Methods And Compositions For Treating Cancer
TERRETT; Jonathan Alexander ; et al.
U.S. patent application number 16/409737 was filed with the patent office on 2020-02-27 for methods and compositions for treating cancer. The applicant listed for this patent is CRISPR Therapeutics AG. Invention is credited to Mary-Lee DEQUEANT, Demetrios KALAITZIDIS, Zinkal Samir PADALIA, Jonathan Alexander TERRETT.
| Application Number | 20200063100 16/409737 |
| Document ID | / |
| Family ID | 67402964 |
| Filed Date | 2020-02-27 |











View All Diagrams
| United States Patent Application | 20200063100 |
| Kind Code | A1 |
| TERRETT; Jonathan Alexander ; et al. | February 27, 2020 |
METHODS AND COMPOSITIONS FOR TREATING CANCER
Abstract
Provided herein, in some embodiments, are methods and compositions (e.g., cell compositions) for the treatment of cancer.
| Inventors: | TERRETT; Jonathan Alexander; (Cambridge, MA) ; KALAITZIDIS; Demetrios; (Cambridge, MA) ; DEQUEANT; Mary-Lee; (Cambridge, MA) ; PADALIA; Zinkal Samir; (Cambridge, MA) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 67402964 | ||||||||||
| Appl. No.: | 16/409737 | ||||||||||
| Filed: | May 10, 2019 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 62826600 | Mar 29, 2019 | |||
| 62773658 | Nov 30, 2018 | |||
| 62756643 | Nov 7, 2018 | |||
| 62701340 | Jul 20, 2018 | |||
| 62670417 | May 11, 2018 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | A61K 39/001112 20180801; C12N 2800/80 20130101; C12N 2310/20 20170501; C07K 16/2875 20130101; C07K 2319/03 20130101; C07K 2319/33 20130101; A61K 2039/86 20180801; C12N 5/0636 20130101; A61K 2039/868 20180801; A61K 2039/892 20180801; C12N 9/22 20130101; A61K 2039/804 20180801; C07K 2317/622 20130101; C12N 2750/14143 20130101; C07K 14/7051 20130101; C12N 15/11 20130101; A61K 39/0011 20130101; A61P 35/00 20180101; A61K 39/001102 20180801; C07K 16/2803 20130101; C12N 15/86 20130101; C07K 2317/565 20130101; C07K 16/2878 20130101; A61K 2039/5156 20130101; A61K 35/17 20130101; A61K 2039/852 20180801 |
| International Class: | C12N 5/0783 20060101 C12N005/0783; C07K 14/725 20060101 C07K014/725; C07K 16/28 20060101 C07K016/28; C12N 15/86 20060101 C12N015/86; C12N 15/11 20060101 C12N015/11; C12N 9/22 20060101 C12N009/22; A61K 35/17 20060101 A61K035/17; A61P 35/00 20060101 A61P035/00 |
Claims
1. An engineered T cell comprising a disrupted CD70 gene and a nucleic acid encoding a chimeric antigen receptor (CAR) that does not bind CD70.
2. The engineered T cell of claim 1, further comprising a disrupted T cell receptor alpha constant region (TRAC) gene, a disrupted beta-2-microglobulin (.beta.2M) gene, or both.
3. (canceled)
4. The engineered T cell of claim 2, wherein the disrupted TRAC gene comprises the nucleic acid encoding the CAR.
5. The engineered T cell of claim 1, wherein the CAR comprises an ectodomain that binds an antigen selected from the group consisting of B cell maturation antigen (BCMA), CD33, and CD19.
6. The engineered T cell of claim 5, wherein the ectodomain comprises an anti-BCMA antibody, an anti-CD33 antibody, or an anti-CD19 antibody.
7. The engineered T cell of claim 5, wherein the ectodomain comprises an anti-BCMA single-chain variable fragment (scFv), an anti-CD33 scFv, or an anti-CD19 scFv.
8. The engineered T cell of claim 7, wherein the ectodomain comprises an anti-BCMA scFv, which comprises the same variable heavy (VH) chain complementarity determining regions (CDRs) and the same variable light (VL) chain CDRs as a reference antibody, and wherein the reference antibody comprises a VH set forth as SEQ ID NO: 60 and a VL set forth as SEQ ID NO: 61.
9. The engineered T cell of claim 8, wherein the anti-BCMA scFv comprises VH and VL chains comprising the amino acid sequences set forth in SEQ ID NOs: 60 and 61, respectively.
10. The engineered T cell of claim 8, wherein the anti-BCMA scFv comprises the amino acid sequence of SEQ ID NO: 59.
11-13. (canceled)
14. The engineered T cell of claim 7, wherein the ectodomain comprises an anti-CD33 scFv, which comprises the same VH CDRs and the same VL chain CDRs as a reference antibody, and wherein the reference antibody comprises a VH set forth as SEQ ID NO: 140 and a VL set forth as SEQ ID NO: 141.
15. The engineered T cell of claim 14, wherein the anti-CD33 scFv comprises VH and VL chains comprising the amino acid sequences set forth in SEQ ID NOs: 140 and 141, respectively.
16. The engineered T cell of claim 14, wherein the anti-CD33 scFv comprises the amino acid sequence of SEQ ID NO: 137.
17-19. (canceled)
20. The engineered T cell of claim 19, wherein the ectodomain comprises an anti-CD19 scFv, which comprises the same VH CDRs and the same VL chain CDRs as a reference antibody, and wherein the reference antibody comprises a VH set forth as SEQ ID NO: 152 and a VL set forth as SEQ ID NO: 153.
21. The engineered T cell of claim 20, wherein the anti-CD19 scFv comprises VH and VL chains comprising the amino acid sequences set forth in SEQ ID NOs: 152 and 153, respectively.
22. The engineered T cell of claim 20, wherein the anti-CD19 scFv comprises the amino acid sequence of SEQ ID NO: 151.
23-42. (canceled)
43. The engineered T cell of claim 1, wherein the engineered T cell further comprises a disrupted programmed cell death-1 (PD-1) gene.
44. The engineered T cell of claim 1, wherein the engineered T cell maintains cytotoxicity following 5 rechallenges or following 10 rechallenges with a target cell, wherein the target cell expresses an antigen specific for the CAR.
45. (canceled)
46. The engineered T cell of claim 44, wherein the target cell is a cancer cell.
47. A population of cells comprising engineered T cells, wherein the engineered T cells comprise a disrupted CD70 gene and a nucleic acid encoding a CAR that does not bind CD70.
48. The population of cells of claim 47, further comprising a disrupted TRAC gene, a disrupted .beta.2M gene, or both.
49. (canceled)
50. The population of cells of claim 48, wherein the disrupted TRAC gene comprises the nucleic acid encoding the CAR.
51. The population of cells of claim 47, wherein the CAR comprises an ectodomain that binds an antigen selected from the group consisting of B cell maturation antigen (BCMA), CD33, and CD19.
52. The population of cells of claim 51, wherein the ectodomain comprises an anti-BCMA antibody, an anti-CD33 antibody, or an anti-CD19 antibody.
53. The population of cells of claim 51, wherein the ectodomain comprises an anti-BCMA single-chain variable fragment (scFv), an anti-CD33 scFv, or an anti-CD19 scFv.
54. The population of cells of claim 53, wherein the ectodomain comprises an anti-BCMA scFv, which comprises the same variable heavy (VH) chain complementarity determining regions (CDRs) and the same variable light (VL) chain CDRs as a reference antibody, wherein the reference antibody comprises a VH set forth as SEQ ID NO: 60 and a VL set forth as SEQ ID NO: 61.
55. The population of cells of claim 54, wherein the anti-BCMA scFv comprises VH and VL chains comprising the amino acid sequences set forth in SEQ ID NOs: 60 and 61, respectively.
56. The population of cells of claim 54, wherein the anti-BCMA scFv comprises the amino acid sequence of SEQ ID NO: 59.
57-59. (canceled)
60. The population of cells of claim 53, wherein the ectodomain comprises an anti-CD33 scFv, which comprises the same VH CDRs and the same VL chain CDRs as a reference antibody, wherein the reference antibody comprises a VH set forth as SEQ ID NO: 140 and a VL set forth as SEQ ID NO: 141.
61. The population of cells of claim 60, wherein the anti-CD33 scFv comprises VH and VL chains comprising the amino acid sequences set forth in SEQ ID NOs: 140 and 141, respectively.
62. The population of cells of claim 60, wherein the anti-CD33 scFv comprises the amino acid sequence of SEQ ID NO: 137.
63-65. (canceled)
66. The population of cells of claim 53, wherein the ectodomain comprises an anti-CD19 scFv, which comprises the same VH CDRs and the same VL chain CDRs as a reference antibody, wherein the reference antibody comprises a VH set forth as SEQ ID NO: 152 and a VL set forth as SEQ ID NO: 153.
67. The population of cells of claim 66, wherein the anti-CD19 scFv comprises VH and VL chains comprising the amino acid sequences set forth in SEQ ID NOs: 152 and 153, respectively.
68. The population of cells of claim 66, wherein the anti-CD19 scFv comprises the amino acid sequence of SEQ ID NO: 151.
69-80. (canceled)
81. The population of cells of claim 48, wherein there is a deletion in the TRAC gene relative to unmodified T cells.
82. The engineered T cell of claim 81, wherein the deletion is 15-30 base pairs.
83. The engineered T cell of claim 81, wherein the deletion is 20 base pairs.
84. The engineered T cell of claim 81, wherein the deletion comprises SEQ ID NO: 86.
85-89. (canceled)
90. The population of cells of claim 47, wherein the engineered T cell maintains cytotoxicity following 5 rechallenges or following 10 rechallenges with a target cell, wherein the target cell expresses an antigen specific for the CAR.
91. (canceled)
92. The population of cells of claim 90, wherein the target cell is a cancer cell.
93. The population of cells of claim 48, wherein the disrupted .beta.2M gene comprises at least one nucleotide sequence selected from any one of SEQ ID NOS: 9-14.
94. The population of cells of claim 47, wherein the disrupted CD70 gene comprises at least one nucleotide sequence selected from any one of SEQ ID NOS: 129-134.
95. The population of cells of claim 48, wherein at least 90% of the engineered T cells do not express a detectable level of TCR surface protein.
96. The population of cells of claim 47, wherein the engineered T cells: (a) exhibit increased cellular proliferative capacity; (b) exhibit increased cell lysis; (c) exhibit reduced cellular exhaustion; (d) maintain cytokine-dependent proliferation; (e) exhibit increased cytokine secretion; or (f) any combination of (a)-(e), relative to control T cells, wherein control T cells express endogenous CD70 protein.
97. A method comprising administering to a subject the population of cells of claim 47.
98. The method of claim 97, wherein the engineered T cells are engineered human T cells.
99. The method of claim 97, wherein the subject has a cancer.
100. The method of claim 99, wherein the cancer expresses BMCA, CD19, CD33 or combinations thereof.
101. (canceled)
102. The method of claim 99, wherein the cancer is a solid tumor malignancy or a hematological malignancy.
103. The method of claim 102, wherein the solid tumor malignancy is selected from the group consisting of: ovarian tumor, pancreatic tumor, kidney tumor, lung tumor, and intestinal tumor.
104-110. (canceled)
111. A method for producing an engineered T cell, the method comprising: (a) delivering to a T cell an RNA-guided nuclease, a gRNA targeting a CD70 gene, and a vector comprising a donor template that comprises a nucleic acid encoding a CAR; and (b) producing an engineered T cell comprising a disrupted CD70 gene and expressing the CAR.
112-114. (canceled)
115. A method for producing an engineered T cell, the method comprising (a) delivering to a T cell an RNA-guided nuclease, a gRNA targeting a TRAC gene, a gRNA targeting a .beta.2M gene, a gRNA targeting a CD70 gene, and a vector comprising a donor template that comprises a nucleic acid encoding a CAR; and (b) producing an engineered T cell.
116-121. (canceled)
122. A method for producing an engineered T cell for immunotherapy against a target cell, comprising: (a) disrupting a CD70 gene in a T cell, and (b) expressing a CAR that binds to an antigen expressed on the target cell, wherein the antigen is not CD70.
123-156. (canceled)
157. A method of increasing proliferation or reducing exhaustion of T cells, comprising disrupting the CD70 gene in the T cells.
158-161. (canceled)
162. The engineered T cell of claim 5, wherein the CAR comprises the amino acid sequence of SEQ ID NO: 57, SEQ ID NO:139, or SEQ ID NO:149.
163-167. (canceled)
168. The population of cells of claim 51, wherein the CAR comprises the amino acid sequence of SEQ ID NO: 57, SEQ ID NO:139, or SEQ ID NO:149.
169-181. (canceled)
Description
RELATED APPLICATIONS
[0001] This application claims the benefit of U.S. Provisional Patent Application Ser. No. 62/670,417 filed May 11, 2018; U.S. Provisional Patent Application Ser. No. 62/701,340 filed Jul. 20, 2018; U.S. Provisional Patent Application Ser. No. 62/756,643 filed Nov. 7, 2018; U.S. Provisional Patent Application Ser. No. 62/773,658 filed Nov. 30, 2018; and U.S. Provisional Patent Application Ser. No. 62/826,600 filed Mar. 29, 2019. The entire contents of the above-referenced patent applications are incorporated herein by this reference.
BACKGROUND
[0002] Chimeric antigen receptor (CAR) T-cell therapy uses genetically-modified T cells to more specifically and efficiently target and kill cancer cells. After T cells have been collected from the blood, the cells are engineered to include CARs on their surface. The CARs may be introduced into the T cells using CRISPR/Cas9 gene editing technology. When these allogeneic CAR T cells are injected into a patient, the receptors enable the T cells to kill cancer cells.
SUMMARY
[0003] In some aspects, the present disclosure provides engineered immune cells (e.g., T cells) and methods of producing immune cells that have been edited using CRISPR/Cas9 gene editing technology to disrupt endogenous CD70 expression (knockout CD70).
[0004] In some aspects of the present disclosure provide an engineered immune cell (e.g., T cell) comprising a disruption in the CD70 gene. In some embodiments, the engineered immune cells are allogeneic T cells comprising a disrupted CD70 gene and a nucleic acid encoding a CAR. In some embodiments, the engineered immune cells are allogeneic T cells comprising a TRAC gene disrupted by insertion of a nucleic acid encoding a CAR, a disrupted .beta.2M gene, and a disrupted CD70 gene. In some embodiments, the T cells are human T cells. In some embodiments, the engineered immune cells (e.g., T cells) comprise a disrupted TRAC gene, a disrupted B2M gene, a disrupted CD70 gene, and a nucleic acid encoding a CAR. In some embodiments, the disrupted TRAC gene comprises the nucleic acid encoding the CAR. In some embodiments the engineered immune cell (e.g., T cell) further comprises a disrupted PD-1 gene. In some embodiments the nucleic acid encoding a CAR target a tumor antigen (e.g., BCMA, CD19, CD33 or CD70).
[0005] In some aspects the engineered immune cell (e.g., T cell) provided exhibits improved T cell function including the prevention of premature exhaustion, enhanced CAR T cell expansion, and increased efficiency of cancer cell killing. In some aspects the engineered immune cell (e.g., T cell) provided exhibit continued, steady cell growth, relative to unedited T cells or relative to edited T cells that express CD70, as well as showing increased cytotoxicity and cytokine (e.g., IL-2 and/or IFN-gamma) secretion.
[0006] In some aspects, the disclosure provides an engineered T cell comprising a disrupted CD70 gene and a nucleic acid encoding a CAR that does not bind CD70. In some aspects, the engineered T cell comprises a disrupted T cell receptor alpha constant region (TRAC) gene. In some aspects, the disrupted TRAC gene comprises the nucleic acid encoding the CAR that does not bind CD70. In some aspects, the engineered T cell comprises a disrupted beta-2-microglobulin (.beta.2M) gene.
[0007] In some aspects, the disclosure provides an engineered T cell comprising: (i) a disrupted TRAC gene; (ii) a disrupted B2M gene; (iii) a disrupted CD70 gene; and (iv) a nucleic acid encoding a CAR that does not bind CD70.
[0008] In some aspects, the disclosure provides a population of cells comprising engineered T cells, wherein the engineered T cells comprise a disrupted CD70 gene and a nucleic acid encoding a CAR that does not bind CD70.
[0009] In some aspects, the engineered T cell in the population of cells comprises a disrupted T cell receptor alpha constant region (TRAC) gene. In some aspects, the disrupted TRAC gene comprises the nucleic acid encoding the CAR that does not bind CD70. In some aspects, the engineered T cell in the population of cells comprises a disrupted beta-2-microglobulin (.beta.2M) gene.
[0010] In some aspects, the disclosure provides a population of cells comprising engineered T cells, wherein the engineered T cells comprise: (i) a disrupted TRAC gene; (ii) a disrupted B2M gene; (iii) a disrupted CD70 gene; and (iv) a nucleic acid encoding a CAR that does not bind CD70.
[0011] In any of the foregoing or related aspects, the CAR comprises an ectodomain that binds i-B cell maturation antigen (BCMA). In some aspects, the ectodomain comprises an anti-BCMA antibody. In some aspects, the ectodomain comprises an anti-BCMA single-chain variable fragment (scFv). In some aspects, the anti-BCMA scFv comprises variable heavy (VH) chain complementarity determining regions (CDRs) and the same variable light (VL) chain CDRs as a reference antibody, wherein the reference antibody comprises a VH set forth as SEQ ID NO: 60 and a VL set forth as SEQ ID NO: 61. In some aspects, the anti-BCMA scFv comprises VH and VL chains comprising the amino acid sequences set forth in SEQ ID NOs: 60 and 61, respectively. In some aspects, the anti-BCMA scFv comprises the amino acid sequence of SEQ ID NO: 59. In some aspects, the anti-BCMA scFv is encoded by a nucleotide sequence having at least 80%, 85%, 90%, 95%, 96%, 97%, 98% or 99% identity to SEQ ID NO: 57.
[0012] In any of the foregoing or related aspects, the anti-BCMA scFv comprises the amino acid sequence of SEQ ID NO: 59. In some embodiments, the anti-BCMA scFv comprises a VH comprising the amino acid sequence of SEQ ID NO: 60. In some embodiments, the anti-BCMA scFv comprises a VL comprising the amino acid sequence of SEQ ID NO: 61. In some embodiments, the anti-BCMA scFv comprises a VH comprising CDR amino acid sequences of (i) SEQ ID NO: 80, SEQ ID NO: 82, and/or SEQ ID NO: 84 or (ii) SEQ ID NO: 81, SEQ ID NO: 83, or SEQ ID NO: 85; and/or the anti-BCMA scFv comprises a VL sequence comprising CDR amino acid sequences of (i) SEQ ID NO: 74, SEQ ID NO: 76, and/or SEQ ID NO: 78.
[0013] In any of the foregoing or related aspects, the CAR comprises an ectodomain that binds CD33. In some the ectodomain comprises an anti-CD33 antibody. In some aspects, the ectodomain comprises an anti-CD33 scFv. In some aspects, the anti-CD33 scFv comprises the same VH CDRs and the same VL chain CDRs as a reference antibody, wherein the reference antibody comprises a VH set forth as SEQ ID NO: 140 and a VL set forth as SEQ ID NO: 141. In some aspects, the anti-CD33 scFv comprises VH and VL chains comprising the amino acid sequences set forth in SEQ ID NOs: 140 and 141, respectively. In some aspects, the anti-CD33 scFv comprises the amino acid sequence of SEQ ID NO: 137.
[0014] In any of the foregoing or related aspects, the CAR comprises an ectodomain that binds CD19. In some aspects, wherein the ectodomain comprises an anti-CD19 antibody. In some aspects, the ectodomain comprises an anti-CD19 scFv. In some aspects, the anti-CD19 scFv comprises the same VH CDRs and the same VL chain CDRs as a reference antibody, wherein the reference antibody comprises a VH set forth as SEQ ID NO: 152 and a VL set forth as SEQ ID NO: 153. In some aspects, the anti-CD19 scFv comprises VH and VL chains comprising the amino acid sequences set forth in SEQ ID NOs: 152 and 153, respectively. In some aspects, the anti-CD19 scFv comprises the amino acid sequence of SEQ ID NO: 151.
[0015] In some aspects, the disclosure provides an engineered T cell comprising: (i) a disrupted TRAC gene; (ii) a disrupted B2M gene; (iii) a disrupted CD70 gene; and (iv) a nucleic acid encoding a CAR that binds CD70. In some aspects, the disrupted TRAC gene comprises the nucleic acid encoding the CAR.
[0016] In some aspects, the disclosure provides a population of cells comprising engineered T cells, wherein the engineered T cells comprise: (i) a disrupted TRAC gene; (ii) a disrupted B2M gene; (iii) a disrupted CD70 gene; and (iv) a nucleic acid encoding a CAR that binds CD70.
[0017] In some aspects, the disclosure provides a population of cells comprising engineered T cells, wherein the engineered T cells comprise:
[0018] (i) a disrupted TRAC gene;
[0019] (ii) a disrupted .beta.2M gene;
[0020] (iii) a disrupted CD70 gene
[0021] (iv) a nucleic acid encoding a CAR comprising (a) an ectodomain that comprises an anti-CD70 scFv, (b) a CD8 transmembrane domain, and (c) an endodomain that comprises a 41BB co-stimulatory domain and a CD3z signaling domain.
[0022] In any of the foregoing or related aspects, the CAR that binds CD70 comprises an ectodomain comprising an anti-CD70 antibody. In some aspects, CAR comprises an ectodomain comprising an anti-CD70 scFv. In some aspects, the anti-CD70 scFv comprises the same VH CDRs and the same VL CDRs as a reference antibody, wherein the reference antibody comprises a VH set forth as SEQ ID NO: 51 and a VL set forth as SEQ ID NO: 52. In some aspects, the anti-CD70 scFv comprises VH and VL chains comprising the amino acid sequences set forth in SEQ ID NOs: 51 and 52, respectively. In some aspects, the anti-CD70 scFv comprises the amino acid sequence of SEQ ID NO: 48 or 50. In some aspects, the anti-CD70 scFv comprises the amino acid sequence of SEQ ID NO: 50.
[0023] In any of the foregoing or related aspects, the anti-CD70 scFv comprises a VH comprising the amino acid sequence of SEQ ID NO: 51. In some embodiments, the anti-CD70 scFv comprises a VL comprising the amino acid sequence of SEQ ID NO: 52. In some embodiments, the anti-CD70 scFv comprises a VH comprising CDR amino acid sequences of (i) SEQ ID NO: 68, SEQ ID NO: 70, and/or SEQ ID NO: 72 or (ii) SEQ ID NO: 69, SEQ ID NO: 71, and/or SEQ ID NO: 73; and/or the anti-CD70 scFv comprises a VL sequence comprising CDR amino acid sequences of (i) SEQ ID NO: 62, SEQ ID NO: 64, and/or SEQ ID NO: 66 or (ii) SEQ ID NO: SEQ ID NO: 63, SEQ ID NO: 65, and/or SEQ ID NO: 67.
[0024] In any of the foregoing or related aspects, the CAR comprises a CD28 or 41BB co-stimulatory domain. In any of the foregoing or related aspects, the CAR comprises a CD3.zeta. signaling domain. In any of the foregoing or related aspects, the CAR comprises a CD8 transmembrane domain.
[0025] In any of the foregoing or related aspects, there is a deletion in the TRAC gene relative to unmodified T cells. In some aspects, the deletion is 15-30 base pairs. In some aspects, the deletion is 20 base pairs. In some aspects, the deletion comprises SEQ ID NO: 86. In some aspects, the deletion is of SEQ ID NO: 86.
[0026] In some aspects, the disclosure provides an engineered T cell comprising a disrupted CD70 gene and a nucleic acid encoding a CAR that binds CD70, wherein the CAR comprises the amino acid sequence set forth in SEQ ID NO: 46. In some aspects, the disclosure provides an engineered T cell comprising a disrupted CD70 gene, and a nucleic acid encoding a CAR that binds CD70, wherein the nucleic acid sequence is at least 90% identical to SEQ ID NO: 45. In some aspects, the disclosure provides an engineered T cell comprising a disrupted CD70 gene, and a nucleic acid encoding a CAR that binds CD70, wherein the nucleic acid sequence is SEQ ID NO: 45.
[0027] In some embodiments, the CD70 gene is disrupted by CRISPR/Cas9 gene editing. In some embodiments, the TRAC gene is disrupted by CRISPR/Cas9 gene editing. In some embodiments, the B2M gene is disrupted by CRISPR/Cas9 gene editing. In some embodiments, the PD-1 gene is disrupted by CRISPR/Cas9 gene editing.
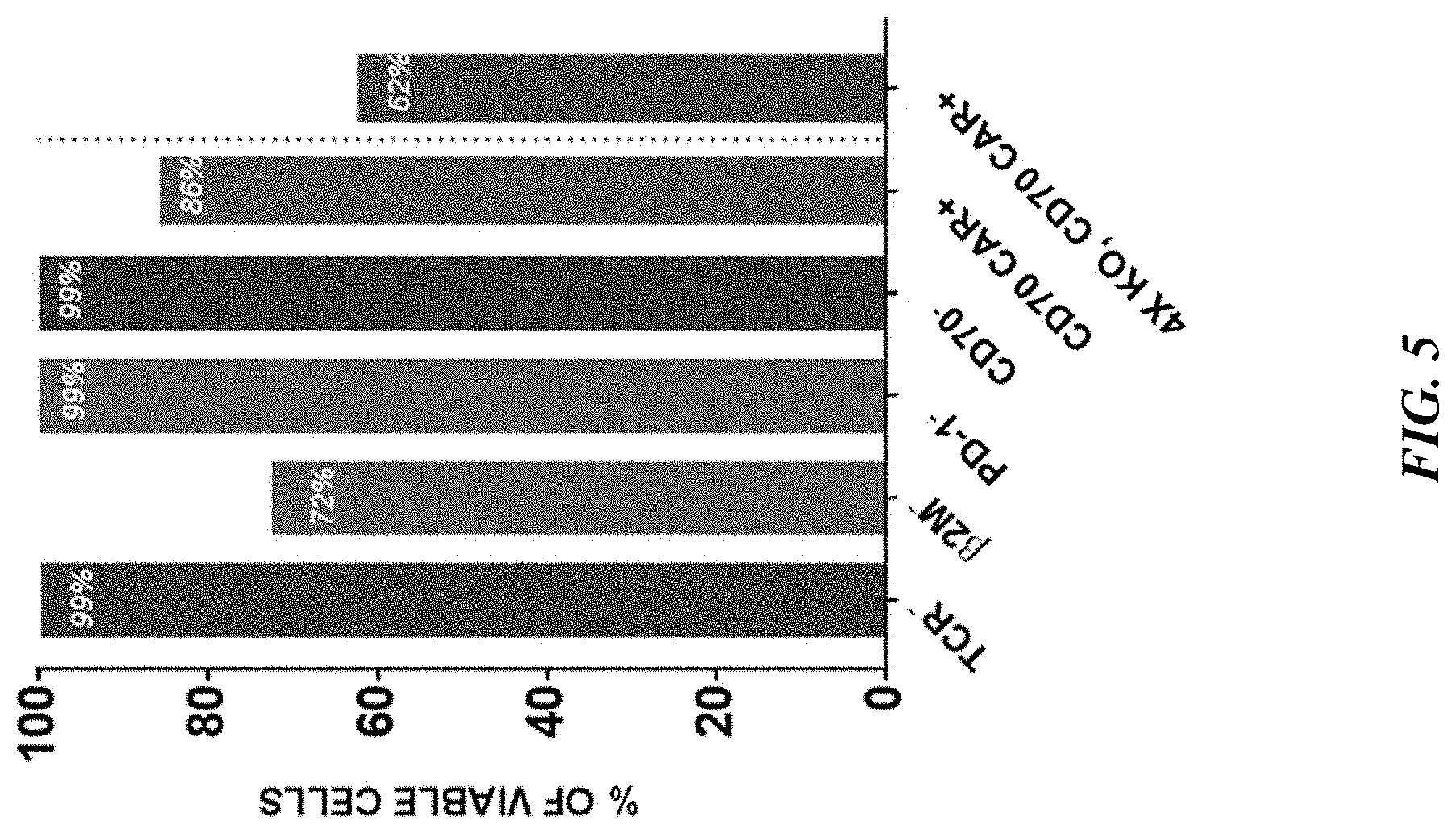
[0028] In some aspects, the disclosure provides an engineered T cell comprising: [0029] (i) a disrupted TRAC gene, wherein the disrupted TRAC gene comprises a nucleic acid encoding a CAR comprising the amino acid sequence set forth in SEQ ID NO: 46; [0030] (ii) a disrupted B2M gene; and [0031] (iii) a disrupted CD70 gene. In some embodiments, the nucleic acid encoding the CAR comprises a sequence at least 80%, at least 90%, at least 95%, at least 96%, at least 97%, at least 98% or at least 99% identical to SEQ ID NO: 45.
[0032] In other aspects, the disclosure provides an engineered T cell comprising: [0033] (i) a disrupted TRAC gene, wherein the disrupted TRAC gene comprises a nucleic acid encoding a CAR, wherein the nucleic acid sequence is at least 90% identical to SEQ ID NO: 45; [0034] (ii) a disrupted B2M gene; and [0035] (iii) a disrupted CD70 gene. In some embodiments, the disrupted TRAC gene comprises a donor sequence comprising the nucleotide sequence set forth in SEQ ID NO: 45 or SEQ ID NO: 44.
[0036] In some aspects, the disclosure provides an engineered T cell comprising: [0037] (i) a disrupted TRAC gene comprising a nucleic acid sequence at least 90% identical to SEQ ID NO: 44; [0038] (ii) a disrupted B2M gene; and [0039] (iii) a disrupted CD70 gene.
[0040] In some aspects, the disclosure provides an engineered T cell comprising: [0041] (i) a disrupted TRAC gene comprising the nucleic acid sequence of SEQ ID NO: 44; [0042] (ii) a disrupted B2M gene; and [0043] (iii) a disrupted CD70 gene.
[0044] In any of the foregoing or related aspects, the engineered T cell comprises a disrupted PD-1 gene.
[0045] In any of the foregoing or related aspects, the engineered T cell maintains cytotoxicity following 5 rechallenges with a target cell, wherein the target cell expresses an antigen specific for the CAR. In some aspects, the engineered T cell maintains cytotoxicity following 10 rechallenges with the target cell. In some aspects, the target cell is a cancer cell. In some aspects, the target cell is a cancer cell of a hematological cancer or solid tumor.
[0046] In any of the foregoing or related aspects, the engineered T cell or population of cells comprises a CAR comprising the amino acid sequence of SEQ ID NO: 57. In some aspects, the CAR is encoded by a nucleic acid sequence having at least 90% identity to SEQ ID NO: 56.
[0047] In any of the foregoing or related aspects, the engineered T cell or population of cells comprises a CAR comprising the amino acid sequence of SEQ ID NO: 139. In some aspects, the CAR is encoded by a nucleic acid sequence having at least 90% identity to SEQ ID NO: 136.
[0048] In any of the foregoing or related aspects, the engineered T cell or population of cells comprises a CAR comprising the amino acid sequence of SEQ ID NO: 149. In some aspects, the CAR is encoded by a nucleic acid sequence having at least 90% identity to SEQ ID NO: 148.
[0049] In any of the foregoing or related aspects, the engineered T cell or population of cells comprises a CAR comprising the amino acid sequence of SEQ ID NO: 46. In some aspects, the CAR is encoded by a nucleic acid sequence having at least 90% identity to SEQ ID NO: 45.
[0050] Other aspects of the present disclosure provide a population of cells comprising any of the engineered immune cells (e.g., T cells) described herein. In some embodiments, a population of cells comprise T cells that comprise a TRAC gene disrupted by insertion of a nucleic acid encoding a CAR, a disrupted .beta.2M gene, and a disrupted CD70 gene. In some embodiments, a population of cells comprise T cells that comprise a disrupted TRAC gene, a disrupted B2M gene, a disrupted CD70 gene, and a nucleic acid encoding a CAR. In some embodiments, a population of cells comprise T cells that comprise a disrupted TRAC gene, wherein the disrupted TRAC gene comprises a nucleic acid encoding a CAR, a disrupted B2M gene, and a disrupted CD70 gene.
[0051] In some aspects, the disclosure provides a population of cells comprising engineered T cells, wherein the engineered T cells comprise: [0052] (i) a disrupted TRAC gene, wherein the disrupted TRAC gene comprises a nucleic acid encoding a CAR comprising (a) an ectodomain that comprises an anti-CD70 antigen-binding fragment, (b) a CD8 transmembrane domain, and (c) an endodomain that comprises a 41BB co-stimulatory domain and a CD3z signaling domain; [0053] (ii) a disrupted beta-2-microglobulin (B2M) gene; and [0054] (iii) a disrupted CD70 gene.
[0055] In some aspects, the disclosure provides a population of cells comprising engineered T cells, wherein the engineered T cells comprise:
[0056] (i) a disrupted TRAC gene, wherein the disrupted TRAC gene comprises a nucleic acid encoding a CAR comprising the amino acid sequence set forth in SEQ ID NO: 46;
[0057] (ii) a disrupted .beta.2M gene; and
[0058] (iii) a disrupted CD70 gene.
[0059] In other aspects, the disclosure provides a population of cells comprising engineered T cells, wherein the engineered T cells comprise:
[0060] (i) a disrupted TRAC gene, wherein the disrupted TRAC gene comprises a nucleic acid encoding a CAR, wherein the nucleic acid sequence is at least 90% identical to SEQ ID NO: 45;
[0061] (ii) a disrupted .beta.2M gene; and
[0062] (iii) a disrupted CD70 gene. In some aspects, the disrupted TRAC gene comprises the nucleic acid sequence set forth in SEQ ID NO: 45.
[0063] In some aspects, the disclosure provides a population of cells comprising engineered T cells, wherein the engineered T cells comprise:
[0064] (i) a disrupted TRAC gene comprising a nucleic acid sequence at least 90% identical to SEQ ID NO: 44;
[0065] (ii) a disrupted .beta.2M gene; and
[0066] (iii) a disrupted CD70 gene. In some aspects, the disrupted TRAC gene comprises the nucleic acid sequence set forth in SEQ ID NO: 44.
[0067] In some embodiments, the CAR comprises a CD3z signaling domain. In some embodiments, the CAR comprises a CD8 transmembrane domain. In some embodiments, the CAR comprises a CD28 or 41BB co-stimulatory domain.
[0068] In any of the foregoing or related aspects of the population of cells, the disrupted .beta.2M gene comprises at least one nucleotide sequence selected from any one of SEQ ID NOS: 9-14. In any of the foregoing or related aspects of the population of cells, the disrupted CD70 gene comprises at least one nucleotide sequence selected from any one of SEQ ID NOS: 129-134.
[0069] In some embodiments, the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 45 and/or the nucleic acid encoding the anti-CD70 CAR comprises the nucleotide sequence of SEQ ID NO: 45. In some embodiments, the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 45. In some embodiments, the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 44. In some embodiments, the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 56 and/or the nucleic acid encoding the anti-BCMA CAR comprises the nucleotide sequence of SEQ ID NO: 56. In some embodiments, the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 56. In some embodiments, the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 55.
[0070] In some embodiments, the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 156 and/or the nucleic acid encoding the anti-CD19 CAR comprises the nucleotide sequence of SEQ ID NO: 148. In some embodiments, the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 148. In some embodiments, the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 156. In some embodiments, the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 135 and/or the nucleic acid encoding the anti-CD33 CAR comprises the nucleotide sequence of SEQ ID NO: 136. In some embodiments, the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 136. In some embodiments, the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 135.
[0071] In any of the foregoing aspects, the engineered T cells:(a) exhibit increased cellular proliferative capacity;
[0072] (b) exhibit increased cell lysis;
[0073] (c) exhibit reduced cellular exhaustion;
[0074] (d) maintain cytokine-dependent proliferation;
[0075] (e) exhibit increased cytokine secretion; or
[0076] (f) any combination of (a)-(e),
[0077] relative to control T cells, wherein control T cells express endogenous CD70 protein.
[0078] In some embodiments, at least 50%, optionally 50%-65%, of the engineered T cells do not express a detectable level of TCR surface protein, do not express a detectable level of .beta.2M surface protein, do not express a detectable level of CD70 surface protein, and/or express a detectable level of the CAR.
[0079] In some embodiments, at least 90%, optionally 90%-100%, of the engineered T cells do not express a detectable level of TCR surface protein. In some embodiments, greater than 99.5% of the engineered T cells do not express a detectable level of TCR surface protein.
[0080] In some embodiments, at least 60%, optionally 60%-75%, of the engineered immune cells (e.g., T cells) do not express a detectable level of .beta.2M surface protein.
[0081] In some embodiments, at least 80%, optionally 80%-100%, of the engineered immune cells (e.g., T cells) do not express a detectable level of CD70 surface protein.
[0082] In some embodiments, at least 80%, optionally 80%-95%, of the engineered immune cells (e.g., T cells) express a detectable level of the CAR (e.g., an anti-CD70 CAR or an anti-BCMA CAR).
[0083] In some embodiments, the engineered immune cells (e.g., T cells) further comprise a disrupted PD-1 gene.
[0084] In some embodiments, at least 50%, optionally 50%-70%, of the engineered T cells do not express a detectable level of TCR surface protein, do not express a detectable level of .beta.2M surface protein, do not express a detectable level of PD-1 surface protein, do not express a detectable level of CD70 surface protein, and/or express a detectable level of the CAR. [0085] In some aspects, the disclosure provides a method for producing an engineered T cell, the method comprising: [0086] (a) delivering to a T cell [0087] an RNA-guided nuclease, [0088] a gRNA targeting a CD70 gene, and [0089] a vector comprising a donor template that comprises a nucleic acid encoding a CAR; and [0090] (b) producing an engineered T cell comprising a disrupted CD70 gene and expressing the CAR.
[0091] In some aspects, the method further comprises delivering to the T cell a gRNA targeting a TRAC gene; wherein the engineered T cell further comprises a disrupted TRAC gene. In some aspects, the nucleic acid encoding the CAR is flanked by left and right homology arms to the TRAC gene; and wherein the engineered T cell comprises the nucleic acid encoding the CAR in the TRAC gene. In some aspects, the method further comprises delivering to the T cell a gRNA targeting a .beta.2M gene; wherein the engineered T cell of further comprises a disrupted .beta.2M gene.
[0092] Also provided herein are methods for producing an engineered T cell, the method comprising (a) delivering to a T cell an RNA-guided nuclease, a gRNA targeting a TRAC gene, a gRNA targeting a .beta.2M gene, a gRNA targeting a CD70 gene, and a vector comprising a donor template that comprises a nucleic acid encoding a CAR, optionally wherein the nucleic acid encoding the CAR is flanked by left and right homology arms to the TRAC gene locus, and (b) producing an engineered T cell.
[0093] In some embodiments, the RNA-guided nuclease is a Cas9 nuclease, optionally a Streptococcus pyogenes Cas9 nuclease. Other RNA-guided nucleases may be used and are described below.
[0094] In some embodiments, wherein the gRNA targeting the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 98 or targets the nucleotide sequence of SEQ ID NO: 118, and optionally wherein the gRNA targeting the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 30. In some embodiments, the gRNA targeting the .beta.2M gene comprises the nucleotide sequence of SEQ ID NO: 99 or targets the nucleotide sequence of SEQ ID NO: 119, and optionally wherein the gRNA targeting the .beta.2M gene comprises the nucleotide sequence of SEQ ID NO: 31. In some embodiments, the gRNA targeting the CD70 gene comprises the nucleotide sequence of SEQ ID NOS: 94 or 95 or targets the nucleotide sequence of SEQ ID NO: 114 or 115, and optionally wherein the gRNA targeting the CD70 gene comprises the nucleotide sequence of SEQ ID NOS: 26 or 27.
[0095] In any of the foregoing aspects, the RNA-guided nuclease and gRNA are complexed in a ribonucleorotein particle (RNP).
[0096] In some embodiments, the methods further comprise delivering to the T cell a gRNA targeting a PD-1 gene. In some aspects, the engineered immune cells are allogeneic T cells comprising a TRAC gene disrupted by insertion of a nucleic acid encoding a CAR, a disrupted .beta.2M gene, and a disrupted PD-1 gene. In some embodiments the engineered immune cell (e.g., T cell) further comprises a disrupted CD70 gene.
[0097] In some embodiments, the gRNA targeting the PD-1 gene comprises the nucleotide sequence of SEQ ID NO: 100 or targets the nucleotide sequence of SEQ ID NO: 120, and optionally wherein the gRNA targeting the PD-1 gene comprises the nucleotide sequence of SEQ ID NO: 32.
[0098] In some aspects, the disclosure provides a method for producing an engineered T cell for immunotherapy against a target cell, comprising:
[0099] (a) disrupting a CD70 gene in a T cell, and
[0100] (b) expressing a CAR that binds to an antigen expressed on the target cell, wherein the antigen is not CD70. In some aspects, the target cell is a cancer cell. In some aspects, the method is ex vivo. In some aspects, the method further comprises comprising disrupting a TRAC gene in the T cell. In some aspects, the method further comprises disrupting a .beta.2M gene in the T cell. In some aspects, the CAR is encoded by a nucleic acid in the disrupted TRAC gene. In some aspects, the CAR is any one of the CARs described herein.
[0101] In some aspects, the disclosure provides a population of engineered T cells produced by any one of the methods described herein.
[0102] In some aspects, the disclosure provides a method of increasing proliferation of T cells, comprising disrupting the CD70 gene in the T cells. In some aspects, the disclosure provides a method of reducing exhaustion of T cells, comprising disrupting the CD70 gene in the T cells. In any of the foregoing aspects, the CD70 gene is disrupted by CRISPR/Cas gene editing. In some aspects, the method further comprises disrupting the TRAC gene, the .beta.2M gene, or both the TRAC and .beta.2M genes in the T cells. In some aspects, the TRAC gene, .beta.2M gene or both TRAC and .beta.2M gene is disrupted by CRISPR/Cas gene editing.
[0103] In some embodiments, the vector comprises a nucleic acid encoding a CAR that comprises the amino acid sequence of SEQ ID NO: 46. In some embodiments, the vector comprises a nucleic acid encoding a CAR that comprises the amino acid sequence of SEQ ID NO: 57. In some embodiments, the vector comprises a nucleic acid encoding a CAR that comprises the amino acid sequence of SEQ ID NO: 149. In some embodiments, the vector comprises a nucleic acid encoding a CAR that comprises the amino acid sequence of SEQ ID NO: 139.
[0104] In some aspects, the disclosure provides methods for administering the population of cells or an engineered T cells described herein to a subject. In some aspects, the engineered T cells are engineered human T cells. In some aspects, the subject has cancer. In some aspects, the cancer expresses CD70, BMCA, CD19, CD33 or combinations thereof. In some aspects, the population of cells is administered to the subject in an amount effective to treat the cancer. In some aspects, the cancer is a solid tumor malignancy or a hematological malignancy. In some aspects, the solid tumor malignancy is selected from the group consisting of: ovarian tumor, pancreatic tumor, kidney tumor, lung tumor, and intestinal tumor. In some aspects, the population of cells is administered to the subject in an amount effective to reduce the volume of a tumor in the subject.
[0105] In some aspects, the disclosure provides a method for treating cancer in a subject, comprising administering the population of cells or an engineered T cells described herein to a subject.
[0106] In some aspects, the disclosure provides a method for treating cancer in a subject, comprising administering to the patient a population of cells comprising engineered T cells, wherein the engineered T cells comprise a disrupted CD70 gene and a nucleic acid encoding a CAR, thereby treating cancer in the subject. In some embodiments, the CAR binds CD70. In some embodiments, the CAR does not bind CD70.
[0107] In other aspects, the disclosure provides a method for treating cancer in a subject, comprising administering to the patient a population of cells comprising engineered T cells, wherein the engineered T cells comprise:
[0108] (i) a disrupted TRAC gene;;
[0109] (ii) a disrupted B2M gene;
[0110] (iii) a disrupted CD70 gene; and
[0111] (iv) a nucleic acid encoding a CAR;
[0112] thereby treating the cancer in the subject.
[0113] In yet other aspects, the disclosure provides a method for treating cancer in a subject, comprising administering to the patient a population of cells comprising engineered T cells, wherein the engineered T cells comprise:
[0114] (i) a disrupted TRAC gene;
[0115] (ii) a disrupted B2M gene;
[0116] (iii) a disrupted CD70 gene; and
[0117] (iv) a nucleic acid encoding a CAR comprising (a) an ectodomain that comprises an anti-CD70 antigen-binding fragment, (b) a CD8 transmembrane domain, and (c) an endodomain that comprises a 41BB co-stimulatory domain and a CD3z signaling domain,
[0118] thereby treating the cancer in the subject. In some embodiments, the CAR comprises the amino acid sequence of SEQ ID NO: 46. In some embodiments, the nucleic acid encoding the CAR comprises the nucleotide sequence of SEQ ID NO: 45. In some embodiments, the disrupted TRAC gene comprises the nucleotide sequence of SEQ ID NO: 45 or SEQ ID NO: 44.
[0119] In some aspects, the disclosure provides a method of treating cancer in a subject, comprising administering to the subject a population of cells comprising engineered T cells, wherein the engineered T cells comprise:
[0120] (i) a disrupted TRAC gene, wherein the disrupted TRAC gene comprises a nucleic acid encoding a CAR comprising the amino acid sequence set forth in SEQ ID NO: 46;
[0121] (ii) a disrupted .beta.2M gene; and
[0122] (iii) a disrupted CD70 gene,
[0123] thereby treating the cancer in the subject.
[0124] In some aspects, the disclosure provides a method of treating cancer in a subject, comprising administering to the subject a population of cells comprising engineered T cells, wherein the engineered T cells comprise:
[0125] (i) a disrupted TRAC gene, wherein the disrupted TRAC gene comprises a nucleic acid encoding a CAR, wherein the nucleic acid sequence is at least 90% identical to SEQ ID NO: 45;
[0126] (ii) a disrupted .beta.2M gene; and
[0127] (iii) a disrupted CD70 gene,
[0128] thereby treating the cancer in the subject. In some aspects, the disrupted TRAC gene comprises the nucleic acid sequence set forth in SEQ ID NO: 45.
[0129] In some aspects, the disclosure provides a method of treating cancer in a subject, comprising administering to the subject a population of cells comprising engineered T cells, wherein the engineered T cells comprise:
[0130] (i) a disrupted TRAC gene comprising a nucleic acid sequence at least 90% identical to SEQ ID NO: 44;
[0131] (ii) a disrupted .beta.2M gene; and
[0132] (iii) a disrupted CD70 gene,
[0133] thereby treating the cancer in the subject. In some aspects, the disrupted TRAC gene comprises the nucleic acid sequence set forth in SEQ ID NO: 44.
[0134] In any of the foregoing or related aspects, the engineered T cells are engineered human T cells. In some embodiments, the engineered T cells are engineered allogeneic T cells.
BRIEF DESCRIPTION OF THE DRAWINGS
[0135] FIG. 1 includes a graph showing highly efficient multiple gene editing in TRAC-/.beta.2M-/PD-1-/CD70- (quadruple knockout) T cells.
[0136] FIG. 2 includes a graph showing similar expansion among multigene-edited cells.
[0137] FIG. 3 includes graphs showing efficient multiple gene editing in TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ (i.e., 3.times. KO (CD70), CD70 CAR.sup.+) T cells.
[0138] FIG. 4 includes a graph showing that normal proportions of CD4+ and CD8+ T cells are maintained among the TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cell population.
[0139] FIG. 5 includes a graph showing efficient multiple gene editing in TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells.
[0140] FIG. 6 includes a graph showing that normal proportions of CD4+ and CD8+ T cells are maintained among the TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+(i.e., 4.times. KO, CD70 CAR.sup.+) T cell population.
[0141] FIGS. 7A-7C include graphs showing data relating to the characterization of anti-BCMA CAR+ T cells with multi-gene edits. Double knockout TRAC.sup.-/.beta.2M.sup.-/anti-BCMA CAR.sup.+ T cells and quadruple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-BCMA CAR.sup.+ T cells were stained for TRAC and .beta.2M (FIG. 7A), PD-1 and CD70 (FIG. 7B), and BCMA CAR (FIG. 7C) expression. The anti-BCMA CAR was expressed at approximately 80% in both the double and quadruple knockout CAR T cells.
[0142] FIG. 8 includes flow cytometry plots showing prevention of loss of CD4+ cells in 3.times. KO (TRAC-/.beta.2M-/CD70-) anti-CD33 CAR T cells compared 2.times. KO (TRAC-/.beta.2M-) anti-CD33 CAR T cells over three weeks.
[0143] FIG. 9 includes a graph showing CD70 KO enhanced cell proliferation in anti-CD33 CAR T cells over two weeks. The total number of viable cells was quantified in 3.times. KO (TRAC-/.beta.2M-/CD70-) and 2.times. KO (TRAC-/.beta.2M-) anti-CD33 CAR T cells.
[0144] FIG. 10 includes a graph showing CD70 KO enhanced cell proliferation in anti-CD19 CAR T cells over two weeks. The total number of viable cells was quantified in 3.times. KO (TRAC-/.beta.2M-/CD70-) and 2.times. KO (TRAC-/.beta.2M-) anti-CD33 CAR T cells.
[0145] FIG. 11 includes graphs showing CD70 KO enhanced cell proliferation in anti-BCMA CAR T cells and rescued the detrimental effect of PD1 KO on BCMA CAR cell proliferation. The total number of viable cells was quantified in 4.times. KO (TRAC-/.beta.2M-/CD70-/PD1-), 3.times. KO (CD70) (TRAC-/.beta.2M-/CD70-), 3.times. KO (PD1) (TRAC-/.beta.2M-/PD1-) and 2.times. KO (TRAC-/.beta.2M-) anti-CD33 CAR T cells.
[0146] FIG. 12 includes graphs showing CD70 KO enhanced cell proliferation in anti-BCMA CAR T cells and rescued the detrimental effect of PD1 KO on BCMA CAR cell proliferation. The total number of viable cells was quantified in 4.times. KO (TRAC-/.beta.2M-/CD70-/PD1-), 3.times. KO (CD70) (TRAC-/.beta.2M-CD70-), 3.times. KO (PD1) (TRAC-/.beta.2M-/PD1-) and 2.times. KO (TRAC-/.beta.2M-) anti-CD33 CAR T cells. The anti-BCMA CAR T cells were derived from a different donor T cells as the CAR T cells shown in FIG. 11.
[0147] FIG. 13 includes a graph showing a comparison of apoptotic cell death due to antigen exposure in 2.times. KO (TRAC-/.beta.2M-) anti-BCMA CAR+ T cells and 3.times. KO (TRAC-/.beta.2M-/CD70-) anti-BCMA CAR+ T cells. CAR+ T cells were exposed to plate-bound BCMA antigen for 24 hours with a re-challenge every 24 hours and apoptosis was assessed following each antigen challenge by flow cytometry. Induction of apoptosis due to antigen challenge was lower in anti-BCMA CAR+ T cells with a CD70 KO compared to those without.
[0148] FIG. 14 includes a graph showing a comparison of CAR T cell expansion following antigen exposure in 2.times. KO (TRAC-/.beta.2M-) anti-BCMA CAR+ T cells and 3.times. KO (TRAC-/.beta.2M-/CD70-) anti-BCMA CAR+ T cells. CAR+ T cells were exposed to plate-bound BCMA antigen for 24 hours with a re-challenge every 24 hours and cell expansion was assessed following each antigen challenge and normalized to the population at time 0 h. Population expansion following antigen challenge was higher in anti-BCMA CAR+ T cells with a CD70 KO compared to those without.
[0149] FIG. 15 includes a graph showing robust cell expansion in TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells. The total number of viable cells was quantified in 3.times. KO (TRAC-/.beta.2M-/CD70-) and 2.times. KO (TRAC-/.beta.2M-) anti-CD70 CAR T cells. 3.times. KO cells were generated with either CD70 sgRNA T7 or T8.
[0150] FIG. 16 includes a graph showing robust cell expansion of TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CA.sup.+ T cells. The total number of viable cells was quantified in 4.times. KO (TRAC-/.beta.2M-/PD1-/CD70-), 3.times. KO (TRAC-/.beta.2M-/PD1-) and 2.times. KO (TRAC-/.beta.2M-) anti-CD70 CAR T cells.
[0151] FIG. 17 includes graphs showing robust cell killing of both Nalm6 (top panel) cells and Raji (bottom panel) cells by anti-CD19 CAR T cells (TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD19 CAR.sup.+ or TRAC.sup.-/.beta.2M.sup.-/anti-CD19 CAR.sup.+ T cells).
[0152] FIG. 18 includes a graph showing robust cell killing of MV411 cells by anti-CD33 CAR T cells (TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD33 CAR.sup.+ or TRAC.sup.-/.beta.2M.sup.-/anti-CD33 CAR.sup.+ T cells).
[0153] FIG. 19 includes a graph showing robust cell killing of A498 cells by 3.times. KO (TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-) anti-CD70 CAR.sup.+ T cells compared to 2.times. KO (TRAC.sup.-/.beta.2M.sup.-) anti-CD70 CAR.sup.+ T cells.
[0154] FIG. 20 includes a graph showing cell expansion of 3.times. KO (TRAC-/.beta.2M-/CD70-) or 2.times. KO (TRAC-/.beta.2M-) anti-CD33 CAR T cells after challenge with MV411 target cells.
[0155] FIG. 21 includes a graph showing cell expansion of 3.times. KO (TRAC-/.beta.2M-/CD70-) or 2.times. KO (TRAC-/.beta.2M-) anti-CD70 CAR T cells after challenge with Nalm6 target cells.
[0156] FIG. 22A includes a graph showing A498 cell killing by anti-CD70 CAR T cells after serial rechallenge. 4.times. KO (TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/PD1.sup.-), 3.times. KO (CD70) (TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-), 3.times. KO (PD1) (TRAC.sup.-/.beta.2M.sup.-/PD1.sup.-) and 2.times. KO (TRAC.sup.-/.beta.2M.sup.-) anti-CD70 CAR+ T cells were utilized. 3.times. KO (CD70), CD70 CAR.sup.+ T cells, and 4.times. KO, CD70 CAR+ T cells were the most effective. FIG. 22B includes a graph showing ACHN cell killing by anti-CD70 CAR T cells after serial rechallenge. The same cells as FIG. 22A were utilized. 3.times. KO (CD70), CD70 CAR.sup.+ T cells and 4.times. KO, CD70 CAR.sup.+ T cells were the most effective. FIG. 23A includes a graph showing ACHN cell killing by anti-CD70 CAR T cells at various effector:target ratios. 4.times. KO (TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/PD1.sup.-), 3.times. KO (CD70) (TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-), 3.times. KO (PD1) (TRAC.sup.-/.beta.2M.sup.-/PD1.sup.-) and 2.times. KO (TRAC.sup.-/.beta.2M.sup.-) anti-CD70 CAR+ T cells were utilized. 3.times. KO (CD70), CD70 CAR.sup.+ T cells and 4.times. KO, CD70 CAR.sup.+ T cells were superior killers following multiple serial rechallenges. FIG. 23B includes a graph showing LAG3 (left) and PD1 (right) expression in the cells from FIG. 23A following eight rechallenges.
[0157] FIGS. 24A-24C include graphs showing that knockout of PD-1 and CD70 enhances cell killing activity of anti-BCMA CAR+ T cells as measured through serial rechallenges with a multiple myeloma cell line (MM.1S). Double knockout (2.times. KO (TRAC.sup.-/.beta.2M.sup.-) anti-BCMA CAR.sup.+ T cells (circles) began to lose their potency towards MM.1S cells after approximately 4 rechallenges, while quadruple knockout (4.times. KO (TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/PD1.sup.-) anti-BCMA CAR.sup.+ T cells (squares) were capable of killing 100% of the MM.1S cells after 10 rechallenges (FIG. 24A). Consistent with this, the quadruple knockout anti-BCMA CAR.sup.+ T cells continued to secrete IFN-g in response to target cells after 10 rechallenges, while the double knockout anti-BCMA CAR.sup.+ T cells showed reduced IFN-g secretion after the third rechallenge (FIG. 24B). The quadruple knockout anti-BCMA CAR.sup.+ T cells also showed higher proliferation in response to exposure to target cells than the double knockout anti-BCMA CAR.sup.+ T cells (FIG. 24C).
[0158] FIGS. 25A-25C include graphs showing highest cell kill activity in A498 PD-L1 kidney cancer cells (which overexpress PD-L1) using quadruple knockout (4.times. KO) TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells and triple knockout (3.times. KO (CD70)) TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells, relative to double knockout (2.times. KO) TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ T cells and triple knockout (3.times. KO (PD1) TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/anti-CD70 CAR.sup.+ T cells. A CAR T cell:A498-PD-L1 cell ratio of 2:1 was used in FIG. 25A, a CAR T cell:A498-PD-L1 cell ratio of 1:1 was used in FIG. 25B, and a CART cell:A498-PD-L1 cell ratio of 0.5:1 was used in FIG. 25C.
[0159] FIG. 26A and FIG. 26B include graphs showing that quadruple knockout (4.times. KO) TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells secrete the highest levels of cytokines IFN-gamma (FIG. 26A) and IL-2 (FIG. 26B), relative to triple knockout (3.times. KO (CD70) TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells, double knockout (2.times. KO) TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ T cells and triple knockout (3.times. KO (PD1) TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/anti-CD70 CAR.sup.+ T cells. A CART cell:A498-PD-L1 cell ratio of 1:1 was used.
[0160] FIG. 27A includes a graph showing results from an experiment designed to assess tumor volume reduction in a subcutaneous A498 renal cell carcinoma model exposed to: 2.times. KO (TRAC.sup.-/.beta.2M.sup.-), CD70 CAR.sup.+ T cells; 3.times. KO (PD-1) (TRAC.sup.-/.beta.2M.sup.-/PD1.sup.-), CD70 CAR.sup.+ T cells; 3.times. KO (CD70) (TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-), CD70 CAR.sup.+ T cells; or 4.times. KO (PD-1, CD70) (TRAC.sup.-/.beta.2M.sup.-/PD1.sup.-/CD70.sup.-), CD70 CAR.sup.+ T cells. FIG. 27B includes a graph showing results from an experiment designed to assess prevention of tumor growth in a subcutaneous A498 renal cell carcinoma rechallenge model. Mice from FIG. 27A were rechallenged with A498 tumor cells on day 25 and tumor volume was assessed over time. FIG. 27C includes a graph showing results from an experiment designed to assess tumor volume reduction in a subcutaneous A498 renal cell carcinoma model (large tumor of .about.150 mm3 at time of CAR-T injection) exposed to: 3.times. KO (CD70) (TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-), CD70 CAR.sup.+ T cells; 4.times. KO (PD-1, CD70) (TRAC.sup.-/.beta.2M.sup.-/PD1.sup.-/CD70.sup.-), CD70 CAR.sup.+ T cells; 2.times. KO (TRAC.sup.-/.beta.2M.sup.-), CD70 CAR.sup.+ T cells; or 3.times. KO (PD-1) (TRAC.sup.-/.beta.2M.sup.-/PD1.sup.-), CD70 CAR.sup.+ T cells.
