Method For Suppressing Brain Metastasis
OCHIYA; Takahiro ; et al.
U.S. patent application number 16/745967 was filed with the patent office on 2020-07-09 for method for suppressing brain metastasis. The applicant listed for this patent is Gene Techno Science Co., Ltd. National Cancer Center Theoria Science Inc.. Invention is credited to Takahiro OCHIYA, Naoomi TOMINAGA.
| Application Number | 20200216912 16/745967 |
| Document ID | / |
| Family ID | 56789258 |
| Filed Date | 2020-07-09 |








View All Diagrams
| United States Patent Application | 20200216912 |
| Kind Code | A1 |
| OCHIYA; Takahiro ; et al. | July 9, 2020 |
METHOD FOR SUPPRESSING BRAIN METASTASIS
Abstract
A method for suppressing brain metastasis includes administering an miR-181c expression inhibitor, an miR-181c activity inhibitor, or an exosome secretion inhibitor as an active ingredient.
| Inventors: | OCHIYA; Takahiro; (Tokyo, JP) ; TOMINAGA; Naoomi; (Tokyo, JP) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 56789258 | ||||||||||
| Appl. No.: | 16/745967 | ||||||||||
| Filed: | January 17, 2020 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 15552766 | Aug 22, 2017 | 10544466 | ||
| PCT/JP2015/000906 | Feb 24, 2015 | |||
| 16745967 | ||||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | A61P 25/16 20180101; C12N 15/113 20130101; C12N 2310/141 20130101; A61P 25/00 20180101; C12Q 1/68 20130101; A61K 31/7088 20130101; C12Q 2600/158 20130101; A61P 25/28 20180101; A61P 25/08 20180101; C12Q 1/6886 20130101; A61P 35/04 20180101; C12Q 2600/178 20130101; C12N 15/09 20130101; A61K 45/00 20130101; C12M 1/34 20130101 |
| International Class: | C12Q 1/6886 20060101 C12Q001/6886; C12Q 1/68 20060101 C12Q001/68; A61K 31/7088 20060101 A61K031/7088; C12M 1/34 20060101 C12M001/34; C12N 15/09 20060101 C12N015/09; A61K 45/00 20060101 A61K045/00; C12N 15/113 20060101 C12N015/113; A61P 35/04 20060101 A61P035/04 |
Claims
1. A method for suppression of brain metastasis, comprising administering one or more agents selected from the group consisting of an miR-181c expression inhibitor, an miR-181c activity inhibitor, and an exosome secretion inhibitor as an active ingredient to a subject in need of the suppression of brain metastasis.
2. The method of claim 1, wherein the one or more agents selected from the group consisting of the miR-181c expression inhibitor, the miR-181c activity inhibitor, and the exosome secretion inhibitor is an agent selected from the following; (i) an antisense oligonucleotide, aptamer, or siRNA to pri-miR-181c or pre-miR-181c; (ii) an antisense oligonucleotide to miR-181c, an aptamer that specifically binds to miR-181c, siRNA against miR-181c, or miRNA mimetics of miR-181c; (iii) an antisense oligonucleotide, aptamer, or siRNA against the nSMase2 and/or RAB27B gene; or (iv) an antibody or an immunoreactive fragment thereof against nSMase2 and/or RAB27B; peptide mimetics or an aptamer that specifically binds to nSMase2 and/or RAB27B; or an antagonist of nSMase2 and/or RAB27B.
Description
INCORPORATION BY REFERENCE TO ANY PRIORITY APPLICATIONS
[0001] Any and all applications for which a foreign or domestic priority claim is identified in the Application Data Sheet as filed with the present application are hereby incorporated by reference under 37 CFR 1.57.
REFERENCE TO ELECTRONIC SEQUENCE LISTING
[0002] The present application is being filed along with an Electronic Sequence Listing. The Electronic Sequence Listing is provided as a file entitled 31839208 1.txt which is 103 kilobytes in size, created on Dec. 9, 2019. The information in the Electronic Sequence Listing is incorporated herein by reference in its entirety.
BACKGROUND OF THE INVENTION
Field of the invention
[0003] The present invention relates to the permeability control of the blood-brain barrier and particularly to a technique for increasing the permeability of the blood-brain barrier. The present invention further relates to the field of brain metastasis of cancer. More specifically, the present invention relates to the fields of diagnosis or risk assessment of brain metastasis and treatment or prevention of brain metastasis.
Description of the Related Art
[0004] Brain metastasis in cancer patient is known to relate to poor prognoses. Moreover, it also is known that passage of cancer cells through the blood-brain barrier (BBB) is an important event in brain metastasis, that main constituent cells of the BBB are brain microvascular endothelial cells (BMECs), and that BMECs are connected each other by tight junctions between cells and thereby have very selective permeability. Moreover, the proteins junctional adhesion molecules (JAM-1, JAM-2 and JAM-3), Occludin, Claudins, and Zonula Occludin proteins (ZO-1 and ZO-2) are known to constitute the tight junction. Furthermore, cyclooxygenase-2 (COX-2), the epidermal growth factor receptor (EGFR) ligand HB-EGF, and a 2,6-sialyltransferase (ST6GALNAC5) have been identified as mediators involved in the BBB invasion of cancer cells. However, the early stage molecule mechanisms of cancer cells passing through the BBB have not been revealed (Non Patent Literature 1). A study using a highly metastatic breast cancer cell line MDA-MB-231 has shown that vascular endothelial growth factor (VEGF) is highly expressed in breast cancer cells and increases the permeability of endothelial cells as well as adhesion of cancer cells onto endothelial cells, and thus VEGF is involved in the brain metastasis (Non Patent Literature 2). Although the expression of VEGF has been reported to be necessary for brain metastasis, it has been also reported that the expression of VEGF is not sufficient for brain metastasis, and the destruction mechanism of the BBB in brain metastasis has not been sufficiently explained by VEGF (Non Patent Literature 3).
[0005] A mechanism of BBB passage of Cryptococcus neoformans which infects the central nervous system (CNS) has been studied. It has been reported that the adhesion of Cryptococcus to human BMECs causes dephosphorylation of Cofilin, which results in actin rearrangement. This report further indicates that the dephosphorylation of Cofilin by Cryptococcus is controlled via the Rho kinase-LIM kinase-Cofilin pathway (Non Patent Literature 4).
[0006] Extracellular vesicles (EVs) including exosomes are known to mediate communication between cells by transporting protein, mRNA, and microRNA (miRNA) contained therein (Non Patent Literature 5). Moreover, it has been reported that extracellular vesicles released from cancer cells are involved in the malignancy of cancer cells in various ways (Non Patent Literature 8), for example, they inhibit the function of natural killer cells (Non Patent Literature 6) and increase the expression of the oncogene protein MET in bone marrow progenitor cells to change the bone marrow progenitor cells toward a pro-metastatic phenotype (Non Patent Literature 7). As seen above, the relation between EVs and cancer metastasis has been suggested. However, little was known about the relation between EVs (and substances contained therein) and the BBB.
CITATION LIST
Non Patent Literature
[0007] Non Patent Literature 1: Arshad, F. et al, Patholog Res Int 2011: 920509 (2010). [0008] Non Patent Literature 2: Lee, T. H. et al, J Biol Chem 278: 5277-5284 (2003). [0009] Non Patent Literature 3: Yano, S. et al, Cancer Res 60: 4959-4967 (2000). [0010] Non Patent Literature 4: Chen, H. M. Steven, et al, Journal of Medical Microbiology 52: 961-970 (2003). [0011] Non Patent Literature 5: Valadi, H. et al, Nat Cell Biol 9:654-659 (2007). [0012] Non Patent Literature 6: Liu, Cunren, et al, The Journal of Immunology 176: 1375-1385 (2006): [0013] Non Patent Literature 7: Peinado, H. et al, Nat Med 18: 883-891 (2012). [0014] Non Patent Literature 8: Yang, C. et al, Clin Dev Immunol 2011: 842-849 (2011).
SUMMARY OF INVENTION
[0015] An object of the present invention is to elucidate mechanisms of the destruction of the BBB in brain metastasis, and to provide new methods for diagnosis/risk assessment and for treatment/prevention of brain metastasis of cancer. Another object of the present invention is to provide a novel method for agent delivery into brain using the mechanism of the BBB destruction. A further object of the present invention is to provide an agent, a composition, and a kit to be used in these methods.
[0016] To elucidate the mechanism of brain metastasis, the present inventors transplanted a highly metastatic human breast cancer cell line to mice, and analyzed EVs obtained from cancer cells metastasized to the brain. As a result, it was found that cancer cell-derived EVs are taken in brain microvascular endothelial cells and destruct the blood-brain barrier (BBB). The present inventors then searched for a substance related to the destruction of the blood-brain barrier from the substances contained in the EVs, and found that miR-181c, a microRNA specifically contained in the EVs derived from the brain metastatic cancer cells, has an effect to destruct the blood-brain barrier. The present inventors further examined the effect of miR-181c in brain microvascular endothelial cells, and found that miR-181c binds to a noncoding region of 3-phosphoinositide dependent protein kinase-1 (hereinafter referred to as "PDPK1") and decreases expression of PDPK1. The present inventors further examined the relation between the decreased expression of PDPK1 and the disruption of the blood-brain barrier (BBB), and found that the inhibition of PDPK1 expression changes the localization of tight junction proteins. Thus, the present inventors have found that cancer cell-derived EVs release miR-181c contained therein in brain microvascular endothelial cells, which inhibits the expression of PDPK1, and thereby change the localization of tight junction proteins to destruct the blood-brain barrier.
[0017] miR-181c has been reported to be involved in the aggravation of cells, but has not been known in relation with the blood-brain barrier.
[0018] Moreover, PDPK1 has been known to mainly target protein kinase B (PKB/AKT1, PKB/AKT2, PKB/AKT3), p70 ribosomal protein S6 kinase (RPS6KB1), p90 ribosomal protein S6 kinase (RPS6KA1, RPS6KA and RPS6KA3), cyclic AMP-dependent protein kinase (PRKACA), protein kinase C (PRKCD and PRKCZ), serum- and glucocorticoid-inducible kinase (SGK1, SGK2 and SGK3), P21 activated kinase-1 (PAK1), and protein kinase PKN (PKN1 and PKN2), and known to be involved in the regulation of cell proliferation and survival as well as intake and storage of glucose and amino acids. However, there has been no report on PDPK1 in relation with the blood-brain barrier.
[0019] Accordingly, the present invention is based on the elucidation of the mechanism for destruction of the blood-brain barrier by brain metastatic cancer cells, and specifically relates to the following inventions: [0020] (1) A method for providing information for diagnosing brain metastasis, comprising measuring miR-181c level in a sample from a subject cancer patient. [0021] (2) A method for providing information for diagnosing brain metastasis in a subject cancer patient, comprising the steps of:
[0022] determining miR-181c level in a sample from the subject cancer patient, and
[0023] judging brain metastasis in the subject cancer patient from the determined miR-181c level,
[0024] wherein when the miR-181c level of the sample from the subject cancer patient is higher than that from a negative control, information that the subject cancer patient has brain metastasis is provided. [0025] (3) The method of (1) or (2), wherein the subject cancer patient is a Stage IV cancer patient. [0026] (4) A method for providing information for diagnosing brain metastasis, comprising measuring miR-181c level in a sample from a subject cancer patient. [0027] (5) A method for providing information for assessing risk of brain metastasis in a subject cancer patient, comprising the steps of:
[0028] determining miR-181c level in a sample from the subject cancer patient,
[0029] assessing risk of brain metastasis in the subject cancer patient from the determined miR-181c level,
[0030] wherein when the miR-181c level of the sample from the subject cancer patient is higher than that from a negative control, information that risk of brain metastasis is high in the subject cancer patient is provided. [0031] (6) The method of (4) or (5), wherein the subject cancer patient is a Stage I to III cancer patient. [0032] (7) The method of any one of (1) to (6), wherein the subject cancer patient is a breast cancer patient. [0033] (8) The method of any one of (1) to (7), wherein the negative control is a cancer patient with no brain metastasis or a healthy subject. [0034] (9) The method of any one of (1) to (8), wherein the sample is blood. [0035] (10) The method of any one of (1) to (9), wherein the miR-181c in the sample is miR-181c extracted from EVs in the sample. [0036] (11) An in vitro diagnostic reagent or an in vitro diagnostic measuring apparatus for diagnosis or risk assessment of brain metastasis by measuring miR181c, which is for use in a method of any one of (1) to (10). [0037] (12) An agent for diagnosis or risk assessment of brain metastasis, comprising a substance that specifically binds to miR-181c. [0038] (13) The agent of (12), wherein the substance that specifically binds to miR-181c is a nucleic acid that specifically binds to miR-181c. [0039] (14) The agent of (13), wherein the nucleic acid that specifically binds to miR-181c is a nucleic acid at least partially having an artificially designed sequence. [0040] (15) The agent of any one of (12) to (14), wherein the substance that specifically binds to miR-181c is labelled. [0041] (16) A pharmaceutical composition for suppressing brain metastasis, comprising one or more agents selected from the group consisting of an miR-181c expression inhibitor, an miR-181c activity inhibitor, and an exosome secretion inhibitor as an active ingredient. [0042] (17) The pharmaceutical composition of (16), wherein the one or more agents selected from the group consisting of an miR-181c expression inhibitor, an miR-181c activity inhibitor, and an exosome secretion inhibitor is an agent selected from: [0043] (i) an antisense oligonucleotide, aptamer, or siRNA against pri-miR-181c or pre-miR-181c; [0044] (ii) an antisense oligonucleotide to miR-181c, an aptamer that specifically binds to miR-181c, siRNA against miR-181c, or miRNA mimetics of miR-181c; [0045] (iii) an antisense oligonucleotide, aptamer, or siRNA against the nSMase2 and/or RAB27B gene; or [0046] (iv) an antibody or an immunoreactive fragment thereof against nSMase2 and/or RAB27B; peptide mimetics or an aptamer that specifically binds to nSMase2 and/or RAB27B; or an antagonist of nSMase2 and/or RAB27B. [0047] (18) A pharmaceutical composition for increasing permeability of blood-brain barrier, comprising a PDPK1 expression inhibitor or PDPK1 activity inhibitor as an active ingredient. [0048] (19) The pharmaceutical composition of (18), wherein the PDPK1 expression inhibitor or activity inhibitor is an antisense oligonucleotide against the PDPK1 gene, an aptamer that specifically binds to the PDPK1 gene, siRNA against the PDPK1 gene, or miRNA capable of suppressing the PDPK1 gene expression; or an antibody against PDPK1 or the immunoreactive fragment thereof; peptide mimetics or an aptamer that specifically binds to PDPK1; or an antagonist of PDPK1. [0049] (20) The pharmaceutical composition of (19), wherein the miRNA capable of suppressing the PDPK1 gene expression is miR-181c. [0050] (21) An agent-delivery composition for delivering a desired active ingredient into brain, comprising a PDPK1 expression inhibitor or PDPK1 activity inhibitor as an active ingredient. [0051] (22) The agent-delivery composition of (21), comprising the desired active ingredient in addition to the PDPK1 expression inhibitor or activity inhibitor. [0052] (23) The agent-delivery composition of (21) or (22), wherein the PDPK1 expression inhibitor or activity inhibitor is an antisense oligonucleotide against the PDPK1 gene, an aptamer that specifically binds to the PDPK1 gene, siRNA against the PDPK1 gene, or miRNA capable of suppressing the PDPK1 gene expression; or an antibody against PDPK1 or the immunoreactive fragment thereof; peptide mimetics or an aptamer that specifically binds to PDPK1; or an antagonist of PDPK1. [0053] (24) The agent-delivery composition of (23), wherein the miRNA capable of suppressing the PDPK1 gene expression is miR-181c. [0054] (25) An apparatus for judging brain metastasis in a subject cancer patient, comprising:
[0055] miR-181c-measuring means for measuring a polynucleotide having a nucleotide sequence of miR-181c or a part thereof in a sample from the subject cancer patient,
[0056] miR-181c level-determining means for determining miR-181c level in the sample from the measurement by the miR-181c-measuring means, and
[0057] metastasis-judging means for judging brain metastasis in the subject cancer patient from the determined miR-181c level by the miR-181c level-determining means;
[0058] wherein the metastasis-judging means judges that the subject cancer patient has brain metastasis when the miR-181c level in the sample from the subject cancer patient is higher than the miR-181c level in a sample from a negative control. [0059] (26) An apparatus for assessing risk of brain metastasis in a subject cancer patient, comprising:
[0060] miR-181c-measuring means for measuring a polynucleotide having a nucleotide sequence of miR-181c or a part thereof in a sample from the subject cancer patient,
[0061] miR-181c level-determining means for determining miR-181c level in the sample from the measurement by the miR-181c-measuring means, and
[0062] risk-assessing means for assessing risk of brain metastasis in the subject cancer patient from the determined miR-181c level by the miR-181c level-determining means;
[0063] wherein the risk-assessing means assesses that the subject cancer patient has a high risk of brain metastasis when the miR-181c level in the sample from the subject cancer patient is higher than the miR-181c level in a sample from a negative control. [0064] (27) A computer program to be installed in an apparatus for providing information for diagnosing brain metastasis in a subject cancer patient,
[0065] wherein the computer program directs the apparatus for diagnosing brain metastasis in a cancer patient to execute:
[0066] an miR-181c-measuring procedure for measuring a polynucleotide having a nucleotide sequence of miR-181c or a part thereof in a sample from the subject cancer patient,
[0067] an miR-181c level-determining procedure for determining an miR-181c level in the sample from the measurement by the miR-181c-measuring procedure,
[0068] a metastasis-judging procedure for judging brain metastasis in the subject cancer patient from the determined miR-181c level by the miR-181c level-determining procedure, and
[0069] a judgment-output procedure of outputting the judgment by the metastasis-judging procedure; and
[0070] the metastasis-judging procedure judges that the subject cancer patient has brain metastasis when the miR-181c level in the sample from the subject cancer patient is higher than the miR-181c level in a sample from a negative control. [0071] (28) A computer program to be installed in an apparatus for assessing risk of brain metastasis in a subject cancer patient,
[0072] wherein the computer program directs the apparatus for diagnosing brain metastasis in a cancer patient to execute:
[0073] an miR-181c-measuring procedure for measuring a polynucleotide having a nucleotide sequence of miR-181c or a part thereof in a sample from the subject cancer patient,
[0074] an miR-181c level-determining procedure for determining an miR-181c level in the sample from the measurement by the miR-181c-measuring procedure,
[0075] a risk-assessing procedure for assessing risk of brain metastasis in the subject cancer patient from the determined miR-181c level by the miR-181c level-determining procedure;
[0076] an assessment-output procedure of outputting the assessment by the risk-assessing procedure; and
[0077] the risk-assessing procedure assesses that the subject cancer patient has a high risk of brain metastasis when the miR-181c level in the sample from the subject cancer patient is higher than the miR-181c level in a sample from a negative control.
[0078] As used herein, the term "cancer" includes epithelial malignant tumors, hematopoietic malignant tumors derived from spinal cord, and the like, and particularly includes ovarian cancer (non-mucinous ovarian cancers and the like), uterine cancer, endometrial cancer, breast cancer, breast adenocarcinoma, prostate cancer, testicular cancer (testicular choriocarcinoma and the like), brain cancer (ependymoma and the like), throat cancer, lung cancer, lung adenocarcinoma, kidney cancer (renal cell cancer and the like), liver cancer, large bowel cancer (colon cancer), pleural mesothelioma, sarcoma, chronic and acute myeloid leukemia, and metastatic cancers such as lung metastatic cancer. As used herein, a "cancer patient" means a patient having cancer (at a site except the brain) with or without metastasis and may be a patient with, for example, hematopoietic cell malignant tumors such as leukemia, lymphomas (Hodgkin's disease and non-Hodgkin's lymphom, and the like), multiple myeloma; breast cancer; endometrial cancer; cervical cancer; ovarian cancer; esophageal cancer; gastric cancer; appendiceal cancer; large bowel cancer (colon cancer, rectal cancer, and the like); liver cancer (hepatocellular cancer, and the like); gallbladder cancer; bile duct cancer; pancreatic cancer; adrenal cancer; gastrointestinal stromal tumor; mesothelioma; head and neck cancers such as laryngeal cancer, oral cancers (oral floor cancer, gingival cancer, tongue cancer, buccal mucosa cancer, and the like); salivary gland cancer; nasal sinus cancers (maxillary sinus cancer, frontal sinus cancer, ethmoid cancer, sphenoidal sinus cancer, and the like); thyroid cancer; kidney cancer; lung cancer; osteosarcoma; prostate cancer; testicular tumor (testicular cancer); renal cell cancer; bladder cancer; rhabdomyosarcoma; skin cancer; or anal cancer.
[0079] The stage of cancer is determined according to a stage classification (clinical advanced stage classification). The detailed criteria for the stage classification of each cancer (organ) has been established, based on, for example, the size of cancer (T), metastasis to nearby lymph nodes (N) and metastasis to a distant organ (M) (TNM classification), which has been widely known in the art (see the website "Cancer staging" of U.S. National Cancer Center, http://www.cancer.gov/cancertopics/factsheet/detection/staging; the latest edition of AJCC Cancer Staging Manual (American Joint Commitee on Cancer)). For example, the staging criteria for breast cancer is as follows. Stage 0: non-invasive cancer: the breast cancer stays in the mammary gland where it has developed (including the Paget's disease); Stage I: lump size of 2 cm or smaller without metastasis to the lymph nodes; Stage IIA: lump size of 2 cm or smaller with metastasis to the axillary lymph nodes, or lump size of 2.1-5 cm without metastasis to the lymph nodes; Stage IIB: lump size of 2.1-5 cm with metastasis to the axillary lymph, or lump size of 5.1 cm or larger without metastasis to the lymph nodes; Stage IIIA: lump size of 5.1 cm or larger with metastasis to the axillary lymph, any size of lump with heavy metastasis to the axillary lymph nodes, or any size of lump without metastasis to the axillary lymph nodes but with metastasis to parasternal lymph nodes; Stage IIIB: any size of lump with infiltration to the chest wall and/or skin; Stage IIIC: any size of lump with metastasis spread to infraclavicular or supraclavicular lymph nodes; Stage IV: metastasis to organs distant from the breast.
[0080] Patients at Stage IV in the cancer staging usually have metastasis to distant organs. Accordingly, in such patients, cancer cells have already invaded into the blood vessels, and therefore brain metastasis is very highly possible if miR-181c level in blood (in EVs contained in the blood) is high and the blood-brain barrier is open. On the other hand, in Stage I to III patients who do not have metastasis to distant organs, since cancer cell is not likely to invade into blood vessels, brain metastasis does not occur immediately even if the miR-181c level in blood (in EVs contained in blood) is high and the blood-brain barrier is open. However, these Stage I to III patients have high possibility (risk) of brain metastasis in the future invasion of cancer cells into the blood. Accordingly, whether a method, agent, apparatus, computer program or the like according to the present invention is used for diagnosis of brain metastasis or for risk assessment of brain metastasis can be determined according to a state of the patient from which the measured sample is derived. When the cancer staging is employed as the state, risk assessment of brain metastasis can be conducted for Stage I to III patients, and diagnosis of brain metastasis can be conducted for Stage IV patients. Instead of cancer staging, the presence or absence of metastasis to a distant organ or the presence or absence of infiltration of cancer cell into the blood may be employed as the state to select either of diagnosis of brain metastasis or risk assessment of brain metastasis. In this case, when there is metastasis to a distant organ or when there is infiltration of cancer cell into the blood, the present invention can be used for a diagnosis of brain metastasis, while when there is no metastasis to a distant organ or when there is no infiltration of cancer cell into the blood, the present invention is used for risk assessment of brain metastasis. Accordingly, as used herein, the "Stage IV patients" may be replaced with patients with metastasis to a distant organ or patients with infiltration of cancer cell into the blood. Also, the "Stage I to III patients" may be replaced with patients with no metastasis to a distant organ or patients with no infiltration of cancer cell into the blood.
[0081] As used herein, the "brain metastasis" and "metastasis to the brain" mean that cancer cells leave the primary lesion developed in other sites than brain, infiltrate into the brain, and grow in the brain, or the state that such infiltrated cancer cells have grown in the brain. Whether there is brain metastasis or not in a cancer patient having a primary lesion other than brain can be determined by a method of diagnostic imaging well known in the art. For example, when tumorigenesis in the brain is confirmed in images such as CT or MRI image for a cancer patient having a primary lesion other than brain to which a gadolinium contrast agent or the like has been administered, it can be determined that there is brain metastasis.
[0082] As used herein, the "healthy subject" means a human having no cancer development. The healthy subject herein is not necessary to have no disease other than cancer, but is preferably a healthy human having no disease even which is not cancer.
[0083] According to the present invention, miR-181c serves to open the BBB and promote brain metastasis in cancer patients. Accordingly, information about brain metastasis in a cancer patient can be obtained by measuring the miR-181c levels in the cancer patient. As used herein, the "miR-181c" is a human miRNA consisting of approximately 22 nucleotides (SEQ ID NO: 1) (Lim L P, et al. (2003) Science, 299: 1540; Landgraf P, et al. (2007) Cell, 129:1401-1414).
[0084] The miR-181c levels are measured by using a substance that specifically binds to miR-181c. As used herein, the "substance that specifically binds to miR-181c" is not particularly limited as long as it is a substance that can specifically bind to miR-181c. Since a nucleic acid usually bind specifically to a nucleic acid having its complementary sequence, a preferable substance that specifically binds to miR-181c is a nucleic acid that specifically binds to miR-181c.
[0085] The "nucleic acid that specifically binds to miR-181c" means a nucleic acid molecule that can specifically bind to miR-181c. Such a nucleic acid molecule usually has a complementary sequence to miR-181c. The "nucleic acid" as used herein comprises a DNA, an RNA, or an artificially generated nucleic acid (including bridged nucleic acids such as locked nucleic acids (2',4'-BNA)), or a combination thereof. For example, the nucleic acid that specifically binds to miR-181c includes, but is not limited to, a primer or a probe. The "primer" is usually a nucleic acid molecule of 10-30 mer (preferably, 17-25 mer, 15-20 mer, or the like) that is used for the amplification of nucleic acid and has, at least in a part (preferably, 7 mer or more, 8 mer or more, 9 mer or more, 10 mer or more, 11 mer or more, 12 mer or more, 13 mer or more, 14 mer or more, 15 mer or more, 20 mer or more, 25 mer or more, or 30 mer or more) thereof, a sequence complementary to a sequence located at an end of the sequence to be amplified. The "probe" is a nucleic acid molecule of 10-200 mer (preferably 10-100 mer, 10-50 mer, 10-30 mer, 10-20 mer, and the like) that has a sequence complementary to a target sequence, and can specifically bind to a target sequence, and has a complementary to a sequence, in at least a part (preferably, 7 mer or more, 8 mer or more, 9 mer or more, 10 mer or more, 11 mer or more, 12 mer or more, 13 mer or more, 14 mer or more, 15 mer or more, 20 mer or more, 25 mer or more, 30 mer or more, 50 mer or more, 100 mer or more) thereof. Methods for designing primers and probes for a target sequence are well known in the art. Preferably, the nucleic acid that specifically binds to miR-181c comprises at least partially an artificially designed sequence (for example, sequences for labeling and tagging).
[0086] As used herein, a substance "specifically binds" means that the substance binds to the target sequence or structure with a substantially higher affinity than binding to other nucleotide sequences or amino acid sequences or structures thereof. The "substantially higher affinity" means a sufficiently high affinity to allow the detection of the target sequence or structure with distinguishing from other sequences or structures in using a desired measuring apparatus or method. For example, the substantially higher affinity may mean that the molecules number of the (miR-181c specifically binding) substances that bind to miR-181c is 3 times or more, 4 times or more, 5 times or more, 6 times or more, 7 times or more, 8 times or more, 9 times or more, 10 times or more, 15 times or more, 20 times or more, 30 times or more, 50 times or more of the molecules number of the (miR-181c specifically binding) substances that bind to other sequences or structures. The binding constant (Ka) in the binding between the miR-181c specifically binding substance of the present invention and miR-181c may be, for example, at least 10.sup.7 M.sup.-1, at least 10.sup.8 M.sup.-1, at least 10.sup.9 M.sup.-1, at least 10.sup.10 M.sup.-1, at least 10.sup.11 M.sup.-1, at least 10.sup.12 M.sup.-1, or at least 10.sup.13 M.sup.-1.
[0087] The "substance that specifically binds to miR-181c (miR-181c specifically binding substances)" (including the nucleic acid that specifically binds to miR-181c) herein may be labelled as needed. Examples of methods for labeling include the radioisotope (RI) labeling, fluorescent labeling, and enzymatic labeling. Examples of a radioisotope used in the RI labelling include 32P, 131I, 35S, 45Ca, 3H, and 14C. Examples of a fluorescent dye used in the fluorescent labeling include DAPI, SYTOX.RTM. Green, SYTO.RTM. 9, TO-PRO(R)-3, Propidium Iodide, Alexa Fluor.RTM. 350, Alexa Fluor.RTM. 647, Oregon Green.RTM., Alexa Fluor.RTM. 405, Alexa Fluor.RTM. 680, Fluorescein (FITC), Alexa Fluor.RTM. 488, Alexa Fluor.RTM. 750, Cy.RTM. 3, Alexa Fluor.RTM. 532, Pacific Blue.TM., Pacific Orange.TM., Alexa Fluor.RTM. 546, Coumarin, Tetramethylrhodamine (TRITC), Alexa Fluor.RTM. 555, BODIPY.RTM. FL, Texas Red.RTM., Alexa Fluor.RTM. 568, Pacific Green.TM., Cy.RTM. 5, and Alexa Fluor.RTM. 594. Available enzymatic labeling include biotin (biotin-16-dUTP, biotin-11-dUTP), digoxigenin (DIG: a steroid based natural product) (deoxyuridine 5'-triphosphate), and alkaline phosphatase.
[0088] According to the present invention, PDPK1 is one of the substances playing major roles in maintaining the structure of the BBB. Therefore, the BBB can be opened by suppressing the expression or inhibiting activity of PDPK1. Accordingly, a PDPK1 expression inhibitor or a PDPK1 activity inhibitor can be used as a DDS agent for an agent intended to be delivered into the brain. As used herein, "PDPK1" is an abbreviation of 3-phosphoinositide dependent protein kinase-1. PDPK1 is a serine/threonine kinase that phosphorylates and activates the AGC family of protein kinases. It is known that there are plural variants of the PDPK1 mRNA and protein. The nucleotide sequence of the mRNA of PDPK1 vulariant1, and the amino acid sequence of the PDPK1 vulariant1 protein are set forth in SEQ ID NO: 2 and SEQ ID NO: 3, respectively. The nucleotide sequence of the mRNA of PDPK1 vulariant2, and the amino acid sequence of the PDPK1 vulariant2 protein are set forth in SEQ ID NO: 4 and SEQ ID NO: 5, respectively. The nucleotide sequence of the mRNA of PDPK1 vulariant3, and the amino acid sequence of the PDPK1 vulariant3 protein is set forth in SEQ ID NO: 6 and SEQ ID NO: 7, respectively. The nucleotide sequence of the mRNA of PDPK1 vulariantX1, and the amino acid sequence of the PDPK1 vulariantX1 protein is set forth in SEQ ID NO: 8 and SEQ ID NO: 9, respectively. The PDPK1 herein may be any of these variants or one of their mutants that may occur in the body.
[0089] The present inventors have also found that, in cancer patients, miR-181c is contained in exosomes during traveling through the blood, and when the exosomes have reached the BBB, the exosomes are incorporated into the BBB constituting cells and open the blood-brain barrier. Accordingly, the inhibition of secretion of exosomes in cancer patients can prevent the opening of the blood-brain barrier and thereby can suppress brain metastasis. Thus, in one aspect, the present invention relates to suppression of brain metastasis using an exosome secretion inhibitor. As used herein, the "exosome secretion inhibitor" is not particularly limited as long as it is an agent that can inhibit the exosome secretion from cancer cells. It is known that neutral sphingomyelinase (hereinafter, referred to as the "nSMase2") (the cDNA sequence is set forth in SEQ ID NO: 10 and the amino acid sequence is set forth in SEQ ID NO: 11) and RAB27B (the cDNA sequence is set forth in SEQ ID NO: 12 and the amino acid sequence is set forth in SEQ ID NO: 13) are involved in the secretion of exosomes. Accordingly, the secretion of exosomes can be inhibited by suppressing the expression or inhibiting the activity of nSMase2 or RAB27B. Thus, an expression or activity inhibitor of nSMase2 or RAB27B can be used as an exosome secretion inhibitor in the present invention.
[0090] As used herein, the "expression inhibitor" of a gene, a protein, or an miRNA is not particularly limited, as long as it is an agent that suppresses the expression of the gene, protein, or miRNA. It is well-understood in the art that an expression inhibitor of a target gene can be obtained and used by designing and/or selecting, for example, an antisense to the gene sequence or to the mRNA sequence generated by transcription of the gene, dsRNA to the mRNA generated by the transcription of the gene, an aptamer to the mRNA generated by the transcription of the gene, or miRNA (including pri-miRNA and pre-miRNA) that can bind to a nucleotide sequence which is located upstream of a coding region and is essential for the expression of the gene.
[0091] It is well-understood in the art that an expression inhibitor of a target protein can be obtained or used by designing and/or selecting, for example, an antisense to a gene sequence encoding the protein or to an mRNA sequence generated by the transcription of the gene, dsRNA to an mRNA generated by the transcription of a gene encoding the protein, an aptamer to a gene sequence encoding the protein or to an mRNA generated by the transcription of the gene, or miRNA (including pri-miRNA and pre-miRNA) that can bind to a nucleotide sequence which is located upstream of a coding region and is essential for the expression of a gene encoding the protein.
[0092] It is well-understood in the art that an expression inhibitor of miRNA can be obtained or used by designing and/or selecting, for example, an antisense to a gene encoding the miRNA, pri-miR of the miRNA, or pre-miR of the miRNA; dsRNA to pri-miR of the miRNA or pre-miR of the miRNA; an aptamer to the pri-miR of the miRNA or pre-miR of the miRNA; or another miRNA (including pri-miRNA and pre-miRNA) that can bind to a nucleotide sequence which is located upstream of a coding region and is essential for the expression of a gene encoding the miRNA.
[0093] For example, the present inventors have found that expression inhibitors of PDPK1 include pri-miR-181c, pre-miR-181c, miR-181c and derivatives thereof.
[0094] As used herein, the derivatives of pri-miRNA, pre-miRNA and miRNA are molecules that have at least a seed sequence (the 7 nucleotides at positions 2 to 8 from the 5' end: ACUUACA) in the miRNA and that act to suppress the expression of the gene (or protein) of interest (PDPK1 in the case of miR-181c). The function of such a derivative is not necessarily quantitatively same as that of the original miRNA, but the derivative may have stronger or weaker activity than the original miRNA as long as it achieves an object of the present invention. Furthermore, the derivatives have a nucleotide sequence having an identity of about 70% or more and less than 100%, about 75% or more and less than 100%, about 80% or more and less than 100%, about 85% or more and less than 100%, about 90% or more and less than 100%, about 95% or more and less than 100% with the pri-miRNA, pre-miRNA and miRNA. The higher sequence identity means the more similarity in the structures between the pri-miRNA, pre-miRNA, or miRNA and the derivative thereof. The derivatives of the pri-miRNA, pre-miRNA, and miRNA may have a sequence that the original pri-miRNA, pre-miRNA, and miRNA do not have. Moreover, the nucleotide lengths of the derivative of pri-miRNA, pre-miRNA, and miRNA may be different from the original nucleotide length as long as they can exhibit the function of the miRNA and may be, for example, 10-50 mer, 15-30 mer, or 20-25 mer.
[0095] Moreover, the derivatives of the pri-miRNA, pre-miRNA, and miRNA include a nucleic acid molecule which has a seed sequence of the pri-miRNA, pre-miRNA and miRNA, and has one or more of addition, substitution, deletion, and modification selected from the following (i) to (iv) (at other than the seed sequence): [0096] (i) addition of one or more (for example, 1-10, 1-5, 1-3, 1-2, or 1) nucleotide(s) to the wildtype sequence of pri-miR-181c, pre-miR-181c, or miR-181c; [0097] (ii) substitution of one or more (for example, 1-10, 1-5, 1-3, 1-2, or 1) nucleotide(s) in the wildtype sequence of pri-miR-181c, pre-miR-181c, or miR-181c with other nucleotide(s); [0098] (iii) deletion of one or more (for example, 1-10, 1-5, 1-3, 1-2, or 1) nucleotide(s) in the wildtype sequence of pri-miR-181c, pre-miR-181c, or miR-181c; and [0099] (iv) modification of one or more (for example, 1-10, 1-5, 1-3, 1-2, or 1) nucleotide(s) in the wildtype sequence of pri-miR-181c, pre-miR-181c, or miR-181c.
