Expansion Of Tumor Infiltrating Lymphocytes (tils) With Tumor Necrosis Factor Receptor Superfamily (tnfrsf) Agonists And Therape
Lotze; Michael T. ; et al.
U.S. patent application number 16/475924 was filed with the patent office on 2020-04-23 for expansion of tumor infiltrating lymphocytes (tils) with tumor necrosis factor receptor superfamily (tnfrsf) agonists and therape. The applicant listed for this patent is Iovance Biotherapeutics, Inc.. Invention is credited to Michael T. Lotze, Krit Ritthipichai.
| Application Number | 20200121719 16/475924 |
| Document ID | / |
| Family ID | 61094591 |
| Filed Date | 2020-04-23 |

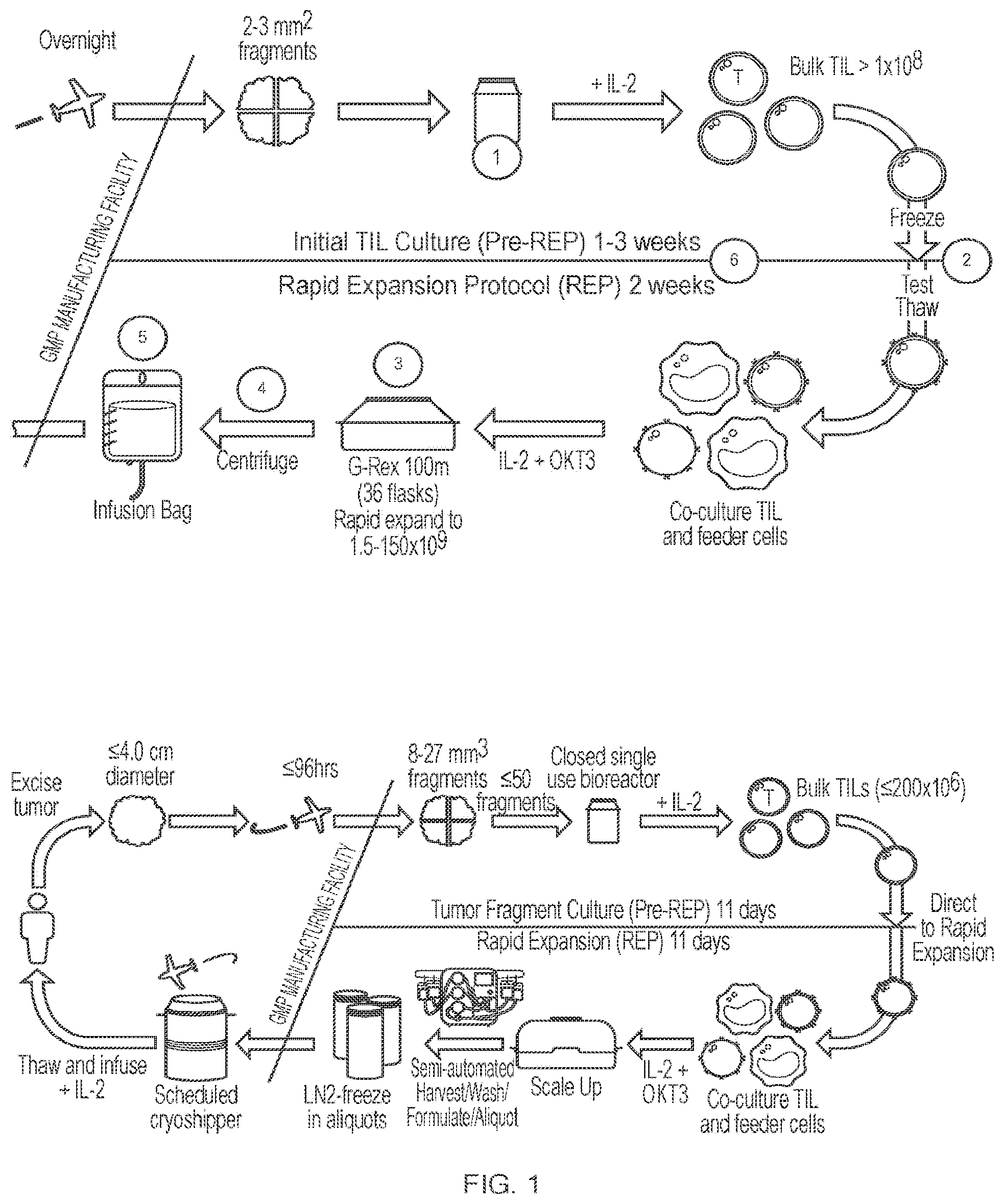



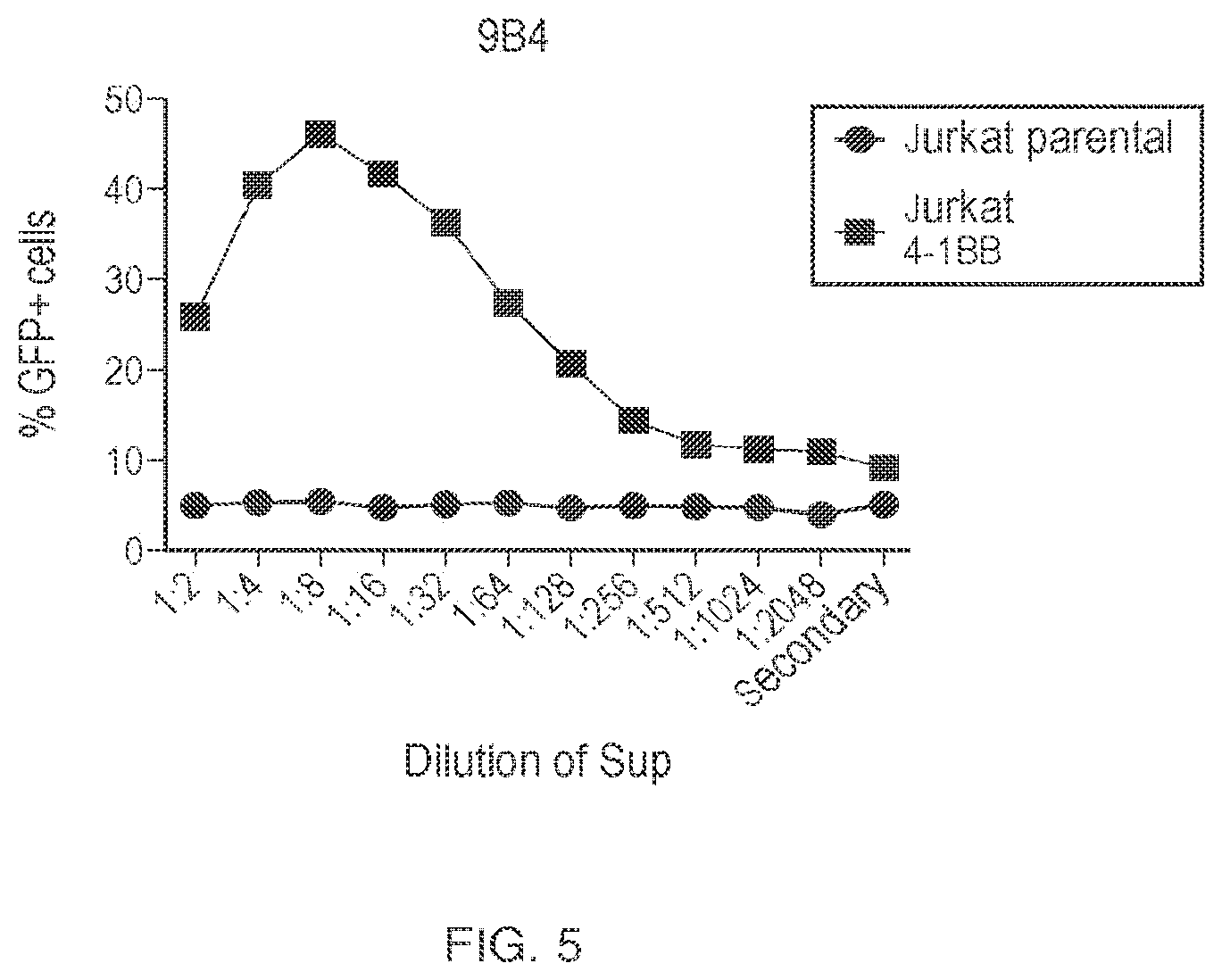
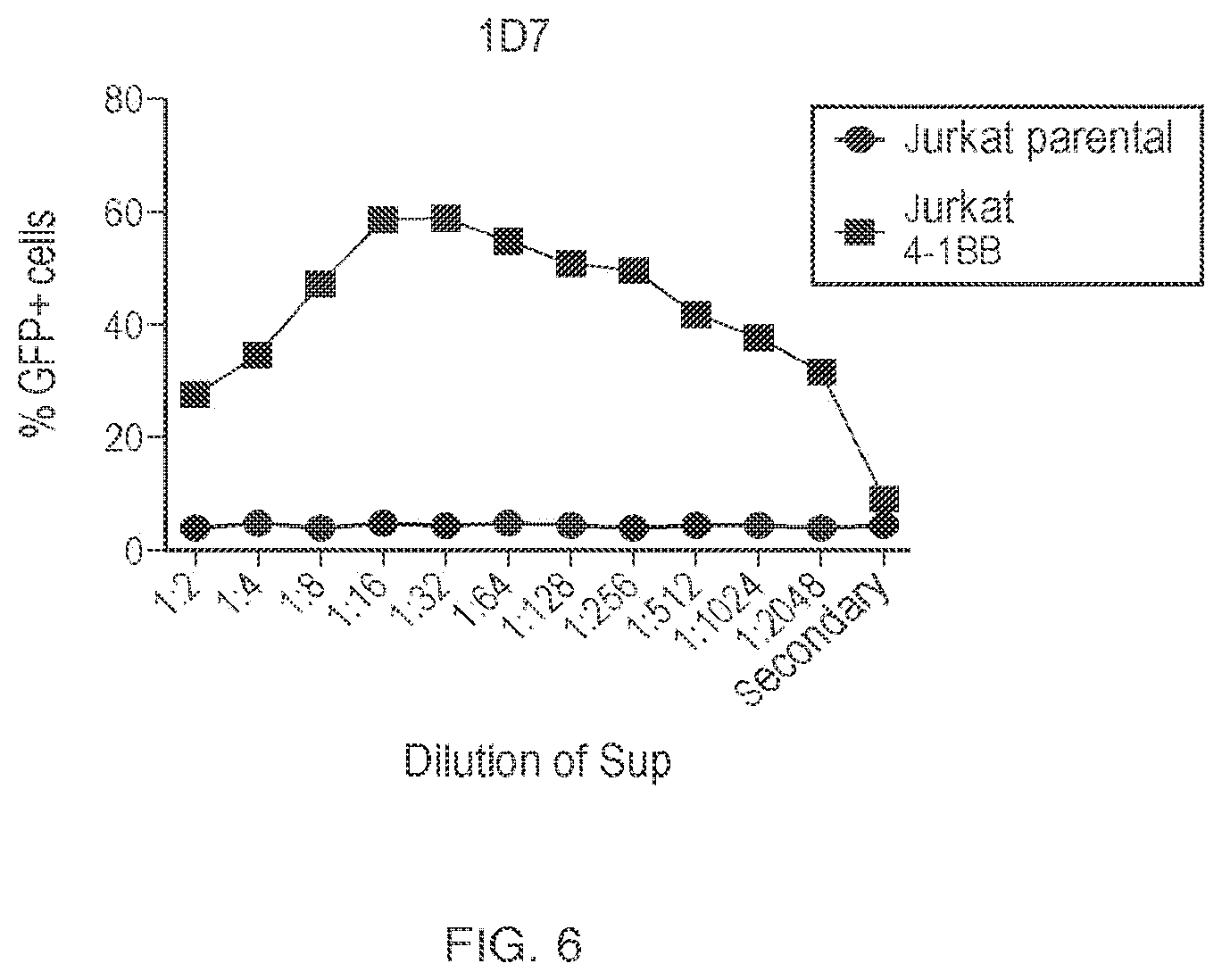
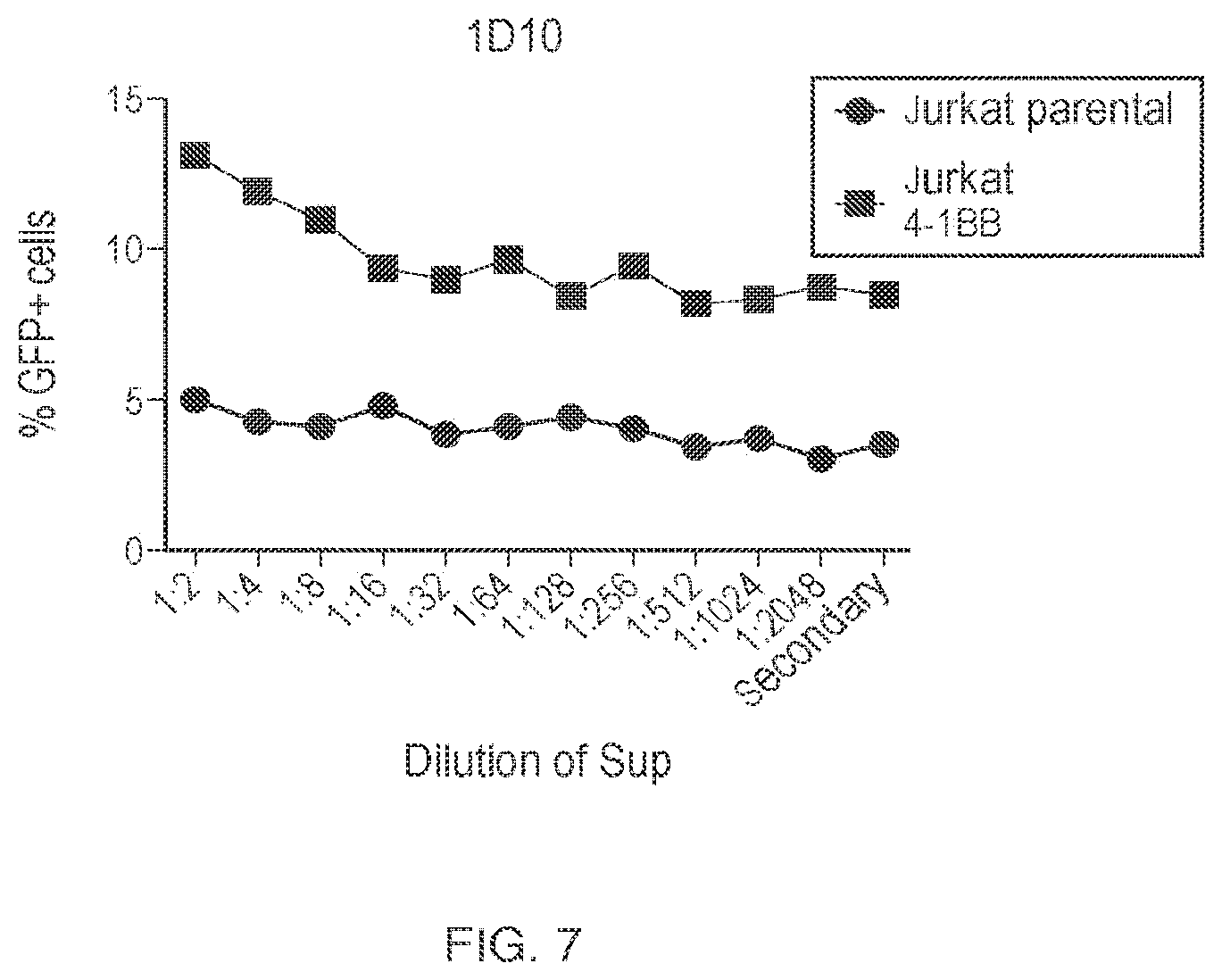