[0161] FIG. 28A includes a graph showing tumor volume reduction in a subcutaneous MM.1S model exposed to: 2.times. KO (TRAC.sup.-/.beta.2M.sup.-), BCMA CAR.sup.+ T cells; 3.times. KO (PD-1) (TRAC.sup.-/.beta.2M.sup.-/PD1.sup.-), BCMA CAR.sup.+ T cells; 3.times. KO (CD70) (TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-), BCMA CAR.sup.+ T cells; or 4.times. KO (PD-1, CD70) (TRAC.sup.-/.beta.2M.sup.-/PD1.sup.-/CD70.sup.-), BCMA CAR.sup.+ T cells.
[0162] FIG. 28B includes a graph showing tumor volume reduction in a subcutaneous MM.1S model following a tumor cell re-challenge. Mice from FIG. 28A were re-challenged with a second inoculation of MM.1S cells on day 45 and tumor volume was assessed over time.
[0163] FIG. 29 includes graphs showing the number of human CD45.sup.+ 2.times. KO (TRAC.sup.-/.beta.2M.sup.-), BCMA CAR.sup.+ T cells; human CD45.sup.+ 3.times. KO (PD-1) (TRAC.sup.-/.beta.2M.sup.-/PD1.sup.-), BCMA CAR.sup.+ T cells; human CD45.sup.+ 3.times. KO (CD70) (TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-), BCMA CAR.sup.+ T cells; and human CD45.sup.+ 4.times. KO (PD-1, CD70) (TRAC.sup.-/.beta.2M.sup.-/PD1.sup.-/CD70.sup.-), BCMA CAR.sup.+ T cells 1 week (right graph), 2 weeks (middle graph), and 3 weeks (left graph) post dosing.
[0164] FIG. 30 includes graphs showing the results from an experiment designed to assess tumor volume reduction in a subcutaneous RPMI-8226 tumor xenograft model exposed to: TRAC.sup.-/.beta.2M/-anti-BCMA CAR.sup.+ T cells (2.times. KO, BCMA CAR.sup.+ T cells); TRAC.sup.-/.beta.2M.sup.-/PD1.sup.-/anti-BCMA CAR.sup.+ T cells (3.times. KO (PD-1), BCMA CAR.sup.+ T cells); TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-BCMA CAR.sup.+ T cells (3.times. KO (CD70), BCMA CAR.sup.+ T cells); or TRAC.sup.-/.beta.2M.sup.-/PD1.sup.-/CD70.sup.-/anti-BCMA CAR.sup.+ T cells (4.times. KO (PD-1, CD70), BCMA CAR.sup.+ T cells), at doses of 1.times.10.sup.5, 3.times.10.sup.5, 1.times.10.sup.6, or 3.times.10.sup.6 cells/mouse.
[0165] FIG. 31 includes a graph showing that TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cell maintain cytokine-dependent proliferation.
[0166] FIG. 32 shows cytokine-dependent growth of the TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells.
[0167] FIG. 33 includes a graph showing 4.times. KO (TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-), BCMA CAR.sup.+ T cells maintain cytokine dependency.
[0168] FIG. 34 includes a graph showing enhanced cytokine (IL-2) release by 3.times. KO (TRAC-/.beta.2M-/CD70-) anti-CD70 CAR+ T cells compared to 2.times. KO (TRAC-/.beta.2M-) anti-CD70 CAR+ T cells when co-cultured with A498 kidney cancer cells at various ratios for 24 hours.
[0169] FIG. 35 includes a graph showing robust cell killing of A498 cells by anti-CD70 CAR T cells (2.times. KO (TRAC.sup.-/.beta.2M.sup.-), CD70 CAR+; 3.times. KO (PD-1) (TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-), CD70 CAR+; and 4.times. KO (TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-) CD70 CAR.sup.+) relative to TCR+ T cells. T cells were co-cultured with A498 cells at various ratios for 24 hours and percentage of cell lysis was measured.
[0170] FIG. 36 includes a graph showing highest cell kill activity in A498 kidney cancer cells using quadruple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR+ T cells (4.times. KO, CD70 CAR+) and triple knockout TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR+ T cells (3.times. KO (CD70), CD70 CAR+), relative to double knockout TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ (i.e., 2.times. KO, CD70 CAR.sup.+) T cells and triple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/anti-CD70 CAR.sup.+ (i.e., 3.times. KO (PD-1), CD70 CAR.sup.+) T cells. A CAR T cell:A498 cell ratio of 0.25:1 was used. Percentage of cell lysis of A498 cells was measured 24 hours after co-culture.
[0171] FIGS. 37A and 37B include graphs showing that quadruple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells (4.times. KO, CD70 CAR+) and triple knockout TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells (3.times. KO (CD70), CD70 CAR+) secrete the highest levels of cytokines IFN-gamma (FIG. 37A) and IL-2 (FIG. 37B), relative to double knockout TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ T cells (2.times. KO, CD70 CAR+) and triple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/anti-CD70 CAR.sup.+ T cells (3.times. KO (PD-1), CD70 CAR+). A CAR T cell:A498 cell ratio of 0.25:1 was used. IFN-gamma and IL-2 secretion was measured 24 hours are co-culture.
[0172] FIG. 38 includes a graph showing that knocking out CD70 in anti-CD70 CAR T cells (3.times. KO (CD70) (TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-), CD70 CAR+; 3.times. KO (PD-1) (TRAC.sup.-/.beta.2M.sup.-/PD1.sup.-), CD70 CAR+; and 4.times. KO (TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/PD-1.sup.-), CD70 CAR+) decreased levels of PD-1 expression in CD4+ T cells relative to anti-CD70 CAR T cells expressing endogenous CD70 (2.times. KO (TRAC.sup.-/.beta.2M.sup.-) CD70 CAR+).
[0173] FIG. 39A and FIG. 39B include graphs showing that knocking out CD70 in anti-CD70 CAR T cells (3.times. KO (CD70) (TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-), CD70 CAR+; 3.times. KO (PD-1) (TRAC.sup.-/.beta.2M.sup.-/PD1.sup.-), CD70 CAR+; and 4.times. KO (TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/PD-1.sup.-), CD70 CAR+) decreased levels of exhaustion marker LAG3 in CD8+ T cells (FIG. 39A) and CD4+ T cells (FIG. 39B) relative to anti-CD70 CAR T cells expressing endogenous CD70 (2.times. KO (TRAC.sup.-/.beta.2M.sup.-) CD70 CAR+).
[0174] FIG. 40A includes graphs showing relative CD70 expression in five different cancer cell lines (left panel) and relative CD70 expression in three different cancel cell lines (right panel). FIG. 40B includes graphs showing relative CD70 expression in nine different cancer cell lines. FIGS. 40C-40D include graphs showing highest cell kill activity in ACHN (ATCC.RTM. CRL-1611.TM.) kidney cancer cells (which express low levels of CD70) using quadruple knockout TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells (4.times. KO, CD70 CAR+) and triple knockout TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells (3.times. KO (CD70), CD70 CAR+), relative to double knockout TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ T cells (2.times. KO, CD70 CAR+) and triple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/anti-CD70 CAR.sup.+ T cells (3.times. KO (PD-1), CD70 CAR+). A CAR T cell:ACHN cell ratio of 0.5:1 was used in FIG. 40C and a CAR T cell:ACHN cell ratio of 0.25:1 was used in FIG. 40D. FIG. 40E and FIG. 40F include graphs showing cell kill activity using quadruple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells (FIG. 40E) and triple knockout TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells (FIG. 40F) against additional solid tumor cell lines with varying levels of CD70 expression (4:1, 1:1, or 0.25:1 effector:target cell ratio). FIG. 40G includes a graph showing cell kill activity using the triple knockout TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells against solid tumor cell lines after a co-culture period of 24 hours or 96 hours. FIGS. 40H-40J include graphs showing cell kill activity using the triple knockout TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells (3KO (CD70), CD70 CAR+) against CD70-deficient chronic myelogenous leukemia (K562) cells (FIG. 40H), CD70-expressing multiple myeloma (MM.1S) cells (FIG. 40I), and CD70-expressing T cell lymphoma (HuT78) cells (FIG. 40J) at various effector:target ratios.
[0175] FIG. 41A and FIG. 41B include graphs showing that quadruple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells (4.times. KO, CD70 CAR+) and triple knockout TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells (3.times. KO (CD70), CD70 CAR+) secrete the highest levels of cytokines IFN-gamma (FIG. 41A) and IL-2 (FIG. 41B), relative to double knockout TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ T cells (2.times. KO, CD70 CAR+) and triple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/anti-CD70 CAR.sup.+ T cells (3.times. KO (PD-1), CD70 CAR+). A CAR T cell:ACHN cell ratio of 0.25:1 was used.
[0176] FIG. 42A includes a graph showing results from an experiment designed to assess tumor volume reduction in a human ovarian tumor xenograft model (e.g., SKOV-3 tumor cells) exposed to 3.times. KO (TRAC-/B2M-/CD70-) anti-CD70 CAR T cells. FIG. 42B includes a graph showing results from an experiment designed to assess tumor volume reduction in a human non-small cell lung tumor xenograft model (e.g., NCI-H1975 tumor cells) exposed to 3.times. KO (TRAC-/B2M-/CD70-) anti-CD70 CAR T cells. FIG. 42C includes a graph showing results from an experiment designed to assess tumor volume reduction in a human pancreatic tumor xenograft model (e.g., Hs766T tumor cells) exposed to 3.times. KO (TRAC-/B2M-/CD70-) anti-CD70 CAR T cells. FIG. 42D includes graphs showing results from an experiment designed to assess tumor volume reduction in a human T-cell lymphoma xenograft model (e.g., HuT78 tumor cells) exposed to 3.times. KO (TRAC-/B2M-/CD70-) anti-CD70 CAR T cells. Tumor volumes of individual mice (left) and mean tumor volumes (right) are shown.
DETAILED DESCRIPTION
[0177] The present disclosure is based, at least in part, on the discovery that disrupting the CD70 gene in immune cells engineered to express an antigen targeting moiety (e.g., a CAR) enhances several characteristics important for cell-based immunotherapy, including anti-tumor efficacy. Specifically, such engineered immune cells showed unexpected superior features, including extended proliferation and in vivo persistence resulting in long-term, enhanced anti-tumor efficacy. Notably, these unexpected features have been demonstrated with targeting moieties specific for various antigens, including BCMA, CD19, CD33 and CD70.
[0178] As demonstrated herein, disrupting the CD70 gene resulted in maintenance of cytotoxicity of immune cells engineered to express an antigen targeting moiety after multiple rounds of challenges by cancer cells in vitro. Without wishing to be bound by theory, this maintenance of cytotoxicity indicates disrupting the CD70 gene makes the engineered immune cells resistant to exhaustion and may result in cells that live longer.
[0179] It was also found that disrupting the CD70 gene in immune cells engineered to express an antigen targeting moiety enhanced anti-tumor efficacy against large tumors and induced a durable anti-cancer memory response. Specifically, the anti-cancer memory response prevented tumor growth upon re-challenge. Further, it has been demonstrated disrupting the CD70 gene results in enhanced cytotoxicity of immune cells engineered to express an antigen targeting moiety at lower ratios of engineered immune cells to target cells, indicating the potential efficacy of low doses of engineered immune cells.
[0180] It has also been shown disruption of the CD70 gene enhances cell proliferation and in vivo persistence of engineered immune cells. Without wishing to be bound by theory, it is believed the superior features of the engineered immune cells described herein allow for more consistent cell populations, larger scale production due to the cells' ability to survive more cell division, and fewer starting cells required to produce the engineered cells. Such features may also prove beneficial in a clinical setting. For example, increased expansion and decreased exhaustion indicates increased efficacy per dose and the ability to obtain efficacy with lower doses.
[0181] It has also been demonstrated that disrupting the CD70 gene in immune cells engineered to express an antigen targeting moiety maintains cytotoxicity against cancer cells expressing highly immune suppressive molecules, i.e., PD-L1. Without wishing to be bound by theory, it is believed the internal negative signal of PD-1 expressed on immune cells when bound to PD-L1 expressed on cancer cells, is overcome by disrupting CD70.
[0182] Accordingly, provided herein are methods and compositions (e.g., cell compositions) for the treatment of cancer, such as BCMA.sup.+, CD19.sup.+, CD33.sup.+, and CD70.sup.+ malignancies, involving the use of the engineered immune cells with increased efficacy and persistence.
CD70 Gene Edit
[0183] Cluster of Differentiation 70 (CD70) is a member of the tumor necrosis factor superfamily and its expression is restricted to activated T and B lymphocytes and mature dendritic cells. CD70 is implicated in tumor cell and regulatory T cell survival through interaction with its ligand, CD27. CD70 and its receptor CD27 have multiple roles in immune function in multiple cell types including T cells (activated and T regs), and B cells. It is unclear exactly how CD70 functions in all of these cell types to control functions such as apoptosis, with publications indicating contradicting roles. For example, it has been reported that CD70 induces apoptosis or survival of T cells depending on the antigenic load (Wensveen, F., et al. J. Immunol, Vol 188: 4256-4267, 2012).
[0184] While CAR T cells have proved to be an effective immunotherapeutic, various challenges remain. For example, over time CAR T cells become exhausted and become ineffective in vivo. With regards to manufacturing, it takes significant time to produce enough cells to dose a patient. To address these limitations, the present disclosure provides CAR T cells that have been engineered to disrupt endogenous CD70 expression while at the same time expressing an antigen targeting moiety (e.g., an scFv).
[0185] Surprisingly, the present disclosure shows disrupting the CD70 gene enables increased CAR T health and function (e.g., extended proliferation, reduced exhaustion) regardless of the antigen being targeted by the scFv in the CAR T. This applies even to antigens expressed on T cells such as CD33 and CD70 where the effects of the disrupted CD70 gene retain CAR T function even where fratricide may be expected. That is, these CD70 knockout cells (e.g., in which the CD70 gene has been edited using CRISPR/Cas9 gene editing technology), independent of the CAR insertion, exhibit continued, steady cell growth, relative to unmodified T cells (or edited T cells that express CD70) and express lower levels of exhaustion markers, such as LAGS. The CAR T cells of the present disclosure, may include any antibody (including whole antibodies and antibody fragments) or other molecule (e.g., receptor or ligand) that specifically binds to a cancer antigen to guide the CAR T cell to a cancer cell. In some embodiments, the antibody is an anti-CD70 antibody (e.g., an anti-CD70 scFv). In other embodiments, the antibody is an anti-CD19 antibody (e.g., an anti-CD19 scFv). In yet other embodiments, the antibody is an anti-BCMA antibody (e.g., an anti-BCMA scFv). In other embodiments, the antibody is an anti-CD33 antibody (e.g.,, an anti-CD33 scFv). Other cancer antigens are encompassed by the present disclosure.
[0186] It should be understood that gene disruption encompasses gene modification through gene editing (e.g., using CRISPR/Cas gene editing to insert or delete one or more nucleotides). In some embodiments, a disrupted gene is a gene that does not encode functional protein. In some embodiments, a cell that comprises a disrupted gene does not express (e.g., at the cell surface) a detectable level (e.g. by antibody, e.g., by flow cytometry) of the protein encoded by the gene. A cell that does not express a detectable level of the protein may be referred to as a knockout cell. For example, a cell having a CD70 gene edit may be considered a CD70 knockout cell if CD70 protein cannot be detected at the cell surface using an antibody that specifically binds CD70 protein.
[0187] Provided herein, in some embodiments, are populations of cells in which a certain percentage of the cells has been edited (e.g., CD70 gene edited), resulting in a certain percentage of cells not expressing a particular gene and/or protein. In some embodiments, at least 50% (e.g., 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, or 85%) of the cells of a gene-edited population of cells are CD70 knockout cells. In some embodiments, at least 50% of the cells (e.g. T cells) of the population do not express detectable levels of CD70 protein. In some embodiments, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, or at least 95% of the cells of a gene-edited population of cells may be CD70 knockout cells.
[0188] In some embodiments, 10%, 15%, 20%, 25%, 30%, 35% or 40% of the engineered T cells of a population do not express a detectable level of CD70 surface protein. In some embodiments, the percent of engineered T cells that do not express a detectable level of CD70 surface protein increases over time. Thus, in some embodiments, at least 50% of the engineered T cells of a population of engineered T cells does not express a detectable level of CD70 surface protein. For example, at least 55%, at least 60%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, or at least 95% of the engineered T cells of a population may not express a detectable level of CD70 surface protein. In some embodiments, 50%-100%, 50%-90%, 50%-80%, 50%-70%, 50%-60%, 60%-100%, 60%-90%, 60%-80%, 60%-70%, 70%-100%, 70%- 90%, 70%-80%, 80%-100%, 80%-90%, or 90%-100% of the engineered T cells of a population does not express a detectable level of CD70 surface protein.
[0189] Non-limiting examples of modified and unmodified CD70 gRNA sequences that may be used as provided herein to create a genomic alteration (e.g., disruption, e.g., deletion, insertion, substitution) in the CD70 gene are listed in Table 5 (e.g., SEQ ID NOS: 23-29 and 33-39). Other gRNA sequences may be designed using the CD70 gene sequence located on Chromosome 19 (GRCh38 coordinates: Chromosome 19: 6,583,183-6,604,103; Ensembl: ENSG00000125726). In certain embodiments, gRNAs targeting the CD70 genomic region create Indels (e.g.: insertions, deletions or substitutions) in, or around, the CD70 gene disrupting expression of the CD70 mRNA and/or protein.
[0190] In some embodiments, a ribonucleoprotein particle (RNP) containing an RNA-guided nuclease (e.g., a Cas nuclease, such as a Cas9 nuclease) and a gRNA targeting the CD70 gene (or any other gene of interest) are delivered to T cells (e.g., primary T cells). In other embodiments, the RNA-guided nuclease and gRNA are delivered separately to T cells. A ribonucleoprotein particle (RNP) is simply an RNA-guided nuclease (e.g., Cas9) pre-complexed/complexed with (bound to) a gRNA.
[0191] In some embodiments, the gRNA targeting the CD70 gene is a synthetic modified gRNA such as but not limited to any one of the gRNAs comprising SEQ ID NO: 33-39. In some embodiments, the gRNA targeting the CD70 gene is a synthetic unmodified gRNA such as but not limited to any one of the gRNAs comprising SEQ ID NO: 23-29.
[0192] In some embodiments, gRNAs targeting the CD70 genomic region and RNA-guided nuclease create double stranded breaks in the CD70 gene. Repair of the break results in Indels in the CD70 gene wherein the CD70 gene sequence may comprises a nucleotide sequence selected from the group consisting of: SEQ ID NOs: 129-134.
Multi-Gene Editing
[0193] The engineered T cells of the present disclosure, in some embodiments, include more than one disrupted gene (e.g.: more than one gene edit), for example, in more than one gene. For example, an engineered T cell may comprise a disrupted CD70 gene, a disrupted T cell receptor alpha chain constant region (TRAC) gene, a disrupted beta-2-microglobulin (.beta.2M) gene, a disrupted programmed cell death-1 (PD-1 or PDCD1) gene, or any combination of two or more of the foregoing disrupted genes. In some embodiments, an engineered T cell comprises a disrupted TRAC gene, a disrupted .beta.2M gene, and a disrupted CD70 gene. In some embodiments, an engineered T cell comprises a disrupted TRAC gene, a disrupted .beta.2M gene, and a disrupted PD-1 gene. In some embodiments, an engineered T cell comprises a disrupted TRAC gene, a disrupted .beta.2M gene, a disrupted CD70 gene and a disrupted PD-1 gene.
[0194] TRAC Gene Edit
[0195] In some embodiments, an engineered T cell comprises a disrupted TRAC gene. This disruption leads to loss of function of the TCR and renders the engineered T cell non-alloreactive and suitable for allogeneic transplantation, minimizing the risk of graft versus host disease. In some embodiments, expression of the endogenous TRAC gene is eliminated to prevent a graft-versus-host response.
[0196] In some embodiments, a disruption in the TRAC gene expression is created by knocking a chimeric antigen receptor (CAR) into the TRAC gene (e.g., using an adeno-associated viral (AAV) vector and donor template). In some embodiments, a disruption in the TRAC gene expression is created with a nuclease and gRNAs targeting the TRAC genomic region. In some embodiments, a genomic deletion in the TRAC gene is created by HDR, wherein a chimeric antigen receptor (CAR) replaces a segment of the TRAC gene (e.g., using an adeno-associated viral (AAV) vector and donor template). In some embodiments, a disruption in the TRAC gene expression is created with a nuclease and gRNAs targeting the TRAC genomic region, and knocking a chimeric antigen receptor (CAR) into the TRAC gene.
[0197] Non-limiting examples of modified and unmodified TRAC gRNA sequences that may be used as provided herein to create a genomic in the TRAC gene are listed in Table 7 (e.g., SEQ ID NOS: 30 and 40). See also International Application No. PCT/US2018/032334, filed May 11, 2018, incorporated herein by reference. Other gRNA sequences may be designed using the TRAC gene sequence located on chromosome 14 (GRCh38: chromosome 14: 22,547,506-22,552,154;. Ensembl; ENSG00000277734). In some embodiments, gRNAs targeting the TRAC genomic region and RNA-guided nuclease create breaks in the TRAC genomic region resulting Indels in the TRAC gene disrupting expression of the mRNA or protein.
[0198] In some embodiments, at least 50% of the engineered T cells of a population do not express a detectable level of T cell receptor (TCR) surface protein. For example, at least 55%, at least 60%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, or at least 95% of the engineered T cells of a population may not express a detectable level of TCR surface protein. In some embodiments, 50%-100%, 50%-90%, 50%-80%, 50%-70%, 50%-60%, 60%-100%, 60%-90%, 60%-80%, 60%-70%, 70%-100%, 70%-90%, 70%-80%, 80%-100%, 80%-90%, or 90%-100% of the engineered T cells of a population do not express a detectable level of TCR surface protein.
[0199] In some embodiments, a ribonucleoprotein particle (RNP) containing an RNA-guided nuclease (e.g., a Cas nuclease, such as a Cas9 nuclease) and a gRNA targeting the TRAC gene (or any other gene of interest) are delivered to T cells (e.g., primary T cells). In other embodiments, the RNA-guided nuclease and gRNA are delivered separately to T cells. A ribonucleoprotein particle (RNP) is simply an RNA-guided nuclease (e.g., Cas9) pre-complexed/complexed with a gRNA.
[0200] In some embodiments, gRNAs and RNA-guided nuclease targeting the TRAC genomic region result Indels in the TRAC gene comprising a nucleotide sequence selected from the following sequences in Table 1:
TABLE-US-00001 TABLE 1 SEQ ID Sequence NO: AAGAGCAACAAATCTGACT 1 AAGAGCAACAGTGCTGTGCCTGGAGCAACAAATCTGACT 2 AAGAGCAACAAATCTGACT AAGAGCAACAGTGCTGGAGCAACAAATCTGACT 3 AAGAGCAACAAATCTGACT AAGAGCAACAGTGCCTGGAGCAACAAATCTGACT 4 AAGAGCAACAAATCTGACT AAGAGCAACAGTGCTGACTAAGAGCAACAAATCTGACT 5 AAGAGCAACAGTGCTGTGGGCCTGGAGCAACAAATCTGACT 6 AAGAGCAACAAATCTGACT AAGAGCAACAGTGCTGGCCTGGAGCAACAAATCTGACT 7 AAGAGCAACAAATCTGACT AAGAGCAACAGTGCTGTGTGCCTGGAGCAACAAATCTGACT 8 AAGAGCAACAAATCTGACT
[0201] In some embodiments, an engineered T cell comprises a deletion in the TRAC gene relative to unmodified T cells. In some embodiments, an engineered T cell comprises a deletion of 15-30 base pairs in the TRAC gene relative to unmodified T cells. In some embodiments, an engineered T cell comprises a deletion of 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29 or 30 base pairs in the TRAC gene relative to unmodified T cells. In some embodiments, an engineered T cell comprises a deletion of more than 30 base pairs in the TRAC gene relative to unmodified T cells. In some embodiments, an engineered T cell comprises a deletion of 20 base pairs in the TRAC gene relative to unmodified T cells. In some embodiments, an engineered T cell comprises a deletion of SEQ ID NO: 86 in the TRAC gene relative to unmodified T cells. In some embodiments, an engineered T cell comprises a deletion comprising SEQ ID NO: 86 in the TRAC gene relative to unmodified T cells. In some embodiments, an engineered T cell comprises a deletion of SEQ ID NO: 118 in the TRAC gene relative to unmodified T cells. In some embodiments, an engineered T cell comprises a deletion comprising SEQ ID NO: 118 in the TRAC gene relative to unmodified T cells.
[0202] .beta.2M Gene Edit
[0203] In some embodiments, an engineered T cell comprises a disrupted .beta.2M gene. .beta.2M is a common (invariant) component of MHC I complexes. Disrupting its expression by gene editing will prevent host versus therapeutic allogeneic T cells responses leading to increased allogeneic T cell persistence. In some embodiments, expression of the endogenous .beta.2M gene is eliminated to prevent a host-versus-graft response.
[0204] Non-limiting examples of modified and unmodified .beta.2M gRNA sequences that may be used as provided herein to create a genomic deletion in the .beta.2M gene are listed in Table 7 (e.g., SEQ ID NOS: 31 and 41). See also International Application No. PCT/US2018/032334, filed May 11, 2018, incorporated herein by reference. Other gRNA sequences may be designed using the .beta.2M gene sequence located on Chromosome 15 (GRCh38 coordinates: Chromosome 15: 44,711,477-44,718,877; Ensembl: ENSG00000166710).
[0205] In some embodiments, gRNAs targeting the .beta.2M genomic region and RNA-guided nuclease create breaks in the .beta.2M genomic region resulting in Indels in the .beta.2M gene disrupting expression of the mRNA or protein.
[0206] In some embodiments, at least 50% of the engineered T cells of a population do not express a detectable level of .beta.2M surface protein. For example, at least 55%, at least 60%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, or at least 95% of the engineered T cells of a population may not express a detectable level of .beta.2M surface protein. In some embodiments, 50%-100%, 50%-90%, 50%-80%, 50%-70%, 50%-60%, 60%-100%, 60%-90%, 60%-80%, 60%-70%, 70%-100%, 70%-90%, 70%-80%, 80%-100%, 80%-90%, or 90%-100% of the engineered T cells of a population do not express a detectable level of .beta.2M surface protein.
[0207] In some embodiments, less than 50% of the engineered T cells of a population of cells express a detectable level of .beta.2M surface protein. In some embodiments, less than 30% of the engineered T cells of a population of cells express a detectable level of .beta.2M surface protein. For example, less than 50%, less than 30%, less than 25%, less than 20%, less than 15%, less than 10%, or less than 5% of the engineered T cells of a population of cells express a detectable level of .beta.2M surface protein. In some embodiments, 40%-30%, 40%-20%, 40% -10%, 40%-5%, 30%-20%, 30%-10%, 30%-5%, 20%-10%, 20%-5%, or 10%-5% of the engineered T cells of a population of cells express a detectable level of .beta.2M surface protein.
[0208] In some embodiments, a ribonucleoprotein particle (RNP) containing an RNA-guided nuclease (e.g., a Cas nuclease, such as a Cas9 nuclease) and a gRNA targeting the B2M gene (or any other gene of interest) are delivered to T cells (e.g., primary T cells). In other embodiments, the RNA-guided nuclease and gRNA are delivered separately to T cells. A ribonucleoprotein particle (RNP) is simply a RNA-guided nuclease (e.g., Cas9) pre-complexed/complexed with a gRNA.
[0209] In some embodiments, an edited .beta.2M gene comprises a nucleotide sequence selected from the following sequences in Table 2.
TABLE-US-00002 TABLE 2 SEQ ID Sequences NO: CGTGGCCTTAGCTGTGCTCGCGCTACTCTCTCTTTCTGCCTGGA 9 GGCTATCCAGCGTGAGTCTCTCCTACCCTCCCGCT CGTGGCCTTAGCTGTGCTCGCGCTACTCTCTCTTTCGCCTGGAG 10 GCTATCCAGCGTGAGTCTCTCCTACCCTCCCGCT CGTGGCCTTAGCTGTGCTCGCGCTACTCTCTCTTTCTGGAGGCT 11 ATCCAGCGTGAGTCTCTCCTACCCTCCCGCT CGTGGCCTTAGCTGTGCTCGCGCTACTCTCTCTTTCTGGATAGC 12 CTGGAGGCTATCCAGCGTGAGTCTCTCCTACCCTCCCGCT CGTGGCCTTAGCTGTGCTCGCGCTATCCAGCGTGAGTCTCTCCT 13 ACCCTCCCGCT CGTGGCCTTAGCTGTGCTCGCGCTACTCTCTCTTTCTGTGGCCT 14 GGAGGCTATCCAGCGTGAGTCTCTCCTACCCTCCCGCT
[0210] PD-1 Gene Edit
[0211] PD-1 is an immune checkpoint molecule that is upregulated in activated T cells and serves to dampen or stop T cell responses. Disrupting PD-1 by gene editing could lead to more persistent and/or potent therapeutic T cell responses and/or reduce immune suppression in a subject. In some embodiments, an engineered T cell comprises a disrupted PD-1 gene. In some embodiments, expression of the endogenous PD-1 gene is eliminated to enhance anti-tumor efficacy of the CAR T cells of the present disclosure.
[0212] Non-limiting examples of modified and unmodified PD-1 gRNA sequences that may be used as provided herein to create a genomic deletion in the PD-1 gene are listed in Table 5 (e.g., SEQ ID NOS: 32 and 42). See also International Application No. PCT/US2018/032334, filed May 11, 2018, incorporated herein by reference. Other gRNA sequences may be designed using the PD-1 gene sequence located on Chromosome 2 (GRCh38 coordinates: Chromosome 2: 241,849,881-241,858,908; Ensembl: ENSG00000188389).
[0213] In some embodiments, gRNAs targeting and RNA-guided nuclease the PD-1 genomic region create breaks in the TRAC genomic region resulting in Indels in the PD-1 gene disrupting expression of the PD-1 mRNA or protein.
[0214] In some embodiments, at least 50% of the engineered T cells of a population do not express a detectable level of PD-1 surface protein. For example, at least 55%, at least 60%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, or at least 95% of the engineered T cells of a population may not express a detectable level of PD-1 surface protein. In some embodiments, 50%-100%, 50%-90%, 50%-80%, 50%-70%, 50%-60%, 60%-100%, 60%-90%, 60%-80%, 60%-70%, 70%-100%, 70%-90%, 70%-80%, 80%-100%, 80%-90%, or 90%-100% of the engineered T cells of a population do not express a detectable level of PD-1 surface protein.
[0215] In some embodiments, a ribonucleoprotein particle (RNP) containing an RNA-guided nuclease (e.g., a Cas nuclease, such as a Cas9 nuclease) and a gRNA targeting the PD-1 gene (or any other gene of interest) are delivered to T cells (e.g., primary T cells). In other embodiments, the RNA-guided nuclease and gRNA are delivered separately to T cells. A ribonucleoprotein particle (RNP) is simply an RNA-guided nuclease (e.g., Cas9) pre-complexed/complexed with a gRNA.
Cellular Phenotypes
[0216] In some embodiments, one or more gene edits within a population of cells results in a phenotype associated with changes in cellular proliferative capacity, cellular exhaustion, cellular viability, cellular lysis capability (e.g., increase cytokine production and/or release), or any combination thereof.
[0217] In some embodiments, engineered T cells of a population comprise a CAR that includes an anti-CD70 scFv ectodomain. In some embodiments, engineered T cells of a population comprise a CAR that includes an anti-BCMA scFv ectodomain. In some embodiments, engineered T cells of a population comprise a CAR that includes an anti-CD19 scFv ectodomain. In some embodiments, engineered T cells of a population comprise a CAR that includes an anti-CD33 scFv ectodomain. Any of the foregoing engineered T cells may also comprise a disruption in one or more of the following genes: TRAC, .beta.2M, PD-1, and/or CD70 (e.g., TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-; TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-; or TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-).
[0218] In some embodiments, engineered T cells of the present disclosure exhibit increased cellular proliferative capacity relative to control cells. In some embodiments, engineered T cells of the present disclosure exhibit at least 20% greater cellular proliferative capacity, relative to control T cells. For example, engineered T cells (e.g., TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-; TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-; or TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-; with or without a CAR) may exhibit at least 25%, at least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, or at least 90% greater cellular proliferative capacity, relative to control T cells. In some embodiments, engineered T cells of the present disclosure exhibit 20%-100%, 20%-90%, 20%-80%, 20%-70%, 20%-60%, 20%-50%, 30%-100%, 30%-90%, 30%-80%, 30%-70%, 30%-60%, 30%-50%, 40%-100%, 40%-90%, 40%-80%, 40%-70%, 40%-60%, 40%-50%, 50%-100%, 50%-90%, 50%-80%, 50%-70%, or 50%-60% greater cellular proliferative capacity, relative to control T cells. Methods of measuring cell proliferation are known to those of skill in the art and described herein.
[0219] In some embodiments, engineered T cells of the present disclosure exhibit reduced exhaustion, relative to control T cells. For example, the engineered T cells may express reduced levels of LAG3 (or other exhaustion markers), relative to control T cells. In some embodiments, the levels of LAG3 expression are reduced by at least 20%, relative to control T cells. For example, the levels of LAG3 expression may be reduced by at least 25%, at least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, or at least 90%, relative to control T cells. In some embodiments, the levels of LAG3 expression are reduced by 20%-100%, 20%-90%, 20%-80%, 20%-70%, 20%-60%, 20%-50%, 30%-100%, 30%-90%, 30%-80%, 30%-70%, 30%-60%, 30%-50%, 40%-100%, 40%-90%, 40%-80%, 40%-70%, 40%-60%, 40%-50%, 50%-100%, 50%-90%, 50%-80%, 50%-70%, or 50%-60%, relative to control T cells. In some embodiments, reduced exhaustion is determined by measuring decreased surface expression of exhaustion markers, including TIGIT, PD-1, LAG-3 or combinations thereof. Methods for measuring surface expression are known to those of skill in the art and described herein.
[0220] In some embodiments, engineered T cells of the present disclosure exhibit increased cellular viability relative to control cells. In some embodiments, engineered T cells of the present disclosure exhibit an at least 20% increase in cellular viability, relative to control cells. For example, engineered T cells of the present disclosure may exhibit at least 25%, at least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, or at least 90% increase in cellular viability, relative to control cells. In some embodiments, engineered T cells of the present disclosure exhibit a 20%-100%, 20%-90%, 20%-80%, 20%-70%, 20%-60%, 20%-50%, 30%-100%, 30%-90%, 30%-80%, 30%-70%, 30%-60%, 30%-50%, 40%-100%, 40%-90%, 40%-80%, 40%-70%, 40%-60%, 40%-50%, 50%-100%, 50%-90%, 50%-80%, 50%-70%, or 50%-60% increase in cellular viability, relative to control cells. Methods of measuring cell viability are known to those of skill in the art and described herein.
[0221] In some embodiments, engineered T cells of the present disclosure exhibit increased cellular lysis capability relative to control cells. In some embodiments, engineered T cells of the present disclosure exhibit an at least 20% increase in cellular lysis capability (kill at least 20% more target cells), relative to control cells. For example, engineered T cells of the present disclosure may exhibit an at least at least 25%, at least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, or at least 90% increase in cellular lysis capability, relative to control cells. In some embodiments, engineered T cells of the present disclosure exhibit a 20%-100%, 20%-90%, 20%-80%, 20%-70%, 20%-60%, 20%-50%, 30%-100%, 30%-90%, 30%-80%, 30%-70%, 30%-60%, 30%-50%, 40%-100%, 40%-90%, 40%-80%, 40%-70%, 40%-60%, 40%-50%, 50%-100%, 50%-90%, 50%-80%, 50%-70%, or 50%-60% increase in cellular lysis capability, relative to control cells.
[0222] In some embodiments, engineered T cells of the present disclosure exhibit increased cytokine secretion relative to control cells. For example, in some embodiments the level of cytokines (e.g., IL-2 and/or IFN-gamma) secreted by the engineered T cells is at least 2-fold (e.g., at least 3-fold, at least 4-fold, or at least 5-fold) greater than the level of cytokines secreted by control T cells.
[0223] Control T cells, in some embodiments, are engineered T cells (e.g., gene edited T cells) that express endogenous CD70 protein (CD70 normally expressed by T cells). In some embodiments, control T cells are engineered T cells that express endogenous CD70 protein and comprise a TRAC gene disrupted by insertion of a nucleic acid encoding a CAR (e.g., an anti-CD70 CAR or anti-BCMA CAR), a disrupted .beta.2M gene, a disrupted PD-1 gene, or any combination of the foregoing disrupted genes. In some embodiments, control T cells are unedited T cells.
[0224] Surprisingly, the multi-gene edited CAR T cells of the present disclosure (e.g., TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.- cells) maintain cytotoxicity (ability to kill cancer cells), following multiple challenges (also referred to as rechallenges(s)) with cancer cells. In some embodiments, the engineered T cells maintain cytotoxicity following at least 1 rechallenge with a target cell, wherein the target cell expresses an antigen recognized by the CAR T cells. In some embodiments, the engineered T cells maintain cytotoxicity following at least 2 rechallenges with a target cell, wherein the target cell expresses an antigen recognized by the CAR T cells. In some embodiments, the engineered T cells maintain cytotoxicity following at least 1 rechallenge with a cancer cell. In some embodiments, the engineered T cells maintain cytotoxicity following at least 2 rechallenges with a cancer cell. In some embodiments, the engineered T cells maintain cytotoxicity following 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 rechallenges with a target cell, wherein the target cell expresses an antigen recognized by the CAR T cells. In some embodiments, the engineered T cells maintain cytotoxicity following 2, 3, 4, 5, 6, 7, 8, 9, or 10 rechallenges with a target cell, wherein the target cell expresses an antigen recognized by the CAR T cells. In some embodiments, the engineered T cells maintain cytotoxicity following 2, 3, 4, 5, 6, 7, 8, 9, or 10 rechallenges with a cancer cell. In some embodiments, the engineered T cells maintain cytotoxicity following 10 or more rechallenges with a a target cell, wherein the target cell expresses an antigen recognized by the CAR T cells. In some embodiments, the engineered T cells maintain cytotoxicity following 10 or more rechallenges with a cancer cell. In some embodiments, the engineered T cells express a CAR specific for CD70 and the target cell (e.g., cancer cell) expresses CD70. In some embodiments, the engineered T cells express a CAR specific for CD19 and the target cell (e.g., cancer cell) expresses CD19. In some embodiments, the engineered T cells express a CAR specific for CD33 and the target cell (e.g., cancer cell) expresses CD33. In some embodiments, the engineered T cells express a CAR specific for BCMA and the target cell (e.g., cancer cell) expresses BCMA.
Gene Editing Methods
[0225] Gene editing (including genomic editing) is a type of genetic engineering in which nucleotide(s)/nucleic acid(s) is/are inserted, deleted, and/or substituted in a DNA sequence, such as in the genome of a targeted cell. Targeted gene editing enables insertion, deletion, and/or substitution at pre-selected sites in the genome of a targeted cell (e.g., in a targeted gene or targeted DNA sequence). When a sequence of an endogenous gene is edited, for example by deletion, insertion or substitution of nucleotide(s)/nucleic acid(s), the endogenous gene comprising the affected sequence may be knocked-out or knocked-down due to the sequence alteration. Therefore, targeted editing may be used to disrupt endogenous gene expression. "Targeted integration" refers to a process involving insertion of one or more exogenous sequences, with or without deletion of an endogenous sequence at the insertion site. Targeted integration can result from targeted gene editing when a donor template containing an exogenous sequence is present. As used herein, a "disrupted gene" refers to a gene comprising an insertion, deletion or substitution relative to an endogenous gene such that expression of a functional protein from the endogenous gene is reduced or inhibited. As used herein, "disrupting a gene" refers to a method of inserting, deleting or substituting at least one nucleotide/nucleic acid in an endogenous gene such that expression of a functional protein from the endogenous gene is reduced or inhibited. Methods of disrupting a gene are known to those of skill in the art and described herein.
[0226] Targeted editing can be achieved either through a nuclease-independent approach, or through a nuclease-dependent approach. In the nuclease-independent targeted editing approach, homologous recombination is guided by homologous sequences flanking an exogenous polynucleotide to be introduced into an endogenous sequence through the enzymatic machinery of the host cell. The exogenous polynucleotide may introduce deletions, insertions or replacement of nucleotides in the endogenous sequence.
[0227] Alternatively, the nuclease-dependent approach can achieve targeted editing with higher frequency through the specific introduction of double strand breaks (DSBs) by specific rare-cutting nucleases (e.g., endonucleases). Such nuclease-dependent targeted editing also utilizes DNA repair mechanisms, for example, non-homologous end joining (NHEJ), which occurs in response to DSBs. DNA repair by NHEJ often leads to random insertions or deletions (indels) of a small number of endogenous nucleotides. In contrast to NHEJ mediated repair, repair can also occur by a homology directed repair (HDR). When a donor template containing exogenous genetic material flanked by a pair of homology arms is present, the exogenous genetic material can be introduced into the genome by HDR, which results in targeted integration of the exogenous genetic material.
[0228] Available endonucleases capable of introducing specific and targeted DSBs include, but not limited to, zinc-finger nucleases (ZFN), transcription activator-like effector nucleases (TALEN), and RNA-guided CRISPR-Cas9 nuclease (CRISPR/Cas9; Clustered Regular Interspaced Short Palindromic Repeats Associated 9). Additionally, DICE (dual integrase cassette exchange) system utilizing phiC31 and Bxb1 integrases may also be used for targeted integration.
[0229] ZFNs are targeted nucleases comprising a nuclease fused to a zinc finger DNA binding domain (ZFBD), which is a polypeptide domain that binds DNA in a sequence-specific manner through one or more zinc fingers. A zinc finger is a domain of about 30 amino acids within the zinc finger binding domain whose structure is stabilized through coordination of a zinc ion. Examples of zinc fingers include, but not limited to, C2H2 zinc fingers, C3H zinc fingers, and C4 zinc fingers. A designed zinc finger domain is a domain not occurring in nature whose design/composition results principally from rational criteria, e.g., application of substitution rules and computerized algorithms for processing information in a database storing information of existing ZFP designs and binding data. See, for example, U.S. Pat. Nos. 6,140,081; 6,453,242; and 6,534,261; see also WO 98/53058; WO 98/53059; WO 98/53060; WO 02/016536 and WO 03/016496. A selected zinc finger domain is a domain not found in nature whose production results primarily from an empirical process such as phage display, interaction trap or hybrid selection. ZFNs are described in greater detail in U.S. Pat. Nos. 7,888,121 and 7,972,854. The most recognized example of a ZFN is a fusion of the Fokl nuclease with a zinc finger DNA binding domain.
[0230] A TALEN is a targeted nuclease comprising a nuclease fused to a TAL effector DNA binding domain. A "transcription activator-like effector DNA binding domain", "TAL effector DNA binding domain", or "TALE DNA binding domain" is a polypeptide domain of TAL effector proteins that is responsible for binding of the TAL effector protein to DNA. TAL effector proteins are secreted by plant pathogens of the genus Xanthomonas during infection. These proteins enter the nucleus of the plant cell, bind effector-specific DNA sequences via their DNA binding domain, and activate gene transcription at these sequences via their transactivation domains. TAL effector DNA binding domain specificity depends on an effector-variable number of imperfect 34 amino acid repeats, which comprise polymorphisms at select repeat positions called repeat variable-diresidues (RVD). TALENs are described in greater detail in US Patent Application No. 2011/0145940. The most recognized example of a TALEN in the art is a fusion polypeptide of the Fokl nuclease to a TAL effector DNA binding domain.
[0231] Additional examples of targeted nucleases suitable for use as provided herein include, but are not limited to, Bxbl, phiC31, R4, PhiBT1, and W.beta./SPBc/TP901-1, whether used individually or in combination.
[0232] Other non-limiting examples of targeted nucleases include naturally-occurring and recombinant nucleases, e.g., CRISPR/Cas9, restriction endonucleases, meganucleases homing endonucleases, and the like.
CRISPR-Cas9 Gene Editing
[0233] The CRISPR-Cas9 system is a naturally-occurring defense mechanism in prokaryotes that has been repurposed as an RNA-guided DNA-targeting platform used for gene editing. It relies on the DNA nuclease Cas9, and two noncoding RNAs, crisprRNA (crRNA) and trans-activating RNA (tracrRNA), to target the cleavage of DNA. CRISPR is an abbreviation for Clustered Regularly Interspaced Short Palindromic Repeats, a family of DNA sequences found in the genomes of bacteria and archaea that contain fragments of DNA (spacer DNA) with similarity to foreign DNA previously exposed to the cell, for example, by viruses that have infected or attacked the prokaryote. These fragments of DNA are used by the prokaryote to detect and destroy similar foreign DNA upon re-introduction, for example, from similar viruses during subsequent attacks. Transcription of the CRISPR locus results in the formation of an RNA molecule comprising the spacer sequence, which associates with and targets Cas (CRISPR-associated) proteins able to recognize and cut the foreign, exogenous DNA. Numerous types and classes of CRISPR/Cas systems have been described (see e.g., Koonin et al., (2017) Curr Opin Microbiol 37:67-78).
[0234] crRNA drives sequence recognition and specificity of the CRISPR-Cas9 complex through Watson-Crick base pairing typically with a 20 nucleotide (nt) sequence in the target DNA. Changing the sequence of the 5' 20nt in the crRNA allows targeting of the CRISPR-Cas9 complex to specific loci. The CRISPR-Cas9 complex only binds DNA sequences that contain a sequence match to the first 20 nt of the crRNA, single-guide RNA (sgRNA), if the target sequence is followed by a specific short DNA motif (with the sequence NGG) referred to as a protospacer adjacent motif (PAM).
[0235] TracrRNA hybridizes with the 3' end of crRNA to form an RNA-duplex structure that is bound by the Cas9 endonuclease to form the catalytically active CRISPR-Cas9 complex, which can then cleave the target DNA.
[0236] Once the CRISPR-Cas9 complex is bound to DNA at a target site, two independent nuclease domains within the Cas9 enzyme each cleave one of the DNA strands upstream of the PAM site, leaving a double-strand break (DSB) where both strands of the DNA terminate in a base pair (a blunt end).
[0237] After binding of CRISPR-Cas9 complex to DNA at a specific target site and formation of the site-specific DSB, the next key step is repair of the DSB. Cells use two main DNA repair pathways to repair the DSB: non-homologous end-joining (NHEJ) and homology-directed repair (HDR).
[0238] NHEJ is a robust repair mechanism that appears highly active in the majority of cell types, including non-dividing cells. NHEJ is error-prone and can often result in the removal or addition of between one and several hundred nucleotides at the site of the DSB, though such modifications are typically <20 nt. The resulting insertions and deletions (indels) can disrupt coding or noncoding regions of genes. Alternatively, HDR uses a long stretch of homologous donor DNA, provided endogenously or exogenously, to repair the DSB with high fidelity. HDR is active only in dividing cells, and occurs at a relatively low frequency in most cell types. In many embodiments of the present disclosure, NHEJ is utilized as the repair operant.
[0239] In some embodiments, the Cas9 (CRISPR associated protein 9) endonuclease is from Streptococcus pyogenes, although other Cas9 homologs may be used. It should be understood, that wild-type Cas9 may be used or modified versions of Cas9 may be used (e.g., evolved versions of Cas9, or Cas9 orthologues or variants), as provided herein. In some embodiments, Cas9 may be substituted with another RNA-guided endonuclease, such as Cpf1 (of a class II CRISPR/Cas system).
[0240] In some embodiments, the CRISPR/Cas system comprise components derived from a Type-I, Type-II, or Type-III system. Updated classification schemes for CRISPR/Cas loci define Class 1 and Class 2 CRISPR/Cas systems, having Types Ito V or VI (Makarova et al., (2015) Nat Rev Microbiol, 13(11):722-36; Shmakov et al., (2015) Mol Cell, 60:385-397). Class 2 CRISPR/Cas systems have single protein effectors. Cas proteins of Types II, V, and VI are single-protein, RNA-guided endonucleases, herein called "Class 2 Cas nucleases." Class 2 Cas nucleases include, for example, Cas9, Cpf1, C2c1, C2c2, and C2c3 proteins. The Cpf1 nuclease (Zetsche et al., (2015) Cell 163:1-13) is homologous to Cas9, and contains a RuvC-like nuclease domain.
[0241] In some embodiments, the Cas nuclease is from a Type-II CRISPR/Cas system (e.g., a Cas9 protein from a CRISPR/Cas9 system). In some embodiments, the Cas nuclease is from a Class 2 CRISPR/Cas system (a single-protein Cas nuclease such as a Cas9 protein or a Cpf1 protein). The Cas9 and Cpf1 family of proteins are enzymes with DNA endonuclease activity, and they can be directed to cleave a desired nucleic acid target by designing an appropriate guide RNA, as described further herein.
[0242] In some embodiments, a Cas nuclease may comprise more than one nuclease domain. For example, a Cas9 nuclease may comprise at least one RuvC-like nuclease domain (e.g. Cpf1) and at least one HNH-like nuclease domain (e.g. Cas9). In some embodiments, the Cas9 nuclease introduces a DSB in the target sequence. In some embodiments, the Cas9 nuclease is modified to contain only one functional nuclease domain. For example, the Cas9 nuclease is modified such that one of the nuclease domains is mutated or fully or partially deleted to reduce its nucleic acid cleavage activity. In some embodiments, the Cas9 nuclease is modified to contain no functional RuvC-like nuclease domain. In other embodiments, the Cas9 nuclease is modified to contain no functional HNH-like nuclease domain. In some embodiments in which only one of the nuclease domains is functional, the Cas9 nuclease is a nickase that is capable of introducing a single-stranded break (a "nick") into the target sequence. In some embodiments, a conserved amino acid within a Cas9 nuclease nuclease domain is substituted to reduce or alter a nuclease activity. In some embodiments, the Cas nuclease nickase comprises an amino acid substitution in the RuvC-like nuclease domain. Exemplary amino acid substitutions in the RuvC-like nuclease domain include D10A (based on the S. pyogenes Cas9 nuclease). In some embodiments, the nickase comprises an amino acid substitution in the HNH-like nuclease domain. Exemplary amino acid substitutions in the HNH-like nuclease domain include E762A, H840A, N863A, H983A, and D986A (based on the S. pyogenes Cas9 nuclease).
[0243] In some embodiments, the Cas nuclease is from a Type-I CRISPR/Cas system. In some embodiments, the Cas nuclease is a component of the Cascade complex of a Type-I CRISPR/Cas system. For example, the Cas nuclease is a Cas3 nuclease. In some embodiments, the Cas nuclease is derived from a Type-III CRISPR/Cas system. In some embodiments, the Cas nuclease is derived from Type-IV CRISPR/Cas system. In some embodiments, the Cas nuclease is derived from a Type-V CRISPR/Cas system. In some embodiments, the Cas nuclease is derived from a Type-VI CRISPR/Cas system.
[0244] Guide RNAs
[0245] The present disclosure provides a genome-targeting nucleic acid that can direct the activities of an associated polypeptide (e.g., a site-directed polypeptide) to a specific target sequence within a target nucleic acid. The genome-targeting nucleic acid can be an RNA. A genome-targeting RNA is referred to as a "guide RNA" or "gRNA" herein. A guide RNA comprises at least a spacer sequence that hybridizes to a target nucleic acid sequence of interest, and a CRISPR repeat sequence. In Type II systems, the gRNA also comprises a second RNA called the tracrRNA sequence. In the Type II gRNA, the CRISPR repeat sequence and tracrRNA sequence hybridize to each other to form a duplex. In the Type V gRNA, the crRNA forms a duplex. In both systems, the duplex binds a site-directed polypeptide, such that the guide RNA and site-direct polypeptide form a complex. In some embodiments, the genome-targeting nucleic acid provides target specificity to the complex by virtue of its association with the site-directed polypeptide. The genome-targeting nucleic acid thus directs the activity of the site-directed polypeptide.
[0246] As is understood by the person of ordinary skill in the art, each guide RNA is designed to include a spacer sequence complementary to its genomic target sequence. See Jinek et al., Science, 337, 816-821 (2012) and Deltcheva et al., Nature, 471, 602-607 (2011).
[0247] In some embodiments, the genome-targeting nucleic acid (e.g., gRNA) is a double-molecule guide RNA. In some embodiments, the genome-targeting nucleic acid (e.g., gRNA) is a single-molecule guide RNA.
[0248] A double-molecule guide RNA comprises two strands of RNA. The first strand comprises in the 5' to 3' direction, an optional spacer extension sequence, a spacer sequence and a minimum CRISPR repeat sequence. The second strand comprises a minimum tracrRNA sequence (complementary to the minimum CRISPR repeat sequence), a 3' tracrRNA sequence and an optional tracrRNA extension sequence.
[0249] A single-molecule guide RNA (referred to as a "sgRNA") in a Type II system comprises, in the 5' to 3' direction, an optional spacer extension sequence, a spacer sequence, a minimum
[0250] CRISPR repeat sequence, a single-molecule guide linker, a minimum tracrRNA sequence, a 3' tracrRNA sequence and an optional tracrRNA extension sequence. The optional tracrRNA extension may comprise elements that contribute additional functionality (e.g., stability) to the guide RNA. The single-molecule guide linker links the minimum CRISPR repeat and the minimum tracrRNA sequence to form a hairpin structure. The optional tracrRNA extension comprises one or more hairpins.
[0251] A single-molecule guide RNA in a Type V system comprises, in the 5' to 3' direction, a minimum CRISPR repeat sequence and a spacer sequence.
[0252] In some embodiments, the sgRNA comprises a 20 nucleotide spacer sequence at the 5' end of the sgRNA sequence. In some embodiments, the sgRNA comprises a less than 20 nucleotide spacer sequence at the 5' end of the sgRNA sequence. In some embodiments, the sgRNA comprises a more than 20 nucleotide spacer sequence at the 5' end of the sgRNA sequence. In some embodiments, the sgRNA comprises a variable length spacer sequence with 17-30 nucleotides at the 5' end of the sgRNA sequence (see Table 3).
[0253] In some embodiments, the sgRNA comprises comprise no uracil at the 3' end of the sgRNA sequence. In some embodiments, the sgRNA comprises comprise one or more uracil at the 3' end of the sgRNA sequence. For example, the sgRNA can comprise 1 uracil (U) at the 3' end of the sgRNA sequence. The sgRNA can comprise 2 uracil (UU) at the 3' end of the sgRNA sequence. The sgRNA can comprise 3 uracil (UUU) at the 3' end of the sgRNA sequence. The sgRNA can comprise 4 uracil (UUUU) at the 3' end of the sgRNA sequence. The sgRNA can comprise 5 uracil (UUUUU) at the 3' end of the sgRNA sequence. The sgRNA can comprise 6 uracil (UUUUUU) at the 3' end of the sgRNA sequence. The sgRNA can comprise 7 uracil (UUUUUUU) at the 3' end of the sgRNA sequence. The sgRNA can comprise 8 uracil (UUUUUUUU) at the 3' end of the sgRNA sequence.
[0254] The sgRNA can be unmodified or modified. For example, modified sgRNAs can comprise one or more 2'-O-methyl phosphorothioate nucleotides.
TABLE-US-00003 TABLE 3 SEQ ID NO. sgRNA sequence 15 nnnnnnnnnnnnnnnnnnnnguuuuagagcuagaaauagcaagu uaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccga gucggugcuuuu 16 nnnnnnnnnnnnnnnnnnnnguuuuagagcuagaaauagcaagu uaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccga gucggugc 17 n.sub.(17-30)guuuuagagcuagaaauagcaaguuaaaauaaggcuagu ccguuaucaacuugaaaaaguggcaccgagucggugcu.sub.(1-8)
[0255] By way of illustration, guide RNAs used in the CRISPR/Cas/Cpf1 system, or other smaller RNAs can be readily synthesized by chemical means, as illustrated below and described in the art. While chemical synthetic procedures are continually expanding, purifications of such RNAs by procedures such as high performance liquid chromatography (HPLC, which avoids the use of gels such as PAGE) tends to become more challenging as polynucleotide lengths increase significantly beyond a hundred or so nucleotides. One approach used for generating RNAs of greater length is to produce two or more molecules that are ligated together. Much longer RNAs, such as those encoding a Cas9 or Cpf1 endonuclease, are more readily generated enzymatically. Various types of RNA modifications can be introduced during or after chemical synthesis and/or enzymatic generation of RNAs, e.g., modifications that enhance stability, reduce the likelihood or degree of innate immune response, and/or enhance other attributes, as described in the art.
[0256] In some embodiments, indel frequency (editing frequency) may be determined using a TIDE analysis, which can be used to identify highly efficient gRNA molecules. In some embodiments, a highly efficient gRNA yields a gene editing frequency of higher than 80%. For example, a gRNA is considered to be highly efficient if it yields a gene editing frequency of at least 80%, at least 85%, at least 90%, at least 95%, or 100%.
[0257] In some embodiments, gene disruption may occur by deletion of a genomic sequence using two guide RNAs. Methods of using CRISPR-Cas gene editing technology to create a genomic deletion in a cell (e.g., to knock out a gene in a cell) are known (Bauer D E et al. Vis. Exp. 2015; 95;e52118).
[0258] Spacer Sequence
[0259] In some embodiments, a gRNA comprises a spacer sequence. A spacer sequence is a sequence (e.g., a 20 nucleotide sequence) that defines the target sequence (e.g., a DNA target sequences, such as a genomic target sequence) of a target nucleic acid of interest. In some embodiments, the spacer sequence is 15 to 30 nucleotides. In some embodiments, the spacer sequence is 15, 16, 17, 18, 19, 29, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30 nucleotides. In some embodiments, a spacer sequence is 20 nucleotides.