[0100] As used herein, the "activity inhibitor" of a protein or miRNA is not particularly limited as long as it is an agent that makes the protein or miRNA incapable of exhibiting its activity. For example, the activity inhibitor may be a substance that binds to the protein or miRNA, or a substance that binds to a substance to which the protein or miRNA binds. Known activity inhibitors of proteins are mimetics, antibodies or immunoreactive fragments thereof, aptamers, receptor derivatives, or antagonists. Known activity inhibitors of miRNA are mimetics, antisense DNA/RNA or dsRNA having a sequence complementary to at least a partial sequence of the miRNA, or aptamers to the miRNA.
[0101] As used herein, the "antisense" is a nucleic acid having a complementary sequence of the target sequence (for example, the sequence of pri-miR-181c, pre-miR-181c, or miR-181c or a DNA encoding pri-miR-181c) and may be DNA or RNA. The antisense is not required to be 100% complementary to the target sequence and may contain non-complementary nucleotides as long as it can specifically hybridize under stringent conditions (Sambrook et al., 1989, Molecular Cloning, A Laboratory Manual, Cold Spring Harbor Press, N.Y., and Ausubel et al. (eds.), 1995, Current Protocols in Molecular Biology, (John Wiley & Sons. N.Y.) at Unit 2.10). Once introduced into cells, the antisense binds to the target sequence and inhibits the transcription, the processing of RNA, the translation, or the stability thereof. The antisense includes antisense polynucleotides as well as polynucleotide mimetics and those that contain a modified back bone. Such an antisense can be designed and produced (for example, chemically synthesized) appropriately using a method well known in the art based on the target sequence information.
[0102] The "dsRNA" is RNA containing double strand RNA structure that has at least partially a sequence complementary to the target sequence. The dsRNA binds to mRNA having the target sequence, and thereby degrades the mRNA to suppress the translation (expression) of the target sequence by RNA interference (RNAi). The dsRNA includes siRNA (short interfering RNA) and shRNA (short hairpin RNA). The dsRNA is not required to have 100% identity with the target sequence as long as it can suppress the expression of the target gene. Moreover, a part of the dsRNA may be substituted with DNA for stabilization or another purpose. The siRNA is preferably double strand RNA having 21-23 nucleotide each. The siRNA can be obtained by a method well known in the art, for example, by chemical synthesis or as an analog of naturally occurring RNA. The shRNA is a RNA short chain having a hairpin turn structure. The shRNA can be obtained by a method well known in the art, for example, by chemical synthesis or introducing a gene encoding the shRNA into cells and expressing the gene.
[0103] The "aptamer" are nucleic acids that bind to a substance such as protein or nucleic acid. The aptamers may be constituted of RNA or DNA. The form of the nucleic acids may be a double strand or a single strand. The length of the aptamer is not particularly limited as long as it can specifically bind to the target molecule, but is for example, 10-200 nucleotides, preferably 10-100 nucleotides, more preferably 15-80 nucleotides, or further preferably 15-50 nucleotides. The aptamers can be selected using a method well known in the art and, for example, the SELEX (Systematic Evolution of Ligands by Exponential Enrichment) method (Tuerk, C. and Gold, L., 1990, Science, 249: 505-510).
[0104] As used herein, the "mimetics" are substances that have a structure similar to that of the protein or the miRNA. The mimetics can bind to the binding partners of the protein or the miRNA or can compete with the protein or miRNA, without exhibiting the activity of the protein or miRNA.
[0105] The "antibody" as used herein may be polyclonal antibody or monoclonal antibody and includes a non-human animal antibody, antibody having both of an amino acid sequence of a non-human animal antibody and an amino acid sequence of a human antibody (chimeric antibody and humanized antibody), and a human antibody. The immune globulin class of the antibodies may be any immune globulin class (isotype), IgG, IgM, IgA, IgE, IgD, or IgY, and, in the case of IgG, may be any subclass (IgG1, IgG2, IgG3, or IgG4). Furthermore, the antibody may be monospecific, bispecific (bispecific antibody), or trispecific (trispecific antibody) (for example, WO1991/003493). The immunoreactive fragments of an antibody mean proteins or peptides containing a part (partial fragment) of the antibody with maintaining the antibody's activity (immunoreactivity, binding activity) to the antigen. Examples of such immunoreactive fragment includes F(ab').sub.2, Fab', Fab, Fab.sub.3, single strand Fv (hereinafter, referred to as the "scFv"), (tandem) bispecific single strand Fv (sc(Fv).sub.2), single strand triple bodies, nanobodies, divalent V.sub.HHs, pentavalent V.sub.HHs, minibodies, (double strand) diabodies, tandem diabodies, bispecific tribodies, bispecific bibodies, dual affinity retargeting molecules (DART), triabodies (or tribodies), tetrabodies (or [sc(Fv).sub.2].sub.2) or (scFv-SA).sub.4), disulfide linked Fv (hereinafter, referred to as "dsFv"), compact IgGs, heavy chain antibodies or polymers thereof (see Nature Biotechnology, 29 (1): 5-6 (2011); Maneesh Jain et al., TRENDS in Biotechnology, 25 (7) (2007): 307-316; and Christoph stein et al., Antibodies (1): 88-123 (2012)). As used herein, the immunoreactive fragments may be any of monopecific, bispecific, trispecific, and multispecific antibodies. The aptamers are nucleic acid molecules capable of binding to a specific nucleic acid or protein. The receptor derivative means a substance having a binding region structure of a receptor to which the protein of interest binds. The receptor derivative includes a conjugate of the constant region of an antibody with a receptor and a membrane protein receptor solubilized by deleting the transmembrane region. The antagonist includes a wide range of substances that competes with the protein or miRNA of interest and inhibits the function thereof.
[0106] Herein, when the activity inhibitor binds to the protein or miRNA or when the activity inhibitor binds to a substance to which the protein or miRNA binds to, the binding is preferably specific and the binding constant (Ka) thereof may be for example, at least 10.sup.7 M.sup.-1, at least 10.sup.8 M.sup.-1, at least 10.sup.9 M.sup.-1, at least 10.sup.10 M.sup.-1, at least 10.sup.11 M.sup.-1, at least 10.sup.12 M.sup.-1, or at least 10.sup.13 M.sup.-1.
[0107] As used herein, the "agent" is a substance mainly used as an active ingredient, and includes a small molecule, a nucleic acid molecule (DNA and/or RNA), a protein, and a fusion thereof. As used herein, the "composition" comprises at least an agent that serves as an active ingredient and optionally comprises water, a solvent, and other additives as needed. The composition herein may comprise only one substance or 2 or more substances as an active ingredient(s). As used herein, the "active ingredient" is not particularly limited, as long as it can exhibit a desired biological activity, and includes a substance capable of exhibiting a therapeutic effect or preventive effect as well as a substance exhibiting a function as DDS. Whether a substance is an active ingredient or not can be determined not only depending on whether the substance has a biological action or not, but also depending on whether the amount of the substance contained in the composition can allow the substance to exhibit the biological action.
[0108] The "composition for diagnosis or risk assessment of brain metastasis" is a composition to be used in a method for diagnosing brain metastasis or a method for assessing risk of brain metastasis as described below, and includes a diagnostic composition (including an extracorporeal diagnostic composition). The composition for diagnosis or risk assessment of brain metastasis may be used for in vivo, ex vivo, or in vitro diagnosis, but preferably is for ex vivo or in vitro use. Compositions for in vivo diagnosis or risk assessment may be formulated to be administered to a patient in accordance with the formulation of pharmaceutical compositions as described below. Examples of the composition for diagnosis or risk assessment herein include an X-ray contrast agent, a reagent for general examination, a reagent for blood test, a reagent for biochemical test, a reagent for immunoserologic examination, an agent for bacteriological examination, and a reagent for functional examination. Since the composition for diagnosis or risk assessment herein needs to determine miR-181c level in a sample, the composition comprises the substance that specifically binds to miR-181c described above.
[0109] In another aspect, the composition for diagnosis or risk assessment herein may be provided as a kit for diagnosis or risk assessment comprising the composition as an active ingredient. The kit can be constituted by any materials as long as miRNA levels can be determined. For example, the kit for diagnosis or risk assessment herein may be a kit comprising a primer that can bind to miR-181c (a kit for measuring miRNA levels by using gene amplification technique such as PCR) or a kit comprising a probe that can bind to miR-181c (a kit for measuring miRNA levels by using a nucleic acid binding detection technique such as hybridization).
[0110] Alternatively, the kit for diagnosis or risk assessment herein may be a kit which determines the miRNA level utilizing an expression inhibitory effect of miRNA as indicator. In this case, the kit may comprise, for example, a nucleic acid encoding a label (luciferase, or the like) having an mRNA 3'UTR that can specifically bind to miR-181c (a kit for measuring miRNA levels by detecting the expression of the label in the reporter assay or the like).
[0111] As used herein, the "pharmaceutical composition" is a composition to be used for treatment or prevention and comprises a therapeutic or preventive agent. The pharmaceutical composition is preferably prepared into a unit dosage form comprising a suitable dose of the active ingredient. Moreover, any oral or parenteral formulation may be employed as long as it can be administered to a patient. Examples of compositions for parenteral administration include injections, nasal drops, suppositories, patches, ointments, and the like, and are preferably injections. Examples of the dosage form of the pharmaceutical composition of the present invention include solutions or lyophilized formulations. When the pharmaceutical composition of the present invention is used as an injection, an excipient (see "Japanese Pharmaceutical Excipients Directory" Yakuji Nippo Limited.; "Handbook of Pharmaceutical Excipients Fifth Edition" APhA Publications) including a solubilizing agent such as propylene glycol or ethylenediamine, a buffer such as phosphate, an isotonizing agent such as sodium chloride or glycerin, a stabilizer such as sulfite, a preservative such as phenol, or a soothing agent such as lidocaine may be added as needed. Moreover, when the pharmaceutical composition of the present invention is used as an injection, a storage container therefor may be an ampoule, a vial, a prefilled syringe, a cartridge for pen type syringes, a bag for infusion, and the like. For example, the pharmaceutical composition may usually contain 5-500 mg, 5-100 mg, 10-250 mg of the active ingredient per unit dosage form.
[0112] As used herein, the pharmaceutical composition for suppressing brain metastasis may comprise one or more agents selected from an miR-181c expression inhibitor, an miR-181c activity inhibitor, and an exosome secretion inhibitor as an active ingredient. Moreover, the pharmaceutical composition for suppressing brain metastasis may comprise one or more agents selected from an miR-181c expression inhibitor, an miR-181c activity inhibitor, and an exosome secretion inhibitor as active ingredient(s), or further comprise other agent(s) to be used in combination as active ingredient(s). Examples of such other agents include other cancer metastasis inhibitors and anticancer agents.
[0113] Examples of the other cancer metastasis inhibitors or anticancer agents that can be included in the pharmaceutical composition for suppressing brain metastasis include alkylating agents including nitrogen mustards such as cyclophosphamide, ifosfamide, melphalan, busulfan, and thiotepa, and nitrosoureas such as nimustine, ranimustine, Dacarbazine, procarbazine, temozolomide, carmustine, Streptozotocin, bendamustine; platinum compounds such as cisplatin, carboplatin, oxaliplatin, and nedaplatin; antimetabolites such as enocitabine, capecitabine, carmofur, cladribine, gemcitabine, cytarabine, cytarabine ocfosphate, tegafur, tegafur/uracil, tegafur/gimeracil/oteracil potassium, doxifluridine, nelarabine, hydroxycarbamide, 5-fluorouracil (5-FU), fludarabine, pemetrexed, pentostatin, mercaptopurine, and methotrexate; plant alkaloids or microtubule inhibitors such as irinotecan, etoposide, eribulin, sobuzoxane, docetaxel, nogitecan, paclitaxel, vinorelbine, vincristine, vindesine, and vinblastine: antitumor antibiotics such as actinomycin D, aclarubicin, amrubicin, idarubicin, epirubicin, zinostatin stimalamer, daunorubicin, doxorubicin, pirarubicin, bleomycin, peplomycin, mitomycin C, mitoxantrone, and liposomal doxorubicin; cancer vaccines such as Sipuleucel-T; molecular target drugs such as ibritumomab tiuxetan, imatinib, everolimus, erlotinib, gefitinib, gemtuzumab ozogamicin, sunitinib, cetuximab, sorafenib, dasatinib, tamibarotene, trastuzumab, tretinoin, panitumumab, bevacizumab, bortezomib, lapatinib, and rituximab; hormone preparations such as anastrozole, exemestane, estramustine, ethinyl estradiol, chlormadinone, goserelin, tamoxifen, dexamethasone, toremifene, bicalutamide, flutamide, predonisolone, fosfestrol, mitotane, methyltestosterone, medroxyprogesterone, mepitiostane, leuprorelin, and letrozole; biological response modifiers such as interferon .alpha., interferon .beta., interferon .gamma., interleukin, ubenimex, freeze-dried BCG, and lentinan.
[0114] As used herein, the pharmaceutical composition for increasing permeability of the blood-brain barrier may comprise one or more PDPK1 expression inhibitors or PDPK1 activity inhibitors as an active ingredient(s). The pharmaceutical composition for increasing permeability of the blood-brain barrier may solely comprise the PDPK1 expression inhibitor and/or PDPK1 activity inhibitor as active ingredient(s), or may further comprise another agent to be used in combination as an active ingredient. Examples of such another agent include drugs that act on and increase the permeability of the blood-brain barrier as well as agents intended to be delivered to the brain. Herein, when the pharmaceutical composition for increasing permeability of the blood-brain barrier is used particularly for delivering a desired agent to the brain, such a pharmaceutical composition is referred to as the "agent-delivery composition". In the agent-delivery composition herein, the "agent intended to be delivered to the brain" may be referred to as the "medicinal component" to be distinguished from the agent (PDPK1 expression inhibitor or PDPK1 activity inhibitor) intended to open the BBB.
[0115] As used herein, the "agent-delivery composition" comprises at least a PDPK1 expression inhibitor or activity inhibitor as an active ingredient. The agent-delivery composition is preferably prepared into a unit dosage form comprising a suitable dose of the medicinal component. An example of such a unit dosage form is an injection (ampoule, vial, prefilled syringe). Usually 5-500 mg, 5-100 mg, or 10-250 mg of the PDPK1 expression inhibitor or activity inhibitor may be contained per unit dosage form.
[0116] Moreover, the agent-delivery composition may comprise, in addition to the PDPK1 expression inhibitor or activity inhibitor, an agent intended to be delivered to the brain, a medicinal component. The PDPK1 expression inhibitor or activity inhibitor and the medicinal component may be contained in one formulation or provided as separate formulations to be used in combination.
[0117] As used herein, the "medicinal component" is a medicine intended to be delivered into the brain, and includes, for example, a therapeutic or preventive agent for a target disease such as neurosurgical diseases such as cerebrovascular disorders (such as cerebral infarction/cerebral hemorrhage/cerebral aneurysm), brain tumor (such as meningioma, pituitary adenoma, including brain metastasis), infectious diseases (such as meningitis), functional brain diseases (such as trigeminal neuralgia), the spinal cord disease (such as disc herniation, spinal stenosis); neurological diseases such as Parkinson's disease, spinocerebellar degeneration, and epilepsy; and mental diseases such as Alzheimer's disease, non-Alzheimer type degenerative dementia, insomnia, and depression. For example, when the target disease is brain tumor, the anticancer agents described above may be used and, particularly when it is glioma, Avastin or Gliadel may be preferably used.
[0118] In another aspect, the agent-delivery composition herein may be provided as a kit for treatment or prevention, comprising the composition as an active ingredient. The constitution of the kit is not limited as long as it comprises the PDPK1 expression inhibitor or activity inhibitor, and the kit may comprise, for example, the PDPK1 expression inhibitor or activity inhibitor and the medicinal component. In this case, the PDPK1 expression inhibitor or activity inhibitor and the medicinal component may be provided in a form to be administered simultaneously or separately.
[0119] Throughout the description, the "level" means a numerical value indicating the quantity of miR-181c, which includes, for example, the concentration, the amount, or any alternative numerically value indicating the quantity to be used instead thereof. Accordingly, the level may be a measurement itself of such as fluorescence or may be a value converted into the concentration or the amount. The level may be absolute numerical value (abundance, abundance per unit area) or a numerical value relative to a control provided as needed. The level may be the mean or median when the same sample is measured plural times (at the same time or at different times).
[0120] As used herein, the "kit" may comprise, in addition to the active ingredient, a package storing components of the kit, such as a paper box or a plastic case, an ampoule, a vial, a tube, a syringe, or the like storing each component, an instruction manual, and the like.
[0121] In another aspect, the present invention involves an apparatus for judging brain metastasis in a cancer patient, an apparatus for assessing risk of brain metastasis in a cancer patient, and a computer program used in these apparatuses (FIG. 23). These apparatuses provide information for diagnosis to a person (usually a doctor) who diagnoses whether there is brain metastasis or not. The apparatus for judging brain metastasis of the present invention comprises miR-181c-measuring means. The "miR-181c-measuring means" is an apparatus that conducts the measurement of a polynucleotide having the nucleotide sequence of or a part of miR-181c (SEQ ID NO: 1) in a sample derived from a subject, which processes the sample from the subject cancer patient as prescribed to convert information about the miR-181c levels in the sample into digits or electrical signals. The miR-181c-measuring means allows to contact the agent for diagnosis or risk assessment of brain metastasis described above and the sample from the subject cancer patient and to react them if necessary, and measures generated parameters such as a strength of a light (fluorescence, luminescence) which reflects the miR-181c levels. The contact between the agent for diagnosis or risk assessment of brain metastasis and the sample from the subject cancer patient usually takes place in a container (reaction vessel) such as a plate or a tube.
[0122] The computer program of the present invention includes a computer program that can make an existing apparatus to be used for judging brain metastasis or for assessing risk of brain metastasis in a cancer patient by installing, even if the existing apparatus is widely used and can be used for other measurement or judgment. The computer program of the present invention is not required to be installed in the apparatus for judging brain metastasis and the apparatus for assessing risk of brain metastasis. The computer program of the present invention may be provided, for example, in such a form to be stored on a recording medium. The "recording medium" is a medium that can carry a program without occupying space, and includes a flexible disk, a hard disk, CD-R, CD-RW, MO (magneto-optical disk), DVD-R, DVD-RW, a flash memory, and the like. Moreover, the computer program of the present invention can be transmitted from the computer program storing computer to other computers or other apparatuses through a communication line. The computer program of the present invention includes those stored on such a computer and being transmitted.
Advantageous Effects of Invention
[0123] miR-181c contributes to brain metastasis by destructing the BBB. Since expression of miR-181c is specifically increased in cancer patients with brain metastasis, miR-181c can be used as indicator for diagnosis or risk assessment of brain metastasis. A brain metastasis can be suppressed by inhibiting the expression or activity of miR-181c or inhibiting secretion of EVs (containing miR-181c). PDPK1 expression inhibitors and PDPK1 activity inhibitors including miR-181c increases the permeability of the BBB, and thus can be used as DDS of agents desired to be delivered into the brain.
BRIEF DESCRIPTION OF THE DRAWINGS
[0124] [FIG. 1] (a) The protocol for in vivo-selection of brain metastatic derivatives. (b) The left photograph shows bioluminescence imaging of a mouse with BMD2a cell brain metastasis; and the right photographs shows bioluminescence imaging of a mouse brain with cancer cell metastasis. (c) Photographs of hematoxylin and eosin (HE)-stained sections from a mouse brain cerebral cortex and midbrain. The upper panels show cerebral cortex and the lower panels show the midbrain. The left panels (Normal) show sections from a mouse without cancer cell metastasis, and the middle panels (Metastasis) show sections from a mouse with cancer cell metastasis. The arrowheads indicate metastatic cancer cells. The right photographs are higher magnification photographs. The bar at the lower right corner in each photograph indicates 100 .mu.m.
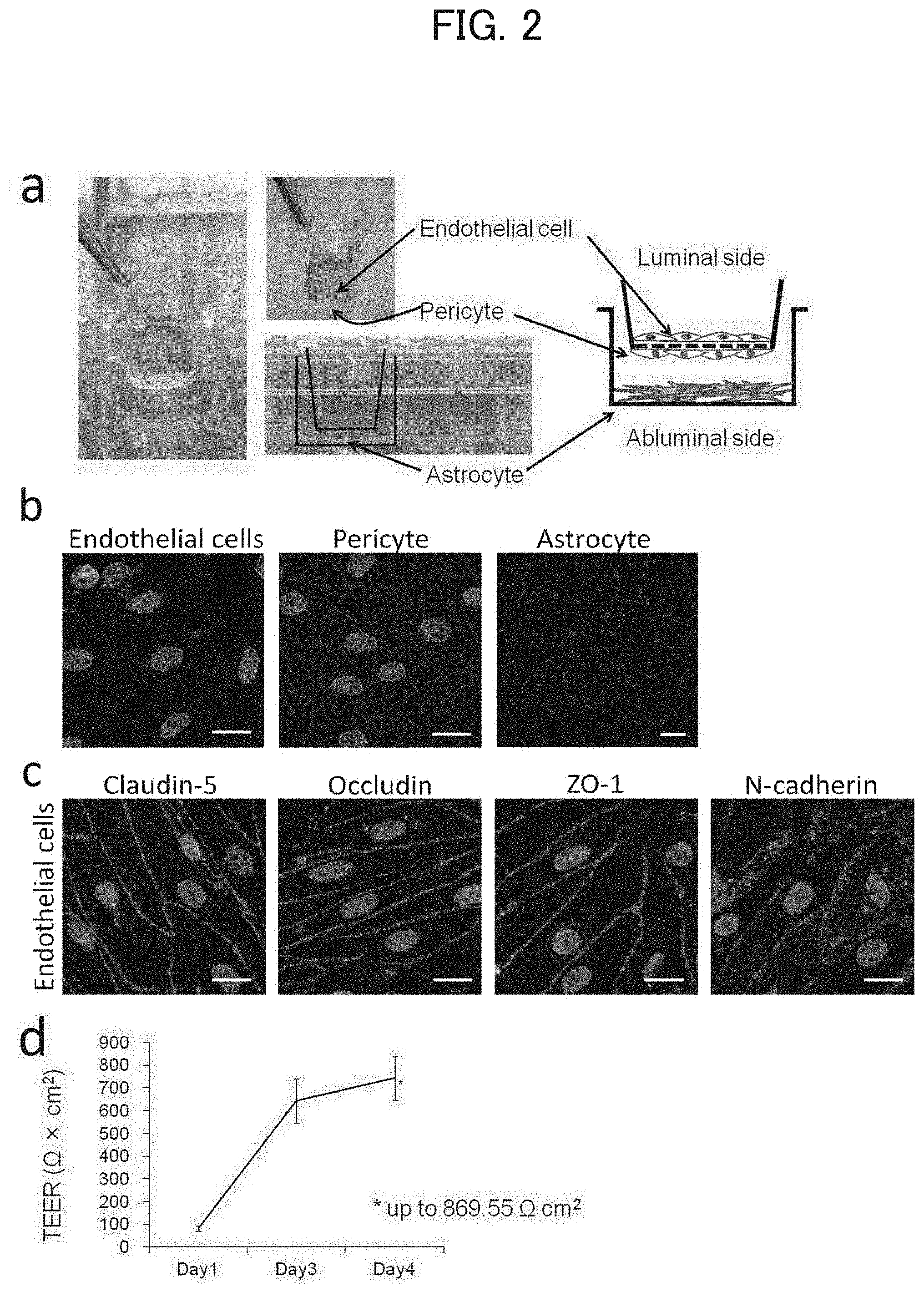
[0125] [FIG. 2] (a) In vitro blood-brain barrier model constructed from primary culture cells of monkey brain capillary endothelial cells, pericytes, and astrocytes. (b) Representative photographs of endothelial cells, pericytes, and astrocytes. The astrocytes were visualized using a fluorescence microscope. The bar at the lower right corner in each photograph indicates 100 .mu.m. (c) Photographs of the results of immunofluorescence staining of tight junction proteins (Claudin-5, Occludin, and ZO-1) and the N-cadherin. The bar at the lower right corner in each photograph indicates 20 .mu.m. (d) A graph of the change of TEER after thawing until the start of the experiment. The value of TEER increased to a maximum of 869.55 .OMEGA..times.cm.sup.2 after thawing the BBB in vitro model. The error bars represent standard deviation (SD).
[0126] [FIG. 3] (a) Representative photographs showing invasiveness of Matrigel by MDA-MB-231-D3H1 cells (hereinafter, D3H1 cells) MDA-MB-231-D3H2LN cells (hereinafter, D3H2LN cells), and BMD2a and BMD2b cells which were established as brain metastasis cell lines. The bar at the lower right corner in each photograph indicates 100 .mu.m. (b) A graph showing the ratio (folds) of the number of D3H1 cells, BMD2a cells, and BMD2b cells invaded Matrigel relative to the number of D3H2LN cells invaded Matrigel. The error bar represents standard deviation (SD), and ** indicates P<0.01. (c) Photographs of representative morphology of D3H1, D3H2LN, BMD2a, and BMD2b cells. The bar at the lower right corner in each photograph indicates 100 .mu.m.
[0127] [FIG. 4] (a) The invasive test of PKH-67 labeled cancer cells using the in vitro BBB model. (b) The left panel is a graph showing the ratio (folds) of the number of D3H2LN, BMD2a, and BMD2b cells that migrated across the in vitro BBB model relative to the number of D3H1 cells that migrated across the BBB model. The error bars represent standard deviation (SD), and * indicates P<0.01 and ** indicates P<0.01. The right panel is photographs of D3H1, D3H2LN, BMD2a, and BMD2b cells that migrated across the in vitro BBB model.
[0128] [FIG. 5] (a) The upper panels are photographs of EVs visualized with a phase-contrast electron microscope. The bars represent 100 nm. The lower panels are graphs of the results of measurements of the EV sizes with NanoSight. The numerical values in the graphs are the mean values of the size of all EVs. (b) A photograph of the result of Western blotting to confirm the expression of CD63 and CD9 as the EV markers and Cytochrome C. In the photograph, from the left to the right, each lane shows D3H1 derived EVs (lane 1), D3H2LN derived EVs (lane 2), BMD2a derived EVs (lane 3), and BMD2b derived EVs (lane 4). (c) A graph of the result of measurement of the number of EVs isolated from D3H1, D3H2LN, BMD2a, and BMD2b cells with NanoSight. The horizontal axis indicates, from the left to the right, D3H1 derived EVs, D3H2LN derived EVs, BMD2a derived EVs, and BMD2b derived EVs. The vertical axis represents the number of EV particles per 1 mL(.times.10.sup.9/mL).
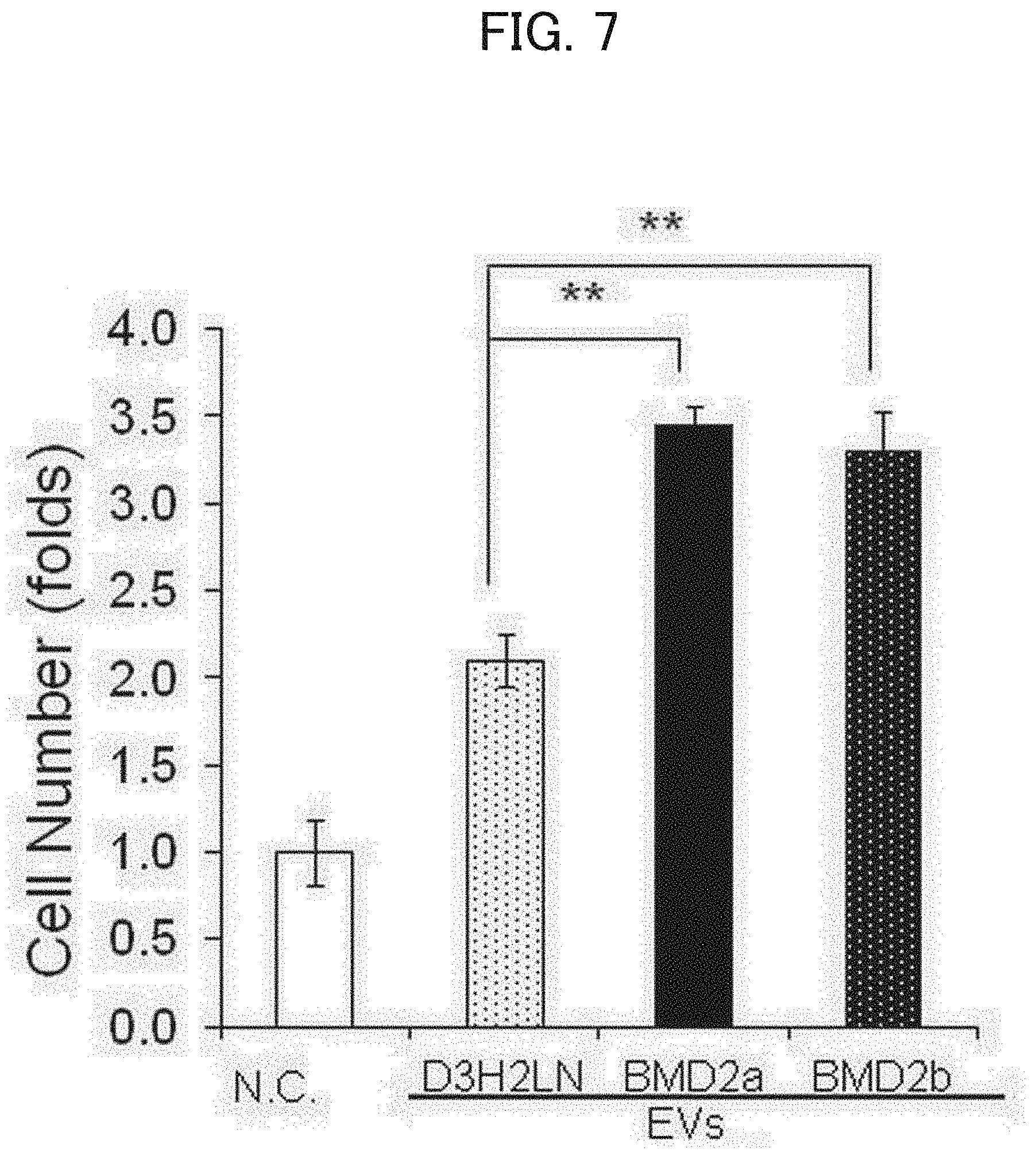
[0129] [FIG. 6] Graphs of the results of treatment of BMD2a cells with siRNAs. In all graphs, the horizontal axis indicates treated siRNAs, which are from the left to the right, negative control cells (N.C.), RAB27B siRNA (RAB27B), nSMase2 siRNA (nSMase2), and RAB27B siRNA and nSMase2 siRNA (S+R). (a) A graph showing the number of EVs released from siRNA-treated BMD2a cells. The vertical axis represents the ratio (folds) of the number of EV particles (unit/mL) from cells treated with siRNA against the EV-release related protein relative to the number of EV particles from the negative control cells. (b) A graph of siRNA-treated BMD2a cells invaded Matrigel. The vertical axis represents the ratio (%) of the number of Matrigel invaded cells treated with siRNA against an EV-release related protein relative to that of the negative control cells. (c) A graph showing the result of the in vitro BBB transmigration activity test of siRNA-treated BMD2a cells. The vertical axis represents the ratio (folds) of the number of BBB transmigrated cells treated with siRNA against an EV-release related protein relative to that of the negative control cells.
[0130] [FIG. 7] A graph of the result of measurement of the BBB transmigrating capacity of D3H1 cells on addition of EVs derived from BMD2a, BMD2b, or D3H2LN cells using the in vitro BBB model. The vertical axis represents the ratio (folds) of the number of D3H1 cells that migrated across the in vitro BBB model on addition of EVs derived from BMD2a, BMD2b, or D3H2LN cells relative to that of the negative control cells. The horizontal axis indicates the cells from which the added EVs are derived, which are from the left to the right, negative control (N.C.), D3H2LN, BMD2a, and BMD2b cells.
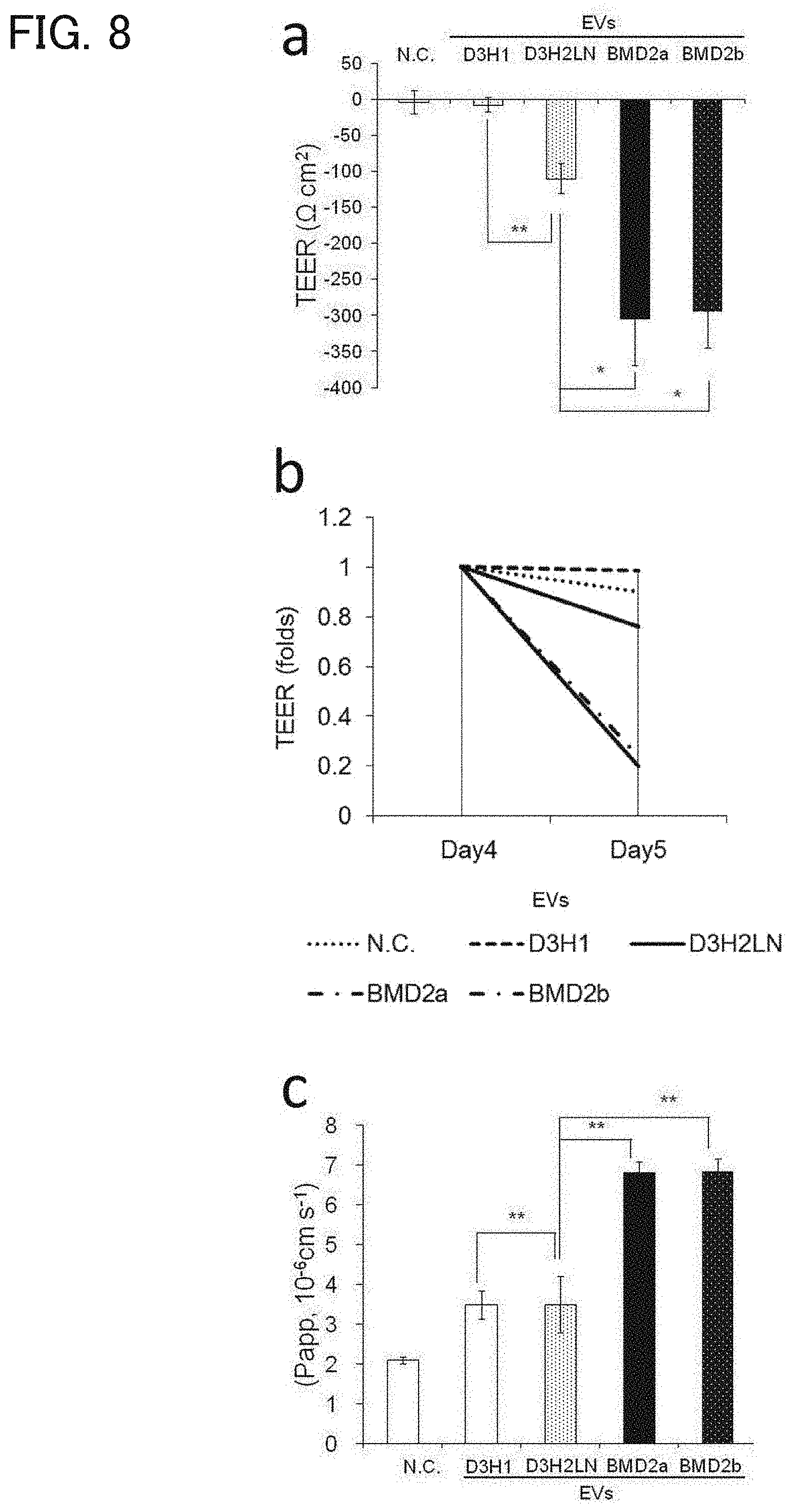
[0131] [FIG. 8] (a) A graph showing the result of TEER measurement at 24 hours after the addition of EVs. The vertical axis represents the measurement of TEER (.OMEGA.cm.sup.2), the horizontal axis indicates the cells from which the added EVs are derived, which are from the left to the right, negative control (N.C.), D3H1, D3H2LN, BMD2a, and BMD2b cells. The error bars represent standard deviation (SD), * indicates P<0.05, and ** indicates P<0.01. (b) A graph showing changes of TEER between before and 24 hours after the EV addition. The horizontal axis represents the ratio (folds) of the TEER value at 24 hours after the EV addition relative to that before the EV addition. (c) A graph representing the result of the permeability test of BBB measured by NaF. The vertical axis represents the apparent permeability coefficient (Papp) (10.sup.-6 cms.sup.-1). The horizontal axis indicates the cells from which the added EVs was derived, which are from the left to right, negative control (N.C.), D3H1, D3H2LN, BMD2a, and BMD2b cells.
