Casz Compositions And Methods Of Use
Doudna; Jennifer A. ; et al.
U.S. patent application number 16/694720 was filed with the patent office on 2020-03-19 for casz compositions and methods of use. The applicant listed for this patent is The Regents of the University of California. Invention is credited to Jillian F. Banfield, David Burstein, Janice S. Chen, Jennifer A. Doudna, Lucas B. Harrington, David Paez-Espino.
| Application Number | 20200087640 16/694720 |
| Document ID | / |
| Family ID | 66332730 |
| Filed Date | 2020-03-19 |












View All Diagrams
| United States Patent Application | 20200087640 |
| Kind Code | A1 |
| Doudna; Jennifer A. ; et al. | March 19, 2020 |
CASZ COMPOSITIONS AND METHODS OF USE
Abstract
Provided are compositions and methods that include one or more of: (1) a "CasZ" protein (also referred to as a CasZ polypeptide), a nucleic acid encoding the CasZ protein, and/or a modified host cell comprising the CasZ protein (and/or a nucleic acid encoding the same); (2) a CasZ guide RNA that binds to and provides sequence specificity to the CasZ protein, a nucleic acid encoding the CasZ guide RNA, and/or a modified host cell comprising the CasZ guide RNA (and/or a nucleic acid encoding the same); and (3) a CasZ transactivating noncoding RNA (trancRNA) (referred to herein as a "CasZ trancRNA"), a nucleic acid encoding the CasZ trancRNA, and/or a modified host cell comprising the CasZ trancRNA (and/or a nucleic acid encoding the same).
| Inventors: | Doudna; Jennifer A.; (Berkeley, CA) ; Burstein; David; (Berkeley, CA) ; Chen; Janice S.; (Berkeley, CA) ; Harrington; Lucas B.; (Berkeley, CA) ; Paez-Espino; David; (Walnut Creek, CA) ; Banfield; Jillian F.; (Berkeley, CA) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 66332730 | ||||||||||
| Appl. No.: | 16/694720 | ||||||||||
| Filed: | November 25, 2019 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| PCT/US2018/058545 | Oct 31, 2018 | |||
| 16694720 | ||||
| 62580395 | Nov 1, 2017 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C12Q 1/6816 20130101; C12Q 1/6809 20130101; C12N 2800/80 20130101; C12N 15/10 20130101; C12N 15/11 20130101; C12N 15/102 20130101; C12N 9/22 20130101; C12N 15/63 20130101; C12N 15/113 20130101; C07K 2319/09 20130101; C12N 2310/20 20170501; C12Q 1/6816 20130101; C12Q 2521/301 20130101 |
| International Class: | C12N 9/22 20060101 C12N009/22; C12N 15/11 20060101 C12N015/11 |
Claims
1-86. (canceled)
87. A composition comprising a programmable nuclease having a length of no more than 900 amino acids, or a nucleic acid encoding the programmable nuclease, and a non-naturally occurring or engineered guide nucleic acid comprising a region that binds to the programmable nuclease and a guide sequence that is complementary to a target sequence of a target nucleic acid, or a nucleic acid encoding the guide nucleic acid.
88. The composition of claim 87, further comprising a transactivating noncoding RNA.
89. The composition of claim 87, further comprising the target nucleic acid.
90. The composition of claim 87, wherein the target nucleic acid is single stranded DNA.
91. The composition of claim 89, wherein the target nucleic acid is single stranded DNA.
92. The composition of claim 90, wherein the target nucleic acid lacks a protospacer adjacent motif (PAM) sequence.
93. The composition of claim 91, wherein the target nucleic acid lacks a PAM sequence.
94. The composition of claim 87, wherein the target nucleic acid is double stranded DNA.
95. The composition of claim 89, wherein the target nucleic acid is double stranded DNA.
96. The composition of claim 87, wherein the target nucleic acid is a eukaryotic target DNA.
97. The composition of claim 89, wherein the target nucleic acid is a prokaryotic target DNA.
98. The composition of claim 87, wherein the target nucleic acid is a prokaryotic target DNA.
99. The composition of claim 87, wherein the target nucleic acid is a viral target DNA
100. The composition of claim 89, wherein the target nucleic acid is a viral target DNA.
101. The composition of claim 87, further comprising a donor polynucleotide.
102. The composition of claim 87, further comprising a cell.
103. The composition of claim 102, wherein the cell comprises the programmable nuclease and the non-naturally occurring or engineered guide nucleic acid.
104. The composition of claim 102, wherein the cell is a eukaryotic cell.
105. The composition of claim 87, wherein the programmable nuclease comprises three partial RuvC domains.
106. The composition of claim 87, wherein the programmable nuclease comprises RuvC-I, RuvC-II, and RuvC-III subdomains.
107. The composition of claim 87, wherein the programmable nuclease is a CasZ protein.
108. The composition of claim 87, wherein the programmable nuclease is selected from a group consisting of a CasZa protein, a CasZb protein, a CasZc protein, a CasZd protein, a CasZe protein, a CasZf protein, a CasZg protein, a CasZh protein, a CasZi protein, a CasZj protein, a CasZk protein, and a CasZl protein.
109. The composition of claim 87, wherein the programmable nuclease is a variant programmable nuclease with reduced nuclease activity compared to a corresponding wild type programmable nuclease.
110. The composition of claim 87, wherein the programmable nuclease is a dead programmable nuclease.
111. The composition of claim 87, wherein the programmable nuclease is conjugated to a heterologous moiety.
112. The composition of claim 111, wherein the heterologous moiety is a heterologous polypeptide.
113. The composition of claim 112, wherein the heterologous polypeptide comprises a nuclear localization signal.
114. The composition of claim 87, wherein the programmable nuclease has a length of from 350 to 900 amino acids.
115. The composition of claim 87, wherein the programmable nuclease has a length of no more than 800 amino acids.
116. The composition of claim 87, wherein the guide nucleic acid is a guide RNA.
Description
CROSS-REFERENCE
[0001] This application claims the benefit of U.S. Provisional Patent Application No. 62/580,395, filed Nov. 1, 2017, which application is incorporated herein by reference in its entirety.
INCORPORATION BY REFERENCE OF SEQUENCE LISTING PROVIDED AS A TEXT FILE
[0002] A Sequence Listing is provided herewith as a text file, "BERK-374WO_SEQLISTING_ST25.txt" created on Oct. 30, 2018 and having a size of 536 KB. The contents of the text file are incorporated by reference herein in their entirety.
INTRODUCTION
[0003] The CRISPR-Cas system, an example of a pathway that was unknown to science prior to the DNA sequencing era, is now understood to confer bacteria and archaea with acquired immunity against phage and viruses. Intensive research has uncovered the biochemistry of this system. CRISPR-Cas systems consist of Cas proteins, which are involved in acquisition, targeting and cleavage of foreign DNA or RNA, and a CRISPR array, which includes direct repeats flanking short spacer sequences that guide Cas proteins to their targets. Class 2 CRISPR-Cas are streamlined versions in which a single Cas protein bound to RNA is responsible for binding to and cleavage of a targeted sequence. The programmable nature of these minimal systems has facilitated their use as a versatile technology that is revolutionizing the field of genome manipulation.
SUMMARY
[0004] The present disclosure provides compositions and methods that include one or more of: (1) a "CasZ" protein (also referred to as a CasZ polypeptide), a nucleic acid encoding the CasZ protein, and/or a modified host cell comprising the CasZ protein (and/or a nucleic acid encoding the same); (2) a CasZ guide RNA that binds to and provides sequence specificity to the CasZ protein, a nucleic acid encoding the CasZ guide RNA, and/or a modified host cell comprising the CasZ guide RNA (and/or a nucleic acid encoding the same); and (3) a CasZ transactivating noncoding RNA (trancRNA) (referred to herein as a "CasZ trancRNA"), a nucleic acid encoding the CasZ trancRNA, and/or a modified host cell comprising the CasZ trancRNA (and/or a nucleic acid encoding the same).
BRIEF DESCRIPTION OF THE DRAWINGS
[0005] FIG. 1 depicts examples of naturally occurring CasZ protein sequences.
[0006] FIG. 2 depicts schematic representations of CasZ loci, which include a Cas1 protein in addition to the CasZ protein.
[0007] FIG. 3 depicts a phylogenetic tree of CasZ sequences in relation to other Class 2 CRISPR/Cas effector protein sequences.
[0008] FIG. 4 depicts a phylogenetic tree of Cas1 sequences from CasZ loci in relation to Cas1 sequences from other Class 2 CRISPR/Cas loci.
[0009] FIG. 5 depicts transcriptomic RNA mapping data demonstrating expression of trancRNA from CasZ loci. The trancRNAs are adjacent to the CasZ repeat array, but do not include the repeat sequence and are not complementary to the repeat sequence. Shown are RNA mapping data for the following loci: CasZa3, CasZb4, CasZc5, CasZd1, and CasZe3. Small repeating aligned arrows represent the repeats of the CRISPR array (indicating the presence of guide RNA-encoding sequence); The peaks outside and adjacent to the repeat arrays represent highly transcribed trancRNAs. FIG. 5 (Cont. 1) nucleotide sequences (Top to Bottom: SEQ ID NOs.:312-331). FIG. 5 (Cont. 3) nucleotide sequences (Top to Bottom: SEQ ID Nos.: 161-177).
[0010] FIG. 6 depicts results for PAM preferences as assayed using PAM depletion assays for CasZc (top) and CasZb (bottom).
[0011] FIG. 7 depict the sequences of Cas14 proteins described herein.
[0012] FIG. 8, Panels A-D depict the architecture and phylogeny of CRISPR-Cas14 genomic loci.
[0013] FIG. 9 depicts a phylogenetic analysis of Cas14 orthologs.
[0014] FIG. 10 depicts a maximum likelihood tree for Cas1 from known CRISPR systems.
[0015] FIG. 11, Panels A-B depict the acquisition of new spacers by CRISPR-Cas14 systems.
[0016] FIG. 12, Panels A-D depict that CRISPR-Cas14a actively adapts and encodes a tracrRNA.
[0017] FIG. 13, Panels A-B depict metatranscriptomics for CRISPR-Cas14 loci.
[0018] FIG. 14, Panels A-B depict RNA processing and heterologous expression by CRISPR-Cas14.
[0019] FIG. 15, Panels A-D depict plasmid depletion by Cas14a1 and SpCas9.
[0020] FIG. 16, Panels A-D depict CRISPR-Cas14a is an RNA-guided DNA-endonuclease.
[0021] FIG. 17, Panels A-E depict degradation of ssDNA by Cas14a1.
[0022] FIG. 18 depicts kinetics of Cas14a1 cleavage of ssDNA with various guide RNA components.
[0023] FIG. 19, Panels A-F depict optimization of Cas14a1 guide RNA components.
[0024] FIG. 20, Panels A-E depict high fidelity ssDNA DNP detection by CRISPR-Cas14a.
[0025] FIG. 20, panel C provides nucleotide sequences (Top to Bottom: SEQ ID NOs:367-370)
[0026] FIG. 21, Panels A-F depict the impact of various activators on Cas14a1 cleavage rate.
[0027] FIG. 22, Panels A-B depict diversity of CRISPR-Cas14 systems.
[0028] FIG. 23, Panels A-C depict a test of Cas14a1 mediated interference in a heterologous host. Diagram of Cas14a1 and LbCas12a constructs to test interference in E. coli.
[0029] FIG. 24 depicts Cas14 nucleotide sequences of plasmids used in the present invention.
[0030] FIG. 25, Panels A-E depict a sequence map of each of the plasmids disclosed in FIG. 24.
DEFINITIONS
[0031] "Heterologous," as used herein, means a nucleotide or polypeptide sequence that is not found in the native nucleic acid or protein, respectively. For example, relative to a CasZ polypeptide, a heterologous polypeptide comprises an amino acid sequence from a protein other than the CasZ polypeptide. In some cases, a portion of a CasZ protein from one species is fused to a portion of a CasZ protein from a different species. The CasZ sequence from each species could therefore be considered heterologous relative to one another. As another example, a CasZ protein (e.g., a dCasZ protein) can be fused to an active domain from a non-CasZ protein (e.g., a histone deacetylase), and the sequence of the active domain could be considered a heterologous polypeptide (it is heterologous to the CasZ protein).
[0032] The terms "polynucleotide" and "nucleic acid," used interchangeably herein, refer to a polymeric form of nucleotides of any length, either ribonucleotides or deoxynucleotides. Thus, this term includes, but is not limited to, single-, double-, or multi-stranded DNA or RNA, genomic DNA, cDNA, DNA-RNA hybrids, or a polymer comprising purine and pyrimidine bases or other natural, chemically or biochemically modified, non-natural, or derivatized nucleotide bases. The terms "polynucleotide" and "nucleic acid" should be understood to include, as applicable to the embodiment being described, single-stranded (such as sense or antisense) and double-stranded polynucleotides.
[0033] The terms "polypeptide," "peptide," and "protein", are used interchangeably herein, refer to a polymeric form of amino acids of any length, which can include genetically coded and non-genetically coded amino acids, chemically or biochemically modified or derivatized amino acids, and polypeptides having modified peptide backbones. The term includes fusion proteins, including, but not limited to, fusion proteins with a heterologous amino acid sequence, fusions with heterologous and homologous leader sequences, with or without N-terminal methionine residues; immunologically tagged proteins; and the like.
[0034] The term "naturally-occurring" as used herein as applied to a nucleic acid, a protein, a cell, or an organism, refers to a nucleic acid, cell, protein, or organism that is found in nature.
[0035] As used herein the term "isolated" is meant to describe a polynucleotide, a polypeptide, or a cell that is in an environment different from that in which the polynucleotide, the polypeptide, or the cell naturally occurs. An isolated genetically modified host cell may be present in a mixed population of genetically modified host cells.
[0036] As used herein, the term "exogenous nucleic acid" refers to a nucleic acid that is not normally or naturally found in and/or produced by a given bacterium, organism, or cell in nature. As used herein, the term "endogenous nucleic acid" refers to a nucleic acid that is normally found in and/or produced by a given bacterium, organism, or cell in nature. An "endogenous nucleic acid" is also referred to as a "native nucleic acid" or a nucleic acid that is "native" to a given bacterium, organism, or cell.
[0037] "Recombinant," as used herein, means that a particular nucleic acid (DNA or RNA) is the product of various combinations of cloning, restriction, and/or ligation steps resulting in a construct having a structural coding or non-coding sequence distinguishable from endogenous nucleic acids found in natural systems. Generally, DNA sequences encoding the structural coding sequence can be assembled from cDNA fragments and short oligonucleotide linkers, or from a series of synthetic oligonucleotides, to provide a synthetic nucleic acid which is capable of being expressed from a recombinant transcriptional unit contained in a cell or in a cell-free transcription and translation system. Such sequences can be provided in the form of an open reading frame uninterrupted by internal non-translated sequences, or introns, which are typically present in eukaryotic genes. Genomic DNA comprising the relevant sequences can also be used in the formation of a recombinant gene or transcriptional unit. Sequences of non-translated DNA may be present 5' or 3' from the open reading frame, where such sequences do not interfere with manipulation or expression of the coding regions, and may indeed act to modulate production of a desired product by various mechanisms (see "DNA regulatory sequences", below).
[0038] Thus, e.g., the term "recombinant" polynucleotide or "recombinant" nucleic acid refers to one which is not naturally occurring, e.g., is made by the artificial combination of two otherwise separated segments of sequence through human intervention. This artificial combination is often accomplished by either chemical synthesis means, or by the artificial manipulation of isolated segments of nucleic acids, e.g., by genetic engineering techniques. Such is usually done to replace a codon with a redundant codon encoding the same or a conservative amino acid, while typically introducing or removing a sequence recognition site. Alternatively, it is performed to join together nucleic acid segments of desired functions to generate a desired combination of functions. This artificial combination is often accomplished by either chemical synthesis means, or by the artificial manipulation of isolated segments of nucleic acids, e.g., by genetic engineering techniques.
[0039] Similarly, the term "recombinant" polypeptide refers to a polypeptide which is not naturally occurring, e.g., is made by the artificial combination of two otherwise separated segments of amino sequence through human intervention. Thus, e.g., a polypeptide that comprises a heterologous amino acid sequence is recombinant.
[0040] By "construct" or "vector" is meant a recombinant nucleic acid, generally recombinant DNA, which has been generated for the purpose of the expression and/or propagation of a specific nucleotide sequence(s), or is to be used in the construction of other recombinant nucleotide sequences.
[0041] The terms "DNA regulatory sequences," "control elements," and "regulatory elements," used interchangeably herein, refer to transcriptional and translational control sequences, such as promoters, enhancers, polyadenylation signals, terminators, protein degradation signals, and the like, that provide for and/or regulate expression of a coding sequence and/or production of an encoded polypeptide in a host cell.
[0042] The term "transformation" is used interchangeably herein with "genetic modification" and refers to a permanent or transient genetic change induced in a cell following introduction of new nucleic acid (e.g., DNA exogenous to the cell) into the cell. Genetic change ("modification") can be accomplished either by incorporation of the new nucleic acid into the genome of the host cell, or by transient or stable maintenance of the new nucleic acid as an episomal element. Where the cell is a eukaryotic cell, a permanent genetic change is generally achieved by introduction of new DNA into the genome of the cell. In prokaryotic cells, permanent changes can be introduced into the chromosome or via extrachromosomal elements such as plasmids and expression vectors, which may contain one or more selectable markers to aid in their maintenance in the recombinant host cell. Suitable methods of genetic modification include viral infection, transfection, conjugation, protoplast fusion, electroporation, particle gun technology, calcium phosphate precipitation, direct microinjection, and the like. The choice of method is generally dependent on the type of cell being transformed and the circumstances under which the transformation is taking place (i.e. in vitro, ex vivo, or in vivo). A general discussion of these methods can be found in Ausubel, et al, Short Protocols in Molecular Biology, 3rd ed., Wiley & Sons, 1995.
[0043] "Operably linked" refers to a juxtaposition wherein the components so described are in a relationship permitting them to function in their intended manner. For instance, a promoter is operably linked to a coding sequence if the promoter affects its transcription or expression. As used herein, the terms "heterologous promoter" and "heterologous control regions" refer to promoters and other control regions that are not normally associated with a particular nucleic acid in nature. For example, a "transcriptional control region heterologous to a coding region" is a transcriptional control region that is not normally associated with the coding region in nature.
[0044] A "host cell," as used herein, denotes an in vivo or in vitro eukaryotic cell, a prokaryotic cell, or a cell from a multicellular organism (e.g., a cell line) cultured as a unicellular entity, which eukaryotic or prokaryotic cells can be, or have been, used as recipients for a nucleic acid (e.g., an expression vector), and include the progeny of the original cell which has been genetically modified by the nucleic acid. It is understood that the progeny of a single cell may not necessarily be completely identical in morphology or in genomic or total DNA complement as the original parent, due to natural, accidental, or deliberate mutation. A "recombinant host cell" (also referred to as a "genetically modified host cell") is a host cell into which has been introduced a heterologous nucleic acid, e.g., an expression vector. For example, a subject prokaryotic host cell is a genetically modified prokaryotic host cell (e.g., a bacterium), by virtue of introduction into a suitable prokaryotic host cell of a heterologous nucleic acid, e.g., an exogenous nucleic acid that is foreign to (not normally found in nature in) the prokaryotic host cell, or a recombinant nucleic acid that is not normally found in the prokaryotic host cell; and a subject eukaryotic host cell is a genetically modified eukaryotic host cell, by virtue of introduction into a suitable eukaryotic host cell of a heterologous nucleic acid, e.g., an exogenous nucleic acid that is foreign to the eukaryotic host cell, or a recombinant nucleic acid that is not normally found in the eukaryotic host cell.
[0045] The term "conservative amino acid substitution" refers to the interchangeability in proteins of amino acid residues having similar side chains. For example, a group of amino acids having aliphatic side chains consists of glycine, alanine, valine, leucine, and isoleucine; a group of amino acids having aliphatic-hydroxyl side chains consists of serine and threonine; a group of amino acids having amide-containing side chains consists of asparagine and glutamine; a group of amino acids having aromatic side chains consists of phenylalanine, tyrosine, and tryptophan; a group of amino acids having basic side chains consists of lysine, arginine, and histidine; and a group of amino acids having sulfur-containing side chains consists of cysteine and methionine. Exemplary conservative amino acid substitution groups are: valine-leucine-isoleucine, phenylalanine-tyrosine, lysine-arginine, alanine-valine, and asparagine-glutamine.
[0046] A polynucleotide or polypeptide has a certain percent "sequence identity" to another polynucleotide or polypeptide, meaning that, when aligned, that percentage of bases or amino acids are the same, and in the same relative position, when comparing the two sequences. Sequence similarity can be determined in a number of different manners. To determine sequence identity, sequences can be aligned using the methods and computer programs, including BLAST, available over the world wide web at ncbi.nlm.nih.gov/BLAST. See, e.g., Altschul et al. (1990), J. Mol. Biol. 215:403-10. Another alignment algorithm is FASTA, available in the Genetics Computing Group (GCG) package, from Madison, Wis., USA, a wholly owned subsidiary of Oxford Molecular Group, Inc. Other techniques for alignment are described in Methods in Enzymology, vol. 266: Computer Methods for Macromolecular Sequence Analysis (1996), ed. Doolittle, Academic Press, Inc., a division of Harcourt Brace & Co., San Diego, Calif., USA. Of particular interest are alignment programs that permit gaps in the sequence. The Smith-Waterman is one type of algorithm that permits gaps in sequence alignments. See Meth. Mol. Biol. 70: 173-187 (1997). Also, the GAP program using the Needleman and Wunsch alignment method can be utilized to align sequences. See J. Mol. Biol. 48: 443-453 (1970).
[0047] As used herein, the terms "treatment," "treating," and the like, refer to obtaining a desired pharmacologic and/or physiologic effect. The effect may be prophylactic in terms of completely or partially preventing a disease or symptom thereof and/or may be therapeutic in terms of a partial or complete cure for a disease and/or adverse effect attributable to the disease. "Treatment," as used herein, covers any treatment of a disease in a mammal, e.g., in a human, and includes: (a) preventing the disease from occurring in a subject which may be predisposed to the disease but has not yet been diagnosed as having it; (b) inhibiting the disease, i.e., arresting its development; and (c) relieving the disease, i.e., causing regression of the disease.
[0048] The terms "individual," "subject," "host," and "patient," used interchangeably herein, refer to an individual organism, e.g., a mammal, including, but not limited to, murines, simians, non-human primates, humans, mammalian farm animals, mammalian sport animals, and mammalian pets.
[0049] Before the present invention is further described, it is to be understood that this invention is not limited to particular embodiments described, as such may, of course, vary. It is also to be understood that the terminology used herein is for the purpose of describing particular embodiments only, and is not intended to be limiting, since the scope of the present invention will be limited only by the appended claims.
[0050] Where a range of values is provided, it is understood that each intervening value, to the tenth of the unit of the lower limit unless the context clearly dictates otherwise, between the upper and lower limit of that range and any other stated or intervening value in that stated range, is encompassed within the invention. The upper and lower limits of these smaller ranges may independently be included in the smaller ranges, and are also encompassed within the invention, subject to any specifically excluded limit in the stated range. Where the stated range includes one or both of the limits, ranges excluding either or both of those included limits are also included in the invention.
[0051] Unless defined otherwise, all technical and scientific terms used herein have the same meaning as commonly understood by one of ordinary skill in the art to which this invention belongs. Although any methods and materials similar or equivalent to those described herein can also be used in the practice or testing of the present invention, the preferred methods and materials are now described. All publications mentioned herein are incorporated herein by reference to disclose and describe the methods and/or materials in connection with which the publications are cited.
[0052] It must be noted that as used herein and in the appended claims, the singular forms "a," "an," and "the" include plural referents unless the context clearly dictates otherwise. Thus, for example, reference to "a CasZ polypeptide" includes a plurality of such polypeptides and reference to "the guide RNA" includes reference to one or more guide RNAs and equivalents thereof known to those skilled in the art, and so forth. It is further noted that the claims may be drafted to exclude any optional element. As such, this statement is intended to serve as antecedent basis for use of such exclusive terminology as "solely," "only" and the like in connection with the recitation of claim elements, or use of a "negative" limitation.
[0053] It is appreciated that certain features of the invention, which are, for clarity, described in the context of separate embodiments, may also be provided in combination in a single embodiment. Conversely, various features of the invention, which are, for brevity, described in the context of a single embodiment, may also be provided separately or in any suitable sub-combination. All combinations of the embodiments pertaining to the invention are specifically embraced by the present invention and are disclosed herein just as if each and every combination was individually and explicitly disclosed. In addition, all sub-combinations of the various embodiments and elements thereof are also specifically embraced by the present invention and are disclosed herein just as if each and every such sub-combination was individually and explicitly disclosed herein.
[0054] The publications discussed herein are provided solely for their disclosure prior to the filing date of the present application. Nothing herein is to be construed as an admission that the present invention is not entitled to antedate such publication by virtue of prior invention. Further, the dates of publication provided may be different from the actual publication dates which may need to be independently confirmed.
DETAILED DESCRIPTION
[0055] The present disclosure provides compositions and methods that include one or more of: (1) a "CasZ" protein (also referred to as a CasZ polypeptide), a nucleic acid encoding the CasZ protein, and/or a modified host cell comprising the CasZ protein (and/or a nucleic acid encoding the same); (2) a CasZ guide RNA that binds to and provides sequence specificity to the CasZ protein, a nucleic acid encoding the CasZ guide RNA, and/or a modified host cell comprising the CasZ guide RNA (and/or a nucleic acid encoding the same); and (3) a CasZ transactivating noncoding RNA (trancRNA) (referred to herein as a "CasZ trancRNA"), a nucleic acid encoding the CasZ trancRNA, and/or a modified host cell comprising the CasZ trancRNA (and/or a nucleic acid encoding the same).
Compositions
[0056] CRISPR/CasZ Proteins, Guide RNAs, and trancRNAs
[0057] Class 2 CRISPR-Cas systems are characterized by effector modules that include a single multidomain protein. In the CasZ system, a CRISPR/Cas endonuclease (e.g., a CasZ protein) interacts with (binds to) a corresponding guide RNA (e.g., a CasZ guide RNA) to form a ribonucleoprotein (RNP) complex that is targeted to a particular site in a target nucleic acid via base pairing between the guide RNA and a target sequence within the target nucleic acid molecule. A guide RNA includes a nucleotide sequence (a guide sequence) that is complementary to a sequence (the target site) of a target nucleic acid. Thus, a CasZ protein forms a complex with a CasZ guide RNA and the guide RNA provides sequence specificity to the RNP complex via the guide sequence. The CasZ protein of the complex provides the site-specific activity. In other words, the CasZ protein is guided to a target site (e.g., stabilized at a target site) within a target nucleic acid (e.g. a target nucleotide sequence within a target chromosomal nucleic acid; or a target nucleotide sequence within a target extrachromosomal nucleic acid, e.g., an episomal nucleic acid, a minicircle nucleic acid, a mitochondrial nucleic acid, a chloroplast nucleic acid, etc.) by virtue of its association with the guide RNA.
[0058] The present disclosure provides compositions comprising a CasZ polypeptide (and/or a nucleic acid encoding the CasZ polypeptide) (e.g., where the CasZ polypeptide can be a naturally existing CasZ protein, a nickase CasZ protein, a dCasZ protein, a chimeric CasZ protein, etc.Xa CasZa, CasZb, CasZc, CasZd, CasZe, CasZf, CasZg, CasZh, CasZi, CasZj, CasZK, or CasZl protein). The present disclosure provides compositions comprising a CasZ guide RNA (and/or a nucleic acid encoding the CasZ guide RNA). For example, the present disclosure provides compositions comprising (a) a CasZ polypeptide (and/or a nucleic acid encoding the CasZ polypeptide) and (b) a CasZ guide RNA (and/or a nucleic acid encoding the CasZ guide RNA). The present disclosure provides a nucleic acid/protein complex (RNP complex) comprising: (a) a CasZ polypeptide; and (b) a CasZ guide RNA. The present disclosure provides compositions comprising a CasZ trancRNA. The present disclosure provides compositions comprising a CasZ trancRNA and one or more of: (a) a CasZ protein, and (b) a CasZ guide RNA (e.g., comprising a CasZ trancRNA and a CasZ protein, a CasZ trancRNA and a CasZ guide RNA, or a CasZ trancRNA and a CasZ protein and a CasZ guide RNA. The present disclosure provides a nucleic acid/protein complex (RNP complex) comprising: (a) a CasZ polypeptide; (b) a CasZ guide RNA; and (c) a CasZ trancRNA. The present disclosure provides compositions comprising a CasZ protein and one or more of: (a) a CasZ trancRNA, and (b) a CasZ guide RNA.
CasZ Protein
[0059] A CasZ polypeptide (this term is used interchangeably with the term "CasZ protein", "Cas14", "Cas14 polypeptide", or "Cas14 protein") can bind and/or modify (e.g., cleave, nick, methylate, demethylate, etc.) a target nucleic acid and/or a polypeptide associated with target nucleic acid (e.g., methylation or acetylation of a histone tail) (e.g., in some cases the CasZ protein includes a fusion partner with an activity, and in some cases the CasZ protein provides nuclease activity). In some cases, the CasZ protein is a naturally-occurring protein (e.g., naturally occurs in prokaryotic cells). In other cases, the CasZ protein is not a naturally-occurring polypeptide (e.g., the CasZ protein is a variant CasZ protein, a chimeric protein, and the like). A CasZ protein includes 3 partial RuvC domains (RuvC-I, RuvC-II, and RuvC-III, also referred to herein as subdomains) that are not contiguous with respect to the primary amino acid sequence of the CasZ protein, but form a RuvC domain once the protein is produced and folds. A naturally occurring CasZ protein functions as an endonuclease that catalyzes cleavage at a specific sequence in a targeted nucleic acid (e.g., a double stranded DNA (dsDNA)). The sequence specificity is provided by the associated guide RNA, which hybridizes to a target sequence within the target DNA. The naturally occurring CasZ guide RNA is a crRNA, where the crRNA includes (i) a guide sequence that hybridizes to a target sequence in the target DNA and (ii) a protein binding segment that binds to the CasZ protein.
[0060] In some embodiments, the CasZ protein of the subject methods and/or compositions is (or is derived from) a naturally occurring (wild type) protein. Examples of naturally occurring CasZ proteins (e.g., CasZa, CasZb, CasZc, CasZd, CasZe, CasZf, CasZg, CasZh, CasZi, CasZj, CasZk, CasZl) are depicted in FIG. 1. In some cases, a subject CasZ protein is a CasZa protein. In some cases, a subject CasZ protein is a CasZb protein. In some cases, a subject CasZ protein is a CasZc protein. In some cases, a subject CasZ protein is a CasZd protein. In some cases, a subject CasZ protein is a CasZe protein. In some cases, a subject CasZ protein is a CasZf protein. In some cases, a subject CasZ protein is a CasZg protein. In some cases, a subject CasZ protein is a CasZh protein. In some cases, a subject CasZ protein is a CasZi protein. In some cases, a subject CasZ protein is a CasZj protein. In some cases, a subject CasZ protein is a CasZk protein. In some cases, a subject CasZ protein is a CasZl protein. In some cases, a subject CasZ protein is a CasZe, CasZf, CasZg, or CasZh protein. In some cases, a subject CasZ protein is a CasZj, CasZk, or CasZl protein.
[0061] It is important to note that this newly discovered protein (CasZ) is short compared to previously identified CRISPR-Cas endonucleases, and thus use of this protein as an alternative provides the advantage that the nucleotide sequence encoding the protein is relatively short. This is useful, for example, in cases where a nucleic acid encoding the CasZ protein is desirable, e.g., in situations that employ a viral vector (e.g., an AAV vector), for delivery to a cell such as a eukaryotic cell (e.g., mammalian cell, human cell, mouse cell, in vitro, ex vivo, in vivo) for research and/or clinical applications. In addition, in their natural context, the CasZ-encoding DNA sequences are present in loci that also have a Cas1 protein.
[0062] In some cases, a subject CasZ protein has a length of 900 amino acids or less (e.g., 850 amino acids or less, 800 amino acids or less, 750 amino acids or less, or 700 amino acids or less). In some cases, a subject CasZ protein has a length of 850 amino acids or less (e.g., 850 amino acids or less). In some cases, a subject CasZ protein length of 800 amino acids or less (e.g., 750 amino acids or less). In some cases, a subject CasZ protein has a length of 700 amino acids or less. In some cases, a subject CasZ protein has a length of 650 amino acids or less.
[0063] In some cases, a subject CasZ protein has a length in a range of from 350-900 amino acids (e.g., 350-850, 350-800, 350-750, 350-700, 400-900, 400-850, 400-800, 400-750, or 400-700 amino acids).
[0064] In some cases, a subject CasZ protein (e.g., CasZa) has a length in a range of from 350-750 amino acids (e.g., 350-700, 350-550, 450-550, 450-750, 450-650, or 450-550 amino acids). In some cases, a subject CasZ protein (e.g., CasZa) has a length in a range of from 450-750 amino acids (e.g., 500-700 amino acids). In some cases, a subject CasZ protein (e.g., CasZa) has a length in a range of from 350-700 amino acids (e.g., 350-650, 350-600, or 350-550 amino acids). In some cases, a subject CasZ protein (e.g., CasZa) has a length in a range of from 500-700 amino acids. In some cases, a subject CasZ protein (e.g., CasZa) has a length in a range of from 450-550 amino acids. In some cases, a subject CasZ protein (e.g., CasZa) has a length in a range of from 350-550 amino acids.
[0065] In some cases, a subject CasZ protein (e.g., CasZb) has a length in a range of from 350-700 amino acids (e.g., 350-650, or 350-620 amino acids). In some cases, a subject CasZ protein (e.g., CasZb) has a length in a range of from 450-700 amino acids (e.g., 450-650, 500-650 or 500-620 amino acids). In some cases, a subject CasZ protein (e.g., CasZb) has a length in a range of from 500-650 amino acids (e.g., 500-620 amino acids). In some cases, a subject CasZ protein (e.g., CasZb) has a length in a range of from 500-620 amino acids.
[0066] In some cases, a subject CasZ protein (e.g., CasZc) has a length in a range of from 600-800 amino acids (e.g., 600-650 or 700-800 amino acids). In some cases, a subject CasZ protein (e.g., CasZc) has a length in a range of from 600-650 amino acids. In some cases, a subject CasZ protein (e.g., CasZc) has a length in a range of from 700-800 amino acids.
[0067] In some cases, a subject CasZ protein (e.g., CasZd) has a length in a range of from 400-650 amino acids (e.g., 400-600, 400-550, 500-650, 500-600 or 500-550 amino acids). In some cases, a subject CasZ protein (e.g., CasZd) has a length in a range of from 500-600 amino acids. In some cases, a subject CasZ protein (e.g., CasZd) has a length in a range of from 500-550 amino acids. In some cases, a subject CasZ protein (e.g., CasZd) has a length in a range of from 400-550 amino acids.
[0068] In some cases, a subject CasZ protein (e.g., CasZe) has a length in a range of from 450-700 amino acids (e.g., 450-650, 450-615, 475-700, 475-650, or 475-615 amino acids). In some cases, a subject CasZ protein (e.g., CasZe) has a length in a range of from 450-675 amino acids. In some cases, a subject CasZ protein (e.g., CasZe) has a length in a range of from 475-675 amino acids.
[0069] In some cases, a subject CasZ protein (e.g., CasZf) has a length in a range of from 400-550 amino acids (e.g., 400-520, 400-500, 400-475, 415-550, 415-520, 415-500, or 415-475 amino acids). In some cases, a subject CasZ protein (e.g., CasZf) has a length in a range of from 400-475 amino acids (e.g., 400-450 amino acids).
[0070] In some cases, a subject CasZ protein (e.g., CasZg) has a length in a range of from 500-750 amino acids (e.g., 550-750 or 500-700 amino acids). In some cases, a subject CasZ protein (e.g., CasZg) has a length in a range of from 700-750 amino acids. In some cases, a subject CasZ protein (e.g., CasZg) has a length in a range of from 550-600 amino acids.
[0071] In some cases, a subject CasZ protein (e.g., CasZh) has a length in a range of from 380-450 amino acids (e.g., 380-420, 400-450, or 400-420 amino acids). In some cases, a subject CasZ protein (e.g., CasZh) has a length in a range of from 400-420 amino acids.
[0072] In some cases, a subject CasZ protein (e.g., CasZi) has a length in a range of from 700-800 amino acids (e.g., 700-750, 720-800, or 720-750 amino acids). In some cases, a subject CasZ protein (e.g., CasZi) has a length in a range of from 720-780 amino acids.
[0073] In some cases, a subject CasZ protein (e.g., CasZj) has a length in a range of from 600-750 amino acids (e.g., 600-700 or 650-700 amino acids). In some cases, a subject CasZ protein (e.g., CasZj) has a length in a range of from 400-420 amino acids.
[0074] In some cases, a subject CasZ protein (e.g., CasZk) has a length in a range of from 450-600 amino acids (e.g., 450-580, 480-600, 480-580, or 500-600 amino acids). In some cases, a subject CasZ protein (e.g., CasZk) has a length in a range of from 480-580 amino acids.
[0075] In some cases, a subject CasZ protein (e.g., CasZl) has a length in a range of from 350-500 amino acids (e.g., 350-450, 380-450, 350-420, or 380-420 amino acids). In some cases a subject CasZ protein (e.g., CasZl) has a length in a range of from 380-420 amino acids.
[0076] In some cases, a subject CasZ protein (of the subject compositions and/or methods) includes an amino acid sequence having 20% or more sequence identity (e.g., 30% or more, 40% or more, 50% or more, 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZa protein of FIG. 1 or FIG. 7. For example, in some cases, a subject CasZ protein includes an amino acid sequence having 50% or more sequence identity (e.g., 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZa protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 80% or more sequence identity (e.g., 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZa protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZa protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZa amino acid sequence of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZa amino acid sequence of FIG. 1 or FIG. 7, with the exception that the sequence includes one or more amino acid substitutions (e.g., 1, 2, or 3 amino acid substitutions) that reduces the naturally occurring catalytic activity of the protein (e.g., such as at one or more catalytic amino acid positions) (e.g., in some cases such that the CasZ protein is a dCasZ). In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZa protein of FIG. 1 or FIG. 7 and has a length in a range of from 350-800 amino acids (e.g., 350-800, 350-750, 350-700, 350-550, 450-550, 450-750, 450-650, or 450-550 amino acids).
[0077] In some cases, a subject CasZ protein (of the subject compositions and/or methods) includes an amino acid sequence having 20% or more sequence identity (e.g., 30% or more, 40% or more, 50% or more, 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZb protein of FIG. 1 or FIG. 7. For example, in some cases, a subject CasZ protein includes an amino acid sequence having 50% or more sequence identity (e.g., 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZb protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 80% or more sequence identity (e.g., 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZb protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZb protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZb amino acid sequence of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZb amino acid sequence of FIG. 1 or FIG. 7, with the exception that the sequence includes an amino acid substitution (e.g., 1, 2, or 3 amino acid substitutions) that reduces the naturally occurring catalytic activity of the protein (e.g., such as at one or more catalytic amino acid positions) (e.g., in some cases such that the CasZ protein is a dCasZ). In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZb protein of FIG. 1 or FIG. 7 and has a length in a range of from 350-700 amino acids (e.g., 350-650, or 350-620 amino acids).
[0078] In some cases, a subject CasZ protein (of the subject compositions and/or methods) includes an amino acid sequence having 20% or more sequence identity (e.g., 30% or more, 40% or more, 50% or more, 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZc protein of FIG. 1 or FIG. 7. For example, in some cases, a subject CasZ protein includes an amino acid sequence having 50% or more sequence identity (e.g., 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZc protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 80% or more sequence identity (e.g., 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZc protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZc protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZc amino acid sequence of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZc amino acid sequence of FIG. 1 or FIG. 7, with the exception that the sequence includes an amino acid substitution (e.g., 1, 2, or 3 amino acid substitutions) that reduces the naturally occurring catalytic activity of the protein (e.g., such as at one or more catalytic amino acid positions) (e.g., in some cases such that the CasZ protein is a dCasZ). In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZc protein of FIG. 1 or FIG. 7 and has a length in a range of from 600-800 amino acids (e.g., 600-650 or 700-800 amino acids).
[0079] In some cases, a subject CasZ protein (of the subject compositions and/or methods) includes an amino acid sequence having 20% or more sequence identity (e.g., 30% or more, 40% or more, 50% or more, 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZd protein of FIG. 1 or FIG. 7. For example, in some cases, a subject CasZ protein includes an amino acid sequence having 50% or more sequence identity (e.g., 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZd protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 80% or more sequence identity (e.g., 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZd protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZd protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZd amino acid sequence of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZd amino acid sequence of FIG. 1 or FIG. 7, with the exception that the sequence includes an amino acid substitution (e.g., 1, 2, or 3 amino acid substitutions) that reduces the naturally occurring catalytic activity of the protein (e.g., such as at one or more catalytic amino acid positions) (e.g., in some cases such that the CasZ protein is a dCasZ). In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZd protein of FIG. 1 or FIG. 7 and has a length in a range of from 400-650 amino acids (e.g., 400-600, 400-550, 500-650, 500-600 or 500-550 amino acids).
[0080] In some cases, a subject CasZ protein (of the subject compositions and/or methods) includes an amino acid sequence having 20% or more sequence identity (e.g., 30% or more, 40% or more, 50% or more, 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZe protein of FIG. 1 or FIG. 7. For example, in some cases, a subject CasZ protein includes an amino acid sequence having 50% or more sequence identity (e.g., 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZe protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 80% or more sequence identity (e.g., 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZe protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZe protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZe amino acid sequence of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZe amino acid sequence of FIG. 1 or FIG. 7, with the exception that the sequence includes an amino acid substitution (e.g., 1, 2, or 3 amino acid substitutions) that reduces the naturally occurring catalytic activity of the protein (e.g., such as at one or more catalytic amino acid positions) (e.g., in some cases such that the CasZ protein is a dCasZ). In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZe protein of FIG. 1 or FIG. 7 and has a length in a range of from 450-700 amino acids (e.g., 450-650, 450-615, 475-700, 475-650, or 475-615 amino acids).
[0081] In some cases, a subject CasZ protein (of the subject compositions and/or methods) includes an amino acid sequence having 20% or more sequence identity (e.g., 30% or more, 40% or more, 50% or more, 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZf protein of FIG. 1 or FIG. 7. For example, in some cases, a subject CasZ protein includes an amino acid sequence having 50% or more sequence identity (e.g., 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZf protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 80% or more sequence identity (e.g., 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZf protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZf protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZf amino acid sequence of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZf amino acid sequence of FIG. 1 or FIG. 7, with the exception that the sequence includes an amino acid substitution (e.g., 1, 2, or 3 amino acid substitutions) that reduces the naturally occurring catalytic activity of the protein (e.g., such as at one or more catalytic amino acid positions) (e.g., in some cases such that the CasZ protein is a dCasZ). In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZf protein of FIG. 1 or FIG. 7 and has a length in a range of from 400-750 amino acids (e.g., 400-700, 700-650, 400-620, 400-600, 400-550, 400-520, 400-500, 400-475, 415-550, 415-520, 415-500, or 415-475 amino acids).
[0082] In some cases, a subject CasZ protein (of the subject compositions and/or methods) includes an amino acid sequence having 20% or more sequence identity (e.g., 30% or more, 40% or more, 50% or more, 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZg protein of FIG. 1 or FIG. 7. For example, in some cases, a subject CasZ protein includes an amino acid sequence having 50% or more sequence identity (e.g., 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZg protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 80% or more sequence identity (e.g., 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZg protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZg protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZg amino acid sequence of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZg amino acid sequence of FIG. 1 or FIG. 7, with the exception that the sequence includes an amino acid substitution (e.g., 1, 2, or 3 amino acid substitutions) that reduces the naturally occurring catalytic activity of the protein (e.g., such as at one or more catalytic amino acid positions) (e.g., in some cases such that the CasZ protein is a dCasZ). In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZg protein of FIG. 1 or FIG. 7 and has a length in a range of from 500-750 amino acids (e.g., 500-750 amino acids (e.g., 550-750 amino acids)).
[0083] In some cases, a subject CasZ protein (of the subject compositions and/or methods) includes an amino acid sequence having 20% or more sequence identity (e.g., 30% or more, 40% or more, 50% or more, 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZh protein of FIG. 1 or FIG. 7. For example, in some cases, a subject CasZ protein includes an amino acid sequence having 50% or more sequence identity (e.g., 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZh protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 80% or more sequence identity (e.g., 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZh protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZh protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZh amino acid sequence of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZh amino acid sequence of FIG. 1 or FIG. 7, with the exception that the sequence includes an amino acid substitution (e.g., 1, 2, or 3 amino acid substitutions) that reduces the naturally occurring catalytic activity of the protein (e.g., such as at one or more catalytic amino acid positions) (e.g., in some cases such that the CasZ protein is a dCasZ). In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZh protein of FIG. 1 or FIG. 7 and has a length in a range of from 380-450 amino acids (e.g., 380-420, 400-450, or 400-420 amino acids).
[0084] In some cases, a subject CasZ protein (of the subject compositions and/or methods) includes an amino acid sequence having 20% or more sequence identity (e.g., 30% or more, 40% or more, 50% or more, 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZi protein of FIG. 1 or FIG. 7. For example, in some cases, a subject CasZ protein includes an amino acid sequence having 50% or more sequence identity (e.g., 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZi protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 80% or more sequence identity (e.g., 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZi protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZi protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZi amino acid sequence of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZi amino acid sequence of FIG. 1 or FIG. 7, with the exception that the sequence includes an amino acid substitution (e.g., 1, 2, or 3 amino acid substitutions) that reduces the naturally occurring catalytic activity of the protein (e.g., such as at one or more catalytic amino acid positions) (e.g., in some cases such that the CasZ protein is a dCasZ). In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZi protein of FIG. 1 or FIG. 7 and has a length in a range of from 700-800 amino acids (e.g., 700-750, 720-800, or 720-750 amino acids).
[0085] In some cases, a subject CasZ protein (of the subject compositions and/or methods) includes an amino acid sequence having 20% or more sequence identity (e.g., 30% or more, 40% or more, 50% or more, 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZj protein of FIG. 1 or FIG. 7. For example, in some cases, a subject CasZ protein includes an amino acid sequence having 50% or more sequence identity (e.g., 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZj protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 80% or more sequence identity (e.g., 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZj protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZj protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZj amino acid sequence of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZj amino acid sequence of FIG. 1 or FIG. 7, with the exception that the sequence includes an amino acid substitution (e.g., 1, 2, or 3 amino acid substitutions) that reduces the naturally occurring catalytic activity of the protein (e.g., such as at one or more catalytic amino acid positions) (e.g., in some cases such that the CasZ protein is a dCasZ). In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZj protein of FIG. 1 or FIG. 7 and has a length in a range of from 600-750 amino acids (e.g., 600-700 or 650-700 amino acids).
[0086] In some cases, a subject CasZ protein (of the subject compositions and/or methods) includes an amino acid sequence having 20% or more sequence identity (e.g., 30% or more, 40% or more, 50% or more, 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZk protein of FIG. 1 or FIG. 7. For example, in some cases, a subject CasZ protein includes an amino acid sequence having 50% or more sequence identity (e.g., 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZk protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 80% or more sequence identity (e.g., 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZk protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZk protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZk amino acid sequence of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZk amino acid sequence of FIG. 1 or FIG. 7, with the exception that the sequence includes an amino acid substitution (e.g., 1, 2, or 3 amino acid substitutions) that reduces the naturally occurring catalytic activity of the protein (e.g., such as at one or more catalytic amino acid positions) (e.g., in some cases such that the CasZ protein is a dCasZ). In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZk protein of FIG. 1 or FIG. 7 and has a length in a range of from 450-600 amino acids (e.g., 450-580, 480-600, 480-580, or 500-600 amino acids).
[0087] In some cases, a subject CasZ protein (of the subject compositions and/or methods) includes an amino acid sequence having 20% or more sequence identity (e.g., 30% or more, 40% or more, 50% or more, 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZl protein of FIG. 1 or FIG. 7. For example, in some cases, a subject CasZ protein includes an amino acid sequence having 50% or more sequence identity (e.g., 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZl protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 80% or more sequence identity (e.g., 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZl protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZl protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZl amino acid sequence of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes a CasZl amino acid sequence of FIG. 1 or FIG. 7, with the exception that the sequence includes an amino acid substitution (e.g., 1, 2, or 3 amino acid substitutions) that reduces the naturally occurring catalytic activity of the protein (e.g., such as at one or more catalytic amino acid positions) (e.g., in some cases such that the CasZ protein is a dCasZ). In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZl protein of FIG. 1 or FIG. 7 and has a length in a range of from 450-600 amino acids (e.g., 450-580, 480-600, 480-580, or 500-600 amino acids).
[0088] In some cases, a subject CasZ protein (of the subject compositions and/or methods) includes an amino acid sequence having 20% or more sequence identity (e.g., 30% or more, 40% or more, 50% or more, 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZe, CasZf, CasZg, or CasZh protein of FIG. 1 or FIG. 7. For example, in some cases, a subject CasZ protein includes an amino acid sequence having 50% or more sequence identity (e.g., 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZe, CasZf, CasZg, or CasZh protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 80% or more sequence identity (e.g., 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZe, CasZf, CasZg, or CasZh protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZe, CasZf, CasZg, or CasZh protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having a CasZe, CasZf, CasZg, or CasZh protein sequence of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having a CasZe, CasZf, CasZg, or CasZh protein sequence of FIG. 1 or FIG. 7, with the exception that the sequence includes an amino acid substitution (e.g., 1, 2, or 3 amino acid substitutions) that reduces the naturally occurring catalytic activity of the protein (e.g., such as at one or more catalytic amino acid positions). In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZe, CasZf, CasZg, or CasZh protein of FIG. 1 or FIG. 7 and has a length in a range of from 350-900 amino acids (e.g., 350-850, 350-800, 400-900, 400-850, or 400-800 amino acids).
[0089] In some cases, a subject CasZ protein (of the subject compositions and/or methods) includes an amino acid sequence having 20% or more sequence identity (e.g., 30% or more, 40% or more, 50% or more, 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZa, CasZb, CasZc, CasZd, CasZe, CasZf, CasZg, CasZh, CasZi, CasZj, CasZK, or CasZl protein of FIG. 1 or FIG. 7. For example, in some cases, a subject CasZ protein includes an amino acid sequence having 50% or more sequence identity (e.g., 60% or more, 70% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZa, CasZb, CasZc, CasZd, CasZe, CasZf, CasZg, CasZh, CasZi, CasZj, CasZK, or CasZl protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 80% or more sequence identity (e.g., 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZa, CasZb, CasZc, CasZd, CasZe, CasZf, CasZg, CasZh, CasZi, CasZj, CasZK, or CasZl protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZa, CasZb, CasZc, CasZd, CasZe, CasZf, CasZg, CasZh, CasZi, CasZj, CasZK, or CasZl protein of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having a CasZa, CasZb, CasZc, CasZd, CasZe, CasZf, CasZg, CasZh, CasZi, CasZj, CasZK, or CasZl protein sequence of FIG. 1 or FIG. 7. In some cases, a subject CasZ protein includes an amino acid sequence having a CasZa, CasZb, CasZc, CasZd, CasZe, CasZf, CasZg, CasZh, CasZi, CasZj, CasZK, or CasZl protein sequence of FIG. 1 or FIG. 7, with the exception that the sequence includes an amino acid substitution (e.g., 1, 2, or 3 amino acid substitutions) that reduces the naturally occurring catalytic activity of the protein (e.g., such as at one or more catalytic amino acid positions). In some cases, a subject CasZ protein includes an amino acid sequence having 90% or more sequence identity (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100% sequence identity) with a CasZa, CasZb, CasZc, CasZd, CasZe, CasZf, CasZg, CasZh, CasZi, CasZj, CasZK, or CasZl protein of FIG. 1 or FIG. 7 and has a length in a range of from 350-900 amino acids (e.g., 350-850, 350-800, 400-900, 400-850, or 400-800 amino acids).
CasZ Variants
[0090] A variant CasZ protein has an amino acid sequence that is different by at least one amino acid (e.g., has a deletion, insertion, substitution, fusion) when compared to the amino acid sequence of the corresponding wild type CasZ protein. A CasZ protein that cleaves one strand but not the other of a double stranded target nucleic acid is referred to herein as a "nickase" (e.g., a "nickase CasZ"). A CasZ protein that has substantially no nuclease activity is referred to herein as a dead CasZ protein ("dCasZ") (with the caveat that nuclease activity can be provided by a heterologous polypeptide--a fusion partner--in the case of a chimeric CasZ protein, which is described in more detail below). For any of the CasZ variant proteins described herein (e.g., nickase CasZ, dCasZ, chimeric CasZ), the CasZ variant can include a CasZ protein sequence with the same parameters described above (e.g., domains that are present, percent identity, length, and the like).
[0091] Variants--Catalytic Activity
[0092] In some cases, the CasZ protein is a variant CasZ protein, e.g., mutated relative to the naturally occurring catalytically active sequence, and exhibits reduced cleavage activity (e.g., exhibits 90%, or less, 80% or less, 70% or less, 60% or less, 50% or less, 40% or less, or 30% or less cleavage activity) when compared to the corresponding naturally occurring sequence. In some cases, such a variant CasZ protein is a catalytically `dead` protein (has substantially no cleavage activity) and can be referred to as a `dCasZ.` In some cases, the variant CasZ protein is a nickase (cleaves only one strand of a double stranded target nucleic acid, e.g., a double stranded target DNA). As described in more detail herein, in some cases, a CasZ protein (in some case a CasZ protein with wild type cleavage activity and in some cases a variant CasZ with reduced cleavage activity, e.g., a dCasZ or a nickase CasZ) is fused (conjugated) to a heterologous polypeptide that has an activity of interest (e.g., a catalytic activity of interest) to form a fusion protein (a chimeric CasZ protein).
[0093] Catalytic residues of CasZ include D405, E586 and D684 when numbered according to CasZi.1 (e.g., see FIG. 1). Thus, in some cases, the CasZ protein has reduced activity and one or more of the above described amino acids (or one or more corresponding amino acids of any CasZ protein) are mutated (e.g., substituted with an alanine). In some cases, the variant CasZ protein is a catalytically `dead` protein (is catalytically inactive) and is referred to as `dCasZ.` A dCasZ protein can be fused to a fusion partner that provides an activity, and in some cases, the dCasZ (e.g., one without a fusion partner that provides catalytic activity--but which can have an NLS when expressed in a eukaryotic cell) can bind to target DNA and can be used for imaging (e.g., the protein can be tagged/labeled) and/or can block RNA polymerase from transcribing from a target DNA. In some cases, the variant CasZ protein is a nickase (cleaves only one strand of a double stranded target nucleic acid, e.g., a double stranded target DNA).
[0094] Variants--Chimeric CasZ (i.e., Fusion Proteins)
[0095] As noted above, in some cases, a CasZ protein (in some cases a CasZ protein with wild type cleavage activity and in some cases a variant CasZ with reduced cleavage activity, e.g., a dCasZ or a nickase CasZ) is fused (conjugated) to a heterologous polypeptide that has an activity of interest (e.g., a catalytic activity of interest) to form a fusion protein (a chimeric CasZ protein). A heterologous polypeptide to which a CasZ protein can be fused is referred to herein as a `fusion partner.`
[0096] In some cases, the fusion partner can modulate transcription (e.g., inhibit transcription, increase transcription) of a target DNA. For example, in some cases the fusion partner is a protein (or a domain from a protein) that inhibits transcription (e.g., a transcriptional repressor, a protein that functions via recruitment of transcription inhibitor proteins, modification of target DNA such as methylation, recruitment of a DNA modifier, modulation of histones associated with target DNA, recruitment of a histone modifier such as those that modify acetylation and/or methylation of histones, and the like). In some cases the fusion partner is a protein (or a domain from a protein) that increases transcription (e.g., a transcription activator, a protein that acts via recruitment of transcription activator proteins, modification of target DNA such as demethylation, recruitment of a DNA modifier, modulation of histones associated with target DNA, recruitment of a histone modifier such as those that modify acetylation and/or methylation of histones, and the like).
[0097] In some cases, a chimeric CasZ protein includes a heterologous polypeptide that has enzymatic activity that modifies a target nucleic acid (e.g., nuclease activity such as FokI nuclease activity, methyltransferase activity, demethylase activity, DNA repair activity, DNA damage activity, deamination activity, dismutase activity, alkylation activity, depurination activity, oxidation activity, pyrimidine dimer forming activity, integrase activity, transposase activity, recombinase activity, polymerase activity, ligase activity, helicase activity, photolyase activity or glycosylase activity).
[0098] In some cases, a chimeric CasZ protein includes a heterologous polypeptide that has enzymatic activity that modifies a polypeptide (e.g., a histone) associated with a target nucleic acid (e.g., methyltransferase activity, demethylase activity, acetyltransferase activity, deacetylase activity, kinase activity, phosphatase activity, ubiquitin ligase activity, deubiquitinating activity, adenylation activity, deadenylation activity, SUMOylating activity, deSUMOylating activity, ribosylation activity, deribosylation activity, myristoylation activity or demyristoylation activity).
[0099] Examples of proteins (or fragments thereof) that can be used in increase transcription include but are not limited to: transcriptional activators such as VP16, VP64, VP48, VP160, p65 subdomain (e.g., from NFkB), and activation domain of EDLL and/or TAL activation domain (e.g., for activity in plants); histone lysine methyltransferases such as SET1A, SET1B, MLL1 to 5, ASH1, SYMD2, NSD1, and the like; histone lysine demethylases such as JHDM2a/b, UTX, JMJD3, and the like; histone acetyltransferases such as GCN5, PCAF, CBP, p300, TAF1, TIP60/PLIP, MOZ/MYST3, MORF/MYST4, SRC1, ACTR, P160, CLOCK, and the like; and DNA demethylases such as Ten-Eleven Translocation (TET) dioxygenase 1 (TET1CD), TET1, DME, DML1, DML2, ROS1, and the like.
[0100] Examples of proteins (or fragments thereof) that can be used in decrease transcription include but are not limited to: transcriptional repressors such as the Kruppel associated box (KRAB or SKD); KOX 1 repression domain; the Mad mSIN3 interaction domain (SID); the ERF repressor domain (ERD), the SRDX repression domain (e.g., for repression in plants), and the like; histone lysine methyltransferases such as Pr-SET7/8, SUV4-20H1, RIZ1, and the like; histone lysine demethylases such as JMJD2A/JHDM3A, JMJD2B, JMJD2C/GASC1, JMJD2D, JARID1A/RBP2, JARID1B/PLU-1, JARID1C/SMCX, JARID1D/SMCY, and the like; histone lysine deacetylases such as HDAC1, HDAC2, HDAC3, HDAC8, HDAC4, HDAC5, HDAC7, HDAC9, SIRT1, SIRT2, HDAC11, and the like; DNA methylases such as HhaI DNA m5c-methyltransferase (M.HhaI), DNA methyltransferase 1 (DNMT1), DNA methyltransferase 3a (DNMT3a), DNA methyltransferase 3b (DNMT3b), METI, DRM3 (plants), ZMET2, CMT1, CMT2 (plants), and the like; and periphery recruitment elements such as Lamin A, Lamin B, and the like.
[0101] In some cases, the fusion partner has enzymatic activity that modifies the target nucleic acid (e.g., ssRNA, dsRNA, ssDNA, dsDNA). Examples of enzymatic activity that can be provided by the fusion partner include but are not limited to: nuclease activity such as that provided by a restriction enzyme (e.g., FokI nuclease), methyltransferase activity such as that provided by a methyltransferase (e.g., HhaI DNA m5c-methyltransferase (M.HhaI), DNA methyltransferase 1 (DNMT1), DNA methyltransferase 3a (DNMT3a), DNA methyltransferase 3b (DNMT3b), METI, DRM3 (plants), ZMET2, CMT1, CMT2 (plants), and the like); demethylase activity such as that provided by a demethylase (e.g., Ten-Eleven Translocation (TET) dioxygenase 1 (TET1CD), TET1, DME, DML1, DML2, ROS1, and the like), DNA repair activity, DNA damage activity, deamination activity such as that provided by a deaminase (e.g., a cytosine deaminase enzyme such as rat APOBEC1), dismutase activity, alkylation activity, depurination activity, oxidation activity, pyrimidine dimer forming activity, integrase activity such as that provided by an integrase and/or resolvase (e.g., Gin invertase such as the hyperactive mutant of the Gin invertase, GinH106Y; human immunodeficiency virus type 1 integrase (IN); Tn3 resolvase; and the like), transposase activity, recombinase activity such as that provided by a recombinase (e.g., catalytic domain of Gin recombinase), polymerase activity, ligase activity, helicase activity, photolyase activity, and glycosylase activity).
[0102] In some cases, the fusion partner has enzymatic activity that modifies a protein associated with the target nucleic acid (e.g., ssRNA, dsRNA, ssDNA, dsDNA) (e.g., a histone, an RNA binding protein, a DNA binding protein, and the like). Examples of enzymatic activity (that modifyies a protein associated with a target nucleic acid) that can be provided by the fusion partner include but are not limited to: methyltransferase activity such as that provided by a histone methyltransferase (HMT) (e.g., suppressor of variegation 3-9 homolog 1 (SUV39H1, also known as KMT1A), euchromatic histone lysine methyltransferase 2 (G9A, also known as KMT1C and EHMT2), SUV39H2, ESET/SETDB1, and the like, SET1A, SET1B, MLL1 to 5, ASH1, SYMD2, NSD1, DOT1L, Pr-SET7/8, SUV4-20H1, EZH2, RIZ1), demethylase activity such as that provided by a histone demethylase (e.g., Lysine Demethylase 1A (KDM1A also known as LSD1), JHDM2a/b, JMJD2A/JHDM3A, JMJD2B, JMJD2C/GASC1, JMJD2D, JARID1A/RBP2, JARID1B/PLU-1, JARID1C/SMCX, JARID1D/SMCY, UTX, JMJD3, and the like), acetyltransferase activity such as that provided by a histone acetylase transferase (e.g., catalytic core/fragment of the human acetyltransferase p300, GCN5, PCAF, CBP, TAF1, TIP60/PLIP, MOZ/MYST3, MORF/MYST4, HBO1/MYST2, HMOF/MYST1, SRC1, ACTR, P160, CLOCK, and the like), deacetylase activity such as that provided by a histone deacetylase (e.g., HDAC1, HDAC2, HDAC3, HDAC8, HDAC4, HDAC5, HDAC7, HDAC9, SIRT1, SIRT2, HDAC11, and the like), kinase activity, phosphatase activity, ubiquitin ligase activity, deubiquitinating activity, adenylation activity, deadenylation activity, SUMOylating activity, deSUMOylating activity, ribosylation activity, deribosylation activity, myristoylation activity, and demyristoylation activity.
[0103] Additional examples of a suitable fusion partners are dihydrofolate reductase (DHFR) destabilization domain (e.g., to generate a chemically controllable chimeric CasZ protein), and a chloroplast transit peptide. Suitable chloroplast transit peptides include, but are not limited to:
TABLE-US-00001 (SEQ ID NO: 101) MASMISSSAVTTVSRASRGQSAAMAPFGGLKSMTGFPVRKVNTDIT SITSNGGRVKCMQVWPPIGKKKFETLSYLPPLTRDSRA; (SEQ ID NO: 102) MASMISSSAVTTVSRASRGQSAAMAPFGGLKSMTGFPVRKVNTDIT SITSNGGRVKS; (SEQ ID NO: 103) MASSMLSSATMVASPAQATMVAPFNGLKSSAAFPATRKANNDITSI TSNGGRVNCMQVWPPIEKKKFETLSYLPDLTDSGGRVNC; (SEQ ID NO: 104) MAQVSRICNGVQNPSLISNLSKSSQRKSPLSVSLKTQQHPRAYPIS SSWGLKKSGMTLIGSELRPLKVMSSVSTAC; (SEQ ID NO: 105) MAQVSRICNGVWNPSLISNLSKSSQRKSPLSVSLKTQQHPRAYPIS SSWGLKKSGMTLIGSELRPLKVMSSVSTAC; (SEQ ID NO: 106) MAQINNMAQGIQTLNPNSNFHKPQVPKSSSFLVFGSKKLKNSANSM LVLKKDSIFMQLFCSFRISASVATAC; (SEQ ID NO: 107) MAALVTSQLATSGTVLSVTDRFRRPGFQGLRPRNPADAALGMRTVG ASAAPKQSRKPHRFDRRCLSMVV; (SEQ ID NO: 108) MAALTTSQLATSATGFGIADRSAPSSLLRHGFQGLKPRSPAGGDAT SLSVTTSARATPKQQRSVQRGSRRFPSVVVC; (SEQ ID NO: 109) MASSVLSSAAVATRSNVAQANMVAPFTGLKSAASFPVSRKQNLDIT SIASNGGRVQC; (SEQ ID NO: 110) MESLAATSVFAPSRVAVPAARALVRAGTVVPTRRTSSTSGTSGVKC SAAVTPQASPVISRSAAAA; and (SEQ ID NO: 111) MGAAATSMQSLKFSNRLVPPSRRLSPVPNNVTCNNLPKSAAPVRTV KCCASSWNSTINGAAATTNGASAASS.
[0104] In some case, a CasZ fusion polypeptide of the present disclosure comprises: a) a CasZ polypeptide of the present disclosure; and b) a chloroplast transit peptide. Thus, for example, a CRISPR-CasZ complex can be targeted to the chloroplast. In some cases, this targeting may be achieved by the presence of an N-terminal extension, called a chloroplast transit peptide (CTP) or plastid transit peptide. Chromosomal transgenes from bacterial sources must have a sequence encoding a CTP sequence fused to a sequence encoding an expressed polypeptide if the expressed polypeptide is to be compartmentalized in the plant plastid (e.g. chloroplast). Accordingly, localization of an exogenous polypeptide to a chloroplast is often 1 accomplished by means of operably linking a polynucleotide sequence encoding a CTP sequence to the 5' region of a polynucleotide encoding the exogenous polypeptide. The CTP is removed in a processing step during translocation into the plastid. Processing efficiency may, however, be affected by the amino acid sequence of the CTP and nearby sequences at the NH 2 terminus of the peptide. Other options for targeting to the chloroplast which have been described are the maize cab-m7 signal sequence (U.S. Pat. No. 7,022,896, WO 97/41228) a pea glutathione reductase signal sequence (WO 97/41228) and the CTP described in US2009029861.
[0105] In some cases, a CasZ fusion polypeptide of the present disclosure can comprise: a) a CasZ polypeptide of the present disclosure; and b) an endosomal escape peptide. In some cases, an endosomal escape polypeptide comprises the amino acid sequence GLFXALLXLLXSLWXLLLXA (SEQ ID NO: 112), wherein each X is independently selected from lysine, histidine, and arginine. In some cases, an endosomal escape polypeptide comprises the amino acid sequence GLFHALLHLLHSLWHLLLHA (SEQ ID NO: 113).
[0106] For examples of some of the above fusion partners (and more) used in the context of fusions with Cas9, Zinc Finger, and/or TALE proteins (for site specific target nucleic modification, modulation of transcription, and/or target protein modification, e.g., histone modification), see, e.g.: Nomura et al, J Am Chem Soc. 2007 Jul. 18; 129(28):8676-7; Rivenbark et al., Epigenetics. 2012 April; 7(4):350-60; Nucleic Acids Res. 2016 Jul. 8; 44(12):5615-28; Gilbert et al., Cell. 2013 Jul. 18; 154(2):442-51; Kearns et al., Nat Methods. 2015 May; 12(5):401-3; Mendenhall et al., Nat Biotechnol. 2013 December; 31(12):1133-6; Hilton et al., Nat Biotechnol. 2015 May; 33(5):510-7; Gordley et al., Proc Natl Acad Sci USA. 2009 Mar. 31; 106(13):5053-8; Akopian et al., Proc Natl Acad Sci USA. 2003 Jul. 22; 100(15):8688-91; Tan et., al., J Virol. 2006 February; 80(4):1939-48; Tan et al., Proc Natl Acad Sci USA. 2003 Oct. 14; 100(21):11997-2002; Papworth et al., Proc Natl Acad Sci USA. 2003 Feb. 18; 100(4):1621-6; Sanjana et al., Nat Protoc. 2012 Jan. 5; 7(1):171-92; Beerli et al., Proc Natl Acad Sci USA. 1998 Dec. 8; 95(25):14628-33; Snowden et al., Curr Biol. 2002 Dec. 23; 12(24):2159-66; Xu et. al., Xu et al., Cell Discov. 2016 May 3; 2:16009; Komor et al., Nature. 2016 Apr. 20; 533(7603):420-4; Chaikind et al., Nucleic Acids Res. 2016 Aug. 11; Choudhury at. al., Oncotarget. 2016 Jun. 23; Du et al., Cold Spring Harb Protoc. 2016 Jan. 4; Pham et al., Methods Mol Biol. 2016; 1358:43-57; Balboa et al., Stem Cell Reports. 2015 Sep. 8; 5(3):448-59; Hara et al., Sci Rep. 2015 Jun. 9; 5:11221; Piatek et al., Plant Biotechnol J. 2015 May; 13(4):578-89; Hu et al., Nucleic Acids Res. 2014 April; 42(7):4375-90; Cheng et al., Cell Res. 2013 October; 23(10):1163-71; and Maeder et al., Nat Methods. 2013 October; 10(10):977-9.
[0107] Additional suitable heterologous polypeptides include, but are not limited to, a polypeptide that directly and/or indirectly provides for increased transcription and/or translation of a target nucleic acid (e.g., a transcription activator or a fragment thereof, a protein or fragment thereof that recruits a transcription activator, a small molecule/drug-responsive transcription and/or translation regulator, a translation-regulating protein, etc.). Non-limiting examples of heterologous polypeptides to accomplish increased or decreased transcription include transcription activator and transcription repressor domains. In some such cases, a chimeric CasZ polypeptide is targeted by the guide nucleic acid (guide RNA) to a specific location (i.e., sequence) in the target nucleic acid and exerts locus-specific regulation such as blocking RNA polymerase binding to a promoter (which selectively inhibits transcription activator function), and/or modifying the local chromatin status (e.g., when a fusion sequence is used that modifies the target nucleic acid or modifies a polypeptide associated with the target nucleic acid). In some cases, the changes are transient (e.g., transcription repression or activation). In some cases, the changes are inheritable (e.g., when epigenetic modifications are made to the target nucleic acid or to proteins associated with the target nucleic acid, e.g., nucleosomal histones).
[0108] Non-limiting examples of heterologous polypeptides for use when targeting ssRNA target nucleic acids include (but are not limited to): splicing factors (e.g., RS domains); protein translation components (e.g., translation initiation, elongation, and/or release factors; e.g., eIF4G); RNA methylases; RNA editing enzymes (e.g., RNA deaminases, e.g., adenosine deaminase acting on RNA (ADAR), including A to I and/or C to U editing enzymes); helicases; RNA-binding proteins; and the like. It is understood that a heterologous polypeptide can include the entire protein or in some cases can include a fragment of the protein (e.g., a functional domain).
[0109] The heterologous polypeptide of a subject chimeric CasZ polypeptide can be any domain capable of interacting with ssRNA (which, for the purposes of this disclosure, includes intramolecular and/or intermolecular secondary structures, e.g., double-stranded RNA duplexes such as hairpins, stem-loops, etc.), whether transiently or irreversibly, directly or indirectly, including but not limited to an effector domain selected from the group comprising; Endonucleases (for example RNase III, the CRR22 DYW domain, Dicer, and PIN (PilT N-terminus) domains from proteins such as SMG5 and SMG6); proteins and protein domains responsible for stimulating RNA cleavage (for example CPSF, CstF, CFIm and CFIIm); Exonucleases (for example XRN-1 or Exonuclease T); Deadenylases (for example HNT3); proteins and protein domains responsible for nonsense mediated RNA decay (for example UPF1, UPF2, UPF3, UPF3b, RNP S1, Y14, DEK, REF2, and SRm160); proteins and protein domains responsible for stabilizing RNA (for example PABP); proteins and protein domains responsible for repressing translation (for example Ago2 and Ago4); proteins and protein domains responsible for stimulating translation (for example Staufen); proteins and protein domains responsible for (e.g., capable of) modulating translation (e.g., translation factors such as initiation factors, elongation factors, release factors, etc., e.g., eIF4G); proteins and protein domains responsible for polyadenylation of RNA (for example PAP1, GLD-2, and Star-PAP); proteins and protein domains responsible for polyuridinylation of RNA (for example CI D1 and terminal uridylate transferase); proteins and protein domains responsible for RNA localization (for example from IMP1, ZBP1, She2p, She3p, and Bicaudal-D); proteins and protein domains responsible for nuclear retention of RNA (for example Rrp6); proteins and protein domains responsible for nuclear export of RNA (for example TAP, NXF1, THO, TREX, REF, and Aly); proteins and protein domains responsible for repression of RNA splicing (for example PTB, Sam68, and hnRNP A1); proteins and protein domains responsible for stimulation of RNA splicing (for example Serine/Arginine-rich (SR) domains); proteins and protein domains responsible for reducing the efficiency of transcription (for example FUS (TLS)); and proteins and protein domains responsible for stimulating transcription (for example CDK7 and HIV Tat). Alternatively, the effector domain may be selected from the group comprising Endonucleases; proteins and protein domains capable of stimulating RNA cleavage; Exonucleases; Deadenylases; proteins and protein domains having nonsense mediated RNA decay activity; proteins and protein domains capable of stabilizing RNA; proteins and protein domains capable of repressing translation; proteins and protein domains capable of stimulating translation; proteins and protein domains capable of modulating translation (e.g., translation factors such as initiation factors, elongation factors, release factors, etc., e.g., eIF4G); proteins and protein domains capable of polyadenylation of RNA; proteins and protein domains capable of polyuridinylation of RNA; proteins and protein domains having RNA localization activity; proteins and protein domains capable of nuclear retention of RNA; proteins and protein domains having RNA nuclear export activity; proteins and protein domains capable of repression of RNA splicing; proteins and protein domains capable of stimulation of RNA splicing; proteins and protein domains capable of reducing the efficiency of transcription; and proteins and protein domains capable of stimulating transcription. Another suitable heterologous polypeptide is a PUF RNA-binding domain, which is described in more detail in WO2012068627, which is hereby incorporated by reference in its entirety.
[0110] Some RNA splicing factors that can be used (in whole or as fragments thereof) as heterologous polypeptides for a chimeric CasZ polypeptide have modular organization, with separate sequence-specific RNA binding modules and splicing effector domains. For example, members of the Serine/Arginine-rich (SR) protein family contain N-terminal RNA recognition motifs (RRMs) that bind to exonic splicing enhancers (ESEs) in pre-mRNAs and C-terminal RS domains that promote exon inclusion. As another example, the hnRNP protein hnRNP Al binds to exonic splicing silencers (ESSs) through its RRM domains and inhibits exon inclusion through a C-terminal Glycine-rich domain. Some splicing factors can regulate alternative use of splice site (ss) by binding to regulatory sequences between the two alternative sites. For example, ASF/SF2 can recognize ESEs and promote the use of intron proximal sites, whereas hnRNP Al can bind to ESSs and shift splicing towards the use of intron distal sites. One application for such factors is to generate ESFs that modulate alternative splicing of endogenous genes, particularly disease associated genes. For example, Bcl-x pre-mRNA produces two splicing isoforms with two alternative 5' splice sites to encode proteins of opposite functions. The long splicing isoform Bcl-xL is a potent apoptosis inhibitor expressed in long-lived postmitotic cells and is up-regulated in many cancer cells, protecting cells against apoptotic signals. The short isoform Bcl-xS is a pro-apoptotic isoform and expressed at high levels in cells with a high turnover rate (e.g., developing lymphocytes). The ratio of the two Bcl-x splicing isoforms is regulated by multiple cw-elements that are located in either the core exon region or the exon extension region (i.e., between the two alternative 5' splice sites). For more examples, see WO2010075303, which is hereby incorporated by reference in its entirety.
[0111] Further suitable fusion partners include, but are not limited to, proteins (or fragments thereof) that are boundary elements (e.g., CTCF), proteins and fragments thereof that provide periphery recruitment (e.g., Lamin A, Lamin B, etc.), protein docking elements (e.g., FKBP/FRB, Pil1/Aby1, etc.).
[0112] Examples of various additional suitable heterologous polypeptide (or fragments thereof) for a subject chimeric CasZ polypeptide include, but are not limited to those described in the following applications (which publications are related to other CRISPR endonucleases such as Cas9, but the described fusion partners can also be used with CasZ instead): PCT patent applications: WO2010075303, WO2012068627, and WO2013155555, and can be found, for example, in U.S. patents and patent applications: U.S. Pat. Nos. 8,906,616; 8,895,308; 8,889,418; 8,889,356; 8,871,445; 8,865,406; 8,795,965; 8,771,945; 8,697,359; 20140068797; 20140170753; 20140179006; 20140179770; 20140186843; 20140186919; 20140186958; 20140189896; 20140227787; 20140234972; 20140242664; 20140242699; 20140242700; 20140242702; 20140248702; 20140256046; 20140273037; 20140273226; 20140273230; 20140273231; 20140273232; 20140273233; 20140273234; 20140273235; 20140287938; 20140295556; 20140295557; 20140298547; 20140304853; 20140309487; 20140310828; 20140310830; 20140315985; 20140335063; 20140335620; 20140342456; 20140342457; 20140342458; 20140349400; 20140349405; 20140356867; 20140356956; 20140356958; 20140356959; 20140357523; 20140357530; 20140364333; and 20140377868; all of which are hereby incorporated by reference in their entirety.
[0113] In some cases, a heterologous polypeptide (a fusion partner) provides for subcellular localization, i.e., the heterologous polypeptide contains a subcellular localization sequence (e.g., a nuclear localization signal (NLS) for targeting to the nucleus, a sequence to keep the fusion protein out of the nucleus, e.g., a nuclear export sequence (NES), a sequence to keep the fusion protein retained in the cytoplasm, a mitochondrial localization signal for targeting to the mitochondria, a chloroplast localization signal for targeting to a chloroplast, an ER retention signal, and the like). In some embodiments, a CasZ fusion polypeptide does not include a NLS so that the protein is not targeted to the nucleus (which can be advantageous, e.g., when the target nucleic acid is an RNA that is present in the cyosol). In some embodiments, the heterologous polypeptide can provide a tag (i.e., the heterologous polypeptide is a detectable label) for ease of tracking and/or purification (e.g., a fluorescent protein, e.g., green fluorescent protein (GFP), YFP, RFP, CFP, mCherry, tdTomato, and the like; a histidine tag, e.g., a 6.times.His tag; a hemagglutinin (HA) tag; a FLAG tag; a Myc tag; and the like).
[0114] In some cases a CasZ protein (e.g., a wild type CasZ protein, a variant CasZ protein, a chimeric CasZ protein, a dCasZ protein, a chimeric CasZ protein where the CasZ portion has reduced nuclease activity--such as a dCasZ protein fused to a fusion partner, and the like) includes (is fused to) a nuclear localization signal (NLS) (e.g, in some cases 2 or more, 3 or more, 4 or more, or 5 or more NLSs). Thus, in some cases, a CasZ polypeptide includes one or more NLSs (e.g., 2 or more, 3 or more, 4 or more, or 5 or more NLSs). In some cases, one or more NLSs (2 or more, 3 or more, 4 or more, or 5 or more NLSs) are positioned at or near (e.g., within 50 amino acids of) the N-terminus and/or the C-terminus. In some cases, one or more NLSs (2 or more, 3 or more, 4 or more, or 5 or more NLSs) are positioned at or near (e.g., within 50 amino acids of) the N-terminus. In some cases, one or more NLSs (2 or more, 3 or more, 4 or more, or 5 or more NLSs) are positioned at or near (e.g., within 50 amino acids of) the C-terminus. In some cases, one or more NLSs (3 or more, 4 or more, or 5 or more NLSs) are positioned at or near (e.g., within 50 amino acids of) both the N-terminus and the C-terminus. In some cases, an NLS is positioned at the N-terminus and an NLS is positioned at the C-terminus.
[0115] In some cases a CasZ protein (e.g., a wild type CasZ protein, a variant CasZ protein, a chimeric CasZ protein, a dCasZ protein, a chimeric CasZ protein where the CasZ portion has reduced nuclease activity--such as a dCasZ protein fused to a fusion partner, and the like) includes (is fused to) between 1 and 10 NLSs (e.g., 1-9, 1-8, 1-7, 1-6, 1-5, 2-10, 2-9, 2-8, 2-7, 2-6, or 2-5 NLSs). In some cases a CasZ protein (e.g., a wild type CasZ protein, a variant CasZ protein, a chimeric CasZ protein, a dCasZ protein, a chimeric CasZ protein where the CasZ portion has reduced nuclease activity--such as a dCasZ protein fused to a fusion partner, and the like) includes (is fused to) between 2 and 5 NLSs (e.g., 2-4, or 2-3 NLSs).
[0116] Non-limiting examples of NLSs include an NLS sequence derived from: the NLS of the SV40 virus large T-antigen, having the amino acid sequence PKKKRKV (SEQ ID NO: 114); the NLS from nucleoplasmin (e.g., the nucleoplasmin bipartite NLS with the sequence KRPAATKKAGQAKKKK (SEQ ID NO: 115)); the c-myc NLS having the amino acid sequence PAAKRVKLD (SEQ ID NO: 116) or RQRRNELKRSP (SEQ ID NO: 117); the hRNPA1 M9 NLS having the sequence NQSSNFGPMKGGNFGGRSSGPYGGGGQYFAKPRNQGGY (SEQ ID NO: 118); the sequence RMRIZFKNKGKDTAELRRRRVEVSVELRKAKKDEQILKRRNV (SEQ ID NO: 119) of the IBB domain from importin-alpha; the sequences VSRKRPRP (SEQ ID NO: 120) and PPKKARED (SEQ ID NO: 121) of the myoma T protein; the sequence PQPKKKPL (SEQ ID NO: 122) of human p53; the sequence SALIKKKKKMAP (SEQ ID NO: 123) of mouse c-abl IV; the sequences DRLRR (SEQ ID NO: 124) and PKQKKRK (SEQ ID NO: 125) of the influenza virus NS1; the sequence RKLKKKIKKL (SEQ ID NO: 126) of the Hepatitis virus delta antigen; the sequence REKKKFLKRR (SEQ ID NO: 127) of the mouse Mx1 protein; the sequence KRKGDEVDGVDEVAKKKSKK (SEQ ID NO: 128) of the human poly(ADP-ribose) polymerase; and the sequence RKCLQAGMNLEARKTKK (SEQ ID NO: 129) of the steroid hormone receptors (human) glucocorticoid. In general, NLS (or multiple NLSs) are of sufficient strength to drive accumulation of the CasZ protein in a detectable amount in the nucleus of a eukaryotic cell. Detection of accumulation in the nucleus may be performed by any suitable technique. For example, a detectable marker may be fused to the CasZ protein such that location within a cell may be visualized. Cell nuclei may also be isolated from cells, the contents of which may then be analyzed by any suitable process for detecting protein, such as immunohistochemistry, Western blot, or enzyme activity assay. Accumulation in the nucleus may also be determined indirectly.
[0117] In some cases, a CasZ fusion polypeptide includes a "Protein Transduction Domain" or PTD (also known as a CPP--cell penetrating peptide), which refers to a polypeptide, polynucleotide, carbohydrate, or organic or inorganic compound that facilitates traversing a lipid bilayer, micelle, cell membrane, organelle membrane, or vesicle membrane. A PTD attached to another molecule, which can range from a small polar molecule to a large macromolecule and/or a nanoparticle, facilitates the molecule traversing a membrane, for example going from extracellular space to intracellular space, or cytosol to within an organelle. In some embodiments, a PTD is covalently linked to the amino terminus a polypeptide (e.g., linked to a wild type CasZ to generate a fusino protein, or linked to a variant CasZ protein such as a dCasZ, nickase CasZ, or chimeric CasZ protein to generate a fusion protein). In some embodiments, a PTD is covalently linked to the carboxyl terminus of a polypeptide (e.g., linked to a wild type CasZ to generate a fusino protein, or linked to a variant CasZ protein such as a dCasZ, nickase CasZ, or chimeric CasZ protein to generate a fusion protein). In some cases, the PTD is inserted interally in the CasZ fusion polypeptide (i.e., is not at the N- or C-terminus of the CasZ fusion polypeptide) at a suitable insertion site. In some cases, a subject CasZ fusion polypeptide includes (is conjugated to, is fused to) one or more PTDs (e.g., two or more, three or more, four or more PTDs). In some cases, a PTD includes a nuclear localization signal (NLS) (e.g, in some cases 2 or more, 3 or more, 4 or more, or 5 or more NLSs). Thus, in some cases, a CasZ fusion polypeptide includes one or more NLSs (e.g., 2 or more, 3 or more, 4 or more, or 5 or more NLSs). In some cases, a PTD is covalently linked to a nucleic acid (e.g., a CasZ guide nucleic acid, a polynucleotide encoding a CasZ guide nucleic acid, a polynucleotide encoding a CasZ fusion polypeptide, a donor polynucleotide, etc.). Examples of PTDs include but are not limited to a minimal undecapeptide protein transduction domain (corresponding to residues 47-57 of HIV-1 TAT comprising YGRKKRRQRRR; SEQ ID NO: 130); a polyarginine sequence comprising a number of arginines sufficient to direct entry into a cell (e.g., 3, 4, 5, 6, 7, 8, 9, 10, or 10-50 arginines); a VP22 domain (Zender et al. (2002) Cancer Gene Ther. 9(6):489-96); an Drosophila Antennapedia protein transduction domain (Noguchi et al. (2003) Diabetes 52(7):1732-1737); a truncated human calcitonin peptide (Trehin et al. (2004) Pharm. Research 21:1248-1256); polylysine (Wender et al. (2000) Proc. Natl. Acad. Sci. USA 97:13003-13008); RRQRRTSKLMKR (SEQ ID NO: 131); Transportan GWTLNSAGYLLGKINLKALAALAKKIL (SEQ ID NO: 132); KALAWEAKLAKALAKALAKHLAKALAKALKCEA (SEQ ID NO: 133); and RQIKIWFQNRRMKWKK (SEQ ID NO: 134). Exemplary PTDs include but are not limited to, YGRKKRRQRRR (SEQ ID NO: 130), RKKRRQRRR (SEQ ID NO: 135); an arginine homopolymer of from 3 arginine residues to 50 arginine residues; Exemplary PTD domain amino acid sequences include, but are not limited to, any of the following: YGRKKRRQRRR (SEQ ID NO: 130); RKKRRQRR (SEQ ID NO: 136); YARAAARQARA (SEQ ID NO: 137); THRLPRRRRRR (SEQ ID NO: 138); and GGRRARRRRRR (SEQ ID NO: 139). In some embodiments, the PTD is an activatable CPP (ACPP) (Aguilera et al. (2009) Integr Biol (Camb) June; 1(5-6): 371-381). ACPPs comprise a polycationic CPP (e.g., Arg9 or "R9") connected via a cleavable linker to a matching polyanion (e.g., Glu9 or "E9"), which reduces the net charge to nearly zero and thereby inhibits adhesion and uptake into cells. Upon cleavage of the linker, the polyanion is released, locally unmasking the polyarginine and its inherent adhesiveness, thus "activating" the ACPP to traverse the membrane.
[0118] Linkers (e.g., for Fusion Partners)
[0119] In some instances, a subject CasZ protein is fused to a fusion partner via a linker polypeptide (e.g., one or more linker polypeptides). The linker polypeptide may have any of a variety of amino acid sequences. Proteins can be joined by a spacer peptide, generally of a flexible nature, although other chemical linkages are not excluded. Suitable linkers include polypeptides of between 4 amino acids and 40 amino acids in length, or between 4 amino acids and 25 amino acids in length. These linkers can be produced by using synthetic, linker-encoding oligonucleotides to couple the proteins, or can be encoded by a nucleic acid sequence encoding the fusion protein. Peptide linkers with a degree of flexibility can be used. The linking peptides may have virtually any amino acid sequence, bearing in mind that the preferred linkers will have a sequence that results in a generally flexible peptide. The use of small amino acids, such as glycine and alanine, are of use in creating a flexible peptide. The creation of such sequences is routine to those of skill in the art. A variety of different linkers are commercially available and are considered suitable for use.
[0120] Examples of linker polypeptides include glycine polymers (G).sub.n, glycine-serine polymers (including, for example, (GS).sub.n, GSGGS.sub.n (SEQ ID NO: 140), GGSGGS.sub.n (SEQ ID NO: 141), and GGGS.sub.n (SEQ ID NO: 142), where n is an integer of at least one), glycine-alanine polymers, alanine-serine polymers. Exemplary linkers can comprise amino acid sequences including, but not limited to, GGSG (SEQ ID NO: 143), GGSGG (SEQ ID NO: 144), GSGSG (SEQ ID NO: 145), GSGGG (SEQ ID NO: 146), GGGSG (SEQ ID NO: 147), GSSSG (SEQ ID NO: 148), and the like. The ordinarily skilled artisan will recognize that design of a peptide conjugated to any desired element can include linkers that are all or partially flexible, such that the linker can include a flexible linker as well as one or more portions that confer less flexible structure.
Detectable Labels
[0121] In some cases, a CasZ polypeptide of the present disclosure comprises (e.g., can be attached/fused to) a detectable label. Suitable detectable labels and/or moieties that can provide a detectable signal can include, but are not limited to, an enzyme, a radioisotope, a member of a specific binding pair; a fluorophore; a fluorescent protein; a quantum dot; and the like.
[0122] Suitable fluorescent proteins include, but are not limited to, green fluorescent protein (GFP) or variants thereof, blue fluorescent variant of GFP (BFP), cyan fluorescent variant of GFP (CFP), yellow fluorescent variant of GFP (YFP), enhanced GFP (EGFP), enhanced CFP (ECFP), enhanced YFP (EYFP), GFPS65T, Emerald, Topaz (TYFP), Venus, Citrine, mCitrine, GFPuv, destabilised EGFP (dEGFP), destabilised ECFP (dECFP), destabilised EYFP (dEYFP), mCFPm, Cerulean, T-Sapphire, CyPet, YPet, mKO, HcRed, t-HcRed, DsRed, DsRed2, DsRed-monomer, J-Red, dimer2, t-dimer2(12), mRFP1, pocilloporin, Renilla GFP, Monster GFP, paGFP, Kaede protein and kindling protein, Phycobiliproteins and Phycobiliprotein conjugates including B-Phycoerythrin, R-Phycoerythrin and Allophycocyanin. Other examples of fluorescent proteins include mHoneydew, mBanana, mOrange, dTomato, tdTomato, mTangerine, mStrawberry, mCherry, mGrape1, mRaspberry, mGrape2, mPlum (Shaner et al. (2005) Nat. Methods 2:905-909), and the like. Any of a variety of fluorescent and colored proteins from Anthozoan species, as described in, e.g., Matz et al. (1999) Nature Biotechnol. 17:969-973, is suitable for use.
[0123] Suitable enzymes include, but are not limited to, horse radish peroxidase (HRP), alkaline phosphatase (AP), beta-galactosidase (GAL), glucose-6-phosphate dehydrogenase, beta-N-acetylglucosaminidase, .beta.-glucuronidase, invertase, Xanthine Oxidase, firefly luciferase, glucose oxidase (GO), and the like.
Protospacer Adjacent Motif (PAM)
[0124] A natural CasZ protein binds to target DNA at a target sequence defined by the region of complementarity between the DNA-targeting RNA and the target DNA. As is the case for many CRISPR endonucleases, site-specific binding (and/or cleavage) of a double stranded target DNA occurs at locations determined by both (i) base-pairing complementarity between the guide RNA and the target DNA; and (ii) a short motif [referred to as the protospacer adjacent motif (PAM)] in the target DNA.
[0125] In some cases, the PAM for a CasZ protein is immediately 5' of the target sequence of the non-complementary strand of the target DNA (also referred to as the non-target strand; the complementary strand hybridizes to the guide sequence of the guide RNA while the non-complementary strand does not directly hybridize with the guide RNA and is the reverse complement of the non-complementary strand). In some cases (e.g., for CasZc), the PAM sequence of the non-complementary strand is 5'-TTA-3'. In some cases (e.g., for CasZb), the PAM sequence of the non-complementary strand is 5'-TTTN-3'. In some cases (e.g., for CasZb), the PAM sequence of the non-complementary strand is 5'-TTTA-3'.
[0126] In some cases, different CasZ proteins (i.e., CasZ proteins from various species) may be advantageous to use in the various provided methods in order to capitalize on various enzymatic characteristics of the different CasZ proteins (e.g., for different PAM sequence preferences; for increased or decreased enzymatic activity; for an increased or decreased level of cellular toxicity; to change the balance between NHEJ, homology-directed repair, single strand breaks, double strand breaks, etc.; to take advantage of a short total sequence; and the like). CasZ proteins from different species may require different PAM sequences in the target DNA. Thus, for a particular CasZ protein of choice, the PAM sequence preference may be different than the sequence(s) described above. Various methods (including in silico and/or wet lab methods) for identification of the appropriate PAM sequence are known in the art and are routine, and any convenient method can be used.
CasZ Guide RNA
[0127] A nucleic acid molecule that binds to a CasZ protein, forming a ribonucleoprotein complex (RNP), and targets the complex to a specific location within a target nucleic acid (e.g., a target DNA) is referred to herein as a "CasZ guide RNA" or simply as a "guide RNA." It is to be understood that in some cases, a hybrid DNA/RNA can be made such that a CasZ guide RNA includes DNA bases in addition to RNA bases, but the term "CasZ guide RNA" is still used to encompass such a molecule herein.
[0128] A CasZ guide RNA can be said to include two segments, a targeting segment and a protein-binding segment. The targeting segment of a CasZ guide RNA includes a nucleotide sequence (a guide sequence) that is complementary to (and therefore hybridizes with) a specific sequence (a target site) within a target nucleic acid (e.g., a target ssRNA, a target ssDNA, the complementary strand of a double stranded target DNA, etc.). The protein-binding segment (or "protein-binding sequence") interacts with (binds to) a CasZ polypeptide. The protein-binding segment of a subject CasZ guide RNA includes two complementary stretches of nucleotides that hybridize to one another to form a double stranded RNA duplex (dsRNA duplex). Site-specific binding and/or cleavage of a target nucleic acid (e.g., genomic DNA) can occur at locations (e.g., target sequence of a target locus) determined by base-pairing complementarity between the CasZ guide RNA (the guide sequence of the CasZ guide RNA) and the target nucleic acid.
[0129] A CasZ guide RNA and a CasZ protein, e.g., a fusion CasZ polypeptide, form a complex (e.g., bind via non-covalent interactions). The CasZ guide RNA provides target specificity to the complex by including a targeting segment, which includes a guide sequence (a nucleotide sequence that is complementary to a sequence of a target nucleic acid). The CasZ protein of the complex provides the site-specific activity (e.g., cleavage activity provided by the CasZ protein and/or an activity provided by the fusion partner in the case of a chimeric CasZ protein). In other words, the CasZ protein is guided to a target nucleic acid sequence (e.g. a target sequence) by virtue of its association with the CasZ guide RNA.
[0130] The "guide sequence" also referred to as the "targeting sequence" of a CasZ guide RNA can be modified so that the CasZ guide RNA can target a CasZ protein (e.g., a naturally occurring CasZ protein, a fusion CasZ polypeptide (chimeric CasZ), and the like) to any desired sequence of any desired target nucleic acid, with the exception (e.g., as described herein) that the PAM sequence can be taken into account. Thus, for example, a CasZ guide RNA can have a guide sequence with complementarity to (e.g., can hybridize to) a sequence in a nucleic acid in a eukaryotic cell, e.g., a viral nucleic acid, a eukaryotic nucleic acid (e.g., a eukaryotic chromosome, chromosomal sequence, a eukaryotic RNA, etc.), and the like.
[0131] In some cases, a CasZ guide RNA has a length of 30 nucleotides (nt) or more (e.g., 35 nt or more, 40 nt or more, 45 nt or more, 50 nt or more, 55 nt or more, or 60 nt or more). In some embodiments, a CasZ guide RNA has a length of 40 nucleotides (nt) or more (e.g., 45 nt or more, 50 nt or more, 55 nt or more, or 60 nt or more). In some cases, a CasZ guide RNA has a length of from 30 nucleotides (nt) to 100 nt (e.g., 30-90, 30-80, 30-75, 30-70, 30-65, 40-100, 40-90, 40-80, 40-75, 40-70, or 40-65 nt). In some cases, a CasZ guide RNA has a length of from 40 nucleotides (nt) to 100 nt (e.g., 40-90, 40-80, 40-75, 40-70, or 40-65 nt).
Guide Sequence of a CasZ Guide RNA
[0132] A subject CasZ guide RNA includes a guide sequence (i.e., a targeting sequence), which is a nucleotide sequence that is complementary to a sequence (a target site) in a target nucleic acid. In other words, the guide sequence of a CasZ guide RNA can interact with a target nucleic acid (e.g., double stranded DNA (dsDNA), single stranded DNA (ssDNA), single stranded RNA (ssRNA), or double stranded RNA (dsRNA)) in a sequence-specific manner via hybridization (i.e., base pairing). The guide sequence of a CasZ guide RNA can be modified (e.g., by genetic engineering)/designed to hybridize to any desired target sequence (e.g., while taking the PAM into account, e.g., when targeting a dsDNA target) within a target nucleic acid (e.g., a eukaryotic target nucleic acid such as genomic DNA).
[0133] In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 60% or more (e.g., 65% or more, 70% or more, 75% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100%). In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 80% or more (e.g., 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100%). In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 90% or more (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100%). In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 100%.
[0134] In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 100% over the seven contiguous 3'-most nucleotides of the target site of the target nucleic acid.
[0135] In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 60% or more (e.g., 70% or more, 75% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100%) over 17 or more (e.g., 18 or more, 19 or more, 20 or more, 21 or more, 22 or more) contiguous nucleotides. In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 80% or more (e.g., 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100%) over 17 or more (e.g., 18 or more, 19 or more, 20 or more, 21 or more, 22 or more) contiguous nucleotides. In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 90% or more (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100%) over 17 or more (e.g., 18 or more, 19 or more, 20 or more, 21 or more, 22 or more) contiguous nucleotides. In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 100% over 17 or more (e.g., 18 or more, 19 or more, 20 or more, 21 or more, 22 or more) contiguous nucleotides.
