Tissue Selective Transgene Expression
TAGLIATELA; Stephanie ; et al.
U.S. patent application number 16/670996 was filed with the patent office on 2020-05-28 for tissue selective transgene expression. The applicant listed for this patent is Encoded Therapeutics, Inc.. Invention is credited to Szu-Ying CHEN, David OBERKOFLER, Kartik RAMAMOORTHI, Stephanie TAGLIATELA, Andrew YOUNG.
| Application Number | 20200165628 16/670996 |
| Document ID | / |
| Family ID | 63713399 |
| Filed Date | 2020-05-28 |









View All Diagrams
| United States Patent Application | 20200165628 |
| Kind Code | A1 |
| TAGLIATELA; Stephanie ; et al. | May 28, 2020 |
TISSUE SELECTIVE TRANSGENE EXPRESSION
Abstract
Provided herein are compositions and methods for selective expression of a transgene. Compositions and methods for selective expression of a transgene comprise one or more human regulatory elements, which, when operably linked to a transgene, can facilitate selective expression of a transgene (e.g., cell-type selective expression) in a target cell as compared to at least one or more non-target cells.
| Inventors: | TAGLIATELA; Stephanie; (South San Francisco, CA) ; YOUNG; Andrew; (South San Francisco, CA) ; CHEN; Szu-Ying; (South San Francisco, CA) ; RAMAMOORTHI; Kartik; (South San Francisco, CA) ; OBERKOFLER; David; (South San Francisco, CA) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 63713399 | ||||||||||
| Appl. No.: | 16/670996 | ||||||||||
| Filed: | October 31, 2019 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 16153433 | Oct 5, 2018 | 10519465 | ||
| 16670996 | ||||
| PCT/US2018/025940 | Apr 3, 2018 | |||
| 16153433 | ||||
| 62480998 | Apr 3, 2017 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | A01K 2217/15 20130101; A61K 48/0058 20130101; C12N 15/86 20130101; C12N 15/113 20130101; A01K 2227/105 20130101; A61P 25/08 20180101; C12N 15/8645 20130101; C07K 14/705 20130101; C12N 15/85 20130101; C12N 2840/007 20130101; C12N 2750/14143 20130101; A61K 48/00 20130101; A01K 2217/052 20130101; C12N 2830/008 20130101; A61P 25/28 20180101; A01K 2267/0312 20130101; C12N 2320/32 20130101 |
| International Class: | C12N 15/85 20060101 C12N015/85; C12N 15/86 20060101 C12N015/86; C12N 15/864 20060101 C12N015/864; A61K 48/00 20060101 A61K048/00; C07K 14/705 20060101 C07K014/705; C12N 15/113 20060101 C12N015/113 |
Claims
1.-13. (canceled)
14. A nucleic acid cassette comprising a regulatory element comprising a nucleic acid sequence with at least 95% sequence identity to SEQ ID NO: 30 operably linked to a transgene that results in selective expression of the transgene in parvalbumin (PV) neurons in the CNS over one or more non-PV cells in the CNS.
15. The nucleic acid cassette of claim 14, wherein the transgene comprises a nucleic acid sequence encoding a DNA binding protein.
16. The nucleic acid cassette of claim 15, wherein the DNA binding protein is a transcriptional modulator of an endogenous gene.
17. The nucleic acid cassette of claim 15, wherein the DNA binding protein comprises a zinc finger.
18. The nucleic acid cassette of claim 15, wherein the DNA binding protein is a nuclease-deactivated zinc finger protein.
19. The nucleic acid cassette of claim 15, wherein the DNA binding protein is linked to a transcriptional activator domain.
20. The nucleic acid cassette of claim 15, wherein the DNA binding protein is linked to a transcriptional repressor domain.
21. The nucleic acid cassette of claim 15, wherein the DNA binding protein is a transcriptional activator that modulates an endogenous SCN1A gene.
22. The nucleic acid cassette of claim 14, wherein the nucleic acid cassette is an adeno-associated virus (AAV) vector.
23. The nucleic acid cassette of claim 22, wherein the AAV vector is AAV9 or scAAV9.
24. The nucleic acid cassette of claim 15, wherein the nucleic acid cassette is an adeno-associated virus (AAV) vector.
25. The nucleic acid cassette of claim 24, wherein the AAV vector is AAV9 or scAAV9.
26. The nucleic acid cassette of claim 16, wherein the nucleic acid cassette is an adeno-associated virus (AAV) vector.
27. The nucleic acid cassette of claim 26, wherein the AAV vector is AAV9 or scAAV9.
28. The nucleic acid cassette of claim 21, wherein the nucleic acid cassette is an adeno-associated virus (AAV) vector.
29. The nucleic acid cassette of claim 28, wherein the AAV vector is AAV9.
30. The nucleic acid cassette of claim 28, wherein the AAV vector is scAAV9.
31. A method for selective expression of a transgene in parvalbumin (PV)-expressing cells, the method comprising: contacting a population of cells comprising PV-expressing cells and non-PV-expressing cells with a nucleic acid cassette comprising a regulatory element comprising a nucleic acid sequence with at least 95% sequence identity to SEQ ID NO: 30 operably linked to the transgene, thereby selectively expressing the transgene in PV-expressing cells.
32. The method of claim 31, wherein the transgene is one or more of SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, and STXBP1.
33. The method of claim 31, wherein the regulatory element is less than 2.5 kb in size.
Description
CROSS-REFERENCE
[0001] This application is a continuation application of U.S. patent application Ser. No. 16/153,433, filed Oct. 5, 2018, which is a continuation application of International Application No. PCT/US2018/025940, filed Apr. 3, 2018, which claims the benefit of U.S. Provisional Application No. 62/480,998, filed Apr. 3, 2017, each of which is incorporated herein by reference in its entirety.
SEQUENCE LISTING
[0002] The instant application contains a Sequence Listing which has been submitted electronically in ASCII format and is hereby incorporated by reference in its entirety. Said ASCII copy, created on Mar. 29, 2018, is named 46482-704_601_SL.txt and is 78,248 bytes in size.
BACKGROUND OF THE DISCLOSURE
[0003] Gene therapy has long been recognized for its enormous potential in how we approach and treat human diseases. Instead of relying on drugs or surgery, patients, especially those with underlying genetic factors, can be treated by directly targeting the underlying cause. Furthermore, by targeting the underlying genetic cause, gene therapy has the potential to effectively cure patients or provide sustained treatment over a longer period of time. Yet, despite this, clinical applications of gene therapy still require improvement in several aspects. One area of concern is off target effects. An attractive approach to address off target effects is to target gene expression of gene therapy to cell type(s) or tissue(s) of interest, or the target cell type(s) or tissue(s). As such, there is a need to identify elements and methods of use thereof for targeting gene therapy or gene expression to a tissue or cell type of interest.
SUMMARY OF THE DISCLOSURE
[0004] There exists a considerable need for targeting gene therapy and gene/transgene expression thereof to the desired tissue and/or cell type in vivo, which can decrease off-target effects, increase therapeutic efficacy in the target tissue and/or cell type, and increase patient safety and tolerance by lowering the effective dose needed to achieve efficacy.
[0005] Provided herein are compositions and methods for selective expression of a transgene in a target tissue or cell type over one or more non-target tissue or cell types. Compositions and methods for selective expression of a transgene comprise one or more regulatory elements (REs) which, when operably linked to a transgene (e.g., an ion channel subunit or a neurotransmitter regulator, or a syntaxin-binding protein), can facilitate or result in selective or preferential expression of the transgene in a target tissue or cell type (e.g., parvalbumin (PV) neurons) as compared to one or more non-target cell types (e.g., non-PV cells). In some cases, the REs are non-naturally occurring sequences. In some cases, the REs are human-derived regulatory elements. In some cases, the REs comprise a sequence from a non-human species, such as a monkey, or a dog, or a rabbit, or a mouse. In some cases, the compositions described herein are delivered into a cell in vivo, ex vivo, or in vitro using a viral vector and/or virus particles, such as adeno-associated virus (AAV) or lentivirus. In some cases, the compositions described herein are delivered into a cell as gene therapy. Also contemplated herein are methods and compositions for treating a neurological condition or disorder associated with a genetic defect in the CNS. In some cases, the relevant cell type or tissue affected by the genetic defect is a PV cell. In some instances, the neurological condition or disease is Dravet syndrome, Alzheimer's disease, epilepsy, and/or seizures. In some cases, the neurological condition or disease is a psychiatric disorder (e.g., schizophrenia, obsessive compulsive disorder, addiction, depression, anxiety, psychosis); an autism spectrum disorder (e.g., Fragile X syndrome, Rett syndrome); epilepsy (e.g., chronic traumatic encephalopathy, generalized epilepsy with febrile seizures plus (GEFS+), epileptic encephalopathy, temporal lobe epilepsy, focal epilepsy, tuberous sclerosis); or neurodegeneration (e.g., Alzheimer's disease, Parkinson's disease). In some cases, the neurological condition or disease is any seizure and/or epilepsy related condition or disease wherein PV neurons are implicated.
[0006] In one aspect, the present disclosure contemplates a nucleic acid cassette comprising one or more regulatory elements operably linked to a transgene that results in selective expression in any target cell type, e.g., PV neurons in the CNS, over one or more non-target cell types, or non-PV cells in the CNS. In some cases, each regulatory element comprises (i) a sequence of SEQ ID NOs: 1-32, (ii) a functional fragment or a combination thereof, or (iii) a sequence with at least 80%, at least 85%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99% sequence identity to (i) or (ii). In some cases, sequence identity is determined using BLAST. In some cases, at least one of the regulatory elements is human derived. In some cases, at least one of the regulatory elements is derived from a non-human mammal. In some cases, the regulatory elements are non-naturally occurring. In some cases, the regulatory elements result in selective expression of the transgene in PV neurons that is greater than expression of the same transgene when operably linked to a non-selective regulatory element, as measured by a co-localization assay. In some cases, the non-selective regulatory element is a constitutive promoter. In some cases, the non-selective regulatory element is any one of CAG, EF1.alpha., SV40, CMV, UBC, PGK, and CBA. In some instances, the regulatory elements result in selective expression of the transgene in PV neurons at a level that is at least 1.5 fold, at least 2 fold, at least 3 fold, at least 4 fold, at least 5 fold, at least 6 fold, at least 7 fold, at least 8 fold, at least 9 fold, at least 10 fold, at least 15 fold, or at least 20 fold as compared to selective expression of the transgene in PV neurons when operably linked to a non-selective regulatory element, as measured by the co-localization assay. In some cases, the regulatory elements result in selective expression in PV neurons that is at least 2%, at least 5%, at least 10%, at least 15%, at least 20%, at least 25%, at least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, or at least 95% higher than expression in PV neurons when the transgene is operably linked to a non-selective regulatory element. In some cases, the regulatory elements result in selective expression in PV neurons that is about 1.5 times, 2 times, 2.5 times, 3 times, 3.5 times, 4 times, 4.5 times, 5 times, 5.5 times, 6 times, 6.5 times, 7 times, 7.5 times, 8 times, 8.5 times, 9 times, 9.5 times, 10 times, 15 times, 20 times, 25 times, 30 times, 40 times, 50 times, or 100 times higher than expected for natural distribution of PV neurons in the CNS. In some cases, the co-localization assay is an immunohistochemical assay. In some cases, the immunohistochemical assay comprises an anti-PV antibody. In some cases, the co-localization assay is performed as shown in Example 5 below. In some cases, the transgene encodes an ion channel subunit, a neurotransmitter regulator, a DNA binding domain, a gene editing protein, or a variant or a functional fragment thereof. In some cases, the ion channel subunit is an alpha subunit or a beta subunit of a sodium ion channel or a subunit of a potassium ion channel. In some cases, the transgene comprises any one of (i) SEQ ID NOs: 37-43; (ii) a functional fragment thereof; or (iii) a sequence having at least 80% sequence identity to (i) or (ii). In some cases, sequence identity is determined using BLAST. In some cases, the transgene comprises (i) SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, or KV3.3; (ii) a functional fragment thereof; or (iii) a sequence having at least 80% sequence identity to (i) or (ii). In some cases, the transgene is a neurotransmitter regulator that comprises (i) STXBP1, (ii) a functional fragment thereof, or (iii) a sequence having at least 80% sequence identity to (i) or (ii). In some cases, the transgene comprises a DNA binding protein that modulates expression of an endogenous gene. In some cases, the endogenous gene is SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.2, KV3.3, or STXBP1. In some cases, the transgene comprises a DNA binding protein that comprises a DNA binding domain of a DNA binding protein or a DNA cleaving protein (e.g., a nuclease, a restriction enzyme, a recombinase, etc.) wherein the DNA cleaving domain or nuclease domain has been deactivated, e.g., a nuclease-deactivated Cas (dCas), a deactivated transcription activator-like effector nuclease, or a nuclease-deactivated zinc finger protein. In some cases, the transgene comprises a DNA binding domain linked to a transcriptional modulating domain (e.g., a transcriptional activator or repressor domain). In some cases, the gene editing protein is a Cas protein. In some cases, the regulatory elements combined are less than 2.5 kb, less than 2 kb, less than 1.5 kb, less than 1 kb, or less than 500 bp in size. In some cases, the non-PV cells comprise one or more of non-PV cell types in the CNS. In some cases, the non-PV cells comprise one or more of excitatory neurons, dopaminergic neurons, astrocytes, microglia, and motor neurons. In some cases, the nucleic acid cassette is a linear construct. In some cases, the nucleic acid cassette is a vector. In some cases, the vector is a plasmid. In some cases, the vector is a viral vector. In some cases, the viral vector is an adeno-associated virus (AAV) vector. In some cases, the AAV vector is AAV1, AAV8, AAV9, scAAV1, scAAV8, or scAAV9. In some cases, the viral vector is a lentiviral vector.
[0007] In one aspect, regulatory elements of any of the nucleic acid cassettes disclosed herein contain less than 600 bp of contiguous sequence from within 10 kb of the transcription start site of GAD2, GAD1, SYN1, NKX2.1, DLX1, DLXS/6, SST, PV, and/or VIP.
[0008] In one aspect, a method of treating a neurological disorder or condition in a subject in need thereof comprises delivering a therapeutically effective amount of any of the nucleic acid cassette disclosed herein. In some cases, the neurological disorder or condition is a psychiatric disorder (e.g., schizophrenia, obsessive compulsive disorder, addiction, depression, anxiety, psychosis); an autism spectrum disorder (e.g., Fragile X syndrome, Rett syndrome); epilepsy (e.g., chronic traumatic encephalopathy, generalized epilepsy with febrile seizures plus (GEFS+), epileptic encephalopathy, temporal lobe epilepsy, focal epilepsy, tuberous sclerosis); or neurodegeneration (e.g., Alzheimer's disease, Parkinson's disease). In some cases, the neurological disorder or condition is Dravet syndrome or Alzheimer's disease. In some cases, the neurological condition or disease is any seizure and/or epilepsy related condition or disease wherein PV neurons are implicated.
[0009] In one aspect, a method of increasing selective expression of a transgene in PV neurons in CNS comprises contacting a cell with a nucleic acid cassette disclosed herein.
[0010] In some aspects, the present disclosure contemplates a method of targeting expression of any transgene to PV neurons in the CNS, the method comprising operably linking one or more of PV neuron selective regulatory elements to a transgene. In some cases, each of the regulatory elements comprises (i) a sequence of SEQ ID NOs: 1-32, (ii) a functional fragment or a combination thereof, or (iii) a sequence with at least 80%, at least 85%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99% sequence identity to (i) or (ii). In some cases, sequence identity is determined using BLAST. In some cases, the regulatory elements result in selective expression of the transgene in PV neurons that is greater than expression of the same transgene when operably linked to a non-selective regulatory element, as measured by a co-localization assay. In some cases, the immunohistochemical assay comprises an anti-PV antibody (e.g., as described in Example 5 below). In some cases, the non-selective regulatory element is a constitutive promoter. In some cases, the non-selective regulatory element is any one of CAG, EF1.alpha., SV40, CMV, UBC, PGK, and CBA. In some cases, the regulatory elements result in selective expression of the transgene in PV neurons at a level that is at least 1.5 fold, at least 2 fold, at least 3 fold, at least 4 fold, at least 5 fold, at least 6 fold, at least 7 fold, at least 8 fold, at least 9 fold, at least 10 fold, at least 15 fold, or at least 20 fold as compared to a non-selective regulatory element when operably linked to the transgene, as measured by a co-localization assay. In some cases, the regulatory elements result in selective expression in PV neurons that is at least 2%, at least 5%, at least 10%, at least 15%, at least 20%, at least 25%, at least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, or at least 95% higher than expression in PV neurons when the transgene is operably linked to a non-selective regulatory element. In some cases, the regulatory elements result in selective expression in PV neurons that is about 1.5 times, 2 times, 2.5 times, 3 times, 3.5 times, 4 times, 4.5 times, 5 times, 5.5 times, 6 times, 6.5 times, 7 times, 7.5 times, 8 times, 8.5 times, 9 times, 9.5 times, 10 times, 15 times, 20 times, 25 times, 30 times, 40 times, 50 times, or 100 times higher than expected for natural distribution of PV neurons in CNS. In some cases, the transgene is any one of SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, STXBP1, a DNA binding protein, a gene editing protein, or a functional fragment thereof. In some cases, the regulatory elements and the transgene are in an AAV. In some cases, the AAV is AAV1, AAV8, AAV9, scAAV1, scAAV8, or scAAV9.
[0011] In another aspect, the present disclosure contemplates a method of treating a neurological condition or disorder in a subject in need thereof, the method comprising contacting a cell with a nucleic acid cassette comprising: one or more regulatory elements operably linked to a transgene that result in selective expression of the transgene in PV neurons over one or more non-PV cells in CNS. In some cases, each of the regulatory elements comprises (i) a sequence of SEQ ID NOs: 1-32, (ii) a functional fragment or a combination thereof, or (iii) a sequence with at least 80%, at least 85%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99% sequence identity to (i) or (ii). In some cases, sequence identity is determined using BLAST. In some cases, the transgene is a voltage-gated ion channel subunit, or a variant or a functional fragment thereof. In some cases, the subunit is a beta subunit of a sodium ion channel. In some cases, the subunit is an alpha subunit of a sodium ion channel. In some cases, the subunit is of a potassium ion channel. In some cases, the transgene is any one of (i) SCN1A, SCN1B, SCN2B, KV3.1, or KV3.3; (ii) a functional fragment thereof; or (iii) a sequence having at least 80% sequence identity to (i) or (ii). In some cases, the transgene is a DNA binding protein. In some cases, the DNA binding protein modulates an endogenous gene. In some cases, the endogenous gene is SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, or STXBP1. In some cases, the transgene is a DNA binding protein that comprises a DNA binding domain of a DNA binding protein or a DNA cleaving protein (e.g., a nuclease, a restriction enzyme, a recombinase, etc.) wherein the DNA cleaving domain or nuclease domain has been deactivated, e.g., a nuclease-deactivated Cas (dCas), a deactivated transcription activator-like effector nuclease, or a nuclease-deactivated zinc finger protein. In some cases, the transgene comprises a DNA binding domain linked to a transcriptional modulating domain (e.g., a transcriptional activator or repressor domain). In some cases, the transgene is a gene editing protein. In some cases, the gene editing protein is a Cas protein, e.g., Cas9. In some cases, the neurological condition or disorder is associated with a haploinsufficiency or a mutation in any of SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, or STXBP1. In some cases, the neurological condition or disorder is epilepsy, neurodegeneration, tauopathy, or neuronal hypoexcitability. In some cases, the neurological condition or disorder is Dravet syndrome. In some cases, the neurological condition or disorder is Alzheimer's disease. In some cases, the neurological condition or disease is a psychiatric disorder (e.g., schizophrenia, obsessive compulsive disorder, addiction, depression, anxiety, psychosis); an autism spectrum disorder (e.g., Fragile X syndrome, Rett syndrome); epilepsy (e.g., chronic traumatic encephalopathy, generalized epilepsy with febrile seizures plus (GEFS+), epileptic encephalopathy, temporal lobe epilepsy, focal epilepsy, tuberous sclerosis); or neurodegeneration (e.g., Alzheimer's disease, Parkinson's disease). In some cases, the neurological condition or disease is any seizure and/or epilepsy related condition or disease wherein PV neurons are implicated. In some cases, the regulatory elements of this disclosure result in selective expression of the transgene in PV neurons that is greater than expression of the same transgene when operably linked to a non-selective regulatory element, as measured by a co-localization assay. In some cases, the non-selective regulatory element is a constitutive promoter. In some cases, the non-selective regulatory element is any one of CAG, EFla, SV40, CMV, UBC, PGK, and CBA. In some cases, the regulatory elements result in selective expression in PV neurons at a level that is at least 1.5 fold, at least 2 fold, at least 3 fold, at least 4 fold, at least 5 fold, at least 6 fold, at least 7 fold, at least 8 fold, at least 9 fold, at least 10 fold, at least 15 fold, or at least 20 fold as compared to a non-selective regulatory element when operably linked to the transgene, as measured by a co-localization assay. In some cases, the regulatory elements result in selective expression in PV neurons that is at least 2%, at least 5%, at least 10%, at least 15%, at least 20%, at least 25%, at least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, or at least 95% higher than expression in PV neurons when the transgene is operably linked to a non-selective regulatory element. In some cases, the regulatory elements result in selective expression in PV neurons that is about 1.5 times, 2 times, 2.5 times, 3 times, 3.5 times, 4 times, 4.5 times, 5 times, 5.5 times, 6 times, 6.5 times, 7 times, 7.5 times, 8 times, 8.5 times, 9 times, 9.5 times, 10 times, 15 times, 20 times, 25 times, 30 times, 40 times, 50 times, or 100 times higher than expected for natural distribution of PV neurons in CNS. In some cases, the nucleic acid cassette is in an AAV. In some cases, the AAV is AAV1, AAV8, AAV9, scAAV1, scAAV8, or scAAV9.
[0012] In one aspect, the present disclosure provides a method of treating Dravet syndrome, comprising contacting a cell with an AAV comprising a transgene, wherein the transgene is any one of (i) SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, or a DNA binding protein, (ii) a functional fragment thereof, or (iii) a sequence having at least 80%, at least 85%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99% sequence identity to (i) or (ii). In some cases, sequence identity is measured using BLAST. In some cases, the DNA binding protein modulates an endogenous gene. In some cases, the DNA binding protein is a transcriptional modulator. In some cases, the transgene is a DNA binding protein that comprises a DNA binding domain of a DNA binding protein or a DNA cleaving protein (e.g., a nuclease, a restriction enzyme, a recombinase, etc.) wherein the DNA cleaving domain or nuclease domain has been deactivated, e.g., a nuclease-deactivated Cas (dCas), a deactivated transcription activator-like effector nuclease, or a nuclease-deactivated zinc finger protein. In some cases, the DNA binding domain is linked to a transcriptional modulating domain (e.g., a transcriptional activator or repressor domain). In some cases, the transgene comprises a gene editing protein, e.g., a Cas protein, Cas9. In some cases, the endogenous gene is SCN1A, SNC2A, SNC8A, SCN1B, or SCN2B. In some cases, the AAV further comprises one or more PV neuron selective regulatory elements or one or more regulatory elements disclosed herein operably linked to the transgene. In some cases, each of the regulatory elements independently comprises (i) a sequence of SEQ ID NOs: 1-32, (ii) a functional fragment or a combination thereof, or (iii) a sequence with at least 80%, at least 85%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99% sequence identity to (i) or (ii).
[0013] In another aspect, the present disclosure provides a method of treating Alzheimer's disease, comprising contacting a cell with an AAV comprising a transgene, wherein the transgene is any one of (i) SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, STXBP1, or a DNA binding protein; (ii) a functional fragment thereof; or (iii) a sequence having at least 80%, at least 85%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99% sequence identity to (i) or (ii). In some cases, sequence identity is measured using BLAST. In some cases, the DNA binding protein modulates an endogenous gene. In some cases, the endogenous gene is SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, or STXBP1. In some cases, the transgene is a DNA binding protein comprising a transcriptional modulator. In some cases, the transgene is a DNA binding protein that comprises a DNA binding domain of a DNA binding protein or a DNA cleaving protein (e.g., a nuclease, a restriction enzyme, a recombinase, etc.) wherein the DNA cleaving domain or nuclease domain has been deactivated, e.g., a nuclease-deactivated Cas (dCas), a deactivated transcription activator-like effector nuclease, or a nuclease-deactivated zinc finger protein. In some cases, the DNA binding domain is linked to a transcriptional modulating domain (e.g., a transcriptional activator or repressor domain). In some cases, the transgene comprises a gene editing protein, e.g., a Cas protein, Cas9. In some cases, the AAV further comprises one or more PV neuron selective regulatory elements or one or more regulatory elements disclosed herein operably linked to the transgene. In some cases, each of the regulatory elements independently comprises each of the regulatory elements independently comprises (i) a sequence of SEQ ID NOs: 1-32, (ii) a functional fragment or a combination thereof, or (iii) a sequence with at least 80%, at least 85%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99% sequence identity to (i) or (ii).
INCORPORATION BY REFERENCE
[0014] All publications, patents, and patent applications mentioned in this specification are herein incorporated by reference to the same extent as if each individual publication, patent, or patent application was specifically and individually indicated to be incorporated by reference.
BRIEF DESCRIPTION OF THE DRAWINGS
[0015] The novel features of the invention are set forth with particularity in the appended claims. A better understanding of the features and advantages of the present invention will be obtained by reference to the following detailed description that sets forth illustrative embodiments, in which the principles of the invention are utilized, and the accompanying drawings of which:
[0016] FIG. 1 illustrates the frequency of seizures (seizures per 12 hr interval) in SCN1A heterozygous mice after treatment with a recombinant AAVDJ vector comprising either SCN1B or eGFP operably linked to a regulatory element comprising a sequence of SEQ ID NO: 32. The graph illustrates the mean values at each day of recording with error bars representing the standard error of the mean.
[0017] FIG. 2 illustrates high gamma power (50-100 Hz) of different mice: wild-type control (WT), untreated transgenic APP/PS1 mice (APP/PS1), or transgenic APP/PS1 mice treated with rAAV comprising SCN1B operably linked to a regulatory element comprising a sequence of SEQ ID NO: 32 (APP/PS1+SCN1B).
[0018] FIG. 3A illustrates immunofluorescence co-localization assay of CNS cells from pups following neonatal systemic injections of AAV9 comprising eGFP transgene operably linked to a regulatory element comprising a sequence of SEQ ID NO: 1 or SEQ ID NO: 8. AAV9 comprising eGFP transgene operably linked to CAG was used as a control. Lower row images illustrate eGFP+ cells. Middle row images illustrate PV+ cells, which were stained with an anti-PV antibody. Top row images (merge) illustrate an overlay of PV+, eGFP+ fluorescence (with representative eGFP+ and PV+ cells which are shown as white or light grey cells indicated by arrowheads) and DAPI+.
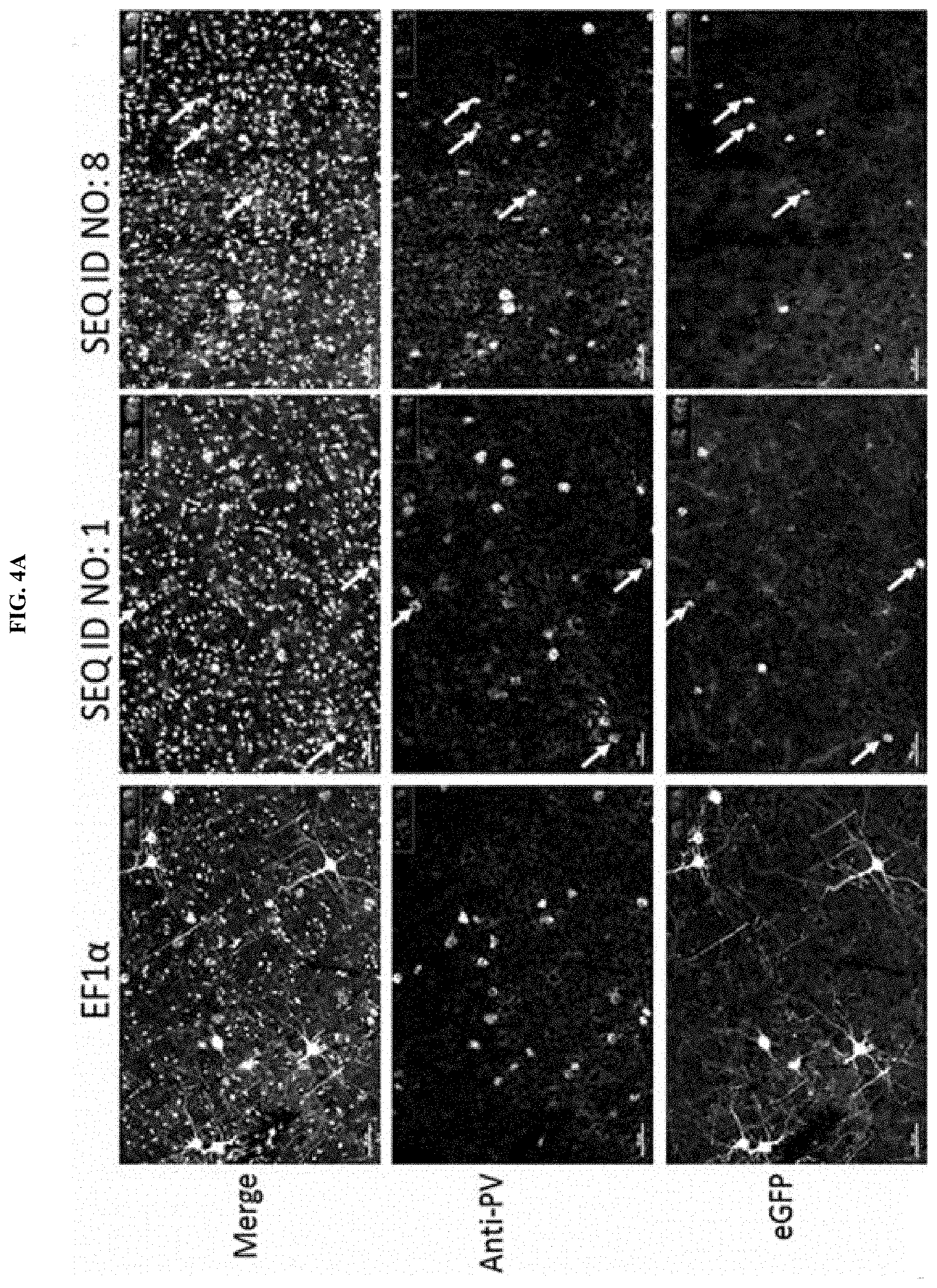
[0019] FIG. 3B illustrates the quantification of immunofluorescence co-localization studies illustrated in FIG. 3A, wherein selective expression in PV cells is expressed as the percentage of eGFP+ cells that were also PV+ in comparison to the CAG control, as measured by the immunofluorescence co-localization assay.
[0020] FIG. 4A illustrates immunofluorescence co-localization assay of CNS cells from adult mice following systemic injections of AAV9 comprising eGFP transgene operably linked to a regulatory element comprising a sequence of SEQ ID NO: 1 or SEQ ID NO: 8. AAV9 comprising eGFP transgene operably linked to EFla was used as a control. Lower row images illustrate eGFP+ cells. Middle row images illustrate PV+ cells, which were stained with an anti-PV antibody. Top row images (merge) illustrate an overlay of PV+eGFP+ fluorescence (with representative eGFP+ and PV+ cells, or the white or light grey cells, indicated by arrowheads) and DAPI+.
[0021] FIG. 4B illustrates the quantification of immunofluorescence co-localization studies illustrated in FIG. 4A, wherein selective expression in PV cells is expressed as the percentage of eGFP+ cells that were also PV+ in comparison to the EF1.alpha. control, as measured by the immunofluorescence co-localization assay.
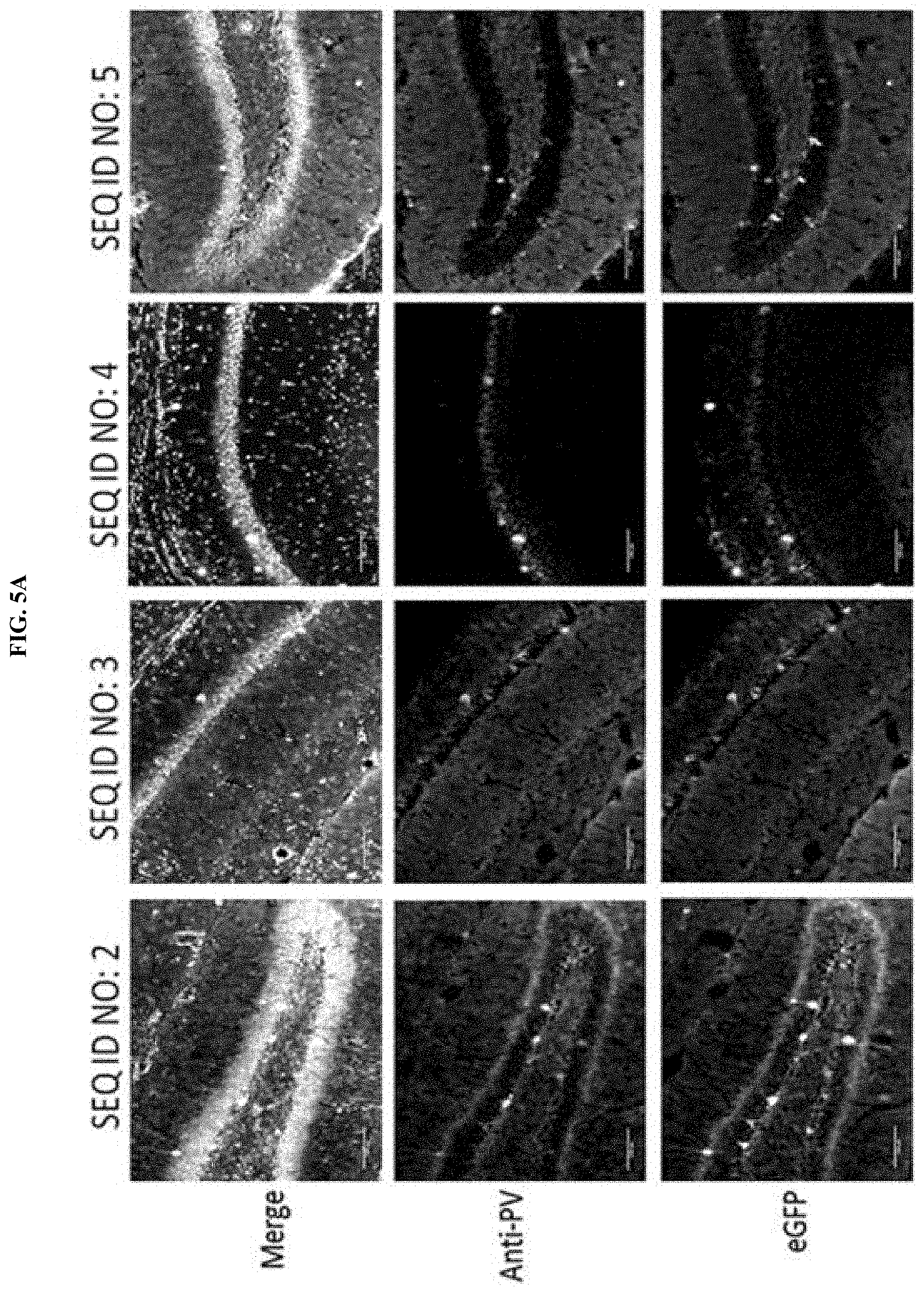
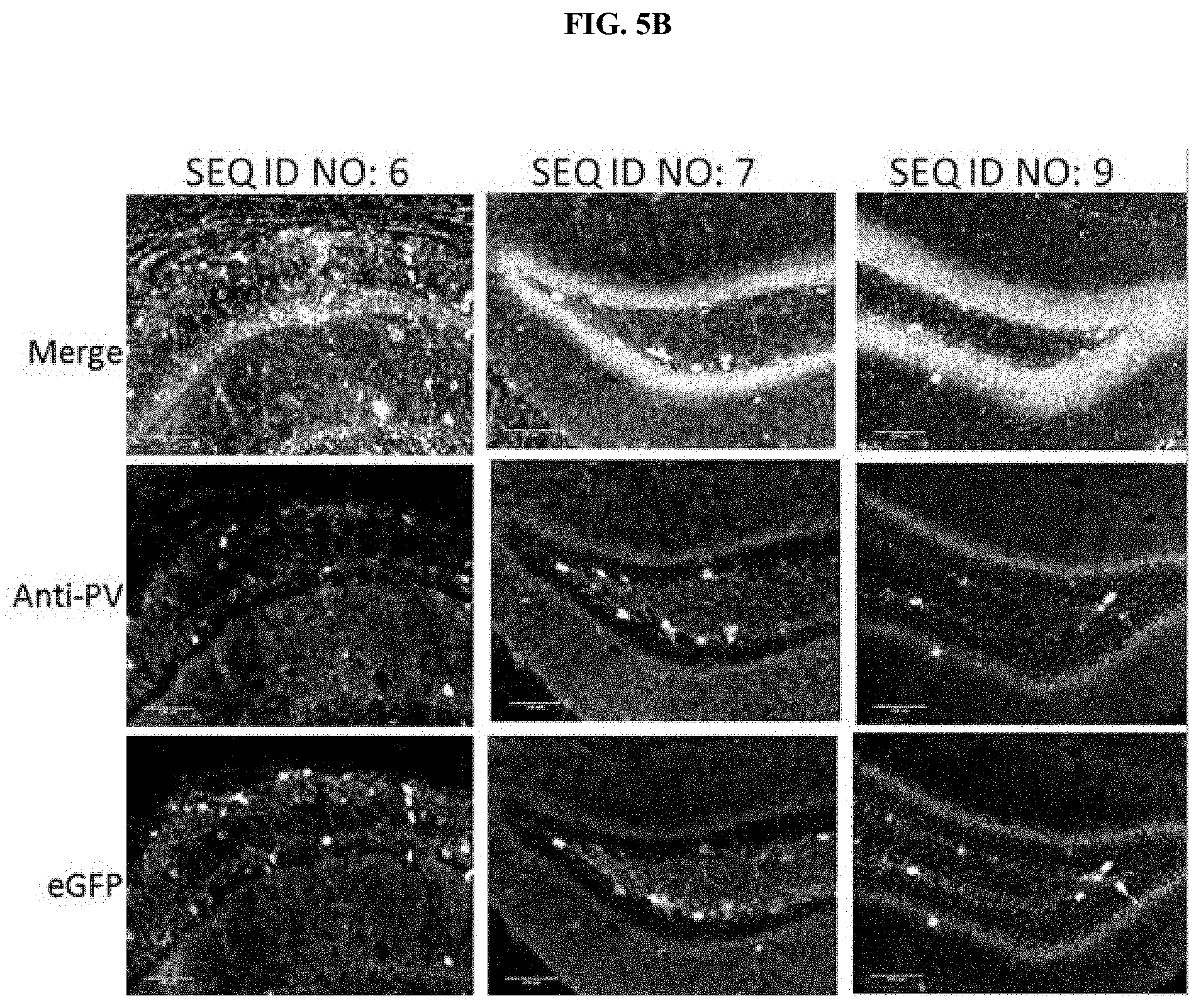
[0022] FIGS. 5A-5F illustrate immunofluorescence co-localization assay of CNS cells from adult mice following direct CNS injections of AAVDJ comprising eGFP transgene operably linked to a regulatory element comprising a sequence of SEQ ID NOs: 2-22. Lower row images illustrate eGFP+ cells. Middle row images illustrate PV cells that were stained with an anti-PV antibody. Top row images (merge) illustrate an overlay of PV+, eGFP+ fluorescence (with representative eGFP+ and PV+ cells, or the white or light grey cells, indicated by arrowheads) and DAPI+. FIG. 5A illustrates the immunofluorescence co-localization assay performed with AAVDJ comprising one of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, and SEQ ID NO: 5 operably linked to eGFP. FIG. 5B illustrates the immunofluorescence co-localization assay performed with AAVDJ comprising one of SEQ ID NO: 6, SEQ ID NO: 7, and SEQ ID NO: 9 operably linked to eGFP. FIG. 5C illustrates the immunofluorescence co-localization assay performed with AAVDJ comprising one of SEQ ID NO: 10, SEQ ID NO: 11, SEQ ID NO: 12, and SEQ ID NO: 13 operably linked to eGFP. FIG. 5D illustrates the immunofluorescence co-localization assay performed with AAVDJ comprising one of SEQ ID NO: 14, SEQ ID NO: 15, SEQ ID NO: 16, and SEQ ID NO: 17 operably linked to eGFP. FIG. 5E illustrates the immunofluorescence co-localization assay performed with AAVDJ comprising one of SEQ ID NO: 18, SEQ ID NO: 19, SEQ ID NO: 20, and SEQ ID NO: 21 operably linked to eGFP. FIG. 5F illustrates the immunofluorescence co-localization assay performed with AAVDJ comprising SEQ ID NO: 22 or SEQ ID NO: 34 operably linked to eGFP, wherein SEQ ID NO: 34 is a previously characterized non-selective regulatory element and was used as a control for comparison.
[0023] FIG. 6 illustrates the quantification of immunofluorescence co-localization studies illustrated in FIGS. 5A-5F, wherein selective expression in PV cells is expressed as the percentage of eGFP+ cells that were also PV+ in comparison to SEQ ID NO: 34, as measured by the immunofluorescence co-localization assay.
[0024] FIG. 7 illustrates a schematic of an example of an expression cassette containing REs of this disclosure, e.g., an enhancer, a promoter, and stability elements. REs can be located upstream and/or downstream of a transgene in an expression cassette, which can be a plasmid, vector, or a viral vector.
DETAILED DESCRIPTION OF THE DISCLOSURE
[0025] The present disclosure contemplates compositions and methods of using such compositions in gene therapy to treat a disease or condition associated with the central nervous system (CNS), e.g., Dravet syndrome, Alzheimer's disease, epilepsy, and/or seizures.
[0026] Gene therapy can replace, modify, delete, or add a gene or a specific nucleic acid sequence, such as an expression cassette, to impart a therapeutic effect in a cell. In some cases, gene therapy is used to deliver an expression cassette into a cell that produces or results in a therapeutic effect. In some cases, a virus, such as AAV, comprising a viral vector that comprises an expression cassette can be used to deliver a transgene into a cell. The expression cassette can contain a transgene that provides a therapeutic effect when expressed in a cell.
[0027] One challenge in gene therapy is ensuring that the transgene is expressed in an appropriate cell type of interest, or the target cell type, to effect or target gene expression. Traditional methods for targeting gene therapy have often relied on delivery methods and/or vehicles (e.g., varying the viruses used or capsid sequences of viruses). In addition to targeting, or selective expression, of an expression cassette in the target cell type over one or more non-target cell types, another challenge in the field is increasing gene expression, especially when the gene is large, in a target cell type or tissue to exert a therapeutic effect.
