Anti-nuclear Antibody Detection And Diagnostics For Systemic And Non-systemic Autoimmune Disorders
SURESH; LAKSHMANAN ; et al.
U.S. patent application number 16/564209 was filed with the patent office on 2020-03-05 for anti-nuclear antibody detection and diagnostics for systemic and non-systemic autoimmune disorders. The applicant listed for this patent is IMMCO DIAGNOSTICS, INC.. Invention is credited to KISHORE MALYAVANTHAM, LAKSHMANAN SURESH.
| Application Number | 20200072845 16/564209 |
| Document ID | / |
| Family ID | 54196278 |
| Filed Date | 2020-03-05 |

| United States Patent Application | 20200072845 |
| Kind Code | A1 |
| SURESH; LAKSHMANAN ; et al. | March 5, 2020 |
ANTI-NUCLEAR ANTIBODY DETECTION AND DIAGNOSTICS FOR SYSTEMIC AND NON-SYSTEMIC AUTOIMMUNE DISORDERS
Abstract
Provided are compositions that contain mammalian cells for use in detecting antibodies. The mammalian cells are modified such that they do not contain LEDGF protein. The mammalian cells are immobilized on a solid substrate. The compositions can also contain mammalian cells that contain the LEDGF protein. Methods for using the cell compositions in diagnostic approaches are included, as are kits for performing diagnostic tests.
| Inventors: | SURESH; LAKSHMANAN; (WILLIAMSVILLE, NY) ; MALYAVANTHAM; KISHORE; (WILLIAMSVILLE, NY) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 54196278 | ||||||||||
| Appl. No.: | 16/564209 | ||||||||||
| Filed: | September 9, 2019 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 15124175 | Sep 7, 2016 | 10451633 | ||
| PCT/US15/22120 | Mar 24, 2015 | |||
| 16564209 | ||||
| 61969771 | Mar 24, 2014 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | G01N 33/564 20130101; C12N 15/85 20130101; G01N 33/5005 20130101; G01N 33/6854 20130101 |
| International Class: | G01N 33/68 20060101 G01N033/68; G01N 33/50 20060101 G01N033/50; G01N 33/564 20060101 G01N033/564; C12N 15/85 20060101 C12N015/85 |
Claims
1-9. (canceled)
10. A method for determining whether a biological sample comprises antibodies that bind to LEDGF protein (anti-LEDGF Abs), the method comprising: i) exposing the biological sample to mammalian cells that are modified such that they do not comprise LEDGF protein (LEDGF- cells), ii) exposing the biological sample to mammalian cells that comprise the LEDGF protein (LEDGF+ cells), and iii) comparing the amount of anti-LEDGF Abs bound to the LEDGF- cells to the amount of anti-LEDGF Abs bound to the LEDGF+ cells, wherein determining a greater amount of anti-LEDGF Abs bound to the LEDGF+ cells relative to the amount of anti-LEDGF Abs bound to the LEDGF- cells is indicative that the biological sample comprised the anti-LEDGF Abs, and wherein the same or less anti-LEDGF Abs bound to the LEDGF+ cells relative to the amount of anti-LEDGF Abs bound to the LEDGF- cells is indicative that the biological sample did not comprise the anti-LEDGF Abs.
11. The method of claim 10 wherein the LEDGF- cells are killed and permeabilized and are immobilized on a solid substrate.
12. The method of claim 11, wherein the LEDGF+ cells are killed and permeabilized, and are immobilized on the same solid substrate as the LEDGF- cells, or are immobilized on a distinct solid substrate.
13. The method of claim 12, wherein the determining the amount of anti-LEDGF Abs is performed using an indirect immunofluorescence (IIF) assay.
14. The method of claim 10, wherein a chromosome in the LEDGF- cells comprises a Cas9 DNA coding sequence, or a DNA sequence encoding a clustered regularly interspaced short palindromic repeats (CRISPR) guide RNA targeted to a DNA sequence encoding the LEDGF protein, or a combination thereof.
15. The method of claim 12, wherein the LEDGF- and LEGDF+ cells are the same mammalian cell type.
16-20. (canceled)
Description
CROSS REFERENCE TO RELATED APPLICATIONS
[0001] This application claims priority to U.S. provisional application No. 61/969,771, filed on Mar. 24, 2014, the disclosure of which is incorporated herein by reference.
FIELD
[0002] The present invention relates generally to autoimmune disease and more particularly to compositions and methods for use in detecting anti-nuclear antibodies
BACKGROUND
[0003] Autoimmune diseases are typically challenging to diagnose, and in general individuals with one autoimmune disease are at higher risk for developing others, such as systemic autoimmune rheumatic diseases (SARDs). The presence of anti-nuclear autoantibodies (ANAs) is considered to be a hallmark of SARDS, and this association has been known for some time. The American College of Rheumatology (ACR) recommends testing for ANAs by indirect immunofluorescence (IIF) assay using HEp-2 cells, as described in their position statement in 2009. This statement explained that HEp-2 cells are able to express 100-150 relevant auto-antigens for use in ANA antibody detection. Thus, immobilized and preserved monolayers of HEp2 cells are the most commonly used substrates in IIF detection of ANAs.
[0004] ANA detection using IIF assay can reveal a multitude of patterns such as homogeneous, fine granular, coarse granular, nucleolar, centromere, nuclear dots, pleomorphic, mitochondrial and a variety of cytoskeletal patterns. Patients can have one or more patterns in combination with varying intensity of reactivity for each pattern. These patterns are a result of specific autoantibody binding to nuclear and cytoplasmic antigens which include but are not necessarily limited to dsDNA, nucleosomes, histones, SS-A Ro52/Ro60, SS-B/La, Ku, Mi-2, RNPs (Ribonucleoproteins: U1SnRNP 68, U1SnRNP A, U1SnRNP C, U2SnRNPs etc,), Scl-70, PM-Scl, Fibrillarin, Th/To, CENP-B, CENP-A, Sp100, PCNA, Ribo-P, Jo1, AMA-M2, Actin, Vimentin, and others.
[0005] Other methodologies have been utilized for screening and confirmation of ANAs. However, due to a variety of reasons, which include but are not limited to prevalence of false negative and false positive results, lack of standardization of test algorithms (i.e., reflex testing), and an inability to detect the diverse arrays of ANAs prevalent in individuals with SARDS, use of HEp-2 cells as the substrate for ANA testing remains the gold standard. Unfortunately, use of HEp-2 cells also involves complex test interpretation, false results and specialized skills, in part because it has been reported that up to 20% of apparently healthy subjects give a positive ANA IIF test result due to the presence of autoantibodies that recognize the so-called "dense fine speckles 70" (DFS70) antigen, which is also referred to herein and in the art as lens epithelium-derived growth factor (LEDGF). PSIP1/LEDGF is also known as AA408851, AU015605, Dfs70, Ledgf, Ledgfa, Ledgfb, mLEDGF, PC4 and SFRS1-interacting protein (PSIP1), Psip2 (isoform), p52, p75, PAIP encoded by the PSIP1 gene. Moreover, the DFS IIF pattern has been reported in up to 20% of ANA positive healthy subjects, but often not in ANA positive sera obtain from SARD patients (Mahler and Fritzler 2012). Since the main objective of the ANA HEp-2 test is to function as a tool for diagnosing and classifying SARD, as well as potentially other autoimmune diseases, the anti-DFS70 antibodies and the DFS pattern they produce reduce the usefulness of the ANA test, such as by increasing false results and otherwise complicating test interpretation. This has important ramifications for a variety of approaches that rely on accurate detection of ANA and treatment decisions for patients who are tested for ANA antibodies. Thus, there is an ongoing and unmet need for improved compositions and methods for detecting ANA. The present disclosure meets these and other needs.
SUMMARY
[0006] The present disclosure comprises in various embodiments compositions and methods for use in detecting ANA autoantibodies, and/or for determining whether a biological sample obtained or derived from an individual comprises antibodies that recognize the LEDGF protein. The disclosure includes compositions and methods that can be used for diagnosing and/or aiding in the diagnosis of autoimmune disorders that are positively correlated with the presence of ANA autoantibodies. Kits/products comprising reagents for use in detection of ANA autoantibodies, such as antibodies to LEDGF protein, are also provided
[0007] In one aspect the present disclosure comprises modified mammalian cells for use in detecting antibodies. The mammalian cells are modified such that they do not express or comprise LEDGF protein (LEDGF- cells). In embodiments, the disclosure includes mixtures of the mammalian LEDGF- cells, and mammalian cells that do express and comprise LEDGF protein (LEDGF+ cells). In embodiments, the mammalian cells are immobilized on a solid substrate, such as a glass, plastic, or other polymer-based substrate. In embodiments, the solid substrate comprises a microscope slide, a diagnostic slide, a microtitre plate, or beads formed of glass or a polymer.