[0136] In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 60% or more (e.g., 70% or more, 75% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100%) over 19 or more (e.g., 20 or more, 21 or more, 22 or more) contiguous nucleotides. In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 80% or more (e.g., 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100%) over 19 or more (e.g., 20 or more, 21 or more, 22 or more) contiguous nucleotides. In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 90% or more (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100%) over 19 or more (e.g., 20 or more, 21 or more, 22 or more) contiguous nucleotides. In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 100% over 19 or more (e.g., 20 or more, 21 or more, 22 or more) contiguous nucleotides.
[0137] In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 60% or more (e.g., 70% or more, 75% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100%) over 17-25 contiguous nucleotides. In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 80% or more (e.g., 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100%) over 17-25 contiguous nucleotides. In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 90% or more (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100%) over 17-25 contiguous nucleotides. In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 100% over 17-25 contiguous nucleotides.
[0138] In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 60% or more (e.g., 70% or more, 75% or more, 80% or more, 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100%) over 19-25 contiguous nucleotides. In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 80% or more (e.g., 85% or more, 90% or more, 95% or more, 97% or more, 98% or more, 99% or more, or 100%) over 19-25 contiguous nucleotides. In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 90% or more (e.g., 95% or more, 97% or more, 98% or more, 99% or more, or 100%) over 19-25 contiguous nucleotides. In some cases, the percent complementarity between the guide sequence and the target site of the target nucleic acid is 100% over 19-25 contiguous nucleotides.
[0139] In some cases, the guide sequence has a length in a range of from 17-30 nucleotides (nt) (e.g., from 17-25, 17-22, 17-20, 19-30, 19-25, 19-22, 19-20, 20-30, 20-25, or 20-22 nt). In some cases, the guide sequence has a length in a range of from 17-25 nucleotides (nt) (e.g., from 17-22, 17-20, 19-25, 19-22, 19-20, 20-25, or 20-22 nt). In some cases, the guide sequence has a length of 17 or more nt (e.g., 18 or more, 19 or more, 20 or more, 21 or more, or 22 or more nt; 19 nt, 20 nt, 21 nt, 22 nt, 23 nt, 24 nt, 25 nt, etc.). In some cases, the guide sequence has a length of 19 or more nt (e.g., 20 or more, 21 or more, or 22 or more nt; 19 nt, 20 nt, 21 nt, 22 nt, 23 nt, 24 nt, 25 nt, etc.). In some cases, the guide sequence has a length of 17 nt. In some cases, the guide sequence has a length of 18 nt. In some cases, the guide sequence has a length of 19 nt. In some cases, the guide sequence has a length of 20 nt. In some cases, the guide sequence has a length of 21 nt. In some cases, the guide sequence has a length of 22 nt. In some cases, the guide sequence has a length of 23 nt.
Protein-Binding Segment of a CasZ Guide RNA
[0140] The protein-binding segment of a subject CasZ guide RNA interacts with a CasZ protein. The CasZ guide RNA guides the bound CasZ protein to a specific nucleotide sequence within target nucleic acid via the above-mentioned guide sequence. The protein-binding segment of a CasZ guide RNA comprises two stretches of nucleotides that are complementary to one another and hybridize to form a double stranded RNA duplex (dsRNA duplex). Thus, the protein-binding segment includes a dsRNA duplex.
[0141] In some cases, the dsRNA duplex region includes a range of from 5-25 base pairs (bp) (e.g., from 5-22, 5-20, 5-18, 5-15, 5-12, 5-10, 5-8, 8-25, 8-22, 8-18, 8-15, 8-12, 12-25, 12-22, 12-18, 12-15, 13-25, 13-22, 13-18, 13-15, 14-25, 14-22, 14-18, 14-15, 15-25, 15-22, 15-18, 17-25, 17-22, or 17-18 bp, e.g., 5 bp, 6 bp, 7 bp, 8 bp, 9 bp, 10 bp, etc.). In some cases, the dsRNA duplex region includes a range of from 6-15 base pairs (bp) (e.g., from 6-12, 6-10, or 6-8 bp, e.g., 6 bp, 7 bp, 8 bp, 9 bp, 10 bp, etc.). In some cases, the duplex region includes 5 or more bp (e.g., 6 or more, 7 or more, or 8 or more bp). In some cases, the duplex region includes 6 or more bp (e.g., 7 or more, or 8 or more bp). In some cases, not all nucleotides of the duplex region are paired, and therefore the duplex forming region can include a bulge. The term "bulge" herein is used to mean a stretch of nucleotides (which can be one nucleotide or multiple nucleotides) that do not contribute to a double stranded duplex, but which are surround 5' and 3' by nucleotides that do contribute, and as such a bulge is considered part of the duplex region. In some cases, the dsRNA includes 1 or more bulges (e.g., 2 or more, 3 or more, 4 or more bulges). In some cases, the dsRNA duplex includes 2 or more bulges (e.g., 3 or more, 4 or more bulges). In some cases, the dsRNA duplex includes 1-5 bulges (e.g., 1-4, 1-3, 2-5, 2-4, or 2-3 bulges).
[0142] Thus, in some cases, the stretches of nucleotides that hybridize to one another to form the dsRNA duplex have 70%-100% complementarity (e.g., 75%-100%, 80%-10%, 85%-100%, 90%-100%, 95%-100% complementarity) with one another. In some cases, the stretches of nucleotides that hybridize to one another to form the dsRNA duplex have 70%-100% complementarity (e.g., 75%-100%, 80%-10%, 85%-100%, 90%-100%, 95%-100% complementarity) with one another. In some cases, the stretches of nucleotides that hybridize to one another to form the dsRNA duplex have 85%-100% complementarity (e.g., 90%-100%, 95%-100% complementarity) with one another. In some cases, the stretches of nucleotides that hybridize to one another to form the dsRNA duplex have 70%-95% complementarity (e.g., 75%-95%, 80%-95%, 85%-95%, 90%-95% complementarity) with one another.
[0143] In other words, in some embodiments, the dsRNA duplex includes two stretches of nucleotides that have 70%-100% complementarity (e.g., 75%-100%, 80%-10%, 85%-100%, 90%-100%, 95%-100% complementarity) with one another. In some cases, the dsRNA duplex includes two stretches of nucleotides that have 85%-100% complementarity (e.g., 90%-100%, 95%-100% complementarity) with one another. In some cases, the dsRNA duplex includes two stretches of nucleotides that have 70%-95% complementarity (e.g., 75%-95%, 80%-95%, 85%-95%, 90%-95% complementarity) with one another.
[0144] The duplex region of a subject CasZ guide RNA can include one or more (1, 2, 3, 4, 5, etc) mutations relative to a naturally occurring duplex region. For example, in some cases a base pair can be maintained while the nucleotides contributing to the base pair from each segment can be different. In some cases, the duplex region of a subject CasZ guide RNA includes more paired bases, less paired bases, a smaller bulge, a larger bulge, fewer bulges, more bulges, or any convenient combination thereof, as compared to a naturally occurring duplex region (of a naturally occurring CasZ guide RNA).
[0145] Examples of various Cas9 guide RNAs and cpf1 guide RNAs can be found in the art, and in some cases variations similar to those introduced into Cas9 guide RNAs can also be introduced into CasZ guide RNAs of the present disclosure (e.g., mutations to the dsRNA duplex region, extension of the 5' or 3' end for added stability for to provide for interaction with another protein, and the like). For example, see Jinek et al., Science. 2012 Aug. 17; 337(6096):816-21; Chylinski et al., RNA Biol. 2013 May; 10(5):726-37; Ma et al., Biomed Res Int. 2013; 2013:270805; Hou et al., Proc Natl Acad Sci USA. 2013 Sep. 24; 110(39):15644-9; Jinek et al., Elife. 2013; 2:e00471; Pattanayak et al., Nat Biotechnol. 2013 September; 31(9):839-43; Qi et al, Cell. 2013 Feb. 28; 152(5):1173-83; Wang et al., Cell. 2013 May 9; 153(4):910-8; Auer et al., Genome Res. 2013 Oct. 31; Chen et al., Nucleic Acids Res. 2013 Nov. 1; 41(20):e19; Cheng et al., Cell Res. 2013 October; 23(10):1163-71; Cho et al., Genetics. 2013 November; 195(3):1177-80; DiCarlo et al., Nucleic Acids Res. 2013 April; 41(7):4336-43; Dickinson et al., Nat Methods. 2013 October; 10(10):1028-34; Ebina et al., Sci Rep. 2013; 3:2510; Fujii et. al, Nucleic Acids Res. 2013 Nov. 1; 41(20):e187; Hu et al., Cell Res. 2013 November; 23(11):1322-5; Jiang et al., Nucleic Acids Res. 2013 Nov. 1; 41(20):e188; Larson et al., Nat Protoc. 2013 November; 8(11):2180-96; Mali et. at., Nat Methods. 2013 October; 10(10):957-63; Nakayama et al., Genesis. 2013 December; 51(12):835-43; Ran et al., Nat Protoc. 2013 November; 8(11):2281-308; Ran et al., Cell. 2013 Sep. 12; 154(6):1380-9; Upadhyay et al., G3 (Bethesda). 2013 Dec. 9; 3(12):2233-8; Walsh et al., Proc Natl Acad Sci USA. 2013 Sep. 24; 110(39):15514-5; Xie et al., Mol Plant. 2013 Oct. 9; Yang et al., Cell. 2013 Sep. 12; 154(6):1370-9; Briner et al., Mol Cell. 2014 Oct. 23; 56(2):333-9; and U.S. patents and patent applications: U.S. Pat. Nos. 8,906,616; 8,895,308; 8,889,418; 8,889,356; 8,871,445; 8,865,406; 8,795,965; 8,771,945; 8,697,359; 20140068797; 20140170753; 20140179006; 20140179770; 20140186843; 20140186919; 20140186958; 20140189896; 20140227787; 20140234972; 20140242664; 20140242699; 20140242700; 20140242702; 20140248702; 20140256046; 20140273037; 20140273226; 20140273230; 20140273231; 20140273232; 20140273233; 20140273234; 20140273235; 20140287938; 20140295556; 20140295557; 20140298547; 20140304853; 20140309487; 20140310828; 20140310830; 20140315985; 20140335063; 20140335620; 20140342456; 20140342457; 20140342458; 20140349400; 20140349405; 20140356867; 20140356956; 20140356958; 20140356959; 20140357523; 20140357530; 20140364333; and 20140377868; all of which are hereby incorporated by reference in their entirety.
[0146] A CasZ guide RNA comprises both the guide sequence and two stretches ("duplex-forming segments") of nucleotides that hybridize to form the dsRNA duplex of the protein-binding segment. The particular sequence of a given CasZ guide RNA can be characteristic of the species in which a crRNA is found. Examples of suitable CasZ guide RNAs are provided herein.
Example Guide RNA Sequences
[0147] Repeat sequences (non-guide sequence portion of a CasZ guide RNA) of crRNAs for naturally existing CasZ proteins (e.g., see FIG. 1 and FIG. 7) are shown in Table 1 and Table 3.
TABLE-US-00002 TABLE 1 crRNA repeat sequences for CasZ proteins CasZ SEQ Protein Repeat sequence ID NO: Za.1 GTTGCATTCCTTCATTCGTCT 51 ATTCGGGTTCTGCAAC Za.2 GTTGCATTCCTTCATTCGTCT 52 ATCCGGGTTCTGCAAG Za.3 GTTGCAGAACCCGAATAGACG 53 AATGAAGGAATGCAAC Za.4 CTATCATATTCAGAACAAAGG 54 GATTAAGGAATGCAAC Za.5 CTTTCATACTCAGAACAAAGG 55 GATTAAGGAATGCAAC Za.6 GTCTACAACTCATTGATAGAA 56 ATCAATGAGTTAGACA Za.7 GTTATAAAGGCGGGGATCGCG 57 ACCGAGCGATTGAAAG Zb.1 GTTGCATTCCTTAATTCATTT 58 TCTCAATATCGGAAAC Zb.2 GTTGCAGAAATAGAATAAAGG 59 AATTAAGGAATGCAAC Zb.3 CTTTCATACTCAGAACAAAGG 55 GATTAAGGAATGCAAC Zb.4 ATTTCATACTCAGAACAAAGG 61 GATTAAGGAATGCAAC Zb.5 GTTTCAGCGCACGAATTAACG 62 AGATGAGAGATGCAACT Zb.6 CTTGCAGAAGCTGAATAGACG 63 AATCAAGGAATGCAAC Zb.7 CACTTGCAGGCCTTGAATAGA 64 GGAGTTAAGGAATGCAAC Zb.8 GTCTCCATGACTGAAAAGTCG 65 TGGCCGAATTGAAAC Zb.9 GTTGCAGCGCCCGAACTGACG 66 AGACGAGAGATGCAAC Zb.10 GTTGCGCGAATAGAATAAAGG 67 AATTAAGGAATGCAAC Zb.11 AGTTGCATTCCTTAATCCCTC 68 TGTTCAGTTTGTGCAAT Zc.1 GTTGCATTCCTAGTTTCTCTA 69 ATTAGCACTGTGCAAC Zc.2 GTTGCGGCGCGCGAATAAACG 70 AGACTAGGAATGCAAC Zc.3 ACTAGTTGCATTCCTTAATCC 71 CTTTGTTCTGAATATGCTAG Zc.4 CTTTCATATTCAGAACAAAGG 72 GATTAAGGAATGCAAC Zc.5 GTTGCAGTCCTTAACCCCTAG 73 TTTCTGAATATGAAAGAT Zc.6 GTTGCAGCCCCCGAACTAACG 74 AGATGAGAGATGCAAC Zc.7 CTTGCAGAACAATCATATATG 75 ACTAATCAGACTGCAAC Zd.1 GTTGCACTCACCGGTGCTCAC 76 GACGTAGGGATGCAAC Zd.2 GTCCCTACTCGCTAGGGAAAC 77 TAATTGAATGGAAAC Ze.1 GTTGCATTCGGGTGCAAAACA 78 GGGAGTAGAGTGTAAC Ze.2 CTTCCAAACTCGAGCCAGTGG 79 GGAGAGAAGTGGCA Ze.3 CCTGTAGACCGGTCTCATTCT 80 GAGAGGGGTATGCAACT Ze.4 GTCTCGAGACCCTACAGATTT 81 TGGAGAGGGGTGGGAC Ze.4b GTCCCACCCCTCTCCAAAATC 82 TGTAGGGTCTCGAGAC Zf.1 GTAGCAGGACTCTCCTCGAGA 83 GAAACAGGGGTATGCT Zf.2 GTACAATACCTCTCCTTTAAG 84 AGAGGGAGGGGTACGCTAC Zf.3 CCCCCTCGTTTCCTTCAGGGG 85 ATTCCTTTCC Zg.1 GGTTCCCCCGGGCGCGGGTGG 86 GGTGGCG Zg.2 GGCTGCTCCGGGTGCGCGTGG 87 AGCGAGG Zh.1 GTTTTATACCCTTTAGAATTT 88 AAACTGTCTAAAAG Zi.1 ATTGCACCGGCCAACGCAAAT 89 CTGATTGATGGACAC Zi.2 GCCGCAGCGGCCGACGCGGCC 90 CTGATCGATGGACAC Zj.1 GTCGAAATGCCCGCGCGGGGG 91 CGTCGTACCCGCGAC Zk.1 GGCTAGCCCGTGCGCGCAGGG 92 ACGAGTGG Zk.2 GCCCGTGCGCGCAGGGACGAG 93 TGG Zk.3 GTTGCAGCGGCCGACGGAGCG 94 CGAGCGTGGATGCCAC Zk.4 CCATCGCCCCGCGCGCACGTG 95 GATGAGCC Zl.1 CTTTAGACTTCTCCGGAAGTC 96 GAATTAATGGAAAC Zl.2 GGGCGCCCCGCGCGAGCGGGG 97 GTTGAAG Za.8 CTTGCAGAACCCGGATAGACG 295 AATGAAGGAATGCAAC Zb.12 CTTGCAGGCCTTGAATAGAGG 296 AGTTAAGGAATGCAAC Zb.13 GTTGCACAGTGCTAATTAGAG 297 AAACTAGGAATGCAAC Zb.14 CTAGCATATTCAGAACAAAGG 298 GATTAAGGAATGCAAC Zb.15 CTTTCATATTCAGAAACTAGG 299 GGTTAAGGACTGCAAC Zc.8 GTTGCATCCCTACGTCGTGAG 300 CACCGGTGAGTGCAAC Ze.5 GGAAAGGAATCCCCTGAAGGA 301 AACGAGGGGG Zg.3 GTGTCCATCAATCAGATTTGC 302 GTTGGCCGGTGCAAT Zb.16 GTTTCAGCGCACGAATTAACG 303 AGATGAGAGATGCAAC Zj.2 CTTTTAGACAGTTTAAATTCT 307 AAAGGGTATAAAAC
[0148] In some cases, a subject CasZ guide RNA comprises (e.g., in addition to a guide sequence, e.g., as part of the protein-binding region) a crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 70% or more identity (e.g., 75% or more, 80% or more, 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 90% or more identity (e.g., 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a crRNA sequence of Table 1 or Table 3.
[0149] In some cases, a subject CasZ guide RNA comprises (e.g., in addition to a guide sequence, e.g., as part of the protein-binding region) a CasZa, CasZb, CasZc, CasZd, CasZe, CasZf, CasZg, CasZh, or CasZi crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 70% or more identity (e.g., 75% or more, 80% or more, 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZa, CasZb, CasZc, CasZd, CasZe, CasZf, CasZg, CasZh, or CasZi crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZa, CasZb, CasZc, CasZd, CasZe, CasZf, CasZg, CasZh, or CasZi crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 90% or more identity (e.g., 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZa, CasZb, CasZc, CasZd, CasZe, CasZf, CasZg, CasZh, or CasZi crRNA sequence of Table 1 or Table 3.
[0150] In some cases, a subject CasZ guide RNA comprises (e.g., in addition to a guide sequence, e.g., as part of the protein-binding region) a CasZa crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 70% or more identity (e.g., 75% or more, 80% or more, 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZa crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZa crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 90% or more identity (e.g., 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZa crRNA sequence of Table 1 or Table 3.
[0151] In some cases, a subject CasZ guide RNA comprises (e.g., in addition to a guide sequence, e.g., as part of the protein-binding region) a CasZb crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 70% or more identity (e.g., 75% or more, 80% or more, 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZb crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZb crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 90% or more identity (e.g., 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZb crRNA sequence of Table 1 or Table 3.
[0152] In some cases, a subject CasZ guide RNA comprises (e.g., in addition to a guide sequence, e.g., as part of the protein-binding region) a CasZc crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 70% or more identity (e.g., 75% or more, 80% or more, 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZc crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZc crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 90% or more identity (e.g., 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZc crRNA sequence of Table 1 or Table 3.
[0153] In some cases, a subject CasZ guide RNA comprises (e.g., in addition to a guide sequence, e.g., as part of the protein-binding region) a CasZd crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 70% or more identity (e.g., 75% or more, 80% or more, 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZd crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZd crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 90% or more identity (e.g., 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZd crRNA sequence of Table 1 or Table 3.
[0154] In some cases, a subject CasZ guide RNA comprises (e.g., in addition to a guide sequence, e.g., as part of the protein-binding region) a CasZe crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 70% or more identity (e.g., 75% or more, 80% or more, 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZe crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZe crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 90% or more identity (e.g., 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZe crRNA sequence of Table 1 or Table 3.
[0155] In some cases, a subject CasZ guide RNA comprises (e.g., in addition to a guide sequence, e.g., as part of the protein-binding region) a CasZf crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 70% or more identity (e.g., 75% or more, 80% or more, 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZf crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZf crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 90% or more identity (e.g., 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZf crRNA sequence of Table 1 or Table 3.
[0156] In some cases, a subject CasZ guide RNA comprises (e.g., in addition to a guide sequence, e.g., as part of the protein-binding region) a CasZg crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 70% or more identity (e.g., 75% or more, 80% or more, 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZg crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZg crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 90% or more identity (e.g., 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZg crRNA sequence of Table 1 or Table 3.
[0157] In some cases, a subject CasZ guide RNA comprises (e.g., in addition to a guide sequence, e.g., as part of the protein-binding region) a CasZh crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 70% or more identity (e.g., 75% or more, 80% or more, 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZh crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZh crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 90% or more identity (e.g., 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZh crRNA sequence of Table 1 or Table 3.
[0158] In some cases, a subject CasZ guide RNA comprises (e.g., in addition to a guide sequence, e.g., as part of the protein-binding region) a CasZi crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 70% or more identity (e.g., 75% or more, 80% or more, 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZi crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZi crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 90% or more identity (e.g., 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZi crRNA sequence of Table 1 or Table 3.
[0159] In some cases, a subject CasZ guide RNA comprises (e.g., in addition to a guide sequence, e.g., as part of the protein-binding region) a CasZj crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 70% or more identity (e.g., 75% or more, 80% or more, 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZj crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZj crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 90% or more identity (e.g., 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZj crRNA sequence of Table 1 or Table 3.
[0160] In some cases, a subject CasZ guide RNA comprises (e.g., in addition to a guide sequence, e.g., as part of the protein-binding region) a CasZk crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 70% or more identity (e.g., 75% or more, 80% or more, 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZk crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZk crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 90% or more identity (e.g., 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZk crRNA sequence of Table 1 or Table 3.
[0161] In some cases, a subject CasZ guide RNA comprises (e.g., in addition to a guide sequence, e.g., as part of the protein-binding region) a CasZl crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 70% or more identity (e.g., 75% or more, 80% or more, 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZl crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZl crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 90% or more identity (e.g., 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZl crRNA sequence of Table 1 or Table 3.
[0162] In some cases, a subject CasZ guide RNA comprises (e.g., in addition to a guide sequence, e.g., as part of the protein-binding region) a CasZj, CasZl, or CasZk crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 70% or more identity (e.g., 75% or more, 80% or more, 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZj, CasZl, or CasZk crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZj, CasZl, or CasZk crRNA sequence of Table 1 or Table 3. In some cases, a subject CasZ guide RNA comprises a nucleotide sequence having 90% or more identity (e.g., 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZj, CasZl, or CasZk crRNA sequence of Table 1 or Table 3.
CasZ Transactivating Noncoding RNA (trancRNA)
[0163] Compositions and methods of the present disclosure include a CasZ transactivating noncoding RNA ("trancRNA"; also referred to herein as a "CasZ trancRNA"). In some cases, a trancRNA forms a complex with a CasZ polypeptide of the present disclosure and a CasZ guide RNA. A trancRNA can be identified as a highly transcribed RNA encoded by a nucleotide sequence present in a CasZ locus. The sequence encoding a trancRNA is usually located between the cas genes and the array of the CasZ locus (the repeats) (e.g., can be located adjacent to the repeat sequences). Examples below demonstrate detection of a CasZ trancRNA. In some cases, a CasZ trancRNA co-immunoprecipitates (forms a complex with) with a CasZ polypeptide. In some cases, the presence of a CasZ trancRNA is required for function of the system. Data related to trancRNAs (e.g., their expression and their location on naturally occurring arrays) is presented in the examples section below.
[0164] In some cases, a CasZ trancRNA has a length of from 60 nucleotides (nt) to 270 nt (e.g., 60-260, 70-270, 70-260, or 75-255 nt). In some cases, a CasZ trancRNA (e.g., a CasZa trancRNA) has a length of from 60-150 nt (e.g., 60-140, 60-130, 65-150, 65-140, 65-130, 70-150, 70-140, or 70-130 nt). In some cases, a CasZ trancRNA (e.g., a CasZa trancRNA) has a length of from 70-130 nt. In some cases, a CasZ trancRNA (e.g., a CasZa trancRNA) has a length of about 80 nt. In some cases, a CasZ trancRNA (e.g., a CasZa trancRNA) has a length of about 90 nt. In some cases, a CasZ trancRNA (e.g., a CasZa trancRNA) has a length of about 120 nt.
[0165] In some cases, a CasZ trancRNA (e.g., a CasZb trancRNA) has a length of from 85-240 nt (e.g., 85-230, 85-220, 85-150, 85-130, 95-240, 95-230, 95-220, 95-150, or 95-130 nt). In some cases, a CasZ trancRNA (e.g., a CasZb trancRNA) has a length of from 95-120 nt. In some cases, a CasZ trancRNA (e.g., a CasZb trancRNA) has a length of about 105 nt. In some cases, a CasZ trancRNA (e.g., a CasZb trancRNA) has a length of about 115 nt. In some cases, a CasZ trancRNA (e.g., a CasZb trancRNA) has a length of about 215 nt.
[0166] In some cases, a CasZ trancRNA (e.g., a CasZc trancRNA) has a length of from 80-275 nt (e.g., 85-260 nt). In some cases, a CasZ trancRNA (e.g., a CasZc trancRNA) has a length of from 80-110 nt (e.g., 85-105 nt). In some cases, a CasZ trancRNA (e.g., a CasZc trancRNA) has a length of from 235-270 nt (e.g., 240-260 nt). In some cases, a CasZ trancRNA (e.g., a CasZc trancRNA) has a length of about 95 nt. In some cases, a CasZ trancRNA (e.g., a CasZc trancRNA) has a length of about 250 nt.
Example trancRNA Sequences
[0167] Examples of trancRNA sequences for naturally existing CasZ proteins are shown in Table 2.
TABLE-US-00003 TABLE 2 CasZ trancRNA sequences CasZ SEQ Protein trancRNA sequence ID NO Za.1 CGATTCCTCCCTACAGTAGTTAGGTAT 151 AGCCGAAAGGTAGAGACTAAATCTGTA GTTGGAGTGGGCCGCTTGCATCGGCC Za.2 TCGTCTCGAGGGTTACCAAAATTGGCA 152 CTTCTCGACTTTAGGCCGATGCAAGCG GCCCACTCCACTACAGATTTAGTCTCT ACCTTGCGGCTATACCTAACTTACTGT AGGGAGGAATCGTG Za.3 CTTCACTGATAAAGTGGAGAACCGCTT 153 CACCAAAAGCTGTCCCTTAGGGGATTA GAACTTGAGTGAAGGTGGGCTGCTTGC ATCAGCCTAA Zb.2 CAGAATAATACTGACTTACTAAGATAT 154 CTTGAGGGTATACCCGAAAAGATTGGC GTTGTTGCAACGCAATAAGATGTAAAT CTGAAAAGGTTTGGAATCATATAAATA ATTTTA Zb.4 AAGCCAAGATATGGAATGCCATTGTAA 155 TATTATGGTGTTGACTTAGTTTAGATT TAAACAATCTTCGATGGCTATATGCGG AAGGTTTGGCGTCGTTGTAACGC Zb.6 CAGTGTGCATAGCTATAACACTACGCA 156 AAGACTGCTAAAGAGCGATGTGCTCTA TCGCAGTCTCACCTTTAATGGACTTAC GGATCTTTTGGAGCACTAAGCTCCGCT GCGGTGCAACACCGCCCTTTTCTTGCC TCTGCTTGCCCTTTCCGGTTATTATAG CCGGGAGAGTGCGGAAGATTACCGCTC TAGCTCGCAGCATGTTACTGAGTC Zc.3 GCAAGTCATTCGGGGACACTTTTTGTT 157 ATTTAAAGTGTTTTAGATAAATCAGTG TCATGCTGAATAACGACCCGACCTATA AATAACATAATCC Zc.5 GTCCTTAAGGTACTACACATTACATGT 158 GAACGTGGAGCTAATAATAGAAATATT ATTAGACTACACCTTATTAATAACGGT AGGAGATCTATATGGTCTTGAATGGAA TAGTAATTGTGAAATTATAATTTCTGT TCTTAGCTACTTAAGATGGCTCGTTGC AAGCCACTCGGGGGCTCTCTTGAAGTC AAAGAGCTTTAGACAAATCAGTGTCAA ACTGAATAACGACCCGACCATGACTTC ATAATCCCG
[0168] In some cases, a subject CasZ trancRNA comprises a CasZa trancRNA sequence above. In some cases, a subject CasZ trancRNA comprises a nucleotide sequence having 70% or more identity (e.g., 75% or more, 80% or more, 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZa trancRNA sequence above. In some cases, a subject CasZ trancRNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZa trancRNA sequence above. In some cases, a subject CasZ trancRNA comprises a nucleotide sequence having 90% or more identity (e.g., 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZa trancRNA sequence above. In some cases, a subject CasZ trancRNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZa trancRNA sequence above, and has a length of from 60-150 nt (e.g., 60-140, 60-130, 65-150, 65-140, 65-130, 70-150, 70-140, or 70-130 nt).
[0169] In some cases, a subject CasZ trancRNA comprises a CasZb trancRNA sequence above. In some cases, a subject CasZ trancRNA comprises a nucleotide sequence having 70% or more identity (e.g., 75% or more, 80% or more, 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZb trancRNA sequence above. In some cases, a subject CasZ trancRNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZb trancRNA sequence above. In some cases, a subject CasZ trancRNA comprises a nucleotide sequence having 90% or more identity (e.g., 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZb trancRNA sequence above. In some cases, a subject CasZ trancRNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZb trancRNA sequence above, and has a length of from 85-240 nt (e.g., 85-230, 85-220, 85-150, 85-130, 95-240, 95-230, 95-220, 95-150, or 95-130 nt).
[0170] In some cases, a subject CasZ trancRNA comprises a CasZc trancRNA sequence above. In some cases, a subject CasZ trancRNA comprises a nucleotide sequence having 70% or more identity (e.g., 75% or more, 80% or more, 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZc trancRNA sequence above. In some cases, a subject CasZ trancRNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZc trancRNA sequence above. In some cases, a subject CasZ trancRNA comprises a nucleotide sequence having 90% or more identity (e.g., 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZc trancRNA sequence above. In some cases, a subject CasZ trancRNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZc trancRNA sequence above, and has a length of from 80-110 nt (e.g., 85-105 nt) or from 235-270 nt (e.g., 240-260 nt).
[0171] In some cases, a subject CasZ trancRNA comprises a CasZa, CasZb, or CasZc trancRNA sequence above. In some cases, a subject CasZ trancRNA comprises a nucleotide sequence having 70% or more identity (e.g., 75% or more, 80% or more, 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZa, CasZb, or CasZc trancRNA sequence above. In some cases, a subject CasZ trancRNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZa, CasZb, or CasZc trancRNA sequence above. In some cases, a subject CasZ trancRNA comprises a nucleotide sequence having 90% or more identity (e.g., 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZa, CasZb, or CasZc trancRNA sequence above. In some cases, a subject CasZ trancRNA comprises a nucleotide sequence having 80% or more identity (e.g., 85% or more, 90% or more, 93% or more, 95% or more, 97% or more, 98% or more, or 100% identity) with a CasZa, CasZb, or CasZc trancRNA sequence above, and has a length of from 60 nucleotides (nt) to 270 nt (e.g., 60-260, 70-270, 70-260, or 75-255 nt).
[0172] In some cases, a CasZ trancRNA comprises a modified nucleotide (e.g., methylated). In some cases, a CasZ trancRNA comprises one or more of: i) a base modification or substitution; ii) a backbone modification; iii) a modified internucleoside linkage; and iv) a modified sugar moiety. Possible nucleic acid modifications are described below.
CasZ Systems
[0173] The present disclosure provides a CasZ system. A CasZ system of the present disclosure can comprise one or more of: (1) a CasZ transactivating noncoding RNA (trancRNA) (referred to herein as a "CasZ trancRNA") or a nucleic acid encoding the CasZ trancRNA (e.g., an expression vector); (2) a CasZ protein (e.g., a wild type protein, a variant, a catalytically compromised variant, a CasZ fusion protein, and the like) or a nucleic acid encoding the CasZ protein (e.g., an RNA, an expression vector, and the like); and (3) a CasZ guide RNA (that binds to and provides sequence specificity to the CasZ protein, e.g., a guide RNA that can bind to a target sequence of a eukaryotic genome) or a nucleic acid encoding the CasZ guide RNA)(e.g., an expression vector). A CasZ system can include a host cell (e.g., a eukaryotic cell, a plant cell, a mammalian cell, a human cell) that comprises one or more of (1), (2), and (3) (in any combination), e.g., in some cases the host cell comprises a trancRNA and/or a nucleic acid encoding the trancRNA. In some cases, a CasZ system includes (e.g., in addition to the above) a donor template nucleic acid. In some cases, the CasZ system is a system of one or more nucleic acids (e.g., one or more expression vectors encoding any combination of the above).
Nucleic Acids
[0174] The present disclosure provides one or more nucleic acids comprising one or more of: a CasZ trancRNA sequence, a nucleotide sequence encoding a CasZ trancRNA, a nucleotide sequence encoding a CasZ polypeptide (e.g., a wild type CasZ protein, a nickase CasZ protein, a dCasZ protein, chimeric CasZ protein/CasZ fusion protein, and the like), a CasZ guide RNA sequence, a nucleotide sequence encoding a CasZ guide RNA, and a donor polynucleotide (donor template, donor DNA) sequence. In some cases, a subject nucleic acid (e.g., the one or more nucleic acids) is a recombinant expression vector (e.g., plasmid, viral vector, minicircle DNA, and the like). In some cases, the nucleotide sequence encoding the CasZ trancRNA, the nucleotide sequence encoding the CasZ protein, and/or the nucleotide sequence encoding the CasZ guide RNA is (are) operably linked to a promoter (e.g., an inducible promoter), e.g., one that is operable in a cell type of choice (e.g., a prokarytoic cell, a eukaryotic cell, a plant cell, an animal cell, a mammalian cell, a primate cell, a rodent cell, a human cell, etc.).
[0175] In some cases, a nucleotide sequence encoding a CasZ polypeptide of the present disclosure is codon optimized. This type of optimization can entail a mutation of a CasZ-encoding nucleotide sequence to mimic the codon preferences of the intended host organism or cell while encoding the same protein. Thus, the codons can be changed, but the encoded protein remains unchanged. For example, if the intended target cell was a human cell, a human codon-optimized CasZ-encoding nucleotide sequence could be used. As another non-limiting example, if the intended host cell were a mouse cell, then a mouse codon-optimized CasZ-encoding nucleotide sequence could be generated. As another non-limiting example, if the intended host cell were a plant cell, then a plant codon-optimized CasZ-encoding nucleotide sequence could be generated. As another non-limiting example, if the intended host cell were an insect cell, then an insect codon-optimized CasZ-encoding nucleotide sequence could be generated.
[0176] The present disclosure provides one or more recombinant expression vectors that include (in different recombinant expression vectors in some cases, and in the same recombinant expression vector in some cases): a CasZ trancRNA sequence, a nucleotide sequence encoding a CasZ trancRNA, a nucleotide sequence encoding a CasZ polypeptide (e.g., a wild type CasZ protein, a nickase CasZ protein, a dCasZ protein, chimeric CasZ protein/CasZ fusion protein, and the like), a CasZ guide RNA sequence, a nucleotide sequence encoding a CasZ guide RNA, and a donor polynucleotide (donor template, donor DNA) sequence. In some cases, a subject nucleic acid (e.g., the one or more nucleic acids) is a recombinant expression vector (e.g., plasmid, viral vector, minicircle DNA, and the like). In some cases, the nucleotide sequence encoding the CasZ trancRNA, the nucleotide sequence encoding the CasZ protein, and/or the nucleotide sequence encoding the CasZ guide RNA is (are) operably linked to a promoter (e.g., an inducible promoter), e.g., one that is operable in a cell type of choice (e.g., a prokarytoic cell, a eukaryotic cell, a plant cell, an animal cell, a mammalian cell, a primate cell, a rodent cell, a human cell, etc.).
[0177] Suitable expression vectors include viral expression vectors (e.g. viral vectors based on vaccinia virus; poliovirus; adenovirus (see, e.g., Li et al., Invest Opthalmol Vis Sci 35:2543 2549, 1994; Borras et al., Gene Ther 6:515 524, 1999; Li and Davidson, PNAS 92:7700 7704, 1995; Sakamoto et al., H Gene Ther 5:1088 1097, 1999; WO 94/12649, WO 93/03769; WO 93/19191; WO 94/28938; WO 95/11984 and WO 95/00655); adeno-associated virus (AAV) (see, e.g., Ali et al., Hum Gene Ther 9:81 86, 1998, Flannery et al., PNAS 94:6916 6921, 1997; Bennett et al., Invest Opthalmol Vis Sci 38:2857 2863, 1997; Jomary et al., Gene Ther 4:683 690, 1997, Rolling et al., Hum Gene Ther 10:641 648, 1999; Ali et al., Hum Mol Genet 5:591 594, 1996; Srivastava in WO 93/09239, Samulski et al., J. Vir. (1989) 63:3822-3828; Mendelson et al., Virol. (1988) 166:154-165; and Flotte et al., PNAS (1993) 90:10613-10617); SV40; herpes simplex virus; human immunodeficiency virus (see, e.g., Miyoshi et al., PNAS 94:10319 23, 1997; Takahashi et al., J Virol 73:7812 7816, 1999); a retroviral vector (e.g., Murine Leukemia Virus, spleen necrosis virus, and vectors derived from retroviruses such as Rous Sarcoma Virus, Harvey Sarcoma Virus, avian leukosis virus, a lentivirus, human immunodeficiency virus, myeloproliferative sarcoma virus, and mammary tumor virus); and the like. In some cases, a recombinant expression vector of the present disclosure is a recombinant adeno-associated virus (AAV) vector. In some cases, a recombinant expression vector of the present disclosure is a recombinant lentivirus vector. In some cases, a recombinant expression vector of the present disclosure is a recombinant retroviral vector.
[0178] Depending on the host/vector system utilized, any of a number of suitable transcription and translation control elements, including constitutive and inducible promoters, transcription enhancer elements, transcription terminators, etc. may be used in the expression vector.
[0179] In some embodiments, a nucleotide sequence encoding a CasZ guide RNA is operably linked to a control element, e.g., a transcriptional control element, such as a promoter. In some embodiments, a nucleotide sequence encoding a CasZ protein or a CasZ fusion polypeptide is operably linked to a control element, e.g., a transcriptional control element, such as a promoter.
[0180] The transcriptional control element can be a promoter. In some cases, the promoter is a constitutively active promoter. In some cases, the promoter is a regulatable promoter. In some cases, the promoter is an inducible promoter. In some cases, the promoter is a tissue-specific promoter. In some cases, the promoter is a cell type-specific promoter. In some cases, the transcriptional control element (e.g., the promoter) is functional in a targeted cell type or targeted cell population. For example, in some cases, the transcriptional control element can be functional in eukaryotic cells, e.g., hematopoietic stem cells (e.g., mobilized peripheral blood (mPB) CD34(+) cell, bone marrow (BM) CD34(+) cell, etc.).
[0181] Non-limiting examples of eukaryotic promoters (promoters functional in a eukaryotic cell) include EF1.alpha., those from cytomegalovirus (CMV) immediate early, herpes simplex virus (HSV) thymidine kinase, early and late SV40, long terminal repeats (LTRs) from retrovirus, and mouse metallothionein-I. Selection of the appropriate vector and promoter is well within the level of ordinary skill in the art. The expression vector may also contain a ribosome binding site for translation initiation and a transcription terminator. The expression vector may also include appropriate sequences for amplifying expression. The expression vector may also include nucleotide sequences encoding protein tags (e.g., 6.times.His tag, hemagglutinin tag, fluorescent protein, etc.) that can be fused to the CasZ protein, thus resulting in a chimeric CasZ polypeptide.
[0182] In some cases, a nucleotide sequence encoding a CasZ guide RNA and/or a CasZ fusion polypeptide is operably linked to an inducible promoter. In some cases, a nucleotide sequence encoding a CasZ guide RNA and/or a CasZ fusion protein is operably linked to a constitutive promoter.
[0183] A promoter can be a constitutively active promoter (i.e., a promoter that is constitutively in an active/"ON" state), it may be an inducible promoter (i.e., a promoter whose state, active/"ON" or inactive/"OFF", is controlled by an external stimulus, e.g., the presence of a particular temperature, compound, or protein.), it may be a spatially restricted promoter (i.e., transcriptional control element, enhancer, etc.) (e.g., tissue specific promoter, cell type specific promoter, etc.), and it may be a temporally restricted promoter (i.e., the promoter is in the "ON" state or "OFF" state during specific stages of embryonic development or during specific stages of a biological process, e.g., hair follicle cycle in mice).
[0184] Suitable promoters can be derived from viruses and can therefore be referred to as viral promoters, or they can be derived from any organism, including prokaryotic or eukaryotic organisms. Suitable promoters can be used to drive expression by any RNA polymerase (e.g., pol I, pol II, pol III). Exemplary promoters include, but are not limited to the SV40 early promoter, mouse mammary tumor virus long terminal repeat (LTR) promoter; adenovirus major late promoter (Ad MLP); a herpes simplex virus (HSV) promoter, a cytomegalovirus (CMV) promoter such as the CMV immediate early promoter region (CMVIE), a rous sarcoma virus (RSV) promoter, a human U6 small nuclear promoter (U6) (Miyagishi et al., Nature Biotechnology 20, 497-500 (2002)), an enhanced U6 promoter (e.g., Xia et al., Nucleic Acids Res. 2003 Sep. 1; 31(17)), a human H1 promoter (H1), and the like.
[0185] In some cases, a nucleotide sequence encoding a CasZ guide RNA is operably linked to (under the control of) a promoter operable in a eukaryotic cell (e.g., a U6 promoter, an enhanced U6 promoter, an H1 promoter, and the like). As would be understood by one of ordinary skill in the art, when expressing an RNA (e.g., a guide RNA) from a nucleic acid (e.g., an expression vector) using a U6 promoter (e.g., in a eukaryotic cell), or another PolIII promoter, the RNA may need to be mutated if there are several Ts in a row (coding for Us in the RNA). This is because a string of Ts (e.g., 5 Ts) in DNA can act as a terminator for polymerase III (PolIII). Thus, in order to ensure transcription of a guide RNA in a eukaryotic cell it may sometimes be necessary to modify the sequence encoding the guide RNA to eliminate runs of Ts. In some cases, a nucleotide sequence encoding a CasZ protein (e.g., a wild type CasZ protein, a nickase CasZ protein, a dCasZ protein, a chimeric CasZ protein and the like) is operably linked to a promoter operable in a eukaryotic cell (e.g., a CMV promoter, an EF1.alpha. promoter, an estrogen receptor-regulated promoter, and the like).
[0186] Examples of inducible promoters include, but are not limited toT7 RNA polymerase promoter, T3 RNA polymerase promoter, Isopropyl-beta-D-thiogalactopyranoside (IPTG)-regulated promoter, lactose induced promoter, heat shock promoter, Tetracycline-regulated promoter, Steroid-regulated promoter, Metal-regulated promoter, estrogen receptor-regulated promoter, etc. Inducible promoters can therefore be regulated by molecules including, but not limited to, doxycycline; estrogen and/or an estrogen analog; IPTG; etc.
[0187] Inducible promoters suitable for use include any inducible promoter described herein or known to one of ordinary skill in the art. Examples of inducible promoters include, without limitation, chemically/biochemically-regulated and physically-regulated promoters such as alcohol-regulated promoters, tetracycline-regulated promoters (e.g., anhydrotetracycline (aTc)-responsive promoters and other tetracycline-responsive promoter systems, which include a tetracycline repressor protein (tetR), a tetracycline operator sequence (tetO) and a tetracycline transactivator fusion protein (tTA)), steroid-regulated promoters (e.g., promoters based on the rat glucocorticoid receptor, human estrogen receptor, moth ecdysone receptors, and promoters from the steroid/retinoid/thyroid receptor superfamily), metal-regulated promoters (e.g., promoters derived from metallothionein (proteins that bind and sequester metal ions) genes from yeast, mouse and human), pathogenesis-regulated promoters (e.g., induced by salicylic acid, ethylene or benzothiadiazole (BTH)), temperature/heat-inducible promoters (e.g., heat shock promoters), and light-regulated promoters (e.g., light responsive promoters from plant cells).
[0188] In some cases, the promoter is a spatially restricted promoter (i.e., cell type specific promoter, tissue specific promoter, etc.) such that in a multi-cellular organism, the promoter is active (i.e., "ON") in a subset of specific cells. Spatially restricted promoters may also be referred to as enhancers, transcriptional control elements, control sequences, etc. Any convenient spatially restricted promoter may be used as long as the promoter is functional in the targeted host cell (e.g., eukaryotic cell; prokaryotic cell).
[0189] In some cases, the promoter is a reversible promoter. Suitable reversible promoters, including reversible inducible promoters are known in the art. Such reversible promoters may be isolated and derived from many organisms, e.g., eukaryotes and prokaryotes. Modification of reversible promoters derived from a first organism for use in a second organism, e.g., a first prokaryote and a second a eukaryote, a first eukaryote and a second a prokaryote, etc., is well known in the art. Such reversible promoters, and systems based on such reversible promoters but also comprising additional control proteins, include, but are not limited to, alcohol regulated promoters (e.g., alcohol dehydrogenase I (alcA) gene promoter, promoters responsive to alcohol transactivator proteins (AlcR), etc.), tetracycline regulated promoters, (e.g., promoter systems including TetActivators, TetON, TetOFF, etc.), steroid regulated promoters (e.g., rat glucocorticoid receptor promoter systems, human estrogen receptor promoter systems, retinoid promoter systems, thyroid promoter systems, ecdysone promoter systems, mifepristone promoter systems, etc.), metal regulated promoters (e.g., metallothionein promoter systems, etc.), pathogenesis-related regulated promoters (e.g., salicylic acid regulated promoters, ethylene regulated promoters, benzothiadiazole regulated promoters, etc.), temperature regulated promoters (e.g., heat shock inducible promoters (e.g., HSP-70, HSP-90, soybean heat shock promoter, etc.), light regulated promoters, synthetic inducible promoters, and the like.
[0190] Methods of introducing a nucleic acid (e.g., DNA or RNA) (e.g., a nucleic acid comprising a donor polynucleotide sequence, one or more nucleic acids encoding a CasZ protein and/or a CasZ guide RNA and/or a CasZ trancRNA, and the like) into a host cell are known in the art, and any convenient method can be used to introduce a nucleic acid (e.g., an expression construct) into a cell. Suitable methods include e.g., viral infection, transfection, lipofection, electroporation, calcium phosphate precipitation, polyethyleneimine (PEI)-mediated transfection, DEAE-dextran mediated transfection, liposome-mediated transfection, particle gun technology, calcium phosphate precipitation, direct microinjection, nanoparticle-mediated nucleic acid delivery, and the like.
[0191] Introducing the recombinant expression vector into cells can occur in any culture media and under any culture conditions that promote the survival of the cells. Introducing the recombinant expression vector into a target cell can be carried out in vivo or ex vivo. Introducing the recombinant expression vector into a target cell can be carried out in vitro.
[0192] In some cases, a CasZ protein can be provided as RNA. The RNA can be provided by direct chemical synthesis or may be transcribed in vitro from a DNA (e.g., encoding the CasZ protein). Once synthesized, the RNA may be introduced into a cell by any of the well-known techniques for introducing nucleic acids into cells (e.g., microinjection, electroporation, transfection, etc.).
[0193] Nucleic acids may be provided to the cells using well-developed transfection techniques; see, e.g. Angel and Yanik (2010) PLoS ONE 5(7): e11756, and the commercially available TransMessenger.RTM. reagents from Qiagen, Stemfect.TM. RNA Transfection Kit from Stemgent, and TransIT.RTM.-mRNA Transfection Kit from Mirus Bio LLC. See also Beumer et al. (2008) PNAS 105(50):19821-19826.
[0194] Vectors may be provided directly to a target host cell. In other words, the cells are contacted with vectors comprising the subject nucleic acids (e.g., recombinant expression vectors having the donor template sequence and encoding the CasZ guide RNA; recombinant expression vectors encoding the CasZ protein; etc.) such that the vectors are taken up by the cells. Methods for contacting cells with nucleic acid vectors that are plasmids, include electroporation, calcium chloride transfection, microinjection, and lipofection are well known in the art. For viral vector delivery, cells can be contacted with viral particles comprising the subject viral expression vectors.
[0195] Retroviruses, for example, lentiviruses, are suitable for use in methods of the present disclosure. Commonly used retroviral vectors are "defective", i.e. unable to produce viral proteins required for productive infection. Rather, replication of the vector requires growth in a packaging cell line. To generate viral particles comprising nucleic acids of interest, the retroviral nucleic acids comprising the nucleic acid are packaged into viral capsids by a packaging cell line. Different packaging cell lines provide a different envelope protein (ecotropic, amphotropic or xenotropic) to be incorporated into the capsid, this envelope protein determining the specificity of the viral particle for the cells (ecotropic for murine and rat; amphotropic for most mammalian cell types including human, dog and mouse; and xenotropic for most mammalian cell types except murine cells). The appropriate packaging cell line may be used to ensure that the cells are targeted by the packaged viral particles. Methods of introducing subject vector expression vectors into packaging cell lines and of collecting the viral particles that are generated by the packaging lines are well known in the art. Nucleic acids can also introduced by direct micro-injection (e.g., injection of RNA).
[0196] Vectors used for providing the nucleic acids encoding CasZ guide RNA and/or a CasZ polypeptide to a target host cell can include suitable promoters for driving the expression, that is, transcriptional activation, of the nucleic acid of interest. In other words, in some cases, the nucleic acid of interest will be operably linked to a promoter. This may include ubiquitously acting promoters, for example, the CMV-D-actin promoter, or inducible promoters, such as promoters that are active in particular cell populations or that respond to the presence of drugs such as tetracycline. By transcriptional activation, it is intended that transcription will be increased above basal levels in the target cell by 10 fold, by 100 fold, more usually by 1000 fold. In addition, vectors used for providing a nucleic acid encoding a CasZ guide RNA and/or a CasZ protein to a cell may include nucleic acid sequences that encode for selectable markers in the target cells, so as to identify cells that have taken up the CasZ guide RNA and/or CasZ protein.
[0197] A nucleic acid comprising a nucleotide sequence encoding a CasZ polypeptide, or a CasZ fusion polypeptide, is in some cases an RNA. Thus, a CasZ fusion protein can be introduced into cells as RNA. Methods of introducing RNA into cells are known in the art and may include, for example, direct injection, transfection, or any other method used for the introduction of DNA. A CasZ protein may instead be provided to cells as a polypeptide. Such a polypeptide may optionally be fused to a polypeptide domain that increases solubility of the product. The domain may be linked to the polypeptide through a defined protease cleavage site, e.g. a TEV sequence, which is cleaved by TEV protease. The linker may also include one or more flexible sequences, e.g. from 1 to 10 glycine residues. In some embodiments, the cleavage of the fusion protein is performed in a buffer that maintains solubility of the product, e.g. in the presence of from 0.5 to 2 M urea, in the presence of polypeptides and/or polynucleotides that increase solubility, and the like. Domains of interest include endosomolytic domains, e.g. influenza HA domain; and other polypeptides that aid in production, e.g. IF2 domain, GST domain, GRPE domain, and the like. The polypeptide may be formulated for improved stability. For example, the peptides may be PEGylated, where the polyethyleneoxy group provides for enhanced lifetime in the blood stream.
[0198] Additionally, or alternatively, a CasZ polypeptide of the present disclosure may be fused to a polypeptide permeant domain to promote uptake by the cell. A number of permeant domains are known in the art and may be used in the non-integrating polypeptides of the present disclosure, including peptides, peptidomimetics, and non-peptide carriers. For example, a permeant peptide may be derived from the third alpha helix of Drosophila melanogaster transcription factor Antennapaedia, referred to as penetratin, which comprises the amino acid sequence RQIKIWFQNRRMKWKK (SEQ ID NO: 134). As another example, the permeant peptide comprises the HIV-1 tat basic region amino acid sequence, which may include, for example, amino acids 49-57 of naturally-occurring tat protein. Other permeant domains include poly-arginine motifs, for example, the region of amino acids 34-56 of HIV-1 rev protein, nona-arginine, octa-arginine, and the like. (See, for example, Futaki et al. (2003) Curr Protein Pept Sci. 2003 April; 4(2): 87-9 and 446; and Wender et al. (2000) Proc. Natl. Acad. Sci. U.S.A 2000 Nov. 21; 97(24):13003-8; published U.S. Patent applications 20030220334; 20030083256; 20030032593; and 20030022831, herein specifically incorporated by reference for the teachings of translocation peptides and peptoids). The nona-arginine (R9) sequence is one of the more efficient PTDs that have been characterized (Wender et al. 2000; Uemura et al. 2002). The site at which the fusion is made may be selected in order to optimize the biological activity, secretion or binding characteristics of the polypeptide. The optimal site will be determined by routine experimentation.
[0199] A CasZ polypeptide of the present disclosure may be produced in vitro or by eukaryotic cells or by prokaryotic cells, and it may be further processed by unfolding, e.g. heat denaturation, dithiothreitol reduction, etc. and may be further refolded, using methods known in the art.
[0200] Modifications of interest that do not alter primary sequence include chemical derivatization of polypeptides, e.g., acylation, acetylation, carboxylation, amidation, etc. Also included are modifications of glycosylation, e.g. those made by modifying the glycosylation patterns of a polypeptide during its synthesis and processing or in further processing steps; e.g. by exposing the polypeptide to enzymes which affect glycosylation, such as mammalian glycosylating or deglycosylating enzymes. Also embraced are sequences that have phosphorylated amino acid residues, e.g. phosphotyrosine, phosphoserine, or phosphothreonine.
[0201] Also suitable for inclusion in embodiments of the present disclosure are nucleic acids (e.g., encoding a CasZ guide RNA, encoding a CasZ fusion protein, etc.) and proteins (e.g., a CasZ fusion protein derived from a wild type protein or a variant protein) that have been modified using ordinary molecular biological techniques and synthetic chemistry so as to improve their resistance to proteolytic degradation, to change the target sequence specificity, to optimize solubility properties, to alter protein activity (e.g., transcription modulatory activity, enzymatic activity, etc.) or to render them more suitable. Analogs of such polypeptides include those containing residues other than naturally occurring L-amino acids, e.g. D-amino acids or non-naturally occurring synthetic amino acids. D-amino acids may be substituted for some or all of the amino acid residues.
[0202] A CasZ polypeptide of the present disclosure may be prepared by in vitro synthesis, using conventional methods as known in the art. Various commercial synthetic apparatuses are available, for example, automated synthesizers by Applied Biosystems, Inc., Beckman, etc. By using synthesizers, naturally occurring amino acids may be substituted with unnatural amino acids. The particular sequence and the manner of preparation will be determined by convenience, economics, purity required, and the like.
[0203] If desired, various groups may be introduced into the peptide during synthesis or during expression, which allow for linking to other molecules or to a surface. Thus cysteines can be used to make thioethers, histidines for linking to a metal ion complex, carboxyl groups for forming amides or esters, amino groups for forming amides, and the like.
[0204] A CasZ polypeptide of the present disclosure may also be isolated and purified in accordance with conventional methods of recombinant synthesis. A lysate may be prepared of the expression host and the lysate purified using high performance liquid chromatography (HPLC), exclusion chromatography, gel electrophoresis, affinity chromatography, or other purification technique. For the most part, the compositions which are used will comprise 20% or more by weight of the desired product, more usually 75% or more by weight, preferably 95% or more by weight, and for therapeutic purposes, usually 99.5% or more by weight, in relation to contaminants related to the method of preparation of the product and its purification. Usually, the percentages will be based upon total protein. Thus, in some cases, a CasZ polypeptide, or a CasZ fusion polypeptide, of the present disclosure is at least 80% pure, at least 85% pure, at least 90% pure, at least 95% pure, at least 98% pure, or at least 99% pure (e.g., free of contaminants, non-CasZ proteins or other macromolecules, etc.).
[0205] To induce cleavage or any desired modification to a target nucleic acid (e.g., genomic DNA), or any desired modification to a polypeptide associated with target nucleic acid, the CasZ guide RNA and/or the CasZ polypeptide and/or the CasZ trancRNA, and/or the donor template sequence, whether they be introduced as nucleic acids or polypeptides, can be provided to the cells for about 30 minutes to about 24 hours, e.g., 1 hour, 1.5 hours, 2 hours, 2.5 hours, 3 hours, 3.5 hours 4 hours, 5 hours, 6 hours, 7 hours, 8 hours, 12 hours, 16 hours, 18 hours, 20 hours, or any other period from about 30 minutes to about 24 hours, which may be repeated with a frequency of about every day to about every 4 days, e.g., every 1.5 days, every 2 days, every 3 days, or any other frequency from about every day to about every four days. The agent(s) may be provided to the subject cells one or more times, e.g. one time, twice, three times, or more than three times, and the cells allowed to incubate with the agent(s) for some amount of time following each contacting event e.g. 16-24 hours, after which time the media is replaced with fresh media and the cells are cultured further.
[0206] In cases in which two or more different targeting complexes are provided to the cell (e.g., two different CasZ guide RNAs that are complementary to different sequences within the same or different target nucleic acid), the complexes may be provided simultaneously (e.g. as two polypeptides and/or nucleic acids), or delivered simultaneously. Alternatively, they may be provided consecutively, e.g. the targeting complex being provided first, followed by the second targeting complex, etc. or vice versa.
[0207] To improve the delivery of a DNA vector into a target cell, the DNA can be protected from damage and its entry into the cell facilitated, for example, by using lipoplexes and polyplexes. Thus, in some cases, a nucleic acid of the present disclosure (e.g., a recombinant expression vector of the present disclosure) can be covered with lipids in an organized structure like a micelle or a liposome. When the organized structure is complexed with DNA it is called a lipoplex. There are three types of lipids, anionic (negatively-charged), neutral, or cationic (positively-charged). Lipoplexes that utilize cationic lipids have proven utility for gene transfer. Cationic lipids, due to their positive charge, naturally complex with the negatively charged DNA. Also as a result of their charge, they interact with the cell membrane. Endocytosis of the lipoplex then occurs, and the DNA is released into the cytoplasm. The cationic lipids also protect against degradation of the DNA by the cell.
[0208] Complexes of polymers with DNA are called polyplexes. Most polyplexes consist of cationic polymers and their production is regulated by ionic interactions. One large difference between the methods of action of polyplexes and lipoplexes is that polyplexes cannot release their DNA load into the cytoplasm, so to this end, co-transfection with endosome-lytic agents (to lyse the endosome that is made during endocytosis) such as inactivated adenovirus must occur. However, this is not always the case; polymers such as polyethylenimine have their own method of endosome disruption as does chitosan and trimethylchitosan.
[0209] Dendrimers, a highly branched macromolecule with a spherical shape, may be also be used to genetically modify stem cells. The surface of the dendrimer particle may be functionalized to alter its properties. In particular, it is possible to construct a cationic dendrimer (i.e., one with a positive surface charge). When in the presence of genetic material such as a DNA plasmid, charge complementarity leads to a temporary association of the nucleic acid with the cationic dendrimer. On reaching its destination, the dendrimer-nucleic acid complex can be taken up into a cell by endocytosis.
[0210] In some cases, a nucleic acid of the disclosure (e.g., an expression vector) includes an insertion site for a guide sequence of interest. For example, a nucleic acid can include an insertion site for a guide sequence of interest, where the insertion site is immediately adjacent to a nucleotide sequence encoding the portion of a CasZ guide RNA that does not change when the guide sequence is changed to hybrized to a desired target sequence (e.g., sequences that contribute to the CasZ binding aspect of the guide RNA, e.g, the sequences that contribute to the dsRNA duplex(es) of the CasZ guide RNA--this portion of the guide RNA can also be referred to as the `scaffold` or `constant region` of the guide RNA). Thus, in some cases, a subject nucleic acid (e.g., an expression vector) includes a nucleotide sequence encoding a CasZ guide RNA, except that the portion encoding the guide sequence portion of the guide RNA is an insertion sequence (an insertion site). An insertion site is any nucleotide sequence used for the insertion of a desired sequence. "Insertion sites" for use with various technologies are known to those of ordinary skill in the art and any convenient insertion site can be used. An insertion site can be for any method for manipulating nucleic acid sequences. For example, in some cases the insertion site is a multiple cloning site (MCS) (e.g., a site including one or more restriction enzyme recognition sequences), a site for ligation independent cloning, a site for recombination-based cloning (e.g., recombination based on att sites), a nucleotide sequence recognized by a CRISPR/Cas (e.g. Cas9) based technology, and the like.
[0211] An insertion site can be any desirable length, and can depend on the type of insertion site (e.g., can depend on whether (and how many) the site includes one or more restriction enzyme recognition sequences, whether the site includes a target site for a CRISPR/Cas protein, etc.). In some cases, an insertion site of a subject nucleic acid is 3 or more nucleotides (nt) in length (e.g., 5 or more, 8 or more, 10 or more, 15 or more, 17 or more, 18 or more, 19 or more, 20 or more or 25 or more, or 30 or more nt in length). In some cases, the length of an insertion site of a subject nucleic acid has a length in a range of from 2 to 50 nucleotides (nt) (e.g., from 2 to 40 nt, from 2 to 30 nt, from 2 to 25 nt, from 2 to 20 nt, from 5 to 50 nt, from 5 to 40 nt, from 5 to 30 nt, from 5 to 25 nt, from 5 to 20 nt, from 10 to 50 nt, from 10 to 40 nt, from 10 to 30 nt, from 10 to 25 nt, from 10 to 20 nt, from 17 to 50 nt, from 17 to 40 nt, from 17 to 30 nt, from 17 to 25 nt). In some cases, the length of an insertion site of a subject nucleic acid has a length in a range of from 5 to 40 nt.
Nucleic Acid Modifications
[0212] In some embodiments, a subject nucleic acid (e.g., a CasZ guide RNA or trancRNA) has one or more modifications, e.g., a base modification, a backbone modification, etc., to provide the nucleic acid with a new or enhanced feature (e.g., improved stability). A nucleoside is a base-sugar combination. The base portion of the nucleoside is normally a heterocyclic base. The two most common classes of such heterocyclic bases are the purines and the pyrimidines. Nucleotides are nucleosides that further include a phosphate group covalently linked to the sugar portion of the nucleoside. For those nucleosides that include a pentofuranosyl sugar, the phosphate group can be linked to the 2', the 3', or the 5' hydroxyl moiety of the sugar. In forming oligonucleotides, the phosphate groups covalently link adjacent nucleosides to one another to form a linear polymeric compound. In turn, the respective ends of this linear polymeric compound can be further joined to form a circular compound, however, linear compounds are suitable. In addition, linear compounds may have internal nucleotide base complementarity and may therefore fold in a manner as to produce a fully or partially double-stranded compound. Within oligonucleotides, the phosphate groups are commonly referred to as forming the internucleoside backbone of the oligonucleotide. The normal linkage or backbone of RNA and DNA is a 3' to 5' phosphodiester linkage.
[0213] Suitable nucleic acid modifications include, but are not limited to: 2'Omethyl modified nucleotides, 2' Fluoro modified nucleotides, locked nucleic acid (LNA) modified nucleotides, peptide nucleic acid (PNA) modified nucleotides, nucleotides with phosphorothioate linkages, and a 5' cap (e.g., a 7-methylguanylate cap (m7G)). Additional details and additional modifications are described below.
[0214] A 2'-O-Methyl modified nucleotide (also referred to as 2'-O-Methyl RNA) is a naturally occurring modification of RNA found in tRNA and other small RNAs that arises as a post-transcriptional modification. Oligonucleotides can be directly synthesized that contain 2'-O-Methyl RNA. This modification increases Tm of RNA:RNA duplexes but results in only small changes in RNA:DNA stability. It is stabile with respect to attack by single-stranded ribonucleases and is typically 5 to 10-fold less susceptible to DNases than DNA. It is commonly used in antisense oligos as a means to increase stability and binding affinity to the target message.
[0215] 2' Fluoro modified nucleotides (e.g., 2' Fluoro bases) have a fluorine modified ribose which increases binding affinity (Tm) and also confers some relative nuclease resistance when compared to native RNA. These modifications are commonly employed in ribozymes and siRNAs to improve stability in serum or other biological fluids.
[0216] LNA bases have a modification to the ribose backbone that locks the base in the C3'-endo position, which favors RNA A-type helix duplex geometry. This modification significantly increases Tm and is also very nuclease resistant. Multiple LNA insertions can be placed in an oligo at any position except the 3'-end. Applications have been described ranging from antisense oligos to hybridization probes to SNP detection and allele specific PCR. Due to the large increase in Tm conferred by LNAs, they also can cause an increase in primer dimer formation as well as self-hairpin formation. In some cases, the number of LNAs incorporated into a single oligo is 10 bases or less.
[0217] The phosphorothioate (PS) bond (i.e., a phosphorothioate linkage) substitutes a sulfur atom for a non-bridging oxygen in the phosphate backbone of a nucleic acid (e.g., an oligo). This modification renders the internucleotide linkage resistant to nuclease degradation. Phosphorothioate bonds can be introduced between the last 3-5 nucleotides at the 5'- or 3'-end of the oligo to inhibit exonuclease degradation. Including phosphorothioate bonds within the oligo (e.g., throughout the entire oligo) can help reduce attack by endonucleases as well.
[0218] In some embodiments, a subject nucleic acid has one or more nucleotides that are 2'-O-Methyl modified nucleotides. In some embodiments, a subject nucleic acid (e.g., a guide RNA, a tranc RNA, etc.) has one or more 2' Fluoro modified nucleotides. In some embodiments, a subject nucleic acid (e.g., a dsRNA, a siNA, etc.) has one or more LNA bases. In some embodiments, a subject nucleic acid (e.g., a dsRNA, a siNA, etc.) has one or more nucleotides that are linked by a phosphorothioate bond (i.e., the subject nucleic acid has one or more phosphorothioate linkages). In some embodiments, a subject nucleic acid (e.g., a dsRNA, a siNA, etc.) has a 5' cap (e.g., a 7-methylguanylate cap (m7G)). In some embodiments, a subject nucleic acid (e.g., a guide RNA, a tranc RNA, etc.) has a combination of modified nucleotides. For example, a subject nucleic acid (e.g., a guide RNA, a tranc RNA, etc.) can have a 5' cap (e.g., a 7-methylguanylate cap (m7G)) in addition to having one or more nucleotides with other modifications (e.g., a 2'-O-Methyl nucleotide and/or a 2' Fluoro modified nucleotide and/or a LNA base and/or a phosphorothioate linkage).
Modified Backbones and Modified Internucleoside Linkages
[0219] Examples of suitable nucleic acids (e.g., a CasZ guide RNA and/or CasZ trancRNA) containing modifications include nucleic acids containing modified backbones or non-natural internucleoside linkages. Nucleic acids having modified backbones include those that retain a phosphorus atom in the backbone and those that do not have a phosphorus atom in the backbone.
[0220] Suitable modified oligonucleotide backbones containing a phosphorus atom therein include, for example, phosphorothioates, chiral phosphorothioates, phosphorodithioates, phosphotriesters, aminoalkylphosphotriesters, methyl and other alkyl phosphonates including 3'-alkylene phosphonates, 5'-alkylene phosphonates and chiral phosphonates, phosphinates, phosphoramidates including 3'-amino phosphoramidate and aminoalkylphosphoramidates, phosphorodiamidates, thionophosphoramidates, thionoalkylphosphonates, thionoalkylphosphotriesters, selenophosphates and boranophosphates having normal 3'-5' linkages, 2'-5' linked analogs of these, and those having inverted polarity wherein one or more internucleotide linkages is a 3' to 3', 5' to 5' or 2' to 2' linkage. Suitable oligonucleotides having inverted polarity comprise a single 3' to 3' linkage at the 3'-most internucleotide linkage i.e. a single inverted nucleoside residue which may be a basic (the nucleobase is missing or has a hydroxyl group in place thereof). Various salts (such as, for example, potassium or sodium), mixed salts and free acid forms are also included.
[0221] In some embodiments, a subject nucleic acid comprises one or more phosphorothioate and/or heteroatom internucleoside linkages, in particular --CH.sub.2--NH--O--CH.sub.2--, --CH.sub.2--N(CH.sub.3)--O--CH.sub.2-- (known as a methylene (methylimino) or MMI backbone), --CH.sub.2--O--N(CH.sub.3)--CH.sub.2--, --CH.sub.2--N(CH.sub.3)--N(CH.sub.3)--CH.sub.2-- and --O--N(CH.sub.3)--CH.sub.2--CH.sub.2-- (wherein the native phosphodiester internucleotide linkage is represented as --O--P(.dbd.O)(OH)--O--CH.sub.2--). MMI type internucleoside linkages are disclosed in the above referenced U.S. Pat. No. 5,489,677, the disclosure of which is incorporated herein by reference in its entirety. Suitable amide internucleoside linkages are disclosed in U.S. Pat. No. 5,602,240, the disclosure of which is incorporated herein by reference in its entirety.
[0222] Also suitable are nucleic acids having morpholino backbone structures as described in, e.g., U.S. Pat. No. 5,034,506. For example, in some embodiments, a subject nucleic acid comprises a 6-membered morpholino ring in place of a ribose ring. In some of these embodiments, a phosphorodiamidate or other non-phosphodiester internucleoside linkage replaces a phosphodiester linkage.
[0223] Suitable modified polynucleotide backbones that do not include a phosphorus atom therein have backbones that are formed by short chain alkyl or cycloalkyl internucleoside linkages, mixed heteroatom and alkyl or cycloalkyl internucleoside linkages, or one or more short chain heteroatomic or heterocyclic internucleoside linkages. These include those having morpholino linkages (formed in part from the sugar portion of a nucleoside); siloxane backbones; sulfide, sulfoxide and sulfone backbones; formacetyl and thioformacetyl backbones; methylene formacetyl and thioformacetyl backbones; riboacetyl backbones; alkene containing backbones; sulfamate backbones; methyleneimino and methylenehydrazino backbones; sulfonate and sulfonamide backbones; amide backbones; and others having mixed N, O, S and CH.sub.2 component parts.
Mimetics
[0224] A subject nucleic acid can be a nucleic acid mimetic. The term "mimetic" as it is applied to polynucleotides is intended to include polynucleotides wherein only the furanose ring or both the furanose ring and the internucleotide linkage are replaced with non-furanose groups, replacement of only the furanose ring is also referred to in the art as being a sugar surrogate. The heterocyclic base moiety or a modified heterocyclic base moiety is maintained for hybridization with an appropriate target nucleic acid. One such nucleic acid, a polynucleotide mimetic that has been shown to have excellent hybridization properties, is referred to as a peptide nucleic acid (PNA). In PNA, the sugar-backbone of a polynucleotide is replaced with an amide containing backbone, in particular an aminoethylglycine backbone. The nucleotides are retained and are bound directly or indirectly to aza nitrogen atoms of the amide portion of the backbone.
[0225] One polynucleotide mimetic that has been reported to have excellent hybridization properties is a peptide nucleic acid (PNA). The backbone in PNA compounds is two or more linked aminoethylglycine units which gives PNA an amide containing backbone. The heterocyclic base moieties are bound directly or indirectly to aza nitrogen atoms of the amide portion of the backbone. Representative U.S. patents that describe the preparation of PNA compounds include, but are not limited to: U.S. Pat. Nos. 5,539,082; 5,714,331; and 5,719,262, the disclosures of which are incorporated herein by reference in their entirety.
[0226] Another class of polynucleotide mimetic that has been studied is based on linked morpholino units (morpholino nucleic acid) having heterocyclic bases attached to the morpholino ring. A number of linking groups have been reported that link the morpholino monomeric units in a morpholino nucleic acid. One class of linking groups has been selected to give a non-ionic oligomeric compound. The non-ionic morpholino-based oligomeric compounds are less likely to have undesired interactions with cellular proteins. Morpholino-based polynucleotides are non-ionic mimics of oligonucleotides which are less likely to form undesired interactions with cellular proteins (Dwaine A. Braasch and David R. Corey, Biochemistry, 2002, 41(14), 4503-4510). Morpholino-based polynucleotides are disclosed in U.S. Pat. No. 5,034,506, the disclosure of which is incorporated herein by reference in its entirety. A variety of compounds within the morpholino class of polynucleotides have been prepared, having a variety of different linking groups joining the monomeric subunits.
[0227] A further class of polynucleotide mimetic is referred to as cyclohexenyl nucleic acids (CeNA). The furanose ring normally present in a DNA/RNA molecule is replaced with a cyclohexenyl ring. CeNA DMT protected phosphoramidite monomers have been prepared and used for oligomeric compound synthesis following classical phosphoramidite chemistry. Fully modified CeNA oligomeric compounds and oligonucleotides having specific positions modified with CeNA have been prepared and studied (see Wang et al., J. Am. Chem. Soc., 2000, 122, 8595-8602, the disclosure of which is incorporated herein by reference in its entirety). In general, the incorporation of CeNA monomers into a DNA chain increases its stability of a DNA/RNA hybrid. CeNA oligoadenylates formed complexes with RNA and DNA complements with similar stability to the native complexes. The study of incorporating CeNA structures into natural nucleic acid structures was shown by NMR and circular dichroism to proceed with easy conformational adaptation.
[0228] A further modification includes Locked Nucleic Acids (LNAs) in which the 2'-hydroxyl group is linked to the 4' carbon atom of the sugar ring thereby forming a 2'-C,4'-C-oxymethylene linkage thereby forming a bicyclic sugar moiety. The linkage can be a methylene (--CH.sub.2--), group bridging the 2' oxygen atom and the 4' carbon atom wherein n is 1 or 2 (Singh et al., Chem. Commun., 1998, 4, 455-456, the disclosure of which is incorporated herein by reference in its entirety). LNA and LNA analogs display very high duplex thermal stabilities with complementary DNA and RNA (Tm=+3 to +10.degree. C.), stability towards 3'-exonucleolytic degradation and good solubility properties. Potent and nontoxic antisense oligonucleotides containing LNAs have been described (e.g., Wahlestedt et al., Proc. Natl. Acad. Sci. U.S.A., 2000, 97, 5633-5638, the disclosure of which is incorporated herein by reference in its entirety).
[0229] The synthesis and preparation of the LNA monomers adenine, cytosine, guanine, 5-methyl-cytosine, thymine and uracil, along with their oligomerization, and nucleic acid recognition properties have been described (e.g., Koshkin et al., Tetrahedron, 1998, 54, 3607-3630, the disclosure of which is incorporated herein by reference in its entirety). LNAs and preparation thereof are also described in WO 98/39352 and WO 99/14226, as well as U.S. applications 20120165514, 20100216983, 20090041809, 20060117410, 20040014959, 20020094555, and 20020086998, the disclosures of which are incorporated herein by reference in their entirety.
Modified Sugar Moieties
[0230] A subject nucleic acid can also include one or more substituted sugar moieties. Suitable polynucleotides comprise a sugar substituent group selected from: OH; F; O-, S-, or N-alkyl; O-, S-, or N-alkenyl; O-, S- or N-alkynyl; or O-alkyl-O-alkyl, wherein the alkyl, alkenyl and alkynyl may be substituted or unsubstituted C.sub.1 to C.sub.10 alkyl or C.sub.2 to C.sub.10 alkenyl and alkynyl. Particularly suitable are O((CH.sub.2).sub.nO).sub.mCH.sub.3, O(CH.sub.2).sub.nOCH.sub.3, O(CH.sub.2).sub.nNH.sub.2, O(CH.sub.2).sub.nCH.sub.3, O(CH.sub.2).sub.nONH.sub.2, and O(CH.sub.2).sub.nON((CH.sub.2).sub.nCH.sub.3).sub.2, where n and m are from 1 to about 10. Other suitable polynucleotides comprise a sugar substituent group selected from: C.sub.1 to C.sub.10 lower alkyl, substituted lower alkyl, alkenyl, alkynyl, alkaryl, aralkyl, O-alkaryl or O-aralkyl, SH, SCH.sub.3, OCN, Cl, Br, CN, CF.sub.3, OCF.sub.3, SOCH.sub.3, SO.sub.2CH.sub.3, ONO.sub.2, NO.sub.2, N.sub.3, NH.sub.2, heterocycloalkyl, heterocycloalkaryl, aminoalkylamino, polyalkylamino, substituted silyl, an RNA cleaving group, a reporter group, an intercalator, a group for improving the pharmacokinetic properties of an oligonucleotide, or a group for improving the pharmacodynamic properties of an oligonucleotide, and other substituents having similar properties. A suitable modification includes 2'-methoxyethoxy (2'-O--CH.sub.2 CH.sub.2OCH.sub.3, also known as 2'-O-(2-methoxyethyl) or 2'-MOE) (Martin et al., Helv. Chim. Acta, 1995, 78, 486-504, the disclosure of which is incorporated herein by reference in its entirety) i.e., an alkoxyalkoxy group. A further suitable modification includes 2'-dimethylaminooxyethoxy, i.e., a O(CH.sub.2).sub.2ON(CH.sub.3).sub.2 group, also known as 2'-DMAOE, as described in examples hereinbelow, and 2'-dimethylaminoethoxyethoxy (also known in the art as 2'-O-dimethyl-amino-ethoxy-ethyl or 2'-DMAEOE), i.e., 2'-O--CH.sub.2--O--CH.sub.2--N(CH.sub.3).sub.2.
[0231] Other suitable sugar substituent groups include methoxy (--O--CH.sub.3), aminopropoxy (--OCH.sub.2 CH.sub.2 CH.sub.2NH.sub.2), allyl (--CH.sub.2--CH.dbd.CH.sub.2), --O-allyl (--O--CH.sub.2--CH.dbd.CH.sub.2) and fluoro (F). 2'-sugar substituent groups may be in the arabino (up) position or ribo (down) position. A suitable 2'-arabino modification is 2'-F. Similar modifications may also be made at other positions on the oligomeric compound, particularly the 3' position of the sugar on the 3' terminal nucleoside or in 2'-5' linked oligonucleotides and the 5' position of 5' terminal nucleotide. Oligomeric compounds may also have sugar mimetics such as cyclobutyl moieties in place of the pentofuranosyl sugar.
Base Modifications and Substitutions
[0232] A subject nucleic acid may also include nucleobase (often referred to in the art simply as "base") modifications or substitutions. As used herein, "unmodified" or "natural" nucleobases include the purine bases adenine (A) and guanine (G), and the pyrimidine bases thymine (T), cytosine (C) and uracil (U). Modified nucleobases include other synthetic and natural nucleobases such as 5-methylcytosine (5-me-C), 5-hydroxymethyl cytosine, xanthine, hypoxanthine, 2-aminoadenine, 6-methyl and other alkyl derivatives of adenine and guanine, 2-propyl and other alkyl derivatives of adenine and guanine, 2-thiouracil, 2-thiothymine and 2-thiocytosine, 5-halouracil and cytosine, 5-propynyl (--C.dbd.C-CH.sub.3) uracil and cytosine and other alkynyl derivatives of pyrimidine bases, 6-azo uracil, cytosine and thymine, 5-uracil (pseudouracil), 4-thiouracil, 8-halo, 8-amino, 8-thiol, 8-thioalkyl, 8-hydroxyl and other 8-substituted adenines and guanines, 5-halo particularly 5-bromo, 5-trifluoromethyl and other 5-substituted uracils and cytosines, 7-methylguanine and 7-methyladenine, 2-F-adenine, 2-amino-adenine, 8-azaguanine and 8-azaadenine, 7-deazaguanine and 7-deazaadenine and 3-deazaguanine and 3-deazaadenine. Further modified nucleobases include tricyclic pyrimidines such as phenoxazine cytidine(1H-pyrimido(5,4-b)(1,4)benzoxazin-2(3H)-one), phenothiazine cytidine (1H-pyrimido(5,4-b)(1,4)benzothiazin-2(3H)-one), G-clamps such as a substituted phenoxazine cytidine (e.g. 9-(2-aminoethoxy)-H-pyrimido(5,4-(b) (1,4)benzoxazin-2(3H)-one), carbazole cytidine (2H-pyrimido(4,5-b)indol-2-one), pyridoindole cytidine (H-pyrido(3',2':4,5)pyrrolo(2,3-d)pyrimidin-2-one).
[0233] Heterocyclic base moieties may also include those in which the purine or pyrimidine base is replaced with other heterocycles, for example 7-deaza-adenine, 7-deazaguanosine, 2-aminopyridine and 2-pyridone. Further nucleobases include those disclosed in U.S. Pat. No. 3,687,808, those disclosed in The Concise Encyclopedia Of Polymer Science And Engineering, pages 858-859, Kroschwitz, J. I., ed. John Wiley & Sons, 1990, those disclosed by Englisch et al., Angewandte Chemie, International Edition, 1991, 30, 613, and those disclosed by Sanghvi, Y. S., Chapter 15, Antisense Research and Applications, pages 289-302, Crooke, S. T. and Lebleu, B., ed., CRC Press, 1993; the disclosures of which are incorporated herein by reference in their entirety. Certain of these nucleobases are useful for increasing the binding affinity of an oligomeric compound. These include 5-substituted pyrimidines, 6-azapyrimidines and N-2, N-6 and O-6 substituted purines, including 2-aminopropyladenine, 5-propynyluracil and 5-propynylcytosine. 5-methylcytosine substitutions have been shown to increase nucleic acid duplex stability by 0.6-1.2.degree. C. (Sanghvi et al., eds., Antisense Research and Applications, CRC Press, Boca Raton, 1993, pp. 276-278; the disclosure of which is incorporated herein by reference in its entirety) and are suitable base substitutions, e.g., when combined with 2'-O-methoxyethyl sugar modifications.
Conjugates
[0234] Another possible modification of a subject nucleic acid involves chemically linking to the polynucleotide one or more moieties or conjugates which enhance the activity, cellular distribution or cellular uptake of the oligonucleotide. These moieties or conjugates can include conjugate groups covalently bound to functional groups such as primary or secondary hydroxyl groups. Conjugate groups include, but are not limited to, intercalators, reporter molecules, polyamines, polyamides, polyethylene glycols, polyethers, groups that enhance the pharmacodynamic properties of oligomers, and groups that enhance the pharmacokinetic properties of oligomers. Suitable conjugate groups include, but are not limited to, cholesterols, lipids, phospholipids, biotin, phenazine, folate, phenanthridine, anthraquinone, acridine, fluoresceins, rhodamines, coumarins, and dyes. Groups that enhance the pharmacodynamic properties include groups that improve uptake, enhance resistance to degradation, and/or strengthen sequence-specific hybridization with the target nucleic acid. Groups that enhance the pharmacokinetic properties include groups that improve uptake, distribution, metabolism or excretion of a subject nucleic acid.
[0235] Conjugate moieties include but are not limited to lipid moieties such as a cholesterol moiety (Letsinger et al., Proc. Natl. Acad. Sci. USA, 1989, 86, 6553-6556), cholic acid (Manoharan et al., Bioorg. Med. Chem. Let., 1994, 4, 1053-1060), a thioether, e.g., hexyl-S-tritylthiol (Manoharan et al., Ann. N.Y. Acad. Sci., 1992, 660, 306-309; Manoharan et al., Bioorg. Med. Chem. Let., 1993, 3, 2765-2770), a thiocholesterol (Oberhauser et al., Nucl. Acids Res., 1992, 20, 533-538), an aliphatic chain, e.g., dodecandiol or undecyl residues (Saison-Behmoaras et al., EMBO J., 1991, 10, 1111-1118; Kabanov et al., FEBS Lett., 1990, 259, 327-330; Svinarchuk et al., Biochimie, 1993, 75, 49-54), a phospholipid, e.g., di-hexadecyl-rac-glycerol or triethylammonium 1,2-di-O-hexadecyl-rac-glycero-3-H-phosphonate (Manoharan et al., Tetrahedron Lett., 1995, 36, 3651-3654; Shea et al., Nucl. Acids Res., 1990, 18, 3777-3783), a polyamine or a polyethylene glycol chain (Manoharan et al., Nucleosides & Nucleotides, 1995, 14, 969-973), or adamantane acetic acid (Manoharan et al., Tetrahedron Lett., 1995, 36, 3651-3654), a palmityl moiety (Mishra et al., Biochim. Biophys. Acta, 1995, 1264, 229-237), or an octadecylamine or hexylamino-carbonyl-oxycholesterol moiety (Crooke et al., J. Pharmacol. Exp. Ther., 1996, 277, 923-937).
[0236] A conjugate may include a "Protein Transduction Domain" or PTD (also known as a CPP--cell penetrating peptide), which may refer to a polypeptide, polynucleotide, carbohydrate, or organic or inorganic compound that facilitates traversing a lipid bilayer, micelle, cell membrane, organelle membrane, or vesicle membrane. A PTD attached to another molecule, which can range from a small polar molecule to a large macromolecule and/or a nanoparticle, facilitates the molecule traversing a membrane, for example going from extracellular space to intracellular space, or cytosol to within an organelle (e.g., the nucleus). In some cases, a PTD is covalently linked to the 3' end of an exogenous polynucleotide. In some cases, a PTD is covalently linked to the 5' end of an exogenous polynucleotide. Exemplary PTDs include but are not limited to a minimal undecapeptide protein transduction domain (corresponding to residues 47-57 of HIV-1 TAT comprising YGRKKRRQRRR; SEQ ID NO: 130); a polyarginine sequence comprising a number of arginines sufficient to direct entry into a cell (e.g., 3, 4, 5, 6, 7, 8, 9, 10, or 10-50 arginines); a VP22 domain (Zender et al. (2002) Cancer Gene Ther. 9(6):489-96); an Drosophila Antennapedia protein transduction domain (Noguchi et al. (2003) Diabetes 52(7):1732-1737); a truncated human calcitonin peptide (Trehin et al. (2004) Pharm. Research 21:1248-1256); polylysine (Wender et al. (2000) Proc. Natl. Acad. Sci. USA 97:13003-13008); RRQRRTSKLMKR SEQ ID NO: 131); Transportan GWTLNSAGYLLGKINLKALAALAKKIL SEQ ID NO: 132); KALAWEAKLAKALAKALAKHLAKALAKALKCEA SEQ ID NO: 133); and RQIKIWFQNRRMKWKK SEQ ID NO: 134). Exemplary PTDs include but are not limited to, YGRKKRRQRRR SEQ ID NO: 130), RKKRRQRRR SEQ ID NO: 135); an arginine homopolymer of from 3 arginine residues to 50 arginine residues; Exemplary PTD domain amino acid sequences include, but are not limited to, any of the following: YGRKKRRQRRR SEQ ID NO: 130); RKKRRQRR SEQ ID NO: 136); YARAAARQARA SEQ ID NO: 137); THRLPRRRRRR SEQ ID NO: 138); and GGRRARRRRRR SEQ ID NO: 139). In some cases, the PTD is an activatable CPP (ACPP) (Aguilera et al. (2009) Integr Biol (Camb) June; 1(5-6): 371-381). ACPPs comprise a polycationic CPP (e.g., Arg9 or "R9") connected via a cleavable linker to a matching polyanion (e.g., Glu9 or "E9"), which reduces the net charge to nearly zero and thereby inhibits adhesion and uptake into cells. Upon cleavage of the linker, the polyanion is released, locally unmasking the polyarginine and its inherent adhesiveness, thus "activating" the ACPP to traverse the membrane.
Introducing Components into a Target Cell
[0237] A CasZ guide RNA (or a nucleic acid comprising a nucleotide sequence encoding same) and/or a CasZ polypeptide (or a nucleic acid comprising a nucleotide sequence encoding same) and/or a CasZ trancRNA (or a nucleic acid that includes a nucleotide sequence encoding same) and/or a donor polynucleotide (donor template) can be introduced into a host cell by any of a variety of well-known methods.
[0238] Any of a variety of compounds and methods can be used to deliver to a target cell a CasZ system of the present disclosure. As a non-limiting example, a CasZ system of the present disclosure can be combined with a lipid. As another non-limiting example, a CasZ system of the present disclosure can be combined with a particle, or formulated into a particle.
[0239] Methods of introducing a nucleic acid into a host cell are known in the art, and any convenient method can be used to introduce a subject nucleic acid (e.g., an expression construct/vector) into a target cell (e.g., prokaryotic cell, eukaryotic cell, plant cell, animal cell, mammalian cell, human cell, and the like). Suitable methods include, e.g., viral infection, transfection, conjugation, protoplast fusion, lipofection, electroporation, calcium phosphate precipitation, polyethyleneimine (PEI)-mediated transfection, DEAE-dextran mediated transfection, liposome-mediated transfection, particle gun technology, calcium phosphate precipitation, direct micro injection, nanoparticle-mediated nucleic acid delivery (see, e.g., Panyam et., al Adv Drug Deliv Rev. 2012 Sep. 13. pii: S0169-409X(12)00283-9. doi: 10.1016/j.addr.2012.09.023), and the like.
[0240] In some cases, a CasZ polypeptide of the present disclosure (e.g., wild type protein, variant protein, chimeric/fusion protein, dCasZ, etc.) is provided as a nucleic acid (e.g., an mRNA, a DNA, a plasmid, an expression vector, a viral vector, etc.) that encodes the CasZ polypeptide. In some cases, the CasZ polypeptide of the present disclosure is provided directly as a protein (e.g., without an associated guide RNA or with an associate guide RNA, i.e., as a ribonucleoprotein complex). A CasZ polypeptide of the present disclosure can be introduced into a cell (provided to the cell) by any convenient method; such methods are known to those of ordinary skill in the art. As an illustrative example, a CasZ polypeptide of the present disclosure can be injected directly into a cell (e.g., with or without a CasZ guide RNA or nucleic acid encoding a CasZ guide RNA, and with or without a donor polynucleotide and with or without a CasZ trancRNA). As another example, a preformed complex of a CasZ polypeptide of the present disclosure and a CasZ guide RNA (an RNP) can be introduced into a cell (e.g, eukaryotic cell) (e.g., via injection, via nucleofection; via a protein transduction domain (PTD) conjugated to one or more components, e.g., conjugated to the CasZ protein, conjugated to a guide RNA, conjugated to a CasZ trancRNA, conjugated to a CasZ polypeptide of the present disclosure and a guide RNA; etc.).
[0241] In some cases, a nucleic acid (e.g., a CasZ guide RNA and/or a nucleic acid encoding it, a nucleic acid encoding a CasZ protein, a CasZ trancRNA and/or a nucleic acid encoding it, and the like) and/or a polypeptide (e.g., a CasZ polypeptide; a CasZ fusion polypeptide) is delivered to a cell (e.g., a target host cell) in a particle, or associated with a particle. In some cases, a CasZ system of the present disclosure is delivered to a cell in a particle, or associated with a particle. The terms "particle" and nanoparticle" can be used interchangeable, as appropriate. For example, a recombinant expression vector comprising a nucleotide sequence encoding a CasZ polypeptide of the present disclosure and/or a CasZ guide RNA, an mRNA comprising a nucleotide sequence encoding a CasZ polypeptide of the present disclosure, and guide RNA may be delivered simultaneously using particles or lipid envelopes; for instance, a CasZ polypeptide and/or a CasZ guide RNA and/or a trancRNA, e.g., as a complex (e.g., a ribonucleoprotein (RNP) complex), can be delivered via a particle, e.g., a delivery particle comprising lipid or lipidoid and hydrophilic polymer, e.g., a cationic lipid and a hydrophilic polymer, for instance wherein the cationic lipid comprises 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) or 1,2-ditetradecanoyl-sn-glycero-3-phosphocholine (DMPC) and/or wherein the hydrophilic polymer comprises ethylene glycol or polyethylene glycol (PEG); and/or wherein the particle further comprises cholesterol (e.g., particle from formulation 1=DOTAP 100, DMPC 0, PEG 0, Cholesterol 0; formulation number 2=DOTAP 90, DMPC 0, PEG 10, Cholesterol 0; formulation number 3=DOTAP 90, DMPC 0, PEG 5, Cholesterol 5). For example, a particle can be formed using a multistep process in which a CasZ polypepide and a CasZ guideRNA are mixed together, e.g., at a 1:1 molar ratio, e.g., at room temperature, e.g., for 30 minutes, e.g., in sterile, nuclease free 1.times. phosphate-buffered saline (PBS); and separately, DOTAP, DMPC, PEG, and cholesterol as applicable for the formulation are dissolved in alcohol, e.g., 100% ethanol; and, the two solutions are mixed together to form particles containing the complexes).
[0242] A CasZ polypeptide of the present disclosure (or an mRNA comprising a nucleotide sequence encoding a CasZ polypeptide of the present disclosure; or a recombinant expression vector comprising a nucleotide sequence encoding a CasZ polypeptide of the present disclosure) and/or CasZ guide RNA (or a nucleic acid such as one or more expression vectors encoding the CasZ guide RNA) may be delivered simultaneously using particles or lipid envelopes. For example, a biodegradable core-shell structured nanoparticle with a poly (.beta.-amino ester) (PBAE) core enveloped by a phospholipid bilayer shell can be used. In some cases, particles/nanoparticles based on self assembling bioadhesive polymers are used; such particles/nanoparticles may be applied to oral delivery of peptides, intravenous delivery of peptides and nasal delivery of peptides, e.g., to the brain. Other embodiments, such as oral absorption and ocular delivery of hydrophobic drugs are also contemplated. A molecular envelope technology, which involves an engineered polymer envelope which is protected and delivered to the site of the disease, can be used. Doses of about 5 mg/kg can be used, with single or multiple doses, depending on various factors, e.g., the target tissue.
[0243] Lipidoid compounds (e.g., as described in US patent application 20110293703) are also useful in the administration of polynucleotides, and can be used to deliver a CasZ polypeptide of the present disclosure, a CasZ fusion polypeptide of the present disclosure, an RNP of the present disclosure, a nucleic acid of the present disclosure, or a CasZ system of the present disclosure. In one aspect, the aminoalcohol lipidoid compounds are combined with an agent to be delivered to a cell or a subject to form microparticles, nanoparticles, liposomes, or micelles. The aminoalcohol lipidoid compounds may be combined with other aminoalcohol lipidoid compounds, polymers (synthetic or natural), surfactants, cholesterol, carbohydrates, proteins, lipids, etc. to form the particles. These particles may then optionally be combined with a pharmaceutical excipient to form a pharmaceutical composition.
[0244] A poly(beta-amino alcohol) (PBAA) can be used to deliver a CasZ polypeptide of the present disclosure, a CasZ fusion polypeptide of the present disclosure, an RNP of the present disclosure, a nucleic acid of the present disclosure (e.g., a CasZ guide RNA and/or a CasZ trancRNA), or a CasZ system of the present disclosure, to a target cell. US Patent Publication No. 20130302401 relates to a class of poly(beta-amino alcohols) (PBAAs) that has been prepared using combinatorial polymerization.
[0245] Sugar-based particles may be used, for example GalNAc, as described with reference to WO2014118272 (incorporated herein by reference) and Nair, J K et al., 2014, Journal of the American Chemical Society 136 (49), 16958-16961) can be used to deliver a CasZ polypeptide of the present disclosure, a CasZ fusion polypeptide of the present disclosure, an RNP of the present disclosure, a nucleic acid of the present disclosure, or a CasZ system of the present disclosure, to a target cell.
[0246] In some cases, lipid nanoparticles (LNPs) are used to deliver a CasZ polypeptide of the present disclosure, a CasZ fusion polypeptide of the present disclosure, an RNP of the present disclosure, a nucleic acid of the present disclosure (e.g., a CasZ guide RNA and/or a CasZ trancRNA), or a CasZ system of the present disclosure, to a target cell. Negatively charged polymers such as RNA may be loaded into LNPs at low pH values (e.g., pH 4) where the ionizable lipids display a positive charge. However, at physiological pH values, the LNPs exhibit a low surface charge compatible with longer circulation times. Four species of ionizable cationic lipids have been focused upon, namely 1,2-dilineoyl-3-dimethylammonium-propane (DLinDAP), 1,2-dilinoleyloxy-3-N,N-dimethylaminopropane (DLinDMA), 1,2-dilinoleyloxy-keto-N,N-dimethyl-3-aminopropane (DLinKDMA), and 1,2-dilinoleyl-4-(2-dimethylaminoethyl)-[1,3]-dioxolane (DLinKC2-DMA). Preparation of LNPs and is described in, e.g., Rosin et al. (2011) Molecular Therapy 19:1286-2200). The cationic lipids 1,2-dilineoyl-3-dimethylammonium-propane (DLinDAP), 1,2-dilinoleyloxy-3-N,N-dimethylaminopropane (DLinDMA), 1,2-dilinoleyloxyketo-N,N-dimethyl-3-aminopropane (DLinK-DMA), 1,2-dilinoleyl-4-(2-dimethylaminoethyl)-[1,3]-dioxolane (DLinKC2-DMA), (3-o-[2''-(methoxypolyethyleneglycol 2000) succinoyl]-1,2-dimyristoyl-sn-glycol (PEG-S-DMG), and R-3-[(.omega.-methoxy-poly(ethylene glycol)2000) carbamoyl]-1,2-dimyristyloxlpropyl-3-amine (PEG-C-DOMG) may be used. A nucleic acid (e.g., a CasZ guide RNA; a nucleic acid of the present disclosure; etc.) may be encapsulated in LNPs containing DLinDAP, DLinDMA, DLinK-DMA, and DLinKC2-DMA (cationic lipid:DSPC:CHOL: PEGS-DMG or PEG-C-DOMG at 40:10:40:10 molar ratios). In some cases, 0.2% SP-DiOC18 is incorporated.
[0247] Spherical Nucleic Acid (SNA.TM.) constructs and other nanoparticles (particularly gold nanoparticles) can be used to deliver a CasZ polypeptide of the present disclosure, a CasZ fusion polypeptide of the present disclosure, an RNP of the present disclosure, a nucleic acid of the present disclosure (e.g., a CasZ guide RNA and/or a CasZ trancRNA), or a CasZ system of the present disclosure, to a target cell. See, e.g., Cutler et al., J. Am. Chem. Soc. 2011 133:9254-9257, Hao et al., Small. 2011 7:3158-3162, Zhang et al., ACS Nano. 2011 5:6962-6970, Cutler et al., J. Am. Chem. Soc. 2012 134:1376-1391, Young et al., Nano Lett. 2012 12:3867-71, Zheng et al., Proc. Natl. Acad. Sci. USA. 2012 109:11975-80, Mirkin, Nanomedicine 2012 7:635-638 Zhang et al., J. Am. Chem. Soc. 2012 134:16488-1691, Weintraub, Nature 2013 495:S14-S16, Choi et al., Proc. Natl. Acad. Sci. USA. 2013 110(19): 7625-7630, Jensen et al., Sci. Transl. Med. 5, 209ra152 (2013) and Mirkin, et al., Small, 10:186-192.
[0248] Self-assembling nanoparticles with RNA may be constructed with polyethyleneimine (PEI) that is PEGylated with an Arg-Gly-Asp (RGD) peptide ligand attached at the distal end of the polyethylene glycol (PEG).
[0249] In general, a "nanoparticle" refers to any particle having a diameter of less than 1000 nm. In some cases, nanoparticles suitable for use in delivering a CasZ polypeptide of the present disclosure, a CasZ fusion polypeptide of the present disclosure, an RNP of the present disclosure, a nucleic acid of the present disclosure (e.g., a CasZ guide RNA and/or a CasZ trancRNA), or a CasZ system of the present disclosure, to a target cell have a diameter of 500 nm or less, e.g., from 25 nm to 35 nm, from 35 nm to 50 nm, from 50 nm to 75 nm, from 75 nm to 100 nm, from 100 nm to 150 nm, from 150 nm to 200 nm, from 200 nm to 300 nm, from 300 nm to 400 nm, or from 400 nm to 500 nm. In some cases, nanoparticles suitable for use in delivering a CasZ polypeptide of the present disclosure, a CasZ fusion polypeptide of the present disclosure, an RNP of the present disclosure, a nucleic acid of the present disclosure, or a CasZ system of the present disclosure, to a target cell have a diameter of from 25 nm to 200 nm. In some cases, nanoparticles suitable for use in delivering a CasZ polypeptide of the present disclosure, a CasZ fusion polypeptide of the present disclosure, an RNP of the present disclosure, a nucleic acid of the present disclosure, or a CasZ system of the present disclosure, to a target cell have a diameter of 100 nm or less In some cases, nanoparticles suitable for use in delivering a CasZ polypeptide of the present disclosure, a CasZ fusion polypeptide of the present disclosure, an RNP of the present disclosure, a nucleic acid of the present disclosure, or a CasZ system of the present disclosure, to a target cell have a diameter of from 35 nm to 60 nm.
[0250] Nanoparticles suitable for use in delivering a CasZ polypeptide of the present disclosure, a CasZ fusion polypeptide of the present disclosure, an RNP of the present disclosure, a nucleic acid of the present disclosure (e.g., a CasZ guide RNA and/or a CasZ trancRNA), or a CasZ system of the present disclosure, to a target cell may be provided in different forms, e.g., as solid nanoparticles (e.g., metal such as silver, gold, iron, titanium), non-metal, lipid-based solids, polymers), suspensions of nanoparticles, or combinations thereof. Metal, dielectric, and semiconductor nanoparticles may be prepared, as well as hybrid structures (e.g., core-shell nanoparticles). Nanoparticles made of semiconducting material may also be labeled quantum dots if they are small enough (typically below 10 nm) that quantization of electronic energy levels occurs. Such nanoscale particles are used in biomedical applications as drug carriers or imaging agents and may be adapted for similar purposes in the present disclosure.
[0251] Semi-solid and soft nanoparticles are also suitable for use in delivering a CasZ polypeptide of the present disclosure, a CasZ fusion polypeptide of the present disclosure, an RNP of the present disclosure, a nucleic acid of the present disclosure (e.g., a CasZ guide RNA and/or a CasZ trancRNA), or a CasZ system of the present disclosure, to a target cell. A prototype nanoparticle of semi-solid nature is the liposome.
[0252] In some cases, an exosome is used to deliver a CasZ polypeptide of the present disclosure, a CasZ fusion polypeptide of the present disclosure, an RNP of the present disclosure, a nucleic acid of the present disclosure (e.g., a CasZ guide RNA and/or a CasZ trancRNA), or a CasZ system of the present disclosure, to a target cell. Exosomes are endogenous nano-vesicles that transport RNAs and proteins, and which can deliver RNA to the brain and other target organs.
[0253] In some cases, a liposome is used to deliver a CasZ polypeptide of the present disclosure, a CasZ fusion polypeptide of the present disclosure, an RNP of the present disclosure, a nucleic acid of the present disclosure (e.g., a CasZ guide RNA and/or a CasZ trancRNA), or a CasZ system of the present disclosure, to a target cell. Liposomes are spherical vesicle structures composed of a uni- or multilamellar lipid bilayer surrounding internal aqueous compartments and a relatively impermeable outer lipophilic phospholipid bilayer. Liposomes can be made from several different types of lipids; however, phospholipids are most commonly used to generate liposomes. Although liposome formation is spontaneous when a lipid film is mixed with an aqueous solution, it can also be expedited by applying force in the form of shaking by using a homogenizer, sonicator, or an extrusion apparatus. Several other additives may be added to liposomes in order to modify their structure and properties. For instance, either cholesterol or sphingomyelin may be added to the liposomal mixture in order to help stabilize the liposomal structure and to prevent the leakage of the liposomal inner cargo. A liposome formulation may be mainly comprised of natural phospholipids and lipids such as 1,2-distearoryl-sn-glycero-3-phosphatidyl choline (DSPC), sphingomyelin, egg phosphatidylcholines and monosialoganglioside.
[0254] A stable nucleic-acid-lipid particle (SNALP) can be used to deliver a CasZ polypeptide of the present disclosure, a CasZ fusion polypeptide of the present disclosure, an RNP of the present disclosure, a nucleic acid of the present disclosure (e.g., a CasZ guide RNA and/or a CasZ trancRNA), or a CasZ system of the present disclosure, to a target cell. The SNALP formulation may contain the lipids 3-N-[(methoxypoly(ethylene glycol) 2000) carbamoyl]-1,2-dimyristyloxy-propylamine (PEG-C-DMA), 1,2-dilinoleyloxy-N,N-dimethyl-3-aminopropane (DLinDMA), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC) and cholesterol, in a 2:40:10:48 molar percent ratio. The SNALP liposomes may be prepared by formulating D-Lin-DMA and PEG-C-DMA with distearoylphosphatidylcholine (DSPC), Cholesterol and siRNA using a 25:1 lipid/siRNA ratio and a 48/40/10/2 molar ratio of Cholesterol/D-Lin-DMA/DSPC/PEG-C-DMA. The resulting SNALP liposomes can be about 80-100 nm in size. A SNALP may comprise synthetic cholesterol (Sigma-Aldrich, St Louis, Mo., USA), dipalmitoylphosphatidylcholine (Avanti Polar Lipids, Alabaster, Ala., USA), 3-N-[(w-methoxy poly(ethylene glycol)2000)carbamoyl]-1,2-dimyrestyloxypropylamine, and cationic 1,2-dilinoleyloxy-3-N,Ndimethylaminopropane. A SNALP may comprise synthetic cholesterol (Sigma-Aldrich), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC; Avanti Polar Lipids Inc.), PEG-cDMA, and 1,2-dilinoleyloxy-3-(N;N-dimethyl)aminopropane (DLinDMA).
[0255] Other cationic lipids, such as amino lipid 2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane (DLin-KC2-DMA) can be used to deliver a CasZ polypeptide of the present disclosure, a CasZ fusion polypeptide of the present disclosure, an RNP of the present disclosure, a nucleic acid of the present disclosure (e.g., a CasZ guide RNA and/or a CasZ trancRNA), or a CasZ system of the present disclosure, to a target cell. A preformed vesicle with the following lipid composition may be contemplated: amino lipid, distearoylphosphatidylcholine (DSPC), cholesterol and (R)-2,3-bis(octadecyloxy) propyl-1-(methoxy poly(ethylene glycol)2000)propylcarbamate (PEG-lipid) in the molar ratio 40/10/40/10, respectively, and a FVII siRNA/total lipid ratio of approximately 0.05 (w/w). To ensure a narrow particle size distribution in the range of 70-90 nm and a low polydispersity index of 0.11.+-0.0.04 (n=56), the particles may be extruded up to three times through 80 nm membranes prior to adding the guide RNA. Particles containing the highly potent amino lipid 16 may be used, in which the molar ratio of the four lipid components 16, DSPC, cholesterol and PEG-lipid (50/10/38.5/1.5) which may be further optimized to enhance in vivo activity.