[0028] The present disclosure provides a plurality of regulatory elements, which are non-coding nucleotide sequences, that can be operably linked to any transgene to increase or to improve selectivity of the transgene expression in the CNS, e.g., in PV neurons. By increasing selectivity of gene expression using one or more regulatory elements disclosed herein, one can improve the efficacy of a gene therapy, decrease the effective dose needed to result in a therapeutic effect, minimize adverse effects or off-target effect, and/or increase patient safety and/or tolerance.
[0029] In one aspect, one or more regulatory elements can be operably linked to any transgene in an expression cassette to modulate gene expression in a cell, such as targeting expression of the transgene in a target cell type or tissue (e.g., PV cells) over one or more non-target cell type or tissue (e.g., non-PV CNS cell-types). In some cases, targeting expression of the transgene in a target cell type or tissue includes increased gene expression in the target cell type or tissue. One or more regulatory elements operably linked to a transgene can be part of an expression cassette, which can be a linear or a circular construct, a plasmid, a vector, a viral vector, e.g., a vector of an adeno-associated virus (AAV). Such expression cassette can be adapted for gene therapy or delivery into a subject (e.g., a human, a patient, or a mammal). In some cases, operably linking one or more regulatory elements to a gene results in targeted expression of the gene in a target tissue or cell type in the CNS, such as a parvalbumin (PV) neuron. In some cases, one or more regulatory elements (e.g., SEQ ID NOs: 1-32, or a functional fragment or a combination thereof, or sequences having at least 80%, at least 90%, at least 95%, or at least 99% sequence identity thereto) increase selectivity of gene expression in a target tissue or cell type in the CNS, such as PV neurons. In some cases, a gene therapy comprises one or more regulatory elements disclosed herein, wherein the regulatory elements are operably linked to a transgene and drive selective expression of the transgene in PV neurons.
[0030] In some cases, selective expression of a gene in PV neurons is used to treat a disease or condition associated with a haploinsufficiency and/or a genetic defect in an endogenous gene, wherein the genetic defect can be a mutation in the gene or dysregulation of the gene. Such genetic defect can result in a reduced level of the gene product and/or a gene product with impaired function and/or activity. In some cases, an expression cassette comprises a gene, a subunit, a variant or a functional fragment thereof, wherein gene expression from the expression cassette is used to treat the disease or condition associated with the genetic defect, impaired function and/or activity, and/or dysregulation of the endogenous gene. In some cases, the disease or condition is Dravet syndrome, Alzheimer's disease, epilepsy, neurodegeneration, tauopathy, neuronal hypoexcitability and/or seizures.
[0031] In some cases, the transgene is an ion channel or a neurotransmitter regulator, a DNA binding protein, or a subunit, variant, or functional fragment thereof. In some cases, the transgene is a sodium ion channel alpha subunit, sodium ion channel beta subunit, or a variant or functional fragment thereof. In some cases, the transgene is a potassium ion channel or a subunit thereof. In some cases, the transgene is SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.2, KV3.3, STXBP1, a DNA binding protein (e.g., a DNA binding protein that modulates expression of an endogenous gene), or a variant or functional fragment thereof. In some cases, the transgene comprises a sequence having at least 80%, at least 85%, at least 90%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99% sequence identity to any one of SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.2, KV3.3, STXBP1, a DNA binding protein, or a variant or functional fragment thereof. In some cases, the transgene is a DNA binding protein that modulates expression of an endogenous gene, such as any one of SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.2, KV3.3, and STXBP1.
[0032] As used herein, the singular forms "a", "an" and "the" are intended to include the plural forms as well, unless the context clearly indicates otherwise. Furthermore, to the extent that the terms "including", "includes", "having", "has", "with", or variants thereof are used in either the detailed description and/or the claims, such terms are intended to be inclusive in a manner similar to the term "comprising".
[0033] The term "about" or "approximately" means within an acceptable error range for the particular value as determined by one of ordinary skill in the art, which will depend in part on how the value is measured or determined, i.e., the limitations of the measurement system. For example, "about" can mean within one or more than one standard deviation, per the practice in the art. Alternatively, "about" can mean a range of up to 20%, up to 15%, up to 10%, up to 5%, or up to 1%) of a given value.
[0034] The terms "determining", "measuring", "evaluating", "assessing", "assaying", "analyzing", and their grammatical equivalents can be used interchangeably herein to refer to any form of measurement, and include determining if an element is present or not (for example, detection). These terms can include both quantitative and/or qualitative determinations. Assessing may be relative or absolute.
[0035] The term "expression" refers to the process by which a nucleic acid sequence or a polynucleotide is transcribed from a DNA template (such as into mRNA or other RNA transcript) and/or the process by which a transcribed mRNA is subsequently translated into peptides, polypeptides, or proteins. Transcripts and encoded polypeptides may be collectively referred to as "gene product." If the polynucleotide is derived from genomic DNA, expression may include splicing of the mRNA in a eukaryotic cell.
[0036] As used herein, "operably linked", "operable linkage", "operatively linked", or grammatical equivalents thereof refer to juxtaposition of genetic elements, e.g., a promoter, an enhancer, a polyadenylation sequence, etc., wherein the elements are in a relationship permitting them to operate in the expected manner. For instance, a regulatory element, which can comprise promoter and/or enhancer sequences, is operatively linked to a coding region if the regulatory element helps initiate transcription of the coding sequence. There may be intervening residues between the regulatory element and coding region so long as this functional relationship is maintained.
[0037] A "vector" as used herein refers to a macromolecule or association of macromolecules that comprises or associates with a polynucleotide and which can be used to mediate delivery of the polynucleotide to a cell. Examples of vectors include plasmids, viral vectors, liposomes, and other gene delivery vehicles. The vector generally comprises genetic elements, e.g., regulatory elements, operatively linked to a gene to facilitate expression of the gene in a target. The combination of regulatory elements and a gene or genes to which they are operably linked for expression is referred to as an "expression cassette".
[0038] The term "AAV" is an abbreviation for adeno-associated virus, and may be used to refer to the virus itself or a derivative thereof. The term covers all serotypes, subtypes, and both naturally occurring and recombinant forms, except where required otherwise. The abbreviation "rAAV" refers to recombinant adeno-associated virus, also referred to as a recombinant AAV vector (or "rAAV vector"). The term "AAV" includes AAV1, AAV2, AAV3, AAV4, AAV5, AAV6, AAV7, AAV8, AAV9, AAV10, AAV11, AAV12, rh10, and hybrids thereof, avian AAV, bovine AAV, canine AAV, equine AAV, primate AAV, non-primate AAV, and ovine AAV. The genomic sequences of various serotypes of AAV, as well as the sequences of the native terminal repeats (TRs), Rep proteins, and capsid subunits are known in the art. Such sequences may be found in the literature or in public databases such as GenBank. An "rAAV vector" as used herein refers to an AAV vector comprising a polynucleotide sequence not of AAV origin (i.e., a polynucleotide heterologous to AAV), typically a sequence of interest for the genetic transformation of a cell. In general, the heterologous polynucleotide is flanked by at least one, and generally by two, AAV inverted terminal repeat sequences (ITRs). The term rAAV vector encompasses both rAAV vector particles and rAAV vector plasmids. An rAAV vector may either be single-stranded (ssAAV) or self-complementary (scAAV). An "AAV virus" or "AAV viral particle" or "rAAV vector particle" refers to a viral particle composed of at least one AAV capsid protein and an encapsidated polynucleotide rAAV vector. If the particle comprises a heterologous polynucleotide (i.e., a polynucleotide other than a wild-type AAV genome such as a transgene to be delivered to a mammalian cell), it is typically referred to as an "rAAV vector particle" or simply an "rAAV vector". Thus, production of rAAV particle necessarily includes production of rAAV vector, as such a vector is contained within an rAAV particle.
[0039] As used herein, the terms "treat", "treatment", "therapy" and the like refer to obtaining a desired pharmacologic and/or physiologic effect, including, but not limited to, alleviating, delaying or slowing the progression, reducing the effects or symptoms, preventing onset, inhibiting, ameliorating the onset of a diseases or disorder, obtaining a beneficial or desired result with respect to a disease, disorder, or medical condition, such as a therapeutic benefit and/or a prophylactic benefit. "Treatment," as used herein, covers any treatment of a disease in a mammal, particularly in a human, and includes: (a) preventing the disease from occurring in a subject which may be predisposed to the disease or at risk of acquiring the disease but has not yet been diagnosed as having it; (b) inhibiting the disease, i.e., arresting its development; and (c) relieving the disease, i.e., causing regression of the disease. A therapeutic benefit includes eradication or amelioration of the underlying disorder being treated. Also, a therapeutic benefit is achieved with the eradication or amelioration of one or more of the physiological symptoms associated with the underlying disorder such that an improvement is observed in the subject, notwithstanding that the subject may still be afflicted with the underlying disorder. In some cases, for prophylactic benefit, the compositions are administered to a subject at risk of developing a particular disease, or to a subject reporting one or more of the physiological symptoms of a disease, even though a diagnosis of this disease may not have been made. The methods of the present disclosure may be used with any mammal. In some cases, the treatment can result in a decrease or cessation of symptoms (e.g., a reduction in the frequency or duration of seizures). A prophylactic effect includes delaying or eliminating the appearance of a disease or condition, delaying or eliminating the onset of symptoms of a disease or condition, slowing, halting, or reversing the progression of a disease or condition, or any combination thereof.
[0040] The term "effective amount" or "therapeutically effective amount" refers to that amount of a composition described herein that is sufficient to affect the intended application, including but not limited to disease treatment, as defined below. The therapeutically effective amount may vary depending upon the intended treatment application (in vivo), or the subject and disease condition being treated, e.g., the weight and age of the subject, the severity of the disease condition, the manner of administration and the like, which can readily be determined by one of ordinary skill in the art. The term also applies to a dose that will induce a particular response in a target cell. The specific dose will vary depending on the particular composition chosen, the dosing regimen to be followed, whether it is administered in combination with other compounds, timing of administration, the tissue to which it is administered, and the physical delivery system in which it is carried.
[0041] A "fragment" of a nucleotide or peptide sequence is meant to refer to a sequence that is less than that believed to be the "full-length" sequence.
[0042] A "variant" of a molecule refers to allelic variations of such sequences, that is, a sequence substantially similar in structure and biological activity to either the entire molecule, or to a fragment thereof.
[0043] The term "functional fragment" is intended to include the "fragments", "variants", "analogues", or "chemical derivatives" of a molecule.
[0044] A "functional fragment" of a DNA or protein sequence possesses at least a biologically active fragment of the sequence, which refers to a fragment that retains a biological activity (either functional or structural) that is substantially similar to a biological activity of the full-length DNA or protein sequence. A biological activity of a DNA sequence can be its ability to influence expression in a manner known to be attributed to the full-length sequence. For example, a functional fragment of a regulatory element will retain the ability to influence transcription as the full-length RE.
[0045] The terms "subject" and "individual" are used interchangeably herein to refer to a vertebrate, preferably a mammal, more preferably a human. "Subject" refers to an animal, such as a mammal, for example a human. The methods described herein can be useful in human therapeutics, veterinary applications, and/or preclinical studies in animal models of a disease or condition. In some case, the subject is a mammal, and in some cases, the subject is human.
[0046] The term "in vivo" refers to an event that takes place in a subject's body.
[0047] The term "in vitro" refers to an event that takes places outside of a subject's body. For example, an in vitro assay encompasses any assay run outside of a subject. In vitro assays encompass cell-based assays in which cells alive or dead are employed. In vitro assays also encompass a cell-free assay in which no intact cells are employed.
[0048] Sequence comparisons, such as for the purpose of assessing identities, mutations, or where one or more positions of a test sequence fall relative to one or more specified positions of a reference sequence, may be performed by any suitable alignment algorithm, including but not limited to the Needleman-Wunsch algorithm (see, e.g., the EMBOSS Needle aligner available at www.ebi.ac.uk/Tools/psa/emboss needle/, optionally with default settings), the BLAST algorithm (see, e.g., the BLAST alignment tool available at blast.ncbi.nlm.nih.gov/Blast.cgi, optionally with default settings), and the Smith-Waterman algorithm (see, e.g., the EMBOSS Water aligner available at www.ebi.ac.uk/Tools/psa/emboss water/, optionally with default settings). Optimal alignment may be assessed using any suitable parameters of a chosen algorithm, including default parameters.
[0049] In general, "sequence identity" or "sequence homology", which can be used interchangeably, refer to an exact nucleotide-to-nucleotide or amino acid-to-amino acid correspondence of two polynucleotides or polypeptide sequences, respectively. Typically, techniques for determining sequence identity include determining the nucleotide sequence of a polynucleotide and/or determining the amino acid sequence encoded thereby, and comparing these sequences to a second nucleotide or amino acid sequence. Two or more sequences (polynucleotide or amino acid) can be compared by determining their "percent identity", also referred to as "percent homology". The percent identity to a reference sequence (e.g., nucleic acid or amino acid sequences), which may be a sequence within a longer molecule (e.g., polynucleotide or polypeptide), may be calculated as the number of exact matches between two optimally aligned sequences divided by the length of the reference sequence and multiplied by 100. Percent identity may also be determined, for example, by comparing sequence information using the advanced BLAST computer program, including version 2.2.9, available from the National Institutes of Health. The BLAST program is based on the alignment method of Karlin and Altschul, Proc. Natl. Acad. Sci. USA 87:2264-2268 (1990) and as discussed in Altschul, et al., J. Mol. Biol. 215:403-410 (1990); Karlin and Altschul, Proc. Natl. Acad. Sci. USA 90:5873-5877 (1993); and Altschul et al., Nucleic Acids Res. 25:3389-3402 (1997). Briefly, the BLAST program defines identity as the number of identical aligned symbols (i.e., nucleotides or amino acids), divided by the total number of symbols in the shorter of the two sequences. The program may be used to determine percent identity over the entire length of the sequences being compared. Default parameters are provided to optimize searches with short query sequences, for example, with the blastp program. The program also allows use of an SEG filter to mask-off segments of the query sequences as determined by the SEG program of Wootton and Federhen, Computers and Chemistry 17: 149-163 (1993). Ranges of desired degrees of sequence identity are approximately 80% to 100% and integer values there between. Typically, the percent identities between a disclosed sequence and a claimed sequence are at least 80%, at least 85%, at least 90%, at least 95%, at least 98%, or at least 99%. In general, an exact match indicates 100% identity over the length of the reference sequence. In some cases, reference to percent sequence identity refers to sequence identity as measured using BLAST (Basic Local Alignment Search Tool). In other cases, ClustalW can be used for multiple sequence alignment.
[0050] Unless otherwise indicated, all terms used herein have the same meaning as they would to one skilled in the art and the practice of the present invention will employ, conventional techniques of molecular biology, microbiology, and recombinant DNA technology, which are within the knowledge of those of skill of the art.
Regulatory Elements
[0051] Regulatory elements are nucleic acid sequences or genetic elements which are capable of influencing (e.g., increasing or decreasing) expression of a gene and/or confer selective expression of a gene (e.g., a reporter gene such as eGFP, a transgene, or a therapeutic gene) in a particular tissue or cell type of interest. In some cases, a regulatory element can be a transgene, an intron, a promoter, an enhancer, UTR, insulator, a repressor, an inverted terminal repeat (ITR) sequence, a long terminal repeat sequence (LTR), stability element, posttranslational response element, or a polyA sequence, or a combination thereof. In some cases, the regulatory element is a promoter or an enhancer, or a combination thereof. In some cases, the regulatory element is derived from a human sequence.
[0052] In some cases, the cell type of interest is a PV neuron. Regulatory elements can function at the DNA and/or the RNA level. Regulatory elements can function to modulate gene expression selectivity in a cell type of interest. Regulatory elements can function to modulate gene expression at the transcriptional phase, post-transcriptional phase, or at the translational phase of gene expression. Regulatory elements include, but are not limited to, promoter, enhancer, repressor, silencer, and insulator sequences. At the RNA level, regulation can occur at the level of translation (e.g., stability elements that stabilize mRNA for translation), RNA cleavage, RNA splicing, and/or transcriptional termination. In some cases, regulatory elements can recruit transcriptional factors to a coding region that increase gene expression selectivity in a cell type of interest. In some cases, regulatory elements can increase the rate at which RNA transcripts are produced, increase the stability of RNA produced, and/or increase the rate of protein synthesis from RNA transcripts. In some cases, regulatory elements can prevent RNA degradation and/or increase its stability to facilitate protein synthesis. In some cases, regulatory elements suppress transcription and/or translation processes in off-target cell-types. In some cases, off-target cell-types include, but are not limited to, excitatory neurons, non-PV CNS cell-types, and non-neuronal CNS cell types.
[0053] Various assays including, but not limited to, DNAase hypersensitivity, ATAC-Seq, and ChIP-Seq can be used to identify putative non-coding regulatory elements (REs). The enzymatic reaction in each of these assays preferentially targets open/accessible chromatin states, a state which is thought to be predictive of regulatory elements. To discover cell-type selective regulatory elements, one can assay for open chromatin sequence for target cell-type of interest (e.g., parvalbumin neurons) and compare that to open chromatin sequences for non-target cell types (e.g., excitatory neurons). Additional filters can be applied to further refine target selection, including proximity to a cell-type selective gene, species conservation, and/or sequence motifs, such as transcription factor binding sites. DNA sequences that are uniquely identified in the target cell type can be synthesized and cloned into an expression vector. The selectivity of a regulatory element can be determined using immunohistochemical methods to quantify co-localization to known cell-type selective proteins.
[0054] For example, one method of isolating a cell-type selective regulatory element includes isolating nuclei from a brain tissue or cell type of interest from an animal model, which can be achieved by using an affinity purification method that isolates the tissue or cell type of interest (e.g., using beads coated to an anti-PV antibody for isolating PV neurons), using high-throughput natural priming and DNA synthesis to generate a pool of sequences from open chromatin regions in the nuclei, sequencing the pool of sequences to identify putative sequences that drive gene expression in the tissue or cell type of interest, and verifying selective expression in a reporter system in a cell line in vitro and/or in an animal model.
[0055] Another method for identifying candidate regulatory elements that are selective in a tissue or cell type of interest include using R26-CAG-LSL-Sunl-sfGFP-Myc knockin mouse for harvesting the tissue or cell type of interest, isolating GFP+/Myc+nuclei from the mouse neocortex of this strain using affinity purification, e.g., using anti-GFP or anti-Myc antibodies and protein G-coated magnetic beads to isolate nuclei from the neocortex. Nuclear RNA from purified nuclei or whole neocortical nuclei can be converted to cDNA and amplified with the Nugen Ovation RNA-seq System V2 (Nugen 7102), followed by sequencing using the Illumina HiSeq 2500. Genomic DNA from purified nuclei can be fragmented and used to make MethylC-seq libraries, which can be sequenced using the Illumina HiSeq 2000. To generate an ATAC-seq library, nuclei bound to beads are transposed using Tn5 transposase (Illumina FC-121-1030). After 9-12 cycles of PCR amplification, libraries are sequenced using an Illumina HiSeq 2500. To generate a ChIP-seq library, excitatory neuron nuclei can be digested to mononucleosomes using micrococcal nuclease, followed by salt extraction of chromatin, and native ChIP and library construction, which can be sequenced on an Illumina HiSeq 2500. After sequencing these libraries, the sequences are mapped to identify, for example, correlation in cell-type-specific hypo-methylation in CG-rich regions, histone modifications, transcriptional factor binding sites, and patterns associated with highly expressed transcriptional factors. Overlapping features and correlations from multiple assays and/or libraries described above provide evidence for identifying candidate sequences within such genomic regions as potential regulatory elements associated with selective expression and/or high expression in the cells isolated from the neocortex. For example, a genomic region characterized by a strong overlap between hypomethylation detected in the methy 1C-seq library, ChIP assay, and an enrichment in transcription factor binding motifs in the same region provide convergent data that indicate the genomic region contains a sequence of a putative regulatory element selective for the tissue or cell type isolated. As another example, to identify candidate PV neuron selective regulatory elements, one can isolate PV neurons and purify nuclei from the isolated PV cells so that genomic sequences that are identified as active in multiple sequencing assays described above have a high likelihood of being PV cell-selective regulatory elements, e.g., a genomic region that is identified as active in an ATAC-seq assay (corresponding to regions of open chromatin), active in RNA-seq (indicative of active gene expression and low DNA methylation patterns in the region), and active in methylC-seq assay (which generates single-base resolution methylome maps from a cell type of interest).
[0056] Once candidate genomic regions are identified as selectively active in a cell type of interest, sequences within the region can be generated using PCR methods and tested in additional assays in vitro and/or in vivo to validate tissue or cell type selectivity of the sequences. Such validation assays include immunohistochemical co-localization assay, wherein an antibody or any detectable marker is used to label the cell type of interest and a second detectable marker, e.g., a fluorescent transgene, is operably linked to the putative regulatory elements. Expression cassettes comprising such elements are delivered into cells in vitro and/or in vivo. Selective expression driven by one or more putative regulatory elements can be validated by measuring the overlap between the cell type of interest (as measured by the detectable signal or fluorescence from its labeled marker, e.g., an anti-PV antibody) and the second detectable marker corresponding to expression of the transgene (e.g., eGFP or RFP) operably linked to the regulatory elements. An overlap in the signals from both detectable markers indicates cell-type selectivity in the labeled cell type if the amount of overlap observed is higher than the overlap observed when the regulatory elements are replaced with a control, such as CAG, EF1.alpha., a constitutive promoter (e.g., SV40, CMV, UBC, PGK, and CBA), a non-selective regulatory element, or a previously characterized non-selective regulatory element. Various mouse strains adapted for expressing a detectable marker in a cell type of interest allows validation of cell type selectivity of a regulatory element in vivo. For example, a number of mouse lines that express Cre in a particular cell type can be used because cell-type selective Cre expression can drive Cre-induced expression of a fluorescent protein, such as RFP, in a cell type of interest. Labeling such cell type of interest in vivo allows one to determine level of cell-type selective expression that is associated with a putative regulatory element operably linked to a fluorescent or reporter transgene in the same mouse. Similar to the co-localization assay, an overlap of the signals from both markers that exceeds the overlap detected for CAG, EF1.alpha., a constitutive promoter (e.g., SV40, CMV, UBC, PGK, and CBA), or a non-selective regulatory element is indicative of cell type selectivity for the regulatory elements tested. In some cases, the mouse strain used is B6 PV-Cre mouse (Jackson Laboratory), which is a B6 PV-Cre knock-in mouse that expresses Cre recombinase in parvalbumin-expressing neurons (e.g., interneurons in the brain and proprioceptive afferent sensory neurons in the dorsal root ganglia), without disrupting endogenous Pvalb expression.
[0057] Upon validation of cell type selectivity of a regulatory element for a particular cell type, sequences of such regulatory elements can be varied using various mutagenesis methods, e.g., error-prone PCR methods, to improve its selectivity. In some cases, two or more regulatory elements having cell selectivity can be combined. In some cases, combined regulatory elements exhibit enhanced cell-type selectivity in driving gene expression in the cell type of interest. In some instances, such regulatory elements are truncated one or more bases at a time to determine the minimal amount of sequence that retains its cell type selectivity. Smaller regulatory elements that retain cell type selectivity are helpful for making gene therapy comprising a large transgene, or where the cloning capacity of a vector or plasmid is limited in view of the size of a transgene that one wishes to deliver using gene therapy.
[0058] The present disclosure provides a plurality of nucleotide sequences that are regulatory elements. In some cases, any one or more of the regulatory elements disclosed herein result in increased selectivity in gene expression in a parvalbumin cell. In some cases, regulatory elements disclosed herein are PV-cell-selective. In some cases, PV cell selective regulatory elements are associated with selective gene expression in PV cells more than expression in non-PV CNS cell-types. In some cases, PV cell selective regulatory elements as associated with reduced gene expression in non-PV CNS cell types.
[0059] Non-limiting examples of regulatory elements include SEQ ID NOs: 1-32, as provided in TABLE 1 below.