[0008] In embodiments, the cells are killed and permeabilized. Those skilled in the art will recognize that permeabilized cells are those cells which have been exposed to organic reagents (commonly referred to in the art as `fixatives`) which can include but are not limited to organic solvents, such as acetone, alcohol, and aldehyde containing solutions, such as formaldehyde, paraformaldehyde, and the like. Cells exposed to such reagents are commonly referred to as "fixed" and the process of treating them with such reagents is referred to as "fixing" the cells. Fixing the mammalian cells such that they are permeabilized is lethal, and thus the fixed/permeabilized cells are also considered to be killed cells.
[0009] The LEDGF- cells are modified using any suitable techniques, reagents and the like such that LEDGF protein is not expressed, or its expression is reduced. In embodiments, mRNA encoding the LEDGF protein is degraded using any of a variety of RNAi-mediated approaches. In another embodiment, the gene encoding the LEDGF protein, which is described more fully below, is disrupted by any suitable technique including but not limited to the use of a clustered regularly interspaced short palindromic repeats (CRISPR) system comprising a CRISPR-associated (Cas) nuclease and a CRISPR guide RNA (gRNA). In embodiments, the modification to the cells comprises integration of a polynucleotide sequence encoding the Cas enzyme and/or the gRNA into at least one chromosome of the cells, such as the LEDGF- cells.
[0010] In embodiments, the compositions and methods use modified LEDGF+ and LEDGF- cells of the same type, i.e., they are both the same type of cancer cell, or they are both of the same cell line, or derived from the same cell line.
[0011] In embodiments the disclosure includes mixtures of LEDGF+ mammalian cells and modified LEDGF- mammalian cells that are useful in diagnostic assays. The mixtures can be such that antibodies that bind to antigens in the modified cells can be used to, for example, establish a background amount of antibody binding that can be compared to antibody binding using the LEDGF+ cells as a comparison substrate. Thus, in certain aspects, a ratio of LEDGF- and LEDGF+ cells are provided. In embodiments, the ratio comprises a LEDGF- cell amount to a LEDGF+ cell amount of 1:1, 1:2, 1:3, 1:4, 1:3, 1:6, 1:7, 1:8, 1:9, 1:10, as well as the reverse ratios.
[0012] In embodiments the disclosure includes modified cells, wherein the modification is such that the cells do not express LEDGF, cell cultures/cell lines derived from such cells, and their progeny.
[0013] In embodiments the disclosure includes LEDF+ cells, wherein the LEDGF protein in the LEGDF+ cells is present in a complex with an antibody, and thus is suitable for use in a variety of immuno-diagnostic tests. In an embodiment, the antibody is a first antibody, such as a primary antibody. In embodiments, the primary antibody that is bound the LEDGF protein the LEDGF+ cells is itself present in a complex with a detectably labeled secondary antibody.
[0014] In embodiments, the LEDGF+ and/or LEDGF- cells comprise one or more nuclear antigens in present in a complex with an antibody.
[0015] In another aspect the disclosure provides a method for determining whether a biological sample comprises antibodies that bind to LEDGF protein (anti-LEDGF Abs). The method generally comprises the steps of: [0016] i) exposing the biological sample to mammalian cells that are modified such that they do not comprise LEDGF protein (LEDGF- cells), [0017] ii) exposing the biological sample to mammalian cells that comprise the LEDGF protein (LEDGF+ cells), and [0018] iii) comparing the amount of anti-LEDGF Abs bound to the LEDGF- cells to the amount of anti-LEDGF Abs bound to the LEDGF+ cells, [0019] wherein determining a greater amount of anti-LEDGF Abs bound to the LEDGF+ cells relative to the amount of anti-LEDGF Abs bound to the LEDGF- cells is indicative that the biological sample comprised the anti-LEDGF Abs, and [0020] wherein the same or less anti-LEDGF Abs bound to the LEDGF+ cells relative to the amount of anti-LEDGF Abs bound to the LEDGF- cells is indicative that the biological sample did not comprise the anti-LEDGF Abs.
[0021] In embodiments, the cells used in the method are killed and permeabilized and are immobilized on a solid substrate. In embodiments, the LEDGF- cells and the LEDGF+ cells are immobilized on the same solid substrate; in embodiments they are immobilized on distinct solid substrates. In embodiments, determining the amount of anti-LEDGF Abs is performed using an indirect immunofluorescence (IIF) assay.
[0022] In another aspect the disclosure comprises a kit comprising modified LEDGF- cells and LEDGF+ cells, wherein the LEDGF- cells and the LEDGF+ cells are immobilized on one or more solid substrates, and wherein the LEDGF- cells and the LEDGF+ cells are killed and permeabilized. In embodiments, the solid substrate(s) and the cells are dried, and are provided in one or more suitable containers.
[0023] In embodiments, the kit further comprises a composition comprising primary antibodies that are capable of binding to anti-nuclear autoantibodies (ANAs). The kit may further comprise detectably labeled secondary antibodies that are capable of binding to the primary antibodies. Any suitable detectable label can be used and many are well known in the art. In embodiments, the detectable label is a florescent label and is thus suitable for use in, for example, an IIF assay.
BRIEF DESCRIPTION OF THE FIGURE
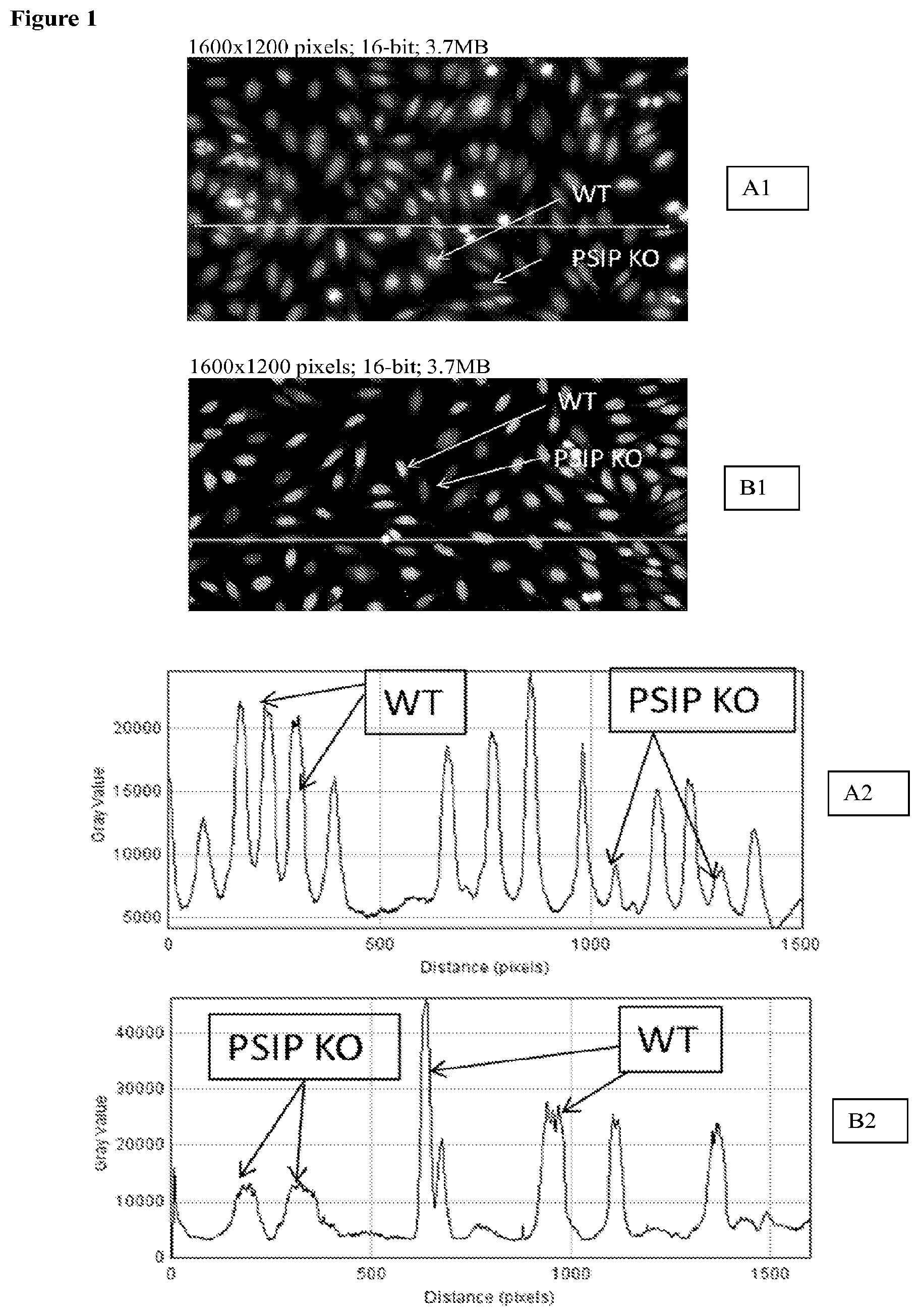
[0024] FIG. 1: Wild Type Hep2 expressing LEDGF and PISP1 disrupted cell line using per Example Sequence 3 and not expressing LEDGF were tested by IIF using confirmed human DFS70 positive anti-sera. Top panel: two examples (A1 and B1) show brightly labeled cells (WT) and cells with background fluorescence signal (PSIP KO). Bottom panel: profile analysis (ImageJ version 1.421, National Institute of Health, USA) plots the intensity of labeled nuclei along the line (A2 is intensity plot for A1; B2 is intensity plot for B1). High peaks correspond to WT cells and low peaks correspond to PSIP-KO cells which do not express any detectable LEDGF protein.
