Increasing Cellular Lipid Production by Increasing the Activity of Diacylglycerol Acyltransferase and Decreasing the Activity of
Tsakraklides; Vasiliki ; et al.
U.S. patent application number 16/560685 was filed with the patent office on 2020-04-09 for increasing cellular lipid production by increasing the activity of diacylglycerol acyltransferase and decreasing the activity of. The applicant listed for this patent is Novogy, Inc.. Invention is credited to Elena E. Brevnova, Vasiliki Tsakraklides.
| Application Number | 20200109377 16/560685 |
| Document ID | / |
| Family ID | 56108108 |
| Filed Date | 2020-04-09 |

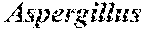








View All Diagrams
| United States Patent Application | 20200109377 |
| Kind Code | A1 |
| Tsakraklides; Vasiliki ; et al. | April 9, 2020 |
Increasing Cellular Lipid Production by Increasing the Activity of Diacylglycerol Acyltransferase and Decreasing the Activity of Triacylglycerol Lipase
Abstract
Disclosed are methods and compositions for increasing the triacylglycerol content of a cell by up-regulating diacylglycerol acyltransferase and down-regulating triacylglycerol lipase. In some embodiments, a DGA1 protein is expressed and a native TGL3 gene is knocked out, thereby increasing the synthesis of triacylglycerol and decreasing its consumption, respectively.
| Inventors: | Tsakraklides; Vasiliki; (Lexington, MA) ; Brevnova; Elena E.; (Winchester, MA) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 56108108 | ||||||||||
| Appl. No.: | 16/560685 | ||||||||||
| Filed: | September 4, 2019 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 15307518 | Oct 28, 2016 | 10443047 | ||
| PCT/US2015/028760 | Jan 15, 2015 | |||
| 16560685 | ||||
| 62090169 | Dec 10, 2014 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C12P 7/6463 20130101; C12Y 203/01199 20150701; Y02E 50/10 20130101; C12N 15/80 20130101; C12Y 203/01158 20130101; C12N 9/1029 20130101; C12Y 203/01015 20130101; C12Y 203/0102 20130101 |
| International Class: | C12N 9/10 20060101 C12N009/10; C12N 15/80 20060101 C12N015/80; C12P 7/64 20060101 C12P007/64 |
Claims
1.-13. (canceled)
14. A method of increasing the lipid content of a cell, comprising transforming a parent cell with a first nucleic acid and a second nucleic acid; wherein said first nucleic acid increases the activity of a native diacylglycerol acyltransferase or comprises a diacylglycerol acyltransferase gene; and said second nucleic acid decreases the activity of a triacylglycerol lipase.
15. The method of any one of claim 14, wherein said first nucleic acid comprises a diacylglycerol acyltransferase gene.
16. The method of claim 15, wherein said diacylglycerol acyltransferase gene encodes a type I diacylglycerol acyltransferase polypeptide, a type II diacylglycerol acyltransferase polypeptide, or a type III diacylglycerol acyltransferase polypeptide.
17. The method of claim 15, wherein said diacylglycerol acyltransferase gene encodes an amino acid sequence selected from the group consisting of: (a) an amino acid sequence set forth in SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 7, SEQ ID NO: 9, SEQ ID NO: 11, SEQ ID NO: 13, SEQ ID NO: 15, SEQ ID NO: 17, SEQ ID NO: 41, SEQ ID NO: 43, SEQ ID NO: 45, SEQ ID NO: 47, SEQ ID NO: 49, SEQ ID NO: 51, SEQ ID NO: 53, SEQ ID NO: 55, SEQ ID NO: 57, SEQ ID NO: 59, SEQ ID NO: 61; SEQ ID NO: 29, SEQ ID NO: 31, SEQ ID NO: 33, SEQ ID NO: 35, SEQ ID NO: 37, SEQ ID NO: 39, SEQ ID NO: 63, SEQ ID NO: 65, SEQ ID NO: 67, SEQ ID NO: 69, SEQ ID NO: 71, SEQ ID NO: 73, SEQ ID NO: 75, SEQ ID NO: 83, or SEQ ID NO: 85, or a biologically-active portion of any one of them; and (b) an amino acid sequence having at least 80% sequence identity to an amino acid sequence forth in SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 7, SEQ ID NO: 9, SEQ ID NO: 11, SEQ ID NO: 13, SEQ ID NO: 15, SEQ ID NO: 17, SEQ ID NO: 41, SEQ ID NO: 43, SEQ ID NO: 45, SEQ ID NO: 47, SEQ ID NO: 49, SEQ ID NO: 51, SEQ ID NO: 53, SEQ ID NO: 55, SEQ ID NO: 57, SEQ ID NO: 59, SEQ ID NO: 61; SEQ ID NO: 29, SEQ ID NO: 31, SEQ ID NO: 33, SEQ ID NO: 35, SEQ ID NO: 37, SEQ ID NO: 39, SEQ ID NO: 63, SEQ ID NO: 65, SEQ ID NO: 67, SEQ ID NO: 69, SEQ ID NO: 71, SEQ ID NO: 73, SEQ ID NO: 75, SEQ ID NO: 83, or SEQ ID NO: 85, or a biologically-active portion of any one of them.
18. The method of claim 14, wherein said second nucleic acid is capable of recombining with a nucleic acid sequence in a triacylglycerol lipase gene and/or a nucleic acid sequence in the regulatory region of a triacylglycerol lipase gene.
19. The method of claim 14, wherein said second nucleic acid is capable of recombining with a nucleic acid sequence in a TGL3, TGL3/4, or TGL4 gene and/or a nucleic acid sequence in the regulatory region of a TGL3, TGL3/4, or TGL4 gene.
20. The method of claim 14, wherein said cell is Yarrowia lipolytica or Arxula adeninivorans.
Description
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] This application is a divisional of U.S. patent application Ser. No. 15/307,518, filed Oct. 28, 2016, now U.S. Pat. No. 10,443,047, which claims the benefit of PCT/US2015/028760 filed May 1, 2015, which claims priority from U.S. Provisional Application No. 61/987,098, filed May 1, 2014, and U.S. Provisional Application No. 62/090,169, filed Dec. 10, 2014, which are incorporated herein by reference in their entireties for all purposes.
BACKGROUND
[0002] Lipids have multiple industrial applications, including in the cosmetic and food industries, as well as serving as precursors for biodiesel and biochemical production. Microbial lipids are produced by many oleaginous organisms, including the well-characterized yeast Yarrowia lipolytica. Lipid yield in oleaginous organisms can be increased by up-regulation or down-regulation or deletion of genes implicated in the lipid pathway. For example, it was reported that up-regulation of native Y. lipolytica DGA1 significantly increased lipid yield and productivity (Metabolic Engineering 15:1-9 (2013)).
[0003] Y. lipolytica DGA1 is a type 2 diacylglycerol acyltransferase encoded by the Y. lipolytica diacylglycerol acyltransferase gene DGAT2. DGA1 is one of the key enzymes in the lipid pathway, and it is involved in the final step of triacylglycerol (TAG) synthesis. Triacylglycerols are the major form of storage lipids in Y. lipolytica. Recent data suggests that DGA1 efficiency may be a significant factor for high levels of lipid accumulation in oleaginous organisms (Metabolic Engineering 15:1-9 (2013)). Additionally, DGA1 genes from other species can be introduced into the host genome and have a significant effect on lipid production and composition. For example, other oleaginous yeast, such as Rhodosporidium toruloides and Lipomyces starkeyi, are able to accumulate significantly more lipids compared to wild type Y. lipolytica strains. It was demonstrated that overexpression in Y. lipolytica of DGA1 from organisms with higher native lipid production levels had a greater effect on Y. lipolytica lipid production than overexpression of native Y. lipolytica DGA1 (U.S. Ser. No. 61/943,664; incorporated by reference). Despite efforts to increase lipid yield in Y. lipolytica by overexpression of DGA1 from Mortierella alpine, no significant effect on lipid production levels has been reported (U.S. Pat. No. 7,198,937; incorporated by reference). The deletion of genes involved in the breakdown of lipids or in pathways that draw flux away from lipid biosynthesis has also been studied. Dulermo et al. demonstrated that the deletion of the triacylglycerol lipase gene TGL3 nearly doubled the total lipid content accumulated by Y. lipolytica (Biochimica Biophysica Acta 1831:1486-95 (2013)). The TGL3 protein is one of two intracellular lipases responsible for the first step of triacylglycerol degradation in Y. lipolytica.
SUMMARY
[0004] In some embodiments, the invention provides a transformed oleaginous cell comprising a first genetic modification, wherein said first genetic modification increases the activity of a native diacylglycerol acyltransferase or encodes at least one copy of a diacylglycerol acyltransferase gene native to the oleaginous cell or from a different species, and a second genetic modification, wherein said second genetic modification decreases the activity of a triacylglycerol lipase in the oleaginous cell.
[0005] In some aspects, the invention provides a method of increasing the lipid content of a cell comprising transforming a parent cell with a first nucleic acid and a second nucleic acid, wherein said first nucleic acid increases the activity of a native diacylglycerol acyltransferase or comprises a diacylglycerol acyltransferase gene and said second nucleic acid decreases the activity of a triacylglycerol lipase. Alternatively, the same nucleic acid could embody both elements described above. Thus, the invention also provides a method of increasing the lipid content of a cell comprising transforming a parent cell with a nucleic acid, wherein said nucleic acid decreases the activity of a triacylglycerol lipase in the cell, and said nucleic acid comprises a diacylglycerol acyltransferase gene or increases the activity of a native diacylglycerol acyltransferase.
[0006] Further, in some aspects, the invention provides a method of increasing the triacylglycerol content of a cell comprising: (a) providing a cell, comprising (i) a first genetic modification, wherein said first genetic modification increases the activity of a native diacylglycerol acyltransferase, or encodes at least one copy of a diacylglycerol acyltransferase gene native to the oleaginous cell or from a different species; and (ii) a second genetic modification that decreases the activity of a triacylglycerol lipase in the cell; (b) growing said cell under conditions whereby the first genetic modification is expressed, thereby producing a triacylglycerol; and (c) optionally recovering the triacylglycerol.
[0007] One skilled in the art will readily appreciate that the present invention is well adapted to carry out the objects and obtain the ends and advantages mentioned, as well as those inherent therein. The embodiments described herein are not intended as limitations on the scope of the invention. These and other features, aspects, and advantages of the present invention will become better understood with reference to the following description, drawings, and claims.
BRIEF DESCRIPTION OF THE FIGURES
[0008] FIG. 1 shows a map of the pNC243 construct used to overexpress the diacylglycerol acyltransferase DGA1 gene NG66 in Y. lipolytica strain NS18 (obtained from ARS Culture Collection, NRRL # YB 392). Vector pNC243 was linearized by PacI/NotI restriction digest before transformation. "2u ori" denotes the S. cerevisiae origin of replication from the 2 .mu.m circle plasmid; "pMB1 ori" denotes the E. coli pMB1 origin of replication from the pBR322 plasmid; "AmpR" denotes the bla gene used as a marker for selection with ampicillin; "PR2" denotes the Y. lipolytica GPD1 promoter -931 to -1; "NG66" denotes the native Rhodosporidium toruloides DGA1 cDNA synthesized by GenScript; "TER1" denotes the Y. lipolytica CYC1 terminator 300 base pairs after stop; "PR22" denotes the S. cerevisiae TEF1 promoter -412 to -1; "NG3" denotes the Streptomyces noursei Nat1 gene used as a marker for selection with nourseothricin; "TER2" denotes the S. cerevisiae CYC1 terminator 275 base pairs after stop; and "Sc URA3" denotes the S. cerevisiae URA3 auxotrophic marker for selection in yeast.
[0009] FIG. 2 is a graph depicting results from a fluorescence-based lipid assay for Y. lipolytica strain NS421, which is a triacylglycerol lipase knockout strain, and Y. lipolytica strain NS377, which is a triacylglycerol lipase knockout strain that overexpresses the diacylglycerol acyltransferase DGA1 gene NG66.
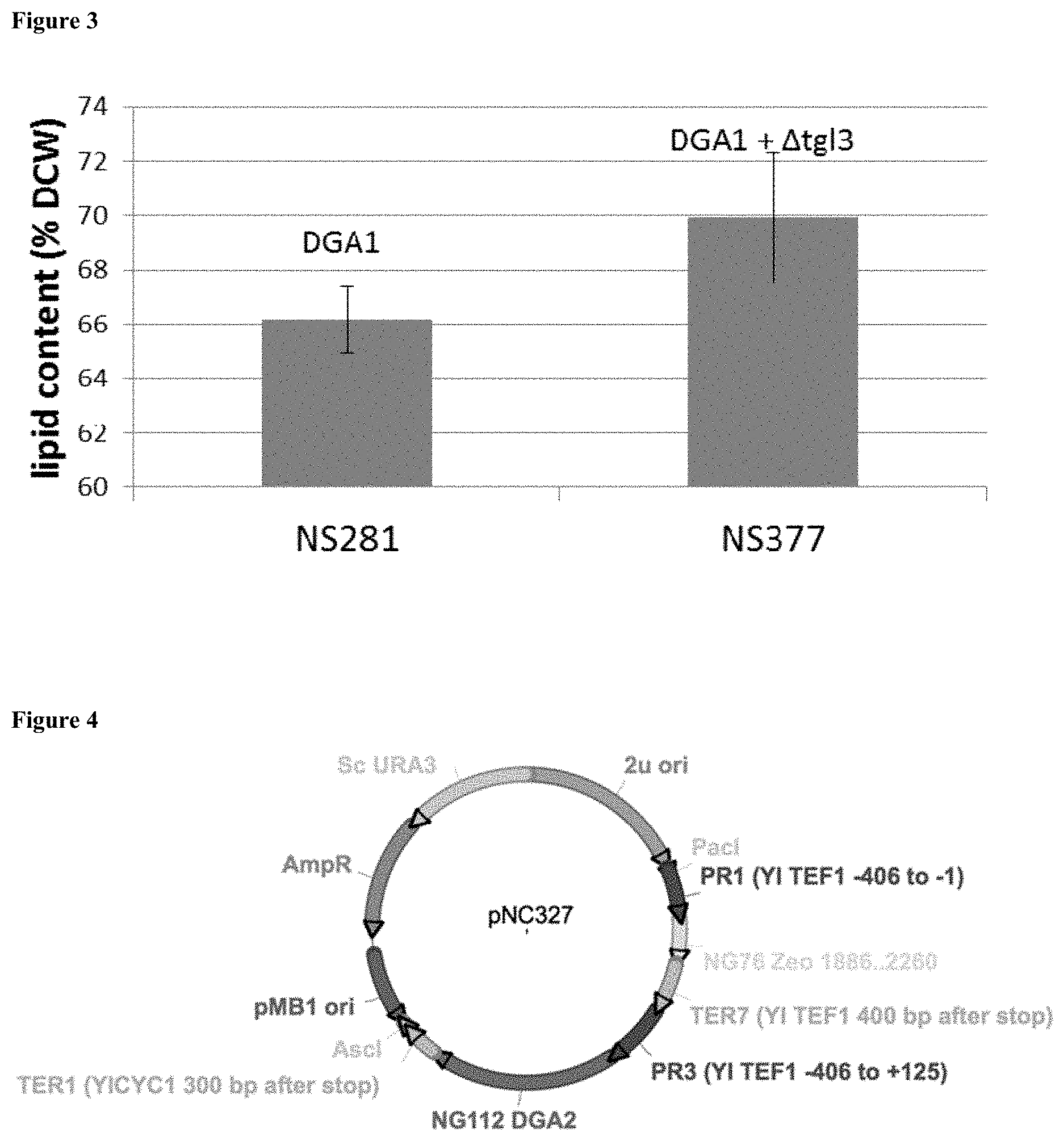
[0010] FIG. 3 is a graph depicting results from a gas chromatography analysis of Y. lipolytica strain NS281, which overexpresses the diacylglycerol acyltransferase DGA1 gene NG66, and Y. lipolytica strain NS377, containing a triacylglycerol lipase knockout and overexpressing the diacylglycerol acyltransferase DGA1 gene NG66.
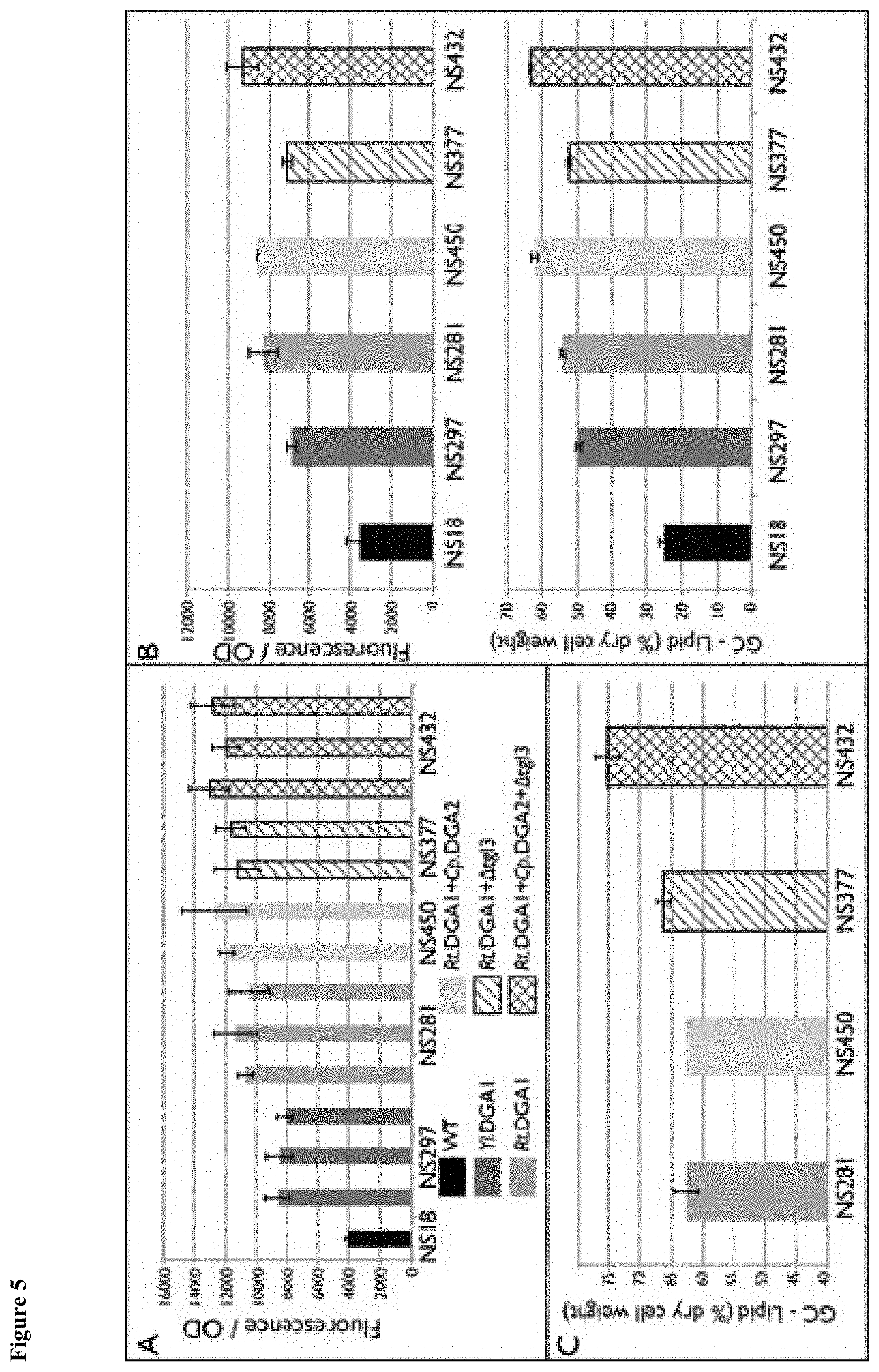
[0011] FIG. 4 depicts a map of the pNC327 construct used to overexpress the NG112 gene (C. purpurea DGA2) in Y. lipolytica. Vector pNC327 was linearized by a PacI/AscI restriction digest before transformation. "2u ori" denotes the S. cerevisiae origin of replication from the 2 .mu.m circle plasmid; "pMB1 ori" denotes the E. coli pMB1 origin of replication from the pBR322 plasmid; "AmpR" denotes the bla gene used as a marker for selection with ampicillin; "PR3" denotes the Y. lipolytica TEF1 promoter -406 to +125; "NG112" denotes the C. purpurea DGA2 gene synthetized by GenScript; "TER1" denotes the Y. lipolytica CYC1 terminator 300 bp after stop; "PR1" denotes the Y. lipolytica TEF1 promoter -406 to -1; "NG76" denotes the Streptoalloteichus hindustanus BLE gene used as marker for selection with Zeocin; "TER7" denotes the Y. lipolytica TEF1 terminator 400 bp after stop; and "Sc URA3" denotes the S. cerevisiae URA3 auxotrophic marker for selection in yeast.
[0012] FIG. 5 comprises three panels, labeled pane (A), (B), and (C). The figure depicts lipid accumulation measured by a fluorescence-based assay or a percentage of the dry cell weight as determined by gas chromatography for Yarrowia lipolytica strains NS297, NS281, NS450, NS377, and NS432. NS297 expresses an additional copy of Y. lipolytica DGA1; NS281 expresses Rhodosporidium toruloides DGA1; NS450 expresses R. toruloides DGA1 and Claviceps purpurea DGA2; NS377 expresses R. toruloides and carries a deletion of Y. lipolytica TGL3; NS432 expresses R. toruloides DGA1 and C. purpurea DGA2 and carries a deletion of Y. lipolytica TGL3. In panel (A), strains were analyzed by fluorescence assay after 96 hours of fermentation in a 48-well plate where two or three transformants were analyzed for each construct. In panel (B), strains were analyzed by fluorescence assay and gas chromatography after 96 hours of fermentation in 50-mL flasks. In panel (C), strains were analyzed by gas chromatography after 140 hours of fermentation in 1-L bioreactors. Data for NS281, NS377, and NS432 are averages obtained from duplicate bioreactor fermentations. Data for NS450 represents the value obtained from a single bioreactor fermentation.
DETAILED DESCRIPTION
Overview
[0013] Disclosed are methods and compositions for creating transformed oleaginous cells with increased triacylglycerol content. Disrupting the yeast triacylglycerol lipase gene TGL3 eliminates a pathway that depletes triacylglycerol content. Additionally, overexpressing the diacylglycerol acyltransferase DGA1 increases the amount of protein that can synthesize triacylglycerol. Therefore, combining DGA1 overexpression with a TGL3 deletion could be an attractive approach to further increase a cell's triacylglycerol content; however, the manipulation of proteins that affect a metabolic pathway is unpredictable at best. Cells that suppress the effects of a genetic modification frequently possess a selective advantage over those that acquire the desired trait. Thus, cells that display a desired phenotype are often difficult to engineer.
[0014] Disclosed is the successful combination of TGL3 deletion and DGA1 overexpression to increase a cell's triacylglycerol content. TGL3 deletion and the concomitant expression of the R. toruloides DGA1 diacylglycerol acyltransferase in Y. lipolytica strains resulted in a higher triacylglycerol content than strains carrying a single genetic modification.
Definitions
[0015] The articles "a" and "an" are used herein to refer to one or to more than one (i.e., to at least one) of the grammatical object of the article. By way of example, "an element" means one element or more than one element.
[0016] The term "activity" refers to the total capacity of a cell to perform a function. For example, a genetic modification that decreases the activity of a triacylglycerol lipase in a cell may reduce the amount of triacylglycerol lipase in a cell or reduce the efficiency of triacylglycerol lipase. A triacylglycerol lipase knockout reduces the amount of triacylglycerol lipase in the cell. Alternatively, a mutation to a triacylglycerol lipase gene may reduce the efficiency of its triacylglycerol lipase protein product with little effect on the amount of cellular triacylglycerol lipase. Mutations that reduce the efficiency of triacylglycerol lipase may affect the active site, for example, by changing one or more active site residues; they may impair the enzyme's kinetics, for example, by sterically blocking substrates or products; they may affect protein folding or dynamics, for example, by reducing the proportion of properly-folded enzymes; they may affect protein localization, for example, by preventing the lipase from localizing to lipid particles; or they may affect protein degradation, for example, by adding one or more protein cleavage sites or by adding one or more residues or amino acid sequences that target the protein for proteolysis. These mutations affect coding regions. Mutations that decrease triacylglycerol lipase activity may instead affect the transcription or translation of the gene. For example, mutation to a triacylglycerol lipase enhancer or promoter can reduce triacylglycerol lipase activity by reducing its expression. Mutating or deleting the non-coding portions of a triacylglycerol lipase gene, such as its introns, may also reduce transcription or translation. Additionally, mutations to the upstream regulators of a triacylglycerol lipase may affect triacylglycerol lipase activity; for example, the over-expression of one or more repressors may decrease triacylglycerol lipase activity, and a knockout or mutation of one or more activators may similarly decrease triacylglycerol lipase activity. A genetic modification that increases the activity of a diacylglycerol acyltransferase in a cell may increase the amount of triacylglycerol acyltransferase in a cell or increase the efficiency of diacylglycerol acyltransferase. For example, the genetic modification may simply insert an additional copy of diacylglycerol acyltransferase into the cell such that the additional copy is transcribed and translated into additional functional diacylglycerol acyltransferase. The added diacylglycerol acyltransferase gene can be native to the host organism or from a different organism. Alternatively, mutating or deleting the non-coding portions of a native diacylglycerol acyltransferase gene, such as its introns, may also increase translation. A native diacylglycerol acyltransferase gene can be altered by adding a new promoter that causes more transcription. Similarly, enhancers may be added to the diacylglycerol acyltransferase gene that increase transcription, or silencers may be mutated or deleted from the diacylglycerol acyltransferase gene to increase transcription. Mutations to a native gene's coding region might also increase diacylglycerol acyltransferase activity, for example, by producing a protein variant that does not interact with inhibitory proteins or molecules. The over-expression of one or more activators may increase diacylglycerol acyltransferase activity by increasing the expression of a diacylglycerol acyltransferase protein, and a knockout or mutation of one or more repressors may similarly increase diacylglycerol acyltransferase activity.
[0017] The term "biologically-active portion" refers to an amino acid sequence that is less than a full-length amino acid sequence, but exhibits at least one activity of the full length sequence. For example, a biologically-active portion of a diacylglycerol acyltransferase may refer to one or more domains of DGA1 or DGA2 having biological activity for converting acyl-CoA and diacylglycerol to triacylglycerol. Biologically-active portions of a DGA1 include peptides or polypeptides comprising amino acid sequences sufficiently identical to or derived from the amino acid sequence of the DGA1 protein, e.g., the amino acid sequence as set forth in SEQ ID NOs: 1, 3, 5, 7, 9, 11, 13, 15, 17, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, or 61 which include fewer amino acids than the full length DGA1, and exhibit at least one activity of a DGA1 protein. Similarly, biologically-active portions of a DGA2 protein include peptides or polypeptides comprising amino acid sequences sufficiently identical to or derived from the amino acid sequence of the DGA2 protein, e.g., the amino acid sequence as set forth in SEQ ID NOs: 29, 31, 33, 35, 37, 39, 63, 65, 67, 69, 71, 73, or 75, which include fewer amino acids than the full length DGA2, and exhibit at least one activity of a DGA2 protein. Biologically-active portions of a DGA3 protein include peptides or polypeptides comprising amino acid sequences sufficiently identical to or derived from the amino acid sequence of the DGA3 protein, e.g., the amino acid sequence as set forth in SEQ ID NOs: 83 or 85, which include fewer amino acids than the full length DGA3, and exhibit at least one activity of a DGA3 protein. A biologically-active portion of a diacylglycerol acyltransferase may comprise, for example, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 141, 142, 143, 144, 145, 146, 147, 148, 149, 150, 151, 152, 153, 154, 155, 156, 157, 158, 159, 160, 161, 162, 163, 164, 165, 166, 167, 168, 169, 170, 171, 172, 173, 174, 175, 176, 177, 178, 179, 180, 181, 182, 183, 184, 185, 186, 187, 188, 189, 190, 191, 192, 193, 194, 195, 196, 197, 198, 199, 200, 201, 202, 203, 204, 205, 206, 207, 208, 209, 210, 211, 212, 213, 214, 215, 216, 217, 218, 219, 220, 221, 222, 223, 224, 225, 226, 227, 228, 229, 230, 231, 232, 233, 234, 235, 236, 237, 238, 239, 240, 241, 242, 243, 244, 245, 246, 247, 248, 249, 250, 251, 252, 253, 254, 255, 256, 257, 258, 259, 260, 261, 262, 263, 264, 265, 266, 267, 268, 269, 270, 271, 272, 273, 274, 275, 276, 277, 278, 279, 280, 281, 282, 283, 284, 285, 286, 287, 288, 289, 290, 291, 292, 293, 294, 295, 296, 297, 298, 299, 300, 301, 302, 303, 304, 305, 306, 307, 308, 309, 310, 311, 312, 313, 314, 315, 316, 317, 318, 319, 320, 321, 322, 323, 324, 325, 326, 327, 328, 329, 330, 331, 332, 333, 334, 335, 336, 337, 338, 339, 340, 341, 342, 343, 344, 345, 346, 347, 348, 349, 350, 351, 352, 353, 354, 355, 356, 357, 358, 359, 360, 361, 362, 363, 364, 365, 366, 367, 368, 369, 370, 371, 372, 373, 374, 375, 376, 377, 378, 379, 380, 381, 382, 383, 384, 385, 386, 387, 388, 389, 390, 391, 392, 393, 394, 395, 396, 397, 398, 399, 400, 401, 402, 403, 404, 405, 406, 407, 408, 409, 410, 411, 412, 413, 414, 415, 416, 417, 418, 419, 420, 421, 422, 423, 424, 425, 426, 427, 428, 429, 430, 431, 432, 433, 434, 435, 436, 437, 438, 439, 440, 441, 442, 443, 444, 445, 446, 447, 448, 449, 450, 451, 452, 453, 454, 455, 456, 457, 458, 459, 460, 461, 462, 463, 464, 465, 466, 467, 468, 469, 470, 471, 472, 473, 474, 475, 476, 477, 478, 479, 480, 481, 482, 483, 484, 485, 486, 487, 488, 489, 490, 491, 492, 493, 494, 495, 496, 497, 498, 499, 500, 501, 502, 503, 504, 505, 506, 507, 508, 509, 510, 511, 512, 513, 514, 515, 516, 517, 518, 519, 520, 521, 522, 523, 524, 525, 526, 527, 528, 529, 530, 531, 532, 533, 534, 535, 536, 537, 538, 539, 540, 541, 542, 543, 544, 545, 546, 547, 548, 549, 550, 551, 552, 553, 554, 555, 556, 557, 558, 559, 560, 561, 562, 563, 564, 565, 566, 567, 568, 569, 570, 571, 572, 573, 574, 575, 576, 577, 578, 579, 580, 581, 582, 583, 584, 585, 586, 587, 588, 589, 590, 591, 592, 593, 594, 595, 596, 597, 598, 599, 600, 601, 602, 603, 604, 605, 606, 607, 608, 609, 610, 611, 612, 613, 614, 615, 616, 617, 618, 619, 620, 621, 622, 623, 624, 625, 626, 627, 628, 629, 630, 631, 632, 633, 634, 635, 636, 637, 638, 639, 640, 641, 642, 643, 644, 645, 646, 647, 648, 649, 650, 651, 652, 653, 654, 655, 656, 657, 658, 659, 660, 661, 662, 663, 664, 665, 666, 667, 668, 669, 670, 671, 672, 673, 674, 675, 676, 677, 678, 679, 680, 681, 682, 683, 684, 685, 686, 687, 688, 689, 690, 691, 692, 693, 694, 695, or 696 amino acids. Typically, biologically active portions comprise a domain or motif having the catalytic activity of converting acyl-CoA and diacylglycerol to triacylglycerol. A biologically active portion of a DGA1 protein can be a polypeptide which is, for example, 278 amino acids in length.
[0018] The term "DGAT1" refers to a gene that encodes a type 1 diacylglycerol acyltransferase protein, such as a gene that encodes a DGA2 protein.
[0019] The term "DGAT2" refers to a gene that encodes a type 2 diacylglycerol acyltransferase protein, such as a gene that encodes a DGA1 protein.
[0020] The term "DGAT3" refers to a gene that encodes a type 2 diacylglycerol acyltransferase protein, such as a gene that encodes a DGA3 protein.
[0021] "Diacylglyceride," "diacylglycerol," and "diglyceride," are esters comprised of glycerol and two fatty acids.
[0022] The terms "diacylglycerol acyltransferase" and "DGA" refer to any protein that catalyzes the formation of triacylglycerides from diacylglycerol. Diacylglycerol acyltransferases include type 1 diacylglycerol acyltransferases (DGA2), type 2 diacylglycerol acyltransferases (DGA1), and all homologs that catalyze the above-mentioned reaction.
[0023] The terms "diacylglycerol acyltransferase, type 1" and "type 1 diacylglycerol acyltransferases" refer to DGA2 and DGA2 orthologs.
[0024] The terms "diacylglycerol acyltransferase, type 2" and "type 2 diacylglycerol acyltransferases" refer to DGA1 and DGA1 orthologs.
[0025] The terms "diacylglycerol acyltransferase, type 3" and "type 3 diacylglycerol acyltransferases" refer to DGA3 and DGA3 orthologs.
[0026] The term "domain" refers to a part of the amino acid sequence of a protein that is able to fold into a stable three-dimensional structure independent of the rest of the protein.
[0027] The term "drug" refers to any molecule that inhibits cell growth or proliferation, thereby providing a selective advantage to cells that contain a gene that confers resistance to the drug. Drugs include antibiotics, antimicrobials, toxins, and pesticides.
[0028] "Dry weight" and "dry cell weight" mean weight determined in the relative absence of water. For example, reference to oleaginous cells as comprising a specified percentage of a particular component by dry weight means that the percentage is calculated based on the weight of the cell after substantially all water has been removed.
[0029] The term "encode" refers to nucleic acids that comprise a coding region, portion of a coding region, or compliments thereof. Both DNA and RNA may encode a gene. Both DNA and RNA may encode a protein.
[0030] The term "exogenous" refers to anything that is introduced into a cell. An "exogenous nucleic acid" is a nucleic acid that entered a cell through the cell membrane. An exogenous nucleic acid may contain a nucleotide sequence that exists in the native genome of a cell and/or nucleotide sequences that did not previously exist in the cell's genome. Exogenous nucleic acids include exogenous genes. An "exogenous gene" is a nucleic acid that codes for the expression of an RNA and/or protein that has been introduced into a cell (e.g., by transformation/transfection), and is also referred to as a "transgene." A cell comprising an exogenous gene may be referred to as a recombinant cell, into which additional exogenous gene(s) may be introduced. The exogenous gene may be from the same or different species relative to the cell being transformed. Thus, an exogenous gene can include a native gene that occupies a different location in the genome of the cell or is under different control, relative to the endogenous copy of the gene. An exogenous gene may be present in more than one copy in the cell. An exogenous gene may be maintained in a cell as an insertion into the genome (nuclear or plastid) or as an episomal molecule.
[0031] The term "expression" refers to the amount of a nucleic acid or amino acid sequence (e.g., peptide, polypeptide, or protein) in a cell. The increased expression of a gene refers to the increased transcription of that gene. The increased expression of an amino acid sequence, peptide, polypeptide, or protein refers to the increased translation of a nucleic acid encoding the amino acid sequence, peptide, polypeptide, or protein.
[0032] The term "gene," as used herein, may encompass genomic sequences that contain exons, particularly polynucleotide sequences encoding polypeptide sequences involved in a specific activity. The term further encompasses synthetic nucleic acids that did not derive from genomic sequence. In certain embodiments, the genes lack introns, as they are synthesized based on the known DNA sequence of cDNA and protein sequence. In other embodiments, the genes are synthesized, non-native cDNA wherein the codons have been optimized for expression in Y. lipolytica based on codon usage. The term can further include nucleic acid molecules comprising upstream, downstream, and/or intron nucleotide sequences.
[0033] The term "genetic modification" refers to the result of a transformation. Every transformation causes a genetic modification by definition.
[0034] The term "homolog", as used herein, refers to (a) peptides, oligopeptides, polypeptides, proteins, and enzymes having amino acid substitutions, deletions and/or insertions relative to the unmodified protein in question and having similar biological and functional activity as the unmodified protein from which they are derived, and (b) nucleic acids which encode peptides, oligopeptides, polypeptides, proteins, and enzymes with the same characteristics described in (a).
[0035] "Inducible promoter" is a promoter that mediates the transcription of an operably linked gene in response to a particular stimulus.
[0036] The term "integrated" refers to a nucleic acid that is maintained in a cell as an insertion into the cell's genome, such as insertion into a chromosome, including insertions into a plastid genome.
[0037] "In operable linkage" refers to a functional linkage between two nucleic acid sequences, such a control sequence (typically a promoter) and the linked sequence (typically a sequence that encodes a protein, also called a coding sequence). A promoter is in operable linkage with a gene if it can mediate transcription of the gene.
[0038] The term "knockout mutation" or "knockout" refers to a genetic modification that prevents a native gene from being transcribed and translated into a functional protein.
[0039] The term "native" refers to the composition of a cell or parent cell prior to a transformation event. A "native gene" refers to a nucleotide sequence that encodes a protein that has not been introduced into a cell by a transformation event. A "native protein" refers to an amino acid sequence that is encoded by a native gene.
[0040] The terms "nucleic acid" refers to a polymeric form of nucleotides of any length, either deoxyribonucleotides or ribonucleotides, or analogs thereof. Polynucleotides may have any three-dimensional structure, and may perform any function. The following are non-limiting examples of polynucleotides: coding or non-coding regions of a gene or gene fragment, loci (locus) defined from linkage analysis, exons, introns, messenger RNA (mRNA), transfer RNA, ribosomal RNA, ribozymes, cDNA, recombinant polynucleotides, branched polynucleotides, plasmids, vectors, isolated DNA of any sequence, isolated RNA of any sequence, nucleic acid probes, and primers. A polynucleotide may comprise modified nucleotides, such as methylated nucleotides and nucleotide analogs. If present, modifications to the nucleotide structure may be imparted before or after assembly of the polymer. A polynucleotide may be further modified, such as by conjugation with a labeling component. In all nucleic acid sequences provided herein, U nucleotides are interchangeable with T nucleotides.
[0041] The term "parent cell" refers to every cell from which a cell descended. The genome of a cell is comprised of the parent cell's genome and any subsequent genetic modifications to the parent cell's genome.
[0042] As used herein, the term "plasmid" refers to a circular DNA molecule that is physically separate from an organism's genomic DNA. Plasmids may be linearized before being introduced into a host cell (referred to herein as a linearized plasmid). Linearized plasmids may not be self-replicating, but may integrate into and be replicated with the genomic DNA of an organism.
[0043] The term "portion" refers to peptides, oligopeptides, polypeptides, protein domains, and proteins. A nucleotide sequence encoding a "portion of a protein" includes both nucleotide sequences that can be transcribed and/or translated and nucleotide sequences that must undergo one or more recombination events to be transcribed and/or translated. For example, a nucleic acid may comprise a nucleotide sequence encoding one or more amino acids of a selectable marker protein. This nucleic acid can be engineered to recombine with one or more different nucleotide sequences that encode the remaining portion of the protein. Such nucleic acids are useful for generating knockout mutations because only recombination with the target sequence is likely to reconstitute the full-length selectable marker gene whereas random-integration events are unlikely to result in a nucleotide sequence that can produce a functional marker protein. A "biologically-active portion" of a polypeptide is any amino acid sequence found in the polypeptide's amino acid sequence that is less than the full amino acid sequence but can perform the same function as the full-length polypeptide. A biologically-active portion of a diacylglycerol acyltransferase includes any amino acid sequence found in a full-length diacylglycerol acyltransferase that can catalyze the formation of triacylglycerol from diacylglycerol and acyl-CoA. A biologically-active portion of a polypeptide includes portions of the polypeptide that have the same activity as the full-length peptide and every portion that has more activity than background. For example, a biologically-active portion of a diacylglycerol acyltransferase may have 0.1, 0.5, 1, 2, 3, 4, 5, 10, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 99.5, 99.6, 99.7, 99.8, 99.9, 100, 100.1, 100.2, 100.3, 100.4, 100.5, 100.6, 100.7, 100.8, 100.9, 101, 105, 110, 115, 120, 125, 130, 135, 140, 145, 150, 160, 170, 180, 190, 200, 220, 240, 260, 280, 300, 320, 340, 360, 380, 400 percent activity relative to the full-length polypeptide or higher. A biologically-active portion of a polypeptide may include portions of a peptide that lack a domain that targets the polypeptide to a cellular compartment.
[0044] A "promoter" is a nucleic acid control sequence that directs the transcription of a nucleic acid. As used herein, a promoter includes the necessary nucleic acid sequences near the start site of transcription. A promoter also optionally includes distal enhancer or repressor elements, which can be located as much as several thousand base pairs from the start site of transcription.
[0045] "Recombinant" refers to a cell, nucleic acid, protein, or vector, which has been modified due to the introduction of an exogenous nucleic acid or the alteration of a native nucleic acid. Thus, e.g., recombinant cells can express genes that are not found within the native (non-recombinant) form of the cell or express native genes differently than those genes are expressed by a non-recombinant cell. Recombinant cells can, without limitation, include recombinant nucleic acids that encode for a gene product or for suppression elements such as mutations, knockouts, antisense, interfering RNA (RNAi), or dsRNA that reduce the levels of active gene product in a cell. A "recombinant nucleic acid" is a nucleic acid originally formed in vitro, in general, by the manipulation of nucleic acid, e.g., using polymerases, ligases, exonucleases, and endonucleases, or otherwise is in a form not normally found in nature. Recombinant nucleic acids may be produced, for example, to place two or more nucleic acids in operable linkage. Thus, an isolated nucleic acid or an expression vector formed in vitro by ligating DNA molecules that are not normally joined in nature, are both considered recombinant for the purposes of this invention. Once a recombinant nucleic acid is made and introduced into a host cell or organism, it may replicate using the in vivo cellular machinery of the host cell; however, such nucleic acids, once produced recombinantly, although subsequently replicated intracellularly, are still considered recombinant for purposes of this invention. Similarly, a "recombinant protein" is a protein made using recombinant techniques, i.e., through the expression of a recombinant nucleic acid.
[0046] The term "regulatory region" refers to nucleotide sequences that affect the transcription or translation of a gene but do not encode an amino acid sequence. Regulatory regions include promoters, operators, enhancers, and silencers.
[0047] "Transformation" refers to the transfer of a nucleic acid into a host organism or the genome of a host organism, resulting in genetically stable inheritance. Host organisms containing the transformed nucleic acid fragments are referred to as "recombinant", "transgenic" or "transformed" organisms. Thus, isolated polynucleotides of the present invention can be incorporated into recombinant constructs, typically DNA constructs, capable of introduction into and replication in a host cell. Such a construct can be a vector that includes a replication system and sequences that are capable of transcription and translation of a polypeptide-encoding sequence in a given host cell. Typically, expression vectors include, for example, one or more cloned genes under the transcriptional control of 5' and 3' regulatory sequences and a selectable marker. Such vectors also can contain a promoter regulatory region (e.g., a regulatory region controlling inducible or constitutive, environmentally- or developmentally-regulated, or location-specific expression), a transcription initiation start site, a ribosome binding site, a transcription termination site, and/or a polyadenylation signal.
[0048] The term "transformed cell" refers to a cell that has undergone a transformation. Thus, a transformed cell comprises the parent's genome and an inheritable genetic modification.
[0049] The terms "triacylglyceride," "triacylglycerol," "triglyceride," and "TAG" are esters comprised of glycerol and three fatty acids.
[0050] The term "triacylglycerol lipase" refers to any protein that can catalyze the removal of a fatty acid chain from a triacylglycerol. Triacylglycerol lipases include TGL3, TLG3/4, and TGL4.
[0051] The term "vector" refers to the means by which a nucleic acid can be propagated and/or transferred between organisms, cells, or cellular components. Vectors include plasmids, linear DNA fragments, viruses, bacteriophage, pro-viruses, phagemids, transposons, and artificial chromosomes, and the like, that may or may not be able to replicate autonomously or integrate into a chromosome of a host cell.
Microbe Engineering
[0052] A. Overview
[0053] In certain embodiments of the invention, a microorganism is genetically modified to increase its triacylglycerol content.
[0054] Genes and gene products may be introduced into microbial host cells. Suitable host cells for expression of the genes and nucleic acid molecules are microbial hosts that can be found broadly within the fungal or bacterial families. Examples of suitable host strains include but are not limited to fungal or yeast species, such as Arxula, Aspergillus, Aurantiochytrium, Candida, Claviceps, Cryptococcus, Cunninghamella, Hansenula, Kluyveromyces, Leucosporidiella, Lipomyces, Mortierella, Ogataea, Pichia, Prototheca, Rhizopus, Rhodosporidium, Rhodotorula, Saccharomyces, Schizosaccharomyces, Tremella, Trichosporon, Yarrowia, or bacterial species, such as members of proteobacteria and actinomycetes, as well as the genera Acinetobacter, Arthrobacter, Brevibacterium, Acidovorax, Bacillus, Clostridia, Streptomyces, Escherichia, Salmonella, Pseudomonas, and Cornyebacterium. Yarrowia lipolytica and Arxula adeninivorans are suited for use as a host microorganism because they can accumulate a large percentage of their weight as triacylglycerols.
[0055] Microbial expression systems and expression vectors containing regulatory sequences that direct high level expression of foreign proteins are known to those skilled in the art. Any of these could be used to construct chimeric genes to produce any one of the gene products of the instant sequences. These chimeric genes could then be introduced into appropriate microorganisms via transformation techniques to provide high-level expression of the enzymes.
[0056] For example, a gene encoding an enzyme can be cloned in a suitable plasmid, and an aforementioned starting parent strain as a host can be transformed with the resulting plasmid. This approach can increase the copy number of each of the genes encoding the enzymes and, as a result, the activities of the enzymes can be increased. The plasmid is not particularly limited so long as it renders a desired genetic modification inheritable to the microorganism's progeny.
[0057] Vectors or cassettes useful for the transformation of suitable host cells are well known in the art. Typically the vector or cassette contains sequences that direct the transcription and translation of the relevant gene, a selectable marker, and sequences that allow autonomous replication or chromosomal integration. Suitable vectors comprise a region 5' of the gene harboring transcriptional initiation controls and a region 3' of the DNA fragment which controls transcriptional termination. It is preferred when both control regions are derived from genes homologous to the transformed host cell, although it is to be understood that such control regions need not be derived from the genes native to the specific species chosen as a production host.