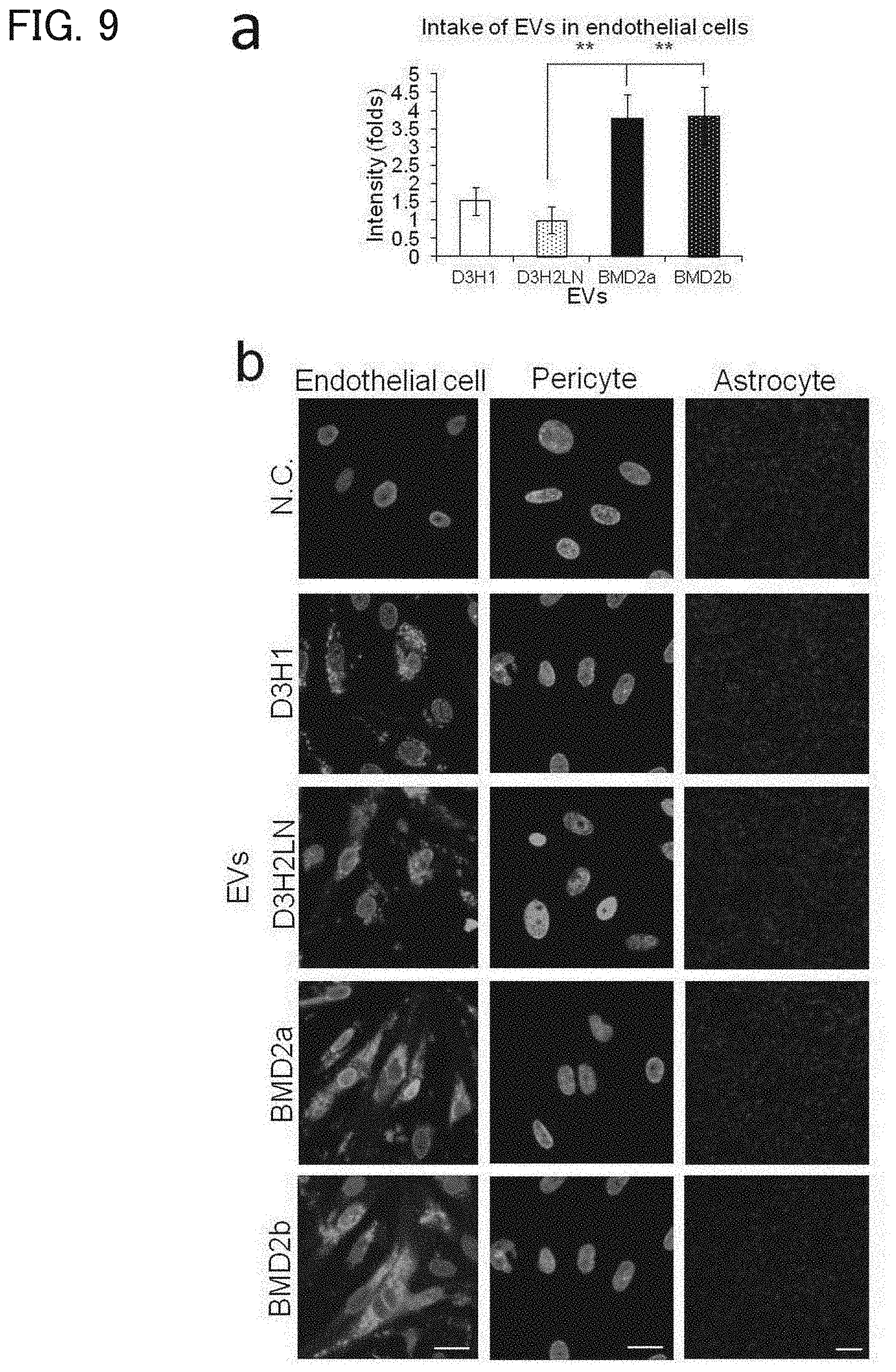
[0132] [FIG. 9] (a) A graph representing the result of measurement of EV uptake to endothelial cells constituting the in vitro BBB model. The vertical axis represents the fluorescence intensity ratio (folds), relative to D3H2LN cell-derived EVs, of other cell-derived EVs taken up by endothelial cells. The horizontal axis indicates the cells from which the EVs used are derived, which are, from the left to the right, D3H1, D3H2LN, BMD2a, and BMD2b cells. The error bars represent standard deviation (SD), and ** indicates P<0.01. (b) Photographs of Hoechst33342 staining (light blue) of endothelial cells, pericytes, and the astrocytes constituting the in vitro BBB model as well as PKH67 (green) of up-taken EVs, observed with a fluorescence microscope. The cells from which the EVs derived are indicated on the left of the photographs. The bars in the photographs of endothelial cells and pericytes indicate 20 .mu.m. The bar in the photographs of astrocytes indicates 100 .mu.m.
[0133] [FIG. 10] (a) Photographs of co-immunofluorescence of tight junction proteins (Claudin-5, Occludin and ZO-1) (red) and actin filaments (green) in vascular endothelial cells constituting the in vitro BBB model after the addition of EVs derived from D3H2LN, BMD2a, and BMD2b cells. The bars indicate 20 .mu.m. (b) Photographs of co-immunofluorescence of N-cadherin (red) and actin filaments (green) in blood vessel cells constituting the in vitro BBB model after the addition of EVs derived from D3H2LN, BMD2a or BMD2b cells. The bars indicate 20 .mu.m. (c) A photograph of Western blot analysis on tight junction proteins, N-cadherin, Actin, and GAPDH (internal control) in proteins recovered from endothelial cells treated with PBS (negative control: N.C.) or EVs derived from D3H2LN, BMD2a, or BMD2b cells.
[0134] [FIG. 11] Fluorescence image photographs of a brain of a mouse administered D3H2LN (control) or BMD2a cell derived EVs. The fluorescence intensity of the upper panels represents DiR-labelled EVs up-taken by a mouse brain. The fluorescence intensity of the lower panels represents the blood vessel permeability of a mouse brain.
[0135] [FIG. 12] (a) A graph showing distribution of photon intensity in the mouse brain. The vertical axis represents photon intensity (.times.10.sup.5). The horizontal axis indicates each mouse group, which are from the left to the right, negative control (N.C.), the D3H2LN-derived EVs administrated group (D3H2LN), and the BMD2a-derived EVs administrated group (BMD2a). * indicates P<0.05 and ** indicates P<0.01. (b) Photographs showing bioluminescence image of cancer cell metastasis in mice into which cancer cells were transplanted after administration of PBS (negative control) (N.C.) (left), EVs derived from D3H2LN (D3H2LN EVs) (middle), and EVs derived from BMD2a (BMD2a EVs) (right). The upper panels are bioluminescence images of the whole body of the mice, and the lower panels are bioluminescence images of a mouse brains. (c) Photographs of sections from a mouse brain cerebral cortex. The upper panels are photographs of hematoxylin and eosin (HE) staining and the arrowheads indicate metastatic cancer cells. The bars at the lower right corner in the photographs indicate 100 .mu.m. The lower panels are image photographs of anti-human vimentin immunofluorescence staining and the bars at the lower right corner in the photographs indicate 20 .mu.m.
[0136] [FIG. 13] (a) A heat map of expression levels of miR-181c in cancer cell-derived EVs. (b) A graph showing the ratio (folds) of the rate of an amount of miR-181c to an amount of total RNA (miR-181c/(total exosome RNA)) in EVs isolated from BMD2a and BMD2b cells as compared with the rate of miR-181c/(total exosome RNA) in EVs derived from D3H2LN cells. The vertical axis represents the ratio (folds) of the rate of the amount of miR-181c to the amount of total RNA in isolated EVs from each cell as compared with the rate (which is set to be 1) of the amount of miR-181c to the amount of total RNA in EVs isolated from D3H2LN cells. For BMD2a cell and BMD2b cell, the error bars represent standard deviation (SD) and ** indicates P<0.01 as compared with EVs derived from D3H2LN cells.
[0137] [FIG. 14] (a) A graph showing a change in amount of miR-181c in vascular endothelial cells of in vitro BBB model. The vertical axis indicates the ratio (folds) of the amount of miR-181c in the RNAs recovered from vascular endothelial cells of the in vitro BBB model after adding cancer cell-derived EVs to the in vitro BBB model, relative to the amount of miR-181c contained in the vascular endothelial cells to which EVs derived from D3H2LN cells were added, wherein the amounts of miR-181c are corrected by that of RNU6. The horizontal axis indicates the cells from which the EVs added to the in vitro BBB model were derived from, which are D3H2LN, BMD2a, and BMD2b cells from the left to the right. For BMD2a and BMD2b cells, the error bars represent standard deviation (SD) and ** indicates P<0.01 as compared with EVs derived from D3H2LN cells. (b) A graph of the TEER measurement at 24 hours after transfecting the vascular endothelial cells of the in vitro BBB model with miR-181c. The vertical axis indicates the measurement of TEER (.OMEGA./cm.sup.2) and the horizontal axis indicates the transfected microRNAs, which are negative control (N.C.) and miR-181c, from the left to the right. The error bars represent standard deviation (S.D.), and ** indicates P<0.01. (c) Photographs of the co-immunofluorescence of tight junction proteins (Claudin-5, Occludin, and ZO-1), N-cadherin (red), and actin filaments (green) in vascular endothelial cells constituting the in vitro BBB model after the transfection with the negative control (N.C.) or miR-181c. The bar indicates 20 .mu.m. (d) A photograph illustrating Western blot analysis of tight junction proteins, N-cadherin, actin, and GAPDH (internal control), using proteins recovered from the vascular endothelial cells constituting the in vitro BBB model, which had been transfected with the negative control (N.C.) or miR-181c.
[0138] [FIG. 15] A graph showing the amount of miR-181c contained in serum EVs derived from patients. The vertical axis indicates the ratio of the amount of miR-181c relative to the amount of miR-16. The left in horizontal axis indicates "without brain metastasis" (n=46) and the right indicates "Brain metastasis" (n=10). * indicates P<0.05. The miR-181c levels were assessed by t-test.
[0139] [FIG. 16] A graph showing the distribution of the amount of miR-181c contained in sera of breast cancer patients. The vertical axis indicates the abundance of miR-181c corrected by the abundance of miR-16 (.DELTA..DELTA.Ct value). The horizontal axis indicates the stage of breast cancer using the TNM staging. * indicates significant differences (P<0.05) in Mann-Whitney U test between the patient group with brain metastasis and the patient groups in Stage 3 and in Stage 4 without brain metastasis.
[0140] [FIG. 17] (a) The ratio of PDPK1 expression amounts in a microarray analysis. After transfecting vascular endothelial cells constituting the in vitro BBB model with the negative control (N.C.) or miR-181c, RNAs recovered from the vascular endothelial cells were analyzed with microarray. The vertical axis indicates a ratio of an amount of PDPK1 expression by each cells relative to an amount of PDPK1 expression by cells transfected with the negative control (N.C.) which is set to be 1. The horizontal axis indicates the micro RNA used for the transfection, N.C. and miR-181c from the left to the right. (b) The expression amount of PDPK1 mRNA expression in vascular endothelial cells constituting the in vitro BBB model. After transfecting the vascular endothelial cells with the negative control (N.C.) or miR-181c, RNAs are recovered from the vascular endothelial cells, and the amount of PDPK1 mRNA contained in the RNA was analyzed by RT-PCR. The vertical axis indicates the value of the amount of PDPK1 mRNA corrected by the amount of GAPDH mRNA (.DELTA..DELTA.Ct value) which is set to be 1. The horizontal axis indicates the microRNAs used for the transfection, N.C. and miR-181c from the left to the right. ** indicates P<0.01. (c) A photograph of Western blot analysis on PDPK1 and GAPDH (internal control), using proteins recovered from the vascular endothelial cells constituting the in vitro BBB model transfected with the negative control (N.C.) or miR-181c. The graph indicates the value of the luminescence intensity of PDPK1 corrected by the luminescence intensity of GAPDH in Western blot analysis. (d) The ratio of PDPK1 expression levels in a microarray analysis. After adding negative control (N.C.), D3H2LN cell-derived EVs, BMD2a cell-derived EVs, or BMD2b cell-derived EVs to the vascular endothelial cells constituting the in vitro BBB model, RNAs were recovered from the vascular endothelial cells and analyzed by microarray analysis. The vertical axis indicates the ratio of the expression amount of PDPK1 relative to the expression amount of PDPK1 by cells treated with the negative control (N.C.), which is set to be 1. The horizontal axis indicates the added EVs, negative control (N.C.), D3H2LN cell-derived EVs, BMD2a cell-derived EVs, and BMD2b cell-derived EVs. (e) The expression levels of PDPK1 mRNA in the vascular endothelial cells constituting the in vitro BBB model. After adding negative control (N.C.), D3H2LN cell-derived EVs, BMD2a cell-derived EVs, or BMD2b cell-derived EVs to the vascular endothelial cells constituting the in vitro BBB model, the amount of PDPK1 mRNA contained in the RNA recovered from the vascular endothelial cells was analyzed by RT-PCR. The vertical axis indicates the ratio of the value (.DELTA..DELTA.Ct value) that is the amount of PDPK1 mRNA corrected by the amount of GAPDH mRNA relative to the value of the negative control which is set to be 1. The horizontal axis indicates the added EVs, negative control (N.C.), D3H2LN cell-derived EVs, BMD2a cell-derived EVs, and BMD2b cell-derived EVs. ** indicates P<0.01. (f) A photograph of Western blot analysis of PDPK1 and GAPDH (internal control) using the proteins recovered from the vascular endothelial cells constituting the in vitro BBB model after adding negative control (N.C.), D3H2LN cell-derived EVs, BMD2a cell-derived EVs, or BMD2b cell-derived EVs. The graph indicates the value of the luminescence intensity of PDPK1 corrected by the luminescence intensity of GAPDH in Western blot analysis.
[0141] [FIG. 18] (a) Photographs of the co-immunofluorescence of tight junction proteins (Claudin-5, Occludin, and ZO-1), N-cadherin (red), and actin filaments (green) in the vascular endothelial cells constituting the in vitro BBB model after transfecting with the negative control (N.C.) or PDPK1 siRNA. The bar indicates 20 .mu.m. (b) A photograph illustrating Western blot analysis of tight junction proteins, N-cadherin, actin, and GAPDH (internal control), using the proteins recovered from the vascular endothelial cells constituting the in vitro BBB model transfected with the negative control (N.C.) or PDPK1 siRNA. (c) A graph of the result of the TEER measurement at 24 hours after the transfection of the vascular endothelial cells of the in vitro BBB model with PDPK1 siRNA. The vertical axis represents the measurement of TEER (.OMEGA., cm.sup.2) and the horizontal axis indicates the microRNAs used for the transfection, which are from the left to the right, the negative control (N.C.) and PDPK1 siRNA. The error bars represent standard deviation (S.D.), and ** indicates P<0.01.
[0142] [FIG. 19] (a) The result of 3'UTR reporter assay, using the PDPK1 mRNA 3'UTR region derived from Human and Macaca. The black bar indicates the result of the negative control (N.C.), and the white bar indicates the result of miR-181c. The values are the ratio relative to the negative control (N.C.) which is set to be 1. * * indicates P<0.01. (b) The PDPK1 mRNA 3'UTR region sequences from Human (SEQ ID NO: 14) and Macaca (SEQ ID NO: 15) and the miR-181c sequence (SEQ ID NO: 1) and the binding region thereof.
[0143] [FIG. 20] (a) A photograph of Western blot analysis of PDPK1, Cofilin, phosphorylated Cofilin (P-cofilin), and GAPDH (internal control) using the proteins recovered from the vascular endothelial cells constituting the in vitro BBB model after adding D3H2LN cell-derived EVs, BMD2a cell-derived EVs, and BMD2b cell-derived EVs. (b) A photograph of Western blot analysis of PDPK1, Cofilin, phosphorylated Cofilin (P-cofilin), and GAPDH (internal control) using the proteins recovered from the vascular endothelial cells constituted the in vitro BBB model transfected with the negative control (N.C.), miR-181c, or PDPK1 siRNA.
[0144] [FIG. 21] A schematic configuration diagram of an apparatus for judging brain metastasis relating to an embodiment of the present invention.
[0145] [FIG. 22] A schematic configuration diagram of an apparatus for assessing risk of brain metastasis relating to an embodiment of the present invention.
[0146] [FIG. 23] A flow of processing that the apparatus for judging brain metastasis performs when the computer program for the apparatus for judging brain metastasis is executed.
[0147] [FIG. 24] A flow of processing that the apparatus for assessing risk of brain metastasis performs when the computer program for the apparatus for assessing risk of brain metastasis is executed.
[0148] [FIG. 25] A schematic diagram of an apparatus of the present invention and a method for judging brain metastasis (solid line) or a method for assessing risk of brain metastasis (dotted line) using the apparatus.
DETAILED DESCRIPTION OF THE PREFERRED EMBODIMENT
1. Method for Diagnosis or Risk Assessment of Brain Metastasis in Cancer Patient
[0149] In one aspect, the present invention relates to a method for diagnosing brain metastasis or a method for assessing risk of brain metastasis, comprising measuring an miR-181c level in a sample from a subject cancer patient. In one aspect, the present invention relates to a method for diagnosing brain metastasis in a subject cancer patient (or a method for assessing risk of brain metastasis), comprising: determining an miR-181c level in a sample from the subject cancer patient, and judging the presence or absence of brain metastasis (or assessing risk of brain metastasis) in the subject cancer patient from the determined miR-181c level, wherein the subject cancer patient is judged to have metastasis to brain (or assessed to have a high risk of brain metastasis) when the miR-181c level in the sample from the subject cancer patient is higher than the miR-181c level in a sample from a negative control.
[0150] Alternatively, in one aspect, the present invention relates to a method for providing information for diagnosing brain metastasis, comprising measuring an miR-181c level in a sample from a subject cancer patient. The present invention relates to a method for providing information for diagnosing brain metastasis (or for assessing risk of brain metastasis) in a subject cancer patient, comprising: a step of determining an miR-181c level in a sample from the subject cancer patient, and a step comprising providing information for judging the presence or absence of brain metastasis (or assessing a risk of brain metastasis) in the subject cancer patient from the determined miR-181c level, wherein information that the subject cancer patient has brain metastasis (or has a high risk of brain metastasis) is provided when the miR-181c level in the sample from the subject cancer patient is higher than the miR-181c level in a sample from a negative control.
[0151] The step of determining an miR-181c level is conducted by measuring a polynucleotide having the nucleotide sequence or a part of miR-181c (SEQ ID NO: 1) in the sample from the subject. The polynucleotide may be measured by a method utilizing detection of nucleic acid binding such as hybridization; a method utilizing amplification of nucleic acids such as PCR; a method utilizing the expression inhibitory activity of miR-181c such as a reporter assay; or decoding the sequence of miRNA or cDNA transcribed from miRNA.
[0152] Examples of the method utilizing hybridization include Southern hybridization, Northern hybridization, dot hybridization, fluorescence in situ hybridization (FISH), microarray, and the ASO method.
[0153] For example, the "step of determining an miR-181c level" in the present invention may comprise the following steps: [0154] (a) a step of contacting a sample from a subject cancer patient with at least one nucleic acid construct (probe) that binds to the nucleotide sequence or a part of miR-181c (SEQ ID NO: 1); [0155] (b) a step of measuring the binding between the probe and miR-181c in the sample; and [0156] (c) a step of determining the miR-181c level in the sample from the measured binding between the probe and miR-181c.
[0157] In the description above, the "binding between the probe and miR-181c" to be measured may be a bound amount, the number of binding, or a percentage of the bound substances. The step of determining the miR-181c level in the sample from the measured binding between the probe and miR-181c may be conducted, for example, by the following procedures. Binding between the probe and miR-181c in a standard sample which is a serial dilution series of miR-181c is measured to generate a standard curve. The miR-181c level in a sample from a subject cancer patient can be calculated by comparing a measurement of the binding between the probe and miR-181c measured in the sample from the subject cancer patient with the standard curve. The standard curve may be determined by measuring standard samples simultaneously with the sample from the subject cancer patient, or alternatively by measuring standard samples separately from (at different time from) the samples from the subject cancer patient. Accordingly, the standard curve may be pre-obtained from the measurements of standard samples which is measured before the measurement of a sample from a subject cancer patient. Instead of directly binding the miRNA with the probe, cDNA may be synthesized from miRNA in a sample from a patient, and the binding of the synthesized cDNA and probe may be measured.
[0158] Examples of the method utilizing PCR include ARMS (amplification refractory mutation system), RT-PCR (reverse transcriptase-PCR), and nested PCR.
[0159] For example, the determination of a miR-181c level in the present invention may comprise the following steps: [0160] (a) a step of amplifying a nucleic acid molecule having miR-181c (SEQ ID NO: 1) or a part thereof in a sample from a subject cancer patient by using at least one nucleic acid construct (primer) that binds to a part of the nucleotide sequence of miR-181c (SEQ ID NO: 1); [0161] (b) a step of measuring the amount of the amplified nucleic acid molecule; and [0162] (c) a step of determining the miR-181c level in the sample from the measured amount of the amplified nucleic acid.
[0163] The above step of determining the miR-181c level in the sample from the measured amount of the amplified nucleic acid may be conducted, for example, by the following procedures. As standard samples, pre-determined copy number of nucleic acids are used to obtain the amplified amount, from which a standard curve is generated. The miR-181c level in a sample from a subject cancer patient can be calculated by comparing the measurement of the amplified nucleic acid in the sample from the subject cancer patient with the standard curve. The standard curve may be determined by measuring standard samples simultaneously with the sample from the subject cancer patient, or alternatively by measuring standard samples separately from (at different time from) the sample from the subject cancer patient. The standard curve may be pre-obtained by measuring standard samples before the measurement of a sample from a subject cancer patient. Instead of directly amplifying the miRNA, cDNA may be synthesized using miRNA obtained from a patient sample, which can be used for amplification.
[0164] As a method for determining the level of miRNA utilizing the expression inhibitory function of miR-181c, a method involving a marker gene of a fluorescence protein (such as luciferase) or the like is widely known. For example, the miR-181c level can be determined by designing a marker gene which generates mRNA having a 3'UTR sequence (for example, SEQ ID NO: 14 or SEQ ID NO: 15) that can bind to the seed sequence (ACUUACA) of miR-181c, and treating the gene transformed cells with a sample under the conditions that allow the expression of the gene.
[0165] For example, the determination of a miR-181c level in the present invention may comprise the following steps: [0166] (a) a step of contacting a sample from a subject cancer patient with cells transformed with a polynucleotide encoding the following (i) and (ii): [0167] (i) a marker gene, [0168] (ii) a 3'UTR sequence that binds to the seed sequence (ACUUACA) of miR-181c in mRNA; [0169] (b) a step of expressing the polynucleotide in the cells; and [0170] (c) a step of determining the miR-181c level in the sample from the amount of the expressed marker gene product.
[0171] In the above method, the step of determining the miR-181c level in the sample from the amount of the expressed marker gene product can be conducted by, for example, generating a standard curve from the measurements of standard samples which contain the pre-determined amount of the marker gene product, and calculating the miR-181c level by applying the measurements for the amount of the marker gene product to the standard curve.
[0172] When the sequence of miRNA or cDNA is decoded, the level of miR-181c can be determined by synthesizing cDNA from the RNA as needed, decoding the sequence of synthesized cDNA with a sequencer, and determining the level of miR-181c from the amount of the decoded miR-181c.
[0173] When cDNA is synthesized from miRNA in the description above, it may be conducted with a cDNA synthesis kit (RNA-Quant cDNA Synthesis Kit, System Biosciences, LLC, San Francisco, USA) or the like.
[0174] The "step of judging or assessing" and the "step comprising providing information for judging or assessing" (collectively, referred to as the "judging step" in this paragraph) in the method for diagnosis of the present invention and the like can be conducted using the determined miR-181c level. The judging step can be conducted by comparing the determined miR-181c level of the subject with the miR-181c level of a negative control. When the miR-181c level in the sample from the subject is higher than the miR-181c level in the sample from the negative control, the miR-181c level is determined to be high and the subject can be judged possibly to have brain metastasis (or assessed possibly to have a high risk of brain metastasis). When the miR-181c level in the sample from the subject is not higher than (that is, equal to or lower than) the miR-181c level in the sample from the negative control, the miR-181c level is determined not high and the subject can be judged possibly not to have brain metastasis (or assessed possibly to have a low risk of brain metastasis).
[0175] As used herein, the "negative control" means healthy subjects or cancer patients having no brain metastasis (for example, Stage 1 and/or Stage 2 cancer patients and/or Stage 3 and/or Stage 4 cancer patients having no brain metastasis). The "miR-181c level of the negative control" can be obtained by measuring the miR-181c level in such a negative control according to the method described above. Alternatively, when there is predetermined information about the miR-181c level already measured for such a negative control, such a level can be used as the miR-181c level of the "negative control". Moreover, a standard comparative sample containing the same amount of miR-181c as the miR-181c level of the "negative control" may be measured simultaneously with the sample from the subject cancer patient.
[0176] Throughout the description, whether the miR-181c level in a subject sample is higher than the miR-181c level in a sample to be compared or not can be determined by a statistical analysis. The statistical significance can be determined by comparing 2 or more samples and determining a confidence interval and/or a p value (Dowdy and Wearden, Statistics for Research, John Wiely & Sons, NewYord, 1983). The confidence interval of the present invention may be, for example, 90%, 95%, 98%, 99%, 99.5%, 99.9%, or 99.99%. Moreover, the p value of the present invention may be, for example, 0.1, 0.05, 0.025, 0.02, 0.01, 0.005, 0.001, 0.0005, 0.0002, or 0.0001.
[0177] Throughout the description, the "method for diagnosis" and the "method for assessing risk" comprises a method for monitoring the state or change of brain metastasis or a method for monitoring the risk of brain metastasis unless such interpretation is inconsistent. Accordingly, the word of "diagnosis" or "risk assessment", as used herein, may be interpreted as monitoring the state or change of brain metastasis or the risk of brain metastasis unless such interpretation in particular is inconsistent. Moreover, when the method for diagnosis or the method for risk assessment herein means monitoring, the diagnosis or risk assessment may be conducted continually or intermittently. Moreover, the method for diagnosis and the method for assessing risk of the present invention may be conducted in vivo, ex vivo, or in vitro, but preferably conducted ex vivo or in vitro.
[0178] As used herein, the "sample" may be selected depending on the purpose of use as appropriate. For example, the sample may be a cell culture supernatant, a cell lysate, a tissue specimen collected from a subject as biopsy, or a liquid collected from a subject. Examples of the sample include tissue, blood, plasma, serum, lymph fluid, urine, stool, serosity, spinal fluid, cerebrospinal fluid, synovial fluid, aqueous humor, lacrimal fluid, saliva, or a fraction or processed product thereof. The sample is preferably blood, plasma, serum, or lymph fluid.
[0179] The risk assessment of brain metastasis means a method for predicting the progress or outcome of the condition of a patient thereby and does not mean that the progress or outcome of the condition can be predicted at 100% of accuracy. The result of assessment of risk of brain metastasis indicates whether the risk of a certain progress or outcome is increased or not, but does not indicates that the certain progress or outcome is more likely to occur as compared with that the certain progress or outcome is not occurring. Thus, the assessment of risk of brain metastasis indicates that the progress or outcome of brain metastasis is more likely to occur in patients with increased the miR-181c level compared to patients exhibiting no such characteristics.
[0180] In the method for diagnosis and the method for risk assessment herein, the "subject cancer patient" is not particularly limited as long as the patient is the cancer patient as described above. The subject cancer patient in the method for diagnosis of brain metastasis is preferably a Stage IV cancer patient, a patient with metastasis to distant organs, or a patient with infiltration of cancer cells into the blood, since brain metastasis occurs mainly on late stages of cancer as described above. On the other hand, the subject in the method for assessing risk of brain metastasis is preferably a Stage I to III patient in which the risk of brain metastasis is high but metastasis itself is not yet likely to occur, a patient with no metastasis to distant organs, or a patient with no infiltration of cancer cells into the blood. Moreover, the subject cancer patient is preferably a breast cancer patient.
2. Method for Treatment or Prevention of Brain Metastasis in Cancer Patient
[0181] According to the mechanism of cancer metastasis found by the present inventors, miR-181c contained in EVs (including exosomes) secreted by cancer cells contributes to brain metastasis. Therefore, in another aspect, the present invention relates to a method for treating or preventing brain metastasis in a cancer patient, comprising inhibiting the expression or activity of miR-181c in the cancer patient. In this method, the inhibition of the expression or activity of miR-181c may be conducted by administering an miR-181c expression inhibitor or activity inhibitor to a patient in need thereof. Accordingly, in a certain embodiment, the present invention relates to a method for treating or preventing brain metastasis in a cancer patient, comprising administering an miR-181c expression inhibitor or activity inhibitor to the cancer patient.
[0182] Alternatively, the inhibition of the expression or activity of miR-181c can be accomplished indirectly by inhibiting the secretion of EVs containing miR-181c. Accordingly, in one aspect, the present invention relates to a method for treating or preventing brain metastasis in a cancer patient, comprising inhibiting secretion of EVs containing miR-181c in the cancer patient. The inhibition of secretion of EVs can be accomplished, for example, by inhibiting the expression or activity of protein (nSMase2 and RAB27B) essential for the secretion of EVs. Accordingly, in another aspect, the present invention relates to a method for treating or preventing brain metastasis in a cancer patient, comprising administering an exosome secretion inhibitor to the cancer patient. In a specific aspect, the present invention relates to a method for treating or preventing brain metastasis in a cancer patient, comprising administering an nSMase2 expression or activity inhibitor and/or an RAB27B expression or activity inhibitor to the cancer patient.
[0183] The administration of the pharmaceutical composition (therapeutic or preventive agent for brain metastasis) may be local or systemic. The mode of administration is not particularly limited and is parenteral or oral administration as described above. Examples of parenteral route of administration include subcutaneous, intraperitoneal, in blood (intravenous or intraarterial), injection or infusion into spinal fluid, or the like and a preferable route is administration into blood. Moreover, the pharmaceutical composition (therapeutic or preventive agent) may be administered at once or administered continuously or intermittently. For example, the administration may be a continuous administration for 1 minute to 2 weeks.
[0184] The dose of the pharmaceutical composition (therapeutic or preventive agent for brain metastasis) is not particularly limited as long as it can exert desired therapeutic effect or preventive effect, and may be determined depending on the stage of the cancer, the presence or absence of brain metastasis, symptoms, sex, age, or the like as appropriate. The dose of the pharmaceutical composition according to the present invention may be determined, for example, by referring a therapeutic effect or preventive effect on brain metastasis as indicator. For example, the pharmaceutical composition may be administered by intravenous injection at a dose of the active ingredient of usually about 0.01-20 mg/kg body weight, preferably about 0.1-10 mg/kg of body weight, further preferably about 0.1-5 mg/kg of body weight as one dose, at about 1-10 times a month, preferably about 1-5 times a month. An amount referring the above may be administered when administered parenterally via another route or orally. When symptoms are particularly severe, the dose or frequency of administration may be increased according to the symptoms.
3. Method for Increasing Permeability of Blood-Brain Barrier and Method for Delivering Desired Active Ingredient Across Blood-Brain Barrier into Brain
[0185] The present inventors have found that the permeability of the blood-brain barrier is improved by decreasing the expression or activity of PDPK1. Accordingly, in another aspect, the present invention relates to a method for increasing permeability of blood-brain barrier, comprising inhibiting expression or activity of PDPK1 in vascular endothelial cells. In a further aspect, the present invention relates to a method for delivering a desired medicinal component across blood-brain barrier into brain, comprising administering the medicinal component with a PDPK1 expression inhibitor or activity inhibitor.
[0186] For example, the method for increasing permeability of blood-brain barrier of the present invention comprises the following method: a method for increasing permeability of blood-brain barrier in a patient, comprising suppressing the expression or inhibiting the activity of PDPK1 in brain microvascular endothelial cells (BMECs) in the patient. The suppression of the expression or inhibition of the activity of PDPK1 can be accomplished by administering a PDPK1 expression inhibitor or activity inhibitor. Accordingly, in another aspect, the present invention relates to a method for increasing the permeability of the blood-brain barrier in a patient, comprising administering a PDPK1 expression inhibitor or activity inhibitor.
[0187] The method for delivering desired medicinal component across blood-brain barrier into brain comprises the following method: a method for delivering a desired medicinal component across the blood-brain barrier of a patient into the brain, comprising suppressing the expression or inhibiting the activity of PDPK1 in brain microvascular endothelial cells (BMECs) in the patient, and administering the desired medicinal component to the patient. In this method, the inhibition of expression or activity of PDPK1 may be conducted by administering a PDPK1 expression inhibitor or activity inhibitor to a patient in need thereof. For example, the inhibition of expression of PDPK1 can be accomplished by administering miR-181c or EVs containing miR-181c. Accordingly, in another aspect, the present invention relates to a method for delivering a desired medicinal component across the blood-brain barrier of a patient into the brain, comprising administering the desired medicinal component and a PDPK1 expression inhibitor or activity inhibitor. Alternatively, the present invention may be a method for delivering a desired medicinal component across the blood-brain barrier of a patient into the brain, comprising increasing the permeability of the blood-brain barrier of the patient by administering a PDPK1 expression inhibitor or activity inhibitor to the patient; and administering the desired medicinal component to the blood-brain barrier permeability increased patient to deliver the medicinal component across the blood-brain barrier into the brain. The desired medicinal component may be administered simultaneously with the PDPK1 expression inhibitor or activity inhibitor or administered separately (for example, before or after the administration of the PDPK1 expression inhibitor or activity inhibitor).
[0188] The administration of the PDPK1 expression inhibitor or activity inhibitor or the agent-delivery composition of the present invention may be local or systemic. The route of administration is not particularly limited and can be parenteral or oral administration as described above. Examples of parenteral route of administration include subcutaneous, intraperitoneal, in blood (intravenous or intraarterial), injection or infusion into spinal fluid, or the like and a preferable route is administration into blood. Moreover, the PDPK1 expression inhibitor or activity inhibitor or the agent-delivery composition may be administered temporarily or administered continuously or intermittently. For example, the administration may be a continuous administration for 1 minute to 2 weeks. Moreover, the PDPK1 expression inhibitor or activity inhibitor or the agent-delivery composition may be administered simultaneously with or separately from the medicinal component, and preferably administered before the administration of the medicinal component or simultaneously with the medicinal component.
[0189] The dose of the PDPK1 expression inhibitor or activity inhibitor or the agent-delivery composition is not particularly limited as long as it is a dose at which a desired permeability of the blood-brain barrier or an intracerebral delivery effect of a desired medicinal component is obtained, and can be determined depending on the disease to be treated or prevented; the type, amount, and route of administration of the desired medicinal component to be administered; the symptoms, sex, and age of the patient; and the like as appropriate. The dose can be determined, for example, by using the amount of medicinal component delivered to the brain or the therapeutic or preventive effect thereby as an indicator. The active ingredient at a dose of usually about 0.01-20 mg/kg body weight, preferably about 0.1-10 mg/kg of body weight, further preferably about 0.1-5 mg/kg of body weight is conveniently administered by intravenous injection about 1-10 times a month, preferably about 1-5 times a month. The above amount may be applied to parenteral administration via another route or oral administration. When symptoms are particularly severe, the dose or frequency of administration may be increased according to the symptoms.
4. Apparatus for Judging Brain Metastasis/Apparatus for Assessing Risk of Brain Metastasis
(Apparatus for Judging Brain Metastasis)
[0190] In one aspect, the present invention relates to an apparatus for judging brain metastasis that measures an miR-181c level. As illustrated in FIG. 21, an apparatus for judging brain metastasis 1 relating to this embodiment is an apparatus for judging brain metastasis in a subject cancer patient. More specifically, the apparatus for judging brain metastasis 1 is an apparatus for judging whether a cancer patient with suspected brain metastasis (usually Stage IV patient) has brain metastasis. The apparatus for judging brain metastasis 1 comprises a measuring unit 21, a level determination unit 31, and a metastasis judgment unit 32, described below. In this embodiment, the apparatus for judging brain metastasis 1 comprises an input device 2 comprising at least the measuring unit 21, an information processing device 3 comprising at least the level determination unit 31 and the metastasis judgment unit 32, and an output device 4. However, the input device 2, the information processing device 3, and the output device 4 does not need to be separate devices and two or more of these devices 2, 3, and 4 may be configured as one device.