[0260] The "target sequence" is adjacent to a PAM sequence and is the sequence modified by an RNA-guided nuclease (e.g., Cas9). The "target nucleic acid" is a double-stranded molecule: one strand comprises the target sequence and is referred to as the "PAM strand," and the other complementary strand is referred to as the "non-PAM strand." One of skill in the art recognizes that the gRNA spacer sequence hybridizes to the reverse complement of the target sequence, which is located in the non-PAM strand of the target nucleic acid of interest. Thus, the gRNA spacer sequence is the RNA equivalent of the target sequence. For example, if the target sequence is 5'-AGAGCAACAGTGCTGTGGCC-3' (SEQ ID NO: 86), then the gRNA spacer sequence is 5'-AGAGCAACAGUGCUGUGGCC-3' (SEQ ID NO: 98). The spacer of a gRNA interacts with a target nucleic acid of interest in a sequence-specific manner via hybridization (i.e., base pairing). The nucleotide sequence of the spacer thus varies depending on the target sequence of the target nucleic acid of interest.
[0261] In a CRISPR/Cas system herein, the spacer sequence is designed to hybridize to a region of the target nucleic acid that is located 5' of a PAM of the Cas9 enzyme used in the system. The spacer may perfectly match the target sequence or may have mismatches. Each Cas9 enzyme has a particular PAM sequence that it recognizes in a target DNA. For example, S. pyogenes recognizes in a target nucleic acid a PAM that comprises the sequence 5'-NRG-3', where R comprises either A or G, where N is any nucleotide and N is immediately 3' of the target nucleic acid sequence targeted by the spacer sequence.
[0262] In some embodiments, the target nucleic acid sequence comprises 20 nucleotides. In some embodiments, the target nucleic acid comprises less than 20 nucleotides. In some embodiments, the target nucleic acid comprises more than 20 nucleotides. In some embodiments, the target nucleic acid comprises at least: 5, 10, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 30 or more nucleotides. In some embodiments, the target nucleic acid comprises at most: 5, 10, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 30 or more nucleotides. In some embodiments, the target nucleic acid sequence comprises 20 bases immediately 5' of the first nucleotide of the PAM. For example, in a sequence comprising 5'-NNNNNNNNNNNNNNNNNNNNNRG-3', the target nucleic acid comprises the sequence that corresponds to the Ns, wherein N is any nucleotide, and the underlined NRG sequence is the S. pyogenes PAM.
[0263] Non-limiting examples of gRNAs that may be used as provided herein are provided in PCT/IB2018/001619, filed May 11, 2018, herein incorporated by this reference.
Methods of Making gRNAs
[0264] The gRNAs of the present disclosure are produced by a suitable means available in the art, including but not limited to in vitro transcription (IVT), synthetic and/or chemical synthesis methods, or a combination thereof. Enzymatic (IVT), solid-phase, liquid-phase, combined synthetic methods, small region synthesis, and ligation methods are utilized. In one embodiment, the gRNAs are made using IVT enzymatic synthesis methods. Methods of making polynucleotides by IVT are known in the art and are described in International Application PCT/US2013/30062. Accordingly, the present disclosure also includes polynucleotides, e.g., DNA, constructs and vectors are used to in vitro transcribe a gRNA described herein.
[0265] In some embodiments, non-natural modified nucleobases are introduced into polynucleotides, e.g., gRNA, during synthesis or post-synthesis. In certain embodiments, modifications are on internucleoside linkages, purine or pyrimidine bases, or sugar. In some embodiments, a modification is introduced at the terminal of a polynucleotide; with chemical synthesis or with a polymerase enzyme. Examples of modified nucleic acids and their synthesis are disclosed in PCT application No. PCT/US2012/058519. Synthesis of modified polynucleotides is also described in Verma and Eckstein, Annual Review of Biochemistry, vol. 76, 99-134 (1998).
[0266] In some embodiments, enzymatic or chemical ligation methods are used to conjugate polynucleotides or their regions with different functional moieties, such as targeting or delivery agents, fluorescent labels, liquids, nanoparticles, etc. Conjugates of polynucleotides and modified polynucleotides are reviewed in Goodchild, Bioconjugate Chemistry, vol. 1(3), 165-187 (1990).
[0267] Certain embodiments of the invention also provide nucleic acids, e.g., vectors, encoding gRNAs described herein. In some embodiments, the nucleic acid is a DNA molecule. In other embodiments, the nucleic acid is an RNA molecule. In some embodiments, the nucleic acid comprises a nucleotide sequence encoding a crRNA. In some embodiments, the nucleotide sequence encoding the crRNA comprises a spacer flanked by all or a portion of a repeat sequence from a naturally-occurring CRISPR/Cas system. In some embodiments, the nucleic acid comprises a nucleotide sequence encoding a tracrRNA. In some embodiments, the crRNA and the tracrRNA is encoded by two separate nucleic acids. In other embodiments, the crRNA and the tracrRNA is encoded by a single nucleic acid. In some embodiments, the crRNA and the tracrRNA is encoded by opposite strands of a single nucleic acid. In other embodiments, the crRNA and the tracrRNA is encoded by the same strand of a single nucleic acid.
[0268] In some embodiments, the gRNAs provided by the disclosure are chemically synthesized by any means described in the art (see e.g., WO/2005/01248). While chemical synthetic procedures are continually expanding, purifications of such RNAs by procedures such as high performance liquid chromatography (HPLC, which avoids the use of gels such as PAGE) tends to become more challenging as polynucleotide lengths increase significantly beyond a hundred or so nucleotides. One approach used for generating RNAs of greater length is to produce two or more molecules that are ligated together.
[0269] In some embodiments, the gRNAs provided by the disclosure are synthesized by enzymatic methods (e.g., in vitro transcription, IVT).
[0270] Various types of RNA modifications can be introduced during or after chemical synthesis and/or enzymatic generation of RNAs, e.g., modifications that enhance stability, reduce the likelihood or degree of innate immune response, and/or enhance other attributes, as described in the art.
[0271] In certain embodiments, more than one guide RNA can be used with a CRISPR/Cas nuclease system. Each guide RNA may contain a different targeting sequence, such that the CRISPR/Cas system cleaves more than one target nucleic acid. In some embodiments, one or more guide RNAs may have the same or differing properties such as activity or stability within the Cas9 RNP complex. Where more than one guide RNA is used, each guide RNA can be encoded on the same or on different vectors. The promoters used to drive expression of the more than one guide RNA is the same or different.
[0272] The guide RNA may target any sequence of interest via the targeting sequence (e.g., spacer sequence) of the crRNA. In some embodiments, the degree of complementarity between the targeting sequence of the guide RNA and the target sequence on the target nucleic acid molecule is about 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 97%, 98%, 99%, or 100%. In some embodiments, the targeting sequence of the guide RNA and the target sequence on the target nucleic acid molecule is 100% complementary. In other embodiments, the targeting sequence of the guide RNA and the target sequence on the target nucleic acid molecule may contain at least one mismatch. For example, the targeting sequence of the guide RNA and the target sequence on the target nucleic acid molecule may contain 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 mismatches. In some embodiments, the targeting sequence of the guide RNA and the target sequence on the target nucleic acid molecule may contain 1-6 mismatches. In some embodiments, the targeting sequence of the guide RNA and the target sequence on the target nucleic acid molecule may contain 5 or 6 mismatches.
[0273] The length of the targeting sequence may depend on the CRISPR/Cas9 system and components used. For example, different Cas9 proteins from different bacterial species have varying optimal targeting sequence lengths. Accordingly, the targeting sequence may comprise 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 35, 40, 45, 50, or more than 50 nucleotides in length. In some embodiments, the targeting sequence may comprise 18-24 nucleotides in length. In some embodiments, the targeting sequence may comprise 19-21 nucleotides in length. In some embodiments, the targeting sequence may comprise 20 nucleotides in length.
[0274] In some embodiments of the present disclosure, a CRISPR/Cas nuclease system includes at least one guide RNA. In some embodiments, the guide RNA and the Cas protein may form a ribonucleoprotein (RNP), e.g., a CRISPR/Cas complex. The guide RNA may guide the Cas protein to a target sequence on a target nucleic acid molecule (e.g., a genomic DNA molecule), where the the Cas protein cleaves the target nucleic acid. In some embodiments, the CRISPR/Cas complex is a Cpf1 /guide RNA complex. In some embodiments, the CRISPR complex is a Type-II CRISPR/Cas9 complex. In some embodiments, the Cas protein is a Cas9 protein. In some embodiments, the CRISPR/Cas9 complex is a Cas9/guide RNA complex.
[0275] Delivery of guide RNA and Nuclease
[0276] In some embodiments, a gRNA and an RNA-guided nuclease are delivered to a cell separately, either simultaneously or sequentially. In some embodiments, a gRNA and an RNA-guided nuclease are delivered to a cell together. In some embodiments, a gRNA and an RNA-guided nuclease are pre-complexed together to form a ribonucleoprotein (RNP).
[0277] RNPs are useful for gene editing, at least because they minimize the risk of promiscuous interactions in a nucleic acid-rich cellular environment and protect the RNA from degradation. Methods for forming RNPs are known in the art. In some embodiments, an RNP containing an RNA-guided nuclease (e.g., a Cas nuclease, such as a Cas9 nuclease) and a gRNA targeting a gene of interest is delivered a cell (e.g.: a T cell). In some embodiments, an RNP is delivered to a T cell by electroporation.
[0278] As used herein, a "TRAC targeting RNP" refers to a gRNA that targets the TRAC gene pre-complexed with an RNA-guided nuclease. As used herein, a ".beta.2M targeting RNP" refers to a gRNA that targets the .beta.2M gene pre-complexed with an RNA-guided nuclease. As used herein, a "CD70 targeting RNP" refers to a gRNA that targets the CD70 gene pre-complexed with an RNA-guided nuclease. As used herein, a "PD-1 targeting RNP" refers to a gRNA that targets the PD-1 gene pre-complexed with an RNA-guided nuclease.
[0279] In some embodiments, a TRAC targeting RNP is delivered to a cell. In some embodiments, a .beta.2M targeting RNP is delivered to a cell. In some embodiments, a CD70 targeting RNP is delivered to a cell. In some embodiments, a PD-1 targeting RNP is delivered to a cell.
[0280] In some embodiments, more than one RNP is delivered to a cell. In some embodiments, more than on RNP is delivered to a cell separately. In some embodiments, more than one RNP is delivered to a cell simultaneously. In some embodiments, at least one of the following RNPs is delivered to a cell:
[0281] (i) a TRAC targeting RNP;
[0282] (ii) a .beta.2M targeting RNP;
[0283] (iii) a CD70 targeting RNP; or
[0284] (iv) a PD-1 targeting RNP. In some embodiments, at least two of the following RNPs are delivered to a cell:
[0285] (i) a TRAC targeting RNP;
[0286] (ii) a .beta.2M targeting RNP;
[0287] (iii) a CD70 targeting RNP; or
[0288] (iv) a PD-1 targeting RNP.
[0289] In some embodiments, an RNA-guided nuclease is delivered to a cell in a DNA vector that expresses the RNA-guided nuclease, an RNA that encodes the RNA-guided nuclease, or a protein. In some embodiments, a gRNA targeting a gene is delivered to a cell as an RNA, or a DNA vector that expresses the gRNA.
[0290] Delivery of an RNA-guided nuclease, gRNA, and/or an RNP may be through direct injection or cell transfection using known methods, for example, electroporation or chemical transfection. Other cell transfection methods may be used.
Chimeric antigen receptor (CAR) T cells
[0291] A chimeric antigen receptor refers to an artificial immune cell receptor that is engineered to recognize and bind to an antigen expressed by tumor cells. Generally, a CAR is designed for a T cell and is a chimera of a signaling domain of the T-cell receptor (TCR) complex and an antigen-recognizing domain (e.g., a single chain fragment (scFv) of an antibody or other antibody fragment) (Enblad et al., Human Gene Therapy. 2015; 26(8):498-505). A T cell that expresses a CAR is referred to as a CAR T cell. CARs have the ability to redirect T-cell specificity and reactivity toward a selected target in a non-MHC-restricted manner. The non-MHC-restricted antigen recognition gives T-cells expressing CARs the ability to recognize an antigen independent of antigen processing, thus bypassing a major mechanism of tumor escape. Moreover, when expressed in T-cells, CARs advantageously do not dimerize with endogenous T-cell receptor (TCR) alpha and beta chains. CARs are often referenced to by the antigen they bind. For example, a "CD19 CAR", a "CD70 CAR", a "CD33 CAR" and a "BCMA CAR" are CARs comprising antigen binding domains that specifically bind to CD19, CD70, CD33 or BCMA, respectively. Accordingly, such terms are interchangeable with anti-CD19 CAR, anti-CD70 CAR, anti-CD33 CAR and anti-BCMA CAR. It will be understood by those of ordinary skill in the art that a CAR that specifically binds an antigen can be referred to with either terminology.
[0292] There are four generations of CARs, each of which contains different components. First generation CARs join an antibody-derived scFv to the CD3zeta or z) intracellular signaling domain of the T-cell receptor through hinge and transmembrane domains. Second generation CARs incorporate an additional domain, e.g., CD28, 4-1BB (41BB), or ICOS, to supply a costimulatory signal. Third-generation CARs contain two costimulatory domains fused with the TCR CD3.zeta. chain. Third-generation costimulatory domains may include, e.g., a combination of CD3.zeta., CD27, CD28, 4-1BB, ICOS, or OX40. CARs, in some embodiments, contain an ectodomain, commonly derived from a single chain variable fragment (scFv), a hinge, a transmembrane domain, and an endodomain with one (first generation), two (second generation), or three (third generation) signaling domains derived from CD3Z and/or co-stimulatory molecules (Maude et al., Blood. 2015; 125(26):4017-4023; Kakarla and Gottschalk, Cancer J. 2014; 20(2):151-155).
[0293] CARs typically differ in their functional properties. The CD3.zeta. signaling domain of the T-cell receptor, when engaged, will activate and induce proliferation of T-cells but can lead to anergy (a lack of reaction by the body's defense mechanisms, resulting in direct induction of peripheral lymphocyte tolerance). Lymphocytes are considered anergic when they fail to respond to a specific antigen. The addition of a costimulatory domain in second-generation CARs improved replicative capacity and persistence of modified T-cells. Similar antitumor effects are observed in vitro with CD28 or 4-1BB CARs, but preclinical in vivo studies suggest that 4-1BB CARs may produce superior proliferation and/or persistence. Clinical trials suggest that both of these second-generation CARs are capable of inducing substantial T-cell proliferation in vivo, but CARs containing the 4-1BB costimulatory domain appear to persist longer. Third generation CARs combine multiple signaling domains (costimulatory) to augment potency.
[0294] In some embodiments, a chimeric antigen receptor is a first generation CAR. In other embodiments, a chimeric antigen receptor is a second generation CAR. In yet other embodiments, a chimeric antigen receptor is a third generation CAR.
[0295] A CAR, in some embodiments, comprises an extracellular (ecto) domain comprising an antigen binding domain (e.g., an antibody, such as an scFv), a transmembrane domain, and a cytoplasmic (endo) domain.
[0296] Ectodomain
[0297] The ectodomain is the region of the CAR that is exposed to the extracellular fluid and, in some embodiments, includes an antigen binding domain, and optionally a signal peptide, a spacer domain, and/or a hinge domain. In some embodiments, the antigen binding domain is a single-chain variable fragment (scFv) that includes the VL and VH of immunoglobulins connected with a short linker peptide. The linker, in some embodiments, includes hydrophilic residues with stretches of glycine and serine for flexibility as well as stretches of glutamate and lysine for added solubility. A single-chain variable fragment (scFv) is not actually a fragment of an antibody, but instead is a fusion protein of the variable regions of the heavy (VH) and light chains (VL) of immunoglobulins, connected with a short linker peptide of ten to about 25 amino acids. The linker is usually rich in glycine for flexibility, as well as serine or threonine for solubility, and can either connect the N-terminus of the VH with the C-terminus of the VL, or vice versa. This protein retains the specificity of the original immunoglobulin, despite removal of the constant regions and the introduction of the linker. In some embodiments, the scFv of the present disclosure is humanized. In other embodiments, the scFv is fully human. In yet other embodiments, the scFv is a chimera (e.g., of mouse and human sequence).
[0298] In some embodiments, the scFv is an anti-CD70 scFv (binds specifically to CD70). Non-limiting examples of anti-CD70 scFv proteins that may be used as provided herein may include the amino acid sequence of SEQ ID NO: 48 or SEQ ID NO: 50.
[0299] In some embodiments, the scFv is an anti-BCMA scFv (binds specifically to BCMA). Non-limiting examples of anti-BCMA scFv proteins that may be used as provided herein may include the amino acid sequence of SEQ ID NO: 59.
[0300] In some embodiments, the scFv is an anti-CD19 scFv (binds specifically to CD19). Non-limiting examples of anti-CD19 scFv proteins that may be used as provided herein may include the amino acid sequence of SEQ ID NO: 151.
[0301] In some embodiments, the scFv is an anti-CD33 scFv (binds specifically to CD33). Non-limiting examples of anti-CD33 scFv proteins that may be used as provided herein may include the amino acid sequence of SEQ ID NO: 137.
[0302] Other scFv proteins may be used.
[0303] The signal peptide can enhance the antigen specificity of CAR binding. Signal peptides can be derived from antibodies, such as, but not limited to, CD8, as well as epitope tags such as, but not limited to, GST or FLAG. Examples of signal peptides include MLLLVTSLLLCELPHPAFLLIP (SEQ ID NO: 88) and MALPVTALLLPLALLLHAARP (SEQ ID NO: 89). Other signal peptides may be used.
[0304] In some embodiments, a spacer domain or hinge domain is located between an extracellular domain (comprising the antigen binding domain) and a transmembrane domain of a CAR, or between a cytoplasmic domain and a transmembrane domain of the CAR. A spacer domain is any oligopeptide or polypeptide that functions to link the transmembrane domain to the extracellular domain and/or the cytoplasmic domain in the polypeptide chain. A hinge domain is any oligopeptide or polypeptide that functions to provide flexibility to the CAR, or domains thereof, or to prevent steric hindrance of the CAR, or domains thereof. In some embodiments, a spacer domain or a hinge domain may comprise up to 300 amino acids (e.g., 10 to 100 amino acids, or 5 to 20 amino acids). In some embodiments, one or more spacer domain(s) may be included in other regions of a CAR. In some embodiments, the hinge domain is a CD8 hinge domain. Other hinge domains may be used.
[0305] Transmembrane Domain
[0306] The transmembrane domain is a hydrophobic alpha helix that spans the membrane. The transmembrane domain provides stability of the CAR. In some embodiments, the transmembrane domain of a CAR as provided herein is a CD8 transmembrane domain. In other embodiments, the transmembrane domain is a CD28 transmembrane domain. In yet other embodiments, the transmembrane domain is a chimera of a CD8 and CD28 transmembrane domain. Other transmembrane domains may be used as provided herein. In some embodiments, the transmembrane domain is a CD8a transmembrane domain: FVPVFLPAKPTTTPAPRPPTPAPTIAS QPLS LRPEACRPAAGG AVHTRGLDFACDIYIWAPLAGTCGVLLLSLVITLYCNHRNR (SEQ ID NO: 90). Other transmembrane domains may be used.
[0307] In some embodiments, the transmembrane domain is a CD8a transmembrane domain comprising the amino acid sequence: IYIWAPLAGTCGVLLLSLVITLY (SEQ ID NO: 126).
[0308] Endodomain
[0309] The endodomain is the functional end of the receptor. Following antigen recognition, receptors cluster and a signal is transmitted to the cell. The most commonly used endodomain component is CD3-zeta, which contains three (3) immunoreceptor tyrosine-based activation motif (ITAM)s. This transmits an activation signal to the T cell after the antigen is bound. In many cases, CD3-zeta may not provide a fully competent activation signal and, thus, a co-stimulatory signaling is used. For example, CD28 and/or 4-1BB may be used with CD3-zeta (CD3.zeta.) to transmit a proliferative/survival signal. Thus, in some embodiments, the co-stimulatory molecule of a CAR as provided herein is a CD28 co-stimulatory molecule. In other embodiments, the co-stimulatory molecule is a 4-1BB co-stimulatory molecule. In some embodiments, a CAR includes CD3.zeta. and CD28. In other embodiments, a CAR includes CD3-zeta and 4-1BB. In still other embodiments, a CAR includes CD3.zeta., CD28, and 4-1BB. Table 4 provides examples of signaling domains derived from 4-1BB, CD28 and CD3-zeta that may be used herein.
TABLE-US-00004 TABLE 4 Name Sequence SEQ ID NO: 4-1BB AAACGGGGCAGAAAGAAACTCCTGTATATATTCAAACAACCATT 18 TATGAGACCAGTACAAACTACTCAAGAGGAAGATGGCTGTAGCT GCCGATTTCCAGAAGAAGAAGAAGGAGGATGTGAACTG KRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCEL 19 CD28 TCAAAGCGGAGTAGGTTGTTGCATTCCGATTACATGAATATGACT 121 CCTCGCCGGCCTGGGCCGACAAGAAAACATTACCAACCCTATGC CCCCCCACGAGACTTCGCTGCGTACAGGTCC SKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRS 20 CD3-zeta CGAGTGAAGTTTTCCCGAAGCGCAGACGCTCCGGCATATCAGCA 21 AGGACAGAATCAGCTGTATAACGAACTGAATTTGGGACGCCGCG AGGAGTATGACGTGCTTGATAAACGCCGGGGGAGAGACCCGGAA ATGGGGGGTAAACCCCGAAGAAAGAATCCCCAAGAAGGACTCTA CAATGAACTCCAGAAGGATAAGATGGCGGAGGCCTACTCAGAAA TAGGTATGAAGGGCGAACGACGACGGGGAAAAGGTCACGATGG CCTCTACCAAGGGTTGAGTACGGCAACCAAAGATACGTACGATG CACTGCATATGCAGGCCCTGCCTCCCAGA RVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEM 22 GGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGL YQGLSTATKDTYDALHMQALPPR
Cancer Antigens
[0310] CD70
[0311] In some embodiments, the T cells of the present disclosure are engineered with a chimeric antigen receptor (CAR) designed to target CD70. CD70 was initially identified as the ligand for CD27, a co-stimulatory receptor involved in T cell proliferation and survival. CD70 is only found on a small percentage of activated T cells and antigen presenting cells in draining lymph nodes during viral infection. Many human tumors also express CD70 including, but not limited to, solid cancers such as clear cell renal cancer, breast cancer, gastric cancer, ovarian cancer, glioblastoma, and hematological malignancies. Due to its restricted expression pattern on normal tissues and overexpression in numerous cancers, CD70 is an attractive therapeutic target.
[0312] Thus, in some embodiments, T cells of the present disclosure are engineered to express a CAR comprising an anti-CD70 antibody (e.g., anti-CD70 scFv). In some embodiments, the anti-CD70 antibody is an anti-CD70 scFv encoded by the sequence of SEQ ID NO: 47 or 49. In some embodiments, the anti-CD70 antibody is an anti-CD70 scFv comprising the sequence of SEQ ID NO: 48 or 50. In some embodiments, the anti-CD70 antibody is an anti-CD70 scFv comprising a VH comprising the sequence of SEQ ID NO: 51. In some embodiments, the anti-CD70 antibody is an anti-CD70 scFv comprising a VL comprising the sequence of SEQ ID NO: 52. In some embodiments, a CAR comprising an anti-CD70 antibody is encoded by the sequence of SEQ ID NO: 45. In some embodiments, a CAR comprising an anti-CD70 antibody comprises the sequence of SEQ ID NO: 46.
[0313] In some embodiments, the anti-CD70 antibody is an anti-CD70 scFv encoded by a nucleotide sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 47 or 49. In some embodiments, the anti-CD70 antibody is an anti-CD70 scFv comprising an amino acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 48 or 50. In some embodiments, the anti-CD70 antibody is an anti-CD70 scFv comprising a VH comprising an amino acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 51. In some embodiments, the anti-CD70 antibody is an anti-CD70 scFv comprising a VL comprising an amino acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 52. In some embodiments, a CAR comprising an anti-CD70 antibody is encoded by a nucleotide sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 45. In some embodiments, a CAR comprising an anti-CD70 antibody comprises an amino acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 46.
[0314] BCMA
[0315] In some embodiments, the T cells of the present disclosure are engineered with a CAR designed to target BCMA. B-cell maturation antigen (BCMA, CD269) is a member of the tumor necrosis factor receptor (TNF) superfamily. BCMA binds B-cell activating factor (BAFF) and a proliferation inducing ligand (APRIL). Among nonmalignant cells, BCMA is expressed primarily by plasma cells and subsets of mature B cells. BCMA is selectively expressed by B-lineage cells including multiple myeloma cells and non-Hodgkin's lymphoma, thus BCMA is also an attractive therapeutic target.
[0316] Thus, in some embodiments, T cells of the present disclosure are engineered to express a CAR comprising an anti-BCMA antibody (e.g., anti-BCMA scFv). In some embodiments, the anti-BCMA antibody is an anti-BCMA scFv encoded by the sequence of SEQ ID NO: 58. In some embodiments, the anti-BCMA antibody is an anti-BCMA scFv comprising the sequence of SEQ ID NO: 59. In some embodiments, the anti-BCMA antibody is an anti-BCMA scFv comprising a VH comprising the sequence of SEQ ID NO: 60. In some embodiments, the anti-BCMA antibody is an anti-BCMA scFv comprising a VL comprising the sequence of SEQ ID NO: 61. In some embodiments, a CAR comprising an anti-BCMA antibody is encoded by the sequence of SEQ ID NO: 56. In some embodiments, a CAR comprising an anti-BCMA antibody comprises the sequence of SEQ ID NO: 57.
[0317] In some embodiments, the anti-BCMA antibody is an anti-BCMA scFv encoded by a nucleotide sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 58. In some embodiments, the anti-BCMA antibody is an anti-BCMA scFv comprising an amino acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 59. In some embodiments, the anti-BCMA antibody is an anti-BCMA scFv comprising a VH comprising an amino acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 60. In some embodiments, the anti-BCMA antibody is an anti-BCMA scFv comprising a VL comprising an amino acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 61. In some embodiments, a CAR comprising an anti-BCMA antibody is encoded by a nucleotide sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 56. In some embodiments, a CAR comprising an anti-BCMA antibody comprises an amino acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 57.
[0318] CD19
[0319] In some embodiments, the T cells of the present disclosure are engineered with a CAR designed to target CD19. Cluster of Differentiation 19 (CD19) is an antigenic determinant detectable on leukemia precursor cells. The human and murine amino acid and nucleic acid sequences can be found in a public database, such as GenBank, UniProt and Swiss-Prot. For example, the amino acid sequence of human CD19 can be found as UniProt/Swiss-Prot Accession No. P15391 and the nucleotide sequence encoding of the human CD19 can be found at Accession No. NM--001178098. CD19 is expressed on most B lineage cancers, including, e.g., acute lymphoblastic leukemia, chronic lymphocyte leukemia and non-Hodgkin's lymphoma. It is also an early marker of B cell progenitors. See, e.g., Nicholson et al. Mol. Immun. 34 (16-17): 1157-1165 (1997).
[0320] Thus, in some embodiments, T cells of the present disclosure are engineered to express a CAR comprising an anti-CD19 antibody (e.g., anti-CD19 scFv). In some embodiments, the anti-CD19 antibody is an anti-CD19 scFv encoded by the sequence of SEQ ID NO: 150. In some embodiments, the anti-CD19 antibody is an anti-CD19 scFv comprising the sequence of SEQ ID NO: 151. In some embodiments, the anti-CD19 antibody is an anti-CD19 scFv comprising a VH comprising the sequence of SEQ ID NO: 152. In some embodiments, the anti-CD19 antibody is an anti-CD19 scFv comprising a VL comprising the sequence of SEQ ID NO: 153. In some embodiments, a CAR comprising an anti-CD19 antibody is encoded by the sequence of SEQ ID NO: 148. In some embodiments, a CAR comprising an anti-CD19 antibody comprises the sequence of SEQ ID NO: 149.
[0321] In some embodiments, the anti-CD19 antibody is an anti-CD19 scFv encoded by a nucleotide sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 150. In some embodiments, the anti-CD19 antibody is an anti-CD19 scFv comprising an amino acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 151. In some embodiments, the anti-CD19 antibody is an anti-CD19 scFv comprising a VH comprising an amino acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 152. In some embodiments, the anti-CD19 antibody is an anti-CD19 scFv comprising a VL comprising an amino acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 153. In some embodiments, a CAR comprising an anti-CD19 antibody is encoded by a nucleotide having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 148. In some embodiments, a CAR comprising an anti-CD19 antibody comprises an amino acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 149.
[0322] CD33
[0323] In some embodiments, the T cells of the present disclosure are engineered with a CAR designed to target CD33. CD33, also known as Siglec3, is a transmembrane receptor expressed on cells of myeloid lineage that is known to bind sialic acids. As CD33 is expressed in cancer cells (e.g., acute myeloid leukemia), it is thought that CD33 represents a cell surface marker for targeting these malignancies.
[0324] Thus, in some embodiments, T cells of the present disclosure are engineered to express a CAR comprising an anti-CD33 antibody (e.g., anti-CD33 scFv). In some embodiments, the anti-CD33 antibody is an anti-CD33 scFv encoded by the sequence of SEQ ID NO: 138. In some embodiments, the anti-CD33 antibody is an anti-CD33 scFv comprising the sequence of SEQ ID NO: 137. In some embodiments, the anti-CD33 antibody is an anti-CD19 scFv comprising a VH comprising the sequence of SEQ ID NO: 140. In some embodiments, the anti-CD33 antibody is an anti-CD33 scFv comprising a VL comprising the sequence of SEQ ID NO: 141. In some embodiments, a CAR comprising an anti-CD33 antibody is encoded by the sequence of SEQ ID NO: 136. In some embodiments, a CAR comprising an anti-CD33 antibody comprises the sequence of SEQ ID NO: 139.
[0325] In some embodiments, the anti-CD33 antibody is an anti-CD33 scFv encoded by a nucleotide sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 138. In some embodiments, the anti-CD33 antibody is an anti-CD33 scFv comprising an amino acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 137. In some embodiments, the anti-CD33 antibody is an anti-CD19 scFv comprising a VH comprising an amino acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 140. In some embodiments, the anti-CD33 antibody is an anti-CD33 scFv comprising a VL comprising an amino acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 141. In some embodiments, a CAR comprising an anti-CD33 antibody is encoded by a nucleotide sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 136. In some embodiments, a CAR comprising an anti-CD33 antibody comprises an amino acid sequence having at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97% 98% or 99% identity to SEQ ID NO: 139.
Antibodies
[0326] An antibody (interchangeably used in plural form) is an immunoglobulin molecule capable of specific binding to a target, such as a carbohydrate, polynucleotide, lipid, polypeptide, etc., through at least one antigen recognition site, located in the variable region of the immunoglobulin molecule. As used herein, the term "antibody" encompasses not only intact (i.e., full-length) monoclonal antibodies, but also antigen-binding fragments (such as Fab, Fab', F(ab')2, Fv), single chain variable fragment (scFv), mutants thereof, fusion proteins comprising an antibody portion, humanized antibodies, chimeric antibodies, diabodies, linear antibodies, single chain antibodies, single domain antibodies (e.g., camel or llama V.sub.HH antibodies), multispecific antibodies (e.g., bispecific antibodies) and any other modified configuration of the immunoglobulin molecule that comprises an antigen recognition site of the required specificity, including glycosylation variants of antibodies, amino acid sequence variants of antibodies, and covalently modified antibodies.
[0327] A typical antibody molecule comprises a heavy chain variable region (VH) and a light chain variable region (VL), which are usually involved in antigen binding. These regions/residues that are responsible for antigen-binding can be identified from amino acid sequences of the VH/VL sequences of a reference antibody (e.g., an anti-CD70 antibody or an anti-BCMA antibody as described herein) by methods known in the art. The VH and VL regions can be further subdivided into regions of hypervariability, also known as "complementarity determining regions" ("CDR"), interspersed with regions that are more conserved, which are known as "framework regions" ("FR"). Each VH and VL is typically composed of three CDRs and four FRs, arranged from amino-terminus to carboxy-terminus in the following order: FR1, CDR1, FR2, CDR2, FR3, CDR3, FR4. The extent of the framework region and CDRs can be precisely identified using methodology known in the art, for example, by the Kabat definition, the Chothia definition, the AbM definition, and/or the contact definition, all of which are well known in the art. As used herein, a CDR may refer to the CDR defined by any method known in the art. Two antibodies having the same CDR means that the two antibodies have the same amino acid sequence of that CDR as determined by the same method. See, e.g., Kabat, E. A., et al. (1991) Sequences of Proteins of Immunological Interest, Fifth Edition, U.S. Department of Health and Human Services, NIH Publication No. 91-3242, Chothia et al., (1989) Nature 342:877; Chothia, C. et al. (1987) J. Mol. Biol. 196:901-917, Al-lazikani et al (1997) J. Molec. Biol. 273:927-948; and Almagro, J. Mol. Recognit. 17:132-143 (2004). See also hgmp.mrc.ac.uk and bioinf.org.uk/abs.
[0328] In some embodiments, an antibody is an scFv, such as an anti-CD70 scFv, an anti-BCMA scFv, an anti-CD19 scFv or an anti-CD33 scFv. An antibody includes an antibody of any class, such as IgD, IgE, IgG, IgA, or IgM (or sub-class thereof), and the antibody need not be of any particular class. Depending on the antibody amino acid sequence of the constant domain of its heavy chains, immunoglobulins can be assigned to different classes. There are five major classes of immunoglobulins: IgA, IgD, IgE, IgG, and IgM, and several of these may be further divided into subclasses (isotypes), e.g., IgG1, IgG2, IgG3, IgG4, IgA1 and IgA2. The heavy-chain constant domains that correspond to the different classes of immunoglobulins are called alpha, delta, epsilon, gamma, and mu, respectively. The subunit structures and three-dimensional configurations of different classes of immunoglobulins are well known.
[0329] The antibodies to be used as provided herein can be murine, rat, human, or any other origin (including chimeric or humanized antibodies). In some examples, the antibody comprises a modified constant region, such as a constant region that is immunologically inert, e.g., does not trigger complement mediated lysis, or does not stimulate antibody-dependent cell mediated cytotoxicity (ADCC).
[0330] In some embodiments, an antibody of the present disclosure is a humanized antibody. Humanized antibodies refer to forms of non-human (e.g., murine) antibodies that are specific chimeric immunoglobulins, immunoglobulin chains, or antigen-binding fragments thereof that contain minimal sequence derived from non-human immunoglobulin. For the most part, humanized antibodies are human immunoglobulins (recipient antibody) in which residues from a complementary determining region (CDR) of the recipient are replaced by residues from a CDR of a non-human species (donor antibody) such as mouse, rat, or rabbit having the desired specificity, affinity, and capacity. In some instances, Fv framework region (FR) residues of the human immunoglobulin are replaced by corresponding non-human residues. Furthermore, the humanized antibody may comprise residues that are found neither in the recipient antibody nor in the imported CDR or framework sequences, but are included to further refine and optimize antibody performance. In general, the humanized antibody will comprise substantially all of at least one, and typically two, variable domains, in which all or substantially all of the CDR regions correspond to those of a non-human immunoglobulin and all or substantially all of the FR regions are those of a human immunoglobulin consensus sequence. A humanized antibody optimally also will comprise at least a portion of an immunoglobulin constant region or domain (Fc), typically that of a human immunoglobulin. Other forms of humanized antibodies have one or more CDRs (one, two, three, four, five, or six) which are altered with respect to the original antibody, which are also termed one or more CDRs "derived from" one or more CDRs from the original antibody. Humanized antibodies may also involve affinity maturation.
[0331] In some embodiments, an antibody of the present disclosure is a chimeric antibody, which can include a heavy constant region and a light constant region from a human antibody.
[0332] Chimeric antibodies refer to antibodies having a variable region or part of variable region from a first species and a constant region from a second species. Typically, in these chimeric antibodies, the variable region of both light and heavy chains mimics the variable regions of antibodies derived from one species of mammals (e.g., a non-human mammal such as mouse, rabbit, and rat), while the constant portions are homologous to the sequences in antibodies derived from another mammal such as human. In some embodiments, amino acid modifications can be made in the variable region and/or the constant region.
[0333] In some embodiments, an antibody of the present disclosure specifically binds a target antigen, such as human CD70, human BCMA, human CD19 or human CD33. An antibody that "specifically binds" to a target or an epitope is a term well understood in the art, and methods to determine such specific binding are also well known in the art. A molecule is said to exhibit "specific binding" if it reacts or associates more frequently, more rapidly, with greater duration and/or with greater affinity with a particular target antigen than it does with alternative targets. An antibody "specifically binds" to a target antigen if it binds with greater affinity, avidity, more readily, and/or with greater duration than it binds to other substances. For example, an antibody that specifically (or preferentially) binds to a CD70, BCMA, CD19 or CD33 epitope is an antibody that binds this CD70, BCMA, CD19 or CD33 epitope with greater affinity, avidity, more readily, and/or with greater duration than it binds to other CD70, BCMA, CD19 or CD33 epitopes or non-CD70, non-BCMA, non-CD19 or non-CD33 epitopes. It is also understood by reading this definition that, for example, an antibody that specifically binds to a first target antigen may or may not specifically or preferentially bind to a second target antigen. As such, "specific binding" or "preferential binding" does not necessarily require (although it can include) exclusive binding. Generally, but not necessarily, reference to binding means preferential binding.
[0334] In some embodiments, the equilibrium dissociation constant (K.sub.D) between the antibody and CD70 is 100 pM to 1 .mu.M. In some embodiments, the K.sub.D between the antibody and CD70 is 1 nM to 100 nM.
[0335] In some embodiments, the equilibrium dissociation constant (K.sub.D) between the antibody and BCMA is 100 pM to 1 .mu.M. In some embodiments, the K.sub.D between the antibody and BCMA is 1 nM to 100 nM.
[0336] In some embodiments, the equilibrium dissociation constant (K.sub.D) between the antibody and CD19 is 100 pM to 1 .mu.M. In some embodiments, the K.sub.D between the antibody and CD19 is 1 nM to 100 nM.
[0337] In some embodiments, the equilibrium dissociation constant (K.sub.D) between the antibody and CD33 is 100 pM to 1 .mu.M. In some embodiments, the K.sub.D between the antibody and CD33 is 1 nM to 100 nM.
[0338] Also within the scope of the present disclosure are functional variants of any of the exemplary antibodies as disclosed herein. A functional variant may contain one or more amino acid residue variations in the VH and/or VL, or in one or more of the VH CDRs and/or one or more of the VL CDRs as relative to a reference antibody, while retaining substantially similar binding and biological activities (e.g., substantially similar binding affinity, binding specificity, inhibitory activity, anti-tumor activity, or a combination thereof) as the reference antibody.
[0339] In some examples, an antibody disclosed herein comprises a VH CDR1, a VH CDR2, and a VH CDR3, which collectively contains no more than 10 amino acid variations (e.g., no more than 9, 8, 7, 6, 5, 4, 3, 2, or 1 amino acid variation) as compared with the VH CDR1, VH CDR2, and VH CDR3 of a reference antibody such as Antibody A (VH: SEQ ID NO: 51; VL: SEQ ID NO: 52) or Antibody B (VH: SEQ ID NO: 60; VL: SEQ ID NO: 61). "Collectively" means that the total number of amino acid variations in all of the three VH CDRs is within the defined range. Alternatively or in addition, antibody may comprise a VL CDR1, a VL CDR2, and a VL CDR3, which collectively contains no more than 10 amino acid variations (e.g., no more than 9, 8, 7, 6, 5, 4, 3, 2 or 1 amino acid variation) as compared with the VL CDR1, VL CDR2, and VL CDR3 of the reference antibody.
[0340] In some examples, an antibody disclosed herein may comprise a VH CDR1, a VH CDR2, and a VH CDR3, at least one of which contains no more than 5 amino acid variations (e.g., no more than 4, 3, 2, or 1 amino acid variation) as the counterpart VH CDR of a reference antibody such as Antibody A (VH: SEQ ID NO: 51; VL: SEQ ID NO: 52) or Antibody B (VH: SEQ ID NO: 60; VL: SEQ ID NO: 61). In specific examples, the antibody comprises a VH CDR3, which contains no more than 5 amino acid variations (e.g., no more than 4, 3, 2, or 1 amino acid variation) as the VH CDR3 of a reference antibody such as Antibody A (VH: SEQ ID NO: 51; VL: SEQ ID NO: 52) or Antibody B (VH: SEQ ID NO: 60; VL: SEQ ID NO: 61). Alternatively or in addition, an antibody may comprise a VL CDR1, a VL CDR2, and a VL CDR3, at least one of which contains no more than 5 amino acid variations (e.g., no more than 4, 3, 2, or 1 amino acid variation) as the counterpart VL CDR of the reference antibody. In specific examples, the antibody comprises a VL CDR3, which contains no more than 5 amino acid variations (e.g., no more than 4, 3, 2, or 1 amino acid variation) as the VL CDR3 of the reference antibody.
[0341] In some instances, the amino acid residue variations can be conservative amino acid residue substitutions. As used herein, a "conservative amino acid substitution" refers to an amino acid substitution that does not alter the relative charge or size characteristics of the protein in which the amino acid substitution is made. Variants can be prepared according to methods for altering polypeptide sequence known to one of ordinary skill in the art such as are found in references which compile such methods, e.g. Molecular Cloning: A Laboratory Manual, J. Sambrook, et al., eds., Second Edition, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., 1989, or Current Protocols in Molecular Biology, F.M. Ausubel, et al., eds., John Wiley & Sons, Inc., New York. Conservative substitutions of amino acids include substitutions made amongst amino acids within the following groups: (a) A.fwdarw.G, S; (b) R.fwdarw.K, H; (c) N.fwdarw.Q, H; (d) D.fwdarw.E, N; (e) C.fwdarw.S, A; (f) Q.fwdarw.N; (g) E.fwdarw.D, Q; (h) G.fwdarw.A; (i) H.fwdarw.N, Q; (j) I.fwdarw.L, V; (k) L.fwdarw.I, V; (l) K.fwdarw.R, H; (m) M.fwdarw.L, I, Y; (n) F.fwdarw.Y, M, L; (o) P.fwdarw.A; (p) S.fwdarw.T; (q) T.fwdarw.S; (r) W.fwdarw.Y, F; (s) Y.fwdarw.W, F; and (t) V.fwdarw.I, L.
[0342] In some embodiments, an antibody disclosed herein may comprise VH CDRs that collectively are at least 80% (e.g., 85%, 90%, 95%, or 98%) identical to the VH CDRs of a reference antibody such as Antibody A (VH: SEQ ID NO: 51; VL: SEQ ID NO: 52) or Antibody B (VH: SEQ ID NO: 60; VL: SEQ ID NO: 61). Alternatively or in addition, the antibody may comprise VL CDRs that collectively are at least 80% (e.g., 85%, 90%, 95%, or 98%) identical to the VL CDRs of the reference antibody. In some embodiments, an antibody may comprise a VH that is at least 80% (e.g., 85%, 90%, 95%, or 98%) identical to the VH of a reference antibody such as Antibody A (VH: SEQ ID NO: 51; VL: SEQ ID NO: 52) or Antibody B (VH: SEQ ID NO: 60; VL: SEQ ID NO: 61) and/or a VL that is at least 80% (e.g., 85%, 90%, 95%, or 98%) identical to the VL of the reference antibody.
[0343] In some embodiments, an anti-CD70 antibody (e.g., anti-CD70 scFv) comprises a VH and a VL comprising the amino acid sequences set forth in SEQ ID NOs: 51 and 52, respectively. In some embodiments, an anti-CD70 antibody (e.g., anti-CD70 scFv) comprises three CDRs (CDR1, CDR2 and CDR2) of the VH set forth in SEQ ID NO: 51, and three CDRs (CDR1, CDR2 and CDR3) of the VL set forth in SEQ ID NO: 52. In some embodiments, an anti-CD70 antibody (e.g., anti-CD70 scFv) comprises three CDRs (CDR1, CDR2 and CDR2) of the VH set forth in SEQ ID NO: 51, and three CDRs (CDR1, CDR2 and CDR3) of the VL set forth in SEQ ID NO: 52, wherein the CDRs are determined according to Kabat. In some embodiments, an anti-CD70 antibody (e.g., anti-CD70 scFv) comprises three CDRs (CDR1, CDR2 and CDR2) of the VH set forth in SEQ ID NO: 51, and three CDRs (CDR1, CDR2 and CDR3) of the VL set forth in SEQ ID NO: 52, wherein the CDRs are determined according to Chothia. In some embodiments, an anti-CD70 antibody (e.g., anti-CD70 scFv) comprises three CDRs (CDR1, CDR2 and CDR2) of the VH set forth in SEQ ID NO: 51, and three CDRs (CDR1, CDR2 and CDR3) of the VL set forth in SEQ ID NO: 52, wherein the CDRs are determined according to AbM. In some embodiments, an anti-CD70 antibody (e.g., anti-CD70 scFv) comprises heavy chain CDR1, CDR2 and CDR3 sequences set forth in SEQ ID NOs: 68, 70 and 72, respectively, and light chain CDR1, CDR2 and CDR3 sequences set forth in SEQ ID NOs: 62, 64 and 66. In some embodiments, an anti-CD70 antibody (e.g., anti-CD70 scFv) comprises heavy chain CDR1, CDR2 and CDR3 sequences set forth in SEQ ID NOs: 69, 71 and 73, respectively, and light chain CDR1, CDR2 and CDR3 sequences set forth in SEQ ID NOs: 63, 65 and 67. In some embodiments, an anti-CD70 antibody is an anti-CD70 scFv comprising the amino acid sequence set forth in SEQ ID NO: 50. In some embodiments, an anti-CD70 antibody is an anti-CD70 scFv encoded by the nucleotide sequence set forth in SEQ ID NO: 49. In some embodiments, an anti-CD70 antibody is an anti-CD70 scFv comprising the amino acid sequence set forth in SEQ ID NO: 48. In some embodiments, an anti-CD70 antibody is an anti-CD70 scFv encoded by the nucleotide sequence set forth in SEQ ID NO: 47.
[0344] In some embodiments, an anti-BCMA antibody (e.g., anti-BCMA scFv) comprises a VH and a VL comprising the amino acid sequences set forth in SEQ ID NOs: 60 and 61, respectively. In some embodiments, an anti-BCMA antibody (e.g., anti-BCMA scFv) comprises three CDRs (CDR1, CDR2 and CDR2) of the VH set forth in SEQ ID NO: 60, and three CDRs (CDR1, CDR2 and CDR3) of the VL set forth in SEQ ID NO: 61. In some embodiments, an anti-BCMA antibody (e.g., anti-BCMA scFv) comprises three CDRs (CDR1, CDR2 and CDR2) of the VH set forth in SEQ ID NO: 60, and three CDRs (CDR1, CDR2 and CDR3) of the VL set forth in SEQ ID NO: 61, wherein the CDRs are determined according to Kabat. In some embodiments, an anti-BCMA antibody (e.g., anti-BCMA scFv) comprises three CDRs (CDR1, CDR2 and CDR2) of the VH set forth in SEQ ID NO: 60, and three CDRs (CDR1, CDR2 and CDR3) of the VL set forth in SEQ ID NO: 61, wherein the CDRs are determined according to Chothia. In some embodiments, an anti-BCMA antibody (e.g., anti-BCMA scFv) comprises three CDRs (CDR1, CDR2 and CDR2) of the VH set forth in SEQ ID NO: 60, and three CDRs (CDR1, CDR2 and CDR3) of the VL set forth in SEQ ID NO: 61, wherein the CDRs are determined according to AbM. In some embodiments, an anti-BCMA antibody (e.g., anti-BCMA scFv) comprises heavy chain CDR1, CDR2 and CDR3 sequences set forth in SEQ ID NOs: 80, 82 and 84, respectively, and light chain CDR1, CDR2 and CDR3 sequences set forth in SEQ ID NOs: 74, 76 and 78. In some embodiments, an anti-BCMA antibody (e.g., anti-BCMA scFv) comprises heavy chain CDR1, CDR2 and CDR3 sequences set forth in SEQ ID NOs: 81, 83 and 85, respectively, and light chain CDR1, CDR2 and CDR3 sequences set forth in SEQ ID NOs: 75, 77 and 79. In some embodiments, an anti-BCMA antibody is an anti-BCMA scFv comprising the amino acid sequence set forth in SEQ ID NO: 59 In some embodiments, an anti-BCMA antibody is an anti-BCMA scFv encoded by the nucleotide sequence set forth in SEQ ID NO: 58.
[0345] In some embodiments, an anti-CD19 antibody (e.g., anti-CD19 scFv) comprises a VH and a VL comprising the amino acid sequences set forth in SEQ ID NOs: 152 and 153, respectively. In some embodiments, an anti-CD19 antibody (e.g., anti-CD19 scFv) comprises three CDRs (CDR1, CDR2 and CDR2) of the VH set forth in SEQ ID NO: 152, and three CDRs (CDR1, CDR2 and CDR3) of the VL set forth in SEQ ID NO: 153. In some embodiments, an anti-CD19 antibody (e.g., anti-CD19 scFv) comprises three CDRs (CDR1, CDR2 and CDR2) of the VH set forth in SEQ ID NO: 152, and three CDRs (CDR1, CDR2 and CDR3) of the VL set forth in SEQ ID NO: 153, wherein the CDRs are determined according to Kabat. In some embodiments, an anti-CD19 antibody (e.g., anti-CD19 scFv) comprises three CDRs (CDR1, CDR2 and CDR2) of the VH set forth in SEQ ID NO: 152, and three CDRs (CDR1, CDR2 and CDR3) of the VL set forth in SEQ ID NO: 153, wherein the CDRs are determined according to Chothia. In some embodiments, an anti-CD19 antibody (e.g., anti-CD19 scFv) comprises three CDRs (CDR1, CDR2 and CDR2) of the VH set forth in SEQ ID NO: 152, and three CDRs (CDR1, CDR2 and CDR3) of the VL set forth in SEQ ID NO: 153, wherein the CDRs are determined according to AbM. In some embodiments, an anti-CD19 antibody (e.g., anti-CD19 scFv) comprises heavy chain CDR1, CDR2 and CDR3 sequences set forth in SEQ ID NOs: 169, 170 and 171, respectively, and light chain CDR1, CDR2 and CDR3 sequences set forth in SEQ ID NOs: 166, 167 and 168, respectively. In some embodiments, an anti-CD19 antibody (e.g., anti-CD19 scFv) comprises heavy chain CDR1, CDR2 and CDR3 sequences set forth in SEQ ID NOs: 175, 176 and 177, respectively, and light chain CDR1, CDR2 and CDR3 sequences set forth in SEQ ID NOs: 172, 173 and 174, respectively. In some embodiments, an anti-CD19 antibody is an anti-CD19 scFv comprising the amino acid sequence set forth in SEQ ID NO: 151. In some embodiments, an anti-CD19 antibody is an anti-CD19 scFv encoded by the nucleotide sequence set forth in SEQ ID NO: 150.
[0346] In some embodiments, an anti-CD33 antibody (e.g., anti-CD33 scFv) comprises a VH and a VL comprising the amino acid sequences set forth in SEQ ID NOs: 140 and 141, respectively. In some embodiments, an anti-CD33 antibody (e.g., anti-CD33 scFv) comprises three CDRs (CDR1, CDR2 and CDR2) of the VH set forth in SEQ ID NO: 140, and three CDRs (CDR1, CDR2 and CDR3) of the VL set forth in SEQ ID NO: 141. In some embodiments, an anti-CD33 antibody (e.g., anti-CD33 scFv) comprises three CDRs (CDR1, CDR2 and CDR2) of the VH set forth in SEQ ID NO: 140, and three CDRs (CDR1, CDR2 and CDR3) of the VL set forth in SEQ ID NO: 141, wherein the CDRs are determined according to Kabat. In some embodiments, an anti-CD33 antibody (e.g., anti-CD33 scFv) comprises three CDRs (CDR1, CDR2 and CDR2) of the VH set forth in SEQ ID NO: 140, and three CDRs (CDR1, CDR2 and CDR3) of the VL set forth in SEQ ID NO: 141, wherein the CDRs are determined according to Chothia. In some embodiments, an anti-CD33 antibody (e.g., anti-CD33 scFv) comprises three CDRs (CDR1, CDR2 and CDR2) of the VH set forth in SEQ ID NO: 140, and three CDRs (CDR1, CDR2 and CDR3) of the VL set forth in SEQ ID NO: 141, wherein the CDRs are determined according to AbM. In some embodiments, an anti-CD33 antibody (e.g., anti-CD33 scFv) comprises heavy chain CDR1, CDR2 and CDR3 sequences set forth in SEQ ID NOs: 142, 143 and 144, respectively, and light chain CDR1, CDR2 and CDR3 sequences set forth in SEQ ID NOs: 145, 146 and 147. In some embodiments, an anti-CD33 antibody (e.g., anti-CD33 scFv) comprises heavy chain CDR1, CDR2 and CDR3 sequences set forth in SEQ ID NOs: 178, 179 and 180, respectively, and light chain CDR1, CDR2 and CDR3 sequences set forth in SEQ ID NOs: 145, 146 and 147. In some embodiments, an anti-CD33 antibody is an anti-CD33 scFv comprising the amino acid sequence set forth in SEQ ID NO: 137. In some embodiments, an anti-CD33 antibody is an anti-CD33 scFv encoded by the nucleotide sequence set forth in SEQ ID NO: 138.
Antigen Targeting Chimeric Antigen Receptor Construct
[0347] In some embodiments, the engineered T cells described herein comprise a tumor antigen targeting CAR. In some embodiments, a tumor antigen is a "tumor associated antigen," referring an immunogenic molecule, such as a protein, that is generally expressed at a higher level in tumor cells than in non-tumor cells, in which it may not be expressed at all, or only at low levels. In some embodiments, tumor-associated structures which are recognized by the immune system of the tumor-harboring host are referred to as tumor-associated antigens. In some embodiments, a tumor-associated antigen is a universal tumor antigen if its broadly expressed by most tumors. In some embodiments, tumor-associated antigens are differentiation antigens, mutational antigens, overexpressed cellular antigens or viral antigens. In some embodiments, a tumor antigen is a "tumor specific antigen" or "TSA," referring to an immunogenic molecule, such as a protein, that is unique to a tumor cell. Tumor specific antigens are exclusively expressed in tumor cells. In some embodiments, the tumor antigen is not CD70.
[0348] In some embodiments, the engineered T cells described herein comprise a non-CD70 targeting CAR (e.g., a CAR that does not bind CD70).
[0349] CD19 CAR
[0350] In some embodiments, the engineered T cells described herein comprise a CD19 targeting CAR, also referred to herein as CD19 CAR, anti-CD19 CAR or anti-CD19 CAR T cells. In some embodiments, the anti-CD19 CAR comprises (i) an ectodomain that comprises an anti-CD19 antigen-binding domain, (ii) a transmembrane domain, and (iii) an endodomain comprising at least one co-stimulatory domain.
[0351] In some embodiments, the anti-CD19 CAR comprises (i) an ectodomain that comprises an anti-CD19 antigen-binding domain, (ii) a CD8 transmembrane domain, and (iii) an endodomain that comprises a CD28 or 41BB co-stimulatory domain, and a CD3-zeta signaling domain. In some embodiments, the anti-CD19 CAR comprises (i) an ectodomain that comprises an anti-CD19 antigen-binding domain, (ii) a CD8 transmembrane domain, and (iii) an endodomain that comprises a CD28 co-stimulatory domain and a CD3-zeta signaling domain. In some embodiments, the anti-CD19 CAR comprises (i) an ectodomain that comprises an anti-CD19 antigen-binding domain, (ii) a CD8 transmembrane domain, and (iii) an endodomain that comprises a 41BB co-stimulatory domain and a CD3-zeta signaling domain.
[0352] In some embodiments, the anti-CD19 CAR comprises (i) an ectodomain that comprises an anti-CD19 antigen-binding domain, (ii) a CD8 transmembrane domain comprising the amino acid sequence set forth in SEQ ID NO: 126, and (iii) an endodomain that comprises a CD28 co-stimulatory domain comprising the amino acid sequence set forth in SEQ ID NO: 20 and a CD3-zeta signaling domain comprising the amino acid sequence set forth in SEQ ID NO: 22.
[0353] In some embodiments, the anti-CD19 CAR comprises (i) an ectodomain that comprises an anti-CD19 scFv comprising the amino acid sequence set forth in SEQ ID NO: 151, (ii) a CD8 transmembrane domain comprising the amino acid sequence set forth in SEQ ID NO: 126, and (iii) an endodomain that comprises a CD28 co-stimulatory domain comprising the amino acid sequence set forth in SEQ ID NO: 20 and a CD3-zeta signaling domain comprising the amino acid sequence set forth in SEQ ID NO: 22.
[0354] In some embodiments, the anti-CD19 CAR comprises (i) an ectodomain that comprises an anti-CD19 scFv comprising variable heavy and light chain regions comprising the amino acid sequences set forth in SEQ ID NOs: 152 and 153, respectively, (ii) a CD8 transmembrane domain comprising the amino acid sequence set forth in SEQ ID NO: 126, and (iii) an endodomain that comprises a CD28 co-stimulatory domain comprising the amino acid sequence set forth in SEQ ID NO: 20 and a CD3-zeta signaling domain comprising the amino acid sequence set forth in SEQ ID NO: 22.
[0355] In some embodiments, the anti-CD19 CAR comprises the amino acid sequence set forth in SEQ ID NO: 149. In some embodiments, the anti-CD19 CAR is encoded by the nucleotide sequence set forth in SEQ ID NO: 148. In some embodiments, the anti-CD19 CAR is encoded by a nucleotide sequence having at least 80%, 85%, 90%, 95%, 96%, 97%, 98%, 99% or 100% identity to the nucleotide sequence set forth in SEQ ID NO: 148.
[0356] CD33 CAR
[0357] In some embodiments, the engineered T cells described herein comprise a CD33 targeting CAR, also referred to herein as CD33 CAR, anti-CD33 CAR or anti-CD33 CAR T cells. In some embodiments, the anti-CD33 CAR comprises (i) an ectodomain that comprises an anti-CD33 antigen-binding domain, (ii) a transmembrane domain, and (iii) an endodomain comprising at least one co-stimulatory domain.
[0358] In some embodiments, the anti-CD33 CAR comprises (i) an ectodomain that comprises an anti-CD33 antigen-binding domain, (ii) a CD8 transmembrane domain, and (iii) an endodomain that comprises a CD28 or 41BB co-stimulatory domain, and a CD3-zeta signaling domain. In some embodiments, the anti-CD33 CAR comprises (i) an ectodomain that comprises an anti-CD33 antigen-binding domain, (ii) a CD8 transmembrane domain, and (iii) an endodomain that comprises a CD28 co-stimulatory domain and a CD3-zeta signaling domain. In some embodiments, the anti-CD33 CAR comprises (i) an ectodomain that comprises an anti-CD33 antigen-binding domain, (ii) a CD8 transmembrane domain, and (iii) an endodomain that comprises a 41BB co-stimulatory domain and a CD3-zeta signaling domain.