[0058] Promoters, cDNAs, and 3'UTRs, as well as other elements of the vectors, can be generated through cloning techniques using fragments isolated from native sources (Green & Sambrook, Molecular Cloning: A Laboratory Manual, (4th ed., 2012); U.S. Pat. No. 4,683,202; incorporated by reference). Alternatively, elements can be generated synthetically using known methods (Gene 164:49-53 (1995)).
[0059] B. Homologous Recombination
[0060] Homologous recombination is the ability of complementary DNA sequences to align and exchange regions of homology. Transgenic DNA ("donor") containing sequences homologous to the genomic sequences being targeted ("template") is introduced into the organism and then undergoes recombination into the genome at the site of the corresponding homologous genomic sequences.
[0061] The ability to carry out homologous recombination in a host organism has many practical implications for what can be carried out at the molecular genetic level and is useful in the generation of a microbe that can produce a desired product. By its nature, homologous recombination is a precise gene targeting event and, hence, most transgenic lines generated with the same targeting sequence will be essentially identical in terms of phenotype, necessitating the screening of far fewer transformation events. Homologous recombination also targets gene insertion events into the host chromosome, potentially resulting in excellent genetic stability, even in the absence of genetic selection. Because different chromosomal loci will likely impact gene expression, even from exogenous promoters/UTRs, homologous recombination can be a method of querying loci in an unfamiliar genome environment and to assess the impact of these environments on gene expression.
[0062] A particularly useful genetic engineering approach using homologous recombination is to co-opt specific host regulatory elements such as promoters/UTRs to drive heterologous gene expression in a highly specific fashion.
[0063] Because homologous recombination is a precise gene targeting event, it can be used to precisely modify any nucleotide(s) within a gene or region of interest, so long as sufficient flanking regions have been identified. Therefore, homologous recombination can be used as a means to modify regulatory sequences impacting gene expression of RNA and/or proteins. It can also be used to modify protein coding regions in an effort to modify enzyme activities such as substrate specificity, affinities and Km, thereby affecting a desired change in the metabolism of the host cell. Homologous recombination provides a powerful means to manipulate the host genome resulting in gene targeting, gene conversion, gene deletion, gene duplication, gene inversion and exchanging gene expression regulatory elements such as promoters, enhancers and 3'UTRs.
[0064] Homologous recombination can be achieved by using targeting constructs containing pieces of endogenous sequences to "target" the gene or region of interest within the endogenous host cell genome. Such targeting sequences can either be located 5' of the gene or region of interest, 3' of the gene/region of interest or even flank the gene/region of interest. Such targeting constructs can be transformed into the host cell either as a supercoiled plasmid DNA with additional vector backbone, a PCR product with no vector backbone, or as a linearized molecule. In some cases, it may be advantageous to first expose the homologous sequences within the transgenic DNA (donor DNA) by cutting the transgenic DNA with a restriction enzyme. This step can increase the recombination efficiency and decrease the occurrence of undesired events. Other methods of increasing recombination efficiency include using PCR to generate transforming transgenic DNA containing linear ends homologous to the genomic sequences being targeted.
[0065] C. Vectors and Vector Components
[0066] Vectors for transforming microorganisms in accordance with the present invention can be prepared by known techniques familiar to those skilled in the art in view of the disclosure herein. A vector typically contains one or more genes, in which each gene codes for the expression of a desired product (the gene product) and is operably linked to one or more control sequences that regulate gene expression or target the gene product to a particular location in the recombinant cell.
[0067] 1. Control Sequences
[0068] Control sequences are nucleic acids that regulate the expression of a coding sequence or direct a gene product to a particular location in or outside a cell. Control sequences that regulate expression include, for example, promoters that regulate transcription of a coding sequence and terminators that terminate transcription of a coding sequence. Another control sequence is a 3' untranslated sequence located at the end of a coding sequence that encodes a polyadenylation signal. Control sequences that direct gene products to particular locations include those that encode signal peptides, which direct the protein to which they are attached to a particular location inside or outside the cell.
[0069] Thus, an exemplary vector design for expression of a gene in a microbe contains a coding sequence for a desired gene product (for example, a selectable marker, or an enzyme) in operable linkage with a promoter active in yeast. Alternatively, if the vector does not contain a promoter in operable linkage with the coding sequence of interest, the coding sequence can be transformed into the cells such that it becomes operably linked to an endogenous promoter at the point of vector integration.
[0070] The promoter used to express a gene can be the promoter naturally linked to that gene or a different promoter.
[0071] A promoter can generally be characterized as constitutive or inducible. Constitutive promoters are generally active or function to drive expression at all times (or at certain times in the cell life cycle) at the same level. Inducible promoters, conversely, are active (or rendered inactive) or are significantly up- or down-regulated only in response to a stimulus. Both types of promoters find application in the methods of the invention. Inducible promoters useful in the invention include those that mediate transcription of an operably linked gene in response to a stimulus, such as an exogenously provided small molecule, temperature (heat or cold), lack of nitrogen in culture media, etc. Suitable promoters can activate transcription of an essentially silent gene or upregulate, preferably substantially, transcription of an operably linked gene that is transcribed at a low level.
[0072] Inclusion of termination region control sequence is optional, and if employed, then the choice is primarily one of convenience, as the termination region is relatively interchangeable. The termination region may be native to the transcriptional initiation region (the promoter), may be native to the DNA sequence of interest, or may be obtainable from another source (See, e.g., Chen & Orozco, Nucleic Acids Research 16:8411 (1988)).
[0073] 2. Genes and Codon Optimization
[0074] Typically, a gene includes a promoter, a coding sequence, and termination control sequences. When assembled by recombinant DNA technology, a gene may be termed an expression cassette and may be flanked by restriction sites for convenient insertion into a vector that is used to introduce the recombinant gene into a host cell. The expression cassette can be flanked by DNA sequences from the genome or other nucleic acid target to facilitate stable integration of the expression cassette into the genome by homologous recombination. Alternatively, the vector and its expression cassette may remain unintegrated (e.g., an episome), in which case, the vector typically includes an origin of replication, which is capable of providing for replication of the vector DNA.
[0075] A common gene present on a vector is a gene that codes for a protein, the expression of which allows the recombinant cell containing the protein to be differentiated from cells that do not express the protein. Such a gene, and its corresponding gene product, is called a selectable marker or selection marker. Any of a wide variety of selectable markers can be employed in a transgene construct useful for transforming the organisms of the invention.
[0076] For optimal expression of a recombinant protein, it is beneficial to employ coding sequences that produce mRNA with codons optimally used by the host cell to be transformed. Thus, proper expression of transgenes can require that the codon usage of the transgene matches the specific codon bias of the organism in which the transgene is being expressed. The precise mechanisms underlying this effect are many, but include the proper balancing of available aminoacylated tRNA pools with proteins being synthesized in the cell, coupled with more efficient translation of the transgenic messenger RNA (mRNA) when this need is met. When codon usage in the transgene is not optimized, available tRNA pools are not sufficient to allow for efficient translation of the transgenic mRNA resulting in ribosomal stalling and termination and possible instability of the transgenic mRNA.
[0077] D. Transformation
[0078] Cells can be transformed by any suitable technique including, e.g., biolistics, electroporation, glass bead transformation, and silicon carbide whisker transformation. Any convenient technique for introducing a transgene into a microorganism can be employed in the present invention. Transformation can be achieved by, for example, the method of D. M. Morrison (Methods in Enzymology 68:326 (1979)), the method by increasing permeability of recipient cells for DNA with calcium chloride (Mandel & Higa, J. Molecular Biology, 53:159 (1970)), or the like.
[0079] Examples of expression of transgenes in oleaginous yeast (e.g., Yarrowia lipolytica) can be found in the literature (Bordes et al., J. Microbiological Methods, 70:493 (2007); Chen et al., Applied Microbiology & Biotechnology 48:232 (1997)). Examples of expression of exogenous genes in bacteria such as E. coli are well known (Green & Sambrook, Molecular Cloning: A Laboratory Manual, (4th ed., 2012)).
[0080] Vectors for transformation of microorganisms in accordance with the present invention can be prepared by known techniques familiar to those skilled in the art. In one embodiment, an exemplary vector design for expression of a gene in a microorganism contains a gene encoding an enzyme in operable linkage with a promoter active in the microorganism. Alternatively, if the vector does not contain a promoter in operable linkage with the gene of interest, the gene can be transformed into the cells such that it becomes operably linked to a native promoter at the point of vector integration. The vector can also contain a second gene that encodes a protein. Optionally, one or both gene(s) is/are followed by a 3' untranslated sequence containing a polyadenylation signal. Expression cassettes encoding the two genes can be physically linked in the vector or on separate vectors. Co-transformation of microbes can also be used, in which distinct vector molecules are simultaneously used to transform cells (Protist 155:381-93 (2004)). The transformed cells can be optionally selected based upon the ability to grow in the presence of the antibiotic or other selectable marker under conditions in which cells lacking the resistance cassette would not grow.
Exemplary Nucleic Acids, Cells, and Methods
[0081] A. Diacylglycerol Acyltransferase Nucleic Acid Molecules and Vectors
[0082] The diacylglycerol acyltransferase may be a type 1 diacylglycerol acyltransferase, type 2 diacylglycerol acyltransferase, type 3 diacylglycerol acyltransferase, or any other protein that catalyzes the conversion of diacylglycerol into a triacylglyceride. In some embodiments, the diacylglycerol acyltransferase is DGA1. For example, the diacylglycerol acyltransferase may be a DGA1 protein encoded by a DGAT2 gene selected from the group consisting of Arxula adeninivorans, Aspergillus terreus, Aurantiochytrium limacinum, Claviceps purpurea, Gloeophyllum trabeum, Lipomyces starkeyi, Microbotryum violaceum, Pichia guilliermondii, Phaeodactylum tricornutum, Puccinia graminis, Rhodosporidium diobovatum, Rhodosporidium toruloides, Rhodotorula graminis, and Yarrowia lipolytica.
[0083] The DGAT2 gene may have a nucleotide sequence set forth in SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, or 62. In other embodiments, the DGAT2 gene is substantially identical to SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, or 62, and the nucleotide sequence encodes a protein that retains the functional activity of a protein encoded by SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, or 62, yet differs in nucleotide sequence due to natural allelic variation or mutagenesis. In another embodiment, the DGAT2 gene comprises an nucleotide sequence at least about 70%, 71%, 72%, 73%, 74%, 75%, 76%, 77%, 78%, 79%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.1%, 99.2%, 99.3%, 99.4%, 99.5%, 99.6%, 99.7%, 99.8%, 99.9% or more identical to SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, or 62.
[0084] The DGA1 may have an amino acid sequence set forth in SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, or 61. In other embodiments, the DGA1 is substantially identical to SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, or 61, and retains the functional activity of the protein of SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, or 61, yet differs in amino acid sequence due to natural allelic variation or mutagenesis. In another embodiment, the DGA1 protein comprises an amino acid sequence at least about 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.1%, 99.2%, 99.3%, 99.4%, 99.5%, 99.6%, 99.7%, 99.8%, 99.9% or more identical to SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, or 61.
[0085] In some embodiments, the diacylglycerol acyltransferase is DGA2. For example, the diacylglycerol acyltransferase may be a DGA2 protein encoded by a DGAT1 gene found in an organism selected from the group consisting of Arxula adeninivorans, Aspergillus terreus, Chaetomium globosum, Claviceps purpurea, Lipomyces starkeyi, Metarhizium acridum, Ophiocordyceps sinensis, Phaeodactylum tricornutum, Pichia guilliermondii, Rhodosporidium toruloides, Rhodotorula graminis, Trichoderma virens, and Yarrowia lipolytica.
[0086] The DGAT1 gene may have a nucleotide sequence set forth in SEQ ID NO: 30, 32, 34, 36, 38, 40, 64, 66, 68, 70, 72, 74, or 76. In other embodiments, the DGAT1 gene is substantially identical to SEQ ID NO: 30, 32, 34, 36, 38, 40, 64, 66, 68, 70, 72, 74, or 76, and the nucleotide sequence encodes a protein that retains the functional activity of a protein encoded by SEQ ID NO: 30, 32, 34, 36, 38, 40, 64, 66, 68, 70, 72, 74, or 76, yet differs in nucleotide sequence due to natural allelic variation or mutagenesis. In another embodiment, the DGAT1 gene comprises an nucleotide sequence at least about 70%, 71%, 72%, 73%, 74%, 75%, 76%, 77%, 78%, 79%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.1%, 99.2%, 99.3%, 99.4%, 99.5%, 99.6%, 99.7%, 99.8%, 99.9% or more identical to SEQ ID NO: 30, 32, 34, 36, 38, 40, 64, 66, 68, 70, 72, 74, or 76.
[0087] The DGA2 may have an amino acid sequence set forth in SEQ ID NO: 29, 31, 33, 35, 37, 39, 63, 65, 67, 69, 71, 73, or 75. In other embodiments, the DGA2 is substantially identical to SEQ ID NO: 29, 31, 33, 35, 37, 39, 63, 65, 67, 69, 71, 73, or 75, and retains the functional activity of the protein of SEQ ID NO: 29, 31, 33, 35, 37, 39, 63, 65, 67, 69, 71, 73, or 75, yet differs in amino acid sequence due to natural allelic variation or mutagenesis. In another embodiment, the DGA2 protein comprises an amino acid sequence at least about 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.1%, 99.2%, 99.3%, 99.4%, 99.5%, 99.6%, 99.7%, 99.8%, 99.9% or more identical to SEQ ID NO: 29, 31, 33, 35, 37, 39, 63, 65, 67, 69, 71, 73, or 75.
[0088] In some embodiments, the diacylglycerol acyltransferase is DGA3. For example, the diacylglycerol acyltransferase may be a DGA3 protein encoded by a DGAT3 gene found in an organism selected from the group consisting of Ricinus communis and Arachis hypogaea.
[0089] The DGAT3 gene may have a nucleotide sequence set forth in SEQ ID NO: 84 or 86. In other embodiments, the DGAT3 gene is substantially identical to SEQ ID NO: 84 or 86, and the nucleotide sequence encodes a protein that retains the functional activity of a protein encoded by SEQ ID NO: 84 or 86, yet differs in nucleotide sequence due to natural allelic variation or mutagenesis. In another embodiment, the DGAT3 gene comprises an nucleotide sequence at least about 70%, 71%, 72%, 73%, 74%, 75%, 76%, 77%, 78%, 79%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.1%, 99.2%, 99.3%, 99.4%, 99.5%, 99.6%, 99.7%, 99.8%, 99.9% or more identical to SEQ ID NO: 84 or 86.
[0090] The DGA3 may have an amino acid sequence set forth in SEQ ID NO: 83 or 85. In other embodiments, the DGA3 is substantially identical to SEQ ID NO: 83 or 85, and retains the functional activity of the protein of SEQ ID NO: 83 or 85, yet differs in amino acid sequence due to natural allelic variation or mutagenesis. In another embodiment, the DGA3 protein comprises an amino acid sequence at least about 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.1%, 99.2%, 99.3%, 99.4%, 99.5%, 99.6%, 99.7%, 99.8%, 99.9% or more identical to SEQ ID NO: 83 or 85.
[0091] The DGAT1, DGAT2, and DGAT3 genes may comprise conservative substitutions, deletions, and/or insertions while still encoding a protein that has functional diacylglycerol acyltransferase activity. For example, the DGAT1, DGAT2, or DGAT3 codons may be optimized for a particular host cell, different codons may be substituted for convenience, such as to introduce a restriction site or to create optimal PCR primers, or codons may be substituted for another purpose. Similarly, the nucleotide sequence may be altered to create conservative amino acid substitutions, deletions, and/or insertions.
[0092] The DGA1, DGA2, and DGA3 polypeptides may comprise conservative substitutions, deletions, and/or insertions while still maintaining functional diacylglycerol acyltransferase activity. Conservative substitution tables are well known in the art (Creighton, Proteins (2d. ed., 1992)).
[0093] Amino acid substitutions, deletions and/or insertions may readily be made using recombinant DNA manipulation techniques. Methods for the manipulation of DNA sequences to produce substitution, insertion or deletion variants of a protein are well known in the art. These methods include M13 mutagenesis, T7-Gen in vitro mutagenesis (USB, Cleveland, Ohio), Quick Change Site Directed mutagenesis (Stratagene, San Diego, Calif.), PCR-mediated site-directed mutagenesis, and other site-directed mutagenesis protocols.
[0094] To determine the percent identity of two amino acid sequences or of two nucleic acid sequences, the sequences can be aligned for optimal comparison purposes (e.g., gaps can be introduced in one or both of a first and a second amino acid or nucleic acid sequence for optimal alignment and non-identical sequences can be disregarded for comparison purposes). The length of a reference sequence aligned for comparison purposes can be at least 95% of the length of the reference sequence. The amino acid residues or nucleotides at corresponding amino acid positions or nucleotide positions can then be compared. When a position in the first sequence is occupied by the same amino acid residue or nucleotide as the corresponding position in the second sequence, then the molecules are identical at that position (as used herein amino acid or nucleic acid "identity" is equivalent to amino acid or nucleic acid "homology"). The percent identity between the two sequences is a function of the number of identical positions shared by the sequences, taking into account the number of gaps, and the length of each gap, which need to be introduced for optimal alignment of the two sequences.
[0095] The comparison of sequences and determination of percent identity between two sequences can be accomplished using a mathematical algorithm. In one embodiment, the percent identity between two amino acid sequences can be determined using the Needleman and Wunsch (J. Molecular Biology 48:444-453 (1970)) algorithm which has been incorporated into the GAP program in the GCG software package (available at http://www.gcg.com), using either a Blosum 62 matrix or a PAM250 matrix, and a gap weight of 16, 14, 12, 10, 8, 6, or 4 and a length weight of 1, 2, 3, 4, 5, or 6. In yet another embodiment, the percent identity between two nucleotide sequences can be determined using the GAP program in the GCG software package (available at http://www.gcg.com), using a NWSgapdna.CMP matrix and a gap weight of 40, 50, 60, 70, or 80 and a length weight of 1, 2, 3, 4, 5, or 6. In another embodiment, the percent identity between two amino acid or nucleotide sequences can be determined using the algorithm of E. Meyers and W. Miller (Computer Applications in the Biosciences 4:11-17 (1988)) which has been incorporated into the ALIGN program (version 2.0 or 2.0U), using a PAM120 weight residue table, a gap length penalty of 12 and a gap penalty of 4.
[0096] Exemplary computer programs which can be used to determine identity between two sequences include, but are not limited to, the suite of BLAST programs, e.g., BLASTN, MEGABLAST, BLASTX, TBLASTN, TBLASTX, and BLASTP, and Clustal programs, e.g., ClustalW, ClustalX, and Clustal Omega.
[0097] Sequence searches are typically carried out using the BLASTN program, when evaluating a given nucleic acid sequence relative to nucleic acid sequences in the GenBank DNA Sequences and other public databases. The BLASTX program is effective for searching nucleic acid sequences that have been translated in all reading frames against amino acid sequences in the GenBank Protein Sequences and other public databases.
[0098] An alignment of selected sequences in order to determine "% identity" between two or more sequences is performed using for example, the CLUSTAL-W program.
[0099] A "coding sequence" or "coding region" refers to a nucleic acid molecule having sequence information necessary to produce a protein product, such as an amino acid or polypeptide, when the sequence is expressed. The coding sequence may comprise and/or consist of untranslated sequences (including introns or 5' or 3' untranslated regions) within translated regions, or may lack such intervening untranslated sequences (e.g., as in cDNA).
[0100] The abbreviation used throughout the specification to refer to nucleic acids comprising and/or consisting of nucleotide sequences are the conventional one-letter abbreviations. Thus when included in a nucleic acid, the naturally occurring encoding nucleotides are abbreviated as follows: adenine (A), guanine (G), cytosine (C), thymine (T) and uracil (U). Also, unless otherwise specified, the nucleic acid sequences presented herein is the 5'.fwdarw.3'direction.
[0101] As used herein, the term "complementary" and derivatives thereof are used in reference to pairing of nucleic acids by the well-known rules that A pairs with T or U and C pairs with G. Complement can be "partial" or "complete". In partial complement, only some of the nucleic acid bases are matched according to the base pairing rules; while in complete or total complement, all the bases are matched according to the pairing rule. The degree of complement between the nucleic acid strands may have significant effects on the efficiency and strength of hybridization between nucleic acid strands as well known in the art. The efficiency and strength of said hybridization depends upon the detection method.
[0102] As used herein, "DGA1" means a diacylglycerol acyltransferase type 2 (DGAT2). DGA1 is an integral membrane protein that catalyzes the final enzymatic step in oil biosynthesis and the production of triacylglycerols in plants, fungi, and mammals. The DGA1 may play a key role in altering the quantity of long-chain polyunsaturated fatty acids produced in oils of oleaginous organisms. DGA1 is related to the acyl-coenzyme A:cholesterol acyltransferase ("ACAT"). This enzyme is responsible for transferring an acyl group from acyl-coenzyme-A to the sn-3 position of 1,2-diacylglycerol ("DAG") to form triacylglycerol ("TAG") (thereby involved in the terminal step of TAG biosynthesis). DGA1 is associated with membrane and lipid body fractions in plants and fungi, particularly, in oilseeds where it contributes to the storage of carbon used as energy reserves. TAG is believed to be an important chemical for storage of energy in cells. DGA1 is known to regulate TAG structure and direct TAG synthesis.
[0103] The DGA1 polynucleotide and polypeptide sequences may be derived from highly oleaginous organisms having very high, native levels of lipid accumulation. (Bioresource Technology 144:360-69 (2013); Progress Lipid Research 52:395-408 (2013); Applied Microbiology & Biotechnology 90:1219-27 (2011); European Journal Lipid Science & Technology 113:1031-51 (2011); Food Technology & Biotechnology 47:215-20 (2009); Advances Applied Microbiology 51:1-51 (2002); Lipids 11:837-44 (1976)). A list of organisms with a reported lipid content of about 50% and higher is shown in Table 1. R. toruloides and L. starkeyi have the highest lipid content. Among the organisms in Table 1, five have publicly accessible sequence for DGA1, R. toruloides, L. starkeyi, A. limacinum, A. terreus, and C. purpurea (bolded in Table 1).
TABLE-US-00001 TABLE 1 List of oleaginous fungi with reported lipid contents of about 50% and above. Organisms with publicly accessible sequences for a DGA1 gene are in bold. Fungi with reported high lipid content Cryptococcus albidus Cryptococcus curvatus Cryptococcus ramirezgomezianus Cryptococcus terreus Cryptococcus wieringae Cunninghamella echinulata Cunninghamella japonica Leucosporidiella creatinivora Lipomyces lipofer Lipomyces tetrasporus Mortierella isabellina Prototheca zopfii Rhizopus arrhizus Rhodosporidium babjevae Rhodosporidium paludigenum Rhodotorula glutinis Rhodotorula mucilaginosa Tremella enchepala Trichosporon cutaneum Trichosporon fermentans
[0104] Nucleic acid constructs for overexpressing the DGA1 gene were described in U.S. Ser. No. 61/943,664 (hereby incorporated by reference). FIG. 1 shows expression construct pNC243 used for overexpression of the R. toruloides DGA1 gene NG66 (SEQ ID No. 6) in Y. lipolytica. DGA1 expression constructs were linearized before transformation by PacI/NotI restriction digest (FIG. 1). The linear expression constructs each included expression cassette for DGA1 gene and for Nat1 gene, used as marker for selection with nourseothricin (NAT).
[0105] Nucleic acid constructs for overexpressing the DGA2 gene and/or other diacylglycerol acyltransferases may be created using the methods described above and/or other methods known in the art.
[0106] B. Triacylglycerol Lipase Nucleic Acid Molecules and Vectors
[0107] Triacylglycerol lipase depletes a cell's triacylglycerol by removing one or more fatty acid chains. Thus, decreasing the net triacylglycerol lipase activity of a cell may increase the cell's triacylglycerol. This decrease may be accomplished by reducing the efficiency of the enzyme, e.g., by mutating amino acids in its active site, or by reducing the expression of the enzyme. For example, a TGL3 knockout mutation will decrease the activity of a triacylglycerol lipase because it prevents the cell from transcribing TGL3.
[0108] In some embodiments, the triacylglycerol lipase is TGL3, TGL3/4, or TGL4.
[0109] The TGL3 gene in Y. lipolytica encodes the triacylglycerol lipase protein TGL3 (SEQ ID No. 19). SEQ ID No. 20 contains the TGL3 nucleotide sequence, 100 upstream nucleotides, and 100 downstream. Thus, the SEQ ID No. 20 nucleotide sequence may be used to design a nucleic acid capable of recombining with a nucleic acid sequence in the native Y. lipolytica triacylglycerol lipase gene.
[0110] Knockout cassettes SEQ ID Nos. 27 and 28 are capable of recombining with the native TGL3 gene in Y. lipolytica. Thus, in some embodiments, the nucleic acids encoded by SEQ ID Nos. 27 and 28 may be used to generate a triacylglycerol lipase knockout mutation in Y. lipolytica. SEQ ID Nos. 27 and 28 each contain portions of a hygromycin resistance gene hph. Neither isolated sequence encodes a functional protein, but the two sequences are capable of encoding a functional kinase that confers hygromycin resistance upon successful recombination. Further, neither SEQ ID No. 27 nor SEQ ID No. 28 contains a promoter or terminator, and thus, they rely on homologous recombination with the Y. lipolytica TGL3 gene in order for the hph gene to be transcribed and translated. In this way, successfully transformed oleaginous cells may be selected by growing the cells on medium containing hygromycin.
[0111] Knockout cassette SEQ ID NO. 27 may be prepared by amplifying a hygromycin resistance gene hph (SEQ ID NO. 22) with primer NP1798 (SEQ ID NO. 23) and primer NP656 (SEQ ID NO. 24). Knockout cassette SEQ ID NO. 28 may be prepared by amplifying a hygromycin resistance gene hph (SEQ ID NO. 22) with primer NP655 (SEQ ID NO. 25) and primer NP1799 (SEQ ID NO. 26).
[0112] Different approaches may be used to design nucleic acids that knockout the TGL3 gene in Y. lipolytica (Biochimica Biophysica Acta 1831:1486-95 (2013)). The methods disclosed herein and other methods known in the art may be used to knockout triacylglycerol different lipase genes in other species. For example, these methods may be used to reduce the activity of the TGL3 gene of Arxula adeninivorans (SEQ ID NO:78), the TGL3/4 gene of Arxula adeninivorans (SEQ ID NO:80), and/or the TGL4 gene of Arxula adeninivorans (SEQ ID NO:82). Similarly, these methods may be used to reduce the activity of the TGL4 gene in Y. lipolytica (SEQ ID NO:88).
[0113] C. Transformed Oleaginous Cell
[0114] In some embodiments, the transformed oleaginous cell is a prokaryotic cell, such as a bacterial cell. In some embodiments, the cell is a eukaryotic cell, such as a mammalian cell, a yeast cell, a filamentous fungi cell, a protist cell, an algae cell, an avian cell, a plant cell, or an insect cell.
[0115] The cell may be selected from the group consisting of Arxula, Aspergillus, Aurantiochytrium, Candida, Claviceps, Cryptococcus, Cunninghamella, Geotrichum, Hansenula, Kluyveromyces, Kodamaea, Leucosporidiella, Lipomyces, Mortierella, Ogataea, Pichia, Prototheca, Rhizopus, Rhodosporidium, Rhodotorula, Saccharomyces, Schizosaccharomyces, Tremella, Trichosporon, Wickerhamomyces, and Yarrowia.
[0116] In some embodiments, the cell is selected from the group of consisting of Arxula adeninivorans, Aspergillus niger, Aspergillus orzyae, Aspergillus terreus, Aurantiochytrium limacinum, Candida utilis, Claviceps purpurea, Cryptococcus albidus, Cryptococcus curvatus, Cryptococcus ramirezgomezianus, Cryptococcus terreus, Cryptococcus wieringae, Cunninghamella echinulata, Cunninghamella japonica, Geotrichum fermentans, Hansenula polymorpha, Kluyveromyces lactis, Kluyveromyces marxianus, Kodamaea ohmeri, Leucosporidiella creatinivora, Lipomyces lipofer, Lipomyces starkeyi, Lipomyces tetrasporus, Mortierella isabellina, Mortierella alpina, Ogataea polymorpha, Pichia ciferrii, Pichia guilliermondii, Pichia pastoris, Pichia stipites, Prototheca zopfii, Rhizopus arrhizus, Rhodosporidium babjevae, Rhodosporidium toruloides, Rhodosporidium paludigenum, Rhodotorula glutinis, Rhodotorula mucilaginosa, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Tremella enchepala, Trichosporon cutaneum, Trichosporon fermentans, Wickerhamomyces ciferrii, and Yarrowia lipolytica.
[0117] In certain embodiments, the transformed oleaginous cell is a high-temperature tolerant yeast cell. In some embodiments the transformed oleaginous cell is Kluyveromyces marxianus.
[0118] In certain embodiments, the cell is Yarrowia lipolytica or Arxula adeninivorans.
[0119] D. Increasing Diacylglycerol Acyltransferase in a Cell
[0120] A protein's activity may be increased by overexpressing the protein. Proteins may be overexpressed in a cell using a variety of genetic modifications. In some embodiments, the genetic modification increases the expression of a native diacylglycerol acyltransferase. A native diacylglycerol acyltransferase may be overexpressed by modifying the upstream transcription regulators of a native diacylglycerol acyltransferase gene, for example, by increasing the expression of a transcription activator or decreasing the expression of a transcription repressor. Alternatively, the promoter of a native diacylglycerol acyltransferase gene may be substituted with a constitutively active or inducible promoter by recombination with an exogenous nucleic acid.
[0121] In some embodiments, the genetic modification encodes at least one copy of a diacylglycerol acyltransferase gene. The diacylglycerol acyltransferase gene may be a gene native to the cell or from a different species. In certain embodiments, the diacylglycerol acyltransferase gene is the type II diacylglycerol acyltransferase gene from Arxula adeninivorans, Aspergillus terreus, Aurantiochytrium limacinum, Claviceps purpurea, Gloeophyllum trabeum, Lipomyces starkeyi, Microbotryum violaceum, Pichia guilliermondii, Phaeodactylum tricornutum, Puccinia graminis, Rhodosporidium diobovatum, Rhodosporidium toruloides, Rhodotorula graminis, or Yarrowia lipolytica. In certain embodiments, the diacylglycerol acyltransferase gene is the type I diacylglycerol acyltransferase gene from Arxula adeninivorans, Aspergillus terreus, Chaetomium globosum, Claviceps purpurea, Lipomyces starkeyi, Metarhizium acridum, Ophiocordyceps sinensis, Phaeodactylum tricornutum, Pichia guilliermondii, Rhodosporidium toruloides, Rhodotorula graminis, Trichoderma virens, or Yarrowia lipolytica. In certain embodiments, the diacylglycerol acyltransferase gene is the type III diacylglycerol acyltransferase gene from Ricinus communis or Arachis hypogaea. In certain embodiments, diacylglycerol acyltransferase is overexpressed by transforming a cell with a gene encoding a diacylglycerol acyltransferase gene. The genetic modification may encode one or more than one copy of a diacylglycerol acyltransferase gene. In certain embodiments, the genetic modification encodes at least one copy of the DGA1 protein from R. toruloides. In some embodiments, the genetic modification encodes at least one copy of the DGA1 protein from R. toruloides and the transformed oleaginous cell is Y. lipolytica.
[0122] In certain embodiments, the diacylglycerol acyltransferase gene is inheritable to the progeny of a transformed oleaginous cell. In some embodiments, the diacylglycerol acyltransferase gene is inheritable because it resides on a plasmid. In certain embodiments, the diacylglycerol acyltransferase gene is inheritable because it is integrated into the genome of the transformed oleaginous cell.
[0123] E. Decreasing Triacylglycerol Lipase Activity in a Cell
[0124] In some embodiments, the transformed oleaginous cell comprises a genetic modification that decreases the activity of a native triacylglycerol lipase. Such genetic modifications may affect a protein that regulates the transcription of a triacylglycerol lipase gene, including modifications that decrease the expression of a transcription activator and/or increase the expression of a transcription repressor. Modifications that affect a regulator protein may both decrease the expression of triacylglycerol lipase and alter other gene expression profiles that shift the cellular equilibrium toward increased triacylglycerol accumulation. Alternatively, the genetic modification may be the introduction of a small interfering RNA, or a nucleic acid that encodes a small interfering RNA. In other embodiments, the genetic modification consists of the homologous recombination of a nucleic acid and the regulatory region of a native triacylglycerol lipase gene, including an operator, promoter, sequences upstream from the promoter, enhancers, and sequences downstream of the gene.
[0125] In some embodiments the transformed oleaginous cell comprises a genetic modification consisting of a homologous recombination event. In certain embodiments, the transformed oleaginous cell comprises a genetic modification consisting of a homologous recombination event between a native triacylglycerol lipase gene and a nucleic acid. Thus, the genetic modification deletes the triacylglycerol lipase gene, prevents its transcription, or prevents the transcription of a gene that can be transcribed into a fully-active protein. A homologous recombination event may mutate or delete a portion of a native triacylglycerol lipase gene. For example, the homologous recombination event may mutate one or more residues in the active site of a native triacylglycerol lipase, thereby reducing the efficiency of the lipase or rendering it inactive. Alternatively, the homologous recombination event may affect post-translational modification, folding, stability, or localization within the cell. In certain embodiments, the genetic modification is a triacylglycerol lipase knockout mutation. Knockout mutations are preferable because they eliminate a pathway that depletes a cell's triacylglycerol content, thereby increasing the triacylglycerol content of a cell.
[0126] A knockout mutation may delete the triacylglycerol lipase gene. Additionally, the knockout mutation may substitute the triacylglycerol lipase gene with a gene that encodes a different protein. The gene may be operably linked to an exogenous promoter. In certain embodiments, the gene is not linked to an exogenous promoter, and instead, the gene is configured to recombine with the triacylglycerol lipase gene such that the triacylglycerol lipase gene's promoter drives transcription of the gene. Thus, the gene is less likely to be expressed if it randomly integrates into the cell's genome. Methods for creating knockouts are well-known in the art (See, e.g., Fickers et al., J. Microbiological Methods 55:727 (2003)).
[0127] In certain embodiments, the genetic modification comprises two homologous recombination events. In the first event, a nucleic acid encoding a portion of a gene recombines with the triacylglycerol lipase gene, and in the second event, a nucleic acid encoding the remaining portion of the gene recombines with the triacylglycerol lipase gene. The two portions of the gene are designed such that neither portion is functional unless they recombine with each other. These two events further reduce the likelihood that the gene can be expressed following random integration events.
[0128] In certain embodiments, the gene encodes a dominant selectable marker. Thus, knockout cells may be selected by screening for the marker. In some embodiments, the dominant selectable marker is a drug resistance marker. A drug resistance marker is a dominant selectable marker that, when expressed by a cell, allows the cell to grow and/or survive in the presence of a drug that would normally inhibit cellular growth and/or survival. Cells expressing a drug resistance marker can be selected by growing the cells in the presence of the drug. In some embodiments, the drug resistance marker is an antibiotic resistance marker. In some embodiments, the drug resistance marker confers resistance to a drug selected from the group consisting of Amphotericin B, Candicidin, Filipin, Hamycin, Natamycin, Nystatin, Rimocidin, Bifonazole, Butoconazole, Clotrimazole, Econazole, Fenticonazole, Isoconazole, Ketoconazole, Luliconazole, Miconazole, Omoconazole, Oxiconazole, Sertaconazole, Sulconazole, Tioconazole, Albaconazole, Fluconazole, Isavuconazole, Itraconazole, Posaconazole, Ravuconazole, Terconazole, Voriconazole, Abafungin, Amorolfin, Butenafine, Naftifine, Terbinafine, Anidulafungin, Caspofungin, Micafungin, Benzoic acid, Ciclopirox, Flucytosine, 5-fluorocytosine, Griseofulvin, Haloprogin, Polygodial, Tolnaftate, Crystal violet, Amikacin, Gentamicin, Kanamycin, Neomycin, Netilmicin, Tobramycin, Paromomycin, Spectinomycin, Geldanamycin, Herbimycin, Rifaximin, Streptomycin, Loracarbef, Ertapenem, Doripenem, Imipenem, Meropenem, Cefadroxil, Cefazolin, Cefalotin, Cefalexin, Cefaclor, Cefamandole, Cefoxitin, Cefprozil, Cefuroxime, Cefixime, Cefdinir, Cefditoren, Cefoperazone, Cefotaxime, Cefpodoxime, Ceftazidime, Ceftibuten, Ceftizoxime, Ceftriaxone, Cefepime, Ceftaroline fosamil, Ceftobiprole, Teicoplanin, Vancomycin, Telavancin, Clindamycin, Lincomycin, Daptomycin, Azithromycin, Clarithromycin, Dirithromycin, Erythromycin, Roxithromycin, Troleandomycin, Telithromycin, Spiramycin, Aztreonam, Furazolidone, Nitrofurantoin, Linezolid, Posizolid, Radezolid, Torezolid, Amoxicillin, Ampicillin, Azlocillin, Carbenicillin, Cloxacillin, Dicloxacillin, Flucloxacillin, Mezlocillin, Methicillin, Nafcillin, Oxacillin, Penicillin G, Penicillin V, Piperacillin, Penicillin G, Temocillin, Ticarcillin, clavulanate, sulbactam, tazobactam, clavulanate, Bacitracin, Colistin, Polymyxin B, Ciprofloxacin, Enoxacin, Gatifloxacin, Levofloxacin, Lomefloxacin, Moxifloxacin, Nalidixic acid, Norfloxacin, Ofloxacin, Trovafloxacin, Grepafloxacin, Sparfloxacin, Temafloxacin, Mafenide, Sulfacetamide, Sulfadiazine, Silver sulfadiazine, Sulfadimethoxine, Sulfamethizole, Sulfamethoxazole, Sulfanilimide, Sulfasalazine, Sulfisoxazole, Trimethoprim-Sulfamethoxazole, Co-trimoxazole, Sulfonamidochrysoidine, Demeclocycline, Doxycycline, Minocycline, Oxytetracycline, Tetracycline, Clofazimine, Dapsone, Capreomycin, Cycloserine, Ethambutol, Ethionamide, Isoniazid, Pyrazinamide, Rifampicin, Rifabutin, Rifapentine, Streptomycin, Arsphenamine, Chloramphenicol, Fosfomycin, Fusidic acid, Metronidazole, Mupirocin, Platensimycin, Quinupristin, Dalfopristin, Thiamphenicol, Tigecycline, Tinidazole, Trimethoprim, Geneticin, Nourseothricin, Hygromycin, Bleomycin, and Puromycin.
[0129] In some embodiments, the dominant selectable marker is a nutritional marker. A nutritional marker is a dominant selectable marker that, when expressed by the cell, enables the cell to grow or survive using one or more particular nutrient sources. Cells expressing a nutritional marker can be selected by growing the cells under limiting nutrient conditions in which cells expressing the nutritional marker can survive and/or grow, but cells lacking the nutrient marker cannot. In some embodiments, the nutritional marker is selected from the group consisting of Orotidine 5-phosphate decarboxylase, Phosphite specific oxidoreductase, Alpha-ketoglutarate-dependent hypophosphite dioxygenase, Alkaline phosphatase, Cyanamide hydratase, Melamine deaminase, Cyanurate amidohydrolase, Biuret hydrolyase, Urea amidolyase, Ammelide aminohydrolase, Guanine deaminase, Phosphodiesterase, Phosphotriesterase, Phosphite hydrogenase, Glycerophosphodiesterase, Parathion hydrolyase, Phosphite dehydrogenase, Dibenzothiophene desulfurization enzyme, Aromatic desulfinase, NADH-dependent FMN reductase, Aminopurine transporter, Hydroxylamine oxidoreductase, Invertase, Beta-glucosidase, Alpha-glucosidase, Beta-galactosidase, Alpha-galactosidase, Amylase, Cellulase, and Pullulonase.
[0130] Different approaches may be used to knockout the TGL3 gene in Y. lipolytica (See, e.g., Dulermo et al., Biochimica Biophysica Acta 1831:1486 (2013)). The methods disclosed herein and other methods known in the art may be used to knockout different triacylglycerol lipase genes in other species. For example, these methods may be used to knockout the TGL3 gene of Arxula adeninivorans (SEQ ID NO:78), the TGL3/4 gene of Arxula adeninivorans (SEQ ID NO:80), or the TGL4 gene of Arxula adeninivorans (SEQ ID NO:82). Similarly, these methods may be used to knockout the TGL4 gene of Y. lipolytica (SEQ ID NO:88).
[0131] In some embodiments, a genetic modification decreases the expression of a native triacylglycerol lipase gene by 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 99.1, 99.2, 99.3, 99.4, 99.5, 99.6, 99.7, 99.8, 99.9, or 100 percent.
[0132] In some embodiments, a genetic modification decreases the efficiency of a native triacylglycerol lipase gene by 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 99.1, 99.2, 99.3, 99.4, 99.5, 99.6, 99.7, 99.8, 99.9, or 100 percent.
[0133] In some embodiments, a genetic modification decreases the activity of a native triacylglycerol lipase gene by 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 99.1, 99.2, 99.3, 99.4, 99.5, 99.6, 99.7, 99.8, 99.9, or 100 percent.
[0134] F. Decreasing Triacylglycerol Lipase Activity in a Cell with Concomitant Overexpression of Diacylglycerol Acyltransferase
[0135] In some embodiments, the transformed oleaginous cell comprises a triacylglycerol lipase knockout mutation and a genetic modification that increase the expression of a native diacylglycerol acyltransferase. In certain embodiments, the transformed oleaginous cell comprises a triacylglycerol lipase knockout mutation and a genetic modification that encodes at least one copy of a diacylglycerol acyltransferase gene that is either native to the cell or from a different species of cell. In other embodiments, a TGL3 gene is disrupted and a DGA1 protein is overexpressed.
[0136] In some embodiments, one nucleic acid increases the expression of a native diacylglycerol acyltransferase or encodes at least one copy of a diacylglycerol acyltransferase gene and a second nucleic acid decreases the activity of a triacylglycerol lipase in the cell. In some embodiments the same nucleic acid encodes at least one copy of a diacylglycerol acyltransferase gene and decreases the activity of a triacylglycerol lipase in the cell. For example, the nucleic acid designed to knock out a triacylglycerol lipase gene may also contain a copy of a diacylglycerol acyltransferase gene.
[0137] G. Triacylglycerol Production
[0138] In certain embodiments, the transformed oleaginous cell are grown in the presence of exogenous fatty acids, glucose, ethanol, xylose, sucrose, starch, starch dextrin, glycerol, cellulose, and/or acetic acid. These substrates may be added during cultivation to increase lipid production. The exogenous fatty acids may include stearate, oleic acid, linoleic acid, .gamma.-linolenic acid, dihomo-.gamma.-linolenic acid, arachidonic acid, .alpha.-linolenic acid, stearidonic acid, eicosatetraenoic acid, eicosapenteaenoic acid, docosapentaenoic acid, eicosadienoic acid, and/or eicosatrienoic acid.
[0139] In certain embodiments, the present invention relates to a product produced by a modified host cell described herein. In certain embodiments, the product is an oil, lipid, or triacylglycerol. In some embodiments, the product is palmitic acid, palmitoleic acid, stearic acid, oleic acid, or linoleic acid. In certain embodiments, the product is a saturated fatty acid. Thus, the product may be caprylic acid, capric acid, lauric acid, myristic acid, palmitic acid, stearic acid, arachidic acid, behenic acid, lignoceric acid, or cerotic acid. In some embodiments, the product is an unsaturated fatty acid. Thus, the product may be myristoleic acid, palmitoleic acid, sapienic acid, oleic acid, elaidic acid, vaccenic acid, linoleic acid, linoelaidic acid, .alpha.-linolenic acid, arachidonic acid, eicosapenteaenoic acid, erucic acid, or docosahexaenoic acid.
[0140] In some embodiments, the product comprises an 18-carbon fatty acid. In some embodiments, the product comprises oleic acid, stearic acid, or linoleic acid. For example, the product may be oleic acid.
[0141] In some embodiments, the present invention relates to a transformed oleaginous cell comprising a first genetic modification and a second genetic modification. The first genetic modification may increase the activity of a native diacylglycerol acyltransferase. Alternatively, the first genetic modification may encode at least one copy of a diacylglycerol acyltransferase gene native to the oleaginous cell or from a different species. The second genetic modification may decrease the activity of a triacylglycerol lipase in the oleaginous cell.
[0142] In some embodiments, the second genetic modification is a triacylglycerol lipase knockout mutation.
[0143] In some embodiments, the first genetic modification encodes at least one copy of a diacylglycerol acyltransferase gene native to the oleaginous cell or from a different species. In some embodiments, a diacylglycerol acyltransferase gene is integrated into the genome of the cell.
[0144] In some embodiments, the transformed oleaginous cell is selected from the group consisting of algae, bacteria, molds, fungi, plants, and yeasts. In certain embodiments, the transformed oleaginous cell is a yeast. The transformed oleaginous cell may be selected from the group consisting of Arxula, Aspergillus, Aurantiochytrium, Candida, Claviceps, Cryptococcus, Cunninghamella, Geotrichum, Hansenula, Kluyveromyces, Kodamaea, Leucosporidiella, Lipomyces, Mortierella, Ogataea, Pichia, Prototheca, Rhizopus, Rhodosporidium, Rhodotorula, Saccharomyces, Schizosaccharomyces, Tremella, Trichosporon, Wickerhamomyces, and Yarrowia. In certain embodiments, the transformed oleaginous cell is selected from the group consisting of Arxula adeninivorans, Aspergillus niger, Aspergillus orzyae, Aspergillus terreus, Aurantiochytrium limacinum, Candida utilis, Claviceps purpurea, Cryptococcus albidus, Cryptococcus curvatus, Cryptococcus ramirezgomezianus, Cryptococcus terreus, Cryptococcus wieringae, Cunninghamella echinulata, Cunninghamella japonica, Geotrichum fermentans, Hansenula polymorpha, Kluyveromyces lactis, Kluyveromyces marxianus, Kodamaea ohmeri, Leucosporidiella creatinivora, Lipomyces lipofer, Lipomyces starkeyi, Lipomyces tetrasporus, Mortierella isabellina, Mortierella alpina, Ogataea polymorpha, Pichia ciferrii, Pichia guilliermondii, Pichia pastoris, Pichia stipites, Prototheca zopfii, Rhizopus arrhizus, Rhodosporidium babjevae, Rhodosporidium toruloides, Rhodosporidium paludigenum, Rhodotorula glutinis, Rhodotorula mucilaginosa, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Tremella enchepala, Trichosporon cutaneum, Trichosporon fermentans, Wickerhamomyces ciferrii, and Yarrowia lipolytica. In other embodiments, the transformed oleaginous cell is Yarrowia lipolytica or Arxula adeninivorans.