View All Diagrams
| United States Patent Application | 20200121719 |
| Kind Code | A1 |
| Lotze; Michael T. ; et al. | April 23, 2020 |
EXPANSION OF TUMOR INFILTRATING LYMPHOCYTES (TILS) WITH TUMOR NECROSIS FACTOR RECEPTOR SUPERFAMILY (TNFRSF) AGONISTS AND THERAPEUTIC COMBINATIONS OF TILS AND TNFRSF AGONISTS
Abstract
Methods of expanding tumor infiltrating lymphocytes (TILs) using a tumor necrosis factor receptor superfamily (TNFRSF) agonist, such as a 4-IBB agonist, a CD27 agonist, a glucocorticoid-induced TNF receptor-related agonist, an OX40 agonist, a HVEM agonist, or a CD95 agonist, and uses of such expanded TILs in the treatment of diseases such as cancer are disclosed herein. In addition, in some embodiments, therapeutic combinations of TILs and TNFRSF agonists useful in the treatment of diseases such as cancer, including compositions, uses, and dosing regimens thereof, are disclosed herein.
| Inventors: | Lotze; Michael T.; (Pittsburgh, PA) ; Ritthipichai; Krit; (Tampa, FL) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 61094591 | ||||||||||
| Appl. No.: | 16/475924 | ||||||||||
| Filed: | January 5, 2018 | ||||||||||
| PCT Filed: | January 5, 2018 | ||||||||||
| PCT NO: | PCT/US18/12605 | ||||||||||
| 371 Date: | July 3, 2019 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 62443556 | Jan 6, 2017 | |||
| 62460477 | Feb 17, 2017 | |||
| 62532807 | Jul 14, 2017 | |||
| 62567151 | Oct 2, 2017 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C12N 2501/515 20130101; C12N 5/0636 20130101; A61K 45/06 20130101; A61P 35/00 20180101; C07K 16/2875 20130101; C12N 2501/25 20130101; A61K 35/17 20130101; C12N 2501/48 20130101; C12N 2501/599 20130101; C12N 5/0634 20130101; C07K 16/2878 20130101; C12N 2501/2302 20130101 |
| International Class: | A61K 35/17 20060101 A61K035/17; C12N 5/0783 20060101 C12N005/0783; C07K 16/28 20060101 C07K016/28; A61P 35/00 20060101 A61P035/00 |
Claims
1. A method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: (a) resecting a tumor from a patient; (b) obtaining a first population of TILs from the tumor; (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3) antibody, peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist and a second TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; (e) harvesting the third population of TILs; and (f) administering a therapeutically effective portion of the third population of TILs to the patient.
2. The method of claim 1, wherein the TNFRSF agonist is selected from the group consisting of a 4-1BB agonist, an OX40 agonist, a CD27 agonist, a GITR agonist, a HVEM agonist, a CD95 agonist, and combinations thereof.
3. The method of claim 1, wherein the TNFRSF agonist is a 4-1BB agonist, and the 4-1BB agonist is selected from the group consisting of urelumab, utomilumab, EU-101, a fusion protein, and fragments, derivatives, variants, biosimilars, and combinations thereof.
4. The method of claim 1, wherein the TNFRSF agonist is an OX40 agonist, and the OX40 agonist is selected from the group consisting of tavolixizumab, GSK3174998, MEDI6469, MEDI6383, MOXR0916, PF 04518600, Creative Biolabs MOM 18455, an OX40 agonist fusion protein, and fragments, derivatives, variants, biosimilars, and combinations thereof.
5. (canceled)
6. (canceled)
7. (canceled)
8. (canceled)
9. The method of claim 1, wherein the TNFRSF agonist is a CD27 agonist, and the CD27 agonist is selected from the group consisting of varlilumab, a CD27 agonist fusion protein, and fragments, derivatives, variants, or biosimilars thereof.
10. (canceled)
11. (canceled)
12. The method of claim 1, wherein the TNFRSF agonist is a GITR agonist, and the GITR agonist is selected from the group consisting of TRX518, 6C8, 36E5, 3D6, 61G6, 6H6, 61F6, 1D8, 17F10, 35D8, 49A1, 9E5, 31H6, 2155, 698, 706, 827, 1649, 1718, 1D7, 33C9, 33F6, 34G4, 35B10, 41E11, 41G5, 42A11, 44C1, 45A8, 46E11, 48H12, 48H7, 49D9, 49E2, 48A9, 5H7, 7A10, 9H6, and fragments, derivatives, variants, biosimilars, and combinations thereof.
13. (canceled)
14. (canceled)
15. (canceled)
16. (canceled)
17. (canceled)
18. (canceled)
19. (canceled)
20. (canceled)
21. The method of claim 1, further comprising the step of treating the patient with the TNFRSF agonist starting on the day after administration of the third population of TILs to the patient, wherein the TNFRSF agonist is administered intravenously at a dose of between 0.1 mg/kg and 50 mg/kg every four weeks for up to eight cycles.
22. (canceled)
23. The method of claim 1, wherein the TNFRSF agonist is selected from the group consisting of urelumab, utomilumab, EU-101, tavolixizumab, Creative Biolabs MOM-18455, and fragments, derivatives, variants, biosimilars, and combinations thereof.
24. The method of claim 1, wherein the first cell culture medium comprises a second TNFRSF agonist.
25. The method of claim 1, wherein the TNFRSF agonist is a 4-1BB agonist, and the second TNFRSF agonist is an OX40 agonist.
26. The method of claim 1, wherein the TNFRSF agonist is added to the first cell culture medium during the initial expansion at an interval selected from the group consisting of every day, every two days, every three days, every four days, every five days, every six days, every seven days, and every two weeks.
27. The method of claim 1, wherein the TNFRSF agonist is added to the second cell culture medium during the rapid expansion at an interval selected from the group consisting of every day, every two days, every three days, every four days, every five days, every six days, every seven days, and every two weeks.
28. The method of claim 1, wherein the TNFRSF agonist is added at a concentration sufficient to achieve a concentration in the cell culture medium of between 0.1 .mu.g/mL and 100 .mu.g/mL.
29. (canceled)
30. The method of claim 1, wherein IL-2 is present at an initial concentration of about 10 to about 6000 IU/mL in the first cell culture medium.
31. (canceled)
32. (canceled)
33. (canceled)
34. The method of claim 1, wherein IL-2 is present at an initial concentration of about 10 to about 6000 IU/mL in the second cell culture medium.
35. (canceled)
36. (canceled)
37. (canceled)
38. (canceled)
39. (canceled)
40. (canceled)
41. (canceled)
42. (canceled)
43. (canceled)
44. (canceled)
45. (canceled)
46. The method of claim 1, wherein OKT-3 antibody is present at an initial concentration of about 10 ng/mL to about 60 ng/mL in the second cell culture medium.
47. (canceled)
48. The method of claim 1, wherein the initial expansion and the rapid expansion each is performed using a gas permeable container.
49. (canceled)
50. The method of claim 1, further comprising the step of treating the patient with a non-myeloablative lymphodepletion regimen prior to administering the third population of TILs to the patient.
51. (canceled)
52. (canceled)
53. (canceled)
54. (canceled)
55. (canceled)
56. The method of claim 1, wherein the cancer is selected from the group consisting of melanoma, cutaneous melanoma, double-refractory melanoma, uveal (ocular) melanoma, ovarian cancer, platinum-resistant ovarian cancer, cervical cancer, lung cancer, non-small cell lung cancer (NSCLC), bladder cancer, breast cancer, triple negative breast cancer, head and neck cancer, head and neck squamous cell cancer, renal cell carcinoma, acute myeloid leukemia, colorectal cancer, pancreatic ductal adenocarcinoma, glioblastoma, cholangiocarcinoma, osteosarcoma, and sarcoma.
57. (canceled)
58. (canceled)
59. (canceled)
60. (canceled)
61. (canceled)
62. The method of claim 1, wherein the patient is treated with a PD-1 inhibitor or PD-L1 inhibitor after administering the third population of TILs to the patient.
63. The method of claim 62, wherein the PD-1 inhibitor or PD-L1 inhibitor is selected from the group consisting of nivolumab, pembrolizumab, durvalumab, atezolizumab, avelumab, and fragments, derivatives, variants, biosimilars, and combinations thereof.
64. (canceled)
65. (canceled)
66. The method of claim 1, wherein the initial expansion is performed over a period of 11 days or less.
67. The method of claim 1, wherein the rapid expansion is performed over a period of 11 days or less.
68. A process for the preparation of a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: (b) obtaining a first population of TILs; (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; and (e) harvesting the third population of TILs.
69. The process according to claim 68 wherein the first population of TILs is obtained from a tumor which tumor has been resected from a patient.
70. The process according to claim 68, wherein the TNFRSF agonist is selected from the group consisting of a 4-1BB agonist, an OX40 agonist, a CD27 agonist, a GITR agonist, a HVEM agonist, a CD95 agonist, and combinations thereof.
71. (canceled)
72. (canceled)
73. (canceled)
74. (canceled)
75. (canceled)
76. (canceled)
77. (canceled)
78. (canceled)
79. (canceled)
80. (canceled)
81. (canceled)
82. (canceled)
83. (canceled)
84. (canceled)
85. (canceled)
86. (canceled)
87. (canceled)
88. (canceled)
89. (canceled)
90. (canceled)
91. (canceled)
92. The process according to claim 68, wherein the TNFRSF agonist is added to the first cell culture medium during the initial expansion at an interval selected from the group consisting of every day, every two days, every three days, every four days, every five days, every six days, every seven days, and every two weeks.
93. The process according to claim 68, wherein the TNFRSF agonist is added to the second cell culture medium during the rapid expansion at an interval selected from the group consisting of every day, every two days, every three days, every four days, every five days, every six days, every seven days, and every two weeks.
94. The process according to claim 68, wherein the TNFRSF agonist is added at a concentration sufficient to achieve a concentration in the cell culture medium of between 0.1 .mu.g/mL and 100 .mu.g/mL.
95. (canceled)
96. The process according to claim 68, wherein IL-2 is present at an initial concentration of about 10 to about 6000 IU/mL in the first cell culture medium.
97. (canceled)
98. (canceled)
99. (canceled)
100. (canceled)
101. (canceled)
102. (canceled)
103. (canceled)
104. (canceled)
105. (canceled)
106. (canceled)
107. (canceled)
108. (canceled)
109. (canceled)
110. (canceled)
111. (canceled)
112. The process according to claim 68, wherein OKT-3 antibody is present at an initial concentration of about 10 ng/mL to about 60 ng/mL in the second cell culture medium.
113. (canceled)
114. The process according to claim 31, wherein the initial expansion and the rapid expansion each is performed using a gas permeable container.
115. (canceled)
116. A population of tumor infiltrating lymphocytes (TILs) obtainable from a process according to claim 68.
117. A pharmaceutical composition comprising a population of tumor infiltrating lymphocytes (TILs) for use in treating a cancer wherein the population of tumor infiltrating lymphocytes (TILs) is obtainable by a process according to claim 68, wherein optionally the pharmaceutical composition comprises the third population of TILs.
118. (canceled)
119. (canceled)
120. (canceled)
121. The pharmaceutical composition for use in the treatment of a cancer according to claim 117 for use in combination with a non-myeloablative lymphodepletion regimen.
122. The pharmaceutical composition for use in the treatment of a cancer according to claim 117 wherein the pharmaceutical composition is for use in combination with a myeloablative lymphodepletion regimen prior to administering the third population of TILs to the patient.
123. (canceled)
124. (canceled)
125. (canceled)
126. (canceled)
127. (canceled)
128. (canceled)
129. (canceled)
130. (canceled)
131. The pharmaceutical composition for use in the treatment of a cancer according to claim 117 wherein the pharmaceutical composition is for use in combination with a PD-1 inhibitor or PD-L1 inhibitor.
132. The pharmaceutical composition for use in the treatment of a cancer according to claim 117 wherein the pharmaceutical composition is for use in combination with a PD-1 inhibitor or PD-L1 inhibitor, wherein the PD-1 inhibitor or PD-L1 inhibitor is selected from the group consisting of nivolumab, pembrolizumab, durvalumab, atezolizumab, avelumab, and fragments, derivatives, variants, biosimilars, and combinations thereof.
133. The pharmaceutical composition for use in the treatment of a cancer according to claim 117, wherein the PD-1 inhibitor or PD-L1 inhibitor is administered prior to or after resection of the tumor from the patient.
134. (canceled)
135. (canceled)
136. (canceled)
137. The pharmaceutical composition for use in the treatment of a cancer according to claim 117 for use in combination with a PD-1 inhibitor or PD-L1 inhibitor, wherein said PD-1 inhibitor or PD-L1 inhibitor is administered after administering the third population of TILs to the patient.
138. (canceled)
139. The pharmaceutical composition for use in the treatment of a cancer according to claim 117, wherein the cancer is selected from the group consisting of melanoma, cutaneous melanoma, double-refractory melanoma, uveal (ocular melanoma), ovarian cancer, platinum-resistant ovarian cancer, cervical cancer, lung cancer, non-small cell lung cancer (NSCLC), bladder cancer, breast cancer, triple-negative breast cancer, head and neck cancer, head and neck squamous cell cancer, renal cell carcinoma, acute myeloid leukemia, colorectal cancer, pancreatic ductal adenocarcinoma, glioblastoma, cholangiocarcinoma, osteosarcoma, and sarcoma.
140. (canceled)
141. (canceled)
142. (canceled)
143. (canceled)
144. (canceled)
145. (canceled)
146. (canceled)
147. (canceled)
148. (canceled)
149. (canceled)
150. (canceled)
151. (canceled)
152. (canceled)
153. (canceled)
154. (canceled)
155. (canceled)
156. (canceled)
157. (canceled)
158. (canceled)
Description
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] This application is a U.S. National Stage application of International Application No. PCT/US2018/012605, which claims priority to U.S. Provisional Application No. 62/443,556, filed Jan. 6, 2017, U.S. Provisional Application No. 62/460,477, filed Feb. 17, 2017, U.S. Provisional Application No. 62/532,807, filed Jul. 14, 2017, and U.S. Provisional Application No. 62/567,151, filed Oct. 2, 2017, the entirety of which are incorporated herein by reference.
FIELD OF THE INVENTION
[0002] Methods of expanding tumor infiltrating lymphocytes (TILs) using a tumor necrosis factor receptor superfamily (TNFRSF) agonist, such as a 4-1BB agonist, a CD27 agonist, a glucocorticoid-induced TNF receptor-related agonist, an OX40 agonist, a HVEM agonist, or a CD95 agonist, and uses of expanded TILs in the treatment of diseases such as cancer are disclosed herein. In addition, therapeutic combinations of TILs and TNFRSF agonists, including compositions and uses thereof in the treatment of diseases such as cancer are disclosed herein.
BACKGROUND OF THE INVENTION
[0003] Treatment of bulky, refractory cancers using adoptive autologous transfer of tumor infiltrating lymphocytes (TILs) represents a powerful approach to therapy for patients with poor prognoses. Gattinoni, et al., Nat. Rev. Immunol. 2006, 6, 383-393. TILs are dominated by T cells, and IL-2-based TIL expansion followed by a "rapid expansion process" (REP) has become a preferred method for TIL expansion because of its speed and efficiency. Dudley, et al., Science 2002, 298, 850-54; Dudley, et al., J. Clin. Oncol. 2005, 23, 2346-57; Dudley, et al., J. Clin. Oncol. 2008, 26, 5233-39; Riddell, et al., Science 1992, 257, 238-41; Dudley, et al., J. Immunother. 2003, 26, 332-42. A number of approaches to improve clinical responses to TIL therapy in melanoma and to expand TIL therapy to other tumor types have been explored with limited success, and the field remains challenging. Goff, et al., J. Clin. Oncol. 2016, 34, 2389-97; Dudley, et al., J. Clin. Oncol. 2008, 26, 5233-39; Rosenberg, et al., Clin. Cancer Res. 2011, 17, 4550-57. Much focus has been placed on selection of TILs during expansion to either select particular subsets (such as CD8.sup.+ T cells) or to target driver mutations such as a mutated ERBB2IP epitope or driver mutations in the KRAS oncogene. Tran, et al., N. Engl. J. Med. 2016, 375, 2255-62; Tran, et al., Science 2014, 344, 641-45. However, such selection approaches, even if they can be developed to show efficacy in larger clinical trials, add significantly to the duration, complexity, and cost of performing TIL therapy and limit the potential for widespread use of TIL therapy in different types of cancers.
[0004] 4-1BB (also known as CD137 and TNFRSF9), which was first identified as an inducible costimulatory receptor expressed on activated T cells, is a membrane spanning glycoprotein member of the TNFRSF. Watts, Annu. Rev. Immunol. 2005, 23, 23-68. 4-1BB is a type 2 transmembrane glycoprotein that is expressed on activated T lymphocytes, and to a larger extent on CD8.sup.+ than CD4.sup.+ T cells. 4-1BB is also expressed on dendritic cells, follicular dendritic cells, natural killer (NK) cells, granulocytes, cells of blood vessel walls at sites of inflammation, tumor vasculature, and atherosclerotic endothelium. The ligand that stimulates 4-1BB (4-1BBL) is expressed on activated antigen-presenting cells (APCs), myeloid progenitor cells and hematopoietic stem cells. 4-1BB is an activation-induced T-cell costimulatory molecule. Signaling through 4-1BB upregulates survival genes, enhances cell division, induces cytokine production, and prevents activation-induced sell death in T cells. Current understanding of 4-1BB indicates that expression is generally activation dependent and encompasses a broad subset of immune cells including activated NK and NK T cells (NKT cells); regulatory T cells; dendritic cells (DC) including follicular DCs; stimulated mast cells, differentiating myeloid cells, monocytes, neutrophils, eosinophils, and activated B cells. 4-1BB strongly enhances the proliferation and effector function of CD8.sup.+ T cells. Crosslinking of 4-1BB enhances T cell proliferation, IL-2 secretion survival and cytolytic activity. Additionally, anti-4-1BB monoclonal antibodies possess strong antitumor properties, which in turn are the result of their powerful CD8+ T-cell activating, IFN-g producing, and cytolytic marker-inducing capabilities. Vinay and Kwon, Mol. Cancer Therapeutics 2012, 11, 1062-70; Lee, et al., PLoS One, 2013, 8, e69677, 1-11.
[0005] Interaction of 4-1BB on activated normal human B cells with its ligand at the time of B cell receptor engagement stimulates proliferation and enhances survival. The potential impact of 4-1BB engagement in B cell lymphoma has been investigated in at least two published studies. Evaluation of several types of human primary NHL samples indicated that 4-1BB was expressed predominantly on infiltrating T cells rather than the lymphoma cells. Houot, et al., Blood, 2009, 114, 3431-38. The addition of 4-1BB agonists to in vitro cultures of B lymphoma cells with, rituximab and NK cells resulted in increased lymphoma killing. Kohrt, et al., Blood, 2011, 117, 2423-32. In addition, B cell immunophenotyping was performed in two experiments using PF-05082566 in cynomolgus monkeys with doses from 0.001-100 mg/kg; in these experiments peripheral blood B cell numbers were either unchanged or decreased, as described in International Patent Application Publication No. WO 2015/119923.
[0006] 4-1BB is undetectable on the surface of naive T cells but expression increases upon activation. Upon 4-1BB activation, two pro-survival members of the TNFR-associated factor (TRAF) family, TRAF1 and TRAF2, are recruited to the 4-1BB cytoplasmic tail, resulting in downstream activation of NFkB and the Mitogen Activated Protein (MAP) kinase cascade including Erk, Jnk, and p38 MAP kinases. NFkB activation leads to upregulation of Bfl-1 and Bel-XL, pro-survival members of the Bcl-2 family. The pro-apoptotic protein Bim is downregulated in a TRAF 1 and Erk dependent manner. Sabbagh, et al., J. Immunol. 2008, 180, 8093-8101. Reports have shown that 4-1BB agonist monoclonal antibodies (mAbs) increase costimulatory molecule expression and markedly enhance cytolytic T lymphocyte responses, resulting in anti-tumor efficacy in various models. 4-1BB agonist mAbs have demonstrated efficacy in prophylactic and therapeutic settings and both monotherapy and combination therapy tumor models and have established durable anti-tumor protective T cell memory responses. Lynch, et al., Immunol Rev., 2008, 222, 277-286. 4-1BB agonists also inhibit autoimmune reactions in a variety of autoimmunity models. Vinay, et al., J. Mol. Med. 2006, 84, 726-36.
[0007] The OX40 receptor (OX40) (also known as TNFRSF4, CD134, ACT-4, and ACT35) is a member of the TNF receptor family which is expressed on activated CD4.sup.+ T cells (see WO 95/12673). Triggering of this receptor via the OX40 ligand, named OX40L, gp34 or ACT-4-ligand, which is present on activated B-cells and dendritic cells, enhances the proliferation of CD4.sup.+ T cells during an immune response and influences the formation of CD4.sup.+ memory T-cells. Furthermore, the OX40-OX40L system mediates adhesion of activated T cells to endothelial cells, thus directing the activated CD4.sup.+ T cells to the site of inflammation.
[0008] It has been shown that OX40.sup.+ T cells are present within tumor lesions containing tumor infiltrating lymphocytes and in tumor cell positive draining lymph nodes. Weinberg, et al., J. Immunol., 2000, 164, 2160-2169. It was shown in several tumor models in mice that engagement of the OX40 receptor in vivo during tumor priming significantly delayed and prevented the appearance of tumors as compared to control treated mice. Weinberg, et al., J. Immunol., 2000, 164, 2160-2169. Hence, it has been contemplated to enhance the immune response of a mammal to an antigen by engaging the OX40-receptor by administering an OX40-receptor binding agent (International Patent Application Publication No. WO 1999/042585; Weinberg, et al., J. Immunol., 2000, 164, 2160-2169). Preclinical studies demonstrated that treatment of tumor bearing hosts with OX40 agonists, including both anti-OX40 monoclonal antibodies and OX40L-Fc fusion proteins, resulted in tumor regression in several preclinical models. Linch, et al., Front. Oncol. 2015, 34, 1-14.
[0009] CD27, also known as TNFRSF7, has overlapping activity with other TNFRSF members including CD40, 4-1BB, and OX40. CD27 plays a critical role in T cell survival, activation, and effector function, and also plays a role in the proliferative and cytotoxic activity of NK cells. CD27 is constitutively expressed on the majority of T cells, including naive T cells. The ligand for CD27 is CD70, which is found on T cells, B cells, and dendritic cells. Oshima, et al., Int. Immunol. 1998, 10, 517-26. CD27 drives the expansion of CD4.sup.+ and CD8.sup.+ T cells, acting after CD28 to sustain T effector cell survival, and influences secondary responses more than primary responses. However, CD27 activation has also been associated with tumor growth through enhancement of the immunosuppressive effects of regulatory T cells. Claus, et al., Cancer Res. 2012, 72, 3664-76. Other data has indicated that the immunostimulatory effects of CD27 may outweigh this tumor promoting effect. Aulwurm, et al., Int. J. Cancer 2006, 118, 1728-35. In mouse models, an agonistic CD27 monoclonal antibody showed antitumor efficiacy and induction of tumor immunity. He, et al., J. Immunol. 2013, 191, 4174-83.
[0010] Glucocorticoid-induced TNFR-related protein (GITR) is a costimulatory checkpoint molecule that is also known as tumor necrosis factor receptor superfamily member 18 (TNFRSF18), activation-inducible TNFR family receptor (AITR), and CD357. GITR is expressed on several cell types, including regulatory T cells (Tregs) and effector T cells, B cells, NK cells, and antigen-presenting cells. Nocentini and Riccardi, Eur. J. Immunol. 2005, 35, 1016-1022. GITR is activated by its conjugate GITR ligand (GITRL). GITR plays a role in stimulating an immune response, and antigen binding proteins to GITR have utility in treating a variety of GITR-related diseases or disorders in which it is desirable to increase an immune response. Ko, et al., J. Exp. Med. 2005, 202, 885-91; Shimizu, et al., Nature Immunology 2002, 3, 135-142; Cohen, et al., Cancer Res. 2006, 66, 4904-12; Azuma, Crit. Rev. Immunol. 2010, 30, 547-57. For example, T cell stimulation through GITR attenuates Treg-mediated suppression and enhances tumor-killing by CD4.sup.+ and CD8.sup.+ T cells. GITR is constitutively expressed at high levels in Tregs (such as CD4+CD25.sup.+ or CD8+CD25.sup.+ cells) and is additionally upregulated upon activation of these cells. Nocentini and Riccardi, Eur. J. Immunol. 2005, 35, 1016-1022. GITR is a co-activating signal to both CD4.sup.+ and CD8.sup.+ naive T cells, and induces and enhances proliferation and effector function, particularly in situations where T cell receptor (TCR) stimulation is suboptimal. Schaer, et al., Curr. Opin. Immunol. 2012, 24, 217-224. The enhanced immune response caused by antigen binding GITR proteins, such as fusion proteins and anti-GITR antibodies (including agonistic antibodies), is of interest in a variety of immunotherapy applications, such as the treatment of cancers, autoimmune diseases, inflammatory diseases, or infections.
[0011] Herpesvirus entry mediator (HVEM), also known as TNFRSF14 and CD270, was first isolated as a receptor for herpes simplex virus-1 (HSV-1). Montgomery, et al., Cell 1996, 87, 427-36. HVEM binds to the TNF family ligands LIGHT and lymphotoxin alpha homotrimer (Lta3). Mauri, et al., Immunity 1998, 8, 21-30. T cell activation can occur through the HVEM-LIGHT interaction, and the interaction provides a costimulatory signal to T cells that is independent of CD28 signaling and can be observed in the presence of suboptimal levels of CD3 antibody (OKT-3). Tamada, et al., J. Immunol. 2000, 165, 4397-404; Harrop, et al., J. Biol. Chem. 1998, 273, 27548-56; Tamada, et al., Nat. Med. 2000, 6, 283-89; Yu, et al., Nat. Immunol. 2004, 5, 141-49. HVEM comprises four cysteine-rich domains (CRDs). del Rio, et al., J. Leukoc. Biol. 2010, 87, 223-35. CRD2 and CRD3 are required for HVEM trimerization with the TNFRSF ligand LIGHT, which delivers a co-stimulatory signal to T cells through HVEM. In contrast, CRD1 and CRD2 bind to the co-inhibitory B and T lymphocyte attenuator (BTLA) receptor and CD160 in a monomeric manner, providing an inhibitory signal to T cells. Studies of the HVEM-LIGHT interaction suggest that it primarily has a CD28-independent costimulatory effect on CD8+ T cells, but also affects CD4+ T cells. Liu, et al., Int. Immunol. 2003, 15, 861-70; Scheu, et al., J. Exp. Med. 2002, 195, 1613-24.
[0012] CD95, also known as Fas, APO-1, and TNFRSF6, is a 45 kDa type-I transmembrane protein which, unlike 4-1BB, OX40, GITR, CD27, and HVEM, contains a death domain. Kischkel, et al., EMBO J. 1995, 14, 5579-88; Krammer, Nature 2000, 407, 789-95. The binding of the inducible CD95 ligand (CD95L) to CD95 on activated T cells leads to apoptotic cell death, and thus it is not normally associated with the same costimulatory function as 4-1BB, OX40, GITR, CD27, and HVEM. Strauss, et al., J. Exp. Med. 2009, 206, 1379-93. However, CD95 also behaves as a dual function receptor that provides for anti-apoptotic and costimulatory effects on T cells under some conditions. Paulsen, et al., Cell Death Differ. 2011, 18, 619-31. CD95 engagement modulates TCR-driven signal initiation in a dose-dependent manner, wherein high doses of CD95 agonists or cellular CD95L silence T cells, while lower doses of these agonists strongly enhance TCR-driven T cell activation and proliferation.
[0013] The present invention provides the unexpected finding that TNFRSF agonists, such as a 4-1BB agonist, a CD27 agonist, a GITR agonist, an OX40 agonist, a HVEM agonist, or a CD95 agonist, are useful in the expansion of TILs from tumors from which it is known to be difficult to obtain TILs and treat the tumor with TILs, and are further useful in the treatment of patients in combination with TIL therapy.
SUMMARY OF THE INVENTION
[0014] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0015] (a) resecting a tumor from a patient; [0016] (b) obtaining a first population of TILs from the tumor; [0017] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0018] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0019] (e) harvesting the third population of TILs; and [0020] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer.
[0021] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0022] (a) resecting a tumor from a patient; [0023] (b) obtaining a first population of TILs from the tumor; [0024] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises L-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0025] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0026] (e) harvesting the third population of TILs; and [0027] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is selected from the group consisting of a 4-1BB agonist, an OX40 agonist, a CD27 agonist, a GITR agonist, a HVEM agonist, a CD95 agonist, and combinations thereof.
[0028] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0029] (a) resecting a tumor from a patient; [0030] (b) obtaining a first population of TILs from the tumor; [0031] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises L-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0032] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0033] (e) harvesting the third population of TILs; and [0034] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is a 4-1BB agonist, and the 4-1BB agonist is selected from the group consisting of urelumab, utomilumab, EU-101 and fragments, derivatives, variants, biosimilars, and combinations thereof.
[0035] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0036] (a) resecting a tumor from a patient; [0037] (b) obtaining a first population of TILs from the tumor; [0038] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises L-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0039] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0040] (e) harvesting the third population of TILs; and [0041] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is a 4-1BB agonist, and the 4-1BB agonist is a 4-1BB agonist fusion protein.
[0042] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0043] (a) resecting a tumor from a patient; [0044] (b) obtaining a first population of TILs from the tumor; [0045] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises L-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0046] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0047] (e) harvesting the third population of TILs; and [0048] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is a 4-1BB agonist fusion protein, and the 4-1BB agonist fusion protein comprises (i) a first soluble 4-1BB binding domain, (ii) a first peptide linker, (iii) a second soluble 4-1BB binding domain, (iv) a second peptide linker, and (v) a third soluble 4-1BB binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, and wherein the additional domain comprises a Fc fragment domain and hinge domain, and wherein the fusion protein is a dimeric structure according to structure I-A or structure I-B.
[0049] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0050] (a) resecting a tumor from a patient; [0051] (b) obtaining a first population of TILs from the tumor; [0052] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises L-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0053] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0054] (e) harvesting the third population of TILs; and [0055] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is a OX40 agonist, and the OX40 agonist is selected from the group consisting of tavolixizumab, GSK3174998, MEDI6469, MEDI6383, MOXR0916, PF-04518600, Creative Biolabs MOM-18455, and fragments, derivatives, variants, biosimilars, and combinations thereof.
[0056] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0057] (a) resecting a tumor from a patient; [0058] (b) obtaining a first population of TILs from the tumor; [0059] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises L-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0060] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0061] (e) harvesting the third population of TILs; and [0062] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is an OX40 agonist, and the OX40 agonist is an OX40 agonist fusion protein.
[0063] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0064] (a) resecting a tumor from a patient; [0065] (b) obtaining a first population of TILs from the tumor; [0066] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises L-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0067] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0068] (e) harvesting the third population of TILs; and [0069] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is an OX40 agonist fusion protein, and the OX40 agonist fusion protein comprises (i) a first soluble OX40 binding domain, (ii) a first peptide linker, (iii) a second soluble OX40 binding domain, (iv) a second peptide linker, and (v) a third soluble OX40 binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, and wherein the additional domain comprises a Fc fragment domain and hinge domain, and wherein the fusion protein is a dimeric structure according to structure I-A or structure I-B.
[0070] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0071] (a) resecting a tumor from a patient; [0072] (b) obtaining a first population of TILs from the tumor; [0073] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0074] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0075] (e) harvesting the third population of TILs; and [0076] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is a CD27 agonist, and the CD27 agonist is varlilumab, or a fragment, derivative, variant, or biosimilar thereof.
[0077] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0078] (a) resecting a tumor from a patient; [0079] (b) obtaining a first population of TILs from the tumor; [0080] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0081] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0082] (e) harvesting the third population of TILs; and [0083] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is a CD27 agonist, and wherein the CD27 agonist is an CD27 agonist fusion protein.
[0084] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0085] (a) resecting a tumor from a patient; [0086] (b) obtaining a first population of TILs from the tumor; [0087] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0088] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0089] (e) harvesting the third population of TILs; and [0090] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is a CD27 agonist, and the CD27 agonist fusion protein comprises (i) a first soluble CD27 binding domain, (ii) a first peptide linker, (iii) a second soluble CD27 binding domain, (iv) a second peptide linker, and (v) a third soluble CD27 binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, and wherein the additional domain comprises a Fc fragment domain and hinge domain, and wherein the fusion protein is a dimeric structure according to structure I-A or structure I-B.
[0091] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0092] (a) resecting a tumor from a patient; [0093] (b) obtaining a first population of TILs from the tumor; [0094] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0095] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0096] (e) harvesting the third population of TILs; and [0097] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is a GITR agonist, and the GITR agonist is selected from the group consisting of TRX518, 6C8, 36E5, 3D6, 61G6, 6H6, 61F6, 1D8, 17F10, 35D8, 49A1, 9E5, 31H6, 2155, 698, 706, 827, 1649, 1718, 1D7, 33C9, 33F6, 34G4, 35B10, 41E11, 41G5, 42A11, 44C1, 45A8, 46E11, 48H12, 48H7, 49D9, 49E2, 48A9, 5H7, 7A10, 9H6, and fragments, derivatives, variants, biosimilars, and combinations thereof.
[0098] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0099] (a) resecting a tumor from a patient; [0100] (b) obtaining a first population of TILs from the tumor; [0101] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0102] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0103] (e) harvesting the third population of TILs; and [0104] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is an GITR agonist, and the GITR agonist is a GITR agonist fusion protein.
[0105] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0106] (a) resecting a tumor from a patient; [0107] (b) obtaining a first population of TILs from the tumor; [0108] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0109] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0110] (e) harvesting the third population of TILs; and [0111] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is a GITR agonist fusion protein, and the GITR agonist fusion protein comprises (i) a first soluble GITR binding domain, (ii) a first peptide linker, (iii) a second soluble GITR binding domain, (iv) a second peptide linker, and (v) a third soluble GITR binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, and wherein the additional domain comprises a Fc fragment domain and hinge domain, and wherein the fusion protein is a dimeric structure according to structure I-A or structure I-B.
[0112] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0113] (a) resecting a tumor from a patient; [0114] (b) obtaining a first population of TILs from the tumor; [0115] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0116] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0117] (e) harvesting the third population of TILs; and [0118] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is a HVEM agonist.
[0119] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0120] (a) resecting a tumor from a patient; [0121] (b) obtaining a first population of TILs from the tumor; [0122] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0123] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0124] (e) harvesting the third population of TILs; and [0125] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is an HVEM agonist, and the HVEM agonist is a HVEM agonist fusion protein.
[0126] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0127] (a) resecting a tumor from a patient; [0128] (b) obtaining a first population of TILs from the tumor; [0129] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0130] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0131] (e) harvesting the third population of TILs; and [0132] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is a HVEM agonist fusion protein, and wherein the HVEM agonist fusion protein comprises (i) a first soluble HVEM binding domain, (ii) a first peptide linker, (iii) a second soluble HVEM binding domain, (iv) a second peptide linker, and (v) a third soluble HVEM binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, and wherein the additional domain comprises a Fc fragment domain and hinge domain, and wherein the fusion protein is a dimeric structure according to structure I-A or structure I-B.
[0133] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0134] (a) resecting a tumor from a patient; [0135] (b) obtaining a first population of TILs from the tumor; [0136] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises L-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0137] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0138] (e) harvesting the third population of TILs; and [0139] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, further comprising the step of treating the patient with the TNFRSF agonist starting on the day after administration of the third population of TILs to the patient, wherein the TNFRSF agonist is administered intravenously at a dose of between 0.1 mg/kg and 50 mg/kg every four weeks for up to eight cycles.
[0140] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0141] (a) resecting a tumor from a patient; [0142] (b) obtaining a first population of TILs from the tumor; [0143] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0144] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0145] (e) harvesting the third population of TILs; and [0146] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, further comprising the step of treating the patient with the TNFRSF agonist prior to the step of resecting of a tumor from the patient, wherein the TNFRSF agonist is administered intravenously at a dose of between 0.1 mg/kg and 50 mg/kg every four weeks for up to eight cycles.
[0147] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0148] (a) resecting a tumor from a patient; [0149] (b) obtaining a first population of TILs from the tumor; [0150] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0151] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0152] (e) harvesting the third population of TILs; and [0153] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is selected from the group consisting of urelumab, utomilumab, EU-101, tavolixizumab, Creative Biolabs MOM-18455, and fragments, derivatives, variants, biosimilars, and combinations thereof.
[0154] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0155] (a) resecting a tumor from a patient; [0156] (b) obtaining a first population of TILs from the tumor; [0157] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0158] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0159] (e) harvesting the third population of TILs; and [0160] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the first cell culture medium comprises a second TNFRSF agonist.
[0161] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0162] (a) resecting a tumor from a patient; [0163] (b) obtaining a first population of TILs from the tumor; [0164] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0165] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0166] (e) harvesting the third population of TILs; and [0167] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is added to the first cell culture medium during the initial expansion at an interval selected from the group consisting of every day, every two days, every three days, every four days, every five days, every six days, every seven days, and every two weeks.
[0168] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0169] (a) resecting a tumor from a patient; [0170] (b) obtaining a first population of TILs from the tumor; [0171] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0172] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0173] (e) harvesting the third population of TILs; and [0174] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is added to the second cell culture medium during the rapid expansion at an interval selected from the group consisting of every day, every two days, every three days, every four days, every five days, every six days, every seven days, and every two weeks.
[0175] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0176] (a) resecting a tumor from a patient; [0177] (b) obtaining a first population of TILs from the tumor; [0178] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0179] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0180] (e) harvesting the third population of TILs; and [0181] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is added at a concentration sufficient to achieve a concentration in the cell culture medium of between 0.1 .mu.g/mL and 100 .mu.g/mL.
[0182] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0183] (a) resecting a tumor from a patient; [0184] (b) obtaining a first population of TILs from the tumor; [0185] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0186] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0187] (e) harvesting the third population of TILs; and [0188] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the TNFRSF agonist is added at a concentration sufficient to achieve a concentration in the cell culture medium of between 20 .mu.g/mL and 40 .mu.g/mL.
[0189] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0190] (a) resecting a tumor from a patient; [0191] (b) obtaining a first population of TILs from the tumor; [0192] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0193] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0194] (e) harvesting the third population of TILs; and [0195] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein IL-2 is present at an initial concentration of about 10 to about 6000 IU/mL in the first cell culture medium.
[0196] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0197] (a) resecting a tumor from a patient; [0198] (b) obtaining a first population of TILs from the tumor; [0199] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0200] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0201] (e) harvesting the third population of TILs; and [0202] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein IL-2 is present at an initial concentration of about 3000 IU/mL in the first cell culture medium.
[0203] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0204] (a) resecting a tumor from a patient; [0205] (b) obtaining a first population of TILs from the tumor; [0206] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises L-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0207] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises L-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0208] (e) harvesting the third population of TILs; and [0209] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, The method of Claim 31, wherein L-2 is present at an initial concentration of about 800 to about 1100 IU/mL in the first cell culture medium.
[0210] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0211] (a) resecting a tumor from a patient; [0212] (b) obtaining a first population of TILs from the tumor; [0213] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises L-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0214] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0215] (e) harvesting the third population of TILs; and [0216] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein IL-2 is present at an initial concentration of about 1000 IU/mL in the first cell culture medium.
[0217] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0218] (a) resecting a tumor from a patient; [0219] (b) obtaining a first population of TILs from the tumor; [0220] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0221] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0222] (e) harvesting the third population of TILs; and [0223] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein IL-2 is present at an initial concentration of about 10 to about 6000 IU/mL in the second cell culture medium.
[0224] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0225] (a) resecting a tumor from a patient; [0226] (b) obtaining a first population of TILs from the tumor; [0227] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0228] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0229] (e) harvesting the third population of TILs; and [0230] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein IL-2 is present at an initial concentration of about 3000 IU/mL in the second cell culture medium.
[0231] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0232] (a) resecting a tumor from a patient; [0233] (b) obtaining a first population of TILs from the tumor; [0234] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0235] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0236] (e) harvesting the third population of TILs; and [0237] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein IL-2 is present at an initial concentration of about 800 to about 1100 IU/mL in the second cell culture medium.
[0238] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0239] (a) resecting a tumor from a patient; [0240] (b) obtaining a first population of TILs from the tumor; [0241] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0242] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0243] (e) harvesting the third population of TILs; and [0244] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein IL-2 is present at an initial concentration of about 1000 IU/mL in the second cell culture medium.
[0245] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0246] (a) resecting a tumor from a patient; [0247] (b) obtaining a first population of TILs from the tumor; [0248] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0249] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0250] (e) harvesting the third population of TILs; and [0251] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein IL-15 is present in the first cell culture medium.
[0252] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0253] (a) resecting a tumor from a patient; [0254] (b) obtaining a first population of TILs from the tumor; [0255] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0256] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0257] (e) harvesting the third population of TILs; and [0258] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein IL-15 is present at an initial concentration of about 5 ng/mL to about 20 ng/mL in the first cell culture medium.
[0259] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0260] (a) resecting a tumor from a patient; [0261] (b) obtaining a first population of TILs from the tumor; [0262] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0263] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0264] (e) harvesting the third population of TILs; and [0265] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein IL-15 is present in the second cell culture medium.
[0266] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0267] (a) resecting a tumor from a patient; [0268] (b) obtaining a first population of TILs from the tumor; [0269] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises L-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0270] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0271] (e) harvesting the third population of TILs; and [0272] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein IL-15 is present at an initial concentration of about 5 ng/mL to about 20 ng/mL in the second cell culture medium.
[0273] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0274] (a) resecting a tumor from a patient; [0275] (b) obtaining a first population of TILs from the tumor; [0276] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises L-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0277] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0278] (e) harvesting the third population of TILs; and [0279] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein IL-21 is present in the first cell culture medium.
[0280] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0281] (a) resecting a tumor from a patient; [0282] (b) obtaining a first population of TILs from the tumor; [0283] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0284] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0285] (e) harvesting the third population of TILs; and [0286] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein IL-21 is present at an initial concentration of about 5 ng/mL to about 20 ng/mL in the first cell culture medium.
[0287] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0288] (a) resecting a tumor from a patient; [0289] (b) obtaining a first population of TILs from the tumor; [0290] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0291] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0292] (e) harvesting the third population of TILs; and [0293] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein IL-21 is present in the second cell culture medium.
[0294] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0295] (a) resecting a tumor from a patient; [0296] (b) obtaining a first population of TILs from the tumor; [0297] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0298] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0299] (e) harvesting the third population of TILs; and [0300] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein IL-21 is present at an initial concentration of about 5 ng/mL to about 20 ng/mL in the second cell culture medium.
[0301] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0302] (a) resecting a tumor from a patient; [0303] (b) obtaining a first population of TILs from the tumor; [0304] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0305] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0306] (e) harvesting the third population of TILs; and [0307] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein OKT-3 antibody is present at an initial concentration of about 10 ng/mL to about 60 ng/mL in the second cell culture medium.
[0308] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0309] (a) resecting a tumor from a patient; [0310] (b) obtaining a first population of TILs from the tumor; [0311] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0312] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0313] (e) harvesting the third population of TILs; and [0314] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein OKT-3 antibody is present at an initial concentration of about 30 ng/mL in the second cell culture medium.
[0315] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0316] (a) resecting a tumor from a patient; [0317] (b) obtaining a first population of TILs from the tumor; [0318] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0319] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0320] (e) harvesting the third population of TILs; and [0321] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the initial expansion is performed using a gas permeable container.
[0322] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0323] (a) resecting a tumor from a patient; [0324] (b) obtaining a first population of TILs from the tumor; [0325] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0326] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0327] (e) harvesting the third population of TILs; and [0328] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the rapid expansion is performed using a gas permeable container.
[0329] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0330] (a) resecting a tumor from a patient; [0331] (b) obtaining a first population of TILs from the tumor; [0332] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0333] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0334] (e) harvesting the third population of TILs; and [0335] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, further comprising the step of treating the patient with a non-myeloablative lymphodepletion regimen prior to administering the third population of TILs to the patient.
[0336] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0337] (a) resecting a tumor from a patient; [0338] (b) obtaining a first population of TILs from the tumor; [0339] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0340] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0341] (e) harvesting the third population of TILs; and [0342] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, further comprising the step of treating the patient with a non-myeloablative lymphodepletion regimen prior to administering the third population of TILs to the patient, wherein the non-myeloablative lymphodepletion regimen comprises the steps of administration of cyclophosphamide at a dose of 60 mg/m.sup.2/day for two days followed by administration of fludarabine at a dose of 25 mg/m.sup.2/day for five days.
[0343] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0344] (a) resecting a tumor from a patient; [0345] (b) obtaining a first population of TILs from the tumor; [0346] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises L-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0347] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0348] (e) harvesting the third population of TILs; and [0349] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, further comprising the step of treating the patient with a decrescendo IL-2 regimen starting on the day after administration of the third population of TILs to the patient, wherein the decrescendo IL-2 regimen comprises aldesleukin administered intravenously at a dose of 18,000,000 IU/m.sup.2 on day 1, 9,000,000 IU/m.sup.2 on day 2, and 4,500,000 IU/m.sup.2 on days 3 and 4.
[0350] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0351] (a) resecting a tumor from a patient; [0352] (b) obtaining a first population of TILs from the tumor; [0353] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0354] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0355] (e) harvesting the third population of TILs; and [0356] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, further comprising the step of treating the patient with pegylated IL-2 after administration of the third population of TILs to the patient at a dose of 0.10 mg/day to 50 mg/day.
[0357] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0358] (a) resecting a tumor from a patient; [0359] (b) obtaining a first population of TILs from the tumor; [0360] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0361] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0362] (e) harvesting the third population of TILs; and [0363] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, further comprising the step of treating the patient with a high-dose IL-2 regimen starting on the day after administration of the third population of TILs to the patient.
[0364] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0365] (a) resecting a tumor from a patient; [0366] (b) obtaining a first population of TILs from the tumor; [0367] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0368] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0369] (e) harvesting the third population of TILs; and [0370] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, further comprising the step of treating the patient with a high-dose IL-2 regimen starting on the day after administration of the third population of TILs to the patient, wherein the high-dose IL-2 regimen comprises 600,000 or 720,000 IU/kg of aldesleukin, or a biosimilar or variant thereof, administered as a 15-minute bolus intravenous infusion every eight hours until tolerance.
[0371] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0372] (a) resecting a tumor from a patient; [0373] (b) obtaining a first population of TILs from the tumor; [0374] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0375] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0376] (e) harvesting the third population of TILs; and [0377] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the cancer is selected from the group consisting of melanoma, ovarian cancer, cervical cancer, lung cancer, bladder cancer, breast cancer, head and neck cancer, renal cell carcinoma, acute myeloid leukemia, colorectal cancer, cholangiocarcinoma, and sarcoma.
[0378] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0379] (a) resecting a tumor from a patient; [0380] (b) obtaining a first population of TILs from the tumor; [0381] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0382] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0383] (e) harvesting the third population of TILs; and [0384] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, wherein the cancer is selected from the group consisting of non-small cell lung cancer (NSCLC), triple negative breast cancer, double-refractory melanoma, and uveal (ocular) melanoma.
[0385] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0386] (a) resecting a tumor from a patient; [0387] (b) obtaining a first population of TILs from the tumor; [0388] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0389] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0390] (e) harvesting the third population of TILs; and [0391] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, further comprising the step of treating the patient with a PD-1 inhibitor or PD-L1 inhibitor prior to resecting the tumor from the patient.
[0392] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0393] (a) resecting a tumor from a patient; [0394] (b) obtaining a first population of TILs from the tumor; [0395] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0396] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0397] (e) harvesting the third population of TILs; and [0398] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, further comprising the step of treating the patient with a PD-1 inhibitor or PD-L1 inhibitor prior to resecting the tumor from the patient, wherein the PD-1 inhibitor or PD-L1 inhibitor is selected from the group consisting of nivolumab, pembrolizumab, durvalumab, atezolizumab, avelumab, and fragments, derivatives, variants, biosimilars, and combinations thereof.
[0399] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0400] (a) resecting a tumor from a patient; [0401] (b) obtaining a first population of TILs from the tumor; [0402] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0403] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0404] (e) harvesting the third population of TILs; and [0405] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, further comprising the step of treating the patient with a PD-1 inhibitor or PD-L1 inhibitor after resecting the tumor from the patient.
[0406] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0407] (a) resecting a tumor from a patient; [0408] (b) obtaining a first population of TILs from the tumor; [0409] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0410] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0411] (e) harvesting the third population of TILs; and [0412] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, further comprising the step of treating the patient with a PD-1 inhibitor or PD-L1 inhibitor after resecting the tumor from the patient, wherein the PD-1 inhibitor or PD-L1 inhibitor is selected from the group consisting of nivolumab, pembrolizumab, durvalumab, atezolizumab, avelumab, and fragments, derivatives, variants, biosimilars, and combinations thereof.
[0413] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0414] (a) resecting a tumor from a patient; [0415] (b) obtaining a first population of TILs from the tumor; [0416] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0417] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0418] (e) harvesting the third population of TILs; and [0419] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, further comprising the step of treating the patient with a PD-1 inhibitor or PD-L1 inhibitor after administering the third population of TILs to the patient.
[0420] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0421] (a) resecting a tumor from a patient; [0422] (b) obtaining a first population of TILs from the tumor; [0423] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0424] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [0425] (e) harvesting the third population of TILs; and [0426] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer, further comprising the step of treating the patient with a PD-1 inhibitor or PD-L1 inhibitor after administering the third population of TILs to the patient, wherein the PD-1 inhibitor or PD-L1 inhibitor is selected from the group consisting of nivolumab, pembrolizumab, durvalumab, atezolizumab, avelumab, and fragments, derivatives, variants, biosimilars, and combinations thereof.
[0427] In an embodiment, the invention provides a process for the preparation of a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0428] (b) obtaining a first population of TILs; [0429] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0430] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; and [0431] (e) harvesting the third population of TILs.
[0432] In an embodiment, the invention provides a population of tumor infiltrating lymphocytes (TILs) obtainable from a process comprising the steps of: [0433] (b) obtaining a first population of TILs; [0434] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0435] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; and [0436] (e) harvesting the third population of TILs.
[0437] In an embodiment, the invention provides a population of TILs is for use in the treatment of cancer. In an embodiment, the invention provides a pharmaceutical composition comprising a population of tumor infiltrating lymphocytes (TILs) for use in treating a cancer wherein the population of tumor infiltrating lymphocytes (TILs) is obtainable by a process comprising the steps of: [0438] (b) obtaining a first population of TILs; [0439] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [0440] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; and [0441] (e) harvesting the third population of TILs.
[0442] In an embodiment, the first population of TILs is obtained from a tumor. In an embodiment, the tumor is firstly resected from a patient. In an embodiment, the first population of TILs is obtained from the tumor which has been resected from a patient. In an embodiment, the population of TILs is for administration in a therapeutically effective amount to a patient with cancer.
[0443] In an embodiment, the invention provides a method of expanding a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0444] (a) resecting a tumor from a patient; [0445] (b) obtaining a first population of TILs from the tumor; [0446] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2, and wherein the initial expansion is performed over a period of 11 days or less; [0447] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3) antibody, peripheral blood mononuclear cells (PBMCs), and a TNFRSF agonist, and wherein the rapid expansion is performed over a period of 11 days or less; [0448] (e) harvesting the third population of TILs; and [0449] (f) optionally cryopreserving the third population of TILs in a dimethylsulfoxide-based media.
[0450] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0451] (a) resecting a tumor from a patient; [0452] (b) obtaining a first population of TILs from the tumor; [0453] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2, and wherein the initial expansion is performed over a period of 11 days or less; [0454] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3) antibody, peripheral blood mononuclear cells (PBMCs), and a TNFRSF agonist, and wherein the rapid expansion is performed over a period of 11 days or less; [0455] (e) harvesting the third population of TILs; and [0456] (f) administering a therapeutically effective portion of the third population of TILs to the patient.
[0457] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0458] (a) resecting a tumor from a patient; [0459] (b) obtaining a first population of TILs from the tumor; [0460] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2, and wherein the initial expansion is performed over a period of 11 days or less; [0461] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3) antibody, peripheral blood mononuclear cells (PBMCs), and a TNFRSF agonist, and wherein the rapid expansion is performed over a period of 11 days or less; [0462] (e) harvesting the third population of TILs; and [0463] (f) administering a therapeutically effective portion of the third population of TILs to the patient, [0464] wherein the TNFRSF agonist is selected from the group consisting of a 4-1BB agonist, an OX40 agonist, and a combination thereof.
[0465] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0466] (a) resecting a tumor from a patient; [0467] (b) obtaining a first population of TILs from the tumor; [0468] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2, and wherein the initial expansion is performed over a period of 11 days or less; [0469] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3) antibody, peripheral blood mononuclear cells (PBMCs), and a TNFRSF agonist, and wherein the rapid expansion is performed over a period of 11 days or less; [0470] (e) harvesting the third population of TILs; and [0471] (f) administering a therapeutically effective portion of the third population of TILs to the patient, [0472] wherein the TNFRSF agonist is selected from the group consisting of a 4-1BB agonist, an OX40 agonist, and a combination thereof, and [0473] wherein the TNFRSF agonist is a 4-1BB agonist, and the 4-1BB agonist is selected from the group consisting of urelumab, utomilumab, EU-101, a fusion protein, and fragments, derivatives, variants, biosimilars, and combinations thereof.
[0474] In an embodiment, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [0475] (a) resecting a tumor from a patient; [0476] (b) obtaining a first population of TILs from the tumor; [0477] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2, and wherein the initial expansion is performed over a period of 11 days or less; [0478] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3) antibody, peripheral blood mononuclear cells (PBMCs), and a TNFRSF agonist, and wherein the rapid expansion is performed over a period of 11 days or less; [0479] (e) harvesting the third population of TILs; and [0480] (f) administering a therapeutically effective portion of the third population of TILs to the patient, [0481] wherein the TNFRSF agonist is selected from the group consisting of a 4-1BB agonist, an OX40 agonist, and a combination thereof, and [0482] wherein the TNFRSF agonist is a OX40 agonist, and the OX40 agonist is selected from the group consisting of tavolixizumab, GSK3174998, MEDI6469, MEDI6383, MOXR0916, PF-04518600, Creative Biolabs MOM-18455, and fragments, derivatives, variants, biosimilars, and combinations thereof. [0483] wherein the OX4 agonist is present at the start of step (d) at a concentration between 1 g/mL and 30 .mu.g/mL.
[0484] In an embodiment, the invention provides a method of any of the foregoing embodiments, wherein the TNFRSF agonist is present at the start of step (d) at a concentration between 5 .mu.g/mL and 20 .mu.g/mL.
[0485] In an embodiment, the invention provides a method of any of the foregoing embodiments, wherein the TNFRSF agonist is present at the start of step (d) at a concentration of about 10 .mu.g/mL.
[0486] In an embodiment, the invention provides a method of any of the foregoing embodiments, wherein the TNFRSF agonist is maintained throughout step (d) at a concentration between 1 .mu.g/mL and 30 .mu.g/mL.
[0487] In an embodiment, the invention provides a method of any of the foregoing embodiments, wherein the TNFRSF agonist is maintained throughout step (d) at a concentration between 5 .mu.g/mL and 20 .mu.g/mL.
[0488] In an embodiment, the invention provides a method of any of the foregoing embodiments, wherein the TNFRSF agonist is maintained throughout step (d) at a concentration of about 10 .mu.g/mL.
[0489] In an embodiment, the invention provides a method of any of the foregoing embodiments, wherein the third population of TILs exhibits an increased ratio of CD8.sup.+ TILs to CD4.sup.+ TILs in comparison to the reference ratio of CD8.sup.+ TILs to CD4.sup.+ TILs in the second population of TILs. In an embodiment, the increased ratio is selected from the group consisting of at least 1% greater than the reference ratio, at least 2% greater than the reference ratio, at least 5% greater than the reference ratio, at least 10% greater than the reference ratio, at least 15% greater than the reference ratio, at least 20% greater than the reference ratio, at least 25% greater than the reference ratio, at least 30% greater than the reference ratio, at least 35% greater than the reference ratio, at least 40% greater than the reference ratio, at least 45% greater than the reference ratio, and at least 50% greater than the reference ratio. In an embodiment, the increased ratio is between 5% and 80% greater than the reference ratio. In an embodiment, the increased ratio is between 10% and 70% greater than the reference ratio. In an embodiment, the increased ratio is between 15% and 60% greater than the reference ratio. In an of the foregoing embodiments, the reference ratio is obtained from a third TIL population that is a responder to the TNFRSF agonist.
[0490] In an embodiment, the invention provides a method of any of the foregoing embodiments, wherein the cancer is selected from the group consisting of melanoma, uveal (ocular) melanoma, ovarian cancer, cervical cancer, lung cancer, bladder cancer, breast cancer, head and neck cancer (head and neck squamous cell cancer), renal cell carcinoma, colorectal cancer, pancreatic cancer, glioblastoma, cholangiocarcinoma, and sarcoma. In an embodiment, the invention provides a method of any of the foregoing embodiments, wherein the cancer is selected from the group consisting of cutaneous melanoma, uveal (ocular) melanoma, platinum-resistant ovarian cancer, pancreatic ductal adenocarcinoma, osteosarcoma, triple-negative breast cancer, and non-small-cell lung cancer.
[0491] In an embodiment, any of the foregoing embodiments may be combined with any of the following embodiments.
[0492] In an embodiment, the process is an in vitro or an ex vivo process.
[0493] In an embodiment, the TNFRSF agonist is selected from the group consisting of a 4-1BB agonist, an OX40 agonist, a CD27 agonist, a GITR agonist, a HVEM agonist, a CD95 agonist, and combinations thereof.
[0494] In an embodiment, the TNFRSF agonist is a 4-1BB agonist.
[0495] In an embodiment, the TNFRSF agonist is a 4-1BB agonist, and the 4-1BB agonist is selected from the group consisting of urelumab, utomilumab, EU-101 and fragments, derivatives, variants, biosimilars, and combinations thereof.
[0496] In an embodiment, the TNFRSF agonist is a 4-1BB agonist, and the 4-1BB agonist is a 4-1BB agonist fusion protein.
[0497] In an embodiment, the TNFRSF agonist is a 4-1BB agonist fusion protein, and the 4-1BB agonist fusion protein comprises (i) a first soluble 4-1BB binding domain, (ii) a first peptide linker, (iii) a second soluble 4-1BB binding domain, (iv) a second peptide linker, and (v) a third soluble 4-1BB binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, and wherein the additional domain comprises a Fc fragment domain and hinge domain, and wherein the fusion protein is a dimeric structure according to structure I-A or structure I-B.
[0498] In an embodiment, the TNFRSF agonist is a OX40 agonist.
[0499] In an embodiment, the TNFRSF agonist is a OX40 agonist, and the OX40 agonist is selected from the group consisting of tavolixizumab, GSK3174998, MEDI6469, MEDI6383, MOXR0916, PF-04518600, Creative Biolabs MOM-18455, and fragments, derivatives, variants, biosimilars, and combinations thereof.
[0500] In an embodiment, the TNFRSF agonist is an OX40 agonist, and the OX40 agonist is an OX40 agonist fusion protein.
[0501] In an embodiment, the TNFRSF agonist is an OX40 agonist fusion protein, and the OX40 agonist fusion protein comprises (i) a first soluble OX40 binding domain, (ii) a first peptide linker, (iii) a second soluble OX40 binding domain, (iv) a second peptide linker, and (v) a third soluble OX40 binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, and wherein the additional domain comprises a Fc fragment domain and hinge domain, and wherein the fusion protein is a dimeric structure according to structure I-A or structure I-B.
[0502] In an embodiment, the TNFRSF agonist is a CD27 agonist.
[0503] In an embodiment, the TNFRSF agonist is a CD27 agonist, and the CD27 agonist is varlilumab, or a fragment, derivative, variant, or biosimilar thereof.
[0504] In an embodiment, the TNFRSF agonist is a CD27 agonist, and wherein the CD27 agonist is an CD27 agonist fusion protein.
[0505] In an embodiment, the TNFRSF agonist is a CD27 agonist, and the CD27 agonist fusion protein comprises (i) a first soluble CD27 binding domain, (ii) a first peptide linker, (iii) a second soluble CD27 binding domain, (iv) a second peptide linker, and (v) a third soluble CD27 binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, and wherein the additional domain comprises a Fc fragment domain and hinge domain, and wherein the fusion protein is a dimeric structure according to structure I-A or structure I-B.
[0506] In an embodiment, the TNFRSF agonist is a GITR agonist.
[0507] In an embodiment, the TNFRSF agonist is a GITR agonist, and the GITR agonist is selected from the group consisting of TRX518, 6C8, 36E5, 3D6, 61G6, 6H6, 61F6, 1D8, 17F10, 35D8, 49A1, 9E5, 31H6, 2155, 698, 706, 827, 1649, 1718, 1D7, 33C9, 33F6, 34G4, 35B10, 41E11, 41G5, 42A11, 44C1, 45A8, 46E11, 48H12, 48H7, 49D9, 49E2, 48A9, 5H7, 7A10, 9H6, and fragments, derivatives, variants, biosimilars, and combinations thereof.
[0508] In an embodiment, the TNFRSF agonist is an GITR agonist, and the GITR agonist is a GITR agonist fusion protein.
[0509] In an embodiment, the TNFRSF agonist is a GITR agonist fusion protein, and the GITR agonist fusion protein comprises (i) a first soluble GITR binding domain, (ii) a first peptide linker, (iii) a second soluble GITR binding domain, (iv) a second peptide linker, and (v) a third soluble GITR binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, and wherein the additional domain comprises a Fc fragment domain and hinge domain, and wherein the fusion protein is a dimeric structure according to structure I-A or structure I-B.
[0510] In an embodiment, the TNFRSF agonist is a HVEM agonist.
[0511] In an embodiment, the TNFRSF agonist is an HVEM agonist, and the HVEM agonist is a HVEM agonist fusion protein.
[0512] In an embodiment, the TNFRSF agonist is a HVEM agonist fusion protein, and wherein the HVEM agonist fusion protein comprises (i) a first soluble HVEM binding domain, (ii) a first peptide linker, (iii) a second soluble HVEM binding domain, (iv) a second peptide linker, and (v) a third soluble HVEM binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, and wherein the additional domain comprises a Fc fragment domain and hinge domain, and wherein the fusion protein is a dimeric structure according to structure I-A or structure I-B.
[0513] In an embodiment, the TNFRSF agonist is selected from the group consisting of urelumab, utomilumab, EU-101, tavolixizumab, Creative Biolabs MOM-18455, and fragments, derivatives, variants, biosimilars, and combinations thereof.
[0514] In an embodiment, the first cell culture medium comprises a second TNFRSF agonist.
[0515] In an embodiment, the TNFRSF agonist is added to the first cell culture medium during the initial expansion at an interval selected from the group consisting of every day, every two days, every three days, every four days, every five days, every six days, every seven days, and every two weeks.
[0516] In an embodiment, the TNFRSF agonist is added to the second cell culture medium during the rapid expansion at an interval selected from the group consisting of every day, every two days, every three days, every four days, every five days, every six days, every seven days, and every two weeks.
[0517] In an embodiment, the TNFRSF agonist is added at a concentration sufficient to achieve a concentration in the cell culture medium of between 0.1 .mu.g/mL and 100 .mu.g/mL.
[0518] In an embodiment, the TNFRSF agonist is added at a concentration sufficient to achieve a concentration in the cell culture medium of between 20 .mu.g/mL and 40 .mu.g/mL.
[0519] Further details of the TNFRSF agonists are provided herein.
[0520] In an embodiment, IL-2 is present at an initial concentration of about 10 to about 6000 IU/mL in the first cell culture medium.
[0521] In an embodiment, IL-2 is present at an initial concentration of about 3000 IU/mL in the first cell culture medium.
[0522] In an embodiment, IL-2 is present at an initial concentration of about 800 to about 1100 IU/mL in the first cell culture medium.
[0523] In an embodiment, IL-2 is present at an initial concentration of about 1000 IU/mL in the first cell culture medium.
[0524] In an embodiment, IL-2 is present at an initial concentration of about 10 to about 6000 IU/mL in the second cell culture medium.
[0525] In an embodiment, IL-2 is present at an initial concentration of about 3000 IU/mL in the second cell culture medium.
[0526] In an embodiment, IL-2 is present at an initial concentration of about 800 to about 1100 IU/mL in the second cell culture medium.
[0527] In an embodiment, IL-2 is present at an initial concentration of about 1000 IU/mL in the second cell culture medium.
[0528] In an embodiment, IL-15 is present in the first cell culture medium.
[0529] In an embodiment, IL-15 is present at an initial concentration of about 5 ng/mL to about 20 ng/mL in the first cell culture medium.
[0530] In an embodiment, IL-15 is present in the second cell culture medium.
[0531] In an embodiment, IL-15 is present at an initial concentration of about 5 ng/mL to about 20 ng/mL in the second cell culture medium.
[0532] In an embodiment, IL-21 is present in the first cell culture medium.
[0533] In an embodiment, IL-21 is present at an initial concentration of about 5 ng/mL to about 20 ng/mL in the first cell culture medium.
[0534] In an embodiment, IL-21 is present in the second cell culture medium.
[0535] In an embodiment, IL-21 is present at an initial concentration of about 5 ng/mL to about 20 ng/mL in the second cell culture medium.
[0536] In an embodiment, OKT-3 antibody is present at an initial concentration of about 10 ng/mL to about 60 ng/mL in the second cell culture medium.
[0537] In an embodiment, OKT-3 antibody is present at an initial concentration of about 30 ng/mL in the second cell culture medium.
[0538] In an embodiment, the initial expansion is performed using a gas permeable container.
[0539] In an embodiment, the rapid expansion is performed using a gas permeable container.
[0540] In an embodiment, the invention provides a population of tumor infiltrating lymphocytes (TILs) for use in treating a cancer wherein the population of tumor infiltrating lymphocytes (TILs) is obtainable by a process of the invention as described herein.
[0541] In an embodiment, the invention provides a pharmaceutical composition comprising a population of tumor infiltrating lymphocytes (TILs) for use in a method of treating a cancer wherein the population of tumor infiltrating lymphocytes (TILs) is obtainable by a process of the invention as described herein.
[0542] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer in combination with a TNFRSF.
[0543] In an embodiment, the invention provides a combination of a population of TILs obtainable by a process of the invention as described herein and a TNFRSF for use in the treatment of cancer.
[0544] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer in combination with a TNFRSF agonist wherein the TNFRSF agonist is for administration on the day after administration of the third population of TILs to the patient, and wherein the TNFRSF agonist is administered intravenously at a dose of between 0.1 mg/kg and 50 mg/kg every four weeks for up to eight cycles.
[0545] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer in combination with a TNFRSF agonist wherein the TNFRSF agonist is for administration prior to the step of resecting of a tumor from the patient, and wherein the TNFRSF agonist for administration intravenously at a dose of between 0.1 mg/kg and 50 mg/kg every four weeks for up to eight cycles.
[0546] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer in combination with a non-myeloablative lymphodepletion regimen.
[0547] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer in combination with a non-myeloablative lymphodepletion regimen prior to administering the third population of TILs and/or a pharmaceutical composition comprising the third population of TILs to the patient.
[0548] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer in combination with a non-myeloablative lymphodepletion regimen prior to administering the third population of TILs and/or a pharmaceutical composition comprising the third population of TILs to the patient, wherein the non-myeloablative lymphodepletion regimen comprises the steps of administration of cyclophosphamide at a dose of 60 mg/m.sup.2/day for two days followed by administration of fludarabine at a dose of 25 mg/m.sup.2/day for five days. Further details of the non-myeloablative lymphodepletion regimen are provided herein, e.g., under the Heading "Non-Myeloablative Lymphodepletion with Chemotherapy".
[0549] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer in combination with a IL-2 regimen.
[0550] In an embodiment, the IL-2 regimen is a decrescendo IL-2 regimen.
[0551] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer in combination with a decrescendo IL-2 regimen starting on the day after administration of the third population of TILs and/or a pharmaceutical composition comprising the third population of TILs to the patient, wherein the decrescendo IL-2 regimen comprises aldesleukin administered intravenously at a dose of 18,000,000 IU/m.sup.2 on day 1, 9,000,000 IU/m.sup.2 on day 2, and 4,500,000 IU/m.sup.2 on days 3 and 4.
[0552] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer in combination with pegylated IL-2.
[0553] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in a method of treating cancer in combination with pegylated IL-2 administered after administration of the third population of TILs and/or a pharmaceutical composition comprising the third population of TILs to the patient at a dose of 0.10 mg/day to 50 mg/day.
[0554] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in a method of treating cancer in combination with a high-dose IL-2 regimen.
[0555] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in a method of treating cancer in combination with a high-dose IL-2 regimen starting on the day after administration of the third population of TILs and/or a pharmaceutical composition comprising the third population of TILs to the patient.
[0556] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer in combination with a high-dose IL-2 regimen starting on the day after administration of the third population of TILs and/or a pharmaceutical composition comprising the third population of TILs to the patient, wherein the high-dose IL-2 regimen comprises 600,000 or 720,000 IU/kg of aldesleukin, or a biosimilar or variant thereof, administered as a 15-minute bolus intravenous infusion every eight hours until tolerance.
[0557] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer, wherein the cancer is selected from the group consisting of melanoma, ovarian cancer, cervical cancer, lung cancer, bladder cancer, breast cancer, head and neck cancer, renal cell carcinoma, acute myeloid leukemia, colorectal cancer, cholangiocarcinoma, and sarcoma.
[0558] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer, wherein the cancer is selected from the group consisting of non-small cell lung cancer (NSCLC), triple negative breast cancer, double-refractory melanoma, and uveal (ocular) melanoma.
[0559] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer in combination with a PD-1 inhibitor or PD-L1 inhibitor.
[0560] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer in combination with a PD-1 inhibitor or PD-L1 inhibitor, wherein the PD-1 inhibitor or PD-L1 inhibitor is selected from the group consisting of nivolumab, pembrolizumab, durvalumab, atezolizumab, avelumab, and fragments, derivatives, variants, biosimilars, and combinations thereof.
[0561] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer in combination with a PD-1 inhibitor or PD-L1 inhibitor, wherein the PD-1 inhibitor or PD-L1 inhibitor is for administration prior to resecting the tumor from the patient.
[0562] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer in combination with a PD-1 inhibitor or PD-L1 inhibitor prior to resecting the tumor from the patient, wherein the PD-1 inhibitor or PD-L1 inhibitor is selected from the group consisting of nivolumab, pembrolizumab, durvalumab, atezolizumab, avelumab, and fragments, derivatives, variants, biosimilars, and combinations thereof.
[0563] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in method of treating cancer in combination with a PD-1 inhibitor or PD-L1 inhibitor.
[0564] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer in combination with a PD-1 inhibitor or PD-L1 inhibitor, wherein the PD-1 inhibitor or PD-L1 inhibitor is selected from the group consisting of nivolumab, pembrolizumab, durvalumab, atezolizumab, avelumab, and fragments, derivatives, variants, biosimilars, and combinations thereof.
[0565] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in a method of treating cancer in combination with a PD-1 inhibitor or PD-L1 inhibitor after resecting the tumor from the patient.
[0566] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer in combination with a PD-1 inhibitor or PD-L1 inhibitor after resecting the tumor from the patient, wherein the PD-1 inhibitor or PD-L1 inhibitor is selected from the group consisting of nivolumab, pembrolizumab, durvalumab, atezolizumab, avelumab, and fragments, derivatives, variants, biosimilars, and combinations thereof.
[0567] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer in combination with a PD-1 inhibitor or PD-L1 inhibitor, wherein the PD-1 or PD-L1 inhibitor is for administration after administering the third population of TILs and/or a pharmaceutical composition comprising the third population of TILs to the patient.
[0568] In an embodiment, the population of TILs and/or the pharmaceutical composition is for use in treating cancer in combination with a PD-1 inhibitor or PD-L1 inhibitor which is for administration after administering the third population of TILs to the patient, wherein the PD-1 inhibitor or PD-L1 inhibitor is selected from the group consisting of nivolumab, pembrolizumab, durvalumab, atezolizumab, avelumab, and fragments, derivatives, variants, biosimilars, and combinations thereof. Further details of the PD-1 inhibitor and the PD-L1 inhibitor are described herein e.g. under the heading "Combinations with PD-1 and PD-L1 Inhibitors". In some embodiments, the population of TILs and/or the pharmaceutical composition comprising a population of TILs further comprise one or more features as described herein, for example, under the headings "Pharmaceutical Compositions, Dosages, and Dosing Regimens for TILs" and "Pharmaceutical Compositions, Dosages, and Dosing Regimens for TNFRSF Agonists".
BRIEF DESCRIPTION OF THE DRAWINGS
[0569] The foregoing summary, as well as the following detailed description of the invention, will be better understood when read in conjunction with the appended drawings.
[0570] FIG. 1 illustrates a TIL expansion and treatment process. TNFRSF agonists of the present disclosure may be used in both the pre-REP stage (top half of figure) or REP stage (bottom half of figure) and may be added when IL-2 is added to each cell culture. Step 1 refers to the addition of 4 tumor fragments into 10 G-Rex 10 flasks. At step 2, approximately 40.times.10.sup.6 TILs or greater are obtained. At step 3, a split occurs into 36 G-Rex 100 flasks for REP. TILs are harvested by centrifugation at step 4. Fresh TIL product is obtained at step 5 after a total process time of approximate 43 days, at which point TILs may be infused into a patient.
[0571] FIG. 2 illustrates a treatment protocol for use with TILs expanded with TNFRSF agonists of the present disclosure. Surgery (and tumor resection) occurs at the start, and lymphodepletion chemo refers to non-myeloablative lymphodepletion with chemotherapy as described elsewhere herein. TNFRSF agonists of the present disclosure may also be used during therapy as described herein after administration of TILs.
[0572] FIG. 3 illustrates the results of an assay to determine if 4-1BB-Fc hybridoma 4B5 activates 4-1BB signaling on Jurkat cells expressing NF-kB using a green fluorescent protein (GFP) reporter in a dose dependent manner. "Secondary" refers to activation of a secondary antibody.
[0573] FIG. 4 illustrates the results of an assay to determine if 4-1BB-Fc hybridoma 1C4 activates 4-1BB signaling on Jurkat cells expressing NF-kB using a GFP reporter in a dose dependent manner. "Secondary" refers to activation of a secondary antibody.
[0574] FIG. 5 illustrates the results of an assay to determine if 4-1BB-Fc hybridoma 9B4 activates 4-1BB signaling on Jurkat cells expressing NF-kB using a GFP reporter in a dose dependent manner. "Secondary" refers to activation of a secondary antibody.
[0575] FIG. 6 illustrates the results of an assay to determine if 4-1BB-Fc hybridoma 1D7 activates 4-1BB signaling on Jurkat cells expressing NF-kB using a GFP reporter in a dose dependent manner. "Secondary" refers to activation of a secondary antibody.
[0576] FIG. 7 illustrates the results of an assay to determine if 4-1BB-Fc hybridoma 1D10 activates 4-1BB signaling on Jurkat cells expressing NF-kB using a GFP reporter in a dose dependent manner. "Secondary" refers to activation of a secondary antibody.
[0577] FIG. 8 illustrates the results of an assay to determine if 4-1BB-Fc hybridoma 3C2 activates 4-1BB signaling on Jurkat cells expressing NF-kB using a GFP reporter in a dose dependent manner. "Secondary" refers to activation of a secondary antibody.
[0578] FIG. 9 illustrates the results of an assay to determine if 4-1BB-Fc hybridoma 10D12 activates 4-1BB signaling on Jurkat cells expressing NF-kB using a GFP reporter in a dose dependent manner. "Secondary" refers to activation of a secondary antibody.
[0579] FIG. 10 illustrates the results of an assay to determine if 4-1BB-Fc hybridoma 8D2 activates 4-1BB signaling on Jurkat cells expressing NF-kB using a GFP reporter in a dose dependent manner. "Secondary" refers to activation of a secondary antibody.
[0580] FIG. 11 illustrates the results of an assay to determine if 4-1BB-Fc hybridoma 4G6 activates 4-1BB signaling on Jurkat cells expressing NF-kB using a GFP reporter in a dose dependent manner. "Secondary" refers to activation of a secondary antibody.
[0581] FIG. 12 illustrates the results of an assay to determine if 4-1BB-Fc hybridoma 8E3 activates 4-1BB signaling on Jurkat cells expressing NF-kB using a GFP reporter in a dose dependent manner. "Secondary" refers to activation of a secondary antibody.
[0582] FIG. 13 illustrates an exemplary TIL expansion and manufacturing protocol (Process 2A).
[0583] FIG. 14 illustrates exemplary method steps undertaken in Process 2A.
[0584] FIG. 15 illustrates an exemplary TIL expansion protocol.
[0585] FIG. 16 illustrates binding affinity for Creative Biolabs (CB) and BPS Biosciences (BPS) 4-1BB agonist antibodies as assessed by percentage of 4-1BB+ cells by flow cytometry. CB 4-1BB agonist exhibited the highest binding affinity.
[0586] FIG. 17 illustrates binding affinity for Creative Biolabs (CB) and BPS Biosciences (BPS) 4-1BB agonist antibodies as assessed by mean fluorescence intensity (MFI). CB 4-1BB agonist exhibited the highest binding affinity.
[0587] FIG. 18 illustrates the results of an assessment of NF-.kappa.B pathway activation of anti-4-1BB agonistic antibodies.
[0588] FIG. 19 illustrates binding affinity for Creative Biolabs OX40 agonist antibody as assessed by percentage of OX40.sup.+ cells by flow cytometry.
[0589] FIG. 20 illustrates binding affinity for Creative Biolabs OX40 agonist antibodies as assessed by mean fluorescence intensity (MFI).
[0590] FIG. 21 illustrates comparable binding affinity between Creative Biolabs anti-OX40 agonist antibody (at five concentrations shown) and a commercial anti-OX40 (clone Ber-ACT35) agonist. The first letter of each tumor designation indicates histology: C=cervical; H=head and neck (head and neck squamous cell carcinoma); L=lung; and M=melanoma.
[0591] FIG. 22 illustrates the results of an assessment of NF-.kappa.B pathway activation of anti-OX40 agonist antibody. OX40 reporter cells were treated with either anti-OX40 alone or Isotype control at the concentrations of 1, 2, 4, 8, and 16 .mu.g/mL with or without PBMC feeder cells for 24 hours. The cells were lysed using One-Step Luciferase reagent, and luciferase activity was measured by luminometer.
[0592] FIG. 23 illustrates the experimental design for 4-1BB and OX40 agonist experiments during pre-REP.
[0593] FIG. 24 illustrates the tumor histologies used in the experimental design of FIG. 23.
[0594] FIG. 25 illustrates the data analysis strategy used to assess the impact of 4-1BB and anti-OX40 agonists used during pre-REP on TIL performance and properties.
[0595] FIG. 26 illustrates total cell count results for cell expansion using CB 4-1BB agonist (N=3). NT=not tested (control). The p value was >0.99.
[0596] FIG. 27 illustrates total cell count results for cell expansion using CB OX40 agonist (N=5). NT=not tested (control). The p value was 0.06.
[0597] FIG. 28 illustrates total cell count results for cell expansion using CB 4-1BB agonist and OX-40 agonist (N=2). NT=not tested (control).
[0598] FIG. 29 illustrates total CD8.sup.+ cell count results for cell expansion using CB 4-1BB agonist (N=3). The p value was 0.5.
[0599] FIG. 30 illustrates total CD8.sup.+ cell count results for cell expansion using CB OX40 agonist (N=5). The p value was 0.03.
[0600] FIG. 31 illustrates total CD8.sup.+ cell count results for cell expansion using CB 4-1BB agonist and OX-40 agonist (N=2). NT=not tested (control).
[0601] FIG. 32 illustrates total CD8.sup.+/CD4.sup.+ cell count ratio results for cell expansion using CB 4-1BB agonist (N=3). The p value was 0.2.
[0602] FIG. 33 illustrates total CD8.sup.+/CD4.sup.+ cell count ratio results for cell expansion using CB OX40 agonist (N=5). The p value was 0.12.
[0603] FIG. 34 illustrates total CD8.sup.+/CD4.sup.+ cell count ratio results for cell expansion using CB 4-1BB agonist and OX-40 agonist (N=2). NT=not tested (control).
[0604] FIG. 35 illustrates the experimental scheme for REP propagation of pre-REP TILs expanded in the presence of 4-1BB or OX40 agonists.
[0605] FIG. 36 illustrates fold expansion of TILs expanded in REP from pre-REP TILs expanded in the presence of CB 4-1BB agonist versus TILs not treated in the pre-REP (NT).
[0606] FIG. 37 illustrates fold expansion of TILs expanded in REP from pre-REP TILs expanded in the presence of CB OX40 agonist versus TILs not treated in the pre-REP (NT).
[0607] FIG. 38 illustrates fold expansion of TILs expanded in REP from pre-REP TILs expanded in the presence of CB 4-1BB agonist and CB OX40 agonist versus TILs not treated in the pre-REP (NT).
[0608] FIG. 39 illustrates the histologies of twenty-one TIL lines used for assessment of CB OX40 agonist during the REP phase.
[0609] FIG. 40 illustrates the experimental scheme for assessment of CB OX40 agonist during the REP phase.
[0610] FIG. 41 illustrates that the presence of an OX40 agonistic antibody preferentially expands CD8.sup.+ TIL during REP (shown as a percentage of CD3+CD4.sup.+ cells).
[0611] FIG. 42 illustrates that the presence of an OX40 agonistic antibody preferentially expands CD8.sup.+ TIL during REP (shown as a percentage of CD3+CD8.sup.+ cells).
[0612] FIG. 43 illustrates that in non-responder TIL lines, down-regulation of OX40 was not observed in CD4.sup.+ subset following anti-OX40 treatment.
[0613] FIG. 44 illustrates experimental details for CB OX40 agonist dose titration in non-responder and responder TIL lines.
[0614] FIG. 45 illustrates the results of CB OX40 agonist dose titration in responder TIL lines.
[0615] FIG. 46 illustrates the results of CB OX40 agonist dose titration in non-responder TIL lines.
[0616] FIG. 47 illustrates comparable TCRvb repertoire profiles for responder L4005.
[0617] FIG. 48 illustrates comparable TCRvb repertoire profiles for responder H3005.
[0618] FIG. 49 illustrates comparable TCRvb repertoire profiles for responder M1022.
[0619] FIG. 50 illustrates the structures I-A and I-B, the cylinders refer to individual polypeptide binding domains. Structures I-A and I-B comprise three linearly-linked TNFRSF binding domains derived from e.g., 4-1BBL or an antibody that binds 4-1BB, which fold to form a trivalent protein, which is then linked to a second trivalent protein through IgG1-Fc (including CH3 and CH2 domains) is then used to link two of the trivalent proteins together through disulfide bonds (small elongated ovals), stabilizing the structure and providing an agonists capable of bringing together the intracellular signaling domains of the six receptors and signaling proteins to form a signaling complex. The TNFRSF binding domains denoted as cylinders may be scFv domains comprising, e.g., a VH and a VL chain connected by a linker that may comprise hydrophilic residues and Gly and Ser sequences for flexibility, as well as Glu and Lys for solubility.
BRIEF DESCRIPTION OF THE SEQUENCE LISTING
[0620] SEQ ID NO: 1 is the amino acid sequence of the heavy chain of muromonab.
[0621] SEQ ID NO:2 is the amino acid sequence of the light chain of muromonab.
[0622] SEQ ID NO:3 is the amino acid sequence of a recombinant human IL-2 protein.
[0623] SEQ ID NO:4 is the amino acid sequence of aldesleukin.
[0624] SEQ ID NO:5 is the amino acid sequence of a recombinant human IL-4 protein.
[0625] SEQ ID NO:6 is the amino acid sequence of a recombinant human IL-7 protein.
[0626] SEQ ID NO:7 is the amino acid sequence of a recombinant human IL-15 protein.
[0627] SEQ ID NO:8 is the amino acid sequence of a recombinant human IL-21 protein.
[0628] SEQ ID NO:9 is the amino acid sequence of human 4-1BB.
[0629] SEQ ID NO:10 is the amino acid sequence of murine 4-1BB.
[0630] SEQ ID NO: 11 is the heavy chain for the 4-1BB agonist monoclonal antibody utomilumab (PF-05082566).
[0631] SEQ ID NO: 12 is the light chain for the 4-1BB agonist monoclonal antibody utomilumab (PF-05082566).
[0632] SEQ ID NO: 13 is the heavy chain variable region (V.sub.H) for the 4-1BB agonist monoclonal antibody utomilumab (PF-05082566).
[0633] SEQ ID NO: 14 is the light chain variable region (V.sub.L) for the 4-1BB agonist monoclonal antibody utomilumab (PF-05082566).
[0634] SEQ ID NO: 15 is the heavy chain CDR1 for the 4-1BB agonist monoclonal antibody utomilumab (PF-05082566).
[0635] SEQ ID NO: 16 is the heavy chain CDR2 for the 4-1BB agonist monoclonal antibody utomilumab (PF-05082566).
[0636] SEQ ID NO: 17 is the heavy chain CDR3 for the 4-1BB agonist monoclonal antibody utomilumab (PF-05082566).
[0637] SEQ ID NO: 18 is the light chain CDR1 for the 4-1BB agonist monoclonal antibody utomilumab (PF-05082566).
[0638] SEQ ID NO: 19 is the light chain CDR2 for the 4-1BB agonist monoclonal antibody utomilumab (PF-05082566).
[0639] SEQ ID NO:20 is the light chain CDR3 for the 4-1BB agonist monoclonal antibody utomilumab (PF-05082566).
[0640] SEQ ID NO:21 is the heavy chain for the 4-1BB agonist monoclonal antibody urelumab (BMS-663513).
[0641] SEQ ID NO:22 is the light chain for the 4-1BB agonist monoclonal antibody urelumab (BMS-663513).
[0642] SEQ ID NO:23 is the heavy chain variable region (V.sub.H) for the 4-1BB agonist monoclonal antibody urelumab (BMS-663513).
[0643] SEQ ID NO:24 is the light chain variable region (V.sub.L) for the 4-1BB agonist monoclonal antibody urelumab (BMS-663513).
[0644] SEQ ID NO:25 is the heavy chain CDR1 for the 4-1BB agonist monoclonal antibody urelumab (BMS-663513).
[0645] SEQ ID NO:26 is the heavy chain CDR2 for the 4-1BB agonist monoclonal antibody urelumab (BMS-663513).
[0646] SEQ ID NO:27 is the heavy chain CDR3 for the 4-1BB agonist monoclonal antibody urelumab (BMS-663513).
[0647] SEQ ID NO:28 is the light chain CDR1 for the 4-1BB agonist monoclonal antibody urelumab (BMS-663513).
[0648] SEQ ID NO:29 is the light chain CDR2 for the 4-1BB agonist monoclonal antibody urelumab (BMS-663513).
[0649] SEQ ID NO:30 is the light chain CDR3 for the 4-1BB agonist monoclonal antibody urelumab (BMS-663513).
[0650] SEQ ID NO:31 is an Fc domain for a TNFRSF agonist fusion protein.
[0651] SEQ ID NO:32 is a linker for a TNFRSF agonist fusion protein.
[0652] SEQ ID NO:33 is a linker for a TNFRSF agonist fusion protein.
[0653] SEQ ID NO:34 is a linker for a TNFRSF agonist fusion protein.
[0654] SEQ ID NO:35 is a linker for a TNFRSF agonist fusion protein.
[0655] SEQ ID NO:36 is a linker for a TNFRSF agonist fusion protein.
[0656] SEQ ID NO:37 is a linker for a TNFRSF agonist fusion protein.
[0657] SEQ ID NO:38 is a linker for a TNFRSF agonist fusion protein.
[0658] SEQ ID NO:39 is a linker for a TNFRSF agonist fusion protein.
[0659] SEQ ID NO:40 is a linker for a TNFRSF agonist fusion protein.
[0660] SEQ ID NO:41 is a linker for a TNFRSF agonist fusion protein.
[0661] SEQ ID NO:42 is an Fc domain for a TNFRSF agonist fusion protein.
[0662] SEQ ID NO:43 is a linker for a TNFRSF agonist fusion protein.
[0663] SEQ ID NO:44 is a linker for a TNFRSF agonist fusion protein.
[0664] SEQ ID NO:45 is a linker for a TNFRSF agonist fusion protein.
[0665] SEQ ID NO:46 is a 4-1BB ligand (4-1BBL) amino acid sequence.
[0666] SEQ ID NO:47 is a soluble portion of 4-1BBL polypeptide.
[0667] SEQ ID NO:48 is a heavy chain variable region (V.sub.H) for the 4-1BB agonist antibody 4B4-1-1 version 1.
[0668] SEQ ID NO:49 is a light chain variable region (V.sub.L) for the 4-1BB agonist antibody 4B4-1-1 version 1.
[0669] SEQ ID NO:50 is a heavy chain variable region (V.sub.H) for the 4-1BB agonist antibody 4B4-1-1 version 2.
[0670] SEQ ID NO:51 is a light chain variable region (V.sub.L) for the 4-1BB agonist antibody 4B4-1-1 version 2.
[0671] SEQ ID NO:52 is a heavy chain variable region (V.sub.H) for the 4-1BB agonist antibody H39E3-2.
[0672] SEQ ID NO:53 is a light chain variable region (V.sub.L) for the 4-1BB agonist antibody H39E3-2.
[0673] SEQ ID NO:54 is the amino acid sequence of human OX40.
[0674] SEQ ID NO:55 is the amino acid sequence of murine OX40.
[0675] SEQ ID NO:56 is the heavy chain for the OX40 agonist monoclonal antibody tavolixizumab (MEDI-0562).
[0676] SEQ ID NO:57 is the light chain for the OX40 agonist monoclonal antibody tavolixizumab (MEDI-0562).
[0677] SEQ ID NO:58 is the heavy chain variable region (V.sub.H) for the OX40 agonist monoclonal antibody tavolixizumab (MEDI-0562).
[0678] SEQ ID NO:59 is the light chain variable region (V.sub.L) for the OX40 agonist monoclonal antibody tavolixizumab (MEDI-0562).
[0679] SEQ ID NO:60 is the heavy chain CDR1 for the OX40 agonist monoclonal antibody tavolixizumab (MEDI-0562).
[0680] SEQ ID NO:61 is the heavy chain CDR2 for the OX40 agonist monoclonal antibody tavolixizumab (MEDI-0562).
[0681] SEQ ID NO:62 is the heavy chain CDR3 for the OX40 agonist monoclonal antibody tavolixizumab (MEDI-0562).
[0682] SEQ ID NO:63 is the light chain CDR1 for the OX40 agonist monoclonal antibody tavolixizumab (MEDI-0562).
[0683] SEQ ID NO:64 is the light chain CDR2 for the OX40 agonist monoclonal antibody tavolixizumab (MEDI-0562).
[0684] SEQ ID NO:65 is the light chain CDR3 for the OX40 agonist monoclonal antibody tavolixizumab (MEDI-0562).
[0685] SEQ ID NO:66 is the heavy chain for the OX40 agonist monoclonal antibody 11D4.
[0686] SEQ ID NO:67 is the light chain for the OX40 agonist monoclonal antibody 11D4.
[0687] SEQ ID NO:68 is the heavy chain variable region (V.sub.H) for the OX40 agonist monoclonal antibody 11D4.
[0688] SEQ ID NO:69 is the light chain variable region (V.sub.L) for the OX40 agonist monoclonal antibody 11D4.
[0689] SEQ ID NO:70 is the heavy chain CDR1 for the OX40 agonist monoclonal antibody 11D4.
[0690] SEQ ID NO:71 is the heavy chain CDR2 for the OX40 agonist monoclonal antibody 11D4.
[0691] SEQ ID NO:72 is the heavy chain CDR3 for the OX40 agonist monoclonal antibody 11D4.
[0692] SEQ ID NO:73 is the light chain CDR1 for the OX40 agonist monoclonal antibody 11D4.
[0693] SEQ ID NO:74 is the light chain CDR2 for the OX40 agonist monoclonal antibody 11D4.
[0694] SEQ ID NO:75 is the light chain CDR3 for the OX40 agonist monoclonal antibody 11D4.
[0695] SEQ ID NO:76 is the heavy chain for the OX40 agonist monoclonal antibody 18D8.
[0696] SEQ ID NO:77 is the light chain for the OX40 agonist monoclonal antibody 18D8.
[0697] SEQ ID NO:78 is the heavy chain variable region (V.sub.H) for the OX40 agonist monoclonal antibody 18D8.
[0698] SEQ ID NO:79 is the light chain variable region (V.sub.L) for the OX40 agonist monoclonal antibody 18D8.
[0699] SEQ ID NO:80 is the heavy chain CDR1 for the OX40 agonist monoclonal antibody 18D8.
[0700] SEQ ID NO:81 is the heavy chain CDR2 for the OX40 agonist monoclonal antibody 18D8.
[0701] SEQ ID NO:82 is the heavy chain CDR3 for the OX40 agonist monoclonal antibody 18D8.
[0702] SEQ ID NO:83 is the light chain CDR1 for the OX40 agonist monoclonal antibody 18D8.
[0703] SEQ ID NO:84 is the light chain CDR2 for the OX40 agonist monoclonal antibody 18D8.
[0704] SEQ ID NO:85 is the light chain CDR3 for the OX40 agonist monoclonal antibody 18D8.
[0705] SEQ ID NO:86 is the heavy chain variable region (V.sub.H) for the OX40 agonist monoclonal antibody Hu119-122.
[0706] SEQ ID NO:87 is the light chain variable region (V.sub.L) for the OX40 agonist monoclonal antibody Hu119-122.
[0707] SEQ ID NO:88 is the heavy chain CDR1 for the OX40 agonist monoclonal antibody Hu119-122.
[0708] SEQ ID NO:89 is the heavy chain CDR2 for the OX40 agonist monoclonal antibody Hu119-122.
[0709] SEQ ID NO:90 is the heavy chain CDR3 for the OX40 agonist monoclonal antibody Hu119-122.
[0710] SEQ ID NO:91 is the light chain CDR1 for the OX40 agonist monoclonal antibody Hu119-122.
[0711] SEQ ID NO:92 is the light chain CDR2 for the OX40 agonist monoclonal antibody Hu119-122.
[0712] SEQ ID NO:93 is the light chain CDR3 for the OX40 agonist monoclonal antibody Hu119-122.
[0713] SEQ ID NO:94 is the heavy chain variable region (V.sub.H) for the OX40 agonist monoclonal antibody Hu106-222.
[0714] SEQ ID NO:95 is the light chain variable region (V.sub.L) for the OX40 agonist monoclonal antibody Hu106-222.
[0715] SEQ ID NO:96 is the heavy chain CDR1 for the OX40 agonist monoclonal antibody Hu106-222.
[0716] SEQ ID NO:97 is the heavy chain CDR2 for the OX40 agonist monoclonal antibody Hu106-222.
[0717] SEQ ID NO:98 is the heavy chain CDR3 for the OX40 agonist monoclonal antibody Hu106-222.
[0718] SEQ ID NO:99 is the light chain CDR1 for the OX40 agonist monoclonal antibody Hu106-222.
[0719] SEQ ID NO: 100 is the light chain CDR2 for the OX40 agonist monoclonal antibody Hu106-222.
[0720] SEQ ID NO: 101 is the light chain CDR3 for the OX40 agonist monoclonal antibody Hu106-222.
[0721] SEQ ID NO:102 is an OX40 ligand (OX40L) amino acid sequence.
[0722] SEQ ID NO:103 is a soluble portion of OX40L polypeptide.
[0723] SEQ ID NO:104 is an alternative soluble portion of OX40L polypeptide.
[0724] SEQ ID NO: 105 is the heavy chain variable region (V.sub.H) for the OX40 agonist monoclonal antibody 008.
[0725] SEQ ID NO: 106 is the light chain variable region (V.sub.L) for the OX40 agonist monoclonal antibody 008.
[0726] SEQ ID NO: 107 is the heavy chain variable region (V.sub.H) for the OX40 agonist monoclonal antibody 011.
[0727] SEQ ID NO: 108 is the light chain variable region (V.sub.L) for the OX40 agonist monoclonal antibody 011.
[0728] SEQ ID NO: 109 is the heavy chain variable region (V.sub.H) for the OX40 agonist monoclonal antibody 021.
[0729] SEQ ID NO: 110 is the light chain variable region (V.sub.L) for the OX40 agonist monoclonal antibody 021.
[0730] SEQ ID NO: 111 is the heavy chain variable region (V.sub.H) for the OX40 agonist monoclonal antibody 023.
[0731] SEQ ID NO: 112 is the light chain variable region (V.sub.L) for the OX40 agonist monoclonal antibody 023.
[0732] SEQ ID NO: 113 is the heavy chain variable region (V.sub.H) for an OX40 agonist monoclonal antibody.
[0733] SEQ ID NO: 114 is the light chain variable region (V.sub.L) for an OX40 agonist monoclonal antibody.
[0734] SEQ ID NO: 115 is the heavy chain variable region (V.sub.H) for an OX40 agonist monoclonal antibody.
[0735] SEQ ID NO: 116 is the light chain variable region (V.sub.L) for an OX40 agonist monoclonal antibody.
[0736] SEQ ID NO: 117 is the heavy chain variable region (V.sub.H) for a humanized OX40 agonist monoclonal antibody.
[0737] SEQ ID NO: 118 is the heavy chain variable region (V.sub.H) for a humanized OX40 agonist monoclonal antibody.
[0738] SEQ ID NO: 119 is the light chain variable region (V.sub.L) for a humanized OX40 agonist monoclonal antibody.
[0739] SEQ ID NO: 120 is the light chain variable region (V.sub.L) for a humanized OX40 agonist monoclonal antibody.
[0740] SEQ ID NO: 121 is the heavy chain variable region (V.sub.H) for a humanized OX40 agonist monoclonal antibody.
[0741] SEQ ID NO: 122 is the heavy chain variable region (V.sub.H) for a humanized OX40 agonist monoclonal antibody.
[0742] SEQ ID NO: 123 is the light chain variable region (V.sub.L) for a humanized OX40 agonist monoclonal antibody.
[0743] SEQ ID NO: 124 is the light chain variable region (V.sub.L) for a humanized OX40 agonist monoclonal antibody.
[0744] SEQ ID NO: 125 is the heavy chain variable region (V.sub.H) for an OX40 agonist monoclonal antibody.
[0745] SEQ ID NO: 126 is the light chain variable region (V.sub.L) for an OX40 agonist monoclonal antibody.
[0746] SEQ ID NO:127 is the amino acid sequence of human CD27.
[0747] SEQ ID NO:128 is the amino acid sequence of macaque CD27.
[0748] SEQ ID NO: 129 is the heavy chain for the CD27 agonist monoclonal antibody varlilumab (CDX-1127).
[0749] SEQ ID NO: 130 is the light chain for the CD27 agonist monoclonal antibody varlilumab (CDX-1127).
[0750] SEQ ID NO: 131 is the heavy chain variable region (V.sub.H) for the CD27 agonist monoclonal antibody varlilumab (CDX-1127).
[0751] SEQ ID NO: 132 is the light chain variable region (V.sub.L) for the CD27 agonist monoclonal antibody varlilumab (CDX-1127).
[0752] SEQ ID NO: 133 is the heavy chain CDR1 for the CD27 agonist monoclonal antibody varlilumab (CDX-1127).
[0753] SEQ ID NO: 134 is the heavy chain CDR2 for the CD27 agonist monoclonal antibody varlilumab (CDX-1127).
[0754] SEQ ID NO: 135 is the heavy chain CDR3 for the CD27 agonist monoclonal antibody varlilumab (CDX-1127).
[0755] SEQ ID NO: 136 is the light chain CDR1 for the CD27 agonist monoclonal antibody varlilumab (CDX-1127).
[0756] SEQ ID NO: 137 is the light chain CDR2 for the CD27 agonist monoclonal antibody varlilumab (CDX-1127).
[0757] SEQ ID NO: 138 is the light chain CDR3 for the CD27 agonist monoclonal antibody varlilumab (CDX-1127).
[0758] SEQ ID NO:139 is an CD27 ligand (CD70) amino acid sequence.
[0759] SEQ ID NO:140 is a soluble portion of CD70 polypeptide.
[0760] SEQ ID NO:141 is an alternative soluble portion of CD70 polypeptide.
[0761] SEQ ID NO:142 is the amino acid sequence of human GITR (human tumor necrosis factor receptor superfamily member 18 (TNFRSF 18) protein).
[0762] SEQ ID NO:143 is the amino acid sequence of murine GITR (murine tumor necrosis factor receptor superfamily member 18 (TNFRSF 18) protein).
[0763] SEQ ID NO: 144 is the amino acid sequence of the heavy chain variant HuN6C8 (glycosylated) of the 6C8 humanized GITR agonist monoclonal antibody, with an N (asparagine) in CDR2, corresponding to SEQ ID NO:60 in U.S. Pat. No. 7,812,135.
[0764] SEQ ID NO: 145 is the amino acid sequence of the heavy chain variant HuN6C8 (aglycosylated) of the 6C8 humanized GITR agonist monoclonal antibody, with an N (asparagine) in CDR2, corresponding to SEQ ID NO:61 in U.S. Pat. No. 7,812,135.
[0765] SEQ ID NO: 146 is the amino acid sequence of the heavy chain variant HuQ6C8 (glycosylated) of the 6C8 humanized GITR agonist monoclonal antibody, with an Q (glutamine) in CDR2, corresponding to SEQ ID NO:62 in U.S. Pat. No. 7,812,135.
[0766] SEQ ID NO: 147 is the amino acid sequence of the heavy chain variant HuQ6C8 (aglycosylated) of the 6C8 humanized GITR agonist monoclonal antibody, with an Q (glutamine) in CDR2, corresponding to SEQ ID NO:63 in U.S. Pat. No. 7,812,135.
[0767] SEQ ID NO:148 is the amino acid sequence of the light chain of the 6C8 humanized GITR agonist monoclonal antibody, corresponding to SEQ ID NO:58 in U.S. Pat. No. 7,812,135.
[0768] SEQ ID NO: 149 is the amino acid sequence of the leader sequence that may optionally be included with the amino acid sequences of SEQ ID NO:144, SEQ ID NO:145, SEQ ID NO: 146, or SEQ ID NO: 147 in GITR agonist monoclonal antibodies.
[0769] SEQ ID NO:150 is the amino acid sequence of the leader sequence that may optionally be included with the amino acid sequence of SEQ ID NO: 148 in GITR agonist monoclonal antibodies.
[0770] SEQ ID NO:151 is the amino acid sequence of the heavy chain variable region of the 6C8 humanized GITR agonist monoclonal antibody, corresponding to SEQ ID NO: 1 in U.S. Pat. No. 7,812,135.
[0771] SEQ ID NO:152 is the amino acid sequence of the heavy chain variable region of the 6C8 humanized GITR agonist monoclonal antibody, corresponding to SEQ ID NO:66 in U.S. Pat. No. 7,812,135.
[0772] SEQ ID NO:153 is the amino acid sequence of the light chain variable region of the 6C8 humanized GITR agonist monoclonal antibody, corresponding to SEQ ID NO:2 in U.S. Pat. No. 7,812,135.
[0773] SEQ ID NO:154 is the amino acid sequence of the heavy chain CDR1 region of the 6C8 humanized GITR agonist monoclonal antibody, corresponding to SEQ ID NO:3 in U.S. Pat. No. 7,812,135.
[0774] SEQ ID NO:155 is the amino acid sequence of the heavy chain CDR2 region of the 6C8 humanized GITR agonist monoclonal antibody, corresponding to SEQ ID NO:4 in U.S. Pat. No. 7,812,135.
[0775] SEQ ID NO:156 is the amino acid sequence of the heavy chain CDR2 region of the 6C8 humanized GITR agonist monoclonal antibody, corresponding to SEQ ID NO: 19 in U.S. Pat. No. 7,812,135.
[0776] SEQ ID NO:157 is the amino acid sequence of the heavy chain CDR3 region of the 6C8 humanized GITR agonist monoclonal antibody, corresponding to SEQ ID NO:5 in U.S. Pat. No. 7,812,135.
[0777] SEQ ID NO:158 is the amino acid sequence of the heavy chain CDR1 region of the 6C8 humanized GITR agonist monoclonal antibody, corresponding to SEQ ID NO:6 in U.S. Pat. No. 7,812,135.
[0778] SEQ ID NO:159 is the amino acid sequence of the heavy chain CDR2 region of the 6C8 humanized GITR agonist monoclonal antibody, corresponding to SEQ ID NO:7 in U.S. Pat. No. 7,812,135.
[0779] SEQ ID NO: 160 is the amino acid sequence of the heavy chain CDR3 region of the 6C8 humanized GITR agonist monoclonal antibody, corresponding to SEQ ID NO:8 in U.S. Pat. No. 7,812,135.
[0780] SEQ ID NO: 161 is the amino acid sequence of the heavy chain variant HuN6C8 (glycosylated) of the 6C8 chimeric GITR agonist monoclonal antibody, with an N (asparagine) in CDR2, corresponding to SEQ ID NO:23 in U.S. Pat. No. 7,812,135.
[0781] SEQ ID NO: 162 is the amino acid sequence of the heavy chain variant HuQ6C8 (aglycosylated) of the 6C8 chimeric GITR agonist monoclonal antibody, with an Q (glutamine) in CDR2, corresponding to SEQ ID NO:24 in U.S. Pat. No. 7,812,135.
[0782] SEQ ID NO: 163 is the amino acid sequence of the light chain of the 6C8 chimeric GITR agonist monoclonal antibody, corresponding to SEQ ID NO:22 in U.S. Pat. No. 7,812,135.
[0783] SEQ ID NO: 164 is the amino acid sequence of the GITR agonist 36E5 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0784] SEQ ID NO: 165 is the amino acid sequence of the GITR agonist 36E5 light chain variable region from U.S. Pat. No. 8,709,424.
[0785] SEQ ID NO: 166 is the amino acid sequence of the GITR agonist 3D6 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0786] SEQ ID NO: 167 is the amino acid sequence of the GITR agonist 3D6 light chain variable region from U.S. Pat. No. 8,709,424.
[0787] SEQ ID NO: 168 is the amino acid sequence of the GITR agonist 61G6 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0788] SEQ ID NO: 169 is the amino acid sequence of the GITR agonist 61G6 light chain variable region from U.S. Pat. No. 8,709,424.
[0789] SEQ ID NO: 170 is the amino acid sequence of the GITR agonist 6H6 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0790] SEQ ID NO: 171 is the amino acid sequence of the GITR agonist 6H6 light chain variable region from U.S. Pat. No. 8,709,424.
[0791] SEQ ID NO: 172 is the amino acid sequence of the GITR agonist 61F6 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0792] SEQ ID NO:173 is the amino acid sequence of the GITR agonist 61F6 light chain variable region from U.S. Pat. No. 8,709,424.
[0793] SEQ ID NO: 174 is the amino acid sequence of the GITR agonist 1D8 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0794] SEQ ID NO: 175 is the amino acid sequence of the GITR agonist 1D8 light chain variable region from U.S. Pat. No. 8,709,424.
[0795] SEQ ID NO: 176 is the amino acid sequence of the GITR agonist 17F10 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0796] SEQ ID NO: 177 is the amino acid sequence of the GITR agonist 17F10 light chain variable region from U.S. Pat. No. 8,709,424.
[0797] SEQ ID NO: 178 is the amino acid sequence of the GITR agonist 35D8 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0798] SEQ ID NO: 179 is the amino acid sequence of the GITR agonist 35D8 light chain variable region from U.S. Pat. No. 8,709,424.
[0799] SEQ ID NO:180 is the amino acid sequence of the GITR agonist 49A1 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0800] SEQ ID NO:181 is the amino acid sequence of the GITR agonist 49A1 light chain variable region from U.S. Pat. No. 8,709,424.
[0801] SEQ ID NO:182 is the amino acid sequence of the GITR agonist 9E5 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0802] SEQ ID NO:183 is the amino acid sequence of the GITR agonist 9E5 light chain variable region from U.S. Pat. No. 8,709,424.
[0803] SEQ ID NO:184 is the amino acid sequence of the GITR agonist 31H6 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0804] SEQ ID NO:185 is the amino acid sequence of the GITR agonist 31H6 light chain variable region from U.S. Pat. No. 8,709,424.
[0805] SEQ ID NO:186 is the amino acid sequence of the humanized GITR agonist 36E5 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0806] SEQ ID NO:187 is the amino acid sequence of the humanized GITR agonist 36E5 light chain variable region from U.S. Pat. No. 8,709,424.
[0807] SEQ ID NO:188 is the amino acid sequence of the humanized GITR agonist 3D6 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0808] SEQ ID NO:189 is the amino acid sequence of the humanized GITR agonist 3D6 light chain variable region from U.S. Pat. No. 8,709,424.
[0809] SEQ ID NO: 190 is the amino acid sequence of the humanized GITR agonist 61G6 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0810] SEQ ID NO: 191 is the amino acid sequence of the humanized GITR agonist 61G6 light chain variable region from U.S. Pat. No. 8,709,424.
[0811] SEQ ID NO: 192 is the amino acid sequence of the humanized GITR agonist 6H6 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0812] SEQ ID NO: 193 is the amino acid sequence of the humanized GITR agonist 6H6 light chain variable region from U.S. Pat. No. 8,709,424.
[0813] SEQ ID NO: 194 is the amino acid sequence of the humanized GITR agonist 61F6 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0814] SEQ ID NO: 195 is the amino acid sequence of the humanized GITR agonist 61F6 light chain variable region from U.S. Pat. No. 8,709,424.
[0815] SEQ ID NO: 196 is the amino acid sequence of the humanized GITR agonist 1D8 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0816] SEQ ID NO: 197 is the amino acid sequence of the humanized GITR agonist 1D8 light chain variable region from U.S. Pat. No. 8,709,424.
[0817] SEQ ID NO:198 is the amino acid sequence of the humanized GITR agonist 17F10 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0818] SEQ ID NO:199 is the amino acid sequence of the humanized GITR agonist 17F10 light chain variable region from U.S. Pat. No. 8,709,424.
[0819] SEQ ID NO:200 is the amino acid sequence of the humanized GITR agonist 35D8 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0820] SEQ ID NO:201 is the amino acid sequence of the humanized GITR agonist 35D8 light chain variable region from U.S. Pat. No. 8,709,424.
[0821] SEQ ID NO:202 is the amino acid sequence of the humanized GITR agonist 49A1 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0822] SEQ ID NO:203 is the amino acid sequence of the humanized GITR agonist 49A1 light chain variable region from U.S. Pat. No. 8,709,424.
[0823] SEQ ID NO:204 is the amino acid sequence of the humanized GITR agonist 9E5 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0824] SEQ ID NO:205 is the amino acid sequence of the humanized GITR agonist 9E5 light chain variable region from U.S. Pat. No. 8,709,424.
[0825] SEQ ID NO:206 is the amino acid sequence of the humanized GITR agonist 31H6 heavy chain variable region from U.S. Pat. No. 8,709,424.
[0826] SEQ ID NO:207 is the amino acid sequence of the humanized GITR agonist 31H6 light chain variable region from U.S. Pat. No. 8,709,424.
[0827] SEQ ID NO:208 is the amino acid sequence of the GITR agonist 2155 variable heavy chain from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0828] SEQ ID NO:209 is the amino acid sequence of the GITR agonist 2155 variable light chain from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0829] SEQ ID NO:210 is the amino acid sequence of the GITR agonist 2155 humanized (HC1) heavy chain from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0830] SEQ ID NO:211 is the amino acid sequence of the GITR agonist 2155 humanized (HC2) heavy chain from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0831] SEQ ID NO:212 is the amino acid sequence of the GITR agonist 2155 humanized (HC3a) heavy chain from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0832] SEQ ID NO:213 is the amino acid sequence of the humanized (HC3b) GITR agonist heavy chain from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0833] SEQ ID NO:214 is the amino acid sequence of the humanized (HC4) GITR agonist heavy chain from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0834] SEQ ID NO:215 is the amino acid sequence of the 2155 humanized (LC1) GITR agonist light chain from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0835] SEQ ID NO:216 is the amino acid sequence of the 2155 humanized (LC2a) GITR agonist light chain from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0836] SEQ ID NO:217 is the amino acid sequence of the 2155 humanized (LC2b) GITR agonist light chain from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0837] SEQ ID NO:218 is the amino acid sequence of the 2155 humanized (LC3) GITR agonist light chain from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0838] SEQ ID NO:219 is the amino acid sequence of the GITR agonist 698 variable heavy chain from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0839] SEQ ID NO:220 is the amino acid sequence of the GITR agonist 698 variable light chain from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0840] SEQ ID NO:221 is the amino acid sequence of the GITR agonist 706 variable heavy chain from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0841] SEQ ID NO:222 is the amino acid sequence of the GITR agonist 706 variable light chain from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0842] SEQ ID NO:223 is the amino acid sequence of the GITR agonist 827 variable heavy chain from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0843] SEQ ID NO:224 is the amino acid sequence of the GITR agonist 827 variable light chain from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0844] SEQ ID NO:225 is the amino acid sequence of the GITR agonist 1718 variable heavy chain from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0845] SEQ ID NO:226 is the amino acid sequence of the GITR agonist 1718 variable light chain from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0846] SEQ ID NO:227 is the amino acid sequence of the GITR agonist 2155 heavy chain CDR3 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0847] SEQ ID NO:228 is the amino acid sequence of the GITR agonist 2155 heavy chain CDR2 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0848] SEQ ID NO:229 is the amino acid sequence of the GITR agonist 2155 heavy chain CDR1 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0849] SEQ ID NO:230 is the amino acid sequence of the GITR agonist 2155 light chain CDR3 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0850] SEQ ID NO:231 is the amino acid sequence of the GITR agonist 2155 light chain CDR2 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0851] SEQ ID NO:232 is the amino acid sequence of the GITR agonist 2155 light chain CDR1 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0852] SEQ ID NO:233 is the amino acid sequence of the GITR agonists 698 and 706 heavy chain CDR3 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0853] SEQ ID NO:234 is the amino acid sequence of the GITR agonists 698 and 706 heavy chain CDR2 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0854] SEQ ID NO:235 is the amino acid sequence of the GITR agonists 698 and 706 heavy chain CDR1 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0855] SEQ ID NO:236 is the amino acid sequence of the GITR agonist 698 light chain CDR3 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0856] SEQ ID NO:237 is the amino acid sequence of the GITR agonists 698, 706, 827, and 1649 light chain CDR2 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0857] SEQ ID NO:238 is the amino acid sequence of the GITR agonists 698, 706, 827, and 1649 light chain CDR1 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0858] SEQ ID NO:239 is the amino acid sequence of the GITR agonists 706, 827, and 1649 light chain CDR3 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0859] SEQ ID NO:240 is the amino acid sequence of the GITR agonists 827 and 1649 heavy chain CDR3 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0860] SEQ ID NO:241 is the amino acid sequence of the GITR agonist 827 heavy chain CDR2 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0861] SEQ ID NO:242 is the amino acid sequence of the GITR agonist 1649 heavy chain CDR2 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0862] SEQ ID NO:243 is the amino acid sequence of the GITR agonist 1718 heavy chain CDR3 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0863] SEQ ID NO:244 is the amino acid sequence of the GITR agonist 1718 heavy chain CDR2 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0864] SEQ ID NO:245 is the amino acid sequence of the GITR agonist 1718 heavy chain CDR1 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0865] SEQ ID NO:246 is the amino acid sequence of the GITR agonist 1718 light chain CDR3 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0866] SEQ ID NO:247 is the amino acid sequence of the GITR agonist 1718 light chain CDR2 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0867] SEQ ID NO:248 is the amino acid sequence of the GITR agonist 1718 light chain CDR1 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0868] SEQ ID NO:249 is the amino acid sequence of the GITR agonists 827 and 1649 heavy chain CDR1 from U.S. Patent Application Publication No. US 2013/0108641 A1.
[0869] SEQ ID NO:250 is the amino acid sequence of the GITR agonist 1D7 heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0870] SEQ ID NO:251 is the amino acid sequence of the GITR agonist 1D7 light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0871] SEQ ID NO:252 is the amino acid sequence of the GITR agonist 1D7 variable heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0872] SEQ ID NO:253 is the amino acid sequence of the GITR agonist 1D7 variable light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0873] SEQ ID NO:254 is the amino acid sequence of the GITR agonist 1D7 heavy chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0874] SEQ ID NO:255 is the amino acid sequence of the GITR agonist 1D7 heavy chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0875] SEQ ID NO:256 is the amino acid sequence of the GITR agonist 1D7 heavy chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0876] SEQ ID NO:257 is the amino acid sequence of the GITR agonist 1D7 light chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0877] SEQ ID NO:258 is the amino acid sequence of the GITR agonist 1D7 light chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0878] SEQ ID NO:259 is the amino acid sequence of the GITR agonist 1D7 light chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0879] SEQ ID NO:260 is the amino acid sequence of the GITR agonist 33C9 heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0880] SEQ ID NO:261 is the amino acid sequence of the GITR agonist 33C9 light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0881] SEQ ID NO:262 is the amino acid sequence of the GITR agonist 33C9 variable heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0882] SEQ ID NO:263 is the amino acid sequence of the GITR agonist 33C9 variable light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0883] SEQ ID NO:264 is the amino acid sequence of the GITR agonist 33C9 heavy chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0884] SEQ ID NO:265 is the amino acid sequence of the GITR agonist 33C9 heavy chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0885] SEQ ID NO:266 is the amino acid sequence of the GITR agonist 33C9 heavy chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0886] SEQ ID NO:267 is the amino acid sequence of the GITR agonist 33C9 light chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0887] SEQ ID NO:268 is the amino acid sequence of the GITR agonist 33C9 light chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0888] SEQ ID NO:269 is the amino acid sequence of the GITR agonist 33C9 light chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0889] SEQ ID NO:270 is the amino acid sequence of the GITR agonist 33F6 heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0890] SEQ ID NO:271 is the amino acid sequence of the GITR agonist 33F6 light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0891] SEQ ID NO:272 is the amino acid sequence of the GITR agonist 33F6 variable heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0892] SEQ ID NO:273 is the amino acid sequence of the GITR agonist 33F6 variable light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0893] SEQ ID NO:274 is the amino acid sequence of the GITR agonist 33F6 heavy chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0894] SEQ ID NO:275 is the amino acid sequence of the GITR agonist 33F6 heavy chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0895] SEQ ID NO:276 is the amino acid sequence of the GITR agonist 33F6 heavy chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0896] SEQ ID NO:277 is the amino acid sequence of the GITR agonist 33F6 light chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0897] SEQ ID NO:278 is the amino acid sequence of the GITR agonist 33F6 light chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0898] SEQ ID NO:279 is the amino acid sequence of the GITR agonist 33F6 light chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0899] SEQ ID NO:280 is the amino acid sequence of the GITR agonist 34G4 heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0900] SEQ ID NO:281 is the amino acid sequence of the GITR agonist 34G4 light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0901] SEQ ID NO:282 is the amino acid sequence of the GITR agonist 34G4 variable heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0902] SEQ ID NO:283 is the amino acid sequence of the GITR agonist 34G4 variable light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0903] SEQ ID NO:284 is the amino acid sequence of the GITR agonist 34G4 heavy chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0904] SEQ ID NO:285 is the amino acid sequence of the GITR agonist 34G4 heavy chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0905] SEQ ID NO:286 is the amino acid sequence of the GITR agonist 34G4 heavy chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0906] SEQ ID NO:287 is the amino acid sequence of the GITR agonist 34G4 light chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0907] SEQ ID NO:288 is the amino acid sequence of the GITR agonist 34G4 light chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0908] SEQ ID NO:289 is the amino acid sequence of the GITR agonist 34G4 light chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0909] SEQ ID NO:290 is the amino acid sequence of the GITR agonist 35B10 heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0910] SEQ ID NO:291 is the amino acid sequence of the GITR agonist 35B10 light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0911] SEQ ID NO:292 is the amino acid sequence of the GITR agonist 35B10 variable heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0912] SEQ ID NO:293 is the amino acid sequence of the GITR agonist 35B10 variable light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0913] SEQ ID NO:294 is the amino acid sequence of the GITR agonist 35B10 heavy chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0914] SEQ ID NO:295 is the amino acid sequence of the GITR agonist 35B10 heavy chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0915] SEQ ID NO:296 is the amino acid sequence of the GITR agonist 35B10 heavy chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0916] SEQ ID NO:297 is the amino acid sequence of the GITR agonist 35B10 light chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0917] SEQ ID NO:298 is the amino acid sequence of the GITR agonist 35B10 light chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0918] SEQ ID NO:299 is the amino acid sequence of the GITR agonist 35B10 light chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0919] SEQ ID NO:300 is the amino acid sequence of the GITR agonist 41E11 heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0920] SEQ ID NO:301 is the amino acid sequence of the GITR agonist 41E11 light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0921] SEQ ID NO:302 is the amino acid sequence of the GITR agonist 41E11 variable heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0922] SEQ ID NO:303 is the amino acid sequence of the GITR agonist 41E11 variable light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0923] SEQ ID NO:304 is the amino acid sequence of the GITR agonist 41E11 heavy chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0924] SEQ ID NO:305 is the amino acid sequence of the GITR agonist 41E11 heavy chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0925] SEQ ID NO:306 is the amino acid sequence of the GITR agonist 41E11 heavy chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0926] SEQ ID NO:307 is the amino acid sequence of the GITR agonist 41E11 light chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0927] SEQ ID NO:308 is the amino acid sequence of the GITR agonist 41E11 light chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0928] SEQ ID NO:309 is the amino acid sequence of the GITR agonist 41E11 light chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0929] SEQ ID NO:310 is the amino acid sequence of the GITR agonist 41G5 heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0930] SEQ ID NO:311 is the amino acid sequence of the GITR agonist 41G5 light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0931] SEQ ID NO:312 is the amino acid sequence of the GITR agonist 41G5 variable heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0932] SEQ ID NO:313 is the amino acid sequence of the GITR agonist 41G5 variable light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0933] SEQ ID NO:314 is the amino acid sequence of the GITR agonist 41G5 heavy chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0934] SEQ ID NO:315 is the amino acid sequence of the GITR agonist 41G5 heavy chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0935] SEQ ID NO:316 is the amino acid sequence of the GITR agonist 41G5 heavy chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0936] SEQ ID NO:317 is the amino acid sequence of the GITR agonist 41G5 light chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0937] SEQ ID NO:318 is the amino acid sequence of the GITR agonist 41G5 light chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0938] SEQ ID NO:319 is the amino acid sequence of the GITR agonist 41G5 light chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0939] SEQ ID NO:320 is the amino acid sequence of the GITR agonist 42A11 heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0940] SEQ ID NO:321 is the amino acid sequence of the GITR agonist 42A11 light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0941] SEQ ID NO:322 is the amino acid sequence of the GITR agonist 42A11 variable heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0942] SEQ ID NO:323 is the amino acid sequence of the GITR agonist 42A11 variable light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0943] SEQ ID NO:324 is the amino acid sequence of the GITR agonist 42A11 heavy chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0944] SEQ ID NO:325 is the amino acid sequence of the GITR agonist 42A11 heavy chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0945] SEQ ID NO:326 is the amino acid sequence of the GITR agonist 42A11 heavy chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0946] SEQ ID NO:327 is the amino acid sequence of the GITR agonist 42A11 light chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0947] SEQ ID NO:328 is the amino acid sequence of the GITR agonist 42A11 light chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0948] SEQ ID NO:329 is the amino acid sequence of the GITR agonist 42A11 light chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0949] SEQ ID NO:330 is the amino acid sequence of the GITR agonist 44C1 heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0950] SEQ ID NO:331 is the amino acid sequence of the GITR agonist 44C1 light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0951] SEQ ID NO:332 is the amino acid sequence of the GITR agonist 44C1 variable heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0952] SEQ ID NO:333 is the amino acid sequence of the GITR agonist 44C1 variable light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0953] SEQ ID NO:334 is the amino acid sequence of the GITR agonist 44C1 heavy chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0954] SEQ ID NO:335 is the amino acid sequence of the GITR agonist 44C1 heavy chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0955] SEQ ID NO:336 is the amino acid sequence of the GITR agonist 44C1 heavy chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0956] SEQ ID NO:337 is the amino acid sequence of the GITR agonist 44C1 light chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0957] SEQ ID NO:338 is the amino acid sequence of the GITR agonist 44C1 light chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0958] SEQ ID NO:339 is the amino acid sequence of the GITR agonist 44C1 light chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0959] SEQ ID NO:340 is the amino acid sequence of the GITR agonist 45A8 heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0960] SEQ ID NO:341 is the amino acid sequence of the GITR agonist 45A8 light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0961] SEQ ID NO:342 is the amino acid sequence of the GITR agonist 45A8 variable heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0962] SEQ ID NO:343 is the amino acid sequence of the GITR agonist 45A8 variable light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0963] SEQ ID NO:344 is the amino acid sequence of the GITR agonist 45A8 heavy chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0964] SEQ ID NO:345 is the amino acid sequence of the GITR agonist 45A8 heavy chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0965] SEQ ID NO:346 is the amino acid sequence of the GITR agonist 45A8 heavy chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0966] SEQ ID NO:347 is the amino acid sequence of the GITR agonist 45A8 light chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0967] SEQ ID NO:348 is the amino acid sequence of the GITR agonist 45A8 light chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0968] SEQ ID NO:349 is the amino acid sequence of the GITR agonist 45A8 light chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0969] SEQ ID NO:350 is the amino acid sequence of the GITR agonist 46E11 heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0970] SEQ ID NO:351 is the amino acid sequence of the GITR agonist 46E11 light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0971] SEQ ID NO:352 is the amino acid sequence of the GITR agonist 46E11 variable heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0972] SEQ ID NO:353 is the amino acid sequence of the GITR agonist 46E11 variable light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0973] SEQ ID NO:354 is the amino acid sequence of the GITR agonist 46E11 heavy chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0974] SEQ ID NO:355 is the amino acid sequence of the GITR agonist 46E11 heavy chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0975] SEQ ID NO:356 is the amino acid sequence of the GITR agonist 46E11 heavy chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0976] SEQ ID NO:357 is the amino acid sequence of the GITR agonist 46E11 light chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0977] SEQ ID NO:358 is the amino acid sequence of the GITR agonist 46E11 light chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0978] SEQ ID NO:359 is the amino acid sequence of the GITR agonist 46E11 light chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0979] SEQ ID NO:360 is the amino acid sequence of the GITR agonist 48H12 heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0980] SEQ ID NO:361 is the amino acid sequence of the GITR agonist 48H12 light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0981] SEQ ID NO:362 is the amino acid sequence of the GITR agonist 48H12 variable heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0982] SEQ ID NO:363 is the amino acid sequence of the GITR agonist 48H12 variable light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0983] SEQ ID NO:364 is the amino acid sequence of the GITR agonist 48H12 heavy chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0984] SEQ ID NO:365 is the amino acid sequence of the GITR agonist 48H12 heavy chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0985] SEQ ID NO:366 is the amino acid sequence of the GITR agonist 48H12 heavy chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0986] SEQ ID NO:367 is the amino acid sequence of the GITR agonist 48H12 light chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0987] SEQ ID NO:368 is the amino acid sequence of the GITR agonist 48H12 light chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0988] SEQ ID NO:369 is the amino acid sequence of the GITR agonist 48H12 light chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0989] SEQ ID NO:370 is the amino acid sequence of the GITR agonist 48H7 heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0990] SEQ ID NO:371 is the amino acid sequence of the GITR agonist 48H7 light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0991] SEQ ID NO:372 is the amino acid sequence of the GITR agonist 48H7 variable heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0992] SEQ ID NO:373 is the amino acid sequence of the GITR agonist 48H7 variable light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0993] SEQ ID NO:374 is the amino acid sequence of the GITR agonist 48H7 heavy chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0994] SEQ ID NO:375 is the amino acid sequence of the GITR agonist 48H7 heavy chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0995] SEQ ID NO:376 is the amino acid sequence of the GITR agonist 48H7 heavy chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0996] SEQ ID NO:377 is the amino acid sequence of the GITR agonist 48H7 light chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0997] SEQ ID NO:378 is the amino acid sequence of the GITR agonist 48H7 light chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0998] SEQ ID NO:379 is the amino acid sequence of the GITR agonist 48H7 light chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[0999] SEQ ID NO:380 is the amino acid sequence of the GITR agonist 49D9 heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1000] SEQ ID NO:381 is the amino acid sequence of the GITR agonist 49D9 light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1001] SEQ ID NO:382 is the amino acid sequence of the GITR agonist 49D9 variable heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1002] SEQ ID NO:383 is the amino acid sequence of the GITR agonist 49D9 variable light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1003] SEQ ID NO:384 is the amino acid sequence of the GITR agonist 49D9 heavy chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1004] SEQ ID NO:385 is the amino acid sequence of the GITR agonist 49D9 heavy chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1005] SEQ ID NO:386 is the amino acid sequence of the GITR agonist 49D9 heavy chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1006] SEQ ID NO:387 is the amino acid sequence of the GITR agonist 49D9 light chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1007] SEQ ID NO:388 is the amino acid sequence of the GITR agonist 49D9 light chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1008] SEQ ID NO:389 is the amino acid sequence of the GITR agonist 49D9 light chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1009] SEQ ID NO:390 is the amino acid sequence of the GITR agonist 49E2 heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1010] SEQ ID NO:391 is the amino acid sequence of the GITR agonist 49E2 light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1011] SEQ ID NO:392 is the amino acid sequence of the GITR agonist 49E2 variable heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1012] SEQ ID NO:393 is the amino acid sequence of the GITR agonist 49E2 variable light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1013] SEQ ID NO:394 is the amino acid sequence of the GITR agonist 49E2 heavy chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1014] SEQ ID NO:395 is the amino acid sequence of the GITR agonist 49E2 heavy chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1015] SEQ ID NO:396 is the amino acid sequence of the GITR agonist 49E2 heavy chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1016] SEQ ID NO:397 is the amino acid sequence of the GITR agonist 49E2 light chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1017] SEQ ID NO:398 is the amino acid sequence of the GITR agonist 49E2 light chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1018] SEQ ID NO:399 is the amino acid sequence of the GITR agonist 49E2 light chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1019] SEQ ID NO:400 is the amino acid sequence of the GITR agonist 48A9 heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1020] SEQ ID NO:401 is the amino acid sequence of the GITR agonist 48A9 light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1021] SEQ ID NO:402 is the amino acid sequence of the GITR agonist 48A9 variable heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1022] SEQ ID NO:403 is the amino acid sequence of the GITR agonist 48A9 variable light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1023] SEQ ID NO:404 is the amino acid sequence of the GITR agonist 48A9 heavy chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1024] SEQ ID NO:405 is the amino acid sequence of the GITR agonist 48A9 heavy chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1025] SEQ ID NO:406 is the amino acid sequence of the GITR agonist 48A9 heavy chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1026] SEQ ID NO:407 is the amino acid sequence of the GITR agonist 48A9 light chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1027] SEQ ID NO:408 is the amino acid sequence of the GITR agonist 48A9 light chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1028] SEQ ID NO:409 is the amino acid sequence of the GITR agonist 48A9 light chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1029] SEQ ID NO:410 is the amino acid sequence of the GITR agonist 5H7 heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1030] SEQ ID NO:411 is the amino acid sequence of the GITR agonist 5H7 light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1031] SEQ ID NO:412 is the amino acid sequence of the GITR agonist 5H7 variable heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1032] SEQ ID NO:413 is the amino acid sequence of the GITR agonist 5H7 variable light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1033] SEQ ID NO:414 is the amino acid sequence of the GITR agonist 5H7 heavy chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1034] SEQ ID NO:415 is the amino acid sequence of the GITR agonist 5H7 heavy chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1035] SEQ ID NO:416 is the amino acid sequence of the GITR agonist 5H7 heavy chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1036] SEQ ID NO:417 is the amino acid sequence of the GITR agonist 5H7 light chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1037] SEQ ID NO:418 is the amino acid sequence of the GITR agonist 5H7 light chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1038] SEQ ID NO:419 is the amino acid sequence of the GITR agonist 5H7 light chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1039] SEQ ID NO:420 is the amino acid sequence of the GITR agonist 7A10 heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1040] SEQ ID NO:421 is the amino acid sequence of the GITR agonist 7A10 light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1041] SEQ ID NO:422 is the amino acid sequence of the GITR agonist 7A10 variable heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1042] SEQ ID NO:423 is the amino acid sequence of the GITR agonist 7A10 variable light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1043] SEQ ID NO:424 is the amino acid sequence of the GITR agonist 7A10 heavy chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1044] SEQ ID NO:425 is the amino acid sequence of the GITR agonist 7A10 heavy chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1045] SEQ ID NO:426 is the amino acid sequence of the GITR agonist 7A10 heavy chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1046] SEQ ID NO:427 is the amino acid sequence of the GITR agonist 7A10 light chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1047] SEQ ID NO:428 is the amino acid sequence of the GITR agonist 7A10 light chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1048] SEQ ID NO:429 is the amino acid sequence of the GITR agonist 7A10 light chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1049] SEQ ID NO:430 is the amino acid sequence of the GITR agonist 9H6 heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1050] SEQ ID NO:431 is the amino acid sequence of the GITR agonist 9H6 light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1051] SEQ ID NO:432 is the amino acid sequence of the GITR agonist 9H6 variable heavy chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1052] SEQ ID NO:433 is the amino acid sequence of the GITR agonist 9H6 variable light chain from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1053] SEQ ID NO:434 is the amino acid sequence of the GITR agonist 9H6 heavy chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1054] SEQ ID NO:435 is the amino acid sequence of the GITR agonist 9H6 heavy chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1055] SEQ ID NO:436 is the amino acid sequence of the GITR agonist 9H6 heavy chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1056] SEQ ID NO:437 is the amino acid sequence of the GITR agonist 9H6 light chain CDR1 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1057] SEQ ID NO:438 is the amino acid sequence of the GITR agonist 9H6 light chain CDR2 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1058] SEQ ID NO:439 is the amino acid sequence of the GITR agonist 9H6 light chain CDR3 from U.S. Patent Application Publication No. US 2015/0064204 A1.
[1059] SEQ ID NO:440 is an GITR ligand (GITRL) amino acid sequence.
[1060] SEQ ID NO:441 is a soluble portion of GITRL polypeptide.
[1061] SEQ ID NO:442 is the amino acid sequence of human HVEM (CD270).
[1062] SEQ ID NO:443 is a HVEM ligand (LIGHT) amino acid sequence.
[1063] SEQ ID NO:444 is a soluble portion of LIGHT polypeptide.
[1064] SEQ ID NO:445 is an alternative soluble portion of LIGHT polypeptide.
[1065] SEQ ID NO:446 is an alternative soluble portion of LIGHT polypeptide.
[1066] SEQ ID NO:447 is the amino acid sequence of human CD95 isoform 1.
[1067] SEQ ID NO:448 is the amino acid sequence of human CD95 isoform 2.
[1068] SEQ ID NO:449 is the amino acid sequence of human CD95 isoform 3.
[1069] SEQ ID NO:450 is the amino acid sequence of human CD95 isoform 4.
[1070] SEQ ID NO:451 is the heavy chain variable region (V.sub.H) for the CD95 agonist monoclonal antibody E09.
[1071] SEQ ID NO:452 is the light chain variable region (V.sub.L) for the CD95 agonist monoclonal antibody E09.
[1072] SEQ ID NO:453 is the heavy chain CDR1 for the CD95 agonist monoclonal antibody E09.
[1073] SEQ ID NO:454 is the heavy chain CDR2 for the CD95 agonist monoclonal antibody E09.
[1074] SEQ ID NO:455 is the heavy chain CDR3 for the CD95 agonist monoclonal antibody E09.
[1075] SEQ ID NO:456 is the light chain CDR1 for the CD95 agonist monoclonal antibody E09.
[1076] SEQ ID NO:457 is the light chain CDR2 for the CD95 agonist monoclonal antibody E09.
[1077] SEQ ID NO:458 is the light chain CDR3 for the CD95 agonist monoclonal antibody E09.
[1078] SEQ ID NO:459 is a CD95 ligand (CD95L) amino acid sequence.
[1079] SEQ ID NO:460 is a soluble portion of CD95L polypeptide.
[1080] SEQ ID NO:461 is an alternative soluble portion of CD95L polypeptide.
[1081] SEQ ID NO:462 is an alternative soluble portion of CD95L polypeptide.
[1082] SEQ ID NO:463 is the heavy chain amino acid sequence of the PD-1 inhibitor nivolumab.
[1083] SEQ ID NO:464 is the light chain amino acid sequence of the PD-1 inhibitor nivolumab.
[1084] SEQ ID NO:465 is the heavy chain variable region (V.sub.H) amino acid sequence of the PD-1 inhibitor nivolumab.
[1085] SEQ ID NO:466 is the light chain variable region (V.sub.L) amino acid sequence of the PD-1 inhibitor nivolumab.
[1086] SEQ ID NO:467 is the heavy chain CDR1 amino acid sequence of the PD-1 inhibitor nivolumab.
[1087] SEQ ID NO:468 is the heavy chain CDR2 amino acid sequence of the PD-1 inhibitor nivolumab.
[1088] SEQ ID NO:469 is the heavy chain CDR3 amino acid sequence of the PD-1 inhibitor nivolumab.
[1089] SEQ ID NO:470 is the light chain CDR1 amino acid sequence of the PD-1 inhibitor nivolumab.
[1090] SEQ ID NO:471 is the light chain CDR2 amino acid sequence of the PD-1 inhibitor nivolumab.
[1091] SEQ ID NO:472 is the light chain CDR3 amino acid sequence of the PD-1 inhibitor nivolumab.
[1092] SEQ ID NO:473 is the heavy chain amino acid sequence of the PD-1 inhibitor pembrolizumab.
[1093] SEQ ID NO:474 is the light chain amino acid sequence of the PD-1 inhibitor pembrolizumab.
[1094] SEQ ID NO:475 is the heavy chain variable region (V.sub.H) amino acid sequence of the PD-1 inhibitor pembrolizumab.
[1095] SEQ ID NO:476 is the light chain variable region (V.sub.L) amino acid sequence of the PD-1 inhibitor pembrolizumab.
[1096] SEQ ID NO:477 is the heavy chain CDR1 amino acid sequence of the PD-1 inhibitor pembrolizumab.
[1097] SEQ ID NO:478 is the heavy chain CDR2 amino acid sequence of the PD-1 inhibitor pembrolizumab.
[1098] SEQ ID NO:479 is the heavy chain CDR3 amino acid sequence of the PD-1 inhibitor pembrolizumab.
[1099] SEQ ID NO:480 is the light chain CDR1 amino acid sequence of the PD-1 inhibitor pembrolizumab.
[1100] SEQ ID NO:481 is the light chain CDR2 amino acid sequence of the PD-1 inhibitor pembrolizumab.
[1101] SEQ ID NO:482 is the light chain CDR3 amino acid sequence of the PD-1 inhibitor pembrolizumab.
[1102] SEQ ID NO:483 is the heavy chain amino acid sequence of the PD-L1 inhibitor durvalumab.
[1103] SEQ ID NO:484 is the light chain amino acid sequence of the PD-L1 inhibitor durvalumab.
[1104] SEQ ID NO:485 is the heavy chain variable region (V.sub.H) amino acid sequence of the PD-L1 inhibitor durvalumab.
[1105] SEQ ID NO:486 is the light chain variable region (V.sub.L) amino acid sequence of the PD-L1 inhibitor durvalumab.
[1106] SEQ ID NO:487 is the heavy chain CDR1 amino acid sequence of the PD-L1 inhibitor durvalumab.
[1107] SEQ ID NO:488 is the heavy chain CDR2 amino acid sequence of the PD-L1 inhibitor durvalumab.
[1108] SEQ ID NO:489 is the heavy chain CDR3 amino acid sequence of the PD-L1 inhibitor durvalumab.
[1109] SEQ ID NO:490 is the light chain CDR1 amino acid sequence of the PD-L1 inhibitor durvalumab.
[1110] SEQ ID NO:491 is the light chain CDR2 amino acid sequence of the PD-L1 inhibitor durvalumab.
[1111] SEQ ID NO:492 is the light chain CDR3 amino acid sequence of the PD-L1 inhibitor durvalumab.
[1112] SEQ ID NO:493 is the heavy chain amino acid sequence of the PD-L1 inhibitor avelumab.
[1113] SEQ ID NO:494 is the light chain amino acid sequence of the PD-L1 inhibitor avelumab.
[1114] SEQ ID NO:495 is the heavy chain variable region (V.sub.H) amino acid sequence of the PD-L1 inhibitor avelumab.
[1115] SEQ ID NO:496 is the light chain variable region (V.sub.L) amino acid sequence of the PD-L1 inhibitor avelumab.
[1116] SEQ ID NO:497 is the heavy chain CDR1 amino acid sequence of the PD-L1 inhibitor avelumab.
[1117] SEQ ID NO:498 is the heavy chain CDR2 amino acid sequence of the PD-L1 inhibitor avelumab.
[1118] SEQ ID NO:499 is the heavy chain CDR3 amino acid sequence of the PD-L1 inhibitor avelumab.
[1119] SEQ ID NO:500 is the light chain CDR1 amino acid sequence of the PD-L1 inhibitor avelumab.
[1120] SEQ ID NO:501 is the light chain CDR2 amino acid sequence of the PD-L1 inhibitor avelumab.
[1121] SEQ ID NO:502 is the light chain CDR3 amino acid sequence of the PD-L1 inhibitor avelumab.
[1122] SEQ ID NO:503 is the heavy chain amino acid sequence of the PD-L1 inhibitor atezolizumab.
[1123] SEQ ID NO:504 is the light chain amino acid sequence of the PD-L1 inhibitor atezolizumab.
[1124] SEQ ID NO:505 is the heavy chain variable region (V.sub.H) amino acid sequence of the PD-L1 inhibitor atezolizumab.
[1125] SEQ ID NO:506 is the light chain variable region (V.sub.L) amino acid sequence of the PD-L1 inhibitor atezolizumab.
[1126] SEQ ID NO:507 is the heavy chain CDR1 amino acid sequence of the PD-L1 inhibitor atezolizumab.
[1127] SEQ ID NO:508 is the heavy chain CDR2 amino acid sequence of the PD-L1 inhibitor atezolizumab.
[1128] SEQ ID NO:509 is the heavy chain CDR3 amino acid sequence of the PD-L1 inhibitor atezolizumab.
[1129] SEQ ID NO:510 is the light chain CDR1 amino acid sequence of the PD-L1 inhibitor atezolizumab.
[1130] SEQ ID NO:511 is the light chain CDR2 amino acid sequence of the PD-L1 inhibitor atezolizumab.
[1131] SEQ ID NO:512 is the light chain CDR3 amino acid sequence of the PD-L1 inhibitor atezolizumab.
DETAILED DESCRIPTION OF THE INVENTION
[1132] Unless defined otherwise, all technical and scientific terms used herein have the same meaning as is commonly understood by one of skill in the art to which this invention belongs. All patents and publications referred to herein are incorporated by reference in their entireties.
Definitions
[1133] The terms "co-administration," "co-administering," "administered in combination with," "administering in combination with," "simultaneous," and "concurrent," as used herein, encompass administration of two or more active pharmaceutical ingredients (in a preferred embodiment of the present invention, for example, at least one TNFRSF agonist and a plurality of TILs) to a subject so that both active pharmaceutical ingredients and/or their metabolites are present in the subject at the same time. Co-administration includes simultaneous administration in separate compositions, administration at different times in separate compositions, or administration in a composition in which two or more active pharmaceutical ingredients are present. Simultaneous administration in separate compositions and administration in a composition in which both agents are present are preferred.
[1134] The term "rapid expansion" means an increase in the number of antigen-specific TILs of at least about 3-fold (or 4-, 5-, 6-, 7-, 8-, or 9-fold) over a period of a week, more preferably at least about 10-fold (or 20-, 30-, 40-, 50-, 60-, 70-, 80-, or 90-fold) over a period of a week, or most preferably at least about 100-fold over a period of a week. A number of rapid expansion protocols are described herein.
[1135] By "tumor infiltrating lymphocytes" or "TILs" herein is meant a population of cells originally obtained as white blood cells that have left the bloodstream of a subject and migrated into a tumor. TILs include, but are not limited to, CD8.sup.+ cytotoxic T cells (lymphocytes), Th1 and Th17 CD4.sup.+ T cells, natural killer cells, dendritic cells and M1 macrophages. TILs include both primary and secondary TILs. "Primary TILs" are those that are obtained from patient tissue samples as outlined herein (sometimes referred to as "freshly harvested"), and "secondary TILs" are any TIL cell populations that have been expanded or proliferated as discussed herein, including, but not limited to bulk TILs and expanded TILs ("REP TILs" or "post-REP TILs").
[1136] By "population of cells" (including TILs) herein is meant a number of cells that share common traits. In general, populations generally range from 1.times.10.sup.6 to 1.times.10.sup.10 in number, with different TIL populations comprising different numbers. For example, initial growth of primary TILs in the presence of IL-2 results in a population of bulk TILs of roughly 1.times.10.sup.8 cells. REP expansion is generally done to provide populations of 1.5.times.10.sup.9 to 1.5.times.10.sup.10 cells for infusion.
[1137] The term "central memory T cell" refers to a subset of T cells that in the human are CD45R0+ and constitutively express CCR7 (CCR7.sup.hi) and CD62L (CD62.sup.hi). The surface phenotype of central memory T cells also includes TCR, CD3, CD127 (IL-7R), and IL-15R. Transcription factors for central memory T cells include BCL-6, BCL-6B, MBD2, and BMI1. Central memory T cells primarily secret IL-2 and CD40L as effector molecules after TCR triggering. Central memory T cells are predominant in the CD4 compartment in blood, and in the human are proportionally enriched in lymph nodes and tonsils.
[1138] The term "anti-CD3 antibody" refers to an antibody or variant thereof, e.g., a monoclonal antibody and including human, humanized, chimeric or murine antibodies which are directed against the CD3 receptor in the T cell antigen receptor of mature T cells. Anti-CD3 antibodies include OKT-3, also known as muromonab. Anti-CD3 antibodies also include the UHCT1 clone, also known as T3 and CD3c. Other anti-CD3 antibodies include, for example, otelixizumab, teplizumab, and visilizumab.
[1139] The term "OKT-3" (also referred to herein as "OKT3") refers to a monoclonal antibody or biosimilar or variant thereof, including human, humanized, chimeric, or murine antibodies, directed against the CD3 receptor in the T cell antigen receptor of mature T cells, and includes commercially-available forms such as OKT-3 (30 ng/mL, MACS GMP CD3 pure, Miltenyi Biotech, Inc., San Diego, Calif., USA) and muromonab or variants, conservative amino acid substitutions, glycoforms, or biosimilars thereof. The amino acid sequences of the heavy and light chains of muromonab are given in Table 1 (SEQ ID NO:1 and SEQ ID NO:2).
TABLE-US-00001 TABLE 1 Amino acid sequences of muromonab. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 1 QVQLQQSGAE LARPGASVKM SCKASGYTFT RYTMHWVKQR PGQGLEWIGY INPSRGYTNY 60 Muromonab NQKFKDKATL TTDKSSSTAY MQLSSLTSED SAVYYCARYY DDHYCLDYWG QGTTLTVSSA 120 heavy KTTAPSVYPL APVCGGTTGS SVTLGCLVKG YFPEPVTLTW NSGSLSSGVH TFPAVLQSDL 180 chain YTLSSSVTVT SSTWPSQSIT CNVAHPASST KVDKKIEPRP KSCDKTHTCP PCPAPELLGG 240 PSVFLFPPKP KDTLMISRTP EVTCVVVDVS HEDPEVKFNW YVDGVEVHNA KTKPREEQYN 300 STYRVVSVLT VLHQDWLNGK EYKCKVSNKA LPAPIEKTIS KAKGQPREPQ VYTLPPSRDE 360 LTKNQVSLTC LVKGFYPSDI AVEWESNGQP ENNYKTTPPV LDSDGSFFLY SKLTVDKSRW 420 QQGNVFSCSV MHEALHNHYT QKSLSLSPGK 450 SEQ ID NO: 2 QIVLTQSPAI MSASPGEKVT MTCSASSSVS YMNWYQQKSG TSPKRWIYDT SKLASGVPAH 60 Muromonab FRGSGSGTSY SLTISGMEAE DAATYYCQQW SSNPFTFGSG TKLEINRADT APTVSIFPPS 120 light SEQLTSGGAS VVCFLNNFYP KDINVKWKID GSERQNGVLN SWTDQDSKDS TYSMSSTLTL 180 chain TKDEYERHNS YTCEATHKTS TSPIVKSFNR NEC 213
[1140] The term "IL-2" (also referred to herein as "IL2") refers to the T cell growth factor known as interleukin-2, and includes all forms of IL-2 including human and mammalian forms, conservative amino acid substitutions, glycoforms, biosimilars, and variants thereof. IL-2 is described, e.g., in Nelson, J. Immunol. 2004, 172, 3983-88 and Malek, Annu. Rev. Immunol. 2008, 26, 453-79, the disclosures of which are incorporated by reference herein. The amino acid sequence of recombinant human IL-2 suitable for use in the invention is given in Table 2 (SEQ ID NO:3). For example, the term IL-2 encompasses human, recombinant forms of IL-2 such as aldesleukin (PROLEUKIN, available commercially from multiple suppliers in 22 million IU per single use vials), as well as the form of recombinant IL-2 commercially supplied by CellGenix, Inc., Portsmouth, N.H., USA (CELLGRO GMP) or ProSpec-Tany TechnoGene Ltd., East Brunswick, N.J., USA (Cat. No. CYT-209-b) and other commercial equivalents from other vendors. Aldesleukin (des-alanyl-1, serine-125 human IL-2) is a nonglycosylated human recombinant form of IL-2 with a molecular weight of approximately 15 kDa. The amino acid sequence of aldesleukin suitable for use in the invention is given in Table 2 (SEQ ID NO:4). The term IL-2 also encompasses pegylated forms of IL-2, as described herein, including the pegylated IL2 prodrug NKTR-214, available from Nektar Therapeutics, South San Francisco, Calif., USA. NKTR-214 and pegylated IL-2 suitable for use in the invention is described in U.S. Patent Application Publication No. US 2014/0328791 A1 and International Patent Application Publication No. WO 2012/065086 A1, the disclosures of which are incorporated by reference herein. Alternative forms of conjugated IL-2 suitable for use in the invention are described in U.S. Pat. Nos. 4,766,106, 5,206,344, 5,089,261 and 4,902,502, the disclosures of which are incorporated by reference herein. Formulations of IL-2 suitable for use in the invention are described in U.S. Pat. No. 6,706,289, the disclosure of which is incorporated by reference herein.
TABLE-US-00002 TABLE 2 Amino acid sequences of interleukins. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 3 MAPTSSSTKK TQLQLEHLLL DLQMILNGIN NYKNPKLTRM LTFKFYMPKK ATELKHLQCL 60 recombinant EEELKPLEEV LNLAQSKNFH LRPRDLISNI NVIVLELKGS ETTFMCEYAD ETATIVEFLN 120 human IL-2 RWITFCQSII STLT 134 (rhIL-2) SEQ ID NO: 4 PTSSSTKKTQ LQLEHLLLDL QMILNGINNY KNPKLTRMLT FKFYMPKKAT ELKHLQCLEE 60 Aldesleukin ELKPLEEVLN LAQSKNFHLR PRDLISNINV IVLELKGSET TFMCEYADET ATIVEFLNRW 120 ITFSQSIIST LT 132 SEQ ID NO: 5 MHKCDITLQE IIKTLNSLTE QKTLCTELTV TDIFAASKNT TEKETFCRAA TVLRQFYSHH 60 recombinant EKDTRCLGAT AQQFHRHKQL IRFLKRLDRN LWGLAGLNSC PVKEANQSTL ENFLERLKTI 120 human IL-4 MREKYSKCSS 130 (rhIL-4) SEQ ID NO: 6 MDCDIEGKDG KQYESVLMVS IDQLLDSMKE IGSNCLNNEF NFFKRHICDA NKEGMFLFRA 60 recombinant ARKLRQFLKM NSTGDFDLHL LKVSEGTTIL LNCTGQVKGR KPAALGEAQP TKSLEENKSL 120 human IL-7 KEQKKLNDLC FLKRLLQEIK TCWNKILMGT KEH 153 (rhIL-7) SEQ ID NO: 7 MNWVNVISDL KKIEDLIQSM HIDATLYTES DVHPSCKVTA MKCFLLELQV ISLESGDASI 60 recombinant HDTVENLIIL ANNSLSSNGN VTESGCKECE ELEEKNIKEF LQSFVHIVQM FINTS 115 human IL-15 (rhIL-15) SEQ ID NO: 8 MQDRHMIRMR QLIDIVDQLK NYVNDLVPEF LPAPEDVETN CEWSAFSCFQ KAQLKSANTG 60 recombinant NNERIINVSI KKLKRKPPST NAGRRQKHRL TCPSCDSYEK KPPKEFLERF KSLLQKMIHQ 120 human IL-21 HLSSRTHGSE DS 132 (rhIL-21)
[1141] The term "IL-4" (also referred to herein as "IL4") refers to the cytokine known as interleukin 4, which is produced by Th2 T cells and by eosinophils, basophils, and mast cells. IL-4 regulates the differentiation of naive helper T cells (Th0 cells) to Th2 T cells. Steinke and Borish, Respir. Res. 2001, 2, 66-70. Upon activation by IL-4, Th2 T cells subsequently produce additional IL-4 in a positive feedback loop. IL-4 also stimulates B cell proliferation and class II MHC expression, and induces class switching to IgE and IgG.sub.1 expression from B cells. Recombinant human IL-4 suitable for use in the invention is commercially available from multiple suppliers, including ProSpec-Tany TechnoGene Ltd., East Brunswick, N.J., USA (Cat. No. CYT-211) and ThermoFisher Scientific, Inc., Waltham, Mass., USA (human IL-15 recombinant protein, Cat. No. Gibco CTP0043). The amino acid sequence of recombinant human IL-4 suitable for use in the invention is given in Table 2 (SEQ ID NO:5).
[1142] The term "IL-7" (also referred to herein as "IL7") refers to a glycosylated tissue-derived cytokine known as interleukin 7, which may be obtained from stromal and epithelial cells, as well as from dendritic cells. Fry and Mackall, Blood 2002, 99, 3892-904. IL-7 can stimulate the development of T cells. IL-7 binds to the IL-7 receptor, a heterodimer consisting of IIL-7 receptor alpha and common gamma chain receptor, which in a series of signals important for T cell development within the thymus and survival within the periphery. Recombinant human IL-7 suitable for use in the invention is commercially available from multiple suppliers, including ProSpec-Tany TechnoGene Ltd., East Brunswick, N.J., USA (Cat. No. CYT-254) and ThermoFisher Scientific, Inc., Waltham, Mass., USA (human IL-7 recombinant protein, Cat. No. Gibco PHC0071). The amino acid sequence of recombinant human IL-7 suitable for use in the invention is given in Table 2 (SEQ ID NO:6).
[1143] The term "IL-15" (also referred to herein as "IL15") refers to the T cell growth factor known as interleukin-15, and includes all forms of IL-15 including human and mammalian forms, conservative amino acid substitutions, glycoforms, biosimilars, and variants thereof. IL-15 is described, e.g., in Fehniger and Caligiuri, Blood 2001, 97, 14-32, the disclosure of which is incorporated by reference herein. IL-15 shares .beta. and .gamma. signaling receptor subunits with IL-2. Recombinant human IL-15 is a single, non-glycosylated polypeptide chain containing 114 amino acids (and an N-terminal methionine) with a molecular mass of 12.8 kDa. Recombinant human IL-15 is commercially available from multiple suppliers, including ProSpec-Tany TechnoGene Ltd., East Brunswick, N.J., USA (Cat. No. CYT-230-b) and ThermoFisher Scientific, Inc., Waltham, Mass., USA (human IL-15 recombinant protein, Cat. No. 34-8159-82). The amino acid sequence of recombinant human IL-15 suitable for use in the invention is given in Table 2 (SEQ ID NO:7).
[1144] The term "IL-21" (also referred to herein as "IL21") refers to the pleiotropic cytokine protein known as interleukin-21, and includes all forms of IL-21 including human and mammalian forms, conservative amino acid substitutions, glycoforms, biosimilars, and variants thereof. IL-21 is described, e.g., in Spolski and Leonard, Nat. Rev. Drug. Disc. 2014, 13, 379-95, the disclosure of which is incorporated by reference herein. IL-21 is primarily produced by natural killer T cells and activated human CD4.sup.+ T cells. Recombinant human IL-21 is a single, non-glycosylated polypeptide chain containing 132 amino acids with a molecular mass of 15.4 kDa. Recombinant human IL-21 is commercially available from multiple suppliers, including ProSpec-Tany TechnoGene Ltd., East Brunswick, N.J., USA (Cat. No. CYT-408-b) and ThermoFisher Scientific, Inc., Waltham, Mass., USA (human IL-21 recombinant protein, Cat. No. 14-8219-80). The amino acid sequence of recombinant human IL-21 suitable for use in the invention is given in Table 2 (SEQ ID NO:8).
[1145] The term "in vivo" refers to an event that takes place in a mammalian subject's body.
[1146] The term "ex vivo" refers to an event that takes place outside of a mammalian subject's body, in an artificial environment.
[1147] The term "in vitro" refers to an event that takes places in a test system. In vitro assays encompass cell-based assays in which alive or dead cells may be are employed and may also encompass a cell-free assay in which no intact cells are employed.
[1148] The term "effective amount" or "therapeutically effective amount" refers to that amount of a compound or combination of compounds as described herein that is sufficient to effect the intended application including, but not limited to, disease treatment. A therapeutically effective amount may vary depending upon the intended application (in vitro or in vivo), or the subject and disease condition being treated (e.g., the weight, age and gender of the subject), the severity of the disease condition, or the manner of administration. The term also applies to a dose that will induce a particular response in target cells (e.g., the reduction of platelet adhesion and/or cell migration). The specific dose will vary depending on the particular compounds chosen, the dosing regimen to be followed, whether the compound is administered in combination with other compounds, timing of administration, the tissue to which it is administered, and the physical delivery system in which the compound is carried.
[1149] A "therapeutic effect" as that term is used herein, encompasses a therapeutic benefit and/or a prophylactic benefit. A prophylactic effect includes delaying or eliminating the appearance of a disease or condition, delaying or eliminating the onset of symptoms of a disease or condition, slowing, halting, or reversing the progression of a disease or condition, or any combination thereof.
[1150] The terms "QD," "qd," or "q.d." mean quaque die, once a day, or once daily. The terms "BID," "bid," or "b.i.d." mean bis in die, twice a day, or twice daily. The terms "TID," "tid," or "t.i.d." mean ter in die, three times a day, or three times daily. The terms "QID," "qid," or "q.i.d." mean quater in die, four times a day, or four times daily.
[1151] The term "pharmaceutically acceptable salt" refers to salts derived from a variety of organic and inorganic counter ions known in the art. Pharmaceutically acceptable acid addition salts can be formed with inorganic acids and organic acids. Preferred inorganic acids from which salts can be derived include, for example, hydrochloric acid, hydrobromic acid, sulfuric acid, nitric acid and phosphoric acid. Preferred organic acids from which salts can be derived include, for example, acetic acid, propionic acid, glycolic acid, pyruvic acid, oxalic acid, maleic acid, malonic acid, succinic acid, fumaric acid, tartaric acid, citric acid, benzoic acid, cinnamic acid, mandelic acid, methanesulfonic acid, ethanesulfonic acid, p-toluenesulfonic acid and salicylic acid. Pharmaceutically acceptable base addition salts can be formed with inorganic and organic bases. Inorganic bases from which salts can be derived include, for example, sodium, potassium, lithium, ammonium, calcium, magnesium, iron, zinc, copper, manganese and aluminum. Organic bases from which salts can be derived include, for example, primary, secondary, and tertiary amines, substituted amines including naturally occurring substituted amines, cyclic amines and basic ion exchange resins. Specific examples include isopropylamine, trimethylamine, diethylamine, triethylamine, tripropylamine, and ethanolamine. In some embodiments, the pharmaceutically acceptable base addition salt is chosen from ammonium, potassium, sodium, calcium, and magnesium salts. The term "cocrystal" refers to a molecular complex derived from a number of cocrystal formers known in the art. Unlike a salt, a cocrystal typically does not involve hydrogen transfer between the cocrystal and the drug, and instead involves intermolecular interactions, such as hydrogen bonding, aromatic ring stacking, or dispersive forces, between the cocrystal former and the drug in the crystal structure.
[1152] The terms "pharmaceutically acceptable carrier" or "pharmaceutically acceptable excipient" are intended to include any and all solvents, dispersion media, coatings, antibacterial and antifungal agents, isotonic and absorption delaying agents, and inert ingredients. The use of such pharmaceutically acceptable carriers or pharmaceutically acceptable excipients for active pharmaceutical ingredients is well known in the art. Except insofar as any conventional pharmaceutically acceptable carrier or pharmaceutically acceptable excipient is incompatible with the active pharmaceutical ingredient, its use in the therapeutic compositions of the invention is contemplated. Additional active pharmaceutical ingredients, such as other drugs, can also be incorporated into the described compositions, processes and methods.
[1153] The term "antigen" refers to a substance that induces an immune response. In some embodiments, an antigen is a molecule capable of being bound by an antibody or a T cell receptor (TCR) if presented by major histocompatibility complex (MHC) molecules. The term "antigen", as used herein, also encompasses T cell epitopes. An antigen is additionally capable of being recognized by the immune system. In some embodiments, an antigen is capable of inducing a humoral immune response or a cellular immune response leading to the activation of B lymphocytes and/or T lynphocytes. In some cases, this may require that the antigen contains or is linked to a Th cell epitope. An antigen can also have one or more epitopes (e.g., B- and T-epitopes). In some embodiments, an antigen will preferably react, typically in a highly specific and selective manner, with its corresponding antibody or TCR and not with the multitude of other antibodies or TCRs which may be induced by their antigens.
[1154] The terms "antibody" and its plural form "antibodies" refer to whole immunoglobulins and any antigen-binding fragment ("antigen-binding portion") or single chains thereof. An "antibody" further refers to a glycoprotein comprising at least two heavy (H) chains and two light (L) chains inter-connected by disulfide bonds, or an antigen-binding portion thereof. Each heavy chain is comprised of a heavy chain variable region (abbreviated herein as V.sub.H) and a heavy chain constant region. The heavy chain constant region is comprised of three domains, CH1, CH2 and CH3. Each light chain is comprised of a light chain variable region (abbreviated herein as V.sub.L) and a light chain constant region. The light chain constant region is comprised of one domain, C.sub.L. The V.sub.H and V.sub.L regions of an antibody may be further subdivided into regions of hypervariability, which are referred to as complementarity determining regions (CDR) or hypervariable regions (HVR), and which can be interspersed with regions that are more conserved, termed framework regions (FR). Each V.sub.H and V.sub.L is composed of three CDRs and four FRs, arranged from amino-terminus to carboxy-terminus in the following order: FR1, CDR1, FR2, CDR2, FR3, CDR3, FR4. The variable regions of the heavy and light chains contain a binding domain that interacts with an antigen epitope or epitopes. The constant regions of the antibodies may mediate the binding of the immunoglobulin to host tissues or factors, including various cells of the immune system (e.g., effector cells) and the first component (Clq) of the classical complement system.
[1155] The terms "monoclonal antibody," "mAb," "monoclonal antibody composition," or their plural forms refer to a preparation of antibody molecules of single molecular composition. A monoclonal antibody composition displays a single binding specificity and affinity for a particular epitope. Monoclonal antibodies specific to TNFRSF receptors can be made using knowledge and skill in the art of injecting test subjects with suitable antigen and then isolating hybridomas expressing antibodies having the desired sequence or functional characteristics. DNA encoding the monoclonal antibodies is readily isolated and sequenced using conventional procedures (e.g., by using oligonucleotide probes that are capable of binding specifically to genes encoding the heavy and light chains of the monoclonal antibodies). The hybridoma cells serve as a preferred source of such DNA. Once isolated, the DNA may be placed into expression vectors, which are then transfected into host cells such as E. coli cells, simian COS cells, Chinese hamster ovary (CHO) cells, or myeloma cells that do not otherwise produce immunoglobulin protein, to obtain the synthesis of monoclonal antibodies in the recombinant host cells. Recombinant production of antibodies will be described in more detail below.
[1156] The terms "antigen-binding portion" or "antigen-binding fragment" of an antibody (or simply "antibody portion" or "fragment"), as used herein, refers to one or more fragments of an antibody that retain the ability to specifically bind to an antigen. It has been shown that the antigen-binding function of an antibody can be performed by fragments of a full-length antibody. Examples of binding fragments encompassed within the term "antigen-binding portion" of an antibody include (i) a Fab fragment, a monovalent fragment consisting of the V.sub.L, V.sub.H, C.sub.L and CH1 domains; (ii) a F(ab')2 fragment, a bivalent fragment comprising two Fab fragments linked by a disulfide bridge at the hinge region; (iii) a Fd fragment consisting of the V.sub.H and CH1 domains; (iv) a Fv fragment consisting of the V.sub.L and V.sub.H domains of a single arm of an antibody, (v) a domain antibody (dAb) fragment (Ward, et al., Nature, 1989, 341, 544-546), which may consist of a V.sub.H or a V.sub.L domain; and (vi) an isolated complementarity determining region (CDR). Furthermore, although the two domains of the Fv fragment, V.sub.L and V.sub.H, are coded for by separate genes, they can be joined, using recombinant methods, by a synthetic linker that enables them to be made as a single protein chain in which the V.sub.L and V.sub.H regions pair to form monovalent molecules known as single chain Fv (scFv); see, e.g., Bird, et al., Science 1988, 242, 423-426; and Huston, et al., Proc. Natl. Acad. Sci. USA 1988, 85, 5879-5883). Such scFv antibodies are also intended to be encompassed within the terms "antigen-binding portion" or "antigen-binding fragment" of an antibody. These antibody fragments are obtained using conventional techniques known to those with skill in the art, and the fragments are screened for utility in the same manner as are intact antibodies.
[1157] The term "human antibody," as used herein, is intended to include antibodies having variable regions in which both the framework and CDR regions are derived from human germline immunoglobulin sequences. Furthermore, if the antibody contains a constant region, the constant region also is derived from human germline immunoglobulin sequences. The human antibodies of the invention may include amino acid residues not encoded by human germline immunoglobulin sequences (e.g., mutations introduced by random or site-specific mutagenesis in vitro or by somatic mutation in vivo). The term "human antibody", as used herein, is not intended to include antibodies in which CDR sequences derived from the germline of another mammalian species, such as a mouse, have been grafted onto human framework sequences.
[1158] The term "human monoclonal antibody" refers to antibodies displaying a single binding specificity which have variable regions in which both the framework and CDR regions are derived from human germline immunoglobulin sequences. In an embodiment, the human monoclonal antibodies are produced by a hybridoma which includes a B cell obtained from a transgenic nonhuman animal, e.g., a transgenic mouse, having a genome comprising a human heavy chain transgene and a light chain transgene fused to an immortalized cell.
[1159] The term "recombinant human antibody", as used herein, includes all human antibodies that are prepared, expressed, created or isolated by recombinant means, such as (a) antibodies isolated from an animal (such as a mouse) that is transgenic or transchromosomal for human immunoglobulin genes or a hybridoma prepared therefrom (described further below), (b) antibodies isolated from a host cell transformed to express the human antibody, e.g., from a transfectoma, (c) antibodies isolated from a recombinant, combinatorial human antibody library, and (d) antibodies prepared, expressed, created or isolated by any other means that involve splicing of human immunoglobulin gene sequences to other DNA sequences. Such recombinant human antibodies have variable regions in which the framework and CDR regions are derived from human germline immunoglobulin sequences. In certain embodiments, however, such recombinant human antibodies can be subjected to in vitro mutagenesis (or, when an animal transgenic for human Ig sequences is used, in vivo somatic mutagenesis) and thus the amino acid sequences of the V.sub.H and V.sub.L regions of the recombinant antibodies are sequences that, while derived from and related to human germline V.sub.H and V.sub.L sequences, may not naturally exist within the human antibody germline repertoire in vivo.
[1160] As used herein, "isotype" refers to the antibody class (e.g., IgM or IgG1) that is encoded by the heavy chain constant region genes.
[1161] The phrases "an antibody recognizing an antigen" and "an antibody specific for an antigen" are used interchangeably herein with the term "an antibody which binds specifically to an antigen."
[1162] The term "human antibody derivatives" refers to any modified form of the human antibody, including a conjugate of the antibody and another active pharmaceutical ingredient or antibody. The terms "conjugate," "antibody-drug conjugate", "ADC," or "immunoconjugate" refers to an antibody, or a fragment thereof, conjugated to another therapeutic moiety, which can be conjugated to antibodies described herein using methods available in the art.
[1163] The terms "humanized antibody," "humanized antibodies," and "humanized" are intended to refer to antibodies in which CDR sequences derived from the germline of another mammalian species, such as a mouse, have been grafted onto human framework sequences. Additional framework region modifications may be made within the human framework sequences. Humanized forms of non-human (for example, murine) antibodies are chimeric antibodies that contain minimal sequence derived from non-human immunoglobulin. For the most part, humanized antibodies are human immunoglobulins (recipient antibody) in which residues from a hypervariable region of the recipient are replaced by residues from a 15 hypervariable region of a non-human species (donor antibody) such as mouse, rat, rabbit or nonhuman primate having the desired specificity, affinity, and capacity. In some instances, Fv framework region (FR) residues of the human immunoglobulin are replaced by corresponding non-human residues. Furthermore, humanized antibodies may comprise residues that are not found in the recipient antibody or in the donor antibody. These modifications are made to further refine antibody performance. In general, the humanized antibody will comprise substantially all of at least one, and typically two, variable domains, in which all or substantially all of the hypervariable loops correspond to those of a non-human immunoglobulin and all or substantially all of the FR regions are those of a human immunoglobulin sequence. The humanized antibody optionally also will comprise at least a portion of an immunoglobulin constant region (Fc), typically that of a human immunoglobulin. For further details, see Jones, et al., Nature 1986, 321, 522-525; Riechmann, et al., Nature 1988, 332, 323-329; and Presta, Curr. Op. Struct. Biol. 1992, 2, 593-596. The TNFRSF agonists described herein may also be modified to employ any Fc variant which is known to impart an improvement (e.g., reduction) in effector function and/or FcR binding. The Fc variants may include, for example, any one of the amino acid substitutions disclosed in International Patent Application Publication Nos. WO 1988/07089 A1, WO 1996/14339 A1, WO 1998/05787 A1, WO 1998/23289 A1, WO 1999/51642 A1, WO 99/58572 A1, WO 2000/09560 A2, WO 2000/32767 A1, WO 2000/42072 A2, WO 2002/44215 A2, WO 2002/060919 A2, WO 2003/074569 A2, WO 2004/016750 A2, WO 2004/029207 A2, WO 2004/035752 A2, WO 2004/063351 A2, WO 2004/074455 A2, WO 2004/099249 A2, WO 2005/040217 A2, WO 2005/070963 A1, WO 2005/077981 A2, WO 2005/092925 A2, WO 2005/123780 A2, WO 2006/019447 A1, WO 2006/047350 A2, and WO 2006/085967 A2; and U.S. Pat. Nos. 5,648,260; 5,739,277; 5,834,250; 5,869,046; 6,096,871; 6,121,022; 6,194,551; 6,242,195; 6,277,375; 6,528,624; 6,538,124; 6,737,056; 6,821,505; 6,998,253; and 7,083,784; the disclosures of which are incorporated by reference herein.
[1164] The term "chimeric antibody" is intended to refer to antibodies in which the variable region sequences are derived from one species and the constant region sequences are derived from another species, such as an antibody in which the variable region sequences are derived from a mouse antibody and the constant region sequences are derived from a human antibody.
[1165] A "diabody" is a small antibody fragment with two antigen-binding sites. The fragments comprises a heavy chain variable domain (V.sub.H) connected to a light chain variable domain (V.sub.L) in the same polypeptide chain (V.sub.H-V.sub.L or V.sub.L-V.sub.H). By using a linker that is too short to allow pairing between the two domains on the same chain, the domains are forced to pair with the complementary domains of another chain and create two antigen-binding sites. Diabodies are described more fully in, e.g., European Patent No. EP 404,097, International Patent Publication No. WO 93/11161; and Bolliger, et al., Proc. Natl. Acad. Sci. USA 1993, 90, 6444-6448.
[1166] The term "glycosylation" refers to a modified derivative of an antibody. An aglycoslated antibody lacks glycosylation. Glycosylation can be altered to, for example, increase the affinity of the antibody for antigen. Such carbohydrate modifications can be accomplished by, for example, altering one or more sites of glycosylation within the antibody sequence. For example, one or more amino acid substitutions can be made that result in elimination of one or more variable region framework glycosylation sites to thereby eliminate glycosylation at that site. Aglycosylation may increase the affinity of the antibody for antigen, as described in U.S. Pat. Nos. 5,714,350 and 6,350,861. Additionally or alternatively, an antibody can be made that has an altered type of glycosylation, such as a hypofucosylated antibody having reduced amounts of fucosyl residues or an antibody having increased bisecting GlcNac structures. Such altered glycosylation patterns have been demonstrated to increase the ability of antibodies. Such carbohydrate modifications can be accomplished by, for example, expressing the antibody in a host cell with altered glycosylation machinery. Cells with altered glycosylation machinery have been described in the art and can be used as host cells in which to express recombinant antibodies of the invention to thereby produce an antibody with altered glycosylation. For example, the cell lines Ms704, Ms705, and Ms709 lack the fucosyltransferase gene, FUT8 (alpha (1,6) fucosyltransferase), such that antibodies expressed in the Ms704, Ms705, and Ms709 cell lines lack fucose on their carbohydrates. The Ms704, Ms705, and Ms709 FUT8-/- cell lines were created by the targeted disruption of the FUT8 gene in CHO/DG44 cells using two replacement vectors (see e.g. U.S. Patent Publication No. 2004/0110704 or Yamane-Ohnuki, et al., Biotechnol. Bioeng., 2004, 87, 614-622). As another example, European Patent No. EP 1,176,195 describes a cell line with a functionally disrupted FUT8 gene, which encodes a fucosyl transferase, such that antibodies expressed in such a cell line exhibit hypofucosylation by reducing or eliminating the alpha 1,6 bond-related enzyme, and also describes cell lines which have a low enzyme activity for adding fucose to the N-acetylglucosamine that binds to the Fc region of the antibody or does not have the enzyme activity, for example the rat myeloma cell line YB2/0 (ATCC CRL 1662). International Patent Publication WO 03/035835 describes a variant CHO cell line, Lec 13 cells, with reduced ability to attach fucose to Asn(297)-linked carbohydrates, also resulting in hypofucosylation of antibodies expressed in that host cell (see also Shields, et al., J. Biol. Chem. 2002, 277, 26733-26740. International Patent Publication WO 99/54342 describes cell lines engineered to express glycoprotein-modifying glycosyl transferases (e.g., beta(1,4)-N-acetylglucosaminyltransferase III (GnTIII)) such that antibodies expressed in the engineered cell lines exhibit increased bisecting GlcNac structures which results in increased ADCC activity of the antibodies (see also Umana, et al., Nat. Biotech. 1999, 17, 176-180). Alternatively, the fucose residues of the antibody may be cleaved off using a fucosidase enzyme. For example, the fucosidase alpha-L-fucosidase removes fucosyl residues from antibodies as described in Tarentino, et al., Biochem. 1975, 14, 5516-5523.
[1167] "Pegylation" refers to a modified antibody or fusion protein, or a fragment thereof, that typically is reacted with polyethylene glycol (PEG), such as a reactive ester or aldehyde derivative of PEG, under conditions in which one or more PEG groups become attached to the antibody or antibody fragment. Pegylation may, for example, increase the biological (e.g., serum) half life of the antibody. Preferably, the pegylation is carried out via an acylation reaction or an alkylation reaction with a reactive PEG molecule (or an analogous reactive water-soluble polymer). As used herein, the term "polyethylene glycol" is intended to encompass any of the forms of PEG that have been used to derivatize other proteins, such as mono (C.sub.1-C.sub.10) alkoxy- or aryloxy-polyethylene glycol or polyethylene glycol-maleimide. The protein or antibody to be pegylated may be an aglycosylated protein or antibody. Methods for pegylation are known in the art and can be applied to the antibodies of the invention, as described for example in European Patent Nos. EP 0154316 and EP 0401384 and U.S. Pat. No. 5,824,778, the disclosures of each of which are incorporated by reference herein.
[1168] The terms "fusion protein" or "fusion polypeptide" refer to proteins that combine the properties of two or more individual proteins. Such proteins have at least two heterologous polypeptides covalently linked either directly or via an amino acid linker. The polypeptides forming the fusion protein are typically linked C-terminus to N-terminus, although they can also be linked C-terminus to C-terminus, N-terminus to N-terminus, or N-terminus to C-terminus. The polypeptides of the fusion protein can be in any order and may include more than one of either or both of the constituent polypeptides. The term encompasses conservatively modified variants, polymorphic variants, alleles, mutants, subsequences, interspecies homologs, and immunogenic fragments of the antigens that make up the fusion protein. Fusion proteins of the disclosure can also comprise additional copies of a component antigen or immunogenic fragment thereof. The fusion protein may contain one or more binding domains linked together and further linked to an Fc domain, such as an IgG Fc domain. Fusion proteins may be further linked together to mimic a monoclonal antibody and provide six or more binding domains. Fusion proteins may be produced by recombinant methods as is known in the art. Preparation of fusion proteins are known in the art and are described, e.g., in International Patent Application Publication Nos. WO 1995/027735 A1, WO 2005/103077 A1, WO 2008/025516 A1, WO 2009/007120 A1, WO 2010/003766 A1, WO 2010/010051 A1, WO 2010/078966 A1, U.S. Patent Application Publication Nos. US 2015/0125419 A1 and US 2016/0272695 A1, and U.S. Pat. No. 8,921,519, the disclosures of each of which are incorporated by reference herein.