[0191] The input device 2 comprises the measuring unit 21 that functions as an miR-181c-measuring means and preferably further comprises a preparation entry/exit unit 22, but the preparation entry/exit unit 22 may be separate from the input device 2. The input device 2 is, for example, a computer comprising a central processing unit (CPU), a read-only memory (ROM), a random access memory (RAM) and a hard disk drive (HDD) or an apparatus containing a computer. Processing in the measuring unit 21 is mainly performed by the CPU reading a computer program in the HDD. The computer program may be, for example, stored in the RAM or the ROM, but not stored in the HDD.
[0192] The measuring unit 21 is a component having functions as miR-181c-measuring means that measures a polynucleotide having the nucleotide sequence of miR-181c or a part thereof in a sample 23. The preparation entry/exit unit 22 preferably comprises a preparation holder 26 for holding a preparation 25 containing the sample 23 and a reagent 24, an illuminator 27 for irradiating excitation light to the preparation 25, and a photodetector 29 for detecting fluorescence 28 emitted from the preparation 25 irradiated by the excitation light. When the preparation 25 itself can emit light without irradiating excitation light from the outside, the preparation entry/exit unit 22 does not need to comprise the illuminator 27 and the photodetector 29 detects the emitted light but not fluorescence.
[0193] The information processing device 3 comprises at least the level determination unit 31 and the metastasis judgment unit 32. The information processing device 3 is preferably a computer with a CPU, a ROM, a RAM, and an HDD. Processing in both of the level judgment unit 31 and the metastasis judgment unit 32 is mainly performed by the CPU reading a computer program in the HDD. The computer program may not be stored in the HDD, for example, but stored in the RAM or the ROM. The input device 2 and the information processing device 3 may comprise a common CPU, common ROM, common RAM, or a common HDD. In this case, the computer program may be stored in an HDD, a RAM, or a ROM that is common to the input device 2 and the information processing device 3.
[0194] The level determination unit 31 is a component that functions as miR-181c level-determining means that receives electrical signals from the measuring unit 21 in the input device 2 and determines the miR-181c level in the measured sample 23. The metastasis judgment unit 32 is a component that functions as metastasis-judging means for judging brain metastasis in the subject cancer patient from the miR-181c level determined in the level determination unit 31. The metastasis judgment unit 32 uses a sample obtained from the subject cancer patient as the sample 23, and judges that the subject cancer patient has brain metastasis if the miR-181c level in the sample is higher than the miR-181c level in a sample from a negative control. The determination by the level determination unit 31 is preferably performed by the CPU reading the information (for example, standard curve data) stored in a memory such as the HDD or the RAM beforehand, and referring to the information. However, the information may not be stored in a memory beforehand, and can be sequentially accumulated by measuring a polynucleotide having the nucleotide sequence of miR-181c or a part thereof with using a concentration predetermined control sample (for example, a sample in a series of serial dilution) as the sample 23. The judgment by the metastasis judgment unit 32 is preferably performed by the CPU reading the information (that is, information on the miR-181c level in the sample from the negative control) stored beforehand in a memory such as the HDD or the RAM and referring to the information. However, the information may not be stored in the memory beforehand, and can be sequentially accumulated by measuring a polynucleotide having the nucleotide sequence of miR-181c or a part thereof in the sample from the negative control used as the sample 23.
[0195] The output device 4 is an apparatus for outputting the judgment by the metastasis judgment unit 32 and, for example, can be a monitor for outputting still image or an animation. The output device 4 may be a computer comprising, for example, a CPU, a ROM, a RAM and an HDD or an apparatus containing the computer, like as the input device 2 or the information processing device 3. In that case, the computer program may be stored in an HDD, a RAM, or ROM which is common to the input device 2, the information processing device 3, and the output device 4, or common to the input device 2 and the output device 4, or common to the information processing device 3 and the output device 4.
[0196] The output device 4 is not limited to a monitor and may be a device that outputs the judgment in any output form such as a speaker that outputs the judgment with sound, a printer that prints the judgment onto a medium such as paper, or a luminescent device that outputs the judgment in the form of luminescence.
(Apparatus for Assessing Risk of Brain Metastasis)
[0197] In another aspect, the present invention relates to an apparatus for assessing risk of brain metastasis by measuring an miR-181c level. As illustrated in FIG. 22, an apparatus for assessing risk of brain metastasis la of this embodiment is an apparatus for assessing the risk of brain metastasis of a subject cancer patient (usually a Stage I, II, or III patient) who does not yet have brain metastasis. The apparatus for assessing risk of brain metastasis la comprises a measuring unit 21, a level determination unit 31, and a risk assessment unit 33 described below. In this embodiment, the apparatus for assessing risk of brain metastasis la has a configuration similar to that of the apparatus for judging brain metastasis 1 described above, and comprises an input device 2, an information processing device 3, and the output device 4. However, the apparatus for assessing risk of brain metastasis la does not need to be an apparatus in such a form that the input device 2, the information processing device 3, and the output device 4 are separated, like the apparatus for judging brain metastasis 1. Two or more of the devices 2, 3, and 4 may be configured as one device.
[0198] The apparatus for assessing risk of brain metastasis la comprises a risk assessment unit 33 instead of the metastasis judgment unit 32 in the apparatus for judging brain metastasis 1. The other configuration of the apparatus for assessing risk of brain metastasis la is common with the configuration of the apparatus for judging brain metastasis 1 except for the metastasis judgment unit 32. Therefore, the redundant descriptions are omitted.
[0199] The risk assessment unit 33 is a component that functions as risk assessment means for judging the risk of brain metastasis in the subject cancer patient from the miR-181c level determined in the level determination unit 31. The risk assessment unit 33 judges that the subject cancer patient has a high risk of brain metastasis if the miR-181c level in a sample 23 from the subject cancer patient is higher than the miR-181c level in a sample from a negative control. The judgment by the risk assessment unit 33 is preferably performed by the CPU reading the information (that is, information on the miR-181c level in the sample from the negative control) stored beforehand in a storage device such as the HDD or the RAM and referring to the information. However, the information may not be stored in the memory beforehand and can be sequentially accumulated by measuring a polynucleotide having the nucleotide sequence of miR-181c or a part thereof in the sample from the negative control used as the sample 23.
5. Computer Program for Brain Metastasis Judgment or Brain Metastasis Risk Assessment
[0200] In one aspect, the present invention relates to a computer program used in the apparatus for judging brain metastasis or the apparatus for assessing risk of brain metastasis.
(Computer Program for Apparatus for Judging Brain Metastasis)
[0201] A computer program of this embodiment is a computer program to be installed in an apparatus for providing information for diagnosing brain metastasis in a subject cancer patient, that is, the above described apparatus for judging brain metastasis 1. The computer program directs the apparatus for judging brain metastasis 1, which is an apparatus for diagnosing brain metastasis in a cancer patient, to execute: an miR-181c-measuring procedure of measuring a polynucleotide having the nucleotide sequence of miR-181c or a part thereof using a sample 23 from the subject cancer patient, an miR-181c level-determining procedure of determining the miR-181c level in the sample 23 measured by the miR-181c-measuring procedure, an metastasis-judging procedure of judging brain metastasis in the subject cancer patient from the miR-181c level determined in the miR-181c level-determining procedure, and a judgement-output procedure of outputting the judgement by the metastasis-judging procedure; wherein the metastasis-judging procedure judges that the subject cancer patient has brain metastasis if the miR-181c level in the sample 23 from the subject cancer patient is higher than the miR-181c level in a sample from a negative control.
[0202] The miR-181c-measuring procedure is executed by the measuring unit 21 in the apparatus for judging brain metastasis 1. The miR-181c level-determining procedure is executed by the level determination unit 31 in the apparatus for judging brain metastasis 1. The metastasis-judging procedure is executed by the metastasis judgment unit 32 in the apparatus for judging brain metastasis 1. The judgment-output procedure is executed by the output device 4. These procedures are executed, for example, by a CPU reading the computer program stored in an HDD. This computer program may be a program that executes, by being read by processing of the CPU, the miR-181c-measuring procedure, the miR-181c level-determining procedure, and the metastasis-judging procedure, but not the subsequent judgment-output procedure.
[0203] This computer program cannot occupy space by itself, but can be circulated in the market by being stored in an information recording medium. The "information recording medium" is for example, a flexible disk, a hard disk, CD-ROM, CD-R, CD-RW, MO (magneto-optical disk), MD, DVD-R, DVD-RW, a flash memory, a chip card, or the like. By connecting these information recording mediums to a data input and output unit (not illustrated) in the apparatus for judging brain metastasis 1, the computer program can be installed in a memory such as an HDD in the apparatus for judging brain metastasis 1. It is also possible to transmit this computer program to the apparatus for judging brain metastasis 1 from a different computer storing the computer program through a communication line and install it in a memory such as an HDD in the apparatus.
[0204] FIG. 23 illustrates a flow of processing that the apparatus for judging brain metastasis performs when the computer program for the apparatus for judging brain metastasis is executed. The CPU in the apparatus for judging brain metastasis 1 reads the computer program for the apparatus for judging brain metastasis stored in a memory such as an HDD in the apparatus 1 and measures the polynucleotide having the nucleotide sequence of miR-181c or a part thereof using a sample 23 from a subject cancer patient (STEP 101: miR-181c-measuring procedure). Then, the CPU reads the computer program and determines the miR-181c level in the sample 23 measured (STEP 102: miR-181c level-determining procedure). Then, the CPU reads the computer program and judges the presence or absence of brain metastasis in the subject cancer patient from the miR-181c level determined in the miR-181c level-determining procedure (STEP 103: metastasis-judging procedure). Then, the CPU reads the computer program and outputs the result of judgement that there is brain metastasis or that there is no brain metastasis from the output device 4 (STEP 104: judgment-output procedure).
(Computer Program for Apparatus for Assessing Risk of Brain Metastasis)
[0205] A computer program relating to another embodiment is a computer program to be installed in an apparatus for assessing risk of brain metastasis in a subject cancer patient, that is, the apparatus for assessing risk of brain metastasis la. The computer program directs the apparatus for assessing risk of brain metastasis la, which is an apparatus for assessing brain metastasis in a cancer patient, to execute: an miR-181c-measuring procedure of measuring a polynucleotide having the nucleotide sequence of miR-181c or a part thereof using a sample 23 from the subject cancer patient, an miR-181c level-determining procedure of determining the miR-181c level in the sample 23 measured by the miR-181c-measuring procedure, a risk-assessing procedure of assessing risk of brain metastasis in the subject cancer patient from the miR-181c level determined in the miR-181c level-determining procedure; an assessment-output procedure of outputting the assessment by the risk-assessing procedure; wherein the risk-assessing procedure judges that the subject cancer patient has a high risk of brain metastasis if the miR-181c level in the sample 23 from the subject cancer patient is higher than the miR-181c level in a sample from a negative control.
[0206] The risk-assessing procedure is executed by the risk assessment unit 33 in the apparatus for assessing risk of brain metastasis la. The miR-181c-measuring procedure, the miR-181c level-determining procedure, and the assessment-output procedure are respectively executed by the measuring unit 21, the level determination unit 31, and the output device 4 as the same manner with the computer program for the apparatus for judging brain metastasis described above. These procedures are executed, for example, by a CPU reading the computer program stored in an HDD. This computer program may be a program that executes, by being read by processing of the CPU, the miR-181c-measuring procedure, the miR-181c level-determining procedure, and the risk-assessing procedure, but not the subsequent assessment-output procedure.
[0207] This computer program can be also stored in an information recording medium illustrated above. By connecting these information recording mediums to a data input and output unit (not illustrated) in the apparatus for assessing risk of brain metastasis 1a, the computer program can be installed in a memory such as an HDD in the apparatus for assessing risk of brain metastasis la. It is also possible to transmit this computer program to the apparatus for assessing risk of brain metastasis la from a different computer storing the computer program through a communication line and install it in a memory such as an HDD in the apparatus.
[0208] FIG. 24 illustrates a flow of processing that the apparatus for assessing risk of brain metastasis performs when the computer program for the apparatus for assessing risk of brain metastasis is executed. The CPU in the apparatus for assessing risk of brain metastasis la reads the computer program for the apparatus for assessing risk of brain metastasis stored in a memory such as an HDD in the apparatus la and measures the polynucleotide having the nucleotide sequence of miR-181c or a part thereof using a sample 23 from a subject cancer patient (STEP 201: miR-181c-measuring procedure). Then, the CPU reads the computer program and determines the miR-181c level in the sample 23 measured (STEP 202: miR-181c level-determining procedure). Then, the CPU reads the computer program and assesses whether the risk of brain metastasis in the subject cancer patient is high or low from the miR-181c level determined in the miR-181c level-determining procedure (STEP 203: risk-assessing procedure). Then, the CPU reads the computer program and outputs the result of assessment that the risk of brain metastasis is high or that the risk of brain metastasis is low from the output device 4 (STEP 204: judgment-output procedure).
[0209] Although the present invention will be hereinafter described in detail with reference to Examples, the present invention is not intended to be limited thereto. The all documents cited throughout the present application are incorporated as it is into the present application by reference.
EXAMPLES
[0210] In the experiments described in Examples below, all experiments using animals were conducted according to the protocols approved by the Institutional Animal Care and Use Committee of National Cancer Center (Independent Administrative Institution). In experiments described below, all numerical values are expressed as mean+/-standard deviation (S.D.). P values less than 0.05 by Student's t-test or Mann-Whitney U test were considered to be statistically significant.
(Cell culture)
[0211] In the experiments described in the Examples, MDA-MB-231-luc-D3H1 cells, MDA-MB-231-luc-D3H2LN cells, BMD2a cells, and BMD2b cells were cultured in the RPMI 1640 medium containing 10% heat inactivated fetal bovine serum (Invitrogen) and antimicrobial-antifungal agents at 37.degree. C., 5% CO.sub.2.
Example 1
Establishment of Brain Metastasis Breast Cancer Cell Line
[0212] Brain metastasized cell lines were newly generated using highly tumorigenic and metastatic human breast cancer cells, MDA-MB-231-luc-D3H2LN cells (hereinafter, referred to as the "D3H2LN cells") (Bos, P. D. et al., Nature, 459:1005-1009 (2009)) (FIG. 1a). Specifically, 100 .mu.L of a cell suspension containing 2.times.10.sup.5 D3H2LN cells was administered by intracardiac injection to the left ventricle in 7 week-old female C.B-17/Icr-scid/scid immunodeficient mice. The mice were observed every week for tumorigenesis by brain metastasis by bioluminescence in vivo imaging after intraperitoneal injection of luciferin using IVIS Spectrum (Caliper Life Science, Hopkinton, Mass., USA). 26-30 days later, brain metastasis of cancer cells was observed (FIG. 1b).
[0213] The brain metastasis sites were examined by histologic analysis after dissection. Cancer cell colonization in brain tissue was confirmed by fixation of half of the tissue with 4% para-formaldehyde and histologic analysis with hematoxylin-eosin (HE) staining (FIG. 1c).
[0214] To collect and culture brain metastatic cancer cells, the remaining half of the tissue was ground and added to the RPMI 1640 medium (Gibco) containing antimicrobial-antifungal agents and 10% FBS. After centrifugation for a short time, the cells were resuspended with 0.025% trypsin-EDTA (Gibco) and incubated at 37.degree. C. for 10 minutes. The cells were resuspended in a medium containing 50 .mu.g/mL of Zeocin (Gibco), then seeded in a 10 cm dish, and cultured and grown to confluent for approximately 30 days.
[0215] Using the grown cell population (brain metastatic derivative 1a, hereinafter, referred to as the "BMD1cells"), the second in vivo selection was conducted in the way same as that described above and two brain metastatic derivative cell populations 2a and 2b (hereinafter, respectively referred to as the "BMD2a cells" and the "BMD2b cells") were obtained.
[0216] The obtained BMD2a and BMD2b cells were administered to mice like the D3H2LN cells and examined their brain metastasis activities. As a result, while brain metastasis was observed only 1 mouse (6.7%) out of the 15 mice in which the D3H2LN cells were injected into the left ventricle, 60% (3 out of 5) of the mice in which the BMD1a cells are injected into the left ventricle had brain metastasis. Accordingly, it was demonstrated that the BMD2a and BMD2b cells have a markedly increased brain metastasis activity compared to the parental D3H2LN cells.
Example 2
Construction of In Vitro Blood-Brain Barrier (BBB) Model
(1) In Vitro Blood-Brain Barrier Model
[0217] An in vitro BBB culture system was constructed to measure effects on BBB easily. Conventional in vitro blood-brain barrier models use monolayer cell culture systems (Zhou, W. at al, J. Biol. Chem., 288:10849-10859). However, BBB consists of three different types of cells and these cells cooperate with each other to maintain the structure of BBB. Therefore, BBB kit.TM. (#MTB-24H, PharmaCo-Cell Company Ltd., Nagasaki, Japan), which is a BBB model system that consists of primary cultures of brain capillary endothelial cells, brain pericytes and astrocytes (FIG. 2a) was used. Brain capillary endothelial cells, brain pericytes, and astrocytes were confirmed by Hoechst33342 staining. As a result, the presence of brain capillary endothelial cells, brain pericytes, and astrocytes in the constructed in vitro blood-brain barrier model was confirmed as illustrated in FIG. 2b.
(2) Confirmation of Localization of Tight Junction Proteins and the Like in Endothelial Cells
[0218] Tight junctions are known to regulate the low permeability of BBB and are formed by specific proteins in endothelial cells (such as Claudin-5, Occludin, and ZO-1). Meanwhile, N-cadherin, which is a calcium-dependent cell-cell adhesion glycoprotein and has five extracellular cadherin repeats to mediate strong cell-cell adhesion, is mostly expressed on the apical and basal membranes. Tight junction proteins and N-cadherin regulate cell polarity through their intimate association with the actin cytoskeletal network. Therefore, the formation of tight junction and adherens junction were confirmed by fluorescently immunostaining Claudin-5, Occludin, ZO-1, or N-cadherin in the endothelial cells in the in vitro BBB model to confirm their localization.
[0219] Specifically, the endothelial cells collected from the in vitro BBB model were fixed with PBS containing 3% PFA at room temperature for 10 minutes, washed with PBS containing Mg.sup.2+ and Ca.sup.2+, and then treated with PBS containing 0.1% Triton-X100 for 10 minutes to permeabilize the cells. After fixation, the cells were incubated with PBS containing 3% BSA for 1 hour to block nonspecific binding of antibody. Subsequently, the endothelial cells were incubated at 37.degree. C. with a rabbit polyclonal antibody to Claudin-5 (Z43.JK, Invitrogen, CA, USA), Occludin (ZMD.467, Invitrogen), ZO-1 (ZMD.437, Invitrogen), or N-cadherin (3B9, Invitrogen) for 1 hour. After washing with PBS containing Mg.sup.2+ and Ca.sup.2+, the cells were incubated at 37.degree. C. with 594 Alexa Fluor-fused antirabbit IgG (Invitrogen) for 1 hour. Actin was stained with ActinGreen.TM. 488 ReadyProbes (TN) Reagent (R37110, Molecular Probes). The stained cells were washed with Mg.sup.2+ and Ca.sup.2+ free PBS, then mounted with VECTASHIELD Mounting Medium (H-1200, Vector Laboratories, CA, USA), and observed with a cofocal microscope (FluoViewFV1000, Olympus, Tokyo, Japan).
[0220] It was demonstrated that all of the tight junction proteins (Claudin-5, Occludin, and ZO-1) and N-cadherin bind to the surface of the cell membrane and constitute intercellular tight junction and adherens junction as illustrated in FIG. 2c.
(3) Measurement of Transendothelial Electrical Resistance (TEER)
[0221] The formation of tight junction by brain capillary endothelial cells was measured by transendothelial electrical resistance (TEER) (Gaillard, P. J. et al., Eur. J. Pharm. Sci., 12:215-222 (2001), Wilhelm, I. et al., Acta. Neurobiol. Exp. (Wars), 71:113-128 (2011)). The resistance values (.OMEGA.) were measured by an ohmmeter (Millicell ERS-2, Millipore). The transendothelial electrical resistance (TEER) values were calculated by means of the unit area resistance by the following calculating formula. R=(A-B)*0.33 cm.sup.2, wherein R denotes TEER (.OMEGA.*cm.sup.2), A denotes measurement resistance value (.OMEGA.), and B denotes blank resistance value (.OMEGA.). It was done for n=12.
[0222] FIG. 2d illustrates the result of measuring the formation of tight junction by the brain capillary endothelial cells by transendothelial electrical resistance (TEER). The transendothelial electrical resistance (TEER) exceeded 150, which indicates that this system can be used as an in vitro BBB model.
(4) Permeability Assay
[0223] NaF does not pass BBB in spite of having a low molecular weight (molecular weight: 376.27) (Nakagawa, S, et al., Neurochem. Int. 54: 253-263 (2009)). Accordingly, permeability of BBB was measured by measuring the concentration of NaF with a fluorescence monochromator. Specifically, a permeability assay modified from a previously described method (B. Kis et al., Adrenomedullin regulates blood-brain barrier functions in vitro. Neuroreport 12, 4139-4142 (2001)) was used. 200 .mu.L of NaF (10 .mu.g/mL, Sigma Aldrich) was added to the upper chamber and 900 .mu.L of DPBS-H (Dulbecco's PBS (Mg+, Ca+) containing 10 mM HEPES and 4.5 mg/mL D-glucose) was added to the lower chamber. The plate was cultured with shaking at 37.degree. C. After 30 min, the DPBS-H of the lower chamber was dispensed into a black plate (n=8) and measured with a multi detection monochrometer microplate reader (485/535 nm, SAFIRE, Tecan). The apparent permeability coefficient (Papp) was calculated by the following calculating formula:
Papp=(VA.times.[C]A).times.A.sup.-1.times.[C]Luminal.sup.-1.times.t.sup.- -1
[0224] The abbreviations in the formula denote the following:
[0225] VA: volume of abluminal chamber (0.9 cm.sup.3)
[0226] A: membrane surface area (0.33 cm.sup.2)
[0227] [C]Luminal: initial luminal tracer concentration (.mu.g/mL)
[0228] [C]A: abluminal tracer concentration (.mu.g/mL)
[0229] t: time of experiment (min)
[0230] The permeability assay conducted by the method described above demonstrated, as a result, that the permeability of NaF is very low.
Example 3
Examination of Invasive Capacity of Brain Metastatic Cancer Cell using Matrigel.TM.
[0231] Brain metastatic cancer cells are also considered to be highly invasive. Therefore, invasion chamber assays using Matrigel.TM. were used to confirm the pathological implications of the metastatic potential of established cell lines. BMD2a, BMD2b, D3H2LN, and D3H1 cells were trypsinized and labelled with PKH26 (Sigma Aldrich). 2.times.10.sup.4 cells of each cancer cell line were suspend in serum-free RPMI1640 medium and plated in the top chamber with a Matrigel(R)-coated membrane (24-well insert, BD Biosciences, NJ, USA). RPMI1640 medium containing 10% serum was added to the lower chamber as a chemoattractant. The cells were cultured for 24 hours and cells that did not migrate or invade through the membrane pores were removed using a cotton swab. Cells on the lower surface of the membrane were stained with the Diff-Quick Staining Set (Sysmex, Hyogo, Japan) and counted. All assays were performed in triplicate. The data were expressed as percentage of the number of cells migrating through the Matrigel.TM. matrix and membrane relative to the number of cells migrating through the control membrane, according to the manufacturer's instructions.
[0232] BMD2a and BMD2b cells were more invasive than D3H2LN cells and low-metastatic D3H1 cells (FIGS. 3a and 3b). However, the morphology of BMD2a and BMD2b cells did not differ from that of D3H2LN cells (FIG. 3c).
Example 4
Examination of Invasive Capacity of Brain Metastatic Cancer Cells using BBB Model
[0233] To confirm the entry of BMD2a and BMD2b cells into the brain parenchyma cell side, BMD2a, BMD2b, D3H2LN and D3H1 cells were trypsinized and labelled with PKH26 (Sigma Aldrich). Subsequently, 2.times.10.sup.4 cancer cells were suspended in serum-free Ham's F-12 medium (Gibco) and added to the upper chamber of the in vitro BBB model prepared in Example 2 and cultured. After 2 days (48 hours), non-invading cells were removed with cotton swabs and the nuclei were stained with Hoechst 33342 (Dojindo Laboratories, Kumamoto, Japan). Subsequently, the PKH26 fluorescence of invading cells that had migrated to the lower surface side was counted with a fluorescence microscope (FIG. 4a). All cells were assayed in triplicate.
[0234] BMD2a and BMD2b cells were more invasive than D3H2LN and D3H1 cells (FIG. 4b). Accordingly, it was confirmed that the established BMD2a and BMD2b cells have a high potential for entry into the brain parenchymal cell side across BBB.
Example 5
Preparation of EVs
[0235] D3H1, D3H2LN, BMD2a, and BMD2b cells were washed three times with Advanced RPMI medium containing antibiotic-antimycotic agents and 2 mM L-glutamine (hereinafter, referred to as the "medium A"). Fresh medium A was added to the washed cells and cultured for 2 days, and then the medium was collected and centrifuged at 2000*g for 10 min at 4.degree. C. To thoroughly remove cellular debris, the culture supernatant was filtered with a 0.22-.mu.m Stericup.RTM. (Millipore, MA, USA). The resultant conditioned medium was ultracentrifuged at 110,000*g for 70 min at 4.degree. C. The resultant pellets were washed with 11 mL of phosphate buffered saline (PBS) and resuspended in PBS.
[0236] The fraction containing the EVs was measured for its protein content using the Micro BCA protein assay kit (Thermo Scientific, MA, USA). The obtained EVs was observed by phase contrast microscopy. Furthermore, the size and the number of the EVs were measured with NanoSight.
[0237] The expression of the standard exosomal markers CD63 and CD9 and the expression of the mitochondrial intermembrane protein Cytochrome C, which is known to be lacking in EVs, was measured by Western blotting. Samples were obtained from each cell line using M-PER (Thermo Scientific, MA, USA) and loaded in Mini-PROTEAN.RTM. TGX Gel (4-12%) (Bio-Rad) at 500 ng/lane (CD63 and CD9) or 3 .mu.g/lane (Cytochrome C) and proteins were separated and then electrotransferred onto PVDF membranes (Millipore). The resultant membranes were blocked in Blocking One (Nacalai Tesque, Kyoto, Japan) and then incubated for 1 hour at room temperature with a primary antibody, anti-CD63 antibody (purified mouse anti-human CD63, H5C6, 1/200, BD), anti-CD9 antibody (ALB6, sc-59140, 1/200, BD), or anti-cytochrome C antibody (purified mouse anti-cytochrome C, 7H8.2C12, 1/200, BD). An HRP-linked anti-mouse IgG secondary antibody (NA931, GE Healthcare) or HRP-linked anti-rabbit IgG secondary antibody (NA934, GE Healthcare) was used at a dilution of 1/2000. The membranes were then made luminescent with ImmunoStar.RTM. LD (Wako, Osaka, Japan).
[0238] As for the protein content, there was no difference in protein concentration among the EVs derived from D3H1 cells, the EVs derived from D3H2LN cells, the EVs derived from BMD2a cells, and the EVs derived from BMD2b cells. FIG. 5a illustrates photographs resulted from the phase contrast microscope observation and the result of the analysis of EV size. The average particle sizes of the EVs derived from D3H1 cells, D3H2LN cells, BMD2a cells, and BMD2b cells were 149 nm, 157 nm, 136 nm, and 148 nm, respectively. All these EVs expressed the standard exosomal markers CD63 and CD9 but not Cytochrome C (FIG. 5b). Furthermore, the number of EV release did not differ among the cells (FIG. 5c).
Example 6
Examination of Correlation between Secretion of EVs and Invasive Capacity of Cancer Cells
[0239] To examine the involvement of EVs in the invasion of brain metastatic cells, the invasive capacity of BMD2a cells in which the EVs secretion was inhibited by siRNAs (Kosaka, N. et al., J. Biol. Chem., 288:10849-10859 (2013); Ostrowski, M et al., Nat. Cell Biol., 12:19-30 (2010)) against nSMase2 and RAB27B, which are proteins involved in the EVs secretion, was assessed in the in vitro BBB model
(1) siRNA Transfection
[0240] 25 nM each of either or both of RAB27B siRNA and nSMase2 siRNA or control siRNA was introduced into PKH26-labelled BMD2a cells using the DharmaFECT transfection reagent (Thermo Scientific) according to the manufacturer's protocol. Cells 24 hours after the transformation were used in assay.
(2) Confirmation of Expression of nSMase2 and RAB27B at mRNA Pevel in siRNA-Introduced Cells
[0241] Total RNA was extracted from cells using QIAzol reagent and RNeasy Mini Kit (both Qiagen). RNU6 was used as an internal control according to a described method (Kosaka, N. et al., Net. Med., 18:883-891 (2012)). The expression level of each mRNA and RNU6 was measured with qRT-PCR performed in 96-well plates using Platinum Quantitative PCR SuperMix (Applied Biosystems) with the 7300 Real-Time PCR System (all Applied Biosystems, CA, USA). All reactions were performed in triplicate. The primers and probes were defined as follows: PAB27B (assay ID: Hs01072206_m1), nSMase2 (assay ID: Hs00920354_m1).
(3) Confirmation of Expression of nSMase2 at Protein Level in siRNA-Introduced Cells
[0242] Expression of nSMase2 in siRNA-introduced cell was confirmed at the protein level by Western blotting. Samples were obtained from each cell line using M-PER (Thermo Scientific, MA, USA) and loaded in Mini-PROTEAN.RTM. TGX Gel (4-12%) (Bio-Rad) and proteins were separated and then electrotransferred onto PVDF membranes (Millipore). The resultant membranes were blocked in Blocking One (Nacalai Tesque, Kyoto, Japan) and then incubated for 1 hour at room temperature with a primary antibody, anti-nSMase2 antibody (H-195, sc-67305, 1/200, Santa Cruz Biotechnology Inc.). An HRP-linked anti-mouse IgG secondary antibody (NA931, GE Healthcare) or HRP-linked anti-rabbit IgG secondary antibody (NA934, GE Healthcare) was used at a dilution of 1/2000. The membranes were then made luminescent with ImmunoStar(R) LD (Wako, Osaka, Japan).
(4) Measurement of Number of EV Release from siRNA-Introduced Cells
[0243] The number of EV release from siRNA-introduced cells were measured with NanoSight.
(5) Examination of Invasive Capacity of Brain Metastatic Cancer Cell using BBB Model
[0244] The Matrigel invasion capacity of generated transformation cells and migration across BBB were examined according to the methods in Example 3 and Example 4, respectively.
(6) Results
[0245] The cells into which either of nSMase2 siRNA and RAB27B siRNA was introduced and the cells into which both of the siRNAs were introduced had a reduced expression of nSMase2 and RAB27B at both mRNA and protein levels compared to the control siRNA-introduced cells (not shown). According to the result of measurement of the number of EV release from siRNA-introduced cells, the cells into which either of nSMase2 siRNA and RAB27B siRNA was introduced and the cells into which both of the siRNAs were introduced had a reduced number of EV release compared to the control siRNA-introduced cells (FIG. 6a). According to the result of test on capacity to migrate across BBB, while the cells treated with control siRNA migrated to the lower side across BBB, the cells in which the production of EVs was inhibited were not found on the lower side (FIG. 6c). In the invasive capacity assay using Matrigel, the inhibition of EVs secretion did not affect the invasive capacity of the cells (FIG. 6b). From this, it was suggested that extravasation is not only dependent on the cells' invasive capacity.
Example 7
Examination of Correlation between EVs and BBB Invasive Capacity of Cancer Cells
[0246] Furthermore, to investigate whether EVs are involved in the extravasation of cancer cells to the brain parenchymal cell side, whether low-metastatic D3H1 cells migrate across BBB and extravasate after the addition of BMD2a cell-, BMD2b cell-, or D3H2LN cell-derived EVs or not was examined.
[0247] The EVs derived from BMD2a, BMD2b, or D3H2LN cells were added to each well and then incubated for 24 hours. Subsequently, PKH26-labelled D3H1 cells were added. After 2 days, the number of cells migrated across the filter was counted.
[0248] The result is illustrated in FIG. 7. D3H1 cells could not migrate across BBB without the addition of EVs. However, the number of cells that migrated across BBB to the lower side was significantly increased by the addition of BMD2a cell- and BMD2b cell-derived EVs. The addition of D3H2LN cell-derived EVs did not efficiently promote the migration of low-metastatic D3H1 cells across BBB compared to BMD2a- and BMD2b cell-derived EVs. These results indicated that the EVs derived from brain metastasis breast cancer cells affected the extravasation of cancer cells to the brain parenchyma side across BBB.
Example 8
Examination of Effect of EVs on Permeability of BBB
[0249] To determine whether EVs derived from brain metastatic cancer cells directly affects BBB disruption, whether addition of EVs in the in vitro BBB model disrupts BBB or not was examined. EVs were prepared according to the description in Example 5. Four days after thawing the in vitro BBB model, purified EVs derived from D3H2LN, BMD2a, and BMD2b cells or PBS (negative control) was added. Before the addition of the EVs and 24 hours after the addition, transendothelial electrical resistance (TEER) was measured to measure the strength of tight junction of the brain capillary endothelial cells, according to the method described in Example 2. All assays were performed in triplicate.
[0250] Furthermore, a permeability test using NaF was conducted according to the method described in Example 2 (2) to measure the permeability of BBB. 24 hours after the addition of purified EVs derived from D3H2LN, BMD2a, and BMD2b cells, NaF was added. NaF moved across BBB was measured with a fluorocytometer. The apparent permeability coefficient (Papp) (10.sup.-6 cms.sup.-1) was calculated according to the calculating formula described in Example 2 (2). All assays were performed in triplicate.
[0251] The measured TEER values are illustrated in FIG. 8a and the relative changes in TEER between before and after the addition of EVs are illustrated in FIG. 8b. TEER values in the BMD2a and BMD2b cell-derived EVs administrated groups were significantly decreased compared to that in the D3H2LN cell-derived EVs administrated group (P<0.05) and the D3H1 cell-derived EVs administrated group (P<0.01).
[0252] The result of the permeability test is illustrated in FIG. 8c. Both BMD2a and BMD2b cell-derived EVs administrated groups exhibited a clearly high (P<0.01) permeability coefficient (Papp) compared to the D3H2LN cell-derived EVs administrated group (P<0.05) and the D3H1 cell-derived EVs administrated group (P<0.01).
Example 9
Analysis of Uptake of EV to BBB Constituent Cells
[0253] To confirm that EVs are taken up to BBB configuration cell, EVs were added to the in vitro BBB model and examined the uptake by the BBB constituent cells. All assays were performed in triplicate.
(1) Labeling of EV with PKH67
[0254] Purified EVs derived from D3H1, D3H2LN, BMD2a, and BMD2b cells prepared by the method in Example 4 were labelled with PKH67 green fluorescent labeling kit (Sigma Aldrich, MO, USA). EVs were reacted with 2 .mu.M PKH67 for 5 minutes and washed using a 100 kDa filter (Microcon YM-100, Millipore) five times to remove excess dye.
(2) Uptake of EVs into Cells in In Vitro BBB Model
[0255] According to the method described in Example 8, the labelled EVs or PBS (negative control) was added to the upper surface side in the in vitro BBB model, which was cultured at 37.degree. C., 5% CO.sub.2 for 24 hours. Brain capillary endothelial cells, brain pericytes, and astrocytes were confirmed by Hoechst33342 staining.
(3) Result
[0256] The result of measurements of uptake of EVs to BBB constituent cells is illustrated in FIG. 9a and FIG. 9b. All cancer cell-derived EVs were taken up by the endothelial cells, but little uptake to pericytes and astrocyte was observed. The observation that EVs derived from BMD led to the highest fluorescence intensity in endothelial cells revealed the tropism of EVs derived from BMD cells to the cerebrovascular endothelial cells.
Example 10
Analysis of Mechanisms of BBB Destruction by EVs
[0257] To examine the mechanisms of BBB destruction by EVs, EVs were added to the in vitro BBB model and examined the change of molecular behaviors in BBB constituent cells. All assays were performed in triplicate.
(1) Localization of Tight Junction Proteins in Endothelial Cells after the EV Addition
[0258] Purified EVs derived from D3H1, D3H2LN, BMD2a, and BMD2b cells prepared by the method in Example 4 or PBS (negative control) was added and, 24 hours later, Claudin-5, Occludin, ZO-1, or N-cadherin in the endothelial cells in the in vitro BBB model was fluorescently immunostained with actin filaments to examine whether the localization is changed using a method similar to that in Example 2 (2).