[0145] In some embodiments, the diacylglycerol acyltransferase gene is a type II diacylglycerol acyltransferase gene. The diacylglycerol acyltransferase gene may be a type II diacylglycerol acyltransferase gene from Arxula adeninivorans, Aspergillus terreus, Aurantiochytrium limacinum, Claviceps purpurea, Gloeophyllum trabeum, Lipomyces starkeyi, Microbotryum violaceum, Pichia guilliermondii, Phaeodactylum tricornutum, Puccinia graminis, Rhodosporidium diobovatum, Rhodosporidium toruloides, Rhodotorula graminis, or Yarrowia lipolytica.
[0146] In some embodiments, the diacylglycerol acyltransferase gene is a type I diacylglycerol acyltransferase gene. The diacylglycerol acyltransferase gene may be a type I diacylglycerol acyltransferase gene from Arxula adeninivorans, Aspergillus terreus, Chaetomium globosum, Claviceps purpurea, Lipomyces starkeyi, Metarhizium acridum, Ophiocordyceps sinensis, Phaeodactylum tricomuturn, Pichia guilliermondii, Rhodosporidium toruloides, Rhodotorula graminis, Trichoderma virens, or Yarrowia lipolytica.
[0147] In some embodiments, the diacylglycerol acyltransferase gene is a type III diacylglycerol acyltransferase gene. The diacylglycerol acyltransferase gene may be a type III diacylglycerol acyltransferase gene from Ricinus communis or Arachis hypogaea.
[0148] In some embodiments, the triacylglycerol lipase is TGL3, TGL3/4, or TGL4.
[0149] In some embodiments, the present invention relates to a product derived from a transformed oleaginous cell. In certain embodiments, the product is an oil, lipid, or triacylglycerol.
[0150] In some embodiments, the present invention relates to a method of increasing the lipid content of a cell, comprising transforming a parent cell with a first nucleic acid and a second nucleic acid. The first nucleic acid may increase the activity of a native diacylglycerol acyltransferase or comprise a diacylglycerol acyltransferase gene. The second nucleic acid may decrease the activity of a triacylglycerol lipase. The first nucleic acid and second nucleic acid may be the same. Thus, in some embodiments, the present invention relates to a method of increasing the lipid content of a cell comprising transforming a parent cell with a nucleic acid, wherein the nucleic acid decreases the activity of a triacylglycerol lipase in the cell and either comprises a diacylglycerol acyltransferase gene or increases the activity of a native diacylglycerol acyltransferase.
[0151] In some embodiments, the present invention relates to a method of increasing the triacylglycerol content of a cell, comprising providing a cell and growing the cell. The cell may comprise a first genetic modification and a second genetic modification. The first genetic modification may increase the activity of a native diacylglycerol acyltransferase or encode at least one copy of a diacylglycerol acyltransferase gene either native to the oleaginous cell or from a different species. The second genetic modification may decrease the activity of a triacylglycerol lipase in the cell. The cell may be grown under conditions whereby the first genetic modification is expressed, thereby producing a triacylglycerol. In some embodiments, the triacylglycerol is recovered.
[0152] In some embodiments, the first nucleic acid comprises a diacylglycerol acyltransferase gene. The diacylglycerol acyltransferase gene may encode a type I diacylglycerol acyltransferase polypeptide, a type II diacylglycerol acyltransferase polypeptide, or a type III diacylglycerol acyltransferase polypeptide.
[0153] In some embodiments, the diacylglycerol acyltransferase gene comprises a nucleotide sequence set forth in SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8, SEQ ID NO: 10, SEQ ID NO: 12, SEQ ID NO: 14, SEQ ID NO: 16, SEQ ID NO: 18, SEQ ID NO: 42, SEQ ID NO: 44, SEQ ID NO: 46, SEQ ID NO: 48, SEQ ID NO: 50, SEQ ID NO: 52, SEQ ID NO: 54, SEQ ID NO: 56, SEQ ID NO: 58, SEQ ID NO: 60, SEQ ID NO: 62, SEQ ID NO: 30, SEQ ID NO: 32, SEQ ID NO: 34, SEQ ID NO: 36, SEQ ID NO: 38, SEQ ID NO: 40, SEQ ID NO: 64, SEQ ID NO: 66, SEQ ID NO: 68, SEQ ID NO: 70, SEQ ID NO: 72, SEQ ID NO: 74, SEQ ID NO: 76, SEQ ID NO: 84, or SEQ ID NO: 86, or a complement of any one of them.
[0154] In some embodiments, the diacylglycerol acyltransferase gene comprises a nucleotide sequence having at least 70%, 75%, 80%, 85%, 90%, 95%, 96%, 97%, 98%, or 99% sequence identity to a nucleotide sequence set forth in SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8, SEQ ID NO: 10, SEQ ID NO: 12, SEQ ID NO: 14, SEQ ID NO: 16, SEQ ID NO: 18, SEQ ID NO: 42, SEQ ID NO: 44, SEQ ID NO: 46, SEQ ID NO: 48, SEQ ID NO: 50, SEQ ID NO: 52, SEQ ID NO: 54, SEQ ID NO: 56, SEQ ID NO: 58, SEQ ID NO: 60, SEQ ID NO: 62, SEQ ID NO: 30, SEQ ID NO: 32, SEQ ID NO: 34, SEQ ID NO: 36, SEQ ID NO: 38, SEQ ID NO: 40, SEQ ID NO: 64, SEQ ID NO: 66, SEQ ID NO: 68, SEQ ID NO: 70, SEQ ID NO: 72, SEQ ID NO: 74, SEQ ID NO: 76, SEQ ID NO: 84, or SEQ ID NO: 86, or a complement of any one of them.
[0155] In some embodiments, the diacylglycerol acyltransferase polypeptide comprises an amino acid sequence set forth in SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 7, SEQ ID NO: 9, SEQ ID NO: 11, SEQ ID NO: 13, SEQ ID NO: 15, SEQ ID NO: 17, SEQ ID NO: 41, SEQ ID NO: 43, SEQ ID NO: 45, SEQ ID NO: 47, SEQ ID NO: 49, SEQ ID NO: 51, SEQ ID NO: 53, SEQ ID NO: 55, SEQ ID NO: 57, SEQ ID NO: 59, SEQ ID NO: 61; SEQ ID NO: 29, SEQ ID NO: 31, SEQ ID NO: 33, SEQ ID NO: 35, SEQ ID NO: 37, SEQ ID NO: 39, SEQ ID NO: 63, SEQ ID NO: 65, SEQ ID NO: 67, SEQ ID NO: 69, SEQ ID NO: 71, SEQ ID NO: 73, SEQ ID NO: 75, SEQ ID NO: 83, or SEQ ID NO: 85, or a biologically-active portion of any one of them.
[0156] In some embodiments, the diacylglycerol acyltransferase polypeptide comprises an amino acid sequence having at least 80%, 85%, 90%, 95%, 96%, 97%, 98%, or 99% sequence identity to an amino acid sequence forth in SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 7, SEQ ID NO: 9, SEQ ID NO: 11, SEQ ID NO: 13, SEQ ID NO: 15, SEQ ID NO: 17, SEQ ID NO: 41, SEQ ID NO: 43, SEQ ID NO: 45, SEQ ID NO: 47, SEQ ID NO: 49, SEQ ID NO: 51, SEQ ID NO: 53, SEQ ID NO: 55, SEQ ID NO: 57, SEQ ID NO: 59, SEQ ID NO: 61; SEQ ID NO: 29, SEQ ID NO: 31, SEQ ID NO: 33, SEQ ID NO: 35, SEQ ID NO: 37, SEQ ID NO: 39, SEQ ID NO: 63, SEQ ID NO: 65, SEQ ID NO: 67, SEQ ID NO: 69, SEQ ID NO: 71, SEQ ID NO: 73, SEQ ID NO: 75, SEQ ID NO: 83, or SEQ ID NO: 85, or a biologically-active portion of any one of them.
[0157] In some embodiments, the second nucleic acid is capable of recombining with a nucleic acid sequence in a triacylglycerol lipase gene and/or a nucleic acid sequence in the regulatory region of a triacylglycerol lipase gene. In certain embodiments, the second nucleic acid is capable of recombining with a nucleic acid sequence in a TGL3, TGL3/4, or TGL4 gene and/or a nucleic acid sequence in the regulatory region of a TGL3, TGL3/4, or TGL4 gene. The second nucleic acid may comprise a gene encoding a protein or a portion of a protein. In certain embodiments, the second nucleic acid comprises a gene encoding a protein that confers resistance to a drug. In other embodiments, the second nucleic acid comprises a gene encoding a protein that enables the cell to grow or proliferate more quickly on one or more particular nutrient sources than a native organism of the same species.
[0158] The present description is further illustrated by the following examples, which should not be construed as limiting in any way. The contents of all cited references (including literature references, issued patents, published patent applications and GenBank Accession numbers as cited throughout this application) are hereby expressly incorporated by reference. When definitions of terms in documents that are incorporated by reference herein conflict with those used herein, the definitions used herein govern.
EXEMPLIFICATION
Example 1: Method to Overexpress DGA1 in Y. lipolytica
[0159] Nucleic acid constructs for overexpressing the DGA1 gene were described in U.S. Ser. No. 61/943,664 (hereby incorporated by reference). FIG. 1 shows expression construct pNC243 used for overexpression of the R. toruloides DGA1 gene NG66 (SEQ ID No. 6) in Y. lipolytica. DGA1 expression constructs were linearized before transformation by a PacI/NotI restriction digest. The linear expression constructs each included an expression cassette for the DGA1 gene and for the Nat1 gene, used as a marker for selection with nourseothricin (NAT).
[0160] DGA1 expression constructs were randomly integrated into the genome of E lipolytica strain NS18 (obtained from ARS Culture Collection, NRRL # YB 392) using a transformation protocol as described in Chen (Applied Microbiology & Biotechnology 48:232-35 (1997)). Transformants were selected on YPD plates with 500 .mu.g/mL NAT and screened for the ability to accumulate lipids by a fluorescent staining lipid assay as described below. For each expression construct, eight transformants were analysed.
[0161] For most constructs, there was significant colony variation between the transformants, likely due to the lack of a functional DGA1 expression cassette in cells that only obtained a functional Nat1 cassette, or due to a negative effect of the site of DGA1 integration on DGA1 expression. Nevertheless, all transformants had a significant increase in lipid content.
[0162] Overexpression of native Y. lipolytica DGA1 (NG15) under a strong promoter increased lipid content as measured by cell fluorescence by about 2-fold compared to the parental strain NS18. Transformants that demonstrated the highest fluorescence (about 3-fold higher compared to NS18) were generated by the overexpression of R. toruloides DGA1 (NG66, NG67) and L. starkeyi DGA1 (NG68).
[0163] In certain experiments, the effect of native R. toruloides DGA1 (NG49) overexpression on lipid production in Y. lipolytica was not as high as the effect of synthetic versions of R. toruloides DGA1 genes that did not contain introns. This result may indicate that the gene splicing of the R. toruloides DGA1 gene in Y. lipolytica was not very efficient. In certain experiments, codon optimization of the R. toruloides DGA1 gene for expression in Y. lipolytica did not have a positive effect on lipid production.
[0164] In order to select strains with the highest lipid production level, Y. lipolytica strain NS18 transformants expressing NG15 (Y. lipolytica DGA1) or NG66 (R. toruloides DGA1) were screened. For NG15, about 50 colonies were screened by lipid assay for the highest lipid accumulation, and the best transformant was named NS249. For NG66, 80 colonies were screened and the 8 best colonies were selected for further analysis. Strain NS249 and the 8 selected NG66 transformants were grown in shake flasks and analysed by the lipid assay for lipid content and by HPLC for glucose consumption. Y. lipolytica strains overexpressing R. toruloides DGA1 have significantly higher lipid content than Y. lipolytica strains with native Y. lipolytica DGA1 gene expressed under the same promoter as R. toruloides DGA1. At the same time, NG66 transformants had significantly less glucose left in the media, demonstrating that NG66 was more efficient in converting glucose to lipids. The difference in efficiency between two DGA1 genes may be attributed to either a higher level of expression of R. toruloides DGA1 in Y. lipolytica or a higher level of R. toruloides DGA1 specific activity, or both. One of the best performing transformants with an integrated NG66 gene was named NS281.
Example 2: Method to Knockout Triacylglycerol Lipase Knockout Gene in Y. lipolytica
[0165] In order to test the idea that combining DGA1 overexpression with a TGL3 deletion leads to higher lipid accumulation in Y. lipolytica, we deleted TGL3 in Y. lipolytica wild-type strain NS18 (obtained from NRLL # YB-392) and its DGA1 overexpressing derivative NS281. NS281 overexpresses the DGA1 gene from Rhodosporidium toruloides as described above. The Y. lipolytica TGL3 gene (YALI0D17534g, SEQ ID NO: 20) was deleted as follows: A two-fragment deletion cassette was amplified by PCR from a plasmid containing the hygromycin resistance gene ("hph," SEQ ID NO: 22) using primer pairs NP1798-NP656 and NP655-NP1799 (SEQ ID NOs: 23-26). The resulting PCR fragments (SEQ ID NOs: 27 & 28) were co-transformed into NS18 and NS281 according to the protocol developed in U.S. Ser. No. 61/819,746 (hereby incorporated by reference). The omission of a promoter and terminator in the hph cassette and the splitting of the hph coding sequence into two PCR fragments reduce the probability that random integration of these pieces will confer hygromycin resistance. The hph gene should only be expressed if it integrates at the TGL3 locus by homologous recombination so that the TGL3 promoter and terminator can direct its transcription. Hygromycin resistant colonies were screened by PCR to confirm the absence of TGL3 and the presence of a tgl3::hyg specific product. Deletion of TGL3 in NS18 resulted in strain NS421. Deletion of TGL3 in NS281 resulted in strain NS377.
Example 3: TGL3 Knockouts that Overexpress DGA1 Accumulate More Lipid than TGL3 Knockouts Alone
[0166] NS421, which contains the TGL3 knockout, and NS377, which contains the TGL3 deletion and also overexpresses R. toruloides DGA1, were grown in a 24-well plate in limiting nitrogen conditions in order to promote lipid accumulation.
[0167] Specifically, each well of an autoclaved, 24-well plate was filled with 1.5 mL of filter-sterilized media containing 0.5 g/L urea, 1.5 g/l yeast extract, 0.85 g/L casamino acids, 1.7 g/L YNB (without amino acids and ammonium sulfate), 100 g/L glucose, and 5.11 g/L potassium hydrogen phthalate (25 mM). Yeast strains that had been incubated for 1-2 days on YPD-agar plates at 30.degree. C. were used to inoculate each well. The 24-well plate was covered with a porous cover and incubated at 30.degree. C., 70-90% humidity, and 900 rpm in an Infors Multitron ATR shaker. After 96 hours, 20 .mu.L of 100% ethanol was added to 20 .mu.L of cells in an analytical microplate and incubated at 4.degree. C. for 30 minutes. 20 .mu.L of cell/ethanol mix was then added to 80 .mu.l of a pre-mixed solution containing 50 .mu.L 1 M potassium iodide, 1 mM .mu.L Bodipy 493/503, 0.5 .mu.L 100% DMSO, 1.5 .mu.L 60% PEG 4000, and 27 .mu.L water in a Costar 96-well, black, clear-bottom plate and covered with a transparent seal. Bodipy fluorescence was monitored with a SpectraMax M2 spectrophotometer (Molecular Devices) kinetic assay at 30.degree. C., and normalized by dividing fluorescence by absorbance at 600 nm. Data was averaged in triplicate growth experiments.
[0168] NS377 accumulated approximately double the amount of lipid as NS421 showing that deletion of TGL3 in combination with DGA1 overexpression increases a cell's lipid content relative to a TGL3 deletion alone (FIG. 2).
Example 4: Cells that Overexpress DGA1 and Contain a TGL3 Deletion Accumulate More TAGs than Cells that Overexpress DGA1 but have No TGL3 Modification
[0169] NS281, which overexpresses R. toruloides DGA1, and NS377, which contains the TGL3 deletion and also overexpresses R. toruloides DGA1, were grown in 1-liter bioreactors using a high cell density fed-batch glucose process. After 118 hours, the cells were analyzed by gas chromatography to measure lipid content as a percentage of dry cell weight. Triplicate chromatography measurements were averaged for the same sample. NS377 reached approximately 4% higher lipid content than NS281, showing that deletion of TGL3 in combination with DGA1 overexpression yields better results than DGA1 overexpression alone (FIG. 3).
Example 5: Cells Comprising Genetic Modifications that Encode a Diacylglycerol Acyltransferase and Decrease the Activity of a Triacylglycerol Lipase Accumulate More TAGs
[0170] In order to test the idea that combining DGA1 and DGA2 overexpression with TGL3 deletion leads to higher lipid accumulation in Y. lipolytica, DGA2 from Claviceps purpurea was overexpressed in strain NS377. Strain NS377 contains a deletion of TGL3 and overexpresses DGA1 from Rhodosporidium toruloides as described in Example 4. DGA2 from Claviceps purpurea was selected based on prior experiments that demonstrate that this gene increases the lipid content of Y. lipolytica in combination with DGA1 from Rhodosporidium toruloides (U.S. Ser. No. 62/004,502, incorporated by reference).
[0171] FIG. 4 shows the map of pNC327, the expression construct used to overexpress C. purpurea DGA2 in NS377. The construct was linearized prior to transformation with a PacI/AscI restriction digest. The linear expression construct included an expression cassette for the C. purpurea DGA2 gene and for the BLE gene used as a marker for selection with Zeocin (ZEO). Transformants were analyzed by the fluorescent lipid assay, and the top lipid producer was designated NS432.
[0172] The lipid production of strains NS297, NS281, NS450, NS377, and NS432 were compared. A subset of these strains were either grown using a batch glucose process (in 48-well plates or 50-mL flasks) or using a high cell density fed-batch glucose process (in 1-L bioreactors). Lipid content was analyzed by fluorescence assay or gas chromatography, and strain NS432 was found to have a higher lipid content than its parent strain NS377 and the strains without the TGL3 knockout (FIG. 5). These results demonstrate the advantage of DGA1 and DGA2 overexpression in a TGL3 knockout.
INCORPORATION BY REFERENCE
[0173] All of the U.S. patents, U.S. patent application publications, foreign patents, foreign patent publications, and non-patent literature cited herein are hereby incorporated by reference.
EQUIVALENTS
[0174] Those skilled in the art will recognize, or be able to ascertain using no more than routine experimentation, many equivalents to the specific embodiments of the invention described herein. Such equivalents are intended to be encompassed by the following claims.
Sequence CWU 1
1
881514PRTYarrowia lipolytica 1Met Thr Ile Asp Ser Gln Tyr Tyr Lys
Ser Arg Asp Lys Asn Asp Thr1 5 10 15Ala Pro Lys Ile Ala Gly Ile Arg
Tyr Ala Pro Leu Ser Thr Pro Leu 20 25 30Leu Asn Arg Cys Glu Thr Phe
Ser Leu Val Trp His Ile Phe Ser Ile 35 40 45Pro Thr Phe Leu Thr Ile
Phe Met Leu Cys Cys Ala Ile Pro Leu Leu 50 55 60Trp Pro Phe Val Ile
Ala Tyr Val Val Tyr Ala Val Lys Asp Asp Ser65 70 75 80Pro Ser Asn
Gly Gly Val Val Lys Arg Tyr Ser Pro Ile Ser Arg Asn 85 90 95Phe Phe
Ile Trp Lys Leu Phe Gly Arg Tyr Phe Pro Ile Thr Leu His 100 105
110Lys Thr Val Asp Leu Glu Pro Thr His Thr Tyr Tyr Pro Leu Asp Val
115 120 125Gln Glu Tyr His Leu Ile Ala Glu Arg Tyr Trp Pro Gln Asn
Lys Tyr 130 135 140Leu Arg Ala Ile Ile Thr Thr Ile Glu Tyr Phe Leu
Pro Ala Phe Met145 150 155 160Lys Arg Ser Leu Ser Ile Asn Glu Gln
Glu Gln Pro Ala Glu Arg Asp 165 170 175Pro Leu Leu Ser Pro Val Ser
Pro Ser Ser Pro Gly Ser Gln Pro Asp 180 185 190Lys Trp Ile Asn His
Asp Ser Arg Tyr Ser Arg Gly Glu Ser Ser Gly 195 200 205Ser Asn Gly
His Ala Ser Gly Ser Glu Leu Asn Gly Asn Gly Asn Asn 210 215 220Gly
Thr Thr Asn Arg Arg Pro Leu Ser Ser Ala Ser Ala Gly Ser Thr225 230
235 240Ala Ser Asp Ser Thr Leu Leu Asn Gly Ser Leu Asn Ser Tyr Ala
Asn 245 250 255Gln Ile Ile Gly Glu Asn Asp Pro Gln Leu Ser Pro Thr
Lys Leu Lys 260 265 270Pro Thr Gly Arg Lys Tyr Ile Phe Gly Tyr His
Pro His Gly Ile Ile 275 280 285Gly Met Gly Ala Phe Gly Gly Ile Ala
Thr Glu Gly Ala Gly Trp Ser 290 295 300Lys Leu Phe Pro Gly Ile Pro
Val Ser Leu Met Thr Leu Thr Asn Asn305 310 315 320Phe Arg Val Pro
Leu Tyr Arg Glu Tyr Leu Met Ser Leu Gly Val Ala 325 330 335Ser Val
Ser Lys Lys Ser Cys Lys Ala Leu Leu Lys Arg Asn Gln Ser 340 345
350Ile Cys Ile Val Val Gly Gly Ala Gln Glu Ser Leu Leu Ala Arg Pro
355 360 365Gly Val Met Asp Leu Val Leu Leu Lys Arg Lys Gly Phe Val
Arg Leu 370 375 380Gly Met Glu Val Gly Asn Val Ala Leu Val Pro Ile
Met Ala Phe Gly385 390 395 400Glu Asn Asp Leu Tyr Asp Gln Val Ser
Asn Asp Lys Ser Ser Lys Leu 405 410 415Tyr Arg Phe Gln Gln Phe Val
Lys Asn Phe Leu Gly Phe Thr Leu Pro 420 425 430Leu Met His Ala Arg
Gly Val Phe Asn Tyr Asp Val Gly Leu Val Pro 435 440 445Tyr Arg Arg
Pro Val Asn Ile Val Val Gly Ser Pro Ile Asp Leu Pro 450 455 460Tyr
Leu Pro His Pro Thr Asp Glu Glu Val Ser Glu Tyr His Asp Arg465 470
475 480Tyr Ile Ala Glu Leu Gln Arg Ile Tyr Asn Glu His Lys Asp Glu
Tyr 485 490 495Phe Ile Asp Trp Thr Glu Glu Gly Lys Gly Ala Pro Glu
Phe Arg Met 500 505 510Ile Glu21545DNAYarrowia lipolytica
2atgactatcg actcacaata ctacaagtcg cgagacaaaa acgacacggc acccaaaatc
60gcgggaatcc gatatgcccc gctatcgaca ccattactca accgatgtga gaccttctct
120ctggtctggc acattttcag cattcccact ttcctcacaa ttttcatgct
atgctgcgca 180attccactgc tctggccatt tgtgattgcg tatgtagtgt
acgctgttaa agacgactcc 240ccgtccaacg gaggagtggt caagcgatac
tcgcctattt caagaaactt cttcatctgg 300aagctctttg gccgctactt
ccccataact ctgcacaaga cggtggatct ggagcccacg 360cacacatact
accctctgga cgtccaggag tatcacctga ttgctgagag atactggccg
420cagaacaagt acctccgagc aatcatcacc accatcgagt actttctgcc
cgccttcatg 480aaacggtctc tttctatcaa cgagcaggag cagcctgccg
agcgagatcc tctcctgtct 540cccgtttctc ccagctctcc gggttctcaa
cctgacaagt ggattaacca cgacagcaga 600tatagccgtg gagaatcatc
tggctccaac ggccacgcct cgggctccga acttaacggc 660aacggcaaca
atggcaccac taaccgacga cctttgtcgt ccgcctctgc tggctccact
720gcatctgatt ccacgcttct taacgggtcc ctcaactcct acgccaacca
gatcattggc 780gaaaacgacc cacagctgtc gcccacaaaa ctcaagccca
ctggcagaaa atacatcttc 840ggctaccacc cccacggcat tatcggcatg
ggagcctttg gtggaattgc caccgaggga 900gctggatggt ccaagctctt
tccgggcatc cctgtttctc ttatgactct caccaacaac 960ttccgagtgc
ctctctacag agagtacctc atgagtctgg gagtcgcttc tgtctccaag
1020aagtcctgca aggccctcct caagcgaaac cagtctatct gcattgtcgt
tggtggagca 1080caggaaagtc ttctggccag acccggtgtc atggacctgg
tgctactcaa gcgaaagggt 1140tttgttcgac ttggtatgga ggtcggaaat
gtcgcccttg ttcccatcat ggcctttggt 1200gagaacgacc tctatgacca
ggttagcaac gacaagtcgt ccaagctgta ccgattccag 1260cagtttgtca
agaacttcct tggattcacc cttcctttga tgcatgcccg aggcgtcttc
1320aactacgatg tcggtcttgt cccctacagg cgacccgtca acattgtggt
tggttccccc 1380attgacttgc cttatctccc acaccccacc gacgaagaag
tgtccgaata ccacgaccga 1440tacatcgccg agctgcagcg aatctacaac
gagcacaagg atgaatattt catcgattgg 1500accgaggagg gcaaaggagc
cccagagttc cgaatgattg agtaa 15453348PRTRhodosporidium toruloides
3Met Gly Gln Gln Ala Thr Pro Glu Glu Leu Tyr Thr Arg Ser Glu Ile1 5
10 15Ser Lys Ile Lys Phe Ala Pro Phe Gly Val Pro Arg Ser Arg Arg
Leu 20 25 30Gln Thr Phe Ser Val Phe Ala Trp Thr Thr Ala Leu Pro Ile
Leu Leu 35 40 45Gly Val Phe Phe Leu Leu Cys Ser Phe Pro Pro Leu Trp
Pro Ala Val 50 55 60Ile Ala Tyr Leu Thr Trp Val Phe Phe Ile Asp Gln
Ala Pro Ile His65 70 75 80Gly Gly Arg Ala Gln Ser Trp Leu Arg Lys
Ser Arg Ile Trp Val Trp 85 90 95Phe Ala Gly Tyr Tyr Pro Val Ser Leu
Ile Lys Ser Ala Asp Leu Pro 100 105 110Pro Asp Arg Lys Tyr Val Phe
Gly Tyr His Pro His Gly Val Ile Gly 115 120 125Met Gly Ala Ile Ala
Asn Phe Ala Thr Asp Ala Thr Gly Phe Ser Thr 130 135 140Leu Phe Pro
Gly Leu Asn Pro His Leu Leu Thr Leu Gln Ser Asn Phe145 150 155
160Lys Leu Pro Leu Tyr Arg Glu Leu Leu Leu Ala Leu Gly Ile Cys Ser
165 170 175Val Ser Met Lys Ser Cys Gln Asn Ile Leu Arg Gln Gly Pro
Gly Ser 180 185 190Ala Leu Thr Ile Val Val Gly Gly Ala Ala Glu Ser
Leu Ser Ala His 195 200 205Pro Gly Thr Ala Asp Leu Thr Leu Lys Arg
Arg Lys Gly Phe Ile Lys 210 215 220Leu Ala Ile Arg Gln Gly Ala Asp
Leu Val Pro Val Phe Ser Phe Gly225 230 235 240Glu Asn Asp Ile Phe
Gly Gln Leu Arg Asn Glu Arg Gly Thr Arg Leu 245 250 255Tyr Lys Leu
Gln Lys Arg Phe Gln Gly Val Phe Gly Phe Thr Leu Pro 260 265 270Leu
Phe Tyr Gly Arg Gly Leu Phe Asn Tyr Asn Val Gly Leu Met Pro 275 280
285Tyr Arg His Pro Ile Val Ser Val Val Gly Arg Pro Ile Ser Val Glu
290 295 300Gln Lys Asp His Pro Thr Thr Ala Asp Leu Glu Glu Val Gln
Ala Arg305 310 315 320Tyr Ile Ala Glu Leu Lys Arg Ile Trp Glu Glu
Tyr Lys Asp Ala Tyr 325 330 335Ala Lys Ser Arg Thr Arg Glu Leu Asn
Ile Ile Ala 340 34541858DNARhodosporidium toruloides 4atgggccagc
aggcgacgcc cgaggagcta tacacacgct cagagatctc caagatcaag 60caagtcgagc
cagctcttct cctcaccacc ccacaacata ccccgcagcc cacgacagcc
120ctcccacagc acctgcagcc tgctgaccag ctcgagaaca cccacagatt
cgcacccttt 180ggcgtcccgc ggtcgcgccg gctgcagacc ttctccgtct
ttgcctggac gacggcactg 240cccatcctac tcggcgtctt cttcctcctc
tggtgcgtca ggcttggcgt gatctgagag 300tagcgggcgg atcatctgac
ctgcttcttc gctgcagctc gttcccaccg ctctggccgg 360ctgtcattgc
ctacctcacc tgggtctttt tcattgacca ggcgccgatt cacggtggac
420gggcgcagtc ttggctgcgg aagagtcgga tatgggtctg gtttgcagga
tactatcccg 480tcaggtgcgt cctctttcca agcctgcgtc tcgaggcctc
gctcacggcc aactcgcccg 540accggctacc tccgaacttt ccgtcaacag
cttgatcaag gtcagtctgc gcgtctctcg 600acttcagtgc tctgtggagg
agctgcgcca ttgggcccga cctgcggagg gcctcaaagg 660acgatgccgc
tgacttcctt tcctccgaca gagcgccgac ttgccgcctg accggaagta
720cgtctttggc taccacccgc acggcgtcat aggcatgggc gccatcgcca
acttcgcgac 780cgacgcaacc ggcttctcga cactcttccc cggcttgaac
cctcacctcc tcaccctcca 840aagcaacttc aagctcccgc tctaccgcga
gttgctgctc gctctcggca tatgctccgt 900ctcgatgaag agctgtcaga
acattctgcg acaaggtgag cggtatgcgc aagacgggcg 960gtcaagcgtg
aacgcagtga acgagaagag ctgaccttcc gccttactcc atccgtgcag
1020gtcctggctc ggctctcact atcgtcgtcg gtggcgccgc cgagagcttg
agtgcgcatc 1080ccggaaccgc cgatcttacg ctcaagcgac gaaaaggctt
catcaaactc gcgatccggc 1140aaggcgccga ccttgtgccc gtcttttcgt
tcggcgagaa cgacgtgcgc acgctctccg 1200agtctctaaa ccggaagcga
atgctgaccg ctgcccaatt ctctctccag atctttggcc 1260agctgcgaaa
cgagcgagga acgcggctgt acaagttgca gaagcgtttc caaggcgtgt
1320ttggcttcac cctccgtacg tctcaccgcg ccgtcttgcc gaactgctcg
ttcagtcgct 1380cacgcagctt tcactcgcgc agctctcttc tacggccggg
gactcttcaa ctgtgcgctc 1440gagttcaccg cttcgccaac agcgaggaat
gcctccgagt acagcccagc tgacgcccca 1500tctcttctca tagacaacgt
cggattgatg ccgtatcgcc atccgatcgt ctctgtcggt 1560gtgaacccgc
tctgtcgctc ctacctgcgt tccttaggct gacaccactc gcgtcaaaca
1620gtcggtcgac caatctcggt agagcagaag gaccacccga ccacggcgga
cctcgaagaa 1680gttcaggcgc ggtatatcgc agaactcaag cggtacgttc
caagtcgtct gcctccgctt 1740gccgcctcaa ataagctgag gcgtgctgac
cgtatctgcc gaaccgtaca gcatctggga 1800agaatacaag gacgcctacg
ccaaaagtcg cacgcgggag ctcaatatta tcgcctga 18585348PRTRhodosporidium
toruloides 5Met Gly Gln Gln Ala Thr Pro Glu Glu Leu Tyr Thr Arg Ser
Glu Ile1 5 10 15Ser Lys Ile Lys Phe Ala Pro Phe Gly Val Pro Arg Ser
Arg Arg Leu 20 25 30Gln Thr Phe Ser Val Phe Ala Trp Thr Thr Ala Leu
Pro Ile Leu Leu 35 40 45Gly Val Phe Phe Leu Leu Cys Ser Phe Pro Pro
Leu Trp Pro Ala Val 50 55 60Ile Ala Tyr Leu Thr Trp Val Phe Phe Ile
Asp Gln Ala Pro Ile His65 70 75 80Gly Gly Arg Ala Gln Ser Trp Leu
Arg Lys Ser Arg Ile Trp Val Trp 85 90 95Phe Ala Gly Tyr Tyr Pro Val
Ser Leu Ile Lys Ser Ala Asp Leu Pro 100 105 110Pro Asp Arg Lys Tyr
Val Phe Gly Tyr His Pro His Gly Val Ile Gly 115 120 125Met Gly Ala
Ile Ala Asn Phe Ala Thr Asp Ala Thr Gly Phe Ser Thr 130 135 140Leu
Phe Pro Gly Leu Asn Pro His Leu Leu Thr Leu Gln Ser Asn Phe145 150
155 160Lys Leu Pro Leu Tyr Arg Glu Leu Leu Leu Ala Leu Gly Ile Cys
Ser 165 170 175Val Ser Met Lys Ser Cys Gln Asn Ile Leu Arg Gln Gly
Pro Gly Ser 180 185 190Ala Leu Thr Ile Val Val Gly Gly Ala Ala Glu
Ser Leu Ser Ala His 195 200 205Pro Gly Thr Ala Asp Leu Thr Leu Lys
Arg Arg Lys Gly Phe Ile Lys 210 215 220Leu Ala Ile Arg Gln Gly Ala
Asp Leu Val Pro Val Phe Ser Phe Gly225 230 235 240Glu Asn Asp Ile
Phe Gly Gln Leu Arg Asn Glu Arg Gly Thr Arg Leu 245 250 255Tyr Lys
Leu Gln Lys Arg Phe Gln Gly Val Phe Gly Phe Thr Leu Pro 260 265
270Leu Phe Tyr Gly Arg Gly Leu Phe Asn Tyr Asn Val Gly Leu Met Pro
275 280 285Tyr Arg His Pro Ile Val Ser Val Val Gly Arg Pro Ile Ser
Val Glu 290 295 300Gln Lys Asp His Pro Thr Thr Ala Asp Leu Glu Glu
Val Gln Ala Arg305 310 315 320Tyr Ile Ala Glu Leu Lys Arg Ile Trp
Glu Glu Tyr Lys Asp Ala Tyr 325 330 335Ala Lys Ser Arg Thr Arg Glu
Leu Asn Ile Ile Ala 340 34561047DNARhodosporidium toruloides
6atgggccagc aggcgacgcc cgaggagcta tacacacgct cagagatctc caagatcaag
60ttcgcaccct ttggcgtccc gcggtcgcgc cggctgcaga ccttctccgt ctttgcctgg
120acgacggcac tgcccatcct actcggcgtc ttcttcctcc tctgctcgtt
cccaccgctc 180tggccggctg tcattgccta cctcacctgg gtctttttca
ttgaccaggc gccgattcac 240ggtggacggg cgcagtcttg gctgcggaag
agtcggatat gggtctggtt tgcaggatac 300tatcccgtca gcttgatcaa
gagcgccgac ttgccgcctg accggaagta cgtctttggc 360taccacccgc
acggcgtcat aggcatgggc gccatcgcca acttcgcgac cgacgcaacc
420ggcttctcga cactcttccc cggcttgaac cctcacctcc tcaccctcca
aagcaacttc 480aagctcccgc tctaccgcga gttgctgctc gctctcggca
tatgctccgt ctcgatgaag 540agctgtcaga acattctgcg acaaggtcct
ggctcggctc tcactatcgt cgtcggtggc 600gccgccgaga gcttgagtgc
gcatcccgga accgccgatc ttacgctcaa gcgacgaaaa 660ggcttcatca
aactcgcgat ccggcaaggc gccgaccttg tgcccgtctt ttcgttcggc
720gagaacgaca tctttggcca gctgcgaaac gagcgaggaa cgcggctgta
caagttgcag 780aagcgtttcc aaggcgtgtt tggcttcacc ctccctctct
tctacggccg gggactcttc 840aactacaacg tcggattgat gccgtatcgc
catccgatcg tctctgtcgt cggtcgacca 900atctcggtag agcagaagga
ccacccgacc acggcggacc tcgaagaagt tcaggcgcgg 960tatatcgcag
aactcaagcg gatctgggaa gaatacaagg acgcctacgc caaaagtcgc
1020acgcgggagc tcaatattat cgcctga 10477348PRTRhodosporidium
toruloides 7Met Gly Gln Gln Ala Thr Pro Glu Glu Leu Tyr Thr Arg Ser
Glu Ile1 5 10 15Ser Lys Ile Lys Phe Ala Pro Phe Gly Val Pro Arg Ser
Arg Arg Leu 20 25 30Gln Thr Phe Ser Val Phe Ala Trp Thr Thr Ala Leu
Pro Ile Leu Leu 35 40 45Gly Val Phe Phe Leu Leu Cys Ser Phe Pro Pro
Leu Trp Pro Ala Val 50 55 60Ile Ala Tyr Leu Thr Trp Val Phe Phe Ile
Asp Gln Ala Pro Ile His65 70 75 80Gly Gly Arg Ala Gln Ser Trp Leu
Arg Lys Ser Arg Ile Trp Val Trp 85 90 95Phe Ala Gly Tyr Tyr Pro Val