[0256] Lipids may be formulated with a CasZ system of the present disclosure or component(s) thereof or nucleic acids encoding the same to form lipid nanoparticles (LNPs). Suitable lipids include, but are not limited to, DLin-KC2-DMA4, C12-200 and colipids disteroylphosphatidyl choline, cholesterol, and PEG-DMG may be formulated with a CasZ system, or component thereof, of the present disclosure, using a spontaneous vesicle formation procedure. The component molar ratio may be about 50/10/38.5/1.5 (DLin-KC2-DMA or C12-200/disteroylphosphatidyl choline/cholesterol/PEG-DMG).
[0257] A CasZ system of the present disclosure, or a component thereof, may be delivered encapsulated in PLGA microspheres such as that further described in US published applications 20130252281 and 20130245107 and 20130244279.
[0258] Supercharged proteins can be used to deliver a CasZ polypeptide of the present disclosure, a CasZ fusion polypeptide of the present disclosure, an RNP of the present disclosure, a nucleic acid of the present disclosure (e.g., a CasZ guide RNA and/or a CasZ trancRNA), or a CasZ system of the present disclosure, to a target cell. Supercharged proteins are a class of engineered or naturally occurring proteins with unusually high positive or negative net theoretical charge. Both supernegatively and superpositively charged proteins exhibit the ability to withstand thermally or chemically induced aggregation. Superpositively charged proteins are also able to penetrate mammalian cells. Associating cargo with these proteins, such as plasmid DNA, RNA, or other proteins, can enable the functional delivery of these macromolecules into mammalian cells both in vitro and in vivo.
[0259] Cell Penetrating Peptides (CPPs) can be used to deliver a CasZ polypeptide of the present disclosure, a CasZ fusion polypeptide of the present disclosure, an RNP of the present disclosure, a nucleic acid of the present disclosure (e.g., a CasZ guide RNA and/or a CasZ trancRNA), or a CasZ system of the present disclosure, to a target cell. CPPs typically have an amino acid composition that either contains a high relative abundance of positively charged amino acids such as lysine or arginine or has sequences that contain an alternating pattern of polar/charged amino acids and non-polar, hydrophobic amino acids.
[0260] An implantable device can be used to deliver a CasZ polypeptide of the present disclosure, a CasZ fusion polypeptide of the present disclosure, an RNP of the present disclosure, a nucleic acid of the present disclosure (e.g., a CasZ guide RNA and/or a CasZ trancRNA) (e.g., a CasZ guide RNA, a nucleic acid encoding a CasZ guide RNA, a nucleic acid encoding CasZ polypeptide, a donor template, and the like), or a CasZ system of the present disclosure, to a target cell (e.g., a target cell in vivo, where the target cell is a target cell in circulation, a target cell in a tissue, a target cell in an organ, etc.). An implantable device suitable for use in delivering a CasZ polypeptide of the present disclosure, a CasZ fusion polypeptide of the present disclosure, an RNP of the present disclosure, a nucleic acid of the present disclosure (e.g., a CasZ guide RNA and/or a CasZ trancRNA), or a CasZ system of the present disclosure, to a target cell (e.g., a target cell in vivo, where the target cell is a target cell in circulation, a target cell in a tissue, a target cell in an organ, etc.) can include a container (e.g., a reservoir, a matrix, etc.) that comprises the CasZ polypeptide, the CasZ fusion polypeptide, the RNP, or the CasZ system (or component thereof, e.g., a nucleic acid of the present disclosure).
[0261] A suitable implantable device can comprise a polymeric substrate, such as a matrix for example, that is used as the device body, and in some cases additional scaffolding materials, such as metals or additional polymers, and materials to enhance visibility and imaging. An implantable delivery device can be advantageous in providing release locally and over a prolonged period, where the polypeptide and/or nucleic acid to be delivered is released directly to a target site, e.g., the extracellular matrix (ECM), the vasculature surrounding a tumor, a diseased tissue, etc. Suitable implantable delivery devices include devices suitable for use in delivering to a cavity such as the abdominal cavity and/or any other type of administration in which the drug delivery system is not anchored or attached, comprising a biostable and/or degradable and/or bioabsorbable polymeric substrate, which may for example optionally be a matrix. In some cases, a suitable implantable drug delivery device comprises degradable polymers, wherein the main release mechanism is bulk erosion. In some cases, a suitable implantable drug delivery device comprises non degradable, or slowly degraded polymers, wherein the main release mechanism is diffusion rather than bulk erosion, so that the outer part functions as membrane, and its internal part functions as a drug reservoir, which practically is not affected by the surroundings for an extended period (for example from about a week to about a few months). Combinations of different polymers with different release mechanisms may also optionally be used. The concentration gradient at the can be maintained effectively constant during a significant period of the total releasing period, and therefore the diffusion rate is effectively constant (termed "zero mode" diffusion). By the term "constant" it is meant a diffusion rate that is maintained above the lower threshold of therapeutic effectiveness, but which may still optionally feature an initial burst and/or may fluctuate, for example increasing and decreasing to a certain degree. The diffusion rate can be so maintained for a prolonged period, and it can be considered constant to a certain level to optimize the therapeutically effective period, for example the effective silencing period.
[0262] In some cases, the implantable delivery system is designed to shield the nucleotide based therapeutic agent from degradation, whether chemical in nature or due to attack from enzymes and other factors in the body of the subject.
[0263] The site for implantation of the device, or target site, can be selected for maximum therapeutic efficacy. For example, a delivery device can be implanted within or in the proximity of a tumor environment, or the blood supply associated with a tumor. The target location can be, e.g.: 1) the brain at degenerative sites like in Parkinson or Alzheimer disease at the basal ganglia, white and gray matter; 2) the spine, as in the case of amyotrophic lateral sclerosis (ALS); 3) uterine cervix; 4) active and chronic inflammatory joints; 5) dermis as in the case of psoriasis; 7) sympathetic and sensoric nervous sites for analgesic effect; 7) a bone; 8) a site of acute or chronic infection; 9) Intra vaginal; 10) Inner ear-auditory system, labyrinth of the inner ear, vestibular system; 11) Intra tracheal; 12) Intra-cardiac; coronary, epicardiac; 13) urinary tract or bladder; 14) biliary system; 15) parenchymal tissue including and not limited to the kidney, liver, spleen; 16) lymph nodes; 17) salivary glands; 18) dental gums; 19) Intra-articular (into joints); 20) Intra-ocular; 21) Brain tissue; 22) Brain ventricles; 23) Cavities, including abdominal cavity (for example but without limitation, for ovary cancer); 24) Intra esophageal; and 25) Intra rectal; and 26) into the vasculature.
[0264] The method of insertion, such as implantation, may optionally already be used for other types of tissue implantation and/or for insertions and/or for sampling tissues, optionally without modifications, or alternatively optionally only with non-major modifications in such methods. Such methods optionally include but are not limited to brachytherapy methods, biopsy, endoscopy with and/or without ultrasound, such as stereotactic methods into the brain tissue, laparoscopy, including implantation with a laparoscope into joints, abdominal organs, the bladder wall and body cavities.
Modified Host Cells
[0265] The present disclosure provides a modified cell comprising a CasZ polypeptide of the present disclosure and/or a nucleic acid comprising a nucleotide sequence encoding a CasZ polypeptide of the present disclosure. The present disclosure provides a modified cell comprising a CasZ polypeptide of the present disclosure, where the modified cell is a cell that does not normally comprise a CasZ polypeptide of the present disclosure. The present disclosure provides a modified cell (e.g., a genetically modified cell) comprising nucleic acid comprising a nucleotide sequence encoding a CasZ polypeptide of the present disclosure. The present disclosure provides a genetically modified cell that is genetically modified with an mRNA comprising a nucleotide sequence encoding a CasZ polypeptide of the present disclosure. The present disclosure provides a genetically modified cell that is genetically modified with a recombinant expression vector comprising a nucleotide sequence encoding a CasZ polypeptide of the present disclosure. The present disclosure provides a genetically modified cell that is genetically modified with a recombinant expression vector comprising: a) a nucleotide sequence encoding a CasZ polypeptide of the present disclosure; and b) a nucleotide sequence encoding a CasZ guide RNA of the present disclosure. The present disclosure provides a genetically modified cell that is genetically modified with a recombinant expression vector comprising: a) a nucleotide sequence encoding a CasZ polypeptide of the present disclosure; b) a nucleotide sequence encoding a CasZ guide RNA of the present disclosure; and c) a nucleotide sequence encoding a donor template.
[0266] A cell that serves as a recipient for a CasZ polypeptide of the present disclosure and/or a nucleic acid comprising a nucleotide sequence encoding a CasZ polypeptide of the present disclosure and/or a CasZ guide RNA of the present disclosure (or a nucleic acid encoding it) and/or a CasZ trancRNA (or a nucleic acid encoding it), can be any of a variety of cells, including, e.g., in vitro cells; in vivo cells; ex vivo cells; primary cells; cancer cells; animal cells; plant cells; algal cells; fungal cells; etc. A cell that serves as a recipient for a CasZ polypeptide of the present disclosure and/or a nucleic acid comprising a nucleotide sequence encoding a CasZ polypeptide of the present disclosure and/or a CasZ guide RNA of the present disclosure is referred to as a "host cell" or a "target cell." A host cell or a target cell can be a recipient of a CasZ system of the present disclosure. A host cell or a target cell can be a recipient of a CasZ RNP of the present disclosure. A host cell or a target cell can be a recipient of a single component of a CasZ system of the present disclosure.
[0267] Non-limiting examples of cells (target cells) include: a prokaryotic cell, eukaryotic cell, a bacterial cell, an archaeal cell, a cell of a single-cell eukaryotic organism, a protozoa cell, a cell from a plant (e.g., cells from plant crops, fruits, vegetables, grains, soy bean, corn, maize, wheat, seeds, tomatos, rice, cassava, sugarcane, pumpkin, hay, potatos, cotton, cannabis, tobacco, flowering plants, conifers, gymnosperms, angiosperms, ferns, clubmosses, hornworts, liverworts, mosses, dicotyledons, monocotyledons, etc.), an algal cell, (e.g., Botryococcus braunii, Chlamydomonas reinhardtii, Nannochloropsis gaditana, Chlorella pyrenoidosa, Sargassum patens, C. agardh, and the like), seaweeds (e.g. kelp) a fungal cell (e.g., a yeast cell, a cell from a mushroom), an animal cell, a cell from an invertebrate animal (e.g., fruit fly, cnidarian, echinoderm, nematode, etc.), a cell from a vertebrate animal (e.g., fish, amphibian, reptile, bird, mammal), a cell from a mammal (e.g., an ungulate (e.g., a pig, a cow, a goat, a sheep); a rodent (e.g., a rat, a mouse); a non-human primate; a human; a feline (e.g., a cat); a canine (e.g., a dog); etc.), and the like. In some cases, the cell is a cell that does not originate from a natural organism (e.g., the cell can be a synthetically made cell; also referred to as an artificial cell).
[0268] A cell can be an in vitro cell (e.g., a cell in culture, e.g., an established cultured cell line). A cell can be an ex vivo cell (cultured cell from an individual). A cell can be and in vivo cell (e.g., a cell in an individual). A cell can be an isolated cell. A cell can be a cell inside of an organism. A cell can be an organism. A cell can be a cell in a cell culture (e.g., in vitro cell culture). A cell can be one of a collection of cells. A cell can be a prokaryotic cell or derived from a prokaryotic cell. A cell can be a bacterial cell or can be derived from a bacterial cell. A cell can be an archaeal cell or derived from an archaeal cell. A cell can be a eukaryotic cell or derived from a eukaryotic cell. A cell can be a plant cell or derived from a plant cell. A cell can be an animal cell or derived from an animal cell. A cell can be an invertebrate cell or derived from an invertebrate cell. A cell can be a vertebrate cell or derived from a vertebrate cell. A cell can be a mammalian cell or derived from a mammalian cell. A cell can be a rodent cell or derived from a rodent cell. A cell can be a human cell or derived from a human cell. A cell can be a microbe cell or derived from a microbe cell. A cell can be a fungi cell or derived from a fungi cell. A cell can be an insect cell. A cell can be an arthropod cell. A cell can be a protozoan cell. A cell can be a helminth cell.
[0269] Suitable cells include a stem cell (e.g. an embryonic stem (ES) cell, an induced pluripotent stem (iPS) cell; a germ cell (e.g., an oocyte, a sperm, an oogonia, a spermatogonia, etc.); a somatic cell, e.g. a fibroblast, an oligodendrocyte, a glial cell, a hematopoietic cell, a neuron, a muscle cell, a bone cell, a hepatocyte, a pancreatic cell, etc.
[0270] Suitable cells include human embryonic stem cells, fetal cardiomyocytes, myofibroblasts, mesenchymal stem cells, autotransplated expanded cardiomyocytes, adipocytes, totipotent cells, pluripotent cells, blood stem cells, myoblasts, adult stem cells, bone marrow cells, mesenchymal cells, embryonic stem cells, parenchymal cells, epithelial cells, endothelial cells, mesothelial cells, fibroblasts, osteoblasts, chondrocytes, exogenous cells, endogenous cells, stem cells, hematopoietic stem cells, bone-marrow derived progenitor cells, myocardial cells, skeletal cells, fetal cells, undifferentiated cells, multi-potent progenitor cells, unipotent progenitor cells, monocytes, cardiac myoblasts, skeletal myoblasts, macrophages, capillary endothelial cells, xenogenic cells, allogenic cells, and post-natal stem cells.
[0271] In some cases, the cell is an immune cell, a neuron, an epithelial cell, and endothelial cell, or a stem cell. In some cases, the immune cell is a T cell, a B cell, a monocyte, a natural killer cell, a dendritic cell, or a macrophage. In some cases, the immune cell is a cytotoxic T cell. In some cases, the immune cell is a helper T cell. In some cases, the immune cell is a regulatory T cell (Treg).
[0272] In some cases, the cell is a stem cell. Stem cells include adult stem cells. Adult stem cells are also referred to as somatic stem cells.
[0273] Adult stem cells are resident in differentiated tissue, but retain the properties of self-renewal and ability to give rise to multiple cell types, usually cell types typical of the tissue in which the stem cells are found. Numerous examples of somatic stem cells are known to those of skill in the art, including muscle stem cells; hematopoietic stem cells; epithelial stem cells; neural stem cells; mesenchymal stem cells; mammary stem cells; intestinal stem cells; mesodermal stem cells; endothelial stem cells; olfactory stem cells; neural crest stem cells; and the like.
[0274] Stem cells of interest include mammalian stem cells, where the term "mammalian" refers to any animal classified as a mammal, including humans; non-human primates; domestic and farm animals; and zoo, laboratory, sports, or pet animals, such as dogs, horses, cats, cows, mice, rats, rabbits, etc. In some cases, the stem cell is a human stem cell. In some cases, the stem cell is a rodent (e.g., a mouse; a rat) stem cell. In some cases, the stem cell is a non-human primate stem cell.
[0275] Stem cells can express one or more stem cell markers, e.g., SOX9, KRT19, KRT7, LGR5, CA9, FXYD2, CDH6, CLDN18, TSPAN8, BPIFB 1, OLFM4, CDH17, and PPARGC1A.
[0276] In some cases, the stem cell is a hematopoietic stem cell (HSC). HSCs are mesoderm-derived cells that can be isolated from bone marrow, blood, cord blood, fetal liver and yolk sac. HSCs are characterized as CD34.sup.+ and CD3.sup.-. HSCs can repopulate the erythroid, neutrophil-macrophage, megakaryocyte and lymphoid hematopoietic cell lineages in vivo. In vitro, HSCs can be induced to undergo at least some self-renewing cell divisions and can be induced to differentiate to the same lineages as is seen in vivo. As such, HSCs can be induced to differentiate into one or more of erythroid cells, megakaryocytes, neutrophils, macrophages, and lymphoid cells.
[0277] In other cases, the stem cell is a neural stem cell (NSC). Neural stem cells (NSCs) are capable of differentiating into neurons, and glia (including oligodendrocytes, and astrocytes). A neural stem cell is a multipotent stem cell which is capable of multiple divisions, and under specific conditions can produce daughter cells which are neural stem cells, or neural progenitor cells that can be neuroblasts or glioblasts, e.g., cells committed to become one or more types of neurons and glial cells respectively. Methods of obtaining NSCs are known in the art.
[0278] In other cases, the stem cell is a mesenchymal stem cell (MSC). MSCs originally derived from the embryonal mesoderm and isolated from adult bone marrow, can differentiate to form muscle, bone, cartilage, fat, marrow stroma, and tendon. Methods of isolating MSC are known in the art; and any known method can be used to obtain MSC. See, e.g., U.S. Pat. No. 5,736,396, which describes isolation of human MSC.
[0279] A cell is in some cases a plant cell. A plant cell can be a cell of a monocotyledon. A cell can be a cell of a dicotyledon.
[0280] In some cases, the cell is a plant cell. For example, the cell can be a cell of a major agricultural plant, e.g., Barley, Beans (Dry Edible), Canola, Corn, Cotton (Pima), Cotton (Upland), Flaxseed, Hay (Alfalfa), Hay (Non-Alfalfa), Oats, Peanuts, Rice, Sorghum, Soybeans, Sugarbeets, Sugarcane, Sunflowers (Oil), Sunflowers (Non-Oil), Sweet Potatoes, Tobacco (Burley), Tobacco (Flue-cured), Tomatoes, Wheat (Durum), Wheat (Spring), Wheat (Winter), and the like. As another example, the cell is a cell of a vegetable crops which include but are not limited to, e.g., alfalfa sprouts, aloe leaves, arrow root, arrowhead, artichokes, asparagus, bamboo shoots, banana flowers, bean sprouts, beans, beet tops, beets, bittermelon, bok choy, broccoli, broccoli rabe (rappini), brussels sprouts, cabbage, cabbage sprouts, cactus leaf (nopales), calabaza, cardoon, carrots, cauliflower, celery, chayote, chinese artichoke (crosnes), chinese cabbage, chinese celery, chinese chives, choy sum, chrysanthemum leaves (tung ho), collard greens, corn stalks, corn-sweet, cucumbers, daikon, dandelion greens, dasheen, dau mue (pea tips), donqua (winter melon), eggplant, endive, escarole, fiddle head ferns, field cress, frisee, gai choy (chinese mustard), gailon, galanga (siam, thai ginger), garlic, ginger root, gobo, greens, hanover salad greens, huauzontle, jerusalem artichokes, jicama, kale greens, kohlrabi, lamb's quarters (quilete), lettuce (bibb), lettuce (boston), lettuce (boston red), lettuce (green leaf), lettuce (iceberg), lettuce (lolla rossa), lettuce (oak leaf--green), lettuce (oak leaf--red), lettuce (processed), lettuce (red leaf), lettuce (romaine), lettuce (ruby romaine), lettuce (russian red mustard), linkok, lo bok, long beans, lotus root, mache, maguey (agave) leaves, malanga, mesculin mix, mizuna, moap (smooth luffa), moo, moqua (fuzzy squash), mushrooms, mustard, nagaimo, okra, ong choy, onions green, opo (long squash), ornamental corn, ornamental gourds, parsley, parsnips, peas, peppers (bell type), peppers, pumpkins, radicchio, radish sprouts, radishes, rape greens, rape greens, rhubarb, romaine (baby red), rutabagas, salicornia (sea bean), sinqua (angled/ridged luffa), spinach, squash, straw bales, sugarcane, sweet potatoes, swiss chard, tamarindo, taro, taro leaf, taro shoots, tatsoi, tepeguaje (guaje), tindora, tomatillos, tomatoes, tomatoes (cherry), tomatoes (grape type), tomatoes (plum type), tumeric, turnip tops greens, turnips, water chestnuts, yampi, yams (names), yu choy, yuca (cassava), and the like.
[0281] A cell is in some cases an arthropod cell. For example, the cell can be a cell of a sub-order, a family, a sub-family, a group, a sub-group, or a species of, e.g., Chelicerata, Myriapodia, Hexipodia, Arachnida, Insecta, Archaeognatha, Thysanura, Palaeoptera, Ephemeroptera, Odonata, Anisoptera, Zygoptera, Neoptera, Exopterygota, Plecoptera, Embioptera, Orthoptera, Zoraptera, Dermaptera, Dictyoptera, Notoptera, Grylloblattidae, Mantophasmatidae, Phasmatodea, Blattaria, Isoptera, Mantodea, Parapneuroptera, Psocoptera, Thysanoptera, Phthiraptera, Hemiptera, Endopterygota or Holometabola, Hymenoptera, Coleoptera, Strepsiptera, Raphidioptera, Megaloptera, Neuroptera, Mecoptera, Siphonaptera, Diptera, Trichoptera, or Lepidoptera.
[0282] A cell is in some cases an insect cell. For example, in some cases, the cell is a cell of a mosquito, a grasshopper, a true bug, a fly, a flea, a bee, a wasp, an ant, a louse, a moth, or a beetle.
Kits
[0283] The present disclosure provides a kit comprising a CasZ system of the present disclosure, or a component of a CasZ system of the present disclosure.
[0284] A kit of the present disclosure can comprise any combination as listed for a CasZ system (e.g., see above). A kit of the present disclosure can comprise: a) a component, as described above, of a CasZ system of the present disclosure, or can comprise a CasZ system of the present disclosure; and b) one or more additional reagents, e.g., i) a buffer; ii) a protease inhibitor; iii) a nuclease inhibitor; iv) a reagent required to develop or visualize a detectable label; v) a positive and/or negative control target DNA; vi) a positive and/or negative control CasZ guide RNA; vii) a CasZ trancRNA; and the like. A kit of the present disclosure can comprise: a) a component, as described above, of a CasZ system of the present disclosure, or can comprise a CasZ system of the present disclosure; and b) a therapeutic agent.
[0285] A kit of the present disclosure can comprise a recombinant expression vector comprising: a) an insertion site for inserting a nucleic acid comprising a nucleotide sequence encoding a portion of a CasZ guide RNA that hybridizes to a target nucleotide sequence in a target nucleic acid; and b) a nucleotide sequence encoding the CasZ-binding portion of a CasZ guide RNA. A kit of the present disclosure can comprise a recombinant expression vector comprising: a) an insertion site for inserting a nucleic acid comprising a nucleotide sequence encoding a portion of a CasZ guide RNA that hybridizes to a target nucleotide sequence in a target nucleic acid; b) a nucleotide sequence encoding the CasZ-binding portion of a CasZ guide RNA; and c) a nucleotide sequence encoding a CasZ polypeptide of the present disclosure. A kit of the present disclosure can comprise a recombinant expression vector comprising a nucleotide sequence encoding a CasZ trancRNA.
Detection of ssDNA
[0286] A CasZ (Cas14) polypeptide of the present disclosure, once activated by detection of a target DNA (double or single stranded), can promiscuously cleave non-targeted single stranded DNA (ssDNA). Once a CasZ (Cas14) is activated by a guide RNA, which occurs when the guide RNA hybridizes to a target sequence of a target DNA (i.e., the sample includes the target DNA, e.g., target ssDNA), the protein becomes a nuclease that promiscuously cleaves ssDNAs (i.e., the nuclease cleaves non-target ssDNAs, i.e., ssDNAs to which the guide sequence of the guide RNA does not hybridize). Thus, when the target DNA is present in the sample (e.g., in some cases above a threshold amount), the result is cleavage of ssDNAs in the sample, which can be detected using any convenient detection method (e.g., using a labeled single stranded detector DNA). In some cases, a CasZ polypeptide requires, in addition to a CasZ guide RNA, a tranc RNA for activation.
[0287] Provided are compositions and methods for detecting a target DNA (double stranded or single stranded) in a sample. In some cases, a detector DNA is used that is single stranded (ssDNA) and does not hybridize with the guide sequence of the guide RNA (i.e., the detector ssDNA is a non-target ssDNA). Such methods can include (a) contacting the sample with: (i) a CasZ polypeptide; (ii) a guide RNA comprising: a region that binds to the CasZ polypeptide, and a guide sequence that hybridizes with the target DNA; and (iii) a detector DNA that is single stranded and does not hybridize with the guide sequence of the guide RNA; and (b) measuring a detectable signal produced by cleavage of the single stranded detector DNA by the CasZ polypeptide, thereby detecting the target DNA. In some cases, the methods include can include (a) contacting the sample with: (i) a CasZ polypeptide; (ii) a guide RNA comprising: a region that binds to the CasZ polypeptide, and a guide sequence that hybridizes with the target DNA; (iii) a CasZ tranc RNA; and (iv) a detector DNA that is single stranded and does not hybridize with the guide sequence of the guide RNA; and (b) measuring a detectable signal produced by cleavage of the single stranded detector DNA by the CasZ polypeptide, thereby detecting the target DNA. As noted above, once a subject CasZ polypeptide protein is activated by a guide RNA, which occurs when the sample includes a target DNA to which the guide RNA hybridizes (i.e., the sample includes the targeted target DNA), the CasZ polypeptide is activated and functions as an endoribonuclease that non-specifically cleaves ssDNAs (including non-target ssDNAs) present in the sample. Thus, when the targeted target DNA is present in the sample (e.g., in some cases above a threshold amount), the result is cleavage of ssDNA (including non-target ssDNA) in the sample, which can be detected using any convenient detection method (e.g., using a labeled detector ssDNA).
[0288] Also provided are compositions and methods for cleaving single stranded DNAs (ssDNAs) (e.g., non-target ssDNAs). Such methods can include contacting a population of nucleic acids, wherein said population comprises a target DNA and a plurality of non-target ssDNAs, with: (i) a CasZ polypeptide; and (ii) a guide RNA comprising: a region that binds to the CasZ polypeptide, and a guide sequence that hybridizes with the target DNA, wherein the CasZ polypeptide cleaves non-target ssDNAs of said plurality. Such methods can include contacting a population of nucleic acids, wherein said population comprises a target DNA and a plurality of non-target ssDNAs, with: (i) a CasZ polypeptide; (ii) a guide RNA comprising: a region that binds to the CasZ polypeptide, and a guide sequence that hybridizes with the target DNA, and (iii) a CasZ tranc RNA, wherein the CasZ polypeptide cleaves non-target ssDNAs of said plurality. Such methods can be used, e.g., to cleave foreign ssDNAs (e.g., viral DNAs) in a cell.
[0289] The contacting step of a subject method can be carried out in a composition comprising divalent metal ions. The contacting step can be carried out in an acellular environment, e.g., outside of a cell. The contacting step can be carried out inside a cell. The contacting step can be carried out in a cell in vitro. The contacting step can be carried out in a cell ex vivo. The contacting step can be carried out in a cell in vivo.
[0290] The guide RNA can be provided as RNA or as a nucleic acid encoding the guide RNA (e.g., a DNA such as a recombinant expression vector). The tranc RNA can be provided as RNA or as a nucleic acid encoding the guide RNA (e.g., a DNA such as a recombinant expression vector). The CasZ polypeptide can be provided as a protein per se or as a nucleic acid encoding the protein (e.g., an mRNA, a DNA such as a recombinant expression vector). In some cases, two or more (e.g., 3 or more, 4 or more, 5 or more, or 6 or more) guide RNAs can be provided. In some cases, a single-molecule RNA comprising: i) a CasZ guide RNA; and ii) a tranc RNA (or a nucleic acid comprising a nucleotide sequence encoding the single-molecule RNA) is used.
[0291] In some cases (e.g., when contacting a sample with a guide RNA and a CasZ polypeptide; or when contacting a sample with a guide RNA, a CasZ polypeptide, and a tranc RNA), the sample is contacted for 2 hours or less (e.g., 1.5 hours or less, 1 hour or less, 40 minutes or less, 30 minutes or less, 20 minutes or less, 10 minutes or less, or 5 minutes or less, or 1 minute or less) prior to the measuring step. For example, in some cases the sample is contacted for 40 minutes or less prior to the measuring step. In some cases, the sample is contacted for 20 minutes or less prior to the measuring step. In some cases, the sample is contacted for 10 minutes or less prior to the measuring step. In some cases, the sample is contacted for 5 minutes or less prior to the measuring step. In some cases, the sample is contacted for 1 minute or less prior to the measuring step. In some cases, the sample is contacted for from 50 seconds to 60 seconds prior to the measuring step. In some cases, the sample is contacted for from 40 seconds to 50 seconds prior to the measuring step. In some cases, the sample is contacted for from 30 seconds to 40 seconds prior to the measuring step. In some cases, the sample is contacted for from 20 seconds to 30 seconds prior to the measuring step. In some cases, the sample is contacted for from 10 seconds to 20 seconds prior to the measuring step.
[0292] In some cases, a method of the present disclosure for detecting a target DNA comprises: a) contacting a sample with a guide RNA, a CasZ polypeptide, and a detector DNA), where the sample is contacted for 2 hours or less (e.g., 1.5 hours or less, 1 hour or less, 40 minutes or less, 30 minutes or less, 20 minutes or less, 10 minutes or less, or 5 minutes or less, or 1 minute or less), under conditions that provide for trans cleavage of the detector DNA; b) maintaining the sample from step (a) for a period of time under conditions that do not provide for trans cleavage of the detector RNA; and c) after the time period of step (b), measuring a detectable signal produced by cleavage of the single stranded detector DNA by the CasZ polypeptide, thereby detecting the target DNA. Conditions that provide for trans cleavage of the detector DNA include temperature conditions such as from 17.degree. C. to about 39.degree. C. (e.g., about 37.degree. C.). Conditions that do not provide for trans cleavage of the detector DNA include temperatures of 10.degree. C. or less, 5.degree. C. or less, 4.degree. C. or less, or 0.degree. C.
[0293] In some cases, a method of the present disclosure for detecting a target DNA comprises: a) contacting a sample with a guide RNA, a tranc RNA, a CasZ polypeptide, and a detector DNA (or contacting a sample with: i) a single-molecule RNA comprising a guide RNA and a tranc RNA; i) a CasZ polypeptide; and iii) a detector DNA), where the sample is contacted for 2 hours or less (e.g., 1.5 hours or less, 1 hour or less, 40 minutes or less, 30 minutes or less, 20 minutes or less, 10 minutes or less, or 5 minutes or less, or 1 minute or less), under conditions that provide for trans cleavage of the detector DNA; b) maintaining the sample from step (a) for a period of time under conditions that do not provide for trans cleavage of the detector RNA; and c) after the time period of step (b), measuring a detectable signal produced by cleavage of the single stranded detector DNA by the CasZ polypeptide, thereby detecting the target DNA. Conditions that provide for trans cleavage of the detector DNA include temperature conditions such as from 17.degree. C. to about 39.degree. C. (e.g., about 37.degree. C.). Conditions that do not provide for trans cleavage of the detector DNA include temperatures of 10.degree. C. or less, 5.degree. C. or less, 4.degree. C. or less, or 0.degree. C.
[0294] In some cases, a detectable signal produced by cleavage of a single-stranded detector DNA is produced for no more than 60 minutes. For example, in some cases, a detectable signal produced by cleavage of a single-stranded detector DNA is produced for no more than 60 minutes, no more than 45 minutes, no more than 30 minutes, no more than 15 minutes, no more than 10 minutes, or no more than 5 minutes. For example, in some cases, a detectable signal produced by cleavage of a single-stranded detector DNA is produced for a period of time of from 1 minute to 60 minutes, e.g., from 1 minute to 5 minutes, from 5 minutes to 10 minutes, from 10 minutes to 15 minutes, from 15 minutes to 30 minutes, from 30 minutes to 45 minutes, or from 45 minutes to 60 minutes. In some cases, after the detectable signal is produced (e.g., produced for no more than 60 minutes), production of the detectable signal can be stopped, e.g., by lowering the temperature of the sample (e.g., lowering the temperature to 10.degree. C. or less, 5.degree. C. or less, 4.degree. C. or less, or 0.degree. C.), by adding an inhibitor to the sample, by lyophilizing the sample, by heating the sample to over 40.degree. C., and the like. The measuring step can occur at any time after production of the detectable signal has been stopped. For example, the measuring step can occur from 5 minutes to 48 hours after production of the detectable signal has been stopped. For example, the measuring step can occur from 5 minutes to 15 minutes, from 15 minutes to 30 minutes, from 30 minutes to 60 minutes, from 1 hour to 4 hours, from 4 hours to 8 hours, from 8 hours to 12 hours, from 12 hours to 24 hours, from 24 hours to 36 hours, or from 36 hours to 48 hours, after production of the detectable signal has been stopped. The measuring step can occur more than 48 hours after production of the detectable signal has been stopped.
[0295] A method of the present disclosure for detecting a target DNA (single-stranded or double-stranded) in a sample can detect a target DNA with a high degree of sensitivity. In some cases, a method of the present disclosure can be used to detect a target DNA present in a sample comprising a plurality of DNAs (including the target DNA and a plurality of non-target DNAs), where the target DNA is present at one or more copies per 10.sup.7 non-target DNAs (e.g., one or more copies per 10.sup.6 non-target DNAs, one or more copies per 10.sup.5 non-target DNAs, one or more copies per 10.sup.4 non-target DNAs, one or more copies per 10.sup.3 non-target DNAs, one or more copies per 10.sup.2 non-target DNAs, one or more copies per 50 non-target DNAs, one or more copies per 20 non-target DNAs, one or more copies per 10 non-target DNAs, or one or more copies per 5 non-target DNAs). In some cases, a method of the present disclosure can be used to detect a target DNA present in a sample comprising a plurality of DNAs (including the target DNA and a plurality of non-target DNAs), where the target DNA is present at one or more copies per 10.sup.18 non-target DNAs (e.g., one or more copies per 10.sup.15 non-target DNAs, one or more copies per 10.sup.12 non-target DNAs, one or more copies per 10.sup.9 non-target DNAs, one or more copies per 10.sup.6 non-target DNAs, one or more copies per 10.sup.5 non-target DNAs, one or more copies per 10.sup.4 non-target DNAs, one or more copies per 10.sup.3 non-target DNAs, one or more copies per 10.sup.2 non-target DNAs, one or more copies per 50 non-target DNAs, one or more copies per 20 non-target DNAs, one or more copies per 10 non-target DNAs, or one or more copies per 5 non-target DNAs).
[0296] In some cases, a method of the present disclosure can detect a target DNA present in a sample, where the target DNA is present at from one copy per 10.sup.7 non-target DNAs to one copy per 10 non-target DNAs (e.g., from 1 copy per 10.sup.7 non-target DNAs to 1 copy per 10.sup.2 non-target DNAs, from 1 copy per 10.sup.7 non-target DNAs to 1 copy per 10.sup.3 non-target DNAs, from 1 copy per 10.sup.7 non-target DNAs to 1 copy per 10.sup.4 non-target DNAs, from 1 copy per 10.sup.7 non-target DNAs to 1 copy per 10.sup.5 non-target DNAs, from 1 copy per 10.sup.7 non-target DNAs to 1 copy per 10.sup.6 non-target DNAs, from 1 copy per 10.sup.6 non-target DNAs to 1 copy per 10 non-target DNAs, from 1 copy per 10.sup.6 non-target DNAs to 1 copy per 10.sup.2 non-target DNAs, from 1 copy per 10.sup.6 non-target DNAs to 1 copy per 10.sup.3 non-target DNAs, from 1 copy per 10.sup.6 non-target DNAs to 1 copy per 10.sup.4 non-target DNAs, from 1 copy per 10.sup.6 non-target DNAs to 1 copy per 10.sup.5 non-target DNAs, from 1 copy per 10.sup.5 non-target DNAs to 1 copy per 10 non-target DNAs, from 1 copy per 10.sup.5 non-target DNAs to 1 copy per 10.sup.2 non-target DNAs, from 1 copy per 10.sup.5 non-target DNAs to 1 copy per 10.sup.3 non-target DNAs, or from 1 copy per 10.sup.5 non-target DNAs to 1 copy per 10.sup.4 non-target DNAs).
[0297] In some cases, a method of the present disclosure can detect a target DNA present in a sample, where the target DNA is present at from one copy per 10.sup.18 non-target DNAs to one copy per 10 non-target DNAs (e.g., from 1 copy per 10.sup.18 non-target DNAs to 1 copy per 10.sup.2 non-target DNAs, from 1 copy per 10.sup.15 non-target DNAs to 1 copy per 10.sup.2 non-target DNAs, from 1 copy per 10.sup.12 non-target DNAs to 1 copy per 10.sup.2 non-target DNAs, from 1 copy per 10.sup.9 non-target DNAs to 1 copy per 10.sup.2 non-target DNAs, from 1 copy per 10.sup.7 non-target DNAs to 1 copy per 10.sup.2 non-target DNAs, from 1 copy per 10.sup.7 non-target DNAs to 1 copy per 10.sup.3 non-target DNAs, from 1 copy per 10.sup.7 non-target DNAs to 1 copy per 10.sup.4 non-target DNAs, from 1 copy per 10.sup.7 non-target DNAs to 1 copy per 10.sup.5 non-target DNAs, from 1 copy per 10.sup.7 non-target DNAs to 1 copy per 10.sup.6 non-target DNAs, from 1 copy per 10.sup.6 non-target DNAs to 1 copy per 10 non-target DNAs, from 1 copy per 10.sup.6 non-target DNAs to 1 copy per 10.sup.2 non-target DNAs, from 1 copy per 10.sup.6 non-target DNAs to 1 copy per 10.sup.3 non-target DNAs, from 1 copy per 10.sup.6 non-target DNAs to 1 copy per 10.sup.4 non-target DNAs, from 1 copy per 10.sup.6 non-target DNAs to 1 copy per 10.sup.5 non-target DNAs, from 1 copy per 10.sup.5 non-target DNAs to 1 copy per 10 non-target DNAs, from 1 copy per 10.sup.5 non-target DNAs to 1 copy per 10.sup.2 non-target DNAs, from 1 copy per 10.sup.5 non-target DNAs to 1 copy per 10.sup.3 non-target DNAs, or from 1 copy per 10.sup.5 non-target DNAs to 1 copy per 10.sup.4 non-target DNAs).
[0298] In some cases, a method of the present disclosure can detect a target DNA present in a sample, where the target DNA is present at from one copy per 10.sup.7 non-target DNAs to one copy per 100 non-target DNAs (e.g., from 1 copy per 10.sup.7 non-target DNAs to 1 copy per 10.sup.2 non-target DNAs, from 1 copy per 10.sup.7 non-target DNAs to 1 copy per 10.sup.3 non-target DNAs, from 1 copy per 10.sup.7 non-target DNAs to 1 copy per 10.sup.4 non-target DNAs, from 1 copy per 10.sup.7 non-target DNAs to 1 copy per 10.sup.5 non-target DNAs, from 1 copy per 10.sup.7 non-target DNAs to 1 copy per 10.sup.6 non-target DNAs, from 1 copy per 10.sup.6 non-target DNAs to 1 copy per 100 non-target DNAs, from 1 copy per 10.sup.6 non-target DNAs to 1 copy per 10.sup.2 non-target DNAs, from 1 copy per 10.sup.6 non-target DNAs to 1 copy per 10.sup.3 non-target DNAs, from 1 copy per 10.sup.6 non-target DNAs to 1 copy per 10.sup.4 non-target DNAs, from 1 copy per 10.sup.6 non-target DNAs to 1 copy per 10.sup.5 non-target DNAs, from 1 copy per 10.sup.5 non-target DNAs to 1 copy per 100 non-target DNAs, from 1 copy per 10.sup.5 non-target DNAs to 1 copy per 10.sup.2 non-target DNAs, from 1 copy per 10.sup.5 non-target DNAs to 1 copy per 10.sup.3 non-target DNAs, or from 1 copy per 10.sup.5 non-target DNAs to 1 copy per 10.sup.4 non-target DNAs).
[0299] In some cases, the threshold of detection, for a subject method of detecting a target DNA in a sample, is 10 nM or less. Thus, e.g., the target DNA can be present in the sample in a concentration of 10 nM or less. The term "threshold of detection" is used herein to describe the minimal amount of target DNA that must be present in a sample in order for detection to occur. Thus, as an illustrative example, when a threshold of detection is 10 nM, then a signal can be detected when a target DNA is present in the sample at a concentration of 10 nM or more. In some cases, a method of the present disclosure has a threshold of detection of 5 nM or less. In some cases, a method of the present disclosure has a threshold of detection of 1 nM or less. In some cases, a method of the present disclosure has a threshold of detection of 0.5 nM or less. In some cases, a method of the present disclosure has a threshold of detection of 0.1 nM or less. In some cases, a method of the present disclosure has a threshold of detection of 0.05 nM or less. In some cases, a method of the present disclosure has a threshold of detection of 0.01 nM or less. In some cases, a method of the present disclosure has a threshold of detection of 0.005 nM or less. In some cases, a method of the present disclosure has a threshold of detection of 0.001 nM or less. In some cases, a method of the present disclosure has a threshold of detection of 0.0005 nM or less. In some cases, a method of the present disclosure has a threshold of detection of 0.0001 nM or less. In some cases, a method of the present disclosure has a threshold of detection of 0.00005 nM or less. In some cases, a method of the present disclosure has a threshold of detection of 0.00001 nM or less. In some cases, a method of the present disclosure has a threshold of detection of 10 pM or less. In some cases, a method of the present disclosure has a threshold of detection of 1 pM or less. In some cases, a method of the present disclosure has a threshold of detection of 500 fM or less. In some cases, a method of the present disclosure has a threshold of detection of 250 fM or less. In some cases, a method of the present disclosure has a threshold of detection of 100 fM or less. In some cases, a method of the present disclosure has a threshold of detection of 50 fM or less. In some cases, a method of the present disclosure has a threshold of detection of 500 aM (attomolar) or less. In some cases, a method of the present disclosure has a threshold of detection of 250 aM or less. In some cases, a method of the present disclosure has a threshold of detection of 100 aM or less. In some cases, a method of the present disclosure has a threshold of detection of 50 aM or less. In some cases, a method of the present disclosure has a threshold of detection of 10 aM or less. In some cases, a method of the present disclosure has a threshold of detection of 1 aM or less.
[0300] In some cases, the threshold of detection (for detecting the target DNA in a subject method), is in a range of from 500 fM to 1 nM (e.g., from 500 fM to 500 pM, from 500 fM to 200 pM, from 500 fM to 100 pM, from 500 fM to 10 pM, from 500 fM to 1 pM, from 800 fM to 1 nM, from 800 fM to 500 pM, from 800 fM to 200 pM, from 800 fM to 100 pM, from 800 fM to 10 pM, from 800 fM to 1 pM, from 1 pM to 1 nM, from 1 pM to 500 pM, from 1 pM to 200 pM, from 1 pM to 100 pM, or from 1 pM to 10 pM) (where the concentration refers to the threshold concentration of target DNA at which the target DNA can be detected). In some cases, a method of the present disclosure has a threshold of detection in a range of from 800 fM to 100 pM. In some cases, a method of the present disclosure has a threshold of detection in a range of from 1 pM to 10 pM. In some cases, a method of the present disclosure has a threshold of detection in a range of from 10 fM to 500 fM, e.g., from 10 fM to 50 fM, from 50 fM to 100 fM, from 100 fM to 250 fM, or from 250 fM to 500 fM.
[0301] In some cases, the minimum concentration at which a target DNA can be detected in a sample is in a range of from 500 fM to 1 nM (e.g., from 500 fM to 500 pM, from 500 fM to 200 pM, from 500 fM to 100 pM, from 500 fM to 10 pM, from 500 fM to 1 pM, from 800 fM to 1 nM, from 800 fM to 500 pM, from 800 fM to 200 pM, from 800 fM to 100 pM, from 800 fM to 10 pM, from 800 fM to 1 pM, from 1 pM to 1 nM, from 1 pM to 500 pM, from 1 pM to 200 pM, from 1 pM to 100 pM, or from 1 pM to 10 pM). In some cases, the minimum concentration at which a target DNA can be detected in a sample is in a range of from 800 fM to 100 pM. In some cases, the minimum concentration at which a target DNA can be detected in a sample is in a range of from 1 pM to 10 pM.
[0302] In some cases, the threshold of detection (for detecting the target DNA in a subject method), is in a range of from 1 aM to 1 nM (e.g., from 1 aM to 500 pM, from 1 aM to 200 pM, from 1 aM to 100 pM, from 1 aM to 10 pM, from 1 aM to 1 pM, from 100 aM to 1 nM, from 100 aM to 500 pM, from 100 aM to 200 pM, from 100 aM to 100 pM, from 100 aM to 10 pM, from 100 aM to 1 pM, from 250 aM to 1 nM, from 250 aM to 500 pM, from 250 aM to 200 pM, from 250 aM to 100 pM, from 250 aM to 10 pM, from 250 aM to 1 pM, from 500 aM to 1 nM, from 500 aM to 500 pM, from 500 aM to 200 pM, from 500 aM to 100 pM, from 500 aM to 10 pM, from 500 aM to 1 pM, from 750 aM to 1 nM, from 750 aM to 500 pM, from 750 aM to 200 pM, from 750 aM to 100 pM, from 750 aM to 10 pM, from 750 aM to 1 pM, from 1 fM to 1 nM, from 1 fM to 500 pM, from 1 fM to 200 pM, from 1 fM to 100 pM, from 1 fM to 10 pM, from 1 fM to 1 pM, from 500 fM to 500 pM, from 500 fM to 200 pM, from 500 fM to 100 pM, from 500 fM to 10 pM, from 500 fM to 1 pM, from 800 fM to 1 nM, from 800 fM to 500 pM, from 800 fM to 200 pM, from 800 fM to 100 pM, from 800 fM to 10 pM, from 800 fM to 1 pM, from 1 pM to 1 nM, from 1 pM to 500 pM, from 1 pM to 200 pM, from 1 pM to 100 pM, or from 1 pM to 10 pM) (where the concentration refers to the threshold concentration of target DNA at which the target DNA can be detected). In some cases, a method of the present disclosure has a threshold of detection in a range of from 1 aM to 800 aM. In some cases, a method of the present disclosure has a threshold of detection in a range of from 50 aM to 1 pM. In some cases, a method of the present disclosure has a threshold of detection in a range of from 50 aM to 500 fM.
[0303] In some cases, a target DNA is present in a sample in a range of from 1 aM to 1 nM (e.g., from 1 aM to 500 pM, from 1 aM to 200 pM, from 1 aM to 100 pM, from 1 aM to 10 pM, from 1 aM to 1 pM, from 100 aM to 1 nM, from 100 aM to 500 pM, from 100 aM to 200 pM, from 100 aM to 100 pM, from 100 aM to 10 pM, from 100 aM to 1 pM, from 250 aM to 1 nM, from 250 aM to 500 pM, from 250 aM to 200 pM, from 250 aM to 100 pM, from 250 aM to 10 pM, from 250 aM to 1 pM, from 500 aM to 1 nM, from 500 aM to 500 pM, from 500 aM to 200 pM, from 500 aM to 100 pM, from 500 aM to 10 pM, from 500 aM to 1 pM, from 750 aM to 1 nM, from 750 aM to 500 pM, from 750 aM to 200 pM, from 750 aM to 100 pM, from 750 aM to 10 pM, from 750 aM to 1 pM, from 1 fM to 1 nM, from 1 fM to 500 pM, from 1 fM to 200 pM, from 1 fM to 100 pM, from 1 fM to 10 pM, from 1 fM to 1 pM, from 500 fM to 500 pM, from 500 fM to 200 pM, from 500 fM to 100 pM, from 500 fM to 10 pM, from 500 fM to 1 pM, from 800 fM to 1 nM, from 800 fM to 500 pM, from 800 fM to 200 pM, from 800 fM to 100 pM, from 800 fM to 10 pM, from 800 fM to 1 pM, from 1 pM to 1 nM, from 1 pM to 500 pM, from 1 pM to 200 pM, from 1 pM to 100 pM, or from 1 pM to 10 pM). In some cases, a target DNA is present in a sample in a range of from 1 aM to 800 aM. In some cases, a target DNA is present in a sample in a range of from 50 aM to 1 pM. In some cases, a target DNA is present in a sample in a range of from 50 aM to 500 fM.
[0304] In some cases, the minimum concentration at which a target DNA can be detected in a sample is in a range of from 1 aM to 1 nM (e.g., from 1 aM to 500 pM, from 1 aM to 200 pM, from 1 aM to 100 pM, from 1 aM to 10 pM, from 1 aM to 1 pM, from 100 aM to 1 nM, from 100 aM to 500 pM, from 100 aM to 200 pM, from 100 aM to 100 pM, from 100 aM to 10 pM, from 100 aM to 1 pM, from 250 aM to 1 nM, from 250 aM to 500 pM, from 250 aM to 200 pM, from 250 aM to 100 pM, from 250 aM to 10 pM, from 250 aM to 1 pM, from 500 aM to 1 nM, from 500 aM to 500 pM, from 500 aM to 200 pM, from 500 aM to 100 pM, from 500 aM to 10 pM, from 500 aM to 1 pM, from 750 aM to 1 nM, from 750 aM to 500 pM, from 750 aM to 200 pM, from 750 aM to 100 pM, from 750 aM to 10 pM, from 750 aM to 1 pM, from 1 fM to 1 nM, from 1 fM to 500 pM, from 1 fM to 200 pM, from 1 fM to 100 pM, from 1 fM to 10 pM, from 1 fM to 1 pM, from 500 fM to 500 pM, from 500 fM to 200 pM, from 500 fM to 100 pM, from 500 fM to 10 pM, from 500 fM to 1 pM, from 800 fM to 1 nM, from 800 fM to 500 pM, from 800 fM to 200 pM, from 800 fM to 100 pM, from 800 fM to 10 pM, from 800 fM to 1 pM, from 1 pM to 1 nM, from 1 pM to 500 pM, from 1 pM to 200 pM, from 1 pM to 100 pM, or from 1 pM to 10 pM). In some cases, the minimum concentration at which a target DNA can be detected in a sample is in a range of from 1 aM to 500 pM. In some cases, the minimum concentration at which a target DNA can be detected in a sample is in a range of from 100 aM to 500 pM.
[0305] In some cases, a target DNA is present in a sample in a range of from 1 aM to 1 nM (e.g., from 1 aM to 500 pM, from 1 aM to 200 pM, from 1 aM to 100 pM, from 1 aM to 10 pM, from 1 aM to 1 pM, from 100 aM to 1 nM, from 100 aM to 500 pM, from 100 aM to 200 pM, from 100 aM to 100 pM, from 100 aM to 10 pM, from 100 aM to 1 pM, from 250 aM to 1 nM, from 250 aM to 500 pM, from 250 aM to 200 pM, from 250 aM to 100 pM, from 250 aM to 10 pM, from 250 aM to 1 pM, from 500 aM to 1 nM, from 500 aM to 500 pM, from 500 aM to 200 pM, from 500 aM to 100 pM, from 500 aM to 10 pM, from 500 aM to 1 pM, from 750 aM to 1 nM, from 750 aM to 500 pM, from 750 aM to 200 pM, from 750 aM to 100 pM, from 750 aM to 10 pM, from 750 aM to 1 pM, from 1 fM to 1 nM, from 1 fM to 500 pM, from 1 fM to 200 pM, from 1 fM to 100 pM, from 1 fM to 10 pM, from 1 fM to 1 pM, from 500 fM to 500 pM, from 500 fM to 200 pM, from 500 fM to 100 pM, from 500 fM to 10 pM, from 500 fM to 1 pM, from 800 fM to 1 nM, from 800 fM to 500 pM, from 800 fM to 200 pM, from 800 fM to 100 pM, from 800 fM to 10 pM, from 800 fM to 1 pM, from 1 pM to 1 nM, from 1 pM to 500 pM, from 1 pM to 200 pM, from 1 pM to 100 pM, or from 1 pM to 10 pM). In some cases, a target DNA is present in a sample in a range of from 1 aM to 500 pM. In some cases, a target DNA is present in a sample in a range of from 100 aM to 500 pM.
[0306] In some cases, a subject composition or method exhibits an attomolar (aM) sensitivity of detection. In some cases, a subject composition or method exhibits a femtomolar (fM) sensitivity of detection. In some cases, a subject composition or method exhibits a picomolar (pM) sensitivity of detection. In some cases, a subject composition or method exhibits a nanomolar (nM) sensitivity of detection.
[0307] Target DNA
[0308] A target DNA can be single stranded (ssDNA) or double stranded (dsDNA). When the target DNA is single stranded, there is no preference or requirement for a PAM sequence in the target DNA. However, when the target DNA is dsDNA, a PAM is usually present adjacent to the target sequence of the target DNA (e.g., see discussion of the PAM elsewhere herein). The source of the target DNA can be the same as the source of the sample, e.g., as described below.
[0309] The source of the target DNA can be any source. In some cases, the target DNA is a viral DNA (e.g., a genomic DNA of a DNA virus). As such, subject method can be for detecting the presence of a viral DNA amongst a population of nucleic acids (e.g., in a sample). A subject method can also be used for the cleavage of non-target ssDNAs in the present of a target DNA. For example, if a method takes place in a cell, a subject method can be used to promiscuously cleave non-target ssDNAs in the cell (ssDNAs that do not hybridize with the guide sequence of the guide RNA) when a particular target DNA is present in the cell (e.g., when the cell is infected with a virus and viral target DNA is detected).
[0310] Examples of possible target DNAs include, but are not limited to, viral DNAs such as: a papovavirus (e.g., human papillomavirus (HPV), polyomavirus); a hepadnavirus (e.g., Hepatitis B Virus (HBV)); a herpesvirus (e.g., herpes simplex virus (HSV), varicella zoster virus (VZV), Epstein-Barr virus (EBV), cytomegalovirus (CMV), herpes lymphotropic virus, Pityriasis Rosea, kaposi's sarcoma-associated herpesvirus); an adenovirus (e.g., atadenovirus, aviadenovirus, ichtadenovirus, mastadenovirus, siadenovirus); a poxvirus (e.g., smallpox, vaccinia virus, cowpox virus, monkeypox virus, orf virus, pseudocowpox, bovine papular stomatitis virus; tanapox virus, yaba monkey tumor virus; molluscum contagiosum virus (MCV)); a parvovirus (e.g., adeno-associated virus (AAV), Parvovirus B19, human bocavirus, bufavirus, human parv4 G1); Geminiviridae; Nanoviridae; Phycodnaviridae; and the like. In some cases, the target DNA is parasite DNA. In some cases, the target DNA is bacterial DNA, e.g., DNA of a pathogenic bacterium.
[0311] Samples
[0312] A subject sample includes nucleic acid (e.g., a plurality of nucleic acids). The term "plurality" is used herein to mean two or more. Thus, in some cases a sample includes two or more (e.g., 3 or more, 5 or more, 10 or more, 20 or more, 50 or more, 100 or more, 500 or more, 1,000 or more, or 5,000 or more) nucleic acids (e.g., DNAs). A subject method can be used as a very sensitive way to detect a target DNA present in a sample (e.g., in a complex mixture of nucleic acids such as DNAs). In some cases, the sample includes 5 or more DNAs (e.g., 10 or more, 20 or more, 50 or more, 100 or more, 500 or more, 1,000 or more, or 5,000 or more DNAs) that differ from one another in sequence. In some cases, the sample includes 10 or more, 20 or more, 50 or more, 100 or more, 500 or more, 10.sup.3 or more, 5.times.10.sup.3 or more, 10.sup.4 or more, 5.times.10.sup.4 or more, 10.sup.5 or more, 5.times.10.sup.5 or more, 10.sup.6 or more 5.times.10.sup.6 or more, or 10.sup.7 or more, DNAs. In some cases, the sample comprises from 10 to 20, from 20 to 50, from 50 to 100, from 100 to 500, from 500 to 10.sup.3, from 10.sup.3 to 5.times.10.sup.3, from 5.times.10.sup.3 to 10.sup.4, from 10.sup.4 to 5.times.10.sup.4, from 5.times.10.sup.4 to 10.sup.5, from 10.sup.5 to 5.times.10.sup.5, from 5.times.10.sup.5 to 10.sup.6, from 10.sup.6 to 5.times.10.sup.6, or from 5.times.10.sup.6 to 10.sup.7, or more than 10.sup.7, DNAs. In some cases, the sample comprises from 5 to 10.sup.7 DNAs (e.g., that differ from one another in sequence) (e.g., from 5 to 10.sup.6, from 5 to 10.sup.5, from 5 to 50,000, from 5 to 30,000, from 10 to 10.sup.6, from 10 to 10.sup.5, from 10 to 50,000, from 10 to 30,000, from 20 to 10.sup.6, from 20 to 10.sup.5, from 20 to 50,000, or from 20 to 30,000 DNAs). In some cases, the sample includes 20 or more DNAs that differ from one another in sequence. In some cases, the sample includes DNAs from a cell lysate (e.g., a eukaryotic cell lysate, a mammalian cell lysate, a human cell lysate, a prokaryotic cell lysate, a plant cell lysate, and the like). For example, in some cases the sample includes DNA from a cell such as a eukaryotic cell, e.g., a mammalian cell such as a human cell.
[0313] The term "sample" is used herein to mean any sample that includes DNA (e.g., in order to determine whether a target DNA is present among a population of DNAs). The sample can be derived from any source, e.g., the sample can be a synthetic combination of purified DNAs; the sample can be a cell lysate, an DNA-enriched cell lysate, or DNAs isolated and/or purified from a cell lysate. The sample can be from a patient (e.g., for the purpose of diagnosis). The sample can be from permeabilized cells. The sample can be from crosslinked cells. The sample can be in tissue sections. The sample can be from tissues prepared by crosslinking followed by delipidation and adjustment to make a uniform refractive index. Examples of tissue preparation by crosslinking followed by delipidation and adjustment to make a uniform refractive index have been described in, for example, Shah et al., Development (2016) 143, 2862-2867 doi:10.1242/dev.138560.
[0314] A "sample" can include a target DNA and a plurality of non-target DNAs. In some cases, the target DNA is present in the sample at one copy per 10 non-target DNAs, one copy per 20 non-target DNAs, one copy per 25 non-target DNAs, one copy per 50 non-target DNAs, one copy per 100 non-target DNAs, one copy per 500 non-target DNAs, one copy per 10.sup.3 non-target DNAs, one copy per 5.times.10.sup.3 non-target DNAs, one copy per 10.sup.4 non-target DNAs, one copy per 5.times.10.sup.4 non-target DNAs, one copy per 10.sup.5 non-target DNAs, one copy per 5.times.10.sup.5 non-target DNAs, one copy per 10.sup.6 non-target DNAs, or less than one copy per 10.sup.6 non-target DNAs. In some cases, the target DNA is present in the sample at from one copy per 10 non-target DNAs to 1 copy per 20 non-target DNAs, from 1 copy per 20 non-target DNAs to 1 copy per 50 non-target DNAs, from 1 copy per 50 non-target DNAs to 1 copy per 100 non-target DNAs, from 1 copy per 100 non-target DNAs to 1 copy per 500 non-target DNAs, from 1 copy per 500 non-target DNAs to 1 copy per 10.sup.3 non-target DNAs, from 1 copy per 10.sup.3 non-target DNAs to 1 copy per 5.times.10.sup.3 non-target DNAs, from 1 copy per 5.times.10.sup.3 non-target DNAs to 1 copy per 10.sup.4 non-target DNAs, from 1 copy per 10.sup.4 non-target DNAs to 1 copy per 10.sup.5 non-target DNAs, from 1 copy per 10.sup.5 non-target DNAs to 1 copy per 10.sup.6 non-target DNAs, or from 1 copy per 10.sup.6 non-target DNAs to 1 copy per 10.sup.7 non-target DNAs.
[0315] Suitable samples include but are not limited to saliva, blood, serum, plasma, urine, aspirate, and biopsy samples. Thus, the term "sample" with respect to a patient encompasses blood and other liquid samples of biological origin, solid tissue samples such as a biopsy specimen or tissue cultures or cells derived therefrom and the progeny thereof. The definition also includes samples that have been manipulated in any way after their procurement, such as by treatment with reagents; washed; or enrichment for certain cell populations, such as cancer cells. The definition also includes sample that have been enriched for particular types of molecules, e.g., DNAs. The term "sample" encompasses biological samples such as a clinical sample such as blood, plasma, serum, aspirate, cerebral spinal fluid (CSF), and also includes tissue obtained by surgical resection, tissue obtained by biopsy, cells in culture, cell supernatants, cell lysates, tissue samples, organs, bone marrow, and the like. A "biological sample" includes biological fluids derived therefrom (e.g., cancerous cell, infected cell, etc.), e.g., a sample comprising DNAs that is obtained from such cells (e.g., a cell lysate or other cell extract comprising DNAs).
[0316] A sample can comprise, or can be obtained from, any of a variety of cells, tissues, organs, or acellular fluids. Suitable sample sources include eukaryotic cells, bacterial cells, and archaeal cells. Suitable sample sources include single-celled organisms and multi-cellular organisms. Suitable sample sources include single-cell eukaryotic organisms; a plant or a plant cell; an algal cell, e.g., Botryococcus braunii, Chlamydomonas reinhardtii, Nannochloropsis gaditana, Sargassum patens, C. agardh, and the like; a fungal cell (e.g., a yeast cell); an animal cell, tissue, or organ; a cell, tissue, or organ from an invertebrate animal (e.g. fruit fly, cnidarian, echinoderm, nematode, an insect, an arachnid, etc.); a cell, tissue, fluid, or organ from a vertebrate animal (e.g., fish, amphibian, reptile, bird, mammal); a cell, tissue, fluid, or organ from a mammal (e.g., a human; a non-human primate; an ungulate; a feline; a bovine; an ovine; a caprine; etc.). Suitable sample sources include nematodes, protozoans, and the like. Suitable sample sources include parasites such as helminths, malarial parasites, etc.
[0317] Suitable sample sources include a cell, tissue, or organism of any of the six kingdoms, e.g., Bacteria (e.g., Eubacteria); Archaebacteria; Protista; Fungi; Plantae; and Animalia. Suitable sample sources include plant-like members of the kingdom Protista, including, but not limited to, algae (e.g., green algae, red algae, glaucophytes, cyanobacteria); fungus-like members of Protista, e.g., slime molds, water molds, etc.; animal-like members of Protista, e.g., flagellates (e.g., Euglena), amoeboids (e.g., amoeba), sporozoans (e.g., Apicomplexa, Myxozoa, Microsporidia), and ciliates (e.g., Paramecium). Suitable sample sources include members of the kingdom Fungi, including, but not limited to, members of any of the phyla: Basidiomycota (club fungi; e.g., members of Agaricus, Amanita, Boletus, Cantherellus, etc.); Ascomycota (sac fungi, including, e.g., Saccharomyces); Mycophycophyta (lichens); Zygomycota (conjugation fungi); and Deuteromycota. Suitable sample sources include members of the kingdom Plantae, including, but not limited to, members of any of the following divisions: Bryophyta (e.g., mosses), Anthocerotophyta (e.g., hornworts), Hepaticophyta (e.g., liverworts), Lycophyta (e.g., club mosses), Sphenophyta (e.g., horsetails), Psilophyta (e.g., whisk ferns), Ophioglossophyta, Pterophyta (e.g., ferns), Cycadophyta, Gingkophyta, Pinophyta, Gnetophyta, and Magnoliophyta (e.g., flowering plants). Suitable sample sources include members of the kingdom Animalia, including, but not limited to, members of any of the following phyla: Porifera (sponges); Placozoa; Orthonectida (parasites of marine invertebrates); Rhombozoa; Cnidaria (corals, anemones, jellyfish, sea pens, sea pansies, sea wasps); Ctenophora (comb jellies); Platyhelminthes (flatworms); Nemertina (ribbon worms); Ngathostomulida (jawed worms)p Gastrotricha; Rotifera; Priapulida; Kinorhyncha; Loricifera; Acanthocephala; Entoprocta; Nemotoda; Nematomorpha; Cycliophora; Mollusca (mollusks); Sipuncula (peanut worms); Annelida (segmented worms); Tardigrada (water bears); Onychophora (velvet worms); Arthropoda (including the subphyla: Chelicerata, Myriapoda, Hexapoda, and Crustacea, where the Chelicerata include, e.g., arachnids, Merostomata, and Pycnogonida, where the Myriapoda include, e.g., Chilopoda (centipedes), Diplopoda (millipedes), Paropoda, and Symphyla, where the Hexapoda include insects, and where the Crustacea include shrimp, krill, barnacles, etc.; Phoronida; Ectoprocta (moss animals); Brachiopoda; Echinodermata (e.g. starfish, sea daisies, feather stars, sea urchins, sea cucumbers, brittle stars, brittle baskets, etc.); Chaetognatha (arrow worms); Hemichordata (acorn worms); and Chordata. Suitable members of Chordata include any member of the following subphyla: Urochordata (sea squirts; including Ascidiacea, Thaliacea, and Larvacea); Cephalochordata (lancelets); Myxini (hagfish); and Vertebrata, where members of Vertebrata include, e.g., members of Petromyzontida (lampreys), Chondrichthyces (cartilaginous fish), Actinopterygii (ray-finned fish), Actinista (coelocanths), Dipnoi (lungfish), Reptilia (reptiles, e.g., snakes, alligators, crocodiles, lizards, etc.), Aves (birds); and Mammalian (mammals). Suitable plants include any monocotyledon and any dicotyledon.
[0318] Suitable sources of a sample include cells, fluid, tissue, or organ taken from an organism; from a particular cell or group of cells isolated from an organism; etc. For example, where the organism is a plant, suitable sources include xylem, the phloem, the cambium layer, leaves, roots, etc. Where the organism is an animal, suitable sources include particular tissues (e.g., lung, liver, heart, kidney, brain, spleen, skin, fetal tissue, etc.), or a particular cell type (e.g., neuronal cells, epithelial cells, endothelial cells, astrocytes, macrophages, glial cells, islet cells, T lymphocytes, B lymphocytes, etc.).
[0319] In some cases, the source of the sample is a (or is suspected of being a diseased cell, fluid, tissue, or organ. In some cases, the source of the sample is a normal (non-diseased) cell, fluid, tissue, or organ. In some cases, the source of the sample is a (or is suspected of being a pathogen-infected cell, tissue, or organ. For example, the source of a sample can be an individual who may or may not be infected--and the sample could be any biological sample (e.g., blood, saliva, biopsy, plasma, serum, bronchoalveolar lavage, sputum, a fecal sample, cerebrospinal fluid, a fine needle aspirate, a swab sample (e.g., a buccal swab, a cervical swab, a nasal swab), interstitial fluid, synovial fluid, nasal discharge, tears, buffy coat, a mucous membrane sample, an epithelial cell sample (e.g., epithelial cell scraping), etc.) collected from the individual. In some cases, the sample is a cell-free liquid sample. In some cases, the sample is a liquid sample that can comprise cells. Pathogens include viruses, fungi, helminths, protozoa, malarial parasites, Plasmodium parasites, Toxoplasma parasites, Schistosoma parasites, and the like. "Helminths" include roundworms, heartworms, and phytophagous nematodes (Nematoda), flukes (Tematoda), Acanthocephala, and tapeworms (Cestoda). Protozoan infections include infections from Giardia spp., Trichomonas spp., African trypanosomiasis, amoebic dysentery, babesiosis, balantidial dysentery, Chaga's disease, coccidiosis, malaria and toxoplasmosis. Examples of pathogens such as parasitic/protozoan pathogens include, but are not limited to: Plasmodium falciparum, Plasmodium vivax, Trypanosoma cruzi and Toxoplasma gondii. Fungal pathogens include, but are not limited to: Cryptococcus neoformans, Histoplasma capsulatum, Coccidioides immitis, Blastomyces dermatitidis, Chlamydia trachomatis, and Candida albicans. Pathogenic viruses include, e.g., immunodeficiency virus (e.g., HIV); influenza virus; dengue; West Nile virus; herpes virus; yellow fever virus; Hepatitis Virus C; Hepatitis Virus A; Hepatitis Virus B; papillomavirus; and the like. Pathogenic viruses can include DNA viruses such as: a papovavirus (e.g., human papillomavirus (HPV), polyomavirus); a hepadnavirus (e.g., Hepatitis B Virus (HBV)); a herpesvirus (e.g., herpes simplex virus (HSV), varicella zoster virus (VZV), epstein-barr virus (EBV), cytomegalovirus (CMV), herpes lymphotropic virus, Pityriasis Rosea, kaposi's sarcoma-associated herpesvirus); an adenovirus (e.g., atadenovirus, aviadenovirus, ichtadenovirus, mastadenovirus, siadenovirus); a poxvirus (e.g., smallpox, vaccinia virus, cowpox virus, monkeypox virus, orf virus, pseudocowpox, bovine papular stomatitis virus; tanapox virus, yaba monkey tumor virus; molluscum contagiosum virus (MCV)); a parvovirus (e.g., adeno-associated virus (AAV), Parvovirus B19, human bocavirus, bufavirus, human parv4 G1); Geminiviridae; Nanoviridae; Phycodnaviridae; and the like. Pathogens can include, e.g., DNAviruses [e.g.: a papovavirus (e.g., human papillomavirus (HPV), polyomavirus); a hepadnavirus (e.g., Hepatitis B Virus (HBV)); a herpesvirus (e.g., herpes simplex virus (HSV), varicella zoster virus (VZV), Epstein-barr virus (EBV), cytomegalovirus (CMV), herpes lymphotropic virus, Pityriasis Rosea, kaposi's sarcoma-associated herpesvirus); an adenovirus (e.g., atadenovirus, aviadenovirus, ichtadenovirus, mastadenovirus, siadenovirus); a poxvirus (e.g., smallpox, vaccinia virus, cowpox virus, monkeypox virus, orf virus, pseudocowpox, bovine papular stomatitis virus; tanapox virus, yaba monkey tumor virus; molluscum contagiosum virus (MCV)); a parvovirus (e.g., adeno-associated virus (AAV), Parvovirus B19, human bocavirus, bufavirus, human parv4 G1); Geminiviridae; Nanoviridae; Phycodnaviridae; and the like], Mycobacterium tuberculosis, Streptococcus agalactiae, methicillin-resistant Staphylococcus aureus, Legionella pneumophila, Streptococcus pyogenes, Escherichia coli, Neisseria gonorrhoeae, Neisseria meningitidis, Pneumococcus, Cryptococcus neoformans, Histoplasma capsulatum, Hemophilus influenzae B, Treponema pallidum, Lyme disease spirochetes, Pseudomonas aeruginosa, Mycobacterium leprae, Brucella abortus, rabies virus, influenza virus, cytomegalovirus, herpes simplex virus I, herpes simplex virus II, human serum parvo-like virus, respiratory syncytial virus, varicella-zoster virus, hepatitis B virus, hepatitis C virus, measles virus, adenovirus, human T-cell leukemia viruses, Epstein-Barr virus, murine leukemia virus, mumps virus, vesicular stomatitis virus, Sindbis virus, lymphocytic choriomeningitis virus, wart virus, blue tongue virus, Sendai virus, feline leukemia virus, Reovirus, polio virus, simian virus 40, mouse mammary tumor virus, dengue virus, rubella virus, West Nile virus, Plasmodium falciparum, Plasmodium vivax, Toxoplasma gondii, Trypanosoma rangeli, Trypanosoma cruzi, Trypanosoma rhodesiense, Trypanosoma brucei, Schistosoma mansoni, Schistosoma japonicum, Babesia bovis, Eimeria tenella, Onchocerca volvulus, Leishmania tropica, Mycobacterium tuberculosis, Trichinella spiralis, Theileria parva, Taenia hydatigena, Taenia ovis, Taenia saginata, Echinococcus granulosus, Mesocestoides corti, Mycoplasma arthritidis, M. hyorhinis, M. orale, M. arginini, Acholeplasma laidlawii, M. salivarium and M. pneumoniae.
[0320] Measuring a Detectable Signal
[0321] In some cases, a subject method includes a step of measuring (e.g., measuring a detectable signal produced by CasZ-mediated ssDNA cleavage). Because a CasZ polypeptide cleaves non-targeted ssDNA once activated, which occurs when a guide RNA hybridizes with a target DNA in the presence of a CasZ polypeptide (and, in some cases, also including a tranc RNA), a detectable signal can be any signal that is produced when ssDNA is cleaved. For example, in some cases the step of measuring can include one or more of: gold nanoparticle based detection (e.g., see Xu et al., Angew Chem Int Ed Engl. 2007; 46(19):3468-70; and Xia et al., Proc Natl Acad Sci USA. 2010 Jun. 15; 107(24):10837-41), fluorescence polarization, colloid phase transition/dispersion (e.g., Baksh et al., Nature. 2004 Jan. 8; 427(6970):139-41), electrochemical detection, semiconductor-based sensing (e.g., Rothberg et al., Nature. 2011 Jul. 20; 475(7356):348-52; e.g., one could use a phosphatase to generate a pH change after ssDNA cleavage reactions, by opening 2'-3' cyclic phosphates, and by releasing inorganic phosphate into solution), and detection of a labeled detector ssDNA (see elsewhere herein for more details). The readout of such detection methods can be any convenient readout. Examples of possible readouts include but are not limited to: a measured amount of detectable fluorescent signal; a visual analysis of bands on a gel (e.g., bands that represent cleaved product versus uncleaved substrate), a visual or sensor based detection of the presence or absence of a color (i.e., color detection method), and the presence or absence of (or a particular amount of) an electrical signal.
[0322] The measuring can in some cases be quantitative, e.g., in the sense that the amount of signal detected can be used to determine the amount of target DNA present in the sample. The measuring can in some cases be qualitative, e.g., in the sense that the presence or absence of detectable signal can indicate the presence or absence of targeted DNA (e.g., virus, SNP, etc.). In some cases, a detectable signal will not be present (e.g., above a given threshold level) unless the targeted DNA(s) (e.g., virus, SNP, etc.) is present above a particular threshold concentration. In some cases, the threshold of detection can be titrated by modifying the amount of CasZ polypeptide, guide RNA, sample volume, and/or detector ssDNA (if one is used). As such, for example, as would be understood by one of ordinary skill in the art, a number of controls can be used if desired in order to set up one or more reactions, each set up to detect a different threshold level of target DNA, and thus such a series of reactions could be used to determine the amount of target DNA present in a sample (e.g., one could use such a series of reactions to determine that a target DNA is present in the sample `at a concentration of at least X`). Non-limiting examples of applications of/uses for the compositions and methods of the disclosure include single-nucleotide polymorphism (SNP) detection, cancer screening, detection of bacterial infection, detection of antibiotic resistance, detection of viral infection, and the like. The compositions and methods of this disclosure can be used to detect any DNA target. For example, any virus that integrates nucleic acid material into the genome can be detected because a subject sample can include cellular genomic DNA--and the guide RNA can be designed to detect integrated nucleotide sequence. A method of the present disclosure in some cases does not include an amplification step. A method of the present disclosure in some cases includes an amplification step.
[0323] In some cases, a method of the present disclosure can be used to determine the amount of a target DNA in a sample (e.g., a sample comprising the target DNA and a plurality of non-target DNAs). Determining the amount of a target DNA in a sample can comprise comparing the amount of detectable signal generated from a test sample to the amount of detectable signal generated from a reference sample. Determining the amount of a target DNA in a sample can comprise: measuring the detectable signal to generate a test measurement; measuring a detectable signal produced by a reference sample to generate a reference measurement; and comparing the test measurement to the reference measurement to determine an amount of target DNA present in the sample.
[0324] For example, in some cases, a method of the present disclosure for determining the amount of a target DNA in a sample comprises: a) contacting the sample (e.g., a sample comprising the target DNA and a plurality of non-target DNAs) with: (i) a guide RNA that hybridizes with the target DNA, (ii) a CasZ polypeptide that cleaves DNAs present in the sample, and (iii) a detector ssDNA; b) measuring a detectable signal produced by CasZ polypeptide-mediated ssDNA cleavage (e.g., cleavage of the detector ssDNA), generating a test measurement; c) measuring a detectable signal produced by a reference sample to generate a reference measurement; and d) comparing the test measurement to the reference measurement to determine an amount of target DNA present in the sample.
[0325] As another example, in some cases, a method of the present disclosure for determining the amount of a target DNA in a sample comprises: a) contacting the sample (e.g., a sample comprising the target DNA and a plurality of non-target DNAs) with: (i) a guide RNA that hybridizes with the target DNA, (ii) a CasZ polypeptide that cleaves DNAs present in the sample, (iii) a tranc RNA; (iv) a detector ssDNA; b) measuring a detectable signal produced by CasZ polypeptide-mediated ssDNA cleavage (e.g., cleavage of the detector ssDNA), generating a test measurement; c) measuring a detectable signal produced by a reference sample to generate a reference measurement; and d) comparing the test measurement to the reference measurement to determine an amount of target DNA present in the sample.
[0326] Amplification of Nucleic Acids in the Sample
[0327] In some embodiments, sensitivity of a subject composition and/or method (e.g., for detecting the presence of a target DNA, such as viral DNA or a SNP, in cellular genomic DNA) can be increased by coupling detection with nucleic acid amplification. In some cases, the nucleic acids in a sample are amplified prior to contact with a CasZ polypeptide that cleaves ssDNA (e.g., amplification of nucleic acids in the sample can begin prior to contact with a CasZ polypeptide). In some cases, the nucleic acids in a sample are amplified simultaneous with contact with a CasZ polypeptide. For example, in some cases a subject method includes amplifying nucleic acids of a sample (e.g., by contacting the sample with amplification components) prior to contacting the amplified sample with a CasZ polypeptide. In some cases, a subject method includes contacting a sample with amplification components at the same time (simultaneous with) that the sample is contacted with a CasZ polypeptide. If all components are added simultaneously (amplification components and detection components such as a CasZ polypeptide, a guide RNA, and a detector DNA), it is possible that the trans-cleavage activity of the CasZ polypeptide, will begin to degrade the nucleic acids of the sample at the same time the nucleic acids are undergoing amplification. However, even if this is the case, amplifying and detecting simultaneously can still increase sensitivity compared to performing the method without amplification.
[0328] In some cases, specific sequences (e.g., sequences of a virus, sequences that include a SNP of interest) are amplified from the sample, e.g., using primers. As such, a sequence to which the guide RNA will hybridize can be amplified in order to increase sensitivity of a subject detection method--this could achieve biased amplification of a desired sequence in order to increase the number of copies of the sequence of interest present in the sample relative to other sequences present in the sample. As one illustrative example, if a subject method is being used to determine whether a given sample includes a particular virus (or a particular SNP), a desired region of viral sequence (or non-viral genomic sequence) can be amplified, and the region amplified will include the sequence that would hybridize to the guide RNA if the viral sequence (or SNP) were in fact present in the sample.
[0329] As noted, in some cases the nucleic acids are amplified (e.g., by contact with amplification components) prior to contacting the amplified nucleic acids with a CasZ polypeptide. In some cases, amplification occurs for 10 seconds or more, (e.g., 30 seconds or more, 45 seconds or more, 1 minute or more, 2 minutes or more, 3 minutes or more, 4 minutes or more, 5 minutes or more, 7.5 minutes or more, 10 minutes or more, etc.) prior to contact with an enzymatically active CasZ polypeptide. In some cases, amplification occurs for 2 minutes or more (e.g., 3 minutes or more, 4 minutes or more, 5 minutes or more, 7.5 minutes or more, 10 minutes or more, etc.) prior to contact with an active CasZ polypeptide. In some cases, amplification occurs for a period of time in a range of from 10 seconds to 60 minutes (e.g., 10 seconds to 40 minutes, 10 seconds to 30 minutes, 10 seconds to 20 minutes, 10 seconds to 15 minutes, 10 seconds to 10 minutes, 10 seconds to 5 minutes, 30 seconds to 40 minutes, 30 seconds to 30 minutes, 30 seconds to 20 minutes, 30 seconds to 15 minutes, 30 seconds to 10 minutes, 30 seconds to 5 minutes, 1 minute to 40 minutes, 1 minute to 30 minutes, 1 minute to 20 minutes, 1 minute to 15 minutes, 1 minute to 10 minutes, 1 minute to 5 minutes, 2 minutes to 40 minutes, 2 minutes to 30 minutes, 2 minutes to 20 minutes, 2 minutes to 15 minutes, 2 minutes to 10 minutes, 2 minutes to 5 minutes, 5 minutes to 40 minutes, 5 minutes to 30 minutes, 5 minutes to 20 minutes, 5 minutes to 15 minutes, or 5 minutes to 10 minutes). In some cases, amplification occurs for a period of time in a range of from 5 minutes to 15 minutes. In some cases, amplification occurs for a period of time in a range of from 7 minutes to 12 minutes.
[0330] In some cases, a sample is contacted with amplification components at the same time as contact with a CasZ polypeptide. In some such cases, the CasZ polypeptide is inactive at the time of contact and is activated once nucleic acids in the sample have been amplified.
[0331] Various amplification methods and components will be known to one of ordinary skill in the art and any convenient method can be used (see, e.g., Zanoli and Spoto, Biosensors (Basel). 2013 March; 3(1): 18-43; Gill and Ghaemi, Nucleosides, Nucleotides, and Nucleic Acids, 2008, 27: 224-243; Craw and Balachandrana, Lab Chip, 2012, 12, 2469-2486; which are herein incorporated by reference in their entirety). Nucleic acid amplification can comprise polymerase chain reaction (PCR), reverse transcription PCR (RT-PCR), quantitative PCR (qPCR), reverse transcription qPCR (RT-qPCR), nested PCR, multiplex PCR, asymmetric PCR, touchdown PCR, random primer PCR, hemi-nested PCR, polymerase cycling assembly (PCA), colony PCR, ligase chain reaction (LCR), digital PCR, methylation specific-PCR (MSP), co-amplification at lower denaturation temperature-PCR (COLD-PCR), allele-specific PCR, intersequence-specific PCR (ISS-PCR), whole genome amplification (WGA), inverse PCR, and thermal asymmetric interlaced PCR (TAIL-PCR).
[0332] In some cases, the amplification is isothermal amplification. The term "isothermal amplification" indicates a method of nucleic acid (e.g., DNA) amplification (e.g., using enzymatic chain reaction) that can use a single temperature incubation thereby obviating the need for a thermal cycler. Isothermal amplification is a form of nucleic acid amplification which does not rely on the thermal denaturation of the target nucleic acid during the amplification reaction and hence may not require multiple rapid changes in temperature. Isothermal nucleic acid amplification methods can therefore be carried out inside or outside of a laboratory environment. By combining with a reverse transcription step, these amplification methods can be used to isothermally amplify RNA.
[0333] Examples of isothermal amplification methods include but are not limited to: loop-mediated isothermal Amplification (LAMP), helicase-dependent Amplification (HDA), recombinase polymerase amplification (RPA), strand displacement amplification (SDA), nucleic acid sequence-based amplification (NASBA), transcription mediated amplification (TMA), nicking enzyme amplification reaction (NEAR), rolling circle amplification (RCA), multiple displacement amplification (MDA), Ramification (RAM), circular helicase-dependent amplification (cHDA), single primer isothermal amplification (SPIA), signal mediated amplification of RNA technology (SMART), self-sustained sequence replication (3SR), genome exponential amplification reaction (GEAR) and isothermal multiple displacement amplification (IMDA).
[0334] In some cases, the amplification is recombinase polymerase amplification (RPA) (see, e.g., U.S. Pat. Nos. 8,030,000; 8,426,134; 8,945,845; 9,309,502; and 9,663,820, which are hereby incorporated by reference in their entirety). Recombinase polymerase amplification (RPA) uses two opposing primers (much like PCR) and employs three enzymes--a recombinase, a single-stranded DNA-binding protein (SSB) and a strand-displacing polymerase. The recombinase pairs oligonucleotide primers with homologous sequence in duplex DNA, SSB binds to displaced strands of DNA to prevent the primers from being displaced, and the strand displacing polymerase begins DNA synthesis where the primer has bound to the target DNA. Adding a reverse transcriptase enzyme to an RPA reaction can facilitate detection RNA as well as DNA, without the need for a separate step to produce cDNA. One example of components for an RPA reaction is as follows (see, e.g., U.S. Pat. Nos. 8,030,000; 8,426,134; 8,945,845; 9,309,502; 9,663,820): 50 mM Tris pH 8.4, 80 mM Potassium actetate, 10 mM Magnesium acetate, 2 mM DTT, 5% PEG compound (Carbowax-20M), 3 mM ATP, 30 mM Phosphocreatine, 100 ng/.mu.l creatine kinase, 420 ng/.mu.l gp32, 140 ng/.mu.l UvsX, 35 ng/.mu.l UvsY, 2000M dNTPs, 300 nM each oligonucleotide, 35 ng/.mu.l Bsu polymerase, and a nucleic acid-containing sample).
[0335] In a transcription mediated amplification (TMA), an RNA polymerase is used to make RNA from a promoter engineered in the primer region, and then a reverse transcriptase synthesizes cDNA from the primer. A third enzyme, e.g., Rnase H can then be used to degrade the RNA target from cDNA without the heat-denatured step. This amplification technique is similar to Self-Sustained Sequence Replication (3SR) and Nucleic Acid Sequence Based Amplification (NASBA), but varies in the enzymes employed. For another example, helicase-dependent amplification (HDA) utilizes a thermostable helicase (Tte-UvrD) rather than heat to unwind dsDNA to create single-strands that are then available for hybridization and extension of primers by polymerase. For yet another example, a loop mediated amplification (LAMP) employs a thermostable polymerase with strand displacement capabilities and a set of four or more specific designed primers. Each primer is designed to have hairpin ends that, once displaced, snap into a hairpin to facilitate self-priming and further polymerase extension. In a LAMP reaction, though the reaction proceeds under isothermal conditions, an initial heat denaturation step is required for double-stranded targets. In addition, amplification yields a ladder pattern of various length products. For yet another example, a strand displacement amplification (SDA) combines the ability of a restriction endonuclease to nick the unmodified strand of its target DNA and an exonuclease-deficient DNA polymerase to extend the 3' end at the nick and displace the downstream DNA strand.
[0336] Detector DNA
[0337] In some cases, a subject method includes contacting a sample (e.g., a sample comprising a target DNA and a plurality of non-target ssDNAs) with: i) a CasZ polypeptide; ii) a guide RNA; and iii) a detector DNA that is single stranded and does not hybridize with the guide sequence of the guide RNA.
[0338] A suitable single-stranded detector DNA has a length of from 7 nucleotides to 25 nucleotides. For example, a suitable single-stranded detector DNA has a length of from 7 nucleotides to 10 nucleotides, from 11 nucleotides to 15 nucleotides, from 15 nucleotides to 20 nucleotides, or from 20 nucleotides to 25 nucleotides. In some cases, a suitable single-stranded detector DNA has a length of from 10 nucleotides to 15 nucleotides. In some cases, a suitable single-stranded detector DNA has a length of 10 nucleotides. In some cases, a suitable single-stranded detector DNA has a length of 11 nucleotides. In some cases, a suitable single-stranded detector DNA has a length of 12 nucleotides. In some cases, a suitable single-stranded detector DNA has a length of 13 nucleotides. In some cases, a suitable single-stranded detector DNA has a length of 14 nucleotides. In some cases, a suitable single-stranded detector DNA has a length of 15 nucleotides.
[0339] In some cases, a subject method includes: a) contacting a sample with a labeled single stranded detector DNA (detector ssDNA) that includes a fluorescence-emitting dye pair; a CasZ polypeptide that cleaves the labeled detector ssDNA after it is activated (by binding to the guide RNA in the context of the guide RNA hybridizing to a target DNA); and b) measuring the detectable signal that is produced by the fluorescence-emitting dye pair. For example, in some cases, a subject method includes contacting a sample with a labeled detector ssDNA comprising a fluorescence resonance energy transfer (FRET) pair or a quencher/fluor pair, or both. In some cases, a subject method includes contacting a sample with a labeled detector ssDNA comprising a FRET pair. In some cases, a subject method includes contacting a sample with a labeled detector ssDNA comprising a fluor/quencher pair.
[0340] Fluorescence-emitting dye pairs comprise a FRET pair or a quencher/fluor pair. In both cases of a FRET pair and a quencher/fluor pair, the emission spectrum of one of the dyes overlaps a region of the absorption spectrum of the other dye in the pair. As used herein, the term "fluorescence-emitting dye pair" is a generic term used to encompass both a "fluorescence resonance energy transfer (FRET) pair" and a "quencher/fluor pair," both of which terms are discussed in more detail below. The term "fluorescence-emitting dye pair" is used interchangeably with the phrase "a FRET pair and/or a quencher/fluor pair."
[0341] In some cases (e.g., when the detector ssDNA includes a FRET pair) the labeled detector ssDNA produces an amount of detectable signal prior to being cleaved, and the amount of detectable signal that is measured is reduced when the labeled detector ssDNA is cleaved. In some cases, the labeled detector ssDNA produces a first detectable signal prior to being cleaved (e.g., from a FRET pair) and a second detectable signal when the labeled detector ssDNA is cleaved (e.g., from a quencher/fluor pair). As such, in some cases, the labeled detector ssDNA comprises a FRET pair and a quencher/fluor pair.
[0342] In some cases, the labeled detector ssDNA comprises a FRET pair. FRET is a process by which radiationless transfer of energy occurs from an excited state fluorophore to a second chromophore in close proximity. The range over which the energy transfer can take place is limited to approximately 10 nanometers (100 angstroms), and the efficiency of transfer is extremely sensitive to the separation distance between fluorophores. Thus, as used herein, the term "FRET" ("fluorescence resonance energy transfer"; also known as "Forster resonance energy transfer") refers to a physical phenomenon involving a donor fluorophore and a matching acceptor fluorophore selected so that the emission spectrum of the donor overlaps the excitation spectrum of the acceptor, and further selected so that when donor and acceptor are in close proximity (usually 10 nm or less) to one another, excitation of the donor will cause excitation of and emission from the acceptor, as some of the energy passes from donor to acceptor via a quantum coupling effect. Thus, a FRET signal serves as a proximity gauge of the donor and acceptor; only when they are in close proximity to one another is a signal generated. The FRET donor moiety (e.g., donor fluorophore) and FRET acceptor moiety (e.g., acceptor fluorophore) are collectively referred to herein as a "FRET pair".
[0343] The donor-acceptor pair (a FRET donor moiety and a FRET acceptor moiety) is referred to herein as a "FRET pair" or a "signal FRET pair." Thus, in some cases, a subject labeled detector ssDNA includes two signal partners (a signal pair), when one signal partner is a FRET donor moiety and the other signal partner is a FRET acceptor moiety. A subject labeled detector ssDNA that includes such a FRET pair (a FRET donor moiety and a FRET acceptor moiety) will thus exhibit a detectable signal (a FRET signal) when the signal partners are in close proximity (e.g., while on the same RNA molecule), but the signal will be reduced (or absent) when the partners are separated (e.g., after cleavage of the RNA molecule by a CasZ polypeptide).
[0344] FRET donor and acceptor moieties (FRET pairs) will be known to one of ordinary skill in the art and any convenient FRET pair (e.g., any convenient donor and acceptor moiety pair) can be used. Examples of suitable FRET pairs include but are not limited to those presented in Table 1. See also: Bajar et al. Sensors (Basel). 2016 Sep. 14; 16(9); and Abraham et al. PLoS One. 2015 Aug. 3; 10(8):e0134436.
TABLE-US-00004 TABLE 6 Examples of FRET pairs (donor and acceptor FRET moieties) Donor Acceptor Tryptophan Dansyl IAEDANS (1) DDPM (2) BFP DsRFP Dansyl Fluorescein isothiocyanate (FITC) Dansyl Octadecylrhodamine Cyan fluorescent Green fluorescent protein protein (CFP) (GFP) CF (3) Texas Red Fluorescein Tetramethylrhodamine Cy3 Cy5 GFP Yellow fluorescent protein (YFP) BODIPY FL (4) BODIPY FL (4) Rhodamine 110 Cy3 Rhodamine 6G Malachite Green FITC Eosin Thiosemicarbazide B-Phycoerythrin Cy5 Cy5 Cy5.5 (1) 5-(2-iodoacetylaminoethyl)aminonaphthalene-1-sulfonic acid (2) N-(4-dimethylamino-3,5-dinitrophenyl)maleimide (3) carboxyfluorescein succinimidyl ester (4) 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene
[0345] In some cases, a detectable signal is produced when the labeled detector ssDNA is cleaved (e.g., in some cases, the labeled detector ssDNA comprises a quencher/fluor pair). One signal partner of a signal quenching pair produces a detectable signal and the other signal partner is a quencher moiety that quenches the detectable signal of the first signal partner (i.e., the quencher moiety quenches the signal of the signal moiety such that the signal from the signal moiety is reduced (quenched) when the signal partners are in proximity to one another, e.g., when the signal partners of the signal pair are in close proximity).
[0346] For example, in some cases, an amount of detectable signal increases when the labeled detector ssDNA is cleaved. For example, in some cases, the signal exhibited by one signal partner (a signal moiety) is quenched by the other signal partner (a quencher signal moiety), e.g., when both are present on the same ssDNA molecule prior to cleavage by a CasZ polypeptide. Such a signal pair is referred to herein as a "quencher/fluor pair", "quenching pair", or "signal quenching pair." For example, in some cases, one signal partner (e.g., the first signal partner) is a signal moiety that produces a detectable signal that is quenched by the second signal partner (e.g., a quencher moiety). The signal partners of such a quencher/fluor pair will thus produce a detectable signal when the partners are separated (e.g., after cleavage of the detector ssDNA by a CasZ polypeptide), but the signal will be quenched when the partners are in close proximity (e.g., prior to cleavage of the detector ssDNA by a CasZ polypeptide).
[0347] A quencher moiety can quench a signal from the signal moiety (e.g., prior to cleavage of the detector ssDNA by a CasZ polypeptide) to various degrees. In some cases, a quencher moiety quenches the signal from the signal moiety where the signal detected in the presence of the quencher moiety (when the signal partners are in proximity to one another) is 95% or less of the signal detected in the absence of the quencher moiety (when the signal partners are separated). For example, in some cases, the signal detected in the presence of the quencher moiety can be 90% or less, 80% or less, 70% or less, 60% or less, 50% or less, 40% or less, 30% or less, 20% or less, 15% or less, 10% or less, or 5% or less of the signal detected in the absence of the quencher moiety. In some cases, no signal (e.g., above background) is detected in the presence of the quencher moiety.
[0348] In some cases, the signal detected in the absence of the quencher moiety (when the signal partners are separated) is at least 1.2 fold greater (e.g., at least 1.3 fold, at least 1.5 fold, at least 1.7 fold, at least 2 fold, at least 2.5 fold, at least 3 fold, at least 3.5 fold, at least 4 fold, at least 5 fold, at least 7 fold, at least 10 fold, at least 20 fold, or at least 50 fold greater) than the signal detected in the presence of the quencher moiety (when the signal partners are in proximity to one another).
[0349] In some cases, the signal moiety is a fluorescent label. In some such cases, the quencher moiety quenches the signal (the light signal) from the fluorescent label (e.g., by absorbing energy in the emission spectra of the label). Thus, when the quencher moiety is not in proximity with the signal moiety, the emission (the signal) from the fluorescent label is detectable because the signal is not absorbed by the quencher moiety. Any convenient donor acceptor pair (signal moiety/quencher moiety pair) can be used and many suitable pairs are known in the art.
[0350] In some cases, the quencher moiety absorbs energy from the signal moiety (also referred to herein as a "detectable label") and then emits a signal (e.g., light at a different wavelength). Thus, in some cases, the quencher moiety is itself a signal moiety (e.g., a signal moiety can be 6-carboxyfluorescein while the quencher moiety can be 6-carboxy-tetramethylrhodamine), and in some such cases, the pair could also be a FRET pair. In some cases, a quencher moiety is a dark quencher. A dark quencher can absorb excitation energy and dissipate the energy in a different way (e.g., as heat). Thus, a dark quencher has minimal to no fluorescence of its own (does not emit fluorescence). Examples of dark quenchers are further described in U.S. Pat. Nos. 8,822,673 and 8,586,718; U.S. patent publications 20140378330, 20140349295, and 20140194611; and international patent applications: WO200142505 and WO200186001, all if which are hereby incorporated by reference in their entirety.
[0351] Examples of fluorescent labels include, but are not limited to: an Alexa Fluor.RTM. dye, an ATTO dye (e.g., ATTO 390, ATTO 425, ATTO 465, ATTO 488, ATTO 495, ATTO 514, ATTO 520, ATTO 532, ATTO Rho6G, ATTO 542, ATTO 550, ATTO 565, ATTO Rho3B, ATTO Rho11, ATTO Rho12, ATTO Thio12, ATTO Rho101, ATTO 590, ATTO 594, ATTO Rho13, ATTO 610, ATTO 620, ATTO Rho14, ATTO 633, ATTO 647, ATTO 647N, ATTO 655, ATTO Oxa12, ATTO 665, ATTO 680, ATTO 700, ATTO 725, ATTO 740), a DyLight dye, a cyanine dye (e.g., Cy2, Cy3, Cy3.5, Cy3b, Cy5, Cy5.5, Cy7, Cy7.5), a FluoProbes dye, a Sulfo Cy dye, a Seta dye, an IRIS Dye, a SeTau dye, an SRfluor dye, a Square dye, fluorescein isothiocyanate (FITC), tetramethylrhodamine (TRITC), Texas Red, Oregon Green, Pacific Blue, Pacific Green, Pacific Orange, quantum dots, and a tethered fluorescent protein.
[0352] In some cases, a detectable label is a fluorescent label selected from: an Alexa Fluor.RTM. dye, an ATTO dye (e.g., ATTO 390, ATTO 425, ATTO 465, ATTO 488, ATTO 495, ATTO 514, ATTO 520, ATTO 532, ATTO Rho6G, ATTO 542, ATTO 550, ATTO 565, ATTO Rho3B, ATTO Rho11, ATTO Rho12, ATTO Thio12, ATTO Rho101, ATTO 590, ATTO 594, ATTO Rho13, ATTO 610, ATTO 620, ATTO Rho14, ATTO 633, ATTO 647, ATTO 647N, ATTO 655, ATTO Oxa12, ATTO 665, ATTO 680, ATTO 700, ATTO 725, ATTO 740), a DyLight dye, a cyanine dye (e.g., Cy2, Cy3, Cy3.5, Cy3b, Cy5, Cy5.5, Cy7, Cy7.5), a FluoProbes dye, a Sulfo Cy dye, a Seta dye, an IRIS Dye, a SeTau dye, an SRfluor dye, a Square dye, fluorescein (FITC), tetramethylrhodamine (TRITC), Texas Red, Oregon Green, Pacific Blue, Pacific Green, and Pacific Orange.
[0353] In some cases, a detectable label is a fluorescent label selected from: an Alexa Fluor.RTM. dye, an ATTO dye (e.g., ATTO 390, ATTO 425, ATTO 465, ATTO 488, ATTO 495, ATTO 514, ATTO 520, ATTO 532, ATTO Rho6G, ATTO 542, ATTO 550, ATTO 565, ATTO Rho3B, ATTO Rho11, ATTO Rho12, ATTO Thio12, ATTO Rho101, ATTO 590, ATTO 594, ATTO Rho13, ATTO 610, ATTO 620, ATTO Rho14, ATTO 633, ATTO 647, ATTO 647N, ATTO 655, ATTO Oxa12, ATTO 665, ATTO 680, ATTO 700, ATTO 725, ATTO 740), a DyLight dye, a cyanine dye (e.g., Cy2, Cy3, Cy3.5, Cy3b, Cy5, Cy5.5, Cy7, Cy7.5), a FluoProbes dye, a Sulfo Cy dye, a Seta dye, an IRIS Dye, a SeTau dye, an SRfluor dye, a Square dye, fluorescein (FITC), tetramethylrhodamine (TRITC), Texas Red, Oregon Green, Pacific Blue, Pacific Green, Pacific Orange, a quantum dot, and a tethered fluorescent protein.
[0354] Examples of ATTO dyes include, but are not limited to: ATTO 390, ATTO 425, ATTO 465, ATTO 488, ATTO 495, ATTO 514, ATTO 520, ATTO 532, ATTO Rho6G, ATTO 542, ATTO 550, ATTO 565, ATTO Rho3B, ATTO Rho11, ATTO Rho12, ATTO Thio12, ATTO Rho101, ATTO 590, ATTO 594, ATTO Rho13, ATTO 610, ATTO 620, ATTO Rho14, ATTO 633, ATTO 647, ATTO 647N, ATTO 655, ATTO Oxa12, ATTO 665, ATTO 680, ATTO 700, ATTO 725, and ATTO 740.
[0355] Examples of AlexaFluor dyes include, but are not limited to: Alexa Fluor.RTM. 350, Alexa Fluor.RTM. 405, Alexa Fluor.RTM. 430, Alexa Fluor.RTM. 488, Alexa Fluor.RTM. 500, Alexa Fluor.RTM. 514, Alexa Fluor.RTM. 532, Alexa Fluor.RTM. 546, Alexa Fluor.RTM. 555, Alexa Fluor.RTM. 568, Alexa Fluor.RTM. 594, Alexa Fluor.RTM. 610, Alexa Fluor.RTM. 633, Alexa Fluor.RTM. 635, Alexa Fluor.RTM. 647, Alexa Fluor.RTM. 660, Alexa Fluor.RTM. 680, Alexa Fluor.RTM. 700, Alexa Fluor.RTM. 750, Alexa Fluor.RTM. 790, and the like.
[0356] Examples of quencher moieties include, but are not limited to: a dark quencher, a Black Hole Quencher.RTM. (BHQ.RTM.) (e.g., BHQ-0, BHQ-1, BHQ-2, BHQ-3), a Qxl quencher, an ATTO quencher (e.g., ATTO 540Q, ATTO 580Q, and ATTO 612Q), dimethylaminoazobenzenesulfonic acid (Dabsyl), Iowa Black RQ, Iowa Black FQ, IRDye QC-1, a QSY dye (e.g., QSY 7, QSY 9, QSY 21), AbsoluteQuencher, Eclipse, and metal clusters such as gold nanoparticles, and the like.
[0357] In some cases, a quencher moiety is selected from: a dark quencher, a Black Hole Quencher.RTM. (BHQ.RTM.) (e.g., BHQ-0, BHQ-1, BHQ-2, BHQ-3), a Qxl quencher, an ATTO quencher (e.g., ATTO 540Q, ATTO 580Q, and ATTO 612Q), dimethylaminoazobenzenesulfonic acid (Dabsyl), Iowa Black RQ, Iowa Black FQ, IRDye QC-1, a QSY dye (e.g., QSY 7, QSY 9, QSY 21), AbsoluteQuencher, Eclipse, and a metal cluster.
[0358] Examples of an ATTO quencher include, but are not limited to: ATTO 540Q, ATTO 580Q, and ATTO 612Q. Examples of a Black Hole Quencher.RTM. (BHQ.RTM.) include, but are not limited to: BHQ-0 (493 nm), BHQ-1 (534 nm), BHQ-2 (579 nm) and BHQ-3 (672 nm).
[0359] For examples of some detectable labels (e.g., fluorescent dyes) and/or quencher moieties, see, e.g., Bao et al., Annu Rev Biomed Eng. 2009; 11:25-47; as well as U.S. Pat. Nos. 8,822,673 and 8,586,718; U.S. patent publications 20140378330, 20140349295, 20140194611, 20130323851, 20130224871, 20110223677, 20110190486, 20110172420, 20060179585 and 20030003486; and international patent applications: WO200142505 and WO200186001, all of which are hereby incorporated by reference in their entirety.
[0360] In some cases, cleavage of a labeled detector ssDNA can be detected by measuring a colorimetric read-out. For example, the liberation of a fluorophore (e.g., liberation from a FRET pair, liberation from a quencher/fluor pair, and the like) can result in a wavelength shift (and thus color shift) of a detectable signal. Thus, in some cases, cleavage of a subject labeled detector ssDNA can be detected by a color-shift. Such a shift can be expressed as a loss of an amount of signal of one color (wavelength), a gain in the amount of another color, a change in the ration of one color to another, and the like.
Kits for Detecting Target DNA
[0361] The present disclosure provides a kit for detecting a target DNA, e.g., in a sample comprising a plurality of DNAs. In some cases, the kit comprises: (a) a labeled detector ssDNA (e.g., a labeled detector ssDNA comprising a fluorescence-emitting dye pair, e.g., a FRET pair and/or a quencher/fluor pair); and (b) one or more of: (i) a guide RNA, and/or a nucleic acid encoding said guide RNA; and ii) a CasZ polypeptide, and/or a nucleic acid encoding said CasZ polypeptide. In some cases, a nucleic acid encoding a guide RNA includes sequence insertion sites for the insertion of guide sequences by a user.
[0362] In some cases, the kit comprises: (a) a labeled detector ssDNA (e.g., a labeled detector ssDNA comprising a fluorescence-emitting dye pair, e.g., a FRET pair and/or a quencher/fluor pair); and (b) one or more of: (i) a guide RNA, and/or a nucleic acid encoding said guide RNA; ii) a tranc RNA and/or a nucleic acid encoding said guide RNA; and iii) a CasZ polypeptide, and/or a nucleic acid encoding said CasZ polypeptide. In some cases, a nucleic acid encoding a guide RNA includes sequence insertion sites for the insertion of guide sequences by a user.
[0363] In some cases, the kit comprises: (a) a labeled detector ssDNA (e.g., a labeled detector ssDNA comprising a fluorescence-emitting dye pair, e.g., a FRET pair and/or a quencher/fluor pair); and (b) one or more of: (i) a single-molecule RNA comprising a guide RNA and a tranc RNA, and/or a nucleic acid encoding single-molecule RNA; and iii) a CasZ polypeptide, and/or a nucleic acid encoding said CasZ polypeptide. In some cases, a nucleic acid encoding a single-molecule RNA includes sequence insertion sites for the insertion of guide sequences by a user.
[0364] In some cases, a subject kit comprises: (a) a labeled detector ssDNA comprising a fluorescence-emitting dye pair, e.g., a FRET pair and/or a quencher/fluor pair; and (b) one or more of: (i) a guide RNA, and/or a nucleic acid encoding said guide RNA; and/or i) a CasZ polypeptide.
[0365] Positive Controls
[0366] A kit of the present disclosure (e.g., one that comprises a labeled detector ssDNA and a CasZ polypeptide) can also include a positive control target DNA. In some cases, the kit also includes a positive control guide RNA that comprises a nucleotide sequence that hybridizes to the control target DNA. In some cases, the positive control target DNA is provided in various amounts, in separate containers. In some cases, the positive control target DNA is provided in various known concentrations, in separate containers, along with control non-target DNAs.
[0367] Nucleic Acids
[0368] While the RNAs of the disclosure (e.g., guide RNAs, tranc RNAs, single-molecule RNAs comprising a guide RNA and a tranc RNA) can be synthesized using any convenient method (e.g., chemical synthesis, in vitro using an RNA polymerase enzyme, e.g., T7 polymerase, T3 polymerase, SP6 polymerase, etc.), nucleic acids encoding such RNAs are also envisioned. Additionally, while a CasZ polypeptide of the disclosure can be provided (e.g., as part of a kit) in protein form, nucleic acids (such as mRNA and/or DNA) encoding the CasZ polypeptide can also be provided.
[0369] In some cases, a kit of the present disclosure comprises a nucleic acid (e.g., a DNA, e.g., a recombinant expression vector) that comprises a nucleotide sequence encoding a single-molecule RNA comprising: i) a guide RNA; and ii) a tranc RNA. In some cases, the nucleotide sequence encodes the guide RNA portion of the single-molecule RNA without a guide sequence. For example, in some cases, the nucleic acid comprises a nucleotide sequence encoding: i) a constant region of a guide RNA (a guide RNA without a guide sequence), and comprises an insertion site for a nucleic acid encoding a guide sequence; and ii) a tranc RNA.