TABLE-US-00001 TABLE 1 List of nucleic acid sequences disclosed herein. SEQ Source/ ID Genomic NO: Nucleic Acid Sequence Location 1 GGAGGAAGCCATCAACTAAACTACAATGACTGTAAGATACAAAATTGGGA Human; ATGGTAACATATTTTGAAGTTCTGTTGACATAAAGAA hg19: chr2: TCATGATATTAATGCCCATGGAAATGAAAGGGCGATCAACACT 171621900- ATGGTTTGAAAAGGGGGAAATTGTAGAGCACAGATGTGTTCGT 171622580 GTGGCAGTGTGCTGTCTCTAGCAATACTCAGAGAAGAGAGAGA ACAATGAAATTCTGATTGGCCCCAGTGTGAGCCCAGATGAGGTT CAGCTGCCAACTTTCTCTTTCACATCTTATGAAAGTCATTTAAGC ACAACTAACTTTTTTTTTTTTTTTTTTTTTTTGAGACAGAGTCTTG CTCTGTTGCCCAGGACAGAGTGCAGTAGTGACTCAATCTCGGCT CACTGCAGCCTCCACCTCCTAGGCTCAAACGGTCCTCCTGCATC AGCCTCCCAAGTAGCTGGAATTACAGGAGTGGCCCACCATGCC CAGCTAATTTTTGTATTTTTAATAGATACGGGGGTTTCACCATAT CACCCAGGCTGGTCTCGAACTCCTGGCCTCAAGTGATCCACCTG CCTCGGCCTCCCAAAGTGCTGGGATTATAGGCGTCAGCCACTAT GCCCAACCCGACCAACCTTTTTTAAAATAAATATTTAAAAAATT GGTATTTCACATATATACTAGT 2 AGTTTGGACAAGAACTATAGTTCTAGCTTTCTCTGGGTCTCCAC Mouse; CTTGCAGAGAATGCAGCTTTCATTATCTCATGAGCCAAACTCTC mm10; chr2: ATCATCTCTTTCCATATATCTGTCGGTGCTCTTCCATGAGTACTC 36053858- TAACACACACAGAAGGAGCACTTACACAGGCTGTTGTTTTTCTC 36054359 TTATTATCATAGCTGTTGTTCAGACATGTGCATTCTGTTCTTGTT GCTTCAATGCTAAAGGAGTCTCAGGATATGAGAACTGTACCAG CCGAGGCATCAGGAAACATGGGTGGAAATTCCCACAGTACTAT TTGTTCACTGTGTGACCTTGGGCCAGTCACATCCCTTTCCTGAG GCTTCGATTCCCCAAGCTATAAAAGAAGCATCTCTTAACCTTTT TTTAGGTCATGAGTCAGGCCCAGCACACTCTCAGGGAGACTCAT GAGAGTACAGATCATTTCCCATAGAAAAACCATAGTTTTATATC CAGAGGCTTTTCTGTAAG 3 GGTTCCAGTTCAGAGGCAGAGCATTTGGGGTTCCCAGTCAGGA Mouse; GCTTTCCTCTCTCCGCTCCTTAGTTTCCTCTCTTTAAAAAAAAAT chr2: 36,09 GGGTGATAGTATAGAAAGGAAGCTCTGGGCTCGGGGACCAGGG 1,144- CCCTGGGATCCCCGCTCCCAGCCACTCGCTCCTGACCCTTCCAG 36,091,966 GGACAAGCTCCCCCCCACCCCGTCCTTTCCAGGCTGCCACTAGA AGAGATGGGGACGCGTGGTCAGCCGCTTCTGTCGCCCCCCAGG GAACGGTCTCACGCTGGAGGGGGCAGTGCCCTCGGAACAGGAC AGTCAGCCCAAGCCAGCCAAGCGCGCGCGGACGTCCTTCACCG CAGAGCAATTGCAGGTACCCCGGGCAAGCCCCGAAGCGTGTGG GCGGGGCTTCGGAGTGGGCGTGGTTGTTCGGGACTTGTGACTCC GCCCCTTGTGCGGGGACCCGCGTGAGGCCGCTCCAAGGATGAA GCTGCCTGGGGCGTGGCCTCGGACCCTGAGCCTCTGATTGGGCG GAGGTCTCAGGGCCCTTCTGCGCCCCACAGGTTATGCAGGCGCA GTTCGCGCAGGACAACAACCCGGACGCGCAGACGCTGCAGAAG CTGGCGGACATGACGGGCCTCAGTCGCAGGGTCATCCAGGTGG GGCTCCGGGGTCTCGGCCTTCAGGTCTAGGGTGAACCTTAGGGA AGCGCTGAAGCTCGTAGTGGTACGGATGGTCGCGCGTGCACGT GGCCGCCCCTCTCCAGTGTGGCCTAAGGACCCCAGTCGGCACG GGTTGACCCTTTTCCTTGATTACTGAGAGTGCAGAGGCTGT 4 TGGTGGGAAGACATGTCCAGGGAAGAAATGGCCTCCAGAGGCC Mouse; TGAGGTGGGGAAATGCTGGAGGTGGAGAGAGGAACAACTGACT chr2: 36,09 GAAAATGAGCTTCCACTGTGGCTTAGTAGCCTATACCAAGTCTA 5,396- GAGTATAGGGTAGGAGAAGATTAGGAAAGCGATGGGTCTGAGA ATGATGTGGCCTGTTGACTTTTGTAAACCCAAAGCACCTTGGAC TAAACCCTATGAACAGTGTGGTGCCACCAAAGACTATAATGAG CTCAGGGAACAGAATTCTGTGTGCATGGTGATTTTTTTTTTTTTT TTCTGCTAACTGCAGTCTGGGTGATGCATTGACAAACCAATCCT GGAAAGTAAGAGGCAAGGGCAGCTGGGACGGTGAGAGGAGCC TGATGGGAACCAGGCCAAGCAGGGCAGCAGAGGCGATGAAGA GGATGTGGTGCATCCAGAGACTCACTTCATTAGCTGGAGGCACT GCTGGATAGGGTCTGAAGGTTCTGGTATCTGAGTTGGCGGGCTG GGTGAGTGGTGGCTCTGCTTCCTGAACAGTGTGTGCAAGAGGA AACAGGGTTAAGGGCTAGGACAGTCACAGGTGAGTCAGCCTCA CAAGAGCAACCTTCCCCTAGTGCAGA 5 GGAGGTCTCCTTTTGCCCCGGTTCCAACAAGAGAATGCAAGGCT Mouse; GTATCTCAATTTCCTTGAGCCTCTCTGTATTATAGAAGAAAAGT mm10: chr2 AGGGAAGCCATACGCCCCTTCTGAGCTTCAGTGTCTCTCTGTCT CTGCAAATGAGGCTGGGGAGGCTGGGGGCGGGCGTGAAAGAG GCCCGCGCCAAGCCGACCCCCACCTCTGCCCCCTCCCCAGGTCA ACAACCTCATCTGGCACGTGCGGTGCCTCGAGTGCTCCGTGTGT CGCACATCGCTGAGGCAGCAGAATAGCTGCTACATCAAGAACA AGGAGATCTACTGCAAGATGGACTACTTCAGGTAGGCAGCGGC CATCCCGCCAGCAAGCGCTGGAGCATGAACGCCTTGCACACGC GTGCCTAGGCCACTTGTGTGGCCTGTGCTCTCCAATTCCTGAGC CCTGCTGTTCAGAGTGCACAACGCGGCTCAGCGCACTGGCCCG GCCCTCCTACTCAGCACGTCTTACACAGAAGGGAGCGCCAGTCT CAGCCTGAGTTCTGGCGGGGGATCTGCCTCGGGTTCCTCCGATC TGACAGGCGCTGGCCACGGGTCTGGTTCCATCTCTGGTCTTTTC TGGCCCCGAGCACCAGTGTGTTCTGTTGAGCTCTGATGTCCGAG GCTCTGGCCCGGATCA 6 CTCTGGCTACCTCTTATCTTGGGCATTCACGACAATTTCTAATTG Mouse; CAGGTAGTTTGTGTGTGTGCGCGTGTTTTTTTTCCCCCTCAGAGG mm10: chr2: CTTGGATTGCAAAGGAACTAAGCGATTACTTCAAGAGCCACGG 36103286- GTTAAGTGCAGGGAGAGGGGGAGAGAGAGGGAAAAAAACCCA 36104328 ATCCAAATTCAAATTGCTTCATTAGAGAGACACCGCTTTTGTGG GGAAGGGCTTTAAATGCCCACTACAAAGTTAGGACTCATTGTTC AGCGCCGGTTTATATAACAGGCGAGGGGAGGCGCTGGGCTCTG ACAGCTCCGAGCCAGTTCAGCAGCCGCCGTCGCCTGCATTCCCT CCCCCTCCCCCAGGTGATGGCCCAGCCAGGGTCCGGCTGCAAA GCGACCACCCGCTGTCTCGAAGGGACCGCTCCGCCTGCCATGGT GAGTCCTTTCGGTCCTGCTTTCGGCCCCGAGTCCCCCCAACAGC ACAGGCCAGGGCTTCTGGCTCAGCCTTCCGGCTACCAACCTCTA CCCCTGCGCTGGAAAACTGCCGATAGGAGCCGCCTCTCGTTGAG CCTTGGTTTTTCTGGCCTGGAATGTGAGCTTTGGCTGCTTCCTGC ACCCAGGATGCGCTGTGTTAAAAGTTGGGGGCCGTCCCTTCTTC TCCAATAGGTCCTTTCATTCTTGTACTCCAGCCTAGGGCGCGAC ATCCCTGGCACATTTCGGTGTCAGTCGGTGCGCGAGGAAACCA GATTCAACTCTGAGTACTCGGCTAAGCGCTTCGCTGTTCCTCTCT CCCATTTCAGGCTCAGTCAGACGCAGAGGCCTTGGCAGGCGCTC TGGACAAGGACGAAGGTAGAGCCTCCCCATGTACGCCCAGCAC ACCGTCTGTCTGCTCGCCGCCCTCTGCTGCCTCTTCCGTGCCGTC TGCCGGCAAGAATATCTGCTCCAGTTGCGGTCTGGAGATCCTGG ACCGGTATCTGCTCAAGGTGAGTCAGGGTAGGTGTGCCTGCTTG CCCACGGGTGTGGTTTGCAGCCCCAAGAGCTGT 7 CAAGACTTTTAAAAGTTTAGATAAATAAACAAACATTTGACGGC Mouse; TTTCCATCACATCTAGACTATAATCCAAAGATCTATATGGTCCC mm10: chr2: AAACGACTTACACTTAACTACCGTCTCCCATATGGCTTCTTCCC 36114311- CCATCAGTCATTGTCCTCAGCCATAGTGGCCTCCCTGTTCCTTTG 36114817 GGTACAAGGGAACAACTCCCTGAGAGGTTCCATTAGCTGCTGTT GCCTGAGATGCTCTTGAGCCCACACCATCTGCTCATTTCTCTCCT CACGTGTCAGTGATTAAGAGGCTGTCCTTGGCCTCCCGTCAAAA TTACATCCCTGCCGCTTTCCACTTCTTGCCTTCTTATTTTCTAAAT AGAACTAACTCACCACTACCCAACATTCTATATAATTGGATATC TGTCCTCTGTTTAAATATAATGTTGACTTCAAGAAAGAACGTTG TCACTGCCCTGTCACCAGACTTTTAAACAGTGCCTATCGTGTGG CACATGCTCAGTGAAATTG 8 TCAACAGGGGGACACTTGGGAAAGAAGGATGGGGACAGAGCC Mouse; GAGAGGACTGTTACACATTAGAGAAACATCAGTGACTGTGCCA mm10: chr15: GCTTTGGGGTAGACTGCACAAAAGCCCTGAGGCAGCACAGGCA 78179109- GGATCCAGTCTGCTGGTCCCAGGAAGCTAACCGTCTCAGACAG 78179610 AGCACAAAGCACCGAGACATGTGCCACAAGGCTTGTGTAGAGA GGTCAGAGGACAGCGTACAGGTCCCAGAGATCAAACTCAACCT CACCAGGCTTGGCAGCAAGCCTTTACCAACCCACCCCCACCCCA CCCACCCTGCACGCGCCCCTCTCCCCTCCCCATGGTCTCCCATG GCTATCTCACTTGGCCCTAAAATGTTTAAGGATGACACTGGCTG CTGAGTGGAAATGAGACAGCAGAAGTCAACAGTAGATTTTAGG AAAGCCAGAGAAAAAGGCTTGTGCTGTTTTTAGAAAGCCAAGG GACAAGCTAAGATAGGGCCCAAGTAAT 9 AAATAGAACTGTGAGATAGGGGGAGAGGGGGCAGGAAGGACA Mouse; AGAGACCCCTGTCTCATTGTGATCCCCACCTGTCTGCTCTGTGG mm10: chr15: GAGGGTACCCATGAGGGCCAGCCCACAGCCCTTAGGTGGACAT 78195347- TGTCTGGTCCTGTCTCACTGTCCCTCCCAGCAGCCCCAGAGGCC 78196134 AGGAGACAGGGGTCTCAGTCCTCACTGAGAGATGTGTAAACTG AGGCCCAGTGAATGTTGAGGGCCAGGGCATGCCCTTGGTGGGA TGTGACCTGGGTCTCCTTCGCACGGGCTTCCTCCCCGAAGCCGA GCTGAGCATTTGGAGTTTGAAATGTTTCCGTACTTAGCAATCTG CTCCTCTATTCCCGGGCGGACTTCCGATAGCTCCGGCCTTATGC TGCACTAGATAAGATGGAGCAGGGAGAGGACACGGCACTACTT ATGTAACCGGCCTCTTGAAAAATGGAGCAGCGGTCAGGGCGGA ACAAGACGTCCTCTCTCTACGCATCCCTCTCCTTTCCCTGCTAAG GCTGCAGCTGGAGTCAGAGGCAGGGCTGTTCCAATCTGTCTTTG ATCAGTAACGCAGCCAGCCTCCAGCCTCCGTCAGCCTCCTCATG GCTGAGACCCGGCCTCAGTTTCCCCCACTTACATCCCGAGGATC AGAGCCTGTGAGGATGAAATGGGATAAGGTAGCTGGAACCGTC TGGCAGAGAGCGAGTCCTCAGGACTGTTGATGCCTGTGGCTGCC TGGCTTGACCCCAAGTGACCCCGCCTCCTCATCCTGCAGCAGGA GAA 10 TCTATAGAATGTGTCCCCAGCCTTGTTTTCCACACTTGATACGC Mouse; AAGGAATGCATACCACAGAGAGGGATGAGGGTAGCATCCAGCC mm10: chr15: TGCTTCCTGTGTGTCGGGGCGCTACAGCCACATCTCCCCAGTCC 78196305- ATCTCAGACCGTCACAGAGCTTCGCCGAATGTATAGCTTTGTTC 78196806 TCTGTGCAGACAGGGAGACAGAGCCTTGGGAAGCATAGGTGCT TGCTTCTTTGCCCACTGAGTCTTAGCTGGACTTGCACACCACAT GCCTCACAGCCGGGCGCACTTGCATTTGTCACCCAGGCCCAGTG ATGATGGCTCTGCTTGCTTTGTGCTTTGTGCCAACTACAGCTCCA GCACCTGTGCCCTGGGTTTTCACTCCTTTAGTTGAACACGTAGTT ACTGGGGTTGTAGGGATGGAGCCTTTCTGCTTCCTTCTGGCAAA GTCCTTAGCGGCCTGCTGCGGGGGTGGGGGGTGTTCAGGGGAG TGGTGATGAAGTATGACAG 11 TCTCCAGTTGGAGAAACAGATGCTGTAACTGGGGCCACAGTAT Mouse; AAAGAGAGCCCAGACATTGAACTGTCAACACAGAAGCCTGGCA mm10: chr15: CACTGGAACTGGCAGTCCAGCTGGGAACAAGGGGTAGAGGCTG 78205234- AGGCCACTAAGTCAACTGAGGCAGGAGACATAGGAGCTAAAGC 78205766 AGCTGAAGGGTGCAGGACAGCTGGGGGGTCTGAAGTGGGCCTC ATGCCCAGAGCTATGAAGTCAGGGGCTGTAGCCTAGGAGCCTT GGAAGCCAGCTGGCAAGCTGTGGCCCAAAGACGCTGACTCACC AGGAGGGGGCAGCTGGAGCCAGGCACTCCTAAGGTTTCCAGGA AGGGCAGCCTTCCAGGGCTCAGCTAGGGGAGACAGTGTTGACA GCAAGTTGTCAGGCAACTTGAGCTACTGGGCAGCTGGGAAGCT GTCCCTTGGTCCCCAGTATCATCATCACCCCAGACGCTGCCCAC CTGCCTCAGGTCCCACACAGTGATCCTCCCATCTTTAACACAAC ACATGACCAGAGAGA 12 GTCACCCTCCCCCCAAACAACCCCTTCTTCTCTGGTTCGAGAAA Mouse; TTACAGGCATGAAAGATATAAATCGGGATGCTTGACTTGGGAA mm10: chr15: TATAAATCACTAAAGCTTGGGGGCAGGGGTGGGCGACCTTTGT 78224841- GACCGTCCTTGTGCGTGCCAGTAAATCCTGTGGTCCAGGGGAGA 78225364 AGAAAAGGCTGTGTGGCTTCTGCTCACAAAGCTGCAGAAACCA TTCTTTAAGCCCAAAAGCACTTCCAGAGAGAGCAGAGCATCCC CAGGCTGCTGGCTCAGCAAGTTCACTGTGCTCAATCTCAGGAAG TGAGGATAAGAGCAGTGCCTGGAGAGTGCCTGGTGCTGAGCTG AGGGTTTCTGAACACATTAAAGCGGGGAGCATGGACCGGGCCT CAGGAGGGGTGTTGAACATCCCTAGGCAGAGGAGTCTAGCTTC CTGGGAAAAGATATCAGGTTAAGCACACACATGTCCTCTGGAA TAAGATAATCTTTCTGATCACACACTATACACACACAAAAGCCT GCTC 13 GCCCTCTAGGCCACCTGACCAGGTCCCCTCAGTCCCCCCCTTCC Mouse; CACACTCCCACACTCAGCCCCCCTCCCCCCCCCCCGACCCCTGC mm10: chr15: AGGATTATCCTGTCTGTGTTCCTGACTCAGCCTGGGAGCCACCT 78241348- GGGCAGCAGGGGCCAAGGGTGTCCTAGAAGGGACCTGGAGTCC 78241856 ACGCTGGGCCAAGCCTGCCCTTTCTCCCTCTGTCTTCCGTCCCTG CTTGCGGTTCTGCTGAATGTGGTTATTTCTCTGGCTCCTTTTACA GAGAATGCTGCTGCTAATTTTATGTGGAGCTCTGAGGCAGTGTA ATTGGAAGCCAGACACCCTGTCAGCAGTGGGCTCCCGTCCTGA GCTGCCATGCTTCCTGCTCTCCTCCCGTCCCGGCTCCTCATTTCA TGCAGCCACCTGTCCCAGGGAGAGAGGAGTCACCCAGGCCCCT CAGTCCGCCCCTTAAATAAGAAAGCCTCCGTTGCTCGGCACACA TACCAAGCAGCCGCTGGTGCAATCT 14 GTGTTCTTCCCTTCCCCTTTGGACCCCCGAGACAAGCCAATAAA Mouse; ATACTCGGCAGGGTGGCTTCTCTCCTTTTTTTGCCAGTAATAAA mm10: chr98- CAGACTCAGAGCAAGTTAAGGGTCTGGTCCAAGGTCATGGCTG 107340928- GGATCAGTGACAGAGCCCAGAAGAGAACCTGAGACTTCTTGCT 107341325 GAGCCAAGCTGGAGAGGACAGAAAGGAATGCGTCTACTCCATG CATGACCCTCTGCCAGCTTTGCTCCTTCCTAAGGGACCATGAAC GATATGTGCACACCGCTCATACGTATGTGCACACCTGCAAGAG GAGGCATCCCATGTACACCTATGAGACGCACAGAGAAACATAT ATGTAGCCATAGGCTAGAAATTCTTTCTCTTTCTAGGTCTGCCCC TCTGCA 15 GGACCACTCAGTGTACACGGAATGTAGAATTGAGTCTGCCATTG Mouse; GTCTTCCCTCAAAGTCTTGGAGGCTTGGGACTGATATTGGGAGC mm10: chr9: ATCTGGGCAGAGAAGGCCACAAAGACAGGGTGGTTTTTCTACA 107349227- CTGGGACATACTCGTGAGCATGCACAGAGGCGTGTCCCCAACTT 107350036 CCCTGTCACCCCTGTCCTCTGCCGGCTAGAGGGGATGCGGGGGT GGACATATGCTGCTATTGGGCAGATATCACATGTTAAGAGGTGG GGGGGGGCTCAAGAGGCGGAGGGCTAGGAGCATCCCATGGGG AGAGGTTCTGGTTTTCTTGCTGCCTCTAGCTGCTATAAATACGTT AGCACTTGAGCAACTGGAAAGCTCTGAGTAATTTAGGATGCAC AAAGCTGTAATTTAACTCCAGCATCTCAGTGTGCGAGAGCATTA AAGATGTAATTAAGATGTTTACACAAAGAGATTGGAGTCTGTG ACACTTGGGGTGCAAAACCCCAGGAAGGGACACAATGGGTGAG GTGAGGATCTGTGGGAGGCCTGGGGACAGTCACTTGGATCCCA GCTATGAGATGGCAGGCCACCCAGCTGTTTCTCCTTGGAAATGT TTTGGCCTGGGGGTTGGGGGTGGGGCATCACACTTTGATATGGA GATGGGGCAACAAAGCCTGCAATATCTGGGGGTGGAGAGGTCA AGTGGATGGAGTCTTTTGAGATCATGTCAGGAAGAGGGCTCGA TCCCCCAAAATCATGGTGACATATGGTGTCTCGGGGTTCACAGG AGCTATGTCTAAAATACAAAAGTAAA 16 TCTGCAGAAGCCTGCCATTCCACCATTTAAACCTGTGACTCCAG Mouse; GCCTTAAGCCTGTTGAAGGTCGAGTCCCAGAAGGGTCATATGTG mm10: chr9: CAACTGCCTAGGGAGAGTTCCCACTCGCAGGGCCAAGAGGAGT 107399438- CCCCCGGTCTGAGGTGTGGGGGCGGGGACGTGCACTGGGCGCT 107399639
GGGACCACGGCTGGGGCTCAGGACTCGC 17 TGCCTCAGTTTCTTCGCCTAGAAAGCCGGGTCTAAGGGTACATG Mouse; CCCTGATTCTTTTCTGGGGTGTCTCGAATTTTAAACAACACATA mm10: chr9: CTGTTCTGGGCTGATGACAAGAGGAAGTACTGGTCGGTGGCTG 107443292- ATGGACATCCACCATGGTGGCAACTGGAGGGAGGGGGAACGGA 107444228 CGTTGAAACCCTGCCCTCCTGGAATCTGTCGCATGCACGCACGT TGACAATGCTTGGCACTGGGGACAGGCTGGGATGGATGGAGCG GAGCGTGAGGAGGAGTGGGCATGCAGGCCCGAGTGTCTGTTTT GCTGATTGCTCCTTTTGCTTTCAAGGAGATTAAACTATTTTTAGT CCATGCCTACTGCTGGTGAGACGCTGGAGGAAGCCTTTCCATCG TTGAGATTTTCTGGAAGCTGCCAAGTGTGGTCTTCAGCTCAATT CTGGGAGCCTCCCAGAGTGGGAGGGAGGAACATTTCCATCTGG GGGCTTCGGGGACAGGCTAAGATCTTCCCTGGGGTCCTTGCTGC GCTGGCCTCCTCAAACCACGCTGCCTCGGCCTGCATAAAGCAGT AATCTGATGTGCCCGATGTTTGTAACGCTGTGTTTAAAAAAAGT AATTTATTTTCTAATTATTCCTTGTCTTGCATAACCATGCATTGC CAAAGTGTCGCTATTTAAAATATTTATCTCTCCACGCCGCAGGA GCAGCTCTGGAGCGTGGAGGGGGAAGAAATAAAAGTCCGCGTG CCAGTCGCAGGCATATTACTTTGACTCGTCCTGGTGGCTTTGAC GTCTCCCTGTAAATACATTTATTTTTCATTAGGACGTTTCTGAGC TTGTGGCCCCCGGAGAGCGGAGTGATTACGCTGTTCATCTGCAA GCGATGCAATAGAGGGGTACTCGCAGAATGACTTCCGCCCAGA GCATCCTGCGCCTGTCT 18 TAAAATACCTTATTTTTTTCCAGTCTCTAAACTGCTAATCTCCCA Mouse; GGCTAAGGGATTCTGGGACAAAGGCAAGGCCTGGAAGTGGAAA mm10: chr9: TCTGTAAAATTAGCTTCAGCGGTATTAGTGTTTGCAGTTGAAGA 107444825- TTGAAAAACTGCTTTCCCAGGGCCTGATTGGAGGCTCCACTCTC 107445746 CTCCAGGAAGAGGCAAGGACTCTGGGCTGGCACTGAGGACAAA TCCTGGGAGGCTGCTATGGGGCCTGGGAGCCAGGCTGCCTTGTG CTAGAGGCCTAGAGAGTGTCTGTGTCCCAAGTCCCAAGCTACCC CCAGCAGCTAACAGCTTTTCCAGTTCTCAGGCACAGCAGGTGCC AAGATCACGCTCTGGAGTCCAGCTGGGCCCCTTCCTCTTCTTTTT TTTTTTTTTTTTTTAAGACCTCCTGGACACTGTTCCTCTCCCCCCC CCCGTGACCCCCCCCCTCAGTTCTCAAACACGTGAGGGTTGGGG GAGGGTTCCACAGCCAGAGAGAGGGGCCAGCTCTGGTGCCTGT GGGTACGCCCGCCCGTATGGCCCATCAGGCCTCTTGTGTGCTTG ATTGCCTCTGATTGGCTGCAGCTGAATTCAGCAAAAGCTATTAT TTGCCCTTGATGAGCCAATCAGATGGCCTCATTGGCCATTCAGA GCAGGCACCGGAACCTGAGGGTGGGGTGGGGGGTGGGGGATG GAGATGGGACTCAGTGAGGGGGTGGGAAGCTCTAAAACAGATG CAGGACCTGAGCCTGTCTGTGTCCACCACGACCTTCACACAGGT CACACCCCCTTCCCCTGACTTGTCACCCCAAACCAGGGCTTGTT GCCCAACCCCACCTCACAATTCCCTCACTCTGTAACACCTTTCC ATATACCTCTGCATGTCTAAACCCAAGACTTGCTCTATGAAATC 19 AGACCCTGCTTAGCACAGCTCTTAGCGGGTCCTTTAGGGGGTCT Mouse; CCCAGCGGGCCCAGTGGGAATGAGATAAGGAAGGACACAGCTG mm10: chr9: TCCATTCTCCCGTGCCTGCTAAGGAGGAAATGGGGCCGCCTTAC 107452080- ATAATTGGGGCAATTTGTTCCACTCTTGTCCTCCTGGTATCATGG 107452718 CTATCACCCCCTCCTTGCTCAGGGAGTCCTTGATTGAGCGAGAA GCTCAGGCCTCCCTCTCTCCCTCCTGCTGGGGGTTGCTGAACAG AGGGTGTAGGAGCCATAGGCTCTGTCACTGCTGAGATCTGCCA GATGTCTAGGCCAGGAGAAAATGGAAAGGGCTAAGTCACAGCA TATGTGGCCACTCAGGCCTATAGCCCCAAATCTGCCTGGTAACC CATTATGTCCCCAGAGAATTTGCATGGGCGGACACCCTCATGCC GGGTCTCAGTAAGGGAAGGGGTGGGAGGCAAAAATATCCCTCC CCACCCTGAATCTCCACCCCCTCCCCCCAGAAACTGACACTTGG CCTTGTCTAAGGATGGGTTTTCCCAAAATCCTTCTGAAAAAAAC AGAATTTCAAGAGTCACTCCCTCCGGGTCTCAGCCTAGAACATA TGCAGTATCCCCTGACGTCCATAGGG 20 AAACTGGCACAGTAATGGCGGGCTGACAGACAAGGGAGTCTGT Mouse; AGCACCCGCTGCCTCCGCCCACCCCTTCTCCGAGCAATTAAAAG mm10: chr9: GTGTTTATGTGGGGCTGGCAGTGGCTTCTGCCTCCCTTCCATTAC 107470414- GAACATTAAGAGATCTTGACCCTTCCACTTTCCCCGCTCTTGAA 107471129 AGGAGCTGCAGACACGTGGAGCCAATTAGGCGCACGCGTGGGC GCCAAGGGCCTGAGCAGCTTTTTCTCCCTGATTGCGGCGTTTAC AGCTGATTATTCTCCCCTCACCCAAACAGTGCTGCTTCCTGGCA AGGTGCCACCCAGAGGAGCCGGCTGGGGGCCCCTGGGGACAGG GGAGGACTGGATTAGTAAATGGGCATCTATCGAATGGCTTTCAT ATGTGTGGCTGGAAGGGAGAAGGGTAGGGCCAGGAATGGTGGC AGCAAGGGCCCAGGTAGCAATGAGGGTTCTTCTAACCCACCAT TTAGGGATAGCGATCAGAAAAGGGCCCTCGAGGAGGTGACCTA AATGTGTGTAGAAGCTGACGGCCACTACACACACACACACACA CACACACACACATACACAAGCATCCTTGTCCTTGGAGTCGGTCA GCATGAGCAAGAGAAGATGTTCCCAGTGGCCATGAGAGTGGA GCCCTCCTCCCTACTTACATCCAGGTTGGATGGCCAGGAGATCC TGAGATCCTTCAAGACTCC 21 AAGCCACATCCTGGGTGGAAATATATGGCTTCAATTCCCACTCT Mouse; TCCGGATGACCTCTGTGGGGAGCCCTGGCTTCACCTTGGTCCAG mm10: chr9: CTTCATCCCTTAGCCTCGCTGCCAGGAAGGCAGTGAGGTCAGAG 107484887- GCTGGTGCTGGCGTG 107485033 22 CCTACCTGGTGCCCGCCAACATCTGGGGGCCATCCTGGCCAGCG Mouse; CCAGCGTGGTGGTGAAGGCACTGTGCGCCGTGGTACTGTTTCTC mm10: chr9: TACCTGCTTTCCTTCGCTGTGGACACGGGCTGCCTGGCCGTCAC 107534490- CCCAGGCTACCTTTTCCCACCCAACTTCTGGATCTGGACCCTGG 107534786 CCACCCACGGGCTCATGGAACAGCACGTGTGGGACGTGGCCAT TAGCCTGGCCACAGTGGTTGTGGCCGGGCGATTACTGGAGCCCC TCTGGGGAGCCTTGGAGCTGCTCATCTTCTTCTC 23 AAACGGACGGGCCTCCGCTGAACCAGTGAGGCCCCAGACGTGC Human; GCATAAATAACCCCTGCGTGCTGCACCACCTGGGGAGAGGGGG hg19: AGGACCACGGTAAAT chr2: 17167 2063- 171672163 24 GGAGCGAGCGCATAGCAAAAGGGACGCGGGGTCCTTTTCTCTG Human; CCGGTGGCACTGGGTAGCTGTGGCCAGGTGTGGTACTTTGATGG hg19: GGCCCAGGGCTGGA chr2: 17167 2697- 171672797 25 GCTCAAGGAAGCGTCGCAGGGTCACAGATCTGGGGGAACCCCG Human; GGGAAAAGCACTGAGGCAAAACCGCCGCTCGTCTCCTACAATA hg19: TATGGGAGGGGGAGG chr2: 17167 2918- 171673018 26 TTGAGTACGTTCTGGATTACTCATAAGACCTTTTTTTTTTCCTTC Human; CGGGCGCAAAACCGTGAGCTGGATTTATAATCGCCCTATAAAG hg19: CTCCAGAGGCGGTCAGGCACCTGCAGAGGAGCCCCGCCGCTCC chr2: 17167 GCCGACTAGCTGCCCCCGCGAGCAACGGCCTCGTGATTTCCCCG 3150- CCGATCCGGTCCCCGCCTCCCCACTCTGCCCCCGCCTACCCCGG 171673696 AGCCGTGCAGCCGCCTCTCCGAATCTCTCTCTTCTCCTGGCGCTC GCGTGCGAGAGGGAACTAGCGAGAACGAGGAAGCAGCTGGAG GTGACGCCGGGCAGATTACGCCTGTCAGGGCCGAGCCGAGCGG ATCGCTGGGCGCTGTGCAGAGGAAAGGCGGGAGTGCCCGGCTC GCTGTCGCAGAGCCGAGGTGGGTAAGCTAGCGACCACCTGGAC TTCCCAGCGCCCAACCGTGGCTTTTCAGCCAGGTCCTCTCCTCC CGCGGCTTCTCAACCAACCCCATCCCAGCGCCGGCCACCCAACC TCCCGAAATGAGTGCTTCCTGCCC 27 CAGCAGCCGAAGGCGCTACTAGGAACGGTAACCTGTTACTTTTC Human; CAGGGGCCGTAGTCGACCCGCTGCCCGAGTTGCTGTGCGACTGC hg19: GCGCGCGGGGCTA chr2: 17167 3900- 171674000 28 GAGTGCAAGGTGACTGTGGTTCTTCTCTGGCCAAGTCCGAGGGA Human; GAACGTAAAGATATGGGCCTTTTTCCCCCTCTCACCTTGTCTCA hg19: CCAAAGTCCCTAGTCCCCGGAGCAGTTAGCCTCTTTCTTTCCAG chr2: 17167 GGAATTAGCCAGACACAACAACGGGAACCAGACACCGAACCA 4400- GACATGCCCGCCCCGTGCGCCCTCCCC 171674600 29 GCTCGCTGCCTTTCCTCCCTCTTGTCTCTCCAGAGCCGGATCTTC Human; AAGGGGAGCCTCCGTGCCCCCGGCTGCTCAGTCCCTCCGGTGTG hg19: CAGGACCCCGGAAGTCCTCCCCGCACAGCTCTCGCTTCTCTTTG chr2: 17167 CAGCCTGTTTCTGCGCCGGACCAGTCGAGGACTCTGGACAGTAG 4903- AGGCCCCGGGACGACCGAGCTG 171675101 30 AAACGGACGGGCCTCCGCTGAACCAGTGAGGCCCCAGACGTGC Human GCATAAATAACCCCTGCGTGCTGCACCACCTGGGGAGAGGGGG AGGACCACGGTAAATGGAGCGAGCGCATAGCAAAAGGGACGC GGGGTCCTTTTCTCTGCCGGTGGCACTGGGTAGCTGTGGCCAGG TGTGGTACTTTGATGGGGCCCAGGGCTGGAGCTCAAGGAAGCG TCGCAGGGTCACAGATCTGGGGGAACCCCGGGGAAAAGCACTG AGGCAAAACCGCCGCTCGTCTCCTACAATATATGGGAGGGGGA GGTTGAGTACGTTCTGGATTACTCATAAGACCTTTTTTTTTTCCT TCCGGGCGCAAAACCGTGAGCTGGATTTATAATCGCCCTATAAA GCTCCAGAGGCGGTCAGGCACCTGCAGAGGAGCCCCGCCGCTC CGCCGACTAGCTGCCCCCGCGAGCAACGGCCTCGTGATTTCCCC GCCGATCCGGTCCCCGCCTCCCCACTCTGCCCCCGCCTACCCCG GAGCCGTGCAGCCGCCTCTCCGAATCTCTCTCTTCTCCTGGCGC TCGCGTGCGAGAGGGAACTAGCGAGAACGAGGAAGCAGCTGG AGGTGACGCCGGGCAGATTACGCCTGTCAGGGCCGAGCCGAGC GGATCGCTGGGCGCTGTGCAGAGGAAAGGCGGGAGTGCCCGGC TCGCTGTCGCAGAGCCGAGGTGGGTAAGCTAGCGACCACCTGG ACTTCCCAGCGCCCAACCGTGGCTTTTCAGCCAGGTCCTCTCCT CCCGCGGCTTCTCAACCAACCCCATCCCAGCGCCGGCCACCCAA CCTCCCGAAATGAGTGCTTCCTGCCCCAGCAGCCGAAGGCGCTA CTAGGAACGGTAACCTGTTACTTTTCCAGGGGCCGTAGTCGACC CGCTGCCCGAGTTGCTGTGCGACTGCGCGCGCGGGGCTAGAGT GCAAGGTGACTGTGGTTCTTCTCTGGCCAAGTCCGAGGGAGAA CGTAAAGATATGGGCCTTTTTCCCCCTCTCACCTTGTCTCACCAA AGTCCCTAGTCCCCGGAGCAGTTAGCCTCTTTCTTTCCAGGGAA TTAGCCAGACACAACAACGGGAACCAGACACCGAACCAGACAT GCCCGCCCCGTGCGCCCTCCCCGCTCGCTGCCTTTCCTCCCTCTT GTCTCTCCAGAGCCGGATCTTCAAGGGGAGCCTCCGTGCCCCCG GCTGCTCAGTCCCTCCGGTGTGCAGGACCCCGGAAGTCCTCCCC GCACAGCTCTCGCTTCTCTTTGCAGCCTGTTTCTGCGCCGGACC AGTCGAGGACTCTGGACAGTAGAGGCCCCGGGACGACCGAGCTG 31 GGAGGAAGCCATCAACTAAACTACAATGACTGTAAGATACAAA Human ATTGGGAATGGTAACATATTTTGAAGTTCTGTTGACATAAAGAA TCATGATATTAATGCCCATGGAAATGAAAGGGCGATCAACACT ATGGTTTGAAAAGGGGGAAATTGTAGAGCACAGATGTGTTCGT GTGGCAGTGTGCTGTCTCTAGCAATACTCAGAGAAGAGAGAGA ACAATGAAATTCTGATTGGCCCCAGTGTGAGCCCAGATGAGGTT CAGCTGCCAACTTTCTCTTTCACATCTTATGAAAGTCATTTAAGC ACAACTAACTTTTTTTTTTTTTTTTTTTTTTTGAGACAGAGTCTTG CTCTGTTGCCCAGGACAGAGTGCAGTAGTGACTCAATCTCGGCT CACTGCAGCCTCCACCTCCTAGGCTCAAACGGTCCTCCTGCATC AGCCTCCCAAGTAGCTGGAATTACAGGAGTGGCCCACCATGCC CAGCTAATTTTTGTATTTTTAATAGATACGGGGGTTTCACCATAT CACCCAGGCTGGTCTCGAACTCCTGGCCTCAAGTGATCCACCTG CCTCGGCCTCCCAAAGTGCTGGGATTATAGGCGTCAGCCACTAT GCCCAACCCGACCAACCTTTTTTAAAATAAATATTTAAAAAATT GGTATTTCACATATATACTAGTATTTACATTTATCCACACAAAA CGGACGGGCCTCCGCTGAACCAGTGAGGCCCCAGACGTGCGCA TAAATAACCCCTGCGTGCTGCACCACCTGGGGAGAGGGGGAGG ACCACGGTAAATGGAGCGAGCGCATAGCAAAAGGGACGCGGG GTCCTTTTCTCTGCCGGTGGCACTGGGTAGCTGTGGCCAGGTG GGTACTTTGATGGGGCCCAGGGCTGGAGCTCAAGGAAGCGTCG CAGGGTCACAGATCTGGGGGAACCCCGGGGAAAAGCACTGAGG CAAAACCGCCGCTCGTCTCCTACAATATATGGGAGGGGGAGGT TGAGTACGTTCTGGATTACTCATAAGACCTTTTTTTTTTCCTTCC GGGCGCAAAACCGTGAGCTGGATTTATAATCGCCCTATAAAGC TCCAGAGGCGGTCAGGCACCTGCAGAGGAGCCCCGCCGCTCCG CCGACTAGCTGCCCCCGCGAGCAACGGCCTCGTGATTTCCCCGC CGATCCGGTCCCCGCCTCCCCACTCTGCCCCCGCCTACCCCGGA GCCGTGCAGCCGCCTCTCCGAATCTCTCTCTTCTCCTGGCGCTCG CGTGCGAGAGGGAACTAGCGAGAACGAGGAAGCAGCTGGAGG TGACGCCGGGCAGATTACGCCTGTCAGGGCCGAGCCGAGCGGA TCGCTGGGCGCTGTGCAGAGGAAAGGCGGGAGTGCCCGGCTCG CTGTCGCAGAGCCGAGGTGGGTAAGCTAGCGACCACCTGGACT TCCCAGCGCCCAACCGTGGCTTTTCAGCCAGGTCCTCTCCTCCC GCGGCTTCTCAACCAACCCCATCCCAGCGCCGGCCACCCAACCT CCCGAAATGAGTGCTTCCTGCCCCAGCAGCCGAAGGCGCTACT AGGAACGGTAACCTGTTACTTTTCCAGGGGCCGTAGTCGACCCG CTGCCCGAGTTGCTGTGCGACTGCGCGCGCGGGGCTAGAGTGC AAGGTGACTGTGGTTCTTCTCTGGCCAAGTCCGAGGGAGAACGT AAAGATATGGGCCTTTTTCCCCCTCTCACCTTGTCTCACCAAAG TCCCTAGTCCCCGGAGCAGTTAGCCTCTTTCTTTCCAGGGAATT AGCCAGACACAACAACGGGAACCAGACACCGAACCAGACATG CCCGCCCCGTGCGCCCTCCCCGCTCGCTGCCTTTCCTCCCTCTTG TCTCTCCAGAGCCGGATCTTCAAGGGGAGCCTCCGTGCCCCCGG CTGCTCAGTCCCTCCGGTGTGCAGGACCCCGGAAGTCCTCCCCG CACAGCTCTCGCTTCTCTTTGCAGCCTGTTTCTGCGCCGGACCA GTCGAGGACTCTGGACAGTAGAGGCCCCGGGACGACCGAGCTG 32 TCAACAGGGGGACACTTGGGAAAGAAGGATGGGGACAGAGCC Human and GAGAGGACTGTTACACATTAGAGAAACATCAGTGACTGTGCCA mouse GCTTTGGGGTAGACTGCACAAAAGCCCTGAGGCAGCACAGGCA GGATCCAGTCTGCTGGTCCCAGGAAGCTAACCGTCTCAGACAG AGCACAAAGCACCGAGACATGTGCCACAAGGCTTGTGTAGAGA GGTCAGAGGACAGCGTACAGGTCCCAGAGATCAAACTCAACCT CACCAGGCTTGGCAGCAAGCCTTTACCAACCCACCCCCACCCCA CCCACCCTGCACGCGCCCCTCTCCCCTCCCCATGGTCTCCCATG GCTATCTCACTTGGCCCTAAAATGTTTAAGGATGACACTGGCTG CTGAGTGGAAATGAGACAGCAGAAGTCAACAGTAGATTTTAGG AAAGCCAGAGAAAAAGGCTTGTGCTGTTTTTAGAAAGCCAAGG GACAAGCTAAGATAGGGCCCAAGTAATGCTAGTATTTACATTTA TCCACACAAAACGGACGGGCCTCCGCTGAACCAGTGAGGCCCC AGACGTGCGCATAAATAACCCCTGCGTGCTGCACCACCTGGGG AGAGGGGGAGGACCACGGTAAATGGAGCGAGCGCATAGCAAA AGGGACGCGGGGTCCTTTTCTCTGCCGGTGGCACTGGGTAGCTG TGGCCAGGTGTGGTACTTTGATGGGGCCCAGGGCTGGAGCTCA AGGAAGCGTCGCAGGGTCACAGATCTGGGGGAACCCCGGGGAA AAGCACTGAGGCAAAACCGCCGCTCGTCTCCTACAATATATGG GAGGGGGAGGTTGAGTACGTTCTGGATTACTCATAAGACCTTTT TTTTTTCCTTCCGGGCGCAAAACCGTGAGCTGGATTTATAATCG CCCTATAAAGCTCCAGAGGCGGTCAGGCACCTGCAGAGGAGCC CCGCCGCTCCGCCGACTAGCTGCCCCCGCGAGCAACGGCCTCGT GATTTCCCCGCCGATCCGGTCCCCGCCTCCCCACTCTGCCCCCG CCTACCCCGGAGCCGTGCAGCCGCCTCTCCGAATCTCTCTCTTC TCCTGGCGCTCGCGTGCGAGAGGGAACTAGCGAGAACGAGGAA GCAGCTGGAGGTGACGCCGGGCAGATTACGCCTGTCAGGGCCG
AGCCGAGCGGATCGCTGGGCGCTGTGCAGAGGAAAGGCGGGA GTGCCCGGCTCGCTGTCGCAGAGCCGAGGTGGGTAAGCTAGCG ACCACCTGGACTTCCCAGCGCCCAACCGTGGCTTTTCAGCCAGG TCCTCTCCTCCCGCGGCTTCTCAACCAACCCCATCCCAGCGCCG GCCACCCAACCTCCCGAAATGAGTGCTTCCTGCCCCAGCAGCCG AAGGCGCTACTAGGAACGGTAACCTGTTACTTTTCCAGGGGCCG TAGTCGACCCGCTGCCCGAGTTGCTGTGCGACTGCGCGCGCGGG GCTAGAGTGCAAGGTGACTGTGGTTCTTCTCTGGCCAAGTCCGA GGGAGAACGTAAAGATATGGGCCTTTTTCCCCCTCTCACCTTGT CTCACCAAAGTCCCTAGTCCCCGGAGCAGTTAGCCTCTTTCTTT CCAGGGAATTAGCCAGACACAACAACGGGAACCAGACACCGA ACCAGACATGCCCGCCCCGTGCGCCCTCCCCGCTCGCTGCCTTT CCTCCCTCTTGTCTCTCCAGAGCCGGATCTTCAAGGGGAGCCTC CGTGCCCCCGGCTGCTCAGTCCCTCCGGTGTGCAGGACCCCGGA AGTCCTCCCCGCACAGCTCTCGCTTCTCTTTGCAGCCTGTTTCTG CGCCGGACCAGTCGAGGACTCTGGACAGTAGAGGCCCCGGGAC GACCGAGCTG 33 ATTTACATTTATCCACACA Human 34 TGCCGCTGGACTCTCTTCCAAGGAACTAGGAGAACCAAGATCC Mouse; GTTTTTCTGCCAAGGGCTGCCCCCCCCACGCCCCCAACCCCCTC chr9; 107,399, ACCCCGATCCCCACAGAAAGAAATCTTGAGGTAGCTGGAGCTT 268- CTTCTGTGGGTGTGACAGGACTGCCATTCTCCTCTGTAGTCTGC 107,400,067 AGAAGCCTGCCATTCCACCATTTAAACCTGTGACTCCAGGCCTT AAGCCTGTTGAAGGTCGAGTCCCAGAAGGGTCATATGTGCAAC TGCCTAGGGAGAGTTCCCACTCGCAGGGCCAAGAGGAGTCCCC CGGTCTGAGGTGTGGGGGCGGGGACGTGCACTGGGCGCTGGGA CCACGGCTGGGGCTCAGGACTCGCGAGCTTGGATTCGGATCGG TTTGCGCGAGCCAGTAGGGCAGGCTCCGGGGTGAACGGGGACG AGGGGCGCGCGGGCACAGGCGGGCGCGTGACCGCGGCGGGGG CGCGCGGAGGCGGGCCGGCCAAGGAGAGGGAGGGAGGGAATG AGGGAGGGAGCGACAGGGGAGGGCGGCGCCGGCAGGTTGGCG GCGGCCGCTATTTGAGCGCAGGTCCCGGGCCAGGCGCTCAAAG CGCTTGGAGCCAGCGCGGCGGGGAGATCGCTGCGCGCAGCCCG CAGAGGCGCTGCGGCCAGTGCAGCCCCGGAGGCCCCGCGCGGA GAAGGAGGTGGAGAAGAGGCCGGCTTTCCGCCCGCCGCCCGCG CCCCCCCACCTCCATCCCGCCGCCGCCGTCCCCCCTCCCTCCCC GCGGCGCCGCATCTTGAATGGAAAC 35 GAGTAATTCATACAAAAGGACTCGCCCCTGCCTTGGGGAATCCC AGGGACCGTCGTTAAACTCCCACTAACGTAGAACCCAGAGATC GCTGCGTTCCCGCCCCCTCACCCGCCCGCTCTCGTCATCACTGA GGTGGAGAAGAGCATGCGTGAGGCTCCGGTGCCCGTCAGTGGG CAGAGCGCACATCGCCCACAGTCCCCGAGAAGTTGGGGGGAGG GGTCGGCAATTGAACCGGTGCCTAGAGAAGGTGGCGCGGGGTA AACTGGGAAAGTGATGTCGTGTACTGGCTCCGCCTTTTTCCCGA GGGTGGGGGAGAACCGTATATAAGTGCAGTAGTCGCCGTGAAC GTTCTTTTTCGCAACGGGTTTGCCGCCAGAACACAGGTAAGTGC CGTGTGTGGTTCCCGCGGGCCTGGCCTCTTTACGGGTTATGGCC CTTGCGTGCCTTGAATTACTTCCACGCCCCTGGCTGCAGTACGT GATTCTTGATCCGAGCTTCGGGTTGGAAGTGGGTGGGAGAGTT CGAGGCCTTGCGCTTAAGGAGCCCCTTCGCCTCGTGCTTGAGTT GAGGCCTGGCTTGGGCGCTGGGGCCGCCGCGTGCGAATCTGGT GGCACCTTCGCGCCTGTCTCGCTGCTTTCGATAAGTCTCTAGCC ATTTAAAATTTTTGATGACCTGCTGCGACGCTTTTTTTCTGGCAA GATAGTCTTGTAAATGCGGGCCAAGATCTGCACACTGGTATTTC GGTTTTTGGGGCCGCGGGCGGCGACGGGGCCCGTGCGTCCCAG CGCACATGTTCGGCGAGGCGGGGCCTGCGAGCGCGGCCACCGA GAATCGGACGGGGGTAGTCTCAAGCTGGCCGGCCTGCTCTGGT GCCTGGCCTCGCGCCGCCGTGTATCGCCCCGCCCTGGGCGGCAA GGCTGGCCCGGTCGGCACCAGTTGCGTGAGCGGAAAGATGGCC GCTTCCCGGCCCTGCTGCAGGGAGCTCAAAATGGAGGACGCGG CGCTCGGGAGAGCGGGCGGGTGAGTCACCCACACAAAGGAAA AGGGCCTTTCCGTCCTCAGCCGTCGCTTCATGTGACTCCACGGA GTACCGGGCGCCGTCCAGGCACCTCGATTAGTTCTCGAGCTTTT GGAGTACGTCGTCTTTAGGTTGGGGGGAGGGGTTTTATGCGATG GAGTTTCCCCACACTGAGTGGGTGGAGACTGAAGTTAGGCCAG CTTGGCACTTGATGTAATTCTCCTTGGAATTTGCCCTTTTTGAGT TTGGATCTTGGTTCATTCTCAAGCCTCAGACAGTGGTTCAAAGT TTTTTTCTTCCATTTCAGGTGTCGTGA
[0060] In one aspect, regulatory elements disclosed herein are cell-type selective. In some cases, regulatory elements disclosed herein are selective for PV neurons. In some cases, regulatory elements disclosed herein are selective for PV neurons in the CNS. In some cases, PV-cell selective regulatory elements or any regulatory elements disclosed herein can result in selective gene expression in PV neurons over at least one, two, three, four, five, or more non-PV CNS cell-types.
[0061] In some cases, any one or more of the regulatory elements disclosed herein are operably linked to a transgene in an expression cassette to result in selective expression in a target cell-type, e.g., a PV neuron. In some cases, a regulatory element of any of the embodiments herein comprises or consists of any one of (i) SEQ ID NOs: 1-33; (ii) a variant, functional fragment, or a combination thereof; or (iii) a sequence having at least 80%, at least 85%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% sequence identity to any one of (i) or (ii). In some cases, a regulatory element comprises any one of SEQ ID NOs: 1-32. In some cases, sequence identity is measured by BLAST.
[0062] In some cases, two or more, three or more, four or more, five or more, six or more, seven or more, eight or more, nine or more, or ten or more of SEQ ID NOs: 1-32, or a functional fragment or a combination thereof, or sequences having at least 80%, at least 90%, at least 95%, or at least 99% sequence identity thereto, are combined to form a larger regulatory element, or are operably linked to a gene in an expression cassette. In some cases, two or more, three or more, four or more, five or more, six or more, seven or more, eight or more, nine or more, or ten or more of SEQ ID NOs: 1-32, or a functional fragment or a combination thereof, or sequences having at least 80%, at least 90%, at least 95%, or at least 99% sequence identity thereto, are combined using a linker sequence of 1-50 nucleotides. In some cases, two or more, three or more, four or more, five or more, six or more, seven or more, eight or more, nine or more, or ten or more of SEQ ID NOs: 1-32, or a functional fragment or a combination thereof, or sequences having at least 80%, at least 90%, at least 95%, or at least 99% sequence identity thereto, are combined without a linker sequence. In some cases, a sequence of SEQ ID NO: 33 is used as a linker between any two regulatory elements. In some cases, a linker sequence between any two regulatory elements comprises SEQ ID NO: 33 or a sequence having at least 80%, at least 85%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% sequence identity to SEQ ID NO: 33. In some cases, sequence identity is measured by BLAST.
[0063] In some cases, when two or more regulatory elements are combined or used in an expression cassette, the regulatory elements need not be adjacent or linked in an expression cassette. For example, one regulatory element can be located upstream of a transgene, while a second regulatory element and/or additional regulatory elements can be located downstream of the transgene. In some cases, one or more regulatory elements can be located upstream of a transgene. In some cases, one or more regulatory elements can be located downstream of a transgene.
[0064] In some cases, any one or more of SEQ ID NOs: 1-22, or a functional fragment or a combination thereof, or sequences having at least 80%, at least 90%, at least 95%, or at least 99% sequence identity thereto, can be combined with any one or more of SEQ ID NO: 23-30, or a functional fragment or a combination thereof, or sequences having at least 80%, at least 90%, at least 95%, or at least 99% sequence identity thereto, to form a larger regulatory element. For example, a regulatory element comprises SEQ ID NO: 1 and SEQ ID NO: 30. A regulatory element comprises SEQ ID NO: 8 and SEQ ID NO: 30. A regulatory element comprises SEQ ID NO: 1 and SEQ ID NOs: 23-29. A regulatory element comprises SEQ ID NO: 8 and SEQ ID NOs: 23-29. In some cases, a regulatory element comprises SEQ ID NO: 30 or any one or more of SEQ ID NOs: 23-29, or a functional fragment or a combination thereof, or sequences having at least 80%, at least 90%, at least 95%, or at least 99% sequence identity thereto.
[0065] In some cases, one or more regulatory elements of the present disclosure result in selective gene expression in a PV cell. In some cases, regulatory elements that show selective activity or function in a target cell type also show minimal activity or function in one or more off-target cell-types, e.g., non-PV CNS cell-types, non-inhibitory neurons or excitatory neurons, non-PV cells.
[0066] In some cases, one or more regulatory elements operably linked to a gene modulates gene expression in a cell, including but not limited to, selective expression in a target cell-type over non-target cell-types. Selective expression in a target cell or cell type can also be referred to as cell-selective expression or cell-type selective expression.
[0067] Selective expression generally refers to expression in a high fraction of cells of the cell type of interest (or the target cell type) as compared to other cells (or non-target cell type). Selective expression can also be viewed as preferential expression in a target cell or target cell type over one or more non-target cells or cell-types. In some cases, selective expression of one or more regulatory elements of this disclosure is compared to CAG, EF1.alpha., a constitutive promoter (e.g., SV40, CMV, UBC, PGK, and CBA), or a non-selective regulatory element that is known to drive expression in any cell or cell type without selectivity. In some cases, selective expression of one or more regulatory elements of this disclosure is compared to CAG, EFla, a constitutive promoter (e.g., SV40, CMV, UBC, PGK, and CBA), a non-selective regulatory element, or an expression cassette without the regulatory elements.
[0068] Non-target cell types can include a different subset, subtype, or type of cells as compared to the target cell or target cell type, or all non-target cell types. In some cases, one or more regulatory elements operably linked to a gene result in selective expression in a target cell type over at least one type of non-target cells, or at least two, at least three, at least four, at least five, or more than five types of non-target cells. In some cases, non-target cell types refer to all other cell types not including the target cell type. In some cases, non-target cell types are all other cell types within a relevant tissue or organ not including the target cell type, e.g., all non-target cell types in the CNS, all non-target cell types in the hippocampus. In some cases, a non-target cell or non-target cell type encompasses a subset or subtype of cells that is not the target cell. For example, non-PV CNS cell-types can include GABAergic cells that express calretinin and/or somatostatin instead of parvalbumin, or all GABAergic cells that do not express parvalbumin. In some cases, cell types are distinguished by having a different cell marker, morphology, phenotype, genotype, function, and/or any other means for classifying cell types.
[0069] Selectivity of expression driven by a regulatory element in a cell or cell type of interest can be measured in a number of ways. Selectivity of gene expression in a target cell type over non-target cell types can be measured by comparing the number of target cells that express a detectable level of a transcript from a gene that is operably linked to one or more regulatory elements to the total number of cells that express the gene. Such measurement, detection, and quantification can be done either in vivo or in vitro.
[0070] In some instances, selectivity for PV neurons can be determined using a co-localization assay. In some cases, the co-localization assay is based on immunohistochemistry. In some cases, a detectable reporter gene is used as a transgene to allow the detection and/or measurement of gene expression in a cell. In some cases, a detectable marker, e.g., a fluorescent marker or an antibody, which specifically labels the target cell is used to detect and/or measure the target cells. In some cases, a co-localization assay employs imaging, e.g., fluorescent imaging, to determine the overlap between different fluorescent labels, e.g., overlap between a fluorescence signal indicative of a target cell and another fluorescence signal indicative of gene expression. In some cases, fluorescent labels used for a co-localization assay include a red fluorescent protein (RFP), such as a tdTomato reporter gene, and a green fluorescent reporter protein, such as eGFP.
[0071] In some instances, a gene operably linked to one or more regulatory elements is a fluorescent protein, e.g., eGFP or RFP, wherein expression of the transgene provides a detectable signal. In some cases, tissue is stained for eGFP or fluorescence from eGFP is detected directly using a fluorescence microscope. A second fluorescent marker or reporter gene having a different fluorescence or detectable signal can be used to indicate the target cells, such as an antibody that identifies the target cells. For example, an anti-PV antibody that interacts specifically with PV neurons can be used to yield a detectable signal that is distinguishable from the fluorescence used to measure gene expression, such as a red fluorescence or a red stain. Thus, in an example wherein eGFP is a transgene operably linked to one or more regulatory elements that drive selective expression in PV neurons, and wherein the PV neurons are labeled with an anti-PV antibody, selectivity of gene expression in PV cells is measured as percentage of eGFP+ cells that are also PV+. In such assay, PV+ cells that are also eGFP+ are indicated by the overlap of both fluorescence signals, i.e., an overlap of the red and green fluorescence. Such measurement, analysis, and/or detection can be done by eye inspection or by a computer.
[0072] In some cases, one can also measure the proportion of a cell type of interest (or target cell type) that expresses a transgene as compared to the proportion of non-target cell types (or other cells) that express the transgene to assess the selectivity of one or more regulatory elements operably linked to the transgene. Similarly, selectivity of expression can also be measured by comparing the number of target cells that express a transgene operably linked to one or more regulatory elements to the total number of all cells that express the transgene. In both approaches, the higher the number of target cells that express the transgene, the more selective are the regulatory elements for the target cells. In some cases, the target cells are PV neurons.
[0073] In some cases, one or more regulatory elements disclosed herein result in increased selectivity in gene expression in a PV neuron. In some cases, one or more regulatory elements disclosed herein result in increased selectivity in gene expression in PV neurons as compared to non-PV CNS cell-types. In some cases, one or more regulatory elements disclosed herein result in increased selectivity in gene expression in PV neurons as compared to non-PV GABAergic cells, wherein non-PV GABAergic cells can be any one or more of GABAergic cells that express calretinin (CR), somatostatin (SOM), cholecystokinin (CCK), neuropeptide Y (NPY), vasointestinal polypeptide (VIP), choline acetyltransferase (ChAT), or a combination thereof. In some cases, one or more regulatory elements disclosed herein result in increased selectivity in gene expression in PV neurons as compared to at least one, at least two, at least three, at least four, at least five, or more than five non-PV GABAergic subtypes. In some cases, one or more regulatory elements disclosed herein result in increased selectivity in gene expression in PV neurons as compared to all other non-PV GABAergic cells, or all other GABAergic cells that do not express PV, or all other CNS cells that do not express PV, or all other neurons that do not express PV. In some cases, one or more regulatory elements disclosed herein result in increased selectivity in gene expression in PV neurons as compared to all non-PV cells in the CNS or all non-PV neurons.
[0074] In some cases, one or more regulatory elements operably linked to a transgene result in selective expression of the transgene in PV cells, wherein the percentage of PV cells expressing the transgene is at a percentage higher than gene expression in PV cells wherein the transgene is operably linked to CAG, EFla, a constitutive promoter (e.g., SV40, CMV, UBC, PGK, and CBA), or a non-selective regulatory element, such as SEQ ID NO: 34, or a functional fragment thereof, or a sequence having at least 80% sequence identity thereto. In some cases, one or more regulatory elements result in selective expression in PV neurons at a level that is at least 1.5 fold, at least 2 hold, at least 3 fold, at least 4 fold, at least 5 fold, at least 6 fold, at least 7 fold, at least 8 fold, at least 9 fold, at least 10 fold, at least 15 fold, at least 20 fold, at least 25 fold, or at least 50 fold as compared to expression of a gene operably linked to CAG, EF1a, a constitutive promoter (e.g., SV40, CMV, UBC, PGK, and CBA), or a non-selective regulatory element such as SEQ ID NO: 34, or a functional fragment thereof, or a sequence having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto.
[0075] In some aspects, a regulatory element is human derived or comprises a sequence that is human derived. In some cases, a regulatory element is mouse derived or comprises a sequence that is mouse derived. In some cases, a regulatory element comprises a non-naturally occurring sequence. In some cases, a regulatory element is non-naturally occurring. In some cases, one or more human derived regulatory elements are combined with another regulatory element to generate a non-naturally occurring regulatory element. In some cases, a human derived regulatory element is combined with a mouse derived regulatory element.