Ser Leu Ile Lys Ser Ala Asp Leu Pro 100 105 110Pro Asp Arg Lys Tyr
Val Phe Gly Tyr His Pro His Gly Val Ile Gly 115 120 125Met Gly Ala
Ile Ala Asn Phe Ala Thr Asp Ala Thr Gly Phe Ser Thr 130 135 140Leu
Phe Pro Gly Leu Asn Pro His Leu Leu Thr Leu Gln Ser Asn Phe145 150
155 160Lys Leu Pro Leu Tyr Arg Glu Leu Leu Leu Ala Leu Gly Ile Cys
Ser 165 170 175Val Ser Met Lys Ser Cys Gln Asn Ile Leu Arg Gln Gly
Pro Gly Ser 180 185 190Ala Leu Thr Ile Val Val Gly Gly Ala Ala Glu
Ser Leu Ser Ala His 195 200 205Pro Gly Thr Ala Asp Leu Thr Leu Lys
Arg Arg Lys Gly Phe Ile Lys 210 215 220Leu Ala Ile Arg Gln Gly Ala
Asp Leu Val Pro Val Phe Ser Phe Gly225 230 235 240Glu Asn Asp Ile
Phe Gly Gln Leu Arg Asn Glu Arg Gly Thr Arg Leu 245 250 255Tyr Lys
Leu Gln Lys Arg Phe Gln Gly Val Phe Gly Phe Thr Leu Pro 260 265
270Leu Phe Tyr Gly Arg Gly Leu Phe Asn Tyr Asn Val Gly Leu Met Pro
275 280 285Tyr Arg His Pro Ile Val Ser Val Val Gly Arg Pro Ile Ser
Val Glu 290 295 300Gln Lys Asp His Pro Thr Thr Ala Asp Leu Glu Glu
Val Gln Ala Arg305 310 315 320Tyr Ile Ala Glu Leu Lys Arg Ile Trp
Glu Glu Tyr Lys Asp Ala Tyr 325 330 335Ala Lys Ser Arg Thr Arg Glu
Leu Asn Ile Ile Ala 340 34581047DNARhodosporidium toruloides
8atgggacagc aggctacccc cgaggagctc tacacccgat ccgagatttc taagattaag
60ttcgcccctt ttggagtgcc ccgatcccga cgactccaga ccttctccgt ttttgcctgg
120accactgctc tgcccattct gctcggcgtc ttctttctgc tctgctcttt
cccccctctc 180tggcccgccg tcatcgctta cctgacctgg gtgttcttta
tcgaccaggc ccctattcac 240ggcggtcgag ctcagtcctg gctgcgaaag
tctcgaattt gggtttggtt cgccggttac 300taccccgtct ctctcatcaa
gtcggctgac ctgccccctg atcgaaagta cgtgttcggc 360taccaccctc
atggtgttat cggtatggga gccattgcta actttgccac cgatgctact
420ggtttctcca ccctctttcc cggactgaac cctcacctgc tcactctcca
gtctaacttc 480aagctccccc tgtaccgaga gctgctcctg gccctgggta
tctgctccgt ctctatgaag 540tcttgtcaga acattctccg acagggacct
ggttcggctc tgaccatcgt cgtgggagga 600gctgctgagt cgctctccgc
ccatcctgga accgctgacc tcactctgaa gcgacgaaag 660ggcttcatca
agctcgccat tcgacagggt gctgacctgg tgcccgtttt ctcctttgga
720gagaacgata ttttcggcca gctgcgaaac gagcgaggaa cccgactcta
caagctgcag 780aagcgatttc agggtgtgtt cggcttcacc ctccctctgt
tctacggacg aggcctcttt
840aactacaacg ttggactgat gccctaccga caccctatcg tctcggttgt
cggccgaccc 900atttccgtgg agcagaagga ccatcctacc actgccgatc
tcgaggaggt gcaggcccga 960tacatcgctg agctgaagcg aatttgggag
gagtacaagg acgcctacgc taagtctcga 1020acccgagagc tgaacatcat tgcctaa
10479410PRTLipomyces starkeyi 9Met Ser Glu Lys Ala Glu Ile Glu Val
Pro Pro Gln Lys Ser Thr Phe1 5 10 15Pro Arg Ser Val His Phe Ala Pro
Leu His Ile Pro Leu Glu Arg Arg 20 25 30Leu Gln Thr Leu Ala Val Leu
Phe His Thr Val Ala Leu Pro Tyr Cys 35 40 45Ile Gly Leu Phe Phe Leu
Met Leu Ala Phe Pro Pro Phe Trp Pro Leu 50 55 60Leu Val Met Tyr Val
Ile Tyr Ala Tyr Gly Phe Asp His Ser Ser Ser65 70 75 80Asn Gly Glu
Ile Ser Arg Arg Arg Ser Pro Leu Phe Arg Arg Leu Pro 85 90 95Leu Phe
Arg Leu Tyr Cys Asp Tyr Phe Pro Ile His Ile His Arg Glu 100 105
110Val Pro Leu Glu Pro Thr Phe Pro Gly Arg Leu Arg Glu Pro Ser Gly
115 120 125Leu Val Glu Arg Trp Ile Ala Lys Met Phe Gly Val Gln Asp
Ala Val 130 135 140Val Glu Gly Asn Glu Ser Asp Val Lys Ala Thr Ala
Asn Gly Asn Gly145 150 155 160Thr Thr Lys Glu Ile Gly Pro Thr Tyr
Val Phe Gly Tyr His Pro His 165 170 175Gly Ile Val Ser Leu Gly Ala
Phe Gly Ala Ile Gly Thr Glu Gly Ala 180 185 190Gly Trp Glu Lys Leu
Phe Pro Gly Ile Pro Val Ser Leu Leu Thr Leu 195 200 205Glu Thr Asn
Phe Ser Leu Pro Phe Tyr Arg Glu Tyr Leu Leu Ser Leu 210 215 220Gly
Ile Ala Ser Val Ser Arg Arg Ser Cys Thr Asn Leu Leu Lys His225 230
235 240Asp Gln Ser Ile Cys Ile Val Ile Gly Gly Ala Gln Glu Ser Leu
Leu 245 250 255Ala Glu Pro Gly Thr Leu Asp Leu Ile Leu Val Lys Arg
Arg Gly Phe 260 265 270Val Lys Leu Ala Met Ser Thr Ala Arg Val Ser
Asp Gln Pro Ile Cys 275 280 285Leu Val Pro Ile Leu Ser Phe Gly Glu
Asn Asp Val Tyr Asp Gln Val 290 295 300Arg Gly Asp Arg Ser Ser Lys
Leu Tyr Lys Ile Gln Thr Phe Ile Lys305 310 315 320Lys Ala Ala Gly
Phe Thr Leu Pro Leu Met Tyr Ala Arg Gly Ile Phe 325 330 335Asn Tyr
Asp Phe Gly Leu Met Pro Tyr Arg Arg Gln Met Thr Leu Val 340 345
350Val Gly Lys Pro Ile Ala Val Pro Tyr Val Ala Gln Pro Thr Glu Ala
355 360 365Glu Ile Glu Val Tyr His Lys Gln Tyr Met Asp Glu Leu Arg
Arg Leu 370 375 380Trp Asp Thr Tyr Lys Asp Asp Tyr Phe Val Asp His
Lys Gly Lys Gly385 390 395 400Val Lys Asn Ser Glu Met Arg Phe Val
Glu 405 410101233DNALipomyces starkeyi 10atgagtgaga aggcagagat
cgaggttccg ccgcaaaaat cgacattccc tcgcagtgtg 60cacttcgctc cacttcatat
tccactggag agacgcctac agactttggc agtcttattc 120cacactgtcg
cgctaccata ctgcatcggt ctgttctttc tcatgctcgc gttccctcct
180ttttggccat tattggtaat gtatgtcata tacgcatacg ggttcgacca
ctcgagctcg 240aacggagaga tctcccgccg gcgatcgccg ctgtttcgaa
gactcccgtt gttcaggctg 300tattgtgatt acttccccat ccacattcac
cgggaggttc cgctcgagcc gacgtttcct 360ggtcgccttc gcgaaccgag
tggccttgtc gagcggtgga ttgcgaagat gttcggcgtg 420caggacgctg
ttgtcgaggg aaatgaatct gacgttaagg ccacggccaa cggcaatggg
480acgacgaaag aaatcggacc gacgtatgtt ttcggctatc atccgcatgg
aattgttagc 540ttgggtgcgt ttggtgctat tggtacggaa ggcgctggat
gggagaagct ctttcctggg 600atcccggtgt cactgctgac tctcgaaaca
aatttcagcc ttccatttta cagagagtat 660ttgctgtcac ttgggattgc
ttcagtatct cgacggtctt gtaccaatct cctcaaacac 720gaccaatcca
tctgcatcgt tatcggcggc gcccaagagt cgctcttagc ggaaccaggc
780actctagatc tgatcctcgt taaacgtcgc ggttttgtca aacttgcaat
gtcaacggcg 840cgggtatctg accaaccgat ttgtcttgtt ccgatcctca
gtttcggcga gaacgacgtg 900tacgaccaag tccgcgggga ccgatcgtcg
aagttgtata agatccagac ttttatcaag 960aaagcggccg ggtttacgct
accattgatg tatgcgcgcg gtatatttaa ttacgacttt 1020gggctgatgc
cgtaccgcag gcaaatgacg ctcgtggtcg gcaagccgat tgcagtgccg
1080tacgtggccc agcctacgga ggctgaaatc gaagtgtatc acaagcagta
catggatgaa 1140ttgaggaggt tatgggacac gtataaggac gactattttg
tagaccacaa gggcaagggg 1200gtcaagaatt ccgagatgcg ttttgtggag taa
123311410PRTLipomyces starkeyi 11Met Ser Glu Lys Ala Glu Ile Glu
Val Pro Pro Gln Lys Ser Thr Phe1 5 10 15Pro Arg Ser Val His Phe Ala
Pro Leu His Ile Pro Leu Glu Arg Arg 20 25 30Leu Gln Thr Leu Ala Val
Leu Phe His Thr Val Ala Leu Pro Tyr Cys 35 40 45Ile Gly Leu Phe Phe
Leu Met Leu Ala Phe Pro Pro Phe Trp Pro Leu 50 55 60Leu Val Met Tyr
Val Ile Tyr Ala Tyr Gly Phe Asp His Ser Ser Ser65 70 75 80Asn Gly
Glu Ile Ser Arg Arg Arg Ser Pro Leu Phe Arg Arg Leu Pro 85 90 95Leu
Phe Arg Leu Tyr Cys Asp Tyr Phe Pro Ile His Ile His Arg Glu 100 105
110Val Pro Leu Glu Pro Thr Phe Pro Gly Arg Leu Arg Glu Pro Ser Gly
115 120 125Leu Val Glu Arg Trp Ile Ala Lys Met Phe Gly Val Gln Asp
Ala Val 130 135 140Val Glu Gly Asn Glu Ser Asp Val Lys Ala Thr Ala
Asn Gly Asn Gly145 150 155 160Thr Thr Lys Glu Ile Gly Pro Thr Tyr
Val Phe Gly Tyr His Pro His 165 170 175Gly Ile Val Ser Leu Gly Ala
Phe Gly Ala Ile Gly Thr Glu Gly Ala 180 185 190Gly Trp Glu Lys Leu
Phe Pro Gly Ile Pro Val Ser Leu Leu Thr Leu 195 200 205Glu Thr Asn
Phe Ser Leu Pro Phe Tyr Arg Glu Tyr Leu Leu Ser Leu 210 215 220Gly
Ile Ala Ser Val Ser Arg Arg Ser Cys Thr Asn Leu Leu Lys His225 230
235 240Asp Gln Ser Ile Cys Ile Val Ile Gly Gly Ala Gln Glu Ser Leu
Leu 245 250 255Ala Glu Pro Gly Thr Leu Asp Leu Ile Leu Val Lys Arg
Arg Gly Phe 260 265 270Val Lys Leu Ala Met Ser Thr Ala Arg Val Ser
Asp Gln Pro Ile Cys 275 280 285Leu Val Pro Ile Leu Ser Phe Gly Glu
Asn Asp Val Tyr Asp Gln Val 290 295 300Arg Gly Asp Arg Ser Ser Lys
Leu Tyr Lys Ile Gln Thr Phe Ile Lys305 310 315 320Lys Ala Ala Gly
Phe Thr Leu Pro Leu Met Tyr Ala Arg Gly Ile Phe 325 330 335Asn Tyr
Asp Phe Gly Leu Met Pro Tyr Arg Arg Gln Met Thr Leu Val 340 345
350Val Gly Lys Pro Ile Ala Val Pro Tyr Val Ala Gln Pro Thr Glu Ala
355 360 365Glu Ile Glu Val Tyr His Lys Gln Tyr Met Asp Glu Leu Arg
Arg Leu 370 375 380Trp Asp Thr Tyr Lys Asp Asp Tyr Phe Val Asp His
Lys Gly Lys Gly385 390 395 400Val Lys Asn Ser Glu Met Arg Phe Val
Glu 405 410121233DNALipomyces starkeyi 12atgtccgaga aggctgagat
tgaggtgccc ccccagaagt ctactttccc tcgatccgtt 60catttcgccc ccctgcatat
ccccctggag cgacgactcc agaccctggc tgtgctcttc 120cacactgttg
ccctgcctta ctgcatcgga ctcttctttc tgatgctcgc tttcccccct
180ttttggcccc tgctcgtgat gtacgttatc tacgcctacg gattcgacca
ttcctcttcg 240aacggcgaga tctctcgacg acgatcgcct ctgttccgac
gactgcccct ctttcgactc 300tactgtgatt acttccctat ccacattcat
cgagaggtcc ccctggagcc tacctttcct 360ggtcgactgc gagagccttc
cggactcgtt gagcgatgga ttgctaagat gttcggtgtc 420caggacgccg
tcgtggaggg aaacgagtct gatgtgaagg ccaccgctaa cggaaacggc
480accactaagg agatcggccc tacttacgtc ttcggatacc acccccatgg
cattgtgtcc 540ctgggagcct ttggcgctat cggtaccgag ggtgctggat
gggagaagct cttccctggt 600attcccgtct cgctgctcac cctggagact
aacttctccc tcccctttta ccgagagtac 660ctgctctctc tgggaatcgc
ctcggtgtcc cgacgatcgt gcaccaacct gctcaagcac 720gaccagtcta
tctgtattgt tatcggaggt gctcaggagt ccctgctcgc tgagcctgga
780accctggacc tcattctggt caagcgacga ggcttcgtga agctggccat
gtccactgct 840cgagtgtctg atcagcctat ttgcctggtt cccatcctct
ctttcggcga gaacgacgtt 900tacgatcagg tccgaggtga ccgatcctct
aagctgtaca agattcagac cttcatcaag 960aaggccgctg gctttactct
ccctctgatg tacgcccgag gcatcttcaa ctacgacttt 1020ggtctgatgc
cctaccgacg acagatgacc ctcgttgtcg gcaagcctat tgccgtcccc
1080tacgtggctc agcccactga ggccgagatc gaggtctacc acaagcagta
catggacgag 1140ctgcgacgac tctgggatac ctacaaggac gattacttcg
ttgaccataa gggcaagggt 1200gtcaagaact ctgagatgcg atttgtggag taa
123313380PRTAspergillus terreus 13Met Pro Arg Asn Thr His Pro Pro
Ala Asn Asn Ala Gly Pro Asn Ala1 5 10 15Ser His Lys Lys Asp Arg Lys
Arg Gln Gly Arg Leu Phe Gln His Thr 20 25 30Val Pro Asn Lys Tyr Ser
Arg Ile Arg Trp Ala Pro Leu Asn Ile Gly 35 40 45Leu Glu Arg Arg Leu
Gln Thr Leu Val Val Leu Cys His Thr Leu Thr 50 55 60Ile Ala Leu Phe
Leu Ala Phe Phe Phe Phe Thr Cys Ala Ile Pro Leu65 70 75 80Thr Trp
Pro Leu Leu Phe Pro Tyr Leu Val Tyr Ile Thr Leu Phe Ser 85 90 95Thr
Ala Pro Thr Ser Gly Thr Leu Lys Gly Arg Ser Asp Phe Leu Arg 100 105
110Ser Leu Pro Ile Trp Lys Leu Tyr Thr Ala Tyr Phe Pro Ala Lys Leu
115 120 125His Arg Ser Glu Pro Leu Leu Pro Thr Arg Lys Tyr Ile Phe
Gly Tyr 130 135 140His Pro His Gly Ile Ile Ser His Gly Ala Phe Ala
Ala Phe Ala Thr145 150 155 160Asp Ala Leu Gly Phe Ser Lys Leu Phe
Pro Gly Ile Thr Asn Thr Leu 165 170 175Leu Thr Leu Asp Ser Asn Phe
Arg Ile Pro Phe Tyr Arg Glu Tyr Ala 180 185 190Met Ala Met Gly Val
Ala Ser Val Ser Arg Glu Ser Cys Glu Asn Leu 195 200 205Leu Thr Lys
Gly Gly Ala Asp Gly Glu Gly Met Gly Arg Ala Ile Thr 210 215 220Ile
Val Val Gly Gly Ala Arg Glu Ser Leu Asp Ala Leu Pro His Thr225 230
235 240Met Arg Leu Val Leu Lys Arg Arg Lys Gly Phe Ile Lys Leu Ala
Ile 245 250 255Arg Thr Gly Ala Asp Leu Val Pro Val Leu Ala Phe Gly
Glu Asn Asp 260 265 270Leu Tyr Glu Gln Val Arg Ser Asp Gln His Pro
Leu Ile Tyr Lys Val 275 280 285Gln Met Leu Val Lys Arg Phe Leu Gly
Phe Thr Val Pro Leu Phe His 290 295 300Ala Arg Gly Ile Phe Asn Tyr
Asp Val Gly Leu Met Pro Tyr Arg Arg305 310 315 320Pro Leu Asn Ile
Val Val Gly Arg Pro Ile Gln Val Val Arg Gln Gln 325 330 335Asp Arg
Asp Lys Ile Asp Asp Glu Tyr Ile Asp Arg Leu His Ala Glu 340 345
350Tyr Val Arg Glu Leu Glu Ser Leu Trp Asp Gln Trp Lys Asp Val Tyr
355 360 365Ala Lys Asp Arg Ile Ser Glu Leu Glu Ile Val Ala 370 375
380141143DNAAspergillus terreus 14atgccccgaa acacccaccc ccccgccaac
aacgccggac ctaacgcctc tcacaagaag 60gaccgaaagc gacagggacg actctttcag
cacaccgttc ctaacaagta ctctcgaatc 120cgatgggccc ccctcaacat
tggcctggag cgacgactgc agaccctcgt cgtgctgtgc 180cataccctca
ctatcgccct gttcctcgct ttctttttct ttacttgtgc cattcccctg
240acctggcctc tgctcttccc ctacctcgtg tacatcaccc tgttttcgac
cgctcctact 300tccggtaccc tgaagggacg atctgacttc ctccgatcgc
tgcctatttg gaagctctac 360actgcctact ttcccgctaa gctgcaccga
tccgagcctc tgctccctac ccgaaagtac 420atcttcggct accaccccca
tggtatcatt tcccatggag ccttcgccgc ttttgccact 480gacgctctcg
gcttctctaa gctgtttcct ggtatcacca acactctgct caccctggat
540tcgaacttcc gaattccctt ttaccgagag tacgccatgg ctatgggagt
ggcttccgtt 600tctcgagagt cgtgcgagaa cctgctcact aagggaggtg
ctgacggaga gggaatgggc 660cgagctatca ccattgttgt cggaggcgcc
cgagagtccc tcgatgctct gcctcacact 720atgcgactgg tcctcaagcg
acgaaagggt ttcatcaagc tggccattcg aaccggagct 780gacctcgttc
ccgtcctggc cttcggcgag aacgacctct acgagcaggt gcgatctgat
840cagcaccctc tgatctacaa ggtccagatg ctcgtgaagc gattcctggg
ttttaccgtg 900cccctgttcc atgctcgagg aatttttaac tacgacgttg
gcctcatgcc ttaccgacga 960cccctgaaca tcgtggttgg tcgacccatt
caggtcgtgc gacagcagga ccgagataag 1020atcgacgatg agtacattga
ccgactccac gccgagtacg tccgagagct cgagtccctg 1080tgggaccagt
ggaaggatgt ttacgccaag gaccgaatct ctgagctgga gattgtcgct 1140taa
114315437PRTClaviceps purpurea 15Met Ala Ala Val Gln Val Ala Arg
Pro Val Pro Pro His His His Asp1 5 10 15Gly Ala Gly Arg Glu His Lys
Gly Glu Arg Ala His Ser Pro Glu Arg 20 25 30Gly Glu Lys Thr Val His
Asn Gly Tyr Gly Leu Ala Glu Thr His Glu 35 40 45Pro Leu Glu Leu Asn
Gly Ser Ala Val Gln Asp Gly Lys His Asp Ser 50 55 60Asp Glu Thr Ile
Thr Asn Gly Asp Tyr Ser Pro Tyr Pro Glu Leu Asp65 70 75 80Cys Gly
Lys Glu Arg Ala Ala His Glu Lys Glu Ala Trp Thr Ala Gly 85 90 95Gly
Val Arg Phe Ala Pro Leu Arg Val Pro Phe Lys Arg Arg Met Gln 100 105
110Thr Ala Ala Val Leu Phe His Cys Met Ser Ile Ile Leu Ile Ser Ser
115 120 125Cys Phe Trp Phe Ser Leu Ala Asn Pro Ile Thr Trp Pro Ile
Leu Val 130 135 140Pro Tyr Leu Val His Leu Ser Leu Ser Asn Ala Ser
Thr Asp Gly Lys145 150 155 160Leu Ser Tyr Arg Ser Glu Trp Leu Arg
Ser Leu Pro Leu Trp Arg Leu 165 170 175Phe Ala Gly Tyr Phe Pro Ala
Lys Leu His Lys Thr Phe Asp Leu Pro 180 185 190Pro Asn Arg Lys Tyr
Ile Phe Gly Tyr His Pro His Gly Ile Ile Ser 195 200 205His Gly Ala
Trp Cys Ala Phe Ala Thr Asn Ala Leu Gly Phe Val Glu 210 215 220Lys
Phe Pro Gly Ile Thr Asn Ser Leu Leu Thr Leu Asp Ser Asn Phe225 230
235 240Arg Val Pro Phe Tyr Arg Asp Trp Ile Leu Ala Met Gly Ile Arg
Ser 245 250 255Val Ser Arg Glu Ser Ile Arg Asn Ile Leu Ser Lys Gly
Gly Pro Asp 260 265 270Ser Asn Gly Gln Gly Arg Ala Val Thr Ile Val
Ile Gly Gly Ala Arg 275 280 285Glu Ser Leu Glu Ala Gln Pro Gly Thr
Leu Arg Leu Ile Leu Gln Gly 290 295 300Arg Lys Gly Phe Ile Lys Val
Ala Leu Arg Ala Gly Ala Asp Leu Val305 310 315 320Pro Val Ile Gly
Phe Gly Glu Asn Asp Leu Tyr Asp Gln Leu Ser Pro 325 330 335Lys Thr
His Pro Leu Val His Lys Ile Gln Met Phe Phe Leu Lys Val 340 345
350Phe Lys Phe Thr Ile Pro Ala Leu His Gly Arg Gly Leu Leu Asn Tyr
355 360 365Asp Val Gly Leu Leu Pro Tyr Arg Arg Ala Val Asn Ile Val
Val Gly 370 375 380Arg Pro Ile Gln Ile Asp Glu Thr Tyr Gly Glu Gln
Pro Pro Gln Glu385 390 395 400Val Ile Asp Arg Tyr His Glu Leu Tyr
Val Gln Glu Val Glu Arg Leu 405 410 415Tyr Ala Ala Tyr Lys Glu Gln
Phe Ser Asn Gly Lys Lys Thr Pro Glu 420 425 430Leu Gln Ile Leu Ser
435161314DNAClaviceps purpurea 16atggctgctg ttcaggttgc ccgacccgtt
cccccccacc accacgatgg cgctggccga 60gagcacaagg gagagcgagc ccattcccct
gagcgaggag agaagaccgt ccacaacggc 120tacggtctgg ccgagactca
tgagcccctg gagctcaacg gttctgctgt gcaggacgga 180aagcacgact
cggatgagac catcactaac ggtgactact ctccctaccc tgagctcgat
240tgcggaaagg agcgagccgc tcatgagaag gaggcttgga ccgctggagg
tgtgcgattc 300gctcctctgc gagttccttt taagcgacga atgcagactg
ccgctgtcct cttccactgc 360atgtccatca ttctgatttc ctcttgtttc
tggttttctc tcgccaaccc catcacctgg 420cctattctcg ttccctacct
ggtccacctg tcgctctcca acgcttctac tgacggcaag 480ctctcctacc
gatctgagtg gctgcgatcc ctgcctctct ggcgactgtt cgccggttac
540tttcccgcta agctccacaa gaccttcgat ctgcccccta accgaaagta
catctttggt 600taccaccccc atggaatcat ttcccatggc gcctggtgtg
ccttcgctac caacgctctg 660ggcttcgttg agaagtttcc tggtattacc
aactcgctgc tcactctcga ctccaacttc 720cgagtgccct tttaccgaga
ttggatcctg gccatgggca ttcgatctgt ttcgcgagag 780tctatccgaa
acattctctc gaagggagga cctgactcca acggacaggg ccgagctgtg
840accatcgtta ttggtggagc ccgagagtct ctggaggctc agcccggaac
tctgcgactc 900attctgcagg gccgaaaggg cttcattaag
gtggctctcc gagctggagc tgacctggtt 960cccgtcatcg gtttcggaga
gaacgacctc tacgatcagc tgtcccctaa gacccacccc 1020ctcgttcata
agatccagat gttctttctg aaggtcttca agtttactat tcctgctctg
1080cacggacgag gtctgctcaa ctacgacgtc ggtctgctcc cttaccgacg
agctgtgaac 1140atcgtcgtgg gacgacccat ccagattgac gagacctacg
gcgagcagcc ccctcaggag 1200gtcatcgatc gataccacga gctctacgtc
caggaggtgg agcgactgta cgccgcttac 1260aaggagcagt tctcgaacgg
aaagaagacc cccgagctcc agatcctgtc ctaa 131417351PRTAurantiochytrium
limacinum 17Met Leu Ala Trp Met Pro Val Leu Ile Ala Leu Pro Arg Arg
Lys Gln1 5 10 15Thr Ala Val Val Leu Leu Phe Val Met Leu Leu Pro Met
Ile Met Val 20 25 30Val Tyr Ser Trp Thr Leu Ile Leu Leu Ile Phe Pro
Leu Thr Thr Leu 35 40 45Pro Thr Leu Ser Tyr Leu Ile Trp Ile Met Tyr
Ile Asp Lys Ser His 50 55 60Glu Thr Gly Lys Arg Lys Pro Phe Met Arg
Tyr Trp Lys Met Trp Arg65 70 75 80His Phe Ala Asn Tyr Phe Pro Leu
Arg Leu Ile Arg Thr Thr Pro Leu 85 90 95Asp Pro Arg Arg Lys Tyr Val
Phe Cys Tyr His Pro His Gly Ile Ile 100 105 110Ser Leu Gly Ala Phe
Gly Asn Phe Ala Thr Asp Ser Thr Gly Phe Ser 115 120 125Arg Lys Phe
Pro Gly Ile Asp Leu Arg Leu Leu Thr Leu Gln Ile Asn 130 135 140Phe
Tyr Cys Pro Ile Ile Arg Glu Leu Leu Leu Tyr Met Gly Leu Cys145 150
155 160Ser Ala Ala Lys Lys Ser Cys Asn Gln Ile Leu Gln Arg Gly Pro
Gly 165 170 175Ser Ala Ile Met Leu Val Val Gly Gly Ala Ala Glu Ser
Leu Asp Ser 180 185 190Gln Pro Gly Thr Tyr Arg Leu Thr Leu Gly Arg
Lys Gly Phe Val Arg 195 200 205Val Ala Leu Asp Asn Gly Ala Asp Leu
Val Pro Val Leu Gly Phe Gly 210 215 220Glu Asn Asp Val Phe Asp Thr
Val Tyr Leu Pro Pro Asn Ser Trp Ala225 230 235 240Arg Asn Val Gln
Glu Phe Val Arg Lys Lys Leu Gly Phe Ala Thr Pro 245 250 255Ile Phe
Ser Gly Arg Gly Ile Phe Gln Tyr Asn Met Gly Leu Met Pro 260 265
270His Arg Lys Pro Ile Ile Val Val Val Gly Lys Pro Ile Lys Ile Pro
275 280 285Lys Ile Pro Asp Glu Leu Lys Gly Arg Ala Leu Ser Thr Thr
Ala Glu 290 295 300Gly Val Ala Leu Val Asp Lys Tyr His Glu Lys Tyr
Val Arg Ala Leu305 310 315 320Arg Glu Leu Trp Asn Leu Tyr Lys Glu
Glu Tyr Ala Thr Glu Pro Lys 325 330 335Ala Ala Tyr Leu Glu Pro Asn
Ser Ile Arg Lys Asn Gln Asn Val 340 345
350181056DNAAurantiochytrium limacinum 18atgctcgcct ggatgcctgt
cctcattgcc ctcccccgac gaaagcagac cgctgttgtt 60ctcctgtttg tgatgctcct
ccctatgatc atggtcgtgt actcctggac cctgatcctg 120ctcattttcc
ccctcaccac tctgcctact ctctcctacc tgatctggat tatgtacatt
180gacaagtctc acgagaccgg aaagcgaaag ccctttatgc gatactggaa
gatgtggcga 240catttcgcca actactttcc tctccgactg atccgaacca
ctcccctgga ccctcgacga 300aagtacgtgt tctgctacca cccccatggc
atcatttccc tcggagcctt cggcaacttt 360gctaccgact cgactggctt
ctcccgaaag tttcccggta tcgatctgcg actgctcacc 420ctccagatta
acttctactg tcctatcatt cgagagctgc tcctgtacat gggtctgtgc
480tctgccgcta agaagtcgtg taaccagatc ctccagcgag gacccggctc
tgctattatg 540ctggttgtcg gcggtgccgc tgagtccctc gactctcagc
ctggcaccta ccgactcact 600ctgggtcgaa agggattcgt gcgagttgcc
ctggacaacg gtgctgatct ggtccccgtg 660ctcggtttcg gagagaacga
cgtgtttgat accgtttacc tgccccctaa ctcgtgggcc 720cgaaacgtcc
aggagttcgt gcgaaagaag ctcggattcg ctacccccat cttttccggc
780cgaggtattt ttcagtacaa catgggtctg atgccccacc gaaagcctat
cattgtggtt 840gtcggaaagc ccatcaagat tcccaagatc cctgacgagc
tgaagggacg agccctctct 900accactgccg agggcgttgc tctggtcgat
aagtaccatg agaagtacgt tcgagccctc 960cgagagctgt ggaacctcta
caaggaggag tacgctaccg agcccaaggc cgcttacctc 1020gagcctaact
cgattcgaaa gaaccagaac gtctaa 105619666PRTYarrowia lipolytica 19Met
Lys Ser Arg Val Ala Val Val Leu Ala Pro Val Leu Ala Pro Phe1 5 10
15Val Ala Ile Leu Lys Asn Leu Trp Val Phe Phe Thr Ala Leu Leu Glu
20 25 30Leu Leu Phe Asp Val Ser Trp His Trp Met Leu Gln Ser Trp His
Trp 35 40 45Trp Cys Ser Thr Asp Gln Lys Thr Leu Leu Gln Leu Gln Leu
Asp Gln 50 55 60Ala Asp Thr Tyr Glu Glu Trp Glu Ser Ile Ala Ser Glu
Leu Asp Glu65 70 75 80Leu Leu Gly Asn Asp Val Trp Arg Gln Thr Ala
Ala Ser Lys Arg Tyr 85 90 95Asp Tyr Arg Leu Ile Ala Gly Arg Leu Arg
Asp Phe Ile Glu Cys Arg 100 105 110Ala Val Gly Asp Ile Ala Thr Leu
Ile Ser Arg Leu Arg Ser Gly Leu 115 120 125Leu Arg Asn Leu Gly Ser
Ile Ser Ser Leu Gln Leu Tyr Thr Arg Ser 130 135 140Tyr Leu Gly Ser
Lys Leu Leu Ile Glu Glu Tyr Ile Thr Glu Val Ile145 150 155 160Asp
Cys Leu Lys Tyr Ile Lys Asp Tyr Gly Thr Thr Gly Gly Leu Asp 165 170
175Thr Lys Gly Val His Phe Phe Pro Lys Ser Glu Gln Arg Gln Leu Asp
180 185 190Ser Glu Gln Leu Thr Arg Gln Lys Lys His Lys Leu Phe Tyr
Asp Thr 195 200 205Arg Gln Ser Phe Gly Arg Thr Ala Leu Val Leu Gln
Gly Gly Thr Ile 210 215 220Phe Gly Leu Thr His Leu Gly Thr Ile Lys
Ala Leu Thr Leu Gln Gly225 230 235 240Leu Leu Pro Gly Ile Val Thr
Gly Phe Lys Glu Gly Ala Phe Ile Ala 245 250 255Ala Leu Thr Gly Ile
Tyr Val Ser Asp Leu Glu Leu Leu Glu Thr Ile 260 265 270Asp Ser Leu
Pro Asp Thr Leu Asn Asp Leu Tyr Gln Lys Tyr Lys Glu 275 280 285Arg
Leu Ala Glu Glu Asn Lys His Lys Asp His Ser Phe Ser Asn Ser 290 295
300Asn Ser Asp Tyr Asp Phe Asp Tyr Ala Phe Asp Phe Glu Gln Phe
Ala305 310 315 320Asn Thr Tyr Asn Val Thr Phe Ser Ser Val Thr Asp
Lys Val Leu Arg 325 330 335Ser Glu Tyr Pro Pro Glu Val Lys Met Tyr
Glu Glu Phe Ile Glu Asn 340 345 350Gln Leu Gly Asp Leu Thr Phe Glu
Glu Ala Phe Asn Lys Ser Asp Arg 355 360 365Val Leu Asn Ile Val Ala
His Ser His Asp Ser Ser Phe Pro Thr Leu 370 375 380Met Asn Tyr Leu
Thr Thr Pro Asn Val Leu Ile Arg Ser Ala Cys Arg385 390 395 400Ala
Ser Met Val Thr Ala His Asp Glu Pro Gln Thr Lys Lys Ala Cys 405 410
415Ala His Leu Leu Val Lys Asp Asp Asp Asn Ser Val Ile Pro Tyr Asp
420 425 430Ala Cys Lys Ser Arg Arg Gly Ser Ser Thr Asp Val Ile Leu
Gly Pro 435 440 445Val Gln Glu Glu Val Asp Pro Leu Asp Ser Thr Ala
Asn Gly Thr Asn 450 455 460Ser Ser Gly Pro Pro Lys Leu Glu Ile Thr
Thr Asp Thr Trp Lys Arg465 470 475 480Asn Asn Ala Asp Asp Glu Asp
His Val Asp Thr Leu Pro Gly Arg Val 485 490 495Ser Ala Leu Pro Thr
Pro Ser Tyr Ser Met Ile Asn Gln Gly Lys Ile 500 505 510Val Ser Pro
Tyr Ala Arg Leu Ser Glu Leu Phe Asn Val Asn His Phe 515 520 525Ile
Val Ser Leu Ser Arg Pro Tyr Leu Ala Pro Leu Leu Ala Ile Glu 530 535
540Gly Arg His Arg Gly Tyr His Gly Trp Arg Val Asn Leu Ile Arg
Val545 550 555 560Leu Lys Leu Glu Phe Glu His Arg Leu Ala Gln Phe
Asp Tyr Ile Gly 565 570 575Leu Leu Pro Thr Ile Phe Arg Arg Phe Phe
Ile Asp Asp Lys Ile Pro 580 585 590Gly Ile Gly Pro Asn Ala Glu Val
Leu Ile Val Pro Glu Leu Ala Ala 595 600 605Gly Met Ile Ser Asp Phe
Lys Lys Ala Phe Ser Asn His Asp Ile Pro 610 615 620Glu Lys Val Arg
Tyr Trp Thr Thr Val Gly Glu Arg Ala Thr Trp Pro625 630 635 640Leu
Val Ala Ala Ile Trp Ala Arg Thr Ala Ile Glu Tyr Thr Leu Asn 645 650
655Asp Met Tyr Asn Gln Thr Lys Arg Gln Asn 660 665202201DNAYarrowia
lipolytica 20cttttacgag tgtgtatcat cacatgatta tgcagcaaga tcagtatcat
ttcggctatc 60cagctctctt cccccgttca gctccttttc taccgcgatt atgaaaagcc
gcgtggccgt 120tgtcttggcg ccggttctgg caccatttgt ggcgattttg
aaaaacctgt gggtcttctt 180cacagctcta ctggagctct tattcgacgt
tagctggcac tggatgttac aatcatggca 240ctggtggtgc tccaccgacc
aaaaaacact gctacaactg cagctggacc aggcagacac 300ctacgaggaa
tgggaaagca ttgcatcgga gctggacgag ctgctgggca acgacgtgtg
360gcgtcagacc gcagcctcga aacgatacga ctaccggctg attgcaggcc
gtctgagaga 420ctttatcgag tgccgggcgg tcggcgacat tgcgacgctg
atttctcgtc tgcgaagcgg 480actgctgcgg aatttgggct cgatttcgtc
gctccagctg tacactcgct cgtacctcgg 540ctctaaactg ctcatcgaag
agtacatcac cgaggtcatt gactgtctca agtacatcaa 600ggactatggg
acgacgggcg gactggacac caagggagtg catttcttcc caaagtccga
660acagcgacaa ctggacagtg aacagctgac tcgacaaaag aaacacaagt
tattctacga 720cacacgacaa tcttttggcc gaacggccct cgtgttgcag
ggaggaacta ttttcggact 780tactcatctc ggaacaatca aggctcttac
tctccagggt ctgctaccgg gtattgtcac 840cggtttcaag gagggggcgt
ttattgccgc tctcacaggc atctacgtat ccgacctgga 900gctgctcgaa
accattgact ctttgccaga cactctcaat gacctgtacc aaaaatacaa
960ggagcgactg gcggaggaaa acaaacacaa ggaccactcg ttcagtaact
ccaattcgga 1020ctacgacttt gactacgcat ttgactttga acagtttgca
aacacctata atgtgacctt 1080ctcgtctgtc actgacaaag tattgcgatc
ggagtacccc ccggaagtca aaatgtacga 1140ggagttcatc gagaatcaac
tcggagacct cacgttcgaa gaggccttca acaaaagcga 1200ccgcgtgctc
aacattgtcg cccattccca tgactcttcc ttcccgacac tgatgaacta
1260cctcaccact cccaatgtgc tcatcagaag cgcatgtaga gcttccatgg
tgaccgccca 1320cgacgagccc caaacgaaaa aggcatgtgc ccatctgctg
gtcaaggatg acgacaacag 1380cgtcattccc tatgacgcct gcaaatccag
gcgaggaagc tcgaccgacg tgattctggg 1440acctgtccag gaggaggtgg
atccattaga ttcaacagct aacggtacta actcttctgg 1500acctcccaaa
ctcgaaatca caactgacac ctggaaacga aacaatgcag acgacgagga
1560ccacgtggat actctcccgg gccgcgtgag tgctctacct acaccttcgt
actccatgat 1620taaccagggc aagattgtct ctccctacgc tcgcctttcc
gaactcttta acgtcaacca 1680cttcatcgtc tctctctcaa gaccctacct
ggcgcctctt ctggccatcg aaggccgaca 1740tagaggctac cacggctgga
gagtgaacct gatccgagta ctgaaactag aattcgaaca 1800cagactcgcc
cagttcgact acataggcct gctgccgacc atcttccgtc ggttcttcat
1860cgacgataag atccctggca tcggtcccaa cgccgaggtg ctcattgttc
ctgagctagc 1920ggctggcatg atctccgact tcaaaaaggc cttttcgaac
cacgacattc ccgagaaggt 1980ccgctactgg accactgtgg gcgaacgagc
cacctggcct ctagtcgccg ccatctgggc 2040cagaacagca atcgagtaca
ccctcaacga catgtacaac cagaccaagc gacaaaacta 2100gaccccgagc
agagcacata actactaacg atgagactaa agtatgtact gtatgtacta
2160aacatacgct cgtaaacagt tgtatttatt ctttttcagc a
220121341PRTEscherichia coli 21Met Lys Lys Pro Glu Leu Thr Ala Thr
Ser Val Glu Lys Phe Leu Ile1 5 10 15Glu Lys Phe Asp Ser Val Ser Asp
Leu Met Gln Leu Ser Glu Gly Glu 20 25 30Glu Ser Arg Ala Phe Ser Phe
Asp Val Gly Gly Arg Gly Tyr Val Leu 35 40 45Arg Val Asn Ser Cys Ala
Asp Gly Phe Tyr Lys Asp Arg Tyr Val Tyr 50 55 60Arg His Phe Ala Ser
Ala Ala Leu Pro Ile Pro Glu Val Leu Asp Ile65 70 75 80Gly Glu Phe
Ser Glu Ser Leu Thr Tyr Cys Ile Ser Arg Arg Ala Gln 85 90 95Gly Val
Thr Leu Gln Asp Leu Pro Glu Thr Glu Leu Pro Ala Val Leu 100 105
110Gln Pro Val Ala Glu Ala Met Asp Ala Ile Ala Ala Ala Asp Leu Ser
115 120 125Gln Thr Ser Gly Phe Gly Pro Phe Gly Pro Gln Gly Ile Gly
Gln Tyr 130 135 140Thr Thr Trp Arg Asp Phe Ile Cys Ala Ile Ala Asp
Pro His Val Tyr145 150 155 160His Trp Gln Thr Val Met Asp Asp Thr
Val Ser Ala Ser Val Ala Gln 165 170 175Ala Leu Asp Glu Leu Met Leu
Trp Ala Glu Asp Cys Pro Glu Val Arg 180 185 190His Leu Val His Ala
Asp Phe Gly Ser Asn Asn Val Leu Thr Asp Asn 195 200 205Gly Arg Ile
Thr Ala Val Ile Asp Trp Ser Glu Ala Met Phe Gly Asp 210 215 220Ser
Gln Tyr Glu Val Ala Asn Ile Phe Phe Trp Arg Pro Trp Leu Ala225 230
235 240Cys Met Glu Gln Gln Thr Arg Tyr Phe Glu Arg Arg His Pro Glu
Leu 245 250 255Ala Gly Ser Pro Arg Leu Arg Ala Tyr Met Leu Arg Ile
Gly Leu Asp 260 265 270Gln Leu Tyr Gln Ser Leu Val Asp Gly Asn Phe
Asp Asp Ala Ala Trp 275 280 285Ala Gln Gly Arg Cys Asp Ala Ile Val
Arg Ser Gly Ala Gly Thr Val 290 295 300Gly Arg Thr Gln Ile Ala Arg
Arg Ser Ala Ala Val Trp Thr Asp Gly305 310 315 320Cys Val Glu Val
Leu Ala Asp Ser Gly Asn Arg Arg Pro Ser Thr Arg 325 330 335Pro Arg
Ala Lys Glu 340221026DNAEscherichia coli 22atgaagaagc ccgagctgac
cgctacctct gttgagaagt tcctgattga gaagtttgat 60tccgtttccg acctgatgca
gctgtccgag ggcgaggagt ctcgagcctt ctcctttgac 120gtgggcggac
gaggttacgt tctgcgagtg aactcgtgtg ccgacggctt ctacaaggat
180cgatacgtct accgacactt tgcttctgcc gctctgccca tccctgaggt
tctcgacatt 240ggcgagttct ctgagtccct cacctactgc atctctcgac
gagctcaggg agtcaccctg 300caggacctcc ctgagactga gctgcctgct
gtcctccagc ctgttgctga ggccatggac 360gctatcgctg ctgctgatct
gtcccagacc tcgggtttcg gcccctttgg acctcaggga 420attggacagt
acaccacttg gcgagacttc atctgtgcta ttgccgatcc tcacgtctac
480cattggcaga ccgttatgga cgatactgtg tcggcttctg tcgctcaggc
tctggacgag 540ctgatgctct gggccgagga ttgccccgag gttcgacacc
tggtgcatgc tgacttcggt 600tccaacaacg ttctcaccga caacggccga
atcactgccg tgattgactg gtccgaggct 660atgtttggcg actcgcagta
cgaggtggcc aacatcttct tttggcgacc ctggctggct 720tgtatggagc
agcagacccg atacttcgag cgacgacatc ctgagctcgc tggatcccct
780cgactgcgag cttacatgct ccgaattggt ctggaccagc tctaccagtc
gctggtggat 840ggcaactttg acgatgctgc ctgggctcag ggacgatgtg
acgccatcgt gcgatctggc 900gctggaaccg tcggacgaac tcagattgcc
cgacgatccg ctgctgtctg gaccgacgga 960tgcgtggagg tcctggctga
ttcgggtaac cgacgaccct ctactcgacc tcgagctaag 1020gagtaa
10262360DNAArtificial SequenceDescription of Artificial Sequence
Synthetic primer 23cagctctctt cccccgttca gctccttttc taccgcgatt
atgaagaagc ccgagctgac 602424DNAArtificial SequenceDescription of
Artificial Sequence Synthetic primer 24agcatcgtca aagttgccat ccac
242524DNAArtificial SequenceDescription of Artificial Sequence
Synthetic primer 25accacttggc gagacttcat ctgt 242660DNAArtificial
SequenceDescription of Artificial Sequence Synthetic primer
26tttagtctca tcgttagtag ttatgtgctc tgctcggggt tactccttag ctcgaggtcg
6027898DNAArtificial SequenceDescription of Artificial Sequence
Synthetic polynucleotide 27cagctctctt cccccgttca gctccttttc
taccgcgatt atgaagaagc ccgagctgac 60cgctacctct gttgagaagt tcctgattga
gaagtttgat tccgtttccg acctgatgca 120gctgtccgag ggcgaggagt
ctcgagcctt ctcctttgac gtgggcggac gaggttacgt 180tctgcgagtg
aactcgtgtg ccgacggctt ctacaaggat cgatacgtct accgacactt
240tgcttctgcc gctctgccca tccctgaggt tctcgacatt ggcgagttct
ctgagtccct 300cacctactgc atctctcgac gagctcaggg agtcaccctg
caggacctcc ctgagactga 360gctgcctgct gtcctccagc ctgttgctga
ggccatggac gctatcgctg ctgctgatct 420gtcccagacc tcgggtttcg
gcccctttgg acctcaggga attggacagt acaccacttg 480gcgagacttc
atctgtgcta ttgccgatcc tcacgtctac cattggcaga ccgttatgga
540cgatactgtg tcggcttctg tcgctcaggc tctggacgag ctgatgctct
gggccgagga 600ttgccccgag gttcgacacc tggtgcatgc tgacttcggt
tccaacaacg ttctcaccga 660caacggccga atcactgccg tgattgactg
gtccgaggct atgtttggcg actcgcagta 720cgaggtggcc aacatcttct
tttggcgacc ctggctggct tgtatggagc agcagacccg 780atacttcgag
cgacgacatc ctgagctcgc tggatcccct cgactgcgag cttacatgct
840ccgaattggt ctggaccagc tctaccagtc gctggtggat ggcaactttg acgatgct
89828633DNAArtificial SequenceDescription of Artificial Sequence
Synthetic polynucleotide 28accacttggc gagacttcat ctgtgctatt
gccgatcctc acgtctacca ttggcagacc 60gttatggacg atactgtgtc ggcttctgtc
gctcaggctc tggacgagct gatgctctgg 120gccgaggatt gccccgaggt
tcgacacctg gtgcatgctg acttcggttc caacaacgtt 180ctcaccgaca
acggccgaat cactgccgtg attgactggt ccgaggctat gtttggcgac
240tcgcagtacg aggtggccaa catcttcttt
tggcgaccct ggctggcttg tatggagcag 300cagacccgat acttcgagcg