[1169] The term "heterologous" when used with reference to portions of a nucleic acid or protein indicates that the nucleic acid or protein comprises two or more subsequences that are not found in the same relationship to each other in nature. For instance, the nucleic acid is typically recombinantly produced, having two or more sequences from unrelated genes arranged to make a new functional nucleic acid, e.g., a promoter from one source and a coding region from another source, or coding regions from different sources. Similarly, a heterologous protein indicates that the protein comprises two or more subsequences that are not found in the same relationship to each other in nature (e.g., a fusion protein).
[1170] The term "conservative amino acid substitutions" means amino acid sequence modifications which do not abrogate the binding of an antibody or fusion protein to the antigen. Conservative amino acid substitutions include the substitution of an amino acid in one class by an amino acid of the same class, where a class is defined by common physicochemical amino acid side chain properties and high substitution frequencies in homologous proteins found in nature, as determined, for example, by a standard Dayhoff frequency exchange matrix or BLOSUM matrix. Six general classes of amino acid side chains have been categorized and include: Class I (Cys); Class II (Ser, Thr, Pro, Ala, Gly); Class III (Asn, Asp, Gln, Glu); Class IV (His, Arg, Lys); Class V (Ile, Leu, Val, Met); and Class VI (Phe, Tyr, Trp). For example, substitution of an Asp for another class III residue such as Asn, Gln, or Glu, is a conservative substitution. Thus, a predicted nonessential amino acid residue in an antibody is preferably replaced with another amino acid residue from the same class. Methods of identifying amino acid conservative substitutions which do not eliminate antigen binding are well-known in the art (see, e.g., Brummell, et al., Biochemistry 1993, 32, 1180-1187; Kobayashi, et al., Protein Eng. 1999, 12, 879-884 (1999); and Burks, et al., Proc. Natl. Acad. Sci. USA 1997, 94, 412-417.
[1171] The terms "sequence identity," "percent identity," and "sequence percent identity" (or synonyms thereof, e.g., "99% identical") in the context of two or more nucleic acids or polypeptides, refer to two or more sequences or subsequences that are the same or have a specified percentage of nucleotides or amino acid residues that are the same, when compared and aligned (introducing gaps, if necessary) for maximum correspondence, not considering any conservative amino acid substitutions as part of the sequence identity. The percent identity can be measured using sequence comparison software or algorithms or by visual inspection. Various algorithms and software are known in the art that can be used to obtain alignments of amino acid or nucleotide sequences. Suitable programs to determine percent sequence identity include for example the BLAST suite of programs available from the U.S. Government's National Center for Biotechnology Information BLAST web site. Comparisons between two sequences can be carried using either the BLASTN or BLASTP algorithm. BLASTN is used to compare nucleic acid sequences, while BLASTP is used to compare amino acid sequences. ALIGN, ALIGN-2 (Genentech, South San Francisco, Calif.) or MegAlign, available from DNASTAR, are additional publicly available software programs that can be used to align sequences. One skilled in the art can determine appropriate parameters for maximal alignment by particular alignment software. In certain embodiments, the default parameters of the alignment software are used.
[1172] Certain embodiments of the present invention comprise a variant of an antibody or fusion protein. As used herein, the term "variant" encompasses but is not limited to antibodies or fusion proteins which comprise an amino acid sequence which differs from the amino acid sequence of a reference antibody by way of one or more substitutions, deletions and/or additions at certain positions within or adjacent to the amino acid sequence of the reference antibody. The variant may comprise one or more conservative substitutions in its amino acid sequence as compared to the amino acid sequence of a reference antibody. Conservative substitutions may involve, e.g., the substitution of similarly charged or uncharged amino acids. The variant retains the ability to specifically bind to the antigen of the reference antibody.
[1173] Nucleic acid sequences implicitly encompass conservatively modified variants thereof (e.g., degenerate codon substitutions) and complementary sequences, as well as the sequence explicitly indicated. Specifically, degenerate codon substitutions may be achieved by generating sequences in which the third position of one or more selected (or all) codons is substituted with mixed-base and/or deoxyinosine residues. Batzer, et al., Nucleic Acid Res. 1991, 19, 5081; Ohtsuka, et al., J. Biol. Chem. 1985, 260, 2605-2608; Rossolini, et al., Mol. Cell. Probes 1994, 8, 91-98. The term nucleic acid is used interchangeably with cDNA, mRNA, oligonucleotide, and polynucleotide.
[1174] The term "biosimilar" means a biological product, including a monoclonal antibody or fusion protein, that is highly similar to a U.S. licensed reference biological product notwithstanding minor differences in clinically inactive components, and for which there are no clinically meaningful differences between the biological product and the reference product in terms of the safety, purity, and potency of the product. Furthermore, a similar biological or "biosimilar" medicine is a biological medicine that is similar to another biological medicine that has already been authorized for use by the European Medicines Agency. The term "biosimilar" is also used synonymously by other national and regional regulatory agencies. Biological products or biological medicines are medicines that are made by or derived from a biological source, such as a bacterium or yeast. They can consist of relatively small molecules such as human insulin or erythropoietin, or complex molecules such as monoclonal antibodies. For example, if the reference monoclonal antibody is rituximab, an biosimilar monoclonal antibody approved by drug regulatory authorities with reference to rituximab is a "biosimilar to" rituximab or is a "biosimilar thereof" of rituximab. In Europe, a similar biological or "biosimilar" medicine is a biological medicine that is similar to another biological medicine that has already been authorized for use by the European Medicines Agency (EMA). The relevant legal basis for similar biological applications in Europe is Article 6 of Regulation (EC) No 726/2004 and Article 10(4) of Directive 2001/83/EC, as amended and therefore in Europe, the biosimilar may be authorised, approved for authorisation or subject of an application for authorisation under Article 6 of Regulation (EC) No 726/2004 and Article 10(4) of Directive 2001/83/EC. The already authorized original biological medicinal product may be referred to as a "reference medicinal product" in Europe. Some of the requirements for a product to be considered a biosimilar are outlined in the CHMP Guideline on Similar Biological Medicinal Products. In addition, product specific guidelines, including guidelines relating to monoclonal antibody biosimilars, are provided on a product-by-product basis by the EMA and published on its website. A biosimilar as described herein may be similar to the reference medicinal product by way of quality characteristics, biological activity, mechanism of action, safety profiles and/or efficacy. In addition, the biosimilar may be used or be intended for use to treat the same conditions as the reference medicinal product. Thus, a biosimilar as described herein may be deemed to have similar or highly similar quality characteristics to a reference medicinal product. Alternatively, or in addition, a biosimilar as described herein may be deemed to have similar or highly similar biological activity to a reference medicinal product. Alternatively, or in addition, a biosimilar as described herein may be deemed to have a similar or highly similar safety profile to a reference medicinal product. Alternatively, or in addition, a biosimilar as described herein may be deemed to have similar or highly similar efficacy to a reference medicinal product. As described herein, a biosimilar in Europe is compared to a reference medicinal product which has been authorised by the EMA. However, in some instances, the biosimilar may be compared to a biological medicinal product which has been authorised outside the European Economic Area (a non-EEA authorised "comparator") in certain studies. Such studies include for example certain clinical and in vivo non-clinical studies. As used herein, the term "biosimilar" also relates to a biological medicinal product which has been or may be compared to a non-EEA authorised comparator. Certain biosimilars are proteins such as antibodies, antibody fragments (for example, antigen binding portions) and fusion proteins. A protein biosimilar may have an amino acid sequence that has minor modifications in the amino acid structure (including for example deletions, additions, and/or substitutions of amino acids) which do not significantly affect the function of the polypeptide. The biosimilar may comprise an amino acid sequence having a sequence identity of 97% or greater to the amino acid sequence of its reference medicinal product, e.g., 97%, 98%, 99% or 100%. The biosimilar may comprise one or more post-translational modifications, for example, although not limited to, glycosylation, oxidation, deamidation, and/or truncation which is/are different to the post-translational modifications of the reference medicinal product, provided that the differences do not result in a change in safety and/or efficacy of the medicinal product. The biosimilar may have an identical or different glycosylation pattern to the reference medicinal product. Particularly, although not exclusively, the biosimilar may have a different glycosylation pattern if the differences address or are intended to address safety concerns associated with the reference medicinal product. Additionally, the biosimilar may deviate from the reference medicinal product in for example its strength, pharmaceutical form, formulation, excipients and/or presentation, providing safety and efficacy of the medicinal product is not compromised. In some embodiments, a biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product. The biosimilar may comprise differences in for example pharmacokinetic (PK) and/or pharmacodynamic (PD) profiles as compared to the reference medicinal product but is still deemed sufficiently similar to the reference medicinal product as to be authorised or considered suitable for authorisation. In certain circumstances, the biosimilar exhibits different binding characteristics as compared to the reference medicinal product, wherein the different binding characteristics are considered by a Regulatory Authority such as the EMA not to be a barrier for authorisation as a similar biological product. The term "biosimilar" is also used synonymously by other national and regional regulatory agencies.
[1175] As used herein, the term "4-1BB agonist" may refer to any antibody or protein that specifically binds to 4-1BB (CD137) antigen. By "specifically binds" it is meant that the binding molecules exhibit essentially background binding to non-4-1BB molecules. The 4-1BB agonist may be any 4-1BB agonist known in the art. In particular, it is one of the 4-1BB agonists described in more detail herein. An isolated binding molecule that specifically binds 4-1BB may, however, have cross-reactivity to 4-1BB molecules from other species. 4-1BB agonistic antibodies and proteins may also specifically bind to e.g., human 4-1BB (h4-1BB or hCD137) on T cells.
[1176] As used herein, the term "OX40 agonist" may refer to any antibody or protein that specifically binds to OX40 (CD134) antigen. By "specifically binds" it is meant that the binding molecules exhibit essentially background binding to non-OX40 molecules. The OX40 agonist may be any OX40 agonist known in the art. In particular, it is one of the OX40 agonists described in more detail herein. An isolated binding molecule that specifically binds OX40 may, however, have cross-reactivity to OX40 molecules from other species. OX40 agonistic antibodies and proteins may also specifically bind to e.g., human OX40 (hOX40 or hCD134) on T cells.
[1177] As used herein, the term "CD27 agonist" may refer to any antibody or protein that specifically binds to CD27 antigen. By "specifically binds" it is meant that the binding molecules exhibit essentially background binding to non-CD27 molecules. The CD27 agonist may be any CD27 agonist known in the art. In particular, it is one of the CD27 agonists described in more detail herein. An isolated binding molecule that specifically binds CD27 may, however, have cross-reactivity to CD27 molecules from other species. CD27 agonistic antibodies and proteins may also specifically bind to e.g., human CD27 (hCD27) on T cells.
[1178] As used herein, the term "GITR agonist" includes molecules that contain at least one antigen binding site that specifically binds to GITR (CD357). By "specifically binds" it is meant that the binding molecules exhibit essentially background binding to non-GITR molecules. The GITR agonist may be any GITR agonist known in the art. In particular, it is one of the GITR agonists described in more detail herein. An isolated binding molecule that specifically binds GITR may, however, have cross-reactivity to GITR molecules from other species. GITR agonistic antibodies and proteins may also specifically bind to e.g., human GITR (hGITR) on T cells and dendritic cells.
[1179] As used herein, the term "HVEM agonist" includes molecules that contain at least one antigen binding site that specifically binds to HVEM (CD270). By "specifically binds" it is meant that the binding molecules exhibit essentially background binding to non-HVEM molecules. The HVEM agonist may be any HVEM agonist known in the art. In particular, it is one of the HVEM agonists described in more detail herein. An isolated binding molecule that specifically binds HVEM may, however, have cross-reactivity to HVEM molecules from other species. HVEM agonistic antibodies and proteins may also specifically bind to e.g., human HVEM (hHVEM) on T cells.
[1180] The term "hematological malignancy" refers to mammalian cancers and tumors of the hematopoietic and lymphoid tissues, including but not limited to tissues of the blood, bone marrow, lymph nodes, and lymphatic system. Hematological malignancies are also referred to as "liquid tumors." Hematological malignancies include, but are not limited to, acute lymphoblastic leukemia (ALL), chronic lymphocytic lymphoma (CLL), small lymphocytic lymphoma (SLL), acute myelogenous leukemia (AML), chronic myelogenous leukemia (CML), acute monocytic leukemia (AMoL), Hodgkin's lymphoma, and non-Hodgkin's lymphomas. The term "B cell hematological malignancy" refers to hematological malignancies that affect B cells.
[1181] The term "solid tumor" refers to an abnormal mass of tissue that usually does not contain cysts or liquid areas. Solid tumors may be benign or malignant. The term "solid tumor cancer" refers to malignant, neoplastic, or cancerous solid tumors. Solid tumor cancers include, but are not limited to, sarcomas, carcinomas, and lymphomas, such as cancers of the lung, breast, prostate, colon, rectum, and bladder. The tissue structure of solid tumors includes interdependent tissue compartments including the parenchyma (cancer cells) and the supporting stromal cells in which the cancer cells are dispersed and which may provide a supporting microenvironment.
[1182] The term "microenvironment," as used herein, may refer to the solid or hematological tumor microenvironment as a whole or to an individual subset of cells within the microenvironment. The tumor microenvironment, as used herein, refers to a complex mixture of "cells, soluble factors, signaling molecules, extracellular matrices, and mechanical cues that promote neoplastic transformation, support tumor growth and invasion, protect the tumor from host immunity, foster therapeutic resistance, and provide niches for dominant metastases to thrive," as described in Swartz, et al., Cancer Res., 2012, 72, 2473. Although tumors express antigens that should be recognized by T cells, tumor clearance by the immune system is rare because of immune suppression by the microenvironment.
[1183] For the avoidance of doubt, it is intended herein that particular features (for example integers, characteristics, values, uses, diseases, formulae, compounds or groups) described in conjunction with a particular aspect, embodiment or example of the invention are to be understood as applicable to any other aspect, embodiment or example described herein unless incompatible therewith. Thus such features may be used where appropriate in conjunction with any of the definition, claims or embodiments defined herein. All of the features disclosed in this specification (including any accompanying claims, abstract and drawings), and/or all of the steps of any method or process so disclosed, may be combined in any combination, except combinations where at least some of the features and/or steps are mutually exclusive. The invention is not restricted to any details of any disclosed embodiments. The invention extends to any novel one, or novel combination, of the features disclosed in this specification (including any accompanying claims, abstract and drawings), or to any novel one, or any novel combination, of the steps of any method or process so disclosed.
[1184] The terms "about" and "approximately" mean within a statistically meaningful range of a value. Such a range can be within an order of magnitude, preferably within 50%, more preferably within 20%, more preferably still within 10%, and even more preferably within 5% of a given value or range. The allowable variation encompassed by the terms "about" or "approximately" depends on the particular system under study, and can be readily appreciated by one of ordinary skill in the art. Moreover, as used herein, the terms "about" and "approximately" mean that dimensions, sizes, formulations, parameters, shapes and other quantities and characteristics are not and need not be exact, but may be approximate and/or larger or smaller, as desired, reflecting tolerances, conversion factors, rounding off, measurement error and the like, and other factors known to those of skill in the art. In general, a dimension, size, formulation, parameter, shape or other quantity or characteristic is "about" or "approximate" whether or not expressly stated to be such. It is noted that embodiments of very different sizes, shapes and dimensions may employ the described arrangements.
[1185] The transitional terms "comprising," "consisting essentially of," and "consisting of," when used in the appended claims, in original and amended form, define the claim scope with respect to what unrecited additional claim elements or steps, if any, are excluded from the scope of the claim(s). The term "comprising" is intended to be inclusive or open-ended and does not exclude any additional, unrecited element, method, step or material. The term "consisting of" excludes any element, step or material other than those specified in the claim and, in the latter instance, impurities ordinary associated with the specified material(s). The term "consisting essentially of" limits the scope of a claim to the specified elements, steps or material(s) and those that do not materially affect the basic and novel characteristic(s) of the claimed invention. All compositions, methods, and kits described herein that embody the present invention can, in alternate embodiments, be more specifically defined by any of the transitional terms "comprising," "consisting essentially of," and "consisting of."
4-1BB (CD137) Agonists
[1186] In an embodiment, the TNFRSF agonist is a 4-1BB (CD137) agonist. The 4-1BB agonist may be any 4-1BB binding molecule known in the art. The 4-1BB binding molecule may be a monoclonal antibody or fusion protein capable of binding to human or mammalian 4-1BB. The 4-1BB agonists or 4-1BB binding molecules may comprise an immunoglobulin heavy chain of any isotype (e.g., IgG, IgE, IgM, IgD, IgA, and IgY), class (e.g., IgG1, IgG2, IgG3, IgG4, IgA1 and IgA2) or subclass of immunoglobulin molecule. The 4-1BB agonist or 4-1BB binding molecule may have both a heavy and a light chain. As used herein, the term binding molecule also includes antibodies (including full length antibodies), monoclonal antibodies (including full length monoclonal antibodies), polyclonal antibodies, multispecific antibodies (e.g., bispecific antibodies), human, humanized or chimeric antibodies, and antibody fragments, e.g., Fab fragments, F(ab') fragments, fragments produced by a Fab expression library, epitope-binding fragments of any of the above, and engineered forms of antibodies, e.g., scFv molecules, that bind to 4-1BB. In an embodiment, the 4-1BB agonist is an antigen binding protein that is a fully human antibody. In an embodiment, the 4-1BB agonist is an antigen binding protein that is a humanized antibody. In some embodiments, 4-1BB agonists for use in the presently disclosed methods and compositions include anti-4-1BB antibodies, human anti-4-1BB antibodies, mouse anti-4-1BB antibodies, mammalian anti-4-1BB antibodies, monoclonal anti-4-1BB antibodies, polyclonal anti-4-1BB antibodies, chimeric anti-4-1BB antibodies, anti-4-1BB adnectins, anti-4-1BB domain antibodies, single chain anti-4-1BB fragments, heavy chain anti-4-1BB fragments, light chain anti-4-1BB fragments, anti-4-1BB fusion proteins, and fragments, derivatives, conjugates, variants, or biosimilars thereof. Agonistic anti-4-1BB antibodies are known to induce strong immune responses. Lee, et al., PLOS One 2013, 8, e69677. In a preferred embodiment, the 4-1BB agonist is an agonistic, anti-4-1BB humanized or fully human monoclonal antibody (i.e., an antibody derived from a single cell line). In an embodiment, the 4-1BB agonist is EU-101 (Eutilex Co. Ltd.), utomilumab, or urelumab, or a fragment, derivative, conjugate, variant, or biosimilar thereof. In a preferred embodiment, the 4-1BB agonist is utomilumab or urelumab, or a fragment, derivative, conjugate, variant, or biosimilar thereof.
[1187] In a preferred embodiment, the 4-1BB agonist or 4-1BB binding molecule may also be a fusion protein. In a preferred embodiment, a multimeric 4-1BB agonist, such as a trimeric or hexameric 4-1BB agonist (with three or six ligand binding domains), may induce superior receptor (4-1BBL) clustering and internal cellular signaling complex formation compared to an agonistic monoclonal antibody, which typically possesses two ligand binding domains. Trimeric (trivalent) or hexameric (or hexavalent) or greater fusion proteins comprising three TNFRSF binding domains and IgG1-Fc and optionally further linking two or more of these fusion proteins are described, e.g., in Gieffers, et al., Mol. Cancer Therapeutics 2013, 12, 2735-47.
[1188] Agonistic 4-1BB antibodies and fusion proteins are known to induce strong immune responses. In a preferred embodiment, the 4-1BB agonist is a monoclonal antibody or fusion protein that binds specifically to 4-1BB antigen in a manner sufficient to reduce toxicity. In some embodiments, the 4-1BB agonist is an agonistic 4-1BB monoclonal antibody or fusion protein that abrogates antibody-dependent cellular toxicity (ADCC), for example NK cell cytotoxicity. In some embodiments, the 4-1BB agonist is an agonistic 4-1BB monoclonal antibody or fusion protein that abrogates antibody-dependent cell phagocytosis (ADCP). In some embodiments, the 4-1BB agonist is an agonistic 4-1BB monoclonal antibody or fusion protein that abrogates complement-dependent cytotoxicity (CDC). In some embodiments, the 4-1BB agonist is an agonistic 4-1BB monoclonal antibody or fusion protein which abrogates Fc region functionality.
[1189] In some embodiments, the 4-1BB agonists are characterized by binding to human 4-1BB (SEQ ID NO:9) with high affinity and agonistic activity. In an embodiment, the 4-1BB agonist is a binding molecule that binds to human 4-1BB (SEQ ID NO:9). In an embodiment, the 4-1BB agonist is a binding molecule that binds to murine 4-1BB (SEQ ID NO: 10). The amino acid sequences of 4-1BB antigen to which a 4-1BB agonist or binding molecule binds are summarized in Table 3.
TABLE-US-00003 TABLE 3 Amino acid sequences of 4-1BB antigens. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 9 MGNSCYNIVA TLLLVLNFER TRSLQDPCSN CPAGTFCDNN RNQICSPCPP NSFSSAGGQR 60 human 4-1BB, TCDICRQCKG VFRTRKECSS TSNAECDCTP GFHCLGAGCS MCEQDCKQGQ ELTKKGCKDC 120 Tumor necrosis CFGTFNDQKR GICRPWTNCS LDGKSVLVNG TKERDVVCGP SPADLSPGAS SVTPPAPARE 180 factor receptor PGHSPQIISF FLALTSTALL FLLFFLTLRF SVVKRGRKKL LYIFKQPFMR PVQTTQEEDG 240 superfamily, CSCRFPEEEE GGCEL 255 member 9 (Homo sapiens) SEQ ID NO: 10 MGNNCYNVVV IVLLLVGCEK VGAVQNSCDN CQPGTFCRKY NPVCKSCPPS TFSSIGGQPN 60 murine 4-1BB, CNICRVCAGY FRFKKFCSST HNAECECIEG FHCLGPQCTR CEKDCRPGQE LTKQGCKTCS 120 Tumor necrosis LGTFNDQNGT GVCRPWTNCS LDGRSVLKTG TTEKDVVCGP PVVSFSPSTT ISVTPEGGPG 180 factor receptor GHSLQVLTLF LALTSALLLA LIFITLLFSV LKWIRKKFPH IFKQPFKKTT GAAQEEDACS 240 superfamily, CRCPQEEEGG GGGYEL 256 member 9 (Mus musculus)
[1190] In some embodiments, the compositions, processes and methods described include a 4-1BB agonist that binds human or murine 4-1BB with a K.sub.D of about 100 pM or lower, binds human or murine 4-1BB with a K.sub.D of about 90 pM or lower, binds human or murine 4-1BB with a K.sub.D of about 80 pM or lower, binds human or murine 4-1BB with a K.sub.D of about 70 pM or lower, binds human or murine 4-1BB with a K.sub.D of about 60 pM or lower, binds human or murine 4-1BB with a K.sub.D of about 50 pM or lower, binds human or murine 4-1BB with a K.sub.D of about 40 pM or lower, or binds human or murine 4-1BB with a K.sub.D of about 30 pM or lower.
[1191] In some embodiments, the compositions, processes and methods described include a 4-1BB agonist that binds to human or murine 4-1BB with a k.sub.assoc of about 7.5.times.10.sup.5 l/Ms or faster, binds to human or murine 4-1BB with a k.sub.assoc of about 7.5.times.10.sup.5 l/Ms or faster, binds to human or murine 4-1BB with a k.sub.assoc of about 8.times.10.sup.5 l/Ms or faster, binds to human or murine 4-1BB with a k.sub.assoc of about 8.5.times.10.sup.5 l/Ms or faster, binds to human or murine 4-1BB with a k.sub.assoc of about 9.times.10.sup.5 l/Ms or faster, binds to human or murine 4-1BB with a k.sub.assoc of about 9.5.times.10.sup.5 l/Ms or faster, or binds to human or murine 4-1BB with a k.sub.assoc of about 1.times.10.sup.6 l/Ms or faster.
[1192] In some embodiments, the compositions, processes and methods described include a 4-1BB agonist that binds to human or murine 4-1BB with a k.sub.dissoc of about 2.times.10.sup.-5 l/s or slower, binds to human or murine 4-1BB with a k.sub.dissoc of about 2.1.times.10.sup.-5 l/s or slower, binds to human or murine 4-1BB with a k.sub.dissoc of about 2.2.times.10.sup.-5 l/s or slower, binds to human or murine 4-1BB with a k.sub.dissoc of about 2.3.times.10.sup.-5 l/s or slower, binds to human or murine 4-1BB with a k.sub.dissoc of about 2.4.times.10.sup.-5 l/s or slower, binds to human or murine 4-1BB with a k.sub.dissoc of about 2.5.times.10.sup.-5 l/s or slower, binds to human or murine 4-1BB with a k.sub.dissoc of about 2.6.times.10.sup.-5 l/s or slower or binds to human or murine 4-1BB with a k.sub.dissoc of about 2.7.times.10.sup.-5 l/s or slower, binds to human or murine 4-1BB with a k.sub.dissoc of about 2.8.times.10.sup.-5 l/s or slower, binds to human or murine 4-1BB with a k.sub.dissoc of about 2.9.times.10.sup.-5 l/s or slower, or binds to human or murine 4-1BB with a k.sub.dissoc of about 3.times.10.sup.-5 l/s or slower.
[1193] In some embodiments, the compositions, processes and methods described include a 4-1BB agonist that binds to human or murine 4-1BB with an IC.sub.50 of about 10 nM or lower, binds to human or murine 4-1BB with an IC.sub.50 of about 9 nM or lower, binds to human or murine 4-1BB with an IC.sub.50 of about 8 nM or lower, binds to human or murine 4-1BB with an IC.sub.50 of about 7 nM or lower, binds to human or murine 4-1BB with an IC.sub.50 of about 6 nM or lower, binds to human or murine 4-1BB with an IC.sub.50 of about 5 nM or lower, binds to human or murine 4-1BB with an IC.sub.50 of about 4 nM or lower, binds to human or murine 4-1BB with an IC.sub.50 of about 3 nM or lower, binds to human or murine 4-1BB with an IC.sub.50 of about 2 nM or lower, or binds to human or murine 4-1BB with an IC.sub.50 of about 1 nM or lower.
[1194] In a preferred embodiment, the 4-1BB agonist is utomilumab, also known as PF-05082566 or MOR-7480, or a fragment, derivative, variant, or biosimilar thereof. Utomilumab is available from Pfizer, Inc. Utomilumab is an immunoglobulin G2-lambda, anti-[Homo sapiens TNFRSF9 (tumor necrosis factor receptor (TNFR) superfamily member 9, 4-1BB, T cell antigen ILA, CD137)], Homo sapiens (fully human) monoclonal antibody. The amino acid sequences of utomilumab are set forth in Table 4. Utomilumab comprises glycosylation sites at Asn59 and Asn292; heavy chain intrachain disulfide bridges at positions 22-96 (V.sub.H-V.sub.L), 143-199 (C.sub.H-C.sub.L), 256-316 (C.sub.H2) and 362-420 (C.sub.H3); light chain intrachain disulfide bridges at positions 22'-87' (V.sub.H-V.sub.L) and 136'-195' (C.sub.H1-C.sub.L); interchain heavy chain-heavy chain disulfide bridges at IgG2A isoform positions 218-218, 219-219, 222-222, and 225-225, at IgG2A/B isoform positions 218-130, 219-219, 222-222, and 225-225, and at IgG2B isoform positions 219-130 (2), 222-222, and 225-225; and interchain heavy chain-light chain disulfide bridges at IgG2A isoform positions 130-213' (2), IgG2A/B isoform positions 218-213' and 130-213', and at IgG2B isoform positions 218-213' (2). The preparation and properties of utomilumab and its variants and fragments are described in U.S. Pat. Nos. 8,821,867; 8,337,850; and 9,468,678, and International Patent Application Publication No. WO 2012/032433 A1, the disclosures of each of which are incorporated by reference herein. Preclinical characteristics of utomilumab are described in Fisher, et al., Cancer Immunolog. & Immunother. 2012, 61, 1721-33. Current clinical trials of utomilumab in a variety of hematological and solid tumor indications include U.S. National Institutes of Health clinicaltrials.gov identifiers NCT02444793, NCT01307267, NCT02315066, and NCT02554812.
[1195] In an embodiment, a 4-1BB agonist comprises a heavy chain given by SEQ ID NO:11 and a light chain given by SEQ ID NO: 12. In an embodiment, a 4-1BB agonist comprises heavy and light chains having the sequences shown in SEQ ID NO: 11 and SEQ ID NO: 12, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a 4-1BB agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO: 11 and SEQ ID NO: 12, respectively. In an embodiment, a 4-1BB agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO: 11 and SEQ ID NO:12, respectively. In an embodiment, a 4-1BB agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO: 11 and SEQ ID NO: 12, respectively. In an embodiment, a 4-1BB agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO: 11 and SEQ ID NO: 12, respectively. In an embodiment, a 4-1BB agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO: 11 and SEQ ID NO: 12, respectively.
[1196] In an embodiment, the 4-1BB agonist comprises the heavy and light chain CDRs or variable regions (VRs) of utomilumab. In an embodiment, the 4-1BB agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO: 13, and the 4-1BB agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO: 14, and conservative amino acid substitutions thereof. In an embodiment, a 4-1BB agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO: 13 and SEQ ID NO:14, respectively. In an embodiment, a 4-1BB agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:13 and SEQ ID NO:14, respectively. In an embodiment, a 4-1BB agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO: 13 and SEQ ID NO: 14, respectively. In an embodiment, a 4-1BB agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:13 and SEQ ID NO:14, respectively. In an embodiment, a 4-1BB agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO: 13 and SEQ ID NO: 14, respectively. In an embodiment, a 4-1BB agonist comprises an scFv antibody comprising V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO: 13 and SEQ ID NO: 14.
[1197] In an embodiment, a 4-1BB agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:15, SEQ ID NO:16, and SEQ ID NO:17, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO: 18, SEQ ID NO: 19, and SEQ ID NO:20, respectively, and conservative amino acid substitutions thereof.
[1198] In an embodiment, the 4-1BB agonist is a 4-1BB agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to utomilumab. In an embodiment, the biosimilar monoclonal antibody comprises an 4-1BB antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is utomilumab. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a 4-1BB agonist antibody authorized or submitted for authorization, wherein the 4-1BB agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is utomilumab. The 4-1BB agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is utomilumab. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is utomilumab.
TABLE-US-00004 TABLE 4 Amino acid sequences for 4-1BB agonist antibodies related to utomilumab. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 11 EVQLVQSGAE VKKPGESLRI SCKGSGYSFS TYWISWVRQM PGKGLEWMGK IYPGDSYTNY 60 heavy chain SPSFQGQVTI SADKSISTAY LQWSSLKASD TAMYYCARGY GIFDYWGQGT LVTVSSASTK 120 for GPSVFPLAPC SRSTSESTAA LGCLVKDYFP EPVTVSWNSG ALTSGVHTFP AVLQSSGLYS 180 utomilumab LSSVVTVPSS NFGTQTYTCN VDHKPSNTKV DKTVERKCCV ECPPCPAPPV AGPSVFLFPP 240 KPKDTLMISR TPEVTCVVVD VSHEDPEVQF NWYVDGVEVH NAKTKPREEQ FNSTFRVVSV 300 LTVVHQDWLN GKEYKCKVSN KGLPAPIEKT ISKTKGQPRE PQVYTLPPSR EEMTKNQVSL 360 TCLVKGFYPS DIAVEWESNG QPENNYKTTP PMLDSDGSFF LYSKLTVDKS RWQQGNVFSC 420 SVMHEALHNH YTQKSLSLSP G 441 SEQ ID NO: 12 SYELTQPPSV SVSPGQTASI TCSGDNIGDQ YAHWYQQKPG QSPVLVIYQD KNRPSGIPER 60 light chain FSGSNSGNTA TLTISGTQAM DEADYYCATY TGFGSLAVFG GGTKLTVLGQ PKAAPSVTLF 120 for PPSSEELQAN KATLVCLISD FYPGAVTVAW KADSSPVKAG VETTTPSKQS NNKYAASSYL 180 utomilumab SLTPEQWKSH RSYSCQVTHE GSTVEKTVAP TECS 214 SEQ ID NO: 13 EVQLVQSGAE VKKPGESLRI SCKGSGYSFS TYWISWVRQM PGKGLEWMG KIYPGDSYTN 60 heavy chain YSPSFQGQVT ISADKSISTA YLQWSSLKAS DTAMYYCARG YGIFDYWGQ GTLVTVSS 118 variable region for utomilumab SEQ ID NO: 14 SYELTQPPSV SVSPGQTASI TCSGDNIGDQ YAHWYQQKPG QSPVLVIYQD KNRPSGIPER 60 light chain FSGSNSGNTA TLTISGTQAM DEADYYCATY TGFGSLAVFG GGTKLTVL 108 variable region for utomilumab SEQ ID NO: 15 STYWIS 6 heavy chain CDR1 for utomilumab SEQ ID NO: 16 KIYPGDSYTN YSPSFQG 17 heavy chain CDR2 for utomilumab SEQ ID NO: 17 RGYGIFDY 8 heavy chain CDR3 for utomilumab SEQ ID NO: 18 SGDNIGDQYA H 11 light chain CDR1 for utomilumab SEQ ID NO: 19 QDKNRPS 7 light chain CDR2 for utomilumab SEQ ID NO: 20 ATYTGFGSLA V 11 light chain CDR3 for utomilumab
[1199] In a preferred embodiment, the 4-1BB agonist is the monoclonal antibody urelumab, also known as BMS-663513 and 20H4.9.h4a, or a fragment, derivative, variant, or biosimilar thereof. Urelumab is available from Bristol-Myers Squibb, Inc., and Creative Biolabs, Inc. Urelumab is an immunoglobulin G4-kappa, anti-[Homo sapiens TNFRSF9 (tumor necrosis factor receptor superfamily member 9, 4-1BB, T cell antigen ILA, CD137)], Homo sapiens (fully human) monoclonal antibody. The amino acid sequences of urelumab are set forth in Table 5. Urelumab comprises N-glycosylation sites at positions 298 (and 298''); heavy chain intrachain disulfide bridges at positions 22-95 (V.sub.H-V.sub.L), 148-204 (C.sub.H1-C.sub.L), 262-322 (C.sub.H2) and 368-426 (C.sub.H3) (and at positions 22''-95'', 148''-204'', 262''-322'', and 368''-426''); light chain intrachain disulfide bridges at positions 23'-88' (V.sub.H-V.sub.L) and 136'-196' (C.sub.H1-C.sub.L) (and at positions 23'''-88''' and 136'''-196'''); interchain heavy chain-heavy chain disulfide bridges at positions 227-227'' and 230-230''; and interchain heavy chain-light chain disulfide bridges at 135-216' and 135''-216'''. The preparation and properties of urelumab and its variants and fragments are described in U.S. Pat. Nos. 7,288,638 and 8,962,804, the disclosures of which are incorporated by reference herein. The preclinical and clinical characteristics of urelumab are described in Segal, et al., Clin. Cancer Res. 2016, available at http:/dx.doi.org/10.1158/1078-0432.CCR-16-1272. Current clinical trials of urelumab in a variety of hematological and solid tumor indications include U.S. National Institutes of Health clinicaltrials.gov identifiers NCT01775631, NCT02110082, NCT02253992, and NCT01471210.
[1200] In an embodiment, a 4-1BB agonist comprises a heavy chain given by SEQ ID NO:21 and a light chain given by SEQ ID NO:22. In an embodiment, a 4-1BB agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:21 and SEQ ID NO:22, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a 4-1BB agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:21 and SEQ ID NO:22, respectively. In an embodiment, a 4-1BB agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:21 and SEQ ID NO:22, respectively. In an embodiment, a 4-1BB agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:21 and SEQ ID NO:22, respectively. In an embodiment, a 4-1BB agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:21 and SEQ ID NO:22, respectively. In an embodiment, a 4-1BB agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:21 and SEQ ID NO:22, respectively.
[1201] In an embodiment, the 4-1BB agonist comprises the heavy and light chain CDRs or variable regions (VRs) of urelumab. In an embodiment, the 4-1BB agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:23, and the 4-1BB agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:24, and conservative amino acid substitutions thereof. In an embodiment, a 4-1BB agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:23 and SEQ ID NO:24, respectively. In an embodiment, a 4-1BB agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:23 and SEQ ID NO:24, respectively. In an embodiment, a 4-1BB agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:23 and SEQ ID NO:24, respectively. In an embodiment, a 4-1BB agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:23 and SEQ ID NO:24, respectively. In an embodiment, a 4-1BB agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:23 and SEQ ID NO:24, respectively. In an embodiment, a 4-1BB agonist comprises an scFv antibody comprising V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:23 and SEQ ID NO:24.
[1202] In an embodiment, a 4-1BB agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:25, SEQ ID NO:26, and SEQ ID NO:27, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:28, SEQ ID NO:29, and SEQ ID NO:30, respectively, and conservative amino acid substitutions thereof.
[1203] In an embodiment, the 4-1BB agonist is a 4-1BB agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to urelumab. In an embodiment, the biosimilar monoclonal antibody comprises an 4-1BB antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is urelumab. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a 4-1BB agonist antibody authorized or submitted for authorization, wherein the 4-1BB agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is urelumab. The 4-1BB agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is urelumab. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is urelumab.
TABLE-US-00005 TABLE 5 Amino acid sequences for 4-1BB agonist antibodies related to urelumab. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 21 QVQLQQWGAG LLKPSETLSL TCAVYGGSFS GYYWSWIRQS PEKGLEWIGE INHGGYVTYN 60 heavy chain PSLESRVTIS VDTSKNQFSL KLSSVTAADT AVYYCARDYG PGNYDWYFDL WGRGTLVTVS 120 for urelumab SASTKGPSVF PLAPCSRSTS ESTAALGCLV KDYFPEPVTV SWNSGALTSG VHTFPAVLQS 180 SGLYSLSSVV TVPSSSLGTK TYTCNVDHKP SNTKVDKRVE SKYGPPCPPC PAPEFLGGPS 240 VFLFPPKPKD TLMISRTPEV TCVVVDVSQE DPEVQFNWYV DGVEVHNAKT KPREEQFNST 300 YRVVSVLTVL HQDWLNGKEY KCKVSNKGLP SSIEKTISKA KGQPREPQVY TLPPSQEEMT 360 KNQVSLTCLV KGFYPSDIAV EWESNGQPEN NYKTTPPVLD SDGSFFLYSR LTVDKSRWQE 420 GNVFSCSVMH EALHNHYTQK SLSLSLGK 448 SEQ ID NO: 22 EIVLTQSPAT LSLSPGERAT LSCRASQSVS SYLAWYQQKP GQAPRLLIYD ASNRATGIPA 60 light chain RFSGSGSGTD FTLTISSLEP EDFAVYYCQQ RSNWPPALTF CGGTKVEIKR TVAAPSVFIF 120 for urelumab PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN SQESVTEQDS KDSTYSLSST 180 LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC 216 SEQ ID NO: 23 MKHLWFFLLL VAAPRWVLSQ VQLQQWGAGL LKPSETLSLT CAVYGGSFSG YYWSWIRQSP 60 variable EKGLEWIGEI NHGGYVTYNP SLESRVTISV DTSKNQFSLK LSSVTAADTA VYYCARDYGP 120 heavy chain for urelumab SEQ ID NO: 24 MEAPAQLLFL LLLWLPDTTG EIVLTQSPAT LSLSPGERAT LSCRASQSVS SYLAWYQQKP 60 variable GQAPRLLIYD ASNRATGIPA RFSGSGSGTD FTLTISSLEP EDFAVYYCQQ 110 light chain for urelumab SEQ ID NO: 25 GYYWS 5 heavy chain CDR1 for urelumab SEQ ID NO: 26 EINHGGYVTY NPSLES 16 heavy chain CDR2 for urelumab SEQ ID NO: 27 DYGPGNYDWY FDL 13 heavy chain CDR3 for urelumab SEQ ID NO: 28 RASQSVSSYL A 11 light chain CDR1 for urelumab SEQ ID NO: 29 DASNRAT 7 light chain CDR2 for urelumab SEQ ID NO: 30 QQRSDWPPAL T 11 light chain CDR3 for urelumab
[1204] In an embodiment, the 4-1BB agonist is selected from the group consisting of 1D8, 3Elor, 4B4 (BioLegend 309809), H4-1BB-M127 (BD Pharmingen 552532), BBK2 (Thermo Fisher MS621PABX), 145501 (Leinco Technologies B591), the antibody produced by cell line deposited as ATCC No. HB-11248 and disclosed in U.S. Pat. No. 6,974,863, 5F4 (BioLegend 31 1503), C65-485 (BD Pharmingen 559446), antibodies disclosed in U.S. Patent Application Publication No. US 2005/0095244, antibodies disclosed in U.S. Pat. No. 7,288,638 (such as 20H4.9-IgG1 (BMS-663031)), antibodies disclosed in U.S. Pat. No. 6,887,673 (such as 4E9 or BMS-554271), antibodies disclosed in U.S. Pat. No. 7,214,493, antibodies disclosed in U.S. Pat. No. 6,303,121, antibodies disclosed in U.S. Pat. No. 6,569,997, antibodies disclosed in U.S. Pat. No. 6,905,685 (such as 4E9 or BMS-554271), antibodies disclosed in U.S. Pat. No. 6,362,325 (such as 1D8 or BMS-469492; 3H3 or BMS-469497; or 3E1), antibodies disclosed in U.S. Pat. No. 6,974,863 (such as 53A2); antibodies disclosed in U.S. Pat. No. 6,210,669 (such as 1D8, 3B8, or 3E1), antibodies described in U.S. Pat. No. 5,928,893, antibodies disclosed in U.S. Pat. No. 6,303,121, antibodies disclosed in U.S. Pat. No. 6,569,997, antibodies disclosed in International Patent Application Publication Nos. WO 2012/177788, WO 2015/119923, and WO 2010/042433, and fragments, derivatives, conjugates, variants, or biosimilars thereof, wherein the disclosure of each of the foregoing patents or patent application publications is incorporated by reference here.
[1205] In an embodiment, the 4-1BB agonist is a 4-1BB agonistic fusion protein described in International Patent Application Publication Nos. WO 2008/025516 A1, WO 2009/007120 A1, WO 2010/003766 A1, WO 2010/010051 A1, and WO 2010/078966 A1; U.S. Patent Application Publication Nos. US 2011/0027218 A1, US 2015/0126709 A1, US 2011/0111494 A1, US 2015/0110734 A1, and US 2015/0126710 A1; and U.S. Pat. Nos. 9,359,420, 9,340,599, 8,921,519, and 8,450,460, the disclosures of which are incorporated by reference herein.
[1206] In an embodiment, the 4-1BB agonist is a 4-1BB agonistic fusion protein as depicted in Structure I-A (C-terminal Fc-antibody fragment fusion protein) or Structure I-B (N-terminal Fc-antibody fragment fusion protein) of FIG. 50, or a fragment, derivative, conjugate, variant, or biosimilar thereof:
In structures I-A and I-B, the cylinders refer to individual polypeptide binding domains. Structures I-A and I-B comprise three linearly-linked TNFRSF binding domains derived from e.g., 4-1BBL or an antibody that binds 4-1BB, which fold to form a trivalent protein, which is then linked to a second triavelent protein through IgG1-Fc (including C.sub.H3 and C.sub.H2 domains) is then used to link two of the trivalent proteins together through disulfide bonds (small elongated ovals), stabilizing the structure and providing an agonists capable of bringing together the intracellular signaling domains of the six receptors and signaling proteins to form a signaling complex. The TNFRSF binding domains denoted as cylinders may be scFv domains comprising, e.g., a V.sub.H and a V.sub.L chain connected by a linker that may comprise hydrophilic residues and Gly and Ser sequences for flexibility, as well as Glu and Lys for solubility. Any scFv domain design may be used, such as those described in de Marco, Microbial Cell Factories, 2011, 10, 44; Ahmad, et al., Clin. & Dev. Immunol. 2012, 980250; Monnier, et al., Antibodies, 2013, 2, 193-208; or in references incorporated elsewhere herein. Fusion protein structures of this form are described in U.S. Pat. Nos. 9,359,420, 9,340,599, 8,921,519, and 8,450,460, the disclosures of which are incorporated by reference herein.
[1207] Amino acid sequences for the other polypeptide domains of structure I-A are given in Table 6. The Fc domain preferably comprises a complete constant domain (amino acids 17-230 of SEQ ID NO:31) the complete hinge domain (amino acids 1-16 of SEQ ID NO:31) or a portion of the hinge domain (e.g., amino acids 4-16 of SEQ ID NO:31). Preferred linkers for connecting a C-terminal Fc-antibody may be selected from the embodiments given in SEQ ID NO:32 to SEQ ID NO:41, including linkers suitable for fusion of additional polypeptides.
TABLE-US-00006 TABLE 6 Amino acid sequences for TNFRSF fusion proteins, including 4-1BB fusion proteins, with C-terminal Fc-antibody fragment fusion protein design (structure I-A). Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 31 KSCDKTHTCP PCPAPELLGG PSVFLFPPKP KDTLMISRTP EVTCVVVDVS HEDPEVKFNW 60 Fc domain YVDGVEVHNA KTKPREEQYN STYRVVSVLT VLHQDWLNGK EYKCKVSNKA LPAPIEKTIS 120 KAKGQPREPQ VYTLPPSREE MTKNQVSLTC LVKGFYPSDI AVEWESNGQP ENNYKTTPPV 180 LDSDGSFFLY SKLTVDKSRW QQGNVFSCSV MHEALHNHYT QKSLSLSPGK 230 SEQ ID NO: 32 GGPGSSKSCD KTHTCPPCPA PE 22 linker SEQ ID NO: 33 GGSGSSKSCD KTHTCPPCPA PE 22 linker SEQ ID NO: 34 GGPGSSSSSS SKSCDKTHTC PPCPAPE 27 linker SEQ ID NO: 35 GGSGSSSSSS SKSCDKTHTC PPCPAPE 27 linker SEQ ID NO: 36 GGPGSSSSSS SSSKSCDKTH TCPPCPAPE 29 linker SEQ ID NO: 37 GGSGSSSSSS SSSKSCDKTH TCPPCPAPE 29 linker SEQ ID NO: 38 GGPGSSGSGS SDKTHTCPPC PAPE 24 linker SEQ ID NO: 39 GGPGSSGSGS DKTHTCPPCP APE 23 linker SEQ ID NO: 40 GGPSSSGSDK THTCPPCPAP E 21 linker SEQ ID NO: 41 GGSSSSSSSS GSDKTHTCPP CPAPE 25 linker
[1208] Amino acid sequences for the other polypeptide domains of structure I-B are given in Table 7. If an Fc antibody fragment is fused to the N-terminus of an TNRFSF fusion protein as in structure I-B, the sequence of the Fc module is preferably that shown in SEQ ID NO:42, and the linker sequences are preferably selected from those embodiments set forth in SEQ ID NO:43 to SEQ ID NO:45.
TABLE-US-00007 TABLE 7 Amino acid sequences for TNFRSF fusion proteins, including 4-1BB fusion proteins, with N-terminal Fc-antibody fragment fusion protein design (structure I-B). Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 42 METDTLLLWV LLLWVPAGNG DKTHTCPPCP APELLGGPSV FLFPPKPKDT LMISRTPEVT 60 Fc domain CVVVDVSHED PEVKFNWYVD GVEVHNAKTK PREEQYNSTY RVVSVLTVLH QDWLNGKEYK 120 CKVSNKALPA PIEKTISKAK GQPREPQVYT LPPSREEMTK NQVSLTCLVK GFYPSDIAVE 180 WESNGQPENN YKTTPPVLDS DGSFFLYSKL TVDKSRWQQG NVFSCSVMHE ALHNHYTQKS 240 LSLSPG 246 SEQ ID NO: 43 SGSGSGSGSG S 11 linker SEQ ID NO: 44 SSSSSSGSGS GS 12 linker SEQ ID NO: 45 SSSSSSGSGS GSGSGS 16 linker
[1209] In an embodiment, a 4-1BB agonist fusion protein according to structures I-A or I-B comprises one or more 4-1BB binding domains selected from the group consisting of a variable heavy chain and variable light chain of utomilumab, a variable heavy chain and variable light chain of urelumab, a variable heavy chain and variable light chain of utomilumab, a variable heavy chain and variable light chain selected from the variable heavy chains and variable light chains described in Table 8, any combination of a variable heavy chain and variable light chain of the foregoing, and fragments, derivatives, conjugates, variants, and biosimilars thereof.
[1210] In an embodiment, a 4-1BB agonist fusion protein according to structures I-A or I-B comprises one or more 4-1BB binding domains comprising a 4-1BBL sequence. In an embodiment, a 4-1BB agonist fusion protein according to structures I-A or I-B comprises one or more 4-1BB binding domains comprising a sequence according to SEQ ID NO:46. In an embodiment, a 4-1BB agonist fusion protein according to structures I-A or I-B comprises one or more 4-1BB binding domains comprising a soluble 4-1BBL sequence. In an embodiment, a 4-1BB agonist fusion protein according to structures I-A or I-B comprises one or more 4-1BB binding domains comprising a sequence according to SEQ ID NO:47.
[1211] In an embodiment, a 4-1BB agonist fusion protein according to structures I-A or I-B comprises one or more 4-1BB binding domains that is a scFv domain comprising V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO: 13 and SEQ ID NO: 14, respectively, wherein the V.sub.H and V.sub.L domains are connected by a linker. In an embodiment, a 4-1BB agonist fusion protein according to structures I-A or I-B comprises one or more 4-1BB binding domains that is a scFv domain comprising V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:23 and SEQ ID NO:24, respectively, wherein the V.sub.H and V.sub.L domains are connected by a linker. In an embodiment, a 4-1BB agonist fusion protein according to structures I-A or I-B comprises one or more 4-1BB binding domains that is a scFv domain comprising V.sub.H and V.sub.L regions that are each at least 95% identical to the V.sub.H and V.sub.L sequences given in Table 8, wherein the V.sub.H and V.sub.L domains are connected by a linker.
TABLE-US-00008 TABLE 8 Additional polypeptide domains useful as 4-1BB binding domains in fusion proteins or as scFv 4-1BB agonist antibodies. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 46 MEYASDASLD PEAPWPPAPR ARACRVLPWA LVAGLLLLLL LAAACAVFLA CPWAVSGARA 60 4-1BBL SPGSAASPRL REGPELSPDD PAGLLDLRQG MFAQLVAQNV LLIDGPLSWY SDPGLAGVSL 120 TGGLSYKEDT KELVVAKAGV YYVFFQLELR RVVAGEGSGS VSLALHLQPL RSAAGAAALA 180 LTVDLPPASS EARNSAFGFQ GRLLHLSAGQ RLGVHLHTEA RARHAWQLTQ GATVLGLFRV 240 TPEIPAGLPS PRSE 254 SEQ ID NO: 47 LRQGMFAQLV AQNVLLIDGP LSWYSDPGLA GVSLTGGLSY KEDTKELVVA KAGVYYVFFQ 60 4-1BBL soluble LELRRVVAGE GSGSVSLALH LQPLRSAAGA AALALTVDLP PASSEARNSA FGFQGRLLHL 120 domain SAGQRLGVHL HTEARARHAW QLTQGATVLG LFRVTPEIPA GLPSPRSE 168 SEQ ID NO: 48 QVQLQQPGAE LVKPGASVKL SCKASGYTFS SYWMHWVKQR PGQVLEWIGE INPGNGHTNY 60 variable heavy NEKFKSKATL TVDKSSSTAY MQLSSLTSED SAVYYCARSF TTARGFAYWG QGTLVTVS 118 chain for 4B4-1- 1 version 1 SEQ ID NO: 49 DIVMTQSPAT QSVTPGDRVS LSCRASQTIS DYLHWYQQKS HESPRLLIKY ASQSISGIPS 60 variable light RFSGSGSGSD FTLSINSVEP EDVGVYYCQD GHSFPPTFGG GTKLEIK 107 chain for 4B4-1- 1 version 1 SEQ ID NO: 50 QVQLQQPGAE LVKPGASVKL SCKASGYTFS SYWMHWVKQR PGQVLEWIGE INPGNGHTNY 60 variable heavy NEKFKSKATL TVDKSSSTAY MQLSSLTSED SAVYYCARSF TTARGFAYWG QGTLVTVSA 119 chain for 4B4-1- 1 version 2 SEQ ID NO: 51 DIVMTQSPAT QSVTPGDRVS LSCRASQTIS DYLHWYQQKS HESPRLLIKY ASQSISGIPS 60 variable light RFSGSGSGSD FTLSINSVEP EDVGVYYCQD GHSFPPTFGG GTKLEIKR 108 chain for 4B4-1- 1 version 2 SEQ ID NO: 52 MDWTWRILFL VAAATGAHSE VQLVESGGGL VQPGGSLRLS CAASGFTFSD YWMSWVRQAP 60 variable heavy GKGLEWVADI KNDGSYTNYA PSLTNRFTIS RDNAKNSLYL QMNSLRAEDT AVYYCARELT 120 chain for H39E3- 2 SEQ ID NO: 53 MEAPAQLLFL LLLWLPDTTG DIVMTQSPDS LAVSLGERAT INCKSSQSLL SSGNQKNYL 60 variable light WYQQKPGQPP KLLIYYASTR QSGVPDRFSG SGSGTDFTLT ISSLQAEDVA 110 chain for H39E3- 2
[1212] In an embodiment, the 4-1BB agonist is a 4-1BB agonistic single-chain fusion polypeptide comprising (i) a first soluble 4-1BB binding domain, (ii) a first peptide linker, (iii) a second soluble 4-1BB binding domain, (iv) a second peptide linker, and (v) a third soluble 4-1BB binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, and wherein the additional domain is a Fab or Fc fragment domain. In an embodiment, the 4-1BB agonist is a 4-1BB agonistic single-chain fusion polypeptide comprising (i) a first soluble 4-1BB binding domain, (ii) a first peptide linker, (iii) a second soluble 4-1BB binding domain, (iv) a second peptide linker, and (v) a third soluble 4-1BB binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, wherein the additional domain is a Fab or Fc fragment domain, wherein each of the soluble 4-1BB domains lacks a stalk region (which contributes to trimerisation and provides a certain distance to the cell membrane, but is not part of the 4-1BB binding domain) and the first and the second peptide linkers independently have a length of 3-8 amino acids.
[1213] In an embodiment, the 4-1BB agonist is a 4-1BB agonistic single-chain fusion polypeptide comprising (i) a first soluble tumor necrosis factor (TNF) superfamily cytokine domain, (ii) a first peptide linker, (iii) a second soluble TNF superfamily cytokine domain, (iv) a second peptide linker, and (v) a third soluble TNF superfamily cytokine domain, wherein each of the soluble TNF superfamily cytokine domains lacks a stalk region and the first and the second peptide linkers independently have a length of 3-8 amino acids, and wherein each TNF superfamily cytokine domain is a 4-1BB binding domain.
[1214] In an embodiment, the 4-1BB agonist is a 4-1BB agonistic scFv antibody comprising any of the foregoing V.sub.H domains linked to any of the foregoing V.sub.L domains.
[1215] In an embodiment, the 4-1BB agonist is BPS Bioscience 4-1BB agonist antibody catalog no. 79097-2, commercially available from BPS Bioscience, San Diego, Calif., USA. In an embodiment, the 4-1BB agonist is Creative Biolabs 4-1BB agonist antibody catalog no. MOM-18179, commercially available from Creative Biolabs, Shirley, N.Y., USA.
OX40 (CD134) Agonists
[1216] In an embodiment, the TNFRSF agonist is an OX40 (CD134) agonist. The OX40 agonist may be any OX40 binding molecule known in the art. The OX40 binding molecule may be a monoclonal antibody or fusion protein capable of binding to human or mammalian OX40. The OX40 agonists or OX40 binding molecules may comprise an immunoglobulin heavy chain of any isotype (e.g., IgG, IgE, IgM, IgD, IgA, and IgY), class (e.g., IgG1, IgG2, IgG3, IgG4, IgA1 and IgA2) or subclass of immunoglobulin molecule. The OX40 agonist or OX40 binding molecule may have both a heavy and a light chain. As used herein, the term binding molecule also includes antibodies (including full length antibodies), monoclonal antibodies (including full length monoclonal antibodies), polyclonal antibodies, multispecific antibodies (e.g., bispecific antibodies), human, humanized or chimeric antibodies, and antibody fragments, e.g., Fab fragments, F(ab') fragments, fragments produced by a Fab expression library, epitope-binding fragments of any of the above, and engineered forms of antibodies, e.g., scFv molecules, that bind to OX40. In an embodiment, the OX40 agonist is an antigen binding protein that is a fully human antibody. In an embodiment, the OX40 agonist is an antigen binding protein that is a humanized antibody. In some embodiments, OX40 agonists for use in the presently disclosed methods and compositions include anti-OX40 antibodies, human anti-OX40 antibodies, mouse anti-OX40 antibodies, mammalian anti-OX40 antibodies, monoclonal anti-OX40 antibodies, polyclonal anti-OX40 antibodies, chimeric anti-OX40 antibodies, anti-OX40 adnectins, anti-OX40 domain antibodies, single chain anti-OX40 fragments, heavy chain anti-OX40 fragments, light chain anti-OX40 fragments, anti-OX40 fusion proteins, and fragments, derivatives, conjugates, variants, or biosimilars thereof. In a preferred embodiment, the OX40 agonist is an agonistic, anti-OX40 humanized or fully human monoclonal antibody (i.e., an antibody derived from a single cell line).
[1217] In a preferred embodiment, the OX40 agonist or OX40 binding molecule may also be a fusion protein. OX40 fusion proteins comprising an Fc domain fused to OX40L are described, for example, in Sadun, et al., J. Immunother. 2009, 182, 1481-89. In a preferred embodiment, a multimeric OX40 agonist, such as a trimeric or hexameric OX40 agonist (with three or six ligand binding domains), may induce superior receptor (OX40L) clustering and internal cellular signaling complex formation compared to an agonistic monoclonal antibody, which typically possesses two ligand binding domains. Trimeric (trivalent) or hexameric (or hexavalent) or greater fusion proteins comprising three TNFRSF binding domains and IgG1-Fc and optionally further linking two or more of these fusion proteins are described, e.g., in Gieffers, et al., Mol. Cancer Therapeutics 2013, 12, 2735-47.
[1218] Agonistic OX40 antibodies and fusion proteins are known to induce strong immune responses. Curti, et al., Cancer Res. 2013, 73, 7189-98. In a preferred embodiment, the OX40 agonist is a monoclonal antibody or fusion protein that binds specifically to OX40 antigen in a manner sufficient to reduce toxicity. In some embodiments, the OX40 agonist is an agonistic OX40 monoclonal antibody or fusion protein that abrogates antibody-dependent cellular toxicity (ADCC), for example NK cell cytotoxicity. In some embodiments, the OX40 agonist is an agonistic OX40 monoclonal antibody or fusion protein that abrogates antibody-dependent cell phagocytosis (ADCP). In some embodiments, the OX40 agonist is an agonistic OX40 monoclonal antibody or fusion protein that abrogates complement-dependent cytotoxicity (CDC). In some embodiments, the OX40 agonist is an agonistic OX40 monoclonal antibody or fusion protein which abrogates Fc region functionality.
[1219] In some embodiments, the OX40 agonists are characterized by binding to human OX40 (SEQ ID NO:54) with high affinity and agonistic activity. In an embodiment, the OX40 agonist is a binding molecule that binds to human OX40 (SEQ ID NO:54). In an embodiment, the OX40 agonist is a binding molecule that binds to murine OX40 (SEQ ID NO:55). The amino acid sequences of OX40 antigen to which an OX40 agonist or binding molecule binds are summarized in Table 9.
TABLE-US-00009 TABLE 9 Amino acid sequences of OX40 antigens. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 54 MCVGARRLGR GPCAALLLLG LGLSTVTGLH CVGDTYPSND RCCHECRPGN GMVSRCSRSQ 60 human OX40 NTVCRPCGPG FYNDVVSSKP CKPCTWCNLR SGSERKQLCT ATQDTVCRCR AGTQPLDSYK 120 (Homo sapiens) PGVDCAPCPP GHFSPGDNQA CKPWTNCTLA GKHTLQPASN SSDAICEDRD PPATQPQETQ 180 GPPARPITVQ PTEAWPRTSQ GPSTRPVEVP GGRAVAAILG LGLVLGLLGP LAILLALYLL 240 RRDQRLPPDA HKPPGGGSFR TPIQEEQADA HSTLAKI 277 SEQ ID NO: 55 MYVWVQQPTA LLLLGLTLGV TARRLNCVKH TYPSGHKCCR ECQPGHGMVS RCDHTRDTLC 60 murine OX40 HPCETGFYNE AMNYDTCKQC TQCNHRSGSE LKQNCTPTQD TVCRCRPGTQ PRQDSGYKLG 120 (Mus musculus) VECVPCPPGH FSPGNNQACK PWTNCTLSGK QTRHPASDSL DAVCEDRSLL ATLLWETQRP 180 TFRPTTVQST TVWPRTSELP SPPTLVTPEG PAFAVLLGLG LGLLAPLTVL LALYLLRKAW 240 RLPNTPKPCW GNSFRTPIQE EHTDAHFTLA KI 272
[1220] In some embodiments, the compositions, processes and methods described include a OX40 agonist that binds human or murine OX40 with a K.sub.D of about 100 pM or lower, binds human or murine OX40 with a K.sub.D of about 90 pM or lower, binds human or murine OX40 with a K.sub.D of about 80 pM or lower, binds human or murine OX40 with a K.sub.D of about 70 pM or lower, binds human or murine OX40 with a K.sub.D of about 60 pM or lower, binds human or murine OX40 with a K.sub.D of about 50 pM or lower, binds human or murine OX40 with a K.sub.D of about 40 pM or lower, or binds human or murine OX40 with a K.sub.D of about 30 pM or lower.
[1221] In some embodiments, the compositions, processes and methods described include a OX40 agonist that binds to human or murine OX40 with a k.sub.assoc of about 7.5.times.10.sup.5 l/Ms or faster, binds to human or murine OX40 with a k.sub.assoc of about 7.5.times.10.sup.5 l/Ms or faster, binds to human or murine OX40 with a k.sub.assoc of about 8.times.10.sup.5 l/Ms or faster, binds to human or murine OX40 with a k.sub.assoc of about 8.5.times.10.sup.5 l/Ms or faster, binds to human or murine OX40 with a k.sub.assoc of about 9.times.10.sup.5 l/Ms or faster, binds to human or murine OX40 with a k.sub.assoc of about 9.5.times.10.sup.5 l/Ms or faster, or binds to human or murine OX40 with a k.sub.assoc of about 1.times.10.sup.6 l/Ms or faster.
[1222] In some embodiments, the compositions, processes and methods described include a OX40 agonist that binds to human or murine OX40 with a k.sub.dissoc of about 2.times.10.sup.-5 l/s or slower, binds to human or murine OX40 with a k.sub.dissoc of about 2.1.times.10.sup.-5 l/s or slower, binds to human or murine OX40 with a k.sub.dissoc of about 2.2.times.10.sup.-5 l/s or slower, binds to human or murine OX40 with a k.sub.dissoc of about 2.3.times.10.sup.-5 l/s or slower, binds to human or murine OX40 with a k.sub.dissoc of about 2.4.times.10.sup.-5 l/s or slower, binds to human or murine OX40 with a k.sub.dissoc of about 2.5.times.10.sup.-5 l/s or slower, binds to human or murine OX40 with a k.sub.dissoc of about 2.6.times.10.sup.-5 l/s or slower or binds to human or murine OX40 with a k.sub.dissoc of about 2.7.times.10.sup.-5 l/s or slower, binds to human or murine OX40 with a k.sub.dissoc of about 2.8.times.10.sup.-5 l/s or slower, binds to human or murine OX40 with a k.sub.dissoc of about 2.9.times.10.sup.-5 l/s or slower, or binds to human or murine OX40 with a k.sub.dissoc of about 3.times.10.sup.-5 l/s or slower.
[1223] In some embodiments, the compositions, processes and methods described include OX40 agonist that binds to human or murine OX40 with an IC.sub.50 of about 10 nM or lower, binds to human or murine OX40 with an IC.sub.50 of about 9 nM or lower, binds to human or murine OX40 with an IC.sub.50 of about 8 nM or lower, binds to human or murine OX40 with an IC.sub.50 of about 7 nM or lower, binds to human or murine OX40 with an IC.sub.50 of about 6 nM or lower, binds to human or murine OX40 with an IC.sub.50 of about 5 nM or lower, binds to human or murine OX40 with an IC.sub.50 of about 4 nM or lower, binds to human or murine OX40 with an IC.sub.50 of about 3 nM or lower, binds to human or murine OX40 with an IC.sub.50 of about 2 nM or lower, or binds to human or murine OX40 with an IC.sub.50 of about 1 nM or lower.
[1224] In some embodiments, the OX40 agonist is tavolixizumab, also known as MEDI0562 or MEDI-0562. Tavolixizumab is available from the MedImmune subsidiary of AstraZeneca, Inc. Tavolixizumab is immunoglobulin G1-kappa, anti-[Homo sapiens TNFRSF4 (tumor necrosis factor receptor (TNFR) superfamily member 4, OX40, CD134)], humanized and chimeric monoclonal antibody. The amino acid sequences of tavolixizumab are set forth in Table 10. Tavolixizumab comprises N-glycosylation sites at positions 301 and 301'', with fucosylated complex bi-antennary CHO-type glycans; heavy chain intrachain disulfide bridges at positions 22-95 (V.sub.H-V.sub.L), 148-204 (C.sub.H1-C.sub.L), 265-325 (C.sub.H2) and 371-429 (C.sub.H3) (and at positions 22''-95'', 148''-204'', 265''-325'', and 371''-429''); light chain intrachain disulfide bridges at positions 23'-88' (V.sub.H-V.sub.L) and 134'-194' (C.sub.H1-C.sub.L) (and at positions 23'''-88''' and 134'''-194'''); interchain heavy chain-heavy chain disulfide bridges at positions 230-230'' and 233-233''; and interchain heavy chain-light chain disulfide bridges at 224-214' and 224''-214'''. Current clinical trials of tavolixizumab in a variety of solid tumor indications include U.S. National Institutes of Health clinicaltrials.gov identifiers NCT02318394 and NCT02705482.
[1225] In an embodiment, a OX40 agonist comprises a heavy chain given by SEQ ID NO:56 and a light chain given by SEQ ID NO:57. In an embodiment, a OX40 agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:56 and SEQ ID NO:57, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a OX40 agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:56 and SEQ ID NO:57, respectively. In an embodiment, a OX40 agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:56 and SEQ ID NO:57, respectively. In an embodiment, a OX40 agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:56 and SEQ ID NO:57, respectively. In an embodiment, a OX40 agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:56 and SEQ ID NO:57, respectively. In an embodiment, a OX40 agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:56 and SEQ ID NO:57, respectively.
[1226] In an embodiment, the OX40 agonist comprises the heavy and light chain CDRs or variable regions (VRs) of tavolixizumab. In an embodiment, the OX40 agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:58, and the OX40 agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:59, and conservative amino acid substitutions thereof. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:58 and SEQ ID NO:59, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:58 and SEQ ID NO:59, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:58 and SEQ ID NO:59, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:58 and SEQ ID NO:59, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:58 and SEQ ID NO:59, respectively. In an embodiment, an OX40 agonist comprises an scFv antibody comprising V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:58 and SEQ ID NO:59.
[1227] In an embodiment, a OX40 agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:60, SEQ ID NO:61, and SEQ ID NO:62, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:63, SEQ ID NO:64, and SEQ ID NO:65, respectively, and conservative amino acid substitutions thereof.
[1228] In an embodiment, the OX40 agonist is a OX40 agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to tavolixizumab. In an embodiment, the biosimilar monoclonal antibody comprises an OX40 antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is tavolixizumab. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a OX40 agonist antibody authorized or submitted for authorization, wherein the OX40 agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is tavolixizumab. The OX40 agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is tavolixizumab. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is tavolixizumab.
TABLE-US-00010 TABLE 10 Amino acid sequences for OX40 agonist antibodies related to tavolixizumab. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 56 QVQLQESGPG LVKPSQTLSL TCAVYGGSFS SGYWNWIRKH PGKGLEYIGY ISYNGITYHN 60 heavy chain for PSLKSRITIN RDTSKNQYSL QLNSVTPEDT AVYYCARYKY DYDGGHAMDY WGQGTLVTVS 120 tavolixizumab SASTKGPSVF PLAPSSKSTS GGTAALGCLV KDYFPEPVTV SWNSGALTSG VHTFPAVLQS 180 SGLYSLSSVV TVPSSSLGTQ TYICNVNHKP SNTKVDKRVE PKSCDKTHTC PPCPAPELLG 240 GPSVFLFPPK PKDTLMISRT PEVTCVVVDV SHEDPEVKFN WYVDGVEVHN AKTKPREEQY 300 NSTYRVVSVL TVLHQDWLNG KEYKCKVSNK ALPAPIEKTI SKAKGQPREP QVYTLPPSRE 360 EMTKNQVSLT CLVKGFYPSD IAVEWESNGQ PENNYKTTPP VLDSDGSFFL YSKLTVDKSR 420 WQQGNVFSCS VMHEALHNHY TQKSLSLSPG K 451 SEQ ID NO: 57 DIQMTQSPSS LSASVGDRVT ITCRASQDIS NYLNWYQQKP GKAPKLLIYY TSKLHSGVPS 60 light chain for RFSGSGSGTD YTLTISSLQP EDFATYYCQQ GSALPWTFGQ GTKVEIKRTV AAPSVFIFPP 120 tavolixizumab SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 58 QVQLQESGPG LVKPSQTLSL TCAVYGGSFS SGYWNWIRKH PGKGLEYIGY ISYNGITYHN 60 heavy chain PSLKSRITIN RDTSKNQYSL QLNSVTPEDT AVYYCARYKY DYDGGHAMDY WGQGTLVT 118 variable region for tavolixizumab SEQ ID NO: 59 DIQMTQSPSS LSASVGDRVT ITCRASQDIS NYLNWYQQKP GKAPKLLIYY TSKLHSGVPS 60 light chain RFSGSGSGTD YTLTISSLQP EDFATYYCQQ GSALPWTFGQ GTKVEIKR 108 variable region for tavolixizumab SEQ ID NO: 60 GSFSSGYWN 9 heavy chain CDR1 for tavolixizumab SEQ ID NO: 61 YIGYISYNGI TYH 13 heavy chain CDR2 for tavolixizumab SEQ ID NO: 62 RYKYDYDGGH AMDY 14 heavy chain CDR3 for tavolixizumab SEQ ID NO: 63 QDISNYLN 8 light chain CDR1 for tavolixizumab SEQ ID NO: 64 LLIYYTSKLH S 11 light chain CDR2 for tavolixizumab SEQ ID NO: 65 QQGSALPW 8 light chain CDR3 for tavolixizumab
[1229] In some embodiments, the OX40 agonist is 11D4, which is a fully human antibody available from Pfizer, Inc. The preparation and properties of 11D4 are described in U.S. Pat. Nos. 7,960,515; 8,236,930; and 9,028,824, the disclosures of which are incorporated by reference herein. The amino acid sequences of 11D4 are set forth in Table 11.
[1230] In an embodiment, a OX40 agonist comprises a heavy chain given by SEQ ID NO:66 and a light chain given by SEQ ID NO:67. In an embodiment, a OX40 agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:66 and SEQ ID NO:67, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a OX40 agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:66 and SEQ ID NO:67, respectively. In an embodiment, a OX40 agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:66 and SEQ ID NO:67, respectively. In an embodiment, a OX40 agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:66 and SEQ ID NO:67, respectively. In an embodiment, a OX40 agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:66 and SEQ ID NO:67, respectively. In an embodiment, a OX40 agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:66 and SEQ ID NO:67, respectively.
[1231] In an embodiment, the OX40 agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 11D4. In an embodiment, the OX40 agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:68, and the OX40 agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:69, and conservative amino acid substitutions thereof. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:68 and SEQ ID NO:69, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:68 and SEQ ID NO:69, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:68 and SEQ ID NO:69, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:68 and SEQ ID NO:69, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:68 and SEQ ID NO:69, respectively.
[1232] In an embodiment, a OX40 agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:70, SEQ ID NO:71, and SEQ ID NO:72, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:73, SEQ ID NO:74, and SEQ ID NO:75, respectively, and conservative amino acid substitutions thereof.
[1233] In an embodiment, the OX40 agonist is a OX40 agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 11D4. In an embodiment, the biosimilar monoclonal antibody comprises an OX40 antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 11D4. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a OX40 agonist antibody authorized or submitted for authorization, wherein the OX40 agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 11D4. The OX40 agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 11D4. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 11D4.
TABLE-US-00011 TABLE 11 Amino acid sequences for OX40 agonist antibodies related to 11D4. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 66 EVQLVESGGG LVQPGGSLRL SCAASGFTFS SYSMNWVRQA PGKGLEWVSY ISSSSSTIDY 60 heavy chain for ADSVKGRFTI SRDNAKNSLY LQMNSLRDED TAVYYCARES GWYLFDYWGQ GTLVTVSSAS 120 11D4 TKGPSVFPLA PCSRSTSEST AALGCLVKDY FPEPVTVSWN SGALTSGVHT FPAVLQSSGL 180 YSLSSVVTVP SSNFGTQTYT CNVDHKPSNT KVDKTVERKC CVECPPCPAP PVAGPSVFLF 240 PPKPKDTLMI SRTPEVTCVV VDVSHEDPEV QFNWYVDGVE VHNAKTKPRE EQFNSTFRVV 300 SVLTVVHQDW LNGKEYKCKV SNKGLPAPIE KTISKTKGQP REPQVYTLPP SREEMTKNQV 360 SLTCLVKGFY PSDIAVEWES NGQPENNYKT TPPMLDSDGS FFLYSKLTVD KSRWQQGNVF 420 SCSVMHEALH NHYTQKSLSL SPGK 444 SEQ ID NO: 67 DIQMTQSPSS LSASVGDRVT ITCRASQGIS SWLAWYQQKP EKAPKSLIYA ASSLQSGVPS 60 light chain for RFSGSGSGTD FTLTISSLQP EDFATYYCQQ YNSYPPTFGG GTKVEIKRTV AAPSVFIFPP 120 11D4 SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 68 EVQLVESGGG LVQPGGSLRL SCAASGFTFS SYSMNWVRQA PGKGLEWVSY ISSSSSTIDY 60 heavy chain ADSVKGRFTI SRDNAKNSLY LQMNSLRDED TAVYYCARES GWYLFDYWGQ GTLVTVSS 118 variable region for 11D4 SEQ ID NO: 69 DIQMTQSPSS LSASVGDRVT ITCRASQGIS SWLAWYQQKP EKAPKSLIYA ASSLQSGVPS 60 light chain RFSGSGSGTD FTLTISSLQP EDFATYYCQQ YNSYPPTFGG GTKVEIK 107 variable region for 11D4 SEQ ID NO: 70 SYSMN 5 heavy chain CDR1 for 11D4 SEQ ID NO: 71 YISSSSSTID YADSVKG 17 heavy chain CDR2 for 11D4 SEQ ID NO: 72 ESGWYLFDY 9 heavy chain CDR3 for 11D4 SEQ ID NO: 73 RASQGISSWL A 11 light chain CDR1 for 11D4 SEQ ID NO: 74 AASSLQS 7 light chain CDR2 for 11D4 SEQ ID NO: 75 QQYNSYPPT 9 light chain CDR3 for 11D4
[1234] In some embodiments, the OX40 agonist is 18D8, which is a fully human antibody available from Pfizer, Inc. The preparation and properties of 18D8 are described in U.S. Pat. Nos. 7,960,515; 8,236,930; and 9,028,824, the disclosures of which are incorporated by reference herein. The amino acid sequences of 18D8 are set forth in Table 12.
[1235] In an embodiment, a OX40 agonist comprises a heavy chain given by SEQ ID NO:76 and a light chain given by SEQ ID NO:77. In an embodiment, a OX40 agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:76 and SEQ ID NO:77, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a OX40 agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:76 and SEQ ID NO:77, respectively. In an embodiment, a OX40 agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:76 and SEQ ID NO:77, respectively. In an embodiment, a OX40 agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:76 and SEQ ID NO:77, respectively. In an embodiment, a OX40 agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:76 and SEQ ID NO:77, respectively. In an embodiment, a OX40 agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:76 and SEQ ID NO:77, respectively.
[1236] In an embodiment, the OX40 agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 18D8. In an embodiment, the OX40 agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:78, and the OX40 agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:79, and conservative amino acid substitutions thereof. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:78 and SEQ ID NO:79, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:78 and SEQ ID NO:79, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:78 and SEQ ID NO:79, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:78 and SEQ ID NO:79, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:78 and SEQ ID NO:79, respectively.
[1237] In an embodiment, a OX40 agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:80, SEQ ID NO:81, and SEQ ID NO:82, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:83, SEQ ID NO:84, and SEQ ID NO:85, respectively, and conservative amino acid substitutions thereof.
[1238] In an embodiment, the OX40 agonist is a OX40 agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 18D8. In an embodiment, the biosimilar monoclonal antibody comprises an OX40 antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 18D8. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a OX40 agonist antibody authorized or submitted for authorization, wherein the OX40 agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 18D8. The OX40 agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 18D8. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 18D8.
TABLE-US-00012 TABLE 12 Amino acid sequences for OX40 agonist antibodies related to 18D8. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 76 EVQLVESGGG LVQPGRSLRL SCAASGFTFD DYAMHWVRQA PGKGLEWVSG ISWNSGSIGY 60 heavy chain for ADSVKGRFTI SRDNAKNSLY LQMNSLRAED TALYYCAKDQ STADYYFYYG MDVWGQGTTV 120 18D8 TVSSASTKGP SVFPLAPCSR STSESTAALG CLVKDYFPEP VTVSWNSGAL TSGVHTFPAV 180 LQSSGLYSLS SVVTVPSSNF GTQTYTCNVD HKPSNTKVDK TVERKCCVEC PPCPAPPVAG 240 PSVFLFPPKP KDTLMISRTP EVTCVVVDVS HEDPEVQFNW YVDGVEVHNA KTKPREEQFN 300 STFRVVSVLT VVHQDWLNGK EYKCKVSNKG LPAPIEKTIS KTKGQPREPQ VYTLPPSREE 360 MTKNQVSLTC LVKGFYPSDI AVEWESNGQP ENNYKTTPPM LDSDGSFFLY SKLTVDKSRW 420 QQGNVFSCSV MHEALHNHYT QKSLSLSPGK 450 SEQ ID NO: 77 EIVVTQSPAT LSLSPGERAT LSCRASQSVS SYLAWYQQKP GQAPRLLIYD ASNRATGIPA 60 light chain for RFSGSGSGTD FTLTISSLEP EDFAVYYCQQ RSNWPTFGQG TKVEIKRTVA APSVFIFPPS 120 18D8 DEQLKSGTAS VVCLLNNFYP REAKVQWKVD NALQSGNSQE SVTEQDSKDS TYSLSSTLTL 180 SKADYEKHKV YACEVTHQGL SSPVTKSFNR GEC 213 SEQ ID NO: 78 EVQLVESGGG LVQPGRSLRL SCAASGFTFD DYAMHWVRQA PGKGLEWVSG ISWNSGSIGY 60 heavy chain ADSVKGRFTI SRDNAKNSLY LQMNSLRAED TALYYCAKDQ STADYYFYYG MDVWGQGTTV 120 variable region TVSS 124 for 18D8 SEQ ID NO: 79 EIVVTQSPAT LSLSPGERAT LSCRASQSVS SYLAWYQQKP GQAPRLLIYD ASNRATGIPA 60 light chain RFSGSGSGTD FTLTISSLEP EDFAVYYCQQ RSNWPTFGQG TKVEIK 106 variable region for 18D8 SEQ ID NO: 80 DYAMH 5 heavy chain CDR1 for 18D8 SEQ ID NO: 81 GISWNSGSIG YADSVKG 17 heavy chain CDR2 for 18D8 SEQ ID NO: 82 DQSTADYYFY YGMDV 15 heavy chain CDR3 for 18D8 SEQ ID NO: 83 RASQSVSSYL A 11 light chain CDR1 for 18D8 SEQ ID NO: 84 DASNRAT 7 light chain CDR2 for 18D8 SEQ ID NO: 85 QQRSNWPT 8 light chain CDR3 for 18D8
[1239] In some embodiments, the OX40 agonist is Hu119-122, which is a humanized antibody available from GlaxoSmithKline plc. The preparation and properties of Hu119-122 are described in U.S. Pat. Nos. 9,006,399 and 9,163,085, and in International Patent Publication No. WO 2012/027328, the disclosures of which are incorporated by reference herein. The amino acid sequences of Hu119-122 are set forth in Table 13.
[1240] In an embodiment, the OX40 agonist comprises the heavy and light chain CDRs or variable regions (VRs) of Hu119-122. In an embodiment, the OX40 agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:86, and the OX40 agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:87, and conservative amino acid substitutions thereof. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO: 86 and SEQ ID NO:87, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:86 and SEQ ID NO:87, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:86 and SEQ ID NO:87, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:86 and SEQ ID NO:87, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:86 and SEQ ID NO:87, respectively.
[1241] In an embodiment, a OX40 agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:88, SEQ ID NO:89, and SEQ ID NO:90, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:91, SEQ ID NO:92, and SEQ ID NO:93, respectively, and conservative amino acid substitutions thereof.
[1242] In an embodiment, the OX40 agonist is a OX40 agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to Hu119-122. In an embodiment, the biosimilar monoclonal antibody comprises an OX40 antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is Hu119-122. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a OX40 agonist antibody authorized or submitted for authorization, wherein the OX40 agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is Hu119-122. The OX40 agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is Hu119-122. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is Hu119-122.
TABLE-US-00013 TABLE 13 Amino acid sequences for OX40 agonist antibodies related to Hu119-122. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 86 EVQLVESGGG LVQPGGSLRL SCAASEYEFP SHDMSWVRQA PGKGLELVAA INSDGGSTYY 60 heavy chain PDTMERRFTI SRDNAKNSLY LQMNSLRAED TAVYYCARHY DDYYAWFAYW GQGTMVTVSS 120 variable region for Hu119-122 SEQ ID NO: 87 EIVLTQSPAT LSLSPGERAT LSCRASKSVS TSGYSYMHWY QQKPGQAPRL LIYLASNLES 60 light chain GVPARFSGSG SGTDFTLTIS SLEPEDFAVY YCQHSRELPL TFGGGTKVEI K 111 variable region for Hu119-122 SEQ ID NO: 88 SHDMS 5 heavy chain CDR1 for Hu119-122 SEQ ID NO: 89 AINSDGGSTY YPDTMER 17 heavy chain CDR2 for Hu119-122 SEQ ID NO: 90 HYDDYYAWFA Y 11 heavy chain CDR3 for Hu119-122 SEQ ID NO: 91 RASKSVSTSG YSYMH 15 light chain CDR1 for Hu119-122 SEQ ID NO: 92 LASNLES 7 light chain CDR2 for Hu119-122 SEQ ID NO: 93 QHSRELPLT 9 light chain CDR3 for Hu119-122
[1243] In some embodiments, the OX40 agonist is Hu106-222, which is a humanized antibody available from GlaxoSmithKline plc. The preparation and properties of Hu106-222 are described in U.S. Pat. Nos. 9,006,399 and 9,163,085, and in International Patent Publication No. WO 2012/027328, the disclosures of which are incorporated by reference herein. The amino acid sequences of Hu106-222 are set forth in Table 14.
[1244] In an embodiment, the OX40 agonist comprises the heavy and light chain CDRs or variable regions (VRs) of Hu106-222. In an embodiment, the OX40 agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:94, and the OX40 agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:95, and conservative amino acid substitutions thereof. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:94 and SEQ ID NO:95, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:94 and SEQ ID NO:95, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:94 and SEQ ID NO:95, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:94 and SEQ ID NO:95, respectively. In an embodiment, a OX40 agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:94 and SEQ ID NO:95, respectively.
[1245] In an embodiment, a OX40 agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:96, SEQ ID NO:97, and SEQ ID NO:98, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:99, SEQ ID NO:100, and SEQ ID NO: 101, respectively, and conservative amino acid substitutions thereof.
[1246] In an embodiment, the OX40 agonist is a OX40 agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to Hu106-222. In an embodiment, the biosimilar monoclonal antibody comprises an OX40 antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is Hu106-222. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a OX40 agonist antibody authorized or submitted for authorization, wherein the OX40 agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is Hu106-222. The OX40 agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is Hu106-222. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is Hu106-222.
TABLE-US-00014 TABLE 14 Amino acid sequences for OX40 agonist antibodies related to Hu106-222. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 94 QVQLVQSGSE LKKPGASVKV SCKASGYTFT DYSMHWVRQA PGQGLKWMGW INTETGEPTY 60 heavy chain ADDFKGRFVF SLDTSVSTAY LQISSLKAED TAVYYCANPY YDYVSYYAMD YWGQGTTVTV 120 variable region SS 122 for Hu106-222 SEQ ID NO: 95 DIQMTQSPSS LSASVGDRVT ITCKASQDVS TAVAWYQQKP GKAPKLLIYS ASYLYTGVPS 60 light chain RFSGSGSGTD FTFTISSLQP EDIATYYCQQ HYSTPRTFGQ GTKLEIK 107 variable region for Hu106-222 SEQ ID NO: 96 DYSMH 5 heavy chain CDR1 for Hu106-222 SEQ ID NO: 97 WINTETGEPT YADDFKG 17 heavy chain CDR2 for Hu106-222 SEQ ID NO: 98 PYYDYVSYYA MDY 13 heavy chain CDR3 for Hu106-222 SEQ ID NO: 99 KASQDVSTAV A 11 light chain CDR1 for Hu106-222 SEQ ID NO: 100 SASYLYT 7 light chain CDR2 for Hu106-222 SEQ ID NO: 101 QQHYSTPRT 9 light chain CDR3 for Hu106-222
[1247] In some embodiments, the OX40 agonist antibody is MEDI6469 (also referred to as 9B 12). MEDI6469 is a murine monoclonal antibody. Weinberg, et al., J. Immunother. 2006, 29, 575-585. In some embodiments the OX40 agonist is an antibody produced by the 9B12 hybridoma, deposited with Biovest Inc. (Malvern, Mass., USA), as described in Weinberg, et al., J. Immunother. 2006, 29, 575-585, the disclosure of which is hereby incorporated by reference in its entirety. In some embodiments, the antibody comprises the CDR sequences of MEDI6469. In some embodiments, the antibody comprises a heavy chain variable region sequence and/or a light chain variable region sequence of MEDI6469.
[1248] In an embodiment, the OX40 agonist is L106 BD (Pharmingen Product #340420). In some embodiments, the OX40 agonist comprises the CDRs of antibody L106 (BD Pharmingen Product #340420). In some embodiments, the OX40 agonist comprises a heavy chain variable region sequence and/or a light chain variable region sequence of antibody L106 (BD Pharmingen Product #340420). In an embodiment, the OX40 agonist is ACT35 (Santa Cruz Biotechnology, Catalog #20073). In some embodiments, the OX40 agonist comprises the CDRs of antibody ACT35 (Santa Cruz Biotechnology, Catalog #20073). In some embodiments, the OX40 agonist comprises a heavy chain variable region sequence and/or a light chain variable region sequence of antibody ACT35 (Santa Cruz Biotechnology, Catalog #20073). In an embodiment, the OX40 agonist is the murine monoclonal antibody anti-mCD134/mOX40 (clone OX86), commercially available from InVivoMAb, BioXcell Inc, West Lebanon, N.H.
[1249] In an embodiment, the OX40 agonist is selected from the OX40 agonists described in International Patent Application Publication Nos. WO 95/12673, WO 95/21925, WO 2006/121810, WO 2012/027328, WO 2013/028231, WO 2013/038191, and WO 2014/148895; European Patent Application EP 0672141; U.S. Patent Application Publication Nos. US 2010/136030, US 2014/377284, US 2015/190506, and US 2015/132288 (including clones 20E5 and 12H3); and U.S. Pat. Nos. 7,504,101, 7,550,140, 7,622,444, 7,696,175, 7,960,515, 7,961,515, 8,133,983, 9,006,399, and 9,163,085, the disclosure of each of which is incorporated herein by reference in its entirety.
[1250] In an embodiment, the OX40 agonist is an OX40 agonistic fusion protein as depicted in Structure I-A (C-terminal Fc-antibody fragment fusion protein) or Structure I-B (N-terminal Fc-antibody fragment fusion protein), or a fragment, derivative, conjugate, variant, or biosimilar thereof. The properties of structures I-A and I-B are described above and in U.S. Pat. Nos. 9,359,420, 9,340,599, 8,921,519, and 8,450,460, the disclosures of which are incorporated by reference herein. Amino acid sequences for the polypeptide domains of structure I-A are given in Table 6. The Fc domain preferably comprises a complete constant domain (amino acids 17-230 of SEQ ID NO:31) the complete hinge domain (amino acids 1-16 of SEQ ID NO:31) or a portion of the hinge domain (e.g., amino acids 4-16 of SEQ ID NO:31). Preferred linkers for connecting a C-terminal Fc-antibody may be selected from the embodiments given in SEQ ID NO:32 to SEQ ID NO:41, including linkers suitable for fusion of additional polypeptides. Likewise, amino acid sequences for the polypeptide domains of structure I-B are given in Table 7. If an Fc antibody fragment is fused to the N-terminus of an TNRFSF fusion protein as in structure I-B, the sequence of the Fc module is preferably that shown in SEQ ID NO:42, and the linker sequences are preferably selected from those embodiments set forth in SEQ ID NO:43 to SEQ ID NO:45.
[1251] In an embodiment, an OX40 agonist fusion protein according to structures I-A or I-B comprises one or more OX40 binding domains selected from the group consisting of a variable heavy chain and variable light chain of tavolixizumab, a variable heavy chain and variable light chain of 11D4, a variable heavy chain and variable light chain of 18D8, a variable heavy chain and variable light chain of Hu119-122, a variable heavy chain and variable light chain of Hu106-222, a variable heavy chain and variable light chain selected from the variable heavy chains and variable light chains described in Table 15, any combination of a variable heavy chain and variable light chain of the foregoing, and fragments, derivatives, conjugates, variants, and biosimilars thereof.
[1252] In an embodiment, an OX40 agonist fusion protein according to structures I-A or I-B comprises one or more OX40 binding domains comprising an OX40L sequence. In an embodiment, an OX40 agonist fusion protein according to structures I-A or I-B comprises one or more OX40 binding domains comprising a sequence according to SEQ ID NO: 102. In an embodiment, an OX40 agonist fusion protein according to structures I-A or I-B comprises one or more OX40 binding domains comprising a soluble OX40L sequence. In an embodiment, a OX40 agonist fusion protein according to structures I-A or I-B comprises one or more OX40 binding domains comprising a sequence according to SEQ ID NO: 103. In an embodiment, a OX40 agonist fusion protein according to structures I-A or I-B comprises one or more OX40 binding domains comprising a sequence according to SEQ ID NO: 104.
[1253] In an embodiment, an OX40 agonist fusion protein according to structures I-A or I-B comprises one or more OX40 binding domains that is a scFv domain comprising V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:58 and SEQ ID NO:59, respectively, wherein the V.sub.H and V.sub.L domains are connected by a linker. In an embodiment, an OX40 agonist fusion protein according to structures I-A or I-B comprises one or more OX40 binding domains that is a scFv domain comprising V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:68 and SEQ ID NO:69, respectively, wherein the V.sub.H and V.sub.L domains are connected by a linker. In an embodiment, an OX40 agonist fusion protein according to structures I-A or I-B comprises one or more OX40 binding domains that is a scFv domain comprising V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:78 and SEQ ID NO:79, respectively, wherein the V.sub.H and V.sub.L domains are connected by a linker. In an embodiment, an OX40 agonist fusion protein according to structures I-A or I-B comprises one or more OX40 binding domains that is a scFv domain comprising V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:86 and SEQ ID NO:87, respectively, wherein the V.sub.H and V.sub.L domains are connected by a linker. In an embodiment, an OX40 agonist fusion protein according to structures I-A or I-B comprises one or more OX40 binding domains that is a scFv domain comprising V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:94 and SEQ ID NO:95, respectively, wherein the V.sub.H and V.sub.L domains are connected by a linker. In an embodiment, an OX40 agonist fusion protein according to structures I-A or I--B comprises one or more OX40 binding domains that is a scFv domain comprising V.sub.H and V.sub.L regions that are each at least 95% identical to the V.sub.H and V.sub.L sequences given in Table 15, wherein the V.sub.H and V.sub.L domains are connected by a linker.
TABLE-US-00015 TABLE 15 Additional polypeptide domains useful as OX40 binding domains in fusion proteins (e.g., structures I-A and I-B) or as scFv OX40 agonist antibodies. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 102 MERVQPLEEN VGNAARPRFE RNKLLLVASV IQGLGLLLCF TYICLHFSAL QVSHRYPRIQ 60 OX40L SIKVQFTEYK KEKGFILTSQ KEDEIMKVQN NSVIINCDGF YLISLKGYFS QEVNISLHYQ 120 KDEEPLFQLK KVRSVNSLMV ASLTYKDKVY LNVTTDNTSL DDFHVNGGEL ILIHQNPGEF 180 CVL 183 SEQ ID NO: 103 SHRYPRIQSI KVQFTEYKKE KGFILTSQKE DEIMKVQNNS VIINCDGFYL ISLKGYFSQE 60 OX40L soluble VNISLHYQKD EEPLFQLKKV RSVNSLMVAS LTYKDKVYLN VTTDNTSLDD FHVNGGELIL 120 domain IHQNPGEFCV L 131 SEQ ID NO: 104 YPRIQSIKVQ FTEYKKEKGF ILTSQKEDEI MKVQNNSVII NCDGFYLISL KGYFSQEVNI 60 OX40L soluble SLHYQKDEEP LFQLKKVRSV NSLMVASLTY KDKVYLNVTT DNTSLDDFHV NGGELILIHQ 120 domain NPGEFCVL 128 (alternative) SEQ ID NO: 105 EVQLVESGGG LVQPGGSLRL SCAASGFTFS NYTMNWVRQA PGKGLEWVSA ISGSGGSTYY 60 variable heavy ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCAKDR YSQVHYALDY WGQGTLVTVS 120 chain for 008 SEQ ID NO: 106 DIVMTQSPDS LPVTPGEPAS ISCRSSQSLL HSNGYNYLDW YLQKAGQSPQ LLIYLGSNRA 60 variable light SGVPDRFSGS GSGTDFTLKI SRVEAEDVGV YYCQQYYNHP TTFGQGTK 108 chain for 008 SEQ ID NO: 107 EVQLVESGGG VVQPGRSLRL SCAASGFTFS DYTMNWVRQA PGKGLEWVSS ISGGSTYYAD 60 variable heavy SRKGRFTISR DNSKNTLYLQ MNNLRAEDTA VYYCARDRYF RQQNAFDYWG QGTLVTVSSA 120 chain for 011 SEQ ID NO: 108 DIVMTQSPDS LPVTPGEPAS ISCRSSQSLL HSNGYNYLDW YLQKAGQSPQ LLIYLGSNRA 60 variable light SGVPDRFSGS GSGTDFTLKI SRVEAEDVGV YYCQQYYNHP TTFGQGTK 108 chain for 011 SEQ ID NO: 109 EVQLVESGGG LVQPRGSLRL SCAASGFTFS SYAMNWVRQA PGKGLEWVAV ISYDGSNKYY 60 variable heavy ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCAKDR YITLPNALDY WGQGTLVTVS 120 chain for 021 SEQ ID NO: 110 DIQMTQSPVS LPVTPGEPAS ISCRSSQSLL HSNGYNYLDW YLQKPGQSPQ LLIYLGSNRA 60 variable light SGVPDRFSGS GSGTDFTLKI SRVEAEDVGV YYCQQYKSNP PTFGQGTK 108 chain for 021 SEQ ID NO: 111 EVQLVESGGG LVHPGGSLRL SCAGSGFTFS SYAMHWVRQA PGKGLEWVSA IGTGGGTYYA 60 variable heavy DSVMGRFTIS RDNSKNTLYL QMNSLRAEDT AVYYCARYDN VMGLYWFDYW GQGTLVTVSS 120 chain for 023 SEQ ID NO: 112 EIVLTQSPAT LSLSPGERAT LSCRASQSVS SYLAWYQQKP GQAPRLLIYD ASNRATGIPA 60 variable light RFSGSGSGTD FTLTISSLEP EDFAVYYCQQ RSNWPPAFGG GTKVEIKR 108 chain for 023 SEQ ID NO: 113 EVQLQQSGPE LVKPGASVKM SCKASGYTFT SYVMHWVKQK PGQGLEWIGY INPYNDGTKY 60 heavy chain NEKFKGKATL TSDKSSSTAY MELSSLTSED SAVYYCANYY GSSLSMDYWG QGTSVTVSS 119 variable region SEQ ID NO: 114 DIQMTQTTSS LSASLGDRVT ISCRASQDIS NYLNWYQQKP DGTVKLLIYY TSRLHSGVPS 60 light chain RFSGSGSGTD YSLTISNLEQ EDIATYFCQQ GNTLPWTFGG GTKLEIKR 108 variable region SEQ ID NO: 115 EVQLQQSGPE LVKPGASVKI SCKTSGYTFK DYTMHWVKQS HGKSLEWIGG IYPNNGGSTY 60 heavy chain NQNFKDKATL TVDKSSSTAY MEFRSLTSED SAVYYCARMG YHGPHLDFDV WGAGTTVTVS 120 variable region P 121 SEQ ID NO: 116 DIVMTQSHKF MSTSLGDRVS ITCKASQDVG AAVAWYQQKP GQSPKLLIYW ASTRHTGVPD 60 light chain RFTGGGSGTD FTLTISNVQS EDLTDYFCQQ YINYPLTFGG GTKLEIKR 108 variable region SEQ ID NO: 117 QIQLVQSGPE LKKPGETVKI SCKASGYTFT DYSMHWVKQA PGKGLKWMGW INTETGEPTY 60 heavy chain ADDFKGRFAF SLETSASTAY LQINNLKNED TATYFCANPY YDYVSYYAMD YWGHGTSVTV 120 variable region SS 122 of humanized antibody SEQ ID NO: 118 QVQLVQSGSE LKKPGASVKV SCKASGYTFT DYSMHWVRQA PGQGLKWMGW INTETGEPTY 60 heavy chain ADDFKGRFVF SLDTSVSTAY LQISSLKAED TAVYYCANPY YDYVSYYAMD YWGQGTTVTV 120 variable region SS 122 of humanized antibody SEQ ID NO: 119 DIVMTQSHKF MSTSVRDRVS ITCKASQDVS TAVAWYQQKP GQSPKLLIYS ASYLYTGVPD 60 light chain RFTGSGSGTD FTFTISSVQA EDLAVYYCQQ HYSTPRTFGG GTKLEIK 107 variable region of humanized antibody SEQ ID NO: 120 DIVMTQSHKF MSTSVRDRVS ITCKASQDVS TAVAWYQQKP GQSPKLLIYS ASYLYTGVPD 60 light chain RFTGSGSGTD FTFTISSVQA EDLAVYYCQQ HYSTPRTFGG GTKLEIK 107 variable region of humanized antibody SEQ ID NO: 121 EVQLVESGGG LVQPGESLKL SCESNEYEFP SHDMSWVRKT PEKRLELVAA INSDGGSTYY 60 heavy chain PDTMERRFII SRDNTKKTLY LQMSSLRSED TALYYCARHY DDYYAWFAYW GQGTLVTVSA 120 variable region of humanized antibody SEQ ID NO: 122 EVQLVESGGG LVQPGGSLRL SCAASEYEFP SHDMSWVRQA PGKGLELVAA INSDGGSTYY 60 heavy chain PDTMERRFTI SRDNAKNSLY LQMNSLRAED TAVYYCARHY DDYYAWFAYW GQGTMVTVSS 120 variable region of humanized antibody SEQ ID NO: 123 DIVLTQSPAS LAVSLGQRAT ISCRASKSVS TSGYSYMHWY QQKPGQPPKL LIYLASNLES 60 light chain GVPARFSGSG SGTDFTLNIH PVEEEDAATY YCQHSRELPL TFGAGTKLEL K 111 variable region of humanized antibody SEQ ID NO: 124 EIVLTQSPAT LSLSPGERAT LSCRASKSVS TSGYSYMHWY QQKPGQAPRL LIYLASNLES 60 light chain GVPARFSGSG SGTDFTLTIS SLEPEDFAVY YCQHSRELPL TFGGGTKVEI K 111 variable region of humanized antibody SEQ ID NO: 125 MYLGLNYVFI VFLLNGVQSE VKLEESGGGL VQPGGSMKLS CAASGFTFSD AWMDWVRQSP 60 heavy chain EKGLEWVAEI RSKANNHATY YAESVNGRFT ISRDDSKSSV YLQMNSLRAE DTGIYYCTWG 120 variable region EVFYFDYWGQ GTTLTVSS 138 SEQ ID NO: 126 MRPSIQFLGL LLFWLHGAQC DIQMTQSPSS LSASLGGKVT ITCKSSQDIN KYIAWYQHKP 60 light chain GKGPRLLIHY TSTLQPGIPS RFSGSGSGRD YSFSISNLEP EDIATYYCLQ YDNLLTFGAG 120 variable region TKLELK 126
[1254] In an embodiment, the OX40 agonist is a OX40 agonistic single-chain fusion polypeptide comprising (i) a first soluble OX40 binding domain, (ii) a first peptide linker, (iii) a second soluble OX40 binding domain, (iv) a second peptide linker, and (v) a third soluble OX40 binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, and wherein the additional domain is a Fab or Fc fragment domain. In an embodiment, the OX40 agonist is a OX40 agonistic single-chain fusion polypeptide comprising (i) a first soluble OX40 binding domain, (ii) a first peptide linker, (iii) a second soluble OX40 binding domain, (iv) a second peptide linker, and (v) a third soluble OX40 binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, wherein the additional domain is a Fab or Fc fragment domain wherein each of the soluble OX40 binding domains lacks a stalk region (which contributes to trimerisation and provides a certain distance to the cell membrane, but is not part of the OX40 binding domain) and the first and the second peptide linkers independently have a length of 3-8 amino acids.
[1255] In an embodiment, the OX40 agonist is an OX40 agonistic single-chain fusion polypeptide comprising (i) a first soluble tumor necrosis factor (TNF) superfamily cytokine domain, (ii) a first peptide linker, (iii) a second soluble TNF superfamily cytokine domain, (iv) a second peptide linker, and (v) a third soluble TNF superfamily cytokine domain, wherein each of the soluble TNF superfamily cytokine domains lacks a stalk region and the first and the second peptide linkers independently have a length of 3-8 amino acids, and wherein the TNF superfamily cytokine domain is an OX40 binding domain.
[1256] In some embodiments, the OX40 agonist is MEDI6383. MEDI6383 is an OX40 agonistic fusion protein and can be prepared as described in U.S. Pat. No. 6,312,700, the disclosure of which is incorporated by reference herein.
[1257] In an embodiment, the OX40 agonist is an OX40 agonistic scFv antibody comprising any of the foregoing V.sub.H domains linked to any of the foregoing V.sub.L domains.
[1258] In an embodiment, the OX40 agonist is Creative Biolabs OX40 agonist monoclonal antibody MOM-18455, commercially available from Creative Biolabs, Inc., Shirley, N.Y., USA.
[1259] In an embodiment, the OX40 agonist is OX40 agonistic antibody clone Ber-ACT35 commercially available from BioLegend, Inc., San Diego, Calif., USA.
CD27 Agonists
[1260] In an embodiment, the TNFRSF agonist is a CD27 agonist. The CD27 agonist may be any CD27 binding molecule known in the art. The CD27 binding molecule may be a monoclonal antibody or fusion protein capable of binding to human or mammalian CD27. The CD27 agonists or CD27 binding molecules may comprise an immunoglobulin heavy chain of any isotype (e.g., IgG, IgE, IgM, IgD, IgA, and IgY), class (e.g., IgG1, IgG2, IgG3, IgG4, IgA1 and IgA2) or subclass of immunoglobulin molecule. The CD27 agonist or CD27 binding molecule may have both a heavy and a light chain. As used herein, the term binding molecule also includes antibodies (including full length antibodies), monoclonal antibodies (including full length monoclonal antibodies), polyclonal antibodies, multispecific antibodies (e.g., bispecific antibodies), human, humanized or chimeric antibodies, and antibody fragments, e.g., Fab fragments, F(ab') fragments, fragments produced by a Fab expression library, epitope-binding fragments of any of the above, and engineered forms of antibodies, e.g., scFv molecules, that bind to CD27. In an embodiment, the CD27 agonist is an antigen binding protein that is a fully human antibody. In an embodiment, the CD27 agonist is an antigen binding protein that is a humanized antibody. In some embodiments, CD27 agonists for use in the presently disclosed methods and compositions include anti-CD27 antibodies, human anti-CD27 antibodies, mouse anti-CD27 antibodies, mammalian anti-CD27 antibodies, monoclonal anti-CD27 antibodies, polyclonal anti-CD27 antibodies, chimeric anti-CD27 antibodies, anti-CD27 adnectins, anti-CD27 domain antibodies, single chain anti-CD27 fragments, heavy chain anti-CD27 fragments, light chain anti-CD27 fragments, anti-CD27 fusion proteins, and fragments, derivatives, conjugates, variants, or biosimilars thereof. In a preferred embodiment, the CD27 agonist is an agonistic, anti-CD27 humanized or fully human monoclonal antibody (i.e., an antibody derived from a single cell line). In a preferred embodiment, the CD27 agonist is varlilumab, or a fragment, derivative, conjugate, variant, or biosimilar thereof.
[1261] In a preferred embodiment, the CD27 agonist or CD27 binding molecule may also be a fusion protein. In a preferred embodiment, a multimeric CD27 agonist, such as a trimeric or hexameric CD27 agonist (with three or six ligand binding domains), may induce superior receptor (CD27L) clustering and internal cellular signaling complex formation compared to an agonistic monoclonal antibody, which typically possesses two ligand binding domains. Trimeric (trivalent) or hexameric (or hexavalent) or greater fusion proteins comprising three TNFRSF binding domains and IgG1-Fc and optionally further linking two or more of these fusion proteins are described, e.g., in Gieffers, et al., Mol. Cancer Therapeutics 2013, 12, 2735-47.
[1262] Agonistic CD27 antibodies and fusion proteins are known to induce strong immune responses. In a preferred embodiment, the CD27 agonist is a monoclonal antibody or fusion protein that binds specifically to CD27 antigen in a manner sufficient to reduce toxicity. In some embodiments, the CD27 agonist is an agonistic CD27 monoclonal antibody or fusion protein that abrogates antibody-dependent cellular toxicity (ADCC), for example NK cell cytotoxicity. In some embodiments, the CD27 agonist is an agonistic CD27 monoclonal antibody or fusion protein that abrogates antibody-dependent cell phagocytosis (ADCP). In some embodiments, the CD27 agonist is an agonistic CD27 monoclonal antibody or fusion protein that abrogates complement-dependent cytotoxicity (CDC). In some embodiments, the CD27 agonist is an agonistic CD27 monoclonal antibody or fusion protein which abrogates Fc region functionality.
[1263] In some embodiments, the CD27 agonists are characterized by binding to human CD27 (SEQ ID NO: 127) with high affinity and agonistic activity. In an embodiment, the CD27 agonist is a binding molecule that binds to human CD27 (SEQ ID NO: 127). In some embodiments, the CD27 agonists are characterized by binding to macaque CD27 (SEQ ID NO: 128) with high affinity and agonistic activity. In an embodiment, the CD27 agonist is a binding molecule that binds to macaque CD27 (SEQ ID NO: 128). The amino acid sequences of CD27 antigens to which a CD27 agonist or binding molecule binds is summarized in Table 16.
TABLE-US-00016 TABLE 16 Amino acid sequences of CD27 antigens. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 127 MARPHPWWLC VLGTLVGLSA TPAPKSCPER HYWAQGKLCC QMCEPGTFLV KDCDQHRKAA 60 human CD27, QCDPCIPGVS FSPDHHTRPH CESCRHCNSG LLVRNCTITA NAECACRNGW QCRDKECTEC 120 Tumor necrosis DPLPNPSLTA RSSQALSPHP QPTHLPYVSE MLEARTAGHM QTLADFRQLP ARTLSTHWPP 180 factor receptor QRSLCSSDFI RILVIFSGMF LVFTLAGALF LHQRRKYRSN KGESPVEPAE PCRYSCPREE 240 superfamily, EGSTIPIQED YRKPEPACSP 260 member 7 (Homo sapiens) SEQ ID NO: 128 MARPHPWWLC FLGTLVGLSA TPAPKSCPER HYWAQGKLCC QMCEPGTFLV KDCDQHRKAA 60 human CD27, QCHPCIPGVS FSPDHHTRPH CESCRHCNSG LLIRNCTITA NAVCACRNGW QCRDKECTEC 120 Tumor necrosis DPPPNPSLTT WPSQALGPHP QPTHLPYVNE MLEARTAGHM QTLADFRHLP ARTLSTHWPP 180 factor receptor QRSLCSSDFI RILVIFSGMF LVFTLAGTLF LHQQRKYRSN KGESPMEPAE PCPYSCPREE 240 superfamily, EGSTIPIQED YRKPEPASSP 260 member 7 (Macaca nemestrina)
[1264] In some embodiments, the compositions, processes and methods described include a CD27 agonist that binds human or murine CD27 with a K.sub.D of about 100 pM or lower, binds human or murine CD27 with a K.sub.D of about 90 pM or lower, binds human or murine CD27 with a K.sub.D of about 80 pM or lower, binds human or murine CD27 with a K.sub.D of about 70 pM or lower, binds human or murine CD27 with a K.sub.D of about 60 pM or lower, binds human or murine CD27 with a K.sub.D of about 50 pM or lower, binds human or murine CD27 with a K.sub.D of about 40 pM or lower, or binds human or murine CD27 with a K.sub.D of about 30 pM or lower.
[1265] In some embodiments, the compositions, processes and methods described include a CD27 agonist that binds to human or murine CD27 with a k.sub.assoc of about 7.5.times.10.sup.5 l/Ms or faster, binds to human or murine CD27 with a k.sub.assoc of about 7.5.times.10.sup.5 l/Ms or faster, binds to human or murine CD27 with a k.sub.assoc of about 8.times.10.sup.5 l/Ms or faster, binds to human or murine CD27 with a k.sub.assoc of about 8.5.times.10.sup.5 l/Ms or faster, binds to human or murine CD27 with a k.sub.assoc of about 9.times.10.sup.5 l/Ms or faster, binds to human or murine CD27 with a k.sub.assoc of about 9.5.times.10.sup.5 l/Ms or faster, or binds to human or murine CD27 with a k.sub.assoc of about 1.times.10.sup.6 l/Ms or faster.
[1266] In some embodiments, the compositions, processes and methods described include a CD27 agonist that binds to human or murine CD27 with a k.sub.dissoc of about 2.times.10.sup.-5 l/s or slower, binds to human or murine CD27 with a k.sub.dissoc of about 2.1.times.10.sup.-5 l/s or slower, binds to human or murine CD27 with a k.sub.dissoc of about 2.2.times.10.sup.-5 l/s or slower, binds to human or murine CD27 with a k.sub.dissoc of about 2.3.times.10.sup.-5 l/s or slower, binds to human or murine CD27 with a k.sub.dissoc of about 2.4.times.10.sup.-5 l/s or slower, binds to human or murine CD27 with a k.sub.dissoc of about 2.5.times.10.sup.-5 l/s or slower, binds to human or murine CD27 with a k.sub.dissoc of about 2.6.times.10.sup.-5 l/s or slower or binds to human or murine CD27 with a k.sub.dissoc of about 2.7.times.10.sup.-5 l/s or slower, binds to human or murine CD27 with a k.sub.dissoc of about 2.8.times.10.sup.-5 l/s or slower, binds to human or murine CD27 with a k.sub.dissoc of about 2.9.times.10.sup.-5 l/s or slower, or binds to human or murine CD27 with a k.sub.dissoc of about 3.times.10.sup.-5 l/s or slower.
[1267] In some embodiments, the compositions, processes and methods described include a CD27 agonist that binds to human or murine CD27 with an IC.sub.50 of about 10 nM or lower, binds to human or murine CD27 with an IC.sub.50 of about 9 nM or lower, binds to human or murine CD27 with an IC.sub.50 of about 8 nM or lower, binds to human or murine CD27 with an IC.sub.50 of about 7 nM or lower, binds to human or murine CD27 with an IC.sub.50 of about 6 nM or lower, binds to human or murine CD27 with an IC.sub.50 of about 5 nM or lower, binds to human or murine CD27 with an IC.sub.50 of about 4 nM or lower, binds to human or murine CD27 with an IC.sub.50 of about 3 nM or lower, binds to human or murine CD27 with an IC.sub.50 of about 2 nM or lower, or binds to human or murine CD27 with an IC.sub.50 of about 1 nM or lower.
[1268] In a preferred embodiment, the CD27 agonist is the monoclonal antibody varlilumab, also known as CDX-1127 or 1F5, or a fragment, derivative, variant, or biosimilar thereof. Varlilumab is available from Celldex Therapeutics, Inc. Varlilumab is an immunoglobulin G1-kappa, anti-[Homo sapiens anti-CD27 (TNFRSF7, tumor necrosis factor receptor superfamily member 7)], Homo sapiens monoclonal antibody. The amino acid sequences of varlilumab are set forth in Table 17. Varlilumab comprises N-glycosylation sites at positions 299 and 299''; heavy chain intrachain disulfide bridges at positions 22-96 (V.sub.H-V.sub.L), 146-202 (C.sub.H1-C.sub.L), 263-323 (C.sub.H2) and 369-427 (C.sub.H3) (and at positions 22''-96'', 146''-202'', 263''-323'', and 369''-427''); light chain intrachain disulfide bridges at positions 23'-88' (V.sub.H-V.sub.L) and 134'-194' (C.sub.H1-C.sub.L) (and at positions 23'''-88''' and 134'''-194'''); interchain heavy chain-heavy chain disulfide bridges at positions 228-228'' and 231-231''; and interchain heavy chain-light chain disulfide bridges at 222-214' and 222''-214'''. The preparation and properties of varlilumab are described in International Patent Application Publication No. WO 2016/145085 A2 and U.S. Patent Application Publication Nos. US 2011/0274685 A1 and US 2012/0213771 A1, the disclosures of which are incorporated by reference herein. Clinical and preclinical studies using varlilumab are known in the art and are described, for example, in Thomas, et al., Oncolmmunology 2014, 3, e27255; Vitale, et al., Clin. Cancer Res. 2012, 18, 3812-21; and He, et al., J. Immunol. 2013, 191, 4174-83. Current clinical trials of varlilumab in a variety of hematological and solid tumor indications include U.S. National Institutes of Health clinicaltrials.gov identifiers NCT01460134, NCT02543645, NCT02413827, NCT02386111, and NCT02335918.
[1269] In an embodiment, a CD27 agonist comprises a heavy chain given by SEQ ID NO: 129 and a light chain given by SEQ ID NO: 130. In an embodiment, a CD27 agonist comprises heavy and light chains having the sequences shown in SEQ ID NO: 129 and SEQ ID NO: 130, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a CD27 agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO: 129 and SEQ ID NO: 130, respectively. In an embodiment, a CD27 agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO: 129 and SEQ ID NO: 130, respectively. In an embodiment, a CD27 agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO: 129 and SEQ ID NO: 130, respectively. In an embodiment, a CD27 agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO: 129 and SEQ ID NO:130, respectively. In an embodiment, a CD27 agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO: 129 and SEQ ID NO:130, respectively.
[1270] In an embodiment, the CD27 agonist comprises the heavy and light chain CDRs or variable regions (VRs) of varlilumab. In an embodiment, the CD27 agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO: 131, and the CD27 agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO: 132, and conservative amino acid substitutions thereof. In an embodiment, a CD27 agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO: 131 and SEQ ID NO: 132, respectively. In an embodiment, a CD27 agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO: 131 and SEQ ID NO: 132, respectively. In an embodiment, a CD27 agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO: 131 and SEQ ID NO: 132, respectively. In an embodiment, a CD27 agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO: 131 and SEQ ID NO: 132, respectively. In an embodiment, a CD27 agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO: 131 and SEQ ID NO: 132, respectively.
[1271] In an embodiment, a CD27 agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO: 133, SEQ ID NO: 134, and SEQ ID NO:135, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO: 136, SEQ ID NO: 137, and SEQ ID NO: 138, respectively, and conservative amino acid substitutions thereof.
[1272] In an embodiment, the CD27 agonist is a CD27 agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to varlilumab. In an embodiment, the biosimilar monoclonal antibody comprises an CD27 antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is varlilumab. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a CD27 agonist antibody authorized or submitted for authorization, wherein the CD27 agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is varlilumab. The CD27 agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is varlilumab. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is varlilumab.
TABLE-US-00017 TABLE 17 Amino acid sequences for CD27 agonist antibodies related to varlilumab. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 129 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYDMHWVRQA PGKGLEWVAV IWYDGSNKYY 60 heavy chain for ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGS GNWGFFDYWG QGTLVTVSSA 120 varlilumab STKGPSVFPL APSSKSTSGG TAALGCLVKD YFPEPVTVSW NSGALTSGVH TFPAVLQSSG 180 LYSLSSVVTV PSSSLGTQTY ICNVNHKPSN TKVDKKVEPK SCDKTHTCPP CPAPELLGGP 240 SVFLFPPKPK DTLMISRTPE VTCVVVDVSH EDPEVKFNWY VDGVEVHNAK TKPREEQYNS 300 TYRVVSVLTV LHQDWLNGKE YKCKVSNKAL PAPIEKTISK AKGQPREPQV YTLPPSRDEL 360 TKNQVSLTCL VKGFYPSDIA VEWESNGQPE NNYKTTPPVL DSDGSFFLYS KLTVDKSRWQ 420 QGNVFSCSVM HEALHNHYTQ KSLSLSPGKG SS 452 SEQ ID NO: 130 DIQMTQSPSS LSASVGDRVT ITCRASQGIS RWLAWYQQKP EKAPKSLIYA ASSLQSGVPS 60 light chain for RFSGSGSGTD FTLTISSLQP EDFATYYCQQ YNTYPRTFGQ GTKVEIKRTV AAPSVFIFPP 120 varlilumab SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 131 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYDMHWVRQA PGKGLEWVAV IWYDGSNKYY 60 heavy chain ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGS GNWGFFDYWG QGTLVTVSS 119 variable region for varlilumab SEQ ID NO: 132 DIQMTQSPSS LSASVGDRVT ITCRASQGIS RWLAWYQQKP EKAPKSLIYA ASSLQSGVPS 60 light chain RFSGSGSGTD FTLTISSLQP EDFATYYCQQ YNTYPRTFGQ GTKVEIK 107 variable region for varlilumab SEQ ID NO: 133 GFTFSSYD 8 heavy chain CDR1 for varlilumab SEQ ID NO: 134 IWYDGSNK 8 heavy chain CDR2 for varlilumab SEQ ID NO: 135 ARGSGNWGFF DY 12 heavy chain CDR3 for varlilumab SEQ ID NO: 136 QGISRW 6 light chain CDR1 for varlilumab SEQ ID NO: 137 AASG 4 light chain CDR2 for varlilumab SEQ ID NO: 138 QQYNTYPRT 9 light chain CDR3 for varlilumab
[1273] In an embodiment, the CD27 agonist is an CD27 agonistic fusion protein as depicted in Structure I-A (C-terminal Fc-antibody fragment fusion protein) or Structure I-B (N-terminal Fc-antibody fragment fusion protein), or a fragment, derivative, conjugate, variant, or biosimilar thereof. The properties of structures I-A and I-B are described above and in U.S. Pat. Nos. 9,359,420, 9,340,599, 8,921,519, and 8,450,460, the disclosures of which are incorporated by reference herein. Amino acid sequences for the polypeptide domains of structure I-A are given in Table 6. The Fc domain preferably comprises a complete constant domain (amino acids 17-230 of SEQ ID NO:31) the complete hinge domain (amino acids 1-16 of SEQ ID NO:31) or a portion of the hinge domain (e.g., amino acids 4-16 of SEQ ID NO:31). Preferred linkers for connecting a C-terminal Fc-antibody may be selected from the embodiments given in SEQ ID NO:32 to SEQ ID NO:41, including linkers suitable for fusion of additional polypeptides. Likewise, amino acid sequences for the polypeptide domains of structure I-B are given in Table 7. If an Fc antibody fragment is fused to the N-terminus of an TNRFSF fusion protein as in structure I-B, the sequence of the Fc module is preferably that shown in SEQ ID NO:42, and the linker sequences are preferably selected from those embodiments set forth in SEQ ID NO:43 to SEQ ID NO:45.
[1274] In an embodiment, an CD27 agonist fusion protein according to structures I-A or I-B comprises one or more CD27 binding domains selected from the group consisting of a variable heavy chain and variable light chain of varlilumab, and fragments, derivatives, conjugates, variants, and biosimilars thereof.
[1275] In an embodiment, an CD27 agonist fusion protein according to structures I-A or I-B comprises one or more CD27 binding domains comprising an CD70 (CD27L) sequence (Table 18). In an embodiment, an CD27 agonist fusion protein according to structures I-A or I-B comprises one or more CD27 binding domains comprising a sequence according to SEQ ID NO: 139. In an embodiment, an CD27 agonist fusion protein according to structures I-A or I-B comprises one or more CD27 binding domains comprising a soluble CD70 sequence. In an embodiment, a CD27 agonist fusion protein according to structures I-A or I-B comprises one or more CD27 binding domains comprising a sequence according to SEQ ID NO: 140. In an embodiment, a CD27 agonist fusion protein according to structures I-A or I-B comprises one or more CD27 binding domains comprising a sequence according to SEQ ID NO:141.
[1276] In an embodiment, an CD27 agonist fusion protein according to structures I-A or I-B comprises one or more CD27 binding domains that is a scFv domain comprising V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO: 131 and SEQ ID NO: 132, respectively, wherein the V.sub.H and V.sub.L domains are connected by a linker.
TABLE-US-00018 TABLE 18 Additional polypeptide domains useful as CD27 binding domains in fusion proteins (e.g., structures I-A and I-B). Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 139 MPEEGSGCSV RRRPYGCVLR AALVPLVAGL VICLVVCIQR FAQAQQQLPL ESLGWDVAEL 60 CD70 (CD27L) QLNHTGPQQD PRLYWQGGPA LGRSFLHGPE LDKGQLRIHR DGIYMVHIQV TLAICSSTTA 120 SRHHPTTLAV GICSPASRSI SLLRLSFHQG CTIASQRLTP LARGDTLCTN LTGTLLPSRN 180 TDETFFGVQW VRP 193 SEQ ID NO: 140 SLGWDVAELQ LNHTGPQQDP RLYWQGGPAL GRSFLHGPEL DKGQLRIHRD GIYMVHIQVT 60 CD70 soluble LAICSSTTAS RHHPTTLAVG ICSPASRSIS LLRLSFHQGC TIASQRLTPL ARGDTLCTNL 120 domain TGTLLPSRNT DETFFGVQWV RP 142 SEQ ID NO: 141 VAELQLNHTG PQQDPRLYWQ GGPALGRSFL HGPELDKGQL RIHRDGIYMV HIQVTLAICS 60 CD70 soluble STTASRHHPT TLAVGICSPA SRSISLLRLS FHQGCTIASQ RLTPLARGDT LCTNLTGTLL 120 domain PSRNTDETFF GVQWVRP 137 (alternative)
[1277] In an embodiment, the CD27 agonist is a CD27 agonistic single-chain fusion polypeptide comprising (i) a first soluble CD27 binding domain, (ii) a first peptide linker, (iii) a second soluble CD27 binding domain, (iv) a second peptide linker, and (v) a third soluble CD27 binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, and wherein the additional domain is a Fab or Fc fragment domain. In an embodiment, the CD27 agonist is a CD27 agonistic single-chain fusion polypeptide comprising (i) a first soluble CD27 binding domain, (ii) a first peptide linker, (iii) a second soluble CD27 binding domain, (iv) a second peptide linker, and (v) a third soluble CD27 binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, wherein the additional domain is a Fab or Fc fragment domain wherein each of the soluble CD27 binding domains lacks a stalk region (which contributes to trimerisation and provides a certain distance to the cell membrane, but is not part of the CD27 binding domain) and the first and the second peptide linkers independently have a length of 3-8 amino acids.
[1278] In an embodiment, the CD27 agonist is an CD27 agonistic single-chain fusion polypeptide comprising (i) a first soluble tumor necrosis factor (TNF) superfamily cytokine domain, (ii) a first peptide linker, (iii) a second soluble TNF superfamily cytokine domain, (iv) a second peptide linker, and (v) a third soluble TNF superfamily cytokine domain, wherein each of the soluble TNF superfamily cytokine domains lacks a stalk region and the first and the second peptide linkers independently have a length of 3-8 amino acids, and wherein the TNF superfamily cytokine domain is an CD27 binding domain.
[1279] In an embodiment, the CD27 agonist is a CD27 agonist described in U.S. Patent Application Publication No. US 2014/0112942 A1, US 2011/0274685 A1, or US 2012/0213771 A1, or International Patent Application Publication No. WO 2012/004367 A1, the disclosures of which are incorporated by reference herein.
[1280] In an embodiment, the CD27 agonist is a CD27 agonistic scFv antibody comprising any of the foregoing V.sub.H domains linked to any of the foregoing V.sub.L domains.
GITR (CD357) Agonists
[1281] In an embodiment, the TNFRSF agonist is a GITR agonist. The GITR agonist may be any GITR binding molecule known in the art. The GITR binding molecule may be a monoclonal antibody or fusion protein capable of binding to human or mammalian GITR. The GITR agonists or GITR binding molecules may comprise an immunoglobulin heavy chain of any isotype (e.g., IgG, IgE, IgM, IgD, IgA, and IgY), class (e.g., IgG1, IgG2, IgG3, IgG4, IgA1 and IgA2) or subclass of immunoglobulin molecule. The GITR agonist or GITR binding molecule may have both a heavy and a light chain. As used herein, the term binding molecule also includes antibodies (including full length antibodies), monoclonal antibodies (including full length monoclonal antibodies), polyclonal antibodies, multispecific antibodies (e.g., bispecific antibodies), human, humanized or chimeric antibodies, and antibody fragments, e.g., Fab fragments, F(ab') fragments, fragments produced by a Fab expression library, epitope-binding fragments of any of the above, and engineered forms of antibodies, e.g., scFv molecules, that bind to OX40. In an embodiment, the GITR agonist is an antigen binding protein that is a fully human antibody. In an embodiment, the GITR agonist is an antigen binding protein that is a humanized antibody. In some embodiments, GITR agonists for use in the presently disclosed methods and compositions include anti-GITR antibodies, human anti-GITR antibodies, mouse anti-OX40 antibodies, mammalian anti-GITR antibodies, monoclonal anti-OX40 antibodies, polyclonal anti-OX40 antibodies, chimeric anti-OX40 antibodies, anti-OX40 adnectins, anti-OX40 domain antibodies, single chain anti-OX40 fragments, heavy chain anti-OX40 fragments, light chain anti-OX40 fragments, anti-OX40 fusion proteins, and fragments, derivatives, conjugates, variants, or biosimilars thereof. In a preferred embodiment, the OX40 agonist is an agonistic, anti-OX40 humanized or fully human monoclonal antibody (i.e., an antibody derived from a single cell line).
[1282] In a preferred embodiment, the GITR agonist or GITR binding molecule may also be a fusion protein. In a preferred embodiment, a multimeric GITR agonist, such as a trimeric or hexameric GITR agonist (with three or six ligand binding domains), may induce superior GITR receptor clustering and internal cellular signaling complex formation compared to an agonistic monoclonal antibody, which typically possesses two ligand binding domains. Trimeric (trivalent) or hexameric (or hexavalent) or greater fusion proteins comprising three TNFRSF binding domains and IgG1-Fc and optionally further linking two or more of these fusion proteins are described, e.g., in Gieffers, et al., Mol. Cancer Therapeutics 2013, 12, 2735-47.
[1283] In some embodiments, the anti-GITR antibodies are characterized by binding to hGITR (SEQ ID NO: 142) with high affinity, in the presence of a stimulating agent, e.g., CD3 antibody (muromonab or OKT3), and are agonistic, and abrogate the suppression of T effector cells by Treg cells. In an embodiment, the GITR binding molecule binds to human GITR (SEQ ID NO: 142). In an embodiment, the GITR binding molecule binds to murine GITR (SEQ ID NO: 143). The amino acid sequences of GITR antigens to which a GITR binding molecule binds are summarized in Table 19.
TABLE-US-00019 TABLE 19 Amino acid sequences of GITR antigens. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 142 MAQHGAMGAF RALCGLALLC ALSLGQRPTG GPGCGPGRLL LGTGTDARCC RVHTTRCCRD 60 human GITR, YPGEECCSEW DCMCVQPEFH CGDPCCTTCR HHPCPPGQGV QSQGKFSFGF QCIDCASGTF 120 tumor necrosis SGGHEGHCKP WTDCTQFGFL TVFPGNKTHN AVCVPGSPPA EPLGWLTVVL LAVAACVLLL 180 factor receptor TSAQLGLHIW QLRSQCMWPR ETQLLLEVPP STEDARSCQF PEEERGERSA EEKGRLGDLW 240 superfamily V 241 member 18 (Homo sapiens) SEQ ID NO: 143 MGAWAMLYGV SMLCVLDLGQ PSVVEEPGCG PGKVQNGSGN NTRCCSLYAP GKEDCPKERC 60 murine GITR, ICVTPEYHCG DPQCKICKHY PCQPGQRVES QGDIVFGFRC VACAMGTFSA GRDGHCRLWT 120 tumor necrosis NCSQFGFLTM FPGNKTHNAV CIPEPLPTEQ YGHLTVIFLV MAACIFFLTT VQLGLHIWQL 180 factor receptor RRQHMCPRET QPFAEVQLSA EDACSFQFPE EERGEQTEEK CHLGGRWP 228 superfamily member 18 (Mus musculus)
[1284] In an embodiment, the GITR agonist is an antigen binding protein that is a fully human antibody. In an embodiment, the GITR agonist is an antigen binding protein that is a humanized antibody. In an embodiment, the GITR agonist is an antigen binding protein that agonizes the activity of human GITR. In an embodiment, the GITR binding molecule is an antigen binding protein that is a fully human IgG1 antibody. In an embodiment, the GITR agonist is an antigen binding protein that is capable of binding Fcgamma receptor (Fc.gamma.R). In an embodiment, the GITR agonist is an antigen binding protein that is capable of binding Fcgamma receptor (Fc.gamma.R) such that a cluster of antigen binding proteins is formed.
[1285] In some embodiments, the compositions, processes and methods described include a GITR agonist that binds human or murine GITR with a K.sub.D of about 100 pM or lower, binds human or murine GITR with a K.sub.D of about 90 pM or lower, binds human or murine GITR with a K.sub.D of about 80 pM or lower, binds human or murine GITR with a K.sub.D of about 70 pM or lower, binds human or murine GITR with a K.sub.D of about 60 pM or lower, binds human or murine GITR with a K.sub.D of about 50 pM or lower, binds human or murine GITR with a K.sub.D of about 40 pM or lower, or binds human or murine GITR with a K.sub.D of about 30 pM or lower.
[1286] In some embodiments, the compositions, processes and methods described include a GITR agonist that binds to human or murine GITR with a k.sub.assoc of about 7.5.times.10.sup.5 l/Ms or faster, binds to human or murine GITR with a k.sub.assoc of about 7.5.times.10.sup.5 l/Ms or faster, binds to human or murine GITR with a k.sub.assoc of about 8.times.10.sup.5 l/Ms or faster, binds to human or murine GITR with a k.sub.assoc of about 8.5.times.10.sup.5 l/Ms or faster, binds to human or murine GITR with a k.sub.assoc of about 9.times.10.sup.5 l/Ms or faster, binds to human or murine GITR with a k.sub.assoc of about 9.5.times.10.sup.5 l/Ms or faster, or binds to human or murine GITR with a k.sub.assoc of about 1.times.10.sup.6 l/Ms or faster.
[1287] In some embodiments, the compositions, processes and methods described include a GITR agonist that binds to human or murine GITR with a k.sub.dissoc of about 2.times.10.sup.-5 l/s or slower, binds to human or murine GITR with a k.sub.dissoc of about 2.1.times.10.sup.-5 l/s or slower, binds to human or murine GITR with a k.sub.dissoc of about 2.2.times.10.sup.-5 l/s or slower, binds to human or murine GITR with a k.sub.dissoc of about 2.3.times.10.sup.-5 l/s or slower, binds to human or murine GITR with a k.sub.dissoc of about 2.4.times.10.sup.-5 l/s or slower, binds to human or murine GITR with a k.sub.dissoc of about 2.5.times.10.sup.-5 l/s or slower, binds to human or murine GITR with a k.sub.dissoc of about 2.6.times.10.sup.-5 l/s or slower or binds to human or murine GITR with a k.sub.dissoc of about 2.7.times.10.sup.-5 l/s or slower, binds to human or murine GITR with a k.sub.dissoc of about 2.8.times.10.sup.-5 l/s or slower, binds to human or murine GITR with a k.sub.dissoc of about 2.9.times.10.sup.-5 l/s or slower, or binds to human or murine GITR with a k.sub.dissoc of about 3.times.10.sup.-5 l/s or slower.
[1288] In some embodiments, the compositions, processes and methods described include a GITR agonist that binds to human or murine GITR with an IC.sub.50 of about 10 nM or lower, binds to human or murine GITR with an IC.sub.50 of about 9 nM or lower, binds to human or murine GITR with an IC.sub.50 of about 8 nM or lower, binds to human or murine GITR with an IC.sub.50 of about 7 nM or lower, binds to human or murine GITR with an IC.sub.50 of about 6 nM or lower, binds to human or murine GITR with an IC.sub.50 of about 5 nM or lower, binds to human or murine GITR with an IC.sub.50 of about 4 nM or lower, binds to human or murine GITR with an IC.sub.50 of about 3 nM or lower, binds to human or murine GITR with an IC.sub.50 of about 2 nM or lower, or binds to human or murine GITR with an IC.sub.50 of about 1 nM or lower.
[1289] In a preferred embodiment, the GITR agonist is an agonistic, anti-GITR monoclonal antibody (i.e., an antibody derived from a single cell line). Agonist anti-GITR antibodies are known to induce strong immune responses. Cohen, et al., Cancer Res. 2006, 66, 4904-12; Schaer, et al., Curr. Opin. Investig. Drugs 2010, 11, 1378-1386. In a preferred embodiment, the GITR agonist is a monoclonal antibody that binds specifically to GITR antigen. In an embodiment, the GITR agonist is a GITR receptor blocker. In some embodiments, the GITR agonist is an agonistic, anti-GITR monoclonal antibody that abrogates antibody-dependent cellular toxicity (ADCC), for example NK cell cytotoxicity. In some embodiments, the GITR agonist is an agonistic, anti-GITR monoclonal antibody that abrogates antibody-dependent cell phagocytosis (ADCP). In some embodiments, the GITR agonist is an agonistic, anti-GITR monoclonal antibody that abrogates complement-dependent cytotoxicity (CDC).
[1290] In an embodiment, the GITR agonist is the agonistic, anti-GITR monoclonal antibody TRX518 (TolerRx, Inc.), also known as 6C8 and Ch-6C8-Agly. TRX518 is a fully-humanized IgG1 anti-human GITR monoclonal antibody in which heavy chain asparagine 297 is substituted with alanine to eliminate N-linked glycosylation, which abrogates Fc region functionality, including ADCC and CDC. Rosenzweig, et al., J. Clin. Oncol. 2010, 28 (supplement; abstract e13028); Jung, et al., Cur. Opin. Biotechnology 2011, 22,858-867. The amino acid sequences of TRX518 are set forth in Table 20. In some embodiments, the GITR binding molecule is the anti-human-GITR monoclonal antibody 6C8, or a variant thereof. The 6C8 antibody is an anti-GITR antibody that binds to human GITR on immune cells, e.g., human T cells and dendritic cells, with high affinity. Preferably, such binding molecules abrogate the suppression of T effector cells by Treg cells and are agonistic to partially activated immune cells in vitro in the presence of a stimulating agent, such as CD3. In some embodiments, the GITR binding molecule is the anti-murine GITR monoclonal antibody 2F8, or a variant thereof. The preparation, properties, and uses of 6C8 and 2F8 antibodies, and their variants, are described in U.S. Pat. Nos. 7,812,135; 8,388,967; and 9,028,823; the disclosures of which are incorporated by reference herein.
[1291] In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a heavy chain selected from the group consisting of SEQ ID NO: 144, SEQ ID NO: 145, SEQ ID NO: 146, and SEQ ID NO: 147, and a light chain comprising SEQ ID NO: 148. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a heavy chain with a sequence identity of greater than 99% to a sequence selected from the group consisting of SEQ ID NO: 144, SEQ ID NO: 145, SEQ ID NO: 146, and SEQ ID NO: 147, and a light chain with a sequence identity of greater than 99% to SEQ ID NO: 148. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a heavy chain with a sequence identity of greater than 98% to a sequence selected from the group consisting of SEQ ID NO: 144, SEQ ID NO: 145, SEQ ID NO: 146, and SEQ ID NO: 147, and a light chain with a sequence identity of greater than 98% to SEQ ID NO: 148. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a heavy chain with a sequence identity of greater than 95% to a sequence selected from the group consisting of SEQ ID NO: 144, SEQ ID NO: 145, SEQ ID NO: 146, and SEQ ID NO: 147, and a light chain with a sequence identity of greater than 95% to SEQ ID NO: 148. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a heavy chain with a sequence identity of greater than 90% to a sequence selected from the group consisting of SEQ ID NO: 144, SEQ ID NO: 145, SEQ ID NO: 146, and SEQ ID NO: 147, and a light chain with a sequence identity of greater than 90% to SEQ ID NO: 148.
[1292] In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a heavy chain that comprises the leader sequence of SEQ ID NO: 149 and further comprises a sequence selected from the group consisting of SEQ ID NO: 144, SEQ ID NO: 145, SEQ ID NO: 146 and SEQ ID NO: 147. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a light chain that comprises the leader sequence of SEQ ID NO: 148 and further comprises a sequence comprising SEQ ID NO:150.
[1293] In an embodiment, the agonistic anti-GITR monoclonal antibody (such as TRX518) comprises a variable heavy chain region (V.sub.H) selected from the group consisting of SEQ ID NO:151 and SEQ ID NO:152, and a variable light chain region (V.sub.L) comprising SEQ ID NO:153. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a variable heavy chain region selected from the group consisting of amino acid residues 20-138 of SEQ ID NO: 151 and amino acid residues 20-138 of SEQ ID NO: 152, and a variable light chain region comprising amino acid residues 20-138 of SEQ ID NO:153. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a variable heavy chain region with a sequence identity of greater than 99% to a sequence selected from the group consisting of amino acid residues 20-138 of SEQ ID NO: 151 and amino acid residues 20-138 of SEQ ID NO: 152, and a variable light chain region with a sequence identity of greater than 99% to a sequence comprising amino acid residues 20-138 of SEQ ID NO:153. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a variable heavy chain region with a sequence identity of greater than 98% to a sequence selected from the group consisting of amino acid residues 20-138 of SEQ ID NO: 151 and amino acid residues 20-138 of SEQ ID NO: 152, and a variable light chain region with a sequence identity of greater than 98% to a sequence comprising amino acid residues 20-138 of SEQ ID NO:153. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a variable heavy chain region with a sequence identity of greater than 95% to a sequence selected from the group consisting of amino acid residues 20-138 of SEQ ID NO: 151 and amino acid residues 20-138 of SEQ ID NO: 152, and a variable light chain region with a sequence identity of greater than 95% to a sequence comprising amino acid residues 20-138 of SEQ ID NO: 153. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a variable heavy chain region with a sequence identity of greater than 90% to a sequence selected from the group consisting of amino acid residues 20-138 of SEQ ID NO: 151 and amino acid residues 20-138 of SEQ ID NO: 152, and a variable light chain region with a sequence identity of greater than 90% to a sequence comprising amino acid residues 20-138 of SEQ ID NO: 153.
[1294] In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a V.sub.H region comprising at least one CDR1 region comprising the amino acid sequence of SEQ ID NO:154; at least one CDR2 region comprising an amino acid sequence selected from the group consisting of SEQ ID NO: 155 and SEQ ID NO: 156; and at least one CDR3 region comprising the amino acid sequence of SEQ ID NO: 157; and a V.sub.L region comprising at least one CDR1 region comprising the amino acid sequence of SEQ ID NO: 158; at least one CDR2 region comprising the amino acid sequence of SEQ ID NO: 159; and at least one CDR3 region comprising the amino acid sequence of SEQ ID NO: 160. In an embodiment, the invention provides isolated nucleic acid molecules encoding a polypeptide sequence comprising a 6C8 CDR, e. g., comprising an amino acid sequence selected from the group consisting of: SEQ ID NO:154, SEQ ID NO:155, SEQ ID NO:156, SEQ ID NO:157, SEQ ID NO:158, SEQ ID NO:159, and SEQ ID NO:160. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises the six CDRs represented by the amino acid sequences of SEQ ID NO: 154, SEQ ID NO:156, SEQ ID NO:157, SEQ ID NO:158, SEQ ID NO:159, and SEQ ID NO:160. In an embodiment, the GITR binding molecule that specifically binds to GITR comprises the six CDRs represented by the amino acid sequences of SEQ ID NO:154, SEQ ID NO:155, SEQ ID NO:157, SEQ ID NO:158, SEQ ID NO:159, and SEQ ID NO:160. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a V.sub.L having at least one CDR domain comprising an amino acid sequence selected from the group consisting of SEQ ID NO:158, SEQ ID NO:159, and SEQ ID NO:160. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a V.sub.L having at least two CDR domains comprising an amino acid sequence selected from the group consisting of SEQ ID NO:158, SEQ ID NO:159, and SEQ ID NO:160. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a V.sub.L having CDR domains comprising the amino acid sequences of SEQ ID NO:158, SEQ ID NO:159, and SEQ ID NO: 160. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a V.sub.L having at least one CDR domain comprising an amino acid sequence selected from the group consisting of SEQ ID NO: 154, SEQ ID NO: 155, and SEQ ID NO: 157. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a V.sub.L having at least two CDR domains comprising an amino acid sequence selected from the group consisting of SEQ ID NO:154, SEQ ID NO:155, and SEQ ID NO:157. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a V.sub.L having CDR domains comprising the amino acid sequences of SEQ ID NO:154, SEQ ID NO:155, and SEQ ID NO:157. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a V.sub.L having at least one CDR domain comprising an amino acid sequence selected from the group consisting of SEQ ID NO:154, SEQ ID NO:156, and SEQ ID NO:157. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a V.sub.L having at least two CDR domains comprising an amino acid sequence selected from the group consisting of SEQ ID NO: 154, SEQ ID NO: 156, and SEQ ID NO: 157. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a V.sub.L having CDR domains comprising the amino acid sequences of SEQ ID NO: 154, SEQ ID NO: 156, and SEQ ID NO: 157. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a V.sub.H domain comprising a CDR set forth in SEQ ID NO:154 (CDR1). In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a V.sub.H domain comprising a CDR set forth in SEQ ID NO: 155 (CDR2, "N" variant). In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a V.sub.H domain comprising a CDR set forth in SEQ ID NO:156 (CDR3, "Q" variant). In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a V.sub.H domain comprising a CDR set forth in SEQ ID NO:157 (CDR3). In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a V.sub.L domain comprising a CDR set forth in SEQ ID NO:158 (CDR1). In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a V.sub.L domain comprising a CDR set forth in SEQ ID NO:159 (CDR2). In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a V.sub.L domain comprising a CDR set forth in SEQ ID NO:160 (CDR3).
[1295] In an embodiment, the agonistic anti-GITR monoclonal antibody is a chimeric 6C8 monoclonal antibody, or an antigen-binding fragment, derivative, conjugate, or variant thereof. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a heavy chain selected from the group consisting of SEQ ID NO: 162 and SEQ ID NO: 163, and a light chain comprising SEQ ID NO: 161. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a heavy chain with a sequence identity of greater than 99% to a sequence selected from the group consisting of SEQ ID NO: 162 and SEQ ID NO: 163, and a light chain with a sequence identity of greater than 99% to SEQ ID NO: 161. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a heavy chain with a sequence identity of greater than 98% to a sequence selected from the group consisting of SEQ ID NO: 162 and SEQ ID NO: 163, and a light chain with a sequence identity of greater than 98% to SEQ ID NO: 161. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a heavy chain with a sequence identity of greater than 95% to a sequence selected from the group consisting of SEQ ID NO: 162 and SEQ ID NO: 163, and a light chain with a sequence identity of greater than 95% to SEQ ID NO: 161. In an embodiment, the agonistic anti-GITR monoclonal antibody comprises a heavy chain with a sequence identity of greater than 90% to a sequence selected from the group consisting of SEQ ID NO: 162 and SEQ ID NO: 163, and a light chain with a sequence identity of greater than 90% to SEQ ID NO:161.
[1296] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to TRX518 or 6C8. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is TRX518 or 6C8. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is TRX518 or 6C8. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is TRX518 or 6C8. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is TRX518 or 6C8.
TABLE-US-00020 TABLE 2 Amino acid sequences for GITR agonist antibodies related to TRX518 and 6C8. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 144 QVTLRESGPA LVKPTQTLTL TCTFSGFSLS TSGMGVGWIR QPPGKALEWL AHIWWDDDKY 60 humanized 6C8 YNPSLKSRLT ISKDTSKNQV VLTMTNMDPV DTATYYCART RRYFPFAYWG QGTLVTVSSA 120 heavy chain STKGPSVFPL APSSKSTSGG TAALGCLVKD YFPEPVTVSW NSGALTSGVH TFPAVLQSSG 180 variant LYSLSSVVTV PSSSLGTQTY ICNVNHKPSN TKVDKKVEPK SCDKTHTCPP CPAPELLGGP 240 SVFLFPPKPK DTLMISRTPE VTCVVVDVSH EDPEVKFNWY VDGVEVHNAK TKPREEQYNS 300 TYRVVSVLTV LHQDWLNGKE YKCKVSNKAL PAPIEKTISK AKGQPREPQV YTLPPSRDEL 360 TKNQVSLTCL VKGFYPSDIA VEWESNGQPE NNYKTTPPVL DSDGSFFLYS KLTVDKSRWQ 420 QGNVFSCSVM HEALHNHYTQ KSLSLSPGK 449 SEQ ID NO: 145 QVTLRESGPA LVKPTQTLTL TCTFSGFSLS TSGMGVGWIR QPPGKALEWL AHIWWDDDKY 60 humanized 6C8 YNPSLKSRLT ISKDTSKNQV VLTMTNMDPV DTATYYCART RRYFPFAYWG QGTLVTVSSA 120 heavy chain STKGPSVFPL APSSKSTSGG TAALGCLVKD YFPEPVTVSW NSGALTSGVH TFPAVLQSSG 180 variant LYSLSSVVTV PSSSLGTQTY ICNVNHKPSN TKVDKKVEPK SCDKTHTCPP CPAPELLGGP 240 SVFLFPPKPK DTLMISRTPE VTCVVVDVSH EDPEVKFNWY VDGVEVHNAK TKPREEQYAS 300 TYRVVSVLTV LHQDWLNGKE YKCKVSNKAL PAPIEKTISK AKGQPREPQV YTLPPSRDEL 360 TKNQVSLTCL VKGFYPSDIA VEWESNGQPE NNYKTTPPVL DSDGSFFLYS KLTVDKSRWQ 420 QGNVFSCSVM HEALHNHYTQ KSLSLSPGK 449 SEQ ID NO: 146 QVTLRESGPA LVKPTQTLTL TCTFSGFSLS TSGMGVGWIR QPPGKALEWL AHIWWDDDKY 60 humanized 6C8 YQPSLKSRLT ISKDTSKNQV VLTMTNMDPV DTATYYCART RRYFPFAYWG QGTLVTVSSA 120 heavy chain STKGPSVFPL APSSKSTSGG TAALGCLVKD YFPEPVTVSW NSGALTSGVH TFPAVLQSSG 180 variant LYSLSSVVTV PSSSLGTQTY ICNVNHKPSN TKVDKKVEPK SCDKTHTCPP CPAPELLGGP 240 SVFLFPPKPK DTLMISRTPE VTCVVVDVSH EDPEVKFNWY VDGVEVHNAK TKPREEQYNS 300 TYRVVSVLTV LHQDWLNGKE YKCKVSNKAL PAPIEKTISK AKGQPREPQV YTLPPSRDEL 360 TKNQVSLTCL VKGFYPSDIA VEWESNGQPE NNYKTTPPVL DSDGSFFLYS KLTVDKSRWQ 420 QGNVFSCSVM HEALHNHYTQ KSLSLSPGK 449 SEQ ID NO: 147 QVTLRESGPA LVKPTQTLTL TCTFSGFSLS TSGMGVGWIR QPPGKALEWL AHIWWDDDKY 60 humanized 6C8 YQPSLKSRLT ISKDTSKNQV VLTMTNMDPV DTATYYCART RRYFPFAYWG QGTLVTVSSA 120 heavy chain STKGPSVFPL APSSKSTSGG TAALGCLVKD YFPEPVTVSW NSGALTSGVH TFPAVLQSSG 180 variant LYSLSSVVTV PSSSLGTQTY ICNVNHKPSN TKVDKKVEPK SCDKTHTCPP CPAPELLGGP 240 SVFLFPPKPK DTLMISRTPE VTCVVVDVSH EDPEVKFNWY VDGVEVHNAK TKPREEQYAS 300 TYRVVSVLTV LHQDWLNGKE YKCKVSNKAL PAPIEKTISK AKGQPREPQV YTLPPSRDEL 360 TKNQVSLTCL VKGFYPSDIA VEWESNGQPE NNYKTTPPVL DSDGSFFLYS KLTVDKSRWQ 420 QGNVFSCSVM HEALHNHYTQ KSLSLSPGK 449 SEQ ID NO: 148 EIVMTQSPAT LSVSPGERAT LSCKASQNVG TNVAWYQQKP GQAPRLLIYS ASYRYSGIPA 60 humanized 6C8 RFSGSGSGTE FTLTISSLQS EDFAVYYCQQ YNTDPLTFGG GTKVEIKRTV AAPSVFIFPP 120 light chain SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 149 MDRLTFSFLL LIVPAYVLS 19 6C8 heavy chain leader SEQ ID NO: 150 METQSQVFVY MLLWLSGVDG 20 6C8 light chain leader SEQ ID NO: 151 MDRLTFSFLL LIVPAYVLSQ VTLKESGPGI LKPSQTLSLT CSFSGFSLST SGMGVGWIRQ 60 humanized 6C8 PSGKGLEWLA HIWWDDDKYY NPSLKSQLTI SKDTSRNQVF LKITSVDTAD AATYYCARTR 120 heavy chain RYFPFAYWGQ GTLVTVSS 138 variable region variant SEQ ID NO: 152 MDRLTFSFLL LIVPAYVLSQ VTLKESGPGI LKPSQTLSLT CSFSGFSLST SGMGVGWIRQ 60 humanized 6C8 PSGKGLEWLA HIWWDDDKYY QPSLKSQLTI SKDTSRNQVF LKITSVDTAD AATYYCARTR 120 heavy chain RYFPFAYWGQ GTLVTVSS 138 variable region variant SEQ ID NO: 153 METQSQVFVY MLLWLSGVDG DIVMTQSQKF MSTSVGDRVS VTCKASQNVG TNVAWYQQKP 60 humanized 6C8 GQSPKALIYS ASYRYSGVPD RFTGSGSGTD FTLTINNVHS EDLAEYFCQQ YNTDPLTFGA 120 light chain GTKLEIK 127 variable region SEQ ID NO: 154 GFSLSTSGMG VG 12 6C8 heavy chain CDR1 SEQ ID NO: 155 HIWWDDDKYY NPSLKS 16 6C8 heavy chain CDR2 variant SEQ ID NO: 156 HIWWDDDKYY QPSLKS 16 6C8 heavy chain CDR2 variant SEQ ID NO: 157 TRRYFPFAY 9 6C8 heavy chain CDR3 SEQ ID NO: 158 KASQNVGTNV A 11 6C8 light chain CDR1 SEQ ID NO: 159 SASYRYS 7 6C8 light chain CDR2 SEQ ID NO: 160 QQYNTDPLT 9 6C8 light chain CDR3 SEQ ID NO: 161 QVTLKESGPG ILKPSQTLSL TCSFSGFSLS TSGMGVGWIR QPSGKGLEWL AHIWWDDDKY 60 chimeric 6C8 YNPSLKSQLT ISKDTSRNQV FLKITSVDTA DAATYYCART RRYFPFAYWG QGTLVTVSSA 120 heavy chain STKGPSVFPL APSSKSTSGG TAALGCLVKD YFPEPVTVSW NSGALTSGVH TFPAVLQSSG 180 variant LYSLSSVVTV PSSSLGTQTY ICNVNHKPSN TKVDKKVEPK SCDKTHTCPP CPAPELLGGP 240 SVFLFPPKPK DTLMISRTPE VTCVVVDVSH EDPEVKFNWY VDGVEVHNAK TKPREEQYNS 300 TYRVVSVLTV LHQDWLNGKE YKCKVSNKAL PAPIEKTISK AKGQPREPQV YTLPPSRDEL 360 TKNQVSLTCL VKGFYPSDIA VEWESNGQPE NNYKTTPPVL DSDGSFFLYS KLTVDKSRWQ 420 QGNVFSCSVM HEALHNHYTQ KSLSLSPGK 449 SEQ ID NO: 162 QVTLKESGPG ILKPSQTLSL TCSFSGFSLS TSGMGVGWIR QPSGKGLEWL AHIWWDDDKY 60 chimeric 6C8 YNPSLKSQLT ISKDTSRNQV FLKITSVDTA DAATYYCART RRYFPFAYWG QGTLVTVSSA 120 heavy chain STKGPSVFPL APSSKSTSGG TAALGCLVKD YFPEPVTVSW NSGALTSGVH TFPAVLQSSG 180 variant LYSLSSVVTV PSSSLGTQTY ICNVNHKPSN TKVDKKVEPK SCDKTHTCPP CPAPELLGGP 240 SVFLFPPKPK DTLMISRTPE VTCVVVDVSH EDPEVKFNWY VDGVEVHNAK TKPREEQYAS 300 TYRVVSVLTV LHQDWLNGKE YKCKVSNKAL PAPIEKTISK AKGQPREPQV YTLPPSRDEL 360 TKNQVSLTCL VKGFYPSDIA VEWESNGQPE NNYKTTPPVL DSDGSFFLYS KLTVDKSRWQ 420 QGNVFSCSVM HEALHNHYTQ KSLSLSPGK 449 SEQ ID NO: 163 DIVMTQSQKF MSTSVGDRVS VTCKASQNVG TNVAWYQQKP GQSPKALIYS ASYRYSGVPD 60 chimeric 6C8 RFTGSGSGTD FTLTINNVHS EDLAEYFCQQ YNTDPLTFGA GTKLEIKRTV AAPSVFIFPP 120 light chain SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 variant LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214
[1297] In an embodiment, the GITR agonist is an agonistic anti-GITR monoclonal antibody with described in U.S. Pat. No. 8,709,424; U.S. Patent Application Publication Nos. US 2012/0189639 A1 and US 2014/0348841 A1, and International Patent Application Publication No. WO 2011/028683 A1 (Merck Sharp & Dohme Corp.), the disclosures of which are incorporated by reference herein. In an embodiment, the GITR agonist is an agonistic, anti-GITR monoclonal antibody selected from the group consisting of 36E5, 3D6, 61G6, 6H6, 61F6, 1D8, 17F10, 35D8, 49A1, 9E5, and 31H6, and fragments, variants, derivatives, or biosimilars thereof. The structure, properties, and preparation of these antibodies are described in U.S. Pat. No. 8,709,424; U.S. Patent Application Publication Nos. US 2012/0189639 A1 and US 2014/0348841 A1, the disclosures of which are incorporated herein by reference.
[1298] In some embodiments, the agonistic, anti-GITR monoclonal antibody comprises a humanized heavy chain variable domain (V.sub.H) comprising a sequence selected from the group consisting of SEQ ID NO:164, SEQ ID NO:166, SEQ ID NO:168, SEQ ID NO:170, SEQ ID NO:172, SEQ ID NO:174, SEQ ID NO:176, SEQ ID NO:178, SEQ ID NO:180, SEQ ID NO:182, SEQ ID NO:184, SEQ ID NO:186, SEQ ID NO:188, SEQ ID NO:190, SEQ ID NO:192, SEQ ID NO:194, SEQ ID NO:196, SEQ ID NO:198, SEQ ID NO:200, SEQ ID NO:202, SEQ ID NO:204, SEQ ID NO:206, or a variant, fragment, or biosimilar thereof, and a humanized heavy chain variable domain (V.sub.H) comprising a sequence selected from the group consisting of SEQ ID NO:165, SEQ ID NO:167, SEQ ID NO:169, SEQ ID NO:171, SEQ ID NO:173, SEQ ID NO:175, SEQ ID NO:177, SEQ ID NO:179, SEQ ID NO:181, SEQ ID NO:183, SEQ ID NO:185, SEQ ID NO:187, SEQ ID NO:189, SEQ ID NO:191, SEQ ID NO:193, SEQ ID NO:195, SEQ ID NO:197, SEQ ID NO:199, SEQ ID NO:201, SEQ ID NO:203, SEQ ID NO:205, SEQ ID NO:207, or a variant, fragment, or biosimilar thereof (Table 21). In some embodiments, the agonistic, anti-GITR monoclonal antibody further comprises a heavy chain constant region, wherein the heavy chain constant region comprises a .gamma.1, .gamma.2, .gamma.3, or .gamma.4 human heavy chain constant region or a variant thereof. In some embodiments, the light chain constant region comprises a lambda or a kappa human light chain constant region.
[1299] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 36E5, 3D6, 61G6, 6H6, 61F6, 1D8, 17F10, 35D8, 49A1, 9E5, and 31H6. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 36E5, 3D6, 61G6, 6H6, 61F6, 1D8, 17F10, 35D8, 49A1, 9E5, and 31H6. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 36E5, 3D6, 61G6, 6H6, 61F6, 1D8, 17F10, 35D8, 49A1, 9E5, and 31H6. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 36E5, 3D6, 61G6, 6H6, 61F6, 1D8, 17F10, 35D8, 49A1, 9E5, and 31H6. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 36E5, 3D6, 61G6, 6H6, 61F6, 1D8, 17F10, 35D8, 49A1, 9E5, and 31H6.
TABLE-US-00021 TABLE 21 Amino acid sequences for GITR agonist antibodies related to the GITR agonists described in International Patent Application Publication No. WO 2011/028683 A1. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 164 EVNLVESGGG LVKPGGSLKV SCAASGFTFS SYAMSWVRQT PEKRLEWVAS ISSGGTTYYP 60 36E5 heavy chain DSVKGRFTIS RDNARNILYL QMSSLRSEDT AMYYCARVGG YYDSMDYWGQ GISVTDSS 118 variable region SEQ ID NO: 165 DIVLTQSPAS LAVSLGQRAT ISCRASESVD NYGVSFMNWF QQKPGQPPKL LIYAASNQGS 60 36E5 light chain GVPARFSGSG SGTDFSLNIH PMEEDDTAMY FCQQTKEVTW TFGGGTKLEI KRA 113 variable region SEQ ID NO: 166 EVQLVESGGG LVQPGRSLKL SCAASGFTFS DYYMAWVRQA PTKGLEWVAY IHANGGSTYY 60 3D6 heavy chain RDSVRGRFSI SRDNGKSTLY LQMDSLRSED TATYYCTTGS FMTAADYTIM DAWGQGASVT 120 variable region VSS 123 SEQ ID NO: 167 DVVMTQTPVS LSVSLGNQAS ISCRSSQSLL HSDGNTFLSW YFQKPGQSPQ LLIYLASNRF 60 3D6 light chain SGVSNRFSGS GSGTDFTLKI SRVEPEDLGV YYCFQHTHLP LTFGSGTKLE IKR 113 variable region SEQ ID NO: 168 DVQLQESGPG LVKPSQSLSL TCTVTGYSIT SDYAWNWIRQ FPGNKLEWMG YISYSGSTRY 60 61G6 heavy chain NPSLKSRISI TRDTSKNQFF LQLNSVTSED TATYYCARQL GLRFFDYWGQ GTTLTVSS 118 variable region SEQ ID NO: 169 QIVLTQSPAL MSASPGEKVT MTCSANSTVN YMYWYQQKPR SSPKPCIYLT SNLASGVPAR 60 61G6 light chain FSGSGSGTSY SLTISSMEAE DAATYYCQQW NSNPPTFGAG TKLELRRA 108 variable region SEQ ID NO: 170 QVQLQQSGAE LMKPGASVKI SCKATGYTFS RYWIEWIKQR PGHGLEWIGE ILPGSGSSNY 60 6H6 heavy chain NEKFKDKATF TADTSSNTAY MQFSSLTSED SAVYYCARKV YYYAMDFWGQ GTSVTVSS 118 variable region SEQ ID NO: 171 QIVLTQSPAI MSVSLGERVT VTCTASSSVS SSYFHWYQQK PGSSPKLWIY STSNLASGVP 60 6H6 light chain ARFSGSGSGT SYSLTISTME AEDAATYYCH QYHRSPRTFG GGTKLEIKRA 110 variable region SEQ ID NO: 172 QVQLQQSGAE LARPGASVKM SCKASGYTFT SYTMHWVKQR PGQGLEWIGY INPRSVYTNY 60 61F6 heavy chain NQKFKDKATL TADKSSSTAY MQLSSLTSED SAVYYCARLG GYYDTMDYWG QGTSVTVSS 119 variable region SEQ ID NO: 173 DIVVTQSPAS LAVSLGQRAT ISCRASESVD NYGISFMNWF QQKPGQPPKL LIYAASNQGS 60 61F6 light chain GVPARFSGSG SGTDFSLNIH PMEEDDTAVY FCQQSKEVPF TFGSGTKLEI KRA 113 variable region SEQ ID NO: 174 QVTLKESGPG ILKPSQTLSL TCSFSGFSLS TSGMGVGWIR QPSGKGLEWL AHIWWDDDKY 60 1D8 heavy chain YSPSLKSQLT ISKDTSRNQV FLKITSLDTA DTATYYCVRS YYYGSSGAMD YWGQGTSVTV 120 variable region SS 122 SEQ ID NO :175 DIVMTQTPLS LPVSLGDQAS ISCRSSQSLV HSDGNTYLHW YLQKPGQSPK LLIYKVSKRF 60 1D8 light chain SGVPDRFSGS GSGTDFTLKI SRVEAEDLGV YFCSQSTHVP PTFGGGTKLE IKRADAAP 118 variable region SEQ ID NO: 176 EVKLVESGGG FVKPGGSLKL SCAASGFTVR NYAMSWVRQT PEKRLEWVAS ISTGDRSYLP 60 17E10 heavy DSMKGRFTIS RDNARNILYL QMSSLRSEDT AIYYCQRYFD FDSFAFWGQG TLVTVSA 117 chain variable region SEQ ID NO: 177 DIQMTQTPSS LSASLGDRVT ISCRASQDIN NFLNWYQQKP DGSLKLLIYY TSKLHSGVPS 60 17E10 light RFSGSGSGTD FSLTISNLDQ EDVATYFCQQ GHTLPPTFGG GTKLEVKRAD AAP 113 chain variable region SEQ ID NO: 178 EVQLQESGPS LVKPSQTLSL TCSVTGDSIT SGYWNWIRKF PGNKLEYMGY ISYSGSTYYN 60 35D8 heavy chain PSLRGRISIT RDTSKSQYYL QLSSVTTEDT ATYYCSRRHL GSGYGWFAYW GQGTLVTVSA 120 variable region SEQ ID NO: 179 DIVMTQSHKF MSTSVGDRVS ITCKASQDVN TAVAWYQQKP GQSPKLLIYW ASTRHTGVPD 60 35D8 light chain RFTGSGSGTD YALTINSVQA EDLALYYCQQ HSYTPPWTFG GGTKLEIRRA DAAP 114 variable region SEQ ID NO: 180 EVQLQESGPS LVKPSQTLSL TCSVTGDSIT SGYWNWIRKF PGNKFEYMGF ISYSGNTYYN 60 49A1 heavy chain PSLRSRISIT RDTSKNQYFL HLNSVTTEDT ATYYCSRRHL ISGYGWFAYW GQGTLVTVSA 120 variable region SEQ ID NO: 181 VIVMTQSHKF MSTSIGDRVN ITCKASQDVI SAVAWYQQKP GQSPKLLIYW ASTRHTGVPD 60 49A1 light chain RFTGSGSGTD FTLTINSVQA EDRALYYCQQ HSYTPPWTFG GGTNLEIKRA DAAP 114 variable region SEQ ID NO: 182 QVTLKESGPG ILQPSQTLSL TCTFSGFSLS TYGVGVGWIR QPSGKGLEWL ANIWWDDDNY 60 9E5 heavy chain YNPSLIHRLT VSKDTSNNQA FLKITNVDTA ETATYYCAQI KEPRDWFFEF WGPGTMVSVS 120 variable region S 121 SEQ ID NO: 183 DIQMTQTPSS MPASLGERVT IFCRASQGVN NFLTWYQQKP DGTIKPLIFY TSNLQSGVPS 60 9E5 light chain RFSGSGSGTD YSLSISSLEP EDFAMYYCQQ YHGFPNTFGA GTKLELKRAD AAP 113 variable region SEQ ID NO: 184 QVTLKESGPG ILQPSQTLSL TCTFSGFSLS TYGVGVGWIR QPSGKGLEWL ANIWWDDDKY 60 31H6 heavy chain YNPSLKNRLT ISKDTSNNQA FLKITNVDTA ETATYYCAQI KEPRDWFFEF WGPGTMVSVS 120 variable region S 121 SEQ ID NO: 185 DIQMTQTPSS MPASLGERVT IFCRASQGVN NYLTWYQQKP DGTIKPLIFY TSNLQSGVPS 60 31H6 light chain RFSGSGSGTD YSLSISSLEP EDFAMYYCQQ YHGFPNTFGA GTKLELKRAD AAP 113 variable region SEQ ID NO: 186 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYAMSWVRQA PGKGLEWVAS ISSGGTTYYP 60 humanized 36E5 DSVKGRFTIS RDNSKNTLYL QMNSLRAEDT AVYYCARVGG YYDSMDYWGQ GTLVTVSS 118 heavy chain variable region SEQ ID NO: 187 EIVLTQSPGT LSLSPGERAT LSCRASESVD XYGVSFMNWY QQKPGQAPRL LIYAASXQGS 60 humanized 36E5 GIPDRFSGSG SGTDFTLTIS RLEPEDFAVY YCQQTKEVTW TFGQGTKVEI KR 112 light chain variable region SEQ ID NO: 188 QVQLVESGGG VVQPGRSLRL SCAASGFTFS DYYMAWVRQA PGKGLEWVAY IHANGGSTYY 60 humanized 3D6 RDSVRGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCXXGS FMTAADYTIM DAWGQGTLVT 120 heavy chain VSS 123 variable region SEQ ID NO: 189 DIVMTQSPLS LPVTPGEPAS ISCRSSQSLL HSDGNTFLSW YLQKPGQSPQ LLIYLASNRF 60 humanized 3D6 SGVPDRFSGS GSGTDFTLKI SRVEAEDVGV YYCFQHTHLP LTFGQGTKVE IKR 113 light chain variable region SEQ ID NO: 190 QVQLQESGPG LVKPSETLSL TCTVSGYSIT SDYAWNWIRQ PPGKGLEWXG YISYSGSTRY 60 humanized 61G6 NPSLKSRXTI SXDTSKNQFS LKLSSVTAAD TAVYYCARQL GLRFFDYWGQ GTLVTVSS 118 heavy chain variable region SEQ ID NO: 191 EIVLTQSPGT LSLSPGERAT LSCSANSTVN YMYWYQQKPG QAPRXXIYLT SNLASGIPDR 60 humanized 61G6 FSGSGSGTDF TLTISRLEPE DFAVYYCQQW NSNPPTFGQG TKVEIKR 107 light chain variable region SEQ ID NO: 192 QVQLVQSGAE VKKPGASVKV SCKASGYTFS RYWIEWVRQA PGQGLEWXGE ILPGSGSSNY 60 humanized 6H6 NEKFKDRXTX TXDTSTSTAY MELRSLRSDD TAVYYCARKV YYYAMDFWGQ GTLVTVSS 118 heavy chain variable region SEQ ID NO: 193 EIVLTQSPGT LSLSPGERAT LSCTASSSVS SSYFHWYQQK PGQAPRLXIY STSNLASGIP 60 humanized 6H6 DRFSGSGSGT DXTLTISRLE PEDFAVYYCH QYHRSPRTFG QGTKVEIKR 109 light chain variable region SEQ ID NO: 194 QVQLVQSGAE VKKPGASVKV SCKASGYTFT SYTMHWVRQA PGQGLEWXGY INPRSVYTNY 60 humanized 61F6 NQKFKDRXTX TXDXSTSTAY MELRSLRSDD TAVYYCARLG GYYDTMDYWG QGTLVTVSS 119 heavy chain variable region SEQ ID NO: 195 DIQMTQSPSS LSASVGDRVT ITCRASESVD NYGISFMNWY QQKPGKAPKL LIYAASNQGS 60 humanized 61F6 GVPSRFSGSG SGTDFTLTIS SLQPEDFATY YCQQSKEVPF TFGQGTKVEI KR 112 light chain variable region SEQ ID NO: 196 QVQLVESGGG VVQPGRSLRL SCAXSGFSLS TSGMGVGWVR QAPGKGLEWV AHIWWDDDKY 60 humanized 1D8 YSPSLKSRXT ISXDXSKNTX YLQMNSLRAE DTAVYYCXRS YYYGSSGAMD YWGQGTLVTV 120 heavy chain SS 122 variable region SEQ ID NO: 197 DIVMTQSPLS LPVTPGEPAS ISCRSSQSLV HSDGNTYLHW YLQKPGQSPQ LLIYKVSKRF 60 humanized 1D8 SGVPDRFSGS GSGTDFTLKI SRVEAEDVGV YYCSQSTHVP PTFGQGTKVE IKR 113 light chain variable region SEQ ID NO: 198 QVQLVESGGG VVQPGRSLRL SCAASGFTVR NYAMSWVRQA PGKGLEWVAS ISTGDRSYLP 60 humanized 17E10 DSMKGRFTIS RDNSKNTLYL QMNSLRAEDT AVYYCXRYFD FDSFAFWGQG TLVTVSS 117 heavy chain variable region SEQ ID NO: 199 DIQMTQSPSS LSASVGDRVT ITCRASQDIN NFLNWYQQKP GKAPKLLIYY TSKLHSGVPS 60 humanized 17E10 RFSGSGSGTD FTLTISSLQP EDFATYYCQQ GHTLPPTFGQ GTKVEIKR 108 light chain variable region SEQ ID NO: 200 QVQLQESGPG LVKPSETLSL TCTVSGDSIT SGYWNWIRQP PGKGLEXXGY ISYSGSTYYN 60 humanized 35D8 PSLRGRVTIS XDTSKNQFSL KLSSVTAADT AVYYCXRRHL GSGYGWFAYW GQGTLVTVSS 120 heavy chain variable region SEQ ID NO: 201 DIVMTQSPDS LAVSLGERAT INCKASQDVN TAVAWYQQKP GQPPKLLIYW ASTRHTGVPD 60 humanized 35D8 RFSGSGSGTD XTLTISSLQA EDVAVYYCQQ HSYTPPWTFG QGTKVEIKR 109 light chain variable region SEQ ID NO: 202 QVQLQESGPG LVKPSETLSL TCTVSGDSIT SGYWNWIRQP PGKGLEXXGF
ISYSGNTYYN 60 humanized 49A1 PSLRSRXTIS XDTSKNQXSL KLSSVTAADT AVYYCXRRHL ISGYGWFAYW GQGTLVTVSS 120 heavy chain variable region SEQ ID NO: 203 XIVMTQSPDS LAVSLGERAT INCKASQDVI SAVAWYQQKP GQPPKLLIYW ASTRHTGVPD 60 humanized 49A1 RFSGSGSGTD FTLTISSLQA EDVAVYYCQQ HSYTPPWTFG QGTKVEIKR 109 light chain variable region SEQ ID NO: 204 QVQLQESGPG LVKPSETLSL TCTXSGFSLS TYGVGVGWIR QPPGKGLEWX XNIWWDDDNY 60 humanized 9E5 YNPSLIHRXT XSXDTSKNQX SLKLSSVTAA DTAVYYCAXI KEPRDWFFEF WGQGTLVTVS 120 heavy chain S 121 variable region SEQ ID NO: 205 DIQMTQSPSS LSASVGDRVT ITCRASQGVN NFLTWYQQKP GKAPKXLIXY TSNLQSGVPS 60 humanized 9E5 RFSGSGSGTD XTLTISSLQP EDFATYYCQQ YHGFPNTFGQ GTKVEIKR 108 light chain variable region SEQ ID NO: 206 QVQLQESGPG LVKPSETLSL TCTXSGFSLS TYGVGVGWIR QPPGKGLEWX XNIWWDDDKY 60 humanized 31H6 YNPSLKNRXT ISXDTSKNQX SLKLSSVTAA DTAVYYCAXI KEPRDWFFEF WGQGTLVTVS 120 heavy chain S 121 variable region SEQ ID NO: 207 DIQMTQSPSS LSASVGDRVT ITCRASQGVN NYLTWYQQKP GKAPKXLIXY TSNLQSGVPS 60 humanized 31H6 RFSGSGSGTD XTLTISSLQP EDFATYYCQQ YHGFPNTFGQ GTKVEIKR 108 light chain variable region
[1300] In an embodiment, the GITR agonist is an agonistic, anti-GITR monoclonal antibody described in U.S. Patent Application Publication No. US 2013/0108641 A1 (Sanofi SA) and International Patent Application Publication No. WO 2011/028683 A1 (Sanofi SA), the disclosures of which are incorporated by reference herein. In an embodiment, a GITR binding molecule includes monoclonal antibodies and variants and fragments thereof, including humanized and chimeric recombinant antibodies, that bind human GITR, comprising a heavy chain variable domain (V.sub.H) selected from the group consisting of SEQ ID NO:208, SEQ ID NO:210, SEQ ID NO:211, SEQ ID NO:212, SEQ ID NO:213, SEQ ID NO:214, SEQ ID NO:219, SEQ ID NO:221, SEQ ID NO:223, and SEQ ID NO:225, and a light chain variable domain (V.sub.L) selected from the group consisting of SEQ ID NO:209, SEQ ID NO:215, SEQ ID NO:216, SEQ ID NO:217, SEQ ID NO:218, SEQ ID NO:220, SEQ ID NO:222, SEQ ID NO:224, and SEQ ID NO:226 (Table 22). In an embodiment, the GITR binding molecule is an agonistic, anti-GITR monoclonal antibody comprising (a) one, two, or three heavy chain CDRs selected from the group consisting of SEQ ID NO:227, SEQ ID NO:228, SEQ ID NO:229, SEQ ID NO:233, SEQ ID NO:234, SEQ ID NO:235, SEQ ID NO:240, SEQ ID NO:241, SEQ ID NO:242, SEQ ID NO:243, SEQ ID NO:244, SEQ ID NO:245, SEQ ID NO:249, and conservative amino acid substitutions thereof, and (b) one, two, or three light chain CDRs selected from the group consisting of SEQ ID NO:230, SEQ ID NO:231, SEQ ID NO:232, SEQ ID NO:236, SEQ ID NO:237, SEQ ID NO:238, SEQ ID NO:239, SEQ ID NO:246, SEQ ID NO:247, SEQ ID NO:248, and conservative amino acid substitutions thereof (Table 22). In an embodiment, the GITR agonist is an agonistic, anti-GITR monoclonal antibody selected from the group consisting of 2155, 698, 706, 827, 1649, and 1718, and fragments, derivatives, variants, biosimilars, and combinations thereof.
[1301] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 2155, 698, 706, 827, 1649, and 1718. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 2155, 698, 706, 827, 1649, and 1718. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 2155, 698, 706, 827, 1649, and 1718. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 2155, 698, 706, 827, 1649, and 1718. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 2155, 698, 706, 827, 1649, and 1718.
TABLE-US-00022 TABLE 22 Amino acid sequences for GITR agonist antibodies related to the GITR agonists described in International Patent Application Publication No. WO 2011/028683 A1. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 208 EVKLVESGGG LVKPGGSLKL SCGASGFTIS SYAMSWVRQS PEKRLEWVAI ISTGGSTYYP 60 2155 variable DSVRGRFTIS RDNARNSLYL QMSSLRSEDT AMYYCARVGG YYDSMDHWGQ GTSVTVSS 118 heavy chain SEQ ID NO: 209 DIVLTQSPAS LAVSLGQRAT ISCRASETVD NYGISFMNWF QQKPGQSPKL LIYAASNQGS 60 2155 variable GVPARFSGSG SGTDFSLNIH PMEEDDTAMY FCQQSKEVPW TFGGGTKLEI K 111 light chain SEQ ID NO: 210 QVTLVESGGG LVKPGGSLTL SCGASGFTIS SYAMSWVRQS PGKALEWVAI ISTGGSTYYP 60 2155 humanized DSVRGRFTIS RDNAKNSLYL TMSSLDSVDT AMYYCARVGG YYDSMDHWGQ GTSVT 115 (HC1) heavy chain SEQ ID NO: 211 QVTLVESGGG LVKPGGSLTL SCGASGFTIS SYAMSWVRQS PGKALEWVAI ISTGGSTYYP 60 2155 humanized DSVRGRFTIS RDNAKNSLYL TMSSLDSVDT ATYYCARVGG YYDSMDHWGQ GTSVT 115 (HC2) heavy chain SEQ ID NO: 212 QVTLVESGGG LVKPGGSLTL SCGASGFTIS SYAMSWVRQS PGKALEWVAI ISTGGSTYYP 60 2155 humanized DKFRGRFTIS RDNAKNSLYL TMSSLRSEDT ATYYCARVGG YYDSMDHWGQ GTSVT 115 (HC3a) heavy chain SEQ ID NO: 213 QVTLKESGGG LVKPGGSLTL SCGASGFTIS SYAMSWVRQS PGKALEWVAI ISTGGSTYYP 60 humanized (HC3b) DKFRGRFTIS RDNAKNSLYL TMSSLRSEDT ATYYCARVGG YYDSMDHWGQ GTSVT 115 heavy chain SEQ ID NO: 214 EVQLVESGGG LIQPGGSLKL SCAASGFTIS SYAMSWVRQA PGKGLEWVAI ISTGGSTYYA 60 humanized (HC4) DSVKGRFTIS RDNSKNTLYL QMNSLRAEDT AVYYCARVGG YYDSMDHWGQ GTSVT 115 heavy chain SEQ ID NO: 215 DIVLTQSPAS LAASVGDRAT ISCRASETVD NYGISFMNWF QQKPGKSPKL LIYAASNQGS 60 2155 humanized GVPARFSGSG SGTDFSLNIH PMQPDDTATY FCQQSKEVPW TFGGGTKLE 109 (LC1) light chain SEQ ID NO: 216 DIVLTQSPAS LSASVGDRAT ISCRASETVD NYGISFMNWF QQKPGQSPKL LIYAASNQGS 60 2155 humanized GVPARFSGSG SGTDFSLTIS PMQPDDTATY YCQQSKEVPW TFGGGTKLE 109 (LC2a) light chain SEQ ID NO: 217 DIVLTQSPAS LSASVGDRAT ISCRASETVD NYGISYMNWF QQKPGQSPKL LIYAASNQGS 60 2155 humanized GVPARFSGSG SGTDFSLTIS PMQPDDTATY YCQQSKEVPW TFGGGTKLE 109 (LC2b) light chain SEQ ID NO: 218 DIVLTQSPAS LAVSPGQRAT ITCRASETVD NYGISFMNWF QQKPGQPPKL LIYAASNQGS 60 2155 humanized GVPARFSGSG SGTDFTLTIN PVEADDTANY YCQQSKEVPW TFGQGTKVE 109 (LC3) light chain SEQ ID NO: 219 EVQLQQSGTV LARPGASVKM SCEASGYSFT TYWMHWIKQR PGQGLEWIGA IYPGNSDTGY 60 698 variable NQKFKGKAKL TAVTSATTAY MELSSLTDED SAVYYCTRTS TYPHFDYWGQ GTTLTVSS 118 heavy chain SEQ ID NO: 220 DILLTQSPAI LSVSPGERVS FSCRASQSIG TSIHWYQQRT NGSPRLLIKY ASESISGIPS 60 698 variable RFSGSGSGTD FTLNINSVES EDIADYYCQQ SNNWPLTFGA GTKLELK 107 light chain SEQ ID NO: 221 EVQLQQSGTV LARPGASVKM SCEASGYSFT TYWMHWIKQR PGQGLEWIGA IYPGNSDTGY 60 706 variable NQKFKGKAKL TAVTSASTAY MELSSLTNED SAVYYCTRTS TYPHFDYWGQ GTTLTVSS 118 heavy chain SEQ ID NO: 222 DILLTQSPAI LSVSPGERVS FSCRASQSIG TSIHWYQQRT NGSPRLLIKY ASESISGIPS 60 706 variable RFSGSGSGTD FTLNINSVES EDIADYYCQQ TNNWPLTFGA GTKLELK 107 light chain SEQ ID NO: 223 EVQLQQSGTV LARPGASVKM SCETSGYSFT TYWIHWIKQR PGQGLEWIAT IYPGNSDAGY 60 827 variable NQKFRGKAKL TAVTSASTAY MELSSLTNED SAVYYCTRSS TYPHFDYWGQ GTTLTVSS 118 heavy chain SEQ ID NO: 224 DILLTQSPAI LSVSPGERVS FSCRASQSIG TSIHWYQQRT NDSPRLLIKY ASESISGIPS 60 827 variable RFSGSGSGTD FTLNINSVES EDIADYYCQQ TNNWPLTFGA GTKLELK 107 light chain SEQ ID NO: 225 QVQVQQSGPE LVKPGASVRI SCKASDYTFT NYYTHWVRQR PGQGLEWLGW IYPGKGYTNY 60 1718 variable NEKFKGKATL TADKSSSTAY MQFSSLTSED SAVYFCASGY GNYYFPYWGQ GTLVTVSA 118 heavy chain SEQ ID NO: 226 IQMTQSSSYL SVSLGGRVTI TCKASDHIKN WLAWYQQKPG NVPRLLMSAA TSLETGFPSR 60 1718 variable FSGSGSGKDF TLTITSLQTE DVATYYCQQY WSTPWTFGGG TKLEIK 106 light chain SEQ ID NO: 227 VGGYYDSMDH 10 2155 heavy chain CDR3 SEQ ID NO: 228 IISTGGSTY 9 2155 heavy chain CDR2 SEQ ID NO: 229 GFTISSYAMS 10 2155 heavy chain CDR1 SEQ ID NO: 230 QQSKEVPWT 9 2155 light chain CDR3 SEQ ID NO: 231 AASNQGS 7 2155 light chain CDR2 SEQ ID NO: 232 RASETVDNYG ISFMN 15 2155 light chain CDR1 SEQ ID NO: 233 TSTYPHFDY 9 698 and 706 heavy chain CDR3 SEQ ID NO: 234 AIYPGNSDTG 10 698 and 706 heavy chain CDR2 SEQ ID NO: 235 GYSFTTYWMH 10 698 and 706 heavy chain CDR1 SEQ ID NO: 236 QQSNNWPLT 9 698 light chain CDR3 SEQ ID NO: 237 KYASESIS 8 698, 706, 827, and 1649 light chain CDR2 SEQ ID NO: 238 RASQSIGTSI H 11 698, 706, 827, and 1649 light chain CDR1 SEQ ID NO: 239 QQTNNWPLT 9 706, 827, and 1649 light chain CDR3 SEQ ID NO: 240 SSTYPHFDY 9 827 and 1649 heavy chain CDR3 SEQ ID NO: 241 TIYPGNSDAG 10 827 heavy chain CDR2 SEQ ID NO: 242 AIYPGNSDAG 10 1649 heavy chain CDR2 SEQ ID NO: 243 GYGNYYFPY 9 1718 heavy chain CDR3 SEQ ID NO: 244 WIYPGKGYTN 10 1718 heavy chain CDR2 SEQ ID NO: 245 DYTFTNYYI 9 1718 heavy chain CDR1 SEQ ID NO: 246 QQTWSTPWT 9 1718 light chain CDR3 SEQ ID NO: 247 AATSLET 7 1718 light chain CDR2 SEQ ID NO: 248 KASDHIKNWL A 11 1718 light chain CDR1 SEQ ID NO: 249 GYSFTTYWIH 10 827 and 1649 heavy chain CDR1
[1302] In a preferred embodiment, the GITR agonist is the monoclonal antibody 1D7, or a fragment, derivative, variant, or biosimilar thereof 1D7 is available from Amgen, Inc. The preparation and properties of 1D7 are described in U.S. Patent Application Publication No. US 2015/0064204 A1, the disclosures of which are incorporated by reference herein. The amino acid sequences of 1D7 are set forth in Table 23.
[1303] In an embodiment, a GITR agonist comprises a heavy chain given by SEQ ID NO:250 and a light chain given by SEQ ID NO:251. In an embodiment, a GITR agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:250 and SEQ ID NO:251, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:250 and SEQ ID NO:251, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:250 and SEQ ID NO:251, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:250 and SEQ ID NO:251, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:250 and SEQ ID NO:251, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:250 and SEQ ID NO:251, respectively.
[1304] In an embodiment, the GITR agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 1D7. In an embodiment, the GITR agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:252, and the GITR agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:253, and conservative amino acid substitutions thereof. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:252 and SEQ ID NO:253, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:252 and SEQ ID NO:253, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:252 and SEQ ID NO:253, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:252 and SEQ ID NO:253, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:252 and SEQ ID NO:253, respectively.
[1305] In an embodiment, a GITR agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:254, SEQ ID NO:255, and SEQ ID NO:256, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:257, SEQ ID NO:258, and SEQ ID NO:259, respectively, and conservative amino acid substitutions thereof.
[1306] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 1D7. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 1D7. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 1D7. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 1D7. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 1D7.
TABLE-US-00023 TABLE 23 Amino acid sequences for GITR agonist antibodies related to 1D7. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 250 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVTV IWYEGSNKYY 60 1D7 heavy chain ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGG QLGKYYYYGM DVWGQGTTVT 120 VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV TVSWNSGALT SGVHTFPAVL 180 QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKR VEPKSCDKTH TCPPCPAPEL 240 LGGPSVFLFP PKPKDTLMIS RTPEVTCVVV DVSHEDPEVK FNWYVDGVEV HNAKTKPREE 300 QYNSTYRVVS VLTVLHQDWL NGKEYKCKVS NKALPAPIEK TISKAKGQPR EPQVYTLPPS 360 REEMTKNQVS LTCLVKGFYP SDIAVEWESN GQPENNYKTT PPVLDSDGSF FLYSKLTVDK 420 SRWQQGNVFS CSVMHEALHN HYTQKSLSLS PGK 453 SEQ ID NO: 251 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYD ASSLQSGVPS 60 1D7 light chain RFSGSGSGTE FTLTISSLQP EDFATYYCLQ HNNYPWTFGQ GTKVEIKRTV AAPSVFIFPP 120 SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 252 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVTV IWYEGSNKYY 60 1D7 variable ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGG QLGKYYYYGM DVWGQGTTVT 120 heavy chain VSS 123 SEQ ID NO: 253 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYD ASSLQSGVPS 60 1D7 variable RFSGSGSGTE FTLTISSLQP EDFATYYCLQ HNNYPWTFGQ GTKVEIKR 108 light chain SEQ ID NO: 254 SYGMH 5 1D7 heavy chain CDR1 SEQ ID NO: 255 VIWYEGSNKY YADSVKG 17 1D7 heavy chain CDR2 SEQ ID NO: 256 GGQLGKYYYY GMDV 14 1D7 heavy chain CDR3 SEQ ID NO: 257 RASQGIRNDL G 11 1D7 light chain CDR1 SEQ ID NO: 258 DASSLQS 7 1D7 light chain CDR2 SEQ ID NO: 259 LQHNNYPWT 9 1D7 light chain CDR3
[1307] In a preferred embodiment, the GITR agonist is the monoclonal antibody 33C9, or a fragment, derivative, variant, or biosimilar thereof. 33C9 is available from Amgen, Inc. The preparation and properties of 33C9 are described in U.S. Patent Application Publication No. US 2015/0064204 A1, the disclosures of which are incorporated by reference herein. The amino acid sequences of 33C9 are set forth in Table 24.
[1308] In an embodiment, a GITR agonist comprises a heavy chain given by SEQ ID NO:260 and a light chain given by SEQ ID NO:261. In an embodiment, a GITR agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:260 and SEQ ID NO:261, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:260 and SEQ ID NO:261, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:260 and SEQ ID NO:261, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:260 and SEQ ID NO:261, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:260 and SEQ ID NO:261, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:260 and SEQ ID NO:261, respectively.
[1309] In an embodiment, the GITR agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 1D7. In an embodiment, the GITR agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:262, and the GITR agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:263, and conservative amino acid substitutions thereof. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:262 and SEQ ID NO:263, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:262 and SEQ ID NO:263, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:262 and SEQ ID NO:263, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:262 and SEQ ID NO:263, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:262 and SEQ ID NO:263, respectively.
[1310] In an embodiment, a GITR agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:264, SEQ ID NO:265, and SEQ ID NO:266, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:267, SEQ ID NO:268, and SEQ ID NO:269, respectively, and conservative amino acid substitutions thereof.
[1311] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 33C9. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 33C9. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 33C9. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 33C9. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 33C9.
TABLE-US-00024 TABLE 24 Amino acid sequences for GITR agonist antibodies related to 33C9. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 260 QVQVVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVSV IWYEGSNKYY 60 33C9 heavy chain ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGG LLGYYYYYGM DVWGQGTTVT 120 VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV TVSWNSGALT SGVHTFPAVL 180 QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKK VEPKSCDKTH TCPPCPAPEL 240 LGGPSVFLFP PKPKDTLMIS RTPEVTCVVV DVSHEDPEVK FNWYVDGVEV HNAKTKPREE 300 QYNSTYRVVS VLTVLHQDWL NGKEYKCKVS NKALPAPIEK TISKAKGQPR EPQVYTLPPS 360 REEMTKNQVS LTCLVKGFYP SDIAVEWESN GQPENNYKTT PPVLDSDGSF FLYSKLTVDK 420 SRWQQGNVFS CSVMHEALHN HYTQKSLSLS PGK 453 SEQ ID NO: 261 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYD ASSLQSGVPS 60 33C9 light chain RFSGSGSGTE FTLTISSLQP EDFATYYCLQ HHSYPWTFGQ GTKVEIKRTV AAPSVFIFPP 120 SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 262 QVQVVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVSV IWYEGSNKYY 60 33C9 variable ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGG LLGYYYYYGM DVWGQGTTVT 120 heavy chain VSS 123 SEQ ID NO: 263 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYD ASSLQSGVPS 60 33C9 variable RFSGSGSGTE FTLTISSLQP EDFATYYCLQ HHSYPWTFGQ GTKVEIKR 108 light chain SEQ ID NO: 264 SYGMH 5 33C9 heavy chain CDR1 SEQ ID NO: 265 VIWYEGSNKY YADSVKG 17 33C9 heavy chain CDR2 SEQ ID NO: 266 GGLLGYYYYY GMDV 14 33C9 heavy chain CDR3 SEQ ID NO: 267 RASQGIRNDL G 11 33C9 light chain CDR1 SEQ ID NO: 268 DASSLQS 7 33C9 light chain CDR2 SEQ ID NO: 269 LQHHSYPWT 9 33C9 light chain CDR3
[1312] In a preferred embodiment, the GITR agonist is the monoclonal antibody 33F6, or a fragment, derivative, variant, or biosimilar thereof. 33F6 is available from Amgen, Inc. The preparation and properties of 33F6 are described in U.S. Patent Application Publication No. US 2015/0064204 A1, the disclosures of which are incorporated by reference herein. The amino acid sequences of 33F6 are set forth in Table 25.
[1313] In an embodiment, a GITR agonist comprises a heavy chain given by SEQ ID NO:270 and a light chain given by SEQ ID NO:271. In an embodiment, a GITR agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:270 and SEQ ID NO:271, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:270 and SEQ ID NO:271, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:270 and SEQ ID NO:271, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:270 and SEQ ID NO:271, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:270 and SEQ ID NO:271, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:270 and SEQ ID NO:271, respectively.
[1314] In an embodiment, the GITR agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 33F6. In an embodiment, the GITR agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:272, and the GITR agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:273, and conservative amino acid substitutions thereof. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:272 and SEQ ID NO:273, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:272 and SEQ ID NO:273, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:272 and SEQ ID NO:273, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:272 and SEQ ID NO:273, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:272 and SEQ ID NO:273, respectively.
[1315] In an embodiment, a GITR agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:274, SEQ ID NO:275, and SEQ ID NO:276, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:277, SEQ ID NO:278, and SEQ ID NO:279, respectively, and conservative amino acid substitutions thereof.
[1316] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 33F6. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 33F6. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 33F6. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 33F6. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 33F6.
TABLE-US-00025 TABLE 25 Amino acid sequences for GITR agonist antibodies related to 33F6. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 270 QVQLVESGGG VVQPGRSLRL SCAASGFTFS NYGMHWVRQA PGKGLEWVAV IWYVGSNKYY 60 33F6 heavy chain ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGG ELRLYYTYGM DVWGQGTTVT 120 VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV TVSWNSGALT SGVHTFPAVL 180 QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKK VEPKSCDKTH TCPPCPAPEL 240 LGGPSVFLFP PKPKDTLMIS RTPEVTCVVV DVSHEDPEVK FNWYVDGVEV HNAKTKPREE 300 QYNSTYRVVS VLTVLHQDWL NGKEYKCKVS NKALPAPIEK TISKAKGQPR EPQVYTLPPS 360 REEMTKNQVS LTCLVKGFYP SDIAVEWESN GQPENNYKTT PPVLDSDGSF FLYSKLTVDK 420 SRWQQGNVFS CSVMHEALHN HYTQKSLSLS PGK 453 SEQ ID NO: 271 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYA ASSLQSGVPS 60 33F6 light chain RFSGSGSGTE FTLTVSSLQP EDFATYYCLQ LNSYPWTFGQ GTKVEIKRTV AAPSVFIFPP 120 SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 272 QVQLVESGGG VVQPGRSLRL SCAASGFTFS NYGMHWVRQA PGKGLEWVAV IWYVGSNKYY 60 33F6 variable ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGG ELRLYYTYGM DVWGQGTTVT 120 heavy chain VSS 123 SEQ ID NO: 273 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYA ASSLQSGVPS 60 33F6 variable RFSGSGSGTE FTLTVSSLQP EDFATYYCLQ LNSYPWTFGQ GTKVEIKR 108 light chain SEQ ID NO: 274 NYGMH 5 33F6 heavy chain CDR1 SEQ ID NO: 275 VIWYVGSNKY YADSVKG 17 33F6 heavy chain CDR2 SEQ ID NO: 276 GGELRLYYTY GMDV 14 33F6 heavy chain CDR3 SEQ ID NO: 277 RASQGIRNDL G 11 33F6 light chain CDR1 SEQ ID NO: 278 AASSLQS 7 33F6 light chain CDR2 SEQ ID NO: 279 LQLNSYPWT 9 33F6 light chain CDR3
[1317] In a preferred embodiment, the GITR agonist is the monoclonal antibody 34G4, or a fragment, derivative, variant, or biosimilar thereof 34G4 is available from Amgen, Inc. The preparation and properties of 34G4 are described in U.S. Patent Application Publication No. US 2015/0064204 A1, the disclosures of which are incorporated by reference herein. The amino acid sequences of 34G4 are set forth in Table 26.
[1318] In an embodiment, a GITR agonist comprises a heavy chain given by SEQ ID NO:280 and a light chain given by SEQ ID NO:281. In an embodiment, a GITR agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:280 and SEQ ID NO:281, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:280 and SEQ ID NO:281, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:280 and SEQ ID NO:281, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:280 and SEQ ID NO:281, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:280 and SEQ ID NO:281, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:280 and SEQ ID NO:281, respectively.
[1319] In an embodiment, the GITR agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 34G4. In an embodiment, the GITR agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:282, and the GITR agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:283, and conservative amino acid substitutions thereof. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:282 and SEQ ID NO:283, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:282 and SEQ ID NO:283, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:282 and SEQ ID NO:283, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:282 and SEQ ID NO:283, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:282 and SEQ ID NO:283, respectively.
[1320] In an embodiment, a GITR agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:284, SEQ ID NO:285, and SEQ ID NO:286, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:287, SEQ ID NO:288, and SEQ ID NO:289, respectively, and conservative amino acid substitutions thereof.
[1321] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 34G4. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 34G4. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 34G4. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 34G4. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 34G4.
TABLE-US-00026 TABLE 26 Amino acid sequences for GITR agonist antibodies related to 34G4. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 280 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVAV IWYEGSNKYY 60 34G4 heavy chain ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGG QLGYYYYYGM DVWGQGTTVT 120 VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV TVSWNSGALT SGVHTFPAVL 180 QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKK VEPKSCDKTH TCPPCPAPEL 240 LGGPSVFLFP PKPKDTLMIS RTPEVTCVVV DVSHEDPEVK FNWYVDGVEV HNAKTKPREE 300 QYNSTYRVVS VLTVLHQDWL NGKEYKCKVS NKALPAPIEK TISKAKGQPR EPQVYTLPPS 360 REEMTKNQVS LTCLVKGFYP SDIAVEWESN GQPENNYKTT PPVLDSDGSF FLYSKLTVDK 420 SRWQQGNVFS CSVMHEALHN HYTQKSLSLS PGK 453 SEQ ID NO: 281 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYD ASSLQSGVPS 60 34G4 light chain RFSGSGSGTD FTLTISSLQP EDFATYYCLQ LNSYPWTFGQ GTKVEIKRTV AAPSVFIFPP 120 SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 282 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVAV IWYEGSNKYY 60 34G4 variable ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGG QLGYYYYYGM DVWGQGTTVT 120 heavy chain VSS 123 SEQ ID NO: 283 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYD ASSLQSGVPS 60 34G4 variable RFSGSGSGTD FTLTISSLQP EDFATYYCLQ LNSYPWTFGQ GTKVEIKR 108 light chain SEQ ID NO: 284 SYGMH 5 34G4 heavy chain CDR1 SEQ ID NO: 285 VIWYEGSNKY YADSVKG 17 34G4 heavy chain CDR2 SEQ ID NO: 286 GGQLGYYYYY GMDV 14 34G4 heavy chain CDR3 SEQ ID NO: 287 RASQGIRNDL G 11 34G4 light chain CDR1 SEQ ID NO: 288 DASSLQS 7 34G4 light chain CDR2 SEQ ID NO: 289 LQLNSYPWT 9 34G4 light chain CDR3
[1322] In a preferred embodiment, the GITR agonist is the monoclonal antibody 35B10, or a fragment, derivative, variant, or biosimilar thereof. 35B10 is available from Amgen, Inc. The preparation and properties of 35B10 are described in U.S. Patent Application Publication No. US 2015/0064204 A1, the disclosures of which are incorporated by reference herein. The amino acid sequences of 35B10 are set forth in Table 27.
[1323] In an embodiment, a GITR agonist comprises a heavy chain given by SEQ ID NO:290 and a light chain given by SEQ ID NO:291. In an embodiment, a GITR agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:290 and SEQ ID NO:291, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:290 and SEQ ID NO:291, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:290 and SEQ ID NO:291, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:290 and SEQ ID NO:291, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:290 and SEQ ID NO:291, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:290 and SEQ ID NO:291, respectively.
[1324] In an embodiment, the GITR agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 35B10. In an embodiment, the GITR agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:292, and the GITR agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:293, and conservative amino acid substitutions thereof. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:292 and SEQ ID NO:293, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:292 and SEQ ID NO:293, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:292 and SEQ ID NO:293, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:292 and SEQ ID NO:293, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:292 and SEQ ID NO:293, respectively.
[1325] In an embodiment, a GITR agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:294, SEQ ID NO:295, and SEQ ID NO:296, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:297, SEQ ID NO:298, and SEQ ID NO:299, respectively, and conservative amino acid substitutions thereof.
[1326] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 35B10. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 35B10. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 35B10. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 35B 10. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 35B10.
TABLE-US-00027 TABLE 27 Amino acid sequences for GITR agonist antibodies related to 35B10. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 290 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVAV IWYAGSNKYY 60 35B10 heavy ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGG ELSFYYYYGM DVWGQGTTVT 120 chain VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV TVSWNSGALT SGVHTFPAVL 180 QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKK VEPKSCDKTH TCPPCPAPEL 240 LGGPSVFLFP PKPKDTLMIS RTPEVTCVVV DVSHEDPEVK FNWYVDGVEV HNAKTKPREE 300 QYNSTYRVVS VLTVLHQDWL NGKEYKCKVS NKALPAPIEK TISKAKGQPR EPQVYTLPPS 360 REEMTKNQVS LTCLVKGFYP SDIAVEWESN GQPENNYKTT PPVLDSDGSF FLYSKLTVDK 420 SRWQQGNVFS CSVMHEALHN HYTQKSLSLS PGK 453 SEQ ID NO: 291 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYA ASTLQSGVPS 60 35B10 light RFSGSGSGTE FTLTISSLQP EDFATYYCLQ HNNYPWTFGQ GTKVEIKRTV AAPSVFIFPP 120 chain SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 292 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVAV IWYAGSNKYY 60 35B10 variable ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGG ELSFYYYYGM DVWGQGTTVT 120 heavy chain VSS 123 SEQ ID NO: 293 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYA ASTLQSGVPS 60 35B10 variable RFSGSGSGTE FTLTISSLQP EDFATYYCLQ HNNYPWTFGQ GTKVEIKR 108 light chain SEQ ID NO: 294 SYGMH 5 35B10 heavy chain CDR1 SEQ ID NO: 295 VIWYAGSNKY YADSVKG 17 35B10 heavy chain CDR2 SEQ ID NO: 296 GGELSFYYYY GMDV 14 35B10 heavy chain CDR3 SEQ ID NO: 297 RASQGIRNDL G 11 35B10 light chain CDR1 SEQ ID NO: 298 AASTLQS 7 35B10 light chain CDR2 SEQ ID NO: 299 LQHNNYPWT 9 35B10 light chain CDR3
[1327] In a preferred embodiment, the GITR agonist is the monoclonal antibody 41E11, or a fragment, derivative, variant, or biosimilar thereof. 41E11 is available from Amgen, Inc. The preparation and properties of 41E11 are described in U.S. Patent Application Publication No. US 2015/0064204 A1, the disclosures of which are incorporated by reference herein. The amino acid sequences of 41E11 are set forth in Table 28.
[1328] In an embodiment, a GITR agonist comprises a heavy chain given by SEQ ID NO:300 and a light chain given by SEQ ID NO:301. In an embodiment, a GITR agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:300 and SEQ ID NO:301, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:300 and SEQ ID NO:301, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:300 and SEQ ID NO:301, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:300 and SEQ ID NO:301, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:300 and SEQ ID NO:301, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:300 and SEQ ID NO:301, respectively.
[1329] In an embodiment, the GITR agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 41E11. In an embodiment, the GITR agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:302, and the GITR agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:303, and conservative amino acid substitutions thereof. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:302 and SEQ ID NO:303, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:302 and SEQ ID NO:303, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:302 and SEQ ID NO:303, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:302 and SEQ ID NO:303, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:302 and SEQ ID NO:303, respectively.
[1330] In an embodiment, a GITR agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:304, SEQ ID NO:305, and SEQ ID NO:306, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:307, SEQ ID NO:308, and SEQ ID NO:309, respectively, and conservative amino acid substitutions thereof.
[1331] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 41E11. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 41E11. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 41E 1. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 41E11. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 41E11.
TABLE-US-00028 TABLE 28 Amino acid sequences for GITR agonist antibodies related to 41E11. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 300 QVQVVESGGG VVQPGRSLRL SCAASGFTFS SYGMYWVRQA PGKGLEWVAV IWYEGSNKYY 60 41E11 heavy ADSVRGRFTI SRDNSKNTLY LQMNSLRAED TALYYCARGG QLGKDYYSGM DVWGQGTTVT 120 chain VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV TVSWNSGALT SGVHTFPAVL 180 QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKK VEPKSCDKTH TCPPCPAPEL 240 LGGPSVFLFP PKPKDTLMIS RTPEVTCVVV DVSHEDPEVK FNWYVDGVEV HNAKTKPREE 300 QYNSTYRVVS VLTVLHQDWL NGKEYKCKVS NKALPAPIEK TISKAKGQPR EPQVYTLPPS 360 REEMTKNQVS LTCLVKGFYP SDIAVEWESN GQPENNYKTT PPVLDSDGSF FLYSKLTVDK 420 SRWQQGNVFS CSVMHEALHN HYTQKSLSLS PGK 453 SEQ ID NO: 301 DIQMTQSPSS LSASVGDRVT ITCRASQVIR NDLGWYQQKP GKAPKRLIYA ASSLQSGVPS 60 41E11 light RFSGSGSGTE FTLTISSLQP EDFATYYCLQ HNSYPLTFGG GTKVEIKRTV AAPSVFIFPP 120 chain SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 302 QVQVVESGGG VVQPGRSLRL SCAASGFTFS SYGMYWVRQA PGKGLEWVAV IWYEGSNKYY 60 41E11 variable ADSVRGRFTI SRDNSKNTLY LQMNSLRAED TALYYCARGG QLGKDYYSGM DVWGQGTTVT 120 heavy chain VSS 123 SEQ ID NO: 303 DIQMTQSPSS LSASVGDRVT ITCRASQVIR NDLGWYQQKP GKAPKRLIYA ASSLQSGVPS 60 41E11 variable RFSGSGSGTE FTLTISSLQP EDFATYYCLQ HNSYPLTFGG GTKVEIKR 108 light chain SEQ ID NO: 304 SYGMY 5 41E11 heavy chain CDR1 SEQ ID NO: 305 VIWYEGSNKY YADSVRG 17 41E11 heavy chain CDR2 SEQ ID NO: 306 GGQLGKDYYS GMDV 14 41E11 heavy chain CDR3 SEQ ID NO: 307 RASQVIRNDL G 11 41E11 light chain CDR1 SEQ ID NO: 308 AASSLQS 7 41E11 light chain CDR2 SEQ ID NO: 309 LQHNSYPLT 9 41E11 light chain CDR3
[1332] In a preferred embodiment, the GITR agonist is the monoclonal antibody 41G5, or a fragment, derivative, variant, or biosimilar thereof. 41G5 is available from Amgen, Inc. The preparation and properties of 41G5 are described in U.S. Patent Application Publication No. US 2015/0064204 A1, the disclosures of which are incorporated by reference herein. The amino acid sequences of 41G5 are set forth in Table 29.
[1333] In an embodiment, a GITR agonist comprises a heavy chain given by SEQ ID NO:310 and a light chain given by SEQ ID NO:311. In an embodiment, a GITR agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:310 and SEQ ID NO:311, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:310 and SEQ ID NO:311, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:310 and SEQ ID NO:311, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:310 and SEQ ID NO:311, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:310 and SEQ ID NO:311, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:310 and SEQ ID NO:311, respectively.
[1334] In an embodiment, the GITR agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 41G5. In an embodiment, the GITR agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:312, and the GITR agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:313, and conservative amino acid substitutions thereof. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:312 and SEQ ID NO:313, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:312 and SEQ ID NO:313, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:312 and SEQ ID NO:313, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:312 and SEQ ID NO:313, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:312 and SEQ ID NO:313, respectively.
[1335] In an embodiment, a GITR agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:314, SEQ ID NO:315, and SEQ ID NO:316, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:317, SEQ ID NO:318, and SEQ ID NO:319, respectively, and conservative amino acid substitutions thereof.
[1336] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 41G5. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 41G5. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 41G5. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 41G5. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 41G5.
TABLE-US-00029 TABLE 29 Amino acid sequences for GITR agonist antibodies related to 41G5. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 310 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVAV IWYPGSNKYY 60 41G5 heavy chain ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGG ELGRYYTYGM DVWGQGTTVT 120 VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV TVSWNSGALT SGVHTFPAVL 180 QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKK VEPKSCDKTH TCPPCPAPEL 240 LGGPSVFLFP PKPKDTLMIS RTPEVTCVVV DVSHEDPEVK FNWYVDGVEV HNAKTKPREE 300 QYNSTYRVVS VLTVLHQDWL NGKEYKCKVS NKALPAPIEK TISKAKGQPR EPQVYTLPPS 360 REEMTKNQVS LTCLVKGFYP SDIAVEWESN GQPENNYKTT PPVLDSDGSF FLYSKLTVDK 420 SRWQQGNVFS CSVMHEALHN HYTQKSLSLS PGK 453 SEQ ID NO: 311 DIQMTQSPSS LSASVGDRVT VTCRASQGIR NDLGWYQQKP GKAPKRLIYA ASSLQSGVPS 60 41G5 light chain RFSGSGSGTE FTLTISSLQP EDFATYYCLQ HNNYPWTFGQ GTKVDIKRTV AAPSVFIFPP 120 SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 312 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVAV IWYPGSNKYY 60 41G5 variable ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGG ELGRYYTYGM DVWGQGTTVT 120 heavy chain VSS 123 SEQ ID NO: 313 DIQMTQSPSS LSASVGDRVT VTCRASQGIR NDLGWYQQKP GKAPKRLIYA ASSLQSGVPS 60 41G5 variable RFSGSGSGTE FTLTISSLQP EDFATYYCLQ HNNYPWTFGQ GTKVDIKR 108 light chain SEQ ID NO: 314 SYGMH 5 41G5 heavy chain CDR1 SEQ ID NO: 315 VIWYPGSNKY YADSVKG 17 41G5 heavy chain CDR2 SEQ ID NO: 316 GGELGRYYTY GMDV 14 41G5 heavy chain CDR3 SEQ ID NO: 317 RASQGIRNDL G 11 41G5 light chain CDR1 SEQ ID NO: 318 AASSLQS 7 41G5 light chain CDR2 SEQ ID NO: 319 LQHNNYPWT 9 41G5 light chain CDR3
[1337] In a preferred embodiment, the GITR agonist is the monoclonal antibody 42A11, or a fragment, derivative, variant, or biosimilar thereof 42A11 is available from Amgen, Inc. The preparation and properties of 42A11 are described in U.S. Patent Application Publication No. US 2015/0064204 A1, the disclosures of which are incorporated by reference herein. The amino acid sequences of 42A11 are set forth in Table 30.
[1338] In an embodiment, a GITR agonist comprises a heavy chain given by SEQ ID NO:320 and a light chain given by SEQ ID NO:321. In an embodiment, a GITR agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:320 and SEQ ID NO:321, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:320 and SEQ ID NO:321, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:320 and SEQ ID NO:321, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:320 and SEQ ID NO:321, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:320 and SEQ ID NO:321, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:320 and SEQ ID NO:321, respectively.
[1339] In an embodiment, the GITR agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 42A11. In an embodiment, the GITR agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:322, and the GITR agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:323, and conservative amino acid substitutions thereof. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:322 and SEQ ID NO:323, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:322 and SEQ ID NO:323, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:322 and SEQ ID NO:323, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:322 and SEQ ID NO:323, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:322 and SEQ ID NO:323, respectively.
[1340] In an embodiment, a GITR agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:324, SEQ ID NO:325, and SEQ ID NO:326, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:327, SEQ ID NO:328, and SEQ ID NO:329, respectively, and conservative amino acid substitutions thereof.
[1341] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 42A11. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 42A11. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 42A11. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 42A11. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 42A11.
TABLE-US-00030 TABLE 30 Amino acid sequences for GITR agonist antibodies related to 42A11. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 320 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVAV IWYEGSNKYY 60 42A11 heavy ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGG QLGYYYYSGM DVWGQGTTVT 120 chain VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV TVSWNSGALT SGVHTFPAVL 180 QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKK VEPKSCDKTH TCPPCPAPEL 240 LGGPSVFLFP PKPKDTLMIS RTPEVTCVVV DVSHEDPEVK FNWYVDGVEV HNAKTKPREE 300 QYNSTYRVVS VLTVLHQDWL NGKEYKCKVS NKALPAPIEK TISKAKGQPR EPQVYTLPPS 360 REEMTKNQVS LTCLVKGFYP SDIAVEWESN GQPENNYKTT PPVLDSDGSF FLYSKLTVDK 420 SRWQQGNVFS CSVMHEALHN HYTQKSLSLS PGK 453 SEQ ID NO: 321 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYD ASSLQSGVPS 60 42A11 light RFSGSGSGTD FTLTISSLQP EEFATYYCLQ HNNYPWTFGQ GTKVEIKRTV AAPSVFIFPP 120 chain SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 322 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVAV IWYEGSNKYY 60 42A11 variable ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGG QLGYYYYSGM DVWGQGTTVT 120 heavy chain VSS 123 SEQ ID NO: 323 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYD ASSLQSGVPS 60 42A11 variable RFSGSGSGTD FTLTISSLQP EEFATYYCLQ HNNYPWTFGQ GTKVEIKR 108 light chain SEQ ID NO: 324 SYGMH 5 42A11 heavy chain CDR1 SEQ ID NO: 325 VIWYEGSNKY YADSVKG 17 42A11 heavy chain CDR2 SEQ ID NO: 326 GGQLGYYYYS GMDV 14 42A11 heavy chain CDR3 SEQ ID NO: 327 RASQGIRNDL G 11 42A11 light chain CDR1 SEQ ID NO: 328 DASSLQS 7 42A11 light chain CDR2 SEQ ID NO: 329 LQHNNYPWT 9 42A11 light chain CDR3
[1342] In a preferred embodiment, the GITR agonist is the monoclonal antibody 44C1, or a fragment, derivative, variant, or biosimilar thereof. 44C1 is available from Amgen, Inc. The preparation and properties of 44C1 are described in U.S. Patent Application Publication No. US 2015/0064204 A1, the disclosures of which are incorporated by reference herein. The amino acid sequences of 44C1 are set forth in Table 31.
[1343] In an embodiment, a GITR agonist comprises a heavy chain given by SEQ ID NO:330 and a light chain given by SEQ ID NO:331. In an embodiment, a GITR agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:330 and SEQ ID NO:331, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:330 and SEQ ID NO:331, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:330 and SEQ ID NO:331, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:330 and SEQ ID NO:331, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:330 and SEQ ID NO:331, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:330 and SEQ ID NO:331, respectively.
[1344] In an embodiment, the GITR agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 44C1. In an embodiment, the GITR agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:332, and the GITR agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:333, and conservative amino acid substitutions thereof. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:332 and SEQ ID NO:333, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:332 and SEQ ID NO:333, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:332 and SEQ ID NO:333, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:332 and SEQ ID NO:333, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:332 and SEQ ID NO:333, respectively.
[1345] In an embodiment, a GITR agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:334, SEQ ID NO:335, and SEQ ID NO:336, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:337, SEQ ID NO:338, and SEQ ID NO:339, respectively, and conservative amino acid substitutions thereof.
[1346] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 44C1. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 44C1. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 44C1. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 44C1. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 44C1.
TABLE-US-00031 TABLE 31 Amino acid sequences for GITR agonist antibodies related to 44C1. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 330 QVQLVESGGG VVQPGRSLRL SCAASGFTLS SYGMHWVRQA PGKGLEWVAV IWYDGSNKYY 60 44C1 heavy chain ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARRG TVTTPDFDYW GQGTLVTVSS 120 ASTKGPSVFP LAPSSKSTSG GTAALGCLVK DYFPEPVTVS WNSGALTSGV HTFPAVLQSS 180 GLYSLSSVVT VPSSSLGTQT YICNVNHKPS NTKVDKKVEP KSCDKTHTCP PCPAPELLGG 240 PSVFLFPPKP KDTLMISRTP EVTCVVVDVS HEDPEVKFNW YVDGVEVHNA KTKPREEQYN 300 STYRVVSVLT VLHQDWLNGK EYKCKVSNKA LPAPIEKTIS KAKGQPREPQ VYTLPPSREE 360 MTKNQVSLTC LVKGFYPSDI AVEWESNGQP ENNYKTTPPV LDSDGSFFLY SKLTVDKSRW 420 QQGNVFSCSV MHEALHNHYT QKSLSLSPGK 450 SEQ ID NO: 331 QSALTQPASV SGSPGQSITI SCTGTSSDVG TYNLVSWYQQ HPGKAPKLMI YEVSKRPSGV 60 44C1 light chain SNRFSGSKSG NTASLTISGL QAEDEADYYC CSYAGFSTWV FGGGTKLTVL GQPKAAPSVT 120 LFPPSSEELQ ANKATLVCLI SDFYPGAVTV AWKADSSPVK AGVETTTPSK QSNNKYAASS 180 YLSLTPEQWK SHRSYSCQVT HEGSTVEKTV APTECS 216 SEQ ID NO: 332 QVQLVESGGG VVQPGRSLRL SCAASGFTLS SYGMHWVRQA PGKGLEWVAV IWYDGSNKYY 60 44C1 variable ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARRG TVTTPDFDYW GQGTLVTVSS 120 heavy chain SEQ ID NO: 333 QSALTQPASV SGSPGQSITI SCTGTSSDVG TYNLVSWYQQ HPGKAPKLMI YEVSKRPSGV 60 44C1 variable SNRFSGSKSG NTASLTISGL QAEDEADYYC CSYAGFSTWV FGGGTKLTVL G 111 light chain SEQ ID NO: 334 SYGMH 5 44C1 heavy chain CDR1 SEQ ID NO: 335 VIWYDGSNKY YADSVKG 17 44C1 heavy chain CDR2 SEQ ID NO: 336 RGTVTTPDFD Y 11 44C1 heavy chain CDR3 SEQ ID NO: 337 TGTSSDVGTY NLVS 14 44C1 light chain CDR1 SEQ ID NO: 338 EVSKRPS 7 44C1 light chain CDR2 SEQ ID NO: 339 CSYAGFSTWV 10 44C1 light chain CDR3
[1347] In a preferred embodiment, the GITR agonist is the monoclonal antibody 45A8, or a fragment, derivative, variant, or biosimilar thereof. 45A8 is available from Amgen, Inc. The preparation and properties of 45A8 are described in U.S. Patent Application Publication No. US 2015/0064204 A1, the disclosures of which are incorporated by reference herein. The amino acid sequences of 45A8 are set forth in Table 32.
[1348] In an embodiment, a GITR agonist comprises a heavy chain given by SEQ ID NO:340 and a light chain given by SEQ ID NO:341. In an embodiment, a GITR agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:340 and SEQ ID NO:341, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:340 and SEQ ID NO:341, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:340 and SEQ ID NO:341, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:340 and SEQ ID NO:341, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:340 and SEQ ID NO:341, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:340 and SEQ ID NO:341, respectively.
