminus (Fc-T2m, C), or the C-terminus of full-size antibodies with heavy and light chain Fabs (FSA-T2m, F), or the C-terminus of IgG Fc regions that have a non-Fab binding/localization moiety at the N-terminus, such as the variable region of a camelid V.sub.HH antibody (V.sub.HH-Fc-T2m, G) or a deca-aspartate motif for bone targeting (D10-Fc-T2m).
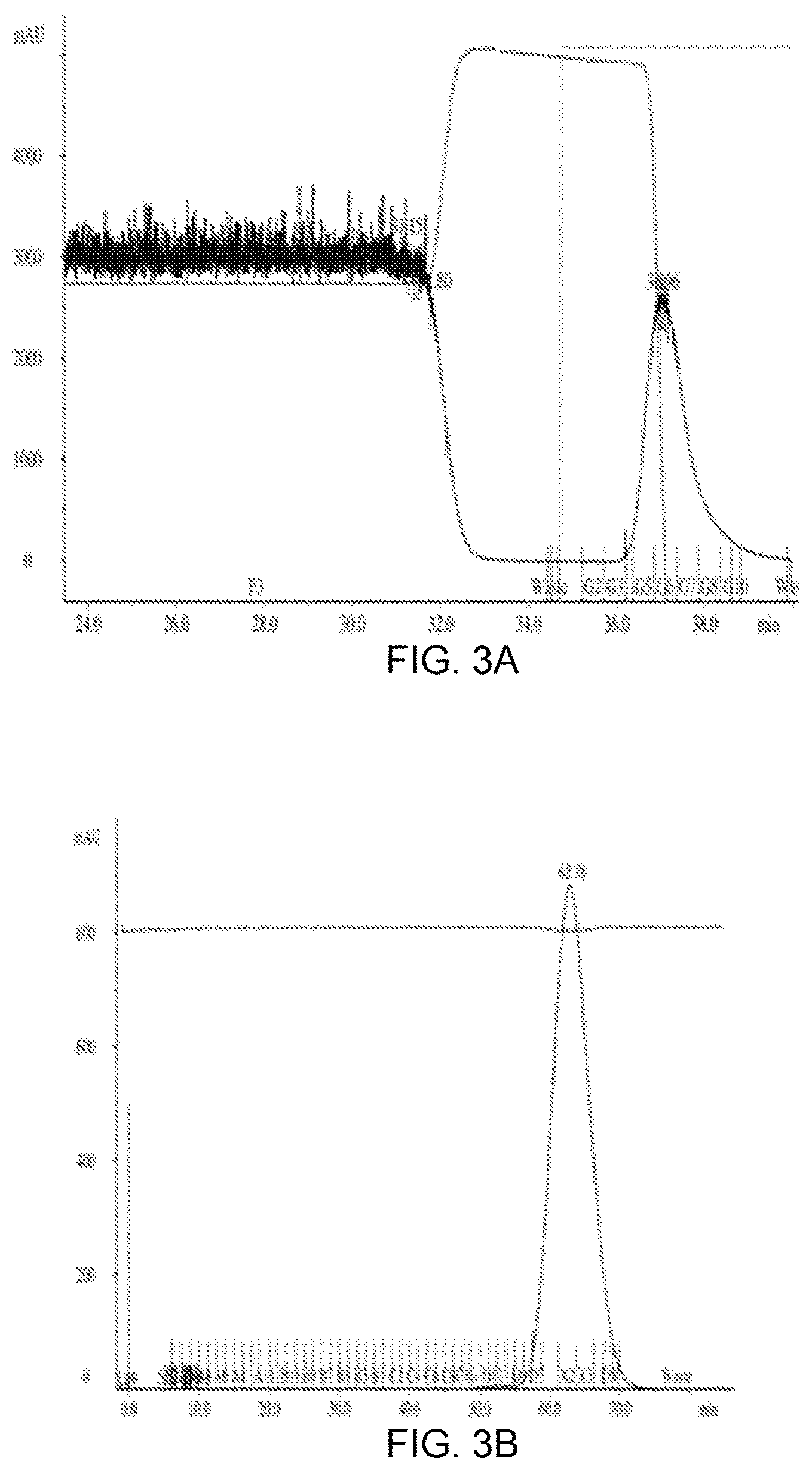
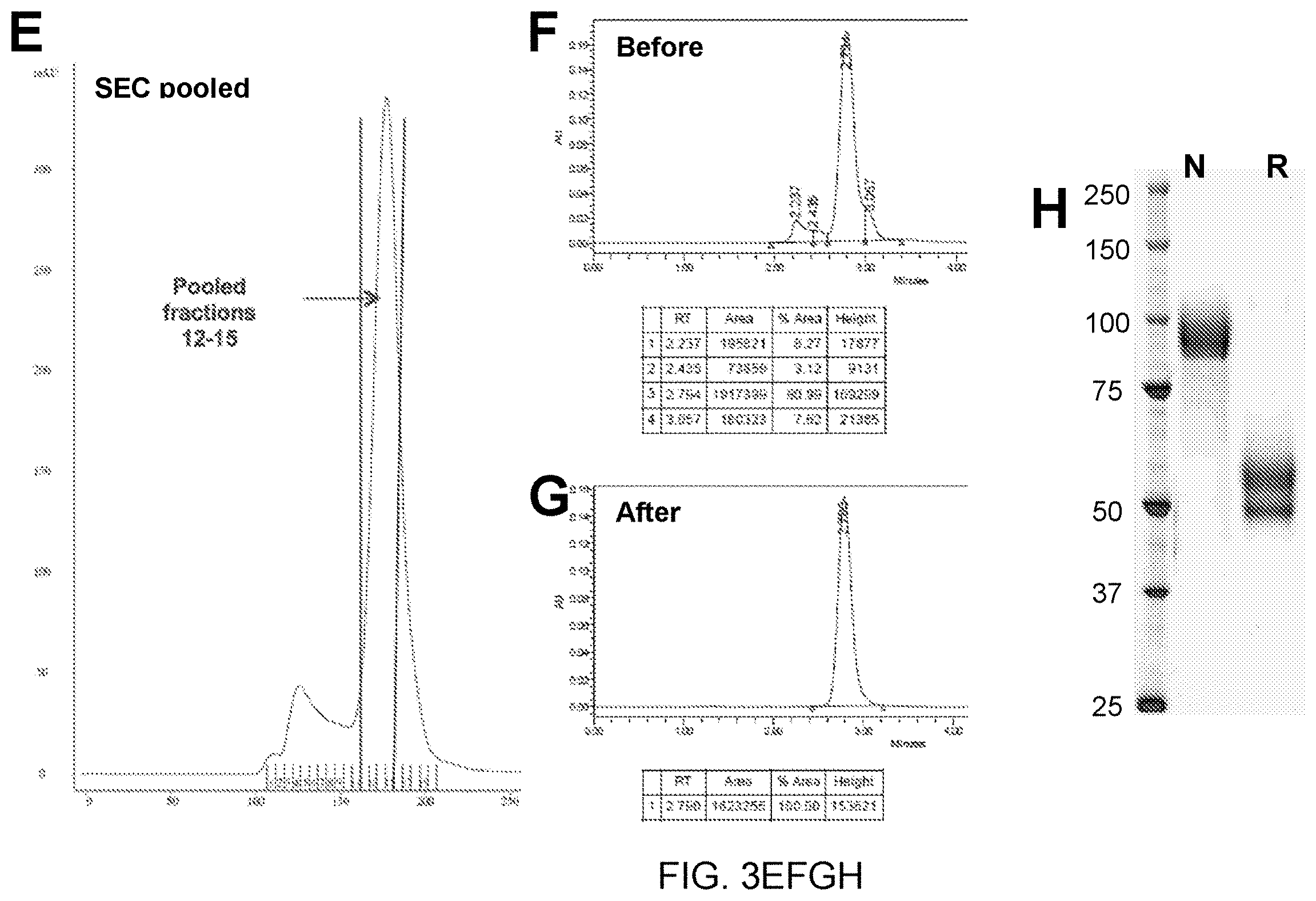
[0060] FIG. 3 presents (ProtA)-affinity column elution profiles, size exclusion (SEC) purification profiles, SDS-PAGE gels, and UPLC-SEC profiles of representatives of constructs type C and D in FIG. 2. FIG. 3A is a (ProtA)-affinity column elution profile for T22d35 fused to the Cetuximab FSA (Cet-T22d35--a representative of construct D in FIG. 2). FIG. 3B is the size exclusion (SEC) purification profile of the Cet-T22d35. FIG. 3C show 4-15% SDS-PAGE gels of ProtA-purified Cet-T22d35 under reducing (left panel) and non-reducing (right panel) conditions (CetHC-T22d35, Cetuximab heavy chain fused to T22d35; CetLC, Cetuximab light chain). Lanes 1 are the pooled Prot-A eluted fractions, while lanes 2 are the pooled SEC fractions. FIG. 3D shows the UPLC-SEC profile of ProtA-purified Cet-T22d35. FIG. 3E shows the (ProtA)-affinity column elution profile for hIgG1Fc.DELTA.K(C)-T2m (a construct with T2m fused to an Fc region with no functional binding moiety at the N-terminus; a representative of construct D in FIG. 2). FIG. 3F, G, H show the UPLC-SEC profile before SEC (F), the UPLC-SEC profile after SEC (G) and the SDS-PAGE (NR & R) (H) of hIgG1Fc.DELTA.K(C)-T2m.
[0061] FIG. 4A shows graphs depicting the efficient inhibition of TGF-.beta.1 (top panel), TGF-.beta.3 (middle panel) and TGF-.beta.2 (bottom panel) signaling in Mv1 Lu luciferase reporter cells by Cet-T2m (a representative of construct F in FIG. 2) and Cet-T22d35 (a representative of construct D in FIG. 2), compared to the significantly lower inhibition potency of non-Fc-fused T22d35.
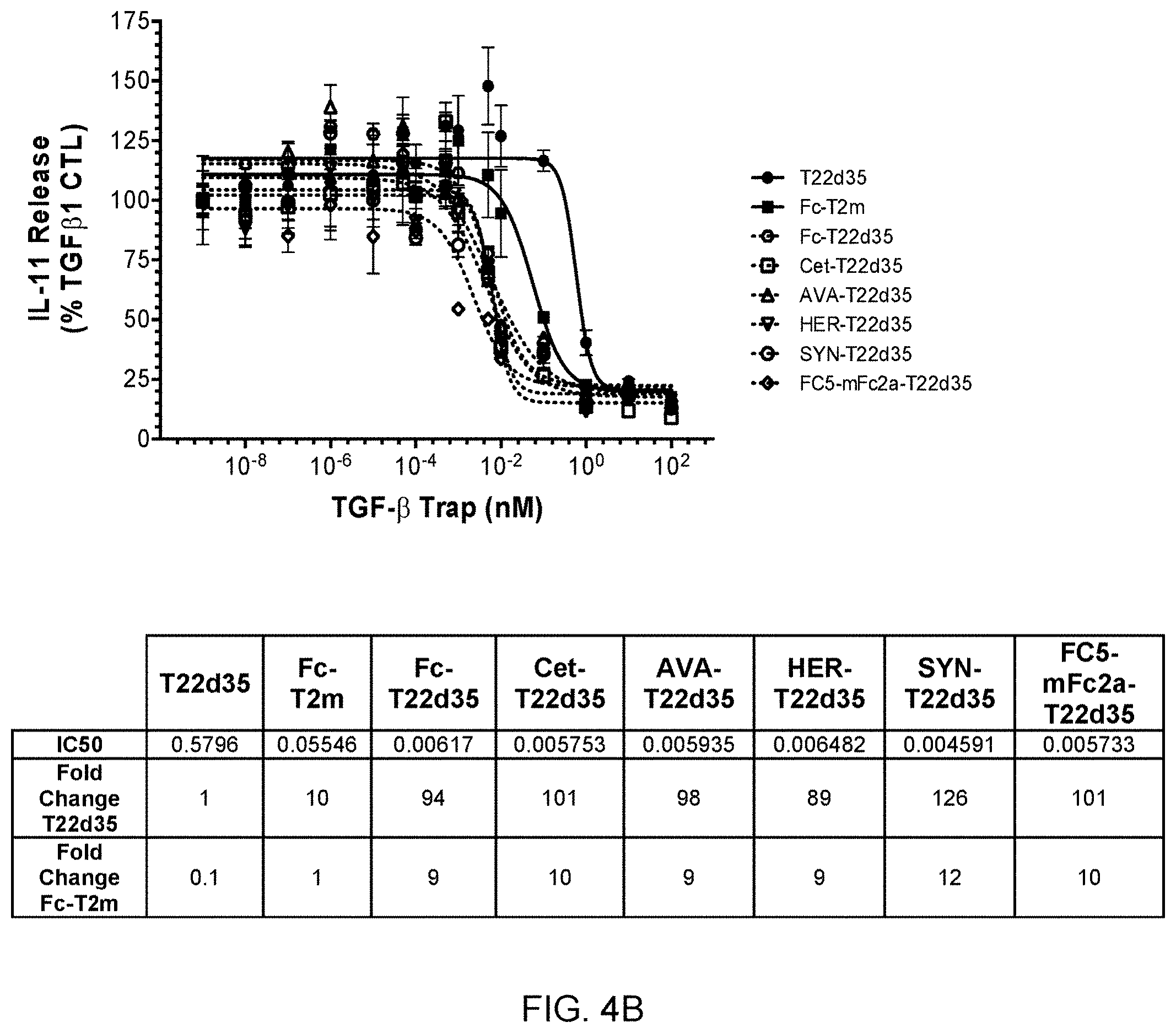
[0062] FIG. 4B shows graphs and a summary table depicting the efficient inhibition of TGF-.beta.1 signaling in an A549/IL-11 cell-based assay by several representatives of FSA-T22d35 constructs (Type D construct from FIG. 2), compared to the lower inhibition potency of Fc-T2m (Type C construct) and non-Fc-fused T22d35.
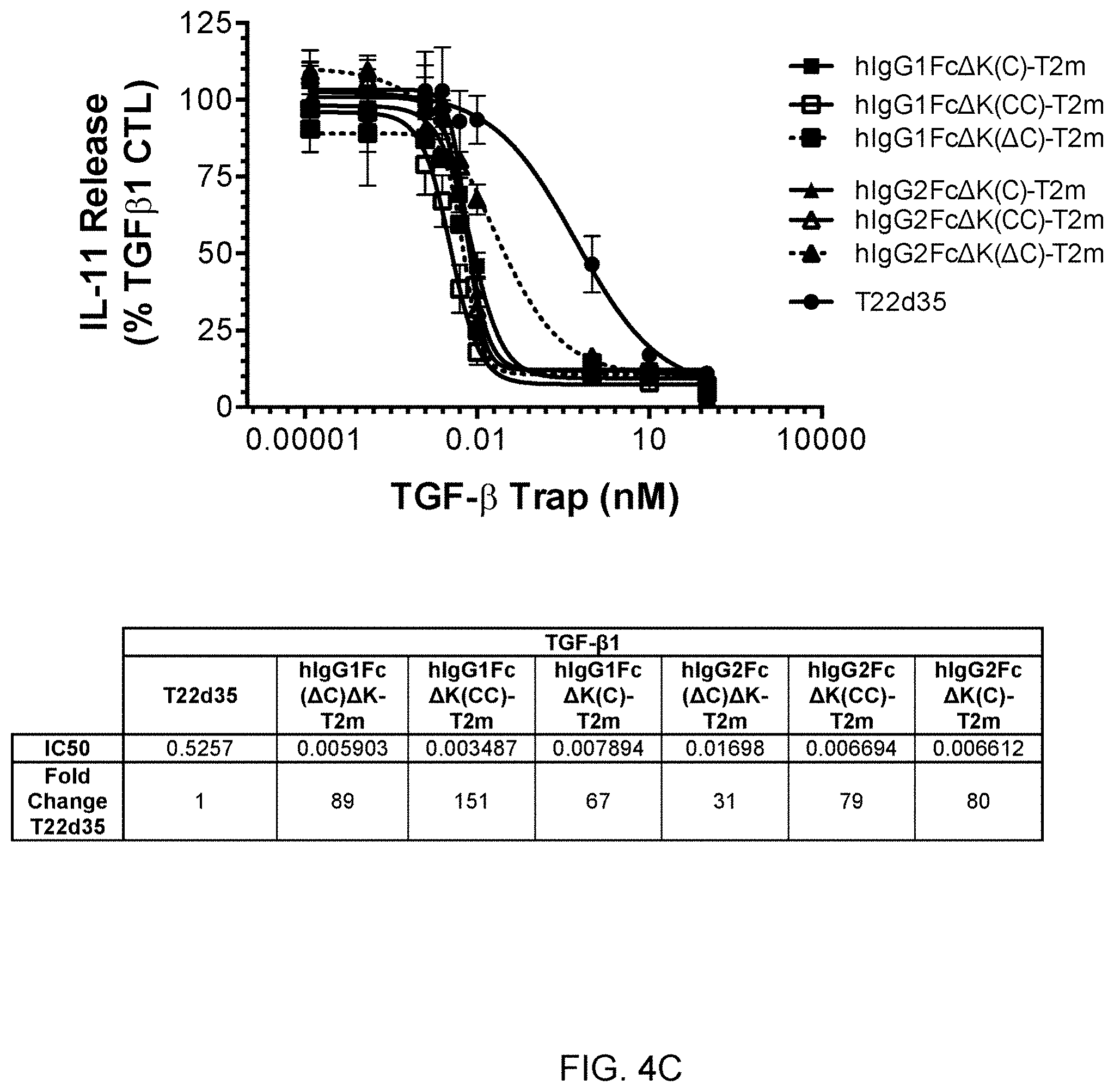
[0063] FIG. 4C shows graphs and a summary table depicting the efficient inhibition of TGF-.beta.1 signaling in an A549/IL-11 release cell-based assay by several representatives of "headless"-T2m constructs (Type C construct from FIG. 2), compared to the lower inhibition potency of non-Fc-fused T22d35.
[0064] FIG. 4D is a graph showing competitive SPR analysis of binding of Cetuximab-fusion constructs to TGF-.beta. isoforms in solution, compared to T22d35.
[0065] FIG. 5A is a SDS-PAGE gel showing the inhibition of EGFR phosphorylation in A549 cells by Cetuximab-fusion constructs. FIG. 5B is a graph showing Cet-T22d35 (triangles) cytotoxicity in MDA-MB-468 and HaCat cells compared to Cetuximab (circles) and T22d35 (squares).
[0066] FIG. 6 is a bar graph showing the apparent permeability coefficient (P.sub.app) values, as a measure of transport of FC5-Fc, FC5-Fc-fusion constructs, T22d35, and T2m across a human brain endothelial cell barrier in vitro, relative to a non-transporting V.sub.HH control (A20.1).
[0067] FIG. 7 demonstrates the Cet-T22d35 inhibition of EGF+TGF-.beta.1 induced EMT in A549 cells. FIG. 7A shows pictures of cultured A549 cells showing their morphologies before treatment (left panel A) and after treatment with EGF+TGF-.beta.1 (right panel B). FIG. 7B shows a western blot of whole cell lysates of A549 cells treated with EGF+TGF-.beta.1 in the presence or absence of various concentrations of Cetuximab (Cetux), Cet-T22d35 or T22d35, probed for the epithelial marker E-Cadherin, while FIG. 7C is the densitometer quantification of the E-cadherin bands in the Western blot. Results show that Cet-T22d35 is much more potent than T22d35 alone or Cetuximab alone in upregulating E-cadherin, i.e. blocking EMT. FIG. 7D shows the inhibition of EGF+TGF-.beta.-induced EMT by Cetuximab (Cetux), Cet-T22d35, T22d35 or Cet-T22d35 plus T22d35 as measured by flow cytometry detection of the epithelial E-Cadherin (top panel) and mesenchymal N-Cadherin (bottom panel) markers.
[0068] FIG. 8A represents the pharmacokinetic (PK) profile of Cet-T22d35 in the serum collected from BALB/C mice that were injected with a single dose of Cet-T22d35. The fusion construct appears to be cleaved in vivo; the terminal half-life of the T22d35 potion of the construct was determined to be 45.8 hours, while the terminal half-life of the Cetuximab portion of the construct was determined to be 262.5 hours. FIG. 8B represents the PK profiles and data table (serum half-lives in bold) for constructs in which the lysine at the C-terminus of the Fc region was removed, i.e. is not present at the fusion joint between the Fc region and T2m (Cet.DELTA.K-T2m, hIgG1Fc.DELTA.K(SS)-T2m, hIgG1Fc.DELTA.K(.DELTA.C)-T2m, and hIg2GFc.DELTA.K(SS)-T2m). The data demonstrate that the removal of the lysine prevents cleavage of the constructs in vivo.
[0069] FIGS. 9A, 9B and 9C present graphs showing the effect of "headless" Fc-T2m constructs (representatives of construct C in FIG. 2) and a FSA-T2m construct (a representative of construct F in FIG. 2) on tumor growth and T-cell function in an immune-competent syngeneic triple negative breast cancer (4T1) model (for comparison, the effects of the 1D11 antibody and non-Fc-fused T22d35 are also shown). The results demonstrate the improved efficacy on T-cell function of two headless-T2m constructs relative to the FSA-T2m construct, and relative to 1D11 and non-Fc-fused T22d35.
[0070] FIG. 10 shows data illustrating enhanced bone localization of two constructs containing a deca-aspartate motif for bone targeting at the N-terminus of the Fc region (D10-hIgG1Fc.DELTA.K(CC)-T2m (SEQ ID NO:136) and D10-GSL-hIgG1Fc.DELTA.K(CC)-T2m (SEQ ID NO:137)--representatives of construct G in FIG. 2). FIGS. 10A and B show results from an A549/IL-11 release cell-based assay demonstrating that the addition of D10 at the N-terminus did not affect TGF-.beta. neutralization potency. FIGS. 10C and D show images demonstrating significant enhancement of the accumulation of the D10-containing constructs in bones relative to a construct without D10.
DETAILED DESCRIPTION OF THE INVENTION
[0071] The present invention relates to TGF-.beta. superfamily receptor ectodomain fusion molecules and uses thereof. More specifically, the present invention relates to TGF-.beta. superfamily receptor ectodomain fusion molecules and their use in TGF-.beta. superfamily ligand neutralization.
[0072] The present invention provides polypeptide constructs, comprising [0073] a first portion comprising the second constant domain (C.sub.H2) and/or third constant domain (C.sub.H3) of an antibody heavy chain, and [0074] a second portion comprising at least one TGF-.beta. superfamily receptor ectodomain (T.beta.SR-ED), wherein the N-terminus of the second portion (ectodomain) is linked to the C-terminus of the first portion (Fc region), and wherein the polypeptide construct neutralizes TGF-.beta. with at least 100-fold more potency than the T.beta.SR-ED alone. The polypeptide construct referred to herein is a synthetic polypeptide produced via protein engineering. It comprises two protein "portions" (or "parts") that are linked to form the chimeric polypeptide construct. When the polypeptide construct is expressed, two polypeptide chains dimerize, such that the C.sub.H2 and C.sub.H3 domains form an antibody Fc region.
[0075] In specific embodiments of the present invention, descriptions of which are elaborated further herein, there are provided polypeptide constructs in which TGF-.beta. superfamily receptor-ectodomains were fused to IgG Fc regions. Specifically, the T2m (single ectodomain) or T22d35 (double ectodomain) moieties were linked (fused) to the C-terminal end of the Fc region. It was observed that fusion constructs of this type have advantages relative to several other versions of receptor-ectodomain based molecules, including non-Fc fused monovalent or multivalent TGF-.beta. receptor ectodomain constructs (such as T2m and T22d35) and constructs in which a receptor ectodomain is fused to the N-terminus of an Fc region. In particular, the constructs have improved manufacturability due to the presence of the Fc region (for example, purification can be accomplished using protein A chromatography). The Fc region also allows for improved circulating half-lives. Importantly, the present constructs have substantially higher TGF-.beta. neutralization potencies compared to T2m and T22d35 alone or to constructs where a receptor ectodomain is fused to the N-terminus of an Fc region. Thus, an advantage of the present invention is unexpected high potency TGF-.beta. superfamily ligand neutralization, including some degree of neutralization of TGF-.beta.2, which was not observed for the non-Fc fused constructs T2m and T22d35. Finally, constructs in accordance with embodiments of the present invention, that is where the ectodomain(s) is/are fused to the C-terminus of an Fc region of an antibody, allows for preservation of the structure and function of the natural N-terminal regions/domains of an antibody; as such, antigen binding to the antibody CDR regions is not perturbed. This leads to the generation of a bifunctional construct able to interact with the target of the antibody while interacting with, and neutralizing, members of the TGF-.beta. superfamily of ligands.
[0076] The invention relates not only to bifunctional constructs, but also to constructs that are monofunctional, and comprise an Fc region that consists only of the CH2 and/or CH3 regions of an antibody constant region. Preferably, the G1, G2 or G4 subclasses are used, and particularly G1 as well as G2. These constructs are monofunctional in the sense that the constant region itself has no particular activity, other than to act as a structure through which dimers of the polypeptide constructs can form. These minimal constant regions can also be altered to provide some benefit, by incorporating the corresponding hinge regions (SEQ ID NO:5-8) and optionally changing the cysteine residue composition. Thus, some or all of the cysteine residues involved in bridging the two Fc fragments or naturally used to bridge between the heavy and light chains of a full-length antibody can be replaced or deleted. These cysteine residues are seen in hinge sequences listed in SEQ ID NO:5-8. First, not all of the naturally-occurring inter-hinge disulfide bonds need to be formed for the Fc homodimerization to occur, while noting that the stability of the Fc homodimer may depend on the number of intermolecular disulphide bridges. Secondly and perhaps more importantly, the presence of hinge-region cysteine residues may become problematic when the Fc region lacks its N-terminal Fab fragment (i.e., is a headless Fc) as in the case of some polypeptide constructs of the present invention. This leads to untethering and exposure of these hinge-region cysteines, and in turn that may result in complex mixtures of high-order polymeric chains, which creates manufacturability issues in addition to potentially diminishing the intended biological activity and efficacy. Because it is practically impossible to predict the outcome of varying the number of inter-hinge disulphide bridges for the "headless" polypeptide constructs of the present invention, we generated a systematic array of N-terminal Fc variants for all four human IgG isotypes either by a deletion approach (in which hinge-region cysteine residues are progressively eliminated by N-terminal truncations) or by a mutagenesis approach (in which hinge-region cysteine residues are progressively mutated to serine from the N-terminus of the hinge region). Non-limiting examples of such N-terminal variants of headless Fc regions are listed in SEQ ID NO:9-34.
[0077] In the present disclosure, an "antibody", also referred to in the art as "immunoglobulin" (Ig), refers to a protein constructed from paired heavy and light polypeptide chains; various Ig isotypes exist, including IgA, IgD, IgE, IgG, and IgM. The structure of an antibody and of each of the domains is well-established and familiar to those of skill in the art, though is summarized herein. When an antibody is correctly folded, each chain folds into a number of distinct globular domains joined by more linear polypeptide sequences; the immunoglobulin light chain folds into a variable (V.sub.L) and a constant (C.sub.L) domain, while the heavy chain folds into a variable (V.sub.H) and three constant (C.sub.H, C.sub.H2, C.sub.H3) domains. Once paired, interaction of the heavy and light chain variable domains (V.sub.H and V.sub.L) and first constant domain (C.sub.L and C.sub.H,) results in the formation of a Fab (Fragment, antigen-binding) containing the binding region (Fv); interaction of two heavy chains results in pairing of C.sub.H2 and C.sub.H3 domains, leading to the formation of a Fc (Fragment, crystallisable). Characteristics described herein for the C.sub.H2 and C.sub.H3 domains also apply to the Fc.
[0078] While the light and heavy chain variable regions show significant sequence diversity between antibodies, the constant regions show less sequence diversity and are responsible for binding a number of natural proteins to elicit important biochemical events. Specifically, and without wishing to be limiting, the Fc fragment binds to endogenous Fc receptors on the surface of lymphocytes.
[0079] The C.sub.H2 and C.sub.H3 domains of the first portion may be of any isotype, including one selected from the group consisting of IgA, IgD, IgE, and IgG. The C.sub.H2 and C.sub.H3 domains may also be from any suitable source. For example and without wishing to be limiting, the C.sub.H2 and C.sub.H3 domains may originate from a human, mouse and other rodents like rats and degu, rabbit, monkey, or other mammalian source. In one example, the C.sub.H2 and C.sub.H3 domains may be of the IgG isotype; in another example, the C.sub.H2 and C.sub.H3 domains are from human.
[0080] In a specific, non-limiting example, the C.sub.H2 and C.sub.H3 domains of the first portion may be of an isotype or comprise a sequence selected from the group consisting of: [0081] a human IgG1, for example but not limited to SEQ ID NO:1 (APELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNA KTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPRE PQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDG SFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK) as comprised in P01857 of the UniProtKB/Swiss-Prot database; [0082] a human IgG2, for example but not limited to SEQ ID NO:2 (APPVAGPSVFLFPPKPKDTLMISRTPEVTCVWDVSHEDPEVQFNWYVDGVEVHNAK TKPREEQFNSTFRVVSVLTVVHQDWLNGKEYKCKVSNKGLPAPIEKTISKTKGQPREP QVYTLPPSREEMTKNQVSLTCLVKGFYPSDISVEWESNGQPENNYKTTPPMLDSDGS FFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK), as comprised in P01859 of the UniProtKB/Swiss-Prot database; [0083] a human IgG3, for example but not limited to SEQ ID NO:3 (APELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVQFKWYVDGVEVHNA KTKPREEQYNSTFRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKTKGQPRE PQVYTLPPSREEMTKNQVSLTCLVKGFYPSDIAVEWESSGQPENNYNTTPPMLDSDG SFFLYSKLTVDKSRWQQGNIFSCSVMHEALHNRFTQKSLSLSPGK), as comprised in P01860 of the UniProtKB/Swiss-Prot database; [0084] a human IgG4, for example but not limited to SEQ ID NO:4 (APEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNA KTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPRE PQVYTLPPSQEEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDG SFFLYSRLTVDKSRWQEGNVFSCSVMHEALHNHYTQKSLSLSLGK), as comprised in P01861 of the UniProtKB/Swiss-Prot database; and [0085] a sequence substantially identical to any of the sequences listed above.
[0086] In the protein constructs of the present invention, the first portion may further comprise a sequence corresponding to the hinge region at the N-terminus of the C.sub.H2 domain. For example, the first portion may further comprise a sequence selected from the group consisting of:
TABLE-US-00001 EPKSCDKTHTCPPCP (SEQ ID NO: 5) for human IgG1; ERKCCVECPPCP (SEQ ID NO: 6) for human IgG2; ELKTPLGDTTHTCPRCPEPKSCDTPPPCPRCPEPKSCDTPPPCPRCPEPK SCDTPPPCPRCP (SEQ ID NO: 7) for human IgG3; ESKYGPPCPSCP (SEQ ID NO: 8) for human IgG4;
[0087] and a sequence substantially identical to any of the sequences listed above.
[0088] Thus, the first portion of the polypeptide construct of the present invention consists of naturally fused Fc and hinge regions for the various IgG isoforms and in embodiments is selected from the group consisting of SEQ ID NO:1-4 for the Fc region and SEQ ID NO:5-8 for the hinge region.
[0089] In specific embodiments, the first portion of the polypeptide construct of the present invention is selected from a group of sequences displaying variation in the N-terminal sequence as exemplified by SEQ ID NO:9-34. These differ in the number of cysteine residues retained from the hinge region as a means to modulating the degree of Fc-region dimerization and hence impacting on both efficacy and manufacturability. Thus, in embodiments, the polypeptide construct comprises a variation in the constant domain, wherein at least one cysteine residue involved in cross-linking is deleted or substituted. Suitable substitutions include serine or alanine, and preferably by serine. A substantially identical sequence may comprise one or more conservative amino acid mutations that still provide for proper folding upon secretion into the culturing medium. It is known in the art that one or more conservative amino acid mutations to a reference sequence may yield a mutant peptide with no substantial change in physiological, chemical, physico-chemical or functional properties compared to the reference sequence; in such a case, the reference and mutant sequences would be considered "substantially identical" polypeptides. A conservative amino acid substitution is defined herein as the substitution of an amino acid residue for another amino acid residue with similar chemical properties (e.g. size, charge, or polarity). These conservative amino acid mutations may be made to the framework regions while maintaining the overall structure of the constant domains; thus the function of the Fc is maintained.
[0090] In a non-limiting example, a conservative mutation may be an amino acid substitution. Such a conservative amino acid substitution may substitute a basic, neutral, hydrophobic, or acidic amino acid for another of the same group. By the term "basic amino acid" it is meant hydrophilic amino acids having a side chain pK value of greater than 7, which are typically positively charged at physiological pH. Basic amino acids include histidine (His or H), arginine (Arg or R), and lysine (Lys or K). By the term "neutral amino acid" (also "polar amino acid"), it is meant hydrophilic amino acids having a side chain that is uncharged at physiological pH, but which has at least one bond in which the pair of electrons shared in common by two atoms is held more closely by one of the atoms. Polar amino acids include serine (Ser or S), threonine (Thr or T), cysteine (Cys or C), tyrosine (Tyr or Y), asparagine (Asn or N), and glutamine (Gln or Q). The term "hydrophobic amino acid" (also "non-polar amino acid") is meant to include amino acids exhibiting a hydrophobicity of greater than zero according to the normalized consensus hydrophobicity scale of (Eisenberg et al, 1984). Hydrophobic amino acids include proline (Pro or P), isoleucine (Ile or I), phenylalanine (Phe or F), valine (Val or V), leucine (Leu or L), tryptophan (Trp or W), methionine (Met or M), alanine (Ala or A), and glycine (Gly or G). "Acidic amino acid" refers to hydrophilic amino acids having a side chain pK value of less than 7, which are typically negatively charged at physiological pH. Acidic amino acids include glutamate (Glu or E), and aspartate (Asp or D).
[0091] In another non-limiting example, a conservative mutation in the C.sub.H2 and/or C.sub.H3 domain may be a substitution that enhances a property selected from the group consisting of the stability, half-life, or Fc properties of C.sub.H2 and/or C.sub.H3 domains or alter glycosylation of the C.sub.H2 and/or C.sub.H3 domain. For example, and without wishing to be limiting in any manner, the mutation may be an alteration at position 228 (EU numbering, 241 according to Kabat) where the serine is substituted by a proline (S228P), which stabilizes the disulfide linkage within the Fc dimer. Another alteration is the mutation at position 409 (EU numbering, 440 according to Kabat) where an arginine is substituted to a lysine for further stabilization of the Fc homodimer at the C.sub.H3-domain level (Yang & Ambrogelly, 2014). Yet another alteration within the C.sub.H2 and/or C.sub.H3 domain may be a substitution of Asn297 (EU numbering, 314 according to Kabat) by glycine or alanine to alter glycosylation of the constant domain. In yet another example, the C.sub.H2 and/or C.sub.H3 domain may be altered by substitution of one or more threonine (T252L, T253S, and/or T256F; see [U.S. 62/777,375]) to increase half-life. Particularly useful are those alterations that enhance Fc properties while remaining silent with respect to conformation, e.g., retaining Fc receptor binding.
[0092] In yet another non-limiting example, the conservative mutations in the C.sub.H2 and/or C.sub.H3 domain may be a substitution that is naturally-occurring. Such mutations may occur in nature as minor sequence differences between species or race.
[0093] Sequence identity is used to evaluate the similarity of two sequences; it is determined by calculating the percent of residues that are the same when the two sequences are aligned for maximum correspondence between residue positions. Any known method may be used to calculate sequence identity; for example, computer software is available to calculate sequence identity. Without wishing to be limiting, sequence identity can be calculated by software such as NCBI BLAST2 service maintained by the Swiss Institute of Bioinformatics (and as found at ca.expasy.org/tools/blast/), or any other appropriate software that is known in the art.
[0094] The substantially identical sequences of the present invention may be at least 90% identical; in another example, the substantially identical sequences may have an identity selected from the group consisting of at least 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, or 100% identical, or any percentage there between, at the amino acid level to sequences described herein. Importantly, the substantially identical sequences retain the activity, specificity, and functionality of the reference sequence. In a non-limiting embodiment, the difference in sequence identity may be due to conservative amino acid mutation(s). In a non-limiting example, the first portion of the polypeptide construct of the present invention may comprise a Fc comprising a sequence selected from the group consisting of a sequence at least 95%, 98% or 99% identical to that of the Fc described herein.
[0095] Accordingly, it will be appreciated that the first portion of a construct will include at least an antibody region that preferably provides for cross-linking of the polypeptide constructs, thereby to provide a dimeric protein. This first portion comprises at least the minimal CH2 and/or CH3 domain. That portion can be altered (i) by substituting or deleting cysteine residues from the hinge regions (SEQ ID NO:5-8) involved in crosslinking between the antibody heavy chains or between the heavy and light chains in order to potentially improve preparation homogeneity and efficacy, and/or (ii) by deleting or suitably replacing (e.g., by mutation to alanine) the terminal lysine residue 447 (EU numbering, 478 according to Kabat) of an IgG heavy chain in order to improve chemical stability of C-terminal fusions to enzymatic proteolysis (e.g., by several serine proteases and typically by trypsin). These changes have a positive impact on potency and/or manufacturability, as revealed herein. The first portion can also be extended to become a full Fc region, by including the CH1 domain. As a full Fc, this portion will provide normal Fc effector functions that include involvement in immune cell recruitment, ADCC, CDC and other antibody functions. Moreover, and in embodiments of the present invention, the first portion can include a complete antibody or any equivalent thereof. In certain embodiments, such as when a construct comprises just one ectodomain that is a TGF-.beta. receptor II ectodomain, there is the proviso that the second portion is not an antibody that binds to an immune checkpoint protein such as PD-L1 (programmed death ligand 1) and is not an antibody that binds to an immunomodulating agent that counteracts immune tolerance of cancer cells, the nature and identity of which is as described in [U.S. Pat. No. 8,815,247], and is not an antibody that binds one of EGFR1, her-2, CD4, CD6, CD20, CD25, MUC-1, IL-2, IL-6, and CTLA-4.
[0096] The second portion of the polypeptide construct of the present invention comprises at least one and preferably two TGF-.beta. superfamily receptor ectodomain/s (T.beta.SR-ED); for example, the second portion may comprise one or two T.beta.SR-ED. The ectodomain of the Transforming Growth Factor-I3 superfamily receptor (T.beta.SR) is the N-terminal extracellular, ligand-binding portion of the receptor. Without wishing to be limiting in any manner, the T.beta.SR ectodomain may bind a molecule selected from the group consisting of TGF-.beta., bone morphogenetic protein (BMP) including BMP2, BMP3, BMP4, BMPS, BMP, BMP6, BMP7, BMP8, BMP9, BMP10, BMP11, BMP12, BMP13, BMP14, an BMP15, activin including activins .beta.A, .beta.B and .beta.C, growth differentiation factor (GDF-1) including GDF-3, GDF-.beta., GDF-9, and GDF-15, nodal, inhibin-.alpha., anti-Mullerian hormone, Lefty-1, Lefty-2, arteman, persephin, neurturin, myostatin, or other known TGF-.beta. superfamily ligands. For example, the T.beta.R ectodomain may be selected from the group consisting of the human TGF-.beta. receptor type II ectodomain (TI.beta.R-II-ED), the human TGF-.beta. receptor type IIb (TI.beta.R-IIb) ectodomain, the human activin receptor type IIa (ActR-IIa) ectodomain, the human activin receptor type IIb (ActR-IIb) ectodomain, or the BMP type Ia (BMPR-Ia) ectodomain.
[0097] In a preferred embodiment the ectodomain binds TGF-.beta.1 and/or TGF-.beta.3. In another preferred embodiment, the ectodomain itself is a human TGF-.beta. receptor type II ectodomain including particularly the TGF-.beta. receptor type IIa (T.beta.RIIa). In one specific, non-limiting example, the T.beta.SR-ED is the TGF-.beta. receptor type II ectodomain (T.beta.RII-ED; SEQ ID NO:35).
[0098] In the second portion as described above, the T.beta.SR ectodomain-based portion may further comprise natural linkers. Appropriate, naturally-derived linkers that can be used to fuse two ectodomains head-to-tail are known to those of skill in the art; for example, and without wishing to be limiting, suitable natural linkers are described in [WO2008/113185].
[0099] In this embodiment, the natural linker, if present, may be selected from the group consisting of
TABLE-US-00002 (SEQ ID NO: 36) IPPHVQKSVNNDMIVTDNNGAVKFP; (SEQ ID NO: 37) IPPHVQKSDVEMEAQKDEIICPSCNRTAHPLRHINNDMIVTDNNGAVKFP; (SEQ ID NO: 39) SEEYNTSNPD; (SEQ ID NO: 40) SEEYNTSNPDIPPHVQKSVNNDMIVTDNNGAVKFP; (SEQ ID NO: 41) SEEYNTSNPDIPPHVQKSDVEMEAQKDEIICPSCNRTAHPLRHINNDMIVT DNNGAVKFP; and a combination thereof.
[0100] In a specific, non-limiting example, the second portion of the polypeptide construct of the present invention may comprise the sequence selected from the group consisting of: [0101] A single TGF-.beta. Type II receptor ectodomain, such as:
TABLE-US-00003 [0101] (SEQ ID NO: 43, also referred to herein as T2m) IPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSIT SICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKE KKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD; (SEQ ID NO: 44) IPPHVQKSDVEMEAQKDEIICPSCNRTAHPLRHINNDMIVTDNNGAVKFPQ LCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETV CHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSE EYNTSNPD;
[0102] A TGF-.beta. Type II receptor ectodomain "doublet", in which a TGF-.beta. Type II receptor ectodomain is linked with another TGF-.beta. Type II receptor ectodomain, which ectodomains can be the same or different TGF-.beta. superfamily receptor ectodomains, such as:
TABLE-US-00004 [0102] (SEQ ID NO: 46, also referred to herein as T22d35) IPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSIT SICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKE KKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSVNNDMIVTD NNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKN DENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDE CNDNIIFSEEYNTSNPD; (SEQ ID NO: 47) IPPHVQKSDVEMEAQKDEIICPSCNRTAHPLRHINNDMIVTDNNGAVKFPQ LCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETV CHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSE EYNTSNPDIPPHVQKSDVEMEAQKDEIICPSCNRTAHPLRHINNDMIVTDN NGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKND ENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDEC NDNIIFSEEYNTSNPD;
and [0103] a sequence substantially identical thereto. "Substantially identical" is as defined above.
[0104] In another specific, non-limiting example, the TGF-.beta. receptor ectodomain is the bone morphogenetic protein receptor la (BMPRIa; SEQ ID NO:69). In this embodiment, the natural linker, if present, may be selected from the group consisting of
TABLE-US-00005 (SEQ ID NO: 70) QNLDSMLHGTGMKSDSDQKKSENGVTLAPED; (SEQ ID NO: 71) PVVIGPFFDGSIR; (SEQ ID NO: 72) PVVIGPFFDGSIRQNLDSMLHGTGMKSDSDQKKSENGVTLAPED;
and [0105] a combination thereof.
[0106] Thus, in a specific, non-limiting example, the second portion of the polypeptide construct of the present invention may comprise the sequence selected from the group consisting of:
TABLE-US-00006 (SEQ ID NO: 74) QNLDSMLHGTGMKSDSDQKKSENGVTLAPEDTLPFLKCYCSGHCPDDAINN TCITNGHCFAIIEEDDQGETTLASGCMKYEGSDFQCKDSPKAQLRRTIECC RTNLCNQYLQPTLPPVVIGPFFDGSIRQNLDSMLHGTGMKSDSDQKKSENG VTLAPEDTLPFLKCYCSGHCPDDAINNTCITNGHCFAIIEEDDQGETTLAS GCMKYEGSDFQCKDSPKAQLRRTIECCRTNLCNQYLQPTLPPVVIGPFFDG SIR;
and [0107] a sequence substantially identical thereto. "Substantially identical" is as defined above.
[0108] In another specific, non-limiting example, the T6SR ectodomain is the activin receptor IIa (ActRIIA; SEQ ID NO:75). In this embodiment, the natural linker, if present, may be selected from the group consisting of
TABLE-US-00007 (SEQ ID NO: 76) AILGRSE; (SEQ ID NO: 77) EMEVTQPTSNPVTPKPPYYNI; (SEQ ID NO: 78) EMEVTQPTSNPVTPKPPYYNIAILGRSE;
and [0109] a combination thereof.
[0110] Thus, another specific non-limiting example of the second portion of the polypeptide construct of the present invention comprises the sequence selected from the group consisting of:
TABLE-US-00008 (SEQ ID NO: 80) AILGRSETQECLFFNANWEKDRTNQTGVEPCYGDKDKRRHCFATWKNISGS IEIVKQGCWLDDINCYDRTDCVEKKDSPEVYFCCCEGNMCNEKFSYFPEME VTQPTSNPVTPKPPYYNIAILGRSETQECLFFNANWEKDRTNQTGVEPCYG DKDKRRHCFATWKNISGSIEIVKQGCWLDDINCYDRTDCVEKKDSPEVYFC CCEGNMCNEKFSYFPEMEVTQPTSNPVTPKPPYYNI;
and [0111] a sequence substantially identical thereto. "Substantially identical" is as defined above.
[0112] In another specific, non-limiting example, the TGF-.beta. receptor ectodomain is the activin receptor IIb (ActRIIb; SEQ ID NO:81). In this embodiment, the natural linker, if present, may be selected from the group consisting of
TABLE-US-00009 (SEQ ID NO: 82) SGRGEAET; (SEQ ID NO: 83) EAGGPEVTYEPPPTAPT; (SEQ ID NO: 84) EAGGPEVTYEPPPTAPTSGRGEAET;
and [0113] a combination thereof.
[0114] Thus, another specific non-limiting example of the second portion of the polypeptide construct of the present invention comprises the sequence selected from the group consisting of:
TABLE-US-00010 (SEQ ID NO: 86) SGRGEAETRECIYYNANWELERTNQSGLERCEGEQDKRLHCYASWRNSSGT IELVKKGCWLDDFNCYDRQECVATEENPQVYFCCCEGNFCNERFTHLPEAG GPEVTYEPPPTAPTSGRGEAETRECIYYNANWELERTNQSGLERCEGEQDK RLHCYASWRNSSGTIELVKKGCWLDDFNCYDRQECVATEENPQVYFCCCEG NFCNERFTHLPEAGGPEVTYEPPPTAPT;
and [0115] a sequence substantially identical thereto. "Substantially identical" is as defined above.
[0116] Thus, in various embodiments of the present invention, the present constructs have an ectodomain comprising an amino acid sequence selected from the group consisting of SEQ ID NO:35, SEQ ID NO:69, SEQ ID NO:75, SEQ ID NO:81, and a sequence substantially identical thereto. In other embodiments, the second portion comprises the entire extracellular portion of a T.beta.SR-ED consisting of a sequence selected from the group consisting of SEQ ID NO:43, SEQ ID NO:44, SEQ ID NO:73, SEQ ID NO:79, SEQ ID NO:85, and a sequence substantially identical thereto.
[0117] The at least two ectodomain portion can have the same or different ectodomains, all belonging to the superfamily. In embodiments, the ectodomains bind the same target. In other embodiments, the ectodomains originate from the same receptor species. In other embodiments, the ectodomains are identical and thus are homomeric. In other embodiments the ectodomains are different and thus are heteromeric. In these embodiments, the ectodomain can be for instance a T.beta.RII-ED that is type a, and another ectodomain can be a T.beta.RII-ED that is type b. A third ectodomain could be the same as either one of these, or different still. For example, when there is more than one ectodomain in the second portion of the polypeptide construct of the present invention, the ectodomains may be all the same (homomers) or all different (heteromers), or any combination of superfamily ectodomains.
[0118] Thus, in embodiments, the second portion of the polypeptide construct of the present invention comprises a repeat of a given T.beta.SR-ED selected from the group consisting of SEQ ID NO:46, 47, 48, 74, 80, 86, and a sequence substantially identical thereto.
[0119] In specific embodiments, the second portion of the polypeptide construct of the present invention comprises heteromeric repeats of two distinct T.beta.SR-EDs genetically fused and selected from the group consisting of SEQ ID NO:61, 62, 63, 88, and a sequence substantially identical thereto.
[0120] In yet other embodiments, the second portion of the polypeptide construct of the present invention comprises homo-multimeric and hetero-multimeric repeats of one or more T.beta.SR-EDs selected for instance from the group consisting of SEQ ID NO:49, 50, 51, 64, 65, 66, 67, 68, and a sequence substantially identical thereto.
[0121] In the protein construct of the present invention, the first and second portions of the polypeptide construct of the present invention are linked. By the term "linked", it is meant that the two portions are covalently bonded. The chemical bond may be achieved by chemical reaction, or may be the product of recombinant expression of the two portions in a single polypeptide chain. In one specific, non-limiting example, the C-terminus of the first portion is linked directly to the N-terminus of the second portion, that is, no additional "linker" amino acids are present between the two portions. In the case where no linker is present, that is to say direct fusion of the two portions, there will be a direct link between the N-terminus of the full ectodomain and the C-terminus of the antibody constant regions C.sub.H2-C.sub.H3. For example, in fusing the Fc variant SEQ ID NO:9 to the SEQ ID NO:43 via the intrinsically disordered linker with SEQ ID NO:36, which is part of the T.beta.RII-ED (i.e., no additional "linker" amino acids added), one connects the glycine at the last position of SEQ ID NO:9 to the isoleucine at the first position of SEQ ID NO:43.
[0122] A common practice when producing fusion constructs is to introduce glycine or glycine-serine linkers (such as GGGGS, or [G.sub.4S].sub.n) between the fused components. As taught in the above paragraph, the polypeptide fusions of the present invention can be produced by direct linkage without use of any additional amino-acid sequence except those present in the Fc portion and in the receptor ectodomain portion. One thus can refrain from utilizing foreign sequences as linkers, providing an advantage due to their potential for undesired immunogenicity and their added molecular weight. Entropic factors are also a potential liability for glycine and glycine-serine linkers, which are highly flexible and may become partially restricted upon target binding, hence causing a loss of entropy unfavourable to binding affinity. Therefore, only the flexible, intrinsically disordered N-terminal regions of the T.beta.SR receptor ectodomains were employed as natural linkers in embodiments of the present invention. However, the particular amino acid compositions and lengths of these intrinsically disordered linkers (e.g., SEQ ID NO:36, 37, 70, 76, 82) precluded accurate prediction of whether the resulting direct-fusion constructs will have the required geometry and favourable molecular interactions for correct binding to their intended dimeric ligands.
[0123] The first and second portions of the polypeptide construct are, in embodiments, connected by natural intrinsically disordered polypeptide linkers selected from the group consisting of SEQ ID NO:36, 37, 38, 53, 70, 76, 82, and a sequence substantially identical thereto.
[0124] In embodiments, when multiple T.beta.SR-ED structured regions are present, these can be fused directly or they can be connected by natural intrinsically disordered polypeptide linkers between the ectodomains, such as SEQ ID NO:40, 41, 42, 55, 58, 59, 60, 72, 78, 84, 87, and a sequence substantially identical thereto.
[0125] Non-limiting examples of full-length polypeptide constructs of the present invention that comprise the two aforementioned portions are selected from the group consisting of SEQ ID NO: 91-120, and a sequence substantially identical thereto.
[0126] It is particularly important to note that, in the polypeptide constructs of the present invention, the N-terminus of the second portion is linked to the C-terminus of the first portion (see for example FIGS. 2A, C, D, E, F, and G).
[0127] Some of the polypeptide constructs of the present invention display significantly greater potency of TGF-.beta. neutralization compared to that of the TGF-.beta. superfamily receptor ectodomain alone; for example, the polypeptide construct may be between at least 50-fold and 1.times.10.sup.6-fold more potent. For example, the polypeptide constructs of the present invention may have a TGF-.beta. neutralization potency selected from the group consisting of at least 50-, 75-, 100-, 150-, 200-300-, 400-, 500-, 600, 1000-1500-, 2000-, 3000-, 4000-, 5000-, 6000-, 7000-, 8000-, 9000, 10000-, 20000-, 30000-, 40000-, 50000-, 60000-, 70000-, 80000-, 90000-, 100000-, 150000-, 200000-, 250000-, 300000-, 350000-, 400000-, 450000-, 500000-, 550000-600000-, 650000-, 700000-, 750000-, 800000-, 850000-, 900000-, 950000, or 1000000-fold, more potent than the T.beta.SR-ED alone, or any amount there between. In one example, the potency of the construct is at least 100-fold greater than the receptor ectodomain alone.
[0128] Additionally, when the polypeptide constructs of the present invention include a T.beta.SR-ED that binds TGF-.beta., the polypeptide construct may neutralize, to varying extents, all three isotypes of TGF-.beta. (that is, TGF-.beta.1, TGF-.beta.2, and TGF-.beta.)
[0129] The polypeptide constructs of the present invention have, as assessed in cell-based assays, TGF-.beta. neutralizing potencies that are significantly higher (100-fold or more) than those of bivalent comparator polypeptides, i.e. non-Fc-fused T22d35 and T2m-Fc. Within the series of polypeptide constructs of the present invention, those that contain two or more copies of the T.beta.RII ectodomain fused to the C-terminus of the Fc constant region have potencies that are higher than those constructs that contain only one copy, as assessed in cell based assays. Additionally, within the series of polypeptide constructs of the present invention, if the first portion within the construct is "headless", i.e. does not contain a Fab region, the potencies of the constructs are increased by engineering (optimizing) the number of cysteines in the hinge region at the "revealed" N-terminus. Engineering of the cysteine residues at the N-termini of "headless" constructs also markedly reduces the aggregation propensity of the constructs. Lastly, within the series of polypeptide constructs of the present invention, in vivo in tumor models, cysteine optimized "headless" constructs exhibit higher anti-tumor immuno-modulatory potencies than constructs in which the first portion is a full-sized antibody.
[0130] The polypeptide construct of the present invention is expressed as a single polypeptide chain. Once expressed, the polypeptide construct of the present invention forms a dimer wherein the C.sub.H2 and C.sub.H3 domains of the respective polypeptide constructs interact to form a properly assembled Fc region such as occurs when the expressed products are secreted into the culturing medium. For example, and without wishing to be limiting, examples of dimerized polypeptide constructs of the present invention are shown in FIGS. 2A and C-G. In one example, homodimers may be formed by identical polypeptide constructs. Alternatively, heterodimers may be formed by two different polypeptide constructs; thus, a heterodimer may be formed by two Fc region polypeptide constructs that have been engineered to induce heterodimerization and inhibit homodimerization.
[0131] The first portion of the polypeptide construct described above may further comprise, at its N-terminus, any suitable antigen-binding antibody fragment known in the art. For example, and without wishing to be limiting in any manner, the first portion of the polypeptide construct may comprise C.sub.H2 and C.sub.H3 domains and one selected from the group consisting of a single-chain Fv (scFv; a molecule consisting of V.sub.L and V.sub.H connected with a peptide linker) and a single-domain antibody (sdAb, a fragment composed of a single V.sub.L or a single V.sub.H; see for example FIG. 1C). In other instances, the antigen-binding fragment may be formed by combining the polypeptide construct with a second polypeptide chain. For example, the first portion of the polypeptide construct may comprise C.sub.H2 and C.sub.H3 domains along with a C.sub.H1 and V.sub.H domains, which when combined with a second polypeptide comprising C.sub.L and V.sub.L form a full-size antibody (i.e., Fc and Fab; see for example FIG. 1D). In another example, the first portion of the polypeptide may comprise C.sub.H2 and C.sub.H3 domains along with V.sub.H, which when combined with a second polypeptide comprising a V.sub.L forms an Fc fused to a Fv.
[0132] The combination of constant domains and antigen-binding fragment may be naturally-occurring, or may be obtained by manipulation of a naturally-occurring antibody or by using recombinant methods. The polypeptide constructs such as those just described may require a sequence selected from the group consisting of linker sequences, disulfide bonds, hinge region sequences, and other type of covalent bond to link them to the C.sub.H2 and C.sub.H3 domains; those of skill in the art will be familiar with various suitable approaches.
[0133] In alternative constructs of the present invention, the polypeptide construct comprises an antibody Fc fragment linked at the C-terminus of each heavy chain to at least one TGF-.beta. superfamily receptor ectodomain (T.beta.SR-ED), as described above and as illustrated in FIG. 2(A,D,E). The construct may further comprise an antigen-binding fragment at the N-terminus of the Fc; the antigen-binding fragment may be selected from the group consisting of a Fv, scFv, Fab, or sdAb, also as described above. In the polypeptide constructs as described above, the TGF-.beta. receptor ectodomain does not interfere in the native function or specificity of the antigen-binding fragment.
[0134] The antigen-binding antibody fragment described above, when present, may be directed to any suitable antigen. In certain limited embodiments, the antigen-binding antibody or fragment does not bind to an antigen that is PD-L1, EGFR1, her-2, CD4, CD6, CD20, CD25, MUC-1, IL-2, IL-6, or CTLA-4.