(2) Change of Tight Junction Protein Levels etc. in Endothelial Cells After EV Addition
[0259] Proteins were isolated from each cell line using M-PER (Thermo Scientific, MA, USA), separated in Mini-PROTEAN.RTM. TGX Gel (4-12%) (Bio-Rad), and electrotransferred onto PVDF membranes (Millipore). The resultant membranes were blocked in Blocking One (Nacalai Tesque, Kyoto, Japan) and then incubated for 1 hour at room temperature with a primary antibody, anti-Claudin-5 antibody (Z43.JK, 1/200, Invitrogen), anti-Occludin antibody (ZMD.481, 1/200, Invitrogen), anti-ZO-1 antibody (H-300, sc-10804, 1/100, Santa Cruz Biotechnology), anti-N-cadherin antibody(3B9, 1/500, Invitrogen), or anti-GAPDH antibody (6C5, 1/1000, Millipore). An HRP-linked anti-mouse IgG secondary antibody (NA931, GE Healthcare) or HRP-linked anti-rabbit IgG secondary antibody (NA934, GE Healthcare) was used at a dilution of 1/2000. The membranes were then made luminescent with ImmunoStar.RTM. LD (Wako, Osaka, Japan).
(3) Results
[0260] The results of localization of the tight junction proteins in the endothelial cells in the in vitro BBB model are illustrated in FIG. 10a and FIG. 10b. The tight junction proteins and the N-cadherin were localized on the cell membrane surface of the endothelial cells treated with PBS or the EVs derived from D3H2LN cells. However, the tight junction proteins and the N-cadherin were found in the cytoplasm in the cells treated with the EVs derived from BMD2a and BMD2b,. Moreover, the treatment of cerebrovascular endothelial cells with the EVs derived from BMD2a and BMD2b had no effect on the expression of the tight junction proteins, N-cadherin, and actin (FIG. 10c). Thus, it was revealed that cancer-derived EVs changes the localization of tight junction proteins, N-cadherin, and actin filament without affecting the expression thereof.
Example 11
Examination of Effect on Blood-Brain Barrier Permeability of EVs in In Vivo
[0261] Since the results of the in vitro BBB model tests demonstrated that cancer cell-derived EVs are taken up by endothelial cells and increase the permeability of BBB, in vivo permeability tests were conducted to examine whether they also have the effect on the blood ability barrier in vivo. Purified EVs derived from D3H2LN cells (control) and BMD2a cells were labelled by XenoLight DiR fluorescent staining. The labelled EVs were injected into the tail vein of mice and, 6 hours later, Tracer-653, a dye for in vivo blood vessel permeability tests, was injected into the mice for monitoring BBB disruption.
[0262] The upper panels in FIG. 11 illustrate the intake of EVs and the lower panels illustrate the permeability of brain blood vessels. As illustrated in the upper panels of FIG. 11, BMD2a cell-derived EVs were more incorporated within the brains of mice than were D3H2LN cell-derived EVs. As illustrated in the lower panels of FIG. 11, BMD2a cell-derived EV-treated mice exhibited greater permeability of brain blood vessels compared to D3H2LN cell-derived EV-treated mice.
Example 12
Effect of EVs on Cancer Metastasis In Vivo
[0263] To examine whether increase in in vivo brain blood vessel permeability by EVs, confirmed in Examples above, actually has an effect on brain metastasis, cancer cells were injected into EV-treated mice and whether brain metastasis occurs or not was examined. Purified EVs (5 .mu.g/animal for each) derived from D3H2LN cells (control) or BMD2a cells or PBS (negative control) were injected in the tail vein of 9 animals per group of C.B-17/Icr-scid/scid mice (Day 0). 24 hours later (Day 1), the D3H2LN breast cancer cell line (2.times.10.sup.5 cells) was injected in the left cardiac ventricle of each mice. 18 days later (Day 19), brain metastasis of cancer cells was observed by intraperitoneal luciferin injection and bioluminescence in vivo imaging using IVIS Spectrum (Caliper Life Science, Hopkinton, Mass., USA). Evaluation was made by measuring photon intensity in the brain from the obtained images. Significant difference was evaluated by Mann Whitney one-tailed testing. Brain metastasis was also confirmed by hematoxylin and eosin (HE) staining and immunofluorescence staining for human vimentin.
[0264] In the BMD2a-derived EVs administrated group, brain metastasis in 5 out of 9 animals (55.6%), which is clearly higher than brain metastasis in the D3H2LN cell-derived EVs administrated group (brain metastasis in 1 out of 9 animals: 11.1%) and the PBS administrated group (brain metastasis in 0 out of 9 animals: 0%), was found. According to the result of fluorescence intensity measurement, there were clearly more invading and metastasizing cancer cells in brains in the BMD2a-derived EVs administrated group compared to those in other groups (FIG. 12a, b, p<0.05). HE staining also confirmed brain metastasis (FIG. 12c). As indicated by these results, it was revealed that increase of the brain blood vessel permeability by EVs increases brain metastasis
Example 13
Elucidation of Molecule Responsible for Change of Blood-Brain Barrier Permeability by EVs
[0265] To elucidate the molecular mechanisms by which EVs changed the permeability of the blood-brain barrier, miRNA contained in EVs derived from D3H2LN, BMD2a, and BMD2b cells was analyzed.
[0266] Total RNA was extracted from EVs using the QIAzol reagent and the miRNeasy Mini Kit (both Qiagen). The quality and quantity of the RNA were determined using NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc.) and Agilent Bioanalyzer (Agilent Technologies) according to the recommended methods. The obtained total RNA was labelled with cyanine 3 (Cy3) using miRNA Complete Labeling and Hyb Kit (Agilent Technologies) according to the manufacturer's instructions. More specifically, 100 ng of total RNA was dephosphorylated by reacting for 30 minutes at 37.degree. C. using Calf Intestinal Alkaline Phosphatase (CIP) Master Mix. The dephosphorylated RNA was denatured with DMSO by incubation for 5 minutes at 100.degree. C. The RNA was then immediately transferred on ice and incubated for 2 minutes. The resultant reaction products were mixed with Ligation master mix for T4 RNA Ligase and Cy3-pCp (Cyanine 3-Cytidine biphosphate) and incubated for 2 hours at 16.degree. C. The labelled RNA was dried using vacuum concentrator at 55.degree. C. for 1.5 hours. Cy3-pCp-labelled RNA was hybridized on Agilent SurePrint G3 Human miRNA 8.times.60K Rel. 19 microarray (design ID: 046064) at 55.degree. C. for 20 hours. There are a total of 2,006 miRNA probes on the microarray without control probes. After washing, the microarray was scanned using an Agilent DNA microarray scanner. Intensity values of each scanned feature were quantified using Agilent Feature Extraction software version 10.7.3.1, which performs background subtractions. Only using features that were flagged as no errors (Detected flags), the expression analysis was performed with Agilent GeneSpring GX version 12.6.1. Features that were not positive, not significant, not uniform, not above background, saturated and population outliers (not detected flags) were excluded. In this experiment, the significant differences of gene expression were identified by applying more than 2-fold change in signal intensity.
[0267] In addition to miRNA, proteins contained in EVs derived from D3H2LN, BMD2a, and BMD2b cells were analyzed. However, any candidate protein characteristic of BMD2a and BMD2b cells was not found (not illustrated). As a result of the miRNA analysis, miR-181c was found to be significantly upregulated in BMD2a cell- and BMD2b cell-derived EVs compared to EVs derived from D3H2LN cells (FIG. 13a, FIG. 13b). Therefore, to confirm whether miR-181c is expressed in D3H2LN, BMD2a, and BMD2b cells, intracellular miR-181c was detected. As a result, any significant difference in expression of miR-181c was not confirmed in any of D3H2LN cells, BMD2a cells, and BMD2b cells (not illustrated).
Example 14
Evaluation of Effect of miR-181c on BBB
[0268] The foregoing results suggested that the BBB destruction by cancer cell-derived EVs involves miR-181c contained in EVs taken up by the endothelial cells. Therefore, the abundance of miR-181c in the endothelial cells after the addition of the EVs was examined to study the effect of miR-181c on BBB. Changes of permeability and behaviors of tight junction proteins were also examined using the in vitro BBB model containing the endothelial cells into which miR-181c had been introduced.
(1) Change of miR-181c Abundance in Endothelial Cells by Addition of EVs
[0269] To evaluate the effect of miR-181c on BBB, EVs isolated from BMD2a and BMD2b was added to endothelial cells and the change of miR-181c abundance was examined. EVs were prepared according to the description in Example 5. Four days after thawing the in vitro BBB model, purified EVs derived from D3H2LN, BMD2a, and BMD2b cells or PBS (negative control) were added. 24 hours after the addition of EVs, the expression of miR-181c in endothelial cells was measured. All assays were performed in triplicate.
(2) Effect of Change of miR-181c Abundancein in Endothelial Cells
[0270] To further examine the effect of change of miR-181c abundance on endothelial cells, synthetic miR-181c was introduced into the endothelial cells constituting the in vitro BBB model. Transendothelial electrical resistance (TEER) was measured according to the method described in Example 2 (3). The localization and expression levels of tight junction proteins, N-cadherin, and actin were detected according to the method described in Example 10. Endothelial cells into which miR-181c was not introduced was used as a negative control.
(3) Results
[0271] The abundance of miR-181c in the endothelial cells was significantly increased by the addition of EVs derived from BMD2a and BMD2b (FIG. 14a). The transfection of miR-181c significantly decreased the value of TEER in the in vitro BBB model (FIG. 14b). While tight junction proteins, N-cadherin and actin localized on the membrane in the negative control group, the localization had been changed to the cytoplasm in miR-181c-transfected cells (FIG. 14c). Furthermore, it was found that the expression of tight junction proteins, N-cadherin, and actin was not affected by the transfection of miR-181c into brain blood vessel endothelial cells (FIG. 14d).
Example 15
Measurement of Serum miR-181c in Brain Metastasis Breast Cancer Patients
[0272] To confirm whether there is actually miR-181c in EVs in the blood of brain metastasis breast cancer patients or not, EVs were purified from sera from brain metastasis breast cancer patients and the expression of miR-181c was examined. Sera from 56 breast cancer patients (10 brain metastasis patients, 46 non-brain metastasis patients) were collected at National Cancer Center Japan (No. 2013-111). Informed consent was obtained from all the patients. Serum was aliquoted and stored at -80.degree. C. until used, and freeze-thawing was avoided as much as possible after that. EVs were isolated from the sera using Total Exosome Isolation (from sera) (Invitrogen), The total RNA was then isolated from the obtained EVs using the RNeasy Mini Kit (Qiagen). The expression of miRNA was measured by qRT-PCT using RNU6 as an internal control by an already reported method (Kosaka, N. et al. Net. Med., 18:883-891 (2012)). More specifically, the expression levels of miR-181c and RNU6 were measured in a 96-well plate by qRT-PCR using TaqMan.RTM. MicroRNA Assays and TaqMan.RTM. Universal PCR Master Mix in 7300 Real-Time PCR System (all from Applied Biosystems, CA, USA). All reactions were performed in triplicate.
[0273] Significantly more miR-181was present in the EVs obtained from sera from the brain metastasis patients compared to the non-brain metastasis patients (P<0.05, T test; FIG. 15). There was no difference in the serum miR-181c abundance among stages in the non-brain metastasis breast cancer patients (FIG. 16). In the comparison between same IV patients, there was a significant difference between the brain metastasis and the non-brain metastasis patients (P<0.05) and the miR-181c level was clearly higher in EVs derived from the sera from brain metastasis patients. This suggested that the secretion of miR-181c to blood contributes to brain metastasis in real patients as well.
[0274] The foregoing results indicated that EVs derived from brain metastatic cancer cells induce the abnormal localization of tight junction proteins by transferring miR-181c into endothelial cells, which results in the destruction of the cell-cell contact.
Example 16
Search of Target of miR-181c in Endothelial Cells
[0275] To elucidate the mechanisms of BBB destruction by miR-181c, search for the target of miR-181c in the endothelial cells was conducted. The gene expression in endothelial cells transfected with a negative control or miR-181c and the gene expression in endothelial cells after the addition of EVs derived from in BMD2a, BMD2b, or D3H2LN cells were analyzed.
[0276] The total RNA from respective endothelial cells was isolated using the QIAzol reagent and the mirRNeasy Mini Kit (both Qiagen). The quality and quantity of the RNA were determined using NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc.) and Agilent Bioanalyzer (Agilent Technologies) according to the recommended methods. The obtained total RNA was labelled with cyanine 3 (Cy3) during amplification using Low Input Quick Amp Labeling Kit, one-color (Agilent Technologies) according to the manufacturer's instructions. More specifically, 100 ng of total RNA was reverse-transcripted into double strand complementary DNA (cDNA) using poly dT-T7 promoter primers. The primers, the template RNA, and a quality control transcription product of known concentration and quality were first denatured at 65.degree. C. for 10 minutes and then incubated for 2 hours at 40.degree. C. with 5.times. first strand buffer, 0.1 M dithiothreitol, 10 mM deoxynucleotide triphosphate mixture, and AffinityScript RNase Block Mix, which was then incubated with AffinityScript enzyme at 70.degree. C. for 15 minutes. Using the cDNA product as template, fluorescent complementary RNA (cRNA) was produced by in vitro transcription. The cDNA product was mixed with transcription master mix in the presence of T7 RNA polymerase and Cy3-labeled CTP (cytidine 5'-triphosphate) and incubated at 40.degree. C. for 2 hours. The labelled cRNA was purified using RNeasy Mini Spin Columns (Qiagen) and eluted with 30 mL of nuclease-free water. After the amplification and labeling, the quantity of the cRNA and incorporation of cyanine were determined using NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc.) and Agilent Bioanalyzer (Agilent Technologies). In each hybridization, 0.6 .mu.g of Cy3-labeled cRNA was fragmented and hybridized with Agilent Cynomolgus macaque Gene Expression Profiling Array (design ID: 028520) at 65.degree. C. for 17 hours. There are a total of 12,243 miRNA probes on the microarray without control probes. After washing, the microarray was scanned using an Agilent DNA microarray scanner. Intensity values of each scanned feature were quantified using Agilent Feature Extraction software version 10.7.3.1, which performs background subtractions. Only using features that were flagged as no errors (Detected flags), the normalization was performed with Agilent GeneSpring GX version 12.6.1 (for each tip: normalization for third quartile shift; for each gene: normalization for the median of all samples). Features that were not positive, not significant, not uniform, not above background, saturated and population outliers (Compromised and not detected flags) were excluded. Changed transcription products were quantified by using the comparison method. In this experiment, the significant differences of gene expression were identified by applying 2-fold change in signal intensity.
[0277] It was confirmed at both mRNA and protein levels that the expression of 3-phosphoinositide dependent protein kinase-1 (PDPK1) is suppressed in the brain blood vessel endothelial cells transfected with miR-181c compared to the negative control (FIG. 17a-c). Furthermore, it was confirmed at both mRNA and protein levels that the expression of PDPK1 is suppressed compared to the control (endothelial cells treated with EVs derived from D3H2LN cells) in the endothelial cells treated with EVs derived from BMD2a and BMD2b cells (FIG. 17d-f). These results indicated that miR-181c suppresses the expression of PDPK1 in brain blood vessel endothelial cells.
Example 17
Effect of PDPK1 on Localization of Tight Junction Proteins etc.
[0278] To examine the effect of PDPK1 on brain blood vessel endothelial cells, transendothelial electrical resistance (TEER) and the localization and expression of tight junction proteins, N-cadherin, and actin filaments after treatment with PDPK1 siRNA were examined.
[0279] 25 nM of PDPK1 siRNA (Ambion, ID: 510275) or 25 nM of control siRNA was introduced into brain blood vessel endothelial cells using DharmaFECT transfection reagent (Thermo Scientific) according to the manufacturer' protocol. Transendothelial electrical resistance (TEER) of the in vitro BBB model using the transfected cells after 24 hours was measured according to the method described in Example 2 (3). Moreover, the localization and expression of tight junction proteins, N-cadherin, and actin filaments in the transfected cells after 24 hours were measured according to the method described in Example 10.
[0280] The introduction of PDPK1 siRNA reduced the expression of PDPK1 protein (see FIG. 20b). While tight junction proteins and N-cadherin were located on the cell membrane in the cells treated with the control siRNA, the localization on the cell membrane had been lost in the PDPK1 siRNA-treated cells (FIG. 18a). Tight junction proteins, N-cadherin, and actin were found in cytoplasm, which is consistent with the results of the observation of the cells treated with BMD2a cell- or BMD2b cell-derived EVs or the miR-181c-transfected cells. Actin condensation was observed in the PDPK1 siRNA-treated endothelial cells. This phenomenon is also consistent with the results of the cells treated with BMD2a cell- or BMD2b cell-derived EVs or the miR-181c-transfected cells (FIG. 10a, FIG. 10b, FIG. 14c, and FIG. 18a). The expression of tight junction proteins, N-cadherin, and actin was not changed with or without PDPK1 siRNA treatment (FIG. 18b). The transendothelial electrical resistance (TEER) was decreased by the introduction of PDPK1 siRNA (FIG. 18c). These results revealed that the inhibition of PDPK1 changes the localization of tight junction proteins and the N-cadherin and increases the permeability of BBB.
[0281] The foregoing results demonstrated that after EVs secreted from brain metastatic cancer cells are taken up by endothelial cells, miR-181c enclosed in the EVs suppresses the expression of PDPK1 in the endothelial cells, thereby changes the localization of tight junction proteins and the N-cadherin, and increases the permeability of BBB.
Example 18
Confirmation of Suppression of PDPK1 Expression by miR-181c
[0282] Furthermore, whether miR-181c can directly suppress the expression of PDPK1 in endothelial cells or not is examined by 3'-untranslated region (3' UTR) luciferase reporter assay using the 3' UTR of the PDPK1 gene (Macaca: SEQ ID NO: 14; Human: SEQ ID NO: 15).
[0283] The 3' UTR for PDPK1 was obtained by PCR amplification using total RNA extracted from brain blood vessel endothelial cells of the Macaca fascicularis as template. The PCR primers used for the 3'UTR amplification are as follows:
TABLE-US-00001 Forward: (SEQ ID NO: 16) AACTCGAGAATGCTGGCTATTGTTGGCCTC Reverse: (SEQ ID NO: 17) AAGCGGCCGCAAGATTAAATCACTGACCCAATAG
[0284] The PCR products were cloned and incorporated into a pGEM-T Easy Vector (Promega, WI, USA). The amplified 3' UTR was cloned downstream of the Renilla luciferase coding region in the psiCHECK-2.TM. (Promega). An alignment analysis confirmed that there is a sequence complementary to miR-181c in the 3' UTR of PDPK1 (FIG. 19b).
[0285] HEK293 cells were seeded at 5.times.10.sup.4 cells/well in 96-well plates and cultured overnight. 100 ng of a reporter plasmid along with 100 nM of pre-miR-181c was co-transfected using DharmaFECT Duo transfection reagent (Thermo Scientific).
[0286] Cells were collected 24 hours after the transfection and assayed for luciferase activity using EnVision (PerkinElmer, MA, USA). To examine the effect of endogenous miRNA, synthetic precursor miRNA (pre-miR; Ambion.RTM., Invitrogen) was co-transfected as a negative control. All experiments were performed in triplicate.
[0287] According to the results obtained by using HEK293 cells, the expression of luciferase, a reporter gene, was decreased by miR-181c (FIG. 19a). These results revealed that PDPK1 is a direct target of miR-181c.
Example 19
Effect on Cofilin Phosphorylation
[0288] PDPK1 is known to be one of the proteins positioning upstream of the pathway that controls Cofilin phosphorylation (Lyle, A. N, et al., Physiology (Bethesda), 21:269-280 (2006); Higuchi, M., et al., Nat. Cell Biol., 10:1356-1364 (2008)). Cofilin is a family of actin-binding proteins, which disassembles actin filaments, that is activated with dephosphorylation. Since actin condensation was observed in the PDPK1 siRNA-treated endothelial cells, whether Cofilin is involve in BBB destruction by miR-181c or not was examined.
[0289] Brain blood vessel endothelial cells were treated with EVs derived from D3H2LN, BMD2a, or BMD2b cells. Moreover, miR-181c was introduced into brain blood vessel endothelial cells. Furthermore, brain blood vessel endothelial cells were treated with PDPK1 siRNA according to the method described in Example 17. The expression of phosphorylated Cofilin in each of these cells was analyzed by Western blot analysis.
[0290] Samples were obtained from each cell line using M-PER (Thermo Scientific, MA, USA) and loaded in Mini-PROTEAN.RTM. TGX Gel (4-12%) (Bio-Rad) and proteins were separated and then electrotransferred onto a PVDF membrane (Millipore). The resultant membranes were blocked in Blocking One (Nacalai Tesque, Kyoto, Japan) and then incubated for 1 hour at room temperature with a primary antibody, anti-PDPK1 antibody (#3062, 1/500, Cell Signaling), anti-Cofilin antibody (D3F9, #5157, 1/1000, Cell Signaling), anti-Phospho-Cofilin antibody (Ser3) (#3311, 1/500, Cell Signaling), or anti-GAPDH antibody (6C5, 1/1000, Millipore). An HRP-linked anti-mouse IgG secondary antibody (NA931, GE Healthcare) or HRP-linked anti-rabbit IgG secondary antibody (NA934, GE Healthcare) was used at a dilution of 1/2000. The membranes were then made luminescent with ImmunoStar.RTM.LD (Wako, Osaka, Japan).
[0291] Phosphorylation of Cofilin in BMD2a cell- or BMD2b cell-derived EVs-treated cells was decreased compared to that in D3H2LN cell-derived EVs-treated cells (FIG. 20a). Furthermore, phosphorylation of Cofilin in miR-181c or PDPK1 siRNA-treated brain endothelial cells was decreased compared to that in control siRNA treated cells (FIG. 20b). These results revealed that miR-181c in EVs reduces the expression of PDPK1 in brain blood vessel endothelial cells and, as a result, phosphorylation of Cofilin is inhibited, which changes behaviors of actin and causes actin condensation and thereby increases permeability of the blood-brain barrier.
Sequence CWU 1
1
17122RNAHomo sapiens 1aacauucaac cugucgguga gu 2227243DNAHomo
sapiensCDS(150)..(1820) 2gcggcgccgg gggcgggggg cggcgggcga
cggggcgggc gcaggatgag ggcggccatt 60gctggggctc cgcttcgggg aggaggacgc
tgaggaggcg ccgagccgcg cagcgctgcg 120ggggaggcgc ccgcgccgac gcggggccc
atg gcc agg acc acc agc cag ctg 173 Met Ala Arg Thr Thr Ser Gln Leu
1 5tat gac gcc gtg ccc atc cag tcc agc gtg gtg tta tgt tcc tgc cca
221Tyr Asp Ala Val Pro Ile Gln Ser Ser Val Val Leu Cys Ser Cys Pro
10 15 20tcc cca tca atg gtg agg acc cag act gag tcc agc acg ccc cct
ggc 269Ser Pro Ser Met Val Arg Thr Gln Thr Glu Ser Ser Thr Pro Pro
Gly25 30 35 40att cct ggt ggc agc agg cag ggc ccc gcc atg gac ggc
act gca gcc 317Ile Pro Gly Gly Ser Arg Gln Gly Pro Ala Met Asp Gly