acgacatcct gagctcgctg gatcccctcg actgcgagct 360tacatgctcc
gaattggtct ggaccagctc taccagtcgc tggtggatgg caactttgac
420gatgctgcct gggctcaggg acgatgtgac gccatcgtgc gatctggcgc
tggaaccgtc 480ggacgaactc agattgcccg acgatccgct gctgtctgga
ccgacggatg cgtggaggtc 540ctggctgatt cgggtaaccg acgaccctct
actcgacctc gagctaagga gtaaccccga 600gcagagcaca taactactaa
cgatgagact aaa 63329526PRTYarrowia lipolytica 29Met Glu Val Arg Arg
Arg Lys Ile Asp Val Leu Lys Ala Gln Lys Asn1 5 10 15Gly Tyr Glu Ser
Gly Pro Pro Ser Arg Gln Ser Ser Gln Pro Ser Ser 20 25 30Arg Ala Ser
Ser Arg Thr Arg Asn Lys His Ser Ser Ser Thr Leu Ser 35 40 45Leu Ser
Gly Leu Thr Met Lys Val Gln Lys Lys Pro Ala Gly Pro Pro 50 55 60Ala
Asn Ser Lys Thr Pro Phe Leu His Ile Lys Pro Val His Thr Cys65 70 75
80Cys Ser Thr Ser Met Leu Ser Arg Asp Tyr Asp Gly Ser Asn Pro Ser
85 90 95Phe Lys Gly Phe Lys Asn Ile Gly Met Ile Ile Leu Ile Val Gly
Asn 100 105 110Leu Arg Leu Ala Phe Glu Asn Tyr Leu Lys Tyr Gly Ile
Ser Asn Pro 115 120 125Phe Phe Asp Pro Lys Ile Thr Pro Ser Glu Trp
Gln Leu Ser Gly Leu 130 135 140Leu Ile Val Val Ala Tyr Ala His Ile
Leu Met Ala Tyr Ala Ile Glu145 150 155 160Ser Ala Ala Lys Leu Leu
Phe Leu Ser Ser Lys His His Tyr Met Ala 165 170 175Val Gly Leu Leu
His Thr Met Asn Thr Leu Ser Ser Ile Ser Leu Leu 180 185 190Ser Tyr
Val Val Tyr Tyr Tyr Leu Pro Asn Pro Val Ala Gly Thr Ile 195 200
205Val Glu Phe Val Ala Val Ile Leu Ser Leu Lys Leu Ala Ser Tyr Ala
210 215 220Leu Thr Asn Ser Asp Leu Arg Lys Ala Ala Ile His Ala Gln
Lys Leu225 230 235 240Asp Lys Thr Gln Asp Asp Asn Glu Lys Glu Ser
Thr Ser Ser Ser Ser 245 250 255Ser Ser Asp Asp Ala Glu Thr Leu Ala
Asp Ile Asp Val Ile Pro Ala 260 265 270Tyr Tyr Ala Gln Leu Pro Tyr
Pro Gln Asn Val Thr Leu Ser Asn Leu 275 280 285Leu Tyr Phe Trp Phe
Ala Pro Thr Leu Val Tyr Gln Pro Val Tyr Pro 290 295 300Lys Thr Glu
Arg Ile Arg Pro Lys His Val Ile Arg Asn Leu Phe Glu305 310 315
320Leu Val Ser Leu Cys Met Leu Ile Gln Phe Leu Ile Phe Gln Tyr Ala
325 330 335Tyr Pro Ile Met Gln Ser Cys Leu Ala Leu Phe Phe Gln Pro
Lys Leu 340 345 350Asp Tyr Ala Asn Ile Ser Glu Arg Leu Met Lys Leu
Ala Ser Val Ser 355 360 365Met Met Val Trp Leu Ile Gly Phe Tyr Ala
Phe Phe Gln Asn Gly Leu 370 375 380Asn Leu Ile Ala Glu Leu Thr Cys
Phe Gly Asn Arg Thr Phe Tyr Gln385 390 395 400Gln Trp Trp Asn Ser
Arg Ser Ile Gly Gln Tyr Trp Thr Leu Trp Asn 405 410 415Lys Pro Val
Asn Gln Tyr Phe Arg His His Val Tyr Val Pro Leu Leu 420 425 430Ala
Arg Gly Met Ser Arg Phe Asn Ala Ser Val Val Val Phe Phe Phe 435 440
445Ser Ala Val Ile His Glu Leu Leu Val Gly Ile Pro Thr His Asn Ile
450 455 460Ile Gly Ala Ala Phe Phe Gly Met Met Ser Gln Val Pro Leu
Ile Met465 470 475 480Ala Thr Glu Asn Leu Gln His Ile Asn Ser Ser
Leu Gly Pro Phe Leu 485 490 495Gly Asn Cys Ala Phe Trp Phe Thr Phe
Phe Leu Gly Gln Pro Thr Cys 500 505 510Ala Phe Leu Tyr Tyr Leu Ala
Tyr Asn Tyr Lys Gln Asn Gln 515 520 525301581DNAYarrowia lipolytica
30atggaagtcc gacgacgaaa aatcgacgtg ctcaaggccc agaaaaacgg ctacgaatcg
60ggcccaccat ctcgacaatc gtcgcagccc tcctcaagag catcgtccag aacccgcaac
120aaacactcct cgtccaccct gtcgctcagc ggactgacca tgaaagtcca
gaagaaacct 180gcgggacccc cggcgaactc caaaacgcca ttcctacaca
tcaagcccgt gcacacgtgc 240tgctccacat caatgctttc gcgcgattat
gacggctcca accccagctt caagggcttc 300aaaaacatcg gcatgatcat
tctcattgtg ggaaatctac ggctcgcatt cgaaaactac 360ctcaaatacg
gcatttccaa cccgttcttc gaccccaaaa ttactccttc cgagtggcag
420ctctcaggct tgctcatagt cgtggcctac gcacatatcc tcatggccta
cgctattgag 480agcgctgcca agctgctgtt cctctctagc aaacaccact
acatggccgt ggggcttctg 540cataccatga acactttgtc gtccatctcg
ttgctgtcct acgtcgtcta ctactacctg 600cccaaccccg tggcaggcac
aatagtcgag tttgtggccg ttattctgtc tctcaaactc 660gcctcatacg
ccctcactaa ctcggatctc cgaaaagccg caattcatgc ccagaagctc
720gacaagacgc aagacgataa cgaaaaggaa tccacctcgt cttcctcttc
ttcagatgac 780gcagagactt tggcagacat tgacgtcatt cctgcatact
acgcacagct gccctacccc 840cagaatgtga cgctgtcgaa cctgctgtac
ttctggtttg ctcccacact ggtctaccag 900cccgtgtacc ccaagacgga
gcgtattcga cccaagcacg tgatccgaaa cctgtttgag 960ctcgtctctc
tgtgcatgct tattcagttt ctcatcttcc agtacgccta ccccatcatg
1020cagtcgtgtc tggctctgtt cttccagccc aagctcgatt atgccaacat
ctccgagcgc 1080ctcatgaagt tggcctccgt gtctatgatg gtctggctca
ttggattcta cgctttcttc 1140cagaacggtc tcaatcttat tgccgagctc
acctgttttg gaaacagaac cttctaccag 1200cagtggtgga attcccgctc
cattggccag tactggactc tatggaacaa gccagtcaac 1260cagtacttta
gacaccacgt ctacgtgcct cttctcgctc ggggcatgtc gcggttcaat
1320gcgtcggtgg tggttttctt tttctccgcc gtcatccatg aactgcttgt
cggcatcccc 1380actcacaaca tcatcggagc cgccttcttc ggcatgatgt
cgcaggtgcc tctgatcatg 1440gctactgaga accttcagca tattaactcc
tctctgggcc ccttccttgg caactgtgca 1500ttctggttca cctttttcct
gggacaaccc acttgtgcat tcctttatta tctggcttac 1560aactacaagc
agaaccagta g 158131697PRTRhodosporidium toruloides 31Met Thr Glu
Arg Ser Leu Pro Val Thr Leu Pro Leu Pro Arg Asn Phe1 5 10 15Ala Leu
Thr Pro His Gln Met Ala Ser Pro Asp Pro Pro Leu Pro Gly 20 25 30Pro
Ala Asn Leu Val Asp Asp Ala Leu Arg His Pro Asp Ser Ala Pro 35 40
45Pro Ile Ser Pro Asp Ser Ala Pro Pro Ser Thr Ala Thr Arg Pro Ser
50 55 60Ala Leu Ser Arg Gly Glu Leu Ser Thr Ala Ser Ser Tyr Ala Ser
Glu65 70 75 80Val Ser Thr Arg Glu Gly Thr Pro Asp Leu Ala Asn Gly
Gln Gly Val 85 90 95Thr Thr Thr Ile Thr Thr Val Thr Gly Lys Gly Gly
Lys Ala Val Thr 100 105 110Gln Thr Leu Thr His Val Gly Ala Ala Ser
Val Asp Ala Arg Phe Ser 115 120 125Ser Thr Thr Asn Ser Ile Thr Leu
Arg Pro Ile Pro Ala Arg Gly Gly 130 135 140Asp Pro Lys Lys Ile Lys
Val Leu Arg Ser Arg Arg Thr His Phe Ala145 150 155 160Pro Arg Thr
Ser His Phe Asp Arg His Asn Leu Thr Ser Ala Ser Asp 165 170 175Pro
Phe Arg Gly Leu Tyr Thr Leu Phe Trp Ile Val Ile Phe Val Gly 180 185
190Ala Leu Lys Thr Val Tyr His Arg Phe Ala Glu Gln Gly Gly Trp Gly
195 200 205Gly Glu Trp Arg Phe Ala Ala Leu Ile Ser Arg Asp Gly Trp
Val Leu 210 215 220Ala Val Ser Asp Ala Val Leu Val Ser Ala Ser Leu
Leu Cys Val Pro225 230 235 240Tyr Ala Lys Leu Leu Val His Gly Trp
Ile Arg Tyr His Gly Ala Gly 245 250 255Val Ile Ile Gln His Ile Cys
Gln Thr Leu Tyr Leu Ala Ile Ala Ile 260 265 270Arg Trp Thr Phe His
Arg Asn Trp Pro Trp Val Gln Ser Gly Phe Met 275 280 285Thr Leu His
Ala Leu Ser Met Leu Met Lys Ile His Ser Tyr Cys Ser 290 295 300Leu
Asn Gly Glu Leu Ser Glu Arg Arg Arg Gln Leu Lys Lys Asp Glu305 310
315 320Lys Arg Leu Glu Glu Val Leu Glu Glu Met Gly Gly Arg Arg Lys
Ala 325 330 335Glu Arg Glu Ala Arg Glu Glu Trp Glu Arg Gln Cys Gly
Glu Ala Ala 340 345 350Arg Ala Lys Glu Gly Glu Ala Gly Val Ser Glu
Gly Glu Lys Glu Ala 355 360 365Ala Ala Thr Leu Ser Ser Thr Asp Ala
Ser Asn Ser Ala Leu Ser Ser 370 375 380Glu Asp Glu Ala Ala Ala Ala
Leu Leu Arg His Arg Gln Pro Thr Ala385 390 395 400Arg Arg Arg Ser
Ile Ser Pro Ser Ala Ser Arg Thr Gly Ser Ser Ser 405 410 415Ala Pro
Ser Ala Thr Leu Ala Pro Ser Arg Ala Glu Glu Pro Gln Glu 420 425
430Gly Val Glu Thr Leu Thr Trp His Pro Ser Asp Gln Val Ser Lys Leu
435 440 445Ala Ile Ala Ile Cys Glu Ala Lys Asp Leu Leu Thr Ser Asn
Gly Lys 450 455 460Lys Pro Val Thr Phe Pro Glu Asn Val Thr Phe Ala
Asn Phe Ile Asp465 470 475 480Tyr Leu Leu Val Pro Thr Leu Val Tyr
Glu Leu Glu Tyr Pro Arg Thr 485 490 495Asp Ser Ile Arg Pro Leu Tyr
Ile Leu Glu Lys Thr Leu Ala Thr Phe 500 505 510Gly Thr Phe Ser Ile
Leu Val Leu Ile Val Asp Ser Phe Ile Leu Pro 515 520 525Val Thr Ser
Arg Thr Asp Thr Pro Leu Phe Gly Phe Val Leu Asp Leu 530 535 540Ala
Leu Pro Phe Thr Leu Ala Tyr Leu Leu Ile Phe Tyr Val Ile Phe545 550
555 560Glu Gly Val Cys Asn Gly Phe Ala Glu Leu Thr Arg Phe Ala Asp
Arg 565 570 575Asn Phe Phe Asp Asp Trp Trp Asn Ser Cys Thr Phe Asp
Glu Phe Ser 580 585 590Arg Lys Trp Asn Arg Pro Val His Ala Phe Leu
Leu Arg His Val Tyr 595 600 605Ala Glu Thr Met Ala Ser Tyr Lys Leu
Ser Lys Leu Ser Ala Ala Phe 610 615 620Val Thr Phe Leu Phe Ser Ala
Cys Val His Glu Leu Val Met Ala Val625 630 635 640Val Thr Lys Lys
Leu Arg Leu Tyr Leu Phe Ser Met Gln Met Ala Gln 645 650 655Leu Pro
Leu Ile Met Val Gly Arg Ala Lys Ile Phe Arg Gln Tyr Pro 660 665
670Ala Leu Gly Asn Leu Phe Phe Trp Leu Ala Leu Leu Ser Gly Phe Pro
675 680 685Leu Leu Gly Thr Leu Tyr Leu Arg Tyr 690
695322094DNARhodosporidium toruloides 32atgacggagc gatcccttcc
agtgacgctc cctcttcctc gaaactttgc gctcacaccg 60caccagatgg cctcgccaga
cccgccactc ccaggcccag ccaacctcgt cgacgacgca 120ctccgacacc
cagactcggc gccgcccatc tcgcccgact ccgcgcctcc ttcgactgcg
180actcggccct ctgctctctc gcgcggagag ctctcgaccg cttcgagcta
cgcgagcgag 240gtgtcgacga gggaggggac accggatctg gcgaatgggc
aaggggttac gacgaccatc 300acgactgtca caggcaaagg cggaaaggcc
gtcacccaga ccctcaccca cgtcggcgcc 360gcctccgtcg acgcccgctt
ctcctccacc acaaactcca tcactctccg ccctatcccc 420gcccgtggcg
gcgacccgaa aaagatcaaa gtcctccgct ctcgtcggac ccacttcgcc
480ccacgcacct cacacttcga ccgtcacaac ctcacctccg cctctgaccc
gttccgcgga 540ctgtacacgt tgttctggat cgtgatcttc gttggggcac
tcaagactgt gtatcatcgg 600tttgcggaac agggtgggtg gggtggagaa
tggaggtttg cggcgttgat tagtcgcgat 660gggtgggttc tggcggttag
tgatgcggtg ttggttagcg cgtcgttgtt gtgcgtgccg 720tatgcaaagc
tcctcgtaca cggctggatc cggtaccacg gcgcaggcgt catcatccaa
780cacatctgtc aaacgctcta cctcgccatc gcgatccgct ggaccttcca
ccgcaactgg 840ccctgggtcc aaagcggttt catgaccctc cacgccctct
cgatgctcat gaagatccat 900agctactgtt ctctgaacgg cgagctttcg
gagcggcgga gacagttgaa gaaggacgag 960aagcggttgg aggaggtgct
ggaggagatg ggtggacgga ggaaggcgga gagggaggcg 1020agggaggagt
gggagaggca gtgtggggag gcggcgaggg ccaaggaggg tgaggcggga
1080gtgagcgagg gggagaagga ggcggcggcg actctatctt cgacggatgc
gtcgaattcg 1140gccctttcgt cggaggacga ggcggctgcg gcgctgttgc
ggcatcgaca gccgactgct 1200cgacgacgat ccatctcgcc ctctgcctca
cgcaccggtt cctcctccgc cccctccgct 1260accctcgccc cctctcgcgc
cgaagaaccc caagaaggcg ttgagacgct cacctggcac 1320ccatccgacc
aagtcagcaa actcgctatc gccatctgcg aggcaaagga cctcctcacg
1380agtaacggca agaagcccgt cacgttcccc gagaacgtca cctttgcgaa
ctttatcgac 1440tacttgcttg tgccgacgtt ggtgtacgag ttggagtacc
ctcggacgga ttccatccgg 1500cccctctaca tcctcgaaaa gaccctcgca
accttcggca ccttctccat tctcgtcctc 1560atcgtcgact cgttcatcct
ccccgtcacc tcgcgcaccg acacgcccct cttcgggttc 1620gtcctcgacc
tcgccctgcc gttcacgctc gcgtacctcc tcatcttcta cgtcatcttt
1680gagggcgtgt gcaatgggtt tgcggagttg acgaggtttg cggatcggaa
tttcttcgac 1740gattggtgga actcgtgcac gttcgacgag ttctcgcgca
agtggaatcg ccccgtccac 1800gccttcctcc tccgccacgt ttacgccgaa
acgatggctt cttacaagct ctcgaagctc 1860tcggctgcgt tcgtcacgtt
cttgttcagc gcctgcgtgc acgaactcgt catggcggtc 1920gtgacgaaga
agcttcggct gtacctgttc tcgatgcaga tggcccagct cccgctcatc
1980atggtgggcc gcgccaagat cttccgacag tatccagcgc tcggcaacct
cttcttctgg 2040ctcgcccttc tctcgggatt cccgcttctc gggacgctgt
atctgcggta ctga 209433555PRTLipomyces starkeyi 33Met Ser Thr Ala
Ala Gln Ser Asp Thr Asp Asn Glu Asp Ile Ser Thr1 5 10 15Val Asp Leu
Val Asp Ser Arg Ala Asp Thr His Thr Ser Ser Asn Val 20 25 30Met Leu
Gln Gln Gln Lys Ser Arg Arg Arg Leu Ile Gly Lys Asp Ala 35 40 45Glu
Pro Arg Thr Gln His Pro Ser Gly Gly Lys Ser Glu Lys Glu Glu 50 55
60Leu Thr Lys Pro Asp Asp Ser Lys Gly Pro Ile Lys Leu Ser His Ile65
70 75 80Tyr Pro Ile His Ala Val Ser Arg Gly Ser Ile Leu Ser Arg Glu
Ser 85 90 95Thr Thr Pro Thr Pro Ser Phe Val Gly Phe Arg Asn Leu Ala
Met Ile 100 105 110Val Leu Gly Lys Leu Gln Tyr Ser Leu Phe Phe Trp
Cys Asp Arg Ala 115 120 125Asn Ile Pro Thr Ala Val Ser Asn Leu Arg
Leu Val Ile Glu Asn Tyr 130 135 140Ser Lys Tyr Gly Val Leu Ile Arg
Phe Ala Arg Leu Gly Ile Ser Gln145 150 155 160Lys Asp Ile Leu Tyr
Cys Ile Phe Leu Thr Ala Thr Ile Pro Leu His 165 170 175Leu Phe Ile
Ala Ile Val Ile Glu Arg Leu Val Ala Ile Pro Thr Val 180 185 190Asn
Tyr Val Ala Ser Leu Ser Glu Ser Glu Asp Lys Lys Arg Ser Asn 195 200
205Pro Lys Met Gly Arg Lys Gly Gly Ser Ile Ser Ile Leu Arg Pro Lys
210 215 220Pro Lys Tyr Met Trp Arg Leu Ile Val Leu Leu His Ser Ile
Asn Ala225 230 235 240Met Ala Cys Leu Trp Val Thr Thr Val Val Val
Tyr Asn Ser Ile Tyr 245 250 255His Pro Leu Ile Gly Thr Ala Cys Glu
Phe His Ala Val Ile Val Cys 260 265 270Leu Lys Val Ala Ser Phe Ala
Leu Thr Asn Arg Asp Leu Arg Glu Ser 275 280 285Met Leu Asn Ser Gln
Pro Val Pro Ala Ile Tyr Asn Leu Ala Pro Tyr 290 295 300Pro Lys Asn
Leu Thr Leu Lys Asn Leu Ser Tyr Phe Trp Trp Ala Pro305 310 315
320Thr Leu Val Tyr Gln Pro Val Tyr Pro Arg Ser Pro Ser Phe Arg Pro
325 330 335Leu Phe Phe Val Lys Arg Ile Leu Glu Met Val Gly Leu Ser
Phe Leu 340 345 350Ile Trp Phe Leu Ser Ala Gln Tyr Ala Val Pro Thr
Leu Glu Asn Ser 355 360 365Leu Val His Phe His Ser Leu Gln Phe Met
Gly Ile Met Glu Arg Leu 370 375 380Met Lys Leu Ala Ser Ile Ser Met
Ala Ile Trp Leu Ala Gly Phe Phe385 390 395 400Cys Ile Phe Gln Ser
Gly Leu Asn Ala Leu Ala Glu Val Met Arg Phe 405 410 415Gly Asp Arg
Ala Phe Tyr Asp Asp Trp Trp Asn Ser Lys Ser Val Gly 420 425 430Glu
Tyr Trp Arg Leu Trp Asn Lys Pro Val Thr Asn Tyr Phe Arg Arg 435 440
445His Ile Tyr Val Pro Leu Val Arg Arg Gly Trp Asn Ser Ala Thr Ala
450 455 460Ser Val Met Val Phe Phe Val Ser Ala Val Leu His Glu Leu
Val Val465 470 475 480Gly Val Pro Thr His Asn Val Ile Gly Val Ala
Phe Ser Ser Met Ile 485 490 495Leu Gln Ile Pro Leu Ile Gln Val Thr
Ala Pro Leu Glu Lys Met His 500 505 510Gly Pro Thr Ser Gly Ile Ile
Gly Asn Cys Ile Phe Trp Phe Ser Phe 515 520 525Phe Ile Gly Gln Pro
Leu Gly Val Leu Leu Tyr Tyr Phe Ala Trp Asn 530 535 540Val Ser Met
Ser Lys Val Lys Met Val Glu Ser545 550
555341668DNALipomyces starkeyi 34atgtcgaccg ctgcacaatc tgatacagac
aacgaggata tatcgactgt cgatttggtt 60gactctcgtg cagatactca cacatcttca
aatgttatgt tgcaacagca aaaatcgcgt 120cggagactaa tcgggaaaga
cgccgagcca agaacacagc atccgtctgg aggcaaatcg 180gagaaggagg
agttgacgaa gccggatgac tcaaagggac ccataaaatt aagtcacata
240tacccgatac atgccgttag ccgaggcagt attctgtcac gagagtcgac
aactcctaca 300ccgagttttg ttgggtttcg aaacttagcc atgatagtgc
tagggaagtt acagtattca 360ttattctttt ggtgcgatcg ggctaacatt
ccgacagccg tcagcaatct tcgattggtg 420attgaaaatt actcaaagta
cggcgttctg atccgattcg cccgactcgg tatttcacaa 480aaggacattc
tgtattgcat attcttgacc gctaccatcc cgctgcacct atttattgct
540attgtcattg aaagactagt tgcgattccg acggtaaact acgtcgcttc
gctcagcgag 600agcgaggata aaaaacgctc caaccccaaa atgggacgga
aggggggcag tatatcgatt 660ttgcgtccta agccaaaata tatgtggcgc
ctgatcgtcc tattgcattc aataaacgca 720atggcttgct tgtgggttac
gactgttgtt gtttacaatt ctatttatca tccccttatt 780gggacagctt
gtgaatttca tgcagtgatt gtgtgtctta aggtcgcatc gtttgcgctt
840accaatcgcg atcttcggga gtcgatgctg aactctcaac ctgtgccagc
catatacaac 900ttggcccctt atccaaaaaa cttaaccctc aagaacttgt
catacttttg gtgggcgccg 960actcttgttt atcaacctgt ctatccgcga
tcgccttcat tccggccttt gttttttgtc 1020aagcggattc tggagatggt
gggcctatca tttttaatat ggttcttgtc agctcaatat 1080gctgtgccga
cgctagaaaa tagtttggtg cattttcaca gtttgcaatt catgggaatt
1140atggagcgac tcatgaagct tgctagcatt agcatggcta tttggcttgc
tggttttttc 1200tgcatttttc agtctggact caatgcgctt gcggaggtaa
tgcggtttgg tgacagagcc 1260ttttacgacg actggtggaa cagcaaatct
gtgggagagt attggcgtct gtggaataag 1320ccggttacga attacttccg
gcgtcatatt tacgtaccgc ttgtgcgccg cgggtggaat 1380tctgcgacag
ccagtgtcat ggtatttttc gtcagcgcgg tgttgcatga gctagttgtt
1440ggagttccga cgcataacgt aattggagtt gcattctcgt cgatgattct
acaaatccca 1500ctcatacaag taaccgcgcc tctggagaag atgcatggac
ctacatctgg aataataggg 1560aactgtatct tttggtttag cttcttcatc
ggtcagcctc tgggcgtgct actttactat 1620tttgcgtgga acgttagtat
gagcaaagta aagatggtcg agagctag 166835517PRTAspergillus terreus
35Met Val Met Asp Thr Gln Thr Thr Ala Ser Ala Thr Ser Thr Ala Leu1
5 10 15Thr Thr Asp His Thr Val Ala Ser Arg Thr Ser Arg Ser Glu Pro
Asn 20 25 30Gly Gly Val His Asn Val Ser Ser Pro Pro Thr Ser Glu Pro
Thr Gly 35 40 45Gly Asn Gly Gly Gly Arg Arg Arg Ser Lys Tyr Arg His
Val Ala Ala 50 55 60Tyr His Ser Glu Val Arg His Ser Ser Leu Ser Arg
Glu Ser Asn Thr65 70 75 80Ser Pro Ser Phe Leu Gly Phe Arg Asn Leu
Met Val Ile Val Leu Gly 85 90 95Glu Cys Pro Ser Ala Leu Leu Arg Phe
Val Asn Pro Thr Glu Asn Ser 100 105 110Tyr Gly Ser Arg Leu Val Ala
Met Asn Leu Arg Leu Val Ile Glu Asn 115 120 125Tyr Val Lys Tyr Gly
Val Leu Ile Cys Ile Arg Cys His Asp Tyr Arg 130 135 140Lys Gln Asp
Val Val Leu Gly Ser Met Leu Phe Ala Leu Val Pro Cys145 150 155
160Gln Leu Phe Ile Ala Tyr Leu Leu Glu Leu Ala Ala Ala Gly Arg Ala
165 170 175Lys Gln Thr Val Gly Arg Lys Lys Lys Asp Gly Ser Ala Glu
Glu Gly 180 185 190Glu Arg Glu Ala Arg Ala Phe Arg His Ile Trp Arg
Phe Ala Leu Ser 195 200 205Phe His Ile Leu Asn Ile Val Leu Asn Leu
Ala Val Thr Ser Phe Val 210 215 220Val Tyr Tyr Tyr Ile His His Pro
Gly Ile Gly Thr Leu Cys Glu Val225 230 235 240His Ala Ile Val Val
Ala Leu Lys Asn Trp Ser Tyr Ala Phe Thr Asn 245 250 255Arg Asp Leu
Arg Glu Ala Met Leu Asn Pro Ser Ala Glu Ser Ala Leu 260 265 270Pro
Glu Ile Tyr Ser Ser Leu Pro Tyr Pro Lys Asn Ile Thr Leu Gly 275 280
285Asn Leu Thr Tyr Phe Trp Leu Ala Pro Thr Leu Leu Tyr Gln Pro Val
290 295 300Tyr Pro Arg Ser Pro Ser Ile Arg Trp Pro Phe Val Ala Lys
Arg Leu305 310 315 320Ser Glu Phe Ala Cys Leu Ser Val Phe Ile Trp
Leu Leu Ser Ala Gln 325 330 335Tyr Ala Ala Pro Val Leu Arg Asn Ser
Ile Asp Lys Ile Arg Asp Met 340 345 350Ala Tyr Ala Ser Ile Phe Glu
Arg Val Met Lys Leu Ser Thr Ile Ser 355 360 365Leu Val Ile Trp Leu
Ala Gly Phe Phe Ala Ile Phe Gln Ser Leu Leu 370 375 380Asn Ala Leu
Ala Glu Ile Met Lys Phe Gly Asp Arg Glu Phe Tyr Thr385 390 395
400Asp Trp Trp Asn Ser Pro Ser Leu Gly Val Tyr Trp Arg Ser Trp Asn
405 410 415Arg Pro Val Tyr Gln Phe Met Lys Arg His Val Tyr Ser Pro
Leu Ile 420 425 430Gly Arg Gly Tyr Ser Pro Phe Val Ala Ser Thr Val
Val Phe Thr Ile 435 440 445Ser Ala Leu Leu His Glu Leu Leu Val Gly
Ile Pro Thr His Asn Met 450 455 460Ile Gly Val Ala Leu Val Gly Met
Leu Phe Gln Leu Pro Leu Ile Ala465 470 475 480Ile Thr Ala Pro Leu
Glu Lys Met Lys Asp Pro Leu Gly Lys Pro Leu 485 490 495Gly Ala Leu
Leu Tyr Phe Phe Ala Trp Gln Ala Lys Tyr Gly Ser Val 500 505 510Ser
Arg Met Gly Asn 515361554DNAAspergillus terreus 36atggtgatgg
acacacaaac cacagcatcc gccaccagca cggcgctcac gaccgaccac 60actgttgcct
ctcggacgtc ccgctctgag ccgaacggtg gtgtgcataa tgtatcgtca
120cctccaacga gcgaaccgac tgggggaaat ggcggaggcc ggcgaaggag
taaataccgg 180catgtcgcag cgtaccattc cgaagtgcgc cattccagtc
tcagtcggga atcgaatact 240tctccgagtt tcctcggatt ccggaacctc
atggtaatcg tattaggtga gtgccctagt 300gctctcctac gttttgtgaa
cccgacggag aactcatacg ggtcgcgact agttgctatg 360aatcttcgat
tggttatcga gaattacgtg aagtatgggg tcttgatctg catcagatgc
420cacgattatc gaaagcagga cgttgtcctg ggctcaatgt tatttgctct
cgtcccatgc 480cagctattca tcgcctacct cctggaattg gccgcagcgg
gtagggccaa acagactgtg 540ggccgaaaga aaaaggacgg atcagccgag
gagggcgaac gtgaagcacg tgcttttcga 600cacatctggc ggtttgcatt
gtcctttcac atcctcaaca ttgttctcaa tctcgccgtc 660acgagcttcg
ttgtgtatta ctacatccac catcccggca ttggtacgct ctgtgaagtg
720catgcgatcg ttgtcgcgtt gaaaaactgg tcctatgcgt tcaccaatcg
ggatctgcga 780gaggcgatgc ttaatccctc ggcggagtcg gcgcttcccg
agatctattc cagcctcccg 840tacccgaaaa acatcacgtt aggaaatcta
acgtacttct ggcttgcacc gacactgttg 900tatcagccag tataccccag
gtcgccttcc atccgatggc cattcgtggc caaacgcttg 960tcggaatttg
cgtgcttgtc ggtgttcatt tggctacttt cggcccaata cgctgcgcca
1020gttttgcgca actccattga caagattcgt gatatggcat atgcatccat
ttttgagcgc 1080gttatgaagc tatccaccat ctctctcgtc atttggctgg
ctgggttctt tgcgattttc 1140caatcactct tgaatgcttt ggcggagatc
atgaagtttg gcgatcggga attctacacc 1200gattggtgga atagcccaag
tctcggtgtt tactggcggt catggaatcg gccagtgtac 1260cagttcatga
agcggcacgt atattctccg ttgatagggc gggggtacag cccgtttgtg
1320gcaagcactg tcgtattcac catctccgct ctccttcatg agctcctcgt
ggggataccc 1380acgcacaaca tgataggcgt cgcgcttgtt ggaatgctgt
tccagctccc gttgatcgcc 1440atcactgccc cattggaaaa gatgaaagat
ccattgggta agcccctggg agcactgctg 1500tatttctttg cctggcaggc
aaaatatggc agtgtgagca ggatgggcaa ctga 155437506PRTClaviceps
purpurea 37Met Ser Ala Thr Gly Val Asp Val Ala Asn Gly Arg Ser Gly
Ala Arg1 5 10 15Arg Arg Asn Asp Thr Ala Val Asp Glu Thr Ile Ser Ala
Val Thr Ala 20 25 30Glu Met Arg Ser Ser Ser His Pro Thr Tyr Arg His
Val Ser Ala Val 35 40 45His Ser Thr Ser Arg Pro Ser Cys Leu Ser His
Asp Ser Asp Ala Ala 50 55 60Pro Ser Phe Ile Gly Phe Arg Asn Leu Met
Val Ile Val Leu Val Val65 70 75 80Gly Asn Val Arg Leu Met Ile Glu
Asn Leu Lys Lys Tyr Gly Val Leu 85 90 95Ile Cys Leu Arg Cys His Ser
Tyr Lys Asn Glu Asp Ile Ile Ile Gly 100 105 110Gly Leu Leu Tyr Phe
Leu Ile Pro Cys His Leu Leu Val Ala Tyr Gly 115 120 125Ile Glu Leu
Ala Ala Ala Arg Gln Ala Arg Glu Ser Arg Thr Arg Pro 130 135 140Pro
Gly Gln Ser Asp Thr Ala Ser Lys Ser Thr Glu Asp Asp Asn Lys145 150
155 160His Phe His Ser Thr Trp Val Leu Ala Ala Trp Ala His Ile Ile
Asn 165 170 175Met Thr Leu Ser Phe Ile Leu Thr Thr Phe Val Val Tyr
Tyr Tyr Val 180 185 190His His Pro Leu Val Gly Thr Leu Thr Glu Met
His Ala Val Ile Val 195 200 205Ser Leu Lys Thr Ala Ser Tyr Ala Phe
Thr Asn Arg Asp Leu Arg His 210 215 220Ala Tyr Leu His Pro Asp Lys
Arg Lys His Ile Pro Glu Leu Tyr Leu225 230 235 240Glu Cys Pro Tyr
Pro Gln Asn Leu Thr Phe Gly Asn Leu Val Tyr Phe 245 250 255Trp Trp
Ala Pro Thr Leu Val Tyr Gln Pro Val Tyr Pro Arg Thr Asp 260 265
270Lys Ile Arg Trp Val Phe Val Phe Lys Arg Leu Gly Glu Val Cys Cys
275 280 285Leu Ser Ala Phe Ile Trp Phe Ala Ser Phe Gln Tyr Ala Ala
Pro Val 290 295 300Leu Arg Asn Ser Leu Asp Lys Ile Ala Ser Leu Asp
Phe Ile Met Ile305 310 315 320Phe Glu Arg Leu Leu Lys Leu Ser Thr
Ile Ser Leu Val Ile Trp Leu 325 330 335Ala Gly Phe Phe Ala Leu Phe
Gln Ser Phe Leu Asn Ala Leu Ala Glu 340 345 350Val Leu Arg Phe Gly
Asp Arg Cys Phe Tyr Asp Asp Trp Trp Asn Ser 355 360 365Glu Ser Leu
Gly Ala Tyr Trp Arg Thr Trp Asn Arg Pro Val Tyr Thr 370 375 380Tyr
Phe Lys Arg His Val Tyr Val Pro Met Ile Gly Arg Gly Trp Ser385 390
395 400Pro Trp Thr Ala Ser Cys Thr Val Phe Phe Val Ser Ala Val Leu
His 405 410 415Glu Val Leu Val Gly Val Pro Thr His Asn Ile Ile Gly
Val Ala Phe 420 425 430Val Gly Met Phe Leu Gln Leu Pro Leu Ile Ala
Leu Thr Ala Pro Met 435 440 445Glu Lys Lys Lys Trp Gly His Thr Gly
Arg Val Met Gly Asn Val Ile 450 455 460Phe Trp Val Ser Phe Thr Ile
Phe Gly Gln Pro Phe Ala Ala Leu Met465 470 475 480Tyr Phe Tyr Ala
Trp Gln Ala Lys Tyr Gly Ser Val Ser Arg Gln Ile 485 490 495Val Leu
Val Asn Pro Val Glu Glu Ala Ser 500 505381521DNAClaviceps purpurea
38atgtccgcca cgggcgttga tgtggccaac ggccgcagcg gcgcgcgacg acgcaacgat
60actgccgtcg acgagactat atccgccgtc acggccgaga tgcgttcctc gtcgcatcca
120acataccgcc atgtgtctgc tgtgcactcc acgagccggc cctcgtgtct
gagccatgat 180tctgacgctg cgccgagctt cattggcttt cgaaatctca
tggtcattgt tctggtcgtt 240ggcaatgttc gattaatgat tgaaaatcta
aaaaagtacg gcgtactgat atgcctccga 300tgtcactcgt ataaaaacga
agacatcatt atcggcggac tgctctactt cctgatcccc 360tgccacttgc
ttgtcgccta cggaatcgag ttagccgccg ccagacaagc acgcgaatct
420cgaactcgtc caccaggcca gtccgacacg gcgtcgaaat caacagaaga
tgacaacaag 480cacttccact caacatgggt gctcgctgcc tgggcacaca
tcatcaacat gacactttcc 540ttcatcctca ccaccttcgt cgtctactac
tacgtgcacc atcccctcgt cggcaccctg 600accgagatgc acgccgtcat
cgtctctctc aaaacagctt cctacgcatt caccaaccga 660gatcttcgcc
acgcatacct ccatcctgac aagcgcaagc acatccccga gctatatctc
720gaatgtccct acccccagaa cctcaccttt ggcaatctcg tgtatttctg
gtgggccccc 780acgctggtat accagcccgt gtatccgcgc accgacaaga
tcagatgggt ttttgttttt 840aaaagactag gcgaagtctg ctgtctcagc
gcattcatct ggttcgccag cttccagtac 900gccgcgcccg tgttgcggaa
ctccctggac aagattgcgt ctctcgactt catcatgatc 960tttgagcgcc
ttctcaagct atccaccatt tctctcgtca tctggctcgc cggcttcttc
1020gccctgttcc agtctttcct gaatgccctg gctgaggtat tgcgctttgg
ggaccggtgc 1080ttctacgacg attggtggaa tagcgagagt ctgggggcgt
attggaggac gtggaacagg 1140cctgtgtata cctacttcaa gcgccatgtg
tatgtgccca tgattgggag gggatggagt 1200ccctggactg ctagttgtac
tgtttttttt gtgtcggcgg tgctgcacga ggttcttgtt 1260ggggtgccca
cccacaatat cattggtgtc gcctttgtgg gcatgtttct gcagcttccc
1320ctaatagccc tcaccgctcc catggaaaag aagaaatggg gccacaccgg
ccgtgtgatg 1380ggcaatgtta ttttctgggt gtcctttaca atctttgggc
agccctttgc agcgctcatg 1440tacttttatg cctggcaggc caagtacggg
agcgtgagtc ggcaaattgt gctggtgaat 1500ccggtggagg aggcgtcttg a
152139551PRTChaetomium globosum 39Met Lys Ala Glu Thr Gly Thr Thr
Met Ala Thr Ser Thr Ser Leu Glu1 5 10 15Thr Ser Gln Val Asn Gly Val
Thr Asn Arg Ala Pro Val Gly Pro Ser 20 25 30His Asp Pro His Ala Thr
Thr Pro Thr His Glu Thr Thr Thr Thr Ile 35 40 45Pro Ser Asp Val Leu
Ala Asn Gly Ser Thr Asn Gly Thr Thr Asn Gly 50 55 60Thr Thr Asp Asp
Ser Leu Asp Ile Ser Glu Leu Arg Lys Ala Phe Arg65 70 75 80Asn Lys
Tyr Arg His Val Glu Ala Val His Ser Glu Ser Lys Pro Ser 85 90 95Cys
Leu Ser His Asp Ala Thr Glu Thr Pro Ser Phe Ile Gly Phe Arg 100 105
110Asn Leu Met Val Ile Val Leu Val Ala Ala Asn Leu Arg Leu Val Ile
115 120 125Glu Asn Ile Gln Lys Tyr Gly Val Leu Ile Cys Ile Lys Cys
His Asp 130 135 140Phe Arg Pro Asn Asp Val Arg Leu Gly Leu Leu Leu
Tyr Ile Leu Ile145 150 155 160Pro Trp His Leu Met Leu Ala Tyr Leu
Ile Glu Leu Val Ala Ala Ala 165 170 175Asn Ala Arg Asn Ser Arg Ala
Lys Ala Lys Lys Arg Asp Gly Ser Thr 180 185 190Ser Pro Thr Glu Asp
Glu Ser Lys Gln Phe Leu Gln Thr Trp Arg Met 195 200 205Leu Arg Ile
Leu His Ala Val Asn Val Thr Ala Ala Leu Ala Val Thr 210 215 220Ser
Tyr Val Val Tyr Tyr Tyr Ile His His Pro Leu Ile Gly Thr Leu225 230
235 240Ser Glu Leu His Ala Ile Ile Val Trp Leu Lys Thr Ala Ser Tyr
Ala 245 250 255Leu Thr Asn Arg Asp Leu Arg His Ala Tyr Leu His Pro
Val Arg Gly 260 265 270Glu Arg Asp Ala Leu Pro Glu Ile Tyr Ala Gln
Cys Pro Tyr Pro Ala 275 280 285Asn Val Thr Phe Ser Asn Leu Thr Tyr
Phe Trp Trp Ala Pro Thr Leu 290 295 300Val Tyr Gln Pro Ala Tyr Pro
Arg Thr Gln Arg Ile Arg Trp Val Phe305 310 315 320Val Ala Lys Arg
Leu Gly Glu Val Val Cys Leu Ser Ala Phe Ile Trp 325 330 335Phe Ala
Ser Ala Gln Tyr Ala Thr Pro Val Leu Arg Asn Ser Leu Asp 340 345
350Lys Ile Ala Thr Leu Asp Tyr Met Ser Ile Val Glu Arg Leu Leu Lys
355 360 365Leu Ser Thr Ile Ser Leu Val Ile Trp Leu Ala Gly Phe Phe
Ala Leu 370 375 380Phe Gln Ser Phe Leu Asn Ala Leu Ala Glu Val Met
Arg Phe Gly Asp385 390 395 400Arg Glu Phe Tyr Glu Ala Trp Trp Asn
Ser Glu Ser Leu Gly Ala Tyr 405 410 415Trp Arg Thr Trp Asn Lys Pro
Val Tyr Gln Phe Phe Arg Arg His Val 420 425 430Tyr Ser Pro Met Arg
Ser Arg Gly Trp Ser His Leu Ser Ala Ser Leu 435 440 445Ala Val Phe
Leu Leu Ser Ala Val Leu His Glu Leu Leu Val Gly Val 450 455 460Pro
Thr His Asn Ile Ile Gly Val Ala Phe Leu Gly Met Phe Leu Gln465 470
475 480Leu Pro Leu Ile Ala Met Thr Ala Arg Leu Gly Gly Arg Arg Gly
Asn 485 490 495Thr Ala His Gly Arg Leu Leu Gly Asn Thr Ile Phe Trp
Val Ser Phe 500 505 510Thr Ile Phe Gly Gln Pro Phe Ala Ala Leu Met
Tyr Phe Tyr Ala Trp 515 520 525Gln Ala Lys Tyr Gly Ser Val Ser Lys
Met Pro Leu Ala Gln Pro Gly 530 535 540Thr Cys Pro Ala Val Val
Val545 550401656DNAChaetomium globosum 40atgaaggcag aaacgggcac
aacgatggca acgtcgacta gtctcgagac ttcccaagtc 60aatggcgtca ccaaccgggc
ccctgttggc cctagtcacg acccccacgc tacaactccg 120actcatgaga
cgacaaccac cataccgtcc gacgtcctcg ccaatggttc tacaaatggg
180actacgaatg ggacgacaga tgattcattg gacatatccg aattgcgcaa
agcgttccgc 240aacaagtatc gccatgtcga ggctgtccac tccgaatcga
aaccatcctg tctgagccat 300gacgctacag agacacccag tttcatcggt
tttaggaatc tcatggtgat tgtgttggtt 360gctgccaatc ttcgcctggt
catcgagaac attcaaaagt atggagttct gatctgcatc 420aaatgccacg
actttcgccc caacgatgta cgcctggggc tcctcctcta
catcctgatc 480ccatggcacc tcatgctcgc ctacctcatt gagctggtcg
ccgccgccaa tgcccgcaac 540tcccgggcca aggcgaagaa gcgggacggc
agtaccagcc cgaccgaaga cgagtccaag 600caattcctgc agacctggcg
gatgctccgc attctccacg ccgtcaacgt cacggccgcc 660ctggccgtca
cctcctacgt ggtctactac tacattcacc acccgctgat cggcacgctc
720tcggagctgc acgccatcat cgtgtggctc aagacggcgt cgtacgcgct
caccaaccgc 780gacctgcgcc acgcctacct acacccggtg cgcggcgagc
gcgacgctct gcccgagatc 840tacgcccagt gcccctaccc ggccaacgtg
accttctcca acttgaccta cttctggtgg 900gcgcccaccc tggtgtacca
gccggcgtac ccgcgcactc agcgcatccg ctgggtcttt 960gtggctaagc
gcctcggcga ggtcgtctgc ttgagcgcct tcatctggtt cgccagcgcc
1020cagtacgcta cccccgtgct gcgaaactcg ctcgacaaga tcgctaccct
ggattacatg 1080tccattgtcg agcgtctgtt gaagctgtcg accatctcgc
tggtcatctg gctggcgggc 1140ttctttgcgc tgtttcagag tttcctgaat
gccttggccg aggtgatgcg gtttggagac 1200cgcgagttct acgaagcatg
gtggaacagc gaaagcctcg gcgcctactg gcgcacctgg 1260aacaaacccg
tgtaccaatt cttccggcgg cacgtctact cgccgatgcg gtcgcgcggg
1320tggagccact tgtcggccag cctcgccgtg tttctgctct cggccgtgct
acacgagctg 1380ctggtggggg tgccgacgca caacatcatc ggcgtcgcct
tcctgggcat gttcctgcag 1440ctgccgctca tcgccatgac ggcgcgcctg
ggcggccgcc gcgggaacac cgcccacggc 1500cgcctgctcg gcaacactat
cttttgggtg tcatttacca tttttggcca gccgtttgcc 1560gcgctgatgt
atttttatgc atggcaggcc aagtatggta gtgtgagcaa gatgccgctg
1620gcgcagccgg ggacgtgtcc ggctgtggtt gtttga 165641388PRTArxula
adeninivorans 41Met Val Arg Phe Ala Pro Leu Asn Val Pro Leu His Arg
Arg Leu Glu1 5 10 15Thr Phe Ala Leu Thr Tyr His Ile Leu Ser Ile Pro
Val Trp Met Ser 20 25 30Phe Phe Leu Leu Cys Cys Ala Ile Pro Leu Met
Trp Pro Leu Val Ile 35 40 45Ile Tyr Leu Leu Tyr Tyr Ala Ser Asp Asn
Ser Ser Glu Asn Gly Gly 50 55 60Val Ala Ser Arg Tyr Ser Pro Lys Phe
Arg Ser Val Pro Leu Trp Lys65 70 75 80Tyr Phe Ala Asn Tyr Phe Pro
Ile Thr Leu His Arg Thr Gln Glu Leu 85 90 95Pro Pro Ala Phe Val Tyr
Gln Gly Glu Asp Leu Asp Pro Glu Thr Pro 100 105 110Asp Asp Ser Asp
Asp Gly His Ala Lys Ser Lys Ser Ile Val Leu Lys 115 120 125Leu Trp
Lys Val Ala Phe Trp Trp Tyr Tyr Leu Pro Lys His Phe Leu 130 135
140Arg Lys Pro Glu Val Arg Pro Thr Gly Arg Arg Tyr Ile Phe Gly
Tyr145 150 155 160His Pro His Gly Ile Ile Gly Met Gly Ala Ile Gly
Ala Ile Ala Thr 165 170 175Glu Gly Ala Gly Trp Ser Lys Leu Phe Pro
Gly Ile Pro Val Ser Leu 180 185 190Leu Thr Leu Ala Asn Asn Phe Arg
Ile Pro Leu Tyr Arg Glu Tyr Leu 195 200 205Met Ser Leu Gly Ile Ala
Ser Val Ser Arg Arg Ser Cys Glu Ala Leu 210 215 220Leu Lys Arg Gly
Gln Ser Ile Cys Ile Val Ile Gly Gly Ala Gln Glu225 230 235 240Ser
Leu Leu Ala His Pro Gly His Met Asp Leu Val Leu Lys Arg Arg 245 250
255Lys Gly Phe Ile Lys Leu Ala Leu Glu Val Gly Asn Thr Asp Leu Val
260 265 270Pro Val Met Ala Phe Gly Glu Asn Asp Leu Tyr Gln Gln Val
Asn Ser 275 280 285Ser Lys Ser Ser Arg Leu Tyr Lys Leu Gln Ser Leu
Val Lys Asn Ala 290 295 300Leu Gly Phe Thr Leu Pro Leu Met His Ala