[0135] The present constructs can further comprise antibody or antibody fragments that target any antigen of interest. They can also comprise the antigen itself, or any other moiety of interest that is genetically encoded. Particular embodiments herein include the EGFR antibody cetuximab and its active fragments, Avastin, Herceptin, Synagis, FC5, or a poly-aspartate bone-localization motif, such a D10, or sequence substantially identical or equivalent thereto.
[0136] The present constructs can comprise a binding protein e.g., antibody and binding fragments thereof, that inhibits a checkpoint protein which may be CTLA-4, PD1, PDL1, PDL2, PDL3, PD1, B7-H3, B7-H4, BTLA, HVEM, TIM3, GAL9, LAG3, VISTA, KIR, 2B4, CD160, CGEN-15049, CHK 1, CHK2, A2aR, CD28, CD86, or one of the B-7 family ligands or a combination thereof.
[0137] Illustrative immune checkpoint inhibitors include Tremelimumab (CTLA-4 blocking antibody), anti-OX40, PD-LI monoclonal Antibody (Anti-B7-HI; MED14736), MK-3475 (PD-1 blocker), Nivolumab (anti-PDI antibody), CT-011 (anti-PDI antibody), BY55 monoclonal antibody, AMP224 (anti-PDLI antibody), BMS-936559 (anti-PDLI antibody), MPLDL3280A (anti-PDLI antibody), MSB0010718C (anti-PDLI antibody) and Yervoy/ipilimumab (anti-CTLA-4 checkpoint inhibitor).
[0138] Other antibodies provided by the present constructs can include rituximab, muromonab-CD3, abciximab, daclizumab, basiliximab, palivizumab, infliximab, trastuzumab, gemtuzumab ozogamicin, alemtuzumab, ibritumomab tiuxetan, adalimumab, omalizumab, tositumomab, I-131 tositumomab, efalizumab, bevacizumab, panitumumab, pertuzumab, natalizumab, etanercept, IGN101, volociximab, Anti-CD80 mAb, Anti-CD23 mAb, CAT-3888, CDP-791, eraptuzumab, MDX-010, MDX-060, MDX-070, matuzumab, CP-675,206, CAL, SGN-30, zanolimumab, adecatumumab, oregovomab, EGFR-binding antibodies cetuximab, nimotuzumab, necitumumab, panitumumab, matuzumab, and zalutumumab, as well as ABT-874, denosumab, AM 108, AMG 714, fontolizumab, daclizumab, golimumab, CNTO 1275, ocrelizumab, HuMax-CD20, belimumab, epratuzumab, MLN1202, visilizumab, tocilizumab, ocrerlizumab, certolizumab, eculizumab, pexelizumab, abciximab, ranibizimumab, mepolizumab, TNX-355, or MYO-029.
[0139] Still other antibodies that can be included in the present constructs are rituximab, zanolimumab, hA20, AME-133, HumaLYM, trastuzumab, pertuzumab, IMC-3G3, ch806, KSB-102, MR1-1, SC100, SC101, SC103, alemtuzumab, muromonab-CD3, OKT4A, ibritumomab, gemtuzumab, alefacept, abciximab, basiliximab, palivizumab, motavizumab, infliximab, adalimumab, CDP-571, etanercept, ABX-CBL, ABX-IL8, ABX-MA1 pemtumomab, Therex, AS1405, natalizumab, HuBC-1, natalizumab, IDEC-131, VLA-1; CAT-152; J695, CAT-192, CAT-213, BR3-Fc, LymphoStat-B, TRAIL-R1mAb, bevacizumab, ranibizumab, omalizumab, efalizumab, MLN-02, zanolimumab, HuMax-IL 15, HuMax-Inflam, HuMax-Cancer, HuMax-Lymphoma, HuMax-TAC, clenoliximab, lumiliximab, BEC2, IMC-1C11, DC101, labetuzumab, arcitumomab, epratuzumab, tacatuzumab, MyelomaCide, LkoCide, ProstaCide, ipilimumab, MDX-060, MDX-070, MDX-018, MDX-1106, MDX-1103, MDX-1333, MDX-214, MDX-1100, MDX-CD4, MDX-1388, MDX-066, MDX-1307, HGS-TR2J, FG-3019, BMS-66513, SGN-30, SGN-40, tocilizumab, CS-1008, IDM-1, golimumab, CNTO 1275, CNTO 95, CNTO 328, mepolizumab, MOR101, MOR102, MOR201, visilizumab, HuZAF, volocixmab, ING-1, MLN2201, daclizumab, HCD122, CDP860, PRO542, C14, oregovomab, edrecolomab, etaracizumab, siplizumab, lintuzumab, Hu1D10, Lym-1, efalizumab, ICM3, galiximab, eculizumab, pexelizumab, LDP-01, huA33, WX-G250, sibrotuzumab, Chimeric KW-2871, hu3S193, huLK26; bivatuzumab, ch14.18, 3F8, BC8, huHMFG1, MORAb-003, MORAb-004, MORAb-009, denosumab, PRO-140, 1D09C3, huMikbeta-1, NI-0401, NI-501, cantuzumab, HuN901, 8H9, chTNT-1/B, bavituximab, huJ591, HeFi-1, Pentacea, abagovomab, tositumomab, 105AD7, GMA161 and GMA321.
[0140] In other embodiments, the constant domain/first portion of the constructs can comprise a polypeptide having medicinal properties, such as agents that stimulate the immune system, in particular in relation to the ability of the immune system to attack tumor cells. These polypeptides can include cytokines (such as interleukin-2) or growth factors that stimulate immune cells directly or indirectly (i.e. act by providing gas to the immune system), as well as ectodomains or other binding agents that neutralize ligands which inhibit immune cells, either directly or indirectly (i.e. act by releasing a brake on the immune system).
[0141] In other embodiments, the constant domain/first portion of the constructs can comprise a polypeptide that does not have active medicinal properties per se, but rather provides a localization signal. This localization motif will serve to focus the intrinsic TGF-.beta. neutralization activity of the second portion of the construct to a particular region of the body. In one example, the first portion comprised a long poly-aspartate bone-localization motif, preferably D10 or an equivalent bone-localization moiety, which acts to enhance localisation of the construct to bone. By increasing the TGF-.beta. neutralization activity of the construct within bone, more favourable dosing levels and schedules may be required for the treatment of bone-related diseases, such as osteogenesis imperfecta, relative to that required for a similar construct without the D10 motif.
[0142] Embodiments exemplifying polypeptide constructs of the present invention that include antigen-binding fragments at the N-terminus of the Fc region (first portion) are selected from the group consisting of SEQ ID NO: 121-131, and a sequence substantially identical thereto.
[0143] In other embodiments, polypeptide constructs of the present invention that include other targeting agents, e.g., a poly-aspartate bone-localization motif, at the N-terminus of the Fc region (first portion), are exemplified by SEQ ID NO: 136-150.
[0144] The polypeptide construct of the present invention may also comprise additional sequences to aid in expression, detection or purification of a recombinant antibody or fragment thereof. Any such sequences or tags known to those of skill in the art may be used. For example, and without wishing to be limiting, the antibody or fragment thereof may comprise a targeting or signal sequence (for example, but not limited to ompA), a detection/purification tag (for example, but not limited to c-Myc, His.sub.s, His.sub.6, or His.sub.8G), or a combination thereof. In another example, the signal peptide may be MVLQTQVFISLLLWISGAYG (SEQ ID NO:89) or MDVVTWRILFLVAAATGTHA (SEQ ID NO:90). In a further example, the additional sequence may be a biotin recognition site such as that described in [WO/1995/04069] or in [WO/2004/076670]. As is also known to those of skill in the art, linker sequences may be used in conjunction with the additional sequences or tags, or may serve as a detection/purification tag.
[0145] The present invention also encompasses nucleic acid sequences encoding the molecules as described herein. Given the degeneracy of the genetic code, a number of nucleotide sequences would have the effect of encoding the desired polypeptide, as would be readily understood by a skilled artisan. The nucleic acid sequence may be codon-optimized for expression in various micro-organisms. The present invention also encompasses vectors comprising the nucleic acids as just described, wherein the vectors typically comprise a promoter and signal sequence that are operably linked to the construct-encoding polynucleotide for driving expression thereof in the selected cellular production host. The vectors can be the same or different provided both result in secretion of the dimeric polypeptide construct.
[0146] Furthermore, the invention encompasses cells, also referred to herein as transgenic cellular host, comprising the nucleic acid and/or vector as described, encoding a first polypeptide construct. The host cells may comprise a second nucleic acid and/or vector encoding a second polypeptide construct different from the first polypeptide construct. The co-expression of the first and second polypeptide constructs may lead to the formation of heterodimers.
[0147] The present invention also encompasses a composition comprising one or more than one polypeptide construct as described herein. The composition may comprise a single polypeptide construct as described above, or may be a mixture of polypeptide constructs. The composition may also comprise one or more than one polypeptide construct of the present invention linked to one or more than one cargo molecule. For example, and without wishing to be limiting in any manner, the composition may comprise one or more than one polypeptide construct of the present invention linked to a cytotoxic drug in order to generate an antibody-drug conjugate (ADC) in accordance with the present invention.
[0148] The composition may also comprise a pharmaceutically acceptable diluent, excipient, or carrier. The diluent, excipient, or carrier may be any suitable diluent, excipient, or carrier known in the art, and must be compatible with other ingredients in the composition, with the method of delivery of the composition, and is not deleterious to the recipient of the composition. The composition may be in any suitable form; for example, the composition may be provided in suspension form, powder form (for example, but limited to lyophilised or encapsulated), capsule or tablet form. For example, and without wishing to be limiting, when the composition is provided in suspension form, the carrier may comprise water, saline, a suitable buffer, or additives to improve solubility and/or stability; reconstitution to produce the suspension is effected in a buffer at a suitable pH to ensure the viability of the antibody or fragment thereof. Dry powders may also include additives to improve stability and/or carriers to increase bulk/volume; for example, and without wishing to be limiting, the dry powder composition may comprise sucrose or trehalose. In a specific, non-limiting example, the composition may be so formulated as to deliver the antibody or fragment thereof to the gastrointestinal tract of the subject. Thus, the composition may comprise encapsulation, time-release, or other suitable technologies for delivery of the antibody or fragment thereof. It would be within the competency of a person of skill in the art to prepare suitable compositions comprising the present compounds.
[0149] The constructs of the present invention may be used to treat diseases or disorders associated with over-expression or over-activation of ligands of the TGF-.beta. superfamily. The disease or disorder can be selected from, but not limited to, cancer, ocular diseases, fibrotic diseases, or genetic disorders of connective tissue.
[0150] In the field of cancer therapy, it has recently been demonstrated that TGF-.beta. is a key factor inhibiting the antitumor response elicited by immunotherapies, such as immune checkpoint inhibitors (ICI's) (Hahn & Akporiaye, 2006). Specifically, therapeutic response to ICI antibodies results primarily from the re-activation of tumor-localized T-cells. Resistance to ICI antibodies is attributed to the presence of immunosuppressive mechanisms that result in a dearth of T-cells in the tumor microenvironment. Thus, it is now recognized that in order to elicit responses in resistant patients, ICI antibodies need to be combined with agents that can activate T-cells and induce their recruitment into the tumor, i.e. reversing of the "non-T-cell-inflamed" tumor phenotype. One publication noted that overcoming the non-T-cell-inflamed tumor microenvironment is the most significant next hurdle in immuno-oncology (Gajewski, 2015).
[0151] We have shown using a proof-of-principle TGF-.beta. trap, T22d35, that blocking of TGF-.beta. effectively reverses the "non-T cell inflamed" tumor phenotype (Zwaagstra et al, 2012). This positions anti-TGF-.beta. molecules as potential synergistic combinations with ICI's and other immunotherapeutics. In support of this, a 2014 study (Holtzhausen et al., ASCO poster presentation) examined effects of a TGF-.beta. blocker when combined an anti-CTLA-4 antibody in a physiologically-relevant transgenic melanoma model. The study demonstrated that while anti-CTLA-4 antibody monotherapy failed to suppress melanoma progression, the combination of the TGF-.beta. antagonist and anti-CTLA-4 antibody significantly and synergistically suppressed both primary melanoma tumor growth as well as melanoma metastasis. These observations correlated with significant increases in effector T-cells in melanoma tissues.
[0152] Fibrotic diseases include those that affect any organ of the body, including, but not limited to kidney, lung, liver, heart, skin and eye. These diseases include, but are not limited to, chronic obstructive pulmonary disease (COPD), glomerulonephritis, liver fibrosis, post-infarction cardiac fibrosis, restenosis, systemic sclerosis, ocular surgery-induced fibrosis, and scarring.
[0153] Genetic disorders of connective tissue include, but are not limited to, Marfan syndrome (MFS) and Osteogenesis imperfecta (01).
[0154] The present invention will be further illustrated in the following examples. However, it is to be understood that these examples are for illustrative purposes only and should not be used to limit the scope of the present invention in any manner.
EXAMPLE 1
Production and Purification of Fusion Molecules
[0155] Several fusion molecules comprising full-size antibody (FSA), V.sub.HH-IgG Fc, D10-Fc or "headless" Fc C-terminally-fused to the T22d35 or T2m ectodomains were constructed (Table 1). All constructs comprising a heavy chain included the signal sequence MDVVTWRILFLVAAATGTHA (SEQ ID NO:89) at the N-terminus, while constructs comprising a light chain included the signal sequence MVLQTQVFISLLLWISGAYG (SEQ ID NO:90) at the N-terminus. The DNA coding for constructs were prepared synthetically (Biobasic Inc. or Genescript USA Inc.). Constructs comprising FSA, D10-Fc and "headless" Fc were cloned into the EcoR1 (5' end) and BamH1 (3' end) sites and those comprising V.sub.HH-IgG Fc were cloned into the HindIII (5' end) and BamH1 (3' end) sites of the pTT5 mammalian expression plasmid vector (Durocher et al, 2002).
TABLE-US-00011 TABLE 1 FSA-, V.sub.HH-IgG Fc-, D10-Fc- and Fc-fusion constructs produced. The letter in brackets in the construct column refers to the type of construct as illustrated in FIG. 2. Fusion Antibody Construct Construct ED Scaffold source SEQ ID NO: Cet-T2m (F) T2m FSA (hIgG1) Cetuximab 121, 123 Cet-T22d35 (D) T22d35 FSA (hIgG1) Cetuximab 121, 122 Her-T22d35 (D) T22d35 FSA (hIgG1) Herceptin 124, 125 Ava-T22d35 (D) T22d35 FSA (hIgG1) Avastin 126, 127 Syn-T22d35 (D) T22d35 FSA (hIgG1) Synagis 128, 129 FC5-Fc-T22d35 (E) T22d35 V.sub.HH-Fc FC5-Fc 130 (mIgG2) FC5-Fc-T2m (G) T2m V.sub.HH-Fc FC5-Fc 131 (mIgG2) D10-Fc-T2m (G) T2m Fc huIgG 136-139 (several variants with differing D10 linkage and IgG isotype) Fc-T22d35 (A) T22d35 Fc huIgG 100, 105 (several variants with differing N-termini and IgG isotype) Fc-T2m (C) T2m Fc huIgG 91-97 (several variants with differing N-termini and IgG isotype) T2m-Fc (R&D) (B) T2m Fc huIgG1 132 T2m-Fc (B) T2m Fc huIgG2 133
[0156] The Cet-T2m and Cet-T22d35 constructs were produced by transient co-transfection of Chinese Hamster Ovary (CHO) cells with the heavy chain (HC)-T2m or (HC)-T22d35 construct combined with the Cetuximab light chain (LC) construct which then assembled as the Cetuximab-T22d35 (Cet-T22d35) or Cetuximab-T2m (Cet-T2m) fusion molecules. Briefly, CetHC-T22d35 (SEQ ID NO:122) and CetLC (SEQ ID NO:121) plasmid DNAs (ratio=3:2) were co-transfected into a 10 L Wavebag culture of CHO-3E7 cells in FreeStyle F17 medium (Invitrogen) containing 4 mM glutamine and 0.1% Kolliphor p-188 (Sigma) in a Wavebag maintained at 37.degree. C. Transfection conditions were: DNA (50% HC+LC plasmids, 30% ssDNA, 15% AKT plasmid, 5% GFP plasmid): PEI(polyethylenimine)pro (Polyplus) (ratio=1:2.5). At 24 hours post-transfection, 10% Tryptone N1feed (TekniScience Inc.) and 0.5 mM Vaporic acid (VPA, Sigma) were added and the temperature was shifted to 32.degree. C. to promote the production and secretion of the fusion proteins and maintained for 15 days post transfection after which the cells were harvested. At final harvest the cell viability was 89.6%. The harvest supernatant (10.8 L) was filtered (0.2 .mu.m) and loaded onto a 55 mL Protein A MabSelect Sure 55 mL column (GE Healthcare). The column was washed with 2 column volumes of PBS and protein was eluted with 3 column volumes of 0.1 M sodium citrate pH 3.6. To maximize the yield, the flow through was reloaded onto the Protein A column and eluted as described above. Eluted fractions were neutralized with 1 M Tris, evaluated by SDS-PAGE and those containing Cet-T22d35 were pooled (FIG. 3A) and subsequently loaded onto a Hi-load Superdex S200 26/60 size exclusion chromatography (SEC) column (GE Healthcare) equilibrated in formulation buffer (DPBS without Ca.sup.2+, without Mg.sup.2+). Protein was eluted using 1 column volume formulation buffer, collected into successive fractions and detected by UV absorbance at 280 nM (FIG. 3B). The main peak SEC fractions containing Cet-T22d35 protein were then pooled and concentrated to a concentration of 7.8 mg/mL. The final yield was 533 mg.
[0157] Similar transfection, production and purification methods were performed for the other FSA-trap examples listed in Table 1. In the case of the V.sub.HH-Fc IgG-, D10 and "headless" Fc-fusion molecules the composition of the transfection mixture was modified as follows: DNA (80% plasmid construct, 15% AKT plasmid, 5% GFP plasmid): PElpro (ratio 1:2.5).
[0158] The integrity of the pooled Prot-A and SEC fractions of Cet-T22d35 protein was analyzed by SDS-PAGE (4-15% polyacrylamide) under reducing and non-reducing conditions (FIG. 3C) and by UPLC-SEC (FIG. 3D). For UPLC-SEC, 2-10 .mu.g of protein in DPBS (Hyclone, minus Ca2+, minus Mg2+) was injected onto a Waters BEH200 SEC column (1.7 .mu.m, 4.6.times.150 mm, SN:01773430816818) and resolved under a flow rate of 0.4 mL/min for 8.5 min at room temperature, using the Waters Acquity UPLC H-Class Bio-System. Protein peaks were detected at 280 nM (Acquity PDA detector). Coomassie brilliant blue (CBB) staining of the gels shows the CetHC-T22d35 (.about.110 Kd) and CetLC bands (-30 Kd) under reducing conditions while under non-reducing conditions a 250 Kd band is detected which represents the fully assembled and highly pure Cet-T22d35 fusion protein. Additional UPLC-SEC analysis of the SEC purified, pooled ProtA sample confirmed the high degree of purity (99.42%) of the Cet-T22d35 protein and the absence of aggregates. Together, these results demonstrate the manufacturability of the Cet-T22d35 fusion protein.
[0159] Similar methods were used to analyse expression levels, purifiability, aggregation levels, and dimeric assembly of several other Fc-ectodomain constructs. The results from these studies are shown in FIG. 3E to L.
[0160] FIG. 3E to 3H show the results from the analysis of the hIgG1Fc.DELTA.K(C)-T2m construct (an example of Type C construct from FIG. 2). FIG. 3E shows the (ProtA)-affinity column elution profile. Fraction 12-15 were pooled, subjected to a UPLC-SEC evaluation (FIG. 3F), further SEC purified to remove aggregates and re-evaluated by UPLC-SEC (FIG. 3G). This confirmed the high degree of purity of the hIgG1Fc.DELTA.K(C)-T2m construct and the absence of aggregates. SDS-PAGE (FIG. 3H) under non-reducing (NR) and reducing (R) conditions shows bands of expected molecular weights, demonstrating the expected assembly of hIgG1Fc.DELTA.K(C)-T2m as a disulphide linked dimer.
[0161] FIGS. 3I and 3J compare the level of aggregation of two "headless" Fc-T2m constructs (examples of Type C in FIG. 2). The Fc-T2m construct is an IgG2-based construct without engineering of the hinge region, thus it contains four cysteine residues, whereas the hIgG2Fc.DELTA.K(CC)-T2m has been engineered by N-terminal truncation of the hinge region to have only two cysteines in the hinge region. It can be seen that the Fc-T2m construct contains a high level of aggregates after Protein A purification, with a doublet peak remaining even after further SEC purification. In contrast, hIgG2Fc.DELTA.K(CC)-T2m, which has an engineered N-terminus, exhibited low levels of aggregates after only Protein A purification. These results demonstrate the advantage of carrying out N-terminal engineering of headless Fc-T2m constructs to reduce aggregation.
[0162] FIGS. 3K and 3L compare the level of aggregation of two "headless" Fc-T22d35 constructs (examples of Type A in FIG. 2). The Fc-T22d35 construct is without engineering of the hinge-region cysteine residues whereas the hIgG1Fc.DELTA.K(C)-T22d35 construct has been engineered by N-terminal truncation of the hinge to have only one cysteine in the hinge region. It can be seen that, similarly to Fc-T2m, the Fc-T22d35 construct contains a high level of aggregates after Protein A purification, with a doublet peak remaining as detected by UPLC-SEC even after further SEC purification. In contrast, hIgG1Fc.DELTA.K(C)-T22d35, which has an engineered N-terminus, exhibited lower levels of aggregates after Protein A purification. Further SEC purification yielded a singlet peak as detected by UPLC-SEC, confirming the absence of aggregates. These results demonstrate the advantage of carrying out N-terminal engineering of headless Fc-T22d35 constructs to reduce aggregation.
EXAMPLE 2
Neutralization and Binding of TGF43 by Fusion Constructs
[0163] The TGF-.beta. neutralization potencies of purified Fc-ectodomain fusion constructs were determined and compared to those of non-Fc-fused T22d35. It should be noted that non-Fc-fused T2m does not neutralize any of TGF-.beta.1, -.beta.2, or -.beta.3 (De Crescenzo et al, 2001).
[0164] TGF-.beta. neutralization potencies for TGF-.beta.1, -.beta.2 and -.beta.3 were determined for purified fusion constructs using two cell-based signaling assays: 1) the Mv1Lu cell luciferase reporter assay with Mv1Lu cells having a PAI-1-luciferase reporter (as described in (Zwaagstra et al, 2012)) and an A549 cell/IL-11 release assay adapted to the MSD (Meso Scale Discovery) platform.
[0165] Mv1Lu cell luciferase reporter assay: Briefly, cells were seeded onto 96-well plates (20,000 cells/well) and then treated with T2m, T22d35, or a fusion construct+25 pM TGF-.beta. at 37.degree. C. for 16 h in DMEM, 1% FBS, 0.1% BSA. Cells were then lysed and luciferase activity was measured (Promega Corp.) using a Synergy 2 plate reader (BioTek Instruments Inc.).
[0166] A549 cell IL-11 release assay: Human A549 lung cancer cells (ATCC-CCL-185, Cedarlane Burlington ON) were seeded in 96-well plates (5.times.103 cells/well). The following day 10 pM TGF-.beta. in complete media in the absence or presence of a serial dilution of fusion protein was incubated for 30 min at RT prior to adding to the cells. After 21 h of incubation (37.degree. C., 5% CO2, humidified atmosphere) conditioned medium was harvested and added to MSD Streptavidin Gold plates (Meso Scale Diagnostics, Gaithersburg, Md.) that were coated with 2 .mu.g/mL biotinylated mouse anti-human IL-11 antibody (MAB618, R&D Systems, Minneapolis, Minn.). After 18 h (4.degree. C.) plates were washed with PBS containing 0.02% Tween 20, a 2 .mu.g/mL SULFO-tagged goat anti-human IL-11 antibody (AF-218-NA, R&D Systems Minneapolis, Minn.) was added and plates were incubated for 1 h at RT. After a final wash, plates were read in a MESO QuickPlex SQ120 machine (Meso Scale Diagnostics, Gaithersburg, Md.). IL-11 readouts were expressed as percent IL-11 release compared to control cells treated with TGF-.beta. alone.
[0167] In one set of experiments, using the Mv1 Lu cell reporter assay, the neutralization potency of Cet-T2m (construct Type F in FIG. 2), Cet-T22d35 (construct Type D in FIG. 2) and T22d35 (non-Fc-fused) were compared. FIG. 4A shows representative TGF-.beta.1 (top panel), TGF-.beta.3 (middle panel) and TGF-.beta.2 (bottom panel) neutralization curves for Cet-T2m, Cet-T22d35 and T22d35 while Table 2 summarizes TGF-.beta.1, -.beta.2 and -.beta.3 neutralization IC.sub.50 values. Unexpectedly, the TGF-.beta.1 and TGF-.beta.3 neutralization curves for the Cetuximab fusion constructs indicated extremely high potencies that lie in the picomolar range (determining a single IC.sub.50 value in these experiments is difficult due to the biphasic nature of the curves). The observed TGF-.beta.1 IC.sub.50 value for Cet-T22d35 was in the picomolar range. In contrast, the TGF-.beta.1 IC.sub.50 for non-Fc-fused T22d35 was approximately 1 nM. This illustrates that there is a large increase in T22d35 potency upon fusion to the C-terminus of the Fc region of Cetuximab (due to the biphasic nature of the Cet-T22d35 curve, the fold difference is difficult to determine in these experiments). The TGF-.beta.1 IC50 value for Cet-T2m was also subpicomolar (but less potent than Cet-T22d35), whereas unfused T2m is not able to detectably neutralize TGF-.beta.1, even at concentrations above 500 nM (De Crescenzo et al, 2001). This demonstrates that, similar to T22d35, a very significant increase in T2m potency occurs upon fusion to the C-terminus of the Fc region of Cetuximab. Both Cet-T22d35 and Cet-T2m neutralized TGF-.beta.2 (IC.sub.50 nM range), whereas T22d35 and T2m (De Crescenzo et al, 2001) alone did not, even at a concentration of 800 nM, again showing the remarkable increase in neutralization potency that occurs upon fusion of T22d35 or T2m to the C-terminus of an Fc region.
[0168] In another set of experiments using the Mv1Lu cell reporter assay, similar extremely high potencies of TGF-.beta. neutralization were observed for other C-terminus Fc fusion constructs (Table 2), e.g. for constructs in which T22d35 or T2m were fused with FSAs such as Herceptin (Her-T22d35), Avastin (Ava-T22d35) or Synagis (Syn-T22d35) [Type F and D constructs in FIG. 2], or with the blood-brain barrier crossing Fc-fused FC5 V.sub.HH antibody (FC5-Fc-T22d35 and FC5-Fc-T2m) [Type E and G constructs in FIG. 2]. In addition, fusion of T22d35 or T2m to the C-terminus of an IgG2-Fc region alone, i.e. an antibody with no Fab region present (Fc-T22d35 and Fc-T2m) [Type A and C constructs in FIG. 2] resulted in fusion proteins with similarly high neutralization potencies. However, fusing T2m to the N-terminus of an IgG2-Fc (T2m-Fc) generates a fusion protein that does not neutralize TGF-.beta.1 and -.beta.3 in the picomolar range, but rather in the range of 1 nanomolar (0.3 to 15 nM in Table 2) and lacks any activity towards TGF-.beta.2. These results are similar to those obtained with commercially available N-terminally IgG1 Fc-fused TGF-.beta. Type II receptor ectodomain (T2m-Fc (R&D Systems)) (0.3 to 0.5 nM in Table 2). Together, these results thus demonstrate that fusion of TGF-.beta. superfamily receptor ectodomains to the C-terminus of an Fc domain in the context of full-size antibodies, a V.sub.HH-Fc, or an Fc region alone, give rise to unexpectedly high TGF-.beta. neutralization potencies.
TABLE-US-00012 TABLE 2 TGF-.beta. neutralization IC.sub.50 of fusion constructs. It should be noted that T2m does not neutralize any of TGF-.beta.1, -.beta.2, or -.beta.3 (De Crescenzo et al, 2001). It should also be noted that the IC.sub.50 values in the table below are estimates due to the biphasic nature of the curves. TGF-.beta.1 TGF-.beta.3 TGF-.beta.2 Av IC.sub.50 (nM) Av IC.sub.50 (nM) Av IC.sub.50 (nM) Cet-T22d35 0.000,001 0.000,002 13.6 Cet-T2m 0.000,1 0.000,0015 129.2 T22d35 1.232 0.033 No neutralization Her-T22d35 0.000,13 0.000,05 7.89 Ava-T22d35 0.000,000,72 0.000,00038 8.60 Syn-T22d35 0.000,042 0.000,0001 35.2 FC5-Fc-T22d35 0.000,001 0.000,001 96.2 FC5-Fc-T2m 0.000,017 0.000,015 432.9 Fc-T22d35 0.001,445 0.000,026 108.4 T2m-Fc (R&D) 0.506 0.323 No neutralization T2m-Fc 14.523 0.276 No neutralization Fc-T2m 0.009,923 0.000,766 460.5
[0169] In order to confirm the relative potencies of Fc-ectodomain constructs, a second cell-based assay, an A549 cell IL-11 release assay, was used. This IL-11 release assay acts as a model of TGF-.beta.-mediated biological responses that contribute to both tumor metastasis and fibrosis. In the set of experiments shown in FIG. 4B, the neutralization potencies of T22d35, Fc-T2m, Fc-T22d35, Cet-T22d35, Her-T22d35, Ava-T22d35, Syn-T22d35 and FC5-Fc-T22d35 were compared. It can be seen that the neutralization curves in this assay are not biphasic, making it less challenging than the Mv1Lu assay to determine and compare IC.sub.50 values. All of the constructs in which T22d35 was fused to the C-terminus of an Fc region exhibited IC.sub.50 values in the range of 5 pM, corroborating the extremely high potency observed for these constructs in the Mv1Lu assay. It can also be seen in FIG. 4B that the IC.sub.50 value for non-Fc-fused T22d35 was .about.0.5 nM. This indicates that a .about.100-fold increase in potency occurs upon fusion of T22d35 to the C-terminus of an Fc region. The IC.sub.50 value for Fc-T2m was 0.05 nM. This indicates that constructs with two ectodomains in the C-terminal portion may be more potent than a construct with one ectodomain in the C-terminal portion (as was observed in the Mv1 Lu assay, FIG. 4A), however, it should be noted that Fc-T2m does not have an optimized N-terminus.
[0170] An additional set of experiments in which the A549 cell IL-11 release assay was used to compare TGF-.beta. neutralization potencies is shown in FIG. 4C. Here, the potencies of several "headless"-T2m constructs were assessed along with that of non-Fc-fused T22d35. The potency of T22d35 was determined to be .about.0.5 nM, consistent with the data shown in FIG. 4B. This is illustrative of the robustness of the A549 cell IL-11 release assay. It can also be seen in FIG. 4C that all of the "headless"-T2m constructs exhibited high potencies with IC.sub.50s in the range of 5 pM (3 to 17 pM). These values are in the same range as those of the T22d35-containing constructs shown in FIG. 4B, and are 10-fold higher potency than that of the "headless"-T2m containing construct also shown in FIG. 4B (Fc-T2m). Since all of the T2m-containing constructs in FIG. 4C have engineered N-termini, whereas Fc-T2m does not, these results indicate that engineering of the cysteine residues of the hinge region of "headless" constructs is able to increase their potency by approximately 10-fold.
[0171] We have also compared the potencies of constructs that include three T.beta.RII structured ectodomains with constructs carrying two ectodomains using the A549 cell IL-11 release assay. We observed that the triple-repeat based constructs (SEQ ID NO:111 and SEQ ID NO:116) are potent in neutralizing TGF-.beta.1 in this assay, and typically have improved 1050 values relative to the corresponding double-repeat based constructs (SEQ ID NO:100 and SEQ ID NO:106, respectively). All constructs involved in this comparative study had the same engineered N-terminus of the Fc portion, hIgG1Fc.DELTA.K(C).
[0172] Binding to TGF-.beta.: Binding of T22d35, Cet-T22d35, and Cet-T2m to TGF-.beta.2 was measured using a competitive SPR binding experiment. In this assay, the molecule of interest was first allowed to bind to a fixed amount of TGF-.beta. in solution. A 2-fold dilution series was prepared in PBS-0.05% Tween, starting with 1000 nM T22d35 trap or 20 nM Cet-T22d35 or Cet-T2m. Each diluted sample was pre-incubated with 1 nM TGF-.beta.2 for 30 min at room temperature to allow binding. The mixture was then flowed over immobilized, pan-specific anti-TGF-.beta. antibody 1D11 (2000 RU 1D11) in order to quantify the amount of ligand left unbound (T.beta.RII ectodomain and 1D11 bind to a similar epitope on TGF-.beta.) using a Biacore T200 instrument. The TGF-.beta.2 binding EC.sub.50 values were determined by plotting the percent free TGF-.beta. versus the protein concentration of the molecule of interest. Binding curves and EC.sub.50 values are shown in FIG. 4D and Table 3. In the case of TGF-.beta.2 binding, a 100-fold increase in binding was observed between Cet-T22d35 and unfused T22d35 (EC.sub.50.about.1 nM versus>100 nM, respectively), indicating that C-terminal fusion of the T22d35 trap to antibody provides a gain in affinity for the TGF-.beta.2 isoform. This correlates with the ability of Cet-T22d35 to neutralize TGF-.beta.2 in the 10 nM range, and the inability of unfused T22d35 to neutralize TGF-.beta.2, as observed in the Mv1Lu-Luc cell reporter assay.
TABLE-US-00013 TABLE 3 EC.sub.50 of Cetuximab-trap binding to TGF-.beta. in solution. EC.sub.50 is the Effective Concentration at which 50% of TGF-.beta. is bound and is given in nM. Trap variant EC.sub.50 for TGF-.beta.2 T22d35 >100 Cetuximab-T2m 1.17 Cetuximab-T22d35 0.50 Note: T2m alone shows an IC50 of greater than 1000 nM (Zwaagstra et al, 2012), and thus is not considered neutralizing.
EXAMPLE 3A
Validation of Antibody Binding
[0173] The ability of an antibody alone or in a fusion construct of Example 1 to bind to its intended target antigen was evaluated using surface plasmon resonance (SPR).
[0174] Direct binding of Cet-T22d35 or Cetuximab to the EGF receptor extracellular domain (EGFR-ED) was quantified by SPR using a BIAcore T200 instrument, performed in the standard manner. Briefly, Cet-T22d35 or Cetuximab alone were captured on the SPR CM5 chip using immobilized anti-human IgG Fc-specific antibody (2000 RU). Variable concentrations of EGFR-ED in PBS-0.05% Tween were then flowed over the capture surface at 100 .mu.l/min and 25.degree. C. The resulting sensorgrams (data not shown) were analyzed using the Biacore T200 evaluation software. The K.sub.D values of Cet-T22d35 and Cetuximab were very similar (847 and 708 pM, respectively), indicating that fusion of T22d35 to the Fc portion of Cetuximab does not appreciably alter binding to EGFR-ED, compared to the non-fused FSA. Similar SPR methods and analyses were performed for other antibody-trap fusion examples, compared with their corresponding target antigens (see Table 4).
[0175] From Table 4 it is evident for each exemplified construct that fusion of a TGF-.beta. superfamily receptor ectodomain(s) to the C-terminus of the Fc region of an antibody did not significantly alter antigen-binding affinities and K.sub.D values of the antibody. This indicates that the ectodomain(s) can be readily fused to any antibody without compromising the ability the antibody to bind its target antigen.
TABLE-US-00014 TABLE 4 SPR determination of antigen-binding affinity of T22d35 fusion constructs or antibodies alone. Antigen K.sub.a (1/Ms) K.sub.d (1/s) K.sub.D (M) Cetuximab- EGFR 1.22 .times. 10.sup.6 8.65 .times. 10.sup.-4 7.08 .times. 10.sup.-10 T22d35 Cetuximab EGFR 1.03 .times. 10.sup.6 8.45 .times. 10.sup.-4 8.47 .times. 10.sup.-10 Her-T22d35 Her2 8.30 .times. 10.sup.4 5.30 .times. 10.sup.-5 6.37 .times. 10.sup.-10 Herceptin Her2 6.88 .times. 10.sup.4 5.03 .times. 10.sup.-5 7.33 .times. 10.sup.-10 Syn-T22d35 RSV-F 3.55 .times. 10.sup.4 1.42 .times. 10.sup.-3 4.10 .times. 10.sup.-9 Synagis RSV-F 2.57 .times. 10.sup.4 1.68 .times. 10.sup.-3 6.60 .times. 10.sup.-9 NOTE: FC-5-Fc, FC5-Fc-T2m, and FC-5-Fc-T22d35 binding affinity was assessed via the transwell functional assay (see Example 3C).
EXAMPLE 3B
Validation of Cetuximab Function
[0176] The ability of Cetuximab to maintain its therapeutic function (i.e. inhibition of EGF-induced EGFR phosphorylation and signaling) when fused to either T2m or T22d35 (Example 1) was evaluated.
[0177] Phosphorylation of EGFR: The ability of Cetuximab-comprising constructs to inhibit EGF-induced phosphorylation of EGFR in human lung cancer A549 cells was evaluated. A549 cells were seeded in 24-well plates (100,000 cells/well) and either mock treated (-) or pre-treated with Cetuximab, Cet-T2m, or Cet-T22d35 (all at 10, 1 or 0.1 nM) or T22d35 (10 nM) at 37.degree. C. for 3 h, then treated with 50 ng/mL EGF at 37.degree. C. for 10 min. Whole cell lysates were prepared and resolved by SDS-PAGE, western blotted and probed with anti-phosphoTyrosine antibody (Clone 4G10, Millipore 05-321). As shown in FIG. 5A, Cetuximab and Cet-T2m and Cet-T22d35 inhibited EGFR phosphorylation to similar extents, whereas T22d35 had no effect, relative to the +EGF control. These results thus confirm that the TGF-.beta. superfamily receptor ectodomain moieties in Cetuximab-T2m and Cetuximab-T22d35 fusion proteins do not interfere with the function of the Cetuximab antibody (i.e. inhibition of EGF induced EGFR signaling).
[0178] Inhibition of EGFR signaling: Inhibition of autocrine EGFR signaling results in varying degrees of cytotoxicity in EGFR-expressing cells treated with Cetuximab. Cytotoxicity of Cet-T22d35 was compared to Cetuximab and T22d35 in human breast cancer cells (MDA-MB-468) and immortalized keratinocyte cells (HaCaT). These cells exhibited significant Cetuximab cytotoxicity, due to their intrinsic dependence on the EGF signaling pathway for basal growth. The cells were seeded onto 96-well plates (MDA-MB-468, 2300 cells/well; HaCaT, 1500 cells/well) and then treated with different doses of inhibitor at 37.degree. C. for 5 days. Cell viability was measured using sulforhodamine reagent to determine the percentage of viable cells relative to mock-treated controls. Results are shown in FIG. 5B and Table 5. The IC.sub.50 values for Cet-T22d35 and Cetuximab were similar in both cell lines (0.2-1.4 nM range), while T22d35 resulted in no cytotoxicity. These results further confirm that the TGF-.beta. superfamily receptor ectodomain moiety in Cetuximab-T22d35 does not interfere with the function of the Cetuximab antibody.
TABLE-US-00015 TABLE 5 Cetuximab-T22d35 cytotoxicity in MDA-MB-468 and HaCat cells. The cytotoxic potency IC.sub.50 values are given in nM. MDA-MB-468 IC.sub.50 HaCaT IC.sub.50 Cetuximab 0.50 0.33 Cetuximab-T22d35 1.42 0.22 T22d35 0 0
EXAMPLE 3C
Validation of FC5 Function
[0179] The ability of FC5 V.sub.HH to maintain its function (i.e. transmigrate the blood-brain barrier) when fused with the T2m or T22d35 moieties (see description in Example 2 and activities in Table 2) was evaluated.
[0180] Briefly, SV40-immortalized Adult Rat Brain Endothelial Cells (Sv-ARBEC) were used to generate an in vitro blood-brain barrier (BBB) model as described (Garberg et al, 2005; Haqqani et al, 2013). Sv-ARBEC cells (80,000 cells/membrane) were seeded on 0.1 mg/mL rat tail collagen type I-coated tissue culture inserts (pore size-1 .mu.m; surface area 0.9 cm.sup.2, Falcon) in 1 ml of growth medium. The bottom chamber of the insert assembly contained 2 ml of growth medium supplemented with the immortalized neonatal rat astrocytes-conditioned medium in a 1:1 (v/v) ratio. Equimolar amounts (5.6 .mu.M) of positive (FC5-Fc) control; negative controls (A20.1); and T22d35, T2m, FC5-Fc-T22d35 or FC5-Fc-T2m were tested for their ability to cross the Sv-ARBEC cell monolayer. Following exposure of equimolar amounts of the proteins to the luminal side of the BBB, samples were taken after 15, 30, and 60 min from the abluminal side. The protein content of each sample was then quantified by mass spectrometry (multiple reaction monitoring-isotype labeled internal standards; MRM-ILIS) as described by (Haqqani et al, 2013) (see method description below).
[0181] Quantified values can be directly plotted or the P.sub.app (apparent permeability coefficient) values can be determined using the following formula
Papp = d Q r / d t A .times. C 0 ##EQU00001##
[0182] The P.sub.app value is commonly used to determine the specific permeability of a molecule, and is a measure of transport across the brain endothelial monolayer. [Qr/dt=cumulative amount in the receiver (bottom) compartment versus time; A=area of the cell monolayer; C0=initial concentration of the dosing solution (top chamber)].
[0183] FIG. 6 shows the results of the experiment. The P.sub.app value of FC5-Fc-T22d35 was similar to the control FC5-Fc, indicating it was transported efficiently and that the fused T22d35 did not interfere with transport. The P.sub.app value for FC5-Fc-T2m was approximately 50% less, compared to FC5-Fc-T22d35 and FC5-Fc, indicating somewhat reduced permeability. Nevertheless, the level of transport of FC5-Fc-T2m was about 4-fold greater than the negative controls (T2m, T22d35, and antibody A20.1).
EXAMPLE 4
Inhibition of Epithelial to Mesenchymal Transition
[0184] Treatment of A549 cells with EGF plus TGF-.beta. results in a strong epithelial to mesenchymal transition (EMT). The EMT is phenotypically characterized by changes in cell morphology (tight cellular junctions with "cobble-stone" appearance converts to elongated cells, see FIG. 7A) and changes in the adherin junction proteins E-cadherin and N-cadherin. The ability of the fusion constructs to block EMT was assessed in A549 cells by western blotting (E-cadherin) and flow cytometry (E-cadherin and N-cadherin).
[0185] Briefly, for the western blot analysis, A549 cells were seeded in 24-well plates (8000 cells/well) and then treated with EGF (50 ng/mL)+TGF-.beta.1 (50 pM) at 37.degree. C. for 3 days in the presence of Cet-T22d35, Cetuximab, or T22d35 (0, 0.05, 0.5, 5, 50, or 500 nM). Whole cell lysates were prepared and resolved by SDS-PAGE. The proteins were transferred to nitrocellulose and then probed with an E-cadherin antibody (BD Transduction laboratories Biosciences) (FIG. 7B). The E-Cadherin positive bands in the Western blot were quantified by densitometer detection and ImageJ analysis (FIG. 7C). EGF+TGF-.beta. treatment resulted in an EMT, as indicated by the disappearance of E-cadherin (compare non-treated and EGF+TGF-.beta. lanes in the absence of inhibitors). Cet-T22d35 blocked the EMT (E-cadherin disappearance) in a dose-dependent manner whereas 500 nM Cetuximab or T22d35 treatments only modestly blocked the EMT (E-cadherin levels .about.20-25% of the non-treated control level).
[0186] The ability of Cet-T22d35, Cetuximab and T22d35 to block the EGF+TGF-.beta. EMT response was further examined by flow cytometry using A549 cells treated with Cet-T22d35 or Cetuximab, (all at 50, 5, 0.5 nM) or the Cetuximab+T22d35 combination (50 nM+100 nM, 5 nM+10 nM, or 0.5 nM+1 nM, respectively) and evaluating the EMT associated changes in E-cadherin and N-cadherin cell surface expression levels (FIG. 7D). In this experiment the molar amounts of the molecules of interest used in `T22d35 alone` and `Cetuximab+T22d35` groups were two-fold higher than for Cet-T22d35 in order to correspond with a 2:1 trap/antibody ratio in the Cet-T22d35 fusion molecule. A549 cells were seeded in 6-well plates (30,000 cells/well) and pre-treated with the inhibitors at 37.degree. C. for 1 h, followed by added treatment with EGF (10 ng/mL)+TGF-.beta.1 (10 pM) and incubation at 37.degree. C. for 3 days. Cells were then dissociated from the plate using 1 mL Dissociation Buffer (Sigma)/well, centrifuged at 2000 rpm for 2 min and re-suspended in 100 .mu.l RPMI-5 media at 4.degree. C. AlexaFluor488-E-cadherin (Santa Cruz, SC21791) and AlexaFluor647-N-cadherin (BD Biosciences, 563434) antibodies (1/25 v/v dilutions) were added and samples were incubated at 4.degree. C. for 1 h. Cells were then centrifuged, washed once in RPMI-5, and re-suspended in 400 .mu.l RPMI-5 containing 15 .mu.g/mL propidium iodide (Life Technologies) at 4.degree. C. Mean fluorescent intensities (MFI) were measured by flow cytometry (BD LS RII flow cytometer, BD Biosciences) to quantify E-cadherin and N-cadherin levels. The results show that Cet-T22d35 was more effective in preventing down-regulation of E-cadherin (FIG. 7D, top panel) and up-regulation of N-cadherin (FIG. 7D, bottom panel), as a measure of blocking EMT, compared to Cetuximab, T22d34 or the Cetuximab+T22d35 combination at each respective dose, and is most notable at the lowest dose used (0.5 nM).
EXAMPLE 5
Pharmacokinetic (PK) Studies on Constructs with and Without a Lysine Residue at the Fusion Site Between the C-Terminus of the Fc Region and the N-Terminus of the Ectodomain
[0187] PK studies were carried out in normal, healthy mice to determine whether fusion of T22d35 to an antibody increased its half-life in vivo, and whether removal of a lysine at the fusion site within constructs reduced the amount of cleavage occurring in vivo.
[0188] Results from Cet-T22d35 (construct Type D in FIG. 2--containing a lysine at the fusion site): A single bolus of Cet-T22d35 protein (10 mgs/Kg) formulated in DPBS was intravenously injected (IV) into the tail vein of normal Balb/c mice and serum samples were collected from the submandibular vein at selected time points (0.5, 1, 2, 4, 8, 14, 24, 48, 96 h). Blood samples were centrifuged at 2000 g at 4.degree. C. for 10 min and the serum supernatant was removed and stored frozen at -80.degree. C., prior to analyses. The samples were thawed at 4.degree. C. and analyzed via mass spectrometry (multiple reaction monitoring-isotype labeled internal standards; MRM-LIS) in order to measure the levels of both the Cetuximab and T22d35 trap moieties. Briefly, 20 .mu.l of sample was thawed and treated with mild detergents (0.1% RapiGest SF, Waters; 5.5 mM TCEP) at 95.degree. C. for 10 min. The sample was cooled to room temperature and Iodoacetamide (IAA) in 50 mM Ammonium Bicarbonate was added to a final concentration of 10 mM IAA, followed by incubation for 40 min in the dark. DTT (10 mM final) was then added and the sample was incubated at room temperature for 15 min, followed by trypsin digestion (Sigma, 0.8 mg/mL final) at 37.degree. C. for 18 h. A mixture of 5 .mu.M each of isotope-labeled trap and cetuximab peptides (formulated in 30% acetonitrile, 0.1% formic acid) were added to final concentrations of 1 .mu.M, as internal standards for quantification. The isotope-labeled peptides were 13C/15N--(H2N-LPYHDFILEDAASPK-OH; SEQ ID NO:134) and 13C/15N--(H2N-ALPAPIEK-OH; SEQ ID NO:135) for T22d35 and Cetuximab, respectively (NewEngland Peptide). Trifluoroacetic acid was then added (0.5% final), followed by incubation at 37.degree. C. for 30 min. The samples were centrifuged at 13000 rpm for 20 min and the supernatant was used analyzed via MRM-ILIS using an Agilent 1260 HPLC system coupled with Agilent QQQ6410B at 55.degree. C. The PK profiles seen in FIG. 8A show that the levels of the Cetuximab and T22d35 moieties diverge at early time points (<10 h) after injection, indicating different kinetics and suggesting possible cleavage of T22d35 from the Cet-T22d35 protein in vivo. Nevertheless, analysis of these curves using a two compartmental model (Phoenix WinNonlin SoftwareVersion 6.3) indicated that the average terminal half-life (T1/2 .beta.) of the T22d35 component was 45.8 h. This represents a 7.6-fold increase compared to the previously determined half-life for unfused T22d35 (T1/2 .beta..about.6 h). As well, the PK profile shows that the Cetuximab moiety was maintained in the blood, with a T1/2 .beta.=262.5 h, indicating that the Cetuximab moiety has a long circulating half-life.
[0189] Results from several constructs with lysine deleted at the fusion site:
[0190] The same methods that were used in the PK study of Cet-T22d35 (FIG. 8A) were applied to assess the PK of Cet.DELTA.K-T2m (Type F in FIG. 2) as well as several "headless" T2m constructs (Type C in FIG. 2; hIgG1Fc.DELTA.K(SS)-T2m, hIgG1Fc.DELTA.K(.DELTA.C)-T2m, and hIgG2Fc.DELTA.K(SS)-T2m), all of which have the lysine deleted at the fusion site. The data shown in FIG. 8B indicate that no detectable cleavage of these constructs is occurring in vivo since the levels of the Fc moieties (closed symbols) and ectodomain moieties (open symbols) do not diverge over time. Additionally, all of the constructs exhibit similar long circulating half-lives with T1/2 .beta.s of approximately 100 h. This represents an improvement to the half-life of the ectodomain moiety of the construct which has a lysine at the fusion site, presented in FIG. 8A (45.8 h).
EXAMPLE 6
Efficacy Studies Comparing the Effect of "Headless" Fc-T2m and FSA-T2M Constructs on Tumor Growth (A) and T-Cell Function (B and C) in an Immune-Competent Syngeneic Triple Negative Breast Cancer (4T1) Model
[0191] A FSA-T2m construct (Cet-T2m--Type F in FIG. 2) and three headless constructs, all with engineered N-termini, (hIgG1Fc.DELTA.K(CC)-T2m, hIgG1Fc.DELTA.K(C)-T2m, and hIgG2Fc.DELTA.K(CC)-T2m--Type C in FIG. 2) were evaluated for their ability to inhibit tumor growth and to affect T-cell function in a syngeneic tumor model derived from 4T1 triple negative breast cancer cells. The results presented in FIG. 9 show the effect on tumor growth (A) and T-cell function (B, C). The effects of these Fc-fused ectodomain constructs were compared to those of a pan-specific neutralizing anti-TGF-.beta. antibody, 1D11 and a non-Fc-fused ectodomain construct, T22d35.
[0192] The protocols used for these syngeneic mouse model studies are described in (Zheng et al, 2013). Briefly, female BALB/c (H-2Kd) mice 6 weeks of age were purchased from The Jackson Laboratories and kept in filter-top cages. The 4T1 breast cancer cells and B16F10 cells were purchased from American type culture collection and cultured in RPMI-1640 supplemented with 2 mmol/L L-glutamine, 100 U/mL penicillin, 100 microgm/L streptomycin, 50 micromol/L 2-mercaptoethanol, and 10% fetal calf serum. Mice were inoculated subcutaneously in the left flank with 100 uL sterile saline containing 5.times.10.sup.4 4T1 cells. Tumors were grown to .about.100 mm.sup.3, as measured by caliper, then mice were randomized and divided into the six treatment groups (8 animals/group) (Day 0). Treatments commenced on Day 1 and continued for 15 days with the animals being dosed at 5 mg/kg twice per week such that they received a total of 4 doses. Tumor growth was monitored by caliper measurements 3.times. per week. Animals were euthanized by exsanguination under anaesthesia on Day 15; T cells were isolated from draining lymph nodes and assessed for their capacity to kill mouse 4T1 and B16F10 tumour cells ex vivo. The capacity of T cells from mice treated with or without test agents to lyse target 4T1 tumor cells was measured using a CytoTox 96 nonradioactive cytotoxicity assay kit (Promega) according to the manufacturer's instructions. Briefly, naive target 4T1 cells or melanoma cell line B16F10 cells are plated and incubated for 4 hr with CD8+ effector T cells isolated from 4T1 tumor-bearing mice using CD8 magnetic MicroBeads (BD Bioscience). The isolated CD8+ cells are confirmed, by flow cytometry, to be over 85% CD8+. A range of ratios of effectors to target cells is tested (100:1, 50:1, 25:1). Lactate dehydrogenase (LDH) release in response to effector T cells is measured in the buffer bathing target cells. Target cells incubated in the absence of effector cells are used as a comparator to control for spontaneous LDH release. Released LDH in culture supernatants is detected after a 30-min incubation using a coupled enzymatic assay. The intensity of the color formed is proportional to the number of lysed cells. Cytotoxic activity of CTL is calculated using the following formula:
Cytotoxic activity %=[(absorbance)-(spontaneous effector cell LDH release)-(spontaneous target cell LDH release)]/[(maximal LDH release)--(spontaneous target cell LDH release)].times.100
[0193] The results presented in FIG. 9A show the effect of the Fc-fused ectodomain constructs listed above on 4T1 tumor growth (the pan-specific neutralizing anti-TGF-.beta. antibody, 1 D11, and a non-Fc-fused ectodomain construct, T22d35, being tested as comparators). As can be seen in FIG. 9A, all of the Fc-fused ectodomain constructs reduced tumor growth relative to the saline treatment when tested for significance by t-test. The 1 D11 and T22d35 comparator treatments were observed to be less effective relative to the Fc-fused ectodomain treatments, and not significantly different from the saline control. These results demonstrate that constructs with an ectodomain fused to the C-terminus of an Fc region have significant anti-tumor potency, with the efficacy being higher than the 1 D11 and T22d35 comparators. There was no significant difference between the FSA-T2m construct (Cet-T2m) and the constructs that have no Fab, i.e. are "headless" (hIgG1Fc.DELTA.K(CC)-T2m, hIgG1Fc.DELTA.K(C)-T2m, and hIgG2Fc.DELTA.K(CC)-T2m) with respect to their effect on tumor growth. The high anti-tumor potency of these Fc-ectodomain fusions relative to comparators likely results from high potency neutralization of TGF-.beta. combined with a favourable circulating half-life.
[0194] To investigate whether these Fc-ectodomain fusions exhibit an immuno-modulatory effect in vivo on cytotoxic T lymphocyte cells (CTLs) present in tumor draining lymph nodes, lymph nodes were removed from mice treated with or without test agents; T-cells were then isolated and tested for their capacity to lyse target 4T1 tumor cells (and B16F10 cells as a test of tumor specificity) using the methods described above.
[0195] As shown in FIG. 9B, treatment of the animals with Fc-ectodomain fusions significantly increased the ability of draining lymph node T-cells to lyse target 4T1 tumor cells ex vivo. It can be seen that this immuno-stimulatory effect is specific to 4T1 cells since the T-cells were not able to effectively lyse B16F10 melanoma cells (FGURE 9C; maximal lysis of .about.15% for B16F10 cells and .about.80% for 4T1 cells). When administered at the same 5 mg/kg dose as the Fc-ectodomain fusions, the non-Fc-fused comparator molecule, T22d35, had no effect above saline. This is consistent with its lack of effect on tumor volume. Although the 1 D11 antibody had no statistically significant effect on tumor volume (FIG. 9A), it did increase the ability of lymph node T-cells to lyse 4T1 tumor cells.
[0196] Interestingly, with respect to the Fc-ectodomain fusions of this invention, the most potent constructs were the hIgG1Fc.DELTA.K(CC)-T2m and hIgG2Fc.DELTA.K(CC)-T2m constructs; both of these constructs containing two cysteines in the engineered hinge region. These constructs were more potent than the construct with one cysteine in the engineered hinge region, hIgG1Fc.DELTA. K(C)-T2m, as well as being more potent than the full-size antibody-T2m construct, Cet-T2m. The difference in potency between constructs with one versus two hinge region cysteines may result from the construct with one cysteine having a lower relative stability. The lower potency of the full size antibody construct relative to the headless constructs with two hinge region cysteines may result from a difference in molecular weight, with the smaller constructs being able to penetrate the tumor microenvironment more effectively.