Thr Ala Ala 45 50 55gag cct cgg ccc ggc gcc ggc tcc ctg cag cat gcc
cag cct ccg ccg 365Glu Pro Arg Pro Gly Ala Gly Ser Leu Gln His Ala
Gln Pro Pro Pro 60 65 70cag cct cgg aag aag cgg cct gag gac ttc aag
ttt ggg aaa atc ctt 413Gln Pro Arg Lys Lys Arg Pro Glu Asp Phe Lys
Phe Gly Lys Ile Leu 75 80 85ggg gaa ggc tct ttt tcc acg gtt gtc ctg
gct cga gaa ctg gca acc 461Gly Glu Gly Ser Phe Ser Thr Val Val Leu
Ala Arg Glu Leu Ala Thr 90 95 100tcc aga gaa tat gcg att aaa att
ctg gag aag cga cat atc ata aaa 509Ser Arg Glu Tyr Ala Ile Lys Ile
Leu Glu Lys Arg His Ile Ile Lys105 110 115 120gag aac aag gtc ccc
tat gta acc aga gag cgg gat gtc atg tcg cgc 557Glu Asn Lys Val Pro
Tyr Val Thr Arg Glu Arg Asp Val Met Ser Arg 125 130 135ctg gat cac
ccc ttc ttt gtt aag ctt tac ttc aca ttt cag gac gac 605Leu Asp His
Pro Phe Phe Val Lys Leu Tyr Phe Thr Phe Gln Asp Asp 140 145 150gag
aag ctg tat ttc ggc ctt agt tat gcc aaa aat gga gaa cta ctt 653Glu
Lys Leu Tyr Phe Gly Leu Ser Tyr Ala Lys Asn Gly Glu Leu Leu 155 160
165aaa tat att cgc aaa atc ggt tca ttc gat gag acc tgt acc cga ttt
701Lys Tyr Ile Arg Lys Ile Gly Ser Phe Asp Glu Thr Cys Thr Arg Phe
170 175 180tac acg gct gag att gtg tct gct tta gag tac ttg cac ggc
aag ggc 749Tyr Thr Ala Glu Ile Val Ser Ala Leu Glu Tyr Leu His Gly
Lys Gly185 190 195 200atc att cac agg gac ctt aaa ccg gaa aac att
ttg tta aat gaa gat 797Ile Ile His Arg Asp Leu Lys Pro Glu Asn Ile
Leu Leu Asn Glu Asp 205 210 215atg cac atc cag atc aca gat ttt gga
aca gca aaa gtc tta tcc cca 845Met His Ile Gln Ile Thr Asp Phe Gly
Thr Ala Lys Val Leu Ser Pro 220 225 230gag agc aaa caa gcc agg gcc
aac tca ttc gtg gga aca gcg cag tac 893Glu Ser Lys Gln Ala Arg Ala
Asn Ser Phe Val Gly Thr Ala Gln Tyr 235 240 245gtt tct cca gag ctg
ctc acg gag aag tcc gcc tgt aag agt tca gac 941Val Ser Pro Glu Leu
Leu Thr Glu Lys Ser Ala Cys Lys Ser Ser Asp 250 255 260ctt tgg gct
ctt gga tgc ata ata tac cag ctt gtg gca gga ctc cca 989Leu Trp Ala
Leu Gly Cys Ile Ile Tyr Gln Leu Val Ala Gly Leu Pro265 270 275
280cca ttc cga gct gga aac gag tat ctt ata ttt cag aag atc att aag
1037Pro Phe Arg Ala Gly Asn Glu Tyr Leu Ile Phe Gln Lys Ile Ile Lys
285 290 295ttg gaa tat gac ttt cca gaa aaa ttc ttc cct aag gca aga
gac ctc 1085Leu Glu Tyr Asp Phe Pro Glu Lys Phe Phe Pro Lys Ala Arg
Asp Leu 300 305 310gtg gag aaa ctt ttg gtt tta gat gcc aca aag cgg
tta ggc tgt gag 1133Val Glu Lys Leu Leu Val Leu Asp Ala Thr Lys Arg
Leu Gly Cys Glu 315 320 325gaa atg gaa gga tac gga cct ctt aaa gca
cac ccg ttc ttc gag tcc 1181Glu Met Glu Gly Tyr Gly Pro Leu Lys Ala
His Pro Phe Phe Glu Ser 330 335 340gtc acg tgg gag aac ctg cac cag
cag acg cct ccg aag ctc acc gct 1229Val Thr Trp Glu Asn Leu His Gln
Gln Thr Pro Pro Lys Leu Thr Ala345 350 355 360tac ctg ccg gct atg
tcg gaa gac gac gag gac tgc tat ggc aat tat 1277Tyr Leu Pro Ala Met
Ser Glu Asp Asp Glu Asp Cys Tyr Gly Asn Tyr 365 370 375gac aat ctc
ctg agc cag ttt ggc tgc atg cag gtg tct tcg tcc tcc 1325Asp Asn Leu
Leu Ser Gln Phe Gly Cys Met Gln Val Ser Ser Ser Ser 380 385 390tcc
tca cac tcc ctg tca gcc tcc gac acg ggc ctg ccc cag agg tca 1373Ser
Ser His Ser Leu Ser Ala Ser Asp Thr Gly Leu Pro Gln Arg Ser 395 400
405ggc agc aac ata gag cag tac att cac gat ctg gac tcg aac tcc ttt
1421Gly Ser Asn Ile Glu Gln Tyr Ile His Asp Leu Asp Ser Asn Ser Phe
410 415 420gaa ctg gac tta cag ttt tcc gaa gat gag aag agg ttg ttg
ttg gag 1469Glu Leu Asp Leu Gln Phe Ser Glu Asp Glu Lys Arg Leu Leu
Leu Glu425 430 435 440aag cag gct ggc gga aac cct tgg cac cag ttt
gta gaa aat aat tta 1517Lys Gln Ala Gly Gly Asn Pro Trp His Gln Phe
Val Glu Asn Asn Leu 445 450 455ata cta aag atg ggc cca gtg gat aag
cgg aag ggt tta ttt gca aga 1565Ile Leu Lys Met Gly Pro Val Asp Lys
Arg Lys Gly Leu Phe Ala Arg 460 465 470cga cga cag ctg ttg ctc aca
gaa gga cca cat tta tat tat gtg gat 1613Arg Arg Gln Leu Leu Leu Thr
Glu Gly Pro His Leu Tyr Tyr Val Asp 475 480 485cct gtc aac aaa gtt
ctg aaa ggt gaa att cct tgg tca caa gaa ctt 1661Pro Val Asn Lys Val
Leu Lys Gly Glu Ile Pro Trp Ser Gln Glu Leu 490 495 500cga cca gag
gcc aag aat ttt aaa act ttc ttt gtc cac acg cct aac 1709Arg Pro Glu
Ala Lys Asn Phe Lys Thr Phe Phe Val His Thr Pro Asn505 510 515
520agg acg tat tat ctg atg gac ccc agc ggg aac gca cac aag tgg tgc
1757Arg Thr Tyr Tyr Leu Met Asp Pro Ser Gly Asn Ala His Lys Trp Cys
525 530 535agg aag atc cag gag gtt tgg agg cag cga tac cag agc cac
ccg gac 1805Arg Lys Ile Gln Glu Val Trp Arg Gln Arg Tyr Gln Ser His
Pro Asp 540 545 550gcc gct gtg cag tga cgtggcctgc ggccgggctg
cccttcgctg ccaggacacc 1860Ala Ala Val Gln 555tgccccagcg cggcttggcc
gccatccggg acgcttccag accacctgcc agccatcaca 1920aggggaacgc
agaggcggaa accttgcagc atttttattt aaaagaaaag aagaaaaaaa
1980acacccaacc acacaaagaa caaaaccagt aacaaacaca aaggaattca
gggtcgcttt 2040gcttgctctc tgtgctccgt ggaggcctcc gtgtgccctc
gttgccgtgg ggacccagct 2100ccatgcacgt caacccagtc ccgcccagac
tagtggacag acctggtgtc accagttttt 2160cctagcatca gtccgaacca
tgcgcccgcc ctgccccaac tgtgtgctgg tcctgctgtg 2220gccgagggga
ccgggtgtgt ttggctcttt atgcccctcc cgctgtggtc ctggaactct
2280tcaccaggga gggagccctg cgggggccgc agctttgtgg agggagccgc
cgtgcttctg 2340tcacctgctc cctttcttgc gtctccctgt gatgggccct
taggcctggc tgggcccatt 2400acatatccct gtggtggctc tggtggcagc
tttctgtggc ccctgctgtg ttggcaggca 2460ggtttgcgtg gtgaggagcg
ggaggggttg gagtggtgcg ggagcaggct gccgagtgga 2520gggtgccatc
gagggctccg gatcccttat cctacttagc agtgttggtc tctggggctg
2580gaagccgagc gcatgctggg agcggtactg tcagaagtga gcccagttag
taccccgctg 2640gctcactgca cgagagagtc ctgccccgag ccctaggtgg
ggccaggagg tgccttggag 2700aagccagcca gagcagagag ggctgctgac
ttccgtgtgg agcagagagg cctgagggcc 2760tcctaaaagg tttaaatgtc
cacgcctctc cagttgctga agtagggtct gagagaaccc 2820tggcatcagc
agacccaggg tgcttctgtc tcctgcagac cacgccaggg agtgcagaca
2880ccaccgtcac acacgcccct tttgtgtttt ggttcaagtt tctcagagcc
cctcagagct 2940tctacatctg tgcatcagaa atctcacagc cttctcatgc
tgccggctca tctgggccca 3000tagagtgggc tttgccagtt gctgttgcac
aggaggcgag aacagcacac ttcaacccca 3060gcttgctggt cggctttcct
ctagagagag ccggttttgg ggccatttcc ctttgatgct 3120ttggtggcct
tgccccgctc tgcagcacag acaggccaga tgcatttgtc ctttgcctag
3180ctactcccca ggtagagagt gctcctggtg gcctggcagg tctgggccct
tctctccctg 3240cccaggttgt ccctggaggg cagccctcac tccctttggg
ggagaggcag acattgctgc 3300ccacagacct gcctctgact caactgtgtc
caccctccct ggtccctacc cccaagtcac 3360aggtgactca gcagtgaccc
tgtgtgccag gccagatcca aactgagagg gaaggtgtcg 3420tttttacact
gctaatgacg agagtggctc tttttagcta ggcgagtaca gacggggcct
3480gggagggggc agagatgttc cccaggccct gcctgtggtt cctgcctggg
ccttggctgc 3540tgctgtgtga gagctgcatg tgagcctgtg accgtgagct
ggggtgagct gggccgcacc 3600taccctgggg ccccagggag caggacgctc
cggggcccag cacgttgccc tgggcctgtg 3660gccggagtcg gagtcctctc
tcctcctcct ggcttttgga aaggcttggc tgtgttgggg 3720agtctctctt
agccctttca ggaatttctg ttcaggcttc ctcctcctca tcagctattt
3780tacccatctc agaacgtcct gtgtctccat gtaggagagt ggctctctca
gatctctcag 3840ggcgtctggt tatagggaaa caagtggagc agggacgtgg
ctttaattgg agcactcggc 3900tgggctgctt ggggagactc ttccgtgcgt
tcttcctctg gatagaacca ccacctcctg 3960ggcgtcactg acaagctcca
tcttaacctc caaagccaca gaactagggg ctcagagcca 4020gagctggcag
ccgccagcca aaatgatgcc attgcctgag ctgacagcca agcccttctg
4080tgggtcacct ttctcctcac ccagcccctt gctcttccct tttgaaaggc
ccgtgtgttt 4140tctttcctta ccctgtgctt gctcatgtct actccggttt
tctctaccac atccttagag 4200ccatcacctg gcacgcaggc gccttacatt
ctacggtaga acgtggggta ctgtgtgtgc 4260acatagacac acttacgtgg
aattacagtt gtgggtttat ccaagatgag gaagatttca 4320cctgctgttt
aatagacttg gggccatgtg cctccccaca catgggcaag gacaggtgga
4380atgtcgggac cacactgtgc ggcttctcgg cacaaagcgg agggaggctg
tggtcgctgc 4440cggcctaggt gtcccaggtg ccccgccttt ctctgggaca
cagttggggg ctggcttctg 4500agggattcct ttctcccctc tttgtgtggc
cccagccagg gcggtgggca gtcctggtgt 4560agagcacaag cctctccacc
ctagagaaat gcctctgtac cacggctacc atgtggaacc 4620ttaacttgca
gaaggcttgt taacaattgt tttgagagag atggctggtc atgccacagc
4680tgctggggac tccgcctact ccagccctct tgggacacac tgtgggattt
gtggcccttc 4740cccagaggaa ttgtggagac tgtcccatgg aacaaaccct
caggcaccag cacagggctc 4800tgggtgactc agtaaaacta acgtttgtct
ctgacaagat cagctgtagg ctcaccggcc 4860agagaagacc actgtgagca
ttttgccgta tatcctgccc tgccatttgt tcacttttta 4920aactaaaata
ggaacatccg acacacaccg tttgcatcgt cttctccctt gatattttaa
4980gcattttccc atgtcatgag tttctcagaa acatgttttt aacaattgta
ctatttagtc 5040attgtccatt tactataatt tatctgacca tttccctact
gtaaaatact taagacggtt 5100tctgattttt ccactattta aataatgctg
tgatgaatat ctttaaaatc ttctgatttc 5160ttactttttt cccccttaga
tgcctggaag tggtattttg aggtgaaaga gtttgttcat 5220tttgaagata
tttctgtctc tctctcgacc tgatgtgtag acgctcactt ccagtagcag
5280aaccacctta gttgtgtctt acagattctg aacaaatcgg tttctgataa
gccatgtgtt 5340ccaaagaatg tctgaataag accgctcttt atttaaatgc
taagaggatg tcactactgc 5400aatccatctg tggccgattt tttccaagag
ccaatttcct tgttttggtt gcaagaacct 5460ggctctgcct gcatgtcagc
tctctgccct ccctgctgcc gtggctttca agcgcttggc 5520agaatcttgt
acttcgtgtc cacaatggta ctgaatttgc atctgcacag tcagcagaga
5580taacaagtgt tgaactgacc ttgccacatg cttagtgagt gatttgtaat
taagtttata 5640gactcagaag gtatattagg acatttggaa tcagtagcag
agcaaagcct ctttgaaaaa 5700aaccacgtag ctgattgggt tttacaagag
tgcatttgtc tcccccttcc acccgtgggg 5760ccccaccttc aggtcttagt
ggttcacaag agcccagcag ccaggctggc tttttcattg 5820tagggcgtgg
ttgtcccagc tggtgtagat ttcaggccgc cccccccaac tccctgccca
5880cagtgttgca gattgcctgg ctggcagcaa gtccagacca cccaaatttg
gttggattct 5940tcatttctcc actgtagttg gggtccattg attgtgcagg
ggaacgtgca ggaggttttt 6000ctaggcaccg tgttcagtgc tgcttcactc
taccagagat tatggccaaa ttgcacggaa 6060tttggtttct tgccctctga
agcctgaggg cccccccttg cctggctggt tgacagaccc 6120ggggtggtca
ctgctgagac ttcagagatc gcagctgctg tgagaatacg gtgaaggtac
6180tttgttctgg aagatgttgt catacacttt tccccagtta ttttcaaact
tgacatgagc 6240ctatgttgac tcactgggtg ggggtccctt cttacgcagc
acacgtggca agtgcctgaa 6300tcggggctgg aggcacttca gagcctctga
ggggccacca cttctggccc aaaattgcag 6360ggttgtagat gaggctgcct
gtggagaact ggtgtgagga ggaagctgtt tccaacaaag 6420agcactttca
tctgttgaga tggctgtggt gagcaactga acgagcctac gtgtgtacct
6480gaattttccc cgtaactcat ttcttccata tgaagaaaca ccaaactatg
tacagagaac 6540tttttacaaa aggcagacct tttttaagct gtgtaaccca
catagcctaa ccacctggca 6600gaatgactac gaataggggt cattgtgctg
gtaaaagcct ctattacgac tgtaagtaag 6660ttggatgttg gcaaaattaa
attgttacag tatttagagc tgctgtagct gttccttcac 6720aacataaaat
aggataaatg actagtacgt ctttcaggtg ggtggcaagc agaacatgcg
6780taatattctc tacctggtct gtagctgtaa ctgtgatgta cagacaaagc
aaaaattaaa 6840agaacttatg aaaacaaatg caatgatact aggatataca
cttttgtatt tttattctta 6900tataaggtta tttgctggct attgttggcc
tctagttcag tctgtgttat ttaaattcta 6960atatatgaat tatttgaatt
gaattcatgt tcggggccac gttgttgtat gtattgatgt 7020acagccttga
atgtgaataa ttattgtaaa ctatatttta caactttttt tctggcttta
7080ttatataaat tttctattgg gtcagtgatt taatcatata atttaatgaa
tctgtttatc 7140cttttttttt ttccaaatac ttgtgcttta ggtgtagtta
ccagatgatg aattttcctc 7200gtatggtcag tagtcttgta ataaaaagca
tgtagagtgt aga 72433556PRTHomo sapiens 3Met Ala Arg Thr Thr Ser Gln
Leu Tyr Asp Ala Val Pro Ile Gln Ser1 5 10 15Ser Val Val Leu Cys Ser
Cys Pro Ser Pro Ser Met Val Arg Thr Gln 20 25 30Thr Glu Ser Ser Thr
Pro Pro Gly Ile Pro Gly Gly Ser Arg Gln Gly 35 40 45Pro Ala Met Asp
Gly Thr Ala Ala Glu Pro Arg Pro Gly Ala Gly Ser 50 55 60Leu Gln His
Ala Gln Pro Pro Pro Gln Pro Arg Lys Lys Arg Pro Glu65 70 75 80Asp
Phe Lys Phe Gly Lys Ile Leu Gly Glu Gly Ser Phe Ser Thr Val 85 90
95Val Leu Ala Arg Glu Leu Ala Thr Ser Arg Glu Tyr Ala Ile Lys Ile
100 105 110Leu Glu Lys Arg His Ile Ile Lys Glu Asn Lys Val Pro Tyr
Val Thr 115 120 125Arg Glu Arg Asp Val Met Ser Arg Leu Asp His Pro
Phe Phe Val Lys 130 135 140Leu Tyr Phe Thr Phe Gln Asp Asp Glu Lys
Leu Tyr Phe Gly Leu Ser145 150 155 160Tyr Ala Lys Asn Gly Glu Leu
Leu Lys Tyr Ile Arg Lys Ile Gly Ser 165 170 175Phe Asp Glu Thr Cys
Thr Arg Phe Tyr Thr Ala Glu Ile Val Ser Ala 180 185 190Leu Glu Tyr
Leu His Gly Lys Gly Ile Ile His Arg Asp Leu Lys Pro 195 200 205Glu
Asn Ile Leu Leu Asn Glu Asp Met His Ile Gln Ile Thr Asp Phe 210 215
220Gly Thr Ala Lys Val Leu Ser Pro Glu Ser Lys Gln Ala Arg Ala
Asn225 230 235 240Ser Phe Val Gly Thr Ala Gln Tyr Val Ser Pro Glu
Leu Leu Thr Glu 245 250 255Lys Ser Ala Cys Lys Ser Ser Asp Leu Trp
Ala Leu Gly Cys Ile Ile 260 265 270Tyr Gln Leu Val Ala Gly Leu Pro
Pro Phe Arg Ala Gly Asn Glu Tyr 275 280 285Leu Ile Phe Gln Lys Ile
Ile Lys Leu Glu Tyr Asp Phe Pro Glu Lys 290 295 300Phe Phe Pro Lys
Ala Arg Asp Leu Val Glu Lys Leu Leu Val Leu Asp305 310 315 320Ala
Thr Lys Arg Leu Gly Cys Glu Glu Met Glu Gly Tyr Gly Pro Leu 325 330
335Lys Ala His Pro Phe Phe Glu Ser Val Thr Trp Glu Asn Leu His Gln
340 345 350Gln Thr Pro Pro Lys Leu Thr Ala Tyr Leu Pro Ala Met Ser
Glu Asp 355 360 365Asp Glu Asp Cys Tyr Gly Asn Tyr Asp Asn Leu Leu
Ser Gln Phe Gly 370 375 380Cys Met Gln Val Ser Ser Ser Ser Ser Ser
His Ser Leu Ser Ala Ser385 390 395 400Asp Thr Gly Leu Pro Gln Arg
Ser Gly Ser Asn Ile Glu Gln Tyr Ile 405 410 415His Asp Leu Asp Ser
Asn Ser Phe Glu Leu Asp Leu Gln Phe Ser Glu 420 425 430Asp Glu Lys
Arg Leu Leu Leu Glu Lys Gln Ala Gly Gly Asn Pro Trp 435 440 445His
Gln Phe Val Glu Asn Asn Leu Ile Leu Lys Met Gly Pro Val Asp 450 455
460Lys Arg Lys Gly Leu Phe Ala Arg Arg Arg Gln Leu Leu Leu Thr
Glu465 470 475 480Gly Pro His Leu Tyr Tyr Val Asp Pro Val Asn Lys
Val Leu Lys Gly 485 490 495Glu Ile Pro Trp Ser Gln Glu Leu Arg Pro
Glu Ala Lys Asn Phe Lys 500 505 510Thr Phe Phe Val His Thr Pro Asn
Arg Thr Tyr Tyr Leu Met Asp Pro 515 520 525Ser Gly Asn Ala His Lys
Trp Cys Arg Lys Ile Gln Glu Val Trp Arg 530 535 540Gln Arg Tyr Gln
Ser His Pro Asp Ala Ala Val Gln545 550 55546862DNAHomo
sapiensCDS(150)..(1439) 4gcggcgccgg gggcgggggg cggcgggcga
cggggcgggc gcaggatgag ggcggccatt 60gctggggctc cgcttcgggg aggaggacgc
tgaggaggcg ccgagccgcg cagcgctgcg 120ggggaggcgc ccgcgccgac gcggggccc
atg gcc agg acc acc agc cag ctg 173 Met Ala Arg Thr Thr Ser Gln Leu
1 5tat gac gcc gtg ccc atc cag tcc agc gtg gtg tta tgt tcc tgc cca
221Tyr Asp Ala Val Pro Ile Gln Ser Ser Val Val Leu Cys Ser Cys Pro
10 15 20tcc cca tca atg gtg agg acc cag
act gag tcc agc acg ccc cct ggc 269Ser Pro Ser Met Val Arg Thr Gln
Thr Glu Ser Ser Thr Pro Pro Gly25 30 35 40att cct ggt ggc agc agg
cag ggc ccc gcc atg gac ggc act gca gcc 317Ile Pro Gly Gly Ser Arg
Gln Gly Pro Ala Met Asp Gly Thr Ala Ala 45 50 55gag cct cgg ccc ggc
gcc ggc tcc ctg cag cat gcc cag cct ccg ccg 365Glu Pro Arg Pro Gly
Ala Gly Ser Leu Gln His Ala Gln Pro Pro Pro 60 65 70cag cct cgg aag
aag cgg cct gag gac ttc aag ttt ggg aaa atc ctt 413Gln Pro Arg Lys
Lys Arg Pro Glu Asp Phe Lys Phe Gly Lys Ile Leu 75 80 85ggg gaa ggc
tct ttt tcc acg gtt gtc ctg gct cga gaa ctg gca acc 461Gly Glu Gly
Ser Phe Ser Thr Val Val Leu Ala Arg Glu Leu Ala Thr 90 95 100tcc
aga gaa tat gcg acc agg gcc aac tca ttc gtg gga aca gcg cag 509Ser
Arg Glu Tyr Ala Thr Arg Ala Asn Ser Phe Val Gly Thr Ala Gln105 110
115 120tac gtt tct cca gag ctg ctc acg gag aag tcc gcc tgt aag agt
tca 557Tyr Val Ser Pro Glu Leu Leu Thr Glu Lys Ser Ala Cys Lys Ser
Ser 125 130 135gac ctt tgg gct ctt gga tgc ata ata tac cag ctt gtg
gca gga ctc 605Asp Leu Trp Ala Leu Gly Cys Ile Ile Tyr Gln Leu Val
Ala Gly Leu 140 145 150cca cca ttc cga gct gga aac gag tat ctt ata
ttt cag aag atc att 653Pro Pro Phe Arg Ala Gly Asn Glu Tyr Leu Ile
Phe Gln Lys Ile Ile 155 160 165aag ttg gaa tat gac ttt cca gaa aaa
ttc ttc cct aag gca aga gac 701Lys Leu Glu Tyr Asp Phe Pro Glu Lys
Phe Phe Pro Lys Ala Arg Asp 170 175 180ctc gtg gag aaa ctt ttg gtt
tta gat gcc aca aag cgg tta ggc tgt 749Leu Val Glu Lys Leu Leu Val
Leu Asp Ala Thr Lys Arg Leu Gly Cys185 190 195 200gag gaa atg gaa
gga tac gga cct ctt aaa gca cac ccg ttc ttc gag 797Glu Glu Met Glu
Gly Tyr Gly Pro Leu Lys Ala His Pro Phe Phe Glu 205 210 215tcc gtc
acg tgg gag aac ctg cac cag cag acg cct ccg aag ctc acc 845Ser Val
Thr Trp Glu Asn Leu His Gln Gln Thr Pro Pro Lys Leu Thr 220 225
230gct tac ctg ccg gct atg tcg gaa gac gac gag gac tgc tat ggc aat
893Ala Tyr Leu Pro Ala Met Ser Glu Asp Asp Glu Asp Cys Tyr Gly Asn
235 240 245tat gac aat ctc ctg agc cag ttt ggc tgc atg cag gtg tct
tcg tcc 941Tyr Asp Asn Leu Leu Ser Gln Phe Gly Cys Met Gln Val Ser
Ser Ser 250 255 260tcc tcc tca cac tcc ctg tca gcc tcc gac acg ggc
ctg ccc cag agg 989Ser Ser Ser His Ser Leu Ser Ala Ser Asp Thr Gly
Leu Pro Gln Arg265 270 275 280tca ggc agc aac ata gag cag tac att
cac gat ctg gac tcg aac tcc 1037Ser Gly Ser Asn Ile Glu Gln Tyr Ile
His Asp Leu Asp Ser Asn Ser 285 290 295ttt gaa ctg gac tta cag ttt
tcc gaa gat gag aag agg ttg ttg ttg 1085Phe Glu Leu Asp Leu Gln Phe
Ser Glu Asp Glu Lys Arg Leu Leu Leu 300 305 310gag aag cag gct ggc
gga aac cct tgg cac cag ttt gta gaa aat aat 1133Glu Lys Gln Ala Gly
Gly Asn Pro Trp His Gln Phe Val Glu Asn Asn 315 320 325tta ata cta
aag atg ggc cca gtg gat aag cgg aag ggt tta ttt gca 1181Leu Ile Leu
Lys Met Gly Pro Val Asp Lys Arg Lys Gly Leu Phe Ala 330 335 340aga
cga cga cag ctg ttg ctc aca gaa gga cca cat tta tat tat gtg 1229Arg
Arg Arg Gln Leu Leu Leu Thr Glu Gly Pro His Leu Tyr Tyr Val345 350
355 360gat cct gtc aac aaa gtt ctg aaa ggt gaa att cct tgg tca caa
gaa 1277Asp Pro Val Asn Lys Val Leu Lys Gly Glu Ile Pro Trp Ser Gln
Glu 365 370 375ctt cga cca gag gcc aag aat ttt aaa act ttc ttt gtc
cac acg cct 1325Leu Arg Pro Glu Ala Lys Asn Phe Lys Thr Phe Phe Val
His Thr Pro 380 385 390aac agg acg tat tat ctg atg gac ccc agc ggg
aac gca cac aag tgg 1373Asn Arg Thr Tyr Tyr Leu Met Asp Pro Ser Gly
Asn Ala His Lys Trp 395 400 405tgc agg aag atc cag gag gtt tgg agg
cag cga tac cag agc cac ccg 1421Cys Arg Lys Ile Gln Glu Val Trp Arg
Gln Arg Tyr Gln Ser His Pro 410 415 420gac gcc gct gtg cag tga
cgtggcctgc ggccgggctg cccttcgctg 1469Asp Ala Ala Val
Gln425ccaggacacc tgccccagcg cggcttggcc gccatccggg acgcttccag
accacctgcc 1529agccatcaca aggggaacgc agaggcggaa accttgcagc
atttttattt aaaagaaaag 1589aagaaaaaaa acacccaacc acacaaagaa
caaaaccagt aacaaacaca aaggaattca 1649gggtcgcttt gcttgctctc
tgtgctccgt ggaggcctcc gtgtgccctc gttgccgtgg 1709ggacccagct
ccatgcacgt caacccagtc ccgcccagac tagtggacag acctggtgtc
1769accagttttt cctagcatca gtccgaacca tgcgcccgcc ctgccccaac
tgtgtgctgg 1829tcctgctgtg gccgagggga ccgggtgtgt ttggctcttt
atgcccctcc cgctgtggtc 1889ctggaactct tcaccaggga gggagccctg
cgggggccgc agctttgtgg agggagccgc 1949cgtgcttctg tcacctgctc
cctttcttgc gtctccctgt gatgggccct taggcctggc 2009tgggcccatt
acatatccct gtggtggctc tggtggcagc tttctgtggc ccctgctgtg
2069ttggcaggca ggtttgcgtg gtgaggagcg ggaggggttg gagtggtgcg
ggagcaggct 2129gccgagtgga gggtgccatc gagggctccg gatcccttat
cctacttagc agtgttggtc 2189tctggggctg gaagccgagc gcatgctggg
agcggtactg tcagaagtga gcccagttag 2249taccccgctg gctcactgca
cgagagagtc ctgccccgag ccctaggtgg ggccaggagg 2309tgccttggag
aagccagcca gagcagagag ggctgctgac ttccgtgtgg agcagagagg
2369cctgagggcc tcctaaaagg tttaaatgtc cacgcctctc cagttgctga
agtagggtct 2429gagagaaccc tggcatcagc agacccaggg tgcttctgtc
tcctgcagac cacgccaggg 2489agtgcagaca ccaccgtcac acacgcccct
tttgtgtttt ggttcaagtt tctcagagcc 2549cctcagagct tctacatctg
tgcatcagaa atctcacagc cttctcatgc tgccggctca 2609tctgggccca
tagagtgggc tttgccagtt gctgttgcac aggaggcgag aacagcacac
2669ttcaacccca gcttgctggt cggctttcct ctagagagag ccggttttgg
ggccatttcc 2729ctttgatgct ttggtggcct tgccccgctc tgcagcacag
acaggccaga tgcatttgtc 2789ctttgcctag ctactcccca ggtagagagt
gctcctggtg gcctggcagg tctgggccct 2849tctctccctg cccaggttgt
ccctggaggg cagccctcac tccctttggg ggagaggcag 2909acattgctgc
ccacagacct gcctctgact caactgtgtc caccctccct ggtccctacc
2969cccaagtcac aggtgactca gcagtgaccc tgtgtgccag gccagatcca
aactgagagg 3029gaaggtgtcg tttttacact gctaatgacg agagtggctc
tttttagcta ggcgagtaca 3089gacggggcct gggagggggc agagatgttc
cccaggccct gcctgtggtt cctgcctggg 3149ccttggctgc tgctgtgtga
gagctgcatg tgagcctgtg accgtgagct ggggtgagct 3209gggccgcacc
taccctgggg ccccagggag caggacgctc cggggcccag cacgttgccc
3269tgggcctgtg gccggagtcg gagtcctctc tcctcctcct ggcttttgga
aaggcttggc 3329tgtgttgggg agtctctctt agccctttca ggaatttctg
ttcaggcttc ctcctcctca 3389tcagctattt tacccatctc agaacgtcct
gtgtctccat gtaggagagt ggctctctca 3449gatctctcag ggcgtctggt
tatagggaaa caagtggagc agggacgtgg ctttaattgg 3509agcactcggc
tgggctgctt ggggagactc ttccgtgcgt tcttcctctg gatagaacca
3569ccacctcctg ggcgtcactg acaagctcca tcttaacctc caaagccaca
gaactagggg 3629ctcagagcca gagctggcag ccgccagcca aaatgatgcc
attgcctgag ctgacagcca 3689agcccttctg tgggtcacct ttctcctcac
ccagcccctt gctcttccct tttgaaaggc 3749ccgtgtgttt tctttcctta
ccctgtgctt gctcatgtct actccggttt tctctaccac 3809atccttagag
ccatcacctg gcacgcaggc gccttacatt ctacggtaga acgtggggta
3869ctgtgtgtgc acatagacac acttacgtgg aattacagtt gtgggtttat
ccaagatgag 3929gaagatttca cctgctgttt aatagacttg gggccatgtg
cctccccaca catgggcaag 3989gacaggtgga atgtcgggac cacactgtgc
ggcttctcgg cacaaagcgg agggaggctg 4049tggtcgctgc cggcctaggt
gtcccaggtg ccccgccttt ctctgggaca cagttggggg 4109ctggcttctg
agggattcct ttctcccctc tttgtgtggc cccagccagg gcggtgggca
4169gtcctggtgt agagcacaag cctctccacc ctagagaaat gcctctgtac
cacggctacc 4229atgtggaacc ttaacttgca gaaggcttgt taacaattgt
tttgagagag atggctggtc 4289atgccacagc tgctggggac tccgcctact
ccagccctct tgggacacac tgtgggattt 4349gtggcccttc cccagaggaa
ttgtggagac tgtcccatgg aacaaaccct caggcaccag 4409cacagggctc
tgggtgactc agtaaaacta acgtttgtct ctgacaagat cagctgtagg
4469ctcaccggcc agagaagacc actgtgagca ttttgccgta tatcctgccc
tgccatttgt 4529tcacttttta aactaaaata ggaacatccg acacacaccg
tttgcatcgt cttctccctt 4589gatattttaa gcattttccc atgtcatgag
tttctcagaa acatgttttt aacaattgta 4649ctatttagtc attgtccatt
tactataatt tatctgacca tttccctact gtaaaatact 4709taagacggtt
tctgattttt ccactattta aataatgctg tgatgaatat ctttaaaatc
4769ttctgatttc ttactttttt cccccttaga tgcctggaag tggtattttg
aggtgaaaga 4829gtttgttcat tttgaagata tttctgtctc tctctcgacc
tgatgtgtag acgctcactt 4889ccagtagcag aaccacctta gttgtgtctt
acagattctg aacaaatcgg tttctgataa 4949gccatgtgtt ccaaagaatg
tctgaataag accgctcttt atttaaatgc taagaggatg 5009tcactactgc
aatccatctg tggccgattt tttccaagag ccaatttcct tgttttggtt
5069gcaagaacct ggctctgcct gcatgtcagc tctctgccct ccctgctgcc
gtggctttca 5129agcgcttggc agaatcttgt acttcgtgtc cacaatggta
ctgaatttgc atctgcacag 5189tcagcagaga taacaagtgt tgaactgacc
ttgccacatg cttagtgagt gatttgtaat 5249taagtttata gactcagaag
gtatattagg acatttggaa tcagtagcag agcaaagcct 5309ctttgaaaaa
aaccacgtag ctgattgggt tttacaagag tgcatttgtc tcccccttcc
5369acccgtgggg ccccaccttc aggtcttagt ggttcacaag agcccagcag
ccaggctggc 5429tttttcattg tagggcgtgg ttgtcccagc tggtgtagat
ttcaggccgc cccccccaac 5489tccctgccca cagtgttgca gattgcctgg
ctggcagcaa gtccagacca cccaaatttg 5549gttggattct tcatttctcc
actgtagttg gggtccattg attgtgcagg ggaacgtgca 5609ggaggttttt
ctaggcaccg tgttcagtgc tgcttcactc taccagagat tatggccaaa
5669ttgcacggaa tttggtttct tgccctctga agcctgaggg cccccccttg
cctggctggt 5729tgacagaccc ggggtggtca ctgctgagac ttcagagatc
gcagctgctg tgagaatacg 5789gtgaaggtac tttgttctgg aagatgttgt
catacacttt tccccagtta ttttcaaact 5849tgacatgagc ctatgttgac
tcactgggtg ggggtccctt cttacgcagc acacgtggca 5909agtgcctgaa
tcggggctgg aggcacttca gagcctctga ggggccacca cttctggccc
5969aaaattgcag ggttgtagat gaggctgcct gtggagaact ggtgtgagga
ggaagctgtt 6029tccaacaaag agcactttca tctgttgaga tggctgtggt
gagcaactga acgagcctac 6089gtgtgtacct gaattttccc cgtaactcat
ttcttccata tgaagaaaca ccaaactatg 6149tacagagaac tttttacaaa
aggcagacct tttttaagct gtgtaaccca catagcctaa 6209ccacctggca
gaatgactac gaataggggt cattgtgctg gtaaaagcct ctattacgac
6269tgtaagtaag ttggatgttg gcaaaattaa attgttacag tatttagagc
tgctgtagct 6329gttccttcac aacataaaat aggataaatg actagtacgt
ctttcaggtg ggtggcaagc 6389agaacatgcg taatattctc tacctggtct
gtagctgtaa ctgtgatgta cagacaaagc 6449aaaaattaaa agaacttatg
aaaacaaatg caatgatact aggatataca cttttgtatt 6509tttattctta
tataaggtta tttgctggct attgttggcc tctagttcag tctgtgttat
6569ttaaattcta atatatgaat tatttgaatt gaattcatgt tcggggccac
gttgttgtat 6629gtattgatgt acagccttga atgtgaataa ttattgtaaa
ctatatttta caactttttt 6689tctggcttta ttatataaat tttctattgg
gtcagtgatt taatcatata atttaatgaa 6749tctgtttatc cttttttttt
ttccaaatac ttgtgcttta ggtgtagtta ccagatgatg 6809aattttcctc
gtatggtcag tagtcttgta ataaaaagca tgtagagtgt aga 68625429PRTHomo
sapiens 5Met Ala Arg Thr Thr Ser Gln Leu Tyr Asp Ala Val Pro Ile
Gln Ser1 5 10 15Ser Val Val Leu Cys Ser Cys Pro Ser Pro Ser Met Val
Arg Thr Gln 20 25 30Thr Glu Ser Ser Thr Pro Pro Gly Ile Pro Gly Gly
Ser Arg Gln Gly 35 40 45Pro Ala Met Asp Gly Thr Ala Ala Glu Pro Arg
Pro Gly Ala Gly Ser 50 55 60Leu Gln His Ala Gln Pro Pro Pro Gln Pro
Arg Lys Lys Arg Pro Glu65 70 75 80Asp Phe Lys Phe Gly Lys Ile Leu