[0370] For example, in some cases, a kit of the present disclosure comprises a nucleic acid (e.g., a DNA, e.g., a recombinant expression vector) that comprises a nucleotide sequence encoding a guide RNA. In some cases, the nucleotide sequence encodes a guide RNA without a guide sequence. For example, in some cases, the nucleic acid comprises a nucleotide sequence encoding a constant region of a guide RNA (a guide RNA without a guide sequence), and comprises an insertion site for a nucleic acid encoding a guide sequence. In some cases, a kit of the present disclosure comprises a nucleic acid (e.g., an mRNA, a DNA, e.g., a recombinant expression vector) that comprises a nucleotide sequence encoding a CasZ polypeptide.
[0371] In some cases, the guide RNA-encoding nucleotide sequence is operably linked to a promoter, e.g., a promoter that is functional in a prokaryotic cell, a promoter that is functional in a eukaryotic cell, a promoter that is functional in a mammalian cell, a promoter that is functional in a human cell, and the like. In some cases, a nucleotide sequence encoding a CasZ polypeptide is operably linked to a promoter, e.g., a promoter that is functional in a prokaryotic cell, a promoter that is functional in a eukaryotic cell, a promoter that is functional in a mammalian cell, a promoter that is functional in a human cell, a cell type-specific promoter, a regulatable promoter, a tissue-specific promoter, and the like.
Utility
[0372] CasZ compositions (e.g., expression vectors, kits, compositions, nucleic acids, and the like) find use in a variety of methods. For example, a CasZ compositions of the present disclosure can be used to (i) modify (e.g., cleave, e.g., nick; methylate; etc.) target nucleic acid (DNA or RNA; single stranded or double stranded); (ii) modulate transcription of a target nucleic acid; (iii) label a target nucleic acid; (iv) bind a target nucleic acid (e.g., for purposes of isolation, labeling, imaging, tracking, etc.); (v) modify a polypeptide (e.g., a histone) associated with a target nucleic acid; and the like. Thus, the present disclosure provides a method of modifying a target nucleic acid. In some cases, a method of the present disclosure for modifying a target nucleic acid comprises contacting the target nucleic acid with: a) a CasZ polypeptide of the present disclosure; and b) one or more (e.g., two) CasZ guide RNAs. In some cases, a method of the present disclosure for modifying a target nucleic acid comprises contacting the target nucleic acid with: a) a CasZ polypeptide, and b) one or more (e.g., two) CasZ guide RNAs, and c) a CasZ trancRNA. In some cases, a method of the present disclosure for modifying a target nucleic acid comprises contacting the target nucleic acid with: a) a CasZ polypeptide of the present disclosure; b) a CasZ guide RNA; and c) a donor nucleic acid (e.g, a donor template). In some cases, a method of the present disclosure for modifying a target nucleic acid comprises contacting the target nucleic acid with: a) a CasZ polypeptide; b) a CasZ guide RNA; c) a CasZ trancRNA, and d) a donor nucleic acid (e.g, a donor template). In some cases, the contacting step is carried out in a cell in vitro. In some cases, the contacting step is carried out in a cell in vivo. In some cases, the contacting step is carried out in a cell ex vivo.
[0373] Because a method that uses a CasZ polypeptide includes binding of the CasZ polypeptide to a particular region in a target nucleic acid (by virtue of being targeted there by an associated CasZ guide RNA), the methods are generally referred to herein as methods of binding (e.g., a method of binding a target nucleic acid). However, it is to be understood that in some cases, while a method of binding may result in nothing more than binding of the target nucleic acid, in other cases, the method can have different final results (e.g., the method can result in modification of the target nucleic acid, e.g., cleavage/methylation/etc., modulation of transcription from the target nucleic acid; modulation of translation of the target nucleic acid; genome editing; modulation of a protein associated with the target nucleic acid; isolation of the target nucleic acid; etc.).
[0374] For examples of suitable methods (e.g., that are used with CRISPR/Cas9 systems), see, for example, Jinek et al., Science. 2012 Aug. 17; 337(6096):816-21; Chylinski et al., RNA Biol. 2013 May; 10(5):726-37; Ma et al., Biomed Res Int. 2013; 2013:270805; Hou et al., Proc Natl Acad Sci USA. 2013 Sep. 24; 110(39):15644-9; Jinek et al., Elife. 2013; 2:e00471; Pattanayak et al., Nat Biotechnol. 2013 September; 31(9):839-43; Qi et al, Cell. 2013 Feb. 28; 152(5):1173-83; Wang et al., Cell. 2013 May 9; 153(4):910-8; Auer et al., Genome Res. 2013 Oct. 31; Chen et al., Nucleic Acids Res. 2013 Nov. 1; 41(20):e19; Cheng et al., Cell Res. 2013 October; 23(10):1163-71; Cho et al., Genetics. 2013 November; 195(3):1177-80; DiCarlo et al., Nucleic Acids Res. 2013 April; 41(7):4336-43; Dickinson et al., Nat Methods. 2013 October; 10(10):1028-34; Ebina et al., Sci Rep. 2013; 3:2510; Fujii et al, Nucleic Acids Res. 2013 Nov. 1; 41(20):e187; Hu et al., Cell Res. 2013 November; 23(11):1322-5; Jiang et al., Nucleic Acids Res. 2013 Nov. 1; 41(20):e188; Larson et al., Nat Protoc. 2013 November; 8(11):2180-96; Mali et. at., Nat Methods. 2013 October; 10(10):957-63; Nakayama et al., Genesis. 2013 December; 51(12):835-43; Ran et al., Nat Protoc. 2013 November; 8(11):2281-308; Ran et al., Cell. 2013 Sep. 12; 154(6):1380-9; Upadhyay et al., G3 (Bethesda). 2013 Dec. 9; 3(12):2233-8; Walsh et al., Proc Natl Acad Sci USA. 2013 Sep. 24; 110(39):15514-5; Xie et al., Mol Plant. 2013 Oct. 9; Yang et al., Cell. 2013 Sep. 12; 154(6):1370-9; and U.S. patents and patent applications: U.S. Pat. Nos. 8,906,616; 8,895,308; 8,889,418; 8,889,356; 8,871,445; 8,865,406; 8,795,965; 8,771,945; 8,697,359; 20140068797; 20140170753; 20140179006; 20140179770; 20140186843; 20140186919; 20140186958; 20140189896; 20140227787; 20140234972; 20140242664; 20140242699; 20140242700; 20140242702; 20140248702; 20140256046; 20140273037; 20140273226; 20140273230; 20140273231; 20140273232; 20140273233; 20140273234; 20140273235; 20140287938; 20140295556; 20140295557; 20140298547; 20140304853; 20140309487; 20140310828; 20140310830; 20140315985; 20140335063; 20140335620; 20140342456; 20140342457; 20140342458; 20140349400; 20140349405; 20140356867; 20140356956; 20140356958; 20140356959; 20140357523; 20140357530; 20140364333; and 20140377868; each of which is hereby incorporated by reference in its entirety.
[0375] For example, the present disclosure provides (but is not limited to) methods of cleaving a target nucleic acid; methods of editing a target nucleic acid; methods of modulating transcription from a target nucleic acid; methods of isolating a target nucleic acid, methods of binding a target nucleic acid, methods of imaging a target nucleic acid, methods of modifying a target nucleic acid, and the like.
[0376] As used herein, the terms/phrases "contact a target nucleic acid" and "contacting a target nucleic acid", for example, with a CasZ polypeptide or with a CasZ fusion polypeptide, etc., encompass all methods for contacting the target nucleic acid. For example, a CasZ polypeptide can be provided to a cell as protein, RNA (encoding the CasZ polypeptide), or DNA (encoding the CasZ polypeptide); while a CasZ guide RNA can be provided as a guide RNA or as a nucleic acid encoding the guide RNA and a CasZ trancRNA can be provided as a trancRNA or as a nucleic acid encoding the trancRNA. As such, when, for example, performing a method in a cell (e.g., inside of a cell in vitro, inside of a cell in vivo, inside of a cell ex vivo), a method that includes contacting the target nucleic acid encompasses the introduction into the cell of any or all of the components in their active/final state (e.g., in the form of a protein(s) for CasZ polypeptide; in the form of a protein for a CasZ fusion polypeptide; in the form of an RNA in some cases for the guide RNA), and also encompasses the introduction into the cell of one or more nucleic acids encoding one or more of the components (e.g., nucleic acid(s) comprising nucleotide sequence(s) encoding a CasZ polypeptide or a CasZ fusion polypeptide, nucleic acid(s) comprising nucleotide sequence(s) encoding guide RNA(s), nucleic acid comprising a nucleotide sequence encoding a donor template, and the like). Because the methods can also be performed in vitro outside of a cell, a method that includes contacting a target nucleic acid, (unless otherwise specified) encompasses contacting outside of a cell in vitro, inside of a cell in vitro, inside of a cell in vivo, inside of a cell ex vivo, etc.
[0377] In some cases, a method of the present disclosure for modifying a target nucleic acid comprises introducing into a target cell a CasZ locus, e.g., a nucleic acid comprising a nucleotide sequence encoding a CasZ polypeptide as well as nucleotide sequences of about 1 kilobase (kb) to 5 kb in length surrounding the CasZ-encoding nucleotide sequence from a cell (e.g., in some cases a cell that in its natural state (the state in which it occurs in nature) comprises a CasZ locus) comprising a CasZ locus, where the target cell does not normally (in its natural state) comprise a CasZ locus (e.g., in some cases the locus includes a CasZ trancRNA. However, one or more spacer sequences, encoding guide sequences for the encoded crRNA(s), can be modified such that one or more target sequences of interest are targeted. Thus, for example, in some cases, a method of the present disclosure for modifying a target nucleic acid comprises introducing into a target cell a CasZ locus, e.g., a nucleic acid obtained from a source cell (e.g., in some cases a cell that in its natural state (the state in which it occurs in nature) comprises a CasZ locus), where the nucleic acid has a length of from 100 nucleotides (nt) to 5 kb in length (e.g., from 100 nt to 500 nt, from 500 nt to 1 kb, from 1 kb to 1.5 kb, from 1.5 kb to 2 kb, from 2 kb to 2.5 kb, from 2.5 kb to 3 kb, from 3 kb to 3.5 kb, from 3.5 kb to 4 kb, or from 4 kb to 5 kb in length) and comprises a nucleotide sequence encoding a CasZ polypeptide. As noted above, in some such cases, one or more spacer sequences, encoding guide sequences for the encoded crRNA(s), can be modified such that one or more target sequences of interest are targeted. In some cases, the method comprises introducing into a target cell: i) a CasZ locus; and ii) a donor DNA template. In some cases, the target nucleic acid is in a cell-free composition in vitro. In some cases, the target nucleic acid is present in a target cell. In some cases, the target nucleic acid is present in a target cell, where the target cell is a prokaryotic cell. In some cases, the target nucleic acid is present in a target cell, where the target cell is a eukaryotic cell. In some cases, the target nucleic acid is present in a target cell, where the target cell is a mammalian cell. In some cases, the target nucleic acid is present in a target cell, where the target cell is a plant cell.
[0378] In some cases, a method of the present disclosure for modifying a target nucleic acid comprises contacting a target nucleic acid with a CasZ polypeptide of the present disclosure, or with a CasZ fusion polypeptide of the present disclosure. In some cases, abmethod of the present disclosure for modifying a target nucleic acid comprises contacting a target nucleic acid with a CasZ polypeptide and a CasZ guide RNA. In some cases, abmethod of the present disclosure for modifying a target nucleic acid comprises contacting a target nucleic acid with a CasZ polypeptide, a CasZ guide RNA, and a CasZ trancRNA. In some cases, abmethod of the present disclosure for modifying a target nucleic acid comprises contacting a target nucleic acid with a CasZ polypeptide, a first CasZ guide RNA, and a second CasZ guide RNA (and in some cases a CasZ trancRNA). In some cases, a method of the present disclosure for modifying a target nucleic acid comprises contacting a target nucleic acid with a CasZ polypeptide of the present disclosure and a CasZ guide RNA and a donor DNA template. In some cases, a method of the present disclosure for modifying a target nucleic acid comprises contacting a target nucleic acid with a CasZ polypeptide of the present disclosure and a CasZ guide RNA and a CasZ trancRNA and a donor DNA template.
[0379] In some cases, the target nucleic acid is in a cell-free composition in vitro. In some cases, the target nucleic acid is present in a target cell. In some cases, the target nucleic acid is present in a target cell, where the target cell is a prokaryotic cell. In some cases, the target nucleic acid is present in a target cell, where the target cell is a eukaryotic cell. In some cases, the target nucleic acid is present in a target cell, where the target cell is a mammalian cell. In some cases, the target nucleic acid is present in a target cell, where the target cell is a plant cell.
Target Nucleic Acids and Target Cells of Interest
[0380] A target nucleic acid can be any nucleic acid (e.g., DNA, RNA), can be double stranded or single stranded, can be any type of nucleic acid (e.g., a chromosome (genomic DNA), derived from a chromosome, chromosomal DNA, plasmid, viral, extracellular, intracellular, mitochondrial, chloroplast, linear, circular, etc.) and can be from any organism (e.g., as long as the CasZ guide RNA comprises a nucleotide sequence that hybridizes to a target sequence in a target nucleic acid, such that the target nucleic acid can be targeted).
[0381] A target nucleic acid can be DNA or RNA. A target nucleic acid can be double stranded (e.g., dsDNA, dsRNA) or single stranded (e.g., ssRNA, ssDNA). In some cases, a target nucleic acid is single stranded. In some cases, a target nucleic acid is a single stranded RNA (ssRNA). In some cases, a target ssRNA (e.g., a target cell ssRNA, a viral ssRNA, etc.) is selected from: mRNA, rRNA, tRNA, non-coding RNA (ncRNA), long non-coding RNA (IncRNA), and microRNA (miRNA). In some cases, a target nucleic acid is a single stranded DNA (ssDNA) (e.g., a viral DNA). As noted above, in some cases, a target nucleic acid is single stranded.
[0382] A target nucleic acid can be located anywhere, for example, outside of a cell in vitro, inside of a cell in vitro, inside of a cell in vivo, inside of a cell ex vivo. Suitable target cells (which can comprise target nucleic acids such as genomic DNA) include, but are not limited to: a bacterial cell; an archaeal cell; a cell of a single-cell eukaryotic organism; a plant cell; an algal cell, e.g., Botryococcus braunii, Chlamydomonas reinhardtii, Nannochloropsis gaditana, Chlorella pyrenoidosa, Sargassum patens, C. agardh, and the like; a fungal cell (e.g., a yeast cell); an animal cell; a cell from an invertebrate animal (e.g. fruit fly, a cnidarian, an echinoderm, a nematode, etc.); a cell of an insect (e.g., a mosquito; a bee; an agricultural pest; etc.); a cell of an arachnid (e.g., a spider; a tick; etc.); a cell from a vertebrate animal (e.g., a fish, an amphibian, a reptile, a bird, a mammal); a cell from a mammal (e.g., a cell from a rodent; a cell from a human; a cell of a non-human mammal; a cell of a rodent (e.g., a mouse, a rat); a cell of a lagomorph (e.g., a rabbit); a cell of an ungulate (e.g., a cow, a horse, a camel, a llama, a vicufla, a sheep, a goat, etc.); a cell of a marine mammal (e.g., a whale, a seal, an elephant seal, a dolphin, a sea lion; etc.) and the like. Any type of cell may be of interest (e.g. a stem cell, e.g. an embryonic stem (ES) cell, an induced pluripotent stem (iPS) cell, a germ cell (e.g., an oocyte, a sperm, an oogonia, a spermatogonia, etc.), an adult stem cell, a somatic cell, e.g. a fibroblast, a hematopoietic cell, a neuron, a muscle cell, a bone cell, a hepatocyte, a pancreatic cell; an in vitro or in vivo embryonic cell of an embryo at any stage, e.g., a 1-cell, 2-cell, 4-cell, 8-cell, etc. stage zebrafish embryo; etc.).
[0383] Cells may be from established cell lines or they may be primary cells, where "primary cells", "primary cell lines", and "primary cultures" are used interchangeably herein to refer to cells and cells cultures that have been derived from a subject and allowed to grow in vitro for a limited number of passages, i.e. splittings, of the culture. For example, primary cultures are cultures that may have been passaged 0 times, 1 time, 2 times, 4 times, 5 times, 10 times, or 15 times, but not enough times go through the crisis stage. Typically, the primary cell lines are maintained for fewer than 10 passages in vitro. Target cells can be unicellular organisms and/or can be grown in culture. If the cells are primary cells, they may be harvest from an individual by any convenient method. For example, leukocytes may be conveniently harvested by apheresis, leukocytapheresis, density gradient separation, etc., while cells from tissues such as skin, muscle, bone marrow, spleen, liver, pancreas, lung, intestine, stomach, etc. can be conveniently harvested by biopsy.
[0384] In some of the above applications, the subject methods may be employed to induce target nucleic acid cleavage, target nucleic acid modification, and/or to bind target nucleic acids (e.g., for visualization, for collecting and/or analyzing, etc.) in mitotic or post-mitotic cells in vivo and/or ex vivo and/or in vitro (e.g., to disrupt production of a protein encoded by a targeted mRNA, to cleave or otherwise modify target DNA, to genetically modify a target cell, and the like). Because the guide RNA provides specificity by hybridizing to target nucleic acid, a mitotic and/or post-mitotic cell of interest in the disclosed methods may include a cell from any organism (e.g. a bacterial cell, an archaeal cell, a cell of a single-cell eukaryotic organism, a plant cell, an algal cell, e.g., Botryococcus braunii, Chlamydomonas reinhardtii, Nannochloropsis gaditana, Chlorella pyrenoidosa, Sargassum patens, C. agardh, and the like, a fungal cell (e.g., a yeast cell), an animal cell, a cell from an invertebrate animal (e.g. fruit fly, cnidarian, echinoderm, nematode, etc.), a cell from a vertebrate animal (e.g., fish, amphibian, reptile, bird, mammal), a cell from a mammal, a cell from a rodent, a cell from a human, etc.). In some cases, a subject CasZ protein (and/or nucleic acid encoding the protein such as DNA and/or RNA), and/or CasZ guide RNA (and/or a DNA encoding the guide RNA), and/or donor template, and/or RNP can be introduced into an individual (i.e., the target cell can be in vivo) (e.g., a mammal, a rat, a mouse, a pig, a primate, a non-human primate, a human, etc.). In some cases, such an administration can be for the purpose of treating and/or preventing a disease, e.g., by editing the genome of targeted cells.
[0385] Plant cells include cells of a monocotyledon, and cells of a dicotyledon. The cells can be root cells, leaf cells, cells of the xylem, cells of the phloem, cells of the cambium, apical meristem cells, parenchyma cells, collenchyma cells, sclerenchyma cells, and the like. Plant cells include cells of agricultural crops such as wheat, corn, rice, sorghum, millet, soybean, etc. Plant cells include cells of agricultural fruit and nut plants, e.g., plant that produce apricots, oranges, lemons, apples, plums, pears, almonds, etc.
[0386] Additional examples of target cells are listed above in the section titled "Modified cells." Non-limiting examples of cells (target cells) include: a prokaryotic cell, eukaryotic cell, a bacterial cell, an archaeal cell, a cell of a single-cell eukaryotic organism, a protozoa cell, a cell from a plant (e.g., cells from plant crops, fruits, vegetables, grains, soy bean, corn, maize, wheat, seeds, tomatos, rice, cassava, sugarcane, pumpkin, hay, potatos, cotton, cannabis, tobacco, flowering plants, conifers, gymnosperms, angiosperms, ferns, clubmosses, hornworts, liverworts, mosses, dicotyledons, monocotyledons, etc.), an algal cell, (e.g., Botryococcus braunii, Chlamydomonas reinhardtii, Nannochloropsis gaditana, Chlorella pyrenoidosa, Sargassum patens, C. agardh, and the like), seaweeds (e.g. kelp) a fungal cell (e.g., a yeast cell, a cell from a mushroom), an animal cell, a cell from an invertebrate animal (e.g., fruit fly, cnidarian, echinoderm, nematode, etc.), a cell from a vertebrate animal (e.g., fish, amphibian, reptile, bird, mammal), a cell from a mammal (e.g., an ungulate (e.g., a pig, a cow, a goat, a sheep); a rodent (e.g., a rat, a mouse); a non-human primate; a human; a feline (e.g., a cat); a canine (e.g., a dog); etc.), and the like. In some cases, the cell is a cell that does not originate from a natural organism (e.g., the cell can be a synthetically made cell; also referred to as an artificial cell).
[0387] A cell can be an in vitro cell (e.g., established cultured cell line). A cell can be an ex vivo cell (cultured cell from an individual). A cell can be and in vivo cell (e.g., a cell in an individual). A cell can be an isolated cell. A cell can be a cell inside of an organism. A cell can be an organism. A cell can be a cell in a cell culture (e.g., in vitro cell culture). A cell can be one of a collection of cells. A cell can be a prokaryotic cell or derived from a prokaryotic cell. A cell can be a bacterial cell or can be derived from a bacterial cell. A cell can be an archaeal cell or derived from an archaeal cell. A cell can be a eukaryotic cell or derived from a eukaryotic cell. A cell can be a plant cell or derived from a plant cell. A cell can be an animal cell or derived from an animal cell. A cell can be an invertebrate cell or derived from an invertebrate cell. A cell can be a vertebrate cell or derived from a vertebrate cell. A cell can be a mammalian cell or derived from a mammalian cell. A cell can be a rodent cell or derived from a rodent cell. A cell can be a human cell or derived from a human cell. A cell can be a microbe cell or derived from a microbe cell. A cell can be a fungi cell or derived from a fungi cell. A cell can be an insect cell. A cell can be an arthropod cell. A cell can be a protozoan cell. A cell can be a helminth cell.
[0388] Suitable cells include a stem cell (e.g. an embryonic stem (ES) cell, an induced pluripotent stem (iPS) cell; a germ cell (e.g., an oocyte, a sperm, an oogonia, a spermatogonia, etc.); a somatic cell, e.g. a fibroblast, an oligodendrocyte, a glial cell, a hematopoietic cell, a neuron, a muscle cell, a bone cell, a hepatocyte, a pancreatic cell, etc.
[0389] Suitable cells include human embryonic stem cells, fetal cardiomyocytes, myofibroblasts, mesenchymal stem cells, autotransplated expanded cardiomyocytes, adipocytes, totipotent cells, pluripotent cells, blood stem cells, myoblasts, adult stem cells, bone marrow cells, mesenchymal cells, embryonic stem cells, parenchymal cells, epithelial cells, endothelial cells, mesothelial cells, fibroblasts, osteoblasts, chondrocytes, exogenous cells, endogenous cells, stem cells, hematopoietic stem cells, bone-marrow derived progenitor cells, myocardial cells, skeletal cells, fetal cells, undifferentiated cells, multi-potent progenitor cells, unipotent progenitor cells, monocytes, cardiac myoblasts, skeletal myoblasts, macrophages, capillary endothelial cells, xenogenic cells, allogenic cells, and post-natal stem cells.
[0390] In some cases, the cell is an immune cell, a neuron, an epithelial cell, and endothelial cell, or a stem cell. In some cases, the immune cell is a T cell, a B cell, a monocyte, a natural killer cell, a dendritic cell, or a macrophage. In some cases, the immune cell is a cytotoxic T cell. In some cases, the immune cell is a helper T cell. In some cases, the immune cell is a regulatory T cell (Treg).
[0391] In some cases, the cell is a stem cell. Stem cells include adult stem cells. Adult stem cells are also referred to as somatic stem cells.
[0392] Adult stem cells are resident in differentiated tissue, but retain the properties of self-renewal and ability to give rise to multiple cell types, usually cell types typical of the tissue in which the stem cells are found. Numerous examples of somatic stem cells are known to those of skill in the art, including muscle stem cells; hematopoietic stem cells; epithelial stem cells; neural stem cells; mesenchymal stem cells; mammary stem cells; intestinal stem cells; mesodermal stem cells; endothelial stem cells; olfactory stem cells; neural crest stem cells; and the like.
[0393] Stem cells of interest include mammalian stem cells, where the term "mammalian" refers to any animal classified as a mammal, including humans; non-human primates; domestic and farm animals; and zoo, laboratory, sports, or pet animals, such as dogs, horses, cats, cows, mice, rats, rabbits, etc. In some cases, the stem cell is a human stem cell. In some cases, the stem cell is a rodent (e.g., a mouse; a rat) stem cell. In some cases, the stem cell is a non-human primate stem cell.
[0394] Stem cells can express one or more stem cell markers, e.g., SOX9, KRT19, KRT7, LGR5, CA9, FXYD2, CDH6, CLDN18, TSPAN8, BPIFB 1, OLFM4, CDH17, and PPARGC1A.
[0395] In some cases, the stem cell is a hematopoietic stem cell (HSC). HSCs are mesoderm-derived cells that can be isolated from bone marrow, blood, cord blood, fetal liver and yolk sac. HSCs are characterized as CD34.sup.+ and CD3.sup.-. HSCs can repopulate the erythroid, neutrophil-macrophage, megakaryocyte and lymphoid hematopoietic cell lineages in vivo. In vitro, HSCs can be induced to undergo at least some self-renewing cell divisions and can be induced to differentiate to the same lineages as is seen in vivo. As such, HSCs can be induced to differentiate into one or more of erythroid cells, megakaryocytes, neutrophils, macrophages, and lymphoid cells.
[0396] In other cases, the stem cell is a neural stem cell (NSC). Neural stem cells (NSCs) are capable of differentiating into neurons, and glia (including oligodendrocytes, and astrocytes). A neural stem cell is a multipotent stem cell which is capable of multiple divisions, and under specific conditions can produce daughter cells which are neural stem cells, or neural progenitor cells that can be neuroblasts or glioblasts, e.g., cells committed to become one or more types of neurons and glial cells respectively. Methods of obtaining NSCs are known in the art.
[0397] In other cases, the stem cell is a mesenchymal stem cell (MSC). MSCs originally derived from the embryonal mesoderm and isolated from adult bone marrow, can differentiate to form muscle, bone, cartilage, fat, marrow stroma, and tendon. Methods of isolating MSC are known in the art; and any known method can be used to obtain MSC. See, e.g., U.S. Pat. No. 5,736,396, which describes isolation of human MSC.
[0398] A cell is in some cases a plant cell. A plant cell can be a cell of a monocotyledon. A cell can be a cell of a dicotyledon.
[0399] In some cases, the cell is a plant cell. For example, the cell can be a cell of a major agricultural plant, e.g., Barley, Beans (Dry Edible), Canola, Corn, Cotton (Pima), Cotton (Upland), Flaxseed, Hay (Alfalfa), Hay (Non-Alfalfa), Oats, Peanuts, Rice, Sorghum, Soybeans, Sugarbeets, Sugarcane, Sunflowers (Oil), Sunflowers (Non-Oil), Sweet Potatoes, Tobacco (Burley), Tobacco (Flue-cured), Tomatoes, Wheat (Durum), Wheat (Spring), Wheat (Winter), and the like. As another example, the cell is a cell of a vegetable crops which include but are not limited to, e.g., alfalfa sprouts, aloe leaves, arrow root, arrowhead, artichokes, asparagus, bamboo shoots, banana flowers, bean sprouts, beans, beet tops, beets, bittermelon, bok choy, broccoli, broccoli rabe (rappini), brussels sprouts, cabbage, cabbage sprouts, cactus leaf (nopales), calabaza, cardoon, carrots, cauliflower, celery, chayote, chinese artichoke (crosnes), chinese cabbage, chinese celery, chinese chives, choy sum, chrysanthemum leaves (tung ho), collard greens, corn stalks, corn-sweet, cucumbers, daikon, dandelion greens, dasheen, dau mue (pea tips), donqua (winter melon), eggplant, endive, escarole, fiddle head ferns, field cress, frisee, gai choy (chinese mustard), gailon, galanga (siam, thai ginger), garlic, ginger root, gobo, greens, hanover salad greens, huauzontle, jerusalem artichokes, jicama, kale greens, kohlrabi, lamb's quarters (quilete), lettuce (bibb), lettuce (boston), lettuce (boston red), lettuce (green leaf), lettuce (iceberg), lettuce (lolla rossa), lettuce (oak leaf--green), lettuce (oak leaf--red), lettuce (processed), lettuce (red leaf), lettuce (romaine), lettuce (ruby romaine), lettuce (russian red mustard), linkok, lo bok, long beans, lotus root, mache, maguey (agave) leaves, malanga, mesculin mix, mizuna, moap (smooth luffa), moo, moqua (fuzzy squash), mushrooms, mustard, nagaimo, okra, ong choy, onions green, opo (long squash), ornamental corn, ornamental gourds, parsley, parsnips, peas, peppers (bell type), peppers, pumpkins, radicchio, radish sprouts, radishes, rape greens, rape greens, rhubarb, romaine (baby red), rutabagas, salicornia (sea bean), sinqua (angled/ridged luffa), spinach, squash, straw bales, sugarcane, sweet potatoes, swiss chard, tamarindo, taro, taro leaf, taro shoots, tatsoi, tepeguaje (guaje), tindora, tomatillos, tomatoes, tomatoes (cherry), tomatoes (grape type), tomatoes (plum type), tumeric, turnip tops greens, turnips, water chestnuts, yampi, yams (names), yu choy, yuca (cassava), and the like.
[0400] A cell is in some cases an arthropod cell. For example, the cell can be a cell of a sub-order, a family, a sub-family, a group, a sub-group, or a species of, e.g., Chelicerata, Myriapodia, Hexipodia, Arachnida, Insecta, Archaeognatha, Thysanura, Palaeoptera, Ephemeroptera, Odonata, Anisoptera, Zygoptera, Neoptera, Exopterygota, Plecoptera, Embioptera, Orthoptera, Zoraptera, Dermaptera, Dictyoptera, Notoptera, Grylloblattidae, Mantophasmatidae, Phasmatodea, Blattaria, Isoptera, Mantodea, Parapneuroptera, Psocoptera, Thysanoptera, Phthiraptera, Hemiptera, Endopterygota or Holometabola, Hymenoptera, Coleoptera, Strepsiptera, Raphidioptera, Megaloptera, Neuroptera, Mecoptera, Siphonaptera, Diptera, Trichoptera, or Lepidoptera.
[0401] A cell is in some cases an insect cell. For example, in some cases, the cell is a cell of a mosquito, a grasshopper, a true bug, a fly, a flea, a bee, a wasp, an ant, a louse, a moth, or a beetle.
Donor Polynucleotide (Donor Template)
[0402] Guided by a CasZ guide RNA, a CasZ protein in some cases generates site-specific double strand breaks (DSBs) or single strand breaks (SSBs) (e.g., when the CasZ protein is a nickase variant) within double-stranded DNA (dsDNA) target nucleic acids, which are repaired either by non-homologous end joining (NHEJ) or homology-directed recombination (HDR).
[0403] In some cases, contacting a target DNA (with a CasZ protein and a CasZ guide RNA) occurs under conditions that are permissive for nonhomologous end joining or homology-directed repair. Thus, in some cases, a subject method includes contacting the target DNA with a donor polynucleotide (e.g., by introducing the donor polynucleotide into a cell), wherein the donor polynucleotide, a portion of the donor polynucleotide, a copy of the donor polynucleotide, or a portion of a copy of the donor polynucleotide integrates into the target DNA. In some cases, the method does not comprise contacting a cell with a donor polynucleotide, and the target DNA is modified such that nucleotides within the target DNA are deleted.
[0404] In some cases, a CasZ trancRNA (or nucleic acid encoding same), a CasZ guide RNA (or nucleic acid encoding same), and/or a CasZ protein (or a nucleic acid encoding same, such as an RNA or a DNA, e.g, one or more expression vectors) are coadministered (e.g., contacted with a target nucleic acid, administered to cells, etc.) with a donor polynucleotide sequence that includes at least a segment with homology to the target DNA sequence, the subject methods may be used to add, i.e. insert or replace, nucleic acid material to a target DNA sequence (e.g. to "knock in" a nucleic acid, e.g., one that encodes for a protein, an siRNA, an miRNA, etc.), to add a tag (e.g., 6.times.His, a fluorescent protein (e.g., a green fluorescent protein; a yellow fluorescent protein, etc.), hemagglutinin (HA), FLAG, etc.), to add a regulatory sequence to a gene (e.g. promoter, polyadenylation signal, internal ribosome entry sequence (IRES), 2A peptide, start codon, stop codon, splice signal, localization signal, etc.), to modify a nucleic acid sequence (e.g., introduce a mutation, remove a disease causing mutation by introducing a correct sequence), and the like. As such, a complex comprising a CasZ guide RNA and CasZ protein (or CasZ guide RNA and CasZ trancRNA and CasZ protein) is useful in any in vitro or in vivo application in which it is desirable to modify DNA in a site-specific, i.e. "targeted", way, for example gene knock-out, gene knock-in, gene editing, gene tagging, etc., as used in, for example, gene therapy, e.g. to treat a disease or as an antiviral, antipathogenic, or anticancer therapeutic, the production of genetically modified organisms in agriculture, the large scale production of proteins by cells for therapeutic, diagnostic, or research purposes, the induction of iPS cells, biological research, the targeting of genes of pathogens for deletion or replacement, etc.
[0405] In applications in which it is desirable to insert a polynucleotide sequence into he genome where a target sequence is cleaved, a donor polynucleotide (a nucleic acid comprising a donor sequence) can also be provided to the cell. By a "donor sequence" or "donor polynucleotide" or "donor template" it is meant a nucleic acid sequence to be inserted at the site cleaved by the CasZ protein (e.g., after dsDNA cleavage, after nicking a target DNA, after dual nicking a target DNA, and the like). The donor polynucleotide can contain sufficient homology to a genomic sequence at the target site, e.g. 70%, 80%, 85%, 90%, 95%, or 100% homology with the nucleotide sequences flanking the target site, e.g. within about 50 bases or less of the target site, e.g. within about 30 bases, within about 15 bases, within about 10 bases, within about 5 bases, or immediately flanking the target site, to support homology-directed repair between it and the genomic sequence to which it bears homology. Approximately 25, 50, 100, or 200 nucleotides, or more than 200 nucleotides, of sequence homology between a donor and a genomic sequence (or any integral value between 10 and 200 nucleotides, or more) can support homology-directed repair. Donor polynucleotides can be of any length, e.g. 10 nucleotides or more, 50 nucleotides or more, 100 nucleotides or more, 250 nucleotides or more, 500 nucleotides or more, 1000 nucleotides or more, 5000 nucleotides or more, etc.
[0406] The donor sequence is typically not identical to the genomic sequence that it replaces. Rather, the donor sequence may contain at least one or more single base changes, insertions, deletions, inversions or rearrangements with respect to the genomic sequence, so long as sufficient homology is present to support homology-directed repair (e.g., for gene correction, e.g., to convert a disease-causing base pair of a non disease-causing base pair). In some embodiments, the donor sequence comprises a non-homologous sequence flanked by two regions of homology, such that homology-directed repair between the target DNA region and the two flanking sequences results in insertion of the non-homologous sequence at the target region. Donor sequences may also comprise a vector backbone containing sequences that are not homologous to the DNA region of interest and that are not intended for insertion into the DNA region of interest. Generally, the homologous region(s) of a donor sequence will have at least 50% sequence identity to a genomic sequence with which recombination is desired. In certain embodiments, 60%, 70%, 80%, 90%, 95%, 98%, 99%, or 99.9% sequence identity is present. Any value between 1% and 100% sequence identity can be present, depending upon the length of the donor polynucleotide.
[0407] The donor sequence may comprise certain sequence differences as compared to the genomic sequence, e.g. restriction sites, nucleotide polymorphisms, selectable markers (e.g., drug resistance genes, fluorescent proteins, enzymes etc.), etc., which may be used to assess for successful insertion of the donor sequence at the cleavage site or in some cases may be used for other purposes (e.g., to signify expression at the targeted genomic locus). In some cases, if located in a coding region, such nucleotide sequence differences will not change the amino acid sequence, or will make silent amino acid changes (i.e., changes which do not affect the structure or function of the protein). Alternatively, these sequences differences may include flanking recombination sequences such as FLPs, loxP sequences, or the like, that can be activated at a later time for removal of the marker sequence.
[0408] In some cases, the donor sequence is provided to the cell as single-stranded DNA. In some cases, the donor sequence is provided to the cell as double-stranded DNA. It may be introduced into a cell in linear or circular form. If introduced in linear form, the ends of the donor sequence may be protected (e.g., from exonucleolytic degradation) by any convenient method and such methods are known to those of skill in the art. For example, one or more dideoxynucleotide residues can be added to the 3' terminus of a linear molecule and/or self-complementary oligonucleotides can be ligated to one or both ends. See, for example, Chang et al. (1987) Proc. Natl. Acad Sci USA 84:4959-4963; Nehls et al. (1996) Science 272:886-889. Additional methods for protecting exogenous polynucleotides from degradation include, but are not limited to, addition of terminal amino group(s) and the use of modified internucleotide linkages such as, for example, phosphorothioates, phosphoramidates, and O-methyl ribose or deoxyribose residues. As an alternative to protecting the termini of a linear donor sequence, additional lengths of sequence may be included outside of the regions of homology that can be degraded without impacting recombination. A donor sequence can be introduced into a cell as part of a vector molecule having additional sequences such as, for example, replication origins, promoters and genes encoding antibiotic resistance. Moreover, donor sequences can be introduced as naked nucleic acid, as nucleic acid complexed with an agent such as a liposome or poloxamer, or can be delivered by viruses (e.g., adenovirus, AAV), as described elsewhere herein for nucleic acids encoding a CasZ guide RNA and/or a CasZ fusion polypeptide and/or donor polynucleotide.
Transgenic, Non-Human Organisms
[0409] As described above, in some cases, a nucleic acid (e.g., a recombinant expression vector) of the present disclosure (e.g., a nucleic acid comprising a nucleotide sequence encoding a CasZ polypeptide; a nucleic acid comprising a nucleotide sequence encoding a CasZ fusion polypeptide; etc.), is used as a transgene to generate a transgenic non-human organism that produces a CasZ polypeptide, or a CasZ fusion polypeptide, of the present disclosure. The present disclosure provides a transgenic-non-human organism comprising a nucleotide sequence encoding a CasZ polypeptide, or a CasZ fusion polypeptide, of the present disclosure.
Transgenic, Non-Human Animals
[0410] The present disclosure provides a transgenic non-human animal, which animal comprises a transgene comprising a nucleic acid comprising a nucleotide sequence encoding a CasZ polypeptide or a CasZ fusion polypeptide. In some embodiments, the genome of the transgenic non-human animal comprises a nucleotide sequence encoding a CasZ polypeptide, or a CasZ fusion polypeptide, of the present disclosure. In some cases, the transgenic non-human animal is homozygous for the genetic modification. In some cases, the transgenic non-human animal is heterozygous for the genetic modification. In some embodiments, the transgenic non-human animal is a vertebrate, for example, a fish (e.g., salmon, trout, zebra fish, gold fish, puffer fish, cave fish, etc.), an amphibian (frog, newt, salamander, etc.), a bird (e.g., chicken, turkey, etc.), a reptile (e.g., snake, lizard, etc.), a non-human mammal (e.g., an ungulate, e.g., a pig, a cow, a goat, a sheep, etc.; a lagomorph (e.g., a rabbit); a rodent (e.g., a rat, a mouse); a non-human primate; etc.), etc. In some cases, the transgenic non-human animal is an invertebrate. In some cases, the transgenic non-human animal is an insect (e.g., a mosquito; an agricultural pest; etc.). In some cases, the transgenic non-human animal is an arachnid.
[0411] Nucleotide sequences encoding a CasZ polypeptide or a CasZ fusion polypeptide, of the present disclosure can be under the control of (i.e., operably linked to) an unknown promoter (e.g., when the nucleic acid randomly integrates into a host cell genome) or can be under the control of (i.e., operably linked to) a known promoter. Suitable known promoters can be any known promoter and include constitutively active promoters (e.g., CMV promoter), inducible promoters (e.g., heat shock promoter, tetracycline-regulated promoter, steroid-regulated promoter, metal-regulated promoter, estrogen receptor-regulated promoter, etc.), spatially restricted and/or temporally restricted promoters (e.g., a tissue specific promoter, a cell type specific promoter, etc.), etc.
Transgenic Plants
[0412] As described above, in some cases, a nucleic acid (e.g., a recombinant expression vector) of the present disclosure (e.g., a nucleic acid comprising a nucleotide sequence encoding a CasZ polypeptide of the present disclosure; a nucleic acid comprising a nucleotide sequence encoding a CasZ fusion polypeptide of the present disclosure; etc.), is used as a transgene to generate a transgenic plant that produces a CasZ polypeptide, or a CasZ fusion polypeptide, of the present disclosure. The present disclosure provides a transgenic plant comprising a nucleotide sequence encoding a CasZ polypeptide, or a CasZ fusion polypeptide, of the present disclosure. In some embodiments, the genome of the transgenic plant comprises a subject nucleic acid. In some embodiments, the transgenic plant is homozygous for the genetic modification. In some embodiments, the transgenic plant is heterozygous for the genetic modification.
[0413] Methods of introducing exogenous nucleic acids into plant cells are well known in the art. Such plant cells are considered "transformed," as defined above. Suitable methods include viral infection (such as double stranded DNA viruses), transfection, conjugation, protoplast fusion, electroporation, particle gun technology, calcium phosphate precipitation, direct microinjection, silicon carbide whiskers technology, Agrobacterium-mediated transformation and the like. The choice of method is generally dependent on the type of cell being transformed and the circumstances under which the transformation is taking place (i.e. in vitro, ex vivo, or in vivo).
[0414] Transformation methods based upon the soil bacterium Agrobacterium tumefaciens are particularly useful for introducing an exogenous nucleic acid molecule into a vascular plant. The wild type form of Agrobacterium contains a Ti (tumor-inducing) plasmid that directs production of tumorigenic crown gall growth on host plants. Transfer of the tumor-inducing T-DNA region of the Ti plasmid to a plant genome requires the Ti plasmid-encoded virulence genes as well as T-DNA borders, which are a set of direct DNA repeats that delineate the region to be transferred. An Agrobacterium-based vector is a modified form of a Ti plasmid, in which the tumor inducing functions are replaced by the nucleic acid sequence of interest to be introduced into the plant host.
[0415] Agrobacterium-mediated transformation generally employs cointegrate vectors or binary vector systems, in which the components of the Ti plasmid are divided between a helper vector, which resides permanently in the Agrobacterium host and carries the virulence genes, and a shuttle vector, which contains the gene of interest bounded by T-DNA sequences. A variety of binary vectors is well known in the art and are commercially available, for example, from Clontech (Palo Alto, Calif.). Methods of coculturing Agrobacterium with cultured plant cells or wounded tissue such as leaf tissue, root explants, hypocotyledons, stem pieces or tubers, for example, also are well known in the art. See, e.g., Glick and Thompson, (eds.), Methods in Plant Molecular Biology and Biotechnology, Boca Raton, Fla.: CRC Press (1993).
[0416] Microprojectile-mediated transformation also can be used to produce a subject transgenic plant. This method, first described by Klein et al. (Nature 327:70-73 (1987)), relies on microprojectiles such as gold or tungsten that are coated with the desired nucleic acid molecule by precipitation with calcium chloride, spermidine or polyethylene glycol. The microprojectile particles are accelerated at high speed into an angiosperm tissue using a device such as the BIOLISTIC PD-1000 (Biorad; Hercules Calif.).
[0417] A nucleic acid of the present disclosure (e.g., a nucleic acid (e.g., a recombinant expression vector) comprising a nucleotide sequence encoding a CasZ polypeptide, or a CasZ fusion polypeptide, of the present disclosure) may be introduced into a plant in a manner such that the nucleic acid is able to enter a plant cell(s), e.g., via an in vivo or ex vivo protocol. By "in vivo," it is meant in the nucleic acid is administered to a living body of a plant e.g. infiltration. By "ex vivo" it is meant that cells or explants are modified outside of the plant, and then such cells or organs are regenerated to a plant. A number of vectors suitable for stable transformation of plant cells or for the establishment of transgenic plants have been described, including those described in Weissbach and Weissbach, (1989) Methods for Plant Molecular Biology Academic Press, and Gelvin et al., (1990) Plant Molecular Biology Manual, Kluwer Academic Publishers. Specific examples include those derived from a Ti plasmid of Agrobacterium tumefaciens, as well as those disclosed by Herrera-Estrella et al. (1983) Nature 303: 209, Bevan (1984) Nucl Acid Res. 12: 8711-8721, Klee (1985) Bio/Technolo 3: 637-642. Alternatively, non-Ti vectors can be used to transfer the DNA into plants and cells by using free DNA delivery techniques. By using these methods transgenic plants such as wheat, rice (Christou (1991) Bio/Technology 9:957-9 and 4462) and corn (Gordon-Kamm (1990) Plant Cell 2: 603-618) can be produced. An immature embryo can also be a good target tissue for monocots for direct DNA delivery techniques by using the particle gun (Weeks et al. (1993) Plant Physiol 102: 1077-1084; Vasil (1993) Bio/Technolo 10: 667-674; Wan and Lemeaux (1994) Plant Physiol 104: 37-48 and for Agrobacterium-mediated DNA transfer (Ishida et al. (1996) Nature Biotech 14: 745-750). Exemplary methods for introduction of DNA into chloroplasts are biolistic bombardment, polyethylene glycol transformation of protoplasts, and microinjection (Danieli et al Nat. Biotechnol 16:345-348, 1998; Staub et al Nat. Biotechnol 18: 333-338, 2000; O'Neill et al Plant J. 3:729-738, 1993; Knoblauch et al Nat. Biotechnol 17: 906-909; U.S. Pat. Nos. 5,451,513, 5,545,817, 5,545,818, and 5,576,198; in Intl. Application No. WO 95/16783; and in Boynton et al., Methods in Enzymology 217: 510-536 (1993), Svab et al., Proc. Natl. Acad. Sci. USA 90: 913-917 (1993), and McBride et al., Proc. Natl. Acad. Sci. USA 91: 7301-7305 (1994)). Any vector suitable for the methods of biolistic bombardment, polyethylene glycol transformation of protoplasts and microinjection will be suitable as a targeting vector for chloroplast transformation. Any double stranded DNA vector may be used as a transformation vector, especially when the method of introduction does not utilize Agrobacterium.
[0418] Plants which can be genetically modified include grains, forage crops, fruits, vegetables, oil seed crops, palms, forestry, and vines. Specific examples of plants which can be modified follow: maize, banana, peanut, field peas, sunflower, tomato, canola, tobacco, wheat, barley, oats, potato, soybeans, cotton, carnations, sorghum, lupin and rice.
[0419] The present disclosure provides transformed plant cells, tissues, plants and products that contain the transformed plant cells. A feature of the subject transformed cells, and tissues and products that include the same is the presence of a subject nucleic acid integrated into the genome, and production by plant cells of a CasZ polypeptide, or a CasZ fusion polypeptide, of the present disclosure. Recombinant plant cells of the present invention are useful as populations of recombinant cells, or as a tissue, seed, whole plant, stem, fruit, leaf, root, flower, stem, tuber, grain, animal feed, a field of plants, and the like.
[0420] Nucleotide sequences encoding a CasZ polypeptide, or a CasZ fusion polypeptide, of the present disclosure can be under the control of (i.e., operably linked to) an unknown promoter (e.g., when the nucleic acid randomly integrates into a host cell genome) or can be under the control of (i.e., operably linked to) a known promoter. Suitable known promoters can be any known promoter and include constitutively active promoters, inducible promoters, spatially restricted and/or temporally restricted promoters, etc.
EXAMPLES OF NON-LIMITING ASPECTS OF THE DISCLOSURE
[0421] Aspects, including embodiments, of the present subject matter described above may be beneficial alone or in combination, with one or more other aspects or embodiments. Without limiting the foregoing description, certain non-limiting aspects of the disclosure, numbered 1-36 are provided below. As will be apparent to those of skill in the art upon reading this disclosure, each of the individually numbered aspects may be used or combined with any of the preceding or following individually numbered aspects. This is intended to provide support for all such combinations of aspects and is not limited to combinations of aspects explicitly provided below:
Aspects
[0422] Aspect 1. A method of guiding a CasZ polypeptide to a target sequence of a target nucleic acid, the method comprising contacting the target nucleic acid with an engineered and/or non-naturally occurring complex comprising: (a) a CasZ polypeptide; and (b) a CasZ guide RNA that comprises a guide sequence that hybridizes to a target sequence of the target nucleic acid, and comprises a region that binds to the CasZ polypeptide.
[0423] Aspect 2. The method of aspect 1, wherein the method results in modification of the target nucleic acid, modulation of transcription from the target nucleic acid, or modification of a polypeptide associated with a target nucleic acid.
[0424] Aspect 3. The method of aspect 2, wherein the target nucleic acid is modified by being cleaved.
[0425] Aspect 4. The method of any one of aspects 1-3, wherein the target nucleic acid is selected from: double stranded DNA, single stranded DNA, RNA, genomic DNA, and extrachromosomal DNA.
[0426] Aspect 5. The method of any one of aspects 1-4, wherein the guide sequence and the region that binds to the CasZ polypeptide are heterologous to one another.
[0427] Aspect 6. The method of any one of aspects 1-5, wherein said contacting results in genome editing.
[0428] Aspect 7. The method of any one of aspects 1-5, wherein said contacting takes place outside of a bacterial cell and outside of an archaeal cell.
[0429] Aspect 8. The method of any one of aspects 1-5, wherein said contacting takes place in vitro outside of a cell.
[0430] Aspect 9. The method of any one of aspects 1-7, wherein said contacting takes place inside of a target cell.
[0431] Aspect 10. The method of aspect 9, wherein said contacting comprises: introducing into the target cell at least one of: (a) the CasZ polypeptide, or a nucleic acid encoding the CasZ polypeptide; and (b) the CasZ guide RNA, or a nucleic acid encoding the CasZ guide RNA.
[0432] Aspect 11. The method of aspect 10, wherein the nucleic acid encoding the CasZ polypeptide is a non-naturally sequence that is codon optimized for expression in the target cell.
[0433] Aspect 12. The method of any one of aspects 9-11, wherein the target cell is a eukaryotic cell.
[0434] Aspect 13. The method of any one of aspects 9-12, wherein the target cell is in culture in vitro.
[0435] Aspect 14. The method of any one of aspects 9-12, wherein the target cell is in vivo.
[0436] Aspect 15. The method of any one of aspects 9-12, wherein the target cell is ex vivo.
[0437] Aspect 16. The method of aspect 12, wherein the eukaryotic cell is selected from the group consisting of: a plant cell, a fungal cell, a single cell eukaryotic organism, a mammalian cell, a reptile cell, an insect cell, an avian cell, a fish cell, a parasite cell, an arthropod cell, an arachnid cell, a cell of an invertebrate, a cell of a vertebrate, a rodent cell, a mouse cell, a rat cell, a primate cell, a non-human primate cell, and a human cell.
[0438] Aspect 17. The method of any one of aspects 9-16, wherein said contacting further comprises: introducing a DNA donor template into the target cell.
[0439] Aspect 18. The method of any one of aspects 1-17, wherein the method comprises contacting the target nucleic acid with a CasZ transactivating noncoding RNA (trancRNA).
[0440] Aspect 19. The method of any one of aspects 9-17, wherein said contacting comprises: introducing a CasZ transactivating noncoding RNA (trancRNA) and/or a nucleic acid encoding the CasZ trancRNA into the target cell.
[0441] Aspect 20. The method of aspect 18 or aspect 19, wherein the trancRNA comprises a nucleotide sequence having 70% or more (at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 95%, at least 98%, at least 99%, or 100%) nucleotide sequence identity with a trancRNA sequence of Table 2.
[0442] Aspect 21. A composition comprising an engineered and/or non-naturally occurring complex comprising: (a) a CasZ polypeptide, or a nucleic acid encoding said CasZ polypeptide; and (b) a CasZ guide RNA, or a nucleic acid encoding said CasZ guide RNA, wherein said CasZ guide RNA comprises a guide sequence that is complementary to a target sequence of a target nucleic acid, and comprises a region that can bind to the CasZ polypeptide.
[0443] Aspect 22. The composition of aspect 21, further comprising a CasZ transactivating noncoding RNA (trancRNA), or a nucleic acid encoding said CasZ trancRNA.
[0444] Aspect 23. A kit comprising an engineered and/or non-naturally occurring complex comprising: (a) a CasZ polypeptide, or a nucleic acid encoding said CasZ polypeptide; (b) a CasZ guide RNA, or a nucleic acid encoding said CasZ guide RNA, wherein said CasZ guide RNA comprises a guide sequence that is complementary to a target sequence of a target nucleic acid, and comprises a region that can bind to the CasZ polypeptide.
[0445] Aspect 24. The kit of aspect 23, further comprising a CasZ transactivating noncoding RNA (trancRNA), or a nucleic acid encoding said CasZ trancRNA.
[0446] Aspect 25. A genetically modified eukaryotic cell, comprising at least one of: (a) a CasZ polypeptide, or a nucleic acid encoding said CasZ polypeptide; (b) a CasZ guide RNA, or a nucleic acid encoding said CasZ guide RNA, wherein said CasZ guide RNA comprises a guide sequence that is complementary to a target sequence of a target nucleic acid, and comprises a region that can bind to the CasZ polypeptide; and (c) a CasZ transactivating noncoding RNA (trancRNA), or a nucleic acid encoding said CasZ trancRNA.
[0447] Aspect 26. The composition, kit, or eukaryotic cell of any one of the preceding aspects, characterized by at least one of: (a) the nucleic acid encoding said CasZ polypeptide comprises a nucleotide sequence that: (i) encodes the CasZ polypeptide and, (ii) is operably linked to a heterologous promoter; (b) the nucleic acid encoding said CasZ guide RNA comprises a nucleotide sequence that: (i) encodes the CasZ guide RNA and, (ii) is operably linked to a heterologous promoter; and (c) the nucleic acid encoding said CasZ trancRNA comprises a nucleotide sequence that: (i) encodes the CasZ trancRNA and, (ii) is operably linked to a heterologous promoter.
[0448] Aspect 27. The composition, kit, or eukaryotic cell of any one of the preceding aspects, for use in a method of therapeutic treatment of a patient.
[0449] Aspect 28. The method, composition, kit, or eukaryotic cell of any one of the preceding aspects, wherein at least one of: the nucleic acid encoding said CasZ polypeptide, the nucleic acid encoding said CasZ guide RNA, and the nucleic acid encoding said CasZ trancRNA, is a recombinant expression vector.
[0450] Aspect 29. The method, composition, kit, or eukaryotic cell of any one of the preceding aspects, wherein the CasZ guide RNA and/or the CasZ trancRNA comprises one or more of: a modified nucleobase, a modified backbone or non-natural internucleoside linkage, a modified sugar moiety, a Locked Nucleic Acid, a Peptide Nucleic Acid, and a deoxyribonucleotide.
[0451] Aspect 30. The method, composition, kit, or eukaryotic cell of any one of the preceding aspects, wherein the CasZ polypeptide is a variant CasZ polypeptide with reduced nuclease activity compared to a corresponding wild type CasZ protein.
[0452] Aspect 31. The method, composition, kit, or eukaryotic cell of any one of the preceding aspects, wherein at least one of: the CasZ polypeptide, the nucleic acid encoding the CasZ polypeptide, the CasZ guide RNA, the nucleic acid encoding the CasZ guide RNA, the CasZ trancRNA, and the nucleic acid encoding the CasZ trancRNA; is conjugated to a heterologous moiety.
[0453] Aspect 32. The method, composition, kit, or eukaryotic cell of aspect 31, wherein the heterologous moiety is a heterologous polypeptide.
[0454] Aspect 33. The method, composition, kit, or eukaryotic cell of any one of the preceding aspects, wherein the CasZ polypeptide has reduced nuclease activity compared to a corresponding wild type CasZ protein, and is fused to a heterologous polypeptide.
[0455] Aspect 34. The method, composition, kit, or eukaryotic cell of aspect 33, wherein the heterologous polypeptide: (i) has DNA modifying activity, (ii) exhibits the ability to increase or decrease transcription, and/or (iii) has enzymatic activity that modifies a polypeptide associated with DNA.
[0456] Aspect 35. The method, composition, kit, or eukaryotic cell of any one of the preceding aspects, wherein the CasZ polypeptide comprises an amino acid sequence having 70% or more (at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 95%, at least 98%, at least 99%, or 100%) amino acid sequence identity with a CasZ protein of FIG. 1 or FIG. 7.
[0457] Aspect 36. The method, composition, kit, or eukaryotic cell of any one of the preceding aspects, wherein the guide sequence and the region that binds to the CasZ polypeptide are heterologous to one another.
[0458] Aspect 37. A method of detecting a target DNA in a sample, the method comprising: (a) contacting the sample with: (i) a CasZ polypeptide; (ii) a guide RNA comprising: a region that binds to the CasZ polypeptide, and a guide sequence that hybridizes with the target DNA; and (iii) a detector DNA that is single stranded and does not hybridize with the guide sequence of the guide RNA; and (b) measuring a detectable signal produced by cleavage of the single stranded detector DNA by the CasZ, thereby detecting the target DNA.
[0459] Aspect 38. The method of aspect 37, wherein the target DNA is single stranded.
[0460] Aspect 39. The method of aspect 37 or 38, wherein the target DNA is double stranded.
[0461] Aspect 40. The method of any one of aspects 37-39, wherein the target DNA is viral DNA.
[0462] Aspect 41. The method of any one of aspects 37-40, wherein the target DNA is papovavirus, hepdnavirus, herpesvirus, adenovirus, poxvirus, or parvovirus DNA.
[0463] Aspect 42. The method of any one of aspects 37-41, wherein the CasZ polypeptide comprises an amino acid sequence having at least 85% amino acid sequence identity to the CasZ amino acid sequence set forth in any one of FIGS. 1 and 7.
[0464] Aspect 43. The method of any one of aspects 37-41, wherein the CasZ polypeptide is a Cas14a polypeptide.
[0465] Aspect 44. The method according to any one of aspects 37-43, wherein the sample comprises DNA molecules from a cell lysate.
[0466] Aspect 45. The method according to any one of aspects 37-44, wherein the sample comprises cells.
[0467] Aspect 46. The method according to any one of aspects 37-45, wherein said contacting is carried out inside of a cell in vitro, ex vivo, or in vivo.
[0468] Aspect 47. The method according to aspect 46, wherein the cell is a eukaryotic cell.
[0469] Aspect 48. The method according to any one of aspects 37-47, wherein the target DNA can be detected at a concentration as low as 10 aM.
[0470] Aspect 49. The method according to any one of aspects 37-48, comprising determining an amount of the target DNA present in the sample.
[0471] Aspect 50. The method according to aspect 49, wherein said determining comprises: measuring the detectable signal to generate a test measurement; measuring a detectable signal produced by a reference sample or cell to generate a reference measurement; and comparing the test measurement to the reference measurement to determine an amount of target DNA present in the sample.
[0472] Aspect 51. The method according to any one of aspects 37-50, wherein measuring a detectable signal comprises one or more of: gold nanoparticle based detection, fluorescence polarization, colloid phase transition/dispersion, electrochemical detection, and semiconductor-based sensing.
[0473] Aspect 52. The method according to any one of aspects 37-51, wherein the single stranded detector DNA comprises a fluorescence-emitting dye pair.
[0474] Aspect 53. The method according to aspect 52, wherein the fluorescence-emitting dye pair produces an amount of detectable signal prior to cleavage of the single stranded detector DNA, and the amount of detectable signal is reduced after cleavage of the single stranded detector DNA.
[0475] Aspect 54. The method according to aspect 52, wherein the single stranded detector DNA produces a first detectable signal prior to being cleaved and a second detectable signal after cleavage of the single stranded detector DNA.
[0476] Aspect 55. The method according to any one of aspects 52-54, wherein the fluorescence-emitting dye pair is a fluorescence resonance energy transfer (FRET) pair.
[0477] Aspect 56. The method according to aspect 18, wherein an amount of detectable signal increases after cleavage of the single stranded detector DNA.
[0478] Aspect 57. The method according to aspect 52 or aspect 56, wherein the fluorescence-emitting dye pair is a quencher/fluor pair.
[0479] Aspect 58. The method according to any one of aspects 52-57, wherein the single stranded detector DNA comprises two or more fluorescence-emitting dye pairs.
[0480] Aspect 59. The method according to aspect 58, wherein said two or more fluorescence-emitting dye pairs include a fluorescence resonance energy transfer (FRET) pair and a quencher/fluor pair.
[0481] Aspect 60. The method according to any one of aspects 37-59, wherein the single stranded detector DNA comprises a modified nucleobase, a modified sugar moiety, and/or a modified nucleic acid linkage.
[0482] Aspect 61. The method according to any one of aspects 37-60, wherein the method comprises amplifying nucleic acids in the sample.
[0483] Aspect 62. The method according to aspect 61, wherein said amplifying comprises isothermal amplification.
[0484] Aspect 63. The method according to aspect 62, wherein the isothermal amplification comprises recombinase polymerase amplification (RPA).
[0485] Aspect 64. The method according to any one of aspects 61-63, wherein said amplifying begins prior to the contacting of step (a).
[0486] Aspect 65. The method according to any one of aspects 61-63, wherein said amplifying begins together with the contacting of step (a).
[0487] Aspect 66. A kit for detecting a target DNA in a sample, the kit comprising: (a) a guide RNA, or a nucleic acid encoding the guide RNA; wherein the guide RNA comprises: a region that binds to a CasZ polypeptide, and a guide sequence that is complementary to a target DNA; and (b) a labeled detector DNA that is single stranded and does not hybridize with the guide sequence of the guide RNA.
[0488] Aspect 67. The kit of aspect 66, further comprising a CasZ polypeptide.
[0489] Aspect 68. The kit of aspect 67, wherein the CasZ polypeptide comprises an amino acid sequence having at least 85% amino acid sequence identity to the CasZ amino acid sequence set forth in any one of FIGS. 1 and 7.
[0490] Aspect 69. The kit of aspect 67, wherein the CasZ polypeptide is a Cas14a polypeptide.
[0491] Aspect 70. The kit of any one of aspects 66-69, wherein the single stranded detector DNA comprises a fluorescence-emitting dye pair.
[0492] Aspect 71. The kit of aspect 70, wherein the fluorescence-emitting dye pair is a FRET pair.
[0493] Aspect 72. The kit of aspect 70, wherein the fluorescence-emitting dye pair is a quencher/fluor pair.
[0494] Aspect 73. The kit of any one of aspects 70-72, wherein the single stranded detector DNA comprises two or more fluorescence-emitting dye pairs.
[0495] Aspect 74. The kit of aspect 73, wherein said two or more fluorescence-emitting dye pairs include a first fluorescence-emitting dye pair that produces a first detectable signal and a second fluorescence-emitting dye pair that produces a second detectable signal.
[0496] Aspect 75. The kit of any one of aspects 66-74, further comprising nucleic acid amplification components.
[0497] Aspect 76. The kit of aspect 75, wherein the nucleic acid amplification components are components for recombinase polymerase amplification (RPA).
[0498] Aspect 77. A method of cleaving single stranded DNAs (ssDNAs), the method comprising: contacting a population of nucleic acids, wherein said population comprises a target DNA and a plurality of non-target ssDNAs, with: (i) a CasZ polypeptide; and (ii) a guide RNA comprising: a region that binds to the CasZ polypeptide, and a guide sequence that hybridizes with the target DNA, wherein the CasZ polypeptide cleaves non-target ssDNAs of said plurality.
[0499] Aspect 78. The method of aspect 77, wherein said contacting is inside of a cell in vitro, ex vivo, or in vivo.
[0500] Aspect 79. The method of aspect 78, wherein the cell is a eukaryotic cell.
[0501] Aspect 80. The method of aspect 79, wherein the eukaryotic cell is a plant cell.
[0502] Aspect 81. The method of any one of aspects 78-80, wherein the non-target ssDNAs are foreign to the cell.
[0503] Aspect 82. The method of aspect 81, wherein the non-target ssDNAs are viral DNAs.
[0504] Aspect 83. The method of any one of aspects 77-82, wherein the target DNA is single stranded.
[0505] Aspect 84. The method of any one of aspects 77-82, wherein the target DNA is double stranded.
[0506] Aspect 85. The method of any one of aspects 77-84, wherein the target DNA is viral DNA.
[0507] Aspect 86. The method of any one of aspects 77-84, wherein the target DNA is papovavirus, hepdnavirus, herpesvirus, adenovirus, poxvirus, or parvovirus DNA.
EXAMPLES
[0508] The following examples are put forth so as to provide those of ordinary skill in the art with a complete disclosure and description of how to make and use the present invention, and are not intended to limit the scope of what the inventors regard as their invention nor are they intended to represent that the experiments below are all or the only experiments performed. Efforts have been made to ensure accuracy with respect to numbers used (e.g. amounts, temperature, etc.) but some experimental errors and deviations should be accounted for. Unless indicated otherwise, parts are parts by weight, molecular weight is weight average molecular weight, temperature is in degrees Celsius, and pressure is at or near atmospheric. Standard abbreviations may be used, e.g., bp, base pair(s); kb, kilobase(s); pl, picoliter(s); s or sec, second(s); min, minute(s); h or hr, hour(s); aa, amino acid(s); kb, kilobase(s); bp, base pair(s); nt, nucleotide(s); i.m., intramuscular(ly); i.p., intraperitoneal(ly); s.c., subcutaneous(ly); and the like.
Materials and Methods
[0509] The following materials and methods generally apply to the results presented in the Examples described herein except where noted otherwise.
[0510] Metagenomics and Metatranscriptomics
[0511] The initial analysis was performed on previously assembled and binned metagenomes from two sites: the Rifle Integrated Field Research (IFRC) site, adjacent to the Colorado River near Rifle, Colo. and Crystal Geyser, a cold, CO.sub.2-driven geyser on the Colorado Plateau in Utah. Metatranscriptomic data from IFRC site was used to detect transcription of non-coding elements in nature. Further mining of CRISPR-Cas14 systems was then performed on public metagenomes from IMG/M.
[0512] CRISPR-Cas Computation Analysis
[0513] The assembled contigs from the various samples were scanned with the HMMer suite for known Cas proteins using Hidden Markov Model (HMMs) profiles. Additional HMMs were constructed for Cas14 proteins based on the MAFFT alignments of putative type V effectors that contained less than 800 aa, and were adjacent to acquisition cas genes and CRISPR arrays. These HMMs were iteratively refined by augmenting them with manually selected novel putative Cas14 sequences that were found using the existing Cas14 HMM models. The sequence of Cas14 repeat sequences are provided in Table 3. CRISPR arrays were identified using a local version of the CrisprFinder software and CRISPRDetect. Phylogenetic trees of Cas1 and type V effector proteins were constructed using RAxML with PROTGAMMALG as the substitution model and 100 bootstrap samplings. Trees were visualized using FigTree 1.4.1 (http://tree.bio.ed.ac.uk/software/figtree/). Metatranscriptomic reads were mapped to assembled contigs using Bowtie2. RNase presence analysis was based on HMMs that were built from alignment of KEGG orthologous groups (KOs) downloaded from KEGG database.
TABLE-US-00005 TABLE 3 Repeat sequences (non-guide sequence portion of a Cas14 guide RNA) of all Cas14 proteins used herein (e.g., see FIG. 7) are shown in Table 3. SEQ Scaffold Cas14 ID Accession Protein Repeat sequence NO: No: Cas14a.1 GTTGCAGAACCCGAATAGACGAATGAAGGAATGCAAC 53 NCBI: (CasZa.3) MK005734 Cas14a.2 CTTGCAGAACCCGGATAGACGAATGAAGGAATGCAAC 295 NCBI: (Za.8) MK005733 Cas14a.3 GTTGCAGAACCCGAATAGACGAATGAAGGAATGCAAC 53 NCBI: (CasZa.3) MK005732 Cas14a.4 CTATCATATTCAGAACAAAGGGATTAAGGAATGCAAC 54 NCBI: (CasZa.4) MK005735 Cas14a.5 CTTTCATACTCAGAACAAAGGGATTAAGGAATGCAAC 55 NCBI: (CasZa.5, MK005736 CasZb.3) Cas14a.6 GTCTACAACTCATTGATAGAAATCAATGAGTTAGACA 56 IMG/M: (CasZa.6) Ga0137385_ 10000156 Cas14b.1 GTTGCAGAAATAGAATAAAGGAATTAAGGAATGCAAC 59 NCBI: (CasZb.2) MK005737 Cas14b.2 CTTTCATACTCAGAACAAAGGGATTAAGGAATGCAAC 55 NCBI: (CasZa.5, MK005738 CasZb.3) Cas14b.3 ATTTCATACTCAGAACAAAGGGATTAAGGAATGCAAC 61 NCBI: (CasZb.4) MK005739 Cas14b.4 GTTTCAGCGCACGAATTAACGAGATGAGAGATGCAAC 303 NCBI: (CasZb.16) MK005740 Cas14b.5 CTTGCAGAAGCTGAATAGACGAATCAAGGAATGCAAC 63 NCBI: (CasZb.6) MK005741 Cas14b.6 CTTGCAGGCCTTGAATAGAGGAGTTAAGGAATGCAAC 296 NCBI: (Za.12) MK005742 Cas14b.7 GTTGCAGCGCCCGAACTGACGAGACGAGAGATGCAAC 66 IMG/M: (CasZb.9) Ga0172369_ 10000737 Cas14b.8 GTTGCGCGAATAGAATAAAGGAATTAAGGAATGCAAC 67 IMG/M: (CasZb.10) Ga0172369_ 10010464 Cas14b.9 AGTTGCATTCCTTAATCCCTCTGTTCAGTTTGTGCAAT 68 IMG/M: (CasZb.11) Ga0172365_ 10004421 Cas14b.10 GTTGCACAGTGCTAATTAGAGAAACTAGGAATGCAAC 297 NCBI: (Zb.13) MK005743 Cas14b.11 GTTGCGGCGCGCGAATAAACGAGACTAGGAATGCAAC 70 NCBI: (CasZc.2) MK005744 Cas14b.12 CTAGCATATTCAGAACAAAGGGATTAAGGAATGCAAC 298 NCBI: (Zb.14) MK005745 Cas14b.13 CTTTCATATTCAGAACAAAGGGATTAAGGAATGCAAC 72 NCBI: (CasZc.4) MK005746 Cas14b.14 CTTTCATATTCAGAAACTAGGGGTTAAGGACTGCAAC 299 NCBI: (Zb.15) MK005747 Cas14b.15 GTTGCAGCCCCCGAACTAACGAGATGAGAGATGCAAC 74 IMG/M: (CasZc.6) Ga0116204_ 1008574 Cas14b.16 CTTGCAGAACAATCATATATGACTAATCAGACTGCAAC 75 IMG/M: (casZc.7) Ga0078972_ 1001015a Cas14c.1 GTTGCATCCCTACGTCGTGAGCACCGGTGAGTGCAAC 300 NCBI: (Zb.8) MK005748 Cas14c.2 GTCCCTACTCGCTAGGGAAACTAATTGAATGGAAAC 77 IMG/M: (CasZd.2) JGI12048 J13642_ 10201286 Cas14d.1 CTTCCAAACTCGAGCCAGTGGGGAGAGAAGTGGCA 79 NCBI: (CasZe.2) MK005750 Cas14d.2 CCTGTAGACCGGTCTCATTCTGAGAGGGGTATGCAACT 80 NCBI: (CasZe.3) MK005751 Cas14d.3 GTCTCGAGACCCTACAGATTTTGGAGAGGGGTGGGAC 81 NCBI: (CasZe.4) MK005752 Cas14e.1 GTAGCAGGACTCTCCTCGAGAGAAACAGGGGTATGCT 83 NCBI: (CasZf.1) MK005753 Cas14e.2 GTACAATACCTCTCCTTTAAGAGAGGGAGGGGTACGCTAC 84 NCBI: (CasZf.2) MK005754 Cas14e.3 GGAAAGGAATCCCCTGAAGGAAACGAGGGGG 301 NCBI: (Zc.5) MK005755 Cas14f.1 GGTTCCCCCGGGCGCGGGTGGGGTGGCG 86 NCBI: (CasZg.1) MK005756 Cas14f.2 GGCTGCTCCGGGTGCGCGTGGAGCGAGG 87 IMG/M: (CasZg.2) Ga0105042_ 100140 Cas14g.1 GTGTCCATCAATCAGATTTGCGTTGGCCGGTGCAAT 302 NCBI: (Ze.3) MK005758 Cas14g.2 GCCGCAGCGGCCGACGCGGCCCTGATCGATGGACAC 90 IMG/M: (CasZi.2) Ga0123330_ 1010394 Cas14h.1 GGCTAGCCCGTGCGCGCAGGGACGAGTGG 92 IMG/M: (CasZk.1) Ga0070762_ 10001740 Cas14h.2 GCCCGTGCGCGCAGGGACGAGTGG 93 IMG/M: (CasZk.2) Ga0070766_ 10011912 Cas14h.3 CCATCGCCCCGCGCGCACGTGGATGAGCC 95 IMG/M: (CasZk.4) Ga0116216_ 10000905 Cas14u.1 GTTATAAAGGCGGGGATCGCGACCGAGCGATTGAAAG 57 IMG/M: (CasZa.7) Ga0066793_ 10010091 Cas14u.2 GTCTCCATGACTGAAAAGTCGTGGCCGAATTGAAAC 65 IMG/M: (CasZb.8) JGI24730 J26740_ 1002785 Cas14u.3 GTTGCATTCGGGTGCAAAACAGGGAGTAGAGTGTAAC 78 NCBI: (CasZe.1) MK005749 Cas14u.4 CTTTTAGACAGTTTAAATTCTAAAGGGTATAAAAC 307 NCBI: (CasZj.2) MK005757 Cas14u.5 GTCGAAATGCCCGCGCGGGGGCGTCGTACCCGCGAC 308 IMG/M: (CasZj.1) Ga0137373_ 10000316 Cas14u.6 GTTGCAGCGGCCGACGGAGCGCGAGCGTGGATGCCAC 309 IMG/M: (CasZk.3) Ga0070717_ 10000077 Cas14u.7 CTTTAGACTTCTCCGGAAGTCGAATTAATGGAAAC 310 IMG/M: (CasZl.1) JGI12210 IMG/M: J13797_ 10004690 Cas14u.8 GGGCGCCCCGCGCGAGCGGGGGTTGAAG 311 IMG/M: (CasZl.2) Ga0073904_ 10021651
[0514] Generation of Expression Plasmids, RNA and DNA Substrates
[0515] Minimal CRISPR loci for putative systems were designed by removing acquisition proteins and generating minimal arrays with a single spacer. These minimal loci were ordered as gBlocks (IDT) and assembled into a plasmid with a tetracycline inducible promoter driving expression of the locus. Plasmid maps were available on Addgene and in the figures. All RNA was in vitro transcribed using T7 polymerase and PCR products as dsDNA template. Resulting IVTs were gel extracted and ethanol precipitated. DNA substrates were obtained from IDT and their sequences are available in Table 4. For radiolabeled cleavage assays DNA oligos were gel extracted from a PAGE gel before radiolabeling. For FQ assays, DNA substrates were used without further purification.
[0516] E. coli RNAseq
[0517] Small RNA sequencing was conducted as described previously with modification in Harrington et al. (2017). E. coli NEB Stable3 was transformed with a plasmid expressing Cas14a1 system with a tetracycline inducible promoter upstream of the Cas14a1 ORF or the same plasmid with an N-terminal 10.times.-histidine tag fused to Cas14. Starters were grown up overnight in SOB, diluted 1:100 in 5 mL fresh SOB containing 214 nM anhydrotetracycline and grown up overnight at 25.degree. C. For sequencing of RNA pulled down with Cas14a, the plasmid containing an N-terminal His-tag fused to Cas14a1 was grown up at 18.degree. C. before lysis and purification as described in "Protein purification", stopping after the Ni-NTA elution. Cells were pelleted and RNA was extracted using hot phenol as previously described. Total nucleic acids were treated with TURBO DNase and phenol extracted. The resulting RNA was treated with rSAP which was heat inactivated before addition of T4 PNK. Adapters were ligated onto the small RNA using the NEBnext small RNA kit and gel-extracted on an 8% native PAGE gel. RNA was sequenced on a MiSeq with single end 300 bp reads. For analysis, the resulting reads were trimmed using Cutadapt, discarding sequences<8 nt and mapped to the plasmid reference using Bowtie2.
[0518] PAM Depletion Assays
[0519] PAM depletion assays were conducted as previously described in Burstein et al. (2017). Randomized plasmid libraries were generated using a primer containing a randomized PAM region adjacent to the target sequence. The randomized primers were hybridized with a primer that was complementary to the 3' end of the primer and the duplex was extended using Klenow Fragment (NEB). The dsDNA containing the target and were digested with EcoRI and NcoI, ligated into pUC 19 backbone and transformed into E. coli DH5a and >107 cells were harvested. Next E. coli NEBstable was transformed with either a CRISPR plasmid or an empty vector control and these transformed E. coli were made electrocompetent by repeated washing with 10% glycerol. These electrocompetent cells were transformed with 200 ng of the target library and plated on bioassay dishes containing selection for the target (carbenicillin, 100 mg 1-1) and CRISPR plasmid (chloramphenicol, 30 mg 1-1). Cells were harvested and prepared for amplicon sequencing on an Illumina MiSeq. The PAM region was extracted using Cutadapt and depletion values were calculated in python. PAMs were visualized using WebLogo.
[0520] Transcriptomic RNA Mapping
[0521] RNA was extracted from 0.2 mm filters using the Invitrogen TRIzol reagent, followed by genomic DNA removal and cleaning using the Qiagen RNase-Free DNase Set kit and the Qiagen Mini RNeasy kit. An Agilent 2100 Bioanalyzer (Agilent Technologies) was used to assess the integrity of the RNA samples. The Applied Biosystems SOLiD Total RNA-Seq kit was used to generate the cDNA template library. The SOLiD EZ Bead system (Life Technologies) was used to perform emulsion clonal bead amplification to generate bead templates for SOLiD platform sequencing. Samples were sequenced at Pacific Northwest National Laboratory on the 5500XL SOLiD platform. The 50 bp single reads were trimmed using Sickle as in Brown et al. (2015).
[0522] Protein Purification
[0523] Cas14a1 was purified as described previously with modification. E. coli BL21(DE3) RIL were transformed with 10.times.His-MBP-Cas14a1 expression plasmid and grown up to OD600=0.5 in Terrific Broth (TB) and induced with 0.5 mM IPTG. Cells were grown overnight at 18.degree. C., collected by centrifugation, resuspended in Lysis Buffer (50 mM Tris-HCl, pH 7.5, 20 mM imidazole, 0.5 mM TCEP, 500 mM NaCl) and broken by sonication. Lysate was batch loaded on to Ni-NTA resin, washed with the above buffer before elution with Elution Buffer (50 mM Tris-HCl, pH 7.5, 300 mM imidazole, 0.5 mM TCEP, 500 mM NaCl). The MBP and His-tag were removed by overnight incubation with TEV at 4.degree. C. The resulting protein exchanged into Buffer A (20 mM HEPES, pH 7.5, 0.5 mM TCEP, 150 mM NaCl) and loaded over tandem MBP, heparin columns (GE, Hi-Trap) and eluted with a linear gradient from Buffer A to Buffer B (20 mM HEPES, pH 7.5, 0.5 mM TCEP, 1250 mM NaCl). The resulting fractions containing Cas14a1 were loaded onto an S200 gel filtration column, flash frozen and stored at -80.degree. C. until use.
[0524] In Vitro Cleavage Assays
[0525] Radiolabeled
[0526] Radiolabeled cleavage assays were conducted in 1.times. Cleavage Buffer (25 mM NaCl, 20 mM HEPES, pH 7.5, 1 mM DTI, 5% glycerol). 100 nM Cas14a1 was complexed with 125 nM crRNA and 125 nM tracrRNA for 10 min at RT..about.1 nM radiolabeled DNA or RNA substrate was added and allowed to react for 30 min at 37.degree. C. The reaction was stopped by adding 2.times. Quench Buffer (90% formamide, 25 mM EDTA and trace bromophenol blue), heated to 95.degree. C. for 2 min and run on a 10% polyacrylamide gel containing 7M Urea and 0.5.times.TBE. Products were visualized by phosphorimaging.
[0527] M13 DNA Cleavage
[0528] M13 DNA cleavage assays were conducted in 100 mM NaCl, 20 mM HEPES, pH 7.5, 1 mM DTT, 5% glycerol. 250 nM Cas14a1 was complexed with 250 nM crRNA and 250 nM tracrRNA and 250 nM ssDNA activator. The reaction was initiated by addition of 5 nM M13 ssDNA plasmid and was quenched by addition of loading buffer supplemented with 10 mM EDTA. Products were separated on a 1.5% agarose TAE gel prestained with SYBR gold (Thermofisher).
[0529] FQ Detection of Trans-Cleavage
[0530] FQ detection was conducted as previously described in Chen et al. (2018) with modification. 100 nM Cas14a1 was complexed with 125 nM crRNA, 125 nM tracrRNA, 50 nM FQ probe and 2 nM ssDNA activator in 1.times. Cleavage Buffer at 37.degree. C. for 10 min. The reaction was then initiated by addition of activator DNA when for all reactions except for the RNA optimization experiments where the variable RNA component was used to initiate. The reaction was monitored in a fluorescence plate reader for up to 120 minutes at 37.degree. C. with fluorescence measurements taken every 1 min (.lamda.ex: 485 nm; .lamda.em: 535 nm). The resulting data were background subtracted using the readings taken in the absence of activator and fit using a single exponential decay curve.
[0531] Data Availability
[0532] Plasmids used herein are available on Addgene (plasmid numbers 112500, 112501, 112502, 112503, 112504, 112505, 112506). Oligonucleotides used herein are provided in Table 4 and Table 5. The plasmids used herein are provided in FIG. 24. The Cas14 protein sequences used herein are provided in FIG. 7.
TABLE-US-00006 TABLE 4 Oligonucleotides and plasmids used herein. SEQ DNA Name Sequence ID NO: Radiolabeld cTACGCCGattatcttctg 220 DNA acaactttcgcaagcggtg activator 1 T taaggtaAAAAAtgCGGGC strand AC Radiolabeld GTGCCCGcaTTTTTtacct 221 DNA tacaccgcttgcgaaagtt activator 1 gtcagaagataatCGGCGT NT strand Ag Radiolabeld tttatatgtttctcctgga 222 DNA gataacgcaatcgtgacaa activator 2 T ctttcgcaagcggtgtaag strand gtaGCAGGCTTCcgaattc cgcgtttttacggc Radiolabeld gccgtaaaaacgcggaatt 223 DNA cgGAAGCCTGCtaccttac activator 2 accgcttgcgaaagttgtc NT strand acgattgcgttatctccag gagaaacatataaa Radiolabeld gatcttcagcTATACATTA 224 Activator 3 TTGCACCAACACTAAGGCA GAGTATGtttacctggac Radiolabeld gatcttcagcTTTGTATTA 225 Activator 4 CTGGAAGGATGCTTGCTTG AGGTGTAaaaacctggac F-Q 5 nt /56-FAM/TTTTT/ 226 3IABkFQ/ F-Q 6 nt /56-FAM/TTTTTT/ 227 3IABkFQ/ F-Q 7 nt /56-FAM/TTTTTTT/ 228 3IABkFQ/ F-Q 8 nt /56-FAM/TTTTTTTT/ 229 3IABkFQ/ F-Q 9 nt /56-FAM/TTTTTTTTT/ 230 3IABkFQ/ F-Q 10 nt /56-FAM/TTTTTTTTTT/ 231 3IABkFQ/ F-Q 11 nt /56-FAM/TTTTTTTTTT 232 T/3IABkFQ/ F-Q 12 nt /56-FAM/TTTTTTTTTTT 233 T/3IABkFQ/ F-Q 10 nt /56-FAM/TATATATATA/ 371 A/T 3IABkFQ/ F-Q 6 nt A/T /56-FAM/TATATA/ 372 3IABkFQ/ F-Q 5 nt A/T /56-FAM/TATAT/ 373 3IABkFQ/ Target 2, GCCGGGGTGGTGCCCATCC 234 Perfect TGGTCGAGCTGGACGGCGA AAAT 3' CGTAAACGGCCACAAGC Target 2, GCCGGGGTGGTGCCCATCC 235 Perfect TGGTCGAGCTGGACGGCGA TCGT 3' CGTGCTCGGCCACAAGC Target 2, GCCGGGGTGGTGCCCATCC 236 1-2 MM TGGTCGAGCTGGACGGCGA GCTAAACGGCCACAAGC Target 2, GCCGGGGTGGTGCCCATCC 237 3-4 MM TGGTCGAGCTGGACGGCCT CGTAAACGGCCACAAGC Target 2, GCCGGGGTGGTGCCCATCC 238 5-6 MM TGGTCGAGCTGGACGCGGA CGTAAACGGCCACAAGC Target 2, GCCGGGGTGGTGCCCATCC 239 7-8 MM TGGTCGAGCTGGAGCGCGA CGTAAACGGCCACAAGC Target 2, GCCGGGGTGGTGCCCATCC 240 9-10 MM TGGTCGAGCTGCTCGGCGA CGTAAACGGCCACAAGC Target 2, GCCGGGGTGGTGCCCATCC 241 11-12 MM TGGTCGAGCACGACGGCGA CGTAAACGGCCACAAGC Target 2, GCCGGGGTGGTGCCCATCC 242 13-14 MM TGGTCGACGTGGACGGCGA CGTAAACGGCCACAAGC Target 2, GCCGGGGTGGTGCCCATCC 243 15-16 MM TGGTCCTGCTGGACGGCGA CGTAAACGGCCACAAGC Target 2, GCCGGGGTGGTGCCCATCC 244 17-18 MM TGGAGGAGCTGGACGGCGA CGTAAACGGCCACAAGC Target 2, GCCGGGGTGGTGCCCATCC 245 19-20 MM TCCTCGAGCTGGACGGCGA CGTAAACGGCCACAAGC HERC2 Amp G*T*G*T*TAATACAAAGG 246 Fwd TACAGGAACAAAGAATTTG HERC2 Amp CAAAGAGAAGCCTCGGCC 247 Rev Target 1, TTTATTCAAGGCAATCACT 248 Perfect ATCAGCTGTGGAACACCCA AAAT 3' GGTAAACTAACACAACT Target 1, TTTATTCAAGGCAATCACT 249 Perfect ATCAGCTGTGGAACACCCA TCGT 3' GGTGCTCTAACACAACT Target 1, TTTATTCAAGGCAATCACT 250 1-2 MM ATCAGCTGTGGAACACCCA CCTAAACTAACACAACT Target 1, TTTATTCAAGGCAATCACT 251 3-4 MM ATCAGCTGTGGAACACCGT GGTAAACTAACACAACT Target 1, TTTATTCAAGGCAATCACT 252 5-6 MM ATCAGCTGTGGAACAGGCA GGTAAACTAACACAACT Target 1, TTTATTCAAGGCAATCACT 253 7-8 MM ATCAGCTGTGGAAGTCCCA GGTAAACTAACACAACT Target 1, TTTATTCAAGGCAATCACT 254 9-10 MM ATCAGCTGTGGTTCACCCA GGTAAACTAACACAACT Target 1, TTTATTCAAGGCAATCACT 255 11-12 MM ATCAGCTGTCCAACACCCA GGTAAACTAACACAACT Target 1, TTTATTCAAGGCAATCACT 256 13-14 MM ATCAGCTCAGGAACACCCA GGTAAACTAACACAACT Target 1, TTTATTCAAGGCAATCACT 257 15-16 MM ATCAGGAGTGGAACACCCA GGTAAACTAACACAACT Target 1, TTTATTCAAGGCAATCACT 258 17-18 MM ATCTCCTGTGGAACACCCA GGTAAACTAACACAACT Target 1, TTTATTCAAGGCAATCACT 259 19-20 MM AAGAGCTGTGGAACACCCA GGTAAACTAACACAACT Target 3, cTACGCCGattatcttctg 220 Perfect acaactttcgcaagcggtg taaggtaAAAAAtgCGGGC AC Target 3, cTACGCCGattatcttctg 260 1-2 MM acaactttcgcaagcggtg taaggCGAAAAAtgCGGGC AC Target 3, cTACGCCGattatcttctg 261 3-4 MM acaactttcgcaagcggtg taaAAtaAAAAAtgCGGGC AC Target 3, cTACGCCGattatcttctg 262 5-6 MM acaactttcgcaagcggtg tGGggtaAAAAAtgCGGGC AC Target 3, cTACGCCGattatcttctg 263 7-8 MM acaactttcgcaagcggtA CaaggtaAAAAAtgCGGGC AC Target 3, cTACGCCGattatcttctg 264 9-10 MM acaactttcgcaagcgACg taaggtaAAAAAtgCGGGC AC Target 3, cTACGCCGattatcttctg 265 11-12 MM acaactttcgcaagTAgtg taaggtaAAAAAtgCGGGC AC Target 3, cTACGCCGattatcttctg 266 13-14 MM acaactttcgcaGAcggtg taaggtaAAAAAtgCGGGC AC Target 3, cTACGCCGattatcttctg 267 15-16 MM acaactttcgTGagcggtg taaggtaAAAAAtgCGGGC AC Target 3, cTACGCCGattatcttctg 268 17-18 MM acaactttTAcaagcggtg taaggtaAAAAAtgCGGGC AC Target 3, cTACGCCGattatcttctg 269 19-20 MM acaactCCcgcaagcggtg taaggtaAAAAAtgCGGGC AC Target 3, cTACGCCGattatcttctg 270 21-22 MM acaaTCttcgcaagcggtg taaggtaAAAAAtgCGGGC AC Target 3, cTACGCCGattatcttctg 271 23-24 MM acGGctttcgcaagcggtg taaggtaAAAAAtgCGGGC AC Target 3, cTACGCCGattatcttctg 272 25-26 MM GTaactttcgcaagcggtg taaggtaAAAAAtgCGGGC AC Full Length cTACGCCGattatcttctg 220 Activator acaactttcgcaagcggtg taaggtaAAAAAtgCGGGC AC -20 5' tatcttctgacaactttcg 273 activator caagcggtgtaaggtaAAA target AAtgCGGGCAC
-25 5' tctgacaactttcgcaagc 274 activator ggtgtaaggtaAAAAAtgC target GGGCAC -30 5' caactttcgcaagcggtgt 275 activator aaggtaAAAAAtgCGGGCA target C -35 5' ttcgcaagcggtgtaaggt 276 activator aAAAAAtgCGGGCAC target -5 3' cTACGCCGattatcttctg 277 activator acaactttcgcaagcggtg target taaggtaAAAAAtgCG -9 3' cTACGCCGattatcttctg 278 activator acaactttcgcaagcggtg target taaggtaAAAA -14 3' cTACGCCGattatcttctg 279 activator acaactttcgcaagcggtg target ta -19 3' cTACGCCGattatcttctg 280 activator acaactttcgcaagcg target -24 3' cTACGCCGattatcttctg 281 activator acaactttcgc target -29 3' cTACGCCGattatcttctg 282 activator acaact target -34 3' cTACGCCGattatcttctg 283 activator a target No loop caactttcgcaagcggtgt 284 aaggtaAAAAAtgCG 0 nt SL atggaatgtggcgaacgct 285 ttcaacGAAAcaactttcg caagcggtgtaaggtaAAA AAtgCG 5 nt SL atggaatgtggcgaacgct 286 tagttgGAAAcaactttcg caagcggtgtaaggtaAAA AAtgCG 10 nt SL atggaatgtggcgaagcga 287 aagttgGAAAcaactttcg caagcggtgtaaggtaAAA AAtgCG 15 nt SL atggaatgtgcgcttgcga 288 aagttgGAAAcaactttcg caagcggtgtaaggtaAAA AAtgCG 20 nt SL atggatacaccgcttgcga 289 aagttgGAAAcaactttcg caagcggtgtaaggtaAAA AAtgCG 25 nt SL taccttacaccgcttgcga 290 aagttgGAAAcaactttcg caagcggtgtaaggtaAAA AAtgCG M13_1 Oligo GTTTTATCTTCTGCTGGTG 291 GTTCGTTCGGTATTTTTAA TG M13_2 Oligo CATTAAAAATACCGAACGA 292 ACCACCAGCAGAAGATAAA AC M13_3 Oligo GACCATTTGCGAAATGTAT 293 CTAATGGTCAAACTAAATC TACTC M13_4 Oligo GAGTAGATTTAGTTTGACC 294 ATTAGATACATTTCGCAAA TGGTC
TABLE-US-00007 TABLE 5 RNA Oligonucleotides. SEQ RNA Name Sequence ID NO: ssRNA cTACGCCGattatcttctgac 332 target aactttcgcaagcggtgtaag gtaAAAAAtgCGGGCACcc crRNA GGAATGCAACtaccttacacc 333 gcttgcgaa tracrRNA CTTCACTGATAAAGTGGAGAA 334 CCGCTTCACCAAAAGCTGTCC CTTAGGGGATTAGAACTTGAG TGAAGGTGGGCTGCTTGCATC AGCCTAATGTCGAGAAGTGCT TTCTTCGGAAAGTAACCCTCG AAACAAATTCATTT crRNA 1 GACGAATGAAGGAATGCAACt 335 accttacaccgcttgcgaa crRNA 2 GACGAATGAAGGAATGCAACc 336 cttacaccgcttgcgaaag crRNA 3 GACGAATGAAGGAATGCAACt 337 tacaccgcttgcgaaagtt crRNA 4 GACGAATGAAGGAATGCAACa 338 caccgcttgcgaaagttgt crRNA MM GACGAATGAAGGAATGCAACC 339 target 2 GTCGCCGTCCAGCTCGACCA crRNA MM GACGAATGAAGGAATGCAACG 340 target 3 ATCGTTACGCTAACTATGA HERC2 A TTCACTGATAAAGTGGAGAAC 341 sgRNA CGCTTCACCAAAAGCTGTCCC TTAGGGGATTAGAACTTGAGT GAAGGTGGGCTGCTTGCATCA GCCTAATGTCGAGAAGTGCTT TCTTCGGAAAGTAACCCTCGA AACAAATTCATTTgaaaGAAT GAAGGAATGCAACacttgaca cttaatgctcaa LbCas12a GGGTAATTTCTACTAAGTGTA 342 HERC2 GATacttgacacttaatgctc crRNA aa 25 nt spacer GACGAATGAAGGAATGCAACt 343 crRNA target accttacaccgcttgcgaaag 4 ttg 20 nt spacer GACGAATGAAGGAATGCAACt 335 crRNA target accttacaccgcttgcgaa 4 18 nt spacer GACGAATGAAGGAATGCAACt 344 crRNA target accttacaccgcttgcg 4 16 nt spacer GACGAATGAAGGAATGCAACt 345 crRNA target accttacaccgcttg 4 14 nt spacer GACGAATGAAGGAATGCAACt 346 crRNA target accttacaccgct 4 12 nt spacer GACGAATGAAGGAATGCAACt 347 crRNA target accttacaccg 4 10 nt spacer GACGAATGAAGGAATGCAACt 348 crRNA target accttacac 4 Full repeat GTTGCAGAACCCGAATAGACG 349 crRNA target AATGAAGGAATGCAACtacct 4 tacaccgcttgcgaa 20 nt repeat GACGAATGAAGGAATGCAACt 335 crRNA target accttacaccgcttgcgaa 4 17 nt repeat GAATGAAGGAATGCAACtacc 350 crRNA target ttacaccgcttgcgaa 4 15 nt repeat ATGAAGGAATGCAACtacctt 351 crRNA target acaccgcttgcgaa 4 10 nt repeat GGAATGCAACtaccttacacc 333 crRNA target gcttgcgaa 4 tracrRNA CTTCACTGATAAAGTGGAGAA 352 +41 nt CCGCTTCACCAAAAGCTGTCC CTTAGGGGATTAGAACTTGAG TGAAGGTGGGCTGCTTGCATC AGCCTAATGTCGAGAAGTGCT TTCTTCGGAAAGTAACCCTCG AAACAAATTCATTTTTCCTCT CCAATTCTGCACAAAAAAAGG TGAGTCCTTAT tracrRNA CTTCACTGATAAAGTGGAGAA 334 +3 nt CCGCTTCACCAAAAGCTGTCC CTTAGGGGATTAGAACTTGAG TGAAGGTGGGCTGCTTGCATC AGCCTAATGTCGAGAAGTGCT TTCTTCGGAAAGTAACCCTCG AAACAAATTCATTT tracrRNA CTTCACTGATAAAGTGGAGAA 353 -26 CCGCTTCACCAAAAGCTGTCC CTTAGGGGATTAGAACTTGAG TGAAGGTGGGCTGCTTGCATC AGCCTAATGTCGAGAAGTGCT TTCTT tracrRNA CTTCACTGATAAAGTGGAGAA 354 -65 CCGCTTCACCAAAAGCTGTCC CTTAGGGGATTAGAACTTGAG TGAAGGTGG tracrRNA TTCACTGATAAAGTGGAGAAC 355 +0 CGCTTCACCAAAAGCTGTCCC TTAGGGGATTAGAACTTGAGT GAAGGTGGGCTGCTTGCATCA GCCTAATGTCGAGAAGTGCTT TCTTCGGAAAGTAACCCTCGA AACAAATTCA tracrRNA ttcacacTTCACTGATAAAGT 356 +90 nt GGAGAACCGCTTCACCAAAAG CTGTCCCTTAGGGGATTAGAA CTTGAGTGAAGGTGGGCTGCT TGCATCAGCCTAATGTCGAGA AGTGCTTTCTTCGGAAAGTAA CCCTCGAAACAAATTCAtttt tcctctccaattctgcacaaa aaaaggtgagtccttataaac cggcgtgcagaacgccggctc accttttttcttcattcgatt tta sgRNA 1 CTTCACTGATAAAGTGGAGAA 357 CCGCTTCACCAAAAGCTGTCC CTTAGGGGATTAGAACTTGAG TGAAGGTGGGCTGCTTGCATC AGCCTAATGTCGAGAAGTGCT TTCTTCGGAAAGTAACCCTCG AAACAAATTCATTTgaaaGAA TGAAGGAATGCAACtacctta caccgcttgcgaa sgRNA 2 CTTCACTGATAAAGTGGAGAA 358 CCGCTTCACCAAAAGCTGTCC CTTAGGGGATTAGAACTTGAG TGAAGGTGGGCTGCTTGCATC AGCCTAATGTCGAGAAGTGCT TTCTTCGGAAAGTAACCCTCG AAACAAATTCATTTTTCCTCT CCAATTCTGCACAAgaaaGTT GCAGAACCCGAATAGACGAAT GAAGGAATGCAACtaccttac accgcttgcgaa M13 target 1 GACGAATGAAGGAATGCAACT 359 crRNA ACCGAACGAACCACCAGCAGA AGA M13 target 2 GACGAATGAAGGAATGCAACT 360 crRNA CTTCTGCTGGTGGTTCGTTCG GTA M13 target 3 GACGAATGAAGGAATGCAACG 361 crRNA TTTGACCATTAGATACATTTC G M13 target 4 GACGAATGAAGGAATGCAACC 362 crRNA GAAATGTATCTAATGGTCAAA C
Example 1
[0533] FIG. 1 depicts examples of naturally occurring CasZ protein sequences.
[0534] FIG. 2 depicts schematic representations of CasZ loci, which include a Cas1 protein in addition to the CasZ protein.
[0535] FIG. 3 depicts a phylogenetic tree of CasZ sequences in relation to other Class 2 CRISPR/Cas effector protein sequences.
[0536] FIG. 4 depicts a phylogenetic tree of Cas1 sequences from CasZ loci in relation to Cas1 sequences from other Class 2 CRISPR/Cas loci.
[0537] FIG. 5 depicts transcriptomic RNA mapping data demonstrating expression of trancRNA from CasZ loci. The trancRNAs are adjacent to the CasZ repeat array, but do not include the repeat sequence and are not complementary to the repeat sequence. Shown are RNA mapping data for the following loci: CasZa3, CasZb4, CasZc5, CasZd1, and CasZe3. Small repeating aligned arrows represent the repeats of the CRISPR array (indicating the presence of guide RNA-encoding sequence); the peaks outside and adjacent to the repeat arrays represent highly transcribed trancRNAs.
[0538] This metatranscriptomic data was not 16S depleted, and hence large portions of the data were mapped to 16S, and mRNA, for example, was almost not represented at all in these reads. Nonetheless, RNA mapping to the predicted trancRNA regions was observed.
Example 2
[0539] A set of CRISPR-Cas systems from uncultivated archaea that contained Cas14, a family of exceptionally compact RNA-guided nucleases of just 400-700 amino acids were disclosed herein, including Cas1 and Cas2 proteins that are responsible for integrating DNA into CRISPR genomic loci and showed evidence of actively adapting their CRISPR arrays to new infections. Despite their small size, Cas14 proteins were capable of RNA-guided single-stranded DNA (ssDNA) cleavage without restrictive sequence requirements. Moreover, target recognition by Cas14 triggered non-specific cutting of ssDNA molecules. Metagenomic data showed that multiple CRISPR-Cas14 systems evolved independently and suggested a potential evolutionary origin of single-effector CRISPR-based adaptive immunity.
[0540] Competition between microbes and viruses stimulated the evolution of CRISPR-based adaptive immunity to provide protection against infectious agents. In class 2 CRISPR-Cas systems, a single 100-200 kilodalton (kD) CRISPR-associated (Cas) protein with multiple functional domains carried out RNA-guided binding and cutting of DNA or RNA substrates. To determine whether simpler, smaller RNA-guided proteins occurred in nature, terabase-scale metagenomic datasets were queried for uncharacterized genes proximal to both a CRISPR array and cas1, the gene that encoded the universal CRISPR integrase. This analysis identified a diverse family of CRISPR-Cas systems that contain cas1, cas2, cas4, and cas14, described herein, encoding a 40-70 kD polypeptide (FIG. 8, Panel A). Twenty-four (24) different cas14 gene variants have been identified that cluster into three subgroups (Cas14a-c) based on comparative sequence analysis (FIG. 8, Panels A-B, FIG. 9, FIG. 10). Cas14 proteins were .about.400-700 amino acids (aa), about half the size of previously known class 2 CRISPR RNA-guided enzymes (FIG. 8, Panels C-D). While the identified Cas14 proteins exhibited considerable sequence diversity, all were united by the presence of a predicted RuvC nuclease domain, whose organization was characteristic of Type V CRISPR-Cas DNA-targeting enzymes (FIG. 8, Panel D).
[0541] The identified Cas14 proteins occurred almost exclusively within DPANN, a super-phylum of symbiotic archaea characterized by small cell and genome sizes. Phylogenetic comparisons showed that Cas14 proteins were widely diverse with similarities to C2c10 and C2c9, families of bacterial RuvC-domain-containing proteins that were sometimes found near a CRISPR array but never together with other cas genes (FIG. 8, Panel B and FIG. 9). This observation and the small size of c2c10 and cas14 genes made it improbable that these systems could function as standalone CRISPR effectors.
[0542] FIG. 8, Panels A-D depict architecture and phylogeny of CRISPR-Cas14 genomic loci. FIG. 8, Panel A depicts a phylogenetic tree of Type V CRISPR systems. Newly identified miniature CRISPR systems are highlighted in orange. FIG. 8, Panel B depicts representative loci architectures for C2c10 and CRISPR-Cas14 systems. FIG. 8, Panel C depicts the length distribution of Cas14a-c systems compared to Cas12a-e and Cas9. FIG. 8, Panel D depicts the domain organization of Cas14a compared to Cas9 and Cas12a. Protein lengths are drawn to scale.
[0543] FIG. 9 depicts the maximum likelihood tree for known Type V CRISPR effectors and class 2 candidates containing a RuvC domain. Inset shows individual orthologs for each newly identified subtype. FIG. 10 depicts a maximum likelihood tree for Cas1 from known CRISPR systems.
Example 3
[0544] Based on their proximity to conserved genes responsible for creating genetic memory of infection (cas1, cas2, cas4) (FIG. 11, Panel A), it was explored whether CRISPR-Cas14 systems actively acquired DNA sequences into their CRISPR arrays. Assembled metagenomic contiguous DNA sequences (contigs) for multiple CRISPR-Cas14 loci revealed that otherwise identical CRISPR systems showed diversity in their CRISPR arrays, suggesting active adaptation to new infections (FIG. 11, Panel B and FIG. 12, Panel A). Without intending to be bound by any particular theory, it is proposed that the active acquisition of new DNA sequences indicated that these CRISPR-Cas14 loci encoded functional enzymes with nucleic acid targeting activity despite their small size. To test this possibility, it was investigated whether RNA components were produced from CRISPR-Cas14 loci. Environmental metatranscriptomic sequencing data were analyzed for the presence of RNA from the native archaeal host that contains CRISPR-Cas14a (FIG. 12, Panel B and FIG. 13, Panel A). In addition to CRISPR RNAs (crRNAs), a highly abundant non-coding RNA was mapped to about a 130-base pair sequence located between cas4a and the adjacent CRISPR array. The 20 nucleotides (nts) at the 3' end of this transcript were mostly complementary to the repeat segment of the crRNA (FIG. 12, Panel C and FIG. 13, Panel B), as observed for trans-activating CRISPR RNAs (tracrRNAs) found in association with Cas9, Cas12b and Cas12e CRISPR systems. In these previously studied systems, the double-stranded-RNA-cutting enzyme Ribonuclease III (RNase III) generated mature tracrRNAs and crRNAs, but no genes encoding RNase III were present in cas14-containing reconstructed genomes (FIG. 14, Panel A). This observation implied that an alternative mechanism for CRISPR-associated RNA processing existed in these hosts.
[0545] To test whether the Cas14a proteins and associated RNA components could assemble in a heterologous organism, a plasmid was introduced into E. coli containing a minimal CRISPR-Cas14a locus that included the Cas14 gene, the CRISPR array and intergenic regions containing the putative tracrRNA. Affinity purification of the Cas14a protein from cell lysate and sequencing of co-purifying RNA revealed a highly abundant mature crRNA as well as the putative tracrRNA, suggesting that Cas14 associated with both crRNA and tracrRNA (FIG. 14, Panel B). The calculated mass of the assembled Cas14a protein-tracrRNA-crRNA particle was 48% RNA by weight compared to just 17% for S. pyogenes Cas9 (SpCas9) (FIG. 12, Panel D) and 8% for F. novicida Cas12a (FnCas12a), hinting at a central role of the RNA in the architecture of the Cas14a complex. Known class 2 CRISPR systems required a short sequence called a protospacer adjacent motif (PAM) to target double-stranded DNA (dsDNA). To test whether Cas14a required a PAM and could conduct dsDNA interference, E. coli was transformed expressing a minimal Cas14a locus with a dsDNA plasmid containing a randomized PAM region next to a sequence matching the target-encoding sequence (spacer) in the Cas14 array. No depletion of a PAM sequence was detected among E. coli transformants, suggesting that the CRISPR-Cas14a system was either unable to target dsDNA, could do so without requiring a PAM, or was inactive in this heterologous host (FIG. 15, Panels A-D).
[0546] FIG. 11, Panels A-B depict acquisition of new spacers by CRISPR-Cas14 systems. FIG. 11, Panel A depicts alignment of Cas14 Cas1 orthologs. Expansion shows conservation of previously implicated active site residues highlighted in red boxes. FIG. 11, Panel B depicts multiple CRISPR arrays assembled for various CRISPR-Cas14 systems revealing spacer diversity for these CRISPR systems. Orange arrows indicate repeats while variously colored boxes indicate unique spacers.
[0547] FIG. 12, Panels A-D depict that CRISPR-Cas14a actively adapts and encodes a tracrRNA. FIG. 12, Panel A depicts pacer diversity for Cas14a and Cas14b with CRISPR repeats diagramed in orange and unique spacers shown in different colors. FIG. 12, Panel B depicts metatranscriptomics reads mapped to Cas14a1 and Cas14a3. Inset shows expansion of most abundant repeat and spacer sequence. FIG. 12, Panel C depicts in silico predicted structure of Cas14a1 crRNA and tracrRNA. RNase III orthologs were not identified in host genomes (FIG. 14, Panel A). FIG. 12, Panel D depicts fraction of various CRISPR complexes mass made up of by RNA and protein.
[0548] FIG. 13, Panels A-B depict metatranscriptomics for CRISPR-Cas14 loci. FIG. 13, Panel A depicts environmental RNA sequencing reads for Cas14a orthologs. Location of Cas14 and the CRISPR array indicated below. RNA structures to the right show the in silico predicted structure of the tracrRNA identified from metatranscriptomics. FIG. 13, Panel B depicts predicted hybridization for Cas14a1 crRNA:tracrRNA duplex.
[0549] FIG. 14, Panels A-B depict RNA processing and heterologous expression by CRISPR-Cas14. FIG. 14, Panel A depicts the presence of common RNase orthologs in Cas14 containing genomes. Light purple represents hits that were significantly shorter than the expected length for the given RNase. Note that RNase III is absent in all investigated genomes. FIG. 14, Panel B depicts small RNAseq reads from heterologous expression of Cas14a1 locus in E. coli (FIG. 14, Panel B, bottom two graphs) compared to metatranscriptomic reads (FIG. 14, Panel B, top graph). Pull down refers to RNA that copurified with Ni-NTA affinity purified Cas14a1.
[0550] FIG. 15, Panels A-D depict plasmid depletion by Cas14a1 and SpCas9. FIG. 15, Panel A depicts a diagram outlining a PAM discovery experiment. E. coli expressing the CRISPR system of interest was challenged with a plasmid containing a randomized PAM sequence flanking the target. The surviving (transformed) cells were harvested and sequenced along with a control harboring an empty vector. The depleted sequences were then sequenced and PAMs depleted more than the PAM Depletion Value Threshold (PDVT) were used to generate a Weblogo. FIG. 15, Panels B-D depict PAM sequences depleted by heterologously expressed Cas14a1 transformed with a target plasmid containing a randomized PAM sequence 5' (FIG. 15, Panel B) or 3' (FIG. 15, Panel C) of the target. "No sequences" indicated that no sequences were found to be depleted at or above the given PDVT.
Example 4
[0551] It was tested whether purified Cas14a-tracrRNA-crRNA complexes were capable of RNA-guided nucleic acid cleavage in vitro. All currently reconstituted DNA-targeting class 2 interference complexes were able to recognize both dsDNA and ssDNA substrates. Purified Cas14a-tracrRNA-crRNA complexes were incubated with radiolabeled target oligonucleotides (ssDNA, dsDNA, and ssRNA) bearing 20-nucleotide sequence complementary to the crRNA guide sequence, or a non-complementary ssDNA, and these substrates were analyzed for Cas14a-mediated cleavage. Only in the presence of a complementary ssDNA substrate was any cleavage product detected (FIG. 16, Panel A and FIG. 17, Panels A-C), and cleavage was dependent on the presence of both tracrRNA and crRNA, which could also be combined into a single-guide RNA (sgRNA) (FIG. 16, Panel B and FIG. 18). The lack of detectable dsDNA cleavage suggested that Cas14a targeted ssDNA selectively, although it was possible that some other factor or sequence requirement could enable dsDNA recognition in the native host. Mutation of the conserved active site residues in the Cas14a RuvC domain eliminated cleavage activity (FIG. 17, Panel A), implicating RuvC as the domain responsible for DNA cutting. Moreover, Cas14a DNA cleavage was sensitive to truncation of the RNA components to lengths shorter than the naturally produced sequences (FIG. 16, Panel B and FIG. 19, Panels A-D). These results established Cas14a as the smallest class 2 CRISPR effector demonstrated to conduct programmable RNA-guided DNA cleavage.
[0552] It was tested whether Cas14a required a PAM for ssDNA cleavage in vitro by tiling Cas14a guides across a ssDNA substrate (FIG. 16, Panel C). Despite sequence variation adjacent to the targets of these different guides, cleavage was observed for all four sequences. The cleavage sites occurred beyond the guide-complementary region of the ssDNA and shifted in response to guide binding position (FIG. 16, Panel C). These data demonstrated Cas14a was an ssDNA-targeting CRISPR endonuclease that did not require a PAM for activation.
[0553] Based on the observation that Cas14a cut outside of the crRNA/DNA targeting heteroduplex, it was proposed that Cas14a may possess target-activated non-specific ssDNA cleavage activity, similar to the RuvC-containing enzyme Cas12a. To test this possibility, Cas14a-tracrRNA-crRNA was incubated with a complementary activator DNA and an aliquot of M13 bacteriophage ssDNA bearing no sequence complementarity to the Cas14a crRNA or activator (FIG. 16, Panel D). The M13 ssDNA was rapidly degraded to small fragments, an activity that was eliminated by mutation of the conserved Cas14a RuvC active site, suggesting that activation of Cas14a resulted in non-specific ssDNA degradation.
[0554] To investigate the specificity of target-dependent non-specific DNA cutting activity by Cas14a, a fluorophore-quencher (FQ) assay was adapted in which cleavage of dye-labeled ssDNA generates a fluorescent signal (FIG. 20, Panel A). When Cas14a was incubated with various guide RNA-target ssDNA pairs, a fluorescent signal was observed only in the presence of the cognate target and showed strong preference for longer FQ-containing substrates (FIG. 19, Panel F and FIG. 20, Panel A). Cas14a mismatch tolerance was tested by tiling 2-nt mismatches across the targeted region in various ssDNA substrates. Mismatches near the middle of the ssDNA target strongly inhibited Cas14a activity, revealing an internal seed sequence that was distinct from the PAM-proximal seed region observed for dsDNA-targeting CRISPR-Cas systems (FIG. 20, Panel B and FIG. 21, Panels A-D). Moreover, DNA substrates containing strong secondary structure resulted in reduced activation of Cas14a (FIG. 21, Panel E). Truncation of ssDNA substrates also resulted in reduced or undetectable trans cleavage (FIG. 21, Panel F). These results suggested a mechanism of fidelity distinct from dsDNA-targeting class 2 CRISPR systems, possibly utilizing a preordered region of the crRNA to gate cleavage activity similarly to the RNA-targeting Cas13a enzymes.