EXAMPLE 7
In Vitro and In Vivo Studies Illustrating Enhanced Bone Localization of Constructs Containing a Deca-Aspartate Motif for Bone Targeting at the N-Terminus of the Fc Region
[0197] In vivo studies were carried out to investigate whether the addition of a 10 amino acid long poly-aspartate bone-localization motif (D10) to the N-terminus of the Fc region of constructs will promote their localization to bone. Optical imaging of D10-hIgG1Fc-T2m fusions: Upon arrival, male Balb/c mice were housed 3 mice/cage. On the day of the experiment, animals were shaved dorsal and ventral and treated with the hair removal cream, NAIR.RTM.. Mice were injected with a single intravenous bolus of 10 mg/kg of two CF770 labeled constructs with a deca-aspartate motif for bone targeting (D10) at their N-termini (D10-hIgG1Fc.DELTA.K(CC)-T2m, D10-GSL-hIgG1Fc.DELTA.K(CC)-T2m) or with a control construct without the D10 motif (hIgG1Fc.DELTA.K(CC)-T2m) and whole body bio-distribution followed using both in vivo and ex-vivo near infrared imaging. Imaging was conducted with a small-animal time-domain eXplore Optix pre-clinical imager MX3 (Advanced Research Technologies, ART) at various time points (prescan, 5 mins, 3 hr, 6 hr, 24 h, 48 h, 72 h, 96 h and 120 h).
[0198] The small animal time-domain eXplore Optix preclinical imagers consists of a 785-nm pulsed laser diode with a repetition frequency of 80 MHz and a time resolution of 12.5 ps light pulse was used for excitation. The fluorescence emission beyond 813 nm was collected by a highly sensitive time-correlated single photon counting system and detected through a fast photomultiplier tube. The data were recorded as temporal point-spread functions (TPSF) and the images were presented as fluorescence intensity maps using ART Optix Optiview analysis software 3.02.
[0199] For in vivo optical imaging, mice were first anesthetized using isofluorane (1.5-2%), positioned on the animal stage within a chamber which allows for gaseous anesthesia and maintenance of animal temperature at 36.degree. C. The scanning of the mouse at each time point lasted up to 20 mins using a 2.5 mm step size and the mouse is placed back in its home cage between imaging time points.
[0200] At the end of the imaging protocol (120 hrs) animals were sacrificed by intracardiac perfusion using heparnized saline with deep anesthesia. The organs (brain, heart, lungs, liver, kidney, spleen and right and left leg bones) were imaged ex-vivo using a 1.0 mm step size using the eXplore Optix pre-clinical imager MX3.
[0201] Data analysis was done using eXplore Optix Optiview analysis software 3.02 (Advanced Research Technologies, Montreal, QC) to estimate the fluorescence total and average fluorescence intensity in region of interest containing the ex-vivo organs.
[0202] The results shown in FIGS. 10A and B demonstrate that the fusion of the deca-aspartate D10 motif on the N-termini of the fusion constructs had no impact on their ability to neutralize TGF-.beta., i.e. the IC.sub.50 of the construct lacking the D10 motif (hIgG1Fc.DELTA.K(CC)-T2m) was 3 nM, which is the same as the value determined in FIG. 4C, while the IC.sub.50s of the D10 containing constructs, D10-hIgG1Fc.DELTA.K(CC)-T2m and D10-GSL-hIgG1Fc.DELTA.K(CC)-T2m, were very similar at 4-5 nM. The results in FIGS. 10A and B also indicate that labeling with the CF770 dye reduced the ability of the constructs to neutralize TGF-.beta. by approximately 4-fold. Since dye conjugation occurs at lysine residues, and since it is know that lysines are at the binding interface between the Type II ectodomain and TGF-.beta., it is not entirely surprising that labeling reduced neutralization potency. In any case, since this is a comparative study of differences in in vivo localization promoted by the D10 peptide, it was felt that partially active constructs would be informative.
[0203] The results shown in FIGS. 10C and D demonstrate that the addition of the D10 peptide to the N-termini of the constructs greatly enhanced bone localization. Images taken 120 h post-injection of the CF770 labeled fusions show a clear accumulation of the D10-fusions in the vertebrae. Further ex vivo imaging of the brain, heart, lungs, liver, kidneys, spleen, and the left and right legs 120 h post-injection confirmed the specific accumulation of the D10-fusions in the bones. The fluorescent signals observed in the kidneys and liver were similar for all fusions indicating that accumulation in these organs was not affected by the presence of the D10 sequence. These results indicate that the TGF-.beta. neutralization activity of constructs may be increased within bone through the addition of the D10 peptide. This could result in more favourable dosing levels and schedules for the treatment of bone-related diseases, such as osteogenesis imperfecta, relative to that required for a similar construct without the D10 motif.
[0204] The embodiments and examples described herein are illustrative and are not meant to limit the scope of the invention as claimed. Variations of the foregoing embodiments, including alternatives, modifications and equivalents, are intended by the inventors to be encompassed by the claims. Furthermore, the discussed combination of features might not be necessary for the inventive solution.
TABLE-US-00016 LISTING OF SEQUENCES SEQ ID NO: Sequence Description 1 APELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDG Human IgG1 Fc VEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPI region EKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAVEWES NGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHN HYTQKSLSLSPGK 2 APPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVQFNWYVDGV Human IgG2 Fc EVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNGKEYKCKVSNKGLPAPIE region KTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDISVEWESN GQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNH YTQKSLSLSPGK 3 APELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVQFKWYVDG Human IgG3 Fc VEVHNAKTKPREEQYNSTFRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPI region EKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDIAVEWES SGQPENNYNTTPPMLDSDGSFFLYSKLTVDKSRWQQGNIFSCSVMHEALHN RFTQKSLSLSPGK 4 APEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDG Human IgG4 Fc VEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSI region EKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKGFYPSDIAVEWES NGQPENNYKTTPPVLDSDGSFFLYSRLTVDKSRWQEGNVFSCSVMHEALHN HYTQKSLSLSLGK 5 EPKSCDKTHTCPPCP Human IgG1 hinge region 6 ERKCCVECPPCP Human IgG2 hinge region 7 ELKTPLGDTTHTCPRCPEPKSCDTPPPCPRCPEPKSCDTPPPCPRCPEPKS Human IgG3 CDTPPPCPRCP hinge region 8 ESKYGPPCPSCP Human IgG4 hinge region 9 APELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDG hIgG1Fc.DELTA.K-.DELTA.C VEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPI Fc variant EKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAVEWES NGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHN HYTQKSLSLSPG 10 PPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNW hIgG1Fc.DELTA.K-C Fc YVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKAL variant PAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAV EWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHE ALHNHYTQKSLSLSPG 11 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDP hIgG1Fc.DELTA.K-CC EVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCK Fc variant VSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFY PSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFS CSVMHEALHNHYTQKSLSLSPG 12 EPKSSDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDV hIgG1Fc.DELTA.K-S Fc SHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGK variant EYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCL VKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQ GNVFSCSVMHEALHNHYTQKSLSLSPG 13 EPKSSDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDV hIgG1.DELTA.K-SS Fc SHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGK variant EYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCL VKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQ GNVFSCSVMHEALHNHYTQKSLSLSPG 14 EPKSSDKTHTSPPSPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDV hIgG1Fc.DELTA.K-SSS SHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGK Fc variant EYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCL VKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQ GNVFSCSVMHEALHNHYTQKSLSLSPG 15 APPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVQFNWYVDGV hIgG2Fc.DELTA.K-.DELTA.C EVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNGKEYKCKVSNKGLPAPIE Fc variant KTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDISVEWESN GQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNH YTQKSLSLSPG 16 PPCPAPPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVQFNWY hIgG2Fc.DELTA.K-C Fc VDGVEVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNGKEYKCKVSNKGLP variant APIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDISVE WESNGQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEA LHNHYTQKSLSLSPG 17 VECPPCPAPPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVQF hIgG2Fc.DELTA.K-CC NWYVDGVEVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNGKEYKCKVSNK Fc variant GLPAPIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDI SVEWESNGQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVM HEALHNHYTQKSLSLSPG 18 ERKCCVECPPCPAPPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHED hIgG2Fc-CCCC PEVQFNWYVDGVEVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNGKEYKC Fc variant KVSNKGLPAPIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGF YPSDISVEWESNGQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQQGNVF SCSVMHEALHNHYTQKSLSLSPGK 19 ERKSSVECPPCPAPPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHED hIgG2Fc.DELTA.K-SS PEVQFNWYVDGVEVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNGKEYKC Fc variant KVSNKGLPAPIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGF YPSDISVEWESNGQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQQGNVF SCSVMHEALHNHYTQKSLSLSPG 20 ERKSSVESPPCPAPPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHED hIgG2Fc.DELTA.K-SSS PEVQFNWYVDGVEVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNGKEYKC Fc variant KVSNKGLPAPIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGF YPSDISVEWESNGQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQQGNVF SCSVMHEALHNHYTQKSLSLSPG 21 ERKSSVESPPSPAPPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHED hIgG2Fc.DELTA.K-SSSS PEVQFNWYVDGVEVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNGKEYKC Fc variant KVSNKGLPAPIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGF YPSDISVEWESNGQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQQGNVF SCSVMHEALHNHYTQKSLSLSPG 22 APELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVQFKWYVDG hIgG3Fc.DELTA.K-.DELTA.C VEVHNAKTKPREEQYNSTFRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPI Fc variant EKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDIAVEWES SGQPENNYNTTPPMLDSDGSFFLYSKLTVDKSRWQQGNIFSCSVMHEALHN RFTQKSLSLSPG 23 PRCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVQFKW hIgG3Fc.DELTA.K-C Fc YVDGVEVHNAKTKPREEQYNSTFRVVSVLTVLHQDWLNGKEYKCKVSNKAL variant PAPIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDIAV EWESSGQPENNYNTTPPMLDSDGSFFLYSKLTVDKSRWQQGNIFSCSVMHE ALHNRFTQKSLSLSPG 24 DTPPPCPRCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDP hIgG3Fc.DELTA.K-CC EVQFKWYVDGVEVHNAKTKPREEQYNSTFRVVSVLTVLHQDWLNGKEYKCK Fc variant VSNKALPAPIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFY PSDIAVEWESSGQPENNYNTTPPMLDSDGSFFLYSKLTVDKSRWQQGNIFS CSVMHEALHNRFTQKSLSLSPG 25 EPKSSDTPPPCPRCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDV hIgG3Fc.DELTA.K-S Fc SHEDPEVQFKWYVDGVEVHNAKTKPREEQYNSTFRVVSVLTVLHQDWLNGK variant EYKCKVSNKALPAPIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCL VKGFYPSDIAVEWESSGQPENNYNTTPPMLDSDGSFFLYSKLTVDKSRWQQ GNIFSCSVMHEALHNRFTQKSLSLSPG 26 EPKSSDTPPPSPRCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDV hIgG3Fc.DELTA.K-SS SHEDPEVQFKWYVDGVEVHNAKTKPREEQYNSTFRVVSVLTVLHQDWLNGK Fc variant EYKCKVSNKALPAPIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCL VKGFYPSDIAVEWESSGQPENNYNTTPPMLDSDGSFFLYSKLTVDKSRWQQ GNIFSCSVMHEALHNRFTQKSLSLSPG 27 EPKSSDTPPPSPRSPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDV hIgG3Fc.DELTA.K-SSS SHEDPEVQFKWYVDGVEVHNAKTKPREEQYNSTFRVVSVLTVLHQDWLNGK Fc variant EYKCKVSNKALPAPIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCL VKGFYPSDIAVEWESSGQPENNYNTTPPMLDSDGSFFLYSKLTVDKSRWQQ GNIFSCSVMHEALHNRFTQKSLSLSPG 28 APEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDG hIgG4Fc.DELTA.K-AC VEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSI Fc variant EKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKGFYPSDIAVEWES NGQPENNYKTTPPVLDSDGSFFLYSRLTVDKSRWQEGNVFSCSVMHEALHN HYTQKSLSLSLG 29 PSCPAPEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNW hIgG4Fc.DELTA.K-C Fc YVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGL variant PSSIEKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKGFYPSDIAV EWESNGQPENNYKTTPPVLDSDGSFFLYSRLTVDKSRWQEGNVFSCSVMHE ALHNHYTQKSLSLSLG 30 ESKYGPPCPSCPAPEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQE hIgG4Fc.DELTA.K-CC DPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYK Fc variant CKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKG FYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSRLTVDKSRWQEGNV FSCSVMHEALHNHYTQKSLSLSLG 31 ESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQE hIgG4Fc.DELTA.K-CC- DPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYK 228P Fc variant CKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKG FYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSRLTVDKSRWQEGNV FSCSVMHEALHNHYTQKSLSLSLG 32 ESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQE hIgG4Fc.DELTA.K-CC- DPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYK 228P-409K Fc CKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKG variant FYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQEGNV FSCSVMHEALHNHYTQKSLSLSLG 33 ESKYGPPSPSCPAPEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQE hIgG4Fc.DELTA.K-S Fc DPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYK variant CKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKG FYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSRLTVDKSRWQEGNV FSCSVMHEALHNHYTQKSLSLSLG 34 ESKYGPPSPSSPAPEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQE hIgG4Fc.DELTA.K-SS DPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYK Fc variant CKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKG FYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSRLTVDKSRWQEGNV FSCSVMHEALHNHYTQKSLSLSLG 35 QLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLET T.beta.RII-ED VCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIF structured domain 36 IPPHVQKSVNNDMIVTDNNGAVKFP T.beta.RII-ED N-term unstructured region and natural linker 37 IPPHVQKSDVEMEAQKDEIICPSCNRTAHPLRHINNDMIVTDNNGAVKFP T.beta.RIIb-ED N-term unstructured region and natural linker 38 IPPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINNDMIVTDNNGAVKFP T.beta.RIIb-ED Cys- mutated N-term unstructured region and natural linker 39 SEEYNTSNPD T.beta.RII-ED C-term unstructured region and natural linker 40 SEEYNTSNPDIPPHVQKSVNNDMIVTDNNGAVKFP T.beta.RII-ED natural
linker 41 SEEYNTSNPDIPPHVQKSDVEMEAQKDEIICPSCNRTAHPLRHINNDMIVT T.beta.RIIb-ED natural DNNGAVKFP linker 42 SEEYNTSNPDIPPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINNDMIVT T.beta.RIIb-ED Cys- DNNGAVKFP mutated linker 43 IPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSIT T.beta.RII-ED SICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKE monomer, also KKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD termed T2 or T2m 44 IPPHVQKSDVEMEAQKDEIICPSCNRTAHPLRHINNDMIVTDNNGAVKFPQ T.beta.bRIIb-ED LCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETV monomer, also CHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSE termed T2b EYNTSNPD 45 IPPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINNDMIVTDNNGAVKFPQ T.beta.RIIb-ED LCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETV monomer Cys- CHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSE mutated in the EYNTSNPD linker region, also termed T2b.sup.AA 46 IPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSIT T.beta.RII-ED dimer, SICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKE also termed T2- KKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSVNNDMIVTD T2 or T22d35 NNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKN DENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDE CNDNIIFSEEYNTSNPD 47 IPPHVQKSDVEMEAQKDEIICPSCNRTAHPLRHINNDMIVTDNNGAVKFPQ T.beta.RIIb-ED dimer, LCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETV also termed T2- CHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSE T2b EYNTSNPDIPPHVQKSDVEMEAQKDEIICPSCNRTAHPLRHINNDMIVTDN NGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKND ENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDEC NDNIIFSEEYNTSNPD 48 IPPHVQKSDVEMEAQKDEIICPSCNRTAHPLRHINNDMIVTDNNGAVKFPQ T.beta.RIIb-ED dimer LCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETV Cys-mutated in CHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSE the linker region, EYNTSNPDIPPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINNDMIVTDN also termed T2- NGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKND T2b.sup.AA ENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDEC NDNIIFSEEYNTSNPD 49 IPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSIT T.beta.RII-ED trimer, SICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKE also termed T2- KKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSVNNDMIVTD T2-T2 NNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKN DENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDE CNDNIIFSEEYNTSNPDIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVR FSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPY HDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 50 IPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSIT T.beta.RIIb-ED trimer, SICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKE also termed T2- KKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSDVEMEAQKD T2b-T2b EIICPSCNRTAHPLRHINNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKS CMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAA SPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSD VEMEAQKDEIICPSCNRTAHPLRHINNDMIVTDNNGAVKFPQLCKFCDVRF STCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYH DFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 51 IPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSIT T.beta.RIIb-ED trimer SICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKE Cys-mutated in KKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSDVEMEAQKD the linker regions, EIIAPSANRTAHPLRHINNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKS also termed T2- CMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAA T2b.sup.AA-T2b.sup.AA SPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSD VEMEAQKDEIIAPSANRTAHPLRHINNDMIVTDNNGAVKFPQLCKFCDVRF STCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYH DFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 52 ALQCFCHLCTKDNFTCVTDGLCFVSVTETTDKVIHNSMCIAEIDLIPRDRP T.beta.RI-ED FVCAPSSKTGSVTTTYCCNQDHCNKIEL structured domain 53 AALLPGAT T.beta.RI-ED N-term unstructured region and natural linker 54 PTTVKSSPGLGPVE T.beta.RI-ED C-term unstructured region and natural linker 55 PTTVKSSPGLGPVEAALLPGAT T.beta.RI-ED natural linker 56 AALLPGATALQCFCHLCTKDNFTCVTDGLCFVSVTETTDKVIHNSMCIAEI T.beta.RI-ED DLIPRDRPFVCAPSSKTGSVTTTYCCNQDHCNKIELPTTVKSSPGLGPVE monomer, also termed T1 or T1m 57 AALLPGATALQCFCHLCTKDNFTCVTDGLCFVSVTETTDKVIHNSMCIAEI T.beta.RI-ED dimer, DLIPRDRPFVCAPSSKTGSVTITYCCNQDHCNKIELPTTVKSSPGLGPVEA also termed T1- ALLPGATALQCFCHLCTKDNFTCVTDGLCFVSVTETTDKVIHNSMCIAEID T1 LIPRDRPFVCAPSSKTGSVTTTYCCNQDHCNKIELPTTVKSSPGLGPVE 58 PTTVKSSPGLGPVEIPPHVQKSVNNDMIVTDNNGAVKFP T.beta.RI-T.beta.RII-ED natural linker 59 PTTVKSSPGLGPVEIPPHVQKSDVEMEAQKDEIICPSCNRTAHPLRHINND T.beta.RI-T.beta.RIIb-ED MIVTDNNGAVKFP natural linker 60 SEEYNTSNPDAALLPGAT T.beta.RII-T.beta.RI-ED natural linker 61 AALLPGATALQCFCHLCTKDNFTCVTDGLCFVSVTETTDKVIHNSMCIAEI T.beta.RI-T.beta.RII-ED DLIPRDRPFVCAPSSKTGSVTTTYCCNQDHCNKIELPTTVKSSPGLGPVEI dimer T1-T2 PPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITS ICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEK KKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 62 AALLPGATALQCFCHLCTKDNFTCVTDGLCFVSVTETTDKVIHNSMCIAEI T.beta.RI-T.beta.RII-ED DLIPRDRPFVCAPSSKTGSVTTTYCCNQDHCNKIELPTTVKSSPGLGPVEI dimer T1-T2b PPHVQKSDVEMEAQKDEIICPSCNRTAHPLRHINNDMIVTDNNGAVKFPQL CKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVC HDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEE YNTSNPD 63 IPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSIT T.beta.RI-T.beta.RII-ED SICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKE dimer T2-T1 KKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDAALLPGATALQCFCHLC TKDNFTCVTDGLCFVSVTETTDKVIHNSMCIAEIDLIPRDRPFVCAPSSKT GSVTTTYCCNQDHCNKIELPTTVKSSPGLGPVE 64 AALLPGATALQCFCHLCTKDNFTCVTDGLCFVSVTETTDKVIHNSMCIAEI T.beta.RI-T.beta.RII-ED DLIPRDRPFVCAPSSKTGSVTTTYCCNQDHCNKIELPTTVKSSPGLGPVEI trimer T1-T2-T2 PPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITS ICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEK KKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSVNNDMIVTDN NGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKND ENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDEC NDNIIFSEEYNTSNPD 65 AALLPGATALQCFCHLCTKDNFTCVTDGLCFVSVTETTDKVIHNSMCIAEI T.beta.RI-T.beta.RII-ED DLIPRDRPFVCAPSSKTGSVTTTYCCNQDHCNKIELPTTVKSSPGLGPVEI trimer T1-T2-T2b PPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITS ICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEK KKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSDVEMEAQKDE IICPSCNRTAHPLRHINNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSC MSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAAS PKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 66 AALLPGATALQCFCHLCTKDNFTCVTDGLCFVSVTETTDKVIHNSMCIAEI T.beta.RI-T.beta.RII-ED DLIPRDRPFVCAPSSKTGSVTTTYCCNQDHCNKIELPTTVKSSPGLGPVEI trimer T1-T2- PPHVQKSDVEMEAQKDEIICPSCNRTAHPLRHINNDMIVTDNNGAVKFPQL T2b.sup.AA CKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVC HDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEE YNTSNPDIPPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINNDMIVTDNN GAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDE NITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECN DNIIFSEEYNTSNPD 67 IPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSIT T.beta.RI-T.beta.RII-ED SICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKE trimer T2-T2-T1 KKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSVNNDMIVTD NNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKN DENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDE CNDNIIFSEEYNTSNPDAALLPGATALQCFCHLCTKDNFTCVTDGLCFVSV TETTDKVIHNSMCIAEIDLIPRDRPFVCAPSSKTGSVTTTYCCNQDHCNKI ELPTTVKSSPGLGPVE 68 IPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSIT T.beta.RI-T.beta.RII-ED SICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKE timer T2-T2b.sup.AA- KKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSDVEMEAQKD T1 EIIAPSANRTAHPLRHINNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKS CMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAA SPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDAALLPGATA LQCFCHLCTKDNFTCVTDGLCFVSVTETTDKVIHNSMCIAEIDLIPRDRPF VCAPSSKTGSVTTTYCCNQDHCNKIELPTTVKSSPGLGPVE 69 TLPFLKCYCSGHCPDDAINNTCITNGHCFAIIEEDDQGETTLASGCMKYEG BMPRIa-ED SDFQCKDSPKAQLRRTIECCRTNLCNQYLQPTLP structured domain 70 QNLDSMLHGTGMKSDSDQKKSENGVTLAPED BMPRIa-ED N- term unstructured region and natural linker 71 PVVIGPFFDGSIR BMPRIa-ED C- term unstructured region and natural linker 72 PVVIGPFFDGSIRQNLDSMLHGTGMKSDSDQKKSENGVTLAPED BMPRIa-ED natural linker 73 QNLDSMLHGTGMKSDSDQKKSENGVTLAPEDTLPFLKCYCSGHCPDDAINN BMPRIa-ED TCITNGHCFAIIEEDDQGETTLASGCMKYEGSDFQCKDSPKAQLRRTIECC monomer RTNLCNQYLQPTLPPVVIGPFFDGSIR 74 QNLDSMLHGTGMKSDSDQKKSENGVTLAPEDTLPFLKCYCSGHCPDDAINN BMPRIa-ED TCITNGHCFAIIEEDDQGETTLASGCMKYEGSDFQCKDSPKAQLRRTIECC dimer RTNLCNQYLQPTLPPVVIGPFFDGSIRQNLDSMLHGTGMKSDSDQKKSENG VTLAPEDTLPFLKCYCSGHCPDDAINNTCITNGHCFAIIEEDDQGETTLAS GCMKYEGSDFQCKDSPKAQLRRTIECCRTNLCNQYLQPTLPPVVIGPFFDG SIR 75 TQECLFFNANWEKDRTNQTGVEPCYGDKDKRRHCFATWKNISGSIEIVKQG ActRIIa-ED CWLDDINCYDRTDCVEKKDSPEVYFCCCEGNMCNEKFSYFP structured domain 76 AILGRSE ActRIIa-ED N- term unstructured region and natural linker 77 EMEVTQPTSNPVTPKPPYYNI ActRIIa-ED C- term unstructured region and natural linker 78 EMEVTQPTSNPVTPKPPYYNIAILGRSE ActRIIa-ED natural linker 79 AILGRSETQECLFFNANWEKDRTNQTGVEPCYGDKDKRRHCFATWKNISGS ActRIIa-ED IEIVKQGCWLDDINCYDRTDCVEKKDSPEVYFCCCEGNMCNEKFSYFPEME monomer VTQPTSNPVTPKPPYYNI 80 AILGRSETQECLFFNANwEKDRTNQTGVEPCYGDKDKRRHCFATWKNISGS ActRIIa-ED dimer IEIVKQGCWLDDINCYDRTDCVEKKDSPEVYFCCCEGNMCNEKFSYFPEME VTQPTSNPVTPKPPYYNIAILGRSETQECLFFNANWEKDRTNQTGVEPCYG DKDKRRHCFATWKNISGSIEIVKQGCWLDDINCYDRTDCVEKKDSPEVYFC CCEGNMCNEKFSYFPEMEVTQPTSNPVTPKPPYYNI 81 RECIYYNANWELERTNQSGLERCEGEQDKRLHCYASWRNSSGTIELVKKGC ActRIIb-ED WLDDFNCYDRQECVATEENPQVYFCCCEGNFCNERFTHLP structured domain 82 SGRGEAET ActRIIb-ED N- term unstructured
region and natural linker 83 EAGGPEVTYEPPPTAPT ActRIIb-ED C- term unstructured region and natural linker 84 EAGGPEVTYEPPPTAPTSGRGEAET ActRIIb-ED natural linker 85 SGRGEAETRECIYYNANWELERTNQSGLERCEGEQDKRLHCYASWRNSSGT ActRIIb-ED IELVKKGCWLDDFNCYDRQECVATEENPQVYFCCCEGNFCNERFTHLPEAG monomer GPEVTYEPPPTAPT 86 SGRGEAETRECIYYNANWELERTNQSGLERCEGEQDRRLHCYASWRNSSGT ActRIIb-ED dimer IELVKKGCWLDDFNCYDRQECVATEENPQVYFCCCEGNFCNERFTHLPEAG GPEVTYEPPPTAPTSGRGEAETRECIYYNANWELERTNQSGLERCEGEQDK RLHCYASWRNSSGTIELVKKGCWLDDFNCYDRQECVATEENPQVYFCCCEG NFCNERFTHLPEAGGPEVTYEPPPTAPT 87 EMEVTQPTSNPVTPKPPYYNIQNLDSMLHGTGMKSDSDQKKSENGVTLAPE ActRIIa-BMPRIa- D ED natural linker 88 AILGRSETQECLFFNANwEKDRTNQTGVEPCYGDKDKRRHCFATWKNISGS ActRIla-BMPRIa- IEIVKQGCWLDDINCYDRTDCVEKKDSPEVYFCCCEGNMCNEKFSYFPEME ED dimer VTQPTSNPVTPKPPYYNIQNLDSMLHGTGMKSDSDQKKSENGVTLAPEDTL PFLKCYCSGHCPDDAINNTCITNGHCFAIIEEDDQGETTLASGCMKYEGSD FQCKDSPKAQLRRTIECCRTNLCNQYLQPTLPPVVIGPFFDGSIR 89 MDWTWRILFLVAAATGTHA Signal peptide 90 MVLQTQVFISLLLWISGAYG Signal peptide 91 APELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDG hIgG1Fc.DELTA.K-.DELTA.C- VEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPI T2m fusion EKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAVEWES NGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHN HYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCD NQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFIL EDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 92 PPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNW hIgG1Fc.DELTA.K-C- YVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKAL T2m fusion PAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAV EWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHE ALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRF STCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYH DFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 93 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDP hIgG1Fc.DELTA.K-CC- EVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCK T2m fusion VSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFY PSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFS CSVMHEALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVKFPQLCK FCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHD PKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYN TSNPD 94 APPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVQFNWYVDGV hIgG2Fc.DELTA.K-.DELTA.C- EVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNGKEYKCKVSNKGLPAPIE T2m fusion KTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDISVEWESN GQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNH YTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDN QKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILE DAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 95 PPCPAPPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVQFNWY hIgG2Fc.DELTA.K-C- VDGVEVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNGKEYKCKVSNKGLP T2m fusion APIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDISVE WESNGQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEA LHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFS TCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHD FILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 96 APPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVQFNWYVDGV hIgG2Fc.DELTA.K-CC- EVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNGKEYKCKVSNKGLPAPIE T2m fusion KTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDISVEWESN GQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNH YTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDN QKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILE DAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 97 MDWTWRILFLVAAATGTHAERKCCVECPPCPAPPVAGPSVFLFPPKPKDTL hIgG2Fc-CCCC- MISRTPEVTCVVVDVSHEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTFRV T2 fusion, also VSVLTVVHQDWLNGKEYKCKVSNKGLPAPIEKTISKTKGQPREPQVYTLPP termed Fc-T2m SREEMTKNQVSLTCLVKGFYPSDISVEWESNGQPENNYKTTPPMLDSDGSF FLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGKIPPHVQKS VNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQE VCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETF FMCSCSSDECNDNIIFSEEYNTSNPD 98 ESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQE hIgG4Fc.DELTA.K-CC- DPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYK 228P-T2m fusion CKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKG FYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSRLTVDKSRWQEGNV FSCSVMHEALHNHYTQKSLSLSLGIPPHVQKSVNNDMIVTDNNGAVKFPQL CKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVC HDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEE YNTSNPD 99 ESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQE hIgG4Fc.DELTA.K-CC- DPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYK 228P-409K-T2m CKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKG fusion FYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQEGNV FSCSVMHEALHNHYTQKSLSLSLGIPPHVQKSVNNDMIVTDNNGAVKFPQL CKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVC HDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEE YNTSNPD 100 PPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNW hIgG1Fc.DELTA.K-C- YVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKAL T22d35 fusion PAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAV EWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHE ALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRF STCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYH DFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDI PPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITS ICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEK KKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 101 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDP hIgG1Fc.DELTA.K-CC- EVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCK T22d35 fusion VSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFY PSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFS CSVMHEALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVKFPQLCK FCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHD PKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYN TSNPDIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMS NCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPK CIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 102 VECPPCPAPPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVQF hIgG2Fc.DELTA.K-CC- NWYVDGVEVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNGKEYKCKVSNK T22d35 fusion GLPAPIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDI SVEWESNGQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVM HEALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDV RFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLP YHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNP DIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSI TSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMK EKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 103 ESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQE hIgG4Fc.DELTA.K-CC- DPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYK 228P-T22d35 CKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKG fusion FYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSRLTVDKSRWQEGNV FSCSVMHEALHNHYTQKSLSLSLGIPPHVQKSVNNDMIVTDNNGAVKFPQL CKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVC HDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEE YNTSNPDIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSC MSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAAS PKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 104 ESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQE hIgG4Fc.DELTA.K-CC- DPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYK 228P-409K- CKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKG T22d35 fusion FYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQEGNV FSCSVMHEALHNHYTQKSLSLSLGIPPHVQKSVNNDMIVTDNNGAVKFPQL CKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVC HDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEE YNTSNPDIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSC MSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAAS PKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 105 MDWTWRILFLVAAATGTHAERKCCVECPPCPAPPVAGPSVFLFPPKPKDTL hIgG2Fc-CCCC- MISRTPEVTCVVVDVSHEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTFRV T22d35 fusion, VSVLTVVHQDWLNGKEYKCKVSNKGLPAPIEKTISKTKGQPREPQVYTLPP also termed Fc- SREEMTKNQVSLTCLVKGFYPSDISVEWESNGQPENNYKTTPPMLDSDGSF T22d35 FLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGKIPPHVQKS VNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQE VCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETF FMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSVNNDMIVTDNNGAVKFP QLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLET VCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFS EEYNTSNPD 106 PPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNW hIgG1Fc.DELTA.K-C-T2- YVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKAL T2b.sup.AA fusion PAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAV EWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHE ALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRF STCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYH DFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDI PPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINNDMIVTDNNGAVKFPQL CKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVC HDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEE YNTSNPD 107 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDP hIgG1Fc.DELTA.K-CC- EVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCK T2-T2b.sup.AA fusion VSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFY PSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFS CSVMHEALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVKFPQLCK FCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHD PKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYN TSNPDIPPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINNDMIVTDNNGA VKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENI TLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDN IIFSEEYNTSNPD 108 VECPPCPAPPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVQF hIgG2Fc.DELTA.K-CC- NWYVDGVEVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNGKEYKCKVSNK T2-T2b.sup.AA fusion GLPAPIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDI SVEWESNGQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVM HEALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDV RFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLP YHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNP DIPPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINNDMIVTDNNGAVKFP QLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLET VCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFS EEYNTSNPD 109 ESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQE hIgG4Fc.DELTA.K-CC- DPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYK 228P-T2-T2b.sup.AA CKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKG fusion FYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSRLTVDKSRWQEGNV FSCSVMHEALHNHYTQKSLSLSLGIPPHVQKSVNNDMIVTDNNGAVKFPQL CKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVC HDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEE YNTSNPDIPPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINNDMIVTDNN GAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDE NITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECN DNIIFSEEYNTSNPD 110 ESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQE hIgG4Fc.DELTA.K-CC- DPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYK 228P-409K-T2- CKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKG T2b.sup.AA fusion FYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQEGNV FSCSVMHEALHNHYTQKSLSLSLGIPPHVQKSVNNDMIVTDNNGAVKFPQL CKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVC HDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEE YNTSNPDIPPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINNDMIVTDNN
GAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDE NITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECN DNIIFSEEYNTSNPD 111 PPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNW hIgG1Fc.DELTA.K-C-T2- YVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKAL T2-T2 fusion PAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAV EWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHE ALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRF STCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYH DFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDI PPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITS ICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEK KKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSVNNDMIVTDN NGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKND ENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDEC NDNIIFSEEYNTSNPD 112 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDP hIgG1Fc.DELTA.K-CC- EVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCK T2-T2-T2 fusion VSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFY PSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFS CSVMHEALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVKFPQLCK FCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHD PKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYN TSNPDIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMS NCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPK CIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSVNND MIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVA VWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCS CSSDECNDNIIFSEEYNTSNPD 113 VECPPCPAPPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVQF hIgG2Fc.DELTA.K-CC- NWYVDGVEVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNGKEYKCKVSNK T2-T2-T2 fusion GLPAPIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDI SVEWESNGQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVM HEALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDV RFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLP YHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNP DIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSI TSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMK EKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSVNNDMIVT DNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRK NDENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSD ECNDNIIFSEEYNTSNPD 114 ESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQE hIgG4Fc.DELTA.K-CC- DPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYK 228P-T2-T2-T2 CKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKG fusion FYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSRLTVDKSRWQEGNV FSCSVMHEALHNHYTQKSLSLSLGIPPHVQKSVNNDMIVTDNNGAVKFPQL CKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVC HDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEE YNTSNPDIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSC MSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAAS PKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSVN NDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVC VAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFM CSCSSDECNDNIIFSEEYNTSNPD 115 ESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQE hIgG4Fc.DELTA.K-CC- DPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYK 228P-409K-T2- CKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKG T2-T2 fusion FYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQEGNV FSCSVMHEALHNHYTQKSLSLSLGIPPHVQKSVNNDMIVTDNNGAVKFPQL CKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVC HDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEE YNTSNPDIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSC MSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAAS PKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSVN NDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVC VAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFM CSCSSDECNDNIIFSEEYNTSNPD 116 PPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNW hIgG1Fc.DELTA.K-C-T2- YVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKAL T2b.sup.AA-T2b.sup.AA PAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAV fusion EWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHE ALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRF STCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYH DFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDI PPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINNDMIVTDNNGAVKFPQL CKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVC HDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEE YNTSNPDIPPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINNDMIVTDNN GAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDE NITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECN DNIIFSEEYNTSNPD 117 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDP hIgG1Fc.DELTA.K-CC- EVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCK T2-T2b.sup.AA-T2b.sup.AA VSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFY fusion PSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFS CSVMHEALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVKFPQLCK FCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHD PKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYN TSNPDIPPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINNDMIVTDNNGA VKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENI TLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDN IIFSEEYNTSNPDIPPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINNDM IVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAV WRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSC SSDECNDNIIFSEEYNTSNPD 118 VECPPCPAPPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVQF hIgG2Fc.DELTA.K-CC- NWYVDGVEVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNGKEYKCKVSNK T2-T2b.sup.AA-T2b.sup.AA GLPAPIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDI fusion SVEWESNGQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVM HEALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDV RFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLP YHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNP DIPPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINNDMIVTDNNGAVKFP QLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLET VCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFS EEYNTSNPDIPPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINNDMIVTD NNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKN DENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDE CNDNIIFSEEYNTSNPD 119 ESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQE hIgG4Fc.DELTA.K-CC- DPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYK 228P-T2-T2b.sup.AA- CKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKG T2b.sup.AA fusion FYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSRLTVDKSRWQEGNV FSCSVMHEALHNHYTQKSLSLSLGIPPHVQKSVNNDMIVTDNNGAVKFPQL CKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVC HDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEE YNTSNPDIPPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINNDMIVTDNN GAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDE NITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECN DNIIFSEEYNTSNPDIPPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINN DMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCV AVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMC SCSSDECNDNIIFSEEYNTSNPD 120 ESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQE hIgG4Fc.DELTA.K-CC- DPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYK 228P-409K-T2- CKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKG T2b.sup.AA-T2b.sup.AA FYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQEGNV fusion FSCSVMHEALHNHYTQKSLSLSLGIPPHVQKSVNNDMIVTDNNGAVKFPQL CKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVC HDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEE YNTSNPDIPPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINNDMIVTDNN GAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDE NITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECN DNIIFSEEYNTSNPDIPPHVQKSDVEMEAQKDEIIAPSANRTAHPLRHINN DMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCV AVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMC SCSSDECNDNIIFSEEYNTSNPD 121 DILLTQSPVILSVSPGERVSFSCRASQSIGTNIHWYQQRTNGSPRLLIKYA Cetuximab LC SESISGIPSRFSGSGSGTDFTLSINSVESEDIADYYCQQNNNWPTTFGAGT KLELKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNA LQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSP VTKSFNRGEC 122 QVQLKQSGPGLVQPSQSLSITCTVSGFSLTNYGVHWVRQSPGKGLEWLGVI Cetuximab HC- WSGGNTDYNTPFTSRLSINKDNSKSQVFFKMNSLQSNDTAIYYCARALTYY T22d35 DYEFAYWGQGTLVTVSAASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFP EPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNV NHKPSNTKVDKRVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMI SRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVS VLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSR DELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFL YSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGKIPPHVQKSVN NDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVC VAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFM CSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSVNNDMIVTDNNGAVKFPQL CKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVC HDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEE YNTSNPD 123 QVQLKQSGPGLVQPSQSLSITCTVSGFSLTNYGVHWVRQSPGKGLEWLGVI Cetuximab HC- WSGGNTDYNTPFTSRLSINKDNSKSQVFFKMNSLQSNDTAIYYCARALTYY T2m DYEFAYWGQGTLVTVSAASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFP EPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNV NHKPSNTKVDKRVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMI SRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVS VLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSR DELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFL YSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGKIPPHVQKSVN NDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVC VAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFM CSCSSDECNDNIIFSEEYNTSNPD 124 DIQMTQSPSSLSASVGDRVTITCRASQDVNTAVAWYQQKPGKAPKLLIYSA Herceptin LC SFLYSGVPSRFSGSRSGTDFTLTISSLQPEDFATYYCQQHYTTPPTFGQGT KVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNA LQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSP VTKSFNRGEC 125 EVQLVESGGGLVQPGGSLRLSCAASGFNIKDTYIHWVRQAPGKGLEWVARI Herceptin HC- YPTNGYTRYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCSRWGGD T22d35 GFYAMDYWGQGTLVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYF PEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICN VNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLM ISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVV SVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPS RDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFF LYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGKIPPHVQKSV NNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEV CVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFF MCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSVNNDMIVTDNNGAVKFPQ LCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETV CHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSE EYNTSNPD 126 DIQMTQSPSSLSASVGDRVTITCSASQDISNYLNWYQQKPGKAPKCLIYFT Avastin LC SSLHSGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQYSTVPWTFGQGT KVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNA LQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSP VTKSFNRGEC 127 EVQLVESGGGLVQPGGSLRLSCAASGYTFTNYGMNWVRQAPGKGLEWVGWI Avastin HC- NTYTGEPTYAADFKRRFTFSLDTSKSTAYLQMNSLRAEDTAVYYCAKYPHY T22d35 YGSSHWYFDVWGQGTLVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVK DYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTY ICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKD TLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTY RVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTL PPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDG SFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGKIPPHVQ KSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKP QEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGE TFFMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSVNNDMIVTDNNGAVK FPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITL ETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNII FSEEYNTSNPD 128 DIQMTQSPSTLSASVGDRVTITCKCQLSVGYMHWYQQKPGKAPKLLIYDTS Synagis LC KLASGVPSRFSGSGSGTEFTLTISSLQPDDFATYYCFQGSGYPFTFGGGTK LEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNAL QSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPV TKSFNRGEC 129 QVTLRESGPALVKPTQTLTLTCTFSGFSLSTSGMSVGWIRQPPGKALEWLA Synagis HC- DIWWDDKKDYNPSLKSRLTISKDTSKNQVVLKVTNMDPADTATYYCARSMI T22d35 TNWYFDVWGAGTTVTVSSASTKGPSVFPLAPSSAAAAGGTAALGCLVKDYF
PEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICN VNHKPSNTKVDKRVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLM ISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVV SVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPS RDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFF LYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGIPPHVQKSVN NDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVC VAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFM CSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSVNNDMIVTDNNGAVKFPQL CKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVC HDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEE YNTSNPD 130 EVQLQASGGGLVQAGGSLRLSCAASGFKITHYTMGWFRQAPGKEREFVSRI FC5-Fc-T22d35 TWGGDNTFYSNSVKGRFTISRDNAKNTVYLQMNSLKPEDTADYYCAAGSTS TATPLRVDYWGKGTQVTVSSASEPRGPTIKPCPPCKCPAPNLLGGPSVFIF PPKIKDVLMISLSPIVTCVVVDVSEDDPDVQISWFVNNVEVHTAQTQTHRE DYNSTLRVVSALPIQHQDWMSGKEFKCKVNNKDLPAPIERTISKPKGSVRA PQVYVLPPPEEEMTKKQVTLTCMVTDFMPEDIYVEWTNNGKTELNYKNTEP VLDSDGSYFMYSKLRVEKKNWVERNSYSCSVVHEGLHNHHTTKSFSRTPGT GIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSI TSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMK EKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQKSVNNDMIVT DNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRK NDENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSD ECNDNIIFSEEYNTSNPD 131 EVQLQASGGGLVQAGGSLRLSCAASGFKITHYTMGWFRQAPGKEREFVSRI FC5-Fc-T2m TWGGDNTFYSNSVKGRFTISRDNAKNTVYLQMNSLKPEDTADYYCAAGSTS TATPLRVDYWGKGTQVTVSSASEPRGPTIKPCPPCKCPAPNLLGGPSVFIF PPKIKDVLMISLSPIVTCVVVDVSEDDPDVQISWFVNNVEVHTAQTQTHRE DYNSTLRVVSALPIQHQDWMSGKEFKCKVNNKDLPAPIERTISKPKGSVRA PQVYVLPPPEEEMTKKQVTLTCMVTDFMPEDIYVEWTNNGKTELNYKNTEP VLDSDGSYFMYSKLRVEKKNWVERNSYSCSVVHEGLHNHHTTKSFSRTPGT GIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSI TSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMK EKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 132 MGRGLLRGLWPLHIVLWTRIASTIPPHVQKSVNNDMIVTDNNGAVKFPQLC T2-hIgG1Fc KFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCH fusion from R&D, DPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEY also termed T2m- NTSNPDMDPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEV Fc (R&D) TCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLH QDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKN QVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTV DKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK 133 MDWTWRILFLVAAATGTHAIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCD T2-hIgG2Fc- VRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKL CCCC fusion, PYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSN also termed T2m- PDERKCCVECPPCPAPPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSH Fc EDPEVQFNWYVDGVEVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNGKEY KCKVSNKGLPAPIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTCLVK GFYPSDISVEWESNGQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQQGN VFSCSVMHEALHNHYTQKSLSLSPG 134 LPYHDFILEDAASPK Isotope-labelled peptide for T22d35 135 ALPAPIEK Isotope-labelled peptide for Cetuximab 136 DDDDDDDDDDDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTC D10-hIgG1Fc.DELTA.K- VVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQD CC-T2m fusion WLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQV SLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDK SRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNN GAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDE NITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECN DNIIFSEEYNTSNPD 137 DDDDDDDDDDGGGSDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTP D10-G.sub.3S- EVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTV hIgG1Fc.DELTA.K-CC- LHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELT T2m fusion KNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKL TVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGIPPHVQKSVNNDMIV TDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWR KNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSS DECNDNIIFSEEYNTSNPD 138 DDDDDDDDDDPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVS D10-hIgG1Fc.DELTA.K- HEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKE C-T2m fusion YKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLV KGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQG NVFSCSVMHEALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVKFP QLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLET VCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFS EEYNTSNPD 139 DDDDDDDDDDGGGSGGGSPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEV D10-(G.sub.3S).sub.2- TCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLH hIgG1Fc.DELTA.K-C- QDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKN T2m fusion QVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTV DKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTD NNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKN DENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDE CNDNIIFSEEYNTSNPD 140 DDDDDDDDDDVECPPCPAPPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVD D10-hIgG2Fc.DELTA.K- VSHEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNG CC-T2m fusion KEYKCKVSNKGLPAPIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTC LVKGFYPSDISVEWESNGQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQ QGNVFSCSVMHEALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVK FPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITL ETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNII FSEEYNTSNPD 141 DDDDDDDDDDESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEV D10-hIgG4Fc.DELTA.K- TCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLH CC-228P-T2m QDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKN fusion QVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSRLTV DKSRWQEGNVFSCSVMHEALHNHYTQKSLSLSLGIPPHVQKSVNNDMIVTD NNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKN DENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDE CNDNIIFSEEYNTSNPD 142 DDDDDDDDDDESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEV D10-hIgG4Fc.DELTA.K- TCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLH CC-228P-409K- QDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKN T2m fusion QVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTV DKSRWQEGNVFSCSVMHEALHNHYTQKSLSLSLGIPPHVQKSVNNDMIVTD NNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKN DENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDE CNDNIIFSEEYNTSNPD 143 DDDDDDDDDDDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTC D10-hIgG1Fc.DELTA.K- VVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQD CC-T22d35 fusion WLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQV SLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDK SRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNN GAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDE NITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECN DNIIFSEEYNTSNPDIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFS TCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHD FILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 144 DDDDDDDDDDVECPPCPAPPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVD D10-hIgG2Fc.DELTA.K- VSHEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNG CC-T22d35 fusion KEYKCKVSNKGLPAPIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTC LVKGFYPSDISVEWESNGQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQ QGNVFSCSVMHEALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVK FPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITL ETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNII FSEEYNTSNPDIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDN QKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILE DAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 145 DDDDDDDDDDESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEV D10-hIgG4Fc.DELTA.K- TCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLH CC-228P-T22d35 QDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKN fusion QVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSRLTV DKSRWQEGNVFSCSVMHEALHNHYTQKSLSLSLGIPPHVQKSVNNDMIVTD NNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKN DENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDE CNDNIIFSEEYNTSNPDIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVR FSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPY HDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 146 DDDDDDDDDDESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEV D10-hIgG4Fc.DELTA.K- TCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKpREEQFNSTYRVVSVLTVLH CC-228P-409K- QDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKN T22d35 fusion QVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTV DKSRWQEGNVFSCSVMHEALHNHYTQKSLSLSLGIPPHVQKSVNNDMIVTD NNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKN DENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDE CNDNIIFSEEYNTSNPDIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVR FSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPY HDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 147 DDDDDDDDDDDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTC D10-hIgG1Fc.DELTA.K- VVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQD CC-T2-T2-T2 WLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQV fusion SLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDK SRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNN GAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDE NITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECN DNIIFSEEYNTSNPDIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFS TCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHD FILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDIP PHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSI CEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEKK KPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 148 DDDDDDDDDDVECPPCPAPPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVD D10-hIgG2Fc.DELTA.K- VSHEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTFRVVSVLTVVHQDWLNG CC-T2-T2-T2 KEYKCKVSNKGLPAPIEKTISKTKGQPREPQVYTLPPSREEMTKNQVSLTC fusion LVKGFYPSDISVEWESNGQPENNYKTTPPMLDSDGSFFLYSKLTVDKSRWQ QGNVFSCSVMHEALHNHYTQKSLSLSPGIPPHVQKSVNNDMIVTDNNGAVK FPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITL ETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNII FSEEYNTSNPDIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDN QKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILE DAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPDIPPHVQ KSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKP QEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGE TFFMCSCSSDECNDNIIFSEEYNTSNPD 149 DDDDDDDDDDESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEV D10-hIgG4Fc.DELTA.K- TCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLH CC-228P-T2-T2- QDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKN T2 fusion QVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSRLTV DKSRWQEGNVFSCSVMHEALHNHYTQKSLSLSLGIPPHVQKSVNNDMIVTD NNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKN DENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDE CNDNIIFSEEYNTSNPDIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVR FSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPY HDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD IPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSIT SICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKE KKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD 150 DDDDDDDDDDESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEV D10-hIgG4Fc.DELTA.K- TCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLH CC-228P-409K- QDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKN T2-T2-T2 fusion QVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTV DKSRWQEGNVFSCSVMHEALHNHYTQKSLSLSLGIPPHVQKSVNNDMIVTD NNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKN DENITLETVCHDPKLPYHDFILEDAASPKCIMKEKKKPGETFFMCSCSSDE CNDNIIFSEEYNTSNPDIPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVR FSTCDNQKSCMSNCSITSICEKPQEVCVAVWRKNDENITLETVCHDPKLPY HDFILEDAASPKCIMKEKKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD IPPHVQKSVNNDMIVTDNNGAVKFPQLCKFCDVRFSTCDNQKSCMSNCSIT SICEKPQEVCVAVWRKNDENITLETVCHDPKLPYHDFILEDAASPKCIMKE KKKPGETFFMCSCSSDECNDNIIFSEEYNTSNPD
REFERENCES
[0205] All patents, patent applications and publications referred to throughout the application are listed below.