Arg Gly Val Phe Asn Tyr305 310 315 320Asp Val Gly Ile Ile Pro Tyr
Arg Arg Pro Ile Asn Val Val Val Gly 325 330 335Lys Pro Ile Pro Ile
Pro His Ile Pro Asn Pro Ser Ala Asp Gln Val 340 345 350Asn Arg Tyr
Gln Ile Gln Tyr Met Thr Glu Leu Lys Glu Leu Tyr Asp 355 360 365Lys
Tyr Lys Asp Lys Cys Ser Asn Lys Asp Leu Pro Val Pro Glu Leu 370 375
380Thr Phe Val Glu385421167DNAArxula adeninivorans 42atggttcggt
tcgctccttt aaatgttcct cttcatcgga ggttagagac gttcgcgctc 60acctaccata
tcctgtcgat tccagtatgg atgtccttct ttttgctatg ctgtgccatt
120cctttaatgt ggccgttggt tatcatctac ctgctgtact atgcttccga
caacagctct 180gagaatggag gggttgcgag caggtattcg ccaaagttca
ggtccgtgcc tctttggaag 240tactttgcaa actactttcc aatcaccctt
caccgtactc aagagctacc gcccgcattc 300gtgtaccaag gcgaagactt
ggaccctgag acgcccgatg acagtgacga cgggcatgca 360aagtcaaagt
ctattgtatt aaagctgtgg aaagttgcat tctggtggta ctacttgccc
420aagcattttc ttcgcaaacc agaggttcgt cctacgggtc gaagatacat
ctttggatat 480cacccccatg gaatcattgg catgggtgcc attggcgcaa
ttgctactga aggtgcgggg 540tggtccaagc tcttccccgg gatccctgtc
agtttgctca ctctggcaaa caactttcga 600atccccctgt accgggaata
tctcatgtct ctgggcattg cctcggtatc tagacggtcc 660tgtgaagctt
tattaaaaag aggacagtca atttgcattg taattggagg cgctcaggaa
720agtcttcttg cacatccagg gcacatggat ttggtgctca agcgacgcaa
gggattcatt 780aaactagctc ttgaagttgg caacaccgac ttggtgccag
ttatggcatt tggagaaaac 840gatctctacc agcaagtgaa cagtagcaaa
tcctcccgtc tatacaagct ccagagccta 900gttaagaatg ccttgggatt
cacgcttccg ctgatgcacg ctcgaggagt gttcaattat 960gacgtgggca
taatacccta tcgaagacca attaacgttg tagtgggcaa gcccatcccc
1020attccacaca ttccaaaccc atctgccgac caggtcaatc ggtaccagat
ccagtacatg 1080actgaactca aagaattgta cgacaagtac aaagacaagt
gcagtaacaa ggatcttcca 1140gttccggagc ttacatttgt agagtag
116743346PRTRhodotorula graminis 43Met Gly Ala Gln Glu Glu Val Asp
Tyr Asp Gln Ser Asp His Thr Lys1 5 10 15Ile Lys Phe Val Pro Phe Val
Val Pro Arg His Arg Arg Leu Gln Thr 20 25 30Phe Ser Val Phe Leu Trp
Thr Thr Ala Leu Pro Ile Ser Leu Gly Ile 35 40 45Phe Cys Ile Leu Cys
Ser Phe Pro Pro Leu Trp Pro Leu Val Ile Gly 50 55 60Tyr Leu Thr Trp
Val Phe Leu Ile Asp Gln Ala Pro Met Arg Gly Gly65 70 75 80Arg Pro
Gln Ala Trp Leu Arg Lys Ser Arg Val Trp Glu Trp Phe Ala 85 90 95Gly
Tyr Tyr Pro Val Ser Leu Ile Lys Ser Ala Asp Leu Pro Pro Asp 100 105
110Gln Arg Tyr Val Phe Gly Tyr His Pro His Gly Val Ile Gly Met Gly
115 120 125Ala Ile Ala Asn Phe Gly Thr Asp Ala Thr Gly Phe Ser Arg
Leu Phe 130 135 140Pro Gly Ile Thr Pro His Leu Leu Thr Leu Ala Ser
Asn Phe Lys Leu145 150 155 160Pro Val Tyr Arg Glu Leu Leu Leu Ala
Leu Gly Ile Ser Ser Val Ser 165 170 175Met Lys Ser Cys Gln Asn Ile
Leu Arg Gln Gly Pro Gly Ser Ser Ile 180 185 190Thr Ile Val Val Gly
Gly Ala Ala Glu Ser Leu Ser Ala His Pro Gly 195 200 205Thr Ala Asp
Leu Thr Leu Lys Arg Arg Lys Gly Phe Ile Lys Leu Ala 210 215 220Ile
Arg Thr Gly Ala Ser Leu Val Pro Val Phe Ser Phe Gly Glu Asn225 230
235 240Asp Ile Phe Asn Gln Leu Ser Asn Glu Arg Gly Thr Arg Leu Tyr
Lys 245 250 255Leu Gln Lys Arg Phe Gln Ala Val Phe Gly Phe Thr Leu
Pro Ile Phe 260 265 270Phe Gly Arg Gly Leu Phe Asn Tyr Asn Met Gly
Leu Met Pro Tyr Arg 275 280 285His Pro Ile Val Ser Val Val Gly Arg
Pro Ile Lys Val Lys Gln Lys 290 295 300Asp His Pro Ser Thr Ala Asp
Leu Glu Glu Val Gln Glu Arg Tyr Ile305 310 315 320Ala Glu Leu Lys
Arg Ile Trp Glu Asp Tyr Lys Glu Val Tyr Ala Lys 325 330 335Ser Arg
Thr Lys Glu Leu Thr Ile Ile Ala 340 345441041DNARhodotorula
graminis 44atgggcgcac aagaagaggt cgactacgac cagtcggacc acaccaagat
caagttcgtg 60ccctttgtcg tcccgcggca ccgtcgcctc cagacgttct cggtcttcct
gtggacgacg 120gccctcccta tctcgctcgg catcttctgc atcctgtgct
ccttccctcc tctttggccg 180ctcgtcatcg ggtacctcac ctgggtcttc
ctcattgacc aggcgccgat gcgcggcggg 240aggccacaag cctggctgcg
aaagtcgcgc gtgtgggagt ggttcgccgg ctactatccc 300gtcagcctca
tcaagagcgc cgacctcccg cccgaccagc gttacgtctt tggctaccac
360cctcacggcg tcatcggcat gggcgccatc gccaactttg gcaccgacgc
gaccgggttc 420tcgcgcctgt tcccgggcat cacgccgcac ctcctcacgc
tcgcgagcaa cttcaagctc 480ccagtctacc gagagctcct cctcgccctc
ggcatctcgt ccgtctcgat gaagagctgc 540cagaacatcc tgcggcaagg
tcccggctcg tccatcacga tcgtcgtcgg cggcgccgcc 600gagagcctga
gcgcgcaccc tggcacggcc gacctgacgc tcaagcgccg caagggcttc
660atcaagctcg ccatccgcac cggcgcctcg ctcgtgcccg tcttttcctt
tggcgagaac 720gacatcttca accagctgtc gaacgagcga gggacgcgcc
tgtacaagct gcagaagcgg 780ttccaggccg tctttggctt cacattgccc
atcttcttcg gccgaggcct gttcaactac 840aacatgggct tgatgccgta
ccgacacccg atcgtctcgg tcgtcggccg cccgatcaag 900gtcaagcaga
aggaccaccc gtcgactgcc gacctcgaag aagtccagga gcggtacatc
960gccgagctca aaaggatctg ggaggactac aaggaggtgt acgccaagag
tcgcaccaag 1020gagctcacca tcatcgcctg a 104145635PRTPichia
guilliermondii 45Met Thr Lys Glu Val Asp Glu Ser Thr Gly Gly Ala
Ser Asp Ile Pro1 5 10 15Asn Met Val Glu Glu Ala Lys Ser Ser Ser Phe
Asp Arg Glu Thr Glu 20 25 30Glu Asn Leu Leu Leu Glu Thr Thr Lys Pro
Asp Glu Asn Leu Val Pro 35 40 45Glu Ser Thr Lys His Asp Glu Lys Leu
Val Pro Glu Ile Thr Lys His 50 55 60Glu Asp Asn Pro Met Glu Asn Asp
Gln Val Ser Gln Asn Thr Ala Thr65 70 75 80Ser Pro Met Thr Gly Ala
Gly Ser Glu Glu Thr Arg Asp Leu Ile Thr 85 90 95Glu Asn Ile Glu Lys
Pro Asp Glu Gly Asp Leu Leu Ile Glu Leu Ile 100 105 110Ser Lys Asp
Asn Asp Gly Asp Gly Asp Asp Gly Leu Lys Asn Arg Lys 115 120 125Gln
Lys Arg Ser Ser Ser Glu Val Lys Arg Leu Arg Met Ser Ser Leu 130 135
140Ala Pro Lys Gly Pro Thr Pro Gln Lys His Glu Arg Pro Lys Tyr
Ile145 150 155 160Asn Val Ala Pro Leu Asn Ile Pro Ile Arg Arg Arg
Leu Glu Met Val 165 170 175Gly Ile Ile Trp His Thr Ile Cys Ile Pro
Thr Phe Val Ser Leu Phe 180 185 190Phe Leu Thr Leu Ser Leu Gly Pro
Phe Ala Trp Val Gly Val Ile Leu 195 200 205Pro Tyr Phe Leu Trp Trp
Tyr Leu Ile Asp Leu His Thr Pro Thr Asn 210 215 220Gly Lys Val Ala
Tyr Arg Ser Arg Asp Trp Met Lys Asn Phe Ile Val225 230 235 240Trp
Asp Trp Phe Val Asp Tyr Phe Pro Ile Arg Val His Lys Ser Cys 245 250
255Glu Leu Glu Pro Thr Phe Ser Asp Val Ile Ile Glu Asp Asp Val Val
260 265 270Pro Asp Asp Glu Glu Asp Leu Ile Ser Glu Gln Ser Arg Thr
Gly Val 275 280 285Asp Lys Leu Phe Lys Phe Leu Gly Leu Arg Lys Arg
Leu Asn Asp Asp 290 295 300Ser Asp Ala Ser Ser Gln Cys Ser Leu Leu
Gln Glu Ser Leu Ser Thr305 310 315 320Arg Arg Lys Val Lys Arg Met
Ser Thr Gly Pro Arg Tyr Ile Phe Gly 325 330 335Tyr His Pro His Gly
Val Ile Ser Met Gly Val Phe Gly Thr Phe Ala 340 345 350Thr Asn Ala
Leu Arg Asn Glu Pro Tyr Glu Pro Pro Leu Arg Leu Leu 355 360 365Lys
Pro Phe Phe His Asp Ser Ser Lys Gly Glu Arg Leu Phe Pro Gly 370 375
380Ile Gly Thr Val Phe Pro Leu Thr Leu Thr Thr Gln Phe Ile Val
Pro385 390 395 400Tyr Tyr Arg Asp Tyr Ile Leu Gly Met Gly Leu Thr
Ser Ala Ser Ala 405 410 415Lys Asn Ile Lys Ser Leu Ile Ser Asn Gly
Asp Asn Ser Ile Cys Val 420 425 430Val Val Gly Gly Ala Gln Glu Ser
Leu Leu Asn Asp Met Val Ala Ala 435 440 445Thr Thr Val Pro Gly Arg
Tyr Gly Lys Ser Asn Leu Pro Asn Asp Ser 450 455 460Asp Thr Asp Ser
Glu Phe Asp Pro Gln Arg Lys Ile Glu Glu Asn Lys465 470 475 480Glu
Glu Thr Gly Val Lys Lys Ile Glu Leu Val Leu Asn Lys Arg Lys 485 490
495Gly Phe Val Lys Ile Ala Ile Glu Leu Gly Asn Val Ser Leu Val Pro
500 505 510Thr Phe Gly Phe Gly Glu Ala Asp Ile Tyr Arg Ile Thr Lys
Pro Lys 515 520 525Pro Gly Ser Phe Gly Glu Met Phe Gln Ser Trp Met
Lys Arg Thr Phe 530 535 540Gln Phe Thr Val Pro Phe Phe Ser Ala Arg
Gly Val Phe Ile Tyr Asp545 550 555 560Phe Gly Phe Leu Pro Tyr Arg
Asn Pro Ile Asn Val Cys Phe Gly Arg 565 570 575Pro Ile His Ile Pro
Ala Gly Leu Leu Asp Gln Tyr Lys Glu Pro Glu 580 585 590Thr Glu Lys
Asp Glu Lys Glu Lys Glu Lys Asn Val Phe Gln Phe Thr 595 600 605Gln
Asp Lys Gln Ala Pro Ala Phe Asn Ile Gln Ser Ile Gln Val Phe 610 615
620Gln Gly Glu Ala Thr Ile Lys Glu Glu Thr Ser625 630
635461908DNAPichia guilliermondii 46atgaccaagg aggttgatga
aagcactggg ggtgccagtg atataccaaa tatggttgaa 60gaagcgaaat catcgagttt
tgaccgtgaa actgaagaga atctgctact ggagaccact 120aaacctgacg
agaatctggt accggagagt actaaacatg acgagaaact tgtaccggag
180atcactaaac atgaagacaa tcccatggaa aatgaccaag tttcccaaaa
cacagccacc 240agtcctatga caggagctgg ttccgaagaa acccgtgatt
tgattacaga gaacattgag 300aaaccagatg agggtgatct gctaattgag
cttatttcca aagataacga tggtgatgga 360gatgatgggt tgaaaaatag
aaaacaaaaa cgatcttctt ctgaagtgaa aaggctgcgc 420atgtcgtctc
tggctcctaa aggtccaact cctcaaaagc atgaacgtcc caagtatata
480aatgtggcac ctcttaatat ccccattcga cggcgcttgg agatggtggg
gataatctgg 540cacaccattt gtattcccac gtttgtcagt ttgtttttct
tgactttgtc gttgggtccg 600tttgcttggg taggggtgat attgccgtac
tttttatggt ggtatcttat cgatttacat 660actcctacaa acggtaaggt
tgcgtatcgg tctcgcgact ggatgaagaa tttcattgtg 720tgggattggt
tcgttgacta ttttcctatc agggtccaca agtcttgtga gttggagcct
780acctttagcg atgttattat tgaagacgat gtggtgcccg atgatgaaga
agaccttatc 840tcagagcaat cacgaactgg agtcgataaa cttttcaaat
ttttggggct tcgaaaacgc 900ttaaatgacg actcggatgc ttcgtcgcag
tgctcactgc tgcaagagtc tttaagcaca 960agacgtaaag tgaaacgtat
gtctactggt cctcgctaca tctttggata ccatccccat 1020ggagtaattt
cgatgggtgt ttttggaact ttcgctacca atgcgttgcg taacgagccg
1080tacgaacctc ccttgcgttt gctaaagcca tttttccacg actcttccaa
gggagaacgg 1140ttgtttcccg gtattggcac cgtctttcca ttgacattga
caacccaatt tattgtgccg 1200tactaccgtg actatatctt gggcatggga
ctcaccagtg cttcggctaa aaacatcaag 1260agccttataa gcaacggaga
caactcgata tgtgtcgttg ttggaggtgc tcaggaatcg 1320ctcctaaacg
atatggtagc cgcaaccaca gttcccggtc gttacggaaa gagcaatttg
1380cccaatgaca gtgataccga tagcgagttt gatcctcagc gtaagattga
agaaaacaag 1440gaagaaaccg gcgtaaagaa aattgaactt gtacttaata
agagaaaggg tttcgtcaag 1500atagcgattg agttgggcaa cgtttcactc
gtgcctacgt ttggttttgg agaagctgac 1560atctacagaa tcaccaaacc
caaaccaggt tcatttggag aaatgttcca atcttggatg 1620aaacgcacat
ttcaattcac ggttccattt ttcagcgcta gaggtgtgtt catttacgac
1680tttgggtttc ttccttacag aaatcccatc aatgtctgct ttggacggcc
cattcatatt 1740ccagccggct tattggatca atacaaagag cccgaaactg
agaaagatga aaaagaaaag 1800gaaaaaaacg tcttccagtt cactcaagac
aaacaagcgc cagccttcaa tatccaatct 1860attcaagttt tccaagggga
agcaaccatc aaagaggaaa cgagttag 190847370PRTMicrobotryum violaceum
47Met Thr Ser Leu Arg Asp Val Asn Pro Thr Ser Thr Gln Ala Ser Leu1
5 10 15Tyr Lys Asp Glu Gly Lys Asp Lys Glu Asp Val Ala Pro Gln Glu
Lys 20 25 30Tyr Thr Gln Ser Leu Arg Thr Asn Ile Lys Phe Ala Pro Leu
Ala Val 35 40 45Pro Arg His Arg Arg Leu Gln Thr Met Ala Val Leu Gly
Trp Thr Thr 50 55 60Ala Leu Pro Leu Met Leu Gly Leu Phe Phe Leu Leu
Cys Ser Ile Pro65 70 75 80Leu Leu Trp Pro Ile Ile Val Pro Tyr Leu
Phe Trp Ile His Leu Ile 85 90 95Asp Asn Ser Pro Thr Gln Gly Gly Arg
Ala Ser Lys Trp Leu Arg Gln 100 105 110Ser Arg Phe Trp Val Trp Phe
Thr Gly Tyr Tyr Pro Ile Ser Leu Val 115 120 125Lys Thr Val Asp Leu
Pro Pro Asp Arg Lys Tyr Val Phe Gly Tyr His 130 135 140Pro His Gly
Ile Ile Gly Met Gly Ala Ile Ala Asn Phe Gly Thr Asp145 150 155
160Ala Thr Gly Phe Ser Glu Leu Phe Pro Gly Leu Asn Pro His Leu Leu
165 170 175Thr Leu Ala Ser Asn Phe Lys Leu Pro Ile Tyr Arg
Asp Phe Leu Leu 180 185 190Ala Leu Gly Ile Cys Ser Val Ser Met Lys
Ser Cys Gln Asn Ile Leu 195 200 205Lys Gln Gly Pro Gly Ser Ala Leu
Thr Ile Val Val Gly Gly Ala Ala 210 215 220Glu Ser Leu Ser Ala His
Pro Gly Thr Ala Asn Leu Thr Leu Arg Arg225 230 235 240Arg Met Gly
Phe Ile Lys Leu Ala Met Arg Gln Gly Ala Asp Leu Val 245 250 255Pro
Val Phe Ser Phe Gly Glu Asn Asp Ile Phe Glu Gln Met Pro Asn 260 265
270Glu Arg Gly Thr Lys Leu Tyr Lys Met Gln Lys Lys Phe Gln Thr Ala
275 280 285Phe Gly Phe Thr Leu Pro Ile Phe His Gly Arg Gly Ile Phe
Asn Tyr 290 295 300Asn Leu Gly Ile Leu Pro Tyr Arg His Pro Ile Val
Ser Val Val Gly305 310 315 320Arg Pro Ile Arg Val Ser Gln Arg Asp
Asn Pro Thr Lys Glu Glu Leu 325 330 335Glu Glu Val Gln Glu Arg Tyr
Ile Glu Glu Leu Lys Arg Ile Trp Asp 340 345 350Asp Tyr Lys Asn Gln
His Ala Ile Lys Arg Lys Gly Glu Leu Arg Ile 355 360 365Ile Ala
370481113DNAMicrobotryum violaceum 48atgacgtcgc tgcgagacgt
gaacccgacc tcgacccaag catcgttgta caaagacgag 60ggcaaggaca aggaggatgt
cgcaccgcag gagaaataca cgcagtcgct ccggaccaac 120atcaaatttg
cacctctagc tgtaccacgc catcgacgac tgcagaccat ggcagtgttg
180ggatggacga ccgcactgcc actcatgctt ggtttgttct ttctattgtg
ctcaatcccc 240cttctatggc ccatcatcgt gccctatctc ttctggatcc
acctcatcga caactcgccg 300acgcagggag gacgagcgag caaatggctt
cgccaaagtc ggttctgggt gtggttcaca 360gggtactatc ctatcagtct
cgtcaagacg gtcgatttac ctccagatcg gaaatacgtc 420ttcggttacc
acccccatgg cataattgga atgggtgcaa ttgccaattt tgggaccgac
480gccactgggt tctctgagct cttcccagga ctcaaccctc atctcctcac
acttgccagc 540aacttcaaat tgccgatata tcgagacttc ttgcttgcgc
tcggcatctg ctcagtcagt 600atgaaatctt gccaaaacat cctcaaacag
ggcccggggt ctgctttgac cattgtcgtc 660ggaggagctg cggaatccct
ttcggcgcat cctggcacag ccaacttgac actccgtcgc 720cgaatgggct
tcatcaagct ggcgatgcgt caaggcgcgg atcttgtacc cgtcttttca
780ttcggagaga acgatatctt cgaacagatg ccgaacgaga gagggacgaa
gctgtacaag 840atgcaaaaga agtttcagac cgcttttgga ttcactctac
cgatcttcca cggccgagga 900atttttaact ataaccttgg catcttgccg
taccgtcatc cgatcgtgtc ggtcgtcggt 960cggcccatcc gcgtttcgca
gcgtgacaac cctactaagg aggaactcga ggaggtgcag 1020gaacgataca
tcgaggagtt gaagagaatc tgggacgatt acaaaaatca acatgccatc
1080aagcgaaagg gcgaacttcg tattattgcc tga 111349365PRTPuccinia
graminis 49Met Lys Asp Asp Ser Arg Ser Pro Ser Gly Ser Glu Pro Glu
Gly Asp1 5 10 15Asn His Lys Lys Glu Lys Arg Pro Ile Trp Ala Pro Ile
Arg Val Pro 20 25 30Pro Tyr Arg Arg Ile Gln Thr Ala Ala Val Leu Leu
Trp Thr Ser Gln 35 40 45Leu Ser Leu Cys Ile Ser Leu Phe Phe Phe Leu
Met Ser Tyr Pro Ile 50 55 60Thr Trp Pro Ile Leu Leu Pro Tyr Val Ile
Trp Ile Leu Val Ile Asp65 70 75 80Pro Ala Pro Glu Lys Gly Gly Arg
Leu Asn Gln Ser Val Arg Thr Trp 85 90 95Lys Phe Trp Asn Leu Phe Ala
Ser Tyr Phe Pro Ile Ser Leu Ile Lys 100 105 110Thr Val Asp Leu Pro
Ser Asp Arg Lys Tyr Val Phe Gly Tyr His Pro 115 120 125His Gly Ile
Ile Gly Met Gly Ala Val Ala Asn Phe Gly Thr Glu Ala 130 135 140Thr
Gly Phe Ser Glu Lys Phe Pro Gly Leu Asn Pro His Leu Leu Thr145 150
155 160Leu Ser Thr Asn Phe Ile Ile Pro Phe Tyr Arg Asp Leu Ile Leu
Ser 165 170 175Leu Gly Ile Cys Ser Val Ser Ile Lys Ser Cys Ile Ser
Ile Leu Lys 180 185 190Ser Lys Asn Lys Arg Ser Ala Asp Val Lys Asn
Asn Lys Gly Glu Gly 195 200 205Asn Cys Leu Val Ile Val Val Gly Gly
Ala Ala Glu Ser Leu Ser Ala 210 215 220His Pro Gly Thr Ala Asp Leu
Thr Leu Lys Arg Arg Leu Gly Phe Ile225 230 235 240Lys Leu Ala Ile
Arg Glu Gly Ala Asp Leu Val Pro Val Phe Ser Phe 245 250 255Gly Glu
Asn Asp Ile Tyr Ala Gln Leu Ser Asn Ser Lys Gly Thr Ala 260 265
270Leu Tyr Ser Leu Gln Lys Arg Phe Gln Ala Val Phe Gly Phe Thr Leu
275 280 285Pro Val Phe His Gly Arg Gly Ile Phe Asn Tyr Ser Leu Gly
Leu Leu 290 295 300Pro Tyr Arg His Pro Ile Val Ser Val Val Gly Lys
Pro Ile Arg Val305 310 315 320Glu Gln Asn Lys Asn Pro Gly Leu Glu
Glu Ile Glu Lys Val Gln Lys 325 330 335Glu Tyr Ile Ala Glu Leu Thr
Ala Val Trp Asp Gln Tyr Lys Asp Leu 340 345 350Tyr Ala Arg Asn Arg
Lys Ser Glu Leu Thr Leu Ile Ala 355 360 365501098DNAPuccinia
graminis 50atgaaggatg actccagaag cccgtctggg tccgaacccg agggcgataa
tcacaagaag 60gagaaaaggc caatctgggc tccgattcgt gtacctcctt acaggcgcat
ccaaacggcc 120gcagtactct tatggacttc tcaactctca ctatgcattt
ccttattctt tttcttaatg 180tcttacccga tcacctggcc gatcctcctt
ccatacgtta tttggatctt ggtcatagat 240cctgctcccg agaagggtgg
ccggttgaat caatctgttc ggacctggaa gttttggaat 300ctatttgcgt
cgtatttccc aatcagttta atcaaaactg ttgatttgcc cagtgaccgc
360aaatatgtct ttggttacca ccctcatggt atcatcggaa tgggcgcggt
ggccaacttt 420ggaacggaag cgacaggatt ttcggaaaaa ttccctggtc
tcaatccaca tctactcaca 480ttgagcacga actttatcat cccattctat
cgagacctga tcctcagtct tggaatctgt 540tcggtgtcga tcaaatcatg
catctcgatc ctcaaatcca aaaacaaacg ctcagctgat 600gtcaagaaca
ataagggcga aggaaattgt ttggttatcg ttgtcggtgg ggctgcggaa
660agtttgtctg ctcatcctgg aacagccgat ctcactctaa aacgacggct
gggtttcatc 720aaactggcca ttcgagaagg agccgatctc gtccctgtgt
tctcctttgg agagaatgac 780atttacgccc aattatcaaa ctcaaaaggc
acggcactct actctcttca aaaacgattt 840caagctgtat ttggctttac
cttacctgtt ttccatggcc gaggtatctt caactactct 900ctcggcttgc
ttccctatcg acacccgatt gtttcagtag ttggtaaacc tattcgagtc
960gagcaaaata aaaaccccgg gctcgaagaa atcgaaaagg ttcagaaaga
atacattgct 1020gaacttaccg cagtatggga tcagtataaa gatttatacg
ctagaaatcg gaagagtgaa 1080ttgactttga ttgcttag
109851373PRTGloeophyllum trabeum 51Met Asp Ala Gly Arg Ala Phe Ser
Ser Ala Ser Arg Ser Leu Ser Ser1 5 10 15Ser Ser Leu Lys Asp Lys Leu
Ser Lys Val Ser Lys Leu Ser Thr Thr 20 25 30Pro Leu Arg Pro Val Ala
Ala His Val Lys Asn Ile Asp Phe Val Pro 35 40 45Ser Lys Ile Pro Arg
Lys Arg Arg Leu Gln Met Leu Ala Val Ala Val 50 55 60Trp Ala Leu Leu
Ile Pro Ile Thr Thr Phe Leu Phe Leu Ile Leu Cys65 70 75 80Ser Phe
Pro Pro Leu Trp Pro Phe Leu Ala Ala Tyr Leu Ile Trp Ile 85 90 95Arg
Trp Ile Asp Arg Ser Pro Glu His Gly Gly Arg Ile Ser Pro Trp 100 105
110Phe Arg Ser Met Arg Phe Trp Arg Tyr Phe Ala Asp Tyr Tyr Pro Ala
115 120 125Ser Phe Leu Lys Glu Cys Asp Leu Pro Pro Asp Arg Pro Tyr
Val Phe 130 135 140Gly Tyr His Pro His Gly Ile Ile Gly Met Gly Ala
Met Ala Thr Phe145 150 155 160Ala Thr Glu Ala Thr Gly Phe Ser Glu
Gln Phe Pro Gly Leu Thr Pro 165 170 175His Leu Leu Thr Leu Ala Thr
Asn Phe Thr Met Pro Ile Tyr Arg Asp 180 185 190Ile Ile Leu Ala Leu
Gly Ile Cys Ser Val Ser Lys Gln Ser Cys Ser 195 200 205Asn Ile Leu
Ser Ser Gly Pro Gly Gln Ala Ile Thr Ile Val Val Gly 210 215 220Gly
Ala Ala Glu Ser Leu Ser Ala Arg Pro Gly Thr Ala Asp Leu Thr225 230
235 240Leu Lys Arg Arg Leu Gly Phe Ile Lys Ile Ala Ile Gln His Gly
Ala 245 250 255Ala Leu Val Pro Val Phe Ser Phe Gly Glu Asn Asp Ile
Tyr Gln Gln 260 265 270Met Pro Asn Glu Lys Gly Thr Thr Ile Tyr Ala
Leu Gln Lys Lys Phe 275 280 285Gln Ser Val Phe Gly Phe Thr Leu Pro
Leu Phe His Gly Arg Gly Met 290 295 300Leu Asn Tyr Asn Leu Gly Leu
Met Pro Tyr Arg Arg Arg Ile Val Ser305 310 315 320Val Ile Gly Arg
Pro Ile Leu Cys Glu Lys Cys Glu Lys Pro Ser Met 325 330 335Glu Glu
Val Thr Arg Val Gln Gln Glu Tyr Ile Ala Glu Leu Leu Arg 340 345
350Ile Trp Asp Thr Tyr Lys Asp Gln Phe Ala Arg Ser Arg Lys Arg Glu
355 360 365Leu Ser Ile Ile Asp 370521122DNAGloeophyllum trabeum
52atggacgctg gtcgcgcctt ctcctctgca tcccgctcgt tatcgtcctc gtccctgaag
60gacaagctgt caaaggtctc gaagctcagc accactcctc tgcgaccggt cgctgcccat
120gtcaagaata tcgacttcgt cccgtccaag atcccccgga aacggaggct
gcagatgctc 180gctgttgcag tatgggcgct cctgataccc atcacgacgt
ttttgttcct catactatgt 240tcttttccac cgctgtggcc atttttagcg
gcgtatctta tatggataag atggatagac 300cggagtcctg agcatggcgg
gaggataagt ccgtggttcc gctcgatgag gttctggaga 360tactttgccg
actactaccc tgcatcgttc ttgaaggaat gcgacctccc cccagaccga
420ccttacgtct tcgggtatca ccctcatggc atcattggca tgggtgccat
ggccactttc 480gccaccgaag ccactggatt cagcgaacag ttccctgggc
tcactcccca cctgctcacc 540ctagccacaa atttcaccat gcccatatac
agagacatca tcctcgccct gggcatatgc 600tccgtcagca agcagtcctg
ctcgaacatc ctcagcagcg gccccgggca ggctatcaca 660atcgtagtag
gaggcgcagc agagagtctt agcgctcggc cgggcacggc cgacctcacg
720ctcaaacgga ggcttggctt catcaagatt gctatacaac acggagcggc
actggtccct 780gtattttctt tcggcgagaa tgatatttat caacaaatgc
ccaacgaaaa gggaaccaca 840atatatgccc tacagaagaa attccagagc
gtcttcggct tcacgttgcc cttgttccac 900ggtcggggca tgctaaatta
taaccttggt ttgatgccgt atcgacggcg gatcgtgtct 960gtcatcggtc
ggcccatatt atgcgagaag tgcgagaagc caagcatgga ggaggttacg
1020cgggtgcaac aggagtacat cgcagagctg ctcagaatat gggacacgta
caaagatcaa 1080tttgctcggt cgcggaagag agaactgagt attattgatt ga
112253346PRTRhodosporidium diobovatum 53Met Gly Ala Leu Asp Ala Gly
Asp His Glu Gly Thr Glu His Pro Lys1 5 10 15Ile Lys Phe Val Pro Phe
Val Val Pro Arg His Arg Arg Leu Gln Thr 20 25 30Phe Ser Val Phe Leu
Trp Thr Thr Ala Leu Pro Leu Ser Leu Gly Ile 35 40 45Phe Cys Ile Leu
Cys Ser Phe Pro Pro Leu Trp Pro Leu Val Ile Gly 50 55 60Tyr Leu Thr
Trp Val Phe Leu Ile Asp Gln Ala Pro Met Arg Gly Gly65 70 75 80Arg
Pro Gln Ala Trp Leu Arg Lys Ser Arg Val Trp Glu Trp Phe Ala 85 90
95Gly Tyr Tyr Pro Val Ser Leu Ile Lys Ser Ala Asp Leu Pro Pro Asp
100 105 110Gln Arg Tyr Val Phe Gly Tyr His Pro His Gly Val Ile Gly
Met Gly 115 120 125Ala Ile Ala Asn Phe Gly Thr Asp Ala Thr Gly Phe
Ser Arg Leu Phe 130 135 140Pro Gly Ile Lys Pro His Leu Leu Thr Leu
Ala Ser Asn Phe Lys Leu145 150 155 160Pro Leu Tyr Arg Glu Leu Leu
Leu Ala Leu Gly Ile Ser Ser Val Ser 165 170 175Met Lys Ser Cys Gln
Asn Ile Leu Arg Gln Gly Pro Gly Ser Ser Ile 180 185 190Thr Ile Val
Val Gly Gly Ala Ala Glu Ser Leu Ser Ala His Pro Gly 195 200 205Thr
Ala Asp Leu Thr Leu Lys Arg Arg Lys Gly Phe Ile Lys Leu Ala 210 215
220Ile Arg Ser Gly Ala Tyr Leu Val Pro Val Phe Ser Phe Gly Glu
Asn225 230 235 240Asp Ile Phe Asn Gln Leu Ser Asn Glu Arg Gly Thr
Arg Leu Tyr Lys 245 250 255Leu Gln Lys Arg Phe Gln Ala Val Phe Gly
Phe Thr Leu Pro Ile Phe 260 265 270Phe Gly Arg Gly Leu Phe Asn Tyr
Asn Met Gly Leu Met Pro Tyr Arg 275 280 285His Pro Ile Val Ser Val
Val Gly Arg Pro Ile Lys Val Thr Gln Lys 290 295 300Asp His Pro Ser
Thr Ala Asp Leu Glu Glu Val Gln Asp Arg Tyr Ile305 310 315 320Ala
Glu Leu Lys Arg Ile Trp Glu Asp Tyr Lys Glu Val Tyr Ala Lys 325 330
335Ser Arg Thr Lys Glu Leu Thr Ile Ile Ala 340
345541041DNARhodosporidium diobovatum 54atgggagcac tagatgcggg
cgaccacgag gggaccgaac accccaagat caagttcgtt 60cctttcgttg tgccgcgaca
ccgcaggctg cagacctttt cggtgtttct gtggacgacc 120gcgctgcctc
tgtcgctcgg catcttctgc attctctgct ccttcccccc actctggccc
180ctcgtcatag ggtatctcac gtgggtattc ctcatcgacc aggcgcccat
gcggggtggc 240aggcctcagg cctggttgcg caagtcgcgt gtgtgggagt
ggttcgccgg ctactaccct 300gtcagcttga tcaagagcgc cgacctcccg
cccgaccagc gctacgtctt tggctaccac 360ccacacggcg tcattgggat
gggcgccatc gccaactttg gtaccgacgc gaccggcttc 420tcgcggctgt
tccccggcat caagccgcac ctcctcacgc tcgccagcaa cttcaagctg
480ccgctctacc gagaactgct cctcgccttg ggcatttcgt ccgtgtcgat
gaagagctgc 540cagaacatcc tgcgccaagg tcccggctcg tcgatcacga
ttgtcgtcgg aggggcagca 600gaaagcctca gcgcgcaccc gggaacggca
gacctgacgc tcaagcggcg gaaggggttc 660atcaagctcg cgatccgctc
aggggcctac ctcgtcccgg tattttcctt tggcgagaat 720gacatcttca
accagctgtc gaatgagcgc ggcacccgac tctacaagct gcaaaagcgg
780ttccaggccg tctttggctt caccttgccc atcttcttcg gtcgcggcct
cttcaactac 840aacatgggct tgatgccata tcgacacccg atcgtctcgg
tcgtcggacg ccccatcaag 900gtcacgcaga aggatcaccc gtcgacggcc
gacctcgaag aggtacagga ccgctacatt 960gccgagttga agaggatctg
ggaggactac aaagaggtgt acgccaagag ccgcaccaag 1020gagctcacca
tcatcgcatg a 104155359PRTPhaeodactylum tricornutum 55Met Lys Glu
Arg Arg Ser Gly Leu Asn Pro Ser Gly Ser Ser Val Tyr1 5 10 15Pro Leu
His Pro Pro Asp Ser Arg Val Leu Val Arg Val Pro Ser Asp 20 25 30Ile
Ser Phe Leu Asp Arg Leu Ile Val Ala Gly Ser Ser Ile Phe Ile 35 40
45Val Gly Ser Leu Val Trp Val Pro Leu Thr Ala Arg Trp Val Tyr Arg
50 55 60Arg Trp Lys Gln Ala Lys Asp Lys Arg Lys Arg Ala Met Tyr Ala
Ser65 70 75 80Leu Leu Val Ile Leu Ala Val Leu Val Ile Gly Gly Pro
His Arg Ser 85 90 95Pro Arg Val Gly Lys Trp Leu Gln Val Arg Lys Trp
Ser Leu Phe Gln 100 105 110Ala Trp Val Lys Phe Ile Ala Met Glu Val
Ile Leu Asp Gln Pro Lys 115 120 125Gly Ile Thr Met Asp Val Gln Gln
Asp Lys Ala Ile Phe Ala Phe Ala 130 135 140Pro His Gly Ile Phe Pro
Phe Ala Phe Ala Phe Gly Val Leu Pro Asp145 150 155 160Ile Ala Thr
Gln Ser Phe Gly Tyr Val Arg Pro Val Val Ala Thr Ala 165 170 175Thr
Arg Leu Phe Pro Val Val Arg Asp Phe Ile Ser Trp Ala Asn Pro 180 185
190Val Asp Ala Ser Lys Asp Ser Val Glu Arg Ala Leu Ala Leu Gly Asp
195 200 205Arg Ile Ala Val Ile Pro Gly Gly Ile Ala Glu Ile Phe Glu
Gly Tyr 210 215 220Pro Lys Pro Asn Thr His Pro Asp Glu Glu Tyr Ala
Ile Val Arg Ser225 230 235 240Gly Phe Leu Arg Leu Ala Ile Lys His
Gly Ile Pro Val Ile Pro Val 245 250 255Tyr Cys Phe Gly Ala Thr Lys
Met Leu Lys Arg Leu Glu Leu Pro Gly 260 265 270Leu Glu Gln Leu Ser
Leu Phe Leu Arg Val Ser Ile Cys Leu Phe Phe 275 280 285Gly Val Gly
Gly Leu Pro Ile Pro Phe Arg Gln Arg Leu Ser Tyr Val 290 295 300Met
Gly Gln Pro Ile Leu Pro Pro Val Arg Thr Thr Gly Ser Asp Ile305 310
315 320Ser Asp Ala His Val Lys Glu Met Gln Asp Arg Phe Cys Ala Glu
Val 325 330 335Gln Arg Leu Phe Asp Arg His Lys Glu Ala Tyr Gly Trp
Ser Tyr Lys 340 345 350Thr Leu Lys Leu Leu Glu Gln
355561080DNAPhaeodactylum tricornutum 56atgaaagaaa gaagatctgg
cctaaatccg tcaggatcct ccgtgtatcc attgcaccct 60cctgacagtc gcgttctcgt
tcgagtcccc tccgatattt cctttcttga tcgtctcatc 120gtcgctggca
gcagtatctt tattgtcggt tcgctagtat gggttccatt gaccgcaaga
180tgggtctaca ggcggtggaa gcaagctaaa gataaacgaa agcgggctat
gtatgcctct 240ctactcgtga ttctggcagt tctcgttatt ggcggacccc
accgatctcc tcgtgtcggc 300aaatggctcc aagtacgaaa gtggtccctc
ttccaagcgt gggtaaagtt tattgccatg 360gaagtgattt tggatcaacc
gaaaggcatt
actatggacg tccaacaaga caaggcgatt 420tttgcattcg cgccacatgg
aatctttccg tttgcgttcg cctttggagt gcttcccgat 480attgccacac
aatcgtttgg ctacgttcgt ccggtcgtgg caaccgccac aaggttgttt
540cctgtagtcc gggatttcat ctcttgggcg aatccggtag acgcttccaa
agattccgtt 600gaacgtgctt tagcattggg cgatcgcatt gctgtaatac
ctggaggaat tgcagaaatt 660ttcgaaggat atccgaaacc gaacacgcat
ccggatgaag agtacgctat cgtacggagt 720ggatttttgc gtttggcaat
aaaacacggt atcccagtga ttcccgtata ctgtttcggc 780gctaccaaaa
tgttgaagcg tctggagctt cccggcctgg agcaactgtc cctgtttcta
840cgcgtgagca tttgcctctt ttttggagtc ggcgggttgc ccatcccttt
ccgacaacga 900ttgtcgtacg taatgggaca accaattttg ccacccgtaa
ggacaacggg cagcgatatt 960tcggacgcac acgtcaaaga aatgcaagat
cgcttttgtg ctgaggtcca gcggctcttt 1020gatcgacata aggaagctta
tggttggtca tacaaaacgc tgaaactatt ggaacagtga
108057329PRTPhaeodactylum tricornutum 57Met Glu Arg Thr Lys Ile Gln
Asp Glu His Lys Ser Pro Pro Asn Pro1 5 10 15Ser Thr Phe Arg Trp Phe
Leu Gly Leu Leu Val Ala Ser Thr Phe Ser 20 25 30Met Val Tyr Phe Val
Ala Pro Phe Tyr Met Leu Thr Val Val Phe Ala 35 40 45Leu Val Phe Lys
Tyr Pro Ser Val Glu Ile Ala Trp Met Tyr Ala Ile 50 55 60Pro Met Ile
Val Ser Ala Ile Leu Pro Pro Met Ala Ser Pro Leu Ala65 70 75 80Leu
Arg Leu Ile Ser Pro Leu Ile Asp Tyr Phe Asp Tyr Glu Glu Ile 85 90
95His Glu Thr Ser Pro Val Asp Val Gln Lys Glu Ile Leu Ser Asn Asn
100 105 110Lys Asn Tyr Leu Leu Val Phe Gln Pro His Gly Ala Leu Ser
Phe Thr 115 120 125Gly Ile Thr Ser Met Val Thr Ala Pro Gln Ala Met
Lys Gly Lys Leu 130 135 140Pro Thr Ala Val Ala Asp Ala Leu Leu Tyr
Thr Pro Ile Leu Lys His145 150 155 160Val Leu Gly Ile Phe Gly Leu
Ile Ser Ala Ser Lys Ser Ser Met Ile 165 170 175Arg Thr Leu Lys Lys
Lys Gly Val Glu Gly Thr Ile Val Leu Tyr Val 180 185 190Gly Gly Ile
Ala Glu Leu Phe Leu Thr Asp Glu Thr Asp Glu Arg Leu 195 200 205Tyr
Leu Arg Lys Arg Lys Gly Phe Ile Lys Leu Ala Leu Gln Gln Gly 210 215
220Val Asp Val Val Pro Val Tyr Leu Phe Gly Asn Thr Asn Ala Leu
Ser225 230 235 240Val Leu Lys Thr Gly Phe Leu Ala Ala Ile Ser Arg
Lys Leu Gln Ile 245 250 255Ser Leu Thr Tyr Ile Trp Gly Lys Trp Tyr
Leu Pro Ile Pro Arg Asp 260 265 270Cys Lys Leu Leu Tyr Ala Ser Gly
Gln Pro Leu Gly Met Pro His Ile 275 280 285Leu Asp Pro Ser Gln Ala
Asp Ile Asp Lys Trp His Glu Lys Tyr Cys 290 295 300Ser Glu Val Met
Arg Ile Phe Glu Lys Tyr Lys Glu Lys Val Pro Glu305 310 315 320Tyr
Lys His Lys Lys Leu Glu Ile Ile 32558990DNAPhaeodactylum
tricornutum 58atggagagaa caaagataca agacgagcac aaaagtcccc
ctaatccgtc gacatttcga 60tggttcctcg gccttctagt ggcgtcgacg ttttccatgg
tctattttgt ggctcccttt 120tacatgctta cagtcgtgtt tgcactagtt
ttcaaatatc cttcggtaga aattgcatgg 180atgtacgcta ttccgatgat
tgtctcggcc attttgccac caatggcttc tccactggcc 240ttgcgactca
tctccccgct cattgactac ttcgattacg aagagatcca cgaaacctca
300ccggtggacg tccagaagga aatactaagc aacaacaaaa actatttgct
agtctttcaa 360ccgcatggag cactgtcgtt tacaggaatc acttcaatgg
tgacagctcc acaagcaatg 420aaaggcaaat tgccaacagc tgtggctgac
gcactcttgt acacacctat actgaaacat 480gtcttaggaa ttttcgggct
gattagtgcc tccaaaagca gcatgatccg aactttaaaa 540aagaagggtg
tggaaggaac cattgttttg tacgttggtg ggattgccga gctctttttg
600accgacgaga cggacgagcg cctctatctg cgaaagcgaa aagggtttat
caaattagct 660ctacaacagg gtgtcgatgt tgtacctgtg tatctatttg
ggaacacaaa cgcgctgtcg 720gtactaaaga cgggatttct cgcggcaatt
tcgcgaaaat tacagatatc tctgacgtac 780atttggggaa agtggtatct
tccgattccc cgtgattgca aattgctgta tgcttccggt 840cagccattag
gaatgcctca tattttagac ccaagccaag ccgacattga taaatggcac
900gaaaagtact gctccgaggt catgcggatc ttcgaaaaat acaaggaaaa
ggttccggaa 960tacaagcaca agaaattaga aattatttga
99059392PRTPhaeodactylum tricornutum 59Met Arg Glu Arg Ser Cys Ala
Asn Ala Ser Asp Asp Asp Ser Ile His1 5 10 15Lys Gln Ser Pro Glu Leu
Glu Ala Glu Phe Leu His Thr Ser Lys Leu 20 25 30Thr Leu Ala Asp Met
Arg Arg Leu Ala His Asp Pro Lys Asp Arg Gly 35 40 45Leu Ala Thr Lys
Pro Ala Ala Gln Ala Thr Lys Glu Asp Val Leu Thr 50 55 60Val Gln Pro
Met Ser Phe Val Glu His Thr Ala Cys Cys Leu Phe Leu65 70 75 80Ala
Phe Gly Val Pro Asn Gly Ala Leu Thr Ile Pro Ile Ala Thr Trp 85 90
95Leu Ile Gly Lys Phe Val Leu Arg Asn Val Phe Leu Ala Phe Leu Leu
100 105 110Ala Gly Cys Ile Leu Leu Pro Leu Ala Ile Leu Pro Gln Glu
Tyr Val 115 120 125Pro Ala Arg Leu Gln Ser Trp Leu Ala Leu Gln Ile
Leu Lys Tyr Phe 130 135 140Ser Phe Ser Leu Val Met Glu Glu Arg Pro
Pro Thr Met Cys Thr Gly145 150 155 160Lys Gln Leu Ile Glu Gln Pro
Ala Arg Pro Arg Ile Val Thr Ala Tyr 165 170 175Pro His Gly Val Phe
Pro Tyr Gly Asn Ala Leu Thr Val Val Thr Trp 180 185 190Pro Leu Leu
Thr Gly His His Ile Val Gly Leu Ala Ala Asn Ala Ala 195 200 205Leu
Arg Thr Pro Ile Phe Lys Gln Ile Leu Arg Ser Ile Gly Val Lys 210 215
220Asp Ala Ser Arg Ala Ser Val Arg Asn Ala Leu Glu Thr Trp Pro
Phe225 230 235 240Thr Val Gly Ile Ser Pro Gly Gly Val Ala Glu Val
Phe Glu Thr Asn 245 250 255His Phe Asn Glu His Ile Leu Leu Lys Glu
Arg Ile Gly Val Ile Lys 260 265 270Met Ala Ile Arg Thr Gly Ala Asp
Leu Val Pro Gly Tyr Met Tyr Gly 275 280 285Asn Thr Asn Leu Tyr Trp
Cys Trp Thr Gly Glu Gly Ile Pro Gly Ala 290 295 300Arg Trp Leu Leu
Glu Tyr Val Ser Arg Lys Ile Leu Gly Phe Ala Leu305 310 315 320Val
Pro Ile Ala Gly Arg Trp Arg Leu Pro Ile Pro Tyr Arg Thr Pro 325 330
335Ile Leu Cys Val Val Gly Lys Pro Ile Pro Thr Ile His Leu Gln Thr
340 345 350Glu Glu Pro Ser Met Glu Gln Ile Val Asp Ile Gln Glu Gln
Leu Ser 355 360 365Thr Glu Leu Lys Ser Met Phe Asp Arg Tyr Lys His
Leu Tyr Gly Trp 370 375 380Glu Asp Arg Met Leu Val Ile Thr385