[0359] In some embodiments, the anti-CD33 CAR comprises (i) an ectodomain that comprises an anti-CD33 antigen-binding domain, (ii) a CD8 transmembrane domain comprising the amino acid sequence set forth in SEQ ID NO: 126, and (iii) an endodomain that comprises a 41BB co-stimulatory domain comprising the amino acid sequence set forth in SEQ ID NO: 19 and a CD3-zeta signaling domain comprising the amino acid sequence set forth in SEQ ID NO: 22.
[0360] In some embodiments, the anti-CD33 CAR comprises (i) an ectodomain that comprises an anti-CD33 scFv comprising the amino acid sequence set forth in SEQ ID NO: 137, (ii) a CD8 transmembrane domain comprising the amino acid sequence set forth in SEQ ID NO: 126, and (iii) an endodomain that comprises a 41BB co-stimulatory domain comprising the amino acid sequence set forth in SEQ ID NO: 19 and a CD3-zeta signaling domain comprising the amino acid sequence set forth in SEQ ID NO: 22.
[0361] In some embodiments, the anti-CD33 CAR comprises (i) an ectodomain that comprises an anti-CD33 scFv comprising variable heavy and light chain regions comprising the amino acid sequences set forth in SEQ ID NOs: 140 and 141, respectively, (ii) a CD8 transmembrane domain comprising the amino acid sequence set forth in SEQ ID NO: 126, and (iii) an endodomain that comprises a 41BB co-stimulatory domain comprising the amino acid sequence set forth in SEQ ID NO: 19 and a CD3-zeta signaling domain comprising the amino acid sequence set forth in SEQ ID NO: 22.
[0362] In some embodiments, the anti-CD33 CAR comprises the amino acid sequence set forth in SEQ ID NO: 139. In some embodiments, the anti-CD33 CAR is encoded by the nucleotide sequence set forth in SEQ ID NO: 136. In some embodiments, the anti-CD33 CAR is encoded by a nucleotide sequence having at least 80%, 85%, 90%, 95%, 96%, 97%, 98%, 99% or 100% identity to the nucleotide sequence set forth in SEQ ID NO: 136.
[0363] BCMA CAR
[0364] In some embodiments, the engineered T cells described herein comprise a BCMA targeting CAR, also referred to herein as BCMA CAR, anti-BCMA CAR or anti-BCMA CAR T cells. In some embodiments, the anti-BCMA CAR comprises (i) an ectodomain that comprises an anti-BCMA antigen-binding domain, (ii) a transmembrane domain, and (iii) an endodomain comprising at least one co-stimulatory domain.
[0365] In some embodiments, the anti-BCMA CAR comprises (i) an ectodomain that comprises an anti-BCMA antigen-binding domain, (ii) a CD8 transmembrane domain, and (iii) an endodomain that comprises a CD28 or 41BB co-stimulatory domain, and a CD3-zeta signaling domain. In some embodiments, the anti-BCMA CAR comprises (i) an ectodomain that comprises an anti-BCMA antigen-binding domain, (ii) a CD8 transmembrane domain, and (iii) an endodomain that comprises a CD28 co-stimulatory domain and a CD3-zeta signaling domain. In some embodiments, the anti-BCMA CAR comprises (i) an ectodomain that comprises an anti-BCMA antigen-binding domain, (ii) a CD8 transmembrane domain, and (iii) an endodomain that comprises a 41BB co-stimulatory domain and a CD3-zeta signaling domain.
[0366] In some embodiments, the anti-BCMA CAR comprises (i) an ectodomain that comprises an anti-BCMA antigen-binding domain, (ii) a CD8 transmembrane domain comprising the amino acid sequence set forth in SEQ ID NO: 126, and (iii) an endodomain that comprises a 41BB co-stimulatory domain comprising the amino acid sequence set forth in SEQ ID NO: 19 and a CD3-zeta signaling domain comprising the amino acid sequence set forth in SEQ ID NO: 22.
[0367] In some embodiments, the anti-BCMA CAR comprises (i) an ectodomain that comprises an anti-BCMA scFv comprising the amino acid sequence set forth in SEQ ID NO: 59, (ii) a CD8 transmembrane domain comprising the amino acid sequence set forth in SEQ ID NO: 126, and (iii) an endodomain that comprises a 41BB co-stimulatory domain comprising the amino acid sequence set forth in SEQ ID NO: 19 and a CD3-zeta signaling domain comprising the amino acid sequence set forth in SEQ ID NO: 22.
[0368] In some embodiments, the anti-BCMA CAR comprises (i) an ectodomain that comprises an anti-BCMA scFv comprising variable heavy and light chain regions comprising the amino acid sequences set forth in SEQ ID NOs: 60 and 61, respectively, (ii) a CD8 transmembrane domain comprising the amino acid sequence set forth in SEQ ID NO: 126, and (iii) an endodomain that comprises a 41BB co-stimulatory domain comprising the amino acid sequence set forth in SEQ ID NO: 19 and a CD3-zeta signaling domain comprising the amino acid sequence set forth in SEQ ID NO: 22.
[0369] In some embodiments, the anti-BCMA CAR comprises the amino acid sequence set forth in SEQ ID NO: 57. In some embodiments, the anti-BCMA CAR is encoded by the nucleotide sequence set forth in SEQ ID NO: 56. In some embodiments, the anti-BCMA CAR is encoded by a nucleotide sequence having at least 80%, 85%, 90%, 95%, 96%, 97%, 98%, 99% or 100% identity to the nucleotide sequence set forth in SEQ ID NO: 56.
[0370] CD70 CAR
[0371] In some embodiments, the engineered T cells described herein comprise a CD70 targeting CAR, also referred to herein as CD70 CAR, anti-CD70 CAR or anti-CD70 CAR T cells. In some embodiments, the anti-CD70 CAR comprises (i) an ectodomain that comprises an anti-CD70 antigen-binding domain, (ii) a transmembrane domain, and (iii) an endodomain comprising at least one co-stimulatory domain.
[0372] In some embodiments, the anti-CD70 CAR comprises (i) an ectodomain that comprises an anti-CD70 antigen-binding domain, (ii) a CD8 transmembrane domain, and (iii) an endodomain that comprises a CD28 or 41BB co-stimulatory domain, and a CD3-zeta signaling domain. In some embodiments, the anti-CD70 CAR comprises (i) an ectodomain that comprises an anti-CD70 antigen-binding domain, (ii) a CD8 transmembrane domain, and (iii) an endodomain that comprises a CD28 co-stimulatory domain and a CD3-zeta signaling domain. In some embodiments, the anti-CD70 CAR comprises (i) an ectodomain that comprises an anti-CD70 antigen-binding domain, (ii) a CD8 transmembrane domain, and (iii) an endodomain that comprises a 41BB co-stimulatory domain and a CD3-zeta signaling domain.
[0373] In some embodiments, the anti-CD70 CAR comprises (i) an ectodomain that comprises an anti-CD70 antigen-binding domain, (ii) a CD8 transmembrane domain comprising the amino acid sequence set forth in SEQ ID NO: 126, and (iii) an endodomain that comprises a 41BB co-stimulatory domain comprising the amino acid sequence set forth in SEQ ID NO: 19 and a CD3-zeta signaling domain comprising the amino acid sequence set forth in SEQ ID NO: 22.
[0374] In some embodiments, the anti-CD70 CAR comprises (i) an ectodomain that comprises an anti-CD70 scFv comprising the amino acid sequence set forth in SEQ ID NO: 50, (ii) a CD8 transmembrane domain comprising the amino acid sequence set forth in SEQ ID NO: 126, and (iii) an endodomain that comprises a 41BB co-stimulatory domain comprising the amino acid sequence set forth in SEQ ID NO: 19 and a CD3-zeta signaling domain comprising the amino acid sequence set forth in SEQ ID NO: 22.
[0375] In some embodiments, the anti-CD70 CAR comprises (i) an ectodomain that comprises an anti-CD70 scFv comprising variable heavy and light chain regions comprising the amino acid sequences set forth in SEQ ID NOs: 51 and 52, respectively, (ii) a CD8 transmembrane domain comprising the amino acid sequence set forth in SEQ ID NO: 126, and (iii) an endodomain that comprises a 41BB co-stimulatory domain comprising the amino acid sequence set forth in SEQ ID NO: 19 and a CD3-zeta signaling domain comprising the amino acid sequence set forth in SEQ ID NO: 22.
[0376] In some embodiments, the anti-CD70 CAR comprises the amino acid sequence set forth in SEQ ID NO: 46. In some embodiments, the anti-CD70 CAR is encoded by the nucleotide sequence set forth in SEQ ID NO: 45. In some embodiments, the anti-CD70 CAR is encoded by a nucleotide sequence having at least 80%, 85%, 90%, 95%, 96%, 97%, 98%, 99% or 100% identity to the nucleotide sequence set forth in SEQ ID NO: 45.
Expression of Chimeric Antigen Receptor Construct
[0377] Donor Template
[0378] The nucleic acid encoding a CAR may be delivered to a T cell that comprises what is referred to herein as a donor template (also referred to as a donor polynucleotide). A donor template can contain a non-homologous sequence, such as the nucleic acid encoding a CAR, flanked by two regions of homology to allow for efficient HDR at a genomic location of interest. Alternatively, a donor template may have no regions of homology to the targeted location in the DNA and may be integrated by NHEJ-dependent end joining following cleavage at the target site.
[0379] A donor template can be DNA or RNA, single-stranded and/or double-stranded, and can be introduced into a cell in linear or circular form. If introduced in linear form, the ends of the donor sequence can be protected (e.g., from exonucleolytic degradation) by methods known to those of skill in the art. For example, one or more dideoxynucleotide residues are added to the 3' terminus of a linear molecule and/or self-complementary oligonucleotides are ligated to one or both ends. See, for example, Chang et al., (1987) Proc. Natl. Acad. Sci. USA 84:4959-4963; Nehls et al., (1996) Science 272:886-889. Additional methods for protecting exogenous polynucleotides from degradation include, but are not limited to, addition of terminal amino group(s) and the use of modified internucleotide linkages such as, for example, phosphorothioates, phosphoramidates, and O-methyl ribose or deoxyribose residues.
[0380] A donor template can be introduced into a cell as part of a vector molecule having additional sequences such as, for example, replication origins, promoters and genes encoding antibiotic resistance. Moreover, a donor template can be introduced as naked nucleic acid, as nucleic acid complexed with an agent such as a liposome or poloxamer, or can be delivered by viruses (e.g., adenovirus, AAV, herpesvirus, retrovirus, lentivirus and integrase defective lentivirus (IDLV)).
[0381] A donor template, in some embodiments, is inserted so that its expression is driven by the endogenous promoter at the integration site, namely the promoter that drives expression of the endogenous gene into which the donor is inserted. However, in some embodiments, the donor template comprises an exogenous promoter and/or enhancer, for example a constitutive promoter, an inducible promoter, or tissue-specific promoter. In some embodiments, the exogenous promoter is an EF1.alpha. promoter comprising a sequence of SEQ ID NO: 123. Other promoters may be used.
[0382] Furthermore, exogenous sequences may also include transcriptional or translational regulatory sequences, for example, promoters, enhancers, insulators, internal ribosome entry sites, sequences encoding 2A peptides and/or polyadenylation signals.
[0383] In some embodiments, the donor template comprises a nucleotide sequence having at least 80%, 85%, 90%, 95%, 96%, 97%, 98% or 98% identity to SEQ ID NO: 44. In some embodiments, the donor template comprises the nucleotide sequence of SEQ ID NO: 44.
[0384] In some embodiments, the donor template comprises a nucleotide sequence having at least 80%, 85%, 90%, 95%, 96%, 97%, 98% or 98% identity to SEQ ID NO: 55. In some embodiments, the donor template comprises the nucleotide sequence of SEQ ID NO: 55.
[0385] In some embodiments, the donor template comprises a nucleotide sequence having at least 80%, 85%, 90%, 95%, 96%, 97%, 98% or 98% identity to SEQ ID NO: 135. In some embodiments, the donor template comprises the nucleotide sequence of SEQ ID NO: 135.
[0386] In some embodiments, the donor template comprises a nucleotide sequence having at least 80%, 85%, 90%, 95%, 96%, 97%, 98% or 98% identity to SEQ ID NO: 156. In some embodiments, the donor template comprises the nucleotide sequence of SEQ ID NO: 156.
[0387] Other Methods
[0388] In some embodiments, a nucleic acid encoding a CAR is introduced into an engineered cell by methods known to those of skill in the art. For example, a CAR may be introduced into an engineered cell by a vector. A variety of different methods known in the art can be used to introduce any of the nucleic acids or expression vectors disclosed herein into an immune effector cell. Non-limiting examples of methods for introducing nucleic acid into a cell include: lipofection, transfection (e.g., calcium phosphate transfection, transfection using highly branched organic compounds, transfection using cationic polymers, dendrimer-based transfection, optical transfection, particle-based transfection (e.g., nanoparticle transfection), or transfection using liposomes (e.g., cationic liposomes)), microinjection, electroporation, cell squeezing, sonoporation, protoplast fusion, impalefection, hydrodynamic delivery, gene gun, magnetofection, viral transfection, and nucleofection.
Delivery Methods and Constructs
[0389] Nucleases and/or donor templates may be delivered using a vector system, including, but not limited to, plasmid vectors, DNA minicircles, retroviral vectors, lentiviral vectors, adenovirus vectors, poxvirus vectors; herpesvirus vectors and adeno-associated virus vectors, and combinations thereof.
[0390] Conventional viral and non-viral based gene transfer methods can be used to introduce nucleic acids encoding nucleases and donor templates in cells (e.g., T cells). Non-viral vector delivery systems include DNA plasmids, DNA minicircles, naked nucleic acid, and nucleic acid complexed with a delivery vehicle such as a liposome or poloxamer. Viral vector delivery systems include DNA and RNA viruses, which have either episomal or integrated genomes after delivery to the cell.
[0391] Methods of non-viral delivery of nucleic acids include electroporation, lipofection, microinjection, biolistics, virosomes, liposomes, immunoliposomes, polycation or lipid:nucleic acid conjugates, naked DNA, naked RNA, capped RNA, artificial virions, and agent-enhanced uptake of DNA. Sonoporation using, e.g., the Sonitron 2000 system (Rich-Mar) can also be used for delivery of nucleic acids. Some specific examples are provided below.
[0392] Adeno-Associated Viral Delivery
[0393] The donor nucleic acid encoding a CAR construct can be delivered to a cell using an adeno-associated virus (AAV). AAVs are small viruses which integrate site-specifically into the host genome and can therefore deliver a transgene, such as CAR. Inverted terminal repeats (ITRs) are present flanking the AAV genome and/or the transgene of interest and serve as origins of replication. Also present in the AAV genome are rep and cap proteins which, when transcribed, form capsids which encapsulate the AAV genome for delivery into target cells. Surface receptors on these capsids which confer AAV serotype, which determines which target organs the capsids will primarily bind and thus what cells the AAV will most efficiently infect. There are twelve currently known human AAV serotypes. In some embodiments, the AAV is AAV serotype 6 (AAV6).
[0394] Adeno-associated viruses are among the most frequently used viruses for gene therapy for several reasons. First, AAVs do not provoke an immune response upon administration to mammals, including humans. Second, AAVs are effectively delivered to target cells, particularly when consideration is given to selecting the appropriate AAV serotype. Finally, AAVs have the ability to infect both dividing and non-dividing cells because the genome can persist in the host cell without integration. This trait makes them an ideal candidate for gene therapy.
[0395] Homology-Directed Repair (HDR)
[0396] The donor nucleic acid encoding a CAR is inserted by homology directed repair (HDR) into the target gene locus. Both strands of the DNA at the target locus are cut by a CRISPR Cas9 enzyme. HDR then occurs to repair the double-strand break (DSB) and insert the donor DNA. For this to occur correctly, the donor sequence is designed with flanking residues which are complementary to the sequence surrounding the DSB site in the target gene (hereinafter "homology arms"). These homology arms serve as the template for DSB repair and allow HDR to be an essentially error-free mechanism. The rate of homology directed repair (HDR) is a function of the distance between the mutation and the cut site so choosing overlapping or nearby target sites is important. Templates can include extra sequences flanked by the homologous regions or can contain a sequence that differs from the genomic sequence, thus allowing sequence editing.
[0397] The target gene can be associated with an immune response in a subject, wherein permanently deleting at least a portion of the target gene will modulate the immune response. For example, to generate a CAR T cell, the target gene can be the TCR.alpha. constant region (TRAC). Disruption of TRAC leads to loss of function of the endogenous TCR.
[0398] In some embodiments, the target gene is in a safe harbor locus.
Engineered T Cells
[0399] Engineered (gene edited) CAR T cells of the present disclosure may be autologous ("self") or non-autologous ("non-self," e.g., allogeneic, syngeneic or xenogeneic). "Autologous" refers to cells from the same subject. "Allogeneic" refers to cells of the same species as a subject, but that differ genetically to the cells in the subject. In some embodiments, the T cells are obtained from a mammal. In some embodiments, the T cells are obtained from a human.
[0400] T cells can be obtained from a number of sources including, but not limited to, peripheral blood mononuclear cells, bone marrow, lymph nodes tissue, cord blood, thymus issue, tissue from a site of infection, ascites, pleural effusion, spleen tissue, and tumors. In certain embodiments, T cells can be obtained from a unit of blood collected from a subject using any number of techniques known to the skilled person, such as sedimentation, e.g., FICOLL.TM. separation.
[0401] In some embodiments, an isolated population of T cells is used. In some embodiments, after isolation of peripheral blood mononuclear cells (PBMC), both cytotoxic and helper T lymphocytes can be sorted into naive, memory, and effector T cell subpopulations either before or after activation, expansion, and/or genetic modification.
[0402] A specific subpopulation of T cells, expressing one or more of the following cell surface markers: TCRab, CD3, CD4, CD8, CD27 CD28, CD38 CD45RA, CD45RO, CD62L, CD127, CD122, CD95, CD197, CCR7, KLRG1, MCH-I proteins and/or MCH-II proteins, can be further isolated by positive or negative selection techniques. In some embodiments, a specific subpopulation of T cells, expressing one or more of the markers selected from the group consisting of TCRab, CD4 and/or CD8, is further isolated by positive or negative selection techniques. In some embodiments, the engineered T cell populations do not express or do not substantially express one or more of the following markers: CD70, CD57, CD244, CD160, PD-1, CTLA4, HM3, and LAG3. In some embodiments, subpopulations of T cells may be isolated by positive or negative selection prior to genetic engineering and/or post genetic engineering.
[0403] In some embodiments, an isolated population of T cells expresses one or more of the markers including, but not limited to a CD3+, CD4+, CD8+, or a combination thereof. In some embodiments, the T cells are isolated from a donor, or subject, and first activated and stimulated to proliferate in vitro prior to undergoing gene editing.
[0404] To achieve sufficient therapeutic doses of T cell compositions, T cells are often subjected to one or more rounds of stimulation, activation and/or expansion. T cells can be activated and expanded generally using methods as described, for example, in U.S. Pat. Nos. 6,352,694; 6,534,055; 6,905,680; 6,692,964; 5,858,358; 6,887,466; 6,905,681; 7,144,575; 7,067,318; 7,172,869; 7,232,566; 7,175,843; 5,883,223; 6,905,874; 6,797,514; and 6,867,041. In some embodiments, T cells are activated and expanded for about 1 day to about 4 days, about 1 day to about 3 days, about 1 day to about 2 days, about 2 days to about 3 days, about 2 days to about 4 days, about 3 days to about 4 days, or about 1 day, about 2 days, about 3 days, or about 4 days prior to introduction of the genome editing compositions into the T cells.
[0405] In some embodiments, T cells are activated and expanded for about 4 hours, about 6 hours, about 12 hours, about 18 hours, about 24 hours, about 36 hours, about 48 hours, about 60 hours, or about 72 hours prior to introduction of the gene editing compositions into the T cells.
[0406] In some embodiments, T cells are activated at the same time that genome editing compositions are introduced into the T cells. T cell populations or isolated T cells generated by any of the gene editing methods described herein are also within the scope of the present disclosure.
[0407] In some embodiments, provided herein is a population of T cells comprising genetically engineered T cells, which comprise a disrupted endogenous CD70 gene and a nucleic acid encoding a chimeric antigen receptor (CAR), e.g., those described herein. In some embodiments, the CAR binds an antigen expressed on a pathological cell. In some embodiments, the CAR binds CD70. In other embodiments, the CAR does not bind CD70. Such a T cell population may further comprise genetically engineered T cells having one or more of the following gene edits: a disrupted endogenous programmed cell death-1 (PD-1) gene, a disrupted endogenous T cell receptor alpha chain constant region (TRAC) gene, and a disrupted endogenous beta-2-microglobulin (.beta.2M) gene. In some examples, the nucleic acid encoding the CAR may be inserted into the TRAC locus. In some embodiments, the population of T cells disclosed herein comprises genetically engineered T cells, which comprise a disrupted CD70 gene and a nucleic acid encoding a chimeric antigen receptor (CAR) that binds an antigen expressed on a pathological cell. In some embodiments, the population of T cells disclosed herein comprises genetically engineered T cells, which comprise a disrupted CD70 gene and a nucleic acid encoding a chimeric antigen receptor (CAR), wherein the CAR binds CD70. In other embodiments, the population of T cells disclosed herein comprises genetically engineered T cells that comprise a disrupted CD70 gene and a nucleic acid encoding a CAR, wherein the CAR does not bind CD70. In some embodiments, the population of T cells disclosed herein comprises genetically engineered T cells, which comprise a disrupted CD70 gene and a nucleic acid encoding a chimeric antigen receptor (CAR) that binds an antigen expressed on a pathological cell, and further comprises a disrupted PD1 gene. In some embodiments, the CAR binds CD70. In some embodiments the CAR does not bind CD70. In some aspects, the CAR binds CD19. In some embodiments, the CAR binds CD33. In some aspects, the CAR binds BCMA. Any of the just-noted engineered T cells may further comprise a disrupted T cell receptor alpha chain constant region (TRAC) gene and/or a disrupted beta-2-microglobulin (.beta.2M) gene.
[0408] In particular examples, provided herein is a population of T cells comprising genetically engineered T cells, which comprise a disrupted CD70 gene, a disrupted T cell receptor alpha chain constant region (TRAC) gene, a disrupted beta-2-microglobulin (.beta.2M) gene, a nucleic acid encoding a chimeric antigen receptor (CAR), e.g., an anti-BCMA CAR, anti-CD19 CAR, anti-CD33 CAR, or anti-CD70 CAR as described herein, and optionally a disrupted programmed cell death-1 (PD-1) gene. Any of the engineered T cells disclosed herein may contain native (undisrupted) HLA genes.
[0409] In some examples, at least 50% (e.g., 60%, 70%, 80%, 90%, or 95%) of the population of T cells express the CAR as disclosed herein and do not express a detectable level of surface CD70. Such cells may further possess the features of not expressing a detectable level of surface TCR, a detectable level of surface .beta.2M, and/or a detectable level of surface PD-1. For example, at least 50% (e.g., 60%, 70%, 80%, 90%, or 95%) of the population of T cells express the CAR as disclosed herein and do not express a detectable level of surface CD70, a detectable level of surface TCR, and a detectable level of surface .beta.2M. In some instances, at least 50% (e.g., 60%, 70%, 80%, 90%, or 95%) of the population of T cells express the CAR as disclosed herein and do not express a detectable level of surface CD70, a detectable level of surface TCR, a detectable level of surface .beta.2M, and a detectable level of PD-1.
[0410] An isolated cell expressing the CAR as described herein and does not express a detectable level of surface CD70 is also within the scope of the present disclosure. Such an isolated cell may not express a detectable level of surface TCR, a detectable level of surface .beta.2M, and/or a detectable level of surface PD-1. In some examples, the isolated cell comprises a nucleic acid encoding the CAR, which is inserted into the TRAC locus.
[0411] Also provided herein are an engineered T cell population comprising engineered T cells comprising an RNA-guided nuclease, e.g., those described herein (for example, a Cas9 nuclease), and a guide RNA (gRNA) targeting a CD70 gene (e.g., those described herein). In some instances, at least 50% (e.g., 60%, 70%, 80%, 90%, or 95%) of the T cells in the T cell population comprise the RNA-guided nuclease and the gRNA targeting the CD70 gene. Such an engineered T cell population may further comprise engineered T cells comprising a gRNA targeting a PD-1 gene, a gRNA targeting a TRAC gene, a gRNA targeting a .beta.2M gene, and/or a nucleic acid (e.g., a vector) comprising a donor template that comprises a nucleotide sequence encoding a CAR (e.g., those described herein), which optionally is flanked by left and right homology arms to the TRAC gene locus. In some examples, at least 50% (e.g., 60%, 70%, 80%, 90%, or 95%) of the T cells in the T cell population comprise the RNA-guided nuclease, the gRNA targeting the CD70 gene, and the nucleic acid coding for the CAR. When the nucleic acid coding for the CAR further comprises the left and right homology arms to the TRAC gene locus, the T cells may also comprise a gRNA targeting the TRAC gene. In addition, the T cells may further comprise a gRNA targeting a PD-1 gene, a gRNA targeting a .beta.2M gene, or a combination thereof.
[0412] Also within the scope of the present disclosure is an isolated engineered T cell comprising the RNA-guided nuclease, the gRNA targeting the CD70 gene, and optionally one or more of a gRNA targeting a PD-1 gene, a gRNA targeting a TRAC gene, a gRNA targeting a .beta.2M gene, and a nucleic acid (e.g., a vector) comprising a donor template that comprises a nucleotide sequence encoding a CAR (e.g., those described herein). The nucleotide sequence encoding the CAR may be flanked by left and right homology arms to the TRAC gene locus.
Generating CAR-T Cells
[0413] In some embodiments, the engineered T cells described herein are generated by modifying the genome of the cells. In some embodiments, a double stranded break (DSB) at a site in a target gene is induced. In some embodiments, the DSB is repaired using one or more endogenous DNA repair pathways. In some embodiments, a DNA repair pathway does not require a homologous sequence (e.g., the non-homologous end joining pathway or NHEJ pathway). In some embodiments, a repair pathway requires a homologous sequence (e.g., the homology-directed pathway or HDR pathway).
[0414] In some embodiments, the engineered T cells described herein are generated by inducing a DSB with CRISPR-Cas9 as an endonuclease, and one or more non-coding RNAs, and repairing the DSB using HDR and a donor polynucleotide template described herein.
[0415] In some embodiments, the engineered T cells described herein are generated using a gRNA complimentary to a sequence of a target gene that is a TRAC. In some embodiments, the engineered T cells described herein are generated using a TRAC gRNA spacer comprising the sequence set forth in SEQ ID NO: 98. In some embodiments, the engineered T cells described herein are generated using a TRAC gRNA comprising the sequence set forth in SEQ ID NO: 30. In some embodiments, the TRAC gRNA comprising the sequence set forth in SEQ ID NO: 98 targets the TRAC sequence set forth in SEQ ID NO: 118. In some embodiments, the TRAC gRNA comprising the sequence set forth in SEQ ID NO: 30 targets the TRAC sequence set forth in SEQ ID NO: 118.
[0416] In some embodiments, the engineered T cells described herein are generated using a TRAC gRNA spacer comprising the sequence set forth in SEQ ID NO: 108. In some embodiments, the engineered T cells described herein are generated using a TRAC gRNA comprising the sequence set forth in SEQ ID NO: 40. In some embodiments, the TRAC gRNA comprising the sequence set forth in SEQ ID NO: 108 targets the TRAC sequence set forth in SEQ ID NO: 118. In some embodiments, the TRAC gRNA comprising the sequence set forth in SEQ ID NO: 40 targets the TRAC sequence set forth in SEQ ID NO: 118.
[0417] In some embodiments, the engineered T cells described herein are generated using a gRNA complimentary to a sequence of a target gene that is a .beta.2M. In some embodiments, the engineered T cells described herein are generated using a .beta.2M gRNA spacer comprising the sequence set forth in SEQ ID NO: 99. In some embodiments, the engineered T cells described herein are generated using a .beta.2M gRNA comprising the sequence set forth in SEQ ID NO: 31. In some embodiments, the .beta.2M gRNA comprising the sequence set forth in SEQ ID NO: 99 targets the .beta.2M sequence set forth in SEQ ID NO: 119. In some embodiments, the .beta.2M gRNA comprising the sequence set forth in SEQ ID NO: 31 targets the .beta.2M sequence set forth in SEQ ID NO: 119.
[0418] In some embodiments, the engineered T cells described herein are generated using a .beta.2M gRNA spacer comprising the sequence set forth in SEQ ID NO: 109. In some embodiments, the engineered T cells described herein are generated using a .beta.2M gRNA comprising the sequence set forth in SEQ ID NO: 41. In some embodiments, the .beta.2M gRNA comprising the sequence set forth in SEQ ID NO: 109 targets the .beta.2M sequence set forth in SEQ ID NO: 119. In some embodiments, the .beta.2M gRNA comprising the sequence set forth in SEQ ID NO: 41 targets the .beta.2M sequence set forth in SEQ ID NO: 119.
[0419] In some embodiments, the engineered T cells described herein are generated using a gRNA complimentary to a sequence of a target gene that is a CD70. In some embodiments, the engineered T cells described herein are generated using a CD70 gRNA spacer comprising the sequence set forth in SEQ ID NO: 94. In some embodiments, the engineered T cells described herein are generated using a CD70 gRNA comprising the sequence set forth in SEQ ID NO: 26. In some embodiments, the CD70 gRNA comprising the sequence set forth in SEQ ID NO: 94 targets the CD70 sequence set forth in SEQ ID NO: 114. In some embodiments, the CD70 gRNA comprising the sequence set forth in SEQ ID NO: 26 targets the CD70 sequence set forth in SEQ ID NO: 114.
[0420] In some embodiments, the engineered T cells described herein are generated using a CD70 gRNA spacer comprising the sequence set forth in SEQ ID NO: 104. In some embodiments, the engineered T cells described herein are generated using a CD70 gRNA comprising the sequence set forth in SEQ ID NO: 36. In some embodiments, the CD70 gRNA comprising the sequence set forth in SEQ ID NO: 104 targets the CD70 sequence set forth in SEQ ID NO: 114. In some embodiments, the CD70 gRNA comprising the sequence set forth in SEQ ID NO: 36 targets the CD70 sequence set forth in SEQ ID NO: 114.
[0421] In some embodiments, the engineered T cells described herein are generated using a gRNA complimentary to a sequence of a target gene that is a CD70. In some embodiments, the engineered T cells described herein are generated using a CD70 gRNA spacer comprising the sequence set forth in SEQ ID NO: 95. In some embodiments, the engineered T cells described herein are generated using a CD70 gRNA comprising the sequence set forth in SEQ ID NO: 27. In some embodiments, the CD70 gRNA comprising the sequence set forth in SEQ ID NO: 95 targets the CD70 sequence set forth in SEQ ID NO: 115. In some embodiments, the CD70 gRNA comprising the sequence set forth in SEQ ID NO: 27 targets the CD70 sequence set forth in SEQ ID NO: 115.
[0422] In some embodiments, the engineered T cells described herein are generated using a CD70 gRNA spacer comprising the sequence set forth in SEQ ID NO: 105. In some embodiments, the engineered T cells described herein are generated using a CD70 gRNA comprising the sequence set forth in SEQ ID NO: 37. In some embodiments, the CD70 gRNA comprising the sequence set forth in SEQ ID NO: 105 targets the CD70 sequence set forth in SEQ ID NO: 115. In some embodiments, the CD70 gRNA comprising the sequence set forth in SEQ ID NO: 37 targets the CD70 sequence set forth in SEQ ID NO: 115.
[0423] In some embodiments, the engineered T cells described herein are generated using a gRNA complimentary to a sequence of a target gene that is a PD-1. In some embodiments, the engineered T cells described herein are generated using a PD-1 gRNA spacer comprising the sequence set forth in SEQ ID NO: 100. In some embodiments, the engineered T cells described herein are generated using a PD-1 gRNA comprising the sequence set forth in SEQ ID NO: 32. In some embodiments, the PD-1 gRNA comprising the sequence set forth in SEQ ID NO: 100 targets the .beta.2M sequence set forth in SEQ ID NO: 120. In some embodiments, the PD-1 gRNA comprising the sequence set forth in SEQ ID NO: 32 targets the PD-1 sequence set forth in SEQ ID NO: 120.
[0424] In some embodiments, the engineered T cells described herein are generated using a PD-1 gRNA spacer comprising the sequence set forth in SEQ ID NO: 110. In some embodiments, the engineered T cells described herein are generated using a PD-1 gRNA comprising the sequence set forth in SEQ ID NO: 42. In some embodiments, the PD-1 gRNA comprising the sequence set forth in SEQ ID NO: 110 targets the PD-1 sequence set forth in SEQ ID NO: 120. In some embodiments, the PD-1 gRNA comprising the sequence set forth in SEQ ID NO: 42 targets the PD-1 sequence set forth in SEQ ID NO: 120.
[0425] In some embodiments, the engineered T cells described herein are generated using a TRAC gRNA comprising the sequence set forth in SEQ ID NO: 98, a .beta.2M gRNA comprising the sequence set forth in SEQ ID NO: 99, a CD70 gRNA comprising the sequence set forth in SEQ ID NO: 94 or 95, and/or a PD-1 gRNA comprising the sequence set forth in SEQ ID NO: 100. In some embodiments, the engineered T cells described herein are generated using a TRAC gRNA comprising the sequence set forth in SEQ ID NO: 108, a .beta.2M gRNA comprising the sequence set forth in SEQ ID NO: 109, a CD70 gRNA comprising the sequence set forth in SEQ ID NO: 104 or 105, and/or a PD-1 gRNA comprising the sequence set forth in SEQ ID NO: 110.
[0426] In some embodiments, the engineered T cells described herein are generated using a TRAC gRNA comprising the sequence set forth in SEQ ID NO: 30, a .beta.2M gRNA comprising the sequence set forth in SEQ ID NO: 31, a CD70 gRNA comprising the sequence set forth in SEQ ID NO: 26 or 27, and/or a PD-1 gRNA comprising the sequence set forth in SEQ ID NO: 32. In some embodiments, the engineered T cells described herein are generated using a TRAC gRNA comprising the sequence set forth in SEQ ID NO: 40, a .beta.2M gRNA comprising the sequence set forth in SEQ ID NO: 41, a CD70 gRNA comprising the sequence set forth in SEQ ID NO: 36 or 27, and/or a PD-1 gRNA comprising the sequence set forth in SEQ ID NO: 42.
[0427] In some embodiments, the engineered T cells are generated using a donor template comprising a non-homologous sequence that is a nucleic acid encoding a CAR. In some embodiments, a donor template is comprised of homology arms that correspond to sequences in a target gene that is a TRAC. In some embodiments, a 5' homology arm (left homology arm) of the donor template comprises the sequence set forth in SEQ ID NO: 122. In some embodiments, a 3' homology arm of the donor template comprises the sequence set forth in SEQ ID NO: 125.
[0428] In some embodiments, an exogenous promoter is an EF1.alpha. promoter comprises the sequence set forth in SEQ ID NO: 123. In some embodiments, a donor template comprises the sequence set forth in SEQ ID NO: 135. In some embodiments, a donor template comprises the sequence set forth in SEQ ID NO: 156. In some embodiments, a donor template comprises the sequence set forth in SEQ ID NO: 44. In some embodiments, a donor template comprises the sequence set forth in SEQ ID NO: 55.
[0429] In some embodiments, polynucleotides encoding gRNAs, nucleases, and donor templates are introduced into cells (e.g., T cells) using conventional viral and non-viral based gene transfer methods.
[0430] In some embodiments, a polynucleotide such as a gRNA, a sgRNA, an mRNA encoding a nuclease, or a donor template are delivered to a cell using a non-viral vector delivery system. Examples of a non-viral vector delivery system include, but are not limited to, a DNA plasmid, a DNA minicircle, a naked nucleic acid, a liposome, a ribonucleoprotein particle (RNP) or a poloxamer. In some embodiments, a method of introducing polynucleotides to a cell using a non-viral vector delivery system includes electroporation, lipofection, microinjection, biolistics, or agent-enhanced uptake.
[0431] In some embodiments, a polynucleotide such as a gRNA, a sgRNA, an mRNA encoding a nuclease, or a donor template are delivered to a cell using a viral vector delivery system. Examples of a viral vector delivery system include, but are not limited to, retroviral vectors, lentiviral vectors, adenovirus vectors, poxvirus vectors, herpesvirus vectors, and adeno-associated virus (AAV) vectors.
[0432] In some embodiments, a donor template encoding a CAR construct is delivered to a cell as one or more polynucleotides. In some embodiments, a donor template encoding a CAR construct is delivered by a viral delivery vehicle. In some embodiments, a viral delivery vehicle is an adeno-associated virus (AAV) vector.
[0433] In some embodiments, an endonuclease (e.g., Cas9) is delivered to a cell as a polypeptide. In some embodiments, an endonuclease (e.g., Cas9) is delivered to a cell separately from a genome-targeting nucleic acid (e.g., a gRNA, a sgRNA). In some embodiments, an endonuclease (e.g., Cas9) is delivered to a cell as a complex with one or more genome-targeting polynucleotides (e.g., a gRNA, a sgRNA). In some embodiments, a endonuclease or a pre-complexed endonuclease is delivered by a non-viral delivery vehicle that includes, but is not limited to, a nanoparticle, a liposome, a ribonucleoprotein, a positively charged peptide, a small molecule RNA-conjugate, an aptamer-RNA chimeras, or an RNA-fusion protein complex. In some embodiments, a method of introducing an endonuclease polypeptide or a pre-complexed endonuclease polypeptide to a cell includes electroporation, lipofection, microinjection, biolistics, or agent-enhanced uptake.
[0434] In some embodiments, a Cas9 polypeptide is pre-complexed with one or more sgRNAs to form a ribonucleoprotein particle (RNP). In some embodiments, a Cas9/sgRNA RNP is formulated using a lipid nanoparticle. In some embodiments, a donor template is formulated using an AAV vector. In some embodiments, delivery to a cell of a formulated Cas9/sgRNA RNP is performed by electroporation of the cell. In some embodiments, a donor template formulated as an AAV vector is delivered prior to electroporation. In some embodiments, a donor template formulated as an AAV vector is delivered during electroporation. In some embodiments, a donor template formulated as an AAV vector is delivered following electroporation.
[0435] In some embodiments, a gene edit performed using a CRISPR/Cas9 endonuclease results in an engineered T cell with a disrupted TRAC gene. In some embodiments, a disruption of a TRAC gene results in eliminated or decreased expression of the TRAC gene. In some embodiments, a disruption of a TRAC gene disrupts or inhibits transcription and translation of an encoded gene product. In some embodiments, a disruption of a TRAC gene results in eliminated or decreased expression of a TRAC gene product. In some embodiments, eliminated or decreased expression of the TRAC gene is associated with loss of function of the TCR. In some embodiments, loss of TCR function renders an engineered T cell suitable to allogeneic transplantation (i.e., minimizing the risk of inducing GvHD). In some embodiments, a disruption of a TRAC gene is created by knocking in a CAR into the TRAC gene (e.g., using an AAV vector and a donor template). In some embodiments, a disruption in the TRAC gene expression is created by gRNAs targeting the TRAC genomic region and knocking in a CAR into the CAR gene. In some embodiments, a knock-in CAR is provided by a donor template with homology arms that correspond to sequences of the TRAC surrounding the site of a DSB.
[0436] In some embodiments, a gene edit performed using a CRISPR/Cas9 endonuclease results in an engineered T cell with a disrupted .beta.2M gene. In some embodiments, gRNAs targeting the B2M genomic region create indels in the .beta.2M gene that disrupt or inhibit transcription and translation of an encoded gene product. In some embodiments, a disruption of a .beta.2M gene results in eliminated or decreased expression of the .beta.2M polypeptide. In some embodiments, eliminated or decreased expression of the B2M polypeptide is associated with loss of function of the MHC I complex. In some embodiments, loss of MHC I function renders an engineered T cell suitable to allogeneic transplantation (i e , minimizing the risk of a host versus allogeneic T cell response). In some embodiments, loss of MHC I function results in increased persistence of an engineered T cell in an allogeneic recipient.
[0437] In some embodiments, a gene edit performed using a CRISPR/Cas9 endonuclease results in an engineered T cell with a disrupted CD70 gene. In some embodiments, gRNAs targeting the CD70 genomic region create indels in the CD70 gene that disrupt or inhibit transcription and translation of an encoded gene product. In some embodiments, a disruption of a CD70 gene results in eliminated or decreased expression of the CD70 polypeptide. In some embodiments, eliminated or decreased expression of the CD70 polypeptide is associated with enhanced cell proliferation, enhanced in vivo persistence, decreased exhaustion, and/or enhanced anti-tumor efficacy.
[0438] In some embodiments, a gene edit performed using a CRISPR/Cas9 endonuclease results in an engineered T cell with a disrupted PD-1 gene. In some embodiments, gRNAs targeting the PD-1 genomic region create indels in the PD-1 gene that disrupt or inhibit transcription and translation of an encoded gene product. In some embodiments, a disruption of a PD-1 gene results in eliminated or decreased expression of the PD-1 polypeptide.
Methods and Compositions
[0439] Provided herein, in some embodiments, are methods for treating cancer. Non-limiting examples of cancers that may be treated as provided herein include multiple myeloma, leukemia (e.g., T cell leukemia, B-cell acute lymphoblastic leukemia (B-ALL), and/or chronic lymphocytic leukemia (C-CLL)), lymphoma (e.g., B-cell non-Hodgkin's lymphoma (B-NHL), Hodgkin's lymphoma, and/or T cell lymphoma), and/or clear cell renal cell carcinoma (ccRCC). In some embodiment, the methods comprise delivering the CAR T cells (e.g., anti-BCMA, anti-CD19, anti-CD33 and/or anti-CD70 CAR T cells) of the present disclosure to a subject having multiple myeloma, leukemia, or lymphoma. Other non-limiting examples of cancers (e.g., solid tumors) that may be treated as provided herein include pancreatic cancer, gastric cancer, ovarian cancer, cervical cancer, breast cancer, renal cancer, thyroid cancer, nasopharyngeal cancer, non-small cell lung (NSCLC), glioblastoma, and/or melanoma.
[0440] CD70 has also been detected on hematological tumors and on carcinomas. The restricted expression pattern of CD70 in normal tissues and its widespread expression in various malignancies makes it an attractive target for antibody-based therapeutics. The use of CAR T cell therapy to target CD70.sup.+ cancers, however, is potentially problematic because of CD70 expression in the T cells. To address this potential problem, the present disclosure also provides CAR T cells that have been engineered to disrupt endogenous CD70 expression while at the same time expressing an anti-CD70 binding moiety (e.g., an anti-CD70 scFv).
[0441] In some embodiments, the cancer is a CD70.sup.+ cancer. In other embodiments, the cancer is a BCMA.sup.+ cancer. In some embodiments, the cancer is a CD19+cancer. In some embodiments, the cancer is a CD33+ cancer. It should be understood that other cancers, expressing other cancer antigens, may be treated using the engineered CD70 knockout CAR T cells of the present disclosure.
[0442] The methods, in some embodiments, comprise administering to a subject (e.g., a patient having a CD70.sup.+ cancer, a BCMA.sup.+ cancer, a CD19+ cancer or a CD33+ cancer) a population of CAR T cells as provided herein. In some embodiments, the methods comprise administering to a subject a population of CAR T cells comprising a CD70 gene knockout. In some embodiments, the methods comprise administering to a subject a population of CAR T cells comprising a CD70 gene knockout and a PD1 gene knockout. In some embodiments, the methods comprise implanting the cells into subject. This implanting step may be accomplished using any method of implantation known in the art. For example, the engineered cells may be injected directly in a subject's blood or otherwise administered to the subject.
[0443] As demonstrated herein, CAR T cells comprising a CD70 gene knockout exhibit extended proliferation and increased in vivo persistence. In some embodiments, CAR T cells comprising a CD70 gene knockout exhibit increased anti-tumor efficacy relative to CAR T cells expressing endogenous CD70. In some embodiments, CAR T cells comprising a CD70 gene knockout exhibit increased anti-tumor efficacy in solid tumors relative to CAR T cells expressing endogenous CD70. Without wishing to be bound by theory, the increased in vivo persistence of CAR T cells comprising a CD70 gene knockout may allow for expansion in solid tumors and therefore provide enhanced anti-tumor efficacy in such tumors relative to CAR T cells expressing endogenous CD70.
[0444] In some embodiments, the disclosure provides a method for treating a solid tumor with the CAR T cells described herein. In some embodiments, the disclosure provides a method for treating a solid tumor with the anti-CD70 CAR T cells described herein.
[0445] The step of administering may include the placement (e.g., transplantation) of cells, e.g., engineered T cells, into a subject, by a method or route that results in at least partial localization of the introduced cells at a desired site, such as tumor, such that a desired effect(s) is produced. Engineered T cells can be administered by any appropriate route that results in delivery to a desired location in the subject where at least a portion of the implanted cells or components of the cells remain viable. The period of viability of the cells after administration to a subject can be as short as a few hours, e.g., twenty-four hours, to a few days, to as long as several years, or even the life time of the subject, i.e., long-term engraftment. For example, in some aspects described herein, an effective amount of engineered T cells is administered via a systemic route of administration, such as an intraperitoneal or intravenous route.
[0446] A subject may be any subject for whom diagnosis, treatment, or therapy is desired. In some embodiments, the subject is a mammal. In some embodiments, the subject is a human.
[0447] A donor is an individual who is not the subject being treated. A donor is an individual who is not the patient. In some embodiments, a donor is an individual who does not have or is not suspected of having the cancer being treated. In some embodiments, multiple donors, e.g., two or more donors, are used.
[0448] In some embodiments, an engineered T cell population being administered according to the methods described herein comprises allogeneic T cells obtained from one or more donors. Allogeneic refers to a cell, cell population, or biological samples comprising cells, obtained from one or more different donors of the same species, where the genes at one or more loci are not identical to the recipient (e.g., subject). For example, an engineered T cell population, being administered to a subject can be derived from one or more unrelated donors, or from one or more non-identical siblings. In some embodiments, syngeneic cell populations may be used, such as those obtained from genetically identical donors, (e.g., identical twins). In some embodiments, the cells are autologous cells; that is, the engineered T cells are obtained or isolated from a subject and administered to the same subject, i.e., the donor and recipient are the same.
[0449] An effective amount refers to the amount of a population of engineered T cells needed to prevent or alleviate at least one or more signs or symptoms of a medical condition (e.g., cancer), and relates to a sufficient amount of a composition to provide the desired effect, e.g., to treat a subject having a medical condition. An effective amount also includes an amount sufficient to prevent or delay the development of a symptom of the disease, alter the course of a symptom of the disease (for example but not limited to, slow the progression of a symptom of the disease), or reverse a symptom of the disease. It is understood that for any given case, an appropriate effective amount can be determined by one of ordinary skill in the art using routine experimentation.
[0450] For use in the various aspects described herein, an effective amount of cells (e.g., engineered T cells) comprises at least 10.sup.2 cells, at least 5.times.10.sup.2 cells, at least 10.sup.3 cells, at least 5.times.10.sup.3 cells, at least 10.sup.4 cells, at least 5.times.10.sup.4 cells, at least 10.sup.5 cells, at least 2.times.10.sup.5 cells, at least 3.times.10.sup.5 cells, at least 4.times.10.sup.5 cells, at least 5.times.10.sup.5 cells, at least 6.times.10.sup.5 cells, at least 7.times.10.sup.5 cells, at least 8.times.10.sup.5 cells, at least 9.times.10.sup.5 cells, at least 1.times.10.sup.6 cells, at least 2.times.10.sup.6 cells, at least 3.times.10.sup.6 cells, at least 4.times.10.sup.6 cells, at least 5.times.10.sup.6 cells, at least 6.times.10.sup.6 cells, at least 7.times.10.sup.6 cells, at least 8.times.10.sup.6 cells, at least 9.times.10.sup.6 cells, or multiples thereof. The cells are derived from one or more donors, or are obtained from an autologous source. In some examples described herein, the cells are expanded in culture prior to administration to a subject in need thereof.
[0451] Modes of administration include injection, infusion, instillation, or ingestion. Injection includes, without limitation, intravenous, intramuscular, intra-arterial, intrathecal, intraventricular, intracapsular, intraorbital, intracardiac, intradermal, intraperitoneal, transtracheal, subcutaneous, subcuticular, intraarticular, sub capsular, subarachnoid, intraspinal, intracerebro spinal, and intrasternal injection and infusion. In some embodiments, the route is intravenous.
[0452] In some embodiments, engineered T cells are administered systemically, which refers to the administration of a population of cells other than directly into a target site, tissue, or organ, such that it enters, instead, the subject's circulatory system and, thus, is subject to metabolism and other like processes.
[0453] The efficacy of a treatment comprising a composition for the treatment of a medical condition can be determined by the skilled clinician. A treatment is considered "effective treatment," if any one or all of the signs or symptoms of, as but one example, levels of functional target are altered in a beneficial manner (e.g., increased by at least 10%), or other clinically accepted symptoms or markers of disease (e.g., cancer) are improved or ameliorated. Efficacy can also be measured by failure of a subject to worsen as assessed by hospitalization or need for medical interventions (e.g., progression of the disease is halted or at least slowed). Methods of measuring these indicators are known to those of skill in the art and/or described herein. Treatment includes any treatment of a disease in subject and includes: (1) inhibiting the disease, e.g., arresting, or slowing the progression of symptoms; or (2) relieving the disease, e.g., causing regression of symptoms; and (3) preventing or reducing the likelihood of the development of symptoms.
[0454] Combination therapies are also encompassed by the present disclosure. For example, CD70 and/or CD27 antibodies can be used to bind and/or modulate the activity of CD70 and/or
[0455] CD27 on CAR T cells and promote a decrease in exhaustion, enhanced CAR T cell expansion and increase efficacy of cancer cell killing. Thus, CD70 and/or CD27 antibodies can be administered with any CAR T cell known in the art to improve the CAR T cell function. For example, any of the engineered T cells provided herein may be administered in combination with anti-CD70 antibodies, anti-CD27 antibodies, or a combination of anti-CD70 antibodies and anti-CD27 antibodies. In some embodiments, TRAC.sup.-/.beta.2M.sup.- CAR.sup.+ T cells (e.g., anti-CD70 CAR or anti-BCMA CAR) are administered in combination with anti-CD70 and/or anti-CD27 antibodies. In some embodiments, TRAC.sup.-/.beta.2M.sup.-/CD70.sup.- CAR.sup.+ T cells (e.g., anti-CD70 CAR or anti-BCMA CAR) are administered in combination with anti-CD70 and/or anti-CD27 antibodies. In some embodiments, TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CAR.sup.+ T cells (e.g., anti-CD70 CAR or anti-BCMA CAR) are administered in combination with anti-CD70 and/or anti-CD27 antibodies. In some embodiments, TRAC.sup.-/.beta.2M.sup.-/CD70.sup.- CAR.sup.+ T cells (e.g., anti-CD70 CAR or anti-BCMA CAR) are administered in combination with anti-CD70 and/or anti-CD27 antibodies. In some embodiments, the antibodies administered in combination can be Varlilumab.
[0456] In some embodiments, the disclosure provides a method of reducing exhaustion of T cells comprising disrupting the CD70 gene in the T cells. In some embodiments, the disclosure provides a method of increasing proliferation of T cells comprising disrupting the CD70 gene in the T cells. In some embodiments, the disclosure provides a method of increasing cytotoxicity of T cells comprising disrupting the CD70 gene in the T cells. In some embodiments, the disclosure provides a method of overcoming inhibitory effect of an immune checkpoint (e.g., PD-1) in T cells comprising disrupting the CD70 gene in the T cells.