[1349] In an embodiment, the GITR agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 45A8. In an embodiment, the GITR agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:342, and the GITR agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:343, and conservative amino acid substitutions thereof. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:342 and SEQ ID NO:343, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:342 and SEQ ID NO:343, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:342 and SEQ ID NO:343, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:342 and SEQ ID NO:343, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:342 and SEQ ID NO:343, respectively.
[1350] In an embodiment, a GITR agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:344, SEQ ID NO:345, and SEQ ID NO:346, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:347, SEQ ID NO:348, and SEQ ID NO:349, respectively, and conservative amino acid substitutions thereof.
[1351] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 45A8. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 45A8. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 45A8. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 45A8. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 45A8.
TABLE-US-00032 TABLE 32 Amino acid sequences for GITR agonist antibodies related to 45A8. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 340 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVAV IWHDGSNKYY 60 45A8 heavy chain ADSVKGRFTI SKDNSKNTLY LQMNSLRAED TAVYYCAREY GGNFDYWGQG TLVTVSSAST 120 KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF PEPVTVSWNS GALTSGVHTF PAVLQSSGLY 180 SLSSVVTVPS SSLGTQTYIC NVNHKPSNTK VDKKVEPKSC DKTHTCPPCP APELLGGPSV 240 FLFPPKPKDT LMISRTPEVT CVVVDVSHED PEVKFNWYVD GVEVHNAKTK PREEQYNSTY 300 RVVSVLTVLH QDWLNGKEYK CKVSNKALPA PIEKTISKAK GQPREPQVYT LPPSREEMTK 360 NQVSLTCLVK GFYPSDIAVE WESNGQPENN YKTTPPVLDS DGSFFLYSKL TVDKSRWQQG 420 NVFSCSVMHE ALHNHYTQKS LSLSPGK 447 SEQ ID NO: 341 QSALTQPASV SGSPGQSITI SCTGTSSDVG TYNLVSWYQQ HPGKAPKLMI YEVSKRPSGI 60 45A8 light chain SNRFSGSKSG NTASLTISGL QAEDEADYYC CSYAGYSTWV FGGGTKLTVL RQPKAAPSVT 120 LFPPSSEELQ ANKATLVCLI SDFYPGAVTV AWKADSSPVK AGVETTTPSK QSNNKYAASS 180 YLSLTPEQWK SHRSYSCQVT HEGSTVEKTV APTECS 216 SEQ ID NO: 342 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVAV IWHDGSNKYY 60 45A8 variable ADSVKGRFTI SKDNSKNTLY LQMNSLRAED TAVYYCAREY GGNFDYWGQG TLVTVSS 117 heavy chain SEQ ID NO: 343 QSALTQPASV SGSPGQSITI SCTGTSSDVG TYNLVSWYQQ HPGKAPKLMI YEVSKRPSGI 60 45A8 variable SNRFSGSKSG NTASLTISGL QAEDEADYYC CSYAGYSTWV FGGGTKLTVL R 111 light chain SEQ ID NO: 344 SYGMH 5 45A8 heavy chain CDR1 SEQ ID NO: 345 VIWHDGSNKY YADSVKG 17 45A8 heavy chain CDR2 SEQ ID NO: 346 EYGGNFDY 8 45A8 heavy chain CDR3 SEQ ID NO: 347 TGTSSDVGTY NLVS 14 45A8 light chain CDR1 SEQ ID NO: 348 EVSKRPS 7 45A8 light chain CDR2 SEQ ID NO: 349 CSYAGYSTWV 10 45A8 light chain CDR3
[1352] In a preferred embodiment, the GITR agonist is the monoclonal antibody 46E11, or a fragment, derivative, variant, or biosimilar thereof 46E11 is available from Amgen, Inc. The preparation and properties of 46E11 are described in U.S. Patent Application Publication No. US 2015/0064204 A1, the disclosures of which are incorporated by reference herein. The amino acid sequences of 46E11 are set forth in Table 33.
[1353] In an embodiment, a GITR agonist comprises a heavy chain given by SEQ ID NO:350 and a light chain given by SEQ ID NO:351. In an embodiment, a GITR agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:350 and SEQ ID NO:351, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:350 and SEQ ID NO:351, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:350 and SEQ ID NO:351, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:350 and SEQ ID NO:351, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:350 and SEQ ID NO:351, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:350 and SEQ ID NO:351, respectively.
[1354] In an embodiment, the GITR agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 46E11. In an embodiment, the GITR agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:352, and the GITR agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:353, and conservative amino acid substitutions thereof. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:352 and SEQ ID NO:353, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:352 and SEQ ID NO:353, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:352 and SEQ ID NO:353, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:352 and SEQ ID NO:353, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:352 and SEQ ID NO:353, respectively.
[1355] In an embodiment, a GITR agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:354, SEQ ID NO:355, and SEQ ID NO:356, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:357, SEQ ID NO:358, and SEQ ID NO:359, respectively, and conservative amino acid substitutions thereof.
[1356] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 46E11. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 46E11. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 46E11. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 46E11. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 46E11.
TABLE-US-00033 TABLE 33 Amino acid sequences for GITR agonist antibodies related to 46E11. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 350 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVAV IWYAGSNKYY 60 46E11 heavy ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGD ILTGYSLYYG MDVWGQGTTV 120 chain TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP VTVSWNSGAL TSGVHTFPAV 180 LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK KVEPKSCDKT HTCPPCPAPE 240 LLGGPSVFLF PPKPKDTLMI SRTPEVTCVV VDVSHEDPEV KFNWYVDGVE VHNAKTKPRE 300 EQYNSTYRVV SVLTVLHQDW LNGKEYKCKV SNKALPAPIE KTISKAKGQP REPQVYTLPP 360 SREEMTKNQV SLTCLVKGFY PSDIAVEWES NGQPENNYKT TPPVLDSDGS FFLYSKLTVD 420 KSRWQQGNVF SCSVMHEALH NHYTQKSLSL SPGK 454 SEQ ID NO: 351 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYA ASSLQSGVPS 60 46E11 light RFSGSGSGAE FTLTISSLQP EDFATYYCLQ HNSYPWTFGQ GTKVEIKRTV AAPSVFIFPP 120 chain SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 352 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVAV IWYAGSNKYY 60 46E11 variable ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGD ILTGYSLYYG MDVWGQGTTV 120 heavy chain TVSS 124 SEQ ID NO: 353 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYA ASSLQSGVPS 60 46E11 variable RFSGSGSGAE FTLTISSLQP EDFATYYCLQ HNSYPWTFGQ GTKVEIKR 108 light chain SEQ ID NO: 354 SYGMH 5 46E11 heavy chain CDR1 SEQ ID NO: 355 VIWYAGSNKY YADSVKG 17 46E11 heavy chain CDR2 SEQ ID NO: 356 GDILTGYSLY YGMDV 15 46E11 heavy chain CDR3 SEQ ID NO: 357 RASQGIRNDL G 11 46E11 light chain CDR1 SEQ ID NO: 358 AASSLQS 7 46E11 light chain CDR2 SEQ ID NO: 359 LQHNSYPWT 9 46E11 light chain CDR3
[1357] In a preferred embodiment, the GITR agonist is the monoclonal antibody 48H12, or a fragment, derivative, variant, or biosimilar thereof. 48H12 is available from Amgen, Inc. The preparation and properties of 48H12 are described in U.S. Patent Application Publication No. US 2015/0064204 A1, the disclosures of which are incorporated by reference herein. The amino acid sequences of 48H12 are set forth in Table 34.
[1358] In an embodiment, a GITR agonist comprises a heavy chain given by SEQ ID NO:360 and a light chain given by SEQ ID NO:361. In an embodiment, a GITR agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:360 and SEQ ID NO:361, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:360 and SEQ ID NO:361, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:360 and SEQ ID NO:361, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:360 and SEQ ID NO:361, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:360 and SEQ ID NO:361, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:360 and SEQ ID NO:361, respectively.
[1359] In an embodiment, the GITR agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 48H12. In an embodiment, the GITR agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:362, and the GITR agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:363, and conservative amino acid substitutions thereof. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:362 and SEQ ID NO:363, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:362 and SEQ ID NO:363, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:362 and SEQ ID NO:363, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:362 and SEQ ID NO:363, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:362 and SEQ ID NO:363, respectively.
[1360] In an embodiment, a GITR agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:364, SEQ ID NO:365, and SEQ ID NO:366, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:367, SEQ ID NO:368, and SEQ ID NO:369, respectively, and conservative amino acid substitutions thereof.
[1361] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 48H12. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 48H12. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 48H12. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 48H12. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 48H12.
TABLE-US-00034 TABLE 34 Amino acid sequences for GITR agonist antibodies related to 48H12. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 360 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVAV IWYAGSNKYY 60 48H12 heavy ADSVKGRFTI SRDNSKNTVY LQMNSLRAED TAVYYCARGG QLALYYYYGM DVWGQGTTVT 120 chain VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV TVSWNSGALT SGVHTFPAVL 180 QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKK VEPKSCDKTH TCPPCPAPEL 240 LGGPSVFLFP PKPKDTLMIS RTPEVTCVVV DVSHEDPEVK FNWYVDGVEV HNAKTKPREE 300 QYNSTYRVVS VLTVLHQDWL NGKEYKCKVS NKALPAPIEK TISKAKGQPR EPQVYTLPPS 360 REEMTKNQVS LTCLVKGFYP SDIAVEWESN GQPENNYKTT PPVLDSDGSF FLYSKLTVDK 420 SRWQQGNVFS CSVMHEALHN HYTQKSLSLS PGK 453 SEQ ID NO: 361 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYA ASSLQSGVPS 60 48H12 light RFSGSGSGTE FTLTISSLQP EDFATYYCLQ HNNYPWTFGQ GTKVEIKRTV AAPSVFIFPP 120 chain SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 362 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVAV IWYAGSNKYY 60 48H12 variable ADSVKGRFTI SRDNSKNTVY LQMNSLRAED TAVYYCARGG QLALYYYYGM DVWGQGTTVT 120 heavy chain VSS 123 SEQ ID NO: 363 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYA ASSLQSGVPS 60 48H12 variable RFSGSGSGTE FTLTISSLQP EDFATYYCLQ HNNYPWTFGQ GTKVEIKR 108 light chain SEQ ID NO: 364 SYGMH 5 48H12 heavy chain CDR1 SEQ ID NO: 365 VIWYAGSNKY YADSVKG 17 48H12 heavy chain CDR2 SEQ ID NO: 366 GGQLALYYYY GMDV 14 48H12 heavy chain CDR3 SEQ ID NO: 367 RASQGIRNDL G 11 48H12 light chain CDR1 SEQ ID NO: 368 AASSLQS 7 48H12 light chain CDR2 SEQ ID NO: 369 LQHNNYPWT 9 48H12 light chain CDR3
[1362] In a preferred embodiment, the GITR agonist is the monoclonal antibody 48H7, or a fragment, derivative, variant, or biosimilar thereof. 48H7 is available from Amgen, Inc. The preparation and properties of 48H7 are described in U.S. Patent Application Publication No. US 2015/0064204 A1, the disclosures of which are incorporated by reference herein. The amino acid sequences of 48H7 are set forth in Table 35.
[1363] In an embodiment, a GITR agonist comprises a heavy chain given by SEQ ID NO:370 and a light chain given by SEQ ID NO:371. In an embodiment, a GITR agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:370 and SEQ ID NO:371, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:370 and SEQ ID NO:371, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:370 and SEQ ID NO:371, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:370 and SEQ ID NO:371, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:370 and SEQ ID NO:371, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:370 and SEQ ID NO:371, respectively.
[1364] In an embodiment, the GITR agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 48H7. In an embodiment, the GITR agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:372, and the GITR agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:373, and conservative amino acid substitutions thereof. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:372 and SEQ ID NO:373, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:372 and SEQ ID NO:373, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:372 and SEQ ID NO:373, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:372 and SEQ ID NO:373, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:372 and SEQ ID NO:373, respectively.
[1365] In an embodiment, a GITR agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:374, SEQ ID NO:375, and SEQ ID NO:376, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:377, SEQ ID NO:378, and SEQ ID NO:379, respectively, and conservative amino acid substitutions thereof.
[1366] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 48H7. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 48H7. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 48H7. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 48H7. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 48H7.
TABLE-US-00035 TABLE 35 Amino acid sequences for GITR agonist antibodies related to 48H7. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 370 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMYWVRQA PGKGLEWVAV IWYEGSNKYY 60 48H7 heavy chain ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYFCARGG ELGRDYYSGM DVWGQGTTVT 120 VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV TVSWNSGALT SGVHTFPAVL 180 QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKK VEPKSCDKTH TCPPCPAPEL 240 LGGPSVFLFP PKPKDTLMIS RTPEVTCVVV DVSHEDPEVK FNWYVDGVEV HNAKTKPREE 300 QYNSTYRVVS VLTVLHQDWL NGKEYKCKVS NKALPAPIEK TISKAKGQPR EPQVYTLPPS 360 REEMTKNQVS LTCLVKGFYP SDIAVEWESN GQPENNYKTT PPVLDSDGSF FLYSKLTVDK 420 SRWQQGNVFS CSVMHEALHN HYTQKSLSLS PGK 453 SEQ ID NO: 371 DIQMTQSPSS LSASVGDRVT ITCRASQVIR NDLGWYQQKP GKAPKRLIYA ASSLQSGVPS 60 48H7 light chain RFSGSGSGTE FTLTISSLQP EDFATYYCLQ HNSYPITFGG GTKVEIKRTV AAPSVFIFPP 120 SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 372 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMYWVRQA PGKGLEWVAV IWYEGSNKYY 60 48H7 variable ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYFCARGG ELGRDYYSGM DVWGQGTTVT 120 heavy chain VSS 123 SEQ ID NO: 373 DIQMTQSPSS LSASVGDRVT ITCRASQVIR NDLGWYQQKP GKAPKRLIYA ASSLQSGVPS 60 48H7 variable RFSGSGSGTE FTLTISSLQP EDFATYYCLQ HNSYPITFGG GTKVEIKR 108 light chain SEQ ID NO: 374 SYGMY 5 48H7 heavy chain CDR1 SEQ ID NO: 375 VIWYEGSNKY YADSVKG 17 48H7 heavy chain CDR2 SEQ ID NO: 376 GGELGRDYYS GMDV 14 48H7 heavy chain CDR3 SEQ ID NO: 377 RASQVIRNDL G 11 48H7 light chain CDR1 SEQ ID NO: 378 AASSLQS 7 48H7 light chain CDR2 SEQ ID NO: 379 LQHNSYPIT 9 48H7 light chain CDR3
[1367] In a preferred embodiment, the GITR agonist is the monoclonal antibody 49D9, or a fragment, derivative, variant, or biosimilar thereof 49D9 is available from Amgen, Inc. The preparation and properties of 49D9 are described in U.S. Patent Application Publication No. US 2015/0064204 A1, the disclosures of which are incorporated by reference herein. The amino acid sequences of 49D9 are set forth in Table 36.
[1368] In an embodiment, a GITR agonist comprises a heavy chain given by SEQ ID NO:380 and a light chain given by SEQ ID NO:381. In an embodiment, a GITR agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:380 and SEQ ID NO:381, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:380 and SEQ ID NO:381, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:380 and SEQ ID NO:381, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:380 and SEQ ID NO:381, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:380 and SEQ ID NO:381, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:380 and SEQ ID NO:381, respectively.
[1369] In an embodiment, the GITR agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 49D9. In an embodiment, the GITR agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:382, and the GITR agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:383, and conservative amino acid substitutions thereof. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:382 and SEQ ID NO:383, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:382 and SEQ ID NO:383, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:382 and SEQ ID NO:383, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:382 and SEQ ID NO:383, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:382 and SEQ ID NO:383, respectively.
[1370] In an embodiment, a GITR agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:384, SEQ ID NO:385, and SEQ ID NO:386, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:387, SEQ ID NO:388, and SEQ ID NO:389, respectively, and conservative amino acid substitutions thereof.
[1371] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 49D9. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 49D9. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 49D9. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 49D9. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 49D9.
TABLE-US-00036 TABLE 36 Amino acid sequences for GITR agonist antibodies related to 49D9. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 380 QMQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVAV IWYAGSNKYY 60 49D9 heavy chain ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGG RLGFYYYYGM DVWGQGTTVT 120 VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV TVSWNSGALT SGVHTFPAVL 180 QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKK VEPKSCDKTH TCPPCPAPEL 240 LGGPSVFLFP PKPKDTLMIS RTPEVTCVVV DVSHEDPEVK FNWYVDGVEV HNAKTKPREE 300 QYNSTYRVVS VLTVLHQDWL NGKEYKCKVS NKALPAPIEK TISKAKGQPR EPQVYTLPPS 360 REEMTKNQVS LTCLVKGFYP SDIAVEWESN GQPENNYKTT PPVLDSDGSF FLYSKLTVDK 420 SRWQQGNVFS CSVMHEALHN HYTQKSLSLS PGK 453 SEQ ID NO: 381 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYA ASSLQSGVPS 60 49D9 light chain RFSGSGSGTE FTLTISSLQP EDFATYYCLQ LNSYPWTFGQ GTKVEIKRTV AAPSVFIFPP 120 SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 382 QMQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVAV IWYAGSNKYY 60 49D9 variable ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGG RLGFYYYYGM DVWGQGTTVT 120 heavy chain VSS 123 SEQ ID NO: 383 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYA ASSLQSGVPS 60 49D9 variable RFSGSGSGTE FTLTISSLQP EDFATYYCLQ LNSYPWTFGQ GTKVEIKR 108 light chain SEQ ID NO: 384 SYGMH 5 49D9 heavy chain CDR1 SEQ ID NO: 385 VIWYAGSNKY YADSVKG 17 49D9 heavy chain CDR2 SEQ ID NO: 386 GGRLGFYYYY GMDV 14 49D9 heavy chain CDR3 SEQ ID NO: 387 RASQGIRNDL G 11 49D9 light chain CDR1 SEQ ID NO: 388 AASSLQS 7 49D9 light chain CDR2 SEQ ID NO: 389 LQLNSYPWT 9 49D9 light chain CDR3
[1372] In a preferred embodiment, the GITR agonist is the monoclonal antibody 49E2, or a fragment, derivative, variant, or biosimilar thereof 49E2 is available from Amgen, Inc. The preparation and properties of 49E2 are described in U.S. Patent Application Publication No. US 2015/0064204 A1, the disclosures of which are incorporated by reference herein. The amino acid sequences of 49E2 are set forth in Table 37.
[1373] In an embodiment, a GITR agonist comprises a heavy chain given by SEQ ID NO:390 and a light chain given by SEQ ID NO:391. In an embodiment, a GITR agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:390 and SEQ ID NO:391, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:390 and SEQ ID NO:391, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:390 and SEQ ID NO:391, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:390 and SEQ ID NO:391, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:390 and SEQ ID NO:391, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:390 and SEQ ID NO:391, respectively.
[1374] In an embodiment, the GITR agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 49E2. In an embodiment, the GITR agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:392, and the GITR agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:393, and conservative amino acid substitutions thereof. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:392 and SEQ ID NO:393, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:392 and SEQ ID NO:393, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:392 and SEQ ID NO:393, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:392 and SEQ ID NO:393, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:392 and SEQ ID NO:393, respectively.
[1375] In an embodiment, a GITR agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:394, SEQ ID NO:395, and SEQ ID NO:396, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:397, SEQ ID NO:398, and SEQ ID NO:399, respectively, and conservative amino acid substitutions thereof.
[1376] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 49E2. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 49E2. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 49E2. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 49E2. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 49E2.
TABLE-US-00037 TABLE 37 Amino acid sequences for GITR agonist antibodies related to 49E2. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 390 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVAV IWSDGNNKYY 60 49E2 heavy chain EDSVKGRFTI SRDSSKNTLF LQMNSLRAED TAVYYCARDT ATPFDYWGQG TLVTVSSAST 120 KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF PEPVTVSWNS GALTSGVHTF PAVLQSSGLY 180 SLSSVVTVPS SSLGTQTYIC NVNHKPSNTK VDKKVEPKSC DKTHTCPPCP APELLGGPSV 240 FLFPPKPKDT LMISRTPEVT CVVVDVSHED PEVKFNWYVD GVEVHNAKTK PREEQYNSTY 300 RVVSVLTVLH QDWLNGKEYK CKVSNKALPA PIEKTISKAK GQPREPQVYT LPPSREEMTK 360 NQVSLTCLVK GFYPSDIAVE WESNGQPENN YKTTPPVLDS DGSFFLYSKL TVDKSRWQQG 420 NVFSCSVMHE ALHNHYTQKS LSLSPGK 447 SEQ ID NO: 391 QSALTQPASV SGSPGQSITI SCTGTSSDVG IYNLVSWYQQ HPGKAPKLMI HEVSKRPSGV 60 49E2 light chain SNRFSGSKSG NTASLTISGL QAEDEADYYC CSYAGISTWV FGGGTKLTVL GQPKAAPSVT 120 LFPPSSEELQ ANKATLVCLI SDFYPGAVTV AWKADSSPVK AGVETTTPSK QSNNKYAASS 180 YLSLTPEQWK SHRSYSCQVT HEGSTVEKTV APTECS 216 SEQ ID NO: 392 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWVAV IWSDGNNKYY 60 49E2 variable EDSVKGRFTI SRDSSKNTLF LQMNSLRAED TAVYYCARDT ATPFDYWGQG TLVTVSS 117 heavy chain SEQ ID NO: 393 QSALTQPASV SGSPGQSITI SCTGTSSDVG IYNLVSWYQQ HPGKAPKLMI HEVSKRPSGV 60 49E2 variable SNRFSGSKSG NTASLTISGL QAEDEADYYC CSYAGISTWV FGGGTKLTVL G 111 light chain SEQ ID NO: 394 SYGMH 5 49E2 heavy chain CDR1 SEQ ID NO: 395 VIWSDGNNKY YEDSVKG 17 49E2 heavy chain CDR2 SEQ ID NO: 396 DTATPFDY 8 49E2 heavy chain CDR3 SEQ ID NO: 397 TGTSSDVGIY NLVS 14 49E2 light chain CDR1 SEQ ID NO: 398 EVSKRPS 7 49E2 light chain CDR2 SEQ ID NO: 399 CSYAGISTWV 10 49E2 light chain CDR3
[1377] In a preferred embodiment, the GITR agonist is the monoclonal antibody 48A9, or a fragment, derivative, variant, or biosimilar thereof. 48A9 is available from Amgen, Inc. The preparation and properties of 48A9 are described in U.S. Patent Application Publication No. US 2015/0064204 A1, the disclosures of which are incorporated by reference herein. The amino acid sequences of 48A9 are set forth in Table 38.
[1378] In an embodiment, a GITR agonist comprises a heavy chain given by SEQ ID NO:400 and a light chain given by SEQ ID NO:401. In an embodiment, a GITR agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:400 and SEQ ID NO:401, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:400 and SEQ ID NO:401, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:400 and SEQ ID NO:401, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:400 and SEQ ID NO:401, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:400 and SEQ ID NO:401, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:400 and SEQ ID NO:401, respectively.
[1379] In an embodiment, the GITR agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 48A9. In an embodiment, the GITR agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:402, and the GITR agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:403, and conservative amino acid substitutions thereof. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:402 and SEQ ID NO:403, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:402 and SEQ ID NO:403, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:402 and SEQ ID NO:403, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:402 and SEQ ID NO:403, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:402 and SEQ ID NO:403, respectively.
[1380] In an embodiment, a GITR agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:404, SEQ ID NO:405, and SEQ ID NO:406, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:407, SEQ ID NO:408, and SEQ ID NO:409, respectively, and conservative amino acid substitutions thereof.
[1381] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 48A9. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 48A9. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 48A9. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 48A9. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 48A9.
TABLE-US-00038 TABLE 38 Amino acid sequences for GITR agonist antibodies related to 48A9. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 400 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SCGMHWVRQA PGKGLEWVAV ISYDGSNKYY 60 48A9 heavy chain ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARDL RYNWNDGGVD YWGQGTLVTV 120 SSASTKGPSV FPLAPSSKST SGGTAALGCL VKDYFPEPVT VSWNSGALTS GVHTFPAVLQ 180 SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNTKVDKKV EPKSCDKTHT CPPCPAPELL 240 GGPSVFLFPP KPKDTLMISR TPEVTCVVVD VSHEDPEVKF NWYVDGVEVH NAKTKPREEQ 300 YNSTYRVVSV LTVLHQDWLN GKEYKCKVSN KALPAPIEKT ISKAKGQPRE PQVYTLPPSR 360 EEMTKNQVSL TCLVKGFYPS DIAVEWESNG QPENNYKTTP PVLDSDGSFF LYSKLTVDKS 420 RWQQGNVFSC SVMHEALHNH YTQKSLSLSP GK 452 SEQ ID NO: 401 DIQMTQSPSS LSASVGDRVI ITCRASQSIS SYLHWYKQKP GKAPKLLIYG ASRLQSGVPS 60 48A9 light chain RFSGSGSGTD FTLTISSLQP EDFATYYCQQ SSSTPLTFGG GTKVEIKRTV AAPSVFIFPP 120 SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 402 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SCGMHWVRQA PGKGLEWVAV ISYDGSNKYY 60 48A9 variable ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARDL RYNWNDGGVD YWGQGTLVTV 120 heavy chain SS 122 SEQ ID NO: 403 DIQMTQSPSS LSASVGDRVI ITCRASQSIS SYLHWYKQKP GKAPKLLIYG ASRLQSGVPS 60 48A9 variable RFSGSGSGTD FTLTISSLQP EDFATYYCQQ SSSTPLTFGG GTKVEIKR 108 light chain SEQ ID NO: 404 SCGMH 5 48A9 heavy chain CDR1 SEQ ID NO: 405 VISYDGSNKY YADSVKG 17 48A9 heavy chain CDR2 SEQ ID NO: 406 DLRYNWNDGG VDY 13 48A9 heavy chain CDR3 SEQ ID NO: 407 RASQSISSYL H 11 48A9 light chain CDR1 SEQ ID NO: 408 GASRLQS 7 48A9 light chain CDR2 SEQ ID NO: 409 QQSSSTPLT 9 48A9 light chain CDR3
[1382] In a preferred embodiment, the GITR agonist is the monoclonal antibody 5H7, or a fragment, derivative, variant, or biosimilar thereof. 5H7 is available from Amgen, Inc. The preparation and properties of 5H7 are described in U.S. Patent Application Publication No. US 2015/0064204 A1, the disclosures of which are incorporated by reference herein. The amino acid sequences of 5H7 are set forth in Table 39.
[1383] In an embodiment, a GITR agonist comprises a heavy chain given by SEQ ID NO:410 and a light chain given by SEQ ID NO:411. In an embodiment, a GITR agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:410 and SEQ ID NO:411, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:410 and SEQ ID NO:411, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:410 and SEQ ID NO:411, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:410 and SEQ ID NO:411, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:410 and SEQ ID NO:411, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:410 and SEQ ID NO:411, respectively.
[1384] In an embodiment, the GITR agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 5H7. In an embodiment, the GITR agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:412, and the GITR agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:413, and conservative amino acid substitutions thereof. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:412 and SEQ ID NO:413, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:412 and SEQ ID NO:413, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:412 and SEQ ID NO:413, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:412 and SEQ ID NO:413, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:412 and SEQ ID NO:413, respectively.
[1385] In an embodiment, a GITR agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:414, SEQ ID NO:415, and SEQ ID NO:416, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:417, SEQ ID NO:418, and SEQ ID NO:419, respectively, and conservative amino acid substitutions thereof.
[1386] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 5H7. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 5H7. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 5H7. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 5H7. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 5H7.
TABLE-US-00039 TABLE 39 Amino acid sequences for GITR agonist antibodies related to 5H7. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 410 QVQLQESGPG LVKPSQTLSL TCTVSGGSIS SGGYFWSWIR QHPGKGLEWI GYIYYSGTTY 60 5H7 heavy chain YNPSLKSRVT ISIDTSKNHF SLKLSSVTAA DTAVYYCARD LFYYDSSGPR GFDPWGQGTL 120 VTVSSASTKG PSVFPLAPSS KSTSGGTAAL GCLVKDYFPE PVTVSWNSGA LTSGVHTFPA 180 VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KRVEPKSCDK THTCPPCPAP 240 ELLGGPSVFL FPPKPKDTLM ISRTPEVTCV VVDVSHEDPE VKFNWYVDGV EVHNAKTKPR 300 EEQYNSTYRV VSVLTVLHQD WLNGKEYKCK VSNKALPAPI EKTISKAKGQ PREPQVYTLP 360 PSREEMTKNQ VSLTCLVKGF YPSDIAVEWE SNGQPENNYK TTPPVLDSDG SFFLYSKLTV 420 DKSRWQQGNV FSCSVMHEAL HNHYTQKSLS LSPGK 455 SEQ ID NO: 411 EIVLTQSPGT LSLSPGERAT LSCRASQTVS SNYLAWYQQK PGQAPRLLIY GSSTRATGIP 60 5H7 light chain DRFSGSGSGT DFTLTISRLE PEDFAVYYCQ QYDSSPWTFG QGTKVEIKRT VAAPSVFIFP 120 PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS QESVTEQDSK DSTYSLSSTL 180 TLSKADYEKH KVYACEVTHQ GLSSPVTKSF NRGEC 215 SEQ ID NO: 412 QVQLQESGPG LVKPSQTLSL TCTVSGGSIS SGGYFWSWIR QHPGKGLEWI GYIYYSGTTY 60 5H7 variable YNPSLKSRVT ISIDTSKNHF SLKLSSVTAA DTAVYYCARD LFYYDSSGPR GFDPWGQGTL 120 heavy chain VTVSS 125 SEQ ID NO: 413 EIVLTQSPGT LSLSPGERAT LSCRASQTVS SNYLAWYQQK PGQAPRLLIY GSSTRATGIP 60 5H7 variable DRFSGSGSGT DFTLTISRLE PEDFAVYYCQ QYDSSPWTFG QGTKVEIKR 109 light chain SEQ ID NO: 414 SGGYFWS 7 5H7 heavy chain CDR1 SEQ ID NO: 415 YIYYSGTTYY NPSLKS 16 5H7 heavy chain CDR2 SEQ ID NO: 416 DLFYYDSSGP RGFDP 15 5H7 heavy chain CDR3 SEQ ID NO: 417 RASQTVSSNY LA 12 5H7 light chain CDR1 SEQ ID NO: 418 GSSTRAT 7 5H7 light chain CDR2 SEQ ID NO: 419 QQYDSSPWT 9 5H7 light chain CDR3
[1387] In a preferred embodiment, the GITR agonist is the monoclonal antibody 7A10, or a fragment, derivative, variant, or biosimilar thereof 7A10 is available from Amgen, Inc. The preparation and properties of 7A10 are described in U.S. Patent Application Publication No. US 2015/0064204 A1, the disclosures of which are incorporated by reference herein. The amino acid sequences of 7A10 are set forth in Table 40.
[1388] In an embodiment, a GITR agonist comprises a heavy chain given by SEQ ID NO:420 and a light chain given by SEQ ID NO:421. In an embodiment, a GITR agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:420 and SEQ ID NO:421, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:420 and SEQ ID NO:421, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:420 and SEQ ID NO:421, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:420 and SEQ ID NO:421, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:420 and SEQ ID NO:421, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:420 and SEQ ID NO:421, respectively.
[1389] In an embodiment, the GITR agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 7A10. In an embodiment, the GITR agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:422, and the GITR agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:423, and conservative amino acid substitutions thereof. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:422 and SEQ ID NO:423, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:422 and SEQ ID NO:423, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:422 and SEQ ID NO:423, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:422 and SEQ ID NO:423, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:422 and SEQ ID NO:423, respectively.
[1390] In an embodiment, a GITR agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:424, SEQ ID NO:425, and SEQ ID NO:426, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:427, SEQ ID NO:428, and SEQ ID NO:429, respectively, and conservative amino acid substitutions thereof.
[1391] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 7A10. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 7A10. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 7A10. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 7A10. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 7A10.
TABLE-US-00040 TABLE 40 Amino acid sequences for GITR agonist antibodies related to 7A10. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 420 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWMAV IWYVGSNKYY 60 7A10 heavy chain ADSVKGRFTI SRDNSKNTLY LQMNSLSAED TAVYYCARGG ELGRDYYSGM DVWGQGTTVT 120 VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV TVSWNSGALT SGVHTFPAVL 180 QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKR VEPKSCDKTH TCPPCPAPEL 240 LGGPSVFLFP PKPKDTLMIS RTPEVTCVVV DVSHEDPEVK FNWYVDGVEV HNAKTKPREE 300 QYNSTYRVVS VLTVLHQDWL NGKEYKCKVS NKALPAPIEK TISKAKGQPR EPQVYTLPPS 360 REEMTKNQVS LTCLVKGFYP SDIAVEWESN GQPENNYKTT PPVLDSDGSF FLYSKLTVDK 420 SRWQQGNVFS CSVMHEALHN HYTQKSLSLS PGK 453 SEQ ID NO: 421 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYA ASSLQSGVPS 60 7A10 light chain RFSGSGSGTE FTLTISSLQP EDFATYYCQQ HNSYPWTFGQ GTKVEIKRTV AAPSVFIFPP 120 SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 422 QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYGMHWVRQA PGKGLEWMAV IWYVGSNKYY 60 7A10 variable ADSVKGRFTI SRDNSKNTLY LQMNSLSAED TAVYYCARGG ELGRDYYSGM DVWGQGTTVT 120 heavy chain VSS 123 SEQ ID NO: 423 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPKRLIYA ASSLQSGVPS 60 7A10 variable RFSGSGSGTE FTLTISSLQP EDFATYYCQQ HNSYPWTFGQ GTKVEIKR 108 light chain SEQ ID NO: 424 SYGMH 5 7A10 heavy chain CDR1 SEQ ID NO: 425 VIWYVGSNKY YADSVKG 17 7A10 heavy chain CDR2 SEQ ID NO: 426 GGELGRDYYS GMDV 14 7A10 heavy chain CDR3 SEQ ID NO: 427 RASQGIRNDL G 11 7A10 light chain CDR1 SEQ ID NO: 428 AASSLQS 7 7A10 light chain CDR2 SEQ ID NO: 429 QQHNSYPWT 9 7A10 light chain CDR3
[1392] In a preferred embodiment, the GITR agonist is the monoclonal antibody 9H6, or a fragment, derivative, variant, or biosimilar thereof. 9H6 is available from Amgen, Inc. The preparation and properties of 9H6 are described in U.S. Patent Application Publication No. US 2015/0064204 A1, the disclosures of which are incorporated by reference herein. The amino acid sequences of 9H6 are set forth in Table 41.
[1393] In an embodiment, a GITR agonist comprises a heavy chain given by SEQ ID NO:430 and a light chain given by SEQ ID NO:431. In an embodiment, a GITR agonist comprises heavy and light chains having the sequences shown in SEQ ID NO:430 and SEQ ID NO:431, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:430 and SEQ ID NO:431, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:430 and SEQ ID NO:431, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:430 and SEQ ID NO:431, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:430 and SEQ ID NO:431, respectively. In an embodiment, a GITR agonist comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:430 and SEQ ID NO:431, respectively.
[1394] In an embodiment, the GITR agonist comprises the heavy and light chain CDRs or variable regions (VRs) of 9H6. In an embodiment, the GITR agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:432, and the GITR agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:433, and conservative amino acid substitutions thereof. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:432 and SEQ ID NO:433, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:432 and SEQ ID NO:433, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:432 and SEQ ID NO:433, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:432 and SEQ ID NO:433, respectively. In an embodiment, a GITR agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:432 and SEQ ID NO:433, respectively.
[1395] In an embodiment, a GITR agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:434, SEQ ID NO:435, and SEQ ID NO:436, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:437, SEQ ID NO:438, and SEQ ID NO:439, respectively, and conservative amino acid substitutions thereof.
[1396] In an embodiment, the GITR agonist is a GITR agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to 9H6. In an embodiment, the biosimilar monoclonal antibody comprises an GITR antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 9H6. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a GITR agonist antibody authorized or submitted for authorization, wherein the GITR agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 9H6. The GITR agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 9H6. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is 9H6.
TABLE-US-00041 TABLE 41 Amino acid sequences for GITR agonist antibodies related to 9H6. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 430 QVQLVESGGG VVQPGRSLRL SCVASGFTFS SYGMHWIRQA PGKGLEWVAV IWYEGSNKYY 60 9H6 heavy chain ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGG RLGKDYYSGM DVWGQGTTVT 120 VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV TVSWNSGALT SGVHTFPAVL 180 QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKR VEPKSCDKTH TCPPCPAPEL 240 LGGPSVFLFP PKPKDTLMIS RTPEVTCVVV DVSHEDPEVK FNWYVDGVEV HNAKTKPREE 300 QYNSTYRVVS VLTVLHQDWL NGKEYKCKVS NKALPAPIEK TISKAKGQPR EPQVYTLPPS 360 REEMTKNQVS LTCLVKGFYP SDIAVEWESN GQPENNYKTT PPVLDSDGSF FLYSKLTVDK 420 SRWQQGNVFS CSVMHEALHN HYTQKSLSLS PGK 453 SEQ ID NO: 431 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPNRLIYA TSSLQSGVPS 60 9H6 light chain RFSGSGSGTE FTLTISSLQP EDFATYYCLQ HNTYPWTFGQ GTKVEIKRTV AAPSVFIFPP 120 SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 432 QVQLVESGGG VVQPGRSLRL SCVASGFTFS SYGMHWIRQA PGKGLEWVAV IWYEGSNKYY 60 9H6 variable ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGG RLGKDYYSGM DVWGQGTTVT 120 heavy chain VSS 123 SEQ ID NO: 433 DIQMTQSPSS LSASVGDRVT ITCRASQGIR NDLGWYQQKP GKAPNRLIYA TSSLQSGVPS 60 9H6 variable RFSGSGSGTE FTLTISSLQP EDFATYYCLQ HNTYPWTFGQ GTKVEIKR 108 light chain SEQ ID NO: 434 SYGMH 5 9H6 heavy chain CDR1 SEQ ID NO: 435 VIWYEGSNKY YADSVKG 17 9H6 heavy chain CDR2 SEQ ID NO: 436 GGRLGKDYYS GMDV 14 9H6 heavy chain CDR3 SEQ ID NO: 437 RASQGIRNDL G 11 9H6 light chain CDR1 SEQ ID NO: 438 ATSSLQS 7 9H6 light chain CDR2 SEQ ID NO: 439 LQHNTYPWT 9 9H6 light chain CDR3
[1397] In an embodiment, the GITR agonist is a GITR agonist described in International Patent Application Publication Nos. WO 2013/039954 A1 and WO 2011/028683 A1; U.S. Patent Application Publication Nos. US 2013/0108641 A1, US 2012/0189639 A1, and US 2014/0348841 A1; and U.S. Pat. Nos. 7,812,135; 8,388,967; and 9,028,823, the disclosures of which are incorporated by reference herein. In an embodiment, the GITR agonist is an agonistic, anti-GITR monoclonal antibody with a structure and preparation described in US Patent Application Publication No. US 2015/0064204 and International Patent Application Publication No. WO 2015/031667 A1 (Amgen, Inc.), the disclosures of which are incorporated by reference herein. In an embodiment, the GITR agonist is a fully-human, agonistic, anti-GITR monoclonal antibody selected from the group consisting of 1D7, 33C9, 33F6, 34G4, 35B10, 41E11, 41G5, 42A11, 44C1, 45A8, 46E11, 48H12, 48H7, 49D9, 49E2, 48A9, 5H7, 7A10, and 9H6. In an embodiment, the GITR agonist is a fully-human, agonistic, anti-GITR monoclonal antibody with an amino acid sequence identity of greater than 99% to the sequence of an antibody selected from the group consisting of 1D7, 33C9, 33F6, 34G4, 35B10, 41E11, 41G5, 42A11, 44C1, 45A8, 46E11, 48H12, 48H7, 49D9, 49E2, 48A9, 5H7, 7A10, and 9H6. In an embodiment, the GITR agonist is a fully-human, agonistic, anti-GITR monoclonal antibody with an amino acid sequence identity of greater than 98% to the sequence of an antibody selected from the group consisting of 1D7, 33C9, 33F6, 34G4, 35B10, 41E11, 41G5, 42A11, 44C1, 45A8, 46E11, 48H12, 48H7, 49D9, 49E2, 48A9, 5H7, 7A10, and 9H6. In an embodiment, the GITR agonist is a fully-human, agonistic, anti-GITR monoclonal antibody selected from the group consisting of 9H6v3, 5H7v2, 33C9v2, 41G5v2, and 7A10v1, as described in US Patent Application Publication No. US 2015/0064204 A1, the disclosure of which is incorporated by reference herein. In an embodiment, the GITR agonist is a fully-human, agonistic, anti-GITR monoclonal antibody selected from the group consisting of 44C1v1, 45A8v1, 49D9v1, 49E2v1, 48A9v1, 5H7v1, 5H7v2, 5H7v3, 5H7v5, 5H7v7, 5H7v9, 5H7v10, 5H7v11, 5H7v13, 5H7v14, 5H7v17, 5H7v18, 5H7v19, 5H7v22, 7A10v1, 7A10v2, 7A10v3, 7A10v4, 7A10v5, 9H6v1, 9H6v2, 9H6v3, 9H6v4, 9H6v5, 9H6v6, 33C9v1, 33C9v2, 33C9v3, 33C9v4, 33C9v5, 41G5v1, 41G5v2, 41G5v3, 41G5v4, and 41G5v5, as described in US Patent Application Publication No. US 2015/0064204 A1, the disclosure of which is incorporated by reference herein.
[1398] In an embodiment, the GITR agonist is an GITR agonistic fusion protein as depicted in Structure I-A (C-terminal Fc-antibody fragment fusion protein) or Structure I-B (N-terminal Fc-antibody fragment fusion protein), or a fragment, derivative, conjugate, variant, or biosimilar thereof. The properties of structures I-A and I-B are described above and in U.S. Pat. Nos. 9,359,420, 9,340,599, 8,921,519, and 8,450,460, the disclosures of which are incorporated by reference herein. Amino acid sequences for the polypeptide domains of structure I-A are given in Table 6. The Fc domain preferably comprises a complete constant domain (amino acids 17-230 of SEQ ID NO:31) the complete hinge domain (amino acids 1-16 of SEQ ID NO:31) or a portion of the hinge domain (e.g., amino acids 4-16 of SEQ ID NO:31). Preferred linkers for connecting a C-terminal Fc-antibody may be selected from the embodiments given in SEQ ID NO:32 to SEQ ID NO:41, including linkers suitable for fusion of additional polypeptides. Likewise, amino acid sequences for the polypeptide domains of structure I-B are given in Table 7. If an Fc antibody fragment is fused to the N-terminus of an TNRFSF fusion protein as in structure I-B, the sequence of the Fc module is preferably that shown in SEQ ID NO:42, and the linker sequences are preferably selected from those embodiments set forth in SEQ ID NO:43 to SEQ ID NO:45.
[1399] In an embodiment, an GITR agonist fusion protein according to structures I-A or I-B comprises one or more GITR binding domains selected from the group consisting of a variable heavy chain and variable light chain of TRX518, 6C8, 36E5, 3D6, 61G6, 6H6, 61F6, 1D8, 17F10, 35D8, 49A1, 9E5, 31H6, 2155, 698, 706, 827, 1649, 1718, 1D7, 33C9, 33F6, 34G4, 35B10, 41E11, 41G5, 42A11, 44C1, 45A8, 46E11, 48H12, 48H7, 49D9, 49E2, 48A9, 5H7, 7A10, 9H6, and fragments, derivatives, conjugates, variants, and biosimilars thereof.
[1400] In an embodiment, a GITR agonist fusion protein according to structures I-A or I-B comprises one or more GITR binding domains comprising an GITRL sequence (Table 42). In an embodiment, an GITR agonist fusion protein according to structures I-A or I-B comprises one or more GITR binding domains comprising a sequence according to SEQ ID NO:440. In an embodiment, an GITR agonist fusion protein according to structures I-A or I-B comprises one or more GITR binding domains comprising a soluble GITRL sequence. In an embodiment, a GITR agonist fusion protein according to structures I-A or I-B comprises one or more GITR binding domains comprising a sequence according to SEQ ID NO:441.
[1401] In an embodiment, an GITR agonist fusion protein according to structures I-A or I-B comprises one or more GITR binding domains that is a scFv domain comprising V.sub.H and V.sub.L regions that are each at least 95% identical to the V.sub.H and V.sub.L GITR sequences shown above in Tables 18 to 39, wherein the V.sub.H and V.sub.L domains are connected by a linker.
TABLE-US-00042 TABLE 42 Additional polypeptide domains useful as GITR binding domains in fusion proteins (e.g., structures I-A and I-B). Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 440 MCLSHLENMP LSHSRTQGAQ RSSWKLWLFC SIVMLLFLCS FSWLIFIFLQ LETAKEPCMA 60 GITRL KFGPLPSKWQ MASSEPPCVN KVSDWKLEIL QNGLYLIYGQ VAPNANYNDV APFEVRLYKN 120 KDMIQTLTNK SKIQNVGGTY ELHVGDTIDL IFNSEHQVLK NNTYWGIILL ANPQFIS 177 SEQ ID NO: 441 TAKEPCMAKF GPLPSKWQMA SSEPPCVNKV SDWKLEILQN GLYLIYGQVA PNANYNDVAP 60 GITRL soluble FEVRLYKNKD MIQTLTNKSK IQNVGGTYEL HVGDTIDLIF NSEHQVLKNN TYWGIILLAN 120 domain PQFIS 125
[1402] In an embodiment, the GITR agonist is a GITR agonistic single-chain fusion polypeptide comprising (i) a first soluble GITR binding domain, (ii) a first peptide linker, (iii) a second soluble GITR binding domain, (iv) a second peptide linker, and (v) a third soluble GITR binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, and wherein the additional domain is a Fab or Fc fragment domain. In an embodiment, the GITR agonist is a GITR agonistic single-chain fusion polypeptide comprising (i) a first soluble GITR binding domain, (ii) a first peptide linker, (iii) a second soluble GITR binding domain, (iv) a second peptide linker, and (v) a third soluble GITR binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, wherein the additional domain is a Fab or Fc fragment domain wherein each of the soluble GITR binding domains lacks a stalk region (which contributes to trimerisation and provides a certain distance to the cell membrane, but is not part of the GITR binding domain) and the first and the second peptide linkers independently have a length of 3-8 amino acids.
[1403] In an embodiment, the GITR agonist is an GITR agonistic single-chain fusion polypeptide comprising (i) a first soluble tumor necrosis factor (TNF) superfamily cytokine domain, (ii) a first peptide linker, (iii) a second soluble TNF superfamily cytokine domain, (iv) a second peptide linker, and (v) a third soluble TNF superfamily cytokine domain, wherein each of the soluble TNF superfamily cytokine domains lacks a stalk region and the first and the second peptide linkers independently have a length of 3-8 amino acids, and wherein the TNF superfamily cytokine domain is an GITR binding domain.
[1404] In an embodiment, the GITR agonist is a GITR agonistic scFv antibody comprising any of the foregoing V.sub.H domains linked to any of the foregoing V.sub.L domains.
HVEM (CD270) Agonists
[1405] In an embodiment, the TNFRSF agonist is a HVEM agonist. HVEM is also known as CD270 and TNFRSF 14. Any HVEM agonist known in the art may be used. The HVEM binding molecule may be a monoclonal antibody or fusion protein capable of binding to human or mammalian HVEM. The HVEM agonists or HVEM binding molecules may comprise an immunoglobulin heavy chain of any isotype (e.g., IgG, IgE, IgM, IgD, IgA, and IgY), class (e.g., IgG1, IgG2, IgG3, IgG4, IgA1 and IgA2) or subclass of immunoglobulin molecule. The HVEM agonist or HVEM binding molecule may have both a heavy and a light chain. As used herein, the term binding molecule also includes antibodies (including full length antibodies), monoclonal antibodies (including full length monoclonal antibodies), polyclonal antibodies, multispecific antibodies (e.g., bispecific antibodies), human, humanized or chimeric antibodies, and antibody fragments, e.g., Fab fragments, F(ab') fragments, fragments produced by a Fab expression library, epitope-binding fragments of any of the above, and engineered forms of antibodies, e.g., scFv molecules, that bind to HVEM. In an embodiment, the HVEM agonist is an antigen binding protein that is a fully human antibody. In an embodiment, the HVEM agonist is an antigen binding protein that is a humanized antibody. In some embodiments, HVEM agonists for use in the presently disclosed methods and compositions include anti-HVEM antibodies, human anti-HVEM antibodies, mouse anti-HVEM antibodies, mammalian anti-HVEM antibodies, monoclonal anti-HVEM antibodies, polyclonal anti-HVEM antibodies, chimeric anti-HVEM antibodies, anti-HVEM adnectins, anti-HVEM domain antibodies, single chain anti-HVEM fragments, heavy chain anti-HVEM fragments, light chain anti-HVEM fragments, anti-HVEM fusion proteins, and fragments, derivatives, conjugates, variants, or biosimilars thereof. In a preferred embodiment, the HVEM agonist is an agonistic, anti-HVEM humanized or fully human monoclonal antibody (i.e., an antibody derived from a single cell line).
[1406] In a preferred embodiment, the HVEM agonist or HVEM binding molecule may also be a fusion protein. In a preferred embodiment, a multimeric HVEM agonist, such as a trimeric or hexameric HVEM agonist (with three or six ligand binding domains), may induce superior receptor (HVEML) clustering and internal cellular signaling complex formation compared to an agonistic monoclonal antibody, which typically possesses two ligand binding domains. Trimeric (trivalent) or hexameric (or hexavalent) or greater fusion proteins comprising three TNFRSF binding domains and IgG1-Fc and optionally further linking two or more of these fusion proteins are described, e.g., in Gieffers, et al., Mol. Cancer Therapeutics 2013, 12, 2735-47.
[1407] Agonistic HVEM antibodies and fusion proteins are known to induce strong immune responses. In a preferred embodiment, the HVEM agonist is a monoclonal antibody or fusion protein that binds specifically to HVEM antigen in a manner sufficient to reduce toxicity. In some embodiments, the HVEM agonist is an agonistic HVEM monoclonal antibody or fusion protein that abrogates antibody-dependent cellular toxicity (ADCC), for example NK cell cytotoxicity. In some embodiments, the HVEM agonist is an agonistic HVEM monoclonal antibody or fusion protein that abrogates antibody-dependent cell phagocytosis (ADCP). In some embodiments, the HVEM agonist is an agonistic HVEM monoclonal antibody or fusion protein that abrogates complement-dependent cytotoxicity (CDC). In some embodiments, the HVEM agonist is an agonistic HVEM monoclonal antibody or fusion protein which abrogates Fc region functionality.
[1408] In some embodiments, the HVEM agonists are characterized by binding to human HVEM (SEQ ID NO:442) with high affinity and agonistic activity. In an embodiment, the HVEM agonist is a binding molecule that binds to human HVEM (SEQ ID NO:442). The amino acid sequence of HVEM antigen to which a HVEM agonist or binding molecule may bind is summarized in Table 43.
TABLE-US-00043 TABLE 43 Amino acid sequence of HVEM (CD270) antigen. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 442 MEPPGDWGPP PWRSTPRTDV LRLVLYLTFL GAPCYAPALP SCKEDEYPVG SECCPKCSPG 60 human CD270, YRVKEACGEL TGTVCEPCPP GTYIAHLNGL SKCLQCQMCD PAMGLRASRN CSRTENAVCG 120 Tumor necrosis CSPGHFCIVQ DGDHCAACRA YATSSPGQRV QKGGTESQDT LCQNCPPGTF SPNGTLEECQ 180 factor receptor HQTKCSWLVT KAGAGTSSSH WVWWFLSGSL VIVIVCSTVG LIICVKRRKP RGDVVKVIVS 240 superfamily, VQRKRQEAEG EATVIEALQA PPDVTTVAVE ETIPSFTGRS PNH 283 member 14 (Homo sapiens)
[1409] In some embodiments, the compositions, processes and methods described include a HVEM agonist that binds human or murine HVEM with a K.sub.D of about 100 pM or lower, binds human or murine HVEM with a K.sub.D of about 90 pM or lower, binds human or murine HVEM with a K.sub.D of about 80 pM or lower, binds human or murine HVEM with a K.sub.D of about 70 pM or lower, binds human or murine HVEM with a K.sub.D of about 60 pM or lower, binds human or murine HVEM with a K.sub.D of about 50 pM or lower, binds human or murine HVEM with a K.sub.D of about 40 pM or lower, or binds human or murine HVEM with a K.sub.D of about 30 pM or lower.
[1410] In some embodiments, the compositions, processes and methods described include a HVEM agonist that binds to human or murine HVEM with a k.sub.assoc of about 7.5.times.10.sup.5 l/Ms or faster, binds to human or murine HVEM with a k.sub.assoc of about 7.5.times.10.sup.5 l/Ms or faster, binds to human or murine HVEM with a k.sub.assoc of about 8.times.10.sup.5 l/Ms or faster, binds to human or murine HVEM with a k.sub.assoc of about 8.5.times.10.sup.5 l/Ms or faster, binds to human or murine HVEM with a k.sub.assoc of about 9.times.10.sup.5 l/Ms or faster, binds to human or murine HVEM with a k.sub.assoc of about 9.5.times.10.sup.5 l/Ms or faster, or binds to human or murine HVEM with a k.sub.assoc of about 1.times.10.sup.6 l/Ms or faster.
[1411] In some embodiments, the compositions, processes and methods described include a HVEM agonist that binds to human or murine HVEM with a k.sub.dissoc of about 2.times.10.sup.-5 l/s or slower, binds to human or murine HVEM with a k.sub.dissoc of about 2.1.times.10.sup.-5 l/s or slower, binds to human or murine HVEM with a k.sub.dissoc of about 2.2.times.10.sup.-5 l/s or slower, binds to human or murine HVEM with a k.sub.dissoc of about 2.3.times.10.sup.-5 l/s or slower, binds to human or murine HVEM with a k.sub.dissoc of about 2.4.times.10.sup.-5 l/s or slower, binds to human or murine HVEM with a k.sub.dissoc of about 2.5.times.10.sup.-5 l/s or slower, binds to human or murine HVEM with a k.sub.dissoc of about 2.6.times.10.sup.-5 l/s or slower or binds to human or murine HVEM with a k.sub.dissoc of about 2.7.times.10.sup.-5 l/s or slower, binds to human or murine HVEM with a k.sub.dissoc of about 2.8.times.10.sup.-5 l/s or slower, binds to human or murine HVEM with a k.sub.dissoc of about 2.9.times.10.sup.-5 l/s or slower, or binds to human or murine HVEM with a k.sub.dissoc of about 3.times.10.sup.-5 l/s or slower.
[1412] In some embodiments, the compositions, processes and methods described include a HVEM agonist that binds to human or murine HVEM with an IC.sub.50 of about 10 nM or lower, binds to human or murine HVEM with an IC.sub.50 of about 9 nM or lower, binds to human or murine HVEM with an IC.sub.50 of about 8 nM or lower, binds to human or murine HVEM with an IC.sub.50 of about 7 nM or lower, binds to human or murine HVEM with an IC.sub.50 of about 6 nM or lower, binds to human or murine HVEM with an IC.sub.50 of about 5 nM or lower, binds to human or murine HVEM with an IC.sub.50 of about 4 nM or lower, binds to human or murine HVEM with an IC.sub.50 of about 3 nM or lower, binds to human or murine HVEM with an IC.sub.50 of about 2 nM or lower, or binds to human or murine HVEM with an IC.sub.50 of about 1 nM or lower.
[1413] In an embodiment, the HVEM agonist is an HVEM agonist described in International Patent Application Publication No. WO 2009/007120 A2 and U.S. Patent Application Publication No. US 2016/0176941 A1, the disclosure of each of which is incorporated by reference herein.
[1414] In an embodiment, the HVEM agonist is the HVEM agonist clone REA247, which is commercially available from Miltenyi Biotech, Inc. (San Diego, Calif. 92121).
[1415] In an embodiment, the HVEM agonist is an HVEM agonistic fusion protein as depicted in Structure I-A (C-terminal Fc-antibody fragment fusion protein) or Structure I-B (N-terminal Fc-antibody fragment fusion protein), or a fragment, derivative, conjugate, variant, or biosimilar thereof. The properties of structures I-A and I-B are described above and in U.S. Pat. Nos. 9,359,420, 9,340,599, 8,921,519, and 8,450,460, the disclosures of which are incorporated by reference herein. Amino acid sequences for the polypeptide domains of structure I-A are given in Table 6. The Fc domain preferably comprises a complete constant domain (amino acids 17-230 of SEQ ID NO:31) the complete hinge domain (amino acids 1-16 of SEQ ID NO:31) or a portion of the hinge domain (e.g., amino acids 4-16 of SEQ ID NO:31). Preferred linkers for connecting a C-terminal Fc-antibody may be selected from the embodiments given in SEQ ID NO:32 to SEQ ID NO:41, including linkers suitable for fusion of additional polypeptides. Likewise, amino acid sequences for the polypeptide domains of structure I-B are given in Table 7. If an Fc antibody fragment is fused to the N-terminus of an TNRFSF fusion protein as in structure I-B, the sequence of the Fc module is preferably that shown in SEQ ID NO:42, and the linker sequences are preferably selected from those embodiments set forth in SEQ ID NO:43 to SEQ ID NO:45.
[1416] In an embodiment, an HVEM agonist fusion protein according to structures I-A or I--B comprises one or more HVEM binding domains comprising an LIGHT (HVEM ligand) sequence (Table 44). In an embodiment, an HVEM agonist fusion protein according to structures I-A or I-B comprises one or more HVEM binding domains comprising a sequence according to SEQ ID NO:443. In an embodiment, an HVEM agonist fusion protein according to structures I-A or I-B comprises one or more HVEM binding domains comprising a soluble LIGHT sequence. In an embodiment, a HVEM agonist fusion protein according to structures I-A or I-B comprises one or more HVEM binding domains comprising a sequence according to SEQ ID NO:444. In an embodiment, a HVEM agonist fusion protein according to structures I-A or I-B comprises one or more HVEM binding domains comprising a sequence according to SEQ ID NO:445. In an embodiment, a HVEM agonist fusion protein according to structures I-A or I--B comprises one or more HVEM binding domains comprising a sequence according to SEQ ID NO:446.
[1417] In an embodiment, an HVEM agonist fusion protein according to structures I-A or I--B comprises one or more HVEM binding domains that is a scFv domain comprising V.sub.H and V.sub.L regions, wherein the V.sub.H and V.sub.L domains are connected by a linker.
TABLE-US-00044 TABLE 44 Additional polypeptide domains useful as HVEM binding domains in fusion proteins (e.g., structures I-A and I-B). Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 443 MEESVVRPSV FVVDGQTDIP FTRLGRSHRR QSCSVARVGL GLLLLLMGAG LAVQGWFLLQ 60 LIGHT (HVEM LHWRLGEMVT RLPDGPAGSW EQLIQERRSH EVNPAAHLTG ANSSLTGSGG PLLWETQLGL 120 ligand) AFLRGLSYHD GALVVTKAGY YYIYSKVQLG GVGCPLGLAS TITHGLYKRT PRYPEELELL 180 VSQQSPCGRA TSSSRVWWDS SFLGGVVHLE AGEKVVVRVL DERLVRLRDG TRSYFGAFMV 240 SEQ ID NO: 444 PAAHLTGANS SLTGSGGPLL WETQLGLAFL RGLSYHDGAL VVTKAGYYYI YSKVQLGGVG 60 LIGHT soluble CPLGLASTIT HGLYKRTPRY PEELELLVSQ QSPCGRATSS SRVWWDSSFL GGVVHLEAGE 120 domain KVVVRVLDER LVRLRDGTRS YFGAFMV 147 SEQ ID NO: 445 AAHLTGANSS LTGSGGPLLW ETQLGLAFLR GLSYHDGALV VTKAGYYYIY SKVQLGGVGC 60 LIGHT soluble PLGLASTITH GLYKRTPRYP EELELLVSQQ SPCGRATSSS RVWWDSSFLG GVVHLEAGEK 120 domain VVVRVLDERL VRLRDGTRSY FGAFMV 146 (alternative) SEQ ID NO: 446 AHLTGANSSL TGSGGPLLWE TQLGLAFLRG LSYHDGALVV TKAGYYYIYS KVQLGGVGCP 60 LIGHT soluble LGLASTITHG LYKRTPRYPE ELELLVSQQS PCGRATSSSR VWWDSSFLGG VVHLEAGEKV 120 domain VVRVLDERLV RLRDGTRSYF GAFMV 145 (alternative)
[1418] In an embodiment, the HVEM agonist is a HVEM agonistic single-chain fusion polypeptide comprising (i) a first soluble HVEM binding domain, (ii) a first peptide linker, (iii) a second soluble HVEM binding domain, (iv) a second peptide linker, and (v) a third soluble HVEM binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, and wherein the additional domain is a Fab or Fc fragment domain. In an embodiment, the HVEM agonist is a HVEM agonistic single-chain fusion polypeptide comprising (i) a first soluble HVEM binding domain, (ii) a first peptide linker, (iii) a second soluble HVEM binding domain, (iv) a second peptide linker, and (v) a third soluble HVEM binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, wherein the additional domain is a Fab or Fc fragment domain wherein each of the soluble HVEM binding domains lacks a stalk region (which contributes to trimerisation and provides a certain distance to the cell membrane, but is not part of the HVEM binding domain) and the first and the second peptide linkers independently have a length of 3-8 amino acids.
[1419] In an embodiment, the HVEM agonist is an HVEM agonistic single-chain fusion polypeptide comprising (i) a first soluble tumor necrosis factor (TNF) superfamily cytokine domain, (ii) a first peptide linker, (iii) a second soluble TNF superfamily cytokine domain, (iv) a second peptide linker, and (v) a third soluble TNF superfamily cytokine domain, wherein each of the soluble TNF superfamily cytokine domains lacks a stalk region and the first and the second peptide linkers independently have a length of 3-8 amino acids, and wherein the TNF superfamily cytokine domain is an HVEM binding domain.
[1420] In an embodiment, the HVEM agonist is a HVEM agonist described in U.S. Pat. No. 7,118,742, the disclosure of which is incorporated by reference herein.
CD95 Agonists
[1421] In an embodiment, the TNFRSF agonist is a CD95 agonist or CD95 binding molecule. CD95 is also known as TNFRSF6, Fas receptor (FasR), and APO-1. Any CD95 agonist or binding molecule known in the art may be used. The CD95 binding molecule may be a monoclonal antibody or fusion protein capable of binding to human or mammalian CD95, and may be used at a concentration appropriate for T cell agonistic activity rather than T cell apoptotic activity, as described elsewhere herein. The CD95 agonists or CD95 binding molecules may comprise an immunoglobulin heavy chain of any isotype (e.g., IgG, IgE, IgM, IgD, IgA, and IgY), class (e.g., IgG1, IgG2, IgG3, IgG4, IgA1 and IgA2) or subclass of immunoglobulin molecule. The CD95 agonist or CD95 binding molecule may have both a heavy and a light chain. As used herein, the term binding molecule also includes antibodies (including full length antibodies), monoclonal antibodies (including full length monoclonal antibodies), polyclonal antibodies, multispecific antibodies (e.g., bispecific antibodies), human, humanized or chimeric antibodies, and antibody fragments, e.g., Fab fragments, F(ab') fragments, fragments produced by a Fab expression library, epitope-binding fragments of any of the above, and engineered forms of antibodies, e.g., scFv molecules, that bind to CD95. In an embodiment, the CD95 agonist is an antigen binding protein that is a fully human antibody. In an embodiment, the CD95 agonist is an antigen binding protein that is a humanized antibody. In some embodiments, CD95 agonists for use in the presently disclosed methods and compositions include anti-CD95 antibodies, human anti-CD95 antibodies, mouse anti-CD95 antibodies, mammalian anti-CD95 antibodies, monoclonal anti-CD95 antibodies, polyclonal anti-CD95 antibodies, chimeric anti-CD95 antibodies, anti-CD95 adnectins, anti-CD95 domain antibodies, single chain anti-CD95 fragments, heavy chain anti-CD95 fragments, light chain anti-CD95 fragments, anti-CD95 fusion proteins, and fragments, derivatives, conjugates, variants, or biosimilars thereof. In a preferred embodiment, the CD95 agonist is an agonistic, anti-CD95 humanized or fully human monoclonal antibody (i.e., an antibody derived from a single cell line).
[1422] In a preferred embodiment, the CD95 agonist or CD95 binding molecule may also be a fusion protein. In a preferred embodiment, a multimeric CD95 agonist, such as a trimeric or hexameric CD95 agonist (with three or six ligand binding domains), may induce superior receptor (CD95L) clustering and internal cellular signaling complex formation compared to an agonistic monoclonal antibody, which typically possesses two ligand binding domains. Trimeric (trivalent) or hexameric (or hexavalent) or greater fusion proteins comprising three TNFRSF binding domains and IgG1-Fc and optionally further linking two or more of these fusion proteins are described, e.g., in Gieffers, et al., Mol. Cancer Therapeutics 2013, 12, 2735-47.
[1423] Agonistic CD95 antibodies and fusion proteins are known to induce strong immune responses. In a preferred embodiment, the CD95 agonist is a monoclonal antibody or fusion protein that binds specifically to CD95 antigen in a manner sufficient to reduce toxicity. In some embodiments, the CD95 agonist is an agonistic CD95 monoclonal antibody or fusion protein that abrogates antibody-dependent cellular toxicity (ADCC), for example NK cell cytotoxicity. In some embodiments, the CD95 agonist is an agonistic CD95 monoclonal antibody or fusion protein that abrogates antibody-dependent cell phagocytosis (ADCP). In some embodiments, the CD95 agonist is an agonistic CD95 monoclonal antibody or fusion protein that abrogates complement-dependent cytotoxicity (CDC). In some embodiments, the CD95 agonist is an agonistic CD95 monoclonal antibody or fusion protein which abrogates Fc region functionality.
[1424] In some embodiments, the CD95 agonists are characterized by binding to human CD95 (SEQ ID NO:447) with high affinity and agonistic activity. In an embodiment, the CD95 agonist is a binding molecule that binds to human CD95 (SEQ ID NO:447). In an embodiment, the CD95 agonist is a binding molecule that binds to human CD95 (SEQ ID NO:448). In an embodiment, the CD95 agonist is a binding molecule that binds to human CD95 (SEQ ID NO:449). In an embodiment, the CD95 agonist is a binding molecule that binds to human CD95 (SEQ ID NO:450). The amino acid sequence of CD95 antigens to which a CD95 agonist or binding molecule may bind is summarized in Table 45.
TABLE-US-00045 TABLE 45 Amino acid sequence of CD95 antigens. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 447 MLGIWTLLPL VLTSVARLSS KSVNAQVTDI NSKGLELRKT VTTVETQNLE GLHHDGQFCH 60 human CD95, KPCPPGERKA RDCTVNGDEP DCVPCQEGKE YTDKAHFSSK CRRCRLCDEG HGLEVEINCT 120 Tumor necrosis RTQNTKCRCK PNFFCNSTVC EHCDPCTKCE HGIIKECTLT SNTKCKEEGS RSNLGWLCLL 180 factor receptor LLPIPLIVWV KRKEVQKTCR KHRKENQGSH ESPTLNPETV AINLSDVDLS KYITTIAGVM 240 superfamily, TLSQVKGFVR KNGVNEAKID EIKNDNVQDT AEQKVQLLRN WHQLHGKKEA YDTLIKDLKK 300 member 6 (Homo ANLCTLAEKI QTIILKDITS DSENSNFRNE IQSLV 335 sapiens), isoform 1 SEQ ID NO: 448 MLGIWTLLPL VLTSVARLSS KSVNAQVTDI NSKGLELRKT VTTVETQNLE GLHHDGQFCH 60 human CD95, KPCPPGERKA RDCTVNGDEP DCVPCQEGKE YTDKAHFSSK CRRCRLCDEG HGLEVEINCT 120 Tumor necrosis RTQNTKCRCK PNFFCNSTVC EHCDPCTKCE HGIIKECTLT SNTKCKEEVK RKEVQKTCRK 180 factor receptor HRKENQGSHE SPTLNPETVA INLSDVDLSK YITTIAGVMT LSQVKGFVRK NGVNEAKIDE 240 superfamily, IKNDNVQDTA EQKVQLLRNW HQLHGKKEAY DTLIKDLKKA NLCTLAEKIQ TIILKDITSD 300 member 6 (Homo SENSNFRNEI QSLV 314 sapiens), isoform 2 SEQ ID NO: 449 MLGIWTLLPL VLTSVARLSS KSVNAQVTDI NSKGLELRKT VTTVETQNLE GLHHDGQFCH 60 human CD95, KPCPPGERKA RDCTVNGDEP DCVPCQEGKE YTDKAHFSSK CRRCRLCDEG HGLEVEINCT 120 Tumor necrosis RTQNTKCRCK PNFFCNSTVC EHCDPCTKCE HGIIKECTLT SNTKCKEEGS RSNLGWLCLL 180 factor receptor LLPIPLIVWV KRKEVQKTCR KHRKENQGSH ESPTLNPMLT 220 superfamily, member 6 (Homo sapiens), isoform 3 SEQ ID NO: 450 MLGIWTLLPL VLTSVARLSS KSVNAQVTDI NSKGLELRKT VTTVETQNLE GLHHDGQFCH 60 human CD95, KPCPPGERKA RDCTVNGDEP DCVPCQEGKE YTDKAHFSSK CRRCRLCDEG HGLEVEINCT 120 Tumor necrosis RTQNTKCRCK PNFFCNSTVC EHCDPCTKCE HGIIKECTLT SNTKCKEEGS RSNLGWLCLL 180 factor receptor LLPIPLIVWG NSGNKFI 197 superfamily, member 6 (Homo sapiens), isoform 4
[1425] In some embodiments, the compositions, processes and methods described include a CD95 agonist that binds human or murine CD95 with a K.sub.D of about 100 pM or lower, binds human or murine CD95 with a K.sub.D of about 90 pM or lower, binds human or murine CD95 with a K.sub.D of about 80 pM or lower, binds human or murine CD95 with a K.sub.D of about 70 pM or lower, binds human or murine CD95 with a K.sub.D of about 60 pM or lower, binds human or murine CD95 with a K.sub.D of about 50 pM or lower, binds human or murine CD95 with a K.sub.D of about 40 pM or lower, or binds human or murine CD95 with a K.sub.D of about 30 pM or lower.
[1426] In some embodiments, the compositions, processes and methods described include a CD95 agonist that binds to human or murine CD95 with a k.sub.assoc of about 7.5.times.10.sup.5 l/Ms or faster, binds to human or murine CD95 with a k.sub.assoc of about 7.5.times.10.sup.5 l/Ms or faster, binds to human or murine CD95 with a k.sub.assoc of about 8.times.10.sup.5 l/Ms or faster, binds to human or murine CD95 with a k.sub.assoc of about 8.5.times.10.sup.5 l/Ms or faster, binds to human or murine CD95 with a k.sub.assoc of about 9.times.10.sup.5 l/Ms or faster, binds to human or murine CD95 with a k.sub.assoc of about 9.5.times.10.sup.5 l/Ms or faster, or binds to human or murine CD95 with a k.sub.assoc of about 1.times.10.sup.6 l/Ms or faster.
[1427] In some embodiments, the compositions, processes and methods described include a CD95 agonist that binds to human or murine CD95 with a k.sub.dissoc of about 2.times.10.sup.-5 l/s or slower, binds to human or murine CD95 with a k.sub.dissoc of about 2.1.times.10.sup.-5 l/s or slower, binds to human or murine CD95 with a k.sub.dissoc of about 2.2.times.10.sup.-5 l/s or slower, binds to human or murine CD95 with a k.sub.dissoc of about 2.3.times.10.sup.-5 l/s or slower, binds to human or murine CD95 with a k.sub.dissoc of about 2.4.times.10.sup.-5 l/s or slower, binds to human or murine CD95 with a k.sub.dissoc of about 2.5.times.10.sup.-5 l/s or slower, binds to human or murine CD95 with a k.sub.dissoc of about 2.6.times.10.sup.-5 l/s or slower or binds to human or murine CD95 with a k.sub.dissoc of about 2.7.times.10.sup.-5 l/s or slower, binds to human or murine CD95 with a k.sub.dissoc of about 2.8.times.10.sup.-5 l/s or slower, binds to human or murine CD95 with a k.sub.dissoc of about 2.9.times.10.sup.-5 l/s or slower, or binds to human or murine CD95 with a k.sub.dissoc of about 3.times.10.sup.-5 l/s or slower.
[1428] In some embodiments, the compositions, processes and methods described include a CD95 agonist that binds to human or murine CD95 with an IC.sub.50 of about 10 nM or lower, binds to human or murine CD95 with an IC.sub.50 of about 9 nM or lower, binds to human or murine CD95 with an IC.sub.50 of about 8 nM or lower, binds to human or murine CD95 with an IC.sub.50 of about 7 nM or lower, binds to human or murine CD95 with an IC.sub.50 of about 6 nM or lower, binds to human or murine CD95 with an IC.sub.50 of about 5 nM or lower, binds to human or murine CD95 with an IC.sub.50 of about 4 nM or lower, binds to human or murine CD95 with an IC.sub.50 of about 3 nM or lower, binds to human or murine CD95 with an IC.sub.50 of about 2 nM or lower, or binds to human or murine CD95 with an IC.sub.50 of about 1 nM or lower.
[1429] In a preferred embodiment, the CD95 agonist is the monoclonal antibody E09, or a fragment, derivative, variant, or biosimilar thereof. The preparation and properties of E09 are described in Chodorge, et al., Cell Death & Differ. 2012, 19, 1187-95. The amino acid sequences of E09 are set forth in Table 46.
[1430] In an embodiment, the CD95 agonist comprises the heavy and light chain CDRs or variable regions (VRs) of E09. In an embodiment, the CD95 agonist heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:451, and the CD95 agonist light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:452, and conservative amino acid substitutions thereof. In an embodiment, a CD95 agonist comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:451 and SEQ ID NO:452, respectively. In an embodiment, a CD95 agonist comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:451 and SEQ ID NO:452, respectively. In an embodiment, a CD95 agonist comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:451 and SEQ ID NO:452, respectively. In an embodiment, a CD95 agonist comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:451 and SEQ ID NO:452, respectively. In an embodiment, a CD95 agonist comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:451 and SEQ ID NO:452, respectively.
[1431] In an embodiment, a CD95 agonist comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:453, SEQ ID NO:454, and SEQ ID NO:455, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:456, SEQ ID NO:457, and SEQ ID NO:458, respectively, and conservative amino acid substitutions thereof.
[1432] In an embodiment, the CD95 agonist is a CD95 agonist biosimilar monoclonal antibody approved by drug regulatory authorities with reference to E09. In an embodiment, the biosimilar monoclonal antibody comprises an CD95 antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is E09. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is a CD95 agonist antibody authorized or submitted for authorization, wherein the CD95 agonist antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is E09. The CD95 agonist antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is E09. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is E09.
TABLE-US-00046 TABLE 46 Amino acid sequences for CD95 agonist antibody E09. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 451 QLQLQESGPG LVKPSETLSL TCTVSGASIS ANSYYGVWVR QSPGKGLEWV GSIAYRGNSN 60 heavy chain SGSTYYNPSL KSRATVSVDT SKNQVSLRLT SVTAADTALY YCARRQLLDD GTGYQWAAFD 120 variable region VWGQGTMVTV SS 132 for E09 SEQ ID NO: 452 QSVLTQPPSV SEAPRQTVTI SCSGNSFNIG RYPVNWYQQL PGKAPKLLIY YNNLRFSGVS 60 light chain DRFSGSKSGT SASLAIRDLL SEDEADYYCS TWDDTLKGWV FGGGTKVTVL 110 variable region for E09 SEQ ID NO: 453 ANSYYGV 7 heavy chain CDR1 for E09 SEQ ID NO: 454 GSIAYRGNSN SGSTYYNPSL KS 22 heavy chain CDR2 for E09 SEQ ID NO: 455 RQLLDDGTGY QWAAFDV 17 heavy chain CDR3 for E09 SEQ ID NO: 456 SGNSFNIGRY PVN 13 light chain CDR1 for E09 SEQ ID NO: 457 YNNLRFS 7 light chain CDR2 for E09 SEQ ID NO: 458 STWDDTLKGW V 11 light chain CDR3 for E09
[1433] In an embodiment, the CD95 agonist is an CD95 agonist described in International Patent Application Publication No. WO 2009/007120 A2 and U.S. Patent Application Publication No. US 2016/0176941 A1, the disclosure of each of which is incorporated by reference herein.
[1434] In an embodiment, the CD95 agonist is an CD95 agonistic fusion protein as depicted in Structure I-A (C-terminal Fc-antibody fragment fusion protein) or Structure I-B (N-terminal Fc-antibody fragment fusion protein), or a fragment, derivative, conjugate, variant, or biosimilar thereof. The properties of structures I-A and I-B are described above and in U.S. Pat. Nos. 9,359,420, 9,340,599, 8,921,519, and 8,450,460, the disclosures of which are incorporated by reference herein. Amino acid sequences for the polypeptide domains of structure I-A are given in Table 6. The Fc domain preferably comprises a complete constant domain (amino acids 17-230 of SEQ ID NO:31) the complete hinge domain (amino acids 1-16 of SEQ ID NO:31) or a portion of the hinge domain (e.g., amino acids 4-16 of SEQ ID NO:31). Preferred linkers for connecting a C-terminal Fc-antibody may be selected from the embodiments given in SEQ ID NO:33 to SEQ ID NO:41, including linkers suitable for fusion of additional polypeptides. Likewise, amino acid sequences for the polypeptide domains of structure I-B are given in Table 7. If an Fc antibody fragment is fused to the N-terminus of an TNRFSF fusion protein as in structure I-B, the sequence of the Fc module is preferably that shown in SEQ ID NO:42, and the linker sequences are preferably selected from those embodiments set forth in SEQ ID NO:43 to SEQ ID NO:45.
[1435] In an embodiment, an CD95 agonist fusion protein according to structures I-A or I-B comprises one or more CD95 binding domains comprising a CD95 ligand sequence (Table 47). In an embodiment, an CD95 agonist fusion protein according to structures I-A or I-B comprises one or more CD95 binding domains comprising a sequence according to SEQ ID NO:459. In an embodiment, an CD95 agonist fusion protein according to structures I-A or I-B comprises one or more CD95 binding domains comprising a soluble LIGHT sequence. In an embodiment, a CD95 agonist fusion protein according to structures I-A or I-B comprises one or more CD95 binding domains comprising a sequence according to SEQ ID NO:460. In an embodiment, a CD95 agonist fusion protein according to structures I-A or I-B comprises one or more CD95 binding domains comprising a sequence according to SEQ ID NO:461. In an embodiment, a CD95 agonist fusion protein according to structures I-A or I-B comprises one or more CD95 binding domains comprising a sequence according to SEQ ID NO:462.
[1436] In an embodiment, an CD95 agonist fusion protein according to structures I-A or I-B comprises one or more CD95 binding domains that is a scFv domain comprising V.sub.H and V.sub.L regions, wherein the V.sub.H and V.sub.L domains are connected by a linker.
TABLE-US-00047 TABLE 47 Additional polypeptide domains useful as CD95 binding domains in fusion proteins (e.g., structures I-A and I-B). Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 459 MQQPFNYPYP QIYWVDSSAS SPWAPPGTVL PCPTSVPRRP GQRRPPPPPP PPPLPPPPPP 60 CD95L (CD95 PPLPPLPLPP LKKRGNHSTG LCLLVMFFMV LVALVGLGLG MFQLFHLQKE LAELRESTSQ 120 ligand) MHTASSLEKQ IGHPSPPPEK KELRKVAHLT GKSNSRSMPL EWEDTYGIVL LSGVKYKKGG 180 LVINETGLYF VYSKVYFRGQ SCNNLPLSHK VYMRNSKYPQ DLVMMEGKMM SYCTTGQMWA 240 RSSYLGAVFN LTSADHLYVN VSELSLVNFE ESQTFFGLYK L 281 SEQ ID NO: 460 VAHLTGKSNS RSMPLEWEDT YGIVLLSGVK YKKGGLVINE TGLYFVYSKV YFRGQSCNNL 60 CD95L soluble PLSHKVYMRN SKYPQDLVMM EGKMMSYCTT GQMWARSSYL GAVFNLTSAD HLYVNVSELS 120 domain LVNFEESQTF FGLYKL 136 SEQ ID NO: 461 AHLTGKSNSR SMPLEWEDTY GIVLLSGVKY KKGGLVINET GLYFVYSKVY FRGQSCNNLP 60 CD95L soluble LSHKVYMRNS KYPQDLVMME GKMMSYCTTG QMWARSSYLG AVFNLTSADH LYVNVSELSL 120 domain VNFEESQTFF GLYKL 135 (alternative) SEQ ID NO: 462 HLTGKSNSRS MPLEWEDTYG IVLLSGVKYK KGGLVINETG LYFVYSKVYF RGQSCNNLPL 60 CD95L soluble SHKVYMRNSK YPQDLVMMEG KMMSYCTTGQ MWARSSYLGA VFNLTSADHL YVNVSELSLV 120 domain NFEESQTFFG LYKL 134 (alternative)
[1437] In an embodiment, the CD95 agonist is a CD95 agonistic single-chain fusion polypeptide comprising (i) a first soluble CD95 binding domain, (ii) a first peptide linker, (iii) a second soluble CD95 binding domain, (iv) a second peptide linker, and (v) a third soluble CD95 binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, and wherein the additional domain is a Fab or Fc fragment domain. In an embodiment, the CD95 agonist is a CD95 agonistic single-chain fusion polypeptide comprising (i) a first soluble CD95 binding domain, (ii) a first peptide linker, (iii) a second soluble CD95 binding domain, (iv) a second peptide linker, and (v) a third soluble CD95 binding domain, further comprising an additional domain at the N-terminal and/or C-terminal end, wherein the additional domain is a Fab or Fc fragment domain wherein each of the soluble CD95 binding domains lacks a stalk region (which contributes to trimerisation and provides a certain distance to the cell membrane, but is not part of the CD95 binding domain) and the first and the second peptide linkers independently have a length of 3-8 amino acids.
[1438] In an embodiment, the CD95 agonist is an CD95 agonistic single-chain fusion polypeptide comprising (i) a first soluble tumor necrosis factor (TNF) superfamily cytokine domain, (ii) a first peptide linker, (iii) a second soluble TNF superfamily cytokine domain, (iv) a second peptide linker, and (v) a third soluble TNF superfamily cytokine domain, wherein each of the soluble TNF superfamily cytokine domains lacks a stalk region and the first and the second peptide linkers independently have a length of 3-8 amino acids, and wherein the TNF superfamily cytokine domain is an CD95 binding domain.
[1439] In an embodiment, the CD95 agonist is a CD95 agonistic scFv antibody comprising any of the foregoing V.sub.H domains linked to any of the foregoing V.sub.L domains.
Methods of Expanding Tumor Infiltrating Lymphocytes
[1440] In an embodiment, the invention provides a method of expanding a population of TILs using any of the TNFRSF agonists of the present disclosure, the method comprising the steps as described in Jin, et al., J. Immunotherapy 2012, 35, 283-292, the disclosure of which is incorporated by reference herein. For example, the tumor may be placed in enzyme media and mechanically dissociated for approximately 1 minute. The mixture may then be incubated for 30 minutes at 37.degree. C. in 5% CO.sub.2 and then mechanically disrupted again for approximately 1 minute. After incubation for 30 minutes at 37.degree. C. in 5% CO.sub.2, the tumor may be mechanically disrupted a third time for approximately 1 minute. If after the third mechanical disruption, large pieces of tissue are present, 1 or 2 additional mechanical dissociations may be applied to the sample, with or without 30 additional minutes of incubation at 37.degree. C. in 5% CO.sub.2. At the end of the final incubation, if the cell suspension contains a large number of red blood cells or dead cells, a density gradient separation using Ficoll may be performed to remove these cells. TIL cultures were initiated in 24-well plates (Costar 24-well cell culture cluster, flat bottom; Corning Incorporated, Corning, N.Y.), each well may be seeded with 1.times.10.sup.6 tumor digest cells or one tumor fragment approximately 1 to 8 mm.sup.3 in size in 2 mL of complete medium (CM) with L-2 (6000 IU/mL; Chiron Corp., Emeryville, Calif.). CM comprises Roswell Park Memorial Institute (RPMI) 1640 buffer with GlutaMAX, supplemented with 10% human AB serum, 25 mM Hepes, and 10 mg/mL gentamicin. Cultures may be initiated in gas-permeable flasks with a 40 mL capacity and a 10 cm.sup.2 gas-permeable silicon bottom (G-Rex 10; Wilson Wolf Manufacturing, New Brighton, each flask may be loaded with 10-40.times.10.sup.6 viable tumor digest cells or 5-30 tumor fragments in 10-40 mL of CM with L-2. G-Rex 10 and 24-well plates may be incubated in a humidified incubator at 37.degree. C. in 5% CO.sub.2 and 5 days after culture initiation, half the media may be removed and replaced with fresh CM and IL-2 and after day 5, half the media may be changed every 2-3 days. Rapid expansion protocol (REP) of TILs may be performed using T-175 flasks and gas-permeable bags or gas-permeable G-Rex flasks, as described elsewhere herein, using the TNFRSF agonists of the present disclosure. For REP in T-175 flasks, 1.times.10.sup.6 TILs may be suspended in 150 mL of media in each flask. The TIL may be cultured with TNFRSF agonists of the present disclosure at a ratio described herein, in a 1 to 1 mixture of CM and AIM-V medium (50/50 medium), supplemented with 3000 IU/mL of IL-2 and 30 ng/mL of anti-CD3 antibody (OKT-3). The T-175 flasks may be incubated at 37.degree. C. in 5% CO.sub.2. Half the media may be changed on day 5 using 50/50 medium with 3000 IU/mL of IL-2. On day 7, cells from 2 T-175 flasks may be combined in a 3 L bag and 300 mL of AIM-V with 5% human AB serum and 3000 IU/mL of IL-2 may be added to the 300 mL of TIL suspension. The number of cells in each bag may be counted every day or two days, and fresh media may be added to keep the cell count between 0.5 and 2.0.times.10.sup.6 cells/mL. For REP in 500 mL capacity flasks with 100 cm.sup.2 gas-permeable silicon bottoms (e.g., G-Rex 100, Wilson Wolf Manufacturing, as described elsewhere herein), 5.times.10.sup.6 or 10.times.10.sup.6 TILs may be cultured with TNFRSF agonists at a ratio described herein (e.g., 1 to 100) in 400 mL of 50/50 medium, supplemented with 3000 IU/mL of IL-2 and 30 ng/mL of anti-CD3 antibody (OKT-3). The G-Rex100 flasks may be incubated at 37.degree. C. in 5% CO.sub.2. On day five, 250 mL of supernatant may be removed and placed into centrifuge bottles and centrifuged at 1500 rpm (491 g) for 10 minutes. The obtained TIL pellets may be resuspended with 150 mL of fresh 50/50 medium with 3000 IU/mL of IL-2 and added back to the G-Rex 100 flasks. When TIL are expanded serially in G-Rex 100 flasks, on day seven the TIL in each G-Rex100 are suspended in the 300 mL of media present in each flask and the cell suspension may be divided into three 100 mL aliquots that may be used to seed 3 G-Rex100 flasks. About 150 mL of AIM-V with 5% human AB serum and 3000 IU/mL of IL-2 may then be added to each flask. G-Rex100 flasks may then be incubated at 37.degree. C. in 5% CO.sub.2, and after four days, 150 mL of AIM-V with 3000 IU/mL of IL-2 may be added to each G-Rex100 flask. After this, the REP may be completed by harvesting cells on day 14 of culture.
[1441] In an embodiment, a method or process of expanding or treating a cancer includes a step wherein TILs are obtained from a patient tumor sample. A patient tumor sample may be obtained using methods known in the art. For example, TILs may be cultured from enzymatic tumor digests and tumor fragments (about 1 to about 8 mm.sup.3 in size) from sharp dissection. Such tumor digests may be produced by incubation in enzymatic media (e.g., Roswell Park Memorial Institute (RPMI) 1640 buffer, 2 mM glutamate, 10 mcg/mL gentamicine, 30 units/mL of DNase and 1.0 mg/mL of collagenase) followed by mechanical dissociation (e.g., using a tissue dissociator). Tumor digests may be produced by placing the tumor in enzymatic media and mechanically dissociating the tumor for approximately 1 minute, followed by incubation for 30 minutes at 37.degree. C. in 5% CO.sub.2, followed by repeated cycles of mechanical dissociation and incubation under the foregoing conditions until only small tissue pieces are present. At the end of this process, if the cell suspension contains a large number of red blood cells or dead cells, a density gradient separation using FICOLL branched hydrophilic polysaccharide may be performed to remove these cells. Alternative methods known in the art may be used, such as those described in U.S. Patent Application Publication No. 2012/0244133 A1, the disclosure of which is incorporated by reference herein. Any of the foregoing methods may be used in any of the embodiments described herein for methods or processes of expanding TILs or methods treating a cancer.
[1442] In an embodiment, a rapid expansion process for TILs may be performed using T-175 flasks and gas permeable bags as previously described (Tran, et al., J. Immunother. 2008, 31, 742-51; Dudley, et al., J. Immunother. 2003, 26, 332-42) or gas permeable cultureware (G-Rex flasks, commercially available from Wilson Wolf Manufacturing Corporation, New Brighton, Minn., USA). For TIL rapid expansion in T-175 flasks, 1.times.10.sup.6 TILs suspended in 150 mL of media may be added to each T-175 flask. The TILs may be cultured with TNFRSF agonists at a ratio of 1 TIL to 100 TNFRSF agonists and the cells were cultured in a 1 to 1 mixture of CM and AIM-V medium, supplemented with 3000 IU (international units) per mL of IL-2 and 30 ng per ml of anti-CD3 antibody (e.g., OKT-3). The T-175 flasks may be incubated at 37.degree. C. in 5% CO.sub.2. Half the media may be exchanged on day 5 using 50/50 medium with 3000 IU per mL of L-2. On day 7 cells from two T-175 flasks may be combined in a 3 L bag and 300 mL of AIM V with 5% human AB serum and 3000 IU per mL of IL-2 was added to the 300 ml of TIL suspension. The number of cells in each bag was counted every day or two and fresh media was added to keep the cell count between 0.5 and 2.0.times.10.sup.6 cells/mL.
[1443] In an embodiment, for TIL rapid expansions in 500 mL capacity gas permeable flasks with 100 cm.sup.2 gas-permeable silicon bottoms (G-Rex 100, commercially available from Wilson Wolf Manufacturing Corporation, New Brighton, Minn., USA), 5.times.10.sup.6 or 10.times.10.sup.6 TIL may be cultured with TNFRSF agonists in 400 mL of 50/50 medium, supplemented with 5% human AB serum, 3000 IU per mL of IL-2 and 30 ng per mL of anti-CD3 (OKT-3). The G-Rex 100 flasks may be incubated at 37.degree. C. in 5% CO.sub.2. On day 5, 250 mL of supernatant may be removed and placed into centrifuge bottles and centrifuged at 1500 rpm (revolutions per minute; 491.times.g) for 10 minutes. The TIL pellets may be re-suspended with 150 mL of fresh medium with 5% human AB serum, 3000 IU per mL of IL-2, and added back to the original G-Rex 100 flasks. When TIL are expanded serially in G-Rex 100 flasks, on day 7 the TIL in each G-Rex 100 may be suspended in the 300 mL of media present in each flask and the cell suspension may be divided into 3 100 mL aliquots that may be used to seed 3 G-Rex 100 flasks. Then 150 mL of AIM-V with 5% human AB serum and 3000 IU per mL of IL-2 may be added to each flask. The G-Rex 100 flasks may be incubated at 37.degree. C. in 5% CO.sub.2 and after 4 days 150 mL of AIM-V with 3000 IU per mL of IL-2 may be added to each G-Rex 100 flask. The cells may be harvested on day 14 of culture.
[1444] In an embodiment, TILs may be prepared as follows. 2 mm.sup.3 tumor fragments are cultured in complete media (CM) comprised of AIM-V medium (Invitrogen Life Technologies, Carlsbad, Calif.) supplemented with 2 mM glutamine (Mediatech, Inc. Manassas, Va.), 100 U/mL penicillin (Invitrogen Life Technologies), 100 .mu.g/mL streptomycin (Invitrogen Life Technologies), 5% heat-inactivated human AB serum (Valley Biomedical, Inc. Winchester, Va.) and 600 IU/mL rhIL-2 (Chiron, Emeryville, Calif.). For enzymatic digestion of solid tumors, tumor specimens are diced into RPMI-1640, washed and centrifuged at 800 rpm for 5 minutes at 15-22.degree. C., and resuspended in enzymatic digestion buffer (0.2 mg/mL Collagenase and 30 units/ml of DNase in RPMI-1640) followed by overnight rotation at room temperature. TILs established from fragments may be grown for 3-4 weeks in CM and expanded fresh or cryopreserved in heat-inactivated HAB serum with 10% dimethylsulfoxide (DMSO) and stored at -180.degree. C. until the time of study. Tumor associated lymphocytes (TAL) obtained from ascites collections were seeded at 3.times.10.sup.6 cells/well of a 24 well plate in CM. TIL growth was inspected about every other day using a low-power inverted microscope.
[1445] In an embodiment, the invention includes a method of expanding tumor infiltrating lymphocytes (TILs), the method comprising contacting a population of TILs comprising at least one TIL with a TNFRSF agonist described herein, wherein said TNFRSF agonist comprises at least one co-stimulatory ligand that specifically binds with a co-stimulatory molecule expressed on the cellular surface of the TILs, wherein binding of said co-stimulatory molecule with said co-stimulatory ligand induces proliferation of the TILs, thereby specifically expanding TILs.
[1446] In an embodiment, the invention provides a method of expanding a population of tumor infiltrating lymphocytes (TILs), the method comprising the steps of contacting the population of TILs with one or more TNFRSF agonists in a cell culture medium.
[1447] In an embodiment, the invention provides a method of expanding a population of tumor infiltrating lymphocytes (TILs), the method comprising the steps of contacting the population of TILs with one or more TNFRSF agonists in a cell culture medium, wherein the concentrations of the one or more TNFRSF agonists in the cell culture medium are independently selected from the group consisting of 50 ng/mL, 100 ng/mL, 500 ng/mL, 1 g/mL, 5 .mu.g/mL, 10 .mu.g/mL, 20 .mu.g/mL, 30 .mu.g/mL, 40 .mu.g/mL, 50 .mu.g/mL, 60 .mu.g/mL, 70 .mu.g/mL, 80 .mu.g/mL, 90 .mu.g/mL, and 100 .mu.g/mL.
[1448] In an embodiment, the invention provides a method of expanding a population of tumor infiltrating lymphocytes (TILs), the method comprising the steps of contacting the population of TILs with one or more TNFRSF agonists in a cell culture medium, wherein the cell culture medium further comprises L-2 at an initial concentration of about 3000 IU/mL and OKT-3 antibody at an initial concentration of about 30 ng/mL.
[1449] In an embodiment, the invention provides a method of expanding a population of tumor infiltrating lymphocytes (TILs), the method comprising the steps of contacting the population of TILs with one or more TNFRSF agonists in a cell culture medium, wherein the cell culture medium further comprises L-2 at an initial concentration of about 3000 IU/mL and OKT-3 antibody at an initial concentration of about 30 ng/mL, and wherein the one or more TNFRSF agonists comprises a 4-1BB agonist.
[1450] In an embodiment, the invention provides a method of expanding a population of tumor infiltrating lymphocytes (TILs), the method comprising the steps of contacting the population of TILs with one or more TNFRSF agonists in a cell culture medium, wherein the cell culture medium further comprises L-2 at an initial concentration of about 3000 IU/mL and OKT-3 antibody at an initial concentration of about 30 ng/mL, and wherein the one or more TNFRSF agonists comprises an OX40 agonist.
[1451] In an embodiment, the invention provides a method of expanding a population of tumor infiltrating lymphocytes (TILs), the method comprising the steps of contacting the population of TILs with one or more TNFRSF agonists in a cell culture medium, wherein the cell culture medium further comprises IL-2 at an initial concentration of about 3000 IU/mL and OKT-3 antibody at an initial concentration of about 30 ng/mL, and wherein the one or more TNFRSF agonists comprises a 4-1BB and an OX40 agonist.
[1452] In an embodiment, the invention provides a method of expanding a population of tumor infiltrating lymphocytes (TILs), the method comprising the steps of contacting the population of TILs with one or more TNFRSF agonists in a cell culture medium, wherein the cell culture medium further comprises IL-2 at an initial concentration of about 3000 IU/mL and OKT-3 antibody at an initial concentration of about 30 ng/mL, and wherein the one or more TNFRSF agonists comprises a CD27 agonist.
[1453] In an embodiment, the invention provides a method of expanding a population of tumor infiltrating lymphocytes (TILs), the method comprising the steps of contacting the population of TILs with one or more TNFRSF agonists in a cell culture medium, wherein the cell culture medium further comprises IL-2 at an initial concentration of about 3000 IU/mL and OKT-3 antibody at an initial concentration of about 30 ng/mL, and wherein the one or more TNFRSF agonists comprises a GITR agonist.
[1454] In an embodiment, the invention provides a method of expanding a population of tumor infiltrating lymphocytes (TILs), the method comprising the steps of contacting the population of TILs with one or more TNFRSF agonists in a cell culture medium, wherein the cell culture medium further comprises IL-2 at an initial concentration of about 3000 IU/mL and OKT-3 antibody at an initial concentration of about 30 ng/mL, and wherein the one or more TNFRSF agonists comprises a HVEM agonist.
[1455] In an embodiment, the invention provides a method of expanding a population of tumor infiltrating lymphocytes (TILs), the method comprising the steps of contacting the population of TILs with one or more TNFRSF agonists in a cell culture medium, wherein the cell culture medium further comprises IL-2 at an initial concentration of about 3000 IU/mL and OKT-3 antibody at an initial concentration of about 30 ng/mL, and wherein the one or more TNFRSF agonists comprises a CD95 agonist.
[1456] In an embodiment, the invention provides a method of expanding a population of tumor infiltrating lymphocytes (TILs), the method comprising the steps of contacting the population of TILs with one or more TNFRSF agonists in a cell culture medium, wherein the population of TILs by at least 50-fold over a period of 7 days in the cell culture medium.
[1457] In an embodiment, the invention provides a method of expanding a population of tumor infiltrating lymphocytes (TILs), the method comprising the steps of contacting the population of TILs with one or more TNFRSF agonists in a cell culture medium, wherein the population of TILs by at least 50-fold over a period of 7 days in the cell culture medium, and wherein the expansion is performed using a gas permeable container.
[1458] In an embodiment, REP can be performed in a gas permeable container using the TNFRSF agonists of the present disclosure by any suitable method. For example, TILs can be rapidly expanded using non-specific T-cell receptor stimulation in the presence of interleukin-2 (IL-2) or interleukin-15 (IL-15). The non-specific T-cell receptor stimulus can include, for example, an anti-CD3 antibody, such as about 30 ng/mL of OKT-3, a monoclonal anti-CD3 antibody (commercially available from Ortho-McNeil, Raritan, N.J. or Miltenyi Biotech, Auburn, Calif.) or UHCT-1 (commercially available from BioLegend, San Diego, Calif., USA). TILs can be rapidly expanded by further stimulation of the TILs in vitro with one or more antigens, including antigenic portions thereof, such as epitope(s), of the cancer, which can be optionally expressed from a vector, such as a human leukocyte antigen A2 (HLA-A2) binding peptide, e.g., 0.3 VM MART-1:26-35 (27 L) or gpl 00:209-217 (210M), optionally in the presence of a T-cell growth factor, such as 300 IU/mL IL-2 or IL-15. Other suitable antigens may include, e.g., NY-ESO-1, TRP-1, TRP-2, tyrosinase cancer antigen, MAGE-A3, SSX-2, and VEGFR2, or antigenic portions thereof. TIL may also be rapidly expanded by re-stimulation with the same antigen(s) of the cancer pulsed onto HLA-A2-expressing antigen-presenting cells. Alternatively, the TILs can be further re-stimulated with, e.g., example, irradiated, autologous lymphocytes or with irradiated HLA-A2+ allogeneic lymphocytes and IL-2.
[1459] In an embodiment, a method for expanding TILs may include using about 5000 mL to about 25000 mL of cell culture medium, about 5000 mL to about 10000 mL of cell culture medium, or about 5800 mL to about 8700 mL of cell culture medium. In an embodiment, a method for expanding TILs may include using about 1000 mL to about 2000 mL of cell medium, about 2000 mL to about 3000 mL of cell culture medium, about 3000 mL to about 4000 mL of cell culture medium, about 4000 mL to about 5000 mL of cell culture medium, about 5000 mL to about 6000 mL of cell culture medium, about 6000 mL to about 7000 mL of cell culture medium, about 7000 mL to about 8000 mL of cell culture medium, about 8000 mL to about 9000 mL of cell culture medium, about 9000 mL to about 10000 mL of cell culture medium, about 10000 mL to about 15000 mL of cell culture medium, about 15000 mL to about 20000 mL of cell culture medium, or about 20000 mL to about 25000 mL of cell culture medium. In an embodiment, expanding the number of TILs uses no more than one type of cell culture medium. Any suitable cell culture medium may be used, e.g., AIM-V cell medium (L-glutamine, 50 .mu.M streptomycin sulfate, and 10 .mu.M gentamicin sulfate) cell culture medium (Invitrogen, Carlsbad Calif.). In this regard, the inventive methods advantageously reduce the amount of medium and the number of types of medium required to expand the number of TIL. In an embodiment, expanding the number of TIL may comprise feeding the cells no more frequently than every third or fourth day. Expanding the number of cells in a gas permeable container simplifies the procedures necessary to expand the number of cells by reducing the feeding frequency necessary to expand the cells.
[1460] In an embodiment, the rapid expansion is performed using a gas permeable container. Such embodiments allow for cell populations to expand from about 5.times.10.sup.5 cells/cm.sup.2 to between 10.times.10.sup.6 and 30.times.10.sup.6 cells/cm.sup.2. In an embodiment, this expansion occurs without feeding. In an embodiment, this expansion occurs without feeding so long as medium resides at a height of about 10 cm in a gas-permeable flask. In an embodiment this is without feeding but with the addition of one or more cytokines. In an embodiment, the cytokine can be added as a bolus without any need to mix the cytokine with the medium. Such containers, devices, and methods are known in the art and have been used to expand TILs, and include those described in U.S. Patent Application Publication No. US 2014/0377739 A1, International Patent Application Publication No. WO 2014/210036 A1, U.S. Patent Application Publication No. US 2013/0115617 A1, International Publication No. WO 2013/188427 A1, U.S. Patent Application Publication No. US 2011/0136228 A1, U.S. Pat. No. 8,809,050, International Patent Application Publication No. WO 2011/072088 A2, U.S. Patent Application Publication No. US 2016/0208216 A1, U.S. Patent Application Publication No. US 2012/0244133 A1, International Patent Application Publication No. WO 2012/129201 A1, U.S. Patent Application Publication No. US 2013/0102075 A1, U.S. Pat. No. 8,956,860, International Patent Application Publication No. WO 2013/173835 A1, and U.S. Patent Application Publication No. US 2015/0175966 A1, the disclosures of which are incorporated herein by reference. Such processes are also described in Jin, et al., J. Immunotherapy 2012, 35, 283-292, the disclosure of which is incorporated by reference herein.
[1461] In an embodiment, the gas permeable container is a G-Rex 10 flask (Wilson Wolf Manufacturing Corporation, New Brighton, Minn., USA). In an embodiment, the gas permeable container includes a 10 cm.sup.2 gas permeable culture surface. In an embodiment, the gas permeable container includes a 40 mL cell culture medium capacity. In an embodiment, the gas permeable container provides 100 to 300 million TILs after 2 medium exchanges.
[1462] In an embodiment, the gas permeable container is a G-Rex 100 flask (Wilson Wolf Manufacturing Corporation, New Brighton, Minn., USA). In an embodiment, the gas permeable container includes a 100 cm.sup.2 gas permeable culture surface. In an embodiment, the gas permeable container includes a 450 mL cell culture medium capacity. In an embodiment, the gas permeable container provides 1 to 3 billion TILs after 2 medium exchanges.
[1463] In an embodiment, the gas permeable container is a G-Rex 100M flask (Wilson Wolf Manufacturing Corporation, New Brighton, Minn., USA). In an embodiment, the gas permeable container includes a 100 cm.sup.2 gas permeable culture surface. In an embodiment, the gas permeable container includes a 1000 mL cell culture medium capacity. In an embodiment, the gas permeable container provides 1 to 3 billion TILs without medium exchange.
[1464] In an embodiment, the gas permeable container is a G-Rex 100 L flask (Wilson Wolf Manufacturing Corporation, New Brighton, Minn., USA). In an embodiment, the gas permeable container includes a 100 cm.sup.2 gas permeable culture surface. In an embodiment, the gas permeable container includes a 2000 mL cell culture medium capacity. In an embodiment, the gas permeable container provides 1 to 3 billion TILs without medium exchange.
[1465] In an embodiment, the gas permeable container is a G-Rex 24 well plate (Wilson Wolf Manufacturing Corporation, New Brighton, Minn., USA). In an embodiment, the gas permeable container includes a plate with wells, wherein each well includes a 2 cm.sup.2 gas permeable culture surface. In an embodiment, the gas permeable container includes a plate with wells, wherein each well includes an 8 mL cell culture medium capacity. In an embodiment, the gas permeable container provides 20 to 60 million cells per well after 2 medium exchanges.
[1466] In an embodiment, the gas permeable container is a G-Rex 6 well plate (Wilson Wolf Manufacturing Corporation, New Brighton, Minn., USA). In an embodiment, the gas permeable container includes a plate with wells, wherein each well includes a 10 cm.sup.2 gas permeable culture surface. In an embodiment, the gas permeable container includes a plate with wells, wherein each well includes a 40 mL cell culture medium capacity. In an embodiment, the gas permeable container provides 100 to 300 million cells per well after 2 medium exchanges.
[1467] In an embodiment, the cell medium in the first and/or second gas permeable container is unfiltered. The use of unfiltered cell medium may simplify the procedures necessary to expand the number of cells. In an embodiment, the cell medium in the first and/or second gas permeable container lacks beta-mercaptoethanol (BME).
[1468] In an embodiment, the duration of the method comprising obtaining a tumor tissue sample from the mammal; culturing the tumor tissue sample in a first gas permeable container containing cell medium therein; obtaining TILs from the tumor tissue sample; expanding the number of TILs in a second gas permeable container containing cell medium therein using TNFRSF agonists for a duration of about 14 to about 42 days, e.g., about 28 days.
[1469] In an embodiment, the ratio of TILs to TNFRSF agonists (cells to moles) in the rapid expansion is about 1 to 25, about 1 to 50, about 1 to 100, about 1 to 125, about 1 to 150, about 1 to 175, about 1 to 200, about 1 to 225, about 1 to 250, about 1 to 275, about 1 to 300, about 1 to 325, about 1 to 350, about 1 to 500, about 1 to 1000, or about 1 to 10000. In an embodiment, the ratio of TILs to TNFRSF agonists in the rapid expansion is between 1 to 50 and 1 to 300. In an embodiment, the ratio of TILs to TNFRSF agonists in the rapid expansion is between 1 to 100 and 1 to 200.
[1470] In an embodiment, the ratio of TILs to TNFRSF agonist (TIL:TNFRSF agonist, cells to moles) is selected from the group consisting of 1:5, 1:10, 1:15, 1:20, 1:25, 1:30, 1:35, 1:40, 1:45, 1:50, 1:55, 1:60, 1:65, 1:70, 1:75, 1:80, 1:85, 1:90, 1:95, 1:100, 1:105, 1:110, 1:115, 1:120, 1:125, 1:130, 1:135, 1:140, 1:145, 1:150, 1:155, 1:160, 1:165, 1:170, 1:175, 1:180, 1:185, 1:190, 1:195, 1:200, 1:225, 1:250, 1:275, 1:300, 1:350, 1:400, 1:450, 1:500, 1:1000, 1:5000, 1:10000, and 1:50000.
[1471] In an embodiment, TILs are expanded in gas-permeable containers. Gas-permeable containers have been used to expand TILs using PBMCs using methods, compositions, and devices known in the art, including those described in U.S. Patent Application Publication No. U.S. Patent Application Publication No. 2005/0106717 A1, the disclosures of which are incorporated herein by reference. In an embodiment, TILs are expanded in gas-permeable bags. In an embodiment, TILs are expanded using a cell expansion system that expands TILs in gas permeable bags, such as the Xuri Cell Expansion System W25 (GE Healthcare). In an embodiment, TILs are expanded using a cell expansion system that expands TILs in gas permeable bags, such as the WAVE Bioreactor System, also known as the Xuri Cell Expansion System W5 (GE Healthcare). In an embodiment, the cell expansion system includes a gas permeable cell bag with a volume selected from the group consisting of about 100 mL, about 200 mL, about 300 mL, about 400 mL, about 500 mL, about 600 mL, about 700 mL, about 800 mL, about 900 mL, about 1 L, about 2 L, about 3 L, about 4 L, about 5 L, about 6 L, about 7 L, about 8 L, about 9 L, about 10 L, about 11 L, about 12 L, about 13 L, about 14 L, about 15 L, about 16 L, about 17 L, about 18 L, about 19 L, about 20 L, about 25 L, and about 30 L. In an embodiment, the cell expansion system includes a gas permeable cell bag with a volume range selected from the group consisting of between 50 and 150 mL, between 150 and 250 mL, between 250 and 350 mL, between 350 and 450 mL, between 450 and 550 mL, between 550 and 650 mL, between 650 and 750 mL, between 750 and 850 mL, between 850 and 950 mL, and between 950 and 1050 mL. In an embodiment, the cell expansion system includes a gas permeable cell bag with a volume range selected from the group consisting of between 1 L and 2 L, between 2 L and 3 L, between 3 L and 4 L, between 4 L and 5 L, between 5 L and 6 L, between 6 L and 7 L, between 7 L and 8 L, between 8 L and 9 L, between 9 L and 10 L, between 10 L and 11 L, between 11 L and 12 L, between 12 L and 13 L, between 13 L and 14 L, between 14 L and 15 L, between 15 L and 16 L, between 16 L and 17 L, between 17 L and 18 L, between 18 L and 19 L, and between 19 L and 20 L. In an embodiment, the cell expansion system includes a gas permeable cell bag with a volume range selected from the group consisting of between 0.5 L and 5 L, between 5 L and 10 L, between 10 L and 15 L, between 15 L and 20 L, between 20 L and 25 L, and between 25 L and 30 L. In an embodiment, the cell expansion system utilizes a rocking time of about 30 minutes, about 1 hour, about 2 hours, about 3 hours, about 4 hours, about 5 hours, about 6 hours, about 7 hours, about 8 hours, about 9 hours, about 10 hours, about 11 hours, about 12 hours, about 24 hours, about 2 days, about 3 days, about 4 days, about 5 days, about 6 days, about 7 days, about 8 days, about 9 days, about 10 days, about 11 days, about 12 days, about 13 days, about 14 days, about 15 days, about 16 days, about 17 days, about 18 days, about 19 days, about 20 days, about 21 days, about 22 days, about 23 days, about 24 days, about 25 days, about 26 days, about 27 days, and about 28 days. In an embodiment, the cell expansion system utilizes a rocking time of between 30 minutes and 1 hour, between 1 hour and 12 hours, between 12 hours and 1 day, between 1 day and 7 days, between 7 days and 14 days, between 14 days and 21 days, and between 21 days and 28 days. In an embodiment, the cell expansion system utilizes a rocking rate of about 2 rocks/minute, about 5 rocks/minute, about 10 rocks/minute, about 20 rocks/minute, about 30 rocks/minute, and about 40 rocks/minute. In an embodiment, the cell expansion system utilizes a rocking rate of between 2 rocks/minute and 5 rocks/minute, 5 rocks/minute and 10 rocks/minute, 10 rocks/minute and 20 rocks/minute, 20 rocks/minute and 30 rocks/minute, and 30 rocks/minute and 40 rocks/minute. In an embodiment, the cell expansion system utilizes a rocking angle of about 2.degree., about 3.degree., about 4.degree., about 5.degree., about 6.degree., about 7.degree., about 8.degree., about 9.degree., about 10.degree., about 11.degree., and about 12.degree.. In an embodiment, the cell expansion system utilizes a rocking angle of between 2.degree. and 3.degree., between 3.degree. and 4.degree., between 4.degree. and 5.degree., between 5.degree. and 6.degree., between 6.degree. and 7.degree., between 7.degree. and 8.degree., between 8.degree. and 9.degree., between 9.degree. and 10.degree., between 10.degree. and 11.degree., and between 11.degree. and 12.degree..
[1472] In an embodiment, a method of expanding TILs using TNFRSF agonists further comprises a step wherein TILs are selected for superior tumor reactivity. Any selection method known in the art may be used. For example, the methods described in U.S. Patent Application Publication No. 2016/0010058 A1, the disclosures of which are incorporated herein by reference, may be used for selection of TILs for superior tumor reactivity.
[1473] In an embodiment, the cell culture medium further comprises OKT-3 antibody. In a preferred embodiment, the cell culture medium comprises about 30 ng/mL of OKT-3 antibody. In an embodiment, the cell culture medium comprises about 0.1 ng/mL, about 0.5 ng/mL, about 1 ng/mL, about 2.5 ng/mL, about 5 ng/mL, about 7.5 ng/mL, about 10 ng/mL, about 15 ng/mL, about 20 ng/mL, about 25 ng/mL, about 30 ng/mL, about 35 ng/mL, about 40 ng/mL, about 50 ng/mL, about 60 ng/mL, about 70 ng/mL, about 80 ng/mL, about 90 ng/mL, about 100 ng/mL, about 200 ng/mL, about 500 ng/mL, and about 1 .mu.g/mL of OKT-3 antibody. In an embodiment, the cell culture medium comprises between 0.1 ng/mL and 1 ng/mL, between 1 ng/mL and 5 ng/mL, between 5 ng/mL and 10 ng/mL, between 10 ng/mL and 20 ng/mL, between 20 ng/mL and 30 ng/mL, between 30 ng/mL and 40 ng/mL, between 40 ng/mL and 50 ng/mL, or between 50 ng/mL and 100 ng/mL of OKT-3 antibody. In an embodiment, the cell culture medium comprises between 10 ng/mL and 60 ng/mL of OKT-3 antibody.
[1474] In an embodiment, the cell culture medium further comprises IL-2. In a preferred embodiment, the cell culture medium comprises about 3000 IU/mL of IL-2. In an embodiment, the cell culture medium comprises about 500 IU/mL, about 700 IU/mL, about 800 IU/mL, about 1000 IU/mL, about 1100 IU/mL, about 1200 IU/mL, about 1500 IU/mL, about 2000 IU/mL, about 2500 IU/mL, about 3000 IU/mL, about 3500 IU/mL, about 4000 IU/mL, about 4500 IU/mL, about 5000 IU/mL, about 5500 IU/mL, about 6000 IU/mL, about 6500 IU/mL, about 7000 IU/mL, about 7500 IU/mL, or about 8000 IU/mL of IL-2. In an embodiment, the cell culture medium comprises between 500 and 1000 IU/mL, 800 and 1200 IU/mL, 1000 and 2000 IU/mL, between 2000 and 3000 IU/mL, between 3000 and 4000 IU/mL, between 4000 and 5000 IU/mL, between 5000 and 6000 IU/mL, between 6000 and 7000 IU/mL, between 7000 and 8000 IU/mL, or between 8000 IU/mL of IL-2. In an embodiment, the cell culture medium comprises between 10 and 6000 IU/mL of IL-2. In an embodiment, the cell culture medium comprises between 500 and 2000 IU/mL of IL-2. In an embodiment, the cell culture medium comprises between 800 and 1100 IU/mL of IL-2.
[1475] In an embodiment, the cell culture medium further comprises IL-15, as described, e.g., in International Patent Application Publication Nos. WO 2015/189356 A1 and WO 2015/189356 A1, the disclosures of each of which are incorporated by reference herein. In an embodiment, the cell culture medium comprises about 0.1 ng/mL, about 0.5 ng/mL, about 1 ng/mL, about 2.5 ng/mL, about 5 ng/mL, about 7.5 ng/mL, about 10 ng/mL, about 15 ng/mL, about 20 ng/mL, about 25 ng/mL, about 30 ng/mL, about 35 ng/mL, about 40 ng/mL, about 50 ng/mL, about 60 ng/mL, about 70 ng/mL, about 80 ng/mL, about 90 ng/mL, about 100 ng/mL, about 200 ng/mL, about 500 ng/mL, or about 1 .mu.g/mL of IL-15. In an embodiment, the cell culture medium comprises between 0.1 ng/mL and 100 ng/mL, between 2 ng/mL and 50 ng/mL, or between 5 ng/mL and 25 ng/mL of IL-15. In an embodiment, the cell culture medium comprises between 10 ng/mL and 20 ng/mL, between 20 ng/mL and 30 ng/mL, between 30 ng/mL and 40 ng/mL, between 40 ng/mL and 50 ng/mL, between 50 ng/mL and 60 ng/mL, between 60 ng/mL and 70 ng/mL, between 70 ng/mL and 80 ng/mL, between 80 ng/mL and 90, ir between 90 ng/mL and 100 ng/mL of IL-15.
[1476] In an embodiment, the cell culture medium further comprises L-21, as described, e.g., in International Patent Application Publication Nos. WO 2015/189356 A1 and WO 2015/189356 A1, the disclosures of each of which are incorporated by reference herein. In an embodiment, the cell culture medium comprises about 0.1 ng/mL, about 0.5 ng/mL, about 1 ng/mL, about 2.5 ng/mL, about 5 ng/mL, about 7.5 ng/mL, about 10 ng/mL, about 15 ng/mL, about 20 ng/mL, about 25 ng/mL, about 30 ng/mL, about 35 ng/mL, about 40 ng/mL, about 50 ng/mL, about 60 ng/mL, about 70 ng/mL, about 80 ng/mL, about 90 ng/mL, about 100 ng/mL, about 200 ng/mL, about 500 ng/mL, or about 1 .mu.g/mL of L-21. In an embodiment, the cell culture medium comprises between 0.1 ng/mL and 100 ng/mL, between 2 ng/mL and 50 ng/mL, or between 5 ng/mL and 25 ng/mL of L-21. In an embodiment, the cell culture medium comprises between 10 ng/mL and 20 ng/mL, between 20 ng/mL and 30 ng/mL, between 30 ng/mL and 40 ng/mL, between 40 ng/mL and 50 ng/mL, between 50 ng/mL and 60 ng/mL, between 60 ng/mL and 70 ng/mL, between 70 ng/mL and 80 ng/mL, between 80 ng/mL and 90, ir between 90 ng/mL and 100 ng/mL of L-21.
[1477] In an embodiment, the cell culture medium further comprises IL-4 and/or IL-7.
[1478] In an embodiment, the TNFRSF agonists of the present invention may be used to expand T cells. Any of the foregoing embodiments of the present invention described for the expansion of TILs may also be applied to the expansion of T cells. In an embodiment, the TNFRSF agonists of the present invention may be used to expand CD8.sup.+ T cells. In an embodiment, the TNFRSF agonists of the present invention may be used to expand CD4.sup.+ T cells. In an embodiment, the TNFRSF agonists of the present invention may be used to expand T cells transduced with a chimeric antigen receptor (CAR-T). In an embodiment, the TNFRSF agonists of the present invention may be used to expand T cells comprising a modified T cell receptor (TCR). The CAR-T cells may be targeted against any suitable antigen, including CD19, as described in the art, e.g., in U.S. Pat. Nos. 7,070,995; 7,446,190; 8,399,645; 8,916,381; and 9,328,156; the disclosures of which are incorporated by reference herein. The modified TCR cells may be targeted against any suitable antigen, including NY-ESO-1, TRP-1, TRP-2, tyrosinase cancer antigen, MAGE-A3, SSX-2, and VEGFR2, or antigenic portions thereof, as described in the art, e.g., in U.S. Pat. Nos. 8,367,804 and 7,569,664, the disclosures of which are incorporated by reference herein.
[1479] In another embodiment, an exemplary TIL manufacturing/expansion process known as process 2A is schematically illustrated in FIG. 13. In certain aspects, the present methods produce TILs which are capable of increased replication cycles upon administration to a subject/patient and as such may provide additional therapeutic benefits over older TILs (i.e., TILs which have further undergone more rounds of replication prior to administration to a subject/patient). Features of younger TILs have been described in the literature, for example Donia, at al., Scandinavian Journal of Immunology, 75:157-167 (2012); Dudley et al., Clin Cancer Res, 16:6122-6131 (2010); Huang et al., J Immunother, 28(3):258-267 (2005); Besser et al., Clin Cancer Res, 19(17):OF1-OF9 (2013); Besser et al., J Immunother 32:415-423 (2009); Robbins, et al., J Immunol 2004; 173:7125-7130; Shen et al., J Immunother, 30:123-129 (2007); Zhou, et al., J Immunother, 28:53-62 (2005); and Tran, et al., J Immunother, 31:742-751 (2008), all of which are incorporated herein by reference in their entireties.
[1480] As discussed herein, the present invention can include a step relating to the restimulation of cyropreserved TILs to increase their metabolic activity and thus relative health prior to transplant into a patient, and methods of testing said metabolic health. As generally outlined herein, TILs are generally taken from a patient sample and manipulated to expand their number prior to transplant into a patient. In some embodiments, the TILs may be optionally genetically manipulated as discussed below.
[1481] In some embodiments, the TILs may be cryopreserved. Once thawed, they may also be restimulated to increase their metabolism prior to infusion into a patient.
[1482] In some embodiments, the first expansion (including processes referred to as the preREP) is shortened in comparison to conventional expansion methods to 7-14 days and the second expansion (including processes referred to as the REP) is shortened to 7-14 days, as discussed in detail below as well as in the examples and figures.
[1483] FIG. 14 illustrates an exemplary 2A process. As illustrated in FIG. 14 and further explained in detail below, in some embodiments, the first expansion (Step B) is shortened to 11 days and the second expansion (Step D) is shortened to 11 days. In some embodiments, the combination of the first and second expansions (Step B and Step D) is shortened to 22 days, as discussed in detail below and in the examples and figures. As will be appreciated, the process illustrated in FIG. 14 and described below is exemplary and the methods described herein encompass alterations and additions to the described steps as well as any combinations.
[1484] In general, TILs are initially obtained from a patient tumor sample ("primary TILs") and then expanded into a larger population for further manipulation as described herein, optionally cyropreserved, restimulated as outlined herein and optionally evaluated for phenotype and metabolic parameters as an indication of TIL health.
[1485] A patient tumor sample may be obtained using methods known in the art, generally via surgical resection, needle biopsy or other means for obtaining a sample that contains a mixture of tumor and TIL cells. In general, the tumor sample may be from any solid tumor, including primary tumors, invasive tumors or metastatic tumors. The tumor sample may also be a liquid tumor, such as a tumor obtained from a hematological malignancy. The solid tumor may be of any cancer type, including, but not limited to, breast, pancreatic, prostate, colorectal, lung, brain, renal, stomach, and skin (including but not limited to squamous cell carcinoma, basal cell carcinoma, and melanoma). In some embodiments, useful TILs are obtained from malignant melanoma tumors, as these have been reported to have particularly high levels of TILs. In some embodiments, the tumor is greater than about 1.5 cm but less than about 4 cm. In some embodiments, the tumor is less than 4 cm.
[1486] Once obtained, the tumor sample is generally fragmented using sharp dissection into small pieces of between 1 to about 8 mm.sup.3, with from about 2-3 mm.sup.3 being particularly useful. The TILs are cultured from these fragments using enzymatic tumor digests. Such tumor digests may be produced by incubation in enzymatic media (e.g., Roswell Park Memorial Institute (RPMI) 1640 buffer, 2 mM glutamate, 10 mcg/mL gentamicine, 30 units/mL of DNase and 1.0 mg/mL of collagenase) followed by mechanical dissociation (e.g., using a tissue dissociator). Tumor digests may be produced by placing the tumor in enzymatic media and mechanically dissociating the tumor for approximately 1 minute, followed by incubation for 30 minutes at 37.degree. C. in 5% CO.sub.2, followed by repeated cycles of mechanical dissociation and incubation under the foregoing conditions until only small tissue pieces are present. At the end of this process, if the cell suspension contains a large number of red blood cells or dead cells, a density gradient separation using FICOLL branched hydrophilic polysaccharide may be performed to remove these cells. Alternative methods known in the art may be used, such as those described in U.S. Patent Application Publication No. 2012/0244133 A1, the disclosure of which is incorporated by reference herein. Any of the foregoing methods may be used in any of the embodiments described herein for methods and processes of expanding TILs or methods treating a cancer.
[1487] In general, the harvested cell suspension is called a "primary cell population" or a "freshly harvested" cell population.
[1488] In an embodiment, TILs can be initially cultured from enzymatic tumor digests and tumor fragments obtained from patients.
[1489] In some embodiments, the TILs, are obtained from tumor fragments. In some embodiments, the tumor fragment is obtained sharp dissection. In some embodiments, the tumor fragment is between about 1 mm.sup.3 and 10 mm.sup.3. In some embodiments, the tumor fragment is between about 1 mm.sup.3 and 8 mm.sup.3. In some embodiments, the tumor fragment is about 1 mm.sup.3. In some embodiments, the tumor fragment is about 2 mm.sup.3. In some embodiments, the tumor fragment is about 3 mm.sup.3. In some embodiments, the tumor fragment is about 4 mm.sup.3. In some embodiments, the tumor fragment is about 5 mm.sup.3. In some embodiments, the tumor fragment is about 6 mm.sup.3. In some embodiments, the tumor fragment is about 7 mm.sup.3. In some embodiments, the tumor fragment is about 8 mm.sup.3. In some embodiments, the tumor fragment is about 9 mm.sup.3. In some embodiments, the tumor fragment is about 10 mm.sup.3. In some embodiments, about the tumor fragment is about 8-27 mm.sup.3. In some embodiments, about the tumor fragment is about 10-25 mm.sup.3. In some embodiments, about the tumor fragment is about 15-25 mm.sup.3. In some embodiments, the tumor fragment is about 8-20 mm.sup.3. In some embodiments, the tumor fragment is about 15-20 mm.sup.3. In some embodiments, the tumor fragment is about 8-15 mm.sup.3. In some embodiments, the tumor fragment is about 8-10 mm.sup.3.
[1490] In some embodiments, the number of tumor fragments is about 40 to about 50 tumor fragments. In some embodiments, the number of tumor fragments is about 40 tumor fragments. In some embodiments, the number of tumor fragments is about 50 tumor fragments. In some embodiments, the tumor fragment size is about 8-27 mm.sup.3 and there are less than about 50 tumor fragments.
[1491] In some embodiments, the TILs, are obtained from tumor digests. In some embodiments, tumor digests were generated by incubation in enzyme media, for example but not limited to RPMI 1640, 2 mM GlutaMAX, 10 mg/mL gentamicin, 30 U/mL DNase, and 1.0 mg/mL collagenase, followed by mechanical dissociation (GentleMACS, Miltenyi Biotec, Auburn, Calif.). After placing the tumor in enzyme media, the tumor can be mechanically dissociated for approximately 1 minute. The solution can then be incubated for 30 minutes at 37.degree. C. in 5% CO.sub.2 and it then mechanically disrupted again for approximately 1 minute. After being incubated again for 30 minutes at 37.degree. C. in 5% CO.sub.2, the tumor can be mechanically disrupted a third time for approximately 1 minute. In some embodiments, after the third mechanical disruption if large pieces of tissue were present, 1 or 2 additional mechanical dissociations were applied to the sample, with or without 30 additional minutes of incubation at 37.degree. C. in 5% CO.sub.2. In some embodiments, at the end of the final incubation if the cell suspension contained a large number of red blood cells or dead cells, a density gradient separation using Ficoll can be performed to remove these cells.
[1492] After dissection or digestion of tumor fragments in Step A, the resulting cells are cultured in serum containing IL-2 under conditions that favor the growth of TILs over tumor and other cells. In some embodiments, the tumor digests are incubated in 2 mL wells in media comprising inactivated human AB serum with 6000 IU/mL of IL-2. This primary cell population is cultured for a period of days, generally from 3 to 14 days, resulting in a bulk TIL population, generally about 1.times.10.sup.8 bulk TIL cells. In some embodiments, this primary cell population is cultured for a period of 7 to 14 days, resulting in a bulk TIL population, generally about 1.times.10.sup.8 bulk TIL cells. In some embodiments, this primary cell population is cultured for a period of 10 to 14 days, resulting in a bulk TIL population, generally about 1.times.10.sup.8 bulk TIL cells. In some embodiments, this primary cell population is cultured for a period of about 11 days, resulting in a bulk TIL population, generally about 1.times.10.sup.8 bulk TIL cells. In some embodiments, this primary cell population is cultured for a period of about 11 days, resulting in a bulk TIL population, generally less than or equal to about 200.times.10.sup.6 bulk TIL cells.
[1493] In a preferred embodiment, expansion of TILs may be performed using an initial bulk TIL expansion step (Step B as pictured in FIG. 14, which can include processes referred to as pre-REP) as described below and herein, followed by a second expansion (Step D, including processes referred to as rapid expansion protocol (REP) steps) as described below under Step D and herein, followed by optional cryopreservation, and followed by a second Step D (including processes referred to as restimulation REP steps) as described below and herein. The TILs obtained from this process may be optionally characterized for phenotypic characteristics and metabolic parameters as described herein.
[1494] In embodiments where TIL cultures are initiated in 24-well plates, for example, using Costar 24-well cell culture cluster, flat bottom (Corning Incorporated, Corning, N.Y., each well can be seeded with 1.times.10.sup.6 tumor digest cells or one tumor fragment in 2 mL of complete medium (CM) with IL-2 (6000 IU/mL; Chiron Corp., Emeryville, Calif.). In some embodiments, the tumor fragment is between about 1 mm.sup.3 and 10 mm.sup.3.
[1495] In some embodiments, CM for Step B consists of RPMI 1640 with GlutaMAX, supplemented with 10% human AB serum, 25 mM HEPES, and 10 mg/mL gentamicin. In embodiments where cultures are initiated in gas-permeable flasks with a 40 mL capacity and a 10 cm.sup.2 gas-permeable silicon bottom (for example, G-Rex10; Wilson Wolf Manufacturing, New Brighton, Minn.) (FIG. 1), each flask was loaded with 10-40.times.10.sup.6 viable tumor digest cells or 5-30 tumor fragments in 10-40 mL of CM with IL-2. Both the G-Rex10 and 24-well plates were incubated in a humidified incubator at 37.degree. C. in 5% CO.sub.2 and 5 days after culture initiation, half the media was removed and replaced with fresh CM and IL-2 and after day 5, half the media was changed every 2-3 days.
[1496] In an embodiment, the cell culture medium further comprises IL-2. In a preferred embodiment, the cell culture medium comprises about 3000 IU/mL of IL-2. In an embodiment, the cell culture medium comprises about 1000 IU/mL, about 1500 IU/mL, about 2000 IU/mL, about 2500 IU/mL, about 3000 IU/mL, about 3500 IU/mL, about 4000 IU/mL, about 4500 IU/mL, about 5000 IU/mL, about 5500 IU/mL, about 6000 IU/mL, about 6500 IU/mL, about 7000 IU/mL, about 7500 IU/mL, or about 8000 IU/mL of IL-2. In an embodiment, the cell culture medium comprises between 1000 and 2000 IU/mL, between 2000 and 3000 IU/mL, between 3000 and 4000 IU/mL, between 4000 and 5000 IU/mL, between 5000 and 6000 IU/mL, between 6000 and 7000 IU/mL, between 7000 and 8000 IU/mL, or between 8000 IU/mL of IL-2.
[1497] In some embodiments, the first expansion (including processes referred to as the pre-REP; Step B) process is shortened to 3-14 days, as discussed in the examples and figures. In some embodiments, the first expansion of Step B is shortened to 7-14 days, as discussed in the Examples and shown in FIGS. 4 and 5. In some embodiments, the first expansion of Step B is shortened to 10-14 days, as discussed in the Examples and shown in FIGS. 4 and 5. In some embodiments, the first expansion of Step B is shortened to 11 days, as discussed in the Examples and shown in FIGS. 4 and 5.
[1498] In some embodiments, IL-2, IL-7, IL-15, and IL-21 as well as combinations thereof can be included during Step B processes as described herein.
[1499] In some embodiments, Step B is performed in a closed system bioreactor. In some embodiments, a closed system is employed for the TIL expansion, as described herein. In some embodiments, a single bioreactor is employed. In some embodiments, the single bioreactor employed is for example a GREX-10 or a GREX-100.
[1500] In some embodiments, the bulk TIL population from Step B can be cryopreserved immediately, using methods known in the art and described herein. Alternatively, the bulk TIL population can be subjected to a second expansion (REP) and then cryopreserved as discussed below.
[1501] In some embodiments, the Step B TILs are not stored and the Step B TILs proceed directly to Step D. In some embodiments, the transition occurs in a closed system, as further described herein.
[1502] In some embodiments, the TIL cell population is expanded in number after harvest and initial bulk processing (i.e., after Step A and Step B). This is referred to herein as the second expansion, which can include expansion processes generally referred to in the art as a rapid expansion process (REP). The second expansion is generally accomplished using culture media comprising a number of components, including feeder cells, a cytokine source, and an anti-CD3 antibody, in a gas-permeable container. In some embodiments, the second expansion can include scaling-up in order to increase the number of TILs obtained in the second expansion.
[1503] In an embodiment, REP and/or the second expansion can be performed in a gas permeable container using the methods of the present disclosure. For example, TILs can be rapidly expanded using non-specific T-cell receptor stimulation in the presence of interleukin-2 (IL-2) or interleukin-15 (IL-15). The non-specific T-cell receptor stimulus can include, for example, about 30 ng/ml of OKT3, a mouse monoclonal anti-CD3 antibody (commercially available from Ortho-McNeil, Raritan, N.J. or Miltenyi Biotech, Auburn, Calif.). TILs can be rapidly expanded further stimulation of the TILs in vitro with one or more antigens, including antigenic portions thereof, such as epitope(s), of the cancer, which can be optionally expressed from a vector, such as a human leukocyte antigen A2 (HLA-A2) binding peptide, e.g., 0.3 VM MART-1:26-35 (27 L) or gpl 00:209-217 (210M), optionally in the presence of a T-cell growth factor, such as 300 IU/mL IL-2 or IL-15. Other suitable antigens may include, e.g., NY-ESO-1, TRP-1, TRP-2, tyrosinase cancer antigen, MAGE-A3, SSX-2, and VEGFR2, or antigenic portions thereof. TIL may also be rapidly expanded by re-stimulation with the same antigen(s) of the cancer pulsed onto HLA-A2-expressing antigen-presenting cells. Alternatively, the TILs can be further re-stimulated with, e.g., example, irradiated, autologous lymphocytes or with irradiated HLA-A2+ allogeneic lymphocytes and IL-2.
[1504] In an embodiment, the cell culture medium further comprises IL-2. In a preferred embodiment, the cell culture medium comprises about 3000 IU/mL of IL-2. In an embodiment, the cell culture medium comprises about 1000 IU/mL, about 1500 IU/mL, about 2000 IU/mL, about 2500 IU/mL, about 3000 IU/mL, about 3500 IU/mL, about 4000 IU/mL, about 4500 IU/mL, about 5000 IU/mL, about 5500 IU/mL, about 6000 IU/mL, about 6500 IU/mL, about 7000 IU/mL, about 7500 IU/mL, or about 8000 IU/mL of IL-2. In an embodiment, the cell culture medium comprises between 1000 and 2000 IU/mL, between 2000 and 3000 IU/mL, between 3000 and 4000 IU/mL, between 4000 and 5000 IU/mL, between 5000 and 6000 IU/mL, between 6000 and 7000 IU/mL, between 7000 and 8000 IU/mL, or between 8000 IU/mL of IL-2.
[1505] In an embodiment, the cell culture medium comprises OKT3 antibody. In a preferred embodiment, the cell culture medium comprises about 30 ng/mL of OKT3 antibody. In an embodiment, the cell culture medium comprises about 0.1 ng/mL, about 0.5 ng/mL, about 1 ng/mL, about 2.5 ng/mL, about 5 ng/mL, about 7.5 ng/mL, about 10 ng/mL, about 15 ng/mL, about 20 ng/mL, about 25 ng/mL, about 30 ng/mL, about 35 ng/mL, about 40 ng/mL, about 50 ng/mL, about 60 ng/mL, about 70 ng/mL, about 80 ng/mL, about 90 ng/mL, about 100 ng/mL, about 200 ng/mL, about 500 ng/mL, and about 1 .mu.g/mL of OKT3 antibody. In an embodiment, the cell culture medium comprises between 0.1 ng/mL and 1 ng/mL, between 1 ng/mL and 5 ng/mL, between 5 ng/mL and 10 ng/mL, between 10 ng/mL and 20 ng/mL, between 20 ng/mL and 30 ng/mL, between 30 ng/mL and 40 ng/mL, between 40 ng/mL and 50 ng/mL, and between 50 ng/mL and 100 ng/mL of OKT3 antibody.
[1506] In some embodiments, IL-2, IL-7, IL-15, and IL-21 as well as combinations thereof can be included during the second expansion in Step D processes as described herein.
[1507] In some embodiments, the second expansion can be conducted in a supplemented cell culture medium comprising IL-2, OKT-3, and antigen-presenting feeder cells.
[1508] In some embodiments the antigen-presenting feeder cells (APCs) are PBMCs. In an embodiment, the ratio of TILs to PBMCs and/or antigen-presenting cells in the rapid expansion and/or the second expansion is about 1 to 25, about 1 to 50, about 1 to 100, about 1 to 125, about 1 to 150, about 1 to 175, about 1 to 200, about 1 to 225, about 1 to 250, about 1 to 275, about 1 to 300, about 1 to 325, about 1 to 350, about 1 to 375, about 1 to 400, or about 1 to 500. In an embodiment, the ratio of TILs to PBMCs in the rapid expansion and/or the second expansion is between 1 to 50 and 1 to 300. In an embodiment, the ratio of TILs to PBMCs in the rapid expansion and/or the second expansion is between 1 to 100 and 1 to 200.
[1509] In an embodiment, REP and/or the second expansion is performed in flasks with the bulk TILs being mixed with a 100- or 200-fold excess of inactivated feeder cells, 30 mg/mL OKT3 anti-CD3 antibody and 3000 IU/mL IL-2 in 150 ml media. Media replacement is done (generally 2/3 media replacement via respiration with fresh media) until the cells are transferred to an alternative growth chamber. Alternative growth chambers include GRex flasks and gas permeable containers as more fully discussed below.
[1510] In some embodiments, the second expansion (also referred to as the REP process) is shortened to 7-14 days, as discussed in the examples and figures. In some embodiments, the second expansion is shortened to 11 days.
[1511] In an embodiment, REP and/or the second expansion may be performed using T-175 flasks and gas permeable bags as previously described (Tran, et al., J. Immunother. 2008, 31, 742-51; Dudley, et al., J. Immunother. 2003, 26, 332-42) or gas permeable cultureware (G-Rex flasks). For TIL rapid expansion and/or second expansion in T-175 flasks, 1.times.10.sup.6 TILs suspended in 150 mL of media may be added to each T-175 flask. The TILs may be cultured in a 1 to 1 mixture of CM and AIM-V medium, supplemented with 3000 IU per mL of IL-2 and 30 ng per ml of anti-CD3. The T-175 flasks may be incubated at 37.degree. C. in 5% CO.sub.2. Half the media may be exchanged on day 5 using 50/50 medium with 3000 IU per mL of IL-2. On day 7 cells from two T-175 flasks may be combined in a 3 L bag and 300 mL of AIM V with 5% human AB serum and 3000 IU per mL of IL-2 was added to the 300 ml of TIL suspension. The number of cells in each bag was counted every day or two and fresh media was added to keep the cell count between 0.5 and 2.0.times.10.sup.6 cells/mL.
[1512] In an embodiment, REP and/or the second expansion may be performed in 500 mL capacity gas permeable flasks with 100 cm gas-permeable silicon bottoms (G-Rex 100, commercially available from Wilson Wolf Manufacturing Corporation, New Brighton, Minn., USA), 5.times.10.sup.6 or 10.times.10.sup.6 TIL may be cultured with PBMCs in 400 mL of 50/50 medium, supplemented with 5% human AB serum, 3000 IU per mL of IL-2 and 30 ng per ml of anti-CD3 (OKT3). The G-Rex 100 flasks may be incubated at 37.degree. C. in 5% CO.sub.2. On day 5, 250 mL of supernatant may be removed and placed into centrifuge bottles and centrifuged at 1500 rpm (491.times.g) for 10 minutes. The TIL pellets may be re-suspended with 150 mL of fresh medium with 5% human AB serum, 3000 IU per mL of IL-2, and added back to the original G-Rex 100 flasks. When TIL are expanded serially in G-Rex 100 flasks, on day 7 the TIL in each G-Rex 100 may be suspended in the 300 mL of media present in each flask and the cell suspension may be divided into 3 100 mL aliquots that may be used to seed 3 G-Rex 100 flasks. Then 150 mL of AIM-V with 5% human AB serum and 3000 IU per mL of IL-2 may be added to each flask. The G-Rex 100 flasks may be incubated at 37.degree. C. in 5% CO.sub.2 and after 4 days 150 mL of AIM-V with 3000 IU per mL of IL-2 may be added to each G-Rexl 00 flask. The cells may be harvested on day 14 of culture.
[1513] In an embodiment, REP and/or the second expansion is performed in flasks with the bulk TILs being mixed with a 100- or 200-fold excess of inactivated feeder cells, 30 mg/mL OKT3 anti-CD3 antibody and 3000 IU/mL IL-2 in 150 ml media. Media replacement is done (generally 2/3 media replacement via respiration with fresh media) until the cells are transferred to an alternative growth chamber. Alternative growth chambers include GRex flasks and gas permeable containers as more fully discussed below.
[1514] In an embodiment, REP and/or the second expansion is performed and further comprises a step wherein TILs are selected for superior tumor reactivity. Any selection method known in the art may be used. For example, the methods described in U.S. Patent Application Publication No. 2016/0010058 A1, the disclosures of which are incorporated herein by reference, may be used for selection of TILs for superior tumor reactivity.
[1515] REP and/or the second expansion of TIL can be performed using T-175 flasks and gas-permeable bags as previously described (Tran K Q, Zhou J, Durflinger K H, et al., 2008, J Immunother., 31:742-751, and Dudley M E, Wunderlich J R, Shelton T E, et al. 2003, J Immunother., 26:332-342) or gas-permeable G-Rex flasks. In some embodiments, REP and/or the second expansion is performed using flasks. In some embodiments, REP is performed using gas-permeable G-Rex flasks. For TIL REP and/or the second expansion in T-175 flasks, about 1.times.10.sup.6 TIL are suspended in about 150 mL of media and this is added to each T-175 flask. The TIL are cultured with irradiated (50 Gy) allogeneic PBMC as "feeder" cells at a ratio of 1 to 100 and the cells were cultured in a 1 to 1 mixture of CM and AIM-V medium (50/50 medium), supplemented with 3000 IU/mL of IL-2 and 30 ng/mL of anti-CD3. The T-175 flasks are incubated at 37.degree. C. in 5% CO.sub.2. In some embodiments, half the media is changed on day 5 using 50/50 medium with 3000 IU/mL of IL-2. In some embodiments, on day 7, cells from 2 T-175 flasks are combined in a 3 L bag and 300 mL of AIM-V with 5% human AB serum and 3000 IU/mL of IL-2 is added to the 300 mL of TIL suspension. The number of cells in each bag can be counted every day or two and fresh media can be added to keep the cell count between about 0.5 and about 2.0.times.10.sup.6 cells/mL.
[1516] For TIL REP and/or the second expansion in 500 mL capacity flasks with 100 cm.sup.2 gas-permeable silicon bottoms (G-Rex100, Wilson Wolf) (FIG. 1), about 5.times.10.sup.6 or 10.times.10.sup.6 TIL are cultured with irradiated allogeneic PBMC at a ratio of 1 to 100 in 400 mL of 50/50 medium, supplemented with 3000 IU/mL of IL-2 and 30 ng/mL of anti-CD3. The G-Rex100 flasks are incubated at 37.degree. C. in 5% CO.sub.2. In some embodiments, on day 5, 250 mL of supernatant is removed and placed into centrifuge bottles and centrifuged at 1500 rpm (491 g) for 10 minutes. The TIL pellets can then be resuspended with 150 mL of fresh 50/50 medium with 3000 IU/mL of IL-2 and added back to the original G-Rex100 flasks. In embodiments where TILs are expanded serially in G-Rex100 flasks, on day 7 the TIL in each G-Rex100 are suspended in the 300 mL of media present in each flask and the cell suspension was divided into three 100 mL aliquots that are used to seed 3 G-Rex100 flasks. Then 150 mL of AIM-V with 5% human AB serum and 3000 IU/mL of IL-2 is added to each flask. The G-Rex100 flasks are incubated at 37.degree. C. in 5% CO.sub.2 and after 4 days 150 mL of AIM-V with 3000 IU/mL of IL-2 is added to each G-Rex100 flask. The cells are harvested on day 14 of culture.
[1517] In an embodiment, the second expansion procedures described herein (Step D, including REP) require an excess of feeder cells during REP TIL expansion and/or during the second expansion. In many embodiments, the feeder cells are peripheral blood mononuclear cells (PBMCs) obtained from standard whole blood units from healthy blood donors. The PBMCs are obtained using standard methods such as Ficoll-Paque gradient separation.
[1518] In general, the allogenic PBMCs are inactivated, either via irradiation or heat treatment, and used in the REP procedures, as described in the examples, in particular example 14, which provides an exemplary protocol for evaluating the replication incompetence of irradiate allogeneic PBMCs.
[1519] In some embodiments, PBMCs are considered replication incompetent and accepted for use in the TIL expansion procedures described herein if the total number of viable cells on day 14 is less than the initial viable cell number put into culture on day 0 of the REP and/or day 0 of the second expansion (i.e., the start day of the second expansion).
[1520] In some embodiments, PBMCs are considered replication incompetent and accepted for use in the TIL expansion procedures described herein if the total number of viable cells, cultured in the presence of OKT3 and IL-2, on day 7 and day 14 has not increased from the initial viable cell number put into culture on day 0 of the REP and/or day 0 of the second expansion (i.e., the start day of the second expansion). In some embodiments, the PBMCs are cultured in the presence of 30 ng/ml OKT3 antibody and 3000 IU/ml IL-2.
[1521] In some embodiments, PBMCs are considered replication incompetent and accepted for use in the TIL expansion procedures described herein if the total number of viable cells, cultured in the presence of OKT3 and IL-2, on day 7 and day 14 has not increased from the initial viable cell number put into culture on day 0 of the REP and/or day 0 of the second expansion (i.e., the start day of the second expansion). In some embodiments, the PBMCs are cultured in the presence of 5-60 ng/ml OKT3 antibody and 1000-6000 IU/ml IL-2. In some embodiments, the PBMCs are cultured in the presence of 10-50 ng/ml OKT3 antibody and 2000-5000 IU/ml IL-2. In some embodiments, the PBMCs are cultured in the presence of 20-40 ng/ml OKT3 antibody and 2000-4000 IU/ml IL-2. In some embodiments, the PBMCs are cultured in the presence of 25-35 ng/ml OKT3 antibody and 2500-3500 IU/ml IL-2.
[1522] In an embodiment, artificial antigen presenting cells are used in the REP stage as a replacement for, or in combination with, PBMCs.
[1523] The expansion methods described herein generally use culture media with high doses of a cytokine, in particular IL-2, as is known in the art.
[1524] Alternatively, using combinations of cytokines for the rapid expansion and or second expansion of TILs is additionally possible, with combinations of two or more of IL-2, IL-15 and IL-21 as is generally outlined in International Publication No. WO 2015/189356 and W International Publication No. WO 2015/189357, hereby expressly incorporated by reference in their entirety. Thus, possible combinations include IL-2 and IL-15, IL-2 and IL-21, IL-15 and IL-21 and IL-2, IL-15 and IL-21, with the latter finding particular use in many embodiments. The use of combinations of cytokines specifically favors the generation of lymphocytes, and in particular T-cells as described therein.
[1525] In some embodiments, the culture media used in expansion methods described herein (including REP) also includes an anti-CD3 antibody. An anti-CD3 antibody in combination with IL-2 induces T cell activation and cell division in the TIL population. This effect can be seen with full length antibodies as well as Fab and F(ab')2 fragments, with the former being generally preferred; see, e.g., Tsoukas et al., J. Immunol. 1985, 135, 1719, hereby incorporated by reference in its entirety.
[1526] As will be appreciated by those in the art, there are a number of suitable anti-human CD3 antibodies that find use in the invention, including anti-human CD3 polyclonal and monoclonal antibodies from various mammals, including, but not limited to, murine, human, primate, rat, and canine antibodies. In particular embodiments, the OKT3 anti-CD3 antibody is used (commercially available from Ortho-McNeil, Raritan, N.J. or Miltenyi Biotech, Auburn, Calif.).
[1527] After the second expansion step, cells can be harvested. In some embodiments the TILs are harvested after one, two, three, four or more second expansion steps.
[1528] TILs can be harvested in any appropriate and sterile manner, including for example by centrifugation. Methods for TIL harvesting are well known in the art and any such know methods can be employed with the present process. In some embodiments, TILs are harvested using an automated system. In some embodiments, TILs are harvest using a semi-automated system. In some embodiments, the TILs from the second expansion are harvested using a semi-automated machine. In some embodiments, the LOVO system is employed (commercially available from Benchmark Electronics, for example). In some embodiments, the harvesting step includes wash the TILs, formulating the TILs, and/or aliquoting the TILs. In some embodiments, the cells are optionally frozen after harvesting or as part of harvesting.
[1529] After Steps A through E are complete, cells are transferred to a container for use in administration to a patient.
[1530] In an embodiment, TILs expanded using APCs of the present disclosure are administered to a patient as a pharmaceutical composition. In an embodiment, the pharmaceutical composition is a suspension of TILs in a sterile buffer. TILs expanded using PBMCs of the present disclosure may be administered by any suitable route as known in the art. In some embodiments, the T-cells are administered as a single intra-arterial or intravenous infusion, which preferably lasts approximately 30 to 60 minutes. Other suitable routes of administration include intraperitoneal, intrathecal, and intralymphatic.
[1531] As will be appreciated, any of the steps A through F described above can be repeated any number of times and may in addition be conducted in different orders than described above.
[1532] In some embodiments, one or more of the expansion steps may be repeated prior to the Final Formulation Step F. Such additional expansion steps may include the elements of the first and/or second expansion steps described above (e.g., include the described components in the cell culture medium). The additional expansion steps may further include additional elements, including additional components in the cell culture medium that are supplemented into the cell culture medium before and/or during the additional expansion steps.
[1533] In further embodiments, any of the expansion steps described in FIG. 14 and in the above paragraphs may be preceded or followed by a cryopreservation step in which the cells produced during an expansion step are preserved using methods known in the art for storage until needed for the remaining steps of the manufacturing/expansion process.
[1534] In an embodiment, the invention includes a kit for expanding TILs according to any of the foregoing methods.
Pharmaceutical Compositions, Dosages, and Dosing Regimens for TILs
[1535] In an embodiment, TILs expanded using processes and methods of the present disclosure are administered to a patient as a pharmaceutical composition. In an embodiment, the pharmaceutical composition is a suspension of TILs in a sterile buffer. TILs expanded using processes and methods of the present disclosure may be administered by any suitable route as known in the art. Preferably, the TILs are administered as a single intra-arterial or intravenous infusion, which preferably lasts approximately 30 to 60 minutes. Other suitable routes of administration include intraperitoneal, intrathecal, and intralymphatic administration.
[1536] Any suitable dose of TILs can be administered. Preferably, from about 2.3.times.10.sup.10 to about 13.7.times.10.sup.10 TILs are administered, with an average of around 7.8.times.10.sup.10 TILs, particularly if the cancer is melanoma. In an embodiment, about 1.2.times.10.sup.10 to about 4.3.times.10.sup.10 of TILs are administered.
[1537] In some embodiments, the number of the TILs provided in the pharmaceutical compositions of the invention is about 1.times.10.sup.6, 2.times.10.sup.6, 3.times.10.sup.6, 4.times.10.sup.6, 5.times.10.sup.6, 6.times.10.sup.6, 7.times.10.sup.6, 8.times.10.sup.6, 9.times.10.sup.6, 1.times.10.sup.7, 2.times.10.sup.7, 3.times.10.sup.7, 4.times.10.sup.7, 5.times.10.sup.7, 6.times.10.sup.7, 7.times.10.sup.7, 8.times.10.sup.7, 9.times.10.sup.7, 1.times.10.sup.8, 2.times.10.sup.8, 3.times.10.sup.8, 4.times.10.sup.8, 5.times.10.sup.8, 6.times.10.sup.8, 7.times.10.sup.8, 8.times.10.sup.8, 9.times.10.sup.8, 1.times.10.sup.9, 2.times.10.sup.9, 3.times.10.sup.9, 4.times.10.sup.9, 5.times.10.sup.9, 6.times.10.sup.9, 7.times.10.sup.9, 8.times.10.sup.9, 9.times.10.sup.9, 1.times.10.sup.10, 2.times.10.sup.10, 3.times.10.sup.10, 4.times.10.sup.10, 5.times.10.sup.10, 6.times.10.sup.10, 7.times.10.sup.10, 8.times.10.sup.10, 9.times.10.sup.10, 1.times.10.sup.11, 2.times.10.sup.11, 3.times.10.sup.11, 4.times.10.sup.11, 5.times.10.sup.11, 6.times.10.sup.11, 7.times.10.sup.118.times.10.sup.11, 9.times.10.sup.11, 1.times.10.sup.12, 2.times.10.sup.12, 3.times.10.sup.124.times.10.sup.12, 5.times.10.sup.12, 6.times.10.sup.12, 7.times.10.sup.12, 8.times.10.sup.12, 9.times.10.sup.12, 1.times.10.sup.13, 2.times.10.sup.13, 3.times.10.sup.13, 4.times.10.sup.13, 5.times.10.sup.13, 6.times.10.sup.13, 7.times.10.sup.13, 8.times.10.sup.13, and 9.times.10.sup.13. In an embodiment, the number of the TILs provided in the pharmaceutical compositions of the invention is in the range of 1.times.10.sup.6 to 5.times.10.sup.6, 5.times.10.sup.6 to 1.times.10.sup.7, 1.times.10.sup.7 to 5.times.10.sup.7, 5.times.10.sup.7 to 1.times.10.sup.8, 1.times.10.sup.8 to 5.times.10.sup.8, 5.times.10.sup.8 to 1.times.10.sup.9, 1.times.10.sup.9 to 5.times.10.sup.9, 5.times.10.sup.9 to 1.times.10.sup.10, 1.times.1010 to 5.times.10.sup.10, 5.times.10.sup.10 to 1.times.10.sup.11, 5.times.10.sup.11 to 1.times.10.sup.12, 1.times.10.sup.12 to 5.times.10.sup.12, and 5.times.10.sup.12 to 1.times.10.sup.13.
[1538] In some embodiments, the concentration of the TILs provided in the pharmaceutical compositions of the invention is less than, for example, 100%, 90%, 80%, 70%, 60%, 50%, 40%, 30%, 20%, 19%, 18%, 17%, 16%, 15%, 14%, 13%, 12%, 11%, 10%, 9%, 8%, 7%, 6%, 5%, 4%, 3%, 2%, 1%, 0.5%, 0.4%, 0.3%, 0.2%, 0.1%, 0.09%, 0.08%, 0.07%, 0.06%, 0.05%, 0.04%, 0.03%, 0.02%, 0.01%, 0.009%, 0.008%, 0.007%, 0.006%, 0.005%, 0.004%, 0.003%, 0.002%, 0.001%, 0.0009%, 0.0008%, 0.0007%, 0.0006%, 0.0005%, 0.0004%, 0.0003%, 0.0002% or 0.0001% w/w, w/v or v/v of the pharmaceutical composition.
[1539] In some embodiments, the concentration of the TILs provided in the pharmaceutical compositions of the invention is greater than 90%, 80%, 70%, 60%, 50%, 40%, 30%, 20%, 19.75%, 19.50%, 19.25% 19%, 18.75%, 18.50%, 18.25% 18%, 17.75%, 17.50%, 17.25% 17%, 16.75%, 16.50%, 16.25% 16%, 15.75%, 15.50%, 15.25% 15%, 14.75%, 14.50%, 14.25% 14%, 13.75%, 13.50%, 13.25% 13%, 12.75%, 12.50%, 12.25% 12%, 11.75%, 11.50%, 11.25% 11%, 10.75%, 10.50%, 10.25% 10%, 9.75%, 9.50%, 9.25% 9%, 8.75%, 8.50%, 8.25% 8%, 7.75%, 7.50%, 7.25% 7%, 6.75%, 6.50%, 6.25% 6%, 5.75%, 5.50%, 5.25% 5%, 4.75%, 4.50%, 4.25%, 4%, 3.75%, 3.50%, 3.25%, 3%, 2.75%, 2.50%, 2.25%, 2%, 1.75%, 1.50%, 125%, 1%, 0.5%, 0.4%, 0.3%, 0.2%, 0.1%, 0.09%, 0.08%, 0.07%, 0.06%, 0.05%, 0.04%, 0.03%, 0.02%, 0.01%, 0.009%, 0.008%, 0.007%, 0.006%, 0.005%, 0.004%, 0.003%, 0.002%, 0.001%, 0.0009%, 0.0008%, 0.0007%, 0.0006%, 0.0005%, 0.0004%, 0.0003%, 0.0002% or 0.0001% w/w, w/v, or v/v of the pharmaceutical composition.
[1540] In some embodiments, the concentration of the TILs provided in the pharmaceutical compositions of the invention is in the range from about 0.0001% to about 50%, about 0.001% to about 40%, about 0.01% to about 30%, about 0.02% to about 29%, about 0.03% to about 28%, about 0.04% to about 27%, about 0.05% to about 26%, about 0.06% to about 25%, about 0.07% to about 24%, about 0.08% to about 23%, about 0.09% to about 22%, about 0.1% to about 21%, about 0.2% to about 20%, about 0.3% to about 19%, about 0.4% to about 18%, about 0.5% to about 17%, about 0.6% to about 16%, about 0.7% to about 15%, about 0.8% to about 14%, about 0.9% to about 12% or about 1% to about 10% w/w, w/v or v/v of the pharmaceutical composition.
[1541] In some embodiments, the concentration of the TILs provided in the pharmaceutical compositions of the invention is in the range from about 0.001% to about 10%, about 0.01% to about 5%, about 0.02% to about 4.5%, about 0.03% to about 4%, about 0.04% to about 3.5%, about 0.05% to about 3%, about 0.06% to about 2.5%, about 0.07% to about 2%, about 0.08% to about 1.5%, about 0.09% to about 1%, about 0.1% to about 0.9% w/w, w/v or v/v of the pharmaceutical composition.
[1542] In some embodiments, the amount of the TILs provided in the pharmaceutical compositions of the invention is equal to or less than 10 g, 9.5 g, 9.0 g, 8.5 g, 8.0 g, 7.5 g, 7.0 g, 6.5 g, 6.0 g, 5.5 g, 5.0 g, 4.5 g, 4.0 g, 3.5 g, 3.0 g, 2.5 g, 2.0 g, 1.5 g, 1.0 g, 0.95 g, 0.9 g, 0.85 g, 0.8 g, 0.75 g, 0.7 g, 0.65 g, 0.6 g, 0.55 g, 0.5 g, 0.45 g, 0.4 g, 0.35 g, 0.3 g, 0.25 g, 0.2 g, 0.15 g, 0.1 g, 0.09 g, 0.08 g, 0.07 g, 0.06 g, 0.05 g, 0.04 g, 0.03 g, 0.02 g, 0.01 g, 0.009 g, 0.008 g, 0.007 g, 0.006 g, 0.005 g, 0.004 g, 0.003 g, 0.002 g, 0.001 g, 0.0009 g, 0.0008 g, 0.0007 g, 0.0006 g, 0.0005 g, 0.0004 g, 0.0003 g, 0.0002 g, or 0.0001 g.
[1543] In some embodiments, the amount of the TILs provided in the pharmaceutical compositions of the invention is more than 0.0001 g, 0.0002 g, 0.0003 g, 0.0004 g, 0.0005 g, 0.0006 g, 0.0007 g, 0.0008 g, 0.0009 g, 0.001 g, 0.0015 g, 0.002 g, 0.0025 g, 0.003 g, 0.0035 g, 0.004 g, 0.0045 g, 0.005 g, 0.0055 g, 0.006 g, 0.0065 g, 0.007 g, 0.0075 g, 0.008 g, 0.0085 g, 0.009 g, 0.0095 g, 0.01 g, 0.015 g, 0.02 g, 0.025 g, 0.03 g, 0.035 g, 0.04 g, 0.045 g, 0.05 g, 0.055 g, 0.06 g, 0.065 g, 0.07 g, 0.075 g, 0.08 g, 0.085 g, 0.09 g, 0.095 g, 0.1 g, 0.15 g, 0.2 g, 0.25 g, 0.3 g, 0.35 g, 0.4 g, 0.45 g, 0.5 g, 0.55 g, 0.6 g, 0.65 g, 0.7 g, 0.75 g, 0.8 g, 0.85 g, 0.9 g, 0.95 g, 1 g, 1.5 g, 2 g, 2.5, 3 g, 3.5, 4 g, 4.5 g, 5 g, 5.5 g, 6 g, 6.5 g, 7 g, 7.5 g, 8 g, 8.5 g, 9 g, 9.5 g, or 10 g.
[1544] The TILs provided in the pharmaceutical compositions of the invention are effective over a wide dosage range. The exact dosage will depend upon the route of administration, the form in which the compound is administered, the gender and age of the subject to be treated, the body weight of the subject to be treated, and the preference and experience of the attending physician. The clinically-established dosages of the TILs may also be used if appropriate. The amounts of the pharmaceutical compositions administered using the methods herein, such as the dosages of TILs, will be dependent on the human or mammal being treated, the severity of the disorder or condition, the rate of administration, the disposition of the active pharmaceutical ingredients and the discretion of the prescribing physician.
[1545] In some embodiments, TILs may be administered in a single dose. Such administration may be by injection, e.g., intravenous injection. In some embodiments, TILs may be administered in multiple doses. Dosing may be once, twice, three times, four times, five times, six times, or more than six times per year. Dosing may be once a month, once every two weeks, once a week, or once every other day. Administration of TILs may continue as long as necessary.
[1546] In some embodiments, an effective dosage of TILs is about 1.times.10.sup.6, 2.times.10.sup.6, 3.times.10.sup.6, 4.times.10.sup.6, 5.times.10.sup.6, 6.times.10.sup.6, 7.times.10.sup.6, 8.times.10.sup.6, 9.times.10.sup.6, 1.times.10.sup.7, 2.times.10.sup.7, 3.times.10.sup.7, 4.times.10.sup.7, 5.times.10.sup.7, 6.times.10.sup.7, 7.times.10.sup.7, 8.times.10.sup.7, 9.times.10.sup.7, 1.times.10.sup.8, 2.times.10.sup.8, 3.times.10.sup.8, 4.times.10.sup.8, 5.times.10.sup.8, 6.times.10.sup.8, 7.times.10.sup.8, 8.times.10.sup.8, 9.times.10.sup.8, 1.times.10.sup.9, 2.times.10.sup.9, 3.times.10.sup.9, 4.times.10.sup.9, 5.times.10.sup.9, 6.times.10.sup.9, 7.times.10.sup.9, 8.times.10.sup.9, 9.times.10.sup.9, 1.times.10.sup.10, 2.times.10.sup.10, 3.times.10.sup.10, 4.times.10.sup.10, 5.times.10.sup.10, 6.times.10.sup.10, 7.times.10.sup.10, 8.times.10.sup.10, 9.times.10.sup.10, 1.times.10.sup.11, 2.times.10.sup.11, 3.times.10.sup.11, 4.times.10.sup.11, 5.times.10.sup.11, 6.times.10.sup.11, 7.times.10.sup.11, 8.times.10.sup.11, 9.times.10.sup.11, 1.times.10.sup.12, 2.times.10.sup.12, 3.times.10.sup.12, 4.times.10.sup.12, 5.times.10.sup.12, 6.times.10.sup.12, 7.times.10.sup.12, 8.times.10.sup.12, 9.times.10.sup.12, 1.times.10.sup.13, 2.times.10.sup.13, 3.times.10.sup.13, 4.times.10.sup.13, 5.times.10.sup.13, 6.times.10.sup.13, 7.times.10.sup.13, 8.times.10.sup.13, and 9.times.10.sup.13. In some embodiments, an effective dosage of TILs is in the range of 1.times.10.sup.6 to 5.times.10.sup.6, 5.times.10.sup.6 to 1.times.10.sup.7, 1.times.10.sup.7 to 5.times.10.sup.7, 5.times.10.sup.7 to 1.times.10.sup.8, 1.times.10.sup.8 to 5.times.10.sup.8, 5.times.10.sup.8 to 1.times.10.sup.9, 1.times.10.sup.9 to 5.times.10.sup.9, 5.times.10.sup.9 to 1.times.10.sup.10, 1.times.10.sup.10 to 5.times.10.sup.10 5.times.10.sup.10 to 1.times.10.sup.11, 5.times.10.sup.11 to 1.times.10.sup.12, 1.times.10.sup.12 to 5.times.10.sup.12, and 5.times.10.sup.12 to 1.times.10.sup.13.
[1547] In some embodiments, an effective dosage of TILs is in the range of about 0.01 mg/kg to about 4.3 mg/kg, about 0.15 mg/kg to about 3.6 mg/kg, about 0.3 mg/kg to about 3.2 mg/kg, about 0.35 mg/kg to about 2.85 mg/kg, about 0.15 mg/kg to about 2.85 mg/kg, about 0.3 mg to about 2.15 mg/kg, about 0.45 mg/kg to about 1.7 mg/kg, about 0.15 mg/kg to about 1.3 mg/kg, about 0.3 mg/kg to about 1.15 mg/kg, about 0.45 mg/kg to about 1 mg/kg, about 0.55 mg/kg to about 0.85 mg/kg, about 0.65 mg/kg to about 0.8 mg/kg, about 0.7 mg/kg to about 0.75 mg/kg, about 0.7 mg/kg to about 2.15 mg/kg, about 0.85 mg/kg to about 2 mg/kg, about 1 mg/kg to about 1.85 mg/kg, about 1.15 mg/kg to about 1.7 mg/kg, about 1.3 mg/kg mg to about 1.6 mg/kg, about 1.35 mg/kg to about 1.5 mg/kg, about 2.15 mg/kg to about 3.6 mg/kg, about 2.3 mg/kg to about 3.4 mg/kg, about 2.4 mg/kg to about 3.3 mg/kg, about 2.6 mg/kg to about 3.15 mg/kg, about 2.7 mg/kg to about 3 mg/kg, about 2.8 mg/kg to about 3 mg/kg, or about 2.85 mg/kg to about 2.95 mg/kg.
[1548] In some embodiments, an effective dosage of TILs is in the range of about 1 mg to about 500 mg, about 10 mg to about 300 mg, about 20 mg to about 250 mg, about 25 mg to about 200 mg, about 1 mg to about 50 mg, about 5 mg to about 45 mg, about 10 mg to about 40 mg, about 15 mg to about 35 mg, about 20 mg to about 30 mg, about 23 mg to about 28 mg, about 50 mg to about 150 mg, about 60 mg to about 140 mg, about 70 mg to about 130 mg, about 80 mg to about 120 mg, about 90 mg to about 110 mg, or about 95 mg to about 105 mg, about 98 mg to about 102 mg, about 150 mg to about 250 mg, about 160 mg to about 240 mg, about 170 mg to about 230 mg, about 180 mg to about 220 mg, about 190 mg to about 210 mg, about 195 mg to about 205 mg, or about 198 to about 207 mg.
[1549] An effective amount of the TILs may be administered in either single or multiple doses by any of the accepted modes of administration of agents having similar utilities, including intranasal and transdermal routes, by intra-arterial injection, intravenously, intraperitoneally, parenterally, intramuscularly, subcutaneously, topically, by transplantation or direct injection into tumor, or by inhalation.
Pharmaceutical Compositions, Dosages, and Dosing Regimens for TNFRSF Agonists
[1550] In one embodiment, the invention provides a pharmaceutical composition for use in the treatment of the diseases and conditions described herein. In a preferred embodiment, the invention provides pharmaceutical compositions, including those described below, for use in the treatment of a hyperproliferative disease. In a preferred embodiment, the invention provides pharmaceutical compositions, including those described below, for use in the treatment of cancer.
[1551] In some embodiments, a TNFRSF agonist antibody formulation comprises one or more excipients selected from tris-hydrochloride, sodium chloride, mannitol, pentetic acid, polysorbate 80, sodium hydroxide, and hydrochloric acid.
[1552] In an embodiment, a TNFRSF agonist is administered to a subject by infusing a dose selected from the group consisting of about 5 mg, about 8 mg, about 10 mg, about 20 mg, about 25 mg, about 50 mg, about 75 mg, 100 mg, about 200 mg, about 300 mg, about 400 mg, about 500 mg, about 600 mg, about 700 mg, about 800 mg, about 900 mg, about 1000 mg, about 1100 mg, about 1200 mg, about 1300 mg, about 1400 mg, about 1500 mg, about 1600 mg, about 1700 mg, about 1800 mg, about 1900 mg, and about 2000 mg. In an embodiment, a TNFRSF agonist is administered weekly. In an embodiment, a TNFRSF agonist is administered every two weeks. In an embodiment, a TNFRSF agonist is administered every three weeks. In an embodiment, a TNFRSF agonist is administered monthly. In an embodiment, a TNFRSF agonist is administered intravenously in a dose of 8 mg given every three weeks for 4 doses over a 12-week period. In an embodiment, a TNFRSF agonist is administered at a lower initial dose, which is escalated when administered at subsequent intervals administered monthly. For example, the first infusion can deliver 300 mg of a TNFRSF agonist, and subsequent weekly doses could deliver 2,000 mg of a TNFRSF agonist for eight weeks, followed by monthly doses of 2,000 mg of a TNFRSF agonist.
[1553] The amounts of TNFRSF agonists administered will be dependent on the human or mammal being treated, the severity of the disorder or condition, the rate of administration, the disposition of the compounds and the discretion of the prescribing physician. However, an effective dosage of each is in the range of about 0.001 to about 100 mg per kg body weight per day, such as about 1 to about 35 mg/kg/day, in single or divided doses. For a 70 kg human, this would amount to about 0.05 to 7 g/day, such as about 0.05 to about 2.5 g/day. In some instances, dosage levels below the lower limit of the aforesaid range may be more than adequate, while in other cases still larger doses may be employed without causing any harmful side effect--e.g., by dividing such larger doses into several small doses for administration throughout the day. The dosage of the TNFRSF agonist(s) may be provided in units of mg/kg of body mass or in mg/m.sup.2 of body surface area. In an embodiment, a TNFRSF agonist and a second TNFRSF agonist are delivered in mg/kg or in mg/m.sup.2 in a ration selected from the group consisting of about 20:1, about 19:1, about 18:1, about 17:1, about 16:1, about 15:1, about 14:1, about 13:1, about 12:1, about 11:1, about 10:1, about 9:1, about 8:1, about 7:1, about 6:1, about 5:1, about 4:1, about 3:1, about 2:1, about 1:1, about 1:2, about 1:3, about 1:4, about 1:5, about 1:6, about 1:7, about 1:8, about 1:9, about 1:10, about 1:11, about 1:12, about 1:13, about 1:14, about 1:15, about 1:16, about 1:17, about 1:18, about 1:19, and about 1:20.
[1554] In some embodiments, the combination of TILs and a TNFRSF agonist is administered in a single dose. Such administration may be by injection, e.g., intravenous injection, in order to introduce the TNFRSF agonist.
[1555] In some embodiments, the combination of TILs and TNFRSF agonists is administered in multiple doses. In a preferred embodiment, the combination of TILs and TNFRSF agonists is administered in multiple doses. Dosing of the TNFRSF agonists may be once, twice, three times, four times, five times, six times, or more than six times per day. Dosing of TILs and TNFRSF agonists may be once a month, once every two weeks, once a week, or once every other day.
[1556] In selected embodiments, the TNFRSF agonists are administered for more than 1, 2, 3, 4, 5, 6, 7, 14, 28 days, 2 months, 3 months, 6 months, 12 months, or 24 months. In some cases, continuous dosing is achieved and maintained as long as necessary.
[1557] In some embodiments, an effective dosage of a TNFRSF agonist disclosed herein is in the range of about 1 mg to about 500 mg, about 10 mg to about 300 mg, about 20 mg to about 250 mg, about 25 mg to about 200 mg, about 10 mg to about 200 mg, about 20 mg to about 150 mg, about 30 mg to about 120 mg, about 10 mg to about 90 mg, about 20 mg to about 80 mg, about 30 mg to about 70 mg, about 40 mg to about 60 mg, about 45 mg to about 55 mg, about 48 mg to about 52 mg, about 50 mg to about 150 mg, about 60 mg to about 140 mg, about 70 mg to about 130 mg, about 80 mg to about 120 mg, about 90 mg to about 110 mg, about 95 mg to about 105 mg, about 150 mg to about 250 mg, about 160 mg to about 240 mg, about 170 mg to about 230 mg, about 180 mg to about 220 mg, about 190 mg to about 210 mg, about 195 mg to about 205 mg, or about 198 to about 202 mg. In some embodiments, an effective dosage of a TNFRSF agonist disclosed herein is about 25 mg, about 50 mg, about 75 mg, about 100 mg, about 125 mg, about 150 mg, about 175 mg, about 200 mg, about 225 mg, or about 250 mg.
[1558] In some embodiments, an effective dosage of a TNFRSF agonist disclosed herein is in the range of about 0.01 mg/kg to about 4.3 mg/kg, about 0.15 mg/kg to about 3.6 mg/kg, about 0.3 mg/kg to about 3.2 mg/kg, about 0.35 mg/kg to about 2.85 mg/kg, about 0.15 mg/kg to about 2.85 mg/kg, about 0.3 mg to about 2.15 mg/kg, about 0.45 mg/kg to about 1.7 mg/kg, about 0.15 mg/kg to about 1.3 mg/kg, about 0.3 mg/kg to about 1.15 mg/kg, about 0.45 mg/kg to about 1 mg/kg, about 0.55 mg/kg to about 0.85 mg/kg, about 0.65 mg/kg to about 0.8 mg/kg, about 0.7 mg/kg to about 0.75 mg/kg, about 0.7 mg/kg to about 2.15 mg/kg, about 0.85 mg/kg to about 2 mg/kg, about 1 mg/kg to about 1.85 mg/kg, about 1.15 mg/kg to about 1.7 mg/kg, about 1.3 mg/kg mg to about 1.6 mg/kg, about 1.35 mg/kg to about 1.5 mg/kg, about 2.15 mg/kg to about 3.6 mg/kg, about 2.3 mg/kg to about 3.4 mg/kg, about 2.4 mg/kg to about 3.3 mg/kg, about 2.6 mg/kg to about 3.15 mg/kg, about 2.7 mg/kg to about 3 mg/kg, about 2.8 mg/kg to about 3 mg/kg, or about 2.85 mg/kg to about 2.95 mg/kg. In some embodiments, an effective dosage of a TNFRSF agonist disclosed herein is about 0.35 mg/kg, about 0.7 mg/kg, about 1 mg/kg, about 1.4 mg/kg, about 1.8 mg/kg, about 2.1 mg/kg, about 2.5 mg/kg, about 2.85 mg/kg, about 3.2 mg/kg, or about 3.6 mg/kg.
[1559] In some embodiments, an effective dosage of a TNFRSF agonist disclosed herein is in the range of about 1 mg to about 500 mg, about 10 mg to about 300 mg, about 20 mg to about 250 mg, about 25 mg to about 200 mg, about 1 mg to about 50 mg, about 5 mg to about 45 mg, about 10 mg to about 40 mg, about 15 mg to about 35 mg, about 20 mg to about 30 mg, about 23 mg to about 28 mg, about 50 mg to about 150 mg, about 60 mg to about 140 mg, about 70 mg to about 130 mg, about 80 mg to about 120 mg, about 90 mg to about 110 mg, or about 95 mg to about 105 mg, about 98 mg to about 102 mg, about 150 mg to about 250 mg, about 160 mg to about 240 mg, about 170 mg to about 230 mg, about 180 mg to about 220 mg, about 190 mg to about 210 mg, about 195 mg to about 205 mg, or about 198 to about 207 mg. In some embodiments, an effective dosage of a TNFRSF agonist disclosed herein is about 25 mg, about 50 mg, about 75 mg, about 100 mg, about 125 mg, about 150 mg, about 175 mg, about 200 mg, about 225 mg, or about 250 mg.
[1560] In some embodiments, an effective dosage of a TNFRSF agonist disclosed herein is in the range of about 0.01 mg/kg to about 4.3 mg/kg, about 0.15 mg/kg to about 3.6 mg/kg, about 0.3 mg/kg to about 3.2 mg/kg, about 0.35 mg/kg to about 2.85 mg/kg, about 0.01 mg/kg to about 0.7 mg/kg, about 0.07 mg/kg to about 0.65 mg/kg, about 0.15 mg/kg to about 0.6 mg/kg, about 0.2 mg/kg to about 0.5 mg/kg, about 0.3 mg/kg to about 0.45 mg/kg, about 0.3 mg/kg to about 0.4 mg/kg, about 0.7 mg/kg to about 2.15 mg/kg, about 0.85 mg/kg to about 2 mg/kg, about 1 mg/kg to about 1.85 mg/kg, about 1.15 mg/kg to about 1.7 mg/kg, about 1.3 mg/kg to about 1.6 mg/kg, about 1.35 mg/kg to about 1.5 mg/kg, about 1.4 mg/kg to about 1.45 mg/kg, about 2.15 mg/kg to about 3.6 mg/kg, about 2.3 mg/kg to about 3.4 mg/kg, about 2.4 mg/kg to about 3.3 mg/kg, about 2.6 mg/kg to about 3.15 mg/kg, about 2.7 mg/kg to about 3 mg/kg, about 2.8 mg/kg to about 3 mg/kg, or about 2.85 mg/kg to about 2.95 mg/kg. In some embodiments, a TNFRSF agonist disclosed herein is about 0.4 mg/kg, about 0.7 mg/kg, about 1 mg/kg, about 1.4 mg/kg, about 1.8 mg/kg, about 2.1 mg/kg, about 2.5 mg/kg, about 2.85 mg/kg, about 3.2 mg/kg, or about 3.6 mg/kg.
[1561] In some embodiments, a TNFRSF agonist is administered at a dosage of 10 to 1000 mg BID, including 10, 20, 30, 40, 50, 60, 70, 80, 90, 100, 150, or 200 mg BID.
[1562] In some embodiments, the concentration of the TNFRSF agonists, and combinations thereof provided in the pharmaceutical compositions of the invention is independently less than, for example, 100%, 90%, 80%, 70%, 60%, 50%, 40%, 30%, 20%, 19%, 18%, 17%, 16%, 15%, 14%, 13%, 12%, 11%, 10%, 9%, 8%, 7%, 6%, 5%, 4%, 3%, 2%, 1%, 0.5%, 0.4%, 0.3%, 0.2%, 0.1%, 0.09%, 0.08%, 0.07%, 0.06%, 0.05%, 0.04%, 0.03%, 0.02%, 0.01%, 0.009%, 0.008%, 0.007%, 0.006%, 0.005%, 0.004%, 0.003%, 0.002%, 0.001%, 0.0009%, 0.0008%, 0.0007%, 0.0006%, 0.0005%, 0.0004%, 0.0003%, 0.0002% or 0.0001% w/w, w/v or v/v of the pharmaceutical composition.
[1563] In some embodiments, the concentration of the TNFRSF agonists, and combinations thereof provided in the pharmaceutical compositions of the invention is independently greater than 90%, 80%, 70%, 60%, 50%, 40%, 30%, 20%, 19.75%, 19.50%, 19.25% 19%, 18.75%, 18.50%, 18.25% 18%, 17.75%, 17.50%, 17.25% 17%, 16.75%, 16.50%, 16.25% 16%, 15.75%, 15.50%, 15.25% 15%, 14.75%, 14.50%, 14.25% 14%, 13.75%, 13.50%, 13.25% 13%, 12.75%, 12.50%, 12.25% 12%, 11.75%, 11.50%, 11.25% 11%, 10.75%, 10.50%, 10.25% 10%, 9.75%, 9.50%, 9.25% 9%, 8.75%, 8.50%, 8.25% 8%, 7.75%, 7.50%, 7.25% 7%, 6.75%, 6.50%, 6.25% 6%, 5.75%, 5.50%, 5.25% 5%, 4.75%, 4.50%, 4.25%, 4%, 3.75%, 3.50%, 3.25%, 3%, 2.75%, 2.50%, 2.25%, 2%, 1.75%, 1.50%, 125%, 1%, 0.5%, 0.4%, 0.3%, 0.2%, 0.1%, 0.09%, 0.08%, 0.07%, 0.06%, 0.05%, 0.04%, 0.03%, 0.02%, 0.01%, 0.009%, 0.008%, 0.007%, 0.006%, 0.005%, 0.004%, 0.003%, 0.002%, 0.001%, 0.0009%, 0.0008%, 0.0007%, 0.0006%, 0.0005%, 0.0004%, 0.0003%, 0.0002% or 0.0001% w/w, w/v, or v/v of the pharmaceutical composition.
[1564] In some embodiments, the concentration of the TNFRSF agonists in pharmaceutical compositions is independently in the range from about 0.0001% to about 50%, about 0.001% to about 40%, about 0.01% to about 30%, about 0.02% to about 29%, about 0.03% to about 28%, about 0.04% to about 27%, about 0.05% to about 26%, about 0.06% to about 25%, about 0.07% to about 24%, about 0.08% to about 23%, about 0.09% to about 22%, about 0.1% to about 21%, about 0.2% to about 20%, about 0.3% to about 19%, about 0.4% to about 18%, about 0.5% to about 17%, about 0.6% to about 16%, about 0.7% to about 15%, about 0.8% to about 14%, about 0.9% to about 12% or about 1% to about 10% w/w, w/v or v/v of the pharmaceutical composition.
[1565] In some embodiments, the concentration of the TNFRSF agonists in pharmaceutical compositions is independently in the range from about 0.001% to about 10%, about 0.01% to about 5%, about 0.02% to about 4.5%, about 0.03% to about 4%, about 0.04% to about 3.5%, about 0.05% to about 3%, about 0.06% to about 2.5%, about 0.07% to about 2%, about 0.08% to about 1.5%, about 0.09% to about 1%, about 0.1% to about 0.9% w/w, w/v or v/v of the pharmaceutical composition.
[1566] In some embodiments, the concentration of the TNFRSF agonists in pharmaceutical compositions is independently equal to or less than 10 g, 9.5 g, 9.0 g, 8.5 g, 8.0 g, 7.5 g, 7.0 g, 6.5 g, 6.0 g, 5.5 g, 5.0 g, 4.5 g, 4.0 g, 3.5 g, 3.0 g, 2.5 g, 2.0 g, 1.5 g, 1.0 g, 0.95 g, 0.9 g, 0.85 g, 0.8 g, 0.75 g, 0.7 g, 0.65 g, 0.6 g, 0.55 g, 0.5 g, 0.45 g, 0.4 g, 0.35 g, 0.3 g, 0.25 g, 0.2 g, 0.15 g, 0.1 g, 0.09 g, 0.08 g, 0.07 g, 0.06 g, 0.05 g, 0.04 g, 0.03 g, 0.02 g, 0.01 g, 0.009 g, 0.008 g, 0.007 g, 0.006 g, 0.005 g, 0.004 g, 0.003 g, 0.002 g, 0.001 g, 0.0009 g, 0.0008 g, 0.0007 g, 0.0006 g, 0.0005 g, 0.0004 g, 0.0003 g, 0.0002 g, or 0.0001 g.
[1567] In some embodiments, the concentration of the TNFRSF agonists in pharmaceutical compositions is independently more than 0.0001 g, 0.0002 g, 0.0003 g, 0.0004 g, 0.0005 g, 0.0006 g, 0.0007 g, 0.0008 g, 0.0009 g, 0.001 g, 0.0015 g, 0.002 g, 0.0025 g, 0.003 g, 0.0035 g, 0.004 g, 0.0045 g, 0.005 g, 0.0055 g, 0.006 g, 0.0065 g, 0.007 g, 0.0075 g, 0.008 g, 0.0085 g, 0.009 g, 0.0095 g, 0.01 g, 0.015 g, 0.02 g, 0.025 g, 0.03 g, 0.035 g, 0.04 g, 0.045 g, 0.05 g, 0.055 g, 0.06 g, 0.065 g, 0.07 g, 0.075 g, 0.08 g, 0.085 g, 0.09 g, 0.095 g, 0.1 g, 0.15 g, 0.2 g, 0.25 g, 0.3 g, 0.35 g, 0.4 g, 0.45 g, 0.5 g, 0.55 g, 0.6 g, 0.65 g, 0.7 g, 0.75 g, 0.8 g, 0.85 g, 0.9 g, 0.95 g, 1 g, 1.5 g, 2 g, 2.5, 3 g, 3.5 g, 4 g, 4.5 g, 5 g, 5.5 g, 6 g, 6.5 g, 7 g, 7.5 g, 8 g, 8.5 g, 9 g, 9.5 g, or 10 g.
[1568] Described below are non-limiting pharmaceutical compositions and methods for preparing the same.
Pharmaceutical Compositions for Injection
[1569] In preferred embodiments, the invention provides a pharmaceutical composition for injection containing the combination of a TIL and at least one TNFRSF agonist, and combinations thereof, and a pharmaceutical excipient suitable for injection, including intratumoral injection or intravenous infusion. Components and amounts of agents in the compositions are as described herein.
[1570] The forms in which the compositions of the present invention may be incorporated for administration by injection include aqueous or oil suspensions, or emulsions, with sesame oil, corn oil, cottonseed oil, or peanut oil, as well as elixirs, mannitol, dextrose, or a sterile aqueous solution, and similar pharmaceutical vehicles.
[1571] Aqueous solutions in saline are also conventionally used for injection. Ethanol, glycerol, propylene glycol and liquid polyethylene glycol (and suitable mixtures thereof), cyclodextrin derivatives, and vegetable oils may also be employed. The proper fluidity can be maintained, for example, by the use of a coating, such as lecithin, for the maintenance of the required particle size in the case of dispersion and by the use of surfactants. The prevention of the action of microorganisms can be brought about by various antibacterial and antifungal agents, for example, parabens, chlorobutanol, phenol, sorbic acid and thimerosal.
[1572] Sterile injectable solutions are prepared by incorporating the combination of the TNFRSF agonists and TILs in the required amounts in the appropriate media with various other ingredients as enumerated above, as required, followed by filtered sterilization. Generally, dispersions are prepared by incorporating the various sterilized active ingredients into a sterile vehicle which contains the basic dispersion medium and the required other ingredients from those enumerated above. In the case of sterile powders for the preparation of sterile injectable solutions, certain desirable methods of preparation are vacuum-drying and freeze-drying techniques which yield a powder of the active ingredient plus any additional desired ingredient from a previously sterile-filtered solution thereof.
Other Pharmaceutical Compositions
[1573] Pharmaceutical compositions may also be prepared from compositions described herein and one or more pharmaceutically acceptable excipients suitable for sublingual, buccal, rectal, intraosseous, intraocular, intranasal, epidural, or intraspinal administration. Preparations for such pharmaceutical compositions are well-known in the art. See, e.g., Anderson, Philip O.; Knoben, James E.; Troutman, William G, eds., Handbook of Clinical Drug Data, Tenth Edition, McGraw-Hill, 2002; and Pratt and Taylor, eds., Principles of Drug Action, Third Edition, Churchill Livingston, N.Y., 1990, each of which is incorporated by reference herein in its entirety.
[1574] Administration of a combination of a TIL and a TNFRSF agonist can be effected by any method that enables delivery of the compounds to the site of action. These methods include oral routes, intraduodenal routes, parenteral injection (including intravenous, intraarterial, subcutaneous, intramuscular, intravascular, intraperitoneal or infusion), topical (e.g., transdermal application), rectal administration, via local delivery by catheter or stent or through inhalation. The combination of compounds can also be administered intraadiposally or intrathecally.
[1575] The invention also provides kits. The kits include a combination of ready-to-administer TILs and a TNFRSF agonist, either alone or in combination in suitable packaging, and written material that can include instructions for use, discussion of clinical studies and listing of side effects. Such kits may also include information, such as scientific literature references, package insert materials, clinical trial results, and/or summaries of these and the like, which indicate or establish the activities and/or advantages of the composition, and/or which describe dosing, administration, side effects, drug interactions, or other information useful to the health care provider. Such information may be based on the results of various studies, for example, studies using experimental animals involving in vivo models and studies based on human clinical trials. The kit may further contain another active pharmaceutical ingredient. In selected embodiments, the TNFRSF agonists and TILs and another active pharmaceutical ingredient are provided as separate compositions in separate containers within the kit. In selected embodiments, the molecule selected from the group consisting of a TNFRSF agonist and the TILs are provided as a single composition within a container in the kit. Suitable packaging and additional articles for use (e.g., measuring cup for liquid preparations, foil wrapping to minimize exposure to air, and the like) are known in the art and may be included in the kit. Kits described herein can be provided, marketed and/or promoted to health providers, including physicians, nurses, pharmacists, formulary officials, and the like. Kits may also, in selected embodiments, be marketed directly to the consumer.
[1576] The kits described above are preferably for use in the treatment of the diseases and conditions described herein. In a preferred embodiment, the kits are for use in the treatment of cancer. In preferred embodiments, the kits are for use in treating solid tumor cancers, lymphomas and leukemias.
[1577] In a preferred embodiment, the kits of the present invention are for use in the treatment of cancer, including any of the cancers described herein.
Methods of Treating Cancers
[1578] The compositions and combinations of TILs and TNFRSF agonists described herein can be used in a method for treating hyperproliferative disorders. In a preferred embodiment, they are for use in treating cancers. In a preferred embodiment, the invention provides a method of treating a cancer and compositions and combinations of TILs and TNFRSF agonists for treating a cancer, wherein the cancer is selected from the group consisting of melanoma, ovarian cancer, cervical cancer, lung cancer, bladder cancer, breast cancer, head and neck cancer, renal cell carcinoma, acute myeloid leukemia, colorectal cancer, and sarcoma. In a preferred embodiment, the invention provides a method of treating a cancer and compositions and combinations of TILs and TNFRSF agonists for treating a cancer, wherein the cancer is selected from the group consisting of non-small cell lung cancer (NSCLC) or triple negative breast cancer, double-refractory melanoma, and uveal (ocular) melanoma. In a preferred embodiment, the invention provides a method of treating a cancer wherein the cancer is selected from the group consisting of melanoma, ovarian cancer, cervical cancer, lung cancer, bladder cancer, breast cancer, head and neck cancer (head and neck squamous cell cancer), renal cell carcinoma, acute myeloid leukemia, colorectal cancer, cholangiocarcinoma, and sarcoma with a combination of TILs and a TNFRSF agonist. In a preferred embodiment, the invention provides compositions and combinations of TILs and TNFRSF agonists for treating a cancer wherein the cancer is selected from the group consisting of melanoma, ovarian cancer, cervical cancer, lung cancer, bladder cancer, breast cancer, head and neck cancer, renal cell carcinoma, acute myeloid leukemia, colorectal cancer, cholangiocarcinoma, and sarcoma. In a preferred embodiment, the invention provides a method of treating a cancer, wherein the cancer is selected from the group consisting of non-small cell lung cancer (NSCLC) or triple negative breast cancer, double-refractory melanoma, and uveal (ocular) melanoma with a combination of TILs and a TNFRSF agonist. In a preferred embodiment, the invention provides compositions and combinations of TILs and TNFRSF agonists for treating a cancer wherein the cancer is selected from the group consisting of non-small cell lung cancer (NSCLC) or triple negative breast cancer, double-refractory melanoma, and uveal (ocular) melanoma. In an embodiment, the TILs are expanded by a process described herein.
[1579] In some embodiments, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [1580] (a) resecting a tumor from a patient; [1581] (b) obtaining a first population of TILs from the tumor; [1582] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [1583] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [1584] (e) harvesting the third population of TILs; and [1585] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer; wherein the cancer is selected from the group consisting of melanoma, ovarian cancer, cervical cancer, lung cancer, bladder cancer, breast cancer, head and neck cancer, renal cell carcinoma, acute myeloid leukemia, colorectal cancer, cholangiocarcinoma, sarcoma, non-small cell lung cancer (NSCLC) or triple negative breast cancer, double-refractory melanoma, and uveal (ocular) melanoma.
[1586] In some embodiments, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [1587] (a) resecting a tumor from a patient; [1588] (b) obtaining a first population of TILs from the tumor; [1589] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2, and wherein the initial expansion is performed over a period of 21 days or less; [1590] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and wherein the rapid expansion is performed over a period of 14 days or less; [1591] (e) harvesting the third population of TILs; and [1592] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer; wherein the cancer is selected from the group consisting of melanoma, ovarian cancer, cervical cancer, lung cancer, bladder cancer, breast cancer, head and neck cancer, renal cell carcinoma, acute myeloid leukemia, colorectal cancer, cholangiocarcinoma, sarcoma, non-small cell lung cancer (NSCLC) or triple negative breast cancer, double-refractory melanoma, and uveal (ocular) melanoma.
[1593] In some embodiments, the invention provides a method of treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [1594] (a) resecting a tumor from a patient; [1595] (b) obtaining a first population of TILs from the tumor; [1596] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises L-2, and wherein the initial expansion is performed over a period of 21 days or less; [1597] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and wherein the rapid expansion is performed over a period of 14 days or less; [1598] (e) harvesting the third population of TILs; and [1599] (f) administering a therapeutically effective portion of the third population of TILs to a patient with the cancer; wherein the cancer is uveal (ocular) melanoma.
[1600] In an embodiment, the invention includes a kit for treating a cancer with a population of TILs according to any of the foregoing methods.
[1601] Efficacy of the methods, compounds, and combinations of compounds described herein in treating, preventing and/or managing the indicated diseases or disorders can be tested using various animal models known in the art. Models for determining efficacy of treatments for pancreatic cancer are described in Herreros-Villanueva, et al., World J. Gastroenterol. 2012, 18, 1286-1294. Models for determining efficacy of treatments for breast cancer are described, e.g., in Fantozzi, Breast Cancer Res. 2006, 8, 212. Models for determining efficacy of treatments for ovarian cancer are described, e.g., in Mullany, et al., Endocrinology 2012, 153, 1585-92; and Fong, et al., J. Ovarian Res. 2009, 2, 12. Models for determining efficacy of treatments for melanoma are described, e.g., in Damsky, et al., Pigment Cell & Melanoma Res. 2010, 23, 853-859. Models for determining efficacy of treatments for lung cancer are described, e.g., in Meuwissen, et al., Genes & Development, 2005, 19, 643-664. Models for determining efficacy of treatments for lung cancer are described, e.g., in Kim, Clin. Exp. Otorhinolaryngol. 2009, 2, 55-60; and Sano, Head Neck Oncol. 2009, 1, 32. Models for determining efficacy of treatments for colorectal cancer, including the CT26 model, are described in Castle, et al., BMC Genomics, 2013, 15, 190; Endo, et al., Cancer Gene Therapy, 2002, 9, 142-148; Roth, et al., Adv. Immunol. 1994, 57, 281-351; Fearon, et al., Cancer Res. 1988, 48, 2975-2980.
Co-Administration of IL-2
[1602] In an embodiment, the invention provides a method treating a cancer with a population of tumor infiltrating lymphocytes (TILs) comprising the steps of: [1603] (a) resecting a tumor from a patient; [1604] (b) obtaining a first population of TILs from the tumor; [1605] (c) performing an initial expansion of the first population of TILs in a first cell culture medium to obtain a second population of TILs, wherein the second population of TILs is at least 5-fold greater in number than the first population of TILs, wherein the first cell culture medium comprises IL-2 and a tumor necrosis factor receptor superfamily (TNFRSF) agonist, and wherein the initial expansion is performed over a period of 21 days or less; [1606] (d) performing a rapid expansion of the second population of TILs in a second cell culture medium to obtain a third population of TILs, wherein the third population of TILs is at least 50-fold greater in number than the second population of TILs after 7 days from the start of the rapid expansion; wherein the second cell culture medium comprises IL-2, OKT-3 (anti-CD3 antibody), peripheral blood mononuclear cells (PBMCs), and optionally the TNFRSF agonist, and wherein the rapid expansion is performed over a period of 14 days or less; [1607] (e) harvesting the third population of TILs; [1608] (f) administering a therapeutically effective portion of the third population of TILs to the patient with the cancer; and [1609] (g) administering an IL-2 regimen to the patient.
[1610] In an embodiment, the IL-2 regimen comprises a high-dose IL-2 regimen, wherein the high-dose IL-2 regimen comprises aldesleukin, or a biosimilar or variant thereof, administered intraveneously starting on the day after administering a therapeutically effective portion of the third population of TILs, wherein the aldesleukin or a biosimilar or variant thereof is administered at a dose of 600,000 or 720,000 IU/kg (patient body mass) using 15-minute bolus intravenous infusions every eight hours until tolerance, for a maximum of 14 doses. Following 9 days of rest, this schedule may be repeated for another 14 doses, for a maximum of 28 doses in total.
[1611] In an embodiment, the IL-2 regimen comprises a high-dose IL-2 regimen, wherein the high-dose IL-2 regimen comprises aldesleukin, or a biosimilar or variant thereof, administered intraveneously starting on the day after administering a therapeutically effective portion of the third population of TILs, wherein the aldesleukin or a biosimilar or variant thereof is administered at a dose of 0.037 mg/kg or 0.044 mg/kg IU/kg (patient body mass) using 15-minute bolus intravenous infusions every eight hours until tolerance, for a maximum of 14 doses. Following 9 days of rest, this schedule may be repeated for another 14 doses, for a maximum of 28 doses in total.
[1612] In an embodiment, the IL-2 regimen comprises a decrescendo IL-2 regimen. Decrescendo IL-2 regimens have been described in O'Day, et al., J. Clin. Oncol. 1999, 17, 2752-61 and Eton, et al., Cancer 2000, 88, 1703-9, the disclosures of which are incorporated herein by reference. In an embodiment, a decrescendo L-2 regimen comprises 18.times.10.sup.6 IU/m.sup.2 administered intraveneously over 6 hours, followed by 18.times.10.sup.6 IU/m.sup.2 administered intraveneously over 12 hours, followed by 18.times.10.sup.6 IU/m.sup.2 administered intraveneously over 24 hrs, followed by 4.5.times.10.sup.6 IU/m.sup.2 administered intraveneously over 72 hours. This treatment cycle may be repeated every 28 days for a maximum of four cycles. In an embodiment, a decrescendo L-2 regimen comprises 18,000,000 IU/m.sup.2 on day 1, 9,000,000 IU/m.sup.2 on day 2, and 4,500,000 IU/m.sup.2 on days 3 and 4.
[1613] In an embodiment, the IL-2 regimen comprises administration of pegylated IL-2 every 1, 2, 4, 6, 7, 14 or 21 days at a dose of 0.10 mg/day to 50 mg/day.
Non-Myeloablative Lymphodepletion with Chemotherapy
[1614] In an embodiment, the invention includes a method of treating a cancer with a population of TILs, wherein a patient is pre-treated with non-myeloablative chemotherapy prior to an infusion of TILs and prior to or concurrent with treatment with a TNFRSF agonist according to the present disclosure. In an embodiment, the non-myeloablative chemotherapy is cyclophosphamide 60 mg/kg/d for 2 days (days 27 and 26 prior to TIL infusion) and fludarabine 25 mg/m.sup.2/d for 5 days (days 27 to 23 prior to TIL infusion). In an embodiment, after non-myeloablative chemotherapy and TIL infusion (at day 0) according to the present disclosure, the patient receives an intravenous infusion of L-2 intravenously at 720,000 IU/kg every 8 hours to physiologic tolerance.
[1615] Experimental findings indicate that lymphodepletion prior to adoptive transfer of tumor-specific T lymphocytes plays a key role in enhancing treatment efficacy by eliminating regulatory T cells and competing elements of the immune system ("cytokine sinks"). Accordingly, some embodiments of the invention utilize a lymphodepletion step (sometimes also referred to as "immunosuppressive conditioning") on the patient prior to the introduction of the reREP TILs of the invention.
[1616] In general, lymphodepletion is achieved using administration of fludarabine or cyclophosphamide (the active form being referred to as mafosfamide) and combinations thereof. Such methods are described in Gassner, et al., Cancer Immunol. Immunother. 2011, 60, 75-85, Muranski, et al., Nat. Clin. Pract. Oncol., 2006, 3, 668-681, Dudley, et al., J. Clin. Oncol. 2008, 26, 5233-5239, and Dudley, et al., J. Clin. Oncol. 2005, 23, 2346-2357, all of which are incorporated by reference herein in their entireties.
[1617] In some embodiments, the fludarabine is administered at a concentration of 0.5 .mu.g/ml-10 .mu.g/ml fludarabine. In some embodiments, the fludarabine is administered at a concentration of 1 .mu.g/ml fludarabine. In some embodiments, the fludarabine treatment is administered for 1 day, 2 days, 3 days, 4 days, 5 days, 6 days, or 7 days or more. In some embodiments, the fludarabine is administered at a dosage of 10 mg/kg/day, 15 mg/kg/day, 20 mg/kg/day, 25 mg/kg/day, 30 mg/kg/day, 35 mg/kg/day, 40 mg/kg/day, or 45 mg/kg/day. In some embodiments, the fludarabine treatment is administered for 2-7 days at 35 mg/kg/day. In some embodiments, the fludarabine treatment is administered for 4-5 days at 35 mg/kg/day. In some embodiments, the fludarabine treatment is administered for 4-5 days at 25 mg/kg/day.
[1618] In some embodiments, the mafosfamide, the active form of cyclophosphamide, is obtained at a concentration of 0.5 .mu.g/mL-10 .mu.g/mL by administration of cyclophosphamide. In some embodiments, mafosfamide, the active form of cyclophosphamide, is obtained at a concentration of 1 .mu.g/mL by administration of cyclophosphamide. In some embodiments, the cyclophosphamide treatment is administered for 1 day, 2 days, 3 days, 4 days, 5 days, 6 days, or 7 days or more. In some embodiments, the cyclophosphamide is administered at a dosage of 100 mg/m.sup.2/day, 150 mg/m.sup.2/day, 175 mg/m.sup.2/day, 200 mg/m.sup.2/day, 225 mg/m.sup.2/day, 250 mg/m.sup.2/day, 275 mg/m.sup.2/day, or 300 mg/m.sup.2/day. In some embodiments, the cyclophosphamide is administered intravenously (i.e., i.v.) In some embodiments, the cyclophosphamide treatment is administered for 2-7 days at 35 mg/kg/day. In some embodiments, the cyclophosphamide treatment is administered for 4-5 days at 250 mg/m.sup.2/day i.v. In some embodiments, the cyclophosphamide treatment is administered for 4 days at 250 mg/m.sup.2/day i.v.
[1619] In some embodiments, lymphodepletion is performed by administering the fludarabine and the cyclophosphamide are together to a patient. In some embodiments, fludarabine is administered at 25 mg/m.sup.2/day i.v. and cyclophosphamide is administered at 250 mg/m.sup.2/day i.v. over 4 days.
[1620] In an embodiment, the lymphodepletion is performed by administration of cyclophosphamide at a dose of 60 mg/m.sup.2/day for two days followed by administration of fludarabine at a dose of 25 mg/m.sup.2/day for five days.
Combinations with PD-1 and PD-L1 Inhibitors
[1621] Programmed death 1 (PD-1) is a 288-amino acid transmembrane immunocheckpoint receptor protein expressed by T cells, B cells, natural killer (NK) T cells, activated monocytes, and dendritic cells. PD-1, which is also known as CD279, belongs to the CD28 family, and in humans is encoded by the Pdcd1 gene on chromosome 2. PD-1 consists of one immunoglobulin (Ig) superfamily domain, a transmembrane region, and an intracellular domain containing an immunoreceptor tyrosine-based inhibitory motif (ITIM) and an immunoreceptor tyrosine-based switch motif (ITSM). PD-1 and its ligands (PD-L1 and PD-L2) are known to play a key role in immune tolerance, as described in Keir, et al., Annu. Rev. Immunol. 2008, 26, 677-704. PD-1 provides inhibitory signals that negatively regulate T cell immune responses. PD-L1 (also known as B7-H1 or CD274) and PD-L2 (also known as B7-DC or CD273) are expressed on tumor cells and stromal cells, which may be encountered by activated T cells expressing PD-1, leading to immunosuppression of the T cells. PD-L1 is a 290 amino acid transmembrane protein encoded by the Cd274 gene on human chromosome 9. Blocking the interaction between PD-1 and its ligands PD-L1 and PD-L2 by use of a PD-1 inhibitor, a PD-L1 inhibitor, and/or a PD-L2 inhibitor can overcome immune resistance, as demonstrated in recent clinical studies, such as that described in Topalian, et al., N. Eng. J. Med. 2012, 366, 2443-54. PD-L1 is expressed on many tumor cell lines, while PD-L2 is expressed is expressed mostly on dendritic cells and a few tumor lines. In addition to T cells (which inducibly express PD-1 after activation), PD-1 is also expressed on B cells, natural killer cells, macrophages, activated monocytes, and dendritic cells.
[1622] The methods, compositions, and combinations of TILs and TNFRSF agonists described herein may also be further combined with programmed death-1 (PD-1), programmed death ligand 1 (PD-L1), and/or programmed death ligand 2 (PD-L2) binding antibodies, antagonists, or inhibitors (i.e., blockers). PD-1, PD-L1, and/or PD-L2 inhibitors may be used in cell culture in conjunction with the TNFRSF agonists described herein during the pre-REP or REP stages of TIL expansion. PD-1, PD-L1, and/or PD-L2 inhibitors may also be used in conjunction with TNFRSF agonists prior to surgical resection of tumor, or during or after infusion of TILs. For example, suitable methods of using PD-1/PD-L1 inhibitors in conjunction with agonistic GITR antibodies and compositions comprising PD-1/PD-L1 antagonists and GITR agonists are described in International Patent Application Publication No. WO 2015/026684 A1, the disclosures of which are incorporated by reference herein.
[1623] In an embodiment, the PD-1 inhibitor may be any PD-1 inhibitor or PD-1 blocker known in the art. In particular, it is one of the PD-1 inhibitors or blockers described in more detail in the following paragraphs. The terms "inhibitor," "antagonist," and "blocker" are used interchangeably herein in reference to PD-1 inhibitors. For avoidance of doubt, references herein to a PD-1 inhibitor that is an antibody may refer to a compound or antigen-binding fragments, variants, conjugates, or biosimilars thereof. For avoidance of doubt, references herein to a PD-1 inhibitor may also refer to a small molecule compound or a pharmaceutically acceptable salt, ester, solvate, hydrate, cocrystal, or prodrug thereof.