[0555] Further investigation of compact Type V systems in metagenomic data revealed a large diversity of systems that, like Cas14a-c, include a gene encoding a short RuvC-containing protein adjacent to acquisition-associated cas genes and a CRISPR array. Twenty (20) additional such systems were found that cluster into five main families (Cas14d-h). These families seemed to have evolved from independent domestication events of TnpB, the transposase-associated protein implicated as the evolutionary parent of type V CRISPR effectors. Excluding cas14g, which was related to cas12b, the cas14-like genes formed separate clades on the type V effector phylogeny (FIG. 22, Panels A-B), and their cas1 genes had different origins (FIG. 10, Panel A). Altogether 38 CRISPR-Cas14 systems belonging to eight families (Cas14a-h) were identified and eight additional systems that could not be clustered with the analysis (termed Cas14u, Table 3).
[0556] The small size of the Cas14 proteins described herein and their resemblance to type V effector proteins suggested that RNA-guided ssDNA cleavage may have existed as an ancestral class 2 CRISPR system. In this scenario, a small, domesticated TnpB-like ssDNA interference complex may have gained additional domains over time, gradually improving dsDNA recognition and cleavage. Smaller Cas9 orthologs exhibited weaker dsDNA-targeting activity than their larger counterparts but retained the ability to robustly cleave ssDNA. Aside from the evolutionary implications, the ability of Cas14 to specifically target ssDNA suggested a role in defense against ssDNA viruses or mobile genetic elements (MGEs) that propagated through ssDNA intermediates. Without intending to be bound by any particular theory, an ssDNA-targeting CRISPR system may be particularly advantageous in certain marine environments where ssDNA viruses comprised the vast majority of viral abundance.
[0557] FIG. 16, Panels A-D depict CRISPR-Cas14a as an RNA-guided DNA-endonuclease. FIG. 16, Panel A depicts cleavage kinetics of Cas14a1 targeting ssDNA, dsDNA, ssRNA and off-target ssDNA. FIG. 16, Panel B depicts a diagram of Cas14a RNP bound to target ssDNA and Cas14a1 cleavage kinetics of radiolabeled ssDNA in the presence of various RNA components. FIG. 16, Panel C depicts tiling of a ssDNA substrate by Cas14a1 guide sequences. FIG. 16, Panel D depicts cleavage of the ssDNA viral M13 genome with activated Cas14a1.
[0558] FIG. 17, Panels A-E depict degradation of ssDNA by Cas14a1. FIG. 17, Panel A depicts SDS-PAGE of purified Cas14a1 and Cas14a1 point mutants. FIG. 17, Panel B depicts optimization of salt, cation and temperature for Cas14a1 cleavage of ssDNA targets. FIG. 17, Panel C depicts radiolabled cleavage of ssDNA by Cas14a1 with spacer sequences of various lengths. FIG. 17, Panel D depicts alignment of Cas14 with previously studied Cas12 proteins to identify RuvC active site residues and FIG. 17, Panel E depicts cleavage of ssDNA by purified Cas14a1 RuvC point mutants.
[0559] FIG. 18 depicts the kinetics of Cas14a1 cleavage of ssDNA with various guide RNA components.
[0560] FIG. 19, Panels A-F depict optimization of Cas14a1 guide RNA components. FIG. 19, Panel A depicts a diagram of Cas14a1 targeting ssDNA. Impact on Cas14a1 cleavage of an FQ ssDNA substrate by varying the spacer length (FIG. 19, Panel B), repeat length (FIG. 19, Panel C), tracrRNA (FIG. 19, Panel D), and fusing the crRNA and tracrRNA together (FIG. 19, Panel E). FIG. 19, Panel F depicts a heat map showing the background subtracted fluorescence resulting from cleavage of an ssDNA FQ reporter in the presence of various guide and target combinations.
[0561] FIG. 20, Panels A-E depict high fidelity ssDNA SNP detection by CRISPR-Cas14a. FIG. 20, Panel A depicts a fluorescent-quencher (FQ) assay for detection of ssDNA by Cas14a1 and the cleavage kinetics for various length FQ substrates. FIG. 20, Panel B depicts cleavage kinetics for Cas14a1 with mismatches tiled across the substrate (individual points represent replicate measurements). FIG. 20, panel C depicts a diagram of Cas14-DETECTR strategy and HERC2 eye color SNP. FIG. 20, panel D depicts titration of T7 exonuclease and impact on Cas14a-DETECTR. FIG. 20, panel E, depicts SNP detection using Cas14a-DETECTR with a blue-eye targeting guide for a blue-eyed and brown-eyed saliva sample compared to ssDNA detection using Cas12a.
[0562] FIG. 21, Panels A-F depict the impact of various activators on Cas14a1 cleavage rate FIG. 21, Panel A depicts a diagram of Cas14a1 targeting of ssDNA with position of mismatches used in panels A-D and raw rates for representative replicates of mismatch (MM) position for Target 1. Cleavage rates for Cas14a targeting substrates with mutations tiled across three different substrates (FIG. 21, Panels B-D). FIG. 21, Panel E depicts trans cleavage rates for substrates with increasing amounts of secondary structure. FIG. 21, Panel F depicts trans leavage rates with truncated substrates. Points represent individual measurements.
[0563] FIG. 22, Panels A-B depict diversity of CRISPR-Cas14 systems. FIG. 22, Panel A depicts representative locus architecture for indicated Cas14 systems. Protein lengths are drawn to scale. FIG. 22, Panel B depicts a maximum likelihood tree for Type V effectors including all eight identified subtypes of Cas14.
[0564] FIG. 23, Panels A-C depict a test of Cas14a1 mediated interference in a heterologous host. Diagram of Cas14a1 and LbCas12a constructs to test interference in E. coli. (B) Plaques of OX 174 spotted on E. coli revealing Cas12a- but not Cas14a1-mediated interference. Each spot represents a 10-fold dilution of the OX 174 stock. (C) Growth curves of E. coli expressing Cas14a1 or LbCas12a infected with OX 174 (T, targeting; NT, non-targeting). FIG. 19, Panel F shows a heat map showing the background-subtracted fluorescence resulting from cleavage of a ssDNA FQ reporter in the presence of various guide and target combinations after a 30-minute incubation.
[0565] While the present invention has been described with reference to the specific embodiments thereof, it should be understood by those skilled in the art that various changes may be made and equivalents may be substituted without departing from the true spirit and scope of the invention. In addition, many modifications may be made to adapt a particular situation, material, composition of matter, process, process step or steps, to the objective, spirit and scope of the present invention. All such modifications are intended to be within the scope of the claims appended hereto.
REFERENCES
[0566] 1. R. Barrangou et al., CRISPR provides acquired resistance against viruses in prokaryotes. Science. 315, 1709-12 (2007). [0567] 2. S. A. Jackson et al., CRISPR-Cas: Adapting to change. Science. 356 (6333), pp. 1-9 (2017). [0568] 3. S. Shmakov et al., Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 15, 169-182 (2017). [0569] 4. J. S. Chen, J. A. Doudna, The chemistry of Cas9 and its CRISPR colleagues. Nat. Rev. Chem. 1, 0078 (2017). [0570] 5. C. T. Brown et al., Unusual biology across a group comprising more than 15% of domain Bacteria. Nature. 523, 208-211 (2015). [0571] 6. K. Anantharaman et al., Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 7, 13219 (2016). [0572] 7. V. M. Markowitz et al., IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res. 42, 568-573 (2014). [0573] 8. V. M. Markowitz et al., IMG: The integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 40, 115-122 (2012). [0574] 9. A. J. Probst et al., Genomic resolution of a cold subsurface aquifer community provides metabolic insights for novel microbes adapted to high CO2concentrations. Environ. Microbiol. 19, 459-474 (2017). [0575] 10. I. Yosef, M. G. Goren, U. Qimron, Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40, 5569-5576 (2012). [0576] 11. J. K. Nunez, A. S. Y. Lee, A. Engelman, J. a. Doudna, Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature. 519, 193-198 (2015). [0577] 12. S. Shmakov et al., Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol. Cell. 60, 385-397 (2015). [0578] 13. D. Burstein et al., New CRISPR-Cas systems from uncultivated microbes. Nature. 542, 237-241 (2017). [0579] 14. C. Rinke et al., Insights into the phylogeny and coding potential of microbial dark matter. Nature. 499, 431-437 (2013). [0580] 15. C. J. Castelle et al., Genomic expansion of domain archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr. Biol. 25, 690-701 (2015). [0581] 16. E. Deltcheva et al., CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 471, 602-607 (2011). [0582] 17. K. E. Savell, J. J. Day, Applications of CRISPR/CAS9 in the mammalian central nervous system. Yale J. Biol. Med. 90 (2017), pp. 567-581. [0583] 18. F. J. M. Mojica, C. Diez-Villasenor, J. Garcia-Martinez, C. Almendros, Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 155, 733-740 (2009). [0584] 19. Y. Zhang, R. Rajan, H. S. Seifert, A. Mondragon, E. J. Sontheimer, DNase H Activity of Neisseria meningitidis Cas9. Mol. Cell. 60, 242-255 (2015). [0585] 20. E. Ma, L. B. Harrington, M. R. O'Connell, K. Zhou, J. A. Doudna, Single-Stranded DNA Cleavage by Divergent CRISPR-Cas9 Enzymes. Mol. Cell. 60, 398-407 (2015). [0586] 21. J. S. Chen et al., CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 360, 436-439 (2018). [0587] 22. B. Zetsche et al., Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 163, 759-771 (2015). [0588] 23. A. East-Seletsky et al., Two distinct RNase activities of CRISPR-C2c2 enable guide--RNA processing and RNA detection. Nature. 538, 270-273 (2016). [0589] 24. L. Liu et al., The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a. Cell. 170, 714-726.e10 (2017). [0590] 25. O. O. Abudayyeh et al., C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science (80-.). 353, 1-9 (2016). [0591] 26. G. J. Knott et al., Guide-bound structures of an RNA-targeting A-cleaving CRISPR-Cas13a enzyme. Nat. Struct. Mol. Biol. 24, 825-833 (2017). [0592] 27. S. Y. Li et al., CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 28, 491-493 (2018). [0593] 28. H. Eiberg et al., Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression. Hum. Genet. 123, 177-187 (2008). [0594] 29. S. Shmakov et al., Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 15, 169-182 (2017). [0595] 30. E. V Koonin, K. S. Makarova, F. Zhang, Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 37, 67-78 (2017). [0596] 31. K. S. Makarova et al., An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol, 1-15 (2015). [0597] 32. O. Barabas et al., Mechanism of IS 200/IS 605 Family DNA Transposases: Activation and Transposon--Directed Target Site Selection, 208-220 (2008). [0598] 33. M. Yoshida et al., Quantitative viral community DNA analysis reveals the dominance of single-stranded DNA viruses in offshore upper bathyal sediment from Tohoku, Japan. Front. Microbiol. 9, 1-10 (2018). [0599] 34. C. T. Brown et al., Unusual biology across a group comprising more than 15% of domain Bacteria. Nature. 523, 208-211 (2015). [0600] 35. K. Anantharaman et al., Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 7, 13219 (2016). [0601] 36. A. J. Probst et al., Genomic resolution of a cold subsurface aquifer community provides metabolic insights for novel microbes adapted to high CO2concentrations. Environ. Microbiol. 19, 459-474 (2017). [0602] 37. V. M. Markowitz et al., IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res. 42, 568-573 (2014). [0603] 38. V. M. Markowitz et al., IMG: The integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 40, 115-122 (2012). [0604] 39. R. D. Finn, J. Clements, S. R. Eddy, HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29-W37 (2011). [0605] 40. D. Burstein et al., New CRISPR-Cas systems from uncultivated microbes. Nature. 542, 237-241 (2017). [0606] 41. I. Grissa, G. Vergnaud, C. Pourcel, CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52-W57 (2007). [0607] 42. A. Biswas, R. H. J. Staals, S. E. Morales, P. C. Fineran, C. M. Brown, CRISPRDetect: A flexible algorithm to define CRISPR arrays. BMC Genomics. 17, 1-14 (2016). [0608] 43. A. Stamatakis, RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30, 1312-1313 (2014). [0609] 44. B. Langmead, S. L. Salzberg, Fast gapped-read alignment with Bowtie 2. Nat Methods. 9, 357-359 (2012). [0610] 45. H. Ogata et al., KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29-34 (1999). [0611] 46. L. B. Harrington et al., A thermostable Cas9 with increased lifetime in human plasma. Nat. Commun. 8, 1-7 (2017). [0612] 47. G. Crooks, G. Hon, J. Chandonia, S. Brenner, NCBI GenBank FTP Site\nWebLogo: a sequence logo generator. Genome Res. 14, 1188-1190 (2004). [0613] 48. L. B. Harrington et al., A Broad-Spectrum Inhibitor of CRISPR-Cas9. Cell. 170, 1224-1233.e15 (2017). [0614] 49. J. S. Chen et al., CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 360, 436-439 (2018). [0615] 50. Brown et al., Unusual biology across a group comprising more than 15% of domain Bacteria. Nature. 523, 208-211 (2015), doi:10.1038/nature14486.
Sequence CWU 1
1
3731500PRTArtificial sequenceSynthetic sequence 1Met Glu Val Gln
Lys Thr Val Met Lys Thr Leu Ser Leu Arg Ile Leu1 5 10 15Arg Pro Leu
Tyr Ser Gln Glu Ile Glu Lys Glu Ile Lys Glu Glu Lys 20 25 30Glu Arg
Arg Lys Gln Ala Gly Gly Thr Gly Glu Leu Asp Gly Gly Phe 35 40 45Tyr
Lys Lys Leu Glu Lys Lys His Ser Glu Met Phe Ser Phe Asp Arg 50 55
60Leu Asn Leu Leu Leu Asn Gln Leu Gln Arg Glu Ile Ala Lys Val Tyr65
70 75 80Asn His Ala Ile Ser Glu Leu Tyr Ile Ala Thr Ile Ala Gln Gly
Asn 85 90 95Lys Ser Asn Lys His Tyr Ile Ser Ser Ile Val Tyr Asn Arg
Ala Tyr 100 105 110Gly Tyr Phe Tyr Asn Ala Tyr Ile Ala Leu Gly Ile
Cys Ser Lys Val 115 120 125Glu Ala Asn Phe Arg Ser Asn Glu Leu Leu
Thr Gln Gln Ser Ala Leu 130 135 140Pro Thr Ala Lys Ser Asp Asn Phe
Pro Ile Val Leu His Lys Gln Lys145 150 155 160Gly Ala Glu Gly Glu
Asp Gly Gly Phe Arg Ile Ser Thr Glu Gly Ser 165 170 175Asp Leu Ile
Phe Glu Ile Pro Ile Pro Phe Tyr Glu Tyr Asn Gly Glu 180 185 190Asn
Arg Lys Glu Pro Tyr Lys Trp Val Lys Lys Gly Gly Gln Lys Pro 195 200
205Val Leu Lys Leu Ile Leu Ser Thr Phe Arg Arg Gln Arg Asn Lys Gly
210 215 220Trp Ala Lys Asp Glu Gly Thr Asp Ala Glu Ile Arg Lys Val
Thr Glu225 230 235 240Gly Lys Tyr Gln Val Ser Gln Ile Glu Ile Asn
Arg Gly Lys Lys Leu 245 250 255Gly Glu His Gln Lys Trp Phe Ala Asn
Phe Ser Ile Glu Gln Pro Ile 260 265 270Tyr Glu Arg Lys Pro Asn Arg
Ser Ile Val Gly Gly Leu Asp Val Gly 275 280 285Ile Arg Ser Pro Leu
Val Cys Ala Ile Asn Asn Ser Phe Ser Arg Tyr 290 295 300Ser Val Asp
Ser Asn Asp Val Phe Lys Phe Ser Lys Gln Val Phe Ala305 310 315
320Phe Arg Arg Arg Leu Leu Ser Lys Asn Ser Leu Lys Arg Lys Gly His
325 330 335Gly Ala Ala His Lys Leu Glu Pro Ile Thr Glu Met Thr Glu
Lys Asn 340 345 350Asp Lys Phe Arg Lys Lys Ile Ile Glu Arg Trp Ala
Lys Glu Val Thr 355 360 365Asn Phe Phe Val Lys Asn Gln Val Gly Ile
Val Gln Ile Glu Asp Leu 370 375 380Ser Thr Met Lys Asp Arg Glu Asp
His Phe Phe Asn Gln Tyr Leu Arg385 390 395 400Gly Phe Trp Pro Tyr
Tyr Gln Met Gln Thr Leu Ile Glu Asn Lys Leu 405 410 415Lys Glu Tyr
Gly Ile Glu Val Lys Arg Val Gln Ala Lys Tyr Thr Ser 420 425 430Gln
Leu Cys Ser Asn Pro Asn Cys Arg Tyr Trp Asn Asn Tyr Phe Asn 435 440
445Phe Glu Tyr Arg Lys Val Asn Lys Phe Pro Lys Phe Lys Cys Glu Lys
450 455 460Cys Asn Leu Glu Ile Ser Ala Asp Tyr Asn Ala Ala Arg Asn
Leu Ser465 470 475 480Thr Pro Asp Ile Glu Lys Phe Val Ala Lys Ala
Thr Lys Gly Ile Asn 485 490 495Leu Pro Glu Lys 5002507PRTArtificial
sequenceSynthetic sequence 2Met Glu Glu Ala Lys Thr Val Ser Lys Thr
Leu Ser Leu Arg Ile Leu1 5 10 15Arg Pro Leu Tyr Ser Ala Glu Ile Glu
Lys Glu Ile Lys Glu Glu Lys 20 25 30Glu Arg Arg Lys Gln Gly Gly Lys
Ser Gly Glu Leu Asp Ser Gly Phe 35 40 45Tyr Lys Lys Leu Glu Lys Lys
His Thr Gln Met Phe Gly Trp Asp Lys 50 55 60Leu Asn Leu Met Leu Ser
Gln Leu Gln Arg Gln Ile Ala Arg Val Phe65 70 75 80Asn Gln Ser Ile
Ser Glu Leu Tyr Ile Glu Thr Val Ile Gln Gly Lys 85 90 95Lys Ser Asn
Lys His Tyr Thr Ser Lys Ile Val Tyr Asn Arg Ala Tyr 100 105 110Ser
Val Phe Tyr Asn Ala Tyr Leu Ala Leu Gly Ile Thr Ser Lys Val 115 120
125Glu Ala Asn Phe Arg Ser Thr Glu Leu Leu Met Gln Lys Ser Ser Leu
130 135 140Pro Thr Ala Lys Ser Asp Asn Phe Pro Ile Leu Leu His Lys
Gln Lys145 150 155 160Gly Val Glu Gly Glu Glu Gly Gly Phe Lys Ile
Ser Ala Asp Gly Asn 165 170 175Asp Leu Ile Phe Glu Ile Pro Ile Pro
Phe Tyr Glu Tyr Asp Ser Ala 180 185 190Asn Lys Lys Glu Pro Phe Lys
Trp Ile Lys Lys Gly Gly Gln Lys Pro 195 200 205Thr Ile Lys Leu Ile
Leu Ser Thr Phe Arg Arg Gln Arg Asn Lys Gly 210 215 220Trp Ala Lys
Asp Glu Gly Thr Asp Ala Glu Ile Arg Lys Val Ile Glu225 230 235
240Gly Lys Tyr Gln Val Ser His Ile Glu Ile Asn Arg Gly Lys Lys Leu
245 250 255Gly Asp His Gln Lys Trp Phe Val Asn Phe Thr Ile Glu Gln
Pro Ile 260 265 270Tyr Glu Arg Lys Leu Asp Lys Asn Ile Ile Gly Gly
Ile Asp Val Gly 275 280 285Ile Lys Ser Pro Leu Val Cys Ala Val Asn
Asn Ser Phe Ala Arg Tyr 290 295 300Ser Val Asp Ser Asn Asp Val Leu
Lys Phe Ser Lys Gln Ala Phe Ala305 310 315 320Phe Arg Arg Arg Leu
Leu Ser Lys Asn Ser Leu Lys Arg Ser Gly His 325 330 335Gly Ser Lys
Asn Lys Leu Asp Pro Ile Thr Arg Met Thr Glu Lys Asn 340 345 350Asp
Arg Phe Arg Lys Lys Ile Ile Glu Arg Trp Ala Lys Glu Val Thr 355 360
365Asn Phe Phe Ile Lys Asn Gln Val Gly Thr Val Gln Ile Glu Asp Leu
370 375 380Ser Thr Met Lys Asp Arg Gln Asp Asn Phe Phe Asn Gln Tyr
Leu Arg385 390 395 400Gly Phe Trp Pro Tyr Tyr Gln Met Gln Asn Leu
Ile Glu Asn Lys Leu 405 410 415Lys Glu Tyr Gly Ile Glu Thr Lys Arg
Ile Lys Ala Arg Tyr Thr Ser 420 425 430Gln Leu Cys Ser Asn Pro Ser
Cys Arg His Trp Asn Ser Tyr Phe Ser 435 440 445Phe Asp His Arg Lys
Thr Asn Asn Phe Pro Lys Phe Lys Cys Glu Lys 450 455 460Cys Ala Leu
Glu Ile Ser Ala Asp Tyr Asn Ala Ala Arg Asn Ile Ser465 470 475
480Thr Pro Asp Ile Glu Lys Phe Val Ala Lys Ala Thr Lys Gly Ile Asn
485 490 495Leu Pro Asp Lys Asn Glu Asn Val Ile Leu Glu 500
5053529PRTArtificial sequenceSynthetic sequence 3Met Ala Lys Asn
Thr Ile Thr Lys Thr Leu Lys Leu Arg Ile Val Arg1 5 10 15Pro Tyr Asn
Ser Ala Glu Val Glu Lys Ile Val Ala Asp Glu Lys Asn 20 25 30Asn Arg
Glu Lys Ile Ala Leu Glu Lys Asn Lys Asp Lys Val Lys Glu 35 40 45Ala
Cys Ser Lys His Leu Lys Val Ala Ala Tyr Cys Thr Thr Gln Val 50 55
60Glu Arg Asn Ala Cys Leu Phe Cys Lys Ala Arg Lys Leu Asp Asp Lys65
70 75 80Phe Tyr Gln Lys Leu Arg Gly Gln Phe Pro Asp Ala Val Phe Trp
Gln 85 90 95Glu Ile Ser Glu Ile Phe Arg Gln Leu Gln Lys Gln Ala Ala
Glu Ile 100 105 110Tyr Asn Gln Ser Leu Ile Glu Leu Tyr Tyr Glu Ile
Phe Ile Lys Gly 115 120 125Lys Gly Ile Ala Asn Ala Ser Ser Val Glu
His Tyr Leu Ser Asp Val 130 135 140Cys Tyr Thr Arg Ala Ala Glu Leu
Phe Lys Asn Ala Ala Ile Ala Ser145 150 155 160Gly Leu Arg Ser Lys
Ile Lys Ser Asn Phe Arg Leu Lys Glu Leu Lys 165 170 175Asn Met Lys
Ser Gly Leu Pro Thr Thr Lys Ser Asp Asn Phe Pro Ile 180 185 190Pro
Leu Val Lys Gln Lys Gly Gly Gln Tyr Thr Gly Phe Glu Ile Ser 195 200
205Asn His Asn Ser Asp Phe Ile Ile Lys Ile Pro Phe Gly Arg Trp Gln
210 215 220Val Lys Lys Glu Ile Asp Lys Tyr Arg Pro Trp Glu Lys Phe
Asp Phe225 230 235 240Glu Gln Val Gln Lys Ser Pro Lys Pro Ile Ser
Leu Leu Leu Ser Thr 245 250 255Gln Arg Arg Lys Arg Asn Lys Gly Trp
Ser Lys Asp Glu Gly Thr Glu 260 265 270Ala Glu Ile Lys Lys Val Met
Asn Gly Asp Tyr Gln Thr Ser Tyr Ile 275 280 285Glu Val Lys Arg Gly
Ser Lys Ile Gly Glu Lys Ser Ala Trp Met Leu 290 295 300Asn Leu Ser
Ile Asp Val Pro Lys Ile Asp Lys Gly Val Asp Pro Ser305 310 315
320Ile Ile Gly Gly Ile Asp Val Gly Val Lys Ser Pro Leu Val Cys Ala
325 330 335Ile Asn Asn Ala Phe Ser Arg Tyr Ser Ile Ser Asp Asn Asp
Leu Phe 340 345 350His Phe Asn Lys Lys Met Phe Ala Arg Arg Arg Ile
Leu Leu Lys Lys 355 360 365Asn Arg His Lys Arg Ala Gly His Gly Ala
Lys Asn Lys Leu Lys Pro 370 375 380Ile Thr Ile Leu Thr Glu Lys Ser
Glu Arg Phe Arg Lys Lys Leu Ile385 390 395 400Glu Arg Trp Ala Cys
Glu Ile Ala Asp Phe Phe Ile Lys Asn Lys Val 405 410 415Gly Thr Val
Gln Met Glu Asn Leu Glu Ser Met Lys Arg Lys Glu Asp 420 425 430Ser
Tyr Phe Asn Ile Arg Leu Arg Gly Phe Trp Pro Tyr Ala Glu Met 435 440
445Gln Asn Lys Ile Glu Phe Lys Leu Lys Gln Tyr Gly Ile Glu Ile Arg
450 455 460Lys Val Ala Pro Asn Asn Thr Ser Lys Thr Cys Ser Lys Cys
Gly His465 470 475 480Leu Asn Asn Tyr Phe Asn Phe Glu Tyr Arg Lys
Lys Asn Lys Phe Pro 485 490 495His Phe Lys Cys Glu Lys Cys Asn Phe
Lys Glu Asn Ala Asp Tyr Asn 500 505 510Ala Ala Leu Asn Ile Ser Asn
Pro Lys Leu Lys Ser Thr Lys Glu Glu 515 520 525Pro4726PRTArtificial
sequenceSynthetic sequence 4Met Glu Arg Gln Lys Val Pro Gln Ile Arg
Lys Ile Val Arg Val Val1 5 10 15Pro Leu Arg Ile Leu Arg Pro Lys Tyr
Ser Asp Val Ile Glu Asn Ala 20 25 30Leu Lys Lys Phe Lys Glu Lys Gly
Asp Asp Thr Asn Thr Asn Asp Phe 35 40 45Trp Arg Ala Ile Arg Asp Arg
Asp Thr Glu Phe Phe Arg Lys Glu Leu 50 55 60Asn Phe Ser Glu Asp Glu
Ile Asn Gln Leu Glu Arg Asp Thr Leu Phe65 70 75 80Arg Val Gly Leu
Asp Asn Arg Val Leu Phe Ser Tyr Phe Asp Phe Leu 85 90 95Gln Glu Lys
Leu Met Lys Asp Tyr Asn Lys Ile Ile Ser Lys Leu Phe 100 105 110Ile
Asn Arg Gln Ser Lys Ser Ser Phe Glu Asn Asp Leu Thr Asp Glu 115 120
125Glu Val Glu Glu Leu Ile Glu Lys Asp Val Thr Pro Phe Tyr Gly Ala
130 135 140Tyr Ile Gly Lys Gly Ile Lys Ser Val Ile Lys Ser Asn Leu
Gly Gly145 150 155 160Lys Phe Ile Lys Ser Val Lys Ile Asp Arg Glu
Thr Lys Lys Val Thr 165 170 175Lys Leu Thr Ala Ile Asn Ile Gly Leu
Met Gly Leu Pro Val Ala Lys 180 185 190Ser Asp Thr Phe Pro Ile Lys
Ile Ile Lys Thr Asn Pro Asp Tyr Ile 195 200 205Thr Phe Gln Lys Ser
Thr Lys Glu Asn Leu Gln Lys Ile Glu Asp Tyr 210 215 220Glu Thr Gly
Ile Glu Tyr Gly Asp Leu Leu Val Gln Ile Thr Ile Pro225 230 235
240Trp Phe Lys Asn Glu Asn Lys Asp Phe Ser Leu Ile Lys Thr Lys Glu
245 250 255Ala Ile Glu Tyr Tyr Lys Leu Asn Gly Val Gly Lys Lys Asp
Leu Leu 260 265 270Asn Ile Asn Leu Val Leu Thr Thr Tyr His Ile Arg
Lys Lys Lys Ser 275 280 285Trp Gln Ile Asp Gly Ser Ser Gln Ser Leu
Val Arg Glu Met Ala Asn 290 295 300Gly Glu Leu Glu Glu Lys Trp Lys
Ser Phe Phe Asp Thr Phe Ile Lys305 310 315 320Lys Tyr Gly Asp Glu
Gly Lys Ser Ala Leu Val Lys Arg Arg Val Asn 325 330 335Lys Lys Ser
Arg Ala Lys Gly Glu Lys Gly Arg Glu Leu Asn Leu Asp 340 345 350Glu
Arg Ile Lys Arg Leu Tyr Asp Ser Ile Lys Ala Lys Ser Phe Pro 355 360
365Ser Glu Ile Asn Leu Ile Pro Glu Asn Tyr Lys Trp Lys Leu His Phe
370 375 380Ser Ile Glu Ile Pro Pro Met Val Asn Asp Ile Asp Ser Asn
Leu Tyr385 390 395 400Gly Gly Ile Asp Phe Gly Glu Gln Asn Ile Ala
Thr Leu Cys Val Lys 405 410 415Asn Ile Glu Lys Asp Asp Tyr Asp Phe
Leu Thr Ile Tyr Gly Asn Asp 420 425 430Leu Leu Lys His Ala Gln Ala
Ser Tyr Ala Arg Arg Arg Ile Met Arg 435 440 445Val Gln Asp Glu Tyr
Lys Ala Arg Gly His Gly Lys Ser Arg Lys Thr 450 455 460Lys Ala Gln
Glu Asp Tyr Ser Glu Arg Met Gln Lys Leu Arg Gln Lys465 470 475
480Ile Thr Glu Arg Leu Val Lys Gln Ile Ser Asp Phe Phe Leu Trp Arg
485 490 495Asn Lys Phe His Met Ala Val Cys Ser Leu Arg Tyr Glu Asp
Leu Asn 500 505 510Thr Leu Tyr Lys Gly Glu Ser Val Lys Ala Lys Arg
Met Arg Gln Phe 515 520 525Ile Asn Lys Gln Gln Leu Phe Asn Gly Ile
Glu Arg Lys Leu Lys Asp 530 535 540Tyr Asn Ser Glu Ile Tyr Val Asn
Ser Arg Tyr Pro His Tyr Thr Ser545 550 555 560Arg Leu Cys Ser Lys
Cys Gly Lys Leu Asn Leu Tyr Phe Asp Phe Leu 565 570 575Lys Phe Arg
Thr Lys Asn Ile Ile Ile Arg Lys Asn Pro Asp Gly Ser 580 585 590Glu
Ile Lys Tyr Met Pro Phe Phe Ile Cys Glu Phe Cys Gly Trp Lys 595 600
605Gln Ala Gly Asp Lys Asn Ala Ser Ala Asn Ile Ala Asp Lys Asp Tyr
610 615 620Gln Asp Lys Leu Asn Lys Glu Lys Glu Phe Cys Asn Ile Arg
Lys Pro625 630 635 640Lys Ser Lys Lys Glu Asp Ile Gly Glu Glu Asn
Glu Glu Glu Arg Asp 645 650 655Tyr Ser Arg Arg Phe Asn Arg Asn Ser
Phe Ile Tyr Asn Ser Leu Lys 660 665 670Lys Asp Asn Lys Leu Asn Gln
Glu Lys Leu Phe Asp Glu Trp Lys Asn 675 680 685Gln Leu Lys Arg Lys
Ile Asp Gly Arg Asn Lys Phe Glu Pro Lys Glu 690 695 700Tyr Lys Asp
Arg Phe Ser Tyr Leu Phe Ala Tyr Tyr Gln Glu Ile Ile705 710 715
720Lys Asn Glu Ser Glu Ser 7255517PRTArtificial sequenceSynthetic
sequence 5Met Val Pro Thr Glu Leu Ile Thr Lys Thr Leu Gln Leu Arg
Val Ile1 5 10 15Arg Pro Leu Tyr Phe Glu Glu Ile Glu Lys Glu Leu Ala
Glu Leu Lys 20 25 30Glu Gln Lys Glu Lys Glu Phe Glu Glu Thr Asn Ser
Leu Leu Leu Glu 35 40 45Ser Lys Lys Ile Asp Ala Lys Ser Leu Lys Lys
Leu Lys Arg Lys Ala 50 55 60Arg Ser Ser Ala Ala Val Glu Phe Trp Lys
Ile Ala Lys Glu Lys Tyr65 70 75 80Pro Asp Ile Leu Thr Lys Pro Glu
Met Glu Phe Ile Phe Ser Glu Met 85 90 95Gln Lys Met Met Ala Arg Phe
Tyr Asn Lys Ser Met Thr Asn Ile Phe 100 105 110Ile Glu Met Asn Asn
Asp Glu Lys Val Asn Pro Leu Ser Leu Ile Ser 115 120 125Lys Ala Ser
Thr Glu Ala Asn Gln Val Ile Lys Cys Ser Ser Ile Ser 130 135 140Ser
Gly Leu Asn Arg Lys Ile Ala Gly Ser Ile Asn Lys Thr Lys Phe145 150
155 160Lys Gln Val Arg Asp Gly Leu Ile Ser Leu Pro Thr Ala Arg Thr
Glu 165 170 175Thr Phe Pro Ile
Ser Phe Tyr Lys Ser Thr Ala Asn Lys Asp Glu Ile 180 185 190Pro Ile
Ser Lys Ile Asn Leu Pro Ser Glu Glu Glu Ala Asp Leu Thr 195 200
205Ile Thr Leu Pro Phe Pro Phe Phe Glu Ile Lys Lys Glu Lys Lys Gly
210 215 220Gln Lys Ala Tyr Ser Tyr Phe Asn Ile Ile Glu Lys Ser Gly
Arg Ser225 230 235 240Asn Asn Lys Ile Asp Leu Leu Leu Ser Thr His
Arg Arg Gln Arg Arg 245 250 255Lys Gly Trp Lys Glu Glu Gly Gly Thr
Ser Ala Glu Ile Arg Arg Leu 260 265 270Met Glu Gly Glu Phe Asp Lys
Glu Trp Glu Ile Tyr Leu Gly Glu Ala 275 280 285Glu Lys Ser Glu Lys
Ala Lys Asn Asp Leu Ile Lys Asn Met Thr Arg 290 295 300Gly Lys Leu
Ser Lys Asp Ile Lys Glu Gln Leu Glu Asp Ile Gln Val305 310 315
320Lys Tyr Phe Ser Asp Asn Asn Val Glu Ser Trp Asn Asp Leu Ser Lys
325 330 335Glu Gln Lys Gln Glu Leu Ser Lys Leu Arg Lys Lys Lys Val
Glu Glu 340 345 350Leu Lys Asp Trp Lys His Val Lys Glu Ile Leu Lys
Thr Arg Ala Lys 355 360 365Ile Gly Trp Val Glu Leu Lys Arg Gly Lys
Arg Gln Arg Asp Arg Asn 370 375 380Lys Trp Phe Val Asn Ile Thr Ile
Thr Arg Pro Pro Phe Ile Asn Lys385 390 395 400Glu Leu Asp Asp Thr
Lys Phe Gly Gly Ile Asp Leu Gly Val Lys Val 405 410 415Pro Phe Val
Cys Ala Val His Gly Ser Pro Ala Arg Leu Ile Ile Lys 420 425 430Glu
Asn Glu Ile Leu Gln Phe Asn Lys Met Val Ser Ala Arg Asn Arg 435 440
445Gln Ile Thr Lys Asp Ser Glu Gln Arg Lys Gly Arg Gly Lys Lys Asn
450 455 460Lys Phe Ile Lys Lys Glu Ile Phe Asn Glu Arg Asn Glu Leu
Phe Arg465 470 475 480Lys Lys Ile Ile Glu Arg Trp Ala Asn Gln Ile
Val Lys Phe Phe Glu 485 490 495Asp Gln Lys Cys Ala Thr Val Gln Ile
Glu Asn Leu Glu Ser Phe Asp 500 505 510Arg Thr Ser Tyr Lys
5156481PRTArtificial sequenceSynthetic sequence 6Met Lys Ser Asp
Thr Lys Asp Lys Lys Ile Ile Ile His Gln Thr Lys1 5 10 15Thr Leu Ser
Leu Arg Ile Val Lys Pro Gln Ser Ile Pro Met Glu Glu 20 25 30Phe Thr
Asp Leu Val Arg Tyr His Gln Met Ile Ile Phe Pro Val Tyr 35 40 45Asn
Asn Gly Ala Ile Asp Leu Tyr Lys Lys Leu Phe Lys Ala Lys Ile 50 55
60Gln Lys Gly Asn Glu Ala Arg Ala Ile Lys Tyr Phe Met Asn Lys Ile65
70 75 80Val Tyr Ala Pro Ile Ala Asn Thr Val Lys Asn Ser Tyr Ile Ala
Leu 85 90 95Gly Tyr Ser Thr Lys Met Gln Ser Ser Phe Ser Gly Lys Arg
Leu Trp 100 105 110Asp Leu Arg Phe Gly Glu Ala Thr Pro Pro Thr Ile
Lys Ala Asp Phe 115 120 125Pro Leu Pro Phe Tyr Asn Gln Ser Gly Phe
Lys Val Ser Ser Glu Asn 130 135 140Gly Glu Phe Ile Ile Gly Ile Pro
Phe Gly Gln Tyr Thr Lys Lys Thr145 150 155 160Val Ser Asp Ile Glu
Lys Lys Thr Ser Phe Ala Trp Asp Lys Phe Thr 165 170 175Leu Glu Asp
Thr Thr Lys Lys Thr Leu Ile Glu Leu Leu Leu Ser Thr 180 185 190Lys
Thr Arg Lys Met Asn Glu Gly Trp Lys Asn Asn Glu Gly Thr Glu 195 200
205Ala Glu Ile Lys Arg Val Met Asp Gly Thr Tyr Gln Val Thr Ser Leu
210 215 220Glu Ile Leu Gln Arg Asp Asp Ser Trp Phe Val Asn Phe Asn
Ile Ala225 230 235 240Tyr Asp Ser Leu Lys Lys Gln Pro Asp Arg Asp
Lys Ile Ala Gly Ile 245 250 255His Met Gly Ile Thr Arg Pro Leu Thr
Ala Val Ile Tyr Asn Asn Lys 260 265 270Tyr Arg Ala Leu Ser Ile Tyr
Pro Asn Thr Val Met His Leu Thr Gln 275 280 285Lys Gln Leu Ala Arg
Ile Lys Glu Gln Arg Thr Asn Ser Lys Tyr Ala 290 295 300Thr Gly Gly
His Gly Arg Asn Ala Lys Val Thr Gly Thr Asp Thr Leu305 310 315
320Ser Glu Ala Tyr Arg Gln Arg Arg Lys Lys Ile Ile Glu Asp Trp Ile
325 330 335Ala Ser Ile Val Lys Phe Ala Ile Asn Asn Glu Ile Gly Thr
Ile Tyr 340 345 350Leu Glu Asp Ile Ser Asn Thr Asn Ser Phe Phe Ala
Ala Arg Glu Gln 355 360 365Lys Leu Ile Tyr Leu Glu Asp Ile Ser Asn
Thr Asn Ser Phe Leu Ser 370 375 380Thr Tyr Lys Tyr Pro Ile Ser Ala
Ile Ser Asp Thr Leu Gln His Lys385 390 395 400Leu Glu Glu Lys Ala
Ile Gln Val Ile Arg Lys Lys Ala Tyr Tyr Val 405 410 415Asn Gln Ile
Cys Ser Leu Cys Gly His Tyr Asn Lys Gly Phe Thr Tyr 420 425 430Gln
Phe Arg Arg Lys Asn Lys Phe Pro Lys Met Lys Cys Gln Gly Cys 435 440
445Leu Glu Ala Thr Ser Thr Glu Phe Asn Ala Ala Ala Asn Val Ala Asn
450 455 460Pro Asp Tyr Glu Lys Leu Leu Ile Lys His Gly Leu Leu Gln
Leu Lys465 470 475 480Lys7358PRTArtificial sequenceSynthetic
sequence 7Met Ser Thr Ile Thr Arg Gln Val Arg Leu Ser Pro Thr Pro
Glu Gln1 5 10 15Ser Arg Leu Leu Met Ala His Cys Gln Gln Tyr Ile Ser
Thr Val Asn 20 25 30Val Leu Val Ala Ala Phe Asp Ser Glu Val Leu Thr
Gly Lys Val Ser 35 40 45Thr Lys Asp Phe Arg Ala Ala Leu Pro Ser Ala
Val Lys Asn Gln Ala 50 55 60Leu Arg Asp Ala Gln Ser Val Phe Lys Arg
Ser Val Glu Leu Gly Cys65 70 75 80Leu Pro Val Leu Lys Lys Pro His
Cys Gln Trp Asn Asn Gln Asn Trp 85 90 95Arg Val Glu Gly Asp Gln Leu
Ile Leu Pro Ile Cys Lys Asp Gly Lys 100 105 110Thr Gln Gln Glu Arg
Phe Arg Cys Ala Ala Val Ala Leu Glu Gly Lys 115 120 125Ala Gly Ile
Leu Arg Ile Lys Lys Lys Arg Gly Lys Trp Ile Ala Asp 130 135 140Leu
Thr Val Thr Gln Glu Asp Ala Pro Glu Ser Ser Gly Ser Ala Ile145 150
155 160Met Gly Val Asp Leu Gly Ile Lys Val Pro Ala Val Ala His Ile
Gly 165 170 175Gly Lys Gly Thr Arg Phe Phe Gly Asn Gly Arg Ser Gln
Arg Ser Met 180 185 190Arg Arg Arg Phe Tyr Ala Arg Arg Lys Thr Leu
Gln Lys Ala Lys Lys 195 200 205Leu Arg Ala Val Arg Lys Ser Lys Gly
Lys Glu Ala Arg Trp Met Lys 210 215 220Thr Ile Asn His Gln Leu Ser
Arg Gln Ile Val Asn His Ala His Ala225 230 235 240Leu Gly Val Gly
Thr Ile Lys Ile Glu Ala Leu Gln Gly Ile Arg Lys 245 250 255Gly Thr
Thr Arg Lys Ser Arg Gly Ala Ala Ala Arg Lys Asn Asn Arg 260 265
270Met Thr Asn Thr Trp Ser Phe Ser Gln Leu Thr Leu Phe Ile Thr Tyr
275 280 285Lys Ala Gln Arg Gln Gly Ile Thr Val Glu Gln Val Asp Pro
Ala Tyr 290 295 300Thr Ser Gln Asp Cys Pro Ala Cys Arg Ala Arg Asn
Gly Ala Gln Asp305 310 315 320Arg Thr Tyr Val Cys Ser Glu Cys Gly
Trp Arg Gly His Arg Asp Thr 325 330 335Val Gly Ala Ile Asn Ile Ser
Arg Arg Ala Gly Leu Ser Gly His Arg 340 345 350Arg Gly Ala Thr Gly
Ala 3558507PRTArtificial sequenceSynthetic sequence 8Met Ile Ala
Gln Lys Thr Ile Lys Ile Lys Leu Asn Pro Thr Lys Glu1 5 10 15Gln Ile
Ile Lys Leu Asn Ser Ile Ile Glu Glu Tyr Ile Lys Val Ser 20 25 30Asn
Phe Thr Ala Lys Lys Ile Ala Glu Ile Gln Glu Ser Phe Thr Asp 35 40
45Ser Gly Leu Thr Gln Gly Thr Cys Ser Glu Cys Gly Lys Glu Lys Thr
50 55 60Tyr Arg Lys Tyr His Leu Leu Lys Lys Asp Asn Lys Leu Phe Cys
Ile65 70 75 80Thr Cys Tyr Lys Arg Lys Tyr Ser Gln Phe Thr Leu Gln
Lys Val Glu 85 90 95Phe Gln Asn Lys Thr Gly Leu Arg Asn Val Ala Lys
Leu Pro Lys Thr 100 105 110Tyr Tyr Thr Asn Ala Ile Arg Phe Ala Ser
Asp Thr Phe Ser Gly Phe 115 120 125Asp Glu Ile Ile Lys Lys Lys Gln
Asn Arg Leu Asn Ser Ile Gln Asn 130 135 140Arg Leu Asn Phe Trp Lys
Glu Leu Leu Tyr Asn Pro Ser Asn Arg Asn145 150 155 160Glu Ile Lys
Ile Lys Val Val Lys Tyr Ala Pro Lys Thr Asp Thr Arg 165 170 175Glu
His Pro His Tyr Tyr Ser Glu Ala Glu Ile Lys Gly Arg Ile Lys 180 185
190Arg Leu Glu Lys Gln Leu Lys Lys Phe Lys Met Pro Lys Tyr Pro Glu
195 200 205Phe Thr Ser Glu Thr Ile Ser Leu Gln Arg Glu Leu Tyr Ser
Trp Lys 210 215 220Asn Pro Asp Glu Leu Lys Ile Ser Ser Ile Thr Asp
Lys Asn Glu Ser225 230 235 240Met Asn Tyr Tyr Gly Lys Glu Tyr Leu
Lys Arg Tyr Ile Asp Leu Ile 245 250 255Asn Ser Gln Thr Pro Gln Ile
Leu Leu Glu Lys Glu Asn Asn Ser Phe 260 265 270Tyr Leu Cys Phe Pro
Ile Thr Lys Asn Ile Glu Met Pro Lys Ile Asp 275 280 285Asp Thr Phe
Glu Pro Val Gly Ile Asp Trp Gly Ile Thr Arg Asn Ile 290 295 300Ala
Val Val Ser Ile Leu Asp Ser Lys Thr Lys Lys Pro Lys Phe Val305 310
315 320Lys Phe Tyr Ser Ala Gly Tyr Ile Leu Gly Lys Arg Lys His Tyr
Lys 325 330 335Ser Leu Arg Lys His Phe Gly Gln Lys Lys Arg Gln Asp
Lys Ile Asn 340 345 350Lys Leu Gly Thr Lys Glu Asp Arg Phe Ile Asp
Ser Asn Ile His Lys 355 360 365Leu Ala Phe Leu Ile Val Lys Glu Ile
Arg Asn His Ser Asn Lys Pro 370 375 380Ile Ile Leu Met Glu Asn Ile
Thr Asp Asn Arg Glu Glu Ala Glu Lys385 390 395 400Ser Met Arg Gln
Asn Ile Leu Leu His Ser Val Lys Ser Arg Leu Gln 405 410 415Asn Tyr
Ile Ala Tyr Lys Ala Leu Trp Asn Asn Ile Pro Thr Asn Leu 420 425
430Val Lys Pro Glu His Thr Ser Gln Ile Cys Asn Arg Cys Gly His Gln
435 440 445Asp Arg Glu Asn Arg Pro Lys Gly Ser Lys Leu Phe Lys Cys
Val Lys 450 455 460Cys Asn Tyr Met Ser Asn Ala Asp Phe Asn Ala Ser
Ile Asn Ile Ala465 470 475 480Arg Lys Phe Tyr Ile Gly Glu Tyr Glu
Pro Phe Tyr Lys Asp Asn Glu 485 490 495Lys Met Lys Ser Gly Val Asn
Ser Ile Ser Met 500 5059534PRTArtificial sequenceSynthetic sequence
9Leu Lys Leu Ser Glu Gln Glu Asn Ile Thr Thr Gly Val Lys Phe Lys1 5
10 15Leu Lys Leu Asp Lys Glu Thr Ser Glu Gly Leu Asn Asp Tyr Phe
Asp 20 25 30Glu Tyr Gly Lys Ala Ile Asn Phe Ala Ile Lys Val Ile Gln
Lys Glu 35 40 45Leu Ala Glu Asp Arg Phe Ala Gly Lys Val Arg Leu Asp
Glu Asn Lys 50 55 60Lys Pro Leu Leu Asn Glu Asp Gly Lys Lys Ile Trp
Asp Phe Pro Asn65 70 75 80Glu Phe Cys Ser Cys Gly Lys Gln Val Asn
Arg Tyr Val Asn Gly Lys 85 90 95Ser Leu Cys Gln Glu Cys Tyr Lys Asn
Lys Phe Thr Glu Tyr Gly Ile 100 105 110Arg Lys Arg Met Tyr Ser Ala
Lys Gly Arg Lys Ala Glu Gln Asp Ile 115 120 125Asn Ile Lys Asn Ser
Thr Asn Lys Ile Ser Lys Thr His Phe Asn Tyr 130 135 140Ala Ile Arg
Glu Ala Phe Ile Leu Asp Lys Ser Ile Lys Lys Gln Arg145 150 155
160Lys Glu Arg Phe Arg Arg Leu Arg Glu Met Lys Lys Lys Leu Gln Glu
165 170 175Phe Ile Glu Ile Arg Asp Gly Asn Lys Ile Leu Cys Pro Lys
Ile Glu 180 185 190Lys Gln Arg Val Glu Arg Tyr Ile His Pro Ser Trp
Ile Asn Lys Glu 195 200 205Lys Lys Leu Glu Asp Phe Arg Gly Tyr Ser
Met Ser Asn Val Leu Gly 210 215 220Lys Ile Lys Ile Leu Asp Arg Asn
Ile Lys Arg Glu Glu Lys Ser Leu225 230 235 240Lys Glu Lys Gly Gln
Ile Asn Phe Lys Ala Arg Arg Leu Met Leu Asp 245 250 255Lys Ser Val
Lys Phe Leu Asn Asp Asn Lys Ile Ser Phe Thr Ile Ser 260 265 270Lys
Asn Leu Pro Lys Glu Tyr Glu Leu Asp Leu Pro Glu Lys Glu Lys 275 280
285Arg Leu Asn Trp Leu Lys Glu Lys Ile Lys Ile Ile Lys Asn Gln Lys
290 295 300Pro Lys Tyr Ala Tyr Leu Leu Arg Lys Asp Asp Asn Phe Tyr
Leu Gln305 310 315 320Tyr Thr Leu Glu Thr Glu Phe Asn Leu Lys Glu
Asp Tyr Ser Gly Ile 325 330 335Val Gly Ile Asp Arg Gly Val Ser His
Ile Ala Val Tyr Thr Phe Val 340 345 350His Asn Asn Gly Lys Asn Glu
Arg Pro Leu Phe Leu Asn Ser Ser Glu 355 360 365Ile Leu Arg Leu Lys
Asn Leu Gln Lys Glu Arg Asp Arg Phe Leu Arg 370 375 380Arg Lys His
Asn Lys Lys Arg Lys Lys Ser Asn Met Arg Asn Ile Glu385 390 395
400Lys Lys Ile Gln Leu Ile Leu His Asn Tyr Ser Lys Gln Ile Val Asp
405 410 415Phe Ala Lys Asn Lys Asn Ala Phe Ile Val Phe Glu Lys Leu
Glu Lys 420 425 430Pro Lys Lys Asn Arg Ser Lys Met Ser Lys Lys Ser
Gln Tyr Lys Leu 435 440 445Ser Gln Phe Thr Phe Lys Lys Leu Ser Asp
Leu Val Asp Tyr Lys Ala 450 455 460Lys Arg Glu Gly Ile Lys Val Leu
Tyr Ile Ser Pro Glu Tyr Thr Ser465 470 475 480Lys Glu Cys Ser His
Cys Gly Glu Lys Val Asn Thr Gln Arg Pro Phe 485 490 495Asn Gly Asn
Ser Ser Leu Phe Lys Cys Asn Lys Cys Gly Val Glu Leu 500 505 510Asn
Ala Asp Tyr Asn Ala Ser Ile Asn Ile Ala Lys Lys Gly Leu Asn 515 520
525Ile Leu Asn Ser Thr Asn 53010537PRTArtificial sequenceSynthetic
sequence 10Met Glu Glu Ser Ile Ile Thr Gly Val Lys Phe Lys Leu Arg
Ile Asp1 5 10 15Lys Glu Thr Thr Lys Lys Leu Asn Glu Tyr Phe Asp Glu
Tyr Gly Lys 20 25 30Ala Ile Asn Phe Ala Val Lys Ile Ile Gln Lys Glu
Leu Ala Asp Asp 35 40 45Arg Phe Ala Gly Lys Ala Lys Leu Asp Gln Asn
Lys Asn Pro Ile Leu 50 55 60Asp Glu Asn Gly Lys Lys Ile Tyr Glu Phe
Pro Asp Glu Phe Cys Ser65 70 75 80Cys Gly Lys Gln Val Asn Lys Tyr
Val Asn Asn Lys Pro Phe Cys Gln 85 90 95Glu Cys Tyr Lys Ile Arg Phe
Thr Glu Asn Gly Ile Arg Lys Arg Met 100 105 110Tyr Ser Ala Lys Gly
Arg Lys Ala Glu His Lys Ile Asn Ile Leu Asn 115 120 125Ser Thr Asn
Lys Ile Ser Lys Thr His Phe Asn Tyr Ala Ile Arg Glu 130 135 140Ala
Phe Ile Leu Asp Lys Ser Ile Lys Lys Gln Arg Lys Lys Arg Asn145 150
155 160Glu Arg Leu Arg Glu Ser Lys Lys Arg Leu Gln Gln Phe Ile Asp
Met 165 170 175Arg Asp Gly Lys Arg Glu Ile Cys Pro Thr Ile Lys Gly
Gln Lys Val 180 185 190Asp Arg Phe Ile His Pro Ser Trp Ile Thr Lys
Asp Lys Lys Leu Glu 195 200 205Asp Phe Arg Gly Tyr Thr Leu Ser Ile
Ile Asn Ser Lys Ile Lys Ile 210 215 220Leu Asp Arg Asn Ile Lys Arg
Glu Glu Lys
Ser Leu Lys Glu Lys Gly225 230 235 240Gln Ile Ile Phe Lys Ala Lys
Arg Leu Met Leu Asp Lys Ser Ile Arg 245 250 255Phe Val Gly Asp Arg
Lys Val Leu Phe Thr Ile Ser Lys Thr Leu Pro 260 265 270Lys Glu Tyr
Glu Leu Asp Leu Pro Ser Lys Glu Lys Arg Leu Asn Trp 275 280 285Leu
Lys Glu Lys Ile Glu Ile Ile Lys Asn Gln Lys Pro Lys Tyr Ala 290 295
300Tyr Leu Leu Arg Lys Asn Ile Glu Ser Glu Lys Lys Pro Asn Tyr
Glu305 310 315 320Tyr Tyr Leu Gln Tyr Thr Leu Glu Ile Lys Pro Glu
Leu Lys Asp Phe 325 330 335Tyr Asp Gly Ala Ile Gly Ile Asp Arg Gly
Ile Asn His Ile Ala Val 340 345 350Cys Thr Phe Ile Ser Asn Asp Gly
Lys Val Thr Pro Pro Lys Phe Phe 355 360 365Ser Ser Gly Glu Ile Leu
Arg Leu Lys Asn Leu Gln Lys Glu Arg Asp 370 375 380Arg Phe Leu Leu
Arg Lys His Asn Lys Asn Arg Lys Lys Gly Asn Met385 390 395 400Arg
Val Ile Glu Asn Lys Ile Asn Leu Ile Leu His Arg Tyr Ser Lys 405 410
415Gln Ile Val Asp Met Ala Lys Lys Leu Asn Ala Ser Ile Val Phe Glu
420 425 430Glu Leu Gly Arg Ile Gly Lys Ser Arg Thr Lys Met Lys Lys
Ser Gln 435 440 445Arg Tyr Lys Leu Ser Leu Phe Ile Phe Lys Lys Leu
Ser Asp Leu Val 450 455 460Asp Tyr Lys Ser Arg Arg Glu Gly Ile Arg
Val Thr Tyr Val Pro Pro465 470 475 480Glu Tyr Thr Ser Lys Glu Cys
Ser His Cys Gly Glu Lys Val Asn Thr 485 490 495Gln Arg Pro Phe Asn
Gly Asn Tyr Ser Leu Phe Lys Cys Asn Lys Cys 500 505 510Gly Ile Gln
Leu Asn Ser Asp Tyr Asn Ala Ser Ile Asn Ile Ala Lys 515 520 525Lys
Gly Leu Lys Ile Pro Asn Ser Thr 530 53511540PRTArtificial
sequenceSynthetic sequence 11Leu Trp Thr Ile Val Ile Gly Asp Phe
Ile Glu Met Pro Lys Gln Asp1 5 10 15Leu Val Thr Thr Gly Ile Lys Phe
Lys Leu Asp Val Asp Lys Glu Thr 20 25 30Arg Lys Lys Leu Asp Asp Tyr
Phe Asp Glu Tyr Gly Lys Ala Ile Asn 35 40 45Phe Ala Val Lys Ile Ile
Gln Lys Asn Leu Lys Glu Asp Arg Phe Ala 50 55 60Gly Lys Ile Ala Leu
Gly Glu Asp Lys Lys Pro Leu Leu Asp Lys Asp65 70 75 80Gly Lys Lys
Ile Tyr Asn Tyr Pro Asn Glu Ser Cys Ser Cys Gly Asn 85 90 95Gln Val
Arg Arg Tyr Val Asn Ala Lys Pro Phe Cys Val Asp Cys Tyr 100 105
110Lys Leu Lys Phe Thr Glu Asn Gly Ile Arg Lys Arg Met Tyr Ser Ala
115 120 125Arg Gly Arg Lys Ala Asp Ser Asp Ile Asn Ile Lys Asn Ser
Thr Asn 130 135 140Lys Ile Ser Lys Thr His Phe Asn Tyr Ala Ile Arg
Glu Gly Phe Ile145 150 155 160Leu Asp Lys Ser Leu Lys Lys Gln Arg
Ser Lys Arg Ile Lys Lys Leu 165 170 175Leu Glu Leu Lys Arg Lys Leu
Gln Glu Phe Ile Asp Ile Arg Gln Gly 180 185 190Gln Met Val Leu Cys
Pro Lys Ile Lys Asn Gln Arg Val Asp Lys Phe 195 200 205Ile His Pro
Ser Trp Leu Lys Arg Asp Lys Lys Leu Glu Glu Phe Arg 210 215 220Gly
Tyr Ser Leu Ser Val Val Glu Gly Lys Ile Lys Ile Phe Asn Arg225 230
235 240Asn Ile Leu Arg Glu Glu Asp Ser Leu Arg Gln Arg Gly His Val
Asn 245 250 255Phe Lys Ala Asn Arg Ile Met Leu Asp Lys Ser Val Arg
Phe Leu Asp 260 265 270Gly Gly Lys Val Asn Phe Asn Leu Asn Lys Gly
Leu Pro Lys Glu Tyr 275 280 285Leu Leu Asp Leu Pro Lys Lys Glu Asn
Lys Leu Ser Trp Leu Asn Glu 290 295 300Lys Ile Ser Leu Ile Lys Leu
Gln Lys Pro Lys Tyr Ala Tyr Leu Leu305 310 315 320Arg Arg Glu Gly
Ser Phe Phe Ile Gln Tyr Thr Ile Glu Asn Val Pro 325 330 335Lys Thr
Phe Ser Asp Tyr Leu Gly Ala Ile Gly Ile Asp Arg Gly Ile 340 345
350Ser His Ile Ala Val Cys Thr Phe Val Ser Lys Asn Gly Val Asn Lys
355 360 365Ala Pro Val Phe Phe Ser Ser Gly Glu Ile Leu Lys Leu Lys
Ser Leu 370 375 380Gln Lys Gln Arg Asp Leu Phe Leu Arg Gly Lys His
Asn Lys Ile Arg385 390 395 400Lys Lys Ser Asn Met Arg Asn Ile Asp
Asn Lys Ile Asn Leu Ile Leu 405 410 415His Lys Tyr Ser Arg Asn Ile
Val Asn Leu Ala Lys Ser Glu Lys Ala 420 425 430Phe Ile Val Phe Glu
Lys Leu Glu Lys Ile Lys Lys Ser Arg Phe Lys 435 440 445Met Ser Lys
Ser Leu Gln Tyr Lys Leu Ser Gln Phe Thr Phe Lys Lys 450 455 460Leu
Ser Asp Leu Val Glu Tyr Lys Ala Lys Ile Glu Gly Ile Lys Val465 470
475 480Asp Tyr Val Pro Pro Glu Tyr Thr Ser Lys Glu Cys Ser His Cys
Gly 485 490 495Glu Lys Val Asp Thr Gln Arg Pro Phe Asn Gly Asn Ser
Ser Leu Phe 500 505 510Lys Cys Asn Lys Cys Arg Val Gln Leu Asn Ala
Asp Tyr Asn Ala Ser 515 520 525Ile Asn Ile Ala Lys Lys Ser Leu Asn
Ile Ser Asn 530 535 54012542PRTArtificial sequenceSynthetic
sequence 12Met Ser Lys Thr Thr Ile Ser Val Lys Leu Lys Ile Ile Asp
Leu Ser1 5 10 15Ser Glu Lys Lys Glu Phe Leu Asp Asn Tyr Phe Asn Glu
Tyr Ala Lys 20 25 30Ala Thr Thr Phe Cys Gln Leu Arg Ile Arg Arg Leu
Leu Arg Asn Thr 35 40 45His Trp Leu Gly Lys Lys Glu Lys Ser Ser Lys
Lys Trp Ile Phe Glu 50 55 60Ser Gly Ile Cys Asp Leu Cys Gly Glu Asn
Lys Glu Leu Val Asn Glu65 70 75 80Asp Arg Asn Ser Gly Glu Pro Ala
Lys Ile Cys Lys Arg Cys Tyr Asn 85 90 95Gly Arg Tyr Gly Asn Gln Met
Ile Arg Lys Leu Phe Val Ser Thr Lys 100 105 110Lys Arg Glu Val Gln
Glu Asn Met Asp Ile Arg Arg Val Ala Lys Leu 115 120 125Asn Asn Thr
His Tyr His Arg Ile Pro Glu Glu Ala Phe Asp Met Ile 130 135 140Lys
Ala Ala Asp Thr Ala Glu Lys Arg Arg Lys Lys Asn Val Glu Tyr145 150
155 160Asp Lys Lys Arg Gln Met Glu Phe Ile Glu Met Phe Asn Asp Glu
Lys 165 170 175Lys Arg Ala Ala Arg Pro Lys Lys Pro Asn Glu Arg Glu
Thr Arg Tyr 180 185 190Val His Ile Ser Lys Leu Glu Ser Pro Ser Lys
Gly Tyr Thr Leu Asn 195 200 205Gly Ile Lys Arg Lys Ile Asp Gly Met
Gly Lys Lys Ile Glu Arg Ala 210 215 220Glu Lys Gly Leu Ser Arg Lys
Lys Ile Phe Gly Tyr Gln Gly Asn Arg225 230 235 240Ile Lys Leu Asp
Ser Asn Trp Val Arg Phe Asp Leu Ala Glu Ser Glu 245 250 255Ile Thr
Ile Pro Ser Leu Phe Lys Glu Met Lys Leu Arg Ile Thr Gly 260 265
270Pro Thr Asn Val His Ser Lys Ser Gly Gln Ile Tyr Phe Ala Glu Trp
275 280 285Phe Glu Arg Ile Asn Lys Gln Pro Asn Asn Tyr Cys Tyr Leu
Ile Arg 290 295 300Lys Thr Ser Ser Asn Gly Lys Tyr Glu Tyr Tyr Leu
Gln Tyr Thr Tyr305 310 315 320Glu Ala Glu Val Glu Ala Asn Lys Glu
Tyr Ala Gly Cys Leu Gly Val 325 330 335Asp Ile Gly Cys Ser Lys Leu
Ala Ala Ala Val Tyr Tyr Asp Ser Lys 340 345 350Asn Lys Lys Ala Gln
Lys Pro Ile Glu Ile Phe Thr Asn Pro Ile Lys 355 360 365Lys Ile Lys
Met Arg Arg Glu Lys Leu Ile Lys Leu Leu Ser Arg Val 370 375 380Lys
Val Arg His Arg Arg Arg Lys Leu Met Gln Leu Ser Lys Thr Glu385 390
395 400Pro Ile Ile Asp Tyr Thr Cys His Lys Thr Ala Arg Lys Ile Val
Glu 405 410 415Met Ala Asn Thr Ala Lys Ala Phe Ile Ser Met Glu Asn
Leu Glu Thr 420 425 430Gly Ile Lys Gln Lys Gln Gln Ala Arg Glu Thr
Lys Lys Gln Lys Phe 435 440 445Tyr Arg Asn Met Phe Leu Phe Arg Lys
Leu Ser Lys Leu Ile Glu Tyr 450 455 460Lys Ala Leu Leu Lys Gly Ile
Lys Ile Val Tyr Val Lys Pro Asp Tyr465 470 475 480Thr Ser Gln Thr
Cys Ser Ser Cys Gly Ala Asp Lys Glu Lys Thr Glu 485 490 495Arg Pro
Ser Gln Ala Ile Phe Arg Cys Leu Asn Pro Thr Cys Arg Tyr 500 505
510Tyr Gln Arg Asp Ile Asn Ala Asp Phe Asn Ala Ala Val Asn Ile Ala
515 520 525Lys Lys Ala Leu Asn Asn Thr Glu Val Val Thr Thr Leu Leu
530 535 54013564PRTArtificial sequenceSynthetic sequence 13Met Ala
Arg Ala Lys Asn Gln Pro Tyr Gln Lys Leu Thr Thr Thr Thr1 5 10 15Gly
Ile Lys Phe Lys Leu Asp Leu Ser Glu Glu Glu Gly Lys Arg Phe 20 25
30Asp Glu Tyr Phe Ser Glu Tyr Ala Lys Ala Val Asn Phe Cys Ala Lys
35 40 45Val Ile Tyr Gln Leu Arg Lys Asn Leu Lys Phe Ala Gly Lys Lys
Glu 50 55 60Leu Ala Ala Lys Glu Trp Lys Phe Glu Ile Ser Asn Cys Asp
Phe Cys65 70 75 80Asn Lys Gln Lys Glu Ile Tyr Tyr Lys Asn Ile Ala
Asn Gly Gln Lys 85 90 95Val Cys Lys Gly Cys His Arg Thr Asn Phe Ser
Asp Asn Ala Ile Arg 100 105 110Lys Lys Met Ile Pro Val Lys Gly Arg
Lys Val Glu Ser Lys Phe Asn 115 120 125Ile His Asn Thr Thr Lys Lys
Ile Ser Gly Thr His Arg His Trp Ala 130 135 140Phe Glu Asp Ala Ala
Asp Ile Ile Glu Ser Met Asp Lys Gln Arg Lys145 150 155 160Glu Lys
Gln Lys Arg Leu Arg Arg Glu Lys Arg Lys Leu Ser Tyr Phe 165 170
175Phe Glu Leu Phe Gly Asp Pro Ala Lys Arg Tyr Glu Leu Pro Lys Val
180 185 190Gly Lys Gln Arg Val Pro Arg Tyr Leu His Lys Ile Ile Asp
Lys Asp 195 200 205Ser Leu Thr Lys Lys Arg Gly Tyr Ser Leu Ser Tyr
Ile Lys Asn Lys 210 215 220Ile Lys Ile Ser Glu Arg Asn Ile Glu Arg
Asp Glu Lys Ser Leu Arg225 230 235 240Lys Ala Ser Pro Ile Ala Phe
Gly Ala Arg Lys Ile Lys Met Ser Lys 245 250 255Leu Asp Pro Lys Arg
Ala Phe Asp Leu Glu Asn Asn Val Phe Lys Ile 260 265 270Pro Gly Lys
Val Ile Lys Gly Gln Tyr Lys Phe Phe Gly Thr Asn Val 275 280 285Ala
Asn Glu His Gly Lys Lys Phe Tyr Lys Asp Arg Ile Ser Lys Ile 290 295
300Leu Ala Gly Lys Pro Lys Tyr Phe Tyr Leu Leu Arg Lys Lys Val
Ala305 310 315 320Glu Ser Asp Gly Asn Pro Ile Phe Glu Tyr Tyr Val
Gln Trp Ser Ile 325 330 335Asp Thr Glu Thr Pro Ala Ile Thr Ser Tyr
Asp Asn Ile Leu Gly Ile 340 345 350Asp Ala Gly Ile Thr Asn Leu Ala
Thr Thr Val Leu Ile Pro Lys Asn 355 360 365Leu Ser Ala Glu His Cys
Ser His Cys Gly Asn Asn His Val Lys Pro 370 375 380Ile Phe Thr Lys
Phe Phe Ser Gly Lys Glu Leu Lys Ala Ile Lys Ile385 390 395 400Lys
Ser Arg Lys Gln Lys Tyr Phe Leu Arg Gly Lys His Asn Lys Leu 405 410
415Val Lys Ile Lys Arg Ile Arg Pro Ile Glu Gln Lys Val Asp Gly Tyr
420 425 430Cys His Val Val Ser Lys Gln Ile Val Glu Met Ala Lys Glu
Arg Asn 435 440 445Ser Cys Ile Ala Leu Glu Lys Leu Glu Lys Pro Lys
Lys Ser Lys Phe 450 455 460Arg Gln Arg Arg Arg Glu Lys Tyr Ala Val
Ser Met Phe Val Phe Lys465 470 475 480Lys Leu Ala Thr Phe Ile Lys
Tyr Lys Ala Ala Arg Glu Gly Ile Glu 485 490 495Ile Ile Pro Val Glu
Pro Glu Gly Thr Ser Tyr Thr Cys Ser His Cys 500 505 510Lys Asn Ala
Gln Asn Asn Gln Arg Pro Tyr Phe Lys Pro Asn Ser Lys 515 520 525Lys
Ser Trp Thr Ser Met Phe Lys Cys Gly Lys Cys Gly Ile Glu Leu 530 535
540Asn Ser Asp Tyr Asn Ala Ala Phe Asn Ile Ala Gln Lys Ala Leu
Asn545 550 555 560Met Thr Ser Ala14610PRTArtificial
sequenceSynthetic sequence 14Met Asp Glu Lys His Phe Phe Cys Ser
Tyr Cys Asn Lys Glu Leu Lys1 5 10 15Ile Ser Lys Asn Leu Ile Asn Lys
Ile Ser Lys Gly Ser Ile Arg Glu 20 25 30Asp Glu Ala Val Ser Lys Ala
Ile Ser Ile His Asn Lys Lys Glu His 35 40 45Ser Leu Ile Leu Gly Ile
Lys Phe Lys Leu Phe Ile Glu Asn Lys Leu 50 55 60Asp Lys Lys Lys Leu
Asn Glu Tyr Phe Asp Asn Tyr Ser Lys Ala Val65 70 75 80Thr Phe Ala
Ala Arg Ile Phe Asp Lys Ile Arg Ser Pro Tyr Lys Phe 85 90 95Ile Gly
Leu Lys Asp Lys Asn Thr Lys Lys Trp Thr Phe Pro Lys Ala 100 105
110Lys Cys Val Phe Cys Leu Glu Glu Lys Glu Val Ala Tyr Ala Asn Glu
115 120 125Lys Asp Asn Ser Lys Ile Cys Thr Glu Cys Tyr Leu Lys Glu
Phe Gly 130 135 140Glu Asn Gly Ile Arg Lys Lys Ile Tyr Ser Thr Arg
Gly Arg Lys Val145 150 155 160Glu Pro Lys Tyr Asn Ile Phe Asn Ser
Thr Lys Glu Leu Ser Ser Thr 165 170 175His Tyr Asn Tyr Ala Ile Arg
Asp Ala Phe Gln Leu Leu Asp Ala Leu 180 185 190Lys Lys Gln Arg Gln
Lys Lys Leu Lys Ser Ile Phe Asn Gln Lys Leu 195 200 205Arg Leu Lys
Glu Phe Glu Asp Ile Phe Ser Asp Pro Gln Lys Arg Ile 210 215 220Glu
Leu Ser Leu Lys Pro His Gln Arg Glu Lys Arg Tyr Ile His Leu225 230
235 240Ser Lys Ser Gly Gln Glu Ser Ile Asn Arg Gly Tyr Thr Leu Arg
Phe 245 250 255Val Arg Gly Lys Ile Lys Ser Leu Thr Arg Asn Ile Glu
Arg Glu Glu 260 265 270Lys Ser Leu Arg Lys Lys Thr Pro Ile His Phe
Lys Gly Asn Arg Leu 275 280 285Met Ile Phe Pro Ala Gly Ile Lys Phe
Asp Phe Ala Ser Asn Lys Val 290 295 300Lys Ile Ser Ile Ser Lys Asn
Leu Pro Asn Glu Phe Asn Phe Ser Gly305 310 315 320Thr Asn Val Lys
Asn Glu His Gly Lys Ser Phe Phe Lys Ser Arg Ile 325 330 335Glu Leu
Ile Lys Thr Gln Lys Pro Lys Tyr Ala Tyr Val Leu Arg Lys 340 345
350Ile Lys Arg Glu Tyr Ser Lys Leu Arg Asn Tyr Glu Ile Glu Lys Ile
355 360 365Arg Leu Glu Asn Pro Asn Ala Asp Leu Cys Asp Phe Tyr Leu
Gln Tyr 370 375 380Thr Ile Glu Thr Glu Ser Arg Asn Asn Glu Glu Ile
Asn Gly Ile Ile385 390 395 400Gly Ile Asp Arg Gly Ile Thr Asn Leu
Ala Cys Leu Val Leu Leu Lys 405 410 415Lys Gly Asp Lys Lys Pro Ser
Gly Val Lys Phe Tyr Lys Gly Asn Lys 420 425 430Ile Leu Gly Met Lys
Ile Ala Tyr Arg Lys His Leu Tyr Leu Leu Lys 435 440 445Gly Lys Arg
Asn Lys Leu Arg Lys Gln Arg Gln Ile Arg Ala Ile Glu 450 455 460Pro
Lys Ile Asn Leu Ile Leu His Gln Ile Ser Lys Asp Ile Val Lys465 470
475 480Ile Ala Lys Glu Lys Asn Phe Ala Ile Ala Leu Glu Gln Leu Glu
Lys 485
490 495Pro Lys Lys Ala Arg Phe Ala Gln Arg Lys Lys Glu Lys Tyr Lys
Leu 500 505 510Ala Leu Phe Thr Phe Lys Asn Leu Ser Thr Leu Ile Glu
Tyr Lys Ser 515 520 525Lys Arg Glu Gly Ile Pro Val Ile Tyr Val Pro
Pro Glu Lys Thr Ser 530 535 540Gln Met Cys Ser His Cys Ala Ile Asn
Gly Asp Glu His Val Asp Thr545 550 555 560Gln Arg Pro Tyr Lys Lys
Pro Asn Ala Gln Lys Pro Ser Tyr Ser Leu 565 570 575Phe Lys Cys Asn
Lys Cys Gly Ile Glu Leu Asn Ala Asp Tyr Asn Ala 580 585 590Ala Phe
Asn Ile Ala Gln Lys Gly Leu Lys Thr Leu Met Leu Asn His 595 600
605Ser His 61015369PRTArtificial sequenceSynthetic sequence 15Met
Leu Gln Thr Leu Leu Val Lys Leu Asp Pro Ser Lys Glu Gln Tyr1 5 10
15Lys Met Leu Tyr Glu Thr Met Glu Arg Phe Asn Glu Ala Cys Asn Gln
20 25 30Ile Ala Glu Thr Val Phe Ala Ile His Ser Ala Asn Lys Ile Glu
Val 35 40 45Gln Lys Thr Val Tyr Tyr Pro Ile Arg Glu Lys Phe Gly Leu
Ser Ala 50 55 60Gln Leu Thr Ile Leu Ala Ile Arg Lys Val Cys Glu Ala
Tyr Lys Arg65 70 75 80Asp Lys Ser Ile Lys Pro Glu Phe Arg Leu Asp
Gly Ala Leu Val Tyr 85 90 95Asp Gln Arg Val Leu Ser Trp Lys Gly Leu
Asp Lys Val Ser Leu Val 100 105 110Thr Leu Gln Gly Arg Gln Ile Ile
Pro Ile Lys Phe Gly Asp Tyr Gln 115 120 125Lys Ala Arg Met Asp Arg
Ile Arg Gly Gln Ala Asp Leu Ile Leu Val 130 135 140Lys Gly Val Phe
Tyr Leu Cys Val Val Val Glu Val Ser Glu Glu Ser145 150 155 160Pro
Tyr Asp Pro Lys Gly Val Leu Gly Val Asp Leu Gly Ile Lys Asn 165 170
175Leu Ala Val Asp Ser Asp Gly Glu Val His Ser Gly Glu Gln Thr Thr
180 185 190Asn Thr Arg Glu Arg Leu Asp Ser Leu Lys Ala Arg Leu Gln
Ser Lys 195 200 205Gly Thr Lys Ser Ala Lys Arg His Leu Lys Lys Leu
Ser Gly Arg Met 210 215 220Ala Lys Phe Ser Lys Asp Val Asn His Cys
Ile Ser Lys Lys Leu Val225 230 235 240Ala Lys Ala Lys Gly Thr Leu
Met Ser Ile Ala Leu Glu Asp Leu Gln 245 250 255Gly Ile Arg Asp Arg
Val Thr Val Arg Lys Ala Gln Arg Arg Asn Leu 260 265 270His Thr Trp
Asn Phe Gly Leu Leu Arg Met Phe Val Asp Tyr Lys Ala 275 280 285Lys
Ile Ala Gly Val Pro Leu Val Phe Val Asp Pro Arg Asn Thr Ser 290 295
300Arg Thr Cys Pro Ser Cys Gly His Val Ala Lys Ala Asn Arg Pro
Thr305 310 315 320Arg Asp Glu Phe Arg Cys Val Ser Cys Gly Phe Ala
Gly Ala Ala Asp 325 330 335His Ile Ala Ala Met Asn Ile Ala Phe Arg
Ala Glu Val Ser Gln Pro 340 345 350Ile Val Thr Arg Phe Phe Val Gln
Ser Gln Ala Pro Ser Phe Arg Val 355 360 365Gly16552PRTArtificial
sequenceSynthetic sequence 16Met Asp Glu Glu Pro Asp Ser Ala Glu
Pro Asn Leu Ala Pro Ile Ser1 5 10 15Val Lys Leu Lys Leu Val Lys Leu
Asp Gly Glu Lys Leu Ala Ala Leu 20 25 30Asn Asp Tyr Phe Asn Glu Tyr
Ala Lys Ala Val Asn Phe Cys Glu Leu 35 40 45Lys Met Gln Lys Ile Arg
Lys Asn Leu Val Asn Ile Arg Gly Thr Tyr 50 55 60Leu Lys Glu Lys Lys
Ala Trp Ile Asn Gln Thr Gly Glu Cys Cys Ile65 70 75 80Cys Lys Lys
Ile Asp Glu Leu Arg Cys Glu Asp Lys Asn Pro Asp Ile 85 90 95Asn Gly
Lys Ile Cys Lys Lys Cys Tyr Asn Gly Arg Tyr Gly Asn Gln 100 105
110Met Ile Arg Lys Leu Phe Val Ser Thr Asn Lys Arg Ala Val Pro Lys
115 120 125Ser Leu Asp Ile Arg Lys Val Ala Arg Leu His Asn Thr His
Tyr His 130 135 140Arg Ile Pro Pro Glu Ala Ala Asp Ile Ile Lys Ala
Ile Glu Thr Ala145 150 155 160Glu Arg Lys Arg Arg Asn Arg Ile Leu
Phe Asp Glu Arg Arg Tyr Asn 165 170 175Glu Leu Lys Asp Ala Leu Glu
Asn Glu Glu Lys Arg Val Ala Arg Pro 180 185 190Lys Lys Pro Lys Glu
Arg Glu Val Arg Tyr Val Pro Ile Ser Lys Lys 195 200 205Asp Thr Pro
Ser Lys Gly Tyr Thr Met Asn Ala Leu Val Arg Lys Val 210 215 220Ser
Gly Met Ala Lys Lys Ile Glu Arg Ala Lys Arg Asn Leu Asn Lys225 230
235 240Arg Lys Lys Ile Glu Tyr Leu Gly Arg Arg Ile Leu Leu Asp Lys
Asn 245 250 255Trp Val Arg Phe Asp Phe Asp Lys Ser Glu Ile Ser Ile
Pro Thr Met 260 265 270Lys Glu Phe Phe Gly Glu Met Arg Phe Glu Ile
Thr Gly Pro Ser Asn 275 280 285Val Met Ser Pro Asn Gly Arg Glu Tyr
Phe Thr Lys Trp Phe Asp Arg 290 295 300Ile Lys Ala Gln Pro Asp Asn
Tyr Cys Tyr Leu Leu Arg Lys Glu Ser305 310 315 320Glu Asp Glu Thr
Asp Phe Tyr Leu Gln Tyr Thr Trp Arg Pro Asp Ala 325 330 335His Pro
Lys Lys Asp Tyr Thr Gly Cys Leu Gly Ile Asp Ile Gly Gly 340 345
350Ser Lys Leu Ala Ser Ala Val Tyr Phe Asp Ala Asp Lys Asn Arg Ala
355 360 365Lys Gln Pro Ile Gln Ile Phe Ser Asn Pro Ile Gly Lys Trp
Lys Thr 370 375 380Lys Arg Gln Lys Val Ile Lys Val Leu Ser Lys Ala
Ala Val Arg His385 390 395 400Lys Thr Lys Lys Leu Glu Ser Leu Arg
Asn Ile Glu Pro Arg Ile Asp 405 410 415Val His Cys His Arg Ile Ala
Arg Lys Ile Val Gly Met Ala Leu Ala 420 425 430Ala Asn Ala Phe Ile
Ser Met Glu Asn Leu Glu Gly Gly Ile Arg Glu 435 440 445Lys Gln Lys
Ala Lys Glu Thr Lys Lys Gln Lys Phe Ser Arg Asn Met 450 455 460Phe
Val Phe Arg Lys Leu Ser Lys Leu Ile Glu Tyr Lys Ala Leu Met465 470
475 480Glu Gly Val Lys Val Val Tyr Ile Val Pro Asp Tyr Thr Ser Gln
Leu 485 490 495Cys Ser Ser Cys Gly Thr Asn Asn Thr Lys Arg Pro Lys
Gln Ala Ile 500 505 510Phe Met Cys Gln Asn Thr Glu Cys Arg Tyr Phe
Gly Lys Asn Ile Asn 515 520 525Ala Asp Phe Asn Ala Ala Ile Asn Ile
Ala Lys Lys Ala Leu Asn Arg 530 535 540Lys Asp Ile Val Arg Glu Leu
Ser545 55017534PRTArtificial sequenceSynthetic sequence 17Met Glu
Lys Asn Asn Ser Glu Gln Thr Ser Ile Thr Thr Gly Ile Lys1 5 10 15Phe
Lys Leu Lys Leu Asp Lys Glu Thr Lys Glu Lys Leu Asn Asn Tyr 20 25
30Phe Asp Glu Tyr Gly Lys Ala Ile Asn Phe Ala Val Arg Ile Ile Gln
35 40 45Met Gln Leu Asn Asp Asp Arg Leu Ala Gly Lys Tyr Lys Arg Asp
Glu 50 55 60Lys Gly Lys Pro Ile Leu Gly Glu Asp Gly Lys Lys Ile Leu
Glu Ile65 70 75 80Pro Asn Asp Phe Cys Ser Cys Gly Asn Gln Val Asn
His Tyr Val Asn 85 90 95Gly Val Ser Phe Cys Gln Glu Cys Tyr Lys Lys
Arg Phe Ser Glu Asn 100 105 110Gly Ile Arg Lys Arg Met Tyr Ser Ala
Lys Gly Arg Lys Ala Glu Gln 115 120 125Asp Ile Asn Ile Lys Asn Ser
Thr Asn Lys Ile Ser Lys Thr His Phe 130 135 140Asn Tyr Ala Ile Arg
Glu Ala Phe Asn Leu Asp Lys Ser Ile Lys Lys145 150 155 160Gln Arg
Glu Lys Arg Phe Lys Lys Leu Lys Asp Met Lys Arg Lys Leu 165 170
175Gln Glu Phe Leu Glu Ile Arg Asp Gly Lys Arg Val Ile Cys Pro Lys
180 185 190Ile Glu Lys Gln Lys Val Glu Arg Tyr Ile His Pro Ser Trp
Ile Asn 195 200 205Lys Glu Lys Lys Leu Glu Glu Phe Arg Gly Tyr Ser
Leu Ser Ile Val 210 215 220Asn Ser Lys Ile Lys Ser Phe Asp Arg Asn
Ile Gln Arg Glu Glu Lys225 230 235 240Ser Leu Lys Glu Lys Gly Gln
Ile Asn Phe Lys Ala Gln Arg Leu Met 245 250 255Leu Asp Lys Ser Val
Lys Phe Leu Lys Asp Asn Lys Val Ser Phe Thr 260 265 270Ile Ser Lys
Glu Leu Pro Lys Thr Phe Glu Leu Asp Leu Pro Lys Lys 275 280 285Glu
Lys Lys Leu Asn Trp Leu Asn Glu Lys Leu Glu Ile Ile Lys Asn 290 295
300Gln Lys Pro Lys Tyr Ala Tyr Leu Leu Arg Lys Glu Asn Asn Ile
Phe305 310 315 320Leu Gln Tyr Thr Leu Asp Ser Ile Pro Glu Ile His
Ser Glu Tyr Ser 325 330 335Gly Ala Val Gly Ile Asp Arg Gly Val Ser
His Ile Ala Val Tyr Thr 340 345 350Phe Leu Asp Lys Asp Gly Lys Asn
Glu Arg Pro Phe Phe Leu Ser Ser 355 360 365Ser Gly Ile Leu Arg Leu
Lys Asn Leu Gln Lys Glu Arg Asp Lys Phe 370 375 380Leu Arg Lys Lys
His Asn Lys Ile Arg Lys Lys Gly Asn Met Arg Asn385 390 395 400Ile
Glu Gln Lys Ile Asn Leu Ile Leu His Glu Tyr Ser Lys Gln Ile 405 410
415Val Asn Phe Ala Lys Asp Lys Asn Ala Phe Ile Val Phe Glu Leu Leu
420 425 430Glu Lys Pro Lys Lys Ser Arg Glu Arg Met Ser Lys Lys Ile
Gln Tyr 435 440 445Lys Leu Ser Gln Phe Thr Phe Lys Lys Leu Ser Asp
Leu Val Asp Tyr 450 455 460Lys Ala Lys Arg Glu Gly Ile Lys Val Ile
Tyr Val Glu Pro Ala Tyr465 470 475 480Thr Ser Lys Asp Cys Ser His
Cys Gly Glu Arg Val Asn Thr Gln Arg 485 490 495Pro Phe Asn Gly Asn
Phe Ser Leu Phe Lys Cys Asn Lys Cys Gly Ile 500 505 510Val Leu Asn
Ser Asp Tyr Asn Ala Ser Leu Asn Ile Ala Arg Lys Gly 515 520 525Leu
Asn Ile Ser Ala Asn 53018577PRTArtificial sequenceSynthetic
sequence 18Met Ala Glu Glu Lys Phe Phe Phe Cys Glu Lys Cys Asn Lys
Asp Ile1 5 10 15Lys Ile Pro Lys Asn Tyr Ile Asn Lys Gln Gly Ala Glu
Glu Lys Ala 20 25 30Arg Ala Lys His Glu His Arg Val His Ala Leu Ile
Leu Gly Ile Lys 35 40 45Phe Lys Ile Tyr Pro Lys Lys Glu Asp Ile Ser
Lys Leu Asn Asp Tyr 50 55 60Phe Asp Glu Tyr Ala Lys Ala Val Thr Phe
Thr Ala Lys Ile Val Asp65 70 75 80Lys Leu Lys Ala Pro Phe Leu Phe
Ala Gly Lys Arg Asp Lys Asp Thr 85 90 95Ser Lys Lys Lys Trp Val Phe
Pro Val Asp Lys Cys Ser Phe Cys Lys 100 105 110Glu Lys Thr Glu Ile
Asn Tyr Arg Thr Lys Gln Gly Lys Asn Ile Cys 115 120 125Asn Ser Cys
Tyr Leu Thr Glu Phe Gly Glu Gln Gly Leu Leu Glu Lys 130 135 140Ile
Tyr Ala Thr Lys Gly Arg Lys Val Ser Ser Ser Phe Asn Leu Phe145 150
155 160Asn Ser Thr Lys Lys Leu Thr Gly Thr His Asn Asn Tyr Val Val
Lys 165 170 175Glu Ser Leu Gln Leu Leu Asp Ala Leu Lys Lys Gln Arg
Ser Lys Arg 180 185 190Leu Lys Lys Leu Ser Asn Thr Arg Arg Lys Leu
Lys Gln Phe Glu Glu 195 200 205Met Phe Glu Lys Glu Asp Lys Arg Phe
Gln Leu Pro Leu Lys Glu Lys 210 215 220Gln Arg Glu Leu Arg Phe Ile
His Val Ser Gln Lys Asp Arg Ala Thr225 230 235 240Glu Phe Lys Gly
Tyr Thr Met Asn Lys Ile Lys Ser Lys Ile Lys Val 245 250 255Leu Arg
Arg Asn Ile Glu Arg Glu Gln Arg Ser Leu Asn Arg Lys Ser 260 265
270Pro Val Phe Phe Arg Gly Thr Arg Ile Arg Leu Ser Pro Ser Val Gln
275 280 285Phe Asp Asp Lys Asp Asn Lys Ile Lys Leu Thr Leu Ser Lys
Glu Leu 290 295 300Pro Lys Glu Tyr Ser Phe Ser Gly Leu Asn Val Ala
Asn Glu His Gly305 310 315 320Arg Lys Phe Phe Ala Glu Lys Leu Lys
Leu Ile Lys Glu Asn Lys Ser 325 330 335Lys Tyr Ala Tyr Leu Leu Arg
Arg Gln Val Asn Lys Asn Asn Lys Lys 340 345 350Pro Ile Tyr Asp Tyr
Tyr Leu Gln Tyr Thr Val Glu Phe Leu Pro Asn 355 360 365Ile Ile Thr
Asn Tyr Asn Gly Ile Leu Gly Ile Asp Arg Gly Ile Asn 370 375 380Thr
Leu Ala Cys Ile Val Leu Leu Glu Asn Lys Lys Glu Lys Pro Ser385 390
395 400Phe Val Lys Phe Phe Ser Gly Lys Gly Ile Leu Asn Leu Lys Asn
Lys 405 410 415Arg Arg Lys Gln Leu Tyr Phe Leu Lys Gly Val His Asn
Lys Tyr Arg 420 425 430Lys Gln Gln Lys Ile Arg Pro Ile Glu Pro Arg
Ile Asp Gln Ile Leu 435 440 445His Asp Ile Ser Lys Gln Ile Ile Asp
Leu Ala Lys Glu Lys Arg Val 450 455 460Ala Ile Ser Leu Glu Gln Leu
Glu Lys Pro Gln Lys Pro Lys Phe Arg465 470 475 480Gln Ser Arg Lys
Ala Lys Tyr Lys Leu Ser Gln Phe Asn Phe Lys Thr 485 490 495Leu Ser
Asn Tyr Ile Asp Tyr Lys Ala Lys Lys Glu Gly Ile Arg Val 500 505
510Ile Tyr Ile Ala Pro Glu Met Thr Ser Gln Asn Cys Ser Arg Cys Ala
515 520 525Met Lys Asn Asp Leu His Val Asn Thr Gln Arg Pro Tyr Lys
Asn Thr 530 535 540Ser Ser Leu Phe Lys Cys Asn Lys Cys Gly Val Glu
Leu Asn Ala Asp545 550 555 560Tyr Asn Ala Ala Phe Asn Ile Ala Gln
Lys Gly Leu Lys Ile Leu Asn 565 570 575Ser19613PRTArtificial
sequenceSynthetic sequence 19Met Ile Ser Leu Lys Leu Lys Leu Leu
Pro Asp Glu Glu Gln Lys Lys1 5 10 15Leu Leu Asp Glu Met Phe Trp Lys
Trp Ala Ser Ile Cys Thr Arg Val 20 25 30Gly Phe Gly Arg Ala Asp Lys
Glu Asp Leu Lys Pro Pro Lys Asp Ala 35 40 45Glu Gly Val Trp Phe Ser
Leu Thr Gln Leu Asn Gln Ala Asn Thr Asp 50 55 60Ile Asn Asp Leu Arg
Glu Ala Met Lys His Gln Lys His Arg Leu Glu65 70 75 80Tyr Glu Lys
Asn Arg Leu Glu Ala Gln Arg Asp Asp Thr Gln Asp Ala 85 90 95Leu Lys
Asn Pro Asp Arg Arg Glu Ile Ser Thr Lys Arg Lys Asp Leu 100 105
110Phe Arg Pro Lys Ala Ser Val Glu Lys Gly Phe Leu Lys Leu Lys Tyr
115 120 125His Gln Glu Arg Tyr Trp Val Arg Arg Leu Lys Glu Ile Asn
Lys Leu 130 135 140Ile Glu Arg Lys Thr Lys Thr Leu Ile Lys Ile Glu
Lys Gly Arg Ile145 150 155 160Lys Phe Lys Ala Thr Arg Ile Thr Leu
His Gln Gly Ser Phe Lys Ile 165 170 175Arg Phe Gly Asp Lys Pro Ala
Phe Leu Ile Lys Ala Leu Ser Gly Lys 180 185 190Asn Gln Ile Asp Ala
Pro Phe Val Val Val Pro Glu Gln Pro Ile Cys 195 200 205Gly Ser Val
Val Asn Ser Lys Lys Tyr Leu Asp Glu Ile Thr Thr Asn 210 215 220Phe
Leu Ala Tyr Ser Val Asn Ala Met Leu Phe Gly Leu Ser Arg Ser225 230
235 240Glu Glu Met Leu Leu Lys Ala Lys Arg Pro Glu Lys Ile Lys Lys
Lys 245 250 255Glu Glu Lys Leu Ala Lys Lys Gln Ser Ala Phe Glu Asn
Lys Lys Lys 260 265 270Glu Leu Gln Lys Leu Leu Gly Arg Glu Leu Thr
Gln Gln Glu Glu Ala 275 280 285Ile Ile Glu Glu Thr Arg Asn Gln Phe
Phe Gln Asp Phe Glu Val Lys 290
295 300Ile Thr Lys Gln Tyr Ser Glu Leu Leu Ser Lys Ile Ala Asn Glu
Leu305 310 315 320Lys Gln Lys Asn Asp Phe Leu Lys Val Asn Lys Tyr
Pro Ile Leu Leu 325 330 335Arg Lys Pro Leu Lys Lys Ala Lys Ser Lys
Lys Ile Asn Asn Leu Ser 340 345 350Pro Ser Glu Trp Lys Tyr Tyr Leu
Gln Phe Gly Val Lys Pro Leu Leu 355 360 365Lys Gln Lys Ser Arg Arg
Lys Ser Arg Asn Val Leu Gly Ile Asp Arg 370 375 380Gly Leu Lys His
Leu Leu Ala Val Thr Val Leu Glu Pro Asp Lys Lys385 390 395 400Thr
Phe Val Trp Asn Lys Leu Tyr Pro Asn Pro Ile Thr Gly Trp Lys 405 410
415Trp Arg Arg Arg Lys Leu Leu Arg Ser Leu Lys Arg Leu Lys Arg Arg
420 425 430Ile Lys Ser Gln Lys His Glu Thr Ile His Glu Asn Gln Thr
Arg Lys 435 440 445Lys Leu Lys Ser Leu Gln Gly Arg Ile Asp Asp Leu
Leu His Asn Ile 450 455 460Ser Arg Lys Ile Val Glu Thr Ala Lys Glu
Tyr Asp Ala Val Ile Val465 470 475 480Val Glu Asp Leu Gln Ser Met
Arg Gln His Gly Arg Ser Lys Gly Asn 485 490 495Arg Leu Lys Thr Leu
Asn Tyr Ala Leu Ser Leu Phe Asp Tyr Ala Asn 500 505 510Val Met Gln
Leu Ile Lys Tyr Lys Ala Gly Ile Glu Gly Ile Gln Ile 515 520 525Tyr
Asp Val Lys Pro Ala Gly Thr Ser Gln Asn Cys Ala Tyr Cys Leu 530 535
540Leu Ala Gln Arg Asp Ser His Glu Tyr Lys Arg Ser Gln Glu Asn
Ser545 550 555 560Lys Ile Gly Val Cys Leu Asn Pro Asn Cys Gln Asn
His Lys Lys Gln 565 570 575Ile Asp Ala Asp Leu Asn Ala Ala Arg Val
Ile Ala Ser Cys Tyr Ala 580 585 590Leu Lys Ile Asn Asp Ser Gln Pro
Phe Gly Thr Arg Lys Arg Phe Lys 595 600 605Lys Arg Thr Thr Asn
61020615PRTArtificial sequenceSynthetic sequence 20Met Glu Thr Leu
Ser Leu Lys Leu Lys Leu Asn Pro Ser Lys Glu Gln1 5 10 15Leu Leu Val
Leu Asp Lys Met Phe Trp Lys Trp Ala Ser Ile Cys Thr 20 25 30Arg Leu
Gly Leu Lys Lys Ala Glu Met Ser Asp Leu Glu Pro Pro Lys 35 40 45Asp
Ala Glu Gly Val Trp Phe Ser Lys Thr Gln Leu Asn Gln Ala Asn 50 55
60Thr Asp Val Asn Asp Leu Arg Lys Ala Met Gln His Gln Gly Lys Arg65
70 75 80Ile Glu Tyr Glu Leu Asp Lys Val Glu Asn Arg Arg Asn Glu Ile
Gln 85 90 95Glu Met Leu Glu Lys Pro Asp Arg Arg Asp Ile Ser Pro Asn
Arg Lys 100 105 110Asp Leu Phe Arg Pro Lys Ala Ala Val Glu Lys Gly
Tyr Leu Lys Leu 115 120 125Lys Tyr His Lys Leu Gly Tyr Trp Ser Lys
Glu Leu Lys Thr Ala Asn 130 135 140Lys Leu Ile Glu Arg Lys Arg Lys
Thr Leu Ala Lys Ile Asp Ala Gly145 150 155 160Lys Met Lys Phe Lys
Pro Thr Arg Ile Ser Leu His Thr Asn Ser Phe 165 170 175Arg Ile Lys
Phe Gly Glu Glu Pro Lys Ile Ala Leu Ser Thr Thr Ser 180 185 190Lys
His Glu Lys Ile Glu Leu Pro Leu Ile Thr Ser Leu Gln Arg Pro 195 200
205Leu Lys Thr Ser Cys Ala Lys Lys Ser Lys Thr Tyr Leu Asp Ala Ala
210 215 220Ile Leu Asn Phe Leu Ala Tyr Ser Thr Asn Ala Ala Leu Phe
Gly Leu225 230 235 240Ser Arg Ser Glu Glu Met Leu Leu Lys Ala Lys
Lys Pro Glu Lys Ile 245 250 255Glu Lys Arg Asp Arg Lys Leu Ala Thr
Lys Arg Glu Ser Phe Asp Lys 260 265 270Lys Leu Lys Thr Leu Glu Lys
Leu Leu Glu Arg Lys Leu Ser Glu Lys 275 280 285Glu Lys Ser Val Phe
Lys Arg Lys Gln Thr Glu Phe Phe Asp Lys Phe 290 295 300Cys Ile Thr
Leu Asp Glu Thr Tyr Val Glu Ala Leu His Arg Ile Ala305 310 315
320Glu Glu Leu Val Ser Lys Asn Lys Tyr Leu Glu Ile Lys Lys Tyr Pro
325 330 335Val Leu Leu Arg Lys Pro Glu Ser Arg Leu Arg Ser Lys Lys
Leu Lys 340 345 350Asn Leu Lys Pro Glu Asp Trp Thr Tyr Tyr Ile Gln
Phe Gly Phe Gln 355 360 365Pro Leu Leu Asp Thr Pro Lys Pro Ile Lys
Thr Lys Thr Val Leu Gly 370 375 380Ile Asp Arg Gly Val Arg His Leu
Leu Ala Val Ser Ile Phe Asp Pro385 390 395 400Arg Thr Lys Thr Phe
Thr Phe Asn Arg Leu Tyr Ser Asn Pro Ile Val 405 410 415Asp Trp Lys
Trp Arg Arg Arg Lys Leu Leu Arg Ser Ile Lys Arg Leu 420 425 430Lys
Arg Arg Leu Lys Ser Glu Lys His Val His Leu His Glu Asn Gln 435 440
445Phe Lys Ala Lys Leu Arg Ser Leu Glu Gly Arg Ile Glu Asp His Phe
450 455 460His Asn Leu Ser Lys Glu Ile Val Asp Leu Ala Lys Glu Asn
Asn Ser465 470 475 480Val Ile Val Val Glu Asn Leu Gly Gly Met Arg
Gln His Gly Arg Gly 485 490 495Arg Gly Lys Trp Leu Lys Ala Leu Asn
Tyr Ala Leu Ser His Phe Asp 500 505 510Tyr Ala Lys Val Met Gln Leu
Ile Lys Tyr Lys Ala Glu Leu Ala Gly 515 520 525Val Phe Val Tyr Asp
Val Ala Pro Ala Gly Thr Ser Ile Asn Cys Ala 530 535 540Tyr Cys Leu
Leu Asn Asp Lys Asp Ala Ser Asn Tyr Thr Arg Gly Lys545 550 555
560Val Ile Asn Gly Lys Lys Asn Thr Lys Ile Gly Glu Cys Lys Thr Cys
565 570 575Lys Lys Glu Phe Asp Ala Asp Leu Asn Ala Ala Arg Val Ile
Ala Leu 580 585 590Cys Tyr Glu Lys Arg Leu Asn Asp Pro Gln Pro Phe
Gly Thr Arg Lys 595 600 605Gln Phe Lys Pro Lys Lys Pro 610
61521775PRTArtificial sequenceSynthetic sequence 21Met Lys Ala Leu
Lys Leu Gln Leu Ile Pro Thr Arg Lys Gln Tyr Lys1 5 10 15Ile Leu Asp
Glu Met Phe Trp Lys Trp Ala Ser Leu Ala Asn Arg Val 20 25 30Ser Gln
Lys Gly Glu Ser Lys Glu Thr Leu Ala Pro Lys Lys Asp Ile 35 40 45Gln
Lys Ile Gln Phe Asn Ala Thr Gln Leu Asn Gln Ile Glu Lys Asp 50 55
60Ile Lys Asp Leu Arg Gly Ala Met Lys Glu Gln Gln Lys Gln Lys Glu65
70 75 80Arg Leu Leu Leu Gln Ile Gln Glu Arg Arg Ser Thr Ile Ser Glu
Met 85 90 95Leu Asn Asp Asp Asn Asn Lys Glu Arg Asp Pro His Arg Pro
Leu Asn 100 105 110Phe Arg Pro Lys Gly Trp Arg Lys Phe His Thr Ser
Lys His Trp Val 115 120 125Gly Glu Leu Ser Lys Ile Leu Arg Gln Glu
Asp Arg Val Lys Lys Thr 130 135 140Ile Glu Arg Ile Val Ala Gly Lys
Ile Ser Phe Lys Pro Lys Arg Ile145 150 155 160Gly Ile Trp Ser Ser
Asn Tyr Lys Ile Asn Phe Phe Lys Arg Lys Ile 165 170 175Ser Ile Asn
Pro Leu Asn Ser Lys Gly Phe Glu Leu Thr Leu Met Thr 180 185 190Glu
Pro Thr Gln Asp Leu Ile Gly Lys Asn Gly Gly Lys Ser Val Leu 195 200
205Asn Asn Lys Arg Tyr Leu Asp Asp Ser Ile Lys Ser Leu Leu Met Phe
210 215 220Ala Leu His Ser Arg Phe Phe Gly Leu Asn Asn Thr Asp Thr
Tyr Leu225 230 235 240Leu Gly Gly Lys Ile Asn Pro Ser Leu Val Lys
Tyr Tyr Lys Lys Asn 245 250 255Gln Asp Met Gly Glu Phe Gly Arg Glu
Ile Val Glu Lys Phe Glu Arg 260 265 270Lys Leu Lys Gln Glu Ile Asn
Glu Gln Gln Lys Lys Ile Ile Met Ser 275 280 285Gln Ile Lys Glu Gln
Tyr Ser Asn Arg Asp Ser Ala Phe Asn Lys Asp 290 295 300Tyr Leu Gly
Leu Ile Asn Glu Phe Ser Glu Val Phe Asn Gln Arg Lys305 310 315
320Ser Glu Arg Ala Glu Tyr Leu Leu Asp Ser Phe Glu Asp Lys Ile Lys
325 330 335Gln Ile Lys Gln Glu Ile Gly Glu Ser Leu Asn Ile Ser Asp
Trp Asp 340 345 350Phe Leu Ile Asp Glu Ala Lys Lys Ala Tyr Gly Tyr
Glu Glu Gly Phe 355 360 365Thr Glu Tyr Val Tyr Ser Lys Arg Tyr Leu
Glu Ile Leu Asn Lys Ile 370 375 380Val Lys Ala Val Leu Ile Thr Asp
Ile Tyr Phe Asp Leu Arg Lys Tyr385 390 395 400Pro Ile Leu Leu Arg
Lys Pro Leu Asp Lys Ile Lys Lys Ile Ser Asn 405 410 415Leu Lys Pro
Asp Glu Trp Ser Tyr Tyr Ile Gln Phe Gly Tyr Asp Ser 420 425 430Ile
Asn Pro Val Gln Leu Met Ser Thr Asp Lys Phe Leu Gly Ile Asp 435 440
445Arg Gly Leu Thr His Leu Leu Ala Tyr Ser Val Phe Asp Lys Glu Lys
450 455 460Lys Glu Phe Ile Ile Asn Gln Leu Glu Pro Asn Pro Ile Met
Gly Trp465 470 475 480Lys Trp Lys Leu Arg Lys Val Lys Arg Ser Leu
Gln His Leu Glu Arg 485 490 495Arg Ile Arg Ala Gln Lys Met Val Lys
Leu Pro Glu Asn Gln Met Lys 500 505 510Lys Lys Leu Lys Ser Ile Glu
Pro Lys Ile Glu Val His Tyr His Asn 515 520 525Ile Ser Arg Lys Ile
Val Asn Leu Ala Lys Asp Tyr Asn Ala Ser Ile 530 535 540Val Val Glu
Ser Leu Glu Gly Gly Gly Leu Lys Gln His Gly Arg Lys545 550 555
560Lys Asn Ala Arg Asn Arg Ser Leu Asn Tyr Ala Leu Ser Leu Phe Asp
565 570 575Tyr Gly Lys Ile Ala Ser Leu Ile Lys Tyr Lys Ala Asp Leu
Glu Gly 580 585 590Val Pro Met Tyr Glu Val Leu Pro Ala Tyr Thr Ser
Gln Gln Cys Ala 595 600 605Lys Cys Val Leu Glu Lys Gly Ser Phe Val
Asp Pro Glu Ile Ile Gly 610 615 620Tyr Val Glu Asp Ile Gly Ile Lys
Gly Ser Leu Leu Asp Ser Leu Phe625 630 635 640Glu Gly Thr Glu Leu
Ser Ser Ile Gln Val Leu Lys Lys Ile Lys Asn 645 650 655Lys Ile Glu
Leu Ser Ala Arg Asp Asn His Asn Lys Glu Ile Asn Leu 660 665 670Ile
Leu Lys Tyr Asn Phe Lys Gly Leu Val Ile Val Arg Gly Gln Asp 675 680
685Lys Glu Glu Ile Ala Glu His Pro Ile Lys Glu Ile Asn Gly Lys Phe
690 695 700Ala Ile Leu Asp Phe Val Tyr Lys Arg Gly Lys Glu Lys Val
Gly Lys705 710 715 720Lys Gly Asn Gln Lys Val Arg Tyr Thr Gly Asn
Lys Lys Val Gly Tyr 725 730 735Cys Ser Lys His Gly Gln Val Asp Ala
Asp Leu Asn Ala Ser Arg Val 740 745 750Ile Ala Leu Cys Lys Tyr Leu
Asp Ile Asn Asp Pro Ile Leu Phe Gly 755 760 765Glu Gln Arg Lys Ser
Phe Lys 770 77522777PRTArtificial sequenceSynthetic sequence 22Met
Val Thr Arg Ala Ile Lys Leu Lys Leu Asp Pro Thr Lys Asn Gln1 5 10
15Tyr Lys Leu Leu Asn Glu Met Phe Trp Lys Trp Ala Ser Leu Ala Asn
20 25 30Arg Phe Ser Gln Lys Gly Ala Ser Lys Glu Thr Leu Ala Pro Lys
Asp 35 40 45Gly Thr Gln Lys Ile Gln Phe Asn Ala Thr Gln Leu Asn Gln
Ile Lys 50 55 60Lys Asp Val Asp Asp Leu Arg Gly Ala Met Glu Lys Gln
Gly Lys Gln65 70 75 80Lys Glu Arg Leu Leu Ile Gln Ile Gln Glu Arg
Leu Leu Thr Ile Ser 85 90 95Glu Ile Leu Arg Asp Asp Ser Lys Lys Glu
Lys Asp Pro His Arg Pro 100 105 110Gln Asn Phe Arg Pro Phe Gly Trp
Arg Arg Phe His Thr Ser Ala Tyr 115 120 125Trp Ser Ser Glu Ala Ser
Lys Leu Thr Arg Gln Val Asp Arg Val Arg 130 135 140Arg Thr Ile Glu
Arg Ile Lys Ala Gly Lys Ile Asn Phe Lys Pro Lys145 150 155 160Arg
Ile Gly Leu Trp Ser Ser Thr Tyr Lys Ile Asn Phe Leu Lys Lys 165 170
175Lys Ile Asn Ile Ser Pro Leu Lys Ser Lys Ser Phe Glu Leu Asp Leu
180 185 190Ile Thr Glu Pro Gln Gln Lys Ile Ile Gly Lys Glu Gly Gly
Lys Ser 195 200 205Val Ala Asn Ser Lys Lys Tyr Leu Asp Asp Ser Ile
Lys Ser Leu Leu 210 215 220Ile Phe Ala Ile Lys Ser Arg Leu Phe Gly
Leu Asn Asn Lys Asp Lys225 230 235 240Pro Leu Phe Glu Asn Ile Ile
Thr Pro Asn Leu Val Arg Tyr His Lys 245 250 255Lys Gly Gln Glu Gln
Glu Asn Phe Lys Lys Glu Val Ile Lys Lys Phe 260 265 270Glu Asn Lys
Leu Lys Lys Glu Ile Ser Gln Lys Gln Lys Glu Ile Ile 275 280 285Phe
Ser Gln Ile Glu Arg Gln Tyr Glu Asn Arg Asp Ala Thr Phe Ser 290 295
300Glu Asp Tyr Leu Arg Ala Ile Ser Glu Phe Ser Glu Ile Phe Asn
Gln305 310 315 320Arg Lys Lys Glu Arg Ala Lys Glu Leu Leu Asn Ser
Phe Asn Glu Lys 325 330 335Ile Arg Gln Leu Lys Lys Glu Val Asn Gly
Asn Ile Ser Glu Glu Asp 340 345 350Leu Lys Ile Leu Glu Val Glu Ala
Glu Lys Ala Tyr Asn Tyr Glu Asn 355 360 365Gly Phe Ile Glu Trp Glu
Tyr Ser Glu Gln Phe Leu Gly Val Leu Glu 370 375 380Lys Ile Ala Arg
Ala Val Leu Ile Ser Asp Asn Tyr Phe Asp Leu Lys385 390 395 400Lys
Tyr Pro Ile Leu Ile Arg Lys Pro Thr Asn Lys Ser Lys Lys Ile 405 410
415Thr Asn Leu Lys Pro Glu Glu Trp Asp Tyr Tyr Ile Gln Phe Gly Tyr
420 425 430Gly Leu Ile Asn Ser Pro Met Lys Ile Glu Thr Lys Asn Phe
Met Gly 435 440 445Ile Asp Arg Gly Leu Thr His Leu Leu Ala Tyr Ser
Ile Phe Asp Arg 450 455 460Asp Ser Glu Lys Phe Thr Ile Asn Gln Leu
Glu Leu Asn Pro Ile Lys465 470 475 480Gly Trp Lys Trp Lys Leu Arg
Lys Val Lys Arg Ser Leu Gln His Leu 485 490 495Glu Arg Arg Met Arg
Ala Gln Lys Gly Val Lys Leu Pro Glu Asn Gln 500 505 510Met Lys Lys
Arg Leu Lys Ser Ile Glu Pro Lys Ile Glu Ser Tyr Tyr 515 520 525His
Asn Leu Ser Arg Lys Ile Val Asn Leu Ala Lys Ala Asn Asn Ala 530 535
540Ser Ile Val Val Glu Ser Leu Glu Gly Gly Gly Leu Lys Gln His
Gly545 550 555 560Arg Lys Lys Asn Ser Arg His Arg Ala Leu Asn Tyr
Ala Leu Ser Leu 565 570 575Phe Asp Tyr Gly Lys Ile Ala Ser Leu Ile
Lys Tyr Lys Ser Asp Leu 580 585 590Glu Gly Val Pro Met Tyr Glu Val
Leu Pro Ala Tyr Thr Ser Gln Gln 595 600 605Cys Ala Lys Cys Val Leu
Lys Lys Gly Ser Phe Val Glu Pro Glu Ile 610 615 620Ile Gly Tyr Ile
Glu Glu Ile Gly Phe Lys Glu Asn Leu Leu Thr Leu625 630 635 640Leu
Phe Glu Asp Thr Gly Leu Ser Ser Val Gln Val Leu Lys Lys Ser 645 650
655Lys Asn Lys Met Thr Leu Ser Ala Arg Asp Lys Glu Gly Lys Met Val
660 665 670Asp Leu Val Leu Lys Tyr Asn Phe Lys Gly Leu Val Ile Ser
Gln Glu 675 680 685Lys Lys Lys Glu Glu Ile Val Glu Phe Pro Ile Lys
Glu Ile Asp Gly 690 695 700Lys Phe Ala Val Leu Asp Ser Ala Tyr Lys
Arg Gly Lys Glu Arg Ile705 710 715 720Ser Lys Lys Gly Asn Gln Lys
Leu Val Tyr Thr Gly Asn Lys Lys Val 725 730 735Gly Tyr Cys Ser Val
His Gly Gln Val Asp Ala Asp Leu Asn Ala Ser 740 745
750Arg Val Ile Ala Leu Cys Lys Tyr Leu Gly Ile Asn Glu Pro Ile Val
755 760 765Phe Gly Glu Gln Arg Lys Ser Phe Lys 770
77523610PRTArtificial sequenceSynthetic sequence 23Leu Asp Leu Ile
Thr Glu Pro Ile Gln Pro His Lys Ser Ser Ser Leu1 5 10 15Arg Ser Lys
Glu Phe Leu Glu Tyr Gln Ile Ser Asp Phe Leu Asn Phe 20 25 30Ser Leu
His Ser Leu Phe Phe Gly Leu Ala Ser Asn Glu Gly Pro Leu 35 40 45Val
Asp Phe Lys Ile Tyr Asp Lys Ile Val Ile Pro Lys Pro Glu Glu 50 55
60Arg Phe Pro Lys Lys Glu Ser Glu Glu Gly Lys Lys Leu Asp Ser Phe65
70 75 80Asp Lys Arg Val Glu Glu Tyr Tyr Ser Asp Lys Leu Glu Lys Lys
Ile 85 90 95Glu Arg Lys Leu Asn Thr Glu Glu Lys Asn Val Ile Asp Arg
Glu Lys 100 105 110Thr Arg Ile Trp Gly Glu Val Asn Lys Leu Glu Glu
Ile Arg Ser Ile 115 120 125Ile Asp Glu Ile Asn Glu Ile Lys Lys Gln
Lys His Ile Ser Glu Lys 130 135 140Ser Lys Leu Leu Gly Glu Lys Trp
Lys Lys Val Asn Asn Ile Gln Glu145 150 155 160Thr Leu Leu Ser Gln
Glu Tyr Val Ser Leu Ile Ser Asn Leu Ser Asp 165 170 175Glu Leu Thr
Asn Lys Lys Lys Glu Leu Leu Ala Lys Lys Tyr Ser Lys 180 185 190Phe
Asp Asp Lys Ile Lys Lys Ile Lys Glu Asp Tyr Gly Leu Glu Phe 195 200
205Asp Glu Asn Thr Ile Lys Lys Glu Gly Glu Lys Ala Phe Leu Asn Pro
210 215 220Asp Lys Phe Ser Lys Tyr Gln Phe Ser Ser Ser Tyr Leu Lys
Leu Ile225 230 235 240Gly Glu Ile Ala Arg Ser Leu Ile Thr Tyr Lys
Gly Phe Leu Asp Leu 245 250 255Asn Lys Tyr Pro Ile Ile Phe Arg Lys
Pro Ile Asn Lys Val Lys Lys 260 265 270Ile His Asn Leu Glu Pro Asp
Glu Trp Lys Tyr Tyr Ile Gln Phe Gly 275 280 285Tyr Glu Gln Ile Asn
Asn Pro Lys Leu Glu Thr Glu Asn Ile Leu Gly 290 295 300Ile Asp Arg
Gly Leu Thr His Ile Leu Ala Tyr Ser Val Phe Glu Pro305 310 315
320Arg Ser Ser Lys Phe Ile Leu Asn Lys Leu Glu Pro Asn Pro Ile Glu
325 330 335Gly Trp Lys Trp Lys Leu Arg Lys Leu Arg Arg Ser Ile Gln
Asn Leu 340 345 350Glu Arg Arg Trp Arg Ala Gln Asp Asn Val Lys Leu
Pro Glu Asn Gln 355 360 365Met Lys Lys Asn Leu Arg Ser Ile Glu Asp
Lys Val Glu Asn Leu Tyr 370 375 380His Asn Leu Ser Arg Lys Ile Val
Asp Leu Ala Lys Glu Lys Asn Ala385 390 395 400Cys Ile Val Phe Glu
Lys Leu Glu Gly Gln Gly Met Lys Gln His Gly 405 410 415Arg Lys Lys
Ser Asp Arg Leu Arg Gly Leu Asn Tyr Lys Leu Ser Leu 420 425 430Phe
Asp Tyr Gly Lys Ile Ala Lys Leu Ile Lys Tyr Lys Ala Glu Ile 435 440
445Glu Gly Ile Pro Ile Tyr Arg Ile Asp Ser Ala Tyr Thr Ser Gln Asn
450 455 460Cys Ala Lys Cys Val Leu Glu Ser Arg Arg Phe Ala Gln Pro
Glu Glu465 470 475 480Ile Ser Cys Leu Asp Asp Phe Lys Glu Gly Asp
Asn Leu Asp Lys Arg 485 490 495Ile Leu Glu Gly Thr Gly Leu Val Glu
Ala Lys Ile Tyr Lys Lys Leu 500 505 510Leu Lys Glu Lys Lys Glu Asp
Phe Glu Ile Glu Glu Asp Ile Ala Met 515 520 525Phe Asp Thr Lys Lys
Val Ile Lys Glu Asn Lys Glu Lys Thr Val Ile 530 535 540Leu Asp Tyr
Val Tyr Thr Arg Arg Lys Glu Ile Ile Gly Thr Asn His545 550 555
560Lys Lys Asn Ile Lys Gly Ile Ala Lys Tyr Thr Gly Asn Thr Lys Ile
565 570 575Gly Tyr Cys Met Lys His Gly Gln Val Asp Ala Asp Leu Asn
Ala Ser 580 585 590Arg Thr Ile Ala Leu Cys Lys Asn Phe Asp Ile Asn
Asn Pro Glu Ile 595 600 605Trp Lys 61024632PRTArtificial
sequenceSynthetic sequence 24Met Ser Asp Glu Ser Leu Val Ser Ser
Glu Asp Lys Leu Ala Ile Lys1 5 10 15Ile Lys Ile Val Pro Asn Ala Glu
Gln Ala Lys Met Leu Asp Glu Met 20 25 30Phe Lys Lys Trp Ser Ser Ile
Cys Asn Arg Ile Ser Arg Gly Lys Glu 35 40 45Asp Ile Glu Thr Leu Arg
Pro Asp Glu Gly Lys Glu Leu Gln Phe Asn 50 55 60Ser Thr Gln Leu Asn
Ser Ala Thr Met Asp Val Ser Asp Leu Lys Lys65 70 75 80Ala Met Ala
Arg Gln Gly Glu Arg Leu Glu Ala Glu Val Ser Lys Leu 85 90 95Arg Gly
Arg Tyr Glu Thr Ile Asp Ala Ser Leu Arg Asp Pro Ser Arg 100 105
110Arg His Thr Asn Pro Gln Lys Pro Ser Ser Phe Tyr Pro Ser Asp Trp
115 120 125Asp Ile Ser Gly Arg Leu Thr Pro Arg Phe His Thr Ala Arg
His Tyr 130 135 140Ser Thr Glu Leu Arg Lys Leu Lys Ala Lys Glu Asp
Lys Met Leu Lys145 150 155 160Thr Ile Asn Lys Ile Lys Asn Gly Lys
Ile Val Phe Lys Pro Lys Arg 165 170 175Ile Thr Leu Trp Pro Ser Ser
Val Asn Met Ala Phe Lys Gly Ser Arg 180 185 190Leu Leu Leu Lys Pro
Phe Ala Asn Gly Phe Glu Met Glu Leu Pro Ile 195 200 205Val Ile Ser
Pro Gln Lys Thr Ala Asp Gly Lys Ser Gln Lys Ala Ser 210 215 220Ala
Glu Tyr Met Arg Asn Ala Leu Leu Gly Leu Ala Gly Tyr Ser Ile225 230
235 240Asn Gln Leu Leu Phe Gly Met Asn Arg Ser Gln Lys Met Leu Ala
Asn 245 250 255Ala Lys Lys Pro Glu Lys Val Glu Lys Phe Leu Glu Gln
Met Lys Asn 260 265 270Lys Asp Ala Asn Phe Asp Lys Lys Ile Lys Ala
Leu Glu Gly Lys Trp 275 280 285Leu Leu Asp Arg Lys Leu Lys Glu Ser
Glu Lys Ser Ser Ile Ala Val 290 295 300Val Arg Thr Lys Phe Phe Lys
Ser Gly Lys Val Glu Leu Asn Glu Asp305 310 315 320Tyr Leu Lys Leu
Leu Lys His Met Ala Asn Glu Ile Leu Glu Arg Asp 325 330 335Gly Phe
Val Asn Leu Asn Lys Tyr Pro Ile Leu Ser Arg Lys Pro Met 340 345
350Lys Arg Tyr Lys Gln Lys Asn Ile Asp Asn Leu Lys Pro Asn Met Trp
355 360 365Lys Tyr Tyr Ile Gln Phe Gly Tyr Glu Pro Ile Phe Glu Arg
Lys Ala 370 375 380Ser Gly Lys Pro Lys Asn Ile Met Gly Ile Asp Arg
Gly Leu Thr His385 390 395 400Leu Leu Ala Val Ala Val Phe Ser Pro
Asp Gln Gln Lys Phe Leu Phe 405 410 415Asn His Leu Glu Ser Asn Pro
Ile Met His Trp Lys Trp Lys Leu Arg 420 425 430Lys Ile Arg Arg Ser
Ile Gln His Met Glu Arg Arg Ile Arg Ala Glu 435 440 445Lys Asn Lys
His Ile His Glu Ala Gln Leu Lys Lys Arg Leu Gly Ser 450 455 460Ile
Glu Glu Lys Thr Glu Gln His Tyr His Ile Val Ser Ser Lys Ile465 470
475 480Ile Asn Trp Ala Ile Glu Tyr Glu Ala Ala Ile Val Leu Glu Ser
Leu 485 490 495Ser His Met Lys Gln Arg Gly Gly Lys Lys Ser Val Arg
Thr Arg Ala 500 505 510Leu Asn Tyr Ala Leu Ser Leu Phe Asp Tyr Glu
Lys Val Ala Arg Leu 515 520 525Ile Thr Tyr Lys Ala Arg Ile Arg Gly
Ile Pro Val Tyr Asp Val Leu 530 535 540Pro Gly Met Thr Ser Lys Thr
Cys Ala Thr Cys Leu Leu Asn Gly Ser545 550 555 560Gln Gly Ala Tyr
Val Arg Gly Leu Glu Thr Thr Lys Ala Ala Gly Lys 565 570 575Ala Thr
Lys Arg Lys Asn Met Lys Ile Gly Lys Cys Met Val Cys Asn 580 585
590Ser Ser Glu Asn Ser Met Ile Asp Ala Asp Leu Asn Ala Ala Arg Val
595 600 605Ile Ala Ile Cys Lys Tyr Lys Asn Leu Asn Asp Pro Gln Pro
Ala Gly 610 615 620Ser Arg Lys Val Phe Lys Arg Phe625
63025625PRTArtificial sequenceSynthetic sequence 25Met Leu Ala Leu
Lys Leu Lys Ile Met Pro Thr Glu Lys Gln Ala Glu1 5 10 15Ile Leu Asp
Ala Met Phe Trp Lys Trp Ala Ser Ile Cys Ser Arg Ile 20 25 30Ala Lys
Met Lys Lys Lys Val Ser Val Lys Glu Asn Lys Lys Glu Leu 35 40 45Ser
Lys Lys Ile Pro Ser Asn Ser Asp Ile Trp Phe Ser Lys Thr Gln 50 55
60Leu Cys Gln Ala Glu Val Asp Val Gly Asp His Lys Lys Ala Leu Lys65
70 75 80Asn Phe Glu Lys Arg Gln Glu Ser Leu Leu Asp Glu Leu Lys Tyr
Lys 85 90 95Val Lys Ala Ile Asn Glu Val Ile Asn Asp Glu Ser Lys Arg
Glu Ile 100 105 110Asp Pro Asn Asn Pro Ser Lys Phe Arg Ile Lys Asp
Ser Thr Lys Lys 115 120 125Gly Asn Leu Asn Ser Pro Lys Phe Phe Thr
Leu Lys Lys Trp Gln Lys 130 135 140Ile Leu Gln Glu Asn Glu Lys Arg
Ile Lys Lys Lys Glu Ser Thr Ile145 150 155 160Glu Lys Leu Lys Arg
Gly Asn Ile Phe Phe Asn Pro Thr Lys Ile Ser 165 170 175Leu His Glu
Glu Glu Tyr Ser Ile Asn Phe Gly Ser Ser Lys Leu Leu 180 185 190Leu
Asn Cys Phe Tyr Lys Tyr Asn Lys Lys Ser Gly Ile Asn Ser Asp 195 200
205Gln Leu Glu Asn Lys Phe Asn Glu Phe Gln Asn Gly Leu Asn Ile Ile
210 215 220Cys Ser Pro Leu Gln Pro Ile Arg Gly Ser Ser Lys Arg Ser
Phe Glu225 230 235 240Phe Ile Arg Asn Ser Ile Ile Asn Phe Leu Met
Tyr Ser Leu Tyr Ala 245 250 255Lys Leu Phe Gly Ile Pro Arg Ser Val
Lys Ala Leu Met Lys Ser Asn 260 265 270Lys Asp Glu Asn Lys Leu Lys
Leu Glu Glu Lys Leu Lys Lys Lys Lys 275 280 285Ser Ser Phe Asn Lys
Thr Val Lys Glu Phe Glu Lys Met Ile Gly Arg 290 295 300Lys Leu Ser
Asp Asn Glu Ser Lys Ile Leu Asn Asp Glu Ser Lys Lys305 310 315
320Phe Phe Glu Ile Ile Lys Ser Asn Asn Lys Tyr Ile Pro Ser Glu Glu
325 330 335Tyr Leu Lys Leu Leu Lys Asp Ile Ser Glu Glu Ile Tyr Asn
Ser Asn 340 345 350Ile Asp Phe Lys Pro Tyr Lys Tyr Ser Ile Leu Ile
Arg Lys Pro Leu 355 360 365Ser Lys Phe Lys Ser Lys Lys Leu Tyr Asn
Leu Lys Pro Thr Asp Tyr 370 375 380Lys Tyr Tyr Leu Gln Leu Ser Tyr
Glu Pro Phe Ser Lys Gln Leu Ile385 390 395 400Ala Thr Lys Thr Ile
Leu Gly Ile Asp Arg Gly Leu Lys His Leu Leu 405 410 415Ala Val Ser
Val Phe Asp Pro Ser Gln Asn Lys Phe Val Tyr Asn Lys 420 425 430Leu
Ile Lys Asn Pro Val Phe Lys Trp Lys Lys Arg Tyr His Asp Leu 435 440
445Lys Arg Ser Ile Arg Asn Arg Glu Arg Arg Ile Arg Ala Leu Thr Gly
450 455 460Val His Ile His Glu Asn Gln Leu Ile Lys Lys Leu Lys Ser
Met Lys465 470 475 480Asn Lys Ile Asn Val Leu Tyr His Asn Val Ser
Lys Asn Ile Val Asp 485 490 495Leu Ala Lys Lys Tyr Glu Ser Thr Ile
Val Leu Glu Arg Leu Glu Asn 500 505 510Leu Lys Gln His Gly Arg Ser
Lys Gly Lys Arg Tyr Lys Lys Leu Asn 515 520 525Tyr Val Leu Ser Asn
Phe Asp Tyr Lys Lys Ile Glu Ser Leu Ile Ser 530 535 540Tyr Lys Ala
Lys Lys Glu Gly Val Pro Val Ser Asn Ile Asn Pro Lys545 550 555
560Tyr Thr Ser Lys Thr Cys Ala Lys Cys Leu Leu Glu Val Asn Gln Leu
565 570 575Ser Glu Leu Lys Asn Glu Tyr Asn Arg Asp Ser Lys Asn Ser
Lys Ile 580 585 590Gly Ile Cys Asn Ile His Gly Gln Ile Asp Ala Asp
Leu Asn Ala Ala 595 600 605Arg Val Ile Ala Leu Cys Tyr Ser Lys Asn
Leu Asn Glu Pro His Phe 610 615 620Lys62526517PRTArtificial
sequenceSynthetic sequence 26Val Ile Asn Leu Phe Gly Tyr Lys Phe
Ala Leu Tyr Pro Asn Lys Thr1 5 10 15Gln Glu Glu Leu Leu Asn Lys His
Leu Gly Glu Cys Gly Trp Leu Tyr 20 25 30Asn Lys Ala Ile Glu Gln Asn
Glu Tyr Tyr Lys Ala Asp Ser Asn Ile 35 40 45Glu Glu Ala Gln Lys Lys
Phe Glu Leu Leu Pro Asp Lys Asn Ser Asp 50 55 60Glu Ala Lys Val Leu
Arg Gly Asn Ile Ser Lys Asp Asn Tyr Val Tyr65 70 75 80Arg Thr Leu
Val Lys Lys Lys Lys Ser Glu Ile Asn Val Gln Ile Arg 85 90 95Lys Ala
Val Val Leu Arg Pro Ala Glu Thr Ile Arg Asn Leu Ala Lys 100 105
110Val Lys Lys Lys Gly Leu Ser Val Gly Arg Leu Lys Phe Ile Pro Ile
115 120 125Arg Glu Trp Asp Val Leu Pro Phe Lys Gln Ser Asp Gln Ile
Arg Leu 130 135 140Glu Glu Asn Tyr Leu Ile Leu Glu Pro Tyr Gly Arg
Leu Lys Phe Lys145 150 155 160Met His Arg Pro Leu Leu Gly Lys Pro
Lys Thr Phe Cys Ile Lys Arg 165 170 175Thr Ala Thr Asp Arg Trp Thr
Ile Ser Phe Ser Thr Glu Tyr Asp Asp 180 185 190Ser Asn Met Arg Lys
Asn Asp Gly Gly Gln Val Gly Ile Asp Val Gly 195 200 205Leu Lys Thr
His Leu Arg Leu Ser Asn Glu Asn Pro Asp Glu Asp Pro 210 215 220Arg
Tyr Pro Asn Pro Lys Ile Trp Lys Arg Tyr Asp Arg Arg Leu Thr225 230
235 240Ile Leu Gln Arg Arg Ile Ser Lys Ser Lys Lys Leu Gly Lys Asn
Arg 245 250 255Thr Arg Leu Arg Leu Arg Leu Ser Arg Leu Trp Glu Lys
Ile Arg Asn 260 265 270Ser Arg Ala Asp Leu Ile Gln Asn Glu Thr Tyr
Glu Ile Leu Ser Glu 275 280 285Asn Lys Leu Ile Ala Ile Glu Asp Leu
Asn Val Lys Gly Met Gln Glu 290 295 300Lys Lys Asp Lys Lys Gly Arg
Lys Gly Arg Thr Arg Ala Gln Glu Lys305 310 315 320Gly Leu His Arg
Ser Ile Ser Asp Ala Ala Phe Ser Glu Phe Arg Arg 325 330 335Val Leu
Glu Tyr Lys Ala Lys Arg Phe Gly Ser Glu Val Lys Pro Val 340 345
350Ser Ala Ile Asp Ser Ser Lys Glu Cys His Asn Cys Gly Asn Lys Lys
355 360 365Gly Met Pro Leu Glu Ser Arg Ile Tyr Glu Cys Pro Lys Cys
Gly Leu 370 375 380Lys Ile Asp Arg Asp Leu Asn Ser Ala Lys Val Ile
Leu Ala Arg Ala385 390 395 400Thr Gly Val Arg Pro Gly Ser Asn Ala
Arg Ala Asp Thr Lys Ile Ser 405 410 415Ala Thr Ala Gly Ala Ser Val
Gln Thr Glu Gly Thr Val Ser Glu Asp 420 425 430Phe Arg Gln Gln Met
Glu Thr Ser Asp Gln Lys Pro Met Gln Gly Glu 435 440 445Gly Ser Lys
Glu Pro Pro Met Asn Pro Glu His Lys Ser Ser Gly Arg 450 455 460Gly
Ser Lys His Val Asn Ile Gly Cys Lys Asn Lys Val Gly Leu Tyr465 470
475 480Asn Glu Asp Glu Asn Ser Arg Ser Thr Glu Lys Gln Ile Met Asp
Glu 485 490 495Asn Arg Ser Thr Thr Glu Asp Met Val Glu Ile Gly Ala
Leu His Ser 500 505 510Pro Val Leu Thr Thr 51527410PRTArtificial
sequenceSynthetic sequence 27Met Ile Ala Ser Ile Asp Tyr Glu Ala
Val Ser Gln Ala Leu Ile Val1 5 10 15Phe Glu Phe Lys Ala Lys Gly Lys
Asp Ser Gln Tyr Gln Ala Ile Asp 20 25 30Glu Ala Ile Arg Ser Tyr
Arg
Phe Ile Arg Asn Ser Cys Leu Arg Tyr 35 40 45Trp Met Asp Asn Lys Lys
Val Gly Lys Tyr Asp Leu Asn Lys Tyr Cys 50 55 60Lys Val Leu Ala Lys
Gln Tyr Pro Phe Ala Asn Lys Leu Asn Ser Gln65 70 75 80Ala Arg Gln
Ser Ala Ala Glu Cys Ser Trp Ser Ala Ile Ser Arg Phe 85 90 95Tyr Asp
Asn Cys Lys Arg Lys Val Ser Gly Lys Lys Gly Phe Pro Lys 100 105
110Phe Lys Lys His Ala Arg Ser Val Glu Tyr Lys Thr Ser Gly Trp Lys
115 120 125Leu Ser Glu Asn Arg Lys Ala Ile Thr Phe Thr Asp Lys Asn
Gly Ile 130 135 140Gly Lys Leu Lys Leu Lys Gly Thr Tyr Asp Leu His
Phe Ser Gln Leu145 150 155 160Glu Asp Met Lys Arg Val Arg Leu Val
Arg Arg Ala Asp Gly Tyr Tyr 165 170 175Val Gln Phe Cys Ile Ser Val
Asp Val Lys Val Glu Thr Glu Pro Thr 180 185 190Gly Lys Ala Ile Gly
Leu Asp Val Gly Ile Lys Tyr Phe Leu Ala Asp 195 200 205Ser Ser Gly
Asn Thr Ile Glu Asn Pro Gln Phe Tyr Arg Lys Ala Glu 210 215 220Lys
Lys Leu Asn Arg Ala Asn Arg Arg Lys Ser Lys Lys Tyr Ile Arg225 230
235 240Gly Val Lys Pro Gln Ser Lys Asn Tyr His Lys Ala Arg Cys Arg
Tyr 245 250 255Ala Arg Lys His Leu Arg Val Ser Arg Gln Arg Lys Glu
Tyr Cys Lys 260 265 270Arg Val Ala Tyr Cys Val Ile His Ser Asn Asp
Val Val Ala Tyr Glu 275 280 285Asp Leu Asn Val Lys Gly Met Val Lys
Asn Arg His Leu Ala Lys Ser 290 295 300Ile Ser Asp Val Ala Trp Ser
Thr Phe Arg His Trp Leu Glu Tyr Phe305 310 315 320Ala Ile Lys Tyr
Gly Lys Leu Thr Ile Pro Val Ala Pro His Asn Thr 325 330 335Ser Gln
Asn Cys Ser Asn Cys Asp Lys Lys Val Pro Lys Ser Leu Ser 340 345
350Thr Arg Thr His Ile Cys His His Cys Gly Tyr Ser Glu Asp Arg Asp
355 360 365Val Asn Ala Ala Lys Asn Ile Leu Lys Lys Ala Leu Ser Thr
Val Gly 370 375 380Gln Thr Gly Ser Leu Lys Leu Gly Glu Ile Glu Pro
Leu Leu Val Leu385 390 395 400Glu Gln Ser Cys Thr Arg Lys Phe Asp
Leu 405 41028486PRTArtificial sequenceSynthetic sequence 28Leu Ala
Glu Glu Asn Thr Leu His Leu Thr Leu Ala Met Ser Leu Pro1 5 10 15Leu
Asn Asp Leu Pro Glu Asn Arg Thr Arg Ser Glu Leu Trp Arg Arg 20 25
30Gln Trp Leu Pro Gln Lys Lys Leu Ser Leu Leu Leu Gly Val Asn Gln
35 40 45Ser Val Arg Lys Ala Ala Ala Asp Cys Leu Arg Trp Phe Glu Pro
Tyr 50 55 60Gln Glu Leu Leu Trp Trp Glu Pro Thr Asp Pro Asp Gly Lys
Lys Leu65 70 75 80Leu Asp Lys Glu Gly Arg Pro Ile Lys Arg Thr Ala
Gly His Met Arg 85 90 95Val Leu Arg Lys Leu Glu Glu Ile Ala Pro Phe
Arg Gly Tyr Gln Leu 100 105 110Gly Ser Ala Val Lys Asn Gly Leu Arg
His Lys Val Ala Asp Leu Leu 115 120 125Leu Ser Tyr Ala Lys Arg Lys
Leu Asp Pro Gln Phe Thr Asp Lys Thr 130 135 140Ser Tyr Pro Ser Ile
Gly Asp Gln Phe Pro Ile Val Trp Thr Gly Ala145 150 155 160Phe Val
Cys Tyr Glu Gln Ser Ile Thr Gly Gln Leu Tyr Leu Tyr Leu 165 170
175Pro Leu Phe Pro Arg Gly Ser His Gln Glu Asp Ile Thr Asn Asn Tyr
180 185 190Asp Pro Asp Arg Gly Pro Ala Leu Gln Val Phe Gly Glu Lys
Glu Ile 195 200 205Ala Arg Leu Ser Arg Ser Thr Ser Gly Leu Leu Leu
Pro Leu Gln Phe 210 215 220Asp Lys Trp Gly Glu Ala Thr Phe Ile Arg
Gly Glu Asn Asn Pro Pro225 230 235 240Thr Trp Lys Ala Thr His Arg
Arg Ser Asp Lys Lys Trp Leu Ser Glu 245 250 255Val Leu Leu Arg Glu
Lys Asp Phe Gln Pro Lys Arg Val Glu Leu Leu 260 265 270Val Arg Asn
Gly Arg Ile Phe Val Asn Val Ala Cys Glu Ile Pro Thr 275 280 285Lys
Pro Leu Leu Glu Val Glu Asn Phe Met Gly Val Ser Phe Gly Leu 290 295
300Glu His Leu Val Thr Val Val Val Ile Asn Arg Asp Gly Asn Val
Val305 310 315 320His Gln Arg Gln Glu Pro Ala Arg Arg Tyr Glu Lys
Thr Tyr Phe Ala 325 330 335Arg Leu Glu Arg Leu Arg Arg Arg Gly Gly
Pro Phe Ser Gln Glu Leu 340 345 350Glu Thr Phe His Tyr Arg Gln Val
Ala Gln Ile Val Glu Glu Ala Leu 355 360 365Arg Phe Lys Ser Val Pro
Ala Val Glu Gln Val Gly Asn Ile Pro Lys 370 375 380Gly Arg Tyr Asn
Pro Arg Leu Asn Leu Arg Leu Ser Tyr Trp Pro Phe385 390 395 400Gly
Lys Leu Ala Asp Leu Thr Ser Tyr Lys Ala Val Lys Glu Gly Leu 405 410
415Pro Lys Pro Tyr Ser Val Tyr Ser Ala Thr Ala Lys Met Leu Cys Ser
420 425 430Thr Cys Gly Ala Ala Asn Lys Glu Gly Asp Gln Pro Ile Ser
Leu Lys 435 440 445Gly Pro Thr Val Tyr Cys Gly Asn Cys Gly Thr Arg
His Asn Thr Gly 450 455 460Phe Asn Thr Ala Leu Asn Leu Ala Arg Arg
Ala Gln Glu Leu Phe Val465 470 475 480Lys Gly Val Val Ala Arg
48529602PRTArtificial sequenceSynthetic sequence 29Met Ser Gln Ser
Leu Leu Lys Trp His Asp Met Ala Gly Arg Asp Lys1 5 10 15Asp Ala Ser
Arg Ser Leu Gln Lys Ser Ala Val Glu Gly Val Leu Leu 20 25 30His Leu
Thr Ala Ser His Arg Val Ala Leu Glu Met Leu Glu Lys Ser 35 40 45Val
Ser Gln Thr Val Ala Val Thr Met Glu Ala Ala Gln Gln Arg Leu 50 55
60Val Ile Val Leu Glu Asp Asp Pro Thr Lys Ala Thr Ser Arg Lys Arg65
70 75 80Val Ile Ser Ala Asp Leu Gln Phe Thr Arg Glu Glu Phe Gly Ser
Leu 85 90 95Pro Asn Trp Ala Gln Lys Leu Ala Ser Thr Cys Pro Glu Ile
Ala Thr 100 105 110Lys Tyr Ala Asp Lys His Ile Asn Ser Ile Arg Ile
Ala Trp Gly Val 115 120 125Ala Lys Glu Ser Thr Asn Gly Asp Ala Val
Glu Gln Lys Leu Gln Trp 130 135 140Gln Ile Arg Leu Leu Asp Val Thr
Met Phe Leu Gln Gln Leu Val Leu145 150 155 160Gln Leu Ala Asp Lys
Ala Leu Leu Glu Gln Ile Pro Ser Ser Ile Arg 165 170 175Gly Gly Ile
Gly Gln Glu Val Ala Gln Gln Val Thr Ser His Ile Gln 180 185 190Leu
Leu Asp Ser Gly Thr Val Leu Lys Ala Glu Leu Pro Thr Ile Ser 195 200
205Asp Arg Asn Ser Glu Leu Ala Arg Lys Gln Trp Glu Asp Ala Ile Gln
210 215 220Thr Val Cys Thr Tyr Ala Leu Pro Phe Ser Arg Glu Arg Ala
Arg Ile225 230 235 240Leu Asp Pro Gly Lys Tyr Ala Ala Glu Asp Pro
Arg Gly Asp Arg Leu 245 250 255Ile Asn Ile Asp Pro Met Trp Ala Arg
Val Leu Lys Gly Pro Thr Val 260 265 270Lys Ser Leu Pro Leu Leu Phe
Val Ser Gly Ser Ser Ile Arg Ile Val 275 280 285Lys Leu Thr Leu Pro
Arg Lys His Ala Ala Gly His Lys His Thr Phe 290 295 300Thr Ala Thr
Tyr Leu Val Leu Pro Val Ser Arg Glu Trp Ile Asn Ser305 310 315
320Leu Pro Gly Thr Val Gln Glu Lys Val Gln Trp Trp Lys Lys Pro Asp
325 330 335Val Leu Ala Thr Gln Glu Leu Leu Val Gly Lys Gly Ala Leu
Lys Lys 340 345 350Ser Ala Asn Thr Leu Val Ile Pro Ile Ser Ala Gly
Lys Lys Arg Phe 355 360 365Phe Asn His Ile Leu Pro Ala Leu Gln Arg
Gly Phe Pro Leu Gln Trp 370 375 380Gln Arg Ile Val Gly Arg Ser Tyr
Arg Arg Pro Ala Thr His Arg Lys385 390 395 400Trp Phe Ala Gln Leu
Thr Ile Gly Tyr Thr Asn Pro Ser Ser Leu Pro 405 410 415Glu Met Ala
Leu Gly Ile His Phe Gly Met Lys Asp Ile Leu Trp Trp 420 425 430Ala
Leu Ala Asp Lys Gln Gly Asn Ile Leu Lys Asp Gly Ser Ile Pro 435 440
445Gly Asn Ser Ile Leu Asp Phe Ser Leu Gln Glu Lys Gly Lys Ile Glu
450 455 460Arg Gln Gln Lys Ala Gly Lys Asn Val Ala Gly Lys Lys Tyr
Gly Lys465 470 475 480Ser Leu Leu Asn Ala Thr Tyr Arg Val Val Asn
Gly Val Leu Glu Phe 485 490 495Ser Lys Gly Ile Ser Ala Glu His Ala
Ser Gln Pro Ile Gly Leu Gly 500 505 510Leu Glu Thr Ile Arg Phe Val
Asp Lys Ala Ser Gly Ser Ser Pro Val 515 520 525Asn Ala Arg His Ser
Asn Trp Asn Tyr Gly Gln Leu Ser Gly Ile Phe 530 535 540Ala Asn Lys
Ala Gly Pro Ala Gly Phe Ser Val Thr Glu Ile Thr Leu545 550 555
560Lys Lys Ala Gln Arg Asp Leu Ser Asp Ala Glu Gln Ala Arg Val Leu
565 570 575Ala Ile Glu Ala Thr Lys Arg Phe Ala Ser Arg Ile Lys Arg
Leu Ala 580 585 590Thr Lys Arg Lys Asp Asp Thr Leu Phe Val 595
60030494PRTArtificial sequenceSynthetic sequence 30Val Glu Pro Val
Glu Lys Glu Arg Phe Tyr Tyr Arg Thr Tyr Thr Phe1 5 10 15Arg Leu Asp
Gly Gln Pro Arg Thr Gln Asn Leu Thr Thr Gln Ser Gly 20 25 30Trp Gly
Leu Leu Thr Lys Ala Val Leu Asp Asn Thr Lys His Tyr Trp 35 40 45Glu
Ile Val His His Ala Arg Ile Ala Asn Gln Pro Ile Val Phe Glu 50 55
60Asn Pro Val Ile Asp Glu Gln Gly Asn Pro Lys Leu Asn Lys Leu Gly65
70 75 80Gln Pro Arg Phe Trp Lys Arg Pro Ile Ser Asp Ile Val Asn Gln
Leu 85 90 95Arg Ala Leu Phe Glu Asn Gln Asn Pro Tyr Gln Leu Gly Ser
Ser Leu 100 105 110Ile Gln Gly Thr Tyr Trp Asp Val Ala Glu Asn Leu
Ala Ser Trp Tyr 115 120 125Ala Leu Asn Lys Glu Tyr Leu Ala Gly Thr
Ala Thr Trp Gly Glu Pro 130 135 140Ser Phe Pro Glu Pro His Pro Leu
Thr Glu Ile Asn Gln Trp Met Pro145 150 155 160Leu Thr Phe Ser Ser
Gly Lys Val Val Arg Leu Leu Lys Asn Ala Ser 165 170 175Gly Arg Tyr
Phe Ile Gly Leu Pro Ile Leu Gly Glu Asn Asn Pro Cys 180 185 190Tyr
Arg Met Arg Thr Ile Glu Lys Leu Ile Pro Cys Asp Gly Lys Gly 195 200
205Arg Val Thr Ser Gly Ser Leu Ile Leu Phe Pro Leu Val Gly Ile Tyr
210 215 220Ala Gln Gln His Arg Arg Met Thr Asp Ile Cys Glu Ser Ile
Arg Thr225 230 235 240Glu Lys Gly Lys Leu Ala Trp Ala Gln Val Ser
Ile Asp Tyr Val Arg 245 250 255Glu Val Asp Lys Arg Arg Arg Met Arg
Arg Thr Arg Lys Ser Gln Gly 260 265 270Trp Ile Gln Gly Pro Trp Gln
Glu Val Phe Ile Leu Arg Leu Val Leu 275 280 285Ala His Lys Ala Pro
Lys Leu Tyr Lys Pro Arg Cys Phe Ala Gly Ile 290 295 300Ser Leu Gly
Pro Lys Thr Leu Ala Ser Cys Val Ile Leu Asp Gln Asp305 310 315
320Glu Arg Val Val Glu Lys Gln Gln Trp Ser Gly Ser Glu Leu Leu Ser
325 330 335Leu Ile His Gln Gly Glu Glu Arg Leu Arg Ser Leu Arg Glu
Gln Ser 340 345 350Lys Pro Thr Trp Asn Ala Ala Tyr Arg Lys Gln Leu
Lys Ser Leu Ile 355 360 365Asn Thr Gln Val Phe Thr Ile Val Thr Phe
Leu Arg Glu Arg Gly Ala 370 375 380Ala Val Arg Leu Glu Ser Ile Ala
Arg Val Arg Lys Ser Thr Pro Ala385 390 395 400Pro Pro Val Asn Phe
Leu Leu Ser His Trp Ala Tyr Arg Gln Ile Thr 405 410 415Glu Arg Leu
Lys Asp Leu Ala Ile Arg Asn Gly Met Pro Leu Thr His 420 425 430Ser
Asn Gly Ser Tyr Gly Val Arg Phe Thr Cys Ser Gln Cys Gly Ala 435 440
445Thr Asn Gln Gly Ile Lys Asp Pro Thr Lys Tyr Lys Val Asp Ile Glu
450 455 460Ser Glu Thr Phe Leu Cys Ser Ile Cys Ser His Arg Glu Ile
Ala Ala465 470 475 480Val Asn Thr Ala Thr Asn Leu Ala Lys Gln Leu
Leu Asp Glu 485 49031526PRTArtificial sequenceSynthetic sequence
31Met Asn Asp Thr Glu Thr Ser Glu Thr Leu Thr Ser His Arg Thr Val1
5 10 15Cys Ala His Leu His Val Val Gly Glu Thr Gly Ser Leu Pro Arg
Leu 20 25 30Val Glu Ala Ala Leu Ala Glu Leu Ile Thr Leu Asn Gly Arg
Ala Thr 35 40 45Gln Ala Leu Leu Ser Leu Ala Lys Asn Gly Leu Val Leu
Arg Arg Asp 50 55 60Lys Glu Glu Asn Leu Ile Ala Ala Glu Leu Thr Leu
Pro Cys Arg Lys65 70 75 80Asn Lys Tyr Ala Asp Val Ala Ala Lys Ala
Gly Glu Pro Ile Leu Ala 85 90 95Thr Arg Ile Asn Asn Lys Gly Lys Leu
Val Thr Lys Lys Trp Tyr Gly 100 105 110Glu Gly Asn Ser Tyr His Ile
Val Arg Phe Thr Pro Glu Thr Gly Met 115 120 125Phe Thr Val Arg Val
Phe Asp Arg Tyr Ala Phe Asp Glu Glu Leu Leu 130 135 140His Leu His
Ser Glu Val Val Phe Gly Ser Asp Leu Pro Lys Gly Ile145 150 155
160Lys Ala Lys Thr Asp Ser Leu Pro Ala Asn Phe Leu Gln Ala Val Phe
165 170 175Thr Ser Phe Leu Glu Leu Pro Phe Gln Gly Phe Pro Asp Ile
Val Val 180 185 190Lys Pro Ala Met Lys Gln Ala Ala Glu Gln Leu Leu
Ser Tyr Val Gln 195 200 205Leu Glu Ala Gly Glu Asn Gln Gln Ala Glu
Tyr Pro Asp Thr Asn Glu 210 215 220Arg Asp Pro Glu Leu Arg Leu Val
Glu Trp Gln Lys Ser Leu His Glu225 230 235 240Leu Ser Val Arg Thr
Glu Pro Phe Glu Phe Val Arg Ala Arg Asp Ile 245 250 255Asp Tyr Tyr
Ala Glu Thr Asp Arg Arg Gly Asn Arg Phe Val Asn Ile 260 265 270Thr
Pro Glu Trp Thr Lys Phe Ala Glu Ser Pro Phe Ala Arg Arg Leu 275 280
285Pro Leu Lys Ile Pro Pro Glu Phe Cys Ile Leu Leu Arg Arg Lys Thr
290 295 300Glu Gly His Ala Lys Ile Pro Asn Arg Ile Tyr Leu Gly Leu
Gln Ile305 310 315 320Phe Asp Gly Val Thr Pro Asp Ser Thr Leu Gly
Val Leu Ala Thr Ala 325 330 335Glu Asp Gly Lys Leu Phe Trp Trp His
Asp His Leu Asp Glu Phe Ser 340 345 350Asn Leu Glu Gly Lys Pro Glu
Pro Lys Leu Lys Asn Lys Pro Gln Leu 355 360 365Leu Met Val Ser Leu
Glu Tyr Asp Arg Glu Gln Arg Phe Glu Glu Ser 370 375 380Val Gly Gly
Asp Arg Lys Ile Cys Leu Val Thr Leu Lys Glu Thr Arg385 390 395
400Asn Phe Arg Arg Gly Trp Asn Gly Arg Ile Leu Gly Ile His Phe Gln
405 410 415His Asn Pro Val Ile Thr Trp Ala Leu Met Asp His Asp Ala
Glu Val 420 425 430Leu Glu Lys Gly Phe Ile Glu Gly Asn Ala Phe Leu
Gly Lys Ala Leu 435 440 445Asp Lys Gln Ala Leu Asn Glu Tyr Leu Gln
Lys Gly Gly Lys Trp Val 450 455 460Gly Asp Arg Ser Phe Gly Asn Lys
Leu Lys Gly Ile Thr His Thr Leu465 470 475 480Ala Ser Leu Ile Val
Arg Leu Ala Arg Glu Lys Asp Ala Trp Ile Ala 485
490 495Leu Glu Glu Ile Ser Trp Val Gln Lys Gln Ser Ala Asp Ser Val
Ala 500 505 510Asn His Glu Ile Val Glu Gln Pro His His Ser Leu Thr
Arg 515 520 52532649PRTArtificial sequenceSynthetic sequence 32Met
Asn Asp Thr Glu Thr Ser Glu Thr Leu Thr Ser His Arg Thr Val1 5 10
15Cys Ala His Leu His Val Val Gly Glu Thr Gly Ser Leu Pro Arg Leu
20 25 30Val Glu Ala Ala Leu Ala Glu Leu Ile Thr Leu Asn Gly Arg Ala
Thr 35 40 45Gln Ala Leu Leu Ser Leu Ala Lys Asn Gly Leu Val Leu Arg
Arg Asp 50 55 60Lys Glu Glu Asn Leu Ile Ala Ala Glu Leu Thr Leu Pro
Cys Arg Lys65 70 75 80Asn Lys Tyr Ala Asp Val Ala Ala Lys Ala Gly
Glu Pro Ile Leu Ala 85 90 95Thr Arg Ile Asn Asn Lys Gly Lys Leu Val
Thr Lys Lys Trp Tyr Gly 100 105 110Glu Gly Asn Ser Tyr His Ile Val
Arg Phe Thr Pro Glu Thr Gly Met 115 120 125Phe Thr Val Arg Val Phe
Asp Arg Tyr Ala Phe Asp Glu Glu Leu Leu 130 135 140His Leu His Ser
Glu Val Val Phe Gly Ser Asp Leu Pro Lys Gly Ile145 150 155 160Lys
Ala Lys Thr Asp Ser Leu Pro Ala Asn Phe Leu Gln Ala Val Phe 165 170
175Thr Ser Phe Leu Glu Leu Pro Phe Gln Gly Phe Pro Asp Ile Val Val
180 185 190Lys Pro Ala Met Lys Gln Ala Ala Glu Gln Leu Leu Ser Tyr
Val Gln 195 200 205Leu Glu Ala Gly Glu Asn Gln Gln Ala Glu Tyr Pro
Asp Thr Asn Glu 210 215 220Arg Asp Pro Glu Leu Arg Leu Val Glu Trp
Gln Lys Ser Leu His Glu225 230 235 240Leu Ser Val Arg Thr Glu Pro
Phe Glu Phe Val Arg Ala Arg Asp Ile 245 250 255Asp Tyr Tyr Ala Glu
Thr Asp Arg Arg Gly Asn Arg Phe Val Asn Ile 260 265 270Thr Pro Glu
Trp Thr Lys Phe Ala Glu Ser Pro Phe Ala Arg Arg Leu 275 280 285Pro
Leu Lys Ile Pro Pro Glu Phe Cys Ile Leu Leu Arg Arg Lys Thr 290 295
300Glu Gly His Ala Lys Ile Pro Asn Arg Ile Tyr Leu Gly Leu Gln
Ile305 310 315 320Phe Asp Gly Val Thr Pro Asp Ser Thr Leu Gly Val
Leu Ala Thr Ala 325 330 335Glu Asp Gly Lys Leu Phe Trp Trp His Asp
His Leu Asp Glu Phe Ser 340 345 350Asn Leu Glu Gly Lys Pro Glu Pro
Lys Leu Lys Asn Lys Pro Gln Leu 355 360 365Leu Met Val Ser Leu Glu
Tyr Asp Arg Glu Gln Arg Phe Glu Glu Ser 370 375 380Val Gly Gly Asp
Arg Lys Ile Cys Leu Val Thr Leu Lys Glu Thr Arg385 390 395 400Asn
Phe Arg Arg Gly Arg His Gly His Thr Arg Thr Asp Arg Leu Pro 405 410
415Ala Gly Asn Thr Leu Trp Arg Ala Asp Phe Ala Thr Ser Ala Glu Val
420 425 430Ala Ala Pro Lys Trp Asn Gly Arg Ile Leu Gly Ile His Phe
Gln His 435 440 445Asn Pro Val Ile Thr Trp Ala Leu Met Asp His Asp
Ala Glu Val Leu 450 455 460Glu Lys Gly Phe Ile Glu Gly Asn Ala Phe
Leu Gly Lys Ala Leu Asp465 470 475 480Lys Gln Ala Leu Asn Glu Tyr
Leu Gln Lys Gly Gly Lys Trp Val Gly 485 490 495Asp Arg Ser Phe Gly
Asn Lys Leu Lys Gly Ile Thr His Thr Leu Ala 500 505 510Ser Leu Ile
Val Arg Leu Ala Arg Glu Lys Asp Ala Trp Ile Ala Leu 515 520 525Glu
Glu Ile Ser Trp Val Gln Lys Gln Ser Ala Asp Ser Val Ala Asn 530 535
540Arg Arg Phe Ser Met Trp Asn Tyr Ser Arg Leu Ala Thr Leu Ile
Glu545 550 555 560Trp Leu Gly Thr Asp Ile Ala Thr Arg Asp Cys Gly
Thr Ala Ala Pro 565 570 575Leu Ala His Lys Val Ser Asp Tyr Leu Thr
His Phe Thr Cys Pro Glu 580 585 590Cys Gly Ala Cys Arg Lys Ala Gly
Gln Lys Lys Glu Ile Ala Asp Thr 595 600 605Val Arg Ala Gly Asp Ile
Leu Thr Cys Arg Lys Cys Gly Phe Ser Gly 610 615 620Pro Ile Pro Asp
Asn Phe Ile Ala Glu Phe Val Ala Lys Lys Ala Leu625 630 635 640Glu
Arg Met Leu Lys Lys Lys Pro Val 64533414PRTArtificial
sequenceSynthetic sequence 33Met Ala Lys Arg Asn Phe Gly Glu Lys
Ser Glu Ala Leu Tyr Arg Ala1 5 10 15Val Arg Phe Glu Val Arg Pro Ser
Lys Glu Glu Leu Ser Ile Leu Leu 20 25 30Ala Val Ser Glu Val Leu Arg
Met Leu Phe Asn Ser Ala Leu Ala Glu 35 40 45Arg Gln Gln Val Phe Thr
Glu Phe Ile Ala Ser Leu Tyr Ala Glu Leu 50 55 60Lys Ser Ala Ser Val
Pro Glu Glu Ile Ser Glu Ile Arg Lys Lys Leu65 70 75 80Arg Glu Ala
Tyr Lys Glu His Ser Ile Ser Leu Phe Asp Gln Ile Asn 85 90 95Ala Leu
Thr Ala Arg Arg Val Glu Asp Glu Ala Phe Ala Ser Val Thr 100 105
110Arg Asn Trp Gln Glu Glu Thr Leu Asp Ala Leu Asp Gly Ala Tyr Lys
115 120 125Ser Phe Leu Ser Leu Arg Arg Lys Gly Asp Tyr Asp Ala His
Ser Pro 130 135 140Arg Ser Arg Asp Ser Gly Phe Phe Gln Lys Ile Pro
Gly Arg Ser Gly145 150 155 160Phe Lys Ile Gly Glu Gly Arg Ile Ala
Leu Ser Cys Gly Ala Gly Arg 165 170 175Lys Leu Ser Phe Pro Ile Pro
Asp Tyr Gln Gln Gly Arg Leu Ala Glu 180 185 190Thr Thr Lys Leu Lys
Lys Phe Glu Leu Tyr Arg Asp Gln Pro Asn Leu 195 200 205Ala Lys Ser
Gly Arg Phe Trp Ile Ser Val Val Tyr Glu Leu Pro Lys 210 215 220Pro
Glu Ala Thr Thr Cys Gln Ser Glu Gln Val Ala Phe Val Ala Leu225 230
235 240Gly Ala Ser Ser Ile Gly Val Val Ser Gln Arg Gly Glu Glu Val
Ile 245 250 255Ala Leu Trp Arg Ser Asp Lys His Trp Val Pro Lys Ile
Glu Ala Val 260 265 270Glu Glu Arg Met Lys Arg Arg Val Lys Gly Ser
Arg Gly Trp Leu Arg 275 280 285Leu Leu Asn Ser Gly Lys Arg Arg Met
His Met Ile Ser Ser Arg Gln 290 295 300His Val Gln Asp Glu Arg Glu
Ile Val Asp Tyr Leu Val Arg Asn His305 310 315 320Gly Ser His Phe
Val Val Thr Glu Leu Val Val Arg Ser Lys Glu Gly 325 330 335Lys Leu
Ala Asp Ser Ser Lys Pro Glu Arg Gly Gly Ser Leu Gly Leu 340 345
350Asn Trp Ala Ala Gln Asn Thr Gly Ser Leu Ser Arg Leu Val Arg Gln
355 360 365Leu Glu Glu Lys Val Lys Glu His Gly Gly Ser Val Arg Lys
His Lys 370 375 380Leu Thr Leu Thr Glu Ala Pro Pro Ala Arg Gly Ala
Glu Asn Lys Leu385 390 395 400Trp Met Ala Arg Lys Leu Arg Glu Ser
Phe Leu Lys Glu Val 405 41034413PRTArtificial sequenceSynthetic
sequence 34Leu Ala Lys Asn Asp Glu Lys Glu Leu Leu Tyr Gln Ser Val
Lys Phe1 5 10 15Glu Ile Tyr Pro Asp Glu Ser Lys Ile Arg Val Leu Thr
Arg Val Ser 20 25 30Asn Ile Leu Val Leu Val Trp Asn Ser Ala Leu Gly
Glu Arg Arg Ala 35 40 45Arg Phe Glu Leu Tyr Ile Ala Pro Leu Tyr Glu
Glu Leu Lys Lys Phe 50 55 60Pro Arg Lys Ser Ala Glu Ser Asn Ala Leu
Arg Gln Lys Ile Arg Glu65 70 75 80Gly Tyr Lys Glu His Ile Pro Thr
Phe Phe Asp Gln Leu Lys Lys Leu 85 90 95Leu Thr Pro Met Arg Lys Glu
Asp Pro Ala Leu Leu Gly Ser Val Pro 100 105 110Arg Ala Tyr Gln Glu
Glu Thr Leu Asn Thr Leu Asn Gly Ser Phe Val 115 120 125Ser Phe Met
Thr Leu Arg Arg Asn Asn Asp Met Asp Ala Lys Pro Pro 130 135 140Lys
Gly Arg Ala Glu Asp Arg Phe His Glu Ile Ser Gly Arg Ser Gly145 150
155 160Phe Lys Ile Asp Gly Ser Glu Phe Val Leu Ser Thr Lys Glu Gln
Lys 165 170 175Leu Arg Phe Pro Ile Pro Asn Tyr Gln Leu Glu Lys Leu
Lys Glu Ala 180 185 190Lys Gln Ile Lys Lys Phe Thr Leu Tyr Gln Ser
Arg Asp Arg Arg Phe 195 200 205Trp Ile Ser Ile Ala Tyr Glu Ile Glu
Leu Pro Asp Gln Arg Pro Phe 210 215 220Asn Pro Glu Glu Val Ile Tyr
Ile Ala Phe Gly Ala Ser Ser Ile Gly225 230 235 240Val Ile Ser Pro
Glu Gly Glu Lys Val Ile Asp Phe Trp Arg Pro Asp 245 250 255Lys His
Trp Lys Pro Lys Ile Lys Glu Val Glu Asn Arg Met Arg Ser 260 265
270Cys Lys Lys Gly Ser Arg Ala Trp Lys Lys Arg Ala Ala Ala Arg Arg
275 280 285Lys Met Tyr Ala Met Thr Gln Arg Gln Gln Lys Leu Asn His
Arg Glu 290 295 300Ile Val Ala Ser Leu Leu Arg Leu Gly Phe His Phe
Val Val Thr Glu305 310 315 320Tyr Thr Val Arg Ser Lys Pro Gly Lys
Leu Ala Asp Gly Ser Asn Pro 325 330 335Lys Arg Gly Gly Ala Pro Gln
Gly Phe Asn Trp Ser Ala Gln Asn Thr 340 345 350Gly Ser Phe Gly Glu
Phe Ile Leu Trp Leu Lys Gln Lys Val Lys Glu 355 360 365Gln Gly Gly
Thr Val Gln Thr Phe Arg Leu Val Leu Gly Gln Ser Glu 370 375 380Arg
Pro Glu Lys Arg Gly Arg Asp Asn Lys Ile Glu Met Val Arg Leu385 390
395 400Leu Arg Glu Lys Tyr Leu Glu Ser Gln Thr Ile Val Val 405
41035449PRTArtificial sequenceSynthetic sequence 35Met Ala Lys Gly
Lys Lys Lys Glu Gly Lys Pro Leu Tyr Arg Ala Val1 5 10 15Arg Phe Glu
Ile Phe Pro Thr Ser Asp Gln Ile Thr Leu Phe Leu Arg 20 25 30Val Ser
Lys Asn Leu Gln Gln Val Trp Asn Glu Ala Trp Gln Glu Arg 35 40 45Gln
Ser Cys Tyr Glu Gln Phe Phe Gly Ser Ile Tyr Glu Arg Ile Gly 50 55
60Gln Ala Lys Lys Arg Ala Gln Glu Ala Gly Phe Ser Glu Val Trp Glu65
70 75 80Asn Glu Ala Lys Lys Gly Leu Asn Lys Lys Leu Arg Gln Gln Glu
Ile 85 90 95Ser Met Gln Leu Val Ser Glu Lys Glu Ser Leu Leu Gln Glu
Leu Ser 100 105 110Ile Ala Phe Gln Glu His Gly Val Thr Leu Tyr Asp
Gln Ile Asn Gly 115 120 125Leu Thr Ala Arg Arg Ile Ile Gly Glu Phe
Ala Leu Ile Pro Arg Asn 130 135 140Trp Gln Glu Glu Thr Leu Asp Ser
Leu Asp Gly Ser Phe Lys Ser Phe145 150 155 160Leu Ala Leu Arg Lys
Asn Gly Asp Pro Asp Ala Lys Pro Pro Arg Gln 165 170 175Arg Val Ser
Glu Asn Ser Phe Tyr Lys Ile Pro Gly Arg Ser Gly Phe 180 185 190Lys
Val Ser Asn Gly Gln Ile Tyr Leu Ser Phe Gly Lys Ile Gly Gln 195 200
205Thr Leu Thr Ser Val Ile Pro Glu Phe Gln Leu Lys Arg Leu Glu Thr
210 215 220Ala Ile Lys Leu Lys Lys Phe Glu Leu Cys Arg Asp Glu Arg
Asp Met225 230 235 240Ala Lys Pro Gly Arg Phe Trp Ile Ser Val Ala
Tyr Glu Ile Pro Lys 245 250 255Pro Glu Lys Val Pro Val Val Ser Lys
Gln Ile Thr Tyr Leu Ala Ile 260 265 270Gly Ala Ser Arg Leu Gly Val
Val Ser Pro Lys Gly Glu Phe Cys Leu 275 280 285Asn Leu Pro Arg Ser
Asp Tyr His Trp Lys Pro Gln Ile Asn Ala Leu 290 295 300Gln Glu Arg
Leu Glu Gly Val Val Lys Gly Ser Arg Lys Trp Lys Lys305 310 315
320Arg Met Ala Ala Cys Thr Arg Met Phe Ala Lys Leu Gly His Gln Gln
325 330 335Lys Gln His Gly Gln Tyr Glu Val Val Lys Lys Leu Leu Arg
His Gly 340 345 350Val His Phe Val Val Thr Glu Leu Lys Val Arg Ser
Lys Pro Gly Ala 355 360 365Leu Ala Asp Ala Ser Lys Ser Asp Arg Lys
Gly Ser Pro Thr Gly Pro 370 375 380Asn Trp Ser Ala Gln Asn Thr Gly
Asn Ile Ala Arg Leu Ile Gln Lys385 390 395 400Leu Thr Asp Lys Ala
Ser Glu His Gly Gly Thr Val Ile Lys Arg Asn 405 410 415Pro Pro Leu
Leu Ser Leu Glu Glu Arg Gln Leu Pro Asp Ala Gln Arg 420 425 430Lys
Ile Phe Ile Ala Lys Lys Leu Arg Glu Glu Phe Leu Ala Asp Gln 435 440
445Lys36711PRTArtificial sequenceSynthetic sequence 36Met Ala Lys
Arg Glu Lys Lys Asp Asp Val Val Leu Arg Gly Thr Lys1 5 10 15Met Arg
Ile Tyr Pro Thr Asp Arg Gln Val Thr Leu Met Asp Met Trp 20 25 30Arg
Arg Arg Cys Ile Ser Leu Trp Asn Leu Leu Leu Asn Leu Glu Thr 35 40
45Ala Ala Tyr Gly Ala Lys Asn Thr Arg Ser Lys Leu Gly Trp Arg Ser
50 55 60Ile Trp Ala Arg Val Val Glu Glu Asn His Ala Lys Ala Leu Ile
Val65 70 75 80Tyr Gln His Gly Lys Cys Lys Lys Asp Gly Ser Phe Val
Leu Lys Arg 85 90 95Asp Gly Thr Val Lys His Pro Pro Arg Glu Arg Phe
Pro Gly Asp Arg 100 105 110Lys Ile Leu Leu Gly Leu Phe Asp Ala Leu
Arg His Thr Leu Asp Lys 115 120 125Gly Ala Lys Cys Lys Cys Asn Val
Asn Gln Pro Tyr Ala Leu Thr Arg 130 135 140Ala Trp Leu Asp Glu Thr
Gly His Gly Ala Arg Thr Ala Asp Ile Ile145 150 155 160Ala Trp Leu
Lys Asp Phe Lys Gly Glu Cys Asp Cys Thr Ala Ile Ser 165 170 175Thr
Ala Ala Lys Tyr Cys Pro Ala Pro Pro Thr Ala Glu Leu Leu Thr 180 185
190Lys Ile Lys Arg Ala Ala Pro Ala Asp Asp Leu Pro Val Asp Gln Ala
195 200 205Ile Leu Leu Asp Leu Phe Gly Ala Leu Arg Gly Gly Leu Lys
Gln Lys 210 215 220Glu Cys Asp His Thr His Ala Arg Thr Val Ala Tyr
Phe Glu Lys His225 230 235 240Glu Leu Ala Gly Arg Ala Glu Asp Ile
Leu Ala Trp Leu Ile Ala His 245 250 255Gly Gly Thr Cys Asp Cys Lys
Ile Val Glu Glu Ala Ala Asn His Cys 260 265 270Pro Gly Pro Arg Leu
Phe Ile Trp Glu His Glu Leu Ala Met Ile Met 275 280 285Ala Arg Leu
Lys Ala Glu Pro Arg Thr Glu Trp Ile Gly Asp Leu Pro 290 295 300Ser
His Ala Ala Gln Thr Val Val Lys Asp Leu Val Lys Ala Leu Gln305 310
315 320Thr Met Leu Lys Glu Arg Ala Lys Ala Ala Ala Gly Asp Glu Ser
Ala 325 330 335Arg Lys Thr Gly Phe Pro Lys Phe Lys Lys Gln Ala Tyr
Ala Ala Gly 340 345 350Ser Val Tyr Phe Pro Asn Thr Thr Met Phe Phe
Asp Val Ala Ala Gly 355 360 365Arg Val Gln Leu Pro Asn Gly Cys Gly
Ser Met Arg Cys Glu Ile Pro 370 375 380Arg Gln Leu Val Ala Glu Leu
Leu Glu Arg Asn Leu Lys Pro Gly Leu385 390 395 400Val Ile Gly Ala
Gln Leu Gly Leu Leu Gly Gly Arg Ile Trp Arg Gln 405 410 415Gly Asp
Arg Trp Tyr Leu Ser Cys Gln Trp Glu Arg Pro Gln Pro Thr 420 425
430Leu Leu Pro Lys Thr Gly Arg Thr Ala Gly Val Lys Ile Ala Ala Ser
435 440 445Ile Val Phe Thr Thr Tyr Asp Asn Arg Gly Gln Thr Lys Glu
Tyr Pro 450 455 460Met Pro Pro Ala Asp Lys Lys Leu Thr Ala Val His
Leu Val Ala Gly465 470 475 480Lys Gln Asn Ser Arg Ala Leu Glu Ala
Gln Lys Glu Lys Glu Lys Lys 485
490 495Leu Lys Ala Arg Lys Glu Arg Leu Arg Leu Gly Lys Leu Glu Lys
Gly 500 505 510His Asp Pro Asn Ala Leu Lys Pro Leu Lys Arg Pro Arg
Val Arg Arg 515 520 525Ser Lys Leu Phe Tyr Lys Ser Ala Ala Arg Leu
Ala Ala Cys Glu Ala 530 535 540Ile Glu Arg Asp Arg Arg Asp Gly Phe
Leu His Arg Val Thr Asn Glu545 550 555 560Ile Val His Lys Phe Asp
Ala Val Ser Val Gln Lys Met Ser Val Ala 565 570 575Pro Met Met Arg
Arg Gln Lys Gln Lys Glu Lys Gln Ile Glu Ser Lys 580 585 590Lys Asn
Glu Ala Lys Lys Glu Asp Asn Gly Ala Ala Lys Lys Pro Arg 595 600
605Asn Leu Lys Pro Val Arg Lys Leu Leu Arg His Val Ala Met Ala Arg
610 615 620Gly Arg Gln Phe Leu Glu Tyr Lys Tyr Asn Asp Leu Arg Gly
Pro Gly625 630 635 640Ser Val Leu Ile Ala Asp Arg Leu Glu Pro Glu
Val Gln Glu Cys Ser 645 650 655Arg Cys Gly Thr Lys Asn Pro Gln Met
Lys Asp Gly Arg Arg Leu Leu 660 665 670Arg Cys Ile Gly Val Leu Pro
Asp Gly Thr Asp Cys Asp Ala Val Leu 675 680 685Pro Arg Asn Arg Asn
Ala Ala Arg Asn Ala Glu Lys Arg Leu Arg Lys 690 695 700His Arg Glu
Ala His Asn Ala705 71037574PRTArtificial sequenceSynthetic sequence
37Met Asn Glu Val Leu Pro Ile Pro Ala Val Gly Glu Asp Ala Ala Asp1
5 10 15Thr Ile Met Arg Gly Ser Lys Met Arg Ile Tyr Pro Ser Val Arg
Gln 20 25 30Ala Ala Thr Met Asp Leu Trp Arg Arg Arg Cys Ile Gln Leu
Trp Asn 35 40 45Leu Leu Leu Glu Leu Glu Gln Ala Ala Tyr Ser Gly Glu
Asn Arg Arg 50 55 60Thr Gln Ile Gly Trp Arg Ser Ile Trp Ala Thr Val
Val Glu Asp Ser65 70 75 80His Ala Glu Ala Val Arg Val Ala Arg Glu