Other Embodiments
[0457] The disclosure relates to the following embodiments. Throughout this section, the term embodiment is abbreviated as `E` followed by an ordinal. For example, E1 is equivalent to Embodiment 1. [0458] E1. An engineered T cell comprising a disrupted CD70 gene, a disrupted programmed cell death-1 (PD-1) gene, and a nucleic acid encoding a chimeric antigen receptor (CAR). [0459] E2. An engineered T cell comprising a disrupted CD70 gene and a nucleic acid encoding a chimeric antigen receptor (CAR) that binds CD70. [0460] E3. An engineered T cell comprising a disrupted CD70 gene and a nucleic acid encoding a chimeric antigen receptor (CAR) that does not bind CD70. [0461] E4. The engineered T cell of embodiment 2 or 3, further comprising a disrupted PD-1 gene. [0462] E5. The engineered T cell of any one of embodiments 1-4 further comprising a disrupted T cell receptor alpha chain constant region (TRAC) gene. [0463] E6. The engineered T cell of any one of embodiments 1-5 further comprising a disrupted beta-2-microglobulin (.beta.2M) gene. [0464] E7. An engineered T cell, comprising
[0465] a disrupted T cell receptor alpha chain constant region (TRAC) gene;
[0466] a disrupted beta-2-microglobulin (.beta.2M) gene;
[0467] a disrupted CD70 gene; and
[0468] a nucleic acid encoding a chimeric antigen receptor (CAR). [0469] E8. The engineered T cell of embodiment 7, wherein the nucleic acid encoding the CAR is inserted into the TRAC gene. [0470] E9. The engineered T cell of embodiment 7 or 8, further comprising a disrupted PD-1 gene. [0471] E10. The engineered T cell of any one of embodiments 1-9, wherein the CAR comprises an ectodomain that comprises an anti-CD70 antibody, optionally wherein the anti-CD70 antibody is an anti-CD70 single-chain variable fragment (scFv). [0472] E11. The engineered T cell of embodiment 10, wherein the anti-CD70 scFv comprises the same heavy chain variable region (VH) complementarity determining regions (CDRs) and the same light chain variable region (VL) CDRs as a reference antibody, wherein the reference antibody comprises a VH set forth as SEQ ID NO: 51 and a VL set forth as SEQ ID NO: 52. [0473] E12. The engineered T cell of embodiment 11, wherein the anti-CD70 scFv comprises the same VH and VL chains as the reference antibody. [0474] E13. The engineered T cell of embodiment 11, wherein the anti-CD70 scFv comprises the amino acid sequence of SEQ ID NO: 48 or 50. [0475] E14. The engineered T cell of embodiment 11, wherein the anti-CD70 scFv comprises the amino acid sequence of SEQ ID NO: 50. [0476] E15. The engineered T cell of any one of embodiments 1-9, wherein the CAR comprises an ectodomain that comprises an anti-BCMA antibody, optionally wherein the anti-BCMA antibody is an anti-BCMA single-chain variable fragment (scFv). [0477] E16. The engineered T cell of embodiment 15, wherein the anti-BCMA scFv comprises the same VH complementarity determining regions (CDRs) and the same VL CDRs as a reference antibody, wherein the reference antibody comprises a VH) set forth as SEQ ID NO: 60 and a VL set forth as SEQ ID NO: 61. [0478] E17. The engineered T cell of embodiment 16, wherein the anti-BCMA scFv comprises the same VH and VL chains as the reference antibody. [0479] E18. The engineered T cell of embodiment 16, wherein the anti-BCMA scFv comprises the amino acid sequence of SEQ ID NO: 59. [0480] E19. The engineered T cell of any one of embodiments 1-18, wherein the CAR comprises a CD28 or 41BB co-stimulatory domain and optionally a CD3t signaling domain. [0481] E20. The engineered T cell of any one of embodiments 5-19, wherein the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 44 or 55 and/or the nucleic acid encoding the CAR comprises the nucleotide sequence of SEQ ID NO: 45 or 56. [0482] E21. The engineered T cell of any one of embodiments 6-20, wherein the disrupted .beta.2M gene comprises gene at least one nucleotide sequence selected from any one of SEQ ID NOS: 9-14. [0483] E22. The engineered T cell of any one of embodiments 1-21, wherein the engineered T cell maintains cytotoxicity following 5 rechallenges with a cancer cell. [0484] E23. The engineered T cell of embodiment 22, wherein the engineered T cell maintains cytotoxicity following 10 rechallenges with a cancer cell. [0485] E24. A population of cells comprising engineered T cells that comprise a disrupted CD70 gene, a disrupted programmed cell death-1 (PD-1) gene, and a nucleic acid encoding a chimeric antigen receptor (CAR). [0486] E25. A population of cells comprising engineered T cells that comprise a disrupted CD70 gene, and a nucleic acid encoding a chimeric antigen receptor (CAR) that binds CD70. [0487] E26. A population of cells comprising engineered T cells that comprise a disrupted CD70 gene and a nucleic acid encoding a chimeric antigen receptor (CAR) that does not bind CD70. [0488] E27. The population of cells of embodiment 25 or 26 further comprising a disrupted programmed cell death-1 (PD-1) gene. [0489] E28. The population of cells of any one of embodiments 24-27 further comprising a disrupted T cell receptor alpha chain constant region (TRAC) gene. [0490] E29. The population of cells of any one of embodiments 24-28 further comprising a disrupted beta-2-microglobulin (.beta.2M) gene. [0491] E30. A population of cells comprising
[0492] engineered T cells that comprise
[0493] a disrupted T cell receptor alpha chain constant region (TRAC) gene;
[0494] a disrupted beta-2-microglobulin (.beta.2M) gene;
[0495] a disrupted CD70 gene; and
[0496] a nucleic acid encoding a chimeric antigen receptor (CAR). [0497] E31. The population of cells of embodiment 30, wherein the nucleic acid encoding the CAR is inserted into the TRAC gene. [0498] E32. The population of cells of embodiment 30 or 31, wherein the engineered T cells further comprise a disrupted programmed cell death-1 (PD-1) gene. [0499] E33. The population of cells of any one of embodiments 24 or 27-32, wherein the CAR comprises an ectodomain that comprises an anti-CD70 antibody, optionally wherein the anti-CD70 antibody is an anti-CD70 single-chain variable fragment (scFv). [0500] E34. The population of cells of embodiment 33, wherein the anti-CD70 scFv comprises the same VH complementarity determining regions (CDRs) and the same VL CDRs as a reference antibody, wherein the reference antibody comprises a VH set forth as SEQ ID NO: 51 and a VL set forth as SEQ ID NO: 52. [0501] E35. The population of cells of embodiment 34, wherein the anti-CD70 scFv comprises the same VH and VL chains as the reference antibody. [0502] E36. The population of cells of embodiment 35, wherein the anti-CD70 scFv comprises the amino acid sequence of SEQ ID NO: 48 or 50. [0503] E37. The population of cells of embodiment 35, wherein the anti-CD70 scFv comprises the amino acid sequence of SEQ ID NO: 50. [0504] E38. The population of cells of any one of embodiments 24-32, wherein the CAR comprises an ectodomain that comprises an anti-BCMA antibody, optionally wherein the anti-BCMA antibody is an anti-BCMA single-chain variable fragment (scFv). [0505] E39. The population of cells of embodiment 38, wherein the anti-BCMA scFv comprises the same VH complementarity determining regions (CDRs) and the same VL CDRs as a reference antibody, wherein the reference antibody comprises a VH set forth as SEQ ID NO: 60 and a VL set forth as SEQ ID NO: 61. [0506] E40. The population of cells of embodiment 39, wherein the anti-BCMA scFv comprises the same VH and VL chains as the reference antibody. [0507] E41. The population of cells of embodiment 39, wherein the anti-BCMA scFv comprises the amino acid sequence of SEQ ID NO: 59. [0508] E42. The population of cells of any one of embodiments 28-41, wherein the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 44 or 55 and/or the nucleic acid encoding the CAR comprises the nucleotide sequence of SEQ ID NO: 45 or 56. [0509] E43. The population of cells of any one of embodiments 29-42, wherein the disrupted .beta.2M gene comprises gene at least one nucleotide sequence selected from any one of SEQ ID NOS: 9-14. [0510] E44. The population of cells of any one of embodiments 24-43, wherein at least 50% of the engineered T cells do not express a detectable level of TCR surface protein, do not express a detectable level of .beta.2M surface protein, do not express a detectable level of CD70 surface protein, do not express a detectable level of PD-1 surface protein, and/or express a detectable level of the CAR. [0511] E45. The population of cells of embodiment 44, wherein 50%-70%, of the engineered T cells do not express a detectable level of TCR surface protein, do not express a detectable level of .beta.2M surface protein, do not express a detectable level of CD70 surface protein, do not express a detectable level of PD-1 surface protein, and/or express a detectable level of the CAR. [0512] E46. The population of cells of any one of embodiments 28-45, wherein at least 90%, optionally 90%-100%, of the engineered T cells do not express a detectable level of TCR surface protein. [0513] E47. The population of cells of any one of embodiments 29-46, wherein at least 60%, optionally 60%-75%, of the engineered T cells do not express a detectable level of .beta.2M surface protein. [0514] E48. The population of cells of any one of embodiments 24-47, wherein at least 80%, optionally 80%-100%, of the engineered T cells do not express a detectable level of CD70 surface protein. [0515] E49. The population of cells of any one of embodiments 1-48, wherein at least 80%, optionally 80%-95%, of the engineered T cells express a detectable level of the CAR. [0516] E50. The population of cells of any one of embodiments 24-49, wherein the engineered T cells exhibit at least 20% greater cellular proliferative capacity, relative to control T cells. [0517] E51. The population of cells of any one of embodiments 24-50, wherein the engineered T cells exhibit at least 20% greater cellular lysis capability, relative to control T cells. [0518] E52. The population of cells of any one of embodiments 24-51, wherein the level of cytokines secreted by the engineered T cells are at least 2-fold greater than the level of cytokines secreted by control T cells. [0519] E53. The population of any one of embodiments 24-52, wherein the engineered T cells exhibit reduced cellular exhaustion, relative to control T cells. [0520] E54. The population of cells of embodiment 53, wherein the engineered T cells express reduced levels of LAG3, relative to control T cells. [0521] E55. The population of cells of any one of embodiments 54, wherein the control T cells are engineered T cells that express endogenous CD70 protein. [0522] E56. The population of cells of any one of embodiments 24-55, wherein the engineered T cells maintain cytokine-dependent proliferation. [0523] E57. The population of cells of any one of embodiments 24-56, wherein the engineered T cells maintain cytotoxicity following 5 rechallenges with a cancer cell. [0524] E58. The population of cells of embodiment 47, wherein the engineered T cells maintain cytotoxicity following 10 rechallenges with a cancer cell. [0525] E59. A method comprising administering to a subject the population of cells of any one of embodiments 24-58. [0526] E60. The method of embodiment 59, wherein the engineered T cells are engineered human T cells. [0527] E61. The method of embodiment 59 or 60, wherein the subject has a cancer. [0528] E62. The method of embodiment 61, wherein the cancer expresses CD70 and/or BCMA. [0529] E63. The method of any one of embodiments 59-62, wherein the population of cells is administered to the subject in an amount effective to treat the cancer. [0530] E64. The method of any one of embodiments 59-63, wherein the cancer is a solid tumor malignancy or a hematological malignancy. [0531] E65. The method embodiment 64, wherein the solid tumor malignancy is selected from the group consisting of: ovarian tumor, pancreatic tumor, kidney tumor, lung tumor, and intestinal tumor. [0532] E66. The method of embodiment 63, wherein the population of cells is administered to the subject in an amount effective to reduce the volume of a tumor in the subject. [0533] E67. A method for producing an engineered T cell, the method comprising
[0534] (a) delivering to a T cell [0535] an RNA-guided nuclease, [0536] a gRNA targeting a CD70 gene, and [0537] a vector comprising a donor template that comprises a nucleic acid encoding a CAR; and
[0538] (b) producing an engineered T cell comprising a disrupted CD70 gene and expressing the CAR. [0539] E68. The method of embodiment 67, further comprising in step (a) delivering to the T cell a gRNA targeting a PD-1 gene; wherein the engineered T cell of step (b) further comprises a disrupted PD-1 gene. [0540] E69. The method of embodiment 67 or embodiment 68, further comprising in step (a) delivering to the T cell a gRNA targeting a TRAC gene; wherein the engineered T cell of step (b) further comprises a disrupted TRAC gene. [0541] E70. The method of embodiment 69, wherein the nucleic acid encoding the CAR is flanked by left and right homology arms to the TRAC gene locus; and wherein the engineered T cell of step (b) comprises the nucleic acid encoding the CAR inserted into the TRAC gene locus. [0542] E71. The method of any one of embodiments embodiment 67-70, further comprising in step
[0543] (a) delivering to the T cell a gRNA targeting a .beta.2M gene; wherein the engineered T cell of step
[0544] (b) further comprises a disrupted .beta.2M gene. [0545] E72. A method for producing an engineered T cell, the method comprising
[0546] (a) delivering to a T cell
[0547] an RNA-guided nuclease,
[0548] a gRNA targeting a TRAC gene,
[0549] a gRNA targeting a .beta.2M gene,
[0550] a gRNA targeting a CD70 gene, and
[0551] a vector comprising a donor template that comprises a nucleic acid encoding a CAR; and
[0552] (b) producing an engineered T cell. [0553] E73. The method of embodiment 72, wherein the nucleic acid encoding the CAR is flanked by left and right homology arms to the TRAC gene locus. [0554] E74. The method of embodiment 72 or 73 further comprising delivering to the T cell a gRNA targeting a PD-1 gene. [0555] E75. The method of any one of embodiments 67-74, wherein the RNA-guided nuclease is a Cas9 nuclease, optionally a S. pyogenes Cas9 nuclease. [0556] E76. The method of any one of embodiments 69-75, wherein the gRNA targeting the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 98 or targets the nucleotide sequence of SEQ ID NO: 118, and optionally wherein the gRNA targeting the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 30. [0557] E77. The method of any one of embodiments 71-76, wherein the gRNA targeting the .beta.2M gene comprises the nucleotide sequence of SEQ ID NO: 99 or targets the nucleotide sequence of SEQ ID NO: 119, and optionally wherein the gRNA targeting the .beta.2M gene comprises the nucleotide sequence of SEQ ID NO: 31. [0558] E78. The method of any one of embodiments 67-77, wherein the gRNA targeting the CD70 gene comprises the nucleotide sequence of SEQ ID NOS: 94 or 95 or targets the nucleotide sequence of SEQ ID NO: 114 or 115, and optionally wherein the gRNA targeting the CD70 gene comprises the nucleotide sequence of SEQ ID NOS: 26 or 27. [0559] E79. The method of any one of embodiments 68-71 and 74-78, wherein the gRNA targeting the PD-1 gene comprises the nucleotide sequence of SEQ ID NO: 100 or targets the nucleotide sequence of SEQ ID NO: 120, and optionally wherein the gRNA targeting the PD-1 gene comprises the nucleotide sequence of SEQ ID NO: 32. [0560] E80. The method of any one of embodiments 67-79, wherein the CAR comprises an ectodomain that comprises an anti-CD70 antibody, optionally wherein the anti-CD70 antibody is an anti-CD70 single-chain variable fragment (scFv). [0561] E81. The method of embodiment 80, wherein the anti-CD70 scFv comprises the same VH complementarity determining regions (CDRs) and the same VL CDRs as a reference antibody, wherein the reference antibody comprises a VH set forth as SEQ ID NO: 51 and a VL set forth as SEQ ID NO: 52. [0562] E82. The method of embodiment 81, wherein the anti-CD70 scFv comprises the same VH and VL chains as the reference antibody. [0563] E83. The method of embodiment 81, wherein the anti-CD70 scFv comprises the amino acid sequence of SEQ ID NO: 48 or 50. [0564] E84. The method of embodiment 81, wherein the anti-CD70 scFv comprises the amino acid sequence of SEQ ID NO: 50. [0565] E85. The method of any one of embodiments 67-79, wherein the CAR comprises an ectodomain that comprises an anti-BCMA antibody, optionally wherein the anti-BCMA antibody is an anti-BCMA single-chain variable fragment (scFv). [0566] E86. The method of embodiment 85, wherein the anti-BCMA scFv comprises the same VH complementarity determining regions (CDRs) and the same VL CDRs as a reference antibody, wherein the reference antibody comprises a VH set forth as SEQ ID NO: 60 and a VL set forth as SEQ ID NO: 61. [0567] E87. The method of embodiment 86, wherein the anti-BCMA scFv comprises the same VH and VL chains as the reference antibody. [0568] E88. The method of embodiment 86, wherein the anti-BCMA scFv comprises the amino acid sequence of SEQ ID NO: 59. [0569] E89. The method of any one of embodiments 67-88, wherein the CAR further comprises a CD28 or 41BB co-stimulatory domain and optionally a CD3z signaling domain. [0570] E90. The method of embodiment 72, wherein the vector comprises a nucleic acid encoding a CAR that comprises the amino acid sequence of SEQ ID NO: 46. [0571] E91. The method of embodiment 72, wherein the vector comprises a nucleic acid encoding a CAR that comprises the amino acid sequence of SEQ ID NO: 57. [0572] E92. An engineered T cell comprising an RNA-guided nuclease and a gRNA targeting a CD70 gene, optionally wherein the gRNA targeting the CD70 gene comprises the nucleotide sequence of SEQ ID NOS: 94 or 95 or targets the nucleotide sequence of SEQ ID NO: 114 or 115, and optionally wherein the gRNA targeting the CD70 gene comprises the nucleotide sequence of SEQ ID NOS: 26 or 27. [0573] E93. The engineered T cell of embodiment 92 further comprising a gRNA targeting a PD-1 gene, optionally wherein the gRNA targeting the PD-1 gene comprises the nucleotide sequence of SEQ ID NO: 100 or targets the nucleotide sequence of SEQ ID NO: 120, and optionally wherein the gRNA targeting the PD-1 gene comprises the nucleotide sequence of SEQ ID NO: 32. [0574] E94. The engineered T cell of embodiment 92 or 93 further comprising a gRNA targeting a TRAC gene, optionally wherein the gRNA targeting the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 98 or targets the nucleotide sequence of SEQ ID NO: 118, and optionally wherein the gRNA targeting the TRAC gene comprises the nucleotide sequence of SEQ ID NO: 30. [0575] E95. The engineered T cell of any one of embodiments 92-94 further comprising a gRNA targeting a .beta.2M gene, optionally wherein the gRNA targeting the .beta.2M gene comprises the nucleotide sequence of SEQ ID NO: 99 or targets the nucleotide sequence of SEQ ID NO: 119, and optionally wherein the gRNA targeting the .beta.2M gene comprises the nucleotide sequence of SEQ ID NO: 31. [0576] E96. The engineered T cell of any one of embodiments 92-95, wherein the RNA-guided nuclease is a Cas9 nuclease, optionally a S. pyogenes Cas9 nuclease. [0577] E97. The engineered T cell of any one of embodiments 92-96 further comprising a vector comprising a donor template that comprises a nucleic acid encoding a CAR, optionally wherein the nucleic acid encoding the CAR is flanked by left and right homology arms to the TRAC gene locus. [0578] E98. The engineered T cell of embodiment 97, wherein the CAR comprises an ectodomain that comprises an anti-CD70 antibody, optionally wherein the anti-CD70 antibody is an anti-CD70 single-chain variable fragment (scFv). [0579] E99. The engineered T cell of embodiment 98, wherein the anti-CD70 scFv comprises the same VH complementarity determining regions (CDRs) and the same VL CDRs as a reference antibody, wherein the reference antibody comprises a VH set forth as SEQ ID NO: 51 and a VL set forth as SEQ ID NO: 52. [0580] E100. The engineered T cell of embodiment 99, wherein the anti-CD70 scFv comprises the same VH and VL chains as the reference antibody. [0581] E101. The engineered T cell of embodiment 99, wherein the anti-CD70 scFv comprises the amino acid sequence of SEQ ID NO: 48 or 50. [0582] E102. The engineered T cell of embodiment 99, wherein the anti-CD70 scFv comprises the amino acid sequence of SEQ ID NO: 50. [0583] E103. The engineered T cell of embodiment 97 wherein the CAR comprises an ectodomain that comprises an anti-BCMA antibody, optionally wherein the anti-BCMA antibody is an anti-BCMA single-chain variable fragment (scFv). [0584] E104. The engineered T cell of embodiment 103, wherein the anti-BCMA scFv comprises the same VH complementarity determining regions (CDRs) and the same VL CDRs as a reference antibody, wherein the reference antibody comprises a VH set forth as SEQ ID NO: 60 and a VL set forth as SEQ ID NO: 61. [0585] E105. The engineered T cell of embodiment 104, wherein the anti-BCMA scFv comprises the same VH and VL chains as the reference antibody. [0586] E106. The engineered T cell of embodiment 104, wherein the anti-BCMA scFv comprises the amino acid sequence of SEQ ID NO: 59. [0587] E107. The engineered T cell of embodiment 97, wherein the vector comprises a nucleic acid encoding a CAR that comprises the amino acid sequence of SEQ ID NO: 46 or 57. [0588] E108. A method of increasing proliferation or reducing exhaustion of T cells, the method comprising disrupting the CD70 gene in the T cells. [0589] E109. The method of embodiment 108 further comprising disrupting in the T cells at least one gene selected from the group consisting of: programmed cell death-1 (PD-1) gene, T cell receptor alpha chain constant region (TRAC) gene, and beta-2-microglobulin (.beta.2M) gene. [0590] E110. The method of any one of embodiments 108-109 further comprising expressing in the T cells a nucleic acid encoding a chimeric antigen receptor (CAR). [0591] E111. The method of any one of embodiments 108-110, wherein the CD70 gene is disrupted by CRISPR/Cas gene editing. [0592] E112. The method of any one of embodiments 110-111, wherein the PD-1, TRAC, and/or .beta.2M gene is disrupted by CRISPR/Cas gene editing. [0593] E113. A method for treating cancer in a subject, comprising administering to the patient a population of cells comprising engineered T cells, wherein the engineered T cells comprise a disrupted CD70 gene and a nucleic acid encoding a CAR, thereby treating cancer in the subject. [0594] 114. The method of embodiment 113, wherein the CAR binds CD70. [0595] E115. The method of embodiment 113, wherein the CAR does not bind CD70. [0596] E116. The method of any one of embodiments 113-115, wherein the engineered T cells further comprise a disrupted TRAC gene. [0597] E117. The method of any one of embodiments 113-116, wherein the engineered T cells further comprise a disrupted B2M gene. [0598] E118. The method of any one of embodiments 113-116, wherein the engineered T cells further comprise a disrupted PD-1 gene. [0599] E119. A method for treating cancer in a subject, comprising administering to the patient a population of cells comprising engineered T cells, wherein the engineered T cells comprise:
[0600] (i) a disrupted TRAC gene;;
[0601] (ii) a disrupted B2M gene;
[0602] (iii) a disrupted CD70 gene; and
[0603] (iv) a nucleic acid encoding a CAR;
[0604] thereby treating the cancer in the subject. [0605] E120. The method of any one of embodiments 113-114 and 116-119, wherein the CAR comprises (a) an ectodomain that comprises an anti-CD70 antigen-binding fragment, (b) a CD8 transmembrane domain, and (c) an endodomain that comprises a 41BB co-stimulatory domain and a CD3z co-stimulatory domain. [0606] E121. The method of embodiment 119 or 120, wherein the disrupted TRAC gene comprises the nucleic acid encoding the CAR. [0607] E122. The method of any one of embodiments 120-121, wherein the anti-CD70 antibody is an anti-CD70 scFv. [0608] E123. The method of embodiment 122, wherein the anti-CD70 scFv comprises the same heavy chain variable region (VH) complementarity determining regions (CDRs) and the same light chain variable region (VL) CDRs as a reference antibody, wherein the reference antibody comprises a VH set forth as SEQ ID NO: 51 and a VL set forth as SEQ ID NO: 52. [0609] E124. The method of embodiment 123, wherein the anti-CD70 scFv comprises the same VH and VL chains as the reference antibody. [0610] E125. The method of embodiment 122, wherein the anti-CD70 scFv comprises the amino acid sequence of SEQ ID NO: 48 or 50. [0611] E126. The method of embodiment 122, wherein the anti-CD70 scFv comprises the amino acid sequence of SEQ ID NO: 50. [0612] E127. The method of any one of embodiments 113 and 115-118, wherein the CAR comprises an ectodomain that comprises an anti-BCMA antibody, optionally wherein the anti-BCMA antibody is an anti-BCMA single-chain variable fragment (scFv). [0613] E128. The method of embodiment 127, wherein the anti-BCMA scFv comprises the same VH complementarity determining regions (CDRs) and the same VL CDRs as a reference antibody, wherein the reference antibody comprises a VH) set forth as SEQ ID NO: 60 and a VL set forth as SEQ ID NO: 61. [0614] E129. The method of embodiment 128, wherein the anti-BCMA scFv comprises the same VH and VL chains as the reference antibody. [0615] E130. The method of embodiment 127, wherein the anti-BCMA scFv comprises the amino acid sequence of SEQ ID NO: 59. [0616] E131. The method of any one of embodiments 113-130, wherein the engineered T cells are engineered human T cells. [0617] E132. The method of any one of embodiments 113-131, wherein the cancer expresses CD70 and/or BCMA. [0618] E133. The method of any one of embodiments 113-132, wherein the population of cells is administered to the subject in an amount effective to treat the cancer. [0619] E134. The method of any one of embodiments 113-133, wherein the cancer is a solid tumor malignancy or a hematological malignancy. [0620] E135. The method embodiment 134, wherein the solid tumor malignancy is selected from the group consisting of: ovarian tumor, pancreatic tumor, kidney tumor, lung tumor, and intestinal tumor. [0621] E136. A population of cells comprising engineered T cells, wherein the engineered T cells comprise:
[0622] (i) a disrupted TRAC gene;;
[0623] (ii) a disrupted beta-2-microglobulin (B2M) gene;
[0624] (iii) a disrupted CD70 gene
[0625] (iv) a nucleic acid encoding a CAR comprising (a) an ectodomain that comprises an anti-CD70 antigen-binding fragment, (b) a CD8 transmembrane domain, and (c) an endodomain that comprises a 41BB co-stimulatory domain and a CD3z co-stimulatory domain. [0626] E137. The population of cells of embodiment 136, wherein the disrupted TRAC gene comprises the nucleic acid encoding the CAR. [0627] E138. The population of cells of any one of embodiments 136-137, wherein the engineered T cells are human T cells. [0628] E139. An engineered T cell comprising:
[0629] (i) a disrupted TRAC gene, wherein the disrupted TRAC gene comprises a nucleic acid encoding a CAR comprising the amino acid sequence set forth in SEQ ID NO: 46;
[0630] (ii) a disrupted B2M gene; and
[0631] (iii) a disrupted CD70 gene. [0632] E140. The engineered T cell of embodiment 139, wherein the nucleic acid encoding the CAR comprises a sequence at least 80%, at least 90%, at least 95%, at least 96%, at least 97%, at least 98% or at least 99% identical to SEQ ID NO: 45. [0633] E141. An engineered T cell comprising:
[0634] (i) a disrupted TRAC gene, wherein the disrupted TRAC gene comprises a nucleic acid encoding a CAR, wherein the nucleic acid sequence is at least 90% identical to SEQ ID NO: 45;
[0635] (ii) a disrupted B2M gene; and
[0636] (iii) a disrupted CD70 gene. [0637] E142. The engineered T cell of any one of embodiments 139-141, wherein the disrupted TRAC gene comprises a donor sequence comprising the nucleotide sequence set forth in SEQ ID NO: 45 or SEQ ID NO: 44. [0638] E143. An engineered T cell comprising:
[0639] (i) a disrupted TRAC gene comprising the nucleic acid sequence of SEQ ID NO: 44;
[0640] (ii) a disrupted B2M gene; and
[0641] (iii) a disrupted CD70 gene. [0642] E144. The engineered T cell of any one of embodiments 139-143, wherein the T cell is a human T cell.
EXAMPLES
Example 1. Efficient Knockout of CD70 by Cas9:sgRNA RNPs in T Cells
[0643] This example describes efficient editing of the CD70 gene in primary human T cells ex vivo using CRISPR/Cas9 gene editing. Genomic segments of the CD70 gene containing the first three (3) protein coding exons were used as input in gRNA design software. The genomic segments also included flanking splice site acceptor/donor sequences. Desired gRNAs were those that would lead to insertions or deletions in the coding sequence, disrupting the amino acid sequence of CD70, leading to out of frame/loss of function allele(s) (referred to as "CD70 knockout" alleles). All seven (7) in silico-identified gRNA spacer sequences targeting the CD70 gene were synthesized, and the gRNAs were specifically modified, as indicated in Table 5. While the gRNAs in Table 5 were modified with 2'-O-methyl phosphorothioate modifications, unmodified gRNAs, or gRNAs with other modifications, may be used. See also PCT/IB2018/001619, filed May 11, 2018, herein incorporated in its entirety by this reference.
TABLE-US-00005 TABLE 5 CD70 gRNA Sequences/Target Sequences gRNA Sequences Name Unmodified Sequence Modified Sequence CD70 sgRNA (E1_T1) UCACCAAGCCCGCGACCAAUg U*C*A*CCAAGCCCGCGACCA uuuuagagcuagaaauagcaaguuaaaaua AUguuuuagagcuagaaauagcaaguuaa aggcuaguccguuaucaacuugaaaaagug aauaaggcuaguccguuaucaacuugaaaa gcaccgagucggugcUUUU aguggcaccgagucggugcU*U*U*U (SEQ ID NO: 23) (SEQ ID NO: 33) CD70 sgRNA (E1_T1) spacer UCACCAAGCCCGCGACCAAU U*C*A*CCAAGCCCGCGACCA (SEQ ID NO: 91) AU (SEQ ID NO: 101) CD70 sgRNA (E1_T3) AUCACCAAGCCCGCGACCAAg A*U*C*ACCAAGCCCGCGACC uuuuagagcuagaaauagcaaguuaaaaua AAguuuuagagcuagaaauagcaaguuaa aggcuaguccguuaucaacuugaaaaagug aauaaggcuaguccguuaucaacuugaaaa gcaccgagucggugcUUUU aguggcaccgagucggugcU*U*U*U (SEQ ID NO: 24) (SEQ ID NO: 34) CD70 sgRNA (E1_T3) spacer AUCACCAAGCCCGCGACCAA A*U*C*ACCAAGCCCGCGACC (SEQ ID NO: 92) AA (SEQ ID NO: 102) CD70 sgRNA (E1_T4) CGGUGCGGCGCAGGCCCUAU C*G*G*UGCGGCGCAGGCCCU guuuuagagcuagaaauagcaaguuaaaau AUguuuuagagcuagaaauagcaaguuaa aaggcuaguccguuaucaacuugaaaaagu aauaaggcuaguccguuaucaacuugaaaa ggcaccgagucggugcUUUU aguggcaccgagucggugcU*U*U*U (SEQ ID NO: 25) (SEQ ID NO: 35) CD70 sgRNA (E1_T4) spacer CGGUGCGGCGCAGGCCCUAU C*G*G*UGCGGCGCAGGCCCU (SEQ ID NO: 93) AU (SEQ ID NO: 103) CD70 sgRNA (E1_T7)); also GCUUUGGUCCCAUUGGUCGC G*C*U*UUGGUCCCAUUGGUC referred to as: T7 guuuuagagcuagaaauagcaaguuaaaau GCguuuuagagcuagaaauagcaaguuaa aaggcuaguccguuaucaacuugaaaaagu aauaaggcuaguccguuaucaacuugaaaa ggcaccgagucggugcUUUU aguggcaccgagucggugcU*U*U*U (SEQ ID NO: 26) (SEQ ID NO: 36) CD70 sgRNA (E1_T7) spacer GCUUUGGUCCCAUUGGUCGC G*C*U*UUGGUCCCAUUGGUC (SEQ ID NO: 94) GC (SEQ ID NO: 104) CD70 sgRNA (E1_T8); also GCCCGCAGGACGCACCCAUAg G*C*C*CGCAGGACGCACCCA referred to as: T8 uuuuagagcuagaaauagcaaguuaaaaua UAguuuuagagcuagaaauagcaaguuaa aggcuaguccguuaucaacuugaaaaagug aauaaggcuaguccguuaucaacuugaaaa gcaccgagucggugcUUUU aguggcaccgagucggugcU*U*U*U (SEQ ID NO: 27) (SEQ ID NO: 37) CD70 sgRNA (E1_T8) spacer GCCCGCAGGACGCACCCAUA G*C*C*CGCAGGACGCACCCA (SEQ ID NO: 95) UA (SEQ ID NO: 105) CD70 sgRNA (E1_T10) GUGCAUCCAGCGCUUCGCAC G*U*G*CAUCCAGCGCUUCGC guuuuagagcuagaaauagcaaguuaaaau ACguuuuagagcuagaaauagcaaguuaa aaggcuaguccguuaucaacuugaaaaagu aauaaggcuaguccguuaucaacuugaaaa ggcaccgagucggugcUUUU aguggcaccgagucggugcU*U*U*U (SEQ ID NO: 28) (SEQ ID NO: 38) CD70 sgRNA (E1_T10) spacer GUGCAUCCAGCGCUUCGCAC G*U*G*CAUCCAGCGCUUCGC (SEQ ID NO: 96) AC (SEQ ID NO: 106) CD70 sgRNA (E3_T1) CAGCUACGUAUCCAUCGUGA C*A*G*CUACGUAUCCAUCGU guuuuagagcuagaaauagcaaguuaaaau GAguuuuagagcuagaaauagcaaguuaa aaggcuaguccguuaucaacuugaaaaagu aauaaggcuaguccguuaucaacuugaaaa ggcaccgagucggugcUUUU aguggcaccgagucggugcU*U*U*U (SEQ ID NO: 29) (SEQ ID NO: 39) CD70 sgRNA (E3_T1) spacer CAGCUACGUAUCCAUCGUGA C*A*G*CUACGUAUCCAUCGU (SEQ ID NO: 97) GA (SEQ ID NO: 107) Target Sequences Name Target Sequence (PAM) CD70 sgRNA (E1_T1) TCACCAAGCCCGCGACCAAT (GGG) (SEQ ID NO: 111) CD70 sgRNA (E1_T3) ATCACCAAGCCCGCGACCAA (TGG) (SEQ ID NO: 112) CD70 sgRNA (E1_T4) CGGTGCGGCGCAGGCCCTAT (GGG) (SEQ ID NO: 113) CD70 sgRNA (E1_T7) GCTTTGGTCCCATTGGTCGC (GGG) (SEQ ID NO: 114) CD70 sgRNA (E1_T8) GCCCGCAGGACGCACCCATA (GGG) (SEQ ID NO: 115) CD70 sgRNA (E1_T10) GTGCATCCAGCGCTTCGCAC (AGG) (SEQ ID NO: 116) CD70 sgRNA (E3_T1) CAGCTACGTATCCATCGTGA (TGG) (SEQ ID NO: 117) TRAC sgRNA AGAGCAACAGTGCTGTGGCC (TGG) (SEQ ID NO: 118) .beta.2M sgRNA GCTACTCTCTCTTTCTGGCC (TGG) (SEQ ID NO: 119) PD-1 sgRNA CTGCAGCTTCTCCAACACAT (CGG) (SEQ ID NO: 120) *: 2'-O-methyl phosphorothioate residue
[0644] Primary human T cells were transfected (electroporated) with a ribonucleoprotein particle (RNP) containing Cas9 nuclease and a synthetic modified sgRNA targeting the CD70 gene (sequences in Table 5) or controls (no Cas9, no gRNA). Four to six (4-6) days post transfection, cells were (1) subjected to a TIDE analysis to assess indel frequency and (2) processed by flow cytometry (primary antibody: FITC anti-human CD70 antibody, clone 113-16, Biolegend) to assess CD70 expression levels at the cell surface.
[0645] Seven (7) gRNAs yielded measurable data by TIDE analysis, as indicated in Table 6. Four (4) gRNA sequences yielded indel percentages (editing frequencies) above 85% with protein expression knockdown above 80% (SEQ ID NOS: 23, 26, 27 and 29), indicating highly efficient gene editing. The data in Table 6 are from one (1) donor. The level of CD70 protein expression (assessed by median fluorescent intensity (MFI)) per test sample was normalized to the level of CD70 protein expression present in control cells.
TABLE-US-00006 TABLE 6 CD70 gRNA sequences, cutting efficiencies, and CD70 surface protein expression in gene edited T cells Protein expression knockdown gRNA Name gRNA Spacer Sequence Indel % R.sup.2 % CD70 EXON1_T1 (E1_T1) UCACCAAGCCCGCGACCAAU 89.3% 0.97 84.8% (SEQ ID NO: 91) CD70 EXON1_T3 (E1_T3) AUCACCAAGCCCGCGACCAA 65.2% 0.93 84.0% (SEQ ID NO: 92) CD70 EXON1_T4 (E1_T4) CGGUGCGGCGCAGGCCCUAU 81.6% 0.83 87.5% (SEQ ID NO: 93) CD70 EXON1_T7 (E1_T7) GCUUUGGUCCCAUUGGUCGC 97.8% 0.98 87.7% (SEQ ID NO: 94) CD70 EXON1_T8 (E1_T8) GCCCGCAGGACGCACCCAUA 90.1% 0.94 88.1% (SEQ ID NO: 95) CD70 EXON1_T10 (E1_T10) GUGCAUCCAGCGCUUCGCAC 28.3% 0.30 83.9% (SEQ ID NO: 96) CD70 EXON3_T1 (E3_T1) CAGCUACGUAUCCAUCGUGA 85.6% 0.93 87.2% (SEQ ID NO: 97)
Analysis of On-Target Indel Profiles in T Cells
[0646] On-target amplicon analysis was conducted at the CD70 locus following gene editing using the T7 guide (SEQ ID NO: 26; SEQ ID NO: 36), targeting the CD70 gene: GCTTTGGTCCCATTGGTCGC (SEQ ID NO: 160; target sequence, with PAM SEQ ID NO: 114).
[0647] Following gene editing, on-target amplicon analysis was conducted around the CD70 locus in TRAC-/.beta.2M-/CD70-/anti-CD70 CAR+ cells (generated as described in Example 3).
[0648] An initial PCR was performed using the KAPA HiFi PCR kit (Kapa Biosystems, Wilmington, Mass.). 100 ng of input gDNA was combined with 10 uM of each primer. The CD70_F and CD70_R primers were paired to amplify the CD70 locus (Table 7).
TABLE-US-00007 TABLE 7 Primers for CD70 amplicon library preparation CD70_F TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGcccaac ttttccatctcaactcaccccaagtg (SEQ ID NO: 127) CD70_R GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGcccct cctgcgctagcgga (SEQ ID NO: 128)
[0649] Analysis of the CD70 locus in a population of T cells following CRISPR/Cas9 gene editing to produce TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR+ T cells results in specific indel frequencies and edited gene sequences at the CD70 locus (Table 8; deletions as dashes and insertions in bold). Two cell populations of edited cells were generated from two different donor T cells (1 and 2). The populations of edited T cells from each donor were analyzed in replicate: 1A/1B and 2A/2B.
TABLE-US-00008 TABLE 8 SEQ ID Std. NO: Gene Edited Sequence 1A 1B 2A 2B Mean Dev. 129 CACACCACGAGGCAGATCACCAAGCCCGCG-- 10.4% 11.1% 14.4% 14.8% 12.7% 0.022 CAATGGGACCAAAGCAGCCCGCAGGACG 130 CACACCACGAGGCAGATCACCAAGCCCGCGA 8.7% 10.0% 11.3% 11.1% 10.3% 0.012 ACCAATGGGACCAAAGCAGCCCGCAGGACG 131 CACACCACGAGGCAGATC------------ 8.2% 7.8% 7.1% 6.8% 7.5% 0.006 ACCAATGGGACCAAAGCAGCCCGCAGGACG 132 CACACCACGAGGCAGATCACCAAGCCCGCG- 3.9% 4.5% 4.2% 4.3% 4.2% 0.002 CCAATGGGACCAAAGCAGCCCGCAGGACG 133 CACACCACGAGGCAGATCACCAAGCCCGC- 2.2% 2.5% 2.4% 2.6% 2.4% 0.002 ACCAATGGGACCAAAGCAGCCCGCAGGACG 134 CACACCACGAGGCAGATCACCA------------------- 2.9% 2.3% 2.0% 2.0% 2.3% 0.004 ------AGCCCGCAGGACG
Example 2. Generation of T Cells with Multiple Gene Knockouts
[0650] This example describes the use of CRISPR/Cas9 gene editing technology to produce human T cells that lack expression of two, three or four genes simultaneously. Specifically, the
[0651] T cell receptor (TCR) gene (gene edited in the TCR Alpha Constant (TRAC) region), the .beta.2-microglobulin (.beta.2M) gene, the Cluster of Differentiation 70 (CD70) gene and/or the programmed cell death 1 (PD-1 or PD1) gene were edited by CRISPR/Cas9 gene editing to produce T cells deficient in two or more of the listed genes. The following abbreviations are used in the Figures for brevity and clarity:
[0652] 2.times. KO: TRAC.sup.-/.beta.2M.sup.-
[0653] 3.times. KO (PD-1): TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-
[0654] 3.times. KO (CD70): TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-
[0655] 4.times. KO: TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-
[0656] Activated primary human T cells were electroporated with Cas9:gRNA RNP complexes. The nucleofection mix contained the Nucleofector.TM. Solution, 5.times.10.sup.6 cells, 1 .mu.M Cas9, and 5 .mu.M gRNA (as described in Hendel et al., Nat Biotechnol. 2015; 33(9):985-989, PMID: 26121415). For the generation of double knockout T cells (2.times. KO), the cells were electroporated with two different RNP complexes, each containing Cas9 protein and one of the following sgRNAs: TRAC (SEQ ID NO: 40) and .beta.2M (SEQ ID NO: 41) at the concentrations indicated above. For the generation of triple knockout T cells (3.times. KO), the cells were electroporated with three different RNP complexes, each RNA complex containing Cas protein and one of the following sgRNAs: (a) TRAC (SEQ ID NO: 40), .beta.2M (SEQ ID NO: 41), and PD-1 (SEQ ID NO: 42) at the concentrations indicated above; or (b) TRAC (SEQ ID NO: 40), .beta.2M (SEQ ID NO: 41), and CD70 (SEQ ID NO: 36 or 37) at the concentrations indicated above. For the generation of quadruple knockout T cells (4.times. KO), the cells were electroporated with four different RNP complexes, each RNA complex containing Cas9 protein and one the following sgRNAs: TRAC (SEQ ID NO: 40), .beta.2M (SEQ ID NO: 41), PD-1 (SEQ ID NO: 42), and CD70 (SEQ ID NO: 36 or 37) at the concentrations indicated above. The unmodified versions (or other modified versions) of the gRNAs may also be used (e.g., SEQ ID NOS: 30, 31, 32, 26, and/or 27). Sequences in Tables 5 and 9.
TABLE-US-00009 TABLE 9 gRNA Sequences/Target Sequences Name Unmodified Sequence Modified Sequence TRAC sgRNA AGAGCAACAGUGCUGUGGCC A*G*A*GCAACAGUGCUGUGG guuuuagagcuagaaauagcaaguuaaaau CCguuuuagagcuagaaauagcaaguuaa aaggcuaguccguuaucaacuugaaaaagu aauaaggcuaguccguuaucaacuugaaaa ggcaccgagucggugcUUUU aguggcaccgagucggugcU*U*U*U (SEQ ID NO: 30) (SEQ ID NO: 40) TRAC sgRNA AGAGCAACAGUGCUGUGGCC A*G*A*GCAACAGUGCUGUGG spacer (SEQ ID NO: 98) CC (SEQ ID NO: 108) .beta.2M sgRNA GCUACUCUCUCUUUCUGGCC G*C*U*ACUCUCUCUUUCUGG guuuuagagcuagaaauagcaaguuaaaau CCguuuuagagcuagaaauagcaaguuaa aaggcuaguccguuaucaacuugaaaaagu aauaaggcuaguccguuaucaacuugaaaa ggcaccgagucggugcUUUU aguggcaccgagucggugcU*U*U*U (SEQ ID NO: 31) (SEQ ID NO: 41) .beta.2M sgRNA GCUACUCUCUCUUUCUGGCC G*C*U*ACUCUCUCUUUCUGG spacer (SEQ ID NO: 99) CC (SEQ ID NO: 109) PD-1 sgRNA CUGCAGCUUCUCCAACACAU C*U*G*CAGCUUCUCCAACAC guuuuagagcuagaaauagcaaguuaaaau AUguuuuagagcuagaaauagcaaguuaa aaggcuaguccguuaucaacuugaaaaagu aauaaggcuaguccguuaucaacuugaaaa ggcaccgagucggugcUUUU (SEQ ID aguggcaccgagucggugcU*U*U*U NO: 32) (SEQ ID NO: 42) PD-1 sgRNA CUGCAGCUUCUCCAACACAU C*U*G*CAGCUUCUCCAACAC spacer (SEQ ID NO: 100) AU (SEQ ID NO: 110)
[0657] About one (1) week post electroporation, cells were either left untreated or treated with phorbol myristate acetate (PMA)/ionomycin overnight. The next day cells were processed for flow cytometry (see, e.g., Kalaitzidis D et al. J Clin Invest 2017; 127(4): 1405-1413) to assess TRAC, .beta.2M, PD-1, and CD70 expression levels at the cell surface of the edited cell population. The following primary antibodies were used (Table 10):
TABLE-US-00010 TABLE 10 Antibodies Anti- body Clone Fluor Catalogue # Dilution For 1 TCR BW242/412 PE 130-091-236 (Miltenyi) 1:100 1 .mu.L .beta.2M 2M2 PE-Cy7 316318 (Biolegend) 1:100 1 .mu.L PD-1 EH12.2H7 PE 329906 (Biolegend) 1:100 1 .mu.L CD70 113-16 FITC 355105 (Biolegend) 1:100 1 .mu.L
[0658] Tables 11 and 12 show highly efficient multiple gene editing. For the double-knock cells (2.times. KO; TRAC.sup.-/.beta.2M.sup.-), 83% of viable cells lacked expression of TCR and .beta.2M (Table 11; 3.times. KO (PD1)). For the triple knockout cells, 70% of viable cells lacked expression of TCR, .beta.2M, and PD-1 (Table 11); and 80% of viable cells lacked expression of TCR, .beta.2M, and CD70 irrespective of the CD70 gRNA used (Table 12). For the quadruple knockout cells (4.times. KO), 78% of viable cells lacked expression of TCR, .beta.2M, PD-1, and CD70 (FIG. 1).
TABLE-US-00011 TABLE 11 % of viable cells lacking expression in 2KO and 3KO (PD1) cell populations TRAC KO .beta.2M KO PD1 KO 2 KO 3 KO (PD1) 2KO 98% 85% NA 83% NA 3 KO (PD1) 98% 73% 99% NA 70%
TABLE-US-00012 TABLE 12 % of viable cells lacking expression in 3KO (CD70) cell populations TRAC KO .beta.2M KO CD70 KO 3KO (CD70) 3KO (CD70) (T7) 99% 79% 99% 80% 3KO (CD70) (T8) 99% 82% 99% 80%
[0659] To assess whether triple and quadruple gene editing in T cells affects cell expansion, cell numbers were enumerated among double, triple, and quadruple gene edited T cells (unedited T cells were used as a control) over a two week period of post editing. 5.times.10.sup.6 cells were generated and plated for each genotype of T cells.
[0660] As shown in FIG. 2, cell proliferation (expansion) continued over the post-electroporation window test. Similar cell proliferation was observed among the double (.beta.2M-/TRAC-), triple (.beta.2M-/TRAC-/PD-1-, or .beta.2M-/TRAC-/CD70-), and quadruple (.beta.2M-/TRAC-/PD-1-/CD70) knockout T cells, as indicated by the number of viable cells. These data suggest that multiple gene editing (up to triple and quadruple, with CD70 and PD-1 genes) does not impact T cell health as measured by T cell proliferation.
Example 3. Generation of CAR T Cells Lacking CD70 and/or PD1
[0661] Generation of Anti-CD70 CAR T Cells with Multiple Knockouts
[0662] This example describes the production of allogeneic human T cells that lack expression of the TCR gene, .beta.2M gene, CD70 gene and/or PD1 gene, and express a chimeric antigen receptor (CAR) targeting CD70. These cells are designated TCR.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ or 3.times. KO (CD70) CD70 CAR.sup.+; TCR.sup.-/.beta.2M.sup.-/PD1.sup.-/anti-CD70 CAR.sup.+ or 3.times. KO (PD1) CD70 CAR.sup.+; TCR.sup.-/.beta.2M.sup.-/PD1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ or 4.times. KO CD70 CAR.sup.+ in the Figures.
[0663] A recombinant adeno-associated adenoviral vector, serotype 6 (AAV6) (MOI 50,000) comprising the nucleotide sequence of SEQ ID NO: 43 (comprising the donor template in SEQ ID NO: 44, encoding anti-CD70 CAR comprising the amino acid sequence of SEQ ID NO: 46) was delivered with Cas9:sgRNA RNPs (1 .mu.M Cas9, 5 .mu.M gRNA) to activated allogeneic human T cells. The following sgRNAs were used: TRAC (SEQ ID NO: 40), .beta.2M (SEQ ID NO: 41), CD70 (SEQ ID NO: 36 or 37) and PD1 (SEQ ID NO: 42). The unmodified versions (or other modified versions) of the gRNAs may also be used (e.g., SEQ ID NOS: 30, 31, 32, 26, and/or 27). About one (1) week post electroporation, cells were processed for flow cytometry to assess TRAC, .beta.2M, CD70, and PD1 expression levels at the cell surface of the edited cell population. The following primary antibodies were used (Table 13):
TABLE-US-00013 TABLE 13 Antibodies Antibody Clone Fluor Catalogue # Dilution TCR BW242/412 PE 130-091-236 (Miltenyi) 1:100 .beta.2M 2M2 PE-Cy7 316318 (Biolegend) 1:100 CD70 113-16 FITC 355105 (Biolegend) 1:100 PD-1 EH12.2H7 PE 329906 (Biolegend) 1:100
[0664] T cell Proportion Assay. The proportions of CD4+ and CD8+ cells were then assessed in the edited T cell populations by flow cytometry using the following antibodies (Table 14):
TABLE-US-00014 TABLE 14 Antibodies Antibody Clone Fluor Catalogue # Dilution CD4 RPA-T4 BV510 300545 (Biolegend) 1:100 CD8 SK1 BV605 344741 (Biolegend) 1:100
High efficiency gene editing and CAR expression was achieved in the edited anti-CD70 CAR T cell populations. In addition, editing did not adversely alter CD4/CD8 T cell populations. FIG. 3 shows highly efficient gene editing and anti-CD70 CAR expression in the triple knockout CAR T cell. More than 55% of viable cells lacked expression of TCR, .beta.2M, and CD70, and also expressed the anti-CD70 CAR. FIG. 4 shows that normal proportions of CD4/CD8 T cell subsets were maintained in the TRAC-/.beta.2M-/CD70-/anti-CD70 CAR+ cells, suggesting that these multiple gene edits do not affect T cell biology as measured by the proportion of CD4/CD8 T cell subsets.
[0665] FIG. 5 shows show highly efficient gene editing and anti-CD70 CAR expression in the quadruple knockout CAR T cell. Greater than 60% of viable cells lacked expression of TCR, .beta.2M, PD-1, and CD70, and expressed the anti-CD70 CAR. FIG. 6 shows that normal proportions of CD4/CD8 T cell subsets were maintained in the TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ cells, suggesting that these multiple gene editing do not affect T cell biology as measured by the proportion of CD4+/CD8+ T cell subsets.
Generation of Anti-BCMA CAR T Cells with Multiple Knockouts
[0666] This example describes the production of allogeneic human T cells that lack expression of the TCR gene, the .beta.2M gene, the PD-1 gene, and/or the CD70 gene, and also express a chimeric antigen receptor (CAR) targeting B-cell maturation antigen (BCMA).
[0667] A recombinant adeno-associated adenoviral vector, serotype 6 (AAV6) comprising the nucleotide sequence of SEQ ID NO: 54 (comprising the donor template in SEQ ID NO: 55, encoding anti-BCMA CAR comprising the amino acid sequence of SEQ ID NO: 57) was delivered with Cas9:gRNA RNPs (1 .mu.M Cas9, and 5 .mu.M gRNA) to activated allogeneic human T cells. The following gRNAs were used: TRAC (SEQ ID NO: 40), .beta.2M (SEQ ID NO: 41), PD-1 (SEQ ID NO: 42), and CD70 (SEQ ID NO: 36 or 37). The unmodified versions (or other modified versions) of the gRNAs may also be used (e.g., SEQ ID NOS: 30, 31, 32, 26 and/or 27). About one (1) week post electroporation, cells were processed for flow cytometry as described above for anti-CD70 CAR+ T cells, with the following difference. Anti-BCMA CAR expression was detected using biotinylated recombinant human BCMA (ACROS Cat# BC7-H82F0). The double and quadruple knockout anti-BCMA CAR.sup.+ cells were then characterized as described herein.
[0668] FIGS. 7A-7B shows highly efficient gene editing of the TRAC gene, .beta.2M gene, the CD70 gene and the PD-1 gene. FIG. 7C shows high expression of the anti-BCMA CAR+ cells in double knockout and quadruple knockout cells.
Generation of Anti-CD19 CAR T Cells with Multiple Knockouts
[0669] Allogeneic human T cells were generated that express a chimeric antigen receptor (CAR) targeting CD19 and lack the expression of the TCR gene , the .beta..beta.2M gene, and optionally the CD70 gene.
[0670] To generate the allogeneic T cells, activated primary human T cells were electroporated with Cas9:gRNA RNP complexes and infected with adeno-associated adenoviral vectors (AAVs) containing anti-CD19 CAR donor template with homology to the TRAC locus. Recombinant AAV serotype 6 (AAV6) comprising the nucleotide sequence of SEQ ID NO: 155 (comprising the donor template in SEQ ID NO: 156, encoding anti-CD19 CAR comprising the amino acid sequence of SEQ ID NO: 149) was delivered with Cas9:sgRNA RNPs (1 .mu.M Cas9, 5 .mu.M gRNA) to activated human T cells. The following sgRNAs were used to knock-out the respective genes: TRAC (SEQ ID NO: 40), .beta.2M (SEQ ID NO: 41), CD70 (SEQ ID NO: 36). The unmodified versions (or other modified versions) of the gRNAs may also be used (e.g., SEQ ID NOs: 30, 21 or 27). About one (1) week post electroporation, cells were processed for flow cytometry as described above for anti-CD70 CAR+ T cells, with the following difference. Anti-CD19 CAR expression was detected using biotinylated recombinant human CD19 (ACROBIOSYSTEMS INC; CD9-H825). The CD70 deficient anti-CD19 CAR.sup.+ T cells were then characterized as described herein.
Generation of anti-CD33 CAR T Cells with Multiple Knockouts
[0671] Allogeneic human T cells were generated that express a chimeric antigen receptor (CAR) targeting CD33 and lack the expression of the T cell receptor (TCR) gene (gene edited in the TCR Alpha Constant (TRAC) region), the .beta.2-microglobulin (.beta.2M) gene, and optionally the CD70 gene.
[0672] To generate the allogeneic T cells, activated primary human T cells were electroporated with Cas9:gRNA RNP complexes and infected with adeno-associated adenoviral vectors (AAVs) containing anti-CD33 CAR donor template with homology to the TRAC locus. Recombinant AAV serotype 6 (AAV6) comprising the nucleotide sequence of SEQ ID NO: 87 (comprising the donor template in SEQ ID NO: 135, encoding anti-CD33 CAR comprising the amino acid sequence of SEQ ID NO: 139was delivered with Cas9:sgRNA RNPs (1 .mu.M Cas9, 5 .mu.M gRNA) to activated human T cells. The following sgRNAs were used to knock-out the respective genes: TRAC (SEQ ID NO: 40), .beta.2M (SEQ ID NO: 41), CD70 (SEQ ID NO: 36). The unmodified versions (or other modified versions) of the gRNAs may also be used (e.g., SEQ ID NOs: 30, 21 or 27).
[0673] Populations of TCR+ T cells (no RNP) and TRAC-/.beta.2M-T cells (TCR and .beta.2M deficient cells without a CAR) were similarly generated for use as controls. About one (1) week post electroporation, cells were processed for flow cytometry as described above for anti-CD70 CAR+ T cells, with the following difference. Anti-CD33 CAR expression was detected using biotinylated recombinant human CD33 (data not shown). The CD70 knockout anti-CD33 CAR.sup.+ T cells were then characterized as described herein.
Characterization of CD4/CD8 Cell Populations in Anti-CD33 CAR T Cells with CD70 Knock-Out
[0674] CD33 can be expressed on T cells with higher levels observed on cultured CD4 cells than CD8 cells. During the course of producing anti-CD33 CAR-T cells CD4 cells become substantially reduced due to fratricide. As shown in FIG. 8, anti-CD33 CAR-T cell cultures with intact CD70 displayed a 97% reduction in CD4+cells over a 3 week culture period, while cultures of cells with disrupted CD70 showed only a 61% reduction over this time course. Thus disrupting the CD70 gene appears to reduce the fratricide observed in the anti-CD33 CAR-T cell cultures. Without wishing to be bound by theory, this effect may occur through an immune stimulatory function which could be potentiated by CD70/CD27 interactions, and genetic disruption of CD70 results in more balanced CD4/CD8 ratios that may be more optimal for therapeutic benefit in malignancy.
Example 4: CD70 KO Improves Cell Proliferation
Effect of CD70 KO on Cell Proliferation of Anti-CD33 CAR T Cells In Vitro
[0675] To assess the ability of cells to expand in cytokine containing media (IL-2+IL-7), anti-CD33 CAR T cells were utilized. Specifically, 5.times.10.sup.6 total anti-CD33 CAR T cells comprising a double knockout (TRAC-/B2M-) or triple knockout (TRAC-/B2M-/CD70-) were generated as described in Example 3, plated and allowed to grow in a 10 mL volume of cytokine containing media. After 1 week cells were counted. 5.times.10.sup.6 cells from the previous culture were then replated in 10 mL volume (fresh cytokine containing media) and 1 week later the total number of cells were enumerated. Allogeneic anti-CD33 CAR-T cells containing a disruption in the CD70 gene expanded to greater levels on the first and second week of replating (FIG. 9). These data show that CD70 knockout can result in greater cell yields in culture.
Effect of CD70 KO on Cell Proliferation of Anti-CD19 CAR T Cells In Vitro
[0676] To further assess the ability of cells to expand in cytokine containing media (IL-2+IL-7), anti-CD19 CAR T cells were utilized. Specifically, 5.times.10.sup.6 total anti-CD19 CAR T cells comprising a double knockout (TRAC-/B2M-) or triple knockout (TRAC-/B2M-/CD70-) were generated as described in Example 3, plated and allowed to grow in a 10 mL volume of cytokine containing media. After 1 week cells were counted. 5.times.10.sup.6 cells from the previous culture were then replated in 10 mL volume (fresh cytokine containing media) and 1 week later the total number of cells were enumerated. Allogeneic anti-CD19 CAR-T cells containing a disruption in the CD70 gene expanded to greater levels on the first and second week of replating as compared to control cells without a CD70 gene distruption (FIG. 10). These data show that CD70 knockout can result in greater cell yields in culture.
Effect of CD70 KO on Cytokine Driven Proliferation and Apoptosis of Anti-BCMA CAR T Cells In Vitro
[0677] Cytokine driven proliferation. To evaluate the effect of CD70 and/or PD1 knockout on cell proliferation, anti-BCMA CAR T cells were utilized. Anti-BCMA CAR T cells were generated as described in Example 3. The following groups of edited T cells were generated:
[0678] TRAC-/B2M-/anti-BCMA CAR+ (Control; 2KO, BCMA CAR+)
[0679] TRAC-/B2M-/CD70-/anti-BCMA CAR+ (3KO (CD70), BCMA CAR+)
[0680] TRAC-/.beta.2M-/PD1-/anti-BCMA CAR+ (3KO (PD1), BCMA CAR+)
[0681] TRAC-/.beta.2M-/CD70-/PD1-/anti-BCMA CAR+ (4KO, BCMA CAR+)
[0682] Edited cells were enriched for TRAC-/B2M- cells by magnetic depletion of CD3+B2M+cells. Briefly, cells were labelled with anti-CD3 Biotin (Biolegend Cat#300404) anti-.beta.2M Biotin (Biolegend Cat #316308) antibodies, each at 0.5 .mu.g per 1 .times.10.sup.6 cells in 100 .mu.l volume at 4.degree. C. for 15 min, washed and incubated with Streptavidin labelled magnetic microbeads (Miltenyi Biotech, 130-048-101) for 15min at 4.degree. C. Cells were resuspended in buffer and passed through LS columns (Miltenyi Biotech, 130-042-401) according to the manufacturer's protocol. To determine the effect of CD70 or PD1 on IL-2/IL-7 driven T cell proliferation, the edited T cells (1E6 cells/ml) were cultured in growth medium (X-vivo medium (04-744, Lonza), supplemented with 5% human AB serum (HP1022, Valley Biomedical)), 50 ng/ml IL-2 (rhlL-2; 130-097-745, Miltenyi Biotech) and 10 ng/ml IL-7 (rhlL-7; Cellgenix 001410-050) for up to four weeks. At indicated days, the cells were counted and re-seeded in fresh medium at 1.5E6 cells/ml in appropriate culture dishes.
[0683] FIGS. 11 and 12 show that knockout of CD70 improved IL-2/IL-7 driven proliferation of anti-BCMA CAR T cells in vitro, as compared to CD70 sufficient controls (i.e. anti-BCMA CAR T cells comprising endogenous CD70). FIG. 11 also shows the CD70 KO can improve health and proliferation competence of anti-BCMA CAR T cells even when the T cells from this donor appear to be in significant decline after 17 days when the CD70 gene is intact (as shown by the reduced cell numbers of donor 1(FIG. 11) compared to donor 2 (FIG. 12)). This property of maintaining T cell health (enabled by KO of the CD70 gene) is broadly applicable to many aspects of CAR T development including: extended expansion during manufacturing increasing yield and consistency, rescue of exhausted/unhealthy T cells enabling potentially lower doses in patients and more robust responses, combination with other KOs that may be more detrimental to T cell health but have other advantages such as overcoming suppression of T cell activity (e.g. PD1 KO). As shown in FIGS. 11 and 12, deleting the PD1 gene by itself shows no benefit to CAR T cell expansion but when combined with a CD70 KO shows synergistic effects.
[0684] Apoptosis. The effect of CD70 KO on apoptotic cell death of anti-BCMA CAR+ T cells following exposure to antigen was evaluated in an antigen rechallenge assay. Briefly, to achieve antigen exposure, anti-BCMA CAR+ T cells were exposed to plate-adhered recombinant BCMA protein. Plates with adhered antigen were prepared by coating 24 well plates with recombinant BCMA protein in 1.times. PBS (1 .mu.g/ml; biotinylated Human BCMA Protein, ACRO Biosystems) overnight at 4.degree. C. and then washing away unbound antigen. Following the wash, antigen-bound plates were then used to challenge anti-BCMA CAR+ T cells either with or without a CD70 knockout. The 2.times. KO (TRAC-/B2M-) anti-BCMA CAR+ T cells and 3.times. KO (TRAC-/B2M-/CD70-) anti-BCMA CAR+ T cells (1.times.10.sup.6 cells/ml) were exposed to plate-bound recombinant BCMA protein (1 .mu.g/ml) for 24 hours in growth medium (X-vivo medium (04-744, Lonza), 5% human AB serum (HP1022, Valley Biomedical)) supplemented with IL-2 (rhlL-2; 130-097-745, Miltenyi Biotech). Cells were then washed, counted and re-challenged (1.times.10.sup.6 cells/ml) with fresh plate-bound antigen every 24 hours for a total of three consecutive re-challenges (24 hr, 48 hr, and 72 hr). At the end of each re-challenge, an aliquot of cells was washed and stained with fluorochrome-conjugated annexin V along with propidium iodide in annexin V binding buffer (BioLegend) for 15 minutes at room temperature. Cells were then washed and resuspend in annexin V binding buffer for analysis by flow cytometry. The cells were counted at each time point and the cell count per ml was derived. For the calculation of fold-expansion at each time point, the initial fold-expansion at time 0 was set at 1. Fold-expansion for all other time points were calculated by multiplying the cell count per ml at each time point by the fold-expansion per ml for the prior time point. For example, the fold expansion at 72 hr was calculated by multiplying the cell count per ml at 72 hr by the fold expansion per ml at 48 hr.
[0685] FIG. 13 demonstrates that the deletion of CD70 (CD70 KO) rescues anti-BCMA CAR+ T cells from apoptosis, as shown by the decrease in the percentage of apoptotic cells following the second (48 hr) and third (72 hr) rechallenge. Furthermore, the absence of CD70 expression in anti-BCMA CAR+ T cells surprisingly enhances the expansion of the anti-BCMA CAR+ T cells in response to antigen exposure (FIG. 14).