[0076] The term "human derived" as used herein refers to sequences that are found in a human genome (or a human genome build), or sequences homologous thereto. A homologous sequence may be a sequence which has a region with at least 80% sequence identity (e.g., as measured by BLAST) as compared to a region of the human genome. For example, a sequence that is at least 80%, at least 85%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99% homologous to a human sequence is deemed human derived. In some cases, a regulatory element contains a human derived sequence and a non-human derived sequence such that overall the regulatory element has low sequence identity to the human genome, while a part of the regulatory element has 100% sequence identity (or local sequence identity) to a sequence in the human genome.
[0077] In some cases, a human-derived regulatory element is a sequence that is 100% identical to a human sequence. In some instances, the sequence of a cell-type selective regulatory element is 100% human derived.
[0078] In other instances, at least 5%, 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 95%, 98%, or 99% of the regulatory element sequence is human derived. For example, a regulatory element can have 50% of its sequence be human derived, and the remaining 50% be non-human derived (e.g., mouse derived or fully synthetic). For further example, a regulatory element that is regarded as 50% human derived and comprises 300 bp may have an overall 45% sequence identity to a sequence in the human genome, while base pairs 1-150 of the RE may have 90% identity (local sequence identity) to a similarly sized region of the human genome.
[0079] In some instances, a sequence that is homologous to a human derived regulatory sequence is at least 90% identical to a human sequence. In some cases, a regulatory element herein comprises a sequence that has at least 80%, at least 85%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99% identity to any one of SEQ ID NOs: 1, 23-31, and 33. In some cases, sequence identity is measured by BLAST. When a regulatory element comprises a sequence that is homologous (e.g., at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity) to any one or more of SEQ ID NOs: 23-29, or a functional fragment or combination thereof, such regulatory element results in higher expression of an operably linked transgene (when a promoter is also present in the expression vector or cassette), as compared to a similar vector without the regulatory element. Such higher expression of a transgene can be observed, e.g., in HEK293T or CHO cells.
[0080] In some cases, one or more regulatory elements comprise any one or more of SEQ ID NO: 23-29 combined with or used in combination with any one or more of SEQ ID NOs: 1-22 with or without a linker sequence such as SEQ ID NO: 33. In some cases, any two regulatory elements of this disclosure are linked together using a polynucleotide linker comprising 1-50 nucleotides, such as SEQ ID NO: 33 or a variant thereof. In some cases, the linker sequence is a human derived sequence. In some cases, the linker sequence is mouse derived or non-naturally occurring. In some cases, two regulatory elements are joined without a linker or without any intervening sequence. In some cases, a linker comprises 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30 nucleotides. In some cases, the linker sequence is a result of restriction enzyme site, ligation, PCR, and/or cloning.
[0081] In some cases, a human derived regulatory element is combined with a mouse derived regulatory element, such as SEQ ID NO: 32, which is a combination of SEQ ID NO: 8 (a mouse derived sequence) and SEQ ID NOs: 23-29 (human derived sequences). In some cases, cell-type selective regulatory elements are combined directly with no additional linker sequence. In other cases, cell-type selective regulatory elements are combined with one or more short linker sequences which can be either deliberate or cloning artifacts. In some cases, a linker sequence comprises 1-50 bases. For example, SEQ ID NO: 31 comprises the sequence of SEQ ID NOs: 1 and 23-29 along with an additional 19 bp of the genomic sequence (SEQ ID NO: 33) immediately following the sequence of SEQ ID NO: 1. SEQ ID NO: 32 also includes these 19 bp but without the sequence of SEQ ID NO: 1. In other examples, the combined cell-type selective regulatory elements can include short sequences, generally less than 50 bp, less than 20 bp, less than 15 bp, or less than 10 bp, from a cloning plasmid or restriction enzyme recognition site.
[0082] In some cases, regulatory elements can be derived from non-coding DNA sequences. In some cases, regulatory elements derived from non-coding DNA are associated with genes, such as upstream sequences, introns, 3' and 5' untranslated regions (UTRs), and/or downstream regions. In other cases, regulatory elements derived from non-coding DNA sequences are not associated with a gene. In some cases, regulatory elements are derived from coding sequences. In some cases, the genomic region from which a regulatory element is derived is distinct from the genomic region from which an operably linked transgene is derived. In some cases, a RE is derived from a distal genomic region or location with respect to the genomic region or location from which the transgene is derived (such as a naturally occurring or an endogenous version of the transgene).
[0083] In one aspect, a regulatory element is any non-coding sequence that modulates gene expression, e.g., selectivity of expression in a target cell. In some cases, the target cell is a PV neuron. In some cases, a regulatory element is derived from a genomic sequence upstream of a transcription initiation site, a 5' UTR sequence, an exonic sequence, an intronic sequence, or a 3' UTR sequence. In some cases, a human derived regulatory element comprises an intronic human derived sequence. In some cases, a regulatory element comprises an enhancer, and its presence in an expression cassette along with a promoter increases expression of an operably linked transgene in the target cell-type (e.g., PV neurons) as compared to expression of the same transgene by the promoter without the enhancer. In some cases, an enhancer increases expression of an operably linked transgene through either a transcriptional mechanism, posttranscriptional mechanism, or both. In some cases, a regulatory element comprises an enhancer sequence, a promoter sequence, or a combination of the enhancer and promoter sequences. In some cases, a regulatory element comprises one or more of a human derived enhancer sequence, a human derived promoter sequence, a human derived intronic sequence, and/or a combination thereof.
[0084] In some cases, a regulatory element comprises one or more of SEQ NOs: 1-32, or a fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto. In some cases, a regulatory element comprises one or more of SEQ ID NOs: 1-22, or a fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto. In some cases, a regulatory element comprises one or more of SEQ ID NOs: 23-29, or a fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto. In some cases, a regulatory element comprises one or more of SEQ ID NOs: 30-32, or a fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto. In some cases, a regulatory element comprises a sequence of SEQ ID NOs: 30-32, or a fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto.
[0085] In some instances, a regulatory element is derived from non-human DNA sequences or both human and non-human genomic sequences. In some cases, cell-type selective regulatory elements, or parts thereof, are homologous to a mammalian genomic sequence. In some cases, the regulatory elements have at least 80%, at least 85%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99% identity to a mammalian genomic sequence. In some cases, a regulatory element is derived from a mouse genomic sequence. In some cases, a regulatory element, or fragments thereof, has at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more than 99% identity to a mouse genomic sequence or a non-human mammalian genomic sequence. In some cases, sequence identity is measured by BLAST. In some cases, the regulatory elements may comprise any of SEQ ID NOs: 1-33, or a fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto.
[0086] In some cases, cell-type selective regulatory elements are short. In some cases, the size of the regulatory elements is compatible with the cloning capacity of a vector, e.g., a viral vector or rAAV, such that the combined size of a transgene and one or more regulatory elements does not exceed the cloning capacity of a vector. In some cases, the cell-type selective regulatory elements have a length of up to about 2050 bp, 2000 bp, 1900 bp, 1800 bp, 1700 bp, 1600 bp, 1500 bp, 1400 bp, 1300 bp, 1200 bp, 1100 bp, 1000 bp, 900 bp, 800 bp, 700 bp, 600 bp, 500 bp, 400 bp, 300 bp, 200 bp, or 100 bp. In some cases, the cell-type selective regulatory elements have a total length of no more than about 20 bp, 30 bp, 40 bp, 50 bp, 60 bp, 70 bp, 80 bp, 90 bp, 100 bp, 200 bp, 300 bp, 400 bp, 500 bp, 600 bp, 700 bp, 800 bp, 900 bp, 1000 bp, 1010 bp, 1020 bp, 1030 bp, 1040 bp, 1050 bp, 1060 bp, 1070 bp, 1080 bp, 1090 bp, 1100 bp, 1200 bp, 1300 bp, 1500 bp, 1600 bp, 1700 bp, 1800 bp, 1900 bp, or 2000 bp. In some cases, the cell-type selective regulatory elements have a length of about 100 bp-1100 bp, 100 bp-1000 bp, 100 bp-900 bp, 200 bp-900 bp, 200 bp-800 bp, 300 bp-600 bp, 400 bp-800 bp, 500 bp-600 bp, or 600 bp-900 bp. In some cases, a regulatory element is between about 400-600 bp, 400-600 bp, 400-700 bp, 400-800 bp, 400-900 bp, 400-1000 bp, or 400-1500 bp. In some cases, a regulatory element is between about 500-600 bp, 500-700 bp, 500-800 bp, 500-900 bp, 500-1000 bp, or 500-1500 bp. In some cases, two or more regulatory elements are combined to form a larger cell-type selective regulatory element of 1300-2500 bp, 1300-2060 bp, about 1350 bp, about 2050 bp, or about 1880 bp.
[0087] In some cases, two or more cell-type selective regulatory elements can be combined. For example, two, three, four, five, six, seven, eight, nine, ten or more cell-type selective regulatory elements can be combined. For example, SEQ ID NO: 30 comprises sequences from seven regulatory elements, i.e., SEQ ID NOs: 23-29, all of which are derived from human genomic sequence. In some cases, cell-type selective regulatory elements refer to PV-neuron selective regulatory elements.
[0088] In some cases, a cell-type selective regulatory element is repeated two or more times to make a combined regulatory element that is also cell-type selective or has enhance cell-type selective property. In some cases, two or more regulatory elements with different cell-type selectivity are combined. In some cases, a cell-type selective regulatory element is combined with a non-selective regulatory element, e.g., a non-selective enhancer element that drives high gene expression. For example, a promoter regulatory element with high selectivity for a target cell can be combined with a regulatory element with high efficiency of expression. In some cases, one or more cell type-selective regulatory elements are combined with one or more high efficiency regulatory elements. For example, any one or more of SEQ ID NOs: 1-32, or a fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto, can be combined with a constitutive promoter, such as a GAD2 promoter, a human synapsin promoter, a minCMV promoter, a TATA box, a super core promoter, or an EF1.alpha. promoter, or a combination thereof.
[0089] In some aspects, the present disclosure provides a list of regulatory elements that can be added to any gene therapy to result in selective gene expression in a target cell type, such as a PV neuron over one or more non-target cell types, such as non-PV CNS cells, including but not limited to excitatory cells and/or non-PV GABAergic cells.
[0090] In some cases, cell-type selective regulatory elements can be combined with other regulatory elements such as a high expressing promoter or a sequence that increases mRNA stability. In some cases, one or more cell-type selective regulatory elements are combined with a human, a non-human, or a non-mammalian sequence, for example a hSyn1 promoter, CBA promoter, a CMV promoter, an EF1.alpha. promoter, a polyA signal (e.g., SV40 polyA signal), or a post-transcriptional regulatory element such as woodchuck hepatitis virus post-transcriptional regulatory element (WPRE).
[0091] In some cases, the combined regulatory elements can come from different species. The combined regulatory elements can come from different genomic regions within a species. In some cases, regulatory elements are derived from distal genomic sequences, e.g., sequences that do not normally or naturally associate with each other or with a cell type of interest, are combined. In some cases, individual regulatory elements used to make a combined regulatory element can come from different human chromosomes.
[0092] In one aspect, a regulatory element of the disclosure comprises a functional fragment of any of SEQ ID NOs: 1-32, or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto. Such functional fragment can increase expression of a transgene in an expression cassette or vector when compared to a similar expression cassette or vector without the regulatory element. Such a functional fragment can function as an enhancer to increase cell-type selective expression when the fragment is operably linked to a transgene as compared to a similar vector or cassette without the functional fragment. A fragment is preferably more than 30, 40, 50, or 60 bp in length.
[0093] In some cases, a PV cell selective regulatory element or any regulatory element of this disclosure comprises any one of SEQ ID NOs: 1-32, (ii) a nucleic acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any one of SEQ ID NOs: 1-32, (iii) a functional fragment of any sequence of (i) or (ii), or (iv) a combination of any sequence of (i), (ii) and/or (iii). In some cases, sequence identity is measured by BLAST. In some cases, two or more of SEQ ID NOs: 1-29, or a functional fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto, are used as a regulatory element to increase transgene expression selectively in PV cells as compared to non-PV CNS cells, or to in any target cell type as compared to non-target cell type. In some cases, a functional fragment is one that results in selective expression in a target cell type over one or more non-target cell types.
[0094] In some cases, two or more copies of a regulatory element can be used to enhance selective expression in a target cell, e.g., two or more copies of any one of SEQ ID NOs: 1-29, or a fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto.
[0095] In other cases, one or more of SEQ ID NOs: 1-32, or a fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto, are operably linked to another regulatory element, such as a promoter or enhancer, to further increase selective expression in a target cell. In some cases, one can enhance any gene therapy by adding one or more regulatory elements as disclosed herein to improve or increase expression from the gene therapy in a target cell as compared to non-target cells. In some cases, the target cell is PV neurons or GABAergic cells that express parvalbumin.
[0096] In some aspects, one or more regulatory elements (e.g., any one or more of SEQ ID NOs: 1-32, or a fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto) disclosed herein show cell-type selectivity for a target cell type over at least one, at least two, at least three, at least four, at least five, or more than five non-target cell types. In some cases, a regulatory element drives selective expression or preferential expression in a target cell subtype over at least one, at least two, at least three, at least four, at least five, or more than five non-target subtypes, or all other known subtypes of the cell. For example, GABAergic cells comprise different subtypes, including PV cells. In some cases, the target cell type is a PV cell. In some cases, one or more regulatory elements are selective for PV cells over at least one, at least two, at least three, at least four, at least five, or more than five non-target cell types. In some instances, one or more regulatory elements are selective for PV cells over all other known CNS cell-types.
[0097] In some cases, any one or more of SEQ ID NOs: 1-32, or a fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto, can be combined with any one or more of SEQ ID NOs: 1-32, or a fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto. In some cases, such combined regulatory elements are linked using a linker of 1-50 nucleotides. In some cases, such combined regulatory elements are not linked.
[0098] In some cases, one or more regulatory elements disclosed herein, when operably linked to any transgene (e.g., a reporter transgene or a therapeutic transgene), drives selective expression or preferential expression in at least one target cell type at a level that is statistically significantly higher than the expression driven by CAG, EF1.alpha., a constitutive promoter (e.g., SV40, CMV, UBC, PGK, and CBA), or a non-selective regulatory element (e.g., SEQ ID NO: 34, or a fragment thereof, or a sequence having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto) when operably linked to the same transgene, or by the same construct without the regulatory elements. In some cases, statistically significantly higher means the regulatory elements drive selective expression in the target cell type at a level that is at least 1.1, 1.2, 1.3, 1.4, 1.5, 2, 2.5, 3, 3.5, 4, 4.5, 5, 5.5, 6, 6.5, 7, 7.5, 8, 8.5, 9, 9.5, 10, 10.5, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95, or 100 times the expression level by CAG, EF1.alpha., a constitutive promoter (e.g., SV40, CMV, UBC, PGK, and CBA), or a non-selective regulatory element when operably linked to the same transgene, or by the same construct without the regulatory elements. In some cases, such cell-type selective expression is assayed using a co-localization assay as described herein. In some cases, the target cell type is a parvalbumin cell. In some cases, such co-localization assay is conducted using an anti-PV antibody. In some cases, such co-localization assay is conducted using a PV-Cre mouse as disclosed herein. In some cases, the non-selective regulatory element is SEQ ID NO: 34, or a fragment thereof, or a sequence having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto.
Expression Cassettes
[0099] The terms "expression cassette" and "nucleic acid cassette" are used interchangeably to refer to a polynucleotide molecule or a nucleic acid sequence. In some cases, an expression cassette comprises one or more regulatory elements disclosed herein operably linked to a transgene. In some cases, an expression cassette comprises one or more regulatory elements. In some cases, an expression cassette comprises one or more cell type selective regulatory elements disclosed herein. In some cases, an expression cassette comprises one or more PV cell selective regulatory elements disclosed herein. In some cases, the expression cassette further comprises a promoter. In some cases, an expression cassette comprises one or more sequences of SEQ ID NOs: 1-32 and/or any combination thereof. In some cases, an expression cassette comprises one or more of SEQ ID NOs: 1-32, (ii) a nucleic acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any one of SEQ ID NOs: 1-32, (iii) a functional fragment of any sequence of (i) or (ii), or (iv) a combination of any sequence of (i), (ii) and/or (iii). In some cases, sequence identity is measured by BLAST. In some cases, a regulatory element is located upstream of a transgene in an expression cassette. In some cases, a regulatory element is located downstream of a transgene in an expression cassette. In some cases, an expression cassette further comprises a promoter, e.g., a hSyn1 promoter, CBA promoter, a CMV promoter, an EF1.alpha. promoter, a polyA signal (e.g., SV40 polyA signal), or a post-transcriptional regulatory element such as woodchuck hepatitis virus post-transcriptional regulatory element (WPRE).
[0100] In some aspects, one or more regulatory elements described herein are operably linked to a transgene in an expression cassette. In some cases, a gene therapy comprises an expression cassette comprising a transgene operably linked to one or more, two or more, three or more, four or more, or five or more regulatory elements of the present disclosure to result in selective expression of the transgene in a target tissue or cell type, such as PV neurons. In some cases, an expression cassette comprises one or more PV cell selective regulatory elements or one or more regulatory elements disclosed herein operably linked to a transgene, e.g., a reporter gene, eGFP, SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.2, KV3.3, STXBP1, a DNA binding protein, or a variant or a fragment thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto.
[0101] In some cases, an expression cassette is adapted for delivery via gene therapy. In some cases, an expression cassette is a linear or a circular construct. In some cases, an expression cassette is part of a plasmid, vector, a viral vector, or rAAV.
[0102] In some cases, a gene therapy is administered directly to the CNS of a subject in need thereof or systematically via injection and/or infusion. In some cases, such subject has been diagnosed with a disease or condition associated with a haploinsufficiency or a genetic mutation, such as a haploinsufficiency or a mutation in any one of the following genes: SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.2, KV3.3, or STXBP1. In some cases, the subject is at risk for or has Dravet syndrome, Alzheimer's disease, epilepsy, neurodegeneration, tauopathy, neuronal hypoexcitability and/or seizures. In some cases, such gene therapy is delivered using a virus or a viral vector, such as rAAV. In some cases, an AAV serotype with a tropism for CNS cells and/or ability to cross the blood brain barrier is used, such as AAV9 or a variant thereof.
[0103] In some cases, one or more regulatory elements (e.g., one or more of SEQ ID NOs: 1-32, or a fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto) operably linked to a transgene in an expression cassette result in selective gene expression in PV cells as compared to non-PV CNS cells, or as compared to a control element, such as a CAG, EFla, a constitutive promoter (e.g., SV40, CMV, UBC, PGK, and CBA), or a non-selective regulatory element (e.g., SEQ ID NO: 34, or a functional fragment thereof, or a sequence having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto). In some cases, regulatory elements result in at least 50%, at least 60%, at least 70%, at least 80%, or at least 90% of all cells expressing the transgene are PV neurons. In some cases, regulatory elements result in selective gene expression in PV neurons that is about 1.5 times, 2 times, 3 times, 4 times, 5 times, 6 times, 7 times, 7.5 times, 8 times, 9 times, or 10 times higher than expected for natural distribution of PV neurons in CNS. In some cases, a regulatory element drives selective expression in PV cells, wherein the percentage of PV cells expressing the transgene is at a percentage that is at least 1.5 fold, at least 2 fold, at least 3 fold, at least 4 fold, at least 5 fold, at least 6 fold, at least 7 fold, at least 8 fold, at least 9 fold, or at least 10 fold higher than the expected distribution of PV cells in the CNS, or at least 1-5%, 5%-10%, 10-15%, 15-20%, 20-25%, 25-30%, 30-35%, 35-40%, 40-45%, 45-50%, 50-55%, 55-60%, 65-70%, 70-75%, 75-80%, 80-85%, 85-90%, or 90-95% higher than the expression in PV cells when the transgene is operably linked to CAG, EF1.alpha., a constitutive promoter (e.g., SV40, CMV, UBC, PGK, and CBA), or a non-selective regulatory element having a sequence of SEQ ID NO: 34, or a functional fragment thereof, or a sequence having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto, and as measured in an immunohistochemical co-localization assay. In some cases, a regulatory element in an expression cassette, or use of such regulatory element in an expression cassette, results in selective gene expression in PV cells, or PV cells in the CNS, or PV neurons, wherein about 10%, 15%, 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, or 95% of the cells expressing the transgene are PV positive.
[0104] In some cases, an expression cassette or a gene therapy comprises one or more, two or more, three or more, four or more, five or more, six or more, seven or more, eight or more, nine or more, or ten or more regulatory elements as described in TABLE 1, e.g., SEQ ID NOs: 1-32, or a functional fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto.
[0105] In some cases, one or more PV cell selective regulatory elements or one or more regulatory elements disclosed herein are operably linked to any transgene in an expression cassette. In some cases, the expression cassette is a gene therapy. In some cases, the expression cassette is part of a vector or a plasmid, e.g., a viral vector or rAAV vector. In some cases, the expression cassette is part of AAV1, AAV8, AAV9, or AAVDJ or a variant or hybrid thereof. In some cases, the expression cassette comprises one or more PV cell selective regulatory elements or one or more regulatory elements disclosed herein operably linked to a transgene, wherein the transgene is SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KAV3.2, KV3.3, STXBP1, DNA binding protein (e.g., transcriptional modulator of an endogenous gene), or a variant or a functional fragment thereof, or a sequence having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto. In some cases, such regulatory elements increase the selective expression of the transgene in PV neurons as compared to non-PV CNS cell-types. In some cases, such regulatory elements increase the expression of the transgene selectively in a target cell type, such as a PV neuron. In some cases, the target cell type is a PV cell.
[0106] Techniques contemplated herein for gene therapy of somatic cells include delivery via a viral vector (e.g., retroviral, adenoviral, AAV, helper-dependent adenoviral systems, hybrid adenoviral systems, herpes simplex, pox virus, lentivirus, and Epstein-Barr virus), and non-viral systems, such as physical systems (naked DNA, DNA bombardment, electroporation, hydrodynamic, ultrasound, and magnetofection), and chemical system (cationic lipids, different cationic polymers, and lipid polymers).
[0107] The cloning capacity of vectors or viral expression vectors is a particular challenge for expression of large transgenes. For example, AA V vectors typically have a packaging capacity of .about.4.8 kb, lentiviruses typically have a capacity of .about.8 kb, adenoviruses typically have a capacity of .about.7.5 kb and alphaviruses typically have a capacity of .about.7.55 kb. Some viruses can have larger packaging capacities, for example herpesvirus can have a capacity of >30 kb and vaccinia a capacity of .about.25 kb. Advantages of using AAV for gene therapy include low pathogenicity, very low frequency of integration into the host genome, and the ability to infect dividing and non-dividing cells.
[0108] To address the size constraints of certain viral vectors or to improve expression from viral vectors, the present disclosure contemplates the use of regulatory elements that are shorter than 2.5 kb, 2 kb, 1.5 kb, 1 kb, 900 bp, 800 bp, 700 bp, 600 bp, 500 bp, 400 bp, 300 bp, 200 bp, 150 bp, or 110 bp, but at least 10 bp, 50 bp or 100 bp in length. In some cases, the size of the combined regulatory element is about 2500 bp, 2000 bp, 1500 bp, 1400 bp, 1300 bp, 1200 bp, 1100 bp, or 1000 bp. In some cases, each combined regulatory element has a total length of about 100 bp, 200 bp, 300 bp, 400 bp, 500 bp, 600 bp, 700 bp, 800 bp, 900 bp, 1000 bp, 1100 bp, 1200 bp, 1300 bp, 1400 bp, 1500 bp, 1600 bp, 1700 bp, 1800 bp, 1900 bp, 2000 bp, 2100 bp, 2200 bp, 2300 bp, 2400 bp, or 2500 bp. In some cases, the size of a combined RE has a total length of about 200 bp-3000 bp, 200 bp-2500 bp, 200 bp-2100 bp, 500 bp-2500 bp, 1000 bp-2500 bp, 1500 bp-2500 bp, 1500b-2000 bp, or 2000 bp-2500 bp.
[0109] In some cases, a regulatory element of the disclosure is preferably (i) one that selectively drives expression in a cell-type of interest, such as PV cells; (ii) includes a human derived sequence, and (iii) is smaller than 2.5 kb, 2 kb, 1.5 kb, or 1 kb.
[0110] Also contemplated herein are expression cassettes, which can be a circular or linear nucleic acid molecule. In some cases, an expression cassette is delivered to cells (e.g., a plurality of different cells or cell types including target cells or cell types and/or non-target cell types) in a vector (e.g., an expression vector). A vector can be an integrating or non-integrating vector, referring to the ability of the vector to integrate the expression cassette and/or transgene into a genome of a cell. Either an integrating vector or a non-integrating vector can be used to deliver an expression cassette containing a transgene operably linked to a regulatory element. Examples of vectors include, but are not limited to, (a) non-viral vectors such as nucleic acid vectors including linear oligonucleotides and circular plasmids; artificial chromosomes such as human artificial chromosomes (HACs), yeast artificial chromosomes (YACs), and bacterial artificial chromosomes (BACs or PACs); episomal vectors; transposons (e.g., PiggyBac); and (b) viral vectors such as retroviral vectors, lentiviral vectors, adenoviral vectors, and AAV vectors. Viruses have several advantages for delivery of nucleic acids, including high infectivity and/or tropism for certain target cells or tissues. In some cases, a virus is used to deliver a nucleic acid molecule or expression cassette comprising one or more regulatory elements, as described herein, operably linked to a transgene.
[0111] Preferred characteristics of viral gene therapy vectors or gene delivery vectors include the ability to be reproducible and stably propagated and purified to high titers; to mediate targeted delivery (e.g., to deliver the transgene specifically to a tissue or organ of interest without widespread vector dissemination elsewhere or off-target delivery); and to mediate gene delivery and/or transgene expression without inducing harmful side effects or off-target effects. To avoid potential harmful side effects, targeted expression or tissue/cell type selective expression can be achieved by placing the transgene under the control of a cell-type-selective regulatory element, e.g., one or more of SEQ ID NOs: 1-32, or a functional fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto, an enhancer, promoter, stability element, UTR, or a combination thereof. For example, viral particles containing a viral vector can be designed to infect many different cell types but expression of the transgene is enhanced and/or optimized in a cell type of interest (e.g. PV neurons), and expression of the transgene is reduced and/or minimized in other non-target cell types (e.g., non-PV CNS cells). The differential expression of the transgene in different cell types can be controlled, engineered, or manipulated using different transcription factors or regulatory elements that are selective for one or more cell types. In some cases, one or more regulatory elements, such as a promoter or enhancer, or a combination thereof, are operably linked to a transgene to drive tissue- or cell-selective expression of the transgene. In some cases, one or more regulatory elements used in a gene therapy or a vector drive gene expression in a cell type selective manner, i.e., confer selective gene expression in a target cell, cell type, or tissue, and/or do not drive gene expression in one or more (e.g., at least one, two, three, or four) off-target cells or cell types. In some cases, one or more regulatory elements operably linked to a transgene enhances selective expression of the transgene in a target cell, cell type, or tissue, while the one or more regulatory elements suppress transgene expression in off-target cells, cell type, or tissue, or confers significantly lower, de minimis, or statistically lower gene expression in one or more off-target cells, cell types, or tissue.
[0112] Several serotypes of AAV, non-pathogenic parvovirus, have been engineered for the purposes of gene delivery, some of which are known to have tropism for certain tissues or cell types. Viruses used for various gene-therapy applications can be engineered to be replication-deficient or to have low toxicity and low pathogenicity in a subject or a host. Such virus-based vectors can be obtained by deleting all, or some, of the coding regions from the viral genome, and leaving intact those sequences (e.g., inverted terminal repeat sequences) that are necessary for functions such as packaging the vector genome into the virus capsid or the integration of vector nucleic acid (e.g., DNA) into the host chromatin. An expression cassette comprising a transgene, for example, can be cloned into a viral backbone such as a modified or engineered viral backbone lacking viral genes, and used in conjunction with additional vectors (e.g., packaging vectors), which can, for example, when co-transfected, produce recombinant viral vector particles. In some cases, an AAV serotype that can cross the blood brain barrier or infect cells of the CNS is preferred. In some cases, AAV9 or a variant thereof is used to deliver an expression cassette of this disclosure, comprising one or more PV selective regulatory elements operably linked to a transgene.
[0113] One advantage of delivering expression cassettes of this disclosure using gene therapy, e.g., rAAV, as described herein, is that such therapies can provide more targeted and sustained therapeutic effects over time. Additionally, viral gene therapies can be engineered to have tropism for a cell type or tissue of interest over non-target cell types or tissues. For example, viral gene therapies can be engineered to infect and deliver a payload or a therapeutic agent, e.g., a transcriptional modulator or a transgene, to one or more regions, tissues, or cell types within the CNS (e.g., PV cells), while having minimal effects on off-target tissues or cell types (e.g., non-CNS tissue or cell types, non-PV CNS cells). In some cases, viral gene therapies can be engineered to deliver a transgene across the blood brain barrier and/or target a specific region or tissue within the CNS (e.g., hippocampus) or a cell type within the CNS, e.g., PV cells.
[0114] In some cases, an AAV vector or an AAV viral particle, or virion, used to deliver one or more regulatory elements and a transgene into a cell, cell type, or tissue, in vivo or in vitro, is preferably replication-deficient. In some cases, an AAV virus is engineered or genetically modified so that it can replicate and generate virions only in the presence of helper factors.
[0115] In some cases, the expression cassette is designed for delivery by an AAV or a recombinant AAV (rAAV). In some cases, an expression cassette is delivered using a lentivirus or a lentiviral vector. In some cases, larger transgenes, i.e., genes that exceed the cloning capacity of AAV, are preferably delivered using a lentivirus or a lentiviral vector.
[0116] The AAV used in the compositions and methods described herein can be of any serotype (e.g., AAV1, AAV2, AAV5, AAV8, AAV9, and AAVDJ), including hybrid or chimeric AAV serotypes. In some cases, AAV is used to deliver and/or express a transgene operably linked to one or more regulatory elements that are selective for PV neurons as compared to non-PV CNS cells. In some cases, an AAV with a high tropism for CNS cells and/or crosses the blood brain barrier is used. In some cases, AAV1, AAV8, AAV9, and/or AAVDJ are used to deliver to deliver an expression cassette described herein.
[0117] In some cases, an expression cassette comprises one or more PV cell selective regulatory elements or one or more regulatory elements disclosed herein operably linked to a transgene that is known to be insufficiently expressed in vivo, such as in a disease or condition associated with haploinsufficiency in the gene. In some cases, the transgene is a voltage-gated ion channel (e.g., a sodium ion channel or a potassium ion channel), a neurotransmitter regulator, or a subunit or functional fragment thereof. In some aspects, the transgene is a DNA binding protein, an ion channel, a neurotransmitter regulator, or a subunit of the ion channel or neurotransmitter regulator. In some cases, the transgene is a DNA binding protein that comprises one or more zinc fingers. In some cases, the DNA binding protein comprises a domain of Cas9, a Cas family protein, nuclease-inactivated Cas9 (or dCas9), a dCas family protein, or a transcriptional activator like effector (TALE). In some cases, the transgene is a DNA binding protein that comprises a DNA binding domain of a DNA binding protein or a DNA cleaving protein (e.g., a nuclease, a restriction enzyme, a recombinase, etc.) wherein the DNA cleaving domain or nuclease domain has been deactivated, e.g., a nuclease-deactivated Cas (dCas), a deactivated transcription activator-like effector nuclease, or a nuclease-deactivated zinc finger protein. In some cases, the DNA binding domain is linked to a transcriptional modulating domain (e.g., a transcriptional activator or repressor domain). In some cases, the transgene comprises a gene editing protein, e.g., a Cas protein, Cas9.
[0118] In some aspects, the transgene is a voltage-gated ion channel or a subunit thereof, such as SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.2, or KV3.3, or a functional fragment or variant thereof. In some cases, the transgene is an alpha subunit of a sodium ion channel. In some cases, the transgene is a beta subunit of a sodium ion channel. In some aspects, the neurotransmitter regulator is STXBP1 or a functional fragment or variant thereof.
[0119] In some aspects, an expression cassette is delivered as a viral vector, such as AAV. In some aspects, the AAV is AAV1, AAV8, AAV9, AAV-DJ, scAAV1, scAAV8, or scAAV9. In some aspects, a gene therapy comprising an expression cassette of this disclosure is administered to a subject in need thereof (e.g., a human patient, a mammal, a transgenic animal, or an animal model). In some cases, the subject in need thereof has symptoms of, has been diagnosed with, or is at risk of developing Alzheimer's disease, Dravet syndrome, epilepsy, neurodegeneration, tauopathy, neuronal hypoexcitability, and/or seizures. In some cases, the subject in need thereof has an insufficient gene expression or a mutation in any one or more of SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.2, KV3.3, and STXBP1.
[0120] In some cases, a gene therapy, such as rAAV9, is used to deliver an expression cassette comprising one or more PV cell selective regulatory elements or one or more regulatory elements disclosed herein operably linked a transgene, wherein the transgene is SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.2, KV3.3, STXBP1, a DNA binding protein, or a functional fragment thereof, or a sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any one of SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.2, KV3.3, STXBP1, a DNA binding protein, or a functional fragment thereof. In some cases, the transgene comprises a sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 8'7%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to any one of SEQ ID NOs: 37-43, or a functional fragment thereof, as provided in TABLE 2 below.
[0121] In some cases, the transgene is any one of SEQ ID NOs: 36-43, or a functional fragment thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto. In some cases, one or more regulatory elements disclosed herein are operably linked to any one of SEQ ID NOs: 36-43 in an expression cassette, or a functional fragment thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto.
TABLE-US-00002 TABLE 2 List of amino acid sequences disclosed herein. SEQ ID NO. Gene Amino Acid Sequence 36 eGFP MVSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKF ICTTGKLPVPWPTLVTTLTYGVQCFSRYPDHMKQHDFFKSAMPEGYV QERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKL EYNYNSHNVYIMADKQKNGIKVNFKIRHNIEDGSVQLADHYQQNTPI GDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLL 37 SCN1B MGTLLALVVGAALVSSAWGGCVEVDSDTEAVYGMTFKILCISCKRRS ETTAETFTEWTFRQKGTEEFVKILRYENEVLQLEEDERFEGRVVWNG SRGTKDLQDLSIFITNVTYNHSGDYECHVYRLLFFDNYEHNTSVVKKI HLEVVDKANRDMASIVSEIMMYVLIVVLTIWLVAEMVYCYKKIAAA TEAAAQENASEYLAITSESKENCTGVQVAE 38 SCN2B MHRDAWLPRPAFSLTGLSLFFSLVPPGRSMEVTVPATLNVLNGSD ARLPCTFNSCYTVNHKQFSLNWTYQECNNCSEEMFLQFRMKIINLKL ERFQDRVEFSGNPSKYDVSVMLRNVQPEDEGIYNCYIMNPPDRHRGH GKIHLQVLMEEPPERDSTVAVIVGASVGGFLAVVILVLMVVKCVRRK KEQKLSTDDLKTEEEGKTDGEGNPDDGAK 39 SCN1A MEQTVLVPPGPDSFNFFTRESLAAIERRIAEEKAKNPKPDKKDDDENG PKPNSDLEAGKNLPFIYGDIPPEMVSEPLEDLDPYYINKKTFIVLNKGK AIFRFSATSALYILTPFNPLRKIAIKILVHSLFSMLIMCTILTNCVFMTMS NPPDWTKNVEYTFTGIYTFESLIKIIARGFCLEDFTFLRDPWNWLDFTV ITFAYVTEFVDLGNVSALRTFRVLRALKTISVIPGLKTIVGALIQSVKK LSDVMILTVFCLSVFALIGLQLFMGNLRNKCIQWPPTNASLEEHSIEKN ITVNYNGTLINETVFEFDWKSYIQDSRYHYFLEGFLDALLCGNSSDAG QCPEGYMCVKAGRNPNYGYTSFDTFSWAFLSLFRLMTQDFWENLYQ LTLRAAGKTYMIFFVLVIFLGSFYLINLILAVVAMAYEEQNQATLEEA EQKEAEFQQMIEQLKKQQEAAQQAATATASEHSREPSAAGRLSDSSS EASKLSSKSAKERRNRRKKRKQKEQSGGEEKDEDEFQKSESEDSIRRK GFRFSIEGNRLTYEKRYSSPHQSLLSIRGSLFSPRRNSRTSLFSFRGRAK DVGSENDFADDEHSTFEDNESRRDSLFVPRRHGERRNSNLSQTSRSSR MLAVFPANGKMHSTVDCNGVVSLVGGPSVPTSPVGQLLPEVIIDKPA TDDNGTTTETEMRKRRSSSFHVSMDFLEDPSQRQRAMSIASILTNTVE ELEESRQKCPPCWYKFSNIFLIWDCSPYWLKVKHVVNLVVMDPFVDL AITICIVLNTLFMAMEHYPMTDHFNNVLTVGNLVFTGIFTAEMFLKIIA MDPYYYFQEGWNIFDGFIVTLSLVELGLANVEGLSVLRSFRLLRVFKL AKSWPTLNMLIKIIGNSVGALGNLTLVLAIIVFIFAVVGMQLFGKSYK DCVCKIASDCQLPRWHMNDFFHSFLIVFRVLCGEWIETMWDCMEVA GQAMCLTVFMMVMVIGNLVVLNLFLALLLSSFSADNLAATDDDNEM NNLQIAVDRMHKGVAYVKRKIYEFIQQSFIRKQKILDEIKPLDDLNNK KDSCMSNHTAEIGKDLDYLKDVNGTTSGIGTGSSVEKYIIDESDYMSF INNPSLTVTVPIAVGESDFENLNTEDFSSESDLEESKEKLNESSSSSEGS TVDIGAPVEEQPVVEPEETLEPEACFTEGCVQRFKCCQINVEEGRGKQ WWNLRRTCFRIVEHNWFETFIVFMILLSSGALAFEDIYIDQRKTIKTML EYADKVFTYIFILEMLLKWVAYGYQTYFTNAWCWLDFLIVDVSLVSL TANALGYSELFAIKSLRTLRALRPLRALSRFEGMRVVVNALLGAIPSI MNVLLVCLIFWLIFSIMGVNLFAGKFYHCINTTTGDRFDIEDVNNHTD CLKLIERNETARWKNVKVNFDNVGFGYLSLLQVATFKGWMDIMYA AVDSRNVELQPKYEESLYMYLYFVIFIIFGSFFTLNLFIGVIIDNFNQQK KKFGGQDIFMTEEQKKYYNAMKKLGSKKPQKPIPRPGNKFQGMVFD FVTRQVFDISIMILICLNMVTMMVETDDQSEYVTTILSRINLVFIVLFTG ECVLKLISLRHYYFTIGWNIFDFVVVILSIVGMFLAELIEKYFVSPTLFR VIRLARIGRILRLIKGAKGIRTLLFALMMSLPALFNIGLLLFLVMFIYAIF GMSNFAYVKREVGIDDMFNFETFGNSMICLFQITTSAGWDGLLAPILN SKPPDCDPNKVNPGSSVKGDCGNPSVGIFFFVSYIIISFLVVVNMYIAVI LENFSVATEESAEPLSEDDFEMFYEVWEKFDPDATQFMEFEKLSQFA AALEPPLNLPQPNKLQLIAMDLPMVSGDRIHCLDILFAFTKRVLGESG EMDALRIQMEERFMASNPSKVSYQPITTTLKRKQEEVSAVIIQRAYRR HLLKRTVKQASFTYNKNKIKGGANLLIKEDMIIDRINENSITEKTDLTM STAACPPSYDRVTKPIVEKHEQEGKDEKAKGK 40 STXBP1 MAPIGLKAVVGEKIMHDVIKKVKKKGEWKVLVVDQLSMRMLSSCCKMTDI MTEGITIVEDINKRREPLPSLEAVYLITPSEKSVHSLISDFKDPPT AKYRAAHVFFTDSCPDALFNELVKSRAAKVIKTLTEINIAFLPYESQV YSLDSADSFQSFYSPHKAQMKNPILERLAEQIATLCATLKEYPAVRYR GEYKDNALLAQLIQDKLDAYKADDPTMGEGPDKARSQLLILDRGFDP SSPVLHELTFQAMSYDLLPIENDVYKYETSGIGEARVKEVLLDEDDDL WIALRHKHIAEVSQEVTRSLKDFSSSKRMNTGEKTTMRDLSQMLKK MPQYQKELSKYSTHLHLAEDCMKHYQGTVDKLCRVEQDLAMGTDA EGEKIKDPMRAIVPILLDANVSTYDKIRIILLYIFLKNGITEENLNKLIQH AQIPPEDSEIITNMAHLGVPIVTDSTLRRRSKPERKERISEQTYQLSRWT PIIKDIMEDTIEDKLDTKHYPYISTRSSASFSTTAVSARYGHWHKNKAP GEYRSGPRLIIFILGGVSLNEMRCAYEVTQANGKWEVLIGSTHILTPTK FLMDLRHPDFRESSRVSFEDQAPTME 41 Kv3.1 MGQGDESERIVINVGGTRHQTYRSTLRTLPGTRLAWLAEPDAHSHFD YDPRADEFFFDRHPGVFAHILNYYRTGKLHCPADVCGPLYEEELAFW GIDETDVEPCCWMTYRQHRDAEEALDSFGGAPLDNSADDADADGPG DSGDGEDELEMTKRLALSDSPDGRPGGFWRRWQPRIWALFEDPYSSR YARYVAFASLFFILVSITTFCLETHERFNPIVNKTEIENVRNGTQVRYY REAETEAFLTYIEGVCVVWFTFEFLMRVIFCPNKVEFIKNSLNIIDFVAI LPFYLEVGLSGLSSKAAKDVLGFLRVVRFVRILRIFKLTRHFVGLRVL GHTLRASTNEFLLLIIFLALGVLIFATMIYYAERIGAQPNDPSASEHTHF KNIPIGFWWAVVTMTTLGYGDMYPQTWSGMLVGALCALAGVLTIA MPVPVIVNNFGMYYSLAMAKQKLPKKKKKHIPRPPQLGSPNYCKSV VNSPHHSTQSDTCPLAQEEILEINRAGRKPLRGMSI 42 Kv3.2 MGKIESNERVILNVGGTRHETYRSTLKTLPGTRLALLASSEPQGDCLT AAGDKLQPLPPPLSPPPRPPPLSPVPSGCFEGGAGNCSSHGGNGGNGG SDHPGGGREFFFDRHPGVFAYVLNYYRTGKLHCPADVCGPLFEEELA FWGIDETDVEPCCWMTYRQHRDAEEALDIFETPDLIGGDPGDDEDLA AKRLGIEDAAGLGGPDGKSGRWRKLQPRMWALFEDPYSSRAARFIAF ASLFFILVSITTFCLETHEAFNIVKNKTEPVINGTSPVLQYEIETDPALTY VEGVCVVWFTFEFLVRIVFSPNKLEFIKNLLNIIDFVAILPFYLEVGLSG LSSKAAKDVLGFLRVVRFVIRLRIFKLTRHFVGLRVLGHTLRASTNEF LLLIIFLALGVLIFATMIYYAERVGAQPNDPSASEHTQFKNIPIGFWWA VVTMTTLGYGDMYPQTWSGMLVGALCALAGVLTIAMPVPVIVNNFG MYYSLAMAKQKLPRKRKKHIPPAPLASSPTFCKTELNMACNSTQSDT CLGKENRLLEHNRSVLSGDDSTGSEPPLSPPERLPIRRSSTRDKNRRGE TCFLLTTGDYTCASDGGIRKASTLEPMESTAQTKGDTRPEAHWNCAH LLNFGCPTGSSFPTL 43 Kv3.3 MLSSVCVSSFRGRQGASKQQPAPPPQPPESPPPPPLPPQQQQPAQPGPA ASPAGPPAPRGPGDRRAEPCPGLPAAAMGRHGGGGGDSGKIVINVGG VRHETYRSTLRTLPGTRLAGLTEPEAAARFDYDPGADEFFFDRHPGVF AYVLNYYRTGKLHCPADVCGPLFEEELGFWGIDETDVEACCWMTYR QHRDAEEALDSFEAPDPAGAANAANAAGAHDGGLDDEAGAGGGGL DGAGGELKRLCFQDAGGGAGGPPGGAGGAGGTWWRRWQPRVWAL FEDPYSSRAARYVAFASLFFILISITTFCLETHEGFIHISNKTVTQASPIP GAPPENITNVEVETEPFLTYVEGVCVVWFTFEFLMRITFCPDKVEFLKS SLNIIDCVAILPFYLEVGLSGLSSKAAKDVLGFLRVVRFVRILRIFKLTR HFVGLRVLGHTLRASTNEFLLLIIFLALGVLIFATMIYYAERIGADPDDI LGSNHTYFKNIPIGFWWAVVTMTTLGYGDMYPKTWSGMLVGALCA LAGVLTIAMPVPVIVNNFGMYYSLAMAKQKLPKKKNKHIPRPPQPGS PNYCKPDPPPPPPPHPHHGSGGISPPPPITPPSMGVTVAGAYPAGPHTH PGLLRGGAGGLGIMGLPPLPAPGEPCPLAQEEVIEINRADPRPNGDPA AAALAHEDCPAIDQPAMSPEDKSPITPGSRGRYSRDRACFLLTDYAPS PDGSIRKATGAPLPPQDWRKPGPPSFLPDLNANAAAWISP
[0122] In some cases, the one or more PV cell selective regulatory elements comprise sequences of SEQ ID NOs: 1-32, a functional fragment or a combination thereof, or sequences comprising at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some cases, sequence identity is measured by BLAST. In some cases, such gene therapy is used to treat epilepsies, neurodegeneration, tauopathy, neuronal hypoexcitability, Dravet syndrome and/or Alzheimer's disease. In some cases, such gene therapy is used to treat epilepsy and/or seizures associated with Dravet syndrome and/or Alzheimer's disease. In some cases, treatment using a gene therapy described herein results in reduced seizure frequency and/or duration. In some cases, treatment using a gene therapy described herein results in increased formation of functional sodium ion channels, functional potassium ion channels, or functional neurotransmitter regulatory in vivo.