[1624] In some embodiments, the compositions and methods described herein include a PD-1 inhibitor. In some embodiments, the PD-1 inhibitor is a small molecule. In a preferred embodiment, the PD-1 inhibitor is an antibody (i.e., an anti-PD-1 antibody), a fragment thereof, including Fab fragments, or a single-chain variable fragment (scFv) thereof. In some embodiments the PD-1 inhibitor is a polyclonal antibody. In a preferred embodiment, the PD-1 inhibitor is a monoclonal antibody. In some embodiments, the PD-1 inhibitor competes for binding with PD-1, and/or binds to an epitope on PD-1. In an embodiment, the antibody competes for binding with PD-1, and/or binds to an epitope on PD-1.
[1625] In some embodiments, the compositions and methods described include a PD-1 inhibitor that binds human PD-1 with a K.sub.D of about 100 pM or lower, binds human PD-1 with a K.sub.D of about 90 pM or lower, binds human PD-1 with a K.sub.D of about 80 pM or lower, binds human PD-1 with a K.sub.D of about 70 pM or lower, binds human PD-1 with a K.sub.D of about 60 pM or lower, binds human PD-1 with a K.sub.D of about 50 pM or lower, binds human PD-1 with a K.sub.D of about 40 pM or lower, binds human PD-1 with a K.sub.D of about 30 pM or lower, binds human PD-1 with a K.sub.D of about 20 pM or lower, binds human PD-1 with a K.sub.D of about 10 pM or lower, or binds human PD-1 with a K.sub.D of about 1 pM or lower.
[1626] In some embodiments, the compositions and methods described include a PD-1 inhibitor that binds to human PD-1 with a k.sub.assoc of about 7.5.times.10.sup.5 l/Ms or faster, binds to human PD-1 with a k.sub.assoc of about 7.5.times.10.sup.5 l/Ms or faster, binds to human PD-1 with a k.sub.assoc of about 8.times.10.sup.5 l/Ms or faster, binds to human PD-1 with a k.sub.assoc of about 8.5.times.10.sup.5 l/Ms or faster, binds to human PD-1 with a k.sub.assoc of about 9.times.10.sup.5 l/Ms or faster, binds to human PD-1 with a k.sub.assoc of about 9.5.times.10.sup.5 l/Ms or faster, or binds to human PD-1 with a k.sub.assoc of about 1.times.10.sup.6 l/Ms or faster.
[1627] In some embodiments, the compositions and methods described include a PD-1 inhibitor that binds to human PD-1 with a k.sub.dissoc of about 2.times.10.sup.-5 l/s or slower, binds to human PD-1 with a k.sub.dissoc of about 2.1.times.10.sup.-5 l/s or slower, binds to human PD-1 with a k.sub.dissoc of about 2.2.times.10.sup.-5 l/s or slower, binds to human PD-1 with a k.sub.dissoc of about 2.3.times.10.sup.-5 l/s or slower, binds to human PD-1 with a k.sub.dissoc of about 2.4.times.10.sup.-5 l/s or slower, binds to human PD-1 with a k.sub.dissoc of about 2.5.times.10.sup.-5 l/s or slower, binds to human PD-1 with a k.sub.dissoc of about 2.6.times.10.sup.-5 l/s or slower or binds to human PD-1 with a k.sub.dissoc of about 2.7.times.10.sup.-5 l/s or slower, binds to human PD-1 with a k.sub.dissoc of about 2.8.times.10.sup.-5 l/s or slower, binds to human PD-1 with a k.sub.dissoc of about 2.9.times.10.sup.-5 l/s or slower, or binds to human PD-1 with a k.sub.dissoc of about 3.times.10.sup.-5 l/s or slower.
[1628] In some embodiments, the compositions and methods described include a PD-1 inhibitor that blocks or inhibits binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 10 nM or lower, blocks or inhibits binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 9 nM or lower, blocks or inhibits binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 8 nM or lower, blocks or inhibits binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 7 nM or lower, blocks or inhibits binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 6 nM or lower, blocks or inhibits binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 5 nM or lower, blocks or inhibits binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 4 nM or lower, blocks or inhibits binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 3 nM or lower, blocks or inhibits binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 2 nM or lower, or blocks or inhibits binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 1 nM or lower.
[1629] In an embodiment, the PD-1 inhibitor is nivolumab (commercially available as OPDIVO from Bristol-Myers Squibb Co.), or biosimilars, antigen-binding fragments, conjugates, or variants thereof. Nivolumab is a fully human IgG4 antibody blocking the PD-1 receptor. In an embodiment, the anti-PD-1 antibody is an immunoglobulin G4 kappa, anti-(human CD274) antibody. Nivolumab is assigned Chemical Abstracts Service (CAS) registry number 946414-94-4 and is also known as 5C4, BMS-936558, MDX-1106, and ONO-4538. The preparation and properties of nivolumab are described in U.S. Pat. No. 8,008,449 and International Patent Publication No. WO 2006/121168, the disclosures of which are incorporated by reference herein. The clinical safety and efficacy of nivolumab in various forms of cancer has been described in Wang, et al., Cancer Immunol Res. 2014, 2, 846-56; Page, et al., Ann. Rev. Med., 2014, 65, 185-202; and Weber, et al., J. Clin. Oncology, 2013, 31, 4311-4318, the disclosures of which are incorporated by reference herein. The amino acid sequences of nivolumab are set forth in Table 48. Nivolumab has intra-heavy chain disulfide linkages at 22-96, 140-196, 254-314, 360-418, 22''-96'', 140''-196'', 254''-314'', and 360''-418''; intra-light chain disulfide linkages at 23'-88', 134'-194', 23'''-88''', and 134'''-194'''; inter-heavy-light chain disulfide linkages at 127-214', 127''-214''', inter-heavy-heavy chain disulfide linkages at 219-219'' and 222-222''; and N-glycosylation sites (H CH.sub.2 84.4) at 290, 290''.
[1630] In an embodiment, a PD-1 inhibitor comprises a heavy chain given by SEQ ID NO:463 and a light chain given by SEQ ID NO:464. In an embodiment, a PD-1 inhibitor comprises heavy and light chains having the sequences shown in SEQ ID NO:463 and SEQ ID NO:464, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a PD-1 inhibitor comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:463 and SEQ ID NO:464, respectively. In an embodiment, a PD-1 inhibitor comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:463 and SEQ ID NO:464, respectively. In an embodiment, a PD-1 inhibitor comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:463 and SEQ ID NO:464, respectively. In an embodiment, a PD-1 inhibitor comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:463 and SEQ ID NO:464, respectively. In an embodiment, a PD-1 inhibitor comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:463 and SEQ ID NO:464, respectively.
[1631] In an embodiment, the PD-1 inhibitor comprises the heavy and light chain CDRs or variable regions (VRs) of nivolumab. In an embodiment, the PD-1 inhibitor heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:465, and the PD-1 inhibitor light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:466, and conservative amino acid substitutions thereof. In an embodiment, a PD-1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:465 and SEQ ID NO:466, respectively. In an embodiment, a PD-1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:465 and SEQ ID NO:466, respectively. In an embodiment, a PD-1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:465 and SEQ ID NO:466, respectively. In an embodiment, a PD-1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:465 and SEQ ID NO:466, respectively. In an embodiment, a PD-1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:465 and SEQ ID NO:466, respectively.
[1632] In an embodiment, a PD-1 inhibitor comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:467, SEQ ID NO:468, and SEQ ID NO:469, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:470, SEQ ID NO:471, and SEQ ID NO:472, respectively, and conservative amino acid substitutions thereof. In an embodiment, the antibody competes for binding with, and/or binds to the same epitope on PD-1 as any of the aforementioned antibodies.
[1633] In an embodiment, the PD-1 inhibitor is an anti-PD-1 biosimilar monoclonal antibody approved by drug regulatory authorities with reference to nivolumab. In an embodiment, the biosimilar comprises an anti-PD-1 antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is nivolumab. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is an anti-PD-1 antibody authorized or submitted for authorization, wherein the anti-PD-1 antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is nivolumab. The anti-PD-1 antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is nivolumab. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is nivolumab.
TABLE-US-00048 TABLE 48 Amino acid sequences for PD-1 inhibitors related to nivolumab. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 463 QVQLVESGGG VVQPGRSLRL DCKASGITFS NSGMHWVRQA PGKGLEWVAV IWYDGSKRYY 60 nivolumab ADSVKGRFTI SRDNSKNTLF LQMNSLRAED TAVYYCATND DYWGQGTLVT VSSASTKGPS 120 heavy chain VFPLAPCSRS TSESTAALGC LVKDYFPEPV TVSWNSGALT SGVHTFPAVL QSSGLYSLSS 180 VVTVPSSSLG TKTYTCNVDH KPSNTKVDKR VESKYGPPCP PCPAPEFLGG PSVFLFPPKP 240 KDTLMISRTP EVTCVVVDVS QEDPEVQFNW YVDGVEVHNA KTKPREEQFN STYRVVSVLT 300 VLHQDWLNGK EYKCKVSNKG LPSSIEKTIS KAKGQPREPQ VYTLPPSQEE MTKNQVSLTC 360 LVKGFYPSDI AVEWESNGQP ENNYKTTPPV LDSDGSFFLY SRLTVDKSRW QEGNVFSCSV 420 MHEALHNHYT QKSLSLSLGK 440 SEQ ID NO: 464 EIVLTQSPAT LSLSPGERAT LSCRASQSVS SYLAWYQQKP GQAPRLLIYD ASNRATGIPA 60 nivolumab RFSGSGSGTD FTLTISSLEP EDFAVYYCQQ SSNWPRTFGQ GTKVEIKRTV AAPSVFIFPP 120 light chain SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 465 QVQLVESGGG VVQPGRSLRL DCKASGITFS NSGMHWVRQA PGKGLEWVAV IWYDGSKRYY 60 nivolumab ADSVKGRFTI SRDNSKNTLF LQMNSLRAED TAVYYCATND DYWGQGTLVT VSS 113 variable heavy chain SEQ ID NO: 466 EIVLTQSPAT LSLSPGERAT LSCRASQSVS SYLAWYQQKP GQAPRLLIYD ASNRATGIPA 60 nivolumab RFSGSGSGTD FTLTISSLEP EDFAVYYCQQ SSNWPRTFGQ GTKVEIK 107 variable light chain SEQ ID NO: 467 NSGMH 5 nivolumab heavy chain CDR1 SEQ ID NO: 468 VIWYDGSKRY YADSVKG 17 nivolumab heavy chain CDR2 SEQ ID NO: 469 NDDY 4 nivolumab heavy chain CDR3 SEQ ID NO: 470 RASQSVSSYL A 11 nivolumab light chain CDR1 SEQ ID NO: 471 DASNRAT 7 nivolumab light chain CDR2 SEQ ID NO: 472 QQSSNWPRT 9 nivolumab light chain CDR3
[1634] In another embodiment, the PD-1 inhibitor comprises pembrolizumab (commercially available as KEYTRUDA from Merck & Co., Inc., Kenilworth, N.J., USA), or antigen-binding fragments, conjugates, or variants thereof. Pembrolizumab is assigned CAS registry number 1374853-91-4 and is also known as lambrolizumab, MK-3475, and SCH-900475. Pembrolizumab has an immunoglobulin G4, anti-(human protein PDCD1 (programmed cell death 1)) (human-Mus musculus monoclonal heavy chain), disulfide with human-Mus musculus monoclonal light chain, dimer structure. The structure of pembrolizumab may also be described as immunoglobulin G4, anti-(human programmed cell death 1); humanized mouse monoclonal [228-L-proline(H10-S>P)].gamma.4 heavy chain (134-218')-disulfide with humanized mouse monoclonal .kappa. light chain dimer (226-226'':229-229'')-bisdisulfide. The properties, uses, and preparation of pembrolizumab are described in International Patent Publication No. WO 2008/156712 A1, U.S. Pat. No. 8,354,509 and U.S. Patent Application Publication Nos. US 2010/0266617 A1, US 2013/0108651 A1, and US 2013/0109843 A2, the disclosures of which are incorporated herein by reference. The clinical safety and efficacy of pembrolizumab in various forms of cancer is described in Fuerst, Oncology Times, 2014, 36, 35-36; Robert, et al., Lancet, 2014, 384, 1109-17; and Thomas, et al., Exp. Opin. Biol. Ther., 2014, 14, 1061-1064. The amino acid sequences of pembrolizumab are set forth in Table 49. Pembrolizumab includes the following disulfide bridges: 22-96, 22''-96'', 23'-92', 23'''-92''', 134-218', 134''-218''', 138'-198', 138'''-198''', 147-203, 147''-203'', 226-226'', 229-229'', 261-321, 261''-321'', 367-425, and 367''-425'', and the following glycosylation sites (N): Asn-297 and Asn-297''. Pembrolizumab is an IgG4/kappa isotype with a stabilizing S228P mutation in the Fc region; insertion of this mutation in the IgG4 hinge region prevents the formation of half molecules typically observed for IgG4 antibodies. Pembrolizumab is heterogeneously glycosylated at Asn297 within the Fc domain of each heavy chain, yielding a molecular weight of approximately 149 kDa for the intact antibody. The dominant glycoform of pembrolizumab is the fucosylated agalacto diantennary glycan form (GOF).
[1635] In an embodiment, a PD-1 inhibitor comprises a heavy chain given by SEQ ID NO:473 and a light chain given by SEQ ID NO:474. In an embodiment, a PD-1 inhibitor comprises heavy and light chains having the sequences shown in SEQ ID NO:473 and SEQ ID NO:474, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a PD-1 inhibitor comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:473 and SEQ ID NO:474, respectively. In an embodiment, a PD-1 inhibitor comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:473 and SEQ ID NO:474, respectively. In an embodiment, a PD-1 inhibitor comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:473 and SEQ ID NO:474, respectively. In an embodiment, a PD-1 inhibitor comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:473 and SEQ ID NO:474, respectively. In an embodiment, a PD-1 inhibitor comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:473 and SEQ ID NO:474, respectively.
[1636] In an embodiment, the PD-1 inhibitor comprises the heavy and light chain CDRs or variable regions (VRs) of pembrolizumab. In an embodiment, the PD-1 inhibitor heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:475, and the PD-1 inhibitor light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:476, and conservative amino acid substitutions thereof. In an embodiment, a PD-1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:475 and SEQ ID NO:476, respectively. In an embodiment, a PD-1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:475 and SEQ ID NO:476, respectively. In an embodiment, a PD-1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:475 and SEQ ID NO:476, respectively. In an embodiment, a PD-1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:475 and SEQ ID NO:476, respectively. In an embodiment, a PD-1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:475 and SEQ ID NO:476, respectively.
[1637] In an embodiment, a PD-1 inhibitor comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:477, SEQ ID NO:478, and SEQ ID NO:479, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:480, SEQ ID NO:481, and SEQ ID NO:482, respectively, and conservative amino acid substitutions thereof. In an embodiment, the antibody competes for binding with, and/or binds to the same epitope on PD-1 as any of the aforementioned antibodies.
[1638] In an embodiment, the PD-1 inhibitor is an anti-PD-1 biosimilar monoclonal antibody approved by drug regulatory authorities with reference to pembrolizumab. In an embodiment, the biosimilar comprises an anti-PD-1 antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is pembrolizumab. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is an anti-PD-1 antibody authorized or submitted for authorization, wherein the anti-PD-1 antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is pembrolizumab. The anti-PD-1 antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is pembrolizumab. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is pembrolizumab.
TABLE-US-00049 TABLE 49 Amino acid sequences for PD-1 inhibitors related to pembrolizumab. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 473 QVQLVQSGVE VKKPGASVKV SCKASGYTFT NYYMYWVRQA PGQGLEWMGG INPSNGGTNF 60 pembrolizumab NEKFKNRVTL TTDSSTTTAY MELKSLQFDD TAVYYCARRD YRFDMGFDYW GQGTTVTVSS 120 heavy chain ASTKGPSVFP LAPCSRSTSE STAALGCLVK DYFPEPVTVS WNSGALTSGV HTFPAVLQSS 180 GLYSLSSVVT VPSSSLGTKT YTCNVDHKPS NTKVDKRVES KYGPPCPPCP APEFLGGPSV 240 FLFPPKPKDT LMISRTPEVT CVVVDVSQED PEVQFNWYVD GVEVHNAKTK PREEQFNSTY 300 RVVSVLTVLH QDWLNGKEYK CKVSNKGLPS SIEKTISKAK GQPREPQVYT LPPSQEEMTK 360 NQVSLTCLVK GFYPSDIAVE WESNGQPENN YKTTPPVLDS DGSFFLYSRL TVDKSRWQEG 420 NVFSCSVMHE ALHNHYTQKS LSLSLGK 447 SEQ ID NO: 474 EIVLTQSPAT LSLSPGERAT LSCRASKGVS TSGYSYLHWY QQKPGQAPRL LIYLASYLES 60 pembrolizumab GVPARFSGSG SGTDFTLTIS SLEPEDFAVY YCQHSRDLPL TFGGGTKVEI KRTVAAPSVF 120 light chain IFPPSDEQLK SGTASVVCLL NNFYPREAKV QWKVDNALQS GNSQESVTEQ DSKDSTYSLS 180 STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRGEC 218 SEQ ID NO: 475 QVQLVQSGVE VKKPGASVKV SCKASGYTFT NYYMYWVRQA PGQGLEWMGG INPSNGGTNF 60 pembrolizumab NEKFKNRVTL TTDSSTTTAY MELKSLQFDD TAVYYCARRD YRFDMGFDYW GQGTTVTVSS 120 variable heavy chain SEQ ID NO: 476 EIVLTQSPAT LSLSPGERAT LSCRASKGVS TSGYSYLHWY QQKPGQAPRL LIYLASYLES 60 pembrolizumab GVPARFSGSG SGTDFTLTIS SLEPEDFAVY YCQHSRDLPL TFGGGTKVEI K 111 variable light chain SEQ ID NO: 477 NYYMY 5 pembrolizumab heavy chain CDR1 SEQ ID NO: 478 GINPSNGGTN FNEKFK 16 pembrolizumab heavy chain CDR2 SEQ ID NO: 479 RDYRFDMGFD Y 11 pembrolizumab heavy chain CDR3 SEQ ID NO: 480 RASKGVSTSG YSYLH 15 pembrolizumab light chain CDR1 SEQ ID NO: 481 LASYLES 7 pembrolizumab light chain CDR2 SEQ ID NO: 482 QHSRDLPLT 9 pembrolizumab light chain CDR3
[1639] In an embodiment, the PD-1 inhibitor is a commercially-available anti-PD-1 monoclonal antibody, such as anti-m-PD-1 clones J43 (Cat # BE0033-2) and RMP1-14 (Cat # BE0146) (Bio X Cell, Inc., West Lebanon, N.H., USA). A number of commercially-available anti-PD-1 antibodies are known to one of ordinary skill in the art.
[1640] In an embodiment, the PD-1 inhibitor is an antibody disclosed in U.S. Pat. No. 8,354,509 or U.S. Patent Application Publication Nos. 2010/0266617 A1, 2013/0108651 A1, 2013/0109843 A2, the disclosures of which are incorporated by reference herein. In an embodiment, the PD-1 inhibitor is an anti-PD-1 antibody described in U.S. Pat. Nos. 8,287,856, 8,580,247, and 8,168,757 and U.S. Patent Application Publication Nos. 2009/0028857 A1, 2010/0285013 A1, 2013/0022600 A1, and 2011/0008369 A1, the teachings of which are hereby incorporated by reference. In another embodiment, the PD-1 inhibitor is an anti-PD-1 antibody disclosed in U.S. Pat. No. 8,735,553 B1, the disclosure of which is incorporated herein by reference. In an embodiment, the PD-1 inhibitor is pidilizumab, also known as CT-011, which is described in U.S. Pat. No. 8,686,119, the disclosure of which is incorporated by reference herein.
[1641] In an embodiment, the PD-1 inhibitor may be a small molecule or a peptide, or a peptide derivative, such as those described in U.S. Pat. Nos. 8,907,053; 9,096,642; and 9,044,442 and U.S. Patent Application Publication No. US 2015/0087581; 1,2,4-oxadiazole compounds and derivatives such as those described in U.S. Patent Application Publication No. 2015/0073024; cyclic peptidomimetic compounds and derivatives such as those described in U.S. Patent Application Publication No. US 2015/0073042; cyclic compounds and derivatives such as those described in U.S. Patent Application Publication No. US 2015/0125491; 1,3,4-oxadiazole and 1,3,4-thiadiazole compounds and derivatives such as those described in International Patent Application Publication No. WO 2015/033301; peptide-based compounds and derivatives such as those described in International Patent Application Publication Nos. WO 2015/036927 and WO 2015/04490, or a macrocyclic peptide-based compounds and derivatives such as those described in U.S. Patent Application Publication No. US 2014/0294898; the disclosures of each of which are hereby incorporated by reference in their entireties.
[1642] In an embodiment, the PD-L1 or PD-L2 inhibitor may be any PD-L1 or PD-L2 inhibitor, antagonist, or blocker known in the art. In particular, it is one of the PD-L1 or PD-L2 inhibitors, antagonist, or blockers described in more detail in the following paragraphs. The terms "inhibitor," "antagonist," and "blocker" are used interchangeably herein in reference to PD-L1 and PD-L2 inhibitors. For avoidance of doubt, references herein to a PD-L1 or PD-L2 inhibitor that is an antibody may refer to a compound or antigen-binding fragments, variants, conjugates, or biosimilars thereof. For avoidance of doubt, references herein to a PD-L1 or PD-L2 inhibitor may refer to a compound or a pharmaceutically acceptable salt, ester, solvate, hydrate, cocrystal, or prodrug thereof.
[1643] In some embodiments, the compositions, processes and methods described herein include a PD-L1 or PD-L2 inhibitor. In some embodiments, the PD-L1 or PD-L2 inhibitor is a small molecule. In a preferred embodiment, the PD-L1 or PD-L2 inhibitor is an antibody (i.e., an anti-PD-1 antibody), a fragment thereof, including Fab fragments, or a single-chain variable fragment (scFv) thereof. In some embodiments the PD-L1 or PD-L2 inhibitor is a polyclonal antibody. In a preferred embodiment, the PD-L1 or PD-L2 inhibitor is a monoclonal antibody. In some embodiments, the PD-L1 or PD-L2 inhibitor competes for binding with PD-L1 or PD-L2, and/or binds to an epitope on PD-L1 or PD-L2. In an embodiment, the antibody competes for binding with PD-L1 or PD-L2, and/or binds to an epitope on PD-L1 or PD-L2.
[1644] In some embodiments, the PD-L1 inhibitors provided herein are selective for PD-L1, in that the compounds bind or interact with PD-L1 at substantially lower concentrations than they bind or interact with other receptors, including the PD-L2 receptor. In certain embodiments, the compounds bind to the PD-L1 receptor at a binding constant that is at least about a 2-fold higher concentration, about a 3-fold higher concentration, about a 5-fold higher concentration, about a 10-fold higher concentration, about a 20-fold higher concentration, about a 30-fold higher concentration, about a 50-fold higher concentration, about a 100-fold higher concentration, about a 200-fold higher concentration, about a 300-fold higher concentration, or about a 500-fold higher concentration than to the PD-L2 receptor.
[1645] In some embodiments, the PD-L2 inhibitors provided herein are selective for PD-L2, in that the compounds bind or interact with PD-L2 at substantially lower concentrations than they bind or interact with other receptors, including the PD-L1 receptor. In certain embodiments, the compounds bind to the PD-L2 receptor at a binding constant that is at least about a 2-fold higher concentration, about a 3-fold higher concentration, about a 5-fold higher concentration, about a 10-fold higher concentration, about a 20-fold higher concentration, about a 30-fold higher concentration, about a 50-fold higher concentration, about a 100-fold higher concentration, about a 200-fold higher concentration, about a 300-fold higher concentration, or about a 500-fold higher concentration than to the PD-L1 receptor.
[1646] Without being bound by any theory, it is believed that tumor cells express PD-L1, and that T cells express PD-1. However, PD-L1 expression by tumor cells is not required for efficacy of PD-1 or PD-L1 inhibitors or blockers. In an embodiment, the tumor cells express PD-L1. In another embodiment, the tumor cells do not express PD-L1. In some embodiments, the methods and compositions described herein include a combination of a PD-1 and a PD-L1 antibody, such as those described herein, in combination with a TIL. The administration of a combination of a PD-1 and a PD-L1 antibody and a TIL may be simultaneous or sequential.
[1647] In some embodiments, the compositions and methods described include a PD-L1 and/or PD-L2 inhibitor that binds human PD-L1 and/or PD-L2 with a K.sub.D of about 100 pM or lower, binds human PD-L1 and/or PD-L2 with a K.sub.D of about 90 pM or lower, binds human PD-L1 and/or PD-L2 with a K.sub.D of about 80 pM or lower, binds human PD-L1 and/or PD-L2 with a K.sub.D of about 70 pM or lower, binds human PD-L1 and/or PD-L2 with a K.sub.D of about 60 pM or lower, a K.sub.D of about 50 pM or lower, binds human PD-L1 and/or PD-L2 with a K.sub.D of about 40 pM or lower, or binds human PD-L1 and/or PD-L2 with a K.sub.D of about 30 pM or lower,
[1648] In some embodiments, the compositions and methods described include a PD-L1 and/or PD-L2 inhibitor that binds to human PD-L1 and/or PD-L2 with a k.sub.assoc of about 7.5.times.10.sup.5 l/Ms or faster, binds to human PD-L1 and/or PD-L2 with a k.sub.assoc of about 8.times.10.sup.5 l/Ms or faster, binds to human PD-L1 and/or PD-L2 with a k.sub.assoc of about 8.5.times.10.sup.5 l/Ms or faster, binds to human PD-L1 and/or PD-L2 with a k.sub.assoc of about 9.times.10.sup.5 l/Ms or faster, binds to human PD-L1 and/or PD-L2 with a k.sub.assoc of about 9.5.times.10.sup.5 l/Ms and/or faster, or binds to human PD-L1 and/or PD-L2 with a k.sub.assoc of about 1.times.10.sup.6 l/Ms or faster.
[1649] In some embodiments, the compositions and methods described include a PD-L1 and/or PD-L2 inhibitor that binds to human PD-L1 or PD-L2 with a k.sub.dissoc of about 2.times.10.sup.-5 l/s or slower, binds to human PD-1 with a k.sub.dissoc of about 2.1.times.10.sup.-5 l/s or slower, binds to human PD-1 with a k.sub.dissoc of about 2.2.times.10.sup.-5 l/s or slower, binds to human PD-1 with a k.sub.dissoc of about 2.3.times.10.sup.-5 l/s or slower, binds to human PD-1 with a k.sub.dissoc of about 2.4.times.10.sup.-5 l/s or slower, binds to human PD-1 with a k.sub.dissoc of about 2.5.times.10.sup.-5 l/s or slower, binds to human PD-1 with a k.sub.dissoc of about 2.6.times.10.sup.-5 l/s or slower, binds to human PD-L1 or PD-L2 with a k.sub.dissoc of about 2.7.times.10.sup.-5 l/s or slower, or binds to human PD-L1 or PD-L2 with a k.sub.dissoc of about 3.times.10.sup.-5 l/s or slower.
[1650] In some embodiments, the compositions and methods described include a PD-L1 and/or PD-L2 inhibitor that blocks or inhibits binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 10 nM or lower; blocks or inhibits binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 9 nM or lower; blocks or inhibits binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 8 nM or lower; blocks or inhibits binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 7 nM or lower; blocks or inhibits binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 6 nM or lower; blocks or inhibits binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 5 nM or lower; blocks or inhibits binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 4 nM or lower; blocks or inhibits binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 3 nM or lower; blocks or inhibits binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 2 nM or lower; or blocks human PD-1, or blocks binding of human PD-L1 or human PD-L2 to human PD-1 with an IC.sub.50 of about 1 nM or lower.
[1651] In an embodiment, the PD-L1 inhibitor is durvalumab, also known as MEDI4736 (which is commercially available from Medimmune, LLC, Gaithersburg, Md., a subsidiary of AstraZeneca plc.), or antigen-binding fragments, conjugates, or variants thereof. In an embodiment, the PD-L1 inhibitor is an antibody disclosed in U.S. Pat. No. 8,779,108 or U.S. Patent Application Publication No. 2013/0034559, the disclosures of which are incorporated by reference herein. The clinical efficacy of durvalumab has been described in Page, et al., Ann. Rev. Med., 2014, 65, 185-202; Brahmer, et al., J. Clin. Oncol. 2014, 32, 5s (supplement, abstract 8021); and McDermott, et al., Cancer Treatment Rev., 2014, 40, 1056-64. The preparation and properties of durvalumab are described in U.S. Pat. No. 8,779,108, the disclosure of which is incorporated by reference herein. The amino acid sequences of durvalumab are set forth in Table 50. The durvalumab monoclonal antibody includes disulfide linkages at 22-96, 22''-96'', 23'-89', 23'''-89''', 135'-195', 135'''-195''', 148-204, 148''-204'', 215'-224, 215'''-224'', 230-230'', 233-233'', 265-325, 265''-325'', 371-429, and 371''-429'; and N-glycosylation sites at Asn-301 and Asn-301''.
[1652] In an embodiment, a PD-L1 inhibitor comprises a heavy chain given by SEQ ID NO:483 and a light chain given by SEQ ID NO:484. In an embodiment, a PD-L1 inhibitor comprises heavy and light chains having the sequences shown in SEQ ID NO:483 and SEQ ID NO:484, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a PD-L1 inhibitor comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:483 and SEQ ID NO:484, respectively. In an embodiment, a PD-L1 inhibitor comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:483 and SEQ ID NO:484, respectively. In an embodiment, a PD-L1 inhibitor comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:483 and SEQ ID NO:484, respectively. In an embodiment, a PD-L1 inhibitor comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:483 and SEQ ID NO:484, respectively. In an embodiment, a PD-L1 inhibitor comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:483 and SEQ ID NO:484, respectively.
[1653] In an embodiment, the PD-L1 inhibitor comprises the heavy and light chain CDRs or variable regions (VRs) of durvalumab. In an embodiment, the PD-L1 inhibitor heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:485, and the PD-L1 inhibitor light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:486, and conservative amino acid substitutions thereof. In an embodiment, a PD-L1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:485 and SEQ ID NO:486, respectively. In an embodiment, a PD-L1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:485 and SEQ ID NO:486, respectively. In an embodiment, a PD-L1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:485 and SEQ ID NO:486, respectively. In an embodiment, a PD-L1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:485 and SEQ ID NO:486, respectively. In an embodiment, a PD-L1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:485 and SEQ ID NO:486, respectively.
[1654] In an embodiment, a PD-L1 inhibitor comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:487, SEQ ID NO:488, and SEQ ID NO:489, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:490, SEQ ID NO:491, and SEQ ID NO:492, respectively, and conservative amino acid substitutions thereof. In an embodiment, the antibody competes for binding with, and/or binds to the same epitope on PD-L1 as any of the aforementioned antibodies.
[1655] In an embodiment, the PD-L1 inhibitor is an anti-PD-L1 biosimilar monoclonal antibody approved by drug regulatory authorities with reference to durvalumab. In an embodiment, the biosimilar comprises an anti-PD-L1 antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is durvalumab. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is an anti-PD-L1 antibody authorized or submitted for authorization, wherein the anti-PD-L1 antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is durvalumab. The anti-PD-L1 antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is durvalumab. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is durvalumab.
TABLE-US-00050 TABLE 50 Amino acid sequences for PD-L1 inhibitors related to durvalumab. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 483 EVQLVESGGG LVQPGGSLRL SCAASGFTFS RYWMSWVRQA PGKGLEWVAN IKQDGSEKYY 60 durvalumab VDSVKGRFTI SRDNAKNSLY LQMNSLRAED TAVYYCAREG GWFGELAFDY WGQGTLVTVS 120 heavy chain SASTKGPSVF PLAPSSKSTS GGTAALGCLV KDYFPEPVTV SWNSGALTSG VHTFPAVLQS 180 SGLYSLSSVV TVPSSSLGTQ TYICNVNHKP SNTKVDKRVE PKSCDKTHTC PPCPAPEFEG 240 GPSVFLFPPK PKDTLMISRT PEVTCVVVDV SHEDPEVKFN WYVDGVEVHN AKTKPREEQY 300 NSTYRVVSVL TVLHQDWLNG KEYKCKVSNK ALPASIEKTI SKAKGQPREP QVYTLPPSRE 360 EMTKNQVSLT CLVKGFYPSD IAVEWESNGQ PENNYKTTPP VLDSDGSFFL YSKLTVDKSR 420 WQQGNVFSCS VMHEALHNHY TQKSLSLSPG K 451 SEQ ID NO: 484 EVQLVESGGG LVQPGGSLRL SCAASGFTFS RYWMSWVRQA PGKGLEWVAN EIVLTQSPGT 60 durvalumab LSLSPGERAT LSCRASQRVS SSYLAWYQQK PGQAPRLLIY DASSRATGIP DRFSGSGSGT 120 light chain DFTLTISRLE PEDFAVYYCQ QYGSLPWTFG QGTKVEIKRT VAAPSVFIFP PSDEQLKSGT 180 ASVVCLLNNF YPREAKVQWK VDNALQSGNS QESVTEQDSK DSTYSLSSTL TLSKADYEKH 240 KVYACEVTHQ GLSSPVTKSF NRGEC 265 SEQ ID NO: 485 EVQLVESGGG LVQPGGSLRL SCAASGFTFS RYWMSWVRQA PGKGLEWVAN IKQDGSEKYY 60 durvalumab VDSVKGRFTI SRDNAKNSLY LQMNSLRAED TAVYYCAREG GWFGELAFDY WGQGTLVTVS 120 variable S 121 heavy chain SEQ ID NO: 486 EIVLTQSPGT LSLSPGERAT LSCRASQRVS SSYLAWYQQK PGQAPRLLIY DASSRATGIP 60 durvalumab DRFSGSGSGT DFTLTISRLE PEDFAVYYCQ QYGSLPWTFG QGTKVEIK 108 variable light chain SEQ ID NO: 487 RYWMS 5 durvalumab heavy chain CDR1 SEQ ID NO: 488 NIKQDGSEKY YVDSVKG 17 durvalumab heavy chain CDR2 SEQ ID NO: 489 EGGWFGELAF DY 12 durvalumab heavy chain CDR3 SEQ ID NO: 490 RASQRVSSSY LA 12 durvalumab light chain CDR1 SEQ ID NO: 491 DASSRAT 7 durvalumab light chain CDR2 SEQ ID NO: 492 QQYGSLPWT 9 durvalumab light chain CDR3
[1656] In an embodiment, the PD-L1 inhibitor is avelumab, also known as MSB0010718C (commercially available from Merck KGaA/EMD Serono), or antigen-binding fragments, conjugates, or variants thereof. The preparation and properties of avelumab are described in U.S. Patent Application Publication No. US 2014/0341917 A1, the disclosure of which is specifically incorporated by reference herein. The amino acid sequences of avelumab are set forth in Table 51. Avelumab has intra-heavy chain disulfide linkages (C23-C104) at 22-96, 147-203, 264-324, 370-428, 22''-96'', 147''-203'', 264''-324'', and 370''-428''; intra-light chain disulfide linkages (C23-C104) at 22'-90', 138'-197', 22'''-90''', and 138'''-197'''; intra-heavy-light chain disulfide linkages (h 5-CL 126) at 223-215' and 223''-215'''; intra-heavy-heavy chain disulfide linkages (h 11, h 14) at 229-229'' and 232-232''; N-glycosylation sites (H CH.sub.2 N84.4) at 300, 300''; fucosylated complex bi-antennary CHO-type glycans; and H CHS K2 C-terminal lysine clipping at 450 and 450'.
[1657] In an embodiment, a PD-L1 inhibitor comprises a heavy chain given by SEQ ID NO:493 and a light chain given by SEQ ID NO:494. In an embodiment, a PD-L1 inhibitor comprises heavy and light chains having the sequences shown in SEQ ID NO:493 and SEQ ID NO:494, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a PD-L1 inhibitor comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:493 and SEQ ID NO:494, respectively. In an embodiment, a PD-L1 inhibitor comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:493 and SEQ ID NO:494, respectively. In an embodiment, a PD-L1 inhibitor comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:493 and SEQ ID NO:494, respectively. In an embodiment, a PD-L1 inhibitor comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:493 and SEQ ID NO:494, respectively. In an embodiment, a PD-L1 inhibitor comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:493 and SEQ ID NO:494, respectively.
[1658] In an embodiment, the PD-L1 inhibitor comprises the heavy and light chain CDRs or variable regions (VRs) of avelumab. In an embodiment, the PD-L1 inhibitor heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:495, and the PD-L1 inhibitor light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:496, and conservative amino acid substitutions thereof. In an embodiment, a PD-L1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:495 and SEQ ID NO:496, respectively. In an embodiment, a PD-L1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:496 and SEQ ID NO:496, respectively. In an embodiment, a PD-L1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:495 and SEQ ID NO:496, respectively. In an embodiment, a PD-L1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:495 and SEQ ID NO:496, respectively. In an embodiment, a PD-L1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:495 and SEQ ID NO:496, respectively.
[1659] In an embodiment, a PD-L1 inhibitor comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:497, SEQ ID NO:498, and SEQ ID NO:499, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:500, SEQ ID NO:501, and SEQ ID NO:502, respectively, and conservative amino acid substitutions thereof. In an embodiment, the antibody competes for binding with, and/or binds to the same epitope on PD-L1 as any of the aforementioned antibodies.
[1660] In an embodiment, the PD-L1 inhibitor is an anti-PD-L1 biosimilar monoclonal antibody approved by drug regulatory authorities with reference to avelumab. In an embodiment, the biosimilar comprises an anti-PD-L1 antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is avelumab. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is an anti-PD-L1 antibody authorized or submitted for authorization, wherein the anti-PD-L1 antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is avelumab. The anti-PD-L1 antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is avelumab. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is avelumab.
TABLE-US-00051 TABLE 51 Amino acid sequences for PD-L1 inhibitors related to avelumab. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 493 EVQLLESGGG LVQPGGSLRL SCAASGFTFS SYIMMWVRQA PGKGLEWVSS IYPSGGITFY 60 avelumab ADTVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARIK LGTVTTVDYW GQGTLVTVSS 120 heavy chain ASTKGPSVFP LAPSSKSTSG GTAALGCLVK DYFPEPVTVS WNSGALTSGV HTFPAVLQSS 180 GLYSLSSVVT VPSSSLGTQT YICNVNHKPS NTKVDKKVEP KSCDKTHTCP PCPAPELLGG 240 PSVFLFPPKP KDTLMISRTP EVTCVVVDVS HEDPEVKFNW YVDGVEVHNA KTKPREEQYN 300 STYRVVSVLT VLHQDWLNGK EYKCKVSNKA LPAPIEKTIS KAKGQPREPQ VYTLPPSRDE 360 LTKNQVSLTC LVKGFYPSDI AVEWESNGQP ENNYKTTPPV LDSDGSFFLY SKLTVDKSRW 420 QQGNVFSCSV MHEALHNHYT QKSLSLSPGK 450 SEQ ID NO: 494 QSALTQPASV SGSPGQSITI SCTGTSSDVG GYNYVSWYQQ HPGKAPKLMI YDVSNRPSGV 60 avelumab SNRFSGSKSG NTASLTISGL QAEDEADYYC SSYTSSSTRV FGTGTKVTVL GQPKANPTVT 120 light chain LFPPSSEELQ ANKATLVCLI SDFYPGAVTV AWKADGSPVK AGVETTKPSK QSNNKYAASS 180 YLSLTPEQWK SHRSYSCQVT HEGSTVEKTV APTECS 216 SEQ ID NO: 495 EVQLLESGGG LVQPGGSLRL SCAASGFTFS SYIMMWVRQA PGKGLEWVSS IYPSGGITFY 60 avelumab ADTVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARIK LGTVTTVDYW GQGTLVTVSS 120 variable heavy chain SEQ ID NO: 496 QSALTQPASV SGSPGQSITI SCTGTSSDVG GYNYVSWYQQ HPGKAPKLMI YDVSNRPSGV 60 avelumab SNRFSGSKSG NTASLTISGL QAEDEADYYC SSYTSSSTRV FGTGTKVTVL 110 variable light chain SEQ ID NO: 497 SYIMM 5 avelumab heavy chain CDR1 SEQ ID NO: 498 SIYPSGGITF YADTVKG 17 avelumab heavy chain CDR2 SEQ ID NO: 499 IKLGTVTTVD Y 11 avelumab heavy chain CDR3 SEQ ID NO: 500 TGTSSDVGGY NYVS 14 avelumab light chain CDR1 SEQ ID NO: 501 DVSNRPS 7 avelumab light chain CDR2 SEQ ID NO: 502 SSYTSSSTRV 10 avelumab light chain CDR3
[1661] In an embodiment, the PD-L1 inhibitor is atezolizumab, also known as MPDL3280A or RG7446 (commercially available as TECENTRIQ from Genentech, Inc., a subsidiary of Roche Holding AG, Basel, Switzerland), or antigen-binding fragments, conjugates, or variants thereof. In an embodiment, the PD-L1 inhibitor is an antibody disclosed in U.S. Pat. No. 8,217,149, the disclosure of which is specifically incorporated by reference herein. In an embodiment, the PD-L1 inhibitor is an antibody disclosed in U.S. Patent Application Publication Nos. 2010/0203056 A1, 2013/0045200 A1, 2013/0045201 A1, 2013/0045202 A1, or 2014/0065135 A1, the disclosures of which are specifically incorporated by reference herein. The preparation and properties of atezolizumab are described in U.S. Pat. No. 8,217,149, the disclosure of which is incorporated by reference herein. The amino acid sequences of atezolizumab are set forth in Table 52. Atezolizumab has intra-heavy chain disulfide linkages (C23-C104) at 22-96, 145-201, 262-322, 368-426, 22''-96'', 145''-201'', 262''-322'', and 368''-426''; intra-light chain disulfide linkages (C23-C104) at 23'-88', 134'-194', 23'''-88''', and 134'''-194'''; intra-heavy-light chain disulfide linkages (h 5-CL 126) at 221-214' and 221''-214'''; intra-heavy-heavy chain disulfide linkages (h 11, h 14) at 227-227'' and 230-230''; and N-glycosylation sites (H CH.sub.2 N84.4>A) at 298 and 298'.
[1662] In an embodiment, a PD-L1 inhibitor comprises a heavy chain given by SEQ ID NO:503 and a light chain given by SEQ ID NO:504. In an embodiment, a PD-L1 inhibitor comprises heavy and light chains having the sequences shown in SEQ ID NO:503 and SEQ ID NO:504, respectively, or antigen binding fragments, Fab fragments, single-chain variable fragments (scFv), variants, or conjugates thereof. In an embodiment, a PD-L1 inhibitor comprises heavy and light chains that are each at least 99% identical to the sequences shown in SEQ ID NO:503 and SEQ ID NO:504, respectively. In an embodiment, a PD-L1 inhibitor comprises heavy and light chains that are each at least 98% identical to the sequences shown in SEQ ID NO:503 and SEQ ID NO:504, respectively. In an embodiment, a PD-L1 inhibitor comprises heavy and light chains that are each at least 97% identical to the sequences shown in SEQ ID NO:503 and SEQ ID NO:504, respectively. In an embodiment, a PD-L1 inhibitor comprises heavy and light chains that are each at least 96% identical to the sequences shown in SEQ ID NO:503 and SEQ ID NO:504, respectively. In an embodiment, a PD-L1 inhibitor comprises heavy and light chains that are each at least 95% identical to the sequences shown in SEQ ID NO:503 and SEQ ID NO:504, respectively.
[1663] In an embodiment, the PD-L1 inhibitor comprises the heavy and light chain CDRs or variable regions (VRs) of atezolizumab. In an embodiment, the PD-L1 inhibitor heavy chain variable region (V.sub.H) comprises the sequence shown in SEQ ID NO:505, and the PD-L1 inhibitor light chain variable region (V.sub.L) comprises the sequence shown in SEQ ID NO:506, and conservative amino acid substitutions thereof. In an embodiment, a PD-L1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 99% identical to the sequences shown in SEQ ID NO:505 and SEQ ID NO:506, respectively. In an embodiment, a PD-L1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 98% identical to the sequences shown in SEQ ID NO:505 and SEQ ID NO:506, respectively. In an embodiment, a PD-L1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 97% identical to the sequences shown in SEQ ID NO:505 and SEQ ID NO:506, respectively. In an embodiment, a PD-L1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 96% identical to the sequences shown in SEQ ID NO:505 and SEQ ID NO:506, respectively. In an embodiment, a PD-L1 inhibitor comprises V.sub.H and V.sub.L regions that are each at least 95% identical to the sequences shown in SEQ ID NO:505 and SEQ ID NO:506, respectively.
[1664] In an embodiment, a PD-L1 inhibitor comprises heavy chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:507, SEQ ID NO:508, and SEQ ID NO:509, respectively, and conservative amino acid substitutions thereof, and light chain CDR1, CDR2 and CDR3 domains having the sequences set forth in SEQ ID NO:510, SEQ ID NO:511, and SEQ ID NO:512, respectively, and conservative amino acid substitutions thereof. In an embodiment, the antibody competes for binding with, and/or binds to the same epitope on PD-L1 as any of the aforementioned antibodies.
[1665] In an embodiment, the anti-PD-L1 antibody is an anti-PD-L1 biosimilar monoclonal antibody approved by drug regulatory authorities with reference to atezolizumab. In an embodiment, the biosimilar comprises an anti-PD-L1 antibody comprising an amino acid sequence which has at least 97% sequence identity, e.g., 97%, 98%, 99% or 100% sequence identity, to the amino acid sequence of a reference medicinal product or reference biological product and which comprises one or more post-translational modifications as compared to the reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is atezolizumab. In some embodiments, the one or more post-translational modifications are selected from one or more of: glycosylation, oxidation, deamidation, and truncation. In some embodiments, the biosimilar is an anti-PD-L1 antibody authorized or submitted for authorization, wherein the anti-PD-L1 antibody is provided in a formulation which differs from the formulations of a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is atezolizumab. The anti-PD-L1 antibody may be authorized by a drug regulatory authority such as the U.S. FDA and/or the European Union's EMA. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is atezolizumab. In some embodiments, the biosimilar is provided as a composition which further comprises one or more excipients, wherein the one or more excipients are the same or different to the excipients comprised in a reference medicinal product or reference biological product, wherein the reference medicinal product or reference biological product is atezolizumab.
TABLE-US-00052 TABLE 52 Amino acid sequences for PD-L1 inhibitors related to atezolizumab. Identifier Sequence (One-Letter Amino Acid Symbols) SEQ ID NO: 503 EVQLVESGGG LVQPGGSLRL SCAASGFTFS DSWIHWVRQA PGKGLEWVAW ISPYGGSTYY 60 atezolizumab ADSVKGRFTI SADTSKNTAY LQMNSLRAED TAVYYCARRH WPGGFDYWGQ GTLVTVSSAS 120 heavy chain TKGPSVFPLA PSSKSTSGGT AALGCLVKDY FPEPVTVSWN SGALTSGVHT FPAVLQSSGL 180 YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KVDKKVEPKS CDKTHTCPPC PAPELLGGPS 240 VFLFPPKPKD TLMISRTPEV TCVVVDVSHE DPEVKFNWYV DGVEVHNAKT KPREEQYAST 300 YRVVSVLTVL HQDWLNGKEY KCKVSNKALP APIEKTISKA KGQPREPQVY TLPPSREEMT 360 KNQVSLTCLV KGFYPSDIAV EWESNGQPEN NYKTTPPVLD SDGSFFLYSK LTVDKSRWQQ 420 GNVFSCSVMH EALHNHYTQK SLSLSPGK 448 SEQ ID NO: 504 DIQMTQSPSS LSASVGDRVT ITCRASQDVS TAVAWYQQKP GKAPKLLIYS ASFLYSGVPS 60 atezolizumab RFSGSGSGTD FTLTISSLQP EDFATYYCQQ YLYHPATFGQ GTKVEIKRTV AAPSVFIFPP 120 light chain SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD STYSLSSTLT 180 LSKADYEKHK VYACEVTHQG LSSPVTKSFN RGEC 214 SEQ ID NO: 505 EVQLVESGGG LVQPGGSLRL SCAASGFTFS DSWIHWVRQA PGKGLEWVAW ISPYGGSTYY 60 atezolizumab ADSVKGRFTI SADTSKNTAY LQMNSLRAED TAVYYCARRH WPGGFDYWGQ GTLVTVSA 118 variable heavy chain SEQ ID NO: 506 DIQMTQSPSS LSASVGDRVT ITCRASQDVS TAVAWYQQKP GKAPKLLIYS ASFLYSGVPS 60 atezolizumab RFSGSGSGTD FTLTISSLQP EDFATYYCQQ YLYHPATFGQ GTKVEIKR 108 variable light chain SEQ ID NO: 507 GFTFSDSWIH 10 atezolizumab heavy chain CDR1 SEQ ID NO: 508 AWISPYGGST YYADSVKG 18 atezolizumab heavy chain CDR2 SEQ ID NO: 509 RHWPGGFDY 9 atezolizumab heavy chain CDR3 SEQ ID NO: 510 RASQDVSTAV A 11 atezolizumab light chain CDR1 SEQ ID NO: 511 SASFLYS 7 atezolizumab light chain CDR2 SEQ ID NO: 512 QQYLYHPAT 9 atezolizumab light chain CDR3
[1666] In an embodiment, PD-L1 inhibitors include those antibodies described in U.S. Patent Application Publication No. US 2014/0341917 A1, the disclosure of which is incorporated by reference herein. In another embodiment, antibodies that compete with any of these antibodies for binding to PD-L1 are also included. In an embodiment, the anti-PD-L1 antibody is MDX-1105, also known as BMS-935559, which is disclosed in U.S. Pat. No. 7,943,743, the disclosures of which are incorporated by reference herein. In an embodiment, the anti-PD-L1 antibody is selected from the anti-PD-L1 antibodies disclosed in U.S. Pat. No. 7,943,743, which are incorporated by reference herein.
[1667] In an embodiment, the PD-L1 inhibitor is a commercially-available monoclonal antibody, such as INVIVOMAB anti-m-PD-L1 clone 10F.9G2 (Catalog # BE0101, Bio X Cell, Inc., West Lebanon, N.H., USA). In an embodiment, the anti-PD-L1 antibody is a commercially-available monoclonal antibody, such as AFFYMETRIX EBIOSCIENCE (MIH1). A number of commercially-available anti-PD-L1 antibodies are known to one of ordinary skill in the art.
[1668] In an embodiment, the PD-L2 inhibitor is a commercially-available monoclonal antibody, such as BIOLEGEND 24F.10C12 Mouse IgG2a, .kappa. isotype (catalog #329602 Biolegend, Inc., San Diego, Calif.), SIGMA anti-PD-L2 antibody (catalog # SAB3500395, Sigma-Aldrich Co., St. Louis, Mo.), or other commercially-available anti-PD-L2 antibodies known to one of ordinary skill in the art.
[1669] While preferred embodiments of the invention are shown and described herein, such embodiments are provided by way of example only and are not intended to otherwise limit the scope of the invention. Various alternatives to the described embodiments of the invention may be employed in practicing the invention.
EXAMPLES
[1670] The embodiments encompassed herein are now described with reference to the following examples. These examples are provided for the purpose of illustration only and the disclosure encompassed herein should in no way be construed as being limited to these examples, but rather should be construed to encompass any and all variations which become evident as a result of the teachings provided herein.
Example 1--Methods of Expanding TILs and Treating Cancer with Expanded TILs
[1671] TILs may be expanded using methods known in the art and any method described herein. For example, methods for expanding TILs are depicted in FIG. 1. A TNFRSF agonist may be added to the method of FIG. 1 as described herein. The TNFRSF agonist may be, for example, a 4-1BB or an OX40 agonist, and may be added during the pre-REP or the REP phases, or during both phases, at concentrations sufficient to enhance TIL growth. The expansion of TILs may be further combined with any method of treating cancer in combination with a TNFRSF agonist in a patient described herein. Methods for expanding TILs and treating a cancer patient with expanded TILs are shown in FIG. 2.
Example 2--Methods of Expanding TILs Using 4-1BB Agonists
[1672] 4-1BB-Fc hybridomas were produced and screened for their ability to activate the 4-1BB signaling pathway by cross-linking with secondary antibody. Hybridoma supernatants were evaluated for their ability to activate 4-1BB signaling on Jurkat cells expressing NF-kB using a green fluorescent protein (GFP) reporter in a dose dependent manner. Supernatants were incubated with Jurkat cells for 20 minutes at room temperature followed by crosslinking with goat anti-mouse secondary antibody (1 mg/mL) overnight at 37.degree. C. Results are shown in FIG. 3 to FIG. 12 for ten clones identified that bound to 4-1BB on Jurkat cells.
[1673] Cells were analyzed on Intellicyte for NKkB Pathway activation revealed by GFP reporter expression. The results indicate that the antibodies activate the 4-1BB pathway on Jurkat cells without secondary antibody crosslinking.
Example 3--Methods of Expanding TILs from Ex-Vivo Cultured Solid Tumor Fragments (Multiple Histologies) Using Utomilumab or Urelumab and 11D4/18D8 and Effect of Activation with Antibodies to 4-1BB and/or OX40 on Expansion and Function of TILs
[1674] TILs are primarily antigen experienced (non-naive) T cells found to varying degrees in all adult tumors associated with immunosuppressive microenvironments in which the local accumulation of damage associated molecular pattern molecules (DAMPs) as well as induced checkpoint receptors including CTLA-4 and PD-1 have often been engaged. Chacon, et al., Clin. Cancer Res. 2015, 21, 611-21; Joseph, et al., Clin. Cancer Res. 2011, 17, 4882-91. These markers, as well as TIM3, LAG3, and TIGIT, define an exhausted phenotype. As such, TIL-expressed co-stimulatory receptors modify TIL fate and expansion. Activation of 4-1BB and or OX40 on TILs enables expansion of TILs from tumor fragments beyond that achievable with IL-2 alone. Activation of other co-stimulatory receptors and/or antagonism of checkpoint receptors will further enhance TIL function (survival, circumvention of tumor immunosuppression), emigration from tumor fragments, and promote in-vitro expansion. Furthermore, these studies in vitro may predict responsiveness to in vivo application of these antibodies alone or in combination with adoptive transfer of TILs. Immunomodulatory mAbs specific for these two activating co-stimulator molecules (e.g., OX40, 11D4 or 18D8, and 4-1BB, utomilumab or urelumab) can be tested for such capacity. It is hypothesized that activation of the costimulator receptors, 4-1BB and OX40, within tumor fragments enhances TIL emigration from fragments of tumor, proliferation, promotion of a memory phenotype and cytotoxicity of emergent T cells. The main goal of this study is to determine whether mAbs specific for (a) 4-1BB and OX40 in combination or (b) anti-4-1BB and (c) anti-OX40 alone augments the outgrowth of cytotoxic and memory phenotype of TIL from tumor fragments.
[1675] 15 mg of each purified mAb specific for (a) OX40 and (b) 4-1BB is used. Tumors of various histologies may be obtained from commercial sources. In total, 20 independent patient tumors will be obtained. Tumors will be shipped in sterile HBSS or another appropriate medium. The tumors will be handled only in a laminar flow hood to maintain sterile conditions. When possible (if tumor >0.5 cm in diameter), a portion of the tumor will be processed for FFPE and/or cryopreserved for downstream IHC and/or DNA/RNA isolation. Biomarker analysis via IHC will include CD3, CD11c, and PD1 and PD-L1. Whenever possible, autologous blood samples (up to 20 mL) will be acquired and PBMCs will be cryopreserved. If whole exome sequencing is performed on the tumors, exome sequences from banked autologous PBMCs will be defined as normal (i.e., no mutations). Alternatively, tumor single cell suspensions may be utilized. The tumors will be washed after receipt and divided into 2-3 mm fragments and placed into cell culture into 24-well plates (1 fragment per well) or 6-well plates (4 fragments per well) with culture medium supplemented with 6,000 IU/mL L-2 (recombinant) only, OX40 agonist, anti-4-1BB agonist, and a combination of OX40 and 4-1BB agonists in triplicates. In some experiments where sufficient tumor is available, titrations of L-2 (6,000; 600; 60; and 0 IU of L-2) will be tested. An excipient control for the IL-2 will be used. The final concentration of each mAb will be 30 .mu.g/mL. Following 24-48 hours of culture, 250 .mu.L of supernatant will be collected from each condition and stored at -20.degree. C. for subsequent analysis of cytokine and chemokine concentrations (pg/10.sup.6 cell/24 hours). TILs will be collected from each condition on day 11, day 21 and/or day of the `pre-REP` (at least 500,000 cells per sample). Two aliquots of TILs will be pelleted and resuspended in <10 .mu.L of PBS and will be frozen in -80 C. If less than <10.sup.6 cells are collected, only gene expression arrays will be performed. Cultures will be fed on day 7 by partial removal of "spent" medium and addition of an equal volume of culture medium plus 6000 IU/mL IL-2. The spent medium will be stored at -20.degree. C. for subsequent cytokine/chemokine analysis using a multiplex assay (e.g., Luminex 100 system). Additional mAb will be added to the culture on day 7 if sufficient tumor fragments are available for initiation of more than 1 replicate of experimental conditions. TIL cultures will be maintained for an additional 14 days. On day 21, the total cell yield, viability, cell surface and intracellular immunophenotype will be determined using flow cytometry. The following markers will be included: CD45RA, CCR7, CD3, TCR-alpha/beta, CD4, CD8, CXCR3, CD56, CD27, CD28, PD-1, PD-L1, BTLA, KLRG1, CD137, CD134, CD33, CD57, CD25, CD127, TIM-3, LAG-3, TIGIT, RAGE, and Ki67. Other biomarkers including CD107a, NKG2D, KIRS, chemokine death receptors (Fas, DR4) and anti-apoptotic/pro-autophagic proteins (BCL-2, BCL-XL, Bim, CD200, and LC3/HMGB1) will also be assessed if sufficient cells are available. Intracellular markers of cytotoxicity and regulatory T cells, Granzyme B, pSTAT3, pSTAT1, and FOXP3, respectively will be assessed. Lytic potency of TILs will be determined using a lysis assay. In cases where additional tumor material is available and (a) a tumor cell suspension generated following enzymatic digestion, (b) an autologous tumor line generated from aforementioned tumor and/or (c) homologous cell line (if available) will be co-cultured with harvested TIL and IFN-.gamma. release measured. If excess cells are obtained, these will be cryopreserved for isolating RNA and DNA for gene expression analysis (including TCR V.beta. clonotyping analysis) which can be performed at a later time using an extended budget. If efficacy (defined below) is observed with anti PDL-1 and anti-CTLA-4 or the combination thereof, the possibility of lowering the concentration of indicated mAb(s) in tumor fragment cultures or performing a more detailed dose response assessment will be explored. If tumor fragments are visible in culture on day 7, they will be harvested and if sufficient cells are available after generation of a single cell suspension, they will be subjected to genetic analysis and flow cytometric phenotypic analysis (in that priority; to be negotiated). Flow cytometric analysis will focus on the phenotype of T cells, dendritic cells, macrophages, B cells, and NK cells after staining using appropriate fluorescent mAb panels. The markers will include: CD11c, CD11b, HLA class II, CD80, CD86, CD83, CD56, CD16, CD19, and CD20.
[1676] The criteria used to assess the efficacy of the addition of 4-1BB agonist, OX40 agonist, and the combination thereof to the tumor fragment cultures are summarized as follows: [1677] Increased number of TIL following expansion (CD4 and/or CD8) [1678] Decreased number of Treg following expansion [1679] Changes in T-cell proliferation markers (Ki67) in effector and Tregs [1680] Changes in effector/memory/differentiated phenotype, CD27, CD28, CD57, CD45RA, HLA-DR, CCR7, OX40, ICOS, CD45RA; telomerase length)) [1681] Increased NK cell (CD3-/CD56.sup.+) numbers, proliferation and activation status [1682] Exploratory changes in intracellular signaling protein or phosphoprotein levels (eg, AKT vs ERK) in T cells [1683] Increased CTL activity/lytic capacity as measured by redirected lysis [1684] Increased IFN-.gamma./HMGB1 production in TIL/tumor lysate, TIL/autologous tumor and/or TL/homologous tumor co-cultures.
[1685] Additional experiments to be performed include (1) whole exome sequencing and RNASeq on FFPE or fresh-frozen tumor material to identify mutated genes and possible neo-epitopes, (2) cytokine and chemokine analysis of culture supernatants collected 24-48 hours following initiation of tumor fragment cultures, (3) gene expression analysis of tumor fragments removed from early culture on day 7, (4) TCR clonotype analysis of the TIL isolated using high-throughput TCR V.beta. CDR3 region sequencing, (5) impact of mAbs on banked TILs for TIL effector function in the presence of IFN-.gamma. induced upregulation of PD-L1 on autologous/homologous tumors (outlined below) and analysis of remaining fragments for residual T-cells by PCR/IHC/digest.
[1686] Differences in assay parameters will be tested for significance using paired and un-paired T-tests (Wilcoxon rank-sum and signed rank tests). Comparison of multiple parameters will use one-way and two-way ANOVA analyses. Spearman regression analysis will be used when applicable to assess correlations between continuous measurements. All data can be tabulated and analyzed.
Example 4--Expansion of TILs Using Hexameric Ligands to 4-1BB, OX40, and Other TNFRSF Members
[1687] The effect of activation with hexameric fusion proteins of structures I-A with binding domains to 4-1BB, OX40, CD27, and other TNFRSF members, on expansion and function of tumor infiltrating lymphocytes (TIL) from ex-vivo cultured solid tumor fragments (multiple histologies) is studied in this example. 100 mg of each hexameric fusion protein (e.g., 4-1BB, OX40, and CD27) would be used with tumors obtained from the following indications: sarcoma, colorectal cancer, acute myeloid leukemia, ovarian cancer, triple negative breast cancer, pancreatic (Ras expressing), renal cancer, and bladder cancer. Tumors of various histologies will be obtained from commercial sources. Approximately 20 independent patient tumors will be obtained (2-3 tumors per indication as listed above). Tumors will be shipped to Lion in sterile HBSS or another appropriate medium. The tumors will be handled only in a laminar flow hood to maintain sterile conditions. Alternatively, tumor single cell suspensions may be utilized. The tumors will be washed after receipt and divided into 2-3 mm (length.times.width.times.height) fragments and placed into cell culture into 24-well plates (1 fragment per well) or 6-well plates (4 fragments per well) with culture medium supplemented with 6,000 IU/mL IL2 (recombinant) only, combination of 4-1BB HERA alone in triplicates will serve as control and three experimental conditions utilized respectively. An excipient control for the IL-2 will be used. The final concentration of HERA will be 30 .mu.g/mL. Following 24-48 hours of culture, 250 .mu.L of supernatant will be collected from each condition and stored at -20.degree. C. for subsequent analysis of cytokine and chemokine concentrations (pg/10.sup.6 cell/24 hr). TILs will be collected from each condition on day 11, day 21 and/or day of the `pre-REP` (at least 500,000 cells per sample). Two aliquots of TILs will be pelleted and resuspended in <10 L of PBS and will be frozen. If less than <10.sup.6 cells are collected, only gene expression arrays will be performed. Cultures will be fed on day 7 by partial removal of "spent" medium and addition of an equal volume of culture medium plus 6000 IU/mL IL-2. The spent medium will be stored at -20.degree. C. for subsequent cytokine/chemokine analysis using a multiplex assay (e.g., Luminex 100 system). Additional ligand will be added to the culture on day 7 if sufficient tumor fragments are available for initiation of more than 1 replicate of experimental conditions. TIL cultures will be maintained for an additional 14 days.
[1688] On day 21, the total cell yield, viability, cell surface and intracellular immunophenotype will be determined using flow cytometry. The following markers will be included: CD45RA, CCR7, CD3, TCR-alpha/beta, CD4, CD8, CXCR3, CD56, CD27, CD28, PD-1, PD-L1, BTLA, KLRG1, CD137, CD134, CD33, CD57, CD25, CD127, TIM-3, LAG-3, TIGIT, RAGE, and Ki67. Other biomarkers including CD107a, NKG2D, KIRS, chemokine death receptors (Fas, DR4) and anti-apoptotic/pro-autophagic proteins (BCL-2, BCL-XL, Bim, CD200, and LC3/HMGB1) will also be assessed if sufficient cells are available. Intracellular markers of cytotoxicity and regulatory T cells, Granzyme B, pSTAT3, pSTAT1, and FOXP3, respectively will be assessed. Lytic potency of TILs will be determined using a lysis assay. In cases where additional tumor material is available and (a) a tumor cell suspension generated following enzymatic digestion, (b) an autologous tumor line generated from aforementioned tumor and/or (c) homologous cell line (if available) will be co-cultured with harvested TIL and IFN-.gamma. release measured. If excess cells are obtained, these will be cryopreserved for isolating RNA and DNA for gene expression analysis by Nanostring Human Immunology Panel (including TCR V.beta. clonotyping analysis). If tumor fragments are visible in culture on day 7, they will be harvested and if sufficient cells are available after generation of a single cell suspension, they will be subjected to genetic analysis and flow cytometric phenotypic analysis. Flow cytometric analysis will focus on the phenotype of T cells, dendritic cells, macrophages, B cells, and NK cells after staining using appropriate fluorescent mAb panels. The markers will include: CD11c, CD11b, HLA class II, CD80, CD86, CD83, CD56, CD16, CD19, and CD20.
[1689] The criteria used to assess the efficacy of the addition of hexameric fusion proteins to the tumor fragment cultures are summarized above in Example 3, and further optional criteria are described in Table 53.
TABLE-US-00053 TABLE 53 Additional criteria for experimental performance for TNFRSF agonist fusion proteins in TIL expansion processes. Increases in T-cell Central .dwnarw.Tregs Memory Threshold of Cell Potency by following (CCR7+, success Count Viability IFN.gamma. Phenotyping expansion CD45RA-) Good >1e6 >75% >500 pg/10.sup.6 >40% CD8s <10% >1% cells/24 hr Very Good >3e6 >80% >1000/10.sup.6 >50% CD8s <3% >3% cells/24 hr Excellent >10e6 >85% >2000/10.sup.6 >75% CD8s <1% >10% cells/24 hr Outstanding >30e6 >90% >4000/10.sup.6 >90% CD8s Not detected >30% cells/24 hr
[1690] Additional experiments include: (1) whole exome sequencing and RNASeq on FFPE or fresh-frozen tumor material to identify mutated genes and possible neo-epitopes, (2) cytokine and chemokine analysis of culture supernatants collected 24-48 hours following initiation of tumor fragment cultures, (3) gene expression analysis of tumor fragments removed from early culture on day 7, (4) TCR clonotype analysis of the TIL isolated using high-throughput TCR V.beta. CDR3 region sequencing, (5) impact of hexameric fusion proteins on Lion banked TILs for TIL effector function in the presence of IFN-.gamma. induced upregulation of PD-L1 on autologous/homologous tumors and analysis of remaining fragments for residual T-cells by PCR/IHC/digest.
[1691] Differences in assay parameters will be tested for significance using paired and un-paired T-tests (Wilcoxon rank-sum and signed rank tests). Comparison of multiple parameters will use one-way and two-way ANOVA analyses. Spearman regression analysis will be used when applicable to assess correlations between continuous measurements.
Example 5--Evaluation of the Impact of 4-1BB and Anti-OX40 Agonistic Antibodies on TIL Expansion and Effector Function
[1692] The objective of this work is to evaluate the impact of 4-1BB (urelumab) and anti-OX40 agonistic antibodies on TIL expansion and effector function and to obtain information on ICOS and GITR expression during expansion.
[1693] In vitro assessment of anti-4-1BB and anti-OX40 agonistic antibodies on TIL expansion and phenotype is performed as follows. Antibody titration is conducted with tumor fragments and aspirates to determine suitable concentration for use with TIL expansion. The impact of anti-4-1BB and anti-OX40 agonists on TIL expansion in both pre-REP and REP (in these specific conditions) is evaluated for (1) IL-2+anti-4-1BB alone, (2) IL-2 anti-OX40 alone, (3) IL-2+anti-41BB+anti-OX40, and (4) IL-2 alone (control). TIL expansion and phenotype will be assessed by (1) expansion of CD3.sup.+ subset, CD3+CD8.sup.+ subset, and CD3+CD4.sup.+ in both percentage and absolute cell counts and viability, and (2) assessment of differentiation and activation status by flow cytometry using 18 color flow; including staining for ICOS and GITR, Ki67, and apoptosis markers.
[1694] In vitro assessment of TCR repertoire and expression profiling of TIL expanded with anti-4-1BB and anti-OX40 agonistic antibodies is performed as follows. TCR repertoire in TILs expanded with IL-2 alone in comparison with treatment conditions is shown by staining with specific anti-TRBV antibodies and using commercially-available TCR repertoire assays from iRepertoire, Inc. Expression profiling on individual TILs is performed using nCounter Vantage.TM. RNA Adaptive Immunity Panel with Nanostring analysis
[1695] In vitro assessment of tumor reactivity and effector function is performed as follows. An autologous tumor cell suspension or tumor cell line is generated (as possible). Tumor reactivity in tumor lysis assay is assessed by co-culturing autologous tumor cells/sorted autologous tumor cell suspension with autologous TIL expanded with IL-2 alone in comparison with treatment conditions described above. In case autologous tumor cell suspensions/tumor cell lines are not available, T cell activation assay by anti-CD3/CD28/CD137 will be conducted to assess effector functions by measuring IFN-gamma production/CD 107a expression instead.
Example 6--Further Evaluation of 4-1BB and OX40 Antibodies on the Ex Vivo Expansion of TIL and their Effector Function Activity
[1696] OX40 and 4-1BB have been found to be expressed by antigen specific CD4.sup.+ and CD8.sup.+ subset, respectively. Activation of co-stimulatory molecules (4-1BB and OX40) on T-cells enhance effector function, cell survival, and cell expansion. Activation of OX40 and 4-1BB receptors was shown to improve TIL expansion and anti-tumor function in murine models. Anti-4-1BB agonistic antibody was shown to increase the yield of melanoma TIL obtained from in vitro expansion. According to the following protocol, the effect of agonistic antibodies against 4-1BB and OX40, alone and in combination, on the ex vivo expansion of TIL and their effector function activity may be studied.
[1697] FIG. 13 describes the TIL expansion protocol used in this study. As illustrated in FIG. 15, tumor tissue was retrieved from the patient, fragmented and subjected to a pre-REP process in the presence of IL-2, as described herein. The tissues were then subjected to a REP process in the presence of IL-2 and anti-CD3 anti-bodies with irradiated PBMCs (FIG. 13).
[1698] The following experimental conditions were implemented in this study:
TABLE-US-00054 No treatment Anti-4-1BB Anti-OX40 Combination Isotype pre- 10 .mu.g/ml 0.5-10 .mu.g/ml 0.5-10 .mu.g/ml 10 .mu.g/ml REP REP 10 .mu.g/ml 0.5-10 .mu.g/ml 0.5-10 .mu.g/ml 10 .mu.g/ml
TABLE-US-00055 Phase Agent Pre-REP REP I anti-4-1BB 3 tumors N/A anti-OX40 3-5 tumors II combo 3-5 tumors
[1699] T-Cell activation, proliferation, and exhaustion may be monitored by flow-cytommetry according to the following list, where Panel 1 illustrates immune cell lineage, T-cell subsets, and T-cell differentiation, and Panel 2 illustrates T-cell activation and exhaustion:
TABLE-US-00056 Panel 1 Panel 2 CD3 CD3 L/D Blue L/D Blue CD19 CD25 CD56 CD95 CD62L PD-1 CD57 2B4/CD244 CD11b CD4 CD123 TIM-3 CD14 CD183 CD8 CD103 CD28 CD8 CD45 TIGIT CD4 CD127 CCR7 CD272 CD27 KLRG1 TCRg/d CD194 CD45RA CD69 CD16 LAG-3
[1700] Without being limited to any one theory of the invention, it is expected that the combination of anti-4-1BB and anti-OX40 agonists, alone or in combination with process 2A, may improve the expansion of pre-REP TILs, particularly in the CD3+CD8.sup.+ TIL subset; improve the success rate of certain tumors; shorten duration pre-REP TIL expansion; and/or enhance multi-functionalities of TIL including effector function and cell survival following antigen re-stimulation.
Example 7--Clinical Study to Assess Efficacy and Safety of Autologous TIL
[1701] This Example describes a Phase 1/2 clinical study for evaluating the efficacy of autologous TIL across multiple tumor types. The objectives of this investigation are to evaluate efficacy using objective response rate (ORR) according to RECIST v1.1 in subjects with ovarian cancer and osteosarcoma. The primary objective for a pancreatic ductal adenocarcinoma (PDAC) cohort is to evaluate efficacy as measured by the 6-month survival rate.
[1702] Secondary objectives may include: (1) evaluating ORR using RECIST v.1.1 in PDAC; (2) determining the disease control rate (DCR) within and across cohorts; (3) determining the duration of response (DOR); (4) determining progression-free survival (PFS) and overall survival (OS); and (5) further characterizing the safety profile of adoptive cell therapy with TIL across multiple tumor types.
Definitions/Abbreviations
[1703] ACT Adoptive Cell Therapy [1704] AE Adverse Event [1705] ALT Alanine Transaminase [1706] ANC Absolute Neutrophil Count [1707] AST Aspartate Transaminase [1708] ASMR Age-standardized Mortality Rate [1709] aPTT Activated Partial Thromboplastin Time [1710] BID Twice Per Day [1711] BSA Body Surface Area [1712] CBC Complete Blood Count [1713] CD4+T CD4+ T Cells [1714] CD8+T CD8+ T Cells [1715] CFR Code of Federal Regulations [1716] CI Confidence Interval [1717] CLS Capillary Leak Syndrome [1718] CMO Contract Manufacturing Organization [1719] COPD Chronic Obstructive Pulmonary Disease [1720] CR Complete Response [1721] CrCl Creatinine Clearance [1722] CT Computed Tomography [1723] CTCAE v4.03 Common Terminology Criteria for Adverse Events Version 4.03 [1724] D5W Dextrose 5% by weight [1725] DCR Disease control rate [1726] DOR Duration of Response [1727] EBV Epstein-Barr Virus [1728] ECHO Echocardiogram [1729] EKG Electrocardiogram [1730] EOC Epithelial Ovarian Cancer [1731] EORTC QLQ-C30 European Organisation for Research and Treatment of Cancer Quality-of-Life Questionnaire--Core 30 instrument [1732] EWV Early Withdrawal Visit [1733] FDA Food and Drug Administration [1734] FEV Forced Expiratory Volume [1735] FVC Forced Vital Capacity [1736] GCP Good Clinical Practice [1737] Hgb Hemoglobin [1738] HIV Human Immunodeficiency Virus [1739] HRQoL Health-related Quality-of-life [1740] ICH International Conference on Harmonization [1741] IL Interleukin [1742] IND Investigational New Drug (Application) [1743] irRECIST Immune-related Response Evaluation Criteria in Solid Tumors [1744] IRB Institutional Review Board [1745] IUD Intrauterine Device [1746] IV Intravenous [1747] IVPB Intravenous Piggyback [1748] LVEF Left Ventricular Ejection Fraction [1749] M1 HLA-DR+CD68+M1 Macrophages [1750] M2 CD 163+ or CD204+M2 Macrophages [1751] MRI Magnetic Resonance Imaging [1752] MUGA Multiple Gated Acquisition Scan [1753] NCI National Cancer Institute [1754] Neu CD66b+ Neutrophils [1755] NMA Nonmyeloablative [1756] NS Normal Saline [1757] OC Ovarian Cancer [1758] ORR Objective Response Rate [1759] OS Overall Survival [1760] PBMC Peripheral Blood Mononuclear Cell [1761] PCR Polymerase Chain Reaction [1762] PD Progressive Disease [1763] PDAC Pancreatic Ductal Adenocarcinoma [1764] PE Physical Exam [1765] PET Positron Emission Tomography [1766] PFS Progression-free survival [1767] PHI Personal Health Information [1768] PI Principal Investigator [1769] PJP Pneumonitis Jiroveci Pneumonia [1770] PO Per Os (by mouth) [1771] PR Partial Response [1772] PS Performance Status [1773] PT Prothrombin Time [1774] QTc Corrected QT Interval [1775] RECIST Response Evaluation Criteria in Solid Tumors [1776] REP Rapid Expansion Protocol [1777] SAE Serious Adverse Event [1778] SAP Statistical Analysis Plan [1779] SD Stable Disease [1780] SGOT Serum Glutamic-oxaloacetic Transaminase [1781] SGPT Serum Glutamic-pyruvic Transaminase [1782] SMX Sulfamethoxazole [1783] STS Soft Tissue Sarcoma [1784] TCR T-cell Receptor [1785] TIL Tumor-infiltrating lymphocyte [1786] TMA Tissue Microarray [1787] TMP Trimethoprim [1788] Treg FOXP3+ Regulatory T Cells [1789] TSH Thyroid Stimulating Hormone [1790] ULN Upper Limit of Normal
[1791] Study Design and Endpoints:
[1792] This study is aimed at evaluating the efficacy of TIL in subjects with: a) osteosarcomas relapsed or refractory to conventional therapy, b) platinum-resistant ovarian cancer, and c) PDAC who have progressed on, or received maximal benefit from, front-line therapy. Each cohort begins with ten subjects in the first stage, and expansion to the second stage is guided by a modified Simon's two stage design.
[1793] The primary endpoint is ORR by RECIST v1.1 for ovarian cancer and osteosarcoma, and the 6-month survival rate in PDAC. The primary endpoint for the PDAC cohort is the 6-month survival rate.
[1794] The secondary efficacy endpoints include ORR (for PDAC) CRR, DCR, DOR, PFS using RECIST v1.1, and OS. DCR includes complete response (CR), partial response (PR), and stable disease (SD). Safety endpoints may include overall assessment of AEs including grade 3 or greater non-hematological toxicities, SAEs and treatment-emergent AEs by grade and relationship to the study treatment. The secondary endpoint for the PDAC cohort is ORR using RECIST v1.1.
[1795] Exploratory endpoints may include: (1) duration of TIL persistence as determined by T cell receptor (TCR) sequencing of infused T cells serially isolated following TIL infusion, or alternatively iRepertoire assessment of mRNA for the TCRs; (2) response as determined by the immune-related response criteria; (3) immunological Phenotype of TIL at the time of infusion by multichannel flow cytometry; (4) baseline and post-treatment tumor assessment via IHC, TCR sequencing, and transcriptional analysis; and (5) HRQOL as assessed per EORTC QLQ-C30 questionnaire.
[1796] Participant Inclusion Criteria.
[1797] Subjects may be between 18 and 70 (subjects aged 16-70 may be enrolled into the osteosarcoma cohort). Subjects should be willing and able to provide informed consent. For patients <18 years of age, their parents or legal guardians should sign a written informed consent. Assent, when appropriate, may be obtained according to institutional guidelines. Clinical performance status of ECOG 0 or 1 at enrollment and within 7 days of initiating lymphodepleting chemotherapy. Subjects should have an area of tumor amenable to excisional biopsy for the generation of TIL separate from, and in addition to, a target lesion to be used for response assessment. Any prior therapy directed at the malignant tumor, including radiation therapy, chemotherapy, and biologic/targeted agents should be discontinued at least 28 days prior to tumor resection for preparing TIL therapy.
[1798] Within 7-14 (e.g., 7 days) days of enrollment and within 12 h to 48 h (e.g., 24 h) of starting lymphodepleting chemotherapy subjects may meet one or more of the following laboratory criteria: (1) absolute neutrophil count (ANC) >1000/mm.sup.3; (2) hemoglobin >8.0 g/dL (transfusion allowed); (3) platelet count >100,000/mm.sup.3; (4) ALT/SGPT and AST/SGOT<2.5.times.the upper limit of normal (ULN) (Patients with liver metastases may have LFT.ltoreq.5.0.times.ULN); (5) calculated creatinine clearance (Cockcroft-Gault) .gtoreq.40.0 mL/min; (6) total bilirubin .ltoreq.1.5.times.ULN; (7) prothrombin Time (PT) & Activated Partial Thromboplastin Time (aPTT) .ltoreq.1.5.times.ULN (correction with vitamin K allowed) unless subject is receiving anticoagulant therapy (which should be managed according to institutional norms prior to and after excisional biopsy); and (8) negative serum pregnancy test (female subjects of childbearing potential).
[1799] Furthermore, subjects should not have a confirmed human immunodeficiency virus (HIV) infection. Subjects should have a 12-lead electrocardiogram (EKG) showing no active ischemia and corrected QT interval (QTc) less than 480 ms. Subjects 40 years of age and older should also have a negative stress cardiac test (i.e. EKG stress test, stress thallium, dobutamine echocardiogram or other stress test that may rule out cardiac ischemia). Stress test may be required of subjects less than 40 years of age if warranted by family history or risk factors by the treating investigator. Subjects of childbearing potential should be willing to practice an approved highly effective method of birth control starting at the time of informed consent and for 1 year after the completion of the lymphodepletion regimen. Subjects should be able to adhere to the study visit schedule and other protocol requirements. Finally, pulmonary function tests (spirometry) demonstrating forced expiratory value (FEV) 1 greater than 65% predicted or forced vital capacity (FVC) greater than 65% of predicted.
[1800] In addition to meeting the above general inclusion criteria, subjects should also meet cohort specific criteria.
[1801] For ovarian cancer, subjects may have high grade non-mucinous histology (carcinosarcomas are allowed). Moreover, subjects may have failed at least two prior lines of chemotherapy (i.e. frontline adjuvant chemotherapy plus one additional line for recurrent/progressive disease).
[1802] For osteosarcoma, subjects may have relapsed or become refractory to conventional therapy and have received a regimen including some combination of high-dose methotrexate, doxorubicin, cisplatin, and/or ifosfamide.
[1803] For pancreatic adenocarcincoma, subjects may have histologically or cytologically documented diagnosis of PDAC with oligo-metastatic disease. Subjects may have progressed on, or received maximal benefit from, front-line therapy. Patients may have received unlimited lines of prior standard of care therapy. Patients with ascites or carcinomatosis are not eligible for the study. Patients may need an albumin of .gtoreq.3.0 mg/dL within 7 days of enrollment.
[1804] Participant Exclusion Criteria.
[1805] A number of criteria may result in exclusion of a participant from the study: [1806] a. Active systemic infections requiring intravenous antibiotics, coagulation disorders or other major medical illnesses of the cardiovascular, respiratory or immune system. PI or his/her designee shall make the final determination regarding appropriateness of enrollment. [1807] b. Patients with active viral hepatitis. [1808] c. Patients who have a left ventricular ejection fraction (LVEF) <45% at Screening. [1809] d. Patients with a history of prior adoptive cell therapies. [1810] e. Persistent prior therapy-related toxicities greater than Grade 2 according to Common Toxicity Criteria for Adverse Events (CTCAE) v4.03, except for peripheral neuropathy, alopecia, or vitiligo prior to enrollment. [1811] f. Primary immunodeficiency. [1812] g. History of organ or hematopoietic stem cell transplant. [1813] h. Chronic steroid therapy, however prednisone or its equivalent is allowed at <10 mg/day. [1814] i. Patients who are pregnant or nursing. [1815] j. Presence of a significant psychiatric disease, which in the opinion of the principal investigator or his/her designee, would prevent adequate informed consent. [1816] k. History of clinically significant autoimmune disease including active, known, or suspected autoimmune disease. Subjects with resolved side effects from prior checkpoint inhibitor therapy, vitiligo, psoriasis, type 1 diabetes or resolved childhood asthma/atopy would be an exception to this rule. Subjects that require intermittent use of bronchodilators or local steroid injections would not be excluded. Subjects with hypothyroidism stable on hormone replacement or Sjorgen's syndrome may not be excluded. [1817] l. History of clinically significant chronic obstructive pulmonary disease (COPD), asthma, or other chronic lung disease. [1818] m. History of a second malignancy (diagnosed in the last 5 years). Exceptions include basal cell carcinoma of the skin, squamous cell carcinoma of the skin, or in situ cervical cancer that has undergone potentially curative therapy. [1819] n. History of known active central nervous system metastases and/or carcinomatous meningitis. Subjects with previously treated brain metastases may participate provided they are stable (without evidence of progression by imaging for at least four weeks prior to the first dose of trial treatment and any neurologic symptoms have returned to baseline), have no evidence of new or enlarging brain metastases, and are not using steroids for at least 7 days prior to initiation of lymphodepletion. [1820] o. Has received a live vaccine within 30 days prior to the initiation of lymphodepletion. [1821] p. Any other condition that in the investigator's judgement would significantly increase the risks of participation.
[1822] Completion or Discontinuation of Treatment.
[1823] Completion of treatment may be defined as having received any volume of TIL infusion followed by at least 1 dose of adjuvant IL-2.
[1824] This study includes a one-time treatment regimen consisting of lymphodepleting chemotherapy, TIL infusion, and adjuvant IL-2 (up to 6 doses). Discontinuation from study treatment should be considered if any of the following criteria are met. However, unless the patient also meets criteria for discontinuation from study participation, every effort may be made to continue follow-up and assessment of all patients, including those that do not complete the full course of therapy, as specified in the Schedule of Events.
[1825] Criteria for early discontinuation from treatment are: [1826] a. Grade 3 or greater autoimmunity that involves vital organs (heart, kidneys, brain, eye, liver, colon, adrenal gland, lungs) with symptoms emerging following TIL infusion; [1827] b. Grade 3 or greater allergic reaction including bronchospasm or generalized urticaria that does not resolve after medical management in the opinion of the Investigator; [1828] c. Grade 3 or greater toxicity due to IL-2 that does not decrease to Grade 2 or less within 96 hours of management; [1829] d. Determination by the Investigator that continued treatment is not in the best interest of the patient; [1830] e. Withdrawal by patient. The patient (or parents/legal guardian for patients <18 years of age) may withdraw consent to treatment but continue consent for follow-up evaluations and/or survival status; [1831] f. Pregnancy; [1832] g. Patient meets criteria for early discontinuation from study; and [1833] h. Patient has become ineligible for study after tumor harvest and prior to TIL or IL-2 administration.
[1834] Criteria for early discontinuation from study are: [1835] a. Withdrawal by patient. The patient (or parents/legal guardian for patients <18 years of age) may withdraw consent. All efforts should be made to continue consent for survival status follow-up; [1836] b. Patient has become ineligible for study after tumor harvest or did not receive any study treatment; [1837] c. Have any complication or delayed healing from excisional procedure that in the investigator's opinion would increase the risks of lymphodepletion, adoptive TIL therapy and adjuvant IL-2; [1838] d. Have a decline in performance status to ECOG>1 (within seven days prior to starting lymphodepletion); [1839] e. Death; and [1840] f. Lost to follow-up after 3 documented attempts to contact the patient.
[1841] Some subjects may undergo tumor harvest and TIL manufacture but may not receive the infusion of investigational product. If TIL is not administered to the patient for whatever reason, even if after lymphodepleting chemotherapy, then the patient should remain on study, but data collection may be reduced to survival status and start of any new anticancer therapy for 3 years. Such subjects may be considered unevaluable for statistical analysis of efficacy and may be replaced.
[1842] If a patient initiates anti-cancer therapy or exhibits disease progression after TIL infusion they may remain in the study, but the data collection may be reduced to response status, survival status and other anti-cancer therapy for 3 years.
[1843] Study Agents.
[1844] The lymphodepletion regimen is scheduled to start on Day -7, after notification that TIL production is expected to be successful for the patient. Patients may receive lymphodepleting chemotherapy as inpatient or outpatient at the discretion of the investigator. Modification of the lymphodepletion regimen is allowed as clinically indicated and should be guided by daily hematological parameters as described below for fludarabine in heavily pre-treated patients or subjects with a history of prolonged myeloid recovery. The regimen comprises 2 daily doses of cyclophosphamide (with mesna) followed by 5 daily doses of fludarabine and should be administered as per institutional protocol/standards for nonmyeloablative chemotherapy. Guidelines for preparation and administration are described below. Subjects should be dosed using actual body weight but not to exceed 140% of Ideal Body Weight as defined below: [1845] a. Ideal Body Weight for Males=50 kg+2.3.times.(number of inches over 60 inches in height). Example: ideal body weight of a 5'10'' male subject 50+2.3.times.10=73 kg [1846] b. Ideal Body Weight for Females=45.5 kg+2.3 (number of inches over 60 inches in height). Example: ideal body weight of a 5'3'' female subject 45.5+2.3.times.3=52.4 kg
[1847] Drugs required for lymphodepletion include cyclophosphamide, fludarabine, and/or mesna.
[1848] Variations from the lymphodepletion (e.g. infusion times; schedule of treatments, etc.) prior to day 0 may be documented in the medical record but may not be considered protocol violations/deviations.
[1849] Cyclophosphamide may be administered at 20 to 80 mg/kg/day (e.g., 60 mg/kg/day) IV in 250 mL normal saline (NS) over approximately 2 hours on Days -7 and -6. Mesna 60 mg/kg with dextrose 5% by weight (D5W) or NS infused intravenously over 24 h on Days -7 and -6. As noted above the dose may be based on the patient's actual body weight, but to prevent undue toxicity, it may not exceed the dose based on 140% of the maximum ideal body weight (defined above). There may be dose adjustments for cyclophosphamide.
[1850] Fludarabine will then be infused at 15 to 50 mg/m.sup.2 (e.g., 25 mg/m.sup.2) IV piggyback (PB) daily over approximately 15-30 minutes on Days -5 to -1. To prevent undue toxicity with fludarabine, the dose may be based on body surface area (BSA), but may not exceed a dose calculated on surface areas based on body weights greater than 140% of the maximum ideal body weight. Hematological parameters (complete blood count [CBC] and differential) are to be reviewed daily during lymphodepletion. If after 3 or 4 doses of fludarabine, the absolute lymphocyte count falls below 100 cells/mm.sup.3 the remaining dose(s) of fludarabine may be omitted following discussion with the PI. Fludarabine dose may be adjusted according to estimated creatinine clearance (CrCl) as follows: (1) CrCl 50-79 mL/min: Reduce dose to 20 mg/m.sup.2; and/or (2) CrCl 40-49 mL/min: Reduce dose to 15 mg/m.sup.2.
[1851] The TIL product that may be used in this protocol is a cellular investigational product comprising a live cell suspension of autologous TIL derived from the patient's own tumor. Each dose may contain up to 150.times.10.sup.9 total viable lymphocytes. The total volume to be infused may be up to 600 mL dependent on total cell dose.
[1852] If not already hospitalized for the lymphodepleting chemotherapy, the patient may be admitted 1-2 days prior to planned TIL administration and prepared with overnight intravenous hydration prior to the TIL administration. Patients may remain hospitalized until the completion of the IL-2 therapy, as per institutional standards.
[1853] The IL-2 infusion may begin 3-24 h after completion of the TIL infusion. IL-2 may be administered at a dose of 200,000 to 1,000,000 IU/kg (e.g., 600,000 IU/kg) (based on total body weight) and may be administered by IV infusion at a frequency of every 8-12 hours as per institutional standard of care and continued for up to a maximum of six doses or as tolerated. IL-2 doses may be skipped if patient experiences a Grade 3 or 4 toxicity due to IL-2 except for reversible Grade 3 toxicities common to IL-2 such as diarrhea, nausea, vomiting, hypotension, skin changes, anorexia, mucositis, dysphagia, or constitutional symptoms and laboratory changes. Management of IL-2 is detailed in Table 54. If these toxicities can be easily reversed within 24 hours by supportive measures, then additional doses may be given. If greater than 2 doses of IL-2 are skipped, IL-2 administration may be stopped. In addition, discretion may be used to hold or stop the dosing.
TABLE-US-00057 TABLE 54 Management of Potential Aldesleukin Toxicities. Expected Expected Supportive Stop toxicity grade Measures Treatment* Chills 3 IV Meperidine 25-50 mg, No IV q1h, prn Fever 3 Acetaminophen 650 mg, No po, q4h; Indomethicin 50- 75 mg, po, q8h Pruritis 3 Hydroxyzine HCL 10-20 No mg po q6h, prn; Diphenhydramine HCL25-50 mg, po, q4h, prn Nausea/Vomiting/ 3 Ondansetron 10 mg, IV, No Anorexia q8h, prn; Granisetron 0.01 mg/kg IV daily prn; Droperidol 1 mg, IV q4- 6 h, prn; Prochlorperazine 25 mg q4h p.r., prn or 10 mg IV q6h prn Diarrhea 3 Loperamide 2 mg, po, If uncontrolled after 24 q3h, prn; Diphenoxylate hours despite all HCl 2.5 mg and atropine supportive measures sulfate 25 mcg, po, q3h, prn; codeine sulfate 30-60 mg, po, q4h, prn Malaise 3 or 4 Bedrest interspersed with If other toxicities occur activity simultaneously Hyperbilirubinemia 3 or 4 Observation If other toxicities occur simultaneously Anemia 3 or 4 Transfusion with PRBCs If uncontrolled despite all supportive measures Thrombocytopenia 3 or 4 Transfusion with platelets If uncontrolled despite all supportive measures Neutropenia 4 Observation No Edema/Weight gain 3 Diuretics prn No Hypotension 3 Fluid resuscitation; If uncontrolled despite all Vasopressor support supportive measures Dyspnea 3 or 4 Oxygen or ventilatory If requires ventilatory support support Oliguria 3 or 4 Fluid boluses or If uncontrolled despite all dopamine at renal doses supportive measures Increased creatinine 3 or 4 Observation Yes (grade 4) Renal failure 3 or 4 Dialysis Yes Pleural effusion 3 Thoracentesis If uncontrolled despite all supportive measures Bowel perforation 3 Surgical intervention Yes Confusion 3 Observation Yes Somnolence 3 or 4 Intubation for airway Yes protection Arrhythmia 3 Correction of fluid and If uncontrolled despite all electrolyte imbalances; supportive measures chemical conversion or electrical conversion therapy Elevated Troponin levels 3 or 4 Observation Yes Myocardial Infarction 4 Supportive care Yes Elevated transaminases 3 or 4 Observation For grade 4 without liver metastases Electrolyte imbalances 3 or 4 Electrolyte replacement If uncontrolled despite all supportive measures *Unless the toxicity is not reversed within 12 hours.
[1854] Study Procedures and Schedule.
[1855] The following procedures may be used in this study.
[1856] Potential subjects may be informed about the study by the investigator. The risks, benefits, and alternatives may be discussed and the Informed Consent Document may be signed before any study related assessments are performed.
[1857] Subjects should meet most, or preferably all, inclusion criteria and preferably do not have any of the conditions specified in the exclusion criteria. Confirmation of general, cohort, specific, and treatment inclusion/exclusion criteria should be documented within seven days of starting lymphodepletion chemotherapy.
[1858] The demographic data may include date of birth (as allowed per local regulations), age, gender, and race/ethnic origin.
[1859] Relevant and significant medical/surgical history and concurrent illnesses may be collected for all patients at Screening (Visit 1) and updated as applicable. Any worsening from pre-existing conditions should be reported as AEs. Patient's prior anti-cancer treatment may also be collected.
[1860] Documentation of cohort-specific diagnosis of cancer may be made and confirmed histologically.
[1861] All medications and therapies (prescription, and non-prescription, including herbal supplements) taken by the patient up to 28 days prior to Screening (Visit 1) may be collected in the database, including the stop dates for medications prohibited in the study, at the time of consent. All medications and therapies being taken by the patients, or changes thereof, at any time during the study, may be recorded in the medical record.
[1862] All baseline grade 2 and higher toxicities may be assessed as per CTCAE v4.03. Any events occurred after screening, but prior to enrollment/tumor resection, may be recorded as Medical History in the database, unless the events are related to protocol mandated procedures. Any events occurring after enrollment/tumor resection may be captured as AEs in the database until the 6 Month visit, subject is taken off the study, or starts other cancer therapy.
[1863] Vital signs shall include height, weight, pulse, respirations, blood pressure and temperature. Height may be measured at Screening (Visit 1) only. All other vital signs may be measured at applicable time points. On Day 0 (Visit 11/TIL infusion), vital signs may be monitored for up to approximately 24 hours post TIL infusion.
[1864] An ECOG performance status may be assessed at Screening (Visit 1) and other time points indicated on the schedule of events.
[1865] Physical examination may be conducted for all visits except for Tumor Resection and shall include vital signs and weight, head and neck, cardiovascular, pulmonary, extremities, and other relevant evaluation. Exams during conducted during follow-up may be symptom directed. Clinically significant changes in the exam findings may be recorded as adverse events as indicated.
[1866] Safety blood and urine tests may be collected and analyzed locally at every visit as indicated in the Schedule of Events.
[1867] Sample collection for high resolution HLA Class I typing may be conducted at Screening (Visit 1).
[1868] Serology for the following diseases may be completed at Screening (Visit 1) to be analyzed locally per institutional standard: HIV, Hepatitis B Virus, Hepatitis C Virus, Cytomegalovirus (CMV), Herpes Simplex Virus; Epstein-Barr virus (EBV) (may be within previous 3 months to Tumor Resection/Visit 2), Chagas Disease, Human T cell Lymphotropic Virus, and West Nile Virus. Sickle Cell Disease may also be screened. Additional testing is to be done as clinically indicated.
[1869] The creatinine clearance may be calculated by site using the Cockcroft-Gault formula at Screening only.
[1870] All subjects can have a baseline 12-lead ECG and assessment of ventricular function by echocardiogram or MUGA. In addition, subjects age 40 or older and those younger than 40 with a history of cardiovascular disease or chest pain may have a stress test documenting absence of ischemia. Patients with an abnormal MUGA or echocardiogram may meet ejection fraction requirements and obtain cardiology clearance prior to enrollment.
[1871] Pulmonary evaluation may be completed within 28 days from Screening (Visit 1). Prior evaluations completed within 6 months prior to Screening (Visit 1) may be accepted. An FEV1 greater than 65% of predicted or FVC greater than 65% of predicted is required. Patients who are unable to conduct reliable PFT spirometry measurements due to abnormal upper airway anatomy (e.g. tracheostomy) may undergo a 6-minute walk test to be evaluate pulmonary function. These patients should, and preferably can, walk a distance of at least 80% predicted for age and sex as well as maintain oxygen saturation greater than 90% throughout.
[1872] Colonoscopy is only required for patients who have had a documented Grade 2 or greater diarrhea or colitis due to previous immunotherapy within six months of Screening. Patients that have been asymptomatic for at least 6 months from Screening or had a normal colonoscopy post anti-PD-1/anti-PD-L1 treatment, with uninflamed mucosa by visual assessment may not need to repeat the colonoscopy.
[1873] A health related quality of life (HRQOL) questionnaire may be conducted in person at baseline Day -21 (Visit 3) and be performed as the first procedure on the subsequent visits. See the Schedules of Events for specific timepoints. Failure to complete any questionnaires may not be considered a deviation requiring reporting.
[1874] Radiographic assessments by computed tomography (CT) scans with contrast of the chest, abdomen and pelvis are required for all patients for tumor assessments. CT scans are performed as indicated in the Schedule of Events until progressive disease by modified RECIST v1.1 is noted (or if the patient withdraws full consent). Response assessments should be evaluated and documented by a qualified radiologist participating in the trial. Magnetic Resonance Imaging (MRI) or positron emission tomography (PET) scans of the chest, abdomen and pelvis in lieu of CT scans may be allowed for patients who have an intolerability to contrast media. The same method of assessment (CT or MRI) and the same technique for acquisition of data should be used consistently throughout the study to characterize each identified and reported lesion. Initial radiographic assessments may be made at 6, 12, 18 and 24 weeks post TIL infusion. Thereafter, Patients may be evaluated for response approximately every 12 weeks. Additional radiological assessments may be performed as clinically indicated.
[1875] Prior to surgical biopsy, subject eligibility may be confirmed, and the PI or designee may provide approval for patient enrollment into the clinical trial and subsequent tumor resection. Subjects may undergo a pre-procedural consultation and a separate informed consent by the team performing the surgical biopsy per institutional standards. Ideally, the targeted tumor should have not been previously irradiated. If the tumor has been previously irradiated a minimum period of 1 to 6 months (e.g., 3 months) may have elapsed between irradiation and resection, during which time additional target-tumor growth may have been demonstrated. If enrolled, tumor resection is expected to occur approximately 1 to 12 weeks (e.g., 6 weeks) prior to the anticipated TIL infusion (Day 0). TIL is an autologous investigational product which is procured and delivered by means that have more in common with autologous blood product delivery than those of traditional drug production. It is imperative that only the patient's own (autologous) study treatment (TIL) be administered to the same individual patient. For these reasons, the patient specimen can be procured and handled per a strict protocol to ensure optimal quality of the specimen and minimum transport time to and from the processing lab facility, as well as to ensure the appropriate identification of the study product at all times including infusion back into the patient.
[1876] In cases where additional or excess tumor tissue can be safely procured at the time of the initial excisional biopsy for TIL harvest, excess tumor tissue for research may be procured. Provision of adequate amount of tumor tissue for TIL manufacturing is priority over the collection of additional tumor tissue that is sent for research. Every effort should be made to obtain adequate tumor tissue for both TIL manufacturing and additional analysis. In addition, a mandatory on-study biopsy may be used to ascertain molecular and immunological changes following treatment and as well as to document presence of infused T cells in the tumor. The tumor tissue analysis may include: 1) immunohistochemistry to identify individual immune cell populations; and/or 2) DNA and RNA analysis, including possible exploratory genomic and transcriptomic evaluation and TCR sequencing to evaluate infused TIL homing to tumor (in the post-treatment biopsy). Provision of adequate amount of tumor tissue for TIL manufacturing is priority over the collection of additional tumor tissue for research. Every effort should be made to obtain adequate tumor tissue for both TIL manufacturing and additional analysis.
[1877] Up to 500.times.10.sup.6 TIL from the infusion product (and genetic material extracted from these samples) may be stored for research. Flow cytometry analysis of the infused TIL may be performed, and DNA from the infusion product may be sent for TCR sequencing. The samples in these research studies may be used to gain further information about the disease and the characteristics of the TIL before and after infusion. Peripheral blood may be collected from the patients for immune monitoring and T cell tracking using TCR sequencing. Blood for Immune Monitoring may be drawn at Tumor Resection (Visit 2) and subsequent collections may be drawn at applicable time points (See Tables 55 and 56).
TABLE-US-00058 TABLE 55 Exemplary Schedule of Events - Pre-Treatment Treatment Phases. Pre-treatment Phase Treatment Phase Visit Number 6,7, 12, 13 1 2 3 4 5 8, 9, 10 11 14, 15 16 17 18 19 Visit Name Baseline Days Days (Day -14 Day Day -5, -4, Day 0 1, 2, Day Day Day 42 Day 84 Screening Biopsy to -21) -7 -6 -3, -2, -1 (TIL Infus.) 3, 4 14 28 (Wk 6) (Wk 12) Visit Window .ltoreq.28 days N/A N/A N/A N/A N/A N/A N/A (.+-.7 days) (.+-.7 days) (.+-.7 days) (.+-.7 days) Written X Informed Consent Medical X History Documentation X of diagnosis Physical Exam X X X X X X X X X X X Vital Signs.sup.a X X X X X X.sup.b X X X X X X ECOG X X X X X X X X performance status CBC, Chem X X X X X X X X X X X X Panel, and urine tests.sup.c .beta.-HCG Serum X X X Pregnancy Test Infection X testing HLA typing X Cardiac X Evaluations Pulmonary X function tests Colonoscopy X Tumor Assessments X X X X (CT/MRI) Response X X X Assessments Concomitant X X X X X X X X X X X X Meds Adverse X X X X X X X X X X X X events Tumor Biopsy X X NMA X X X lymphodepletion.sup.d TIL Infusion.sup.e X IL-2.sup.f X Immune X X X X X X Monitoring HRQOL X X Questionnaire Prophylactic Medications PJP X X X X X X X X X Filgrastim X Fungal X X X X X X Prophylaxis Herpes Virus X X X X X X Prophylaxis .sup.aVital signs may include height, weight, heart rate, respiratory rate, blood pressure, and temperature. Height at may be measured at Screening only. BSA and BMI may be Calculated Day -7 (Visit 4) only. .sup.bOn Day 0 (TIL infusion), vital signs may be monitored every 30 minutes during infusion then hourly (+/-15 minutes) for four hours and then routinely (every four to six hours), unless otherwise clinically indicated, for up to approximately 24 hours post TIL infusion. .sup.cChemistry: sodium, potassium, chloride, total CO2, or bicarbonate, creatinine, glucose, BUN, albumin, calcium, magnesium, phosphorus, alkaline phosphatase, ALT/SGPT, AST/SGOT, total bilirubin, direct bilirubin, LDH, total protein, total CK, uric acid, and serum creatinine. Thyroid panel (to include TSH and Free T4) is to be done at Visits 1 and 19 and as clinically indicated. Coagulations: PT, PTT, and INR. Hematology: CBC with differential; Urinalysis: Bilirubin, Blood, Glucose, Ketones, pH, Protein, Specific gravity, Color and Apperance. .sup.dCyclophosphamide with mesna for 2 days at Day -7 and Day -6 (Visits 4 thru 5) followed by 5 days of fludarabine at Day -5 thru Day -1 (Visits 6 thru 10). .sup.eTIL infusion is to be done 1 to 2 days after the last dose of agent in the NMA lymphodepletion regimen. .sup.fInitiate IL-2 at 600,000 IU/kg within approximately 3 to 24 hours after TIL infusion and continue every 8-12 hours for up to six doses.
TABLE-US-00059 TABLE 56 Exemplary Schedule of Events: Post-Treatment and Long-Term Follow-Up Long-term Post-treatment Follow-up Follow-up Visit Number 20 21 22 23 24 25 EWV Visit Name Day 126 Early (Month 4.5/ Day 16 Day 25 Day 336 Day 504 Day 672 Withdrawal Quarterly Week 18) (Month 6) (Month 9) (Month 12) (Month 18) (Month 24) Visit Contact Visit Window (.+-.14 days) (.+-.14 days) (.+-.14 days) (.+-.14 days) (.+-.21 days) (.+-.21 days) (.+-.21 days) Physical Exam.sup.a X X X X X X X Vital Signs.sup.b X X X X X ECOG performance status X X X X X X X CBC, Chem Panel, and X X X urine tests.sup.c Tumor Assessments X X X X X X X (CT/MRI).sup.d Response Assessments X X X X X X X Concomitant Meds X X X X X Adverse events.sup.e X X X Immune Monitoring.sup.f X X X X HRQOL Questionnaire X X X Survival Follow-up X Prophylactic Medications PJP X X Herpes Virus Prophylaxis X X .sup.aPE may include gastrointestinal (abdomen, liver), cardiovascular, extremities, head, eyes, ears, nose, and throat, respiratory system, skin, psychiatric (mental status), general nutrition. PE conducted during follow-up may be symptom directed. .sup.bVital signs may include weight, heart rate, respiratory rate, blood pressure, and temperature. .sup.cChemistry: sodium, potassium, chloride, total CO2, or bicarbonate, creatinine, glucose, BUN, albumin, calcium, magnesium, phosphorus, alkaline phosphatase, ALT/SGPT, AST/SGOT, total bilirubin, direct bilirubin, LDH, total protein, total CK, uric acid, and serum creatinine. Thyroid panel (to include TSH and Free T4) is to be done as clinically indicated. Coagulations: PT/PTT/INR. Hematology: CBC with differential; Urinalysis: Bilirubin, Blood, Glucose, Ketones, pH, Protein, Specific gravity, Color and Appearance. .sup.dCT Scans of the chest, abdomen and pelvis, are required at the indicated time points. Additional radiological assessments may be performed per Investigator's discretion. MRI may be used if patients are intolerable to contrast media. .sup.eAny AEs occurred after Screening, but prior to enrollment/tumor resection, may be recorded as Medical History in the database. Any AEs occurred after enrollment/tumor resection may be captured as AEs through Day 168 (Visit 21/Month 6) and as clinically indicated, or until the first dose of the subsequent anti-cancer therapy, whichever occurs first. All AEs attributed to protocol-required procedures or treatment may be collected through Day 672 (Visit 25/Month 24). .sup.fBlood draw for Immune Monitoring is to be collected at visits between Day 168 (Visit 21/Month 6) through Day 336 (Visit 23/Month 12) and ETV.
[1878] Concomitant Medications, Treatments, and Procedures.
[1879] Medications for medical problems other than antineoplastic agents are permitted. Those with conditions requiring anti-inflammatory drugs for chronic conditions potentially affecting TIL administration may be considered only with approval of the PI. Palliative radiation therapy is permitted between tumor resection and lymphodepletion as long as it does not affect target and non-target lesions. Use of systemic steroid therapy .ltoreq.10 mg/day of prednisone or equivalent is permitted. Use of >10 mg/day of prednisone or equivalent is permitted in cases of exacerbation of known disease or for treatment of new symptoms on study per Investigator's discretion. Any changes in concomitant medications may be recorded only in the patient's medical record throughout the trial. For subject who have CT IV contrast allergy, radiologic evaluation using MRI or PET-CT (without intravenous contrast is the preferred management. Every attempt should be made to maintain consistency in imaging modality for each patient.
[1880] All other anti-neoplastic drugs and radiation are prohibited. Subjects are also discouraged from using over-the-counter supplements and homeopathic products, especially those with purported anti-inflammatory properties, such as boswelia.
[1881] Patients treated with lymphodepletion are subject to opportunistic infections and appropriate infectious agent prophylaxis is required. The prophylaxis regimens and duration listed below may be modified as clinically indicated in consultation with an Infectious Diseases specialist.
[1882] Patients may receive levofloxacin at 100 to 1000 mg (e.g., 500 mg) daily (or an equivalent antibiotic) until ANC recovers to greater than 500/mm.sup.3.
[1883] Patients may receive the fixed combination of trimethoprim (TMP) and sulfamethoxazole (SMX) as double strength (DS) tablet [DS tabs=TMP 160 mg/tab and SMX 800 mg/tab] PO BID twice a week. TMP/SMX-DS may be taken by patients beginning on Day -7 and continuing for a minimum of 6 months after lymphodepletion. For patients with sulfa allergies, Pentamidine may be given (once discharged from the hospital) 300 mg IV every 21 days for 6 months after lymphodepletion. If IV Pentamidine is not feasible after discharge, PCP prophylaxis can be substituted with oral antimicrobials such as Atovaquone as per standard of care for 6 months after lymphodepletion. Patients may be given prophylactic antibiotics intravenously during high dose IL-2 therapy.
[1884] Starting on the day of TIL infusion subjects may be administered valacylcovir 100 to 1000 mg (e.g., 500 mg) PO daily if patient is able to take oral medications or acyclovir 5 mg/kg IVPB every 8 hours if patient needs intravenous medications, which is continued for 6 months (or at the discretion of the treating physician). Reversible renal insufficiency has been reported with IV administered acyclovir but not with oral acyclovir. Neurologic toxicity including delirium, tremors, coma, acute psychiatric disturbances, and abnormal electroencephalograms has been reported with higher doses of acyclovir. If symptoms occur, a dosage adjustment may be made or the drug be discontinued. Acyclovir may not be used concomitantly with other nucleoside analogs (e.g. ganciclovir), which interfere with DNA synthesis. In patients with renal disease, the dose is adjusted as per product labeling.
[1885] Patients may begin Fluconazole 50 to 500 mg (e.g., 200 mg) PO daily with the T cell infusion (Day 0) and continue for 6 months (or at the discretion of the treating physician).
[1886] To reduce the duration of neutropenia following NMA lymphodepletion chemotherapy, filgrastim (G-CSF) may be given at 1 to 10 .mu.g/kg/day (e.g., 5 .mu.g/kg/day) daily subcutaneously until ANC>500/mm.sup.3 for at least 2 consecutive days. Approximate dosing to correspond to the 300 mcg or 480 mcg dosage forms is allowed.
[1887] Ondansetron may be used to control nausea and vomiting during the chemotherapy preparative regimen. It can cause headache, dizziness, myalgias, drowsiness, malaise, and weakness. Less common side effects include chest pain, hypotension, pruritus, constipation and urinary retention. Consult the package insert for a complete list of side effects and specific dose instructions.
[1888] Furosemide may be used to enhance urine output during the chemotherapy preparative regimen with cyclophosphamide. Adverse effects include dizziness, vertigo, paresthesias, weakness, orthostatic hypotension, photosensitivity, rash and pruritus. Consult the package insert for a complete list of side effects and specific dose instructions.
[1889] Patients may start on broad-spectrum antibiotics, either a 3.sup.rd or 4.sup.th generation cephalosporin with adequate pseudomonas coverage as per local antibiogram or a quinolone for temperature .gtoreq.38.5.degree. C. with an ANC less than 500/mm.sup.3. Aminoglycosides should be avoided if possible. Infectious disease consultation may be obtained from all patients with unexplained fever or any infectious complications.
[1890] Using daily CBC values as a guide, the patient may also receive platelets and packed red blood cells as needed. Attempts may be made to keep Hgb>8.0 g/dL, and platelets >20,000/mL guided by the clinical scenario. Leukocyte filters may be utilized for all blood and platelet transfusions to decrease sensitization to transfused WBC's and decrease the risk of CMV infection. Irradiated blood and blood products should be used.
[1891] Description of Statistical Methods.
[1892] The primary endpoint for ovarian cancer and osteosarcoma cohorts is the ORR as assessed by investigators using RECIST 1.1 criteria. The ORR is derived as the sum of the number of patients with a confirmed CR or partial response (PR) divided by the number of patients in the All-Treated analysis set x 100%. The primary endpoint for the cohort of PDAC is the percentage of patients who survive for 183 days. The 6-month landmark survival rate may be calculated based on the Kaplan Meier method.
[1893] PFS is defined as the time (in months) from the start date of lymphodepletion to PD or death due to any cause, whichever event is earlier. Patients not experiencing PD or death at the time of data cut or end of study (i.e., database lock) may have their event times censored on the last adequate tumor assessment. DOR is measured from the first time measurement criteria are met for a CR or PR, whichever response is observed first, until the first date that progressive disease (PD) or death occurs. Patients not experiencing PD or death prior to the time of data cut or end of study may have their event times censored on the last adequate tumor assessment. The analysis of DOR is based on responders only as assessed by investigators per RECIST v1.1. DCR is derived as the sum of the number of patients who achieved PR/CR or SD per the RECIST v1.1 divided by the number of patients in the All-Treated analysis set x 100%. OS is defined as the time (in months) from the start date of the lymphodepletion to death due to any cause. Patients not having expired at the time of data cut or end of study may have their event times censored on the last date of their known survival status.
[1894] All exploratory analyses may be descriptive and performed by cohort. Some analysis results may be reported separated from the final clinical study report. T-cell repertoire analysis may be used to determine TIL persistence. Molecular and immunological features of tumors before and after TIL therapy may be determined using exome sequencing and immunohistochemistry/immunofluorescence analyses. Sensitivity analyses on ORR, DCR, DOR, and PFS as measured by investigators using the irRECIST criteria may be performed. Pearson correlation coefficient and linear regression, when appropriate, may be used to quantify the relationship between phenotypic attributes (CD8%, CD27 and CD28 expression, etc.) and tumor response to therapy. Baseline CA19-9 of patients with PDAC and baseline CA-125 of patients with ovarian cancer may be assessed for potential correlations with the efficacy outcome.
[1895] Grade 3 or higher treatment-emergent AEs and their incidence rates may be compared descriptively to historical data of TIL in other cancer disease types. AE incidence rates may be estimated with 95% CIs per cohort and all cohorts combined. The treatment-emergent AEs start from the first dose of cyclophosphamide and up to 6 months from the last dose of IL-2.
[1896] A study disposition summary may display number and percentages of patients who exit the study early by the primary reason in 2 parts: (1) After the tumor harvest prior to lymphodepletion; and (2) On or after the first dose of cyclophosphamide. Patients who are treated and being followed for the survival status at the time of study termination (i.e., completers) are not a part of this summary. Patients who did not receive planned full study treatment doses may also be summarized by its primary reason.
[1897] Summary of tumor response data per cohort may be based on the best overall response as assessed by investigators per RECIST 1.1. The summary may display percentages with 80% confidence intervals (CIs) for ORR and 95% CIs for DCR by the Wilcoxon score method among patients in the All-Treated analysis set. The median time-to-event and the landmark rate may also be measured with 80% CIs for the 6-month survival rate and 95% CIs for DOR, PFS, OS, and other landmark rates by the KM method.
[1898] All exploratory analyses may be descriptive and performed by cohort. The analysis may be defined separately from the statistical analysis plan for this study and reported independently outside the clinical study report (CSR). HRQOL may be assessed using the EORTC QLQ-C30 instrument and analyzed per the published evaluation manual.
[1899] Sample Size.
[1900] For ovarian cancer and osteosarcoma, the Simon's two stage minimax design may be used to monitor the efficacy of each cohort independently. The null hypothesis that the historical response rate of 5% to be tested against the estimated experimental cohort response rate of 20%. In the first stage, 10 patients may be treated per cohort. If there is no confirmed response in these 10 patients, so long as the patient are evaluable, the cohort may be terminated. Other efficacy estimates including maximum % reductions in target lesion sum of diameters and/or time to PD/death may be considered for termination. A confirmed response shall be determined by RECIST 1,1 criteria with first assessment at 6 weeks and second confirmatory scan at 12 weeks. If the study moves forward to Stage II, an additional 8 patients may be treated leading to a total of 18 patients for that cohort. Three or more responders out of 18 treated patients for the cohort may be considered clinically relevant to justify further investigation. The power of this design is >=70% under the 1-sided type I error rate of 10%.
[1901] For PDAC, the Simon's minimax two-stage design may also be used to monitor the 6-month survival rate. The null hypothesis that the historical 6-month survival rate of 35% to be tested against the estimated experimental cohort survival rate of 50% (ASCO January 2016). In the first stage, 11 patients may be treated and followed for .gtoreq.6 months without holding further enrollment. If there are 8 or more deaths among first 11 patients within 183 days counting from the first study drug administration, this cohort may be considered termination.
[1902] Otherwise, an additional 11 patients may be treated for a total of 22. The final result for the cohort may be clinically meaningful if .gtoreq.10 patients survive at least for 183 days. The power of this design is approximately 70% under the 1-sided type I error rate of 10%.
Example 8--Methods of Expanding TILs Using TNFRSF Agonists During Pre-REP and REP Steps
[1903] The antibodies used in this Example are described elsewhere herein and are further described in Table 57.
TABLE-US-00060 TABLE 57 4-1BB and 0X40 Agonistic Monoclonal Antibodies. Short 4-1BB OX40 name CB 4-1BB BPS4-1BB CB OX40 Source Creative BPS Biosciences Creative Biolabs (San Diego, CA, Biolabs (Shirley, NY, USA) (Shirley, NY, USA) USA) Clone and Urelumab; Unknown; Unknown; Product Catalog no. TAB- Catalog no. Catalog no. Information 179 79097-2, Lot no. MOM-18455 170718 Isotype IgG4 IgG1 IgG Formulation PBS PBS and 20% PBS glycerol Purity >95% Unknown >98% Sequence Available Unknown Unknown and epitope Publication N/A Wilcox, R. A., et N/A al., J. Clin. Invest. 2002; Foell, J. et. al., J. Clin. Invest. 2003.
[1904] In addition to the monoclonal antibodies described above, the OX40 agonistic antibody clone Ber-ACT35 (BioLegend, San Diego, Calif., USA) was also used in selected experiments described herein.
[1905] The overall experimental strategy included the following steps: reagent procurement and validation; ex vivo expansion experimental design; adding anti-4-1BB or anti-OX40 at day 0 of pre-REP experiments, using fresh melanoma, lung, cervical tumor samples; assessing the anti-OX40 in 21 mini-REP carried out on thawed head & neck, lung, melanoma, triple-negative breast cancer, and breast cancer pre-REP TIL samples; and assessment of TIL yield and cell lineage phenotype (CD4:CD8), T-cell subsets/extended phenotype, and functional assays.
[1906] The comparability of anti-4-1BB binding affinity for two 4-1BB agonists was assessed. 4-1BB reporter cells were stained with anti-4-1BB antibody (Creative Biolabs) or anti-4-1BB (BPS Biosciences) at concentrations of 0.01, 0.03, 0.1, 0.3, 1, and 3 .mu.g/ml together with FITC-conjugated mouse anti-human IgG and analyzed by flow cytometry. The results are shown in FIG. 16 and FIG. 17 (for % of 4-1BB.sup.+ cells and mean fluorescence intensity (MFI) of 4-1BB.sup.+ cells, respectively) and indicate that the Creative Biolabs (CB) 4-1BB urelumab antibody has the highest binding affinity.
[1907] An assessment of NF-.kappa.B pathway activation of 4-1BB agonistic antibodies was also performed. 4-1BB reporter cells were treated with either anti-4-1BB (CB or BPS antibodies) at a concentration of 1, 2, 4, and 8 .mu.g/mL for 24 hours. The cells were lysed using One-Step Luciferase reagent, and luciferase activity was measured by a luminometer. The results are shown in FIG. 18. Log EC50 for the CB antibody was determined to be 3.9 .mu.g/mL and for the BPS antibody was determined to be 2.13 .mu.g/mL. Both CB and BPS anti-4-1BB agonists had similar Log EC50 values even though the BPS antibody exhibited greater NF-kB signaling activation.
[1908] The binding affinity of the CB OX40 agonist was also assessed. OX40 reporter cells were stained with anti-OX40 Creative Biolabs (CB) agonist at the concentrations of 0.01, 0.03, 0.1, 0.3, 1, and 3 .mu.g/ml together with FITC-conjugated mouse anti-human IgG and analyzed by flow cytometry. Results are shown in FIG. 19 and FIG. 20 for % of OX40.sup.+ cells and MFI of OX40.sup.+ cells, respectively, and indicate that the CB OX40 has a high binding affinity.
[1909] The comparability of OX40 binding affinity for two OX40 agonists, the CB OX40 agonist and the OX40 agonistic antibody clone Ber-ACT35 (BioLegend, San Diego, Calif., USA), was assessed. Five different histologic TIL lines (including cervical, head and neck, lung, and melanoma) were stained with either anti-OX40 agonistic antibody at concentration of 0.1, 0.3, 1, 3, 10 (g/mL) together with anti-human IgG secondary antibody or anti-OX40 (clone Ber-ACT35) alone. The results are shown in FIG. 21, and indicate comparable binding affinity for the two agonists.
[1910] An assessment of NF-kB pathway activation of the CB OX40 agonist antibody was also performed, with results shown in FIG. 22. OX40 reporter cells were treated with either anti-OX40 alone or isotype control at concentrations of 1, 2, 4, 8, and 16 .mu.g/mL with or without feeder cells for 24 hours. The cells were lysed using One-Step Luciferase reagent, and luciferase activity was measured by luminometer. The use of PBMC feeders initiated NF-.kappa.B activation using the OX40 reporter cell line, suggesting that clustering is involved in activation.
[1911] The experimental design for use of 4-1BB and OX40 agonists during the pre-REP step is shown in FIG. 23. The tumor histologies explored are shown in FIG. 24. The data analysis strategy is shown in FIG. 25. No treatment (IL-2 alone), anti-4-1BB, and anti-OX40 were analyzed in matched-pair manner. Using this approach, the samples were assigned into three different groups including: Group 1, No treatment and anti-4-1BB (n=3); Group 2, No treatment and anti-OX40 (n=5); and Group 3, No treatment and anti-4-1BB and anti-OX40 (n=2). Total cell count results from expansions are shown in FIG. 26 (CB 4-1BB agonist versus not tested, N=3); FIG. 27 (CB OX40 agonist versus not tested, N=5); and FIG. 28 (CB 4-1BB agonist and OX-40 agonist, N=2). CD8.sup.+ cell count results for cell expansion are shown in FIG. 29 (CB 4-1BB agonist versus not tested, N=3); FIG. 30 (CB OX40 agonist versus not tested, N=5); and FIG. 31 (CB 4-1BB agonist and OX-40 agonist, N=2). Total CD8.sup.+/CD4.sup.+ cell count ratio results for cell expansions are shown in FIG. 32 (CB 4-1BB agonist versus not tested, N=3); FIG. 33 (CB OX40 agonist versus not tested, N=5); and FIG. 34 (CB 4-1BB agonist and OX-40 agonist, N=2).
[1912] REP propagation of pre-REP TILs expanded in the presence of 4-1BB or OX40 agonists was also explored using the scheme shown in FIG. 35. Pre-REP TILs were expanded with either CB 4-1BB agonist or CB OX40 agonist were further propagated in a REP protocol in the presence of irradiated PBMCs, anti-CD3 antibody (30 ng/mL), and IL-2 (3000 IU/mL) for 11 days. TILs were harvested and counted, and fold expansion determined. Results are shown in FIG. 36, FIG. 37, and FIG. 38.
[1913] Assessment of OX40 during the REP phase was also tested. Twenty-one TIL lines from different histologies (FIG. 39) were propagated with REP with addition of CB OX40 agonist or isotype control antibody at concentration of 5 .mu.g/mL. The experimental scheme is shown in FIG. 40. Results are shown in FIG. 41, FIG. 42, and FIG. 43. Surprisingly, the OX40 agonist preferentially expands CD8.sup.+ TILs during REP. TILs treated with OX40 agonist were classified as responders or non-responders.
[1914] Anti-OX40 dose titration in non-responder and responder TIL lines was performed to further study this effect and to define the optimal concentration of OX40 agonist in responders and non-responders. TIL lines were categorized into two groups (responder and non-responder) based on enhanced CD8.sup.+ skewness following anti-OX40 treatment. Three non-responders (L4005, H3005, and M1022) and responders (T6001, T6003, and L4002) were propagated with REP in the presence of OX40 agonist or isotype control antibody following the conditions shown in FIG. 44. FIG. 45 and FIG. 46 illustrates that a dose-dependent manner of CD8.sup.+ skewness following anti-OX40 treatment was observed in responders, with concentrations in the 1 to 10 g/mL range promoting skewness. Non-responders did not exhibit CD8.sup.+ skewness following anti-OX40 treatment even at high concentration (30 .mu.g/mL).
[1915] The impact of OX40 agonist on TCRvb repertoire in responders was also investigated. To determine whether anti-OX40, previously shown to skew CD8.sup.+ population, preferentially expand certain TCR vb repertoire. Responder TIL lines were propagated with REP in 24-well plates with either IL-2 alone or IL-2 with CD OX40 agonist monoclonal antibody (5 .mu.g/mL). On day 11, TIL were harvested and stained with anti-CD3, anti-CD8, anti-CD4, and TCRvb repertoire antibodies, and analyzed by flow cytometry. Results are shown for three responders with three histologies in FIG. 47, FIG. 48, and FIG. 49. Minimal changes in TCRvb repertoire was observed, indicating that the high degree of polyclonality exhibited by the shortened 22-day process in an embodiment of FIG. 1 or FIG. 2 is surprisingly preserved in conjunction with the use of OX40 agonists during REP.
[1916] In conclusion, use of CB anti-OX40 antibody significantly enhanced pre-REP CD8.sup.+ TIL expansion, while use of CB anti-4-1BB antibody also demonstrated a promising trend. REP-fold expansion was comparable regardless of pre-treatment condition. Surprisingly, OX40 agonistic antibody increased CD8.sup.+/CD4.sup.+ ratio in REP TIL previously grown with IL-2 alone. In non-responder TILs, down-regulation of OX-40 was not observed in the CD4.sup.+ subset following anti-OX40 treatment. The dose-dependent manner of CD8.sup.+ skewness following anti-OX40 treatment was observed in responders. The change in TCRvb repertoire was very subtle even though significant CD8.sup.+ skewness was observed.
Sequence CWU 1
1
5121450PRTArtificial SequenceMuromonab heavy chain 1Gln Val Gln Leu
Gln Gln Ser Gly Ala Glu Leu Ala Arg Pro Gly Ala1 5 10 15Ser Val Lys
Met Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Arg Tyr 20 25 30Thr Met
His Trp Val Lys Gln Arg Pro Gly Gln Gly Leu Glu Trp Ile 35 40 45Gly
Tyr Ile Asn Pro Ser Arg Gly Tyr Thr Asn Tyr Asn Gln Lys Phe 50 55
60Lys Asp Lys Ala Thr Leu Thr Thr Asp Lys Ser Ser Ser Thr Ala Tyr65
70 75 80Met Gln Leu Ser Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Tyr
Cys 85 90 95Ala Arg Tyr Tyr Asp Asp His Tyr Cys Leu Asp Tyr Trp Gly
Gln Gly 100 105 110Thr Thr Leu Thr Val Ser Ser Ala Lys Thr Thr Ala
Pro Ser Val Tyr 115 120 125Pro Leu Ala Pro Val Cys Gly Gly Thr Thr
Gly Ser Ser Val Thr Leu 130 135 140Gly Cys Leu Val Lys Gly Tyr Phe
Pro Glu Pro Val Thr Leu Thr Trp145 150 155 160Asn Ser Gly Ser Leu
Ser Ser Gly Val His Thr Phe Pro Ala Val Leu 165 170 175Gln Ser Asp
Leu Tyr Thr Leu Ser Ser Ser Val Thr Val Thr Ser Ser 180 185 190Thr
Trp Pro Ser Gln Ser Ile Thr Cys Asn Val Ala His Pro Ala Ser 195 200
205Ser Thr Lys Val Asp Lys Lys Ile Glu Pro Arg Pro Lys Ser Cys Asp
210 215 220Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu
Gly Gly225 230 235 240Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys
Asp Thr Leu Met Ile 245 250 255Ser Arg Thr Pro Glu Val Thr Cys Val
Val Val Asp Val Ser His Glu 260 265 270Asp Pro Glu Val Lys Phe Asn
Trp Tyr Val Asp Gly Val Glu Val His 275 280 285Asn Ala Lys Thr Lys
Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg 290 295 300Val Val Ser
Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys305 310 315
320Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu
325 330 335Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln
Val Tyr 340 345 350Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn
Gln Val Ser Leu 355 360 365Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser
Asp Ile Ala Val Glu Trp 370 375 380Glu Ser Asn Gly Gln Pro Glu Asn
Asn Tyr Lys Thr Thr Pro Pro Val385 390 395 400Leu Asp Ser Asp Gly
Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp 405 410 415Lys Ser Arg
Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His 420 425 430Glu
Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro 435 440
445Gly Lys 4502213PRTArtificial SequenceMuromonab light chain 2Gln
Ile Val Leu Thr Gln Ser Pro Ala Ile Met Ser Ala Ser Pro Gly1 5 10
15Glu Lys Val Thr Met Thr Cys Ser Ala Ser Ser Ser Val Ser Tyr Met
20 25 30Asn Trp Tyr Gln Gln Lys Ser Gly Thr Ser Pro Lys Arg Trp Ile
Tyr 35 40 45Asp Thr Ser Lys Leu Ala Ser Gly Val Pro Ala His Phe Arg
Gly Ser 50 55 60Gly Ser Gly Thr Ser Tyr Ser Leu Thr Ile Ser Gly Met
Glu Ala Glu65 70 75 80Asp Ala Ala Thr Tyr Tyr Cys Gln Gln Trp Ser
Ser Asn Pro Phe Thr 85 90 95Phe Gly Ser Gly Thr Lys Leu Glu Ile Asn
Arg Ala Asp Thr Ala Pro 100 105 110Thr Val Ser Ile Phe Pro Pro Ser
Ser Glu Gln Leu Thr Ser Gly Gly 115 120 125Ala Ser Val Val Cys Phe
Leu Asn Asn Phe Tyr Pro Lys Asp Ile Asn 130 135 140Val Lys Trp Lys
Ile Asp Gly Ser Glu Arg Gln Asn Gly Val Leu Asn145 150 155 160Ser
Trp Thr Asp Gln Asp Ser Lys Asp Ser Thr Tyr Ser Met Ser Ser 165 170
175Thr Leu Thr Leu Thr Lys Asp Glu Tyr Glu Arg His Asn Ser Tyr Thr
180 185 190Cys Glu Ala Thr His Lys Thr Ser Thr Ser Pro Ile Val Lys
Ser Phe 195 200 205Asn Arg Asn Glu Cys 2103134PRTArtificial
Sequencerecombinant human IL-2 (rhIL-2) 3Met Ala Pro Thr Ser Ser
Ser Thr Lys Lys Thr Gln Leu Gln Leu Glu1 5 10 15His Leu Leu Leu Asp
Leu Gln Met Ile Leu Asn Gly Ile Asn Asn Tyr 20 25 30Lys Asn Pro Lys
Leu Thr Arg Met Leu Thr Phe Lys Phe Tyr Met Pro 35 40 45Lys Lys Ala
Thr Glu Leu Lys His Leu Gln Cys Leu Glu Glu Glu Leu 50 55 60Lys Pro
Leu Glu Glu Val Leu Asn Leu Ala Gln Ser Lys Asn Phe His65 70 75
80Leu Arg Pro Arg Asp Leu Ile Ser Asn Ile Asn Val Ile Val Leu Glu
85 90 95Leu Lys Gly Ser Glu Thr Thr Phe Met Cys Glu Tyr Ala Asp Glu
Thr 100 105 110Ala Thr Ile Val Glu Phe Leu Asn Arg Trp Ile Thr Phe
Cys Gln Ser 115 120 125Ile Ile Ser Thr Leu Thr 1304132PRTArtificial
SequenceAldesleukin 4Pro Thr Ser Ser Ser Thr Lys Lys Thr Gln Leu
Gln Leu Glu His Leu1 5 10 15Leu Leu Asp Leu Gln Met Ile Leu Asn Gly
Ile Asn Asn Tyr Lys Asn 20 25 30Pro Lys Leu Thr Arg Met Leu Thr Phe
Lys Phe Tyr Met Pro Lys Lys 35 40 45Ala Thr Glu Leu Lys His Leu Gln
Cys Leu Glu Glu Glu Leu Lys Pro 50 55 60Leu Glu Glu Val Leu Asn Leu
Ala Gln Ser Lys Asn Phe His Leu Arg65 70 75 80Pro Arg Asp Leu Ile
Ser Asn Ile Asn Val Ile Val Leu Glu Leu Lys 85 90 95Gly Ser Glu Thr
Thr Phe Met Cys Glu Tyr Ala Asp Glu Thr Ala Thr 100 105 110Ile Val
Glu Phe Leu Asn Arg Trp Ile Thr Phe Ser Gln Ser Ile Ile 115 120
125Ser Thr Leu Thr 1305130PRTArtificial Sequencerecombinant human
IL-4 (rhIL-4) 5Met His Lys Cys Asp Ile Thr Leu Gln Glu Ile Ile Lys
Thr Leu Asn1 5 10 15Ser Leu Thr Glu Gln Lys Thr Leu Cys Thr Glu Leu
Thr Val Thr Asp 20 25 30Ile Phe Ala Ala Ser Lys Asn Thr Thr Glu Lys
Glu Thr Phe Cys Arg 35 40 45Ala Ala Thr Val Leu Arg Gln Phe Tyr Ser
His His Glu Lys Asp Thr 50 55 60Arg Cys Leu Gly Ala Thr Ala Gln Gln
Phe His Arg His Lys Gln Leu65 70 75 80Ile Arg Phe Leu Lys Arg Leu
Asp Arg Asn Leu Trp Gly Leu Ala Gly 85 90 95Leu Asn Ser Cys Pro Val
Lys Glu Ala Asn Gln Ser Thr Leu Glu Asn 100 105 110Phe Leu Glu Arg
Leu Lys Thr Ile Met Arg Glu Lys Tyr Ser Lys Cys 115 120 125Ser Ser
1306153PRTArtificial Sequencerecombinant human IL-7 (rhIL-7) 6Met
Asp Cys Asp Ile Glu Gly Lys Asp Gly Lys Gln Tyr Glu Ser Val1 5 10
15Leu Met Val Ser Ile Asp Gln Leu Leu Asp Ser Met Lys Glu Ile Gly
20 25 30Ser Asn Cys Leu Asn Asn Glu Phe Asn Phe Phe Lys Arg His Ile
Cys 35 40 45Asp Ala Asn Lys Glu Gly Met Phe Leu Phe Arg Ala Ala Arg
Lys Leu 50 55 60Arg Gln Phe Leu Lys Met Asn Ser Thr Gly Asp Phe Asp
Leu His Leu65 70 75 80Leu Lys Val Ser Glu Gly Thr Thr Ile Leu Leu
Asn Cys Thr Gly Gln 85 90 95Val Lys Gly Arg Lys Pro Ala Ala Leu Gly
Glu Ala Gln Pro Thr Lys 100 105 110Ser Leu Glu Glu Asn Lys Ser Leu
Lys Glu Gln Lys Lys Leu Asn Asp 115 120 125Leu Cys Phe Leu Lys Arg
Leu Leu Gln Glu Ile Lys Thr Cys Trp Asn 130 135 140Lys Ile Leu Met
Gly Thr Lys Glu His145 1507115PRTArtificial Sequencerecombinant
human IL-15 (rhIL-15) 7Met Asn Trp Val Asn Val Ile Ser Asp Leu Lys
Lys Ile Glu Asp Leu1 5 10 15Ile Gln Ser Met His Ile Asp Ala Thr Leu
Tyr Thr Glu Ser Asp Val 20 25 30His Pro Ser Cys Lys Val Thr Ala Met
Lys Cys Phe Leu Leu Glu Leu 35 40 45Gln Val Ile Ser Leu Glu Ser Gly
Asp Ala Ser Ile His Asp Thr Val 50 55 60Glu Asn Leu Ile Ile Leu Ala
Asn Asn Ser Leu Ser Ser Asn Gly Asn65 70 75 80Val Thr Glu Ser Gly
Cys Lys Glu Cys Glu Glu Leu Glu Glu Lys Asn 85 90 95Ile Lys Glu Phe
Leu Gln Ser Phe Val His Ile Val Gln Met Phe Ile 100 105 110Asn Thr
Ser 1158132PRTArtificial Sequencerecombinant human IL-21 (rhIL-21)
8Met Gln Asp Arg His Met Ile Arg Met Arg Gln Leu Ile Asp Ile Val1 5
10 15Asp Gln Leu Lys Asn Tyr Val Asn Asp Leu Val Pro Glu Phe Leu
Pro 20 25 30Ala Pro Glu Asp Val Glu Thr Asn Cys Glu Trp Ser Ala Phe
Ser Cys 35 40 45Phe Gln Lys Ala Gln Leu Lys Ser Ala Asn Thr Gly Asn
Asn Glu Arg 50 55 60Ile Ile Asn Val Ser Ile Lys Lys Leu Lys Arg Lys
Pro Pro Ser Thr65 70 75 80Asn Ala Gly Arg Arg Gln Lys His Arg Leu
Thr Cys Pro Ser Cys Asp 85 90 95Ser Tyr Glu Lys Lys Pro Pro Lys Glu
Phe Leu Glu Arg Phe Lys Ser 100 105 110Leu Leu Gln Lys Met Ile His
Gln His Leu Ser Ser Arg Thr His Gly 115 120 125Ser Glu Asp Ser
1309255PRTHomo Sapiens 9Met Gly Asn Ser Cys Tyr Asn Ile Val Ala Thr
Leu Leu Leu Val Leu1 5 10 15Asn Phe Glu Arg Thr Arg Ser Leu Gln Asp
Pro Cys Ser Asn Cys Pro 20 25 30Ala Gly Thr Phe Cys Asp Asn Asn Arg
Asn Gln Ile Cys Ser Pro Cys 35 40 45Pro Pro Asn Ser Phe Ser Ser Ala
Gly Gly Gln Arg Thr Cys Asp Ile 50 55 60Cys Arg Gln Cys Lys Gly Val
Phe Arg Thr Arg Lys Glu Cys Ser Ser65 70 75 80Thr Ser Asn Ala Glu
Cys Asp Cys Thr Pro Gly Phe His Cys Leu Gly 85 90 95Ala Gly Cys Ser
Met Cys Glu Gln Asp Cys Lys Gln Gly Gln Glu Leu 100 105 110Thr Lys
Lys Gly Cys Lys Asp Cys Cys Phe Gly Thr Phe Asn Asp Gln 115 120
125Lys Arg Gly Ile Cys Arg Pro Trp Thr Asn Cys Ser Leu Asp Gly Lys
130 135 140Ser Val Leu Val Asn Gly Thr Lys Glu Arg Asp Val Val Cys
Gly Pro145 150 155 160Ser Pro Ala Asp Leu Ser Pro Gly Ala Ser Ser
Val Thr Pro Pro Ala 165 170 175Pro Ala Arg Glu Pro Gly His Ser Pro
Gln Ile Ile Ser Phe Phe Leu 180 185 190Ala Leu Thr Ser Thr Ala Leu
Leu Phe Leu Leu Phe Phe Leu Thr Leu 195 200 205Arg Phe Ser Val Val
Lys Arg Gly Arg Lys Lys Leu Leu Tyr Ile Phe 210 215 220Lys Gln Pro
Phe Met Arg Pro Val Gln Thr Thr Gln Glu Glu Asp Gly225 230 235
240Cys Ser Cys Arg Phe Pro Glu Glu Glu Glu Gly Gly Cys Glu Leu 245
250 25510256PRTMus Musculus 10Met Gly Asn Asn Cys Tyr Asn Val Val
Val Ile Val Leu Leu Leu Val1 5 10 15Gly Cys Glu Lys Val Gly Ala Val
Gln Asn Ser Cys Asp Asn Cys Gln 20 25 30Pro Gly Thr Phe Cys Arg Lys
Tyr Asn Pro Val Cys Lys Ser Cys Pro 35 40 45Pro Ser Thr Phe Ser Ser
Ile Gly Gly Gln Pro Asn Cys Asn Ile Cys 50 55 60Arg Val Cys Ala Gly
Tyr Phe Arg Phe Lys Lys Phe Cys Ser Ser Thr65 70 75 80His Asn Ala
Glu Cys Glu Cys Ile Glu Gly Phe His Cys Leu Gly Pro 85 90 95Gln Cys
Thr Arg Cys Glu Lys Asp Cys Arg Pro Gly Gln Glu Leu Thr 100 105
110Lys Gln Gly Cys Lys Thr Cys Ser Leu Gly Thr Phe Asn Asp Gln Asn
115 120 125Gly Thr Gly Val Cys Arg Pro Trp Thr Asn Cys Ser Leu Asp
Gly Arg 130 135 140Ser Val Leu Lys Thr Gly Thr Thr Glu Lys Asp Val
Val Cys Gly Pro145 150 155 160Pro Val Val Ser Phe Ser Pro Ser Thr
Thr Ile Ser Val Thr Pro Glu 165 170 175Gly Gly Pro Gly Gly His Ser
Leu Gln Val Leu Thr Leu Phe Leu Ala 180 185 190Leu Thr Ser Ala Leu
Leu Leu Ala Leu Ile Phe Ile Thr Leu Leu Phe 195 200 205Ser Val Leu
Lys Trp Ile Arg Lys Lys Phe Pro His Ile Phe Lys Gln 210 215 220Pro
Phe Lys Lys Thr Thr Gly Ala Ala Gln Glu Glu Asp Ala Cys Ser225 230
235 240Cys Arg Cys Pro Gln Glu Glu Glu Gly Gly Gly Gly Gly Tyr Glu
Leu 245 250 25511441PRTArtificial Sequenceheavy chain for
utomilumab 11Glu Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys
Pro Gly Glu1 5 10 15Ser Leu Arg Ile Ser Cys Lys Gly Ser Gly Tyr Ser
Phe Ser Thr Tyr 20 25 30Trp Ile Ser Trp Val Arg Gln Met Pro Gly Lys
Gly Leu Glu Trp Met 35 40 45Gly Lys Ile Tyr Pro Gly Asp Ser Tyr Thr
Asn Tyr Ser Pro Ser Phe 50 55 60Gln Gly Gln Val Thr Ile Ser Ala Asp
Lys Ser Ile Ser Thr Ala Tyr65 70 75 80Leu Gln Trp Ser Ser Leu Lys
Ala Ser Asp Thr Ala Met Tyr Tyr Cys 85 90 95Ala Arg Gly Tyr Gly Ile
Phe Asp Tyr Trp Gly Gln Gly Thr Leu Val 100 105 110Thr Val Ser Ser
Ala Ser Thr Lys Gly Pro Ser Val Phe Pro Leu Ala 115 120 125Pro Cys
Ser Arg Ser Thr Ser Glu Ser Thr Ala Ala Leu Gly Cys Leu 130 135
140Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp Asn Ser
Gly145 150 155 160Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val
Leu Gln Ser Ser 165 170 175Gly Leu Tyr Ser Leu Ser Ser Val Val Thr
Val Pro Ser Ser Asn Phe 180 185 190Gly Thr Gln Thr Tyr Thr Cys Asn
Val Asp His Lys Pro Ser Asn Thr 195 200 205Lys Val Asp Lys Thr Val
Glu Arg Lys Cys Cys Val Glu Cys Pro Pro 210 215 220Cys Pro Ala Pro
Pro Val Ala Gly Pro Ser Val Phe Leu Phe Pro Pro225 230 235 240Lys
Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys 245 250
255Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Gln Phe Asn Trp
260 265 270Tyr Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys Pro
Arg Glu 275 280 285Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser Val
Leu Thr Val Val 290 295 300His Gln Asp Trp Leu Asn Gly Lys Glu Tyr
Lys Cys Lys Val Ser Asn305 310 315 320Lys Gly Leu Pro Ala Pro Ile
Glu Lys Thr Ile Ser Lys Thr Lys Gly 325 330 335Gln Pro Arg Glu Pro
Gln Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu 340 345 350Met Thr Lys
Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr 355 360 365Pro
Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn 370 375
380Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser Asp Gly Ser Phe
Phe385 390 395 400Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp
Gln Gln Gly Asn 405 410 415Val Phe Ser Cys Ser Val Met His Glu Ala
Leu His Asn His Tyr Thr 420 425 430Gln Lys Ser Leu Ser Leu Ser Pro
Gly 435
44012214PRTArtificial Sequencelight chain for utomilumab 12Ser Tyr
Glu Leu Thr Gln Pro Pro Ser Val Ser Val Ser Pro Gly Gln1 5 10 15Thr
Ala Ser Ile Thr Cys Ser Gly Asp Asn Ile Gly Asp Gln Tyr Ala 20 25
30His Trp Tyr Gln Gln Lys Pro Gly Gln Ser Pro Val Leu Val Ile Tyr
35 40 45Gln Asp Lys Asn Arg Pro Ser Gly Ile Pro Glu Arg Phe Ser Gly
Ser 50 55 60Asn Ser Gly Asn Thr Ala Thr Leu Thr Ile Ser Gly Thr Gln
Ala Met65 70 75 80Asp Glu Ala Asp Tyr Tyr Cys Ala Thr Tyr Thr Gly
Phe Gly Ser Leu 85 90 95Ala Val Phe Gly Gly Gly Thr Lys Leu Thr Val
Leu Gly Gln Pro Lys 100 105 110Ala Ala Pro Ser Val Thr Leu Phe Pro
Pro Ser Ser Glu Glu Leu Gln 115 120 125Ala Asn Lys Ala Thr Leu Val
Cys Leu Ile Ser Asp Phe Tyr Pro Gly 130 135 140Ala Val Thr Val Ala
Trp Lys Ala Asp Ser Ser Pro Val Lys Ala Gly145 150 155 160Val Glu
Thr Thr Thr Pro Ser Lys Gln Ser Asn Asn Lys Tyr Ala Ala 165 170
175Ser Ser Tyr Leu Ser Leu Thr Pro Glu Gln Trp Lys Ser His Arg Ser
180 185 190Tyr Ser Cys Gln Val Thr His Glu Gly Ser Thr Val Glu Lys
Thr Val 195 200 205Ala Pro Thr Glu Cys Ser 21013116PRTArtificial
Sequenceheavy chain variable region for utomilumab 13Glu Val Gln
Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Glu1 5 10 15Ser Leu
Arg Ile Ser Cys Lys Gly Ser Gly Tyr Ser Phe Ser Thr Tyr 20 25 30Trp
Ile Ser Trp Val Arg Gln Met Pro Gly Lys Gly Leu Glu Trp Met 35 40
45Gly Lys Ile Tyr Pro Gly Asp Ser Tyr Thr Asn Tyr Ser Pro Ser Phe
50 55 60Gln Gly Gln Val Thr Ile Ser Ala Asp Lys Ser Ile Ser Thr Ala
Tyr65 70 75 80Leu Gln Trp Ser Ser Leu Lys Ala Ser Asp Thr Ala Met
Tyr Tyr Cys 85 90 95Ala Arg Gly Tyr Gly Ile Phe Asp Tyr Trp Gly Gln
Gly Thr Leu Val 100 105 110Thr Val Ser Ser 11514108PRTArtificial
Sequencelight chain variable region for utomilumab 14Ser Tyr Glu
Leu Thr Gln Pro Pro Ser Val Ser Val Ser Pro Gly Gln1 5 10 15Thr Ala
Ser Ile Thr Cys Ser Gly Asp Asn Ile Gly Asp Gln Tyr Ala 20 25 30His
Trp Tyr Gln Gln Lys Pro Gly Gln Ser Pro Val Leu Val Ile Tyr 35 40
45Gln Asp Lys Asn Arg Pro Ser Gly Ile Pro Glu Arg Phe Ser Gly Ser
50 55 60Asn Ser Gly Asn Thr Ala Thr Leu Thr Ile Ser Gly Thr Gln Ala
Met65 70 75 80Asp Glu Ala Asp Tyr Tyr Cys Ala Thr Tyr Thr Gly Phe
Gly Ser Leu 85 90 95Ala Val Phe Gly Gly Gly Thr Lys Leu Thr Val Leu
100 105156PRTArtificial Sequenceheavy chain CDR1 for utomilumab
15Ser Thr Tyr Trp Ile Ser1 51617PRTArtificial Sequenceheavy chain
CDR2 for utomilumab 16Lys Ile Tyr Pro Gly Asp Ser Tyr Thr Asn Tyr
Ser Pro Ser Phe Gln1 5 10 15Gly178PRTArtificial Sequenceheavy chain
CDR3 for utomilumab 17Arg Gly Tyr Gly Ile Phe Asp Tyr1
51811PRTArtificial Sequencelight chain CDR1 for utomilumab 18Ser
Gly Asp Asn Ile Gly Asp Gln Tyr Ala His1 5 10197PRTArtificial
Sequencelight chain CDR2 for utomilumab 19Gln Asp Lys Asn Arg Pro
Ser1 52011PRTArtificial Sequencelight chain CDR3 for utomilumab
20Ala Thr Tyr Thr Gly Phe Gly Ser Leu Ala Val1 5
1021448PRTArtificial Sequenceheavy chain for urelumab 21Gln Val Gln
Leu Gln Gln Trp Gly Ala Gly Leu Leu Lys Pro Ser Glu1 5 10 15Thr Leu
Ser Leu Thr Cys Ala Val Tyr Gly Gly Ser Phe Ser Gly Tyr 20 25 30Tyr
Trp Ser Trp Ile Arg Gln Ser Pro Glu Lys Gly Leu Glu Trp Ile 35 40
45Gly Glu Ile Asn His Gly Gly Tyr Val Thr Tyr Asn Pro Ser Leu Glu
50 55 60Ser Arg Val Thr Ile Ser Val Asp Thr Ser Lys Asn Gln Phe Ser
Leu65 70 75 80Lys Leu Ser Ser Val Thr Ala Ala Asp Thr Ala Val Tyr
Tyr Cys Ala 85 90 95Arg Asp Tyr Gly Pro Gly Asn Tyr Asp Trp Tyr Phe
Asp Leu Trp Gly 100 105 110Arg Gly Thr Leu Val Thr Val Ser Ser Ala
Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro Leu Ala Pro Cys Ser
Arg Ser Thr Ser Glu Ser Thr Ala 130 135 140Ala Leu Gly Cys Leu Val
Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150 155 160Ser Trp Asn
Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala 165 170 175Val
Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val 180 185
190Pro Ser Ser Ser Leu Gly Thr Lys Thr Tyr Thr Cys Asn Val Asp His
195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys Arg Val Glu Ser Lys
Tyr Gly 210 215 220Pro Pro Cys Pro Pro Cys Pro Ala Pro Glu Phe Leu
Gly Gly Pro Ser225 230 235 240Val Phe Leu Phe Pro Pro Lys Pro Lys
Asp Thr Leu Met Ile Ser Arg 245 250 255Thr Pro Glu Val Thr Cys Val
Val Val Asp Val Ser Gln Glu Asp Pro 260 265 270Glu Val Gln Phe Asn
Trp Tyr Val Asp Gly Val Glu Val His Asn Ala 275 280 285Lys Thr Lys
Pro Arg Glu Glu Gln Phe Asn Ser Thr Tyr Arg Val Val 290 295 300Ser
Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr305 310
315 320Lys Cys Lys Val Ser Asn Lys Gly Leu Pro Ser Ser Ile Glu Lys
Thr 325 330 335Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val
Tyr Thr Leu 340 345 350Pro Pro Ser Gln Glu Glu Met Thr Lys Asn Gln
Val Ser Leu Thr Cys 355 360 365Leu Val Lys Gly Phe Tyr Pro Ser Asp
Ile Ala Val Glu Trp Glu Ser 370 375 380Asn Gly Gln Pro Glu Asn Asn
Tyr Lys Thr Thr Pro Pro Val Leu Asp385 390 395 400Ser Asp Gly Ser
Phe Phe Leu Tyr Ser Arg Leu Thr Val Asp Lys Ser 405 410 415Arg Trp
Gln Glu Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala 420 425
430Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Leu Gly Lys
435 440 44522216PRTArtificial Sequencelight chain for urelumab
22Glu Ile Val Leu Thr Gln Ser Pro Ala Thr Leu Ser Leu Ser Pro Gly1
5 10 15Glu Arg Ala Thr Leu Ser Cys Arg Ala Ser Gln Ser Val Ser Ser
Tyr 20 25 30Leu Ala Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro Arg Leu
Leu Ile 35 40 45Tyr Asp Ala Ser Asn Arg Ala Thr Gly Ile Pro Ala Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Glu Pro65 70 75 80Glu Asp Phe Ala Val Tyr Tyr Cys Gln Gln
Arg Ser Asn Trp Pro Pro 85 90 95Ala Leu Thr Phe Cys Gly Gly Thr Lys
Val Glu Ile Lys Arg Thr Val 100 105 110Ala Ala Pro Ser Val Phe Ile
Phe Pro Pro Ser Asp Glu Gln Leu Lys 115 120 125Ser Gly Thr Ala Ser
Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg 130 135 140Glu Ala Lys
Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn145 150 155
160Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser
165 170 175Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys
His Lys 180 185 190Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser
Ser Pro Val Thr 195 200 205Lys Ser Phe Asn Arg Gly Glu Cys 210
21523120PRTArtificial Sequencevariable heavy chain for urelumab
23Met Lys His Leu Trp Phe Phe Leu Leu Leu Val Ala Ala Pro Arg Trp1
5 10 15Val Leu Ser Gln Val Gln Leu Gln Gln Trp Gly Ala Gly Leu Leu
Lys 20 25 30Pro Ser Glu Thr Leu Ser Leu Thr Cys Ala Val Tyr Gly Gly
Ser Phe 35 40 45Ser Gly Tyr Tyr Trp Ser Trp Ile Arg Gln Ser Pro Glu
Lys Gly Leu 50 55 60Glu Trp Ile Gly Glu Ile Asn His Gly Gly Tyr Val
Thr Tyr Asn Pro65 70 75 80Ser Leu Glu Ser Arg Val Thr Ile Ser Val
Asp Thr Ser Lys Asn Gln 85 90 95Phe Ser Leu Lys Leu Ser Ser Val Thr
Ala Ala Asp Thr Ala Val Tyr 100 105 110Tyr Cys Ala Arg Asp Tyr Gly
Pro 115 12024110PRTArtificial Sequencevariable light chain for
urelumab 24Met Glu Ala Pro Ala Gln Leu Leu Phe Leu Leu Leu Leu Trp
Leu Pro1 5 10 15Asp Thr Thr Gly Glu Ile Val Leu Thr Gln Ser Pro Ala
Thr Leu Ser 20 25 30Leu Ser Pro Gly Glu Arg Ala Thr Leu Ser Cys Arg
Ala Ser Gln Ser 35 40 45Val Ser Ser Tyr Leu Ala Trp Tyr Gln Gln Lys
Pro Gly Gln Ala Pro 50 55 60Arg Leu Leu Ile Tyr Asp Ala Ser Asn Arg
Ala Thr Gly Ile Pro Ala65 70 75 80Arg Phe Ser Gly Ser Gly Ser Gly
Thr Asp Phe Thr Leu Thr Ile Ser 85 90 95Ser Leu Glu Pro Glu Asp Phe
Ala Val Tyr Tyr Cys Gln Gln 100 105 110255PRTArtificial
Sequenceheavy chain CDR1 for urelumab 25Gly Tyr Tyr Trp Ser1
52616PRTArtificial Sequenceheavy chain CDR2 for urelumab 26Glu Ile
Asn His Gly Gly Tyr Val Thr Tyr Asn Pro Ser Leu Glu Ser1 5 10
152713PRTArtificial Sequenceheavy chain CDR3 for urelumab 27Asp Tyr
Gly Pro Gly Asn Tyr Asp Trp Tyr Phe Asp Leu1 5 102811PRTArtificial
Sequencelight chain CDR1 for urelumab 28Arg Ala Ser Gln Ser Val Ser
Ser Tyr Leu Ala1 5 10297PRTArtificial Sequencelight chain CDR2 for
urelumab 29Asp Ala Ser Asn Arg Ala Thr1 53011PRTArtificial
Sequencelight chain CDR3 for urelumab 30Gln Gln Arg Ser Asp Trp Pro
Pro Ala Leu Thr1 5 1031230PRTArtificial SequenceFc domain 31Lys Ser
Cys Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu1 5 10 15Leu
Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp 20 25
30Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp
35 40 45Val Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp
Gly 50 55 60Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln
Tyr Asn65 70 75 80Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu
His Gln Asp Trp 85 90 95Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser
Asn Lys Ala Leu Pro 100 105 110Ala Pro Ile Glu Lys Thr Ile Ser Lys
Ala Lys Gly Gln Pro Arg Glu 115 120 125Pro Gln Val Tyr Thr Leu Pro
Pro Ser Arg Glu Glu Met Thr Lys Asn 130 135 140Gln Val Ser Leu Thr
Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile145 150 155 160Ala Val
Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr 165 170
175Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys
180 185 190Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe
Ser Cys 195 200 205Ser Val Met His Glu Ala Leu His Asn His Tyr Thr
Gln Lys Ser Leu 210 215 220Ser Leu Ser Pro Gly Lys225
2303222PRTArtificial Sequencelinker 32Gly Gly Pro Gly Ser Ser Lys
Ser Cys Asp Lys Thr His Thr Cys Pro1 5 10 15Pro Cys Pro Ala Pro Glu
203322PRTArtificial Sequencelinker 33Gly Gly Ser Gly Ser Ser Lys
Ser Cys Asp Lys Thr His Thr Cys Pro1 5 10 15Pro Cys Pro Ala Pro Glu
203427PRTArtificial Sequencelinker 34Gly Gly Pro Gly Ser Ser Ser
Ser Ser Ser Ser Lys Ser Cys Asp Lys1 5 10 15Thr His Thr Cys Pro Pro
Cys Pro Ala Pro Glu 20 253527PRTArtificial Sequencelinker 35Gly Gly
Ser Gly Ser Ser Ser Ser Ser Ser Ser Lys Ser Cys Asp Lys1 5 10 15Thr
His Thr Cys Pro Pro Cys Pro Ala Pro Glu 20 253629PRTArtificial
Sequencelinker 36Gly Gly Pro Gly Ser Ser Ser Ser Ser Ser Ser Ser
Ser Lys Ser Cys1 5 10 15Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala
Pro Glu 20 253729PRTArtificial Sequencelinker 37Gly Gly Ser Gly Ser
Ser Ser Ser Ser Ser Ser Ser Ser Lys Ser Cys1 5 10 15Asp Lys Thr His
Thr Cys Pro Pro Cys Pro Ala Pro Glu 20 253824PRTArtificial
Sequencelinker 38Gly Gly Pro Gly Ser Ser Gly Ser Gly Ser Ser Asp
Lys Thr His Thr1 5 10 15Cys Pro Pro Cys Pro Ala Pro Glu
203923PRTArtificial Sequencelinker 39Gly Gly Pro Gly Ser Ser Gly
Ser Gly Ser Asp Lys Thr His Thr Cys1 5 10 15Pro Pro Cys Pro Ala Pro
Glu 204021PRTArtificial Sequencelinker 40Gly Gly Pro Ser Ser Ser
Gly Ser Asp Lys Thr His Thr Cys Pro Pro1 5 10 15Cys Pro Ala Pro Glu
204125PRTArtificial Sequencelinker 41Gly Gly Ser Ser Ser Ser Ser
Ser Ser Ser Gly Ser Asp Lys Thr His1 5 10 15Thr Cys Pro Pro Cys Pro
Ala Pro Glu 20 2542246PRTArtificial SequenceFc domain 42Met Glu Thr
Asp Thr Leu Leu Leu Trp Val Leu Leu Leu Trp Val Pro1 5 10 15Ala Gly
Asn Gly Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro 20 25 30Glu
Leu Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys 35 40
45Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val
50 55 60Asp Val Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val
Asp65 70 75 80Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu
Glu Gln Tyr 85 90 95Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val
Leu His Gln Asp 100 105 110Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys
Val Ser Asn Lys Ala Leu 115 120 125Pro Ala Pro Ile Glu Lys Thr Ile
Ser Lys Ala Lys Gly Gln Pro Arg 130 135 140Glu Pro Gln Val Tyr Thr
Leu Pro Pro Ser Arg Glu Glu Met Thr Lys145 150 155 160Asn Gln Val
Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp 165 170 175Ile
Ala Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys 180 185
190Thr Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser
195 200 205Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val
Phe Ser 210 215 220Cys Ser Val Met His Glu Ala Leu His Asn His Tyr
Thr Gln Lys Ser225 230 235 240Leu Ser Leu Ser Pro Gly
2454311PRTArtificial Sequencelinker 43Ser Gly Ser Gly Ser Gly Ser
Gly Ser Gly Ser1 5 104412PRTArtificial Sequencelinker 44Ser Ser Ser
Ser Ser Ser Gly Ser Gly Ser Gly Ser1 5 104516PRTArtificial
Sequencelinker 45Ser Ser Ser Ser Ser Ser Gly Ser Gly Ser Gly Ser
Gly Ser Gly Ser1 5 10 1546254PRTArtificial Sequence4-1BBL 46Met Glu
Tyr Ala Ser Asp Ala Ser Leu Asp Pro Glu Ala Pro Trp Pro1 5 10 15Pro
Ala Pro Arg Ala Arg Ala Cys Arg Val Leu Pro Trp Ala Leu
Val 20 25 30Ala Gly Leu Leu Leu Leu Leu Leu Leu Ala Ala Ala Cys Ala
Val Phe 35 40 45Leu Ala Cys Pro Trp Ala Val Ser Gly Ala Arg Ala Ser
Pro Gly Ser 50 55 60Ala Ala Ser Pro Arg Leu Arg Glu Gly Pro Glu Leu
Ser Pro Asp Asp65 70 75 80Pro Ala Gly Leu Leu Asp Leu Arg Gln Gly
Met Phe Ala Gln Leu Val 85 90 95Ala Gln Asn Val Leu Leu Ile Asp Gly
Pro Leu Ser Trp Tyr Ser Asp 100 105 110Pro Gly Leu Ala Gly Val Ser
Leu Thr Gly Gly Leu Ser Tyr Lys Glu 115 120 125Asp Thr Lys Glu Leu
Val Val Ala Lys Ala Gly Val Tyr Tyr Val Phe 130 135 140Phe Gln Leu
Glu Leu Arg Arg Val Val Ala Gly Glu Gly Ser Gly Ser145 150 155
160Val Ser Leu Ala Leu His Leu Gln Pro Leu Arg Ser Ala Ala Gly Ala
165 170 175Ala Ala Leu Ala Leu Thr Val Asp Leu Pro Pro Ala Ser Ser
Glu Ala 180 185 190Arg Asn Ser Ala Phe Gly Phe Gln Gly Arg Leu Leu
His Leu Ser Ala 195 200 205Gly Gln Arg Leu Gly Val His Leu His Thr
Glu Ala Arg Ala Arg His 210 215 220Ala Trp Gln Leu Thr Gln Gly Ala
Thr Val Leu Gly Leu Phe Arg Val225 230 235 240Thr Pro Glu Ile Pro
Ala Gly Leu Pro Ser Pro Arg Ser Glu 245 25047168PRTArtificial
Sequence4-1BBL soluble domain 47Leu Arg Gln Gly Met Phe Ala Gln Leu
Val Ala Gln Asn Val Leu Leu1 5 10 15Ile Asp Gly Pro Leu Ser Trp Tyr
Ser Asp Pro Gly Leu Ala Gly Val 20 25 30Ser Leu Thr Gly Gly Leu Ser
Tyr Lys Glu Asp Thr Lys Glu Leu Val 35 40 45Val Ala Lys Ala Gly Val
Tyr Tyr Val Phe Phe Gln Leu Glu Leu Arg 50 55 60Arg Val Val Ala Gly
Glu Gly Ser Gly Ser Val Ser Leu Ala Leu His65 70 75 80Leu Gln Pro
Leu Arg Ser Ala Ala Gly Ala Ala Ala Leu Ala Leu Thr 85 90 95Val Asp
Leu Pro Pro Ala Ser Ser Glu Ala Arg Asn Ser Ala Phe Gly 100 105
110Phe Gln Gly Arg Leu Leu His Leu Ser Ala Gly Gln Arg Leu Gly Val
115 120 125His Leu His Thr Glu Ala Arg Ala Arg His Ala Trp Gln Leu
Thr Gln 130 135 140Gly Ala Thr Val Leu Gly Leu Phe Arg Val Thr Pro
Glu Ile Pro Ala145 150 155 160Gly Leu Pro Ser Pro Arg Ser Glu
16548118PRTArtificial Sequencevariable heavy chain for 4B4-1-1
version 1 48Gln Val Gln Leu Gln Gln Pro Gly Ala Glu Leu Val Lys Pro
Gly Ala1 5 10 15Ser Val Lys Leu Ser Cys Lys Ala Ser Gly Tyr Thr Phe
Ser Ser Tyr 20 25 30Trp Met His Trp Val Lys Gln Arg Pro Gly Gln Val
Leu Glu Trp Ile 35 40 45Gly Glu Ile Asn Pro Gly Asn Gly His Thr Asn
Tyr Asn Glu Lys Phe 50 55 60Lys Ser Lys Ala Thr Leu Thr Val Asp Lys
Ser Ser Ser Thr Ala Tyr65 70 75 80Met Gln Leu Ser Ser Leu Thr Ser
Glu Asp Ser Ala Val Tyr Tyr Cys 85 90 95Ala Arg Ser Phe Thr Thr Ala
Arg Gly Phe Ala Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val Thr Val
Ser 11549107PRTArtificial Sequencevariable light chain for 4B4-1-1
version 1 49Asp Ile Val Met Thr Gln Ser Pro Ala Thr Gln Ser Val Thr
Pro Gly1 5 10 15Asp Arg Val Ser Leu Ser Cys Arg Ala Ser Gln Thr Ile
Ser Asp Tyr 20 25 30Leu His Trp Tyr Gln Gln Lys Ser His Glu Ser Pro
Arg Leu Leu Ile 35 40 45Lys Tyr Ala Ser Gln Ser Ile Ser Gly Ile Pro
Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Ser Asp Phe Thr Leu Ser
Ile Asn Ser Val Glu Pro65 70 75 80Glu Asp Val Gly Val Tyr Tyr Cys
Gln Asp Gly His Ser Phe Pro Pro 85 90 95Thr Phe Gly Gly Gly Thr Lys
Leu Glu Ile Lys 100 10550119PRTArtificial Sequencevariable heavy
chain for 4B4-1-1 version 2 50Gln Val Gln Leu Gln Gln Pro Gly Ala
Glu Leu Val Lys Pro Gly Ala1 5 10 15Ser Val Lys Leu Ser Cys Lys Ala
Ser Gly Tyr Thr Phe Ser Ser Tyr 20 25 30Trp Met His Trp Val Lys Gln
Arg Pro Gly Gln Val Leu Glu Trp Ile 35 40 45Gly Glu Ile Asn Pro Gly
Asn Gly His Thr Asn Tyr Asn Glu Lys Phe 50 55 60Lys Ser Lys Ala Thr
Leu Thr Val Asp Lys Ser Ser Ser Thr Ala Tyr65 70 75 80Met Gln Leu
Ser Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Tyr Cys 85 90 95Ala Arg
Ser Phe Thr Thr Ala Arg Gly Phe Ala Tyr Trp Gly Gln Gly 100 105
110Thr Leu Val Thr Val Ser Ala 11551108PRTArtificial
Sequencevariable light chain for 4B4-1-1 version 2 51Asp Ile Val
Met Thr Gln Ser Pro Ala Thr Gln Ser Val Thr Pro Gly1 5 10 15Asp Arg
Val Ser Leu Ser Cys Arg Ala Ser Gln Thr Ile Ser Asp Tyr 20 25 30Leu
His Trp Tyr Gln Gln Lys Ser His Glu Ser Pro Arg Leu Leu Ile 35 40
45Lys Tyr Ala Ser Gln Ser Ile Ser Gly Ile Pro Ser Arg Phe Ser Gly
50 55 60Ser Gly Ser Gly Ser Asp Phe Thr Leu Ser Ile Asn Ser Val Glu
Pro65 70 75 80Glu Asp Val Gly Val Tyr Tyr Cys Gln Asp Gly His Ser
Phe Pro Pro 85 90 95Thr Phe Gly Gly Gly Thr Lys Leu Glu Ile Lys Arg
100 10552120PRTArtificial Sequencevariable heavy chain for H39E3-2
52Met Asp Trp Thr Trp Arg Ile Leu Phe Leu Val Ala Ala Ala Thr Gly1
5 10 15Ala His Ser Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val
Gln 20 25 30Pro Gly Gly Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe
Thr Phe 35 40 45Ser Asp Tyr Trp Met Ser Trp Val Arg Gln Ala Pro Gly
Lys Gly Leu 50 55 60Glu Trp Val Ala Asp Ile Lys Asn Asp Gly Ser Tyr
Thr Asn Tyr Ala65 70 75 80Pro Ser Leu Thr Asn Arg Phe Thr Ile Ser
Arg Asp Asn Ala Lys Asn 85 90 95Ser Leu Tyr Leu Gln Met Asn Ser Leu
Arg Ala Glu Asp Thr Ala Val 100 105 110Tyr Tyr Cys Ala Arg Glu Leu
Thr 115 12053109PRTArtificial Sequencevariable light chain for
H39E3-2 53Met Glu Ala Pro Ala Gln Leu Leu Phe Leu Leu Leu Leu Trp
Leu Pro1 5 10 15Asp Thr Thr Gly Asp Ile Val Met Thr Gln Ser Pro Asp
Ser Leu Ala 20 25 30Val Ser Leu Gly Glu Arg Ala Thr Ile Asn Cys Lys
Ser Ser Gln Ser 35 40 45Leu Leu Ser Ser Gly Asn Gln Lys Asn Tyr Leu
Trp Tyr Gln Gln Lys 50 55 60Pro Gly Gln Pro Pro Lys Leu Leu Ile Tyr
Tyr Ala Ser Thr Arg Gln65 70 75 80Ser Gly Val Pro Asp Arg Phe Ser
Gly Ser Gly Ser Gly Thr Asp Phe 85 90 95Thr Leu Thr Ile Ser Ser Leu
Gln Ala Glu Asp Val Ala 100 10554277PRTHomo Sapiens 54Met Cys Val
Gly Ala Arg Arg Leu Gly Arg Gly Pro Cys Ala Ala Leu1 5 10 15Leu Leu
Leu Gly Leu Gly Leu Ser Thr Val Thr Gly Leu His Cys Val 20 25 30Gly
Asp Thr Tyr Pro Ser Asn Asp Arg Cys Cys His Glu Cys Arg Pro 35 40
45Gly Asn Gly Met Val Ser Arg Cys Ser Arg Ser Gln Asn Thr Val Cys
50 55 60Arg Pro Cys Gly Pro Gly Phe Tyr Asn Asp Val Val Ser Ser Lys
Pro65 70 75 80Cys Lys Pro Cys Thr Trp Cys Asn Leu Arg Ser Gly Ser
Glu Arg Lys 85 90 95Gln Leu Cys Thr Ala Thr Gln Asp Thr Val Cys Arg
Cys Arg Ala Gly 100 105 110Thr Gln Pro Leu Asp Ser Tyr Lys Pro Gly
Val Asp Cys Ala Pro Cys 115 120 125Pro Pro Gly His Phe Ser Pro Gly
Asp Asn Gln Ala Cys Lys Pro Trp 130 135 140Thr Asn Cys Thr Leu Ala
Gly Lys His Thr Leu Gln Pro Ala Ser Asn145 150 155 160Ser Ser Asp
Ala Ile Cys Glu Asp Arg Asp Pro Pro Ala Thr Gln Pro 165 170 175Gln
Glu Thr Gln Gly Pro Pro Ala Arg Pro Ile Thr Val Gln Pro Thr 180 185
190Glu Ala Trp Pro Arg Thr Ser Gln Gly Pro Ser Thr Arg Pro Val Glu
195 200 205Val Pro Gly Gly Arg Ala Val Ala Ala Ile Leu Gly Leu Gly
Leu Val 210 215 220Leu Gly Leu Leu Gly Pro Leu Ala Ile Leu Leu Ala
Leu Tyr Leu Leu225 230 235 240Arg Arg Asp Gln Arg Leu Pro Pro Asp
Ala His Lys Pro Pro Gly Gly 245 250 255Gly Ser Phe Arg Thr Pro Ile
Gln Glu Glu Gln Ala Asp Ala His Ser 260 265 270Thr Leu Ala Lys Ile
27555272PRTMus Musculus 55Met Tyr Val Trp Val Gln Gln Pro Thr Ala
Leu Leu Leu Leu Gly Leu1 5 10 15Thr Leu Gly Val Thr Ala Arg Arg Leu
Asn Cys Val Lys His Thr Tyr 20 25 30Pro Ser Gly His Lys Cys Cys Arg
Glu Cys Gln Pro Gly His Gly Met 35 40 45Val Ser Arg Cys Asp His Thr
Arg Asp Thr Leu Cys His Pro Cys Glu 50 55 60Thr Gly Phe Tyr Asn Glu
Ala Val Asn Tyr Asp Thr Cys Lys Gln Cys65 70 75 80Thr Gln Cys Asn
His Arg Ser Gly Ser Glu Leu Lys Gln Asn Cys Thr 85 90 95Pro Thr Gln
Asp Thr Val Cys Arg Cys Arg Pro Gly Thr Gln Pro Arg 100 105 110Gln
Asp Ser Gly Tyr Lys Leu Gly Val Asp Cys Val Pro Cys Pro Pro 115 120
125Gly His Phe Ser Pro Gly Asn Asn Gln Ala Cys Lys Pro Trp Thr Asn
130 135 140Cys Thr Leu Ser Gly Lys Gln Thr Arg His Pro Ala Ser Asp
Ser Leu145 150 155 160Asp Ala Val Cys Glu Asp Arg Ser Leu Leu Ala
Thr Leu Leu Trp Glu 165 170 175Thr Gln Arg Pro Thr Phe Arg Pro Thr
Thr Val Gln Ser Thr Thr Val 180 185 190Trp Pro Arg Thr Ser Glu Leu
Pro Ser Pro Pro Thr Leu Val Thr Pro 195 200 205Glu Gly Pro Ala Phe
Ala Val Leu Leu Gly Leu Gly Leu Gly Leu Leu 210 215 220Ala Pro Leu
Thr Val Leu Leu Ala Leu Tyr Leu Leu Arg Lys Ala Trp225 230 235
240Arg Leu Pro Asn Thr Pro Lys Pro Cys Trp Gly Asn Ser Phe Arg Thr
245 250 255Pro Ile Gln Glu Glu His Thr Asp Ala His Phe Thr Leu Ala
Lys Ile 260 265 27056451PRTArtificial Sequenceheavy chain for
tavolixizumab 56Gln Val Gln Leu Gln Glu Ser Gly Pro Gly Leu Val Lys
Pro Ser Gln1 5 10 15Thr Leu Ser Leu Thr Cys Ala Val Tyr Gly Gly Ser
Phe Ser Ser Gly 20 25 30Tyr Trp Asn Trp Ile Arg Lys His Pro Gly Lys
Gly Leu Glu Tyr Ile 35 40 45Gly Tyr Ile Ser Tyr Asn Gly Ile Thr Tyr
His Asn Pro Ser Leu Lys 50 55 60Ser Arg Ile Thr Ile Asn Arg Asp Thr
Ser Lys Asn Gln Tyr Ser Leu65 70 75 80Gln Leu Asn Ser Val Thr Pro
Glu Asp Thr Ala Val Tyr Tyr Cys Ala 85 90 95Arg Tyr Lys Tyr Asp Tyr
Asp Gly Gly His Ala Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr Leu
Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe
Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135
140Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr
Val145 150 155 160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His
Thr Phe Pro Ala 165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu
Ser Ser Val Val Thr Val 180 185 190Pro Ser Ser Ser Leu Gly Thr Gln
Thr Tyr Ile Cys Asn Val Asn His 195 200 205Lys Pro Ser Asn Thr Lys
Val Asp Lys Arg Val Glu Pro Lys Ser Cys 210 215 220Asp Lys Thr His
Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly225 230 235 240Gly
Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 245 250
255Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His
260 265 270Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val
Glu Val 275 280 285His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr
Asn Ser Thr Tyr 290 295 300Arg Val Val Ser Val Leu Thr Val Leu His
Gln Asp Trp Leu Asn Gly305 310 315 320Lys Glu Tyr Lys Cys Lys Val
Ser Asn Lys Ala Leu Pro Ala Pro Ile 325 330 335Glu Lys Thr Ile Ser
Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val 340 345 350Tyr Thr Leu
Pro Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser 355 360 365Leu
Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu 370 375
380Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro
Pro385 390 395 400Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser
Lys Leu Thr Val 405 410 415Asp Lys Ser Arg Trp Gln Gln Gly Asn Val
Phe Ser Cys Ser Val Met 420 425 430His Glu Ala Leu His Asn His Tyr
Thr Gln Lys Ser Leu Ser Leu Ser 435 440 445Pro Gly Lys
45057214PRTArtificial Sequencelight chain for tavolixizumab 57Asp
Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10
15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Asp Ile Ser Asn Tyr
20 25 30Leu Asn Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu
Ile 35 40 45Tyr Tyr Thr Ser Lys Leu His Ser Gly Val Pro Ser Arg Phe
Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Tyr Thr Leu Thr Ile Ser Ser
Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Gly
Ser Ala Leu Pro Trp 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile
Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro
Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys
Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp
Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu
Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170
175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr
180 185 190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr
Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys 21058118PRTArtificial
Sequenceheavy chain variable region for tavolixizumab 58Gln Val Gln
Leu Gln Glu Ser Gly Pro Gly Leu Val Lys Pro Ser Gln1 5 10 15Thr Leu
Ser Leu Thr Cys Ala Val Tyr Gly Gly Ser Phe Ser Ser Gly 20 25 30Tyr
Trp Asn Trp Ile Arg Lys His Pro Gly Lys Gly Leu Glu Tyr Ile 35 40
45Gly Tyr Ile Ser Tyr Asn Gly Ile Thr Tyr His Asn Pro Ser Leu Lys
50 55 60Ser Arg Ile Thr Ile Asn Arg Asp Thr Ser Lys Asn Gln Tyr Ser
Leu65 70 75 80Gln Leu Asn Ser Val Thr Pro Glu Asp Thr Ala Val Tyr
Tyr Cys Ala 85 90 95Arg Tyr Lys Tyr Asp Tyr Asp Gly Gly His Ala
Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr
11559108PRTArtificial Sequencelight chain variable region for
tavolixizumab 59Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala
Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Asp
Ile Ser Asn Tyr 20 25 30Leu Asn Trp Tyr Gln Gln Lys Pro Gly Lys Ala
Pro Lys Leu Leu Ile 35 40 45Tyr Tyr Thr Ser Lys Leu His Ser Gly Val
Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Tyr Thr Leu
Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr
Cys Gln Gln Gly Ser Ala Leu Pro Trp 85 90 95Thr Phe Gly Gln Gly Thr
Lys Val Glu Ile Lys Arg 100 105609PRTArtificial Sequenceheavy chain
CDR1 for tavolixizumab 60Gly Ser Phe Ser Ser Gly Tyr Trp Asn1
56113PRTArtificial Sequenceheavy chain CDR2 for tavolixizumab 61Tyr
Ile Gly Tyr Ile Ser Tyr Asn Gly Ile Thr Tyr His1 5
106214PRTArtificial Sequenceheavy chain CDR3 for tavolixizumab
62Arg Tyr Lys Tyr Asp Tyr Asp Gly Gly His Ala Met Asp Tyr1 5
10638PRTArtificial Sequencelight chain CDR1 for tavolixizumab 63Gln
Asp Ile Ser Asn Tyr Leu Asn1 56411PRTArtificial Sequencelight chain
CDR2 for tavolixizumab 64Leu Leu Ile Tyr Tyr Thr Ser Lys Leu His
Ser1 5 10658PRTArtificial Sequencelight chain CDR3 for
tavolixizumab 65Gln Gln Gly Ser Ala Leu Pro Trp1
566444PRTArtificial Sequenceheavy chain for 11D4 66Glu Val Gln Leu
Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg
Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30Ser Met
Asn Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ser
Tyr Ile Ser Ser Ser Ser Ser Thr Ile Asp Tyr Ala Asp Ser Val 50 55
60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn Ser Leu Tyr65
70 75 80Leu Gln Met Asn Ser Leu Arg Asp Glu Asp Thr Ala Val Tyr Tyr
Cys 85 90 95Ala Arg Glu Ser Gly Trp Tyr Leu Phe Asp Tyr Trp Gly Gln
Gly Thr 100 105 110Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro
Ser Val Phe Pro 115 120 125Leu Ala Pro Cys Ser Arg Ser Thr Ser Glu
Ser Thr Ala Ala Leu Gly 130 135 140Cys Leu Val Lys Asp Tyr Phe Pro
Glu Pro Val Thr Val Ser Trp Asn145 150 155 160Ser Gly Ala Leu Thr
Ser Gly Val His Thr Phe Pro Ala Val Leu Gln 165 170 175Ser Ser Gly
Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser Ser 180 185 190Asn
Phe Gly Thr Gln Thr Tyr Thr Cys Asn Val Asp His Lys Pro Ser 195 200
205Asn Thr Lys Val Asp Lys Thr Val Glu Arg Lys Cys Cys Val Glu Cys
210 215 220Pro Pro Cys Pro Ala Pro Pro Val Ala Gly Pro Ser Val Phe
Leu Phe225 230 235 240Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser
Arg Thr Pro Glu Val 245 250 255Thr Cys Val Val Val Asp Val Ser His
Glu Asp Pro Glu Val Gln Phe 260 265 270Asn Trp Tyr Val Asp Gly Val
Glu Val His Asn Ala Lys Thr Lys Pro 275 280 285Arg Glu Glu Gln Phe
Asn Ser Thr Phe Arg Val Val Ser Val Leu Thr 290 295 300Val Val His
Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val305 310 315
320Ser Asn Lys Gly Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Thr
325 330 335Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro
Ser Arg 340 345 350Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys
Leu Val Lys Gly 355 360 365Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp
Glu Ser Asn Gly Gln Pro 370 375 380Glu Asn Asn Tyr Lys Thr Thr Pro
Pro Met Leu Asp Ser Asp Gly Ser385 390 395 400Phe Phe Leu Tyr Ser
Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln 405 410 415Gly Asn Val
Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn His 420 425 430Tyr
Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 435
44067214PRTArtificial Sequencelight chain for 11D4 67Asp Ile Gln
Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg
Val Thr Ile Thr Cys Arg Ala Ser Gln Gly Ile Ser Ser Trp 20 25 30Leu
Ala Trp Tyr Gln Gln Lys Pro Glu Lys Ala Pro Lys Ser Leu Ile 35 40
45Tyr Ala Ala Ser Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly
50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln
Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Tyr Asn Ser
Tyr Pro Pro 85 90 95Thr Phe Gly Gly Gly Thr Lys Val Glu Ile Lys Arg
Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp
Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu Leu
Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys Val
Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser Val
Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser
Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185
190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser
195 200 205Phe Asn Arg Gly Glu Cys 21068118PRTArtificial
Sequenceheavy chain variable region for 11D4 68Glu Val Gln Leu Val
Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu
Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30Ser Met Asn
Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ser Tyr
Ile Ser Ser Ser Ser Ser Thr Ile Asp Tyr Ala Asp Ser Val 50 55 60Lys
Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn Ser Leu Tyr65 70 75
80Leu Gln Met Asn Ser Leu Arg Asp Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Arg Glu Ser Gly Trp Tyr Leu Phe Asp Tyr Trp Gly Gln Gly
Thr 100 105 110Leu Val Thr Val Ser Ser 11569107PRTArtificial
Sequencelight chain variable region for 11D4 69Asp Ile Gln Met Thr
Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr
Ile Thr Cys Arg Ala Ser Gln Gly Ile Ser Ser Trp 20 25 30Leu Ala Trp
Tyr Gln Gln Lys Pro Glu Lys Ala Pro Lys Ser Leu Ile 35 40 45Tyr Ala
Ala Ser Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser
Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75
80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Tyr Asn Ser Tyr Pro Pro
85 90 95Thr Phe Gly Gly Gly Thr Lys Val Glu Ile Lys 100
105705PRTArtificial Sequenceheavy chain CDR1 for 11D4 70Ser Tyr Ser
Met Asn1 57117PRTArtificial Sequenceheavy chain CDR2 for 11D4 71Tyr
Ile Ser Ser Ser Ser Ser Thr Ile Asp Tyr Ala Asp Ser Val Lys1 5 10
15Gly729PRTArtificial Sequenceheavy chain CDR3 for 11D4 72Glu Ser
Gly Trp Tyr Leu Phe Asp Tyr1 57311PRTArtificial Sequencelight chain
CDR1 for 11D4 73Arg Ala Ser Gln Gly Ile Ser Ser Trp Leu Ala1 5
10747PRTArtificial Sequencelight chain CDR2 for 11D4 74Ala Ala Ser
Ser Leu Gln Ser1 5759PRTArtificial Sequencelight chain CDR3 for
11D4 75Gln Gln Tyr Asn Ser Tyr Pro Pro Thr1 576450PRTArtificial
Sequenceheavy chain for 18D8 76Glu Val Gln Leu Val Glu Ser Gly Gly
Gly Leu Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala
Ser Gly Phe Thr Phe Asp Asp Tyr 20 25 30Ala Met His Trp Val Arg Gln
Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ser Gly Ile Ser Trp Asn
Ser Gly Ser Ile Gly Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr
Ile Ser Arg Asp Asn Ala Lys Asn Ser Leu Tyr65 70 75 80Leu Gln Met
Asn Ser Leu Arg Ala Glu Asp Thr Ala Leu Tyr Tyr Cys 85 90 95Ala Lys
Asp Gln Ser Thr Ala Asp Tyr Tyr Phe Tyr Tyr Gly Met Asp 100 105
110Val Trp Gly Gln Gly Thr Thr Val Thr Val Ser Ser Ala Ser Thr Lys
115 120 125Gly Pro Ser Val Phe Pro Leu Ala Pro Cys Ser Arg Ser Thr
Ser Glu 130 135 140Ser Thr Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr
Phe Pro Glu Pro145 150 155 160Val Thr Val Ser Trp Asn Ser Gly Ala
Leu Thr Ser Gly Val His Thr 165 170 175Phe Pro Ala Val Leu Gln Ser
Ser Gly Leu Tyr Ser Leu Ser Ser Val 180 185 190Val Thr Val Pro Ser
Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys Asn 195 200 205Val Asp His
Lys Pro Ser Asn Thr Lys Val Asp Lys Thr Val Glu Arg 210 215 220Lys
Cys Cys Val Glu Cys Pro Pro Cys Pro Ala Pro Pro Val Ala Gly225 230
235 240Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met
Ile 245 250 255Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val
Ser His Glu 260 265 270Asp Pro Glu Val Gln Phe Asn Trp Tyr Val Asp
Gly Val Glu Val His 275 280 285Asn Ala Lys Thr Lys Pro Arg Glu Glu
Gln Phe Asn Ser Thr Phe Arg 290 295 300Val Val Ser Val Leu Thr Val
Val His Gln Asp Trp Leu Asn Gly Lys305 310 315 320Glu Tyr Lys Cys
Lys Val Ser Asn Lys Gly Leu Pro Ala Pro Ile Glu 325 330 335Lys Thr
Ile Ser Lys Thr Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr 340 345
350Thr Leu Pro Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser Leu
355 360 365Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val
Glu Trp 370 375 380Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr
Thr Pro Pro Met385 390 395 400Leu Asp Ser Asp Gly Ser Phe Phe Leu
Tyr Ser Lys Leu Thr Val Asp 405 410 415Lys Ser Arg Trp Gln Gln Gly
Asn Val Phe Ser Cys Ser Val Met His 420 425 430Glu Ala Leu His Asn
His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro 435 440 445Gly Lys
45077213PRTArtificial Sequencelight chain for 18D8 77Glu Ile Val
Val Thr Gln Ser Pro Ala Thr Leu Ser Leu Ser Pro Gly1 5 10 15Glu Arg
Ala Thr Leu Ser Cys Arg Ala Ser Gln Ser Val Ser Ser Tyr 20 25 30Leu
Ala Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro Arg Leu Leu Ile 35 40
45Tyr Asp Ala Ser Asn Arg Ala Thr Gly Ile Pro Ala Arg Phe Ser Gly
50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Glu
Pro65 70 75 80Glu Asp Phe Ala Val Tyr Tyr Cys Gln Gln Arg Ser Asn
Trp Pro Thr 85 90 95Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg Thr
Val Ala Ala Pro 100 105 110Ser Val Phe Ile Phe Pro Pro Ser Asp Glu
Gln Leu Lys Ser Gly Thr 115 120 125Ala Ser Val Val Cys Leu Leu Asn
Asn Phe Tyr Pro Arg Glu Ala Lys 130 135 140Val Gln Trp Lys Val Asp
Asn Ala Leu Gln Ser Gly Asn Ser Gln Glu145 150 155 160Ser Val Thr
Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser Ser 165 170 175Thr
Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr Ala 180 185
190Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser Phe
195 200 205Asn Arg Gly Glu Cys 21078124PRTArtificial Sequenceheavy
chain variable region for 18D8 78Glu Val Gln Leu Val Glu Ser Gly
Gly Gly Leu Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala
Ala Ser Gly Phe Thr Phe Asp Asp Tyr 20 25 30Ala Met His Trp Val Arg
Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ser Gly Ile Ser Trp
Asn Ser Gly Ser Ile Gly Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe
Thr Ile Ser Arg Asp Asn Ala Lys Asn Ser Leu Tyr65 70 75 80Leu Gln
Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Leu Tyr Tyr Cys 85 90 95Ala
Lys Asp Gln Ser Thr Ala Asp Tyr Tyr Phe Tyr Tyr Gly Met Asp 100 105
110Val Trp Gly Gln Gly Thr Thr Val Thr Val Ser Ser 115
12079106PRTArtificial Sequencelight chain variable region for 18D8
79Glu Ile Val Val Thr Gln Ser Pro Ala Thr Leu Ser Leu Ser Pro Gly1
5 10 15Glu Arg Ala Thr Leu Ser Cys Arg Ala Ser Gln Ser Val Ser Ser
Tyr 20 25 30Leu Ala Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro Arg Leu
Leu Ile 35 40 45Tyr Asp Ala Ser Asn Arg Ala Thr Gly Ile Pro Ala Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Glu Pro65 70 75 80Glu Asp Phe Ala Val Tyr Tyr Cys Gln Gln
Arg Ser Asn Trp Pro Thr 85 90 95Phe Gly Gln Gly Thr Lys Val Glu Ile
Lys 100 105805PRTArtificial Sequenceheavy chain CDR1 for 18D8 80Asp
Tyr Ala Met His1 58117PRTArtificial Sequenceheavy chain CDR2 for
18D8 81Gly Ile Ser Trp Asn Ser Gly Ser Ile Gly Tyr Ala Asp Ser Val
Lys1 5 10 15Gly8215PRTArtificial Sequenceheavy chain CDR3 for 18D8
82Asp Gln Ser Thr Ala Asp Tyr Tyr Phe Tyr Tyr Gly Met Asp Val1 5 10
158311PRTArtificial Sequencelight chain CDR1 for 18D8 83Arg Ala Ser
Gln Ser Val Ser Ser Tyr Leu Ala1 5 10847PRTArtificial Sequencelight
chain CDR2 for 18D8 84Asp Ala Ser Asn Arg Ala Thr1
5858PRTArtificial Sequencelight chain CDR3 for 18D8 85Gln Gln Arg
Ser Asn Trp Pro Thr1 586120PRTArtificial Sequenceheavy chain
variable region for Hu119-122 86Glu Val Gln Leu Val Glu Ser Gly Gly
Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala
Ser Glu Tyr Glu Phe Pro Ser His 20 25 30Asp Met Ser Trp Val Arg Gln
Ala Pro Gly Lys Gly Leu Glu Leu Val 35 40 45Ala Ala Ile Asn Ser Asp
Gly Gly Ser Thr Tyr Tyr Pro Asp Thr Met 50 55 60Glu Arg Arg Phe Thr
Ile Ser Arg Asp Asn Ala Lys Asn Ser Leu Tyr65 70 75 80Leu Gln Met
Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg
His Tyr Asp Asp Tyr Tyr Ala Trp Phe Ala Tyr Trp Gly Gln 100 105
110Gly Thr Met Val Thr Val Ser Ser 115 12087111PRTArtificial
Sequencelight chain variable region for Hu119-122 87Glu Ile Val Leu
Thr Gln Ser Pro Ala Thr Leu Ser Leu Ser Pro Gly1 5 10 15Glu Arg Ala
Thr Leu Ser Cys Arg Ala Ser Lys Ser Val Ser Thr Ser 20 25 30Gly Tyr
Ser Tyr Met His Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro 35 40 45Arg
Leu Leu Ile Tyr Leu Ala Ser Asn Leu Glu Ser Gly Val Pro
Ala 50 55 60Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr
Ile Ser65 70 75 80Ser Leu Glu Pro Glu Asp Phe Ala Val Tyr Tyr Cys
Gln His Ser Arg 85 90 95Glu Leu Pro Leu Thr Phe Gly Gly Gly Thr Lys
Val Glu Ile Lys 100 105 110885PRTArtificial Sequenceheavy chain
CDRl for Hu119-122 88Ser His Asp Met Ser1 58917PRTArtificial
Sequenceheavy chain CDR2 for Hu119-122 89Ala Ile Asn Ser Asp Gly
Gly Ser Thr Tyr Tyr Pro Asp Thr Met Glu1 5 10
15Arg9011PRTArtificial Sequenceheavy chain CDR3 for Hu119-122 90His
Tyr Asp Asp Tyr Tyr Ala Trp Phe Ala Tyr1 5 109115PRTArtificial
Sequencelight chain CDR1 for Hu119-122 91Arg Ala Ser Lys Ser Val
Ser Thr Ser Gly Tyr Ser Tyr Met His1 5 10 15927PRTArtificial
Sequencelight chain CDR2 for Hu119-122 92Leu Ala Ser Asn Leu Glu
Ser1 5939PRTArtificial Sequencelight chain CDR3 for Hu119-122 93Gln
His Ser Arg Glu Leu Pro Leu Thr1 594122PRTArtificial Sequenceheavy
chain variable region for Hu106-222 94Gln Val Gln Leu Val Gln Ser
Gly Ser Glu Leu Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys
Lys Ala Ser Gly Tyr Thr Phe Thr Asp Tyr 20 25 30Ser Met His Trp Val
Arg Gln Ala Pro Gly Gln Gly Leu Lys Trp Met 35 40 45Gly Trp Ile Asn
Thr Glu Thr Gly Glu Pro Thr Tyr Ala Asp Asp Phe 50 55 60Lys Gly Arg
Phe Val Phe Ser Leu Asp Thr Ser Val Ser Thr Ala Tyr65 70 75 80Leu
Gln Ile Ser Ser Leu Lys Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90
95Ala Asn Pro Tyr Tyr Asp Tyr Val Ser Tyr Tyr Ala Met Asp Tyr Trp
100 105 110Gly Gln Gly Thr Thr Val Thr Val Ser Ser 115
12095107PRTArtificial Sequencelight chain variable region for
Hu106-222 95Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser
Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Lys Ala Ser Gln Asp Val
Ser Thr Ala 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro
Lys Leu Leu Ile 35 40 45Tyr Ser Ala Ser Tyr Leu Tyr Thr Gly Val Pro
Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Phe Thr
Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Ile Ala Thr Tyr Tyr Cys
Gln Gln His Tyr Ser Thr Pro Arg 85 90 95Thr Phe Gly Gln Gly Thr Lys
Leu Glu Ile Lys 100 105965PRTArtificial Sequenceheavy chain CDR1
for Hu106-222 96Asp Tyr Ser Met His1 59717PRTArtificial
Sequenceheavy chain CDR2 for Hu106-222 97Trp Ile Asn Thr Glu Thr
Gly Glu Pro Thr Tyr Ala Asp Asp Phe Lys1 5 10
15Gly9813PRTArtificial Sequenceheavy chain CDR3 for Hu106-222 98Pro
Tyr Tyr Asp Tyr Val Ser Tyr Tyr Ala Met Asp Tyr1 5
109911PRTArtificial Sequencelight chain CDR1 for Hu106-222 99Lys
Ala Ser Gln Asp Val Ser Thr Ala Val Ala1 5 101007PRTArtificial
Sequencelight chain CDR2 for Hu106-222 100Ser Ala Ser Tyr Leu Tyr
Thr1 51019PRTArtificial Sequencelight chain CDR3 for Hu106-222
101Gln Gln His Tyr Ser Thr Pro Arg Thr1 5102183PRTArtificial
SequenceOX40L 102Met Glu Arg Val Gln Pro Leu Glu Glu Asn Val Gly
Asn Ala Ala Arg1 5 10 15Pro Arg Phe Glu Arg Asn Lys Leu Leu Leu Val
Ala Ser Val Ile Gln 20 25 30Gly Leu Gly Leu Leu Leu Cys Phe Thr Tyr
Ile Cys Leu His Phe Ser 35 40 45Ala Leu Gln Val Ser His Arg Tyr Pro
Arg Ile Gln Ser Ile Lys Val 50 55 60Gln Phe Thr Glu Tyr Lys Lys Glu
Lys Gly Phe Ile Leu Thr Ser Gln65 70 75 80Lys Glu Asp Glu Ile Met
Lys Val Gln Asn Asn Ser Val Ile Ile Asn 85 90 95Cys Asp Gly Phe Tyr
Leu Ile Ser Leu Lys Gly Tyr Phe Ser Gln Glu 100 105 110Val Asn Ile
Ser Leu His Tyr Gln Lys Asp Glu Glu Pro Leu Phe Gln 115 120 125Leu
Lys Lys Val Arg Ser Val Asn Ser Leu Met Val Ala Ser Leu Thr 130 135
140Tyr Lys Asp Lys Val Tyr Leu Asn Val Thr Thr Asp Asn Thr Ser
Leu145 150 155 160Asp Asp Phe His Val Asn Gly Gly Glu Leu Ile Leu
Ile His Gln Asn 165 170 175Pro Gly Glu Phe Cys Val Leu
180103131PRTArtificial SequenceOX40L soluble domain 103Ser His Arg
Tyr Pro Arg Ile Gln Ser Ile Lys Val Gln Phe Thr Glu1 5 10 15Tyr Lys
Lys Glu Lys Gly Phe Ile Leu Thr Ser Gln Lys Glu Asp Glu 20 25 30Ile
Met Lys Val Gln Asn Asn Ser Val Ile Ile Asn Cys Asp Gly Phe 35 40
45Tyr Leu Ile Ser Leu Lys Gly Tyr Phe Ser Gln Glu Val Asn Ile Ser
50 55 60Leu His Tyr Gln Lys Asp Glu Glu Pro Leu Phe Gln Leu Lys Lys
Val65 70 75 80Arg Ser Val Asn Ser Leu Met Val Ala Ser Leu Thr Tyr
Lys Asp Lys 85 90 95Val Tyr Leu Asn Val Thr Thr Asp Asn Thr Ser Leu
Asp Asp Phe His 100 105 110Val Asn Gly Gly Glu Leu Ile Leu Ile His
Gln Asn Pro Gly Glu Phe 115 120 125Cys Val Leu
130104128PRTArtificial SequenceOX40L soluble domain (alternative)
104Tyr Pro Arg Ile Gln Ser Ile Lys Val Gln Phe Thr Glu Tyr Lys Lys1
5 10 15Glu Lys Gly Phe Ile Leu Thr Ser Gln Lys Glu Asp Glu Ile Met
Lys 20 25 30Val Gln Asn Asn Ser Val Ile Ile Asn Cys Asp Gly Phe Tyr
Leu Ile 35 40 45Ser Leu Lys Gly Tyr Phe Ser Gln Glu Val Asn Ile Ser
Leu His Tyr 50 55 60Gln Lys Asp Glu Glu Pro Leu Phe Gln Leu Lys Lys
Val Arg Ser Val65 70 75 80Asn Ser Leu Met Val Ala Ser Leu Thr Tyr
Lys Asp Lys Val Tyr Leu 85 90 95Asn Val Thr Thr Asp Asn Thr Ser Leu
Asp Asp Phe His Val Asn Gly 100 105 110Gly Glu Leu Ile Leu Ile His
Gln Asn Pro Gly Glu Phe Cys Val Leu 115 120 125105120PRTArtificial
Sequencevariable heavy chain for 008 105Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys
Ala Ala Ser Gly Phe Thr Phe Ser Asn Tyr 20 25 30Thr Met Asn Trp Val
Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ser Ala Ile Ser
Gly Ser Gly Gly Ser Thr Tyr Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg
Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr65 70 75 80Leu
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90
95Ala Lys Asp Arg Tyr Ser Gln Val His Tyr Ala Leu Asp Tyr Trp Gly
100 105 110Gln Gly Thr Leu Val Thr Val Ser 115
120106108PRTArtificial Sequencevariable light chain for 008 106Asp
Ile Val Met Thr Gln Ser Pro Asp Ser Leu Pro Val Thr Pro Gly1 5 10
15Glu Pro Ala Ser Ile Ser Cys Arg Ser Ser Gln Ser Leu Leu His Ser
20 25 30Asn Gly Tyr Asn Tyr Leu Asp Trp Tyr Leu Gln Lys Ala Gly Gln
Ser 35 40 45Pro Gln Leu Leu Ile Tyr Leu Gly Ser Asn Arg Ala Ser Gly
Val Pro 50 55 60Asp Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr
Leu Lys Ile65 70 75 80Ser Arg Val Glu Ala Glu Asp Val Gly Val Tyr
Tyr Cys Gln Gln Tyr 85 90 95Tyr Asn His Pro Thr Thr Phe Gly Gln Gly
Thr Lys 100 105107120PRTArtificial Sequencevariable heavy chain for
011 107Glu Val Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly
Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser
Asp Tyr 20 25 30Thr Met Asn Trp Val Arg Gln Ala Pro Gly Lys Gly Leu
Glu Trp Val 35 40 45Ser Ser Ile Ser Gly Gly Ser Thr Tyr Tyr Ala Asp
Ser Arg Lys Gly 50 55 60Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn
Thr Leu Tyr Leu Gln65 70 75 80Met Asn Asn Leu Arg Ala Glu Asp Thr
Ala Val Tyr Tyr Cys Ala Arg 85 90 95Asp Arg Tyr Phe Arg Gln Gln Asn
Ala Phe Asp Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val Thr Val Ser
Ser Ala 115 120108108PRTArtificial Sequencevariable light chain for
011 108Asp Ile Val Met Thr Gln Ser Pro Asp Ser Leu Pro Val Thr Pro
Gly1 5 10 15Glu Pro Ala Ser Ile Ser Cys Arg Ser Ser Gln Ser Leu Leu
His Ser 20 25 30Asn Gly Tyr Asn Tyr Leu Asp Trp Tyr Leu Gln Lys Ala
Gly Gln Ser 35 40 45Pro Gln Leu Leu Ile Tyr Leu Gly Ser Asn Arg Ala
Ser Gly Val Pro 50 55 60Asp Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp
Phe Thr Leu Lys Ile65 70 75 80Ser Arg Val Glu Ala Glu Asp Val Gly
Val Tyr Tyr Cys Gln Gln Tyr 85 90 95Tyr Asn His Pro Thr Thr Phe Gly
Gln Gly Thr Lys 100 105109120PRTArtificial Sequencevariable heavy
chain for 021 109Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val
Gln Pro Arg Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe
Thr Phe Ser Ser Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro Gly
Lys Gly Leu Glu Trp Val 35 40 45Ala Val Ile Ser Tyr Asp Gly Ser Asn
Lys Tyr Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg
Asp Asn Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu
Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Lys Asp Arg Tyr
Ile Thr Leu Pro Asn Ala Leu Asp Tyr Trp Gly 100 105 110Gln Gly Thr
Leu Val Thr Val Ser 115 120110108PRTArtificial Sequencevariable
light chain for 021 110Asp Ile Gln Met Thr Gln Ser Pro Val Ser Leu
Pro Val Thr Pro Gly1 5 10 15Glu Pro Ala Ser Ile Ser Cys Arg Ser Ser
Gln Ser Leu Leu His Ser 20 25 30Asn Gly Tyr Asn Tyr Leu Asp Trp Tyr
Leu Gln Lys Pro Gly Gln Ser 35 40 45Pro Gln Leu Leu Ile Tyr Leu Gly
Ser Asn Arg Ala Ser Gly Val Pro 50 55 60Asp Arg Phe Ser Gly Ser Gly
Ser Gly Thr Asp Phe Thr Leu Lys Ile65 70 75 80Ser Arg Val Glu Ala
Glu Asp Val Gly Val Tyr Tyr Cys Gln Gln Tyr 85 90 95Lys Ser Asn Pro
Pro Thr Phe Gly Gln Gly Thr Lys 100 105111120PRTArtificial
Sequencevariable heavy chain for 023 111Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val His Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys
Ala Gly Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30Ala Met His Trp Val
Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ser Ala Ile Gly
Thr Gly Gly Gly Thr Tyr Tyr Ala Asp Ser Val Met 50 55 60Gly Arg Phe
Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr Leu65 70 75 80Gln
Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys Ala 85 90
95Arg Tyr Asp Asn Val Met Gly Leu Tyr Trp Phe Asp Tyr Trp Gly Gln
100 105 110Gly Thr Leu Val Thr Val Ser Ser 115
120112108PRTArtificial Sequencevariable light chain for 023 112Glu
Ile Val Leu Thr Gln Ser Pro Ala Thr Leu Ser Leu Ser Pro Gly1 5 10
15Glu Arg Ala Thr Leu Ser Cys Arg Ala Ser Gln Ser Val Ser Ser Tyr
20 25 30Leu Ala Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro Arg Leu Leu
Ile 35 40 45Tyr Asp Ala Ser Asn Arg Ala Thr Gly Ile Pro Ala Arg Phe
Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser
Leu Glu Pro65 70 75 80Glu Asp Phe Ala Val Tyr Tyr Cys Gln Gln Arg
Ser Asn Trp Pro Pro 85 90 95Ala Phe Gly Gly Gly Thr Lys Val Glu Ile
Lys Arg 100 105113119PRTArtificial Sequenceheavy chain variable
region 113Glu Val Gln Leu Gln Gln Ser Gly Pro Glu Leu Val Lys Pro
Gly Ala1 5 10 15Ser Val Lys Met Ser Cys Lys Ala Ser Gly Tyr Thr Phe
Thr Ser Tyr 20 25 30Val Met His Trp Val Lys Gln Lys Pro Gly Gln Gly
Leu Glu Trp Ile 35 40 45Gly Tyr Ile Asn Pro Tyr Asn Asp Gly Thr Lys
Tyr Asn Glu Lys Phe 50 55 60Lys Gly Lys Ala Thr Leu Thr Ser Asp Lys
Ser Ser Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Thr Ser
Glu Asp Ser Ala Val Tyr Tyr Cys 85 90 95Ala Asn Tyr Tyr Gly Ser Ser
Leu Ser Met Asp Tyr Trp Gly Gln Gly 100 105 110Thr Ser Val Thr Val
Ser Ser 115114108PRTArtificial Sequencelight chain variable region
114Asp Ile Gln Met Thr Gln Thr Thr Ser Ser Leu Ser Ala Ser Leu Gly1
5 10 15Asp Arg Val Thr Ile Ser Cys Arg Ala Ser Gln Asp Ile Ser Asn
Tyr 20 25 30Leu Asn Trp Tyr Gln Gln Lys Pro Asp Gly Thr Val Lys Leu
Leu Ile 35 40 45Tyr Tyr Thr Ser Arg Leu His Ser Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Tyr Ser Leu Thr Ile Ser
Asn Leu Glu Gln65 70 75 80Glu Asp Ile Ala Thr Tyr Phe Cys Gln Gln
Gly Asn Thr Leu Pro Trp 85 90 95Thr Phe Gly Gly Gly Thr Lys Leu Glu
Ile Lys Arg 100 105115121PRTArtificial Sequenceheavy chain variable
region 115Glu Val Gln Leu Gln Gln Ser Gly Pro Glu Leu Val Lys Pro
Gly Ala1 5 10 15Ser Val Lys Ile Ser Cys Lys Thr Ser Gly Tyr Thr Phe
Lys Asp Tyr 20 25 30Thr Met His Trp Val Lys Gln Ser His Gly Lys Ser
Leu Glu Trp Ile 35 40 45Gly Gly Ile Tyr Pro Asn Asn Gly Gly Ser Thr
Tyr Asn Gln Asn Phe 50 55 60Lys Asp Lys Ala Thr Leu Thr Val Asp Lys
Ser Ser Ser Thr Ala Tyr65 70 75 80Met Glu Phe Arg Ser Leu Thr Ser
Glu Asp Ser Ala Val Tyr Tyr Cys 85 90 95Ala Arg Met Gly Tyr His Gly
Pro His Leu Asp Phe Asp Val Trp Gly 100 105 110Ala Gly Thr Thr Val
Thr Val Ser Pro 115 120116108PRTArtificial Sequencelight chain
variable region 116Asp Ile Val Met Thr Gln Ser His Lys Phe Met Ser
Thr Ser Leu Gly1 5 10 15Asp Arg Val Ser Ile Thr Cys Lys Ala Ser Gln
Asp Val Gly Ala Ala 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Gln
Ser Pro Lys Leu Leu Ile 35 40 45Tyr Trp Ala Ser Thr Arg His Thr Gly
Val Pro Asp Arg Phe Thr Gly 50 55 60Gly Gly Ser Gly Thr Asp Phe Thr
Leu Thr Ile Ser Asn Val Gln Ser65 70 75 80Glu Asp Leu Thr Asp Tyr
Phe Cys Gln Gln Tyr Ile Asn Tyr Pro Leu 85 90 95Thr Phe Gly Gly Gly
Thr Lys Leu Glu Ile Lys Arg 100 105117122PRTArtificial
Sequenceheavy chain variable region of humanized antibody 117Gln
Ile Gln Leu Val Gln Ser Gly Pro Glu Leu Lys Lys Pro Gly Glu1 5 10
15Thr Val
Lys Ile Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Asp Tyr 20 25 30Ser
Met His Trp Val Lys Gln Ala Pro Gly Lys Gly Leu Lys Trp Met 35 40
45Gly Trp Ile Asn Thr Glu Thr Gly Glu Pro Thr Tyr Ala Asp Asp Phe
50 55 60Lys Gly Arg Phe Ala Phe Ser Leu Glu Thr Ser Ala Ser Thr Ala
Tyr65 70 75 80Leu Gln Ile Asn Asn Leu Lys Asn Glu Asp Thr Ala Thr
Tyr Phe Cys 85 90 95Ala Asn Pro Tyr Tyr Asp Tyr Val Ser Tyr Tyr Ala
Met Asp Tyr Trp 100 105 110Gly His Gly Thr Ser Val Thr Val Ser Ser
115 120118122PRTArtificial Sequenceheavy chain variable region of
humanized antibody 118Gln Val Gln Leu Val Gln Ser Gly Ser Glu Leu
Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly
Tyr Thr Phe Thr Asp Tyr 20 25 30Ser Met His Trp Val Arg Gln Ala Pro
Gly Gln Gly Leu Lys Trp Met 35 40 45Gly Trp Ile Asn Thr Glu Thr Gly
Glu Pro Thr Tyr Ala Asp Asp Phe 50 55 60Lys Gly Arg Phe Val Phe Ser
Leu Asp Thr Ser Val Ser Thr Ala Tyr65 70 75 80Leu Gln Ile Ser Ser
Leu Lys Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Asn Pro Tyr
Tyr Asp Tyr Val Ser Tyr Tyr Ala Met Asp Tyr Trp 100 105 110Gly Gln
Gly Thr Thr Val Thr Val Ser Ser 115 120119107PRTArtificial
Sequencelight chain variable region of humanized antibody 119Asp
Ile Val Met Thr Gln Ser His Lys Phe Met Ser Thr Ser Val Arg1 5 10
15Asp Arg Val Ser Ile Thr Cys Lys Ala Ser Gln Asp Val Ser Thr Ala
20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Gln Ser Pro Lys Leu Leu
Ile 35 40 45Tyr Ser Ala Ser Tyr Leu Tyr Thr Gly Val Pro Asp Arg Phe
Thr Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Phe Thr Ile Ser Ser
Val Gln Ala65 70 75 80Glu Asp Leu Ala Val Tyr Tyr Cys Gln Gln His
Tyr Ser Thr Pro Arg 85 90 95Thr Phe Gly Gly Gly Thr Lys Leu Glu Ile
Lys 100 105120107PRTArtificial Sequencelight chain variable region
of humanized antibody 120Asp Ile Val Met Thr Gln Ser His Lys Phe
Met Ser Thr Ser Val Arg1 5 10 15Asp Arg Val Ser Ile Thr Cys Lys Ala
Ser Gln Asp Val Ser Thr Ala 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro
Gly Gln Ser Pro Lys Leu Leu Ile 35 40 45Tyr Ser Ala Ser Tyr Leu Tyr
Thr Gly Val Pro Asp Arg Phe Thr Gly 50 55 60Ser Gly Ser Gly Thr Asp
Phe Thr Phe Thr Ile Ser Ser Val Gln Ala65 70 75 80Glu Asp Leu Ala
Val Tyr Tyr Cys Gln Gln His Tyr Ser Thr Pro Arg 85 90 95Thr Phe Gly
Gly Gly Thr Lys Leu Glu Ile Lys 100 105121120PRTArtificial
Sequenceheavy chain variable region of humanized antibody 121Glu
Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Glu1 5 10
15Ser Leu Lys Leu Ser Cys Glu Ser Asn Glu Tyr Glu Phe Pro Ser His
20 25 30Asp Met Ser Trp Val Arg Lys Thr Pro Glu Lys Arg Leu Glu Leu
Val 35 40 45Ala Ala Ile Asn Ser Asp Gly Gly Ser Thr Tyr Tyr Pro Asp
Thr Met 50 55 60Glu Arg Arg Phe Ile Ile Ser Arg Asp Asn Thr Lys Lys
Thr Leu Tyr65 70 75 80Leu Gln Met Ser Ser Leu Arg Ser Glu Asp Thr
Ala Leu Tyr Tyr Cys 85 90 95Ala Arg His Tyr Asp Asp Tyr Tyr Ala Trp
Phe Ala Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ala
115 120122120PRTArtificial Sequenceheavy chain variable region of
humanized antibody 122Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu
Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Glu
Tyr Glu Phe Pro Ser His 20 25 30Asp Met Ser Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Leu Val 35 40 45Ala Ala Ile Asn Ser Asp Gly Gly
Ser Thr Tyr Tyr Pro Asp Thr Met 50 55 60Glu Arg Arg Phe Thr Ile Ser
Arg Asp Asn Ala Lys Asn Ser Leu Tyr65 70 75 80Leu Gln Met Asn Ser
Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg His Tyr
Asp Asp Tyr Tyr Ala Trp Phe Ala Tyr Trp Gly Gln 100 105 110Gly Thr
Met Val Thr Val Ser Ser 115 120123111PRTArtificial Sequencelight
chain variable region of humanized antibody 123Asp Ile Val Leu Thr
Gln Ser Pro Ala Ser Leu Ala Val Ser Leu Gly1 5 10 15Gln Arg Ala Thr
Ile Ser Cys Arg Ala Ser Lys Ser Val Ser Thr Ser 20 25 30Gly Tyr Ser
Tyr Met His Trp Tyr Gln Gln Lys Pro Gly Gln Pro Pro 35 40 45Lys Leu
Leu Ile Tyr Leu Ala Ser Asn Leu Glu Ser Gly Val Pro Ala 50 55 60Arg
Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Asn Ile His65 70 75
80Pro Val Glu Glu Glu Asp Ala Ala Thr Tyr Tyr Cys Gln His Ser Arg
85 90 95Glu Leu Pro Leu Thr Phe Gly Ala Gly Thr Lys Leu Glu Leu Lys
100 105 110124111PRTArtificial Sequencelight chain variable region
of humanized antibody 124Glu Ile Val Leu Thr Gln Ser Pro Ala Thr
Leu Ser Leu Ser Pro Gly1 5 10 15Glu Arg Ala Thr Leu Ser Cys Arg Ala
Ser Lys Ser Val Ser Thr Ser 20 25 30Gly Tyr Ser Tyr Met His Trp Tyr
Gln Gln Lys Pro Gly Gln Ala Pro 35 40 45Arg Leu Leu Ile Tyr Leu Ala
Ser Asn Leu Glu Ser Gly Val Pro Ala 50 55 60Arg Phe Ser Gly Ser Gly
Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser65 70 75 80Ser Leu Glu Pro
Glu Asp Phe Ala Val Tyr Tyr Cys Gln His Ser Arg 85 90 95Glu Leu Pro
Leu Thr Phe Gly Gly Gly Thr Lys Val Glu Ile Lys 100 105
110125138PRTArtificial Sequenceheavy chain variable region 125Met
Tyr Leu Gly Leu Asn Tyr Val Phe Ile Val Phe Leu Leu Asn Gly1 5 10
15Val Gln Ser Glu Val Lys Leu Glu Glu Ser Gly Gly Gly Leu Val Gln
20 25 30Pro Gly Gly Ser Met Lys Leu Ser Cys Ala Ala Ser Gly Phe Thr
Phe 35 40 45Ser Asp Ala Trp Met Asp Trp Val Arg Gln Ser Pro Glu Lys
Gly Leu 50 55 60Glu Trp Val Ala Glu Ile Arg Ser Lys Ala Asn Asn His
Ala Thr Tyr65 70 75 80Tyr Ala Glu Ser Val Asn Gly Arg Phe Thr Ile
Ser Arg Asp Asp Ser 85 90 95Lys Ser Ser Val Tyr Leu Gln Met Asn Ser
Leu Arg Ala Glu Asp Thr 100 105 110Gly Ile Tyr Tyr Cys Thr Trp Gly
Glu Val Phe Tyr Phe Asp Tyr Trp 115 120 125Gly Gln Gly Thr Thr Leu
Thr Val Ser Ser 130 135126126PRTArtificial Sequencelight chain
variable region 126Met Arg Pro Ser Ile Gln Phe Leu Gly Leu Leu Leu
Phe Trp Leu His1 5 10 15Gly Ala Gln Cys Asp Ile Gln Met Thr Gln Ser
Pro Ser Ser Leu Ser 20 25 30Ala Ser Leu Gly Gly Lys Val Thr Ile Thr
Cys Lys Ser Ser Gln Asp 35 40 45Ile Asn Lys Tyr Ile Ala Trp Tyr Gln
His Lys Pro Gly Lys Gly Pro 50 55 60Arg Leu Leu Ile His Tyr Thr Ser
Thr Leu Gln Pro Gly Ile Pro Ser65 70 75 80Arg Phe Ser Gly Ser Gly
Ser Gly Arg Asp Tyr Ser Phe Ser Ile Ser 85 90 95Asn Leu Glu Pro Glu
Asp Ile Ala Thr Tyr Tyr Cys Leu Gln Tyr Asp 100 105 110Asn Leu Leu
Thr Phe Gly Ala Gly Thr Lys Leu Glu Leu Lys 115 120
125127260PRTHomo Sapiens 127Met Ala Arg Pro His Pro Trp Trp Leu Cys
Val Leu Gly Thr Leu Val1 5 10 15Gly Leu Ser Ala Thr Pro Ala Pro Lys
Ser Cys Pro Glu Arg His Tyr 20 25 30Trp Ala Gln Gly Lys Leu Cys Cys
Gln Met Cys Glu Pro Gly Thr Phe 35 40 45Leu Val Lys Asp Cys Asp Gln
His Arg Lys Ala Ala Gln Cys Asp Pro 50 55 60Cys Ile Pro Gly Val Ser
Phe Ser Pro Asp His His Thr Arg Pro His65 70 75 80Cys Glu Ser Cys
Arg His Cys Asn Ser Gly Leu Leu Val Arg Asn Cys 85 90 95Thr Ile Thr
Ala Asn Ala Glu Cys Ala Cys Arg Asn Gly Trp Gln Cys 100 105 110Arg
Asp Lys Glu Cys Thr Glu Cys Asp Pro Leu Pro Asn Pro Ser Leu 115 120
125Thr Ala Arg Ser Ser Gln Ala Leu Ser Pro His Pro Gln Pro Thr His
130 135 140Leu Pro Tyr Val Ser Glu Met Leu Glu Ala Arg Thr Ala Gly
His Met145 150 155 160Gln Thr Leu Ala Asp Phe Arg Gln Leu Pro Ala
Arg Thr Leu Ser Thr 165 170 175His Trp Pro Pro Gln Arg Ser Leu Cys
Ser Ser Asp Phe Ile Arg Ile 180 185 190Leu Val Ile Phe Ser Gly Met
Phe Leu Val Phe Thr Leu Ala Gly Ala 195 200 205Leu Phe Leu His Gln
Arg Arg Lys Tyr Arg Ser Asn Lys Gly Glu Ser 210 215 220Pro Val Glu
Pro Ala Glu Pro Cys Arg Tyr Ser Cys Pro Arg Glu Glu225 230 235
240Glu Gly Ser Thr Ile Pro Ile Gln Glu Asp Tyr Arg Lys Pro Glu Pro
245 250 255Ala Cys Ser Pro 260128260PRTMacaca Nemestrina 128Met Ala
Arg Pro His Pro Trp Trp Leu Cys Phe Leu Gly Thr Leu Val1 5 10 15Gly
Leu Ser Ala Thr Pro Ala Pro Lys Ser Cys Pro Glu Arg His Tyr 20 25
30Trp Ala Gln Gly Lys Leu Cys Cys Gln Met Cys Glu Pro Gly Thr Phe
35 40 45Leu Val Lys Asp Cys Asp Gln His Arg Lys Ala Ala Gln Cys His
Pro 50 55 60Cys Ile Pro Gly Val Ser Phe Ser Pro Asp His His Thr Arg
Pro His65 70 75 80Cys Glu Ser Cys Arg His Cys Asn Ser Gly Leu Leu
Ile Arg Asn Cys 85 90 95Thr Ile Thr Ala Asn Ala Val Cys Ala Cys Arg
Asn Gly Trp Gln Cys 100 105 110Arg Asp Lys Glu Cys Thr Glu Cys Asp
Pro Pro Pro Asn Pro Ser Leu 115 120 125Thr Thr Trp Pro Ser Gln Ala
Leu Gly Pro His Pro Gln Pro Thr His 130 135 140Leu Pro Tyr Val Asn
Glu Met Leu Glu Ala Arg Thr Ala Gly His Met145 150 155 160Gln Thr
Leu Ala Asp Phe Arg His Leu Pro Ala Arg Thr Leu Ser Thr 165 170
175His Trp Pro Pro Gln Arg Ser Leu Cys Ser Ser Asp Phe Ile Arg Ile
180 185 190Leu Val Ile Phe Ser Gly Met Phe Leu Val Phe Thr Leu Ala
Gly Thr 195 200 205Leu Phe Leu His Gln Gln Arg Lys Tyr Arg Ser Asn
Lys Gly Glu Ser 210 215 220Pro Met Glu Pro Ala Glu Pro Cys Pro Tyr
Ser Cys Pro Arg Glu Glu225 230 235 240Glu Gly Ser Thr Ile Pro Ile
Gln Glu Asp Tyr Arg Lys Pro Glu Pro 245 250 255Ala Ser Ser Pro
260129452PRTArtificial Sequenceheavy chain for varlilumab 129Gln
Val Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10
15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr
20 25 30Asp Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Val 35 40 45Ala Val Ile Trp Tyr Asp Gly Ser Asn Lys Tyr Tyr Ala Asp
Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn
Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr
Ala Val Tyr Tyr Cys 85 90 95Ala Arg Gly Ser Gly Asn Trp Gly Phe Phe
Asp Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val Thr Val Ser Ser Ala
Ser Thr Lys Gly Pro Ser Val Phe 115 120 125Pro Leu Ala Pro Ser Ser
Lys Ser Thr Ser Gly Gly Thr Ala Ala Leu 130 135 140Gly Cys Leu Val
Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp145 150 155 160Asn
Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val Leu 165 170
175Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser
180 185 190Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His
Lys Pro 195 200 205Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys
Ser Cys Asp Lys 210 215 220Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Glu Leu Leu Gly Gly Pro225 230 235 240Ser Val Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser 245 250 255Arg Thr Pro Glu Val
Thr Cys Val Val Val Asp Val Ser His Glu Asp 260 265 270Pro Glu Val
Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn 275 280 285Ala
Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val 290 295
300Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys
Glu305 310 315 320Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala
Pro Ile Glu Lys 325 330 335Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg
Glu Pro Gln Val Tyr Thr 340 345 350Leu Pro Pro Ser Arg Asp Glu Leu
Thr Lys Asn Gln Val Ser Leu Thr 355 360 365Cys Leu Val Lys Gly Phe
Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu 370 375 380Ser Asn Gly Gln
Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu385 390 395 400Asp
Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys 405 410
415Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu
420 425 430Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
Pro Gly 435 440 445Lys Gly Ser Ser 450130214PRTArtificial
Sequencelight chain for varlilumab 130Asp Ile Gln Met Thr Gln Ser
Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr
Cys Arg Ala Ser Gln Gly Ile Ser Arg Trp 20 25 30Leu Ala Trp Tyr Gln
Gln Lys Pro Glu Lys Ala Pro Lys Ser Leu Ile 35 40 45Tyr Ala Ala Ser
Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser
Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu
Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Tyr Asn Thr Tyr Pro Arg 85 90
95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala Ala
100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys
Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr
Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn Ala Leu
Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu Gln Asp
Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr Leu
Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala Cys Glu
Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200 205Phe