Effect of CD70 KO on Cell Proliferation of Anti-CD70 CAR T Cells In Vitro
[0686] To further assess the impact of disrupting the CD70 gene in CAR T cells, anti-CD70 CAR T cells were generated as described in Example 3. Specifically, 3.times. KO (TRAC-/.beta.2M-/CD70-) anti-CD70 CAR T cells were generated using two different gRNAs (T7 (SEQ ID NO: 36 and T8 (SEQ ID NO: 37)). After electroporation, cell expansion was assessed as described in Example 2 by counting viable cells. FIG. 15 shows that triple knockout TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells generated with either T7 or T8 gRNAs exhibited greater cell expansion relative to double knockout TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ T cells. These data suggest that knocking-out the CD70 gene gives a cell proliferation advantage to anti-CD70 CAR+ T cells.
[0687] Cell expansion was also assessed in the quadruple knockout, TRAC.sup.-/.beta.2M.sup.-/PD-1/CD70.sup.-/anti-CD70 CAR.sup.+ T cells. These cells exhibited greater expansion relative to triple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/anti-CD70 CAR.sup.+ T cells and to double knockout TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ T cells (FIG. 16).
Example 5. CD70 KO Increases Durability and Potency of CAR T Cells In Vitro
[0688] Cell Killing Function of Anti-CD19 CAR T Cells with CD70 Knock-Out
[0689] Following preparation of edited anti-CD19 CAR T cells as described in Example 3, the functional activity of the CAR T cells was verified using a flow cytometry-based cytotoxicity assay. The anti-CD19 CAR T cells (TRAC-/.beta.2M-/CD19 CAR+ and TRAC-/.beta.2M-/CD70-/CD19 CAR+) were co-cultured with one of two CD19-expressing cancer cell lines (target cells): Nalm6 (ATCC cr13273) or Raji (ATCC cc1-86). The target cells were labeled with 5 .mu.M efluor670 (eBiosciences), washed and incubated in co-cultures with the TRAC-/.beta.2M-/anti-CD19 CAR+, or TRAC-/.beta.2M-/CD70-/anti-CD19 CAR+ at varying ratios (0.01, 0.05, 0.1, 0.5, 1:1 T cells:target cells). The target cells were seeded at 50,000 cells per well in a 96-well, U-bottom plate. The co-culture was incubated overnight. After incubation, wells were washed and media was replaced with 200 .mu.L of media containing a 1:500 dilution of 5 mg/mL DAPI (Molecular Probes). 25 .mu.L of CountBright beads (Life Technologies) were then added to each well and the cell cultures were analyzed for cell viability by flow cytometry (i.e., viable cells being negative for DAPI staining).
[0690] Percent cell lysis of the target cells (e.g.: Nalm6 or Raji cells) was then determined using the following formula:
Percent cell lysis=(1-((total number of target cells in a test sample)/(total number of target cells in a control sample)).times.100;
[0691] wherein a test sample was target cells (e.g.: Nalm6 or Raji cells) co-cultured with 1) TRAC-/.beta.2M-/CD19 CAR+ T cells or 2) TRAC-/.beta.2M-/CD70-/CD19 CAR+ T cells; and
[0692] a control sample was target cells alone that had not been co-cultured.
[0693] Disruption of the CD70 gene led to enhanced cytolytic activity of the anti-CD19 CAR-T cells against the Raji cell line at low CAR-T to target ratios (FIG. 17, bottom panel). Disruption of CD70 did not enhance anti-CD19 CAR-T activity against the Nalm6 cell line (FIG. 17, top panel). Of note, the Nalm6 cell line is relatively easier to lyse by these CAR-T cells (>80% lyses at 0.1:1 CAR-T cell to target ratio for Nalm6 vs 0% for Raji at this ratio for the wild-type cells) likely explaining the lack of resulting increased efficacy due to CD70 disruption in this assay against Nalm6 cells. The increased activity conferred by CD70 loss against the Raji cell line indicates that in challenging tumor environments, particularly when CAR-T to tumor ratios are low, CD70 loss may have substantial benefit to the CAR-T cells in eradicating tumor cells.
Cell Killing Function of Anti-CD33 CAR T Cells with CD70 Knock-Out
[0694] Following preparation of the edited anti-CD33 CAR+ T cells as described in Example 3, the functional activity of the CAR T cells was verified using a flow cytometry-based cytotoxicity assay. The anti-CD33 CAR T cells (TRAC-/.beta.2M-/CD33 CAR+ and TRAC-/.beta.2M-/CD70-/CD33 CAR+) or control T cells (no RNP) were co-cultured with the CD33-expressing cancer cell line MV4-11 (ATCC CRL-9591). The target cells were labeled with 5 .mu.M efluor670 (eBiosciences), washed and incubated in co-cultures with the TRAC-/B2M-/anti-CD33 CAR+, TRAC-/.beta.2M-/CD70-/anti-CD33 CAR+, or controls at varying ratios (0.01:1, 0.03:1, 0.06:1, 0.125:1, 0.25:1, 0.5:1, or 1:1 T cells:target cells). The target cells were seeded at 50,000 cells per well in a 96-well, U-bottom plate. The co-culture was incubated overnight. After 48 hrs, wells were washed and media was replaced with 200 .mu.L, of media containing a 1:500 dilution of 5 mg/mL DAPI (Molecular Probes). 25 .mu.L of CountBright beads (Life Technologies) were then added to each well and the cell cultures were analyzed for cell viability by flow cytometry (i.e., viable cells being negative for DAPI staining).
[0695] Percent cell lysis of the target cells (e.g.: MV4-11) was then determined using the following formula:
Percent cell lysis=(1-((total number of target cells in a test sample)/(total number of target cells in a control sample)).times.100;
[0696] wherein a test sample was target cells (e.g.: MV4-11 cells) co-cultured with 1) TRAC-/.beta.2M-/CD33 CAR+ T cells; or 2) TRAC-/.beta.2M-/CD70-/CD33 CAR+ T cells, and
[0697] a control sample was target cells alone that had not been co-cultured.
[0698] Although both populations of anti-CD33 CAR T cells effectively killed MV4-11 cells, reaching nearly 100% cells kill at ratios of 0.5 CAR T cell: MV4-11 cell, the TRAC-/B2M-/CD70-/CD33 CAR+ T cells demonstrated higher cell killing at lower CAR T to cancer cell rations (FIG. 18). These data demonstrate that allogeneic anti-CD33 CAR T cells with the additional CD70 knock-out are more efficacious at lower CAR T cell to target cell ratios.
Cell Killing Function of Anti-CD70 CAR T Cells with CD70 Knock-Out
[0699] A cell killing assay was used to assess the ability of the TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ cells and TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ cells to kill a CD70.sup.+ adherent renal cell carcinoma (RCC)-derived cell line (A498 cells). Adherent cells were seeded in 96-well plates at 50,000 cells per well and left overnight at 37.degree. C. The next day edited anti-CD70 CAR T cells were added to the wells containing target cells at the indicated ratios. After the indicated incubation period, CAR T cells were removed from the culture by aspiration and 100 .mu.L Cell titer-Glo (Promega) was added to each well of the plate to assess the number of remaining viable cells. The amount of light emitted per well was then quantified using a plate reader. The cells exhibited potent cell killing of RCC-derived cells following 24-hour co-incubation (FIG. 19). The anti-CD70 CAR T cells demonstrated higher potency when CD70 was knocked out, which is clearly visible at low T cell: A498 ratios (1:1 and 0.5:1) where cell lysis remains above 90% for TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+, while cells lysis drops below 90% for the TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+. This suggests that knocking-out the CD70 gene gives a higher cell kill potency to anti-CD70 CAR+ T cells.
Example 6. Rechallenge of CD70 Deficient CAR T Cells In Vitro
CD70 Knockout Improves Anti-CD33 CAR+ T Cell Killing Upon Serial Rechallenge
[0700] To assess the ability of cells to expand after challenge and rechallenge with antigen-expressing cells (e.g.: target cells) anti-CD33 CAR T cells were generated as described in Example 3 and utilized. Specifically, 5.times.10.sup.6 total T cells were plated in the presence of 5.times.10.sup.6 irradiated target cells (MV-4-11) and allowed to grow in a 10 mL volume. After 1 week, cells were counted, 5.times.10.sup.6 cells from the previous culture were then replated in 10 mL volume along with a fresh aliquot of 5.times.10.sup.6 irradiated target cells and 1 week later the total number of cells were enumerated. The process was repeated as indicated, each rechallenge started with 5.times.10.sup.6 cells. The number of viable cells were counted as described in Example 2. Allogeneic anti-CD33 CAR-T cells containing a disruption in the CD70 gene expanded to greater levels on the second week of after 2 challenges with MV-4-11 cells (FIG. 20). These data show that CD70 can limit T-cell expansion in the presence of antigen expressing cells and its loss can result in greater cell expansion after antigen stimulation.
CD70 Knockout Improves Anti-CD19 CAR+ T Cell Killing Upon Serial Rechallenge
[0701] To further assess the ability of cells to expand after challenge and rechallenge with antigen-expressing cells (e.g.: target cells) anti-CD19 CAR T cells were generated as described in Example 3 and utilized. Specifically, 5.times.10.sup.6 total T cells were plated in the presence of 5.times.10.sup.6 irradiated target cells (Nalm6) and allowed to grow in a 10 mL volume. After 1 week, cells were counted, 5.times.10.sup.6 cells from the previous culture were then replated in 10 mL volume along with a fresh aliquot of 5.times.10.sup.6 irradiated target cells and 1 week later the total number of cells were enumerated. The process was repeated as indicated, each rechallenge started with 5.times.10.sup.6 cells. The number of viable cells were counted as described in Example 2. Allogeneic anti-CD19 CAR-T cells containing a disruption in the CD70 gene expanded to similar amounts during the first 2 challenges. However, at three challenges allogeneic anti-CD19 CAR-T cells containing a disruption in the CD70 gene expanded to greater level on the third challenge with Nalm6 cells (FIG. 21). These data show that the presence of CD70 can limit T-cell expansion in the presence of antigen expressing cells and its loss can result in greater cell expansion after repeated antigen stimulation.
Knockout of CD70, or PD-1 plus CD70, Maintain Anti-CD70 CAR.sup.+ T Cell Killing Upon Serial Rechallenge
[0702] The anti-CD70 CAR+ T cells generated above were serially rechallenged with CD70+ kidney cancer cell line, A498, and evaluated for their ability to kill the CD70+ kidney cancer cell lines A498 or ACHN.
[0703] A498 cells were plated in a T25 flask and mixed at a ratio of 2:1 (T-cell to A498) with 10.times.10.sup.6 anti-CD70 CAR.sup.+ T cells containing either two (TRAC.sup.-/.beta.2M.sup.-), three (TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-) or (TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-)), or four (TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-) gRNA edits.
[0704] Two or three days after each challenge, cells were counted, washed, resuspended in fresh T cell media, and re-challenged the next day with the same ratio of two anti-CD70 CAR.sup.+ T cell per one A498 cell (2:1, CAR.sup.+ T:target). Challenging of anti-CD70 CAR.sup.+ T cells with CD70+ A498 cells was repeated 13 times. Three to four days following each exposure to A498 cells (and prior to the next rechallenge), aliquots of the culture were taken and analyzed for the ability of the CAR T Cells to kill A498 or ACHN target cells at a ratio of 2:1 (CAR T cell: Target cell). Cell kill was measured using Cell titer-glo (Promega). Prior to the first challenge with A498, anti-CD70 CAR+ T cells with 2.times. KO (TRAC.sup.-/.beta.2M.sup.-), 3.times. KO (TRAC.sup.-/.beta.2M.sup.-/CD70), 3.times. KO (TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-), and 4.times. KO (TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-) each exhibited a target cell killing of A498 cells approaching 100%. By challenge nine however, the 2.times. KO (TRAC.sup.-/.beta.2M.sup.-) and 3.times. KO (TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-) anti-CD70 CAR+ T cells induced target cell killing of A498 cells below 40%, while 3.times. KO (TRAC.sup.-/.beta.2M.sup.-/CD70) and 4.times. KO (TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-) anti-CD70 CAR.sup.+ T cells exhibited target cell killing above 60% (FIG. 22A). The target cell killing for 3.times. KO (TRAC.sup.-/.beta.2M.sup.-/CD70) and 4.times. KO (TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-) anti-CD70 CAR.sup.+ T cells remained above 60% even following 13 re-challenges with A498 cells, demonstrating that these CAR+ T cells were resistant to exhaustion.
[0705] Anti-CD70 CAR T cells were also evaluated for their ability to kill ACHN cells at a ratio of 2:1 (T-cell to ACHN) following serial rechallenge with A498 renal carcinoma cells (FIG. 22B). Prior to the first challenge with A498, the double knockout TRAC.sup.-/.beta.2M.sup.-anti-CD70 CAR.sup.+ T cells, the triple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-anti-CD70 CAR.sup.+ T, the triple knockout TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells and the quadruple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T exhibited a cell kill efficiency above 62%, 47%, 73% and 81%, respectively.
[0706] After challenge five, the triple knockout TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells and the quadruple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells still efficiently killed above 55% of ACHN cells at a ratio of 2:1 (T-cell to ACHN), while the double knockout TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ T cells and the triple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/anti-CD70 CAR.sup.+ T cell kill dropped below 11% of ACHN cells. This trend continued, wherein the double knockout TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ T cells and the triple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/anti-CD70 CAR.sup.+ T cells failed to survive beyond 10 rechallenges. In contrast, the triple knockout TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells and the quadruple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells continued to expand in culture and to kill greater than 30% of ACHN cells at a ratio of 2:1 (T-cell to ACHN) following two rechallenges.
[0707] The data demonstrate that the 4.times. KO, CD70 CAR.sup.+ T cells and the 3.times. KO (CD70), CD70 CAR.sup.+ cells are more potent than the 2.times. KO, CD70 CAR.sup.+ T or 3.times. KO (PD1), CD70 CAR+ T cells. In addition, the 3.times. (CD70) KO and 4.times. KO prevents T cell exhaustion.
[0708] After 5 rechallenges the cells were evaluated for their ability to kill cancer cells. Surprisingly, the 3KO and 4KO anti-CD70 CAR+ T cells remained highly effective at killing cancer cells (FIG. 23A) even after multiple cancer cell challenges. The cell killing effect of the anti-CD70 CAR+ T cells on ACHN cells is reproducible at even at reduced effector to target cell ratios of 1:1, 0.5:1, and 0.25:1. (FIG. 23A).
[0709] To ensure long-term benefit upon CAR T treatment, CAR T cells should be able to identify and eradicate their target cells over a long period of time, to rule out the possibility of cancer cell escape from CAR-T mediated cell kill. The in vitro re-challenge assay mimics a recurrent encounter of CAR-T cells with target cells over several cycles of CAR-T cell activation. These data demonstrate the superiority of the triple knockout TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells and of the quadruple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells, in sustaining multiple challenges with kidney cancer cells, without showing reduction of their target cell killing ability, as compared to the double knockout TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ T cells and the triple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/anti-CD70 CAR.sup.+ T cells.
[0710] Exhaustion and activation markers were also measured by flow cytometry in the anti-CD70 CAR+ T cells following rechallenge. After 8 challenges, the Triple (TRAC-/.beta.2M-/CD70-) and Quadruple (TRAC-/.beta.2M-/PD1-/CD70-) KO anti-CD70 CAR+ T cells exhibited higher activation marker LAG3 expression than the Double (TRAC-/.beta.2M-) and Quadruple (TRAC-/.beta.2M-/PD1-/CD70-) KO anti-CD70 CAR+ T cells, consistent with their level of high cell kill activity. It was observed that PD1 expression was lower in the Triple (TRAC-/.beta.2M-/CD70-) anti-CD70 CAR+T cells (similar to Triple (TRAC-/.beta.2M-/PD1-) and Quadruple (TRAC-/.beta.2M-/PD1-/CD70-) KO anti-CD70 CAR+T cells) compared to the Double (TRAC-/.beta.2M-) anti-CD70 CAR+T cells, suggesting that knocking-out CD70 has an effect on the downregulation of the exhaustion marker PD1 expression in the Anti-CD70 CAR+T cells. (FIG. 23B).
Knockout of PD-1 and CD70 Maintains Anti-BCMA CAR.sup.+ T Cell Killing Upon Serial Rechallenge
[0711] The anti-BCMA CAR+ T cells generated as described in Example 3 were serially rechallenged with and evaluated for their ability to kill the BCMA+ multiple myeloma cell line MM.1S (ATCC CRL-2974). The ability to secrete cytokines upon serial T cell activation through CAR engagement was also measured after each rechallenge. MM.1S cells were labeled with 5 .mu.M eFlour670 and mixed at a ratio of 2:1 (MM.1S to T-cell) in a 6 well tissue culture dish with 1.times.10.sup.6 anti-BCMA CAR.sup.+ T cells containing either two (TRAC.sup.-/.beta.2M.sup.-) or four ((TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-) gRNA edits. One day following exposure to MM.1S cells, an aliquot of the culture was taken and analyzed for both target cell kill & IFN-g secretion by CAR-T cells. To measure cytokine release, T cells and target cells were co-incubated for 24 hours at the ratios indicated. Supernatant media was collected for use in IL-2 or IFN.gamma. ELISAs (RD Systems) on a new plate following the manufacturer's instructions (RD Systems). To quantify cell killing, cells were washed, media was replaced with 200 mL of media containing a 1:500 dilution of 5 mg/mL DAPI (Molecular Probes) (to enumerate dead/dying cells). Finally, 25 mL of CountBright beads (Life Technologies) was added to each well. Cells were then processed by flow cytometry. [0712] 1) Cells/mL=((number of live target cell events)/(number of bead events)).times.((Assigned bead count of lot (beads/50 .mu.L))/(volume of sample)) [0713] 2) Total target cells were calculated by multiplying cells/mL.times.the total volume of cells. [0714] 3) The percent cell lysis was then calculated with the following equation:
[0714] % Cell lysis=(1-((Total Number of Target Cells in Test Sample)/(Total Number of Target Cells in Control Sample)).times.100
[0715] Two or three days after each challenge, cells were counted, washed, resuspended in fresh T cell media, and rechallenged with the same ratio of one anti-BCMA CAR.sup.+ T cell per two eFlour670 labeled MM.15 cells. Challenging of anti-BCMA CAR.sup.+ T cells with BCMA+ MM.1S cells was repeated 10 sequential times. Prior to any challenge with MM.1S cells, co-incubation of either 2.times. KO (TRAC-/B2M-) or 4.times. KO (TRAC-/B2M-/CD70-/PD-1-) anti-BCMA CAR+ T cells with MM.1S cells resulted in complete killing of target cells. Additionally, IFN.gamma. production by both 2.times. KO and 4.times. KO anti-BCMA CAR+ T cells was similar. Following a 4th rechallenge with MM.1S cells however, target cell killing and IFN.gamma. production by 2.times. KO anti-BCMA CAR+ T cells decreased relative to that induced by 4.times. KO anti-BCMA CAR+ T cells. By the 8th rechallenge, target cell killing was only approximately 20% for 2.times. KO anti-BCMA CAR+ T cells, while both IFNg and target cell killing by 4.times. KO anti-BCMA CAR+ T cells remained comparable to that seen prior to any challenge with MM.1S cells (FIGS. 24A-24B). In addition, the quadruple knockout anti-BCMA CAR T cells showed higher proliferation in response to exposure to target cells (FIG. 24C).
[0716] To ensure long-term benefit upon CAR T treatment, CAR T cells should be able to identify and eradicate their target cells over long period of time, to rule out the possibility of cancer cell escape from CAR-T mediated cell killing. The in vitro rechallenge assay mimics a recurrent encounter of CAR-T cells with target cells over several cycles of CAR-T cell activation, therefore demonstrating the superiority of the 4.times. knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-BCMA CAR.sup.+ T cells, in sustaining multiple challenges with target cells, without showing reduction of cell killing ability, as compared to the double knockout TRAC.sup.-/.beta.2M.sup.-/anti-BCMA CAR+ T cells (FIGS. 24A-24C).
Example 7. CD70 KO Overcomes Challenge of Excess Inhibitory Molecules
[0717] Comparison of the Effects of Multi-Knockout Anti-CD70 CAR+ T cells on A498-PD-L1 Renal Carcinoma Cells
[0718] Cell Kill Assay. The ability of multi-gene edited anti-CD70 CAR.sup.+ cells to kill A498 renal carcinoma cells overexpressing PD-L1 was determined using the cell kill assay described herein. To create cells overexpressing PD-L1 (CD274), A498 cells were infected with lentivirus encoding a PD-L1 cDNA and a puromycin resistance gene (Genecopoeia). After selection with puromycin, cells were stained with an anti-PD-L1 antibody to assess expression of PD-L1. The A498 cells expressing PD-L1 are referred to as A498-PD-L1 and were used in the functional assays described.
[0719] The TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ (2.times. KO, CD70 CAR.sup.+), TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/anti-CD70 CAR.sup.+ (3.times. KO (PD-1), CD70 CAR.sup.+), TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ (3.times. KO (CD70), CD70 CAR.sup.+) and TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ (4.times. KO, CD70 CAR.sup.+) T cells were incubated with the A498-PD-L1 cells at a CAR T cell:A498-PD-L1 target cells ratio of 2:1 (FIG. 25A), 1:1 (FIG. 25B), or 0.5:1 (FIG. 25C). The CD70 knockout cells exhibited potent cell killing of RCC-derived cells following 24-hour co-incubation (FIGS. 25A-25C). The cells with PD1 knockout alone did not effectively lyse cells in the presence of PD-L1 overexpression. However, the CD70 knockout was able to rescue the PD1 knockout and enhanced cell lysis was observed in the CART cells with CD70 KO and PD1 KO. These data demonstrate that the loss of CD70 on the surface of these CAR-T cells enhances their function even in the presence of highly immune suppressive molecules expressed by tumor cells such as PD-L1.
[0720] Cytokine Release Assay. A cytokine release assay was performed as described herein. The ability of the double knockout, triple knockout, and quadruple knockout anti-CD70 CAR.sup.+ T cells to produce IL-2 and IFN-g when co-cultured in the presence of A948-PD-L1 cells following 24-hour co-incubation at a ratio (CAR T cell:A948-PD-L1 target cell) of 1:1 was assessed using an ELISA assay. IL-2 and IFN-g from supernatants of cell co-cultures were measured. The quadruple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells secreted the highest levels of IFN-g (FIG. 26A) and IL-2 (FIG. 26B) when cultured with A948-PD-L1 cells. These data demonstrate the knock-out of CD70 enhances CAR-T cells secretion of cytokines even in the presence of highly immune suppressive molecules expressed by tumor cells such as PD-L1. The knockout of CD70 together with a knockout of PD-1 in CAR-T cells further enhances the effect. Without wishing to be bound by theory, it is believed knocking-out CD70 in anti-CD70 CAR+ T cells can rescue the detrimental phenotypes of other cell knockouts (e.g.: PD1). These data demonstrate that knocking-out CD70 in CAR T cells enhances target cell killing and CAR T cell function in a highly immune suppressive context.
Example 8. CD70 Knockout Improves In Vivo Efficacy
[0721] Efficacy of CD70 and PD1 knockout in Anti-CD70 CART cells: the Subcutaneous Renal Cell Carcinoma Tumor Xenograft Model in NOG Mice
[0722] Treatment in Small Tumor Model
[0723] The ability of T cells expressing a CD70 CAR to eliminate kidney carcinoma cells that express high levels of CD70 was evaluated in in vivo using a subcutaneous renal cell carcinoma (A498) tumor xenograft model in mice.
[0724] CRISPR/Cas9 and AAV6 were used as above (see for example, Example 3) to create human T cells that lack expression of the TCR, .beta.2M, CD70 and/or PD1 with concomitant expression from the TRAC locus using a CAR construct targeting CD70 (SEQ ID NO: 45; SEQ ID NO: 46). In this example activated T cells were first electroporated with 2, 3 or 4 distinct Cas9:sgRNA RNP complexes containing sgRNAs targeting TRAC (SEQ ID NO: 40), .beta.2M (SEQ ID NO: 41), PD1 (SEQ ID NO: 42), and CD70 (SEQ ID NO: 36 or 37). The DNA double stranded break at the TRAC locus was repaired by homology directed repair with an AAV6-delivered DNA template comprising a donor template (SEQ ID NO: 44; SEQ ID NO: 45) (encoding anti-CD70 CAR comprising the amino acid sequence of SEQ ID NO: 45) containing right and left homology arms to the TRAC locus flanking a chimeric antigen receptor cassette (-/+ regulatory elements for gene expression).
[0725] The resulting modified T cells are 2.times. KO (TRAC-/.beta.2M-), 3.times. KO (TRAC-/.beta.2M-/PD1- or TRAC-/.beta.2M-/CD70-) and 4.times. KO (TRAC-/.beta.2M-/PD1-/CD70-) anti-CD70 CAR+ (with 41BB costimulatory domain) T cells. The ability of these anti-CD70 CAR+ T cells to ameliorate disease caused by a CD70+ renal carcinoma cell line was evaluated in NOG mice using methods employed by Translational Drug Development, LLC (Scottsdale, Ariz.). In brief, 12, 5-8 week old female, CIEA NOG (NOD.Cg-Prkdc.sup.scidI12rg.sup.tm1Sug/JicTac) mice were individually housed in ventilated microisolator cages, maintained under pathogen-free conditions, 5-7 days prior to the start of the study. Mice received a subcutaneous inoculation of 5.times.10.sup.6 A498 renal carcinoma cells/mouse in the right hind flank. When mean tumor size reached 25-75 mm.sup.3 (target of .about.50 mm.sup.3), the mice were further divided into 5 treatment groups as shown in Table 15. On Day 1, treatment group 2 to 5 received a single 200 .mu.l intravenous dose of anti-CD70 CAR+ T cells according to Table 15.
TABLE-US-00015 TABLE 15 Treatment groups T cell treatment Group CAR-T A498 cells (i.v.) N 1 None 5 .times. 10.sup.6 None 5 cells/mouse 2 2X KO, anti-CD70 5 .times. 10.sup.6 1 .times. 10.sup.7 5 CAR+ T cells cells/mouse cells/mouse 3 3X KO (PD1), anti- 5 .times. 10.sup.6 1 .times. 10.sup.7 5 CD70 CAR+ T cells cells/mouse cells/mouse 4 3X KO (CD70,) anti- 5 .times. 10.sup.6 1 .times. 10.sup.7 5 CD70 CAR+ T cells cells/mouse cells/mouse 5 4X KO (CD70, PD1), 5 .times. 10.sup.6 1 .times. 10.sup.7 5 anti-CD70 CAR+ T cells cells/mouse cells/mouse
[0726] Tumor volume was measured 2 times weekly from day of treatment initiation. By day 5 treatment with all four types of anti-CD70 CAR T cells began to show a decrease in tumor volume and by day 22, all four types of anti-CD70 CAR T cells completely eliminated CD70+ kidney cancer tumors during the duration of the study until day 91 (FIG. 27A). These data demonstrate that all four anti-CD70 CAR T cells can regress CD70+ kidney cancer tumors in vivo.
[0727] To test the activity of the anti-CD70 CAR T cells after rechallenge, surviving mice were inoculated in the subcutaneous left hind flank with 5.times.10.sup.6 A498 renal carcinoma cells/mouse on day 25. (Table 16). Sustained efficacy was evaluated from day 46 onward. Results are shown in FIG. 27B. At day 56, 5 out of 5 mice treated with 2.times., CD70 CAR+ T cells exhibited tumors regrowth post rechallenge, 4 out of 5 mice treated with 3.times. (PD1), CD70 CAR+ T cells exhibited tumors regrowth post rechallenge, 2 out of 5 mice treated with 4.times. (CD70,PD1), CD70 CAR+ T cells exhibited tumors regrowth post rechallenge, while none of the mice treated with 3.times. (CD70), CD70 CAR+ T cells exhibited tumors regrowth post rechallenge. This trend continued, at day 70, 4 out of 5 mice treated with 3.times. (PD1), CD70 CAR.sup.+ T cells exhibited tumors regrowth post rechallenge, 4 out of 5 mice treated with 4.times. (CD70, PD1), CD70 CAR.sup.+ T cells exhibited tumors regrowth post rechallenge, while only one of the mice treated with 3.times. (CD70), CD70 CAR.sup.+ T cells exhibited a small tumor regrowth that began to appear at 34 days post rechallenge. Even out to day 91, only 1 of 5 mice treated with 3.times. (CD70), CD70 CAR.sup.+ T cells was starting to exhibit tumor regrowth, indicating that TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells retain a higher in vivo efficacy after re-exposure to tumor cells.
TABLE-US-00016 TABLE 16 Size of rechallenge tumors in untreated mice or mice treated with CD70 CAR T cells Tumor volume (mm.sup.3) at day post CAR T cell dosing Day Day CAR T treatment Subject Day 77 81 Day 84 88 Day 91 2KO, CD70 CAR T 1 1044 1265 1853 1927 2150 2 653 927 1040 1123 1256 3 899 1267 1603 1678 2167 4 701 1287 1672 1817 2490 5 689 1146 1423 1525 1901 3KO (PD1), CD70 1 385 692 983 1172 1369 CAR T 2 0 0 0 0 0 3 566 738 1030 1537 1740 4 712 1111 1337 1482 1832 5 632 778 1017 1129 1289 3KO (CD70), CD70 1 0 0 0 0 0 CAR T 2 34 56 75 104 135 3 0 0 0 0 0 4 0 0 0 0 0 5 0 0 0 0 0 4KO, CD70 CAR T 1 66 91 182 215 304 2 56 85 119 126 155 3 0 0 0 0 0 4 76 175 218 256 316 5 35 30 51 58 63 No treatment 1 567 1263 1673 1751 2020 2 882 1214 1535 1609 2047 3 1158 1304 1676 1924 2389 4 295 391 667 789 1078 5 707 1213 1676 1766 2056
Treatment in Large Tumor Model
[0728] The in vivo efficacy of anti-CD70 CAR T cells against larger renal cell carcinoma tumors was investigated. As above, CRISPR/Cas9 and AAV6 were used to create human T cells that lack expression of the TCR, .beta.2M, CD70 and/or PD1 with concomitant expression from the TRAC locus using a CAR construct targeting CD70 (SEQ ID NO: 45). In this example activated T cells were first electroporated with 2, 3 or 4 distinct Cas9:sgRNA RNP complexes containing sgRNAs targeting TRAC (SEQ ID NO: 40), .beta.2M (SEQ ID NO: 41), PD1 (SEQ ID NO: 42), and CD70 (SEQ ID NO: 36 or 37). The DNA double stranded break at the TRAC locus was repaired by homology directed repair with an AAV6-delivered DNA template (SEQ ID NO: 43) (encoding anti-CD70 CAR comprising the amino acid sequence of SEQ ID NO: 5) containing right and left homology arms to the TRAC locus flanking a chimeric antigen receptor cassette (-/+ regulatory elements for gene expression).
[0729] The resulting modified T cells are 2.times. KO (TRAC-/.beta.2M-), 3.times. KO (TRAC-/.beta.2M-/PD1- or TRAC-/.beta.2M-/CD70-) and 4.times. KO (TRAC-/.beta.2M-/PD1-/CD70-) anti-CD70 CAR+ (with 41BB costimulatory domain) T cells. The ability of these anti-CD70 CAR+ T cells to ameliorate disease caused by a CD70+renal carcinoma cell line was evaluated in NOG mice using methods employed by Translational Drug Development, LLC (Scottsdale, Ariz.). In brief, 12, 5-8 week old female mice, CIEA NOG (NOD.Cg-Prkdc.sup.scidI12rg.sup.tm1Sug/JicTac) were individually housed in ventilated microisolator cages, maintained under pathogen-free conditions, 5-7 days prior to the start of the study. Mice received a subcutaneous inoculation of 5.times.10.sup.6 A498 renal carcinoma cells/mouse. When mean tumor size reached 125-175 mm.sup.3 (target of .about.150 mm.sup.3), the mice were further divided into 5 treatment groups as shown in Table 17. On day 1, treatment group 2 to 5 received a single 200 .mu.l intravenous dose of anti-CD70 CAR+ T cells according to Table 17.
TABLE-US-00017 TABLE 17 Treatment groups T cell treatment Group CAR-T A498 cells (i.v) N 1 None 5 .times. 10.sup.6 None 5 cells/mouse 2 2X KO, CD70 5 .times. 10.sup.6 1 .times. 10.sup.7 5 CAR+ T cells cells/mouse cells/mouse 3 3X KO (PD1), 5 .times. 10.sup.6 1 .times. 10.sup.7 4 CD70 CAR+ T cells cells/mouse cells/mouse 4 3X KO (CD70,) 5 .times. 10.sup.6 1 .times. 10.sup.7 5 CD70 CAR+ T cells cells/mouse cells/mouse 5 4X KO (CD70, PD1), 5 .times. 10.sup.6 1 .times. 10.sup.7 5 CD70 CAR+ T cells cells/mouse cells/mouse
[0730] Tumor volume was measured 2 times weekly from day of treatment initiation. By day 4 treatment only the 3.times. KO (TRAC-/.beta.2M-/CD70-) anti-CD70 CAR+ T cells and 4.times. KO (TRAC-/.beta.2M-/PD1-/CD70-) anti-CD70 CAR+ T cells began to show a decrease in tumor volume (FIG. 27C). In contrast, the tumor growth for animals treated with 2.times. KO (TRAC-/.beta.2M-) anti-CD70 CAR+ T cells or 3.times. KO (TRAC-/.beta.2M-/PD1-) anti-CD70 CAR+ T cells was similar to the no treatment group. By day 23 treatment, the 3.times. KO (TRAC-/.beta.2M-/CD70-) anti-CD70 CAR+ T cells completely eliminated CD70+ kidney cancer tumors in vivo. By day 23 treatment, elimination of the tumors in response to the 4.times. KO (TRAC-/.beta.2M-/PD1-/CD70-) anti-CD70 CAR+ T cells was almost complete, with 4 of 5 mice exhibiting no detectable kidney cancer tumors in vivo.
[0731] These data show that 3.times. KO (TRAC-/.beta.2M-/CD70-) anti-CD70 CAR+ T cells and 4.times. KO (TRAC-/.beta.2M-/PD1-/CD70-) anti-CD70 CAR+ T cells can significantly regress large CD70+ kidney cancer tumors in vivo.
In Vivo Tumor Model for anti-BCMA CAR in context of PD1, CD70, and PD1 with CD70 Knock Outs.
[0732] The efficacy of TRAC-/.beta.2M-/anti-BCMA (4-1BB co-stim) CAR+ T cells, TRAC-/.beta.2M-/PD-1-/anti-BCMA (4-1BB co-stim), TRAC-/.beta.2M-/CD70-/anti-BCMA (4-1BB co-stim), and TRAC-/.beta.2M-/PD-1-/CD70-/anti-BCMA (4-1BB co-stim) CAR+ T cells against the subcutaneous MM.1S tumor xenograft model in NOG mice was evaluated. In brief, 25, 5-8 week old female, CIEA NOG (NOD.Cg-Prkdc.sup.scidI12rg.sup.tm1Sug/JicTac) mice were individually housed in ventilated microisolator cages, maintained under pathogen-free conditions, 5-7 days prior to the start of the study. On day 1, 25 mice received a subcutaneous inoculation in the right flank of 5.times.10.sup.6 MM.1S cells in 50% Matrigel/mouse. When the mean tumor volume reached between 75 and 125 mm.sup.3, the mice were divided into 5 treatment groups (N=5) and dosed with T cell populations comprising .about.50% anti-BCMA CAR.sup.+ T cells, as indicated in Table 18.
TABLE-US-00018 TABLE 18 Dosing # of T Cells Anti-BCMA Group CAR T Cell injected CAR+ T cells N 1 N/A N/A N/A 5 2 TRAC-/.beta.2M-/ 1 .times. 10.sup.7 5 .times. 10.sup.6 5 anti-BCMA cells/mouse (5 million) 3 TRAC-/.beta.2M-/ 1 .times. 10.sup.7 5 .times. 10.sup.6 5 PD-1-/anti-BCMA cells/mouse (5 million) 4 TRAC-/.beta.2M-/ 1 .times. 10.sup.7 5 .times. 10.sup.6 5 CD70-/anti-BCMA cells/mouse (5 million) 5 TRAC-/.beta.2M-/ 1 .times. 10.sup.7 5 .times. 10.sup.6 5 PD-1-/CD70-/ cells/mouse (5 million) anti-BCMA
[0733] Tumor volume and body weights were measured twice weekly and individual mice were euthanized when their tumor volume reached .gtoreq.2000 mm.sup.3.
[0734] By day 16, all treatment groups showed tumor regression from the starting volumes while animals in the control group had tumors averaging greater than 1500 mm.sup.3. By day 27, all animals in the control group had reached the tumor volume endpoint of .gtoreq.2000 mm.sup.3 while all treatment groups had an average tumor volume less than 20 mm.sup.3 (FIG. 28A). On day 45, all mice from each treatment group (Groups 2-5) were further subjected to a secondary tumor challenge (re-challenge). The mice received a second subcutaneous inoculation in the left flank of 5.times.10.sup.6 MM.1S cells in 50% Matrigel/mouse. A new group of control mice were entered (N=5) and also received an inoculation of 5.times.10.sup.6 MM.1S cells in 50% Matrigel/mouse in the left flank.
[0735] All mice were monitored for tumor growth in both the initial right flank tumor and the rechallenge tumor in the left flank. All treatment groups successfully inhibited tumor growth in the initial right flank tumor in most subjects (FIG. 28A; Table 19). Tumor growth was inhibited by all treatments both before and after tumor re-challenge for the duration of the experiment to day 77. Only one subject treated with TRAC-/.beta.2M-/CD70-/PD1-/anti-BCMA CAR.sup.+ T cells exhibited tumor growth from the initial cancer cell challenge (Table 19).
[0736] Surprisingly, tumor growth after re-challenge in the left flank was also significantly inhibited by all treatment groups from the date of re-challenge (day 45) to day 77 (FIG. 28B; Table 19). These data demonstrate that the CAR+ T cells persist in vivo to inhibit initial tumor growth, as well as inhibiting growth of new tumors following a re-challenge with additional cancer cells even though no further CAR-T cells were delivered to these mice. For example, three of the four mice initially treated with populations of TRAC-/.beta.2M-/anti-BCMA CAR.sup.+ T cells, three of the five mice initially treated with TRAC-/.beta.2M-/CD70-/anti-BCMA CAR.sup.+ T cells, and three of the five mice initially treated with TRAC-/.beta.2M-/PD1-/anti-BCMA CAR.sup.+ T cells, exhibited no new tumor growth despite a second challenge (re-challenge) to new cancer cells. These data demonstrate that, unexpectedly, anti-BCMA CAR.sup.+ T cells are capable of persisting for long periods of time in vivo, e.g., up to at least 77 days following injection, and retain their ability to inhibit tumor cell growth and reduce tumor volumes for long periods in vivo. These surprising results indicate that use of such TRAC-/.beta.2M-/anti-BCMA CAR.sup.+ T cells would achieve superior long-term anti-cancer effect in vivo.
[0737] Human CD45+ cells were quantified from mouse blood using BD Trucount tubes following the manufacturers protocol and detected using Brilliant Violet 786 conjugated anti-human CD45 (Biolegend Cat #368528). All groups showed values of less than 100 huCD45+ cells/.mu.l at 1 week. Two weeks post dosing, the number of circulating CD45+ in all groups peaked before falling to pre-week 1 values by week 3 (FIG. 29). Upon re-challenge at Day 45, the anti-BCMA CAR+ T cell treated subjects were able to eliminate or inhibit tumor growth without subsequent expansion of circulating CAR T cells following cancer rechallenge. These data further demonstrate that, unexpectedly, anti-BCMA CAR+ T cells are capable of persisting for long periods of time in vivo, e.g., up to at least 77 days following injection, and retain their ability to inhibit tumor cell growth and reduce tumor volumes for long periods in vivo. These surprising results indicate that use of TRAC-/.beta.2M-/anti-BCMA CAR+ T cells, TRAC-/.beta.2M-/CD70-/anti-BCMA CAR+ T cells and TRAC-/.beta.2M-/PD1-/anti-BCMA CAR+ T cells would achieve superior long-term anti-cancer effect in vivo.
TABLE-US-00019 TABLE 19 Size of tumors in untreated mice or mice treated with anti-BCMA CAR T cells Tumor volume Tumor volume at Day 45 (mm.sup.3) at Day 77 (mm.sup.3) Treatment Mouse Right Flank Right Flank Left Flank No Treatment 1 TS TS TS 2 TS TS TS 3 TS TS TS 4 TS TS TS 5 TS TS TS TRAC-/.beta.2M-/ 1 0 0 (MS at 0 (MS at anti-BCMA Day 59) Day 59:) 2 0 0 0 3 0 0 0 4 0 0 297 5 FD-T at FD-T at FD-T at day 16 day 16 day 16 TRAC-/ 1 0 0 1820 .beta.2M-/PD1-/ 2 0 0 0 anti-BCMA 3 0 0 487 4 0 0 0 5 0 0 0 TRAC-/ 1 10 0 0 .beta.2M-CD70-/ 2 0 0 0 anti-BCMA 3 0 0 (TS at 2349 (TS at day 77) day 77) 4 0 0 0 5 0 0 258 TRAC-/.beta.2M-/ 1 0 0 1157 PD1-/CD70-/ 2 0 0 1842 anti-BCMA 3 0 0 1664 4 0 0 0 5 89 1583 (TS at 1560 (TS at day 73) day 73) TS = sacrificed because of tumor volume; MS = Moribund sacrifice; FD-T = animal found dead.
[0738] In vivo Tumor Model for Anti-BCMA CAR in Context of CD70 Knockout: Effect of CD70 KO on Moderate CAR T Dosing
[0739] The efficacy of several anti-BCMA CAR.sup.+ T cell genotypes, both with and without CD70 knockouts, was evaluated against the subcutaneous RPMI-8226 tumor xenograft model in NOG mice. In brief, eighty five (85), 5-8 week old female, CIEA NOG (NOD.Cg-Prkdc.sup.scidI12rg.sup.tm1Sug/JicTac) mice were individually housed in ventilated microisolator cages, maintained under pathogen-free conditions, 5-7 days prior to the start of the study. On day 1 mice received a subcutaneous inoculation of 10.times.10.sup.6 RPMI-8226 cells/mouse. Ten (10) days post inoculation with RPMI-8226 cells, the mice were divided into 17 treatment groups (N=5) and dosed with T cell populations comprising .about.80% anti-BCMA CAR.sup.+ T cells, as indicated in Table 20.
TABLE-US-00020 TABLE 20 Anti-BCMA # of T Cells Anti-BCMA Group CAR T Cell injected CAR+ T cells N 1 N/A N/A N/A 5 2 TRAC-/.beta.2M-/ 3 .times. 10.sup.6 2.4 .times. 10.sup.6 5 anti-BCMA cells/mouse (2.4 million) 3 TRAC-/.beta.2M-/ 1 .times. 10.sup.6 .sup. 8 .times. 10.sup.5 5 anti-BCMA cells/mouse (0.8 million) 4 TRAC-/.beta.2M-/ 3 .times. 10.sup.5 2.4 .times. 10.sup.5 5 anti-BCMA cells/mouse (0.24 million) 5 TRAC-/.beta.2M-/ 1 .times. 10.sup.5 .sup. 8 .times. 10.sup.4 5 anti-BCMA cells/mouse (0.08 million) 6 TRAC-/.beta.2M-/ 3 .times. 10.sup.6 2.4 .times. 10.sup.6 5 PD1-/anti-BCMA cells/mouse (2.4 million) 7 TRAC-/.beta.2M-/ 1 .times. 10.sup.6 .sup. 8 .times. 10.sup.5 5 PD1-/anti-BCMA cells/mouse (0.8 million) 8 TRAC-/.beta.2M-/ 3 .times. 10.sup.5 2.4 .times. 10.sup.5 5 PD1-/anti-BCMA cells/mouse (0.24 million) 9 TRAC-/.beta.2M-/ 1 .times. 10.sup.5 .sup. 8 .times. 10.sup.4 5 PD1-/anti-BCMA cells/mouse (0.08 million) 10 TRAC-/.beta.2M-/ 3 .times. 10.sup.6 2.4 .times. 10.sup.6 5 CD70-/anti-BCMA cells/mouse (2.4 million) 11 TRAC-/.beta.2M-/ 1 .times. 10.sup.6 .sup. 8 .times. 10.sup.5 5 CD70-/anti-BCMA cells/mouse (0.8 million) 12 TRAC-/.beta.2M-/ 3 .times. 10.sup.5 2.4 .times. 10.sup.5 5 CD70-/anti-BCMA cells/mouse (0.24 million) 13 TRAC-/.beta.2M-/ 1 .times. 10.sup.5 .sup. 8 .times. 10.sup.4 5 CD70-/anti-BCMA cells/mouse (0.08 million) 14 TRAC-/.beta.2M-/ 3 .times. 10.sup.6 2.4 .times. 10.sup.6 5 PD1-/CD70-/ cells/mouse (2.4 million) anti-BCMA 15 TRAC-/.beta.2M-/ 1 .times. 10.sup.6 .sup. 8 .times. 10.sup.5 5 PD1-/CD70-/ cells/mouse (0.8 million) anti-BCMA 16 TRAC-/.beta.2M-/ 3 .times. 10.sup.5 2.4 .times. 10.sup.5 5 PD1-/CD70-/ cells/mouse (0.24 million) anti-BCMA 17 TRAC-/.beta.2M-/ 1 .times. 10.sup.5 .sup. 8 .times. 10.sup.4 5 PD1-/CD70-/ cells/mouse (0.08 million) anti-BCMA
[0740] Tumor volume and body weight was measured twice weekly, and individual mice were euthanized when tumor volume was .gtoreq.2000 mm.sup.3. By day 22, the data show a statistically significant decrease in the tumor volume in response to higher doses of anti-BCMA CAR T cells (1.times.10.sup.5-3.times.10.sup.6 cell doses) compared to any anti-BCMA CART cell genotype dosed at 100,000 cells (groups 5, 9, 13 and 17) (FIG. 30; Table 21).
[0741] At day 36, the TRAC-/.beta.2M-/CD70-/anti-BCMA CAR+ T cells dosed at a moderate dose of 3.times.10.sup.5 cells exhibited a greater effect on decreasing tumor volume than the anti-BCMA CAR+ T cells without a CD70 KO (e.g., TRAC-/.beta.2M-/anti-BCMA CAR+ T cells, TRAC-/.beta.2M-/PD1-/anti-BCMA CAR+ T cells, or TRAC-/.beta.2M-/PD1-/CD70-/anti-BCMA CAR+ T cells) (FIG. 30). All of the higher doses of 1.times.10.sup.6 or greater all anti-BCMA CAR+ T cells (FIG. 30; 1 Mil, 3 Mil), showed complete regression in tumor volume. This trend continued out to Day 57 of the study.
[0742] These results demonstrate that inhibiting the activity of CD70 (e.g., by knocking out CD70) increases the efficacy and potency of CAR.sup.+ T cells in vivo. This effect is independent of the presence of an anti-CD70 CAR.
TABLE-US-00021 TABLE 21 Anti- BCMA CAR+ T cells/ Tumor Volume (mm.sup.3) Tumor Volume (mm.sup.3) Group Treatment dose at Day 36 at Day 57 1 No N/A 220 220 186 173 278 1790 1794 938 2055 2029 Treatment 2 TRAC-/ 2.4 .times. 0 0 0 0 0 0 0 0 0 0 .beta.2M-/anti- 10.sup.6 BCMA (2.4 million) 3 8 .times. 0 0 0 0 0 0 0 0 0 0 10.sup.5 (0.8 million) 4 2.4 .times. 65 65 77 56 0 516 441 257 97 337 10.sup.5 (0.24 million) 5 8 .times. 264 264 386 276 185 2391 2283 2058 2147 2359 10.sup.4 (0.08 million) 6 TRAC-/ 2.4 .times. 0 0 0 0 0 0 0 0 0 0 .beta.2M-/ 10.sup.6 PD1-/anti- (2.4 BCMA million) 7 8 .times. 0 0 0 0 0 0 0 0 0 0 10.sup.5 (0.8 million) 8 2.4 .times. 135 135 59 57 28 764 518 280 181 79 10.sup.5 (0.24 million) 9 8 .times. 10.sup.4 261 261 265 287 312 1532 1557 2218 2010 2098 (0.08 million) 10 TRAC-/ 2.4 .times. 0 0 0 0 0 0 0 0 0 0 .beta.2M-/ 10.sup.6 CD70-/ (2.4 anti- million) 11 BCMA 8 .times. 0 0 0 0 0 0 0 0 0 0 10.sup.5 (0.8 million) 12 2.4 .times. 47 47 0 0 0 526 58 47 0 0 10.sup.5 (0.24 million) 13 8 .times. 10.sup.4 292 292 267 313 235 2075 2127 2096 1365 2354 (0.08 million) 14 TRAC-/ 2.4 .times. 0 0 0 0 0 0 0 0 0 0 .beta.2M-/ 10.sup.6 PD1-/ (2.4 CD70-/ million) 15 anti- 8 .times. 0 0 0 0 0 0 0 0 0 0 BCMA 10.sup.5 (0.8 million) 16 2.4 .times. 100 100 91 19 20 478 576 82 131 289 10.sup.5 (0.24 million) 17 8 .times. 310 310 319 345 451 1528 2160 2843 2557 1499 10.sup.4 (0.08 million)
Example 9. Multi Knockout CAR T Cells Retain Cytokine Dependency
[0743] Cytokine Dependency. To determine whether gene editing resulted in unwanted off-target editing that could generate cells with adverse properties, such as uncontrolled cell growth, the ability gene edited CAR T cells to grow in the absence of cytokines and/or serum was assessed.
[0744] Anti-CD70 CAR T cells: The ability of TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ cells to grow in the absence of cytokines and/or serum was assessed. 5.times.10.sup.6 TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ cells were plated .about.2 weeks post cell production (Day 0). The number of viable cells were enumerated 7 and 14 days post plating in either full media, 5% human serum without cytokines (IL-2 and IL-7), or base media lacking serum and cytokines. No cells were detected at 14 days plated in the cultures that lacked cytokines, suggesting that any potential off-target effects due to genome editing did not induce growth factor independent growth/proliferation to the cells (FIG. 31). The cells only proliferated in the presence of cytokines (full media that contains cytokines) and did not proliferate in the presence of serum alone. Thus, in vivo, the cells would likely not grow in the absence of cytokine, growth factor or antigen stimulation due to any off-target genome editing.
[0745] The ability of TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/PD1.sup.- anti-CD70 CAR.sup.+ cells to grow in the absence of cytokines and/or serum was also assessed. 2.times.10.sup.6 cells were plated .about.2 weeks post cell production (Day 0). The number of viable cells were enumerated until 26 days post plating in either full media, 5% human serum without cytokines (IL-2 and IL-7), or base media lacking serum and cytokines. No cells were detected at 26 days plated in the cultures that lacked cytokines, suggesting that any potential off-target effects due to genome editing did not induce growth factor independent growth/proliferation to the cells (FIG. 32). The cells only proliferated in the presence of cytokines (full media that contains cytokines) and did not proliferate in the presence of serum alone. Thus, genome editing did not induce any adverse events that allow the cells to grow in the absence of cytokine, growth factor or antigen stimulation.
[0746] Anti-BCMA CAR+ T cells: The ability of TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/PD-1.sup.-/anti-BCMA CAR.sup.+ cells to grow in the absence of cytokines and/or serum was assessed. TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/PD-1.sup.-/anti-BCMA CAR.sup.+ cells are also referred to as 4.times. KO, BCMA CAR.sup.+ cells. 1.times.10.sup.6 4.times. KO, BCMA CAR+ cells were plated following the 10 rechallenges described in Example 6. The number of viable cells were enumerated 7 and 14 days post plating in either full media, 5% human serum without cytokines (IL-2 and IL-7), or base media lacking serum and cytokines. No cells were detected at 13 days plated in the cultures that lacked cytokines, suggesting that any potential off-target effects due to genome editing did not induce growth factor independent growth/proliferation to the cells (FIG. 33). The cells only proliferated in the presence of cytokines (full media that contains cytokines) and did not proliferate in the presence of serum alone. Thus, in vivo, the cells would likely not grow in an uncontrolled way.
[0747] Other CAR T cells: It has previously been shown that the anti-CD33 CAR+ T cells and anti-CD19 CAR+ T cells exemplified herein only proliferated in the presence of cytokines and do not proliferate in the presence of serum alone. Thus, in vivo, these cells would likely not grow in an uncontrolled way.
[0748] Cytokine Release Assay. To measure cytokine release, T cells and target cells were co-incubated for 24 hours at the ratios indicated. Supernatant media was collected for use in IL-2 or IFN.gamma. ELISAs (RD Systems) on a new plate following the manufacturer's instructions (RD Systems).
[0749] The ability of the TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ cells to produce interleukin-2 (IL-2) when co-cultured in the presence of A498 cells was analyzed using the ELISA assay. Both the triple knockout TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells and double knockout TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ T cells secreted high levels of IL-2. Strikingly, the TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ cells secreted higher levels of IL-2 than the TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ cells when cultured with A498 cells (FIG. 34). These results suggest that knocking-out the CD70 gene gives an advantage to anti-CD70 CAR+ T cells to secrete more IL-2.
Example 10. Effect of Multiple Knockout on Anti-CD70 CAR+ T cells on A498 Renal Carcinoma Cells
Effect of Multi Knock-Out on the Function of Anti-CD70 CAR+ T Cells.
[0750] Cell Killing Assay. The ability of multi-gene editing to kill A498 renal carcinoma cellswas determined using the cell kill assay described above. In brief, the TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ (2.times. KO, CD70 CAR.sup.+), TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/anti-CD70 CAR.sup.+ (3.times. KO (PD-1), CD70 CAR.sup.+), TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ (3.times. KO (CD70), CD70 CAR.sup.+) and TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ (4.times. KO, CD70 CAR.sup.+) cells were incubated with a CD70.sup.+ adherent RCC-derived cell line (A498 cells) at various CAR T cell:A498 target cells ratios.
[0751] The TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ cells exhibited potent cell killing of RCC-derived cells following 24-hour co-incubation (FIG. 35). The quadruple TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T demonstrated higher cell kill potency than triple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/anti-CD70 CAR.sup.+ T cells that demonstrated higher potency than double knockout TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ T cells (visible at low T-cell: A498 Ratio of 0.5:1 and 0.25:1). The results demonstrate knocking out both the CD70 and PD-1 genes gave the anti-CD70 CAR+ cells higher cell kill potency.
[0752] The gene edited cells also exhibited potent cell killing of RCC-derived cells following 24-hour co-incubation at a CAR T cell:A948 target cell ratio of 0.24:1. (FIG. 36). Specifically, the triple knockout TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells and the quadruple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells demonstrated higher potency than the double knockout TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ T cells or the triple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/anti-CD70 CAR.sup.+ T cells. These data indicate that knockout of CD70 in the context of an anti-CD70 CAR improved the cell killing ability of the anti-CD70 CAR.sup.+ T cells.
[0753] Cytokine Release Assay. A cytokine release assay was performed as described above. The ability of the double knockout, triple knockout, and quadruple knockout anti-CD70 CAR.sup.+ T cells to produce IL-2 and interferon gamma (IFN-gamma (IFN-g)) when co-cultured in the presence of A498 cells following 24-hour co-incubation at a ratio (CAR T cell:A498 target cell) of 0.25:1 was assessed using an ELISA assay. IL-2 and IFN-g from supernatants of cell co-cultures were measured. The triple knockout TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells and quadruple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells secreted the highest levels of IFN-g (FIG. 37A) and IL-2 (FIG. 37B) when cultured with A498 cells.
Effect of CD70 Knockout on Exhaustion Marker Expression
[0754] The levels of the exhaustion markers PD-1 and LAG3 were assessed on the TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ (2.times. KO, CD70 CAR.sup.+), TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/anti-CD70 CAR.sup.+ (3.times. KO (PD-1), CD70 CAR.sup.+), TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ (3.times. KO (CD70), CD70 CAR.sup.+) and TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ (4.times. KO, CD70 CAR.sup.+) T cells used in the Examples above. CD4.sup.+ T cells were assessed for PD-1 expression (FIG. 38) and both CD8.sup.+ T cells and CD4.sup.+ T cells were assessed for LAG3 expression (FIG. 39A and FIG. 39B, respectively) by flow cytometry.
[0755] The data demonstrate that CD70 KO reduces exhaustion marker expression in CAR T cells. The data in FIG. 38 shows that PD-1 expression is decreased, as expected, when PD-1 is knocked out, and it is also decreased when CD70 is knocked out.
[0756] The data in FIGS. 39A and 39B show that knocking out CD70, reduces the LAG3 expression marker in CD4 and CD8 cells.
[0757] The data demonstrate that knocking out CD70, specifically, could reduce the potential exhaustion of the CD8.sup.+ and CD4.sup.+ gene edited populations of CAR+ T cells leading to better therapeutics.
Example 11. CD70 KO Improves Cell Kill in Multiple Cell Types
[0758] CD70 Expression in Various Cancer Cell Lines. Relative CD70 expression was measured in various cancer cell lines to further evaluate the ability of anti-CD70 CAR.sup.+ T cells to kill various cancer types. CD70 expression was measured by FACS analysis using Alexa Fluor 647 anti-human CD70 antibody (BioLegend Cat. No. 355115). FIG. 40A (left graph) shows the relative expression of CD70 in ACHN cells, as measured by FACS, compared to other kidney cancer cell lines A498, 786-O, cacki-1 and Caki-2. Additionally, non-kidney cancer cell lines were evaluated for CD70 expression by FACS analysis (Table 22, FIG. 40A and FIG. 40B) using either an Alexa Fluor 647 anti-human CD70 antibody (BioLegend Cat. No. 355115; FIG. 40A, right panel) or a FITC anti-human CD70 antibody (BioLegend Cat. No. 355105) in FIG. 40B. SNU-1 (intestinal cancer cells) exhibited high levels of CD70 expression that were similar to A498 (FIG. 40A, right panel). SKOV-3 (ovarian), HuT78 (lymphoma), NCI-H1975 (lung) and Hs-766T (pancreatic) cell lines exhibited levels of CD70 expression that were similar or higher than ACHN but lower than A498 (Table 22, FIG. 40B).