[0123] In some cases, AAV serotypes 1, 8, and/or 9, or a hybrid thereof can be used with an expression cassettes described herein to target selective expression in PV cells. In some cases, an expression cassette designed for delivery by an AAV comprises a 5' ITR, one or more cell-type selective regulatory elements, an optional enhancer, an optional minimal promoter, a transgene, optionally one or more introns, an optional polyA signal, and a 3' ITR. In some instances, an expression cassette can contain a 5' ITR, two cell-type selective REs, a basal promoter, a transgene, one or more post-transcriptional RNA regulatory elements, and a 3'ITR.
[0124] An exemplary AAV expression cassette is illustrated in FIG. 7. In some cases, the expression cassette contains a 5' AAV ITR, an enhancer (e.g., PV cell selective enhancer or one or more combined regulatory elements), a promoter (e.g., one or more PV cell selective promoters or regulatory elements), a transgene (e.g., SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.2, KV3.3, STXBP1, or a DNA binding protein), a post-transcriptional regulatory element, and a 3' AAV ITR. The promoter can be PV cell selective, or a constitutive promoter. In some cases, the transgene is a reporter gene, e.g., a coding sequence for eGFP, RFP, or a fluorescent marker. In other cases, the transgene is a DNA binding protein that modulates gene expression.
[0125] In some cases, the transgene is a therapeutic transgene, e.g., a coding sequence for SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.2, KV3.3, STXBP1, or a DNA binding protein, or a functional fragment or a sequence having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto. The post-transcriptional regulatory element can be any sequence which influences the expression of a protein from an mRNA or stability of an RNA, for example, an intron, an internal ribosome entry site (IRES), or a woodchuck hepatitis virus post-transcriptional regulatory element. In some cases, the post-transcriptional regulatory element is a combination of two or more post-transcriptional regulatory elements.
[0126] The expression cassette can be designed for delivery by an optimized therapeutic retroviral vector, e.g., a lentiviral vector. The retroviral vector can be a lentiviral vector comprising a left (5') LTR; sequences which aid packaging and/or nuclear import of the virus, at least one cell-type selective regulatory element, optionally a lentiviral reverse response element (RRE); optionally a promoter or active portion thereof; a transgene operably linked to one or more regulatory elements; optionally an insulator; and a right (3') retroviral LTR.
[0127] In some cases, the expression cassette comprises one or more cell-type selective regulatory elements disclosed herein. In some cases, the expression cassette comprises two or more regulatory elements combined. In some examples, the expression cassette comprises two or more regulatory elements that are not combined, for example, a promoter upstream of the transgene and an enhancer or stability element located downstream of the transgene.
[0128] In some cases, the expression cassette contains a putative cell-type selective regulatory element that has selective activity in a cell type of interest, for example, a putative PV cell selective regulatory element. The expression cassette containing the putative regulatory element can be packaged in a viral vector and transfected into an animal model to assess the activity of the putative cell-type selective regulatory element. In some cases, a putative cell type selective regulatory element can be assessed in vitro or ex vivo by delivering a vector containing the putative cell type selective regulatory element into a plurality of cells or cell types that include a target cell or cell type, and then comparing the cell type selective activity of the putative regulatory element to a control regulatory element, such as a constitutive promoter or regulatory element, or a previously known regulatory element.
[0129] In some cases, selective expression is used to selectively express a therapeutic moiety or a transgene in a cell-type of interest (or tissue-type of interest), such as PV neurons in the CNS. In some cases, a vector comprising a cell-type selective regulatory element operably linked to a transgene results in an increased selective expression of the transgene in the cell-type of interest as compared to one or more (e.g., at least two, three, four, or five) other cells, cell types, tissues, or tissue types, or results in a preferred expression of the transgene in the cell-type of interest as compared to one or more cells or cell types, e.g., at least one, two, three, four, or five non-target cell types.
[0130] Any known technique can be used to deliver the regulatory elements and a transgene, or compositions comprising regulatory elements and a transgene, to cells of interest (or a target cell or cell type) to confer or induce in vitro, in vivo, or ex vivo expression of the transgene in a cell-type selective manner.
[0131] The expression cassettes containing cell-type selective regulatory elements of this disclosure further comprise one or more transgenes. The transgenes can be protein-coding genes. In some cases, the expression cassette contains a transgene. The transgene can replace an absent or defective gene, or compensate for deficient expression of a protein inside a cell. The transgene can be involved in a cell signaling pathway. In some cases, a transgene can encode a wild-type protein, a functional fragment thereof, a variant or mutant protein having enhanced therapeutic properties, e.g., enhanced activity. In some cases, the transgene can encode a DNA binding protein comprising one or zinc finger or a domain of dCas9, an ion channel, such as a potassium ion channel or a sodium ion channel, or a subunit thereof, a neurotransmitter factor or a neurotransmitter regulator. In some cases, a transgene can encode an ion channel subunit, a variant, or a mutant thereof. In some cases, the transgene is a DNA binding protein that comprises a DNA binding domain of a DNA binding protein or a DNA cleaving protein (e.g., a nuclease, a restriction enzyme, a recombinase, etc.) wherein the DNA cleaving domain or nuclease domain has been deactivated, e.g., a nuclease-deactivated Cas (dCas), a deactivated transcription activator-like effector nuclease, or a nuclease-deactivated zinc finger protein. In some cases, the DNA binding domain is linked to a transcriptional modulating domain (e.g., a transcriptional activator or repressor domain). In some cases, the transgene comprises a gene editing protein, e.g., a Cas protein, Cas9.
[0132] The regulatory elements disclosed herein can be located at any position within an expression vector or cassette. For example, the regulatory elements can be positioned upstream of an enhancer, downstream of an enhancer but upstream of a promoter, within the 5' UTR of a transgene, within an intron in the transgene, in the 3' UTR of the transgene, or downstream of the transgene. In some cases, one or more regulatory elements are positioned upstream or downstream of the operably linked transgene.
[0133] In some examples, a regulatory element of this disclosure results in selective expression of an operably linked transgene ata level that is at least 0.5, 1.0, 1.1, 1.2, 1.3, 1.4, 1.5, 1.6, 1.7, 1.8, 1.9, 2.0, 2.5, or 3 IU/ml in a target cell type (e.g., PV cells) as measured by ELISA. In some cases, a regulatory element's ability to increase transgene expression can be assessed in a mouse wherein the total amount of transgene expression in the whole mouse and/or the total number of cell types or tissue types having transgene expression are measured.
[0134] When assessing the activity of an expression cassette or vector, the activity or expression can be represented as an activity or expression level per unit dose, or normalized to a dose of expression cassette or vector administered or delivered to a cell, mouse, or a subject. In some cases, expression or activity of a transgene is normalized to an amount of plasmid or DNA (e.g., .mu.g/kg per mouse), or viral particles (e.g., normalized to an amount of genome copies/kg per mouse or subject) used to allow comparison across different expression vectors or cassettes with or without a regulatory element. For example, when assessing a regulatory element's activity in a mouse, selective expression or activity in PV cells assayed can be normalized to a dose of about 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, or greater than 10 or 720 .mu.g of expression vector, cassette, or plasmid per mouse. In some cases, the expression level or activity can be normalized to 10.sup.10, 10.sup.11, 10.sup.12, 10.sup.13, 10.sup.14, or 10.sup.15 gc/kg of viral particles containing an expression vector or cassette as disclosed herein per mouse.
[0135] In some aspects, an expression cassette comprises one or more regulatory elements (e.g., any one or more of SEQ ID NOs: 1-32, or a functional fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto) disclosed herein operably linked to a transgene to result in cell-type selective expression, or preferential expression, of the transgene in a target cell type over at least one, at least two, at least three, at least four, at least five, or more than five non-target cell types. In some cases, an expression cassette comprises one or more regulatory elements operably linked to a transgene to result in cell-type selective expression or preferential expression in a target cell subtype over at least one, at least two, at least three, at least four, at least five, or more than five non-target subtypes, or all other known subtypes of the cell. In some cases, the transgene is any one of SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, and STXBP1, or a functional fragment thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto. In some cases the transgene is a DNA binding protein that modulates an endogenous gene (e.g., an endogenous SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.2, KV3.3, or STXBP1). In some cases, the transgene is any one of SEQ ID NOs: 36-43, or a functional fragment thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto. In some cases, the transgene is a transcriptional modulator. In some cases, the transgene is a DNA binding protein that comprises a DNA binding domain of a DNA binding protein or a DNA cleaving protein (e.g., a nuclease, a restriction enzyme, a recombinase, etc.) wherein the DNA cleaving domain or nuclease domain has been deactivated, e.g., a nuclease-deactivated Cas (dCas), a deactivated transcription activator-like effector nuclease, or a nuclease-deactivated zinc finger protein. In some cases, the DNA binding domain is linked to a transcriptional modulating domain (e.g., a transcriptional activator or repressor domain). In some cases, the transgene is a gene editing protein, such as a Cas family protein, Cas9, a zinc finger nuclease, a zinc finger nuclease, or a transcription activator-like effector nuclease. In some cases, the transgene is a reporter gene or a fluorescent marker. In some cases, an expression cassette disclosed herein is in a viral vector. In some cases, an expression cassette disclosed herein is packaged in an rAAV, such as rAAV9 or rAAVDJ. In some cases, an expression cassette disclosed herein is delivered into a cell as a gene therapy. In some cases, a gene therapy disclosed herein is delivered into a subject, preferably a human or a mammal. In some cases, an expression cassette disclosed herein is used to treat a neurological condition or disease, such as epilepsy, a neurodegenerative disease, tauopathy, neuronal hypoexcitability, Dravet syndrome or Alzheimer's disease.
[0136] In some cases, an expression cassette (e.g., gene therapy, viral vector, vector, or plasmid) comprises any one or more of SEQ ID NOs: 1-32, or a functional fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto, operably linked to a transgene. In some cases, such combined regulatory elements are linked using a linker of 1-50 nucleotides. In some cases, such combined regulatory elements are not linked. In some cases, two or more regulatory elements are located upstream and/or downstream of the promoter. In some cases, two or more regulatory elements are located upstream and/or downstream of the transgene.
[0137] In some cases, an expression cassette comprises one or more regulatory elements disclosed herein, when operably linked to any transgene (e.g., a reporter transgene or a therapeutic transgene), drives selective expression or preferential expression in at least one target cell type at a level that is statistically significantly higher than the expression driven by CAG, EF1.alpha., a constitutive promoter, or a non-selective regulatory element when operably linked to the same transgene, or by the same construct without the regulatory elements. In some cases, statistically significantly higher means the regulatory elements drive selective expression in the target cell type at a level that is at least 1.1, 1.2, 1.3, 1.4, 1.5, 2, 2.5, 3, 3.5, 4, 4.5, 5, 5.5, 6, 6.5, 7, 7.5, 8, 8.5, 9, 9.5, 10, 10.5, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95, or 100 times the expression level by CAG, EF1.alpha., a constitutive promoter, or a non-selective regulatory element when operably linked to the same transgene in the target cell type, or by the same construct without the regulatory elements. In some cases, such cell-type selective expression is assayed using a co-localization assay as described herein.
[0138] In other aspects, an expression cassette comprising one or more regulatory elements disclosed herein operably linked to any transgene disclosed herein results in selective expression or preferential expression of the transgene in a target cell type over at least one, at least two, at least three, at least four, at least five, or more than five non-target cell types or non-target subtypes.
[0139] In some instances, the target cell type is a PV cell. In some cases, the non-target cell subtypes are at least one, at least two, at least three, or at least four of the non-PV GABAergic subtypes disclosed herein. In some cases, an expression cassette comprising a regulatory element disclosed herein is selective for PV cells over all non-PV GABAergic cells or all non-PV CNS cells. In some cases, cell-type selectivity is measured according to a co-localization assay disclosed herein. In some cases, cell-type selectivity is measured using a mouse that expresses Cre in the target cell type.
Parvalbumin (PV) Neurons
[0140] GABAergic neurons produce gamma aminobutyric acid (GABA), the main inhibitory neurotransmitter in the CNS. GABA is important for reducing neural excitability throughout the nervous system. GABA acts at inhibitory synapses by binding specific transmembrane receptors and causing the opening of ion channels which negatively change the membrane polarization. This generally results in hyperpolarization of the cell and increases the signal required to trigger an action potential. Defects in GABAergic neurons can result in an imbalance between excitatory and inhibitory signaling, and have been implicated in many neurological diseases, including Dravet syndrome, epilepsy, neurodegeneration, tauopathies and Alzheimer's disease. Other neurological conditions or diseases implicated include a psychiatric disorder (e.g., schizophrenia, obsessive compulsive disorder, addiction, depression, anxiety, psychosis); an autism spectrum disorder (e.g., Fragile X syndrome, Rett syndrome); epilepsy (e.g., chronic traumatic encephalopathy, generalized epilepsy with febrile seizures plus (GEFS+), epileptic encephalopathy, temporal lobe epilepsy, focal epilepsy, tuberous sclerosis); and/or neurodegeneration (e.g., Alzheimer's disease, Parkinson's disease). In some cases, the neurological condition or disease is any seizure and/or epilepsy related condition or disease wherein PV neurons are implicated.
[0141] Parvalbumin is a calcium-binding protein, which is expressed in about 40% of total GABAergic interneurons in the somatosensory cortex. Within the CNS, PV cells are generally considered GABAergic cells. Various studies have also identified GABAergic cells to include distinct subtypes of cells, including cells that express PV, SOM, CR, CCK, NPY, VIP, or a combination thereof.
[0142] PV neurons are particularly relevant for various neurological diseases or conditions, such as Dravet syndrome, Alzheimer's disease, epilepsy, neurodegeneration, tauopathies and/or seizures. In some cases, a PV neuron-associated neurological condition or disease is a psychiatric disorder (e.g., schizophrenia, obsessive compulsive disorder, addiction, depression, anxiety, psychosis); an autism spectrum disorder (e.g., Fragile X syndrome, Rett syndrome); epilepsy (e.g., chronic traumatic encephalopathy, generalized epilepsy with febrile seizures plus (GEFS+), epileptic encephalopathy, temporal lobe epilepsy, focal epilepsy, tuberous sclerosis); or neurodegeneration (e.g., Alzheimer's disease, Parkinson's disease). In some cases, the neurological condition or disease is any seizure and/or epilepsy related condition or disease wherein PV neurons are implicated. In various aspects, the target cell is PV cells in the CNS, or GABAergic cells that express PV.
[0143] In various aspects, PV-expressing interneurons are also called basket cells, which can be further subdivided by size of the cell body (e.g., large basket cell, small basket cell, and nest basket cell), and dendritic and axonal projection. Physiologically, PV-expressing basket cells are often fast-spiking (FS), characterized by a high-frequency train of action potentials (APs) with little adaptation. It is widely accepted that PV basket neurons innervate the soma and proximal dendrites of excitatory pyramidal neurons. Feedforward inhibition mediated through FS PV-expressing basket neurons can be found in several cortical networks including thalamocortical, translaminar, and interareal circuits. FS PV basket neurons strongly inhibit neighboring excitatory pyramidal neurons. It has been shown that PV basket neurons and pyramidal neurons that share common excitatory inputs tend to be reciprocally connected (feedback inhibition). These connections can serve to regulate the precise time window in which the excitatory neurons can generate spikes in response to excitatory drives. In addition, thalamocortical and intracortical excitatory inputs onto FS PV basket neurons are depressed by high frequency stimulation, which mediates activity-dependent feedforward inhibition. PV-expressing basket cells also innervate other interneurons including other basket cells, and are electrically coupled with each other through gap junctions. It has been proposed that this feature may help to generate and maintain cortical network synchronization and oscillation.
[0144] In some cases, one or more regulatory elements disclosed herein result in increased selectivity in gene expression in PV neurons as compared to at least one, at least two, at least three, at least four, or at least five non-PV expressing neurons. In some cases, non-PV cells include all non-PV GABAergic cells. In some cases, non-PV GABAergic neurons include, but are not limited to, calretinin (CR), somatostatin (SOM), cholecystokinin (CCK), CR+SOM, CR+neuropeptide Y (NPY), CR+vasointestinal polypeptide (VIP), SOM+NPY, SOM+VIP, VIP+choline acetyltransferase (ChAT), CCK+NPY, CR+SOM+NPY, and CR+SOM+VIP expressing cells.
[0145] In some cases, any one of CAG, EF1.alpha., a constitutive promoter (e.g., SV40, CMV, UBC, PGK, and CBA), or a non-selective regulatory element that drives gene expression in a non-cell type selective manner can be used for comparison with PV selective regulatory elements, or any cell type selective regulatory elements, disclosed herein. In some cases, a regulatory element that results in selective expression in PV cells at a level above the expression of a gene operably linked to CAG or EF1.alpha. control is indicative of selectivity to PV cells. In some cases, a regulatory element disclosed herein shows selective gene expression in PV cells that is at least 2%, at least 5%, at least 10%, at least 15%, at least 20%, at least 25%, at least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, or at least 95% higher than the PV expression level from a transgene that is operably linked to CAG, EFla, a constitutive promoter (e.g., SV40, CMV, UBC, PGK, and CBA), or a non-selective regulatory element (e.g., SEQ ID NO: 34, or a functional fragment thereof, or a sequence having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto), and as measured in a co-localization assay. In some cases, a regulatory element disclosed herein shows a selective gene expression in PV cells that is at least 1.5 fold, at least 2 fold, at least 3 fold, at least 4 fold, at least 5 fold, at least 6 fold, at least 7 fold, at least 8 fold, at least 9 fold, or at least 10 fold the expression level under CAG, EF1.alpha., a constitutive promoter (e.g., SV40, CMV, UBC, PGK, and CBA), or a non-selective regulatory element (e.g., SEQ ID NO: 34, or a functional fragment thereof, or a sequence having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto) and as measured in a co-localization assay described herein.
[0146] In preferred cases, one or more regulatory elements described herein selectively drive expression of a transgene in a GABAergic cell, such as a GABAergic cell that expresses parvalbumin over at least one other CNS cell type (e.g., at least two, at least three, at least four, at least five non-PV cells, or two or more, three or more, or four or more non-PV cells and/or non-PV GABAergic neurons).
[0147] In some cases, a target cell type is a GABAergic neuron that expresses parvalbumin, or PV cells.
[0148] One way of selectively expressing a transgene within a subpopulation of cells in the brain is to use a viral vector comprising a transgene operably linked to a cell-type selective regulatory element, or a regulatory element that is selective (or has selective activity) in the subpopulation of cells in the brain, e.g., PV cells. A viral vector can be selected to have high infectivity without selectivity for a particular cell type, while the regulatory element confers selectivity. For example, a cell-type selective regulatory element can drive expression of a transgene in PV neurons and not in other neurons.
[0149] In some cases, the present disclosure involves the use of regulatory elements (i.e., PV cell selective regulatory elements) that selectively drive expression in PV neurons.
[0150] GABAergic cells are inhibitory neurons which produce gamma-aminobutyric acid.
[0151] GABAergic cells can be identified by the expression of glutamic acid decarboxylase 2 (GAD2). Other markers of GABAergic cells include GAD1, NKX2.1, DLX1, DLXS, SST, PV and VIP.
[0152] In some instances, a non-PV CNS cell is an excitatory neuron, a dopaminergic neuron, an astrocyte, a microglia, a motor neuron or a vascular cell. In some instances, a non-GABAergic neuron is a cell that does not express one or more of GAD2, GAD1, NKX2.1, DLX1, DLXS, SST and VIP. In some instances, a non-PV neuron is a GABAergic neuron that does not express parvalbumin. In some instances, other CNS cells refer to CNS cell types that have never expressed any of PV, GAD2, GAD1, NKX2.1, DLX1, DLXS, SST and VIP.
[0153] In some instances, a regulatory element disclosed herein is selective for a PV expressing cell over at least one, two, three, four, five, or more than five non-PV CNS cell-types. In some cases, non-PV cell types include non-PV GABAergic cells. In some cases, the cell type of interest is a PV cell. In some cases, REs selective for PV cells are referred to as PV cell selective regulatory elements.
[0154] In some cases, the PV cell selective regulatory elements disclosed herein include the sequences of SEQ ID NOs: 1-32, or any combination thereof.
[0155] In some cases, one or more PV cell selective regulatory elements or one or more regulatory elements disclosed herein are used to increase expression of a transgene in PV-expressing cells by at least 2, 5, 10, 15, 20, 30, 40, 50, 60, 70, 80, 90, 100, or more fold as compared to expression without the regulatory element. In some cases, a RE in an expression cassette increases gene expression by at least 1.5%, 2%, 5%, 10%, 15%, 20%, or 50%, or more than 1.5%, 2%, 5%, 10%, 15%, 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, or 100% as compared to expression without the regulatory elements. In some cases, compositions and methods of use thereof comprise expression cassettes containing one or more regulatory elements that result in a 10-500% increase in transgene expression, e.g., expression of SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KCNC1 (also known as KV3.1), KCNC3 (also known as KV3.3), STXBP1, a DNA binding protein, or a variant or functional fragment thereof, or a protein thereof, as compared to the level without the regulatory elements or as compared to a non-selective regulatory element (e.g., CAG, EFla, a constitutive promoter, or SEQ ID NO: 34, or a functional fragment thereof, or a sequence having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto). In some cases, the increase in gene expression and/or protein level of any one of SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KCNC1, KCNC3, and STXBP1 is 1.5-5%, 5%-10%, 10-15%, 15-20%, 20-30%, 30-40%, 40-50%, 50-60%, 60-70%, 70-80%, 80-90%, 90-100%, 100-150%, 150-200%, 250-300%, 300-350%, 350-400%, 400-450%, 450-500%, or 1.5-20%, 20%-50%, 50%-100%, 100-200%, 200-300%, 300-400%, or 400-500% as compared to the level without the expression cassette or regulatory elements. In some cases, such gene or protein expression is selective in PV cells as compared to an expression cassette comprising a control (e.g., CAG, EF1.alpha., a constitutive promoter (e.g., SV40, CMV, UBC, PGK, and CBA), or a non-selective regulatory element) or a non-cell type selective regulatory element (e.g., SEQ ID NO: 34, or a functional fragment thereof, or a sequence having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto).
[0156] In some cases, selectivity of expression in PV cells can be calculated by dividing the number of cells that express both PV and eGFP (the transgene operably linked to one or more regulatory elements) by the total number of cells that express eGFP, and multiplying by 100 to convert into a percentage. PV cell selective regulatory elements as described herein can be highly selective for expression in PV cells. For example, PV cell selective regulatory elements or one or more regulatory elements disclosed herein can exhibit about 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or greater than about 99% selectivity for PV neurons.
[0157] In some cases, a PV cell selective regulatory element or any regulatory element disclosed herein confers selectivity in expressing a transgene in PV neurons at a level that is statistically higher than a control regulatory element, e.g., EF1.alpha. or a previously known regulatory element. In some instances, the statistical difference between a PV cell selective regulatory element and a control regulatory element is at least 2 fold, 5 fold, 10 fold, 20 fold, or more than 2 fold difference, or more than 5 fold, 10 fold, or 20 fold difference as determined by any one of the methods described herein, such as a co-localization assay.
[0158] The present disclosure includes regulatory elements that are selective for PV cells. These PV cell selective REs or any cell type selective REs are preferably short, preferably less than about 1100 base pairs, 1000 bp, 900 bp, 800 bp, 700 bp, 600 bp, 500 bp, 400 bp, 300 bp, 200 bp, or less than about 110 bp. The PV cell selective REs or any cell type selective REs can be between 1050 bp and 100 bp, between 100 bp and 500 bp, or between 500 bp and 1050 bp. Some examples of PV cell selective regulatory elements are provided by SEQ ID NOs 1-32, or a functional fragment or combination thereof. Other PV cell selective regulatory elements contemplated by the present disclosure include sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity to any of these sequences described herein, or a part or fragment of one of the sequences described herein.
[0159] In some cases, a PV cell selective regulatory element or any regulatory element disclosed herein has at least about 70%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more than 99% identity to a sequence described herein, or a fragment of a sequence described herein. In some cases, a PV cell selective regulatory element has at least about 80% identity to at least about 30%, 40%, 50%, 60%, 70%, 80%, 90% or 95% of a sequence described herein, or a functional fragment thereof.
[0160] In some cases, a PV cell selective regulatory element comprises at least 80% identity to any one or more of SEQ ID NOs: 1-32, or a functional fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto. In some cases, a PV cell selective regulatory element has 90% identity to 50% or more of a sequence of SEQ ID NOs: 1-22. In some instances, a PV-selective regulatory element is a functional fragment of any of SEQ ID NOs: 1-32 or a combination thereof. In some cases, the functional fragment is able to selectively express a transgene in PV cells with at least 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, or greater than 95% selectivity of expression in PV cells.
[0161] In some cases, two or more PV cell selective regulatory elements of this disclosure or any two or more regulatory elements disclosed herein are combined to form combination regulatory elements. In some instances, two or more, three or more, four or more, five or more, six or more, seven or more, eight or more, nine or more, or ten or more PV cell selective regulatory elements, or a plurality of regulatory elements disclosed herein, are combined. For example, SEQ ID NO: 31 is a combination of SEQ ID NOs 1 and 23-29. As another example, SEQ ID NO: 32 is a combination of SEQ ID NO: 8 and SEQ ID NO: 23-29. In some cases, fragments of two or more PV-selective regulatory elements can be combined to form a combination regulatory element. For example, 50% of SEQ ID NO: 1 can be combined with 30% of SEQ ID NO: 8 and 90% of SEQ ID NO: 30 to form a combination regulatory element.
[0162] In some cases, one or more PV cell selective regulatory elements of this disclosure or any one or more regulatory elements disclosed herein selectively express an operably linked transgene in PV neurons as compared to one or more other CNS cell types. This selective expression can be quantified by counting the number of PV neurons which express detectable levels of the linked transgene as a percentage of the total number of cells expression the transgene, including the number of non-PV neurons which express the transgene. In other words, selectivity of a PV regulatory element in a particular cell type or target cell can be determined by measuring and/or comparing the number of PV neurons (or target cells) expressing the transgene that is operably linked to the regulatory element relative to the number of non-target cell types that express the transgene (or the total number of cells expressing the transgene).
[0163] In some cases, PV cell selective regulatory elements can exhibit about 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or greater than about 99% selectivity for PV neurons, PV neurons in the CNS, or GABAergic neurons that also express PV. In some cases, one or more regulatory elements of this disclosure can exhibit about 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or greater than about 99% selectivity for PV neurons as compared to CAG or EF1.alpha. or non-cell-type selective element, or as compared to non-PV CNS cells, or as compared to at least one, at least two, at least three, at least four, or at least five other non-PV GABAergic neuronal sub-types in the CNS.
[0164] In some cases, a PV selective regulatory element confers selectivity in expressing a transgene in PV neurons at a level that is statistically higher than a control regulatory element, e.g., CAG, EF1.alpha., a constitutive promoter (e.g., SV40, CMV, UBC, PGK, and CBA), a non-selective regulatory element, or a previously known regulatory element. In some instances, the statistical difference between a PV cell selective RE and a control element is at least 2 fold, 5 fold, 10 fold, 20 fold, or more than 2 fold difference, or more than 5 fold, 10 fold, or 20 fold difference as determined by any one of the methods described herein. In some cases, the selectivity in PV is measured using a co-localization assay as described herein.
[0165] In some aspects, the cell-type selective regulatory elements described herein are useful for selectively modulating expression of a transgene in a CNS cell type compared to other CNS cell types. For example, the cell-type selective regulatory elements described herein can be useful for selectively modulating expression of a transgene in PV cells over other CNS cells, including other types of neurons. For gene therapy, selective expression of a transgene in a target cell type and/or minimized expression of the transgene in a non-target cell type can be desired. Expression of the transgene in an unintended cell-type (e.g., non-target cell type) can result in an adverse effect to the subject. Expression of the transgene in an unintended cell-type can counteract the therapeutic effect of the transgene in the intended cell type. For example, a transgene intended for expression in PV cells can have a negative effect for the subject if expressed in glutamatergic neurons. The cell-type selective regulatory elements described herein can be used in expression cassettes to ensure appropriate expression of a transgene and/or to reduce off-target effects of gene therapy.
[0166] The cell-type selective regulatory elements herein can be used in gene expression cassettes whereby they are operably linked to one or more transgenes. Such gene expression cassettes are used to deliver transgenes into cells for expression. The expression cassette can contain a cell-type selective regulatory element as described herein, a combination of cell-type selective regulatory elements, or a fragment of a cell-type selective regulatory element as described herein operably linked to a transgene.
[0167] Preferably, the expression cassettes herein include one or more cell-type selective regulatory elements operably linked to a transgene, whereby the two do not function together in their endogenous context in vivo. For example, a transgene for sodium ion channel beta subunit, such as SCN1B, can be operably linked to one or more regulatory elements that do not function in the same context in vivo, or do not detectably drive expression of SCN1B endogenously. Similarly, a nucleic acid cassette can include a neurotransmitter regulator, such as STXBP1, operably linked to a regulatory element that does not function in the same context endogenously or in vivo, or is not in the same open reading frame, or is not on the same human chromosome, or does not detectably drive expression of STM3P1 in vivo. In some cases, a cell-type selective regulatory element is linked to a transgene, wherein the cell-type selective regulatory element does not regulate the endogenous gene corresponding to the transgene in vivo.
[0168] In some aspects, cell-type selective regulatory elements disclosed herein are derived from sequences isolated from a human chromosomal locus different from a locus of a native gene corresponding to the transgene. Thus, in some instances, an expression cassette comprises cell-type selective regulatory element(s) having a sequence derived from a chromosome different from the chromosome corresponding to the transgene on the same cassette. In other instances, regulatory element(s) of the disclosure and a transgene that are operably linked in an expression cassette are derived from sequences located more than 20 kb apart in the human genome, or at distal genomic locations. When two or more human derived regulatory elements are utilized on an expression cassette, the two or more regulatory elements can have sequences located more than 5 kb apart, more than 10 kb apart, more than 15 kb apart, or more than 20 kb apart in the human genome, or wherein the two or more regulatory elements do not interact with each other naturally in the genome.
[0169] In some cases, an expression cassette comprising PV-selective regulatory elements can exclude known sequences derived from hSyn1 or GAD2 promoter sequences. In some instances, the PV-selective regulatory elements do not comprise the full promoter sequence of any one of the GAD2, GAD1, SYN1, NKX2.1, DLX1, DLX5, SST and VIP promoters. In some instances, the PV-selective regulatory elements do not comprise more than 500 contiguous base pairs of sequence derived from the promoter or one or more of GAD2, GAD1, SYN1, NKX2.1, DLX1, DLX5, SST and VIP. In some instances, the PV-selective regulatory elements do not comprise sequences which are within 1 kb, 2 kb, 3 kb, 4 kb, 5 kb, 6 kb, 7 kb, 8 kb, 9 kb, or 10 kb of the transcription start site of any one of GAD2, SYN1, NKX2.1, DLX1, DLX5, SST, and VIP.
[0170] In some cases, a transgene is useful to treat a disease associated with a specific cell type of interest. In some cases, a cell type of interest is a neuron, an inhibitory neuron, a GABAergic neuron, or a PV neuron. In some cases, a transgene is any one or more of SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, and STXBP1. In some cases, a transgene is a DNA binding protein that modulates expression of a gene (e.g., a transcriptional activator or a transcriptional repressor that modulates expression of an endogenous gene). In some cases, a transgene is a gene editing protein, such as a zinc finger nuclease, a transcription activator-like effector nuclease, a Cas family protein. In some cases, a transgene is a reporter gene or a detectable marker, such as eGFP, tdTomato, or RFP. In some cases, a transgene is a Cas protein, such as Cas9.
[0171] Transgenes useful to treat a condition associated with PV neuron cells can be incorporated in a vector, nucleic acid cassette, or method as described herein. Transgenes used herein generally do not contain introns, or do not contain more than one intron. A transgene can be obtained from a cDNA sequence rather than from genomic sequence. In some cases, transgenes can contain some, or all, of their endogenous introns. In some examples, such a transgene encodes for a DNA binding domain or an ion channel. Examples of DNA binding domains that can be encoded for in the expression cassettes of this disclosure include zinc fingers, Cas9, a Cas family protein, dCas9, a dCas family protein or a transcriptional activator like effector (TALE). In some cases, the transgene is a DNA binding protein that comprises a DNA binding domain of a DNA binding protein or a DNA cleaving protein (e.g., a nuclease, a restriction enzyme, a recombinase, etc.) wherein the DNA cleaving domain or nuclease domain has been deactivated, e.g., a nuclease-deactivated Cas (dCas), a deactivated transcription activator-like effector nuclease, or a nuclease-deactivated zinc finger protein. In some cases, the DNA binding domain is linked to a transcriptional modulating domain (e.g., a transcriptional activator or repressor domain). In some cases, the transgene comprises a gene editing protein, e.g., a Cas protein, Cas9. In some cases, the transgene is a subunit or a component of an ion channel or a membrane protein, or a gene associated with a neurological condition or disease disclosed herein. Examples of ion channel transgenes which can be used in the expression cassettes of this disclosure include voltage gated and ligand gated ion channels. Voltage gated ion channels include sodium channels, calcium channels, potassium channels, and proton channels. In some instances, the transgene codes for a subunit of a voltage gated sodium channel. Examples of voltage gated sodium channel subunits include SCN1B (NM_001037.4), SCN1A (NM_001165963.1), and SCN2B, (NM_004588.4).
[0172] In some instances, the transgene codes for a subunit of a voltage gated potassium channel. Examples of voltage gated sodium channel subunits include KCNC1 (NM_001112741.1), and KCNC3 (NM_004977.2). In some cases, a transgene is any one or more of SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, STXBP1, a variant and a functional fragment thereof. In some cases, a transgene is a sequence having at least 80%, at least 85%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, at least 99%, or at least 99.5% sequence identity to any one of SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, STXBP1, a variant or a functional fragment thereof. In some cases, such sequence identity is measured using BLAST.
[0173] In some cases, an expression cassette disclosed herein comprises one or more PV-selective regulatory elements or one or more regulatory elements of this disclosure operably linked to a transgene having at least 80%, at least 85%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, at least 99%, or at least 99.5% sequence identity to a sequence encoding any one of SEQ ID NOs: 37-43, or a functional fragment or variant thereof, or the GenBank sequences corresponding to SEQ ID NOs: 37-43. In some cases, an expression cassette disclosed herein comprises a transgene having a sequence according to (i) a sequence of SEQ ID NOs: 37-43, or (ii) a functional fragment thereof, or (iii) a sequence having at least 80%, at least 85%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, at least 99%, or at least 99.5% sequence identity to (i) or (ii).
[0174] In some examples, the transgene is a neurotransmitter regulator, or a variant or functional fragment thereof. A neurotransmitter regulator may be involved in regulating production or release of a neurotransmitter in the CNS. For example, a neurotransmitter regulator may assist with synaptic fusion to release neurotransmitters. An example of a neurotransmitter regulator is STXBP1 (NM_001032221.3) or a functional fragment thereof, or a sequence having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto. The transgene may also be a subunit of a neurotransmitter regulator.
[0175] In some case, an expression cassette of this disclosure can contain an AAV2 5' ITR, a PV-selective enhancer, a PV-selective promoter, or a combination of one or more PV-selective promoters and enhancers, a cDNA of SCN1B, a WPRE, a hGH polyA signal, PV-selective regulatory element, and an AAV2 3'ITR. In one example, the expression cassette comprises an AmpR promoter, and AmpR coding sequence, a bacterial origin of replication, an AAV2 ITR, SEQ ID NO: 8, SEQ ID NOs: 23-29, a transgene (coding sequence, a WPRE, a human growth hormone poly A signal, a AAV2 ITR, and an f1 origin.
[0176] As another example, an expression cassette of this disclosure can contain an AAV2 5' ITR, an enhancer, a promoter, a transcriptional activator of the endogenous SCN1A gene, a WPRE, a hGH polyA signal, a regulatory element, and an AAV2 3'ITR. In some cases, an expression cassette comprises AAV2 5' ITR, a promoter, an intronic element, transcriptional modifier, synthetic polyA, and an AAV2 3' ITR. In some cases, an expression cassette of this disclosure can contain an AAV2 5' ITR, a PV-selective enhancer, a PV-selective promoter, a sequence encoding a transcriptional activator of SCN1A or SCN1B, a WPRE, a hGH polyA signal, a PV-selective regulatory element or any of the regulatory elements disclosed herein, and an AAV2 3'ITR.
[0177] An expression cassette comprising one or more regulatory element of this disclosure can be used to treat a medical condition. In some cases, an expression cassette containing a regulatory element of this disclosure is used to treat a neurological condition or a neurodegenerative condition. The neurological condition can be caused by a known genetic event or may have an unknown cause.
[0178] The neurological condition can be a disease associated with PV neurons. The neurological condition can be a disease associated with inhibitory neurons, such as PV neurons. Diseases or conditions associated with PV neurons can be treated by delivering an expression cassette carrying a transgene and one or more PV cell-selective regulatory elements or any of the regulatory elements as described herein to a cell in vivo. In some aspects, expression cassettes comprising a transgene operably linked to one or more PV cell-selective regulatory elements or any of the regulatory elements disclosed herein can be used to treat Dravet syndrome, Alzheimer's disease, epilepsy, a neurodegenerative disorder, tauopathy, neuronal hypoexcitability and/or seizures. In some cases, an expression cassette of this disclosure is used to treat a psychiatric disorder (e.g., schizophrenia, obsessive compulsive disorder, addiction, depression, anxiety, psychosis); an autism spectrum disorder (e.g., Fragile X syndrome, Rett syndrome); epilepsy (e.g., Dravet syndrome, chronic traumatic encephalopathy, generalized epilepsy with febrile seizures plus (GEFS+), epileptic encephalopathy, temporal lobe epilepsy, focal epilepsy, tuberous sclerosis); and/or neurodegeneration (e.g., Alzheimer's disease, Parkinson's disease). In some cases, the neurological condition or disease is any seizure and/or epilepsy related condition or disease wherein PV neurons are implicated.
[0179] Majority of Dravet syndrome cases is associated with mutations in the SCN1A and/or SCN2A genes. Mutations or abnormalities in SCN1A has also been associated with seizure disorders, epilepsy, autism, familial hemiplegic migraine type 3 (FHM3), genetic epilepsy with febrile seizures plus (GEFS+), and effectiveness of certain anti-seizure medications. For instance, ICS5N+5G>A mutation in SCN1A is associated with the maximum safe amount (dose) of the anti-seizure drugs phenytoin and carbamazepine.
[0180] In some Alzheimer's patients, production of amyloid .beta. (A.beta.) involving many peptides and proteases that can affect excitability of neurons, causing seizures and downregulation of the Nav1.1 sodium channel in PV neurons.
[0181] Diseases associated with dysfunctional PV neurons such as those due to loss of function mutations in SCN1A or Nav1.1 include: Dravet syndrome, Ohtahara syndrome, epilepsy, early infantile epileptic encephalopathy 6 (EIEE6), familial febrile seizures 3A (FEB3A), intractable childhood epilepsy with generalized tonic-clonic seizures (ICEGTC), migraine, familial hemiplegic 3 (FHM3), Panayiotopoulos syndrome, familial atrial fibrillation 13 (ATFB13), generalized epilepsy with febrile seizures plus type 1 (gefs+type 1), Brugada syndrome, nonspecific cardiac conduction defect, generalized epilepsy with febrile seizures plus, benign familial infantile seizures, early infantile epileptic encephalopathy11 (EIEE11), benign familial infantile epilepsy, neurodegeneration, tauopathies and Alzheimer's disease. In some cases, the neurological condition is Dravet syndrome. Dravet syndrome is associated with mutations in the SCN1A and/or SCN2A genes. In some cases, one or more regulatory elements of this disclosure are used in a gene therapy or an expression cassette to treat a neurological condition or disease associated with PV neurons, e.g., a psychiatric disorder (e.g., schizophrenia, obsessive compulsive disorder, addiction, depression, anxiety, psychosis); an autism spectrum disorder (e.g., Fragile X syndrome, Rett syndrome); epilepsy (e.g., Dravet syndrome, chronic traumatic encephalopathy, generalized epilepsy with febrile seizures plus (GEFS+), epileptic encephalopathy, temporal lobe epilepsy, focal epilepsy, tuberous sclerosis); or neurodegeneration (e.g., Alzheimer's disease, Parkinson's disease). In some cases, one or more regulatory elements of this disclosure (e.g., PV neuron selective regulatory elements) are used to treat Dravet syndrome and/or Alzheimer's disease (e.g., in an expression cassette, a vector, or a gene therapy). In some cases, the neurological condition or disease is any seizure and/or epilepsy related condition or disease wherein PV neurons are implicated.