Gly Glu Gly Ser Phe Ser Thr Val 85 90 95Val Leu Ala Arg Glu Leu Ala
Thr Ser Arg Glu Tyr Ala Thr Arg Ala 100 105 110Asn Ser Phe Val Gly
Thr Ala Gln Tyr Val Ser Pro Glu Leu Leu Thr 115 120 125Glu Lys Ser
Ala Cys Lys Ser Ser Asp Leu Trp Ala Leu Gly Cys Ile 130 135 140Ile
Tyr Gln Leu Val Ala Gly Leu Pro Pro Phe Arg Ala Gly Asn Glu145 150
155 160Tyr Leu Ile Phe Gln Lys Ile Ile Lys Leu Glu Tyr Asp Phe Pro
Glu 165 170 175Lys Phe Phe Pro Lys Ala Arg Asp Leu Val Glu Lys Leu
Leu Val Leu 180 185 190Asp Ala Thr Lys Arg Leu Gly Cys Glu Glu Met
Glu Gly Tyr Gly Pro 195 200 205Leu Lys Ala His Pro Phe Phe Glu Ser
Val Thr Trp Glu Asn Leu His 210 215 220Gln Gln Thr Pro Pro Lys Leu
Thr Ala Tyr Leu Pro Ala Met Ser Glu225 230 235 240Asp Asp Glu Asp
Cys Tyr Gly Asn Tyr Asp Asn Leu Leu Ser Gln Phe 245 250 255Gly Cys
Met Gln Val Ser Ser Ser Ser Ser Ser His Ser Leu Ser Ala 260 265
270Ser Asp Thr Gly Leu Pro Gln Arg Ser Gly Ser Asn Ile Glu Gln Tyr
275 280 285Ile His Asp Leu Asp Ser Asn Ser Phe Glu Leu Asp Leu Gln
Phe Ser 290 295 300Glu Asp Glu Lys Arg Leu Leu Leu Glu Lys Gln Ala
Gly Gly Asn Pro305 310 315 320Trp His Gln Phe Val Glu Asn Asn Leu
Ile Leu Lys Met Gly Pro Val 325 330 335Asp Lys Arg Lys Gly Leu Phe
Ala Arg Arg Arg Gln Leu Leu Leu Thr 340 345 350Glu Gly Pro His Leu
Tyr Tyr Val Asp Pro Val Asn Lys Val Leu Lys 355 360 365Gly Glu Ile
Pro Trp Ser Gln Glu Leu Arg Pro Glu Ala Lys Asn Phe 370 375 380Lys
Thr Phe Phe Val His Thr Pro Asn Arg Thr Tyr Tyr Leu Met Asp385 390
395 400Pro Ser Gly Asn Ala His Lys Trp Cys Arg Lys Ile Gln Glu Val
Trp 405 410 415Arg Gln Arg Tyr Gln Ser His Pro Asp Ala Ala Val Gln
420 42567032DNAHomo sapiensCDS(150)..(1514) 6gcggcgccgg gggcgggggg
cggcgggcga cggggcgggc gcaggatgag ggcggccatt 60gctggggctc cgcttcgggg
aggaggacgc tgaggaggcg ccgagccgcg cagcgctgcg 120ggggaggcgc
ccgcgccgac gcggggccc atg gcc agg acc acc agc cag ctg 173 Met Ala
Arg Thr Thr Ser Gln Leu 1 5tat gac gcc gtg ccc atc cag tcc agc gtg
gtg tta tgt tcc tgc cca 221Tyr Asp Ala Val Pro Ile Gln Ser Ser Val
Val Leu Cys Ser Cys Pro 10 15 20tcc cca tca atg gtg agg acc cag act
gag tcc agc acg ccc cct ggc 269Ser Pro Ser Met Val Arg Thr Gln Thr
Glu Ser Ser Thr Pro Pro Gly25 30 35 40att cct ggt ggc agc agg cag
ggc ccc gcc atg gac ggc act gca gcc 317Ile Pro Gly Gly Ser Arg Gln
Gly Pro Ala Met Asp Gly Thr Ala Ala 45 50 55gag cct cgg ccc ggc gcc
ggc tcc ctg cag cat gcc cag cct ccg ccg 365Glu Pro Arg Pro Gly Ala
Gly Ser Leu Gln His Ala Gln Pro Pro Pro 60 65 70cag cct cgg aag aag
cgg cct gag gac ttc aag ttt ggg aaa atc ctt 413Gln Pro Arg Lys Lys
Arg Pro Glu Asp Phe Lys Phe Gly Lys Ile Leu 75 80 85ggg gaa ggc tct
ttt tcc acg gtt gtc ctg gct cga gaa ctg gca acc 461Gly Glu Gly Ser
Phe Ser Thr Val Val Leu Ala Arg Glu Leu Ala Thr 90 95 100tcc aga
gaa tat gcg att aaa att ctg gag aag cga cat atc ata aaa 509Ser Arg
Glu Tyr Ala Ile Lys Ile Leu Glu Lys Arg His Ile Ile Lys105 110 115
120gag aac aag gtc ccc tat gta acc aga gag cgg gat gtc atg tcg cgc
557Glu Asn Lys Val Pro Tyr Val Thr Arg Glu Arg Asp Val Met Ser Arg
125 130 135ctg gat cac ccc ttc ttt gtt aag ctt tac ttc aca ttt cag
gac gac 605Leu Asp His Pro Phe Phe Val Lys Leu Tyr Phe Thr Phe Gln
Asp Asp 140 145 150gag aag ctg tat ttc ggc ctt agt tat gcc aaa aat
gga gaa cta ctt 653Glu Lys Leu Tyr Phe Gly Leu Ser Tyr Ala Lys Asn
Gly Glu Leu Leu 155 160 165aaa tat att cgc aaa atc ggt tca ttc gat
gag acc tgt acc cga ttt 701Lys Tyr Ile Arg Lys Ile Gly Ser Phe Asp
Glu Thr Cys Thr Arg Phe 170 175 180tac acg gct gag att gtg tct gct
tta gag tac ttg cac ggc aag ggc 749Tyr Thr Ala Glu Ile Val Ser Ala
Leu Glu Tyr Leu His Gly Lys Gly185 190 195 200atc att cac agg gac
ctt aaa ccg gaa aac att ttg tta aat gaa gat 797Ile Ile His Arg Asp
Leu Lys Pro Glu Asn Ile Leu Leu Asn Glu Asp 205 210 215atg cac atc
cag atc aca gat ttt gga aca gca aaa gtc tta tcc cca 845Met His Ile
Gln Ile Thr Asp Phe Gly Thr Ala Lys Val Leu Ser Pro 220 225 230gag
agc aaa caa gcc agg gcc aac tca ttc gtg gga aca gcg cag tac 893Glu
Ser Lys Gln Ala Arg Ala Asn Ser Phe Val Gly Thr Ala Gln Tyr 235 240
245gtt tct cca gag ctg ctc acg gag aag tcc gcc tgt aag agt tca gac
941Val Ser Pro Glu Leu Leu Thr Glu Lys Ser Ala Cys Lys Ser Ser Asp
250 255 260ctt tgg gct ctt gga tgc ata ata tac cag ctt gtg gca gga
ctc cca 989Leu Trp Ala Leu Gly Cys Ile Ile Tyr Gln Leu Val Ala Gly
Leu Pro265 270 275 280cca ttc cga gct gga aac gag tat ctt ata ttt
cag aag atc att aag 1037Pro Phe Arg Ala Gly Asn Glu Tyr Leu Ile
Phe Gln Lys Ile Ile Lys 285 290 295ttg gaa tat gac ttt cca gaa aaa
ttc ttc cct aag gca aga gac ctc 1085Leu Glu Tyr Asp Phe Pro Glu Lys
Phe Phe Pro Lys Ala Arg Asp Leu 300 305 310gtg gag aaa ctt ttg gtt
tta gat gcc aca aag cgg tta ggc tgt gag 1133Val Glu Lys Leu Leu Val
Leu Asp Ala Thr Lys Arg Leu Gly Cys Glu 315 320 325gaa atg gaa gga
tac gga cct ctt aaa gca cac ccg ttc ttc gag tcc 1181Glu Met Glu Gly
Tyr Gly Pro Leu Lys Ala His Pro Phe Phe Glu Ser 330 335 340gtc acg
tgg gag aac ctg cac cag cag acg cct ccg aag ctc acc gct 1229Val Thr
Trp Glu Asn Leu His Gln Gln Thr Pro Pro Lys Leu Thr Ala345 350 355
360tac ctg ccg gct atg tcg gaa gac gac gag gac tgc tat ggc aat tat
1277Tyr Leu Pro Ala Met Ser Glu Asp Asp Glu Asp Cys Tyr Gly Asn Tyr
365 370 375gac aat ctc ctg agc cag ttt ggc tgc atg cag gtg tct tcg
tcc tcc 1325Asp Asn Leu Leu Ser Gln Phe Gly Cys Met Gln Val Ser Ser
Ser Ser 380 385 390tcc tca cac tcc ctg tca gcc tcc gac acg ggc ctg
ccc cag agg tca 1373Ser Ser His Ser Leu Ser Ala Ser Asp Thr Gly Leu
Pro Gln Arg Ser 395 400 405ggc agc aac ata gag cag tac att cac gat
ctg gac tcg aac tcc ttt 1421Gly Ser Asn Ile Glu Gln Tyr Ile His Asp
Leu Asp Ser Asn Ser Phe 410 415 420gaa ctg gac tta cag ttt tcc gaa
gat gag aag agg ttg ttg ttg gag 1469Glu Leu Asp Leu Gln Phe Ser Glu
Asp Glu Lys Arg Leu Leu Leu Glu425 430 435 440aag cag gct ggc gga
aac cct tgc cta aca gga cgt att atc tga 1514Lys Gln Ala Gly Gly Asn
Pro Cys Leu Thr Gly Arg Ile Ile 445 450tggaccccag cgggaacgca
cacaagtggt gcaggaagat ccaggaggtt tggaggcagc 1574gataccagag
ccacccggac gccgctgtgc agtgacgtgg cctgcggccg ggctgccctt
1634cgctgccagg acacctgccc cagcgcggct tggccgccat ccgggacgct
tccagaccac 1694ctgccagcca tcacaagggg aacgcagagg cggaaacctt
gcagcatttt tatttaaaag 1754aaaagaagaa aaaaaacacc caaccacaca
aagaacaaaa ccagtaacaa acacaaagga 1814attcagggtc gctttgcttg
ctctctgtgc tccgtggagg cctccgtgtg ccctcgttgc 1874cgtggggacc
cagctccatg cacgtcaacc cagtcccgcc cagactagtg gacagacctg
1934gtgtcaccag tttttcctag catcagtccg aaccatgcgc ccgccctgcc
ccaactgtgt 1994gctggtcctg ctgtggccga ggggaccggg tgtgtttggc
tctttatgcc cctcccgctg 2054tggtcctgga actcttcacc agggagggag
ccctgcgggg gccgcagctt tgtggaggga 2114gccgccgtgc ttctgtcacc
tgctcccttt cttgcgtctc cctgtgatgg gcccttaggc 2174ctggctgggc
ccattacata tccctgtggt ggctctggtg gcagctttct gtggcccctg
2234ctgtgttggc aggcaggttt gcgtggtgag gagcgggagg ggttggagtg
gtgcgggagc 2294aggctgccga gtggagggtg ccatcgaggg ctccggatcc
cttatcctac ttagcagtgt 2354tggtctctgg ggctggaagc cgagcgcatg
ctgggagcgg tactgtcaga agtgagccca 2414gttagtaccc cgctggctca
ctgcacgaga gagtcctgcc ccgagcccta ggtggggcca 2474ggaggtgcct
tggagaagcc agccagagca gagagggctg ctgacttccg tgtggagcag
2534agaggcctga gggcctccta aaaggtttaa atgtccacgc ctctccagtt
gctgaagtag 2594ggtctgagag aaccctggca tcagcagacc cagggtgctt
ctgtctcctg cagaccacgc 2654cagggagtgc agacaccacc gtcacacacg
ccccttttgt gttttggttc aagtttctca 2714gagcccctca gagcttctac
atctgtgcat cagaaatctc acagccttct catgctgccg 2774gctcatctgg
gcccatagag tgggctttgc cagttgctgt tgcacaggag gcgagaacag
2834cacacttcaa ccccagcttg ctggtcggct ttcctctaga gagagccggt
tttggggcca 2894tttccctttg atgctttggt ggccttgccc cgctctgcag
cacagacagg ccagatgcat 2954ttgtcctttg cctagctact ccccaggtag
agagtgctcc tggtggcctg gcaggtctgg 3014gcccttctct ccctgcccag
gttgtccctg gagggcagcc ctcactccct ttgggggaga 3074ggcagacatt
gctgcccaca gacctgcctc tgactcaact gtgtccaccc tccctggtcc
3134ctacccccaa gtcacaggtg actcagcagt gaccctgtgt gccaggccag
atccaaactg 3194agagggaagg tgtcgttttt acactgctaa tgacgagagt
ggctcttttt agctaggcga 3254gtacagacgg ggcctgggag ggggcagaga
tgttccccag gccctgcctg tggttcctgc 3314ctgggccttg gctgctgctg
tgtgagagct gcatgtgagc ctgtgaccgt gagctggggt 3374gagctgggcc
gcacctaccc tggggcccca gggagcagga cgctccgggg cccagcacgt
3434tgccctgggc ctgtggccgg agtcggagtc ctctctcctc ctcctggctt
ttggaaaggc 3494ttggctgtgt tggggagtct ctcttagccc tttcaggaat
ttctgttcag gcttcctcct 3554cctcatcagc tattttaccc atctcagaac
gtcctgtgtc tccatgtagg agagtggctc 3614tctcagatct ctcagggcgt
ctggttatag ggaaacaagt ggagcaggga cgtggcttta 3674attggagcac
tcggctgggc tgcttgggga gactcttccg tgcgttcttc ctctggatag
3734aaccaccacc tcctgggcgt cactgacaag ctccatctta acctccaaag
ccacagaact 3794aggggctcag agccagagct ggcagccgcc agccaaaatg
atgccattgc ctgagctgac 3854agccaagccc ttctgtgggt cacctttctc
ctcacccagc cccttgctct tcccttttga 3914aaggcccgtg tgttttcttt
ccttaccctg tgcttgctca tgtctactcc ggttttctct 3974accacatcct
tagagccatc acctggcacg caggcgcctt acattctacg gtagaacgtg
4034gggtactgtg tgtgcacata gacacactta cgtggaatta cagttgtggg
tttatccaag 4094atgaggaaga tttcacctgc tgtttaatag acttggggcc
atgtgcctcc ccacacatgg 4154gcaaggacag gtggaatgtc gggaccacac
tgtgcggctt ctcggcacaa agcggaggga 4214ggctgtggtc gctgccggcc
taggtgtccc aggtgccccg cctttctctg ggacacagtt 4274gggggctggc
ttctgaggga ttcctttctc ccctctttgt gtggccccag ccagggcggt
4334gggcagtcct ggtgtagagc acaagcctct ccaccctaga gaaatgcctc
tgtaccacgg 4394ctaccatgtg gaaccttaac ttgcagaagg cttgttaaca
attgttttga gagagatggc 4454tggtcatgcc acagctgctg gggactccgc
ctactccagc cctcttggga cacactgtgg 4514gatttgtggc ccttccccag
aggaattgtg gagactgtcc catggaacaa accctcaggc 4574accagcacag
ggctctgggt gactcagtaa aactaacgtt tgtctctgac aagatcagct
4634gtaggctcac cggccagaga agaccactgt gagcattttg ccgtatatcc
tgccctgcca 4694tttgttcact ttttaaacta aaataggaac atccgacaca
caccgtttgc atcgtcttct 4754cccttgatat tttaagcatt ttcccatgtc
atgagtttct cagaaacatg tttttaacaa 4814ttgtactatt tagtcattgt
ccatttacta taatttatct gaccatttcc ctactgtaaa 4874atacttaaga
cggtttctga tttttccact atttaaataa tgctgtgatg aatatcttta
4934aaatcttctg atttcttact tttttccccc ttagatgcct ggaagtggta
ttttgaggtg 4994aaagagtttg ttcattttga agatatttct gtctctctct
cgacctgatg tgtagacgct 5054cacttccagt agcagaacca ccttagttgt
gtcttacaga ttctgaacaa atcggtttct 5114gataagccat gtgttccaaa
gaatgtctga ataagaccgc tctttattta aatgctaaga 5174ggatgtcact
actgcaatcc atctgtggcc gattttttcc aagagccaat ttccttgttt
5234tggttgcaag aacctggctc tgcctgcatg tcagctctct gccctccctg
ctgccgtggc 5294tttcaagcgc ttggcagaat cttgtacttc gtgtccacaa
tggtactgaa tttgcatctg 5354cacagtcagc agagataaca agtgttgaac
tgaccttgcc acatgcttag tgagtgattt 5414gtaattaagt ttatagactc
agaaggtata ttaggacatt tggaatcagt agcagagcaa 5474agcctctttg
aaaaaaacca cgtagctgat tgggttttac aagagtgcat ttgtctcccc
5534cttccacccg tggggcccca ccttcaggtc ttagtggttc acaagagccc
agcagccagg 5594ctggcttttt cattgtaggg cgtggttgtc ccagctggtg
tagatttcag gccgcccccc 5654ccaactccct gcccacagtg ttgcagattg
cctggctggc agcaagtcca gaccacccaa 5714atttggttgg attcttcatt
tctccactgt agttggggtc cattgattgt gcaggggaac 5774gtgcaggagg
tttttctagg caccgtgttc agtgctgctt cactctacca gagattatgg
5834ccaaattgca cggaatttgg tttcttgccc tctgaagcct gagggccccc
ccttgcctgg 5894ctggttgaca gacccggggt ggtcactgct gagacttcag
agatcgcagc tgctgtgaga 5954atacggtgaa ggtactttgt tctggaagat
gttgtcatac acttttcccc agttattttc 6014aaacttgaca tgagcctatg
ttgactcact gggtgggggt cccttcttac gcagcacacg 6074tggcaagtgc
ctgaatcggg gctggaggca cttcagagcc tctgaggggc caccacttct
6134ggcccaaaat tgcagggttg tagatgaggc tgcctgtgga gaactggtgt
gaggaggaag 6194ctgtttccaa caaagagcac tttcatctgt tgagatggct
gtggtgagca actgaacgag 6254cctacgtgtg tacctgaatt ttccccgtaa
ctcatttctt ccatatgaag aaacaccaaa 6314ctatgtacag agaacttttt
acaaaaggca gacctttttt aagctgtgta acccacatag 6374cctaaccacc
tggcagaatg actacgaata ggggtcattg tgctggtaaa agcctctatt
6434acgactgtaa gtaagttgga tgttggcaaa attaaattgt tacagtattt
agagctgctg 6494tagctgttcc ttcacaacat aaaataggat aaatgactag
tacgtctttc aggtgggtgg 6554caagcagaac atgcgtaata ttctctacct
ggtctgtagc tgtaactgtg atgtacagac 6614aaagcaaaaa ttaaaagaac
ttatgaaaac aaatgcaatg atactaggat atacactttt 6674gtatttttat
tcttatataa ggttatttgc tggctattgt tggcctctag ttcagtctgt
6734gttatttaaa ttctaatata tgaattattt gaattgaatt catgttcggg
gccacgttgt 6794tgtatgtatt gatgtacagc cttgaatgtg aataattatt
gtaaactata ttttacaact 6854ttttttctgg ctttattata taaattttct
attgggtcag tgatttaatc atataattta 6914atgaatctgt ttatcctttt
tttttttcca aatacttgtg ctttaggtgt agttaccaga 6974tgatgaattt
tcctcgtatg gtcagtagtc ttgtaataaa aagcatgtag agtgtaga
70327454PRTHomo sapiens 7Met Ala Arg Thr Thr Ser Gln Leu Tyr Asp
Ala Val Pro Ile Gln Ser1 5 10 15Ser Val Val Leu Cys Ser Cys Pro Ser
Pro Ser Met Val Arg Thr Gln 20 25 30Thr Glu Ser Ser Thr Pro Pro Gly
Ile Pro Gly Gly Ser Arg Gln Gly 35 40 45Pro Ala Met Asp Gly Thr Ala
Ala Glu Pro Arg Pro Gly Ala Gly Ser 50 55 60Leu Gln His Ala Gln Pro
Pro Pro Gln Pro Arg Lys Lys Arg Pro Glu65 70 75 80Asp Phe Lys Phe
Gly Lys Ile Leu Gly Glu Gly Ser Phe Ser Thr Val 85 90 95Val Leu Ala
Arg Glu Leu Ala Thr Ser Arg Glu Tyr Ala Ile Lys Ile 100 105 110Leu
Glu Lys Arg His Ile Ile Lys Glu Asn Lys Val Pro Tyr Val Thr 115 120
125Arg Glu Arg Asp Val Met Ser Arg Leu Asp His Pro Phe Phe Val Lys
130 135 140Leu Tyr Phe Thr Phe Gln Asp Asp Glu Lys Leu Tyr Phe Gly
Leu Ser145 150 155 160Tyr Ala Lys Asn Gly Glu Leu Leu Lys Tyr Ile
Arg Lys Ile Gly Ser 165 170 175Phe Asp Glu Thr Cys Thr Arg Phe Tyr
Thr Ala Glu Ile Val Ser Ala 180 185 190Leu Glu Tyr Leu His Gly Lys
Gly Ile Ile His Arg Asp Leu Lys Pro 195 200 205Glu Asn Ile Leu Leu
Asn Glu Asp Met His Ile Gln Ile Thr Asp Phe 210 215 220Gly Thr Ala
Lys Val Leu Ser Pro Glu Ser Lys Gln Ala Arg Ala Asn225 230 235
240Ser Phe Val Gly Thr Ala Gln Tyr Val Ser Pro Glu Leu Leu Thr Glu
245 250 255Lys Ser Ala Cys Lys Ser Ser Asp Leu Trp Ala Leu Gly Cys
Ile Ile 260 265 270Tyr Gln Leu Val Ala Gly Leu Pro Pro Phe Arg Ala
Gly Asn Glu Tyr 275 280 285Leu Ile Phe Gln Lys Ile Ile Lys Leu Glu
Tyr Asp Phe Pro Glu Lys 290 295 300Phe Phe Pro Lys Ala Arg Asp Leu
Val Glu Lys Leu Leu Val Leu Asp305 310 315 320Ala Thr Lys Arg Leu
Gly Cys Glu Glu Met Glu Gly Tyr Gly Pro Leu 325 330 335Lys Ala His
Pro Phe Phe Glu Ser Val Thr Trp Glu Asn Leu His Gln 340 345 350Gln
Thr Pro Pro Lys Leu Thr Ala Tyr Leu Pro Ala Met Ser Glu Asp 355 360
365Asp Glu Asp Cys Tyr Gly Asn Tyr Asp Asn Leu Leu Ser Gln Phe Gly
370 375 380Cys Met Gln Val Ser Ser Ser Ser Ser Ser His Ser Leu Ser
Ala Ser385 390 395 400Asp Thr Gly Leu Pro Gln Arg Ser Gly Ser Asn
Ile Glu Gln Tyr Ile 405 410 415His Asp Leu Asp Ser Asn Ser Phe Glu
Leu Asp Leu Gln Phe Ser Glu 420 425 430Asp Glu Lys Arg Leu Leu Leu
Glu Lys Gln Ala Gly Gly Asn Pro Cys 435 440 445Leu Thr Gly Arg Ile
Ile 45087337DNAHomo sapiensCDS(327)..(1916) 8attgctgggg ctccgcttcg
gggaggagga cgctgaggag gcgccgagcc gcgcagcgct 60gcgggggagg cgcccgcgcc
gacgcggggc ccatggccag gaccaccagc cagctgattc 120ttgatgacag
tggtcgggaa aactcgcctt tcacggcccg gccaaggggc atttgggtgc
180ttttgctgcc cgtggctgtg ctcagcgttg tgggaagccc ctgggaggcc
gaatgtgcag 240gatcaccgag gggaaagtga gcttgacagt atgacgccgt
gcccatccag tccagcgtgg 300tgttatgttc ctgcccatcc ccatca atg gtg agg
acc cag act gag tcc agc 353 Met Val Arg Thr Gln Thr Glu Ser Ser 1
5acg ccc cct ggc att cct ggt ggc agc agg cag ggc ccc gcc atg gac
401Thr Pro Pro Gly Ile Pro Gly Gly Ser Arg Gln Gly Pro Ala Met
Asp10 15 20 25ggc act gca gcc gag cct cgg ccc ggc gcc ggc tcc ctg
cag cat gcc 449Gly Thr Ala Ala Glu Pro Arg Pro Gly Ala Gly Ser Leu
Gln His Ala 30 35 40cag cct ccg ccg cag cct cgg aag aag cgg cct gag
gac ttc aag ttt 497Gln Pro Pro Pro Gln Pro Arg Lys Lys Arg Pro Glu
Asp Phe Lys Phe 45 50 55ggg aaa atc ctt ggg gaa ggc tct ttt tcc acg
gtt gtc ctg gct cga 545Gly Lys Ile Leu Gly Glu Gly Ser Phe Ser Thr
Val Val Leu Ala Arg 60 65 70gaa ctg gca acc tcc aga gaa tat gcg att
aaa att ctg gag aag cga 593Glu Leu Ala Thr Ser Arg Glu Tyr Ala Ile
Lys Ile Leu Glu Lys Arg 75 80 85cat atc ata aaa gag aac aag gtc ccc
tat gta acc aga gag cgg gat 641His Ile Ile Lys Glu Asn Lys Val Pro
Tyr Val Thr Arg Glu Arg Asp90 95 100 105gtc atg tcg cgc ctg gat cac
ccc ttc ttt gtt aag ctt tac ttc aca 689Val Met Ser Arg Leu Asp His
Pro Phe Phe Val Lys Leu Tyr Phe Thr 110 115 120ttt cag gac gac gag
aag ctg tat ttc ggc ctt agt tat gcc aaa aat 737Phe Gln Asp Asp Glu
Lys Leu Tyr Phe Gly Leu Ser Tyr Ala Lys Asn 125 130 135gga gaa cta
ctt aaa tat att cgc aaa atc ggt tca ttc gat gag acc 785Gly Glu Leu
Leu Lys Tyr Ile Arg Lys Ile Gly Ser Phe Asp Glu Thr 140 145 150tgt
acc cga ttt tac acg gct gag att gtg tct gct tta gag tac ttg 833Cys
Thr Arg Phe Tyr Thr Ala Glu Ile Val Ser Ala Leu Glu Tyr Leu 155 160
165cac ggc aag ggc atc att cac agg gac ctt aaa ccg gaa aac att ttg
881His Gly Lys Gly Ile Ile His Arg Asp Leu Lys Pro Glu Asn Ile
Leu170 175 180 185tta aat gaa gat atg cac atc cag atc aca gat ttt
gga aca gca aaa 929Leu Asn Glu Asp Met His Ile Gln Ile Thr Asp Phe
Gly Thr Ala Lys 190 195 200gtc tta tcc cca gag agc aaa caa gcc agg
gcc aac tca ttc gtg gga 977Val Leu Ser Pro Glu Ser Lys Gln Ala Arg
Ala Asn Ser Phe Val Gly 205 210 215aca gcg cag tac gtt tct cca gag
ctg ctc acg gag aag tcc gcc tgt 1025Thr Ala Gln Tyr Val Ser Pro Glu
Leu Leu Thr Glu Lys Ser Ala Cys 220 225 230aag agt tca gac ctt tgg
gct ctt gga tgc ata ata tac cag ctt gtg 1073Lys Ser Ser Asp Leu Trp
Ala Leu Gly Cys Ile Ile Tyr Gln Leu Val 235 240 245gca gga ctc cca
cca ttc cga gct gga aac gag tat ctt ata ttt cag 1121Ala Gly Leu Pro
Pro Phe Arg Ala Gly Asn Glu Tyr Leu Ile Phe Gln250 255 260 265aag
atc att aag ttg gaa tat gac ttt cca gaa aaa ttc ttc cct aag 1169Lys
Ile Ile Lys Leu Glu Tyr Asp Phe Pro Glu Lys Phe Phe Pro Lys 270 275
280gca aga gac ctc gtg gag aaa ctt ttg gtt tta gat gcc aca aag cgg
1217Ala Arg Asp Leu Val Glu Lys Leu Leu Val Leu Asp Ala Thr Lys Arg
285 290 295tta ggc tgt gag gaa atg gaa gga tac gga cct ctt aaa gca
cac ccg 1265Leu Gly Cys Glu Glu Met Glu Gly Tyr Gly Pro Leu Lys Ala
His Pro 300 305 310ttc ttc gag tcc gtc acg tgg gag aac ctg cac cag
cag acg cct ccg 1313Phe Phe Glu Ser Val Thr Trp Glu Asn Leu His Gln
Gln Thr Pro Pro 315 320 325aag ctc acc gct tac ctg ccg gct atg tcg
gaa gac gac gag gac tgc 1361Lys Leu Thr Ala Tyr Leu Pro Ala Met Ser
Glu Asp Asp Glu Asp Cys330 335 340 345tat ggc aat tat gac aat ctc
ctg agc cag ttt ggc tgc atg cag gtg 1409Tyr Gly Asn Tyr Asp Asn Leu
Leu Ser Gln Phe Gly Cys Met Gln Val 350 355 360tct tcg tcc tcc tcc
tca cac tcc ctg tca gcc tcc gac acg ggc ctg 1457Ser Ser Ser Ser Ser
Ser His Ser Leu Ser Ala Ser Asp Thr Gly Leu 365 370 375ccc cag agg
tca ggc agc aac ata gag cag tac att cac gat ctg gac 1505Pro Gln Arg
Ser Gly Ser Asn Ile Glu Gln Tyr Ile His Asp Leu Asp 380 385 390tcg
aac tcc ttt gaa ctg gac tta cag ttt tcc gaa gat gag aag agg 1553Ser
Asn Ser Phe Glu Leu Asp Leu Gln Phe Ser Glu Asp Glu Lys Arg 395 400
405ttg ttg ttg gag aag cag gct ggc gga aac cct tgg cac cag ttt gta
1601Leu Leu Leu Glu Lys Gln Ala Gly Gly Asn Pro Trp His Gln Phe
Val410 415 420 425gaa aat aat tta ata cta aag atg ggc cca gtg gat
aag cgg aag ggt 1649Glu Asn Asn Leu Ile Leu Lys Met Gly Pro Val Asp
Lys Arg Lys Gly 430 435 440tta ttt gca aga cga cga cag ctg ttg ctc
aca gaa gga cca cat tta 1697Leu Phe Ala Arg Arg Arg Gln Leu Leu Leu
Thr Glu Gly Pro His Leu 445 450 455tat tat gtg gat cct gtc aac aaa
gtt ctg aaa ggt gaa att cct tgg 1745Tyr Tyr Val Asp Pro Val Asn Lys
Val Leu Lys Gly Glu Ile Pro Trp 460 465 470tca caa gaa ctt cga cca
gag gcc aag aat ttt aaa act ttc ttt gtc 1793Ser Gln Glu Leu Arg Pro
Glu Ala Lys Asn Phe Lys Thr Phe Phe Val 475 480
485cac acg cct aac agg acg tat tat ctg atg gac ccc agc ggg aac gca
1841His Thr Pro Asn Arg Thr Tyr Tyr Leu Met Asp Pro Ser Gly Asn
Ala490 495 500 505cac aag tgg tgc agg aag atc cag gag gtt tgg agg
cag cga tac cag 1889His Lys Trp Cys Arg Lys Ile Gln Glu Val Trp Arg
Gln Arg Tyr Gln 510 515 520agc cac ccg gac gcc gct gtg cag tga
cgtggcctgc ggccgggctg 1936Ser His Pro Asp Ala Ala Val Gln
525cccttcgctg ccaggacacc tgccccagcg cggcttggcc gccatccggg
acgcttccag 1996accacctgcc agccatcaca aggggaacgc agaggcggaa
accttgcagc atttttattt 2056aaaagaaaag aagaaaaaaa acacccaacc
acacaaagaa caaaaccagt aacaaacaca 2116aaggaattca gggtcgcttt
gcttgctctc tgtgctccgt ggaggcctcc gtgtgccctc 2176gttgccgtgg
ggacccagct ccatgcacgt caacccagtc ccgcccagac tagtggacag
2236acctggtgtc accagttttt cctagcatca gtccgaacca tgcgcccgcc
ctgccccaac 2296tgtgtgctgg tcctgctgtg gccgagggga ccgggtgtgt
ttggctcttt atgcccctcc 2356cgctgtggtc ctggaactct tcaccaggga
gggagccctg cgggggccgc agctttgtgg 2416agggagccgc cgtgcttctg
tcacctgctc cctttcttgc gtctccctgt gatgggccct 2476taggcctggc
tgggcccatt acatatccct gtggtggctc tggtggcagc tttctgtggc
2536ccctgctgtg ttggcaggca ggtttgcgtg gtgaggagcg ggaggggttg
gagtggtgcg 2596ggagcaggct gccgagtgga gggtgccatc gagggctccg
gatcccttat cctacttagc 2656agtgttggtc tctggggctg gaagccgagc
gcatgctggg agcggtactg tcagaagtga 2716gcccagttag taccccgctg
gctcactgca cgagagagtc ctgccccgag ccctaggtgg 2776ggccaggagg
tgccttggag aagccagcca gagcagagag ggctgctgac ttccgtgtgg
2836agcagagagg cctgagggcc tcctaaaagg tttaaatgtc cacgcctctc
cagttgctga 2896agtagggtct gagagaaccc tggcatcagc agacccaggg
tgcttctgtc tcctgcagac 2956cacgccaggg agtgcagaca ccaccgtcac
acacgcccct tttgtgtttt ggttcaagtt 3016tctcagagcc cctcagagct
tctacatctg tgcatcagaa atctcacagc cttctcatgc 3076tgccggctca
tctgggccca tagagtgggc tttgccagtt gctgttgcac aggaggcgag
3136aacagcacac ttcaacccca gcttgctggt cggctttcct ctagagagag
ccggttttgg 3196ggccatttcc ctttgatgct ttggtggcct tgccccgctc
tgcagcacag acaggccaga 3256tgcatttgtc ctttgcctag ctactcccca
ggtagagagt gctcctggtg gcctggcagg 3316tctgggccct tctctccctg
cccaggttgt ccctggaggg cagccctcac tccctttggg 3376ggagaggcag
acattgctgc ccacagacct gcctctgact caactgtgtc caccctccct
3436ggtccctacc cccaagtcac aggtgactca gcagtgaccc tgtgtgccag
gccagatcca 3496aactgagagg gaaggtgtcg tttttacact gctaatgacg
agagtggctc tttttagcta 3556ggcgagtaca gacggggcct gggagggggc
agagatgttc cccaggccct gcctgtggtt 3616cctgcctggg ccttggctgc
tgctgtgtga gagctgcatg tgagcctgtg accgtgagct 3676ggggtgagct
gggccgcacc taccctgggg ccccagggag caggacgctc cggggcccag
3736cacgttgccc tgggcctgtg gccggagtcg gagtcctctc tcctcctcct
ggcttttgga 3796aaggcttggc tgtgttgggg agtctctctt agccctttca
ggaatttctg ttcaggcttc 3856ctcctcctca tcagctattt tacccatctc
agaacgtcct gtgtctccat gtaggagagt 3916ggctctctca gatctctcag
ggcgtctggt tatagggaaa caagtggagc agggacgtgg 3976ctttaattgg
agcactcggc tgggctgctt ggggagactc ttccgtgcgt tcttcctctg
4036gatagaacca ccacctcctg ggcgtcactg acaagctcca tcttaacctc
caaagccaca 4096gaactagggg ctcagagcca gagctggcag ccgccagcca
aaatgatgcc attgcctgag 4156ctgacagcca agcccttctg tgggtcacct
ttctcctcac ccagcccctt gctcttccct 4216tttgaaaggc ccgtgtgttt
tctttcctta ccctgtgctt gctcatgtct actccggttt 4276tctctaccac
atccttagag ccatcacctg gcacgcaggc gccttacatt ctacggtaga
4336acgtggggta ctgtgtgtgc acatagacac acttacgtgg aattacagtt
gtgggtttat 4396ccaagatgag gaagatttca cctgctgttt aatagacttg
gggccatgtg cctccccaca 4456catgggcaag gacaggtgga atgtcgggac
cacactgtgc ggcttctcgg cacaaagcgg 4516agggaggctg tggtcgctgc
cggcctaggt gtcccaggtg ccccgccttt ctctgggaca 4576cagttggggg
ctggcttctg agggattcct ttctcccctc tttgtgtggc cccagccagg
4636gcggtgggca gtcctggtgt agagcacaag cctctccacc ctagagaaat
gcctctgtac 4696cacggctacc atgtggaacc ttaacttgca gaaggcttgt
taacaattgt tttgagagag 4756atggctggtc atgccacagc tgctggggac
tccgcctact ccagccctct tgggacacac 4816tgtgggattt gtggcccttc
cccagaggaa ttgtggagac tgtcccatgg aacaaaccct 4876caggcaccag
cacagggctc tgggtgactc agtaaaacta acgtttgtct ctgacaagat
4936cagctgtagg ctcaccggcc agagaagacc actgtgagca ttttgccgta
tatcctgccc 4996tgccatttgt tcacttttta aactaaaata ggaacatccg
acacacaccg tttgcatcgt 5056cttctccctt gatattttaa gcattttccc
atgtcatgag tttctcagaa acatgttttt 5116aacaattgta ctatttagtc
attgtccatt tactataatt tatctgacca tttccctact 5176gtaaaatact
taagacggtt tctgattttt ccactattta aataatgctg tgatgaatat
5236ctttaaaatc ttctgatttc ttactttttt cccccttaga tgcctggaag
tggtattttg 5296aggtgaaaga gtttgttcat tttgaagata tttctgtctc
tctctcgacc tgatgtgtag 5356acgctcactt ccagtagcag aaccacctta
gttgtgtctt acagattctg aacaaatcgg 5416tttctgataa gccatgtgtt
ccaaagaatg tctgaataag accgctcttt atttaaatgc 5476taagaggatg
tcactactgc aatccatctg tggccgattt tttccaagag ccaatttcct
5536tgttttggtt gcaagaacct ggctctgcct gcatgtcagc tctctgccct
ccctgctgcc 5596gtggctttca agcgcttggc agaatcttgt acttcgtgtc
cacaatggta ctgaatttgc 5656atctgcacag tcagcagaga taacaagtgt
tgaactgacc ttgccacatg cttagtgagt 5716gatttgtaat taagtttata
gactcagaag gtatattagg acatttggaa tcagtagcag 5776agcaaagcct
ctttgaaaaa aaccacgtag ctgattgggt tttacaagag tgcatttgtc
5836tcccccttcc acccgtgggg ccccaccttc aggtcttagt ggttcacaag