Gly Lys Lys Arg Lys Asp 85 90 95Gly Thr Phe Arg Lys Ala Pro Ser Gly
Lys Glu Ile Pro Pro Leu Asp 100 105 110Pro Ala Met Leu Ala Lys Ile
Gln Arg Gln Met Asn Gly Ala Val Asp 115 120 125Val Asp Pro Lys Thr
Gly Glu Val Thr Pro Ala Gln Pro Arg Leu Phe 130 135 140Met Trp Glu
His Glu Leu Gln Lys Ile Met Ala Arg Leu Lys Gln Ala145 150 155
160Pro Arg Thr His Trp Ile Asp Asp Leu Pro Ser His Ala Ala Gln Ser
165 170 175Val Val Lys Asp Leu Ile Lys Ala Leu Gln Ala Met Leu Arg
Glu Arg 180 185 190Lys Lys Arg Ala Ser Gly Ile Gly Gly Arg Asp Thr
Gly Phe Pro Lys 195 200 205Phe Lys Lys Asn Arg Tyr Ala Ala Gly Ser
Val Tyr Phe Ala Asn Thr 210 215 220Gln Leu Arg Phe Glu Ala Lys Arg
Gly Lys Ala Gly Asp Pro Asp Ala225 230 235 240Val Arg Gly Glu Phe
Ala Arg Val Lys Leu Pro Asn Gly Val Gly Trp 245 250 255Met Glu Cys
Arg Met Pro Arg His Ile Asn Ala Ala His Ala Tyr Ala 260 265 270Gln
Ala Thr Leu Met Gly Gly Arg Ile Trp Arg Gln Gly Glu Asn Trp 275 280
285Tyr Leu Ser Cys Gln Trp Lys Met Pro Lys Pro Ala Pro Leu Pro Arg
290 295 300Ala Gly Arg Thr Ala Ala Ile Lys Ile Ala Ala Ala Ile Pro
Ile Thr305 310 315 320Thr Val Asp Asn Arg Gly Gln Thr Arg Glu Tyr
Ala Met Pro Pro Ile 325 330 335Asp Arg Glu Arg Ile Ala Ala His Ala
Ala Ala Gly Arg Ala Gln Ser 340 345 350Arg Ala Leu Glu Ala Arg Lys
Arg Arg Ala Lys Lys Arg Glu Ala Tyr 355 360 365Ala Lys Lys Arg His
Ala Lys Lys Leu Glu Arg Gly Ile Ala Ala Lys 370 375 380Pro Pro Gly
Arg Ala Arg Ile Lys Leu Ser Pro Gly Phe Tyr Ala Ala385 390 395
400Ala Ala Lys Leu Ala Lys Leu Glu Ala Glu Asp Ala Asn Ala Arg Glu
405 410 415Ala Trp Leu His Glu Ile Thr Thr Gln Ile Val Arg Asn Phe
Asp Val 420 425 430Ile Ala Val Pro Arg Met Glu Val Ala Lys Leu Met
Lys Lys Pro Glu 435 440 445Pro Pro Glu Glu Lys Glu Glu Gln Val Lys
Ala Pro Trp Gln Gly Lys 450 455 460Arg Arg Ser Leu Lys Ala Ala Arg
Val Met Met Arg Arg Thr Ala Met465 470 475 480Ala Leu Ile Gln Thr
Thr Leu Lys Tyr Lys Ala Val Asp Leu Arg Gly 485 490 495Pro Gln Ala
Tyr Glu Glu Ile Ala Pro Leu Asp Val Thr Ala Ala Ala 500 505 510Cys
Ser Gly Cys Gly Val Leu Lys Pro Glu Trp Lys Met Ala Arg Ala 515 520
525Lys Gly Arg Glu Ile Met Arg Cys Gln Glu Pro Leu Pro Gly Gly Lys
530 535 540Thr Cys Asn Thr Val Leu Thr Tyr Thr Arg Asn Ser Ala Arg
Val Ile545 550 555 560Gly Arg Glu Leu Ala Val Arg Leu Ala Glu Arg
Gln Lys Ala 565 57038400PRTArtificial sequenceSynthetic sequence
38Met Thr Thr Gln Lys Thr Tyr Asn Phe Cys Phe Tyr Asp Gln Arg Phe1
5 10 15Phe Glu Leu Ser Lys Glu Ala Gly Glu Val Tyr Ser Arg Ser Leu
Glu 20 25 30Glu Phe Trp Lys Ile Tyr Asp Glu Thr Gly Val Trp Leu Ser
Lys Phe 35 40 45Asp Leu Gln Lys His Met Arg Asn Lys Leu Glu Arg Lys
Leu Leu His 50 55 60Ser Asp Ser Phe Leu Gly Ala Met Gln Gln Val His
Ala Asn Leu Ala65 70 75 80Ser Trp Lys Gln Ala Lys Lys Val Val Pro
Asp Ala Cys Pro Pro Arg 85 90 95Lys Pro Lys Phe Leu Gln Ala Ile Leu
Phe Lys Lys Ser Gln Ile Lys 100 105 110Tyr Lys Asn Gly Phe Leu Arg
Leu Thr Leu Gly Thr Glu Lys Glu Phe 115 120 125Leu Tyr Leu Lys Trp
Asp Ile Asn Ile Pro Leu Pro Ile Tyr Gly Ser 130 135 140Val Thr Tyr
Ser Lys Thr Arg Gly Trp Lys Ile Asn Leu Cys Leu Glu145 150 155
160Thr Glu Val Glu Gln Lys Asn Leu Ser Glu Asn Lys Tyr Leu Ser Ile
165 170 175Asp Leu Gly Val Lys Arg Val Ala Thr Ile Phe Asp Gly Glu
Asn Thr 180 185 190Ile Thr Leu Ser Gly Lys Lys Phe Met Gly Leu Met
His Tyr Arg Asn 195 200 205Lys Leu Asn Gly Lys Thr Gln Ser Arg Leu
Ser His Lys Lys Lys Gly 210 215 220Ser Asn Asn Tyr Lys Lys Ile Gln
Arg Ala Lys Arg Lys Thr Thr Asp225 230 235 240Arg Leu Leu Asn Ile
Gln Lys Glu Met Leu His Lys Tyr Ser Ser Phe 245 250 255Ile Val Asn
Tyr Ala Ile Arg Asn Asp Ile Gly Asn Ile Ile Ile Gly 260 265 270Asp
Asn Ser Ser Thr His Asp Ser Pro Asn Met Arg Gly Lys Thr Asn 275 280
285Gln Lys Ile Ser Gln Asn Pro Glu Gln Lys Leu Lys Asn Tyr Ile Lys
290 295 300Tyr Lys Phe Glu Ser Ile Ser Gly Arg Val Asp Ile Val Pro
Glu Pro305 310 315 320Tyr Thr Ser Arg Lys Cys Pro His Cys Lys Asn
Ile Lys Lys Ser Ser 325 330 335Pro Lys Gly Arg Thr Tyr Lys Cys Lys
Lys Cys Gly Phe Ile Phe Asp 340 345 350Arg Asp Gly Val Gly Ala Ile
Asn Ile Tyr Asn Glu Asn Val Ser Phe 355 360 365Gly Gln Ile Ile Ser
Pro Gly Arg Ile Arg Ser Leu Thr Glu Pro Ile 370 375 380Gly Met Lys
Phe His Asn Glu Ile Tyr Phe Lys Ser Tyr Val Ala Ala385 390 395
40039743PRTArtificial sequenceSynthetic sequence 39Met Ser Val Arg
Ser Phe Gln Ala Arg Val Glu Cys Asp Lys Gln Thr1 5 10 15Met Glu His
Leu Trp Arg Thr His Lys Val Phe Asn Glu Arg Leu Pro 20 25 30Glu Ile
Ile Lys Ile Leu Phe Lys Met Lys Arg Gly Glu Cys Gly Gln 35 40 45Asn
Asp Lys Gln Lys Ser Leu Tyr Lys Ser Ile Ser Gln Ser Ile Leu 50 55
60Glu Ala Asn Ala Gln Asn Ala Asp Tyr Leu Leu Asn Ser Val Ser Ile65
70 75 80Lys Gly Trp Lys Pro Gly Thr Ala Lys Lys Tyr Arg Asn Ala Ser
Phe 85 90 95Thr Trp Ala Asp Asp Ala Ala Lys Leu Ser Ser Gln Gly Ile
His Val 100 105 110Tyr Asp Lys Lys Gln Val Leu Gly Asp Leu Pro Gly
Met Met Ser Gln 115 120 125Met Val Cys Arg Gln Ser Val Glu Ala Ile
Ser Gly His Ile Glu Leu 130 135 140Thr Lys Lys Trp Glu Lys Glu His
Asn Glu Trp Leu Lys Glu Lys Glu145 150 155 160Lys Trp Glu Ser Glu
Asp Glu His Lys Lys Tyr Leu Asp Leu Arg Glu 165 170 175Lys Phe Glu
Gln Phe Glu Gln Ser Ile Gly Gly Lys Ile Thr Lys Arg 180 185 190Arg
Gly Arg Trp His Leu Tyr Leu Lys Trp Leu Ser Asp Asn Pro Asp 195 200
205Phe Ala Ala Trp Arg Gly Asn Lys Ala Val Ile Asn Pro Leu Ser Glu
210 215 220Lys Ala Gln Ile Arg Ile Asn Lys Ala Lys Pro Asn Lys Lys
Asn Ser225 230 235 240Val Glu Arg Asp Glu Phe Phe Lys Ala Asn Pro
Glu Met Lys Ala Leu 245 250 255Asp Asn Leu His Gly Tyr Tyr Glu Arg
Asn Phe Val Arg Arg Arg Lys 260 265 270Thr Lys Lys Asn Pro Asp Gly
Phe Asp His Lys Pro Thr Phe Thr Leu 275 280 285Pro His Pro Thr Ile
His Pro Arg Trp Phe Val Phe Asn Lys Pro Lys 290 295 300Thr Asn Pro
Glu Gly Tyr Arg Lys Leu Ile Leu Pro Lys Lys Ala Gly305 310 315
320Asp Leu Gly Ser Leu Glu Met Arg Leu Leu Thr Gly Glu Lys Asn Lys
325 330 335Gly Asn Tyr Pro Asp Asp Trp Ile Ser Val Lys Phe Lys Ala
Asp Pro 340 345 350Arg Leu Ser Leu Ile Arg Pro Val Lys Gly Arg Arg
Val Val Arg Lys 355 360 365Gly Lys Glu Gln Gly Gln Thr Lys Glu Thr
Asp Ser Tyr Glu Phe Phe 370 375 380Asp Lys His Leu Lys Lys Trp Arg
Pro Ala Lys Leu Ser Gly Val Lys385 390 395 400Leu Ile Phe Pro Asp
Lys Thr Pro Lys Ala Ala Tyr Leu Tyr Phe Thr 405 410 415Cys Asp Ile
Pro Asp Glu Pro Leu Thr Glu Thr Ala Lys Lys Ile Gln 420 425 430Trp
Leu Glu Thr Gly Asp Val Thr Lys Lys Gly Lys Lys Arg Lys Lys 435 440
445Lys Val Leu Pro His Gly Leu Val Ser Cys Ala Val Asp Leu Ser Met
450 455 460Arg Arg Gly Thr Thr Gly Phe Ala Thr Leu Cys Arg Tyr Glu
Asn Gly465 470 475 480Lys Ile His Ile Leu Arg Ser Arg Asn Leu Trp
Val Gly Tyr Lys Glu 485 490 495Gly Lys Gly Cys His Pro Tyr Arg Trp
Thr Glu Gly Pro Asp Leu Gly 500 505 510His Ile Ala Lys His Lys Arg
Glu Ile Arg Ile Leu Arg Ser Lys Arg 515 520 525Gly Lys Pro Val Lys
Gly Glu Glu Ser His Ile Asp Leu Gln Lys His 530 535 540Ile Asp Tyr
Met Gly Glu Asp Arg Phe Lys Lys Ala Ala Arg Thr Ile545 550 555
560Val Asn Phe Ala Leu Asn Thr Glu Asn Ala Ala Ser Lys Asn Gly Phe
565 570 575Tyr Pro Arg Ala Asp Val Leu Leu Leu Glu Asn Leu Glu Gly
Leu Ile 580 585 590Pro Asp Ala Glu Lys Glu Arg Gly Ile Asn Arg Ala
Leu Ala Gly Trp 595 600 605Asn Arg Arg His Leu Val Glu Arg Val Ile
Glu Met Ala Lys Asp Ala 610 615 620Gly Phe Lys Arg Arg Val Phe Glu
Ile Pro Pro Tyr Gly Thr Ser Gln625 630 635 640Val Cys Ser Lys Cys
Gly Ala Leu Gly Arg Arg Tyr Ser Ile Ile Arg 645 650 655Glu Asn Asn
Arg Arg Glu Ile Arg Phe Gly Tyr Val Glu Lys Leu Phe 660 665 670Ala
Cys Pro Asn Cys Gly Tyr Cys Ala Asn Ala Asp His Asn Ala Ser 675 680
685Val Asn Leu Asn Arg Arg Phe Leu Ile Glu Asp Ser Phe Lys Ser Tyr
690 695 700Tyr Asp Trp Lys Arg Leu Ser Glu Lys Lys Gln Lys Glu Glu
Ile Glu705 710 715 720Thr Ile Glu Ser Lys Leu Met Asp Lys Leu Cys
Ala Met His Lys Ile 725 730 735Ser Arg Gly Ser Ile Ser Lys
74040769PRTArtificial sequenceSynthetic sequence 40Met His Leu Trp
Arg Thr His Cys Val Phe Asn Gln Arg Leu Pro Ala1 5 10 15Leu Leu Lys
Arg Leu Phe Ala Met Arg Arg Gly Glu Val Gly Gly Asn 20 25 30Glu Ala
Gln Arg Gln Val Tyr Gln Arg Val Ala Gln Phe Val Leu Ala 35 40 45Arg
Asp Ala Lys Asp Ser Val Asp Leu Leu Asn Ala Val Ser Leu Arg 50 55
60Lys Arg Ser Ala Asn Ser Ala Phe Lys Lys Lys Ala Thr Ile Ser Cys65
70 75 80Asn Gly Gln Ala Arg Glu Val Thr Gly Glu Glu Val Phe Ala Glu
Ala 85 90 95Val Ala Leu Ala Ser Lys Gly Val Phe Ala Tyr Asp Lys Asp
Asp Met 100 105 110Arg Ala Gly Leu Pro Asp Ser Leu Phe Gln Pro Leu
Thr Arg Asp Ala 115 120 125Val Ala Cys Met Arg Ser His Glu Glu Leu
Val Ala Thr Trp Lys Lys 130 135 140Glu Tyr Arg Glu Trp Arg Asp Arg
Lys Ser Glu Trp Glu Ala Glu Pro145 150 155 160Glu His Ala Leu Tyr
Leu Asn Leu Arg Pro Lys Phe Glu Glu Gly Glu 165 170 175Ala Ala Arg
Gly Gly Arg Phe Arg Lys Arg Ala Glu Arg Asp His Ala 180 185 190Tyr
Leu Asp Trp Leu Glu Ala Asn Pro Gln Leu Ala Ala Trp Arg Arg 195 200
205Lys Ala Pro Pro Ala Val Val Pro Ile Asp Glu Ala Gly Lys Arg Arg
210 215 220Ile Ala Arg Ala Lys Ala Trp Lys Gln Ala Ser Val Arg Ala
Glu Glu225 230 235 240Phe Trp Lys Arg Asn Pro Glu Leu His Ala Leu
His Lys Ile His Val 245 250 255Gln Tyr Leu Arg Glu Phe Val Arg Pro
Arg Arg Thr Arg Arg Asn Lys 260 265 270Arg Arg Glu Gly Phe Lys Gln
Arg Pro Thr Phe Thr Met Pro Asp Pro 275 280 285Val Arg His Pro Arg
Trp Cys Leu Phe Asn Ala Pro Gln Thr Ser Pro 290 295 300Gln Gly Tyr
Arg Leu Leu Arg Leu Pro Gln Ser Arg Arg Thr Val Gly305 310 315
320Ser Val Glu Leu Arg Leu Leu Thr Gly Pro Ser Asp Gly Ala Gly Phe
325 330 335Pro Asp Ala Trp Val Asn Val Arg Phe Lys Ala Asp Pro Arg
Leu Ala 340 345 350Gln Leu Arg Pro Val Lys Val Pro Arg Thr Val Thr
Arg Gly Lys Asn 355 360 365Lys Gly Ala Lys Val Glu Ala Asp Gly Phe
Arg Tyr Tyr Asp Asp Gln 370 375 380Leu Leu Ile Glu Arg Asp Ala Gln
Val Ser Gly Val Lys Leu Leu Phe385 390 395 400Arg Asp Ile Arg Met
Ala Pro Phe Ala Asp Lys Pro Ile Glu Asp Arg 405 410 415Leu Leu Ser
Ala Thr Pro Tyr Leu Val Phe Ala Val Glu Ile Lys Asp 420 425 430Glu
Ala Arg Thr Glu Arg Ala Lys Ala Ile Arg Phe Asp Glu Thr Ser 435 440
445Glu Leu Thr Lys Ser Gly Lys Lys Arg Lys Thr Leu Pro Ala Gly Leu
450 455 460Val Ser Val Ala Val Asp Leu Asp Thr Arg Gly Val Gly Phe
Leu Thr465 470 475 480Arg Ala Val Ile Gly Val Pro Glu Ile Gln Gln
Thr His His Gly Val 485 490 495Arg Leu Leu Gln Ser Arg Tyr Val Ala
Val Gly Gln Val Glu Ala Arg 500 505 510Ala Ser Gly Glu Ala Glu
Trp Ser Pro Gly Pro Asp Leu Ala His Ile 515 520 525Ala Arg His Lys
Arg Glu Ile Arg Arg Leu Arg Gln Leu Arg Gly Lys 530 535 540Pro Val
Lys Gly Glu Arg Ser His Val Arg Leu Gln Ala His Ile Asp545 550 555
560Arg Met Gly Glu Asp Arg Phe Lys Lys Ala Ala Arg Lys Ile Val Asn
565 570 575Glu Ala Leu Arg Gly Ser Asn Pro Ala Ala Gly Asp Pro Tyr
Thr Arg 580 585 590Ala Asp Val Leu Leu Tyr Glu Ser Leu Glu Thr Leu
Leu Pro Asp Ala 595 600 605Glu Arg Glu Arg Gly Ile Asn Arg Ala Leu
Leu Arg Trp Asn Arg Ala 610 615 620Lys Leu Ile Glu His Leu Lys Arg
Met Cys Asp Asp Ala Gly Ile Arg625 630 635 640His Phe Pro Val Ser
Pro Phe Gly Thr Ser Gln Val Cys Ser Lys Cys 645 650 655Gly Ala Leu
Gly Arg Arg Tyr Ser Leu Ala Arg Glu Asn Gly Arg Ala 660 665 670Val
Ile Arg Phe Gly Trp Val Glu Arg Leu Phe Ala Cys Pro Asn Pro 675 680
685Glu Cys Pro Gly Arg Arg Pro Asp Arg Pro Asp Arg Pro Phe Thr Cys
690 695 700Asn Ser Asp His Asn Ala Ser Val Asn Leu His Arg Val Phe
Ala Leu705 710 715 720Gly Asp Gln Ala Val Ala Ala Phe Arg Ala Leu
Ala Pro Arg Asp Ser 725 730 735Pro Ala Arg Thr Leu Ala Val Lys Arg
Val Glu Asp Thr Leu Arg Pro 740 745 750Gln Leu Met Arg Val His Lys
Leu Ala Asp Ala Gly Val Asp Ser Pro 755 760
765Phe41666PRTArtificial sequenceSynthetic sequence 41Met Ala Thr
Leu Val Tyr Arg Tyr Gly Val Arg Ala His Gly Ser Ala1 5 10 15Arg Gln
Gln Asp Ala Val Val Ser Asp Pro Ala Met Leu Glu Gln Leu 20 25 30Arg
Leu Gly His Glu Leu Arg Asn Ala Leu Val Gly Val Gln His Arg 35 40
45Tyr Glu Asp Gly Lys Arg Ala Val Trp Ser Gly Phe Ala Ser Val Ala
50 55 60Ala Ala Asp His Arg Val Thr Thr Gly Glu Thr Ala Val Ala Glu
Leu65 70 75 80Glu Lys Gln Ala Arg Ala Glu His Ser Ala Asp Arg Thr
Ala Ala Thr 85 90 95Arg Gln Gly Thr Ala Glu Ser Leu Lys Ala Ala Arg
Ala Ala Val Lys 100 105 110Gln Ala Arg Ala Asp Arg Lys Ala Ala Met
Ala Ala Val Ala Glu Gln 115 120 125Ala Lys Pro Lys Ile Gln Ala Leu
Gly Asp Asp Arg Asp Ala Glu Ile 130 135 140Lys Asp Leu Tyr Arg Arg
Phe Cys Gln Asp Gly Val Leu Leu Pro Arg145 150 155 160Cys Gly Arg
Cys Ala Gly Asp Leu Arg Ser Asp Gly Asp Cys Thr Asp 165 170 175Cys
Gly Ala Ala His Glu Pro Arg Lys Leu Tyr Trp Ala Thr Tyr Asn 180 185
190Ala Ile Arg Glu Asp His Gln Thr Ala Val Lys Leu Val Glu Ala Lys
195 200 205Arg Lys Ala Gly Gln Pro Ala Arg Leu Arg Phe Arg Arg Trp
Thr Gly 210 215 220Asp Gly Thr Leu Thr Val Gln Leu Gln Arg Met His
Gly Pro Ala Cys225 230 235 240Arg Cys Val Thr Cys Ala Glu Lys Leu
Thr Arg Arg Ala Arg Lys Thr 245 250 255Asp Pro Gln Ala Pro Ala Val
Ala Ala Asp Pro Ala Tyr Pro Pro Thr 260 265 270Asp Pro Pro Arg Asp
Pro Ala Leu Leu Ala Ser Gly Gln Gly Lys Trp 275 280 285Arg Asn Val
Leu Gln Leu Gly Thr Trp Ile Pro Pro Gly Glu Trp Ser 290 295 300Ala
Met Ser Arg Ala Glu Arg Arg Arg Val Gly Arg Ser His Ile Gly305 310
315 320Trp Gln Leu Gly Gly Gly Arg Gln Leu Thr Leu Pro Val Gln Leu
His 325 330 335Arg Gln Met Pro Ala Asp Ala Asp Val Ala Met Ala Gln
Leu Thr Arg 340 345 350Val Arg Val Gly Gly Arg His Arg Met Ser Val
Ala Leu Thr Ala Lys 355 360 365Leu Pro Asp Pro Pro Gln Val Gln Gly
Leu Pro Pro Val Ala Leu His 370 375 380Leu Gly Trp Arg Gln Arg Pro
Asp Gly Ser Leu Arg Val Ala Thr Trp385 390 395 400Ala Cys Pro Gln
Pro Leu Asp Leu Pro Pro Ala Val Ala Asp Val Val 405 410 415Val Ser
His Gly Gly Arg Trp Gly Glu Val Ile Met Pro Ala Arg Trp 420 425
430Leu Ala Asp Ala Glu Val Pro Pro Arg Leu Leu Gly Arg Arg Asp Lys
435 440 445Ala Met Glu Pro Val Leu Glu Ala Leu Ala Asp Trp Leu Glu
Ala His 450 455 460Thr Glu Ala Cys Thr Ala Arg Met Thr Pro Ala Leu
Val Arg Arg Trp465 470 475 480Arg Ser Gln Gly Arg Leu Ala Gly Leu
Thr Asn Arg Trp Arg Gly Gln 485 490 495Pro Pro Thr Gly Ser Ala Glu
Ile Leu Thr Tyr Leu Glu Ala Trp Arg 500 505 510Ile Gln Asp Lys Leu
Leu Trp Glu Arg Glu Ser His Leu Arg Arg Arg 515 520 525Leu Ala Ala
Arg Arg Asp Asp Ala Trp Arg Arg Val Ala Ser Trp Leu 530 535 540Ala
Arg His Ala Gly Val Leu Val Val Asp Asp Ala Asp Ile Ala Glu545 550
555 560Leu Arg Arg Arg Asp Asp Pro Ala Asp Thr Asp Pro Thr Met Pro
Ala 565 570 575Ser Ala Ala Gln Ala Ala Arg Ala Arg Ala Ala Leu Ala
Ala Pro Gly 580 585 590Arg Leu Arg His Leu Ala Thr Ile Thr Ala Thr
Arg Asp Gly Leu Gly 595 600 605Val His Thr Val Ala Ser Ala Gly Leu
Thr Arg Leu His Arg Lys Cys 610 615 620Gly His Gln Ala Gln Pro Asp
Pro Arg Tyr Ala Ala Ser Ala Val Val625 630 635 640Thr Cys Pro Gly
Cys Gly Asn Gly Tyr Asp Gln Asp Tyr Asn Ala Ala 645 650 655Met Leu
Met Leu Asp Arg Gln Gln Gln Pro 660 66542564PRTArtificial
sequenceSynthetic sequence 42Met Ser Arg Val Glu Leu His Arg Ala
Tyr Lys Phe Arg Leu Tyr Pro1 5 10 15Thr Pro Ala Gln Val Ala Glu Leu
Ala Glu Trp Glu Arg Gln Leu Arg 20 25 30Arg Leu Tyr Asn Leu Ala His
Ser Gln Arg Leu Ala Ala Met Gln Arg 35 40 45His Val Arg Pro Lys Ser
Pro Gly Val Leu Lys Ser Glu Cys Leu Ser 50 55 60Cys Gly Ala Val Ala
Val Ala Glu Ile Gly Thr Asp Gly Lys Ala Lys65 70 75 80Lys Thr Val
Lys His Ala Val Gly Cys Ser Val Leu Glu Cys Arg Ser 85 90 95Cys Gly
Gly Ser Pro Asp Ala Glu Gly Arg Thr Ala His Thr Ala Ala 100 105
110Cys Ser Phe Val Asp Tyr Tyr Arg Gln Gly Arg Glu Met Thr Gln Leu
115 120 125Leu Glu Glu Asp Asp Gln Leu Ala Arg Val Val Cys Ser Ala
Arg Gln 130 135 140Glu Thr Leu Arg Asp Leu Glu Lys Ala Trp Gln Arg
Trp His Lys Met145 150 155 160Pro Gly Phe Gly Lys Pro His Phe Lys
Lys Arg Ile Asp Ser Cys Arg 165 170 175Ile Tyr Phe Ser Thr Pro Lys
Ser Trp Ala Val Asp Leu Gly Tyr Leu 180 185 190Ser Phe Thr Gly Val
Ala Ser Ser Val Gly Arg Ile Lys Ile Arg Gln 195 200 205Asp Arg Val
Trp Pro Gly Asp Ala Lys Phe Ser Ser Cys His Val Val 210 215 220Arg
Asp Val Asp Glu Trp Tyr Ala Val Phe Pro Leu Thr Phe Thr Lys225 230
235 240Glu Ile Glu Lys Pro Lys Gly Gly Ala Val Gly Ile Asn Arg Gly
Ala 245 250 255Val His Ala Ile Ala Asp Ser Thr Gly Arg Val Val Asp
Ser Pro Lys 260 265 270Phe Tyr Ala Arg Ser Leu Gly Val Ile Arg His
Arg Ala Arg Leu Leu 275 280 285Asp Arg Lys Val Pro Phe Gly Arg Ala
Val Lys Pro Ser Pro Thr Lys 290 295 300Tyr His Gly Leu Pro Lys Ala
Asp Ile Asp Ala Ala Ala Ala Arg Val305 310 315 320Asn Ala Ser Pro
Gly Arg Leu Val Tyr Glu Ala Arg Ala Arg Gly Ser 325 330 335Ile Ala
Ala Ala Glu Ala His Leu Ala Ala Leu Val Leu Pro Ala Pro 340 345
350Arg Gln Thr Ser Gln Leu Pro Ser Glu Gly Arg Asn Arg Glu Arg Ala
355 360 365Arg Arg Phe Leu Ala Leu Ala His Gln Arg Val Arg Arg Gln
Arg Glu 370 375 380Trp Phe Leu His Asn Glu Ser Ala His Tyr Ala Gln
Ser Tyr Thr Lys385 390 395 400Ile Ala Ile Glu Asp Trp Ser Thr Lys
Glu Met Thr Ser Ser Glu Pro 405 410 415Arg Asp Ala Glu Glu Met Lys
Arg Val Thr Arg Ala Arg Asn Arg Ser 420 425 430Ile Leu Asp Val Gly
Trp Tyr Glu Leu Gly Arg Gln Ile Ala Tyr Lys 435 440 445Ser Glu Ala
Thr Gly Ala Glu Phe Ala Lys Val Asp Pro Gly Leu Arg 450 455 460Glu
Thr Glu Thr His Val Pro Glu Ala Ile Val Arg Glu Arg Asp Val465 470
475 480Asp Val Ser Gly Met Leu Arg Gly Glu Ala Gly Ile Ser Gly Thr
Cys 485 490 495Ser Arg Cys Gly Gly Leu Leu Arg Ala Ser Ala Ser Gly
His Ala Asp 500 505 510Ala Glu Cys Glu Val Cys Leu His Val Glu Val
Gly Asp Val Asn Ala 515 520 525Ala Val Asn Val Leu Lys Arg Ala Met
Phe Pro Gly Ala Ala Pro Pro 530 535 540Ser Lys Glu Lys Ala Lys Val
Thr Ile Gly Ile Lys Gly Arg Lys Lys545 550 555 560Lys Arg Ala
Ala43565PRTArtificial sequenceSynthetic sequence 43Met Ser Arg Val
Glu Leu His Arg Ala Tyr Lys Phe Arg Leu Tyr Pro1 5 10 15Thr Pro Val
Gln Val Ala Glu Leu Ser Glu Trp Glu Arg Gln Leu Arg 20 25 30Arg Leu
Tyr Asn Leu Gly His Glu Gln Arg Leu Leu Thr Leu Thr Arg 35 40 45His
Leu Arg Pro Lys Ser Pro Gly Val Leu Lys Gly Glu Cys Leu Ser 50 55
60Cys Asp Ser Thr Gln Val Gln Glu Val Gly Ala Asp Gly Arg Pro Lys65
70 75 80Thr Thr Val Arg His Ala Glu Gln Cys Pro Thr Leu Ala Cys Arg
Ser 85 90 95Cys Gly Ala Leu Arg Asp Ala Glu Gly Arg Thr Ala His Thr
Val Ala 100 105 110Cys Ala Phe Val Asp Tyr Tyr Arg Gln Gly Arg Glu
Met Thr Glu Leu 115 120 125Leu Ala Ala Asp Asp Gln Leu Ala Arg Val
Val Cys Ser Ala Arg Gln 130 135 140Glu Val Leu Arg Asp Leu Asp Lys
Ala Trp Gln Arg Trp Arg Lys Met145 150 155 160Pro Gly Phe Gly Lys
Pro Arg Phe Lys Arg Arg Thr Asp Ser Cys Arg 165 170 175Ile Tyr Phe
Ser Thr Pro Lys Ala Trp Lys Leu Glu Gly Gly His Leu 180 185 190Ser
Phe Thr Gly Ala Ala Thr Thr Val Gly Ala Ile Lys Met Arg Gln 195 200
205Asp Arg Asn Trp Pro Ala Ser Val Gln Phe Ser Ser Cys His Val Val
210 215 220Arg Asp Val Asp Glu Trp Tyr Ala Val Phe Pro Leu Thr Phe
Val Ala225 230 235 240Glu Val Ala Arg Pro Lys Gly Gly Ala Val Gly
Ile Asn Arg Gly Ala 245 250 255Val His Ala Ile Ala Asp Ser Thr Gly
Arg Val Val Asp Ser Pro Arg 260 265 270Tyr Tyr Ala Arg Ala Leu Gly
Val Ile Arg His Arg Ala Arg Leu Phe 275 280 285Asp Arg Lys Val Pro
Ser Gly His Ala Val Lys Pro Ser Pro Thr Lys 290 295 300Tyr Arg Gly
Leu Ser Ala Ile Glu Val Asp Arg Val Ala Arg Ala Thr305 310 315
320Gly Phe Thr Pro Gly Arg Val Val Thr Glu Ala Leu Asn Arg Gly Gly
325 330 335Val Ala Tyr Ala Glu Cys Ala Leu Ala Ala Ile Ala Val Leu
Gly His 340 345 350Gly Pro Glu Arg Pro Leu Thr Ser Asp Gly Arg Asn
Arg Glu Lys Ala 355 360 365Arg Lys Phe Leu Ala Leu Ala His Gln Arg
Val Arg Arg Gln Arg Glu 370 375 380Trp Phe Leu His Asn Glu Ser Ala
His Tyr Ala Arg Thr Tyr Ser Lys385 390 395 400Ile Ala Ile Glu Asp
Trp Ser Thr Lys Glu Met Thr Ala Ser Glu Pro 405 410 415Gln Gly Glu
Glu Thr Arg Arg Val Thr Arg Ser Arg Asn Arg Ser Ile 420 425 430Leu
Asp Val Gly Trp Tyr Glu Leu Gly Arg Gln Leu Ala Tyr Lys Thr 435 440
445Glu Ala Thr Gly Ala Glu Phe Ala Gln Val Asp Pro Gly Leu Lys Glu
450 455 460Thr Glu Thr Asn Val Pro Lys Ala Ile Ala Asp Ala Arg Asp
Val Asp465 470 475 480Val Ser Gly Met Leu Arg Gly Glu Ala Gly Ile
Ser Gly Thr Cys Ser 485 490 495Lys Cys Gly Gly Leu Leu Arg Ala Pro
Ala Ser Gly His Ala Asp Ala 500 505 510Glu Cys Glu Ile Cys Leu Asn
Val Glu Val Gly Asp Val Asn Ala Ala 515 520 525Val Asn Val Leu Lys
Arg Ala Met Phe Pro Gly Asp Ala Pro Pro Ala 530 535 540Ser Gly Glu
Lys Pro Lys Val Ser Ile Gly Ile Lys Gly Arg Gln Lys545 550 555
560Lys Lys Lys Ala Ala 56544499PRTArtificial sequenceSynthetic
sequence 44Met Glu Ala Ile Ala Thr Gly Met Ser Pro Glu Arg Arg Val
Glu Leu1 5 10 15Gly Ile Leu Pro Gly Ser Val Glu Leu Lys Arg Ala Tyr
Lys Phe Arg 20 25 30Leu Tyr Pro Met Lys Val Gln Gln Ala Glu Leu Ser
Glu Trp Glu Arg 35 40 45Gln Leu Arg Arg Leu Tyr Asn Leu Ala His Glu
Gln Arg Leu Ala Ala 50 55 60Leu Leu Arg Tyr Arg Asp Trp Asp Phe Gln
Lys Gly Ala Cys Pro Ser65 70 75 80Cys Arg Val Ala Val Pro Gly Val
His Thr Ala Ala Cys Asp His Val 85 90 95Asp Tyr Phe Arg Gln Ala Arg
Glu Met Thr Gln Leu Leu Glu Val Asp 100 105 110Ala Gln Leu Ser Arg
Val Ile Cys Cys Ala Arg Gln Glu Val Leu Arg 115 120 125Asp Leu Asp
Lys Ala Trp Gln Arg Trp Arg Lys Lys Leu Gly Gly Arg 130 135 140Pro
Arg Phe Lys Arg Arg Thr Asp Ser Cys Arg Ile Tyr Leu Ser Thr145 150
155 160Pro Lys His Trp Glu Ile Ala Gly Arg Tyr Leu Arg Leu Ser Gly
Leu 165 170 175Ala Ser Ser Val Gly Glu Ile Arg Ile Glu Gln Asp Arg
Ala Phe Pro 180 185 190Glu Gly Ala Leu Leu Ser Ser Cys Ser Ile Val
Arg Asp Val Asp Glu 195 200 205Trp Tyr Ala Cys Leu Pro Leu Thr Phe
Thr Gln Pro Ile Glu Arg Ala 210 215 220Pro His Arg Ser Val Gly Leu
Asn Arg Gly Val Val His Ala Leu Ala225 230 235 240Asp Ser Asp Gly
Arg Val Val Asp Ser Pro Lys Phe Phe Glu Arg Ala 245 250 255Leu Ala
Thr Val Gln Lys Arg Ser Arg Asp Leu Ala Arg Lys Val Ser 260 265
270Gly Ser Arg Asn Ala His Lys Ala Arg Ile Lys Leu Ala Lys Ala His
275 280 285Gln Arg Val Arg Arg Gln Arg Ala Ala Phe Leu His Gln Glu
Ser Ala 290 295 300Tyr Tyr Ser Lys Gly Phe Asp Leu Val Ala Leu Glu
Asp Met Ser Val305 310 315 320Arg Lys Met Thr Ala Thr Ala Gly Glu
Ala Pro Glu Met Gly Arg Gly 325 330 335Ala Gln Arg Asp Leu Asn Arg
Gly Ile Leu Asp Val Gly Trp Tyr Glu 340 345 350Leu Ala Arg Gln Ile
Asp Tyr Lys Arg Leu Ala His Gly Gly Glu Leu 355 360 365Leu Arg Val
Asp Pro Gly Gln Thr Thr Pro Leu Ala Cys Val Thr Glu 370 375 380Glu
Gln Pro Ala Arg Gly Ile Ser Ser Ala Cys Ala Val Cys Gly Ile385 390
395 400Pro Leu
Ala Arg Pro Ala Ser Gly Asn Ala Arg Met Arg Cys Thr Ala 405 410
415Cys Gly Ser Ser Gln Val Gly Asp Val Asn Ala Ala Glu Asn Val Leu
420 425 430Thr Arg Ala Leu Ser Ser Ala Pro Ser Gly Pro Lys Ser Pro
Lys Ala 435 440 445Ser Ile Lys Ile Lys Gly Arg Gln Lys Arg Leu Gly
Thr Pro Ala Asn 450 455 460Arg Ala Gly Glu Ala Ser Gly Gly Asp Pro
Pro Val Arg Gly Pro Val465 470 475 480Glu Gly Gly Thr Leu Ala Tyr
Val Val Glu Pro Val Ser Glu Ser Gln 485 490 495Ser Asp
Thr45560PRTArtificial sequenceSynthetic sequence 45Met Thr Val Arg
Thr Tyr Lys Tyr Arg Ala Tyr Pro Thr Pro Glu Gln1 5 10 15Ala Glu Ala
Leu Thr Ser Trp Leu Arg Phe Ala Ser Gln Leu Tyr Asn 20 25 30Ala Ala
Leu Glu His Arg Lys Asn Ala Trp Gly Arg His Asp Ala His 35 40 45Gly
Arg Gly Phe Arg Phe Trp Asp Gly Asp Ala Ala Pro Arg Lys Lys 50 55
60Ser Asp Pro Pro Gly Arg Trp Val Tyr Arg Gly Gly Gly Gly Ala His65
70 75 80Ile Ser Lys Asn Asp Gln Gly Lys Leu Leu Thr Glu Phe Arg Arg
Glu 85 90 95His Ala Glu Leu Leu Pro Pro Gly Met Pro Ala Leu Val Gln
His Glu 100 105 110Val Leu Ala Arg Leu Glu Arg Ser Met Ala Ala Phe
Phe Gln Arg Ala 115 120 125Thr Lys Gly Gln Lys Ala Gly Tyr Pro Arg
Trp Arg Ser Glu His Arg 130 135 140Tyr Asp Ser Leu Thr Phe Gly Leu
Thr Ser Pro Ser Lys Glu Arg Phe145 150 155 160Asp Pro Glu Thr Gly
Glu Ser Leu Gly Arg Gly Lys Thr Val Gly Ala 165 170 175Gly Thr Tyr
His Asn Gly Asp Leu Arg Leu Thr Gly Leu Gly Glu Leu 180 185 190Arg
Ile Leu Glu His Arg Arg Ile Pro Met Gly Ala Ile Pro Lys Ser 195 200
205Val Ile Val Arg Arg Ser Gly Lys Arg Trp Phe Val Ser Ile Ala Met
210 215 220Glu Met Pro Ser Val Glu Pro Ala Ala Ser Gly Arg Pro Ala
Val Gly225 230 235 240Leu Asp Met Gly Val Val Thr Trp Gly Thr Ala
Phe Thr Ala Asp Thr 245 250 255Ser Ala Ala Ala Ala Leu Val Ala Asp
Leu Arg Arg Met Ala Thr Asp 260 265 270Pro Ser Asp Cys Arg Arg Leu
Glu Glu Leu Glu Arg Glu Ala Ala Gln 275 280 285Leu Ser Glu Val Leu
Ala His Cys Arg Ala Arg Gly Leu Asp Pro Ala 290 295 300Arg Pro Arg
Arg Cys Pro Lys Glu Leu Thr Lys Leu Tyr Arg Arg Ser305 310 315
320Leu His Arg Leu Gly Glu Leu Asp Arg Ala Cys Ala Arg Ile Arg Arg
325 330 335Arg Leu Gln Ala Ala His Asp Ile Ala Glu Pro Val Pro Asp
Glu Ala 340 345 350Gly Ser Ala Val Leu Ile Glu Gly Ser Asn Ala Gly
Met Arg His Ala 355 360 365Arg Arg Val Ala Arg Thr Gln Arg Arg Val
Ala Arg Arg Thr Arg Ala 370 375 380Gly His Ala His Ser Asn Arg Arg
Lys Lys Ala Val Gln Ala Tyr Ala385 390 395 400Arg Ala Lys Glu Arg
Glu Arg Ser Ala Arg Gly Asp His Arg His Lys 405 410 415Val Ser Arg
Ala Leu Val Arg Gln Phe Glu Glu Ile Ser Val Glu Ala 420 425 430Leu
Asp Ile Lys Gln Leu Thr Val Ala Pro Glu His Asn Pro Asp Pro 435 440
445Gln Pro Asp Leu Pro Ala His Val Gln Arg Arg Arg Asn Arg Gly Glu
450 455 460Leu Asp Ala Ala Trp Gly Ala Phe Phe Ala Ala Leu Asp Tyr
Lys Ala465 470 475 480Ala Asp Ala Gly Gly Arg Val Ala Arg Lys Pro
Ala Pro His Thr Thr 485 490 495Gln Glu Cys Ala Arg Cys Gly Thr Leu
Val Pro Lys Pro Ile Ser Leu 500 505 510Arg Val His Arg Cys Pro Ala
Cys Gly Tyr Thr Ala Pro Arg Thr Val 515 520 525Asn Ser Ala Arg Asn
Val Leu Gln Arg Pro Leu Glu Glu Pro Gly Arg 530 535 540Ala Gly Pro
Ser Gly Ala Asn Gly Arg Gly Val Pro His Ala Val Ala545 550 555
56046404PRTArtificial sequenceSynthetic sequence 46Met Asn Cys Arg
Tyr Arg Tyr Arg Ile Tyr Pro Thr Pro Gly Gln Arg1 5 10 15Gln Ser Leu
Ala Arg Leu Phe Gly Cys Val Arg Val Val Trp Asn Asp 20 25 30Ala Leu
Phe Leu Cys Arg Gln Ser Glu Lys Leu Pro Lys Asn Ser Glu 35 40 45Leu
Gln Lys Leu Cys Ile Thr Gln Ala Lys Lys Thr Glu Ala Arg Gly 50 55
60Trp Leu Gly Gln Val Ser Ala Ile Pro Leu Gln Gln Ser Val Ala Asp65
70 75 80Leu Gly Val Ala Phe Lys Asn Phe Phe Gln Ser Arg Ser Gly Lys
Arg 85 90 95Lys Gly Lys Lys Val Asn Pro Pro Arg Val Lys Arg Arg Asn
Asn Arg 100 105 110Gln Gly Ala Arg Phe Thr Arg Gly Gly Phe Lys Val
Lys Thr Ser Lys 115 120 125Val Tyr Leu Ala Arg Ile Gly Asp Ile Lys
Ile Lys Trp Ser Arg Pro 130 135 140Leu Pro Ser Glu Pro Ser Ser Val
Thr Val Ile Lys Asp Cys Ala Gly145 150 155 160Gln Tyr Phe Leu Ser
Phe Val Val Glu Val Lys Pro Glu Ile Lys Pro 165 170 175Pro Lys Asn
Pro Ser Ile Gly Ile Asp Leu Gly Leu Lys Thr Phe Ala 180 185 190Ser
Cys Ser Asn Gly Glu Lys Ile Asp Ser Pro Asp Tyr Ser Arg Leu 195 200
205Tyr Arg Lys Leu Lys Arg Cys Gln Arg Arg Leu Ala Lys Arg Gln Arg
210 215 220Gly Ser Lys Arg Arg Glu Arg Met Arg Val Lys Val Ala Lys
Leu Asn225 230 235 240Ala Gln Ile Arg Asp Lys Arg Lys Asp Phe Leu
His Lys Leu Ser Thr 245 250 255Lys Val Val Asn Glu Asn Gln Val Ile
Ala Leu Glu Asp Leu Asn Val 260 265 270Gly Gly Met Leu Lys Asn Arg
Lys Leu Ser Arg Ala Ile Ser Gln Ala 275 280 285Gly Trp Tyr Glu Phe
Arg Ser Leu Cys Glu Gly Lys Ala Glu Lys His 290 295 300Asn Arg Asp
Phe Arg Val Ile Ser Arg Trp Glu Pro Thr Ser Gln Val305 310 315
320Cys Ser Glu Cys Gly Tyr Arg Trp Gly Lys Ile Asp Leu Ser Val Arg
325 330 335Ser Ile Val Cys Ile Asn Cys Gly Val Glu His Asp Arg Asp
Asp Asn 340 345 350Ala Ser Val Asn Ile Glu Gln Ala Gly Leu Lys Val
Gly Val Gly His 355 360 365Thr His Asp Ser Lys Arg Thr Gly Ser Ala
Cys Lys Thr Ser Asn Gly 370 375 380Ala Val Cys Val Glu Pro Ser Thr
His Arg Glu Tyr Val Gln Leu Thr385 390 395 400Leu Phe Asp
Trp47392PRTArtificial sequenceSynthetic sequence 47Met Lys Ser Arg
Trp Thr Phe Arg Cys Tyr Pro Thr Pro Glu Gln Glu1 5 10 15Gln His Leu
Ala Arg Thr Phe Gly Cys Val Arg Phe Val Trp Asn Trp 20 25 30Ala Leu
Arg Ala Arg Thr Asp Ala Phe Arg Ala Gly Glu Arg Ile Gly 35 40 45Tyr
Pro Ala Thr Asp Lys Ala Leu Thr Leu Leu Lys Gln Gln Pro Glu 50 55
60Thr Val Trp Leu Asn Glu Val Ser Ser Val Cys Leu Gln Gln Ala Leu65
70 75 80Arg Asp Leu Gln Val Ala Phe Ser Asn Phe Phe Asp Lys Arg Ala
Ala 85 90 95His Pro Ser Phe Lys Arg Lys Glu Ala Arg Gln Ser Ala Asn
Tyr Thr 100 105 110Glu Arg Gly Phe Ser Phe Asp His Glu Arg Arg Ile
Leu Lys Leu Ala 115 120 125Lys Ile Gly Ala Ile Lys Val Lys Trp Ser
Arg Lys Ala Ile Pro His 130 135 140Pro Ser Ser Ile Arg Leu Ile Arg
Thr Ala Ser Gly Lys Tyr Phe Val145 150 155 160Ser Leu Val Val Glu
Thr Gln Pro Ala Pro Met Pro Glu Thr Gly Glu 165 170 175Ser Val Gly
Val Asp Phe Gly Val Ala Arg Leu Ala Thr Leu Ser Asn 180 185 190Gly
Glu Arg Ile Ser Asn Pro Lys His Gly Ala Lys Trp Gln Arg Arg 195 200
205Leu Ala Phe Tyr Gln Lys Arg Leu Ala Arg Ala Thr Lys Gly Ser Lys
210 215 220Arg Arg Met Arg Ile Lys Arg His Val Ala Arg Ile His Glu
Lys Ile225 230 235 240Gly Asn Ser Arg Ser Asp Thr Leu His Lys Leu
Ser Thr Asp Leu Val 245 250 255Thr Arg Phe Asp Leu Ile Cys Val Glu
Asp Leu Asn Leu Arg Gly Met 260 265 270Val Lys Asn His Ser Leu Ala
Arg Ser Leu His Asp Ala Ser Ile Gly 275 280 285Ser Ala Ile Arg Met
Ile Glu Glu Lys Ala Glu Arg Tyr Gly Lys Asn 290 295 300Val Val Lys
Ile Asp Arg Trp Phe Pro Ser Ser Lys Thr Cys Ser Asp305 310 315
320Cys Gly His Ile Val Glu Gln Leu Pro Leu Asn Val Arg Glu Trp Thr
325 330 335Cys Pro Glu Cys Gly Thr Thr His Asp Arg Asp Ala Asn Ala
Ala Ala 340 345 350Asn Ile Leu Ala Val Gly Gln Thr Val Ser Ala His
Gly Gly Thr Val 355 360 365Arg Arg Ser Arg Ala Lys Ala Ser Glu Arg
Lys Ser Gln Arg Ser Ala 370 375 380Asn Arg Gln Gly Val Asn Arg
Ala385 3904810PRTArtificial sequenceSynthetic sequence 48Ala Ala
Ala Ala Ala Ala Ala Ala Ala Ala1 5 104910PRTArtificial
sequenceSynthetic sequence 49Ala Ala Ala Ala Ala Ala Ala Ala Ala
Ala1 5 105010PRTArtificial sequenceSynthetic sequence 50Ala Ala Ala
Ala Ala Ala Ala Ala Ala Ala1 5 105137DNAArtificial
sequenceSynthetic sequence 51gttgcattcc ttcattcgtc tattcgggtt
ctgcaac 375237DNAArtificial sequenceSynthetic sequence 52gttgcattcc
ttcattcgtc tatccgggtt ctgcaag 375337DNAArtificial sequenceSynthetic
sequence 53gttgcagaac ccgaatagac gaatgaagga atgcaac
375437DNAArtificial sequenceSynthetic sequence 54ctatcatatt
cagaacaaag ggattaagga atgcaac 375537DNAArtificial sequenceSynthetic
sequence 55ctttcatact cagaacaaag ggattaagga atgcaac
375637DNAArtificial sequenceSynthetic sequence 56gtctacaact
cattgataga aatcaatgag ttagaca 375737DNAArtificial sequenceSynthetic
sequence 57gttataaagg cggggatcgc gaccgagcga ttgaaag
375837DNAArtificial sequenceSynthetic sequence 58gttgcattcc
ttaattcatt ttctcaatat cggaaac 375937DNAArtificial sequenceSynthetic
sequence 59gttgcagaaa tagaataaag gaattaagga atgcaac
376037DNAArtificial sequenceSynthetic sequence 60ctttcatact
cagaacaaag ggattaagga atgcaac 376137DNAArtificial sequenceSynthetic
sequence 61atttcatact cagaacaaag ggattaagga atgcaac
376238DNAArtificial sequenceSynthetic sequence 62gtttcagcgc
acgaattaac gagatgagag atgcaact 386337DNAArtificial
sequenceSynthetic sequence 63cttgcagaag ctgaatagac gaatcaagga
atgcaac 376439DNAArtificial sequenceSynthetic sequence 64cacttgcagg
ccttgaatag aggagttaag gaatgcaac 396536DNAArtificial
sequenceSynthetic sequence 65gtctccatga ctgaaaagtc gtggccgaat
tgaaac 366637DNAArtificial sequenceSynthetic sequence 66gttgcagcgc
ccgaactgac gagacgagag atgcaac 376737DNAArtificial sequenceSynthetic
sequence 67gttgcgcgaa tagaataaag gaattaagga atgcaac
376838DNAArtificial sequenceSynthetic sequence 68agttgcattc
cttaatccct ctgttcagtt tgtgcaat 386937DNAArtificial
sequenceSynthetic sequence 69gttgcattcc tagtttctct aattagcact
gtgcaac 377037DNAArtificial sequenceSynthetic sequence 70gttgcggcgc
gcgaataaac gagactagga atgcaac 377141DNAArtificial sequenceSynthetic
sequence 71actagttgca ttccttaatc cctttgttct gaatatgcta g
417237DNAArtificial sequenceSynthetic sequence 72ctttcatatt
cagaacaaag ggattaagga atgcaac 377339DNAArtificial sequenceSynthetic
sequence 73gttgcagtcc ttaaccccta gtttctgaat atgaaagat
397437DNAArtificial sequenceSynthetic sequence 74gttgcagccc
ccgaactaac gagatgagag atgcaac 377538DNAArtificial sequenceSynthetic
sequence 75cttgcagaac aatcatatat gactaatcag actgcaac
387637DNAArtificial sequenceSynthetic sequence 76gttgcactca
ccggtgctca cgacgtaggg atgcaac 377736DNAArtificial sequenceSynthetic
sequence 77gtccctactc gctagggaaa ctaattgaat ggaaac
367837DNAArtificial sequenceSynthetic sequence 78gttgcattcg
ggtgcaaaac agggagtaga gtgtaac 377935DNAArtificial sequenceSynthetic
sequence 79cttccaaact cgagccagtg gggagagaag tggca
358038DNAArtificial sequenceSynthetic sequence 80cctgtagacc
ggtctcattc tgagaggggt atgcaact 388137DNAArtificial
sequenceSynthetic sequence 81gtctcgagac cctacagatt ttggagaggg
gtgggac 378237DNAArtificial sequenceSynthetic sequence 82gtcccacccc
tctccaaaat ctgtagggtc tcgagac 378337DNAArtificial sequenceSynthetic
sequence 83gtagcaggac tctcctcgag agaaacaggg gtatgct
378440DNAArtificial sequenceSynthetic sequence 84gtacaatacc
tctcctttaa gagagggagg ggtacgctac 408531DNAArtificial
sequenceSynthetic sequence 85ccccctcgtt tccttcaggg gattcctttc c
318628DNAArtificial sequenceSynthetic sequence 86ggttcccccg
ggcgcgggtg gggtggcg 288728DNAArtificial sequenceSynthetic sequence
87ggctgctccg ggtgcgcgtg gagcgagg 288835DNAArtificial
sequenceSynthetic sequence 88gttttatacc ctttagaatt taaactgtct aaaag
358936DNAArtificial sequenceSynthetic sequence 89attgcaccgg
ccaacgcaaa tctgattgat ggacac 369036DNAArtificial sequenceSynthetic
sequence 90gccgcagcgg ccgacgcggc cctgatcgat ggacac
369136DNAArtificial sequenceSynthetic sequence 91gtcgaaatgc
ccgcgcgggg gcgtcgtacc cgcgac 369229DNAArtificial sequenceSynthetic
sequence 92ggctagcccg tgcgcgcagg gacgagtgg 299324DNAArtificial
sequenceSynthetic sequence 93gcccgtgcgc gcagggacga gtgg
249437DNAArtificial sequenceSynthetic sequence 94gttgcagcgg
ccgacggagc gcgagcgtgg atgccac 379529DNAArtificial sequenceSynthetic
sequence 95ccatcgcccc gcgcgcacgt ggatgagcc 299635DNAArtificial
sequenceSynthetic sequence 96ctttagactt ctccggaagt cgaattaatg gaaac
359728DNAArtificial sequenceSynthetic sequence 97gggcgccccg
cgcgagcggg ggttgaag 289810PRTArtificial sequenceSynthetic sequence
98Ala Ala Ala Ala Ala Ala Ala Ala Ala Ala1 5 109910PRTArtificial
sequenceSynthetic sequence 99Ala Ala Ala Ala Ala Ala Ala Ala Ala
Ala1 5 1010010PRTArtificial sequenceSynthetic sequence 100Ala Ala
Ala Ala Ala Ala Ala Ala Ala Ala1 5 1010184PRTArtificial
sequenceSynthetic sequence 101Met Ala Ser Met Ile Ser Ser Ser Ala
Val Thr Thr Val Ser Arg Ala1 5 10 15Ser Arg Gly Gln Ser Ala Ala Met
Ala Pro Phe Gly Gly Leu Lys Ser 20 25 30Met Thr Gly Phe Pro Val Arg
Lys Val Asn Thr Asp Ile Thr Ser Ile 35 40 45Thr Ser Asn Gly Gly Arg
Val Lys Cys Met Gln Val Trp Pro Pro Ile 50 55 60Gly Lys Lys Lys Phe
Glu Thr Leu Ser Tyr Leu Pro Pro Leu Thr Arg65 70 75 80Asp Ser Arg
Ala10257PRTArtificial sequenceSynthetic sequence 102Met Ala Ser Met
Ile Ser Ser Ser Ala Val Thr Thr Val Ser Arg Ala1 5 10 15Ser Arg Gly
Gln Ser Ala Ala Met Ala Pro Phe Gly Gly Leu Lys Ser 20 25
30Met Thr Gly Phe Pro Val Arg Lys Val Asn Thr Asp Ile Thr Ser Ile
35 40 45Thr Ser Asn Gly Gly Arg Val Lys Ser 50 5510385PRTArtificial
sequenceSynthetic sequence 103Met Ala Ser Ser Met Leu Ser Ser Ala
Thr Met Val Ala Ser Pro Ala1 5 10 15Gln Ala Thr Met Val Ala Pro Phe
Asn Gly Leu Lys Ser Ser Ala Ala 20 25 30Phe Pro Ala Thr Arg Lys Ala
Asn Asn Asp Ile Thr Ser Ile Thr Ser 35 40 45Asn Gly Gly Arg Val Asn
Cys Met Gln Val Trp Pro Pro Ile Glu Lys 50 55 60Lys Lys Phe Glu Thr
Leu Ser Tyr Leu Pro Asp Leu Thr Asp Ser Gly65 70 75 80Gly Arg Val
Asn Cys 8510476PRTArtificial sequenceSynthetic sequence 104Met Ala
Gln Val Ser Arg Ile Cys Asn Gly Val Gln Asn Pro Ser Leu1 5 10 15Ile
Ser Asn Leu Ser Lys Ser Ser Gln Arg Lys Ser Pro Leu Ser Val 20 25
30Ser Leu Lys Thr Gln Gln His Pro Arg Ala Tyr Pro Ile Ser Ser Ser
35 40 45Trp Gly Leu Lys Lys Ser Gly Met Thr Leu Ile Gly Ser Glu Leu
Arg 50 55 60Pro Leu Lys Val Met Ser Ser Val Ser Thr Ala Cys65 70
7510576PRTArtificial sequenceSynthetic sequence 105Met Ala Gln Val
Ser Arg Ile Cys Asn Gly Val Trp Asn Pro Ser Leu1 5 10 15Ile Ser Asn
Leu Ser Lys Ser Ser Gln Arg Lys Ser Pro Leu Ser Val 20 25 30Ser Leu
Lys Thr Gln Gln His Pro Arg Ala Tyr Pro Ile Ser Ser Ser 35 40 45Trp
Gly Leu Lys Lys Ser Gly Met Thr Leu Ile Gly Ser Glu Leu Arg 50 55
60Pro Leu Lys Val Met Ser Ser Val Ser Thr Ala Cys65 70
7510672PRTArtificial sequenceSynthetic sequence 106Met Ala Gln Ile
Asn Asn Met Ala Gln Gly Ile Gln Thr Leu Asn Pro1 5 10 15Asn Ser Asn
Phe His Lys Pro Gln Val Pro Lys Ser Ser Ser Phe Leu 20 25 30Val Phe
Gly Ser Lys Lys Leu Lys Asn Ser Ala Asn Ser Met Leu Val 35 40 45Leu
Lys Lys Asp Ser Ile Phe Met Gln Leu Phe Cys Ser Phe Arg Ile 50 55
60Ser Ala Ser Val Ala Thr Ala Cys65 7010769PRTArtificial
sequenceSynthetic sequence 107Met Ala Ala Leu Val Thr Ser Gln Leu
Ala Thr Ser Gly Thr Val Leu1 5 10 15Ser Val Thr Asp Arg Phe Arg Arg
Pro Gly Phe Gln Gly Leu Arg Pro 20 25 30Arg Asn Pro Ala Asp Ala Ala
Leu Gly Met Arg Thr Val Gly Ala Ser 35 40 45Ala Ala Pro Lys Gln Ser
Arg Lys Pro His Arg Phe Asp Arg Arg Cys 50 55 60Leu Ser Met Val
Val6510877PRTArtificial sequenceSynthetic sequence 108Met Ala Ala
Leu Thr Thr Ser Gln Leu Ala Thr Ser Ala Thr Gly Phe1 5 10 15Gly Ile
Ala Asp Arg Ser Ala Pro Ser Ser Leu Leu Arg His Gly Phe 20 25 30Gln
Gly Leu Lys Pro Arg Ser Pro Ala Gly Gly Asp Ala Thr Ser Leu 35 40
45Ser Val Thr Thr Ser Ala Arg Ala Thr Pro Lys Gln Gln Arg Ser Val
50 55 60Gln Arg Gly Ser Arg Arg Phe Pro Ser Val Val Val Cys65 70
7510957PRTArtificial sequenceSynthetic sequence 109Met Ala Ser Ser
Val Leu Ser Ser Ala Ala Val Ala Thr Arg Ser Asn1 5 10 15Val Ala Gln
Ala Asn Met Val Ala Pro Phe Thr Gly Leu Lys Ser Ala 20 25 30Ala Ser
Phe Pro Val Ser Arg Lys Gln Asn Leu Asp Ile Thr Ser Ile 35 40 45Ala
Ser Asn Gly Gly Arg Val Gln Cys 50 5511065PRTArtificial
sequenceSynthetic sequence 110Met Glu Ser Leu Ala Ala Thr Ser Val
Phe Ala Pro Ser Arg Val Ala1 5 10 15Val Pro Ala Ala Arg Ala Leu Val
Arg Ala Gly Thr Val Val Pro Thr 20 25 30Arg Arg Thr Ser Ser Thr Ser
Gly Thr Ser Gly Val Lys Cys Ser Ala 35 40 45Ala Val Thr Pro Gln Ala
Ser Pro Val Ile Ser Arg Ser Ala Ala Ala 50 55
60Ala6511172PRTArtificial sequenceSynthetic sequence 111Met Gly Ala
Ala Ala Thr Ser Met Gln Ser Leu Lys Phe Ser Asn Arg1 5 10 15Leu Val
Pro Pro Ser Arg Arg Leu Ser Pro Val Pro Asn Asn Val Thr 20 25 30Cys
Asn Asn Leu Pro Lys Ser Ala Ala Pro Val Arg Thr Val Lys Cys 35 40
45Cys Ala Ser Ser Trp Asn Ser Thr Ile Asn Gly Ala Ala Ala Thr Thr
50 55 60Asn Gly Ala Ser Ala Ala Ser Ser65 7011220PRTArtificial
sequenceSynthetic sequencemisc_feature(4)..(4)Xaa can be any
naturally occurring amino acidmisc_feature(8)..(8)Xaa can be any
naturally occurring amino acidmisc_feature(11)..(11)Xaa can be any
naturally occurring amino acidmisc_feature(15)..(15)Xaa can be any
naturally occurring amino acidmisc_feature(19)..(19)Xaa can be any
naturally occurring amino acid 112Gly Leu Phe Xaa Ala Leu Leu Xaa
Leu Leu Xaa Ser Leu Trp Xaa Leu1 5 10 15Leu Leu Xaa Ala
2011320PRTArtificial sequenceSynthetic sequence 113Gly Leu Phe His
Ala Leu Leu His Leu Leu His Ser Leu Trp His Leu1 5 10 15Leu Leu His
Ala 201147PRTArtificial sequenceSynthetic sequence 114Pro Lys Lys
Lys Arg Lys Val1 511516PRTArtificial sequenceSynthetic sequence
115Lys Arg Pro Ala Ala Thr Lys Lys Ala Gly Gln Ala Lys Lys Lys Lys1
5 10 151169PRTArtificial sequenceSynthetic sequence 116Pro Ala Ala
Lys Arg Val Lys Leu Asp1 511711PRTArtificial sequenceSynthetic
sequence 117Arg Gln Arg Arg Asn Glu Leu Lys Arg Ser Pro1 5
1011838PRTArtificial sequenceSynthetic sequence 118Asn Gln Ser Ser
Asn Phe Gly Pro Met Lys Gly Gly Asn Phe Gly Gly1 5 10 15Arg Ser Ser
Gly Pro Tyr Gly Gly Gly Gly Gln Tyr Phe Ala Lys Pro 20 25 30Arg Asn
Gln Gly Gly Tyr 3511942PRTArtificial sequenceSynthetic sequence
119Arg Met Arg Ile Glx Phe Lys Asn Lys Gly Lys Asp Thr Ala Glu Leu1
5 10 15Arg Arg Arg Arg Val Glu Val Ser Val Glu Leu Arg Lys Ala Lys
Lys 20 25 30Asp Glu Gln Ile Leu Lys Arg Arg Asn Val 35
401208PRTArtificial sequenceSynthetic sequence 120Val Ser Arg Lys
Arg Pro Arg Pro1 51218PRTArtificial sequenceSynthetic sequence
121Pro Pro Lys Lys Ala Arg Glu Asp1 51228PRTArtificial
sequenceSynthetic sequence 122Pro Gln Pro Lys Lys Lys Pro Leu1
512312PRTArtificial sequenceSynthetic sequence 123Ser Ala Leu Ile
Lys Lys Lys Lys Lys Met Ala Pro1 5 101245PRTArtificial
sequenceSynthetic sequence 124Asp Arg Leu Arg Arg1
51257PRTArtificial sequenceSynthetic sequence 125Pro Lys Gln Lys
Lys Arg Lys1 512610PRTArtificial sequenceSynthetic sequence 126Arg
Lys Leu Lys Lys Lys Ile Lys Lys Leu1 5 1012710PRTArtificial
sequenceSynthetic sequence 127Arg Glu Lys Lys Lys Phe Leu Lys Arg
Arg1 5 1012820PRTArtificial sequenceSynthetic sequence 128Lys Arg
Lys Gly Asp Glu Val Asp Gly Val Asp Glu Val Ala Lys Lys1 5 10 15Lys
Ser Lys Lys 2012917PRTArtificial sequenceSynthetic sequence 129Arg
Lys Cys Leu Gln Ala Gly Met Asn Leu Glu Ala Arg Lys Thr Lys1 5 10
15Lys13011PRTArtificial sequenceSynthetic sequence 130Tyr Gly Arg
Lys Lys Arg Arg Gln Arg Arg Arg1 5 1013112PRTArtificial
sequenceSynthetic sequence 131Arg Arg Gln Arg Arg Thr Ser Lys Leu
Met Lys Arg1 5 1013227PRTArtificial sequenceSynthetic sequence
132Gly Trp Thr Leu Asn Ser Ala Gly Tyr Leu Leu Gly Lys Ile Asn Leu1
5 10 15Lys Ala Leu Ala Ala Leu Ala Lys Lys Ile Leu 20
2513333PRTArtificial sequenceSynthetic sequence 133Lys Ala Leu Ala
Trp Glu Ala Lys Leu Ala Lys Ala Leu Ala Lys Ala1 5 10 15Leu Ala Lys
His Leu Ala Lys Ala Leu Ala Lys Ala Leu Lys Cys Glu 20 25
30Ala13416PRTArtificial sequenceSynthetic sequence 134Arg Gln Ile
Lys Ile Trp Phe Gln Asn Arg Arg Met Lys Trp Lys Lys1 5 10
151359PRTArtificial sequenceSynthetic sequence 135Arg Lys Lys Arg
Arg Gln Arg Arg Arg1 51368PRTArtificial sequenceSynthetic sequence
136Arg Lys Lys Arg Arg Gln Arg Arg1 513711PRTArtificial
sequenceSynthetic sequence 137Tyr Ala Arg Ala Ala Ala Arg Gln Ala
Arg Ala1 5 1013811PRTArtificial sequenceSynthetic sequence 138Thr
His Arg Leu Pro Arg Arg Arg Arg Arg Arg1 5 1013911PRTArtificial
sequenceSynthetic sequence 139Gly Gly Arg Arg Ala Arg Arg Arg Arg
Arg Arg1 5 101405PRTArtificial sequenceSynthetic sequence 140Gly
Ser Gly Gly Ser1 51416PRTArtificial sequenceSynthetic sequence
141Gly Gly Ser Gly Gly Ser1 51424PRTArtificial sequenceSynthetic
sequence 142Gly Gly Gly Ser11434PRTArtificial sequenceSynthetic
sequence 143Gly Gly Ser Gly11445PRTArtificial sequenceSynthetic
sequence 144Gly Gly Ser Gly Gly1 51455PRTArtificial
sequenceSynthetic sequence 145Gly Ser Gly Ser Gly1
51465PRTArtificial sequenceSynthetic sequence 146Gly Ser Gly Gly
Gly1 51475PRTArtificial sequenceSynthetic sequence 147Gly Gly Gly
Ser Gly1 51485PRTArtificial sequenceSynthetic sequence 148Gly Ser
Ser Ser Gly1 51494PRTArtificial sequenceSynthetic sequence 149Ala
Ala Ala Ala11504PRTArtificial sequenceSynthetic sequence 150Ala Ala
Ala Ala115180DNAArtificial sequenceSynthetic sequence 151cgattcctcc
ctacagtagt taggtatagc cgaaaggtag agactaaatc tgtagttgga 60gtgggccgct
tgcatcggcc 80152122DNAArtificial sequenceSynthetic sequence
152tcgtctcgag ggttaccaaa attggcactt ctcgacttta ggccgatgca
agcggcccac 60tccactacag atttagtctc taccttgcgg ctatacctaa cttactgtag
ggaggaatcg 120tg 12215391DNAArtificial sequenceSynthetic sequence
153cttcactgat aaagtggaga accgcttcac caaaagctgt cccttagggg
attagaactt 60gagtgaaggt gggctgcttg catcagccta a
91154114DNAArtificial sequenceSynthetic sequence 154cagaataata
ctgacttact aagatatctt gagggtatac ccgaaaagat tggcgttgtt 60gcaacgcaat
aagatgtaaa tctgaaaagg tttggaatca tataaataat ttta
114155104DNAArtificial sequenceSynthetic sequence 155aagccaagat
atggaatgcc attgtaatat tatggtgttg acttagttta gatttaaaca 60atcttcgatg
gctatatgcg gaaggtttgg cgtcgttgta acgc 104156213DNAArtificial
sequenceSynthetic sequence 156cagtgtgcat agctataaca ctacgcaaag
actgctaaag agcgatgtgc tctatcgcag 60tctcaccttt aatggactta cggatctttt
ggagcactaa gctccgctgc ggtgcaacac 120cgcccttttc ttgcctctgc
ttgccctttc cggttattat agccgggaga gtgcggaaga 180ttaccgctct
agctcgcagc atgttactga gtc 21315794DNAArtificial sequenceSynthetic
sequence 157gcaagtcatt cggggacact ttttgttatt taaagtgttt tagataaatc
agtgtcatgc 60tgaataacga cccgacctat aaataacata atcc
94158252DNAArtificial sequenceSynthetic sequence 158gtccttaagg
tactacacat tacatgtgaa cgtggagcta ataatagaaa tattattaga 60ctacacctta
ttaataacgg taggagatct atatggtctt gaatggaata gtaattgtga
120aattataatt tctgttctta gctacttaag atggctcgtt gcaagccact
cgggggctct 180cttgaagtca aagagcttta gacaaatcag tgtcaaactg
aataacgacc cgaccatgac 240ttcataatcc cg 252159152DNAArtificial
sequenceSynthetic sequence 159ctcgaggcta tttcatactc agaacaaagg
gattaaggaa tgcaacccat ttcaaaatct 60gtggaattat cacaaccatt aacatttcat
actcagaaca aagggattaa ggaatgcaac 120ggacaccttg ctatgtcctt
ttgatgtatg tg 15216061DNAArtificial sequenceSynthetic sequence
160ctcgaggcta tttcatactc agaacaaagc tcagaacaaa gggaataagg
aatgcaacgg 60a 61161101DNAArtificial sequenceSynthetic sequence
161ctcagaaaca aagggattaa ggaatgcaac ccatttcaaa atctgtggaa
actcagaaca 60aagggattaa ggaatgcaac ggacaccttg ctatgtcctt c
10116286DNAArtificial sequenceSynthetic sequence 162ctcagaacaa
agggattaag gaatgcaacc catttcctca gaacaaaggg attaaggaat 60gcaacggaca
ccttgctatg tccttc 8616376DNAArtificial sequenceSynthetic sequence
163ctcagaacaa agggattaag gaatgcaacc catttctcag aacaaaggga
ttaaggaatg 60caacggacac cttgct 7616482DNAArtificial
sequenceSynthetic sequence 164ctcagaacaa agggattaag gaatgcaacc
catttcaaaa ctcagaacaa agggattaag 60gaatgcaacg gacaccttgc ta
8216572DNAArtificial sequenceSynthetic sequence 165ctcagaacaa
agggattaag gaatgcaacc catttcctca gaacaaaggg attaaggaat 60gcaacggaca
cc 7216659DNAArtificial sequenceSynthetic sequence 166ctcagaacaa
agggattaag gaatgcaacc cgggattaag gaatgcaacg gacaccttg
5916750DNAArtificial sequenceSynthetic sequence 167ctcagaacaa
agggattaag gaatgcaacc aacggacacc ttgctatgtc 5016845DNAArtificial
sequenceSynthetic sequence 168ctcagaacaa agggattaag gaatgcaacc
catttcaaaa tctgt 4516941DNAArtificial sequenceSynthetic sequence
169ctcagaacaa agggattaag gaatgcaacc catttcaaaa t
4117036DNAArtificial sequenceSynthetic sequence 170ctcagaacaa
agggattaag gaatgcaacc catttc 3617136DNAArtificial sequenceSynthetic
sequence 171ctcagaacaa agggattaag gaatgcaacc catttc
3617236DNAArtificial sequenceSynthetic sequence 172ctcagaacaa
agggattaag gaatgcaacc catttc 3617337DNAArtificial sequenceSynthetic
sequence 173ctcagaacaa agggattaag gaatgcaacc catttca
3717438DNAArtificial sequenceSynthetic sequence 174ctcagaacaa
agggattaag gaatgcaacc catttcaa 3817546DNAArtificial
sequenceSynthetic sequence 175ctcagaacaa agggattaag gaatgcaacc
catttcaaaa tctgtg 4617646DNAArtificial sequenceSynthetic sequence
176ctcagaacaa agggattaag gaatgcaacc catttcaaaa tctgtg
4617744DNAArtificial sequenceSynthetic sequence 177ctcagaacaa
agggattaag gaatgcaacc catttcaaaa tctg 44178726PRTArtificial
sequenceSynthetic sequence 178Ser Glu Ser Glu Asn Lys Ile Ile Glu
Gln Tyr Tyr Ala Phe Leu Tyr1 5 10 15Ser Phe Arg Asp Lys Tyr Glu Lys
Pro Glu Phe Lys Asn Arg Gly Asp 20 25 30Ile Lys Arg Lys Leu Gln Asn
Lys Trp Glu Asp Phe Leu Lys Glu Gln 35 40 45Asn Leu Lys Asn Asp Lys
Lys Leu Ser Asn Tyr Ile Phe Ser Asn Arg 50 55 60Asn Phe Arg Arg Ser
Tyr Asp Arg Glu Glu Glu Asn Glu Glu Gly Ile65 70 75 80Asp Glu Lys
Lys Ser Lys Pro Lys Arg Ile Asn Cys Phe Glu Lys Glu 85 90 95Lys Asn
Leu Lys Asp Gln Tyr Asp Lys Asp Ala Ile Asn Ala Ser Ala 100 105
110Asn Lys Asp Gly Ala Gln Lys Trp Gly Cys Phe Glu Cys Ile Phe Phe
115 120 125Pro Met Tyr Lys Ile Glu Ser Gly Asp Pro Asn Lys Arg Ile
Ile Ile 130 135 140Asn Lys Thr Arg Phe Lys Leu Phe Asp Phe Tyr Leu
Asn Leu Lys Gly145 150 155 160Cys Lys Ser Cys Leu Arg Ser Thr Tyr
His Pro Tyr Arg Ser Asn Val 165 170 175Tyr Ile Glu Ser Asn Tyr Asp
Lys Leu Lys Arg Glu Ile Gly Asn Phe 180 185 190Leu Gln Gln Lys Asn
Ile Phe Gln Arg Met Arg Lys Ala Lys Val Ser 195 200 205Glu Gly Lys
Tyr Leu Thr Asn Leu Asp Glu Tyr Arg Leu Ser Cys Val 210 215 220Ala
Met His Phe Lys Asn Arg Trp Leu Phe Phe Asp Ser Ile Gln Lys225 230
235 240Val Leu Arg Glu Thr Ile Lys Gln Arg Leu Lys Gln Met Arg Glu
Ser 245 250 255Tyr Asp Glu Gln Ala Lys Thr Lys Arg Ser Lys Gly His
Gly Arg Ala 260 265 270Lys Tyr Glu Asp Gln Val Arg Met Ile Arg Arg
Arg Ala Tyr Ser Ala 275 280 285Gln Ala His Lys Leu Leu Asp Asn Gly
Tyr Ile Thr Leu Phe Asp Tyr 290
295 300Asp Asp Lys Glu Ile Asn Lys Val Cys Leu Thr Ala Ile Asn Gln
Glu305 310 315 320Gly Phe Asp Ile Gly Gly Tyr Leu Asn Ser Asp Ile
Asp Asn Val Met 325 330 335Pro Pro Ile Glu Ile Ser Phe His Leu Lys
Trp Lys Tyr Asn Glu Pro 340 345 350Ile Leu Asn Ile Glu Ser Pro Phe
Ser Lys Ala Lys Ile Ser Asp Tyr 355 360 365Leu Arg Lys Ile Arg Glu
Asp Leu Asn Leu Glu Arg Gly Lys Glu Gly 370 375 380Lys Ala Arg Ser
Lys Lys Asn Val Arg Arg Lys Val Leu Ala Ser Lys385 390 395 400Gly
Glu Asp Gly Tyr Lys Lys Ile Phe Thr Asp Phe Phe Ser Lys Trp 405 410
415Lys Glu Glu Leu Glu Gly Asn Ala Met Glu Arg Val Leu Ser Gln Ser
420 425 430Ser Gly Asp Ile Gln Trp Ser Lys Lys Lys Arg Ile His Tyr
Thr Thr 435 440 445Leu Val Leu Asn Ile Asn Leu Leu Asp Lys Lys Gly
Val Gly Asn Leu 450 455 460Lys Tyr Tyr Glu Ile Ala Glu Lys Thr Lys
Ile Leu Ser Phe Asp Lys465 470 475 480Asn Glu Asn Lys Phe Trp Pro
Ile Thr Ile Gln Val Leu Leu Asp Gly 485 490 495Tyr Glu Ile Gly Thr
Glu Tyr Asp Glu Ile Lys Gln Leu Asn Glu Lys 500 505 510Thr Ser Lys
Gln Phe Thr Ile Tyr Asp Pro Asn Thr Lys Ile Ile Lys 515 520 525Ile
Pro Phe Thr Asp Ser Lys Ala Val Pro Leu Gly Met Leu Gly Ile 530 535
540Asn Ile Ala Thr Leu Lys Thr Val Lys Lys Thr Glu Arg Asp Ile
Lys545 550 555 560Val Ser Lys Ile Phe Lys Gly Gly Leu Asn Ser Lys
Ile Val Ser Lys 565 570 575Ile Gly Lys Gly Ile Tyr Ala Gly Tyr Phe
Pro Thr Val Asp Lys Glu 580 585 590Ile Leu Glu Glu Val Glu Glu Asp
Thr Leu Asp Asn Glu Phe Ser Ser 595 600 605Lys Ser Gln Arg Asn Ile
Phe Leu Lys Ser Ile Ile Lys Asn Tyr Asp 610 615 620Lys Met Leu Lys
Glu Gln Leu Phe Asp Phe Tyr Ser Phe Leu Val Arg625 630 635 640Asn
Asp Leu Gly Val Arg Phe Leu Thr Asp Arg Glu Leu Gln Asn Ile 645 650
655Glu Asp Glu Ser Phe Asn Leu Glu Lys Arg Phe Phe Glu Thr Asp Arg
660 665 670Asp Arg Ile Ala Arg Trp Phe Asp Asn Thr Asn Thr Asp Asp
Gly Lys 675 680 685Glu Lys Phe Lys Lys Leu Ala Asn Glu Ile Val Asp
Ser Tyr Lys Pro 690 695 700Arg Leu Ile Arg Leu Pro Val Val Arg Val
Ile Lys Arg Ile Gln Pro705 710 715 720Val Lys Gln Arg Glu Met
725179517PRTArtificial sequenceSynthetic sequence 179Lys Tyr Ser
Thr Arg Asp Phe Ser Glu Leu Asn Glu Ile Gln Val Thr1 5 10 15Ala Cys
Lys Gln Asp Glu Phe Phe Lys Val Ile Gln Asn Ala Trp Arg 20 25 30Glu
Ile Ile Lys Lys Arg Phe Leu Glu Asn Arg Glu Asn Phe Ile Glu 35 40
45Lys Lys Ile Phe Lys Asn Lys Lys Gly Arg Gly Lys Arg Gln Glu Ser
50 55 60Asp Lys Thr Ile Gln Arg Asn Arg Ala Ser Val Met Lys Asn Phe
Gln65 70 75 80Leu Ile Glu Asn Glu Lys Ile Ile Leu Arg Ala Pro Ser
Gly His Val 85 90 95Ala Cys Val Phe Pro Val Lys Val Gly Leu Asp Ile
Gly Gly Phe Lys 100 105 110Thr Asp Asp Leu Glu Lys Asn Ile Phe Pro
Pro Arg Thr Ile Thr Ile 115 120 125Asn Val Phe Trp Lys Asn Arg Asp
Arg Gln Arg Lys Gly Arg Lys Leu 130 135 140Glu Val Trp Gly Ile Lys
Ala Arg Thr Lys Leu Ile Glu Lys Val His145 150 155 160Lys Trp Asp
Lys Leu Glu Glu Val Lys Lys Lys Arg Leu Lys Ser Leu 165 170 175Glu
Gln Lys Gln Glu Lys Ser Leu Asp Asn Trp Ser Glu Val Asn Asn 180 185
190Asp Ser Phe Tyr Lys Val Gln Ile Asp Glu Leu Gln Glu Lys Ile Asp
195 200 205Lys Ser Leu Lys Gly Arg Thr Met Asn Lys Ile Leu Asp Asn
Lys Ala 210 215 220Lys Glu Ser Lys Glu Ala Glu Gly Leu Tyr Ile Glu
Trp Glu Lys Asp225 230 235 240Phe Glu Gly Glu Met Leu Arg Arg Ile
Glu Ala Ser Thr Gly Gly Glu 245 250 255Glu Lys Trp Gly Lys Arg Arg
Gln Arg Arg His Thr Ser Leu Leu Leu 260 265 270Asp Ile Lys Asn Asn
Ser Arg Gly Ser Lys Glu Ile Ile Asn Phe Tyr 275 280 285Ser Tyr Ala
Lys Gln Gly Lys Lys Glu Lys Lys Ile Glu Phe Phe Pro 290 295 300Phe
Pro Leu Thr Ile Thr Leu Asp Ala Glu Glu Glu Ser Pro Leu Asn305 310
315 320Ile Lys Ser Ile Pro Ile Glu Asp Lys Asn Ala Thr Ser Lys Tyr
Phe 325 330 335Ser Ile Pro Phe Thr Glu Thr Arg Ala Thr Pro Leu Ser
Ile Leu Gly 340 345 350Asp Arg Val Gln Lys Phe Lys Thr Lys Asn Ile
Ser Gly Ala Ile Lys 355 360 365Arg Asn Leu Gly Ser Ser Ile Ser Ser
Cys Lys Ile Val Gln Asn Ala 370 375 380Glu Thr Ser Ala Lys Ser Ile
Leu Ser Leu Pro Asn Val Lys Glu Asp385 390 395 400Asn Asn Met Glu
Ile Phe Ile Asn Thr Met Ser Lys Asn Tyr Phe Arg 405 410 415Ala Met
Met Lys Gln Met Glu Ser Phe Ile Phe Glu Met Glu Pro Lys 420 425
430Thr Leu Ile Asp Pro Tyr Lys Glu Lys Ala Ile Lys Trp Phe Glu Val
435 440 445Ala Ala Ser Ser Arg Ala Lys Arg Lys Leu Lys Lys Leu Ser
Lys Ala 450 455 460Asp Ile Lys Lys Ser Glu Leu Leu Leu Ser Asn Thr
Glu Glu Phe Glu465 470 475 480Lys Glu Lys Gln Glu Lys Leu Glu Ala
Leu Glu Lys Glu Ile Glu Glu 485 490 495Phe Tyr Leu Pro Arg Ile Val
Arg Leu Gln Leu Thr Lys Thr Ile Leu 500 505 510Glu Thr Pro Val Met
515180481PRTArtificial sequenceSynthetic sequence 180Lys Lys Leu
Gln Leu Leu Gly His Lys Ile Leu Leu Lys Glu Tyr Asp1 5 10 15Pro Asn
Ala Val Asn Ala Ala Ala Asn Phe Glu Thr Ser Thr Ala Glu 20 25 30Leu
Cys Gly Gln Cys Lys Met Lys Pro Phe Lys Asn Lys Arg Arg Phe 35 40
45Gln Tyr Thr Phe Gly Lys Asn Tyr His Gly Cys Leu Ser Cys Ile Gln
50 55 60Asn Val Tyr Tyr Ala Lys Lys Arg Ile Val Gln Ile Ala Lys Glu
Glu65 70 75 80Leu Lys His Gln Leu Thr Asp Ser Ile Ala Ser Ile Pro
Tyr Lys Tyr 85 90 95Thr Ser Leu Phe Ser Asn Thr Asn Ser Ile Asp Glu
Leu Tyr Ile Leu 100 105 110Lys Gln Glu Arg Ala Ala Phe Phe Ser Asn
Thr Asn Ser Ile Asp Glu 115 120 125Leu Tyr Ile Thr Gly Ile Glu Asn
Asn Ile Ala Phe Lys Val Ile Ser 130 135 140Ala Ile Trp Asp Glu Ile
Ile Lys Lys Arg Arg Gln Arg Tyr Ala Glu145 150 155 160Ser Leu Thr
Asp Thr Gly Thr Val Lys Ala Asn Arg Gly His Gly Gly 165 170 175Thr
Ala Tyr Lys Ser Asn Thr Arg Gln Glu Lys Ile Arg Ala Leu Gln 180 185
190Lys Gln Thr Leu His Met Val Thr Asn Pro Tyr Ile Ser Leu Ala Arg
195 200 205Tyr Lys Asn Asn Tyr Ile Val Ala Thr Leu Pro Arg Thr Ile
Gly Met 210 215 220His Ile Gly Ala Ile Lys Asp Arg Asp Pro Gln Lys
Lys Leu Ser Asp225 230 235 240Tyr Ala Ile Asn Phe Asn Val Phe Trp
Ser Asp Asp Arg Gln Leu Ile 245 250 255Glu Leu Ser Thr Val Gln Tyr
Thr Gly Asp Met Val Arg Lys Ile Glu 260 265 270Ala Glu Thr Gly Glu
Asn Asn Lys Trp Gly Glu Asn Met Lys Arg Thr 275 280 285Lys Thr Ser
Leu Leu Leu Glu Ile Leu Thr Lys Lys Thr Thr Asp Glu 290 295 300Leu
Thr Phe Lys Asp Trp Ala Phe Ser Thr Lys Lys Glu Ile Asp Ser305 310
315 320Val Thr Lys Lys Thr Tyr Gln Gly Phe Pro Ile Gly Ile Ile Phe
Glu 325 330 335Gly Asn Glu Ser Ser Val Lys Phe Gly Ser Gln Asn Tyr
Phe Pro Leu 340 345 350Pro Phe Asp Ala Lys Ile Thr Pro Pro Thr Ala
Glu Gly Phe Arg Leu 355 360 365Asp Trp Leu Arg Lys Gly Ser Phe Ser
Ser Gln Met Lys Thr Ser Tyr 370 375 380Gly Leu Ala Ile Tyr Ser Asn
Lys Val Thr Asn Ala Ile Pro Ala Tyr385 390 395 400Val Ile Lys Asn
Met Phe Tyr Lys Ile Ala Arg Ala Glu Asn Gly Lys 405 410 415Gln Ile
Lys Ala Lys Phe Leu Lys Lys Tyr Leu Asp Ile Ala Gly Asn 420 425
430Asn Tyr Val Pro Phe Ile Ile Met Gln His Tyr Arg Val Leu Asp Thr
435 440 445Phe Glu Glu Met Pro Ile Ser Gln Pro Lys Val Ile Arg Leu
Ser Leu 450 455 460Thr Lys Thr Gln His Ile Ile Ile Lys Lys Asp Lys
Thr Asp Ser Lys465 470 475 480Met181534PRTArtificial
sequenceSynthetic sequence 181Asn Thr Ser Asn Leu Ile Asn Leu Gly
Lys Lys Ala Ile Asn Ile Ser1 5 10 15Ala Asn Tyr Asp Ala Asn Leu Glu
Val Gly Cys Lys Asn Cys Lys Phe 20 25 30Leu Ser Ser Asn Gly Asn Phe
Pro Arg Gln Thr Asn Val Lys Glu Gly 35 40 45Cys His Ser Cys Glu Lys
Ser Thr Tyr Glu Pro Ser Ile Tyr Leu Val 50 55 60Lys Ile Gly Glu Arg
Lys Ala Lys Tyr Asp Val Leu Asp Ser Leu Lys65 70 75 80Lys Phe Thr
Phe Gln Ser Leu Lys Tyr Gln Ser Lys Lys Ser Met Lys 85 90 95Ser Arg
Asn Lys Lys Pro Lys Glu Leu Lys Glu Phe Val Ile Phe Ala 100 105
110Asn Lys Asn Lys Ala Phe Asp Val Ile Gln Lys Ser Tyr Asn His Leu
115 120 125Ile Leu Gln Ile Lys Lys Glu Ile Asn Arg Met Asn Ser Lys
Lys Arg 130 135 140Lys Lys Asn His Lys Arg Arg Leu Phe Arg Asp Arg
Glu Lys Gln Leu145 150 155 160Asn Lys Leu Arg Leu Ile Glu Ser Ser
Asn Leu Phe Leu Pro Arg Glu 165 170 175Asn Lys Gly Asn Asn His Val
Phe Thr Tyr Val Ala Ile His Ser Val 180 185 190Gly Arg Asp Ile Gly
Val Ile Gly Ser Tyr Asp Glu Lys Leu Asn Phe 195 200 205Glu Thr Glu
Leu Thr Tyr Gln Leu Tyr Phe Asn Asp Asp Lys Arg Leu 210 215 220Leu
Tyr Ala Tyr Lys Pro Lys Gln Asn Lys Ile Ile Lys Ile Lys Glu225 230
235 240Lys Leu Trp Asn Leu Arg Lys Glu Lys Glu Pro Leu Asp Leu Glu
Tyr 245 250 255Glu Lys Pro Leu Asn Lys Ser Ile Thr Phe Ser Ile Lys
Asn Asp Asn 260 265 270Leu Phe Lys Val Ser Lys Asp Leu Met Leu Arg
Arg Ala Lys Phe Asn 275 280 285Ile Gln Gly Lys Glu Lys Leu Ser Lys
Glu Glu Arg Lys Ile Asn Arg 290 295 300Asp Leu Ile Lys Ile Lys Gly
Leu Val Asn Ser Met Ser Tyr Gly Arg305 310 315 320Phe Asp Glu Leu
Lys Lys Glu Lys Asn Ile Trp Ser Pro His Ile Tyr 325 330 335Arg Glu
Val Arg Gln Lys Glu Ile Lys Pro Cys Leu Ile Lys Asn Gly 340 345
350Asp Arg Ile Glu Ile Phe Glu Gln Leu Lys Lys Lys Met Glu Arg Leu
355 360 365Arg Arg Phe Arg Glu Lys Arg Gln Lys Lys Ile Ser Lys Asp
Leu Ile 370 375 380Phe Ala Glu Arg Ile Ala Tyr Asn Phe His Thr Lys
Ser Ile Lys Asn385 390 395 400Thr Ser Asn Lys Ile Asn Ile Asp Gln
Glu Ala Lys Arg Gly Lys Ala 405 410 415Ser Tyr Met Arg Lys Arg Ile
Gly Tyr Glu Thr Phe Lys Asn Lys Tyr 420 425 430Cys Glu Gln Cys Leu
Ser Lys Gly Asn Val Tyr Arg Asn Val Gln Lys 435 440 445Gly Cys Ser
Cys Phe Glu Asn Pro Phe Asp Trp Ile Lys Lys Gly Asp 450 455 460Glu
Asn Leu Leu Pro Lys Lys Asn Glu Asp Leu Arg Val Lys Gly Ala465 470
475 480Phe Arg Asp Glu Ala Leu Glu Lys Gln Ile Val Lys Ile Ala Phe
Asn 485 490 495Ile Ala Lys Gly Tyr Glu Asp Phe Tyr Asp Asn Leu Gly
Glu Ser Thr 500 505 510Glu Lys Asp Leu Lys Leu Lys Phe Lys Val Gly
Thr Thr Ile Asn Glu 515 520 525Gln Glu Ser Leu Lys Leu
530182537PRTArtificial sequenceSynthetic sequence 182Thr Ser Asn
Pro Ile Lys Leu Gly Lys Lys Ala Ile Asn Ile Ser Ala1 5 10 15Asn Tyr
Asp Ser Asn Leu Gln Ile Gly Cys Lys Asn Cys Lys Phe Leu 20 25 30Ser
Tyr Asn Gly Asn Phe Pro Arg Gln Thr Asn Val Lys Glu Gly Cys 35 40
45His Ser Cys Glu Lys Ser Thr Tyr Glu Pro Pro Val Tyr Thr Val Arg
50 55 60Ile Gly Glu Arg Arg Ser Lys Tyr Asp Val Leu Asp Ser Leu Lys
Lys65 70 75 80Phe Ile Phe Leu Ser Leu Lys Tyr Arg Gln Ser Lys Lys
Met Lys Thr 85 90 95Arg Ser Lys Gly Ile Arg Gly Leu Glu Glu Phe Val
Ile Ser Ala Asn 100 105 110Leu Lys Lys Ala Met Asp Val Ile Gln Lys
Ser Tyr Arg His Leu Ile 115 120 125Leu Asn Ile Lys Asn Glu Ile Val
Arg Met Asn Gly Lys Lys Arg Asn 130 135 140Lys Asn His Lys Arg Leu
Leu Phe Arg Asp Arg Glu Lys Gln Leu Asn145 150 155 160Lys Leu Arg
Leu Ile Glu Gly Ser Ser Phe Phe Lys Pro Pro Thr Val 165 170 175Lys
Gly Asp Asn Ser Ile Phe Thr Cys Val Ala Ile His Asn Ile Gly 180 185
190Arg Asp Ile Gly Ile Ala Gly Asp Tyr Phe Asp Lys Leu Glu Pro Lys
195 200 205Ile Glu Leu Thr Tyr Gln Leu Tyr Tyr Glu Tyr Asn Pro Lys
Lys Glu 210 215 220Ser Glu Ile Asn Lys Arg Leu Leu Tyr Ala Tyr Lys
Pro Lys Gln Asn225 230 235 240Lys Ile Ile Glu Ile Lys Glu Lys Leu
Trp Asn Leu Arg Lys Glu Lys 245 250 255Ser Pro Leu Asp Leu Glu Tyr
Glu Lys Pro Leu Thr Lys Ser Ile Thr 260 265 270Phe Leu Val Lys Arg
Asp Gly Val Phe Arg Ile Ser Lys Asp Leu Met 275 280 285Leu Arg Lys
Ala Lys Phe Ile Ile Gln Gly Lys Glu Lys Leu Ser Lys 290 295 300Glu
Glu Arg Lys Ile Asn Arg Asp Leu Ile Lys Ile Lys Ser Asn Ile305 310
315 320Ile Ser Leu Thr Tyr Gly Arg Phe Asp Glu Leu Lys Lys Asp Lys
Thr 325 330 335Ile Trp Ser Pro His Ile Phe Arg Asp Val Lys Gln Gly
Lys Ile Thr 340 345 350Pro Cys Ile Glu Arg Lys Gly Asp Arg Met Asp
Ile Phe Gln Gln Leu 355 360 365Arg Lys Lys Ser Glu Arg Leu Arg Glu
Asn Arg Lys Lys Arg Gln Lys 370 375 380Lys Ile Ser Lys Asp Leu Ile
Phe Ala Glu Arg Ile Ala Tyr Asn Phe385 390 395 400His Thr Lys Ser
Ile Lys Asn Thr Ser Asn Leu Ile Asn Ile Lys His 405 410 415Glu Ala
Lys Arg Gly Lys Ala Ser Tyr Met Arg Lys Arg Ile Gly Asn 420 425
430Glu Thr Phe Arg Ile Lys Tyr Cys Glu Gln Cys Phe Pro Lys Asn Asn
435 440 445Val Tyr Lys Asn Val Gln Lys Gly Cys Ser Cys Phe Glu Asp
Pro Phe 450 455 460Glu Tyr Ile Lys Lys Gly Asn Glu Asp Leu Ile Pro
Asn Lys Asn Gln465 470 475 480Asp Leu Lys Ala Lys Gly Ala Phe Arg
Asp Asp Ala Leu Glu Lys Gln 485
490 495Ile Ile Lys Val Ala Phe Asn Ile Ala Lys Gly Tyr Glu Asp Phe
Tyr 500 505 510Glu Asn Leu Lys Lys Thr Thr Glu Lys Asp Ile Arg Leu
Lys Phe Lys 515 520 525Val Gly Thr Ile Ile Ser Glu Glu Met 530
535183541PRTArtificial sequenceSynthetic sequence 183Asn Asn Ser
Ile Asn Leu Ser Lys Lys Ala Ile Asn Ile Ser Ala Asn1 5 10 15Tyr Asp
Ala Asn Leu Gln Val Arg Cys Lys Asn Cys Lys Phe Leu Ser 20 25 30Ser
Asn Gly Asn Phe Pro Arg Gln Thr Asp Val Lys Glu Gly Cys His 35 40
45Ser Cys Glu Lys Ser Thr Tyr Glu Pro Pro Val Tyr Asp Val Lys Ile
50 55 60Gly Glu Ile Lys Ala Lys Tyr Glu Val Leu Asp Ser Leu Lys Lys
Phe65 70 75 80Thr Phe Gln Ser Leu Lys Tyr Gln Leu Ser Lys Ser Met
Lys Phe Arg 85 90 95Ser Lys Lys Ile Lys Glu Leu Lys Glu Phe Val Ile
Phe Ala Lys Glu 100 105 110Ser Lys Ala Leu Asn Val Ile Asn Arg Ser
Tyr Lys His Leu Ile Leu 115 120 125Asn Ile Lys Asn Asp Ile Asn Arg
Met Asn Ser Lys Lys Arg Ile Lys 130 135 140Asn His Lys Gly Arg Leu
Phe Leu Asp Arg Gln Lys Gln Leu Ser Lys145 150 155 160Leu Lys Leu
Ile Glu Gly Ser Ser Phe Phe Val Pro Ala Lys Asn Val 165 170 175Gly
Asn Lys Ser Val Phe Thr Cys Val Ala Ile His Ser Ile Gly Arg 180 185
190Asp Ile Gly Ile Ala Gly Leu Tyr Asp Ser Phe Thr Lys Pro Val Asn
195 200 205Glu Ile Thr Tyr Gln Ile Phe Phe Ser Gly Glu Arg Arg Leu
Leu Tyr 210 215 220Ala Tyr Lys Pro Lys Gln Leu Lys Ile Leu Ser Ile
Lys Glu Asn Leu225 230 235 240Trp Ser Leu Lys Asn Glu Lys Lys Pro
Leu Asp Leu Leu Tyr Glu Lys 245 250 255Pro Leu Gly Lys Asn Leu Asn
Phe Asn Val Lys Gly Gly Asp Leu Phe 260 265 270Arg Val Ser Lys Asp
Leu Met Ile Arg Asn Ala Lys Phe Asn Val His 275 280 285Gly Arg Gln
Arg Leu Ser Asp Glu Glu Arg Leu Ile Asn Arg Asn Phe 290 295 300Ile
Lys Ile Lys Gly Glu Val Val Ser Leu Ser Tyr Gly Arg Phe Glu305 310
315 320Glu Leu Lys Lys Asp Arg Lys Leu Trp Ser Pro His Ile Phe Lys
Asp 325 330 335Val Arg Gln Asn Lys Ile Lys Pro Cys Leu Val Met Gln
Gly Gln Arg 340 345 350Ile Asp Ile Phe Glu Gln Leu Lys Arg Lys Leu
Glu Leu Leu Lys Lys 355 360 365Ile Arg Lys Ser Arg Gln Lys Lys Leu
Ser Lys Asp Leu Ile Phe Gly 370 375 380Glu Arg Ile Ala Tyr Asn Phe
His Thr Lys Ser Ile Lys Asn Thr Ser385 390 395 400Asn Lys Ile Asn
Ile Asp Ser Asp Ala Lys Arg Gly Arg Ala Ser Tyr 405 410 415Met Arg
Lys Arg Ile Gly Asn Glu Thr Phe Lys Leu Lys Tyr Cys Asp 420 425
430Val Cys Phe Pro Lys Ala Asn Val Tyr Arg Arg Val Gln Asn Gly Cys
435 440 445Ser Cys Ser Glu Asn Pro Tyr Asn Tyr Ile Lys Lys Gly Asp
Lys Asp 450 455 460Leu Leu Pro Lys Lys Asp Glu Gly Leu Ala Ile Lys
Gly Ala Phe Arg465 470 475 480Asp Glu Lys Leu Asn Lys Gln Ile Ile
Lys Val Ala Phe Asn Ile Ala 485 490 495Lys Gly Tyr Glu Asp Phe Tyr
Asp Asp Leu Lys Lys Arg Thr Glu Lys 500 505 510Asp Val Asp Leu Lys
Phe Lys Ile Gly Thr Thr Val Leu Asp Gln Lys 515 520 525Pro Met Glu
Ile Phe Asp Gly Ile Val Ile Thr Trp Leu 530 535
540184542PRTArtificial sequenceSynthetic sequence 184Leu Leu Thr
Thr Val Val Glu Thr Asn Asn Leu Ala Lys Lys Ala Ile1 5 10 15Asn Val
Ala Ala Asn Phe Asp Ala Asn Ile Asp Arg Gln Tyr Tyr Arg 20 25 30Cys
Thr Pro Asn Leu Cys Arg Phe Ile Ala Gln Ser Pro Arg Glu Thr 35 40
45Lys Glu Lys Asp Ala Gly Cys Ser Ser Cys Thr Gln Ser Thr Tyr Asp
50 55 60Pro Lys Val Tyr Val Ile Lys Ile Gly Lys Leu Leu Ala Lys Tyr
Glu65 70 75 80Ile Leu Lys Ser Leu Lys Arg Phe Leu Phe Met Asn Arg
Tyr Phe Lys 85 90 95Gln Lys Lys Thr Glu Arg Ala Gln Gln Lys Gln Lys
Ile Gly Thr Glu 100 105 110Leu Asn Glu Met Ser Ile Phe Ala Lys Ala
Thr Asn Ala Met Glu Val 115 120 125Ile Lys Arg Ala Thr Lys His Cys
Thr Tyr Asp Ile Ile Pro Glu Thr 130 135 140Lys Ser Leu Gln Met Leu
Lys Arg Arg Arg His Arg Val Lys Val Arg145 150 155 160Ser Leu Leu
Lys Ile Leu Lys Glu Arg Arg Met Lys Ile Lys Lys Ile 165 170 175Pro
Asn Thr Phe Ile Glu Ile Pro Lys Gln Ala Lys Lys Asn Lys Ser 180 185
190Asp Tyr Tyr Val Ala Ala Ala Leu Lys Ser Cys Gly Ile Asp Val Gly
195 200 205Leu Cys Gly Ala Tyr Glu Lys Asn Ala Glu Val Glu Ala Glu
Tyr Thr 210 215 220Tyr Gln Leu Tyr Tyr Glu Tyr Lys Gly Asn Ser Ser
Thr Lys Arg Ile225 230 235 240Leu Tyr Cys Tyr Asn Asn Pro Gln Lys
Asn Ile Arg Glu Phe Trp Glu 245 250 255Ala Phe Tyr Ile Gln Gly Ser
Lys Ser His Val Asn Thr Pro Gly Thr 260 265 270Ile Arg Leu Lys Met
Glu Lys Phe Leu Ser Pro Ile Thr Ile Glu Ser 275 280 285Glu Ala Leu
Asp Phe Arg Val Trp Asn Ser Asp Leu Lys Ile Arg Asn 290 295 300Gly
Gln Tyr Gly Phe Ile Lys Lys Arg Ser Leu Gly Lys Glu Ala Arg305 310
315 320Glu Ile Lys Lys Gly Met Gly Asp Ile Lys Arg Lys Ile Gly Asn
Leu 325 330 335Thr Tyr Gly Lys Ser Pro Ser Glu Leu Lys Ser Ile His
Val Tyr Arg 340 345 350Thr Glu Arg Glu Asn Pro Lys Lys Pro Arg Ala
Ala Arg Lys Lys Glu 355 360 365Asp Asn Phe Met Glu Ile Phe Glu Met
Gln Arg Lys Lys Asp Tyr Glu 370 375 380Val Asn Lys Lys Arg Arg Lys
Glu Ala Thr Asp Ala Ala Lys Ile Met385 390 395 400Asp Phe Ala Glu
Glu Pro Ile Arg His Tyr His Thr Asn Asn Leu Lys 405 410 415Ala Val
Arg Arg Ile Asp Met Asn Glu Gln Val Glu Arg Lys Lys Thr 420 425
430Ser Val Phe Leu Lys Arg Ile Met Gln Asn Gly Tyr Arg Gly Asn Tyr
435 440 445Cys Arg Lys Cys Ile Lys Ala Pro Glu Gly Ser Asn Arg Asp
Glu Asn 450 455 460Val Leu Glu Lys Asn Glu Gly Cys Leu Asp Cys Ile
Gly Ser Glu Phe465 470 475 480Ile Trp Lys Lys Ser Ser Lys Glu Lys
Lys Gly Leu Trp His Thr Asn 485 490 495Arg Leu Leu Arg Arg Ile Arg
Leu Gln Cys Phe Thr Thr Ala Lys Ala 500 505 510Tyr Glu Asn Phe Tyr
Asn Asp Leu Phe Glu Lys Lys Glu Ser Ser Leu 515 520 525Asp Ile Ile
Lys Leu Lys Val Ser Ile Thr Thr Lys Ser Met 530 535
540185564PRTArtificial sequenceSynthetic sequence 185Ala Ser Thr
Met Asn Leu Ala Lys Gln Ala Ile Asn Phe Ala Ala Asn1 5 10 15Tyr Asp
Ser Asn Leu Glu Ile Gly Cys Lys Gly Cys Lys Phe Met Ser 20 25 30Thr
Trp Ser Lys Lys Ser Asn Pro Lys Phe Tyr Pro Arg Gln Asn Asn 35 40
45Gln Ala Asn Lys Cys His Ser Cys Thr Tyr Ser Thr Gly Glu Pro Glu
50 55 60Val Pro Ile Ile Glu Ile Gly Glu Arg Ala Ala Lys Tyr Lys Ile
Phe65 70 75 80Thr Ala Leu Lys Lys Phe Val Phe Met Ser Val Ala Tyr
Lys Glu Arg 85 90 95Arg Arg Gln Arg Phe Lys Ser Lys Lys Pro Lys Glu
Leu Lys Glu Leu 100 105 110Ala Ile Cys Ser Asn Arg Glu Lys Ala Met
Glu Val Ile Gln Lys Ser 115 120 125Val Val His Cys Tyr Gly Asp Val
Lys Gln Glu Ile Pro Arg Ile Arg 130 135 140Lys Ile Lys Val Leu Lys
Asn His Lys Gly Arg Leu Phe Tyr Lys Gln145 150 155 160Lys Arg Ser
Lys Ile Lys Ile Ala Lys Leu Glu Lys Gly Ser Phe Phe 165 170 175Lys
Thr Phe Ile Pro Lys Val His Asn Asn Gly Cys His Ser Cys His 180 185
190Glu Ala Ser Leu Asn Lys Pro Ile Leu Val Thr Thr Ala Leu Asn Thr
195 200 205Ile Gly Ala Asp Ile Gly Leu Ile Asn Asp Tyr Ser Thr Ile
Ala Pro 210 215 220Thr Glu Thr Asp Ile Ser Trp Gln Val Tyr Tyr Glu
Phe Ile Pro Asn225 230 235 240Gly Asp Ser Glu Ala Val Lys Lys Arg
Leu Leu Tyr Phe Tyr Lys Pro 245 250 255Lys Gly Ala Leu Ile Lys Ser
Ile Arg Asp Lys Tyr Phe Lys Lys Gly 260 265 270His Glu Asn Ala Val
Asn Thr Gly Phe Phe Lys Tyr Gln Gly Lys Ile 275 280 285Val Lys Gly
Pro Ile Lys Phe Val Asn Asn Glu Leu Asp Phe Ala Arg 290 295 300Lys
Pro Asp Leu Lys Ser Met Lys Ile Lys Arg Ala Gly Phe Ala Ile305 310
315 320Pro Ser Ala Lys Arg Leu Ser Lys Glu Asp Arg Glu Ile Asn Arg
Glu 325 330 335Ser Ile Lys Ile Lys Asn Lys Ile Tyr Ser Leu Ser Tyr
Gly Arg Lys 340 345 350Lys Thr Leu Ser Asp Lys Asp Ile Ile Lys His
Leu Tyr Arg Pro Val 355 360 365Arg Gln Lys Gly Val Lys Pro Leu Glu
Tyr Arg Lys Ala Pro Asp Gly 370 375 380Phe Leu Glu Phe Phe Tyr Ser
Leu Lys Arg Lys Glu Arg Arg Leu Arg385 390 395 400Lys Gln Lys Glu
Lys Arg Gln Lys Asp Met Ser Glu Ile Ile Asp Ala 405 410 415Ala Asp
Glu Phe Ala Trp His Arg His Thr Gly Ser Ile Lys Lys Thr 420 425
430Thr Asn His Ile Asn Phe Lys Ser Glu Val Lys Arg Gly Lys Val Pro
435 440 445Ile Met Lys Lys Arg Ile Ala Asn Asp Ser Phe Asn Thr Arg
His Cys 450 455 460Gly Lys Cys Val Lys Gln Gly Asn Ala Ile Asn Lys
Tyr Tyr Ile Glu465 470 475 480Lys Gln Lys Asn Cys Phe Asp Cys Asn
Ser Ile Glu Phe Lys Trp Glu 485 490 495Lys Ala Ala Leu Glu Lys Lys
Gly Ala Phe Lys Leu Asn Lys Arg Leu 500 505 510Gln Tyr Ile Val Lys
Ala Cys Phe Asn Val Ala Lys Ala Tyr Glu Ser 515 520 525Phe Tyr Glu
Asp Phe Arg Lys Gly Glu Glu Glu Ser Leu Asp Leu Lys 530 535 540Phe
Lys Ile Gly Thr Thr Thr Thr Leu Lys Gln Tyr Pro Gln Asn Lys545 550
555 560Ala Arg Ala Met186610PRTArtificial sequenceSynthetic
sequence 186His Ser His Asn Leu Met Leu Thr Lys Leu Gly Lys Gln Ala
Ile Asn1 5 10 15Phe Ala Ala Asn Tyr Asp Ala Asn Leu Glu Ile Gly Cys
Lys Asn Cys 20 25 30Lys Phe Leu Ser Tyr Ser Pro Lys Gln Ala Asn Pro
Lys Lys Tyr Pro 35 40 45Arg Gln Thr Asp Val His Glu Asp Gly Asn Ile
Ala Cys His Ser Cys 50 55 60Met Gln Ser Thr Lys Glu Pro Pro Val Tyr
Ile Val Pro Ile Gly Glu65 70 75 80Arg Lys Ser Lys Tyr Glu Ile Leu
Thr Ser Leu Asn Lys Phe Thr Phe 85 90 95Leu Ala Leu Lys Tyr Lys Glu
Lys Lys Arg Gln Ala Phe Arg Ala Lys 100 105 110Lys Pro Lys Glu Leu
Gln Glu Leu Ala Ile Ala Phe Asn Lys Glu Lys 115 120 125Ala Ile Lys
Val Ile Asp Lys Ser Ile Gln His Leu Ile Leu Asn Ile 130 135 140Lys
Pro Glu Ile Ala Arg Ile Gln Arg Gln Lys Arg Leu Lys Asn Arg145 150
155 160Lys Gly Lys Leu Leu Tyr Leu His Lys Arg Tyr Ala Ile Lys Met
Gly 165 170 175Leu Ile Lys Asn Gly Lys Tyr Phe Lys Val Gly Ser Pro
Lys Lys Asp 180 185 190Gly Lys Lys Leu Leu Val Leu Cys Ala Leu Asn
Thr Ile Gly Arg Asp 195 200 205Ile Gly Ile Ile Gly Asn Ile Glu Glu
Asn Asn Arg Ser Glu Thr Glu 210 215 220Ile Thr Tyr Gln Leu Tyr Phe
Asp Cys Leu Asp Ala Asn Pro Asn Glu225 230 235 240Leu Arg Ile Lys
Glu Ile Glu Tyr Asn Arg Leu Lys Ser Tyr Glu Arg 245 250 255Lys Ile
Lys Arg Leu Val Tyr Ala Tyr Lys Pro Lys Gln Thr Lys Ile 260 265
270Leu Glu Ile Arg Ser Lys Phe Phe Ser Lys Gly His Glu Asn Lys Val
275 280 285Asn Thr Gly Ser Phe Asn Phe Glu Asn Pro Leu Asn Lys Ser
Ile Ser 290 295 300Ile Lys Val Lys Asn Ser Ala Phe Asp Phe Lys Ile
Gly Ala Pro Phe305 310 315 320Ile Met Leu Arg Asn Gly Lys Phe His
Ile Pro Thr Lys Lys Arg Leu 325 330 335Ser Lys Glu Glu Arg Glu Ile
Asn Arg Thr Leu Ser Lys Ile Lys Gly 340 345 350Arg Val Phe Arg Leu
Thr Tyr Gly Arg Asn Ile Ser Glu Gln Gly Ser 355 360 365Lys Ser Leu
His Ile Tyr Arg Lys Glu Arg Gln His Pro Lys Leu Ser 370 375 380Leu
Glu Ile Arg Lys Gln Pro Asp Ser Phe Ile Asp Glu Phe Glu Lys385 390
395 400Leu Arg Leu Lys Gln Asn Phe Ile Ser Lys Leu Lys Lys Gln Arg
Gln 405 410 415Lys Lys Leu Ala Asp Leu Leu Gln Phe Ala Asp Arg Ile
Ala Tyr Asn 420 425 430Tyr His Thr Ser Ser Leu Glu Lys Thr Ser Asn
Phe Ile Asn Tyr Lys 435 440 445Pro Glu Val Lys Arg Gly Arg Thr Ser
Tyr Ile Lys Lys Arg Ile Gly 450 455 460Asn Glu Gly Phe Glu Lys Leu
Tyr Cys Glu Thr Cys Ile Lys Ser Asn465 470 475 480Asp Lys Glu Asn
Ala Tyr Ala Val Glu Lys Glu Glu Leu Cys Phe Val 485 490 495Cys Lys
Ala Lys Pro Phe Thr Trp Lys Lys Thr Asn Lys Asp Lys Leu 500 505
510Gly Ile Phe Lys Tyr Pro Ser Arg Ile Lys Asp Phe Ile Arg Ala Ala
515 520 525Phe Thr Val Ala Lys Ser Tyr Asn Asp Phe Tyr Glu Asn Leu
Lys Lys 530 535 540Lys Asp Leu Lys Asn Glu Ile Phe Leu Lys Phe Lys
Ile Gly Leu Ile545 550 555 560Leu Ser His Glu Lys Lys Asn His Ile
Ser Ile Ala Lys Ser Val Ala 565 570 575Glu Asp Glu Arg Ile Ser Gly
Lys Ser Ile Lys Asn Ile Leu Asn Lys 580 585 590Ser Ile Lys Leu Glu
Lys Asn Cys Tyr Ser Cys Phe Phe His Lys Glu 595 600 605Asp Met
610187552PRTArtificial sequenceSynthetic sequence 187Ser Leu Glu
Arg Val Ile Asp Lys Arg Asn Leu Ala Lys Lys Ala Ile1 5 10 15Asn Ile
Ala Ala Asn Phe Asp Ala Asn Ile Asn Lys Gly Phe Tyr Arg 20 25 30Cys
Glu Thr Asn Gln Cys Met Phe Ile Ala Gln Lys Pro Arg Lys Thr 35 40
45Asn Asn Thr Gly Cys Ser Ser Cys Leu Gln Ser Thr Tyr Asp Pro Val
50 55 60Ile Tyr Val Val Lys Val Gly Glu Met Leu Ala Lys Tyr Glu Ile
Leu65 70 75 80Lys Ser Leu Lys Arg Phe Val Phe Met Asn Arg Ser Phe
Lys Gln Lys 85 90 95Lys Thr Glu Lys Ala Lys Gln Lys Glu Arg Ile Gly
Gly Glu Leu Asn 100 105 110Glu Met Ser Ile Phe Ala Asn Ala Ala Leu
Ala Met Gly Val Ile Lys 115 120 125Arg Ala Ile Arg His Cys His Val
Asp Ile Arg Pro Glu Ile Asn Arg 130 135
140Leu Ser Glu Leu Lys Lys Thr Lys His Arg Val Ala Ala Lys Ser
Leu145 150 155 160Val Lys Ile Val Lys Gln Arg Lys Thr Lys Trp Lys
Gly Ile Pro Asn 165 170 175Ser Phe Ile Gln Ile Pro Gln Lys Ala Arg
Asn Lys Asp Ala Asp Phe 180 185 190Tyr Val Ala Ser Ala Leu Lys Ser
Gly Gly Ile Asp Ile Gly Leu Cys 195 200 205Gly Thr Tyr Asp Lys Lys
Pro His Ala Asp Pro Arg Trp Thr Tyr Gln 210 215 220Leu Tyr Phe Asp
Thr Glu Asp Glu Ser Glu Lys Arg Leu Leu Tyr Cys225 230 235 240Tyr
Asn Asp Pro Gln Ala Lys Ile Arg Asp Phe Trp Lys Thr Phe Tyr 245 250
255Glu Arg Gly Asn Pro Ser Met Val Asn Ser Pro Gly Thr Ile Glu Phe
260 265 270Arg Met Glu Gly Phe Phe Glu Lys Met Thr Pro Ile Ser Ile
Glu Ser 275 280 285Lys Asp Phe Asp Phe Arg Val Trp Asn Lys Asp Leu
Leu Ile Arg Arg 290 295 300Gly Leu Tyr Glu Ile Lys Lys Arg Lys Asn
Leu Asn Arg Lys Ala Arg305 310 315 320Glu Ile Lys Lys Ala Met Gly
Ser Val Lys Arg Val Leu Ala Asn Met 325 330 335Thr Tyr Gly Lys Ser
Pro Thr Asp Lys Lys Ser Ile Pro Val Tyr Arg 340 345 350Val Glu Arg
Glu Lys Pro Lys Lys Pro Arg Ala Val Arg Lys Glu Glu 355 360 365Asn
Glu Leu Ala Asp Lys Leu Glu Asn Tyr Arg Arg Glu Asp Phe Leu 370 375
380Ile Arg Asn Arg Arg Lys Arg Glu Ala Thr Glu Ile Ala Lys Ile
Ile385 390 395 400Asp Ala Ala Glu Pro Pro Ile Arg His Tyr His Thr
Asn His Leu Arg 405 410 415Ala Val Lys Arg Ile Asp Leu Ser Lys Pro
Val Ala Arg Lys Asn Thr 420 425 430Ser Val Phe Leu Lys Arg Ile Met
Gln Asn Gly Tyr Arg Gly Asn Tyr 435 440 445Cys Lys Lys Cys Ile Lys
Gly Asn Ile Asp Pro Asn Lys Asp Glu Cys 450 455 460Arg Leu Glu Asp
Ile Lys Lys Cys Ile Cys Cys Glu Gly Thr Gln Asn465 470 475 480Ile
Trp Ala Lys Lys Glu Lys Leu Tyr Thr Gly Arg Ile Asn Val Leu 485 490
495Asn Lys Arg Ile Lys Gln Met Lys Leu Glu Cys Phe Asn Val Ala Lys
500 505 510Ala Tyr Glu Asn Phe Tyr Asp Asn Leu Ala Ala Leu Lys Glu
Gly Asp 515 520 525Leu Lys Val Leu Lys Leu Lys Val Ser Ile Pro Ala
Leu Asn Pro Glu 530 535 540Ala Ser Asp Pro Glu Glu Asp Met545
550188534PRTArtificial sequenceSynthetic sequence 188Asn Ala Ser
Ile Asn Leu Gly Lys Arg Ala Ile Asn Leu Ser Ala Asn1 5 10 15Tyr Asp
Ser Asn Leu Val Ile Gly Cys Lys Asn Cys Lys Phe Leu Ser 20 25 30Phe
Asn Gly Asn Phe Pro Arg Gln Thr Asn Val Arg Glu Gly Cys His 35 40
45Ser Cys Asp Lys Ser Thr Tyr Ala Pro Glu Val Tyr Ile Val Lys Ile
50 55 60Gly Glu Arg Lys Ala Lys Tyr Asp Val Leu Asp Ser Leu Lys Lys
Phe65 70 75 80Thr Phe Gln Ser Leu Lys Tyr Gln Ile Lys Lys Ser Met
Arg Glu Arg 85 90 95Ser Lys Lys Pro Lys Glu Leu Leu Glu Phe Val Ile
Phe Ala Asn Lys 100 105 110Asp Lys Ala Phe Asn Val Ile Gln Lys Ser
Tyr Glu His Leu Ile Leu 115 120 125Asn Ile Lys Gln Glu Ile Asn Arg
Met Asn Gly Lys Lys Arg Ile Lys 130 135 140Asn His Lys Lys Arg Leu
Phe Lys Asp Arg Glu Lys Gln Leu Asn Lys145 150 155 160Leu Arg Leu
Ile Gly Ser Ser Ser Leu Phe Phe Pro Arg Glu Asn Lys 165 170 175Gly
Asp Lys Asp Leu Phe Thr Tyr Val Ala Ile His Ser Val Gly Arg 180 185
190Asp Ile Gly Val Ala Gly Ser Tyr Glu Ser His Ile Glu Pro Ile Ser
195 200 205Asp Leu Thr Tyr Gln Leu Phe Ile Asn Asn Glu Lys Arg Leu
Leu Tyr 210 215 220Ala Tyr Lys Pro Lys Gln Asn Lys Ile Ile Glu Leu
Lys Glu Asn Leu225 230 235 240Trp Asn Leu Lys Lys Glu Lys Lys Pro
Leu Asp Leu Glu Phe Thr Lys 245 250 255Pro Leu Glu Lys Ser Ile Thr
Phe Ser Val Lys Asn Asp Lys Leu Phe 260 265 270Lys Val Ser Lys Asp
Leu Met Leu Arg Gln Ala Lys Phe Asn Ile Gln 275 280 285Gly Lys Glu
Lys Leu Ser Lys Glu Glu Arg Gln Ile Asn Arg Asp Phe 290 295 300Ser
Lys Ile Lys Ser Asn Val Ile Ser Leu Ser Tyr Gly Arg Phe Glu305 310
315 320Glu Leu Lys Lys Glu Lys Asn Ile Trp Ser Pro His Ile Tyr Arg
Glu 325 330 335Val Lys Gln Lys Glu Ile Lys Pro Cys Ile Val Arg Lys
Gly Asp Arg 340 345 350Ile Glu Leu Phe Glu Gln Leu Lys Arg Lys Met
Asp Lys Leu Lys Lys 355 360 365Phe Arg Lys Glu Arg Gln Lys Lys Ile
Ser Lys Asp Leu Asn Phe Ala 370 375 380Glu Arg Ile Ala Tyr Asn Phe
His Thr Lys Ser Ile Lys Asn Thr Ser385 390 395 400Asn Lys Ile Asn
Ile Asp Gln Glu Ala Lys Arg Gly Lys Ala Ser Tyr 405 410 415Met Arg
Lys Arg Ile Gly Asn Glu Ser Phe Arg Lys Lys Tyr Cys Glu 420 425
430Gln Cys Phe Ser Val Gly Asn Val Tyr His Asn Val Gln Asn Gly Cys
435 440 445Ser Cys Phe Asp Asn Pro Ile Glu Leu Ile Lys Lys Gly Asp
Glu Gly 450 455 460Leu Ile Pro Lys Gly Lys Glu Asp Arg Lys Tyr Lys
Gly Ala Leu Arg465 470 475 480Asp Asp Asn Leu Gln Met Gln Ile Ile
Arg Val Ala Phe Asn Ile Ala 485 490 495Lys Gly Tyr Glu Asp Phe Tyr
Asn Asn Leu Lys Glu Lys Thr Glu Lys 500 505 510Asp Leu Lys Leu Lys
Phe Lys Ile Gly Thr Thr Ile Ser Thr Gln Glu 515 520 525Ser Asn Asn
Lys Glu Met 530189577PRTArtificial sequenceSynthetic sequence
189Ser Asn Leu Ile Lys Leu Gly Lys Gln Ala Ile Asn Phe Ala Ala Asn1
5 10 15Tyr Asp Ala Asn Leu Glu Val Gly Cys Lys Asn Cys Lys Phe Leu
Ser 20 25 30Ser Thr Asn Lys Tyr Pro Arg Gln Thr Asn Val His Leu Asp
Asn Lys 35 40 45Met Ala Cys Arg Ser Cys Asn Gln Ser Thr Met Glu Pro
Ala Ile Tyr 50 55 60Ile Val Arg Ile Gly Glu Lys Lys Ala Lys Tyr Asp
Ile Tyr Asn Ser65 70 75 80Leu Thr Lys Phe Asn Phe Gln Ser Leu Lys
Tyr Lys Ala Lys Arg Ser 85 90 95Gln Arg Phe Lys Pro Lys Gln Pro Lys
Glu Leu Gln Glu Leu Ser Ile 100 105 110Ala Val Arg Lys Glu Lys Ala
Leu Asp Ile Ile Gln Lys Ser Ile Asp 115 120 125His Leu Ile Gln Asp
Ile Arg Pro Glu Ile Pro Arg Ile Lys Gln Gln 130 135 140Lys Arg Tyr
Lys Asn His Val Gly Lys Leu Phe Tyr Leu Gln Lys Arg145 150 155
160Arg Lys Asn Lys Leu Asn Leu Ile Gly Lys Gly Ser Phe Phe Lys Val
165 170 175Phe Ser Pro Lys Glu Lys Lys Asn Glu Leu Leu Val Ile Cys
Ala Leu 180 185 190Thr Asn Ile Gly Arg Asp Ile Gly Leu Ile Gly Asn
Tyr Asn Thr Ile 195 200 205Ile Asn Pro Leu Phe Glu Val Thr Tyr Gln
Leu Tyr Tyr Asp Tyr Ile 210 215 220Pro Lys Lys Asn Asn Lys Asn Val
Gln Arg Arg Leu Leu Tyr Ala Tyr225 230 235 240Lys Ser Lys Asn Glu
Lys Ile Leu Lys Leu Lys Glu Ala Phe Phe Lys 245 250 255Arg Gly His
Glu Asn Ala Val Asn Leu Gly Ser Phe Ser Tyr Glu Lys 260 265 270Pro
Leu Glu Lys Ser Leu Thr Leu Lys Ile Lys Asn Asp Lys Asp Asp 275 280
285Phe Gln Val Ser Pro Ser Leu Arg Ile Arg Thr Gly Arg Phe Phe Val
290 295 300Pro Ser Lys Arg Asn Leu Ser Arg Gln Glu Arg Glu Ile Asn
Arg Arg305 310 315 320Leu Val Lys Ile Lys Ser Lys Ile Lys Asn Met
Thr Tyr Gly Lys Phe 325 330 335Glu Thr Ala Arg Asp Lys Gln Ser Val
His Ile Phe Arg Leu Glu Arg 340 345 350Gln Lys Glu Lys Leu Pro Leu
Gln Phe Arg Lys Asp Glu Lys Glu Phe 355 360 365Met Glu Glu Phe Gln
Lys Leu Lys Arg Arg Thr Asn Ser Leu Lys Lys 370 375 380Leu Arg Lys
Ser Arg Gln Lys Lys Leu Ala Asp Leu Leu Gln Leu Ser385 390 395
400Glu Lys Val Val Tyr Asn Asn His Thr Gly Thr Leu Lys Lys Thr Ser
405 410 415Asn Phe Leu Asn Phe Ser Ser Ser Val Lys Arg Gly Lys Thr
Ala Tyr 420 425 430Ile Lys Glu Leu Leu Gly Gln Glu Gly Phe Glu Thr
Leu Tyr Cys Ser 435 440 445Asn Cys Ile Asn Lys Gly Gln Lys Thr Arg
Tyr Asn Ile Glu Thr Lys 450 455 460Glu Lys Cys Phe Ser Cys Lys Asp
Val Pro Phe Val Trp Lys Lys Lys465 470 475 480Ser Thr Asp Lys Asp
Arg Lys Gly Ala Phe Leu Phe Pro Ala Lys Leu 485 490 495Lys Asp Val
Ile Lys Ala Thr Phe Thr Val Ala Lys Ala Tyr Glu Asp 500 505 510Phe
Tyr Asp Asn Leu Lys Ser Ile Asp Glu Lys Lys Pro Tyr Ile Lys 515 520
525Phe Lys Ile Gly Leu Ile Leu Ala His Val Arg His Glu His Lys Ala
530 535 540Arg Ala Lys Glu Glu Ala Gly Gln Lys Asn Ile Tyr Asn Lys
Pro Ile545 550 555 560Lys Ile Asp Lys Asn Cys Lys Glu Cys Phe Phe
Phe Lys Glu Glu Ala 565 570 575Met190613PRTArtificial
sequenceSynthetic sequence 190Asn Thr Thr Arg Lys Lys Phe Arg Lys
Arg Thr Gly Phe Pro Gln Ser1 5 10 15Asp Asn Ile Lys Leu Ala Tyr Cys
Ser Ala Ile Val Arg Ala Ala Asn 20 25 30Leu Asp Ala Asp Ile Gln Lys
Lys His Asn Gln Cys Asn Pro Asn Leu 35 40 45Cys Val Gly Ile Lys Ser
Asn Glu Gln Ser Arg Lys Tyr Glu His Ser 50 55 60Asp Arg Gln Ala Leu
Leu Cys Tyr Ala Cys Asn Gln Ser Thr Gly Ala65 70 75 80Pro Lys Val
Asp Tyr Ile Gln Ile Gly Glu Ile Gly Ala Lys Tyr Lys 85 90 95Ile Leu
Gln Met Val Asn Ala Tyr Asp Phe Leu Ser Leu Ala Tyr Asn 100 105
110Leu Thr Lys Leu Arg Asn Gly Lys Ser Arg Gly His Gln Arg Met Ser
115 120 125Gln Leu Asp Glu Val Val Ile Val Ala Asp Tyr Glu Lys Ala
Thr Glu 130 135 140Val Ile Lys Arg Ser Ile Asn His Leu Leu Asp Asp
Ile Arg Gly Gln145 150 155 160Leu Ser Lys Leu Lys Lys Arg Thr Gln
Asn Glu His Ile Thr Glu His 165 170 175Lys Gln Ser Lys Ile Arg Arg
Lys Leu Arg Lys Leu Ser Arg Leu Leu 180 185 190Lys Arg Arg Arg Trp
Lys Trp Gly Thr Ile Pro Asn Pro Tyr Leu Lys 195 200 205Asn Trp Val
Phe Thr Lys Lys Asp Pro Glu Leu Val Thr Val Ala Leu 210 215 220Leu
His Lys Leu Gly Arg Asp Ile Gly Leu Val Asn Arg Ser Lys Arg225 230
235 240Arg Ser Lys Gln Lys Leu Leu Pro Lys Val Gly Phe Gln Leu Tyr
Tyr 245 250 255Lys Trp Glu Ser Pro Ser Leu Asn Asn Ile Lys Lys Ser
Lys Ala Lys 260 265 270Lys Leu Pro Lys Arg Leu Leu Ile Pro Tyr Lys
Asn Val Lys Leu Phe 275 280 285Asp Asn Lys Gln Lys Leu Glu Asn Ala
Ile Lys Ser Leu Leu Glu Ser 290 295 300Tyr Gln Lys Thr Ile Lys Val
Glu Phe Asp Gln Phe Phe Gln Asn Arg305 310 315 320Thr Glu Glu Ile
Ile Ala Glu Glu Gln Gln Thr Leu Glu Arg Gly Leu 325 330 335Leu Lys
Gln Leu Glu Lys Lys Lys Asn Glu Phe Ala Ser Gln Lys Lys 340 345
350Ala Leu Lys Glu Glu Lys Lys Lys Ile Lys Glu Pro Arg Lys Ala Lys
355 360 365Leu Leu Met Glu Glu Ser Arg Ser Leu Gly Phe Leu Met Ala
Asn Val 370 375 380Ser Tyr Ala Leu Phe Asn Thr Thr Ile Glu Asp Leu
Tyr Lys Lys Ser385 390 395 400Asn Val Val Ser Gly Cys Ile Pro Gln
Glu Pro Val Val Val Phe Pro 405 410 415Ala Asp Ile Gln Asn Lys Gly
Ser Leu Ala Lys Ile Leu Phe Ala Pro 420 425 430Lys Asp Gly Phe Arg
Ile Lys Phe Ser Gly Gln His Leu Thr Ile Arg 435 440 445Thr Ala Lys
Phe Lys Ile Arg Gly Lys Glu Ile Lys Ile Leu Thr Lys 450 455 460Thr
Lys Arg Glu Ile Leu Lys Asn Ile Glu Lys Leu Arg Arg Val Trp465 470
475 480Tyr Arg Glu Gln His Tyr Lys Leu Lys Leu Phe Gly Lys Glu Val
Ser 485 490 495Ala Lys Pro Arg Phe Leu Asp Lys Arg Lys Thr Ser Ile
Glu Arg Arg 500 505 510Asp Pro Asn Lys Leu Ala Asp Gln Thr Asp Asp
Arg Gln Ala Glu Leu 515 520 525Arg Asn Lys Glu Tyr Glu Leu Arg His
Lys Gln His Lys Met Ala Glu 530 535 540Arg Leu Asp Asn Ile Asp Thr
Asn Ala Gln Asn Leu Gln Thr Leu Ser545 550 555 560Phe Trp Val Gly
Glu Ala Asp Lys Pro Pro Lys Leu Asp Glu Lys Asp 565 570 575Ala Arg
Gly Phe Gly Val Arg Thr Cys Ile Ser Ala Trp Lys Trp Phe 580 585
590Met Glu Asp Leu Leu Lys Lys Gln Glu Glu Asp Pro Leu Leu Lys Leu
595 600 605Lys Leu Ser Ile Met 610191615PRTArtificial
sequenceSynthetic sequence 191Pro Lys Lys Pro Lys Phe Gln Lys Arg
Thr Gly Phe Pro Gln Pro Asp1 5 10 15Asn Leu Arg Lys Glu Tyr Cys Leu
Ala Ile Val Arg Ala Ala Asn Leu 20 25 30Asp Ala Asp Phe Glu Lys Lys
Cys Thr Lys Cys Glu Gly Ile Lys Thr 35 40 45Asn Lys Lys Gly Asn Ile
Val Lys Gly Arg Thr Tyr Asn Ser Ala Asp 50 55 60Lys Asp Asn Leu Leu
Cys Tyr Ala Cys Asn Ile Ser Thr Gly Ala Pro65 70 75 80Ala Val Asp
Tyr Val Phe Val Gly Ala Leu Glu Ala Lys Tyr Lys Ile 85 90 95Leu Gln
Met Val Lys Ala Tyr Asp Phe His Ser Leu Ala Tyr Asn Leu 100 105
110Ala Lys Leu Trp Lys Gly Arg Gly Arg Gly His Gln Arg Met Gly Gly
115 120 125Leu Asn Glu Val Val Ile Val Ser Asn Asn Glu Lys Ala Leu
Asp Val 130 135 140Ile Glu Lys Ser Leu Asn His Phe His Asp Glu Ile
Arg Gly Glu Leu145 150 155 160Ser Arg Leu Lys Ala Lys Phe Gln Asn
Glu His Leu His Val His Lys 165 170 175Glu Ser Lys Leu Arg Arg Lys
Leu Arg Lys Ile Ser Arg Leu Leu Lys 180 185 190Arg Arg Arg Trp Lys
Trp Asp Val Ile Pro Asn Ser Tyr Leu Arg Asn 195 200 205Phe Thr Phe
Thr Lys Thr Arg Pro Asp Phe Ile Ser Val Ala Leu Leu 210 215 220His
Arg Val Gly Arg Asp Ile Gly Leu Val Thr Lys Thr Lys Ile Pro225 230
235 240Lys Pro Thr Asp Leu Leu Pro Gln Phe Gly Phe Gln Ile Tyr Tyr
Thr 245 250 255Trp Asp Glu Pro Lys Leu Asn Lys Leu Lys Lys Ser Arg
Leu Arg Ser 260 265 270Glu Pro Lys Arg Leu Leu Val Pro Tyr Lys Lys
Ile Glu Leu Tyr Lys 275 280 285Asn Lys Ser Val Leu Glu Glu Ala Ile
Arg His Leu Ala Glu Val Tyr 290 295 300Thr Glu Asp Leu Thr Ile Cys
Phe Lys Asp Phe Phe Glu Thr Gln Lys305 310 315
320Arg Lys Phe Val Ser Lys Glu Lys Glu Ser Leu Lys Arg Glu Leu Leu
325 330 335Lys Glu Leu Thr Lys Leu Lys Lys Asp Phe Ser Glu Arg Lys
Thr Ala 340 345 350Leu Lys Arg Asp Arg Lys Glu Ile Lys Glu Pro Lys
Lys Ala Lys Leu 355 360 365Leu Met Glu Glu Ser Arg Ser Leu Gly Phe
Leu Ala Ala Asn Thr Ser 370 375 380Tyr Ala Leu Phe Asn Leu Ile Ala
Ala Asp Leu Tyr Thr Lys Ser Lys385 390 395 400Lys Ala Cys Ser Thr
Lys Leu Pro Arg Gln Leu Ser Thr Ile Leu Pro 405 410 415Leu Glu Ile
Lys Glu His Lys Ser Thr Thr Ser Leu Ala Ile Lys Pro 420 425 430Glu
Glu Gly Phe Lys Ile Arg Phe Ser Asn Thr His Leu Ser Ile Arg 435 440
445Thr Pro Lys Phe Lys Met Lys Gly Ala Asp Ile Lys Ala Leu Thr Lys
450 455 460Arg Lys Arg Glu Ile Leu Lys Asn Ala Thr Lys Leu Glu Lys
Ser Trp465 470 475 480Tyr Gly Leu Lys His Tyr Lys Leu Lys Leu Tyr
Gly Lys Glu Val Ala 485 490 495Ala Lys Pro Arg Phe Leu Asp Lys Arg
Asn Pro Ser Ile Asp Arg Arg 500 505 510Asp Pro Lys Glu Leu Met Glu
Gln Ile Glu Asn Arg Arg Asn Glu Val 515 520 525Lys Asp Leu Glu Tyr
Glu Ile Arg Lys Gly Gln His Gln Met Ala Lys 530 535 540Arg Leu Asp
Asn Val Asp Thr Asn Ala Gln Asn Leu Gln Thr Lys Ser545 550 555
560Phe Trp Val Gly Glu Ala Asp Lys Pro Pro Glu Leu Asp Ser Met Glu
565 570 575Ala Lys Lys Leu Gly Leu Arg Thr Cys Ile Ser Ala Trp Lys
Trp Phe 580 585 590Met Lys Asp Leu Val Leu Leu Gln Glu Lys Ser Pro
Asn Leu Lys Leu 595 600 605Lys Leu Ser Leu Thr Glu Met 610
615192775PRTArtificial sequenceSynthetic sequence 192Lys Phe Ser
Lys Arg Gln Glu Gly Phe Leu Ile Pro Asp Asn Ile Asp1 5 10 15Leu Tyr
Lys Cys Leu Ala Ile Val Arg Ser Ala Asn Leu Asp Ala Asp 20 25 30Val
Gln Gly His Lys Ser Cys Tyr Gly Val Lys Lys Asn Gly Thr Tyr 35 40
45Arg Val Lys Gln Asn Gly Lys Lys Gly Val Lys Glu Lys Gly Arg Lys
50 55 60Tyr Val Phe Asp Leu Ile Ala Phe Lys Gly Asn Ile Glu Lys Ile
Pro65 70 75 80His Glu Ala Ile Glu Glu Lys Asp Gln Gly Arg Val Ile
Val Leu Gly 85 90 95Lys Phe Asn Tyr Lys Leu Ile Leu Asn Ile Glu Lys
Asn His Asn Asp 100 105 110Arg Ala Ser Leu Glu Ile Lys Asn Lys Ile
Lys Lys Leu Val Gln Ile 115 120 125Ser Ser Leu Glu Thr Gly Glu Phe
Leu Ser Asp Leu Leu Ser Gly Lys 130 135 140Ile Gly Ile Asp Glu Val
Tyr Gly Ile Ile Glu Pro Asp Val Phe Ser145 150 155 160Gly Lys Glu
Leu Val Cys Lys Ala Cys Gln Gln Ser Thr Tyr Ala Pro 165 170 175Leu
Val Glu Tyr Met Pro Val Gly Glu Leu Asp Ala Lys Tyr Lys Ile 180 185
190Leu Ser Ala Ile Lys Gly Tyr Asp Phe Leu Ser Leu Ala Tyr Asn Leu
195 200 205Ser Arg Asn Arg Ala Asn Lys Lys Arg Gly His Gln Lys Leu
Gly Gly 210 215 220Gly Glu Leu Ser Glu Val Val Ile Ser Ala Asn Tyr
Asp Lys Ala Leu225 230 235 240Asn Val Ile Lys Arg Ser Ile Asn His
Tyr His Val Glu Ile Lys Pro 245 250 255Glu Ile Ser Lys Leu Lys Lys
Lys Met Gln Asn Glu Pro Leu Lys Val 260 265 270Met Lys Gln Ala Arg
Ile Arg Arg Glu Leu His Gln Leu Ser Arg Lys 275 280 285Val Lys Arg
Leu Lys Trp Lys Trp Gly Met Ile Pro Asn Pro Glu Leu 290 295 300Gln
Asn Ile Ile Phe Glu Lys Lys Glu Lys Asp Phe Val Ser Tyr Ala305 310
315 320Leu Leu His Thr Leu Gly Arg Asp Ile Gly Leu Phe Lys Asp Thr
Ser 325 330 335Met Leu Gln Val Pro Asn Ile Ser Asp Tyr Gly Phe Gln
Ile Tyr Tyr 340 345 350Ser Trp Glu Asp Pro Lys Leu Asn Ser Ile Lys
Lys Ile Lys Asp Leu 355 360 365Pro Lys Arg Leu Leu Ile Pro Tyr Lys
Arg Leu Asp Phe Tyr Ile Asp 370 375 380Thr Ile Leu Val Ala Lys Val
Ile Lys Asn Leu Ile Glu Leu Tyr Arg385 390 395 400Lys Ser Tyr Val
Tyr Glu Thr Phe Gly Glu Glu Tyr Gly Tyr Ala Lys 405 410 415Lys Ala
Glu Asp Ile Leu Phe Asp Trp Asp Ser Ile Asn Leu Ser Glu 420 425
430Gly Ile Glu Gln Lys Ile Gln Lys Ile Lys Asp Glu Phe Ser Asp Leu
435 440 445Leu Tyr Glu Ala Arg Glu Ser Lys Arg Gln Asn Phe Val Glu
Ser Phe 450 455 460Glu Asn Ile Leu Gly Leu Tyr Asp Lys Asn Phe Ala
Ser Asp Arg Asn465 470 475 480Ser Tyr Gln Glu Lys Ile Gln Ser Met
Ile Ile Lys Lys Gln Gln Glu 485 490 495Asn Ile Glu Gln Lys Leu Lys
Arg Glu Phe Lys Glu Val Ile Glu Arg 500 505 510Gly Phe Glu Gly Met
Asp Gln Asn Lys Lys Tyr Tyr Lys Val Leu Ser 515 520 525Pro Asn Ile
Lys Gly Gly Leu Leu Tyr Thr Asp Thr Asn Asn Leu Gly 530 535 540Phe
Phe Arg Ser His Leu Ala Phe Met Leu Leu Ser Lys Ile Ser Asp545 550
555 560Asp Leu Tyr Arg Lys Asn Asn Leu Val Ser Lys Gly Gly Asn Lys
Gly 565 570 575Ile Leu Asp Gln Thr Pro Glu Thr Met Leu Thr Leu Glu
Phe Gly Lys 580 585 590Ser Asn Leu Pro Asn Ile Ser Ile Lys Arg Lys
Phe Phe Asn Ile Lys 595 600 605Tyr Asn Ser Ser Trp Ile Gly Ile Arg
Lys Pro Lys Phe Ser Ile Lys 610 615 620Gly Ala Val Ile Arg Glu Ile
Thr Lys Lys Val Arg Asp Glu Gln Arg625 630 635 640Leu Ile Lys Ser
Leu Glu Gly Val Trp His Lys Ser Thr His Phe Lys 645 650 655Arg Trp
Gly Lys Pro Arg Phe Asn Leu Pro Arg His Pro Asp Arg Glu 660 665
670Lys Asn Asn Asp Asp Asn Leu Met Glu Ser Ile Thr Ser Arg Arg Glu
675 680 685Gln Ile Gln Leu Leu Leu Arg Glu Lys Gln Lys Gln Gln Glu
Lys Met 690 695 700Ala Gly Arg Leu Asp Lys Ile Asp Lys Glu Ile Gln
Asn Leu Gln Thr705 710 715 720Ala Asn Phe Gln Ile Lys Gln Ile Asp
Lys Lys Pro Ala Leu Thr Glu 725 730 735Lys Ser Glu Gly Lys Gln Ser
Val Arg Asn Ala Leu Ser Ala Trp Lys 740 745 750Trp Phe Met Glu Asp
Leu Ile Lys Tyr Gln Lys Arg Thr Pro Ile Leu 755 760 765Gln Leu Lys
Leu Ala Lys Met 770 775193777PRTArtificial sequenceSynthetic
sequence 193Lys Phe Ser Lys Arg Gln Glu Gly Phe Val Ile Pro Glu Asn
Ile Gly1 5 10 15Leu Tyr Lys Cys Leu Ala Ile Val Arg Ser Ala Asn Leu
Asp Ala Asp 20 25 30Val Gln Gly His Val Ser Cys Tyr Gly Val Lys Lys
Asn Gly Thr Tyr 35 40 45Val Leu Lys Gln Asn Gly Lys Lys Ser Ile Arg
Glu Lys Gly Arg Lys 50 55 60Tyr Ala Ser Asp Leu Val Ala Phe Lys Gly
Asp Ile Glu Lys Ile Pro65 70 75 80Phe Glu Val Ile Glu Glu Lys Lys
Lys Glu Gln Ser Ile Val Leu Gly 85 90 95Lys Phe Asn Tyr Lys Leu Val
Leu Asp Val Met Lys Gly Glu Lys Asp 100 105 110Arg Ala Ser Leu Thr
Met Lys Asn Lys Ser Lys Lys Leu Val Gln Val 115 120 125Ser Ser Leu
Gly Thr Asp Glu Phe Leu Leu Thr Leu Leu Asn Glu Lys 130 135 140Phe
Gly Ile Glu Glu Ile Tyr Gly Ile Ile Glu Pro Glu Val Phe Ser145 150
155 160Gly Lys Lys Leu Val Cys Lys Ala Cys Gln Gln Ser Thr Tyr Ala
Pro 165 170 175Leu Val Glu Tyr Met Pro Val Gly Glu Leu Asp Ser Lys
Tyr Lys Ile 180 185 190Leu Ser Ala Ile Lys Gly Tyr Asp Phe Leu Ser
Leu Ala Tyr Asn Leu 195 200 205Ala Arg His Arg Ser Asn Lys Lys Arg
Gly His Gln Lys Leu Gly Gly 210 215 220Gly Glu Leu Ser Glu Val Val
Ile Ser Ala Asn Asn Ala Lys Ala Leu225 230 235 240Asn Val Ile Lys
Arg Ser Leu Asn His Tyr Tyr Ser Glu Ile Lys Pro 245 250 255Glu Ile
Ser Lys Leu Arg Lys Lys Met Gln Asn Glu Pro Leu Lys Val 260 265
270Gly Lys Gln Ala Arg Met Arg Arg Glu Leu His Gln Leu Ser Arg Lys
275 280 285Val Lys Arg Leu Lys Trp Lys Trp Gly Lys Ile Pro Asn Leu
Glu Leu 290 295 300Gln Asn Ile Thr Phe Lys Glu Ser Asp Arg Asp Phe
Ile Ser Tyr Ala305 310 315 320Leu Leu His Thr Leu Gly Arg Asp Ile
Gly Met Phe Asn Lys Thr Glu 325 330 335Ile Lys Met Pro Ser Asn Ile
Leu Gly Tyr Gly Phe Gln Ile Tyr Tyr 340 345 350Asp Trp Glu Glu Pro
Lys Leu Asn Thr Ile Lys Lys Ser Lys Asn Thr 355 360 365Pro Lys Arg
Ile Leu Ile Pro Tyr Lys Lys Leu Asp Phe Tyr Asn Asp 370 375 380Ser
Ile Leu Val Ala Arg Ala Ile Lys Glu Leu Val Gly Leu Phe Gln385 390
395 400Glu Ser Tyr Glu Trp Glu Ile Phe Gly Asn Glu Tyr Asn Tyr Ala
Lys 405 410 415Glu Ala Glu Val Glu Leu Ile Lys Leu Asp Glu Glu Ser
Ile Asn Gly 420 425 430Asn Val Glu Lys Lys Leu Gln Arg Ile Lys Glu
Asn Phe Ser Asn Leu 435 440 445Leu Glu Lys Ala Arg Glu Lys Lys Arg
Gln Asn Phe Ile Glu Ser Phe 450 455 460Glu Ser Ile Ala Arg Leu Tyr
Asp Glu Ser Phe Thr Ala Asp Arg Asn465 470 475 480Glu Tyr Gln Arg
Glu Ile Gln Ser Phe Ile Ile Glu Lys Gln Lys Gln 485 490 495Ser Ile
Glu Lys Lys Leu Lys Asn Glu Phe Lys Lys Ile Val Glu Lys 500 505
510Lys Phe Asn Glu Gln Glu Gln Gly Lys Lys His Tyr Arg Val Leu Asn
515 520 525Pro Thr Ile Ile Asn Glu Phe Leu Pro Lys Asp Lys Asn Asn
Leu Gly 530 535 540Phe Leu Arg Ser Lys Ile Ala Phe Ile Leu Leu Ser
Lys Ile Ser Asp545 550 555 560Asp Leu Tyr Lys Lys Ser Asn Ala Val
Ser Lys Gly Gly Glu Lys Gly 565 570 575Ile Ile Lys Gln Gln Pro Glu
Thr Ile Leu Asp Leu Glu Phe Ser Lys 580 585 590Ser Lys Leu Pro Ser
Ile Asn Ile Lys Lys Lys Leu Phe Asn Ile Lys 595 600 605Tyr Thr Ser
Ser Trp Leu Gly Ile Arg Lys Pro Lys Phe Asn Ile Lys 610 615 620Gly
Ala Lys Ile Arg Glu Ile Thr Arg Arg Val Arg Asp Val Gln Arg625 630
635 640Thr Leu Lys Ser Ala Glu Ser Ser Trp Tyr Ala Ser Thr His Phe
Arg 645 650 655Arg Trp Gly Phe Pro Arg Phe Asn Gln Pro Arg His Pro
Asp Lys Glu 660 665 670Lys Lys Ser Asp Asp Arg Leu Ile Glu Ser Ile
Thr Leu Leu Arg Glu 675 680 685Gln Ile Gln Ile Leu Leu Arg Glu Lys
Gln Lys Gly Gln Lys Glu Met 690 695 700Ala Gly Arg Leu Asp Asp Val
Asp Lys Lys Ile Gln Asn Leu Gln Thr705 710 715 720Ala Asn Phe Gln
Ile Lys Gln Thr Gly Asp Lys Pro Ala Leu Thr Glu 725 730 735Lys Ser
Ala Gly Lys Gln Ser Phe Arg Asn Ala Leu Ser Ala Trp Lys 740 745
750Trp Phe Met Glu Asn Leu Leu Lys Tyr Gln Asn Lys Thr Pro Asp Leu
755 760 765Lys Leu Lys Ile Ala Arg Thr Val Met 770
775194610PRTArtificial sequenceSynthetic sequence 194Lys Trp Ile
Glu Pro Asn Asn Ile Asp Phe Asn Lys Cys Leu Ala Ile1 5 10 15Thr Arg
Ser Ala Asn Leu Asp Ala Asp Val Gln Gly His Lys Met Cys 20 25 30Tyr
Gly Ile Lys Thr Asn Gly Thr Tyr Lys Ala Ile Gly Lys Ile Asn 35 40
45Lys Lys His Asn Thr Gly Ile Ile Glu Lys Arg Arg Thr Tyr Val Tyr
50 55 60Asp Leu Ile Val Thr Lys Glu Lys Asn Glu Lys Ile Val Lys Lys
Thr65 70 75 80Asp Phe Met Ala Ile Asp Glu Glu Ile Glu Phe Asp Glu
Lys Lys Glu 85 90 95Lys Leu Leu Lys Lys Tyr Ile Lys Ala Glu Val Leu
Gly Thr Gly Glu 100 105 110Leu Ile Arg Lys Asp Leu Asn Asp Gly Glu
Lys Phe Asp Asp Leu Cys 115 120 125Ser Ile Glu Glu Pro Gln Ala Phe
Arg Arg Ser Glu Leu Val Cys Lys 130 135 140Ala Cys Asn Gln Ser Thr
Tyr Ala Ser Asp Ile Arg Tyr Ile Pro Ile145 150 155 160Gly Glu Ile
Glu Ala Lys Tyr Lys Ile Leu Lys Ala Ile Lys Gly Tyr 165 170 175Asp
Phe Leu Ser Leu Lys Tyr Asn Leu Gly Arg Leu Arg Asp Ser Lys 180 185
190Lys Arg Gly His Gln Lys Met Gly Gln Gly Glu Leu Lys Glu Phe Val
195 200 205Ile Cys Ala Asn Lys Glu Lys Ala Leu Asp Val Ile Lys Arg
Ser Leu 210 215 220Asn His Tyr Leu Asn Glu Val Lys Asp Glu Ile Ser
Arg Leu Asn Lys225 230 235 240Lys Met Gln Asn Glu Pro Leu Lys Val
Asn Asp Gln Ala Arg Trp Arg 245 250 255Arg Glu Leu Asn Gln Ile Ser
Arg Arg Leu Lys Arg Leu Lys Trp Lys 260 265 270Trp Gly Glu Ile Pro
Asn Pro Glu Leu Lys Asn Leu Ile Phe Lys Ser 275 280 285Ser Arg Pro
Glu Phe Val Ser Tyr Ala Leu Ile His Thr Leu Gly Arg 290 295 300Asp
Ile Gly Leu Ile Asn Glu Thr Glu Leu Lys Pro Asn Asn Ile Gln305 310
315 320Glu Tyr Gly Phe Gln Ile Tyr Tyr Lys Trp Glu Asp Pro Glu Leu
Asn 325 330 335His Ile Lys Lys Val Lys Asn Ile Pro Lys Arg Phe Ile
Ile Pro Tyr 340 345 350Lys Asn Leu Asp Leu Phe Gly Lys Tyr Thr Ile
Leu Ser Arg Ala Ile 355 360 365Glu Gly Ile Leu Lys Leu Tyr Ser Ser
Ser Phe Gln Tyr Lys Ser Phe 370 375 380Lys Asp Pro Asn Leu Phe Ala
Lys Glu Gly Glu Lys Lys Ile Thr Asn385 390 395 400Glu Asp Phe Glu
Leu Gly Tyr Asp Glu Lys Ile Lys Lys Ile Lys Asp 405 410 415Asp Phe
Lys Ser Tyr Lys Lys Ala Leu Leu Glu Lys Lys Lys Asn Thr 420 425
430Leu Glu Asp Ser Leu Asn Ser Ile Leu Ser Val Tyr Glu Gln Ser Leu
435 440 445Leu Thr Glu Gln Ile Asn Asn Val Lys Lys Trp Lys Glu Gly
Leu Leu 450 455 460Lys Ser Lys Glu Ser Ile His Lys Gln Lys Lys Ile
Glu Asn Ile Glu465 470 475 480Asp Ile Ile Ser Arg Ile Glu Glu Leu
Lys Asn Val Glu Gly Trp Ile 485 490 495Arg Thr Lys Glu Arg Asp Ile
Val Asn Lys Glu Glu Thr Asn Leu Lys 500 505 510Arg Glu Ile Lys Lys
Glu Leu Lys Asp Ser Tyr Tyr Glu Glu Val Arg 515 520 525Lys Asp Phe
Ser Asp Leu Lys Lys Gly Glu Glu Ser Glu Lys Lys Pro 530 535 540Phe
Arg Glu Glu Pro Lys Pro Ile Val Ile Lys Asp Tyr Ile Lys Phe545 550
555 560Asp Val Leu Pro Gly Glu Asn Ser Ala Leu Gly Phe Phe Leu Ser
His 565 570 575Leu Ser Phe Asn Leu Phe Asp Ser Ile Gln Tyr Glu Leu
Phe Glu Lys 580 585 590Ser Arg Leu Ser Ser Ser Lys His Pro Gln Ile
Pro Glu Thr Ile Leu 595 600 605Asp
Leu 610195632PRTArtificial sequenceSynthetic sequence 195Phe Arg
Lys Phe Val Lys Arg Ser Gly Ala Pro Gln Pro Asp Asn Leu1 5 10 15Asn
Lys Tyr Lys Cys Ile Ala Ile Val Arg Ala Ala Asn Leu Asp Ala 20 25
30Asp Ile Met Ser Asn Glu Ser Ser Asn Cys Val Met Cys Lys Gly Ile
35 40 45Lys Met Asn Lys Arg Lys Thr Ala Lys Gly Ala Ala Lys Thr Thr
Glu 50 55 60Leu Gly Arg Val Tyr Ala Gly Gln Ser Gly Asn Leu Leu Cys
Thr Ala65 70 75 80Cys Thr Lys Ser Thr Met Gly Pro Leu Val Asp Tyr
Val Pro Ile Gly 85 90 95Arg Ile Arg Ala Lys Tyr Thr Ile Leu Arg Ala
Val Lys Glu Tyr Asp 100 105 110Phe Leu Ser Leu Ala Tyr Asn Leu Ala
Arg Thr Arg Val Ser Lys Lys 115 120 125Gly Gly Arg Gln Lys Met His
Ser Leu Ser Glu Leu Val Ile Ala Ala 130 135 140Glu Tyr Glu Ile Ala
Trp Asn Ile Ile Lys Ser Ser Val Ile His Tyr145 150 155 160His Gln
Glu Thr Lys Glu Glu Ile Ser Gly Leu Arg Lys Lys Leu Gln 165 170
175Ala Glu His Ile His Lys Asn Lys Glu Ala Arg Ile Arg Arg Glu Met
180 185 190His Gln Ile Ser Arg Arg Ile Lys Arg Leu Lys Trp Lys Trp
His Met 195 200 205Ile Pro Asn Ser Glu Leu His Asn Phe Leu Phe Lys
Gln Gln Asp Pro 210 215 220Ser Phe Val Ala Val Ala Leu Leu His Thr
Leu Gly Arg Asp Ile Gly225 230 235 240Met Ile Asn Lys Pro Lys Gly
Ser Ala Lys Arg Glu Phe Ile Pro Glu 245 250 255Tyr Gly Phe Gln Ile
Tyr Tyr Lys Trp Met Asn Pro Lys Leu Asn Asp 260 265 270Ile Asn Lys
Gln Lys Tyr Arg Lys Met Pro Lys Arg Ser Leu Ile Pro 275 280 285Tyr
Lys Asn Leu Asn Val Phe Gly Asp Arg Glu Leu Ile Glu Asn Ala 290 295
300Met His Lys Leu Leu Lys Leu Tyr Asp Glu Asn Leu Glu Val Lys
Gly305 310 315 320Ser Lys Phe Phe Lys Thr Arg Val Val Ala Ile Ser
Ser Lys Glu Ser 325 330 335Glu Lys Leu Lys Arg Asp Leu Leu Trp Lys
Gly Glu Leu Ala Lys Ile 340 345 350Lys Lys Asp Phe Asn Ala Asp Lys
Asn Lys Met Gln Glu Leu Phe Lys 355 360 365Glu Val Lys Glu Pro Lys
Lys Ala Asn Ala Leu Met Lys Gln Ser Arg 370 375 380Asn Met Gly Phe
Leu Leu Gln Asn Ile Ser Tyr Gly Ala Leu Gly Leu385 390 395 400Leu
Ala Asn Arg Met Tyr Glu Ala Ser Ala Lys Gln Ser Lys Gly Asp 405 410
415Ala Thr Lys Gln Pro Ser Ile Val Ile Pro Leu Glu Met Glu Phe Gly
420 425 430Asn Ala Phe Pro Lys Leu Leu Leu Arg Ser Gly Lys Phe Ala
Met Asn 435 440 445Val Ser Ser Pro Trp Leu Thr Ile Arg Lys Pro Lys
Phe Val Ile Lys 450 455 460Gly Asn Lys Ile Lys Asn Ile Thr Lys Leu
Met Lys Asp Glu Lys Ala465 470 475 480Lys Leu Lys Arg Leu Glu Thr
Ser Tyr His Arg Ala Thr His Phe Arg 485 490 495Pro Thr Leu Arg Gly
Ser Ile Asp Trp Asp Ser Pro Tyr Phe Ser Ser 500 505 510Pro Lys Gln
Pro Asn Thr His Arg Arg Ser Pro Asp Arg Leu Ser Ala 515 520 525Asp
Ile Thr Glu Tyr Arg Gly Arg Leu Lys Ser Val Glu Ala Glu Leu 530 535
540Arg Glu Gly Gln Arg Ala Met Ala Lys Lys Leu Asp Ser Val Asp
Met545 550 555 560Thr Ala Ser Asn Leu Gln Thr Ser Asn Phe Gln Leu
Glu Lys Gly Glu 565 570 575Asp Pro Arg Leu Thr Glu Ile Asp Glu Lys
Gly Arg Ser Ile Arg Asn 580 585 590Cys Ile Ser Ser Trp Lys Lys Phe
Met Glu Asp Leu Met Lys Ala Gln 595 600 605Glu Ala Asn Pro Val Ile
Lys Ile Lys Ile Ala Leu Lys Asp Glu Ser 610 615 620Ser Val Leu Ser
Glu Asp Ser Met625 630196625PRTArtificial sequenceSynthetic
sequence 196Lys Phe His Pro Glu Asn Leu Asn Lys Ser Tyr Cys Leu Ala
Ile Val1 5 10 15Arg Ala Ala Asn Leu Asp Ala Asp Ile Gln Gly His Ile
Asn Cys Ile 20 25 30Gly Ile Lys Ser Asn Lys Ser Asp Arg Asn Tyr Glu
Asn Lys Leu Glu 35 40 45Ser Leu Gln Asn Val Glu Leu Leu Cys Lys Ala
Cys Thr Lys Ser Thr 50 55 60Tyr Lys Pro Asn Ile Asn Ser Val Pro Val
Gly Glu Lys Lys Ala Lys65 70 75 80Tyr Ser Ile Leu Ser Glu Ile Lys
Lys Tyr Asp Phe Asn Ser Leu Val 85 90 95Tyr Asn Leu Lys Lys Tyr Arg
Lys Gly Lys Ser Arg Gly His Gln Lys 100 105 110Leu Asn Glu Leu Arg
Glu Leu Val Ile Thr Ser Glu Tyr Lys Lys Ala 115 120 125Leu Asp Val
Ile Asn Lys Ser Val Asn His Tyr Leu Val Asn Ile Lys 130 135 140Asn
Lys Met Ser Lys Leu Lys Lys Ile Leu Gln Asn Glu His Ile His145 150
155 160Val Gly Thr Leu Ala Arg Ile Arg Arg Glu Arg Asn Arg Ile Ser
Arg 165 170 175Lys Leu Asp His Tyr Arg Lys Lys Trp Lys Phe Val Pro
Asn Lys Ile 180 185 190Leu Lys Asn Tyr Val Phe Lys Asn Gln Ser Pro
Asp Phe Val Ser Val 195 200 205Ala Leu Leu His Lys Leu Gly Arg Asp
Ile Gly Leu Ile Thr Lys Thr 210 215 220Ala Ile Leu Gln Lys Ser Phe
Pro Glu Tyr Ser Leu Gln Leu Tyr Tyr225 230 235 240Lys Tyr Asp Thr
Pro Lys Leu Asn Tyr Leu Lys Lys Ser Lys Phe Lys 245 250 255Ser Leu
Pro Lys Arg Ile Leu Ile Ser Tyr Lys Tyr Pro Lys Phe Asp 260 265
270Ile Asn Ser Asn Tyr Ile Glu Glu Ser Ile Asp Lys Leu Leu Lys Leu
275 280 285Tyr Glu Glu Ser Pro Ile Tyr Lys Asn Asn Ser Lys Ile Ile
Glu Phe 290 295 300Phe Lys Lys Ser Glu Asp Asn Leu Ile Lys Ser Glu
Asn Asp Ser Leu305 310 315 320Lys Arg Gly Ile Met Lys Glu Phe Glu
Lys Val Thr Lys Asn Phe Ser 325 330 335Ser Lys Lys Lys Lys Leu Lys
Glu Glu Leu Lys Leu Lys Asn Glu Asp 340 345 350Lys Asn Ser Lys Met
Leu Ala Lys Val Ser Arg Pro Ile Gly Phe Leu 355 360 365Lys Ala Tyr
Leu Ser Tyr Met Leu Phe Asn Ile Ile Ser Asn Arg Ile 370 375 380Phe
Glu Phe Ser Arg Lys Ser Ser Gly Arg Ile Pro Gln Leu Pro Ser385 390
395 400Cys Ile Ile Asn Leu Gly Asn Gln Phe Glu Asn Phe Lys Asn Glu
Leu 405 410 415Gln Asp Ser Asn Ile Gly Ser Lys Lys Asn Tyr Lys Tyr
Phe Cys Asn 420 425 430Leu Leu Leu Lys Ser Ser Gly Phe Asn Ile Ser
Tyr Glu Glu Glu His 435 440 445Leu Ser Ile Lys Thr Pro Asn Phe Phe
Ile Asn Gly Arg Lys Leu Lys 450 455 460Glu Ile Thr Ser Glu Lys Lys
Lys Ile Arg Lys Glu Asn Glu Gln Leu465 470 475 480Ile Lys Gln Trp
Lys Lys Leu Thr Phe Phe Lys Pro Ser Asn Leu Asn 485 490 495Gly Lys
Lys Thr Ser Asp Lys Ile Arg Phe Lys Ser Pro Asn Asn Pro 500 505
510Asp Ile Glu Arg Lys Ser Glu Asp Asn Ile Val Glu Asn Ile Ala Lys
515 520 525Val Lys Tyr Lys Leu Glu Asp Leu Leu Ser Glu Gln Arg Lys
Glu Phe 530 535 540Asn Lys Leu Ala Lys Lys His Asp Gly Val Asp Val
Glu Ala Gln Cys545 550 555 560Leu Gln Thr Lys Ser Phe Trp Ile Asp
Ser Asn Ser Pro Ile Lys Lys 565 570 575Ser Leu Glu Lys Lys Asn Glu
Lys Val Ser Val Lys Lys Lys Met Lys 580 585 590Ala Ile Arg Ser Cys
Ile Ser Ala Trp Lys Trp Phe Met Ala Asp Leu 595 600 605Ile Glu Ala
Gln Lys Glu Thr Pro Met Ile Lys Leu Lys Leu Ala Leu 610 615
620Met625197517PRTArtificial sequenceSynthetic sequence 197Thr Thr
Leu Val Pro Ser His Leu Ala Gly Ile Glu Val Met Asp Glu1 5 10 15Thr
Thr Ser Arg Asn Glu Asp Met Ile Gln Lys Glu Thr Ser Arg Ser 20 25
30Asn Glu Asp Glu Asn Tyr Leu Gly Val Lys Asn Lys Cys Gly Ile Asn
35 40 45Val His Lys Ser Gly Arg Gly Ser Ser Lys His Glu Pro Asn Met
Pro 50 55 60Pro Glu Lys Ser Gly Glu Gly Gln Met Pro Lys Gln Asp Ser
Thr Glu65 70 75 80Met Gln Gln Arg Phe Asp Glu Ser Val Thr Gly Glu
Thr Gln Val Ser 85 90 95Ala Gly Ala Thr Ala Ser Ile Lys Thr Asp Ala
Arg Ala Asn Ser Gly 100 105 110Pro Arg Val Gly Thr Ala Arg Ala Leu
Ile Val Lys Ala Ser Asn Leu 115 120 125Asp Arg Asp Ile Lys Leu Gly
Cys Lys Pro Cys Glu Tyr Ile Arg Ser 130 135 140Glu Leu Pro Met Gly
Lys Lys Asn Gly Cys Asn His Cys Glu Lys Ser145 150 155 160Ser Asp
Ile Ala Ser Val Pro Lys Val Glu Ser Gly Phe Arg Lys Ala 165 170
175Lys Tyr Glu Leu Val Arg Arg Phe Glu Ser Phe Ala Ala Asp Ser Ile
180 185 190Ser Arg His Leu Gly Lys Glu Gln Ala Arg Thr Arg Gly Lys
Arg Gly 195 200 205Lys Lys Asp Lys Lys Glu Gln Met Gly Lys Val Asn
Leu Asp Glu Ile 210 215 220Ala Ile Leu Lys Asn Glu Ser Leu Ile Glu
Tyr Thr Glu Asn Gln Ile225 230 235 240Leu Asp Ala Arg Ser Asn Arg
Ile Lys Glu Trp Leu Arg Ser Leu Arg 245 250 255Leu Arg Leu Arg Thr
Arg Asn Lys Gly Leu Lys Lys Ser Lys Ser Ile 260 265 270Arg Arg Gln
Leu Ile Thr Leu Arg Arg Asp Tyr Arg Lys Trp Ile Lys 275 280 285Pro
Asn Pro Tyr Arg Pro Asp Glu Asp Pro Asn Glu Asn Ser Leu Arg 290 295
300Leu His Thr Lys Leu Gly Val Asp Ile Gly Val Gln Gly Gly Asp
Asn305 310 315 320Lys Arg Met Asn Ser Asp Asp Tyr Glu Thr Ser Phe
Ser Ile Thr Trp 325 330 335Arg Asp Thr Ala Thr Arg Lys Ile Cys Phe
Thr Lys Pro Lys Gly Leu 340 345 350Leu Pro Arg His Met Lys Phe Lys
Leu Arg Gly Tyr Pro Glu Leu Ile 355 360 365Leu Tyr Asn Glu Glu Leu
Arg Ile Gln Asp Ser Gln Lys Phe Pro Leu 370 375 380Val Asp Trp Glu
Arg Ile Pro Ile Phe Lys Leu Arg Gly Val Ser Leu385 390 395 400Gly
Lys Lys Lys Val Lys Ala Leu Asn Arg Ile Thr Glu Ala Pro Arg 405 410
415Leu Val Val Ala Lys Arg Ile Gln Val Asn Ile Glu Ser Lys Lys Lys
420 425 430Lys Val Leu Thr Arg Tyr Val Tyr Asn Asp Lys Ser Ile Asn
Gly Arg 435 440 445Leu Val Lys Ala Glu Asp Ser Asn Lys Asp Pro Leu
Leu Glu Phe Lys 450 455 460Lys Gln Ala Glu Glu Ile Asn Ser Asp Ala
Lys Tyr Tyr Glu Asn Gln465 470 475 480Glu Ile Ala Lys Asn Tyr Leu
Trp Gly Cys Glu Gly Leu His Lys Asn 485 490 495Leu Leu Glu Glu Gln
Thr Lys Asn Pro Tyr Leu Ala Phe Lys Tyr Gly 500 505 510Phe Leu Asn
Ile Val 515198410PRTArtificial sequenceSynthetic sequence 198Leu
Asp Phe Lys Arg Thr Cys Ser Gln Glu Leu Val Leu Leu Pro Glu1 5 10
15Ile Glu Gly Leu Lys Leu Ser Gly Thr Gln Gly Val Thr Ser Leu Ala
20 25 30Lys Lys Leu Ile Asn Lys Ala Ala Asn Val Asp Arg Asp Glu Ser
Tyr 35 40 45Gly Cys His His Cys Ile His Thr Arg Thr Ser Leu Ser Lys
Pro Val 50 55 60Lys Lys Asp Cys Asn Ser Cys Asn Gln Ser Thr Asn His
Pro Ala Val65 70 75 80Pro Ile Thr Leu Lys Gly Tyr Lys Ile Ala Phe
Tyr Glu Leu Trp His 85 90 95Arg Phe Thr Ser Trp Ala Val Asp Ser Ile
Ser Lys Ala Leu His Arg 100 105 110Asn Lys Val Met Gly Lys Val Asn
Leu Asp Glu Tyr Ala Val Val Asp 115 120 125Asn Ser His Ile Val Cys
Tyr Ala Val Arg Lys Cys Tyr Glu Lys Arg 130 135 140Gln Arg Ser Val
Arg Leu His Lys Arg Ala Tyr Arg Cys Arg Ala Lys145 150 155 160His
Tyr Asn Lys Ser Gln Pro Lys Val Gly Arg Ile Tyr Lys Lys Ser 165 170
175Lys Arg Arg Asn Ala Arg Asn Leu Lys Lys Glu Ala Lys Arg Tyr Phe
180 185 190Gln Pro Asn Glu Ile Thr Asn Gly Ser Ser Asp Ala Leu Phe
Tyr Lys 195 200 205Ile Gly Val Asp Leu Gly Ile Ala Lys Gly Thr Pro
Glu Thr Glu Val 210 215 220Lys Val Asp Val Ser Ile Cys Phe Gln Val
Tyr Tyr Gly Asp Ala Arg225 230 235 240Arg Val Leu Arg Val Arg Lys
Met Asp Glu Leu Gln Ser Phe His Leu 245 250 255Asp Tyr Thr Gly Lys
Leu Lys Leu Lys Gly Ile Gly Asn Lys Asp Thr 260 265 270Phe Thr Ile
Ala Lys Arg Asn Glu Ser Leu Lys Trp Gly Ser Thr Lys 275 280 285Tyr
Glu Val Ser Arg Ala His Lys Lys Phe Lys Pro Phe Gly Lys Lys 290 295
300Gly Ser Val Lys Arg Lys Cys Asn Asp Tyr Phe Arg Ser Ile Ala
Ser305 310 315 320Trp Ser Cys Glu Ala Ala Ser Gln Arg Ala Gln Ser
Asn Leu Lys Asn 325 330 335Ala Phe Pro Tyr Gln Lys Ala Leu Val Lys
Cys Tyr Lys Asn Leu Asp 340 345 350Tyr Lys Gly Val Lys Lys Asn Asp
Met Trp Tyr Arg Leu Cys Ser Asn 355 360 365Arg Ile Phe Arg Tyr Ser
Arg Ile Ala Glu Asp Ile Ala Gln Tyr Gln 370 375 380Ser Asp Lys Gly
Lys Ala Lys Phe Glu Phe Val Ile Leu Ala Gln Ser385 390 395 400Val
Ala Glu Tyr Asp Ile Ser Ala Ile Met 405 410199602PRTArtificial
sequenceSynthetic sequence 199Val Phe Leu Thr Asp Asp Lys Arg Lys
Thr Ala Leu Arg Lys Ile Arg1 5 10 15Ser Ala Phe Arg Lys Thr Ala Glu
Ile Ala Leu Val Arg Ala Gln Glu 20 25 30Ala Asp Ser Leu Asp Arg Gln
Ala Lys Lys Leu Thr Ile Glu Thr Val 35 40 45Ser Phe Gly Ala Pro Gly
Ala Lys Asn Ala Phe Ile Gly Ser Leu Gln 50 55 60Gly Tyr Asn Trp Asn
Ser His Arg Ala Asn Val Pro Ser Ser Gly Ser65 70 75 80Ala Lys Asp
Val Phe Arg Ile Thr Glu Leu Gly Leu Gly Ile Pro Gln 85 90 95Ser Ala
His Glu Ala Ser Ile Gly Lys Ser Phe Glu Leu Val Gly Asn 100 105
110Val Val Arg Tyr Thr Ala Asn Leu Leu Ser Lys Gly Tyr Lys Lys Gly
115 120 125Ala Val Asn Lys Gly Ala Lys Gln Gln Arg Glu Ile Lys Gly
Lys Glu 130 135 140Gln Leu Ser Phe Asp Leu Ile Ser Asn Gly Pro Ile
Ser Gly Asp Lys145 150 155 160Leu Ile Asn Gly Gln Lys Asp Ala Leu
Ala Trp Trp Leu Ile Asp Lys 165 170 175Met Gly Phe His Ile Gly Leu
Ala Met Glu Pro Leu Ser Ser Pro Asn 180 185 190Thr Tyr Gly Ile Thr
Leu Gln Ala Phe Trp Lys Arg His Thr Ala Pro 195 200 205Arg Arg Tyr
Ser Arg Gly Val Ile Arg Gln Trp Gln Leu Pro Phe Gly 210 215 220Arg
Gln Leu Ala Pro Leu Ile His Asn Phe Phe Arg Lys Lys Gly Ala225 230
235 240Ser Ile Pro Ile Val Leu Thr Asn Ala Ser Lys Lys Leu Ala Gly
Lys 245 250 255Gly Val Leu Leu Glu
Gln Thr Ala Leu Val Asp Pro Lys Lys Trp Trp 260 265 270Gln Val Lys
Glu Gln Val Thr Gly Pro Leu Ser Asn Ile Trp Glu Arg 275 280 285Ser
Val Pro Leu Val Leu Tyr Thr Ala Thr Phe Thr His Lys His Gly 290 295
300Ala Ala His Lys Arg Pro Leu Thr Leu Lys Val Ile Arg Ile Ser
Ser305 310 315 320Gly Ser Val Phe Leu Leu Pro Leu Ser Lys Val Thr
Pro Gly Lys Leu 325 330 335Val Arg Ala Trp Met Pro Asp Ile Asn Ile
Leu Arg Asp Gly Arg Pro 340 345 350Asp Glu Ala Ala Tyr Lys Gly Pro
Asp Leu Ile Arg Ala Arg Glu Arg 355 360 365Ser Phe Pro Leu Ala Tyr
Thr Cys Val Thr Gln Ile Ala Asp Glu Trp 370 375 380Gln Lys Arg Ala
Leu Glu Ser Asn Arg Asp Ser Ile Thr Pro Leu Glu385 390 395 400Ala
Lys Leu Val Thr Gly Ser Asp Leu Leu Gln Ile His Ser Thr Val 405 410
415Gln Gln Ala Val Glu Gln Gly Ile Gly Gly Arg Ile Ser Ser Pro Ile
420 425 430Gln Glu Leu Leu Ala Lys Asp Ala Leu Gln Leu Val Leu Gln
Gln Leu 435 440 445Phe Met Thr Val Asp Leu Leu Arg Ile Gln Trp Gln
Leu Lys Gln Glu 450 455 460Val Ala Asp Gly Asn Thr Ser Glu Lys Ala
Val Gly Trp Ala Ile Arg465 470 475 480Ile Ser Asn Ile His Lys Asp
Ala Tyr Lys Thr Ala Ile Glu Pro Cys 485 490 495Thr Ser Ala Leu Lys
Gln Ala Trp Asn Pro Leu Ser Gly Phe Glu Glu 500 505 510Arg Thr Phe
Gln Leu Asp Ala Ser Ile Val Arg Lys Arg Ser Thr Ala 515 520 525Lys
Thr Pro Asp Asp Glu Leu Val Ile Val Leu Arg Gln Gln Ala Ala 530 535
540Glu Met Thr Val Ala Val Thr Gln Ser Val Ser Lys Glu Leu Met
Glu545 550 555 560Leu Ala Val Arg His Ser Ala Thr Leu His Leu Leu
Val Gly Glu Val 565 570 575Ala Ser Lys Gln Leu Ser Arg Ser Ala Asp
Lys Asp Arg Gly Ala Met 580 585 590Asp His Trp Lys Leu Leu Ser Gln
Ser Met 595 600200494PRTArtificial sequenceSynthetic sequence
200Glu Asp Leu Leu Gln Lys Ala Leu Asn Thr Ala Thr Asn Val Ala Ala1
5 10 15Ile Glu Arg His Ser Cys Ile Ser Cys Leu Phe Thr Glu Ser Glu
Ile 20 25 30Asp Val Lys Tyr Lys Thr Pro Asp Lys Ile Gly Gln Asn Thr
Ala Gly 35 40 45Cys Gln Ser Cys Thr Phe Arg Val Gly Tyr Ser Gly Asn