[0182] Methods and compositions of this disclosure can be used to treat a subject who has been diagnosed with a disease, for example, a neurological or neurodegenerative disease. The subject can be a patient suffering from a form of epilepsy. In some instances, the subject is a patient with Dravet syndrome. The subject can be a patient suffering from a neurodegenerative disease, for example, a patient with Alzheimer's disease. In some instances, epilepsy, encephalopathy, and/or seizures are associated with a genetic mutation in SCN8A. In some cases, a genetic mutation in SCN8A can give rise to epilepsy syndromes, e.g., Dravet syndrome. In some instances, a genetic mutation in STXBP1 is associated with encephalopathy with epilepsy, characterized by recurrent seizures.
[0183] In some instances, a subject treated with one or more compositions described herein is one diagnosed with a mutation or genetic aberration in an ion channel or a neurotransmitter regulator (e.g., a syntaxin binding protein). Examples of such mutations include mutations in SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KCNC1, KCNC3, and/or STXBP1, or combination thereof. The expression cassette containing a cell-type selective regulatory element as described herein can be delivered to a subject to treat or prevent a disease with symptoms associated with a specific cell type. For example, an expression cassette comprising a transgene operably linked to one or more PV cell selective regulatory element is delivered to a subject who has symptoms, or is at risk of developing symptoms, associated with PV neurons.
[0184] In some cases, the treatment can be administered to a subject with, or at risk of developing, Dravet syndrome. Symptoms associated with Dravet syndrome include seizures, memory defects, developmental delay, poor muscle tone and/or cognitive problems. Treatment with an expression cassette of this disclosure can result in an improvement of one or more symptoms, such as a reduction in number, duration, and/or intensity of seizures. Administration of a gene therapy as described herein to a subject at risk of developing Dravet syndrome can prevent the development of or slow the progression of one or more symptoms.
[0185] In another example the treatment may be administered to a subject suffering from Alzheimer's disease. Symptoms associated with Alzheimer's disease include short term memory loss, cognitive difficulties, seizures, and difficulties with language, executive functions, perception (agnosia), and execution of movements (apraxia). Treatment with an expression cassette of this disclosure can result in an improvement of one or more Alzheimer's disease symptoms, such as a reduction in progression of memory loss, or the prevention of one or more symptoms. In some cases, the treatment can result in a correction of high gamma power brain activity. The treatment can result in a decrease in seizure frequency and/or seizure severity, or a decrease in high gamma power activity by 10%, 20%, 30%, 40%, 50%, 60%, or 70%. In some cases, the treatment can result in an improvement in cognitive function. Learning and/or memory can be improved by 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 100% or more than 100%.
[0186] Methods and compositions of this disclosure can be used to treat a subject who is at risk of developing a disease. The subject can be known to be predisposed to a disease, for example, a neurological disease or a disease associated with epilepsy, seizures, and/or encephalopathy. The subject can be predisposed to a disease due to a genetic event, or due to known risk factors. For example, a subject can carry a mutation in SCN1A which is associated with Dravet syndrome. In some cases the subject can be predisposed to a disease such as Alzheimer's disease due to the age of the subject.
[0187] The treatment can result in a decrease or cessation of symptoms. For example, treatment can improve learning, memory, cognitive function, and/or motor function; reduce frequency and/or duration of seizures; and/or reduce temperature sensitivity (or increase the temperature threshold for triggering a seizure).
[0188] In some instances, the target cell type of a gene therapy or expression cassette disclosed herein is a PV cell. In some cases, the non-target cell subtypes are at least one, at least two, at least three, or at least four of the non-PV GABAergic subtypes disclosed herein. In some cases, an expression cassette comprising a regulatory element disclosed herein is selective for PV cells over all non-PV CNS cells. In some cases, cell-type selectivity is measured according to a co-localization assay disclosed herein. In some cases, cell-type selectivity is measured using a mouse that expresses Cre in the target cell type.
[0189] In some instances, the treatment does not result in an adverse reaction for the subject. Treatment with a gene therapy containing a PV-selective regulatory element can cause fewer, or less severe, adverse reactions in a subject than treatment with a similar gene therapy containing the same transgene linked to a non-selective regulatory element.
[0190] In various aspects, any expression cassette disclosed herein can be adapted for or used in a gene therapy (e.g., rAAV or rAAV9 gene therapy) to treat any one or more of Dravet syndrome, Alzheimer's disease, epilepsy, neurodegeneration, tauopathy, neuronal hypoexcitability and/or seizure. In some cases, a gene therapy comprises any one or more, two or more, three or more, four or more, five or more, six or more, seven or more, eight or more, nine or more, or ten or more of SEQ ID NOs: 1-32, or a functional fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto, operably linked to a transgene. In some cases, a gene therapy comprises an expression cassette of this disclosure. In some cases, a gene therapy comprises one or more regulatory elements disclosed herein operably linked to any transgene (e.g., a reporter transgene or a therapeutic transgene) such that the regulatory elements drive selective expression or preferential expression in at least one target cell type at a level that is statistically significantly higher than the expression driven by CAG or EF1.alpha. or a non-selective regulatory element when operably linked to the same transgene, or by the same construct without the regulatory elements. In some cases, statistically significantly higher means the regulatory elements drive selective expression in the target cell type ata level that is at least 1.1, 1.2, 1.3, 1.4, 1.5, 2, 2.5, 3, 3.5, 4, 4.5, 5, 5.5, 6, 6.5, 7, 7.5, 8, 8.5, 9, 9.5, 10, 10.5, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95, or 100 times the expression level by the CAG, EF1.alpha., a constitutive promoter, or a non-selective regulatory element when operably linked to the same transgene in the target cell type, or by the same construct without the regulatory elements. In some cases, such cell-type selective expression is assayed using a co-localization assay as described herein. In some cases, the transgene is any one or more of SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, STXBP1, and a functional fragment thereof. In some cases, the transgene is a DNA binding protein that modulates expression of an endogenous gene, such as a transcriptional modulator, a transcriptional activator, or a transcriptional repressor. In some cases, the transgene is a DNA binding protein that comprises a DNA binding domain of a DNA binding protein or a DNA cleaving protein (e.g., a nuclease, a restriction enzyme, a recombinase, etc.) wherein the DNA cleaving domain or nuclease domain has been deactivated, e.g., a nuclease-deactivated Cas (dCas), a deactivated transcription activator-like effector nuclease, or a nuclease-deactivated zinc finger protein. In some cases, the DNA binding domain is linked to a transcriptional modulating domain (e.g., a transcriptional activator or repressor domain). In some cases, the transgene is a gene editing protein, such as a zinc finger nuclease or a transcription activator-like effector nuclease. In some cases, a transgene is a reporter gene or a detectable marker, such as eGFP, tdTomato, or RFP. In some cases, a transgene is a Cas protein, such as Cas9.
[0191] In some cases, a gene therapy comprising an expression cassette disclosed herein is used to treat a neurological condition or disease. In some cases, a gene therapy comprising an expression cassette disclosed herein is used to treat a neurological condition or disease, wherein the expression cassette comprises any one or more, two or more, three or more, four or more, five or more, six or more, seven or more, eight or more, nine or more, or ten or more of SEQ ID NOs: 1-32, or a functional fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto, operably linked to a transgene. In some cases, the transgene is any one or more of SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, STXBP1, a DNA binding protein and a functional fragment thereof. In some cases, a gene therapy comprising an expression cassette disclosed herein is used to treat Dravet syndrome. In some aspects, a gene therapy comprising an expression cassette disclosed herein is used to treat Alzheimer's disease. In some cases, a gene therapy comprising an expression cassette disclosed herein is used to treat epilepsy and/or seizure symptoms associated with Dravet syndrome and/or Alzheimer's disease. In some cases, treating any one of Dravet syndrome, Alzheimer's disease, epilepsy, neurodegeneration, tauopathy, neuronal hypoexcitability and/or seizures comprises delivering or administering a gene therapy of this disclosure to a cell of a subject in need thereof. In some cases, the subject in need thereof is at risk for or has any one of Dravet syndrome, Alzheimer's disease, epilepsy, and/or seizures. In some cases, the subject is a child or a minor. In some cases, a gene therapy comprising an expression cassette disclosed herein is used to treat an infant, a child, or a minor diagnosed with or is at risk of developing Dravet syndrome. In some cases, a gene therapy comprising an expression cassette disclosed herein is used to treat a subject comprising a mutation or a genetic defect in any one or more of SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, and STXBP1.
[0192] In some aspects, the present disclosure provides a method of treating any one of Dravet syndrome, Alzheimer's disease, epilepsy, neurodegeneration, tauopathy, neuronal hypoexcitability and/or seizures, comprising administering a gene therapy into a cell of a subject, wherein the gene therapy comprises an expression cassette disclosed herein. In some cases, such expression cassette comprises any one or more of SEQ ID NOs: 1-32, or a functional fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto, operably linked to a transgene, wherein the transgene is any one of SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, STXBP1, and a functional fragment thereof. In some cases, the transgene is a subunit of a sodium ion channel or a potassium ion channel. In some cases, the transgene is a syntaxin binding protein. In some cases, the transgene is a transcriptional modulator, e.g., a transcriptional activator or a transcriptional repressor. In some cases, the transgene is a transcriptional modulator that modulates the expression of an endogenous gene (e.g., SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, or STXBP1). In some cases, a transgene is a gene editing protein, such as a zinc finger nuclease or a transcription activator-like effector nuclease. In some cases, the transgene is a Cas protein, such as Cas9.
[0193] In other aspects, the present disclosure provides a method for modifying any gene therapy designed for treating Dravet syndrome, Alzheimer's disease, epilepsy, neurodegeneration, tauopathy, neuronal hypoexcitability and/or seizures by adding one or more regulatory elements disclosed herein to improve the cell-type selectivity of the gene therapy. In some cases, the gene therapy is an rAAV gene therapy.
[0194] In some cases, treatment with an expression cassette disclosed herein reduces seizure duration and/or frequency, e.g., seizures associated with Dravet syndrome, by at least 1%, 2%, 3%, 4%, 5%, 6%, 7%, 8%, 9%, 10%, 11%, 12%, 13%, 14%, 15%, 16%, 17%, 18%, 19%, 20%, 21%, 22%, 23%, 24%, 25%, 26%, 27%, 28%, 29%, 30%, 31%, 32%, 33%, 34%, 35%, 36%, 37%, 38%, 39%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, or 95% as compared to an untreated control or as compared to the level before treatment.
[0195] In some cases, treatment with an expression cassette disclosed herein reduces high gamma power activity (e.g., high gamma power activity associated with Alzheimer's disease) by at least 1%, 2%, 3%, 4%, 5%, 6%, 7%, 8%, 9%, 10%, 11%, 12%, 13%, 14%, 15%, 16%, 17%, 18%, 19%, 20%, 21%, 22%, 23%, 24%, 25%, 26%, 27%, 28%, 29%, 30%, 31%, 32%, 33%, 34%, 35%, 36%, 37%, 38%, 39%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, or 95% as compared to an untreated control or as compared to the level before treatment.
[0196] In one aspect, the present disclosure provides a nucleic acid cassette of this disclosure comprises one or more regulatory elements operably linked to a transgene that result in selective expression in a target cell type over one or more non-target cell types, e.g., selective expression in PV neurons in the CNS over one or more non-PV CNS cell types. In some cases, each of the regulatory elements comprises (i) a sequence of SEQ ID NOs: 1-32, (ii) a functional fragment or a combination thereof, or (iii) a sequence with at least 80% sequence identity to (i) or (ii). In some cases, the percent sequence identity can be measured using BLAST. In some cases, at least one of the regulatory elements is human derived. In some cases, at least one of the regulatory elements is derived from a non-human mammal. In some cases, the regulatory elements are non-naturally occurring. In some cases, the regulatory elements result in a selectivity of expression in PV cells that is greater than expression of the transgene operably linked to CAG or EFla or a non-selective regulatory element as measured by a co-localization assay. In some cases, a transgene is any one or more of SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, and STXBP1, or a functional fragment hereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto. In some cases, a transgene is a DNA binding protein that modulates expression of a gene (e.g., an endogenous gene such as SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, and STXBP1), such as transcriptional modulator, transcriptional activator, or transcriptional repressor. In some cases, a transgene is a gene editing protein, such as a zinc finger nuclease or a transcription activator-like effector nuclease. In some cases, a transgene is a reporter gene or a detectable marker, such as eGFP, tdTomato, or RFP. In some cases, a transgene is a Cas protein, such as Cas9. In some cases, the regulatory elements result in selective expression in PV cells at a level that is at least 0.5 fold, at least 0.6 fold, at least 0.7 fold, at least 0.8 fold, at least 0.9 fold, at least 1.1 fold, at least 1.2 fold, at least 1.3 fold, at least 1.4 fold, at least 1.5 fold, at least 2 fold, at least 3 fold, at least 4 fold, at least 5 fold, at least 6 fold, at least 7 fold, at least 8 fold, at least 9 fold, at least 10 fold, at least 11 fold, at least 12 fold, at least 13 fold, at least 14 fold, at least 15 fold, at least 16 fold, at least 17 fold, at least 18 fold, at least 19 fold, at least 20 fold, at least 25 fold, at least 30 fold, at least 40 fold, at least 50 fold, at least 60 fold, at least 70 fold, at least 80 fold, at least 90 fold, at least 100 fold as compared to that of a CAG or EF1.alpha. or a non-selective regulatory element, as measured by the co-localization assay. In some cases, a fold difference refers to the fold difference between the percentage of eGFP+, PV+ cells that result from one or more regulatory elements and that of a non-selective regulatory element. In some cases, the co-localization assay is an immunohistochemical assay, as described below in Example 5. In some instances, a co-localization assay is performed using a commercially available anti-PV antibody. In some cases, the transgene encodes an ion channel subunit, a neurotransmitter regulator, or a variant or a functional fragment thereof. In some cases, the ion channel subunit is an alpha subunit or a beta subunit of a sodium ion channel or a subunit of a potassium ion channel. In some cases, the transgene is any one of (i) SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, or a DNA binding protein; (ii) a functional fragment thereof or (iii) a sequence having at least 80% sequence identity to (i) or (ii). In some cases, the neurotransmitter regulator is (i) STXBP1, (ii) a functional fragment thereof, or (iii) a sequence having at least 80% sequence identity to (i) or (ii). In some cases, the regulatory elements and the operably linked transgene are located on different chromosomes. In some cases, the regulatory elements combined are less than 2.5 kb, less than 1.5 kb, less than 1 kb, or less than 500 bp in size. In some cases, the non-PV cells comprise any one or more of non-PV CNS cell types, including but not limited to excitatory neurons, dopaminergic neurons, astrocytes, microglia, or motor neurons. In some cases, the nucleic acid cassette is a linear construct. In some cases, the nucleic acid cassette is a vector. In some cases, the nucleic acid cassette is a plasmid. In some cases, the vector is a viral vector. In some cases, the viral vector is an adeno-associated virus (AAV) vector. In some cases, the AAV vector is AAV1, AAV8, AAV9, scAAV1, scAAV8, or scAAV9. In some cases, the viral vector is a lentiviral vector. In some cases, the regulatory elements contain less than 600 bp of contiguous sequence from within 10 kb of the transcription start site of GAD2, GAD1, SYN1, NKX2.1, DLX1, DLX5/6, SST, PV, and/or VIP.
[0197] In various embodiments disclosed herein, a regulatory element is less than 2050 bp, 2000 bp, 1900 bp, 1800 bp, 1700 bp, 1600 bp, 1500 bp, 1400 bp, 1300 bp, 1200 bp, 1100 bp, 1000 bp, 900 bp, 800 bp, 700 bp, 600 bp, 500 bp, 400 bp, 300 bp, 200 bp, 100 bp, 90 bp, 80 bp, 70 bp, 60 bp, 50 bp, 40 bp, 30 bp, 20 bp, 10 bp, or 5 bp. In various embodiments disclosed herein, an expression cassette comprises a transgene that is larger than a typical transgene size in a conventional viral vector, e.g., AAV. In some aspects, an expression cassette of any embodiment disclosed herein comprises a transgene that is at least 1 kb, 1.5 kb, 2 kb, 2.5 kb, 3 kb, 3.5 kb, 4 kb, 4.5 kb, 5 kb, 5.5 kb, 6 kb, 6.5 kb, 7 kb, 7.5 kb, or 8 kb. In some aspects, any embodiment disclosed herein comprises an expression cassette (e.g., AAV) that comprises a transgene that is more than 1, 1.2, 1.3, 1.4, 1.5, 1.6, 1.7, 1.8, 1.9, 2, 2.1, 2.2, 2.3, 2.4, 2.5, 2.6, 2.7, 2.8, 2.9, 3, 3.1, 3.2, 3.3, 3.4, 3.5, 3.6, 3.7, 3.8, 3.9, or 4 kb in size. In some aspects, any embodiment disclosed herein can further comprise one or more heterologous nucleic acid sequence or element.
[0198] In one aspect, a method of treating a neurological disorder or condition in a subject in need thereof comprises delivering a therapeutically effective amount of a nucleic acid cassette described herein. In another aspect, a method of increasing selective expression of a transgene in PV neurons comprises contacting a cell with a nucleic acid cassette described herein. In some cases, a method of any embodiment disclosed herein is used to treat a neurological condition or disease, e.g., a psychiatric disorder (e.g., schizophrenia, obsessive compulsive disorder, addiction, depression, anxiety, psychosis); an autism spectrum disorder (e.g., Fragile X syndrome, Rett syndrome); epilepsy (e.g., Dravet syndrome, chronic traumatic encephalopathy, generalized epilepsy with febrile seizures plus (GEFS+), epileptic encephalopathy, temporal lobe epilepsy, focal epilepsy, tuberous sclerosis); or neurodegeneration (e.g., Alzheimer's disease, Parkinson's disease). In some cases, a method of any embodiment disclosed herein can be used to treat Dravet syndrome. A method of any embodiment disclosed herein can be used to treat Alzheimer's disease. In some cases, methods and/or compositions of this disclosure can be used to treat any neurological condition or disease associated with seizure and/or epilepsy, and/or wherein PV neurons are implicated.
[0199] In one aspect, a neurological condition described herein is treated with a gene therapy, preferably one that results in preferential expression in one tissue type or cell type over another, e.g., a PV neuron as determined via a co-localization assay. In some cases, the gene therapy is an AAV.
[0200] In one aspect, a method of targeting expression of any transgene to PV neurons in the CNS comprises operably linking one or more of PV neuron selective regulatory elements to a transgene. In some cases, the regulatory elements comprise one or more sequences of SEQ ID NOs: 1-32, or sequences with at least 80% sequence identity to SEQ ID NOs: 1-32, or a functional fragment thereof. In some cases, the regulatory elements result in selective expression in PV neurons at a level that is at least 2 fold, at least 5 fold, or at least 7 fold, or at least 10 fold as compared to CAG or EF1.alpha. or a non-selective regulatory element operably linked to the transgene, as measured by a co-localization assay. In some cases, the transgene is SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, STXBP1, a DNA binding protein, or a functional fragment thereof. In some cases, the regulatory elements and the transgene are in an AAV. In some cases, the AAV is AAV9.
[0201] In one aspect, a method of treating a neurological condition or disorder in a subject in need thereof comprises contacting a cell with a nucleic acid cassette comprising one or more regulatory elements operably linked to a transgene that results in selective expression in PV neurons over one or more non-PV CNS cells. In some cases, the regulatory elements comprise one or more of SEQ ID NOs: 1-32, or a functional fragment or a combination thereof, or sequences having at least 80%, at least 85%, at least 90%, at least 95%, or at least 99% sequence identity thereto. In some cases, the transgene is a voltage-gated ion channel subunit, or a variant or a functional fragment thereof. In some cases, the subunit is a beta subunit of a sodium ion channel. In some cases, the subunit is an alpha subunit of a sodium ion channel. In some cases, the subunit is of a potassium ion channel. In some cases, the transgene is any one of (i) SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, a DNA binding protein or STXBP1; (ii) a functional fragment thereof; or (iii) a sequence having at least 80% sequence identity to (i) or (ii). In some cases, the neurological condition or disorder is associated with a haploinsufficiency or a mutation in any of SCN1A, SCN1B, SCN2B, KV3.1, and KV3.3. In some cases, the neurological condition or disorder is Dravet syndrome. In some cases, the neurological condition or disorder is Alzheimer's disease. In some cases, the neurological condition or disease is a psychiatric disorder (e.g., schizophrenia, obsessive compulsive disorder, addiction, depression, anxiety, psychosis); an autism spectrum disorder (e.g., Fragile X syndrome, Rett syndrome); epilepsy (e.g., chronic traumatic encephalopathy, generalized epilepsy with febrile seizures plus (GEFS+), epileptic encephalopathy, temporal lobe epilepsy, focal epilepsy, tuberous sclerosis); or neurodegeneration (e.g., Alzheimer's disease, Parkinson's disease). In some cases, the neurological condition or disease is any seizure and/or epilepsy related condition or disease wherein PV neurons are implicated. In some cases, the nucleic acid cassette results in selective expression in PV neurons at a level that is at least 2 fold, at least 5 fold, or at least 7 fold, or at least 10 fold as compared to CAG or EFla or a non-selective regulatory element operably linked to the transgene, as measured by a co-localization assay. In some cases, the nucleic acid cassette is in an AAV. In some cases, the AAV is AAV9.
[0202] In one aspect, a method of treating Dravet syndrome comprises contacting a cell with an AAV comprising a transgene that is any one of (i) SCN1A, SCN1B, SCN2B, a DNA binding protein (ii) a functional fragment thereof, or (iii) a sequence having at least 80% sequence identity to (i) or (ii). In some cases, the AAV further comprises one or more PV neuron selective regulatory elements or any of the regulatory elements disclosed herein operably linked to the transgene. In some cases, each of the regulatory elements independently comprises a sequence comprising any one of SEQ ID NOs: 1-32, or any functional fragment or combination thereof, or a sequence comprising at least 80% sequence identity to any one of SEQ ID NOs: 1-32.
[0203] In another aspect, a method of treating Alzheimer's disease comprises contacting a cell with an AAV comprising a transgene is any one of (i) SCN1A, SCN2B, KV3.1, KV3.3, and STXBP1, (ii) a functional fragment thereof, and (iii) a sequence having at least 80% sequence identity to (i) or (ii). In some cases, the AAV further comprises one or more PV neuron selective regulatory elements operably linked to the transgene. In some cases, each of the regulatory elements independently comprises a sequence comprising any one of SEQ ID NOs: 1-32, or any functional fragment or combination thereof, or a sequence comprising at least 80% sequence identity to any one of SEQ ID NOs: 1-32.
EXAMPLES
[0204] These examples are provided for illustrative purposes only and not to limit the scope of the claims provided herein.
Example 1
Identifying Putative PV-Selective Regulatory Elements
[0205] To identify and screen putative regulatory element that are selective for PV cells, one can harvest PV cells from a R26-CAG-LSL-Sunl-sfGFP-Myc knockin mouse using affinity purification, e.g., using anti-GFP or anti-Myc antibodies and protein G-coated magnetic beads. PV cells can be enriched by using anti-PV antibody coated beads or affinity purification matrix. Nuclei are then isolated from the PV cells. Nuclear RNA can be purified from the nuclei and converted to cDNA, and amplified with the Nugen Ovation RNA-seq System V2 (Nugen 7102), followed by sequencing using the Illumina HiSeq 2500. Genomic DNA can be purified from nuclei, fragmented, and used to make methylC-seq libraries, which can be sequenced using the Illumina HiSeq 2000. To generate an ATAC-seq library, nuclei bound to beads are transposed using Tn5 transposase (Illumina FC-121-1030). After 9-12 cycles of PCR amplification, libraries are sequenced using an Illumina HiSeq 2500. To generate a ChIP-seq library, nuclei of PV cells are digested to mononucleosomes using micrococcal nuclease, followed by salt extraction of chromatin, and native ChIP and library construction, which can be sequenced on an Illumina HiSeq 2500. After sequencing these libraries, the sequences are mapped to identify correlations and patterns in hypo-methylation in CG-rich regions, histone modifications, transcriptional factor binding sites, and patterns associated with highly expressed transcriptional factors in PV cells. Overlapping features and correlations from multiple assays and/or libraries described above provide convergent evidence for identifying candidate sequences that are putative PV-selective regulatory elements. Putative PV-selective regulatory elements can be further tested using a co-localization assay as described in Example 5 below. Putative PV-selective regulatory elements can also be tested in B6 PV-Cre mouse (Jackson Laboratory), which is a B6 PV-Cre knock-in mouse that expresses Cre recombinase in parvalbumin-expressing, as described in Example 2 below. After validating PV-selectivity of the regulatory elements, the regulatory elements can be operably linked to a transgene to target expression selectively to PV cells over at least one, two, three, four, five, or more than five non-PV cells.
Example 2
Selectivity for PV Neurons in PV-Cre Mouse
[0206] Selectivity for PV neurons can be determined using fluorescent imaging. AAV9 vectors containing eGFP operably linked to (i) a control promoter (EF1.alpha.); or (ii) a PV-selective RE identified in Example 1 above; or (iii) a PV-selective RE selected SEQ ID NOs: 1-32; and AAV9 vectors containing a Cre dependent tdTomato are co-injected into a B6 PV-Cre mouse (Jackson Labs). PV-Cre is a knock-in mouse that expresses Cre recombinase in parvalbumin-expressing neurons (such as interneurons in the brain and proprioceptive afferent sensory neurons in the dorsal root ganglia), without disrupting endogenous Pvalb expression.
[0207] Mice are infused bilaterally with 1.5 .mu.L of AAV9 vector (5.sup.12 to 1.sup.13 gc/ml) into the dorsal and ventral hippocampus at a rate of 0.3 .mu.L/min with a 4 min rest period following injection. Mice are anesthetized for the injection. The animals are placed in a stereotaxic frame (Kopf instruments, USA), using the following coordinates for the dorsal hippocampus (AP -2.0 mm, lateral .+-.1.5, DV -1.4 mm from dura) and the ventral hippocampus (AP -3.1 mm, lateral .+-.2.8, DV -3.8 mm from dura). A Hamilton syringe (model #80308; 10 .mu.L syringe with corresponding 30 ga blunt tip needle) can be used with the stereotactic micromanipulator, to designate and drill the bur holes. The drill is only used to penetrate the bone. Following drilling, the infusion cannula is lowered into the brain to the depth of the desired location for injection, e.g., injection volume: 1.5 .mu.L; injection rate: 0.3 .mu.L/min. Prior to infusion, the needle is allowed to equilibrate for 1 minute. Once delivery is completed, the needle is left for 4 min and then withdrawn over approximately 1 min. Once all infusions are complete, the skin incision is closed with sutures and administered post-surgery analgesics. The treated mice undergo daily health checks for the remainder of the study and are weighed once weekly to monitor body weight.
[0208] For tissue collection, mice are euthanized via isoflurane overdose and perfused with 4% Paraformaldehyde (PFA). A piece of brain tissue containing the hippocampus is extracted and placed in 4% PFA at 4.degree. C. for at least 12 hours. The brain tissue is then dehydrated in 30% sucrose (in phosphate buffered saline) at 4.degree. C. until the tissue sinks to the bottom of the tube. Brain tissue is embedded in Tissue-Tek OCT for sectioning in a cryostat. Sectioned brain tissue is stained for eGFP and tdTomato using standard immunohistochemistry procedures with anti-RFP polyclonal rabbit antibody (Rockland Antibodies and Assay) and anti-eGFP polyclonal chicken antibody (Ayes Labs). Fluorescence microscope imaging is used to visualize the cells eGFP, or green fluorescence, corresponds to all gene expression. Red fluorescence from tdTomato corresponds to PV+ cells. An overlap of the two fluorescence signals, which can be visualized as yellow or white cells, represents PV+ cells that express the eGFP transgene. AAV9 vectors comprising a PV-selective regulatory element is expected to yield higher number of cells that are eGFP+ and PV+ as compared to the control promoter (EF1a). For example, fluorescence imaging of cells from mice injected with AAV9s comprising any one of PV-selective REs (e.g., SEQ ID NOs: 1-32 or putative REs identified in Example 1) are expected to show higher number of eGFP+ cells that are also PV+. Selectivity for PV cells can be quantified as percentage of all eGFP+ cells that are also PV+.
Example 3
Reduction of Seizures in Dravet Mouse Model
[0209] B6(Cg)-Scn1a.sup.tm1.1Dsf/J mice were obtained from the Dravet syndrome European Federation via the Jackson Laboratories. These mice contain a Dravet syndrome associated mutation in exon 24 of SCN1A (A to V at position 1783). The mice also contain a floxed exon 24 with wildtype sequence. When not manipulated, this strain of mice expresses two copies of the WT allele of SCN1A. However, upon delivery of an AAV expressing Cre recombinase, any cell targeted by the AAV will switch to expressing one copy of the mutant allele. Upon expression of the mutant SCN1A subunit, mice develop spontaneous seizures within 10 days.
[0210] B6(Cg)-Scn1a.sup.tm1.1Dsf/J and control C57B16 mice were injected, as in Example 2, with AAVs expressing CRE recombinase under the control of the EF1.alpha. promoter and an AAV comprising PV cell selective regulatory element SEQ ID NO: 32 driving expression of either eGFP (SEQ ID NO: 36) or SCN1B (SEQ ID NO: 37). Once all four infusions were complete, telemetry implantation was performed immediately, (F20-EET, Data Sciences International). Electrocorticogram data was monitored continuously for 14 days from 10 days after the surgery. Electrocorticogram data was analyzed and all seizure events were recorded, annotated with date, time start, time stop, duration of the seizures, and severity score. FIG. 1 illustrates the frequency of seizures in 12 hour windows over 14 days following treatment. The mice treated with SCN1B showed a trend towards lower seizure frequency compared to the control animals.
[0211] This observation was consistent with the notion that the beta unit of the sodium ion channel, e.g, SCN1B, can contribute to the trafficking and assembly of the sodium ion channel and that increasing the expression of the beta unit selectively in PV neurons can result in increased trafficking and assembly of the Nav1.1 channel, thus leading to a trend towards lower seizure frequency and duration in the mice treated with SCN1B gene therapy.
Example 4
Treating Alzheimer's Disease in a Mouse Model
[0212] Female APP/PS1 and WT mice bred at PsychoGenics were used in the study. APP/PS1 mice contain human transgenes for both Amyloid Beta Precursor Protein (APP) bearing the Swedish mutation (670 G-T and 671 A-C) and Presenilin 1 (PSEN1) containing an L166P mutation, both under the control of the Thyl promoter. These mice develop symptoms of Alzheimer's disease, including amyloid plaques and memory defects. Further description of these mice can be found in Radde et al, 2006 (Radde, Rebecca, et al. "A.beta.42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology." EMBO reports 7.9 (2006): 940-946).
[0213] APP/PS1 mice were used as a model to determine the effect of treatment with SCN1B under the control of a RE on symptoms of Alzheimer's disease. APP/PS1 mice and non-transgenic controls were injected with either a control vector expressing eGFP or a treatment vector expressing SCN1B, both under the control of SEQ ID NO: 32; and implanted with an EET transmitter as in Example 3. Brain activity was assessed over 24 hours at 4 weeks after surgery. Electrocorticogram data was automatically analyzed and power levels in the different frequency bands were compared. FIG. 2 illustrates the high gamma power (50-100 Hz) in non-transgenic controls (WT), APP/PS1, and APP/PS1 mice treated with SCN1B. Increased high gamma power activity is associated with seizures in Alzheimer's patients and epilepsy patients. The APP/PS1 mice showed a higher level of high gamma power activity than the control mice. However, the increase was absent in the treated mice indicating effective treatment with the vector.
Example 5
Selectivity for PV Neurons in C57BL/6J (WT) Mouse
[0214] The selectivity of various REs disclosed herein were tested for selective gene expression in PV neurons using immunohistochemical methods. C57BL/6J (WT) mouse line was used for the PV immunohistochemical assays. Expression cassettes comprising reporter transgene eGFP operably linked to a regulatory element (SEQ ID NO: 1 or SEQ ID NO: 8) or a CAG promoter in an AAV9 construct.
[0215] Pup Systemic Infusions:
[0216] Postnatal day 1 C57BL/6J mice were infused via facial vein injection with AA9 vector (1 E.sup.12 to 3 E.sup.12) using a 300 U insulin syringe with a 31 G needle. For tissue collection, mice were euthanized 21 days post-infusion via overdose of sodium pentobarbital (i.p.) and perfused with heparinized (2.5 IU/ml) saline followed by perfusion with 4% formaldehyde. Brains were removed and subsequently immersion-fixed in 4% formaldehyde for 24-48 hours at 4 degrees Celsius. The brain was then placed into PBS containing 30% sucrose and allowed to sink at 4 degrees Celsius (.about.2-3 days). Upon sinking the individual brain hemispheres were frozen in Tissue-Tek OCT with the midline facing down. Frozen brains were processed for sagittal sections on a cryostat and placed free-floating into PBS. Sections were stained for eGFP and parvalbumin (PV) using standard immunohistochemistry procedures with chicken anti-GFP (Ayes Lab, GFP-1020) and mouse anti-PV (Sigma, P3088).
[0217] Adult Systemic Infusions:
[0218] 4-week-old, C57BL/6 mice were infused via tail vein injection with 604, of AAV9 vector (4.9.sup.13 to 1.sup.14 gc/ml) expressing eGFP. For tissue collection mice were euthanized 21 days post-infusion via isoflurane overdose and whole brains were extracted, washed with PBS and placed into separate 5m1 tubes containing ice cold 4% formaldehyde. Tissue was fixed at 4 degrees Celsius overnight. The following day, the brain was placed into PBS containing 30% sucrose and allowed to sink at 4 degrees Celsius. Upon sinking the individual brain hemispheres were frozen in Tissue-Tek OCT with the midline facing down. Frozen brains were processed for sagittal sections on a cryostat and placed free-floating into PBS. Sections were stained for EGFP and parvalbumin (PV) using standard immunohistochemistry procedures with chicken anti-GFP (Ayes Lab, GFP-1020) and mouse anti-PV (Sigma, P3088).
[0219] Immunohistochemistry Protocol:
[0220] Immunohistochemistry was used to analyze the co-localization of eGFP signal and PV signal using the anti-PV antibody, wherein overlay of the signals exhibited as white or light gray spots in the top panel images (merge), wherein representative overlay was indicated by arrowheads. Overlay of the eGFP and PV fluorescence is indicative of expression in PV cells. Such experiments can be used to determine the selective expression of expression in PV cells. To perform the immunohistochemical experiments, tissues obtained from each mouse were blocked with a Blocking Buffer Solution (comprising 3% BSA, 3% NGS, 0.3% Triton X-100, 0.2% Tween-20 in 1.times.PBS) for 1 hour at room temperature. The tissues were then incubated with primary antibodies in blocking buffer overnight at 4C, washed three times with 1 mL 1.times.PBS, each with 5 minutes interval. Then the tissues were incubated with secondary antibodies in blocking buffer for 1 hour at room temperature, followed by washing three times, each time with 1 mL 1.times.PBS and with 5 minutes interval. The tissues were incubated DAPI (1:1000) in PBS buffer for 5 minute and wash twice with 1 mL 1.times.PBS. Tissues were mounted onto slides, imaged, and analyzed using a fluorescence microscope. Images were taken using a Vectra 3 imaging system (Perkin Elmer) and quantified for co-labeling of eGFP and PV staining using inform-Tissue finder, advanced image analysis software or hand scored. At least 80 GFP positive cells were counted in each panel before determining the percentage of co-localization.
[0221] FIGS. 3A-3C illustrates the results of the immunohistochemistry experiments performed in pups after systemic AAV9 injections. FIGS. 4A-4C illustrates results of similar immunohistochemistry experiments performed in adult mice following AAV9 injections.
[0222] FIG. 3A illustrates the overlay of the immunohistochemistry experiments performed in pups after systemic AAV9 injections. FIG. 3B illustrates the quantification of the co-localization of the immunohistochemistry experiments, wherein selectivity for PV cells was measured as percentage of GFP+ cells that were also PV+, as compared to eGFP expression under the control of the CAG promoter.
[0223] FIG. 4A illustrates the overlay of the immunohistochemistry experiments performed in adult mice after systemic AAV9 injections. FIG. 4B illustrates the quantification of the co-localization of the immunohistochemistry experiments, wherein selectivity for PV cells was measured as percentage of GFP+ cells that were also PV+, as compared to eGFP expression under the control of the EF1.alpha..
[0224] It is estimated that GABAergic neurons constitute about 20% of CNS, while PV cells constitute about 40% of GABAergic neurons, which means that PV cells make up approximately 8% of all neurons in the CNS. See Pelkey, K A et al., 2017; and Lee, S. et al., 2010. Thus, one would predict that about 8% of the cells labeled by a non-selective regulatory element (e.g., CAG, EF1.alpha., or a constitutive promoter) would be PV positive, or within this range. Therefore, expression in PV cells above 8% is indicative of increased selectivity in PV cells. Notably, AAV9 injections comprising regulatory element SEQ ID NO: 8 resulted in about 60% of cells as PV positive, which was 7.5 times higher than what was expected by the distribution of PV cells.
[0225] Similar immunohistochemistry experiments as described above were performed to determine the selective expression of additional regulatory elements, SEQ ID NOs: 2-7 and 9-22 as compared to a non-selective regulatory element having a sequence of SEQ ID NO: 34, except AAVDJ viral vector was used to deliver eGFP operably linked to a regulatory element into C57BL/6J (WT) mice. Such AAVDJ virus was injected directly into the CNS in the hippocampus of adult mice. At least 80 GFP positive cells were counted in each experiment before calculating the percentage of co-localization, or selectivity, as percentage of GFP positive cells that were also PV positive. FIGS. 5A-5F illustrate the fluorescence imaging used for determining co-localization, or selectivity, measured as percentage of eGFP positive cells that were also PV positive and in comparison to the signal of non-selective regulatory element SEQ ID NO: 34. Cells that were positive for a marker appear as white/gray cells in the images. Merge images illustrate the overlap between the corresponding eGFP and anti-PV images. Cells that were positive for both eGFP and PV appear as white/light gray cells in the merge image. FIG. 6 illustrates the quantification of the co-localization analysis, measured as percentage of eGFP+ cells that were also PV+.
Example 6
Treatment of Dravet Syndrome in Different Mouse Lines
[0226] Treatment of Dravet syndrome and/or symptoms thereof using the expression cassettes described herein can be tested in various mouse lines, such as B6(Cg)-Scn1a.sup.tm1.1Dsf/J as described above, Scn1a.sup.tm1Kea, and Scn1a-R1470X mouse lines. These mouse lines are established mouse models for Dravet syndrome. Scn1a.sup.tm1Kea and Scn1a-R1470X mouse lines do not require CRE recombinase.
[0227] The Sen1a.sup.tm1Kea mouse (available from the Jackson Laboratory; described in Hawkins et al., Scientific Reports, vol. 7: 1532.7 (2017)) comprises a deletion of the first coding exon of SCN1A. Mice homozygous for the SCN1A knockout allele are characterized by tremors, ataxia, seizures, and die by postnatal day 16. Heterozygous mice on the C57BL/6 background develop spontaneous seizures and die within weeks. Such mouse strain can be used to study safety and efficacy of treatment of epilepsy and Dravet syndrome. See Miller et al., Genes Brain Behav. 2014 February; 13(2):163-72 for additional information.
[0228] The Scn1a-R1470X mouse is a knock-in mouse carrying a premature stop codon, R1407X, in exon 21 of the SCN1A gene. The same mutation has been identified as a pathogenic mutation in three unrelated SMEI patients. Scn1a.sup.RX/RX pups are characterized by recurrent spontaneous seizures at 12 postnatal days, including tonic-clonic and clonic seizures at 12-16 postnatal days, and rhythmic jerking movements and involuntary muscle contraction. See Ogiwara et al., Journal of Neuroscience, May 30, 2007, 27 (22) 5903-5914 for additional information.
[0229] To test the compositions described herein, such as AAV gene therapy and treatment using such gene therapy, Dravet mice of each of the mouse strains described above and control mice (e.g., a wild-type mouse or an untreated Dravet mouse for the strain) are injected (e.g., administered by intraperitoneal injection) with AAVs expressing either eGFP or another reporter gene, or an expression cassette comprising one or more PV-selective REs (e.g., SEQ ID NOs: 1-32) as described herein operably linked to a transgene disclosed herein, such as SCN1A, SCN1B, or SCN2B, or any of SEQ ID NOs: 37-39, or a variant or functional fragment thereof. Following AAV injections, mouse survival is monitored over time. All mice are monitored daily for general health (e.g. weight, hydration, grooming, and mobility) and deaths were recorded. Telemetry implantation can be performed immediately after AAV injections (F20-EET, Data Sciences International). Electrocorticogram data can be recorded and monitored continuously for at least 14 days from 10 days after the surgery. All seizure events can be recorded for at least 14 days following AAV treatment, annotated with date, time start, time stop, duration, and severity score. A reduction in the frequency and/or duration of seizures following treatment with an AAV as described above as compared to the eGFP control or an untreated control is indicative of the efficacy of the gene therapy in reducing the symptoms and/or severity of Dravet syndrome.
[0230] After treatment of the mice with AAV, the expression levels of the transgene (e.g., SCN1A; SCN1B; SCN2B; a DNA binding protein, such as a transcriptional activator, that modulates an endogenous SCN1A, SCN1B, or SCN2B; any of SEQ ID NOs: 37-39; or any variant or functional fragment thereof) can be monitored over time using various PCR and/or sequencing methods to show AAV treatment can result in an increase in gene expression in PV cells. Northern blot analysis and in situ hybridization can also be used to analyze transgene expression in vivo. The level of the protein expressed from the protein can also be monitored after treatment to show an increase in transgene expression correlates with an increase in the corresponding protein in vivo. Protein levels can be assayed using various methods, including, but not limited to, Western blot analysis, immunohistochemistry, immunofluorescence histochemistry, and/or ELISA assays. Formation of functional voltage-gated sodium ion channels can also be assayed using current-clamp analysis.
[0231] Hyperthermia-induced seizures can be evaluated to compare the wild-type mice and/or untreated Dravet mice with Dravet mice treated with AAV gene therapy comprising an expression cassette described herein (e.g., an expression cassette comprising one or more REs of this disclosure operably linked to a transgene of this disclosure, such as SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, a functional fragment thereof, or a DNA binding protein that modulates an endogenous SCN1A, SNC2A, SNC8A, SCN1B, or SCN2B). In such experiments, the core body temperature is monitored with a RET-3 rectal temperature probe (Physitemp Instruments, Inc, New Jersey, USA) and controlled by a heat lamp connected to a rodent temperature regulator (TCAT-2DF, Physitemp) reconfigured with a Partlow 1160+controller (West Control Solutions, Brighton, UK). Body temperature is raised 0.5.degree. C. every two minutes until the onset of the first clonic convulsion. As compared to the untreated Dravet mice, Dravet mice treated with an AAV gene therapy are expected to have a higher threshold temperature before the onset of first clonic convulsion and/or have a higher proportion of mice that remain seizure free at the maximum temperature tested.