[0206] Arteaga C L (2006) Inhibition of TGFbeta signaling in cancer therapy. Curr Opin Genet Dev 16: 30-37
[0207] De Crescenzo G, Grothe S, Zwaagstra J, Tsang M, O'Connor-McCourt M D (2001) Real-time monitoring of the interactions of transforming growth factor-beta (TGF-beta) isoforms with latency-associated protein and the ectodomains of the TGF-beta type II and III receptors reveals different kinetic models and stoichiometries of binding. J Biol Chem 276: 29632-29643
[0208] Durocher Y, Perret S, Kamen A (2002) High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res 30: E9
[0209] Economides A N, Carpenter L R, Rudge J S, Wong V, Koehler-Stec E M, Hartnett C, Pyles E A, Xu X, Daly T J, Young M R, Fandl J P, Lee F, Carver S, McNay J, Bailey K, Ramakanth S, Hutabarat R, Huang T T, Radziejewski C, Yancopoulos G D, Stahl N (2003) Cytokine traps: multi-component, high-affinity blockers of cytokine action. Nat Med 9: 47-52
[0210] Eisenberg D, Schwarz E, Komaromy M, Wall R (1984) Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol 179: 125-142
[0211] Gajewski T F (2015) The Next Hurdle in Cancer Immunotherapy: Overcoming the Non-T-Cell-Inflamed Tumor Microenvironment. Semin Oncol 42: 663-671
[0212] Garberg P, Ball M, Borg N, Cecchelli R, Fenart L, Hurst R D, Lindmark T, Mabondzo A, Nilsson J E, Raub T J, Stanimirovic D, Terasaki T, Oberg J O, Osterberg T (2005) In vitro models for the blood-brain barrier. Toxicol In Vitro 19: 299-334
[0213] Hahn T, Akporiaye E T (2006) Targeting transforming growth factor beta to enhance cancer immunotherapy. Curr Oncol 13: 141-143
[0214] Haqqani A S, Caram-Salas N, Ding W, Brunette E, Delaney C E, Baumann E, Boileau E, Stanimirovic D (2013) Multiplexed evaluation of serum and CSF pharmacokinetics of brain-targeting single-domain antibodies using a NanoLC-SRM-ILIS method. Mol Pharm 10: 1542-1556
[0215] Hawinkels L J, Ten Dijke P (2011) Exploring anti-TGF-beta therapies in cancer and fibrosis. Growth Factors 29: 140-152
[0216] Holash J, Davis S, Papadopoulos N, Croll S D, Ho L, Russell M, Boland P, Leidich R, Hylton D, Burova E, loffe E, Huang T, Radziejewski C, Bailey K, Fandl J P, Daly T, Wiegand S J, Yancopoulos G D, Rudge J S (2002) VEGF-Trap: a VEGF blocker with potent antitumor effects. Proc Natl Acad Sci USA 99: 11393-11398
[0217] Jin P, Zhang J, Beryt M, Turin L, Brdlik C, Feng Y, Bai X, Liu J, Jorgensen B, Shepard H M (2009) Rational optimization of a bispecific ligand trap targeting EGF receptor family ligands. Mol Med 15: 11-20
[0218] Li M O, Wan Y Y, Sanjabi S, Robertson A K, Flavell R A (2006) Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol 24: 99-146
[0219] Massague J, Blain S W, Lo R S (2000) TGFbeta signaling in growth control, cancer, and heritable disorders. Cell 103: 295-309
[0220] Mourskaia A A, Northey J J, Siegel P M (2007) Targeting aberrant TGF-beta signaling in pre-clinical models of cancer. Anticancer Agents Med Chem 7: 504-514
[0221] Rodgarkia-Dara C, Vejda S, Erlach N, Losert A, Bursch W, Berger W, Schulte-Hermann R, Grusch M (2006) The activin axis in liver biology and disease. Mutat Res 613: 123-137
[0222] Santarpia M, Gonzalez-Cao M, Viteri S, Karachaliou N, Altavilla G, Rosell R (2015) Programmed cell death protein-1/programmed cell death ligand-1 pathway inhibition and predictive biomarkers: understanding transforming growth factor-beta role. Transl Lung Cancer Res 4: 728-742
[0223] Thiery J P, Acloque H, Huang R Y, Nieto M A (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139: 871-890
[0224] Wojtowicz-Praga S (2003) Reversal of tumor-induced immunosuppression by TGF-beta inhibitors. Invest New Drugs 21: 21-32
[0225] Yang L, Pang Y, Moses H L (2010) TGF-beta and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol 31: 220-227
[0226] Yang X, Ambrogelly A (2014) Enlarging the repertoire of therapeutic monoclonal antibodies platforms: domesticating half molecule exchange to produce stable IgG4 and IgG1 bispecific antibodies. Curr Opin Biotechnol 30: 225-229
[0227] Zheng X, Koropatnick J, Chen D, Velenosi T, Ling H, Zhang X, Jiang N, Navarro B, Ichim T E, Urquhart B, Min W (2013) Silencing IDO in dendritic cells: a novel approach to enhance cancer immunotherapy in a murine breast cancer model. Int J Cancer 132: 967-977
[0228] Zwaagstra J C, Sulea T, Baardsnes J, Lenferink A E, Collins C, Cantin C, Paul-Roc B, Grothe S, Hossain S, Richer L P, L'Abbe D, Tom R, Cass B, Durocher Y, O'Connor-McCourt M D (2012) Engineering and therapeutic application of single-chain bivalent TGF-beta family traps. Mol Cancer Ther 11: 1477-1487
[0229] WO/1995/04069
[0230] WO/2004/076670
[0231] WO 2008/113185
[0232] WO 2010/031168
[0233] U.S. Pat. No. 8,815,247
[0234] U.S. Pat. No. 6,277,7375
[0235] US2015/0225483
Sequence CWU 1
1
1501217PRTArtificial SequenceHuman IgG1 Fc region 1Ala Pro Glu Leu
Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys1 5 10 15Pro Lys Asp
Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val 20 25 30Val Val
Asp Val Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr 35 40 45Val
Asp Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu 50 55
60Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His65
70 75 80Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn
Lys 85 90 95Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys
Gly Gln 100 105 110Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser
Arg Asp Glu Leu 115 120 125Thr Lys Asn Gln Val Ser Leu Thr Cys Leu
Val Lys Gly Phe Tyr Pro 130 135 140Ser Asp Ile Ala Val Glu Trp Glu
Ser Asn Gly Gln Pro Glu Asn Asn145 150 155 160Tyr Lys Thr Thr Pro
Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu 165 170 175Tyr Ser Lys
Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val 180 185 190Phe
Ser Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln 195 200
205Lys Ser Leu Ser Leu Ser Pro Gly Lys 210 2152216PRTArtificial
SequenceHuman IgG2 Fc region 2Ala Pro Pro Val Ala Gly Pro Ser Val
Phe Leu Phe Pro Pro Lys Pro1 5 10 15Lys Asp Thr Leu Met Ile Ser Arg
Thr Pro Glu Val Thr Cys Val Val 20 25 30Val Asp Val Ser His Glu Asp
Pro Glu Val Gln Phe Asn Trp Tyr Val 35 40 45Asp Gly Val Glu Val His
Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln 50 55 60Phe Asn Ser Thr Phe
Arg Val Val Ser Val Leu Thr Val Val His Gln65 70 75 80Asp Trp Leu
Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Gly 85 90 95Leu Pro
Ala Pro Ile Glu Lys Thr Ile Ser Lys Thr Lys Gly Gln Pro 100 105
110Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu Met Thr
115 120 125Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr
Pro Ser 130 135 140Asp Ile Ser Val Glu Trp Glu Ser Asn Gly Gln Pro
Glu Asn Asn Tyr145 150 155 160Lys Thr Thr Pro Pro Met Leu Asp Ser
Asp Gly Ser Phe Phe Leu Tyr 165 170 175Ser Lys Leu Thr Val Asp Lys
Ser Arg Trp Gln Gln Gly Asn Val Phe 180 185 190Ser Cys Ser Val Met
His Glu Ala Leu His Asn His Tyr Thr Gln Lys 195 200 205Ser Leu Ser
Leu Ser Pro Gly Lys 210 2153217PRTArtificial SequenceHuman IgG3 Fc
region 3Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro
Lys1 5 10 15Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr
Cys Val 20 25 30Val Val Asp Val Ser His Glu Asp Pro Glu Val Gln Phe
Lys Trp Tyr 35 40 45Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys
Pro Arg Glu Glu 50 55 60Gln Tyr Asn Ser Thr Phe Arg Val Val Ser Val
Leu Thr Val Leu His65 70 75 80Gln Asp Trp Leu Asn Gly Lys Glu Tyr
Lys Cys Lys Val Ser Asn Lys 85 90 95Ala Leu Pro Ala Pro Ile Glu Lys
Thr Ile Ser Lys Thr Lys Gly Gln 100 105 110Pro Arg Glu Pro Gln Val
Tyr Thr Leu Pro Pro Ser Arg Glu Glu Met 115 120 125Thr Lys Asn Gln
Val Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro 130 135 140Ser Asp
Ile Ala Val Glu Trp Glu Ser Ser Gly Gln Pro Glu Asn Asn145 150 155
160Tyr Asn Thr Thr Pro Pro Met Leu Asp Ser Asp Gly Ser Phe Phe Leu
165 170 175Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly
Asn Ile 180 185 190Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn
Arg Phe Thr Gln 195 200 205Lys Ser Leu Ser Leu Ser Pro Gly Lys 210
2154217PRTArtificial SequenceHuman IgG4 Fc region 4Ala Pro Glu Phe
Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys1 5 10 15Pro Lys Asp
Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val 20 25 30Val Val
Asp Val Ser Gln Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr 35 40 45Val
Asp Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu 50 55
60Gln Phe Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His65
70 75 80Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn
Lys 85 90 95Gly Leu Pro Ser Ser Ile Glu Lys Thr Ile Ser Lys Ala Lys
Gly Gln 100 105 110Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser
Gln Glu Glu Met 115 120 125Thr Lys Asn Gln Val Ser Leu Thr Cys Leu
Val Lys Gly Phe Tyr Pro 130 135 140Ser Asp Ile Ala Val Glu Trp Glu
Ser Asn Gly Gln Pro Glu Asn Asn145 150 155 160Tyr Lys Thr Thr Pro
Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu 165 170 175Tyr Ser Arg
Leu Thr Val Asp Lys Ser Arg Trp Gln Glu Gly Asn Val 180 185 190Phe
Ser Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln 195 200
205Lys Ser Leu Ser Leu Ser Leu Gly Lys 210 215515PRTArtificial
SequenceHuman IgG1 hinge region 5Glu Pro Lys Ser Cys Asp Lys Thr
His Thr Cys Pro Pro Cys Pro1 5 10 15612PRTArtificial SequenceHuman
IgG2 hinge region 6Glu Arg Lys Cys Cys Val Glu Cys Pro Pro Cys Pro1
5 10762PRTArtificial SequenceHuman IgG3 hinge region 7Glu Leu Lys
Thr Pro Leu Gly Asp Thr Thr His Thr Cys Pro Arg Cys1 5 10 15Pro Glu
Pro Lys Ser Cys Asp Thr Pro Pro Pro Cys Pro Arg Cys Pro 20 25 30Glu
Pro Lys Ser Cys Asp Thr Pro Pro Pro Cys Pro Arg Cys Pro Glu 35 40
45Pro Lys Ser Cys Asp Thr Pro Pro Pro Cys Pro Arg Cys Pro 50 55
60812PRTArtificial SequenceHuman IgG4 hinge region 8Glu Ser Lys Tyr
Gly Pro Pro Cys Pro Ser Cys Pro1 5 109216PRTArtificial
SequencehIgG1FcdeltaK-deltaC Fc variant 9Ala Pro Glu Leu Leu Gly
Gly Pro Ser Val Phe Leu Phe Pro Pro Lys1 5 10 15Pro Lys Asp Thr Leu
Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val 20 25 30Val Val Asp Val
Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr 35 40 45Val Asp Gly
Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu 50 55 60Gln Tyr
Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His65 70 75
80Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys
85 90 95Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly
Gln 100 105 110Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg
Asp Glu Leu 115 120 125Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val
Lys Gly Phe Tyr Pro 130 135 140Ser Asp Ile Ala Val Glu Trp Glu Ser
Asn Gly Gln Pro Glu Asn Asn145 150 155 160Tyr Lys Thr Thr Pro Pro
Val Leu Asp Ser Asp Gly Ser Phe Phe Leu 165 170 175Tyr Ser Lys Leu
Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val 180 185 190Phe Ser
Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln 195 200
205Lys Ser Leu Ser Leu Ser Pro Gly 210 21510220PRTArtificial
SequencehIgG1FcdeltaK-C Fc variant 10Pro Pro Cys Pro Ala Pro Glu
Leu Leu Gly Gly Pro Ser Val Phe Leu1 5 10 15Phe Pro Pro Lys Pro Lys
Asp Thr Leu Met Ile Ser Arg Thr Pro Glu 20 25 30Val Thr Cys Val Val
Val Asp Val Ser His Glu Asp Pro Glu Val Lys 35 40 45Phe Asn Trp Tyr
Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys 50 55 60Pro Arg Glu
Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu65 70 75 80Thr
Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys 85 90
95Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys
100 105 110Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro
Pro Ser 115 120 125Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr
Cys Leu Val Lys 130 135 140Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu
Trp Glu Ser Asn Gly Gln145 150 155 160Pro Glu Asn Asn Tyr Lys Thr
Thr Pro Pro Val Leu Asp Ser Asp Gly 165 170 175Ser Phe Phe Leu Tyr
Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln 180 185 190Gln Gly Asn
Val Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn 195 200 205His
Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly 210 215
22011226PRTArtificial SequencehIgG1FcdeltaK-CC Fc variant 11Asp Lys
Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly1 5 10 15Gly
Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 20 25
30Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His
35 40 45Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu
Val 50 55 60His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser
Thr Tyr65 70 75 80Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp
Trp Leu Asn Gly 85 90 95Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala
Leu Pro Ala Pro Ile 100 105 110Glu Lys Thr Ile Ser Lys Ala Lys Gly
Gln Pro Arg Glu Pro Gln Val 115 120 125Tyr Thr Leu Pro Pro Ser Arg
Asp Glu Leu Thr Lys Asn Gln Val Ser 130 135 140Leu Thr Cys Leu Val
Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu145 150 155 160Trp Glu
Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro 165 170
175Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val
180 185 190Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser
Val Met 195 200 205His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser
Leu Ser Leu Ser 210 215 220Pro Gly22512231PRTArtificial
SequencehIgG1FcdeltaK-S Fc variant 12Glu Pro Lys Ser Ser Asp Lys
Thr His Thr Cys Pro Pro Cys Pro Ala1 5 10 15Pro Glu Leu Leu Gly Gly
Pro Ser Val Phe Leu Phe Pro Pro Lys Pro 20 25 30Lys Asp Thr Leu Met
Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val 35 40 45Val Asp Val Ser
His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val 50 55 60Asp Gly Val
Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln65 70 75 80Tyr
Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln 85 90
95Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala
100 105 110Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly
Gln Pro 115 120 125Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg
Asp Glu Leu Thr 130 135 140Lys Asn Gln Val Ser Leu Thr Cys Leu Val
Lys Gly Phe Tyr Pro Ser145 150 155 160Asp Ile Ala Val Glu Trp Glu
Ser Asn Gly Gln Pro Glu Asn Asn Tyr 165 170 175Lys Thr Thr Pro Pro
Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr 180 185 190Ser Lys Leu
Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe 195 200 205Ser
Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys 210 215
220Ser Leu Ser Leu Ser Pro Gly225 23013231PRTArtificial
SequencehIgG1deltaK-SS Fc variant 13Glu Pro Lys Ser Ser Asp Lys Thr
His Thr Cys Pro Pro Cys Pro Ala1 5 10 15Pro Glu Leu Leu Gly Gly Pro
Ser Val Phe Leu Phe Pro Pro Lys Pro 20 25 30Lys Asp Thr Leu Met Ile
Ser Arg Thr Pro Glu Val Thr Cys Val Val 35 40 45Val Asp Val Ser His
Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val 50 55 60Asp Gly Val Glu
Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln65 70 75 80Tyr Asn
Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln 85 90 95Asp
Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala 100 105
110Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro
115 120 125Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg Asp Glu
Leu Thr 130 135 140Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly
Phe Tyr Pro Ser145 150 155 160Asp Ile Ala Val Glu Trp Glu Ser Asn
Gly Gln Pro Glu Asn Asn Tyr 165 170 175Lys Thr Thr Pro Pro Val Leu
Asp Ser Asp Gly Ser Phe Phe Leu Tyr 180 185 190Ser Lys Leu Thr Val
Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe 195 200 205Ser Cys Ser
Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys 210 215 220Ser
Leu Ser Leu Ser Pro Gly225 23014231PRTArtificial
SequencehIgG1FcdeltaK-SSS Fc variant 14Glu Pro Lys Ser Ser Asp Lys
Thr His Thr Ser Pro Pro Ser Pro Ala1 5 10 15Pro Glu Leu Leu Gly Gly
Pro Ser Val Phe Leu Phe Pro Pro Lys Pro 20 25 30Lys Asp Thr Leu Met
Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val 35 40 45Val Asp Val Ser
His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val 50 55 60Asp Gly Val
Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln65 70 75 80Tyr
Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln 85 90
95Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala
100 105 110Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly
Gln Pro 115 120 125Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg
Asp Glu Leu Thr 130 135 140Lys Asn Gln Val Ser Leu Thr Cys Leu Val
Lys Gly Phe Tyr Pro Ser145 150 155 160Asp Ile Ala Val Glu Trp Glu
Ser Asn Gly Gln Pro Glu Asn Asn Tyr 165 170 175Lys Thr Thr Pro Pro
Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr 180 185 190Ser Lys Leu
Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe 195 200 205Ser
Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys 210 215
220Ser Leu Ser Leu Ser Pro Gly225 23015215PRTArtificial
SequencehIgG2FcdeltaK-deltaC Fc variant 15Ala Pro Pro Val Ala Gly
Pro Ser Val Phe Leu Phe Pro Pro Lys Pro1 5 10 15Lys Asp Thr Leu Met
Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val 20 25 30Val Asp Val Ser
His Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr Val 35 40 45Asp Gly Val
Glu Val His Asn Ala Lys Thr Lys
Pro Arg Glu Glu Gln 50 55 60Phe Asn Ser Thr Phe Arg Val Val Ser Val
Leu Thr Val Val His Gln65 70 75 80Asp Trp Leu Asn Gly Lys Glu Tyr
Lys Cys Lys Val Ser Asn Lys Gly 85 90 95Leu Pro Ala Pro Ile Glu Lys
Thr Ile Ser Lys Thr Lys Gly Gln Pro 100 105 110Arg Glu Pro Gln Val
Tyr Thr Leu Pro Pro Ser Arg Glu Glu Met Thr 115 120 125Lys Asn Gln
Val Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser 130 135 140Asp
Ile Ser Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr145 150
155 160Lys Thr Thr Pro Pro Met Leu Asp Ser Asp Gly Ser Phe Phe Leu
Tyr 165 170 175Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly
Asn Val Phe 180 185 190Ser Cys Ser Val Met His Glu Ala Leu His Asn
His Tyr Thr Gln Lys 195 200 205Ser Leu Ser Leu Ser Pro Gly 210
21516219PRTArtificial SequencehIgG2FcdeltaK-C Fc variant 16Pro Pro
Cys Pro Ala Pro Pro Val Ala Gly Pro Ser Val Phe Leu Phe1 5 10 15Pro
Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val 20 25
30Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Gln Phe
35 40 45Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys
Pro 50 55 60Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser Val
Leu Thr65 70 75 80Val Val His Gln Asp Trp Leu Asn Gly Lys Glu Tyr
Lys Cys Lys Val 85 90 95Ser Asn Lys Gly Leu Pro Ala Pro Ile Glu Lys
Thr Ile Ser Lys Thr 100 105 110Lys Gly Gln Pro Arg Glu Pro Gln Val
Tyr Thr Leu Pro Pro Ser Arg 115 120 125Glu Glu Met Thr Lys Asn Gln
Val Ser Leu Thr Cys Leu Val Lys Gly 130 135 140Phe Tyr Pro Ser Asp
Ile Ser Val Glu Trp Glu Ser Asn Gly Gln Pro145 150 155 160Glu Asn
Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser Asp Gly Ser 165 170
175Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln
180 185 190Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu His
Asn His 195 200 205Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly 210
21517222PRTArtificial SequencehIgG2FcdeltaK-CC Fc variant 17Val Glu
Cys Pro Pro Cys Pro Ala Pro Pro Val Ala Gly Pro Ser Val1 5 10 15Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 20 25
30Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu
35 40 45Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala
Lys 50 55 60Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val
Val Ser65 70 75 80Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly
Lys Glu Tyr Lys 85 90 95Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro
Ile Glu Lys Thr Ile 100 105 110Ser Lys Thr Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr Leu Pro 115 120 125Pro Ser Arg Glu Glu Met Thr
Lys Asn Gln Val Ser Leu Thr Cys Leu 130 135 140Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ser Val Glu Trp Glu Ser Asn145 150 155 160Gly Gln
Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser 165 170
175Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg
180 185 190Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu
Ala Leu 195 200 205His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
Pro Gly 210 215 22018228PRTArtificial SequencehIgG2Fc-CCCC Fc
variant 18Glu Arg Lys Cys Cys Val Glu Cys Pro Pro Cys Pro Ala Pro
Pro Val1 5 10 15Ala Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys
Asp Thr Leu 20 25 30Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val
Val Asp Val Ser 35 40 45His Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr
Val Asp Gly Val Glu 50 55 60Val His Asn Ala Lys Thr Lys Pro Arg Glu
Glu Gln Phe Asn Ser Thr65 70 75 80Phe Arg Val Val Ser Val Leu Thr
Val Val His Gln Asp Trp Leu Asn 85 90 95Gly Lys Glu Tyr Lys Cys Lys
Val Ser Asn Lys Gly Leu Pro Ala Pro 100 105 110Ile Glu Lys Thr Ile
Ser Lys Thr Lys Gly Gln Pro Arg Glu Pro Gln 115 120 125Val Tyr Thr
Leu Pro Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val 130 135 140Ser
Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ser Val145 150
155 160Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr
Pro 165 170 175Pro Met Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser
Lys Leu Thr 180 185 190Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val
Phe Ser Cys Ser Val 195 200 205Met His Glu Ala Leu His Asn His Tyr
Thr Gln Lys Ser Leu Ser Leu 210 215 220Ser Pro Gly
Lys22519227PRTArtificial SequencehIgG2FcdeltaK-SS Fc variant 19Glu
Arg Lys Ser Ser Val Glu Cys Pro Pro Cys Pro Ala Pro Pro Val1 5 10
15Ala Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu
20 25 30Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val
Ser 35 40 45His Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr Val Asp Gly
Val Glu 50 55 60Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Phe
Asn Ser Thr65 70 75 80Phe Arg Val Val Ser Val Leu Thr Val Val His
Gln Asp Trp Leu Asn 85 90 95Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn
Lys Gly Leu Pro Ala Pro 100 105 110Ile Glu Lys Thr Ile Ser Lys Thr
Lys Gly Gln Pro Arg Glu Pro Gln 115 120 125Val Tyr Thr Leu Pro Pro
Ser Arg Glu Glu Met Thr Lys Asn Gln Val 130 135 140Ser Leu Thr Cys
Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ser Val145 150 155 160Glu
Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro 165 170
175Pro Met Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr
180 185 190Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys
Ser Val 195 200 205Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys
Ser Leu Ser Leu 210 215 220Ser Pro Gly22520227PRTArtificial
SequencehIgG2FcdeltaK-SSS Fc variant 20Glu Arg Lys Ser Ser Val Glu
Ser Pro Pro Cys Pro Ala Pro Pro Val1 5 10 15Ala Gly Pro Ser Val Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu 20 25 30Met Ile Ser Arg Thr
Pro Glu Val Thr Cys Val Val Val Asp Val Ser 35 40 45His Glu Asp Pro
Glu Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu 50 55 60Val His Asn
Ala Lys Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr65 70 75 80Phe
Arg Val Val Ser Val Leu Thr Val Val His Gln Asp Trp Leu Asn 85 90
95Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro
100 105 110Ile Glu Lys Thr Ile Ser Lys Thr Lys Gly Gln Pro Arg Glu
Pro Gln 115 120 125Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu Met Thr
Lys Asn Gln Val 130 135 140Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ser Val145 150 155 160Glu Trp Glu Ser Asn Gly Gln
Pro Glu Asn Asn Tyr Lys Thr Thr Pro 165 170 175Pro Met Leu Asp Ser
Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr 180 185 190Val Asp Lys
Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val 195 200 205Met
His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu 210 215
220Ser Pro Gly22521227PRTArtificial SequencehIgG2FcdeltaK-SSSS Fc
variant 21Glu Arg Lys Ser Ser Val Glu Ser Pro Pro Ser Pro Ala Pro
Pro Val1 5 10 15Ala Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys
Asp Thr Leu 20 25 30Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val
Val Asp Val Ser 35 40 45His Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr
Val Asp Gly Val Glu 50 55 60Val His Asn Ala Lys Thr Lys Pro Arg Glu
Glu Gln Phe Asn Ser Thr65 70 75 80Phe Arg Val Val Ser Val Leu Thr
Val Val His Gln Asp Trp Leu Asn 85 90 95Gly Lys Glu Tyr Lys Cys Lys
Val Ser Asn Lys Gly Leu Pro Ala Pro 100 105 110Ile Glu Lys Thr Ile
Ser Lys Thr Lys Gly Gln Pro Arg Glu Pro Gln 115 120 125Val Tyr Thr
Leu Pro Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val 130 135 140Ser
Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ser Val145 150
155 160Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr
Pro 165 170 175Pro Met Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser
Lys Leu Thr 180 185 190Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val
Phe Ser Cys Ser Val 195 200 205Met His Glu Ala Leu His Asn His Tyr
Thr Gln Lys Ser Leu Ser Leu 210 215 220Ser Pro
Gly22522216PRTArtificial SequencehIgG3FcdeltaK-deltaC Fc variant
22Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys1
5 10 15Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys
Val 20 25 30Val Val Asp Val Ser His Glu Asp Pro Glu Val Gln Phe Lys
Trp Tyr 35 40 45Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys Pro
Arg Glu Glu 50 55 60Gln Tyr Asn Ser Thr Phe Arg Val Val Ser Val Leu
Thr Val Leu His65 70 75 80Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys
Cys Lys Val Ser Asn Lys 85 90 95Ala Leu Pro Ala Pro Ile Glu Lys Thr
Ile Ser Lys Thr Lys Gly Gln 100 105 110Pro Arg Glu Pro Gln Val Tyr
Thr Leu Pro Pro Ser Arg Glu Glu Met 115 120 125Thr Lys Asn Gln Val
Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro 130 135 140Ser Asp Ile
Ala Val Glu Trp Glu Ser Ser Gly Gln Pro Glu Asn Asn145 150 155
160Tyr Asn Thr Thr Pro Pro Met Leu Asp Ser Asp Gly Ser Phe Phe Leu
165 170 175Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly
Asn Ile 180 185 190Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn
Arg Phe Thr Gln 195 200 205Lys Ser Leu Ser Leu Ser Pro Gly 210
21523220PRTArtificial SequencehIgG3FcdeltaK-C Fc variant 23Pro Arg
Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe Leu1 5 10 15Phe
Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu 20 25
30Val Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Gln
35 40 45Phe Lys Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys Thr
Lys 50 55 60Pro Arg Glu Glu Gln Tyr Asn Ser Thr Phe Arg Val Val Ser
Val Leu65 70 75 80Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu
Tyr Lys Cys Lys 85 90 95Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu
Lys Thr Ile Ser Lys 100 105 110Thr Lys Gly Gln Pro Arg Glu Pro Gln
Val Tyr Thr Leu Pro Pro Ser 115 120 125Arg Glu Glu Met Thr Lys Asn
Gln Val Ser Leu Thr Cys Leu Val Lys 130 135 140Gly Phe Tyr Pro Ser
Asp Ile Ala Val Glu Trp Glu Ser Ser Gly Gln145 150 155 160Pro Glu
Asn Asn Tyr Asn Thr Thr Pro Pro Met Leu Asp Ser Asp Gly 165 170
175Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln
180 185 190Gln Gly Asn Ile Phe Ser Cys Ser Val Met His Glu Ala Leu
His Asn 195 200 205Arg Phe Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly
210 215 22024226PRTArtificial SequencehIgG3FcdeltaK-CC Fc variant
24Asp Thr Pro Pro Pro Cys Pro Arg Cys Pro Ala Pro Glu Leu Leu Gly1
5 10 15Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu
Met 20 25 30Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val
Ser His 35 40 45Glu Asp Pro Glu Val Gln Phe Lys Trp Tyr Val Asp Gly
Val Glu Val 50 55 60His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr
Asn Ser Thr Phe65 70 75 80Arg Val Val Ser Val Leu Thr Val Leu His
Gln Asp Trp Leu Asn Gly 85 90 95Lys Glu Tyr Lys Cys Lys Val Ser Asn
Lys Ala Leu Pro Ala Pro Ile 100 105 110Glu Lys Thr Ile Ser Lys Thr
Lys Gly Gln Pro Arg Glu Pro Gln Val 115 120 125Tyr Thr Leu Pro Pro
Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser 130 135 140Leu Thr Cys
Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu145 150 155
160Trp Glu Ser Ser Gly Gln Pro Glu Asn Asn Tyr Asn Thr Thr Pro Pro
165 170 175Met Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu
Thr Val 180 185 190Asp Lys Ser Arg Trp Gln Gln Gly Asn Ile Phe Ser
Cys Ser Val Met 195 200 205His Glu Ala Leu His Asn Arg Phe Thr Gln
Lys Ser Leu Ser Leu Ser 210 215 220Pro Gly22525231PRTArtificial
SequencehIgG3FcdeltaK-S Fc variant 25Glu Pro Lys Ser Ser Asp Thr
Pro Pro Pro Cys Pro Arg Cys Pro Ala1 5 10 15Pro Glu Leu Leu Gly Gly
Pro Ser Val Phe Leu Phe Pro Pro Lys Pro 20 25 30Lys Asp Thr Leu Met
Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val 35 40 45Val Asp Val Ser
His Glu Asp Pro Glu Val Gln Phe Lys Trp Tyr Val 50 55 60Asp Gly Val
Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln65 70 75 80Tyr
Asn Ser Thr Phe Arg Val Val Ser Val Leu Thr Val Leu His Gln 85 90
95Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala
100 105 110Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Thr Lys Gly
Gln Pro 115 120 125Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg
Glu Glu Met Thr 130 135 140Lys Asn Gln Val Ser Leu Thr Cys Leu Val
Lys Gly Phe Tyr Pro Ser145 150 155 160Asp Ile Ala Val Glu Trp Glu
Ser Ser Gly Gln Pro Glu Asn Asn Tyr 165 170 175Asn Thr Thr Pro Pro
Met Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr 180 185 190Ser Lys Leu
Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Ile Phe 195 200 205Ser
Cys Ser Val Met His Glu Ala Leu His Asn Arg Phe Thr Gln Lys 210 215
220Ser Leu Ser Leu Ser Pro Gly225 23026231PRTArtificial
SequencehIgG3FcdeltaK-SS Fc variant 26Glu Pro Lys Ser Ser Asp Thr
Pro Pro
Pro Ser Pro Arg Cys Pro Ala1 5 10 15Pro Glu Leu Leu Gly Gly Pro Ser
Val Phe Leu Phe Pro Pro Lys Pro 20 25 30Lys Asp Thr Leu Met Ile Ser
Arg Thr Pro Glu Val Thr Cys Val Val 35 40 45Val Asp Val Ser His Glu
Asp Pro Glu Val Gln Phe Lys Trp Tyr Val 50 55 60Asp Gly Val Glu Val
His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln65 70 75 80Tyr Asn Ser
Thr Phe Arg Val Val Ser Val Leu Thr Val Leu His Gln 85 90 95Asp Trp
Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala 100 105
110Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Thr Lys Gly Gln Pro
115 120 125Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu
Met Thr 130 135 140Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly
Phe Tyr Pro Ser145 150 155 160Asp Ile Ala Val Glu Trp Glu Ser Ser
Gly Gln Pro Glu Asn Asn Tyr 165 170 175Asn Thr Thr Pro Pro Met Leu
Asp Ser Asp Gly Ser Phe Phe Leu Tyr 180 185 190Ser Lys Leu Thr Val
Asp Lys Ser Arg Trp Gln Gln Gly Asn Ile Phe 195 200 205Ser Cys Ser
Val Met His Glu Ala Leu His Asn Arg Phe Thr Gln Lys 210 215 220Ser
Leu Ser Leu Ser Pro Gly225 23027231PRTArtificial
SequencehIgG3FcdeltaK-SSS Fc variant 27Glu Pro Lys Ser Ser Asp Thr
Pro Pro Pro Ser Pro Arg Ser Pro Ala1 5 10 15Pro Glu Leu Leu Gly Gly
Pro Ser Val Phe Leu Phe Pro Pro Lys Pro 20 25 30Lys Asp Thr Leu Met
Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val 35 40 45Val Asp Val Ser
His Glu Asp Pro Glu Val Gln Phe Lys Trp Tyr Val 50 55 60Asp Gly Val
Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln65 70 75 80Tyr
Asn Ser Thr Phe Arg Val Val Ser Val Leu Thr Val Leu His Gln 85 90
95Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala
100 105 110Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Thr Lys Gly
Gln Pro 115 120 125Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg
Glu Glu Met Thr 130 135 140Lys Asn Gln Val Ser Leu Thr Cys Leu Val
Lys Gly Phe Tyr Pro Ser145 150 155 160Asp Ile Ala Val Glu Trp Glu
Ser Ser Gly Gln Pro Glu Asn Asn Tyr 165 170 175Asn Thr Thr Pro Pro
Met Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr 180 185 190Ser Lys Leu
Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Ile Phe 195 200 205Ser
Cys Ser Val Met His Glu Ala Leu His Asn Arg Phe Thr Gln Lys 210 215
220Ser Leu Ser Leu Ser Pro Gly225 23028216PRTArtificial
SequencehIgG4FcdeltaK-deltaC Fc variant 28Ala Pro Glu Phe Leu Gly
Gly Pro Ser Val Phe Leu Phe Pro Pro Lys1 5 10 15Pro Lys Asp Thr Leu
Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val 20 25 30Val Val Asp Val
Ser Gln Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr 35 40 45Val Asp Gly
Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu 50 55 60Gln Phe
Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His65 70 75
80Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys
85 90 95Gly Leu Pro Ser Ser Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly
Gln 100 105 110Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Gln
Glu Glu Met 115 120 125Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val
Lys Gly Phe Tyr Pro 130 135 140Ser Asp Ile Ala Val Glu Trp Glu Ser
Asn Gly Gln Pro Glu Asn Asn145 150 155 160Tyr Lys Thr Thr Pro Pro
Val Leu Asp Ser Asp Gly Ser Phe Phe Leu 165 170 175Tyr Ser Arg Leu
Thr Val Asp Lys Ser Arg Trp Gln Glu Gly Asn Val 180 185 190Phe Ser
Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln 195 200
205Lys Ser Leu Ser Leu Ser Leu Gly 210 21529220PRTArtificial
SequencehIgG4FcdeltaK-C Fc variant 29Pro Ser Cys Pro Ala Pro Glu
Phe Leu Gly Gly Pro Ser Val Phe Leu1 5 10 15Phe Pro Pro Lys Pro Lys
Asp Thr Leu Met Ile Ser Arg Thr Pro Glu 20 25 30Val Thr Cys Val Val
Val Asp Val Ser Gln Glu Asp Pro Glu Val Gln 35 40 45Phe Asn Trp Tyr
Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys 50 55 60Pro Arg Glu
Glu Gln Phe Asn Ser Thr Tyr Arg Val Val Ser Val Leu65 70 75 80Thr
Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys 85 90
95Val Ser Asn Lys Gly Leu Pro Ser Ser Ile Glu Lys Thr Ile Ser Lys
100 105 110Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro
Pro Ser 115 120 125Gln Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr
Cys Leu Val Lys 130 135 140Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu
Trp Glu Ser Asn Gly Gln145 150 155 160Pro Glu Asn Asn Tyr Lys Thr
Thr Pro Pro Val Leu Asp Ser Asp Gly 165 170 175Ser Phe Phe Leu Tyr
Ser Arg Leu Thr Val Asp Lys Ser Arg Trp Gln 180 185 190Glu Gly Asn
Val Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn 195 200 205His
Tyr Thr Gln Lys Ser Leu Ser Leu Ser Leu Gly 210 215
22030228PRTArtificial SequencehIgG4FcdeltaK-CC Fc variant 30Glu Ser
Lys Tyr Gly Pro Pro Cys Pro Ser Cys Pro Ala Pro Glu Phe1 5 10 15Leu
Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr 20 25
30Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val
35 40 45Ser Gln Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr Val Asp Gly
Val 50 55 60Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Phe
Asn Ser65 70 75 80Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His
Gln Asp Trp Leu 85 90 95Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn
Lys Gly Leu Pro Ser 100 105 110Ser Ile Glu Lys Thr Ile Ser Lys Ala
Lys Gly Gln Pro Arg Glu Pro 115 120 125Gln Val Tyr Thr Leu Pro Pro
Ser Gln Glu Glu Met Thr Lys Asn Gln 130 135 140Val Ser Leu Thr Cys
Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala145 150 155 160Val Glu
Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr 165 170
175Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Arg Leu
180 185 190Thr Val Asp Lys Ser Arg Trp Gln Glu Gly Asn Val Phe Ser
Cys Ser 195 200 205Val Met His Glu Ala Leu His Asn His Tyr Thr Gln
Lys Ser Leu Ser 210 215 220Leu Ser Leu Gly22531228PRTArtificial
SequencehIgG4FcdeltaK-CC-228P Fc variant 31Glu Ser Lys Tyr Gly Pro
Pro Cys Pro Pro Cys Pro Ala Pro Glu Phe1 5 10 15Leu Gly Gly Pro Ser
Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr 20 25 30Leu Met Ile Ser
Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val 35 40 45Ser Gln Glu
Asp Pro Glu Val Gln Phe Asn Trp Tyr Val Asp Gly Val 50 55 60Glu Val
His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser65 70 75
80Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu
85 90 95Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Gly Leu Pro
Ser 100 105 110Ser Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro
Arg Glu Pro 115 120 125Gln Val Tyr Thr Leu Pro Pro Ser Gln Glu Glu
Met Thr Lys Asn Gln 130 135 140Val Ser Leu Thr Cys Leu Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala145 150 155 160Val Glu Trp Glu Ser Asn
Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr 165 170 175Pro Pro Val Leu
Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Arg Leu 180 185 190Thr Val
Asp Lys Ser Arg Trp Gln Glu Gly Asn Val Phe Ser Cys Ser 195 200
205Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser
210 215 220Leu Ser Leu Gly22532228PRTArtificial
SequencehIgG4FcdeltaK-CC-228P-409K Fc variant 32Glu Ser Lys Tyr Gly
Pro Pro Cys Pro Pro Cys Pro Ala Pro Glu Phe1 5 10 15Leu Gly Gly Pro
Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr 20 25 30Leu Met Ile
Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val 35 40 45Ser Gln
Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr Val Asp Gly Val 50 55 60Glu
Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser65 70 75
80Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu
85 90 95Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Gly Leu Pro
Ser 100 105 110Ser Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro
Arg Glu Pro 115 120 125Gln Val Tyr Thr Leu Pro Pro Ser Gln Glu Glu
Met Thr Lys Asn Gln 130 135 140Val Ser Leu Thr Cys Leu Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala145 150 155 160Val Glu Trp Glu Ser Asn
Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr 165 170 175Pro Pro Val Leu
Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu 180 185 190Thr Val
Asp Lys Ser Arg Trp Gln Glu Gly Asn Val Phe Ser Cys Ser 195 200
205Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser
210 215 220Leu Ser Leu Gly22533228PRTArtificial
SequencehIgG4FcdeltaK-S Fc variant 33Glu Ser Lys Tyr Gly Pro Pro
Ser Pro Ser Cys Pro Ala Pro Glu Phe1 5 10 15Leu Gly Gly Pro Ser Val
Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr 20 25 30Leu Met Ile Ser Arg
Thr Pro Glu Val Thr Cys Val Val Val Asp Val 35 40 45Ser Gln Glu Asp
Pro Glu Val Gln Phe Asn Trp Tyr Val Asp Gly Val 50 55 60Glu Val His
Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser65 70 75 80Thr
Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu 85 90
95Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Gly Leu Pro Ser
100 105 110Ser Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg
Glu Pro 115 120 125Gln Val Tyr Thr Leu Pro Pro Ser Gln Glu Glu Met
Thr Lys Asn Gln 130 135 140Val Ser Leu Thr Cys Leu Val Lys Gly Phe
Tyr Pro Ser Asp Ile Ala145 150 155 160Val Glu Trp Glu Ser Asn Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr 165 170 175Pro Pro Val Leu Asp
Ser Asp Gly Ser Phe Phe Leu Tyr Ser Arg Leu 180 185 190Thr Val Asp
Lys Ser Arg Trp Gln Glu Gly Asn Val Phe Ser Cys Ser 195 200 205Val
Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser 210 215
220Leu Ser Leu Gly22534228PRTArtificial SequencehIgG4FcdeltaK-SS Fc
variant 34Glu Ser Lys Tyr Gly Pro Pro Ser Pro Ser Ser Pro Ala Pro
Glu Phe1 5 10 15Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro
Lys Asp Thr 20 25 30Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val
Val Val Asp Val 35 40 45Ser Gln Glu Asp Pro Glu Val Gln Phe Asn Trp
Tyr Val Asp Gly Val 50 55 60Glu Val His Asn Ala Lys Thr Lys Pro Arg
Glu Glu Gln Phe Asn Ser65 70 75 80Thr Tyr Arg Val Val Ser Val Leu
Thr Val Leu His Gln Asp Trp Leu 85 90 95Asn Gly Lys Glu Tyr Lys Cys
Lys Val Ser Asn Lys Gly Leu Pro Ser 100 105 110Ser Ile Glu Lys Thr
Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro 115 120 125Gln Val Tyr
Thr Leu Pro Pro Ser Gln Glu Glu Met Thr Lys Asn Gln 130 135 140Val
Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala145 150
155 160Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr
Thr 165 170 175Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr
Ser Arg Leu 180 185 190Thr Val Asp Lys Ser Arg Trp Gln Glu Gly Asn
Val Phe Ser Cys Ser 195 200 205Val Met His Glu Ala Leu His Asn His
Tyr Thr Gln Lys Ser Leu Ser 210 215 220Leu Ser Leu
Gly22535101PRTArtificial SequenceTbetaRII-ED structured domain
35Gln Leu Cys Lys Phe Cys Asp Val Arg Phe Ser Thr Cys Asp Asn Gln1
5 10 15Lys Ser Cys Met Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys
Pro 20 25 30Gln Glu Val Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn
Ile Thr 35 40 45Leu Glu Thr Val Cys His Asp Pro Lys Leu Pro Tyr His
Asp Phe Ile 50 55 60Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys
Glu Lys Lys Lys65 70 75 80Pro Gly Glu Thr Phe Phe Met Cys Ser Cys
Ser Ser Asp Glu Cys Asn 85 90 95Asp Asn Ile Ile Phe
1003625PRTArtificial SequenceTbetaRII-ED N-term unstructured region
and natural linker 36Ile Pro Pro His Val Gln Lys Ser Val Asn Asn
Asp Met Ile Val Thr1 5 10 15Asp Asn Asn Gly Ala Val Lys Phe Pro 20
253750PRTArtificial SequenceTbetaRIIb-ED N-term unstructured region
and natural linker 37Ile Pro Pro His Val Gln Lys Ser Asp Val Glu
Met Glu Ala Gln Lys1 5 10 15Asp Glu Ile Ile Cys Pro Ser Cys Asn Arg
Thr Ala His Pro Leu Arg 20 25 30His Ile Asn Asn Asp Met Ile Val Thr
Asp Asn Asn Gly Ala Val Lys 35 40 45Phe Pro 503850PRTArtificial
SequenceTbetaRIIb-ED Cys-mutated N-term unstructured region and
natural linker 38Ile Pro Pro His Val Gln Lys Ser Asp Val Glu Met
Glu Ala Gln Lys1 5 10 15Asp Glu Ile Ile Ala Pro Ser Ala Asn Arg Thr
Ala His Pro Leu Arg 20 25 30His Ile Asn Asn Asp Met Ile Val Thr Asp
Asn Asn Gly Ala Val Lys 35 40 45Phe Pro 503910PRTArtificial
SequenceTbetaRII-ED C-term unstructured region and natural linker
39Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp1 5 104035PRTArtificial
SequenceTbetaRII-ED natural linker 40Ser Glu Glu Tyr Asn Thr Ser
Asn Pro Asp Ile Pro Pro His Val Gln1 5 10 15Lys Ser Val Asn Asn Asp
Met Ile Val Thr Asp Asn Asn Gly Ala Val 20 25 30Lys Phe Pro
354160PRTArtificial SequenceTbetaRIIb-ED natural linker 41Ser Glu
Glu Tyr Asn Thr Ser Asn Pro Asp Ile Pro Pro His Val Gln1 5 10 15Lys
Ser Asp Val Glu Met Glu Ala Gln Lys Asp Glu Ile Ile Cys Pro 20 25
30Ser Cys Asn Arg Thr Ala His Pro Leu Arg His Ile Asn Asn Asp Met
35 40
45Ile Val Thr Asp Asn Asn Gly Ala Val Lys Phe Pro 50 55
604260PRTArtificial SequenceTbetaRIIb-ED Cys-mutated linker 42Ser
Glu Glu Tyr Asn Thr Ser Asn Pro Asp Ile Pro Pro His Val Gln1 5 10
15Lys Ser Asp Val Glu Met Glu Ala Gln Lys Asp Glu Ile Ile Ala Pro
20 25 30Ser Ala Asn Arg Thr Ala His Pro Leu Arg His Ile Asn Asn Asp
Met 35 40 45Ile Val Thr Asp Asn Asn Gly Ala Val Lys Phe Pro 50 55
6043136PRTArtificial SequenceTbetaRII-ED monomer, also termed T2 or
T2m 43Ile Pro Pro His Val Gln Lys Ser Val Asn Asn Asp Met Ile Val
Thr1 5 10 15Asp Asn Asn Gly Ala Val Lys Phe Pro Gln Leu Cys Lys Phe
Cys Asp 20 25 30Val Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys Met
Ser Asn Cys 35 40 45Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val
Cys Val Ala Val 50 55 60Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu
Thr Val Cys His Asp65 70 75 80Pro Lys Leu Pro Tyr His Asp Phe Ile
Leu Glu Asp Ala Ala Ser Pro 85 90 95Lys Cys Ile Met Lys Glu Lys Lys
Lys Pro Gly Glu Thr Phe Phe Met 100 105 110Cys Ser Cys Ser Ser Asp
Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu 115 120 125Glu Tyr Asn Thr
Ser Asn Pro Asp 130 13544161PRTArtificial SequenceTbetabRIIb-ED
monomer, also termed T2b 44Ile Pro Pro His Val Gln Lys Ser Asp Val
Glu Met Glu Ala Gln Lys1 5 10 15Asp Glu Ile Ile Cys Pro Ser Cys Asn
Arg Thr Ala His Pro Leu Arg 20 25 30His Ile Asn Asn Asp Met Ile Val
Thr Asp Asn Asn Gly Ala Val Lys 35 40 45Phe Pro Gln Leu Cys Lys Phe
Cys Asp Val Arg Phe Ser Thr Cys Asp 50 55 60Asn Gln Lys Ser Cys Met
Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu65 70 75 80Lys Pro Gln Glu
Val Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn 85 90 95Ile Thr Leu
Glu Thr Val Cys His Asp Pro Lys Leu Pro Tyr His Asp 100 105 110Phe
Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys 115 120
125Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu
130 135 140Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser
Asn Pro145 150 155 160Asp45161PRTArtificial SequenceTbetaRIIb-ED
monomer Cys-mutated in the linker region, also termed T2bAA 45Ile
Pro Pro His Val Gln Lys Ser Asp Val Glu Met Glu Ala Gln Lys1 5 10
15Asp Glu Ile Ile Ala Pro Ser Ala Asn Arg Thr Ala His Pro Leu Arg
20 25 30His Ile Asn Asn Asp Met Ile Val Thr Asp Asn Asn Gly Ala Val
Lys 35 40 45Phe Pro Gln Leu Cys Lys Phe Cys Asp Val Arg Phe Ser Thr
Cys Asp 50 55 60Asn Gln Lys Ser Cys Met Ser Asn Cys Ser Ile Thr Ser
Ile Cys Glu65 70 75 80Lys Pro Gln Glu Val Cys Val Ala Val Trp Arg
Lys Asn Asp Glu Asn 85 90 95Ile Thr Leu Glu Thr Val Cys His Asp Pro
Lys Leu Pro Tyr His Asp 100 105 110Phe Ile Leu Glu Asp Ala Ala Ser
Pro Lys Cys Ile Met Lys Glu Lys 115 120 125Lys Lys Pro Gly Glu Thr
Phe Phe Met Cys Ser Cys Ser Ser Asp Glu 130 135 140Cys Asn Asp Asn
Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn Pro145 150 155
160Asp46272PRTArtificial SequenceTbetaRII-ED dimer, also termed
T2-T2 or T22d35 46Ile Pro Pro His Val Gln Lys Ser Val Asn Asn Asp
Met Ile Val Thr1 5 10 15Asp Asn Asn Gly Ala Val Lys Phe Pro Gln Leu
Cys Lys Phe Cys Asp 20 25 30Val Arg Phe Ser Thr Cys Asp Asn Gln Lys
Ser Cys Met Ser Asn Cys 35 40 45Ser Ile Thr Ser Ile Cys Glu Lys Pro
Gln Glu Val Cys Val Ala Val 50 55 60Trp Arg Lys Asn Asp Glu Asn Ile
Thr Leu Glu Thr Val Cys His Asp65 70 75 80Pro Lys Leu Pro Tyr His
Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro 85 90 95Lys Cys Ile Met Lys
Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met 100 105 110Cys Ser Cys
Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu 115 120 125Glu
Tyr Asn Thr Ser Asn Pro Asp Ile Pro Pro His Val Gln Lys Ser 130 135
140Val Asn Asn Asp Met Ile Val Thr Asp Asn Asn Gly Ala Val Lys
Phe145 150 155 160Pro Gln Leu Cys Lys Phe Cys Asp Val Arg Phe Ser