Ser His Thr 50 55 60Leu Pro Met Gly Asn Arg Ile Ala Leu Asp Lys Leu
Arg Glu Thr Ile65 70 75 80Gln Arg Tyr Ala Trp His Ser Leu Leu Phe
Asn Val Pro Pro Ala Pro 85 90 95Thr Ser Lys Arg Val Arg Ala Ile Ser
Glu Leu Arg Val Ala Ala Gly 100 105 110Arg Glu Arg Leu Phe Thr Val
Ile Thr Phe Val Gln Thr Asn Ile Leu 115 120 125Ser Lys Leu Gln Lys
Arg Tyr Ala Ala Asn Trp Thr Pro Lys Ser Gln 130 135 140Glu Arg Leu
Ser Arg Leu Arg Glu Glu Gly Gln His Ile Leu Ser Leu145 150 155
160Leu Glu Ser Gly Ser Trp Gln Gln Lys Glu Val Val Arg Glu Asp Gln
165 170 175Asp Leu Ile Val Cys Ser Ala Leu Thr Lys Pro Gly Leu Ser
Ile Gly 180 185 190Ala Phe Cys Arg Pro Lys Tyr Leu Lys Pro Ala Lys
His Ala Leu Val 195 200 205Leu Arg Leu Ile Phe Val Glu Gln Trp Pro
Gly Gln Ile Trp Gly Gln 210 215 220Ser Lys Arg Thr Arg Arg Met Arg
Arg Arg Lys Asp Val Glu Arg Val225 230 235 240Tyr Asp Ile Ser Val
Gln Ala Trp Ala Leu Lys Gly Lys Glu Thr Arg 245 250 255Ile Ser Glu
Cys Ile Asp Thr Met Arg Arg His Gln Gln Ala Tyr Ile 260 265 270Gly
Val Leu Pro Phe Leu Ile Leu Ser Gly Ser Thr Val Arg Gly Lys 275 280
285Gly Asp Cys Pro Ile Leu Lys Glu Ile Thr Arg Met Arg Tyr Cys Pro
290 295 300Asn Asn Glu Gly Leu Ile Pro Leu Gly Ile Phe Tyr Arg Gly
Ser Ala305 310 315 320Asn Lys Leu Leu Arg Val Val Lys Gly Ser Ser
Phe Thr Leu Pro Met 325 330 335Trp Gln Asn Ile Glu Thr Leu Pro His
Pro Glu Pro Phe Ser Pro Glu 340 345 350Gly Trp Thr Ala Thr Gly Ala
Leu Tyr Glu Lys Asn Leu Ala Tyr Trp 355 360 365Ser Ala Leu Asn Glu
Ala Val Asp Trp Tyr Thr Gly Gln Ile Leu Ser 370 375 380Ser Gly Leu
Gln Tyr Pro Asn Gln Asn Glu Phe Leu Ala Arg Leu Gln385 390 395
400Asn Val Ile Asp Ser Ile Pro Arg Lys Trp Phe Arg Pro Gln Gly Leu
405 410 415Lys Asn Leu Lys Pro Asn Gly Gln Glu Asp Ile Val Pro Asn
Glu Phe 420 425 430Val Ile Pro Gln Asn Ala Ile Arg Ala His His Val
Ile Glu Trp Tyr 435 440 445His Lys Thr Asn Asp Leu Val Ala Lys Thr
Leu Leu Gly Trp Gly Ser 450 455 460Gln Thr Thr Leu Asn Gln Thr Arg
Pro Gln Gly Asp Leu Arg Phe Thr465 470 475 480Tyr Thr Arg Tyr Tyr
Phe Arg Glu Lys Glu Val Pro Glu Val 485 490201649PRTArtificial
sequenceSynthetic sequence 201Val Pro Lys Lys Lys Leu Met Arg Glu
Leu Ala Lys Lys Ala Val Phe1 5 10 15Glu Ala Ile Phe Asn Asp Pro Ile
Pro Gly Ser Phe Gly Cys Lys Arg 20 25 30Cys Thr Leu Ile Asp Gly Ala
Arg Val Thr Asp Ala Ile Glu Lys Lys 35 40 45Gln Gly Ala Lys Arg Cys
Ala Gly Cys Glu Pro Cys Thr Phe His Thr 50 55 60Leu Tyr Asp Ser Val
Lys His Ala Leu Pro Ala Ala Thr Gly Cys Asp65 70 75 80Arg Thr Ala
Ile Asp Thr Gly Leu Trp Glu Ile Leu Thr Ala Leu Arg 85 90 95Ser Tyr
Asn Trp Met Ser Phe Arg Arg Asn Ala Val Ser Asp Ala Ser 100 105
110Gln Lys Gln Val Trp Ser Ile Glu Glu Leu Ala Ile Trp Ala Asp Lys
115 120 125Glu Arg Ala Leu Arg Val Ile Leu Ser Ala Leu Thr His Thr
Ile Gly 130 135 140Lys Leu Lys Asn Gly Phe Ser Arg Asp Gly Val Trp
Lys Gly Gly Lys145 150 155 160Gln Leu Tyr Glu Asn Leu Ala Gln Lys
Asp Leu Ala Lys Gly Leu Phe 165 170 175Ala Asn Gly Glu Ile Phe Gly
Lys Glu Leu Val Glu Ala Asp His Asp 180 185 190Met Leu Ala Trp Thr
Ile Val Pro Asn His Gln Phe His Ile Gly Leu 195 200 205Ile Arg Gly
Asn Trp Lys Pro Ala Ala Val Glu Ala Ser Thr Ala Phe 210 215 220Asp
Ala Arg Trp Leu Thr Asn Gly Ala Pro Leu Arg Asp Thr Arg Thr225 230
235 240His Gly His Arg Gly Arg Arg Phe Asn Arg Thr Glu Lys Leu Thr
Val 245 250 255Leu Cys Ile Lys Arg Asp Gly Gly Val Ser Glu Glu Phe
Arg Gln Glu 260 265 270Arg Asp Tyr Glu Leu Ser Val Met Leu Leu Gln
Pro Lys Asn Lys Leu 275 280 285Lys Pro Glu Pro Lys Gly Glu Leu Asn
Ser Phe Glu Asp Leu His Asp 290 295 300His Trp Trp Phe Leu Lys Gly
Asp Glu Ala Thr Ala Leu Val Gly Leu305 310 315 320Thr Ser Asp Pro
Thr Val Gly Asp Phe Ile Gln Leu Gly Leu Tyr Ile 325 330 335Arg Asn
Pro Ile Lys Ala His Gly Glu Thr Lys Arg Arg Leu Leu Ile 340 345
350Cys Phe Glu Pro Pro Ile Lys Leu Pro Leu Arg Arg Ala Phe Pro Ser
355 360 365Glu Ala Phe Lys Thr Trp Glu Pro Thr Ile Asn Val Phe Arg
Asn Gly 370 375 380Arg Arg Asp Thr Glu Ala Tyr Tyr Asp Ile Asp Arg
Ala Arg Val Phe385 390 395 400Glu Phe Pro Glu Thr Arg Val Ser Leu
Glu His Leu Ser Lys Gln Trp 405 410 415Glu Val Leu Arg Leu Glu Pro
Asp Arg Glu Asn Thr Asp Pro Tyr Glu 420 425 430Ala Gln Gln Asn Glu
Gly Ala Glu Leu Gln Val Tyr Ser Leu Leu Gln 435 440 445Glu Ala Ala
Gln Lys Met Ala Pro Lys Val Val Ile Asp Pro Phe Gly 450 455 460Gln
Phe Pro Leu Glu Leu Phe Ser Thr Phe Val Ala Gln Leu Phe Asn465 470
475 480Ala Pro Leu Ser Asp Thr Lys Ala Lys Ile Gly Lys Pro Leu Asp
Ser 485 490 495Gly Phe Val Val Glu Ser His Leu His Leu Leu Glu Glu
Asp Phe Ala 500 505 510Tyr Arg Asp Phe Val Arg Val Thr Phe Met Gly
Thr Glu Pro Thr Phe 515 520 525Arg Val Ile His Tyr Ser Asn Gly Glu
Gly Tyr Trp Lys Lys Thr Val 530 535 540Leu Lys Gly Lys Asn Asn Ile
Arg Thr Ala Leu Ile Pro Glu Gly Ala545 550 555 560Lys Ala Ala Val
Asp Ala Tyr Lys Asn Lys Arg Cys Pro Leu Thr Leu 565 570 575Glu Ala
Ala Ile Leu Asn Glu Glu Lys Asp Arg Arg Leu Val Leu Gly 580 585
590Asn Lys Ala Leu Ser Leu Leu Ala Gln Thr Ala Arg Gly Asn Leu Thr
595 600 605Ile Leu Glu Ala Leu Ala Ala Glu Val Leu Arg Pro Leu Ser
Gly Thr 610 615 620Glu Gly Val Val His Leu His Ala Cys Val Thr Arg
His Ser Thr Leu625 630 635 640Thr Glu Ser Thr Glu Thr Asp Asn Met
645202414PRTArtificial sequenceSynthetic sequence 202Val Glu Lys
Leu Phe Ser Glu Arg Leu Lys Arg Ala Met Trp Leu Lys1 5 10 15Asn Glu
Ala Gly Arg Ala Pro Pro Ala Glu Thr Leu Thr Leu Lys His 20 25 30Lys
Arg Val Ser Gly Gly His Glu Lys Val Lys Glu Glu Leu Gln Arg 35 40
45Val Leu Arg Ser Leu Ser Gly Thr Asn Gln Ala Ala Trp Asn Leu Gly
50 55 60Leu Ser Gly Gly Arg Glu Pro Lys Ser Ser Asp Ala Leu Lys Gly
Glu65 70 75 80Lys Ser Arg Val Val Leu Glu Thr Val Val Phe His Ser
Gly His Asn 85 90 95Arg Val Leu Tyr Asp Val Ile Glu Arg Glu Asp Gln
Val His Gln Arg 100 105 110Ser Ser Ile Met His Met Arg Arg Lys Gly
Ser Asn Leu Leu Arg Leu 115 120 125Trp Gly Arg Ser Gly Lys Val Arg
Arg Lys Met Arg Glu Glu Val Ala 130 135 140Glu Ile Lys Pro Val Trp
His Lys Asp Ser Arg Trp Leu Ala Ile Val145 150 155 160Glu Glu Gly
Arg Gln Ser Val Val Gly Ile Ser Ser Ala Gly Leu Ala 165 170 175Val
Phe Ala Val Gln Glu Ser Gln Cys Thr Thr Ala Glu Pro Lys Pro 180 185
190Leu Glu Tyr Val Val Ser Ile Trp Phe Arg Gly Ser Lys Ala Leu Asn
195 200 205Pro Gln Asp Arg Tyr Leu Glu Phe Lys Lys Leu Lys Thr Thr
Glu Ala 210 215 220Leu Arg Gly Gln Gln Tyr Asp Pro Ile Pro Phe Ser
Leu Lys Arg Gly225 230 235 240Ala Gly Cys Ser Leu Ala Ile Arg Gly
Glu Gly Ile Lys Phe Gly Ser 245 250 255Arg Gly Pro Ile Lys Gln Phe
Phe Gly Ser Asp Arg Ser Arg Pro Ser 260 265 270His Ala Asp Tyr Asp
Gly Lys Arg Arg Leu Ser Leu Phe Ser Lys Tyr 275 280 285Ala Gly Asp
Leu Ala Asp Leu Thr Glu Glu Gln Trp Asn Arg Thr Val 290 295 300Ser
Ala Phe Ala Glu Asp Glu Val Arg Arg Ala Thr Leu Ala Asn Ile305 310
315 320Gln Asp Phe Leu Ser Ile Ser His Glu Lys Tyr Ala Glu Arg Leu
Lys 325 330 335Lys Arg Ile Glu Ser Ile Glu Glu Pro Val Ser Ala Ser
Lys Leu Glu 340 345 350Ala Tyr Leu Ser Ala Ile Phe Glu Thr Phe Val
Gln Gln Arg Glu Ala 355 360 365Leu Ala Ser Asn Phe Leu Met Arg Leu
Val Glu Ser Val Ala Leu Leu 370 375 380Ile Ser Leu Glu Glu Lys Ser
Pro Arg Val Glu Phe Arg Val Ala Arg385 390 395 400Tyr Leu Ala Glu
Ser Lys Glu Gly Phe Asn Arg Lys Ala Met 405 410203413PRTArtificial
sequenceSynthetic sequence 203Val Val Ile Thr Gln Ser Glu Leu Tyr
Lys Glu Arg Leu Leu Arg Val1 5 10 15Met Glu Ile Lys Asn Asp Arg Gly
Arg Lys Glu Pro Arg Glu Ser Gln 20 25 30Gly Leu Val Leu Arg Phe Thr
Gln Val Thr Gly Gly Gln Glu Lys Val 35 40 45Lys Gln Lys Leu Trp Leu
Ile Phe Glu Gly Phe Ser Gly Thr Asn Gln 50 55 60Ala Ser Trp Asn Phe
Gly Gln Pro Ala Gly Gly Arg Lys Pro Asn Ser65 70 75 80Gly Asp Ala
Leu Lys Gly Pro Lys Ser Arg Val Thr Tyr Glu Thr Val 85 90 95Val Phe
His Phe Gly Leu Arg Leu Leu Ser Ala Val Ile Glu Arg His 100 105
110Asn Leu Lys Gln Gln Arg Gln Thr Met Ala Tyr Met Lys Arg Arg Ala
115 120 125Ala Ala Arg Lys Lys Trp Ala Arg Ser Gly Lys Lys Cys Ser
Arg Met 130 135 140Arg Asn Glu Val Glu Lys Ile Lys Pro Lys Trp His
Lys Asp Pro Arg145 150 155 160Trp Phe Asp Ile Val Lys Glu Gly Glu
Pro Ser Ile Val Gly Ile Ser 165 170 175Ser Ala Gly Phe Ala Ile Tyr
Ile Val Glu Glu Pro Asn Phe Pro Arg 180 185 190Gln Asp Pro Leu Glu
Ile Glu Tyr Ala Ile Ser Ile Trp Phe Arg Arg 195 200 205Asp Arg Ser
Gln Tyr Leu Thr Phe Lys Lys Ile Gln Lys Ala Glu Lys 210 215 220Leu
Lys Glu Leu Gln Tyr Asn Pro Ile Pro Phe Arg Leu Lys Gln Glu225 230
235 240Lys Thr Ser Leu Val Phe Glu Ser Gly Asp Ile Lys Phe Gly Ser
Arg 245 250 255Gly Ser Ile Glu His Phe Arg Asp Glu Ala Arg Gly Lys
Pro Pro Lys 260 265 270Ala Asp Met Asp Asn Asn Arg Arg Leu Thr Met
Phe Ser Val Phe Ser 275 280 285Gly Asn Leu Thr Asn Leu Thr Glu Glu
Gln Tyr Ala Arg Pro Val Ser 290 295 300Gly Leu Leu Ala Pro Asp Glu
Lys Arg Met Pro Thr Leu Leu Lys Lys305 310 315 320Leu Gln Asp Phe
Phe Thr Pro Ile His Glu Lys Tyr Gly Glu Arg Ile 325 330 335Lys Gln
Arg Leu Ala Asn Ser Glu Ala Ser Lys Arg Pro Phe Lys Lys 340 345
350Leu Glu Glu Tyr Leu Pro Ala Ile Tyr Leu Glu Phe Arg Ala Arg Arg
355 360 365Glu Gly Leu Ala Ser Asn Trp Val Leu Val Leu Ile Asn Ser
Val Arg 370 375 380Thr Leu Val Arg Ile Lys Ser Glu Asp Pro Tyr Ile
Glu Phe Lys Val385 390 395 400Ser Gln Tyr Leu Leu Glu Lys Glu Asp
Asn Lys Ala Leu 405 410204449PRTArtificial sequenceSynthetic
sequence 204Lys Gln Asp Ala Leu Phe Glu Glu Arg Leu Lys Lys Ala Ile
Phe Ile1 5 10 15Lys Arg Gln Ala Asp Pro Leu Gln Arg Glu Glu Leu Ser
Leu Leu Pro 20 25 30Pro Asn Arg Lys Ile Val Thr Gly Gly His Glu Ser
Ala Lys Asp Thr 35 40 45Leu Lys Gln Ile Leu Arg Ala Ile Asn Gly Thr
Asn Gln Ala Ser Trp 50 55 60Asn Pro Gly Thr Pro Ser Gly Lys Arg Asp
Ser Lys Ser Ala Asp Ala65 70 75 80Leu Ala Gly Pro Lys Ser Arg Val
Lys Leu Glu Thr Val Val Phe His 85 90 95Val Gly His Arg Leu Leu Lys
Lys Val Val Glu Tyr Gln Gly His Gln 100 105 110Lys Gln Gln His Gly
Leu Lys Ala Phe Met Arg Thr Cys Ala Ala Met 115 120 125Arg Lys Lys
Trp Lys Arg Ser Gly Lys Val Val Gly Glu Leu Arg Glu
130 135 140Gln Leu Ala Asn Ile Gln Pro Lys Trp His Tyr Asp Ser Arg
Pro Leu145 150 155 160Asn Leu Cys Phe Glu Gly Lys Pro Ser Val Val
Gly Leu Arg Ser Ala 165 170 175Gly Ile Ala Leu Tyr Thr Ile Gln Lys
Ser Val Val Pro Val Lys Glu 180 185 190Pro Lys Pro Ile Glu Tyr Ala
Val Ser Ile Trp Phe Arg Gly Pro Lys 195 200 205Ala Met Asp Arg Glu
Asp Arg Cys Leu Glu Phe Lys Lys Leu Lys Ile 210 215 220Ala Thr Glu
Leu Arg Lys Leu Gln Phe Glu Pro Ile Val Ser Thr Leu225 230 235
240Thr Gln Gly Ile Lys Gly Phe Ser Leu Tyr Ile Gln Gly Asn Ser Val
245 250 255Lys Phe Gly Ser Arg Gly Pro Ile Lys Tyr Phe Ser Asn Glu
Ser Val 260 265 270Arg Gln Arg Pro Pro Lys Ala Asp Pro Asp Gly Asn
Lys Arg Leu Ala 275 280 285Leu Phe Ser Lys Phe Ser Gly Asp Leu Ser
Asp Leu Thr Glu Glu Gln 290 295 300Trp Asn Arg Pro Ile Leu Ala Phe
Glu Gly Ile Ile Arg Arg Ala Thr305 310 315 320Leu Gly Asn Ile Gln
Asp Tyr Leu Thr Val Gly His Glu Gln Phe Ala 325 330 335Ile Ser Leu
Glu Gln Leu Leu Ser Glu Lys Glu Ser Val Leu Gln Met 340 345 350Ser
Ile Glu Gln Gln Arg Leu Lys Lys Asn Leu Gly Lys Lys Ala Glu 355 360
365Asn Glu Trp Val Glu Ser Phe Gly Ala Glu Gln Ala Arg Lys Lys Ala
370 375 380Gln Gly Ile Arg Glu Tyr Ile Ser Gly Phe Phe Gln Glu Tyr
Cys Ser385 390 395 400Gln Arg Glu Gln Trp Ala Glu Asn Trp Val Gln
Gln Leu Asn Lys Ser 405 410 415Val Arg Leu Phe Leu Thr Ile Gln Asp
Ser Thr Pro Phe Ile Glu Phe 420 425 430Arg Val Ala Arg Tyr Leu Pro
Lys Gly Glu Lys Lys Lys Gly Lys Ala 435 440
445Met205711PRTArtificial sequenceSynthetic sequence 205Ala Asn His
Ala Glu Arg His Lys Arg Leu Arg Lys Glu Ala Asn Arg1 5 10 15Ala Ala
Asn Arg Asn Arg Pro Leu Val Ala Asp Cys Asp Thr Gly Asp 20 25 30Pro
Leu Val Gly Ile Cys Arg Leu Leu Arg Arg Gly Asp Lys Met Gln 35 40
45Pro Asn Lys Thr Gly Cys Arg Ser Cys Glu Gln Val Glu Pro Glu Leu
50 55 60Arg Asp Ala Ile Leu Val Ser Gly Pro Gly Arg Leu Asp Asn Tyr
Lys65 70 75 80Tyr Glu Leu Phe Gln Arg Gly Arg Ala Met Ala Val His
Arg Leu Leu 85 90 95Lys Arg Val Pro Lys Leu Asn Arg Pro Lys Lys Ala
Ala Gly Asn Asp 100 105 110Glu Lys Lys Ala Glu Asn Lys Lys Ser Glu
Ile Gln Lys Glu Lys Gln 115 120 125Lys Gln Arg Arg Met Met Pro Ala
Val Ser Met Lys Gln Val Ser Val 130 135 140Ala Asp Phe Lys His Val
Ile Glu Asn Thr Val Arg His Leu Phe Gly145 150 155 160Asp Arg Arg
Asp Arg Glu Ile Ala Glu Cys Ala Ala Leu Arg Ala Ala 165 170 175Ser
Lys Tyr Phe Leu Lys Ser Arg Arg Val Arg Pro Arg Lys Leu Pro 180 185
190Lys Leu Ala Asn Pro Asp His Gly Lys Glu Leu Lys Gly Leu Arg Leu
195 200 205Arg Glu Lys Arg Ala Lys Leu Lys Lys Glu Lys Glu Lys Gln
Ala Glu 210 215 220Leu Ala Arg Ser Asn Gln Lys Gly Ala Val Leu His
Val Ala Thr Leu225 230 235 240Lys Lys Asp Ala Pro Pro Met Pro Tyr
Glu Lys Thr Gln Gly Arg Asn 245 250 255Asp Tyr Thr Thr Phe Val Ile
Ser Ala Ala Ile Lys Val Gly Ala Thr 260 265 270Arg Gly Thr Lys Pro
Leu Leu Thr Pro Gln Pro Arg Glu Trp Gln Cys 275 280 285Ser Leu Tyr
Trp Arg Asp Gly Gln Arg Trp Ile Arg Gly Gly Leu Leu 290 295 300Gly
Leu Gln Ala Gly Ile Val Leu Gly Pro Lys Leu Asn Arg Glu Leu305 310
315 320Leu Glu Ala Val Leu Gln Arg Pro Ile Glu Cys Arg Met Ser Gly
Cys 325 330 335Gly Asn Pro Leu Gln Val Arg Gly Ala Ala Val Asp Phe
Phe Met Thr 340 345 350Thr Asn Pro Phe Tyr Val Ser Gly Ala Ala Tyr
Ala Gln Lys Lys Phe 355 360 365Lys Pro Phe Gly Thr Lys Arg Ala Ser
Glu Asp Gly Ala Ala Ala Lys 370 375 380Ala Arg Glu Lys Leu Met Thr
Gln Leu Ala Lys Val Leu Asp Lys Val385 390 395 400Val Thr Gln Ala
Ala His Ser Pro Leu Asp Gly Ile Trp Glu Thr Arg 405 410 415Pro Glu
Ala Lys Leu Arg Ala Met Ile Met Ala Leu Glu His Glu Trp 420 425
430Ile Phe Leu Arg Pro Gly Pro Cys His Asn Ala Ala Glu Glu Val Ile
435 440 445Lys Cys Asp Cys Thr Gly Gly His Ala Ile Leu Trp Ala Leu
Ile Asp 450 455 460Glu Ala Arg Gly Ala Leu Glu His Lys Glu Phe Tyr
Ala Val Thr Arg465 470 475 480Ala His Thr His Asp Cys Glu Lys Gln
Lys Leu Gly Gly Arg Leu Ala 485 490 495Gly Phe Leu Asp Leu Leu Ile
Ala Gln Asp Val Pro Leu Asp Asp Ala 500 505 510Pro Ala Ala Arg Lys
Ile Lys Thr Leu Leu Glu Ala Thr Pro Pro Ala 515 520 525Pro Cys Tyr
Lys Ala Ala Thr Ser Ile Ala Thr Cys Asp Cys Glu Gly 530 535 540Lys
Phe Asp Lys Leu Trp Ala Ile Ile Asp Ala Thr Arg Ala Gly His545 550
555 560Gly Thr Glu Asp Leu Trp Ala Arg Thr Leu Ala Tyr Pro Gln Asn
Val 565 570 575Asn Cys Lys Cys Lys Ala Gly Lys Asp Leu Thr His Arg
Leu Ala Asp 580 585 590Phe Leu Gly Leu Leu Ile Lys Arg Asp Gly Pro
Phe Arg Glu Arg Pro 595 600 605Pro His Lys Val Thr Gly Asp Arg Lys
Leu Val Phe Ser Gly Asp Lys 610 615 620Lys Cys Lys Gly His Gln Tyr
Val Ile Leu Ala Lys Ala His Asn Glu625 630 635 640Glu Val Val Arg
Ala Trp Ile Ser Arg Trp Gly Leu Lys Ser Arg Thr 645 650 655Asn Lys
Ala Gly Tyr Ala Ala Thr Glu Leu Asn Leu Leu Leu Asn Trp 660 665
670Leu Ser Ile Cys Arg Arg Arg Trp Met Asp Met Leu Thr Val Gln Arg
675 680 685Asp Thr Pro Tyr Ile Arg Met Lys Thr Gly Arg Leu Val Val
Asp Asp 690 695 700Lys Lys Glu Arg Lys Ala Met705
710206574PRTArtificial sequenceSynthetic sequence 206Ala Lys Gln
Arg Glu Ala Leu Arg Val Ala Leu Glu Arg Gly Ile Val1 5 10 15Arg Ala
Ser Asn Arg Thr Tyr Thr Leu Val Thr Asn Cys Thr Lys Gly 20 25 30Gly
Pro Leu Pro Glu Gln Cys Arg Met Ile Glu Arg Gly Lys Ala Arg 35 40
45Ala Met Lys Trp Glu Pro Lys Leu Val Gly Cys Gly Ser Cys Ala Ala
50 55 60Ala Thr Val Asp Leu Pro Ala Ile Glu Glu Tyr Ala Gln Pro Gly
Arg65 70 75 80Leu Asp Val Ala Lys Tyr Lys Leu Thr Thr Gln Ile Leu
Ala Met Ala 85 90 95Thr Arg Arg Met Met Val Arg Ala Ala Lys Leu Ser
Arg Arg Lys Gly 100 105 110Gln Trp Pro Ala Lys Val Gln Glu Glu Lys
Glu Glu Pro Pro Glu Pro 115 120 125Lys Lys Met Leu Lys Ala Val Glu
Met Arg Pro Val Ala Ile Val Asp 130 135 140Phe Asn Arg Val Ile Gln
Thr Thr Ile Glu His Leu Trp Ala Glu Arg145 150 155 160Ala Asn Ala
Asp Glu Ala Glu Leu Lys Ala Leu Lys Ala Ala Ala Ala 165 170 175Tyr
Phe Gly Pro Ser Leu Lys Ile Arg Ala Arg Gly Pro Pro Lys Ala 180 185
190Ala Ile Gly Arg Glu Leu Lys Lys Ala His Arg Lys Lys Ala Tyr Ala
195 200 205Glu Arg Lys Lys Ala Arg Arg Lys Arg Ala Glu Leu Ala Arg
Ser Gln 210 215 220Ala Arg Gly Ala Ala Ala His Ala Ala Ile Arg Glu
Arg Asp Ile Pro225 230 235 240Pro Met Ala Tyr Glu Arg Thr Gln Gly
Arg Asn Asp Val Thr Thr Ile 245 250 255Pro Ile Ala Ala Ala Ile Lys
Ile Ala Ala Thr Arg Gly Ala Arg Pro 260 265 270Leu Pro Ala Pro Lys
Pro Met Lys Trp Gln Cys Ser Leu Tyr Trp Asn 275 280 285Glu Gly Gln
Arg Trp Ile Arg Gly Gly Met Leu Thr Ala Gln Ala Tyr 290 295 300Ala
His Ala Ala Asn Ile His Arg Pro Met Arg Cys Glu Met Trp Gly305 310
315 320Val Gly Asn Pro Leu Lys Val Arg Ala Phe Glu Gly Arg Val Ala
Asp 325 330 335Pro Asp Gly Ala Lys Gly Arg Lys Ala Glu Phe Arg Leu
Gln Thr Asn 340 345 350Ala Phe Tyr Val Ser Gly Ala Ala Tyr Arg Asn
Lys Lys Phe Lys Pro 355 360 365Phe Gly Thr Asp Arg Gly Gly Ile Gly
Ser Ala Arg Lys Lys Arg Glu 370 375 380Arg Leu Met Ala Gln Leu Ala
Lys Ile Leu Asp Lys Val Val Ser Gln385 390 395 400Ala Ala His Ser
Pro Leu Asp Asp Ile Trp His Thr Arg Pro Ala Gln 405 410 415Lys Leu
Arg Ala Met Ile Lys Gln Leu Glu His Glu Trp Met Phe Leu 420 425
430Arg Pro Gln Ala Pro Thr Val Glu Gly Thr Lys Pro Asp Val Asp Val
435 440 445Ala Gly Asn Met Gln Arg Gln Ile Lys Ala Leu Met Ala Pro
Asp Leu 450 455 460Pro Pro Ile Glu Lys Gly Ser Pro Ala Lys Arg Phe
Thr Gly Asp Lys465 470 475 480Arg Lys Lys Gly Glu Arg Ala Val Arg
Val Ala Glu Ala His Ser Asp 485 490 495Glu Val Val Thr Ala Trp Ile
Ser Arg Trp Gly Ile Gln Thr Arg Arg 500 505 510Asn Glu Gly Ser Tyr
Ala Ala Gln Glu Leu Glu Leu Leu Leu Asn Trp 515 520 525Leu Gln Ile
Cys Arg Arg Arg Trp Leu Asp Met Thr Ala Ala Gln Arg 530 535 540Val
Ser Pro Tyr Ile Arg Met Lys Ser Gly Arg Met Ile Thr Asp Ala545 550
555 560Ala Asp Glu Gly Val Ala Pro Ile Pro Leu Val Glu Asn Met 565
570207743PRTArtificial sequenceSynthetic sequence 207Lys Ser Ile
Ser Gly Arg Ser Ile Lys His Met Ala Cys Leu Lys Asp1 5 10 15Met Leu
Lys Ser Glu Ile Thr Glu Ile Glu Glu Lys Gln Lys Lys Glu 20 25 30Ser
Leu Arg Lys Trp Asp Tyr Tyr Ser Lys Phe Ser Asp Glu Ile Leu 35 40
45Phe Arg Arg Asn Leu Asn Val Ser Ala Asn His Asp Ala Asn Ala Cys
50 55 60Tyr Gly Cys Asn Pro Cys Ala Phe Leu Lys Glu Val Tyr Gly Phe
Arg65 70 75 80Ile Glu Arg Arg Asn Asn Glu Arg Ile Ile Ser Tyr Arg
Arg Gly Leu 85 90 95Ala Gly Cys Lys Ser Cys Val Gln Ser Thr Gly Tyr
Pro Pro Ile Glu 100 105 110Phe Val Arg Arg Lys Phe Gly Ala Asp Lys
Ala Met Glu Ile Val Arg 115 120 125Glu Val Leu His Arg Arg Asn Trp
Gly Ala Leu Ala Arg Asn Ile Gly 130 135 140Arg Glu Lys Glu Ala Asp
Pro Ile Leu Gly Glu Leu Asn Glu Leu Leu145 150 155 160Leu Val Asp
Ala Arg Pro Tyr Phe Gly Asn Lys Ser Ala Ala Asn Glu 165 170 175Thr
Asn Leu Ala Phe Asn Val Ile Thr Arg Ala Ala Lys Lys Phe Arg 180 185
190Asp Glu Gly Met Tyr Asp Ile His Lys Gln Leu Asp Ile His Ser Glu
195 200 205Glu Gly Lys Val Pro Lys Gly Arg Lys Ser Arg Leu Ile Arg
Ile Glu 210 215 220Arg Lys His Lys Ala Ile His Gly Leu Asp Pro Gly
Glu Thr Trp Arg225 230 235 240Tyr Pro His Cys Gly Lys Gly Glu Lys
Tyr Gly Val Trp Leu Asn Arg 245 250 255Ser Arg Leu Ile His Ile Lys
Gly Asn Glu Tyr Arg Cys Leu Thr Ala 260 265 270Phe Gly Thr Thr Gly
Arg Arg Met Ser Leu Asp Val Ala Cys Ser Val 275 280 285Leu Gly His
Pro Leu Val Lys Lys Lys Arg Lys Lys Gly Lys Lys Thr 290 295 300Val
Asp Gly Thr Glu Leu Trp Gln Ile Lys Lys Ala Thr Glu Thr Leu305 310
315 320Pro Glu Asp Pro Ile Asp Cys Thr Phe Tyr Leu Tyr Ala Ala Lys
Pro 325 330 335Thr Lys Asp Pro Phe Ile Leu Lys Val Gly Ser Leu Lys
Ala Pro Arg 340 345 350Trp Lys Lys Leu His Lys Asp Phe Phe Glu Tyr
Ser Asp Thr Glu Lys 355 360 365Thr Gln Gly Gln Glu Lys Gly Lys Arg
Val Val Arg Arg Gly Lys Val 370 375 380Pro Arg Ile Leu Ser Leu Arg
Pro Asp Ala Lys Phe Lys Val Ser Ile385 390 395 400Trp Asp Asp Pro
Tyr Asn Gly Lys Asn Lys Glu Gly Thr Leu Leu Arg 405 410 415Met Glu
Leu Ser Gly Leu Asp Gly Ala Lys Lys Pro Leu Ile Leu Lys 420 425
430Arg Tyr Gly Glu Pro Asn Thr Lys Pro Lys Asn Phe Val Phe Trp Arg
435 440 445Pro His Ile Thr Pro His Pro Leu Thr Phe Thr Pro Lys His
Asp Phe 450 455 460Gly Asp Pro Asn Lys Lys Thr Lys Arg Arg Arg Val
Phe Asn Arg Glu465 470 475 480Tyr Tyr Gly His Leu Asn Asp Leu Ala
Lys Met Glu Pro Asn Ala Lys 485 490 495Phe Phe Glu Asp Arg Glu Val
Ser Asn Lys Lys Asn Pro Lys Ala Lys 500 505 510Asn Ile Arg Ile Gln
Ala Lys Glu Ser Leu Pro Asn Ile Val Ala Lys 515 520 525Asn Gly Arg
Trp Ala Ala Phe Asp Pro Asn Asp Ser Leu Trp Lys Leu 530 535 540Tyr
Leu His Trp Arg Gly Arg Arg Lys Thr Ile Lys Gly Gly Ile Ser545 550
555 560Gln Glu Phe Gln Glu Phe Lys Glu Arg Leu Asp Leu Tyr Lys Lys
His 565 570 575Glu Asp Glu Ser Glu Trp Lys Glu Lys Glu Lys Leu Trp
Glu Asn His 580 585 590Glu Lys Glu Trp Lys Lys Thr Leu Glu Ile His
Gly Ser Ile Ala Glu 595 600 605Val Ser Gln Arg Cys Val Met Gln Ser
Met Met Gly Pro Leu Asp Gly 610 615 620Leu Val Gln Lys Lys Asp Tyr
Val His Ile Gly Gln Ser Ser Leu Lys625 630 635 640Ala Ala Asp Asp
Ala Trp Thr Phe Ser Ala Asn Arg Tyr Lys Lys Ala 645 650 655Thr Gly
Pro Lys Trp Gly Lys Ile Ser Val Ser Asn Leu Leu Tyr Asp 660 665
670Ala Asn Gln Ala Asn Ala Glu Leu Ile Ser Gln Ser Ile Ser Lys Tyr
675 680 685Leu Ser Lys Gln Lys Asp Asn Gln Gly Cys Glu Gly Arg Lys
Met Lys 690 695 700Phe Leu Ile Lys Ile Ile Glu Pro Leu Arg Glu Asn
Phe Val Lys His705 710 715 720Thr Arg Trp Leu His Glu Met Thr Gln
Lys Asp Cys Glu Val Arg Ala 725 730 735Gln Phe Ser Arg Val Ser Met
740208769PRTArtificial sequenceSynthetic sequence 208Phe Pro Ser
Asp Val Gly Ala Asp Ala Leu Lys His Val Arg Met Leu1 5 10 15Gln Pro
Arg Leu Thr Asp Glu Val Arg Lys Val Ala Leu Thr Arg Ala 20 25 30Pro
Ser Asp Arg Pro Ala Leu Ala Arg Phe Ala Ala Val Ala Gln Asp 35 40
45Gly Leu Ala Phe Val Arg His Leu Asn Val Ser Ala Asn His Asp Ser
50 55 60Asn Cys Thr Phe Pro Arg Asp Pro Arg Asp Pro Arg Arg Gly Pro
Cys65 70 75 80Glu Pro Asn Pro Cys Ala Phe Leu Arg Glu Val Trp Gly
Phe Arg Ile 85 90 95Val Ala Arg Gly Asn Glu Arg Ala Leu Ser Tyr Arg
Arg Gly Leu Ala 100 105
110Gly Cys Lys Ser Cys Val Gln Ser Thr Gly Phe Pro Ser Val Pro Phe
115 120 125His Arg Ile Gly Ala Asp Asp Cys Met Arg Lys Leu His Glu
Ile Leu 130 135 140Lys Ala Arg Asn Trp Arg Leu Leu Ala Arg Asn Ile
Gly Arg Glu Arg145 150 155 160Glu Ala Asp Pro Leu Leu Thr Glu Leu
Ser Glu Tyr Leu Leu Val Asp 165 170 175Ala Arg Thr Tyr Pro Asp Gly
Ala Ala Pro Asn Ser Gly Arg Leu Ala 180 185 190Glu Asn Val Ile Lys
Arg Ala Ala Lys Lys Phe Arg Asp Glu Gly Met 195 200 205Arg Asp Ile
His Ala Gln Leu Arg Val His Ser Arg Glu Gly Lys Val 210 215 220Pro
Lys Gly Arg Leu Gln Arg Leu Arg Arg Ile Glu Arg Lys His Arg225 230
235 240Ala Ile His Ala Leu Asp Pro Gly Pro Ser Trp Glu Ala Glu Gly
Ser 245 250 255Ala Arg Ala Glu Val Gln Gly Val Ala Val Tyr Arg Ser
Gln Leu Leu 260 265 270Arg Val Gly His His Thr Gln Gln Ile Glu Pro
Val Gly Ile Val Ala 275 280 285Arg Thr Leu Phe Gly Val Gly Arg Thr
Asp Leu Asp Val Ala Val Ser 290 295 300Val Leu Gly Ala Pro Leu Thr
Lys Arg Lys Lys Gly Ser Lys Thr Leu305 310 315 320Glu Ser Thr Glu
Asp Phe Arg Ile Ala Lys Ala Arg Glu Thr Arg Ala 325 330 335Glu Asp
Lys Ile Glu Val Ala Phe Val Leu Tyr Pro Thr Ala Ser Leu 340 345
350Leu Arg Asp Glu Ile Pro Lys Asp Ala Phe Pro Ala Met Arg Ile Asp
355 360 365Arg Phe Leu Leu Lys Val Gly Ser Val Gln Ala Asp Arg Glu
Ile Leu 370 375 380Leu Gln Asp Asp Tyr Tyr Arg Phe Gly Asp Ala Glu
Val Lys Ala Gly385 390 395 400Lys Asn Lys Gly Arg Thr Val Thr Arg
Pro Val Lys Val Pro Arg Leu 405 410 415Gln Ala Leu Arg Pro Asp Ala
Lys Phe Arg Val Asn Val Trp Ala Asp 420 425 430Pro Phe Gly Ala Gly
Asp Ser Pro Gly Thr Leu Leu Arg Leu Glu Val 435 440 445Ser Gly Val
Thr Arg Arg Ser Gln Pro Leu Arg Leu Leu Arg Tyr Gly 450 455 460Gln
Pro Ser Thr Gln Pro Ala Asn Phe Leu Cys Trp Arg Pro His Arg465 470
475 480Val Pro Asp Pro Met Thr Phe Thr Pro Arg Gln Lys Phe Gly Glu
Arg 485 490 495Arg Lys Asn Arg Arg Thr Arg Arg Pro Arg Val Phe Glu
Arg Leu Tyr 500 505 510Gln Val His Ile Lys His Leu Ala His Leu Glu
Pro Asn Arg Lys Trp 515 520 525Phe Glu Glu Ala Arg Val Ser Ala Gln
Lys Trp Ala Lys Ala Arg Ala 530 535 540Ile Arg Arg Lys Gly Ala Glu
Asp Ile Pro Val Val Ala Pro Pro Ala545 550 555 560Lys Arg Arg Trp
Ala Ala Leu Gln Pro Asn Ala Glu Leu Trp Asp Leu 565 570 575Tyr Ala
His Asp Arg Glu Ala Arg Lys Arg Phe Arg Gly Gly Arg Ala 580 585
590Ala Glu Gly Glu Glu Phe Lys Pro Arg Leu Asn Leu Tyr Leu Ala His
595 600 605Glu Pro Glu Ala Glu Trp Glu Ser Lys Arg Asp Arg Trp Glu
Arg Tyr 610 615 620Glu Lys Lys Trp Thr Ala Val Leu Glu Glu His Ser
Arg Met Cys Ala625 630 635 640Val Ala Asp Arg Thr Leu Pro Gln Phe
Leu Ser Asp Pro Leu Gly Ala 645 650 655Arg Met Asp Asp Lys Asp Tyr
Ala Phe Val Gly Lys Ser Ala Leu Ala 660 665 670Val Ala Glu Ala Phe
Val Glu Glu Gly Thr Val Glu Arg Ala Gln Gly 675 680 685Asn Cys Ser
Ile Thr Ala Lys Lys Lys Phe Ala Ser Asn Ala Ser Arg 690 695 700Lys
Arg Leu Ser Val Ala Asn Leu Leu Asp Val Ser Asp Lys Ala Asp705 710
715 720Arg Ala Leu Val Phe Gln Ala Val Arg Gln Tyr Val Gln Arg Gln
Ala 725 730 735Glu Asn Gly Gly Val Glu Gly Arg Arg Met Ala Phe Leu
Arg Lys Leu 740 745 750Leu Ala Pro Leu Arg Gln Asn Phe Val Cys His
Thr Arg Trp Leu His 755 760 765Met209564PRTArtificial
sequenceSynthetic sequence 209Ala Ala Arg Lys Lys Lys Arg Gly Lys
Ile Gly Ile Thr Val Lys Ala1 5 10 15Lys Glu Lys Ser Pro Pro Ala Ala
Gly Pro Phe Met Ala Arg Lys Leu 20 25 30Val Asn Val Ala Ala Asn Val
Asp Gly Val Glu Val His Leu Cys Val 35 40 45Glu Cys Glu Ala Asp Ala
His Gly Ser Ala Ser Ala Arg Leu Leu Gly 50 55 60Gly Cys Arg Ser Cys
Thr Gly Ser Ile Gly Ala Glu Gly Arg Leu Met65 70 75 80Gly Ser Val
Asp Val Asp Arg Glu Arg Val Ile Ala Glu Pro Val His 85 90 95Thr Glu
Thr Glu Arg Leu Gly Pro Asp Val Lys Ala Phe Glu Ala Gly 100 105
110Thr Ala Glu Ser Lys Tyr Ala Ile Gln Arg Gly Leu Glu Tyr Trp Gly
115 120 125Val Asp Leu Ile Ser Arg Asn Arg Ala Arg Thr Val Arg Lys
Met Glu 130 135 140Glu Ala Asp Arg Pro Glu Ser Ser Thr Met Glu Lys
Thr Ser Trp Asp145 150 155 160Glu Ile Ala Ile Lys Thr Tyr Ser Gln
Ala Tyr His Ala Ser Glu Asn 165 170 175His Leu Phe Trp Glu Arg Gln
Arg Arg Val Arg Gln His Ala Leu Ala 180 185 190Leu Phe Arg Arg Ala
Arg Glu Arg Asn Arg Gly Glu Ser Pro Leu Gln 195 200 205Ser Thr Gln
Arg Pro Ala Pro Leu Val Leu Ala Ala Leu His Ala Glu 210 215 220Ala
Ala Ala Ile Ser Gly Arg Ala Arg Ala Glu Tyr Val Leu Arg Gly225 230
235 240Pro Ser Ala Asn Val Arg Ala Ala Ala Ala Asp Ile Asp Ala Lys
Pro 245 250 255Leu Gly His Tyr Lys Thr Pro Ser Pro Lys Val Ala Arg
Gly Phe Pro 260 265 270Val Lys Arg Asp Leu Leu Arg Ala Arg His Arg
Ile Val Gly Leu Ser 275 280 285Arg Ala Tyr Phe Lys Pro Ser Asp Val
Val Arg Gly Thr Ser Asp Ala 290 295 300Ile Ala His Val Ala Gly Arg
Asn Ile Gly Val Ala Gly Gly Lys Pro305 310 315 320Lys Glu Ile Glu
Lys Thr Phe Thr Leu Pro Phe Val Ala Tyr Trp Glu 325 330 335Asp Val
Asp Arg Val Val His Cys Ser Ser Phe Lys Ala Asp Gly Pro 340 345
350Trp Val Arg Asp Gln Arg Ile Lys Ile Arg Gly Val Ser Ser Ala Val
355 360 365Gly Thr Phe Ser Leu Tyr Gly Leu Asp Val Ala Trp Ser Lys
Pro Thr 370 375 380Ser Phe Tyr Ile Arg Cys Ser Asp Ile Arg Lys Lys
Phe His Pro Lys385 390 395 400Gly Phe Gly Pro Met Lys His Trp Arg
Gln Trp Ala Lys Glu Leu Asp 405 410 415Arg Leu Thr Glu Gln Arg Ala
Ser Cys Val Val Arg Ala Leu Gln Asp 420 425 430Asp Glu Glu Leu Leu
Gln Thr Met Glu Arg Gly Gln Arg Tyr Tyr Asp 435 440 445Val Phe Ser
Cys Ala Ala Thr His Ala Thr Arg Gly Glu Ala Asp Pro 450 455 460Ser
Gly Gly Cys Ser Arg Cys Glu Leu Val Ser Cys Gly Val Ala His465 470
475 480Lys Val Thr Lys Lys Ala Lys Gly Asp Thr Gly Ile Glu Ala Val
Ala 485 490 495Val Ala Gly Cys Ser Leu Cys Glu Ser Lys Leu Val Gly
Pro Ser Lys 500 505 510Pro Arg Val His Arg Gln Met Ala Ala Leu Arg
Gln Ser His Ala Leu 515 520 525Asn Tyr Leu Arg Arg Leu Gln Arg Glu
Trp Glu Ala Leu Glu Ala Val 530 535 540Gln Ala Pro Thr Pro Tyr Leu
Arg Phe Lys Tyr Ala Arg His Leu Glu545 550 555 560Val Arg Ser
Met210565PRTArtificial sequenceSynthetic sequence 210Ala Ala Lys
Lys Lys Lys Gln Arg Gly Lys Ile Gly Ile Ser Val Lys1 5 10 15Pro Lys
Glu Gly Ser Ala Pro Pro Ala Asp Gly Pro Phe Met Ala Arg 20 25 30Lys
Leu Val Asn Val Ala Ala Asn Val Asp Gly Val Glu Val Asn Leu 35 40
45Cys Ile Glu Cys Glu Ala Asp Ala His Gly Ser Ala Pro Ala Arg Leu
50 55 60Leu Gly Gly Cys Lys Ser Cys Thr Gly Ser Ile Gly Ala Glu Gly
Arg65 70 75 80Leu Met Gly Ser Val Asp Val Asp Arg Ala Asp Ala Ile
Ala Lys Pro 85 90 95Val Asn Thr Glu Thr Glu Lys Leu Gly Pro Asp Val
Gln Ala Phe Glu 100 105 110Ala Gly Thr Ala Glu Thr Lys Tyr Ala Leu
Gln Arg Gly Leu Glu Tyr 115 120 125Trp Gly Val Asp Leu Ile Ser Arg
Asn Arg Ser Arg Thr Val Arg Arg 130 135 140Thr Glu Glu Gly Gln Pro
Glu Ser Ala Thr Met Glu Lys Thr Ser Trp145 150 155 160Asp Glu Ile
Ala Ile Lys Ser Tyr Thr Arg Ala Tyr His Ala Ser Glu 165 170 175Asn
His Leu Phe Trp Glu Arg Gln Arg Arg Val Arg Gln His Ala Leu 180 185
190Ala Leu Phe Lys Arg Ala Lys Glu Arg Asn Arg Gly Asp Ser Thr Leu
195 200 205Pro Arg Glu Pro Gly His Gly Leu Val Ala Ile Ala Ala Leu
Ala Cys 210 215 220Glu Ala Tyr Ala Val Gly Gly Arg Asn Leu Ala Glu
Thr Val Val Arg225 230 235 240Gly Pro Thr Phe Gly Thr Ala Arg Ala
Val Arg Asp Val Glu Ile Ala 245 250 255Ser Leu Gly Arg Tyr Lys Thr
Pro Ser Pro Lys Val Ala His Gly Ser 260 265 270Pro Val Lys Arg Asp
Phe Leu Arg Ala Arg His Arg Ile Val Gly Leu 275 280 285Ala Arg Ala
Tyr Tyr Arg Pro Ser Asp Val Val Arg Gly Thr Ser Asp 290 295 300Ala
Ile Ala His Val Ala Gly Arg Asn Ile Gly Val Ala Gly Gly Lys305 310
315 320Pro Arg Ala Val Glu Ala Val Phe Thr Leu Pro Phe Val Ala Tyr
Trp 325 330 335Glu Asp Val Asp Arg Val Val His Cys Ser Ser Phe Gln
Val Ser Ala 340 345 350Pro Trp Asn Arg Asp Gln Arg Met Lys Ile Ala
Gly Val Thr Thr Ala 355 360 365Ala Gly Thr Phe Ser Leu His Gly Gly
Glu Leu Lys Trp Ala Lys Pro 370 375 380Thr Ser Phe Tyr Ile Arg Cys
Ser Asp Thr Arg Arg Lys Phe Arg Pro385 390 395 400Lys Gly Phe Gly
Pro Met Lys Arg Trp Arg Gln Trp Ala Lys Asp Leu 405 410 415Asp Arg
Leu Val Glu Gln Arg Ala Ser Cys Val Val Arg Ala Leu Gln 420 425
430Asp Asp Ala Ala Leu Leu Glu Thr Met Glu Arg Gly Gln Arg Tyr Tyr
435 440 445Asp Val Phe Ala Cys Ala Val Thr His Ala Thr Arg Gly Glu
Ala Asp 450 455 460Arg Leu Ala Gly Cys Ser Arg Cys Ala Leu Thr Pro
Cys Gln Glu Ala465 470 475 480His Arg Val Thr Thr Lys Pro Arg Gly
Asp Ala Gly Val Glu Gln Val 485 490 495Gln Thr Ser Asp Cys Ser Leu
Cys Glu Gly Lys Leu Val Gly Pro Ser 500 505 510Lys Pro Arg Leu His
Arg Thr Leu Thr Leu Leu Arg Gln Glu His Gly 515 520 525Leu Asn Tyr
Leu Arg Arg Leu Gln Arg Glu Trp Glu Ser Leu Glu Ala 530 535 540Val
Gln Val Pro Thr Pro Tyr Leu Arg Phe Lys Tyr Ala Arg His Leu545 550
555 560Glu Val Arg Ser Met 565211499PRTArtificial sequenceSynthetic
sequence 211Thr Asp Ser Gln Ser Glu Ser Val Pro Glu Val Val Tyr Ala
Leu Thr1 5 10 15Gly Gly Glu Val Pro Gly Arg Val Pro Pro Asp Gly Gly
Ser Ala Glu 20 25 30Gly Ala Arg Asn Ala Pro Thr Gly Leu Arg Lys Gln
Arg Gly Lys Ile 35 40 45Lys Ile Ser Ala Lys Pro Ser Lys Pro Gly Ser
Pro Ala Ser Ser Leu 50 55 60Ala Arg Thr Leu Val Asn Glu Ala Ala Asn
Val Asp Gly Val Gln Ser65 70 75 80Ser Gly Cys Ala Thr Cys Arg Met
Arg Ala Asn Gly Ser Ala Pro Arg 85 90 95Ala Leu Pro Ile Gly Cys Val
Ala Cys Ala Ser Ser Ile Gly Arg Ala 100 105 110Pro Gln Glu Glu Thr
Val Cys Ala Leu Pro Thr Thr Gln Gly Pro Asp 115 120 125Val Arg Leu
Leu Glu Gly Gly His Ala Leu Arg Lys Tyr Asp Ile Gln 130 135 140Arg
Ala Leu Glu Tyr Trp Gly Val Asp Leu Ile Gly Arg Asn Leu Asp145 150
155 160Arg Gln Ala Gly Arg Gly Met Glu Pro Ala Glu Gly Ala Thr Ala
Thr 165 170 175Met Lys Arg Val Ser Met Asp Glu Leu Ala Val Leu Asp
Phe Gly Lys 180 185 190Ser Tyr Tyr Ala Ser Glu Gln His Leu Phe Ala
Ala Arg Gln Arg Arg 195 200 205Val Arg Gln His Ala Lys Ala Leu Lys
Ile Arg Ala Lys His Ala Asn 210 215 220Arg Ser Gly Ser Val Lys Arg
Ala Leu Asp Arg Ser Arg Lys Gln Val225 230 235 240Thr Ala Leu Ala
Arg Glu Phe Phe Lys Pro Ser Asp Val Val Arg Gly 245 250 255Asp Ser
Asp Ala Leu Ala His Val Val Gly Arg Asn Leu Gly Val Ser 260 265
270Arg His Pro Ala Arg Glu Ile Pro Gln Thr Phe Thr Leu Pro Leu Cys
275 280 285Ala Tyr Trp Glu Asp Val Asp Arg Val Ile Ser Cys Ser Ser
Leu Leu 290 295 300Ala Gly Glu Pro Phe Ala Arg Asp Gln Glu Ile Arg
Ile Glu Gly Val305 310 315 320Ser Ser Ala Leu Gly Ser Leu Arg Leu
Tyr Arg Gly Ala Ile Glu Trp 325 330 335His Lys Pro Thr Ser Leu Tyr
Ile Arg Cys Ser Asp Thr Arg Arg Lys 340 345 350Phe Arg Pro Arg Gly
Gly Leu Lys Lys Arg Trp Arg Gln Trp Ala Lys 355 360 365Asp Leu Asp
Arg Leu Val Glu Gln Arg Ala Cys Cys Ile Val Arg Ser 370 375 380Leu
Gln Ala Asp Val Glu Leu Leu Gln Thr Met Glu Arg Ala Gln Arg385 390
395 400Phe Tyr Asp Val His Asp Cys Ala Ala Thr His Val Gly Pro Val
Ala 405 410 415Val Arg Cys Ser Pro Cys Ala Gly Lys Gln Phe Asp Trp
Asp Arg Tyr 420 425 430Arg Leu Leu Ala Ala Leu Arg Gln Glu His Ala
Leu Asn Tyr Leu Arg 435 440 445Arg Leu Gln Arg Glu Trp Glu Ser Leu
Glu Ala Gln Gln Val Lys Met 450 455 460Pro Tyr Leu Arg Phe Lys Tyr
Ala Arg Lys Leu Glu Val Ser Gly Pro465 470 475 480Leu Ile Gly Leu
Glu Val Arg Arg Glu Pro Ser Met Gly Thr Ala Ile 485 490 495Ala Glu
Met212358PRTArtificial sequenceSynthetic sequence 212Ala Gly Thr
Ala Gly Arg Arg His Gly Ser Leu Gly Ala Arg Arg Ser1 5 10 15Ile Asn
Ile Ala Gly Val Thr Asp Arg His Gly Arg Trp Gly Cys Glu 20 25 30Ser
Cys Val Tyr Thr Arg Asp Gln Ala Gly Asn Arg Ala Arg Cys Ala 35 40
45Pro Cys Asp Gln Ser Thr Tyr Ala Pro Asp Val Gln Glu Val Thr Ile
50 55 60Gly Gln Arg Gln Ala Lys Tyr Thr Ile Phe Leu Thr Leu Gln Ser
Phe65 70 75 80Ser Trp Thr Asn Thr Met Arg Asn Asn Lys Arg Ala Ala
Ala Gly Arg 85 90 95Ser Lys Arg Thr Thr Gly Lys Arg Ile Gly Gln Leu
Ala Glu Ile Lys 100 105 110Ile Thr Gly Val Gly Leu Ala His Ala His
Asn Val Ile Gln Arg Ser 115 120 125Leu Gln His Asn Ile Thr Lys Met
Trp Arg Ala Glu Lys Gly Lys Ser 130 135 140Lys Arg Val Ala Arg Leu
Lys Lys Ala Lys Gln Leu Thr Lys Arg Arg145 150 155 160Ala Tyr Phe
Arg Arg Arg Met Ser Arg Gln Ser Arg
Gly Asn Gly Phe 165 170 175Phe Arg Thr Gly Lys Gly Gly Ile His Ala
Val Ala Pro Val Lys Ile 180 185 190Gly Leu Asp Val Gly Met Ile Ala
Ser Gly Ser Ser Glu Pro Ala Asp 195 200 205Glu Gln Thr Val Thr Leu
Asp Ala Ile Trp Lys Gly Arg Lys Lys Lys 210 215 220Ile Arg Leu Ile
Gly Ala Lys Gly Glu Leu Ala Val Ala Ala Cys Arg225 230 235 240Phe
Arg Glu Gln Gln Thr Lys Gly Asp Lys Cys Ile Pro Leu Ile Leu 245 250
255Gln Asp Gly Glu Val Arg Trp Asn Gln Asn Asn Trp Gln Cys His Pro
260 265 270Lys Lys Leu Val Pro Leu Cys Gly Leu Glu Val Ser Arg Lys
Phe Val 275 280 285Ser Gln Ala Asp Arg Leu Ala Gln Asn Lys Val Ala
Ser Pro Leu Ala 290 295 300Ala Arg Phe Asp Lys Thr Ser Val Lys Gly
Thr Leu Val Glu Ser Asp305 310 315 320Phe Ala Ala Val Leu Val Asn
Val Thr Ser Ile Tyr Gln Gln Cys His 325 330 335Ala Met Leu Leu Arg
Ser Gln Glu Pro Thr Pro Ser Leu Arg Val Gln 340 345 350Arg Thr Ile
Thr Ser Met 355213369PRTArtificial sequenceSynthetic sequence
213Gly Val Arg Phe Ser Pro Ala Gln Ser Gln Val Phe Phe Arg Thr Val1
5 10 15Ile Pro Gln Ser Val Glu Ala Arg Phe Ala Ile Asn Met Ala Ala
Ile 20 25 30His Asp Ala Ala Gly Ala Phe Gly Cys Ser Val Cys Arg Phe
Glu Asp 35 40 45Arg Thr Pro Arg Asn Ala Lys Ala Val His Gly Cys Ser
Pro Cys Thr 50 55 60Arg Ser Thr Asn Arg Pro Asp Val Phe Val Leu Pro
Val Gly Ala Ile65 70 75 80Lys Ala Lys Tyr Asp Val Phe Met Arg Leu
Leu Gly Phe Asn Trp Thr 85 90 95His Leu Asn Arg Arg Gln Ala Lys Arg
Val Thr Val Arg Asp Arg Ile 100 105 110Gly Gln Leu Asp Glu Leu Ala
Ile Ser Met Leu Thr Gly Lys Ala Lys 115 120 125Ala Val Leu Lys Lys
Ser Ile Cys His Asn Val Asp Lys Ser Phe Lys 130 135 140Ala Met Arg
Gly Ser Leu Lys Lys Leu His Arg Lys Ala Ser Lys Thr145 150 155
160Gly Lys Ser Gln Leu Arg Ala Lys Leu Ser Asp Leu Arg Glu Arg Thr
165 170 175Asn Thr Thr Gln Glu Gly Ser His Val Glu Gly Asp Ser Asp
Val Ala 180 185 190Leu Asn Lys Ile Gly Leu Asp Val Gly Leu Val Gly
Lys Pro Asp Tyr 195 200 205Pro Ser Glu Glu Ser Val Glu Val Val Val
Cys Leu Tyr Phe Val Gly 210 215 220Lys Val Leu Ile Leu Asp Ala Gln
Gly Arg Ile Arg Asp Met Arg Ala225 230 235 240Lys Gln Tyr Asp Gly
Phe Lys Ile Pro Ile Ile Gln Arg Gly Gln Leu 245 250 255Thr Val Leu
Ser Val Lys Asp Leu Gly Lys Trp Ser Leu Val Arg Gln 260 265 270Asp
Tyr Val Leu Ala Gly Asp Leu Arg Phe Glu Pro Lys Ile Ser Lys 275 280
285Asp Arg Lys Tyr Ala Glu Cys Val Lys Arg Ile Ala Leu Ile Thr Leu
290 295 300Gln Ala Ser Leu Gly Phe Lys Glu Arg Ile Pro Tyr Tyr Val
Thr Lys305 310 315 320Gln Val Glu Ile Lys Asn Ala Ser His Ile Ala
Phe Val Thr Glu Ala 325 330 335Ile Gln Asn Cys Ala Glu Asn Phe Arg
Glu Met Thr Glu Tyr Leu Met 340 345 350Lys Tyr Gln Glu Lys Ser Pro
Asp Leu Lys Val Leu Leu Thr Gln Leu 355 360
365Met214486PRTArtificial sequenceSynthetic sequence 214Arg Ala Val
Val Gly Lys Val Phe Leu Glu Gln Ala Arg Arg Ala Leu1 5 10 15Asn Leu
Ala Thr Asn Phe Gly Thr Asn His Arg Thr Gly Cys Asn Gly 20 25 30Cys
Tyr Val Thr Pro Gly Lys Leu Ser Ile Pro Gln Asp Gly Glu Lys 35 40
45Asn Ala Ala Gly Cys Thr Ser Cys Leu Met Lys Ala Thr Ala Ser Tyr
50 55 60Val Ser Tyr Pro Lys Pro Leu Gly Glu Lys Val Ala Lys Tyr Ser
Thr65 70 75 80Leu Asp Ala Leu Lys Gly Phe Pro Trp Tyr Ser Leu Arg
Leu Asn Leu 85 90 95Arg Pro Asn Tyr Arg Gly Lys Pro Ile Asn Gly Val
Gln Glu Val Ala 100 105 110Pro Val Ser Lys Phe Arg Leu Ala Glu Glu
Val Ile Gln Ala Val Gln 115 120 125Arg Tyr His Phe Thr Glu Leu Glu
Gln Ser Phe Pro Gly Gly Arg Arg 130 135 140Arg Leu Arg Glu Leu Arg
Ala Phe Tyr Thr Lys Glu Tyr Arg Arg Ala145 150 155 160Pro Glu Gln
Arg Gln His Val Val Asn Gly Asp Arg Asn Ile Val Val 165 170 175Val
Thr Val Leu His Glu Leu Gly Phe Ser Val Gly Met Phe Asn Glu 180 185
190Val Glu Leu Leu Pro Lys Thr Pro Ile Glu Cys Ala Val Asn Val Phe
195 200 205Ile Arg Gly Asn Arg Val Leu Leu Glu Val Arg Lys Pro Gln
Phe Asp 210 215 220Lys Glu Arg Leu Leu Val Glu Ser Leu Trp Lys Lys
Asp Ser Arg Arg225 230 235 240His Thr Ala Lys Trp Thr Pro Pro Asn
Asn Glu Gly Arg Ile Phe Thr 245 250 255Ala Glu Gly Trp Lys Asp Phe
Gln Leu Pro Leu Leu Leu Gly Ser Thr 260 265 270Ser Arg Ser Leu Arg
Ala Ile Glu Lys Glu Gly Phe Val Gln Leu Ala 275 280 285Pro Gly Arg
Asp Pro Asp Tyr Asn Asn Thr Ile Asp Glu Gln His Ser 290 295 300Gly
Arg Pro Phe Leu Pro Leu Tyr Leu Tyr Leu Gln Gly Thr Ile Ser305 310
315 320Gln Glu Tyr Cys Val Phe Ala Gly Thr Trp Val Ile Pro Phe Gln
Asp 325 330 335Gly Ile Ser Pro Tyr Ser Thr Lys Asp Thr Phe Gln Pro
Asp Leu Lys 340 345 350Arg Lys Ala Tyr Ser Leu Leu Leu Asp Ala Val
Lys His Arg Leu Gly 355 360 365Asn Lys Val Ala Ser Gly Leu Gln Tyr
Gly Arg Phe Pro Ala Ile Glu 370 375 380Glu Leu Lys Arg Leu Val Arg
Met His Gly Ala Thr Arg Lys Ile Pro385 390 395 400Arg Gly Glu Lys
Asp Leu Leu Lys Lys Gly Asp Pro Asp Thr Pro Glu 405 410 415Trp Trp
Leu Leu Glu Gln Tyr Pro Glu Phe Trp Arg Leu Cys Asp Ala 420 425
430Ala Ala Lys Arg Val Ser Gln Asn Val Gly Leu Leu Leu Ser Leu Lys
435 440 445Lys Gln Pro Leu Trp Gln Arg Arg Trp Leu Glu Ser Arg Thr
Arg Asn 450 455 460Glu Pro Leu Asp Asn Leu Pro Leu Ser Met Ala Leu
Thr Leu His Leu465 470 475 480Thr Asn Glu Glu Ala Leu
485215400PRTArtificial sequenceSynthetic sequence 215Ala Ala Val
Tyr Ser Lys Phe Tyr Ile Glu Asn His Phe Lys Met Gly1 5 10 15Ile Pro
Glu Thr Leu Ser Arg Ile Arg Gly Pro Ser Ile Ile Gln Gly 20 25 30Phe
Ser Val Asn Glu Asn Tyr Ile Asn Ile Ala Gly Val Gly Asp Arg 35 40
45Asp Phe Ile Phe Gly Cys Lys Lys Cys Lys Tyr Thr Arg Gly Lys Pro
50 55 60Ser Ser Lys Lys Ile Asn Lys Cys His Pro Cys Lys Arg Ser Thr
Tyr65 70 75 80Pro Glu Pro Val Ile Asp Val Arg Gly Ser Ile Ser Glu
Phe Lys Tyr 85 90 95Lys Ile Tyr Asn Lys Leu Lys Gln Glu Pro Asn Gln
Ser Ile Lys Gln 100 105 110Asn Thr Lys Gly Arg Met Asn Pro Ser Asp
His Thr Ser Ser Asn Asp 115 120 125Gly Ile Ile Ile Asn Gly Ile Asp
Asn Arg Ile Ala Tyr Asn Val Ile 130 135 140Phe Ser Ser Tyr Lys His
Leu Met Glu Lys Gln Ile Asn Leu Leu Arg145 150 155 160Asp Thr Thr
Lys Arg Lys Ala Arg Gln Ile Lys Lys Tyr Asn Asn Ser 165 170 175Gly
Lys Lys Lys His Ser Leu Arg Ser Gln Thr Lys Gly Asn Leu Lys 180 185
190Asn Arg Tyr His Met Leu Gly Met Phe Lys Lys Gly Ser Leu Thr Ile
195 200 205Thr Asn Glu Gly Asp Phe Ile Thr Ala Val Arg Lys Val Gly
Leu Asp 210 215 220Ile Ser Leu Tyr Lys Asn Glu Ser Leu Asn Lys Gln
Glu Val Glu Thr225 230 235 240Glu Leu Cys Leu Asn Ile Lys Trp Gly
Arg Thr Lys Ser Tyr Thr Val 245 250 255Ser Gly Tyr Ile Pro Leu Pro
Ile Asn Ile Asp Trp Lys Leu Tyr Leu 260 265 270Phe Glu Lys Glu Thr
Gly Leu Thr Leu Arg Leu Phe Gly Asn Lys Tyr 275 280 285Lys Ile Gln
Ser Lys Lys Phe Leu Ile Ala Gln Leu Phe Lys Pro Lys 290 295 300Arg
Pro Pro Cys Ala Asp Pro Val Val Lys Lys Ala Gln Lys Trp Ser305 310
315 320Ala Leu Asn Ala His Val Gln Gln Met Ala Gly Leu Phe Ser Asp
Ser 325 330 335His Leu Leu Lys Arg Glu Leu Lys Asn Arg Met His Lys
Gln Leu Asp 340 345 350Phe Lys Ser Leu Trp Val Gly Thr Glu Asp Tyr
Ile Lys Trp Phe Glu 355 360 365Glu Leu Ser Arg Ser Tyr Val Glu Gly
Ala Glu Lys Ser Leu Glu Phe 370 375 380Phe Arg Gln Asp Tyr Phe Cys
Phe Asn Tyr Thr Lys Gln Thr Thr Met385 390 395
400216666PRTArtificial sequenceSynthetic sequence 216Pro Gln Gln
Gln Arg Asp Leu Met Leu Met Ala Ala Asn Tyr Asp Gln1 5 10 15Asp Tyr
Gly Asn Gly Cys Gly Pro Cys Thr Val Val Ala Ser Ala Ala 20 25 30Tyr
Arg Pro Asp Pro Gln Ala Gln His Gly Cys Lys Arg His Leu Arg 35 40
45Thr Leu Gly Ala Ser Ala Val Thr His Val Gly Leu Gly Asp Arg Thr
50 55 60Ala Thr Ile Thr Ala Leu His Arg Leu Arg Gly Pro Ala Ala Leu
Ala65 70 75 80Ala Arg Ala Arg Ala Ala Gln Ala Ala Ser Ala Pro Met
Thr Pro Asp 85 90 95Thr Asp Ala Pro Asp Asp Arg Arg Arg Leu Glu Ala
Ile Asp Ala Asp 100 105 110Asp Val Val Leu Val Gly Ala His Arg Ala
Leu Trp Ser Ala Val Arg 115 120 125Arg Trp Ala Asp Asp Arg Arg Ala
Ala Leu Arg Arg Arg Leu His Ser 130 135 140Glu Arg Glu Trp Leu Leu
Lys Asp Gln Ile Arg Trp Ala Glu Leu Tyr145 150 155 160Thr Leu Ile
Glu Ala Ser Gly Thr Pro Pro Gln Gly Arg Trp Arg Asn 165 170 175Thr
Leu Gly Ala Leu Arg Gly Gln Ser Arg Trp Arg Arg Val Leu Ala 180 185
190Pro Thr Met Arg Ala Thr Cys Ala Glu Thr His Ala Glu Leu Trp Asp
195 200 205Ala Leu Ala Glu Leu Val Pro Glu Met Ala Lys Asp Arg Arg
Gly Leu 210 215 220Leu Arg Pro Pro Val Glu Ala Asp Ala Leu Trp Arg
Ala Pro Met Ile225 230 235 240Val Glu Gly Trp Arg Gly Gly His Ser
Val Val Val Asp Ala Val Ala 245 250 255Pro Pro Leu Asp Leu Pro Gln
Pro Cys Ala Trp Thr Ala Val Arg Leu 260 265 270Ser Gly Asp Pro Arg
Gln Arg Trp Gly Leu His Leu Ala Val Pro Pro 275 280 285Leu Gly Gln
Val Gln Pro Pro Asp Pro Leu Lys Ala Thr Leu Ala Val 290 295 300Ser
Met Arg His Arg Gly Gly Val Arg Val Arg Thr Leu Gln Ala Met305 310
315 320Ala Val Asp Ala Asp Ala Pro Met Gln Arg His Leu Gln Val Pro
Leu 325 330 335Thr Leu Gln Arg Gly Gly Gly Leu Gln Trp Gly Ile His
Ser Arg Gly 340 345 350Val Arg Arg Arg Glu Ala Arg Ser Met Ala Ser
Trp Glu Gly Pro Pro 355 360 365Ile Trp Thr Gly Leu Gln Leu Val Asn
Arg Trp Lys Gly Gln Gly Ser 370 375 380Ala Leu Leu Ala Pro Asp Arg
Pro Pro Asp Thr Pro Pro Tyr Ala Pro385 390 395 400Asp Ala Ala Val
Ala Pro Ala Gln Pro Asp Thr Lys Arg Ala Arg Arg 405 410 415Thr Leu
Lys Glu Ala Cys Thr Val Cys Arg Cys Ala Pro Gly His Met 420 425
430Arg Gln Leu Gln Val Thr Leu Thr Gly Asp Gly Thr Trp Arg Arg Phe
435 440 445Arg Leu Arg Ala Pro Gln Gly Ala Lys Arg Lys Ala Glu Val
Leu Lys 450 455 460Val Ala Thr Gln His Asp Glu Arg Ile Ala Asn Tyr
Thr Ala Trp Tyr465 470 475 480Leu Lys Arg Pro Glu His Ala Ala Gly
Cys Asp Thr Cys Asp Gly Asp 485 490 495Ser Arg Leu Asp Gly Ala Cys
Arg Gly Cys Arg Pro Leu Leu Val Gly 500 505 510Asp Gln Cys Phe Arg
Arg Tyr Leu Asp Lys Ile Glu Ala Asp Arg Asp 515 520 525Asp Gly Leu
Ala Gln Ile Lys Pro Lys Ala Gln Glu Ala Val Ala Ala 530 535 540Met
Ala Ala Lys Arg Asp Ala Arg Ala Gln Lys Val Ala Ala Arg Ala545 550
555 560Ala Lys Leu Ser Glu Ala Thr Gly Gln Arg Thr Ala Ala Thr Arg
Asp 565 570 575Ala Ser His Glu Ala Arg Ala Gln Lys Glu Leu Glu Ala
Val Ala Thr 580 585 590Glu Gly Thr Thr Val Arg His Asp Ala Ala Ala
Val Ser Ala Phe Gly 595 600 605Ser Trp Val Ala Arg Lys Gly Asp Glu
Tyr Arg His Gln Val Gly Val 610 615 620Leu Ala Asn Arg Leu Glu His
Gly Leu Arg Leu Gln Glu Leu Met Ala625 630 635 640Pro Asp Ser Val
Val Ala Asp Gln Gln Arg Ala Ser Gly His Ala Arg 645 650 655Val Gly
Tyr Arg Tyr Val Leu Thr Ala Met 660 665217560PRTArtificial
sequenceSynthetic sequence 217Ala Val Ala His Pro Val Gly Arg Gly
Asn Ala Gly Ser Pro Gly Ala1 5 10 15Arg Gly Pro Glu Glu Leu Pro Arg
Gln Leu Val Asn Arg Ala Ser Asn 20 25 30Val Thr Arg Pro Ala Thr Tyr
Gly Cys Ala Pro Cys Arg His Val Arg 35 40 45Leu Ser Ile Pro Lys Pro
Val Leu Thr Gly Cys Arg Ala Cys Glu Gln 50 55 60Thr Thr His Pro Ala
Pro Lys Arg Ala Val Arg Gly Gly Ala Asp Ala65 70 75 80Ala Lys Tyr
Asp Leu Ala Ala Phe Phe Ala Gly Trp Ala Ala Asp Leu 85 90 95Glu Gly
Arg Asn Arg Arg Arg Gln Val His Ala Pro Leu Asp Pro Gln 100 105
110Pro Asp Pro Asn His Glu Pro Ala Val Thr Leu Gln Lys Ile Asp Leu
115 120 125Ala Glu Val Ser Ile Glu Glu Phe Gln Arg Val Leu Ala Arg
Ser Val 130 135 140Lys His Arg His Asp Gly Arg Ala Ser Arg Glu Arg
Glu Lys Ala Arg145 150 155 160Ala Tyr Ala Gln Val Ala Lys Lys Arg
Arg Asn Ser His Ala His Gly 165 170 175Ala Arg Thr Arg Arg Ala Val
Arg Arg Gln Thr Arg Ala Val Arg Arg 180 185 190Ala His Arg Met Gly
Ala Asn Ser Gly Glu Ile Leu Val Ala Ser Gly 195 200 205Ala Glu Asp
Pro Val Pro Glu Ala Ile Asp His Ala Ala Gln Leu Arg 210 215 220Arg
Arg Ile Arg Ala Cys Ala Arg Asp Leu Glu Gly Leu Arg His Leu225 230
235 240Ser Arg Arg Tyr Leu Lys Thr Leu Glu Lys Pro Cys Arg Arg Pro
Arg 245 250 255Ala Pro Asp Leu Gly Arg Ala Arg Cys His Ala Leu Val
Glu Ser Leu 260 265 270Gln Ala Ala Glu Arg Glu Leu Glu Glu Leu Arg
Arg Cys Asp Ser Pro 275 280 285Asp Thr Ala Met Arg Arg Leu Asp Ala
Val Leu Ala Ala Ala Ala Ser 290 295 300Thr Asp Ala Thr Phe Ala Thr
Gly Trp Thr Val Val Gly Met Asp Leu305 310 315 320Gly Val Ala Pro
Arg Gly Ser Ala Ala Pro Glu Val
Ser Pro Met Glu 325 330 335Met Ala Ile Ser Val Phe Trp Arg Lys Gly
Ser Arg Arg Val Ile Val 340 345 350Ser Lys Pro Ile Ala Gly Met Pro
Ile Arg Arg His Glu Leu Ile Arg 355 360 365Leu Glu Gly Leu Gly Thr
Leu Arg Leu Asp Gly Asn His Tyr Thr Gly 370 375 380Ala Gly Val Thr
Lys Gly Arg Gly Leu Ser Glu Gly Thr Glu Pro Asp385 390 395 400Phe
Arg Glu Lys Ser Pro Ser Thr Leu Gly Phe Thr Leu Ser Asp Tyr 405 410
415Arg His Glu Ser Arg Trp Arg Pro Tyr Gly Ala Lys Gln Gly Lys Thr
420 425 430Ala Arg Gln Phe Phe Ala Ala Met Ser Arg Glu Leu Arg Ala
Leu Val 435 440 445Glu His Gln Val Leu Ala Pro Met Gly Pro Pro Leu
Leu Glu Ala His 450 455 460Glu Arg Arg Phe Glu Thr Leu Leu Lys Gly
Gln Asp Asn Lys Ser Ile465 470 475 480His Ala Gly Gly Gly Gly Arg
Tyr Val Trp Arg Gly Pro Pro Asp Ser 485 490 495Lys Lys Arg Pro Ala
Ala Asp Gly Asp Trp Phe Arg Phe Gly Arg Gly 500 505 510His Ala Asp
His Arg Gly Trp Ala Asn Lys Arg His Glu Leu Ala Ala 515 520 525Asn
Tyr Leu Gln Ser Ala Phe Arg Leu Trp Ser Thr Leu Ala Glu Ala 530 535
540Gln Glu Pro Thr Pro Tyr Ala Arg Tyr Lys Tyr Thr Arg Val Thr
Met545 550 555 560218404PRTArtificial sequenceSynthetic sequence
218Trp Asp Phe Leu Thr Leu Gln Val Tyr Glu Arg His Thr Ser Pro Glu1
5 10 15Val Cys Val Ala Gly Asn Ser Thr Lys Cys Ala Ser Gly Thr Arg
Lys 20 25 30Ser Asp His Thr His Gly Val Gly Val Lys Leu Gly Ala Gln
Glu Ile 35 40 45Asn Val Ser Ala Asn Asp Asp Arg Asp His Glu Val Gly
Cys Asn Ile 50 55 60Cys Val Ile Ser Arg Val Ser Leu Asp Ile Lys Gly
Trp Arg Tyr Gly65 70 75 80Cys Glu Ser Cys Val Gln Ser Thr Pro Glu
Trp Arg Ser Ile Val Arg 85 90 95Phe Asp Arg Asn His Lys Glu Ala Lys
Gly Glu Cys Leu Ser Arg Phe 100 105 110Glu Tyr Trp Gly Ala Gln Ser
Ile Ala Arg Ser Leu Lys Arg Asn Lys 115 120 125Leu Met Gly Gly Val
Asn Leu Asp Glu Leu Ala Ile Val Gln Asn Glu 130 135 140Asn Val Val
Lys Thr Ser Leu Lys His Leu Phe Asp Lys Arg Lys Asp145 150 155
160Arg Ile Gln Ala Asn Leu Lys Ala Val Lys Val Arg Met Arg Glu Arg
165 170 175Arg Lys Ser Gly Arg Gln Arg Lys Ala Leu Arg Arg Gln Cys
Arg Lys 180 185 190Leu Lys Arg Tyr Leu Arg Ser Tyr Asp Pro Ser Asp
Ile Lys Glu Gly 195 200 205Asn Ser Cys Ser Ala Phe Thr Lys Leu Gly
Leu Asp Ile Gly Ile Ser 210 215 220Pro Asn Lys Pro Pro Lys Ile Glu
Pro Lys Val Glu Val Val Phe Ser225 230 235 240Leu Phe Tyr Gln Gly
Ala Cys Asp Lys Ile Val Thr Val Ser Ser Pro 245 250 255Glu Ser Pro
Leu Pro Arg Ser Trp Lys Ile Lys Ile Asp Gly Ile Arg 260 265 270Ala
Leu Tyr Val Lys Ser Thr Lys Val Lys Phe Gly Gly Arg Thr Phe 275 280
285Arg Ala Gly Gln Arg Asn Asn Arg Arg Lys Val Arg Pro Pro Asn Val
290 295 300Lys Lys Gly Lys Arg Lys Gly Ser Arg Ser Gln Phe Phe Asn
Lys Phe305 310 315 320Ala Val Gly Leu Asp Ala Val Ser Gln Gln Leu
Pro Ile Ala Ser Val 325 330 335Gln Gly Leu Trp Gly Arg Ala Glu Thr
Lys Lys Ala Gln Thr Ile Cys 340 345 350Leu Lys Gln Leu Glu Ser Asn
Lys Pro Leu Lys Glu Ser Gln Arg Cys 355 360 365Leu Phe Leu Ala Asp
Asn Trp Val Val Arg Val Cys Gly Phe Leu Arg 370 375 380Ala Leu Ser
Gln Arg Gln Gly Pro Thr Pro Tyr Ile Arg Tyr Arg Tyr385 390 395
400Arg Cys Asn Met219392PRTArtificial sequenceSynthetic sequence
219Ala Arg Asn Val Gly Gln Arg Asn Ala Ser Arg Gln Ser Lys Arg Glu1
5 10 15Ser Ala Lys Ala Arg Ser Arg Arg Val Thr Gly Gly His Ala Ser
Val 20 25 30Thr Gln Gly Val Ala Leu Ile Asn Ala Ala Ala Asn Ala Asp
Arg Asp 35 40 45His Thr Thr Gly Cys Glu Pro Cys Thr Trp Glu Arg Val
Asn Leu Pro 50 55 60Leu Gln Glu Val Ile His Gly Cys Asp Ser Cys Thr
Lys Ser Ser Pro65 70 75 80Phe Trp Arg Asp Ile Lys Val Val Asn Lys
Gly Tyr Arg Glu Ala Lys 85 90 95Glu Glu Ile Met Arg Ile Ala Ser Gly
Ile Ser Ala Asp His Leu Ser 100 105 110Arg Ala Leu Ser His Asn Lys
Val Met Gly Arg Leu Asn Leu Asp Glu 115 120 125Val Cys Ile Leu Asp
Phe Arg Thr Val Leu Asp Thr Ser Leu Lys His 130 135 140Leu Thr Asp
Ser Arg Ser Asn Gly Ile Lys Glu His Ile Arg Ala Val145 150 155
160His Arg Lys Ile Arg Met Arg Arg Lys Ser Gly Lys Thr Ala Arg Ala
165 170 175Leu Arg Lys Gln Tyr Phe Ala Leu Arg Arg Gln Trp Lys Ala
Gly His 180 185 190Lys Pro Asn Ser Ile Arg Glu Gly Asn Ser Leu Thr
Ala Leu Arg Ala 195 200 205Val Gly Phe Asp Val Gly Val Ser Glu Gly
Thr Glu Pro Met Pro Ala 210 215 220Pro Gln Thr Glu Val Val Leu Ser
Val Phe Tyr Lys Gly Ser Ala Thr225 230 235 240Arg Ile Leu Arg Ile
Ser Ser Pro His Pro Ile Ala Lys Arg Ser Trp 245 250 255Lys Val Lys
Ile Ala Gly Ile Lys Ala Leu Lys Leu Ile Arg Arg Glu 260 265 270His
Asp Phe Ser Phe Gly Arg Glu Thr Tyr Asn Ala Ser Gln Arg Ala 275 280
285Glu Lys Arg Lys Phe Ser Pro His Ala Ala Arg Lys Asp Phe Phe Asn
290 295 300Ser Phe Ala Val Gln Leu Asp Arg Leu Ala Gln Gln Leu Cys
Val Ser305 310 315 320Ser Val Glu Asn Leu Trp Val Thr Glu Pro Gln
Gln Lys Leu Leu Thr 325 330 335Leu Ala Lys Asp Thr Ala Pro Tyr Gly
Ile Arg Glu Gly Ala Arg Phe 340 345 350Ala Asp Thr Arg Ala Arg Leu
Ala Trp Asn Trp Val Phe Arg Val Cys 355 360 365Gly Phe Thr Arg Ala
Leu His Gln Glu Gln Glu Pro Thr Pro Tyr Cys 370 375 380Arg Phe Thr
Trp Arg Ser Lys Met385 39022059DNAArtificial sequenceSynthetic
sequence 220ctacgccgat tatcttctga caactttcgc aagcggtgta aggtaaaaaa
tgcgggcac 5922159DNAArtificial sequenceSynthetic sequence
221gtgcccgcat tttttacctt acaccgcttg cgaaagttgt cagaagataa tcggcgtag
5922290DNAArtificial sequenceSynthetic sequence 222tttatatgtt
tctcctggag ataacgcaat cgtgacaact ttcgcaagcg gtgtaaggta 60gcaggcttcc
gaattccgcg tttttacggc 9022390DNAArtificial sequenceSynthetic
sequence 223gccgtaaaaa cgcggaattc ggaagcctgc taccttacac cgcttgcgaa
agttgtcacg 60attgcgttat ctccaggaga aacatataaa 9022456DNAArtificial
sequenceSynthetic sequence 224gatcttcagc tatacattat tgcaccaaca
ctaaggcaga gtatgtttac ctggac 5622556DNAArtificial sequenceSynthetic
sequence 225gatcttcagc tttgtattac tggaaggatg cttgcttgag gtgtaaaaac
ctggac 562265DNAArtificial sequenceSynthetic sequence 226ttttt
52276DNAArtificial sequenceSynthetic sequence 227tttttt
62287DNAArtificial sequenceSynthetic sequence 228ttttttt
72298DNAArtificial sequenceSynthetic sequence 229tttttttt
82309DNAArtificial sequenceSynthetic sequence 230ttttttttt
923110DNAArtificial sequenceSynthetic sequence 231tttttttttt
1023211DNAArtificial sequenceSynthetic sequence 232tttttttttt t
1123312DNAArtificial sequenceSynthetic sequence 233tttttttttt tt
1223455DNAArtificial sequenceSynthetic sequence 234gccggggtgg
tgcccatcct ggtcgagctg gacggcgacg taaacggcca caagc
5523555DNAArtificial sequenceSynthetic sequence 235gccggggtgg
tgcccatcct ggtcgagctg gacggcgacg tgctcggcca caagc
5523655DNAArtificial sequenceSynthetic sequence 236gccggggtgg
tgcccatcct ggtcgagctg gacggcgagc taaacggcca caagc
5523755DNAArtificial sequenceSynthetic sequence 237gccggggtgg
tgcccatcct ggtcgagctg gacggcctcg taaacggcca caagc
5523855DNAArtificial sequenceSynthetic sequence 238gccggggtgg
tgcccatcct ggtcgagctg gacgcggacg taaacggcca caagc
5523955DNAArtificial sequenceSynthetic sequence 239gccggggtgg
tgcccatcct ggtcgagctg gagcgcgacg taaacggcca caagc
5524055DNAArtificial sequenceSynthetic sequence 240gccggggtgg
tgcccatcct ggtcgagctg ctcggcgacg taaacggcca caagc
5524155DNAArtificial sequenceSynthetic sequence 241gccggggtgg
tgcccatcct ggtcgagcac gacggcgacg taaacggcca caagc
5524255DNAArtificial sequenceSynthetic sequence 242gccggggtgg
tgcccatcct ggtcgacgtg gacggcgacg taaacggcca caagc
5524355DNAArtificial sequenceSynthetic sequence 243gccggggtgg
tgcccatcct ggtcctgctg gacggcgacg taaacggcca caagc
5524455DNAArtificial sequenceSynthetic sequence 244gccggggtgg
tgcccatcct ggaggagctg gacggcgacg taaacggcca caagc
5524555DNAArtificial sequenceSynthetic sequence 245gccggggtgg
tgcccatcct cctcgagctg gacggcgacg taaacggcca caagc
5524634DNAArtificial sequenceSynthetic sequence 246gtgttaatac
aaaggtacag gaacaaagaa tttg 3424718DNAArtificial sequenceSynthetic
sequence 247caaagagaag cctcggcc 1824855DNAArtificial
sequenceSynthetic sequence 248tttattcaag gcaatcacta tcagctgtgg
aacacccagg taaactaaca caact 5524955DNAArtificial sequenceSynthetic
sequence 249tttattcaag gcaatcacta tcagctgtgg aacacccagg tgctctaaca
caact 5525055DNAArtificial sequenceSynthetic sequence 250tttattcaag
gcaatcacta tcagctgtgg aacacccacc taaactaaca caact
5525155DNAArtificial sequenceSynthetic sequence 251tttattcaag
gcaatcacta tcagctgtgg aacaccgtgg taaactaaca caact
5525255DNAArtificial sequenceSynthetic sequence 252tttattcaag
gcaatcacta tcagctgtgg aacaggcagg taaactaaca caact
5525355DNAArtificial sequenceSynthetic sequence 253tttattcaag
gcaatcacta tcagctgtgg aagtcccagg taaactaaca caact
5525455DNAArtificial sequenceSynthetic sequence 254tttattcaag
gcaatcacta tcagctgtgg ttcacccagg taaactaaca caact
5525555DNAArtificial sequenceSynthetic sequence 255tttattcaag
gcaatcacta tcagctgtcc aacacccagg taaactaaca caact
5525655DNAArtificial sequenceSynthetic sequence 256tttattcaag
gcaatcacta tcagctcagg aacacccagg taaactaaca caact
5525755DNAArtificial sequenceSynthetic sequence 257tttattcaag
gcaatcacta tcaggagtgg aacacccagg taaactaaca caact
5525855DNAArtificial sequenceSynthetic sequence 258tttattcaag
gcaatcacta tctcctgtgg aacacccagg taaactaaca caact
5525955DNAArtificial sequenceSynthetic sequence 259tttattcaag
gcaatcacta agagctgtgg aacacccagg taaactaaca caact
5526059DNAArtificial sequenceSynthetic sequence 260ctacgccgat
tatcttctga caactttcgc aagcggtgta aggcgaaaaa tgcgggcac
5926159DNAArtificial sequenceSynthetic sequence 261ctacgccgat
tatcttctga caactttcgc aagcggtgta aaataaaaaa tgcgggcac
5926259DNAArtificial sequenceSynthetic sequence 262ctacgccgat
tatcttctga caactttcgc aagcggtgtg gggtaaaaaa tgcgggcac
5926359DNAArtificial sequenceSynthetic sequence 263ctacgccgat
tatcttctga caactttcgc aagcggtaca aggtaaaaaa tgcgggcac
5926459DNAArtificial sequenceSynthetic sequence 264ctacgccgat
tatcttctga caactttcgc aagcgacgta aggtaaaaaa tgcgggcac
5926559DNAArtificial sequenceSynthetic sequence 265ctacgccgat
tatcttctga caactttcgc aagtagtgta aggtaaaaaa tgcgggcac
5926659DNAArtificial sequenceSynthetic sequence 266ctacgccgat
tatcttctga caactttcgc agacggtgta aggtaaaaaa tgcgggcac
5926759DNAArtificial sequenceSynthetic sequence 267ctacgccgat
tatcttctga caactttcgt gagcggtgta aggtaaaaaa tgcgggcac
5926859DNAArtificial sequenceSynthetic sequence 268ctacgccgat
tatcttctga caacttttac aagcggtgta aggtaaaaaa tgcgggcac
5926959DNAArtificial sequenceSynthetic sequence 269ctacgccgat
tatcttctga caactcccgc aagcggtgta aggtaaaaaa tgcgggcac
5927059DNAArtificial sequenceSynthetic sequence 270ctacgccgat
tatcttctga caatcttcgc aagcggtgta aggtaaaaaa tgcgggcac
5927159DNAArtificial sequenceSynthetic sequence 271ctacgccgat
tatcttctga cggctttcgc aagcggtgta aggtaaaaaa tgcgggcac
5927259DNAArtificial sequenceSynthetic sequence 272ctacgccgat
tatcttctgg taactttcgc aagcggtgta aggtaaaaaa tgcgggcac
5927349DNAArtificial sequenceSynthetic sequence 273tatcttctga
caactttcgc aagcggtgta aggtaaaaaa tgcgggcac 4927444DNAArtificial
sequenceSynthetic sequence 274tctgacaact ttcgcaagcg gtgtaaggta
aaaaatgcgg gcac 4427539DNAArtificial sequenceSynthetic sequence
275caactttcgc aagcggtgta aggtaaaaaa tgcgggcac 3927634DNAArtificial
sequenceSynthetic sequence 276ttcgcaagcg gtgtaaggta aaaaatgcgg gcac
3427754DNAArtificial sequenceSynthetic sequence 277ctacgccgat
tatcttctga caactttcgc aagcggtgta aggtaaaaaa tgcg
5427849DNAArtificial sequenceSynthetic sequence 278ctacgccgat
tatcttctga caactttcgc aagcggtgta aggtaaaaa 4927940DNAArtificial
sequenceSynthetic sequence 279ctacgccgat tatcttctga caactttcgc
aagcggtgta 4028035DNAArtificial sequenceSynthetic sequence
280ctacgccgat tatcttctga caactttcgc aagcg 3528130DNAArtificial
sequenceSynthetic sequence 281ctacgccgat tatcttctga caactttcgc
3028225DNAArtificial sequenceSynthetic sequence 282ctacgccgat
tatcttctga caact 2528320DNAArtificial sequenceSynthetic sequence
283ctacgccgat tatcttctga 2028434DNAArtificial sequenceSynthetic
sequence 284caactttcgc aagcggtgta aggtaaaaaa tgcg
3428563DNAArtificial sequenceSynthetic sequence 285atggaatgtg
gcgaacgctt tcaacgaaac aactttcgca agcggtgtaa ggtaaaaaat 60gcg
6328663DNAArtificial sequenceSynthetic sequence 286atggaatgtg
gcgaacgctt agttggaaac aactttcgca agcggtgtaa ggtaaaaaat 60gcg
6328763DNAArtificial sequenceSynthetic sequence 287atggaatgtg
gcgaagcgaa agttggaaac aactttcgca agcggtgtaa ggtaaaaaat 60gcg
6328863DNAArtificial sequenceSynthetic sequence 288atggaatgtg
cgcttgcgaa agttggaaac aactttcgca agcggtgtaa ggtaaaaaat 60gcg
6328963DNAArtificial sequenceSynthetic sequence 289atggatacac
cgcttgcgaa agttggaaac aactttcgca agcggtgtaa ggtaaaaaat 60gcg
6329063DNAArtificial sequenceSynthetic sequence 290taccttacac
cgcttgcgaa agttggaaac aactttcgca agcggtgtaa ggtaaaaaat 60gcg
6329140DNAArtificial sequenceSynthetic sequence 291gttttatctt
ctgctggtgg ttcgttcggt atttttaatg 4029240DNAArtificial
sequenceSynthetic sequence 292cattaaaaat accgaacgaa ccaccagcag
aagataaaac 4029343DNAArtificial sequenceSynthetic sequence
293gaccatttgc gaaatgtatc taatggtcaa actaaatcta ctc
4329443DNAArtificial sequenceSynthetic sequence 294gagtagattt
agtttgacca ttagatacat ttcgcaaatg gtc 4329537DNAArtificial
sequenceSynthetic sequence 295cttgcagaac ccggatagac gaatgaagga
atgcaac 3729637DNAArtificial sequenceSynthetic sequence
296cttgcaggcc ttgaatagag gagttaagga atgcaac 3729737DNAArtificial
sequenceSynthetic sequence 297gttgcacagt gctaattaga gaaactagga
atgcaac 3729837DNAArtificial sequenceSynthetic sequence
298ctagcatatt cagaacaaag ggattaagga atgcaac 3729937DNAArtificial
sequenceSynthetic sequence 299ctttcatatt cagaaactag gggttaagga
ctgcaac 3730037DNAArtificial sequenceSynthetic sequence
300gttgcatccc tacgtcgtga gcaccggtga gtgcaac 3730131DNAArtificial
sequenceSynthetic sequence 301ggaaaggaat cccctgaagg aaacgagggg g
3130236DNAArtificial sequenceSynthetic sequence
302gtgtccatca atcagatttg cgttggccgg tgcaat 3630337DNAArtificial
sequenceSynthetic sequence 303gtttcagcgc acgaattaac gagatgagag
atgcaac 37304500PRTArtificial sequenceSynthetic sequence 304Lys Glu
Pro Leu Asn Ile Gly Lys Thr Ala Lys Ala Val Phe Lys Glu1 5 10 15Ile
Asp Pro Thr Ser Leu Asn Arg Ala Ala Asn Tyr Asp Ala Ser Ile 20 25
30Glu Leu Asn Cys Lys Glu Cys Lys Phe Lys Pro Phe Lys Asn Val Lys
35 40 45Arg Tyr Glu Phe Asn Phe Tyr Asn Asn Trp Tyr Arg Cys Asn Pro
Asn 50 55 60Ser Cys Leu Gln Ser Thr Tyr Lys Ala Gln Val Arg Lys Val
Glu Ile65 70 75 80Gly Tyr Glu Lys Leu Lys Asn Glu Ile Leu Thr Gln
Met Gln Tyr Tyr 85 90 95Pro Trp Phe Gly Arg Leu Tyr Gln Asn Phe Phe
His Asp Glu Arg Asp 100 105 110Lys Met Thr Ser Leu Asp Glu Ile Gln
Val Ile Gly Val Gln Asn Lys 115 120 125Val Phe Phe Asn Thr Val Glu
Lys Ala Trp Arg Glu Ile Ile Lys Lys 130 135 140Arg Phe Lys Asp Asn
Lys Glu Thr Met Glu Thr Ile Pro Glu Leu Lys145 150 155 160His Ala
Ala Gly His Gly Lys Arg Lys Leu Ser Asn Lys Ser Leu Leu 165 170
175Arg Arg Arg Phe Ala Phe Val Gln Lys Ser Phe Lys Phe Val Asp Asn
180 185 190Ser Asp Val Ser Tyr Arg Ser Phe Ser Asn Asn Ile Ala Cys
Val Leu 195 200 205Pro Ser Arg Ile Gly Val Asp Leu Gly Gly Val Ile
Ser Arg Asn Pro 210 215 220Lys Arg Glu Tyr Ile Pro Gln Glu Ile Ser
Phe Asn Ala Phe Trp Lys225 230 235 240Gln His Glu Gly Leu Lys Lys
Gly Arg Asn Ile Glu Ile Gln Ser Val 245 250 255Gln Tyr Lys Gly Glu
Thr Val Lys Arg Ile Glu Ala Asp Thr Gly Glu 260 265 270Asp Lys Ala
Trp Gly Lys Asn Arg Gln Arg Arg Phe Thr Ser Leu Ile 275 280 285Leu
Lys Leu Val Pro Lys Gln Gly Gly Lys Lys Val Trp Lys Tyr Pro 290 295
300Glu Lys Arg Asn Glu Gly Asn Tyr Glu Tyr Phe Pro Ile Pro Ile
Glu305 310 315 320Phe Ile Leu Asp Ser Gly Glu Thr Ser Ile Arg Phe
Gly Gly Asp Glu 325 330 335Gly Glu Ala Gly Lys Gln Lys His Leu Val
Ile Pro Phe Asn Asp Ser 340 345 350Lys Ala Thr Pro Leu Ala Ser Gln
Gln Thr Leu Leu Glu Asn Ser Arg 355 360 365Phe Asn Ala Glu Val Lys
Ser Cys Ile Gly Leu Ala Ile Tyr Ala Asn 370 375 380Tyr Phe Tyr Gly
Tyr Ala Arg Asn Tyr Val Ile Ser Ser Ile Tyr His385 390 395 400Lys
Asn Ser Lys Asn Gly Gln Ala Ile Thr Ala Ile Tyr Leu Glu Ser 405 410
415Ile Ala His Asn Tyr Val Lys Ala Ile Glu Arg Gln Leu Gln Asn Leu
420 425 430Leu Leu Asn Leu Arg Asp Phe Ser Phe Met Glu Ser His Lys
Lys Glu 435 440 445Leu Lys Lys Tyr Phe Gly Gly Asp Leu Glu Gly Thr
Gly Gly Ala Gln 450 455 460Lys Arg Arg Glu Lys Glu Glu Lys Ile Glu
Lys Glu Ile Glu Gln Ser465 470 475 480Tyr Leu Pro Arg Leu Ile Arg
Leu Ser Leu Thr Lys Met Val Thr Lys 485 490 495Gln Val Glu Met
500305507PRTArtificial sequenceSynthetic sequence 305Glu Leu Ile
Val Asn Glu Asn Lys Asp Pro Leu Asn Ile Gly Lys Thr1 5 10 15Ala Lys
Ala Val Phe Lys Glu Ile Asp Pro Thr Ser Ile Asn Arg Ala 20 25 30Ala
Asn Tyr Asp Ala Ser Ile Glu Leu Ala Cys Lys Glu Cys Lys Phe 35 40
45Lys Pro Phe Asn Asn Thr Lys Arg His Asp Phe Ser Phe Tyr Ser Asn
50 55 60Trp His Arg Cys Ser Pro Asn Ser Cys Leu Gln Ser Thr Tyr Arg
Ala65 70 75 80Lys Ile Arg Lys Thr Glu Ile Gly Tyr Glu Lys Leu Lys
Asn Glu Ile 85 90 95Leu Asn Gln Met Gln Tyr Tyr Pro Trp Phe Gly Arg
Leu Tyr Gln Asn 100 105 110Phe Phe Asn Asp Gln Arg Asp Lys Met Thr
Ser Leu Asp Glu Ile Gln 115 120 125Val Thr Gly Val Gln Asn Lys Ile
Phe Phe Asn Thr Val Glu Lys Ala 130 135 140Trp Arg Glu Ile Ile Lys
Lys Arg Phe Arg Asp Asn Lys Glu Thr Met145 150 155 160Arg Thr Ile
Pro Asp Leu Lys Asn Lys Ser Gly His Gly Ser Arg Lys 165 170 175Leu
Ser Asn Lys Ser Leu Leu Arg Arg Arg Phe Ala Phe Ala Gln Lys 180 185
190Ser Phe Lys Leu Val Asp Asn Ser Asp Val Ser Tyr Arg Ala Phe Ser
195 200 205Asn Asn Val Ala Cys Val Leu Pro Ser Lys Ile Gly Val Asp
Ile Gly 210 215 220Gly Ile Ile Asn Lys Asp Leu Lys Arg Glu Tyr Ile
Pro Gln Glu Ile225 230 235 240Thr Phe Asn Val Phe Trp Lys Gln His
Asp Gly Leu Lys Lys Gly Arg 245 250 255Asn Ile Glu Ile His Ser Val
Gln Tyr Lys Gly Glu Ile Val Lys Arg 260 265 270Ile Glu Ala Asp Thr
Gly Glu Asp Lys Ala Trp Gly Lys Asn Arg Gln 275 280 285Arg Arg Phe
Thr Ser Leu Ile Leu Lys Ile Thr Pro Lys Gln Gly Gly 290 295 300Lys
Lys Ile Trp Lys Phe Pro Glu Lys Lys Asn Ala Ser Asp Tyr Glu305 310
315 320Tyr Phe Pro Ile Pro Ile Glu Phe Ile Leu Asp Asn Gly Asp Ala
Ser 325 330 335Ile Lys Phe Gly Gly Glu Glu Gly Glu Val Gly Lys Gln
Lys His Leu 340 345 350Leu Ile Pro Phe Asn Asp Ser Lys Ala Thr Pro
Leu Ser Ser Lys Gln 355 360 365Met Leu Leu Glu Thr Ser Arg Phe Asn
Ala Glu Val Lys Ser Thr Ile 370 375 380Gly Leu Ala Leu Tyr Ala Asn
Tyr Phe Val Ser Tyr Ala Arg Asn Tyr385 390 395 400Val Ile Lys Ser
Thr Tyr His Lys Asn Ser Lys Lys Gly Gln Ile Val 405 410 415Thr Glu
Ile Tyr Leu Glu Ser Ile Ser Gln Asn Phe Val Arg Ala Ile 420 425
430Gln Arg Gln Leu Gln Ser Leu Met Leu Asn Leu Lys Asp Trp Gly Phe
435 440 445Met Gln Thr His Lys Lys Glu Leu Lys Lys Tyr Phe Gly Ser
Asp Leu 450 455 460Glu Gly Ser Lys Gly Gly Gln Lys Arg Arg Glu Lys
Glu Glu Lys Ile465 470 475 480Glu Lys Glu Ile Glu Ala Ser Tyr Leu
Pro Arg Leu Ile Arg Leu Ser 485 490 495Leu Thr Lys Ser Val Thr Lys
Ala Glu Glu Met 500 505306529PRTArtificial sequenceSynthetic
sequence 306Pro Glu Glu Lys Thr Ser Lys Leu Lys Pro Asn Ser Ile Asn
Leu Ala1 5 10 15Ala Asn Tyr Asp Ala Asn Glu Lys Phe Asn Cys Lys Glu
Cys Lys Phe 20 25 30His Pro Phe Lys Asn Lys Lys Arg Tyr Glu Phe Asn
Phe Tyr Asn Asn 35 40 45Leu His Gly Cys Lys Ser Cys Thr Lys Ser Thr
Asn Asn Pro Ala Val 50 55 60Lys Arg Ile Glu Ile Gly Tyr Gln Lys Leu
Lys Phe Glu Ile Lys Asn65 70 75 80Gln Met Glu Ala Tyr Pro Trp Phe
Gly Arg Leu Arg Ile Asn Phe Tyr 85 90 95Ser Asp Glu Lys Arg Lys Met
Ser Glu Leu Asn Glu Met Gln Val Thr 100 105 110Gly Val Lys Asn Lys
Ile Phe Phe Asp Ala Ile Glu Cys Ala Trp Arg 115 120 125Glu Ile Leu
Lys Lys Arg Phe Arg Glu Ser Lys Glu Thr Leu Ile Thr 130 135 140Ile
Pro Lys Leu Lys Asn Lys Ala Gly His Gly Ala Arg Lys His Arg145 150
155 160Asn Lys Lys Leu Leu Ile Arg Arg Arg Ala Phe Met Lys Lys Asn
Phe 165 170 175His Phe Leu Asp Asn Asp Ser Ile Ser Tyr Arg Ser Phe
Ala Asn Asn 180 185 190Ile Ala Cys Val Leu Pro Ser Lys Val Gly Val
Asp Ile Gly Gly Ile 195 200 205Ile Ser Pro Asp Val Gly Lys Asp Ile
Lys Pro Val Asp Ile Ser Leu 210 215 220Asn Leu Met Trp Ala Ser Lys
Glu Gly Ile Lys Ser Gly Arg Lys Val225 230 235 240Glu Ile Tyr Ser
Thr Gln Tyr Asp Gly Asn Met Val Lys Lys Ile Glu 245 250 255Ala Glu
Thr Gly Glu Asp Lys Ser Trp Gly Lys Asn Arg Lys Arg Arg 260 265
270Gln Thr Ser Leu Leu Leu Ser Ile Pro Lys Pro Ser Lys Gln Val Gln
275 280 285Glu Phe Asp Phe Lys Glu Trp Pro Arg Tyr Lys Asp Ile Glu
Lys Lys 290 295 300Val Gln Trp Arg Gly Phe Pro Ile Lys Ile Ile Phe
Asp Ser Asn His305 310 315 320Asn Ser Ile Glu Phe Gly Thr Tyr Gln
Gly Gly Lys Gln Lys Val Leu 325 330 335Pro Ile Pro Phe Asn Asp Ser
Lys Thr Thr Pro Leu Gly Ser Lys Met 340 345 350Asn Lys Leu Glu Lys
Leu Arg Phe Asn Ser Lys Ile Lys Ser Arg Leu 355 360 365Gly Ser Ala
Ile Ala Ala Asn Lys Phe Leu Glu Ala Ala Arg Thr Tyr 370 375 380Cys
Val Asp Ser Leu Tyr His Glu Val Ser Ser Ala Asn Ala Ile Gly385 390
395 400Lys Gly Lys Ile Phe Ile Glu Tyr Tyr Leu Glu Ile Leu Ser Gln
Asn 405 410 415Tyr Ile Glu Ala Ala Gln Lys Gln Leu Gln Arg Phe Ile
Glu Ser Ile 420 425 430Glu Gln Trp Phe Val Ala Asp Pro Phe Gln Gly
Arg Leu Lys Gln Tyr 435 440 445Phe Lys Asp Asp Leu Lys Arg Ala Lys
Cys Phe Leu Cys Ala Asn Arg 450 455 460Glu Val Gln Thr Thr Cys Tyr
Ala Ala Val Lys Leu His Lys Ser Cys465 470 475 480Ala Glu Lys Val
Lys Asp Lys Asn Lys Glu Leu Ala Ile Lys Glu Arg 485 490 495Asn Asn
Lys Glu Asp Ala Val Ile Lys Glu Val Glu Ala Ser Asn Tyr 500 505
510Pro Arg Val Ile Arg Leu Lys Leu Thr Lys Thr Ile Thr Asn Lys Ala
515 520 525Met30735DNAArtificial sequenceSynthetic sequence
307cttttagaca gtttaaattc taaagggtat aaaac 3530836DNAArtificial
sequenceSynthetic sequence 308gtcgaaatgc ccgcgcgggg gcgtcgtacc
cgcgac 3630937DNAArtificial sequenceSynthetic sequence
309gttgcagcgg ccgacggagc gcgagcgtgg atgccac 3731035DNAArtificial
sequenceSynthetic sequence 310ctttagactt ctccggaagt cgaattaatg
gaaac 3531128DNAArtificial sequenceSynthetic sequence 311gggcgccccg
cgcgagcggg ggttgaag 28312136DNAArtificial SequenceSynthetic
sequence 312gaaggatgct tgcttgaggt gtagttgtca ttccttcatt cgtctattcg
ggttctgcaa 60ctatacatta ttgcaccaac actaaggcag agtatggttg cattccttca
ttcgtctatt 120cgggttctgc aacggg 136313102DNAArtificial
SequenceSynthetic sequence 313agaatgcttg cttgaggtgt agttgcattc
cttcattcgt ctattcgggt aattgcacca 60acactaaggc agagtatggt tgcattcctt
cattcgtcta gg 10231475DNAArtificial SequenceSynthetic sequence
314cgatgcttgc ttgaggtgta gttgcattgc accaacacta aggcagagta
tggttgcatt 60ccttcattcg tctag 7531590DNAArtificial
SequenceSynthetic sequence 315gatgcttgct tgaggtgtag ttgcattcct
tcattctgca ccaacactaa ggcagagtat 60ggttgcattc cttttcgggt tctgcaacgg
9031673DNAArtificial SequenceSynthetic sequence 316gcttgcttga
ggtgtagttg cattccttca ttccacctac actaaggcag agtatggttg 60cattccttca
ttc 7331773DNAArtificial SequenceSynthetic sequence 317gcttgcttga
ggtgtagttg cattccttca ttccacctac actaaggcag agtatggttg 60cattccttca
ttc 7331863DNAArtificial SequenceSynthetic sequence 318gcttgcttga
ggtgtagttg cattcgcaac actaaggcag agtatggttg cattccttca 60ttc
6331966DNAArtificial SequenceSynthetic sequence 319cttgcttgag
gtgtagttgc cgacactaag gcagagtatg gttgcattat tcgggttctg 60caacgg
6632069DNAArtificial SequenceSynthetic sequence 320cttgcttgag
gtgtagttgc attccttcat tcaaacacta aggcagagta tggttgcatt 60ccttcattc
6932173DNAArtificial SequenceSynthetic sequence 321ttgcttgagg
tgtagttgca ttccttcatt ctaacactaa ggcagagtat ggttgcattc 60cttcattcgt
cta 7332258DNAArtificial SequenceSynthetic sequence 322ttgcttgagg
tgtagttgca tccacactaa ggcagagtat ggttgcattc cttcattc
5832367DNAArtificial SequenceSynthetic sequence 323tgcttaaggt
gtagttgcat tccttcattc caacactaag gcagagtatg gttgcattcc 60ttcattc
6732466DNAArtificial SequenceSynthetic sequence 324tgcttgaggt
gtagttgcat tccttcattc cacactaagg cagagtatgg ttgcattcct 60tcattc
6632572DNAArtificial SequenceSynthetic sequence 325tgcttgaggt
gtagttgcat tccttcattc aacactaagg cagagtatgg ttgcattcct 60tcattcgtct
at 7232664DNAArtificial SequenceSynthetic sequence 326cttgaggtgt
agttgcattc cttcattcaa cactaaggca gagtatggtt gcattccttc 60attc
6432759DNAArtificial SequenceSynthetic sequence 327cttgaggtgt
agttgcattc cttaacacta aggcagagta tggttgcatt ccttcattc
5932863DNAArtificial SequenceSynthetic sequence 328ttgaggtgta
gttgcattcc ttcattcaac actaaggcag agtatggttg cattccttca 60ttc
6332962DNAArtificial SequenceSynthetic sequence 329tgaggtgtag
ttgcattcct tcattcaaca ctaaggcaga gtatggttgc attccttcat 60tc
6233061DNAArtificial SequenceSynthetic sequence 330tgaggtgtag
ttgcattcct tcgttcacac taaggcagag tatggttgca ttccttcatt 60c
6133161DNAArtificial SequenceSynthetic sequence 331tgaggtgtag
ttgcattcct tcattcacac taaggcagag tatggttgca ttccttcatt 60c
6133261DNAArtificial SequenceSynthetic sequence 332ctacgccgat
tatcttctga caactttcgc aagcggtgta aggtaaaaaa tgcgggcacc 60c
6133330DNAArtificial SequenceSynthetic sequence 333ggaatgcaac
taccttacac cgcttgcgaa 30334140DNAArtificial SequenceSynthetic
sequence 334cttcactgat aaagtggaga accgcttcac caaaagctgt cccttagggg
attagaactt 60gagtgaaggt gggctgcttg catcagccta atgtcgagaa gtgctttctt
cggaaagtaa 120ccctcgaaac aaattcattt 14033540DNAArtificial
SequenceSynthetic sequence 335gacgaatgaa ggaatgcaac taccttacac
cgcttgcgaa 4033640DNAArtificial SequenceSynthetic sequence
336gacgaatgaa ggaatgcaac ccttacaccg cttgcgaaag 4033740DNAArtificial
SequenceSynthetic sequence 337gacgaatgaa ggaatgcaac ttacaccgct
tgcgaaagtt 4033840DNAArtificial SequenceSynthetic sequence
338gacgaatgaa ggaatgcaac acaccgcttg cgaaagttgt 4033941DNAArtificial
SequenceSynthetic sequence 339gacgaatgaa ggaatgcaac cgtcgccgtc
cagctcgacc a 4134040DNAArtificial SequenceSynthetic sequence
340gacgaatgaa ggaatgcaac gatcgttacg ctaactatga
40341180DNAArtificial SequenceSynthetic sequence 341ttcactgata
aagtggagaa ccgcttcacc aaaagctgtc ccttagggga ttagaacttg 60agtgaaggtg
ggctgcttgc atcagcctaa tgtcgagaag tgctttcttc ggaaagtaac
120cctcgaaaca aattcatttg aaagaatgaa ggaatgcaac acttgacact
taatgctcaa 18034244DNAArtificial SequenceSynthetic sequence
342gggtaatttc tactaagtgt agatacttga cacttaatgc tcaa
4434345DNAArtificial SequenceSynthetic sequence 343gacgaatgaa
ggaatgcaac taccttacac cgcttgcgaa agttg 4534438DNAArtificial
SequenceSynthetic sequence 344gacgaatgaa ggaatgcaac taccttacac
cgcttgcg 3834536DNAArtificial SequenceSynthetic sequence
345gacgaatgaa ggaatgcaac taccttacac cgcttg 3634634DNAArtificial
SequenceSynthetic sequence 346gacgaatgaa ggaatgcaac
taccttacac cgct 3434732DNAArtificial SequenceSynthetic sequence
347gacgaatgaa ggaatgcaac taccttacac cg 3234830DNAArtificial
SequenceSynthetic sequence 348gacgaatgaa ggaatgcaac taccttacac
3034957DNAArtificial SequenceSynthetic sequence 349gttgcagaac
ccgaatagac gaatgaagga atgcaactac cttacaccgc ttgcgaa
5735037DNAArtificial SequenceSynthetic sequence 350gaatgaagga
atgcaactac cttacaccgc ttgcgaa 3735135DNAArtificial
SequenceSynthetic sequence 351atgaaggaat gcaactacct tacaccgctt
gcgaa 35352179DNAArtificial SequenceSynthetic sequence
352cttcactgat aaagtggaga accgcttcac caaaagctgt cccttagggg
attagaactt 60gagtgaaggt gggctgcttg catcagccta atgtcgagaa gtgctttctt
cggaaagtaa 120ccctcgaaac aaattcattt ttcctctcca attctgcaca
aaaaaaggtg agtccttat 179353110DNAArtificial SequenceSynthetic
sequence 353cttcactgat aaagtggaga accgcttcac caaaagctgt cccttagggg
attagaactt 60gagtgaaggt gggctgcttg catcagccta atgtcgagaa gtgctttctt
11035472DNAArtificial SequenceSynthetic sequence 354cttcactgat
aaagtggaga accgcttcac caaaagctgt cccttagggg attagaactt 60gagtgaaggt
gg 72355136DNAArtificial SequenceSynthetic sequence 355ttcactgata
aagtggagaa ccgcttcacc aaaagctgtc ccttagggga ttagaacttg 60agtgaaggtg
ggctgcttgc atcagcctaa tgtcgagaag tgctttcttc ggaaagtaac
120cctcgaaaca aattca 136356234DNAArtificial SequenceSynthetic
sequence 356ttcacacttc actgataaag tggagaaccg cttcaccaaa agctgtccct
taggggatta 60gaacttgagt gaaggtgggc tgcttgcatc agcctaatgt cgagaagtgc
tttcttcgga 120aagtaaccct cgaaacaaat tcatttttcc tctccaattc
tgcacaaaaa aaggtgagtc 180cttataaacc ggcgtgcaga acgccggctc
accttttttc ttcattcgat ttta 234357181DNAArtificial SequenceSynthetic
sequence 357cttcactgat aaagtggaga accgcttcac caaaagctgt cccttagggg
attagaactt 60gagtgaaggt gggctgcttg catcagccta atgtcgagaa gtgctttctt
cggaaagtaa 120ccctcgaaac aaattcattt gaaagaatga aggaatgcaa
ctaccttaca ccgcttgcga 180a 181358222DNAArtificial SequenceSynthetic
sequence 358cttcactgat aaagtggaga accgcttcac caaaagctgt cccttagggg
attagaactt 60gagtgaaggt gggctgcttg catcagccta atgtcgagaa gtgctttctt
cggaaagtaa 120ccctcgaaac aaattcattt ttcctctcca attctgcaca
agaaagttgc agaacccgaa 180tagacgaatg aaggaatgca actaccttac
accgcttgcg aa 22235945DNAArtificial SequenceSynthetic sequence
359gacgaatgaa ggaatgcaac taccgaacga accaccagca gaaga
4536045DNAArtificial SequenceSynthetic sequence 360gacgaatgaa
ggaatgcaac tcttctgctg gtggttcgtt cggta 4536143DNAArtificial
SequenceSynthetic sequence 361gacgaatgaa ggaatgcaac gtttgaccat
tagatacatt tcg 4336243DNAArtificial SequenceSynthetic sequence
362gacgaatgaa ggaatgcaac cgaaatgtat ctaatggtca aac
433637580DNAArtificial sequenceSynthetic sequence 363gcaaccgcac
ctgtggcgcc ggtgatgccg gccacgatgc gtccggcgta gaggatcgag 60atctcgatcc
cgcgaaatta atacgactca ctatagggag accacaacgg tttccctcta
120gtgccggctc cggagagctc tttaattaag cggccgccct gcaggactcg
agttctagaa 180ataattttgt ttaactttaa gaaggagata tacatatgaa
atcttctcac catcaccatc 240accatcacca tcaccatggt tcttctatga
aaatcgaaga aggtaaactg gtaatctgga 300ttaacggcga taaaggctat
aacggtctcg ctgaagtcgg taagaaattc gagaaagata 360ccggaattaa
agtcaccgtt gagcatccgg ataaactgga agagaaattc ccacaggttg
420cggcaactgg cgatggccct gacattatct tctgggcaca cgaccgcttt
ggtggctacg 480ctcaatctgg cctgttggct gaaatcaccc cggacaaagc
gttccaggac aagctgtatc 540cgtttacctg ggatgccgta cgttacaacg
gcaagctgat tgcttacccg atcgctgttg 600aagcgttatc gctgatttat
aacaaagatc tgctgccgaa cccgccaaaa acctgggaag 660agatcccggc
gctggataaa gaactgaaag cgaaaggtaa gagcgcgctg atgttcaacc
720tgcaagaacc gtacttcacc tggccgctga ttgctgctga cgggggttat
gcgttcaagt 780atgaaaacgg caagtacgac attaaagacg tgggcgtgga
taacgctggc gcgaaagcgg 840gtctgacctt cctggttgac ctgattaaaa
acaaacacat gaatgcagac accgattact 900ccatcgcaga agctgccttt
aataaaggcg aaacagcgat gaccatcaac ggcccgtggg 960catggtccaa
catcgacacc agcaaagtga attatggtgt aacggtactg ccgaccttca
1020agggtcaacc atccaaaccg ttcgttggcg tgctgagcgc aggtattaac
gccgccagtc 1080cgaacaaaga gctggcaaaa gagttcctcg aaaactatct
gctgactgat gaaggtctgg 1140aagcggttaa taaagacaaa ccgctgggtg
ccgtagcgct gaagtcttac gaggaagagt 1200tggcgaaaga tccacgtatt
gccgccacta tggaaaacgc ccagaaaggt gaaatcatgc 1260cgaacatccc
gcagatgtcc gctttctggt atgccgtgcg tactgcggtg atcaacgccg
1320ccagcggtcg tcagactgtc gatgaagccc tgaaagacgc gcagactaat
tcgagctcga 1380acaacaacaa caataacaat aacaacaacc tcgggatcga
ggaaaacctg tacttccaat 1440ccaatgcaat ggaagaaagc attattaccg
gtgtgaaatt caaactgcgc atcgataaag 1500aaaccaccaa aaaactgaac
gagtacttcg atgaatatgg caaagcaatt aacttcgccg 1560tgaagatcat
tcagaaagaa ctggcagatg atcgttttgc aggtaaagca aaactggacc
1620agaataaaaa cccgatcctg gatgaaaacg gcaaaaaaat ctatgaattc
ccggatgaat 1680tttgcagctg tggtaaacag gttaacaagt acgttaacaa
caaaccgttt tgccaagagt 1740gctataaaat ccgctttacc gaaaatggta
ttcgcaaacg tatgtatagc gccaaaggtc 1800gtaaagccga acataaaatc
aatatcctga acagcaccaa caagatcagc aaaacccatt 1860ttaactatgc
cattcgcgaa gccttcattc tggataaaag catcaaaaag cagcgcaaaa
1920aacgtaatga acgtctgcgt gaaagtaaaa aacgtctgca gcagtttatc
gatatgcgtg 1980atggtaaacg tgaaatttgc ccgaccatta aaggtcagaa
agtggatcgt tttattcatc 2040cgagctggat caccaaagat aaaaagctgg
aagattttcg cggttatacc ctgagcatta 2100tcaacagcaa aattaagatt
ctggatcgca acatcaaacg cgaagaaaaa agcctgaaag 2160aaaaaggcca
gatcatcttt aaagccaaac gtctgatgct ggataaatcc attcgttttg
2220ttggtgatcg caaagtgctg tttacaatta gtaaaaccct gccgaaagag
tatgaactgg 2280atctgccgag caaagaaaaa cggctgaatt ggctgaaaga
gaagatcgag attatcaaga 2340accagaaacc gaaatatgcc tatctgctgc
gcaaaaacat tgagagcgaa aaaaaaccga 2400actatgagta ctatctgcag
tacaccctgg aaattaaacc ggaactgaaa gatttttatg 2460atggtgccat
tggtattgac cgtggcatta atcatattgc cgtttgcacc tttattagca
2520acgatggtaa agttacccct ccgaaatttt tcagcagcgg tgaaattctg
cgtctgaaaa 2580atctgcagaa agagcgtgat cgctttctgc tgcgtaaaca
caacaaaaat cgcaaaaaag 2640gcaacatgcg cgtgatcgaa aacaaaatca
atctgatcct gcaccgttat agcaagcaga 2700ttgttgatat ggccaaaaag
ctgaatgcca gcattgtttt tgaagaactg ggtcgtattg 2760gtaaaagccg
caccaaaatg aaaaaaagcc agcgttataa actgagcctg ttcatcttca
2820agaaactgag cgatctggtt gattacaaaa gccgtcgtga aggtattcgt
gttacctatg 2880ttccgcctga atataccagc aaagaatgta gccattgcgg
tgaaaaagtt aatacccagc 2940gtccgtttaa tggcaactat agcctgttta
aatgcaacaa atgtggcatc cagctgaaca 3000gcgattataa tgcaagcatc
aacattgcga aaaagggcct gaaaattccg aatagcacct 3060aataacattg
gaagtggata acggatccgc gatcgcggcg cgccacctgg tggccggccg
3120gtaccacgcg tgcgcgctga tccggctgct aacaaagccc gaaaggaagc
tgagttggct 3180gctgccaccg ctgagcaata actagcataa ccccttgggg
cctctaaacg ggtcttgagg 3240ggttttttgc tgaaaggagg aactatatcc
ggatatccac aggacgggtg tggtcgccat 3300gatcgcgtag tcgatagtgg
ctccaagtag cgaagcgagc aggactgggc ggcggccaaa 3360gcggtcggac
agtgctccga gaacgggtgc gcatagaaat tgcatcaacg catatagcgc
3420tagcagcacg ccatagtgac tggcgatgct gtcggaatgg acgatatccc
gcaagaggcc 3480cggcagtacc ggcataacca agcctatgcc tacagcatcc
agggtgacgg tgccgaggat 3540gacgatgagc gcattgttag atttcataca
cggtgcctga ctgcgttagc aatttaactg 3600tgataaacta ccgcattaaa
gcttatcgat gataagctgt caaacatgag aattcttgaa 3660gacgaaaggg
cctcgtgata cgcctatttt tataggttaa tgtcatgata ataatggttt
3720cttagacgtc aggtggcact tttcggggaa atgtgcgcgg aacccctatt
tgtttatttt 3780tctaaataca ttcaaatatg tatccgctca tgagacaata
accctgataa atgcttcaat 3840aacattgaaa aaggaagagt atgagtattc
aacatttccg tgtcgccctt attccctttt 3900ttgcggcatt ttgccttcct
gtttttgctc acccagaaac gctggtgaaa gtaaaagatg 3960ctgaagatca
gttgggtgca cgagtgggtt acatcgaact ggatctcaac agcggtaaga
4020tccttgagag ttttcgcccc gaagaacgtt ttccaatgat gagcactttt
aaagttctgc 4080tatgtggcgc ggtattatcc cgtgttgacg ccgggcaaga
gcaactcggt cgccgcatac 4140actattctca gaatgacttg gttgagtact
caccagtcac agaaaagcat cttacggatg 4200gcatgacagt aagagaatta
tgcagtgctg ccataaccat gagtgataac actgcggcca 4260acttacttct
gacaacgatc ggaggaccga aggagctaac cgcttttttg cacaacatgg
4320gggatcatgt aactcgcctt gatcgttggg aaccggagct gaatgaagcc
ataccaaacg 4380acgagcgtga caccacgatg cctgcagcaa tggcaacaac
gttgcgcaaa ctattaactg 4440gcgaactact tactctagct tcccggcaac
aattaataga ctggatggag gcggataaag 4500ttgcaggacc acttctgcgc
tcggcccttc cggctggctg gtttattgct gataaatctg 4560gagccggtga
gcgtgggtct cgcggtatca ttgcagcact ggggccagat ggtaagccct
4620cccgtatcgt agttatctac acgacgggga gtcaggcaac tatggatgaa
cgaaatagac 4680agatcgctga gataggtgcc tcactgatta agcattggta
actgtcagac caagtttact 4740catatatact ttagattgat ttaaaacttc
atttttaatt taaaaggatc taggtgaaga 4800tcctttttga taatctcatg
accaaaatcc cttaacgtga gttttcgttc cactgagcgt 4860cagaccccgt
agaaaagatc aaaggatctt cttgagatcc tttttttctg cgcgtaatct
4920gctgcttgca aacaaaaaaa ccaccgctac cagcggtggt ttgtttgccg
gatcaagagc 4980taccaactct ttttccgaag gtaactggct tcagcagagc
gcagatacca aatactgtcc 5040ttctagtgta gccgtagtta ggccaccact
tcaagaactc tgtagcaccg cctacatacc 5100tcgctctgct aatcctgtta
ccagtggctg ctgccagtgg cgataagtcg tgtcttaccg 5160ggttggactc
aagacgatag ttaccggata aggcgcagcg gtcgggctga acggggggtt
5220cgtgcacaca gcccagcttg gagcgaacga cctacaccga actgagatac
ctacagcgtg 5280agctatgaga aagcgccacg cttcccgaag ggagaaaggc
ggacaggtat ccggtaagcg 5340gcagggtcgg aacaggagag cgcacgaggg
agcttccagg gggaaacgcc tggtatcttt 5400atagtcctgt cgggtttcgc
cacctctgac ttgagcgtcg atttttgtga tgctcgtcag 5460gggggcggag
cctatggaaa aacgccagca acgcggcctt tttacggttc ctggcctttt
5520gctggccttt tgctcacatg ttctttcctg cgttatcccc tgattctgtg
gataaccgta 5580ttaccgcctt tgagtgagct gataccgctc gccgcagccg
aacgaccgag cgcagcgagt 5640cagtgagcga ggaagcggaa gagcgcctga
tgcggtattt tctccttacg catctgtgcg 5700gtatttcaca ccgcaatggt
gcactctcag tacaatctgc tctgatgccg catagttaag 5760ccagtataca
ctccgctatc gctacgtgac tgggtcatgg ctgcgccccg acacccgcca
5820acacccgctg acgcgccctg acgggcttgt ctgctcccgg catccgctta
cagacaagct 5880gtgaccgtct ccgggagctg catgtgtcag aggttttcac
cgtcatcacc gaaacgcgcg 5940aggcagctgc ggtaaagctc atcagcgtgg
tcgtgaagcg attcacagat gtctgcctgt 6000tcatccgcgt ccagctcgtt
gagtttctcc agaagcgtta atgtctggct tctgataaag 6060cgggccatgt
taagggcggt tttttcctgt ttggtcactg atgcctccgt gtaaggggga
6120tttctgttca tgggggtaat gataccgatg aaacgagaga ggatgctcac
gatacgggtt 6180actgatgatg aacatgcccg gttactggaa cgttgtgagg
gtaaacaact ggcggtatgg 6240atgcggcggg accagagaaa aatcactcag
ggtcaatgcc agcgcttcgt taatacagat 6300gtaggtgttc cacagggtag
ccagcagcat cctgcgatgc agatccggaa cataatggtg 6360cagggcgctg
acttccgcgt ttccagactt tacgaaacac ggaaaccgaa gaccattcat
6420gttgttgctc aggtcgcaga cgttttgcag cagcagtcgc ttcacgttcg
ctcgcgtatc 6480ggtgattcat tctgctaacc agtaaggcaa ccccgccagc
ctagccgggt cctcaacgac 6540aggagcacga tcatgcgcac ccgtggccag
gacccaacgc tgcccgagat gcgccgcgtg 6600cggctgctgg agatggcgga
cgcgatggat atgttctgcc aagggttggt ttgcgcattc 6660acagttctcc
gcaagaattg attggctcca attcttggag tggtgaatcc gttagcgagg
6720tgccgccggc ttccattcag gtcgaggtgg cccggctcca tgcaccgcga
cgcaacgcgg 6780ggaggcagac aaggtatagg gcggcgccta caatccatgc
caacccgttc catgtgctcg 6840ccgaggcggc ataaatcgcc gtgacgatca
gcggtccaat gatcgaagtt aggctggtaa 6900gagccgcgag cgatccttga
agctgtccct gatggtcgtc atctacctgc ctggacagca 6960tggcctgcaa
cgcgggcatc ccgatgccgc cggaagcgag aagaatcata atggggaagg
7020ccatccagcc tcgcgtcgcg aacgccagca agacgtagcc cagcgcgtcg
gccgccatgc 7080cggcgataat ggcctgcttc tcgccgaaac gtttggtggc
gggaccagtg acgaaggctt 7140gagcgagggc gtgcaagatt ccgaataccg
caagcgacag gccgatcatc gtcgcgctcc 7200agcgaaagcg gtcctcgccg
aaaatgaccc agagcgctgc cggcacctgt cctacgagtt 7260gcatgataaa
gaagacagtc ataagtgcgg cgacgatagt catgccccgc gcccaccgga
7320aggagctgac tgggttgaag gctctcaagg gcatcggtcg acgctctccc
ttatgcgact 7380cctgcattag gaagcagccc agtagtaggt tgaggccgtt
gagcaccgcc gccgcaagga 7440atggtgcatg caaggagatg gcgcccaaca
gtcccccggc cacggggcct gccaccatac 7500ccacgccgaa acaagcgctc
atgagcccga agtggcgagc ccgatcttcc ccatcggtga 7560tgtcggcgat
ataggcgcca 75803644715DNAArtificial sequenceSynthetic sequence
364atcgttgata gagttatttt accactccct atcagtgata gagaaaagaa
ttcaaaagat 60ctaaagagga gaaaggatct atggaagaaa gcattattac cggtgtgaaa
ttcaaactgc 120gcatcgataa agaaaccacc aaaaaactga acgagtactt
cgatgaatat ggcaaagcaa 180ttaacttcgc cgtgaagatc attcagaaag
aactggcaga tgatcgtttt gcaggtaaag 240caaaactgga ccagaataaa
aacccgatcc tggatgaaaa cggcaaaaaa atctatgaat 300tcccggatga
attttgcagc tgtggtaaac aggttaacaa gtacgttaac aacaaaccgt
360tttgccaaga gtgctataaa atccgcttta ccgaaaatgg tattcgcaaa
cgtatgtata 420gcgccaaagg tcgtaaagcc gaacataaaa tcaatatcct
gaacagcacc aacaagatca 480gcaaaaccca ttttaactat gccattcgcg
aagccttcat tctggataaa agcatcaaaa 540agcagcgcaa aaaacgtaat
gaacgtctgc gtgaaagtaa aaaacgtctg cagcagttta 600tcgatatgcg
tgatggtaaa cgtgaaattt gcccgaccat taaaggtcag aaagtggatc
660gttttattca tccgagctgg atcaccaaag ataaaaagct ggaagatttt
cgcggttata 720ccctgagcat tatcaacagc aaaattaaga ttctggatcg
caacatcaaa cgcgaagaaa 780aaagcctgaa agaaaaaggc cagatcatct
ttaaagccaa acgtctgatg ctggataaat 840ccattcgttt tgttggtgat
cgcaaagtgc tgtttacaat tagtaaaacc ctgccgaaag 900agtatgaact
ggatctgccg agcaaagaaa aacggctgaa ttggctgaaa gagaagatcg
960agattatcaa gaaccagaaa ccgaaatatg cctatctgct gcgcaaaaac
attgagagcg 1020aaaaaaaacc gaactatgag tactatctgc agtacaccct
ggaaattaaa ccggaactga 1080aagattttta tgatggtgcc attggtattg
accgtggcat taatcatatt gccgtttgca 1140cctttattag caacgatggt
aaagttaccc ctccgaaatt tttcagcagc ggtgaaattc 1200tgcgtctgaa
aaatctgcag aaagagcgtg atcgctttct gctgcgtaaa cacaacaaaa
1260atcgcaaaaa aggcaacatg cgcgtgatcg aaaacaaaat caatctgatc
ctgcaccgtt 1320atagcaagca gattgttgat atggccaaaa agctgaatgc
cagcattgtt tttgaagaac 1380tgggtcgtat tggtaaaagc cgcaccaaaa
tgaaaaaaag ccagcgttat aaactgagcc 1440tgttcatctt caagaaactg
agcgatctgg ttgattacaa aagccgtcgt gaaggtattc 1500gtgttaccta
tgttccgcct gaatatacca gcaaagaatg tagccattgc ggtgaaaaag
1560ttaataccca gcgtccgttt aatggcaact atagcctgtt taaatgcaac
aaatgtggca 1620tccagctgaa cagcgattat aatgcaagca tcaacattgc
gaaaaagggc ctgaaaattc 1680cgaatagcac ctaataatgt tggttaagcc
acaatatgga atattgttct tatggacagt 1740attgacttaa attaataatc
ttcgaaggct atatgcggaa gatttggcgt tgttgtaacg 1800caataagggg
taaccctgaa aaggtttgaa atcatataaa cctagtttta tttgagttta
1860ggctcagata aaatgaacag accaatcttt aattccgttc tgatttaaaa
aatcagaatc 1920tctttataaa tagtattaca aaaagtgtac attccaaaat
ccgaaagcag aattgacctt 1980tttaagccta aaaaagccaa atttcaaggc
tctttcatac tcagaacaaa gggattaagg 2040aatgcaacta ccttacaccg
cttgcgaaag ttgtcagaag ataatctttc atactcagaa 2100caaagggatt
aaggaatgca actatcttat ccatttcttg acatcaaatt ttcttgcagc
2160atctgaattg cttaattgct ttccttgctt cagcaggaaa tagccaagat
tttccagttc 2220tgctggcgtt attgcaagca cggggatttt gtgcttgctg
tagattttgt tcttcaggac 2280cttctttttc catgctagag tcacactggc
tcaccttcgg gtgggccttt ctgcgtttat 2340acctagggat atattccgct
tcctcgctca ctgactcgct acgctcggtc gttcgactgc 2400ggcgagcgga
aatggcttac gaacggggcg gagatttcct ggaagatgcc aggaagatac
2460ttaacaggga agtgagaggg ccgcggcaaa gccgtttttc cataggctcc
gcccccctga 2520caagcatcac gaaatctgac gctcaaatca gtggtggcga
aacccgacag gactataaag 2580ataccaggcg tttccccctg gcggctccct
cgtgcgctct cctgttcctg cctttcggtt 2640taccggtgtc attccgctgt
tatggccgcg tttgtctcat tccacgcctg acactcagtt 2700ccgggtaggc
agttcgctcc aagctggact gtatgcacga accccccgtt cagtccgacc
2760gctgcgcctt atccggtaac tatcgtcttg agtccaaccc ggaaagacat
gcaaaagcac 2820cactggcagc agccactggt aattgattta gaggagttag
tcttgaagtc atgcgccggt 2880taaggctaaa ctgaaaggac aagttttggt
gactgcgctc ctccaagcca gttacctcgg 2940ttcaaagagt tggtagctca
gagaaccttc gaaaaaccgc cctgcaaggc ggttttttcg 3000ttttcagagc
aagagattac gcgcagacca aaacgatctc aagaagatca tcttattaat
3060cagataaaat atttctagat ttcagtgcaa tttatctctt caaatgtagc
acctgaagtc 3120agccccatac gatataagtt gttactagtg cttggattct
caccaataaa aaacgcccgg 3180cggcaaccga gcgttctgaa caaatccaga
tggagttctg aggtcattac tggatctatc 3240aacaggagtc caagcgagct
cgatatcaaa ttacgccccg ccctgccact catcgcagta 3300ctgttgtaat
tcattaagca ttctgccgac atggaagcca tcacaaacgg catgatgaac
3360ctgaatcgcc agcggcatca gcaccttgtc gccttgcgta taatatttgc
ccatggtgaa 3420aacgggggcg aagaagttgt ccatattggc cacgtttaaa
tcaaaactgg tgaaactcac 3480ccagggattg gctgagacga aaaacatatt
ctcaataaac cctttaggga aataggccag 3540gttttcaccg taacacgcca
catcttgcga atatatgtgt agaaactgcc ggaaatcgtc 3600gtggtattca
ctccagagcg atgaaaacgt ttcagtttgc tcatggaaaa cggtgtaaca
3660agggtgaaca ctatcccata tcaccagctc accgtctttc attgccatac
gaaattccgg 3720atgagcattc atcaggcggg caagaatgtg aataaaggcc
ggataaaact tgtgcttatt 3780tttctttacg gtctttaaaa aggccgtaat
atccagctga acggtctggt tataggtaca 3840ttgagcaact gactgaaatg
cctcaaaatg ttctttacga tgccattggg atatatcaac 3900ggtggtatat
ccagtgattt ttttctccat tttagcttcc ttagctcctg aaaatctcga
3960taactcaaaa aatacgcccg gtagtgatct tatttcatta tggtgaaagt
tggaacctct 4020tacgtgccga tcaacgtctc attttcgcca gatatcgacg
tcttaagacc cactttcaca 4080tttaagttgt ttttctaatc cgcatatgat
caattcaagg ccgaataaga aggctggctc 4140tgcaccttgg tgatcaaata
attcgatagc ttgtcgtaat aatggcggca tactatcagt 4200agtaggtgtt
tccctttctt ctttagcgac ttgatgctct tgatcttcca atacgcaacc
4260taaagtaaaa tgccccacag cgctgagtgc atataatgca ttctctagtg
aaaaaccttg 4320ttggcataaa aaggctaatt gattttcgag agtttcatac
tgtttttctg taggccgtgt 4380acctaaatgt acttttgctc catcgcgatg
acttagtaaa gcacatctaa aacttttagc 4440gttattacgt aaaaaatctt
gccagctttc cccttctaaa gggcaaaagt gagtatggtg 4500cctatctaac
atctcaatgg ctaaggcgtc gagcaaagcc cgcttatttt ttacatgcca
4560atacaatgta ggctgctcta cacctagctt ctgggcgagt ttacgggttg
ttaaaccttc 4620gattccgacc tcattaagca gctctaatgc gctgttaatc
actttacttt tatctaatct 4680agacatcatt aattcctaat ttttgttgac actct
47153654670DNAArtificial sequenceSynthetic sequence
365taattcctaa
tttttgttga cactctatcg ttgatagagt tattttacca ctccctatca 60gtgatagaga
aaagaattca aaagatctaa agaggagaaa ggatctatga aatctcacca
120tcaccatcac catgaaaacc tgtacttcca atccaatatt ggaagtggaa
tggccaaaaa 180caccattacc aaaacactga aactgcgtat tgtgcgtccg
tataatagcg cagaagtgga 240aaaaattgtt gccgacgaaa aaaacaaccg
cgaaaaaatc gcactggaaa agaacaaaga 300caaagtgaaa gaagcctgca
gcaaacatct gaaagttgca gcatattgta ccacacaggt 360tgaacgtaat
gcatgcctgt tttgtaaagc acgtaaactg gatgacaaat tctaccaaaa
420actgcgtggt cagtttccgg atgcagtttt ttggcaagaa atcagcgaaa
tttttcgcca 480gctgcagaaa caggcagcag aaatctataa tcagagcctg
atcgaactgt actacgagat 540ttttatcaaa ggcaaaggta ttgcaaatgc
cagcagcgtt gaacattatc tgagtgatgt 600ttgttatacc cgtgcagcag
aactgtttaa aaacgcagca attgcaagcg gtctgcgtag 660caaaatcaaa
agcaattttc gtctgaaaga actgaaaaac atgaaaagtg gtctgccgac
720caccaaaagc gataattttc cgattccgct ggttaaacag aaaggtggtc
agtataccgg 780ttttgaaatt agcaatcata atagcgactt catcatcaag
attccgtttg gtcgttggca 840ggtcaaaaaa gagattgata aatatcgtcc
gtgggagaaa tttgactttg aacaggttca 900gaaaagcccg aaaccgatta
gcctgctgct gagcacccag cgtcgtaaac gtaataaagg 960ttggagcaaa
gatgaaggca ccgaagccga aatcaaaaaa gttatgaatg gcgattatca
1020gaccagctac attgaagtta aacgtggcag caaaatcggt gaaaaaagcg
catggatgct 1080gaatctgagc attgatgttc cgaaaattga taaaggtgtg
gatccgagca ttattggtgg 1140tattgatgtt ggtgttaaat caccgctggt
ttgcgcaatt aacaatgcat ttagccgtta 1200tagcatcagc gataacgacc
tgtttcactt caacaagaaa atgtttgcac gtcgtcgtat 1260cctgctgaaa
aaaaaccgtc ataaacgtgc aggtcatggt gcaaaaaaca aactgaaacc
1320gatcaccatt ctgaccgaaa aaagtgaacg ttttcgcaaa aagctgattg
aacgttgggc 1380atgtgaaatc gcggatttct tcattaaaaa caaagttggc
accgtgcaga tggaaaatct 1440ggaaagcatg aaacgtaaag aggacagcta
ttttaacatt cgcctgcgtg gcttttggcc 1500gtatgcagaa atgcagaaca
aaatcgaatt caaactgaag cagtatggca tcgaaattcg 1560taaagttgca
ccgaataata ccagcaaaac ctgtagcaaa tgtggccatc tgaacaacta
1620tttcaacttc gagtaccgca agaaaaacaa attcccgcac tttaaatgcg
aaaaatgcaa 1680cttcaaagaa aacgccgatt ataatgcagc cctgaatatt
tcaaacccga aactgaaaag 1740caccaaagag gaaccgtaaa tatttatact
ttattatcct tcattgacaa aaatgagaat 1800gttatcccag ataacatttg
atgtacacag attcacactt cactgataaa gtggagaacc 1860gcttcaccaa
aagctgtccc ttaggggatt agaacttgag tgaaggtggg ctgcttgcat
1920cagcctaatg tcgagaagtg ctttcttcgg aaagtaaccc tcgaaacaaa
ttcatttttc 1980ctctccaatt ctgcacaaaa aaaggtgagt ccttataaac
cggcgtgcag aacgccggct 2040cacctttttt cttcattcga ttttatgctt
aaaagccgta aaaacgcgga attcggcgcc 2100gttgcagaac ccgaatagac
gaatgaagga atgcaactac cttacaccgc ttgcgaaagt 2160tgtcagaaga
taatcttgca gaacccgaat agacgaatga aggaatgcaa tcttgacaga
2220gcccgattgc gttatctcca ggagaaacat ataaaagcat caaccgctga
tcggactaga 2280gtcacactgg ctcaccttcg ggtgggcctt tctgcgttta
tacctaggga tatattccgc 2340ttcctcgctc actgactcgc tacgctcggt
cgttcgactg cggcgagcgg aaatggctta 2400cgaacggggc ggagatttcc
tggaagatgc caggaagata cttaacaggg aagtgagagg 2460gccgcggcaa
agccgttttt ccataggctc cgcccccctg acaagcatca cgaaatctga
2520cgctcaaatc agtggtggcg aaacccgaca ggactataaa gataccaggc
gtttccccct 2580ggcggctccc tcgtgcgctc tcctgttcct gcctttcggt
ttaccggtgt cattccgctg 2640ttatggccgc gtttgtctca ttccacgcct
gacactcagt tccgggtagg cagttcgctc 2700caagctggac tgtatgcacg
aaccccccgt tcagtccgac cgctgcgcct tatccggtaa 2760ctatcgtctt
gagtccaacc cggaaagaca tgcaaaagca ccactggcag cagccactgg
2820taattgattt agaggagtta gtcttgaagt catgcgccgg ttaaggctaa
actgaaagga 2880caagttttgg tgactgcgct cctccaagcc agttacctcg
gttcaaagag ttggtagctc 2940agagaacctt cgaaaaaccg ccctgcaagg
cggttttttc gttttcagag caagagatta 3000cgcgcagacc aaaacgatct
caagaagatc atcttattaa tcagataaaa tatttctaga 3060tttcagtgca
atttatctct tcaaatgtag cacctgaagt cagccccata cgatataagt
3120tgttactagt gcttggattc tcaccaataa aaaacgcccg gcggcaaccg
agcgttctga 3180acaaatccag atggagttct gaggtcatta ctggatctat
caacaggagt ccaagcgagc 3240tcgatatcaa attacgcccc gccctgccac
tcatcgcagt actgttgtaa ttcattaagc 3300attctgccga catggaagcc
atcacaaacg gcatgatgaa cctgaatcgc cagcggcatc 3360agcaccttgt
cgccttgcgt ataatatttg cccatggtga aaacgggggc gaagaagttg
3420tccatattgg ccacgtttaa atcaaaactg gtgaaactca cccagggatt
ggctgagacg 3480aaaaacatat tctcaataaa ccctttaggg aaataggcca
ggttttcacc gtaacacgcc 3540acatcttgcg aatatatgtg tagaaactgc
cggaaatcgt cgtggtattc actccagagc 3600gatgaaaacg tttcagtttg
ctcatggaaa acggtgtaac aagggtgaac actatcccat 3660atcaccagct
caccgtcttt cattgccata cgaaattccg gatgagcatt catcaggcgg
3720gcaagaatgt gaataaaggc cggataaaac ttgtgcttat ttttctttac
ggtctttaaa 3780aaggccgtaa tatccagctg aacggtctgg ttataggtac
attgagcaac tgactgaaat 3840gcctcaaaat gttctttacg atgccattgg
gatatatcaa cggtggtata tccagtgatt 3900tttttctcca ttttagcttc
cttagctcct gaaaatctcg ataactcaaa aaatacgccc 3960ggtagtgatc
ttatttcatt atggtgaaag ttggaacctc ttacgtgccg atcaacgtct
4020cattttcgcc agatatcgac gtcttaagac ccactttcac atttaagttg
tttttctaat 4080ccgcatatga tcaattcaag gccgaataag aaggctggct
ctgcaccttg gtgatcaaat 4140aattcgatag cttgtcgtaa taatggcggc
atactatcag tagtaggtgt ttccctttct 4200tctttagcga cttgatgctc
ttgatcttcc aatacgcaac ctaaagtaaa atgccccaca 4260gcgctgagtg
catataatgc attctctagt gaaaaacctt gttggcataa aaaggctaat
4320tgattttcga gagtttcata ctgtttttct gtaggccgtg tacctaaatg
tacttttgat 4380ccatcgcgat gacttagtaa agcacatcta aaacttttag
cgttattacg taaaaaatct 4440tgccagcttt ccccttctaa agggcaaaag
tgagtatggt gcctatctaa catctcaatg 4500gctaaggcgt cgagcaaagc
ccgcttattt tttacatgcc aatacaatgt aggctgctct 4560acacctagct
tctgggcgag tttacgggtt gttaaacctt cgattccgac ctcattaagc
4620agctctaatg cgctgttaat cactttactt ttatctaatc tagacatcat
46703664607DNAArtificial sequenceSynthetic sequence 366ttacttttat
ctaatctaga catcattaat tcctaatttt tgttgacact ctatcgttga 60tagagttatt
ttaccactcc ctatcagtga tagagaaaag aattcaaaag atctaaagag
120gagaaaggat ctatggccaa aaacaccatt accaaaacac tgaaactgcg
tattgtgcgt 180ccgtataata gcgcagaagt ggaaaaaatt gttgccgacg
aaaaaaacaa ccgcgaaaaa 240atcgcactgg aaaagaacaa agacaaagtg
aaagaagcct gcagcaaaca tctgaaagtt 300gcagcatatt gtaccacaca
ggttgaacgt aatgcatgcc tgttttgtaa agcacgtaaa 360ctggatgaca
aattctacca aaaactgcgt ggtcagtttc cggatgcagt tttttggcaa
420gaaatcagcg aaatttttcg ccagctgcag aaacaggcag cagaaatcta
taatcagagc 480ctgatcgaac tgtactacga gatttttatc aaaggcaaag
gtattgcaaa tgccagcagc 540gttgaacatt atctgagtga tgtttgttat
acccgtgcag cagaactgtt taaaaacgca 600gcaattgcaa gcggtctgcg
tagcaaaatc aaaagcaatt ttcgtctgaa agaactgaaa 660aacatgaaaa
gtggtctgcc gaccaccaaa agcgataatt ttccgattcc gctggttaaa
720cagaaaggtg gtcagtatac cggttttgaa attagcaatc ataatagcga
cttcatcatc 780aagattccgt ttggtcgttg gcaggtcaaa aaagagattg
ataaatatcg tccgtgggag 840aaatttgact ttgaacaggt tcagaaaagc
ccgaaaccga ttagcctgct gctgagcacc 900cagcgtcgta aacgtaataa
aggttggagc aaagatgaag gcaccgaagc cgaaatcaaa 960aaagttatga
atggcgatta tcagaccagc tacattgaag ttaaacgtgg cagcaaaatc
1020ggtgaaaaaa gcgcatggat gctgaatctg agcattgatg ttccgaaaat
tgataaaggt 1080gtggatccga gcattattgg tggtattgat gttggtgtta
aatcaccgct ggtttgcgca 1140attaacaatg catttagccg ttatagcatc
agcgataacg acctgtttca cttcaacaag 1200aaaatgtttg cacgtcgtcg
tatcctgctg aaaaaaaacc gtcataaacg tgcaggtcat 1260ggtgcaaaaa
acaaactgaa accgatcacc attctgaccg aaaaaagtga acgttttcgc
1320aaaaagctga ttgaacgttg ggcatgtgaa atcgcggatt tcttcattaa
aaacaaagtt 1380ggcaccgtgc agatggaaaa tctggaaagc atgaaacgta
aagaggacag ctattttaac 1440attcgcctgc gtggcttttg gccgtatgca
gaaatgcaga acaaaatcga attcaaactg 1500aagcagtatg gcatcgaaat
tcgtaaagtt gcaccgaata ataccagcaa aacctgtagc 1560aaatgtggcc
atctgaacaa ctatttcaac ttcgagtacc gcaagaaaaa caaattcccg
1620cactttaaat gcgaaaaatg caacttcaaa gaaaacgccg attataatgc
agccctgaat 1680atttcaaacc cgaaactgaa aagcaccaaa gaggaaccgt
aaatatttat actttattat 1740ccttcattga caaaaatgag aatgttatcc
cagataacat ttgatgtaca cagattcaca 1800cttcactgat aaagtggaga
accgcttcac caaaagctgt cccttagggg attagaactt 1860gagtgaaggt
gggctgcttg catcagccta atgtcgagaa gtgctttctt cggaaagtaa
1920ccctcgaaac aaattcattt ttcctctcca attctgcaca aaaaaaggtg
agtccttata 1980aaccggcgtg cagaacgccg gctcaccttt tttcttcatt
cgattttatg cttaaaagcc 2040gtaaaaacgc ggaattcggc gccgttgcag
aacccgaata gacgaatgaa ggaatgcaac 2100taccttacac cgcttgcgaa
agttgtcaga agataatctt gcagaacccg aatagacgaa 2160tgaaggaatg
caatcttgac agagcccgat tgcgttatct ccaggagaaa catataaaag
2220catcaaccgc tgatcggact agagtcacac tggctcacct tcgggtgggc
ctttctgcgt 2280ttatacctag ggatatattc cgcttcctcg ctcactgact
cgctacgctc ggtcgttcga 2340ctgcggcgag cggaaatggc ttacgaacgg
ggcggagatt tcctggaaga tgccaggaag 2400atacttaaca gggaagtgag
agggccgcgg caaagccgtt tttccatagg ctccgccccc 2460ctgacaagca
tcacgaaatc tgacgctcaa atcagtggtg gcgaaacccg acaggactat
2520aaagatacca ggcgtttccc cctggcggct ccctcgtgcg ctctcctgtt
cctgcctttc 2580ggtttaccgg tgtcattccg ctgttatggc cgcgtttgtc
tcattccacg cctgacactc 2640agttccgggt aggcagttcg ctccaagctg
gactgtatgc acgaaccccc cgttcagtcc 2700gaccgctgcg ccttatccgg
taactatcgt cttgagtcca acccggaaag acatgcaaaa 2760gcaccactgg
cagcagccac tggtaattga tttagaggag ttagtcttga agtcatgcgc
2820cggttaaggc taaactgaaa ggacaagttt tggtgactgc gctcctccaa
gccagttacc 2880tcggttcaaa gagttggtag ctcagagaac cttcgaaaaa
ccgccctgca aggcggtttt 2940ttcgttttca gagcaagaga ttacgcgcag
accaaaacga tctcaagaag atcatcttat 3000taatcagata aaatatttct
agatttcagt gcaatttatc tcttcaaatg tagcacctga 3060agtcagcccc
atacgatata agttgttact agtgcttgga ttctcaccaa taaaaaacgc
3120ccggcggcaa ccgagcgttc tgaacaaatc cagatggagt tctgaggtca
ttactggatc 3180tatcaacagg agtccaagcg agctcgatat caaattacgc
cccgccctgc cactcatcgc 3240agtactgttg taattcatta agcattctgc
cgacatggaa gccatcacaa acggcatgat 3300gaacctgaat cgccagcggc
atcagcacct tgtcgccttg cgtataatat ttgcccatgg 3360tgaaaacggg
ggcgaagaag ttgtccatat tggccacgtt taaatcaaaa ctggtgaaac
3420tcacccaggg attggctgag acgaaaaaca tattctcaat aaacccttta
gggaaatagg 3480ccaggttttc accgtaacac gccacatctt gcgaatatat
gtgtagaaac tgccggaaat 3540cgtcgtggta ttcactccag agcgatgaaa
acgtttcagt ttgctcatgg aaaacggtgt 3600aacaagggtg aacactatcc
catatcacca gctcaccgtc tttcattgcc atacgaaatt 3660ccggatgagc
attcatcagg cgggcaagaa tgtgaataaa ggccggataa aacttgtgct
3720tatttttctt tacggtcttt aaaaaggccg taatatccag ctgaacggtc
tggttatagg 3780tacattgagc aactgactga aatgcctcaa aatgttcttt
acgatgccat tgggatatat 3840caacggtggt atatccagtg atttttttct
ccattttagc ttccttagct cctgaaaatc 3900tcgataactc aaaaaatacg
cccggtagtg atcttatttc attatggtga aagttggaac 3960ctcttacgtg
ccgatcaacg tctcattttc gccagatatc gacgtcttaa gacccacttt
4020cacatttaag ttgtttttct aatccgcata tgatcaattc aaggccgaat
aagaaggctg 4080gctctgcacc ttggtgatca aataattcga tagcttgtcg
taataatggc ggcatactat 4140cagtagtagg tgtttccctt tcttctttag
cgacttgatg ctcttgatct tccaatacgc 4200aacctaaagt aaaatgcccc
acagcgctga gtgcatataa tgcattctct agtgaaaaac 4260cttgttggca
taaaaaggct aattgatttt cgagagtttc atactgtttt tctgtaggcc
4320gtgtacctaa atgtactttt gctccatcgc gatgacttag taaagcacat
ctaaaacttt 4380tagcgttatt acgtaaaaaa tcttgccagc tttccccttc
taaagggcaa aagtgagtat 4440ggtgcctatc taacatctca atggctaagg
cgtcgagcaa agcccgctta ttttttacat 4500gccaatacaa tgtaggctgc
tctacaccta gcttctgggc gagtttacgg gttgttaaac 4560cttcgattcc
gacctcatta agcagctcta atgcgctgtt aatcact 46073677556DNAArtificial
sequenceSynthetic sequence 367acccacgccg aaacaagcgc tcatgagccc
gaagtggcga gcccgatctt ccccatcggt 60gatgtcggcg atataggcgc cagcaaccgc
acctgtggcg ccggtgatgc cggccacgat 120gcgtccggcg tagaggatcg
agatctcgat cccgcgaaat taatacgact cactataggg 180agaccacaac
ggtttccctc tagtgccggc tccggagagc tctttaatta agcggccgcc
240ctgcaggact cgagttctag aaataatttt gtttaacttt aagaaggaga
tatacatatg 300aaatcttctc accatcacca tcaccatcac catcaccatg
gttcttctat gaaaatcgaa 360gaaggtaaac tggtaatctg gattaacggc
gataaaggct ataacggtct cgctgaagtc 420ggtaagaaat tcgagaaaga
taccggaatt aaagtcaccg ttgagcatcc ggataaactg 480gaagagaaat
tcccacaggt tgcggcaact ggcgatggcc ctgacattat cttctgggca
540cacgaccgct ttggtggcta cgctcaatct ggcctgttgg ctgaaatcac
cccggacaaa 600gcgttccagg acaagctgta tccgtttacc tgggatgccg
tacgttacaa cggcaagctg 660attgcttacc cgatcgctgt tgaagcgtta
tcgctgattt ataacaaaga tctgctgccg 720aacccgccaa aaacctggga
agagatcccg gcgctggata aagaactgaa agcgaaaggt 780aagagcgcgc
tgatgttcaa cctgcaagaa ccgtacttca cctggccgct gattgctgct
840gacgggggtt atgcgttcaa gtatgaaaac ggcaagtacg acattaaaga
cgtgggcgtg 900gataacgctg gcgcgaaagc gggtctgacc ttcctggttg
acctgattaa aaacaaacac 960atgaatgcag acaccgatta ctccatcgca
gaagctgcct ttaataaagg cgaaacagcg 1020atgaccatca acggcccgtg
ggcatggtcc aacatcgaca ccagcaaagt gaattatggt 1080gtaacggtac
tgccgacctt caagggtcaa ccatccaaac cgttcgttgg cgtgctgagc
1140gcaggtatta acgccgccag tccgaacaaa gagctggcaa aagagttcct
cgaaaactat 1200ctgctgactg atgaaggtct ggaagcggtt aataaagaca
aaccgctggg tgccgtagcg 1260ctgaagtctt acgaggaaga gttggcgaaa
gatccacgta ttgccgccac tatggaaaac 1320gcccagaaag gtgaaatcat
gccgaacatc ccgcagatgt ccgctttctg gtatgccgtg 1380cgtactgcgg
tgatcaacgc cgccagcggt cgtcagactg tcgatgaagc cctgaaagac
1440gcgcagacta attcgagctc gaacaacaac aacaataaca ataacaacaa
cctcgggatc 1500gaggaaaacc tgtacttcca atccaatgca atggccaaaa
acaccattac caaaacactg 1560aaactgcgta ttgtgcgtcc gtataatagc
gcagaagtgg aaaaaattgt tgccgacgaa 1620aaaaacaacc gcgaaaaaat
cgcactggaa aagaacaaag acaaagtgaa agaagcctgc 1680agcaaacatc
tgaaagttgc agcatattgt accacacagg ttgaacgtaa tgcatgcctg
1740ttttgtaaag cacgtaaact ggatgacaaa ttctaccaaa aactgcgtgg
tcagtttccg 1800gatgcagttt tttggcaaga aatcagcgaa atttttcgcc
agctgcagaa acaggcagca 1860gaaatctata atcagagcct gatcgaactg
tactacgaga tttttatcaa aggcaaaggt 1920attgcaaatg ccagcagcgt
tgaacattat ctgagtgatg tttgttatac ccgtgcagca 1980gaactgttta
aaaacgcagc aattgcaagc ggtctgcgta gcaaaatcaa aagcaatttt
2040cgtctgaaag aactgaaaaa catgaaaagt ggtctgccga ccaccaaaag
cgataatttt 2100ccgattccgc tggttaaaca gaaaggtggt cagtataccg
gttttgaaat tagcaatcat 2160aatagcgact tcatcatcaa gattccgttt
ggtcgttggc aggtcaaaaa agagattgat 2220aaatatcgtc cgtgggagaa
atttgacttt gaacaggttc agaaaagccc gaaaccgatt 2280agcctgctgc
tgagcaccca gcgtcgtaaa cgtaataaag gttggagcaa agatgaaggc
2340accgaagccg aaatcaaaaa agttatgaat ggcgattatc agaccagcta
cattgaagtt 2400aaacgtggca gcaaaatctg tgaaaaaagc gcatggatgc
tgaatctgag cattgatgtt 2460ccgaaaattg ataaaggtgt ggatccgagc
attattggtg gtattgatgt tggtgttaaa 2520tcaccgctgg tttgcgcaat
taacaatgca tttagccgtt atagcatcag cgataacgac 2580ctgtttcact
tcaacaagaa aatgtttgca cgtcgtcgta tcctgctgaa aaaaaaccgt
2640cataaacgtg caggtcatgg tgcaaaaaac aaactgaaac cgatcaccat
tctgaccgaa 2700aaaagtgaac gttttcgcaa aaagctgatt gaacgttggg
catgtgaaat cgcggatttc 2760ttcattaaaa acaaagttgg caccgtgcag
atggaaaatc tggaaagcat gaaacgtaaa 2820gaggacagct attttaacat
tcgcctgcgt ggcttttggc cgtatgcaga aatgcagaac 2880aaaatcgaat
tcaaactgaa gcagtatggc atcgaaattc gtaaagttgc accgaataat
2940accagcaaaa cctgtagcaa atgtggccat ctgaacaact atttcaactt
cgagtaccgc 3000aagaaaaaca aattcccgca ctttaaatgc gaaaaatgca
acttcaaaga aaacgccgat 3060tataatgcag ccctgaatat ttcaaacccg
aaactgaaaa gcaccaaaga ggaaccgtaa 3120taacattgga agtggataac
ggatccgcga tcgcggcgcg ccacctggtg gccggccggt 3180accacgcgtg
cgcgctgatc cggctgctaa caaagcccga aaggaagctg agttggctgc
3240tgccaccgct gagcaataac tagcataacc ccttggggcc tctaaacggg
tcttgagggg 3300ttttttgctg aaaggaggaa ctatatccgg atatccacag
gacgggtgtg gtcgccatga 3360tcgcgtagtc gatagtggct ccaagtagcg
aagcgagcag gactgggcgg cggccaaagc 3420ggtcggacag tgctccgaga
acgggtgcgc atagaaattg catcaacgca tatagcgcta 3480gcagcacgcc
atagtgactg gcgatgctgt cggaatggac gatatcccgc aagaggcccg
3540gcagtaccgg cataaccaag cctatgccta cagcatccag ggtgacggtg
ccgaggatga 3600cgatgagcgc attgttagat ttcatacacg gtgcctgact
gcgttagcaa tttaactgtg 3660ataaactacc gcattaaagc ttatcgatga
taagctgtca aacatgagaa ttcttgaaga 3720cgaaagggcc tcgtgatacg
cctattttta taggttaatg tcatgataat aatggtttct 3780tagacgtcag
gtggcacttt tcggggaaat gtgcgcggaa cccctatttg tttatttttc
3840taaatacatt caaatatgta tccgctcatg agacaataac cctgataaat
gcttcaataa 3900cattgaaaaa ggaagagtat gagtattcaa catttccgtg
tcgcccttat tccctttttt 3960gcggcatttt gccttcctgt ttttgctcac
ccagaaacgc tggtgaaagt aaaagatgct 4020gaagatcagt tgggtgcacg
agtgggttac atcgaactgg atctcaacag cggtaagatc 4080cttgagagtt
ttcgccccga agaacgtttt ccaatgatga gcacttttaa agttctgcta
4140tgtggcgcgg tattatcccg tgttgacgcc gggcaagagc aactcggtcg
ccgcatacac 4200tattctcaga atgacttggt tgagtactca ccagtcacag
aaaagcatct tacggatggc 4260atgacagtaa gagaattatg cagtgctgcc
ataaccatga gtgataacac tgcggccaac 4320ttacttctga caacgatcgg
aggaccgaag gagctaaccg cttttttgca caacatgggg 4380gatcatgtaa
ctcgccttga tcgttgggaa ccggagctga atgaagccat accaaacgac
4440gagcgtgaca ccacgatgcc tgcagcaatg gcaacaacgt tgcgcaaact
attaactggc 4500gaactactta ctctagcttc ccggcaacaa ttaatagact
ggatggaggc ggataaagtt 4560gcaggaccac ttctgcgctc ggcccttccg
gctggctggt ttattgctga taaatctgga 4620gccggtgagc gtgggtctcg
cggtatcatt gcagcactgg ggccagatgg taagccctcc 4680cgtatcgtag
ttatctacac gacggggagt caggcaacta tggatgaacg aaatagacag
4740atcgctgaga taggtgcctc actgattaag cattggtaac tgtcagacca
agtttactca 4800tatatacttt agattgattt aaaacttcat ttttaattta
aaaggatcta ggtgaagatc 4860ctttttgata atctcatgac caaaatccct
taacgtgagt tttcgttcca ctgagcgtca 4920gaccccgtag aaaagatcaa
aggatcttct tgagatcctt tttttctgcg cgtaatctgc 4980tgcttgcaaa
caaaaaaacc accgctacca gcggtggttt gtttgccgga tcaagagcta
5040ccaactcttt ttccgaaggt aactggcttc agcagagcgc agataccaaa
tactgtcctt 5100ctagtgtagc cgtagttagg ccaccacttc aagaactctg
tagcaccgcc tacatacctc 5160gctctgctaa tcctgttacc agtggctgct
gccagtggcg ataagtcgtg tcttaccggg 5220ttggactcaa gacgatagtt
accggataag gcgcagcggt cgggctgaac ggggggttcg 5280tgcacacagc
ccagcttgga gcgaacgacc tacaccgaac tgagatacct acagcgtgag
5340ctatgagaaa gcgccacgct tcccgaaggg agaaaggcgg acaggtatcc
ggtaagcggc 5400agggtcggaa caggagagcg cacgagggag cttccagggg
gaaacgcctg gtatctttat 5460agtcctgtcg ggtttcgcca cctctgactt
gagcgtcgat ttttgtgatg ctcgtcaggg 5520gggcggagcc tatggaaaaa
cgccagcaac gcggcctttt tacggttcct ggccttttgc 5580tggccttttg
ctcacatgtt ctttcctgcg ttatcccctg attctgtgga taaccgtatt
5640accgcctttg
agtgagctga taccgctcgc cgcagccgaa cgaccgagcg cagcgagtca
5700gtgagcgagg aagcggaaga gcgcctgatg cggtattttc tccttacgca
tctgtgcggt 5760atttcacacc gcaatggtgc actctcagta caatctgctc
tgatgccgca tagttaagcc 5820agtatacact ccgctatcgc tacgtgactg
ggtcatggct gcgccccgac acccgccaac 5880acccgctgac gcgccctgac
gggcttgtct gctcccggca tccgcttaca gacaagctgt 5940gaccgtctcc
gggagctgca tgtgtcagag gttttcaccg tcatcaccga aacgcgcgag
6000gcagctgcgg taaagctcat cagcgtggtc gtgaagcgat tcacagatgt
ctgcctgttc 6060atccgcgtcc agctcgttga gtttctccag aagcgttaat
gtctggcttc tgataaagcg 6120ggccatgtta agggcggttt tttcctgttt
ggtcactgat gcctccgtgt aagggggatt 6180tctgttcatg ggggtaatga
taccgatgaa acgagagagg atgctcacga tacgggttac 6240tgatgatgaa
catgcccggt tactggaacg ttgtgagggt aaacaactgg cggtatggat
6300gcggcgggac cagagaaaaa tcactcaggg tcaatgccag cgcttcgtta
atacagatgt 6360aggtgttcca cagggtagcc agcagcatcc tgcgatgcag
atccggaaca taatggtgca 6420gggcgctgac ttccgcgttt ccagacttta
cgaaacacgg aaaccgaaga ccattcatgt 6480tgttgctcag gtcgcagacg
ttttgcagca gcagtcgctt cacgttcgct cgcgtatcgg 6540tgattcattc
tgctaaccag taaggcaacc ccgccagcct agccgggtcc tcaacgacag
6600gagcacgatc atgcgcaccc gtggccagga cccaacgctg cccgagatgc
gccgcgtgcg 6660gctgctggag atggcggacg cgatggatat gttctgccaa
gggttggttt gcgcattcac 6720agttctccgc aagaattgat tggctccaat
tcttggagtg gtgaatccgt tagcgaggtg 6780ccgccggctt ccattcaggt
cgaggtggcc cggctccatg caccgcgacg caacgcgggg 6840aggcagacaa
ggtatagggc ggcgcctaca atccatgcca acccgttcca tgtgctcgcc
6900gaggcggcat aaatcgccgt gacgatcagc ggtccaatga tcgaagttag
gctggtaaga 6960gccgcgagcg atccttgaag ctgtccctga tggtcgtcat
ctacctgcct ggacagcatg 7020gcctgcaacg cgggcatccc gatgccgccg
gaagcgagaa gaatcataat ggggaaggcc 7080atccagcctc gcgtcgcgaa
cgccagcaag acgtagccca gcgcgtcggc cgccatgccg 7140gcgataatgg
cctgcttctc gccgaaacgt ttggtggcgg gaccagtgac gaaggcttga
7200gcgagggcgt gcaagattcc gaataccgca agcgacaggc cgatcatcgt
cgcgctccag 7260cgaaagcggt cctcgccgaa aatgacccag agcgctgccg
gcacctgtcc tacgagttgc 7320atgataaaga agacagtcat aagtgcggcg
acgatagtca tgccccgcgc ccaccggaag 7380gagctgactg ggttgaaggc
tctcaagggc atcggtcgac gctctccctt atgcgactcc 7440tgcattagga
agcagcccag tagtaggttg aggccgttga gcaccgccgc cgcaaggaat
7500ggtgcatgca aggagatggc gcccaacagt cccccggcca cggggcctgc caccat
755636830DNAArtificial sequenceSynthetic sequence 368ttcatttgag
cattaaatgt caagttctgc 3036930DNAArtificial sequenceSynthetic
sequence 369ttcatttgag cattaagtgt caagttctgc 3037021DNAArtificial
sequenceSynthetic sequence 370aaactcgtaa ttcacagttc a
2137110DNAArtificial SequenceSynthetic sequence 371tatatatata
103726DNAArtificial SequenceSynthetic sequence 372tatata
63735DNAArtificial SequenceSynthetic sequence 373tatat 5
References
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014