[0232] Different doses of AAV comprising an expression cassette can also be administered to mice to determine the safety and efficacy profile of each gene therapy treatment. These preclinical studies can also inform the optimal dose(s) of the gene therapy to use for treating Dravet syndrome.
Example 7
Treatment of Alzheimer's Disease in Mouse
[0233] Female APP/PS1 and wild-type (WT) mice, which are bred at PsychoGenics and are established mouse model of Alzheimer's disease, can be used to study the safety and efficacy of the compositions described herein in treating Alzheimer's disease, comprising one or more PV-selective REs. APP/PS1 mice is describe above in Example 4.
[0234] APP/PS1 mice and non-transgenic controls are injected with either a control AAV vector expressing eGFP or a treatment AAV vector comprising one or more PV-selective REs disclosed herein, e.g., SEQ ID NOs: 1-32, operably linked to a transgene that is deficient or impaired in Alzheimer's disease, such as SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, STXBP1, a DNA binding protein that modulates an endogenous gene (e.g., SCN1A, SNC2A, SNC8A, SCN1B, SCN2B, KV3.1, KV3.3, or STXBP1), or any one of SEQ ID NOs: 37-43, or a functional fragment thereof.
[0235] Following AAV injections, mouse survival is monitored over time. All mice are monitored daily for general health (e.g. weight, hydration, grooming, and mobility) and deaths were recorded. After injections of the AAVs, mice are also implanted with an EET transmitter as described in Example 3 above. Brain activity can be recorded and monitored over 24 hours for at least 4 weeks after surgery. Electrocorticogram data can be automatically analyzed, and power levels in the different frequency bands (50-100 Hz) can be compared across different groups: WT mice, untreated APP/PS1 mice, and AAV-treated APP/PS1 mice, each treated with an AAV gene therapy as described above, Increased high gamma power activity is associated with seizures in Alzheimer's patients and epilepsy patients. Thus, the untreated APP/PS1 mice are expected to show a higher level of high gamma power activity than the control mice, while this increase is expected to be absent or reduced in the treated mice, indicating an effective treatment with an AAV gene therapy.
[0236] After treatment of the mice with AAVs, the expression levels of the transgene can be monitored over time using various PCR and/or sequencing methods to show AAV treatment can result in an increase in endogenous expression of the transgene. Northern blot analysis and in situ hybridization can also be used to analyze gene expression in vivo. The level of the protein expressed from the transgene can also be monitored after treatment to show an increase in gene expression correlates with an increase in protein levels. Protein level can be assayed using various methods, including, but not limited to, Western blot analysis, immunohistochemistry, and/or ELBA assays. Formation of functional voltage-gated sodium or potassium ion channels can also be assayed using current-clamp analysis.
[0237] Different doses of AAV comprising an expression cassette can also be administered to mice to determine the safety and efficacy profile of each gene therapy treatment. These preclinical studies can also inform the optimal dose(s) of the gene therapy to use for treating Alzheimer's disease.
Sequence CWU 1
1
431681DNAHomo sapiens 1ggaggaagcc atcaactaaa ctacaatgac tgtaagatac
aaaattggga atggtaacat 60attttgaagt tctgttgaca taaagaatca tgatattaat
gcccatggaa atgaaagggc 120gatcaacact atggtttgaa aagggggaaa
ttgtagagca cagatgtgtt cgtgtggcag 180tgtgctgtct ctagcaatac
tcagagaaga gagagaacaa tgaaattctg attggcccca 240gtgtgagccc
agatgaggtt cagctgccaa ctttctcttt cacatcttat gaaagtcatt
300taagcacaac taactttttt tttttttttt tttttttgag acagagtctt
gctctgttgc 360ccaggacaga gtgcagtagt gactcaatct cggctcactg
cagcctccac ctcctaggct 420caaacggtcc tcctgcatca gcctcccaag
tagctggaat tacaggagtg gcccaccatg 480cccagctaat ttttgtattt
ttaatagata cgggggtttc accatatcac ccaggctggt 540ctcgaactcc
tggcctcaag tgatccacct gcctcggcct cccaaagtgc tgggattata
600ggcgtcagcc actatgccca acccgaccaa ccttttttaa aataaatatt
taaaaaattg 660gtatttcaca tatatactag t 6812502DNAMus musculus
2agtttggaca agaactatag ttctagcttt ctctgggtct ccaccttgca gagaatgcag
60ctttcattat ctcatgagcc aaactctcat catctctttc catatatctg tcggtgctct
120tccatgagta ctctaacaca cacagaagga gcacttacac aggctgttgt
ttttctctta 180ttatcatagc tgttgttcag acatgtgcat tctgttcttg
ttgcttcaat gctaaaggag 240tctcaggata tgagaactgt accagccgag
gcatcaggaa acatgggtgg aaattcccac 300agtactattt gttcactgtg
tgaccttggg ccagtcacat ccctttcctg aggcttcgat 360tccccaagct
ataaaagaag catctcttaa ccttttttta ggtcatgagt caggcccagc
420acactctcag ggagactcat gagagtacag atcatttccc atagaaaaac
catagtttta 480tatccagagg cttttctgta ag 5023823DNAMus musculus
3ggttccagtt cagaggcaga gcatttgggg ttcccagtca ggagctttcc tctctccgct
60ccttagtttc ctctctttaa aaaaaaatgg gtgatagtat agaaaggaag ctctgggctc
120ggggaccagg gccctgggat ccccgctccc agccactcgc tcctgaccct
tccagggaca 180agctcccccc caccccgtcc tttccaggct gccactagaa
gagatgggga cgcgtggtca 240gccgcttctg tcgcccccca gggaacggtc
tcacgctgga gggggcagtg ccctcggaac 300aggacagtca gcccaagcca
gccaagcgcg cgcggacgtc cttcaccgca gagcaattgc 360aggtaccccg
ggcaagcccc gaagcgtgtg ggcggggctt cggagtgggc gtggttgttc
420gggacttgtg actccgcccc ttgtgcgggg acccgcgtga ggccgctcca
aggatgaagc 480tgcctggggc gtggcctcgg accctgagcc tctgattggg
cggaggtctc agggcccttc 540tgcgccccac aggttatgca ggcgcagttc
gcgcaggaca acaacccgga cgcgcagacg 600ctgcagaagc tggcggacat
gacgggcctc agtcgcaggg tcatccaggt ggggctccgg 660ggtctcggcc
ttcaggtcta gggtgaacct tagggaagcg ctgaagctcg tagtggtacg
720gatggtcgcg cgtgcacgtg gccgcccctc tccagtgtgg cctaaggacc
ccagtcggca 780cgggttgacc cttttccttg attactgaga gtgcagaggc tgt
8234633DNAMus musculus 4tggtgggaag acatgtccag ggaagaaatg gcctccagag
gcctgaggtg gggaaatgct 60ggaggtggag agaggaacaa ctgactgaaa atgagcttcc
actgtggctt agtagcctat 120accaagtcta gagtataggg taggagaaga
ttaggaaagc gatgggtctg agaatgatgt 180ggcctgttga cttttgtaaa
cccaaagcac cttggactaa accctatgaa cagtgtggtg 240ccaccaaaga
ctataatgag ctcagggaac agaattctgt gtgcatggtg attttttttt
300tttttttctg ctaactgcag tctgggtgat gcattgacaa accaatcctg
gaaagtaaga 360ggcaagggca gctgggacgg tgagaggagc ctgatgggaa
ccaggccaag cagggcagca 420gaggcgatga agaggatgtg gtgcatccag
agactcactt cattagctgg aggcactgct 480ggatagggtc tgaaggttct
ggtatctgag ttggcgggct gggtgagtgg tggctctgct 540tcctgaacag
tgtgtgcaag aggaaacagg gttaagggct aggacagtca caggtgagtc
600agcctcacaa gagcaacctt cccctagtgc aga 6335670DNAMus musculus
5ggaggtctcc ttttgccccg gttccaacaa gagaatgcaa ggctgtatct caatttcctt
60gagcctctct gtattataga agaaaagtag ggaagccata cgccccttct gagcttcagt
120gtctctctgt ctctgcaaat gaggctgggg aggctggggg cgggcgtgaa
agaggcccgc 180gccaagccga cccccacctc tgccccctcc ccaggtcaac
aacctcatct ggcacgtgcg 240gtgcctcgag tgctccgtgt gtcgcacatc
gctgaggcag cagaatagct gctacatcaa 300gaacaaggag atctactgca
agatggacta cttcaggtag gcagcggcca tcccgccagc 360aagcgctgga
gcatgaacgc cttgcacacg cgtgcctagg ccacttgtgt ggcctgtgct
420ctccaattcc tgagccctgc tgttcagagt gcacaacgcg gctcagcgca
ctggcccggc 480cctcctactc agcacgtctt acacagaagg gagcgccagt
ctcagcctga gttctggcgg 540gggatctgcc tcgggttcct ccgatctgac
aggcgctggc cacgggtctg gttccatctc 600tggtcttttc tggccccgag
caccagtgtg ttctgttgag ctctgatgtc cgaggctctg 660gcccggatca
67061043DNAMus musculus 6ctctggctac ctcttatctt gggcattcac
gacaatttct aattgcaggt agtttgtgtg 60tgtgcgcgtg ttttttttcc ccctcagagg
cttggattgc aaaggaacta agcgattact 120tcaagagcca cgggttaagt
gcagggagag ggggagagag agggaaaaaa acccaatcca 180aattcaaatt
gcttcattag agagacaccg cttttgtggg gaagggcttt aaatgcccac
240tacaaagtta ggactcattg ttcagcgccg gtttatataa caggcgaggg
gaggcgctgg 300gctctgacag ctccgagcca gttcagcagc cgccgtcgcc
tgcattccct ccccctcccc 360caggtgatgg cccagccagg gtccggctgc
aaagcgacca cccgctgtct cgaagggacc 420gctccgcctg ccatggtgag
tcctttcggt cctgctttcg gccccgagtc cccccaacag 480cacaggccag
ggcttctggc tcagccttcc ggctaccaac ctctacccct gcgctggaaa
540actgccgata ggagccgcct ctcgttgagc cttggttttt ctggcctgga
atgtgagctt 600tggctgcttc ctgcacccag gatgcgctgt gttaaaagtt
gggggccgtc ccttcttctc 660caataggtcc tttcattctt gtactccagc
ctagggcgcg acatccctgg cacatttcgg 720tgtcagtcgg tgcgcgagga
aaccagattc aactctgagt actcggctaa gcgcttcgct 780gttcctctct
cccatttcag gctcagtcag acgcagaggc cttggcaggc gctctggaca
840aggacgaagg tagagcctcc ccatgtacgc ccagcacacc gtctgtctgc
tcgccgccct 900ctgctgcctc ttccgtgccg tctgccggca agaatatctg
ctccagttgc ggtctggaga 960tcctggaccg gtatctgctc aaggtgagtc
agggtaggtg tgcctgcttg cccacgggtg 1020tggtttgcag ccccaagagc tgt
10437507DNAMus musculus 7caagactttt aaaagtttag ataaataaac
aaacatttga cggctttcca tcacatctag 60actataatcc aaagatctat atggtcccaa
acgacttaca cttaactacc gtctcccata 120tggcttcttc ccccatcagt
cattgtcctc agccatagtg gcctccctgt tcctttgggt 180acaagggaac
aactccctga gaggttccat tagctgctgt tgcctgagat gctcttgagc
240ccacaccatc tgctcatttc tctcctcacg tgtcagtgat taagaggctg
tccttggcct 300cccgtcaaaa ttacatccct gccgctttcc acttcttgcc
ttcttatttt ctaaatagaa 360ctaactcacc actacccaac attctatata
attggatatc tgtcctctgt ttaaatataa 420tgttgacttc aagaaagaac
gttgtcactg ccctgtcacc agacttttaa acagtgccta 480tcgtgtggca
catgctcagt gaaattg 5078502DNAMus musculus 8tcaacagggg gacacttggg
aaagaaggat ggggacagag ccgagaggac tgttacacat 60tagagaaaca tcagtgactg
tgccagcttt ggggtagact gcacaaaagc cctgaggcag 120cacaggcagg
atccagtctg ctggtcccag gaagctaacc gtctcagaca gagcacaaag
180caccgagaca tgtgccacaa ggcttgtgta gagaggtcag aggacagcgt
acaggtccca 240gagatcaaac tcaacctcac caggcttggc agcaagcctt
taccaaccca cccccacccc 300acccaccctg cacgcgcccc tctcccctcc
ccatggtctc ccatggctat ctcacttggc 360cctaaaatgt ttaaggatga
cactggctgc tgagtggaaa tgagacagca gaagtcaaca 420gtagatttta
ggaaagccag agaaaaaggc ttgtgctgtt tttagaaagc caagggacaa
480gctaagatag ggcccaagta at 5029788DNAMus musculus 9aaatagaact
gtgagatagg gggagagggg gcaggaagga caagagaccc ctgtctcatt 60gtgatcccca
cctgtctgct ctgtgggagg gtacccatga gggccagccc acagccctta
120ggtggacatt gtctggtcct gtctcactgt ccctcccagc agccccagag
gccaggagac 180aggggtctca gtcctcactg agagatgtgt aaactgaggc
ccagtgaatg ttgagggcca 240gggcatgccc ttggtgggat gtgacctggg
tctccttcgc acgggcttcc tccccgaagc 300cgagctgagc atttggagtt
tgaaatgttt ccgtacttag caatctgctc ctctattccc 360gggcggactt
ccgatagctc cggccttatg ctgcactaga taagatggag cagggagagg
420acacggcact acttatgtaa ccggcctctt gaaaaatgga gcagcggtca
gggcggaaca 480agacgtcctc tctctacgca tccctctcct ttccctgcta
aggctgcagc tggagtcaga 540ggcagggctg ttccaatctg tctttgatca
gtaacgcagc cagcctccag cctccgtcag 600cctcctcatg gctgagaccc
ggcctcagtt tcccccactt acatcccgag gatcagagcc 660tgtgaggatg
aaatgggata aggtagctgg aaccgtctgg cagagagcga gtcctcagga
720ctgttgatgc ctgtggctgc ctggcttgac cccaagtgac cccgcctcct
catcctgcag 780caggagaa 78810502DNAMus musculus 10tctatagaat
gtgtccccag ccttgttttc cacacttgat acgcaaggaa tgcataccac 60agagagggat
gagggtagca tccagcctgc ttcctgtgtg tcggggcgct acagccacat
120ctccccagtc catctcagac cgtcacagag cttcgccgaa tgtatagctt
tgttctctgt 180gcagacaggg agacagagcc ttgggaagca taggtgcttg
cttctttgcc cactgagtct 240tagctggact tgcacaccac atgcctcaca
gccgggcgca cttgcatttg tcacccaggc 300ccagtgatga tggctctgct
tgctttgtgc tttgtgccaa ctacagctcc agcacctgtg 360ccctgggttt
tcactccttt agttgaacac gtagttactg gggttgtagg gatggagcct
420ttctgcttcc ttctggcaaa gtccttagcg gcctgctgcg ggggtggggg
gtgttcaggg 480gagtggtgat gaagtatgac ag 50211533DNAMus musculus
11tctccagttg gagaaacaga tgctgtaact ggggccacag tataaagaga gcccagacat
60tgaactgtca acacagaagc ctggcacact ggaactggca gtccagctgg gaacaagggg
120tagaggctga ggccactaag tcaactgagg caggagacat aggagctaaa
gcagctgaag 180ggtgcaggac agctgggggg tctgaagtgg gcctcatgcc
cagagctatg aagtcagggg 240ctgtagccta ggagccttgg aagccagctg
gcaagctgtg gcccaaagac gctgactcac 300caggaggggg cagctggagc
caggcactcc taaggtttcc aggaagggca gccttccagg 360gctcagctag
gggagacagt gttgacagca agttgtcagg caacttgagc tactgggcag
420ctgggaagct gtcccttggt ccccagtatc atcatcaccc cagacgctgc
ccacctgcct 480caggtcccac acagtgatcc tcccatcttt aacacaacac
atgaccagag aga 53312524DNAMus musculus 12gtcaccctcc ccccaaacaa
ccccttcttc tctggttcga gaaattacag gcatgaaaga 60tataaatcgg gatgcttgac
ttgggaatat aaatcactaa agcttggggg caggggtggg 120cgacctttgt
gaccgtcctt gtgcgtgcca gtaaatcctg tggtccaggg gagaagaaaa
180ggctgtgtgg cttctgctca caaagctgca gaaaccattc tttaagccca
aaagcacttc 240cagagagagc agagcatccc caggctgctg gctcagcaag
ttcactgtgc tcaatctcag 300gaagtgagga taagagcagt gcctggagag
tgcctggtgc tgagctgagg gtttctgaac 360acattaaagc ggggagcatg
gaccgggcct caggaggggt gttgaacatc cctaggcaga 420ggagtctagc
ttcctgggaa aagatatcag gttaagcaca cacatgtcct ctggaataag
480ataatctttc tgatcacaca ctatacacac acaaaagcct gctc 52413509DNAMus
musculus 13gccctctagg ccacctgacc aggtcccctc agtccccccc ttcccacact
cccacactca 60gcccccctcc cccccccccg acccctgcag gattatcctg tctgtgttcc
tgactcagcc 120tgggagccac ctgggcagca ggggccaagg gtgtcctaga
agggacctgg agtccacgct 180gggccaagcc tgccctttct ccctctgtct
tccgtccctg cttgcggttc tgctgaatgt 240ggttatttct ctggctcctt
ttacagagaa tgctgctgct aattttatgt ggagctctga 300ggcagtgtaa
ttggaagcca gacaccctgt cagcagtggg ctcccgtcct gagctgccat
360gcttcctgct ctcctcccgt cccggctcct catttcatgc agccacctgt
cccagggaga 420gaggagtcac ccaggcccct cagtccgccc cttaaataag
aaagcctccg ttgctcggca 480cacataccaa gcagccgctg gtgcaatct
50914398DNAMus musculus 14gtgttcttcc cttccccttt ggacccccga
gacaagccaa taaaatactc ggcagggtgg 60cttctctcct ttttttgcca gtaataaaca
gactcagagc aagttaaggg tctggtccaa 120ggtcatggct gggatcagtg
acagagccca gaagagaacc tgagacttct tgctgagcca 180agctggagag
gacagaaagg aatgcgtcta ctccatgcat gaccctctgc cagctttgct
240ccttcctaag ggaccatgaa cgatatgtgc acaccgctca tacgtatgtg
cacacctgca 300agaggaggca tcccatgtac acctatgaga cgcacagaga
aacatatatg tagccatagg 360ctagaaattc tttctctttc taggtctgcc cctctgca
39815810DNAMus musculus 15ggaccactca gtgtacacgg aatgtagaat
tgagtctgcc attggtcttc cctcaaagtc 60ttggaggctt gggactgata ttgggagcat
ctgggcagag aaggccacaa agacagggtg 120gtttttctac actgggacat
actcgtgagc atgcacagag gcgtgtcccc aacttccctg 180tcacccctgt
cctctgccgg ctagagggga tgcgggggtg gacatatgct gctattgggc
240agatatcaca tgttaagagg tggggggggg ctcaagaggc ggagggctag
gagcatccca 300tggggagagg ttctggtttt cttgctgcct ctagctgcta
taaatacgtt agcacttgag 360caactggaaa gctctgagta atttaggatg
cacaaagctg taatttaact ccagcatctc 420agtgtgcgag agcattaaag
atgtaattaa gatgtttaca caaagagatt ggagtctgtg 480acacttgggg
tgcaaaaccc caggaaggga cacaatgggt gaggtgagga tctgtgggag
540gcctggggac agtcacttgg atcccagcta tgagatggca ggccacccag
ctgtttctcc 600ttggaaatgt tttggcctgg gggttggggg tggggcatca
cactttgata tggagatggg 660gcaacaaagc ctgcaatatc tgggggtgga
gaggtcaagt ggatggagtc ttttgagatc 720atgtcaggaa gagggctcga
tcccccaaaa tcatggtgac atatggtgtc tcggggttca 780caggagctat
gtctaaaata caaaagtaaa 81016202DNAMus musculus 16tctgcagaag
cctgccattc caccatttaa acctgtgact ccaggcctta agcctgttga 60aggtcgagtc
ccagaagggt catatgtgca actgcctagg gagagttccc actcgcaggg
120ccaagaggag tcccccggtc tgaggtgtgg gggcggggac gtgcactggg
cgctgggacc 180acggctgggg ctcaggactc gc 20217937DNAMus musculus
17tgcctcagtt tcttcgccta gaaagccggg tctaagggta catgccctga ttcttttctg
60gggtgtctcg aattttaaac aacacatact gttctgggct gatgacaaga ggaagtactg
120gtcggtggct gatggacatc caccatggtg gcaactggag ggagggggaa
cggacgttga 180aaccctgccc tcctggaatc tgtcgcatgc acgcacgttg
acaatgcttg gcactgggga 240caggctggga tggatggagc ggagcgtgag
gaggagtggg catgcaggcc cgagtgtctg 300ttttgctgat tgctcctttt
gctttcaagg agattaaact atttttagtc catgcctact 360gctggtgaga
cgctggagga agcctttcca tcgttgagat tttctggaag ctgccaagtg
420tggtcttcag ctcaattctg ggagcctccc agagtgggag ggaggaacat
ttccatctgg 480gggcttcggg gacaggctaa gatcttccct ggggtccttg
ctgcgctggc ctcctcaaac 540cacgctgcct cggcctgcat aaagcagtaa
tctgatgtgc ccgatgtttg taacgctgtg 600tttaaaaaaa gtaatttatt
ttctaattat tccttgtctt gcataaccat gcattgccaa 660agtgtcgcta
tttaaaatat ttatctctcc acgccgcagg agcagctctg gagcgtggag
720ggggaagaaa taaaagtccg cgtgccagtc gcaggcatat tactttgact
cgtcctggtg 780gctttgacgt ctccctgtaa atacatttat ttttcattag
gacgtttctg agcttgtggc 840ccccggagag cggagtgatt acgctgttca
tctgcaagcg atgcaataga ggggtactcg 900cagaatgact tccgcccaga
gcatcctgcg cctgtct 93718922DNAMus musculus 18taaaatacct tatttttttc
cagtctctaa actgctaatc tcccaggcta agggattctg 60ggacaaaggc aaggcctgga
agtggaaatc tgtaaaatta gcttcagcgg tattagtgtt 120tgcagttgaa
gattgaaaaa ctgctttccc agggcctgat tggaggctcc actctcctcc
180aggaagaggc aaggactctg ggctggcact gaggacaaat cctgggaggc
tgctatgggg 240cctgggagcc aggctgcctt gtgctagagg cctagagagt
gtctgtgtcc caagtcccaa 300gctaccccca gcagctaaca gcttttccag
ttctcaggca cagcaggtgc caagatcacg 360ctctggagtc cagctgggcc
ccttcctctt cttttttttt tttttttttt aagacctcct 420ggacactgtt
cctctccccc cccccgtgac cccccccctc agttctcaaa cacgtgaggg
480ttgggggagg gttccacagc cagagagagg ggccagctct ggtgcctgtg
ggtacgcccg 540cccgtatggc ccatcaggcc tcttgtgtgc ttgattgcct
ctgattggct gcagctgaat 600tcagcaaaag ctattatttg cccttgatga
gccaatcaga tggcctcatt ggccattcag 660agcaggcacc ggaacctgag
ggtggggtgg ggggtggggg atggagatgg gactcagtga 720gggggtggga
agctctaaaa cagatgcagg acctgagcct gtctgtgtcc accacgacct
780tcacacaggt cacaccccct tcccctgact tgtcacccca aaccagggct
tgttgcccaa 840ccccacctca caattccctc actctgtaac acctttccat
atacctctgc atgtctaaac 900ccaagacttg ctctatgaaa tc 92219639DNAMus
musculus 19agaccctgct tagcacagct cttagcgggt cctttagggg gtctcccagc
gggcccagtg 60ggaatgagat aaggaaggac acagctgtcc attctcccgt gcctgctaag
gaggaaatgg 120ggccgcctta cataattggg gcaatttgtt ccactcttgt
cctcctggta tcatggctat 180caccccctcc ttgctcaggg agtccttgat
tgagcgagaa gctcaggcct ccctctctcc 240ctcctgctgg gggttgctga
acagagggtg taggagccat aggctctgtc actgctgaga 300tctgccagat
gtctaggcca ggagaaaatg gaaagggcta agtcacagca tatgtggcca
360ctcaggccta tagccccaaa tctgcctggt aacccattat gtccccagag
aatttgcatg 420ggcggacacc ctcatgccgg gtctcagtaa gggaaggggt
gggaggcaaa aatatccctc 480cccaccctga atctccaccc cctcccccca
gaaactgaca cttggccttg tctaaggatg 540ggttttccca aaatccttct
gaaaaaaaca gaatttcaag agtcactccc tccgggtctc 600agcctagaac
atatgcagta tcccctgacg tccataggg 63920716DNAMus musculus
20aaactggcac agtaatggcg ggctgacaga caagggagtc tgtagcaccc gctgcctccg
60cccacccctt ctccgagcaa ttaaaaggtg tttatgtggg gctggcagtg gcttctgcct
120cccttccatt acgaacatta agagatcttg acccttccac tttccccgct
cttgaaagga 180gctgcagaca cgtggagcca attaggcgca cgcgtgggcg
ccaagggcct gagcagcttt 240ttctccctga ttgcggcgtt tacagctgat
tattctcccc tcacccaaac agtgctgctt 300cctggcaagg tgccacccag
aggagccggc tgggggcccc tggggacagg ggaggactgg 360attagtaaat
gggcatctat cgaatggctt tcatatgtgt ggctggaagg gagaagggta
420gggccaggaa tggtggcagc aagggcccag gtagcaatga gggttcttct
aacccaccat 480ttagggatag cgatcagaaa agggccctcg aggaggtgac
ctaaatgtgt gtagaagctg 540acggccacta cacacacaca cacacacaca
cacacacata cacaagcatc cttgtccttg 600gagtcggtca gcatgagcaa
gagaaagatg ttcccagtgg ccatgagagt ggagccctcc 660tccctactta
catccaggtt ggatggccag gagatcctga gatccttcaa gactcc 71621147DNAMus
musculus 21aagccacatc ctgggtggaa atatatggct tcaattccca ctcttccgga
tgacctctgt 60ggggagccct ggcttcacct tggtccagct tcatccctta gcctcgctgc
caggaaggca 120gtgaggtcag aggctggtgc tggcgtg 14722297DNAMus musculus
22cctacctggt gcccgccaac atctgggggc catcctggcc agcgccagcg tggtggtgaa
60ggcactgtgc gccgtggtac tgtttctcta cctgctttcc ttcgctgtgg acacgggctg
120cctggccgtc accccaggct accttttccc acccaacttc tggatctgga
ccctggccac 180ccacgggctc atggaacagc acgtgtggga cgtggccatt
agcctggcca cagtggttgt 240ggccgggcga ttactggagc ccctctgggg
agccttggag ctgctcatct tcttctc 29723101DNAHomo sapiens 23aaacggacgg
gcctccgctg aaccagtgag gccccagacg tgcgcataaa taacccctgc 60gtgctgcacc
acctggggag agggggagga ccacggtaaa t 10124101DNAHomo sapiens
24ggagcgagcg catagcaaaa gggacgcggg gtccttttct ctgccggtgg cactgggtag
60ctgtggccag gtgtggtact ttgatggggc ccagggctgg a 10125101DNAHomo
sapiens 25gctcaaggaa gcgtcgcagg gtcacagatc tgggggaacc ccggggaaaa
gcactgaggc 60aaaaccgccg ctcgtctcct acaatatatg ggagggggag g
10126547DNAHomo sapiens 26ttgagtacgt tctggattac tcataagacc
tttttttttt ccttccgggc gcaaaaccgt 60gagctggatt tataatcgcc ctataaagct
ccagaggcgg tcaggcacct gcagaggagc 120cccgccgctc cgccgactag
ctgcccccgc gagcaacggc ctcgtgattt ccccgccgat 180ccggtccccg
cctccccact ctgcccccgc ctaccccgga gccgtgcagc cgcctctccg
240aatctctctc ttctcctggc
gctcgcgtgc gagagggaac tagcgagaac gaggaagcag 300ctggaggtga
cgccgggcag attacgcctg tcagggccga gccgagcgga tcgctgggcg
360ctgtgcagag gaaaggcggg agtgcccggc tcgctgtcgc agagccgagg
tgggtaagct 420agcgaccacc tggacttccc agcgcccaac cgtggctttt
cagccaggtc ctctcctccc 480gcggcttctc aaccaacccc atcccagcgc
cggccaccca acctcccgaa atgagtgctt 540cctgccc 54727101DNAHomo sapiens
27cagcagccga aggcgctact aggaacggta acctgttact tttccagggg ccgtagtcga
60cccgctgccc gagttgctgt gcgactgcgc gcgcggggct a 10128201DNAHomo
sapiens 28gagtgcaagg tgactgtggt tcttctctgg ccaagtccga gggagaacgt
aaagatatgg 60gcctttttcc ccctctcacc ttgtctcacc aaagtcccta gtccccggag
cagttagcct 120ctttctttcc agggaattag ccagacacaa caacgggaac
cagacaccga accagacatg 180cccgccccgt gcgccctccc c 20129199DNAHomo
sapiens 29gctcgctgcc tttcctccct cttgtctctc cagagccgga tcttcaaggg
gagcctccgt 60gcccccggct gctcagtccc tccggtgtgc aggaccccgg aagtcctccc
cgcacagctc 120tcgcttctct ttgcagcctg tttctgcgcc ggaccagtcg
aggactctgg acagtagagg 180ccccgggacg accgagctg 199301351DNAHomo
sapiens 30aaacggacgg gcctccgctg aaccagtgag gccccagacg tgcgcataaa
taacccctgc 60gtgctgcacc acctggggag agggggagga ccacggtaaa tggagcgagc
gcatagcaaa 120agggacgcgg ggtccttttc tctgccggtg gcactgggta
gctgtggcca ggtgtggtac 180tttgatgggg cccagggctg gagctcaagg
aagcgtcgca gggtcacaga tctgggggaa 240ccccggggaa aagcactgag
gcaaaaccgc cgctcgtctc ctacaatata tgggaggggg 300aggttgagta
cgttctggat tactcataag accttttttt tttccttccg ggcgcaaaac
360cgtgagctgg atttataatc gccctataaa gctccagagg cggtcaggca
cctgcagagg 420agccccgccg ctccgccgac tagctgcccc cgcgagcaac
ggcctcgtga tttccccgcc 480gatccggtcc ccgcctcccc actctgcccc
cgcctacccc ggagccgtgc agccgcctct 540ccgaatctct ctcttctcct
ggcgctcgcg tgcgagaggg aactagcgag aacgaggaag 600cagctggagg
tgacgccggg cagattacgc ctgtcagggc cgagccgagc ggatcgctgg
660gcgctgtgca gaggaaaggc gggagtgccc ggctcgctgt cgcagagccg
aggtgggtaa 720gctagcgacc acctggactt cccagcgccc aaccgtggct
tttcagccag gtcctctcct 780cccgcggctt ctcaaccaac cccatcccag
cgccggccac ccaacctccc gaaatgagtg 840cttcctgccc cagcagccga
aggcgctact aggaacggta acctgttact tttccagggg 900ccgtagtcga
cccgctgccc gagttgctgt gcgactgcgc gcgcggggct agagtgcaag
960gtgactgtgg ttcttctctg gccaagtccg agggagaacg taaagatatg
ggcctttttc 1020cccctctcac cttgtctcac caaagtccct agtccccgga
gcagttagcc tctttctttc 1080cagggaatta gccagacaca acaacgggaa
ccagacaccg aaccagacat gcccgccccg 1140tgcgccctcc ccgctcgctg
cctttcctcc ctcttgtctc tccagagccg gatcttcaag 1200gggagcctcc
gtgcccccgg ctgctcagtc cctccggtgt gcaggacccc ggaagtcctc
1260cccgcacagc tctcgcttct ctttgcagcc tgtttctgcg ccggaccagt
cgaggactct 1320ggacagtaga ggccccggga cgaccgagct g 1351312051DNAHomo
sapiens 31ggaggaagcc atcaactaaa ctacaatgac tgtaagatac aaaattggga
atggtaacat 60attttgaagt tctgttgaca taaagaatca tgatattaat gcccatggaa
atgaaagggc 120gatcaacact atggtttgaa aagggggaaa ttgtagagca
cagatgtgtt cgtgtggcag 180tgtgctgtct ctagcaatac tcagagaaga
gagagaacaa tgaaattctg attggcccca 240gtgtgagccc agatgaggtt
cagctgccaa ctttctcttt cacatcttat gaaagtcatt 300taagcacaac
taactttttt tttttttttt tttttttgag acagagtctt gctctgttgc
360ccaggacaga gtgcagtagt gactcaatct cggctcactg cagcctccac
ctcctaggct 420caaacggtcc tcctgcatca gcctcccaag tagctggaat
tacaggagtg gcccaccatg 480cccagctaat ttttgtattt ttaatagata
cgggggtttc accatatcac ccaggctggt 540ctcgaactcc tggcctcaag
tgatccacct gcctcggcct cccaaagtgc tgggattata 600ggcgtcagcc
actatgccca acccgaccaa ccttttttaa aataaatatt taaaaaattg
660gtatttcaca tatatactag tatttacatt tatccacaca aaacggacgg
gcctccgctg 720aaccagtgag gccccagacg tgcgcataaa taacccctgc
gtgctgcacc acctggggag 780agggggagga ccacggtaaa tggagcgagc
gcatagcaaa agggacgcgg ggtccttttc 840tctgccggtg gcactgggta
gctgtggcca ggtgtggtac tttgatgggg cccagggctg 900gagctcaagg
aagcgtcgca gggtcacaga tctgggggaa ccccggggaa aagcactgag
960gcaaaaccgc cgctcgtctc ctacaatata tgggaggggg aggttgagta
cgttctggat 1020tactcataag accttttttt tttccttccg ggcgcaaaac
cgtgagctgg atttataatc 1080gccctataaa gctccagagg cggtcaggca
cctgcagagg agccccgccg ctccgccgac 1140tagctgcccc cgcgagcaac
ggcctcgtga tttccccgcc gatccggtcc ccgcctcccc 1200actctgcccc
cgcctacccc ggagccgtgc agccgcctct ccgaatctct ctcttctcct
1260ggcgctcgcg tgcgagaggg aactagcgag aacgaggaag cagctggagg
tgacgccggg 1320cagattacgc ctgtcagggc cgagccgagc ggatcgctgg
gcgctgtgca gaggaaaggc 1380gggagtgccc ggctcgctgt cgcagagccg
aggtgggtaa gctagcgacc acctggactt 1440cccagcgccc aaccgtggct
tttcagccag gtcctctcct cccgcggctt ctcaaccaac 1500cccatcccag
cgccggccac ccaacctccc gaaatgagtg cttcctgccc cagcagccga
1560aggcgctact aggaacggta acctgttact tttccagggg ccgtagtcga
cccgctgccc 1620gagttgctgt gcgactgcgc gcgcggggct agagtgcaag
gtgactgtgg ttcttctctg 1680gccaagtccg agggagaacg taaagatatg
ggcctttttc cccctctcac cttgtctcac 1740caaagtccct agtccccgga
gcagttagcc tctttctttc cagggaatta gccagacaca 1800acaacgggaa
ccagacaccg aaccagacat gcccgccccg tgcgccctcc ccgctcgctg
1860cctttcctcc ctcttgtctc tccagagccg gatcttcaag gggagcctcc
gtgcccccgg 1920ctgctcagtc cctccggtgt gcaggacccc ggaagtcctc
cccgcacagc tctcgcttct 1980ctttgcagcc tgtttctgcg ccggaccagt
cgaggactct ggacagtaga ggccccggga 2040cgaccgagct g
2051321878DNAUnknownDescription of Unknown RE sequence 32tcaacagggg
gacacttggg aaagaaggat ggggacagag ccgagaggac tgttacacat 60tagagaaaca
tcagtgactg tgccagcttt ggggtagact gcacaaaagc cctgaggcag
120cacaggcagg atccagtctg ctggtcccag gaagctaacc gtctcagaca
gagcacaaag 180caccgagaca tgtgccacaa ggcttgtgta gagaggtcag
aggacagcgt acaggtccca 240gagatcaaac tcaacctcac caggcttggc
agcaagcctt taccaaccca cccccacccc 300acccaccctg cacgcgcccc
tctcccctcc ccatggtctc ccatggctat ctcacttggc 360cctaaaatgt
ttaaggatga cactggctgc tgagtggaaa tgagacagca gaagtcaaca
420gtagatttta ggaaagccag agaaaaaggc ttgtgctgtt tttagaaagc
caagggacaa 480gctaagatag ggcccaagta atgctagtat ttacatttat
ccacacaaaa cggacgggcc 540tccgctgaac cagtgaggcc ccagacgtgc
gcataaataa cccctgcgtg ctgcaccacc 600tggggagagg gggaggacca
cggtaaatgg agcgagcgca tagcaaaagg gacgcggggt 660ccttttctct
gccggtggca ctgggtagct gtggccaggt gtggtacttt gatggggccc
720agggctggag ctcaaggaag cgtcgcaggg tcacagatct gggggaaccc
cggggaaaag 780cactgaggca aaaccgccgc tcgtctccta caatatatgg
gagggggagg ttgagtacgt 840tctggattac tcataagacc tttttttttt
ccttccgggc gcaaaaccgt gagctggatt 900tataatcgcc ctataaagct
ccagaggcgg tcaggcacct gcagaggagc cccgccgctc 960cgccgactag
ctgcccccgc gagcaacggc ctcgtgattt ccccgccgat ccggtccccg
1020cctccccact ctgcccccgc ctaccccgga gccgtgcagc cgcctctccg
aatctctctc 1080ttctcctggc gctcgcgtgc gagagggaac tagcgagaac
gaggaagcag ctggaggtga 1140cgccgggcag attacgcctg tcagggccga
gccgagcgga tcgctgggcg ctgtgcagag 1200gaaaggcggg agtgcccggc
tcgctgtcgc agagccgagg tgggtaagct agcgaccacc 1260tggacttccc
agcgcccaac cgtggctttt cagccaggtc ctctcctccc gcggcttctc
1320aaccaacccc atcccagcgc cggccaccca acctcccgaa atgagtgctt
cctgccccag 1380cagccgaagg cgctactagg aacggtaacc tgttactttt
ccaggggccg tagtcgaccc 1440gctgcccgag ttgctgtgcg actgcgcgcg
cggggctaga gtgcaaggtg actgtggttc 1500ttctctggcc aagtccgagg
gagaacgtaa agatatgggc ctttttcccc ctctcacctt 1560gtctcaccaa
agtccctagt ccccggagca gttagcctct ttctttccag ggaattagcc
1620agacacaaca acgggaacca gacaccgaac cagacatgcc cgccccgtgc
gccctccccg 1680ctcgctgcct ttcctccctc ttgtctctcc agagccggat
cttcaagggg agcctccgtg 1740cccccggctg ctcagtccct ccggtgtgca
ggaccccgga agtcctcccc gcacagctct 1800cgcttctctt tgcagcctgt
ttctgcgccg gaccagtcga ggactctgga cagtagaggc 1860cccgggacga ccgagctg
18783319DNAHomo sapiens 33atttacattt atccacaca 1934800DNAMus
musculus 34tgccgctgga ctctcttcca aggaactagg agaaccaaga tccgtttttc
tgccaagggc 60tgcccccccc acgcccccaa ccccctcacc ccgatcccca cagaaagaaa
tcttgaggta 120gctggagctt cttctgtggg tgtgacagga ctgccattct