390601179DNAPhaeodactylum tricornutum 60atgcgtgagc gaagctgcgc
caacgcttct gacgatgaca gcattcacaa gcagtcgcca 60gaattggagg ctgagtttct
tcataccagc aagttgacct tagccgacat gcgacgattg 120gcgcacgatc
cgaaggatcg ggggttggca acaaaacctg cggcgcaagc tacgaaagaa
180gacgtcttga cggtacaacc catgagtttc gtagaacaca ctgcttgctg
tctgtttctc 240gcgtttggag tgcccaatgg cgctctgacg attcccatag
caacgtggct gatcggaaaa 300ttcgtgttgc gcaacgtttt cttggcgttt
ctgttagcag gctgtatact tctaccgctt 360gcgatactgc cgcaagaata
tgtgcccgcc cgattgcaat cgtggcttgc tttgcagata 420ctgaaatatt
tttctttctc tttggtcatg gaggaacgcc ctccgacaat gtgtactggc
480aagcagctga tcgagcagcc cgctcggcca cgaatcgtca cagcctatcc
gcacggagtt 540ttcccatacg gaaacgcgtt gactgtagtc acatggccgt
tgttgacggg acaccatatt 600gtgggtttgg cagcaaatgc cgctttgcgg
acaccgatct ttaaacaaat cttgcggagc 660attggcgtca aggacgcctc
tcgagcgtcg gtacggaatg cgctggaaac atggcctttc 720accgtcggga
tttcgccagg tggcgtggcg gaagtttttg aaacaaacca cttcaatgag
780cacattctgt tgaaagaacg tattggtgtc atcaagatgg ccattcgcac
cggtgcggat 840cttgtaccag gctatatgta tggtaatact aatctgtact
ggtgctggac aggggaaggt 900attcctggag ctcggtggct attggagtat
gtttcgcgta aaatcctagg ttttgccctc 960gtgcctatag cgggtagatg
gagactacca ataccgtaca ggactccgat attgtgtgtc 1020gtgggcaagc
caataccaac cattcatttg caaaccgaag aaccatcaat ggagcaaatc
1080gtggacattc aggaacaatt gtcaacagaa ttgaaatcaa tgttcgaccg
ctataagcac 1140ctgtacggat gggaagatcg aatgctagtg atcacataa
117961320PRTPhaeodactylum tricornutum 61Met Thr Arg Ser Lys Phe Ile
Gly Ser Ala Gly Ala Ile Gly Leu Phe1 5 10 15Cys Leu Met Ile Ile Pro
Asn Val Gly Ile Leu Ile Ala Thr Phe Leu 20 25 30Tyr Pro Lys Val Leu
Gly Leu Tyr Phe Leu Ile Pro Tyr Tyr Ala Tyr 35 40 45Asn Leu Ser Ile
Gly Lys His Glu Ala Arg Asp Gly Asn Gly Trp Asn 50 55 60Trp Phe Ser
Glu Asn Phe Phe Val Phe Asn Ile Val Arg Gly Tyr Leu65 70 75 80Asn
Leu Lys Ile Glu Ala Asp Ser Glu Leu Lys Glu Ala Glu Ala Lys 85 90
95Glu Gly Ala Gln Phe Val Phe Ala Val Ser Pro His Gly Thr Asn Ala
100 105 110Asp Tyr Arg Val Phe Ile Asp Gly Met Leu His Glu Ala Leu
Pro Gln 115 120 125Thr Ala Ser Lys Ile Arg Thr Leu Ala Ala Thr Val
Leu Phe His Ile 130 135 140Pro Leu Val Arg Glu Ile Ala Leu Trp Thr
Gly Cys Val Asp Ala Ser145 150 155 160Arg Ala Val Ala Val Glu Arg
Leu Lys Glu Glu Gly Gly Ser Leu Leu 165 170 175Val Ile Pro Gly Gly
Gln Ala Glu Gln Met Tyr Thr Gln Tyr Gly Arg 180 185 190Glu Arg Val
Tyr Leu Lys Arg Arg Lys Gly Phe Leu Lys Leu Cys Leu 195 200 205Lys
Tyr Glu Ile Pro Val Val Pro Ala Tyr Val Phe Gly Val Ser Asp 210 215
220Tyr Tyr Phe Thr Ser Ala Lys Leu Phe Gly Leu Arg Met Trp Leu
Val225 230 235 240Gln Asn Leu Gly Ile Ala Leu Pro Leu Cys Trp Gly
Arg Tyr Gly Leu 245 250 255Pro Ile Cys Pro Arg Pro Val Asp Thr Thr
Leu Val Phe Asp Lys Pro 260 265 270Leu Tyr Leu Ser Cys Gln Asn Pro
Ser Asn Pro Ser Glu Asp Glu Val 275 280 285Asp Lys Ala His Leu Gln
Phe Cys Gln Ala Leu Glu Lys Leu Phe Asp 290 295 300Thr His Lys Glu
Arg Leu Gly Tyr Gly Asp Arg Lys Leu Glu Ile Ile305 310 315
32062963DNAPhaeodactylum tricornutum 62atgaccagat cgaagtttat
aggaagtgct ggagctattg gcttattttg tttgatgatc 60ataccgaatg tgggaattct
gatcgcaaca tttctttatc ccaaagtact tgggctctac 120tttctgattc
cgtactacgc atacaacttg tccattggca aacacgaagc tcgagacggc
180aacggctgga attggttcag cgagaatttc tttgtcttta acattgtgag
gggatatcta 240aatcttaaga ttgaagctga ctccgagctc aaggaagccg
aagcgaaaga aggcgcccaa 300tttgtgttcg ccgttagccc tcacggaacg
aacgcagact atcgagtttt tattgacggt 360atgctacatg aggcactccc
acagactgca agcaagatca gaacactagc ggcgacagta 420ctgttccaca
ttcccttggt tcgtgaaatc gcactttgga caggatgtgt cgatgccagc
480cgcgcagttg ctgtcgagag attaaaagaa gaaggtggtt cactgcttgt
gattcccggt 540ggccaagcag aacaaatgta cacccaatat ggacgtgaaa
gagtatatct gaaacggcgc 600aaaggatttt tgaagctttg cttgaagtac
gagattccgg tcgtcccagc ttatgttttt 660ggcgtatctg actattactt
cacgtccgca aagctctttg gtctgcgaat gtggctcgtt 720cagaatcttg
gcattgctct tccactgtgc tggggaagat atggtctacc aatctgtcct
780agaccagtcg ataccaccct tgtctttgac aaacctttat acctatcctg
ccagaatccg 840tcgaatccct cggaagacga ggttgacaag gctcatctgc
aattttgcca agccctcgag 900aagctgtttg atacacacaa agagaggctt
gggtacggcg atcgaaagct ggaaataatt 960tag 96363663PRTRhodotorula
graminis 63Met Ser Thr Ala Asp Leu Pro Pro Gly Pro Ala Gln Leu Leu
Glu Asp1 5 10 15Ala Leu Arg Gln Pro Asp Gly Pro Pro Leu Leu Ser Thr
Ser Ala Ala 20 25 30Asp Pro Ser Ser Pro Leu Gln Leu Asp His Asp His
Arg Pro Gly Met 35 40 45Ala Ala Asp Ala Ala Ser Ser Ala Ser Asp Ser
Ser Ile Ser Thr Val 50 55 60Ser Ser Val Leu Arg Gly Gln Gln Ala Thr
Thr Thr Val Thr Thr Asn65 70 75 80Arg Gly Glu Gly Gly Arg Glu Thr
Thr Glu Thr Phe Thr His Val Gly 85 90 95Ala Ala Asn Val Asp Ala Glu
Tyr Ser Ser Ser Thr Gly His Ile Thr 100 105 110Leu Arg Pro Val Val
Ala Lys Gly Gly Asp Pro Arg Arg Ile Arg Leu 115 120 125Val Arg Ser
Arg Arg Thr His Phe Glu Pro Arg Ile Ser His Phe Asp 130 135 140Arg
His Asn Lys Thr Ser Ala Glu Asp Thr Phe Arg Gly Phe Phe Ser145 150
155 160Leu Phe Trp Ile Val Ile Ala Val Gly Gly Thr Arg Thr Ile Tyr
Asn 165 170 175Arg Val Ala Glu Thr Gly Gly Leu Leu Gly Gly Trp Gln
Phe Ala Ala 180 185 190Leu Ile Ser Glu Asp Ala Trp Ala Leu Ala Leu
Ser Asp Ala Val Leu 195 200 205Val Gly Ser Thr Ile Leu Cys Val Pro
Phe Val Lys Leu Ile Val Asn 210 215 220Gly Trp Val Arg Tyr Tyr Tyr
Thr Gly Leu Val Leu Gln His Leu Ala225 230 235 240Gln Thr Leu Tyr
Leu Gly Ile Ala Val Arg Trp Thr Phe His Arg His 245 250 255Trp Pro
Trp Val Gln Ser Gly Phe Met Thr Leu His Ala Leu Ser Met 260 265
270Leu Met Lys Ile His Ser Tyr Cys Ser Leu Asn Gly Glu Leu Ser Glu
275 280 285Arg Val Arg Gln Leu Glu Lys Asp Glu Arg Lys Leu His Glu
Ala Val 290 295 300Glu Glu Leu Gly Gly Gln Asp Ala Leu Glu Arg Glu
Gly Arg Val Ala305 310 315 320Trp Glu Lys Ala Cys Ala Glu Ala Ala
Glu Gln Lys Ala Ala Glu Glu 325 330 335Ala Ala Gly Gly Arg Gly Lys
Ala Ser Ala Ser Ser Leu Ala Pro Pro 340 345 350Pro Ala Thr Gly Pro
Gln Pro Ser Ser Asp Glu Glu Ala Val Ser Thr 355 360 365Thr Leu Arg
Gln Arg Pro Ser Ala Ala Arg Arg Arg Ser Leu Ser Pro 370 375 380Ser
Ala Ala Arg Thr His Val Thr Pro Pro Ser Arg Lys Ala Glu Pro385 390
395 400His Asp Val Glu Thr Leu Thr Trp Ser Pro Asn Glu Arg Val Ser
His 405 410 415Leu Ala Ile Ala Ile Cys Glu Ala Arg Glu Ala Leu Ser
Ser Ser Gly 420 425 430Ala Ala Lys Val Ser Phe Pro Asp Asn Val Thr
Val Leu Asn Phe Val 435 440 445Asp Tyr Leu Leu Val Pro Thr Leu Val
Tyr Glu Leu Glu Tyr Pro Arg 450 455 460Thr Asp Ser Ile Arg Pro Leu
Tyr Ile Leu Glu Lys Thr Leu Ala Thr465 470 475 480Phe Gly Thr Phe
Ser Val Leu Leu Leu Ile Val Glu His Phe Ile Tyr 485 490 495Pro Val
Met Pro Gly Pro Asp Ser Ser Phe Ile Ser Ser Leu Leu Asp 500 505
510Leu Ala Leu Pro Phe Thr Ile Cys Tyr Leu Leu Ile Phe Tyr Ile Ile
515 520 525Phe Glu Cys Ile Cys Asn Ala Phe Ala Glu Ile Thr Arg Phe
Ser Asp 530 535 540Arg Ala Phe Tyr Ser Asp Trp Trp Asn Ser Ile Ser
Phe Asp Glu Phe545 550 555 560Ser Arg Lys Trp Asn Arg Pro Val His
Thr Phe Leu Leu Arg His Val 565 570 575Tyr Ala Thr Thr Ile Ser Thr
Tyr Lys Leu Ser Lys Phe Ser Ala Ala 580 585 590Phe Val Thr Phe Leu
Leu Ser Ala Leu Val His Glu Leu Val Met Val 595 600 605Val Val Thr
His Lys Ile Arg Met Tyr Leu Phe Met Ala Gln Leu Pro 610 615 620Leu
Ile Met Leu Gly Arg Ala Ser Ile Phe Lys Arg His Pro Ala Leu625 630
635 640Gly Asn Leu Phe Phe Trp Phe Gly Leu Leu Ser Gly Phe Pro Leu
Leu 645 650 655Ala Val Ala Tyr Leu Lys Phe 660641992DNARhodotorula
graminis 64atgagcaccg ccgatcttcc accaggtcct gcccagctgc tcgaagacgc
cctgcgccag 60ccagacggcc cccctctcct gtcgacctcc gccgccgatc cctcctcccc
acttcaactc 120gaccacgacc accgccccgg catggctgca gacgccgcca
gctcagcttc agacagctct 180atcagcacgg tgtccagtgt cctgcgcggt
cagcaagcca cgacaacggt gacgaccaac 240aggggagaag gcgggcgaga
aacgaccgag accttcaccc acgtcggcgc cgccaatgtc 300gacgccgagt
actcgtcctc gaccggccac atcacgctcc gacccgtcgt ggcaaagggc
360ggtgaccctc gccggatccg cctcgtccgc tcgcgccgca cccacttcga
gccgcgcatc 420tcgcacttcg accgccacaa caagacgtcg gccgaggaca
cgttccgcgg cttcttctcg 480ctcttctgga tcgtcatcgc cgtcggcggc
acgaggacca tctacaaccg cgtcgccgag 540acgggcggtc tcctcggcgg
gtggcagttt gcggcgctca tctccgagga cgcatgggct 600ctggcgctga
gcgatgcggt cctcgtcggg tcgacgatac tctgcgtccc gttcgtcaag
660ctcatcgtca acggctgggt ccggtactac tacacgggcc tcgtcctcca
gcacctcgcc 720cagacgctct acctcggcat cgccgtccga tggacgttcc
accgtcactg gccctgggtc 780cagagcggct tcatgacgct gcacgccctg
agcatgctca tgaagatcca ctcgtactgc 840tcgctcaacg gcgagctgtc
cgagcgcgtg cggcagctcg agaaggacga gcgcaagctg 900cacgaggcgg
tcgaggagct tggcggccag gacgcgctcg agcgcgaggg gcgcgtggcg
960tgggagaagg cgtgcgccga ggcggccgag cagaaggcgg ccgaggaggc
ggcaggcggt 1020cgcggcaaag cttcggcgtc ctcgctcgcc ccgccgccgg
cgacagggcc gcagccctcg 1080tccgacgagg aggccgtctc gacgacgctc
cgacagcgac cgtcggccgc tcgccgccgc 1140tcgctctcgc cgtcggccgc
acggacccac gtcacgccgc cgtcgcgcaa ggccgagccg 1200cacgacgtcg
agacgctcac ctggtcgccc aacgagcgcg tgtcgcacct cgccatcgcc
1260atctgcgagg cacgcgaggc cctgtcgtcg agcggcgccg ccaaggtctc
gttcccggac 1320aacgtcacgg tcctcaactt tgtcgactac cttctcgtcc
cgacgctcgt gtacgagctt 1380gagtacccga ggaccgactc tatccgaccc
ttgtacatcc tcgagaagac cctcgccacg 1440ttcggcacat tctcggtcct
cctcctcatc gtcgagcact tcatctaccc ggtcatgccc 1500gggcccgaca
gctcgttcat ctcgtccctc ctcgacctcg ccctcccatt caccatctgc
1560tacctcctca tcttctacat catcttcgag tgtatctgca acgccttcgc
cgagatcacg 1620cgcttctcgg accgggcctt ctacagcgac tggtggaact
cgatctcgtt cgacgagttc 1680tcgcgcaagt ggaaccggcc cgtgcacacg
ttcctcctgc gccacgtgta cgcgacgacc 1740atctcgacct acaagctcag
caagttctcg gccgcctttg tcacgttcct cctgagcgcg 1800ctcgtgcacg
agctcgtcat ggtagtcgtg acgcacaaga tccgcatgta tctctttatg
1860gcgcagctcc ccctcatcat gctcggccga gcaagcatct tcaagcgtca
ccctgcgctc 1920ggcaacctct tcttctggtt cggcctcttg agcggtttcc
ctctgctagc tgtagcgtac 1980ctcaagttct ag 199265597PRTPichia
guilliermondii 65Met Ser Lys Glu Asn Leu Leu Lys Ile Ser Gln Tyr
Asn Thr Glu Arg1 5 10 15Arg Pro Ser Leu Ala Thr Asp Val Asp Tyr Ser
Ser Thr Asp Leu Ser 20 25 30Ser Arg Leu Asp Ser Ala Asn Thr Thr Asn
Gly Thr Pro Thr Val Thr 35 40 45Leu His Lys Arg Gln Ser Ser Thr Glu
Leu Leu Ser Glu Ser Pro Glu 50 55 60Gln Lys Arg Phe Leu Lys Thr Ile
Asp Thr Leu Asn Arg Thr Thr Asn65 70 75 80Ser Arg Leu Arg Gln Arg
Leu Asn Arg Glu Gly Asp Lys His Lys Lys 85 90 95Glu His Lys Glu His
Glu Lys His Lys Asp Asp His Ser Lys Tyr Lys 100 105 110Ser Arg Phe
Gly Asp Ile His Phe Tyr Ser Asn Met Thr Thr Ile Phe 115 120 125Asp
Ala Asp Tyr Phe Lys Glu Ser Gln Phe Phe Gly Val Tyr Ile Leu 130 135
140Phe Trp Leu Gly Thr Ala Phe Leu Ile Leu Asn Asn Leu Val His
Thr145 150 155 160Phe Leu Glu Asn Gly Asp Asn Leu Leu Asp Gly Pro
Val Val Arg Thr 165 170 175Phe Lys Lys Asp Leu Leu Lys Ile Ala Leu
Thr Asp Leu Gly Met Tyr 180 185 190Leu Thr Met Tyr Val Ser Val Phe
Ile Gln Leu Gly Ile Arg Lys Gly 195 200 205Trp Tyr Ser Trp Ser Ser
Thr Gly Ala Thr Leu Gln Asn Ile Tyr Ser 210 215 220Phe Val Tyr Phe
Phe Ala Trp Ser Tyr Phe Ala Ser Pro Lys Tyr Met225 230 235 240Asp
Tyr Pro Trp Ile Gly Lys Val Phe Leu Ala Leu His Ser Leu Val 245 250
255Phe Leu Met Lys Met His Ser Tyr Ala Thr Tyr Asn Gly Tyr Leu Trp
260 265 270Asn Ile Phe Asn Glu Leu Gln Val Ser Arg Lys Tyr Leu Lys
Ile Leu 275 280 285Asp Glu Thr Asp Glu Ser Met Ile Glu Gly Lys Ser
Val Ser Asp Leu 290 295 300Arg Lys Ala Leu Val Asp Ser Ile Gly Phe
Cys Ser Tyr Glu Leu Glu305 310 315 320Tyr Gln Ser Lys Ser Thr Ser
Val Asn Thr Asp Val Glu Ile Thr Gly 325 330 335Asp Lys Asn Lys Leu
Asn Thr Thr Lys Ser Thr Ser Ser Leu Asp Asp 340 345 350Asp Tyr Val
Ser Phe Pro Asn Asn Ile Thr Phe Phe Asp Phe Phe Arg 355 360 365Tyr
Ser Met Phe Pro Thr Val Val Tyr Ser Leu Lys Phe Pro Arg Thr 370 375
380Lys Arg Ile Arg Trp Gly Tyr Val Met Glu Lys Ser Phe Ala Val
Phe385 390 395 400Gly Ile Ile Phe Leu Met Ile Thr Val Ala Gln Asn
Trp Met Tyr Pro 405 410 415Ile Val Val Arg Ala Gln Glu Ala Ser Lys
Leu Pro Met Ser Arg Glu 420 425 430Lys Val Leu Gln Tyr Cys Leu Val
Leu Leu Asp Met Ile Pro Pro Phe 435 440 445Leu Met Glu Tyr Leu Phe
Thr Phe Phe Leu Ile Trp Asp Val Ile Leu 450 455 460Asn Ala Ile Ala
Glu Leu Ser Arg Phe Ala Asp Arg Asp Phe Tyr Gly465 470 475 480Pro
Trp Trp Ser Cys Thr Asp Trp Ser Glu Phe Ala Arg Ile Trp Asn 485 490
495Arg Pro Val His Lys Phe Leu Leu Arg His Val Tyr Gln Ser Thr Ile
500 505 510Ser Thr Phe Lys Leu Asn Lys Asn Gln Ala Ser Leu Val Thr
Phe Ile 515 520 525Ile Leu Ser Phe Val His Glu Phe Val Met Phe Val
Ile Phe Arg Lys 530 535 540Val Arg Phe Tyr Met Leu Ala Leu Gln Met
Ser Gln Leu Pro Leu Ile545 550 555 560Met Ile Ser Arg Thr Lys Phe
Met Arg Asp Lys Lys Val Leu Gly Asn 565 570 575Val Ile Cys Trp Val
Gly Phe Ile Ser Gly Pro Ser Met Ile Cys Thr 580 585 590Leu Tyr Leu
Val Phe 595661794DNAPichia guilliermondii 66atgtccaagg aaaacttact
taagatcagc cagtataata ctgagagaag accgtcgttg 60gccacagacg ttgactactc
ttccaccgat ttatccagtc gtctggattc ggccaacacg 120acaaacggaa
caccgaccgt aactcttcac aagaggcaat cgtctacaga gctcttgtct
180gagtcacctg aacagaaaag gttcttgaaa acgatagaca ctttgaatcg
aaccacaaat 240tctagattac gccagaggtt aaaccgtgag ggcgataagc
ataaaaagga acacaaagaa 300catgaaaaac ataaagatga ccattctaaa
tacaagtctc ggtttggaga tatccatttc 360tactcaaaca tgacaaccat
cttcgatgct gattacttta aggaatcgca gttctttgga 420gtttacattc
tcttttggct cggaacggca ttcttaattc tcaacaactt ggtccataca
480tttttggaga acggagacaa tcttctcgat ggaccagttg tcagaacgtt
taaaaaggac 540ttacttaaaa ttgctcttac agacttggga atgtacttga
cgatgtacgt ctctgtcttt 600attcaattgg gcatccgcaa aggatggtat
agctggagct caacaggagc caccttgcaa 660aacatatact cattcgtgta
cttctttgcc tggagttact ttgcgtcgcc aaagtacatg 720gactaccctt
ggattggaaa ggtgtttctt gcacttcaca gcttggtgtt tctcatgaaa
780atgcattctt atgccacata caacggctat ctttggaaca tcttcaacga
gcttcaagtg 840tcacgaaagt acttgaagat attggacgag accgatgaat
ccatgattga gggtaagagt 900gtttccgatt tgcgaaaggc tttggtagac
agcattggtt tctgctcata cgagttggag 960taccagtcca aatcaacgag
cgtgaacacg gatgtcgaaa tcaccggcga caagaacaaa 1020ttgaacacaa
ccaagtctac cagttcactc gatgacgact atgtgagttt ccccaataac
1080attacgtttt tcgatttttt caggtattca atgtttccaa cagtggtgta
ttctctcaag 1140ttcccacgta caaagcgtat tagatggggt tacgtcatgg
aaaagtcatt tgcagtgttt 1200ggcatcatct tcttgatgat caccgtcgct
caaaactgga tgtatcctat cgttgtacga 1260gcacaagagg ctagcaaact
cccaatgtca agagaaaagg tattgcagta ctgtttggtt 1320ttactagaca
tgattccacc atttctcatg gaatatcttt tcaccttttt cttgatttgg
1380gacgtgatcc taaatgcgat agccgaattg agtaggtttg cagatcggga
cttttatggt 1440ccttggtggt cttgtaccga ttggtcggaa tttgcaagaa
tttggaatcg tcctgttcac 1500aaatttttgc ttcgtcatgt gtaccagtca
actatcagta ctttcaaact caataaaaac 1560caagcgtcgt tggtgacgtt
tatcattctg agttttgttc atgagtttgt catgtttgtc 1620atttttagaa
aggtgagatt ctacatgttg gcgctccaga tgtctcagct tccattgata
1680atgattagtc gaacaaaatt catgagagac aaaaaagtgt tgggaaatgt
tatctgctgg 1740gtaggattca tttctggacc atcgatgatc tgtactttgt
atttagtatt ttaa 179467515PRTArxula adeninivorans 67Met Ala Thr Ala
Thr Ala Ile Ala Thr Val Thr Glu Gly Leu Gly Leu1 5 10 15Asp Lys Val
Leu Ser Lys Glu Gln Pro Gly Leu Ser Lys Leu Ala Pro 20 25 30Arg Ala
Asn Thr Asn Val Gln Pro Thr Gln Leu Gln Ser Pro Ser Pro 35 40 45Pro
Gln Ser Arg Ser Ser Ser Pro Ile Ser Ala Ser Ser Ser Ser Glu 50 55
60Ser Leu Glu Leu Lys Val Pro Lys Ala Lys Ser Pro Ser Ser Ser Lys65
70 75 80His Lys Pro His Tyr Arg Pro Val His Val Arg Ser Thr Ala Ser
Ile 85 90 95Leu Ser Arg Asp Pro Ala Ala Arg Thr Glu Pro Pro Ser Tyr
Ser Gly 100 105 110Phe Arg Asn Leu Ala Met Ile Ala Leu Ala Val Ser
Asn Met Arg Leu 115 120 125Leu Leu Glu Asp Tyr Gln Asn Tyr Gly Val
Phe His Thr Leu Asn Ile 130 135 140Met Gly Leu Ser Ala His Asp Val
Arg Leu Thr Leu Ala Leu Thr Ala145 150 155 160Ser Val Pro Phe His
Leu Phe Val Ala Leu Ala Ile Glu Arg Ile Ala 165 170 175Val Leu Thr
Met Pro Ser Lys Ser Thr Ala His Asn His Arg Ser Lys 180 185 190His
Leu Trp Gly Leu Phe Ala Val Leu His Ala Leu Asn Ala Ala Ala 195 200
205Val Leu Ala Ile Ser Ser Tyr Thr Val Tyr Ser Arg Met Trp Ser Pro
210 215 220Ala Val Gly Thr Leu Cys Glu Cys His Ala Ile Val Val Cys
Phe Lys225 230 235 240Val Ala Ser Tyr Ala Leu Thr Asn Arg Asp Leu
Arg Asp Ala Ala Ile 245 250 255Asp Gly Leu Glu Thr Thr Asp Pro Leu
Leu Ser Lys Leu Pro Tyr Pro 260 265 270Ser Asn Leu Thr Leu Ser Asn
Leu Val Tyr Phe Trp Trp Ala Pro Thr 275 280 285Leu Val Tyr Gln Pro
Ile Tyr Pro Arg Trp Pro Leu His Arg Arg Trp 290 295 300Gly Phe Ile
Phe Ser Arg Leu Leu Glu Ile Met Gly Ser Met Val Leu305 310 315
320Ile Trp Phe Ile Ser Thr Gln Tyr Ala Asn Pro Ile Leu Glu Ser Ser
325 330 335Leu Gly His Phe Glu Gln Phe Asn Val Val Lys Ile Ser Glu
Cys Leu 340 345 350Leu Lys Leu Ala Ser Val Ser Met Ala Ile Trp Leu
Leu Gly Phe Phe 355 360 365Cys Leu Phe Gln Ser Phe Leu Asn Leu Leu
Ala Glu Leu Val Arg Phe 370 375 380Gly Asp Arg Glu Phe Tyr Gln Asp
Trp Trp Asn Ala Gly Ser Val Gly385 390 395 400Thr Tyr Trp Arg Lys
Trp Asn Arg Pro Val His Asn Tyr Phe Leu Arg 405 410 415His Phe Tyr
Ile Pro Met Leu Lys Arg Gly Tyr Ser Gln Arg Thr Ala 420 425 430Ser
Val Ile Val Phe Phe Leu Ser Ala Ile Leu His Glu Val Ala Val 435 440
445Gly Val Pro Thr Gln Ser Leu Ile Gly Val Ala Phe Val Gly Met Gly
450 455 460Ala Gln Ile Pro Leu Val Leu Ala Thr Ser Pro Leu Glu Lys
Met Gly465 470 475 480Glu Thr Gly Ala Thr Ile Gly Asn Cys Ile Phe
Trp Leu Ser Phe Phe 485 490 495Leu Gly Gln Pro Met Gly Val Leu Leu
Tyr Tyr Phe Ala Trp Asn Met 500 505 510Lys His Gln
515681548DNAArxula adeninivorans 68atggccaccg ctactgctat cgctacggtc
acggagggcc tgggactaga taaggtgcta 60tccaaggagc agccaggctt gtcgaagcta
gctcctcgag cgaatacaaa tgtacaaccg 120acccagttgc agtccccgtc
tccaccacaa tctcgatctt cgtctccaat ttcggcctcc 180tcatcatcag
agtccctgga gctcaaggtg cccaaggcca aatcgccatc atcttccaaa
240cacaaaccac actaccgccc cgtgcatgtg cggtcaacag catccatcct
gtccagagac 300ccggccgcca gaaccgagcc tccctcttac tctgggttca
ggaacctagc catgattgca 360ttggcggttt ctaatatgcg cctccttctc
gaggactatc aaaactatgg cgtgttccac 420actctcaaca ttatgggctt
gagcgcacac gacgttcgcc tcacactggc attgacagct 480tcggttccgt
tccatctgtt tgtggccctg gccattgagc gcatcgcagt cctcactatg
540ccctccaaat ctacagcaca caaccaccgc tcaaagcatc tctggggctt
gtttgcagtt 600ctgcatgctc tcaacgccgc tgctgtgcta gcaatcagct
catacaccgt atacagtcgc 660atgtggagtc ctgctgtggg aacattgtgc
gaatgccacg caatcgtggt atgctttaag 720gtggcatcgt atgcgcttac
caaccgagac ttacgagatg ctgccattga tgggctagag 780acaactgacc
ctctgttgtc caagttgccc tacccatcca accttacctt gtcaaatctc
840gtgtatttct ggtgggcccc aaccctagtg tatcagccaa tttaccctcg
atggcccctg 900catcgacgat ggggcttcat cttttctcgc ctgctcgaga
ttatgggatc tatggtacta 960atctggttca tttccaccca atacgccaac
cccattttgg aatcatcctt ggggcacttt 1020gaacagttta acgtggttaa
aatctcagaa tgtctcctca aattagcatc ggtctccatg 1080gccatctggc
ttttgggttt cttttgtctc tttcaatcgt ttttgaactt gctggcagaa
1140ttggttcgtt ttggcgaccg cgagttctac caagactggt ggaacgccgg
ctcagtaggt 1200acctactggc gcaaatggaa ccgaccagtg cacaactatt
tcttgcgcca tttctacatc 1260ccaatgctca agcgaggtta ttcacagcgc
actgcctcgg tcattgtatt ctttttatct 1320gccattctcc atgaagttgc
tgttggcgtg cctactcagt ccttgattgg agttgcgttt 1380gtaggcatgg
gtgcccagat tcctctagtg ctggccacta gtcctttgga aaagatgggc
1440gaaactggcg caactattgg caactgcatc ttttggctct ctttcttcct
gggccagcca 1500atgggggtac tgctttacta ctttgcgtgg aatatgaagc accagtag
154869564PRTPhaeodactylum tricornutum 69Met Asp Glu Thr Glu Ile Thr
Pro Leu Leu Arg Phe Ser Thr Pro Ser1 5 10 15Arg Ala Glu His Ser Ser
Trp Ile Lys Leu Ala Ser Glu Ser Cys Ala 20 25 30Tyr Ser Glu Thr Asp
Glu Phe Leu Ala Asp Glu Ala Ala Arg Ala Thr 35 40 45Gln Arg Ala Leu
Gln His Gln Glu Ala Leu Gln Met Ala Gln Ala Met 50 55 60Pro Gly Ala
Lys Pro Gly Thr Leu Pro Pro Leu Tyr Phe Ala Pro Thr65 70 75 80Ile
Lys Arg Ser Arg Ser Phe Ala Lys Leu Gln Glu His His Gly Asp 85 90
95Gly Met Pro Arg Val Asn Met Arg Arg Thr Lys Ser Arg Asp Phe Asn
100 105 110Ala Asp Lys Leu Asp Ala Arg Ser Thr Lys Gly Tyr Pro Pro
Ser Lys 115 120 125Pro Met His Arg Ala Ala Glu Pro Ser Tyr Leu Ser
Ala Asp Ala Pro 130 135 140Ile Gln Asn Tyr Arg Gly Phe Leu Asn Leu
Gly Val Ile Ile Leu Ile145 150 155 160Val Ser Asn Phe Arg Leu Ile
Leu Gly Thr Ile Arg Ser Asn Gly Phe 165 170 175Val Leu Thr Thr Ala
Val Lys His Tyr Lys Asn Leu Asn His Leu Lys 180 185 190Glu Asp Pro
Trp Gln Glu Phe Pro Phe Val Ser Gly Phe Leu Leu Gln 195 200 205Leu
Val Phe Val Ser Ile Ala Phe Gly Ile Glu Trp Met Leu Cys Arg 210 215
220Lys Tyr Phe Asn Glu Asn Phe Gly Met Ile Leu His His Phe Asn
Ala225 230 235 240His Ser Ala Leu Leu Ile Pro Leu Gly Ile Val Trp
Asn Leu Ile Asp 245 250 255Arg Pro Ala Val Gly Ala Ile Leu Leu Leu
His Ala Thr Ile Thr Trp 260 265 270Met Lys Leu Ile Ser Tyr Met Leu
Ala Asn Glu Asp Tyr Arg Leu Ser 275 280 285Ser Arg Arg Val Gly Gly
Asn Pro His Leu Ala Thr Leu Ala Leu Val 290 295 300Glu Asn Leu Asp
Ser Asp Glu Ala Asn Ile Asn Tyr Pro Gln Asn Val305 310 315 320Thr
Leu Arg Asn Ile Phe Tyr Phe Trp Cys Ala Pro Thr Leu Thr Tyr 325 330
335Gln Ile Ala Phe Pro Lys Ser Pro Arg Val Arg Tyr Trp Lys Ile Ala
340 345 350Asp Ile Leu Met Arg Met Thr Val Ser Ile Ala Leu Phe Thr
Phe Leu 355 360 365Leu Ala Gln Ile Val Gln Pro Ala Leu Glu Glu Leu
Val Ser Asp Leu 370 375 380Asp Glu Thr Asn Gly Ser Tyr Thr Ala Ala
Ile Phe Ala Glu Tyr Trp385 390 395 400Leu Lys Leu Ser Ile Ala Asn
Thr Tyr Leu Trp Leu Leu Met Phe Tyr 405 410 415Thr Tyr Phe His Leu
Tyr Leu Asn Leu Phe Ala Glu Leu Leu Arg Phe 420 425 430Gly Asp Arg
Val Phe Tyr Lys Asp Trp Trp Asn Ser Ser Glu Val Ser 435 440 445Ala
Tyr Trp Arg Leu Trp Asn Met Pro Val His Tyr Trp Leu Ile Arg 450 455
460His Val Tyr Phe Pro Cys Val Arg Leu Lys Met Pro Lys Val Ala
Ala465 470 475 480Thr Phe Val Val Phe Phe Leu Ser Ala Val Met His
Glu Val Leu Val 485 490 495Ser Val Pro Phe His Ile Ile Arg Pro Trp
Ser Phe Ile Gly Met Met 500 505 510Met Gln Ile Pro Leu Val Ala Phe
Thr Lys Tyr Leu Tyr Arg Lys Phe 515 520 525Pro Gly Gly Ser Ile Gly
Asn Val Leu Phe Trp Met Thr Phe Cys Val 530 535
540Ile Gly Gln Pro Met Ala Ile Leu Leu Tyr Tyr His Asp Ile Met
Asn545 550 555 560Arg Lys Gly Asn701695DNAPhaeodactylum tricornutum
70atggatgaga ccgaaattac acctttgttg cgtttttcga caccttcccg agccgaacac
60tcgtcctgga taaagcttgc ctcggaatcc tgtgcttaca gcgaaacgga cgagtttctc
120gctgacgagg ccgctcgcgc aacccagcgt gctttgcaac atcaagaagc
gctgcaaatg 180gcccaagcca tgcctggggc aaagccagga acgctgccgc
cactctactt cgcgcctacc 240ataaagcgtt cgcgttcctt tgctaagcta
caagaacatc atggagatgg gatgcctcgg 300gtaaatatgc gtcggaccaa
atcgcgagat tttaacgcgg ataagttgga tgcgcgaagt 360accaagggct
atcccccttc caagccgatg catcgtgcgg cagagccctc atacctcagc
420gcggatgctc ccattcaaaa ctaccgagga tttctgaatt taggcgttat
tattttgatt 480gtttctaact ttcggctgat cttgggcaca atccgtagca
acggatttgt cttgacgact 540gcagtgaagc actacaagaa cctaaatcac
ctcaaggaag atccctggca ggaatttcct 600tttgtatcag gatttcttct
ccagctcgtc tttgtttcga ttgcgtttgg gatcgaatgg 660atgttgtgcc
ggaaatactt caacgaaaac ttcggcatga tccttcatca cttcaatgcc
720cactcagcct tgctgatacc tttaggtatt gtttggaatc tcatcgatag
acctgcggtt 780ggtgcaattt tgcttttaca cgctacgata acatggatga
aactcatttc ttacatgttg 840gcgaacgaag attaccggct atcatcgcgt
cgcgttgggg gcaacccaca cctagctacg 900ctcgcattag tcgaaaatct
agattcagat gaggcgaaca ttaactaccc ccaaaatgtt 960actctccgca
acatttttta tttttggtgt gctccgacgt tgacttacca gattgccttc
1020ccgaagtccc cgcgagttcg ctattggaaa atcgcggata tcctgatgcg
catgacggtg 1080tccatcgcac tattcacctt tttgctggca caaattgttc
agcctgcatt ggaagagcta 1140gtgagcgacc tggacgagac caatggatcc
tacaccgcag caatatttgc cgagtactgg 1200ctgaaacttt cgattgctaa
cacatattta tggcttctta tgttctatac atatttccat 1260ttgtatctga
acctctttgc tgagcttctg cgatttggag atcgtgtgtt ctacaaagat
1320tggtggaatt cgtcggaagt atctgcatat tggaggcttt ggaatatgcc
tgttcactat 1380tggttgatcc gacatgtgta tttcccctgc gtgcgactga
agatgccgaa ggtcgctgca 1440acctttgtcg tttttttcct ctccgccgtt
atgcacgagg tgcttgtcag cgtacccttt 1500catattattc gtccgtggtc
ttttatcggg atgatgatgc agattccttt ggttgcgttc 1560acaaagtatc
tctatcgcaa attcccgggc ggctcgattg gtaatgtcct gttctggatg
1620acattttgcg tcattggcca gccaatggcg attctcttgt actatcatga
tattatgaat 1680cgaaaaggaa attga 169571503PRTMetarhizium acridum
71Met Ser Thr Ala Thr Thr Thr Ser Val Ser Pro Ala Asn Gly Thr Val1
5 10 15Ser Lys Arg Asn Ala Thr Lys Arg Arg Asn Gly Asn Ala Ser Pro
Gly 20 25 30Pro Val Glu Glu Glu Ser Glu Asp Ala Ala Ala Ala Glu Lys
Pro Arg 35 40 45Ala Ser Val Ala Gln Lys Asn Tyr Arg His Val Ala Ala
Val His Ser 50 55 60Lys Ser Arg Pro Ser Cys Leu Ser His Asp Ser Asp
Ala Thr Pro Ser65 70 75 80Phe Ile Gly Phe Arg Asn Leu Met Val Ile
Val Leu Asp Val Leu Ile 85 90 95Gly Gly Leu Leu Tyr Phe Leu Ile Pro
Cys His Leu Leu Val Ala Tyr 100 105 110Leu Ile Glu Leu Ala Ala Ala
Lys Gln Ala Arg Gly Ser Arg Lys Arg 115 120 125Leu Lys Pro Gly Ser
Thr Val Pro Ser Glu Gln Asp Asn Ser Lys Phe 130 135 140His Ser Thr
Trp Val Leu Val Ala Trp Ala His Gly Ile Asn Met Thr145 150 155
160Leu Ala Leu Ala Leu Thr Thr Phe Met Val Tyr Phe Tyr Ile His His
165 170 175Pro Leu Val Gly Thr Leu Thr Glu Met His Ala Val Ile Val
Ser Leu 180 185 190Lys Thr Ala Ser Tyr Ala Phe Thr Asn Arg Asp Leu
Arg His Ala Tyr 195 200 205Leu His Pro Val Lys Gly Glu Phe Ile Pro
Glu Leu Tyr Ser Lys Cys 210 215 220Pro Tyr Pro Asn Asn Ile Thr Phe
Gly Asn Leu Ala Tyr Phe Trp Trp225 230 235 240Ala Pro Thr Leu Val
Tyr Gln Pro Val Tyr Pro Arg Thr Asp Lys Ile 245 250 255Arg Trp Val
Phe Val Phe Lys Arg Leu Gly Glu Val Cys Cys Leu Ser 260 265 270Ala
Phe Ile Trp Phe Ala Ser Phe Gln Tyr Ala Ala Pro Val Leu Gln 275 280
285Asn Ser Leu Asp Lys Ile Ala Ser Leu Asp Leu Leu Met Ile Leu Glu
290 295 300Arg Leu Leu Lys Leu Ser Thr Ile Ser Leu Val Ile Trp Leu
Ala Gly305 310 315 320Phe Phe Ala Leu Phe Gln Ser Phe Leu Asn Ala
Leu Ala Glu Val Leu 325 330 335Arg Phe Gly Asp Arg Ser Phe Tyr Asp
Asp Trp Trp Asn Ser Glu Ser 340 345 350Leu Gly Ala Tyr Trp Arg Thr
Trp Asn Arg Pro Val Tyr Thr Tyr Phe 355 360 365Lys Arg His Val Tyr
Val Pro Met Ile Gly Arg Gly Trp Ser Pro Trp 370 375 380Ala Ala Ser
Cys Ala Val Phe Phe Val Ser Ala Val Leu His Glu Val385 390 395
400Leu Val Gly Val Pro Thr His Asn Ile Ile Gly Thr Leu Ser Ser Val
405 410 415Leu Ser Ile Val Leu Thr Leu Val Pro Asn Leu Tyr Ser Gly
Val Ala 420 425 430Phe Leu Gly Met Phe Leu Gln Leu Pro Leu Ile Ala
Ile Thr Ala Pro 435 440 445Leu Glu Lys Met Lys Trp Gly His Thr Gly
Arg Val Met Gly Asn Val 450 455 460Ile Phe Trp Val Ser Phe Thr Ile
Phe Gly Gln Pro Phe Ala Ala Leu465 470 475 480Met Tyr Phe Tyr Ala
Trp Gln Ala Lys Tyr Gly Ser Val Ser Lys Glu 485 490 495Pro Ile Leu
Ala Leu Gln Thr 500721512DNAMetarhizium acridum 72atgagcacgg
ccaccaccac cagtgtcagc ccagcgaatg gcaccgtgag caagagaaat 60gccaccaagc
gtcgcaacgg caatgcatct cccggcccgg tggaagaaga atccgaagac
120gcagccgcag ccgagaagcc cagagcctct gtcgcccaga agaactatcg
ccacgtagca 180gcagtgcatt ccaagagccg cccgtcgtgc ctaagccacg
actccgatgc cacgccaagc 240tttatcgggt ttcgaaatct catggtcatt
gttttggatg tcctcatcgg cggacttctc 300tactttctca ttccctgcca
tctgttggtt gcctacttga tcgaattggc cgccgcaaaa 360caggctcgag
gatcccgaaa gcgcctcaaa ccaggctcta ctgtaccgtc ggaacaagac
420aattccaagt tccattcaac atgggttctg gtggcctggg ctcatggtat
caatatgacg 480cttgctttag ccctcacaac ctttatggtt tacttttaca
tccaccaccc gctcgttggg 540accctgaccg agatgcatgc cgtcattgtg
tcgttgaaga cagcctcgta cgcattcacc 600aaccgagatc ttcgccacgc
ttacctgcac cccgttaaag gagagtttat tcctgaactc 660tactcgaaat
gcccgtaccc gaataacatc acctttggca acctcgccta cttctggtgg
720gcgccgacgc tggtctatca gcccgtatac ccgcgcaccg acaagatcag
atgggtcttt 780gtttttaaga ggctgggcga agtatgctgt ttgagcgcat
tcatctggtt cgccagcttc 840caatacgccg cgccggttct gcagaattcg
ctcgacaaga ttgcttcgtt ggacttactc 900atgatcctag agcggctgct
gaagctgtca accatttctc tggttatttg gctggcagga 960ttctttgccc
tattccagtc cttcttaaac gcacttgccg aagtgctgcg gttcggcgac
1020cgatcatttt acgacgactg gtggaacagc gagagtctcg gagcctactg
gagaacgtgg 1080aacaggcccg tatatacgta ctttaagcgc catgtgtatg
tacccatgat tgggcgtgga 1140tggagcccat gggctgcaag ttgcgccgtc
ttttttgtgt ctgccgtgtt acacgaggtt 1200cttgttggtg ttcccaccca
caacattatc ggtacgctat cctccgtctt atccatcgtc 1260ttgaccctcg
ttcctaacct atattcaggc gttgcttttc taggcatgtt cttgcagctt
1320cctctcatcg ccatcacggc ccctctagag aaaatgaaat gggggcatac
cggcagagta 1380atgggaaacg taatcttttg ggtgtccttt accatcttcg
gtcagccatt tgcggcattg 1440atgtactttt acgcatggca ggccaagtac
ggtagcgtca gtaaagaacc gattcttgcg 1500ttgcagacat ga
151273516PRTOphiocordyceps sinensis 73Met Ala Ala Thr Gly Thr Ser
Val Glu Pro Ser Thr Gly Thr Ala Thr1 5 10 15Gln Arg His Ser Gly Lys
Asp Gln Thr Gly Val Glu Pro Arg Thr Gly 20 25 30Thr Val Lys Thr Ser
Gln Lys Lys Tyr Arg His Val Val Val Val His 35 40 45Ser Gln Val Arg
Pro Ser Cys Leu Ser His Asp Ser Asp Ala Ala Pro 50 55 60Ser Phe Ile
Gly Phe Arg Asn Leu Met Val Ile Val Leu Val Val Gly65 70 75 80Asn
Leu Arg Leu Met Ile Glu Asn Ile Gln Lys Ala Arg Ser Tyr Leu 85 90
95Ser Phe Ile Pro Gly Gln Cys Ala Pro Gly Tyr Gly Val Leu Ile Cys
100 105 110Ile Arg Cys His Ala Tyr Ser Arg Gln Asp Ile Leu Val Gly
Gly Leu 115 120 125Leu Tyr Ile Leu Ile Pro Cys His Leu Leu Ala Ala
Tyr Leu Ile Glu 130 135 140Leu Ala Ala Ala Gln Gln Ala Leu Gly Ser
Arg Lys Arg Leu Lys Asp145 150 155 160Gly Ala Ala Ser Pro Glu Glu
Glu Asp Arg Asn Ser Asn Lys Phe His 165 170 175Ala Thr Trp Leu Ile
Val Ala Trp Val His Ala Val Asn Ile Thr Leu 180 185 190Ala Leu Val
Val Thr Ser Ala Val Val Tyr Phe Tyr Ile His His Pro 195 200 205Leu
Ile Gly Thr Leu Thr Glu Met His Ala Ile Ile Val Trp Leu Lys 210 215
220Thr Ala Ser Tyr Ala Phe Thr Asn Arg Asp Leu Arg His Ala Tyr
Leu225 230 235 240His Pro Val Glu Gly Glu Leu Val Pro Asp Met Tyr
Ala Lys Cys Pro 245 250 255Tyr Pro Gln Asn Ile Thr Phe Gly Asn Leu
Val Tyr Phe Trp Trp Ala 260 265 270Pro Thr Leu Val Tyr Gln Pro Val
Tyr Pro Arg Thr Asp Lys Ile Arg 275 280 285Trp Leu Phe Val Ala Lys
Arg Leu Gly Glu Val Phe Cys Leu Ser Ala 290 295 300Phe Ile Trp Phe
Ala Ser Phe Gln Tyr Ala Ala Pro Val Leu Arg Asn305 310 315 320Ser
Leu Asp Lys Ile Ala Ser Leu Asp Phe Ala Ser Ile Phe Glu Arg 325 330
335Leu Val Lys Leu Ser Thr Ile Ser Leu Val Ile Trp Leu Ala Gly Phe
340 345 350Phe Ala Leu Phe Gln Ser Phe Leu Asn Ala Leu Ala Glu Val
Leu Arg 355 360 365Phe Gly Asp Arg Ala Phe Tyr Asp Asp Trp Trp Asn
Ser Glu Ser Leu 370 375 380Gly Ala Tyr Trp Arg Thr Trp Asn Lys Pro
Val Tyr Thr Tyr Phe Lys385 390 395 400Arg His Val Tyr Met Pro Met