TABLE-US-00022 TABLE 22 Relative CD70 Cell Line Cancer type expression A498 Kidney Carcinoma High ACHN Kidney (derived from Medium-Low metastasis) SK-OV-3 Ovarian Adenocarcinoma Medium NCI-H1975 Lung Adenocarcinoma (NSCLC) Medium Calu-1 Lung Carcinoma Low DU 145 Prostate Carcinoma Low SNU-1 Gastric Carcinoma High Hs 766T Pancreatic Carcinoma Medium MJ T cell Lymphoma High HuT78 T cell Lymphoma Medium HuT102 T cell Lymphoma Medium PANC-1 Pancreatic Carcinoma Low U937 AML No expression K562 chronic myelogenous leukemia No expression (Negative Control)
[0759] Cell Kill Assay. The ability of multi-gene edited anti-CD70 CAR+ cells to kill ACHN renal carcinoma cells was determined using the cell kill assay described above. The TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ (2.times. KO, CD70 CAR.sup.+), TRAC.sup.-/.beta.2M.sup.-/anti-CD70 CAR.sup.+ (3.times. KO (PD-1), CD70 CAR.sup.+), TRAC.sup.-/.beta.2M.sup.-/anti-CD70.sup.-/anti-CD70 CAR.sup.+ (3.times. KO (CD70), CD70 CAR.sup.+) and TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ (4.times. KO, CD70 CAR.sup.+) cells were incubated with an adherent RCC-derived cell line expressing low levels of CD70 antigen (ACHN cells) (FIG. 40A shows the relative expression of CD70 in ACHN cells, as measured by FACS, compared to other kidney cancer cell lines A498, 786-O,l cacki-1 and Caki-2) at a CAR T cell:ACHN target cells ratio of 0.5:1 (FIG. 40C) and 0.25:1 (FIG. 40D). The gene edited cells exhibited potent cell killing of RCC-derived cells following 24-hour co-incubation (FIGS. 40C and 40D). The cells demonstrated higher potency when PD-1 was knocked out, when CD70 was knocked out, and even slightly higher potency when both PD-1 and CD70 were knocked out. In conclusion, knockout of PD-1 or CD70 or of both PD-1 and CD70 together improves the cell killing ability of the anti-CD70 CAR+ cells in ACHN cells.
[0760] Although ACHN cells were found to express moderate to low levels of CD70, they were surprisingly susceptible to killing by 3.times. KO (PD-1), CD70 CAR+ T cells, 3.times. KO (CD70), CD70 CAR+ T cells, and 4.times. KO CD70 CAR+ T cells (FIG. 40C and 40D). This indicates that high CD70 expression is not a requirement for effective killing of a target cell by gene-edited T cells that express an anti-CD70 CAR. Additionally, given that the levels of CD70 expression on SNU-1, SK-OV-3, NCI-H1975 and HS-766T cell lines were found to be similar or higher than ACHN, it was expected that anti-CD70 CAR.sup.+ T cells would be especially efficient at killing these cancer cell types as well. Indeed, it was found that TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ (4.times. KO, CD70 CAR.sup.+) and TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ (3.times. KO (CD70), CD70 CAR.sup.+) exhibited surprisingly potent cell killing of numerous solid tumor cell lines after only 24 hours of co-culture (FIG. 40E shows killing by 4.times. KO CAR+ T cells and FIG. 40F shows killing by 3.times. KO CAR+ T cells). Both 3.times. KO, CD70 CAR+ and 4.times. KO, CD70 CAR+ T cells killed >60% of kidney, pancreatic, and ovarian tumor cells (A498, ACHN, SK-OV-3, and Hs-766T) at a 4:1 effector:target cell ratio and >50% at a 1:1 effector:target cell ratio. Cell killing of cancer cell lines that had medium to low CD70 expression (NCI-H1975, Calu-1 and DU 145) was still effective with >30% killing at an effector:target cell ratio of 4:1 within 24 hours of co-culture (FIGS. 40E and 40F). Longer exposure (i.e., 96 hours) to either 3.times. KO or 4.times. KO, CD70 CAR+ T cells resulted in an increase in cancer cell killing across all cell types, particularly for SKOV-3, Hs-766T, and NIC-H1975 cells wherein killing was >80% at an effector:target cell ratio of 1:1 (FIG. 40G).
[0761] SNU-1 Cell Kill by was Assessed by Visual Assessment.
[0762] Target cell killing following long exposure to CAR+ T cells was also assessed by microscopy for SNU-1 cancer cells. SNU-1 cells were plated at a density of 1 million cells per well in a 6 well plate and mixed at an effector:target ratio of 4:1 with 3.times. KO (CD70), anti-CD70 CAR.sup.+ T cells. The co-culture was incubated for six (6) days and the presence of viable cancer cells was assessed by microscope. All gastric carcinoma target cells (SNU-1) were eliminated in wells containing TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells, as compared to control wells, indicating cancer cells were completely eliminated by anti-CD70 CAR.sup.+ T cells with an extended co-culture.
[0763] The ability of anti-CD70 CAR+ T cells to selectively kill CD70-expressing cells was determined. A flow cytometry assay was designed to test killing of cancer cell suspension lines (e.g., K562, MM.1S and HuT78 cancer cells that are referred to as "target cells") by 3.times. KO (CD70) (TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-) anti-CD70 CAR+ T cells. Two of the target cell lines that were used were CD70-expressing cancer cells (e.g., MM.1S and HuT78), while a third that was used as negative control cancer cells lack CD70 expression (e.g., K562). The TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR+ T cells were co-cultured with either the CD70-expressing MM.1S or HuT78 cell lines or the CD70-negative K562 cell line. The target cells were labeled with 5 .mu.M efluor670 (eBiosciences), washed and seeded at a density of 50,000 target cells per well in a 96-well U-bottom plate. The target cells were co-cultured with TRAC.sup.-/.beta.2M.sup.-/CD70.sup.- anti-CD70 CAR+ T cells at varying ratios (0.5:1, 1:1, 2:1 and 4:1 CAR+ T cells to target cells) and incubated overnight. Target cell killing was determined following a 24 hour co-culture. The cells were washed and 200 .mu.L of media containing a 1:500 dilution of 5 mg/mL DAPI (Molecular Probes) (to enumerate dead/dying cells) was added to each well. Cells were then analyzed by flow cytometry and the amount of remaining live target cells was quantified.
[0764] FIG. 40H, FIG. 40I, and FIG. 40J demonstrate selective target cell killing by TRAC-/B2M-/CD70- anti-CD70 CAR+ T cells. A 24 hour co-culture with 3.times. KO (CD70) CAR+ T cells resulted in nearly complete killing of T cell lymphoma cells (HuT78), even at a low CAR+ T cell to CD70-expressing target cell ratio of 0.5:1 (FIG. 40J) Likewise, a 24 hour co-culture resulted in nearly complete killing of multiple myeloma cells (MM.1S) at all CAR+ T cell to target cell ratios tested (FIG. 40I). Killing of target cells was found to be selective in that TRAC-/B2M-/anti-CD70 CAR+ T cells induced no killing of CD70-deficient K562 cells that was above the level of control samples (e.g., either cancer cells alone or co-culture with no RNP T cells) at any effector:target cell ratio tested (FIG. 40H).
[0765] Cytokine Release Assay. A cytokine release assay was performed as described above. The ability of the double knockout, triple knockout, and quadruple knockout anti-CD70 CAR.sup.+ T cells to produce IL-2 and IFN-g when co-cultured in the presence of ACHN cells following 24-hour co-incubation at a ratio (CAR T cell:ACHN target cell) of 0.25:1 was assessed using an ELISA assay. IL-2 and IFN-g from supernatants of cell co-cultures were measured. The triple knockout TRAC.sup.-/.beta.2M.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells and quadruple knockout TRAC.sup.-/.beta.2M.sup.-/PD-1.sup.-/CD70.sup.-/anti-CD70 CAR.sup.+ T cells secreted the highest levels of IFN-g (FIG. 41) and IL-2 (FIG. 41B) when cultured with ACHN cells. In conclusion, knockout of CD70 or of both PD-1 and CD70 together improves the cell killing ability of the anti-CD70 CAR+ cells in ACHN cells.
Example 12. Efficacy of CD70 KO in Anti-CD70 CAR+ T Cells: The Tumor Xenograft Model in NOG Mice
Treatment in the Ovarian Tumor Model
[0766] The ability of T cells expressing an anti-CD70 CAR to eliminate ovarian adenocarcinoma cells that express moderate levels of CD70 was evaluated in vivo using a subcutaneous ovarian carcinoma (SKOV-3) tumor xenograft model in mice.
[0767] CRISPR/Cas9 and AAV6 were used as above (see for example, Example 3) to generate human T cells that lack expression of the TCR, .beta.2M, CD70 with concomitant expression from the TRAC locus using a CAR construct targeting CD70 (SEQ ID NO: 45; SEQ ID NO: 46. In this example activated T cells were first electroporated with 3 distinct Cas9:sgRNA RNP complexes containing sgRNAs targeting TRAC (SEQ ID NO: 40), .beta.2M (SEQ ID NO: 41), and CD70 (SEQ ID NO: 36 or 37). The DNA double stranded break at the TRAC locus was repaired by homology directed repair with an AAV6-delivered DNA template comprising a donor template (SEQ ID NO: 44; SEQ ID NO: 45) (encoding anti-CD70 CAR comprising the amino acid sequence of SEQ ID NO: 45) containing right and left homology arms to the TRAC locus flanking a chimeric antigen receptor cassette (-/+ regulatory elements for gene expression).
[0768] The resulting modified T cells are 3.times. KO (TRAC-/.beta.2M-/CD70-) anti-CD70 CAR+ T cells. The ability of these anti-CD70 CAR+ T cells to ameliorate disease caused by a CD70+ ovarian carcinoma cell line was evaluated in NOG mice using methods employed by Translational Drug Development, LLC (Scottsdale, Ariz.). In brief, 12 5-8 week old female, CIEA NOG (NOD.Cg-Prkd.sup.scidI12rg.sup.tm1Sug/JicTac) mice were individually housed in ventilated microisolator cages, maintained under pathogen-free conditions, 5-7 days prior to the start of the study. Mice received a subcutaneous inoculation of 5.times.10.sup.6 SKOV-3 ovarian carcinoma cells/mouse in the right hind flank. When mean tumor size reached 25-75 mm.sup.3 (target of .about.50 mm.sup.3), the mice were further divided into two treatment groups as shown in Table 23. On Day 1, treatment group 2 received a single 200 .mu.l intravenous dose of anti-CD7OCAR+ T cells according to Table 23.
TABLE-US-00023 TABLE 23 Treatment groups T cell SKOV-3 treatment Group CAR-T cells (i.v.) N 1 None 5 .times. 10.sup.6 None 5 cells/mouse 2 3X KO (CD70,) 5 .times. 10.sup.6 1 .times. 10.sup.7 5 anti-CD70 CAR+ T cells cells/mouse cells/mouse
[0769] Tumor volume was measured 2 times weekly from day of treatment initiation. By day 9 post-injection, tumors treated with anti-CD70 CART cells began to show a decrease in tumor volume relative to tumors in untreated animals. By day 17 post-injection, CD70+ ovarian cancer tumors in mice treated with anti-CD70 CAR T cells were completely eliminated. This complete regression of tumor growth was sustained in treated animals through day 44 post-injection, whereupon 4 out of 5 mice treated with anti-CD70 CART cells remained tumor-free until the end-of-observation (day 69) (FIG. 42A). These data demonstrate that 3.times. KO (TRAC-/.beta.2M-/CD70-) anti-CD70 CAR+ cells are highly potent in vivo for treating human ovarian tumors.
Treatment in the Non-Small Cell Lung Carcinoma (NSCLC) Tumor Model
[0770] The ability of T cells expressing a CD70 CAR to eliminate lung adenocarcionma cells that express moderate levels of CD70 was evaluated in in vivo using a subcutaneous lung carcinoma (NCI-H1975) tumor xenograft model in mice.
[0771] CRISPR/Cas9 and AAV6 were used as above (see for example, Example 3) to create human T cells that lack expression of the TCR, .beta.2M, CD70 with concomitant expression from the TRAC locus using a CAR construct targeting CD70 (SEQ ID NO: 43; SEQ ID NO: 44). In this example activated T cells were first electroporated with 3 distinct Cas9:sgRNA RNP complexes containing sgRNAs targeting TRAC (SEQ ID NO: 40), .beta. 2M (SEQ ID NO: 41), and CD70 (SEQ ID NO: 36 or 37). The DNA double stranded break at the TRAC locus was repaired by homology directed repair with an AAV6-delivered DNA template (SEQ ID NO: 43; SEQ ID NO: 44) (encoding anti-CD70 CAR comprising the amino acid sequence of SEQ ID NO: 45) containing right and left homology arms to the TRAC locus flanking a chimeric antigen receptor cassette (-/+ regulatory elements for gene expression).
[0772] The resulting modified T cells are 3.times. KO (TRAC-/(.beta.2M-/CD70-) anti-CD70 CAR+ (with 41BB costimulatory domain) T cells. The ability of these anti-CD70 CAR+ T cells to ameliorate disease caused by a CD70+ lung carcinoma cell line was evaluated in NOG mice using methods employed by Translational Drug Development, LLC (Scottsdale, Ariz.). In brief, 12, 5-8 week old female, CIEA NOG (NOD.Cg-Prkdc.sup.scidI12rg.sup.tm1Sug/JicTac) mice were individually housed in ventilated microisolator cages, maintained under pathogen-free conditions, 5-7 days prior to the start of the study. Mice received a subcutaneous inoculation of 5.times.10.sup.6 NCI-H1975 lung carcinoma cells/mouse in the right hind flank. When mean tumor size reached 25-75 mm.sup.3 (target of .about.50 mm.sup.3), the mice were further divided into 2 treatment groups as shown in Table 24. On Day 1, treatment group 2 received a single 200 .mu.l intravenous dose of anti-CD7OCAR+ T cells according to Table 24.
TABLE-US-00024 TABLE 24 Treatment groups T cell NCI-H1975 treatment Group CAR-T cells (i.v.) N 1 None 5 .times. 10.sup.6 None 5 cells/mouse 2 3X KO (CD70,) 5 .times. 10.sup.6 1 .times. 10.sup.7 5 anti-CD70 CAR+ T cells cells/mouse cells/mouse
[0773] Tumor volume was measured 2 times weekly from day of treatment initiation. By day 12 post-injection, tumors treated with anti-CD70 CAR T cells began to show a decrease in tumor volume relative to tumors in untreated animals. This complete regression of tumors in treated animals continue through day 33 post injection. Treatment with anti-CD70 CAR T cells resulted in potent activity against established H1975 lung cancer xenografts through 40 days post injection (tumor regrowth was suppressed in all mice up to day 40 with tumor size <100 mm.sup.3), whereupon tumors began to grow. (FIG. 42B). These data demonstrate that 3.times. KO (TRAC-/.beta.2M-/CD70-) anti-CD70 CAR+ cells have potent activity against human CD70+lung cancer tumors in vivo.
Treatment in the Pancreatic Tumor Model
[0774] The ability of T cells expressing a CD70 CAR to eliminate pancreatic carcinoma cells that express moderate levels of CD70 was evaluated in in vivo using a subcutaneous pancreatic (Hs 766T) tumor xenograft model in mice.
[0775] CRISPR/Cas9 and AAV6 were used as above (see for example, Example 3) to create human T cells that lack expression of the TCR, .beta.2M, CD70 with concomitant expression from the TRAC locus using a CAR construct targeting CD70 (SEQ ID NO: 43; SEQ ID NO: 44). In this example activated T cells were first electroporated with 3 distinct Cas9:sgRNA RNP complexes containing sgRNAs targeting TRAC (SEQ ID NO: 40), .beta.2M (SEQ ID NO: 41), and CD70 (SEQ ID NO: 36 or 37). The DNA double stranded break at the TRAC locus was repaired by homology directed repair with an AAV6-delivered DNA template (SEQ ID NO: 43; SEQ ID NO: 44) (encoding anti-CD70 CAR comprising the amino acid sequence of SEQ ID NO: 45) containing right and left homology arms to the TRAC locus flanking a chimeric antigen receptor cassette (-/+ regulatory elements for gene expression).
[0776] The resulting modified T cells are 3.times. KO (TRAC-/.beta.2M-/CD70-) anti-CD70 CAR+ T cells. The ability of these anti-CD70 CAR+ T cells to ameliorate disease caused by a CD70+ pancreatic carcinoma cell line was evaluated in NOG mice using methods employed by Translational Drug Development, LLC (Scottsdale, Ariz.). In brief, 12, 5-8 week old female, CIEA NOG (NOD.Cg-Prkdc.sup.scidI12rg.sup.tm1Sug/JicTac) mice were individually housed in ventilated microisolator cages, maintained under pathogen-free conditions, 5-7 days prior to the start of the study. Mice received a subcutaneous inoculation of 5.times.10.sup.6 Hs766T pancreatic carcinoma cells in the right hind flank. When mean tumor size reached 25-75 mm.sup.3 (target of .about.50 mm.sup.3), the mice were further divided into 2 treatment groups as shown in Table 25. On Day 1, treatment group 2 received a single 200 .mu.l intravenous dose of anti-CD70 CAR+ T cells according to Table 25.
TABLE-US-00025 TABLE 25 Treatment groups T cell Hs766T treatment Group CAR-T cells (i.v.) N 1 None 5 .times. 10.sup.6 None 5 cells/mouse 2 3X KO (CD70,) 5 .times. 10.sup.6 1 .times. 10.sup.7 5 anti-CD70 CAR+ T cells cells/mouse cells/mouse
[0777] Tumor volume was measured 2 times weekly from day of treatment initiation. By Day 15 post-injection, tumors treated with anti-CD70 CAR T cells began to show a decrease in tumor volume in all treated mice. Treatment with anti-CD70 CAR+ T cells effectively reduced the size of the CD70+ pancreatic cancer tumors, in all mice tested (<37 mm.sup.3) with no evidence of further growth for the duration of the study (through Day 67) (FIG. 42C). These data demonstrate that 3.times. KO (TRAC-/.beta.2M-/CD70-) anti-CD70 CAR+ cells induce regression of human CD70+ pancreatic cancer tumors in vivo, with potent activity against established Hs766T pancreatic cancer xenografts and durable responses beyond 60 days following treatment initiation.
Treatment in the cutaneous T-cell Lymphoma Tumor Xenograft Model
[0778] The ability of T cells expressing an anti-CD70 CAR to eliminate T cell lymphoma was evaluated in in vivo using a subcutaneous T-cell lymphoma (Hu T78) tumor xenograft model in mice.
[0779] CRISPR/Cas9 and AAV6 were used as above (see for example, Example 3) to create human T cells that lack expression of the TCR, .beta.2M, CD70 with concomitant expression from the TRAC locus using a CAR construct targeting CD70 (SEQ ID NO: 43; SEQ ID NO: 44). In this example activated T cells were first electroporated with 3 distinct Cas9:sgRNA RNP complexes containing sgRNAs targeting TRAC (SEQ ID NO: 40), .beta.2M (SEQ ID NO: 41), and CD70 (SEQ ID NO: 36 or 37). The DNA double stranded break at the TRAC locus was repaired by homology directed repair with an AAV6-delivered DNA template (SEQ ID NO: 43; SEQ ID NO: 44) (encoding anti-CD70 CAR comprising the amino acid sequence of SEQ ID NO: 45) containing right and left homology arms to the TRAC locus flanking a chimeric antigen receptor cassette (-/+ regulatory elements for gene expression).
[0780] The resulting modified T cells are 3.times. KO (TRAC-/.beta.2M-/CD70-) anti-CD70 CAR+ T cells. The ability of these anti-CD70 CAR+ T cells to ameliorate disease caused by a CD70+ T-cell lymphoma cell line was evaluated in NOG mice using methods employed by Translational Drug Development, LLC (Scottsdale, Ariz.). In brief, 12, 5-8 week old female, CIEA NOG (NOD.Cg-Prkdc.sup.scidI12rg.sup.tm1Sig/JicTac) mice were individually housed in ventilated microisolator cages, maintained under pathogen-free conditions, 5-7 days prior to the start of the study. Mice received a subcutaneous inoculation of 3.times.10.sup.6 HuT78 T-cell lymphoma cells in the right hind flank. When mean tumor size reached 25-75 mm.sup.3 (target of .about.50 mm.sup.3), the mice were further divided into 2 treatment groups as shown in Table 26. On Day 1, treatment group 2 received a single 200 .mu.l intravenous dose of anti-CD70 CAR+ T cells according to Table 26.
TABLE-US-00026 TABLE 26 Treatment groups T cell HuT78 treatment Group CAR-T cells (i.v.) N 1 None 3 .times. 10.sup.6 None 5 cells/mouse 2 3X KO (CD70,) 3 .times. 10.sup.6 1 .times. 10.sup.7 4 anti-CD70 CAR+ T cells cells/mouse cells/mouse
[0781] Tumor volume was measured 2 times weekly from day of treatment initiation. By Day 12 post-injection, tumors treated with anti-CD70 CAR T cells began to show a decrease in tumor volume in all treated mice. Treatment with anti-CD70 CAR+ T cells effectively reduced the size of the CD70+ T-cell lymphoma tumors, in all mice tested at Day 15 (FIG. 42C). These data demonstrate that 3.times. KO (TRAC-/.beta.2M-/CD70-) anti-CD70 CAR+ cells induce regression of human CD70+T-cell lymphoma tumors in vivo, with potent activity against established HuT78 T-cell lymphoma xenografts.
TABLE-US-00027 Summary of Sequences SEQ ID NO Description Sequence 1 TRAC Indel AAGAGCAACAAATCTGACT 2 TRAC Indel AAGAGCAACAGTGCTGTGCCTGGAGCAACAAATCTGACTAAGAGCA ACAAATCTGACT 3 TRAC Indel AAGAGCAACAGTGCTGGAGCAACAAATCTGACTAAGAGCAACAAAT CTGACT 4 TRAC Indel AAGAGCAACAGTGCCTGGAGCAACAAATCTGACTAAGAGCAACAAA TCTGACT 5 TRAC Indel AAGAGCAACAGTGCTGACTAAGAGCAACAAATCTGACT 6 TRAC Indel AAGAGCAACAGTGCTGTGGGCCTGGAGCAACAAATCTGACTAAGAG CAACAAATCTGACT 7 TRAC Indel AAGAGCAACAGTGCTGGCCTGGAGCAACAAATCTGACTAAGAGCAA CAAATCTGACT 8 TRAC Indel AAGAGCAACAGTGCTGTGTGCCTGGAGCAACAAATCTGACTAAGAG CAACAAATCTGACT 9 B2M Indel CGTGGCCTTAGCTGTGCTCGCGCTACTCTCTCTTTCTGCCTGGAGGCT ATCCAGCGTGAGTCTCTCCTACCCTCCCGCT 10 B2M Indel CGTGGCCTTAGCTGTGCTCGCGCTACTCTCTCTTTCGCCTGGAGGCTA TCCAGCGTGAGTCTCTCCTACCCTCCCGCT 11 B2M Indel CGTGGCCTTAGCTGTGCTCGCGCTACTCTCTCTTTCTGGAGGCTATCC AGCGTGAGTCTCTCCTACCCTCCCGCT 12 B2M Indel CGTGGCCTTAGCTGTGCTCGCGCTACTCTCTCTTTCTGGATAGCCTGG AGGCTATCCAGCGTGAGTCTCTCCTACCCTCCCGCT 13 B2M Indel CGTGGCCTTAGCTGTGCTCGCGCTATCCAGCGTGAGTCTCTCCTACC CTCCCGCT 14 B2M Indel CGTGGCCTTAGCTGTGCTCGCGCTACTCTCTCTTTCTGTGGCCTGGAG GCTATCCAGCGTGAGTCTCTCCTACCCTCCCGCT 15 sgRNA nnnnnnnnnnnnnnnnnnnnguuuuagagcuagaaauagcaaguuaaaauaaggcuaguccguua- u caacuugaaaaaguggcaccgagucggugcuuuu 16 sgRNA nnnnnnnnnnnnnnnnnnnnguuuuagagcuagaaauagcaaguuaaaauaaggcuaguccguua- u caacuugaaaaaguggcaccgagucggugc 17 sgRNA n.sub.(17-30)guuuuagagcuagaaauagcaaguuaaaauaaggcuaguccguuaucaacuu- gaaa aaguggcaccgagucggugcu.sub.(1-8) 18 4-1BB AAACGGGGCAGAAAGAAACTCCTGTATATATTCAAACAACCATTTA nucleotide TGAGACCAGTACAAACTACTCAAGAGGAAGATGGCTGTAGCTGCCG sequence ATTTCCAGAAGAAGAAGAAGGAGGATGTGAACTG 19 4-1BB KRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCEL amino acid sequence 20 CD28 amino SKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAYRS acid sequence 21 CD3-z CGAGTGAAGTTTTCCCGAAGCGCAGACGCTCCGGCATATCAGCAAG nucleotide GACAGAATCAGCTGTATAACGAACTGAATTTGGGACGCCGCGAGGA sequence GTATGACGTGCTTGATAAACGCCGGGGGAGAGACCCGGAAATGGGG GGTAAACCCCGAAGAAAGAATCCCCAAGAAGGACTCTACAATGAAC TCCAGAAGGATAAGATGGCGGAGGCCTACTCAGAAATAGGTATGAA GGGCGAACGACGACGGGGAAAAGGTCACGATGGCCTCTACCAAGG GTTGAGTACGGCAACCAAAGATACGTACGATGCACTGCATATGCAG GCCCTGCCTCCCAGA 22 CD3-z RVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGG amino acid KPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLS sequence TATKDTYDALHMQALPPR 23 CD70 UCACCAAGCCCGCGACCAAUguuuuagagcuagaaauagcaaguuaaaauaaggcuag sgRNA uccguuaucaacuugaaaaaguggcaccgagucggugcUUUU (E1_T1) 24 CD70 AUCACCAAGCCCGCGACCAAguuuuagagcuagaaauagcaaguuaaaauaaggcuag sgRNA uccguuaucaacuugaaaaaguggcaccgagucggugcUUUU (E1_T3) 25 CD70 CGGUGCGGCGCAGGCCCUAUguuuuagagcuagaaauagcaaguuaaaauaaggcuag sgRNA uccguuaucaacuugaaaaaguggcaccgagucggugcUUUU (E1_T4) 26 CD70 GCUUUGGUCCCAUUGGUCGCguuuuagagcuagaaauagcaaguuaaaauaaggcuag sgRNA uccguuaucaacuugaaaaaguggcaccgagucggugcUUUU (E1_T7) 27 CD70 GCCCGCAGGACGCACCCAUAguuuuagagcuagaaauagcaaguuaaaauaaggcuag sgRNA uccguuaucaacuugaaaaaguggcaccgagucggugcUUUU (E1_T8) 28 CD70 GUGCAUCCAGCGCUUCGCACguuuuagagcuagaaauagcaaguuaaaauaaggcuag sgRNA uccguuaucaacuugaaaaaguggcaccgagucggugcUUUU (E1_T10) 29 CD70 CAGCUACGUAUCCAUCGUGAguuuuagagcuagaaauagcaaguuaaaauaaggcuag sgRNA uccguuaucaacuugaaaaaguggcaccgagucggugcUUUU (E3_T1) 30 TRAC AGAGCAACAGUGCUGUGGCCguuuuagagcuagaaauagcaaguuaaaauaaggcuag sgRNA uccguuaucaacuugaaaaaguggcaccgagucggugcUUUU 31 .beta.2M GCUACUCUCUCUUUCUGGCCguuuuagagcuagaaauagcaaguuaaaauaaggcuag sgRNA uccguuaucaacuugaaaaaguggcaccgagucggugcUUUU 32 PD-1 CUGCAGCUUCUCCAACACAUguuuuagagcuagaaauagcaaguuaaaauaaggcuag sgRNA uccguuaucaacuugaaaaaguggcaccgagucggugcUUUU 33 CD70 U*C*A*CCAAGCCCGCGACCAAUguuuuagagcuagaaauagcaaguuaaaauaaggc sgRNA uaguccguuaucaacuugaaaaaguggcaccgagucggugcU*U*U*U (E1_T1) 34 CD70 A*U*C*ACCAAGCCCGCGACCAAguuuuagagcuagaaauagcaaguuaaaauaaggc sgRNA uaguccguuaucaacuugaaaaaguggcaccgagucggugcU*U*U*U (E1_T3) 35 CD70 C*G*G*UGCGGCGCAGGCCCUAUguuuuagagcuagaaauagcaaguuaaaauaaggc sgRNA uaguccguuaucaacuugaaaaaguggcaccgagucggugcU*U*U*U (E1_T4) 36 CD70 G*C*U*UUGGUCCCAUUGGUCGCguuuuagagcuagaaauagcaaguuaaaauaaggc sgRNA uaguccguuaucaacuugaaaaaguggcaccgagucggugcU*U*U*U (E1_T7) 37 CD70 G*C*C*CGCAGGACGCACCCAUAguuuuagagcuagaaauagcaaguuaaaauaaggc sgRNA uaguccguuaucaacuugaaaaaguggcaccgagucggugcU*U*U*U (E1_T8) 38 CD70 G*U*G*CAUCCAGCGCUUCGCACguuuuagagcuagaaauagcaaguuaaaauaaggc sgRNA uaguccguuaucaacuugaaaaaguggcaccgagucggugcU*U*U*U (E1_T10) 39 CD70 C*A*G*CUACGUAUCCAUCGUGAguuuuagagcuagaaauagcaaguuaaaauaaggc sgRNA uaguccguuaucaacuugaaaaaguggcaccgagucggugcU*U*U*U (E3_T1) 40 TRAC A*G*A*GCAACAGUGCUGUGGCCguuuuagagcuagaaauagcaaguuaaaauaagg sgRNA cuaguccguuaucaacuugaaaaaguggcaccgagucggugcU*U*U*U 41 .beta.2M G*C*U*ACUCUCUCUUUCUGGCCguuuuagagcuagaaauagcaaguuaaaauaaggc sgRNA uaguccguuaucaacuugaaaaaguggcaccgagucggugcU*U*U*U 42 PD-1 C*U*G*CAGCUUCUCCAACACAUguuuuagagcuagaaauagcaaguuaaaauaaggc sgRNA uaguccguuaucaacuugaaaaaguggcaccgagucggugcU*U*U*U 43 CD70 CCTGCAGGCAGCTGCGCGCTCGCTCGCTCACTGAGGCCGCCCGGGC rAAV GTCGGGCGACCTTTGGTCGCCCGGCCTCAGTGAGCGAGCGAGCGCG (CD70B CAGAGAGGGAGTGGCCAACTCCATCACTAGGGGTTCCTGCGGCCGC scFV with ACGCGTGAGATGTAAGGAGCTGCTGTGACTTGCTCAAGGCCTTATAT 41BB) CGAGTAAACGGTAGTGCTGGGGCTTAGACGCAGGTGTTCTGATTTAT AGTTCAAAACCTCTATCAATGAGAGAGCAATCTCCTGGTAATGTGAT AGATTTCCCAACTTAATGCCAACATACCATAAACCTCCCATTCTGCT AATGCCCAGCCTAAGTTGGGGAGACCACTCCAGATTCCAAGATGTA CAGTTTGCTTTGCTGGGCCTTTTTCCCATGCCTGCCTTTACTCTGCCA GAGTTATATTGCTGGGGTTTTGAAGAAGATCCTATTAAATAAAAGA ATAAGCAGTATTATTAAGTAGCCCTGCATTTCAGGTTTCCTTGAGTG GCAGGCCAGGCCTGGCCGTGAACGTTCACTGAAATCATGGCCTCTT GGCCAAGATTGATAGCTTGTGCCTGTCCCTGAGTCCCAGTCCATCAC GAGCAGCTGGTTTCTAAGATGCTATTTCCCGTATAAAGCATGAGACC GTGACTTGCCAGCCCCACAGAGCCCCGCCCTTGTCCATCACTGGCAT CTGGACTCCAGCCTGGGTTGGGGCAAAGAGGGAAATGAGATCATGT CCTAACCCTGATCCTCTTGTCCCACAGATATCCAGAACCCTGACCCT GCCGTGTACCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCT GCCTATTCACCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAG GATTCTGATGTGTATATCACAGACAAAACTGTGCTAGACATGAGGT CTATGGACTTCAGGCTCCGGTGCCCGTCAGTGGGCAGAGCGCACAT CGCCCACAGTCCCCGAGAAGTTGGGGGGAGGGGTCGGCAATTGAAC CGGTGCCTAGAGAAGGTGGCGCGGGGTAAACTGGGAAAGTGATGTC GTGTACTGGCTCCGCCTTTTTCCCGAGGGTGGGGGAGAACCGTATAT AAGTGCAGTAGTCGCCGTGAACGTTCTTTTTCGCAACGGGTTTGCCG CCAGAACACAGGTAAGTGCCGTGTGTGGTTCCCGCGGGCCTGGCCT CTTTACGGGTTATGGCCCTTGCGTGCCTTGAATTACTTCCACTGGCT GCAGTACGTGATTCTTGATCCCGAGCTTCGGGTTGGAAGTGGGTGG GAGAGTTCGAGGCCTTGCGCTTAAGGAGCCCCTTCGCCTCGTGCTTG AGTTGAGGCCTGGCCTGGGCGCTGGGGCCGCCGCGTGCGAATCTGG TGGCACCTTCGCGCCTGTCTCGCTGCTTTCGATAAGTCTCTAGCCATT TAAAATTTTTGATGACCTGCTGCGACGCTTTTTTTCTGGCAAGATAG TCTTGTAAATGCGGGCCAAGATCTGCACACTGGTATTTCGGTTTTTG GGGCCGCGGGCGGCGACGGGGCCCGTGCGTCCCAGCGCACATGTTC GGCGAGGCGGGGCCTGCGAGCGCGGCCACCGAGAATCGGACGGGG GTAGTCTCAAGCTGGCCGGCCTGCTCTGGTGCCTGGCCTCGCGCCGC CGTGTATCGCCCCGCCCTGGGCGGCAAGGCTGGCCCGGTCGGCACC AGTTGCGTGAGCGGAAAGATGGCCGCTTCCCGGCCCTGCTGCAGGG AGCTCAAAATGGAGGACGCGGCGCTCGGGAGAGCGGGCGGGTGAG TCACCCACACAAAGGAAAAGGGCCTTTCCGTCCTCAGCCGTCGCTTC ATGTGACTCCACGGAGTACCGGGCGCCGTCCAGGCACCTCGATTAG TTCTCGAGCTTTTGGAGTACGTCGTCTTTAGGTTGGGGGGAGGGGTT TTATGCGATGGAGTTTCCCCACACTGAGTGGGTGGAGACTGAAGTT AGGCCAGCTTGGCACTTGATGTAATTCTCCTTGGAATTTGCCCTTTTT GAGTTTGGATCTTGGTTCATTCTCAAGCCTCAGACAGTGGTTCAAAG TTTTTTTCTTCCATTTCAGGTGTCGTGACCACCATGGCGCTTCCGGTG ACAGCACTGCTCCTCCCCTTGGCGCTGTTGCTCCACGCAGCAAGGCC GCAGGTCCAGTTGGTGCAAAGCGGGGCGGAGGTGAAAAAACCCGG CGCTTCCGTGAAGGTGTCCTGTAAGGCGTCCGGTTATACGTTCACGA ACTACGGGATGAATTGGGTTCGCCAAGCGCCGGGGCAGGGACTGAA ATGGATGGGGTGGATAAATACCTACACCGGCGAACCTACATACGCC GACGCTTTTAAAGGGCGAGTCACTATGACGCGCGATACCAGCATAT CCACCGCATACATGGAGCTGTCCCGACTCCGGTCAGACGACACGGC TGTCTACTATTGTGCTCGGGACTATGGCGATTATGGCATGGACTACT GGGGTCAGGGTACGACTGTAACAGTTAGTAGTGGTGGAGGCGGCAG TGGCGGGGGGGGAAGCGGAGGAGGGGGTTCTGGTGACATAGTTATG ACCCAATCCCCAGATAGTTTGGCGGTTTCTCTGGGCGAGAGGGCAA CGATTAATTGTCGCGCATCAAAGAGCGTTTCAACGAGCGGATATTCT TTTATGCATTGGTACCAGCAAAAACCCGGACAACCGCCGAAGCTGC TGATCTACTTGGCTTCAAATCTTGAGTCTGGGGTGCCGGACCGATTT TCTGGTAGTGGAAGCGGAACTGACTTTACGCTCACGATCAGTTCACT GCAGGCTGAGGATGTAGCGGTCTATTATTGCCAGCACAGTAGAGAA GTCCCCTGGACCTTCGGTCAAGGCACGAAAGTAGAAATTAAAAGTG CTGCTGCCTTTGTCCCGGTATTTCTCCCAGCCAAACCGACCACGACT CCCGCCCCGCGCCCTCCGACACCCGCTCCCACCATCGCCTCTCAACC TCTTAGTCTTCGCCCCGAGGCATGCCGACCCGCCGCCGGGGGTGCTG TTCATACGAGGGGCTTGGACTTCGCTTGTGATATTTACATTTGGGCT CCGTTGGCGGGTACGTGCGGCGTCCTTTTGTTGTCACTCGTTATTACT TTGTATTGTAATCACAGGAATCGCAAACGGGGCAGAAAGAAACTCC TGTATATATTCAAACAACCATTTATGAGACCAGTACAAACTACTCAA GAGGAAGATGGCTGTAGCTGCCGATTTCCAGAAGAAGAAGAAGGA GGATGTGAACTGCGAGTGAAGTTTTCCCGAAGCGCAGACGCTCCGG CATATCAGCAAGGACAGAATCAGCTGTATAACGAACTGAATTTGGG ACGCCGCGAGGAGTATGACGTGCTTGATAAACGCCGGGGGAGAGAC CCGGAAATGGGGGGTAAACCCCGAAGAAAGAATCCCCAAGAAGGA CTCTACAATGAACTCCAGAAGGATAAGATGGCGGAGGCCTACTCAG AAATAGGTATGAAGGGCGAACGACGACGGGGAAAAGGTCACGATG GCCTCTACCAAGGGTTGAGTACGGCAACCAAAGATACGTACGATGC ACTGCATATGCAGGCCCTGCCTCCCAGATAATAATAAAATCGCTATC CATCGAAGATGGATGTGTGTTGGTTTTTTGTGTGTGGAGCAACAAAT CTGACTTTGCATGTGCAAACGCCTTCAACAACAGCATTATTCCAGAA GACACCTTCTTCCCCAGCCCAGGTAAGGGCAGCTTTGGTGCCTTCGC AGGCTGTTTCCTTGCTTCAGGAATGGCCAGGTTCTGCCCAGAGCTCT GGTCAATGATGTCTAAAACTCCTCTGATTGGTGGTCTCGGCCTTATC CATTGCCACCAAAACCCTCTTTTTACTAAGAAACAGTGAGCCTTGTT CTGGCAGTCCAGAGAATGACACGGGAAAAAAGCAGATGAAGAGAA GGTGGCAGGAGAGGGCACGTGGCCCAGCCTCAGTCTCTCCAACTGA GTTCCTGCCTGCCTGCCTTTGCTCAGACTGTTTGCCCCTTACTGCTCT TCTAGGCCTCATTCTAAGCCCCTTCTCCAAGTTGCCTCTCCTTATTTC TCCCTGTCTGCCAAAAAATCTTTCCCAGCTCACTAAGTCAGTCTCAC GCAGTCACTCATTAACCCACCAATCACTGATTGTGCCGGCACATGA ATGCACCAGGTGTTGAAGTGGAGGAATTAAAAAGTCAGATGAGGGG TGTGCCCAGAGGAAGCACCATTCTAGTTGGGGGAGCCCATCTGTCA