agcccagcag 5896ccaggctggc tttttcattg tagggcgtgg ttgtcccagc
tggtgtagat ttcaggccgc 5956cccccccaac tccctgccca cagtgttgca
gattgcctgg ctggcagcaa gtccagacca 6016cccaaatttg gttggattct
tcatttctcc actgtagttg gggtccattg attgtgcagg 6076ggaacgtgca
ggaggttttt ctaggcaccg tgttcagtgc tgcttcactc taccagagat
6136tatggccaaa ttgcacggaa tttggtttct tgccctctga agcctgaggg
cccccccttg 6196cctggctggt tgacagaccc ggggtggtca ctgctgagac
ttcagagatc gcagctgctg 6256tgagaatacg gtgaaggtac tttgttctgg
aagatgttgt catacacttt tccccagtta 6316ttttcaaact tgacatgagc
ctatgttgac tcactgggtg ggggtccctt cttacgcagc 6376acacgtggca
agtgcctgaa tcggggctgg aggcacttca gagcctctga ggggccacca
6436cttctggccc aaaattgcag ggttgtagat gaggctgcct gtggagaact
ggtgtgagga 6496ggaagctgtt tccaacaaag agcactttca tctgttgaga
tggctgtggt gagcaactga 6556acgagcctac gtgtgtacct gaattttccc
cgtaactcat ttcttccata tgaagaaaca 6616ccaaactatg tacagagaac
tttttacaaa aggcagacct tttttaagct gtgtaaccca 6676catagcctaa
ccacctggca gaatgactac gaataggggt cattgtgctg gtaaaagcct
6736ctattacgac tgtaagtaag ttggatgttg gcaaaattaa attgttacag
tatttagagc 6796tgctgtagct gttccttcac aacataaaat aggataaatg
actagtacgt ctttcaggtg 6856ggtggcaagc agaacatgcg taatattctc
tacctggtct gtagctgtaa ctgtgatgta 6916cagacaaagc aaaaattaaa
agaacttatg aaaacaaatg caatgatact aggatataca 6976cttttgtatt
tttattctta tataaggtta tttgctggct attgttggcc tctagttcag
7036tctgtgttat ttaaattcta atatatgaat tatttgaatt gaattcatgt
tcggggccac 7096gttgttgtat gtattgatgt acagccttga atgtgaataa
ttattgtaaa ctatatttta 7156caactttttt tctggcttta ttatataaat
tttctattgg gtcagtgatt taatcatata 7216atttaatgaa tctgtttatc
cttttttttt ttccaaatac ttgtgcttta ggtgtagtta 7276ccagatgatg
aattttcctc gtatggtcag tagtcttgta ataaaaagca tgtagagtgt 7336a
73379529PRTHomo sapiens 9Met Val Arg Thr Gln Thr Glu Ser Ser Thr
Pro Pro Gly Ile Pro Gly1 5 10 15Gly Ser Arg Gln Gly Pro Ala Met Asp
Gly Thr Ala Ala Glu Pro Arg 20 25 30Pro Gly Ala Gly Ser Leu Gln His
Ala Gln Pro Pro Pro Gln Pro Arg 35 40 45Lys Lys Arg Pro Glu Asp Phe
Lys Phe Gly Lys Ile Leu Gly Glu Gly 50 55 60Ser Phe Ser Thr Val Val
Leu Ala Arg Glu Leu Ala Thr Ser Arg Glu65 70 75 80Tyr Ala Ile Lys
Ile Leu Glu Lys Arg His Ile Ile Lys Glu Asn Lys 85 90 95Val Pro Tyr
Val Thr Arg Glu Arg Asp Val Met Ser Arg Leu Asp His 100 105 110Pro
Phe Phe Val Lys Leu Tyr Phe Thr Phe Gln Asp Asp Glu Lys Leu 115 120
125Tyr Phe Gly Leu Ser Tyr Ala Lys Asn Gly Glu Leu Leu Lys Tyr Ile
130 135 140Arg Lys Ile Gly Ser Phe Asp Glu Thr Cys Thr Arg Phe Tyr
Thr Ala145 150 155 160Glu Ile Val Ser Ala Leu Glu Tyr Leu His Gly
Lys Gly Ile Ile His 165 170 175Arg Asp Leu Lys Pro Glu Asn Ile Leu
Leu Asn Glu Asp Met His Ile 180 185 190Gln Ile Thr Asp Phe Gly Thr
Ala Lys Val Leu Ser Pro Glu Ser Lys 195 200 205Gln Ala Arg Ala Asn
Ser Phe Val Gly Thr Ala Gln Tyr Val Ser Pro 210 215 220Glu Leu Leu
Thr Glu Lys Ser Ala Cys Lys Ser Ser Asp Leu Trp Ala225 230 235
240Leu Gly Cys Ile Ile Tyr Gln Leu Val Ala Gly Leu Pro Pro Phe Arg
245 250 255Ala Gly Asn Glu Tyr Leu Ile Phe Gln Lys Ile Ile Lys Leu
Glu Tyr 260 265 270Asp Phe Pro Glu Lys Phe Phe Pro Lys Ala Arg Asp
Leu Val Glu Lys 275 280 285Leu Leu Val Leu Asp Ala Thr Lys Arg Leu
Gly Cys Glu Glu Met Glu 290 295 300Gly Tyr Gly Pro Leu Lys Ala His
Pro Phe Phe Glu Ser Val Thr Trp305 310 315 320Glu Asn Leu His Gln
Gln Thr Pro Pro Lys Leu Thr Ala Tyr Leu Pro 325 330 335Ala Met Ser
Glu Asp Asp Glu Asp Cys Tyr Gly Asn Tyr Asp Asn Leu 340 345 350Leu
Ser Gln Phe Gly Cys Met Gln Val Ser Ser Ser Ser Ser Ser His 355 360
365Ser Leu Ser Ala Ser Asp Thr Gly Leu Pro Gln Arg Ser Gly Ser Asn
370 375 380Ile Glu Gln Tyr Ile His Asp Leu Asp Ser Asn Ser Phe Glu
Leu Asp385 390 395 400Leu Gln Phe Ser Glu Asp Glu Lys Arg Leu Leu
Leu Glu Lys Gln Ala 405 410 415Gly Gly Asn Pro Trp His Gln Phe Val
Glu Asn Asn Leu Ile Leu Lys 420 425 430Met Gly Pro Val Asp Lys Arg
Lys Gly Leu Phe Ala Arg Arg Arg Gln 435 440 445Leu Leu Leu Thr Glu
Gly Pro His Leu Tyr Tyr Val Asp Pro Val Asn 450 455 460Lys Val Leu
Lys Gly Glu Ile Pro Trp Ser Gln Glu Leu Arg Pro Glu465 470 475
480Ala Lys Asn Phe Lys Thr Phe Phe Val His Thr Pro Asn Arg Thr Tyr
485 490 495Tyr Leu Met Asp Pro Ser Gly Asn Ala His Lys Trp Cys Arg
Lys Ile 500 505 510Gln Glu Val Trp Arg Gln Arg Tyr Gln Ser His Pro
Asp Ala Ala Val 515 520 525Gln101968DNAHomo sapiensCDS(1)..(1968)
10atg gtt ttg tac acg acc ccc ttt cct aac agc tgt ctg tcc gcc ctg
48Met Val Leu Tyr Thr Thr Pro Phe Pro Asn Ser Cys Leu Ser Ala Leu1
5 10 15cac tgt gtg tcc tgg gcc ctt atc ttt cca tgc tac tgg ctg gtg
gac 96His Cys Val Ser Trp Ala Leu Ile Phe Pro Cys Tyr Trp Leu Val
Asp 20 25 30cgg ctc gct gcc tcc ttc ata ccc acc acc tac gag aag cgc
cag cgg 144Arg Leu Ala Ala Ser Phe Ile Pro Thr Thr Tyr Glu Lys Arg
Gln Arg 35 40 45gca gac gac ccg tgc tgc ctg cag ctg ctc tgc act gcc
ctc ttc acg 192Ala Asp Asp Pro Cys Cys Leu Gln Leu Leu Cys Thr Ala
Leu Phe Thr 50 55 60ccc atc tac ctg gcc ctc ctg gtg gcc tcg ctg ccc
ttt gcg ttt ctc 240Pro Ile Tyr Leu Ala Leu Leu Val Ala Ser Leu Pro
Phe Ala Phe Leu65 70 75 80ggc ttt ctc ttc tgg tcc cca ctg cag tcg
gcc cgc cgg ccc tac atc 288Gly Phe Leu Phe Trp Ser Pro Leu Gln Ser
Ala Arg Arg Pro Tyr Ile 85 90 95tat tca cgg ctg gaa gac aag ggc ctg
gcc ggt ggg gca gcc ctg ctc 336Tyr Ser Arg Leu Glu Asp Lys Gly Leu
Ala Gly Gly Ala Ala Leu Leu 100 105 110agt gaa tgg aag ggc acg ggg
cct ggc aaa agc ttc tgc ttt gcc act 384Ser Glu Trp Lys Gly Thr Gly
Pro Gly Lys Ser Phe Cys Phe Ala Thr 115 120 125gcc aac gtc tgc ctc
ctg ccc gac tca ctc gcc agg gtc aac aac ctt 432Ala Asn Val Cys Leu
Leu Pro Asp Ser Leu Ala Arg Val Asn Asn Leu 130 135 140ttt aac acc
caa gcg cgg gcc aag gag atc ggg cag aga atc cgc aat 480Phe Asn Thr
Gln Ala Arg Ala Lys Glu Ile Gly Gln Arg Ile Arg Asn145 150 155
160ggg gcc gcc cgg ccc cag atc aaa att tac atc gac tcc ccc acc aat
528Gly Ala Ala Arg Pro Gln Ile Lys Ile Tyr Ile Asp Ser Pro Thr Asn
165 170 175acc tcc atc agc gcc gct agc ttc agc agc ctg gtg tca cca
cag ggc 576Thr Ser Ile Ser Ala Ala Ser Phe Ser Ser Leu Val Ser Pro
Gln Gly 180 185 190ggc gat ggg gtg gcc cgg gcc gtc ccc ggg agc att
aag agg aca gcc 624Gly Asp Gly Val Ala Arg Ala Val Pro Gly Ser Ile
Lys Arg Thr Ala 195 200 205tct gtg gag tac aag ggt gac ggt ggg cgg
cac ccc ggt gac gag gct 672Ser Val Glu Tyr Lys Gly Asp Gly Gly Arg
His Pro Gly Asp Glu Ala 210 215 220gcc aac ggc cca gcc tct ggg gac
cct gtc gac agc agc agc ccg gag 720Ala Asn Gly Pro Ala Ser Gly Asp
Pro Val Asp Ser Ser Ser Pro Glu225 230 235 240gat gcc tgc atc gtg
cgc atc ggt ggc gag gag ggc ggc cgg cca cct 768Asp Ala Cys Ile Val
Arg Ile Gly Gly Glu Glu Gly Gly Arg Pro Pro 245 250 255gaa gct gac
gac cct gtg cct ggg ggc cag gcc agg aac gga gct ggc 816Glu Ala Asp
Asp Pro Val Pro Gly Gly Gln Ala Arg Asn Gly Ala Gly 260 265 270ggg
ggc cca agg ggc cag acg ccc aac cat aat cag cag gac ggg gat 864Gly
Gly Pro Arg Gly Gln Thr Pro Asn His Asn Gln Gln Asp Gly Asp 275 280
285tca ggg agc ctg ggc agc ccc tcg gcc tcc cgg gag tcc ctg gtg aag
912Ser Gly Ser Leu Gly Ser Pro Ser Ala Ser Arg Glu Ser Leu Val Lys
290 295 300ggg cga gct ggg cca gac acc agt gcc agc ggg gag cca ggt
gcc aac 960Gly Arg Ala Gly Pro Asp Thr Ser Ala Ser Gly Glu Pro Gly
Ala Asn305 310 315 320agc aag ctc ctg tac aag gcc tcg gtg gtg aag
aag gcg gct gca cgc 1008Ser Lys Leu Leu Tyr Lys Ala Ser Val Val Lys
Lys Ala Ala Ala Arg 325 330 335agg agg cgg cac ccc gac gag gcc ttc
gac cat gag gtc tcc gcc ttc 1056Arg Arg Arg His Pro Asp Glu Ala Phe
Asp His Glu Val Ser Ala Phe 340 345 350ttc ccc gcc aac ctg gac ttc
ctg tgc ctg cag gag gtg ttt gac aag 1104Phe Pro Ala Asn Leu Asp Phe
Leu Cys Leu Gln Glu Val Phe Asp Lys 355 360 365cga gca gcc acc aaa
ttg aaa gag cag ctg cac ggc tac ttc gag tac 1152Arg Ala Ala Thr Lys
Leu Lys Glu Gln Leu His Gly Tyr Phe Glu Tyr 370 375 380atc ctg tac
gac gtc ggg gtc tac ggc tgc cag ggc tgc tgc agc ttc 1200Ile Leu Tyr
Asp Val Gly Val Tyr Gly Cys Gln Gly Cys Cys Ser Phe385 390 395
400aag tgt ctc aac agc ggc ctc ctc ttt gcc agc cgc tac ccc atc atg
1248Lys Cys Leu Asn Ser Gly Leu Leu Phe Ala Ser Arg Tyr Pro Ile Met
405 410 415gac gtg gcc tat cac tgt tac ccc aac aag tgt aac gac gat
gcc ctg 1296Asp Val Ala Tyr His Cys Tyr Pro Asn Lys Cys Asn Asp Asp
Ala Leu 420 425 430gcc tct aag gga gct ctg ttt ctc aag gtg cag gtg
gga agc aca cct 1344Ala Ser Lys Gly Ala Leu Phe Leu Lys Val Gln Val
Gly Ser Thr Pro 435 440 445cag gac caa aga atc gtc ggg tac atc gcc
tgc aca cac ctg cat gcc 1392Gln Asp Gln Arg Ile Val Gly Tyr Ile Ala
Cys Thr His Leu His Ala 450 455 460cca caa gag gac agc gcc atc cgg
tgt ggg cag ctg gac ctg ctt cag 1440Pro Gln Glu Asp Ser Ala Ile Arg
Cys Gly Gln Leu Asp Leu Leu Gln465 470 475 480gac tgg ctg gct gat
ttc cga aaa tct acc tcc tcg tcc agc gca gcc 1488Asp Trp Leu Ala Asp
Phe Arg Lys Ser Thr Ser Ser Ser Ser Ala Ala 485 490 495aac ccc gag
gag ctg gtg gca ttt gac gtc gtc tgt gga gat ttc aac 1536Asn Pro Glu
Glu Leu Val Ala Phe Asp Val Val Cys Gly Asp Phe Asn 500 505 510ttt
gat aac tgc tcc tct gac gac aag ctg gag cag caa cac tcc ctg 1584Phe
Asp Asn Cys Ser Ser Asp Asp Lys Leu Glu Gln Gln His Ser Leu 515 520
525ttc acc cac tac agg gac ccc tgc cgc ctg ggg cct ggt gag gag aag
1632Phe Thr His Tyr Arg Asp Pro Cys Arg Leu Gly Pro Gly Glu Glu Lys
530 535 540ccg tgg gcc atc ggt act ctg ctg gac acg aac ggc ctg tac
gat gag 1680Pro Trp Ala Ile Gly Thr Leu Leu Asp Thr Asn Gly Leu Tyr
Asp Glu545 550 555 560gat gtg tgc acc ccc gac aac ctg cag aag gtc
ctg gag agt gag gag 1728Asp Val Cys Thr Pro Asp Asn Leu Gln Lys Val
Leu Glu Ser Glu Glu 565 570 575ggc cgc agg gag tac ctg gcg ttt ccc
acc agc aag agc tcg ggc cag 1776Gly Arg Arg Glu Tyr Leu Ala Phe Pro
Thr Ser Lys Ser Ser Gly Gln 580 585 590aag ggg cgg aag gag ctg ctg
aag ggc aac ggc cgg cgc atc gac tac 1824Lys Gly Arg Lys Glu Leu Leu
Lys Gly Asn Gly Arg Arg Ile Asp Tyr
595 600 605atg ctg cat gca gag gag ggg ctg tgc cca gac tgg aag gcc
gag gtg 1872Met Leu His Ala Glu Glu Gly Leu Cys Pro Asp Trp Lys Ala
Glu Val 610 615 620gaa gaa ttc agt ttt atc acc cag ctg tcc ggc ctg
acg gac cac ctg 1920Glu Glu Phe Ser Phe Ile Thr Gln Leu Ser Gly Leu
Thr Asp His Leu625 630 635 640cca gta gcc atg cga ctg atg gtg tct
tcg ggg gag gag gag gca tag 1968Pro Val Ala Met Arg Leu Met Val Ser
Ser Gly Glu Glu Glu Ala 645 650 65511655PRTHomo sapiens 11Met Val
Leu Tyr Thr Thr Pro Phe Pro Asn Ser Cys Leu Ser Ala Leu1 5 10 15His
Cys Val Ser Trp Ala Leu Ile Phe Pro Cys Tyr Trp Leu Val Asp 20 25
30Arg Leu Ala Ala Ser Phe Ile Pro Thr Thr Tyr Glu Lys Arg Gln Arg
35 40 45Ala Asp Asp Pro Cys Cys Leu Gln Leu Leu Cys Thr Ala Leu Phe
Thr 50 55 60Pro Ile Tyr Leu Ala Leu Leu Val Ala Ser Leu Pro Phe Ala
Phe Leu65 70 75 80Gly Phe Leu Phe Trp Ser Pro Leu Gln Ser Ala Arg
Arg Pro Tyr Ile 85 90 95Tyr Ser Arg Leu Glu Asp Lys Gly Leu Ala Gly
Gly Ala Ala Leu Leu 100 105 110Ser Glu Trp Lys Gly Thr Gly Pro Gly
Lys Ser Phe Cys Phe Ala Thr 115 120 125Ala Asn Val Cys Leu Leu Pro
Asp Ser Leu Ala Arg Val Asn Asn Leu 130 135 140Phe Asn Thr Gln Ala
Arg Ala Lys Glu Ile Gly Gln Arg Ile Arg Asn145 150 155 160Gly Ala
Ala Arg Pro Gln Ile Lys Ile Tyr Ile Asp Ser Pro Thr Asn 165 170
175Thr Ser Ile Ser Ala Ala Ser Phe Ser Ser Leu Val Ser Pro Gln Gly
180 185 190Gly Asp Gly Val Ala Arg Ala Val Pro Gly Ser Ile Lys Arg
Thr Ala 195 200 205Ser Val Glu Tyr Lys Gly Asp Gly Gly Arg His Pro
Gly Asp Glu Ala 210 215 220Ala Asn Gly Pro Ala Ser Gly Asp Pro Val
Asp Ser Ser Ser Pro Glu225 230 235 240Asp Ala Cys Ile Val Arg Ile
Gly Gly Glu Glu Gly Gly Arg Pro Pro 245 250 255Glu Ala Asp Asp Pro
Val Pro Gly Gly Gln Ala Arg Asn Gly Ala Gly 260 265 270Gly Gly Pro
Arg Gly Gln Thr Pro Asn His Asn Gln Gln Asp Gly Asp 275 280 285Ser
Gly Ser Leu Gly Ser Pro Ser Ala Ser Arg Glu Ser Leu Val Lys 290 295
300Gly Arg Ala Gly Pro Asp Thr Ser Ala Ser Gly Glu Pro Gly Ala
Asn305 310 315 320Ser Lys Leu Leu Tyr Lys Ala Ser Val Val Lys Lys
Ala Ala Ala Arg 325 330 335Arg Arg Arg His Pro Asp Glu Ala Phe Asp
His Glu Val Ser Ala Phe 340 345 350Phe Pro Ala Asn Leu Asp Phe Leu
Cys Leu Gln Glu Val Phe Asp Lys 355 360 365Arg Ala Ala Thr Lys Leu
Lys Glu Gln Leu His Gly Tyr Phe Glu Tyr 370 375 380Ile Leu Tyr Asp
Val Gly Val Tyr Gly Cys Gln Gly Cys Cys Ser Phe385 390 395 400Lys
Cys Leu Asn Ser Gly Leu Leu Phe Ala Ser Arg Tyr Pro Ile Met 405 410
415Asp Val Ala Tyr His Cys Tyr Pro Asn Lys Cys Asn Asp Asp Ala Leu
420 425 430Ala Ser Lys Gly Ala Leu Phe Leu Lys Val Gln Val Gly Ser
Thr Pro 435 440 445Gln Asp Gln Arg Ile Val Gly Tyr Ile Ala Cys Thr
His Leu His Ala 450 455 460Pro Gln Glu Asp Ser Ala Ile Arg Cys Gly
Gln Leu Asp Leu Leu Gln465 470 475 480Asp Trp Leu Ala Asp Phe Arg
Lys Ser Thr Ser Ser Ser Ser Ala Ala 485 490 495Asn Pro Glu Glu Leu
Val Ala Phe Asp Val Val Cys Gly Asp Phe Asn 500 505 510Phe Asp Asn
Cys Ser Ser Asp Asp Lys Leu Glu Gln Gln His Ser Leu 515 520 525Phe
Thr His Tyr Arg Asp Pro Cys Arg Leu Gly Pro Gly Glu Glu Lys 530 535
540Pro Trp Ala Ile Gly Thr Leu Leu Asp Thr Asn Gly Leu Tyr Asp
Glu545 550 555 560Asp Val Cys Thr Pro Asp Asn Leu Gln Lys Val Leu
Glu Ser Glu Glu 565 570 575Gly Arg Arg Glu Tyr Leu Ala Phe Pro Thr
Ser Lys Ser Ser Gly Gln 580 585 590Lys Gly Arg Lys Glu Leu Leu Lys
Gly Asn Gly Arg Arg Ile Asp Tyr 595 600 605Met Leu His Ala Glu Glu
Gly Leu Cys Pro Asp Trp Lys Ala Glu Val 610 615 620Glu Glu Phe Ser
Phe Ile Thr Gln Leu Ser Gly Leu Thr Asp His Leu625 630 635 640Pro
Val Ala Met Arg Leu Met Val Ser Ser Gly Glu Glu Glu Ala 645 650
655127003DNAHomo sapiensCDS(244)..(900) 12actcgcagtc ctgacgggca
ggggctgcgg accgcccggc cttggaccca tccggagcca 60caggttggag gagataagta
gctgtccccg tgctcatcgc cctgtggagc agatcctgtc 120tccttgctga
cggtggagcc cgggagttcc agggcttggg aaggggaagg aaacctctct
180gaaatctgac acctgctctc ccggcaagga aacttcgcag gctgaccgac
caagaccatc 240act atg acc gat gga gac tat gat tat ctg atc aaa ctc
ctg gcc ctc 288 Met Thr Asp Gly Asp Tyr Asp Tyr Leu Ile Lys Leu Leu
Ala Leu 1 5 10 15ggg gat tca ggg gtg ggg aag aca aca ttt ctt tat
aga tac aca gat 336Gly Asp Ser Gly Val Gly Lys Thr Thr Phe Leu Tyr
Arg Tyr Thr Asp 20 25 30aat aaa ttc aat ccc aaa ttc atc act aca gta
gga ata gac ttt cgg 384Asn Lys Phe Asn Pro Lys Phe Ile Thr Thr Val
Gly Ile Asp Phe Arg 35 40 45gaa aaa cgt gtg gtt tat aat gca caa gga
ccg aat gga tct tca ggg 432Glu Lys Arg Val Val Tyr Asn Ala Gln Gly
Pro Asn Gly Ser Ser Gly 50 55 60aaa gca ttt aaa gtg cat ctt cag ctt
tgg gac act gcg gga caa gag 480Lys Ala Phe Lys Val His Leu Gln Leu
Trp Asp Thr Ala Gly Gln Glu 65 70 75cgg ttc cgg agt ctc acc act gca
ttt ttc aga gac gcc atg ggc ttc 528Arg Phe Arg Ser Leu Thr Thr Ala
Phe Phe Arg Asp Ala Met Gly Phe80 85 90 95tta tta atg ttt gac ctc
acc agt caa cag agc ttc tta aat gtc aga 576Leu Leu Met Phe Asp Leu
Thr Ser Gln Gln Ser Phe Leu Asn Val Arg 100 105 110aac tgg atg agc
caa ctg caa gca aat gct tat tgt gaa aat cca gat 624Asn Trp Met Ser
Gln Leu Gln Ala Asn Ala Tyr Cys Glu Asn Pro Asp 115 120 125ata gta
tta att ggc aac aag gca gac cta cca gat cag agg gaa gtc 672Ile Val
Leu Ile Gly Asn Lys Ala Asp Leu Pro Asp Gln Arg Glu Val 130 135
140aat gaa cgg caa gct cgg gaa ctg gct gac aaa tat ggc ata cca tat
720Asn Glu Arg Gln Ala Arg Glu Leu Ala Asp Lys Tyr Gly Ile Pro Tyr
145 150 155ttt gaa aca agt gca gca act gga cag aat gtg gag aaa gct
gta gaa 768Phe Glu Thr Ser Ala Ala Thr Gly Gln Asn Val Glu Lys Ala
Val Glu160 165 170 175acc ctt ttg gac tta atc atg aag cga atg gaa
cag tgt gtg gag aag 816Thr Leu Leu Asp Leu Ile Met Lys Arg Met Glu
Gln Cys Val Glu Lys 180 185 190aca caa atc cct gat act gtc aat ggt
gga aat tct gga aac ttg gat 864Thr Gln Ile Pro Asp Thr Val Asn Gly
Gly Asn Ser Gly Asn Leu Asp 195 200 205ggg gaa aag cca cca gag aag
aaa tgt atc tgc tag actctacata 910Gly Glu Lys Pro Pro Glu Lys Lys
Cys Ile Cys 210 215gaaactgaac atcaagaacc ccaccaaaat attactttta
aaaacaatga caaaccacac 970aattgttgtt gagtaaacca cgcacaatgg
catgtctttc tttttctgcc agaaaatcta 1030ttttaagaaa ccagaatagt
caacagtgtt caaaagaatt gactagttat ccctgaggcc 1090ctttcaaaca
tgatcaaaga tttcccaatg tgatctcatc atcatggata ctcaatttgt
1150tttttcttat agagaaaatg agtatataag acaatataca agaagaaata
tcagtgagtt 1210ttaaatcaga acaagttacc tgtcacattg aagaaaaggg
taggcactaa agggagaaca 1270cagaaagaag aatttctaaa atattggatt
tacttcttat attgagtcag atgcatactt 1330ttagatttgc attggggaaa
atgtactagc taaaaatgga tacacaatga agaattctat 1390ttggctaatt
aagaatgata tactatgtac acccaataag ctgtactaga atgaataaat
1450tactgataag gttacaaata ggtaaatgtc acacttctgt taaaatgcag
gaggtagtgt 1510cataatgccg tctttatatt cttaataaat agcactttga
caagaacagg actgtaaatg 1570atgaagtaca agacaaatac cctgggaaaa
aaaatgaaag tatgagaaat tggcatttct 1630acagctgaaa ttcaatgtat
ctgttagaga tgtctggaag ggttacttag ccaaatttta 1690ctcaagccaa
ttaggagctg atattatcag ttggaattaa gagaactcca gaggtttcca
1750tttcaaacaa aattttagaa attggtttgg tgttcagctt cacatttcat
tttttcttag 1810cacatgttga taaaatagtc acaaggagaa attaccagtt
acggtttatt aaatctcttt 1870taaaatgcag tcaaggaaaa ctagccttga
atttttttta gataaaataa gatggtgata 1930tgaaacaaaa agtggcaatt
attgcaggtt tccttttagt ttacaaaagt actggaaact 1990aaatcatatt
tcttccctcc aaatttcacc cattcctgac tttgaatcaa ttgcagaaat
2050gcaggtgtgt tactttgttg atcaataact ttggaacaat tatggatcaa
ttctatggtc 2110actctgaatt ttcatgtcat taatcacata aaaattgata
atacctcatt ctgtattaca 2170atatgatttt attttgccaa aggcaagaca
cctatagttg agctgtattt tgggggattg 2230ggtgaggaag gacttctgat
cttatctcaa caaaaaactg gccagtattt ttgttaatgt 2290aaagcttcct
tttctttcta aaaaatagta acaaaattat ttttcattgg cctattctgt
2350tcttgtgtct aaactaacat tacattaatt tttaatctta gtttctgata
aacacaagcc 2410attcctatca aaatattatt tatttcagtc aattttacca
aataacaaag acaatatatt 2470ttcgtttttt tttattatga gcatatgatt
ttttgacagg ctgtttcctc gtcgtataga 2530ttttttccaa tcaaacctac
tttttccata ctctgtgcat attttttgtg aagttataca 2590cattgaagac
cctaaaaatc ccagtccatc attcagctta cctctgcgaa cttctatctg
2650gtattgaatc agtttcagaa acacagacag atccaaggaa atgtctcttt
ataatgttct 2710taggatggac tagacccata aatgtgccat gaatcaaaat
attaataatt tgaaagcttt 2770catgctgtta gcccctgatg aaattctcag
cattaactgg ccagctcctc tgatttctgc 2830agcatcgcaa caggttcgaa
gatgggttgt ggctgggtat tccctcccat ggtgtttcct 2890ctgggatgct
cttcattatc tcaatgcctg tgccatgaag atagaaaact gtaagctaac
2950atttaagatg tttcttctgg aaggaaagtg agcaggaaca agttatattg
ccactgctgt 3010ggcaaatttt ggtgaacttt tggggtcatt atatcaattt
tttctttgga ttcaaattgt 3070aatgtcccct gcatttcctt aatagggaat
gtgaaacctt tataaaactc taaaagtatt 3130ctgttttgat atgtcttttt
gtttctattc attttcagtt atatgattga tttacttatg 3190ccaagattct
gtcactgtca gttatttaat gagtgttttt tcagggtctg ttttaagatc
3250attatttgat agctgtagca tgaagcagag gttgatgatg cccataattg
caagactatt 3310cctgtaaaaa taacaattat tgggtaataa cttcaagagg
aatgagaagt gacaaaattg 3370atttaaaata ttgttctact tataaataaa
tgcttgatat aaaaaatttt ctccataaag 3430tttgacatct gaccccagat
tctatgtaat cattattaga aattccttct ctcattattt 3490caggattagt
agttctgtgt aattcatttt acaatttcaa attgttctgg tgccataaag
3550tatacagact actttaaaga tttccaaatc ccctaattta ccccacaaca
gcatgtaatt 3610ttagccaaga tatgtcctgt tactaagtat ctcccaatgc
tttagtaaaa cgtatttagg 3670agaaatgttg aaaatgtaca tgaagctcct
ttctgatata gaaaccattt ctggagtatt 3730tacactggtt tgatgtttac
attgctctaa ctcggtgcct cagatacctc tgtgaccaaa 3790tttgtctcca
accacatagc tcatttccta taatgttata tcataggaag ccctcacaga
3850gacactaaca cagctaaaga tcttctgata ttatcagcaa gggatgcaag
gactttattg 3910gaatctggag agtttaactg ccttctcttg gtctcctcac
ttacttctta tgaagttggc 3970attacctgag actcttagct gtgattaggt
acaagcttac cttttagggt agaaaaagaa 4030agatcatttg aaaaatgtat
ctaaaataat ccagagaaca taatgtttgt cttggtctga 4090taatgataag
aagtcaagga ttggcagaga aaatactaaa cgccaagagt tgagcctgtg
4150ggtctctcca taagagtttt aaaactcttg ccagttacca ctttatccaa
tttgctatca 4210ttttcgtatt atcagctatc gccctgtaaa atattcaaaa
ctagctattt ctaaagtaaa 4270cattttatct gttactttta accagatagg
tgtctttgtc atccttctac tataaattgt 4330tctttgccaa cctgtacagg
tagatgaacc aggcgagagt tttaatcagc cttttcttgt 4390cccctttgta
agaaagagat gcttgccata gagaaggaca tgagtacatt aaaaataatt
4450taatagccac aatatgatgt tctttaagct gcaaattgag tacactggga
atcaacaaat 4510ttgatgaagc ctgtctgtct cttcaccagt ggagtgagtg
cagcagttag aaagagaagc 4570aatattgtgc aactggtgca gtggtgagtt
aatcatagtg tataaccttg tgttcatgaa 4630acaggttgtt cattgttctg
catctctctt catttaaaaa ggatacacaa ttctttcctc 4690attgcatatt
acaccaaacg tttgagggaa aaatcctcat tcgtaaagga ttttggatgt
4750ataatctaaa actcaacaat aaagaaataa tattccaagt ctctggtttc
ctaagataca 4810taataactgt ttataaagaa ggtctaagag ctgatatttg
ccaaagtgat agaagagttg 4870ttttttcctc tctactacca agctttaaga
cattaaaaga agtctagtgt atttgaatat 4930tttagagaaa gctttatcat
tttttaagat gccaagatgc tgcctacgtt tgcaaaagtt 4990gtctaagaat
tcaccatgag ctatattttc ttctggatct ttgaccaagg tgatgtcagc
5050ttatttctgg ggaaggtgtt gagctcttat acatgaaaat ggatataggc
tattctctgg 5110gatgagtgtc atttcaatgc tttataaatc catgaagctg
cttgtctcat aaagtagaac 5170tgatacaaat tttggttgga tatatagaga
attttataaa tgtattgcct tagaatttct 5230gggtggagac ccaactacaa
tgacattgtc atgccagaac tataaagata attagagtta 5290aaagttgttt
aaattgtgcc cttaaataca gcagaacctg gagaaggtca tacttcaaag
5350gtcgattttg agtccgaata aagaaagacc tagtaacaga tagttttttt
ttgttcattt 5410tcttctacca agtagaggtt tatgccctca gaactaaact
agtaaaaata tctgaacaaa 5470aaacctttcg ttgttggcat aaaaatgtga
tacacttaga gacattttgt ttattgcata 5530taaatctaat ttttccataa
attagattta tgatattttc ataaagcact tgattagttt 5590ttcaaggcgt
accatcacaa agatgctttc ctgcagagtt ctttgtatca acagcctatg
5650gttgagatgt tttctcattt cctgtagaga gagaatacca ctaacaaaca
aacaaaaact 5710ttagtgccaa aatagtggaa ctattttgtc atcttttgag
aaaaaaatat acaaagaagt 5770catcttttca ttaagtggat tccctggttc
ctttccagct ggttgtggaa gtaatggcta 5830acatccttca gctgactttg
tctacaagga ttattagcaa attctgtagg agcaagcatg 5890tctgacctta
acttaatgga tcccttattc aatcagtggc ttctgtcttt atgtctgttg
5950gcatatcaaa atggtttctg ttcctagaaa agtaataaca tatgcttatc
tttattcttt 6010ttccaggtga ttttgttttc aaatgctcct tgtgaaaaca
cctagtgttg tagaaaggaa 6070agtggccaga aagaacaact tgggaccatg
agtaggtcat taaatagctt agtgatttat 6130cctcatatag ggcttataaa
ccctgtatgt gtttatatgt gcttcacaga gttcgtgtca 6190ggctcaaagg
agatatgtat aagaaagtgg tttgtaaatt atgttccatt tcataaatag
6250acactattca caaactaaaa tctaataaaa aaccacagtt gtaatttaaa
ctgcttgata 6310taaaaagagg tatcatagca gggaaaacac actaattttc
atacagtaga ggtattgaaa 6370actgaaaatg ggaaggcaac ttgaagtcat
tgtatttgat tgaaaatgtt taatacatct 6430cattattgac aaaatatgtc
atcttgtatt tatttcaagg aaaccaatga attctaggta 6490gtatattaca
agttggtcaa aatattccat gtacaaatag ggcttctgtg tccatagcct
6550tgtaagagat actgattgta tctgaaatta ttttttaaaa aaataaatta
tcctgcttta 6610gttagtgtgt taaaagtaga cgatgttcta atataacact
gaagtgcttc attgtatccc 6670aacagtttac cttcaagtaa tattatcttt
atttttaggc taagcacgtt tgattatttt 6730gtctgtctcc tatatagatc
tgttttgtct agtgctatga atgtaactta aaactataaa 6790cttgaagttt
ttattctata tgccccttaa tagactgtgg ttcctgacgc acactgttag
6850gtcattattt tgttgtacca aagttctagt ggcttcagaa atcatagcat
ccaatgattt 6910tttggtgtct ggctatgaat actatggttg agaattgtat
tcagtgattg tttctgcaca 6970cttttcaaat aaaaaatgaa tttttatcaa tta
700313218PRTHomo sapiens 13Met Thr Asp Gly Asp Tyr Asp Tyr Leu Ile
Lys Leu Leu Ala Leu Gly1 5 10 15Asp Ser Gly Val Gly Lys Thr Thr Phe
Leu Tyr Arg Tyr Thr Asp Asn 20 25 30Lys Phe Asn Pro Lys Phe Ile Thr
Thr Val Gly Ile Asp Phe Arg Glu 35 40 45Lys Arg Val Val Tyr Asn Ala
Gln Gly Pro Asn Gly Ser Ser Gly Lys 50 55 60Ala Phe Lys Val His Leu
Gln Leu Trp Asp Thr Ala Gly Gln Glu Arg65 70 75 80Phe Arg Ser Leu
Thr Thr Ala Phe Phe Arg Asp Ala Met Gly Phe Leu 85 90 95Leu Met Phe
Asp Leu Thr Ser Gln Gln Ser Phe Leu Asn Val Arg Asn 100 105 110Trp
Met Ser Gln Leu Gln Ala Asn Ala Tyr Cys Glu Asn Pro Asp Ile 115 120
125Val Leu Ile Gly Asn Lys Ala Asp Leu Pro Asp Gln Arg Glu Val Asn
130 135 140Glu Arg Gln Ala Arg Glu Leu Ala Asp Lys Tyr Gly Ile Pro
Tyr Phe145 150 155 160Glu Thr Ser Ala Ala Thr Gly Gln Asn Val Glu
Lys Ala Val Glu Thr 165 170 175Leu Leu Asp Leu Ile Met Lys Arg Met
Glu Gln Cys Val Glu Lys Thr 180 185 190Gln Ile Pro Asp Thr Val Asn
Gly Gly Asn Ser Gly Asn Leu Asp Gly 195 200 205Glu Lys Pro Pro Glu
Lys Lys Cys Ile Cys 210 2151443DNAMacaca arctoides 14ttgatgtaca
gccttgaatg tgaataatta ttgtaaacta tat 431543DNAHomo sapiens
15ttgatgtaca gccttgaatg tgaataatta ttgtaaacta tat
431630DNAArtificial Sequenceforward primer 16aactcgagaa tgctggctat
tgttggcctc 301734DNAArtificial SequenceReverse primer 17aagcggccgc
aagattaaat cactgaccca atag 34
References
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014

D00015

D00016
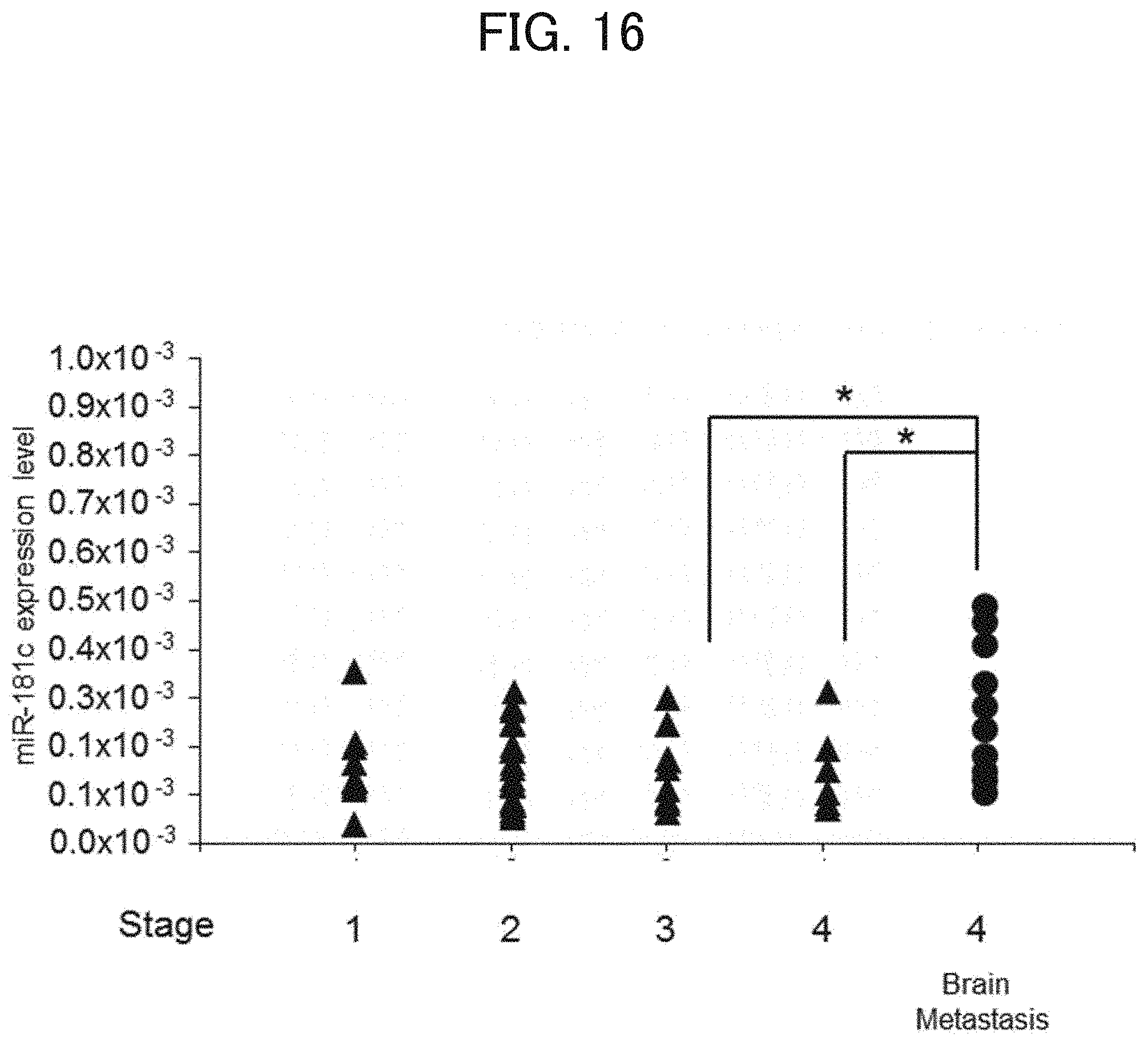
D00017

D00018

D00019

D00020

D00021

D00022

D00023
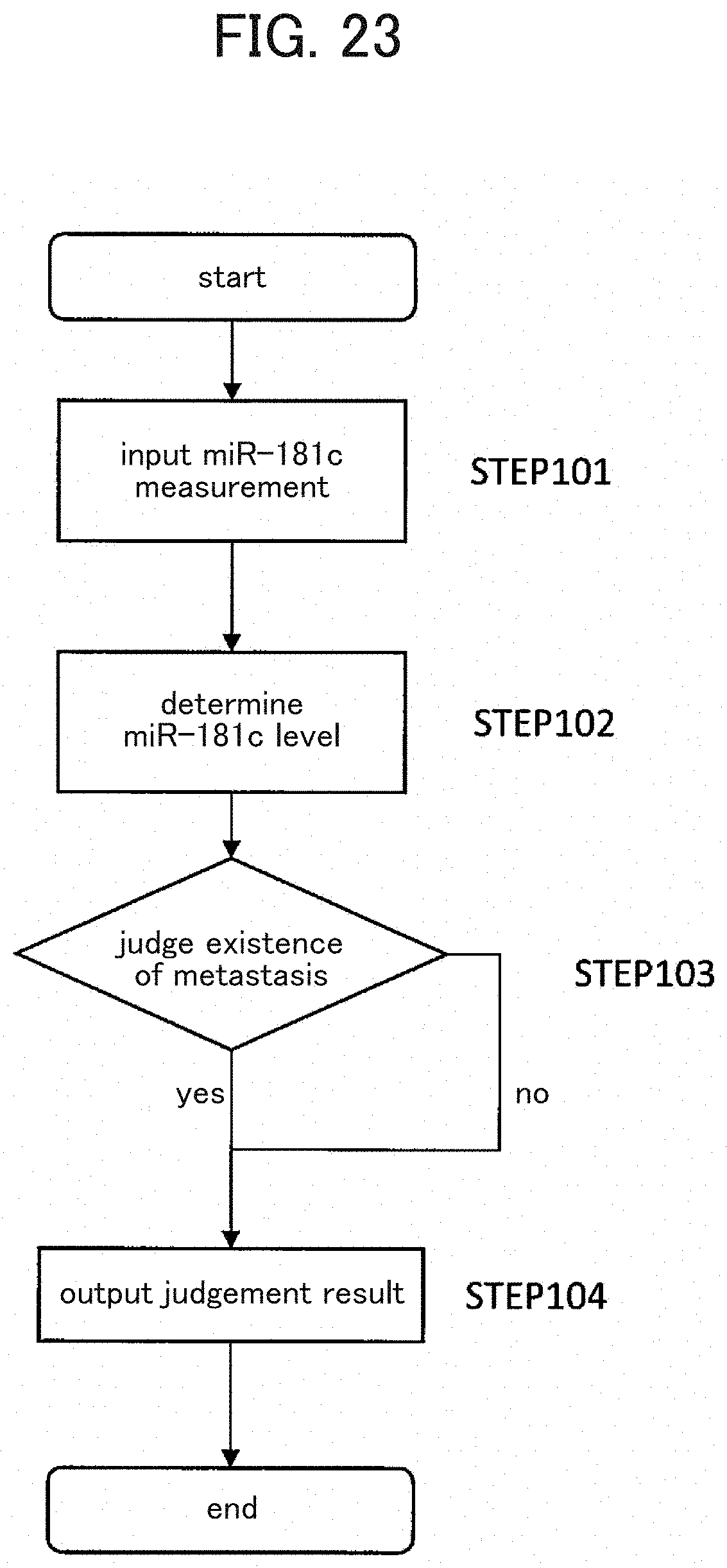
D00024

D00025

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.