DETAILED DESCRIPTION
[0025] The present disclosure provides compositions and methods for using the compositions in detecting ANA autoantibodies, and/or for determining whether a biological sample obtained or derived from an individual comprises antibodies that recognize the LEDGF protein, and for diagnosing and/or aiding in the diagnosis of autoimmune disorders that are positively correlated with the presence of ANA autoantibodies. Such disorders include but not necessarily limited to systemic autoimmune rheumatic diseases (SARDs). Kits/products comprising reagents for use in detection of ANA autoantibodies are also provided.
[0026] In general, the disclosure provides approaches to reducing and/or eliminating the DFS pattern that is frequently characteristic of IIF analysis of ANAs that in current testing typically rely on HEp-2 cells as a substrate, and encompasses in vitro compositions comprising modified mammalian cells that have less LEDGF expression relative to unmodified HEp-2 cells, and methods of using such modified cells to detect ANA autoantibodies. However, rather than being limited to modified HEp-2 cells, the present disclosure provides compositions comprising mammalian cells of any origin, wherein the cells have been modified to be improved substrates for ANA and/or LEDGF testing. Thus, in various embodiments, the present disclosure involves mammalian cells that comprise the PSIP1 gene, and to at least some degree express the LEDGF protein encoded by the PSIP1 gene, but subsequent to being modified as more fully described below, express less LEDGF protein relative to unmodified cells of the same type, or do not express any detectable LEDGF protein. It will be recognized by those skilled in the art that most mammalian cells express LEDGF, and thus it is expected that cells used in compositions and methods of this disclosure can include cells of any mammalian cell line, or cells derived from any mammalian cell line, or any other suitable source. In embodiments, the cells are immortalized. In embodiments, the cells are progeny of a cell line derived from cancer, such as a tumor. In embodiments, the cells are multiploid and as such have more than two copies of at least one chromosome. In embodiments, the cells comprise more than two copies of a chromosome that comprises the PSIP1 gene, which as described further below encodes the LEDGF protein. In embodiments, the cells are aneuploid, and may be pseudo hypotripoloid. In embodiments, the cells are derived from human cells and comprise more than 23 distinct chromosomes. In embodiments, the disclosure provides modified cell lines that are altered such that expression of the LEDGF protein is reduced or eliminated. In embodiments, the disclosure provides modifications of known cell lines, such as cell lines that can be grown in monolayers and fixed, including but not necessarily limited to HEp-2 and HeLa cells. In embodiments, cell types that can be modified for use with the present disclosure are commercially available from sources, such as the American Type Culture Collection (ATCC). In non-limiting embodiments the cells are HEp2 or HeLa cells. These cells are available from ATCC as catalog #CCL-23 for HEp2, and CCL-2 or CCL-2.2 for HeLa adherent and suspension cultures respectively. As described above, in embodiments, the disclosure includes cells that are modified such that they express no detectable LEDGF protein, or express less LEDGF protein than cells of the same type. Thus, it will be recognized that cells of the same type can comprise, as one non-limiting example, HEp2 cells, wherein the modified HEp2 cells express less LEDGF protein than unmodified HEp2 cells, wherein the HEp2 cells are the "type" of cells that are described. The same applies to any other mammalian cells, wherein the unmodified cells express detectable LEDGF, including but not limited to HeLa cells, and other cell lines derived from, for example, blood plasma cells, monocytes, neutrophils, T-lymphocytes, platelets, T-cell leukemia cells, myeloid leukemia cells, lymphoblastic leukemia cells, kidney cells, kidney cancer cells, liver cells, liver cancer cells, lung cells, lung cancer cells, colon cells, colon cancer cells, heart cells, bone cells, bone cancer cells, brain cells, brain cancer cells, ovary cells, ovarian cancer cells, prostate cells, prostate cancer cells, cervical cells, cervical cancer cells, melanoma, breast tissue cells, breast cancer cells, skin cells, melanoma cells, pancreatic cells and pancreatic cancer cells, and others.
[0027] Approaches to immunodiagnostic assays provided in this disclosure involve modifying mammalian cells to cause down-regulation or elimination of the PSIP1/LEDGF gene product, and methods of using the modified cells in autoimmune assays. The disclosure also includes the modified cells, and compositions comprising the modified, such as cell cultures. Kits for use in the assays are also provided.
[0028] As described above, the DFS pattern is well known in the art and comprises a dense fine speckled pattern resulting from autoantibodies that specifically bind to LEDGF protein (Ayaki, Sueno et al. 1999). Autoantibodies to LEDGF protein were first reported in association with atopic dermatitis and other conditions such as Asthma, interstitial cystitis (Ochs, Muro et al. 2000), alopecia areata (Okamoto, Ogawa et al. 2004) and in 0-20% of healthy individuals (Watanabe, Kodera et al. 2004, Mahler, Parker et al. 2012). LEDGF belongs to a selected group of autoantigens that are targeted for cleavage during cell death, and it has been proposed that the caspase-induced LEDGF cleavage and the generation of autoantibodies to the protein might contribute to the pathogenesis of various human atopic and inflammatory disorders associated with deregulated apoptosis (Ganapathy, Daniels et al. 2003, Ganapathy and Casiano 2004). LEDGF protein has also been implicated in HIV integration, and LEDGF has been knocked down both transiently (using siRNA) and stably (using shRNA followed by selection) resulting in a 3-5 fold reduction in HIV-1 replication in HeLaP4 cells (Vandekerckhove, Christ et al. 2006).
[0029] The amino acid sequence of the LEDGF protein is known in the art and the canonical sequence is provided here as SEQ ID NO:1: MTRDFKPGDLIFAKMKGYPHWPARVDEVPDGAVKPPTNKLPIFFFGTHETAFLGPKDIF PYSENKEKYGKPNKRKGFNEGLWEIDNNPKVKFSSQQAATKQSNASSDVEVEEKETSV SKEDTDHEEKASNEDVTKAVDITTPKAARRGRKRKAEKQVETEEAGVVTTATASVNLK VSPKRGRPAATEVKIPKPRGRPKMVKQPCPSESDIITEEDKSKKKGQEEKQPKKQPKKD EEGQKEEDKPRKEPDKKEGKKEVESKRKNLAKTGVTSTSDSEEEGDDQEGEKKRKGGR NFQTAHRRNMLKGQHEKEAADRKRKQEEQMETEQQNKDEGKKPEVKKVEKKRETSM DSRLQRIHAEIKNSLKIDNLDVNRCIEALDELASLQVTMQQAQKHTEMITTLKKIRRFKV SQVIMEKSTMLYNKFKNMFLVGEGDSVITQVLNKSLAEQRQHEEANKTKDQGKKGPN KKLEKEQTGSKTLNGGSDAQDGNQPQHNGESNEDSKDNHEASTKKKPSSEERETEISLK DSTLDN (SEQ ID NO:1). Other isoforms and truncated versions which comprise mutations that differ from the canonical sequence are known in the art. In particular, GenBank (NCBI) entries NP_001121689.1, NM_001128217.1, [O75475-1], NP_066967.3, NM_021144.3, [O75475-2], NP_150091.2, NM_033222.3, [O75475-1], XP_005251413.1 provide LEDGF, and the polynucleotide and amino acid sequences described in these database entries are incorporated herein by reference as they exist as of the filing of this application or patent XM_005251356.1, [O75475-2], XP_005251415.1, XM_005251358.1 and [O75475-3].