D00015

D00016

D00017

D00018

D00019
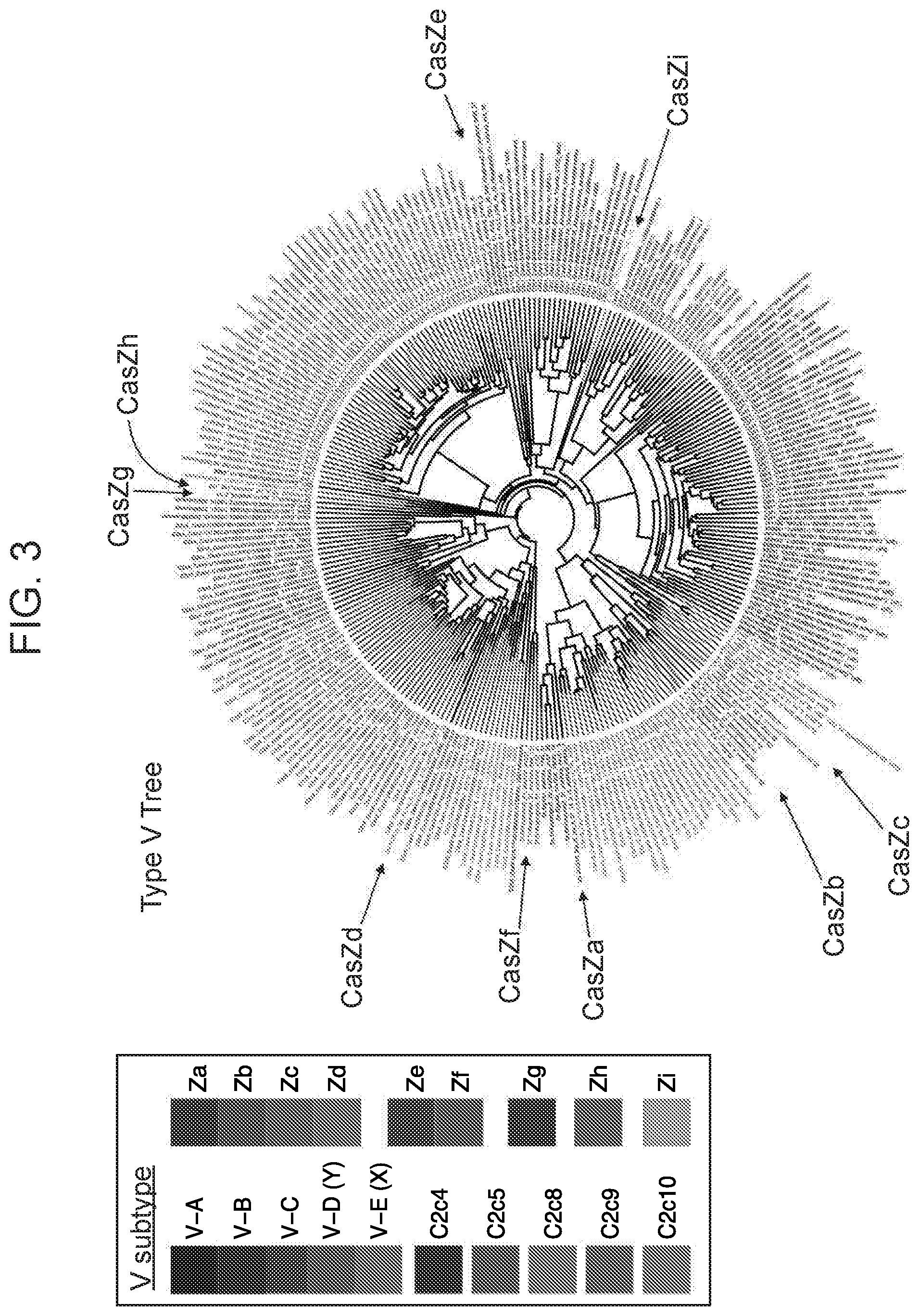
D00020

D00021

D00022
D00023

D00024

D00025

D00026

D00027

D00028

D00029

D00030

D00031

D00032

D00033

D00034

D00035

D00036

D00037

D00038

D00039

D00040

D00041

D00042

D00043
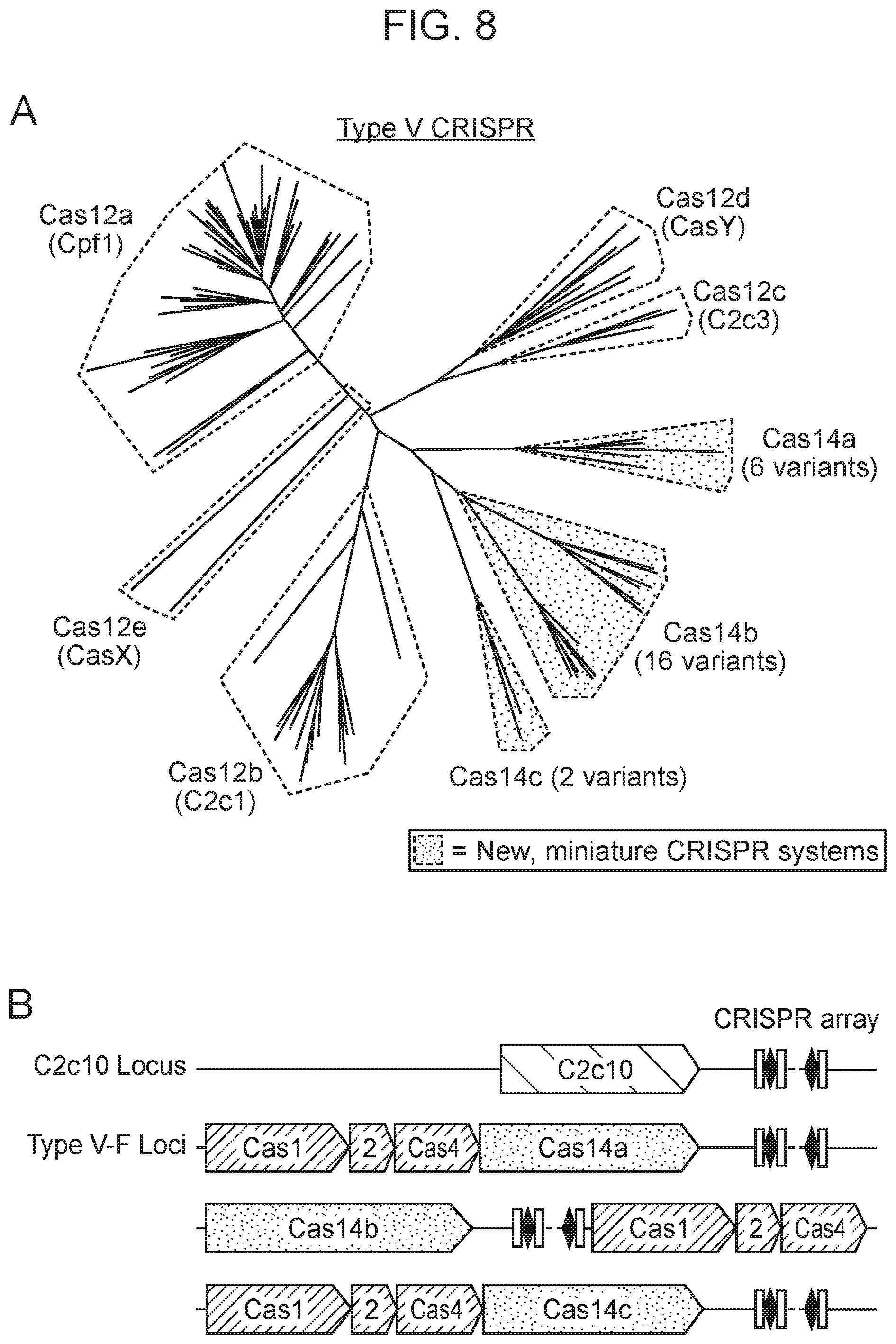
D00044
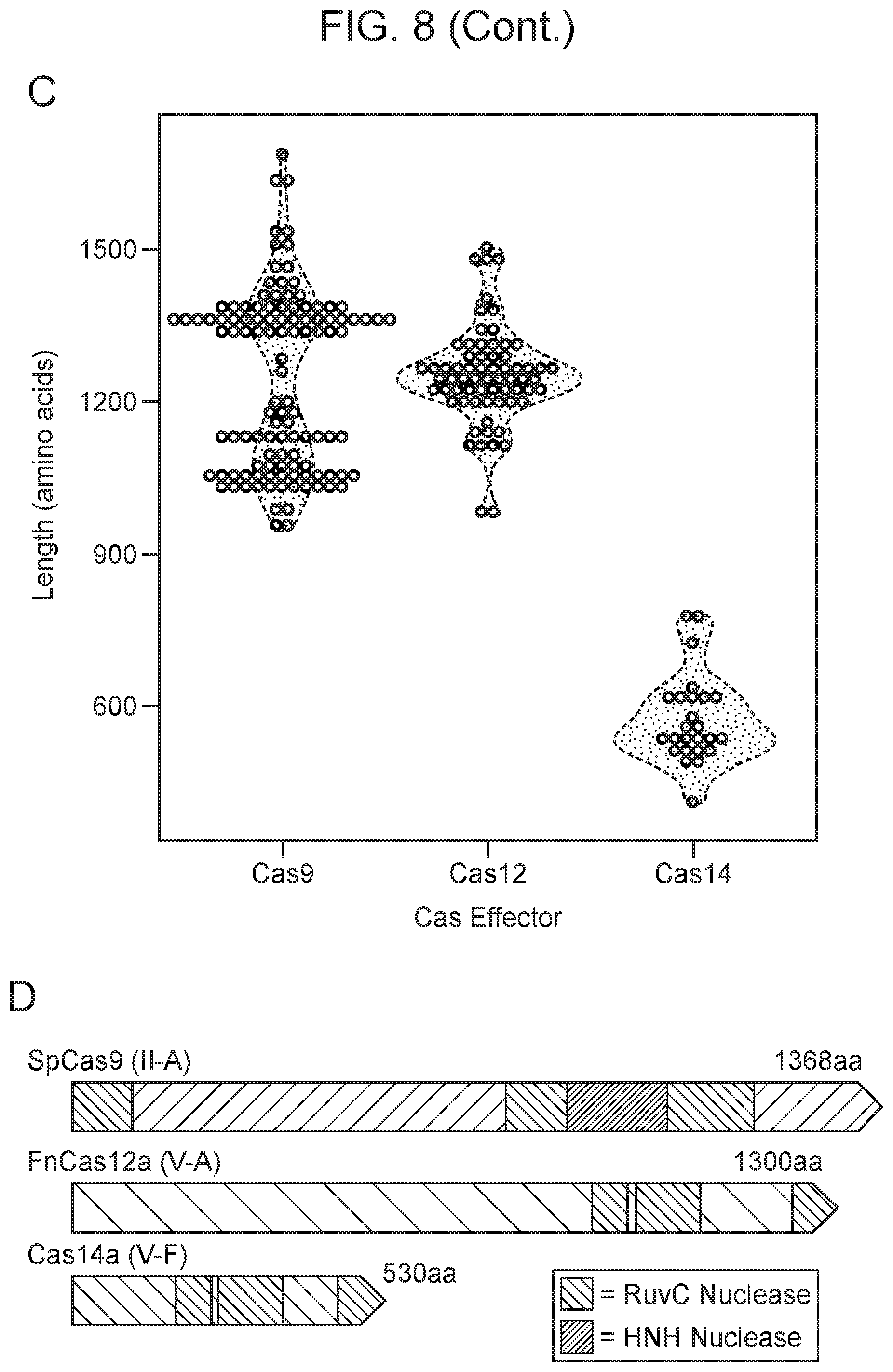
D00045

D00046
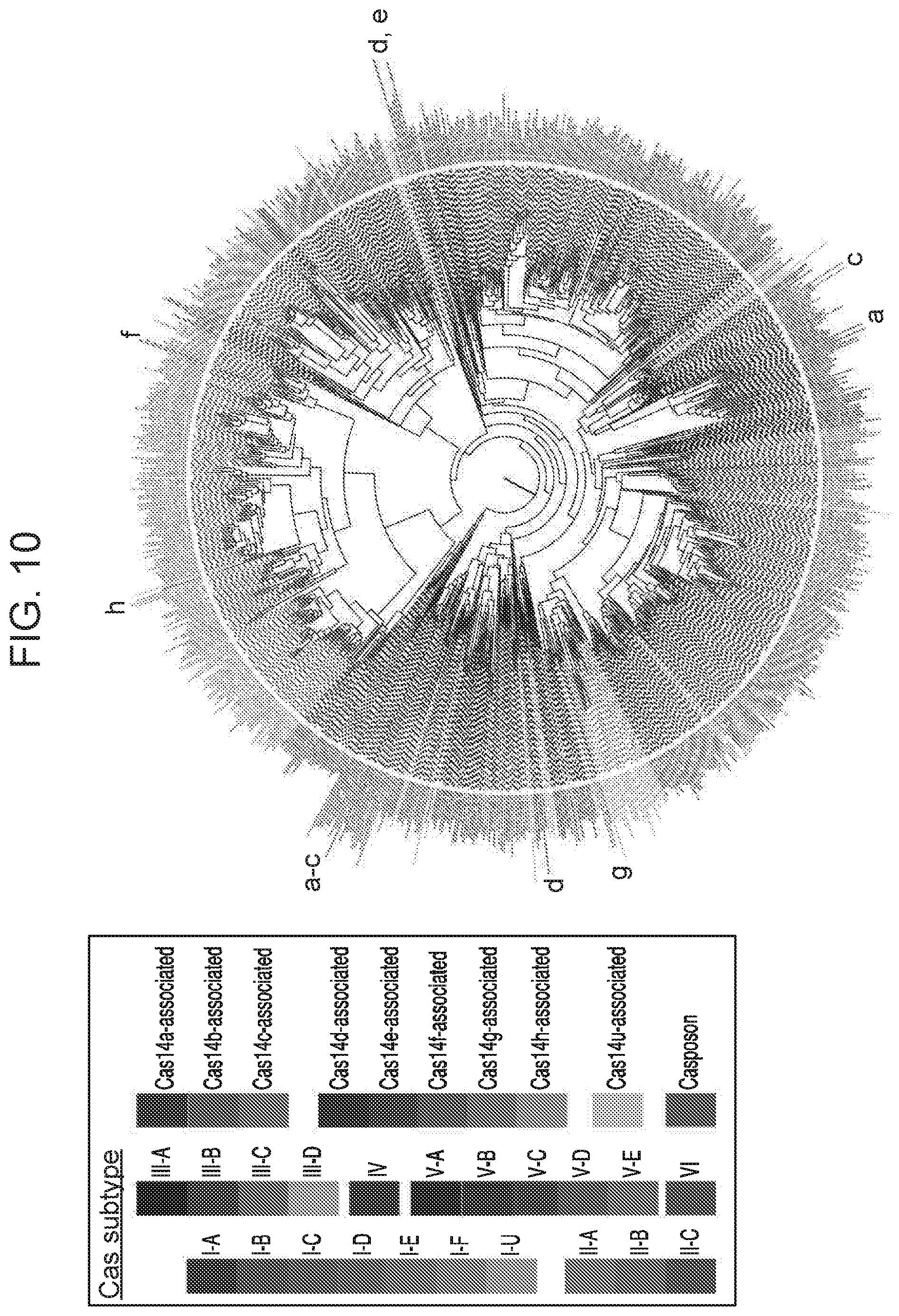
D00047
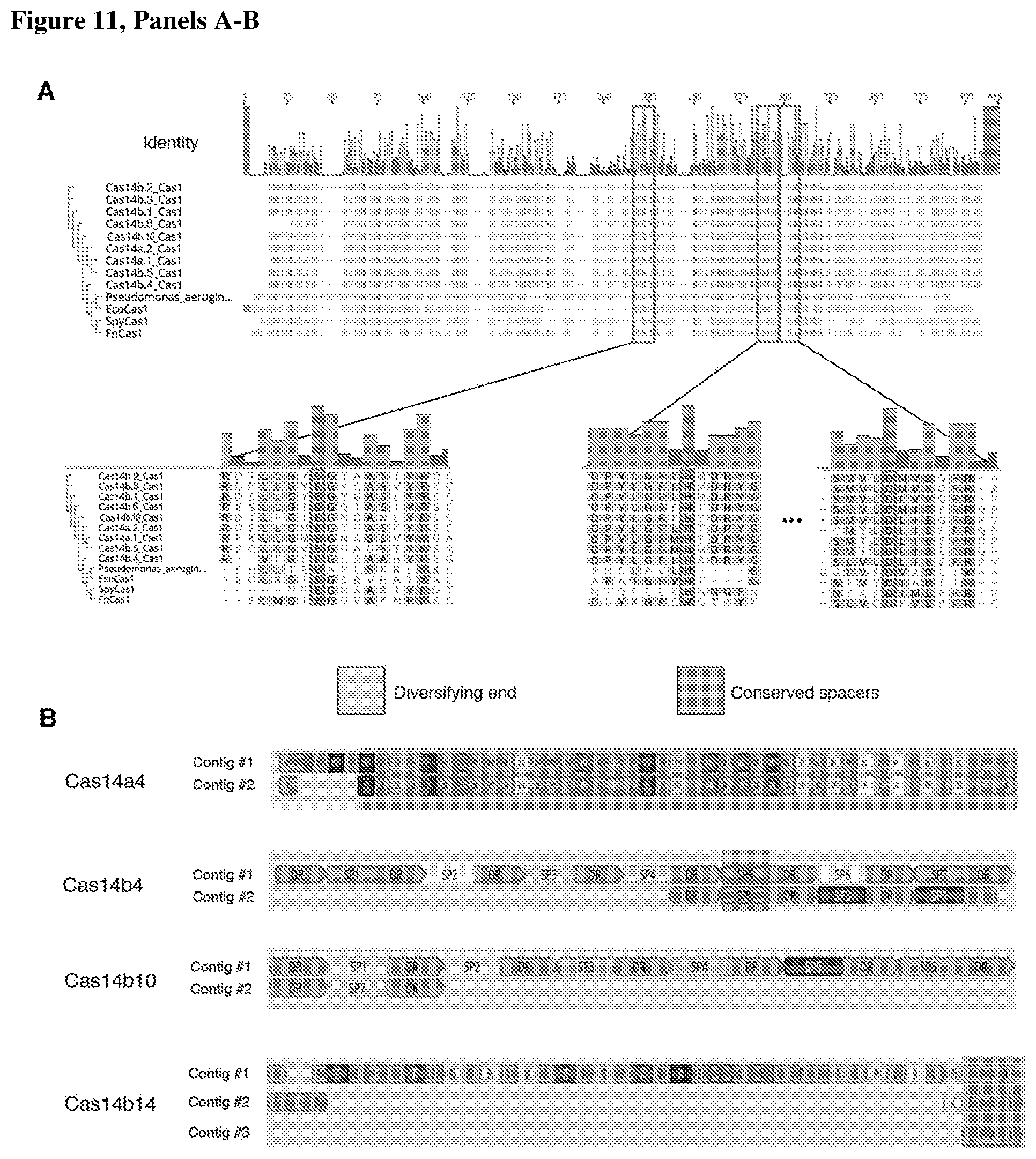
D00048
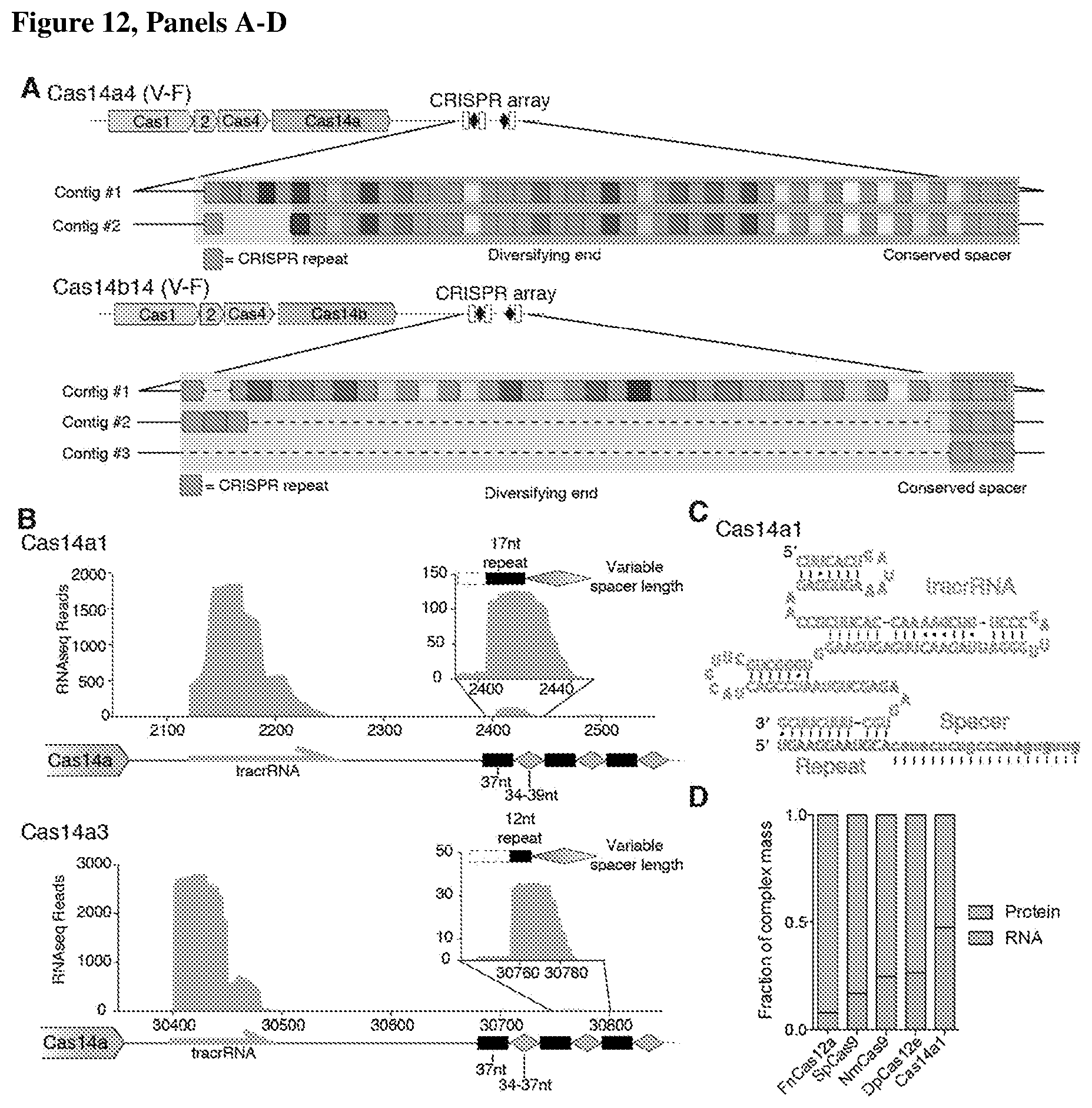
D00049

D00050
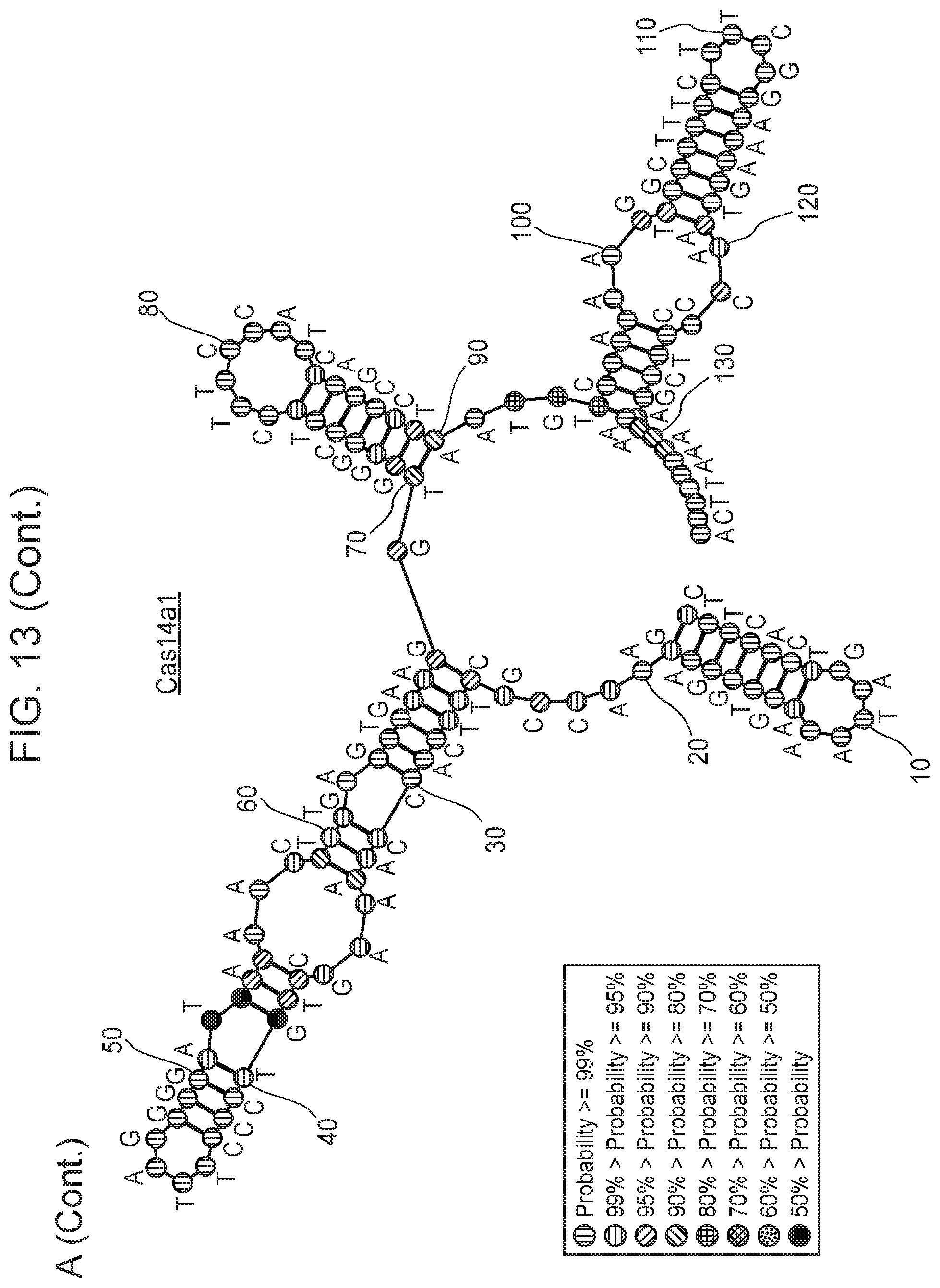
D00051

D00052
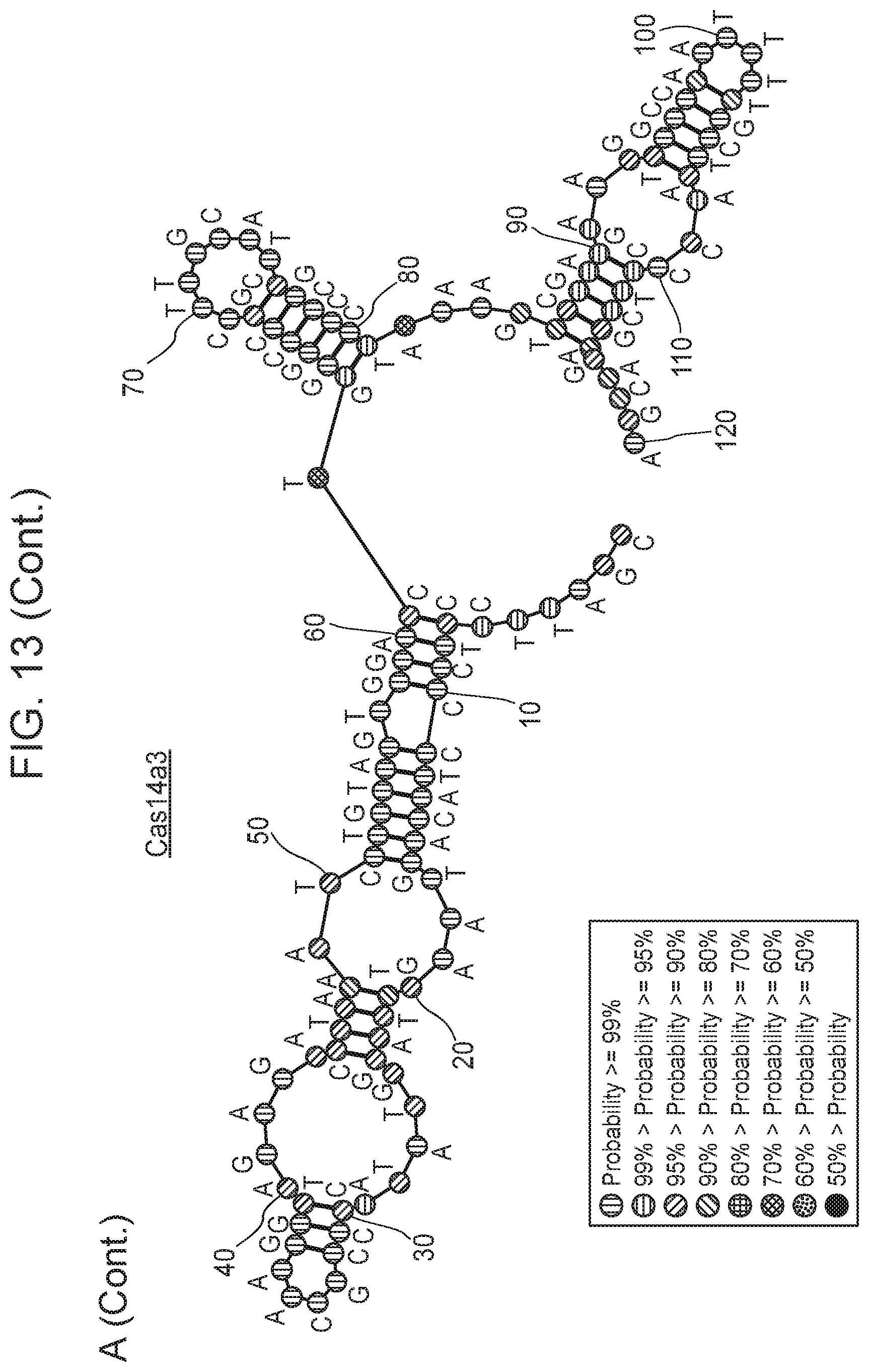
D00053

D00054
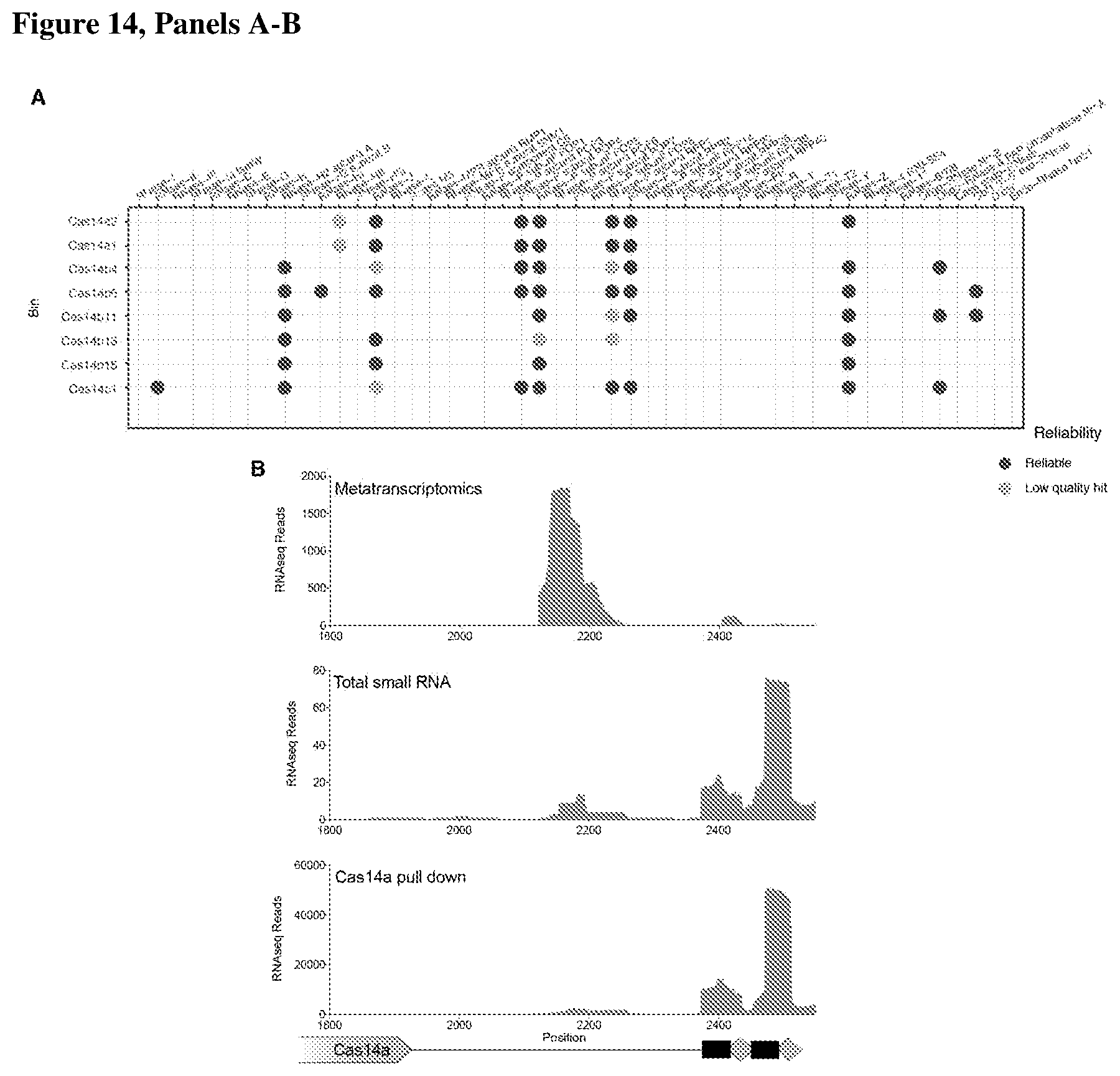
D00055
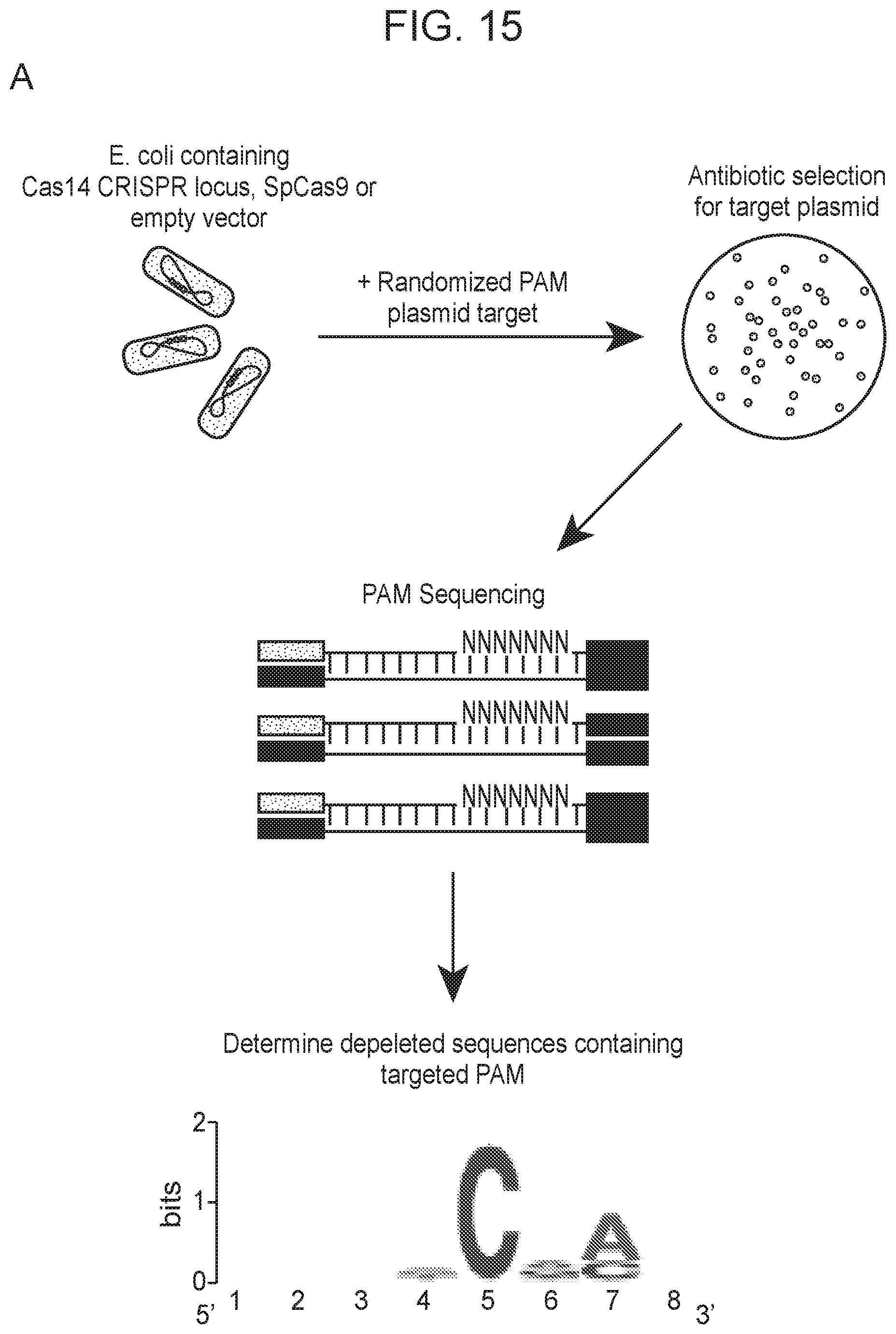
D00056
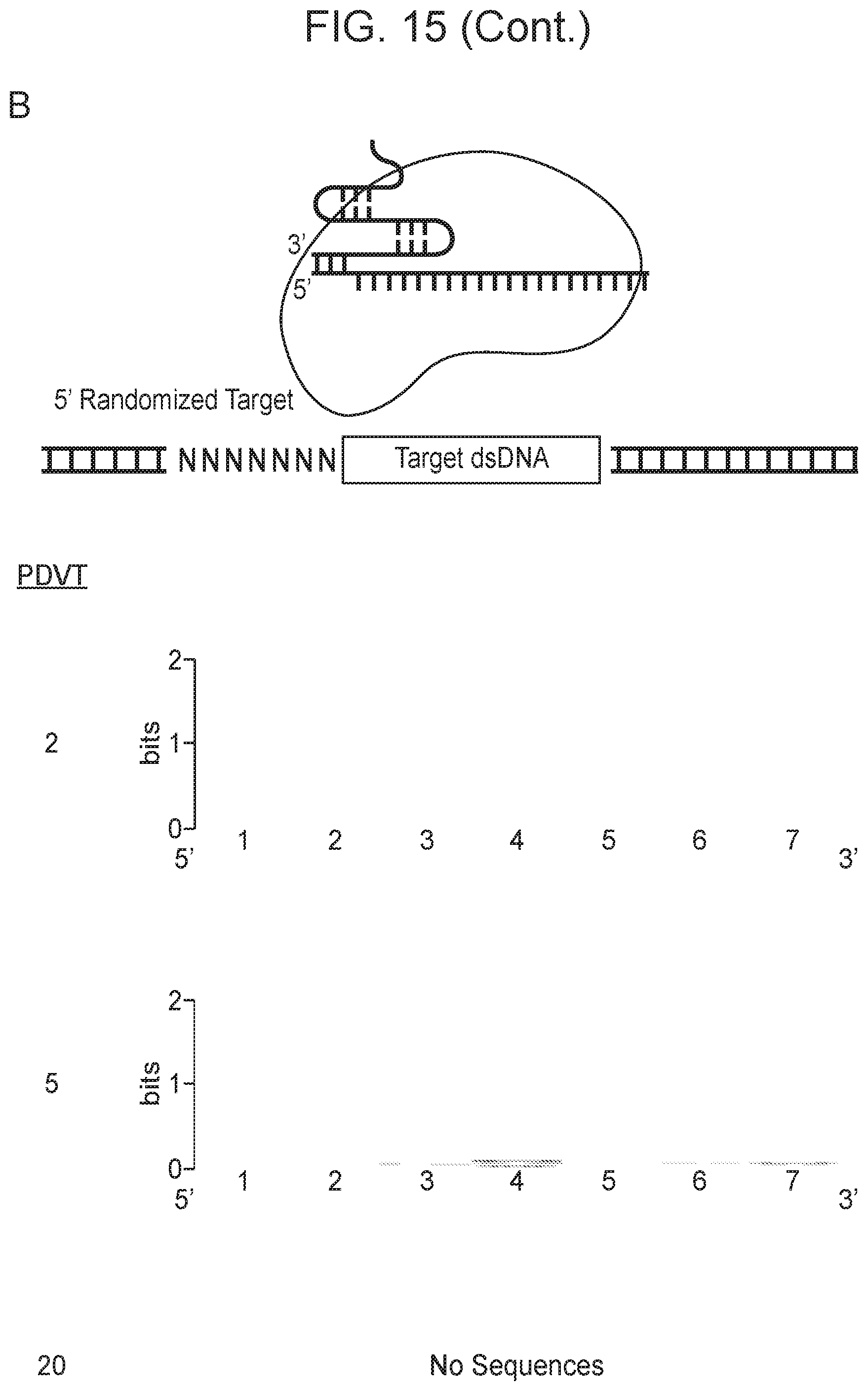
D00057

D00058
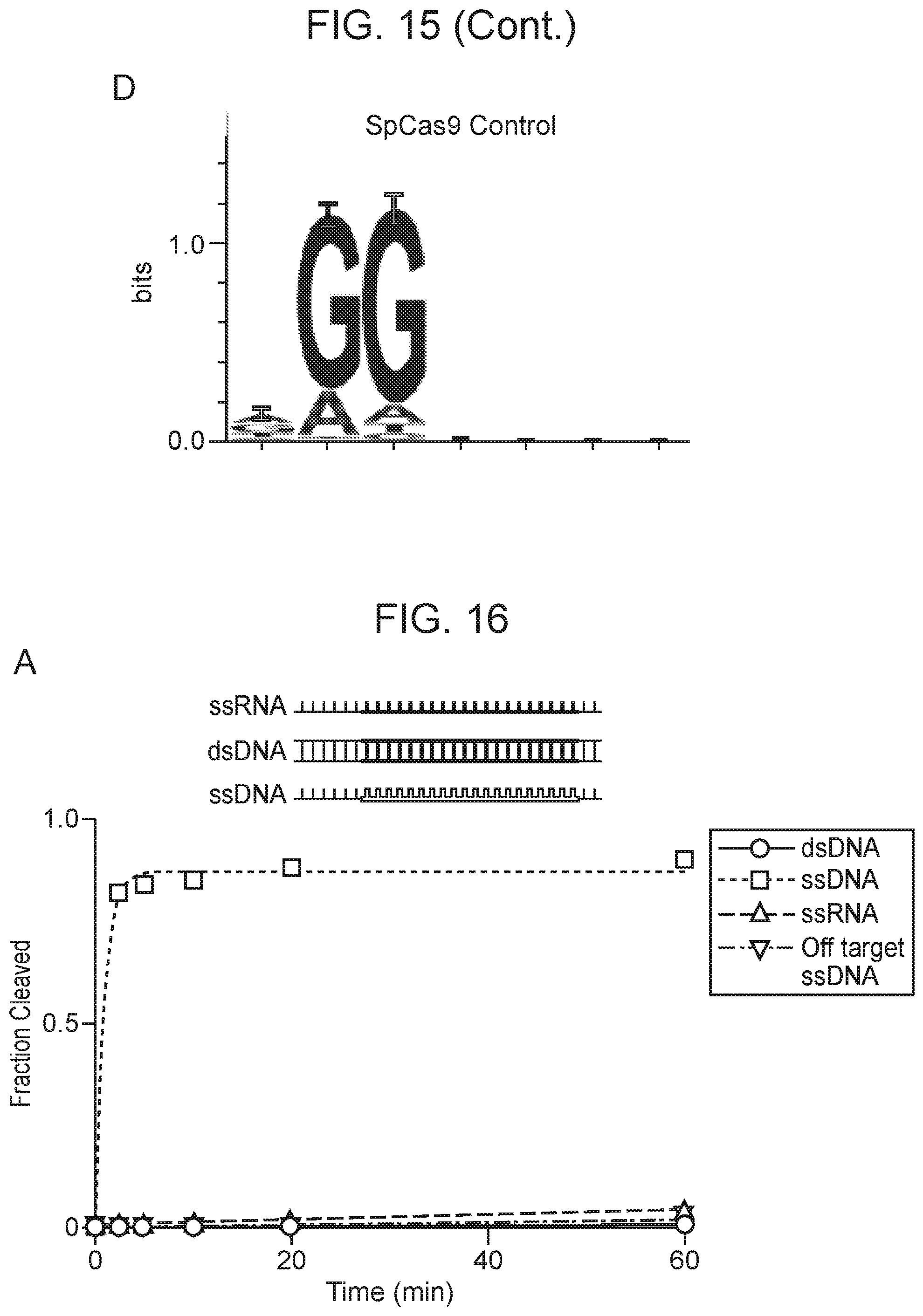
D00059
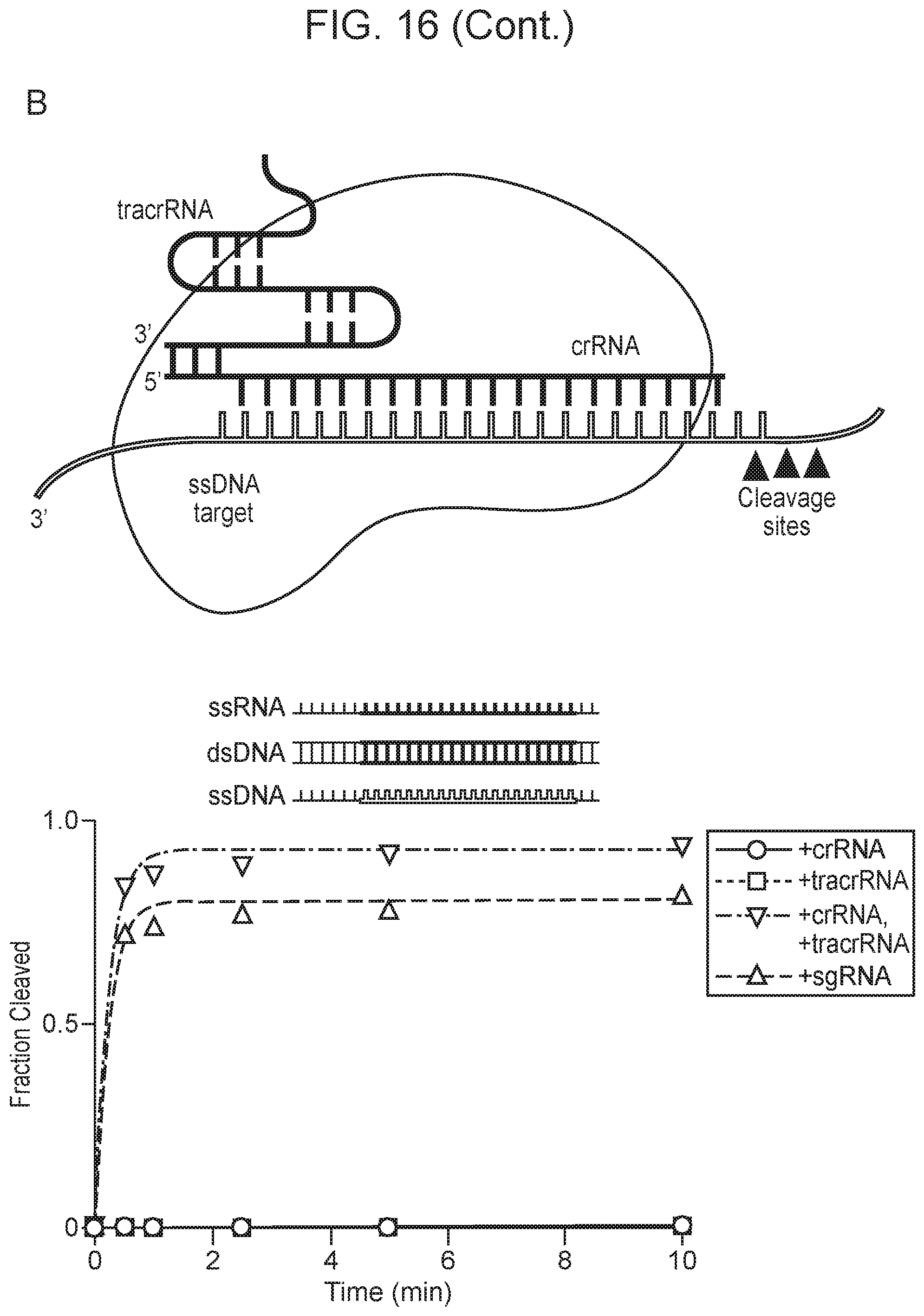
D00060

D00061

D00062
D00063

D00064

D00065

D00066
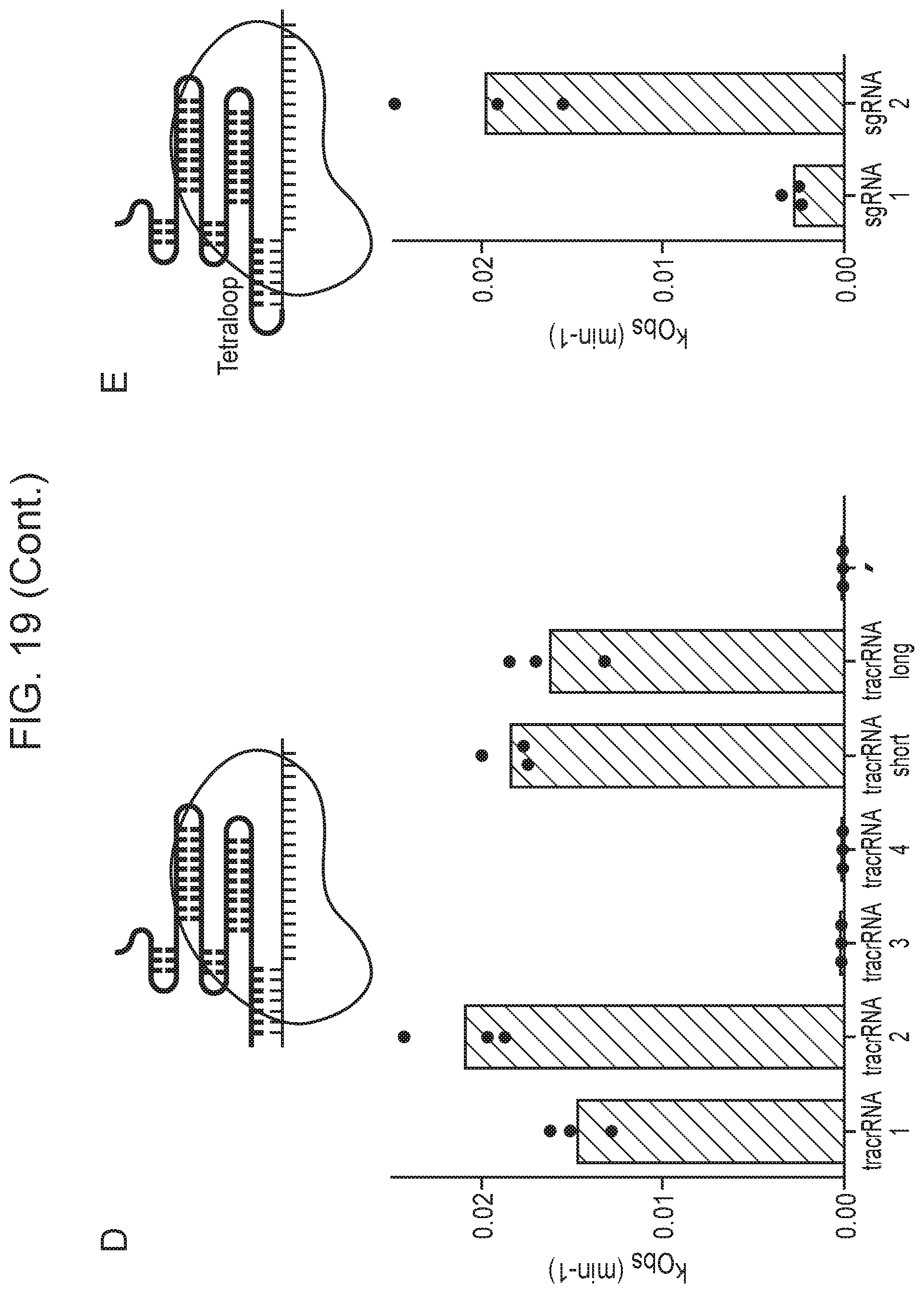
D00067
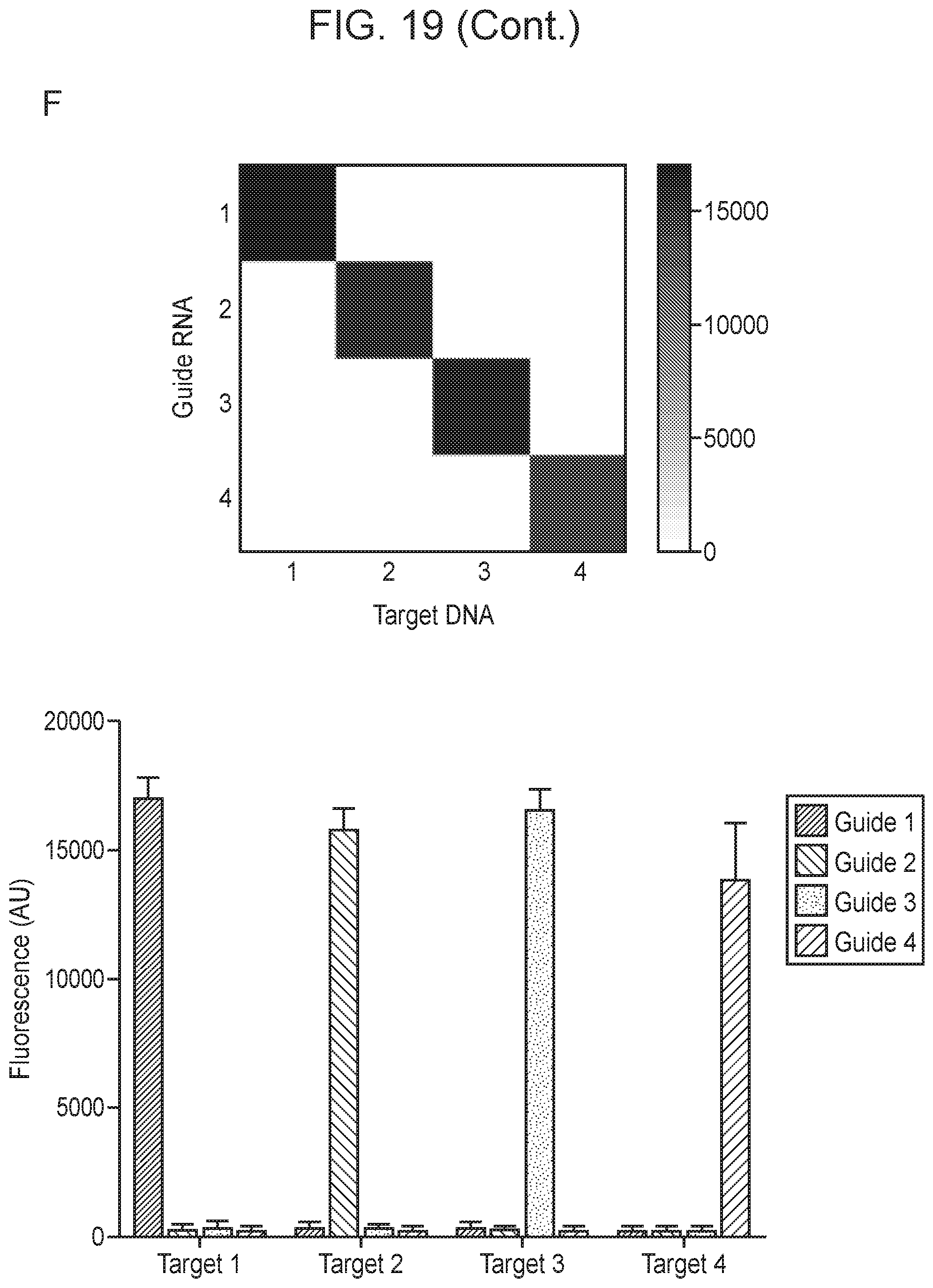
D00068
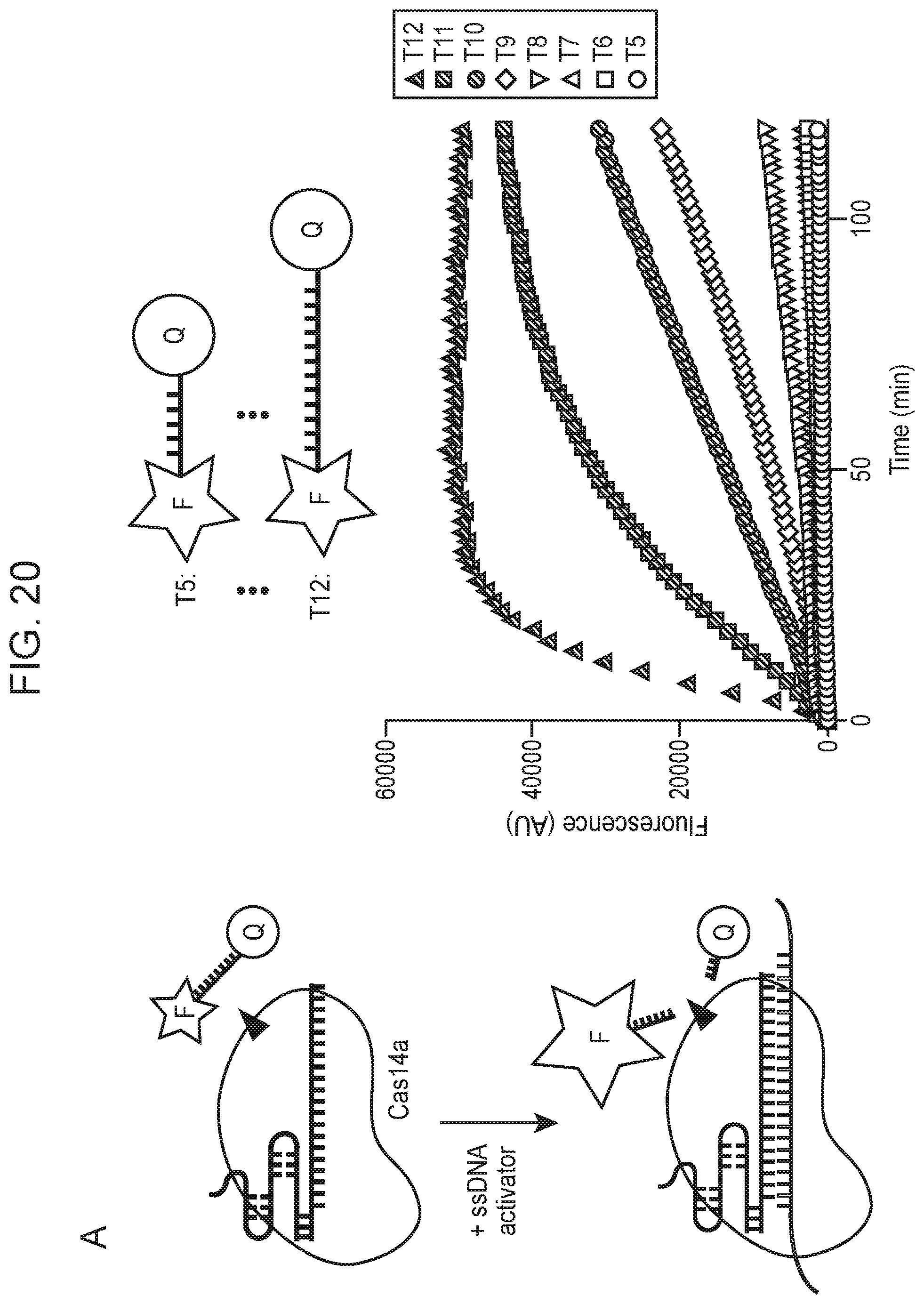
D00069

D00070
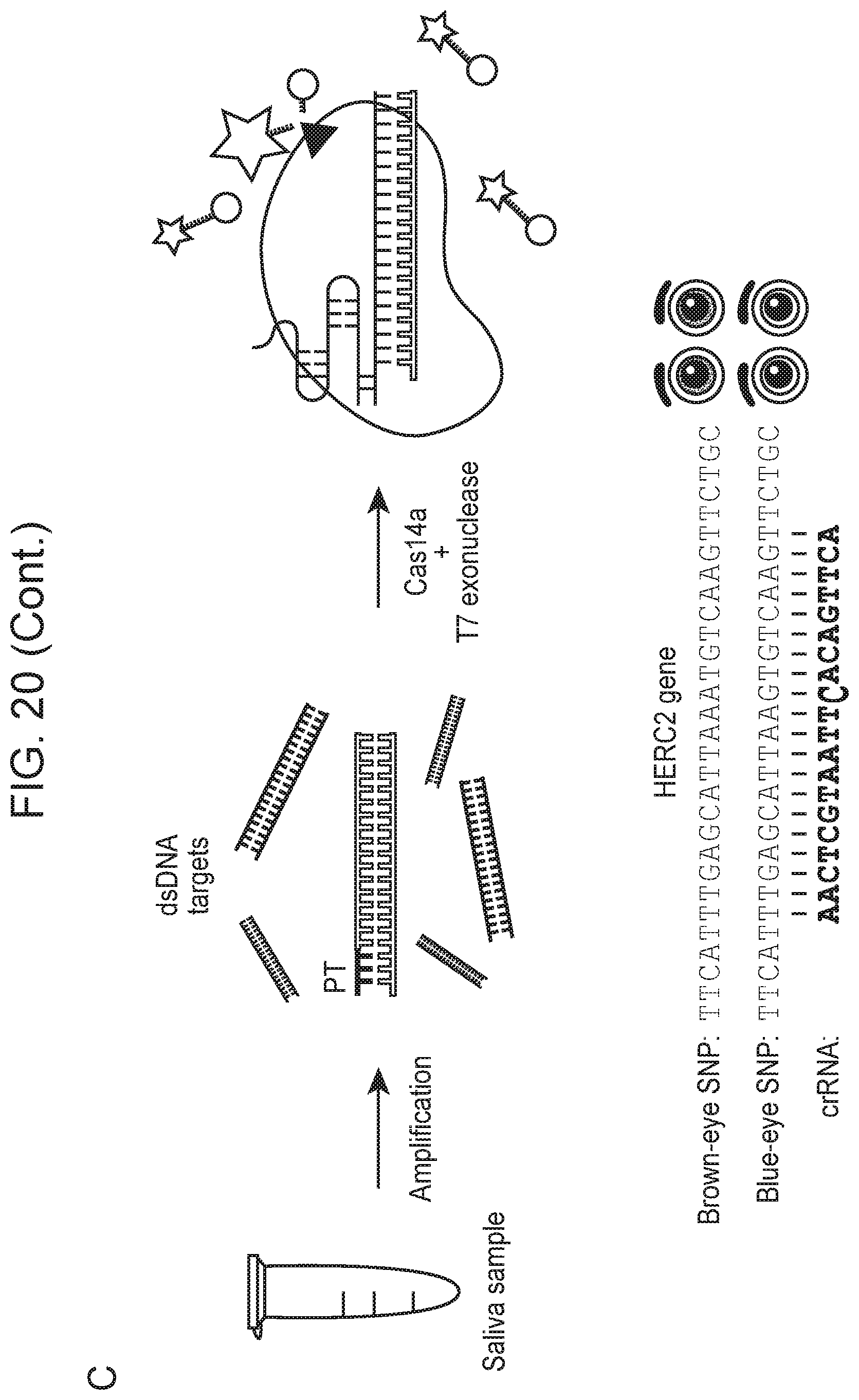
D00071

D00072

D00073
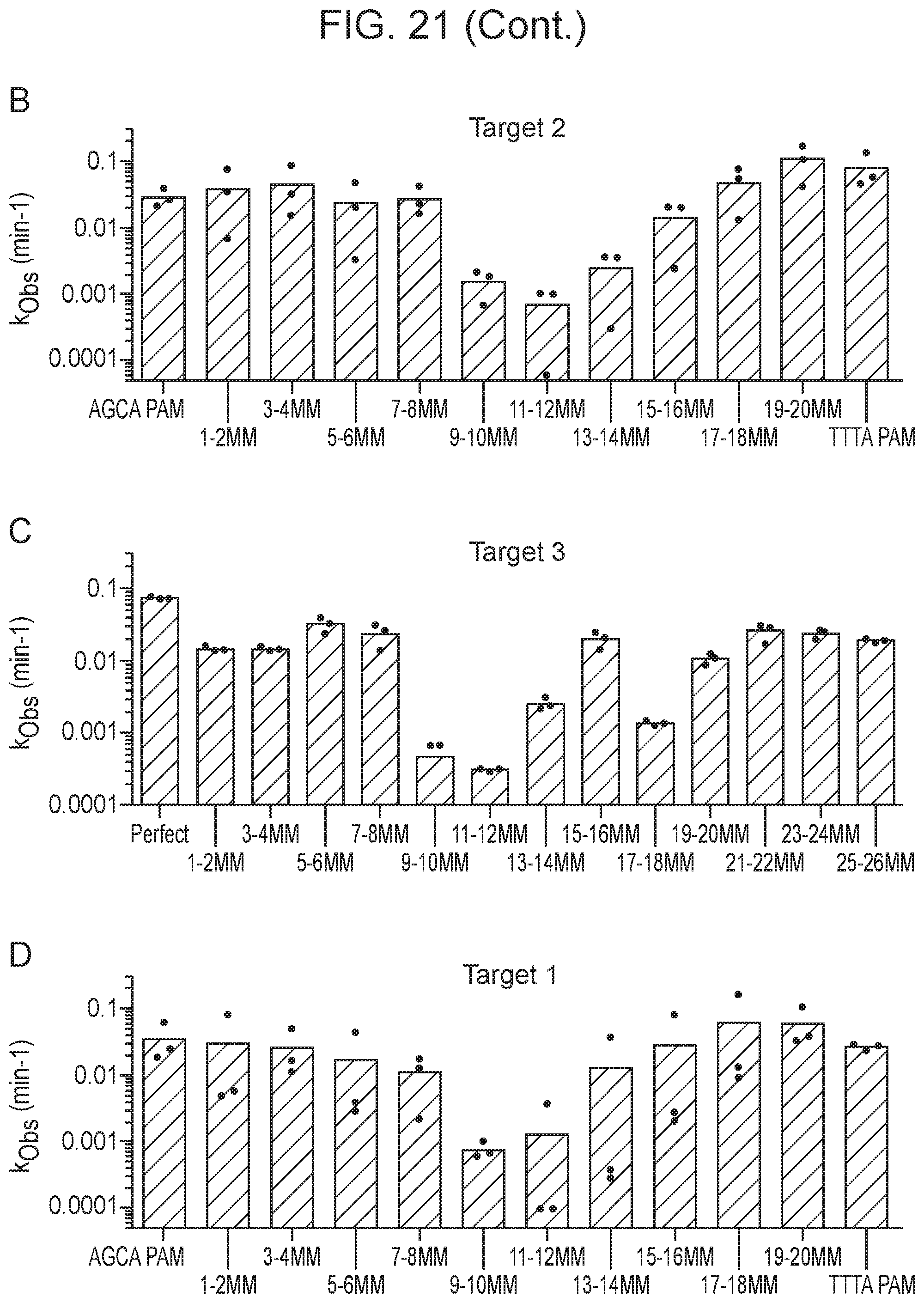
D00074
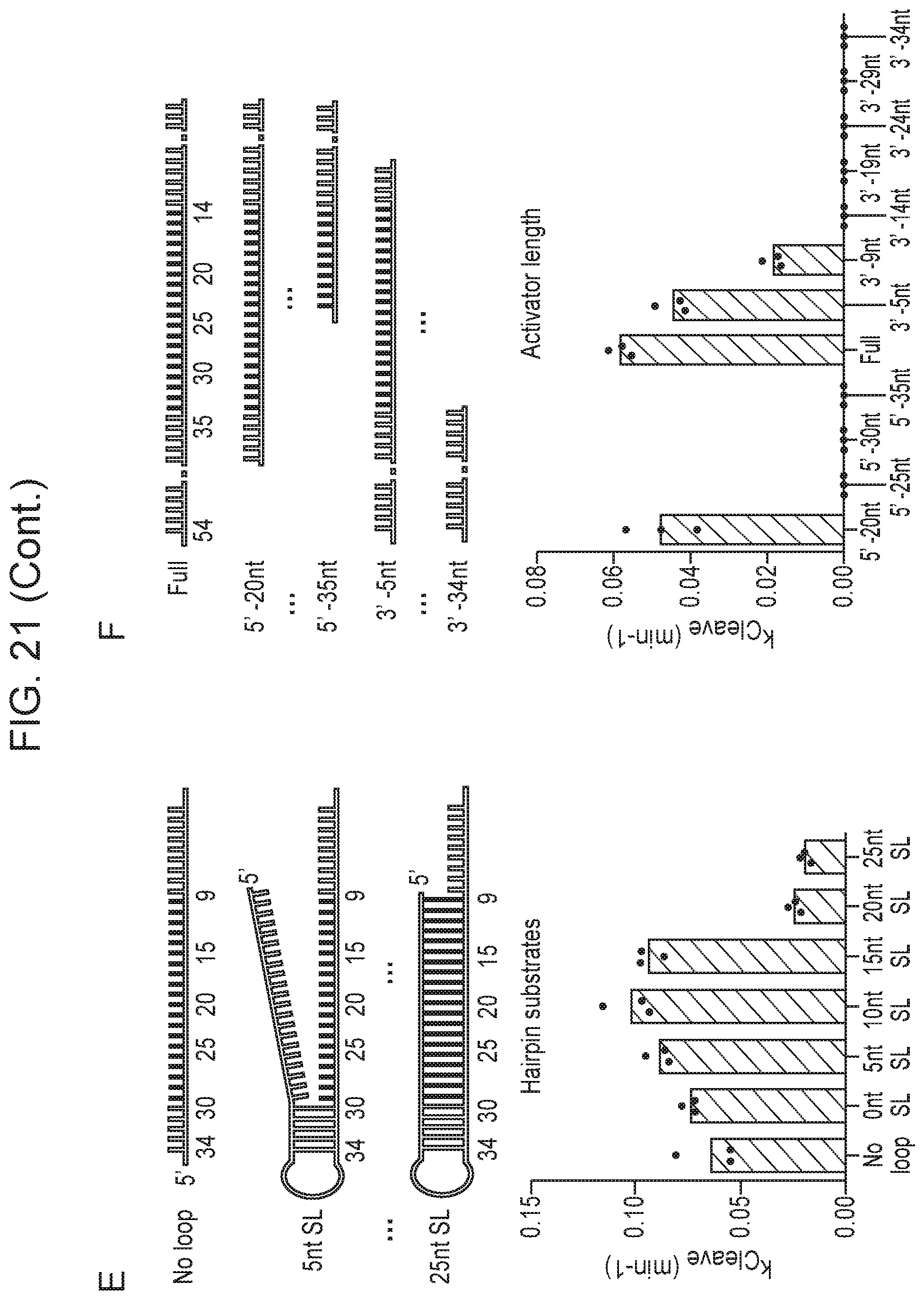
D00075

D00076
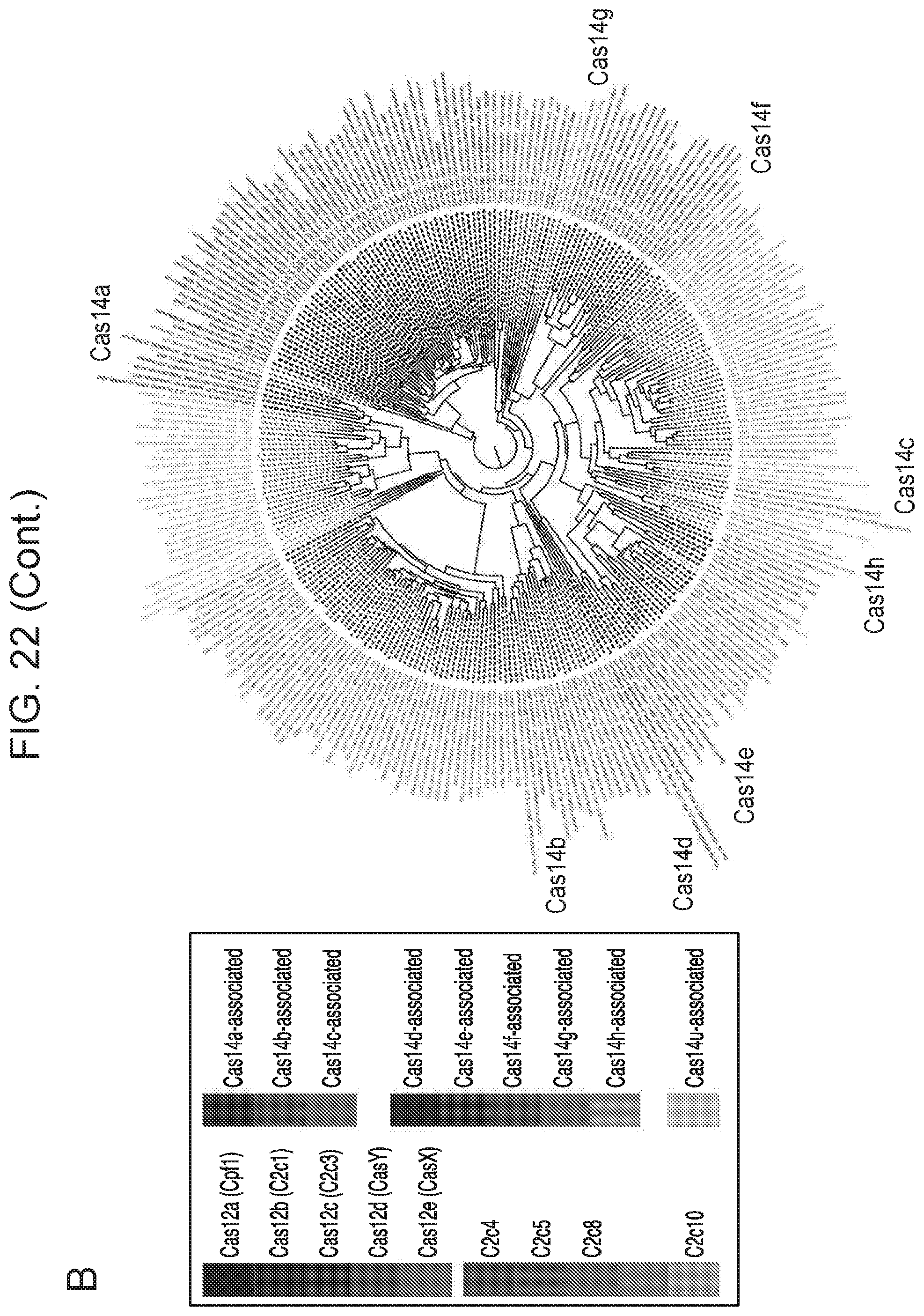
D00077
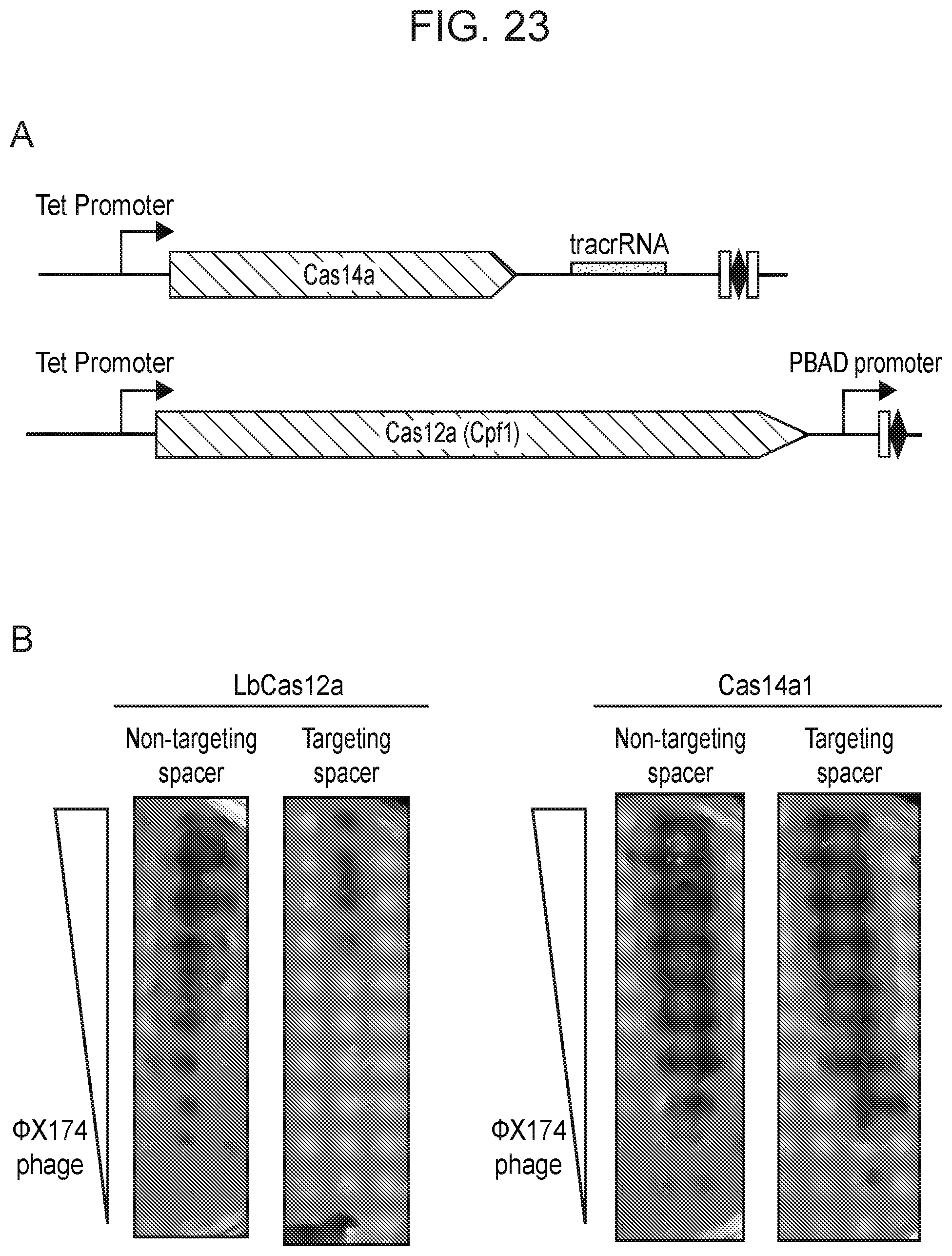
D00078

D00079

D00080

D00081

D00082

D00083

D00084

D00085

D00086

D00087

D00088
D00089

D00090

D00091
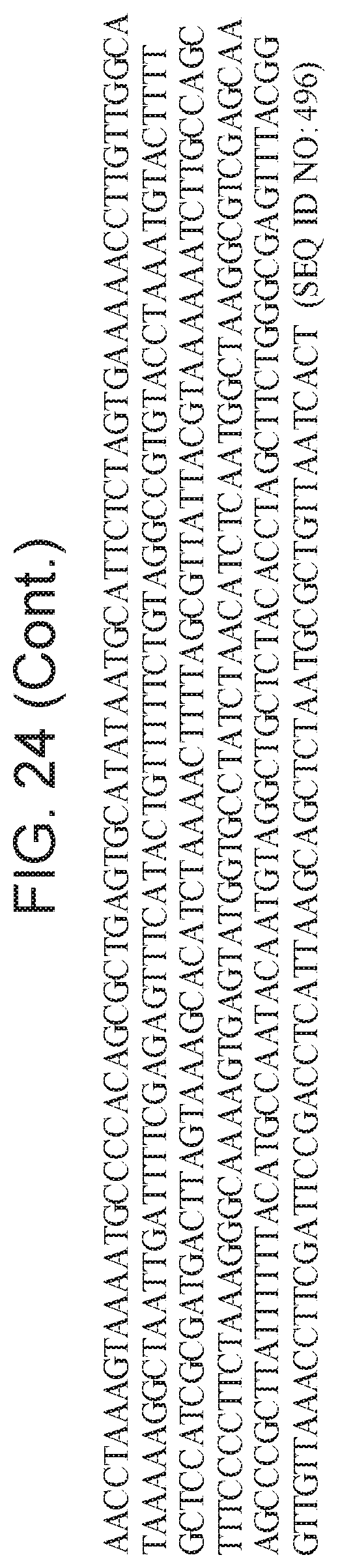
D00092

D00093

D00094

D00095

D00096
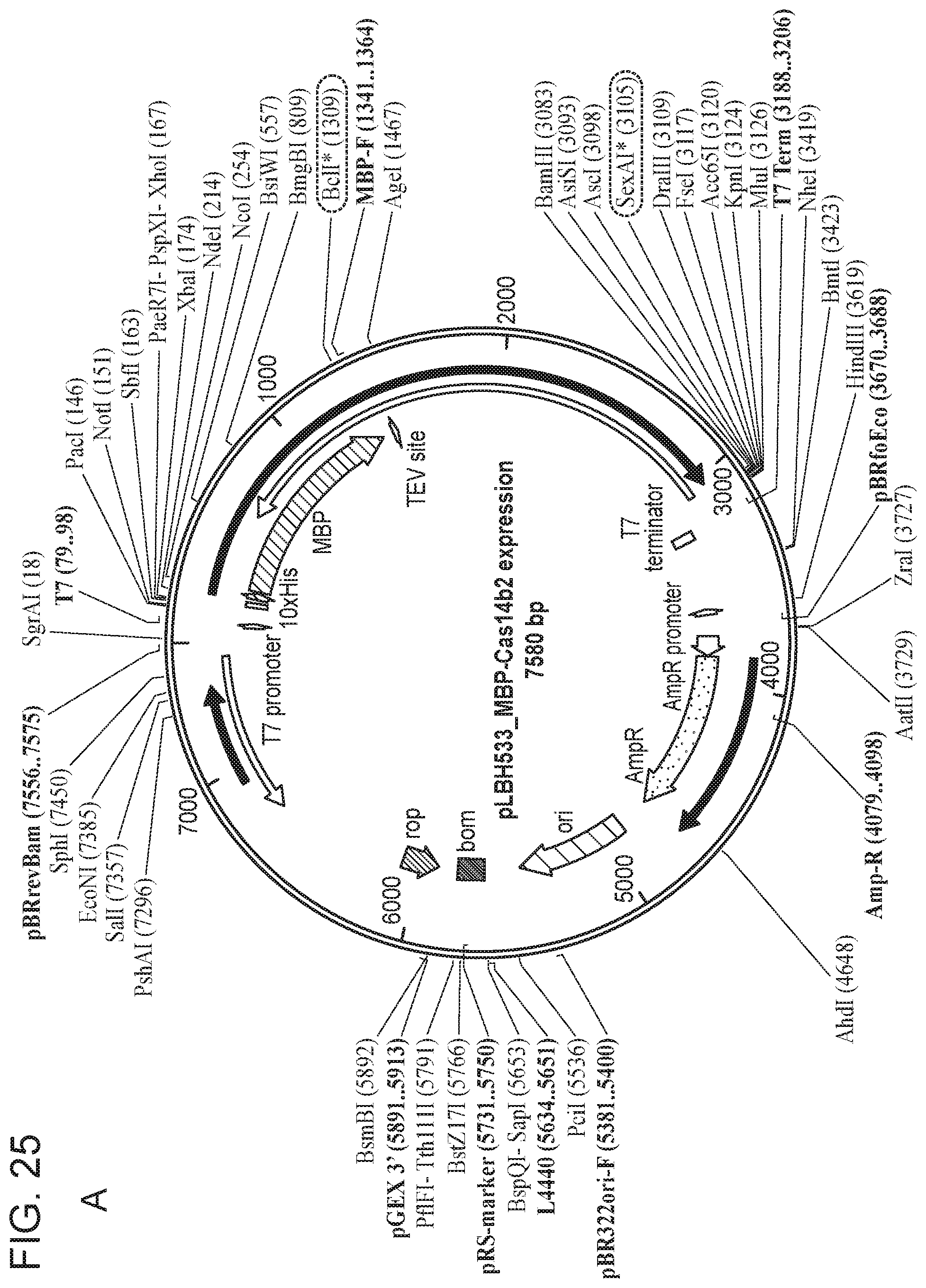
D00097

D00098
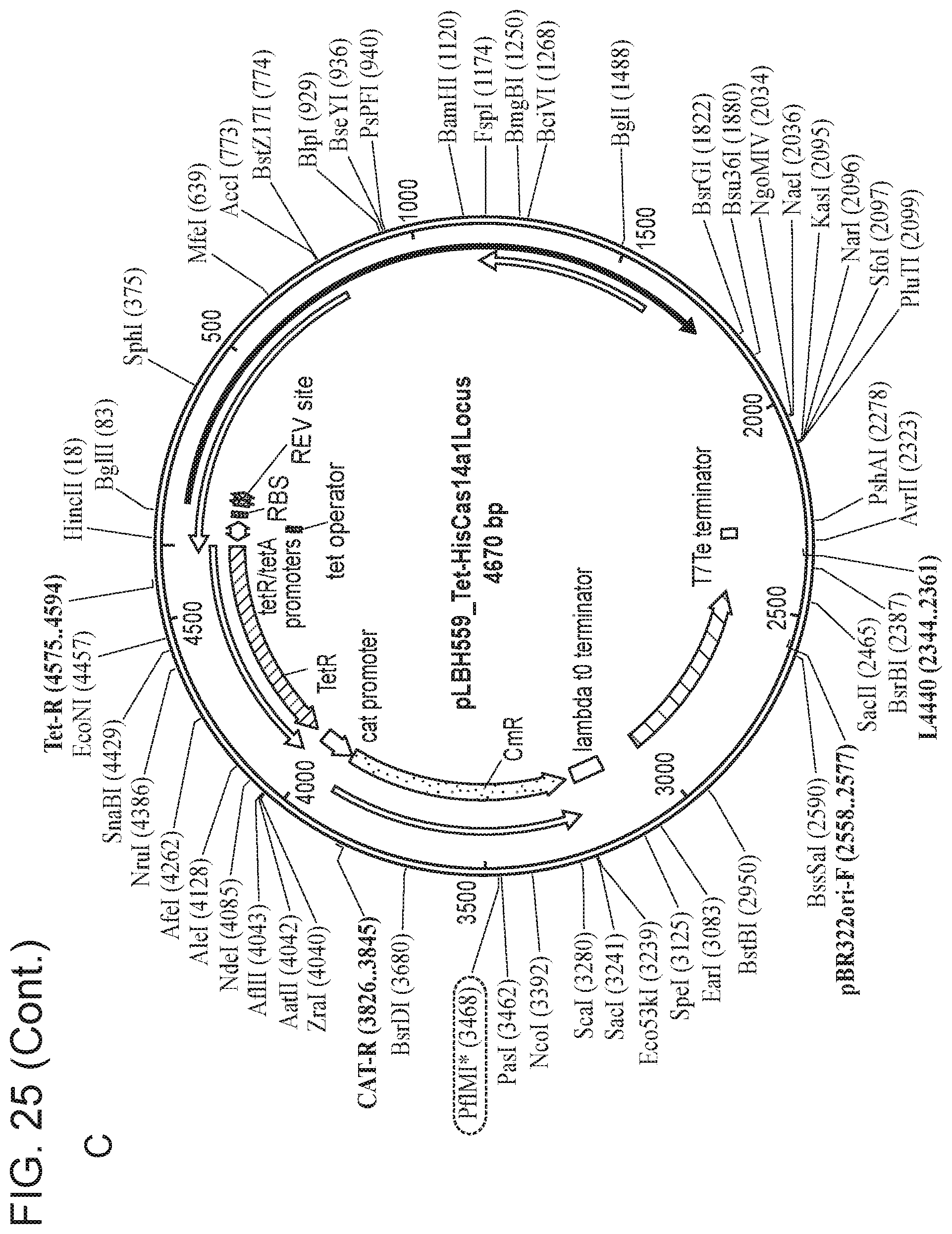
D00099

D00100
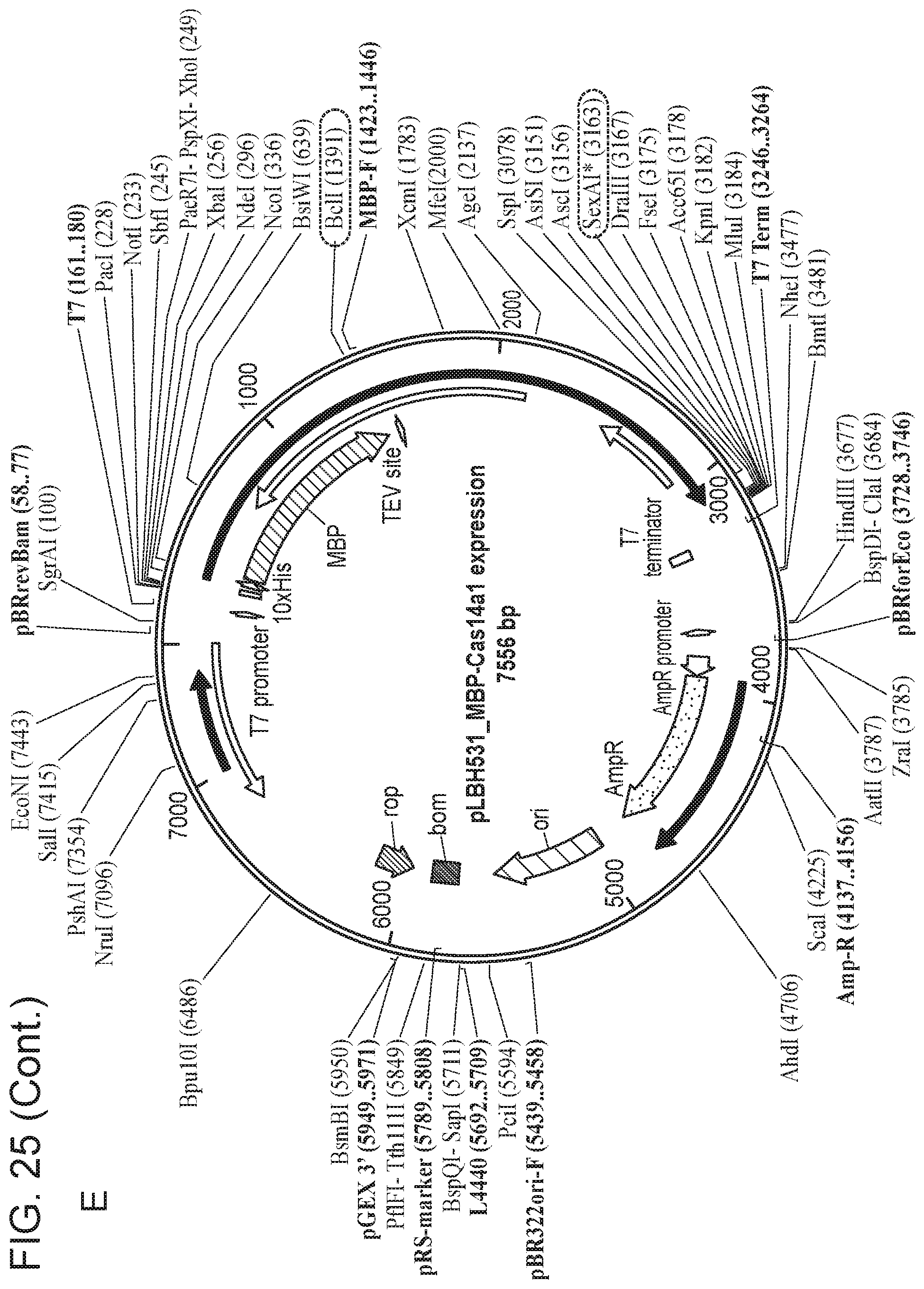
S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.