Asn Arg Gly Glu Cys 210131119PRTArtificial Sequenceheavy chain
variable
region for varlilumab 131Gln Val Gln Leu Val Glu Ser Gly Gly Gly
Val Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser
Gly Phe Thr Phe Ser Ser Tyr 20 25 30Asp Met His Trp Val Arg Gln Ala
Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Val Ile Trp Tyr Asp Gly
Ser Asn Lys Tyr Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile
Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn
Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Gly
Ser Gly Asn Trp Gly Phe Phe Asp Tyr Trp Gly Gln Gly 100 105 110Thr
Leu Val Thr Val Ser Ser 115132107PRTArtificial Sequencelight chain
variable region for varlilumab 132Asp Ile Gln Met Thr Gln Ser Pro
Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys
Arg Ala Ser Gln Gly Ile Ser Arg Trp 20 25 30Leu Ala Trp Tyr Gln Gln
Lys Pro Glu Lys Ala Pro Lys Ser Leu Ile 35 40 45Tyr Ala Ala Ser Ser
Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly
Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp
Phe Ala Thr Tyr Tyr Cys Gln Gln Tyr Asn Thr Tyr Pro Arg 85 90 95Thr
Phe Gly Gln Gly Thr Lys Val Glu Ile Lys 100 1051338PRTArtificial
Sequenceheavy chain CDR1 for varlilumab 133Gly Phe Thr Phe Ser Ser
Tyr Asp1 51348PRTArtificial Sequenceheavy chain CDR2 for varlilumab
134Ile Trp Tyr Asp Gly Ser Asn Lys1 513512PRTArtificial
Sequenceheavy chain CDR3 for varlilumab 135Ala Arg Gly Ser Gly Asn
Trp Gly Phe Phe Asp Tyr1 5 101366PRTArtificial Sequencelight chain
CDR1 for varlilumab 136Gln Gly Ile Ser Arg Trp1 51374PRTArtificial
Sequencelight chain CDR2 for varlilumab 137Ala Ala Ser
Gly11389PRTArtificial Sequencelight chain CDR3 for varlilumab
138Gln Gln Tyr Asn Thr Tyr Pro Arg Thr1 5139193PRTArtificial
SequenceCD70 (CD27L) 139Met Pro Glu Glu Gly Ser Gly Cys Ser Val Arg
Arg Arg Pro Tyr Gly1 5 10 15Cys Val Leu Arg Ala Ala Leu Val Pro Leu
Val Ala Gly Leu Val Ile 20 25 30Cys Leu Val Val Cys Ile Gln Arg Phe
Ala Gln Ala Gln Gln Gln Leu 35 40 45Pro Leu Glu Ser Leu Gly Trp Asp
Val Ala Glu Leu Gln Leu Asn His 50 55 60Thr Gly Pro Gln Gln Asp Pro
Arg Leu Tyr Trp Gln Gly Gly Pro Ala65 70 75 80Leu Gly Arg Ser Phe
Leu His Gly Pro Glu Leu Asp Lys Gly Gln Leu 85 90 95Arg Ile His Arg
Asp Gly Ile Tyr Met Val His Ile Gln Val Thr Leu 100 105 110Ala Ile
Cys Ser Ser Thr Thr Ala Ser Arg His His Pro Thr Thr Leu 115 120
125Ala Val Gly Ile Cys Ser Pro Ala Ser Arg Ser Ile Ser Leu Leu Arg
130 135 140Leu Ser Phe His Gln Gly Cys Thr Ile Ala Ser Gln Arg Leu
Thr Pro145 150 155 160Leu Ala Arg Gly Asp Thr Leu Cys Thr Asn Leu
Thr Gly Thr Leu Leu 165 170 175Pro Ser Arg Asn Thr Asp Glu Thr Phe
Phe Gly Val Gln Trp Val Arg 180 185 190Pro140142PRTArtificial
SequenceCD70 soluble domain 140Ser Leu Gly Trp Asp Val Ala Glu Leu
Gln Leu Asn His Thr Gly Pro1 5 10 15Gln Gln Asp Pro Arg Leu Tyr Trp
Gln Gly Gly Pro Ala Leu Gly Arg 20 25 30Ser Phe Leu His Gly Pro Glu
Leu Asp Lys Gly Gln Leu Arg Ile His 35 40 45Arg Asp Gly Ile Tyr Met
Val His Ile Gln Val Thr Leu Ala Ile Cys 50 55 60Ser Ser Thr Thr Ala
Ser Arg His His Pro Thr Thr Leu Ala Val Gly65 70 75 80Ile Cys Ser
Pro Ala Ser Arg Ser Ile Ser Leu Leu Arg Leu Ser Phe 85 90 95His Gln
Gly Cys Thr Ile Ala Ser Gln Arg Leu Thr Pro Leu Ala Arg 100 105
110Gly Asp Thr Leu Cys Thr Asn Leu Thr Gly Thr Leu Leu Pro Ser Arg
115 120 125Asn Thr Asp Glu Thr Phe Phe Gly Val Gln Trp Val Arg Pro
130 135 140141137PRTArtificial SequenceCD70 soluble domain
(alternative) 141Val Ala Glu Leu Gln Leu Asn His Thr Gly Pro Gln
Gln Asp Pro Arg1 5 10 15Leu Tyr Trp Gln Gly Gly Pro Ala Leu Gly Arg
Ser Phe Leu His Gly 20 25 30Pro Glu Leu Asp Lys Gly Gln Leu Arg Ile
His Arg Asp Gly Ile Tyr 35 40 45Met Val His Ile Gln Val Thr Leu Ala
Ile Cys Ser Ser Thr Thr Ala 50 55 60Ser Arg His His Pro Thr Thr Leu
Ala Val Gly Ile Cys Ser Pro Ala65 70 75 80Ser Arg Ser Ile Ser Leu
Leu Arg Leu Ser Phe His Gln Gly Cys Thr 85 90 95Ile Ala Ser Gln Arg
Leu Thr Pro Leu Ala Arg Gly Asp Thr Leu Cys 100 105 110Thr Asn Leu
Thr Gly Thr Leu Leu Pro Ser Arg Asn Thr Asp Glu Thr 115 120 125Phe
Phe Gly Val Gln Trp Val Arg Pro 130 135142241PRTHomo Sapiens 142Met
Ala Gln His Gly Ala Met Gly Ala Phe Arg Ala Leu Cys Gly Leu1 5 10
15Ala Leu Leu Cys Ala Leu Ser Leu Gly Gln Arg Pro Thr Gly Gly Pro
20 25 30Gly Cys Gly Pro Gly Arg Leu Leu Leu Gly Thr Gly Thr Asp Ala
Arg 35 40 45Cys Cys Arg Val His Thr Thr Arg Cys Cys Arg Asp Tyr Pro
Gly Glu 50 55 60Glu Cys Cys Ser Glu Trp Asp Cys Met Cys Val Gln Pro
Glu Phe His65 70 75 80Cys Gly Asp Pro Cys Cys Thr Thr Cys Arg His
His Pro Cys Pro Pro 85 90 95Gly Gln Gly Val Gln Ser Gln Gly Lys Phe
Ser Phe Gly Phe Gln Cys 100 105 110Ile Asp Cys Ala Ser Gly Thr Phe
Ser Gly Gly His Glu Gly His Cys 115 120 125Lys Pro Trp Thr Asp Cys
Thr Gln Phe Gly Phe Leu Thr Val Phe Pro 130 135 140Gly Asn Lys Thr
His Asn Ala Val Cys Val Pro Gly Ser Pro Pro Ala145 150 155 160Glu
Pro Leu Gly Trp Leu Thr Val Val Leu Leu Ala Val Ala Ala Cys 165 170
175Val Leu Leu Leu Thr Ser Ala Gln Leu Gly Leu His Ile Trp Gln Leu
180 185 190Arg Ser Gln Cys Met Trp Pro Arg Glu Thr Gln Leu Leu Leu
Glu Val 195 200 205Pro Pro Ser Thr Glu Asp Ala Arg Ser Cys Gln Phe
Pro Glu Glu Glu 210 215 220Arg Gly Glu Arg Ser Ala Glu Glu Lys Gly
Arg Leu Gly Asp Leu Trp225 230 235 240Val143228PRTMus Musculus
143Met Gly Ala Trp Ala Met Leu Tyr Gly Val Ser Met Leu Cys Val Leu1
5 10 15Asp Leu Gly Gln Pro Ser Val Val Glu Glu Pro Gly Cys Gly Pro
Gly 20 25 30Lys Val Gln Asn Gly Ser Gly Asn Asn Thr Arg Cys Cys Ser
Leu Tyr 35 40 45Ala Pro Gly Lys Glu Asp Cys Pro Lys Glu Arg Cys Ile
Cys Val Thr 50 55 60Pro Glu Tyr His Cys Gly Asp Pro Gln Cys Lys Ile
Cys Lys His Tyr65 70 75 80Pro Cys Gln Pro Gly Gln Arg Val Glu Ser
Gln Gly Asp Ile Val Phe 85 90 95Gly Phe Arg Cys Val Ala Cys Ala Met
Gly Thr Phe Ser Ala Gly Arg 100 105 110Asp Gly His Cys Arg Leu Trp
Thr Asn Cys Ser Gln Phe Gly Phe Leu 115 120 125Thr Met Phe Pro Gly
Asn Lys Thr His Asn Ala Val Cys Ile Pro Glu 130 135 140Pro Leu Pro
Thr Glu Gln Tyr Gly His Leu Thr Val Ile Phe Leu Val145 150 155
160Met Ala Ala Cys Ile Phe Phe Leu Thr Thr Val Gln Leu Gly Leu His
165 170 175Ile Trp Gln Leu Arg Arg Gln His Met Cys Pro Arg Glu Thr
Gln Pro 180 185 190Phe Ala Glu Val Gln Leu Ser Ala Glu Asp Ala Cys
Ser Phe Gln Phe 195 200 205Pro Glu Glu Glu Arg Gly Glu Gln Thr Glu
Glu Lys Cys His Leu Gly 210 215 220Gly Arg Trp
Pro225144449PRTArtificial Sequencehumanized 6C8 heavy chain variant
144Gln Val Thr Leu Arg Glu Ser Gly Pro Ala Leu Val Lys Pro Thr Gln1
5 10 15Thr Leu Thr Leu Thr Cys Thr Phe Ser Gly Phe Ser Leu Ser Thr
Ser 20 25 30Gly Met Gly Val Gly Trp Ile Arg Gln Pro Pro Gly Lys Ala
Leu Glu 35 40 45Trp Leu Ala His Ile Trp Trp Asp Asp Asp Lys Tyr Tyr
Asn Pro Ser 50 55 60Leu Lys Ser Arg Leu Thr Ile Ser Lys Asp Thr Ser
Lys Asn Gln Val65 70 75 80Val Leu Thr Met Thr Asn Met Asp Pro Val
Asp Thr Ala Thr Tyr Tyr 85 90 95Cys Ala Arg Thr Arg Arg Tyr Phe Pro
Phe Ala Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val Thr Val Ser Ser
Ala Ser Thr Lys Gly Pro Ser Val Phe 115 120 125Pro Leu Ala Pro Ser
Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala Leu 130 135 140Gly Cys Leu
Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp145 150 155
160Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val Leu
165 170 175Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val
Pro Ser 180 185 190Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val
Asn His Lys Pro 195 200 205Ser Asn Thr Lys Val Asp Lys Lys Val Glu
Pro Lys Ser Cys Asp Lys 210 215 220Thr His Thr Cys Pro Pro Cys Pro
Ala Pro Glu Leu Leu Gly Gly Pro225 230 235 240Ser Val Phe Leu Phe
Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser 245 250 255Arg Thr Pro
Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp 260 265 270Pro
Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn 275 280
285Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val
290 295 300Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly
Lys Glu305 310 315 320Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro
Ala Pro Ile Glu Lys 325 330 335Thr Ile Ser Lys Ala Lys Gly Gln Pro
Arg Glu Pro Gln Val Tyr Thr 340 345 350Leu Pro Pro Ser Arg Asp Glu
Leu Thr Lys Asn Gln Val Ser Leu Thr 355 360 365Cys Leu Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu 370 375 380Ser Asn Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu385 390 395
400Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys
405 410 415Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met
His Glu 420 425 430Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser
Leu Ser Pro Gly 435 440 445Lys145449PRTArtificial Sequencehumanized
6C8 heavy chain variant 145Gln Val Thr Leu Arg Glu Ser Gly Pro Ala
Leu Val Lys Pro Thr Gln1 5 10 15Thr Leu Thr Leu Thr Cys Thr Phe Ser
Gly Phe Ser Leu Ser Thr Ser 20 25 30Gly Met Gly Val Gly Trp Ile Arg
Gln Pro Pro Gly Lys Ala Leu Glu 35 40 45Trp Leu Ala His Ile Trp Trp
Asp Asp Asp Lys Tyr Tyr Asn Pro Ser 50 55 60Leu Lys Ser Arg Leu Thr
Ile Ser Lys Asp Thr Ser Lys Asn Gln Val65 70 75 80Val Leu Thr Met
Thr Asn Met Asp Pro Val Asp Thr Ala Thr Tyr Tyr 85 90 95Cys Ala Arg
Thr Arg Arg Tyr Phe Pro Phe Ala Tyr Trp Gly Gln Gly 100 105 110Thr
Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser Val Phe 115 120
125Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala Leu
130 135 140Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val
Ser Trp145 150 155 160Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr
Phe Pro Ala Val Leu 165 170 175Gln Ser Ser Gly Leu Tyr Ser Leu Ser
Ser Val Val Thr Val Pro Ser 180 185 190Ser Ser Leu Gly Thr Gln Thr
Tyr Ile Cys Asn Val Asn His Lys Pro 195 200 205Ser Asn Thr Lys Val
Asp Lys Lys Val Glu Pro Lys Ser Cys Asp Lys 210 215 220Thr His Thr
Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro225 230 235
240Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser
245 250 255Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His
Glu Asp 260 265 270Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val
Glu Val His Asn 275 280 285Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr
Ala Ser Thr Tyr Arg Val 290 295 300Val Ser Val Leu Thr Val Leu His
Gln Asp Trp Leu Asn Gly Lys Glu305 310 315 320Tyr Lys Cys Lys Val
Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys 325 330 335Thr Ile Ser
Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr 340 345 350Leu
Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr 355 360
365Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu
370 375 380Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro
Val Leu385 390 395 400Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys
Leu Thr Val Asp Lys 405 410 415Ser Arg Trp Gln Gln Gly Asn Val Phe
Ser Cys Ser Val Met His Glu 420 425 430Ala Leu His Asn His Tyr Thr
Gln Lys Ser Leu Ser Leu Ser Pro Gly 435 440
445Lys146449PRTArtificial Sequencehumanized 6C8 heavy chain variant
146Gln Val Thr Leu Arg Glu Ser Gly Pro Ala Leu Val Lys Pro Thr Gln1
5 10 15Thr Leu Thr Leu Thr Cys Thr Phe Ser Gly Phe Ser Leu Ser Thr
Ser 20 25 30Gly Met Gly Val Gly Trp Ile Arg Gln Pro Pro Gly Lys Ala
Leu Glu 35 40 45Trp Leu Ala His Ile Trp Trp Asp Asp Asp Lys Tyr Tyr
Gln Pro Ser 50 55 60Leu Lys Ser Arg Leu Thr Ile Ser Lys Asp Thr Ser
Lys Asn Gln Val65 70 75 80Val Leu Thr Met Thr Asn Met Asp Pro Val
Asp Thr Ala Thr Tyr Tyr 85 90 95Cys Ala Arg Thr Arg Arg Tyr Phe Pro
Phe Ala Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val Thr Val Ser Ser
Ala Ser Thr Lys Gly Pro Ser Val Phe 115 120 125Pro Leu Ala Pro Ser
Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala Leu 130 135 140Gly Cys Leu
Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp145 150 155
160Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val Leu
165 170 175Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val
Pro Ser 180 185 190Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val
Asn His Lys Pro 195 200 205Ser Asn Thr Lys Val Asp Lys Lys Val Glu
Pro Lys Ser Cys Asp Lys 210 215 220Thr His Thr Cys Pro Pro Cys Pro
Ala Pro Glu Leu Leu Gly Gly Pro225 230 235 240Ser Val Phe Leu Phe
Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser 245 250 255Arg Thr Pro
Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp
260 265 270Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val
His Asn 275 280 285Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser
Thr Tyr Arg Val 290 295 300Val Ser Val Leu Thr Val Leu His Gln Asp
Trp Leu Asn Gly Lys Glu305 310 315 320Tyr Lys Cys Lys Val Ser Asn
Lys Ala Leu Pro Ala Pro Ile Glu Lys 325 330 335Thr Ile Ser Lys Ala
Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr 340 345 350Leu Pro Pro
Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr 355 360 365Cys
Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu 370 375
380Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val
Leu385 390 395 400Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu
Thr Val Asp Lys 405 410 415Ser Arg Trp Gln Gln Gly Asn Val Phe Ser
Cys Ser Val Met His Glu 420 425 430Ala Leu His Asn His Tyr Thr Gln
Lys Ser Leu Ser Leu Ser Pro Gly 435 440 445Lys147449PRTArtificial
Sequencehumanized 6C8 heavy chain variant 147Gln Val Thr Leu Arg
Glu Ser Gly Pro Ala Leu Val Lys Pro Thr Gln1 5 10 15Thr Leu Thr Leu
Thr Cys Thr Phe Ser Gly Phe Ser Leu Ser Thr Ser 20 25 30Gly Met Gly
Val Gly Trp Ile Arg Gln Pro Pro Gly Lys Ala Leu Glu 35 40 45Trp Leu
Ala His Ile Trp Trp Asp Asp Asp Lys Tyr Tyr Gln Pro Ser 50 55 60Leu
Lys Ser Arg Leu Thr Ile Ser Lys Asp Thr Ser Lys Asn Gln Val65 70 75
80Val Leu Thr Met Thr Asn Met Asp Pro Val Asp Thr Ala Thr Tyr Tyr
85 90 95Cys Ala Arg Thr Arg Arg Tyr Phe Pro Phe Ala Tyr Trp Gly Gln
Gly 100 105 110Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro
Ser Val Phe 115 120 125Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly
Gly Thr Ala Ala Leu 130 135 140Gly Cys Leu Val Lys Asp Tyr Phe Pro
Glu Pro Val Thr Val Ser Trp145 150 155 160Asn Ser Gly Ala Leu Thr
Ser Gly Val His Thr Phe Pro Ala Val Leu 165 170 175Gln Ser Ser Gly
Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser 180 185 190Ser Ser
Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro 195 200
205Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp Lys
210 215 220Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly
Gly Pro225 230 235 240Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp
Thr Leu Met Ile Ser 245 250 255Arg Thr Pro Glu Val Thr Cys Val Val
Val Asp Val Ser His Glu Asp 260 265 270Pro Glu Val Lys Phe Asn Trp
Tyr Val Asp Gly Val Glu Val His Asn 275 280 285Ala Lys Thr Lys Pro
Arg Glu Glu Gln Tyr Ala Ser Thr Tyr Arg Val 290 295 300Val Ser Val
Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu305 310 315
320Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys
325 330 335Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val
Tyr Thr 340 345 350Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln
Val Ser Leu Thr 355 360 365Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp
Ile Ala Val Glu Trp Glu 370 375 380Ser Asn Gly Gln Pro Glu Asn Asn
Tyr Lys Thr Thr Pro Pro Val Leu385 390 395 400Asp Ser Asp Gly Ser
Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys 405 410 415Ser Arg Trp
Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu 420 425 430Ala
Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly 435 440
445Lys148214PRTArtificial Sequencehumanized 6C8 light chain 148Glu
Ile Val Met Thr Gln Ser Pro Ala Thr Leu Ser Val Ser Pro Gly1 5 10
15Glu Arg Ala Thr Leu Ser Cys Lys Ala Ser Gln Asn Val Gly Thr Asn
20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro Arg Leu Leu
Ile 35 40 45Tyr Ser Ala Ser Tyr Arg Tyr Ser Gly Ile Pro Ala Arg Phe
Ser Gly 50 55 60Ser Gly Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser
Leu Gln Ser65 70 75 80Glu Asp Phe Ala Val Tyr Tyr Cys Gln Gln Tyr
Asn Thr Asp Pro Leu 85 90 95Thr Phe Gly Gly Gly Thr Lys Val Glu Ile
Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro
Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys
Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp
Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu
Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170
175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr
180 185 190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr
Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys 21014919PRTArtificial
Sequence6C8 heavy chain leader 149Met Asp Arg Leu Thr Phe Ser Phe
Leu Leu Leu Ile Val Pro Ala Tyr1 5 10 15Val Leu
Ser15020PRTArtificial Sequence6C8 light chain leader 150Met Glu Thr
Gln Ser Gln Val Phe Val Tyr Met Leu Leu Trp Leu Ser1 5 10 15Gly Val
Asp Gly 20151138PRTArtificial Sequencehumanized 6C8 heavy chain
variable region variant 151Met Asp Arg Leu Thr Phe Ser Phe Leu Leu
Leu Ile Val Pro Ala Tyr1 5 10 15Val Leu Ser Gln Val Thr Leu Lys Glu
Ser Gly Pro Gly Ile Leu Lys 20 25 30Pro Ser Gln Thr Leu Ser Leu Thr
Cys Ser Phe Ser Gly Phe Ser Leu 35 40 45Ser Thr Ser Gly Met Gly Val
Gly Trp Ile Arg Gln Pro Ser Gly Lys 50 55 60Gly Leu Glu Trp Leu Ala
His Ile Trp Trp Asp Asp Asp Lys Tyr Tyr65 70 75 80Asn Pro Ser Leu
Lys Ser Gln Leu Thr Ile Ser Lys Asp Thr Ser Arg 85 90 95Asn Gln Val
Phe Leu Lys Ile Thr Ser Val Asp Thr Ala Asp Ala Ala 100 105 110Thr
Tyr Tyr Cys Ala Arg Thr Arg Arg Tyr Phe Pro Phe Ala Tyr Trp 115 120
125Gly Gln Gly Thr Leu Val Thr Val Ser Ser 130
135152138PRTArtificial Sequencehumanized 6C8 heavy chain variable
region variant 152Met Asp Arg Leu Thr Phe Ser Phe Leu Leu Leu Ile
Val Pro Ala Tyr1 5 10 15Val Leu Ser Gln Val Thr Leu Lys Glu Ser Gly
Pro Gly Ile Leu Lys 20 25 30Pro Ser Gln Thr Leu Ser Leu Thr Cys Ser
Phe Ser Gly Phe Ser Leu 35 40 45Ser Thr Ser Gly Met Gly Val Gly Trp
Ile Arg Gln Pro Ser Gly Lys 50 55 60Gly Leu Glu Trp Leu Ala His Ile
Trp Trp Asp Asp Asp Lys Tyr Tyr65 70 75 80Gln Pro Ser Leu Lys Ser
Gln Leu Thr Ile Ser Lys Asp Thr Ser Arg 85 90 95Asn Gln Val Phe Leu
Lys Ile Thr Ser Val Asp Thr Ala Asp Ala Ala 100 105 110Thr Tyr Tyr
Cys Ala Arg Thr Arg Arg Tyr Phe Pro Phe Ala Tyr Trp 115 120 125Gly
Gln Gly Thr Leu Val Thr Val Ser Ser 130 135153127PRTArtificial
Sequencehumanized 6C8 light chain variable region 153Met Glu Thr
Gln Ser Gln Val Phe Val Tyr Met Leu Leu Trp Leu Ser1 5 10 15Gly Val
Asp Gly Asp Ile Val Met Thr Gln Ser Gln Lys Phe Met Ser 20 25 30Thr
Ser Val Gly Asp Arg Val Ser Val Thr Cys Lys Ala Ser Gln Asn 35 40
45Val Gly Thr Asn Val Ala Trp Tyr Gln Gln Lys Pro Gly Gln Ser Pro
50 55 60Lys Ala Leu Ile Tyr Ser Ala Ser Tyr Arg Tyr Ser Gly Val Pro
Asp65 70 75 80Arg Phe Thr Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu
Thr Ile Asn 85 90 95Asn Val His Ser Glu Asp Leu Ala Glu Tyr Phe Cys
Gln Gln Tyr Asn 100 105 110Thr Asp Pro Leu Thr Phe Gly Ala Gly Thr
Lys Leu Glu Ile Lys 115 120 12515412PRTArtificial Sequence6C8 heavy
chain CDR1 154Gly Phe Ser Leu Ser Thr Ser Gly Met Gly Val Gly1 5
1015516PRTArtificial Sequence6C8 heavy chain CDR2 variant 155His
Ile Trp Trp Asp Asp Asp Lys Tyr Tyr Asn Pro Ser Leu Lys Ser1 5 10
1515616PRTArtificial Sequence6C8 heavy chain CDR2 variant 156His
Ile Trp Trp Asp Asp Asp Lys Tyr Tyr Gln Pro Ser Leu Lys Ser1 5 10
151579PRTArtificial Sequence6C8 heavy chain CDR3 157Thr Arg Arg Tyr
Phe Pro Phe Ala Tyr1 515811PRTArtificial Sequence6C8 light chain
CDR1 158Lys Ala Ser Gln Asn Val Gly Thr Asn Val Ala1 5
101597PRTArtificial Sequence6C8 light chain CDR2 159Ser Ala Ser Tyr
Arg Tyr Ser1 51609PRTArtificial Sequence6C8 light chain CDR3 160Gln
Gln Tyr Asn Thr Asp Pro Leu Thr1 5161449PRTArtificial
Sequencechimeric 6C8 heavy chain variant 161Gln Val Thr Leu Lys Glu
Ser Gly Pro Gly Ile Leu Lys Pro Ser Gln1 5 10 15Thr Leu Ser Leu Thr
Cys Ser Phe Ser Gly Phe Ser Leu Ser Thr Ser 20 25 30Gly Met Gly Val
Gly Trp Ile Arg Gln Pro Ser Gly Lys Gly Leu Glu 35 40 45Trp Leu Ala
His Ile Trp Trp Asp Asp Asp Lys Tyr Tyr Asn Pro Ser 50 55 60Leu Lys
Ser Gln Leu Thr Ile Ser Lys Asp Thr Ser Arg Asn Gln Val65 70 75
80Phe Leu Lys Ile Thr Ser Val Asp Thr Ala Asp Ala Ala Thr Tyr Tyr
85 90 95Cys Ala Arg Thr Arg Arg Tyr Phe Pro Phe Ala Tyr Trp Gly Gln
Gly 100 105 110Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro
Ser Val Phe 115 120 125Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly
Gly Thr Ala Ala Leu 130 135 140Gly Cys Leu Val Lys Asp Tyr Phe Pro
Glu Pro Val Thr Val Ser Trp145 150 155 160Asn Ser Gly Ala Leu Thr
Ser Gly Val His Thr Phe Pro Ala Val Leu 165 170 175Gln Ser Ser Gly
Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser 180 185 190Ser Ser
Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro 195 200
205Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp Lys
210 215 220Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly
Gly Pro225 230 235 240Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp
Thr Leu Met Ile Ser 245 250 255Arg Thr Pro Glu Val Thr Cys Val Val
Val Asp Val Ser His Glu Asp 260 265 270Pro Glu Val Lys Phe Asn Trp
Tyr Val Asp Gly Val Glu Val His Asn 275 280 285Ala Lys Thr Lys Pro
Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val 290 295 300Val Ser Val
Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu305 310 315
320Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys
325 330 335Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val
Tyr Thr 340 345 350Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln
Val Ser Leu Thr 355 360 365Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp
Ile Ala Val Glu Trp Glu 370 375 380Ser Asn Gly Gln Pro Glu Asn Asn
Tyr Lys Thr Thr Pro Pro Val Leu385 390 395 400Asp Ser Asp Gly Ser
Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys 405 410 415Ser Arg Trp
Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu 420 425 430Ala
Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly 435 440
445Lys162449PRTArtificial Sequencechimeric 6C8 heavy chain variant
162Gln Val Thr Leu Lys Glu Ser Gly Pro Gly Ile Leu Lys Pro Ser Gln1
5 10 15Thr Leu Ser Leu Thr Cys Ser Phe Ser Gly Phe Ser Leu Ser Thr
Ser 20 25 30Gly Met Gly Val Gly Trp Ile Arg Gln Pro Ser Gly Lys Gly
Leu Glu 35 40 45Trp Leu Ala His Ile Trp Trp Asp Asp Asp Lys Tyr Tyr
Asn Pro Ser 50 55 60Leu Lys Ser Gln Leu Thr Ile Ser Lys Asp Thr Ser
Arg Asn Gln Val65 70 75 80Phe Leu Lys Ile Thr Ser Val Asp Thr Ala
Asp Ala Ala Thr Tyr Tyr 85 90 95Cys Ala Arg Thr Arg Arg Tyr Phe Pro
Phe Ala Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val Thr Val Ser Ser
Ala Ser Thr Lys Gly Pro Ser Val Phe 115 120 125Pro Leu Ala Pro Ser
Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala Leu 130 135 140Gly Cys Leu
Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp145 150 155
160Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val Leu
165 170 175Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val
Pro Ser 180 185 190Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val
Asn His Lys Pro 195 200 205Ser Asn Thr Lys Val Asp Lys Lys Val Glu
Pro Lys Ser Cys Asp Lys 210 215 220Thr His Thr Cys Pro Pro Cys Pro
Ala Pro Glu Leu Leu Gly Gly Pro225 230 235 240Ser Val Phe Leu Phe
Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser 245 250 255Arg Thr Pro
Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp 260 265 270Pro
Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn 275 280
285Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Ala Ser Thr Tyr Arg Val
290 295 300Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly
Lys Glu305 310 315 320Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro
Ala Pro Ile Glu Lys 325 330 335Thr Ile Ser Lys Ala Lys Gly Gln Pro
Arg Glu Pro Gln Val Tyr Thr 340 345 350Leu Pro Pro Ser Arg Asp Glu
Leu Thr Lys Asn Gln Val Ser Leu Thr 355 360 365Cys Leu Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu 370 375 380Ser Asn Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu385 390 395
400Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys
405 410 415Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met
His Glu 420 425 430Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser
Leu Ser Pro Gly 435 440 445Lys163214PRTArtificial Sequencechimeric
6C8 light chain variant 163Asp Ile Val Met Thr Gln Ser Gln Lys Phe
Met Ser Thr Ser Val Gly1 5 10 15Asp Arg Val Ser Val Thr Cys Lys Ala
Ser Gln Asn Val Gly Thr Asn 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro
Gly Gln Ser Pro Lys Ala Leu Ile 35 40 45Tyr Ser Ala Ser Tyr Arg Tyr
Ser Gly Val Pro Asp Arg Phe Thr Gly 50 55 60Ser Gly Ser Gly Thr Asp
Phe Thr Leu Thr Ile Asn Asn Val His Ser65
70 75 80Glu Asp Leu Ala Glu Tyr Phe Cys Gln Gln Tyr Asn Thr Asp Pro
Leu 85 90 95Thr Phe Gly Ala Gly Thr Lys Leu Glu Ile Lys Arg Thr Val
Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln
Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn
Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn
Ala Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu
Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu
Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala
Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200
205Phe Asn Arg Gly Glu Cys 210164118PRTArtificial Sequence36E5
heavy chain variable region 164Glu Val Asn Leu Val Glu Ser Gly Gly
Gly Leu Val Lys Pro Gly Gly1 5 10 15Ser Leu Lys Val Ser Cys Ala Ala
Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30Ala Met Ser Trp Val Arg Gln
Thr Pro Glu Lys Arg Leu Glu Trp Val 35 40 45Ala Ser Ile Ser Ser Gly
Gly Thr Thr Tyr Tyr Pro Asp Ser Val Lys 50 55 60Gly Arg Phe Thr Ile
Ser Arg Asp Asn Ala Arg Asn Ile Leu Tyr Leu65 70 75 80Gln Met Ser
Ser Leu Arg Ser Glu Asp Thr Ala Met Tyr Tyr Cys Ala 85 90 95Arg Val
Gly Gly Tyr Tyr Asp Ser Met Asp Tyr Trp Gly Gln Gly Ile 100 105
110Ser Val Thr Asp Ser Ser 115165113PRTArtificial Sequence36E5
light chain variable region 165Asp Ile Val Leu Thr Gln Ser Pro Ala
Ser Leu Ala Val Ser Leu Gly1 5 10 15Gln Arg Ala Thr Ile Ser Cys Arg
Ala Ser Glu Ser Val Asp Asn Tyr 20 25 30Gly Val Ser Phe Met Asn Trp
Phe Gln Gln Lys Pro Gly Gln Pro Pro 35 40 45Lys Leu Leu Ile Tyr Ala
Ala Ser Asn Gln Gly Ser Gly Val Pro Ala 50 55 60Arg Phe Ser Gly Ser
Gly Ser Gly Thr Asp Phe Ser Leu Asn Ile His65 70 75 80Pro Met Glu
Glu Asp Asp Thr Ala Met Tyr Phe Cys Gln Gln Thr Lys 85 90 95Glu Val
Thr Trp Thr Phe Gly Gly Gly Thr Lys Leu Glu Ile Lys Arg 100 105
110Ala166123PRTArtificial Sequence3D6 heavy chain variable region
166Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Arg1
5 10 15Ser Leu Lys Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Asp
Tyr 20 25 30Tyr Met Ala Trp Val Arg Gln Ala Pro Thr Lys Gly Leu Glu
Trp Val 35 40 45Ala Tyr Ile His Ala Asn Gly Gly Ser Thr Tyr Tyr Arg
Asp Ser Val 50 55 60Arg Gly Arg Phe Ser Ile Ser Arg Asp Asn Gly Lys
Ser Thr Leu Tyr65 70 75 80Leu Gln Met Asp Ser Leu Arg Ser Glu Asp
Thr Ala Thr Tyr Tyr Cys 85 90 95Thr Thr Gly Ser Phe Met Tyr Ala Ala
Asp Tyr Tyr Ile Met Asp Ala 100 105 110Trp Gly Gln Gly Ala Ser Val
Thr Val Ser Ser 115 120167113PRTArtificial Sequence3D6 light chain
variable region 167Asp Val Val Met Thr Gln Thr Pro Val Ser Leu Ser
Val Ser Leu Gly1 5 10 15Asn Gln Ala Ser Ile Ser Cys Arg Ser Ser Gln
Ser Leu Leu His Ser 20 25 30Asp Gly Asn Thr Phe Leu Ser Trp Tyr Phe
Gln Lys Pro Gly Gln Ser 35 40 45Pro Gln Leu Leu Ile Tyr Leu Ala Ser
Asn Arg Phe Ser Gly Val Ser 50 55 60Asn Arg Phe Ser Gly Ser Gly Ser
Gly Thr Asp Phe Thr Leu Lys Ile65 70 75 80Ser Arg Val Glu Pro Glu
Asp Leu Gly Val Tyr Tyr Cys Phe Gln His 85 90 95Thr His Leu Pro Leu
Thr Phe Gly Ser Gly Thr Lys Leu Glu Ile Lys 100 105
110Arg168118PRTArtificial Sequence61G6 heavy chain variable region
168Asp Val Gln Leu Gln Glu Ser Gly Pro Gly Leu Val Lys Pro Ser Gln1
5 10 15Ser Leu Ser Leu Thr Cys Thr Val Thr Gly Tyr Ser Ile Thr Ser
Asp 20 25 30Tyr Ala Trp Asn Trp Ile Arg Gln Phe Pro Gly Asn Lys Leu
Glu Trp 35 40 45Met Gly Tyr Ile Ser Tyr Ser Gly Ser Thr Arg Tyr Asn
Pro Ser Leu 50 55 60Lys Ser Arg Ile Ser Ile Thr Arg Asp Thr Ser Lys
Asn Gln Phe Phe65 70 75 80Leu Gln Leu Asn Ser Val Thr Ser Glu Asp
Thr Ala Thr Tyr Tyr Cys 85 90 95Ala Arg Gln Leu Gly Leu Arg Phe Phe
Asp Tyr Trp Gly Gln Gly Thr 100 105 110Thr Leu Thr Val Ser Ser
115169108PRTArtificial Sequence61G6 light chain variable region
169Gln Ile Val Leu Thr Gln Ser Pro Ala Leu Met Ser Ala Ser Pro Gly1
5 10 15Glu Lys Val Thr Met Thr Cys Ser Ala Asn Ser Thr Val Asn Tyr
Met 20 25 30Tyr Trp Tyr Gln Gln Lys Pro Arg Ser Ser Pro Lys Pro Cys
Ile Tyr 35 40 45Leu Thr Ser Asn Leu Ala Ser Gly Val Pro Ala Arg Phe
Ser Gly Ser 50 55 60Gly Ser Gly Thr Ser Tyr Ser Leu Thr Ile Ser Ser
Met Glu Ala Glu65 70 75 80Asp Ala Ala Thr Tyr Tyr Cys Gln Gln Trp
Asn Ser Asn Pro Pro Thr 85 90 95Phe Gly Ala Gly Thr Lys Leu Glu Leu
Arg Arg Ala 100 105170118PRTArtificial Sequence6H6 heavy chain
variable region 170Gln Val Gln Leu Gln Gln Ser Gly Ala Glu Leu Met
Lys Pro Gly Ala1 5 10 15Ser Val Lys Ile Ser Cys Lys Ala Thr Gly Tyr
Thr Phe Ser Arg Tyr 20 25 30Trp Ile Glu Trp Ile Lys Gln Arg Pro Gly
His Gly Leu Glu Trp Ile 35 40 45Gly Glu Ile Leu Pro Gly Ser Gly Ser
Ser Asn Tyr Asn Glu Lys Phe 50 55 60Lys Asp Lys Ala Thr Phe Thr Ala
Asp Thr Ser Ser Asn Thr Ala Tyr65 70 75 80Met Gln Phe Ser Ser Leu
Thr Ser Glu Asp Ser Ala Val Tyr Tyr Cys 85 90 95Ala Arg Lys Val Tyr
Tyr Tyr Ala Met Asp Phe Trp Gly Gln Gly Thr 100 105 110Ser Val Thr
Val Ser Ser 115171110PRTArtificial Sequence6H6 light chain variable
region 171Gln Ile Val Leu Thr Gln Ser Pro Ala Ile Met Ser Val Ser
Leu Gly1 5 10 15Glu Arg Val Thr Val Thr Cys Thr Ala Ser Ser Ser Val
Ser Ser Ser 20 25 30Tyr Phe His Trp Tyr Gln Gln Lys Pro Gly Ser Ser
Pro Lys Leu Trp 35 40 45Ile Tyr Ser Thr Ser Asn Leu Ala Ser Gly Val
Pro Ala Arg Phe Ser 50 55 60Gly Ser Gly Ser Gly Thr Ser Tyr Ser Leu
Thr Ile Ser Thr Met Glu65 70 75 80Ala Glu Asp Ala Ala Thr Tyr Tyr
Cys His Gln Tyr His Arg Ser Pro 85 90 95Arg Thr Phe Gly Gly Gly Thr
Lys Leu Glu Ile Lys Arg Ala 100 105 110172119PRTArtificial
Sequence61F6 heavy chain variable region 172Gln Val Gln Leu Gln Gln
Ser Gly Ala Glu Leu Ala Arg Pro Gly Ala1 5 10 15Ser Val Lys Met Ser
Cys Lys Ala Ser Gly Tyr Thr Phe Thr Ser Tyr 20 25 30Thr Met His Trp
Val Lys Gln Arg Pro Gly Gln Gly Leu Glu Trp Ile 35 40 45Gly Tyr Ile
Asn Pro Arg Ser Val Tyr Thr Asn Tyr Asn Gln Lys Phe 50 55 60Lys Asp
Lys Ala Thr Leu Thr Ala Asp Lys Ser Ser Ser Thr Ala Tyr65 70 75
80Met Gln Leu Ser Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Tyr Cys
85 90 95Ala Arg Leu Gly Gly Tyr Tyr Asp Thr Met Asp Tyr Trp Gly Gln
Gly 100 105 110Thr Ser Val Thr Val Ser Ser 115173113PRTArtificial
Sequence61F6 light chain variable region 173Asp Ile Val Val Thr Gln
Ser Pro Ala Ser Leu Ala Val Ser Leu Gly1 5 10 15Gln Arg Ala Thr Ile
Ser Cys Arg Ala Ser Glu Ser Val Asp Asn Tyr 20 25 30Gly Ile Ser Phe
Met Asn Trp Phe Gln Gln Lys Pro Gly Gln Pro Pro 35 40 45Lys Leu Leu
Ile Tyr Ala Ala Ser Asn Gln Gly Ser Gly Val Pro Ala 50 55 60Arg Phe
Ser Gly Ser Gly Ser Gly Thr Asp Phe Ser Leu Asn Ile His65 70 75
80Pro Met Glu Glu Asp Asp Thr Ala Val Tyr Phe Cys Gln Gln Ser Lys
85 90 95Glu Val Pro Phe Thr Phe Gly Ser Gly Thr Lys Leu Glu Ile Lys
Arg 100 105 110Ala174122PRTArtificial Sequence1D8 heavy chain
variable region 174Gln Val Thr Leu Lys Glu Ser Gly Pro Gly Ile Leu
Lys Pro Ser Gln1 5 10 15Thr Leu Ser Leu Thr Cys Ser Phe Ser Gly Phe
Ser Leu Ser Thr Ser 20 25 30Gly Met Gly Val Gly Trp Ile Arg Gln Pro
Ser Gly Lys Gly Leu Glu 35 40 45Trp Leu Ala His Ile Trp Trp Asp Asp
Asp Lys Tyr Tyr Ser Pro Ser 50 55 60Leu Lys Ser Gln Leu Thr Ile Ser
Lys Asp Thr Ser Arg Asn Gln Val65 70 75 80Phe Leu Lys Ile Thr Ser
Leu Asp Thr Ala Asp Thr Ala Thr Tyr Tyr 85 90 95Cys Val Arg Ser Tyr
Tyr Tyr Gly Ser Ser Gly Ala Met Asp Tyr Trp 100 105 110Gly Gln Gly
Thr Ser Val Thr Val Ser Ser 115 120175118PRTArtificial Sequence1D8
light chain variable region 175Asp Ile Val Met Thr Gln Thr Pro Leu
Ser Leu Pro Val Ser Leu Gly1 5 10 15Asp Gln Ala Ser Ile Ser Cys Arg
Ser Ser Gln Ser Leu Val His Ser 20 25 30Asp Gly Asn Thr Tyr Leu His
Trp Tyr Leu Gln Lys Pro Gly Gln Ser 35 40 45Pro Lys Leu Leu Ile Tyr
Lys Val Ser Lys Arg Phe Ser Gly Val Pro 50 55 60Asp Arg Phe Ser Gly
Ser Gly Ser Gly Thr Asp Phe Thr Leu Lys Ile65 70 75 80Ser Arg Val
Glu Ala Glu Asp Leu Gly Val Tyr Phe Cys Ser Gln Ser 85 90 95Thr His
Val Pro Pro Thr Phe Gly Gly Gly Thr Lys Leu Glu Ile Lys 100 105
110Arg Ala Asp Ala Ala Pro 115176117PRTArtificial Sequence17F10
heavy chain variable region 176Glu Val Lys Leu Val Glu Ser Gly Gly
Gly Phe Val Lys Pro Gly Gly1 5 10 15Ser Leu Lys Leu Ser Cys Ala Ala
Ser Gly Phe Thr Val Arg Asn Tyr 20 25 30Ala Met Ser Trp Val Arg Gln
Thr Pro Glu Lys Arg Leu Glu Trp Val 35 40 45Ala Ser Ile Ser Thr Gly
Asp Arg Ser Tyr Leu Pro Asp Ser Met Lys 50 55 60Gly Arg Phe Thr Ile
Ser Arg Asp Asn Ala Arg Asn Ile Leu Tyr Leu65 70 75 80Gln Met Ser
Ser Leu Arg Ser Glu Asp Thr Ala Ile Tyr Tyr Cys Gln 85 90 95Arg Tyr
Phe Asp Phe Asp Ser Phe Ala Phe Trp Gly Gln Gly Thr Leu 100 105
110Val Thr Val Ser Ala 115177113PRTArtificial Sequence17F10 light
chain variable region 177Asp Ile Gln Met Thr Gln Thr Pro Ser Ser
Leu Ser Ala Ser Leu Gly1 5 10 15Asp Arg Val Thr Ile Ser Cys Arg Ala
Ser Gln Asp Ile Asn Asn Phe 20 25 30Leu Asn Trp Tyr Gln Gln Lys Pro
Asp Gly Ser Leu Lys Leu Leu Ile 35 40 45Tyr Tyr Thr Ser Lys Leu His
Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp
Phe Ser Leu Thr Ile Ser Asn Leu Asp Gln65 70 75 80Glu Asp Val Ala
Thr Tyr Phe Cys Gln Gln Gly His Thr Leu Pro Pro 85 90 95Thr Phe Gly
Gly Gly Thr Lys Leu Glu Val Lys Arg Ala Asp Ala Ala 100 105
110Pro178120PRTArtificial Sequence35D8 heavy chain variable region
178Glu Val Gln Leu Gln Glu Ser Gly Pro Ser Leu Val Lys Pro Ser Gln1
5 10 15Thr Leu Ser Leu Thr Cys Ser Val Thr Gly Asp Ser Ile Thr Ser
Gly 20 25 30Tyr Trp Asn Trp Ile Arg Lys Phe Pro Gly Asn Lys Leu Glu
Tyr Met 35 40 45Gly Tyr Ile Ser Tyr Ser Gly Ser Thr Tyr Tyr Asn Pro
Ser Leu Arg 50 55 60Gly Arg Ile Ser Ile Thr Arg Asp Thr Ser Lys Ser
Gln Tyr Tyr Leu65 70 75 80Gln Leu Ser Ser Val Thr Thr Glu Asp Thr
Ala Thr Tyr Tyr Cys Ser 85 90 95Arg Arg His Leu Gly Ser Gly Tyr Gly
Trp Phe Ala Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser
Ala 115 120179114PRTArtificial Sequence35D8 light chain variable
region 179Asp Ile Val Met Thr Gln Ser His Lys Phe Met Ser Thr Ser
Val Gly1 5 10 15Asp Arg Val Ser Ile Thr Cys Lys Ala Ser Gln Asp Val
Asn Thr Ala 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Gln Ser Pro
Lys Leu Leu Ile 35 40 45Tyr Trp Ala Ser Thr Arg His Thr Gly Val Pro
Asp Arg Phe Thr Gly 50 55 60Ser Gly Ser Gly Thr Asp Tyr Ala Leu Thr
Ile Asn Ser Val Gln Ala65 70 75 80Glu Asp Leu Ala Leu Tyr Tyr Cys
Gln Gln His Ser Tyr Thr Pro Pro 85 90 95Trp Thr Phe Gly Gly Gly Thr
Lys Leu Glu Ile Arg Arg Ala Asp Ala 100 105 110Ala
Pro180120PRTArtificial Sequence49A1 heavy chain variable region
180Glu Val Gln Leu Gln Glu Ser Gly Pro Ser Leu Val Lys Pro Ser Gln1
5 10 15Thr Leu Ser Leu Thr Cys Ser Val Thr Gly Asp Ser Ile Thr Ser
Gly 20 25 30Tyr Trp Asn Trp Ile Arg Lys Phe Pro Gly Asn Lys Phe Glu
Tyr Met 35 40 45Gly Phe Ile Ser Tyr Ser Gly Asn Thr Tyr Tyr Asn Pro
Ser Leu Arg 50 55 60Ser Arg Ile Ser Ile Thr Arg Asp Thr Ser Lys Asn
Gln Tyr Phe Leu65 70 75 80His Leu Asn Ser Val Thr Thr Glu Asp Thr
Ala Thr Tyr Tyr Cys Ser 85 90 95Arg Arg His Leu Ile Ser Gly Tyr Gly
Trp Phe Ala Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser
Ala 115 120181114PRTArtificial Sequence49A1 light chain variable
region 181Val Ile Val Met Thr Gln Ser His Lys Phe Met Ser Thr Ser
Ile Gly1 5 10 15Asp Arg Val Asn Ile Thr Cys Lys Ala Ser Gln Asp Val
Ile Ser Ala 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Gln Ser Pro
Lys Leu Leu Ile 35 40 45Tyr Trp Ala Ser Thr Arg His Thr Gly Val Pro
Asp Arg Phe Thr Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr
Ile Asn Ser Val Gln Ala65 70 75 80Glu Asp Arg Ala Leu Tyr Tyr Cys
Gln Gln His Ser Tyr Thr Pro Pro 85 90 95Trp Thr Phe Gly Gly Gly Thr
Asn Leu Glu Ile Lys Arg Ala Asp Ala 100 105 110Ala
Pro182121PRTArtificial Sequence9E5 heavy chain variable region
182Gln Val Thr Leu Lys Glu Ser Gly Pro Gly Ile Leu Gln Pro Ser Gln1
5 10 15Thr Leu Ser Leu Thr Cys Thr Phe Ser Gly Phe Ser Leu Ser Thr
Tyr 20 25 30Gly Val Gly Val Gly Trp Ile Arg Gln Pro Ser Gly Lys Gly
Leu Glu 35 40 45Trp Leu Ala Asn Ile Trp Trp Asp Asp Asp Asn Tyr Tyr
Asn Pro Ser 50 55 60Leu Ile His Arg Leu Thr Val Ser Lys Asp Thr Ser
Asn Asn Gln Ala65 70 75
80Phe Leu Lys Ile Thr Asn Val Asp Thr Ala Glu Thr Ala Thr Tyr Tyr
85 90 95Cys Ala Gln Ile Lys Glu Pro Arg Asp Trp Phe Phe Glu Phe Trp
Gly 100 105 110Pro Gly Thr Met Val Ser Val Ser Ser 115
120183113PRTArtificial Sequence9E5 light chain variable region
183Asp Ile Gln Met Thr Gln Thr Pro Ser Ser Met Pro Ala Ser Leu Gly1
5 10 15Glu Arg Val Thr Ile Phe Cys Arg Ala Ser Gln Gly Val Asn Asn
Phe 20 25 30Leu Thr Trp Tyr Gln Gln Lys Pro Asp Gly Thr Ile Lys Pro
Leu Ile 35 40 45Phe Tyr Thr Ser Asn Leu Gln Ser Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Tyr Ser Leu Ser Ile Ser
Ser Leu Glu Pro65 70 75 80Glu Asp Phe Ala Met Tyr Tyr Cys Gln Gln
Tyr His Gly Phe Pro Asn 85 90 95Thr Phe Gly Ala Gly Thr Lys Leu Glu
Leu Lys Arg Ala Asp Ala Ala 100 105 110Pro184121PRTArtificial
Sequence31H6 heavy chain variable region 184Gln Val Thr Leu Lys Glu
Ser Gly Pro Gly Ile Leu Gln Pro Ser Gln1 5 10 15Thr Leu Ser Leu Thr
Cys Thr Phe Ser Gly Phe Ser Leu Ser Thr Tyr 20 25 30Gly Val Gly Val
Gly Trp Ile Arg Gln Pro Ser Gly Lys Gly Leu Glu 35 40 45Trp Leu Ala
Asn Ile Trp Trp Asp Asp Asp Lys Tyr Tyr Asn Pro Ser 50 55 60Leu Lys
Asn Arg Leu Thr Ile Ser Lys Asp Thr Ser Asn Asn Gln Ala65 70 75
80Phe Leu Lys Ile Thr Asn Val Asp Thr Ala Glu Thr Ala Thr Tyr Tyr
85 90 95Cys Ala Gln Ile Lys Glu Pro Arg Asp Trp Phe Phe Glu Phe Trp
Gly 100 105 110Pro Gly Thr Met Val Ser Val Ser Ser 115
120185113PRTArtificial Sequence31H6 light chain variable region
185Asp Ile Gln Met Thr Gln Thr Pro Ser Ser Met Pro Ala Ser Leu Gly1
5 10 15Glu Arg Val Thr Ile Phe Cys Arg Ala Ser Gln Gly Val Asn Asn
Tyr 20 25 30Leu Thr Trp Tyr Gln Gln Lys Pro Asp Gly Thr Ile Lys Pro
Leu Ile 35 40 45Phe Tyr Thr Ser Asn Leu Gln Ser Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Tyr Ser Leu Ser Ile Ser
Ser Leu Glu Pro65 70 75 80Glu Asp Phe Ala Met Tyr Tyr Cys Gln Gln
Tyr His Gly Phe Pro Asn 85 90 95Thr Phe Gly Ala Gly Thr Lys Leu Glu
Leu Lys Arg Ala Asp Ala Ala 100 105 110Pro186118PRTArtificial
Sequencehumanized 36E5 heavy chain variable region 186Gln Val Gln
Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30Ala
Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ala Ser Ile Ser Ser Gly Gly Thr Thr Tyr Tyr Pro Asp Ser Val Lys
50 55 60Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr
Leu65 70 75 80Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr
Tyr Cys Ala 85 90 95Arg Val Gly Gly Tyr Tyr Asp Ser Met Asp Tyr Trp
Gly Gln Gly Thr 100 105 110Leu Val Thr Val Ser Ser
115187112PRTArtificial Sequencehumanized 36E5 light chain variable
regionmisc_feature(31)..(31)Xaa can be any naturally occurring
amino acidmisc_feature(57)..(57)Xaa can be any naturally occurring
amino acid 187Glu Ile Val Leu Thr Gln Ser Pro Gly Thr Leu Ser Leu
Ser Pro Gly1 5 10 15Glu Arg Ala Thr Leu Ser Cys Arg Ala Ser Glu Ser
Val Asp Xaa Tyr 20 25 30Gly Val Ser Phe Met Asn Trp Tyr Gln Gln Lys
Pro Gly Gln Ala Pro 35 40 45Arg Leu Leu Ile Tyr Ala Ala Ser Xaa Gln
Gly Ser Gly Ile Pro Asp 50 55 60Arg Phe Ser Gly Ser Gly Ser Gly Thr
Asp Phe Thr Leu Thr Ile Ser65 70 75 80Arg Leu Glu Pro Glu Asp Phe
Ala Val Tyr Tyr Cys Gln Gln Thr Lys 85 90 95Glu Val Thr Trp Thr Phe
Gly Gln Gly Thr Lys Val Glu Ile Lys Arg 100 105
110188123PRTArtificial Sequencehumanized 3D6 heavy chain variable
regionmisc_feature(97)..(98)Xaa can be any naturally occurring
amino acid 188Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln
Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr
Phe Ser Asp Tyr 20 25 30Tyr Met Ala Trp Val Arg Gln Ala Pro Gly Lys
Gly Leu Glu Trp Val 35 40 45Ala Tyr Ile His Ala Asn Gly Gly Ser Thr
Tyr Tyr Arg Asp Ser Val 50 55 60Arg Gly Arg Phe Thr Ile Ser Arg Asp
Asn Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg
Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Xaa Xaa Gly Ser Phe Met
Tyr Ala Ala Asp Tyr Tyr Ile Met Asp Ala 100 105 110Trp Gly Gln Gly
Thr Leu Val Thr Val Ser Ser 115 120189113PRTArtificial
Sequencehumanized 3D6 light chain variable region 189Asp Ile Val
Met Thr Gln Ser Pro Leu Ser Leu Pro Val Thr Pro Gly1 5 10 15Glu Pro
Ala Ser Ile Ser Cys Arg Ser Ser Gln Ser Leu Leu His Ser 20 25 30Asp
Gly Asn Thr Phe Leu Ser Trp Tyr Leu Gln Lys Pro Gly Gln Ser 35 40
45Pro Gln Leu Leu Ile Tyr Leu Ala Ser Asn Arg Phe Ser Gly Val Pro
50 55 60Asp Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Lys
Ile65 70 75 80Ser Arg Val Glu Ala Glu Asp Val Gly Val Tyr Tyr Cys
Phe Gln His 85 90 95Thr His Leu Pro Leu Thr Phe Gly Gln Gly Thr Lys
Val Glu Ile Lys 100 105 110Arg190118PRTArtificial Sequencehumanized
61G6 heavy chain variable regionmisc_feature(49)..(49)Xaa can be
any naturally occurring amino acidmisc_feature(68)..(68)Xaa can be
any naturally occurring amino acidmisc_feature(72)..(72)Xaa can be
any naturally occurring amino acid 190Gln Val Gln Leu Gln Glu Ser
Gly Pro Gly Leu Val Lys Pro Ser Glu1 5 10 15Thr Leu Ser Leu Thr Cys
Thr Val Ser Gly Tyr Ser Ile Thr Ser Asp 20 25 30Tyr Ala Trp Asn Trp
Ile Arg Gln Pro Pro Gly Lys Gly Leu Glu Trp 35 40 45Xaa Gly Tyr Ile
Ser Tyr Ser Gly Ser Thr Arg Tyr Asn Pro Ser Leu 50 55 60Lys Ser Arg
Xaa Thr Ile Ser Xaa Asp Thr Ser Lys Asn Gln Phe Ser65 70 75 80Leu
Lys Leu Ser Ser Val Thr Ala Ala Asp Thr Ala Val Tyr Tyr Cys 85 90
95Ala Arg Gln Leu Gly Leu Arg Phe Phe Asp Tyr Trp Gly Gln Gly Thr
100 105 110Leu Val Thr Val Ser Ser 115191107PRTArtificial
Sequencehumanized 61G6 light chain variable
regionmisc_feature(45)..(46)Xaa can be any naturally occurring
amino acid 191Glu Ile Val Leu Thr Gln Ser Pro Gly Thr Leu Ser Leu
Ser Pro Gly1 5 10 15Glu Arg Ala Thr Leu Ser Cys Ser Ala Asn Ser Thr
Val Asn Tyr Met 20 25 30Tyr Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro
Arg Xaa Xaa Ile Tyr 35 40 45Leu Thr Ser Asn Leu Ala Ser Gly Ile Pro
Asp Arg Phe Ser Gly Ser 50 55 60Gly Ser Gly Thr Asp Phe Thr Leu Thr
Ile Ser Arg Leu Glu Pro Glu65 70 75 80Asp Phe Ala Val Tyr Tyr Cys
Gln Gln Trp Asn Ser Asn Pro Pro Thr 85 90 95Phe Gly Gln Gly Thr Lys
Val Glu Ile Lys Arg 100 105192118PRTArtificial Sequencehumanized
6H6 heavy chain variable regionmisc_feature(48)..(48)Xaa can be any
naturally occurring amino acidmisc_feature(68)..(68)Xaa can be any
naturally occurring amino acidmisc_feature(70)..(70)Xaa can be any
naturally occurring amino acidmisc_feature(72)..(72)Xaa can be any
naturally occurring amino acid 192Gln Val Gln Leu Val Gln Ser Gly
Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys
Ala Ser Gly Tyr Thr Phe Ser Arg Tyr 20 25 30Trp Ile Glu Trp Val Arg
Gln Ala Pro Gly Gln Gly Leu Glu Trp Xaa 35 40 45Gly Glu Ile Leu Pro
Gly Ser Gly Ser Ser Asn Tyr Asn Glu Lys Phe 50 55 60Lys Asp Arg Xaa
Thr Xaa Thr Xaa Asp Thr Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu
Leu Arg Ser Leu Arg Ser Asp Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala
Arg Lys Val Tyr Tyr Tyr Ala Met Asp Phe Trp Gly Gln Gly Thr 100 105
110Leu Val Thr Val Ser Ser 115193109PRTArtificial Sequencehumanized
6H6 light chain variable regionmisc_feature(48)..(48)Xaa can be any
naturally occurring amino acidmisc_feature(72)..(72)Xaa can be any
naturally occurring amino acid 193Glu Ile Val Leu Thr Gln Ser Pro
Gly Thr Leu Ser Leu Ser Pro Gly1 5 10 15Glu Arg Ala Thr Leu Ser Cys
Thr Ala Ser Ser Ser Val Ser Ser Ser 20 25 30Tyr Phe His Trp Tyr Gln
Gln Lys Pro Gly Gln Ala Pro Arg Leu Xaa 35 40 45Ile Tyr Ser Thr Ser
Asn Leu Ala Ser Gly Ile Pro Asp Arg Phe Ser 50 55 60Gly Ser Gly Ser
Gly Thr Asp Xaa Thr Leu Thr Ile Ser Arg Leu Glu65 70 75 80Pro Glu
Asp Phe Ala Val Tyr Tyr Cys His Gln Tyr His Arg Ser Pro 85 90 95Arg
Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg 100
105194119PRTArtificial Sequencehumanized 61F6 heavy chain variable
regionmisc_feature(48)..(48)Xaa can be any naturally occurring
amino acidmisc_feature(68)..(68)Xaa can be any naturally occurring
amino acidmisc_feature(70)..(70)Xaa can be any naturally occurring
amino acidmisc_feature(72)..(72)Xaa can be any naturally occurring
amino acidmisc_feature(74)..(74)Xaa can be any naturally occurring
amino acid 194Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys
Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr
Phe Thr Ser Tyr 20 25 30Thr Met His Trp Val Arg Gln Ala Pro Gly Gln
Gly Leu Glu Trp Xaa 35 40 45Gly Tyr Ile Asn Pro Arg Ser Val Tyr Thr
Asn Tyr Asn Gln Lys Phe 50 55 60Lys Asp Arg Xaa Thr Xaa Thr Xaa Asp
Xaa Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu Arg Ser Leu Arg
Ser Asp Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Leu Gly Gly Tyr
Tyr Asp Thr Met Asp Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val Thr
Val Ser Ser 115195112PRTArtificial Sequencehumanized 61F6 light
chain variable region 195Asp Ile Gln Met Thr Gln Ser Pro Ser Ser
Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala
Ser Glu Ser Val Asp Asn Tyr 20 25 30Gly Ile Ser Phe Met Asn Trp Tyr
Gln Gln Lys Pro Gly Lys Ala Pro 35 40 45Lys Leu Leu Ile Tyr Ala Ala
Ser Asn Gln Gly Ser Gly Val Pro Ser 50 55 60Arg Phe Ser Gly Ser Gly
Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser65 70 75 80Ser Leu Gln Pro
Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Ser Lys 85 90 95Glu Val Pro
Phe Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg 100 105
110196122PRTArtificial Sequencehumanized 1D8 heavy chain variable
regionmisc_feature(24)..(24)Xaa can be any naturally occurring
amino acidmisc_feature(69)..(69)Xaa can be any naturally occurring
amino acidmisc_feature(73)..(73)Xaa can be any naturally occurring
amino acidmisc_feature(75)..(75)Xaa can be any naturally occurring
amino acidmisc_feature(80)..(80)Xaa can be any naturally occurring
amino acidmisc_feature(98)..(98)Xaa can be any naturally occurring
amino acid 196Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln
Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Xaa Ser Gly Phe Ser
Leu Ser Thr Ser 20 25 30Gly Met Gly Val Gly Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu 35 40 45Trp Val Ala His Ile Trp Trp Asp Asp Asp
Lys Tyr Tyr Ser Pro Ser 50 55 60Leu Lys Ser Arg Xaa Thr Ile Ser Xaa
Asp Xaa Ser Lys Asn Thr Xaa65 70 75 80Tyr Leu Gln Met Asn Ser Leu
Arg Ala Glu Asp Thr Ala Val Tyr Tyr 85 90 95Cys Xaa Arg Ser Tyr Tyr
Tyr Gly Ser Ser Gly Ala Met Asp Tyr Trp 100 105 110Gly Gln Gly Thr
Leu Val Thr Val Ser Ser 115 120197113PRTArtificial
Sequencehumanized 1D8 light chain variable region 197Asp Ile Val
Met Thr Gln Ser Pro Leu Ser Leu Pro Val Thr Pro Gly1 5 10 15Glu Pro
Ala Ser Ile Ser Cys Arg Ser Ser Gln Ser Leu Val His Ser 20 25 30Asp
Gly Asn Thr Tyr Leu His Trp Tyr Leu Gln Lys Pro Gly Gln Ser 35 40
45Pro Gln Leu Leu Ile Tyr Lys Val Ser Lys Arg Phe Ser Gly Val Pro
50 55 60Asp Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Lys
Ile65 70 75 80Ser Arg Val Glu Ala Glu Asp Val Gly Val Tyr Tyr Cys
Ser Gln Ser 85 90 95Thr His Val Pro Pro Thr Phe Gly Gln Gly Thr Lys
Val Glu Ile Lys 100 105 110Arg198117PRTArtificial Sequencehumanized
17F10 heavy chain variable regionmisc_feature(96)..(96)Xaa can be
any naturally occurring amino acid 198Gln Val Gln Leu Val Glu Ser
Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys
Ala Ala Ser Gly Phe Thr Val Arg Asn Tyr 20 25 30Ala Met Ser Trp Val
Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Ser Ile Ser
Thr Gly Asp Arg Ser Tyr Leu Pro Asp Ser Met Lys 50 55 60Gly Arg Phe
Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr Leu65 70 75 80Gln
Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys Xaa 85 90
95Arg Tyr Phe Asp Phe Asp Ser Phe Ala Phe Trp Gly Gln Gly Thr Leu
100 105 110Val Thr Val Ser Ser 115199108PRTArtificial
Sequencehumanized 17F10 light chain variable region 199Asp Ile Gln
Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg
Val Thr Ile Thr Cys Arg Ala Ser Gln Asp Ile Asn Asn Phe 20 25 30Leu
Asn Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40
45Tyr Tyr Thr Ser Lys Leu His Ser Gly Val Pro Ser Arg Phe Ser Gly
50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln
Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Gly His Thr
Leu Pro Pro 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg
100 105200120PRTArtificial Sequencehumanized 35D8 heavy chain
variable regionmisc_feature(47)..(48)Xaa can be any naturally
occurring amino acidmisc_feature(71)..(71)Xaa can be any naturally
occurring amino acidmisc_feature(96)..(96)Xaa can be any naturally
occurring amino acid 200Gln Val Gln Leu Gln Glu Ser Gly Pro Gly Leu
Val Lys Pro Ser Glu1 5 10 15Thr Leu Ser Leu Thr Cys Thr Val Ser Gly
Asp Ser Ile Thr Ser Gly 20 25
30Tyr Trp Asn Trp Ile Arg Gln Pro Pro Gly Lys Gly Leu Glu Xaa Xaa
35 40 45Gly Tyr Ile Ser Tyr Ser Gly Ser Thr Tyr Tyr Asn Pro Ser Leu
Arg 50 55 60Gly Arg Val Thr Ile Ser Xaa Asp Thr Ser Lys Asn Gln Phe
Ser Leu65 70 75 80Lys Leu Ser Ser Val Thr Ala Ala Asp Thr Ala Val
Tyr Tyr Cys Xaa 85 90 95Arg Arg His Leu Gly Ser Gly Tyr Gly Trp Phe
Ala Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser 115
120201109PRTArtificial Sequencehumanized 35D8 light chain variable
regionmisc_feature(71)..(71)Xaa can be any naturally occurring
amino acid 201Asp Ile Val Met Thr Gln Ser Pro Asp Ser Leu Ala Val
Ser Leu Gly1 5 10 15Glu Arg Ala Thr Ile Asn Cys Lys Ala Ser Gln Asp
Val Asn Thr Ala 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Gln Pro
Pro Lys Leu Leu Ile 35 40 45Tyr Trp Ala Ser Thr Arg His Thr Gly Val
Pro Asp Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Xaa Thr Leu
Thr Ile Ser Ser Leu Gln Ala65 70 75 80Glu Asp Val Ala Val Tyr Tyr
Cys Gln Gln His Ser Tyr Thr Pro Pro 85 90 95Trp Thr Phe Gly Gln Gly
Thr Lys Val Glu Ile Lys Arg 100 105202120PRTArtificial
Sequencehumanized 49A1 heavy chain variable
regionmisc_feature(47)..(48)Xaa can be any naturally occurring
amino acidmisc_feature(67)..(67)Xaa can be any naturally occurring
amino acidmisc_feature(71)..(71)Xaa can be any naturally occurring
amino acidmisc_feature(78)..(78)Xaa can be any naturally occurring
amino acidmisc_feature(96)..(96)Xaa can be any naturally occurring
amino acid 202Gln Val Gln Leu Gln Glu Ser Gly Pro Gly Leu Val Lys
Pro Ser Glu1 5 10 15Thr Leu Ser Leu Thr Cys Thr Val Ser Gly Asp Ser
Ile Thr Ser Gly 20 25 30Tyr Trp Asn Trp Ile Arg Gln Pro Pro Gly Lys
Gly Leu Glu Xaa Xaa 35 40 45Gly Phe Ile Ser Tyr Ser Gly Asn Thr Tyr
Tyr Asn Pro Ser Leu Arg 50 55 60Ser Arg Xaa Thr Ile Ser Xaa Asp Thr
Ser Lys Asn Gln Xaa Ser Leu65 70 75 80Lys Leu Ser Ser Val Thr Ala
Ala Asp Thr Ala Val Tyr Tyr Cys Xaa 85 90 95Arg Arg His Leu Ile Ser
Gly Tyr Gly Trp Phe Ala Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val
Thr Val Ser Ser 115 120203109PRTArtificial Sequencehumanized 49A1
light chain variable regionmisc_feature(1)..(1)Xaa can be any
naturally occurring amino acid 203Xaa Ile Val Met Thr Gln Ser Pro
Asp Ser Leu Ala Val Ser Leu Gly1 5 10 15Glu Arg Ala Thr Ile Asn Cys
Lys Ala Ser Gln Asp Val Ile Ser Ala 20 25 30Val Ala Trp Tyr Gln Gln
Lys Pro Gly Gln Pro Pro Lys Leu Leu Ile 35 40 45Tyr Trp Ala Ser Thr
Arg His Thr Gly Val Pro Asp Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly
Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Ala65 70 75 80Glu Asp
Val Ala Val Tyr Tyr Cys Gln Gln His Ser Tyr Thr Pro Pro 85 90 95Trp
Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg 100
105204121PRTArtificial Sequencehumanized 9E5 heavy chain variable
regionmisc_feature(24)..(24)Xaa can be any naturally occurring
amino acidmisc_feature(50)..(51)Xaa can be any naturally occurring
amino acidmisc_feature(69)..(69)Xaa can be any naturally occurring
amino acidmisc_feature(71)..(71)Xaa can be any naturally occurring
amino acidmisc_feature(73)..(73)Xaa can be any naturally occurring
amino acidmisc_feature(80)..(80)Xaa can be any naturally occurring
amino acidmisc_feature(99)..(99)Xaa can be any naturally occurring
amino acid 204Gln Val Gln Leu Gln Glu Ser Gly Pro Gly Leu Val Lys
Pro Ser Glu1 5 10 15Thr Leu Ser Leu Thr Cys Thr Xaa Ser Gly Phe Ser
Leu Ser Thr Tyr 20 25 30Gly Val Gly Val Gly Trp Ile Arg Gln Pro Pro
Gly Lys Gly Leu Glu 35 40 45Trp Xaa Xaa Asn Ile Trp Trp Asp Asp Asp
Asn Tyr Tyr Asn Pro Ser 50 55 60Leu Ile His Arg Xaa Thr Xaa Ser Xaa
Asp Thr Ser Lys Asn Gln Xaa65 70 75 80Ser Leu Lys Leu Ser Ser Val
Thr Ala Ala Asp Thr Ala Val Tyr Tyr 85 90 95Cys Ala Xaa Ile Lys Glu
Pro Arg Asp Trp Phe Phe Glu Phe Trp Gly 100 105 110Gln Gly Thr Leu
Val Thr Val Ser Ser 115 120205108PRTArtificial Sequencehumanized
9E5 light chain variable regionmisc_feature(46)..(46)Xaa can be any
naturally occurring amino acidmisc_feature(49)..(49)Xaa can be any
naturally occurring amino acidmisc_feature(71)..(71)Xaa can be any
naturally occurring amino acid 205Asp Ile Gln Met Thr Gln Ser Pro
Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys
Arg Ala Ser Gln Gly Val Asn Asn Phe 20 25 30Leu Thr Trp Tyr Gln Gln
Lys Pro Gly Lys Ala Pro Lys Xaa Leu Ile 35 40 45Xaa Tyr Thr Ser Asn
Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly
Thr Asp Xaa Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp
Phe Ala Thr Tyr Tyr Cys Gln Gln Tyr His Gly Phe Pro Asn 85 90 95Thr
Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg 100
105206121PRTArtificial Sequencehumanized 31H6 heavy chain variable
regionmisc_feature(24)..(24)Xaa can be any naturally occurring
amino acidmisc_feature(50)..(51)Xaa can be any naturally occurring
amino acidmisc_feature(69)..(69)Xaa can be any naturally occurring
amino acidmisc_feature(73)..(73)Xaa can be any naturally occurring
amino acidmisc_feature(80)..(80)Xaa can be any naturally occurring
amino acidmisc_feature(99)..(99)Xaa can be any naturally occurring
amino acid 206Gln Val Gln Leu Gln Glu Ser Gly Pro Gly Leu Val Lys
Pro Ser Glu1 5 10 15Thr Leu Ser Leu Thr Cys Thr Xaa Ser Gly Phe Ser
Leu Ser Thr Tyr 20 25 30Gly Val Gly Val Gly Trp Ile Arg Gln Pro Pro
Gly Lys Gly Leu Glu 35 40 45Trp Xaa Xaa Asn Ile Trp Trp Asp Asp Asp
Lys Tyr Tyr Asn Pro Ser 50 55 60Leu Lys Asn Arg Xaa Thr Ile Ser Xaa
Asp Thr Ser Lys Asn Gln Xaa65 70 75 80Ser Leu Lys Leu Ser Ser Val
Thr Ala Ala Asp Thr Ala Val Tyr Tyr 85 90 95Cys Ala Xaa Ile Lys Glu
Pro Arg Asp Trp Phe Phe Glu Phe Trp Gly 100 105 110Gln Gly Thr Leu
Val Thr Val Ser Ser 115 120207108PRTArtificial Sequencehumanized
31H6 light chain variable regionmisc_feature(46)..(46)Xaa can be
any naturally occurring amino acidmisc_feature(49)..(49)Xaa can be
any naturally occurring amino acidmisc_feature(71)..(71)Xaa can be
any naturally occurring amino acid 207Asp Ile Gln Met Thr Gln Ser
Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr
Cys Arg Ala Ser Gln Gly Val Asn Asn Tyr 20 25 30Leu Thr Trp Tyr Gln
Gln Lys Pro Gly Lys Ala Pro Lys Xaa Leu Ile 35 40 45Xaa Tyr Thr Ser
Asn Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser
Gly Thr Asp Xaa Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu
Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Tyr His Gly Phe Pro Asn 85 90
95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg 100
105208118PRTArtificial Sequence2155 variable heavy chain 208Glu Val
Lys Leu Val Glu Ser Gly Gly Gly Leu Val Lys Pro Gly Gly1 5 10 15Ser
Leu Lys Leu Ser Cys Gly Ala Ser Gly Phe Thr Ile Ser Ser Tyr 20 25
30Ala Met Ser Trp Val Arg Gln Ser Pro Glu Lys Arg Leu Glu Trp Val
35 40 45Ala Ile Ile Ser Thr Gly Gly Ser Thr Tyr Tyr Pro Asp Ser Val
Arg 50 55 60Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Arg Asn Ser Leu
Tyr Leu65 70 75 80Gln Met Ser Ser Leu Arg Ser Glu Asp Thr Ala Met
Tyr Tyr Cys Ala 85 90 95Arg Val Gly Gly Tyr Tyr Asp Ser Met Asp His
Trp Gly Gln Gly Thr 100 105 110Ser Val Thr Val Ser Ser
115209111PRTArtificial Sequence2155 variable light chain 209Asp Ile
Val Leu Thr Gln Ser Pro Ala Ser Leu Ala Val Ser Leu Gly1 5 10 15Gln
Arg Ala Thr Ile Ser Cys Arg Ala Ser Glu Thr Val Asp Asn Tyr 20 25
30Gly Ile Ser Phe Met Asn Trp Phe Gln Gln Lys Pro Gly Gln Ser Pro
35 40 45Lys Leu Leu Ile Tyr Ala Ala Ser Asn Gln Gly Ser Gly Val Pro
Ala 50 55 60Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Ser Leu Asn
Ile His65 70 75 80Pro Met Glu Glu Asp Asp Thr Ala Met Tyr Phe Cys
Gln Gln Ser Lys 85 90 95Glu Val Pro Trp Thr Phe Gly Gly Gly Thr Lys
Leu Glu Ile Lys 100 105 110210115PRTArtificial Sequence2155
humanized (HC1) heavy chain 210Gln Val Thr Leu Val Glu Ser Gly Gly
Gly Leu Val Lys Pro Gly Gly1 5 10 15Ser Leu Thr Leu Ser Cys Gly Ala
Ser Gly Phe Thr Ile Ser Ser Tyr 20 25 30Ala Met Ser Trp Val Arg Gln
Ser Pro Gly Lys Ala Leu Glu Trp Val 35 40 45Ala Ile Ile Ser Thr Gly
Gly Ser Thr Tyr Tyr Pro Asp Ser Val Arg 50 55 60Gly Arg Phe Thr Ile
Ser Arg Asp Asn Ala Lys Asn Ser Leu Tyr Leu65 70 75 80Thr Met Ser
Ser Leu Asp Ser Val Asp Thr Ala Met Tyr Tyr Cys Ala 85 90 95Arg Val
Gly Gly Tyr Tyr Asp Ser Met Asp His Trp Gly Gln Gly Thr 100 105
110Ser Val Thr 115211115PRTArtificial Sequence2155 humanized (HC2)
heavy chain 211Gln Val Thr Leu Val Glu Ser Gly Gly Gly Leu Val Lys
Pro Gly Gly1 5 10 15Ser Leu Thr Leu Ser Cys Gly Ala Ser Gly Phe Thr
Ile Ser Ser Tyr 20 25 30Ala Met Ser Trp Val Arg Gln Ser Pro Gly Lys
Ala Leu Glu Trp Val 35 40 45Ala Ile Ile Ser Thr Gly Gly Ser Thr Tyr
Tyr Pro Asp Ser Val Arg 50 55 60Gly Arg Phe Thr Ile Ser Arg Asp Asn
Ala Lys Asn Ser Leu Tyr Leu65 70 75 80Thr Met Ser Ser Leu Asp Ser
Val Asp Thr Ala Thr Tyr Tyr Cys Ala 85 90 95Arg Val Gly Gly Tyr Tyr
Asp Ser Met Asp His Trp Gly Gln Gly Thr 100 105 110Ser Val Thr
115212115PRTArtificial Sequence2155 humanized (HC3a) heavy chain
212Gln Val Thr Leu Val Glu Ser Gly Gly Gly Leu Val Lys Pro Gly Gly1
5 10 15Ser Leu Thr Leu Ser Cys Gly Ala Ser Gly Phe Thr Ile Ser Ser
Tyr 20 25 30Ala Met Ser Trp Val Arg Gln Ser Pro Gly Lys Ala Leu Glu
Trp Val 35 40 45Ala Ile Ile Ser Thr Gly Gly Ser Thr Tyr Tyr Pro Asp
Lys Phe Arg 50 55 60Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn
Ser Leu Tyr Leu65 70 75 80Thr Met Ser Ser Leu Arg Ser Glu Asp Thr
Ala Thr Tyr Tyr Cys Ala 85 90 95Arg Val Gly Gly Tyr Tyr Asp Ser Met
Asp His Trp Gly Gln Gly Thr 100 105 110Ser Val Thr
115213115PRTArtificial Sequencehumanized (HC3b) heavy chain 213Gln
Val Thr Leu Lys Glu Ser Gly Gly Gly Leu Val Lys Pro Gly Gly1 5 10
15Ser Leu Thr Leu Ser Cys Gly Ala Ser Gly Phe Thr Ile Ser Ser Tyr
20 25 30Ala Met Ser Trp Val Arg Gln Ser Pro Gly Lys Ala Leu Glu Trp
Val 35 40 45Ala Ile Ile Ser Thr Gly Gly Ser Thr Tyr Tyr Pro Asp Lys
Phe Arg 50 55 60Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn Ser
Leu Tyr Leu65 70 75 80Thr Met Ser Ser Leu Arg Ser Glu Asp Thr Ala
Thr Tyr Tyr Cys Ala 85 90 95Arg Val Gly Gly Tyr Tyr Asp Ser Met Asp
His Trp Gly Gln Gly Thr 100 105 110Ser Val Thr
115214115PRTArtificial Sequencehumanized (HC4) heavy chain 214Glu
Val Gln Leu Val Glu Ser Gly Gly Gly Leu Ile Gln Pro Gly Gly1 5 10
15Ser Leu Lys Leu Ser Cys Ala Ala Ser Gly Phe Thr Ile Ser Ser Tyr
20 25 30Ala Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Val 35 40 45Ala Ile Ile Ser Thr Gly Gly Ser Thr Tyr Tyr Ala Asp Ser
Val Lys 50 55 60Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr
Leu Tyr Leu65 70 75 80Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala
Val Tyr Tyr Cys Ala 85 90 95Arg Val Gly Gly Tyr Tyr Asp Ser Met Asp
His Trp Gly Gln Gly Thr 100 105 110Ser Val Thr
115215109PRTArtificial Sequence2155 humanized (LC1) light chain
215Asp Ile Val Leu Thr Gln Ser Pro Ala Ser Leu Ala Ala Ser Val Gly1
5 10 15Asp Arg Ala Thr Ile Ser Cys Arg Ala Ser Glu Thr Val Asp Asn
Tyr 20 25 30Gly Ile Ser Phe Met Asn Trp Phe Gln Gln Lys Pro Gly Lys
Ser Pro 35 40 45Lys Leu Leu Ile Tyr Ala Ala Ser Asn Gln Gly Ser Gly
Val Pro Ala 50 55 60Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Ser
Leu Asn Ile His65 70 75 80Pro Met Gln Pro Asp Asp Thr Ala Thr Tyr
Phe Cys Gln Gln Ser Lys 85 90 95Glu Val Pro Trp Thr Phe Gly Gly Gly
Thr Lys Leu Glu 100 105216109PRTArtificial Sequence2155 humanized
(LC2a) light chain 216Asp Ile Val Leu Thr Gln Ser Pro Ala Ser Leu
Ser Ala Ser Val Gly1 5 10 15Asp Arg Ala Thr Ile Ser Cys Arg Ala Ser
Glu Thr Val Asp Asn Tyr 20 25 30Gly Ile Ser Phe Met Asn Trp Phe Gln
Gln Lys Pro Gly Gln Ser Pro 35 40 45Lys Leu Leu Ile Tyr Ala Ala Ser
Asn Gln Gly Ser Gly Val Pro Ala 50 55 60Arg Phe Ser Gly Ser Gly Ser
Gly Thr Asp Phe Ser Leu Thr Ile Ser65 70 75 80Pro Met Gln Pro Asp
Asp Thr Ala Thr Tyr Tyr Cys Gln Gln Ser Lys 85 90 95Glu Val Pro Trp
Thr Phe Gly Gly Gly Thr Lys Leu Glu 100 105217109PRTArtificial
Sequence2155 humanized (LC2b) light chain 217Asp Ile Val Leu Thr
Gln Ser Pro Ala Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Ala Thr
Ile Ser Cys Arg Ala Ser Glu Thr Val Asp Asn Tyr 20 25 30Gly Ile Ser
Tyr Met Asn Trp Phe Gln Gln Lys Pro Gly Gln Ser Pro 35 40 45Lys Leu
Leu Ile Tyr Ala Ala Ser Asn Gln Gly Ser Gly Val Pro Ala 50 55 60Arg
Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Ser Leu Thr Ile Ser65 70 75
80Pro Met Gln Pro Asp Asp Thr Ala Thr Tyr Tyr Cys Gln Gln Ser Lys
85 90 95Glu Val Pro Trp Thr Phe Gly Gly Gly Thr Lys Leu Glu 100
105218109PRTArtificial Sequence2155 humanized (LC3) light chain
218Asp Ile Val Leu Thr Gln Ser Pro Ala Ser Leu Ala Val Ser Pro Gly1
5 10 15Gln Arg Ala Thr Ile Thr Cys Arg Ala Ser Glu Thr Val Asp Asn
Tyr 20 25 30Gly Ile Ser Phe Met Asn Trp Phe Gln Gln Lys Pro Gly Gln
Pro Pro 35 40 45Lys Leu Leu Ile
Tyr Ala Ala Ser Asn Gln Gly Ser Gly Val Pro Ala 50 55 60Arg Phe Ser
Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Asn65 70 75 80Pro
Val Glu Ala Asp Asp Thr Ala Asn Tyr Tyr Cys Gln Gln Ser Lys 85 90
95Glu Val Pro Trp Thr Phe Gly Gln Gly Thr Lys Val Glu 100
105219118PRTArtificial Sequence698 variable heavy chain 219Glu Val
Gln Leu Gln Gln Ser Gly Thr Val Leu Ala Arg Pro Gly Ala1 5 10 15Ser
Val Lys Met Ser Cys Glu Ala Ser Gly Tyr Ser Phe Thr Thr Tyr 20 25
30Trp Met His Trp Ile Lys Gln Arg Pro Gly Gln Gly Leu Glu Trp Ile
35 40 45Gly Ala Ile Tyr Pro Gly Asn Ser Asp Thr Gly Tyr Asn Gln Lys
Phe 50 55 60Lys Gly Lys Ala Lys Leu Thr Ala Val Thr Ser Ala Thr Thr
Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Thr Asp Glu Asp Ser Ala
Val Tyr Tyr Cys 85 90 95Thr Arg Thr Ser Thr Tyr Pro His Phe Asp Tyr
Trp Gly Gln Gly Thr 100 105 110Thr Leu Thr Val Ser Ser
115220107PRTArtificial Sequence698 variable light chain 220Asp Ile
Leu Leu Thr Gln Ser Pro Ala Ile Leu Ser Val Ser Pro Gly1 5 10 15Glu
Arg Val Ser Phe Ser Cys Arg Ala Ser Gln Ser Ile Gly Thr Ser 20 25
30Ile His Trp Tyr Gln Gln Arg Thr Asn Gly Ser Pro Arg Leu Leu Ile
35 40 45Lys Tyr Ala Ser Glu Ser Ile Ser Gly Ile Pro Ser Arg Phe Ser
Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Asn Ile Asn Ser Val
Glu Ser65 70 75 80Glu Asp Ile Ala Asp Tyr Tyr Cys Gln Gln Ser Asn
Asn Trp Pro Leu 85 90 95Thr Phe Gly Ala Gly Thr Lys Leu Glu Leu Lys
100 105221118PRTArtificial Sequence706 variable heavy chain 221Glu
Val Gln Leu Gln Gln Ser Gly Thr Val Leu Ala Arg Pro Gly Ala1 5 10
15Ser Val Lys Met Ser Cys Glu Ala Ser Gly Tyr Ser Phe Thr Thr Tyr
20 25 30Trp Met His Trp Ile Lys Gln Arg Pro Gly Gln Gly Leu Glu Trp
Ile 35 40 45Gly Ala Ile Tyr Pro Gly Asn Ser Asp Thr Gly Tyr Asn Gln
Lys Phe 50 55 60Lys Gly Lys Ala Lys Leu Thr Ala Val Thr Ser Ala Ser
Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Thr Asn Glu Asp Ser
Ala Val Tyr Tyr Cys 85 90 95Thr Arg Thr Ser Thr Tyr Pro His Phe Asp
Tyr Trp Gly Gln Gly Thr 100 105 110Thr Leu Thr Val Ser Ser
115222107PRTArtificial Sequence706 variable light chain 222Asp Ile
Leu Leu Thr Gln Ser Pro Ala Ile Leu Ser Val Ser Pro Gly1 5 10 15Glu
Arg Val Ser Phe Ser Cys Arg Ala Ser Gln Ser Ile Gly Thr Ser 20 25
30Ile His Trp Tyr Gln Gln Arg Thr Asn Gly Ser Pro Arg Leu Leu Ile
35 40 45Lys Tyr Ala Ser Glu Ser Ile Ser Gly Ile Pro Ser Arg Phe Ser
Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Asn Ile Asn Ser Val
Glu Ser65 70 75 80Glu Asp Ile Ala Asp Tyr Tyr Cys Gln Gln Thr Asn
Asn Trp Pro Leu 85 90 95Thr Phe Gly Ala Gly Thr Lys Leu Glu Leu Lys
100 105223118PRTArtificial Sequence827 variable heavy chain 223Glu
Val Gln Leu Gln Gln Ser Gly Thr Val Leu Ala Arg Pro Gly Ala1 5 10
15Ser Val Lys Met Ser Cys Glu Thr Ser Gly Tyr Ser Phe Thr Thr Tyr
20 25 30Trp Ile His Trp Ile Lys Gln Arg Pro Gly Gln Gly Leu Glu Trp
Ile 35 40 45Ala Thr Ile Tyr Pro Gly Asn Ser Asp Ala Gly Tyr Asn Gln
Lys Phe 50 55 60Arg Gly Lys Ala Lys Leu Thr Ala Val Thr Ser Ala Ser
Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Thr Asn Glu Asp Ser
Ala Val Tyr Tyr Cys 85 90 95Thr Arg Ser Ser Thr Tyr Pro His Phe Asp
Tyr Trp Gly Gln Gly Thr 100 105 110Thr Leu Thr Val Ser Ser
115224107PRTArtificial Sequence827 variable light chain 224Asp Ile
Leu Leu Thr Gln Ser Pro Ala Ile Leu Ser Val Ser Pro Gly1 5 10 15Glu
Arg Val Ser Phe Ser Cys Arg Ala Ser Gln Ser Ile Gly Thr Ser 20 25
30Ile His Trp Tyr Gln Gln Arg Thr Asn Asp Ser Pro Arg Leu Leu Ile
35 40 45Lys Tyr Ala Ser Glu Ser Ile Ser Gly Ile Pro Ser Arg Phe Ser
Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Asn Ile Asn Ser Val
Glu Ser65 70 75 80Glu Asp Ile Ala Asp Tyr Tyr Cys Gln Gln Thr Asn
Asn Trp Pro Leu 85 90 95Thr Phe Gly Ala Gly Thr Lys Leu Glu Leu Lys
100 105225118PRTArtificial Sequence1718 variable heavy chain 225Gln
Val Gln Val Gln Gln Ser Gly Pro Glu Leu Val Lys Pro Gly Ala1 5 10
15Ser Val Arg Ile Ser Cys Lys Ala Ser Asp Tyr Thr Phe Thr Asn Tyr
20 25 30Tyr Ile His Trp Val Arg Gln Arg Pro Gly Gln Gly Leu Glu Trp
Leu 35 40 45Gly Trp Ile Tyr Pro Gly Lys Gly Tyr Thr Asn Tyr Asn Glu
Lys Phe 50 55 60Lys Gly Lys Ala Thr Leu Thr Ala Asp Lys Ser Ser Ser
Thr Ala Tyr65 70 75 80Met Gln Phe Ser Ser Leu Thr Ser Glu Asp Ser
Ala Val Tyr Phe Cys 85 90 95Ala Ser Gly Tyr Gly Asn Tyr Tyr Phe Pro
Tyr Trp Gly Gln Gly Thr 100 105 110Leu Val Thr Val Ser Ala
115226106PRTArtificial Sequence1718 variable light chain 226Ile Gln
Met Thr Gln Ser Ser Ser Tyr Leu Ser Val Ser Leu Gly Gly1 5 10 15Arg
Val Thr Ile Thr Cys Lys Ala Ser Asp His Ile Lys Asn Trp Leu 20 25
30Ala Trp Tyr Gln Gln Lys Pro Gly Asn Val Pro Arg Leu Leu Met Ser
35 40 45Ala Ala Thr Ser Leu Glu Thr Gly Phe Pro Ser Arg Phe Ser Gly
Ser 50 55 60Gly Ser Gly Lys Asp Phe Thr Leu Thr Ile Thr Ser Leu Gln
Thr Glu65 70 75 80Asp Val Ala Thr Tyr Tyr Cys Gln Gln Tyr Trp Ser
Thr Pro Trp Thr 85 90 95Phe Gly Gly Gly Thr Lys Leu Glu Ile Lys 100
10522710PRTArtificial Sequence2155 heavy chain CDR3 227Val Gly Gly
Tyr Tyr Asp Ser Met Asp His1 5 102289PRTArtificial Sequence2155
heavy chain CDR2 228Ile Ile Ser Thr Gly Gly Ser Thr Tyr1
522910PRTArtificial Sequence2155 heavy chain CDR1 229Gly Phe Thr
Ile Ser Ser Tyr Ala Met Ser1 5 102309PRTArtificial Sequence2155
light chain CDR3 230Gln Gln Ser Lys Glu Val Pro Trp Thr1
52317PRTArtificial Sequence2155 light chain CDR2 231Ala Ala Ser Asn
Gln Gly Ser1 523215PRTArtificial Sequence2155 light chain CDR1
232Arg Ala Ser Glu Thr Val Asp Asn Tyr Gly Ile Ser Phe Met Asn1 5
10 152339PRTArtificial Sequence698 and 706 heavy chain CDR3 233Thr
Ser Thr Tyr Pro His Phe Asp Tyr1 523410PRTArtificial Sequence698
and 706 heavy chain CDR2 234Ala Ile Tyr Pro Gly Asn Ser Asp Thr
Gly1 5 1023510PRTArtificial Sequence698 and 706 heavy chain CDR1
235Gly Tyr Ser Phe Thr Thr Tyr Trp Met His1 5 102369PRTArtificial
Sequence698 light chain CDR3 236Gln Gln Ser Asn Asn Trp Pro Leu
Thr1 52378PRTArtificial Sequence698, 706, 827, and 1649 light chain
CDR2 237Lys Tyr Ala Ser Glu Ser Ile Ser1 523811PRTArtificial
Sequence698, 706, 827, and 1649 light chain CDR1 238Arg Ala Ser Gln
Ser Ile Gly Thr Ser Ile His1 5 102399PRTArtificial Sequence706,
827, and 1649 light chain CDR3 239Gln Gln Thr Asn Asn Trp Pro Leu
Thr1 52409PRTArtificial Sequence827 and 1649 heavy chain CDR3
240Ser Ser Thr Tyr Pro His Phe Asp Tyr1 524110PRTArtificial
Sequence827 heavy chain CDR2 241Thr Ile Tyr Pro Gly Asn Ser Asp Ala
Gly1 5 1024210PRTArtificial Sequence1649 heavy chain CDR2 242Ala
Ile Tyr Pro Gly Asn Ser Asp Ala Gly1 5 102439PRTArtificial
Sequence1718 heavy chain CDR3 243Gly Tyr Gly Asn Tyr Tyr Phe Pro
Tyr1 524410PRTArtificial Sequence1718 heavy chain CDR2 244Trp Ile
Tyr Pro Gly Lys Gly Tyr Thr Asn1 5 102459PRTArtificial Sequence1718
heavy chain CDR1 245Asp Tyr Thr Phe Thr Asn Tyr Tyr Ile1
52469PRTArtificial Sequence1718 light chain CDR3 246Gln Gln Thr Trp
Ser Thr Pro Trp Thr1 52477PRTArtificial Sequence1718 light chain
CDR2 247Ala Ala Thr Ser Leu Glu Thr1 524811PRTArtificial
Sequence1718 light chain CDR1 248Lys Ala Ser Asp His Ile Lys Asn
Trp Leu Ala1 5 1024910PRTArtificial Sequence827 and 1649 heavy
chain CDR1 249Gly Tyr Ser Phe Thr Thr Tyr Trp Ile His1 5
10250453PRTArtificial Sequence1D7 heavy chain 250Gln Val Gln Leu
Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg
Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30Gly Met
His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Thr
Val Ile Trp Tyr Glu Gly Ser Asn Lys Tyr Tyr Ala Asp Ser Val 50 55
60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr65
70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr
Cys 85 90 95Ala Arg Gly Gly Gln Leu Gly Lys Tyr Tyr Tyr Tyr Gly Met
Asp Val 100 105 110Trp Gly Gln Gly Thr Thr Val Thr Val Ser Ser Ala
Ser Thr Lys Gly 115 120 125Pro Ser Val Phe Pro Leu Ala Pro Ser Ser
Lys Ser Thr Ser Gly Gly 130 135 140Thr Ala Ala Leu Gly Cys Leu Val
Lys Asp Tyr Phe Pro Glu Pro Val145 150 155 160Thr Val Ser Trp Asn
Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe 165 170 175Pro Ala Val
Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val 180 185 190Thr
Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val 195 200
205Asn His Lys Pro Ser Asn Thr Lys Val Asp Lys Arg Val Glu Pro Lys
210 215 220Ser Cys Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Glu Leu225 230 235 240Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr 245 250 255Leu Met Ile Ser Arg Thr Pro Glu Val
Thr Cys Val Val Val Asp Val 260 265 270Ser His Glu Asp Pro Glu Val
Lys Phe Asn Trp Tyr Val Asp Gly Val 275 280 285Glu Val His Asn Ala
Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser 290 295 300Thr Tyr Arg
Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu305 310 315
320Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala
325 330 335Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg
Glu Pro 340 345 350Gln Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu Met
Thr Lys Asn Gln 355 360 365Val Ser Leu Thr Cys Leu Val Lys Gly Phe
Tyr Pro Ser Asp Ile Ala 370 375 380Val Glu Trp Glu Ser Asn Gly Gln
Pro Glu Asn Asn Tyr Lys Thr Thr385 390 395 400Pro Pro Val Leu Asp
Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu 405 410 415Thr Val Asp
Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser 420 425 430Val
Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser 435 440
445Leu Ser Pro Gly Lys 450251214PRTArtificial Sequence1D7 light
chain 251Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser
Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Gly Ile
Arg Asn Asp 20 25 30Leu Gly Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro
Lys Arg Leu Ile 35 40 45Tyr Asp Ala Ser Ser Leu Gln Ser Gly Val Pro
Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Glu Phe Thr Leu Thr
Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys
Leu Gln His Asn Asn Tyr Pro Trp 85 90 95Thr Phe Gly Gln Gly Thr Lys
Val Glu Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile
Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser
Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys
Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150
155 160Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu
Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His
Lys Val Tyr 180 185 190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser
Pro Val Thr Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys
210252123PRTArtificial Sequence1D7 variable heavy chain 252Gln Val
Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser
Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25
30Gly Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val
35 40 45Thr Val Ile Trp Tyr Glu Gly Ser Asn Lys Tyr Tyr Ala Asp Ser
Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr
Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala
Val Tyr Tyr Cys 85 90 95Ala Arg Gly Gly Gln Leu Gly Lys Tyr Tyr Tyr
Tyr Gly Met Asp Val 100 105 110Trp Gly Gln Gly Thr Thr Val Thr Val
Ser Ser 115 120253108PRTArtificial Sequence1D7 variable light chain
253Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1
5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Gly Ile Arg Asn
Asp 20 25 30Leu Gly Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Arg
Leu Ile 35 40 45Tyr Asp Ala Ser Ser Leu Gln Ser Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Leu Gln
His Asn Asn Tyr Pro Trp 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu
Ile Lys Arg 100 1052545PRTArtificial Sequence1D7 heavy chain CDR1
254Ser Tyr Gly Met His1 525517PRTArtificial Sequence1D7 heavy chain
CDR2 255Val Ile Trp Tyr Glu Gly Ser Asn Lys Tyr Tyr Ala Asp Ser Val
Lys1 5 10 15Gly25614PRTArtificial Sequence1D7 heavy chain CDR3
256Gly Gly Gln Leu Gly Lys Tyr Tyr Tyr Tyr Gly Met Asp Val1 5
1025711PRTArtificial Sequence1D7 light chain CDR1 257Arg Ala Ser
Gln Gly Ile Arg Asn Asp Leu Gly1 5 102587PRTArtificial Sequence1D7
light chain CDR2 258Asp Ala Ser Ser Leu Gln Ser1 52599PRTArtificial
Sequence1D7 light chain CDR3 259Leu Gln His Asn Asn Tyr Pro Trp
Thr1 5260453PRTArtificial Sequence33C9 heavy chain 260Gln Val Gln
Val Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30Gly
Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ser
Val Ile Trp Tyr Glu Gly Ser Asn Lys Tyr Tyr Ala Asp Ser Val 50 55
60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr65
70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr
Cys 85 90 95Ala Arg Gly Gly Leu Leu Gly Tyr Tyr Tyr Tyr Tyr Gly Met
Asp Val 100 105 110Trp Gly Gln Gly Thr Thr Val Thr Val Ser Ser Ala
Ser Thr Lys Gly 115 120 125Pro Ser Val Phe Pro Leu Ala Pro Ser Ser
Lys Ser Thr Ser Gly Gly 130 135 140Thr Ala Ala Leu Gly Cys Leu Val
Lys Asp Tyr Phe Pro Glu Pro Val145 150 155 160Thr Val Ser Trp Asn
Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe 165 170 175Pro Ala Val
Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val 180 185 190Thr
Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val 195 200
205Asn His Lys Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys
210 215 220Ser Cys Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Glu Leu225 230 235 240Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr 245 250 255Leu Met Ile Ser Arg Thr Pro Glu Val
Thr Cys Val Val Val Asp Val 260 265 270Ser His Glu Asp Pro Glu Val
Lys Phe Asn Trp Tyr Val Asp Gly Val 275 280 285Glu Val His Asn Ala
Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser 290 295 300Thr Tyr Arg
Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu305 310 315
320Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala
325 330 335Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg
Glu Pro 340 345 350Gln Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu Met
Thr Lys Asn Gln 355 360 365Val Ser Leu Thr Cys Leu Val Lys Gly Phe
Tyr Pro Ser Asp Ile Ala 370 375 380Val Glu Trp Glu Ser Asn Gly Gln
Pro Glu Asn Asn Tyr Lys Thr Thr385 390 395 400Pro Pro Val Leu Asp
Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu 405 410 415Thr Val Asp
Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser 420 425 430Val
Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser 435 440
445Leu Ser Pro Gly Lys 450261214PRTArtificial Sequence33C9 light
chain 261Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser
Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Gly Ile
Arg Asn Asp 20 25 30Leu Gly Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro
Lys Arg Leu Ile 35 40 45Tyr Asp Ala Ser Ser Leu Gln Ser Gly Val Pro
Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Glu Phe Thr Leu Thr
Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys
Leu Gln His His Ser Tyr Pro Trp 85 90 95Thr Phe Gly Gln Gly Thr Lys
Val Glu Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile
Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser
Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys
Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150
155 160Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu
Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His
Lys Val Tyr 180 185 190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser
Pro Val Thr Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys
210262123PRTArtificial Sequence33C9 variable heavy chain 262Gln Val
Gln Val Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser
Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25
30Gly Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val
35 40 45Ser Val Ile Trp Tyr Glu Gly Ser Asn Lys Tyr Tyr Ala Asp Ser
Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr
Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala
Val Tyr Tyr Cys 85 90 95Ala Arg Gly Gly Leu Leu Gly Tyr Tyr Tyr Tyr
Tyr Gly Met Asp Val 100 105 110Trp Gly Gln Gly Thr Thr Val Thr Val
Ser Ser 115 120263108PRTArtificial Sequence33C9 variable light
chain 263Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser
Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Gly Ile
Arg Asn Asp 20 25 30Leu Gly Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro
Lys Arg Leu Ile 35 40 45Tyr Asp Ala Ser Ser Leu Gln Ser Gly Val Pro
Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Glu Phe Thr Leu Thr
Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys
Leu Gln His His Ser Tyr Pro Trp 85 90 95Thr Phe Gly Gln Gly Thr Lys
Val Glu Ile Lys Arg 100 1052645PRTArtificial Sequence33C9 heavy
chain CDR1 264Ser Tyr Gly Met His1 526517PRTArtificial Sequence33C9
heavy chain CDR2 265Val Ile Trp Tyr Glu Gly Ser Asn Lys Tyr Tyr Ala
Asp Ser Val Lys1 5 10 15Gly26614PRTArtificial Sequence33C9 heavy
chain CDR3 266Gly Gly Leu Leu Gly Tyr Tyr Tyr Tyr Tyr Gly Met Asp
Val1 5 1026711PRTArtificial Sequence33C9 light chain CDR1 267Arg
Ala Ser Gln Gly Ile Arg Asn Asp Leu Gly1 5 102687PRTArtificial
Sequence33C9 light chain CDR2 268Asp Ala Ser Ser Leu Gln Ser1
52699PRTArtificial Sequence33C9 light chain CDR3 269Leu Gln His His
Ser Tyr Pro Trp Thr1 5270453PRTArtificial Sequence33F6 heavy chain
270Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1
5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Asn
Tyr 20 25 30Gly Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu
Trp Val 35 40 45Ala Val Ile Trp Tyr Val Gly Ser Asn Lys Tyr Tyr Ala
Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys
Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Gly Gly Glu Leu Arg Leu Tyr
Tyr Tyr Tyr Gly Met Asp Val 100 105 110Trp Gly Gln Gly Thr Thr Val
Thr Val Ser Ser Ala Ser Thr Lys Gly 115 120 125Pro Ser Val Phe Pro
Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly 130 135 140Thr Ala Ala
Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val145 150 155
160Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe
165 170 175Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser
Val Val 180 185 190Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr
Ile Cys Asn Val 195 200 205Asn His Lys Pro Ser Asn Thr Lys Val Asp
Lys Lys Val Glu Pro Lys 210 215 220Ser Cys Asp Lys Thr His Thr Cys
Pro Pro Cys Pro Ala Pro Glu Leu225 230 235 240Leu Gly Gly Pro Ser
Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr 245 250 255Leu Met Ile
Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val 260 265 270Ser
His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val 275 280
285Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser
290 295 300Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp
Trp Leu305 310 315 320Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn
Lys Ala Leu Pro Ala 325 330 335Pro Ile Glu Lys Thr Ile Ser Lys Ala
Lys Gly Gln Pro Arg Glu Pro 340 345 350Gln Val Tyr Thr Leu Pro Pro
Ser Arg Glu Glu Met Thr Lys Asn Gln 355 360 365Val Ser Leu Thr Cys
Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala 370 375 380Val Glu Trp
Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr385 390 395
400Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu
405 410 415Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser
Cys Ser 420 425 430Val Met His Glu Ala Leu His Asn His Tyr Thr Gln
Lys Ser Leu Ser 435 440 445Leu Ser Pro Gly Lys
450271214PRTArtificial Sequence33F6 light chain 271Asp Ile Gln Met
Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val
Thr Ile Thr Cys Arg Ala Ser Gln Gly Ile Arg Asn Asp 20 25 30Leu Gly
Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr
Ala Ala Ser Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55
60Ser Gly Ser Gly Thr Glu Phe Thr Leu Thr Val Ser Ser Leu Gln Pro65
70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Leu Gln Leu Asn Ser Tyr Pro
Trp 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg Thr Val
Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln
Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn
Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn
Ala Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu
Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu
Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala
Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200
205Phe Asn Arg Gly Glu Cys 210272123PRTArtificial Sequence33F6
variable heavy chain 272Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val
Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Ser Asn Tyr 20 25 30Gly Met His Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Val Ile Trp Tyr Val Gly Ser
Asn Lys Tyr Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser
Arg Asp Asn Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser
Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Gly Gly
Glu Leu Arg Leu Tyr Tyr Tyr Tyr Gly Met Asp Val 100 105 110Trp Gly
Gln Gly Thr Thr Val Thr Val Ser Ser 115 120273108PRTArtificial
Sequence33F6 variable light chain 273Asp Ile Gln Met Thr Gln Ser
Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr
Cys Arg Ala Ser Gln Gly Ile Arg Asn Asp 20 25 30Leu Gly Trp Tyr Gln
Gln Lys Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr Ala Ala Ser
Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser
Gly Thr Glu Phe Thr Leu Thr Val Ser Ser Leu Gln Pro65 70 75 80Glu
Asp Phe Ala Thr Tyr Tyr Cys Leu Gln Leu Asn Ser Tyr Pro Trp 85 90
95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg 100
1052745PRTArtificial Sequence33F6 heavy chain CDR1 274Asn Tyr Gly
Met His1 527517PRTArtificial Sequence33F6 heavy chain CDR2 275Val
Ile Trp Tyr Val Gly Ser Asn Lys Tyr Tyr Ala Asp Ser Val Lys1 5 10
15Gly27614PRTArtificial Sequence33F6 heavy chain CDR3 276Gly Gly
Glu Leu Arg Leu Tyr Tyr Tyr Tyr Gly Met Asp Val1 5
1027711PRTArtificial Sequence33F6 light chain CDR1 277Arg Ala Ser
Gln Gly Ile Arg Asn Asp Leu Gly1 5 102787PRTArtificial Sequence33F6
light chain CDR2 278Ala Ala Ser Ser Leu Gln Ser1 52799PRTArtificial
Sequence33F6 light chain CDR3 279Leu Gln Leu Asn Ser Tyr Pro Trp
Thr1 5280453PRTArtificial Sequence34G4 heavy chain 280Gln Val Gln
Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30Gly
Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ala Val Ile Trp Tyr Glu Gly Ser Asn Lys Tyr Tyr Ala Asp Ser Val
50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu
Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val
Tyr Tyr Cys 85 90 95Ala Arg Gly Gly Gln Leu Gly Tyr Tyr Tyr Tyr Tyr
Gly Met Asp Val 100 105 110Trp Gly Gln Gly Thr Thr Val Thr Val Ser
Ser Ala Ser Thr Lys Gly 115 120 125Pro Ser Val Phe Pro Leu Ala Pro
Ser Ser Lys Ser Thr Ser Gly Gly 130 135 140Thr Ala Ala Leu Gly Cys
Leu Val Lys Asp Tyr Phe Pro Glu Pro Val145 150 155 160Thr Val Ser
Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe 165 170 175Pro
Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val 180 185
190Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val
195 200 205Asn His Lys Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu
Pro Lys 210 215 220Ser Cys Asp Lys Thr His Thr Cys Pro Pro Cys Pro
Ala Pro Glu Leu225 230 235 240Leu Gly Gly Pro Ser Val Phe Leu Phe
Pro Pro Lys Pro Lys Asp Thr 245 250 255Leu Met Ile Ser Arg Thr Pro
Glu Val Thr Cys Val Val Val Asp Val 260 265 270Ser His Glu Asp Pro
Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val 275 280 285Glu Val His
Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser 290 295 300Thr
Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu305 310
315 320Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro
Ala 325 330 335Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro
Arg Glu Pro 340 345 350Gln Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu
Met Thr Lys Asn Gln 355 360 365Val Ser Leu Thr Cys Leu Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala 370 375 380Val Glu Trp Glu Ser Asn Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr385 390 395 400Pro Pro Val Leu
Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu 405 410 415Thr Val
Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser 420 425
430Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser
435 440 445Leu Ser Pro Gly Lys 450281214PRTArtificial Sequence34G4
light chain 281Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala
Ser Val Gly1 5
10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Gly Ile Arg Asn
Asp 20 25 30Leu Gly Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Arg
Leu Ile 35 40 45Tyr Asp Ala Ser Ser Leu Gln Ser Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Leu Gln
Leu Asn Ser Tyr Pro Trp 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu
Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro
Pro Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val
Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln
Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150 155
160Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser
165 170 175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys
Val Tyr 180 185 190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro
Val Thr Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys
210282123PRTArtificial Sequence34G4 variable heavy chain 282Gln Val
Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser
Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25
30Gly Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val
35 40 45Ala Val Ile Trp Tyr Glu Gly Ser Asn Lys Tyr Tyr Ala Asp Ser
Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr
Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala
Val Tyr Tyr Cys 85 90 95Ala Arg Gly Gly Gln Leu Gly Tyr Tyr Tyr Tyr
Tyr Gly Met Asp Val 100 105 110Trp Gly Gln Gly Thr Thr Val Thr Val
Ser Ser 115 120283108PRTArtificial Sequence34G4 variable light
chain 283Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser
Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Gly Ile
Arg Asn Asp 20 25 30Leu Gly Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro
Lys Arg Leu Ile 35 40 45Tyr Asp Ala Ser Ser Leu Gln Ser Gly Val Pro
Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr
Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys
Leu Gln Leu Asn Ser Tyr Pro Trp 85 90 95Thr Phe Gly Gln Gly Thr Lys
Val Glu Ile Lys Arg 100 1052845PRTArtificial Sequence34G4 heavy
chain CDR1 284Ser Tyr Gly Met His1 528517PRTArtificial Sequence34G4
heavy chain CDR2 285Val Ile Trp Tyr Glu Gly Ser Asn Lys Tyr Tyr Ala
Asp Ser Val Lys1 5 10 15Gly28614PRTArtificial Sequence34G4 heavy
chain CDR3 286Gly Gly Gln Leu Gly Tyr Tyr Tyr Tyr Tyr Gly Met Asp
Val1 5 1028711PRTArtificial Sequence34G4 light chain CDR1 287Arg
Ala Ser Gln Gly Ile Arg Asn Asp Leu Gly1 5 102887PRTArtificial
Sequence34G4 light chain CDR2 288Asp Ala Ser Ser Leu Gln Ser1
52899PRTArtificial Sequence34G4 light chain CDR3 289Leu Gln Leu Asn
Ser Tyr Pro Trp Thr1 5290453PRTArtificial Sequence35B10 heavy chain
290Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1
5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser
Tyr 20 25 30Gly Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu
Trp Val 35 40 45Ala Val Ile Trp Tyr Ala Gly Ser Asn Lys Tyr Tyr Ala
Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys
Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Gly Gly Glu Leu Ser Phe Tyr
Tyr Tyr Tyr Gly Met Asp Val 100 105 110Trp Gly Gln Gly Thr Thr Val
Thr Val Ser Ser Ala Ser Thr Lys Gly 115 120 125Pro Ser Val Phe Pro
Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly 130 135 140Thr Ala Ala
Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val145 150 155
160Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe
165 170 175Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser
Val Val 180 185 190Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr
Ile Cys Asn Val 195 200 205Asn His Lys Pro Ser Asn Thr Lys Val Asp
Lys Lys Val Glu Pro Lys 210 215 220Ser Cys Asp Lys Thr His Thr Cys
Pro Pro Cys Pro Ala Pro Glu Leu225 230 235 240Leu Gly Gly Pro Ser
Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr 245 250 255Leu Met Ile
Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val 260 265 270Ser
His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val 275 280
285Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser
290 295 300Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp
Trp Leu305 310 315 320Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn
Lys Ala Leu Pro Ala 325 330 335Pro Ile Glu Lys Thr Ile Ser Lys Ala
Lys Gly Gln Pro Arg Glu Pro 340 345 350Gln Val Tyr Thr Leu Pro Pro
Ser Arg Glu Glu Met Thr Lys Asn Gln 355 360 365Val Ser Leu Thr Cys
Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala 370 375 380Val Glu Trp
Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr385 390 395
400Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu
405 410 415Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser
Cys Ser 420 425 430Val Met His Glu Ala Leu His Asn His Tyr Thr Gln
Lys Ser Leu Ser 435 440 445Leu Ser Pro Gly Lys
450291214PRTArtificial Sequence35B10 light chain 291Asp Ile Gln Met
Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val
Thr Ile Thr Cys Arg Ala Ser Gln Gly Ile Arg Asn Asp 20 25 30Leu Gly
Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr
Ala Ala Ser Thr Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55
60Ser Gly Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65
70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Leu Gln His Asn Asn Tyr Pro
Trp 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg Thr Val
Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln
Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn
Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn
Ala Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu
Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu
Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala
Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200
205Phe Asn Arg Gly Glu Cys 210292123PRTArtificial Sequence35B10
variable heavy chain 292Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val
Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Ser Ser Tyr 20 25 30Gly Met His Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Val Ile Trp Tyr Ala Gly Ser
Asn Lys Tyr Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser
Arg Asp Asn Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser
Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Gly Gly
Glu Leu Ser Phe Tyr Tyr Tyr Tyr Gly Met Asp Val 100 105 110Trp Gly
Gln Gly Thr Thr Val Thr Val Ser Ser 115 120293108PRTArtificial
Sequence35B10 variable light chain 293Asp Ile Gln Met Thr Gln Ser
Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr
Cys Arg Ala Ser Gln Gly Ile Arg Asn Asp 20 25 30Leu Gly Trp Tyr Gln
Gln Lys Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr Ala Ala Ser
Thr Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser
Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu
Asp Phe Ala Thr Tyr Tyr Cys Leu Gln His Asn Asn Tyr Pro Trp 85 90
95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg 100
1052945PRTArtificial Sequence35B10 heavy chain CDR1 294Ser Tyr Gly
Met His1 529517PRTArtificial Sequence35B10 heavy chain CDR2 295Val
Ile Trp Tyr Ala Gly Ser Asn Lys Tyr Tyr Ala Asp Ser Val Lys1 5 10
15Gly29614PRTArtificial Sequence35B10 heavy chain CDR3 296Gly Gly
Glu Leu Ser Phe Tyr Tyr Tyr Tyr Gly Met Asp Val1 5
1029711PRTArtificial Sequence35B10 light chain CDR1 297Arg Ala Ser
Gln Gly Ile Arg Asn Asp Leu Gly1 5 102987PRTArtificial
Sequence35B10 light chain CDR2 298Ala Ala Ser Thr Leu Gln Ser1
52999PRTArtificial Sequence35B10 light chain CDR3 299Leu Gln His
Asn Asn Tyr Pro Trp Thr1 5300453PRTArtificial Sequence41E11 heavy
chain 300Gln Val Gln Val Val Glu Ser Gly Gly Gly Val Val Gln Pro
Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe
Ser Ser Tyr 20 25 30Gly Met Tyr Trp Val Arg Gln Ala Pro Gly Lys Gly
Leu Glu Trp Val 35 40 45Ala Val Ile Trp Tyr Glu Gly Ser Asn Lys Tyr
Tyr Ala Asp Ser Val 50 55 60Arg Gly Arg Phe Thr Ile Ser Arg Asp Asn
Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala
Glu Asp Thr Ala Leu Tyr Tyr Cys 85 90 95Ala Arg Gly Gly Gln Leu Gly
Lys Asp Tyr Tyr Ser Gly Met Asp Val 100 105 110Trp Gly Gln Gly Thr
Thr Val Thr Val Ser Ser Ala Ser Thr Lys Gly 115 120 125Pro Ser Val
Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly 130 135 140Thr
Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val145 150
155 160Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr
Phe 165 170 175Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser
Ser Val Val 180 185 190Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr
Tyr Ile Cys Asn Val 195 200 205Asn His Lys Pro Ser Asn Thr Lys Val
Asp Lys Lys Val Glu Pro Lys 210 215 220Ser Cys Asp Lys Thr His Thr
Cys Pro Pro Cys Pro Ala Pro Glu Leu225 230 235 240Leu Gly Gly Pro
Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr 245 250 255Leu Met
Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val 260 265
270Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val
275 280 285Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr
Asn Ser 290 295 300Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His
Gln Asp Trp Leu305 310 315 320Asn Gly Lys Glu Tyr Lys Cys Lys Val
Ser Asn Lys Ala Leu Pro Ala 325 330 335Pro Ile Glu Lys Thr Ile Ser
Lys Ala Lys Gly Gln Pro Arg Glu Pro 340 345 350Gln Val Tyr Thr Leu
Pro Pro Ser Arg Glu Glu Met Thr Lys Asn Gln 355 360 365Val Ser Leu
Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala 370 375 380Val
Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr385 390
395 400Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys
Leu 405 410 415Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe
Ser Cys Ser 420 425 430Val Met His Glu Ala Leu His Asn His Tyr Thr
Gln Lys Ser Leu Ser 435 440 445Leu Ser Pro Gly Lys
450301214PRTArtificial Sequence41E11 light chain 301Asp Ile Gln Met
Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val
Thr Ile Thr Cys Arg Ala Ser Gln Val Ile Arg Asn Asp 20 25 30Leu Gly
Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr
Ala Ala Ser Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55
60Ser Gly Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65
70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Leu Gln His Asn Ser Tyr Pro
Leu 85 90 95Thr Phe Gly Gly Gly Thr Lys Val Glu Ile Lys Arg Thr Val
Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln
Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn
Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn
Ala Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu
Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu
Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala
Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200
205Phe Asn Arg Gly Glu Cys 210302123PRTArtificial Sequence41E11
variable heavy chain 302Gln Val Gln Val Val Glu Ser Gly Gly Gly Val
Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Ser Ser Tyr 20 25 30Gly Met Tyr Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Val Ile Trp Tyr Glu Gly Ser
Asn Lys Tyr Tyr Ala Asp Ser Val 50 55 60Arg Gly Arg Phe Thr Ile Ser
Arg Asp Asn Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser
Leu Arg Ala Glu Asp Thr Ala Leu Tyr Tyr Cys 85 90 95Ala Arg Gly Gly
Gln Leu Gly Lys Asp Tyr Tyr Ser Gly Met Asp Val 100 105 110Trp Gly
Gln Gly Thr Thr Val Thr Val Ser Ser 115 120303108PRTArtificial
Sequence41E11 variable light chain 303Asp Ile Gln Met Thr Gln Ser
Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr
Cys Arg Ala Ser Gln Val Ile Arg Asn Asp 20 25 30Leu Gly Trp Tyr Gln
Gln Lys Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr Ala Ala Ser
Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser
Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu
Asp Phe Ala Thr Tyr Tyr Cys Leu Gln His Asn Ser Tyr Pro
Leu 85 90 95Thr Phe Gly Gly Gly Thr Lys Val Glu Ile Lys Arg 100
1053045PRTArtificial Sequence41E11 heavy chain CDR1 304Ser Tyr Gly
Met Tyr1 530517PRTArtificial Sequence41E11 heavy chain CDR2 305Val
Ile Trp Tyr Glu Gly Ser Asn Lys Tyr Tyr Ala Asp Ser Val Arg1 5 10
15Gly30614PRTArtificial Sequence41E11 heavy chain CDR3 306Gly Gly
Gln Leu Gly Lys Asp Tyr Tyr Ser Gly Met Asp Val1 5
1030711PRTArtificial Sequence41E11 light chain CDR1 307Arg Ala Ser
Gln Val Ile Arg Asn Asp Leu Gly1 5 103087PRTArtificial
Sequence41E11 light chain CDR2 308Ala Ala Ser Ser Leu Gln Ser1
53099PRTArtificial Sequence41E11 light chain CDR3 309Leu Gln His
Asn Ser Tyr Pro Leu Thr1 5310453PRTArtificial Sequence41G5 heavy
chain 310Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro
Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe
Ser Ser Tyr 20 25 30Gly Met His Trp Val Arg Gln Ala Pro Gly Lys Gly
Leu Glu Trp Val 35 40 45Ala Val Ile Trp Tyr Pro Gly Ser Asn Lys Tyr
Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn
Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala
Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Gly Gly Glu Leu Gly
Arg Tyr Tyr Tyr Tyr Gly Met Asp Val 100 105 110Trp Gly Gln Gly Thr
Thr Val Thr Val Ser Ser Ala Ser Thr Lys Gly 115 120 125Pro Ser Val
Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly 130 135 140Thr
Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val145 150
155 160Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr
Phe 165 170 175Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser
Ser Val Val 180 185 190Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr
Tyr Ile Cys Asn Val 195 200 205Asn His Lys Pro Ser Asn Thr Lys Val
Asp Lys Lys Val Glu Pro Lys 210 215 220Ser Cys Asp Lys Thr His Thr
Cys Pro Pro Cys Pro Ala Pro Glu Leu225 230 235 240Leu Gly Gly Pro
Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr 245 250 255Leu Met
Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val 260 265
270Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val
275 280 285Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr
Asn Ser 290 295 300Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His
Gln Asp Trp Leu305 310 315 320Asn Gly Lys Glu Tyr Lys Cys Lys Val
Ser Asn Lys Ala Leu Pro Ala 325 330 335Pro Ile Glu Lys Thr Ile Ser
Lys Ala Lys Gly Gln Pro Arg Glu Pro 340 345 350Gln Val Tyr Thr Leu
Pro Pro Ser Arg Glu Glu Met Thr Lys Asn Gln 355 360 365Val Ser Leu
Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala 370 375 380Val
Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr385 390
395 400Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys
Leu 405 410 415Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe
Ser Cys Ser 420 425 430Val Met His Glu Ala Leu His Asn His Tyr Thr
Gln Lys Ser Leu Ser 435 440 445Leu Ser Pro Gly Lys
450311214PRTArtificial Sequence41G5 light chain 311Asp Ile Gln Met
Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val
Thr Val Thr Cys Arg Ala Ser Gln Gly Ile Arg Asn Asp 20 25 30Leu Gly
Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr
Ala Ala Ser Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55
60Ser Gly Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65
70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Leu Gln His Asn Asn Tyr Pro
Trp 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Asp Ile Lys Arg Thr Val
Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln
Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn
Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn
Ala Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu
Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu
Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala
Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200
205Phe Asn Arg Gly Glu Cys 210312123PRTArtificial Sequence41G5
variable heavy chain 312Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val
Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Ser Ser Tyr 20 25 30Gly Met His Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Val Ile Trp Tyr Pro Gly Ser
Asn Lys Tyr Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser
Arg Asp Asn Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser
Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Gly Gly
Glu Leu Gly Arg Tyr Tyr Tyr Tyr Gly Met Asp Val 100 105 110Trp Gly
Gln Gly Thr Thr Val Thr Val Ser Ser 115 120313108PRTArtificial
Sequence41G5 variable light chain 313Asp Ile Gln Met Thr Gln Ser
Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Val Thr
Cys Arg Ala Ser Gln Gly Ile Arg Asn Asp 20 25 30Leu Gly Trp Tyr Gln
Gln Lys Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr Ala Ala Ser
Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser
Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu
Asp Phe Ala Thr Tyr Tyr Cys Leu Gln His Asn Asn Tyr Pro Trp 85 90
95Thr Phe Gly Gln Gly Thr Lys Val Asp Ile Lys Arg 100
1053145PRTArtificial Sequence41G5 heavy chain CDR1 314Ser Tyr Gly
Met His1 531517PRTArtificial Sequence41G5 heavy chain CDR2 315Val
Ile Trp Tyr Pro Gly Ser Asn Lys Tyr Tyr Ala Asp Ser Val Lys1 5 10
15Gly31614PRTArtificial Sequence41G5 heavy chain CDR3 316Gly Gly
Glu Leu Gly Arg Tyr Tyr Tyr Tyr Gly Met Asp Val1 5
1031711PRTArtificial Sequence41G5 light chain CDR1 317Arg Ala Ser
Gln Gly Ile Arg Asn Asp Leu Gly1 5 103187PRTArtificial Sequence41G5
light chain CDR2 318Ala Ala Ser Ser Leu Gln Ser1 53199PRTArtificial
Sequence41G5 light chain CDR3 319Leu Gln His Asn Asn Tyr Pro Trp
Thr1 5320453PRTArtificial Sequence42A11 heavy chain 320Gln Val Gln
Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30Gly
Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ala Val Ile Trp Tyr Glu Gly Ser Asn Lys Tyr Tyr Ala Asp Ser Val
50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu
Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val
Tyr Tyr Cys 85 90 95Ala Arg Gly Gly Gln Leu Gly Tyr Tyr Tyr Tyr Ser
Gly Met Asp Val 100 105 110Trp Gly Gln Gly Thr Thr Val Thr Val Ser
Ser Ala Ser Thr Lys Gly 115 120 125Pro Ser Val Phe Pro Leu Ala Pro
Ser Ser Lys Ser Thr Ser Gly Gly 130 135 140Thr Ala Ala Leu Gly Cys
Leu Val Lys Asp Tyr Phe Pro Glu Pro Val145 150 155 160Thr Val Ser
Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe 165 170 175Pro
Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val 180 185
190Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val
195 200 205Asn His Lys Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu
Pro Lys 210 215 220Ser Cys Asp Lys Thr His Thr Cys Pro Pro Cys Pro
Ala Pro Glu Leu225 230 235 240Leu Gly Gly Pro Ser Val Phe Leu Phe
Pro Pro Lys Pro Lys Asp Thr 245 250 255Leu Met Ile Ser Arg Thr Pro
Glu Val Thr Cys Val Val Val Asp Val 260 265 270Ser His Glu Asp Pro
Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val 275 280 285Glu Val His
Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser 290 295 300Thr
Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu305 310
315 320Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro
Ala 325 330 335Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro
Arg Glu Pro 340 345 350Gln Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu
Met Thr Lys Asn Gln 355 360 365Val Ser Leu Thr Cys Leu Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala 370 375 380Val Glu Trp Glu Ser Asn Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr385 390 395 400Pro Pro Val Leu
Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu 405 410 415Thr Val
Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser 420 425
430Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser
435 440 445Leu Ser Pro Gly Lys 450321214PRTArtificial Sequence42A11
light chain 321Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala
Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Gly
Ile Arg Asn Asp 20 25 30Leu Gly Trp Tyr Gln Gln Lys Pro Gly Lys Ala
Pro Lys Arg Leu Ile 35 40 45Tyr Asp Ala Ser Ser Leu Gln Ser Gly Val
Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu
Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Glu Phe Ala Thr Tyr Tyr
Cys Leu Gln His Asn Asn Tyr Pro Trp 85 90 95Thr Phe Gly Gln Gly Thr
Lys Val Glu Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe
Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala
Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135
140Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser
Gln145 150 155 160Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr
Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr
Glu Lys His Lys Val Tyr 180 185 190Ala Cys Glu Val Thr His Gln Gly
Leu Ser Ser Pro Val Thr Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys
210322123PRTArtificial Sequence42A11 variable heavy chain 322Gln
Val Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10
15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr
20 25 30Gly Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Val 35 40 45Ala Val Ile Trp Tyr Glu Gly Ser Asn Lys Tyr Tyr Ala Asp
Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn
Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr
Ala Val Tyr Tyr Cys 85 90 95Ala Arg Gly Gly Gln Leu Gly Tyr Tyr Tyr
Tyr Ser Gly Met Asp Val 100 105 110Trp Gly Gln Gly Thr Thr Val Thr
Val Ser Ser 115 120323108PRTArtificial Sequence42A11 variable light
chain 323Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser
Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Gly Ile
Arg Asn Asp 20 25 30Leu Gly Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro
Lys Arg Leu Ile 35 40 45Tyr Asp Ala Ser Ser Leu Gln Ser Gly Val Pro
Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr
Ile Ser Ser Leu Gln Pro65 70 75 80Glu Glu Phe Ala Thr Tyr Tyr Cys
Leu Gln His Asn Asn Tyr Pro Trp 85 90 95Thr Phe Gly Gln Gly Thr Lys
Val Glu Ile Lys Arg 100 1053245PRTArtificial Sequence42A11 heavy
chain CDR1 324Ser Tyr Gly Met His1 532517PRTArtificial
Sequence42A11 heavy chain CDR2 325Val Ile Trp Tyr Glu Gly Ser Asn
Lys Tyr Tyr Ala Asp Ser Val Lys1 5 10 15Gly32614PRTArtificial
Sequence42A11 heavy chain CDR3 326Gly Gly Gln Leu Gly Tyr Tyr Tyr
Tyr Ser Gly Met Asp Val1 5 1032711PRTArtificial Sequence42A11 light
chain CDR1 327Arg Ala Ser Gln Gly Ile Arg Asn Asp Leu Gly1 5
103287PRTArtificial Sequence42A11 light chain CDR2 328Asp Ala Ser
Ser Leu Gln Ser1 53299PRTArtificial Sequence42A11 light chain CDR3
329Leu Gln His Asn Asn Tyr Pro Trp Thr1 5330450PRTArtificial
Sequence44C1 heavy chain 330Gln Val Gln Leu Val Glu Ser Gly Gly Gly
Val Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser
Gly Phe Thr Leu Ser Ser Tyr 20 25 30Gly Met His Trp Val Arg Gln Ala
Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Val Ile Trp Tyr Asp Gly
Ser Asn Lys Tyr Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile
Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn
Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Arg
Gly Thr Val Thr Thr Pro Asp Phe Asp Tyr Trp Gly Gln 100 105 110Gly
Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser Val 115 120
125Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala
130 135 140Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr
Val Ser145 150 155 160Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His
Thr Phe Pro Ala Val 165 170 175Leu Gln Ser Ser Gly Leu Tyr Ser Leu
Ser Ser Val Val Thr Val Pro 180 185 190Ser Ser Ser Leu Gly Thr Gln
Thr Tyr Ile Cys Asn Val Asn His Lys 195 200 205Pro Ser Asn Thr Lys
Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp 210 215 220Lys Thr His
Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly225 230 235
240Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile
245 250 255Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser
His Glu 260 265 270Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly
Val Glu Val His 275 280 285Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln
Tyr Asn
Ser Thr Tyr Arg 290 295 300Val Val Ser Val Leu Thr Val Leu His Gln
Asp Trp Leu Asn Gly Lys305 310 315 320Glu Tyr Lys Cys Lys Val Ser
Asn Lys Ala Leu Pro Ala Pro Ile Glu 325 330 335Lys Thr Ile Ser Lys
Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr 340 345 350Thr Leu Pro
Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser Leu 355 360 365Thr
Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp 370 375
380Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro
Val385 390 395 400Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys
Leu Thr Val Asp 405 410 415Lys Ser Arg Trp Gln Gln Gly Asn Val Phe
Ser Cys Ser Val Met His 420 425 430Glu Ala Leu His Asn His Tyr Thr
Gln Lys Ser Leu Ser Leu Ser Pro 435 440 445Gly Lys
450331216PRTArtificial Sequence44C1 light chain 331Gln Ser Ala Leu
Thr Gln Pro Ala Ser Val Ser Gly Ser Pro Gly Gln1 5 10 15Ser Ile Thr
Ile Ser Cys Thr Gly Thr Ser Ser Asp Val Gly Thr Tyr 20 25 30Asn Leu
Val Ser Trp Tyr Gln Gln His Pro Gly Lys Ala Pro Lys Leu 35 40 45Met
Ile Tyr Glu Val Ser Lys Arg Pro Ser Gly Val Ser Asn Arg Phe 50 55
60Ser Gly Ser Lys Ser Gly Asn Thr Ala Ser Leu Thr Ile Ser Gly Leu65
70 75 80Gln Ala Glu Asp Glu Ala Asp Tyr Tyr Cys Cys Ser Tyr Ala Gly
Phe 85 90 95Ser Thr Trp Val Phe Gly Gly Gly Thr Lys Leu Thr Val Leu
Gly Gln 100 105 110Pro Lys Ala Ala Pro Ser Val Thr Leu Phe Pro Pro
Ser Ser Glu Glu 115 120 125Leu Gln Ala Asn Lys Ala Thr Leu Val Cys
Leu Ile Ser Asp Phe Tyr 130 135 140Pro Gly Ala Val Thr Val Ala Trp
Lys Ala Asp Ser Ser Pro Val Lys145 150 155 160Ala Gly Val Glu Thr
Thr Thr Pro Ser Lys Gln Ser Asn Asn Lys Tyr 165 170 175Ala Ala Ser
Ser Tyr Leu Ser Leu Thr Pro Glu Gln Trp Lys Ser His 180 185 190Arg
Ser Tyr Ser Cys Gln Val Thr His Glu Gly Ser Thr Val Glu Lys 195 200
205Thr Val Ala Pro Thr Glu Cys Ser 210 215332120PRTArtificial
Sequence44C1 variable heavy chain 332Gln Val Gln Leu Val Glu Ser
Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys
Ala Ala Ser Gly Phe Thr Leu Ser Ser Tyr 20 25 30Gly Met His Trp Val
Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Val Ile Trp
Tyr Asp Gly Ser Asn Lys Tyr Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg
Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr65 70 75 80Leu
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90
95Ala Arg Arg Gly Thr Val Thr Thr Pro Asp Phe Asp Tyr Trp Gly Gln
100 105 110Gly Thr Leu Val Thr Val Ser Ser 115
120333111PRTArtificial Sequence44C1 variable light chain 333Gln Ser
Ala Leu Thr Gln Pro Ala Ser Val Ser Gly Ser Pro Gly Gln1 5 10 15Ser
Ile Thr Ile Ser Cys Thr Gly Thr Ser Ser Asp Val Gly Thr Tyr 20 25
30Asn Leu Val Ser Trp Tyr Gln Gln His Pro Gly Lys Ala Pro Lys Leu
35 40 45Met Ile Tyr Glu Val Ser Lys Arg Pro Ser Gly Val Ser Asn Arg
Phe 50 55 60Ser Gly Ser Lys Ser Gly Asn Thr Ala Ser Leu Thr Ile Ser
Gly Leu65 70 75 80Gln Ala Glu Asp Glu Ala Asp Tyr Tyr Cys Cys Ser
Tyr Ala Gly Phe 85 90 95Ser Thr Trp Val Phe Gly Gly Gly Thr Lys Leu
Thr Val Leu Gly 100 105 1103345PRTArtificial Sequence44C1 heavy
chain CDR1 334Ser Tyr Gly Met His1 533517PRTArtificial Sequence44C1
heavy chain CDR2 335Val Ile Trp Tyr Asp Gly Ser Asn Lys Tyr Tyr Ala
Asp Ser Val Lys1 5 10 15Gly33611PRTArtificial Sequence44C1 heavy
chain CDR3 336Arg Gly Thr Val Thr Thr Pro Asp Phe Asp Tyr1 5
1033714PRTArtificial Sequence44C1 light chain CDR1 337Thr Gly Thr
Ser Ser Asp Val Gly Thr Tyr Asn Leu Val Ser1 5 103387PRTArtificial
Sequence44C1 light chain CDR2 338Glu Val Ser Lys Arg Pro Ser1
533910PRTArtificial Sequence44C1 light chain CDR3 339Cys Ser Tyr
Ala Gly Phe Ser Thr Trp Val1 5 10340447PRTArtificial Sequence45A8
heavy chain 340Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln
Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr
Phe Ser Ser Tyr 20 25 30Gly Met His Trp Val Arg Gln Ala Pro Gly Lys
Gly Leu Glu Trp Val 35 40 45Ala Val Ile Trp His Asp Gly Ser Asn Lys
Tyr Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Lys Asp
Asn Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg
Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Glu Tyr Gly Gly
Asn Phe Asp Tyr Trp Gly Gln Gly Thr Leu 100 105 110Val Thr Val Ser
Ser Ala Ser Thr Lys Gly Pro Ser Val Phe Pro Leu 115 120 125Ala Pro
Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala Leu Gly Cys 130 135
140Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp Asn
Ser145 150 155 160Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala
Val Leu Gln Ser 165 170 175Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
Thr Val Pro Ser Ser Ser 180 185 190Leu Gly Thr Gln Thr Tyr Ile Cys
Asn Val Asn His Lys Pro Ser Asn 195 200 205Thr Lys Val Asp Lys Lys
Val Glu Pro Lys Ser Cys Asp Lys Thr His 210 215 220Thr Cys Pro Pro
Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser Val225 230 235 240Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250
255Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu
260 265 270Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn
Ala Lys 275 280 285Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr
Arg Val Val Ser 290 295 300Val Leu Thr Val Leu His Gln Asp Trp Leu
Asn Gly Lys Glu Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Ala
Leu Pro Ala Pro Ile Glu Lys Thr Ile 325 330 335Ser Lys Ala Lys Gly
Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Arg
Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val
Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375
380Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp
Ser385 390 395 400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val
Asp Lys Ser Arg 405 410 415Trp Gln Gln Gly Asn Val Phe Ser Cys Ser
Val Met His Glu Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser
Leu Ser Leu Ser Pro Gly Lys 435 440 445341216PRTArtificial
Sequence45A8 light chain 341Gln Ser Ala Leu Thr Gln Pro Ala Ser Val
Ser Gly Ser Pro Gly Gln1 5 10 15Ser Ile Thr Ile Ser Cys Thr Gly Thr
Ser Ser Asp Val Gly Thr Tyr 20 25 30Asn Leu Val Ser Trp Tyr Gln Gln
His Pro Gly Lys Ala Pro Lys Leu 35 40 45Met Ile Tyr Glu Val Ser Lys
Arg Pro Ser Gly Ile Ser Asn Arg Phe 50 55 60Ser Gly Ser Lys Ser Gly
Asn Thr Ala Ser Leu Thr Ile Ser Gly Leu65 70 75 80Gln Ala Glu Asp
Glu Ala Asp Tyr Tyr Cys Cys Ser Tyr Ala Gly Tyr 85 90 95Ser Thr Trp
Val Phe Gly Gly Gly Thr Lys Leu Thr Val Leu Arg Gln 100 105 110Pro
Lys Ala Ala Pro Ser Val Thr Leu Phe Pro Pro Ser Ser Glu Glu 115 120
125Leu Gln Ala Asn Lys Ala Thr Leu Val Cys Leu Ile Ser Asp Phe Tyr
130 135 140Pro Gly Ala Val Thr Val Ala Trp Lys Ala Asp Ser Ser Pro
Val Lys145 150 155 160Ala Gly Val Glu Thr Thr Thr Pro Ser Lys Gln
Ser Asn Asn Lys Tyr 165 170 175Ala Ala Ser Ser Tyr Leu Ser Leu Thr
Pro Glu Gln Trp Lys Ser His 180 185 190Arg Ser Tyr Ser Cys Gln Val
Thr His Glu Gly Ser Thr Val Glu Lys 195 200 205Thr Val Ala Pro Thr
Glu Cys Ser 210 215342117PRTArtificial Sequence45A8 variable heavy
chain 342Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro
Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe
Ser Ser Tyr 20 25 30Gly Met His Trp Val Arg Gln Ala Pro Gly Lys Gly
Leu Glu Trp Val 35 40 45Ala Val Ile Trp His Asp Gly Ser Asn Lys Tyr
Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Lys Asp Asn
Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala
Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Glu Tyr Gly Gly Asn
Phe Asp Tyr Trp Gly Gln Gly Thr Leu 100 105 110Val Thr Val Ser Ser
115343111PRTArtificial Sequence45A8 variable light chain 343Gln Ser
Ala Leu Thr Gln Pro Ala Ser Val Ser Gly Ser Pro Gly Gln1 5 10 15Ser
Ile Thr Ile Ser Cys Thr Gly Thr Ser Ser Asp Val Gly Thr Tyr 20 25
30Asn Leu Val Ser Trp Tyr Gln Gln His Pro Gly Lys Ala Pro Lys Leu
35 40 45Met Ile Tyr Glu Val Ser Lys Arg Pro Ser Gly Ile Ser Asn Arg
Phe 50 55 60Ser Gly Ser Lys Ser Gly Asn Thr Ala Ser Leu Thr Ile Ser
Gly Leu65 70 75 80Gln Ala Glu Asp Glu Ala Asp Tyr Tyr Cys Cys Ser
Tyr Ala Gly Tyr 85 90 95Ser Thr Trp Val Phe Gly Gly Gly Thr Lys Leu
Thr Val Leu Arg 100 105 1103445PRTArtificial Sequence45A8 heavy
chain CDR1 344Ser Tyr Gly Met His1 534517PRTArtificial Sequence45A8
heavy chain CDR2 345Val Ile Trp His Asp Gly Ser Asn Lys Tyr Tyr Ala
Asp Ser Val Lys1 5 10 15Gly3468PRTArtificial Sequence45A8 heavy
chain CDR3 346Glu Tyr Gly Gly Asn Phe Asp Tyr1 534714PRTArtificial
Sequence45A8 light chain CDR1 347Thr Gly Thr Ser Ser Asp Val Gly
Thr Tyr Asn Leu Val Ser1 5 103487PRTArtificial Sequence45A8 light
chain CDR2 348Glu Val Ser Lys Arg Pro Ser1 534910PRTArtificial
Sequence45A8 light chain CDR3 349Cys Ser Tyr Ala Gly Tyr Ser Thr
Trp Val1 5 10350454PRTArtificial Sequence46E11 heavy chain 350Gln
Val Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10
15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr
20 25 30Gly Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Val 35 40 45Ala Val Ile Trp Tyr Ala Gly Ser Asn Lys Tyr Tyr Ala Asp
Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn
Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr
Ala Val Tyr Tyr Cys 85 90 95Ala Arg Gly Asp Ile Leu Thr Gly Tyr Ser
Leu Tyr Tyr Gly Met Asp 100 105 110Val Trp Gly Gln Gly Thr Thr Val
Thr Val Ser Ser Ala Ser Thr Lys 115 120 125Gly Pro Ser Val Phe Pro
Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly 130 135 140Gly Thr Ala Ala
Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro145 150 155 160Val
Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr 165 170
175Phe Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val
180 185 190Val Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile
Cys Asn 195 200 205Val Asn His Lys Pro Ser Asn Thr Lys Val Asp Lys
Lys Val Glu Pro 210 215 220Lys Ser Cys Asp Lys Thr His Thr Cys Pro
Pro Cys Pro Ala Pro Glu225 230 235 240Leu Leu Gly Gly Pro Ser Val
Phe Leu Phe Pro Pro Lys Pro Lys Asp 245 250 255Thr Leu Met Ile Ser
Arg Thr Pro Glu Val Thr Cys Val Val Val Asp 260 265 270Val Ser His
Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly 275 280 285Val
Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn 290 295
300Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp
Trp305 310 315 320Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn
Lys Ala Leu Pro 325 330 335Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala
Lys Gly Gln Pro Arg Glu 340 345 350Pro Gln Val Tyr Thr Leu Pro Pro
Ser Arg Glu Glu Met Thr Lys Asn 355 360 365Gln Val Ser Leu Thr Cys
Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile 370 375 380Ala Val Glu Trp
Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr385 390 395 400Thr
Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys 405 410
415Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys
420 425 430Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys
Ser Leu 435 440 445Ser Leu Ser Pro Gly Lys 450351214PRTArtificial
Sequence46E11 light chain 351Asp Ile Gln Met Thr Gln Ser Pro Ser
Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg
Ala Ser Gln Gly Ile Arg Asn Asp 20 25 30Leu Gly Trp Tyr Gln Gln Lys
Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr Ala Ala Ser Ser Leu
Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Ala
Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe
Ala Thr Tyr Tyr Cys Leu Gln His Asn Ser Tyr Pro Trp 85 90 95Thr Phe
Gly Gln Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala Ala 100 105
110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly
115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg
Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser
Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu Gln Asp Ser Lys
Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys
Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala Cys Glu Val Thr
His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200 205Phe Asn Arg
Gly Glu Cys 210352124PRTArtificial Sequence46E11 variable heavy
chain 352Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro
Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe
Ser Ser
Tyr 20 25 30Gly Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu
Trp Val 35 40 45Ala Val Ile Trp Tyr Ala Gly Ser Asn Lys Tyr Tyr Ala
Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys
Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Gly Asp Ile Leu Thr Gly Tyr
Ser Leu Tyr Tyr Gly Met Asp 100 105 110Val Trp Gly Gln Gly Thr Thr
Val Thr Val Ser Ser 115 120353108PRTArtificial Sequence46E11
variable light chain 353Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu
Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser
Gln Gly Ile Arg Asn Asp 20 25 30Leu Gly Trp Tyr Gln Gln Lys Pro Gly
Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr Ala Ala Ser Ser Leu Gln Ser
Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Ala Glu Phe
Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr
Tyr Tyr Cys Leu Gln His Asn Ser Tyr Pro Trp 85 90 95Thr Phe Gly Gln
Gly Thr Lys Val Glu Ile Lys Arg 100 1053545PRTArtificial
Sequence46E11 heavy chain CDR1 354Ser Tyr Gly Met His1
535517PRTArtificial Sequence46E11 heavy chain CDR2 355Val Ile Trp
Tyr Ala Gly Ser Asn Lys Tyr Tyr Ala Asp Ser Val Lys1 5 10
15Gly35615PRTArtificial Sequence46E11 heavy chain CDR3 356Gly Asp
Ile Leu Thr Gly Tyr Ser Leu Tyr Tyr Gly Met Asp Val1 5 10
1535711PRTArtificial Sequence46E11 light chain CDR1 357Arg Ala Ser
Gln Gly Ile Arg Asn Asp Leu Gly1 5 103587PRTArtificial
Sequence46E11 light chain CDR2 358Ala Ala Ser Ser Leu Gln Ser1
53599PRTArtificial Sequence46E11 light chain CDR3 359Leu Gln His
Asn Ser Tyr Pro Trp Thr1 5360453PRTArtificial Sequence48H12 heavy
chain 360Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro
Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe
Ser Ser Tyr 20 25 30Gly Met His Trp Val Arg Gln Ala Pro Gly Lys Gly
Leu Glu Trp Val 35 40 45Ala Val Ile Trp Tyr Ala Gly Ser Asn Lys Tyr
Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn
Ser Lys Asn Thr Val Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala
Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Gly Gly Gln Leu Ala
Leu Tyr Tyr Tyr Tyr Gly Met Asp Val 100 105 110Trp Gly Gln Gly Thr
Thr Val Thr Val Ser Ser Ala Ser Thr Lys Gly 115 120 125Pro Ser Val
Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly 130 135 140Thr
Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val145 150
155 160Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr
Phe 165 170 175Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser
Ser Val Val 180 185 190Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr
Tyr Ile Cys Asn Val 195 200 205Asn His Lys Pro Ser Asn Thr Lys Val
Asp Lys Lys Val Glu Pro Lys 210 215 220Ser Cys Asp Lys Thr His Thr
Cys Pro Pro Cys Pro Ala Pro Glu Leu225 230 235 240Leu Gly Gly Pro
Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr 245 250 255Leu Met
Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val 260 265
270Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val
275 280 285Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr
Asn Ser 290 295 300Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His
Gln Asp Trp Leu305 310 315 320Asn Gly Lys Glu Tyr Lys Cys Lys Val
Ser Asn Lys Ala Leu Pro Ala 325 330 335Pro Ile Glu Lys Thr Ile Ser
Lys Ala Lys Gly Gln Pro Arg Glu Pro 340 345 350Gln Val Tyr Thr Leu
Pro Pro Ser Arg Glu Glu Met Thr Lys Asn Gln 355 360 365Val Ser Leu
Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala 370 375 380Val
Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr385 390
395 400Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys
Leu 405 410 415Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe
Ser Cys Ser 420 425 430Val Met His Glu Ala Leu His Asn His Tyr Thr
Gln Lys Ser Leu Ser 435 440 445Leu Ser Pro Gly Lys
450361214PRTArtificial Sequence48H12 light chain 361Asp Ile Gln Met
Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val
Thr Ile Thr Cys Arg Ala Ser Gln Gly Ile Arg Asn Asp 20 25 30Leu Gly
Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr
Ala Ala Ser Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55
60Ser Gly Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65
70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Leu Gln His Asn Asn Tyr Pro
Trp 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg Thr Val
Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln
Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn
Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn
Ala Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu
Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu
Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala
Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200
205Phe Asn Arg Gly Glu Cys 210362123PRTArtificial Sequence48H12
variable heavy chain 362Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val
Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Ser Ser Tyr 20 25 30Gly Met His Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Val Ile Trp Tyr Ala Gly Ser
Asn Lys Tyr Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser
Arg Asp Asn Ser Lys Asn Thr Val Tyr65 70 75 80Leu Gln Met Asn Ser
Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Gly Gly
Gln Leu Ala Leu Tyr Tyr Tyr Tyr Gly Met Asp Val 100 105 110Trp Gly
Gln Gly Thr Thr Val Thr Val Ser Ser 115 120363108PRTArtificial
Sequence48H12 variable light chain 363Asp Ile Gln Met Thr Gln Ser
Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr
Cys Arg Ala Ser Gln Gly Ile Arg Asn Asp 20 25 30Leu Gly Trp Tyr Gln
Gln Lys Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr Ala Ala Ser
Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser
Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu
Asp Phe Ala Thr Tyr Tyr Cys Leu Gln His Asn Asn Tyr Pro Trp 85 90
95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg 100
1053645PRTArtificial Sequence48H12 heavy chain CDR1 364Ser Tyr Gly
Met His1 536517PRTArtificial Sequence48H12 heavy chain CDR2 365Val
Ile Trp Tyr Ala Gly Ser Asn Lys Tyr Tyr Ala Asp Ser Val Lys1 5 10
15Gly36614PRTArtificial Sequence48H12 heavy chain CDR3 366Gly Gly
Gln Leu Ala Leu Tyr Tyr Tyr Tyr Gly Met Asp Val1 5
1036711PRTArtificial Sequence48H12 light chain CDR1 367Arg Ala Ser
Gln Gly Ile Arg Asn Asp Leu Gly1 5 103687PRTArtificial
Sequence48H12 light chain CDR2 368Ala Ala Ser Ser Leu Gln Ser1
53699PRTArtificial Sequence48H12 light chain CDR3 369Leu Gln His
Asn Asn Tyr Pro Trp Thr1 5370453PRTArtificial Sequence48H7 heavy
chain 370Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro
Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe
Ser Ser Tyr 20 25 30Gly Met Tyr Trp Val Arg Gln Ala Pro Gly Lys Gly
Leu Glu Trp Val 35 40 45Ala Val Ile Trp Tyr Glu Gly Ser Asn Lys Tyr
Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn
Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala
Glu Asp Thr Ala Val Tyr Phe Cys 85 90 95Ala Arg Gly Gly Glu Leu Gly
Arg Asp Tyr Tyr Ser Gly Met Asp Val 100 105 110Trp Gly Gln Gly Thr
Thr Val Thr Val Ser Ser Ala Ser Thr Lys Gly 115 120 125Pro Ser Val
Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly 130 135 140Thr
Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val145 150
155 160Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr
Phe 165 170 175Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser
Ser Val Val 180 185 190Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr
Tyr Ile Cys Asn Val 195 200 205Asn His Lys Pro Ser Asn Thr Lys Val
Asp Lys Lys Val Glu Pro Lys 210 215 220Ser Cys Asp Lys Thr His Thr
Cys Pro Pro Cys Pro Ala Pro Glu Leu225 230 235 240Leu Gly Gly Pro
Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr 245 250 255Leu Met
Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val 260 265
270Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val
275 280 285Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr
Asn Ser 290 295 300Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His
Gln Asp Trp Leu305 310 315 320Asn Gly Lys Glu Tyr Lys Cys Lys Val
Ser Asn Lys Ala Leu Pro Ala 325 330 335Pro Ile Glu Lys Thr Ile Ser
Lys Ala Lys Gly Gln Pro Arg Glu Pro 340 345 350Gln Val Tyr Thr Leu
Pro Pro Ser Arg Glu Glu Met Thr Lys Asn Gln 355 360 365Val Ser Leu
Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala 370 375 380Val
Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr385 390
395 400Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys
Leu 405 410 415Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe
Ser Cys Ser 420 425 430Val Met His Glu Ala Leu His Asn His Tyr Thr
Gln Lys Ser Leu Ser 435 440 445Leu Ser Pro Gly Lys
450371214PRTArtificial Sequence48H7 light chain 371Asp Ile Gln Met
Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val
Thr Ile Thr Cys Arg Ala Ser Gln Val Ile Arg Asn Asp 20 25 30Leu Gly
Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr
Ala Ala Ser Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55
60Ser Gly Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65
70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Leu Gln His Asn Ser Tyr Pro
Ile 85 90 95Thr Phe Gly Gly Gly Thr Lys Val Glu Ile Lys Arg Thr Val
Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln
Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn
Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn
Ala Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu
Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu
Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala
Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200
205Phe Asn Arg Gly Glu Cys 210372123PRTArtificial Sequence48H7
variable heavy chain 372Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val
Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Ser Ser Tyr 20 25 30Gly Met Tyr Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Val Ile Trp Tyr Glu Gly Ser
Asn Lys Tyr Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser
Arg Asp Asn Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser
Leu Arg Ala Glu Asp Thr Ala Val Tyr Phe Cys 85 90 95Ala Arg Gly Gly
Glu Leu Gly Arg Asp Tyr Tyr Ser Gly Met Asp Val 100 105 110Trp Gly
Gln Gly Thr Thr Val Thr Val Ser Ser 115 120373108PRTArtificial
Sequence48H7 variable light chain 373Asp Ile Gln Met Thr Gln Ser
Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr
Cys Arg Ala Ser Gln Val Ile Arg Asn Asp 20 25 30Leu Gly Trp Tyr Gln
Gln Lys Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr Ala Ala Ser
Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser
Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu
Asp Phe Ala Thr Tyr Tyr Cys Leu Gln His Asn Ser Tyr Pro Ile 85 90
95Thr Phe Gly Gly Gly Thr Lys Val Glu Ile Lys Arg 100
1053745PRTArtificial Sequence48H7 heavy chain CDR1 374Ser Tyr Gly
Met Tyr1 537517PRTArtificial Sequence48H7 heavy chain CDR2 375Val
Ile Trp Tyr Glu Gly Ser Asn Lys Tyr Tyr Ala Asp Ser Val Lys1 5 10
15Gly37614PRTArtificial Sequence48H7 heavy chain CDR3 376Gly Gly
Glu Leu Gly Arg Asp Tyr Tyr Ser Gly Met Asp Val1 5
1037711PRTArtificial Sequence48H7 light chain CDR1 377Arg Ala Ser
Gln Val Ile Arg Asn Asp Leu Gly1 5 103787PRTArtificial Sequence48H7
light chain CDR2 378Ala Ala Ser Ser Leu Gln Ser1 53799PRTArtificial
Sequence48H7 light chain CDR3 379Leu Gln His Asn Ser Tyr Pro Ile
Thr1 5380453PRTArtificial Sequence49D9 heavy chain 380Gln Met Gln
Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30Gly
Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ala Val Ile Trp Tyr Ala Gly Ser Asn Lys Tyr Tyr Ala Asp Ser Val
50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu
Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val
Tyr Tyr Cys 85 90 95Ala Arg Gly
Gly Arg Leu Gly Phe Tyr Tyr Tyr Tyr Gly Met Asp Val 100 105 110Trp
Gly Gln Gly Thr Thr Val Thr Val Ser Ser Ala Ser Thr Lys Gly 115 120
125Pro Ser Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly
130 135 140Thr Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu
Pro Val145 150 155 160Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser
Gly Val His Thr Phe 165 170 175Pro Ala Val Leu Gln Ser Ser Gly Leu
Tyr Ser Leu Ser Ser Val Val 180 185 190Thr Val Pro Ser Ser Ser Leu
Gly Thr Gln Thr Tyr Ile Cys Asn Val 195 200 205Asn His Lys Pro Ser
Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys 210 215 220Ser Cys Asp
Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu225 230 235
240Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr
245 250 255Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val
Asp Val 260 265 270Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr
Val Asp Gly Val 275 280 285Glu Val His Asn Ala Lys Thr Lys Pro Arg
Glu Glu Gln Tyr Asn Ser 290 295 300Thr Tyr Arg Val Val Ser Val Leu
Thr Val Leu His Gln Asp Trp Leu305 310 315 320Asn Gly Lys Glu Tyr
Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala 325 330 335Pro Ile Glu
Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro 340 345 350Gln
Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu Met Thr Lys Asn Gln 355 360
365Val Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala
370 375 380Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys
Thr Thr385 390 395 400Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe
Leu Tyr Ser Lys Leu 405 410 415Thr Val Asp Lys Ser Arg Trp Gln Gln
Gly Asn Val Phe Ser Cys Ser 420 425 430Val Met His Glu Ala Leu His
Asn His Tyr Thr Gln Lys Ser Leu Ser 435 440 445Leu Ser Pro Gly Lys
450381214PRTArtificial Sequence49D9 light chain 381Asp Ile Gln Met
Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val
Thr Ile Thr Cys Arg Ala Ser Gln Gly Ile Arg Asn Asp 20 25 30Leu Gly
Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr
Ala Ala Ser Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55
60Ser Gly Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65
70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Leu Gln Leu Asn Ser Tyr Pro
Trp 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg Thr Val
Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln
Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn
Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn
Ala Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu
Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu
Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala
Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200
205Phe Asn Arg Gly Glu Cys 210382123PRTArtificial Sequence49D9
variable heavy chain 382Gln Met Gln Leu Val Glu Ser Gly Gly Gly Val
Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Ser Ser Tyr 20 25 30Gly Met His Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Val Ile Trp Tyr Ala Gly Ser
Asn Lys Tyr Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser
Arg Asp Asn Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser
Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Gly Gly
Arg Leu Gly Phe Tyr Tyr Tyr Tyr Gly Met Asp Val 100 105 110Trp Gly
Gln Gly Thr Thr Val Thr Val Ser Ser 115 120383108PRTArtificial
Sequence49D9 variable light chain 383Asp Ile Gln Met Thr Gln Ser
Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr
Cys Arg Ala Ser Gln Gly Ile Arg Asn Asp 20 25 30Leu Gly Trp Tyr Gln
Gln Lys Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr Ala Ala Ser
Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser
Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu
Asp Phe Ala Thr Tyr Tyr Cys Leu Gln Leu Asn Ser Tyr Pro Trp 85 90
95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg 100
1053845PRTArtificial Sequence49D9 heavy chain CDR1 384Ser Tyr Gly
Met His1 538517PRTArtificial Sequence49D9 heavy chain CDR2 385Val
Ile Trp Tyr Ala Gly Ser Asn Lys Tyr Tyr Ala Asp Ser Val Lys1 5 10
15Gly38614PRTArtificial Sequence49D9 heavy chain CDR3 386Gly Gly
Arg Leu Gly Phe Tyr Tyr Tyr Tyr Gly Met Asp Val1 5
1038711PRTArtificial Sequence49D9 light chain CDR1 387Arg Ala Ser
Gln Gly Ile Arg Asn Asp Leu Gly1 5 103887PRTArtificial Sequence49D9
light chain CDR2 388Ala Ala Ser Ser Leu Gln Ser1 53899PRTArtificial
Sequence49D9 light chain CDR3 389Leu Gln Leu Asn Ser Tyr Pro Trp
Thr1 5390447PRTArtificial Sequence49E2 heavy chain 390Gln Val Gln
Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30Gly
Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ala Val Ile Trp Ser Asp Gly Asn Asn Lys Tyr Tyr Glu Asp Ser Val
50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Ser Ser Lys Asn Thr Leu
Phe65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val
Tyr Tyr Cys 85 90 95Ala Arg Asp Thr Ala Thr Pro Phe Asp Tyr Trp Gly
Gln Gly Thr Leu 100 105 110Val Thr Val Ser Ser Ala Ser Thr Lys Gly
Pro Ser Val Phe Pro Leu 115 120 125Ala Pro Ser Ser Lys Ser Thr Ser
Gly Gly Thr Ala Ala Leu Gly Cys 130 135 140Leu Val Lys Asp Tyr Phe
Pro Glu Pro Val Thr Val Ser Trp Asn Ser145 150 155 160Gly Ala Leu
Thr Ser Gly Val His Thr Phe Pro Ala Val Leu Gln Ser 165 170 175Ser
Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser Ser Ser 180 185
190Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro Ser Asn
195 200 205Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp Lys
Thr His 210 215 220Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly
Gly Pro Ser Val225 230 235 240Phe Leu Phe Pro Pro Lys Pro Lys Asp
Thr Leu Met Ile Ser Arg Thr 245 250 255Pro Glu Val Thr Cys Val Val
Val Asp Val Ser His Glu Asp Pro Glu 260 265 270Val Lys Phe Asn Trp
Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 275 280 285Thr Lys Pro
Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser 290 295 300Val
Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys305 310
315 320Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr
Ile 325 330 335Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr
Thr Leu Pro 340 345 350Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val
Ser Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr Pro Ser Asp Ile
Ala Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro Glu Asn Asn Tyr
Lys Thr Thr Pro Pro Val Leu Asp Ser385 390 395 400Asp Gly Ser Phe
Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg 405 410 415Trp Gln
Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu 420 425
430His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 435
440 445391216PRTArtificial Sequence49E2 light chain 391Gln Ser Ala
Leu Thr Gln Pro Ala Ser Val Ser Gly Ser Pro Gly Gln1 5 10 15Ser Ile
Thr Ile Ser Cys Thr Gly Thr Ser Ser Asp Val Gly Ile Tyr 20 25 30Asn
Leu Val Ser Trp Tyr Gln Gln His Pro Gly Lys Ala Pro Lys Leu 35 40
45Met Ile His Glu Val Ser Lys Arg Pro Ser Gly Val Ser Asn Arg Phe
50 55 60Ser Gly Ser Lys Ser Gly Asn Thr Ala Ser Leu Thr Ile Ser Gly
Leu65 70 75 80Gln Ala Glu Asp Glu Ala Asp Tyr Tyr Cys Cys Ser Tyr
Ala Gly Ile 85 90 95Ser Thr Trp Val Phe Gly Gly Gly Thr Lys Leu Thr
Val Leu Gly Gln 100 105 110Pro Lys Ala Ala Pro Ser Val Thr Leu Phe
Pro Pro Ser Ser Glu Glu 115 120 125Leu Gln Ala Asn Lys Ala Thr Leu
Val Cys Leu Ile Ser Asp Phe Tyr 130 135 140Pro Gly Ala Val Thr Val
Ala Trp Lys Ala Asp Ser Ser Pro Val Lys145 150 155 160Ala Gly Val
Glu Thr Thr Thr Pro Ser Lys Gln Ser Asn Asn Lys Tyr 165 170 175Ala
Ala Ser Ser Tyr Leu Ser Leu Thr Pro Glu Gln Trp Lys Ser His 180 185
190Arg Ser Tyr Ser Cys Gln Val Thr His Glu Gly Ser Thr Val Glu Lys
195 200 205Thr Val Ala Pro Thr Glu Cys Ser 210
215392117PRTArtificial Sequence49E2 variable heavy chain 392Gln Val
Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser
Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25
30Gly Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val
35 40 45Ala Val Ile Trp Ser Asp Gly Asn Asn Lys Tyr Tyr Glu Asp Ser
Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Ser Ser Lys Asn Thr
Leu Phe65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala
Val Tyr Tyr Cys 85 90 95Ala Arg Asp Thr Ala Thr Pro Phe Asp Tyr Trp
Gly Gln Gly Thr Leu 100 105 110Val Thr Val Ser Ser
115393111PRTArtificial Sequence49E2 variable light chain 393Gln Ser
Ala Leu Thr Gln Pro Ala Ser Val Ser Gly Ser Pro Gly Gln1 5 10 15Ser
Ile Thr Ile Ser Cys Thr Gly Thr Ser Ser Asp Val Gly Ile Tyr 20 25
30Asn Leu Val Ser Trp Tyr Gln Gln His Pro Gly Lys Ala Pro Lys Leu
35 40 45Met Ile His Glu Val Ser Lys Arg Pro Ser Gly Val Ser Asn Arg
Phe 50 55 60Ser Gly Ser Lys Ser Gly Asn Thr Ala Ser Leu Thr Ile Ser
Gly Leu65 70 75 80Gln Ala Glu Asp Glu Ala Asp Tyr Tyr Cys Cys Ser
Tyr Ala Gly Ile 85 90 95Ser Thr Trp Val Phe Gly Gly Gly Thr Lys Leu
Thr Val Leu Gly 100 105 1103945PRTArtificial Sequence49E2 heavy
chain CDR1 394Ser Tyr Gly Met His1 539517PRTArtificial Sequence49E2
heavy chain CDR2 395Val Ile Trp Ser Asp Gly Asn Asn Lys Tyr Tyr Glu
Asp Ser Val Lys1 5 10 15Gly3968PRTArtificial Sequence49E2 heavy
chain CDR3 396Asp Thr Ala Thr Pro Phe Asp Tyr1 539714PRTArtificial
Sequence49E2 light chain CDR1 397Thr Gly Thr Ser Ser Asp Val Gly
Ile Tyr Asn Leu Val Ser1 5 103987PRTArtificial Sequence49E2 light
chain CDR2 398Glu Val Ser Lys Arg Pro Ser1 539910PRTArtificial
Sequence49E2 light chain CDR3 399Cys Ser Tyr Ala Gly Ile Ser Thr
Trp Val1 5 10400452PRTArtificial Sequence48A9 heavy chain 400Gln
Val Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10
15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Cys
20 25 30Gly Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Val 35 40 45Ala Val Ile Ser Tyr Asp Gly Ser Asn Lys Tyr Tyr Ala Asp
Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn
Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr
Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Leu Arg Tyr Asn Trp Asn Asp
Gly Gly Val Asp Tyr Trp 100 105 110Gly Gln Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro 115 120 125Ser Val Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr 130 135 140Ala Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr145 150 155 160Val
Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro 165 170
175Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr
180 185 190Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn
Val Asn 195 200 205His Lys Pro Ser Asn Thr Lys Val Asp Lys Lys Val
Glu Pro Lys Ser 210 215 220Cys Asp Lys Thr His Thr Cys Pro Pro Cys
Pro Ala Pro Glu Leu Leu225 230 235 240Gly Gly Pro Ser Val Phe Leu
Phe Pro Pro Lys Pro Lys Asp Thr Leu 245 250 255Met Ile Ser Arg Thr
Pro Glu Val Thr Cys Val Val Val Asp Val Ser 260 265 270His Glu Asp
Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu 275 280 285Val
His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr 290 295
300Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu
Asn305 310 315 320Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala
Leu Pro Ala Pro 325 330 335Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly
Gln Pro Arg Glu Pro Gln 340 345 350Val Tyr Thr Leu Pro Pro Ser Arg
Glu Glu Met Thr Lys Asn Gln Val 355 360 365Ser Leu Thr Cys Leu Val
Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val 370 375 380Glu Trp Glu Ser
Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro385 390 395 400Pro
Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr 405 410
415Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val
420 425 430Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu
Ser Leu 435 440 445Ser Pro Gly Lys 450401214PRTArtificial
Sequence48A9 light chain 401Asp Ile Gln Met Thr Gln Ser Pro Ser Ser
Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Ile Ile Thr Cys Arg Ala
Ser Gln Ser Ile Ser Ser Tyr 20 25 30Leu His Trp Tyr Lys Gln Lys Pro
Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Gly Ala Ser Arg Leu Gln
Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp
Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln
Ser Ser Ser Thr Pro Leu 85 90 95Thr Phe Gly Gly Gly Thr Lys Val Glu
Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro
Pro Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val
Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln
Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150 155
160Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser
165 170 175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys
Val Tyr 180 185 190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro
Val Thr Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys
210402122PRTArtificial Sequence48A9 variable heavy chain 402Gln Val
Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser
Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Cys 20 25
30Gly Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val
35 40 45Ala Val Ile Ser Tyr Asp Gly Ser Asn Lys Tyr Tyr Ala Asp Ser
Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr
Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala
Val Tyr Tyr Cys 85 90 95Ala Arg Asp Leu Arg Tyr Asn Trp Asn Asp Gly
Gly Val Asp Tyr Trp 100 105 110Gly Gln Gly Thr Leu Val Thr Val Ser
Ser 115 120403108PRTArtificial Sequence48A9 variable light chain
403Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1
5 10 15Asp Arg Val Ile Ile Thr Cys Arg Ala Ser Gln Ser Ile Ser Ser
Tyr 20 25 30Leu His Trp Tyr Lys Gln Lys Pro Gly Lys Ala Pro Lys Leu
Leu Ile 35 40 45Tyr Gly Ala Ser Arg Leu Gln Ser Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln
Ser Ser Ser Thr Pro Leu 85 90 95Thr Phe Gly Gly Gly Thr Lys Val Glu
Ile Lys Arg 100 1054045PRTArtificial Sequence48A9 heavy chain CDR1
404Ser Cys Gly Met His1 540517PRTArtificial Sequence48A9 heavy
chain CDR2 405Val Ile Ser Tyr Asp Gly Ser Asn Lys Tyr Tyr Ala Asp
Ser Val Lys1 5 10 15Gly40613PRTArtificial Sequence48A9 heavy chain
CDR3 406Asp Leu Arg Tyr Asn Trp Asn Asp Gly Gly Val Asp Tyr1 5
1040711PRTArtificial Sequence48A9 light chain CDR1 407Arg Ala Ser
Gln Ser Ile Ser Ser Tyr Leu His1 5 104087PRTArtificial Sequence48A9
light chain CDR2 408Gly Ala Ser Arg Leu Gln Ser1 54099PRTArtificial
Sequence48A9 light chain CDR3 409Gln Gln Ser Ser Ser Thr Pro Leu
Thr1 5410455PRTArtificial Sequence5H7 heavy chain 410Gln Val Gln
Leu Gln Glu Ser Gly Pro Gly Leu Val Lys Pro Ser Gln1 5 10 15Thr Leu
Ser Leu Thr Cys Thr Val Ser Gly Gly Ser Ile Ser Ser Gly 20 25 30Gly
Tyr Phe Trp Ser Trp Ile Arg Gln His Pro Gly Lys Gly Leu Glu 35 40
45Trp Ile Gly Tyr Ile Tyr Tyr Ser Gly Thr Thr Tyr Tyr Asn Pro Ser
50 55 60Leu Lys Ser Arg Val Thr Ile Ser Ile Asp Thr Ser Lys Asn His
Phe65 70 75 80Ser Leu Lys Leu Ser Ser Val Thr Ala Ala Asp Thr Ala
Val Tyr Tyr 85 90 95Cys Ala Arg Asp Leu Phe Tyr Tyr Asp Ser Ser Gly
Pro Arg Gly Phe 100 105 110Asp Pro Trp Gly Gln Gly Thr Leu Val Thr
Val Ser Ser Ala Ser Thr 115 120 125Lys Gly Pro Ser Val Phe Pro Leu
Ala Pro Ser Ser Lys Ser Thr Ser 130 135 140Gly Gly Thr Ala Ala Leu
Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu145 150 155 160Pro Val Thr
Val Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His 165 170 175Thr
Phe Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser 180 185
190Val Val Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys
195 200 205Asn Val Asn His Lys Pro Ser Asn Thr Lys Val Asp Lys Arg
Val Glu 210 215 220Pro Lys Ser Cys Asp Lys Thr His Thr Cys Pro Pro
Cys Pro Ala Pro225 230 235 240Glu Leu Leu Gly Gly Pro Ser Val Phe
Leu Phe Pro Pro Lys Pro Lys 245 250 255Asp Thr Leu Met Ile Ser Arg
Thr Pro Glu Val Thr Cys Val Val Val 260 265 270Asp Val Ser His Glu
Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp 275 280 285Gly Val Glu
Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr 290 295 300Asn
Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp305 310
315 320Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala
Leu 325 330 335Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly
Gln Pro Arg 340 345 350Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg
Glu Glu Met Thr Lys 355 360 365Asn Gln Val Ser Leu Thr Cys Leu Val
Lys Gly Phe Tyr Pro Ser Asp 370 375 380Ile Ala Val Glu Trp Glu Ser
Asn Gly Gln Pro Glu Asn Asn Tyr Lys385 390 395 400Thr Thr Pro Pro
Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser 405 410 415Lys Leu
Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser 420 425
430Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser
435 440 445Leu Ser Leu Ser Pro Gly Lys 450 455411215PRTArtificial
Sequence5H7 light chain 411Glu Ile Val Leu Thr Gln Ser Pro Gly Thr
Leu Ser Leu Ser Pro Gly1 5 10 15Glu Arg Ala Thr Leu Ser Cys Arg Ala
Ser Gln Thr Val Ser Ser Asn 20 25 30Tyr Leu Ala Trp Tyr Gln Gln Lys
Pro Gly Gln Ala Pro Arg Leu Leu 35 40 45Ile Tyr Gly Ser Ser Thr Arg
Ala Thr Gly Ile Pro Asp Arg Phe Ser 50 55 60Gly Ser Gly Ser Gly Thr
Asp Phe Thr Leu Thr Ile Ser Arg Leu Glu65 70 75 80Pro Glu Asp Phe
Ala Val Tyr Tyr Cys Gln Gln Tyr Asp Ser Ser Pro 85 90 95Trp Thr Phe
Gly Gln Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala 100 105 110Ala
Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser 115 120
125Gly Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu
130 135 140Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly
Asn Ser145 150 155 160Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp
Ser Thr Tyr Ser Leu 165 170 175Ser Ser Thr Leu Thr Leu Ser Lys Ala
Asp Tyr Glu Lys His Lys Val 180 185 190Tyr Ala Cys Glu Val Thr His
Gln Gly Leu Ser Ser Pro Val Thr Lys 195 200 205Ser Phe Asn Arg Gly
Glu Cys 210 215412125PRTArtificial Sequence5H7 variable heavy chain
412Gln Val Gln Leu Gln Glu Ser Gly Pro Gly Leu Val Lys Pro Ser Gln1
5 10 15Thr Leu Ser Leu Thr Cys Thr Val Ser Gly Gly Ser Ile Ser Ser
Gly 20 25 30Gly Tyr Phe Trp Ser Trp Ile Arg Gln His Pro Gly Lys Gly
Leu Glu 35 40 45Trp Ile Gly Tyr Ile Tyr Tyr Ser Gly Thr Thr Tyr Tyr
Asn Pro Ser 50 55 60Leu Lys Ser Arg Val Thr Ile Ser Ile Asp Thr Ser
Lys Asn His Phe65 70 75 80Ser Leu Lys Leu Ser Ser Val Thr Ala Ala
Asp Thr Ala Val Tyr Tyr 85 90 95Cys Ala Arg Asp Leu Phe Tyr Tyr Asp
Ser Ser Gly Pro Arg Gly Phe 100 105 110Asp Pro Trp Gly Gln Gly Thr
Leu Val Thr Val Ser Ser 115 120 125413109PRTArtificial Sequence5H7
variable light chain 413Glu Ile Val Leu Thr Gln Ser Pro Gly Thr Leu
Ser Leu Ser Pro Gly1 5 10 15Glu Arg Ala Thr Leu Ser Cys Arg Ala Ser
Gln Thr Val Ser Ser Asn 20 25 30Tyr Leu Ala Trp Tyr Gln Gln Lys Pro
Gly Gln Ala Pro Arg Leu Leu 35 40 45Ile Tyr Gly Ser Ser Thr Arg Ala
Thr Gly Ile Pro Asp Arg Phe Ser 50 55 60Gly Ser Gly Ser Gly Thr Asp
Phe Thr Leu Thr Ile Ser Arg Leu Glu65 70 75 80Pro Glu Asp Phe Ala
Val Tyr Tyr Cys Gln Gln Tyr Asp Ser Ser Pro 85 90 95Trp Thr Phe Gly
Gln Gly Thr Lys Val Glu Ile Lys Arg 100 1054147PRTArtificial
Sequence5H7 heavy chain CDR1 414Ser Gly Gly Tyr Phe Trp Ser1
541516PRTArtificial Sequence5H7 heavy chain CDR2 415Tyr Ile Tyr Tyr
Ser Gly Thr Thr Tyr Tyr Asn Pro Ser Leu Lys Ser1 5 10
1541615PRTArtificial Sequence5H7 heavy chain CDR3 416Asp Leu Phe
Tyr Tyr Asp Ser Ser Gly Pro Arg Gly Phe Asp Pro1 5 10
1541712PRTArtificial Sequence5H7 light chain CDR1 417Arg Ala Ser
Gln Thr Val Ser Ser Asn Tyr Leu Ala1 5 104187PRTArtificial
Sequence5H7 light chain CDR2 418Gly Ser Ser Thr Arg Ala Thr1
54199PRTArtificial Sequence5H7 light chain CDR3 419Gln Gln Tyr Asp
Ser Ser Pro Trp Thr1 5420453PRTArtificial Sequence7A10 heavy chain
420Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1
5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser
Tyr 20 25 30Gly Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu
Trp Met 35 40 45Ala Val Ile Trp Tyr Val Gly Ser Asn Lys Tyr Tyr Ala
Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys
Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Ser Ala Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Gly Gly Glu Leu Gly Arg Asp
Tyr Tyr Ser Gly Met Asp Val 100 105 110Trp Gly Gln Gly Thr Thr Val
Thr Val Ser Ser Ala Ser Thr Lys Gly 115 120 125Pro Ser Val Phe Pro
Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly 130 135 140Thr Ala Ala
Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val145 150 155
160Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe
165 170 175Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser
Val Val 180 185 190Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr
Ile Cys Asn Val 195 200 205Asn His Lys Pro Ser Asn Thr Lys Val Asp
Lys Arg Val Glu Pro Lys 210 215 220Ser Cys Asp Lys Thr His Thr Cys
Pro Pro Cys Pro Ala Pro Glu Leu225 230 235 240Leu Gly Gly Pro Ser
Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr 245 250 255Leu Met Ile
Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val 260 265 270Ser
His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val 275 280
285Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser
290 295 300Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp
Trp Leu305 310 315 320Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn
Lys Ala Leu Pro Ala 325 330 335Pro Ile Glu Lys Thr Ile Ser Lys Ala
Lys Gly Gln Pro Arg Glu Pro 340 345 350Gln Val Tyr Thr Leu Pro Pro
Ser Arg Glu Glu Met Thr Lys Asn Gln 355 360 365Val Ser Leu Thr Cys
Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala 370 375 380Val Glu Trp
Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr385 390 395
400Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu
405 410 415Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser
Cys Ser 420 425 430Val Met His Glu Ala Leu His Asn His Tyr Thr Gln
Lys Ser Leu Ser 435 440 445Leu Ser Pro Gly Lys
450421214PRTArtificial Sequence7A10 light chain 421Asp Ile Gln Met
Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val
Thr Ile Thr Cys Arg Ala Ser Gln Gly Ile Arg Asn Asp 20 25 30Leu Gly
Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr
Ala Ala Ser Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55
60Ser Gly Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65
70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln His Asn Ser Tyr Pro
Trp 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg Thr Val
Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln
Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn
Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn
Ala Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu
Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu
Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala
Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200
205Phe Asn Arg Gly Glu Cys 210422123PRTArtificial Sequence7A10
variable heavy chain 422Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val
Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Ser Ser Tyr 20 25 30Gly Met His Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Trp Met 35 40 45Ala Val Ile Trp Tyr Val Gly Ser
Asn Lys Tyr Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser
Arg Asp Asn Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser
Leu Ser Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Gly Gly
Glu Leu Gly Arg Asp Tyr Tyr Ser Gly Met Asp Val 100 105 110Trp Gly
Gln Gly Thr Thr Val Thr Val Ser Ser 115 120423108PRTArtificial
Sequence7A10 variable light chain 423Asp Ile Gln Met Thr Gln Ser
Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr
Cys Arg Ala Ser Gln Gly Ile Arg Asn Asp 20 25 30Leu Gly Trp Tyr Gln
Gln Lys Pro Gly Lys Ala Pro Lys Arg Leu Ile 35 40 45Tyr Ala Ala Ser
Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser
Gly Thr Glu Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu
Asp Phe Ala Thr Tyr Tyr Cys Gln Gln His Asn Ser Tyr Pro Trp 85 90
95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Arg 100
1054245PRTArtificial Sequence7A10 heavy chain CDR1 424Ser Tyr Gly
Met His1 542517PRTArtificial Sequence7A10 heavy chain CDR2 425Val
Ile Trp Tyr Val Gly Ser Asn Lys Tyr Tyr Ala Asp Ser Val Lys1
5 10 15Gly42614PRTArtificial Sequence7A10 heavy chain CDR3 426Gly
Gly Glu Leu Gly Arg Asp Tyr Tyr Ser Gly Met Asp Val1 5
1042711PRTArtificial Sequence7A10 light chain CDR1 427Arg Ala Ser
Gln Gly Ile Arg Asn Asp Leu Gly1 5 104287PRTArtificial Sequence7A10
light chain CDR2 428Ala Ala Ser Ser Leu Gln Ser1 54299PRTArtificial
Sequence7A10 light chain CDR3 429Gln Gln His Asn Ser Tyr Pro Trp
Thr1 5430453PRTArtificial Sequence9H6 heavy chain 430Gln Val Gln
Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser Leu
Arg Leu Ser Cys Val Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30Gly
Met His Trp Ile Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ala Val Ile Trp Tyr Glu Gly Ser Asn Lys Tyr Tyr Ala Asp Ser Val
50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu
Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val
Tyr Tyr Cys 85 90 95Ala Arg Gly Gly Arg Leu Gly Lys Asp Tyr Tyr Ser
Gly Met Asp Val 100 105 110Trp Gly Gln Gly Thr Thr Val Thr Val Ser
Ser Ala Ser Thr Lys Gly 115 120 125Pro Ser Val Phe Pro Leu Ala Pro
Ser Ser Lys Ser Thr Ser Gly Gly 130 135 140Thr Ala Ala Leu Gly Cys
Leu Val Lys Asp Tyr Phe Pro Glu Pro Val145 150 155 160Thr Val Ser
Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe 165 170 175Pro
Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val 180 185
190Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val
195 200 205Asn His Lys Pro Ser Asn Thr Lys Val Asp Lys Arg Val Glu
Pro Lys 210 215 220Ser Cys Asp Lys Thr His Thr Cys Pro Pro Cys Pro
Ala Pro Glu Leu225 230 235 240Leu Gly Gly Pro Ser Val Phe Leu Phe
Pro Pro Lys Pro Lys Asp Thr 245 250 255Leu Met Ile Ser Arg Thr Pro
Glu Val Thr Cys Val Val Val Asp Val 260 265 270Ser His Glu Asp Pro
Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val 275 280 285Glu Val His
Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser 290 295 300Thr
Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu305 310
315 320Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro
Ala 325 330 335Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro
Arg Glu Pro 340 345 350Gln Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu
Met Thr Lys Asn Gln 355 360 365Val Ser Leu Thr Cys Leu Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala 370 375 380Val Glu Trp Glu Ser Asn Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr385 390 395 400Pro Pro Val Leu
Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu 405 410 415Thr Val
Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser 420 425
430Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser
435 440 445Leu Ser Pro Gly Lys 450431214PRTArtificial Sequence9H6
light chain 431Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala
Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Gly
Ile Arg Asn Asp 20 25 30Leu Gly Trp Tyr Gln Gln Lys Pro Gly Lys Ala
Pro Asn Arg Leu Ile 35 40 45Tyr Ala Thr Ser Ser Leu Gln Ser Gly Val
Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Glu Phe Thr Leu
Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr
Cys Leu Gln His Asn Thr Tyr Pro Trp 85 90 95Thr Phe Gly Gln Gly Thr
Lys Val Glu Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe
Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala
Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135
140Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser
Gln145 150 155 160Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr
Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr
Glu Lys His Lys Val Tyr 180 185 190Ala Cys Glu Val Thr His Gln Gly
Leu Ser Ser Pro Val Thr Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys
210432123PRTArtificial Sequence9H6 variable heavy chain 432Gln Val
Gln Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser
Leu Arg Leu Ser Cys Val Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25
30Gly Met His Trp Ile Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val
35 40 45Ala Val Ile Trp Tyr Glu Gly Ser Asn Lys Tyr Tyr Ala Asp Ser
Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr
Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala
Val Tyr Tyr Cys 85 90 95Ala Arg Gly Gly Arg Leu Gly Lys Asp Tyr Tyr
Ser Gly Met Asp Val 100 105 110Trp Gly Gln Gly Thr Thr Val Thr Val
Ser Ser 115 120433108PRTArtificial Sequence9H6 variable light chain
433Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1
5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Gly Ile Arg Asn
Asp 20 25 30Leu Gly Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Asn Arg
Leu Ile 35 40 45Tyr Ala Thr Ser Ser Leu Gln Ser Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Leu Gln
His Asn Thr Tyr Pro Trp 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu
Ile Lys Arg 100 1054345PRTArtificial Sequence9H6 heavy chain CDR1
434Ser Tyr Gly Met His1 543517PRTArtificial Sequence9H6 heavy chain
CDR2 435Val Ile Trp Tyr Glu Gly Ser Asn Lys Tyr Tyr Ala Asp Ser Val
Lys1 5 10 15Gly43614PRTArtificial Sequence9H6 heavy chain CDR3
436Gly Gly Arg Leu Gly Lys Asp Tyr Tyr Ser Gly Met Asp Val1 5
1043711PRTArtificial Sequence9H6 light chain CDR1 437Arg Ala Ser
Gln Gly Ile Arg Asn Asp Leu Gly1 5 104387PRTArtificial Sequence9H6
light chain CDR2 438Ala Thr Ser Ser Leu Gln Ser1 54399PRTArtificial
Sequence9H6 light chain CDR3 439Leu Gln His Asn Thr Tyr Pro Trp
Thr1 5440177PRTArtificial SequenceGITRL 440Met Cys Leu Ser His Leu
Glu Asn Met Pro Leu Ser His Ser Arg Thr1 5 10 15Gln Gly Ala Gln Arg
Ser Ser Trp Lys Leu Trp Leu Phe Cys Ser Ile 20 25 30Val Met Leu Leu
Phe Leu Cys Ser Phe Ser Trp Leu Ile Phe Ile Phe 35 40 45Leu Gln Leu
Glu Thr Ala Lys Glu Pro Cys Met Ala Lys Phe Gly Pro 50 55 60Leu Pro
Ser Lys Trp Gln Met Ala Ser Ser Glu Pro Pro Cys Val Asn65 70 75
80Lys Val Ser Asp Trp Lys Leu Glu Ile Leu Gln Asn Gly Leu Tyr Leu
85 90 95Ile Tyr Gly Gln Val Ala Pro Asn Ala Asn Tyr Asn Asp Val Ala
Pro 100 105 110Phe Glu Val Arg Leu Tyr Lys Asn Lys Asp Met Ile Gln
Thr Leu Thr 115 120 125Asn Lys Ser Lys Ile Gln Asn Val Gly Gly Thr
Tyr Glu Leu His Val 130 135 140Gly Asp Thr Ile Asp Leu Ile Phe Asn
Ser Glu His Gln Val Leu Lys145 150 155 160Asn Asn Thr Tyr Trp Gly
Ile Ile Leu Leu Ala Asn Pro Gln Phe Ile 165 170
175Ser441125PRTArtificial SequenceGITRL soluble domain 441Thr Ala
Lys Glu Pro Cys Met Ala Lys Phe Gly Pro Leu Pro Ser Lys1 5 10 15Trp
Gln Met Ala Ser Ser Glu Pro Pro Cys Val Asn Lys Val Ser Asp 20 25
30Trp Lys Leu Glu Ile Leu Gln Asn Gly Leu Tyr Leu Ile Tyr Gly Gln
35 40 45Val Ala Pro Asn Ala Asn Tyr Asn Asp Val Ala Pro Phe Glu Val
Arg 50 55 60Leu Tyr Lys Asn Lys Asp Met Ile Gln Thr Leu Thr Asn Lys
Ser Lys65 70 75 80Ile Gln Asn Val Gly Gly Thr Tyr Glu Leu His Val
Gly Asp Thr Ile 85 90 95Asp Leu Ile Phe Asn Ser Glu His Gln Val Leu
Lys Asn Asn Thr Tyr 100 105 110Trp Gly Ile Ile Leu Leu Ala Asn Pro
Gln Phe Ile Ser 115 120 125442283PRTHomo Sapiens 442Met Glu Pro Pro
Gly Asp Trp Gly Pro Pro Pro Trp Arg Ser Thr Pro1 5 10 15Arg Thr Asp
Val Leu Arg Leu Val Leu Tyr Leu Thr Phe Leu Gly Ala 20 25 30Pro Cys
Tyr Ala Pro Ala Leu Pro Ser Cys Lys Glu Asp Glu Tyr Pro 35 40 45Val
Gly Ser Glu Cys Cys Pro Lys Cys Ser Pro Gly Tyr Arg Val Lys 50 55
60Glu Ala Cys Gly Glu Leu Thr Gly Thr Val Cys Glu Pro Cys Pro Pro65
70 75 80Gly Thr Tyr Ile Ala His Leu Asn Gly Leu Ser Lys Cys Leu Gln
Cys 85 90 95Gln Met Cys Asp Pro Ala Met Gly Leu Arg Ala Ser Arg Asn
Cys Ser 100 105 110Arg Thr Glu Asn Ala Val Cys Gly Cys Ser Pro Gly
His Phe Cys Ile 115 120 125Val Gln Asp Gly Asp His Cys Ala Ala Cys
Arg Ala Tyr Ala Thr Ser 130 135 140Ser Pro Gly Gln Arg Val Gln Lys
Gly Gly Thr Glu Ser Gln Asp Thr145 150 155 160Leu Cys Gln Asn Cys
Pro Pro Gly Thr Phe Ser Pro Asn Gly Thr Leu 165 170 175Glu Glu Cys
Gln His Gln Thr Lys Cys Ser Trp Leu Val Thr Lys Ala 180 185 190Gly
Ala Gly Thr Ser Ser Ser His Trp Val Trp Trp Phe Leu Ser Gly 195 200
205Ser Leu Val Ile Val Ile Val Cys Ser Thr Val Gly Leu Ile Ile Cys
210 215 220Val Lys Arg Arg Lys Pro Arg Gly Asp Val Val Lys Val Ile
Val Ser225 230 235 240Val Gln Arg Lys Arg Gln Glu Ala Glu Gly Glu
Ala Thr Val Ile Glu 245 250 255Ala Leu Gln Ala Pro Pro Asp Val Thr
Thr Val Ala Val Glu Glu Thr 260 265 270Ile Pro Ser Phe Thr Gly Arg
Ser Pro Asn His 275 280443240PRTArtificial SequenceLIGHT (HVEM
ligand) 443Met Glu Glu Ser Val Val Arg Pro Ser Val Phe Val Val Asp
Gly Gln1 5 10 15Thr Asp Ile Pro Phe Thr Arg Leu Gly Arg Ser His Arg
Arg Gln Ser 20 25 30Cys Ser Val Ala Arg Val Gly Leu Gly Leu Leu Leu
Leu Leu Met Gly 35 40 45Ala Gly Leu Ala Val Gln Gly Trp Phe Leu Leu
Gln Leu His Trp Arg 50 55 60Leu Gly Glu Met Val Thr Arg Leu Pro Asp
Gly Pro Ala Gly Ser Trp65 70 75 80Glu Gln Leu Ile Gln Glu Arg Arg
Ser His Glu Val Asn Pro Ala Ala 85 90 95His Leu Thr Gly Ala Asn Ser
Ser Leu Thr Gly Ser Gly Gly Pro Leu 100 105 110Leu Trp Glu Thr Gln
Leu Gly Leu Ala Phe Leu Arg Gly Leu Ser Tyr 115 120 125His Asp Gly
Ala Leu Val Val Thr Lys Ala Gly Tyr Tyr Tyr Ile Tyr 130 135 140Ser
Lys Val Gln Leu Gly Gly Val Gly Cys Pro Leu Gly Leu Ala Ser145 150
155 160Thr Ile Thr His Gly Leu Tyr Lys Arg Thr Pro Arg Tyr Pro Glu
Glu 165 170 175Leu Glu Leu Leu Val Ser Gln Gln Ser Pro Cys Gly Arg
Ala Thr Ser 180 185 190Ser Ser Arg Val Trp Trp Asp Ser Ser Phe Leu
Gly Gly Val Val His 195 200 205Leu Glu Ala Gly Glu Lys Val Val Val
Arg Val Leu Asp Glu Arg Leu 210 215 220Val Arg Leu Arg Asp Gly Thr
Arg Ser Tyr Phe Gly Ala Phe Met Val225 230 235
240444147PRTArtificial SequenceLIGHT soluble domain 444Pro Ala Ala
His Leu Thr Gly Ala Asn Ser Ser Leu Thr Gly Ser Gly1 5 10 15Gly Pro
Leu Leu Trp Glu Thr Gln Leu Gly Leu Ala Phe Leu Arg Gly 20 25 30Leu
Ser Tyr His Asp Gly Ala Leu Val Val Thr Lys Ala Gly Tyr Tyr 35 40
45Tyr Ile Tyr Ser Lys Val Gln Leu Gly Gly Val Gly Cys Pro Leu Gly
50 55 60Leu Ala Ser Thr Ile Thr His Gly Leu Tyr Lys Arg Thr Pro Arg
Tyr65 70 75 80Pro Glu Glu Leu Glu Leu Leu Val Ser Gln Gln Ser Pro
Cys Gly Arg 85 90 95Ala Thr Ser Ser Ser Arg Val Trp Trp Asp Ser Ser
Phe Leu Gly Gly 100 105 110Val Val His Leu Glu Ala Gly Glu Lys Val
Val Val Arg Val Leu Asp 115 120 125Glu Arg Leu Val Arg Leu Arg Asp
Gly Thr Arg Ser Tyr Phe Gly Ala 130 135 140Phe Met
Val145445146PRTArtificial SequenceLIGHT soluble domain
(alternative) 445Ala Ala His Leu Thr Gly Ala Asn Ser Ser Leu Thr
Gly Ser Gly Gly1 5 10 15Pro Leu Leu Trp Glu Thr Gln Leu Gly Leu Ala
Phe Leu Arg Gly Leu 20 25 30Ser Tyr His Asp Gly Ala Leu Val Val Thr
Lys Ala Gly Tyr Tyr Tyr 35 40 45Ile Tyr Ser Lys Val Gln Leu Gly Gly
Val Gly Cys Pro Leu Gly Leu 50 55 60Ala Ser Thr Ile Thr His Gly Leu
Tyr Lys Arg Thr Pro Arg Tyr Pro65 70 75 80Glu Glu Leu Glu Leu Leu
Val Ser Gln Gln Ser Pro Cys Gly Arg Ala 85 90 95Thr Ser Ser Ser Arg
Val Trp Trp Asp Ser Ser Phe Leu Gly Gly Val 100 105 110Val His Leu
Glu Ala Gly Glu Lys Val Val Val Arg Val Leu Asp Glu 115 120 125Arg
Leu Val Arg Leu Arg Asp Gly Thr Arg Ser Tyr Phe Gly Ala Phe 130 135
140Met Val145446145PRTArtificial SequenceLIGHT soluble domain
(alternative) 446Ala His Leu Thr Gly Ala Asn Ser Ser Leu Thr Gly
Ser Gly Gly Pro1 5 10 15Leu Leu Trp Glu Thr Gln Leu Gly Leu Ala Phe
Leu Arg Gly Leu Ser 20 25 30Tyr His Asp Gly Ala Leu Val Val Thr Lys
Ala Gly Tyr Tyr Tyr Ile 35 40 45Tyr Ser Lys Val Gln Leu Gly Gly Val
Gly Cys Pro Leu Gly Leu Ala 50 55 60Ser Thr Ile Thr His Gly Leu Tyr
Lys Arg Thr Pro Arg Tyr Pro Glu65 70 75 80Glu Leu Glu Leu Leu Val
Ser Gln Gln Ser Pro Cys Gly Arg Ala Thr 85 90 95Ser Ser Ser Arg Val
Trp Trp Asp Ser Ser Phe Leu Gly Gly Val Val 100 105 110His Leu Glu
Ala Gly Glu Lys Val Val Val Arg Val Leu Asp Glu Arg 115 120 125Leu
Val Arg Leu Arg Asp Gly Thr Arg Ser Tyr Phe Gly Ala Phe Met 130 135
140Val145447335PRTHomo Sapiens 447Met Leu Gly Ile Trp Thr Leu Leu
Pro Leu Val Leu Thr Ser Val Ala1 5 10 15Arg Leu Ser Ser Lys Ser Val
Asn Ala Gln Val Thr Asp Ile Asn Ser 20 25 30Lys Gly Leu Glu Leu Arg
Lys Thr Val Thr Thr Val Glu Thr Gln Asn 35 40 45Leu Glu Gly Leu His
His Asp Gly Gln Phe Cys His Lys Pro Cys Pro 50 55 60Pro Gly Glu Arg
Lys Ala Arg Asp Cys Thr Val Asn Gly Asp Glu Pro65 70
75 80Asp Cys Val Pro Cys Gln Glu Gly Lys Glu Tyr Thr Asp Lys Ala
His 85 90 95Phe Ser Ser Lys Cys Arg Arg Cys Arg Leu Cys Asp Glu Gly
His Gly 100 105 110Leu Glu Val Glu Ile Asn Cys Thr Arg Thr Gln Asn
Thr Lys Cys Arg 115 120 125Cys Lys Pro Asn Phe Phe Cys Asn Ser Thr
Val Cys Glu His Cys Asp 130 135 140Pro Cys Thr Lys Cys Glu His Gly
Ile Ile Lys Glu Cys Thr Leu Thr145 150 155 160Ser Asn Thr Lys Cys
Lys Glu Glu Gly Ser Arg Ser Asn Leu Gly Trp 165 170 175Leu Cys Leu
Leu Leu Leu Pro Ile Pro Leu Ile Val Trp Val Lys Arg 180 185 190Lys
Glu Val Gln Lys Thr Cys Arg Lys His Arg Lys Glu Asn Gln Gly 195 200
205Ser His Glu Ser Pro Thr Leu Asn Pro Glu Thr Val Ala Ile Asn Leu
210 215 220Ser Asp Val Asp Leu Ser Lys Tyr Ile Thr Thr Ile Ala Gly
Val Met225 230 235 240Thr Leu Ser Gln Val Lys Gly Phe Val Arg Lys
Asn Gly Val Asn Glu 245 250 255Ala Lys Ile Asp Glu Ile Lys Asn Asp
Asn Val Gln Asp Thr Ala Glu 260 265 270Gln Lys Val Gln Leu Leu Arg
Asn Trp His Gln Leu His Gly Lys Lys 275 280 285Glu Ala Tyr Asp Thr
Leu Ile Lys Asp Leu Lys Lys Ala Asn Leu Cys 290 295 300Thr Leu Ala
Glu Lys Ile Gln Thr Ile Ile Leu Lys Asp Ile Thr Ser305 310 315
320Asp Ser Glu Asn Ser Asn Phe Arg Asn Glu Ile Gln Ser Leu Val 325
330 335448314PRTHomo Sapiens 448Met Leu Gly Ile Trp Thr Leu Leu Pro
Leu Val Leu Thr Ser Val Ala1 5 10 15Arg Leu Ser Ser Lys Ser Val Asn
Ala Gln Val Thr Asp Ile Asn Ser 20 25 30Lys Gly Leu Glu Leu Arg Lys
Thr Val Thr Thr Val Glu Thr Gln Asn 35 40 45Leu Glu Gly Leu His His
Asp Gly Gln Phe Cys His Lys Pro Cys Pro 50 55 60Pro Gly Glu Arg Lys
Ala Arg Asp Cys Thr Val Asn Gly Asp Glu Pro65 70 75 80Asp Cys Val
Pro Cys Gln Glu Gly Lys Glu Tyr Thr Asp Lys Ala His 85 90 95Phe Ser
Ser Lys Cys Arg Arg Cys Arg Leu Cys Asp Glu Gly His Gly 100 105
110Leu Glu Val Glu Ile Asn Cys Thr Arg Thr Gln Asn Thr Lys Cys Arg
115 120 125Cys Lys Pro Asn Phe Phe Cys Asn Ser Thr Val Cys Glu His
Cys Asp 130 135 140Pro Cys Thr Lys Cys Glu His Gly Ile Ile Lys Glu
Cys Thr Leu Thr145 150 155 160Ser Asn Thr Lys Cys Lys Glu Glu Val
Lys Arg Lys Glu Val Gln Lys 165 170 175Thr Cys Arg Lys His Arg Lys
Glu Asn Gln Gly Ser His Glu Ser Pro 180 185 190Thr Leu Asn Pro Glu
Thr Val Ala Ile Asn Leu Ser Asp Val Asp Leu 195 200 205Ser Lys Tyr
Ile Thr Thr Ile Ala Gly Val Met Thr Leu Ser Gln Val 210 215 220Lys
Gly Phe Val Arg Lys Asn Gly Val Asn Glu Ala Lys Ile Asp Glu225 230
235 240Ile Lys Asn Asp Asn Val Gln Asp Thr Ala Glu Gln Lys Val Gln
Leu 245 250 255Leu Arg Asn Trp His Gln Leu His Gly Lys Lys Glu Ala
Tyr Asp Thr 260 265 270Leu Ile Lys Asp Leu Lys Lys Ala Asn Leu Cys
Thr Leu Ala Glu Lys 275 280 285Ile Gln Thr Ile Ile Leu Lys Asp Ile
Thr Ser Asp Ser Glu Asn Ser 290 295 300Asn Phe Arg Asn Glu Ile Gln
Ser Leu Val305 310449220PRTHomo Sapiens 449Met Leu Gly Ile Trp Thr
Leu Leu Pro Leu Val Leu Thr Ser Val Ala1 5 10 15Arg Leu Ser Ser Lys
Ser Val Asn Ala Gln Val Thr Asp Ile Asn Ser 20 25 30Lys Gly Leu Glu
Leu Arg Lys Thr Val Thr Thr Val Glu Thr Gln Asn 35 40 45Leu Glu Gly
Leu His His Asp Gly Gln Phe Cys His Lys Pro Cys Pro 50 55 60Pro Gly
Glu Arg Lys Ala Arg Asp Cys Thr Val Asn Gly Asp Glu Pro65 70 75
80Asp Cys Val Pro Cys Gln Glu Gly Lys Glu Tyr Thr Asp Lys Ala His
85 90 95Phe Ser Ser Lys Cys Arg Arg Cys Arg Leu Cys Asp Glu Gly His
Gly 100 105 110Leu Glu Val Glu Ile Asn Cys Thr Arg Thr Gln Asn Thr
Lys Cys Arg 115 120 125Cys Lys Pro Asn Phe Phe Cys Asn Ser Thr Val
Cys Glu His Cys Asp 130 135 140Pro Cys Thr Lys Cys Glu His Gly Ile
Ile Lys Glu Cys Thr Leu Thr145 150 155 160Ser Asn Thr Lys Cys Lys
Glu Glu Gly Ser Arg Ser Asn Leu Gly Trp 165 170 175Leu Cys Leu Leu
Leu Leu Pro Ile Pro Leu Ile Val Trp Val Lys Arg 180 185 190Lys Glu
Val Gln Lys Thr Cys Arg Lys His Arg Lys Glu Asn Gln Gly 195 200
205Ser His Glu Ser Pro Thr Leu Asn Pro Met Leu Thr 210 215
220450197PRTHomo Sapiens 450Met Leu Gly Ile Trp Thr Leu Leu Pro Leu
Val Leu Thr Ser Val Ala1 5 10 15Arg Leu Ser Ser Lys Ser Val Asn Ala
Gln Val Thr Asp Ile Asn Ser 20 25 30Lys Gly Leu Glu Leu Arg Lys Thr
Val Thr Thr Val Glu Thr Gln Asn 35 40 45Leu Glu Gly Leu His His Asp
Gly Gln Phe Cys His Lys Pro Cys Pro 50 55 60Pro Gly Glu Arg Lys Ala
Arg Asp Cys Thr Val Asn Gly Asp Glu Pro65 70 75 80Asp Cys Val Pro
Cys Gln Glu Gly Lys Glu Tyr Thr Asp Lys Ala His 85 90 95Phe Ser Ser
Lys Cys Arg Arg Cys Arg Leu Cys Asp Glu Gly His Gly 100 105 110Leu
Glu Val Glu Ile Asn Cys Thr Arg Thr Gln Asn Thr Lys Cys Arg 115 120
125Cys Lys Pro Asn Phe Phe Cys Asn Ser Thr Val Cys Glu His Cys Asp
130 135 140Pro Cys Thr Lys Cys Glu His Gly Ile Ile Lys Glu Cys Thr
Leu Thr145 150 155 160Ser Asn Thr Lys Cys Lys Glu Glu Gly Ser Arg
Ser Asn Leu Gly Trp 165 170 175Leu Cys Leu Leu Leu Leu Pro Ile Pro
Leu Ile Val Trp Gly Asn Ser 180 185 190Gly Asn Lys Phe Ile
195451132PRTArtificial Sequenceheavy chain variable region for E09
451Gln Leu Gln Leu Gln Glu Ser Gly Pro Gly Leu Val Lys Pro Ser Glu1
5 10 15Thr Leu Ser Leu Thr Cys Thr Val Ser Gly Ala Ser Ile Ser Ala
Asn 20 25 30Ser Tyr Tyr Gly Val Trp Val Arg Gln Ser Pro Gly Lys Gly
Leu Glu 35 40 45Trp Val Gly Ser Ile Ala Tyr Arg Gly Asn Ser Asn Ser
Gly Ser Thr 50 55 60Tyr Tyr Asn Pro Ser Leu Lys Ser Arg Ala Thr Val
Ser Val Asp Thr65 70 75 80Ser Lys Asn Gln Val Ser Leu Arg Leu Thr
Ser Val Thr Ala Ala Asp 85 90 95Thr Ala Leu Tyr Tyr Cys Ala Arg Arg
Gln Leu Leu Asp Asp Gly Thr 100 105 110Gly Tyr Gln Trp Ala Ala Phe
Asp Val Trp Gly Gln Gly Thr Met Val 115 120 125Thr Val Ser Ser
130452110PRTArtificial Sequencelight chain variable region for E09
452Gln Ser Val Leu Thr Gln Pro Pro Ser Val Ser Glu Ala Pro Arg Gln1
5 10 15Thr Val Thr Ile Ser Cys Ser Gly Asn Ser Phe Asn Ile Gly Arg
Tyr 20 25 30Pro Val Asn Trp Tyr Gln Gln Leu Pro Gly Lys Ala Pro Lys
Leu Leu 35 40 45Ile Tyr Tyr Asn Asn Leu Arg Phe Ser Gly Val Ser Asp
Arg Phe Ser 50 55 60Gly Ser Lys Ser Gly Thr Ser Ala Ser Leu Ala Ile
Arg Asp Leu Leu65 70 75 80Ser Glu Asp Glu Ala Asp Tyr Tyr Cys Ser
Thr Trp Asp Asp Thr Leu 85 90 95Lys Gly Trp Val Phe Gly Gly Gly Thr
Lys Val Thr Val Leu 100 105 1104537PRTArtificial Sequenceheavy
chain CDRl for E09 453Ala Asn Ser Tyr Tyr Gly Val1
545422PRTArtificial Sequenceheavy chain CDR2 for E09 454Gly Ser Ile
Ala Tyr Arg Gly Asn Ser Asn Ser Gly Ser Thr Tyr Tyr1 5 10 15Asn Pro
Ser Leu Lys Ser 2045517PRTArtificial Sequenceheavy chain CDR3 for
E09 455Arg Gln Leu Leu Asp Asp Gly Thr Gly Tyr Gln Trp Ala Ala Phe
Asp1 5 10 15Val45613PRTArtificial Sequencelight chain CDR1 for E09
456Ser Gly Asn Ser Phe Asn Ile Gly Arg Tyr Pro Val Asn1 5
104577PRTArtificial Sequencelight chain CDR2 for E09 457Tyr Asn Asn
Leu Arg Phe Ser1 545811PRTArtificial Sequencelight chain CDR3 for
E09 458Ser Thr Trp Asp Asp Thr Leu Lys Gly Trp Val1 5
10459281PRTArtificial SequenceCD95L (CD95 ligand) 459Met Gln Gln
Pro Phe Asn Tyr Pro Tyr Pro Gln Ile Tyr Trp Val Asp1 5 10 15Ser Ser
Ala Ser Ser Pro Trp Ala Pro Pro Gly Thr Val Leu Pro Cys 20 25 30Pro
Thr Ser Val Pro Arg Arg Pro Gly Gln Arg Arg Pro Pro Pro Pro 35 40
45Pro Pro Pro Pro Pro Leu Pro Pro Pro Pro Pro Pro Pro Pro Leu Pro
50 55 60Pro Leu Pro Leu Pro Pro Leu Lys Lys Arg Gly Asn His Ser Thr
Gly65 70 75 80Leu Cys Leu Leu Val Met Phe Phe Met Val Leu Val Ala
Leu Val Gly 85 90 95Leu Gly Leu Gly Met Phe Gln Leu Phe His Leu Gln
Lys Glu Leu Ala 100 105 110Glu Leu Arg Glu Ser Thr Ser Gln Met His
Thr Ala Ser Ser Leu Glu 115 120 125Lys Gln Ile Gly His Pro Ser Pro
Pro Pro Glu Lys Lys Glu Leu Arg 130 135 140Lys Val Ala His Leu Thr
Gly Lys Ser Asn Ser Arg Ser Met Pro Leu145 150 155 160Glu Trp Glu
Asp Thr Tyr Gly Ile Val Leu Leu Ser Gly Val Lys Tyr 165 170 175Lys
Lys Gly Gly Leu Val Ile Asn Glu Thr Gly Leu Tyr Phe Val Tyr 180 185
190Ser Lys Val Tyr Phe Arg Gly Gln Ser Cys Asn Asn Leu Pro Leu Ser
195 200 205His Lys Val Tyr Met Arg Asn Ser Lys Tyr Pro Gln Asp Leu
Val Met 210 215 220Met Glu Gly Lys Met Met Ser Tyr Cys Thr Thr Gly
Gln Met Trp Ala225 230 235 240Arg Ser Ser Tyr Leu Gly Ala Val Phe
Asn Leu Thr Ser Ala Asp His 245 250 255Leu Tyr Val Asn Val Ser Glu
Leu Ser Leu Val Asn Phe Glu Glu Ser 260 265 270Gln Thr Phe Phe Gly
Leu Tyr Lys Leu 275 280460136PRTArtificial SequenceCD95L soluble
domain 460Val Ala His Leu Thr Gly Lys Ser Asn Ser Arg Ser Met Pro
Leu Glu1 5 10 15Trp Glu Asp Thr Tyr Gly Ile Val Leu Leu Ser Gly Val
Lys Tyr Lys 20 25 30Lys Gly Gly Leu Val Ile Asn Glu Thr Gly Leu Tyr
Phe Val Tyr Ser 35 40 45Lys Val Tyr Phe Arg Gly Gln Ser Cys Asn Asn
Leu Pro Leu Ser His 50 55 60Lys Val Tyr Met Arg Asn Ser Lys Tyr Pro
Gln Asp Leu Val Met Met65 70 75 80Glu Gly Lys Met Met Ser Tyr Cys
Thr Thr Gly Gln Met Trp Ala Arg 85 90 95Ser Ser Tyr Leu Gly Ala Val
Phe Asn Leu Thr Ser Ala Asp His Leu 100 105 110Tyr Val Asn Val Ser
Glu Leu Ser Leu Val Asn Phe Glu Glu Ser Gln 115 120 125Thr Phe Phe
Gly Leu Tyr Lys Leu 130 135461135PRTArtificial SequenceCD95L
soluble domain (alternative) 461Ala His Leu Thr Gly Lys Ser Asn Ser
Arg Ser Met Pro Leu Glu Trp1 5 10 15Glu Asp Thr Tyr Gly Ile Val Leu
Leu Ser Gly Val Lys Tyr Lys Lys 20 25 30Gly Gly Leu Val Ile Asn Glu
Thr Gly Leu Tyr Phe Val Tyr Ser Lys 35 40 45Val Tyr Phe Arg Gly Gln
Ser Cys Asn Asn Leu Pro Leu Ser His Lys 50 55 60Val Tyr Met Arg Asn
Ser Lys Tyr Pro Gln Asp Leu Val Met Met Glu65 70 75 80Gly Lys Met
Met Ser Tyr Cys Thr Thr Gly Gln Met Trp Ala Arg Ser 85 90 95Ser Tyr
Leu Gly Ala Val Phe Asn Leu Thr Ser Ala Asp His Leu Tyr 100 105
110Val Asn Val Ser Glu Leu Ser Leu Val Asn Phe Glu Glu Ser Gln Thr
115 120 125Phe Phe Gly Leu Tyr Lys Leu 130 135462134PRTArtificial
SequenceCD95L soluble domain (alternative) 462His Leu Thr Gly Lys
Ser Asn Ser Arg Ser Met Pro Leu Glu Trp Glu1 5 10 15Asp Thr Tyr Gly
Ile Val Leu Leu Ser Gly Val Lys Tyr Lys Lys Gly 20 25 30Gly Leu Val
Ile Asn Glu Thr Gly Leu Tyr Phe Val Tyr Ser Lys Val 35 40 45Tyr Phe
Arg Gly Gln Ser Cys Asn Asn Leu Pro Leu Ser His Lys Val 50 55 60Tyr
Met Arg Asn Ser Lys Tyr Pro Gln Asp Leu Val Met Met Glu Gly65 70 75
80Lys Met Met Ser Tyr Cys Thr Thr Gly Gln Met Trp Ala Arg Ser Ser
85 90 95Tyr Leu Gly Ala Val Phe Asn Leu Thr Ser Ala Asp His Leu Tyr
Val 100 105 110Asn Val Ser Glu Leu Ser Leu Val Asn Phe Glu Glu Ser
Gln Thr Phe 115 120 125Phe Gly Leu Tyr Lys Leu
130463440PRTArtificial Sequencenivolumab heavy chain 463Gln Val Gln
Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser Leu
Arg Leu Asp Cys Lys Ala Ser Gly Ile Thr Phe Ser Asn Ser 20 25 30Gly
Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ala Val Ile Trp Tyr Asp Gly Ser Lys Arg Tyr Tyr Ala Asp Ser Val
50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu
Phe65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val
Tyr Tyr Cys 85 90 95Ala Thr Asn Asp Asp Tyr Trp Gly Gln Gly Thr Leu
Val Thr Val Ser 100 105 110Ser Ala Ser Thr Lys Gly Pro Ser Val Phe
Pro Leu Ala Pro Cys Ser 115 120 125Arg Ser Thr Ser Glu Ser Thr Ala
Ala Leu Gly Cys Leu Val Lys Asp 130 135 140Tyr Phe Pro Glu Pro Val
Thr Val Ser Trp Asn Ser Gly Ala Leu Thr145 150 155 160Ser Gly Val
His Thr Phe Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr 165 170 175Ser
Leu Ser Ser Val Val Thr Val Pro Ser Ser Ser Leu Gly Thr Lys 180 185
190Thr Tyr Thr Cys Asn Val Asp His Lys Pro Ser Asn Thr Lys Val Asp
195 200 205Lys Arg Val Glu Ser Lys Tyr Gly Pro Pro Cys Pro Pro Cys
Pro Ala 210 215 220Pro Glu Phe Leu Gly Gly Pro Ser Val Phe Leu Phe
Pro Pro Lys Pro225 230 235 240Lys Asp Thr Leu Met Ile Ser Arg Thr
Pro Glu Val Thr Cys Val Val 245 250 255Val Asp Val Ser Gln Glu Asp
Pro Glu Val Gln Phe Asn Trp Tyr Val 260 265 270Asp Gly Val Glu Val
His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln 275 280 285Phe Asn Ser
Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln 290 295 300Asp
Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Gly305 310
315 320Leu Pro Ser Ser Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln
Pro 325 330 335Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Gln Glu
Glu Met Thr 340 345 350Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys
Gly Phe Tyr Pro Ser 355 360 365Asp Ile Ala Val Glu Trp Glu Ser Asn
Gly Gln Pro Glu Asn Asn Tyr 370 375
380Lys Thr Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu
Tyr385 390 395 400Ser Arg Leu Thr Val Asp Lys Ser Arg Trp Gln Glu
Gly Asn Val Phe 405 410 415Ser Cys Ser Val Met His Glu Ala Leu His
Asn His Tyr Thr Gln Lys 420 425 430Ser Leu Ser Leu Ser Leu Gly Lys
435 440464214PRTArtificial Sequencenivolumab light chain 464Glu Ile
Val Leu Thr Gln Ser Pro Ala Thr Leu Ser Leu Ser Pro Gly1 5 10 15Glu
Arg Ala Thr Leu Ser Cys Arg Ala Ser Gln Ser Val Ser Ser Tyr 20 25
30Leu Ala Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro Arg Leu Leu Ile
35 40 45Tyr Asp Ala Ser Asn Arg Ala Thr Gly Ile Pro Ala Arg Phe Ser
Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu
Glu Pro65 70 75 80Glu Asp Phe Ala Val Tyr Tyr Cys Gln Gln Ser Ser
Asn Trp Pro Arg 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys
Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser
Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu
Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys
Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser
Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170
175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr
180 185 190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr
Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys 210465113PRTArtificial
Sequencenivolumab variable heavy chain 465Gln Val Gln Leu Val Glu
Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Asp
Cys Lys Ala Ser Gly Ile Thr Phe Ser Asn Ser 20 25 30Gly Met His Trp
Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Val Ile
Trp Tyr Asp Gly Ser Lys Arg Tyr Tyr Ala Asp Ser Val 50 55 60Lys Gly
Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Phe65 70 75
80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Thr Asn Asp Asp Tyr Trp Gly Gln Gly Thr Leu Val Thr Val
Ser 100 105 110Ser466107PRTArtificial Sequencenivolumab variable
light chain 466Glu Ile Val Leu Thr Gln Ser Pro Ala Thr Leu Ser Leu
Ser Pro Gly1 5 10 15Glu Arg Ala Thr Leu Ser Cys Arg Ala Ser Gln Ser
Val Ser Ser Tyr 20 25 30Leu Ala Trp Tyr Gln Gln Lys Pro Gly Gln Ala
Pro Arg Leu Leu Ile 35 40 45Tyr Asp Ala Ser Asn Arg Ala Thr Gly Ile
Pro Ala Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu
Thr Ile Ser Ser Leu Glu Pro65 70 75 80Glu Asp Phe Ala Val Tyr Tyr
Cys Gln Gln Ser Ser Asn Trp Pro Arg 85 90 95Thr Phe Gly Gln Gly Thr
Lys Val Glu Ile Lys 100 1054675PRTArtificial Sequencenivolumab
heavy chain CDR1 467Asn Ser Gly Met His1 546817PRTArtificial
Sequencenivolumab heavy chain CDR2 468Val Ile Trp Tyr Asp Gly Ser
Lys Arg Tyr Tyr Ala Asp Ser Val Lys1 5 10 15Gly4694PRTArtificial
Sequencenivolumab heavy chain CDR3 469Asn Asp Asp
Tyr147011PRTArtificial Sequencenivolumab light chain CDR1 470Arg
Ala Ser Gln Ser Val Ser Ser Tyr Leu Ala1 5 104717PRTArtificial
Sequencenivolumab light chain CDR2 471Asp Ala Ser Asn Arg Ala Thr1
54729PRTArtificial Sequencenivolumab light chain CDR3 472Gln Gln
Ser Ser Asn Trp Pro Arg Thr1 5473447PRTArtificial
Sequencepembrolizumab heavy chain 473Gln Val Gln Leu Val Gln Ser
Gly Val Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys
Lys Ala Ser Gly Tyr Thr Phe Thr Asn Tyr 20 25 30Tyr Met Tyr Trp Val
Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Gly Ile Asn
Pro Ser Asn Gly Gly Thr Asn Phe Asn Glu Lys Phe 50 55 60Lys Asn Arg
Val Thr Leu Thr Thr Asp Ser Ser Thr Thr Thr Ala Tyr65 70 75 80Met
Glu Leu Lys Ser Leu Gln Phe Asp Asp Thr Ala Val Tyr Tyr Cys 85 90
95Ala Arg Arg Asp Tyr Arg Phe Asp Met Gly Phe Asp Tyr Trp Gly Gln
100 105 110Gly Thr Thr Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro
Ser Val 115 120 125Phe Pro Leu Ala Pro Cys Ser Arg Ser Thr Ser Glu
Ser Thr Ala Ala 130 135 140Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro
Glu Pro Val Thr Val Ser145 150 155 160Trp Asn Ser Gly Ala Leu Thr
Ser Gly Val His Thr Phe Pro Ala Val 165 170 175Leu Gln Ser Ser Gly
Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro 180 185 190Ser Ser Ser
Leu Gly Thr Lys Thr Tyr Thr Cys Asn Val Asp His Lys 195 200 205Pro
Ser Asn Thr Lys Val Asp Lys Arg Val Glu Ser Lys Tyr Gly Pro 210 215
220Pro Cys Pro Pro Cys Pro Ala Pro Glu Phe Leu Gly Gly Pro Ser
Val225 230 235 240Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met
Ile Ser Arg Thr 245 250 255Pro Glu Val Thr Cys Val Val Val Asp Val
Ser Gln Glu Asp Pro Glu 260 265 270Val Gln Phe Asn Trp Tyr Val Asp
Gly Val Glu Val His Asn Ala Lys 275 280 285Thr Lys Pro Arg Glu Glu
Gln Phe Asn Ser Thr Tyr Arg Val Val Ser 290 295 300Val Leu Thr Val
Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys305 310 315 320Cys
Lys Val Ser Asn Lys Gly Leu Pro Ser Ser Ile Glu Lys Thr Ile 325 330
335Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro
340 345 350Pro Ser Gln Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr
Cys Leu 355 360 365Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu
Trp Glu Ser Asn 370 375 380Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr
Pro Pro Val Leu Asp Ser385 390 395 400Asp Gly Ser Phe Phe Leu Tyr
Ser Arg Leu Thr Val Asp Lys Ser Arg 405 410 415Trp Gln Glu Gly Asn
Val Phe Ser Cys Ser Val Met His Glu Ala Leu 420 425 430His Asn His
Tyr Thr Gln Lys Ser Leu Ser Leu Ser Leu Gly Lys 435 440
445474218PRTArtificial Sequencepembrolizumab light chain 474Glu Ile
Val Leu Thr Gln Ser Pro Ala Thr Leu Ser Leu Ser Pro Gly1 5 10 15Glu
Arg Ala Thr Leu Ser Cys Arg Ala Ser Lys Gly Val Ser Thr Ser 20 25
30Gly Tyr Ser Tyr Leu His Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro
35 40 45Arg Leu Leu Ile Tyr Leu Ala Ser Tyr Leu Glu Ser Gly Val Pro
Ala 50 55 60Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr
Ile Ser65 70 75 80Ser Leu Glu Pro Glu Asp Phe Ala Val Tyr Tyr Cys
Gln His Ser Arg 85 90 95Asp Leu Pro Leu Thr Phe Gly Gly Gly Thr Lys
Val Glu Ile Lys Arg 100 105 110Thr Val Ala Ala Pro Ser Val Phe Ile
Phe Pro Pro Ser Asp Glu Gln 115 120 125Leu Lys Ser Gly Thr Ala Ser
Val Val Cys Leu Leu Asn Asn Phe Tyr 130 135 140Pro Arg Glu Ala Lys
Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser145 150 155 160Gly Asn
Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr 165 170
175Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys
180 185 190His Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser
Ser Pro 195 200 205Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 210
215475120PRTArtificial Sequencepembrolizumab variable heavy chain
475Gln Val Gln Leu Val Gln Ser Gly Val Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Asn
Tyr 20 25 30Tyr Met Tyr Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Gly Ile Asn Pro Ser Asn Gly Gly Thr Asn Phe Asn
Glu Lys Phe 50 55 60Lys Asn Arg Val Thr Leu Thr Thr Asp Ser Ser Thr
Thr Thr Ala Tyr65 70 75 80Met Glu Leu Lys Ser Leu Gln Phe Asp Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Arg Asp Tyr Arg Phe Asp Met
Gly Phe Asp Tyr Trp Gly Gln 100 105 110Gly Thr Thr Val Thr Val Ser
Ser 115 120476111PRTArtificial Sequencepembrolizumab variable light
chain 476Glu Ile Val Leu Thr Gln Ser Pro Ala Thr Leu Ser Leu Ser
Pro Gly1 5 10 15Glu Arg Ala Thr Leu Ser Cys Arg Ala Ser Lys Gly Val
Ser Thr Ser 20 25 30Gly Tyr Ser Tyr Leu His Trp Tyr Gln Gln Lys Pro
Gly Gln Ala Pro 35 40 45Arg Leu Leu Ile Tyr Leu Ala Ser Tyr Leu Glu
Ser Gly Val Pro Ala 50 55 60Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp
Phe Thr Leu Thr Ile Ser65 70 75 80Ser Leu Glu Pro Glu Asp Phe Ala
Val Tyr Tyr Cys Gln His Ser Arg 85 90 95Asp Leu Pro Leu Thr Phe Gly
Gly Gly Thr Lys Val Glu Ile Lys 100 105 1104775PRTArtificial
Sequencepembrolizumab heavy chain CDR1 477Asn Tyr Tyr Met Tyr1
547816PRTArtificial Sequencepembrolizumab heavy chain CDR2 478Gly
Ile Asn Pro Ser Asn Gly Gly Thr Asn Phe Asn Glu Lys Phe Lys1 5 10
1547911PRTArtificial Sequencepembrolizumab heavy chain CDR3 479Arg
Asp Tyr Arg Phe Asp Met Gly Phe Asp Tyr1 5 1048015PRTArtificial
Sequencepembrolizumab light chain CDR1 480Arg Ala Ser Lys Gly Val
Ser Thr Ser Gly Tyr Ser Tyr Leu His1 5 10 154817PRTArtificial
Sequencepembrolizumab light chain CDR2 481Leu Ala Ser Tyr Leu Glu
Ser1 54829PRTArtificial Sequencepembrolizumab light chain CDR3
482Gln His Ser Arg Asp Leu Pro Leu Thr1 5483451PRTArtificial
Sequencedurvalumab heavy chain 483Glu Val Gln Leu Val Glu Ser Gly
Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala
Ala Ser Gly Phe Thr Phe Ser Arg Tyr 20 25 30Trp Met Ser Trp Val Arg
Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Asn Ile Lys Gln
Asp Gly Ser Glu Lys Tyr Tyr Val Asp Ser Val 50 55 60Lys Gly Arg Phe
Thr Ile Ser Arg Asp Asn Ala Lys Asn Ser Leu Tyr65 70 75 80Leu Gln
Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala
Arg Glu Gly Gly Trp Phe Gly Glu Leu Ala Phe Asp Tyr Trp Gly 100 105
110Gln Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser
115 120 125Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly
Thr Ala 130 135 140Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu
Pro Val Thr Val145 150 155 160Ser Trp Asn Ser Gly Ala Leu Thr Ser
Gly Val His Thr Phe Pro Ala 165 170 175Val Leu Gln Ser Ser Gly Leu
Tyr Ser Leu Ser Ser Val Val Thr Val 180 185 190Pro Ser Ser Ser Leu
Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His 195 200 205Lys Pro Ser
Asn Thr Lys Val Asp Lys Arg Val Glu Pro Lys Ser Cys 210 215 220Asp
Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Phe Glu Gly225 230
235 240Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu
Met 245 250 255Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp
Val Ser His 260 265 270Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val
Asp Gly Val Glu Val 275 280 285His Asn Ala Lys Thr Lys Pro Arg Glu
Glu Gln Tyr Asn Ser Thr Tyr 290 295 300Arg Val Val Ser Val Leu Thr
Val Leu His Gln Asp Trp Leu Asn Gly305 310 315 320Lys Glu Tyr Lys
Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Ser Ile 325 330 335Glu Lys
Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val 340 345
350Tyr Thr Leu Pro Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser
355 360 365Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala
Val Glu 370 375 380Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys
Thr Thr Pro Pro385 390 395 400Val Leu Asp Ser Asp Gly Ser Phe Phe
Leu Tyr Ser Lys Leu Thr Val 405 410 415Asp Lys Ser Arg Trp Gln Gln
Gly Asn Val Phe Ser Cys Ser Val Met 420 425 430His Glu Ala Leu His
Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser 435 440 445Pro Gly Lys
450484265PRTArtificial Sequencedurvalumab light chain 484Glu Val
Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser
Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Arg Tyr 20 25
30Trp Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val
35 40 45Ala Asn Glu Ile Val Leu Thr Gln Ser Pro Gly Thr Leu Ser Leu
Ser 50 55 60Pro Gly Glu Arg Ala Thr Leu Ser Cys Arg Ala Ser Gln Arg
Val Ser65 70 75 80Ser Ser Tyr Leu Ala Trp Tyr Gln Gln Lys Pro Gly
Gln Ala Pro Arg 85 90 95Leu Leu Ile Tyr Asp Ala Ser Ser Arg Ala Thr
Gly Ile Pro Asp Arg 100 105 110Phe Ser Gly Ser Gly Ser Gly Thr Asp
Phe Thr Leu Thr Ile Ser Arg 115 120 125Leu Glu Pro Glu Asp Phe Ala
Val Tyr Tyr Cys Gln Gln Tyr Gly Ser 130 135 140Leu Pro Trp Thr Phe
Gly Gln Gly Thr Lys Val Glu Ile Lys Arg Thr145 150 155 160Val Ala
Ala Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu 165 170
175Lys Ser Gly Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro
180 185 190Arg Glu Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln
Ser Gly 195 200 205Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys
Asp Ser Thr Tyr 210 215 220Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys
Ala Asp Tyr Glu Lys His225 230 235 240Lys Val Tyr Ala Cys Glu Val
Thr His Gln Gly Leu Ser Ser Pro Val 245 250 255Thr Lys Ser Phe Asn
Arg Gly Glu Cys 260 265485121PRTArtificial Sequencedurvalumab
variable heavy chain 485Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu
Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Ser Arg Tyr 20 25 30Trp Met Ser Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Asn Ile Lys Gln Asp Gly Ser
Glu Lys Tyr Tyr Val Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser
Arg Asp Asn Ala Lys Asn Ser Leu Tyr65 70 75 80Leu
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90
95Ala Arg Glu Gly Gly Trp Phe Gly Glu Leu Ala Phe Asp Tyr Trp Gly
100 105 110Gln Gly Thr Leu Val Thr Val Ser Ser 115
120486108PRTArtificial Sequencedurvalumab variable light chain
486Glu Ile Val Leu Thr Gln Ser Pro Gly Thr Leu Ser Leu Ser Pro Gly1
5 10 15Glu Arg Ala Thr Leu Ser Cys Arg Ala Ser Gln Arg Val Ser Ser
Ser 20 25 30Tyr Leu Ala Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro Arg
Leu Leu 35 40 45Ile Tyr Asp Ala Ser Ser Arg Ala Thr Gly Ile Pro Asp
Arg Phe Ser 50 55 60Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile
Ser Arg Leu Glu65 70 75 80Pro Glu Asp Phe Ala Val Tyr Tyr Cys Gln
Gln Tyr Gly Ser Leu Pro 85 90 95Trp Thr Phe Gly Gln Gly Thr Lys Val
Glu Ile Lys 100 1054875PRTArtificial Sequencedurvalumab heavy chain
CDR1 487Arg Tyr Trp Met Ser1 548817PRTArtificial Sequencedurvalumab
heavy chain CDR2 488Asn Ile Lys Gln Asp Gly Ser Glu Lys Tyr Tyr Val
Asp Ser Val Lys1 5 10 15Gly48912PRTArtificial Sequencedurvalumab
heavy chain CDR3 489Glu Gly Gly Trp Phe Gly Glu Leu Ala Phe Asp
Tyr1 5 1049012PRTArtificial Sequencedurvalumab light chain CDR1
490Arg Ala Ser Gln Arg Val Ser Ser Ser Tyr Leu Ala1 5
104917PRTArtificial Sequencedurvalumab light chain CDR2 491Asp Ala
Ser Ser Arg Ala Thr1 54929PRTArtificial Sequencedurvalumab light
chain CDR3 492Gln Gln Tyr Gly Ser Leu Pro Trp Thr1
5493450PRTArtificial Sequenceavelumab heavy chain 493Glu Val Gln
Leu Leu Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30Ile
Met Met Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ser Ser Ile Tyr Pro Ser Gly Gly Ile Thr Phe Tyr Ala Asp Thr Val
50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu
Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val
Tyr Tyr Cys 85 90 95Ala Arg Ile Lys Leu Gly Thr Val Thr Thr Val Asp
Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser Ala Ser
Thr Lys Gly Pro Ser Val 115 120 125Phe Pro Leu Ala Pro Ser Ser Lys
Ser Thr Ser Gly Gly Thr Ala Ala 130 135 140Leu Gly Cys Leu Val Lys
Asp Tyr Phe Pro Glu Pro Val Thr Val Ser145 150 155 160Trp Asn Ser
Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val 165 170 175Leu
Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro 180 185
190Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys
195 200 205Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser
Cys Asp 210 215 220Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu
Leu Leu Gly Gly225 230 235 240Pro Ser Val Phe Leu Phe Pro Pro Lys
Pro Lys Asp Thr Leu Met Ile 245 250 255Ser Arg Thr Pro Glu Val Thr
Cys Val Val Val Asp Val Ser His Glu 260 265 270Asp Pro Glu Val Lys
Phe Asn Trp Tyr Val Asp Gly Val Glu Val His 275 280 285Asn Ala Lys
Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg 290 295 300Val
Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys305 310
315 320Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile
Glu 325 330 335Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro
Gln Val Tyr 340 345 350Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys
Asn Gln Val Ser Leu 355 360 365Thr Cys Leu Val Lys Gly Phe Tyr Pro
Ser Asp Ile Ala Val Glu Trp 370 375 380Glu Ser Asn Gly Gln Pro Glu
Asn Asn Tyr Lys Thr Thr Pro Pro Val385 390 395 400Leu Asp Ser Asp
Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp 405 410 415Lys Ser
Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His 420 425
430Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro
435 440 445Gly Lys 450494216PRTArtificial Sequenceavelumab light
chain 494Gln Ser Ala Leu Thr Gln Pro Ala Ser Val Ser Gly Ser Pro
Gly Gln1 5 10 15Ser Ile Thr Ile Ser Cys Thr Gly Thr Ser Ser Asp Val
Gly Gly Tyr 20 25 30Asn Tyr Val Ser Trp Tyr Gln Gln His Pro Gly Lys
Ala Pro Lys Leu 35 40 45Met Ile Tyr Asp Val Ser Asn Arg Pro Ser Gly
Val Ser Asn Arg Phe 50 55 60Ser Gly Ser Lys Ser Gly Asn Thr Ala Ser
Leu Thr Ile Ser Gly Leu65 70 75 80Gln Ala Glu Asp Glu Ala Asp Tyr
Tyr Cys Ser Ser Tyr Thr Ser Ser 85 90 95Ser Thr Arg Val Phe Gly Thr
Gly Thr Lys Val Thr Val Leu Gly Gln 100 105 110Pro Lys Ala Asn Pro
Thr Val Thr Leu Phe Pro Pro Ser Ser Glu Glu 115 120 125Leu Gln Ala
Asn Lys Ala Thr Leu Val Cys Leu Ile Ser Asp Phe Tyr 130 135 140Pro
Gly Ala Val Thr Val Ala Trp Lys Ala Asp Gly Ser Pro Val Lys145 150
155 160Ala Gly Val Glu Thr Thr Lys Pro Ser Lys Gln Ser Asn Asn Lys
Tyr 165 170 175Ala Ala Ser Ser Tyr Leu Ser Leu Thr Pro Glu Gln Trp
Lys Ser His 180 185 190Arg Ser Tyr Ser Cys Gln Val Thr His Glu Gly
Ser Thr Val Glu Lys 195 200 205Thr Val Ala Pro Thr Glu Cys Ser 210
215495120PRTArtificial Sequenceavelumab variable heavy chain 495Glu
Val Gln Leu Leu Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10
15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr
20 25 30Ile Met Met Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Val 35 40 45Ser Ser Ile Tyr Pro Ser Gly Gly Ile Thr Phe Tyr Ala Asp
Thr Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn
Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr
Ala Val Tyr Tyr Cys 85 90 95Ala Arg Ile Lys Leu Gly Thr Val Thr Thr
Val Asp Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ser
115 120496110PRTArtificial Sequenceavelumab variable light chain
496Gln Ser Ala Leu Thr Gln Pro Ala Ser Val Ser Gly Ser Pro Gly Gln1
5 10 15Ser Ile Thr Ile Ser Cys Thr Gly Thr Ser Ser Asp Val Gly Gly
Tyr 20 25 30Asn Tyr Val Ser Trp Tyr Gln Gln His Pro Gly Lys Ala Pro
Lys Leu 35 40 45Met Ile Tyr Asp Val Ser Asn Arg Pro Ser Gly Val Ser
Asn Arg Phe 50 55 60Ser Gly Ser Lys Ser Gly Asn Thr Ala Ser Leu Thr
Ile Ser Gly Leu65 70 75 80Gln Ala Glu Asp Glu Ala Asp Tyr Tyr Cys
Ser Ser Tyr Thr Ser Ser 85 90 95Ser Thr Arg Val Phe Gly Thr Gly Thr
Lys Val Thr Val Leu 100 105 1104975PRTArtificial Sequenceavelumab
heavy chain CDR1 497Ser Tyr Ile Met Met1 549817PRTArtificial
Sequenceavelumab heavy chain CDR2 498Ser Ile Tyr Pro Ser Gly Gly
Ile Thr Phe Tyr Ala Asp Thr Val Lys1 5 10 15Gly49911PRTArtificial
Sequenceavelumab heavy chain CDR3 499Ile Lys Leu Gly Thr Val Thr
Thr Val Asp Tyr1 5 1050014PRTArtificial Sequenceavelumab light
chain CDR1 500Thr Gly Thr Ser Ser Asp Val Gly Gly Tyr Asn Tyr Val
Ser1 5 105017PRTArtificial Sequenceavelumab light chain CDR2 501Asp
Val Ser Asn Arg Pro Ser1 550210PRTArtificial Sequenceavelumab light
chain CDR3 502Ser Ser Tyr Thr Ser Ser Ser Thr Arg Val1 5
10503448PRTArtificial Sequenceatezolizumab heavy chain 503Glu Val
Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser
Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Asp Ser 20 25
30Trp Ile His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val
35 40 45Ala Trp Ile Ser Pro Tyr Gly Gly Ser Thr Tyr Tyr Ala Asp Ser
Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Ala Asp Thr Ser Lys Asn Thr
Ala Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala
Val Tyr Tyr Cys 85 90 95Ala Arg Arg His Trp Pro Gly Gly Phe Asp Tyr
Trp Gly Gln Gly Thr 100 105 110Leu Val Thr Val Ser Ser Ala Ser Thr
Lys Gly Pro Ser Val Phe Pro 115 120 125Leu Ala Pro Ser Ser Lys Ser
Thr Ser Gly Gly Thr Ala Ala Leu Gly 130 135 140Cys Leu Val Lys Asp
Tyr Phe Pro Glu Pro Val Thr Val Ser Trp Asn145 150 155 160Ser Gly
Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val Leu Gln 165 170
175Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser Ser
180 185 190Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys
Pro Ser 195 200 205Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser
Cys Asp Lys Thr 210 215 220His Thr Cys Pro Pro Cys Pro Ala Pro Glu
Leu Leu Gly Gly Pro Ser225 230 235 240Val Phe Leu Phe Pro Pro Lys
Pro Lys Asp Thr Leu Met Ile Ser Arg 245 250 255Thr Pro Glu Val Thr
Cys Val Val Val Asp Val Ser His Glu Asp Pro 260 265 270Glu Val Lys
Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala 275 280 285Lys
Thr Lys Pro Arg Glu Glu Gln Tyr Ala Ser Thr Tyr Arg Val Val 290 295
300Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu
Tyr305 310 315 320Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro
Ile Glu Lys Thr 325 330 335Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr Leu 340 345 350Pro Pro Ser Arg Glu Glu Met Thr
Lys Asn Gln Val Ser Leu Thr Cys 355 360 365Leu Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu Trp Glu Ser 370 375 380Asn Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp385 390 395 400Ser
Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser 405 410
415Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala
420 425 430Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro
Gly Lys 435 440 445504214PRTArtificial Sequenceatezolizumab light
chain 504Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser
Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Asp Val
Ser Thr Ala 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro
Lys Leu Leu Ile 35 40 45Tyr Ser Ala Ser Phe Leu Tyr Ser Gly Val Pro
Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr
Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys
Gln Gln Tyr Leu Tyr His Pro Ala 85 90 95Thr Phe Gly Gln Gly Thr Lys
Val Glu Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile
Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser
Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys
Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150
155 160Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu
Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His
Lys Val Tyr 180 185 190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser
Pro Val Thr Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys
210505118PRTArtificial Sequenceatezolizumab variable heavy chain
505Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1
5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Asp
Ser 20 25 30Trp Ile His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu
Trp Val 35 40 45Ala Trp Ile Ser Pro Tyr Gly Gly Ser Thr Tyr Tyr Ala
Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Ala Asp Thr Ser Lys
Asn Thr Ala Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Ala Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Arg His Trp Pro Gly Gly Phe
Asp Tyr Trp Gly Gln Gly Thr 100 105 110Leu Val Thr Val Ser Ala
115506108PRTArtificial Sequenceatezolizumab variable light chain
506Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1
5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Asp Val Ser Thr
Ala 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu
Leu Ile 35 40 45Tyr Ser Ala Ser Phe Leu Tyr Ser Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln
Tyr Leu Tyr His Pro Ala 85 90 95Thr Phe Gly Gln Gly Thr Lys Val Glu
Ile Lys Arg 100 10550710PRTArtificial Sequenceatezolizumab heavy
chain CDR1 507Gly Phe Thr Phe Ser Asp Ser Trp Ile His1 5
1050818PRTArtificial Sequenceatezolizumab heavy chain CDR2 508Ala
Trp Ile Ser Pro Tyr Gly Gly Ser Thr Tyr Tyr Ala Asp Ser Val1 5 10
15Lys Gly5099PRTArtificial Sequenceatezolizumab heavy chain CDR3
509Arg His Trp Pro Gly Gly Phe Asp Tyr1 551011PRTArtificial
Sequenceatezolizumab light chain CDR1 510Arg Ala Ser Gln Asp Val
Ser Thr Ala Val Ala1 5 105117PRTArtificial Sequenceatezolizumab
light chain CDR2 511Ser Ala Ser Phe Leu Tyr Ser1 55129PRTArtificial
Sequenceatezolizumab light chain CDR3 512Gln Gln Tyr Leu Tyr His
Pro Ala Thr1 5
References
D00000