[0030] The immunoreactive sequence of the SEQ ID NO:1 has been reported (Ogawa, Sugiura et al. 2004) to be a polypeptide from amino acid number 349-455 described in SEQ ID NO:2.
TABLE-US-00001 (SEQ ID NO: 2) DSRLQRIHAEIKNSLKIDNLDVNRCIEALDELASLQVTMQQAQKHTEMIT TLKKIRRFKVSQVIMEKSTMLYNKFKNMFLVGEGDSVITQVLNKSLAEQR QHEEANK.
[0031] The cDNA sequence encoding LEDGF is also known in the art and is provided here as SEQ ID NO:3.
TABLE-US-00002 (SEQ ID NO: 3) ATGACTCGCGATTTCAAACCTGGAGACCTCATCTTCGCCAAGATGAAAGG TTATCCCCATTGGCCAGCTCGAGTAGACGAAGTTCCTGATGGAGCTGTAA AGCCACCCACAAACAAACTACCCATTTTCTTTTTTGGAACTCATGAGACT GCTTTTTTAGGACCAAAGGATATATTTCCTTACTCAGAAAATAAGGAAAA GTATGGCAAACCAAATAAAAGAAAAGGTTTTAATGAAGGTTTATGGGAGA TAGATAACAATCCAAAAGTGAAATTTTCAAGTCAACAGGCAGCAACTAAA CAATCAAATGCATCATCTGATGTTGAAGTTGAAGAAAAGGAAACTAGTGT TTCAAAGGAAGATACCGACCATGAAGAAAAAGCCAGCAATGAGGATGTGA CTAAAGCAGTTGACATAACTACTCCAAAAGCTGCCAGAAGGGGGAGAAAG AGAAAGGCAGAAAAACAAGTAGAAACTGAGGAGGCAGGAGTAGTGACAAC AGCAACAGCATCTGTTAATCTAAAAGTGAGTCCTAAAAGAGGACGACCTG CAGCTACAGAAGTCAAGATTCCAAAACCAAGAGGCAGACCCAAAATGGTA AAACAGCCCTGTCCTTCAGAGAGTGACATCATTACTGAAGAGGACAAAAG TAAGAAAAAGGGGCAAGAGGAAAAACAACCTAAAAAGCAGCCTAAGAAGG ATGAAGAGGGCCAGAAGGAAGAAGATAAGCCAAGAAAAGAGCCGGATAAA AAAGAGGGGAAGAAAGAAGTTGAATCAAAAAGGAAAAATTTAGCTAAAAC AGGGGTTACTTCAACCTCCGATTCTGAAGAAGAAGGAGATGATCAAGAAG GTGAAAAGAAGAGAAAAGGTGGGAGGAACTTTCAGACTGCTCACAGAAGG AATATGCTGAAAGGCCAACATGAGAAAGAAGCAGCAGATCGAAAACGCAA GCAAGAGGAACAAATGGAAACTGAGCAGCAGAATAAAGATGAAGGAAAGA AGCCAGAAGTTAAGAAAGTGGAGAAGAAGCGAGAAACATCAATGGATTCT CGACTTCAAAGGATACATGCTGAGATTAAAAATTCACTCAAAATTGATAA TCTTGATGTGAACAGATGCATTGAGGCCTTGGATGAACTTGCTTCACTTC AGGTCACAATGCAACAAGCTCAGAAACACACAGAGATGATTACTACACTG AAAAAAATACGGCGATTCAAAGTTAGTCAGGTAATCATGGAAAAGTCTAC AATGTTGTATAACAAGTTTAAGAACATGTTCTTGGTTGGTGAAGGAGATT CCGTGATCACCCAAGTGCTGAATAAATCTCTTGCTGAACAAAGACAGCAT GAGGAAGCGAATAAAACCAAAGATCAAGGGAAGAAAGGGCCAAACAAAAA GCTAGAGAAGGAACAAACAGGGTCAAAGACTCTAAATGGAGGATCTGATG CTCAAGATGGTAATCAGCCACAACATAACGGGGAGAGCAATGAAGACAGC AAAGACAACCATGAAGCCAGCACGAAGAAAAAGCCATCCAGTGAAGAGAG AGAGACTGAAATATCTCTGAAGGATTCTACACTAGATAACTAG
[0032] To provide compositions and methods for improved ANA and/or LEDGF antibody detection for systemic and non-systemic autoimmune diseases (organ specific autoimmune diseases, atopic dermatitis, alopecia etc.,) and differentiation from disease free human population, any suitable mammalian cells, including but not limited to HEp-2 HeLa, HEK293, or a cell line suitable for culturing as adherent (monolayer) or suspension format can be modified in a variety of ways, given the benefit of the present disclosure. In various embodiments LEDGF protein is reduced in the modified cells by reducing mRNA encoding it. In another approach the disclosure includes disrupting the PSIP1 gene from making a protein by via knock-out or targeted mutation. The disclosure also includes making and using modified cells characterized by reduced or eliminated LEDGF protein. In embodiments, the modified cells are also engineered to express a detectable marker, such as a fluorescent protein or an immunoreactive protein that can further be detected using a specific secondary antibody (including but not necessarily limited to a Poly histidine tag, c-Myc tag, FLAG tag etc., which are well characterized in the art).
[0033] In one aspect, the disclosure includes reducing LEDGF mRNA, and as a result reducing the LEDGF protein, in modified mammalian cells. In one approach this aspect comprises introducing into the suitable mammalian cells a polynucleotide that can inhibit translation of LEDGF mRNA, and/or can participate in and/or facilitate RNAi-mediated reduction of LEDGF mRNA. In one embodiment, an antisense polynucleotide is used to inhibit translation of LEDGF mRNA. Antisense nucleic acids can be DNA or RNA molecules that are complementary to at least a portion of the LEDGF mRNA. In embodiments, oligomers of about fifteen nucleotides, and/or those that hybridize to the AUG initiation codon will be particularly efficient. The polynucleotides described herein for use in targeting LEDGF mRNA can in certain embodiments be modified, such as to be resistant to nucleases.
[0034] In embodiments, the present disclosure provides for replacement of the PSIP1 gene with a sequence encoding a detectable marker, such as a fluorescent protein, or integrating such a sequence into the PSIP1 gene, thereby disrupting it, or integrating such a sequence elsewhere in the genome of the cells. By replacing PSIP1 or integrating a sequence encoding a detectable protein into it the disclosure provides for marking modified mammalian cells which do not express LEDGF. This is valuable in that those cells which express the detectable protein can be selected for use in the immunoassays of the invention, and for including in products that are intended to be used in such immunoassay. In embodiments, disrupting the PSIP1 gene with a sequence encoding a fluorescent protein will allow for enriching a cell population with cells that contain the LEDGF disruption, such as by using FACS to separate cells that contain the disruption from those that do not, thereby providing an isolated and/or purified population of modified cells that do not express LEDGF. The detectable marker can be any protein that can be detected, and is preferably a fluorescent protein. Any fluorescent protein can be used. In embodiment, the fluorescent protein is selected from GFP, eGFP, Red Fluorescent protein or variants thereof such as tRFP, dsRED, mCherry, tdTomato etc.,) or any fluorescent protein that does not interfere with the conjugates used in, for example, an IIF method to detect autoantibodies. Thus, in embodiments, the present disclosure includes cells characterized by having the PSIP1 gene disrupted or knocked out. In embodiments, the knock out comprises a disruption of the gene by introducing (a knock in) of a detectable protein.
[0035] In embodiments, the disclosure includes introducing an expression vector which can inhibit LEDGF protein, and which may also express a detectable marker. For example, an expression vector with two distinct promoters or a bidirectional promoter can be used to express shRNA targeted to PSIP1 and express the detectable marker. In alternative embodiments, two distinct expression vectors can be used for this purpose. In embodiments, the vectors are stably or transiently present in the cells. In embodiments, one or both vectors, or the single vector encoding the shRNA and the detectable marker, is integrated into at least one position in a chromosome in a mammalian cell.