cctctgtagt ctgcagaagc 180ctgccattcc accatttaaa cctgtgactc
caggccttaa gcctgttgaa ggtcgagtcc 240cagaagggtc atatgtgcaa
ctgcctaggg agagttccca ctcgcagggc caagaggagt 300cccccggtct
gaggtgtggg ggcggggacg tgcactgggc gctgggacca cggctggggc
360tcaggactcg cgagcttgga ttcggatcgg tttgcgcgag ccagtagggc
aggctccggg 420gtgaacgggg acgaggggcg cgcgggcaca ggcgggcgcg
tgaccgcggc gggggcgcgc 480ggaggcgggc cggccaagga gagggaggga
gggaatgagg gagggagcga caggggaggg 540cggcgccggc aggttggcgg
cggccgctat ttgagcgcag gtcccgggcc aggcgctcaa 600agcgcttgga
gccagcgcgg cggggagatc gctgcgcgca gcccgcagag gcgctgcggc
660cagtgcagcc ccggaggccc cgcgcggaga aggaggtgga gaagaggccg
gctttccgcc 720cgccgcccgc gcccccccac ctccatcccg ccgccgccgt
cccccctccc tccccgcggc 780gccgcatctt gaatggaaac
800351335DNAArtificial SequenceDescription of Artificial Sequence
Synthetic polynucleotide 35gagtaattca tacaaaagga ctcgcccctg
ccttggggaa tcccagggac cgtcgttaaa 60ctcccactaa cgtagaaccc agagatcgct
gcgttcccgc cccctcaccc gcccgctctc 120gtcatcactg aggtggagaa
gagcatgcgt gaggctccgg tgcccgtcag tgggcagagc 180gcacatcgcc
cacagtcccc gagaagttgg ggggaggggt cggcaattga accggtgcct
240agagaaggtg gcgcggggta aactgggaaa gtgatgtcgt gtactggctc
cgcctttttc 300ccgagggtgg gggagaaccg tatataagtg cagtagtcgc
cgtgaacgtt ctttttcgca 360acgggtttgc cgccagaaca caggtaagtg
ccgtgtgtgg ttcccgcggg cctggcctct 420ttacgggtta tggcccttgc
gtgccttgaa ttacttccac gcccctggct gcagtacgtg 480attcttgatc
ccgagcttcg ggttggaagt gggtgggaga gttcgaggcc ttgcgcttaa
540ggagcccctt cgcctcgtgc ttgagttgag gcctggcttg ggcgctgggg
ccgccgcgtg 600cgaatctggt ggcaccttcg cgcctgtctc gctgctttcg
ataagtctct agccatttaa 660aatttttgat gacctgctgc gacgcttttt
ttctggcaag atagtcttgt aaatgcgggc 720caagatctgc acactggtat
ttcggttttt ggggccgcgg gcggcgacgg ggcccgtgcg 780tcccagcgca
catgttcggc gaggcggggc ctgcgagcgc ggccaccgag aatcggacgg
840gggtagtctc aagctggccg gcctgctctg gtgcctggcc tcgcgccgcc
gtgtatcgcc 900ccgccctggg cggcaaggct ggcccggtcg gcaccagttg
cgtgagcgga aagatggccg 960cttcccggcc ctgctgcagg gagctcaaaa
tggaggacgc ggcgctcggg agagcgggcg 1020ggtgagtcac ccacacaaag
gaaaagggcc tttccgtcct cagccgtcgc ttcatgtgac 1080tccacggagt
accgggcgcc gtccaggcac ctcgattagt tctcgagctt ttggagtacg
1140tcgtctttag gttgggggga ggggttttat gcgatggagt ttccccacac
tgagtgggtg 1200gagactgaag ttaggccagc ttggcacttg atgtaattct
ccttggaatt tgcccttttt 1260gagtttggat cttggttcat tctcaagcct
cagacagtgg ttcaaagttt ttttcttcca 1320tttcaggtgt cgtga
133536222PRTArtificial SequenceDescription of Artificial Sequence
Synthetic polypeptide 36Met Val Ser Lys Gly Glu Glu Leu Phe Thr Gly
Val Val Pro Ile Leu1 5 10 15Val Glu Leu Asp Gly Asp Val Asn Gly His
Lys Phe Ser Val Ser Gly 20 25 30Glu Gly Glu Gly Asp Ala Thr Tyr Gly
Lys Leu Thr Leu Lys Phe Ile 35 40 45Cys Thr Thr Gly Lys Leu Pro Val
Pro Trp Pro Thr Leu Val Thr Thr 50 55 60Leu Thr Tyr Gly Val Gln Cys
Phe Ser Arg Tyr Pro Asp His Met Lys65 70 75 80Gln His Asp Phe Phe
Lys Ser Ala Met Pro Glu Gly Tyr Val Gln Glu 85 90 95Arg Thr Ile Phe
Phe Lys Asp Asp Gly Asn Tyr Lys Thr Arg Ala Glu 100 105 110Val Lys
Phe Glu Gly Asp Thr Leu Val Asn Arg Ile Glu Leu Lys Gly 115 120
125Ile Asp Phe Lys Glu Asp Gly Asn Ile Leu Gly His Lys Leu Glu Tyr
130 135 140Asn Tyr Asn Ser His Asn Val Tyr Ile Met Ala Asp Lys Gln
Lys Asn145 150 155 160Gly Ile Lys Val Asn Phe Lys Ile Arg His Asn
Ile Glu Asp Gly Ser 165 170 175Val Gln Leu Ala Asp His Tyr Gln Gln
Asn Thr Pro Ile Gly Asp Gly 180 185 190Pro Val Leu Leu Pro Asp Asn
His Tyr Leu Ser Thr Gln Ser Ala Leu 195 200 205Ser Lys Asp Pro Asn
Glu Lys Arg Asp His Met Val Leu Leu 210 215
22037218PRTUnknownDescription of Unknown SCN1B sequence 37Met Gly
Thr Leu Leu Ala Leu Val Val Gly Ala Ala Leu Val Ser Ser1 5 10 15Ala
Trp Gly Gly Cys Val Glu Val Asp Ser Asp Thr Glu Ala Val Tyr 20 25
30Gly Met Thr Phe Lys Ile Leu Cys Ile Ser Cys Lys Arg Arg Ser Glu
35 40 45Thr Thr Ala Glu Thr Phe Thr Glu Trp Thr Phe Arg Gln Lys Gly
Thr 50 55 60Glu Glu Phe Val Lys Ile Leu Arg Tyr Glu Asn Glu Val Leu
Gln Leu65 70 75 80Glu Glu Asp Glu Arg Phe Glu Gly Arg Val Val Trp
Asn Gly Ser Arg 85 90 95Gly Thr Lys Asp Leu Gln Asp Leu Ser Ile Phe
Ile Thr Asn Val Thr 100 105 110Tyr Asn His Ser Gly Asp Tyr Glu Cys
His Val Tyr Arg Leu Leu Phe 115 120 125Phe Asp Asn Tyr Glu His Asn
Thr Ser Val Val Lys Lys Ile His Leu 130 135 140Glu Val Val Asp Lys
Ala Asn Arg Asp Met Ala Ser Ile Val Ser Glu145 150 155 160Ile Met
Met Tyr Val Leu Ile Val Val Leu Thr Ile Trp Leu Val Ala 165 170
175Glu Met Val Tyr Cys Tyr Lys Lys Ile Ala Ala Ala Thr Glu Ala Ala
180 185 190Ala Gln Glu Asn Ala Ser Glu Tyr Leu Ala Ile Thr Ser Glu
Ser Lys 195 200 205Glu Asn Cys Thr Gly Val Gln Val Ala Glu 210
21538215PRTUnknownDescription of Unknown SCN2B sequence 38Met His
Arg Asp Ala Trp Leu Pro Arg Pro Ala Phe Ser Leu Thr Gly1 5 10 15Leu
Ser Leu Phe Phe Ser Leu Val Pro Pro Gly Arg Ser Met Glu Val 20 25
30Thr Val Pro Ala Thr Leu Asn Val Leu Asn Gly Ser Asp Ala Arg Leu
35 40 45Pro Cys Thr Phe Asn Ser Cys Tyr Thr Val Asn His Lys Gln Phe
Ser 50 55 60Leu Asn Trp Thr Tyr Gln Glu Cys Asn Asn Cys Ser Glu Glu
Met Phe65 70 75 80Leu Gln Phe Arg Met Lys Ile Ile Asn Leu Lys Leu
Glu Arg Phe Gln 85 90 95Asp Arg Val Glu Phe Ser Gly Asn Pro Ser Lys
Tyr Asp Val Ser Val 100 105 110Met Leu Arg Asn Val Gln Pro Glu Asp
Glu Gly Ile Tyr Asn Cys Tyr 115 120 125Ile Met Asn Pro Pro Asp Arg
His Arg Gly His Gly Lys Ile His Leu 130 135 140Gln Val Leu Met Glu
Glu Pro Pro Glu Arg Asp Ser Thr Val Ala Val145 150 155 160Ile Val
Gly Ala Ser Val Gly Gly Phe Leu Ala Val Val Ile Leu Val 165 170
175Leu Met Val Val Lys Cys Val Arg Arg Lys Lys Glu Gln Lys Leu Ser
180 185 190Thr Asp Asp Leu Lys Thr Glu Glu Glu Gly Lys Thr Asp Gly
Glu Gly 195 200 205Asn Pro Asp Asp Gly Ala Lys 210
215392009PRTUnknownDescription of Unknown SCN1A sequence 39Met Glu
Gln Thr Val Leu Val Pro Pro Gly Pro Asp Ser Phe Asn Phe1 5 10 15Phe
Thr Arg Glu Ser Leu Ala Ala Ile Glu Arg Arg Ile Ala Glu Glu 20 25
30Lys Ala Lys Asn Pro Lys Pro Asp Lys Lys Asp Asp Asp Glu Asn Gly
35 40 45Pro Lys Pro Asn Ser Asp Leu Glu Ala Gly Lys Asn Leu Pro Phe
Ile 50 55 60Tyr Gly Asp Ile Pro Pro Glu Met Val Ser Glu Pro Leu Glu
Asp Leu65 70 75 80Asp Pro Tyr Tyr Ile Asn Lys Lys Thr Phe Ile Val
Leu Asn Lys Gly 85 90 95Lys Ala Ile Phe Arg Phe Ser Ala Thr Ser Ala
Leu Tyr Ile Leu Thr 100 105 110Pro Phe Asn Pro Leu Arg Lys Ile Ala
Ile Lys Ile Leu Val His Ser 115 120 125Leu Phe Ser Met Leu Ile Met
Cys Thr Ile Leu Thr Asn Cys Val Phe 130 135 140Met Thr Met Ser Asn
Pro Pro Asp Trp Thr Lys Asn Val Glu Tyr Thr145 150 155 160Phe Thr
Gly Ile Tyr Thr Phe Glu Ser Leu Ile Lys Ile Ile Ala Arg 165 170
175Gly Phe Cys Leu Glu Asp Phe Thr Phe Leu Arg Asp Pro Trp Asn Trp
180 185 190Leu Asp Phe Thr Val Ile Thr Phe Ala Tyr Val Thr Glu Phe
Val Asp 195 200 205Leu Gly Asn Val Ser Ala Leu Arg Thr Phe Arg Val
Leu Arg Ala Leu 210 215 220Lys Thr Ile Ser Val Ile Pro Gly Leu Lys
Thr Ile Val Gly Ala Leu225 230 235 240Ile Gln Ser Val Lys Lys Leu
Ser Asp Val Met Ile Leu Thr Val Phe 245 250 255Cys Leu Ser Val Phe
Ala Leu Ile Gly Leu Gln Leu Phe Met Gly Asn 260 265 270Leu Arg Asn
Lys Cys Ile Gln Trp Pro Pro Thr Asn Ala Ser Leu Glu 275 280 285Glu
His Ser Ile Glu Lys Asn Ile Thr Val Asn Tyr Asn Gly Thr Leu 290 295
300Ile Asn Glu Thr Val Phe Glu Phe Asp Trp Lys Ser Tyr Ile Gln
Asp305 310 315
320Ser Arg Tyr His Tyr Phe Leu Glu Gly Phe Leu Asp Ala Leu Leu Cys
325 330 335Gly Asn Ser Ser Asp Ala Gly Gln Cys Pro Glu Gly Tyr Met
Cys Val 340 345 350Lys Ala Gly Arg Asn Pro Asn Tyr Gly Tyr Thr Ser
Phe Asp Thr Phe 355 360 365Ser Trp Ala Phe Leu Ser Leu Phe Arg Leu
Met Thr Gln Asp Phe Trp 370 375 380Glu Asn Leu Tyr Gln Leu Thr Leu
Arg Ala Ala Gly Lys Thr Tyr Met385 390 395 400Ile Phe Phe Val Leu
Val Ile Phe Leu Gly Ser Phe Tyr Leu Ile Asn 405 410 415Leu Ile Leu
Ala Val Val Ala Met Ala Tyr Glu Glu Gln Asn Gln Ala 420 425 430Thr
Leu Glu Glu Ala Glu Gln Lys Glu Ala Glu Phe Gln Gln Met Ile 435 440
445Glu Gln Leu Lys Lys Gln Gln Glu Ala Ala Gln Gln Ala Ala Thr Ala
450 455 460Thr Ala Ser Glu His Ser Arg Glu Pro Ser Ala Ala Gly Arg
Leu Ser465 470 475 480Asp Ser Ser Ser Glu Ala Ser Lys Leu Ser Ser
Lys Ser Ala Lys Glu 485 490 495Arg Arg Asn Arg Arg Lys Lys Arg Lys
Gln Lys Glu Gln Ser Gly Gly 500 505 510Glu Glu Lys Asp Glu Asp Glu
Phe Gln Lys Ser Glu Ser Glu Asp Ser 515 520 525Ile Arg Arg Lys Gly
Phe Arg Phe Ser Ile Glu Gly Asn Arg Leu Thr 530 535 540Tyr Glu Lys
Arg Tyr Ser Ser Pro His Gln Ser Leu Leu Ser Ile Arg545 550 555
560Gly Ser Leu Phe Ser Pro Arg Arg Asn Ser Arg Thr Ser Leu Phe Ser
565 570 575Phe Arg Gly Arg Ala Lys Asp Val Gly Ser Glu Asn Asp Phe
Ala Asp 580 585 590Asp Glu His Ser Thr Phe Glu Asp Asn Glu Ser Arg
Arg Asp Ser Leu 595 600 605Phe Val Pro Arg Arg His Gly Glu Arg Arg
Asn Ser Asn Leu Ser Gln 610 615 620Thr Ser Arg Ser Ser Arg Met Leu
Ala Val Phe Pro Ala Asn Gly Lys625 630 635 640Met His Ser Thr Val
Asp Cys Asn Gly Val Val Ser Leu Val Gly Gly 645 650 655Pro Ser Val
Pro Thr Ser Pro Val Gly Gln Leu Leu Pro Glu Val Ile 660 665 670Ile
Asp Lys Pro Ala Thr Asp Asp Asn Gly Thr Thr Thr Glu Thr Glu 675 680
685Met Arg Lys Arg Arg Ser Ser Ser Phe His Val Ser Met Asp Phe Leu
690 695 700Glu Asp Pro Ser Gln Arg Gln Arg Ala Met Ser Ile Ala Ser
Ile Leu705 710 715 720Thr Asn Thr Val Glu Glu Leu Glu Glu Ser Arg
Gln Lys Cys Pro Pro 725 730 735Cys Trp Tyr Lys Phe Ser Asn Ile Phe
Leu Ile Trp Asp Cys Ser Pro 740 745 750Tyr Trp Leu Lys Val Lys His
Val Val Asn Leu Val Val Met Asp Pro 755 760 765Phe Val Asp Leu Ala
Ile Thr Ile Cys Ile Val Leu Asn Thr Leu Phe 770 775 780Met Ala Met
Glu His Tyr Pro Met Thr Asp His Phe Asn Asn Val Leu785 790 795
800Thr Val Gly Asn Leu Val Phe Thr Gly Ile Phe Thr Ala Glu Met Phe
805 810 815Leu Lys Ile Ile Ala Met Asp Pro Tyr Tyr Tyr Phe Gln Glu
Gly Trp 820 825 830Asn Ile Phe Asp Gly Phe Ile Val Thr Leu Ser Leu
Val Glu Leu Gly 835 840 845Leu Ala Asn Val Glu Gly Leu Ser Val Leu
Arg Ser Phe Arg Leu Leu 850 855 860Arg Val Phe Lys Leu Ala Lys Ser
Trp Pro Thr Leu Asn Met Leu Ile865 870 875 880Lys Ile Ile Gly Asn
Ser Val Gly Ala Leu Gly Asn Leu Thr Leu Val 885 890 895Leu Ala Ile
Ile Val Phe Ile Phe Ala Val Val Gly Met Gln Leu Phe 900 905 910Gly
Lys Ser Tyr Lys Asp Cys Val Cys Lys Ile Ala Ser Asp Cys Gln 915 920
925Leu Pro Arg Trp His Met Asn Asp Phe Phe His Ser Phe Leu Ile Val
930 935 940Phe Arg Val Leu Cys Gly Glu Trp Ile Glu Thr Met Trp Asp
Cys Met945 950 955 960Glu Val Ala Gly Gln Ala Met Cys Leu Thr Val
Phe Met Met Val Met 965 970 975Val Ile Gly Asn Leu Val Val Leu Asn
Leu Phe Leu Ala Leu Leu Leu 980 985 990Ser Ser Phe Ser Ala Asp Asn
Leu Ala Ala Thr Asp Asp Asp Asn Glu 995 1000 1005Met Asn Asn Leu
Gln Ile Ala Val Asp Arg Met His Lys Gly Val 1010 1015 1020Ala Tyr
Val Lys Arg Lys Ile Tyr Glu Phe Ile Gln Gln Ser Phe 1025 1030
1035Ile Arg Lys Gln Lys Ile Leu Asp Glu Ile Lys Pro Leu Asp Asp
1040 1045 1050Leu Asn Asn Lys Lys Asp Ser Cys Met Ser Asn His Thr
Ala Glu 1055 1060 1065Ile Gly Lys Asp Leu Asp Tyr Leu Lys Asp Val
Asn Gly Thr Thr 1070 1075 1080Ser Gly Ile Gly Thr Gly Ser Ser Val
Glu Lys Tyr Ile Ile Asp 1085 1090 1095Glu Ser Asp Tyr Met Ser Phe
Ile Asn Asn Pro Ser Leu Thr Val 1100 1105 1110Thr Val Pro Ile Ala
Val Gly Glu Ser Asp Phe Glu Asn Leu Asn 1115 1120 1125Thr Glu Asp
Phe Ser Ser Glu Ser Asp Leu Glu Glu Ser Lys Glu 1130 1135 1140Lys
Leu Asn Glu Ser Ser Ser Ser Ser Glu Gly Ser Thr Val Asp 1145 1150
1155Ile Gly Ala Pro Val Glu Glu Gln Pro Val Val Glu Pro Glu Glu
1160 1165 1170Thr Leu Glu Pro Glu Ala Cys Phe Thr Glu Gly Cys Val
Gln Arg 1175 1180 1185Phe Lys Cys Cys Gln Ile Asn Val Glu Glu Gly
Arg Gly Lys Gln 1190 1195 1200Trp Trp Asn Leu Arg Arg Thr Cys Phe
Arg Ile Val Glu His Asn 1205 1210 1215Trp Phe Glu Thr Phe Ile Val
Phe Met Ile Leu Leu Ser Ser Gly 1220 1225 1230Ala Leu Ala Phe Glu
Asp Ile Tyr Ile Asp Gln Arg Lys Thr Ile 1235 1240 1245Lys Thr Met
Leu Glu Tyr Ala Asp Lys Val Phe Thr Tyr Ile Phe 1250 1255 1260Ile
Leu Glu Met Leu Leu Lys Trp Val Ala Tyr Gly Tyr Gln Thr 1265 1270
1275Tyr Phe Thr Asn Ala Trp Cys Trp Leu Asp Phe Leu Ile Val Asp
1280 1285 1290Val Ser Leu Val Ser Leu Thr Ala Asn Ala Leu Gly Tyr
Ser Glu 1295 1300 1305Leu Gly Ala Ile Lys Ser Leu Arg Thr Leu Arg
Ala Leu Arg Pro 1310 1315 1320Leu Arg Ala Leu Ser Arg Phe Glu Gly
Met Arg Val Val Val Asn 1325 1330 1335Ala Leu Leu Gly Ala Ile Pro
Ser Ile Met Asn Val Leu Leu Val 1340 1345 1350Cys Leu Ile Phe Trp
Leu Ile Phe Ser Ile Met Gly Val Asn Leu 1355 1360 1365Phe Ala Gly
Lys Phe Tyr His Cys Ile Asn Thr Thr Thr Gly Asp 1370 1375 1380Arg
Phe Asp Ile Glu Asp Val Asn Asn His Thr Asp Cys Leu Lys 1385 1390
1395Leu Ile Glu Arg Asn Glu Thr Ala Arg Trp Lys Asn Val Lys Val
1400 1405 1410Asn Phe Asp Asn Val Gly Phe Gly Tyr Leu Ser Leu Leu
Gln Val 1415 1420 1425Ala Thr Phe Lys Gly Trp Met Asp Ile Met Tyr
Ala Ala Val Asp 1430 1435 1440Ser Arg Asn Val Glu Leu Gln Pro Lys
Tyr Glu Glu Ser Leu Tyr 1445 1450 1455Met Tyr Leu Tyr Phe Val Ile
Phe Ile Ile Phe Gly Ser Phe Phe 1460 1465 1470Thr Leu Asn Leu Phe
Ile Gly Val Ile Ile Asp Asn Phe Asn Gln 1475 1480 1485Gln Lys Lys
Lys Phe Gly Gly Gln Asp Ile Phe Met Thr Glu Glu 1490 1495 1500Gln
Lys Lys Tyr Tyr Asn Ala Met Lys Lys Leu Gly Ser Lys Lys 1505 1510
1515Pro Gln Lys Pro Ile Pro Arg Pro Gly Asn Lys Phe Gln Gly Met
1520 1525 1530Val Phe Asp Phe Val Thr Arg Gln Val Phe Asp Ile Ser
Ile Met 1535 1540 1545Ile Leu Ile Cys Leu Asn Met Val Thr Met Met
Val Glu Thr Asp 1550 1555 1560Asp Gln Ser Glu Tyr Val Thr Thr Ile
Leu Ser Arg Ile Asn Leu 1565 1570 1575Val Phe Ile Val Leu Phe Thr
Gly Glu Cys Val Leu Lys Leu Ile 1580 1585 1590Ser Leu Arg His Tyr
Tyr Phe Thr Ile Gly Trp Asn Ile Phe Asp 1595 1600 1605Phe Val Val
Val Ile Leu Ser Ile Val Gly Met Phe Leu Ala Glu 1610 1615 1620Leu
Ile Glu Lys Tyr Phe Val Ser Pro Thr Leu Phe Arg Val Ile 1625 1630
1635Arg Leu Ala Arg Ile Gly Arg Ile Leu Arg Leu Ile Lys Gly Ala
1640 1645 1650Lys Gly Ile Arg Thr Leu Leu Phe Ala Leu Met Met Ser
Leu Pro 1655 1660 1665Ala Leu Phe Asn Ile Gly Leu Leu Leu Phe Leu
Val Met Phe Ile 1670 1675 1680Tyr Ala Ile Phe Gly Met Ser Asn Phe
Ala Tyr Val Lys Arg Glu 1685 1690 1695Val Gly Ile Asp Asp Met Phe
Asn Phe Glu Thr Phe Gly Asn Ser 1700 1705 1710Met Ile Cys Leu Phe
Gln Ile Thr Thr Ser Ala Gly Trp Asp Gly 1715 1720 1725Leu Leu Ala
Pro Ile Leu Asn Ser Lys Pro Pro Asp Cys Asp Pro 1730 1735 1740Asn
Lys Val Asn Pro Gly Ser Ser Val Lys Gly Asp Cys Gly Asn 1745 1750
1755Pro Ser Val Gly Ile Phe Phe Phe Val Ser Tyr Ile Ile Ile Ser
1760 1765 1770Phe Leu Val Val Val Asn Met Tyr Ile Ala Val Ile Leu
Glu Asn 1775 1780 1785Phe Ser Val Ala Thr Glu Glu Ser Ala Glu Pro
Leu Ser Glu Asp 1790 1795 1800Asp Phe Glu Met Phe Tyr Glu Val Trp
Glu Lys Phe Asp Pro Asp 1805 1810 1815Ala Thr Gln Phe Met Glu Phe
Glu Lys Leu Ser Gln Phe Ala Ala 1820 1825 1830Ala Leu Glu Pro Pro
Leu Asn Leu Pro Gln Pro Asn Lys Leu Gln 1835 1840 1845Leu Ile Ala
Met Asp Leu Pro Met Val Ser Gly Asp Arg Ile His 1850 1855 1860Cys
Leu Asp Ile Leu Phe Ala Phe Thr Lys Arg Val Leu Gly Glu 1865 1870
1875Ser Gly Glu Met Asp Ala Leu Arg Ile Gln Met Glu Glu Arg Phe
1880 1885 1890Met Ala Ser Asn Pro Ser Lys Val Ser Tyr Gln Pro Ile
Thr Thr 1895 1900 1905Thr Leu Lys Arg Lys Gln Glu Glu Val Ser Ala
Val Ile Ile Gln 1910 1915 1920Arg Ala Tyr Arg Arg His Leu Leu Lys
Arg Thr Val Lys Gln Ala 1925 1930 1935Ser Phe Thr Tyr Asn Lys Asn
Lys Ile Lys Gly Gly Ala Asn Leu 1940 1945 1950Leu Ile Lys Glu Asp
Met Ile Ile Asp Arg Ile Asn Glu Asn Ser 1955 1960 1965Ile Thr Glu
Lys Thr Asp Leu Thr Met Ser Thr Ala Ala Cys Pro 1970 1975 1980Pro
Ser Tyr Asp Arg Val Thr Lys Pro Ile Val Glu Lys His Glu 1985 1990
1995Gln Glu Gly Lys Asp Glu Lys Ala Lys Gly Lys 2000
200540603PRTUnknownDescription of Unknown STXBP1 sequence 40Met Ala
Pro Ile Gly Leu Lys Ala Val Val Gly Glu Lys Ile Met His1 5 10 15Asp
Val Ile Lys Lys Val Lys Lys Lys Gly Glu Trp Lys Val Leu Val 20 25
30Val Asp Gln Leu Ser Met Arg Met Leu Ser Ser Cys Cys Lys Met Thr
35 40 45Asp Ile Met Thr Glu Gly Ile Thr Ile Val Glu Asp Ile Asn Lys
Arg 50 55 60Arg Glu Pro Leu Pro Ser Leu Glu Ala Val Tyr Leu Ile Thr
Pro Ser65 70 75 80Glu Lys Ser Val His Ser Leu Ile Ser Asp Phe Lys
Asp Pro Pro Thr 85 90 95Ala Lys Tyr Arg Ala Ala His Val Phe Phe Thr
Asp Ser Cys Pro Asp 100 105 110Ala Leu Phe Asn Glu Leu Val Lys Ser
Arg Ala Ala Lys Val Ile Lys 115 120 125Thr Leu Thr Glu Ile Asn Ile
Ala Phe Leu Pro Tyr Glu Ser Gln Val 130 135 140Tyr Ser Leu Asp Ser
Ala Asp Ser Phe Gln Ser Phe Tyr Ser Pro His145 150 155 160Lys Ala
Gln Met Lys Asn Pro Ile Leu Glu Arg Leu Ala Glu Gln Ile 165 170
175Ala Thr Leu Cys Ala Thr Leu Lys Glu Tyr Pro Ala Val Arg Tyr Arg
180 185 190Gly Glu Tyr Lys Asp Asn Ala Leu Leu Ala Gln Leu Ile Gln
Asp Lys 195 200 205Leu Asp Ala Tyr Lys Ala Asp Asp Pro Thr Met Gly
Glu Gly Pro Asp 210 215 220Lys Ala Arg Ser Gln Leu Leu Ile Leu Asp
Arg Gly Phe Asp Pro Ser225 230 235 240Ser Pro Val Leu His Glu Leu
Thr Phe Gln Ala Met Ser Tyr Asp Leu 245 250 255Leu Pro Ile Glu Asn
Asp Val Tyr Lys Tyr Glu Thr Ser Gly Ile Gly 260 265 270Glu Ala Arg
Val Lys Glu Val Leu Leu Asp Glu Asp Asp Asp Leu Trp 275 280 285Ile
Ala Leu Arg His Lys His Ile Ala Glu Val Ser Gln Glu Val Thr 290 295
300Arg Ser Leu Lys Asp Phe Ser Ser Ser Lys Arg Met Asn Thr Gly
Glu305 310 315 320Lys Thr Thr Met Arg Asp Leu Ser Gln Met Leu Lys
Lys Met Pro Gln 325 330 335Tyr Gln Lys Glu Leu Ser Lys Tyr Ser Thr
His Leu His Leu Ala Glu 340 345 350Asp Cys Met Lys His Tyr Gln Gly
Thr Val Asp Lys Leu Cys Arg Val 355 360 365Glu Gln Asp Leu Ala Met
Gly Thr Asp Ala Glu Gly Glu Lys Ile Lys 370 375 380Asp Pro Met Arg
Ala Ile Val Pro Ile Leu Leu Asp Ala Asn Val Ser385 390 395 400Thr
Tyr Asp Lys Ile Arg Ile Ile Leu Leu Tyr Ile Phe Leu Lys Asn 405 410
415Gly Ile Thr Glu Glu Asn Leu Asn Lys Leu Ile Gln His Ala Gln Ile
420 425 430Pro Pro Glu Asp Ser Glu Ile Ile Thr Asn Met Ala His Leu
Gly Val 435 440 445Pro Ile Val Thr Asp Ser Thr Leu Arg Arg Arg Ser
Lys Pro Glu Arg 450 455 460Lys Glu Arg Ile Ser Glu Gln Thr Tyr Gln
Leu Ser Arg Trp Thr Pro465 470 475 480Ile Ile Lys Asp Ile Met Glu
Asp Thr Ile Glu Asp Lys Leu Asp Thr 485 490 495Lys His Tyr Pro Tyr
Ile Ser Thr Arg Ser Ser Ala Ser Phe Ser Thr 500 505 510Thr Ala Val
Ser Ala Arg Tyr Gly His Trp His Lys Asn Lys Ala Pro 515 520 525Gly
Glu Tyr Arg Ser Gly Pro Arg Leu Ile Ile Phe Ile Leu Gly Gly 530 535
540Val Ser Leu Asn Glu Met Arg Cys Ala Tyr Glu Val Thr Gln Ala
Asn545 550 555 560Gly Lys Trp Glu Val Leu Ile Gly Ser Thr His Ile
Leu Thr Pro Thr 565 570 575Lys Phe Leu Met Asp Leu Arg His Pro Asp
Phe Arg Glu Ser Ser Arg 580 585 590Val Ser Phe Glu Asp Gln Ala Pro
Thr Met Glu 595 60041511PRTUnknownDescription of Unknown Kv3.1
sequence 41Met Gly Gln Gly Asp Glu Ser Glu Arg Ile Val Ile Asn Val
Gly Gly1 5 10 15Thr Arg His Gln Thr Tyr Arg Ser Thr Leu Arg Thr Leu
Pro Gly Thr 20 25 30Arg Leu Ala Trp Leu Ala Glu Pro Asp Ala His Ser
His Phe Asp Tyr 35 40 45Asp Pro Arg Ala Asp Glu Phe Phe Phe Asp Arg
His Pro Gly Val Phe 50 55 60Ala His Ile Leu Asn Tyr Tyr Arg Thr Gly
Lys Leu His Cys Pro Ala65 70 75 80Asp Val Cys Gly Pro Leu Tyr Glu
Glu Glu Leu Ala Phe Trp Gly Ile 85 90 95Asp Glu Thr Asp Val Glu Pro
Cys Cys Trp Met Thr Tyr Arg Gln His 100 105 110Arg Asp Ala Glu Glu
Ala Leu Asp Ser Phe Gly Gly Ala Pro Leu Asp 115 120 125Asn Ser Ala
Asp Asp Ala Asp Ala Asp Gly Pro Gly Asp Ser Gly Asp 130 135 140Gly
Glu Asp Glu Leu Glu Met Thr Lys Arg Leu Ala
Leu Ser Asp Ser145 150 155 160Pro Asp Gly Arg Pro Gly Gly Phe Trp
Arg Arg Trp Gln Pro Arg Ile 165 170 175Trp Ala Leu Phe Glu Asp Pro
Tyr Ser Ser Arg Tyr Ala Arg Tyr Val 180 185 190Ala Phe Ala Ser Leu
Phe Phe Ile Leu Val Ser Ile Thr Thr Phe Cys 195 200 205Leu Glu Thr
His Glu Arg Phe Asn Pro Ile Val Asn Lys Thr Glu Ile 210 215 220Glu
Asn Val Arg Asn Gly Thr Gln Val Arg Tyr Tyr Arg Glu Ala Glu225 230
235 240Thr Glu Ala Phe Leu Thr Tyr Ile Glu Gly Val Cys Val Val Trp
Phe 245 250 255Thr Phe Glu Phe Leu Met Arg Val Ile Phe Cys Pro Asn
Lys Val Glu 260 265 270Phe Ile Lys Asn Ser Leu Asn Ile Ile Asp Phe
Val Ala Ile Leu Pro 275 280 285Phe Tyr Leu Glu Val Gly Leu Ser Gly
Leu Ser Ser Lys Ala Ala Lys 290 295 300Asp Val Leu Gly Phe Leu Arg
Val Val Arg Phe Val Arg Ile Leu Arg305 310 315 320Ile Phe Lys Leu
Thr Arg His Phe Val Gly Leu Arg Val Leu Gly His 325 330 335Thr Leu
Arg Ala Ser Thr Asn Glu Phe Leu Leu Leu Ile Ile Phe Leu 340 345
350Ala Leu Gly Val Leu Ile Phe Ala Thr Met Ile Tyr Tyr Ala Glu Arg
355 360 365Ile Gly Ala Gln Pro Asn Asp Pro Ser Ala Ser Glu His Thr
His Phe 370 375 380Lys Asn Ile Pro Ile Gly Phe Trp Trp Ala Val Val
Thr Met Thr Thr385 390 395 400Leu Gly Tyr Gly Asp Met Tyr Pro Gln
Thr Trp Ser Gly Met Leu Val 405 410 415Gly Ala Leu Cys Ala Leu Ala
Gly Val Leu Thr Ile Ala Met Pro Val 420 425 430Pro Val Ile Val Asn
Asn Phe Gly Met Tyr Tyr Ser Leu Ala Met Ala 435 440 445Lys Gln Lys
Leu Pro Lys Lys Lys Lys Lys His Ile Pro Arg Pro Pro 450 455 460Gln
Leu Gly Ser Pro Asn Tyr Cys Lys Ser Val Val Asn Ser Pro His465 470
475 480His Ser Thr Gln Ser Asp Thr Cys Pro Leu Ala Gln Glu Glu Ile
Leu 485 490 495Glu Ile Asn Arg Ala Gly Arg Lys Pro Leu Arg Gly Met
Ser Ile 500 505 51042639PRTUnknownDescription of Unknown Kv3.2
sequence 42Met Gly Lys Ile Glu Ser Asn Glu Arg Val Ile Leu Asn Val
Gly Gly1 5 10 15Thr Arg His Glu Thr Tyr Arg Ser Thr Leu Lys Thr Leu
Pro Gly Thr 20 25 30Arg Leu Ala Leu Leu Ala Ser Ser Glu Pro Gln Gly
Asp Cys Leu Thr 35 40 45Ala Ala Gly Asp Lys Leu Gln Pro Leu Pro Pro
Pro Leu Ser Pro Pro 50 55 60Pro Arg Pro Pro Pro Leu Ser Pro Val Pro
Ser Gly Cys Phe Glu Gly65 70 75 80Gly Ala Gly Asn Cys Ser Ser His
Gly Gly Asn Gly Gly Asn Gly Gly 85 90 95Ser Asp His Pro Gly Gly Gly
Arg Glu Phe Phe Phe Asp Arg His Pro 100 105 110Gly Val Phe Ala Tyr
Val Leu Asn Tyr Tyr Arg Thr Gly Lys Leu His 115 120 125Cys Pro Ala
Asp Val Cys Gly Pro Leu Phe Glu Glu Glu Leu Ala Phe 130 135 140Trp
Gly Ile Asp Glu Thr Asp Val Glu Pro Cys Cys Trp Met Thr Tyr145 150
155 160Arg Gln His Arg Asp Ala Glu Glu Ala Leu Asp Ile Phe Glu Thr
Pro 165 170 175Asp Leu Ile Gly Gly Asp Pro Gly Asp Asp Glu Asp Leu
Ala Ala Lys 180 185 190Arg Leu Gly Ile Glu Asp Ala Ala Gly Leu Gly
Gly Pro Asp Gly Lys 195 200 205Ser Gly Arg Trp Arg Lys Leu Gln Pro
Arg Met Trp Ala Leu Phe Glu 210 215 220Asp Pro Tyr Ser Ser Arg Ala
Ala Arg Phe Ile Ala Phe Ala Ser Leu225 230 235 240Phe Phe Ile Leu
Val Ser Ile Thr Thr Phe Cys Leu Glu Thr His Glu 245 250 255Ala Phe
Asn Ile Val Lys Asn Lys Thr Glu Pro Val Ile Asn Gly Thr 260 265
270Ser Pro Val Leu Gln Tyr Glu Ile Glu Thr Asp Pro Ala Leu Thr Tyr
275 280 285Val Glu Gly Val Cys Val Val Trp Phe Thr Phe Glu Phe Leu
Val Arg 290 295 300Ile Val Phe Ser Pro Asn Lys Leu Glu Phe Ile Lys
Asn Leu Leu Asn305 310 315 320Ile Ile Asp Phe Val Ala Ile Leu Pro
Phe Tyr Leu Glu Val Gly Leu 325 330 335Ser Gly Leu Ser Ser Lys Ala
Ala Lys Asp Val Leu Gly Phe Leu Arg 340 345 350Val Val Arg Phe Val
Arg Ile Leu Arg Ile Phe Lys Leu Thr Arg His 355 360 365Phe Val Gly
Leu Arg Val Leu Gly His Thr Leu Arg Ala Ser Thr Asn 370 375 380Glu
Phe Leu Leu Leu Ile Ile Phe Leu Ala Leu Gly Val Leu Ile Phe385 390
395 400Ala Thr Met Ile Tyr Tyr Ala Glu Arg Val Gly Ala Gln Pro Asn
Asp 405 410 415Pro Ser Ala Ser Glu His Thr Gln Phe Lys Asn Ile Pro
Ile Gly Phe 420 425 430Trp Trp Ala Val Val Thr Met Thr Thr Leu Gly
Tyr Gly Asp Met Tyr 435 440 445Pro Gln Thr Trp Ser Gly Met Leu Val
Gly Ala Leu Cys Ala Leu Ala 450 455 460Gly Val Leu Thr Ile Ala Met
Pro Val Pro Val Ile Val Asn Asn Phe465 470 475 480Gly Met Tyr Tyr
Ser Leu Ala Met Ala Lys Gln Lys Leu Pro Arg Lys 485 490 495Arg Lys
Lys His Ile Pro Pro Ala Pro Leu Ala Ser Ser Pro Thr Phe 500 505
510Cys Lys Thr Glu Leu Asn Met Ala Cys Asn Ser Thr Gln Ser Asp Thr
515 520 525Cys Leu Gly Lys Glu Asn Arg Leu Leu Glu His Asn Arg Ser
Val Leu 530 535 540Ser Gly Asp Asp Ser Thr Gly Ser Glu Pro Pro Leu
Ser Pro Pro Glu545 550 555 560Arg Leu Pro Ile Arg Arg Ser Ser Thr
Arg Asp Lys Asn Arg Arg Gly 565 570 575Glu Thr Cys Phe Leu Leu Thr
Thr Gly Asp Tyr Thr Cys Ala Ser Asp 580 585 590Gly Gly Ile Arg Lys
Ala Ser Thr Leu Glu Pro Met Glu Ser Thr Ala 595 600 605Gln Thr Lys
Gly Asp Thr Arg Pro Glu Ala His Trp Asn Cys Ala His 610 615 620Leu
Leu Asn Phe Gly Cys Pro Thr Gly Ser Ser Phe Pro Thr Leu625 630
63543757PRTUnknownDescription of Unknown Kv3.3 sequence 43Met Leu
Ser Ser Val Cys Val Ser Ser Phe Arg Gly Arg Gln Gly Ala1 5 10 15Ser
Lys Gln Gln Pro Ala Pro Pro Pro Gln Pro Pro Glu Ser Pro Pro 20 25
30Pro Pro Pro Leu Pro Pro Gln Gln Gln Gln Pro Ala Gln Pro Gly Pro
35 40 45Ala Ala Ser Pro Ala Gly Pro Pro Ala Pro Arg Gly Pro Gly Asp
Arg 50 55 60Arg Ala Glu Pro Cys Pro Gly Leu Pro Ala Ala Ala Met Gly
Arg His65 70 75 80Gly Gly Gly Gly Gly Asp Ser Gly Lys Ile Val Ile
Asn Val Gly Gly 85 90 95Val Arg His Glu Thr Tyr Arg Ser Thr Leu Arg
Thr Leu Pro Gly Thr 100 105 110Arg Leu Ala Gly Leu Thr Glu Pro Glu
Ala Ala Ala Arg Phe Asp Tyr 115 120 125Asp Pro Gly Ala Asp Glu Phe
Phe Phe Asp Arg His Pro Gly Val Phe 130 135 140Ala Tyr Val Leu Asn
Tyr Tyr Arg Thr Gly Lys Leu His Cys Pro Ala145 150 155 160Asp Val
Cys Gly Pro Leu Phe Glu Glu Glu Leu Gly Phe Trp Gly Ile 165 170
175Asp Glu Thr Asp Val Glu Ala Cys Cys Trp Met Thr Tyr Arg Gln His
180 185 190Arg Asp Ala Glu Glu Ala Leu Asp Ser Phe Glu Ala Pro Asp
Pro Ala 195 200 205Gly Ala Ala Asn Ala Ala Asn Ala Ala Gly Ala His
Asp Gly Gly Leu 210 215 220Asp Asp Glu Ala Gly Ala Gly Gly Gly Gly
Leu Asp Gly Ala Gly Gly225 230 235 240Glu Leu Lys Arg Leu Cys Phe
Gln Asp Ala Gly Gly Gly Ala Gly Gly 245 250 255Pro Pro Gly Gly Ala
Gly Gly Ala Gly Gly Thr Trp Trp Arg Arg Trp 260 265 270Gln Pro Arg
Val Trp Ala Leu Phe Glu Asp Pro Tyr Ser Ser Arg Ala 275 280 285Ala
Arg Tyr Val Ala Phe Ala Ser Leu Phe Phe Ile Leu Ile Ser Ile 290 295
300Thr Thr Phe Cys Leu Glu Thr His Glu Gly Phe Ile His Ile Ser
Asn305 310 315 320Lys Thr Val Thr Gln Ala Ser Pro Ile Pro Gly Ala
Pro Pro Glu Asn 325 330 335Ile Thr Asn Val Glu Val Glu Thr Glu Pro
Phe Leu Thr Tyr Val Glu 340 345 350Gly Val Cys Val Val Trp Phe Thr
Phe Glu Phe Leu Met Arg Ile Thr 355 360 365Phe Cys Pro Asp Lys Val
Glu Phe Leu Lys Ser Ser Leu Asn Ile Ile 370 375 380Asp Cys Val Ala
Ile Leu Pro Phe Tyr Leu Glu Val Gly Leu Ser Gly385 390 395 400Leu
Ser Ser Lys Ala Ala Lys Asp Val Leu Gly Phe Leu Arg Val Val 405 410
415Arg Phe Val Arg Ile Leu Arg Ile Phe Lys Leu Thr Arg His Phe Val
420 425 430Gly Leu Arg Val Leu Gly His Thr Leu Arg Ala Ser Thr Asn
Glu Phe 435 440 445Leu Leu Leu Ile Ile Phe Leu Ala Leu Gly Val Leu
Ile Phe Ala Thr 450 455 460Met Ile Tyr Tyr Ala Glu Arg Ile Gly Ala
Asp Pro Asp Asp Ile Leu465 470 475 480Gly Ser Asn His Thr Tyr Phe
Lys Asn Ile Pro Ile Gly Phe Trp Trp 485 490 495Ala Val Val Thr Met
Thr Thr Leu Gly Tyr Gly Asp Met Tyr Pro Lys 500 505 510Thr Trp Ser
Gly Met Leu Val Gly Ala Leu Cys Ala Leu Ala Gly Val 515 520 525Leu
Thr Ile Ala Met Pro Val Pro Val Ile Val Asn Asn Phe Gly Met 530 535
540Tyr Tyr Ser Leu Ala Met Ala Lys Gln Lys Leu Pro Lys Lys Lys
Asn545 550 555 560Lys His Ile Pro Arg Pro Pro Gln Pro Gly Ser Pro
Asn Tyr Cys Lys 565 570 575Pro Asp Pro Pro Pro Pro Pro Pro Pro His
Pro His His Gly Ser Gly 580 585 590Gly Ile Ser Pro Pro Pro Pro Ile
Thr Pro Pro Ser Met Gly Val Thr 595 600 605Val Ala Gly Ala Tyr Pro
Ala Gly Pro His Thr His Pro Gly Leu Leu 610 615 620Arg Gly Gly Ala
Gly Gly Leu Gly Ile Met Gly Leu Pro Pro Leu Pro625 630 635 640Ala
Pro Gly Glu Pro Cys Pro Leu Ala Gln Glu Glu Val Ile Glu Ile 645 650
655Asn Arg Ala Asp Pro Arg Pro Asn Gly Asp Pro Ala Ala Ala Ala Leu
660 665 670Ala His Glu Asp Cys Pro Ala Ile Asp Gln Pro Ala Met Ser
Pro Glu 675 680 685Asp Lys Ser Pro Ile Thr Pro Gly Ser Arg Gly Arg
Tyr Ser Arg Asp 690 695 700Arg Ala Cys Phe Leu Leu Thr Asp Tyr Ala
Pro Ser Pro Asp Gly Ser705 710 715 720Ile Arg Lys Ala Thr Gly Ala
Pro Pro Leu Pro Pro Gln Asp Trp Arg 725 730 735Lys Pro Gly Pro Pro
Ser Phe Leu Pro Asp Leu Asn Ala Asn Ala Ala 740 745 750Ala Trp Ile
Ser Pro 755
References
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.