D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012
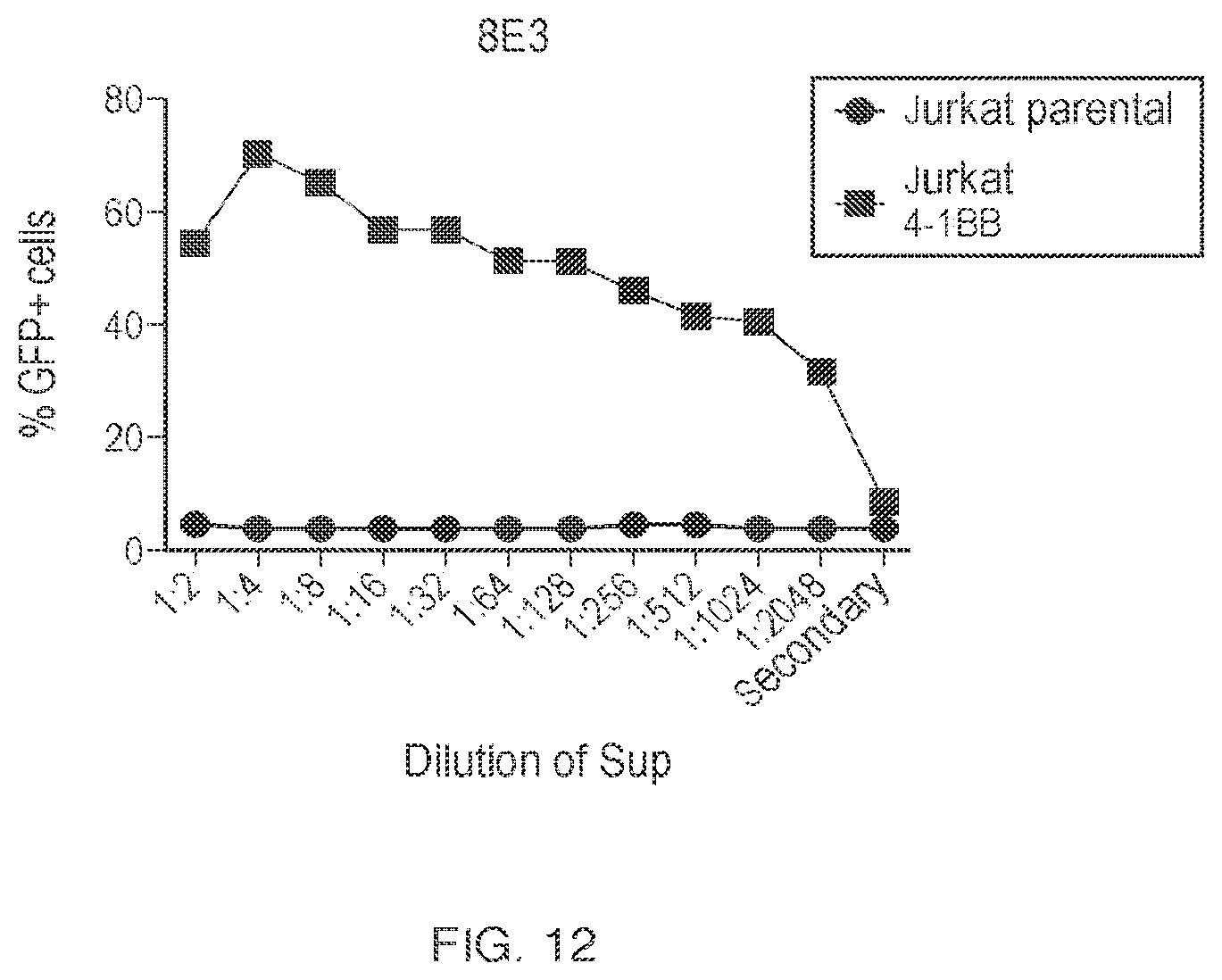
D00013