Thr Cys Asp Asn 165 170 175Gln Lys Ser Cys Met Ser Asn Cys Ser Ile
Thr Ser Ile Cys Glu Lys 180 185 190Pro Gln Glu Val Cys Val Ala Val
Trp Arg Lys Asn Asp Glu Asn Ile 195 200 205Thr Leu Glu Thr Val Cys
His Asp Pro Lys Leu Pro Tyr His Asp Phe 210 215 220Ile Leu Glu Asp
Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys225 230 235 240Lys
Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu Cys 245 250
255Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp
260 265 27047322PRTArtificial SequenceTbetaRIIb-ED dimer, also
termed T2-T2b 47Ile Pro Pro His Val Gln Lys Ser Asp Val Glu Met Glu
Ala Gln Lys1 5 10 15Asp Glu Ile Ile Cys Pro Ser Cys Asn Arg Thr Ala
His Pro Leu Arg 20 25 30His Ile Asn Asn Asp Met Ile Val Thr Asp Asn
Asn Gly Ala Val Lys 35 40 45Phe Pro Gln Leu Cys Lys Phe Cys Asp Val
Arg Phe Ser Thr Cys Asp 50 55 60Asn Gln Lys Ser Cys Met Ser Asn Cys
Ser Ile Thr Ser Ile Cys Glu65 70 75 80Lys Pro Gln Glu Val Cys Val
Ala Val Trp Arg Lys Asn Asp Glu Asn 85 90 95Ile Thr Leu Glu Thr Val
Cys His Asp Pro Lys Leu Pro Tyr His Asp 100 105 110Phe Ile Leu Glu
Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys 115 120 125Lys Lys
Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu 130 135
140Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn
Pro145 150 155 160Asp Ile Pro Pro His Val Gln Lys Ser Asp Val Glu
Met Glu Ala Gln 165 170 175Lys Asp Glu Ile Ile Cys Pro Ser Cys Asn
Arg Thr Ala His Pro Leu 180 185 190Arg His Ile Asn Asn Asp Met Ile
Val Thr Asp Asn Asn Gly Ala Val 195 200 205Lys Phe Pro Gln Leu Cys
Lys Phe Cys Asp Val Arg Phe Ser Thr Cys 210 215 220Asp Asn Gln Lys
Ser Cys Met Ser Asn Cys Ser Ile Thr Ser Ile Cys225 230 235 240Glu
Lys Pro Gln Glu Val Cys Val Ala Val Trp Arg Lys Asn Asp Glu 245 250
255Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys Leu Pro Tyr His
260 265 270Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met
Lys Glu 275 280 285Lys Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser
Cys Ser Ser Asp 290 295 300Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu
Glu Tyr Asn Thr Ser Asn305 310 315 320Pro Asp48322PRTArtificial
SequenceTbetaRIIb-ED dimer Cys-mutated in the linker region, also
termed T2-T2bAA 48Ile Pro Pro His Val Gln Lys Ser Asp Val Glu Met
Glu Ala Gln Lys1 5 10 15Asp Glu Ile Ile Cys Pro Ser Cys Asn Arg Thr
Ala His Pro Leu Arg 20 25 30His Ile Asn Asn Asp Met Ile Val Thr Asp
Asn Asn Gly Ala Val Lys 35 40 45Phe Pro Gln Leu Cys Lys Phe Cys Asp
Val Arg Phe Ser Thr Cys Asp 50 55 60Asn Gln Lys Ser Cys Met Ser Asn
Cys Ser Ile Thr Ser Ile Cys Glu65 70 75 80Lys Pro Gln Glu Val Cys
Val Ala Val Trp Arg Lys Asn Asp Glu Asn 85 90 95Ile Thr Leu Glu Thr
Val Cys His Asp Pro Lys Leu Pro Tyr His Asp 100 105 110Phe Ile Leu
Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys 115 120 125Lys
Lys Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu 130 135
140Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn
Pro145 150 155 160Asp Ile Pro Pro His Val Gln Lys Ser Asp Val Glu
Met Glu Ala Gln 165 170 175Lys Asp Glu Ile Ile Ala Pro Ser Ala Asn
Arg Thr Ala His Pro Leu 180 185 190Arg His Ile Asn Asn Asp Met Ile
Val Thr Asp Asn Asn Gly Ala Val 195 200 205Lys Phe Pro Gln Leu Cys
Lys Phe Cys Asp Val Arg Phe Ser Thr Cys 210 215 220Asp Asn Gln Lys
Ser Cys Met Ser Asn Cys Ser Ile Thr Ser Ile Cys225 230 235 240Glu
Lys Pro Gln Glu Val Cys Val Ala Val Trp Arg Lys Asn Asp Glu 245 250
255Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys Leu Pro Tyr His
260 265 270Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met
Lys Glu 275 280 285Lys Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser
Cys Ser Ser Asp 290 295 300Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu
Glu Tyr Asn Thr Ser Asn305 310 315 320Pro Asp49408PRTArtificial
SequenceTbetaRII-ED trimer, also termed T2-T2-T2 49Ile Pro Pro His
Val Gln Lys Ser Val Asn Asn Asp Met Ile Val Thr1 5 10 15Asp Asn Asn
Gly Ala Val Lys Phe Pro Gln Leu Cys Lys Phe Cys Asp 20 25 30Val Arg
Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys Met Ser Asn Cys 35 40 45Ser
Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val 50 55
60Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His Asp65
70 75 80Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser
Pro 85 90 95Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe
Phe Met 100 105 110Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile
Ile Phe Ser Glu 115 120 125Glu Tyr Asn Thr Ser Asn Pro Asp Ile Pro
Pro His Val Gln Lys Ser 130 135 140Val Asn Asn Asp Met Ile Val Thr
Asp Asn Asn Gly Ala Val Lys Phe145 150 155 160Pro Gln Leu Cys Lys
Phe Cys Asp Val Arg Phe Ser Thr Cys Asp Asn 165 170 175Gln Lys Ser
Cys Met Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys 180 185 190Pro
Gln Glu Val Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile 195 200
205Thr Leu Glu Thr Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe
210 215 220Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu
Lys Lys225 230 235 240Lys Pro Gly Glu Thr Phe Phe Met Cys Ser Cys
Ser Ser Asp Glu Cys 245 250 255Asn Asp Asn Ile Ile Phe Ser Glu Glu
Tyr Asn Thr Ser Asn Pro Asp 260 265 270Ile Pro Pro His Val Gln Lys
Ser Val Asn Asn Asp Met Ile Val Thr 275 280 285Asp Asn Asn Gly Ala
Val Lys Phe Pro Gln Leu Cys Lys Phe Cys Asp 290 295 300Val Arg Phe
Ser Thr Cys Asp Asn Gln Lys Ser Cys Met Ser Asn Cys305 310 315
320Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val
325 330 335Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys
His Asp 340 345 350Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp
Ala Ala Ser Pro 355 360 365Lys Cys Ile Met Lys Glu Lys Lys Lys Pro
Gly Glu Thr Phe Phe Met 370 375 380Cys Ser Cys Ser Ser Asp Glu Cys
Asn Asp Asn Ile Ile Phe Ser Glu385 390 395 400Glu Tyr Asn Thr Ser
Asn Pro Asp 40550458PRTArtificial SequenceTbetaRIIb-ED trimer, also
termed T2-T2b-T2b 50Ile Pro Pro His Val Gln Lys Ser Val Asn Asn Asp
Met Ile Val Thr1 5 10 15Asp Asn Asn Gly Ala Val Lys Phe Pro Gln Leu
Cys Lys Phe Cys Asp 20 25 30Val Arg Phe Ser Thr Cys Asp Asn Gln Lys
Ser Cys Met Ser Asn Cys 35 40 45Ser Ile Thr Ser Ile Cys Glu Lys Pro
Gln Glu Val Cys Val Ala Val 50 55 60Trp Arg Lys Asn Asp Glu Asn Ile
Thr Leu Glu Thr Val Cys His Asp65 70 75 80Pro Lys Leu Pro Tyr His
Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro 85 90 95Lys Cys Ile Met Lys
Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met 100 105 110Cys Ser Cys
Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu 115 120 125Glu
Tyr Asn Thr Ser Asn Pro Asp Ile Pro Pro His Val Gln Lys Ser 130 135
140Asp Val Glu Met Glu Ala Gln Lys Asp Glu Ile Ile Cys Pro Ser
Cys145 150 155 160Asn Arg Thr Ala His Pro Leu Arg His Ile Asn Asn
Asp Met Ile Val 165 170 175Thr Asp Asn Asn Gly Ala Val Lys Phe Pro
Gln Leu Cys Lys Phe Cys 180 185 190Asp Val Arg Phe Ser Thr Cys Asp
Asn Gln Lys Ser Cys Met Ser Asn 195 200 205Cys Ser Ile Thr Ser Ile
Cys Glu Lys Pro Gln Glu Val Cys Val Ala 210 215 220Val Trp Arg Lys
Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His225 230 235 240Asp
Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser 245 250
255Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe
260 265 270Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile
Phe Ser 275 280 285Glu Glu Tyr Asn Thr Ser Asn Pro Asp Ile Pro Pro
His Val Gln Lys 290 295 300Ser Asp Val Glu Met Glu Ala Gln Lys Asp
Glu Ile Ile Cys Pro Ser305 310 315 320Cys Asn Arg Thr Ala His Pro
Leu Arg His Ile Asn Asn Asp Met Ile 325 330 335Val Thr Asp Asn Asn
Gly Ala Val Lys Phe Pro Gln Leu Cys Lys Phe 340 345 350Cys Asp Val
Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys Met Ser 355 360 365Asn
Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys Val 370 375
380Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val
Cys385 390 395 400His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu
Glu Asp Ala Ala 405 410 415Ser Pro Lys Cys Ile Met Lys Glu Lys Lys
Lys Pro Gly Glu Thr Phe 420 425 430Phe Met Cys Ser Cys Ser Ser Asp
Glu Cys Asn Asp Asn Ile Ile Phe 435 440 445Ser Glu Glu Tyr Asn Thr
Ser Asn Pro Asp 450 45551458PRTArtificial SequenceTbetaRIIb-ED
trimer Cys-mutated in the linker regions, also termed
T2-T2bAA-T2bAA 51Ile Pro Pro His Val Gln Lys Ser Val Asn Asn Asp
Met Ile Val Thr1 5 10 15Asp Asn Asn Gly Ala Val Lys Phe Pro Gln Leu
Cys Lys Phe Cys Asp 20 25 30Val Arg Phe Ser Thr Cys Asp Asn Gln Lys
Ser Cys Met Ser Asn Cys 35 40 45Ser Ile Thr Ser Ile Cys Glu Lys
Pro
Gln Glu Val Cys Val Ala Val 50 55 60Trp Arg Lys Asn Asp Glu Asn Ile
Thr Leu Glu Thr Val Cys His Asp65 70 75 80Pro Lys Leu Pro Tyr His
Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro 85 90 95Lys Cys Ile Met Lys
Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met 100 105 110Cys Ser Cys
Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu 115 120 125Glu
Tyr Asn Thr Ser Asn Pro Asp Ile Pro Pro His Val Gln Lys Ser 130 135
140Asp Val Glu Met Glu Ala Gln Lys Asp Glu Ile Ile Ala Pro Ser
Ala145 150 155 160Asn Arg Thr Ala His Pro Leu Arg His Ile Asn Asn
Asp Met Ile Val 165 170 175Thr Asp Asn Asn Gly Ala Val Lys Phe Pro
Gln Leu Cys Lys Phe Cys 180 185 190Asp Val Arg Phe Ser Thr Cys Asp
Asn Gln Lys Ser Cys Met Ser Asn 195 200 205Cys Ser Ile Thr Ser Ile
Cys Glu Lys Pro Gln Glu Val Cys Val Ala 210 215 220Val Trp Arg Lys
Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His225 230 235 240Asp
Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser 245 250
255Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe
260 265 270Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile
Phe Ser 275 280 285Glu Glu Tyr Asn Thr Ser Asn Pro Asp Ile Pro Pro
His Val Gln Lys 290 295 300Ser Asp Val Glu Met Glu Ala Gln Lys Asp
Glu Ile Ile Ala Pro Ser305 310 315 320Ala Asn Arg Thr Ala His Pro
Leu Arg His Ile Asn Asn Asp Met Ile 325 330 335Val Thr Asp Asn Asn
Gly Ala Val Lys Phe Pro Gln Leu Cys Lys Phe 340 345 350Cys Asp Val
Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys Met Ser 355 360 365Asn
Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys Val 370 375
380Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val
Cys385 390 395 400His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu
Glu Asp Ala Ala 405 410 415Ser Pro Lys Cys Ile Met Lys Glu Lys Lys
Lys Pro Gly Glu Thr Phe 420 425 430Phe Met Cys Ser Cys Ser Ser Asp
Glu Cys Asn Asp Asn Ile Ile Phe 435 440 445Ser Glu Glu Tyr Asn Thr
Ser Asn Pro Asp 450 4555279PRTArtificial SequenceTbetaRI-ED
structured domain 52Ala Leu Gln Cys Phe Cys His Leu Cys Thr Lys Asp
Asn Phe Thr Cys1 5 10 15Val Thr Asp Gly Leu Cys Phe Val Ser Val Thr
Glu Thr Thr Asp Lys 20 25 30Val Ile His Asn Ser Met Cys Ile Ala Glu
Ile Asp Leu Ile Pro Arg 35 40 45Asp Arg Pro Phe Val Cys Ala Pro Ser
Ser Lys Thr Gly Ser Val Thr 50 55 60Thr Thr Tyr Cys Cys Asn Gln Asp
His Cys Asn Lys Ile Glu Leu65 70 75538PRTArtificial
SequenceTbetaRI-ED N-term unstructured region and natural linker
53Ala Ala Leu Leu Pro Gly Ala Thr1 55414PRTArtificial
SequenceTbetaRI-ED C-term unstructured region and natural linker
54Pro Thr Thr Val Lys Ser Ser Pro Gly Leu Gly Pro Val Glu1 5
105522PRTArtificial SequenceTbetaRI-ED natural linker 55Pro Thr Thr
Val Lys Ser Ser Pro Gly Leu Gly Pro Val Glu Ala Ala1 5 10 15Leu Leu
Pro Gly Ala Thr 2056101PRTArtificial SequenceTbetaRI-ED monomer,
also termed T1 or T1m 56Ala Ala Leu Leu Pro Gly Ala Thr Ala Leu Gln
Cys Phe Cys His Leu1 5 10 15Cys Thr Lys Asp Asn Phe Thr Cys Val Thr
Asp Gly Leu Cys Phe Val 20 25 30Ser Val Thr Glu Thr Thr Asp Lys Val
Ile His Asn Ser Met Cys Ile 35 40 45Ala Glu Ile Asp Leu Ile Pro Arg
Asp Arg Pro Phe Val Cys Ala Pro 50 55 60Ser Ser Lys Thr Gly Ser Val
Thr Thr Thr Tyr Cys Cys Asn Gln Asp65 70 75 80His Cys Asn Lys Ile
Glu Leu Pro Thr Thr Val Lys Ser Ser Pro Gly 85 90 95Leu Gly Pro Val
Glu 10057202PRTArtificial SequenceTbetaRI-ED dimer, also termed
T1-T1 57Ala Ala Leu Leu Pro Gly Ala Thr Ala Leu Gln Cys Phe Cys His
Leu1 5 10 15Cys Thr Lys Asp Asn Phe Thr Cys Val Thr Asp Gly Leu Cys
Phe Val 20 25 30Ser Val Thr Glu Thr Thr Asp Lys Val Ile His Asn Ser
Met Cys Ile 35 40 45Ala Glu Ile Asp Leu Ile Pro Arg Asp Arg Pro Phe
Val Cys Ala Pro 50 55 60Ser Ser Lys Thr Gly Ser Val Thr Thr Thr Tyr
Cys Cys Asn Gln Asp65 70 75 80His Cys Asn Lys Ile Glu Leu Pro Thr
Thr Val Lys Ser Ser Pro Gly 85 90 95Leu Gly Pro Val Glu Ala Ala Leu
Leu Pro Gly Ala Thr Ala Leu Gln 100 105 110Cys Phe Cys His Leu Cys
Thr Lys Asp Asn Phe Thr Cys Val Thr Asp 115 120 125Gly Leu Cys Phe
Val Ser Val Thr Glu Thr Thr Asp Lys Val Ile His 130 135 140Asn Ser
Met Cys Ile Ala Glu Ile Asp Leu Ile Pro Arg Asp Arg Pro145 150 155
160Phe Val Cys Ala Pro Ser Ser Lys Thr Gly Ser Val Thr Thr Thr Tyr
165 170 175Cys Cys Asn Gln Asp His Cys Asn Lys Ile Glu Leu Pro Thr
Thr Val 180 185 190Lys Ser Ser Pro Gly Leu Gly Pro Val Glu 195
2005839PRTArtificial SequenceTbetaRI-TbetaRII-ED natural linker
58Pro Thr Thr Val Lys Ser Ser Pro Gly Leu Gly Pro Val Glu Ile Pro1
5 10 15Pro His Val Gln Lys Ser Val Asn Asn Asp Met Ile Val Thr Asp
Asn 20 25 30Asn Gly Ala Val Lys Phe Pro 355964PRTArtificial
SequenceTbetaRI-TbetaRIIb-ED natural linker 59Pro Thr Thr Val Lys
Ser Ser Pro Gly Leu Gly Pro Val Glu Ile Pro1 5 10 15Pro His Val Gln
Lys Ser Asp Val Glu Met Glu Ala Gln Lys Asp Glu 20 25 30Ile Ile Cys
Pro Ser Cys Asn Arg Thr Ala His Pro Leu Arg His Ile 35 40 45Asn Asn
Asp Met Ile Val Thr Asp Asn Asn Gly Ala Val Lys Phe Pro 50 55
606018PRTArtificial SequenceTbetaRII-TbetaRI-ED natural linker
60Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp Ala Ala Leu Leu Pro Gly1
5 10 15Ala Thr61237PRTArtificial SequenceTbetaRI-TbetaRII-ED dimer
T1-T2 61Ala Ala Leu Leu Pro Gly Ala Thr Ala Leu Gln Cys Phe Cys His
Leu1 5 10 15Cys Thr Lys Asp Asn Phe Thr Cys Val Thr Asp Gly Leu Cys
Phe Val 20 25 30Ser Val Thr Glu Thr Thr Asp Lys Val Ile His Asn Ser
Met Cys Ile 35 40 45Ala Glu Ile Asp Leu Ile Pro Arg Asp Arg Pro Phe
Val Cys Ala Pro 50 55 60Ser Ser Lys Thr Gly Ser Val Thr Thr Thr Tyr
Cys Cys Asn Gln Asp65 70 75 80His Cys Asn Lys Ile Glu Leu Pro Thr
Thr Val Lys Ser Ser Pro Gly 85 90 95Leu Gly Pro Val Glu Ile Pro Pro
His Val Gln Lys Ser Val Asn Asn 100 105 110Asp Met Ile Val Thr Asp
Asn Asn Gly Ala Val Lys Phe Pro Gln Leu 115 120 125Cys Lys Phe Cys
Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser 130 135 140Cys Met
Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu145 150 155
160Val Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu
165 170 175Thr Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile
Leu Glu 180 185 190Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys
Lys Lys Pro Gly 195 200 205Glu Thr Phe Phe Met Cys Ser Cys Ser Ser
Asp Glu Cys Asn Asp Asn 210 215 220Ile Ile Phe Ser Glu Glu Tyr Asn
Thr Ser Asn Pro Asp225 230 23562262PRTArtificial
SequenceTbetaRI-TbetaRII-ED dimer T1-T2b 62Ala Ala Leu Leu Pro Gly
Ala Thr Ala Leu Gln Cys Phe Cys His Leu1 5 10 15Cys Thr Lys Asp Asn
Phe Thr Cys Val Thr Asp Gly Leu Cys Phe Val 20 25 30Ser Val Thr Glu
Thr Thr Asp Lys Val Ile His Asn Ser Met Cys Ile 35 40 45Ala Glu Ile
Asp Leu Ile Pro Arg Asp Arg Pro Phe Val Cys Ala Pro 50 55 60Ser Ser
Lys Thr Gly Ser Val Thr Thr Thr Tyr Cys Cys Asn Gln Asp65 70 75
80His Cys Asn Lys Ile Glu Leu Pro Thr Thr Val Lys Ser Ser Pro Gly
85 90 95Leu Gly Pro Val Glu Ile Pro Pro His Val Gln Lys Ser Asp Val
Glu 100 105 110Met Glu Ala Gln Lys Asp Glu Ile Ile Cys Pro Ser Cys
Asn Arg Thr 115 120 125Ala His Pro Leu Arg His Ile Asn Asn Asp Met
Ile Val Thr Asp Asn 130 135 140Asn Gly Ala Val Lys Phe Pro Gln Leu
Cys Lys Phe Cys Asp Val Arg145 150 155 160Phe Ser Thr Cys Asp Asn
Gln Lys Ser Cys Met Ser Asn Cys Ser Ile 165 170 175Thr Ser Ile Cys
Glu Lys Pro Gln Glu Val Cys Val Ala Val Trp Arg 180 185 190Lys Asn
Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys 195 200
205Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys
210 215 220Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met
Cys Ser225 230 235 240Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile
Phe Ser Glu Glu Tyr 245 250 255Asn Thr Ser Asn Pro Asp
26063237PRTArtificial SequenceTbetaRI-TbetaRII-ED dimer T2-T1 63Ile
Pro Pro His Val Gln Lys Ser Val Asn Asn Asp Met Ile Val Thr1 5 10
15Asp Asn Asn Gly Ala Val Lys Phe Pro Gln Leu Cys Lys Phe Cys Asp
20 25 30Val Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys Met Ser Asn
Cys 35 40 45Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys Val
Ala Val 50 55 60Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val
Cys His Asp65 70 75 80Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu
Asp Ala Ala Ser Pro 85 90 95Lys Cys Ile Met Lys Glu Lys Lys Lys Pro
Gly Glu Thr Phe Phe Met 100 105 110Cys Ser Cys Ser Ser Asp Glu Cys
Asn Asp Asn Ile Ile Phe Ser Glu 115 120 125Glu Tyr Asn Thr Ser Asn
Pro Asp Ala Ala Leu Leu Pro Gly Ala Thr 130 135 140Ala Leu Gln Cys
Phe Cys His Leu Cys Thr Lys Asp Asn Phe Thr Cys145 150 155 160Val
Thr Asp Gly Leu Cys Phe Val Ser Val Thr Glu Thr Thr Asp Lys 165 170
175Val Ile His Asn Ser Met Cys Ile Ala Glu Ile Asp Leu Ile Pro Arg
180 185 190Asp Arg Pro Phe Val Cys Ala Pro Ser Ser Lys Thr Gly Ser
Val Thr 195 200 205Thr Thr Tyr Cys Cys Asn Gln Asp His Cys Asn Lys
Ile Glu Leu Pro 210 215 220Thr Thr Val Lys Ser Ser Pro Gly Leu Gly
Pro Val Glu225 230 23564373PRTArtificial
SequenceTbetaRI-TbetaRII-ED trimer T1-T2-T2 64Ala Ala Leu Leu Pro
Gly Ala Thr Ala Leu Gln Cys Phe Cys His Leu1 5 10 15Cys Thr Lys Asp
Asn Phe Thr Cys Val Thr Asp Gly Leu Cys Phe Val 20 25 30Ser Val Thr
Glu Thr Thr Asp Lys Val Ile His Asn Ser Met Cys Ile 35 40 45Ala Glu
Ile Asp Leu Ile Pro Arg Asp Arg Pro Phe Val Cys Ala Pro 50 55 60Ser
Ser Lys Thr Gly Ser Val Thr Thr Thr Tyr Cys Cys Asn Gln Asp65 70 75
80His Cys Asn Lys Ile Glu Leu Pro Thr Thr Val Lys Ser Ser Pro Gly
85 90 95Leu Gly Pro Val Glu Ile Pro Pro His Val Gln Lys Ser Val Asn
Asn 100 105 110Asp Met Ile Val Thr Asp Asn Asn Gly Ala Val Lys Phe
Pro Gln Leu 115 120 125Cys Lys Phe Cys Asp Val Arg Phe Ser Thr Cys
Asp Asn Gln Lys Ser 130 135 140Cys Met Ser Asn Cys Ser Ile Thr Ser
Ile Cys Glu Lys Pro Gln Glu145 150 155 160Val Cys Val Ala Val Trp
Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu 165 170 175Thr Val Cys His
Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu 180 185 190Asp Ala
Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly 195 200
205Glu Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn
210 215 220Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp Ile
Pro Pro225 230 235 240His Val Gln Lys Ser Val Asn Asn Asp Met Ile
Val Thr Asp Asn Asn 245 250 255Gly Ala Val Lys Phe Pro Gln Leu Cys
Lys Phe Cys Asp Val Arg Phe 260 265 270Ser Thr Cys Asp Asn Gln Lys
Ser Cys Met Ser Asn Cys Ser Ile Thr 275 280 285Ser Ile Cys Glu Lys
Pro Gln Glu Val Cys Val Ala Val Trp Arg Lys 290 295 300Asn Asp Glu
Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys Leu305 310 315
320Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile
325 330 335Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met Cys
Ser Cys 340 345 350Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser
Glu Glu Tyr Asn 355 360 365Thr Ser Asn Pro Asp
37065398PRTArtificial SequenceTbetaRI-TbetaRII-ED trimer T1-T2-T2b
65Ala Ala Leu Leu Pro Gly Ala Thr Ala Leu Gln Cys Phe Cys His Leu1
5 10 15Cys Thr Lys Asp Asn Phe Thr Cys Val Thr Asp Gly Leu Cys Phe
Val 20 25 30Ser Val Thr Glu Thr Thr Asp Lys Val Ile His Asn Ser Met
Cys Ile 35 40 45Ala Glu Ile Asp Leu Ile Pro Arg Asp Arg Pro Phe Val
Cys Ala Pro 50 55 60Ser Ser Lys Thr Gly Ser Val Thr Thr Thr Tyr Cys
Cys Asn Gln Asp65 70 75 80His Cys Asn Lys Ile Glu Leu Pro Thr Thr
Val Lys Ser Ser Pro Gly 85 90 95Leu Gly Pro Val Glu Ile Pro Pro His
Val Gln Lys Ser Val Asn Asn 100 105 110Asp Met Ile Val Thr Asp Asn
Asn Gly Ala Val Lys Phe Pro Gln Leu 115 120 125Cys Lys Phe Cys Asp
Val Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser 130 135 140Cys Met Ser
Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu145 150 155
160Val Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu
165 170 175Thr Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile
Leu Glu 180 185 190Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys
Lys Lys Pro Gly 195 200 205Glu Thr Phe Phe Met Cys Ser Cys Ser Ser
Asp Glu Cys Asn Asp Asn 210 215 220Ile Ile Phe Ser Glu Glu Tyr Asn
Thr Ser Asn Pro Asp Ile Pro Pro225 230 235 240His Val Gln Lys Ser
Asp Val Glu Met Glu Ala Gln Lys Asp Glu Ile 245 250 255Ile Cys Pro
Ser Cys Asn Arg Thr Ala His Pro Leu Arg His Ile Asn 260 265 270Asn
Asp Met Ile Val Thr Asp Asn Asn Gly Ala Val Lys Phe Pro Gln 275 280
285Leu Cys Lys Phe Cys Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys
290 295 300Ser Cys Met
Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln305 310 315
320Glu Val Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu
325 330 335Glu Thr Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe
Ile Leu 340 345 350Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu
Lys Lys Lys Pro 355 360 365Gly Glu Thr Phe Phe Met Cys Ser Cys Ser
Ser Asp Glu Cys Asn Asp 370 375 380Asn Ile Ile Phe Ser Glu Glu Tyr
Asn Thr Ser Asn Pro Asp385 390 39566423PRTArtificial
SequenceTbetaRI-TbetaRII-ED trimer T1-T2-T2bAA 66Ala Ala Leu Leu
Pro Gly Ala Thr Ala Leu Gln Cys Phe Cys His Leu1 5 10 15Cys Thr Lys
Asp Asn Phe Thr Cys Val Thr Asp Gly Leu Cys Phe Val 20 25 30Ser Val
Thr Glu Thr Thr Asp Lys Val Ile His Asn Ser Met Cys Ile 35 40 45Ala
Glu Ile Asp Leu Ile Pro Arg Asp Arg Pro Phe Val Cys Ala Pro 50 55
60Ser Ser Lys Thr Gly Ser Val Thr Thr Thr Tyr Cys Cys Asn Gln Asp65
70 75 80His Cys Asn Lys Ile Glu Leu Pro Thr Thr Val Lys Ser Ser Pro
Gly 85 90 95Leu Gly Pro Val Glu Ile Pro Pro His Val Gln Lys Ser Asp
Val Glu 100 105 110Met Glu Ala Gln Lys Asp Glu Ile Ile Cys Pro Ser
Cys Asn Arg Thr 115 120 125Ala His Pro Leu Arg His Ile Asn Asn Asp
Met Ile Val Thr Asp Asn 130 135 140Asn Gly Ala Val Lys Phe Pro Gln
Leu Cys Lys Phe Cys Asp Val Arg145 150 155 160Phe Ser Thr Cys Asp
Asn Gln Lys Ser Cys Met Ser Asn Cys Ser Ile 165 170 175Thr Ser Ile
Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val Trp Arg 180 185 190Lys
Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys 195 200
205Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys
210 215 220Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met
Cys Ser225 230 235 240Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile
Phe Ser Glu Glu Tyr 245 250 255Asn Thr Ser Asn Pro Asp Ile Pro Pro
His Val Gln Lys Ser Asp Val 260 265 270Glu Met Glu Ala Gln Lys Asp
Glu Ile Ile Ala Pro Ser Ala Asn Arg 275 280 285Thr Ala His Pro Leu
Arg His Ile Asn Asn Asp Met Ile Val Thr Asp 290 295 300Asn Asn Gly
Ala Val Lys Phe Pro Gln Leu Cys Lys Phe Cys Asp Val305 310 315
320Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys Met Ser Asn Cys Ser
325 330 335Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala
Val Trp 340 345 350Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val
Cys His Asp Pro 355 360 365Lys Leu Pro Tyr His Asp Phe Ile Leu Glu
Asp Ala Ala Ser Pro Lys 370 375 380Cys Ile Met Lys Glu Lys Lys Lys
Pro Gly Glu Thr Phe Phe Met Cys385 390 395 400Ser Cys Ser Ser Asp
Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu 405 410 415Tyr Asn Thr
Ser Asn Pro Asp 42067373PRTArtificial SequenceTbetaRI-TbetaRII-ED
trimer T2-T2-T1 67Ile Pro Pro His Val Gln Lys Ser Val Asn Asn Asp
Met Ile Val Thr1 5 10 15Asp Asn Asn Gly Ala Val Lys Phe Pro Gln Leu
Cys Lys Phe Cys Asp 20 25 30Val Arg Phe Ser Thr Cys Asp Asn Gln Lys
Ser Cys Met Ser Asn Cys 35 40 45Ser Ile Thr Ser Ile Cys Glu Lys Pro
Gln Glu Val Cys Val Ala Val 50 55 60Trp Arg Lys Asn Asp Glu Asn Ile
Thr Leu Glu Thr Val Cys His Asp65 70 75 80Pro Lys Leu Pro Tyr His
Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro 85 90 95Lys Cys Ile Met Lys
Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met 100 105 110Cys Ser Cys
Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu 115 120 125Glu
Tyr Asn Thr Ser Asn Pro Asp Ile Pro Pro His Val Gln Lys Ser 130 135
140Val Asn Asn Asp Met Ile Val Thr Asp Asn Asn Gly Ala Val Lys
Phe145 150 155 160Pro Gln Leu Cys Lys Phe Cys Asp Val Arg Phe Ser
Thr Cys Asp Asn 165 170 175Gln Lys Ser Cys Met Ser Asn Cys Ser Ile
Thr Ser Ile Cys Glu Lys 180 185 190Pro Gln Glu Val Cys Val Ala Val
Trp Arg Lys Asn Asp Glu Asn Ile 195 200 205Thr Leu Glu Thr Val Cys
His Asp Pro Lys Leu Pro Tyr His Asp Phe 210 215 220Ile Leu Glu Asp
Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys225 230 235 240Lys
Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu Cys 245 250
255Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp
260 265 270Ala Ala Leu Leu Pro Gly Ala Thr Ala Leu Gln Cys Phe Cys
His Leu 275 280 285Cys Thr Lys Asp Asn Phe Thr Cys Val Thr Asp Gly
Leu Cys Phe Val 290 295 300Ser Val Thr Glu Thr Thr Asp Lys Val Ile
His Asn Ser Met Cys Ile305 310 315 320Ala Glu Ile Asp Leu Ile Pro
Arg Asp Arg Pro Phe Val Cys Ala Pro 325 330 335Ser Ser Lys Thr Gly
Ser Val Thr Thr Thr Tyr Cys Cys Asn Gln Asp 340 345 350His Cys Asn
Lys Ile Glu Leu Pro Thr Thr Val Lys Ser Ser Pro Gly 355 360 365Leu
Gly Pro Val Glu 37068398PRTArtificial SequenceTbetaRI-TbetaRII-ED
trimer T2-T2bAA-T1 68Ile Pro Pro His Val Gln Lys Ser Val Asn Asn
Asp Met Ile Val Thr1 5 10 15Asp Asn Asn Gly Ala Val Lys Phe Pro Gln
Leu Cys Lys Phe Cys Asp 20 25 30Val Arg Phe Ser Thr Cys Asp Asn Gln
Lys Ser Cys Met Ser Asn Cys 35 40 45Ser Ile Thr Ser Ile Cys Glu Lys
Pro Gln Glu Val Cys Val Ala Val 50 55 60Trp Arg Lys Asn Asp Glu Asn
Ile Thr Leu Glu Thr Val Cys His Asp65 70 75 80Pro Lys Leu Pro Tyr
His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro 85 90 95Lys Cys Ile Met
Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met 100 105 110Cys Ser
Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu 115 120
125Glu Tyr Asn Thr Ser Asn Pro Asp Ile Pro Pro His Val Gln Lys Ser
130 135 140Asp Val Glu Met Glu Ala Gln Lys Asp Glu Ile Ile Ala Pro
Ser Ala145 150 155 160Asn Arg Thr Ala His Pro Leu Arg His Ile Asn
Asn Asp Met Ile Val 165 170 175Thr Asp Asn Asn Gly Ala Val Lys Phe
Pro Gln Leu Cys Lys Phe Cys 180 185 190Asp Val Arg Phe Ser Thr Cys
Asp Asn Gln Lys Ser Cys Met Ser Asn 195 200 205Cys Ser Ile Thr Ser
Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala 210 215 220Val Trp Arg
Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His225 230 235
240Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser
245 250 255Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr
Phe Phe 260 265 270Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn
Ile Ile Phe Ser 275 280 285Glu Glu Tyr Asn Thr Ser Asn Pro Asp Ala
Ala Leu Leu Pro Gly Ala 290 295 300Thr Ala Leu Gln Cys Phe Cys His
Leu Cys Thr Lys Asp Asn Phe Thr305 310 315 320Cys Val Thr Asp Gly
Leu Cys Phe Val Ser Val Thr Glu Thr Thr Asp 325 330 335Lys Val Ile
His Asn Ser Met Cys Ile Ala Glu Ile Asp Leu Ile Pro 340 345 350Arg
Asp Arg Pro Phe Val Cys Ala Pro Ser Ser Lys Thr Gly Ser Val 355 360
365Thr Thr Thr Tyr Cys Cys Asn Gln Asp His Cys Asn Lys Ile Glu Leu
370 375 380Pro Thr Thr Val Lys Ser Ser Pro Gly Leu Gly Pro Val
Glu385 390 3956985PRTArtificial SequenceBMPRIa-ED structured domain
69Thr Leu Pro Phe Leu Lys Cys Tyr Cys Ser Gly His Cys Pro Asp Asp1
5 10 15Ala Ile Asn Asn Thr Cys Ile Thr Asn Gly His Cys Phe Ala Ile
Ile 20 25 30Glu Glu Asp Asp Gln Gly Glu Thr Thr Leu Ala Ser Gly Cys
Met Lys 35 40 45Tyr Glu Gly Ser Asp Phe Gln Cys Lys Asp Ser Pro Lys
Ala Gln Leu 50 55 60Arg Arg Thr Ile Glu Cys Cys Arg Thr Asn Leu Cys
Asn Gln Tyr Leu65 70 75 80Gln Pro Thr Leu Pro 857031PRTArtificial
SequenceBMPRIa-ED N-term unstructured region and natural linker
70Gln Asn Leu Asp Ser Met Leu His Gly Thr Gly Met Lys Ser Asp Ser1
5 10 15Asp Gln Lys Lys Ser Glu Asn Gly Val Thr Leu Ala Pro Glu Asp
20 25 307113PRTArtificial SequenceBMPRIa-ED C-term unstructured
region and natural linker 71Pro Val Val Ile Gly Pro Phe Phe Asp Gly
Ser Ile Arg1 5 107244PRTArtificial SequenceBMPRIa-ED natural linker
72Pro Val Val Ile Gly Pro Phe Phe Asp Gly Ser Ile Arg Gln Asn Leu1
5 10 15Asp Ser Met Leu His Gly Thr Gly Met Lys Ser Asp Ser Asp Gln
Lys 20 25 30Lys Ser Glu Asn Gly Val Thr Leu Ala Pro Glu Asp 35
4073129PRTArtificial SequenceBMPRIa-ED monomer 73Gln Asn Leu Asp
Ser Met Leu His Gly Thr Gly Met Lys Ser Asp Ser1 5 10 15Asp Gln Lys
Lys Ser Glu Asn Gly Val Thr Leu Ala Pro Glu Asp Thr 20 25 30Leu Pro
Phe Leu Lys Cys Tyr Cys Ser Gly His Cys Pro Asp Asp Ala 35 40 45Ile
Asn Asn Thr Cys Ile Thr Asn Gly His Cys Phe Ala Ile Ile Glu 50 55
60Glu Asp Asp Gln Gly Glu Thr Thr Leu Ala Ser Gly Cys Met Lys Tyr65
70 75 80Glu Gly Ser Asp Phe Gln Cys Lys Asp Ser Pro Lys Ala Gln Leu
Arg 85 90 95Arg Thr Ile Glu Cys Cys Arg Thr Asn Leu Cys Asn Gln Tyr
Leu Gln 100 105 110Pro Thr Leu Pro Pro Val Val Ile Gly Pro Phe Phe
Asp Gly Ser Ile 115 120 125Arg74258PRTArtificial SequenceBMPRIa-ED
dimer 74Gln Asn Leu Asp Ser Met Leu His Gly Thr Gly Met Lys Ser Asp
Ser1 5 10 15Asp Gln Lys Lys Ser Glu Asn Gly Val Thr Leu Ala Pro Glu
Asp Thr 20 25 30Leu Pro Phe Leu Lys Cys Tyr Cys Ser Gly His Cys Pro
Asp Asp Ala 35 40 45Ile Asn Asn Thr Cys Ile Thr Asn Gly His Cys Phe
Ala Ile Ile Glu 50 55 60Glu Asp Asp Gln Gly Glu Thr Thr Leu Ala Ser
Gly Cys Met Lys Tyr65 70 75 80Glu Gly Ser Asp Phe Gln Cys Lys Asp
Ser Pro Lys Ala Gln Leu Arg 85 90 95Arg Thr Ile Glu Cys Cys Arg Thr
Asn Leu Cys Asn Gln Tyr Leu Gln 100 105 110Pro Thr Leu Pro Pro Val
Val Ile Gly Pro Phe Phe Asp Gly Ser Ile 115 120 125Arg Gln Asn Leu
Asp Ser Met Leu His Gly Thr Gly Met Lys Ser Asp 130 135 140Ser Asp
Gln Lys Lys Ser Glu Asn Gly Val Thr Leu Ala Pro Glu Asp145 150 155
160Thr Leu Pro Phe Leu Lys Cys Tyr Cys Ser Gly His Cys Pro Asp Asp
165 170 175Ala Ile Asn Asn Thr Cys Ile Thr Asn Gly His Cys Phe Ala
Ile Ile 180 185 190Glu Glu Asp Asp Gln Gly Glu Thr Thr Leu Ala Ser
Gly Cys Met Lys 195 200 205Tyr Glu Gly Ser Asp Phe Gln Cys Lys Asp
Ser Pro Lys Ala Gln Leu 210 215 220Arg Arg Thr Ile Glu Cys Cys Arg
Thr Asn Leu Cys Asn Gln Tyr Leu225 230 235 240Gln Pro Thr Leu Pro
Pro Val Val Ile Gly Pro Phe Phe Asp Gly Ser 245 250 255Ile
Arg7592PRTArtificial SequenceActRIIa-ED structured domain 75Thr Gln
Glu Cys Leu Phe Phe Asn Ala Asn Trp Glu Lys Asp Arg Thr1 5 10 15Asn
Gln Thr Gly Val Glu Pro Cys Tyr Gly Asp Lys Asp Lys Arg Arg 20 25
30His Cys Phe Ala Thr Trp Lys Asn Ile Ser Gly Ser Ile Glu Ile Val
35 40 45Lys Gln Gly Cys Trp Leu Asp Asp Ile Asn Cys Tyr Asp Arg Thr
Asp 50 55 60Cys Val Glu Lys Lys Asp Ser Pro Glu Val Tyr Phe Cys Cys
Cys Glu65 70 75 80Gly Asn Met Cys Asn Glu Lys Phe Ser Tyr Phe Pro
85 90767PRTArtificial SequenceActRIIa-ED N-term unstructured region
and natural linker 76Ala Ile Leu Gly Arg Ser Glu1
57721PRTArtificial SequenceActRIIa-ED C-term unstructured region
and natural linker 77Glu Met Glu Val Thr Gln Pro Thr Ser Asn Pro
Val Thr Pro Lys Pro1 5 10 15Pro Tyr Tyr Asn Ile 207828PRTArtificial
SequenceActRIIa-ED natural linker 78Glu Met Glu Val Thr Gln Pro Thr
Ser Asn Pro Val Thr Pro Lys Pro1 5 10 15Pro Tyr Tyr Asn Ile Ala Ile
Leu Gly Arg Ser Glu 20 2579120PRTArtificial SequenceActRIIa-ED
monomer 79Ala Ile Leu Gly Arg Ser Glu Thr Gln Glu Cys Leu Phe Phe
Asn Ala1 5 10 15Asn Trp Glu Lys Asp Arg Thr Asn Gln Thr Gly Val Glu
Pro Cys Tyr 20 25 30Gly Asp Lys Asp Lys Arg Arg His Cys Phe Ala Thr
Trp Lys Asn Ile 35 40 45Ser Gly Ser Ile Glu Ile Val Lys Gln Gly Cys
Trp Leu Asp Asp Ile 50 55 60Asn Cys Tyr Asp Arg Thr Asp Cys Val Glu
Lys Lys Asp Ser Pro Glu65 70 75 80Val Tyr Phe Cys Cys Cys Glu Gly
Asn Met Cys Asn Glu Lys Phe Ser 85 90 95Tyr Phe Pro Glu Met Glu Val
Thr Gln Pro Thr Ser Asn Pro Val Thr 100 105 110Pro Lys Pro Pro Tyr
Tyr Asn Ile 115 12080240PRTArtificial SequenceActRIIa-ED dimer
80Ala Ile Leu Gly Arg Ser Glu Thr Gln Glu Cys Leu Phe Phe Asn Ala1
5 10 15Asn Trp Glu Lys Asp Arg Thr Asn Gln Thr Gly Val Glu Pro Cys
Tyr 20 25 30Gly Asp Lys Asp Lys Arg Arg His Cys Phe Ala Thr Trp Lys
Asn Ile 35 40 45Ser Gly Ser Ile Glu Ile Val Lys Gln Gly Cys Trp Leu
Asp Asp Ile 50 55 60Asn Cys Tyr Asp Arg Thr Asp Cys Val Glu Lys Lys
Asp Ser Pro Glu65 70 75 80Val Tyr Phe Cys Cys Cys Glu Gly Asn Met
Cys Asn Glu Lys Phe Ser 85 90 95Tyr Phe Pro Glu Met Glu Val Thr Gln
Pro Thr Ser Asn Pro Val Thr 100 105 110Pro Lys Pro Pro Tyr Tyr Asn
Ile Ala Ile Leu Gly Arg Ser Glu Thr 115 120 125Gln Glu Cys Leu Phe
Phe Asn Ala Asn Trp Glu Lys Asp Arg Thr Asn 130 135 140Gln Thr Gly
Val Glu Pro Cys Tyr Gly Asp Lys Asp Lys Arg Arg His145 150 155
160Cys Phe Ala Thr Trp Lys Asn Ile Ser Gly Ser Ile Glu Ile Val Lys
165 170 175Gln Gly Cys Trp Leu Asp Asp Ile Asn Cys Tyr Asp Arg Thr
Asp Cys 180 185 190Val Glu Lys Lys Asp Ser Pro Glu Val Tyr Phe Cys
Cys Cys Glu Gly 195 200 205Asn Met Cys Asn Glu Lys Phe Ser Tyr Phe
Pro Glu Met Glu Val Thr 210 215 220Gln Pro Thr Ser Asn Pro Val Thr
Pro Lys Pro Pro
Tyr Tyr Asn Ile225 230 235 2408191PRTArtificial SequenceActRIIb-ED
structured domain 81Arg Glu Cys Ile Tyr Tyr Asn Ala Asn Trp Glu Leu
Glu Arg Thr Asn1 5 10 15Gln Ser Gly Leu Glu Arg Cys Glu Gly Glu Gln
Asp Lys Arg Leu His 20 25 30Cys Tyr Ala Ser Trp Arg Asn Ser Ser Gly
Thr Ile Glu Leu Val Lys 35 40 45Lys Gly Cys Trp Leu Asp Asp Phe Asn
Cys Tyr Asp Arg Gln Glu Cys 50 55 60Val Ala Thr Glu Glu Asn Pro Gln
Val Tyr Phe Cys Cys Cys Glu Gly65 70 75 80Asn Phe Cys Asn Glu Arg
Phe Thr His Leu Pro 85 90828PRTArtificial SequenceActRIIb-ED N-term
unstructured region and natural linker 82Ser Gly Arg Gly Glu Ala
Glu Thr1 58317PRTArtificial SequenceActRIIb-ED C-term unstructured
region and natural linker 83Glu Ala Gly Gly Pro Glu Val Thr Tyr Glu
Pro Pro Pro Thr Ala Pro1 5 10 15Thr8425PRTArtificial
SequenceActRIIb-ED natural linker 84Glu Ala Gly Gly Pro Glu Val Thr
Tyr Glu Pro Pro Pro Thr Ala Pro1 5 10 15Thr Ser Gly Arg Gly Glu Ala
Glu Thr 20 2585116PRTArtificial SequenceActRIIb-ED monomer 85Ser
Gly Arg Gly Glu Ala Glu Thr Arg Glu Cys Ile Tyr Tyr Asn Ala1 5 10
15Asn Trp Glu Leu Glu Arg Thr Asn Gln Ser Gly Leu Glu Arg Cys Glu
20 25 30Gly Glu Gln Asp Lys Arg Leu His Cys Tyr Ala Ser Trp Arg Asn
Ser 35 40 45Ser Gly Thr Ile Glu Leu Val Lys Lys Gly Cys Trp Leu Asp
Asp Phe 50 55 60Asn Cys Tyr Asp Arg Gln Glu Cys Val Ala Thr Glu Glu
Asn Pro Gln65 70 75 80Val Tyr Phe Cys Cys Cys Glu Gly Asn Phe Cys
Asn Glu Arg Phe Thr 85 90 95His Leu Pro Glu Ala Gly Gly Pro Glu Val
Thr Tyr Glu Pro Pro Pro 100 105 110Thr Ala Pro Thr
11586232PRTArtificial SequenceActRIIb-ED dimer 86Ser Gly Arg Gly
Glu Ala Glu Thr Arg Glu Cys Ile Tyr Tyr Asn Ala1 5 10 15Asn Trp Glu
Leu Glu Arg Thr Asn Gln Ser Gly Leu Glu Arg Cys Glu 20 25 30Gly Glu
Gln Asp Lys Arg Leu His Cys Tyr Ala Ser Trp Arg Asn Ser 35 40 45Ser
Gly Thr Ile Glu Leu Val Lys Lys Gly Cys Trp Leu Asp Asp Phe 50 55
60Asn Cys Tyr Asp Arg Gln Glu Cys Val Ala Thr Glu Glu Asn Pro Gln65
70 75 80Val Tyr Phe Cys Cys Cys Glu Gly Asn Phe Cys Asn Glu Arg Phe
Thr 85 90 95His Leu Pro Glu Ala Gly Gly Pro Glu Val Thr Tyr Glu Pro
Pro Pro 100 105 110Thr Ala Pro Thr Ser Gly Arg Gly Glu Ala Glu Thr
Arg Glu Cys Ile 115 120 125Tyr Tyr Asn Ala Asn Trp Glu Leu Glu Arg
Thr Asn Gln Ser Gly Leu 130 135 140Glu Arg Cys Glu Gly Glu Gln Asp
Lys Arg Leu His Cys Tyr Ala Ser145 150 155 160Trp Arg Asn Ser Ser
Gly Thr Ile Glu Leu Val Lys Lys Gly Cys Trp 165 170 175Leu Asp Asp
Phe Asn Cys Tyr Asp Arg Gln Glu Cys Val Ala Thr Glu 180 185 190Glu
Asn Pro Gln Val Tyr Phe Cys Cys Cys Glu Gly Asn Phe Cys Asn 195 200
205Glu Arg Phe Thr His Leu Pro Glu Ala Gly Gly Pro Glu Val Thr Tyr
210 215 220Glu Pro Pro Pro Thr Ala Pro Thr225 2308752PRTArtificial
SequenceActRIIa-BMPRIa-ED natural linker 87Glu Met Glu Val Thr Gln
Pro Thr Ser Asn Pro Val Thr Pro Lys Pro1 5 10 15Pro Tyr Tyr Asn Ile
Gln Asn Leu Asp Ser Met Leu His Gly Thr Gly 20 25 30Met Lys Ser Asp
Ser Asp Gln Lys Lys Ser Glu Asn Gly Val Thr Leu 35 40 45Ala Pro Glu
Asp 5088249PRTArtificial SequenceActRIIa-BMPRIa-ED dimer 88Ala Ile
Leu Gly Arg Ser Glu Thr Gln Glu Cys Leu Phe Phe Asn Ala1 5 10 15Asn
Trp Glu Lys Asp Arg Thr Asn Gln Thr Gly Val Glu Pro Cys Tyr 20 25
30Gly Asp Lys Asp Lys Arg Arg His Cys Phe Ala Thr Trp Lys Asn Ile
35 40 45Ser Gly Ser Ile Glu Ile Val Lys Gln Gly Cys Trp Leu Asp Asp
Ile 50 55 60Asn Cys Tyr Asp Arg Thr Asp Cys Val Glu Lys Lys Asp Ser
Pro Glu65 70 75 80Val Tyr Phe Cys Cys Cys Glu Gly Asn Met Cys Asn
Glu Lys Phe Ser 85 90 95Tyr Phe Pro Glu Met Glu Val Thr Gln Pro Thr
Ser Asn Pro Val Thr 100 105 110Pro Lys Pro Pro Tyr Tyr Asn Ile Gln
Asn Leu Asp Ser Met Leu His 115 120 125Gly Thr Gly Met Lys Ser Asp
Ser Asp Gln Lys Lys Ser Glu Asn Gly 130 135 140Val Thr Leu Ala Pro
Glu Asp Thr Leu Pro Phe Leu Lys Cys Tyr Cys145 150 155 160Ser Gly
His Cys Pro Asp Asp Ala Ile Asn Asn Thr Cys Ile Thr Asn 165 170
175Gly His Cys Phe Ala Ile Ile Glu Glu Asp Asp Gln Gly Glu Thr Thr
180 185 190Leu Ala Ser Gly Cys Met Lys Tyr Glu Gly Ser Asp Phe Gln
Cys Lys 195 200 205Asp Ser Pro Lys Ala Gln Leu Arg Arg Thr Ile Glu
Cys Cys Arg Thr 210 215 220Asn Leu Cys Asn Gln Tyr Leu Gln Pro Thr
Leu Pro Pro Val Val Ile225 230 235 240Gly Pro Phe Phe Asp Gly Ser
Ile Arg 2458919PRTArtificial SequenceSignal peptide 89Met Asp Trp
Thr Trp Arg Ile Leu Phe Leu Val Ala Ala Ala Thr Gly1 5 10 15Thr His
Ala9020PRTArtificial SequenceSignal peptide 90Met Val Leu Gln Thr
Gln Val Phe Ile Ser Leu Leu Leu Trp Ile Ser1 5 10 15Gly Ala Tyr Gly
2091352PRTArtificial SequencehIgG1FcdeltaK-deltaC-T2m fusion 91Ala
Pro Glu Leu Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys1 5 10
15Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val
20 25 30Val Val Asp Val Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp
Tyr 35 40 45Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg
Glu Glu 50 55 60Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr
Val Leu His65 70 75 80Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys
Lys Val Ser Asn Lys 85 90 95Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile
Ser Lys Ala Lys Gly Gln 100 105 110Pro Arg Glu Pro Gln Val Tyr Thr
Leu Pro Pro Ser Arg Asp Glu Leu 115 120 125Thr Lys Asn Gln Val Ser
Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro 130 135 140Ser Asp Ile Ala
Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn145 150 155 160Tyr
Lys Thr Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu 165 170
175Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val
180 185 190Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn His Tyr
Thr Gln 195 200 205Lys Ser Leu Ser Leu Ser Pro Gly Ile Pro Pro His
Val Gln Lys Ser 210 215 220Val Asn Asn Asp Met Ile Val Thr Asp Asn
Asn Gly Ala Val Lys Phe225 230 235 240Pro Gln Leu Cys Lys Phe Cys
Asp Val Arg Phe Ser Thr Cys Asp Asn 245 250 255Gln Lys Ser Cys Met
Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys 260 265 270Pro Gln Glu
Val Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile 275 280 285Thr
Leu Glu Thr Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe 290 295
300Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys
Lys305 310 315 320Lys Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser
Ser Asp Glu Cys 325 330 335Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr
Asn Thr Ser Asn Pro Asp 340 345 35092356PRTArtificial
SequencehIgG1FcdeltaK-C-T2m fusion 92Pro Pro Cys Pro Ala Pro Glu
Leu Leu Gly Gly Pro Ser Val Phe Leu1 5 10 15Phe Pro Pro Lys Pro Lys
Asp Thr Leu Met Ile Ser Arg Thr Pro Glu 20 25 30Val Thr Cys Val Val
Val Asp Val Ser His Glu Asp Pro Glu Val Lys 35 40 45Phe Asn Trp Tyr
Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys 50 55 60Pro Arg Glu
Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu65 70 75 80Thr
Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys 85 90
95Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys
100 105 110Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro
Pro Ser 115 120 125Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr
Cys Leu Val Lys 130 135 140Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu
Trp Glu Ser Asn Gly Gln145 150 155 160Pro Glu Asn Asn Tyr Lys Thr
Thr Pro Pro Val Leu Asp Ser Asp Gly 165 170 175Ser Phe Phe Leu Tyr
Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln 180 185 190Gln Gly Asn
Val Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn 195 200 205His
Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Ile Pro Pro His 210 215
220Val Gln Lys Ser Val Asn Asn Asp Met Ile Val Thr Asp Asn Asn
Gly225 230 235 240Ala Val Lys Phe Pro Gln Leu Cys Lys Phe Cys Asp
Val Arg Phe Ser 245 250 255Thr Cys Asp Asn Gln Lys Ser Cys Met Ser
Asn Cys Ser Ile Thr Ser 260 265 270Ile Cys Glu Lys Pro Gln Glu Val
Cys Val Ala Val Trp Arg Lys Asn 275 280 285Asp Glu Asn Ile Thr Leu
Glu Thr Val Cys His Asp Pro Lys Leu Pro 290 295 300Tyr His Asp Phe
Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met305 310 315 320Lys
Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser 325 330
335Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr
340 345 350Ser Asn Pro Asp 35593362PRTArtificial
SequencehIgG1FcdeltaK-CC-T2m fusion 93Asp Lys Thr His Thr Cys Pro
Pro Cys Pro Ala Pro Glu Leu Leu Gly1 5 10 15Gly Pro Ser Val Phe Leu
Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 20 25 30Ile Ser Arg Thr Pro
Glu Val Thr Cys Val Val Val Asp Val Ser His 35 40 45Glu Asp Pro Glu
Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val 50 55 60His Asn Ala
Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr65 70 75 80Arg
Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly 85 90
95Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile
100 105 110Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro
Gln Val 115 120 125Tyr Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys
Asn Gln Val Ser 130 135 140Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro
Ser Asp Ile Ala Val Glu145 150 155 160Trp Glu Ser Asn Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro 165 170 175Val Leu Asp Ser Asp
Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val 180 185 190Asp Lys Ser
Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met 195 200 205His
Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser 210 215
220Pro Gly Ile Pro Pro His Val Gln Lys Ser Val Asn Asn Asp Met
Ile225 230 235 240Val Thr Asp Asn Asn Gly Ala Val Lys Phe Pro Gln
Leu Cys Lys Phe 245 250 255Cys Asp Val Arg Phe Ser Thr Cys Asp Asn
Gln Lys Ser Cys Met Ser 260 265 270Asn Cys Ser Ile Thr Ser Ile Cys
Glu Lys Pro Gln Glu Val Cys Val 275 280 285Ala Val Trp Arg Lys Asn
Asp Glu Asn Ile Thr Leu Glu Thr Val Cys 290 295 300His Asp Pro Lys
Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala305 310 315 320Ser
Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe 325 330
335Phe Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe
340 345 350Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp 355
36094351PRTArtificial SequencehIgG2FcdeltaK-deltaC-T2m fusion 94Ala
Pro Pro Val Ala Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro1 5 10
15Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val
20 25 30Val Asp Val Ser His Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr
Val 35 40 45Asp Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu
Glu Gln 50 55 60Phe Asn Ser Thr Phe Arg Val Val Ser Val Leu Thr Val
Val His Gln65 70 75 80Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys
Val Ser Asn Lys Gly 85 90 95Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser
Lys Thr Lys Gly Gln Pro 100 105 110Arg Glu Pro Gln Val Tyr Thr Leu
Pro Pro Ser Arg Glu Glu Met Thr 115 120 125Lys Asn Gln Val Ser Leu
Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser 130 135 140Asp Ile Ser Val
Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr145 150 155 160Lys
Thr Thr Pro Pro Met Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr 165 170
175Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe
180 185 190Ser Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr
Gln Lys 195 200 205Ser Leu Ser Leu Ser Pro Gly Ile Pro Pro His Val
Gln Lys Ser Val 210 215 220Asn Asn Asp Met Ile Val Thr Asp Asn Asn
Gly Ala Val Lys Phe Pro225 230 235 240Gln Leu Cys Lys Phe Cys Asp
Val Arg Phe Ser Thr Cys Asp Asn Gln 245 250 255Lys Ser Cys Met Ser
Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro 260 265 270Gln Glu Val
Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr 275 280 285Leu
Glu Thr Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile 290 295
300Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys
Lys305 310 315 320Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser Ser
Asp Glu Cys Asn 325 330 335Asp Asn Ile Ile Phe Ser Glu Glu Tyr Asn
Thr Ser Asn Pro Asp 340 345 35095355PRTArtificial
SequencehIgG2FcdeltaK-C-T2m fusion 95Pro Pro Cys Pro Ala Pro Pro
Val Ala Gly Pro Ser Val Phe Leu Phe1 5 10 15Pro Pro Lys Pro Lys Asp
Thr Leu Met Ile Ser Arg Thr Pro Glu Val 20 25 30Thr Cys Val Val Val
Asp Val Ser His Glu Asp Pro Glu Val Gln Phe 35 40 45Asn Trp Tyr Val
Asp Gly Val Glu Val His Asn Ala Lys Thr Lys Pro 50 55 60Arg Glu Glu
Gln Phe Asn Ser Thr Phe Arg Val Val Ser Val Leu Thr65 70 75 80Val
Val His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val 85
90
95Ser Asn Lys Gly Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Thr
100 105 110Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro
Ser Arg 115 120 125Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys
Leu Val Lys Gly 130 135 140Phe Tyr Pro Ser Asp Ile Ser Val Glu Trp
Glu Ser Asn Gly Gln Pro145 150 155 160Glu Asn Asn Tyr Lys Thr Thr
Pro Pro Met Leu Asp Ser Asp Gly Ser 165 170 175Phe Phe Leu Tyr Ser
Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln 180 185 190Gly Asn Val
Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn His 195 200 205Tyr
Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Ile Pro Pro His Val 210 215
220Gln Lys Ser Val Asn Asn Asp Met Ile Val Thr Asp Asn Asn Gly
Ala225 230 235 240Val Lys Phe Pro Gln Leu Cys Lys Phe Cys Asp Val
Arg Phe Ser Thr 245 250 255Cys Asp Asn Gln Lys Ser Cys Met Ser Asn
Cys Ser Ile Thr Ser Ile 260 265 270Cys Glu Lys Pro Gln Glu Val Cys
Val Ala Val Trp Arg Lys Asn Asp 275 280 285Glu Asn Ile Thr Leu Glu
Thr Val Cys His Asp Pro Lys Leu Pro Tyr 290 295 300His Asp Phe Ile
Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys305 310 315 320Glu
Lys Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser Ser 325 330
335Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser
340 345 350Asn Pro Asp 35596351PRTArtificial
SequencehIgG2FcdeltaK-CC-T2m fusion 96Ala Pro Pro Val Ala Gly Pro
Ser Val Phe Leu Phe Pro Pro Lys Pro1 5 10 15Lys Asp Thr Leu Met Ile
Ser Arg Thr Pro Glu Val Thr Cys Val Val 20 25 30Val Asp Val Ser His
Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr Val 35 40 45Asp Gly Val Glu
Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln 50 55 60Phe Asn Ser
Thr Phe Arg Val Val Ser Val Leu Thr Val Val His Gln65 70 75 80Asp
Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Gly 85 90
95Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Thr Lys Gly Gln Pro
100 105 110Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu
Met Thr 115 120 125Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly
Phe Tyr Pro Ser 130 135 140Asp Ile Ser Val Glu Trp Glu Ser Asn Gly
Gln Pro Glu Asn Asn Tyr145 150 155 160Lys Thr Thr Pro Pro Met Leu
Asp Ser Asp Gly Ser Phe Phe Leu Tyr 165 170 175Ser Lys Leu Thr Val
Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe 180 185 190Ser Cys Ser
Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys 195 200 205Ser
Leu Ser Leu Ser Pro Gly Ile Pro Pro His Val Gln Lys Ser Val 210 215
220Asn Asn Asp Met Ile Val Thr Asp Asn Asn Gly Ala Val Lys Phe
Pro225 230 235 240Gln Leu Cys Lys Phe Cys Asp Val Arg Phe Ser Thr
Cys Asp Asn Gln 245 250 255Lys Ser Cys Met Ser Asn Cys Ser Ile Thr
Ser Ile Cys Glu Lys Pro 260 265 270Gln Glu Val Cys Val Ala Val Trp
Arg Lys Asn Asp Glu Asn Ile Thr 275 280 285Leu Glu Thr Val Cys His
Asp Pro Lys Leu Pro Tyr His Asp Phe Ile 290 295 300Leu Glu Asp Ala
Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys305 310 315 320Pro
Gly Glu Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu Cys Asn 325 330
335Asp Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp 340
345 35097383PRTArtificial SequencehIgG2Fc-CCCC-T2 fusion, also
termed Fc-T2m 97Met Asp Trp Thr Trp Arg Ile Leu Phe Leu Val Ala Ala
Ala Thr Gly1 5 10 15Thr His Ala Glu Arg Lys Cys Cys Val Glu Cys Pro
Pro Cys Pro Ala 20 25 30Pro Pro Val Ala Gly Pro Ser Val Phe Leu Phe
Pro Pro Lys Pro Lys 35 40 45Asp Thr Leu Met Ile Ser Arg Thr Pro Glu
Val Thr Cys Val Val Val 50 55 60Asp Val Ser His Glu Asp Pro Glu Val
Gln Phe Asn Trp Tyr Val Asp65 70 75 80Gly Val Glu Val His Asn Ala
Lys Thr Lys Pro Arg Glu Glu Gln Phe 85 90 95Asn Ser Thr Phe Arg Val
Val Ser Val Leu Thr Val Val His Gln Asp 100 105 110Trp Leu Asn Gly
Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Gly Leu 115 120 125Pro Ala
Pro Ile Glu Lys Thr Ile Ser Lys Thr Lys Gly Gln Pro Arg 130 135
140Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu Met Thr
Lys145 150 155 160Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly Phe
Tyr Pro Ser Asp 165 170 175Ile Ser Val Glu Trp Glu Ser Asn Gly Gln
Pro Glu Asn Asn Tyr Lys 180 185 190Thr Thr Pro Pro Met Leu Asp Ser
Asp Gly Ser Phe Phe Leu Tyr Ser 195 200 205Lys Leu Thr Val Asp Lys
Ser Arg Trp Gln Gln Gly Asn Val Phe Ser 210 215 220Cys Ser Val Met
His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser225 230 235 240Leu
Ser Leu Ser Pro Gly Lys Ile Pro Pro His Val Gln Lys Ser Val 245 250
255Asn Asn Asp Met Ile Val Thr Asp Asn Asn Gly Ala Val Lys Phe Pro
260 265 270Gln Leu Cys Lys Phe Cys Asp Val Arg Phe Ser Thr Cys Asp
Asn Gln 275 280 285Lys Ser Cys Met Ser Asn Cys Ser Ile Thr Ser Ile
Cys Glu Lys Pro 290 295 300Gln Glu Val Cys Val Ala Val Trp Arg Lys
Asn Asp Glu Asn Ile Thr305 310 315 320Leu Glu Thr Val Cys His Asp
Pro Lys Leu Pro Tyr His Asp Phe Ile 325 330 335Leu Glu Asp Ala Ala
Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys 340 345 350Pro Gly Glu
Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu Cys Asn 355 360 365Asp
Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp 370 375
38098364PRTArtificial SequencehIgG4Fc K-CC-228P-T2m fusion 98Glu
Ser Lys Tyr Gly Pro Pro Cys Pro Pro Cys Pro Ala Pro Glu Phe1 5 10
15Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr
20 25 30Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp
Val 35 40 45Ser Gln Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr Val Asp
Gly Val 50 55 60Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln
Phe Asn Ser65 70 75 80Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu
His Gln Asp Trp Leu 85 90 95Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser
Asn Lys Gly Leu Pro Ser 100 105 110Ser Ile Glu Lys Thr Ile Ser Lys
Ala Lys Gly Gln Pro Arg Glu Pro 115 120 125Gln Val Tyr Thr Leu Pro
Pro Ser Gln Glu Glu Met Thr Lys Asn Gln 130 135 140Val Ser Leu Thr
Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala145 150 155 160Val
Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr 165 170
175Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Arg Leu
180 185 190Thr Val Asp Lys Ser Arg Trp Gln Glu Gly Asn Val Phe Ser
Cys Ser 195 200 205Val Met His Glu Ala Leu His Asn His Tyr Thr Gln
Lys Ser Leu Ser 210 215 220Leu Ser Leu Gly Ile Pro Pro His Val Gln
Lys Ser Val Asn Asn Asp225 230 235 240Met Ile Val Thr Asp Asn Asn
Gly Ala Val Lys Phe Pro Gln Leu Cys 245 250 255Lys Phe Cys Asp Val
Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys 260 265 270Met Ser Asn
Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val 275 280 285Cys
Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr 290 295
300Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu
Asp305 310 315 320Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys
Lys Pro Gly Glu 325 330 335Thr Phe Phe Met Cys Ser Cys Ser Ser Asp
Glu Cys Asn Asp Asn Ile 340 345 350Ile Phe Ser Glu Glu Tyr Asn Thr
Ser Asn Pro Asp 355 36099364PRTArtificial
SequencehIgG4FcdeltaK-CC-228P-409K-T2m fusion 99Glu Ser Lys Tyr Gly
Pro Pro Cys Pro Pro Cys Pro Ala Pro Glu Phe1 5 10 15Leu Gly Gly Pro
Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr 20 25 30Leu Met Ile
Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val 35 40 45Ser Gln
Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr Val Asp Gly Val 50 55 60Glu
Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser65 70 75
80Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu
85 90 95Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Gly Leu Pro
Ser 100 105 110Ser Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro
Arg Glu Pro 115 120 125Gln Val Tyr Thr Leu Pro Pro Ser Gln Glu Glu
Met Thr Lys Asn Gln 130 135 140Val Ser Leu Thr Cys Leu Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala145 150 155 160Val Glu Trp Glu Ser Asn
Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr 165 170 175Pro Pro Val Leu
Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu 180 185 190Thr Val
Asp Lys Ser Arg Trp Gln Glu Gly Asn Val Phe Ser Cys Ser 195 200
205Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser
210 215 220Leu Ser Leu Gly Ile Pro Pro His Val Gln Lys Ser Val Asn
Asn Asp225 230 235 240Met Ile Val Thr Asp Asn Asn Gly Ala Val Lys
Phe Pro Gln Leu Cys 245 250 255Lys Phe Cys Asp Val Arg Phe Ser Thr
Cys Asp Asn Gln Lys Ser Cys 260 265 270Met Ser Asn Cys Ser Ile Thr
Ser Ile Cys Glu Lys Pro Gln Glu Val 275 280 285Cys Val Ala Val Trp
Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr 290 295 300Val Cys His
Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp305 310 315
320Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu
325 330 335Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp
Asn Ile 340 345 350Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp
355 360100492PRTArtificial SequencehIgG1FcdeltaK-C-T22d35 fusion
100Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe Leu1
5 10 15Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro
Glu 20 25 30Val Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu
Val Lys 35 40 45Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala
Lys Thr Lys 50 55 60Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val
Val Ser Val Leu65 70 75 80Thr Val Leu His Gln Asp Trp Leu Asn Gly
Lys Glu Tyr Lys Cys Lys 85 90 95Val Ser Asn Lys Ala Leu Pro Ala Pro
Ile Glu Lys Thr Ile Ser Lys 100 105 110Ala Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr Leu Pro Pro Ser 115 120 125Arg Asp Glu Leu Thr
Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys 130 135 140Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln145 150 155
160Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser Asp Gly
165 170 175Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg
Trp Gln 180 185 190Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu
Ala Leu His Asn 195 200 205His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
Pro Gly Ile Pro Pro His 210 215 220Val Gln Lys Ser Val Asn Asn Asp
Met Ile Val Thr Asp Asn Asn Gly225 230 235 240Ala Val Lys Phe Pro
Gln Leu Cys Lys Phe Cys Asp Val Arg Phe Ser 245 250 255Thr Cys Asp
Asn Gln Lys Ser Cys Met Ser Asn Cys Ser Ile Thr Ser 260 265 270Ile
Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val Trp Arg Lys Asn 275 280
285Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys Leu Pro
290 295 300Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys
Ile Met305 310 315 320Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe
Met Cys Ser Cys Ser 325 330 335Ser Asp Glu Cys Asn Asp Asn Ile Ile
Phe Ser Glu Glu Tyr Asn Thr 340 345 350Ser Asn Pro Asp Ile Pro Pro
His Val Gln Lys Ser Val Asn Asn Asp 355 360 365Met Ile Val Thr Asp
Asn Asn Gly Ala Val Lys Phe Pro Gln Leu Cys 370 375 380Lys Phe Cys
Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys385 390 395
400Met Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val
405 410 415Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu
Glu Thr 420 425 430Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe
Ile Leu Glu Asp 435 440 445Ala Ala Ser Pro Lys Cys Ile Met Lys Glu
Lys Lys Lys Pro Gly Glu 450 455 460Thr Phe Phe Met Cys Ser Cys Ser
Ser Asp Glu Cys Asn Asp Asn Ile465 470 475 480Ile Phe Ser Glu Glu
Tyr Asn Thr Ser Asn Pro Asp 485 490101498PRTArtificial
SequencehIgG1FcdeltaK-CC-T22d35 fusion 101Asp Lys Thr His Thr Cys
Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly1 5 10 15Gly Pro Ser Val Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 20 25 30Ile Ser Arg Thr
Pro Glu Val Thr Cys Val Val Val Asp Val Ser His 35 40 45Glu Asp Pro
Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val 50 55 60His Asn
Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr65 70 75
80Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly
85 90 95Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro
Ile 100 105 110Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu
Pro Gln Val 115 120 125Tyr Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr
Lys Asn Gln Val Ser 130 135 140Leu Thr Cys Leu Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu145 150 155 160Trp Glu Ser Asn Gly Gln
Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro 165 170 175Val Leu Asp Ser
Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val 180 185 190Asp Lys
Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met 195
200 205His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu
Ser 210 215 220Pro Gly Ile Pro Pro His Val Gln Lys Ser Val Asn Asn
Asp Met Ile225 230 235 240Val Thr Asp Asn Asn Gly Ala Val Lys Phe
Pro Gln Leu Cys Lys Phe 245 250 255Cys Asp Val Arg Phe Ser Thr Cys
Asp Asn Gln Lys Ser Cys Met Ser 260 265 270Asn Cys Ser Ile Thr Ser
Ile Cys Glu Lys Pro Gln Glu Val Cys Val 275 280 285Ala Val Trp Arg
Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys 290 295 300His Asp
Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala305 310 315
320Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe
325 330 335Phe Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile
Ile Phe 340 345 350Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp Ile Pro
Pro His Val Gln 355 360 365Lys Ser Val Asn Asn Asp Met Ile Val Thr
Asp Asn Asn Gly Ala Val 370 375 380Lys Phe Pro Gln Leu Cys Lys Phe
Cys Asp Val Arg Phe Ser Thr Cys385 390 395 400Asp Asn Gln Lys Ser
Cys Met Ser Asn Cys Ser Ile Thr Ser Ile Cys 405 410 415Glu Lys Pro
Gln Glu Val Cys Val Ala Val Trp Arg Lys Asn Asp Glu 420 425 430Asn
Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys Leu Pro Tyr His 435 440
445Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu
450 455 460Lys Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser
Ser Asp465 470 475 480Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu
Tyr Asn Thr Ser Asn 485 490 495Pro Asp102494PRTArtificial
SequencehIgG2FcdeltaK-CC-T22d35 fusion 102Val Glu Cys Pro Pro Cys
Pro Ala Pro Pro Val Ala Gly Pro Ser Val1 5 10 15Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 20 25 30Pro Glu Val Thr
Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu 35 40 45Val Gln Phe
Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 50 55 60Thr Lys
Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser65 70 75
80Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys
85 90 95Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro Ile Glu Lys Thr
Ile 100 105 110Ser Lys Thr Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr
Thr Leu Pro 115 120 125Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val
Ser Leu Thr Cys Leu 130 135 140Val Lys Gly Phe Tyr Pro Ser Asp Ile
Ser Val Glu Trp Glu Ser Asn145 150 155 160Gly Gln Pro Glu Asn Asn
Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser 165 170 175Asp Gly Ser Phe
Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg 180 185 190Trp Gln
Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu 195 200
205His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Ile Pro
210 215 220Pro His Val Gln Lys Ser Val Asn Asn Asp Met Ile Val Thr
Asp Asn225 230 235 240Asn Gly Ala Val Lys Phe Pro Gln Leu Cys Lys
Phe Cys Asp Val Arg 245 250 255Phe Ser Thr Cys Asp Asn Gln Lys Ser
Cys Met Ser Asn Cys Ser Ile 260 265 270Thr Ser Ile Cys Glu Lys Pro
Gln Glu Val Cys Val Ala Val Trp Arg 275 280 285Lys Asn Asp Glu Asn
Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys 290 295 300Leu Pro Tyr
His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys305 310 315
320Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser
325 330 335Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu
Glu Tyr 340 345 350Asn Thr Ser Asn Pro Asp Ile Pro Pro His Val Gln
Lys Ser Val Asn 355 360 365Asn Asp Met Ile Val Thr Asp Asn Asn Gly
Ala Val Lys Phe Pro Gln 370 375 380Leu Cys Lys Phe Cys Asp Val Arg
Phe Ser Thr Cys Asp Asn Gln Lys385 390 395 400Ser Cys Met Ser Asn
Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln 405 410 415Glu Val Cys
Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu 420 425 430Glu
Thr Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu 435 440
445Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro
450 455 460Gly Glu Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu Cys
Asn Asp465 470 475 480Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser
Asn Pro Asp 485 490103500PRTArtificial
SequencehIgG4FcdeltaK-CC-228P-T22d35 fusion 103Glu Ser Lys Tyr Gly
Pro Pro Cys Pro Pro Cys Pro Ala Pro Glu Phe1 5 10 15Leu Gly Gly Pro
Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr 20 25 30Leu Met Ile
Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val 35 40 45Ser Gln
Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr Val Asp Gly Val 50 55 60Glu
Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser65 70 75
80Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu
85 90 95Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Gly Leu Pro
Ser 100 105 110Ser Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro
Arg Glu Pro 115 120 125Gln Val Tyr Thr Leu Pro Pro Ser Gln Glu Glu
Met Thr Lys Asn Gln 130 135 140Val Ser Leu Thr Cys Leu Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala145 150 155 160Val Glu Trp Glu Ser Asn
Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr 165 170 175Pro Pro Val Leu
Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Arg Leu 180 185 190Thr Val
Asp Lys Ser Arg Trp Gln Glu Gly Asn Val Phe Ser Cys Ser 195 200
205Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser
210 215 220Leu Ser Leu Gly Ile Pro Pro His Val Gln Lys Ser Val Asn
Asn Asp225 230 235 240Met Ile Val Thr Asp Asn Asn Gly Ala Val Lys
Phe Pro Gln Leu Cys 245 250 255Lys Phe Cys Asp Val Arg Phe Ser Thr
Cys Asp Asn Gln Lys Ser Cys 260 265 270Met Ser Asn Cys Ser Ile Thr
Ser Ile Cys Glu Lys Pro Gln Glu Val 275 280 285Cys Val Ala Val Trp
Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr 290 295 300Val Cys His
Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp305 310 315
320Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu
325 330 335Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp
Asn Ile 340 345 350Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp
Ile Pro Pro His 355 360 365Val Gln Lys Ser Val Asn Asn Asp Met Ile
Val Thr Asp Asn Asn Gly 370 375 380Ala Val Lys Phe Pro Gln Leu Cys
Lys Phe Cys Asp Val Arg Phe Ser385 390 395 400Thr Cys Asp Asn Gln
Lys Ser Cys Met Ser Asn Cys Ser Ile Thr Ser 405 410 415Ile Cys Glu
Lys Pro Gln Glu Val Cys Val Ala Val Trp Arg Lys Asn 420 425 430Asp
Glu Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys Leu Pro 435 440
445Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met
450 455 460Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser
Cys Ser465 470 475 480Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser
Glu Glu Tyr Asn Thr 485 490 495Ser Asn Pro Asp
500104500PRTArtificial SequencehIgG4FcdeltaK-CC-228P-409K-T22d35
fusion 104Glu Ser Lys Tyr Gly Pro Pro Cys Pro Pro Cys Pro Ala Pro
Glu Phe1 5 10 15Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro
Lys Asp Thr 20 25 30Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val
Val Val Asp Val 35 40 45Ser Gln Glu Asp Pro Glu Val Gln Phe Asn Trp
Tyr Val Asp Gly Val 50 55 60Glu Val His Asn Ala Lys Thr Lys Pro Arg
Glu Glu Gln Phe Asn Ser65 70 75 80Thr Tyr Arg Val Val Ser Val Leu
Thr Val Leu His Gln Asp Trp Leu 85 90 95Asn Gly Lys Glu Tyr Lys Cys
Lys Val Ser Asn Lys Gly Leu Pro Ser 100 105 110Ser Ile Glu Lys Thr
Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro 115 120 125Gln Val Tyr
Thr Leu Pro Pro Ser Gln Glu Glu Met Thr Lys Asn Gln 130 135 140Val
Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala145 150
155 160Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr
Thr 165 170 175Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr
Ser Lys Leu 180 185 190Thr Val Asp Lys Ser Arg Trp Gln Glu Gly Asn
Val Phe Ser Cys Ser 195 200 205Val Met His Glu Ala Leu His Asn His
Tyr Thr Gln Lys Ser Leu Ser 210 215 220Leu Ser Leu Gly Ile Pro Pro
His Val Gln Lys Ser Val Asn Asn Asp225 230 235 240Met Ile Val Thr
Asp Asn Asn Gly Ala Val Lys Phe Pro Gln Leu Cys 245 250 255Lys Phe
Cys Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys 260 265
270Met Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val
275 280 285Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu
Glu Thr 290 295 300Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe
Ile Leu Glu Asp305 310 315 320Ala Ala Ser Pro Lys Cys Ile Met Lys
Glu Lys Lys Lys Pro Gly Glu 325 330 335Thr Phe Phe Met Cys Ser Cys
Ser Ser Asp Glu Cys Asn Asp Asn Ile 340 345 350Ile Phe Ser Glu Glu
Tyr Asn Thr Ser Asn Pro Asp Ile Pro Pro His 355 360 365Val Gln Lys
Ser Val Asn Asn Asp Met Ile Val Thr Asp Asn Asn Gly 370 375 380Ala
Val Lys Phe Pro Gln Leu Cys Lys Phe Cys Asp Val Arg Phe Ser385 390
395 400Thr Cys Asp Asn Gln Lys Ser Cys Met Ser Asn Cys Ser Ile Thr
Ser 405 410 415Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val Trp
Arg Lys Asn 420 425 430Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His
Asp Pro Lys Leu Pro 435 440 445Tyr His Asp Phe Ile Leu Glu Asp Ala
Ala Ser Pro Lys Cys Ile Met 450 455 460Lys Glu Lys Lys Lys Pro Gly
Glu Thr Phe Phe Met Cys Ser Cys Ser465 470 475 480Ser Asp Glu Cys
Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr 485 490 495Ser Asn
Pro Asp 500105519PRTArtificial SequencehIgG2Fc-CCCC-T22d35 fusion,
also termed Fc- T22d35 105Met Asp Trp Thr Trp Arg Ile Leu Phe Leu
Val Ala Ala Ala Thr Gly1 5 10 15Thr His Ala Glu Arg Lys Cys Cys Val
Glu Cys Pro Pro Cys Pro Ala 20 25 30Pro Pro Val Ala Gly Pro Ser Val
Phe Leu Phe Pro Pro Lys Pro Lys 35 40 45Asp Thr Leu Met Ile Ser Arg
Thr Pro Glu Val Thr Cys Val Val Val 50 55 60Asp Val Ser His Glu Asp
Pro Glu Val Gln Phe Asn Trp Tyr Val Asp65 70 75 80Gly Val Glu Val
His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Phe 85 90 95Asn Ser Thr
Phe Arg Val Val Ser Val Leu Thr Val Val His Gln Asp 100 105 110Trp
Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Gly Leu 115 120
125Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Thr Lys Gly Gln Pro Arg
130 135 140Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu Met
Thr Lys145 150 155 160Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly
Phe Tyr Pro Ser Asp 165 170 175Ile Ser Val Glu Trp Glu Ser Asn Gly
Gln Pro Glu Asn Asn Tyr Lys 180 185 190Thr Thr Pro Pro Met Leu Asp
Ser Asp Gly Ser Phe Phe Leu Tyr Ser 195 200 205Lys Leu Thr Val Asp
Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser 210 215 220Cys Ser Val
Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser225 230 235
240Leu Ser Leu Ser Pro Gly Lys Ile Pro Pro His Val Gln Lys Ser Val
245 250 255Asn Asn Asp Met Ile Val Thr Asp Asn Asn Gly Ala Val Lys
Phe Pro 260 265 270Gln Leu Cys Lys Phe Cys Asp Val Arg Phe Ser Thr
Cys Asp Asn Gln 275 280 285Lys Ser Cys Met Ser Asn Cys Ser Ile Thr
Ser Ile Cys Glu Lys Pro 290 295 300Gln Glu Val Cys Val Ala Val Trp
Arg Lys Asn Asp Glu Asn Ile Thr305 310 315 320Leu Glu Thr Val Cys
His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile 325 330 335Leu Glu Asp
Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys 340 345 350Pro
Gly Glu Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu Cys Asn 355 360
365Asp Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp Ile
370 375 380Pro Pro His Val Gln Lys Ser Val Asn Asn Asp Met Ile Val
Thr Asp385 390 395 400Asn Asn Gly Ala Val Lys Phe Pro Gln Leu Cys
Lys Phe Cys Asp Val 405 410 415Arg Phe Ser Thr Cys Asp Asn Gln Lys
Ser Cys Met Ser Asn Cys Ser 420 425 430Ile Thr Ser Ile Cys Glu Lys
Pro Gln Glu Val Cys Val Ala Val Trp 435 440 445Arg Lys Asn Asp Glu
Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro 450 455 460Lys Leu Pro
Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys465 470 475
480Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met Cys
485 490 495Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser
Glu Glu 500 505 510Tyr Asn Thr Ser Asn Pro Asp
515106517PRTArtificial SequencehIgG1FcdeltaK-C-T2-T2bAA fusion
106Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe Leu1
5 10 15Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro
Glu 20 25 30Val Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu
Val Lys 35 40 45Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala
Lys Thr Lys 50 55 60Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val
Val Ser Val Leu65 70 75 80Thr Val Leu His Gln Asp Trp Leu Asn Gly
Lys Glu Tyr Lys Cys Lys 85 90 95Val Ser Asn Lys Ala Leu Pro Ala Pro
Ile Glu Lys Thr Ile Ser Lys 100 105 110Ala
Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser 115 120
125Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys
130 135 140Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn
Gly Gln145 150 155 160Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val
Leu Asp Ser Asp Gly 165 170 175Ser Phe Phe Leu Tyr Ser Lys Leu Thr
Val Asp Lys Ser Arg Trp Gln 180 185 190Gln Gly Asn Val Phe Ser Cys
Ser Val Met His Glu Ala Leu His Asn 195 200 205His Tyr Thr Gln Lys
Ser Leu Ser Leu Ser Pro Gly Ile Pro Pro His 210 215 220Val Gln Lys
Ser Val Asn Asn Asp Met Ile Val Thr Asp Asn Asn Gly225 230 235
240Ala Val Lys Phe Pro Gln Leu Cys Lys Phe Cys Asp Val Arg Phe Ser
245 250 255Thr Cys Asp Asn Gln Lys Ser Cys Met Ser Asn Cys Ser Ile
Thr Ser 260 265 270Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val
Trp Arg Lys Asn 275 280 285Asp Glu Asn Ile Thr Leu Glu Thr Val Cys
His Asp Pro Lys Leu Pro 290 295 300Tyr His Asp Phe Ile Leu Glu Asp
Ala Ala Ser Pro Lys Cys Ile Met305 310 315 320Lys Glu Lys Lys Lys
Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser 325 330 335Ser Asp Glu
Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr 340 345 350Ser
Asn Pro Asp Ile Pro Pro His Val Gln Lys Ser Asp Val Glu Met 355 360
365Glu Ala Gln Lys Asp Glu Ile Ile Ala Pro Ser Ala Asn Arg Thr Ala
370 375 380His Pro Leu Arg His Ile Asn Asn Asp Met Ile Val Thr Asp
Asn Asn385 390 395 400Gly Ala Val Lys Phe Pro Gln Leu Cys Lys Phe
Cys Asp Val Arg Phe 405 410 415Ser Thr Cys Asp Asn Gln Lys Ser Cys
Met Ser Asn Cys Ser Ile Thr 420 425 430Ser Ile Cys Glu Lys Pro Gln
Glu Val Cys Val Ala Val Trp Arg Lys 435 440 445Asn Asp Glu Asn Ile
Thr Leu Glu Thr Val Cys His Asp Pro Lys Leu 450 455 460Pro Tyr His
Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile465 470 475
480Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser Cys
485 490 495Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu
Tyr Asn 500 505 510Thr Ser Asn Pro Asp 515107523PRTArtificial
SequencehIgG1FcdeltaK-CC-T2-T2bAA fusion 107Asp Lys Thr His Thr Cys
Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly1 5 10 15Gly Pro Ser Val Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 20 25 30Ile Ser Arg Thr
Pro Glu Val Thr Cys Val Val Val Asp Val Ser His 35 40 45Glu Asp Pro
Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val 50 55 60His Asn
Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr65 70 75
80Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly
85 90 95Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro
Ile 100 105 110Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu
Pro Gln Val 115 120 125Tyr Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr
Lys Asn Gln Val Ser 130 135 140Leu Thr Cys Leu Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu145 150 155 160Trp Glu Ser Asn Gly Gln
Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro 165 170 175Val Leu Asp Ser
Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val 180 185 190Asp Lys
Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met 195 200
205His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
210 215 220Pro Gly Ile Pro Pro His Val Gln Lys Ser Val Asn Asn Asp
Met Ile225 230 235 240Val Thr Asp Asn Asn Gly Ala Val Lys Phe Pro
Gln Leu Cys Lys Phe 245 250 255Cys Asp Val Arg Phe Ser Thr Cys Asp
Asn Gln Lys Ser Cys Met Ser 260 265 270Asn Cys Ser Ile Thr Ser Ile
Cys Glu Lys Pro Gln Glu Val Cys Val 275 280 285Ala Val Trp Arg Lys
Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys 290 295 300His Asp Pro
Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala305 310 315
320Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe
325 330 335Phe Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile
Ile Phe 340 345 350Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp Ile Pro
Pro His Val Gln 355 360 365Lys Ser Asp Val Glu Met Glu Ala Gln Lys
Asp Glu Ile Ile Ala Pro 370 375 380Ser Ala Asn Arg Thr Ala His Pro
Leu Arg His Ile Asn Asn Asp Met385 390 395 400Ile Val Thr Asp Asn
Asn Gly Ala Val Lys Phe Pro Gln Leu Cys Lys 405 410 415Phe Cys Asp
Val Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys Met 420 425 430Ser
Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys 435 440
445Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val
450 455 460Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu
Asp Ala465 470 475 480Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys
Lys Pro Gly Glu Thr 485 490 495Phe Phe Met Cys Ser Cys Ser Ser Asp
Glu Cys Asn Asp Asn Ile Ile 500 505 510Phe Ser Glu Glu Tyr Asn Thr
Ser Asn Pro Asp 515 520108519PRTArtificial
SequencehIgG2FcdeltaK-CC-T2-T2bAA fusion 108Val Glu Cys Pro Pro Cys
Pro Ala Pro Pro Val Ala Gly Pro Ser Val1 5 10 15Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 20 25 30Pro Glu Val Thr
Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu 35 40 45Val Gln Phe
Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 50 55 60Thr Lys
Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser65 70 75
80Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys
85 90 95Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro Ile Glu Lys Thr
Ile 100 105 110Ser Lys Thr Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr
Thr Leu Pro 115 120 125Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val
Ser Leu Thr Cys Leu 130 135 140Val Lys Gly Phe Tyr Pro Ser Asp Ile
Ser Val Glu Trp Glu Ser Asn145 150 155 160Gly Gln Pro Glu Asn Asn
Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser 165 170 175Asp Gly Ser Phe
Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg 180 185 190Trp Gln
Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu 195 200
205His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Ile Pro
210 215 220Pro His Val Gln Lys Ser Val Asn Asn Asp Met Ile Val Thr
Asp Asn225 230 235 240Asn Gly Ala Val Lys Phe Pro Gln Leu Cys Lys
Phe Cys Asp Val Arg 245 250 255Phe Ser Thr Cys Asp Asn Gln Lys Ser
Cys Met Ser Asn Cys Ser Ile 260 265 270Thr Ser Ile Cys Glu Lys Pro
Gln Glu Val Cys Val Ala Val Trp Arg 275 280 285Lys Asn Asp Glu Asn
Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys 290 295 300Leu Pro Tyr
His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys305 310 315
320Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser
325 330 335Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu
Glu Tyr 340 345 350Asn Thr Ser Asn Pro Asp Ile Pro Pro His Val Gln
Lys Ser Asp Val 355 360 365Glu Met Glu Ala Gln Lys Asp Glu Ile Ile
Ala Pro Ser Ala Asn Arg 370 375 380Thr Ala His Pro Leu Arg His Ile
Asn Asn Asp Met Ile Val Thr Asp385 390 395 400Asn Asn Gly Ala Val
Lys Phe Pro Gln Leu Cys Lys Phe Cys Asp Val 405 410 415Arg Phe Ser
Thr Cys Asp Asn Gln Lys Ser Cys Met Ser Asn Cys Ser 420 425 430Ile
Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val Trp 435 440
445Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro
450 455 460Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser
Pro Lys465 470 475 480Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu
Thr Phe Phe Met Cys 485 490 495Ser Cys Ser Ser Asp Glu Cys Asn Asp
Asn Ile Ile Phe Ser Glu Glu 500 505 510Tyr Asn Thr Ser Asn Pro Asp
515109525PRTArtificial SequencehIgG4FcdeltaK-CC-228P-T2-T2bAA
fusion 109Glu Ser Lys Tyr Gly Pro Pro Cys Pro Pro Cys Pro Ala Pro
Glu Phe1 5 10 15Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro
Lys Asp Thr 20 25 30Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val
Val Val Asp Val 35 40 45Ser Gln Glu Asp Pro Glu Val Gln Phe Asn Trp
Tyr Val Asp Gly Val 50 55 60Glu Val His Asn Ala Lys Thr Lys Pro Arg
Glu Glu Gln Phe Asn Ser65 70 75 80Thr Tyr Arg Val Val Ser Val Leu
Thr Val Leu His Gln Asp Trp Leu 85 90 95Asn Gly Lys Glu Tyr Lys Cys
Lys Val Ser Asn Lys Gly Leu Pro Ser 100 105 110Ser Ile Glu Lys Thr
Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro 115 120 125Gln Val Tyr
Thr Leu Pro Pro Ser Gln Glu Glu Met Thr Lys Asn Gln 130 135 140Val
Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala145 150
155 160Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr
Thr 165 170 175Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr
Ser Arg Leu 180 185 190Thr Val Asp Lys Ser Arg Trp Gln Glu Gly Asn
Val Phe Ser Cys Ser 195 200 205Val Met His Glu Ala Leu His Asn His
Tyr Thr Gln Lys Ser Leu Ser 210 215 220Leu Ser Leu Gly Ile Pro Pro
His Val Gln Lys Ser Val Asn Asn Asp225 230 235 240Met Ile Val Thr
Asp Asn Asn Gly Ala Val Lys Phe Pro Gln Leu Cys 245 250 255Lys Phe
Cys Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys 260 265
270Met Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val
275 280 285Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu
Glu Thr 290 295 300Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe
Ile Leu Glu Asp305 310 315 320Ala Ala Ser Pro Lys Cys Ile Met Lys
Glu Lys Lys Lys Pro Gly Glu 325 330 335Thr Phe Phe Met Cys Ser Cys
Ser Ser Asp Glu Cys Asn Asp Asn Ile 340 345 350Ile Phe Ser Glu Glu
Tyr Asn Thr Ser Asn Pro Asp Ile Pro Pro His 355 360 365Val Gln Lys
Ser Asp Val Glu Met Glu Ala Gln Lys Asp Glu Ile Ile 370 375 380Ala
Pro Ser Ala Asn Arg Thr Ala His Pro Leu Arg His Ile Asn Asn385 390
395 400Asp Met Ile Val Thr Asp Asn Asn Gly Ala Val Lys Phe Pro Gln
Leu 405 410 415Cys Lys Phe Cys Asp Val Arg Phe Ser Thr Cys Asp Asn
Gln Lys Ser 420 425 430Cys Met Ser Asn Cys Ser Ile Thr Ser Ile Cys
Glu Lys Pro Gln Glu 435 440 445Val Cys Val Ala Val Trp Arg Lys Asn
Asp Glu Asn Ile Thr Leu Glu 450 455 460Thr Val Cys His Asp Pro Lys
Leu Pro Tyr His Asp Phe Ile Leu Glu465 470 475 480Asp Ala Ala Ser
Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly 485 490 495Glu Thr
Phe Phe Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn 500 505
510Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp 515 520
525110525PRTArtificial SequencehIgG4FcdeltaK-CC-228P-409K-T2-T2bAA
fusion 110Glu Ser Lys Tyr Gly Pro Pro Cys Pro Pro Cys Pro Ala Pro
Glu Phe1 5 10 15Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro
Lys Asp Thr 20 25 30Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val
Val Val Asp Val 35 40 45Ser Gln Glu Asp Pro Glu Val Gln Phe Asn Trp
Tyr Val Asp Gly Val 50 55 60Glu Val His Asn Ala Lys Thr Lys Pro Arg
Glu Glu Gln Phe Asn Ser65 70 75 80Thr Tyr Arg Val Val Ser Val Leu
Thr Val Leu His Gln Asp Trp Leu 85 90 95Asn Gly Lys Glu Tyr Lys Cys
Lys Val Ser Asn Lys Gly Leu Pro Ser 100 105 110Ser Ile Glu Lys Thr
Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro 115 120 125Gln Val Tyr
Thr Leu Pro Pro Ser Gln Glu Glu Met Thr Lys Asn Gln 130 135 140Val
Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala145 150
155 160Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr
Thr 165 170 175Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr
Ser Lys Leu 180 185 190Thr Val Asp Lys Ser Arg Trp Gln Glu Gly Asn
Val Phe Ser Cys Ser 195 200 205Val Met His Glu Ala Leu His Asn His
Tyr Thr Gln Lys Ser Leu Ser 210 215 220Leu Ser Leu Gly Ile Pro Pro
His Val Gln Lys Ser Val Asn Asn Asp225 230 235 240Met Ile Val Thr
Asp Asn Asn Gly Ala Val Lys Phe Pro Gln Leu Cys 245 250 255Lys Phe
Cys Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys 260 265
270Met Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val
275 280 285Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu
Glu Thr 290 295 300Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe
Ile Leu Glu Asp305 310 315 320Ala Ala Ser Pro Lys Cys Ile Met Lys
Glu Lys Lys Lys Pro Gly Glu 325 330 335Thr Phe Phe Met Cys Ser Cys
Ser Ser Asp Glu Cys Asn Asp Asn Ile 340 345 350Ile Phe Ser Glu Glu
Tyr Asn Thr Ser Asn Pro Asp Ile Pro Pro His 355 360 365Val Gln Lys
Ser Asp Val Glu Met Glu Ala Gln Lys Asp Glu Ile Ile 370 375 380Ala
Pro Ser Ala Asn Arg Thr Ala His Pro Leu Arg His Ile Asn Asn385 390
395 400Asp Met Ile Val Thr Asp Asn Asn Gly Ala Val Lys Phe Pro Gln
Leu 405 410 415Cys Lys Phe Cys Asp Val Arg Phe Ser Thr Cys Asp Asn
Gln Lys Ser 420 425 430Cys Met Ser Asn Cys Ser Ile Thr Ser Ile Cys
Glu Lys Pro Gln Glu 435 440 445Val Cys Val Ala Val Trp Arg Lys Asn
Asp Glu Asn Ile Thr Leu Glu 450 455
460Thr Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu
Glu465 470 475 480Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys
Lys Lys Pro Gly 485 490 495Glu Thr Phe Phe Met Cys Ser Cys Ser Ser
Asp Glu Cys Asn Asp Asn 500 505 510Ile Ile Phe Ser Glu Glu Tyr Asn
Thr Ser Asn Pro Asp 515 520 525111628PRTArtificial
SequencehIgG1FcdeltaK-C-T2-T2-T2 fusion 111Pro Pro Cys Pro Ala Pro
Glu Leu Leu Gly Gly Pro Ser Val Phe Leu1 5 10 15Phe Pro Pro Lys Pro
Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu 20 25 30Val Thr Cys Val
Val Val Asp Val Ser His Glu Asp Pro Glu Val Lys 35 40 45Phe Asn Trp
Tyr Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys 50 55 60Pro Arg
Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu65 70 75
80Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys
85 90 95Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser
Lys 100 105 110Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu
Pro Pro Ser 115 120 125Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu
Thr Cys Leu Val Lys 130 135 140Gly Phe Tyr Pro Ser Asp Ile Ala Val
Glu Trp Glu Ser Asn Gly Gln145 150 155 160Pro Glu Asn Asn Tyr Lys
Thr Thr Pro Pro Val Leu Asp Ser Asp Gly 165 170 175Ser Phe Phe Leu
Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln 180 185 190Gln Gly
Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn 195 200
205His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Ile Pro Pro His
210 215 220Val Gln Lys Ser Val Asn Asn Asp Met Ile Val Thr Asp Asn
Asn Gly225 230 235 240Ala Val Lys Phe Pro Gln Leu Cys Lys Phe Cys
Asp Val Arg Phe Ser 245 250 255Thr Cys Asp Asn Gln Lys Ser Cys Met
Ser Asn Cys Ser Ile Thr Ser 260 265 270Ile Cys Glu Lys Pro Gln Glu
Val Cys Val Ala Val Trp Arg Lys Asn 275 280 285Asp Glu Asn Ile Thr
Leu Glu Thr Val Cys His Asp Pro Lys Leu Pro 290 295 300Tyr His Asp
Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met305 310 315
320Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser
325 330 335Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr
Asn Thr 340 345 350Ser Asn Pro Asp Ile Pro Pro His Val Gln Lys Ser
Val Asn Asn Asp 355 360 365Met Ile Val Thr Asp Asn Asn Gly Ala Val
Lys Phe Pro Gln Leu Cys 370 375 380Lys Phe Cys Asp Val Arg Phe Ser
Thr Cys Asp Asn Gln Lys Ser Cys385 390 395 400Met Ser Asn Cys Ser
Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val 405 410 415Cys Val Ala
Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr 420 425 430Val
Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp 435 440
445Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu
450 455 460Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp
Asn Ile465 470 475 480Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn Pro
Asp Ile Pro Pro His 485 490 495Val Gln Lys Ser Val Asn Asn Asp Met
Ile Val Thr Asp Asn Asn Gly 500 505 510Ala Val Lys Phe Pro Gln Leu
Cys Lys Phe Cys Asp Val Arg Phe Ser 515 520 525Thr Cys Asp Asn Gln
Lys Ser Cys Met Ser Asn Cys Ser Ile Thr Ser 530 535 540Ile Cys Glu
Lys Pro Gln Glu Val Cys Val Ala Val Trp Arg Lys Asn545 550 555
560Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys Leu Pro
565 570 575Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys
Ile Met 580 585 590Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met
Cys Ser Cys Ser 595 600 605Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe
Ser Glu Glu Tyr Asn Thr 610 615 620Ser Asn Pro
Asp625112634PRTArtificial SequencehIgG1FcdeltaK-CC-T2-T2-T2 fusion
112Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly1
5 10 15Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu
Met 20 25 30Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val
Ser His 35 40 45Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly
Val Glu Val 50 55 60His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr
Asn Ser Thr Tyr65 70 75 80Arg Val Val Ser Val Leu Thr Val Leu His
Gln Asp Trp Leu Asn Gly 85 90 95Lys Glu Tyr Lys Cys Lys Val Ser Asn
Lys Ala Leu Pro Ala Pro Ile 100 105 110Glu Lys Thr Ile Ser Lys Ala
Lys Gly Gln Pro Arg Glu Pro Gln Val 115 120 125Tyr Thr Leu Pro Pro
Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser 130 135 140Leu Thr Cys
Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu145 150 155
160Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro
165 170 175Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu
Thr Val 180 185 190Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser
Cys Ser Val Met 195 200 205His Glu Ala Leu His Asn His Tyr Thr Gln
Lys Ser Leu Ser Leu Ser 210 215 220Pro Gly Ile Pro Pro His Val Gln
Lys Ser Val Asn Asn Asp Met Ile225 230 235 240Val Thr Asp Asn Asn
Gly Ala Val Lys Phe Pro Gln Leu Cys Lys Phe 245 250 255Cys Asp Val
Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys Met Ser 260 265 270Asn
Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys Val 275 280
285Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys
290 295 300His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp
Ala Ala305 310 315 320Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys
Pro Gly Glu Thr Phe 325 330 335Phe Met Cys Ser Cys Ser Ser Asp Glu
Cys Asn Asp Asn Ile Ile Phe 340 345 350Ser Glu Glu Tyr Asn Thr Ser
Asn Pro Asp Ile Pro Pro His Val Gln 355 360 365Lys Ser Val Asn Asn
Asp Met Ile Val Thr Asp Asn Asn Gly Ala Val 370 375 380Lys Phe Pro
Gln Leu Cys Lys Phe Cys Asp Val Arg Phe Ser Thr Cys385 390 395
400Asp Asn Gln Lys Ser Cys Met Ser Asn Cys Ser Ile Thr Ser Ile Cys
405 410 415Glu Lys Pro Gln Glu Val Cys Val Ala Val Trp Arg Lys Asn
Asp Glu 420 425 430Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys
Leu Pro Tyr His 435 440 445Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro
Lys Cys Ile Met Lys Glu 450 455 460Lys Lys Lys Pro Gly Glu Thr Phe
Phe Met Cys Ser Cys Ser Ser Asp465 470 475 480Glu Cys Asn Asp Asn
Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn 485 490 495Pro Asp Ile
Pro Pro His Val Gln Lys Ser Val Asn Asn Asp Met Ile 500 505 510Val
Thr Asp Asn Asn Gly Ala Val Lys Phe Pro Gln Leu Cys Lys Phe 515 520
525Cys Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys Met Ser
530 535 540Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val
Cys Val545 550 555 560Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr
Leu Glu Thr Val Cys 565 570 575His Asp Pro Lys Leu Pro Tyr His Asp
Phe Ile Leu Glu Asp Ala Ala 580 585 590Ser Pro Lys Cys Ile Met Lys
Glu Lys Lys Lys Pro Gly Glu Thr Phe 595 600 605Phe Met Cys Ser Cys
Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe 610 615 620Ser Glu Glu
Tyr Asn Thr Ser Asn Pro Asp625 630113630PRTArtificial
SequencehIgG2FcdeltaK-CC-T2-T2-T2 fusion 113Val Glu Cys Pro Pro Cys
Pro Ala Pro Pro Val Ala Gly Pro Ser Val1 5 10 15Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 20 25 30Pro Glu Val Thr
Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu 35 40 45Val Gln Phe
Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 50 55 60Thr Lys
Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser65 70 75
80Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys
85 90 95Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro Ile Glu Lys Thr
Ile 100 105 110Ser Lys Thr Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr
Thr Leu Pro 115 120 125Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val
Ser Leu Thr Cys Leu 130 135 140Val Lys Gly Phe Tyr Pro Ser Asp Ile
Ser Val Glu Trp Glu Ser Asn145 150 155 160Gly Gln Pro Glu Asn Asn
Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser 165 170 175Asp Gly Ser Phe
Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg 180 185 190Trp Gln
Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu 195 200
205His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Ile Pro
210 215 220Pro His Val Gln Lys Ser Val Asn Asn Asp Met Ile Val Thr
Asp Asn225 230 235 240Asn Gly Ala Val Lys Phe Pro Gln Leu Cys Lys
Phe Cys Asp Val Arg 245 250 255Phe Ser Thr Cys Asp Asn Gln Lys Ser
Cys Met Ser Asn Cys Ser Ile 260 265 270Thr Ser Ile Cys Glu Lys Pro
Gln Glu Val Cys Val Ala Val Trp Arg 275 280 285Lys Asn Asp Glu Asn
Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys 290 295 300Leu Pro Tyr
His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys305 310 315
320Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser
325 330 335Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu
Glu Tyr 340 345 350Asn Thr Ser Asn Pro Asp Ile Pro Pro His Val Gln
Lys Ser Val Asn 355 360 365Asn Asp Met Ile Val Thr Asp Asn Asn Gly
Ala Val Lys Phe Pro Gln 370 375 380Leu Cys Lys Phe Cys Asp Val Arg
Phe Ser Thr Cys Asp Asn Gln Lys385 390 395 400Ser Cys Met Ser Asn
Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln 405 410 415Glu Val Cys
Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu 420 425 430Glu
Thr Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu 435 440
445Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro
450 455 460Gly Glu Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu Cys
Asn Asp465 470 475 480Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser
Asn Pro Asp Ile Pro 485 490 495Pro His Val Gln Lys Ser Val Asn Asn
Asp Met Ile Val Thr Asp Asn 500 505 510Asn Gly Ala Val Lys Phe Pro
Gln Leu Cys Lys Phe Cys Asp Val Arg 515 520 525Phe Ser Thr Cys Asp
Asn Gln Lys Ser Cys Met Ser Asn Cys Ser Ile 530 535 540Thr Ser Ile
Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val Trp Arg545 550 555
560Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys
565 570 575Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro
Lys Cys 580 585 590Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe
Phe Met Cys Ser 595 600 605Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile
Ile Phe Ser Glu Glu Tyr 610 615 620Asn Thr Ser Asn Pro Asp625
630114636PRTArtificial SequencehIgG4FcdeltaK-CC-228P-T2-T2-T2
fusion 114Glu Ser Lys Tyr Gly Pro Pro Cys Pro Pro Cys Pro Ala Pro
Glu Phe1 5 10 15Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro
Lys Asp Thr 20 25 30Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val
Val Val Asp Val 35 40 45Ser Gln Glu Asp Pro Glu Val Gln Phe Asn Trp
Tyr Val Asp Gly Val 50 55 60Glu Val His Asn Ala Lys Thr Lys Pro Arg
Glu Glu Gln Phe Asn Ser65 70 75 80Thr Tyr Arg Val Val Ser Val Leu
Thr Val Leu His Gln Asp Trp Leu 85 90 95Asn Gly Lys Glu Tyr Lys Cys
Lys Val Ser Asn Lys Gly Leu Pro Ser 100 105 110Ser Ile Glu Lys Thr
Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro 115 120 125Gln Val Tyr
Thr Leu Pro Pro Ser Gln Glu Glu Met Thr Lys Asn Gln 130 135 140Val
Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala145 150
155 160Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr
Thr 165 170 175Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr
Ser Arg Leu 180 185 190Thr Val Asp Lys Ser Arg Trp Gln Glu Gly Asn
Val Phe Ser Cys Ser 195 200 205Val Met His Glu Ala Leu His Asn His
Tyr Thr Gln Lys Ser Leu Ser 210 215 220Leu Ser Leu Gly Ile Pro Pro
His Val Gln Lys Ser Val Asn Asn Asp225 230 235 240Met Ile Val Thr
Asp Asn Asn Gly Ala Val Lys Phe Pro Gln Leu Cys 245 250 255Lys Phe
Cys Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys 260 265
270Met Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val
275 280 285Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu
Glu Thr 290 295 300Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe
Ile Leu Glu Asp305 310 315 320Ala Ala Ser Pro Lys Cys Ile Met Lys
Glu Lys Lys Lys Pro Gly Glu 325 330 335Thr Phe Phe Met Cys Ser Cys
Ser Ser Asp Glu Cys Asn Asp Asn Ile 340 345 350Ile Phe Ser Glu Glu
Tyr Asn Thr Ser Asn Pro Asp Ile Pro Pro His 355 360 365Val Gln Lys
Ser Val Asn Asn Asp Met Ile Val Thr Asp Asn Asn Gly 370 375 380Ala
Val Lys Phe Pro Gln Leu Cys Lys Phe Cys Asp Val Arg Phe Ser385 390
395 400Thr Cys Asp Asn Gln Lys Ser Cys Met Ser Asn Cys Ser Ile Thr
Ser 405 410 415Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val Trp
Arg Lys Asn 420 425 430Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His
Asp Pro Lys Leu Pro 435 440 445Tyr His Asp Phe Ile Leu Glu Asp Ala
Ala Ser Pro Lys Cys Ile Met 450 455 460Lys Glu Lys Lys Lys Pro Gly
Glu Thr Phe Phe Met Cys Ser Cys Ser465 470 475 480Ser Asp Glu Cys
Asn Asp Asn Ile Ile Phe Ser Glu
Glu Tyr Asn Thr 485 490 495Ser Asn Pro Asp Ile Pro Pro His Val Gln
Lys Ser Val Asn Asn Asp 500 505 510Met Ile Val Thr Asp Asn Asn Gly
Ala Val Lys Phe Pro Gln Leu Cys 515 520 525Lys Phe Cys Asp Val Arg
Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys 530 535 540Met Ser Asn Cys
Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val545 550 555 560Cys
Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr 565 570
575Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp
580 585 590Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro
Gly Glu 595 600 605Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu Cys
Asn Asp Asn Ile 610 615 620Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn
Pro Asp625 630 635115636PRTArtificial
SequencehIgG4FcdeltaK-CC-228P-409K-T2-T2-T2 fusion 115Glu Ser Lys
Tyr Gly Pro Pro Cys Pro Pro Cys Pro Ala Pro Glu Phe1 5 10 15Leu Gly
Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr 20 25 30Leu
Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val 35 40
45Ser Gln Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr Val Asp Gly Val
50 55 60Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Phe Asn
Ser65 70 75 80Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln
Asp Trp Leu 85 90 95Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys
Gly Leu Pro Ser 100 105 110Ser Ile Glu Lys Thr Ile Ser Lys Ala Lys
Gly Gln Pro Arg Glu Pro 115 120 125Gln Val Tyr Thr Leu Pro Pro Ser
Gln Glu Glu Met Thr Lys Asn Gln 130 135 140Val Ser Leu Thr Cys Leu
Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala145 150 155 160Val Glu Trp
Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr 165 170 175Pro
Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu 180 185
190Thr Val Asp Lys Ser Arg Trp Gln Glu Gly Asn Val Phe Ser Cys Ser
195 200 205Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser
Leu Ser 210 215 220Leu Ser Leu Gly Ile Pro Pro His Val Gln Lys Ser
Val Asn Asn Asp225 230 235 240Met Ile Val Thr Asp Asn Asn Gly Ala
Val Lys Phe Pro Gln Leu Cys 245 250 255Lys Phe Cys Asp Val Arg Phe
Ser Thr Cys Asp Asn Gln Lys Ser Cys 260 265 270Met Ser Asn Cys Ser
Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val 275 280 285Cys Val Ala
Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr 290 295 300Val
Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp305 310
315 320Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly
Glu 325 330 335Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu Cys Asn
Asp Asn Ile 340 345 350Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn Pro
Asp Ile Pro Pro His 355 360 365Val Gln Lys Ser Val Asn Asn Asp Met
Ile Val Thr Asp Asn Asn Gly 370 375 380Ala Val Lys Phe Pro Gln Leu
Cys Lys Phe Cys Asp Val Arg Phe Ser385 390 395 400Thr Cys Asp Asn
Gln Lys Ser Cys Met Ser Asn Cys Ser Ile Thr Ser 405 410 415Ile Cys
Glu Lys Pro Gln Glu Val Cys Val Ala Val Trp Arg Lys Asn 420 425
430Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys Leu Pro
435 440 445Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys
Ile Met 450 455 460Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met
Cys Ser Cys Ser465 470 475 480Ser Asp Glu Cys Asn Asp Asn Ile Ile
Phe Ser Glu Glu Tyr Asn Thr 485 490 495Ser Asn Pro Asp Ile Pro Pro
His Val Gln Lys Ser Val Asn Asn Asp 500 505 510Met Ile Val Thr Asp
Asn Asn Gly Ala Val Lys Phe Pro Gln Leu Cys 515 520 525Lys Phe Cys
Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys 530 535 540Met
Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val545 550
555 560Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu
Thr 565 570 575Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile
Leu Glu Asp 580 585 590Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys
Lys Lys Pro Gly Glu 595 600 605Thr Phe Phe Met Cys Ser Cys Ser Ser
Asp Glu Cys Asn Asp Asn Ile 610 615 620Ile Phe Ser Glu Glu Tyr Asn
Thr Ser Asn Pro Asp625 630 635116678PRTArtificial
SequencehIgG1FcdeltaK-C-T2-T2bAA-T2bAA fusion 116Pro Pro Cys Pro
Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe Leu1 5 10 15Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu 20 25 30Val Thr
Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Lys 35 40 45Phe
Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys 50 55
60Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu65
70 75 80Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys
Lys 85 90 95Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile
Ser Lys 100 105 110Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr
Leu Pro Pro Ser 115 120 125Arg Asp Glu Leu Thr Lys Asn Gln Val Ser
Leu Thr Cys Leu Val Lys 130 135 140Gly Phe Tyr Pro Ser Asp Ile Ala
Val Glu Trp Glu Ser Asn Gly Gln145 150 155 160Pro Glu Asn Asn Tyr
Lys Thr Thr Pro Pro Val Leu Asp Ser Asp Gly 165 170 175Ser Phe Phe
Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln 180 185 190Gln
Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn 195 200
205His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Ile Pro Pro His
210 215 220Val Gln Lys Ser Val Asn Asn Asp Met Ile Val Thr Asp Asn
Asn Gly225 230 235 240Ala Val Lys Phe Pro Gln Leu Cys Lys Phe Cys
Asp Val Arg Phe Ser 245 250 255Thr Cys Asp Asn Gln Lys Ser Cys Met
Ser Asn Cys Ser Ile Thr Ser 260 265 270Ile Cys Glu Lys Pro Gln Glu
Val Cys Val Ala Val Trp Arg Lys Asn 275 280 285Asp Glu Asn Ile Thr
Leu Glu Thr Val Cys His Asp Pro Lys Leu Pro 290 295 300Tyr His Asp
Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met305 310 315
320Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser
325 330 335Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr
Asn Thr 340 345 350Ser Asn Pro Asp Ile Pro Pro His Val Gln Lys Ser
Asp Val Glu Met 355 360 365Glu Ala Gln Lys Asp Glu Ile Ile Ala Pro
Ser Ala Asn Arg Thr Ala 370 375 380His Pro Leu Arg His Ile Asn Asn
Asp Met Ile Val Thr Asp Asn Asn385 390 395 400Gly Ala Val Lys Phe
Pro Gln Leu Cys Lys Phe Cys Asp Val Arg Phe 405 410 415Ser Thr Cys
Asp Asn Gln Lys Ser Cys Met Ser Asn Cys Ser Ile Thr 420 425 430Ser
Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val Trp Arg Lys 435 440
445Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys Leu
450 455 460Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys
Cys Ile465 470 475 480Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe
Phe Met Cys Ser Cys 485 490 495Ser Ser Asp Glu Cys Asn Asp Asn Ile
Ile Phe Ser Glu Glu Tyr Asn 500 505 510Thr Ser Asn Pro Asp Ile Pro
Pro His Val Gln Lys Ser Asp Val Glu 515 520 525Met Glu Ala Gln Lys
Asp Glu Ile Ile Ala Pro Ser Ala Asn Arg Thr 530 535 540Ala His Pro
Leu Arg His Ile Asn Asn Asp Met Ile Val Thr Asp Asn545 550 555
560Asn Gly Ala Val Lys Phe Pro Gln Leu Cys Lys Phe Cys Asp Val Arg
565 570 575Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys Met Ser Asn Cys
Ser Ile 580 585 590Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys Val
Ala Val Trp Arg 595 600 605Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr
Val Cys His Asp Pro Lys 610 615 620Leu Pro Tyr His Asp Phe Ile Leu
Glu Asp Ala Ala Ser Pro Lys Cys625 630 635 640Ile Met Lys Glu Lys
Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser 645 650 655Cys Ser Ser
Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr 660 665 670Asn
Thr Ser Asn Pro Asp 675117684PRTArtificial
SequencehIgG1FcdeltaK-CC-T2-T2bAA-T2bAA fusion 117Asp Lys Thr His
Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly1 5 10 15Gly Pro Ser
Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 20 25 30Ile Ser
Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His 35 40 45Glu
Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val 50 55
60His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr65
70 75 80Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn
Gly 85 90 95Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala
Pro Ile 100 105 110Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg
Glu Pro Gln Val 115 120 125Tyr Thr Leu Pro Pro Ser Arg Asp Glu Leu
Thr Lys Asn Gln Val Ser 130 135 140Leu Thr Cys Leu Val Lys Gly Phe
Tyr Pro Ser Asp Ile Ala Val Glu145 150 155 160Trp Glu Ser Asn Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro 165 170 175Val Leu Asp
Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val 180 185 190Asp
Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met 195 200
205His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
210 215 220Pro Gly Ile Pro Pro His Val Gln Lys Ser Val Asn Asn Asp
Met Ile225 230 235 240Val Thr Asp Asn Asn Gly Ala Val Lys Phe Pro
Gln Leu Cys Lys Phe 245 250 255Cys Asp Val Arg Phe Ser Thr Cys Asp
Asn Gln Lys Ser Cys Met Ser 260 265 270Asn Cys Ser Ile Thr Ser Ile
Cys Glu Lys Pro Gln Glu Val Cys Val 275 280 285Ala Val Trp Arg Lys
Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys 290 295 300His Asp Pro
Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala305 310 315
320Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe
325 330 335Phe Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile
Ile Phe 340 345 350Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp Ile Pro
Pro His Val Gln 355 360 365Lys Ser Asp Val Glu Met Glu Ala Gln Lys
Asp Glu Ile Ile Ala Pro 370 375 380Ser Ala Asn Arg Thr Ala His Pro
Leu Arg His Ile Asn Asn Asp Met385 390 395 400Ile Val Thr Asp Asn
Asn Gly Ala Val Lys Phe Pro Gln Leu Cys Lys 405 410 415Phe Cys Asp
Val Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys Met 420 425 430Ser
Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys 435 440
445Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val
450 455 460Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu
Asp Ala465 470 475 480Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys
Lys Pro Gly Glu Thr 485 490 495Phe Phe Met Cys Ser Cys Ser Ser Asp
Glu Cys Asn Asp Asn Ile Ile 500 505 510Phe Ser Glu Glu Tyr Asn Thr
Ser Asn Pro Asp Ile Pro Pro His Val 515 520 525Gln Lys Ser Asp Val
Glu Met Glu Ala Gln Lys Asp Glu Ile Ile Ala 530 535 540Pro Ser Ala
Asn Arg Thr Ala His Pro Leu Arg His Ile Asn Asn Asp545 550 555
560Met Ile Val Thr Asp Asn Asn Gly Ala Val Lys Phe Pro Gln Leu Cys
565 570 575Lys Phe Cys Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys
Ser Cys 580 585 590Met Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys
Pro Gln Glu Val 595 600 605Cys Val Ala Val Trp Arg Lys Asn Asp Glu
Asn Ile Thr Leu Glu Thr 610 615 620Val Cys His Asp Pro Lys Leu Pro
Tyr His Asp Phe Ile Leu Glu Asp625 630 635 640Ala Ala Ser Pro Lys
Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu 645 650 655Thr Phe Phe
Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile 660 665 670Ile
Phe Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp 675
680118680PRTArtificial SequencehIgG2FcdeltaK-CC-T2-T2bAA-T2bAA
fusion 118Val Glu Cys Pro Pro Cys Pro Ala Pro Pro Val Ala Gly Pro
Ser Val1 5 10 15Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile
Ser Arg Thr 20 25 30Pro Glu Val Thr Cys Val Val Val Asp Val Ser His
Glu Asp Pro Glu 35 40 45Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu
Val His Asn Ala Lys 50 55 60Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser
Thr Phe Arg Val Val Ser65 70 75 80Val Leu Thr Val Val His Gln Asp
Trp Leu Asn Gly Lys Glu Tyr Lys 85 90 95Cys Lys Val Ser Asn Lys Gly
Leu Pro Ala Pro Ile Glu Lys Thr Ile 100 105 110Ser Lys Thr Lys Gly
Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro 115 120 125Pro Ser Arg
Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu 130 135 140Val
Lys Gly Phe Tyr Pro Ser Asp Ile Ser Val Glu Trp Glu Ser Asn145 150
155 160Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp
Ser 165 170 175Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp
Lys Ser Arg 180 185 190Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val
Met His Glu Ala Leu 195 200 205His Asn His Tyr Thr Gln Lys Ser Leu
Ser Leu Ser Pro Gly Ile Pro 210 215 220Pro His Val Gln Lys Ser Val
Asn Asn Asp Met Ile Val Thr Asp Asn225 230 235 240Asn Gly Ala Val
Lys Phe Pro Gln Leu Cys Lys Phe Cys Asp Val Arg 245 250 255Phe Ser
Thr Cys Asp Asn Gln Lys Ser Cys Met Ser Asn Cys Ser Ile 260 265
270Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val Trp Arg
275 280
285Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys
290 295 300Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro
Lys Cys305 310 315 320Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr
Phe Phe Met Cys Ser 325 330 335Cys Ser Ser Asp Glu Cys Asn Asp Asn
Ile Ile Phe Ser Glu Glu Tyr 340 345 350Asn Thr Ser Asn Pro Asp Ile
Pro Pro His Val Gln Lys Ser Asp Val 355 360 365Glu Met Glu Ala Gln
Lys Asp Glu Ile Ile Ala Pro Ser Ala Asn Arg 370 375 380Thr Ala His
Pro Leu Arg His Ile Asn Asn Asp Met Ile Val Thr Asp385 390 395
400Asn Asn Gly Ala Val Lys Phe Pro Gln Leu Cys Lys Phe Cys Asp Val
405 410 415Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys Met Ser Asn
Cys Ser 420 425 430Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys
Val Ala Val Trp 435 440 445Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu
Thr Val Cys His Asp Pro 450 455 460Lys Leu Pro Tyr His Asp Phe Ile
Leu Glu Asp Ala Ala Ser Pro Lys465 470 475 480Cys Ile Met Lys Glu
Lys Lys Lys Pro Gly Glu Thr Phe Phe Met Cys 485 490 495Ser Cys Ser
Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu 500 505 510Tyr
Asn Thr Ser Asn Pro Asp Ile Pro Pro His Val Gln Lys Ser Asp 515 520
525Val Glu Met Glu Ala Gln Lys Asp Glu Ile Ile Ala Pro Ser Ala Asn
530 535 540Arg Thr Ala His Pro Leu Arg His Ile Asn Asn Asp Met Ile
Val Thr545 550 555 560Asp Asn Asn Gly Ala Val Lys Phe Pro Gln Leu
Cys Lys Phe Cys Asp 565 570 575Val Arg Phe Ser Thr Cys Asp Asn Gln
Lys Ser Cys Met Ser Asn Cys 580 585 590Ser Ile Thr Ser Ile Cys Glu
Lys Pro Gln Glu Val Cys Val Ala Val 595 600 605Trp Arg Lys Asn Asp
Glu Asn Ile Thr Leu Glu Thr Val Cys His Asp 610 615 620Pro Lys Leu
Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro625 630 635
640Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met
645 650 655Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe
Ser Glu 660 665 670Glu Tyr Asn Thr Ser Asn Pro Asp 675
680119686PRTArtificial SequencehIgG4FcdeltaK-CC-228P-T2-T2bAA-T2bAA
fusion 119Glu Ser Lys Tyr Gly Pro Pro Cys Pro Pro Cys Pro Ala Pro
Glu Phe1 5 10 15Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro
Lys Asp Thr 20 25 30Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val
Val Val Asp Val 35 40 45Ser Gln Glu Asp Pro Glu Val Gln Phe Asn Trp
Tyr Val Asp Gly Val 50 55 60Glu Val His Asn Ala Lys Thr Lys Pro Arg
Glu Glu Gln Phe Asn Ser65 70 75 80Thr Tyr Arg Val Val Ser Val Leu
Thr Val Leu His Gln Asp Trp Leu 85 90 95Asn Gly Lys Glu Tyr Lys Cys
Lys Val Ser Asn Lys Gly Leu Pro Ser 100 105 110Ser Ile Glu Lys Thr
Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro 115 120 125Gln Val Tyr
Thr Leu Pro Pro Ser Gln Glu Glu Met Thr Lys Asn Gln 130 135 140Val
Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala145 150
155 160Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr
Thr 165 170 175Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr
Ser Arg Leu 180 185 190Thr Val Asp Lys Ser Arg Trp Gln Glu Gly Asn
Val Phe Ser Cys Ser 195 200 205Val Met His Glu Ala Leu His Asn His
Tyr Thr Gln Lys Ser Leu Ser 210 215 220Leu Ser Leu Gly Ile Pro Pro
His Val Gln Lys Ser Val Asn Asn Asp225 230 235 240Met Ile Val Thr
Asp Asn Asn Gly Ala Val Lys Phe Pro Gln Leu Cys 245 250 255Lys Phe
Cys Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys 260 265
270Met Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val
275 280 285Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu
Glu Thr 290 295 300Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe
Ile Leu Glu Asp305 310 315 320Ala Ala Ser Pro Lys Cys Ile Met Lys
Glu Lys Lys Lys Pro Gly Glu 325 330 335Thr Phe Phe Met Cys Ser Cys
Ser Ser Asp Glu Cys Asn Asp Asn Ile 340 345 350Ile Phe Ser Glu Glu
Tyr Asn Thr Ser Asn Pro Asp Ile Pro Pro His 355 360 365Val Gln Lys
Ser Asp Val Glu Met Glu Ala Gln Lys Asp Glu Ile Ile 370 375 380Ala
Pro Ser Ala Asn Arg Thr Ala His Pro Leu Arg His Ile Asn Asn385 390
395 400Asp Met Ile Val Thr Asp Asn Asn Gly Ala Val Lys Phe Pro Gln
Leu 405 410 415Cys Lys Phe Cys Asp Val Arg Phe Ser Thr Cys Asp Asn
Gln Lys Ser 420 425 430Cys Met Ser Asn Cys Ser Ile Thr Ser Ile Cys
Glu Lys Pro Gln Glu 435 440 445Val Cys Val Ala Val Trp Arg Lys Asn
Asp Glu Asn Ile Thr Leu Glu 450 455 460Thr Val Cys His Asp Pro Lys
Leu Pro Tyr His Asp Phe Ile Leu Glu465 470 475 480Asp Ala Ala Ser
Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly 485 490 495Glu Thr
Phe Phe Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn 500 505
510Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp Ile Pro Pro
515 520 525His Val Gln Lys Ser Asp Val Glu Met Glu Ala Gln Lys Asp
Glu Ile 530 535 540Ile Ala Pro Ser Ala Asn Arg Thr Ala His Pro Leu
Arg His Ile Asn545 550 555 560Asn Asp Met Ile Val Thr Asp Asn Asn
Gly Ala Val Lys Phe Pro Gln 565 570 575Leu Cys Lys Phe Cys Asp Val
Arg Phe Ser Thr Cys Asp Asn Gln Lys 580 585 590Ser Cys Met Ser Asn
Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln 595 600 605Glu Val Cys
Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu 610 615 620Glu
Thr Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu625 630
635 640Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys
Pro 645 650 655Gly Glu Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu
Cys Asn Asp 660 665 670Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser
Asn Pro Asp 675 680 685120686PRTArtificial
SequencehIgG4FcdeltaK-CC-228P-409K-T2-T2bAA-T2bAA fusion 120Glu Ser
Lys Tyr Gly Pro Pro Cys Pro Pro Cys Pro Ala Pro Glu Phe1 5 10 15Leu
Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr 20 25
30Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val
35 40 45Ser Gln Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr Val Asp Gly
Val 50 55 60Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Phe
Asn Ser65 70 75 80Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His
Gln Asp Trp Leu 85 90 95Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn
Lys Gly Leu Pro Ser 100 105 110Ser Ile Glu Lys Thr Ile Ser Lys Ala
Lys Gly Gln Pro Arg Glu Pro 115 120 125Gln Val Tyr Thr Leu Pro Pro
Ser Gln Glu Glu Met Thr Lys Asn Gln 130 135 140Val Ser Leu Thr Cys
Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala145 150 155 160Val Glu
Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr 165 170
175Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu
180 185 190Thr Val Asp Lys Ser Arg Trp Gln Glu Gly Asn Val Phe Ser
Cys Ser 195 200 205Val Met His Glu Ala Leu His Asn His Tyr Thr Gln
Lys Ser Leu Ser 210 215 220Leu Ser Leu Gly Ile Pro Pro His Val Gln
Lys Ser Val Asn Asn Asp225 230 235 240Met Ile Val Thr Asp Asn Asn
Gly Ala Val Lys Phe Pro Gln Leu Cys 245 250 255Lys Phe Cys Asp Val
Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys 260 265 270Met Ser Asn
Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val 275 280 285Cys
Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr 290 295
300Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu
Asp305 310 315 320Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys
Lys Pro Gly Glu 325 330 335Thr Phe Phe Met Cys Ser Cys Ser Ser Asp
Glu Cys Asn Asp Asn Ile 340 345 350Ile Phe Ser Glu Glu Tyr Asn Thr
Ser Asn Pro Asp Ile Pro Pro His 355 360 365Val Gln Lys Ser Asp Val
Glu Met Glu Ala Gln Lys Asp Glu Ile Ile 370 375 380Ala Pro Ser Ala
Asn Arg Thr Ala His Pro Leu Arg His Ile Asn Asn385 390 395 400Asp
Met Ile Val Thr Asp Asn Asn Gly Ala Val Lys Phe Pro Gln Leu 405 410
415Cys Lys Phe Cys Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser
420 425 430Cys Met Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro
Gln Glu 435 440 445Val Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn
Ile Thr Leu Glu 450 455 460Thr Val Cys His Asp Pro Lys Leu Pro Tyr
His Asp Phe Ile Leu Glu465 470 475 480Asp Ala Ala Ser Pro Lys Cys
Ile Met Lys Glu Lys Lys Lys Pro Gly 485 490 495Glu Thr Phe Phe Met
Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn 500 505 510Ile Ile Phe
Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp Ile Pro Pro 515 520 525His
Val Gln Lys Ser Asp Val Glu Met Glu Ala Gln Lys Asp Glu Ile 530 535
540Ile Ala Pro Ser Ala Asn Arg Thr Ala His Pro Leu Arg His Ile
Asn545 550 555 560Asn Asp Met Ile Val Thr Asp Asn Asn Gly Ala Val
Lys Phe Pro Gln 565 570 575Leu Cys Lys Phe Cys Asp Val Arg Phe Ser
Thr Cys Asp Asn Gln Lys 580 585 590Ser Cys Met Ser Asn Cys Ser Ile
Thr Ser Ile Cys Glu Lys Pro Gln 595 600 605Glu Val Cys Val Ala Val
Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu 610 615 620Glu Thr Val Cys
His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu625 630 635 640Glu
Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro 645 650
655Gly Glu Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp
660 665 670Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp
675 680 685121214PRTArtificial SequenceCetuximab LC 121Asp Ile Leu
Leu Thr Gln Ser Pro Val Ile Leu Ser Val Ser Pro Gly1 5 10 15Glu Arg
Val Ser Phe Ser Cys Arg Ala Ser Gln Ser Ile Gly Thr Asn 20 25 30Ile
His Trp Tyr Gln Gln Arg Thr Asn Gly Ser Pro Arg Leu Leu Ile 35 40
45Lys Tyr Ala Ser Glu Ser Ile Ser Gly Ile Pro Ser Arg Phe Ser Gly
50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Ser Ile Asn Ser Val Glu
Ser65 70 75 80Glu Asp Ile Ala Asp Tyr Tyr Cys Gln Gln Asn Asn Asn
Trp Pro Thr 85 90 95Thr Phe Gly Ala Gly Thr Lys Leu Glu Leu Lys Arg
Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp
Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu Leu
Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys Val
Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser Val
Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser
Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185
190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser
195 200 205Phe Asn Arg Gly Glu Cys 210122721PRTArtificial
SequenceCetuximab HC-T22d35 122Gln Val Gln Leu Lys Gln Ser Gly Pro
Gly Leu Val Gln Pro Ser Gln1 5 10 15Ser Leu Ser Ile Thr Cys Thr Val
Ser Gly Phe Ser Leu Thr Asn Tyr 20 25 30Gly Val His Trp Val Arg Gln
Ser Pro Gly Lys Gly Leu Glu Trp Leu 35 40 45Gly Val Ile Trp Ser Gly
Gly Asn Thr Asp Tyr Asn Thr Pro Phe Thr 50 55 60Ser Arg Leu Ser Ile
Asn Lys Asp Asn Ser Lys Ser Gln Val Phe Phe65 70 75 80Lys Met Asn
Ser Leu Gln Ser Asn Asp Thr Ala Ile Tyr Tyr Cys Ala 85 90 95Arg Ala
Leu Thr Tyr Tyr Asp Tyr Glu Phe Ala Tyr Trp Gly Gln Gly 100 105
110Thr Leu Val Thr Val Ser Ala Ala Ser Thr Lys Gly Pro Ser Val Phe
115 120 125Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala
Ala Leu 130 135 140Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val
Thr Val Ser Trp145 150 155 160Asn Ser Gly Ala Leu Thr Ser Gly Val
His Thr Phe Pro Ala Val Leu 165 170 175Gln Ser Ser Gly Leu Tyr Ser
Leu Ser Ser Val Val Thr Val Pro Ser 180 185 190Ser Ser Leu Gly Thr
Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro 195 200 205Ser Asn Thr
Lys Val Asp Lys Arg Val Glu Pro Lys Ser Cys Asp Lys 210 215 220Thr
His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro225 230
235 240Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile
Ser 245 250 255Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser
His Glu Asp 260 265 270Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly
Val Glu Val His Asn 275 280 285Ala Lys Thr Lys Pro Arg Glu Glu Gln
Tyr Asn Ser Thr Tyr Arg Val 290 295 300Val Ser Val Leu Thr Val Leu
His Gln Asp Trp Leu Asn Gly Lys Glu305 310 315 320Tyr Lys Cys Lys
Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys 325 330 335Thr Ile
Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr 340 345
350Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr
355 360 365Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu
Trp Glu 370 375 380Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr
Pro Pro Val Leu385 390 395 400Asp Ser Asp Gly Ser Phe Phe Leu Tyr
Ser Lys Leu Thr Val Asp Lys 405 410 415Ser Arg Trp Gln Gln Gly Asn
Val Phe Ser Cys Ser Val Met His Glu 420 425 430Ala Leu His Asn His
Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly 435 440 445Lys Ile Pro
Pro His Val Gln Lys Ser Val Asn Asn Asp Met
Ile Val 450 455 460Thr Asp Asn Asn Gly Ala Val Lys Phe Pro Gln Leu
Cys Lys Phe Cys465 470 475 480Asp Val Arg Phe Ser Thr Cys Asp Asn
Gln Lys Ser Cys Met Ser Asn 485 490 495Cys Ser Ile Thr Ser Ile Cys
Glu Lys Pro Gln Glu Val Cys Val Ala 500 505 510Val Trp Arg Lys Asn
Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His 515 520 525Asp Pro Lys
Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser 530 535 540Pro
Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe545 550
555 560Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe
Ser 565 570 575Glu Glu Tyr Asn Thr Ser Asn Pro Asp Ile Pro Pro His
Val Gln Lys 580 585 590Ser Val Asn Asn Asp Met Ile Val Thr Asp Asn
Asn Gly Ala Val Lys 595 600 605Phe Pro Gln Leu Cys Lys Phe Cys Asp
Val Arg Phe Ser Thr Cys Asp 610 615 620Asn Gln Lys Ser Cys Met Ser
Asn Cys Ser Ile Thr Ser Ile Cys Glu625 630 635 640Lys Pro Gln Glu
Val Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn 645 650 655Ile Thr
Leu Glu Thr Val Cys His Asp Pro Lys Leu Pro Tyr His Asp 660 665
670Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys
675 680 685Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser Ser
Asp Glu 690 695 700Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr Asn
Thr Ser Asn Pro705 710 715 720Asp123585PRTArtificial
SequenceCetuximab HC-T2m 123Gln Val Gln Leu Lys Gln Ser Gly Pro Gly
Leu Val Gln Pro Ser Gln1 5 10 15Ser Leu Ser Ile Thr Cys Thr Val Ser
Gly Phe Ser Leu Thr Asn Tyr 20 25 30Gly Val His Trp Val Arg Gln Ser
Pro Gly Lys Gly Leu Glu Trp Leu 35 40 45Gly Val Ile Trp Ser Gly Gly
Asn Thr Asp Tyr Asn Thr Pro Phe Thr 50 55 60Ser Arg Leu Ser Ile Asn
Lys Asp Asn Ser Lys Ser Gln Val Phe Phe65 70 75 80Lys Met Asn Ser
Leu Gln Ser Asn Asp Thr Ala Ile Tyr Tyr Cys Ala 85 90 95Arg Ala Leu
Thr Tyr Tyr Asp Tyr Glu Phe Ala Tyr Trp Gly Gln Gly 100 105 110Thr
Leu Val Thr Val Ser Ala Ala Ser Thr Lys Gly Pro Ser Val Phe 115 120
125Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala Leu
130 135 140Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val
Ser Trp145 150 155 160Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr
Phe Pro Ala Val Leu 165 170 175Gln Ser Ser Gly Leu Tyr Ser Leu Ser
Ser Val Val Thr Val Pro Ser 180 185 190Ser Ser Leu Gly Thr Gln Thr
Tyr Ile Cys Asn Val Asn His Lys Pro 195 200 205Ser Asn Thr Lys Val
Asp Lys Arg Val Glu Pro Lys Ser Cys Asp Lys 210 215 220Thr His Thr
Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro225 230 235
240Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser
245 250 255Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His
Glu Asp 260 265 270Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val
Glu Val His Asn 275 280 285Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr
Asn Ser Thr Tyr Arg Val 290 295 300Val Ser Val Leu Thr Val Leu His
Gln Asp Trp Leu Asn Gly Lys Glu305 310 315 320Tyr Lys Cys Lys Val
Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys 325 330 335Thr Ile Ser
Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr 340 345 350Leu
Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr 355 360
365Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu
370 375 380Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro
Val Leu385 390 395 400Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys
Leu Thr Val Asp Lys 405 410 415Ser Arg Trp Gln Gln Gly Asn Val Phe
Ser Cys Ser Val Met His Glu 420 425 430Ala Leu His Asn His Tyr Thr
Gln Lys Ser Leu Ser Leu Ser Pro Gly 435 440 445Lys Ile Pro Pro His
Val Gln Lys Ser Val Asn Asn Asp Met Ile Val 450 455 460Thr Asp Asn
Asn Gly Ala Val Lys Phe Pro Gln Leu Cys Lys Phe Cys465 470 475
480Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys Met Ser Asn
485 490 495Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys
Val Ala 500 505 510Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu
Thr Val Cys His 515 520 525Asp Pro Lys Leu Pro Tyr His Asp Phe Ile
Leu Glu Asp Ala Ala Ser 530 535 540Pro Lys Cys Ile Met Lys Glu Lys
Lys Lys Pro Gly Glu Thr Phe Phe545 550 555 560Met Cys Ser Cys Ser
Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser 565 570 575Glu Glu Tyr
Asn Thr Ser Asn Pro Asp 580 585124214PRTArtificial
SequenceHerceptin LC 124Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu
Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser
Gln Asp Val Asn Thr Ala 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly
Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ser Ala Ser Phe Leu Tyr Ser
Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Arg Ser Gly Thr Asp Phe
Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr
Tyr Tyr Cys Gln Gln His Tyr Thr Thr Pro Pro 85 90 95Thr Phe Gly Gln
Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser
Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly 115 120
125Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala
130 135 140Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn
Ser Gln145 150 155 160Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser
Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys Ala Asp
Tyr Glu Lys His Lys Val Tyr 180 185 190Ala Cys Glu Val Thr His Gln
Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200 205Phe Asn Arg Gly Glu
Cys 210125722PRTArtificial SequenceHerceptin HC-T22d35 125Glu Val
Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser
Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Asn Ile Lys Asp Thr 20 25
30Tyr Ile His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val
35 40 45Ala Arg Ile Tyr Pro Thr Asn Gly Tyr Thr Arg Tyr Ala Asp Ser
Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Ala Asp Thr Ser Lys Asn Thr
Ala Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala
Val Tyr Tyr Cys 85 90 95Ser Arg Trp Gly Gly Asp Gly Phe Tyr Ala Met
Asp Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser Ala
Ser Thr Lys Gly Pro Ser Val 115 120 125Phe Pro Leu Ala Pro Ser Ser
Lys Ser Thr Ser Gly Gly Thr Ala Ala 130 135 140Leu Gly Cys Leu Val
Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser145 150 155 160Trp Asn
Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val 165 170
175Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro
180 185 190Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn
His Lys 195 200 205Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro
Lys Ser Cys Asp 210 215 220Lys Thr His Thr Cys Pro Pro Cys Pro Ala
Pro Glu Leu Leu Gly Gly225 230 235 240Pro Ser Val Phe Leu Phe Pro
Pro Lys Pro Lys Asp Thr Leu Met Ile 245 250 255Ser Arg Thr Pro Glu
Val Thr Cys Val Val Val Asp Val Ser His Glu 260 265 270Asp Pro Glu
Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His 275 280 285Asn
Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg 290 295
300Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly
Lys305 310 315 320Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro
Ala Pro Ile Glu 325 330 335Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro
Arg Glu Pro Gln Val Tyr 340 345 350Thr Leu Pro Pro Ser Arg Asp Glu
Leu Thr Lys Asn Gln Val Ser Leu 355 360 365Thr Cys Leu Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp 370 375 380Glu Ser Asn Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val385 390 395 400Leu
Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp 405 410
415Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His
420 425 430Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu
Ser Pro 435 440 445Gly Lys Ile Pro Pro His Val Gln Lys Ser Val Asn
Asn Asp Met Ile 450 455 460Val Thr Asp Asn Asn Gly Ala Val Lys Phe
Pro Gln Leu Cys Lys Phe465 470 475 480Cys Asp Val Arg Phe Ser Thr
Cys Asp Asn Gln Lys Ser Cys Met Ser 485 490 495Asn Cys Ser Ile Thr
Ser Ile Cys Glu Lys Pro Gln Glu Val Cys Val 500 505 510Ala Val Trp
Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys 515 520 525His
Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala 530 535
540Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr
Phe545 550 555 560Phe Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp
Asn Ile Ile Phe 565 570 575Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp
Ile Pro Pro His Val Gln 580 585 590Lys Ser Val Asn Asn Asp Met Ile
Val Thr Asp Asn Asn Gly Ala Val 595 600 605Lys Phe Pro Gln Leu Cys
Lys Phe Cys Asp Val Arg Phe Ser Thr Cys 610 615 620Asp Asn Gln Lys
Ser Cys Met Ser Asn Cys Ser Ile Thr Ser Ile Cys625 630 635 640Glu
Lys Pro Gln Glu Val Cys Val Ala Val Trp Arg Lys Asn Asp Glu 645 650
655Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys Leu Pro Tyr His
660 665 670Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met
Lys Glu 675 680 685Lys Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser
Cys Ser Ser Asp 690 695 700Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu
Glu Tyr Asn Thr Ser Asn705 710 715 720Pro Asp126214PRTArtificial
SequenceAvastin LC 126Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu
Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Ser Ala Ser
Gln Asp Ile Ser Asn Tyr 20 25 30Leu Asn Trp Tyr Gln Gln Lys Pro Gly
Lys Ala Pro Lys Val Leu Ile 35 40 45Tyr Phe Thr Ser Ser Leu His Ser
Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe
Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr
Tyr Tyr Cys Gln Gln Tyr Ser Thr Val Pro Trp 85 90 95Thr Phe Gly Gln
Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser
Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly 115 120
125Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala
130 135 140Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn
Ser Gln145 150 155 160Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser
Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys Ala Asp
Tyr Glu Lys His Lys Val Tyr 180 185 190Ala Cys Glu Val Thr His Gln
Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200 205Phe Asn Arg Gly Glu
Cys 210127725PRTArtificial SequenceAvastin HC-T22d35 127Glu Val Gln
Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Tyr Thr Phe Thr Asn Tyr 20 25 30Gly
Met Asn Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Gly Trp Ile Asn Thr Tyr Thr Gly Glu Pro Thr Tyr Ala Ala Asp Phe
50 55 60Lys Arg Arg Phe Thr Phe Ser Leu Asp Thr Ser Lys Ser Thr Ala
Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val
Tyr Tyr Cys 85 90 95Ala Lys Tyr Pro His Tyr Tyr Gly Ser Ser His Trp
Tyr Phe Asp Val 100 105 110Trp Gly Gln Gly Thr Leu Val Thr Val Ser
Ser Ala Ser Thr Lys Gly 115 120 125Pro Ser Val Phe Pro Leu Ala Pro
Ser Ser Lys Ser Thr Ser Gly Gly 130 135 140Thr Ala Ala Leu Gly Cys
Leu Val Lys Asp Tyr Phe Pro Glu Pro Val145 150 155 160Thr Val Ser
Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe 165 170 175Pro
Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val 180 185
190Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val
195 200 205Asn His Lys Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu
Pro Lys 210 215 220Ser Cys Asp Lys Thr His Thr Cys Pro Pro Cys Pro
Ala Pro Glu Leu225 230 235 240Leu Gly Gly Pro Ser Val Phe Leu Phe
Pro Pro Lys Pro Lys Asp Thr 245 250 255Leu Met Ile Ser Arg Thr Pro
Glu Val Thr Cys Val Val Val Asp Val 260 265 270Ser His Glu Asp Pro
Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val 275 280 285Glu Val His
Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser 290 295 300Thr
Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu305 310
315 320Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro
Ala 325 330 335Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro
Arg Glu Pro 340 345 350Gln Val Tyr Thr Leu Pro Pro Ser Arg Asp Glu
Leu Thr Lys Asn Gln 355 360 365Val Ser Leu Thr Cys Leu Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala 370 375 380Val Glu Trp Glu Ser Asn Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr385 390 395 400Pro Pro Val Leu
Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu 405 410 415Thr Val
Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser 420 425
430Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser
435 440
445Leu Ser Pro Gly Lys Ile Pro Pro His Val Gln Lys Ser Val Asn Asn
450 455 460Asp Met Ile Val Thr Asp Asn Asn Gly Ala Val Lys Phe Pro
Gln Leu465 470 475 480Cys Lys Phe Cys Asp Val Arg Phe Ser Thr Cys
Asp Asn Gln Lys Ser 485 490 495Cys Met Ser Asn Cys Ser Ile Thr Ser
Ile Cys Glu Lys Pro Gln Glu 500 505 510Val Cys Val Ala Val Trp Arg
Lys Asn Asp Glu Asn Ile Thr Leu Glu 515 520 525Thr Val Cys His Asp
Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu 530 535 540Asp Ala Ala
Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly545 550 555
560Glu Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn
565 570 575Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp Ile
Pro Pro 580 585 590His Val Gln Lys Ser Val Asn Asn Asp Met Ile Val
Thr Asp Asn Asn 595 600 605Gly Ala Val Lys Phe Pro Gln Leu Cys Lys
Phe Cys Asp Val Arg Phe 610 615 620Ser Thr Cys Asp Asn Gln Lys Ser
Cys Met Ser Asn Cys Ser Ile Thr625 630 635 640Ser Ile Cys Glu Lys
Pro Gln Glu Val Cys Val Ala Val Trp Arg Lys 645 650 655Asn Asp Glu
Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys Leu 660 665 670Pro
Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile 675 680
685Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser Cys
690 695 700Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu
Tyr Asn705 710 715 720Thr Ser Asn Pro Asp 725128213PRTArtificial
SequenceSynagis LC 128Asp Ile Gln Met Thr Gln Ser Pro Ser Thr Leu
Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Lys Cys Gln
Leu Ser Val Gly Tyr Met 20 25 30His Trp Tyr Gln Gln Lys Pro Gly Lys
Ala Pro Lys Leu Leu Ile Tyr 35 40 45Asp Thr Ser Lys Leu Ala Ser Gly
Val Pro Ser Arg Phe Ser Gly Ser 50 55 60Gly Ser Gly Thr Glu Phe Thr
Leu Thr Ile Ser Ser Leu Gln Pro Asp65 70 75 80Asp Phe Ala Thr Tyr
Tyr Cys Phe Gln Gly Ser Gly Tyr Pro Phe Thr 85 90 95Phe Gly Gly Gly
Thr Lys Leu Glu Ile Lys Arg Thr Val Ala Ala Pro 100 105 110Ser Val
Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly Thr 115 120
125Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala Lys
130 135 140Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser
Gln Glu145 150 155 160Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr
Tyr Ser Leu Ser Ser 165 170 175Thr Leu Thr Leu Ser Lys Ala Asp Tyr
Glu Lys His Lys Val Tyr Ala 180 185 190Cys Glu Val Thr His Gln Gly
Leu Ser Ser Pro Val Thr Lys Ser Phe 195 200 205Asn Arg Gly Glu Cys
210129721PRTArtificial SequenceSynagis HC-T22d35 129Gln Val Thr Leu
Arg Glu Ser Gly Pro Ala Leu Val Lys Pro Thr Gln1 5 10 15Thr Leu Thr
Leu Thr Cys Thr Phe Ser Gly Phe Ser Leu Ser Thr Ser 20 25 30Gly Met
Ser Val Gly Trp Ile Arg Gln Pro Pro Gly Lys Ala Leu Glu 35 40 45Trp
Leu Ala Asp Ile Trp Trp Asp Asp Lys Lys Asp Tyr Asn Pro Ser 50 55
60Leu Lys Ser Arg Leu Thr Ile Ser Lys Asp Thr Ser Lys Asn Gln Val65
70 75 80Val Leu Lys Val Thr Asn Met Asp Pro Ala Asp Thr Ala Thr Tyr
Tyr 85 90 95Cys Ala Arg Ser Met Ile Thr Asn Trp Tyr Phe Asp Val Trp
Gly Ala 100 105 110Gly Thr Thr Val Thr Val Ser Ser Ala Ser Thr Lys
Gly Pro Ser Val 115 120 125Phe Pro Leu Ala Pro Ser Ser Ala Ala Ala
Ala Gly Gly Thr Ala Ala 130 135 140Leu Gly Cys Leu Val Lys Asp Tyr
Phe Pro Glu Pro Val Thr Val Ser145 150 155 160Trp Asn Ser Gly Ala
Leu Thr Ser Gly Val His Thr Phe Pro Ala Val 165 170 175Leu Gln Ser
Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro 180 185 190Ser
Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys 195 200
205Pro Ser Asn Thr Lys Val Asp Lys Arg Val Glu Pro Lys Ser Cys Asp
210 215 220Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu
Gly Gly225 230 235 240Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys
Asp Thr Leu Met Ile 245 250 255Ser Arg Thr Pro Glu Val Thr Cys Val
Val Val Asp Val Ser His Glu 260 265 270Asp Pro Glu Val Lys Phe Asn
Trp Tyr Val Asp Gly Val Glu Val His 275 280 285Asn Ala Lys Thr Lys
Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg 290 295 300Val Val Ser
Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys305 310 315
320Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu
325 330 335Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln
Val Tyr 340 345 350Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn
Gln Val Ser Leu 355 360 365Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser
Asp Ile Ala Val Glu Trp 370 375 380Glu Ser Asn Gly Gln Pro Glu Asn
Asn Tyr Lys Thr Thr Pro Pro Val385 390 395 400Leu Asp Ser Asp Gly
Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp 405 410 415Lys Ser Arg
Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His 420 425 430Glu
Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro 435 440
445Gly Ile Pro Pro His Val Gln Lys Ser Val Asn Asn Asp Met Ile Val
450 455 460Thr Asp Asn Asn Gly Ala Val Lys Phe Pro Gln Leu Cys Lys
Phe Cys465 470 475 480Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys
Ser Cys Met Ser Asn 485 490 495Cys Ser Ile Thr Ser Ile Cys Glu Lys
Pro Gln Glu Val Cys Val Ala 500 505 510Val Trp Arg Lys Asn Asp Glu
Asn Ile Thr Leu Glu Thr Val Cys His 515 520 525Asp Pro Lys Leu Pro
Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser 530 535 540Pro Lys Cys
Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe545 550 555
560Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser
565 570 575Glu Glu Tyr Asn Thr Ser Asn Pro Asp Ile Pro Pro His Val
Gln Lys 580 585 590Ser Val Asn Asn Asp Met Ile Val Thr Asp Asn Asn
Gly Ala Val Lys 595 600 605Phe Pro Gln Leu Cys Lys Phe Cys Asp Val
Arg Phe Ser Thr Cys Asp 610 615 620Asn Gln Lys Ser Cys Met Ser Asn
Cys Ser Ile Thr Ser Ile Cys Glu625 630 635 640Lys Pro Gln Glu Val
Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn 645 650 655Ile Thr Leu
Glu Thr Val Cys His Asp Pro Lys Leu Pro Tyr His Asp 660 665 670Phe
Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys 675 680
685Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu
690 695 700Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser
Asn Pro705 710 715 720Asp130630PRTArtificial SequenceFC5-Fc-T22d35
130Glu Val Gln Leu Gln Ala Ser Gly Gly Gly Leu Val Gln Ala Gly Gly1
5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Lys Ile Thr His
Tyr 20 25 30Thr Met Gly Trp Phe Arg Gln Ala Pro Gly Lys Glu Arg Glu
Phe Val 35 40 45Ser Arg Ile Thr Trp Gly Gly Asp Asn Thr Phe Tyr Ser
Asn Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys
Asn Thr Val Tyr65 70 75 80Leu Gln Met Asn Ser Leu Lys Pro Glu Asp
Thr Ala Asp Tyr Tyr Cys 85 90 95Ala Ala Gly Ser Thr Ser Thr Ala Thr
Pro Leu Arg Val Asp Tyr Trp 100 105 110Gly Lys Gly Thr Gln Val Thr
Val Ser Ser Ala Ser Glu Pro Arg Gly 115 120 125Pro Thr Ile Lys Pro
Cys Pro Pro Cys Lys Cys Pro Ala Pro Asn Leu 130 135 140Leu Gly Gly
Pro Ser Val Phe Ile Phe Pro Pro Lys Ile Lys Asp Val145 150 155
160Leu Met Ile Ser Leu Ser Pro Ile Val Thr Cys Val Val Val Asp Val