[0036] In another aspect the disclosure includes RNAi-mediated reduction in LEDGF mRNA. RNAi-based inhibition can be achieved using any suitable RNA polynucleotide that is targeted to LEDGF mRNA. In embodiments, a single stranded or double stranded RNA, wherein at least one strand is complementary to the LEDGF mRNA, can be introduced into the cell to promote RNAi-based degradation of LEDGF mRNA. In another embodiment, microRNA (miRNA) targeted to the LEDGF mRNA can be used. In another embodiment, a ribozyme that can specifically cleave LEDGF mRNA can be used. In yet another embodiment, small interfering RNA (siRNA) can be used. siRNA (or ribozymes) can be introduced directly, for example, as a double stranded siRNA complex, or by using a modified expression vector, such as a lentiviral vector, to produce an shRNA. As is known in the art, shRNAs adopt a typical hairpin secondary structure that contains a paired sense and antisense portion, and a short loop sequence between the paired sense and antisense portions. shRNA is delivered to the cytoplasm where it is processed by DICER into siRNAs. siRNA is recognized by RNA-induced silencing complex (RISC), and once incorporated into RISC, siRNAs facilitate cleavage and degradation of targeted mRNA. In embodiments, an shRNA polynucleotide used to suppress LEDGF expression can comprise or consist of between 45-100 nucleotides, inclusive, and including all integers between 45 and 100. The portion of the shRNA that is complementary to the LEDGF mRNA mRNA can be from 21-29 nucleotides, inclusive, and including all integers between 21 and 29.
[0037] For delivering siRNA via shRNA, modified lentiviral vectors can be made and used according to standard techniques, given the benefit of the present disclosure. Further, lentiviral vectors expressing shRNAs targeted to many human mRNAs are commercially available. Additionally, custom siRNAs or shRNA can be obtained from, for example Thermo-Dharmacon for transient transfection resulting in temporary reduction in LEDGF levels. Alternatively, lentiviral constructs expressing human PSIP11 targeted shRNA can be obtained from Thermo Dharmacon. These lentiviruses are capable of stably and permanently infecting target cells, such as by integrating into a chromosome in the cells. However, as will be apparent from the following description, RNAi-mediated approaches for disrupting expression of the LEDGF protein may not be optimal. For example, we introduced DNA sequences encoding shRNAs designed against the PSIP1 gene and cloned them into lenti-viral vectors downstream of a U6 promoter. Lentivirus with the DNA insert capable of producing either target shRNA or negative control were used to infect HEp2 cells. Viral infectivity and titer was measured by an integrated RFP (Red fluorescent protein) marker that is expressed in all the infected cells. The RFP marker was also fused to puromycin (antibiotic) resistance factor which is used for selection of cells that stably incorporated the construct into the genome of HEp2 cells. Examples of tested sequences are below, where the DNA equivalent of the
TABLE-US-00003 shRNA sequence is provided: shRNA(h PSIP1) example #1 sequence: (SEQ ID NO: 4) AGACAGCATGAGGAAGCGA. Cloned shRNA hairpin sequence: (SEQ ID NO: 5) AGACAGCATGAGGAAGCGAttcaagagaTCGCTTCCTCATGCTGTCT shRNA(h PSIP1) example #2 sequence: (SEQ ID NO: 6) AGTTCCTGATGGAGCTGTAAA Cloned shRNA hairpin sequence: (SEQ ID NO: 7) AGTTCCTGATGGAGCTGTAAAcgagTTTACAGCTCCATCAGGAACT hPSIP1) example #3 sequence: (SEQ ID NO: 8) GCAATGAAGACAGCAAAGACA Cloned shRNA hairpin sequence: (SEQ ID NO: 9) GCAATGAAGACAGCAAAGACAcgagTGTCTTTGCTGTCTTCATTGC shRNA-Neg-Control: (SEQ ID NO: 10) GTCTCCACGCGCAGTACATTT Cloned shRNA-Neg hairpin sequence: (SEQ ID NO: 11) GTCTCCACGCGCAGTACATTTcgagAAATGTACTGCGCGTGGAGAC
[0038] IIF analysis using the above sequences (lentiviral transduction procedure followed by selection for resistant colonies) using DFS70 specific antisera indicated a low level decrease in PSIP1/LEDGF levels. Negative controls did not show any reduction in the PSIP1/LEDGF levels using the same procedure. Further, while siRNA can produce an intense reduction in mRNA levels, the effects are usually transient. Thus, even though shRNA technology is compatible with selection processes and allows the isolation of colonies stably expressing the short hairpin RNA, which further aids in the degradation of specific complementary mRNA in subsequent generation of cells, as observed in the aforementioned approaches, the reduction of PSIP1 levels at the mRNA level were not adequate to provide an improved substrate for IIF analysis for use in detecting autoantibodies directed to ANA. Thus, the disclosure includes alternative approaches for disrupting LEDGF protein production. In this regard, the disclosure also includes disrupting the PSIP1 gene with a mutation such that LEDGF mRNA and protein are not expressed. In one embodiment, the PSIP1 gene can be disrupted by targeted mutagenesis. In embodiments, targeted mutagenesis can be achieved by, for example, targeting a CRISPR (clustered regularly interspaced short palindromic repeats) site in the PSIP1 gene. So-called CRISPR systems designed for targeting specific genomic sequences are known in the art and can be adapted to disrupt the PSIP1 gene for making modified cells encompassed by this disclosure. In general, the CRISPR system includes one or more expression vectors encoding at least a targeting RNA and a polynucleotide sequence encoding a CRISPR-associated nuclease, such as Cas9, but other Cas nucleases can be used. CRISPR systems for targeted disruption of mammalian chromosomal sequences are commercially available and can be adapted to disrupt the PSIP1 gene in HEp-2 cells given the benefit of this disclosure.
[0039] In embodiments, a targeting RNA encoded by the CRISPR system can be a CRISPR RNA (crRNA) or a guide RNA, such as sgRNA. The sequence of the targeting RNA has a segment that is the same as or complementarity to any CRISPR site in the PSIP1 gene. In this regard, the target sequence comprises a specific sequence on its 3' end referred to as a protospacer adjacent motif or "PAM". In an embodiment a CRISPR Type II system is used, and the target sequences therefore conform to the well-known N12-20NGG motif, wherein the NGG is the PAM sequence. Thus, in embodiments, a target RNA will comprise or consist of a segment that is from 12-20 nucleotides in length which is the same as or complementary to a DNA target sequence (a spacer) in the PSIP1 gene. The 12-20 nucleotides directed to the spacer sequence will be present in the targeting RNA, regardless of whether the targeting RNA is a crRNA or a guide RNA. In embodiments, a separate trans-activating crRNA (tracrRNA) can be used to assist in maturation of a crRNA targeted to the PSIP1 gene. Introduction a CRISPR system into HEp-2 cells will result in binding of a targeting RNA/Cas9 complex to the PSIP1 target sequence so that the Cas9 can cut both strands of DNA causing a double strand break. The double stranded break can be repaired by non-homologous end joining DNA repair, or by a homology directed repair pathway, which will result in either insertions or deletions at the break site, or by using a repair template to introduce mutations, respectively. Double-stranded breaks can also be introduced into the PSIP1 gene by expressing Transcription activator-like effector nucleases (TALENs) in the cells. TALENs are artificial restriction enzymes generated by fusing a TAL effector DNA binding domain to a DNA cleavage domain and are known in the art and can be adapted for use in embodiments of this disclosure. In yet another approach, zinc-finger nucleases (ZFNs) can be expressed in the cells to target the PSIP1 gene. ZFNs are artificial restriction enzymes produced by fusing a zinc finger DNA-binding domain to a DNA-cleavage domain. ZF domains can be designed to target PSIP1 gene DNA sequences where the zinc-finger nucleases cleave the sequence, thereby disrupting the gene. In another embodiment, site-specific gene integration or targeted integration of a sequence into specific integration sites within the gene can be accomplished by using commercial systems such as Jump-In.TM. or Flp-In.TM. systems commercially available from Thermo Fisher Scientific Inc. Multiple integration sites may be targeted by PhiC31 in the Jump-In.TM. Fast system. As will be recognized by those skilled in the art, a FRT site (34 bp) in the target genome is needed for gene integration, and is provided by specific commercial cell lines derived from Flp-In.TM. technology.
[0040] In a non-limiting reduction to practice, we used a CRISPR-CAS-9 system to design specific constructs with guide RNA (gRNA) and a complimentary region upstream of Protospacer Adjacent Motif (PAM) sequences to create double strand breaks in the target PSIP1 gene. We then selected colonies with homozygous disruption of the PSIP1 gene at the break site. A cell line such as HEp2 may have multiple copies of PSIP1 gene and it is thus important to isolate a clone where all copies of the PSIP1 gene have been disrupted, thereby eliminating the LEDGF protein from cells. Five CRISPR-CAS9 examples for PSIP1 gene disruption are described below. There are numerous PAM sites spanning across the exons and introns of the PSIP1 gene, but exons are preferred targets for CRISPR-CAS9 induced mutations or disruptions in the coding region.