D00014

D00015

D00016

D00017

D00018
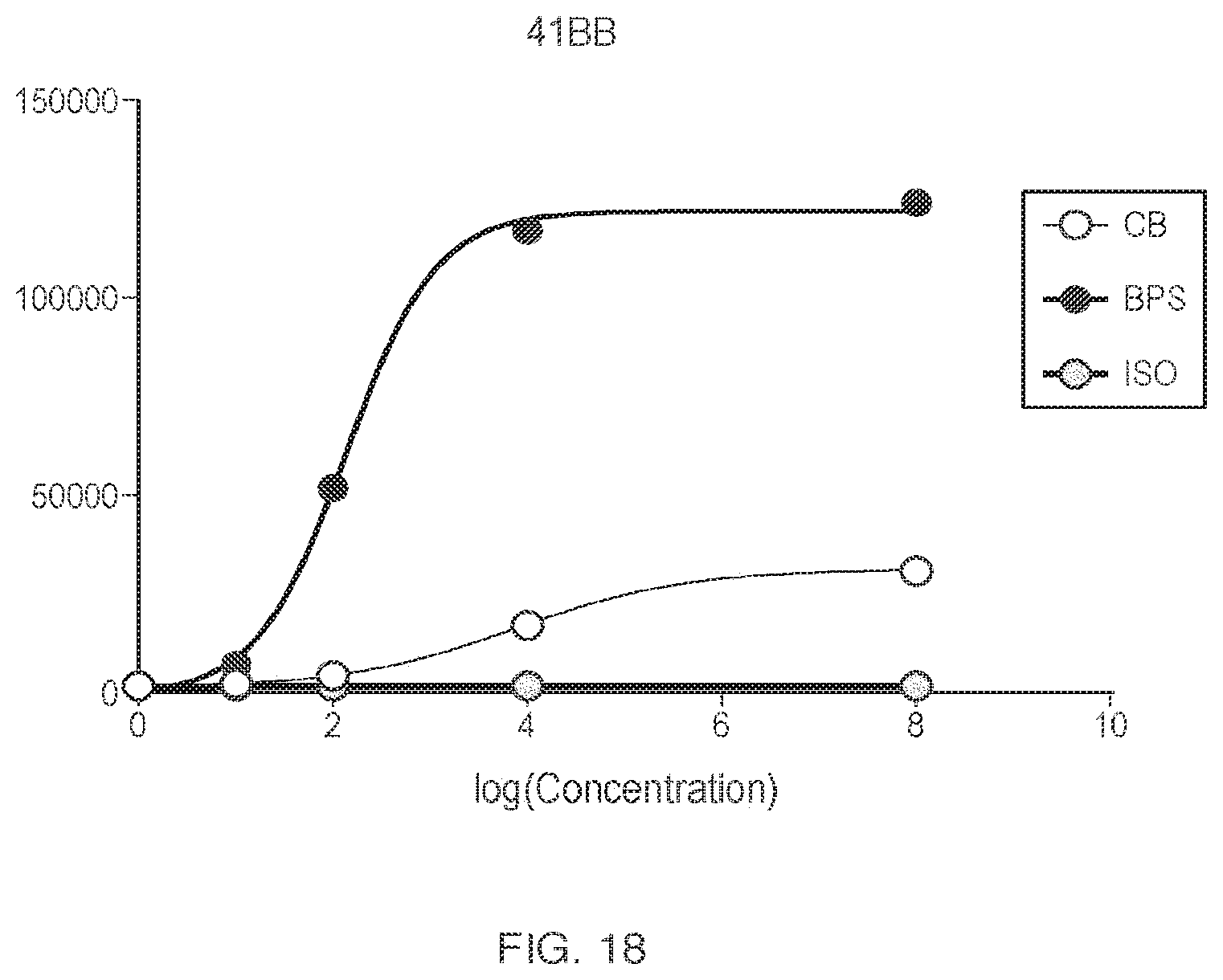
D00019

D00020

D00021
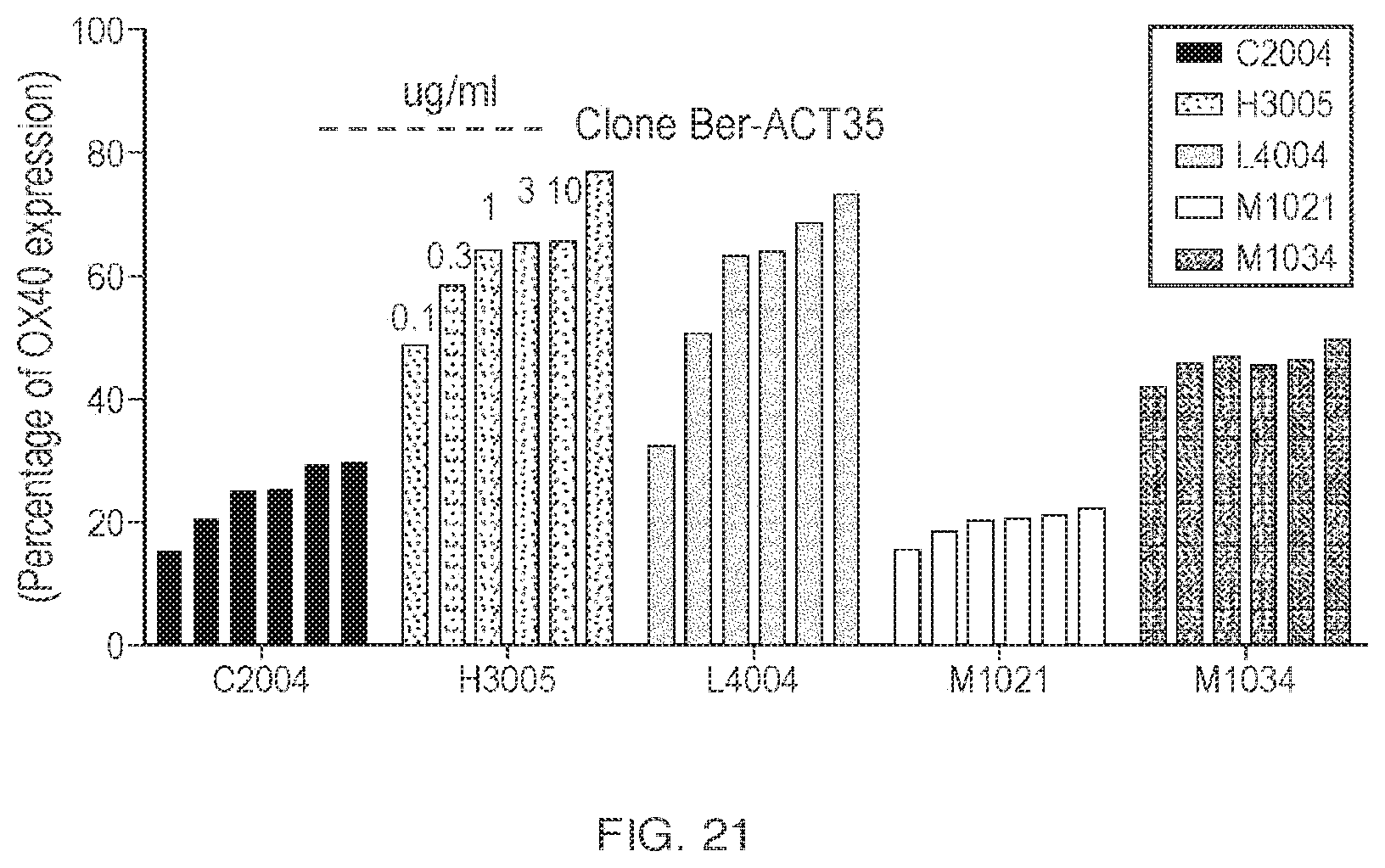
D00022

D00023

D00024
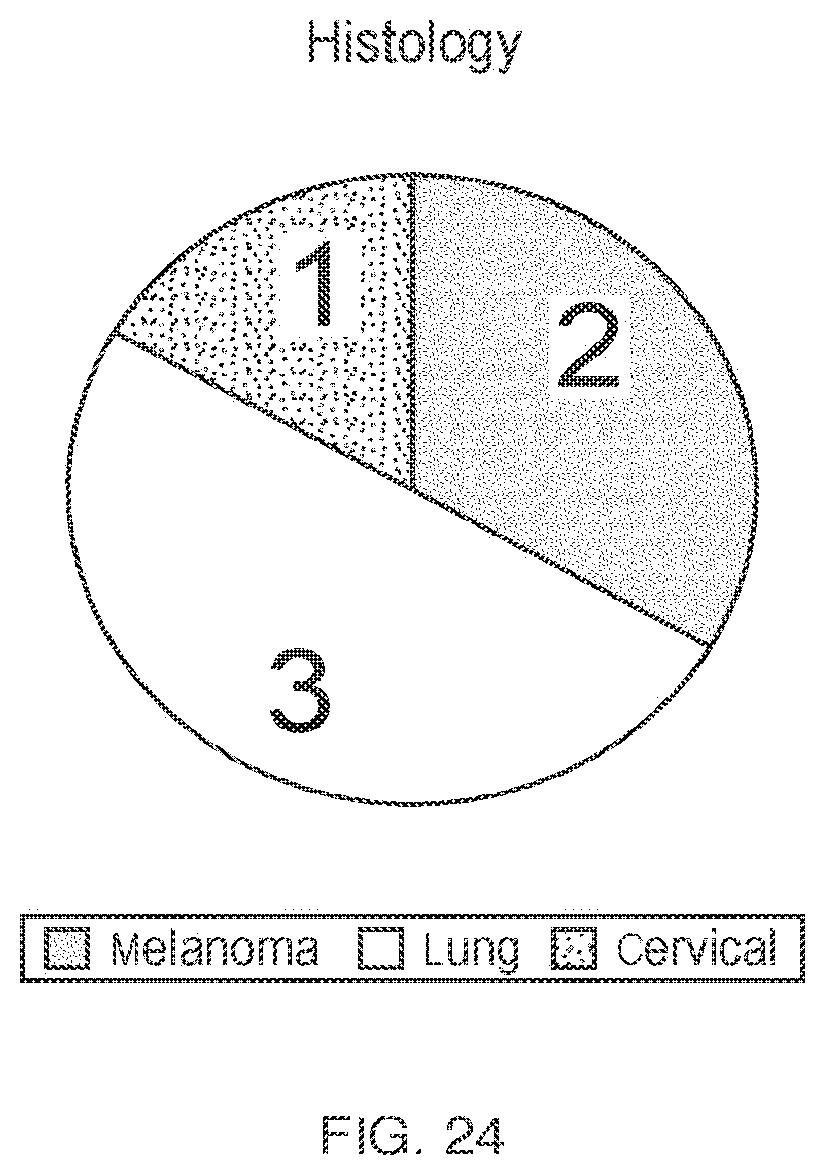
D00025

D00026

D00027
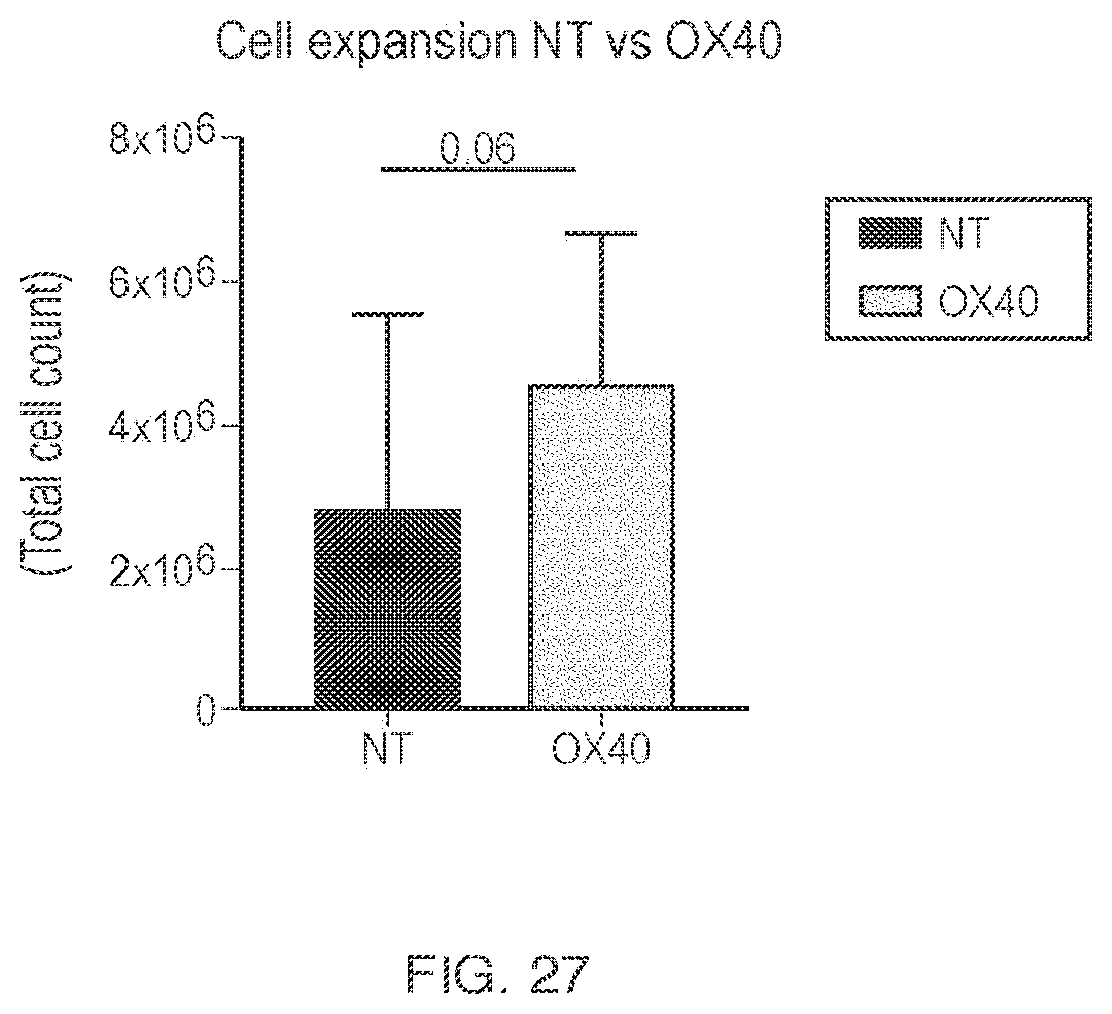
D00028
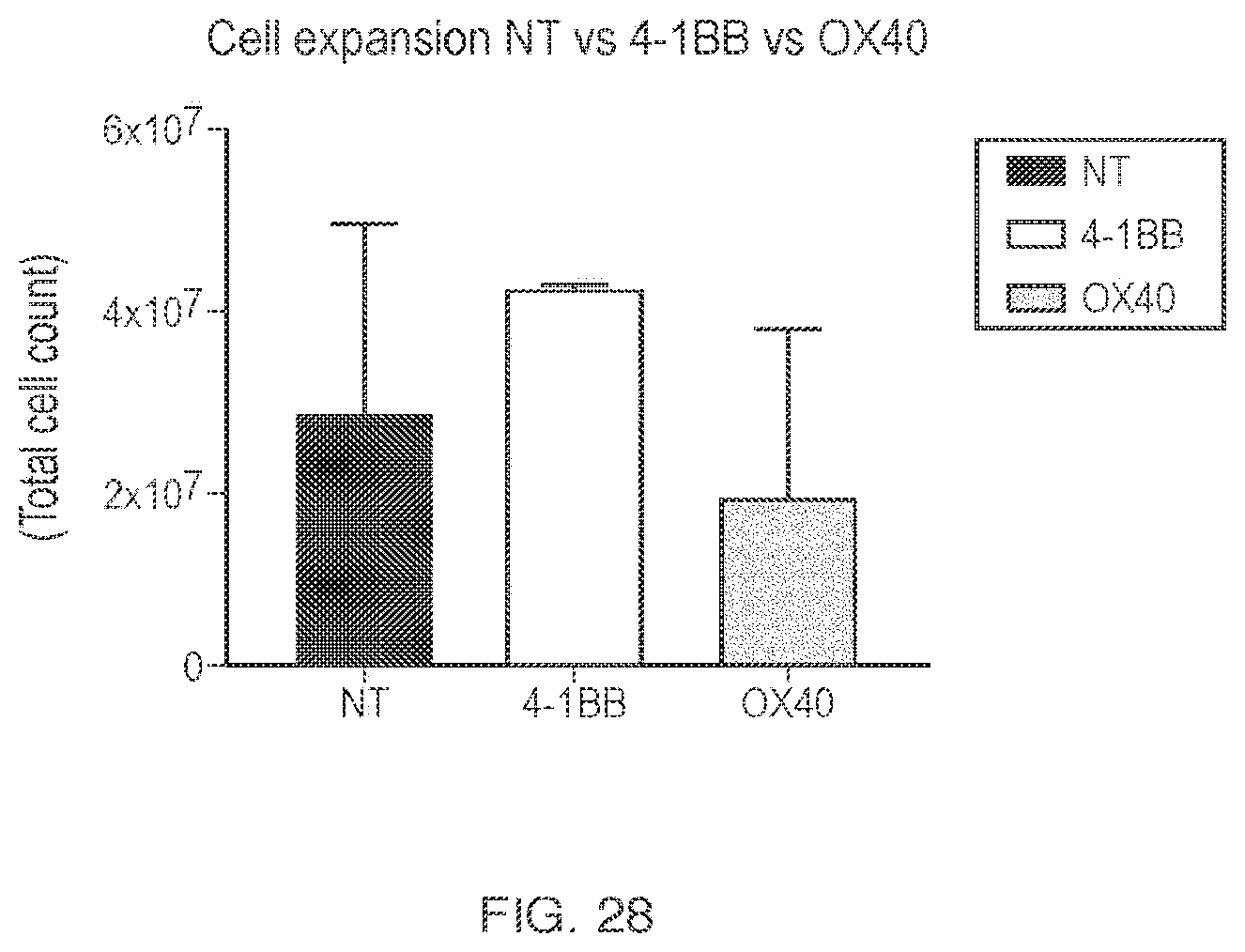
D00029
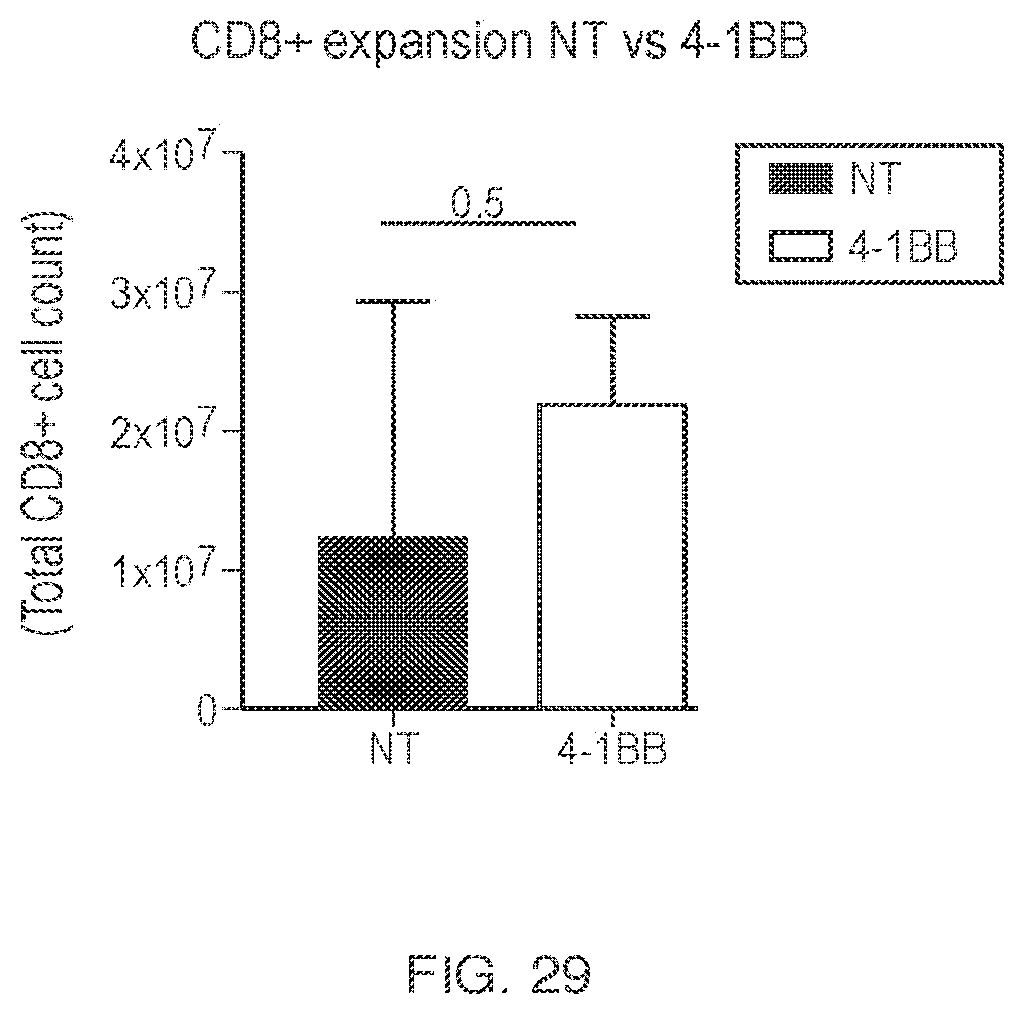
D00030

D00031
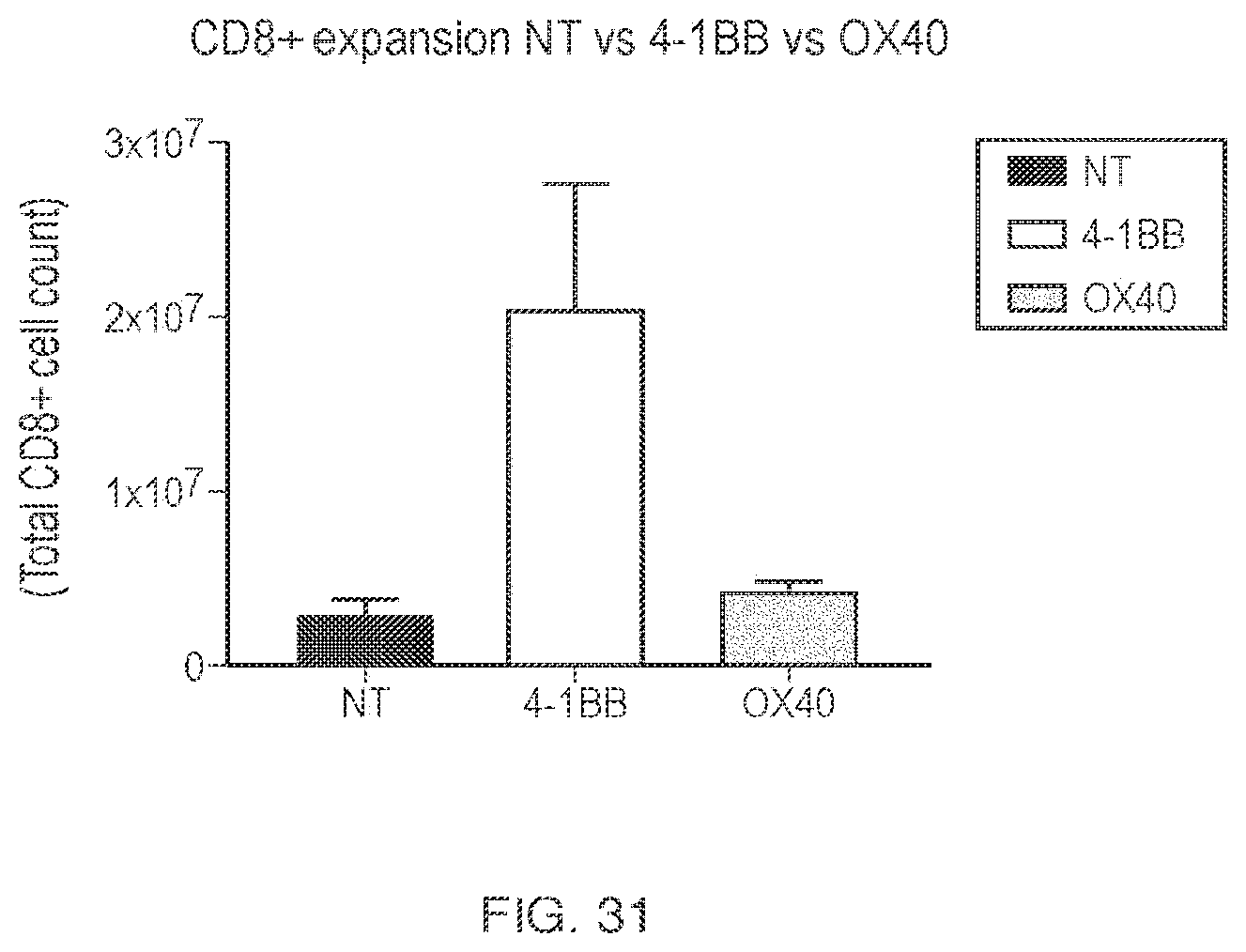
D00032
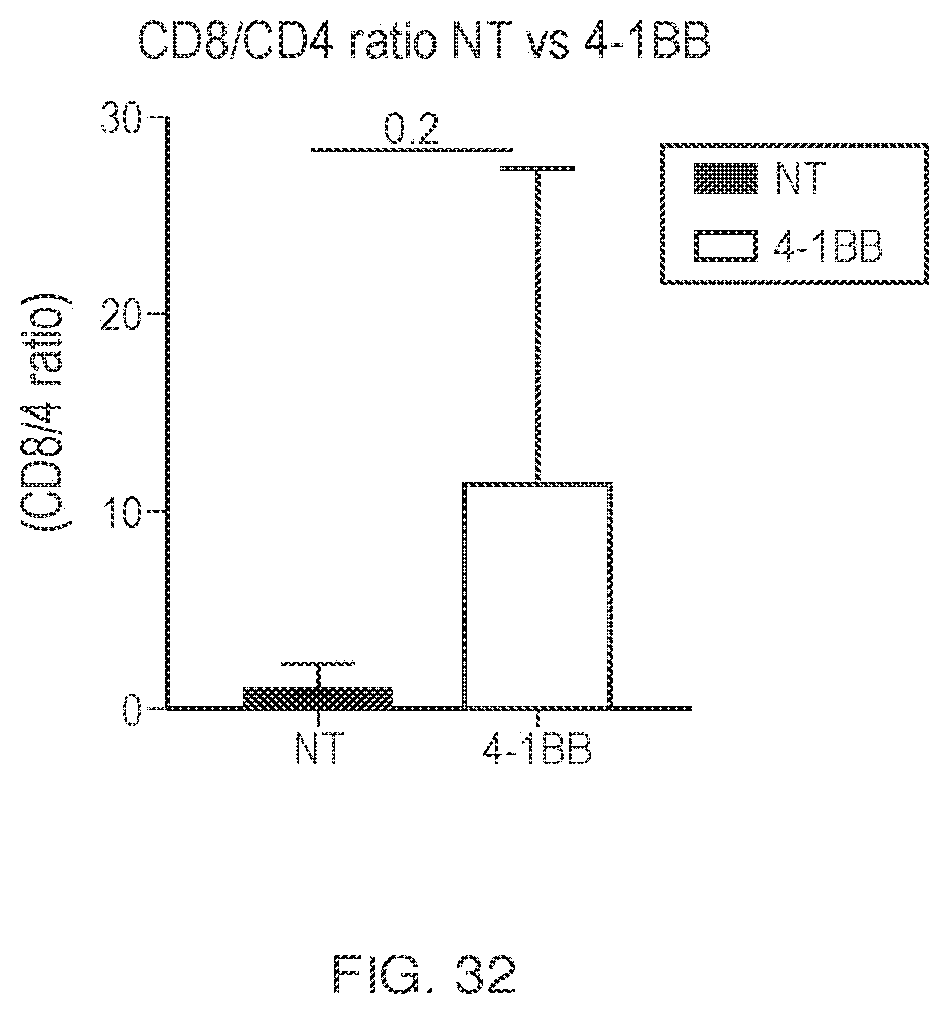
D00033
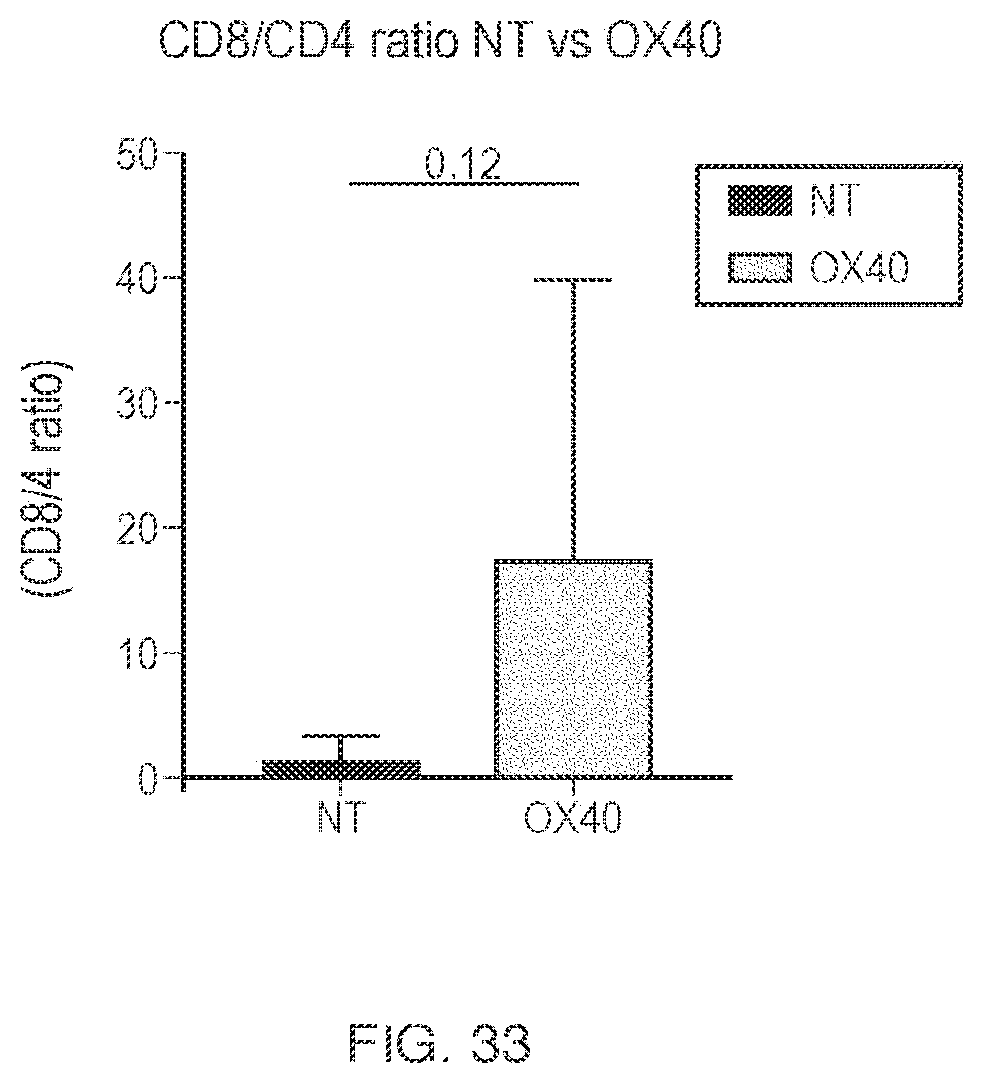
D00034

D00035
D00036

D00037

D00038
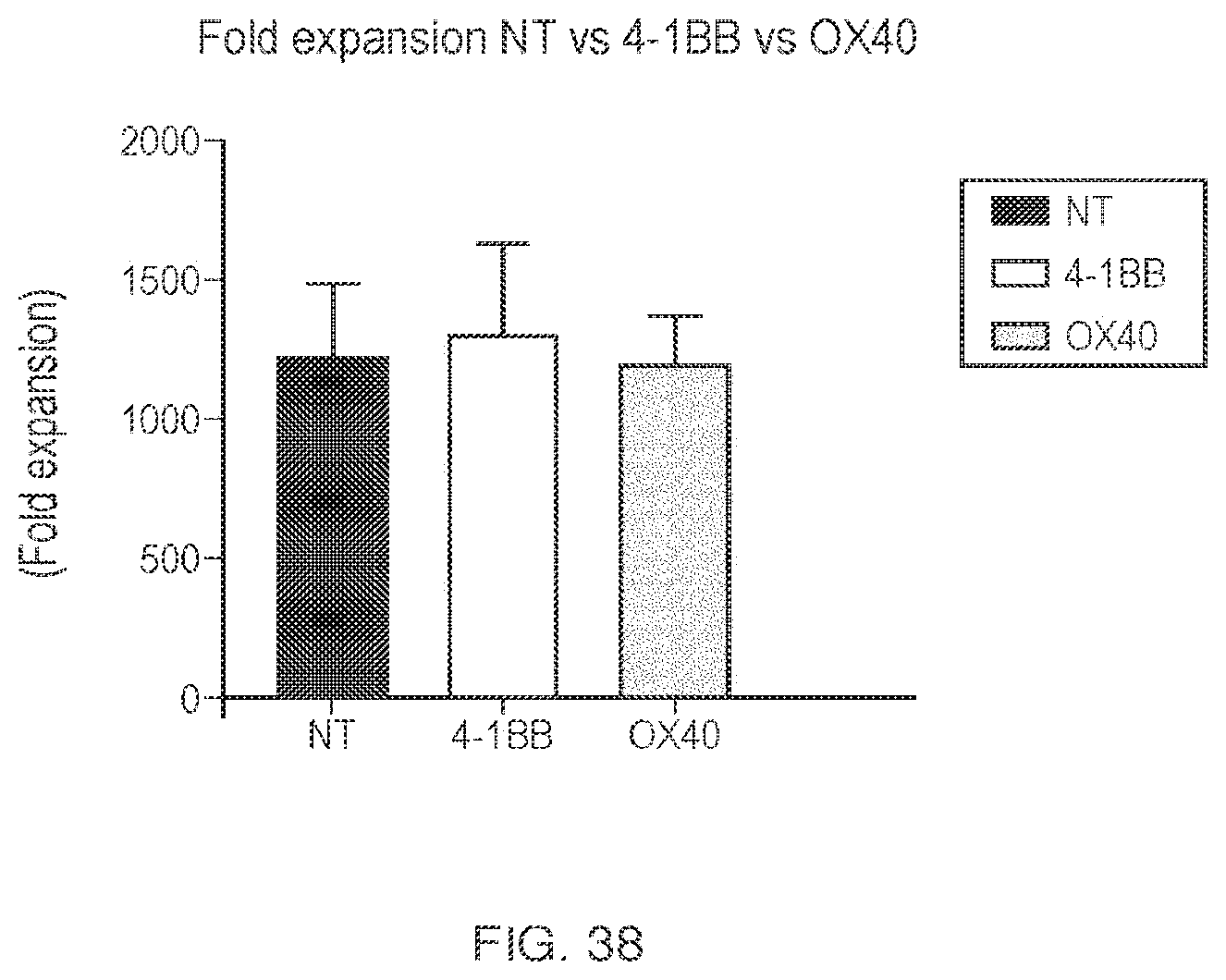
D00039

D00040
D00041
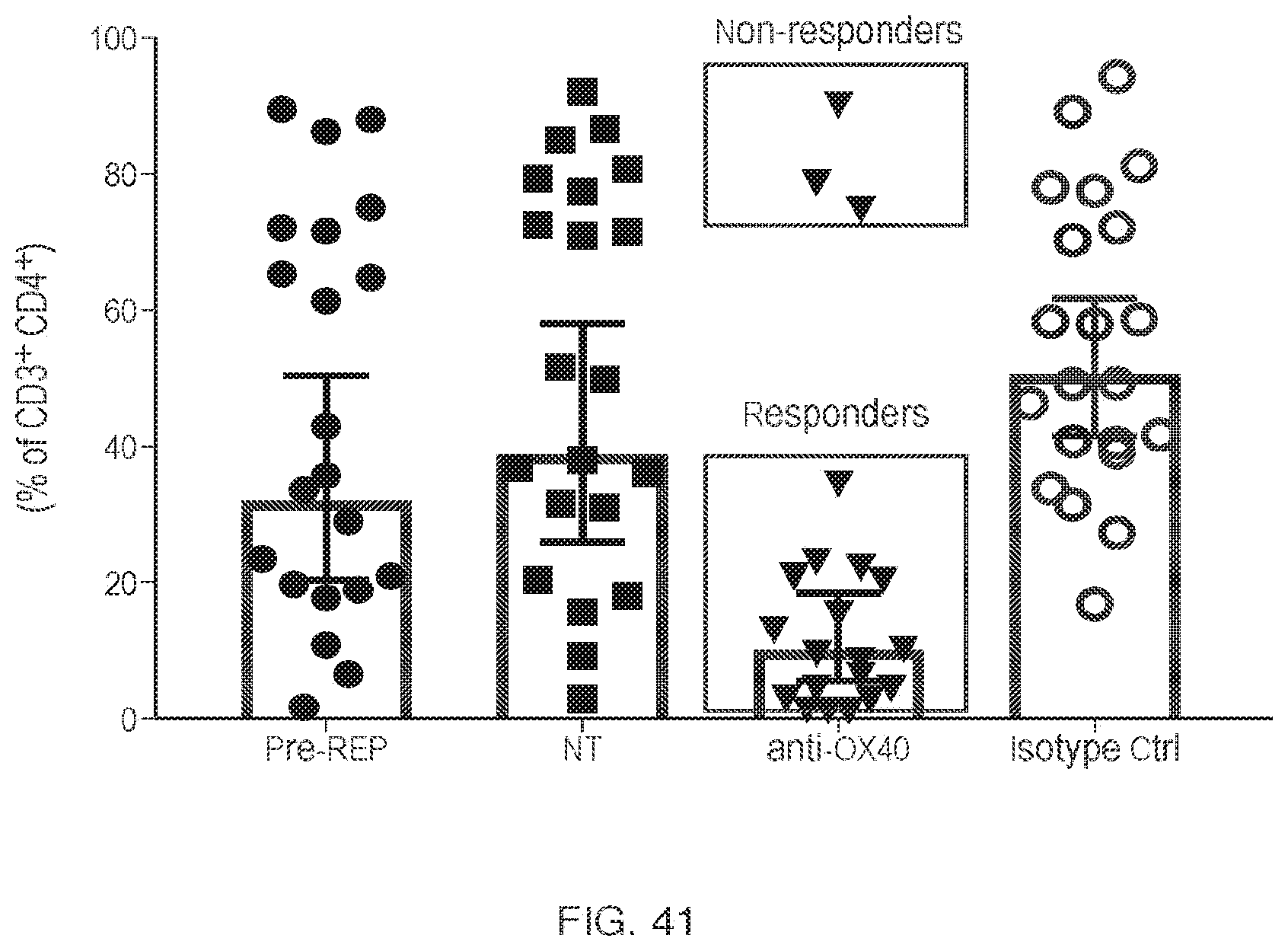
D00042

D00043

D00044

D00045

D00046

D00047
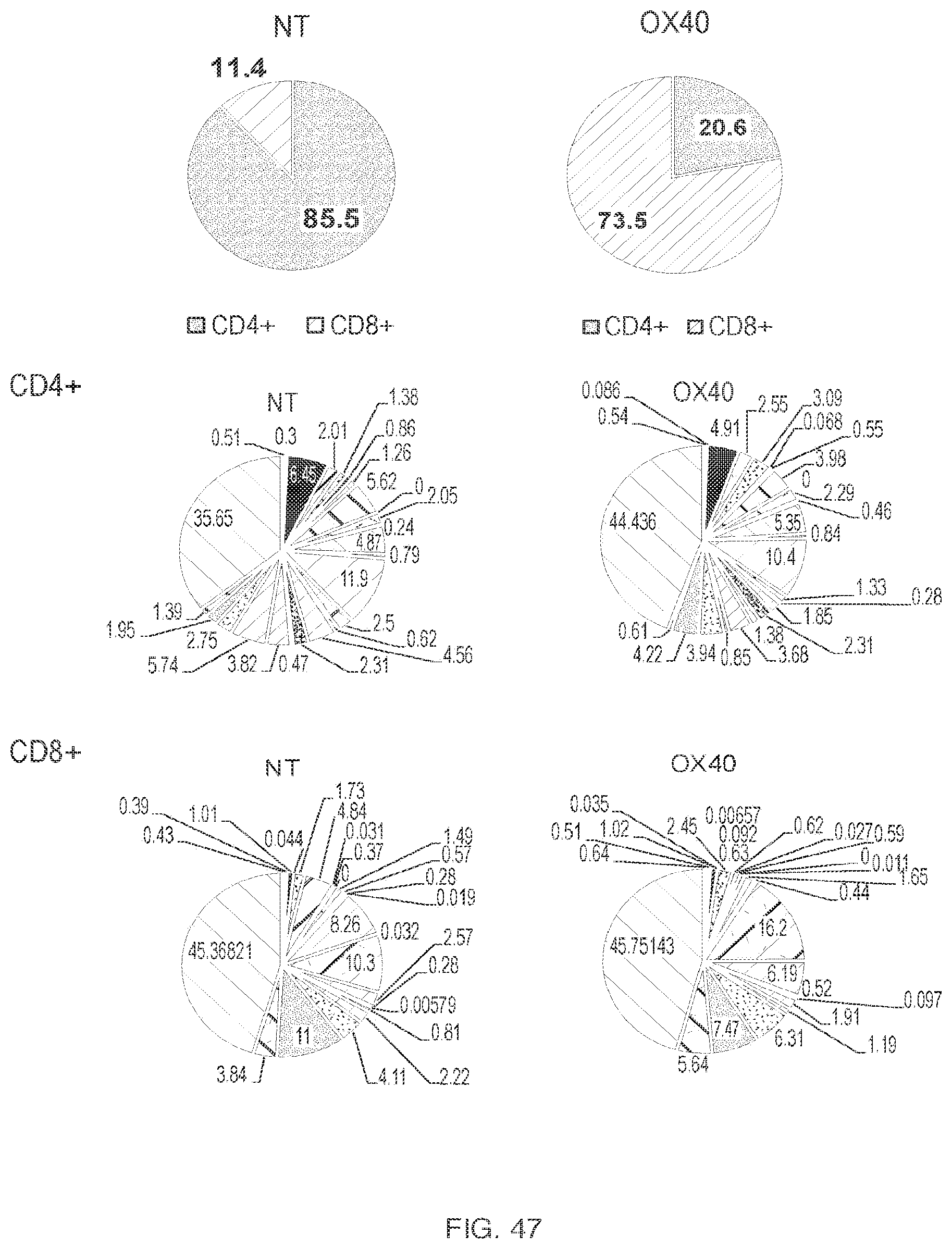
D00048

D00049

D00050

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.