165 170 175Ser Glu Asp Asp Pro Asp Val Gln Ile Ser Trp Phe Val Asn
Asn Val 180 185 190Glu Val His Thr Ala Gln Thr Gln Thr His Arg Glu
Asp Tyr Asn Ser 195 200 205Thr Leu Arg Val Val Ser Ala Leu Pro Ile
Gln His Gln Asp Trp Met 210 215 220Ser Gly Lys Glu Phe Lys Cys Lys
Val Asn Asn Lys Asp Leu Pro Ala225 230 235 240Pro Ile Glu Arg Thr
Ile Ser Lys Pro Lys Gly Ser Val Arg Ala Pro 245 250 255Gln Val Tyr
Val Leu Pro Pro Pro Glu Glu Glu Met Thr Lys Lys Gln 260 265 270Val
Thr Leu Thr Cys Met Val Thr Asp Phe Met Pro Glu Asp Ile Tyr 275 280
285Val Glu Trp Thr Asn Asn Gly Lys Thr Glu Leu Asn Tyr Lys Asn Thr
290 295 300Glu Pro Val Leu Asp Ser Asp Gly Ser Tyr Phe Met Tyr Ser
Lys Leu305 310 315 320Arg Val Glu Lys Lys Asn Trp Val Glu Arg Asn
Ser Tyr Ser Cys Ser 325 330 335Val Val His Glu Gly Leu His Asn His
His Thr Thr Lys Ser Phe Ser 340 345 350Arg Thr Pro Gly Thr Gly Ile
Pro Pro His Val Gln Lys Ser Val Asn 355 360 365Asn Asp Met Ile Val
Thr Asp Asn Asn Gly Ala Val Lys Phe Pro Gln 370 375 380Leu Cys Lys
Phe Cys Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys385 390 395
400Ser Cys Met Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln
405 410 415Glu Val Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile
Thr Leu 420 425 430Glu Thr Val Cys His Asp Pro Lys Leu Pro Tyr His
Asp Phe Ile Leu 435 440 445Glu Asp Ala Ala Ser Pro Lys Cys Ile Met
Lys Glu Lys Lys Lys Pro 450 455 460Gly Glu Thr Phe Phe Met Cys Ser
Cys Ser Ser Asp Glu Cys Asn Asp465 470 475 480Asn Ile Ile Phe Ser
Glu Glu Tyr Asn Thr Ser Asn Pro Asp Ile Pro 485 490 495Pro His Val
Gln Lys Ser Val Asn Asn Asp Met Ile Val Thr Asp Asn 500 505 510Asn
Gly Ala Val Lys Phe Pro Gln Leu Cys Lys Phe Cys Asp Val Arg 515 520
525Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys Met Ser Asn Cys Ser Ile
530 535 540Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val
Trp Arg545 550 555 560Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val
Cys His Asp Pro Lys 565 570 575Leu Pro Tyr His Asp Phe Ile Leu Glu
Asp Ala Ala Ser Pro Lys Cys 580 585 590Ile Met Lys Glu Lys Lys Lys
Pro Gly Glu Thr Phe Phe Met Cys Ser 595 600 605Cys Ser Ser Asp Glu
Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr 610 615 620Asn Thr Ser
Asn Pro Asp625 630131494PRTArtificial SequenceFC5-Fc-T2m 131Glu Val
Gln Leu Gln Ala Ser Gly Gly Gly Leu Val Gln Ala Gly Gly1 5 10 15Ser
Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Lys Ile Thr His Tyr 20 25
30Thr Met Gly Trp Phe Arg Gln Ala Pro Gly Lys Glu Arg Glu Phe Val
35 40 45Ser Arg Ile Thr Trp Gly Gly Asp Asn Thr Phe Tyr Ser Asn Ser
Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn Thr
Val Tyr65 70 75 80Leu Gln Met Asn Ser Leu Lys Pro Glu Asp Thr Ala
Asp Tyr Tyr Cys 85 90 95Ala Ala Gly Ser Thr Ser Thr Ala Thr Pro Leu
Arg Val Asp Tyr Trp 100 105 110Gly Lys Gly Thr Gln Val Thr Val Ser
Ser Ala Ser Glu Pro Arg Gly 115 120 125Pro Thr Ile Lys Pro Cys Pro
Pro Cys Lys Cys Pro Ala Pro Asn Leu 130 135 140Leu Gly Gly Pro Ser
Val Phe Ile Phe Pro Pro Lys Ile Lys Asp Val145 150 155 160Leu Met
Ile Ser Leu Ser Pro Ile Val Thr Cys Val Val Val Asp Val 165 170
175Ser Glu Asp Asp Pro Asp Val Gln Ile Ser Trp Phe Val Asn Asn Val
180 185 190Glu Val His Thr Ala Gln Thr Gln Thr His Arg Glu Asp Tyr
Asn Ser 195 200 205Thr Leu Arg Val Val Ser Ala Leu Pro Ile Gln His
Gln Asp Trp Met 210 215 220Ser Gly Lys Glu Phe Lys Cys Lys Val Asn
Asn Lys Asp Leu Pro Ala225 230 235 240Pro Ile Glu Arg Thr Ile Ser
Lys Pro Lys Gly Ser Val Arg Ala Pro 245 250 255Gln Val Tyr Val Leu
Pro Pro Pro Glu Glu Glu Met Thr Lys Lys Gln 260 265 270Val Thr Leu
Thr Cys Met Val Thr Asp Phe Met Pro Glu Asp Ile Tyr 275 280 285Val
Glu Trp Thr Asn Asn Gly Lys Thr Glu Leu Asn Tyr Lys Asn Thr 290 295
300Glu Pro Val Leu Asp Ser Asp Gly Ser Tyr Phe Met Tyr Ser Lys
Leu305 310 315 320Arg Val Glu Lys Lys Asn Trp Val Glu Arg Asn Ser
Tyr Ser Cys Ser 325 330 335Val Val His Glu Gly Leu His Asn His His
Thr Thr Lys Ser Phe Ser 340 345 350Arg Thr Pro Gly Thr Gly Ile Pro
Pro His Val Gln Lys Ser Val Asn 355 360 365Asn Asp Met Ile Val Thr
Asp Asn Asn Gly Ala Val Lys Phe Pro Gln 370 375 380Leu Cys Lys Phe
Cys Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys385 390 395 400Ser
Cys Met Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln 405 410
415Glu Val Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu
420 425 430Glu Thr Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe
Ile Leu 435 440 445Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu
Lys Lys Lys Pro 450 455 460Gly Glu Thr Phe Phe Met Cys Ser Cys Ser
Ser Asp Glu Cys Asn Asp465 470 475 480Asn Ile Ile Phe Ser Glu Glu
Tyr Asn Thr Ser Asn Pro Asp 485 490132392PRTArtificial
SequenceT2-hIgG1Fc fusion from R&D, also termed T2m-Fc
(R&D) 132Met Gly Arg Gly Leu Leu Arg Gly Leu Trp Pro Leu His
Ile Val Leu1 5 10 15Trp Thr Arg Ile Ala Ser Thr Ile Pro Pro His Val
Gln Lys Ser Val 20 25 30Asn Asn Asp Met Ile Val Thr Asp Asn Asn Gly
Ala Val Lys Phe Pro 35 40 45Gln Leu Cys Lys Phe Cys Asp Val Arg Phe
Ser Thr Cys Asp Asn Gln 50 55 60Lys Ser Cys Met Ser Asn Cys Ser Ile
Thr Ser Ile Cys Glu Lys Pro65 70 75 80Gln Glu Val Cys Val Ala Val
Trp Arg Lys Asn Asp Glu Asn Ile Thr 85 90 95Leu Glu Thr Val Cys His
Asp Pro Lys Leu Pro Tyr His Asp Phe Ile
100 105 110Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys
Lys Lys 115 120 125Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser Ser
Asp Glu Cys Asn 130 135 140Asp Asn Ile Ile Phe Ser Glu Glu Tyr Asn
Thr Ser Asn Pro Asp Met145 150 155 160Asp Pro Lys Ser Cys Asp Lys
Thr His Thr Cys Pro Pro Cys Pro Ala 165 170 175Pro Glu Leu Leu Gly
Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro 180 185 190Lys Asp Thr
Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val 195 200 205Val
Asp Val Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val 210 215
220Asp Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu
Gln225 230 235 240Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr
Val Leu His Gln 245 250 255Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys
Lys Val Ser Asn Lys Ala 260 265 270Leu Pro Ala Pro Ile Glu Lys Thr
Ile Ser Lys Ala Lys Gly Gln Pro 275 280 285Arg Glu Pro Gln Val Tyr
Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr 290 295 300Lys Asn Gln Val
Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser305 310 315 320Asp
Ile Ala Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr 325 330
335Lys Thr Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr
340 345 350Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn
Val Phe 355 360 365Ser Cys Ser Val Met His Glu Ala Leu His Asn His
Tyr Thr Gln Lys 370 375 380Ser Leu Ser Leu Ser Pro Gly Lys385
390133382PRTArtificial SequenceT2-hIgG2Fc-CCCC fusion, also termed
T2m-Fc 133Met Asp Trp Thr Trp Arg Ile Leu Phe Leu Val Ala Ala Ala
Thr Gly1 5 10 15Thr His Ala Ile Pro Pro His Val Gln Lys Ser Val Asn
Asn Asp Met 20 25 30Ile Val Thr Asp Asn Asn Gly Ala Val Lys Phe Pro
Gln Leu Cys Lys 35 40 45Phe Cys Asp Val Arg Phe Ser Thr Cys Asp Asn
Gln Lys Ser Cys Met 50 55 60Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu
Lys Pro Gln Glu Val Cys65 70 75 80Val Ala Val Trp Arg Lys Asn Asp
Glu Asn Ile Thr Leu Glu Thr Val 85 90 95Cys His Asp Pro Lys Leu Pro
Tyr His Asp Phe Ile Leu Glu Asp Ala 100 105 110Ala Ser Pro Lys Cys
Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr 115 120 125Phe Phe Met
Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile 130 135 140Phe
Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp Glu Arg Lys Cys Cys145 150
155 160Val Glu Cys Pro Pro Cys Pro Ala Pro Pro Val Ala Gly Pro Ser
Val 165 170 175Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile
Ser Arg Thr 180 185 190Pro Glu Val Thr Cys Val Val Val Asp Val Ser
His Glu Asp Pro Glu 195 200 205Val Gln Phe Asn Trp Tyr Val Asp Gly
Val Glu Val His Asn Ala Lys 210 215 220Thr Lys Pro Arg Glu Glu Gln
Phe Asn Ser Thr Phe Arg Val Val Ser225 230 235 240Val Leu Thr Val
Val His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys 245 250 255Cys Lys
Val Ser Asn Lys Gly Leu Pro Ala Pro Ile Glu Lys Thr Ile 260 265
270Ser Lys Thr Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro
275 280 285Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr
Cys Leu 290 295 300Val Lys Gly Phe Tyr Pro Ser Asp Ile Ser Val Glu
Trp Glu Ser Asn305 310 315 320Gly Gln Pro Glu Asn Asn Tyr Lys Thr
Thr Pro Pro Met Leu Asp Ser 325 330 335Asp Gly Ser Phe Phe Leu Tyr
Ser Lys Leu Thr Val Asp Lys Ser Arg 340 345 350Trp Gln Gln Gly Asn
Val Phe Ser Cys Ser Val Met His Glu Ala Leu 355 360 365His Asn His
Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly 370 375
38013415PRTArtificial SequenceIsotope-labelled peptide for T22d35
134Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys1 5
10 151358PRTArtificial SequenceIsotope-labelled peptide for
Cetuximab 135Ala Leu Pro Ala Pro Ile Glu Lys1 5136372PRTArtificial
SequenceD10-hIgG1FcdeltaK-CC-T2m fusion 136Asp Asp Asp Asp Asp Asp
Asp Asp Asp Asp Asp Lys Thr His Thr Cys1 5 10 15Pro Pro Cys Pro Ala
Pro Glu Leu Leu Gly Gly Pro Ser Val Phe Leu 20 25 30Phe Pro Pro Lys
Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu 35 40 45Val Thr Cys
Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Lys 50 55 60Phe Asn
Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys65 70 75
80Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu
85 90 95Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys
Lys 100 105 110Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr
Ile Ser Lys 115 120 125Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr
Thr Leu Pro Pro Ser 130 135 140Arg Asp Glu Leu Thr Lys Asn Gln Val
Ser Leu Thr Cys Leu Val Lys145 150 155 160Gly Phe Tyr Pro Ser Asp
Ile Ala Val Glu Trp Glu Ser Asn Gly Gln 165 170 175Pro Glu Asn Asn
Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser Asp Gly 180 185 190Ser Phe
Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln 195 200
205Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn
210 215 220His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Ile Pro
Pro His225 230 235 240Val Gln Lys Ser Val Asn Asn Asp Met Ile Val
Thr Asp Asn Asn Gly 245 250 255Ala Val Lys Phe Pro Gln Leu Cys Lys
Phe Cys Asp Val Arg Phe Ser 260 265 270Thr Cys Asp Asn Gln Lys Ser
Cys Met Ser Asn Cys Ser Ile Thr Ser 275 280 285Ile Cys Glu Lys Pro
Gln Glu Val Cys Val Ala Val Trp Arg Lys Asn 290 295 300Asp Glu Asn
Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys Leu Pro305 310 315
320Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met
325 330 335Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser
Cys Ser 340 345 350Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu
Glu Tyr Asn Thr 355 360 365Ser Asn Pro Asp 370137376PRTArtificial
SequenceD10-G3S-hIgG1FcdeltaK-CC-T2m fusion 137Asp Asp Asp Asp Asp
Asp Asp Asp Asp Asp Gly Gly Gly Ser Asp Lys1 5 10 15Thr His Thr Cys
Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro 20 25 30Ser Val Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser 35 40 45Arg Thr
Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp 50 55 60Pro
Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn65 70 75
80Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val
85 90 95Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys
Glu 100 105 110Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro
Ile Glu Lys 115 120 125Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr 130 135 140Leu Pro Pro Ser Arg Asp Glu Leu Thr
Lys Asn Gln Val Ser Leu Thr145 150 155 160Cys Leu Val Lys Gly Phe
Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu 165 170 175Ser Asn Gly Gln
Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu 180 185 190Asp Ser
Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys 195 200
205Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu
210 215 220Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
Pro Gly225 230 235 240Ile Pro Pro His Val Gln Lys Ser Val Asn Asn
Asp Met Ile Val Thr 245 250 255Asp Asn Asn Gly Ala Val Lys Phe Pro
Gln Leu Cys Lys Phe Cys Asp 260 265 270Val Arg Phe Ser Thr Cys Asp
Asn Gln Lys Ser Cys Met Ser Asn Cys 275 280 285Ser Ile Thr Ser Ile
Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val 290 295 300Trp Arg Lys
Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His Asp305 310 315
320Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro
325 330 335Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe
Phe Met 340 345 350Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile
Ile Phe Ser Glu 355 360 365Glu Tyr Asn Thr Ser Asn Pro Asp 370
375138366PRTArtificial SequenceD10-hIgG1FcdeltaK-C-T2m fusion
138Asp Asp Asp Asp Asp Asp Asp Asp Asp Asp Pro Pro Cys Pro Ala Pro1
5 10 15Glu Leu Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro
Lys 20 25 30Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val
Val Val 35 40 45Asp Val Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp
Tyr Val Asp 50 55 60Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg
Glu Glu Gln Tyr65 70 75 80Asn Ser Thr Tyr Arg Val Val Ser Val Leu
Thr Val Leu His Gln Asp 85 90 95Trp Leu Asn Gly Lys Glu Tyr Lys Cys
Lys Val Ser Asn Lys Ala Leu 100 105 110Pro Ala Pro Ile Glu Lys Thr
Ile Ser Lys Ala Lys Gly Gln Pro Arg 115 120 125Glu Pro Gln Val Tyr
Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys 130 135 140Asn Gln Val
Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp145 150 155
160Ile Ala Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys
165 170 175Thr Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu
Tyr Ser 180 185 190Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly
Asn Val Phe Ser 195 200 205Cys Ser Val Met His Glu Ala Leu His Asn
His Tyr Thr Gln Lys Ser 210 215 220Leu Ser Leu Ser Pro Gly Ile Pro
Pro His Val Gln Lys Ser Val Asn225 230 235 240Asn Asp Met Ile Val
Thr Asp Asn Asn Gly Ala Val Lys Phe Pro Gln 245 250 255Leu Cys Lys
Phe Cys Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys 260 265 270Ser
Cys Met Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln 275 280
285Glu Val Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu
290 295 300Glu Thr Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe
Ile Leu305 310 315 320Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys
Glu Lys Lys Lys Pro 325 330 335Gly Glu Thr Phe Phe Met Cys Ser Cys
Ser Ser Asp Glu Cys Asn Asp 340 345 350Asn Ile Ile Phe Ser Glu Glu
Tyr Asn Thr Ser Asn Pro Asp 355 360 365139374PRTArtificial
SequenceD10-(G3S)2-hIgG1FcdeltaK-C-T2m fusion 139Asp Asp Asp Asp
Asp Asp Asp Asp Asp Asp Gly Gly Gly Ser Gly Gly1 5 10 15Gly Ser Pro
Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser Val 20 25 30Phe Leu
Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 35 40 45Pro
Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu 50 55
60Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys65
70 75 80Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val
Ser 85 90 95Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu
Tyr Lys 100 105 110Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile
Glu Lys Thr Ile 115 120 125Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro
Gln Val Tyr Thr Leu Pro 130 135 140Pro Ser Arg Asp Glu Leu Thr Lys
Asn Gln Val Ser Leu Thr Cys Leu145 150 155 160Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 165 170 175Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser 180 185 190Asp
Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg 195 200
205Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu
210 215 220His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly
Ile Pro225 230 235 240Pro His Val Gln Lys Ser Val Asn Asn Asp Met
Ile Val Thr Asp Asn 245 250 255Asn Gly Ala Val Lys Phe Pro Gln Leu
Cys Lys Phe Cys Asp Val Arg 260 265 270Phe Ser Thr Cys Asp Asn Gln
Lys Ser Cys Met Ser Asn Cys Ser Ile 275 280 285Thr Ser Ile Cys Glu
Lys Pro Gln Glu Val Cys Val Ala Val Trp Arg 290 295 300Lys Asn Asp
Glu Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys305 310 315
320Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys
325 330 335Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met
Cys Ser 340 345 350Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe
Ser Glu Glu Tyr 355 360 365Asn Thr Ser Asn Pro Asp
370140368PRTArtificial SequenceD10-hIgG2FcdeltaK-CC-T2m fusion
140Asp Asp Asp Asp Asp Asp Asp Asp Asp Asp Val Glu Cys Pro Pro Cys1
5 10 15Pro Ala Pro Pro Val Ala Gly Pro Ser Val Phe Leu Phe Pro Pro
Lys 20 25 30Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr
Cys Val 35 40 45Val Val Asp Val Ser His Glu Asp Pro Glu Val Gln Phe
Asn Trp Tyr 50 55 60Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys
Pro Arg Glu Glu65 70 75 80Gln Phe Asn Ser Thr Phe Arg Val Val Ser
Val Leu Thr Val Val His 85 90 95Gln Asp Trp Leu Asn Gly Lys Glu Tyr
Lys Cys Lys Val Ser Asn Lys 100 105 110Gly Leu Pro Ala Pro Ile Glu
Lys Thr Ile Ser Lys Thr Lys Gly Gln 115 120 125Pro Arg Glu Pro Gln
Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu Met 130 135 140Thr Lys Asn
Gln Val Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro145 150 155
160Ser Asp Ile Ser Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn
165 170 175Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser Asp Gly Ser Phe
Phe Leu 180 185 190Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln
Gln Gly Asn Val 195 200 205Phe Ser Cys Ser Val Met His Glu Ala Leu
His Asn His Tyr Thr Gln 210 215
220Lys Ser Leu Ser Leu Ser Pro Gly Ile Pro Pro His Val Gln Lys
Ser225 230 235 240Val Asn Asn Asp Met Ile Val Thr Asp Asn Asn Gly
Ala Val Lys Phe 245 250 255Pro Gln Leu Cys Lys Phe Cys Asp Val Arg
Phe Ser Thr Cys Asp Asn 260 265 270Gln Lys Ser Cys Met Ser Asn Cys
Ser Ile Thr Ser Ile Cys Glu Lys 275 280 285Pro Gln Glu Val Cys Val
Ala Val Trp Arg Lys Asn Asp Glu Asn Ile 290 295 300Thr Leu Glu Thr
Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe305 310 315 320Ile
Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys 325 330
335Lys Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu Cys
340 345 350Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn
Pro Asp 355 360 365141374PRTArtificial
SequenceD10-hIgG4FcdeltaK-CC-228P-T2m fusion 141Asp Asp Asp Asp Asp
Asp Asp Asp Asp Asp Glu Ser Lys Tyr Gly Pro1 5 10 15Pro Cys Pro Pro
Cys Pro Ala Pro Glu Phe Leu Gly Gly Pro Ser Val 20 25 30Phe Leu Phe
Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 35 40 45Pro Glu
Val Thr Cys Val Val Val Asp Val Ser Gln Glu Asp Pro Glu 50 55 60Val
Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys65 70 75
80Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Tyr Arg Val Val Ser
85 90 95Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr
Lys 100 105 110Cys Lys Val Ser Asn Lys Gly Leu Pro Ser Ser Ile Glu
Lys Thr Ile 115 120 125Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln
Val Tyr Thr Leu Pro 130 135 140Pro Ser Gln Glu Glu Met Thr Lys Asn
Gln Val Ser Leu Thr Cys Leu145 150 155 160Val Lys Gly Phe Tyr Pro
Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 165 170 175Gly Gln Pro Glu
Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser 180 185 190Asp Gly
Ser Phe Phe Leu Tyr Ser Arg Leu Thr Val Asp Lys Ser Arg 195 200
205Trp Gln Glu Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu
210 215 220His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Leu Gly
Ile Pro225 230 235 240Pro His Val Gln Lys Ser Val Asn Asn Asp Met
Ile Val Thr Asp Asn 245 250 255Asn Gly Ala Val Lys Phe Pro Gln Leu
Cys Lys Phe Cys Asp Val Arg 260 265 270Phe Ser Thr Cys Asp Asn Gln
Lys Ser Cys Met Ser Asn Cys Ser Ile 275 280 285Thr Ser Ile Cys Glu
Lys Pro Gln Glu Val Cys Val Ala Val Trp Arg 290 295 300Lys Asn Asp
Glu Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys305 310 315
320Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys
325 330 335Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met
Cys Ser 340 345 350Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe
Ser Glu Glu Tyr 355 360 365Asn Thr Ser Asn Pro Asp
370142374PRTArtificial SequenceD10-hIgG4FcdeltaK-CC-228P-409K-T2m
fusion 142Asp Asp Asp Asp Asp Asp Asp Asp Asp Asp Glu Ser Lys Tyr
Gly Pro1 5 10 15Pro Cys Pro Pro Cys Pro Ala Pro Glu Phe Leu Gly Gly
Pro Ser Val 20 25 30Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met
Ile Ser Arg Thr 35 40 45Pro Glu Val Thr Cys Val Val Val Asp Val Ser
Gln Glu Asp Pro Glu 50 55 60Val Gln Phe Asn Trp Tyr Val Asp Gly Val
Glu Val His Asn Ala Lys65 70 75 80Thr Lys Pro Arg Glu Glu Gln Phe
Asn Ser Thr Tyr Arg Val Val Ser 85 90 95Val Leu Thr Val Leu His Gln
Asp Trp Leu Asn Gly Lys Glu Tyr Lys 100 105 110Cys Lys Val Ser Asn
Lys Gly Leu Pro Ser Ser Ile Glu Lys Thr Ile 115 120 125Ser Lys Ala
Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro 130 135 140Pro
Ser Gln Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu145 150
155 160Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser
Asn 165 170 175Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val
Leu Asp Ser 180 185 190Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr
Val Asp Lys Ser Arg 195 200 205Trp Gln Glu Gly Asn Val Phe Ser Cys
Ser Val Met His Glu Ala Leu 210 215 220His Asn His Tyr Thr Gln Lys
Ser Leu Ser Leu Ser Leu Gly Ile Pro225 230 235 240Pro His Val Gln
Lys Ser Val Asn Asn Asp Met Ile Val Thr Asp Asn 245 250 255Asn Gly
Ala Val Lys Phe Pro Gln Leu Cys Lys Phe Cys Asp Val Arg 260 265
270Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys Met Ser Asn Cys Ser Ile
275 280 285Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val
Trp Arg 290 295 300Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys
His Asp Pro Lys305 310 315 320Leu Pro Tyr His Asp Phe Ile Leu Glu
Asp Ala Ala Ser Pro Lys Cys 325 330 335Ile Met Lys Glu Lys Lys Lys
Pro Gly Glu Thr Phe Phe Met Cys Ser 340 345 350Cys Ser Ser Asp Glu
Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr 355 360 365Asn Thr Ser
Asn Pro Asp 370143508PRTArtificial
SequenceD10-hIgG1FcdeltaK-CC-T22d35 fusion 143Asp Asp Asp Asp Asp
Asp Asp Asp Asp Asp Asp Lys Thr His Thr Cys1 5 10 15Pro Pro Cys Pro
Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe Leu 20 25 30Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu 35 40 45Val Thr
Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Lys 50 55 60Phe
Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys65 70 75
80Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu
85 90 95Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys
Lys 100 105 110Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr
Ile Ser Lys 115 120 125Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr
Thr Leu Pro Pro Ser 130 135 140Arg Asp Glu Leu Thr Lys Asn Gln Val
Ser Leu Thr Cys Leu Val Lys145 150 155 160Gly Phe Tyr Pro Ser Asp
Ile Ala Val Glu Trp Glu Ser Asn Gly Gln 165 170 175Pro Glu Asn Asn
Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser Asp Gly 180 185 190Ser Phe
Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln 195 200
205Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn
210 215 220His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Ile Pro
Pro His225 230 235 240Val Gln Lys Ser Val Asn Asn Asp Met Ile Val
Thr Asp Asn Asn Gly 245 250 255Ala Val Lys Phe Pro Gln Leu Cys Lys
Phe Cys Asp Val Arg Phe Ser 260 265 270Thr Cys Asp Asn Gln Lys Ser
Cys Met Ser Asn Cys Ser Ile Thr Ser 275 280 285Ile Cys Glu Lys Pro
Gln Glu Val Cys Val Ala Val Trp Arg Lys Asn 290 295 300Asp Glu Asn
Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys Leu Pro305 310 315
320Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met
325 330 335Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe Met Cys Ser
Cys Ser 340 345 350Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser Glu
Glu Tyr Asn Thr 355 360 365Ser Asn Pro Asp Ile Pro Pro His Val Gln
Lys Ser Val Asn Asn Asp 370 375 380Met Ile Val Thr Asp Asn Asn Gly
Ala Val Lys Phe Pro Gln Leu Cys385 390 395 400Lys Phe Cys Asp Val
Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys 405 410 415Met Ser Asn
Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val 420 425 430Cys
Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr 435 440
445Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp
450 455 460Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys Lys Pro
Gly Glu465 470 475 480Thr Phe Phe Met Cys Ser Cys Ser Ser Asp Glu
Cys Asn Asp Asn Ile 485 490 495Ile Phe Ser Glu Glu Tyr Asn Thr Ser
Asn Pro Asp 500 505144504PRTArtificial
SequenceD10-hIgG2FcdeltaK-CC-T22d35 fusion 144Asp Asp Asp Asp Asp
Asp Asp Asp Asp Asp Val Glu Cys Pro Pro Cys1 5 10 15Pro Ala Pro Pro
Val Ala Gly Pro Ser Val Phe Leu Phe Pro Pro Lys 20 25 30Pro Lys Asp
Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val 35 40 45Val Val
Asp Val Ser His Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr 50 55 60Val
Asp Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu65 70 75
80Gln Phe Asn Ser Thr Phe Arg Val Val Ser Val Leu Thr Val Val His
85 90 95Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn
Lys 100 105 110Gly Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Thr
Lys Gly Gln 115 120 125Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro
Ser Arg Glu Glu Met 130 135 140Thr Lys Asn Gln Val Ser Leu Thr Cys
Leu Val Lys Gly Phe Tyr Pro145 150 155 160Ser Asp Ile Ser Val Glu
Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn 165 170 175Tyr Lys Thr Thr
Pro Pro Met Leu Asp Ser Asp Gly Ser Phe Phe Leu 180 185 190Tyr Ser
Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val 195 200
205Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln
210 215 220Lys Ser Leu Ser Leu Ser Pro Gly Ile Pro Pro His Val Gln
Lys Ser225 230 235 240Val Asn Asn Asp Met Ile Val Thr Asp Asn Asn
Gly Ala Val Lys Phe 245 250 255Pro Gln Leu Cys Lys Phe Cys Asp Val
Arg Phe Ser Thr Cys Asp Asn 260 265 270Gln Lys Ser Cys Met Ser Asn
Cys Ser Ile Thr Ser Ile Cys Glu Lys 275 280 285Pro Gln Glu Val Cys
Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile 290 295 300Thr Leu Glu
Thr Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe305 310 315
320Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys
325 330 335Lys Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser Ser Asp
Glu Cys 340 345 350Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr
Ser Asn Pro Asp 355 360 365Ile Pro Pro His Val Gln Lys Ser Val Asn
Asn Asp Met Ile Val Thr 370 375 380Asp Asn Asn Gly Ala Val Lys Phe
Pro Gln Leu Cys Lys Phe Cys Asp385 390 395 400Val Arg Phe Ser Thr
Cys Asp Asn Gln Lys Ser Cys Met Ser Asn Cys 405 410 415Ser Ile Thr
Ser Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val 420 425 430Trp
Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His Asp 435 440
445Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro
450 455 460Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe
Phe Met465 470 475 480Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn
Ile Ile Phe Ser Glu 485 490 495Glu Tyr Asn Thr Ser Asn Pro Asp
500145510PRTArtificial SequenceD10-hIgG4FcdeltaK-CC-228P-T22d35
fusion 145Asp Asp Asp Asp Asp Asp Asp Asp Asp Asp Glu Ser Lys Tyr
Gly Pro1 5 10 15Pro Cys Pro Pro Cys Pro Ala Pro Glu Phe Leu Gly Gly
Pro Ser Val 20 25 30Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met
Ile Ser Arg Thr 35 40 45Pro Glu Val Thr Cys Val Val Val Asp Val Ser
Gln Glu Asp Pro Glu 50 55 60Val Gln Phe Asn Trp Tyr Val Asp Gly Val
Glu Val His Asn Ala Lys65 70 75 80Thr Lys Pro Arg Glu Glu Gln Phe
Asn Ser Thr Tyr Arg Val Val Ser 85 90 95Val Leu Thr Val Leu His Gln
Asp Trp Leu Asn Gly Lys Glu Tyr Lys 100 105 110Cys Lys Val Ser Asn
Lys Gly Leu Pro Ser Ser Ile Glu Lys Thr Ile 115 120 125Ser Lys Ala
Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro 130 135 140Pro
Ser Gln Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu145 150
155 160Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser
Asn 165 170 175Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val
Leu Asp Ser 180 185 190Asp Gly Ser Phe Phe Leu Tyr Ser Arg Leu Thr
Val Asp Lys Ser Arg 195 200 205Trp Gln Glu Gly Asn Val Phe Ser Cys
Ser Val Met His Glu Ala Leu 210 215 220His Asn His Tyr Thr Gln Lys
Ser Leu Ser Leu Ser Leu Gly Ile Pro225 230 235 240Pro His Val Gln
Lys Ser Val Asn Asn Asp Met Ile Val Thr Asp Asn 245 250 255Asn Gly
Ala Val Lys Phe Pro Gln Leu Cys Lys Phe Cys Asp Val Arg 260 265
270Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys Met Ser Asn Cys Ser Ile
275 280 285Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val
Trp Arg 290 295 300Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys
His Asp Pro Lys305 310 315 320Leu Pro Tyr His Asp Phe Ile Leu Glu
Asp Ala Ala Ser Pro Lys Cys 325 330 335Ile Met Lys Glu Lys Lys Lys
Pro Gly Glu Thr Phe Phe Met Cys Ser 340 345 350Cys Ser Ser Asp Glu
Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr 355 360 365Asn Thr Ser
Asn Pro Asp Ile Pro Pro His Val Gln Lys Ser Val Asn 370 375 380Asn
Asp Met Ile Val Thr Asp Asn Asn Gly Ala Val Lys Phe Pro Gln385 390
395 400Leu Cys Lys Phe Cys Asp Val Arg Phe Ser Thr Cys Asp Asn Gln
Lys 405 410 415Ser Cys Met Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu
Lys Pro Gln 420 425 430Glu Val Cys Val Ala Val Trp Arg Lys Asn Asp
Glu Asn Ile Thr Leu 435 440 445Glu Thr Val Cys His Asp Pro Lys Leu
Pro Tyr His Asp Phe Ile Leu 450 455 460Glu Asp Ala Ala Ser Pro Lys
Cys Ile Met Lys Glu Lys Lys Lys Pro465 470 475 480Gly Glu Thr Phe
Phe Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp 485 490 495Asn Ile
Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp 500 505
510146510PRTArtificial
SequenceD10-hIgG4FcdeltaK-CC-228P-409K-T22d35
fusion 146Asp Asp Asp Asp Asp Asp Asp Asp Asp Asp Glu Ser Lys Tyr
Gly Pro1 5 10 15Pro Cys Pro Pro Cys Pro Ala Pro Glu Phe Leu Gly Gly
Pro Ser Val 20 25 30Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met
Ile Ser Arg Thr 35 40 45Pro Glu Val Thr Cys Val Val Val Asp Val Ser
Gln Glu Asp Pro Glu 50 55 60Val Gln Phe Asn Trp Tyr Val Asp Gly Val
Glu Val His Asn Ala Lys65 70 75 80Thr Lys Pro Arg Glu Glu Gln Phe
Asn Ser Thr Tyr Arg Val Val Ser 85 90 95Val Leu Thr Val Leu His Gln
Asp Trp Leu Asn Gly Lys Glu Tyr Lys 100 105 110Cys Lys Val Ser Asn
Lys Gly Leu Pro Ser Ser Ile Glu Lys Thr Ile 115 120 125Ser Lys Ala
Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro 130 135 140Pro
Ser Gln Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu145 150
155 160Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser
Asn 165 170 175Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val
Leu Asp Ser 180 185 190Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr
Val Asp Lys Ser Arg 195 200 205Trp Gln Glu Gly Asn Val Phe Ser Cys
Ser Val Met His Glu Ala Leu 210 215 220His Asn His Tyr Thr Gln Lys
Ser Leu Ser Leu Ser Leu Gly Ile Pro225 230 235 240Pro His Val Gln
Lys Ser Val Asn Asn Asp Met Ile Val Thr Asp Asn 245 250 255Asn Gly
Ala Val Lys Phe Pro Gln Leu Cys Lys Phe Cys Asp Val Arg 260 265
270Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys Met Ser Asn Cys Ser Ile
275 280 285Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val
Trp Arg 290 295 300Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys
His Asp Pro Lys305 310 315 320Leu Pro Tyr His Asp Phe Ile Leu Glu
Asp Ala Ala Ser Pro Lys Cys 325 330 335Ile Met Lys Glu Lys Lys Lys
Pro Gly Glu Thr Phe Phe Met Cys Ser 340 345 350Cys Ser Ser Asp Glu
Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr 355 360 365Asn Thr Ser
Asn Pro Asp Ile Pro Pro His Val Gln Lys Ser Val Asn 370 375 380Asn
Asp Met Ile Val Thr Asp Asn Asn Gly Ala Val Lys Phe Pro Gln385 390
395 400Leu Cys Lys Phe Cys Asp Val Arg Phe Ser Thr Cys Asp Asn Gln
Lys 405 410 415Ser Cys Met Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu
Lys Pro Gln 420 425 430Glu Val Cys Val Ala Val Trp Arg Lys Asn Asp
Glu Asn Ile Thr Leu 435 440 445Glu Thr Val Cys His Asp Pro Lys Leu
Pro Tyr His Asp Phe Ile Leu 450 455 460Glu Asp Ala Ala Ser Pro Lys
Cys Ile Met Lys Glu Lys Lys Lys Pro465 470 475 480Gly Glu Thr Phe
Phe Met Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp 485 490 495Asn Ile
Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn Pro Asp 500 505
510147644PRTArtificial SequenceD10-hIgG1FcdeltaK-CC-T2-T2-T2 fusion
147Asp Asp Asp Asp Asp Asp Asp Asp Asp Asp Asp Lys Thr His Thr Cys1
5 10 15Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe
Leu 20 25 30Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr
Pro Glu 35 40 45Val Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro
Glu Val Lys 50 55 60Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn
Ala Lys Thr Lys65 70 75 80Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr
Arg Val Val Ser Val Leu 85 90 95Thr Val Leu His Gln Asp Trp Leu Asn
Gly Lys Glu Tyr Lys Cys Lys 100 105 110Val Ser Asn Lys Ala Leu Pro
Ala Pro Ile Glu Lys Thr Ile Ser Lys 115 120 125Ala Lys Gly Gln Pro
Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser 130 135 140Arg Asp Glu
Leu Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys145 150 155
160Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln
165 170 175Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser
Asp Gly 180 185 190Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys
Ser Arg Trp Gln 195 200 205Gln Gly Asn Val Phe Ser Cys Ser Val Met
His Glu Ala Leu His Asn 210 215 220His Tyr Thr Gln Lys Ser Leu Ser
Leu Ser Pro Gly Ile Pro Pro His225 230 235 240Val Gln Lys Ser Val
Asn Asn Asp Met Ile Val Thr Asp Asn Asn Gly 245 250 255Ala Val Lys
Phe Pro Gln Leu Cys Lys Phe Cys Asp Val Arg Phe Ser 260 265 270Thr
Cys Asp Asn Gln Lys Ser Cys Met Ser Asn Cys Ser Ile Thr Ser 275 280
285Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val Trp Arg Lys Asn
290 295 300Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys
Leu Pro305 310 315 320Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser
Pro Lys Cys Ile Met 325 330 335Lys Glu Lys Lys Lys Pro Gly Glu Thr
Phe Phe Met Cys Ser Cys Ser 340 345 350Ser Asp Glu Cys Asn Asp Asn
Ile Ile Phe Ser Glu Glu Tyr Asn Thr 355 360 365Ser Asn Pro Asp Ile
Pro Pro His Val Gln Lys Ser Val Asn Asn Asp 370 375 380Met Ile Val
Thr Asp Asn Asn Gly Ala Val Lys Phe Pro Gln Leu Cys385 390 395
400Lys Phe Cys Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys
405 410 415Met Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln
Glu Val 420 425 430Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile
Thr Leu Glu Thr 435 440 445Val Cys His Asp Pro Lys Leu Pro Tyr His
Asp Phe Ile Leu Glu Asp 450 455 460Ala Ala Ser Pro Lys Cys Ile Met
Lys Glu Lys Lys Lys Pro Gly Glu465 470 475 480Thr Phe Phe Met Cys
Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile 485 490 495Ile Phe Ser
Glu Glu Tyr Asn Thr Ser Asn Pro Asp Ile Pro Pro His 500 505 510Val
Gln Lys Ser Val Asn Asn Asp Met Ile Val Thr Asp Asn Asn Gly 515 520
525Ala Val Lys Phe Pro Gln Leu Cys Lys Phe Cys Asp Val Arg Phe Ser
530 535 540Thr Cys Asp Asn Gln Lys Ser Cys Met Ser Asn Cys Ser Ile
Thr Ser545 550 555 560Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala
Val Trp Arg Lys Asn 565 570 575Asp Glu Asn Ile Thr Leu Glu Thr Val
Cys His Asp Pro Lys Leu Pro 580 585 590Tyr His Asp Phe Ile Leu Glu
Asp Ala Ala Ser Pro Lys Cys Ile Met 595 600 605Lys Glu Lys Lys Lys
Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser 610 615 620Ser Asp Glu
Cys Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr625 630 635
640Ser Asn Pro Asp148640PRTArtificial
SequenceD10-hIgG2FcdeltaK-CC-T2-T2-T2 fusion 148Asp Asp Asp Asp Asp
Asp Asp Asp Asp Asp Val Glu Cys Pro Pro Cys1 5 10 15Pro Ala Pro Pro
Val Ala Gly Pro Ser Val Phe Leu Phe Pro Pro Lys 20 25 30Pro Lys Asp
Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val 35 40 45Val Val
Asp Val Ser His Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr 50 55 60Val
Asp Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu65 70 75
80Gln Phe Asn Ser Thr Phe Arg Val Val Ser Val Leu Thr Val Val His
85 90 95Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn
Lys 100 105 110Gly Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Thr
Lys Gly Gln 115 120 125Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro
Ser Arg Glu Glu Met 130 135 140Thr Lys Asn Gln Val Ser Leu Thr Cys
Leu Val Lys Gly Phe Tyr Pro145 150 155 160Ser Asp Ile Ser Val Glu
Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn 165 170 175Tyr Lys Thr Thr
Pro Pro Met Leu Asp Ser Asp Gly Ser Phe Phe Leu 180 185 190Tyr Ser
Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val 195 200
205Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln
210 215 220Lys Ser Leu Ser Leu Ser Pro Gly Ile Pro Pro His Val Gln
Lys Ser225 230 235 240Val Asn Asn Asp Met Ile Val Thr Asp Asn Asn
Gly Ala Val Lys Phe 245 250 255Pro Gln Leu Cys Lys Phe Cys Asp Val
Arg Phe Ser Thr Cys Asp Asn 260 265 270Gln Lys Ser Cys Met Ser Asn
Cys Ser Ile Thr Ser Ile Cys Glu Lys 275 280 285Pro Gln Glu Val Cys
Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile 290 295 300Thr Leu Glu
Thr Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe305 310 315
320Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu Lys Lys
325 330 335Lys Pro Gly Glu Thr Phe Phe Met Cys Ser Cys Ser Ser Asp
Glu Cys 340 345 350Asn Asp Asn Ile Ile Phe Ser Glu Glu Tyr Asn Thr
Ser Asn Pro Asp 355 360 365Ile Pro Pro His Val Gln Lys Ser Val Asn
Asn Asp Met Ile Val Thr 370 375 380Asp Asn Asn Gly Ala Val Lys Phe
Pro Gln Leu Cys Lys Phe Cys Asp385 390 395 400Val Arg Phe Ser Thr
Cys Asp Asn Gln Lys Ser Cys Met Ser Asn Cys 405 410 415Ser Ile Thr
Ser Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val 420 425 430Trp
Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His Asp 435 440
445Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro
450 455 460Lys Cys Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe
Phe Met465 470 475 480Cys Ser Cys Ser Ser Asp Glu Cys Asn Asp Asn
Ile Ile Phe Ser Glu 485 490 495Glu Tyr Asn Thr Ser Asn Pro Asp Ile
Pro Pro His Val Gln Lys Ser 500 505 510Val Asn Asn Asp Met Ile Val
Thr Asp Asn Asn Gly Ala Val Lys Phe 515 520 525Pro Gln Leu Cys Lys
Phe Cys Asp Val Arg Phe Ser Thr Cys Asp Asn 530 535 540Gln Lys Ser
Cys Met Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys545 550 555
560Pro Gln Glu Val Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile
565 570 575Thr Leu Glu Thr Val Cys His Asp Pro Lys Leu Pro Tyr His
Asp Phe 580 585 590Ile Leu Glu Asp Ala Ala Ser Pro Lys Cys Ile Met
Lys Glu Lys Lys 595 600 605Lys Pro Gly Glu Thr Phe Phe Met Cys Ser
Cys Ser Ser Asp Glu Cys 610 615 620Asn Asp Asn Ile Ile Phe Ser Glu
Glu Tyr Asn Thr Ser Asn Pro Asp625 630 635 640149646PRTArtificial
SequenceD10-hIgG4FcdeltaK-CC-228P-T2-T2-T2 fusion 149Asp Asp Asp
Asp Asp Asp Asp Asp Asp Asp Glu Ser Lys Tyr Gly Pro1 5 10 15Pro Cys
Pro Pro Cys Pro Ala Pro Glu Phe Leu Gly Gly Pro Ser Val 20 25 30Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 35 40
45Pro Glu Val Thr Cys Val Val Val Asp Val Ser Gln Glu Asp Pro Glu
50 55 60Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala
Lys65 70 75 80Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Tyr Arg
Val Val Ser 85 90 95Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly
Lys Glu Tyr Lys 100 105 110Cys Lys Val Ser Asn Lys Gly Leu Pro Ser
Ser Ile Glu Lys Thr Ile 115 120 125Ser Lys Ala Lys Gly Gln Pro Arg
Glu Pro Gln Val Tyr Thr Leu Pro 130 135 140Pro Ser Gln Glu Glu Met
Thr Lys Asn Gln Val Ser Leu Thr Cys Leu145 150 155 160Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 165 170 175Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser 180 185
190Asp Gly Ser Phe Phe Leu Tyr Ser Arg Leu Thr Val Asp Lys Ser Arg
195 200 205Trp Gln Glu Gly Asn Val Phe Ser Cys Ser Val Met His Glu
Ala Leu 210 215 220His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
Leu Gly Ile Pro225 230 235 240Pro His Val Gln Lys Ser Val Asn Asn
Asp Met Ile Val Thr Asp Asn 245 250 255Asn Gly Ala Val Lys Phe Pro
Gln Leu Cys Lys Phe Cys Asp Val Arg 260 265 270Phe Ser Thr Cys Asp
Asn Gln Lys Ser Cys Met Ser Asn Cys Ser Ile 275 280 285Thr Ser Ile
Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val Trp Arg 290 295 300Lys
Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys305 310
315 320Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys
Cys 325 330 335Ile Met Lys Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe
Met Cys Ser 340 345 350Cys Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile
Phe Ser Glu Glu Tyr 355 360 365Asn Thr Ser Asn Pro Asp Ile Pro Pro
His Val Gln Lys Ser Val Asn 370 375 380Asn Asp Met Ile Val Thr Asp
Asn Asn Gly Ala Val Lys Phe Pro Gln385 390 395 400Leu Cys Lys Phe
Cys Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys 405 410 415Ser Cys
Met Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln 420 425
430Glu Val Cys Val Ala Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu
435 440 445Glu Thr Val Cys His Asp Pro Lys Leu Pro Tyr His Asp Phe
Ile Leu 450 455 460Glu Asp Ala Ala Ser Pro Lys Cys Ile Met Lys Glu
Lys Lys Lys Pro465 470 475 480Gly Glu Thr Phe Phe Met Cys Ser Cys
Ser Ser Asp Glu Cys Asn Asp 485 490 495Asn Ile Ile Phe Ser Glu Glu
Tyr Asn Thr Ser Asn Pro Asp Ile Pro 500 505 510Pro His Val Gln Lys
Ser Val Asn Asn Asp Met Ile Val Thr Asp Asn 515 520 525Asn Gly Ala
Val Lys Phe Pro Gln Leu Cys Lys Phe Cys Asp Val Arg 530 535 540Phe
Ser Thr Cys Asp Asn Gln Lys Ser Cys Met Ser Asn Cys Ser Ile545 550
555 560Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala Val Trp
Arg 565 570 575Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys His
Asp Pro Lys 580 585 590Leu Pro Tyr His Asp Phe Ile Leu Glu Asp Ala
Ala Ser Pro Lys Cys 595 600 605Ile Met Lys Glu Lys Lys Lys Pro Gly
Glu Thr Phe Phe Met Cys Ser 610 615 620Cys Ser Ser Asp Glu Cys Asn
Asp Asn Ile Ile Phe Ser Glu Glu Tyr625 630 635 640Asn Thr Ser Asn
Pro Asp 645150649PRTArtificial
SequenceD10-hIgG4FcdeltaK-CC-228P-409K-T2-T2-T2 fusion 150Asp Asp
Asp Asp Asp Asp Asp Asp Asp Asp Glu Ser Lys Tyr Gly Pro1 5 10 15Pro
Cys Pro Pro Cys Pro Ala Pro Glu Phe Leu Gly Gly Pro Ser Val 20 25
30Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr
35 40 45Pro Glu Val Thr Cys Val Val Val Asp Val Ser Gln Glu Asp Pro
Glu 50 55 60Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn
Ala Lys65 70 75 80Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Tyr
Arg Val Val Ser 85 90 95Val Leu Thr Val Leu His Gln Asp Trp Leu Asn
Gly Lys Glu Tyr Lys 100 105 110Cys Lys Val Ser Asn Lys Gly Leu Pro
Ser Ser Ile Ser Glu Gln Glu 115 120 125Lys Thr Ile Ser Lys Ala Lys
Gly Gln Pro Arg Glu Pro Gln Val Tyr 130 135 140Thr Leu Pro Pro Ser
Gln Glu Glu Met Thr Lys Asn Gln Val Ser Leu145 150 155 160Thr Cys
Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp 165 170
175Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val
180 185 190Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr
Val Asp 195 200 205Lys Ser Arg Trp Gln Glu Gly Asn Val Phe Ser Cys
Ser Val Met His 210 215 220Glu Ala Leu His Asn His Tyr Thr Gln Lys
Ser Leu Ser Leu Ser Leu225 230 235 240Gly Ile Pro Pro His Val Gln
Lys Ser Val Asn Asn Asp Met Ile Val 245 250 255Thr Asp Asn Asn Gly
Ala Val Lys Phe Pro Gln Leu Cys Lys Phe Cys 260 265 270Asp Val Arg
Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys Met Ser Asn 275 280 285Cys
Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu Val Cys Val Ala 290 295
300Val Trp Arg Lys Asn Asp Glu Asn Ile Thr Leu Glu Thr Val Cys
His305 310 315 320Asp Pro Lys Leu Pro Tyr His Asp Phe Ile Leu Glu
Asp Ala Ala Ser 325 330 335Pro Lys Cys Ile Met Lys Glu Lys Lys Lys
Pro Gly Glu Thr Phe Phe 340 345 350Met Cys Ser Cys Ser Ser Asp Glu
Cys Asn Asp Asn Ile Ile Phe Ser 355 360 365Glu Glu Tyr Asn Thr Ser
Asn Pro Asp Ile Pro Pro His Val Gln Lys 370 375 380Ser Val Asn Asn
Asp Met Ile Val Thr Asp Asn Asn Gly Ala Val Lys385 390 395 400Phe
Pro Gln Leu Cys Lys Phe Cys Asp Val Arg Phe Ser Thr Cys Asp 405 410
415Asn Gln Lys Ser Cys Met Ser Asn Cys Ser Ile Thr Ser Ile Cys Glu
420 425 430Lys Pro Gln Glu Val Cys Val Ala Val Trp Arg Lys Asn Asp
Glu Asn 435 440 445Ile Thr Leu Glu Thr Val Cys His Asp Pro Lys Leu
Pro Tyr His Asp 450 455 460Phe Ile Leu Glu Asp Ala Ala Ser Pro Lys
Cys Ile Met Lys Glu Lys465 470 475 480Lys Lys Pro Gly Glu Thr Phe
Phe Met Cys Ser Cys Ser Ser Asp Glu 485 490 495Cys Asn Asp Asn Ile
Ile Phe Ser Glu Glu Tyr Asn Thr Ser Asn Pro 500 505 510Asp Ile Pro
Pro His Val Gln Lys Ser Val Asn Asn Asp Met Ile Val 515 520 525Thr
Asp Asn Asn Gly Ala Val Lys Phe Pro Gln Leu Cys Lys Phe Cys 530 535
540Asp Val Arg Phe Ser Thr Cys Asp Asn Gln Lys Ser Cys Met Ser
Asn545 550 555 560Cys Ser Ile Thr Ser Ile Cys Glu Lys Pro Gln Glu
Val Cys Val Ala 565 570 575Val Trp Arg Lys Asn Asp Glu Asn Ile Thr
Leu Glu Thr Val Cys His 580 585 590Asp Pro Lys Leu Pro Tyr His Asp
Phe Ile Leu Glu Asp Ala Ala Ser 595 600 605Pro Lys Cys Ile Met Lys
Glu Lys Lys Lys Pro Gly Glu Thr Phe Phe 610 615 620Met Cys Ser Cys
Ser Ser Asp Glu Cys Asn Asp Asn Ile Ile Phe Ser625 630 635 640Glu
Glu Tyr Asn Thr Ser Asn Pro Asp 645
D00000

D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012
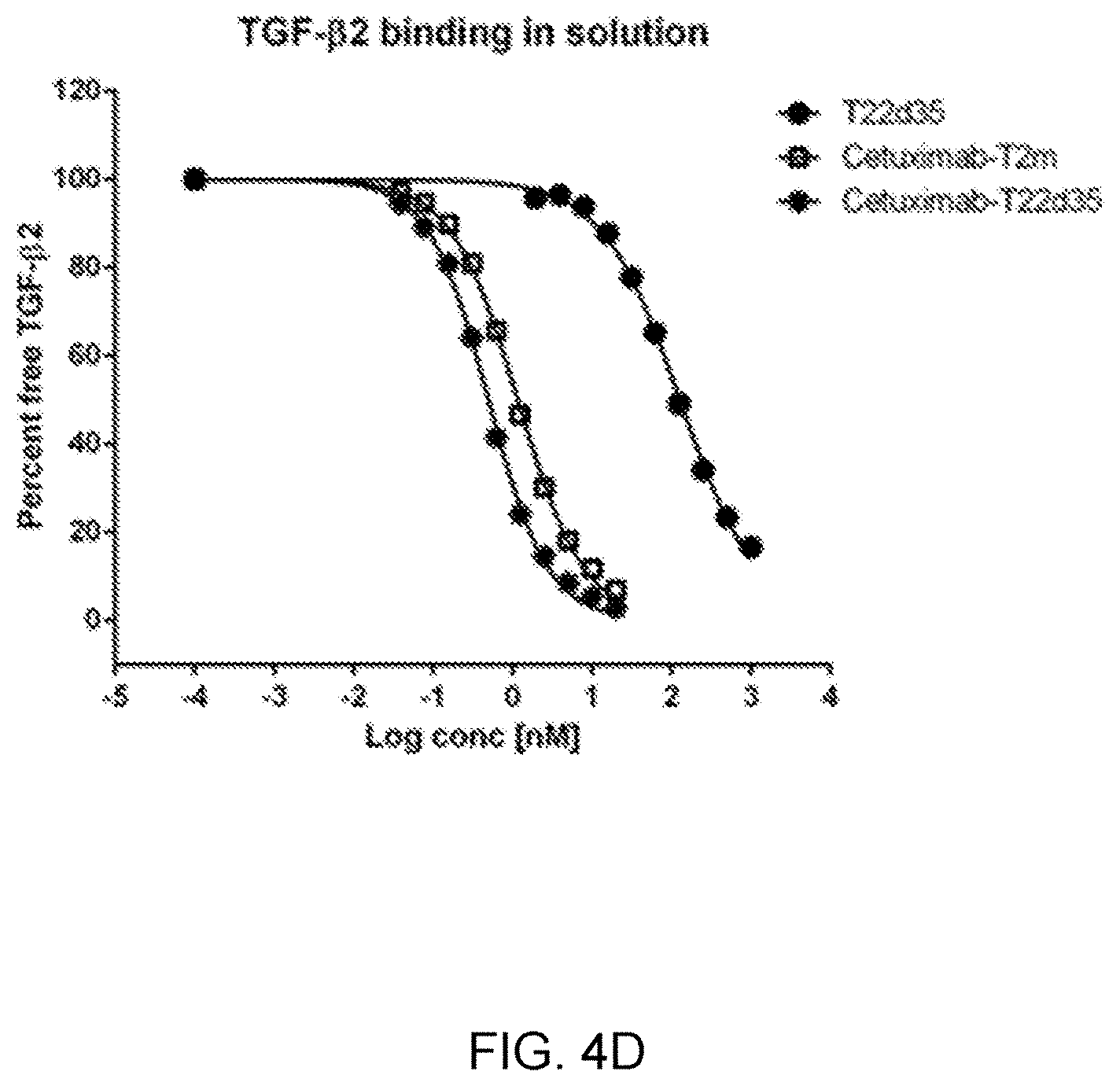
D00013

D00014
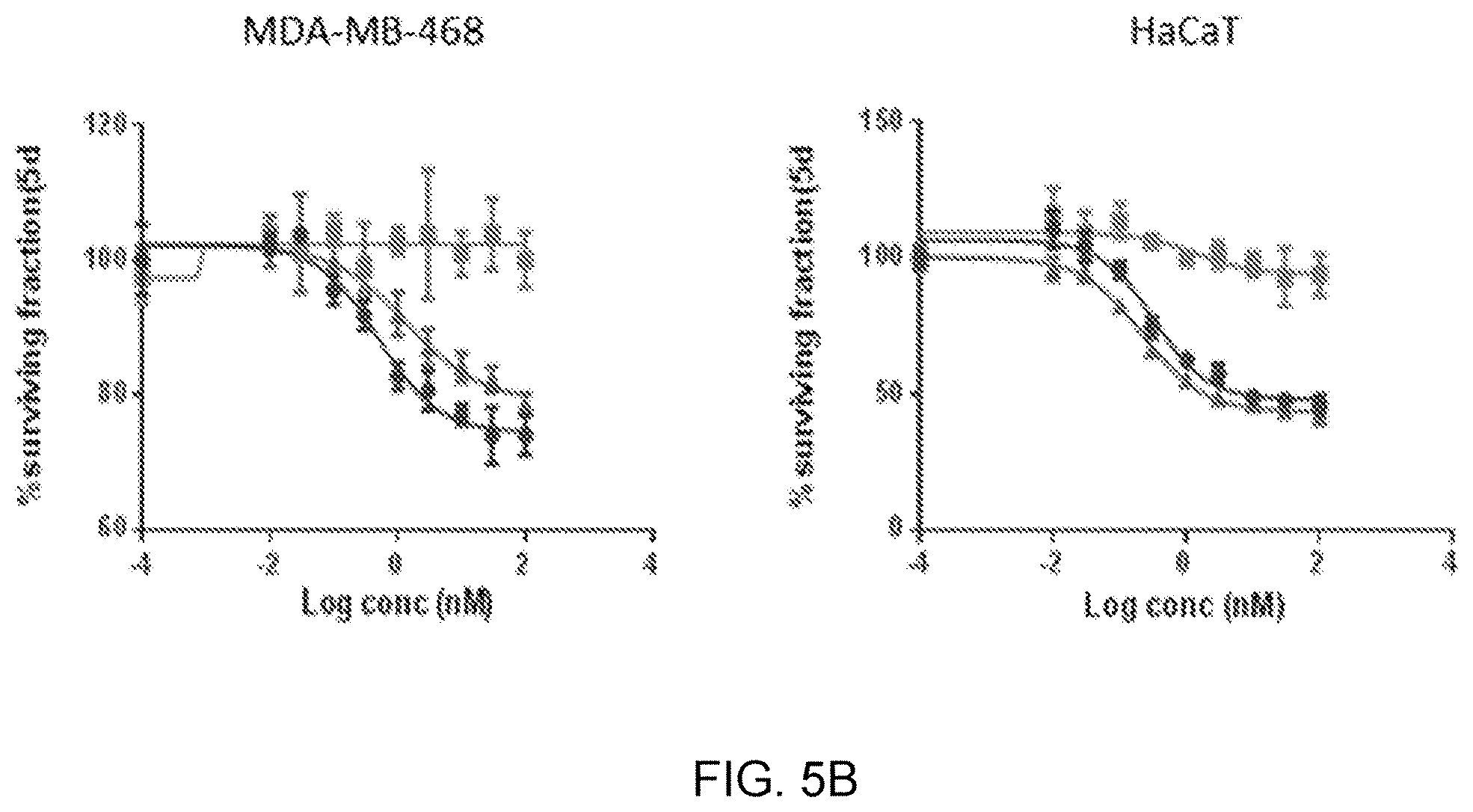
D00015
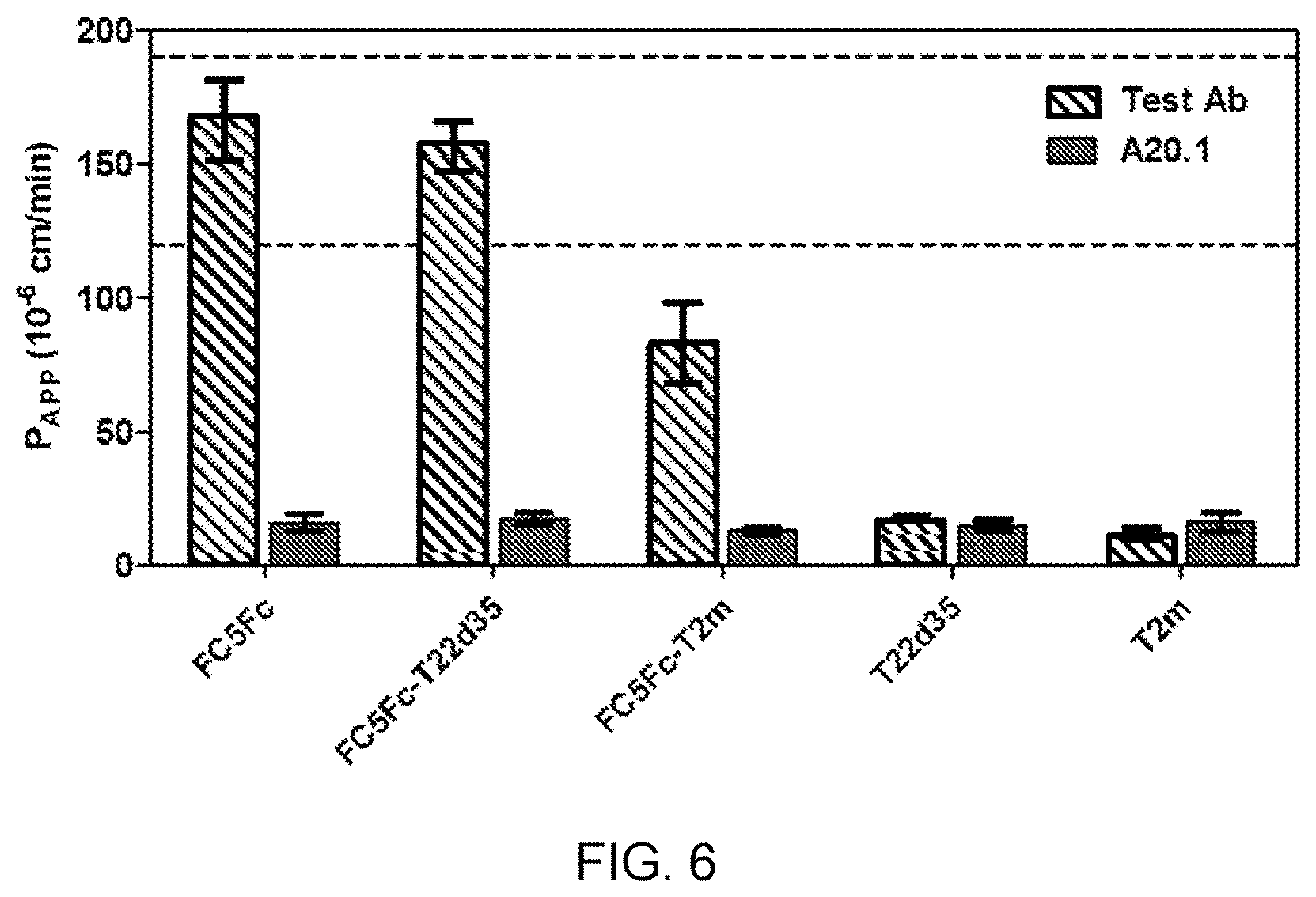
D00016
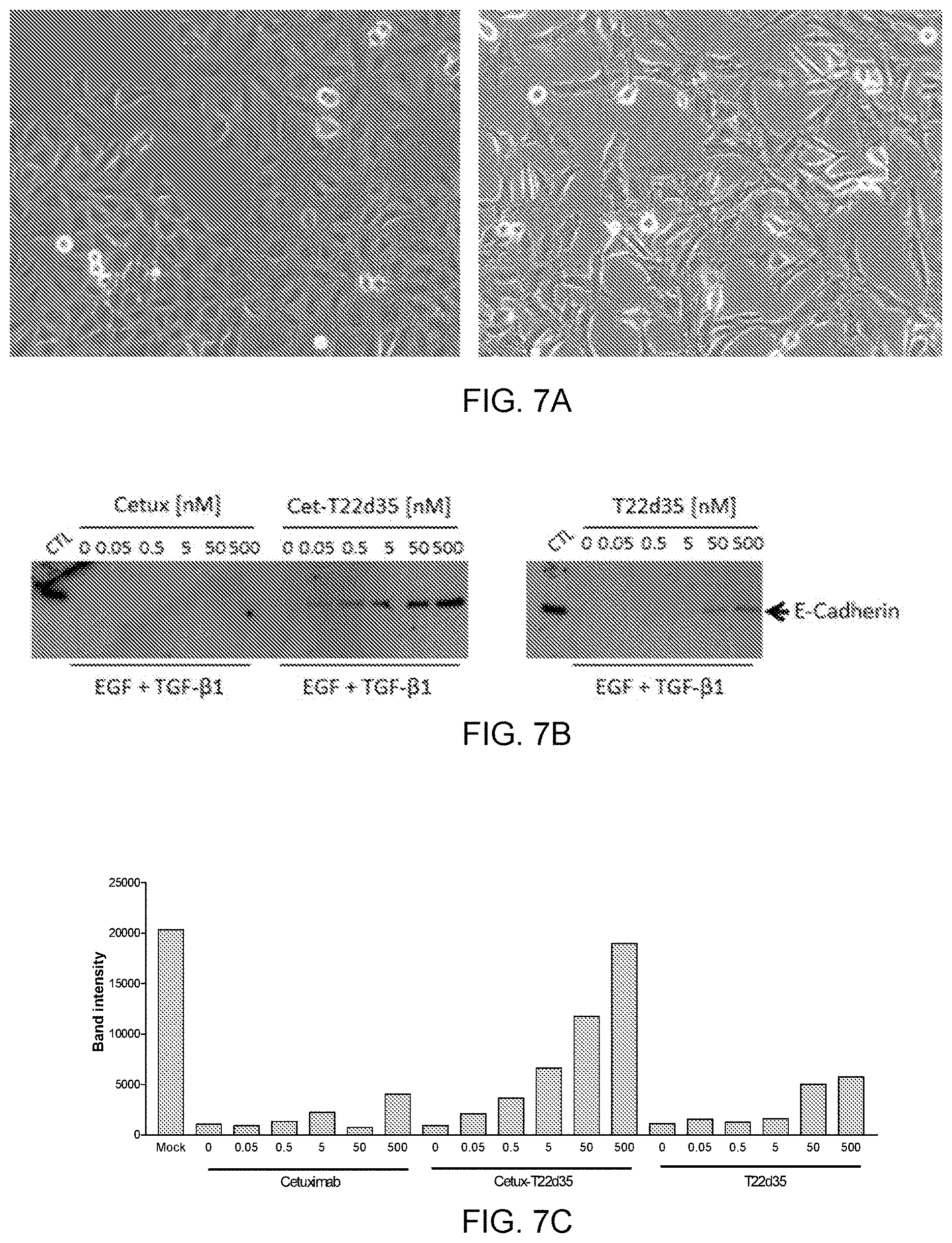
D00017

D00018
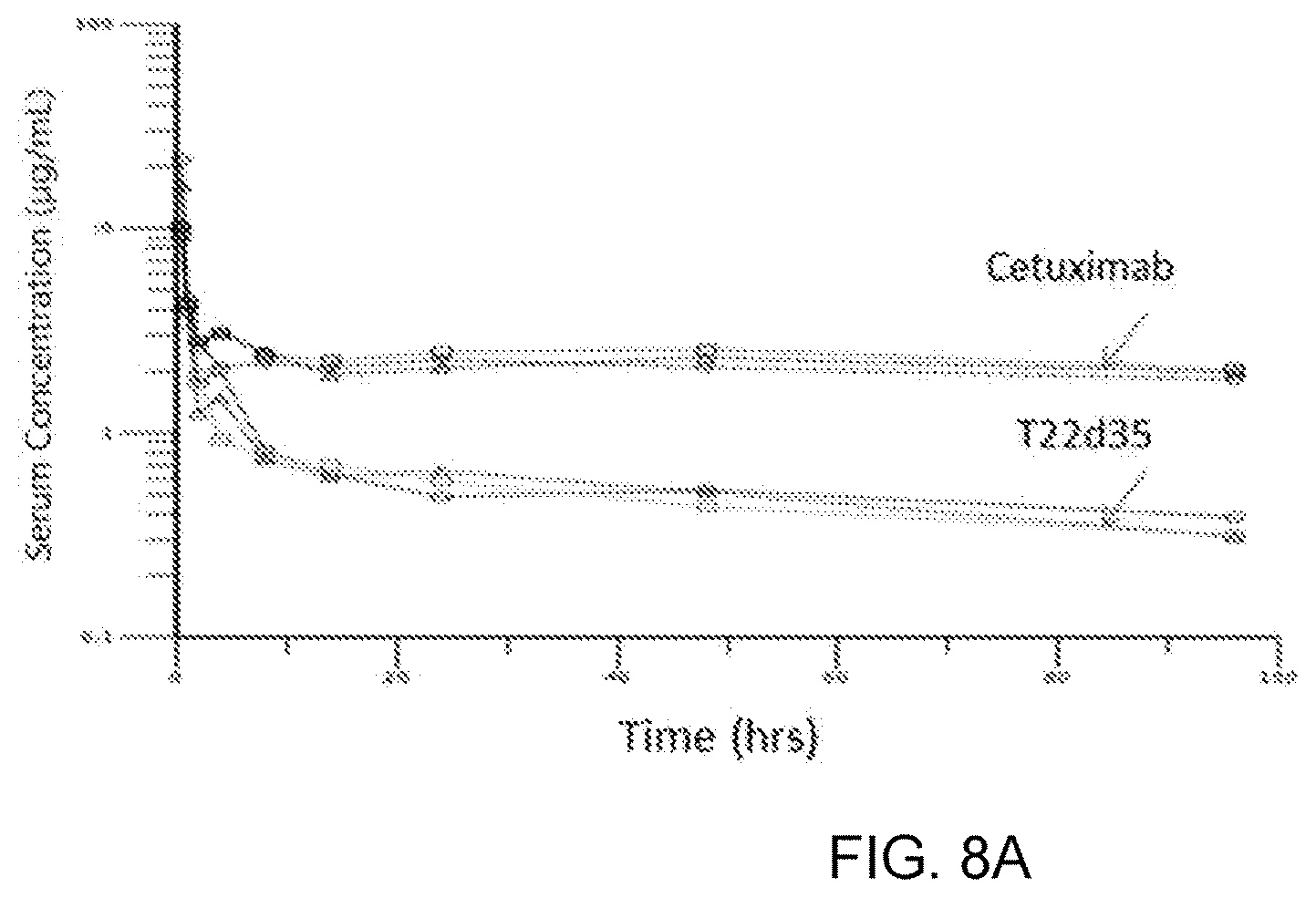
D00019
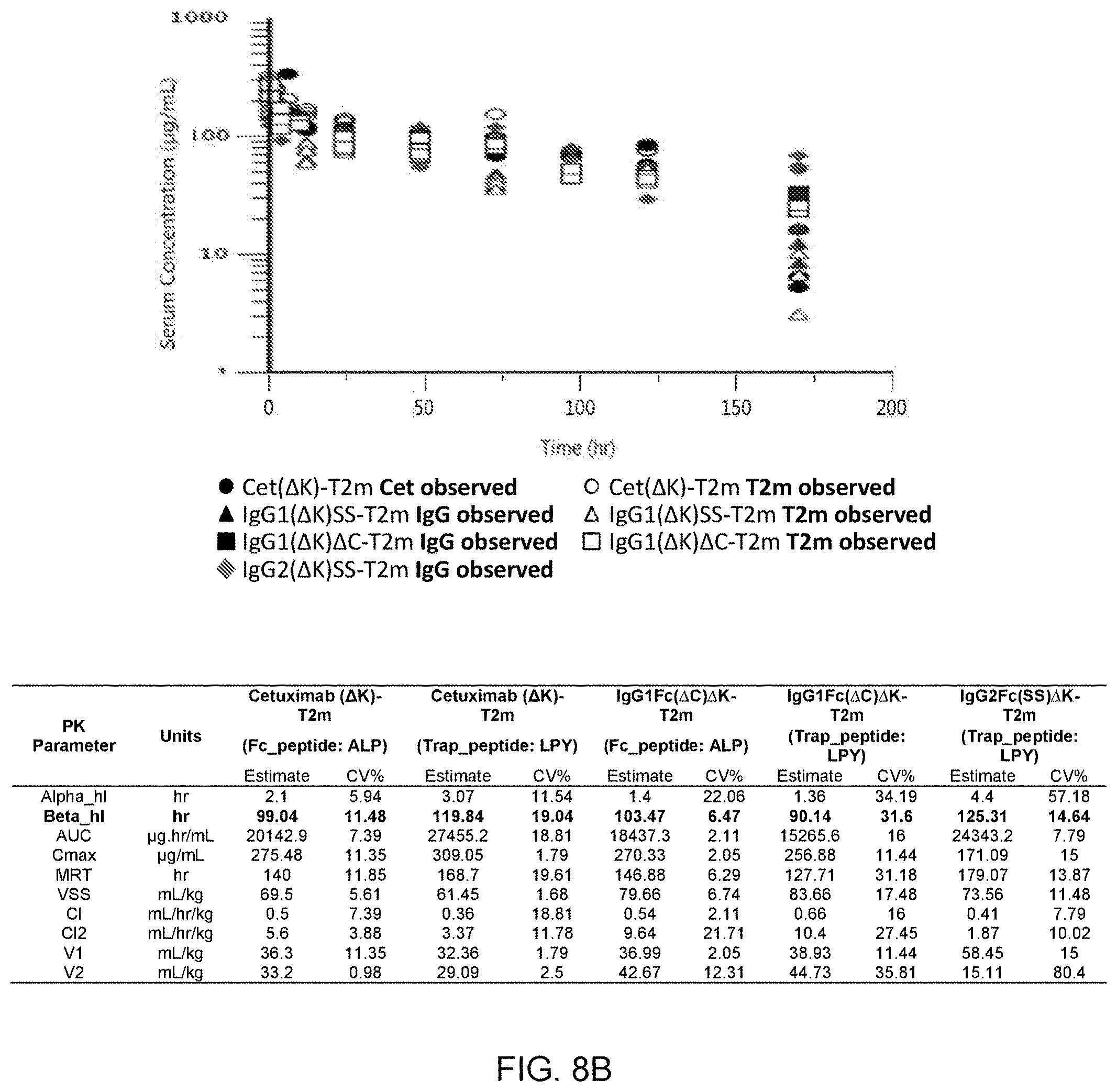
D00020
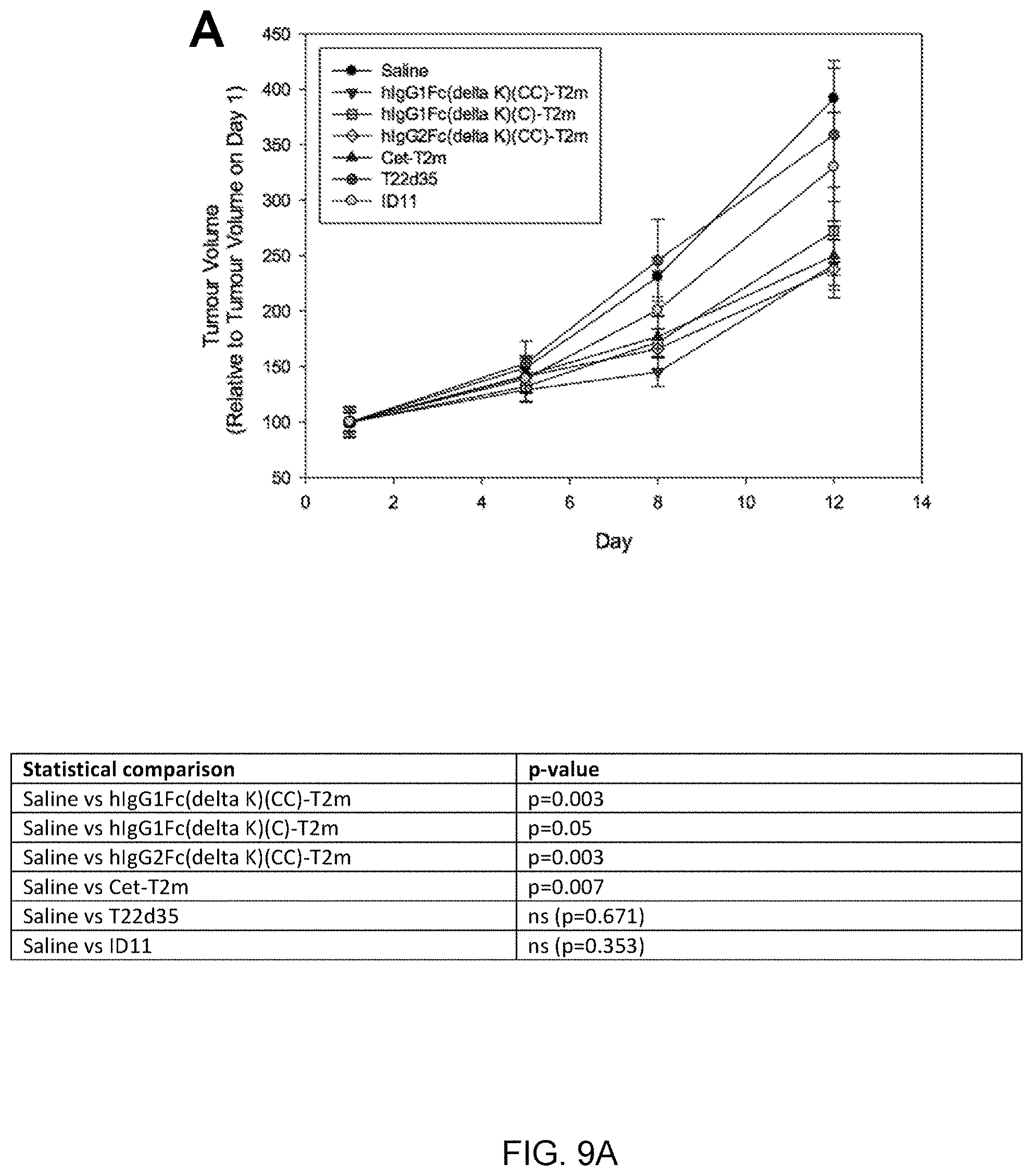
D00021

D00022

D00023
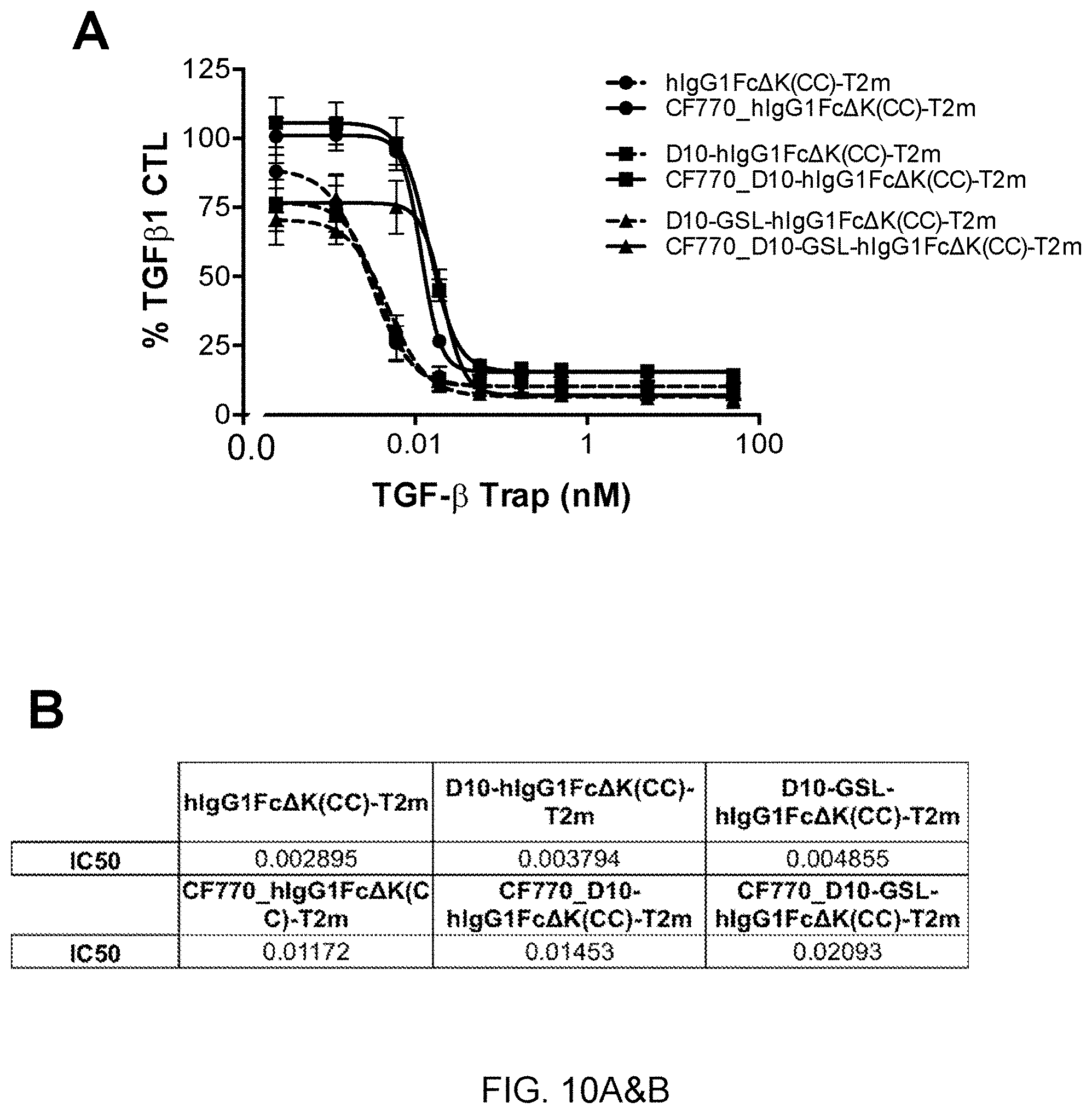
D00024
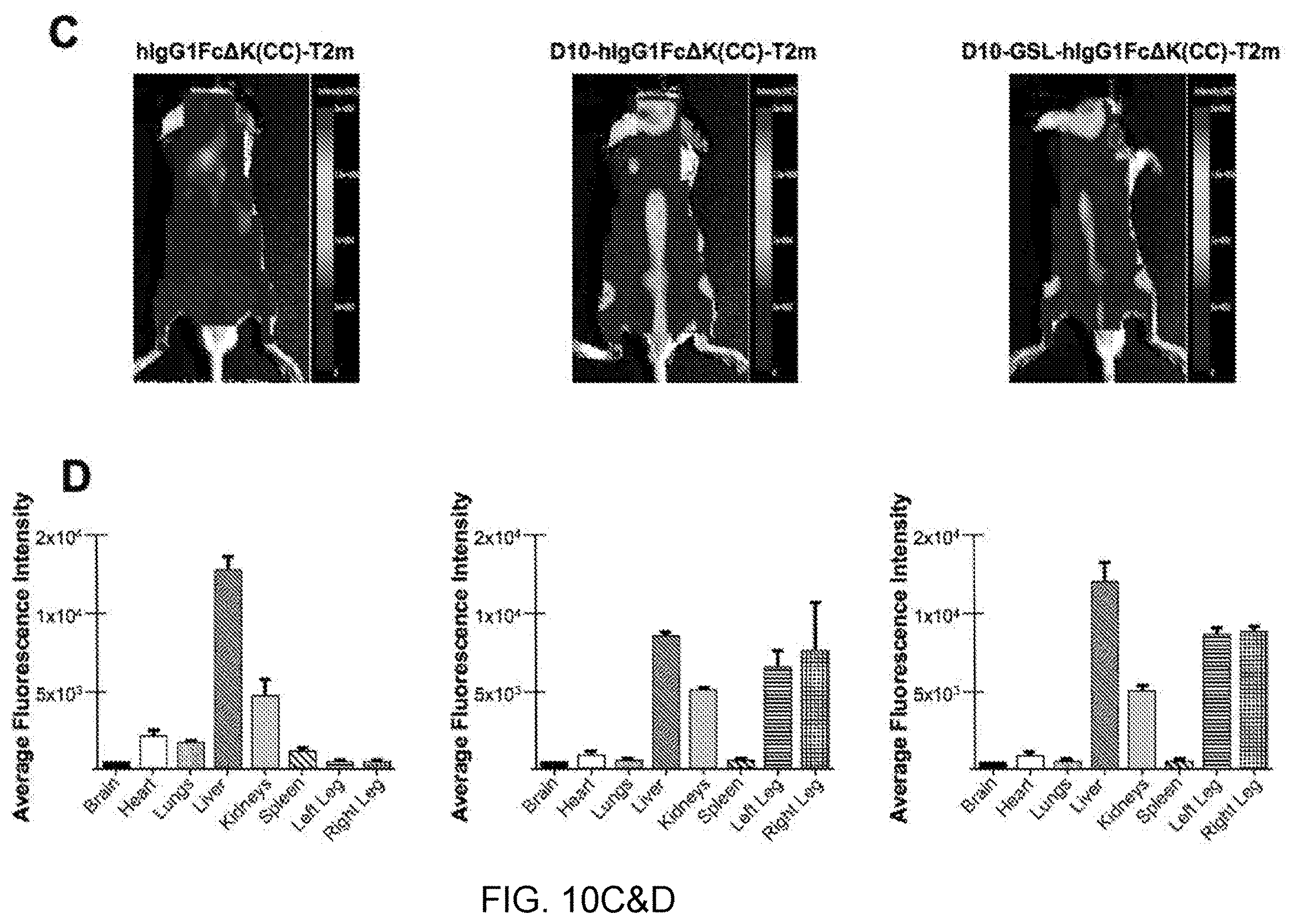

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.