Ile Gly Arg Gly Trp Ser Pro Arg Val 405 410 415Ala Ser Leu Val Val
Phe Phe Ile Ser Ala Val Leu His Glu Ile Leu 420 425 430Val Gly Leu
Pro Thr His Asn Val Ile Gly Val Ala Phe Leu Gly Met 435 440 445Phe
Leu Gln Leu Pro Leu Ile Ala Ile Thr Ala Pro Met Glu Lys Met 450 455
460Arg Leu Gly Lys Gly Gly Lys Leu Val Gly Asn Val Ile Phe Trp
Val465 470 475 480Ser Phe Thr Ile Phe Gly Gln Pro Phe Ala Thr Leu
Met Tyr Phe Tyr 485 490 495Ala Trp Gln Ala Lys Tyr Gly Ser Val Ser
Arg Glu Met Gln Gln Ala 500 505 510Ala Ser Ile Lys
515741551DNAOphiocordyceps sinensis 74atggcggcta cggggaccag
cgtcgagccc tcgactggta ccgcgacaca acgccactcc 60ggcaaggatc agactggggt
cgagccacgc accggcacgg tcaagacatc ccagaaaaag 120tatcgccatg
tcgttgtcgt ccactcccag gtccggccct cgtgcctcag ccacgattca
180gatgccgccc ccagcttcat tggcttccgc aatctcatgg ttattgtcct
ggtcgtcggc 240aacttgcgat tgatgattga aaacatccaa aaggctcgtt
catacctgtc gttcatacct 300ggccaatgcg cccccggcta cggagtcttg
atctgcatcc gctgccacgc ctacagccgc 360caagacattc tcgtcggcgg
gctgctgtac atcctcattc cctgccatct cctggccgcc 420tatctcatcg
agctcgccgc cgcccagcag gcactggggt cgagaaagcg cctcaaggat
480ggcgccgcca gcccggagga ggaggaccgc aacagcaaca agtttcacgc
gacatggctc 540atcgtcgcct gggtccatgc cgtcaacatc accctggccc
tggtcgtgac ctcggccgtc 600gtctactttt acatccacca cccactcatc
ggcaccctca ccgaaatgca cgccatcatc 660gtctggctca agacggcctc
gtacgccttt actaaccgcg acctgcgcca cgcgtacctg 720caccccgtcg
agggcgagct cgtcccggac atgtacgcca agtgcccgta tccgcaaaac
780atcacctttg gcaacctcgt ctacttctgg tgggccccga cgctcgtcta
ccagcccgtc 840tatccccgga ccgacaagat caggtggctc tttgtcgcca
agcggctggg agaggtcttt 900tgcttgagcg ccttcatctg gttcgccagc
ttccagtatg ccgcgcccgt cctgcgcaac 960tctctcgaca aaattgcttc
gctcgacttt gcctccatct ttgagcggct ggtgaagctg 1020tccaccatct
ccctcgtcat ctggctcgcc ggcttcttcg ccctcttcca gtcctttctc
1080aacgccctcg ccgaggtgct tcggttcggc gaccgggctt tctacgatga
ctggtggaac 1140agcgagagcc taggcgccta ctggcggacc tggaacaagc
ccgtctacac ctacttcaag 1200cgccacgtgt acatgcccat gatcgggcgt
ggctggagtc ccagggtggc cagtctggtc 1260gtcttcttca tctcagccgt
cctccacgag atccttgtcg ggctacccac tcacaacgtc 1320atcggcgtcg
cctttctcgg catgtttctc cagctgcctc tcatcgccat cacggcgccc
1380atggagaaga tgaggctcgg caaaggcggc aagctcgtag gcaacgtcat
cttctgggtg 1440tcgtttacca tctttggcca gccctttgcg acattgatgt
acttttatgc ttggcaggcc 1500aaatacggga gcgtgagcag ggagatgcag
caagcggcaa gcatcaagta a 155175510PRTTrichoderma virens 75Met Ala
Pro Pro Ala Glu Ser Ser Thr Thr Thr Ser Val Glu Ala Ser1 5 10 15Thr
Gly Ser Val Ser Arg Arg His Ala Ser Gln Ser Glu Ala Asp Leu 20 25
30Thr Ser Val Glu Pro Val Asn Gly Thr Thr Lys Asn Arg Leu Ser Lys
35 40 45Thr Pro Pro Lys Lys Tyr Arg His Val Ala Ala Val His Ser Gln
Thr 50 55 60Arg Pro Ser Cys Leu Ser His Asp Ser Pro Ala Ala Pro Ser
Phe Leu65 70 75 80Gly Phe Arg Asn Leu Met Val Ile Val Leu Val Val
Gly Asn Leu Arg 85 90 95Leu Met Ile Glu Asn Ile Gln Lys Tyr Gly Val
Leu Ile Cys Ile Arg 100 105 110Cys His Asp Tyr Arg Arg Gln Asp Val
Leu Leu Gly Leu Leu Leu Tyr 115 120 125Phe Leu Ile Pro Cys His Leu
Phe Ala Ala Tyr Leu Ile Glu Leu Val 130 135 140Ala Ala Lys Gln Ala
Glu Gly Ser Arg Lys Arg Ile Lys Asp Asn Asn145 150 155 160Ser Gly
Pro Ser Glu Ala Glu Arg Lys Lys Phe His Ser Ile Trp Val 165 170
175Leu Ala Ala Leu Ala His Gly Ile Asn Ile Thr Leu Ala Leu Ala Ile
180 185 190Thr Thr Val Val Val Tyr Phe Tyr Val Tyr His Pro Leu Ile
Gly Thr 195 200 205Leu Thr Glu Met His Ala Ile Ile Val Trp Leu Lys
Thr Ala Ser Tyr 210 215 220Ala Phe Thr Asn Arg Asp Leu Arg His Ala
Tyr Leu His Pro Val Glu225 230 235 240Gly Glu Glu Val Pro Asp Leu
Tyr Lys Ser Cys Pro Tyr Pro Gln Asn 245 250 255Val Thr Met Lys Asn
Leu Val Tyr Phe Trp Trp Ala Pro Thr Leu Val 260 265 270Tyr Gln Pro
Val Tyr Pro Arg Thr Asp Lys Ile Arg Trp Val Phe Val 275 280 285Phe
Lys Arg Leu Gly Glu Ile Phe Cys Leu Ala Val Phe Ile Trp Val 290 295
300Ala Ser Ala Gln Tyr Ala Thr Pro Val Leu Arg Asn Ser Leu Asp
Lys305 310 315 320Ile Ala Ser Leu Asp Leu Pro Asn Ile Leu Glu Arg
Leu Met Lys Leu 325 330 335Ser Thr Ile Ser Leu Val Ile Trp Leu Ala
Gly Phe Phe Ala Leu Phe 340 345 350Gln Ser Phe Leu Asn Ala Leu Ala
Glu Ile Met Arg Phe Gly Asp Arg 355 360 365Ser Phe Tyr Asp Asp Trp
Trp Asn Ser Glu Ser Leu Gly Ala Tyr Trp 370 375 380Arg Thr Trp Asn
Lys Pro Val Tyr Thr Tyr Phe Lys Arg His Val Tyr385 390 395 400Met
Pro Met Ile Gly Arg Gly Trp Ser Pro Ala Ala Ala Ser Phe Ala 405 410
415Val Phe Phe Val Ser Ala Val Leu His Glu Ile Leu Val Gly Val Pro
420 425 430Thr His Asn Ile Ile Gly Val Ala Phe Phe Gly Met Phe Leu
Gln Leu 435 440 445Pro Leu Ile Ala Ile Thr Thr Pro Leu Glu Lys Met
Lys Leu Gly His 450 455 460Gly Gly Arg Ile Leu Gly Asn Val Ile Phe
Trp Val Ser Phe Thr Ile465 470 475 480Phe Gly Gln Pro Phe Ala Ala
Leu Met Tyr Phe Tyr Ala Trp Gln Ala 485 490 495Lys Tyr Gly Ser Val
Ser Arg Leu Pro Gln Met Val His His 500 505 510761533DNATrichoderma
virens 76atggcgcctc ctgcagagtc ctccacgacg acaagcgtcg aggcctctac
cggctccgtg 60tctcgccgcc acgcctcaca aagtgaagca gatctaacgt cggtggagcc
cgtcaacggc 120acgaccaaga accggctctc caagacaccg ccgaagaaat
atcgccatgt cgctgcggtg 180cattcccaga cgcggccgtc gtgcctgagc
catgattccc ctgcggctcc cagctttctc 240ggattccgca atctcatggt
cattgtgctg gttgttggca atctccgatt gatgattgag 300aatattcaaa
agtacggcgt cttaatttgc atcaggtgtc acgactacag acgtcaagat
360gtgctcttgg gtcttttgct ttattttctt atcccctgcc atttgtttgc
agcatacctg 420atagagctgg tcgctgccaa gcaggctgag ggatccagga
agcgaatcaa ggacaacaac 480tctggcccgt cagaggcaga gcgcaagaag
ttccactcaa tctgggttct tgcggctttg 540gcccatggaa
tcaacatcac tcttgccctt gcaattacca ccgttgtggt ctacttttac
600gtctatcatc cgctgattgg cactttgacc gagatgcatg ccatcattgt
gtggctcaag 660acggcatcat atgcattcac caaccgagat cttcgtcacg
cctatctgca tccagttgag 720ggagaggaag tgcctgattt gtacaaatcc
tgcccctatc cacaaaacgt gacgatgaag 780aacttggtat acttctggtg
ggctccgact ctggtgtacc aacctgttta tccgcggacc 840gacaagattc
gatgggtgtt cgtgtttaag cgactaggag agatcttttg ccttgctgtg
900ttcatttggg ttgccagtgc ccaatatgcc acccccgttt tgcgcaactc
tctcgacaag 960attgcctctc ttgatttgcc caacatcttg gagcggctta
tgaaactctc gacaatctct 1020ttggtcatct ggctggccgg cttctttgcg
ctcttccaat ctttcttaaa cgcccttgcc 1080gagataatga ggtttggcga
taggtcattc tacgacgact ggtggaacag tgagagcttg 1140ggcgcctact
ggaggacgtg gaacaagcct gtttatactt acttcaagcg ccatgtctat
1200atgcccatga tcggacgagg ctggagcccg gccgctgcca gtttcgcagt
cttttttgtt 1260tctgccgttc ttcatgaaat tcttgttggt gttccaacac
ataacattat cggcgtcgct 1320ttcttcggca tgttccttca gcttcctctc
atcgccatta ctactccgct ggagaagatg 1380aaactcggtc atggtggccg
cattcttgga aatgtcatat tttgggtttc gtttacaatc 1440tttggacagc
cattcgcggc cctgatgtat ttctacgctt ggcaggccaa gtatggcagc
1500gtgagtaggt tacctcagat ggtgcaccac taa 153377579PRTArxula
adeninivorans 77Met Ile Arg Ala Ala Tyr Gly Ser Val Ser Arg Ala Arg
Asp Ser Leu1 5 10 15Thr Leu Arg Ala Pro Ser Phe Pro Thr Thr Ala Val
Glu Val Arg Asp 20 25 30Lys Ile Leu Trp Ile Leu Tyr Ala Trp Ile Glu
Met Phe Thr Asp Val 35 40 45Phe Ser Phe Trp Thr Glu Lys Val Trp Gly
Tyr Val Ser Thr Pro Thr 50 55 60Lys Glu Ser Ile Leu Arg Lys Gln Leu
Asp Glu Ala Lys Ser Tyr His65 70 75 80Glu Trp Glu Glu Leu Ser Tyr
Lys Leu Asp Ser Ile Leu Gly Asn Asp 85 90 95Ile Trp Arg Gln Asn Pro
Val Ser Arg Lys Tyr Asp Tyr Arg Leu Ile 100 105 110Ser Thr Arg Leu
Lys Glu Leu Val Ala Ala Arg Asp Asn Arg Asn Ile 115 120 125Glu Leu
Leu Met Asp Arg Leu Arg Ser Gly Leu Leu Arg Asn Ile Gly 130 135
140Ser Ile Ala Ser Thr His Leu Tyr Asn Arg Ala Tyr Ser Gly Thr
Lys145 150 155 160Leu Leu Ile Glu Asp Tyr Ile Asn Val Val Ile Gln
Cys Leu Glu Tyr 165 170 175Val Glu Arg Gly Gly Arg Pro Leu Thr Ala
Ser Ala Ser Lys Ile Pro 180 185 190Asn Gly Gly Glu Pro Pro Ser Pro
Arg Thr Tyr His Lys Pro Met Ile 195 200 205Thr Arg Gln Arg Lys Leu
Asn Phe Phe Asn Asp Thr Arg Gln Ser Phe 210 215 220Gly Ser Thr Ala
Val Val Leu His Gly Gly Ser Leu Phe Gly Leu Cys225 230 235 240His
Ile Gly Met Ile Lys Thr Leu Phe Asn Gln Gly Leu Leu Pro Arg 245 250
255Ile Val Cys Gly Ser Thr Val Gly Ala Leu Val Ala Ser Leu Val Cys
260 265 270Ser Cys Val Asp Glu Glu Val Tyr Glu Thr Leu Asp Asn Val
Ser Ser 275 280 285Glu Met Ser Pro Leu Arg Gln Gly Tyr Thr Asp Ile
Lys Tyr His Ser 290 295 300Val Ala Glu Gly Val Ile Ser Ser Met Cys
Pro Pro Glu Ile Leu Ile305 310 315 320Phe Glu Gln Tyr Ile Arg Glu
Lys Leu Gly Asp Leu Thr Phe Glu Glu 325 330 335Ala Tyr Gln Arg Thr
Gly Arg Ile Leu Asn Ile Pro Val Thr Pro Lys 340 345 350Ala Lys Pro
Gly Gln Val Ala Pro Pro Val Pro Thr Leu Leu Asn Tyr 355 360 365Leu
Ser Ser Pro Asn Val Val Val Trp Ser Ala Ala Gln Cys Ser Ile 370 375
380Gly Thr Gly Ile Ile His Lys Lys Val Glu Leu Leu Val Lys Gly
Leu385 390 395 400Asp Gly Gln Leu Lys Pro Tyr Leu Asp Ala Asp Asp
Ile Glu Tyr Thr 405 410 415Pro Ala Asn Gln Ala Val Tyr Ala Ala Asp
Arg Glu Ser Pro Tyr Thr 420 425 430Arg Leu Ser Glu Leu Phe Asn Val
Asn Asn Tyr Ile Val Ser Val Ala 435 440 445Arg Pro Tyr Phe Ala Pro
Ile Leu Leu Ser Asp Phe Lys Tyr Arg Ala 450 455 460Ala Lys Ser Phe
Lys Thr Arg Phe Leu Lys Leu Thr Arg Leu Glu Leu465 470 475 480Gln
Tyr Arg Leu Asn Gln Leu Ser Gln Leu Gly Leu Val Pro Pro Met 485 490
495Ile Gln Gln Trp Phe Val Asp Gly Asn Ile Pro Ala Gly Phe Gln Val
500 505 510Thr Val Val Pro Glu Leu Pro Ser Leu Ile Arg Asp Ile Gly
Lys Val 515 520 525Phe Asp Ser Asp Asn Ile Lys Glu Lys Val Asp Tyr
Trp Ile Lys Ile 530 535 540Gly Glu Arg Ser Val Trp Pro Val Leu Asn
Ile Ile Trp Ala Arg Cys545 550 555 560Ala Ile Glu Phe Val Leu Asp
Asp Leu Tyr His Ser Arg Arg Lys Asp 565 570 575Glu Leu
Asp781740DNAArxula adeninivorans 78atgattaggg ctgcctacgg gtcagtgtcc
agggcccgag attctttaac gttgagggct 60ccatcttttc ctaccactgc tgtggaggtc
cgtgacaaga ttctatggat tctgtatgcc 120tggattgaaa tgttcacgga
cgtctttagc ttctggacgg agaaggtgtg gggttatgtt 180tctactccta
ctaaagaaag cattcttaga aagcaactcg acgaggcaaa atcataccat
240gaatgggagg agctcagcta caaactagac tcaattttag ggaacgatat
ttggcgacag 300aaccctgtta gccgaaagta tgactatcgc ctgatttcta
cccgcctcaa ggaattggtt 360gctgctaggg ataatcgcaa cattgaattg
ctaatggatc ggctaaggtc aggcctgctt 420cgtaatattg gatcgattgc
aagtactcat ctctacaacc gagcgtattc gggcacaaaa 480ctgttaattg
aggattacat taatgtagtg attcaatgcc tggagtatgt tgaacggggc
540ggcaggccat tgactgcttc agcatccaag attcccaatg gcggtgaacc
cccttctcca 600cgaacctacc ataagcccat gattaccaga cagcgcaagc
tcaacttctt caatgataca 660cgccagtcgt ttggaagtac agctgtggta
cttcacggcg ggtccttgtt tggactttgc 720catattggca tgattaaaac
attgttcaac cagggtctac ttcctcgcat agtctgtggc 780tccacagtgg
gagcactagt agcgagtcta gtatgctcct gtgtggatga agaggtgtat
840gagactttgg ataatgtgtc ttcggaaatg tctcctctcc gccaaggata
cactgatata 900aagtaccatt cggtagccga aggggtcatt tcatcaatgt
gtccgccaga gattttgatt 960tttgaacagt acatccgaga aaaactcgga
gacctgacat ttgaagaagc atatcaacgc 1020accggccgca ttcttaatat
cccagtgaca ccaaaggcaa aaccaggtca ggtagcacca 1080ccagtcccga
cgctcctgaa ttatttgtcg agcccgaatg ttgtagtatg gtcagcagcg
1140caatgcagca ttggaacggg gattattcac aagaaggttg aacttttagt
aaaaggtctg 1200gatggtcaat taaaacctta tttggatgcg gatgatattg
aatacactcc tgcaaatcaa 1260gctgtatacg ctgctgatcg cgagagtccc
tatacaagat tgtctgagct gttcaatgtg 1320aacaattaca ttgtatcagt
agctcgcccc tactttgccc caattctgct ttcggatttc 1380aagtaccgtg
cagctaaaag cttcaagacc cggttcctca aactaacccg tctggagtta
1440cagtatcgtc tcaatcagct gtctcaattg gggctggttc cgcccatgat
tcaacaatgg 1500tttgtggacg gtaacattcc cgccgggttc caagttaccg
tggtgcctga attaccctca 1560cttattagag acatcggcaa ggttttcgat
tcggataata taaaggagaa ggtcgactac 1620tggattaaga tcggtgagcg
cagtgtgtgg ccagtgctga atattatctg ggcaaggtgc 1680gcaattgagt
ttgtgctcga cgatctatat cacagccgac gtaaagacga actcgactag
174079633PRTArxula adeninivorans 79Met Asn Pro Phe Asp Val Asp Tyr
Thr Asn Arg Asp His Leu Val Asp1 5 10 15Phe Glu Arg Ala Leu His Glu
Asp Glu Ala Ser His Ile Ile Ser Val 20 25 30Asn Asp Trp Ala Pro Val
His Ala Pro Leu Lys Arg Arg Leu Arg Arg 35 40 45Lys Pro Thr Asp Ser
Asp Pro Gly Thr Gly Leu Gly Tyr Thr Leu Leu 50 55 60Arg Trp Pro Ile
Leu Val Ala Ile Ala Leu Trp Leu Ala Leu Leu Ala65 70 75 80Phe Val
Tyr Ala Ile Val Arg Phe Trp Val Ala Leu Phe Glu Tyr Phe 85 90 95Val
Thr Trp Arg Gly Pro Arg Arg Asn Leu Arg Glu Lys Leu Arg Ser 100 105
110Ala Arg Ser Tyr Glu Glu Trp Ile Ser Ala Ala Lys Val Leu Asp Asp
115 120 125His Leu Gly Asn Thr Ser Trp Lys His Asn Pro Lys Phe Ser
Arg Tyr 130 135 140Asp Tyr Arg Thr Ile Asp Arg Ile Thr Asn Ser Leu
Arg Gln Leu Arg145 150 155 160Asn Gln Asn Lys Ala Glu Glu Val Gly
Ser Ile Leu Gln Gly Cys Val 165 170 175Lys His Asn Phe Ala Gly Thr
Gln Gly Gln Pro Leu Tyr Ser Gln Cys 180 185 190Tyr Tyr Gly Thr Lys
Asp Leu Val Glu Glu Phe Asn Ser Glu Ile Val 195 200 205Lys Ser Leu
Asp Tyr Leu Ala Thr His Pro Asp Leu Ser Pro Gln Ser 210 215 220Arg
Arg Leu Leu Phe Lys Met Phe Ser Lys Asn Phe Gly Lys Thr Ala225 230
235 240Leu Cys Leu Ser Gly Gly Ala Thr Phe Ala Tyr Arg His Phe Gly
Val 245 250 255Val Lys Ala Leu Leu Glu Gln Gly Leu Leu Pro Asn Ile
Ile Ser Gly 260 265 270Thr Ser Gly Gly Gly Leu Val Ala Ala Leu Val
Gly Thr Arg Thr Asn 275 280 285Ser Glu Leu Arg Glu Leu Leu Thr Pro
Gln Leu Ala Asp Lys Ile Thr 290 295 300Ala Cys Trp Glu Lys Phe Pro
Lys Trp Val Tyr Arg Phe Tyr Ser Thr305 310 315 320Gly Ala Arg Phe
Asp Ala Val Asp Trp Ala Glu Arg Ser Cys Trp Phe 325 330 335Thr Leu
Gly Ser Leu Thr Phe Arg Glu Ala Tyr Asp Arg Thr Gly Lys 340 345
350Ile Leu Asn Ile Ser Thr Val Pro Ala Asp Pro Asn Ser Pro Ser Ile
355 360 365Leu Cys Asn Tyr Ile Thr Ser Pro Asp Cys Val Ile Trp Ser
Ala Leu 370 375 380Leu Ala Ser Ala Ala Val Pro Gly Ile Leu Asn Pro
Val Val Leu Met385 390 395 400Met Lys Thr Lys Lys Gly Asn Leu Val
Pro Tyr Ser Phe Gly Asn Lys 405 410 415Trp Lys Asp Gly Ser Leu Arg
Thr Asp Ile Pro Val His Ala Leu Asn 420 425 430Val Tyr Phe Asn Val
Asn Phe Thr Ile Val Ser Gln Val Asn Pro His 435 440 445Ile Ser Leu
Phe Met Tyr Ala Pro Arg Gly Thr Val Gly Arg Pro Val 450 455 460Ser
His Arg Gln Gly Lys Gly Trp Arg Gly Gly Phe Leu Gly Ser Ala465 470
475 480Leu Glu Asp Met Leu Lys Leu Glu Ile Arg Lys Trp Leu Lys Leu
Met 485 490 495Lys Asn Leu Ser Leu Met Pro Arg Phe Phe Asn Gln Asp
Trp Ser Ser 500 505 510Val Trp Leu Gln Thr Phe Glu Gly Ser Val Thr
Leu Trp Pro Arg Ile 515 520 525Arg Leu Lys Asp Phe Tyr Tyr Ile Leu
Ser Asp Pro Thr Arg Glu Gln 530 535 540Met Glu Thr Met Ile Ile Ser
Gly Gln Arg Cys Thr Phe Pro Lys Leu545 550 555 560Leu Phe Ile Lys
His Gln Val Asn Ile Glu Arg Ala Ile Asp Arg Gly 565 570 575Arg Lys
His Asn Ala Lys Ala Arg Glu Glu Asn Gly Pro Gln Leu Arg 580 585
590Arg Val Asn Pro Phe Leu His Asp Leu Asp Asp Arg Val Tyr His Ser
595 600 605Ser Ser Ser Val Asp Pro Arg Glu Phe Gln Asp Asp His Asp
Asp Glu 610 615 620Asp Asp Asp Ser Thr Asp Ser Ser Met625
630801902DNAArxula adeninivorans 80atgaacccgt ttgatgtaga ttacacaaac
agggaccatc tggtcgactt tgaacgagct 60ttgcacgaag atgaggcttc ccatattata
tcggtaaacg actgggctcc agtgcatgct 120cctctcaagc gacggttgag
acgcaagccg acagattcgg atcctgggac aggattagga 180tacactttgc
ttagatggcc tattctggtg gcaattgcgc tgtggctggc cctgttagca
240tttgtgtacg ccatagtgag gttttgggtc gctctgtttg agtactttgt
tacctggcga 300ggaccccggc gcaatcttcg tgaaaagcta cgcagcgctc
gtagttacga ggaatggatt 360agtgctgcca aagttcttga tgaccatcta
ggaaatactt cttggaagca caacccaaag 420ttctctcgat acgactaccg
tactattgat cgcatcacta actcactgcg gcaactgcga 480aaccagaaca
aggccgagga ggttggctct attctacaag gatgcgtcaa gcacaacttt
540gctggaactc agggccaacc tttgtactct cagtgctact atggcacaaa
ggacctggta 600gaggagttca attctgaaat tgtgaaatcg ctcgattacc
tggcaaccca tccagacctg 660agtcctcaat ctagacgtct tttgttcaaa
atgttttcca agaattttgg aaagacggca 720ttgtgcctct ctggaggggc
aacatttgcc tatagacatt tcggagttgt taaagcgctc 780ttggaacagg
gcttgctgcc taatattatt tctggtactt ctggcggagg attggtagct
840gcgctagttg gtaccagaac aaatagtgaa ctccgtgagc ttctcactcc
tcaactggcc 900gacaagatca ccgcctgctg ggaaaagttc ccaaaatggg
tttatagatt ctacagcacc 960ggcgctcgat tcgatgccgt cgactgggct
gaacggtctt gctggtttac actaggaagc 1020ctgactttta gagaggccta
cgatcgaact ggaaagatcc tcaacatttc cactgttcct 1080gctgacccta
attccccttc aatcctctgc aattacatta cttctcccga ctgtgtcatc
1140tggtcggctt tacttgcttc tgctgcagta ccgggaattc tgaacccagt
ggtgctcatg 1200atgaagacga aaaagggcaa tctggtacct tacagctttg
gtaacaagtg gaaggatggt 1260tctctccgaa ctgatattcc tgtccacgca
ctcaacgtgt actttaacgt caacttcacc 1320atcgtgtccc aggtcaaccc
tcacatttct ctgttcatgt atgccccgcg gggaactgtg 1380ggtaggccag
tatctcaccg tcagggtaaa ggctggcgag gtgggttcct aggctcagct
1440ttggaagaca tgctgaagct ggaaattcgt aaatggctca aactcatgaa
aaaccttagt 1500cttatgccac ggtttttcaa tcaagattgg tcttcagtat
ggcttcaaac gttcgaggga 1560tccgtcacct tgtggccaag gatcaggcta
aaggactttt attatattct gtctgatccc 1620actcgggaac aaatggaaac
catgatcatt agtggacagc gatgcacatt cccaaagctc 1680ttgttcatca
agcaccaagt caacatagag cgggcaattg accgtggaag aaagcacaat
1740gcaaaagcca gggaggaaaa tggtccccag cttagacggg taaacccatt
cctgcacgac 1800ttggatgacc gtgtatacca ttccagctct agcgtggacc
ctcgcgagtt tcaggatgat 1860cacgatgatg aagacgacga cagcactgat
tctagcatgt aa 190281662PRTArxula adeninivorans 81Met Gln Ser Leu
Asp Leu Leu Asp Asp Arg Ser Trp Val Pro Asn Tyr1 5 10 15Ala Arg Val
Gly Leu Lys Ser Leu Lys Glu Tyr Leu Val Ser His Arg 20 25 30Tyr Gln
Ser Glu Glu Ala Arg Lys His Ala Glu Ala Leu Glu Arg Trp 35 40 45Thr
Lys Ser Gln Ala Gln Ala Glu Thr Tyr Glu Gln Trp Leu Phe Ala 50 55
60Ser Glu Gln Leu Asp Lys Leu Ser Gly Asn Asp Lys Trp Lys Glu Asp65
70 75 80Pro Val Ser Pro Tyr Tyr Asp Ser Val Leu Val Gln Gln Arg Leu
Gln 85 90 95Gln Leu Arg Asp Ala Arg Val Asn Ser Asn Met Asp Glu Leu
Leu Tyr 100 105 110Leu Val Arg Thr Ser Leu Gln Arg Asn Leu Gly Asn
Met Gly Asp Pro 115 120 125Arg Leu Tyr Val Arg Thr His Thr Gly Ser
Lys Thr Leu Ile Glu Gln 130 135 140Tyr Ile Ala Glu Val Glu Leu Ala
Leu Asp Thr Leu Leu Ser Cys Gly145 150 155 160Pro Gly Thr Phe Ser
Pro Lys Val Leu Leu Ser Asn Leu Ile Gln Thr 165 170 175Arg Lys Ala
Phe Gly Arg Thr Ala Leu Val Leu Ser Gly Gly Ser Thr 180 185 190Phe
Gly Ile Leu His Ile Gly Val Met Arg Glu Leu His Arg Ala His 195 200
205Leu Leu Pro Gln Val Ile Ser Gly Ser Ser Ala Gly Ser Ile Phe Ala
210 215 220Ser Met Leu Cys Ile His Leu Glu Asp Glu Ile Glu Glu Leu
Leu Gln225 230 235 240Leu Pro Leu His Lys Glu Ser Phe Glu Ile Phe
Glu Pro Ala Gly Glu 245 250 255Arg Glu Gly Leu Met Val Arg Leu Ala
Arg Phe Leu Lys His Gly Thr 260 265 270Trp Phe Asp Asn Lys Tyr Leu
Ser Thr Thr Met Arg Glu Leu Leu Gly 275 280 285Asp Leu Thr Phe Gln
Glu Ala Tyr Tyr Arg Thr Gln Arg Ile Leu Asn 290 295 300Val Thr Val
Ser Pro Ser Ser Met His Glu Met Pro Lys Ile Leu Asn305 310 315
320Tyr Leu Thr Ala Pro Asn Val Leu Ile Trp Ser Ala Val Cys Ala Ser
325 330 335Cys Ser Val Pro Phe Val Phe Asp Ser His Asp Ile Leu Ala
Lys Asn 340 345 350Pro Arg Thr Gly Glu Phe Tyr Ser Trp Asn Ala Ser
Thr Phe Ile Asp 355 360 365Gly Ser Val Tyr Asn Asp Leu Pro Leu Ser
Arg Leu Ala Glu Met Phe 370 375 380Asn Val Asn His Phe Ile Ala Cys
Gln Val Asn Pro His Val Val Pro385 390 395 400Phe Val Lys Phe Ala
Glu Thr Met Ser Leu Val Glu Ala Arg Pro Thr 405 410 415Thr Thr Glu
Pro Gly Ser Leu Thr Lys Leu Trp His Ser Thr Gln Leu 420 425 430Ala
Leu Ser Ser Glu Ile Ser His Tyr Leu Asp Leu Ala Ala Glu Met 435 440
445Gly Leu Phe Lys Asn Ile Ser Ser Lys Leu Arg Ser Val Leu Asp Gln
450 455 460Gln Tyr Ser Gly Asp Ile Thr Ile Leu Pro Glu Leu Tyr Leu
Ser Glu465
470 475 480Phe Gly Gln Ile Phe Lys Asn Pro Ser Lys Glu Phe Phe Gln
Lys Ala 485 490 495Glu Leu Arg Ala Ala Arg Ala Thr Trp Pro Lys Met
Ser His Ile His 500 505 510Asn Arg Val Ala Ile Glu Leu Ala Leu Val
Lys Ala Ile His Lys Leu 515 520 525Arg Ala Arg Ile Val Ser Gln Ser
Val His Glu Pro Gly Ser Ser Leu 530 535 540Gln Val His Ala Ala Asn
Asp Glu Gly Thr Leu Ala Pro Ile Arg Arg545 550 555 560Arg His Ser
Ser Thr Lys Leu His His Arg Arg Gln Arg Ser Asp Gly 565 570 575Met
Ala Val Lys Tyr Leu Val Arg Arg His Ser Leu Gln Tyr Phe Gly 580 585
590Thr Glu Gly Pro Gly Pro Ala Ala Leu Ser Arg Lys Lys Ser Ser Ala
595 600 605Gly Leu Thr Gln Ala His Thr Pro Thr Pro Ser Leu Thr Asn
Ser Val 610 615 620Ser Val Gly Gly Ser Pro Arg His Arg Arg Phe Thr
Thr Ser Ser Arg625 630 635 640Gln Ser Ser Gly Asp His Leu Glu Met
Phe Ser Gln Asn His Pro Leu 645 650 655Glu Arg Ile Ser Thr Gly
660821989DNAArxula adeninivorans 82atgcaatccc tggacctatt agacgacagg
tcctgggtcc ccaattatgc gcgtgtgggc 60ctgaaatcgc taaaagaata cttggttagc
catagatatc agtctgaaga agctcgaaag 120catgccgaag cgttagaaag
atggacaaag tctcaggctc aggcggagac atacgaacag 180tggctatttg
cttcggagca gctcgacaag ctgtctggga acgacaagtg gaaagaggac
240ccggtgtccc catattatga cagtgtgcta gtacaacagc ggttacagca
gctccgagat 300gctagggtga atagtaacat ggacgagctg ctgtatttgg
tccgcactag cttgcaaaga 360aacttgggta acatgggtga tcctcgacta
tacgtgagga cccatactgg ctctaagacg 420ctcattgaac aatatattgc
tgaggtagaa ctggcattag acactctgct gagctgcgga 480ccggggacgt
tttcacccaa agttctgtta tccaatctta ttcagacaag aaaggcgttt
540ggacgaacag ccctggtgct ttctggaggt agtacgtttg gaattttaca
tattggtgta 600atgcgagagc ttcaccgagc ccatctgtta ccgcaggtca
tttctggatc gtcggccgga 660tccatctttg cgtccatgct atgtattcac
ttagaagacg agattgaaga actactgcaa 720ctgcctctac acaaggaaag
ctttgaaatc ttcgaacctg ctggagaacg agaaggacta 780atggttcggc
tggcacggtt cctcaaacat ggcacttggt tcgacaacaa gtatcttagc
840acaactatgc gagagcttct aggagacctc actttccagg aggcctacta
ccgaacgcag 900cgaattctaa atgtcactgt gtctccttcg agtatgcacg
aaatgccgaa gattctcaac 960tatctgaccg ctcctaacgt gctcatttgg
tcggcagtgt gtgcatcgtg ctcagtacca 1020tttgtgtttg attctcacga
cattctggca aaaaaccctc gaactgggga gttttattca 1080tggaacgctt
ctactttcat cgacgggagt gtgtataatg atctgccatt gtctcgacta
1140gcggaaatgt ttaacgtgaa ccattttatt gcgtgccagg taaacccgca
tgtggttcca 1200ttcgtcaaat ttgccgagac aatgtcattg gtggaagctc
gtcccactac tactgaaccg 1260ggatcgttga caaagctatg gcacagtact
cagctcgcgc tttctagtga gatctcacac 1320tatctggatt tggctgctga
aatgggcttg ttcaagaaca ttagttccaa gctgcgatcg 1380gtgctagatc
aacaatattc cggcgacatt actattcttc ccgaattata cctgtctgag
1440tttggtcaga ttttcaaaaa cccatcaaag gagttcttcc agaaggcaga
gcttcgagct 1500gccagagcga catggcccaa gatgtcccac attcacaacc
gtgtggccat cgagttggct 1560ttagtaaagg caattcacaa gcttcgtgcc
cgtattgtat ctcagagcgt ccatgagcct 1620ggcagttctc tacaagtaca
tgctgctaat gacgaaggca ccctagcacc tattcgccgt 1680cgccattctt
cgaccaagct tcaccataga cgacaacggt ccgatggaat ggccgtgaaa
1740tacttggtcc gcagacattc gctacagtac tttggcactg agggccctgg
tcccgctgcg 1800ctatctcgta aaaagagttc ggccgggctt acccaggctc
atactcctac gccttcactg 1860accaacagcg ttagtgtagg gggcagtcca
aggcaccgtc gcttcactac tagctctaga 1920cagtcctcag gagaccattt
ggaaatgttc tctcaaaatc atccgctaga acgtatctct 1980accggctga
198983332PRTRicinus communis 83Met Glu Val Ser Gly Leu Gly Cys Phe
Ser Ser Ala Ala Thr Pro Ser1 5 10 15Leu Cys Gly Ala Val Asp Ser Gly
Gly Val Ser Ser Leu Arg Pro Arg 20 25 30Lys Ala Phe His Arg Val Ser
Asp Ser Cys Leu Gly Phe Arg Asp Asn 35 40 45Gly His Leu Gln Tyr Tyr
Cys Gln Gly Gly Phe Val Arg Cys Gly Gly 50 55 60Gly Asn Lys Lys Ser
Ile Lys Lys Lys Leu Lys Leu Val Lys Ser Leu65 70 75 80Ser Glu Asp
Phe Ser Met Phe Pro His Asn Asn Ala Leu Leu His Gln 85 90 95Pro Gln
Ser Ile Ser Leu Gln Glu Ala Ala Gln Gly Leu Met Lys Gln 100 105
110Leu Gln Glu Leu Arg Ala Lys Glu Lys Glu Leu Lys Arg Gln Lys Lys
115 120 125Gln Glu Lys Lys Ala Lys Leu Lys Ser Glu Ser Ser Ser Ser
Ser Ser 130 135 140Ser Glu Ser Ser Ser Asp Ser Glu Arg Gly Glu Val
Ile His Met Ser145 150 155 160Arg Phe Arg Asp Glu Thr Ile Pro Ala
Ala Leu Pro Gln Leu His Pro 165 170 175Leu Thr His His His Pro Thr
Ser Thr Leu Pro Val Ser Pro Thr Gln 180 185 190Glu Cys Asn Pro Met
Asp Tyr Thr Ser Thr His His Glu Lys Arg Cys 195 200 205Cys Val Gly
Pro Ser Thr Gly Ala Asp Asn Ala Val Gly Asp Cys Cys 210 215 220Asn
Asp Arg Asn Ser Ser Met Thr Glu Glu Leu Ser Ala Asn Arg Ile225 230
235 240Glu Val Cys Met Gly Asn Lys Cys Lys Lys Ser Gly Gly Ala Ala
Leu 245 250 255Leu Glu Glu Phe Gln Arg Val Leu Gly Val Glu Ala Ala
Val Val Gly 260 265 270Cys Lys Cys Met Gly Asn Cys Arg Asp Gly Pro
Asn Val Arg Val Arg 275 280 285Asn Ser Val Gln Asp Arg Asn Thr Asp
Asp Ser Val Arg Thr Pro Ser 290 295 300Asn Pro Leu Cys Ile Gly Val
Gly Leu Glu Asp Val Asp Val Ile Val305 310 315 320Ala Asn Phe Phe
Gly Leu Gly Leu Ala Pro Ala Ser 325 33084999DNARicinus communis
84atggaagtct caggcctggg ctgcttctcc tcggctgcaa cgccatcttt gtgtggggcg
60gtggattcag gcggagtatc ctctttgaga ccgaggaagg cattccatag ggtttctgat
120tcttgtttag ggtttagaga taatggacat ctgcagtatt attgtcaagg
aggatttgtc 180aggtgcggag gagggaacaa gaaatctatc aagaaaaagt
tgaaattagt gaagtccttg 240tctgaggact tttccatgtt tcctcataac
aatgctttgc tccatcaacc tcaatccatc 300tccctccagg aagctgcaca
aggattaatg aaacagctcc aagaattgcg agcaaaggag 360aaggaattaa
agaggcagaa gaaacaagag aaaaaagcca agctaaaatc tgaatcatcc
420tcatcctcat cctctgaatc cagtagtgat agcgaacgtg gggaggttat
tcacatgagc 480cgcttcagag atgaaactat tcctgccgca ctacctcaat
tgcacccact tactcatcac 540cacccaactt ccaccctacc agtctcccca
acccaagaat gcaacccgat ggattacact 600tcaacacatc atgaaaaacg
atgctgcgtt ggaccaagca ccggtgccga taacgcagtc 660ggtgactgtt
gcaatgatag gaatagctcg atgacagagg aattgtcagc aaacagaatt
720gaggtgtgca tgggtaataa gtgcaagaag tcgggaggtg cagcgttatt
ggaggaattt 780cagagggttt tgggtgtaga ggctgcagtt gttgggtgca
agtgcatggg gaactgcagg 840gacggtccta atgtaagggt caggaattct
gtccaagaca gaaacacaga tgactctgtt 900cgaaccccct ccaatcctct
ctgcattggt gttggtttgg aggatgtgga tgttattgtg 960gccaatttct
ttgggttggg tctggcccct gcatcttaa 99985345PRTArachis hypogaea 85Met
Glu Val Ser Gly Ala Val Leu Arg Asn Val Thr Cys Pro Ser Phe1 5 10
15Ser Val His Val Ser Ser Arg Arg Arg Gly Gly Asp Ser Cys Val Thr
20 25 30Val Pro Val Arg Met Arg Lys Lys Ala Val Val Arg Cys Cys Cys
Gly 35 40 45Phe Ser Asp Ser Gly His Val Gln Tyr Tyr Gly Asp Glu Lys
Lys Lys 50 55 60Glu Asn Gly Thr Ala Met Leu Ser Thr Lys Lys Lys Leu
Lys Met Leu65 70 75 80Lys Lys Arg Val Leu Phe Asp Asp Leu Gln Gly
Asn Leu Thr Trp Asp 85 90 95Ala Ala Met Val Leu Met Lys Gln Leu Glu
Gln Val Arg Ala Glu Glu 100 105 110Lys Glu Leu Lys Lys Lys Arg Lys
Gln Glu Lys Lys Glu Ala Lys Leu 115 120 125Lys Ala Ser Lys Met Asn
Thr Asn Pro Asp Cys Glu Ser Ser Ser Ser 130 135 140Ser Ser Ser Ser
Glu Ser Glu Ser Glu Ser Ser Glu Ser Glu Cys Asp145 150 155 160Asn
Glu Val Val Asp Met Lys Lys Asn Ile Lys Val Gly Val Ala Val 165 170
175Ala Val Ala Asp Ser Pro Arg Lys Ala Glu Thr Met Ile Leu Tyr Thr
180 185 190Ser Leu Val Ala Arg Asp Val Ser Ala Asn His His His His
Asn Ala 195 200 205Val Glu Leu Phe Ser Arg Asn Asn Asp Ile Ser Val
Gly Ser Ile Asn 210 215 220Gly Gly Leu Lys Asn Glu Asn Thr Ala Val
Ile Thr Thr Glu Ala Ile225 230 235 240Pro Gln Lys Arg Ile Glu Val
Cys Met Gly Asn Lys Cys Lys Lys Ser 245 250 255Gly Ser Ile Ala Leu
Leu Gln Glu Phe Glu Arg Val Val Gly Ala Glu 260 265 270Gly Gly Ala
Ala Ala Ala Val Val Gly Cys Lys Cys Met Gly Lys Cys 275 280 285Lys
Ser Ala Pro Asn Val Arg Ile Gln Asn Ser Thr Ala Asp Lys Ile 290 295
300Ala Glu Gly Phe Asn Asp Ser Val Lys Val Pro Ala Asn Pro Leu
Cys305 310 315 320Ile Gly Val Ala Trp Arg Met Leu Lys Pro Leu Trp
Leu Arg Phe Leu 325 330 335Gly Glu Asn Gln Glu Ser Thr Asn Glu 340
345861038DNAArachis hypogaea 86atggaggttt caggcgccgt tctaaggaat
gtcacgtgcc cttccttttc tgtgcacgtg 60agttcccgtc gtcgtggtgg tgatagttgt
gttacagtgc cggtgaggat gagaaaaaag 120gcggtggtgc gttgttgctg
cgggttcagt gattcggggc atgtgcagta ttacggggac 180gagaagaaga
aggagaatgg aaccgctatg ttgagcacca agaagaagct caagatgctg
240aagaaacgtg tccttttcga tgatcttcaa ggaaacctga cttgggatgc
tgctatggtt 300ttgatgaagc agctagagca agtaagggca gaggagaagg
aattgaagaa aaaaaggaag 360caagagaaga aggaggcaaa actcaaagcc
tctaagatga acaccaatcc tgattgcgaa 420tcgtcatcgt catcgtcatc
atctgaatct gaatctgaat caagtgagag tgaatgtgac 480aatgaggtgg
ttgacatgaa gaagaacatt aaggttggtg ttgccgttgc tgttgccgat
540tccccacgaa aggcggaaac catgattcta tacacctccc ttgttgcccg
agatgttagt 600gctaatcatc atcatcataa tgccgtggaa ttattctcta
gaaacaatga catatcagtt 660ggaagcatta atggtggcct taagaatgag
aatactgcgg ttattaccac tgaagctatt 720cctcagaaga ggattgaggt
atgcatggga aacaagtgca agaaatccgg atctattgca 780ttgttgcaag
aatttgagag agtggttggt gctgaaggag gtgctgctgc tgcagttgtt
840ggatgcaagt gcatggggaa gtgcaagagt gcacctaatg tgaggattca
gaactctact 900gcagataaaa tagctgaggg gttcaatgat tcagttaagg
ttccagctaa ccctctttgc 960attggggttg catggaggat gttgaaacca
ttgtggctta gattcttggg cgagaatcag 1020gaaagtacta atgaataa
103887816PRTYarrowia lipolytica 87Met Phe Thr Ser Arg Val Ser Glu
Ala Ser Thr Thr Asn Phe Ile Arg1 5 10 15Pro Thr Ala Arg Ser His Ile
His Phe Phe Phe Ala Phe Ile Ala Ala 20 25 30Thr Val His Gln Leu Leu
Leu Met Leu Tyr Gln Leu Leu Gly Asp Gly 35 40 45Tyr Leu Lys Ser Phe
Val Asp Thr Gly Ile Thr Leu Ala Gln Gln Ser 50 55 60Gly Leu Ser Gly
Ile Val Asn Ala Leu Thr Ser Glu Ala Lys Leu Arg65 70 75 80Ile Asp
Lys Arg Ser Ile Ile Lys Lys Leu Leu Glu Asp Gln Glu Asn 85 90 95Ala
Glu Ser Tyr Phe Asp Trp Leu Lys Ala Ser Ser Glu Leu Asp Tyr 100 105
110Leu Leu Gly Asn Gln Glu Trp Lys Glu Arg Asp Glu Cys Pro Ala Tyr
115 120 125Asp Tyr Glu Tyr Val Arg Leu Arg Leu Asp Glu Leu Arg His
Ala Arg 130 135 140Thr Asn Asn Asp Thr Thr Arg Leu Leu Tyr Leu Val
Arg Thr Thr Trp145 150 155 160Ser Arg Asn Leu Gly Asn Leu Gly Asp
Val Lys Leu Tyr His Asn Ser 165 170 175Phe Thr Gly Thr Lys Arg Leu
Ile Glu Asp Tyr Ile Leu Glu Cys Glu 180 185 190Leu Ala Leu Asn Ala
Leu Leu Ala Ala Gly Asn Asp Lys Ile Pro Asp 195 200 205Gln Glu Leu
Leu Thr Glu Leu Leu Asn Thr Arg Lys Ala Phe Gly Arg 210 215 220Thr
Ala Leu Leu Leu Ser Gly Gly Gly Cys Leu Gly Leu Leu His Thr225 230
235 240Gly Val Leu Gln Ala Leu Ser Asp Thr Ser Leu Leu Pro His Val
Ile 245 250 255Ser Gly Ser Ser Ala Gly Ser Ile Met Ala Ala Gly Leu
Cys Ile His 260 265 270Lys Asp Glu Glu His Glu Ala Phe Ile Thr Glu
Leu Met Glu Arg Asp 275 280 285Phe Asp Ile Phe Glu Glu Ser Gly Asn
Glu Asp Thr Val Leu Glu Arg 290 295 300Val Ser Arg Met Leu Lys His
Gly Ser Leu Leu Asp Asn Arg Tyr Met305 310 315 320Gln Asp Thr Met
Arg Glu Leu Phe Gly Asp Met Thr Phe Leu Glu Ala 325 330 335Tyr Asn
Arg Thr Arg Arg Ile Leu Asn Val Thr Val Ser Ser Ala Gly
References
D00001

D00002

D00003

P00001
P00002

P00003

P00004

P00005

P00006

P00007

P00008

P00009

P00010
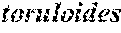
S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.