[0041] Each representative sequence below includes U6 promoter sequence, gRNA targeting site and a gRNA scaffold upstream of PAM sequence which in combination with a CAS9 enzyme supplied to the cell either as part of the same vector or a different vector will create a functional CRISPR complex capable of creating a double stranded break in the targeted area of the genome.
TABLE-US-00004 Example Sequence 1: (SEQ ID NO: 12): 5'GTACAAAAAAGCAGGCTTTAAAGGAACCAATTCAGTCGACTGGATCCG GTACCAAGGTCGGGCAGGAAGAGGGCCTATTTCCCATGATTCCTTCATAT TTGCATATACGATACAAGGCTGTTAGAGAGATAATTAGAATTAATTTGAC TGTAAACACAAAGATATTAGTACAAAATACGTGACGTAGAAAGTAATAAT TTCTTGGGTAGTTTGCAGTTTTAAAATTATGTTTTAAAATGGACTATCAT ATGCTTACCGTAACTTGAAAGTATTTCGATTTCTTGGCTTTATATATCTT GTGGAAAGGACGAAACACCGTAATCAGCCACAACATAACGTTTTAGAGCT AGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTG GCACCGAGTCGGTGCTTTTTTTCTAGACCCAGCTTTCTTGTACAAAGTTG GCATTA 3' Example Sequence 2: (SEQ ID NO: 13): 5'TGTACAAAAAAGCAGGCTTTAAAGGAACCAATTCAGTCGACTGGATCC GGTACCAAGGTCGGGCAGGAAGAGGGCCTATTTCCCATGATTCCTTCATA TTTGCATATACGATACAAGGCTGTTAGAGAGATAATTAGAATTAATTTGA CTGTAAACACAAAGATATTAGTACAAAATACGTGACGTAGAAAGTAATAA TTTCTTGGGTAGTTTGCAGTTTTAAAATTATGTTTTAAAATGGACTATCA TATGCTTACCGTAACTTGAAAGTATTTCGATTTCTTGGCTTTATATATCT TGTGGAAAGGACGAAACACCGACGCCTCTGCGGCAGCTGGGTTTTAGAGC TAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGT GGCACCGAGTCGGTGCTTTTTTTCTAGACCCAGCTTTCTTGTACAAAGTT GGCATTA 3' Example Sequence 3: (SEQ ID NO: 14) 5'TGTACAAAAAAGCAGGCTTTAAAGGAACCAATTCAGTCGACTGGATCC GGTACCAAGGTCGGGCAGGAAGAGGGCCTATTTCCCATGATTCCTTCATA TTTGCATATACGATACAAGGCTGTTAGAGAGATAATTAGAATTAATTTGA CTGTAAACACAAAGATATTAGTACAAAATACGTGACGTAGAAAGTAATAA TTTCTTGGGTAGTTTGCAGTTTTAAAATTATGTTTTAAAATGGACTATCA TATGCTTACCGTAACTTGAAAGTATTTCGATTTCTTGGCTTTATATATCT TGTGGAAAGGACGAAACACCGAGGTAGACGAAGTTCCTGAGTTTTAGAGC TAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGT GGCACCGAGTCGGTGCTTTTTTTCTAGACCCAGCTTTCTTGTACAAAGTT GGCATTA 3' Example Sequence 4 (SEQ ID NO: 15): 5'TGTACAAAAAAGCAGGCTTTAAAGGAACCAATTCAGTCGACTGGATCC GGTACCAAGGTCGGGCAGGAAGAGGGCCTATTTCCCATGATTCCTTCATA TTTGCATATACGATACAAGGCTGTTAGAGAGATAATTAGAATTAATTTGA CTGTAAACACAAAGATATTAGTACAAAATACGTGACGTAGAAAGTAATAA TTTCTTGGGTAGTTTGCAGTTTTAAAATTATGTTTTAAAATGGACTATCA TATGCTTACCGTAACTTGAAAGTATTTCGATTTCTTGGCTTTATATATCT TGTGGAAAGGACGAAACACCGAACTACCCATTTTCTTTTTGTTTTAGAGC TAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGT GGCACCGAGTCGGTGCTTTTTTTCTAGACCCAGCTTTCTTGTACAAAGTT GGCATTA 3' Example Sequence 5: (SEQ ID NO: 16) 5'TGTACAAAAAAGCAGGCTTTAAAGGAACCAATTCAGTCGACTGGATCC GGTACCAAGGTCGGGCAGGAAGAGGGCCTATTTCCCATGATTCCTTCATA TTTGCATATACGATACAAGGCTGTTAGAGAGATAATTAGAATTAATTTGA CTGTAAACACAAAGATATTAGTACAAAATACGTGACGTAGAAAGTAATAA TTTCTTGGGTAGTTTGCAGTTTTAAAATTATGTTTTAAAATGGACTATCA TATGCTTACCGTAACTTGAAAGTATTTCGATTTCTTGGCTTTATATATCT TGTGGAAAGGACGAAACACCGAGTGCTTTTTTAGGACCAAGTTTTAGAGC TAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGT GGCACCGAGTCGGTGCTTTTTTTCTAGACCCAGCTTTCTTGTACAAAGTT GGCATTA 3'
[0042] For Example Sequence 3, the disruption was targeted into the exon 1 of the PSIP1 gene, thereby eliminating of the potential to make a partial LEDGF protein. Further, we isolated a single colony of HEp2 cells where all copies of PSIP1 gene were disrupted as confirmed by DNA sequencing. Following confirmation by DNA sequencing, the cells from the colony were mixed with WT cells in 1:1 ratio and IIF analysis was performed using a panel of anti-sera that were specifically positive for DFS70 pattern and confirmed by LIA (Line Immunoassay) or Line blot assay for DFS70 antisera (ImmcoStripe ANA-Advanced LIA, Immco Diagnostics Inc., Buffalo, N.Y.). The results are depicted in FIG. 1. Thus, the disclosure includes mammalian cell cultures which comprise mammalian cells wherein every copy of the PSIP1 gene in the cells is disrupted, and thus the cells do not express LEDGF protein. In an embodiment, the cells do not express detectable LEDGF protein, wherein the detection is by IIF. In embodiments, when the PSIP1 gene is disrupted using a CRISPR approach, the cells can further comprise a Cas9 protein coding region integrated into one or more locations in the chromosome(s) of the cells, and can further comprise a sequence encoding the gRNA integrated in the chromosome(s) of the cells.
[0043] In another aspect the disclosure includes a method for detecting ANA antibodies, and/or LEDGF antibodies. The method comprises obtaining a biological sample from an individual, mixing the sample with modified cells described herein, and performing an immuno-assay, such as an IIF assay to determine the antibodies. The presence of the antibodies is a diagnosis or aids in the diagnosis of an autoimmune disease, such as SARDS, and the absence of the ANA antibodies indicate the lack of an autoimmune disease. Thus, the disclosure provides diagnostic methods using novel agents in the steps of the method. In embodiments, the presence of antibodies to LEDGF in LEDGF+ cells, is indicative that further diagnostic testing of the individual is warranted.
[0044] As noted above, IIF assays using HEp-2 cells as a substrate to detect ANA antibodies are well known in the art and can be used with the modified cells of the present disclosure without modifying such well known protocols. The biological sample that is used in the assay can be any biological sample, including but not limited to blood, serum, semen, pleural fluid, cerebrospinal fluid, saliva, urine, exosomes, or tissue. The biological sample can be used directly, or it can be subjected to a processing step before being exposed to the cells. The amount of antibodies, if any, can be compared to any suitable reference for, for instance, correcting for background, or for staging the degree and/or severity of an autoimmune disease that is positively correlated with the antibodies. In embodiments, the disclosure includes testing combinations of LEDGF+ and LEDGF- cells to determine whether or not a sample comprises antibodies that bind to LEDGF, and thus can provide for correction of background that complicates previously available approaches which frequently result in false positive results for ANA autoantibodies.
[0045] In another aspect, the disclosure includes kits and articles of manufacture for use in detecting ANA antibodies. The kit can comprise at least one container in which the modified cells of this disclosure are kept. The cells can be preserved using any suitable reagents, and can be provided, for example, in the form of a pellet. The kit can include reagents for use in IIF assays, and instruction which describe the modified cells, such as by providing a description of how or that they have been modified to reduce DFS, and instructions for using the cells in the IIF assays. In embodiments, the kits comprise LEDGF+ and LEDGF- cells which are fixed to one or more suitable solid substrates. The cells may be permeabilized using any suitable approach, many of which are well known in the art, and are thus killed cells. In embodiments, the fixed cells that are immobilized on the solid substrate are dried.
[0046] It will be apparent form the foregoing that the present disclosure includes the modified cells described herein, the methods for making the modified cells, cell cultures comprising the modified cells, and all methods for using the modified cells in any assay designed to detect any one, or any combination of the antibodies that are comprised by the ANA antibody profile.
REFERENCES
[0047] Ayaki, M., T. Sueno, D. P. Singh, L. T. Chylack, Jr. and T. Shinohara (1999). "Antibodies to lens epithelium-derived growth factor (LEDGF) kill epithelial cells of whole lenses in organ culture." Exp Eye Res 69(1): 139-142.
[0048] Ganapathy, V. and C. A. Casiano (2004). "Autoimmunity to the nuclear autoantigen DFS70 (LEDGF): what exactly are the autoantibodies trying to tell us?" Arthritis Rheum 50(3): 684-688.
[0049] Ganapathy, V., T. Daniels and C. A. Casiano (2003). "LEDGF/p75: a novel nuclear autoantigen at the crossroads of cell survival and apoptosis." Autoimmun Rev 2(5): 290-297.
[0050] Mahler, M. and M. J. Fritzler (2012). "The Clinical Significance of the Dense Fine Speckled Immunofluorescence Pattern on HEp-2 Cells for the Diagnosis of Systemic Autoimmune Diseases." Clin Dev Immunol 2012: 494356.
[0051] Mahler, M., T. Parker, C. L. Peebles, L. E. Andrade, A. Swart, Y. Carbone, D. J. Ferguson, D. Villalta, N. Bizzaro, J. G. Hanly and M. J. Fritzler (2012). "Anti-DFS70/LEDGF Antibodies Are More Prevalent in Healthy Individuals Compared to Patients with Systemic Autoimmune Rheumatic Diseases." J Rheumatol.
[0052] Ochs, R. L., Y. Muro, Y. Si, H. Ge, E. K. Chan and E. M. Tan (2000). "Autoantibodies to DFS 70 kd/transcription coactivator p75 in atopic dermatitis and other conditions." J Allergy Clin Immunol 105(6 Pt 1): 1211-1220.
[0053] Ogawa, Y., K. Sugiura, A. Watanabe, M. Kunimatsu, M. Mishima, Y. Tomita and Y. Muro (2004). "Autoantigenicity of DFS70 is restricted to the conformational epitope of C-terminal alpha-helical domain." J Autoimmun 23(3): 221-231.
[0054] Okamoto, M., Y. Ogawa, A. Watanabe, K. Sugiura, Y. Shimomura, N. Aoki, T. Nagasaka, Y. Tomita and Y. Muro (2004). "Autoantibodies to DFS70/LEDGF are increased in alopecia areata patients." J Autoimmun 23(3): 257-266.
[0055] Vandekerckhove, L., F. Christ, B. Van Maele, J. De Rijck, R. Gijsbers, C. Van den Haute, M. Witvrouw and Z. Debyser (2006). "Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus." J Virol 80(4): 1886-1896.
[0056] Watanabe, A., M. Kodera, K. Sugiura, T. Usuda, E. M. Tan, Y. Takasaki, Y. Tomita and Y. Muro (2004). "Anti-DFS70 antibodies in 597 healthy hospital workers." Arthritis Rheum 50(3): 892-900.
[0057] While the disclosure has been particularly shown and described with reference to specific embodiments, it should be understood by those having skill in the art that various changes in form and detail may be made therein without departing from the spirit and scope of the present disclosure as disclosed herein.
Sequence CWU 1
1
161530PRTHomo sapien 1Met Thr Arg Asp Phe Lys Pro Gly Asp Leu Ile
Phe Ala Lys Met Lys1 5 10 15Gly Tyr Pro His Trp Pro Ala Arg Val Asp
Glu Val Pro Asp Gly Ala 20 25 30Val Lys Pro Pro Thr Asn Lys Leu Pro
Ile Phe Phe Phe Gly Thr His 35 40 45Glu Thr Ala Phe Leu Gly Pro Lys
Asp Ile Phe Pro Tyr Ser Glu Asn 50 55 60Lys Glu Lys Tyr Gly Lys Pro
Asn Lys Arg Lys Gly Phe Asn Glu Gly65 70 75 80Leu Trp Glu Ile Asp
Asn Asn Pro Lys Val Lys Phe Ser Ser Gln Gln 85 90 95Ala Ala Thr Lys
Gln Ser Asn Ala Ser Ser Asp Val Glu Val Glu Glu 100 105 110Lys Glu
Thr Ser Val Ser Lys Glu Asp Thr Asp His Glu Glu Lys Ala 115 120
125Ser Asn Glu Asp Val Thr Lys Ala Val Asp Ile Thr Thr Pro Lys Ala
130 135 140Ala Arg Arg Gly Arg Lys Arg Lys Ala Glu Lys Gln Val Glu
Thr Glu145 150 155 160Glu Ala Gly Val Val Thr Thr Ala Thr Ala Ser
Val Asn Leu Lys Val 165 170 175Ser Pro Lys Arg Gly Arg Pro Ala Ala
Thr Glu Val Lys Ile Pro Lys 180 185 190Pro Arg Gly Arg Pro Lys Met
Val Lys Gln Pro Cys Pro Ser Glu Ser 195 200 205Asp Ile Ile Thr Glu
Glu Asp Lys Ser Lys Lys Lys Gly Gln Glu Glu 210 215 220Lys Gln Pro
Lys Lys Gln Pro Lys Lys Asp Glu Glu Gly Gln Lys Glu225 230 235
240Glu Asp Lys Pro Arg Lys Glu Pro Asp Lys Lys Glu Gly Lys Lys Glu
245 250 255Val Glu Ser Lys Arg Lys Asn Leu Ala Lys Thr Gly Val Thr
Ser Thr 260 265 270Ser Asp Ser Glu Glu Glu Gly Asp Asp Gln Glu Gly
Glu Lys Lys Arg 275 280 285Lys Gly Gly Arg Asn Phe Gln Thr Ala His
Arg Arg Asn Met Leu Lys 290 295 300Gly Gln His Glu Lys Glu Ala Ala
Asp Arg Lys Arg Lys Gln Glu Glu305 310 315 320Gln Met Glu Thr Glu
Gln Gln Asn Lys Asp Glu Gly Lys Lys Pro Glu 325 330 335Val Lys Lys
Val Glu Lys Lys Arg Glu Thr Ser Met Asp Ser Arg Leu 340 345 350Gln
Arg Ile His Ala Glu Ile Lys Asn Ser Leu Lys Ile Asp Asn Leu 355 360
365Asp Val Asn Arg Cys Ile Glu Ala Leu Asp Glu Leu Ala Ser Leu Gln
370 375 380Val Thr Met Gln Gln Ala Gln Lys His Thr Glu Met Ile Thr
Thr Leu385 390 395 400Lys Lys Ile Arg Arg Phe Lys Val Ser Gln Val
Ile Met Glu Lys Ser 405 410 415Thr Met Leu Tyr Asn Lys Phe Lys Asn
Met Phe Leu Val Gly Glu Gly 420 425 430Asp Ser Val Ile Thr Gln Val
Leu Asn Lys Ser Leu Ala Glu Gln Arg 435 440 445Gln His Glu Glu Ala
Asn Lys Thr Lys Asp Gln Gly Lys Lys Gly Pro 450 455 460Asn Lys Lys
Leu Glu Lys Glu Gln Thr Gly Ser Lys Thr Leu Asn Gly465 470 475
480Gly Ser Asp Ala Gln Asp Gly Asn Gln Pro Gln His Asn Gly Glu Ser
485 490 495Asn Glu Asp Ser Lys Asp Asn His Glu Ala Ser Thr Lys Lys
Lys Pro 500 505 510Ser Ser Glu Glu Arg Glu Thr Glu Ile Ser Leu Lys
Asp Ser Thr Leu 515 520 525Asp Asn 5302107PRTHomo sapien 2Asp Ser
Arg Leu Gln Arg Ile His Ala Glu Ile Lys Asn Ser Leu Lys1 5 10 15Ile
Asp Asn Leu Asp Val Asn Arg Cys Ile Glu Ala Leu Asp Glu Leu 20 25
30Ala Ser Leu Gln Val Thr Met Gln Gln Ala Gln Lys His Thr Glu Met
35 40 45Ile Thr Thr Leu Lys Lys Ile Arg Arg Phe Lys Val Ser Gln Val
Ile 50 55 60Met Glu Lys Ser Thr Met Leu Tyr Asn Lys Phe Lys Asn Met
Phe Leu65 70 75 80Val Gly Glu Gly Asp Ser Val Ile Thr Gln Val Leu
Asn Lys Ser Leu 85 90 95Ala Glu Gln Arg Gln His Glu Glu Ala Asn Lys
100 10531593DNAHomo sapien 3atgactcgcg atttcaaacc tggagacctc
atcttcgcca agatgaaagg ttatccccat 60tggccagctc gagtagacga agttcctgat
ggagctgtaa agccacccac aaacaaacta 120cccattttct tttttggaac
tcatgagact gcttttttag gaccaaagga tatatttcct 180tactcagaaa
ataaggaaaa gtatggcaaa ccaaataaaa gaaaaggttt taatgaaggt
240ttatgggaga tagataacaa tccaaaagtg aaattttcaa gtcaacaggc
agcaactaaa 300caatcaaatg catcatctga tgttgaagtt gaagaaaagg
aaactagtgt ttcaaaggaa 360gataccgacc atgaagaaaa agccagcaat
gaggatgtga ctaaagcagt tgacataact 420actccaaaag ctgccagaag
ggggagaaag agaaaggcag aaaaacaagt agaaactgag 480gaggcaggag
tagtgacaac agcaacagca tctgttaatc taaaagtgag tcctaaaaga
540ggacgacctg cagctacaga agtcaagatt ccaaaaccaa gaggcagacc
caaaatggta 600aaacagccct gtccttcaga gagtgacatc attactgaag
aggacaaaag taagaaaaag 660gggcaagagg aaaaacaacc taaaaagcag
cctaagaagg atgaagaggg ccagaaggaa 720gaagataagc caagaaaaga
gccggataaa aaagagggga agaaagaagt tgaatcaaaa 780aggaaaaatt
tagctaaaac aggggttact tcaacctccg attctgaaga agaaggagat
840gatcaagaag gtgaaaagaa gagaaaaggt gggaggaact ttcagactgc
tcacagaagg 900aatatgctga aaggccaaca tgagaaagaa gcagcagatc
gaaaacgcaa gcaagaggaa 960caaatggaaa ctgagcagca gaataaagat
gaaggaaaga agccagaagt taagaaagtg 1020gagaagaagc gagaaacatc
aatggattct cgacttcaaa ggatacatgc tgagattaaa 1080aattcactca
aaattgataa tcttgatgtg aacagatgca ttgaggcctt ggatgaactt
1140gcttcacttc aggtcacaat gcaacaagct cagaaacaca cagagatgat
tactacactg 1200aaaaaaatac ggcgattcaa agttagtcag gtaatcatgg
aaaagtctac aatgttgtat 1260aacaagttta agaacatgtt cttggttggt
gaaggagatt ccgtgatcac ccaagtgctg 1320aataaatctc ttgctgaaca
aagacagcat gaggaagcga ataaaaccaa agatcaaggg 1380aagaaagggc
caaacaaaaa gctagagaag gaacaaacag ggtcaaagac tctaaatgga
1440ggatctgatg ctcaagatgg taatcagcca caacataacg gggagagcaa
tgaagacagc 1500aaagacaacc atgaagccag cacgaagaaa aagccatcca
gtgaagagag agagactgaa 1560atatctctga aggattctac actagataac tag
1593419DNAartificial sequenceDNA equivalent of shRNA sequences
4agacagcatg aggaagcga 19547DNAartificial sequenceDNA equivalent of
shRNA sequences 5agacagcatg aggaagcgat tcaagagatc gcttcctcat
gctgtct 47621DNAartificial sequenceDNA equivalent of shRNA
sequences 6agttcctgat ggagctgtaa a 21746DNAartificial sequenceDNA
equivalent of shRNA sequences 7agttcctgat ggagctgtaa acgagtttac
agctccatca ggaact 46821DNAartificial sequenceDNA equivalent of
shRNA sequences 8gcaatgaaga cagcaaagac a 21946DNAartificial
sequenceDNA equivalent of shRNA sequences 9gcaatgaaga cagcaaagac
acgagtgtct ttgctgtctt cattgc 461021DNAartificial sequenceDNA
equivalent of shRNA sequences 10gtctccacgc gcagtacatt t
211146DNAartificial sequenceDNA equivalent of shRNA sequences
11gtctccacgc gcagtacatt tcgagaaatg tactgcgcgt ggagac 4612454DNAHomo
sapien 12gtacaaaaaa gcaggcttta aaggaaccaa ttcagtcgac tggatccggt
accaaggtcg 60ggcaggaaga gggcctattt cccatgattc cttcatattt gcatatacga
tacaaggctg 120ttagagagat aattagaatt aatttgactg taaacacaaa
gatattagta caaaatacgt 180gacgtagaaa gtaataattt cttgggtagt
ttgcagtttt aaaattatgt tttaaaatgg 240actatcatat gcttaccgta
acttgaaagt atttcgattt cttggcttta tatatcttgt 300ggaaaggacg
aaacaccgta atcagccaca acataacgtt ttagagctag aaatagcaag
360ttaaaataag gctagtccgt tatcaacttg aaaaagtggc accgagtcgg
tgcttttttt 420ctagacccag ctttcttgta caaagttggc atta 45413455DNAHomo
sapien 13tgtacaaaaa agcaggcttt aaaggaacca attcagtcga ctggatccgg
taccaaggtc 60gggcaggaag agggcctatt tcccatgatt ccttcatatt tgcatatacg
atacaaggct 120gttagagaga taattagaat taatttgact gtaaacacaa
agatattagt acaaaatacg 180tgacgtagaa agtaataatt tcttgggtag
tttgcagttt taaaattatg ttttaaaatg 240gactatcata tgcttaccgt
aacttgaaag tatttcgatt tcttggcttt atatatcttg 300tggaaaggac
gaaacaccga cgcctctgcg gcagctgggt tttagagcta gaaatagcaa
360gttaaaataa ggctagtccg ttatcaactt gaaaaagtgg caccgagtcg
gtgctttttt 420tctagaccca gctttcttgt acaaagttgg catta
45514455DNAHomo sapien 14tgtacaaaaa agcaggcttt aaaggaacca
attcagtcga ctggatccgg taccaaggtc 60gggcaggaag agggcctatt tcccatgatt
ccttcatatt tgcatatacg atacaaggct 120gttagagaga taattagaat
taatttgact gtaaacacaa agatattagt acaaaatacg 180tgacgtagaa
agtaataatt tcttgggtag tttgcagttt taaaattatg ttttaaaatg
240gactatcata tgcttaccgt aacttgaaag tatttcgatt tcttggcttt
atatatcttg 300tggaaaggac gaaacaccga ggtagacgaa gttcctgagt
tttagagcta gaaatagcaa 360gttaaaataa ggctagtccg ttatcaactt
gaaaaagtgg caccgagtcg gtgctttttt 420tctagaccca gctttcttgt
acaaagttgg catta 45515455DNAHomo sapien 15tgtacaaaaa agcaggcttt
aaaggaacca attcagtcga ctggatccgg taccaaggtc 60gggcaggaag agggcctatt
tcccatgatt ccttcatatt tgcatatacg atacaaggct 120gttagagaga
taattagaat taatttgact gtaaacacaa agatattagt acaaaatacg
180tgacgtagaa agtaataatt tcttgggtag tttgcagttt taaaattatg
ttttaaaatg 240gactatcata tgcttaccgt aacttgaaag tatttcgatt
tcttggcttt atatatcttg 300tggaaaggac gaaacaccga actacccatt
ttctttttgt tttagagcta gaaatagcaa 360gttaaaataa ggctagtccg
ttatcaactt gaaaaagtgg caccgagtcg gtgctttttt 420tctagaccca
gctttcttgt acaaagttgg catta 45516455DNAHomo sapien 16tgtacaaaaa
agcaggcttt aaaggaacca attcagtcga ctggatccgg taccaaggtc 60gggcaggaag
agggcctatt tcccatgatt ccttcatatt tgcatatacg atacaaggct
120gttagagaga taattagaat taatttgact gtaaacacaa agatattagt
acaaaatacg 180tgacgtagaa agtaataatt tcttgggtag tttgcagttt
taaaattatg ttttaaaatg 240gactatcata tgcttaccgt aacttgaaag
tatttcgatt tcttggcttt atatatcttg 300tggaaaggac gaaacaccga
gtgctttttt aggaccaagt tttagagcta gaaatagcaa 360gttaaaataa
ggctagtccg ttatcaactt gaaaaagtgg caccgagtcg gtgctttttt
420tctagaccca gctttcttgt acaaagttgg catta 455
D00000

D00001

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.