GCTGGGAAAAGTCCAAATAACTTCAGATTGGAATGTGTTTTAACTCA GGGTTGAGAAAACAGCTACCTTCAGGACAAAAGTCAGGGAAGGGCT CTCTGAAGAAATGCTACTTGAAGATACCAGCCCTACCAAGGGCAGG GAGAGGACCCTATAGAGGCCTGGGACAGGAGCTCAATGAGAAAGG TAACCACGTGCGGACCGAGGCTGCAGCGTCGTCCTCCCTAGGAACC CCTAGTGATGGAGTTGGCCACTCCCTCTCTGCGCGCTCGCTCGCTCA CTGAGGCCGGGCGACCAAAGGTCGCCCGACGCCCGGGCTTTGCCCG GGCGGCCTCAGTGAGCGAGCGAGCGCGCAGCTGCCTGCAGG 44 CD70 GAGATGTAAGGAGCTGCTGTGACTTGCTCAAGGCCTTATATCGAGTA LHA to AACGGTAGTGCTGGGGCTTAGACGCAGGTGTTCTGATTTATAGTTCA RHA AAACCTCTATCAATGAGAGAGCAATCTCCTGGTAATGTGATAGATTT (CD70B CCCAACTTAATGCCAACATACCATAAACCTCCCATTCTGCTAATGCC scFV with CAGCCTAAGTTGGGGAGACCACTCCAGATTCCAAGATGTACAGTTTG 41BB) CTTTGCTGGGCCTTTTTCCCATGCCTGCCTTTACTCTGCCAGAGTTAT ATTGCTGGGGTTTTGAAGAAGATCCTATTAAATAAAAGAATAAGCA GTATTATTAAGTAGCCCTGCATTTCAGGTTTCCTTGAGTGGCAGGCC AGGCCTGGCCGTGAACGTTCACTGAAATCATGGCCTCTTGGCCAAGA TTGATAGCTTGTGCCTGTCCCTGAGTCCCAGTCCATCACGAGCAGCT GGTTTCTAAGATGCTATTTCCCGTATAAAGCATGAGACCGTGACTTG CCAGCCCCACAGAGCCCCGCCCTTGTCCATCACTGGCATCTGGACTC CAGCCTGGGTTGGGGCAAAGAGGGAAATGAGATCATGTCCTAACCC TGATCCTCTTGTCCCACAGATATCCAGAACCCTGACCCTGCCGTGTA CCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCCTATTCA CCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGATTCTGAT GTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTATGGACTT CAGGCTCCGGTGCCCGTCAGTGGGCAGAGCGCACATCGCCCACAGT CCCCGAGAAGTTGGGGGGAGGGGTCGGCAATTGAACCGGTGCCTAG AGAAGGTGGCGCGGGGTAAACTGGGAAAGTGATGTCGTGTACTGGC TCCGCCTTTTTCCCGAGGGTGGGGGAGAACCGTATATAAGTGCAGTA GTCGCCGTGAACGTTCTTTTTCGCAACGGGTTTGCCGCCAGAACACA GGTAAGTGCCGTGTGTGGTTCCCGCGGGCCTGGCCTCTTTACGGGTT ATGGCCCTTGCGTGCCTTGAATTACTTCCACTGGCTGCAGTACGTGA TTCTTGATCCCGAGCTTCGGGTTGGAAGTGGGTGGGAGAGTTCGAGG CCTTGCGCTTAAGGAGCCCCTTCGCCTCGTGCTTGAGTTGAGGCCTG GCCTGGGCGCTGGGGCCGCCGCGTGCGAATCTGGTGGCACCTTCGC GCCTGTCTCGCTGCTTTCGATAAGTCTCTAGCCATTTAAAATTTTTGA TGACCTGCTGCGACGCTTTTTTTCTGGCAAGATAGTCTTGTAAATGC GGGCCAAGATCTGCACACTGGTATTTCGGTTTTTGGGGCCGCGGGCG GCGACGGGGCCCGTGCGTCCCAGCGCACATGTTCGGCGAGGCGGGG CCTGCGAGCGCGGCCACCGAGAATCGGACGGGGGTAGTCTCAAGCT GGCCGGCCTGCTCTGGTGCCTGGCCTCGCGCCGCCGTGTATCGCCCC GCCCTGGGCGGCAAGGCTGGCCCGGTCGGCACCAGTTGCGTGAGCG GAAAGATGGCCGCTTCCCGGCCCTGCTGCAGGGAGCTCAAAATGGA GGACGCGGCGCTCGGGAGAGCGGGCGGGTGAGTCACCCACACAAA GGAAAAGGGCCTTTCCGTCCTCAGCCGTCGCTTCATGTGACTCCACG GAGTACCGGGCGCCGTCCAGGCACCTCGATTAGTTCTCGAGCTTTTG GAGTACGTCGTCTTTAGGTTGGGGGGAGGGGTTTTATGCGATGGAGT TTCCCCACACTGAGTGGGTGGAGACTGAAGTTAGGCCAGCTTGGCA CTTGATGTAATTCTCCTTGGAATTTGCCCTTTTTGAGTTTGGATCTTG GTTCATTCTCAAGCCTCAGACAGTGGTTCAAAGTTTTTTTCTTCCATT TCAGGTGTCGTGACCACCATGGCGCTTCCGGTGACAGCACTGCTCCT CCCCTTGGCGCTGTTGCTCCACGCAGCAAGGCCGCAGGTCCAGTTGG TGCAAAGCGGGGCGGAGGTGAAAAAACCCGGCGCTTCCGTGAAGGT GTCCTGTAAGGCGTCCGGTTATACGTTCACGAACTACGGGATGAATT GGGTTCGCCAAGCGCCGGGGCAGGGACTGAAATGGATGGGGTGGAT AAATACCTACACCGGCGAACCTACATACGCCGACGCTTTTAAAGGG CGAGTCACTATGACGCGCGATACCAGCATATCCACCGCATACATGG AGCTGTCCCGACTCCGGTCAGACGACACGGCTGTCTACTATTGTGCT CGGGACTATGGCGATTATGGCATGGACTACTGGGGTCAGGGTACGA CTGTAACAGTTAGTAGTGGTGGAGGCGGCAGTGGCGGGGGGGGAAG CGGAGGAGGGGGTTCTGGTGACATAGTTATGACCCAATCCCCAGAT AGTTTGGCGGTTTCTCTGGGCGAGAGGGCAACGATTAATTGTCGCGC ATCAAAGAGCGTTTCAACGAGCGGATATTCTTTTATGCATTGGTACC AGCAAAAACCCGGACAACCGCCGAAGCTGCTGATCTACTTGGCTTC AAATCTTGAGTCTGGGGTGCCGGACCGATTTTCTGGTAGTGGAAGCG GAACTGACTTTACGCTCACGATCAGTTCACTGCAGGCTGAGGATGTA GCGGTCTATTATTGCCAGCACAGTAGAGAAGTCCCCTGGACCTTCGG TCAAGGCACGAAAGTAGAAATTAAAAGTGCTGCTGCCTTTGTCCCG GTATTTCTCCCAGCCAAACCGACCACGACTCCCGCCCCGCGCCCTCC GACACCCGCTCCCACCATCGCCTCTCAACCTCTTAGTCTTCGCCCCG AGGCATGCCGACCCGCCGCCGGGGGTGCTGTTCATACGAGGGGCTT GGACTTCGCTTGTGATATTTACATTTGGGCTCCGTTGGCGGGTACGT GCGGCGTCCTTTTGTTGTCACTCGTTATTACTTTGTATTGTAATCACA GGAATCGCAAACGGGGCAGAAAGAAACTCCTGTATATATTCAAACA ACCATTTATGAGACCAGTACAAACTACTCAAGAGGAAGATGGCTGT AGCTGCCGATTTCCAGAAGAAGAAGAAGGAGGATGTGAACTGCGAG TGAAGTTTTCCCGAAGCGCAGACGCTCCGGCATATCAGCAAGGACA GAATCAGCTGTATAACGAACTGAATTTGGGACGCCGCGAGGAGTAT GACGTGCTTGATAAACGCCGGGGGAGAGACCCGGAAATGGGGGGTA AACCCCGAAGAAAGAATCCCCAAGAAGGACTCTACAATGAACTCCA GAAGGATAAGATGGCGGAGGCCTACTCAGAAATAGGTATGAAGGGC GAACGACGACGGGGAAAAGGTCACGATGGCCTCTACCAAGGGTTGA GTACGGCAACCAAAGATACGTACGATGCACTGCATATGCAGGCCCT GCCTCCCAGATAATAATAAAATCGCTATCCATCGAAGATGGATGTGT GTTGGTTTTTTGTGTGTGGAGCAACAAATCTGACTTTGCATGTGCAA ACGCCTTCAACAACAGCATTATTCCAGAAGACACCTTCTTCCCCAGC CCAGGTAAGGGCAGCTTTGGTGCCTTCGCAGGCTGTTTCCTTGCTTC AGGAATGGCCAGGTTCTGCCCAGAGCTCTGGTCAATGATGTCTAAA ACTCCTCTGATTGGTGGTCTCGGCCTTATCCATTGCCACCAAAACCC TCTTTTTACTAAGAAACAGTGAGCCTTGTTCTGGCAGTCCAGAGAAT GACACGGGAAAAAAGCAGATGAAGAGAAGGTGGCAGGAGAGGGCA CGTGGCCCAGCCTCAGTCTCTCCAACTGAGTTCCTGCCTGCCTGCCTT TGCTCAGACTGTTTGCCCCTTACTGCTCTTCTAGGCCTCATTCTAAGC CCCTTCTCCAAGTTGCCTCTCCTTATTTCTCCCTGTCTGCCAAAAAAT CTTTCCCAGCTCACTAAGTCAGTCTCACGCAGTCACTCATTAACCCA CCAATCACTGATTGTGCCGGCACATGAATGCACCAGGTGTTGAAGTG GAGGAATTAAAAAGTCAGATGAGGGGTGTGCCCAGAGGAAGCACC ATTCTAGTTGGGGGAGCCCATCTGTCAGCTGGGAAAAGTCCAAATA ACTTCAGATTGGAATGTGTTTTAACTCAGGGTTGAGAAAACAGCTAC CTTCAGGACAAAAGTCAGGGAAGGGCTCTCTGAAGAAATGCTACTT GAAGATACCAGCCCTACCAAGGGCAGGGAGAGGACCCTATAGAGGC CTGGGACAGGAGCTCAATGAGAAAGG 45 CD70 CAR ATGGCGCTTCCGGTGACAGCACTGCTCCTCCCCTTGGCGCTGTTGCT nucleotide CCACGCAGCAAGGCCGCAGGTCCAGTTGGTGCAAAGCGGGGCGGAG sequence GTGAAAAAACCCGGCGCTTCCGTGAAGGTGTCCTGTAAGGCGTCCG (CD70B GTTATACGTTCACGAACTACGGGATGAATTGGGTTCGCCAAGCGCCG scFV with GGGCAGGGACTGAAATGGATGGGGTGGATAAATACCTACACCGGCG 41BB) AACCTACATACGCCGACGCTTTTAAAGGGCGAGTCACTATGACGCG CGATACCAGCATATCCACCGCATACATGGAGCTGTCCCGACTCCGGT CAGACGACACGGCTGTCTACTATTGTGCTCGGGACTATGGCGATTAT GGCATGGACTACTGGGGTCAGGGTACGACTGTAACAGTTAGTAGTG GTGGAGGCGGCAGTGGCGGGGGGGGAAGCGGAGGAGGGGGTTCTG GTGACATAGTTATGACCCAATCCCCAGATAGTTTGGCGGTTTCTCTG GGCGAGAGGGCAACGATTAATTGTCGCGCATCAAAGAGCGTTTCAA CGAGCGGATATTCTTTTATGCATTGGTACCAGCAAAAACCCGGACAA CCGCCGAAGCTGCTGATCTACTTGGCTTCAAATCTTGAGTCTGGGGT GCCGGACCGATTTTCTGGTAGTGGAAGCGGAACTGACTTTACGCTCA CGATCAGTTCACTGCAGGCTGAGGATGTAGCGGTCTATTATTGCCAG CACAGTAGAGAAGTCCCCTGGACCTTCGGTCAAGGCACGAAAGTAG AAATTAAAAGTGCTGCTGCCTTTGTCCCGGTATTTCTCCCAGCCAAA CCGACCACGACTCCCGCCCCGCGCCCTCCGACACCCGCTCCCACCAT CGCCTCTCAACCTCTTAGTCTTCGCCCCGAGGCATGCCGACCCGCCG CCGGGGGTGCTGTTCATACGAGGGGCTTGGACTTCGCTTGTGATATT TACATTTGGGCTCCGTTGGCGGGTACGTGCGGCGTCCTTTTGTTGTC ACTCGTTATTACTTTGTATTGTAATCACAGGAATCGCAAACGGGGCA GAAAGAAACTCCTGTATATATTCAAACAACCATTTATGAGACCAGTA CAAACTACTCAAGAGGAAGATGGCTGTAGCTGCCGATTTCCAGAAG AAGAAGAAGGAGGATGTGAACTGCGAGTGAAGTTTTCCCGAAGCGC AGACGCTCCGGCATATCAGCAAGGACAGAATCAGCTGTATAACGAA CTGAATTTGGGACGCCGCGAGGAGTATGACGTGCTTGATAAACGCC GGGGGAGAGACCCGGAAATGGGGGGTAAACCCCGAAGAAAGAATC CCCAAGAAGGACTCTACAATGAACTCCAGAAGGATAAGATGGCGGA GGCCTACTCAGAAATAGGTATGAAGGGCGAACGACGACGGGGAAA AGGTCACGATGGCCTCTACCAAGGGTTGAGTACGGCAACCAAAGAT ACGTACGATGCACTGCATATGCAGGCCCTGCCTCCCAGATAA 46 CD70 CAR MALPVTALLLPLALLLHAARPQVQLVQSGAEVKKPGASVKVSCKASG amino acid YTFTNYGMNWVRQAPGQGLKWMGWINTYTGEPTYADAFKGRVTMT sequence RDTSISTAYMELSRLRSDDTAVYYCARDYGDYGMDYWGQGTTVTVSS (CD70B GGGGSGGGGSGGGGSGDIVMTQSPDSLAVSLGERATINCRASKSVSTSG scFV with YSFMHWYQQKPGQPPKLLIYLASNLESGVPDRFSGSGSGTDFTLTISSLQ 41BB) AEDVAVYYCQHSREVPWTFGQGTKVEIKSAAAFVPVFLPAKPTTTPAP RPPTPAPTIASQPLSLRPEACRPAAGGAVHTRGLDFACDIYIWAPLAGTC GVLLLSLVITLYCNHRNRKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCR FPEEEEGGCELRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDK RRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGK GHDGLYQGLSTATKDTYDALHMQALPPR 47 CD70A GATATAGTTATGACCCAATCACCCGATAGTCTTGCGGTAAGCCTGGG scFv GGAGCGAGCAACAATAAACTGTCGGGCATCAAAATCCGTCAGTACA nucleotide AGCGGGTATTCATTCATGCACTGGTATCAACAGAAACCCGGTCAGCC sequence ACCCAAGCTCCTGATTTATCTTGCGTCTAATCTTGAGTCCGGCGTCCC AGACCGGTTTTCCGGCTCCGGGAGCGGCACGGATTTTACTCTTACTA TTTCTAGCCTTCAGGCCGAAGATGTGGCGGTATACTACTGCCAGCAT TCAAGGGAAGTTCCTTGGACGTTCGGTCAGGGCACGAAAGTGGAAA TTAAAGGCGGGGGGGGATCCGGCGGGGGAGGGTCTGGAGGAGGTG GCAGTGGTCAGGTCCAACTGGTGCAGTCCGGGGCAGAGGTAAAAAA ACCCGGCGCGTCTGTTAAGGTTTCATGCAAGGCCAGTGGATATACTT TCACCAATTACGGAATGAACTGGGTGAGGCAGGCCCCTGGTCAAGG CCTGAAATGGATGGGATGGATAAACACGTACACCGGTGAACCTACC TATGCCGATGCCTTTAAGGGTCGGGTTACGATGACGAGAGACACCTC CATATCAACAGCCTACATGGAGCTCAGCAGATTGAGGAGTGACGAT ACGGCAGTCTATTACTGTGCAAGAGACTACGGCGATTATGGCATGG ATTACTGGGGCCAGGGCACTACAGTAACCGTTTCCAGC 48 CD70A DIVMTQSPDSLAVSLGERATINCRASKSVSTSGYSFMHWYQQKPGQPP scFv amino KLLIYLASNLESGVPDRFSGSGSGTDFTLTISSLQAEDVAVYYCQHSREV acid PWTFGQGTKVEIKGGGGSGGGGSGGGGSGQVQLVQSGAEVKKPGASV sequence KVSCKASGYTFTNYGMNWVRQAPGQGLKWMGWINTYTGEPTYADAF (linker KGRVTMTRDTSISTAYMELSRLRSDDTAVYYCARDYGDYGMDYWGQ underlined) GTTVTVSS 49 CD70B CAGGTCCAGTTGGTGCAAAGCGGGGCGGAGGTGAAAAAACCCGGCG scFv CTTCCGTGAAGGTGTCCTGTAAGGCGTCCGGTTATACGTTCACGAAC nucleotide TACGGGATGAATTGGGTTCGCCAAGCGCCGGGGCAGGGACTGAAAT sequence GGATGGGGTGGATAAATACCTACACCGGCGAACCTACATACGCCGA CGCTTTTAAAGGGCGAGTCACTATGACGCGCGATACCAGCATATCCA CCGCATACATGGAGCTGTCCCGACTCCGGTCAGACGACACGGCTGTC TACTATTGTGCTCGGGACTATGGCGATTATGGCATGGACTACTGGGG TCAGGGTACGACTGTAACAGTTAGTAGTGGTGGAGGCGGCAGTGGC GGGGGGGGAAGCGGAGGAGGGGGTTCTGGTGACATAGTTATGACCC AATCCCCAGATAGTTTGGCGGTTTCTCTGGGCGAGAGGGCAACGATT AATTGTCGCGCATCAAAGAGCGTTTCAACGAGCGGATATTCTTTTAT GCATTGGTACCAGCAAAAACCCGGACAACCGCCGAAGCTGCTGATC TACTTGGCTTCAAATCTTGAGTCTGGGGTGCCGGACCGATTTTCTGG TAGTGGAAGCGGAACTGACTTTACGCTCACGATCAGTTCACTGCAGG CTGAGGATGTAGCGGTCTATTATTGCCAGCACAGTAGAGAAGTCCCC TGGACCTTCGGTCAAGGCACGAAAGTAGAAATTAAA 50 CD70B QVQLVQSGAEVKKPGASVKVSCKASGYTFTNYGMNWVRQAPGQGLK scFv amino WMGWINTYTGEPTYADAFKGRVTMTRDTSISTAYMELSRLRSDDTAV acid YYCARDYGDYGMDYWGQGTTVTVSSGGGGSGGGGSGGGGSGDIVMT sequence QSPDSLAVSLGERATINCRASKSVSTSGYSFMHWYQQKPGQPPKWYL (linker ASNLESGVPDRFSGSGSGTDFTLTISSLQAEDVAVYYCQHSREVPWTFG underlined) QGTKVEIK 51 CD70 VH QVQLVQSGAEVKKPGASVKVSCKASGYTFTNYGMNWVRQAPGQGLK WMGWINTYTGEPTYADAFKGRVTMTRDTSISTAYMELSRLRSDDTAV YYCARDYGDYGMDYWGQGTTVTVSS 52 CD70 VL DIVMTQSPDSLAVSLGERATINCRASKSVSTSGYSFMHWYQQKPGQPP KLLIYLASNLESGVPDRFSGSGSGTDFTLTISSLQAEDVAVYYCQHSREV PWTFGQGTKVEIK 53 Linker GGGGSGGGGSGGGGSG 54 BCMA CCTGCAGGCAGCTGCGCGCTCGCTCGCTCACTGAGGCCGCCCGGGC rAAV GTCGGGCGACCTTTGGTCGCCCGGCCTCAGTGAGCGAGCGAGCGCG CAGAGAGGGAGTGGCCAACTCCATCACTAGGGGTTCCTGCGGCCGC ACGCGTGAGATGTAAGGAGCTGCTGTGACTTGCTCAAGGCCTTATAT CGAGTAAACGGTAGTGCTGGGGCTTAGACGCAGGTGTTCTGATTTAT AGTTCAAAACCTCTATCAATGAGAGAGCAATCTCCTGGTAATGTGAT AGATTTCCCAACTTAATGCCAACATACCATAAACCTCCCATTCTGCT AATGCCCAGCCTAAGTTGGGGAGACCACTCCAGATTCCAAGATGTA CAGTTTGCTTTGCTGGGCCTTTTTCCCATGCCTGCCTTTACTCTGCCA GAGTTATATTGCTGGGGTTTTGAAGAAGATCCTATTAAATAAAAGAA TAAGCAGTATTATTAAGTAGCCCTGCATTTCAGGTTTCCTTGAGTGG CAGGCCAGGCCTGGCCGTGAACGTTCACTGAAATCATGGCCTCTTGG CCAAGATTGATAGCTTGTGCCTGTCCCTGAGTCCCAGTCCATCACGA GCAGCTGGTTTCTAAGATGCTATTTCCCGTATAAAGCATGAGACCGT GACTTGCCAGCCCCACAGAGCCCCGCCCTTGTCCATCACTGGCATCT GGACTCCAGCCTGGGTTGGGGCAAAGAGGGAAATGAGATCATGTCC TAACCCTGATCCTCTTGTCCCACAGATATCCAGAACCCTGACCCTGC CGTGTACCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCC TATTCACCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGAT TCTGATGTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTAT GGACTTCAGGCTCCGGTGCCCGTCAGTGGGCAGAGCGCACATCGCC CACAGTCCCCGAGAAGTTGGGGGGAGGGGTCGGCAATTGAACCGGT GCCTAGAGAAGGTGGCGCGGGGTAAACTGGGAAAGTGATGTCGTGT ACTGGCTCCGCCTTTTTCCCGAGGGTGGGGGAGAACCGTATATAAGT GCAGTAGTCGCCGTGAACGTTCTTTTTCGCAACGGGTTTGCCGCCAG AACACAGGTAAGTGCCGTGTGTGGTTCCCGCGGGCCTGGCCTCTTTA CGGGTTATGGCCCTTGCGTGCCTTGAATTACTTCCACTGGCTGCAGT ACGTGATTCTTGATCCCGAGCTTCGGGTTGGAAGTGGGTGGGAGAGT TCGAGGCCTTGCGCTTAAGGAGCCCCTTCGCCTCGTGCTTGAGTTGA GGCCTGGCCTGGGCGCTGGGGCCGCCGCGTGCGAATCTGGTGGCAC CTTCGCGCCTGTCTCGCTGCTTTCGATAAGTCTCTAGCCATTTAAAAT TTTTGATGACCTGCTGCGACGCTTTTTTTCTGGCAAGATAGTCTTGTA AATGCGGGCCAAGATCTGCACACTGGTATTTCGGTTTTTGGGGCCGC GGGCGGCGACGGGGCCCGTGCGTCCCAGCGCACATGTTCGGCGAGG CGGGGCCTGCGAGCGCGGCCACCGAGAATCGGACGGGGGTAGTCTC AAGCTGGCCGGCCTGCTCTGGTGCCTGGCCTCGCGCCGCCGTGTATC GCCCCGCCCTGGGCGGCAAGGCTGGCCCGGTCGGCACCAGTTGCGT GAGCGGAAAGATGGCCGCTTCCCGGCCCTGCTGCAGGGAGCTCAAA ATGGAGGACGCGGCGCTCGGGAGAGCGGGCGGGTGAGTCACCCACA CAAAGGAAAAGGGCCTTTCCGTCCTCAGCCGTCGCTTCATGTGACTC CACGGAGTACCGGGCGCCGTCCAGGCACCTCGATTAGTTCTCGAGCT TTTGGAGTACGTCGTCTTTAGGTTGGGGGGAGGGGTTTTATGCGATG GAGTTTCCCCACACTGAGTGGGTGGAGACTGAAGTTAGGCCAGCTT
GGCACTTGATGTAATTCTCCTTGGAATTTGCCCTTTTTGAGTTTGGAT CTTGGTTCATTCTCAAGCCTCAGACAGTGGTTCAAAGTTTTTTTCTTC CATTTCAGGTGTCGTGACCACCATGGCGCTTCCGGTGACAGCACTGC TCCTCCCCTTGGCGCTGTTGCTCCACGCAGCAAGGCCGCAGGTGCAG CTGGTGCAGAGCGGAGCCGAGCTCAAGAAGCCCGGAGCCTCCGTGA AGGTGAGCTGCAAGGCCAGCGGCAACACCCTGACCAACTACGTGAT CCACTGGGTGAGACAAGCCCCCGGCCAAAGGCTGGAGTGGATGGGC TACATCCTGCCCTACAACGACCTGACCAAGTACAGCCAGAAGTTCCA GGGCAGGGTGACCATCACCAGGGATAAGAGCGCCTCCACCGCCTAT ATGGAGCTGAGCAGCCTGAGGAGCGAGGACACCGCTGTGTACTACT GTACAAGGTGGGACTGGGACGGCTTCTTTGACCCCTGGGGCCAGGG CACAACAGTGACCGTCAGCAGCGGCGGCGGAGGCAGCGGCGGCGG CGGCAGCGGCGGAGGCGGAAGCGAAATCGTGATGACCCAGAGCCCC GCCACACTGAGCGTGAGCCCTGGCGAGAGGGCCAGCATCTCCTGCA GGGCTAGCCAAAGCCTGGTGCACAGCAACGGCAACACCCACCTGCA CTGGTACCAGCAGAGACCCGGACAGGCTCCCAGGCTGCTGATCTAC AGCGTGAGCAACAGGTTCTCCGAGGTGCCTGCCAGGTTTAGCGGCA GCGGAAGCGGCACCGACTTTACCCTGACCATCAGCAGCGTGGAGTC CGAGGACTTCGCCGTGTATTACTGCAGCCAGACCAGCCACATCCCTT ACACCTTCGGCGGCGGCACCAAGCTGGAGATCAAAAGTGCTGCTGC CTTTGTCCCGGTATTTCTCCCAGCCAAACCGACCACGACTCCCGCCC CGCGCCCTCCGACACCCGCTCCCACCATCGCCTCTCAACCTCTTAGT CTTCGCCCCGAGGCATGCCGACCCGCCGCCGGGGGTGCTGTTCATAC GAGGGGCTTGGACTTCGCTTGTGATATTTACATTTGGGCTCCGTTGG CGGGTACGTGCGGCGTCCTTTTGTTGTCACTCGTTATTACTTTGTATT GTAATCACAGGAATCGCAAACGGGGCAGAAAGAAACTCCTGTATAT ATTCAAACAACCATTTATGAGACCAGTACAAACTACTCAAGAGGAA GATGGCTGTAGCTGCCGATTTCCAGAAGAAGAAGAAGGAGGATGTG AACTGCGAGTGAAGTTTTCCCGAAGCGCAGACGCTCCGGCATATCA GCAAGGACAGAATCAGCTGTATAACGAACTGAATTTGGGACGCCGC GAGGAGTATGACGTGCTTGATAAACGCCGGGGGAGAGACCCGGAAA TGGGGGGTAAACCCCGAAGAAAGAATCCCCAAGAAGGACTCTACAA TGAACTCCAGAAGGATAAGATGGCGGAGGCCTACTCAGAAATAGGT ATGAAGGGCGAACGACGACGGGGAAAAGGTCACGATGGCCTCTACC AAGGGTTGAGTACGGCAACCAAAGATACGTACGATGCACTGCATAT GCAGGCCCTGCCTCCCAGATAATAATAAAATCGCTATCCATCGAAG ATGGATGTGTGTTGGTTTTTTGTGTGTGGAGCAACAAATCTGACTTT GCATGTGCAAACGCCTTCAACAACAGCATTATTCCAGAAGACACCTT CTTCCCCAGCCCAGGTAAGGGCAGCTTTGGTGCCTTCGCAGGCTGTT TCCTTGCTTCAGGAATGGCCAGGTTCTGCCCAGAGCTCTGGTCAATG ATGTCTAAAACTCCTCTGATTGGTGGTCTCGGCCTTATCCATTGCCAC CAAAACCCTCTTTTTACTAAGAAACAGTGAGCCTTGTTCTGGCAGTC CAGAGAATGACACGGGAAAAAAGCAGATGAAGAGAAGGTGGCAGG AGAGGGCACGTGGCCCAGCCTCAGTCTCTCCAACTGAGTTCCTGCCT GCCTGCCTTTGCTCAGACTGTTTGCCCCTTACTGCTCTTCTAGGCCTC ATTCTAAGCCCCTTCTCCAAGTTGCCTCTCCTTATTTCTCCCTGTCTG CCAAAAAATCTTTCCCAGCTCACTAAGTCAGTCTCACGCAGTCACTC ATTAACCCACCAATCACTGATTGTGCCGGCACATGAATGCACCAGGT GTTGAAGTGGAGGAATTAAAAAGTCAGATGAGGGGTGTGCCCAGAG GAAGCACCATTCTAGTTGGGGGAGCCCATCTGTCAGCTGGGAAAAG TCCAAATAACTTCAGATTGGAATGTGTTTTAACTCAGGGTTGAGAAA ACAGCTACCTTCAGGACAAAAGTCAGGGAAGGGCTCTCTGAAGAAA TGCTACTTGAAGATACCAGCCCTACCAAGGGCAGGGAGAGGACCCT ATAGAGGCCTGGGACAGGAGCTCAATGAGAAAGGTAACCACGTGCG GACCGAGGCTGCAGCGTCGTCCTCCCTAGGAACCCCTAGTGATGGA GTTGGCCACTCCCTCTCTGCGCGCTCGCTCGCTCACTGAGGCCGGGC GACCAAAGGTCGCCCGACGCCCGGGCTTTGCCCGGGCGGCCTCAGT GAGCGAGCGAGCGCGCAGCTGCCTGCAGG 55 BCMA GAGATGTAAGGAGCTGCTGTGACTTGCTCAAGGCCTTATATCGAGTA RHA to AACGGTAGTGCTGGGGCTTAGACGCAGGTGTTCTGATTTATAGTTCA LHA AAACCTCTATCAATGAGAGAGCAATCTCCTGGTAATGTGATAGATTT CCCAACTTAATGCCAACATACCATAAACCTCCCATTCTGCTAATGCC CAGCCTAAGTTGGGGAGACCACTCCAGATTCCAAGATGTACAGTTTG CTTTGCTGGGCCTTTTTCCCATGCCTGCCTTTACTCTGCCAGAGTTAT ATTGCTGGGGTTTTGAAGAAGATCCTATTAAATAAAAGAATAAGCA GTATTATTAAGTAGCCCTGCATTTCAGGTTTCCTTGAGTGGCAGGCC AGGCCTGGCCGTGAACGTTCACTGAAATCATGGCCTCTTGGCCAAGA TTGATAGCTTGTGCCTGTCCCTGAGTCCCAGTCCATCACGAGCAGCT GGTTTCTAAGATGCTATTTCCCGTATAAAGCATGAGACCGTGACTTG CCAGCCCCACAGAGCCCCGCCCTTGTCCATCACTGGCATCTGGACTC CAGCCTGGGTTGGGGCAAAGAGGGAAATGAGATCATGTCCTAACCC TGATCCTCTTGTCCCACAGATATCCAGAACCCTGACCCTGCCGTGTA CCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCCTATTCA CCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGATTCTGAT GTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTATGGACTT CAGGCTCCGGTGCCCGTCAGTGGGCAGAGCGCACATCGCCCACAGT CCCCGAGAAGTTGGGGGGAGGGGTCGGCAATTGAACCGGTGCCTAG AGAAGGTGGCGCGGGGTAAACTGGGAAAGTGATGTCGTGTACTGGC TCCGCCTTTTTCCCGAGGGTGGGGGAGAACCGTATATAAGTGCAGTA GTCGCCGTGAACGTTCTTTTTCGCAACGGGTTTGCCGCCAGAACACA GGTAAGTGCCGTGTGTGGTTCCCGCGGGCCTGGCCTCTTTACGGGTT ATGGCCCTTGCGTGCCTTGAATTACTTCCACTGGCTGCAGTACGTGA TTCTTGATCCCGAGCTTCGGGTTGGAAGTGGGTGGGAGAGTTCGAGG CCTTGCGCTTAAGGAGCCCCTTCGCCTCGTGCTTGAGTTGAGGCCTG GCCTGGGCGCTGGGGCCGCCGCGTGCGAATCTGGTGGCACCTTCGC GCCTGTCTCGCTGCTTTCGATAAGTCTCTAGCCATTTAAAATTTTTGA TGACCTGCTGCGACGCTTTTTTTCTGGCAAGATAGTCTTGTAAATGC GGGCCAAGATCTGCACACTGGTATTTCGGTTTTTGGGGCCGCGGGCG GCGACGGGGCCCGTGCGTCCCAGCGCACATGTTCGGCGAGGCGGGG CCTGCGAGCGCGGCCACCGAGAATCGGACGGGGGTAGTCTCAAGCT GGCCGGCCTGCTCTGGTGCCTGGCCTCGCGCCGCCGTGTATCGCCCC GCCCTGGGCGGCAAGGCTGGCCCGGTCGGCACCAGTTGCGTGAGCG GAAAGATGGCCGCTTCCCGGCCCTGCTGCAGGGAGCTCAAAATGGA GGACGCGGCGCTCGGGAGAGCGGGCGGGTGAGTCACCCACACAAA GGAAAAGGGCCTTTCCGTCCTCAGCCGTCGCTTCATGTGACTCCACG GAGTACCGGGCGCCGTCCAGGCACCTCGATTAGTTCTCGAGCTTTTG GAGTACGTCGTCTTTAGGTTGGGGGGAGGGGTTTTATGCGATGGAGT TTCCCCACACTGAGTGGGTGGAGACTGAAGTTAGGCCAGCTTGGCA CTTGATGTAATTCTCCTTGGAATTTGCCCTTTTTGAGTTTGGATCTTG GTTCATTCTCAAGCCTCAGACAGTGGTTCAAAGTTTTTTTCTTCCATT TCAGGTGTCGTGACCACCATGGCGCTTCCGGTGACAGCACTGCTCCT CCCCTTGGCGCTGTTGCTCCACGCAGCAAGGCCGCAGGTGCAGCTGG TGCAGAGCGGAGCCGAGCTCAAGAAGCCCGGAGCCTCCGTGAAGGT GAGCTGCAAGGCCAGCGGCAACACCCTGACCAACTACGTGATCCAC TGGGTGAGACAAGCCCCCGGCCAAAGGCTGGAGTGGATGGGCTACA TCCTGCCCTACAACGACCTGACCAAGTACAGCCAGAAGTTCCAGGG CAGGGTGACCATCACCAGGGATAAGAGCGCCTCCACCGCCTATATG GAGCTGAGCAGCCTGAGGAGCGAGGACACCGCTGTGTACTACTGTA CAAGGTGGGACTGGGACGGCTTCTTTGACCCCTGGGGCCAGGGCAC AACAGTGACCGTCAGCAGCGGCGGCGGAGGCAGCGGCGGCGGCGG CAGCGGCGGAGGCGGAAGCGAAATCGTGATGACCCAGAGCCCCGCC ACACTGAGCGTGAGCCCTGGCGAGAGGGCCAGCATCTCCTGCAGGG CTAGCCAAAGCCTGGTGCACAGCAACGGCAACACCCACCTGCACTG GTACCAGCAGAGACCCGGACAGGCTCCCAGGCTGCTGATCTACAGC GTGAGCAACAGGTTCTCCGAGGTGCCTGCCAGGTTTAGCGGCAGCG GAAGCGGCACCGACTTTACCCTGACCATCAGCAGCGTGGAGTCCGA GGACTTCGCCGTGTATTACTGCAGCCAGACCAGCCACATCCCTTACA CCTTCGGCGGCGGCACCAAGCTGGAGATCAAAAGTGCTGCTGCCTTT GTCCCGGTATTTCTCCCAGCCAAACCGACCACGACTCCCGCCCCGCG CCCTCCGACACCCGCTCCCACCATCGCCTCTCAACCTCTTAGTCTTCG CCCCGAGGCATGCCGACCCGCCGCCGGGGGTGCTGTTCATACGAGG GGCTTGGACTTCGCTTGTGATATTTACATTTGGGCTCCGTTGGCGGG TACGTGCGGCGTCCTTTTGTTGTCACTCGTTATTACTTTGTATTGTAA TCACAGGAATCGCAAACGGGGCAGAAAGAAACTCCTGTATATATTC AAACAACCATTTATGAGACCAGTACAAACTACTCAAGAGGAAGATG GCTGTAGCTGCCGATTTCCAGAAGAAGAAGAAGGAGGATGTGAACT GCGAGTGAAGTTTTCCCGAAGCGCAGACGCTCCGGCATATCAGCAA GGACAGAATCAGCTGTATAACGAACTGAATTTGGGACGCCGCGAGG AGTATGACGTGCTTGATAAACGCCGGGGGAGAGACCCGGAAATGGG GGGTAAACCCCGAAGAAAGAATCCCCAAGAAGGACTCTACAATGAA CTCCAGAAGGATAAGATGGCGGAGGCCTACTCAGAAATAGGTATGA AGGGCGAACGACGACGGGGAAAAGGTCACGATGGCCTCTACCAAG GGTTGAGTACGGCAACCAAAGATACGTACGATGCACTGCATATGCA GGCCCTGCCTCCCAGATAATAATAAAATCGCTATCCATCGAAGATGG ATGTGTGTTGGTTTTTTGTGTGTGGAGCAACAAATCTGACTTTGCAT GTGCAAACGCCTTCAACAACAGCATTATTCCAGAAGACACCTTCTTC CCCAGCCCAGGTAAGGGCAGCTTTGGTGCCTTCGCAGGCTGTTTCCT TGCTTCAGGAATGGCCAGGTTCTGCCCAGAGCTCTGGTCAATGATGT CTAAAACTCCTCTGATTGGTGGTCTCGGCCTTATCCATTGCCACCAA AACCCTCTTTTTACTAAGAAACAGTGAGCCTTGTTCTGGCAGTCCAG AGAATGACACGGGAAAAAAGCAGATGAAGAGAAGGTGGCAGGAGA GGGCACGTGGCCCAGCCTCAGTCTCTCCAACTGAGTTCCTGCCTGCC TGCCTTTGCTCAGACTGTTTGCCCCTTACTGCTCTTCTAGGCCTCATT CTAAGCCCCTTCTCCAAGTTGCCTCTCCTTATTTCTCCCTGTCTGCCA AAAAATCTTTCCCAGCTCACTAAGTCAGTCTCACGCAGTCACTCATT AACCCACCAATCACTGATTGTGCCGGCACATGAATGCACCAGGTGTT GAAGTGGAGGAATTAAAAAGTCAGATGAGGGGTGTGCCCAGAGGA AGCACCATTCTAGTTGGGGGAGCCCATCTGTCAGCTGGGAAAAGTC CAAATAACTTCAGATTGGAATGTGTTTTAACTCAGGGTTGAGAAAAC AGCTACCTTCAGGACAAAAGTCAGGGAAGGGCTCTCTGAAGAAATG CTACTTGAAGATACCAGCCCTACCAAGGGCAGGGAGAGGACCCTAT AGAGGCCTGGGACAGGAGCTCAATGAGAAAGG 56 BCMA ATGGCGCTTCCGGTGACAGCACTGCTCCTCCCCTTGGCGCTGTTGCT CAR CCACGCAGCAAGGCCGCAGGTGCAGCTGGTGCAGAGCGGAGCCGAG nucleotide CTCAAGAAGCCCGGAGCCTCCGTGAAGGTGAGCTGCAAGGCCAGCG sequence GCAACACCCTGACCAACTACGTGATCCACTGGGTGAGACAAGCCCC CGGCCAAAGGCTGGAGTGGATGGGCTACATCCTGCCCTACAACGAC CTGACCAAGTACAGCCAGAAGTTCCAGGGCAGGGTGACCATCACCA GGGATAAGAGCGCCTCCACCGCCTATATGGAGCTGAGCAGCCTGAG GAGCGAGGACACCGCTGTGTACTACTGTACAAGGTGGGACTGGGAC GGCTTCTTTGACCCCTGGGGCCAGGGCACAACAGTGACCGTCAGCA GCGGCGGCGGAGGCAGCGGCGGCGGCGGCAGCGGCGGAGGCGGAA GCGAAATCGTGATGACCCAGAGCCCCGCCACACTGAGCGTGAGCCC TGGCGAGAGGGCCAGCATCTCCTGCAGGGCTAGCCAAAGCCTGGTG CACAGCAACGGCAACACCCACCTGCACTGGTACCAGCAGAGACCCG GACAGGCTCCCAGGCTGCTGATCTACAGCGTGAGCAACAGGTTCTCC GAGGTGCCTGCCAGGTTTAGCGGCAGCGGAAGCGGCACCGACTTTA CCCTGACCATCAGCAGCGTGGAGTCCGAGGACTTCGCCGTGTATTAC TGCAGCCAGACCAGCCACATCCCTTACACCTTCGGCGGCGGCACCA AGCTGGAGATCAAAAGTGCTGCTGCCTTTGTCCCGGTATTTCTCCCA GCCAAACCGACCACGACTCCCGCCCCGCGCCCTCCGACACCCGCTCC CACCATCGCCTCTCAACCTCTTAGTCTTCGCCCCGAGGCATGCCGAC CCGCCGCCGGGGGTGCTGTTCATACGAGGGGCTTGGACTTCGCTTGT GATATTTACATTTGGGCTCCGTTGGCGGGTACGTGCGGCGTCCTTTT GTTGTCACTCGTTATTACTTTGTATTGTAATCACAGGAATCGCAAAC GGGGCAGAAAGAAACTCCTGTATATATTCAAACAACCATTTATGAG ACCAGTACAAACTACTCAAGAGGAAGATGGCTGTAGCTGCCGATTT CCAGAAGAAGAAGAAGGAGGATGTGAACTGCGAGTGAAGTTTTCCC GAAGCGCAGACGCTCCGGCATATCAGCAAGGACAGAATCAGCTGTA TAACGAACTGAATTTGGGACGCCGCGAGGAGTATGACGTGCTTGAT AAACGCCGGGGGAGAGACCCGGAAATGGGGGGTAAACCCCGAAGA AAGAATCCCCAAGAAGGACTCTACAATGAACTCCAGAAGGATAAGA TGGCGGAGGCCTACTCAGAAATAGGTATGAAGGGCGAACGACGACG GGGAAAAGGTCACGATGGCCTCTACCAAGGGTTGAGTACGGCAACC AAAGATACGTACGATGCACTGCATATGCAGGCCCTGCCTCCCAGA 57 BCMA MALPVTALLLPLALLLHAARPQVQLVQSGAELKKPGASVKVSCKASGN CAR amino TLTNYVIHWVRQAPGQRLEWMGYILPYNDLTKYSQKFQGRVTITRDKS acid ASTAYMELSSLRSEDTAVYYCTRWDWDGFFDPWGQGTTVTVSSGGGG sequence SGGGGSGGGGSEIVMTQSPATLSVSPGERASISCRASQSLVHSNGNTHL HWYQQRPGQAPRLLIYSVSNRFSEVPARFSGSGSGTDFTLTISSVESEDF AVYYCSQTSHIPYTFGGGTKLEIKSAAAFVPVFLPAKPTTTPAPRPPTPA PTIASQPLSLRPEACRPAAGGAVHTRGLDFACDIYIWAPLAGTCGVLLLS LVITLYCNHRNRKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEE GGCELRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRD PEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGL YQGLSTATKDTYDALHMQALPPR 58 BCMA CAGGTGCAGCTGGTGCAGAGCGGAGCCGAGCTCAAGAAGCCCGGAG scFv CCTCCGTGAAGGTGAGCTGCAAGGCCAGCGGCAACACCCTGACCAA nucleotide CTACGTGATCCACTGGGTGAGACAAGCCCCCGGCCAAAGGCTGGAG sequence TGGATGGGCTACATCCTGCCCTACAACGACCTGACCAAGTACAGCC AGAAGTTCCAGGGCAGGGTGACCATCACCAGGGATAAGAGCGCCTC CACCGCCTATATGGAGCTGAGCAGCCTGAGGAGCGAGGACACCGCT GTGTACTACTGTACAAGGTGGGACTGGGACGGCTTCTTTGACCCCTG GGGCCAGGGCACAACAGTGACCGTCAGCAGCGGCGGCGGAGGCAG CGGCGGCGGCGGCAGCGGCGGAGGCGGAAGCGAAATCGTGATGAC CCAGAGCCCCGCCACACTGAGCGTGAGCCCTGGCGAGAGGGCCAGC ATCTCCTGCAGGGCTAGCCAAAGCCTGGTGCACAGCAACGGCAACA CCCACCTGCACTGGTACCAGCAGAGACCCGGACAGGCTCCCAGGCT GCTGATCTACAGCGTGAGCAACAGGTTCTCCGAGGTGCCTGCCAGGT TTAGCGGCAGCGGAAGCGGCACCGACTTTACCCTGACCATCAGCAG CGTGGAGTCCGAGGACTTCGCCGTGTATTACTGCAGCCAGACCAGCC ACATCCCTTACACCTTCGGCGGCGGCACCAAGCTGGAGATCAAA 59 BCMA QVQLVQSGAELKKPGASVKVSCKASGNTLTNYVIHWVRQAPGQRLEW scFv amino MGYILPYNDLTKYSQKFQGRVTITRDKSASTAYMELSSLRSEDTAVYY acid CTRWDWDGFFDPWGQGTTVTVSSGGGGSGGGGSGGGGSEIVMTQSPA sequence TLSVSPGERASISCRASQSLVHSNGNTHLHWYQQRPGQAPRLLIYSVSN (linker RFSEVPARFSGSGSGTDFTLTISSVESEDFAVYYCSQTSHIPYTFGGGTKL underlined) EIK 60 BCMA VH QVQLVQSGAELKKPGASVKVSCKASGNTLTNYVIHWVRQAPGQRLEW MGYILPYNDLTKYSQKFQGRVTITRDKSASTAYMELSSLRSEDTAVYY CTRWDWDGFFDPWGQGTTVTVSS 61 BCMA VL EIVMTQSPATLSVSPGERASISCRASQSLVHSNGNTHLHWYQQRPGQAP RLLIYSVSNRFSEVPARFSGSGSGTDFTLTISSVESEDFAVYYCSQTSHIP YTFGGGTKLEIK 62 CD70 VL RASKSVSTSGYSFMH CDR1 (Kabat) 63 CD70 VL SKSVSTSGYSF CDR1 (Chothia) 64 CD70 VL LASNLES CDR2 (Kabat) 65 CD70 VL LAS CDR2 (Chothia) 66 CD70 VL QHSREVPWT CDR3 (Kabat)
67 CD70 VL SREVPW CDR3 (Chothia) 68 CD70 VH NYGMN CDR1 (Kabat) 69 CD70 VH GYTFTNYGMN CDR1 (Chothia) 70 CD70 VH WINTYTGEPTYADAFKG CDR2 (Kabat) 71 CD70 VH NTYTGE CDR2 (Chothia) 72 CD70 VH DYGDYGMDY CDR3 (Kabat) 73 CD70 VH CARDYGDYGMDYWG CDR3 (Chothia) 74 BCMA VL RASQSLVHSNGNTHLH CDR1 (Kabat) 75 BCMA VL RASQSLVHSNGNTHLH CDR1 (Chothia) 76 BCMA VL SVSNR CDR2 (Kabat) 77 BCMA VL SVSNR CDR2 (Chothia) 78 BCMA VL SQTSHIPYT CDR3 (Kabat) 79 BCMA VL SQTSHIPYT CDR3 (Chothia) 80 BCMA VH NYVIH CDR1 (Kabat) 81 BCMA VH GNTLTNY CDR1 (Chothia) 82 BCMA VH YILPYNDLTKYSQKFQG CDR2 (Kabat) 83 BCMA VH LPYNDL CDR2 (Chothia) 84 BCMA VH WDWDGFFDP CDR3 (Kabat) 85 BCMA VH WDWDGFFDP CDR3 (Chothia) 86 TRAC AGAGCAACAGTGCTGTGGCC target sequence 87 anti-CD33 CCTGCAGGCAGCTGCGCGCTCGCTCGCTCACTGAGGCCGCCCGGGC CAR rAAV GTCGGGCGACCTTTGGTCGCCCGGCCTCAGTGAGCGAGCGAGCGCG CAGAGAGGGAGTGGCCAACTCCATCACTAGGGGTTCCTGCGGCCGC ACGCGTGAGATGTAAGGAGCTGCTGTGACTTGCTCAAGGCCTTATAT CGAGTAAACGGTAGTGCTGGGGCTTAGACGCAGGTGTTCTGATTTAT AGTTCAAAACCTCTATCAATGAGAGAGCAATCTCCTGGTAATGTGAT AGATTTCCCAACTTAATGCCAACATACCATAAACCTCCCATTCTGCT AATGCCCAGCCTAAGTTGGGGAGACCACTCCAGATTCCAAGATGTA CAGTTTGCTTTGCTGGGCCTTTTTCCCATGCCTGCCTTTACTCTGCCA GAGTTATATTGCTGGGGTTTTGAAGAAGATCCTATTAAATAAAAGAA TAAGCAGTATTATTAAGTAGCCCTGCATTTCAGGTTTCCTTGAGTGG CAGGCCAGGCCTGGCCGTGAACGTTCACTGAAATCATGGCCTCTTGG CCAAGATTGATAGCTTGTGCCTGTCCCTGAGTCCCAGTCCATCACGA GCAGCTGGTTTCTAAGATGCTATTTCCCGTATAAAGCATGAGACCGT GACTTGCCAGCCCCACAGAGCCCCGCCCTTGTCCATCACTGGCATCT GGACTCCAGCCTGGGTTGGGGCAAAGAGGGAAATGAGATCATGTCC TAACCCTGATCCTCTTGTCCCACAGATATCCAGAACCCTGACCCTGC CGTGTACCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCC TATTCACCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGAT TCTGATGTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTAT GGACTTCAggctccggtgcccgtcagtgggcagagcgcacatcgcccacagtccccgagaagttgggg ggaggggtcggcaattgaaccggtgcctagagaaggtggcgcggggtaaactgggaaagtgatgtcgtgtac- t ggctccgcctttttcccgagggtgggggagaaccgtatataagtgcagtagtcgccgtgaacgttctttttc- gcaac gggtttgccgccagaacacaggtaagtgccgtgtgtggttcccgcgggcctggcctctttacgggttatggc- cctt gcgtgccttgaattacttccactggctgcagtacgtgattcttgatcccgagcttcgggttggaagtgggtg- ggaga gttcgaggccttgcgcttaaggagccccttcgcctcgtgcttgagttgaggcctggcctgggcgctggggcc- gcc gcgtgcgaatctggtggcaccttcgcgcctgtctcgctgctttcgataagtctctagccatttaaaattttt- gatgacct gctgcgacgctttttttctggcaagatagtcttgtaaatgcgggccaagatctgcacactggtatttcggtt- tttgggg ccgcgggcggcgacggggcccgtgcgtcccagcgcacatgttcggcgaggcggggcctgcgagcgcggcc accgagaatcggacgggggtagtctcaagctggccggcctgctctggtgcctggcctcgcgccgccgtgtat- cg ccccgccctgggcggcaaggctggcccggtcggcaccagttgcgtgagcggaaagatggccgcttcccggcc ctgctgcagggagctcaaaatggaggacgcggcgctcgggagagcgggcgggtgagtcacccacacaaagg aaaagggcctttccgtcctcagccgtcgcttcatgtgactccacggagtaccgggcgccgtccaggcacctc- gat tagttctcgagcttttggagtacgtcgtctttaggttggggggaggggttttatgcgatggagtttccccac- actgagt gggtggagactgaagttaggccagcttggcacttgatgtaattctccttggaatttgccctttttgagtttg- gatcttgg ttcattctcaagcctcagacagtggttcaaagtttttttcttccatttcaggtgtcgtgaCCACCATGGCGC- T TCCGGTGACAGCACTGCTCCTCCCCTTGGCGCTGTTGCTCCACGCAG CAAGGCCGGAAATCGTCCTCACACAATCCCCGGGGAGCCTCGCAGT CAGTCCTGGGGAACGAGTCACTATGAGCTGCAAATCCAGTCAGAGT GTTTTTTTCTCAAGTAGCCAGAAGAACTACCTCGCATGGTACCAACA AATACCGGGGCAATCTCCCCGCTTGCTTATATACTGGGCAAGTACCC GCGAATCCGGCGTACCGGATCGATTCACGGGATCTGGGTCAGGTAC TGATTTCACTTTGACTATCAGCTCTGTTCAGCCTGAAGATTTGGCAAT TTACTACTGTCACCAATACTTGAGTAGCCGAACTTTCGGCCAGGGCA CGAAGCTCGAAATCAAGGGCGGAGGGGGAGGTTCTGGTGGGGGCG GTTCTGGCGGTGGAGGAAGCCAAGTACAGTTGCAACAGCCAGGGGC GGAGGTCGTAAAACCTGGGGCGTCTGTCAAGATGAGCTGTAAAGCA AGTGGATACACCTTCACCTCCTACTATATACATTGGATTAAGCAAAC TCCGGGTCAGGGGCTGGAATGGGTTGGCGTTATATACCCCGGGAAC GATGATATATCATACAACCAAAAATTTCAAGGCAAGGCGACTCTGA CTGCCGATAAGAGTAGCACAACAGCTTACATGCAGCTTTCTTCCCTG ACCAGCGAAGATTCAGCAGTTTACTACTGCGCTCGGGAAGTGCGCCT GCGATACTTTGATGTCTGGGGTCAAGGAACTACAGTTACTGTATCAA GCAGTGCTGCTGCCTTTGTCCCGGTATTTCTCCCAGCCAAACCGACC ACGACTCCCGCCCCGCGCCCTCCGACACCCGCTCCCACCATCGCCTC TCAACCTCTTAGTCTTCGCCCCGAGGCATGCCGACCCGCCGCCGGGG GTGCTGTTCATACGAGGGGCTTGGACTTCGCTTGTGATATTTACATTT GGGCTCCGTTGGCGGGTACGTGCGGCGTCCTTTTGTTGTCACTCGTT ATTACTTTGTATTGTAATCACAGGAATCGCAAACGGGGCAGAAAGA AACTCCTGTATATATTCAAACAACCATTTATGAGACCAGTACAAACT ACTCAAGAGGAAGATGGCTGTAGCTGCCGATTTCCAGAAGAAGAAG AAGGAGGATGTGAACTGCGAGTGAAGTTTTCCCGAAGCGCAGACGC TCCGGCATATCAGCAAGGACAGAATCAGCTGTATAACGAACTGAAT TTGGGACGCCGCGAGGAGTATGACGTGCTTGATAAACGCCGGGGGA GAGACCCGGAAATGGGGGGTAAACCCCGAAGAAAGAATCCCCAAG AAGGACTCTACAATGAACTCCAGAAGGATAAGATGGCGGAGGCCTA CTCAGAAATAGGTATGAAGGGCGAACGACGACGGGGAAAAGGTCA CGATGGCCTCTACCAAGGGTTGAGTACGGCAACCAAAGATACGTAC GATGCACTGCATATGCAGGCCCTGCCTCCCAGATAATAATAAAATCG CTATCCATCGAAGATGGATGTGTGTTGGTTTTTTGTGTGTGGAGCAA CAAATCTGACTTTGCATGTGCAAACGCCTTCAACAACAGCATTATTC CAGAAGACACCTTCTTCCCCAGCCCAGGTAAGGGCAGCTTTGGTGCC TTCGCAGGCTGTTTCCTTGCTTCAGGAATGGCCAGGTTCTGCCCAGA GCTCTGGTCAATGATGTCTAAAACTCCTCTGATTGGTGGTCTCGGCC TTATCCATTGCCACCAAAACCCTCTTTTTACTAAGAAACAGTGAGCC TTGTTCTGGCAGTCCAGAGAATGACACGGGAAAAAAGCAGATGAAG AGAAGGTGGCAGGAGAGGGCACGTGGCCCAGCCTCAGTCTCTCCAA CTGAGTTCCTGCCTGCCTGCCTTTGCTCAGACTGTTTGCCCCTTACTG CTCTTCTAGGCCTCATTCTAAGCCCCTTCTCCAAGTTGCCTCTCCTTA TTTCTCCCTGTCTGCCAAAAAATCTTTCCCAGCTCACTAAGTCAGTCT CACGCAGTCACTCATTAACCCACCAATCACTGATTGTGCCGGCACAT GAATGCACCAGGTGTTGAAGTGGAGGAATTAAAAAGTCAGATGAGG GGTGTGCCCAGAGGAAGCACCATTCTAGTTGGGGGAGCCCATCTGT CAGCTGGGAAAAGTCCAAATAACTTCAGATTGGAATGTGTTTTAACT CAGGGTTGAGAAAACAGCTACCTTCAGGACAAAAGTCAGGGAAGGG CTCTCTGAAGAAATGCTACTTGAAGATACCAGCCCTACCAAGGGCA GGGAGAGGACCCTATAGAGGCCTGGGACAGGAGCTCAATGAGAAA GGTAACCACGTGCGGACCGAGGCTGCAGCGTCGTCCTCCCTAGGAA CCCCTAGTGATGGAGTTGGCCACTCCCTCTCTGCGCGCTCGCTCGCT CACTGAGGCCGGGCGACCAAAGGTCGCCCGACGCCCGGGCTTTGCC CGGGCGGCCTCAGTGAGCGAGCGAGCGCGCAGCTGCCTGCAGG 88 signal MLLLVTSLLLCELPHPAFLLIP peptide 89 signal MALPVTALLLPLALLLHAARP peptide 90 CD8a FVPVFLPAKPTTTPAPRPPTPAPTIASQPLSLRPEACRPAAGG trans- AVHTRGLDFACDIYIWAPLAGTCGVLLLSLVITLYCNHRNR membrane domain 91 CD70 UCACCAAGCCCGCGACCAAU sgRNA (E1_T1) spacer 92 CD70 AUCACCAAGCCCGCGACCAA sgRNA (E1_T3) spacer 93 CD70 CGGUGCGGCGCAGGCCCUAU sgRNA (E1_T4) spacer 94 CD70 GCUUUGGUCCCAUUGGUCGC sgRNA (E1_T7) spacer 95 CD70 GCCCGCAGGACGCACCCAUA sgRNA (E1_T8) spacer 96 CD70 GUGCAUCCAGCGCUUCGCAC sgRNA (E1_T10) spacer 97 CD70 CAGCUACGUAUCCAUCGUGA sgRNA (E3_T1) spacer 98 TRAC AGAGCAACAGUGCUGUGGCC spacer 99 .beta.2M GCUACUCUCUCUUUCUGGCC sgRNA spacer 100 PD-1 CUGCAGCUUCUCCAACACAU sgRNA spacer 101 CD70 U*C*A*CCAAGCCCGCGACCAAU sgRNA (E1_T3) spacer 102 CD70 A*U*C*ACCAAGCCCGCGACCAA sgRNA (E1_T4)
spacer 103 CD70 C*G*G*UGCGGCGCAGGCCCUAU sgRNA (E1_T7) spacer 104 CD70 G*C*U*UUGGUCCCAUUGGUCGC sgRNA (E1_T8) spacer 105 CD70 G*C*C*CGCAGGACGCACCCAUA sgRNA (E1_T10) spacer 106 CD70 G*U*G*CAUCCAGCGCUUCGCAC sgRNA (E3_T10) spacer 107 CD70 C*A*G*CUACGUAUCCAUCGUGA sgRNA (E1_T3) spacer 108 TRAC A*G*A*GCAACAGUGCUGUGGCC spacer 109 .beta.2M G*C*U*ACUCUCUCUUUCUGGCC sgRNA spacer 110 PD-1 C*U*G*CAGCUUCUCCAACACAU sgRNA spacer 111 CD70 TCACCAAGCCCGCGACCAATGGG sgRNA (E1_T1) with PAM 112 CD70 ATCACCAAGCCCGCGACCAATGG sgRNA (E1_T3) with PAM 113 CD70 CGGTGCGGCGCAGGCCCTATGGG sgRNA (E1_T4) with PAM 114 CD70 GCTTTGGTCCCATTGGTCGCGGG sgRNA (E1_T7) with PAM 115 CD70 GCCCGCAGGACGCACCCATAGGG sgRNA (E1_T8) with PAM 116 CD70 GTGCATCCAGCGCTTCGCACAGG sgRNA (E1_T10) with PAM 117 CD70 CAGCTACGTATCCATCGTGATGG sgRNA (E3_T1) with PAM 118 TRAC AGAGCAACAGTGCTGTGGCCTGG sgRNA with PAM 119 .beta.2M GCTACTCTCTCTTTCTGGCCTGG sgRNA with PAM 120 PD-1 CTGCAGCTTCTCCAACACATCGG sgRNA with PAM 121 CD28 TCAAAGCGGAGTAGGTTGTTGCATTCCGATTACATGAATATGACTCC nucleotide TCGCCGGCCTGGGCCGACAAGAAAACATTACCAACCCTATGCCCCC sequence CCACGAGACTTCGCTGCGTACAGGTCC 122 TRAC- GAGATGTAAGGAGCTGCTGTGACTTGCTCAAGGCCTTATATCGAGTA LHA AACGGTAGTGCTGGGGCTTAGACGCAGGTGTTCTGATTTATAGTTCA AAACCTCTATCAATGAGAGAGCAATCTCCTGGTAATGTGATAGATTT CCCAACTTAATGCCAACATACCATAAACCTCCCATTCTGCTAATGCC CAGCCTAAGTTGGGGAGACCACTCCAGATTCCAAGATGTACAGTTTG CTTTGCTGGGCCTTTTTCCCATGCCTGCCTTTACTCTGCCAGAGTTAT ATTGCTGGGGTTTTGAAGAAGATCCTATTAAATAAAAGAATAAGCA GTATTATTAAGTAGCCCTGCATTTCAGGTTTCCTTGAGTGGCAGGCC AGGCCTGGCCGTGAACGTTCACTGAAATCATGGCCTCTTGGCCAAGA TTGATAGCTTGTGCCTGTCCCTGAGTCCCAGTCCATCACGAGCAGCT GGTTTCTAAGATGCTATTTCCCGTATAAAGCATGAGACCGTGACTTG CCAGCCCCACAGAGCCCCGCCCTTGTCCATCACTGGCATCTGGACTC CAGCCTGGGTTGGGGCAAAGAGGGAAATGAGATCATGTCCTAACCC TGATCCTCTTGTCCCACAGATATCCAGAACCCTGACCCTGCCGTGTA CCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCCTATTCA CCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGATTCTGAT GTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTATGGACTT CA 123 EF1.alpha. GGCTCCGGTGCCCGTCAGTGGGCAGAGCGCACATCGCCCACAGTCC promoter CCGAGAAGTTGGGGGGAGGGGTCGGCAATTGAACCGGTGCCTAGAG AAGGTGGCGCGGGGTAAACTGGGAAAGTGATGTCGTGTACTGGCTC CGCCTTTTTCCCGAGGGTGGGGGAGAACCGTATATAAGTGCAGTAGT CGCCGTGAACGTTCTTTTTCGCAACGGGTTTGCCGCCAGAACACAGG TAAGTGCCGTGTGTGGTTCCCGCGGGCCTGGCCTCTTTACGGGTTAT GGCCCTTGCGTGCCTTGAATTACTTCCACTGGCTGCAGTACGTGATT CTTGATCCCGAGCTTCGGGTTGGAAGTGGGTGGGAGAGTTCGAGGC CTTGCGCTTAAGGAGCCCCTTCGCCTCGTGCTTGAGTTGAGGCCTGG CCTGGGCGCTGGGGCCGCCGCGTGCGAATCTGGTGGCACCTTCGCGC CTGTCTCGCTGCTTTCGATAAGTCTCTAGCCATTTAAAATTTTTGATG ACCTGCTGCGACGCTTTTTTTCTGGCAAGATAGTCTTGTAAATGCGG GCCAAGATCTGCACACTGGTATTTCGGTTTTTGGGGCCGCGGGCGGC GACGGGGCCCGTGCGTCCCAGCGCACATGTTCGGCGAGGCGGGGCC TGCGAGCGCGGCCACCGAGAATCGGACGGGGGTAGTCTCAAGCTGG CCGGCCTGCTCTGGTGCCTGGCCTCGCGCCGCCGTGTATCGCCCCGC CCTGGGCGGCAAGGCTGGCCCGGTCGGCACCAGTTGCGTGAGCGGA AAGATGGCCGCTTCCCGGCCCTGCTGCAGGGAGCTCAAAATGGAGG ACGCGGCGCTCGGGAGAGCGGGCGGGTGAGTCACCCACACAAAGG AAAAGGGCCTTTCCGTCCTCAGCCGTCGCTTCATGTGACTCCACGGA GTACCGGGCGCCGTCCAGGCACCTCGATTAGTTCTCGAGCTTTTGGA GTACGTCGTCTTTAGGTTGGGGGGAGGGGTTTTATGCGATGGAGTTT CCCCACACTGAGTGGGTGGAGACTGAAGTTAGGCCAGCTTGGCACT TGATGTAATTCTCCTTGGAATTTGCCCTTTTTGAGTTTGGATCTTGGT TCATTCTCAAGCCTCAGACAGTGGTTCAAAGTTTTTTTCTTCCATTTC AGGTGTCGTGA 124 Synthetic AATAAAATCGCTATCCATCGAAGATGGATGTGTGTTGGTTTTTTGTG poly(A) TG signal 125 TRAC- TGGAGCAACAAATCTGACTTTGCATGTGCAAACGCCTTCAACAACA RHA GCATTATTCCAGAAGACACCTTCTTCCCCAGCCCAGGTAAGGGCAGC TTTGGTGCCTTCGCAGGCTGTTTCCTTGCTTCAGGAATGGCCAGGTTC TGCCCAGAGCTCTGGTCAATGATGTCTAAAACTCCTCTGATTGGTGG TCTCGGCCTTATCCATTGCCACCAAAACCCTCTTTTTACTAAGAAAC AGTGAGCCTTGTTCTGGCAGTCCAGAGAATGACACGGGAAAAAAGC AGATGAAGAGAAGGTGGCAGGAGAGGGCACGTGGCCCAGCCTCAG TCTCTCCAACTGAGTTCCTGCCTGCCTGCCTTTGCTCAGACTGTTTGC CCCTTACTGCTCTTCTAGGCCTCATTCTAAGCCCCTTCTCCAAGTTGC CTCTCCTTATTTCTCCCTGTCTGCCAAAAAATCTTTCCCAGCTCACTA AGTCAGTCTCACGCAGTCACTCATTAACCCACCAATCACTGATTGTG CCGGCACATGAATGCACCAGGTGTTGAAGTGGAGGAATTAAAAAGT CAGATGAGGGGTGTGCCCAGAGGAAGCACCATTCTAGTTGGGGGAG CCCATCTGTCAGCTGGGAAAAGTCCAAATAACTTCAGATTGGAATGT GTTTTAACTCAGGGTTGAGAAAACAGCTACCTTCAGGACAAAAGTC AGGGAAGGGCTCTCTGAAGAAATGCTACTTGAAGATACCAGCCCTA CCAAGGGCAGGGAGAGGACCCTATAGAGGCCTGGGACAGGAGCTC AATGAGAAAGG 126 CD8a IYIWAPLAGTCGVLLLSLVITLY trans- membrane 127 CD70 TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGcccaacttttccatctcaactca forward ccccaagtg primer 128 CD70 GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGcccctcctgcgctagcgga reverse primer 129 CD70 Indel CACACCACGAGGCAGATCACCAAGCCCGCG- CAATGGGACCAAAGCAGCCCGCAGGACG 130 CD70 Indel CACACCACGAGGCAGATCACCAAGCCCGCGAACCAATGGGACCAAA GCAGCCCGCAGGACG 131 CD70 Indel CACACCACGAGGCAGATC------------ ACCAATGGGACCAAAGCAGCCCGCAGGACG 132 CD70 Indel CACACCACGAGGCAGATCACCAAGCCCGCG- CCAATGGGACCAAAGCAGCCCGCAGGACG 133 CD70 Indel CACACCACGAGGCAGATCACCAAGCCCGC- ACCAATGGGACCAAAGCAGCCCGCAGGACG 134 CD70 Indel CACACCACGAGGCAGATCACCA------------------------- AGCCCGCAGGACG 135 Anti-CD33 GAGATGTAAGGAGCTGCTGTGACTTGCTCAAGGCCTTATATCGAGT CAR AAACGGTAGTGCTGGGGCTTAGACGCAGGTGTTCTGATTTATAGTTC Donor AAAACCTCTATCAATGAGAGAGCAATCTCCTGGTAATGTGATAGAT LHA to TTCCCAACTTAATGCCAACATACCATAAACCTCCCATTCTGCTAATG RHA CCCAGCCTAAGTTGGGGAGACCACTCCAGATTCCAAGATGTACAGT 41BB TTGCTTTGCTGGGCCTTTTTCCCATGCCTGCCTTTACTCTGCCAGAGT costim. TATATTGCTGGGGTTTTGAAGAAGATCCTATTAAATAAAAGAATAA GCAGTATTATTAAGTAGCCCTGCATTTCAGGTTTCCTTGAGTGGCAG GCCAGGCCTGGCCGTGAACGTTCACTGAAATCATGGCCTCTTGGCC AAGATTGATAGCTTGTGCCTGTCCCTGAGTCCCAGTCCATCACGAGC AGCTGGTTTCTAAGATGCTATTTCCCGTATAAAGCATGAGACCGTGA CTTGCCAGCCCCACAGAGCCCCGCCCTTGTCCATCACTGGCATCTGG ACTCCAGCCTGGGTTGGGGCAAAGAGGGAAATGAGATCATGTCCTA ACCCTGATCCTCTTGTCCCACAGATATCCAGAACCCTGACCCTGCCG TGTACCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCCTA TTCACCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGATTC TGATGTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTATG GACTTCAggctccggtgcccgtcagtgggcagagcgcacatcgcccacagtccccgagaagttggggg gaggggtcggcaattgaaccggtgcctagagaaggtggcgcggggtaaactgggaaagtgatgtcgtgtact- g gctccgcctttttcccgagggtgggggagaaccgtatataagtgcagtagtcgccgtgaacgttctttttcg- caacg ggtttgccgccagaacacaggtaagtgccgtgtgtggttcccgcgggcctggcctctttacgggttatggcc- cttg cgtgccttgaattacttccactggctgcagtacgtgattcttgatcccgagcttcgggttggaagtgggtgg- gagag ttcgaggccttgcgcttaaggagccccttcgcctcgtgcttgagttgaggcctggcctgggcgctggggccg- cc gcgtgcgaatctggtggcaccttcgcgcctgtctcgctgctttcgataagtctctagccatttaaaattttt- gatgacc tgctgcgacgctttttttctggcaagatagtcttgtaaatgcgggccaagatctgcacactggtatttcggt- ttttggg gccgcgggcggcgacggggcccgtgcgtcccagcgcacatgttcggcgaggcggggcctgcgagcgcggc caccgagaatcggacgggggtagtctcaagctggccggcctgctctggtgcctggcctcgcgccgccgtgta- tc gccccgccctgggcggcaaggctggcccggtcggcaccagttgcgtgagcggaaagatggccgcttcccgg ccctgctgcagggagctcaaaatggaggacgcggcgctcgggagagcgggcgggtgagtcacccacacaaa ggaaaagggcctttccgtcctcagccgtcgcttcatgtgactccacggagtaccgggcgccgtccaggcacc- tc gattagttctcgagcttttggagtacgtcgtctttaggttggggggaggggttttatgcgatggagtttccc- cacact gagtgggtggagactgaagttaggccagcttggcacttgatgtaattctccttggaatttgccctttttgag- tttggat cttggttcattctcaagcctcagacagtggttcaaagtttttttcttccatttcaggtgtcgtgaCCACCAT- GG CGCTTCCGGTGACAGCACTGCTCCTCCCCTTGGCGCTGTTGCTCCAC GCAGCAAGGCCGGAAATCGTCCTCACACAATCCCCGGGGAGCCTCG CAGTCAGTCCTGGGGAACGAGTCACTATGAGCTGCAAATCCAGTCA GAGTGTTTTTTTCTCAAGTAGCCAGAAGAACTACCTCGCATGGTACC AACAAATACCGGGGCAATCTCCCCGCTTGCTTATATACTGGGCAAU ACCCGCGAATCCGGCGTACCGGATCGATTCACGGGATCTGGGTCAG GTACTGATTTCACTTTGACTATCAGCTCTGTTCAGCCTGAAGATTTG GCAATTTACTACTGTCACCAATACTTGAGTAGCCGAACTTTCGGCCA GGGCACGAAGCTCGAAATCAAGGGCGGAGGGGGAGGTTCTGGTGG GGGCGGTTCTGGCGGTGGAGGAAGCCAAGTACAGTTGCAACAGCCA GGGGCGGAGGTCGTAAAACCTGGGGCGTCTGTCAAGATGAGCTGTA AAGCAAGTGGATACACCTTCACCTCCTACTATATACATTGGATTAAC CAAACTCCGGGTCAGGGGCTGGAATGGGTTGGCGTTATATACCCCG GGAACGATGATATATCATACAACCAAAAATTTCAAGGCAAGGCGAC TCTGACTGCCGATAAGAGTAGCACAACAGCTTACATGCAGCTTTCT1 CCCTGACCAGCGAAGATTCAGCAGTTTACTACTGCGCTCGGGAAGT GCGCCTGCGATACTTTGATGTCTGGGGTCAAGGAACTACAGTTACT( TATCAAGCAGTGCTGCTGCCTTTGTCCCGGTATTTCTCCCAGCCAAA
CCGACCACGACTCCCGCCCCGCGCCCTCCGACACCCGCTCCCACCAl CGCCTCTCAACCTCTTAGTCTTCGCCCCGAGGCATGCCGACCCGCCG CCGGGGGTGCTGTTCATACGAGGGGCTTGGACTTCGCTTGTGATATT TACATTTGGGCTCCGTTGGCGGGTACGTGCGGCGTCCTTTTGTTGTC ACTCGTTATTACTTTGTATTGTAATCACAGGAATCGCAAACGGGGCA GAAAGAAACTCCTGTATATATTCAAACAACCATTTATGAGACCAGT ACAAACTACTCAAGAGGAAGATGGCTGTAGCTGCCGATTTCCAGAA GAAGAAGAAGGAGGATGTGAACTGCGAGTGAAGTTTTCCCGAAGCC CAGACGCTCCGGCATATCAGCAAGGACAGAATCAGCTGTATAACGA ACTGAATTTGGGACGCCGCGAGGAGTATGACGTGCTTGATAAACGC CGGGGGAGAGACCCGGAAATGGGGGGTAAACCCCGAAGAAAGAAT CCCCAAGAAGGACTCTACAATGAACTCCAGAAGGATAAGATGGCG( AGGCCTACTCAGAAATAGGTATGAAGGGCGAACGACGACGGGGAA AAGGTCACGATGGCCTCTACCAAGGGTTGAGTACGGCAACCAAAGA TACGTACGATGCACTGCATATGCAGGCCCTGCCTCCCAGATAATAA1 AAAATCGCTATCCATCGAAGATGGATGTGTGTTGGTTTTTTGTGTGT GGAGCAACAAATCTGACTTTGCATGTGCAAACGCCTTCAACAACAG CATTATTCCAGAAGACACCTTCTTCCCCAGCCCAGGTAAGGGCAGC1 TTGGTGCCTTCGCAGGCTGTTTCCTTGCTTCAGGAATGGCCAGGTTC TGCCCAGAGCTCTGGTCAATGATGTCTAAAACTCCTCTGATTGGTGG TCTCGGCCTTATCCATTGCCACCAAAACCCTCTTTTTACTAAGAAAC AGTGAGCCTTGTTCTGGCAGTCCAGAGAATGACACGGGAAAAAAGC AGATGAAGAGAAGGTGGCAGGAGAGGGCACGTGGCCCAGCCTCAG TCTCTCCAACTGAGTTCCTGCCTGCCTGCCTTTGCTCAGACTGTTTGC CCCTTACTGCTCTTCTAGGCCTCATTCTAAGCCCCTTCTCCAAGTTGC CTCTCCTTATTTCTCCCTGTCTGCCAAAAAATCTTTCCCAGCTCACTA AGTCAGTCTCACGCAGTCACTCATTAACCCACCAATCACTGATTGTG CCGGCACATGAATGCACCAGGTGTTGAAGTGGAGGAATTAAAAAGI CAGATGAGGGGTGTGCCCAGAGGAAGCACCATTCTAGTTGGGGGAC CCCATCTGTCAGCTGGGAAAAGTCCAAATAACTTCAGATTGGAATG TGTTTTAACTCAGGGTTGAGAAAACAGCTACCTTCAGGACAAAAGT CAGGGAAGGGCTCTCTGAAGAAATGCTACTTGAAGATACCAGCCCT ACCAAGGGCAGGGAGAGGACCCTATAGAGGCCTGGGACAGGAGCT CAATGAGAAAGG 136 Anti-CD33 CCACCATGGCGCTTCCGGTGACAGCACTGCTCCTCCCCTTGGCGCTG CAR TTGCTCCACGCAGCAAGGCCGGAAATCGTCCTCACACAATCCCCGG 41BB GGAGCCTCGCAGTCAGTCCTGGGGAACGAGTCACTATGAGCTGCAA costim ATCCAGTCAGAGTGTTTTTTTCTCAAGTAGCCAGAAGAACTACCTCG CATGGTACCAACAAATACCGGGGCAATCTCCCCGCTTGCTTATATAC TGGGCAAGTACCCGCGAATCCGGCGTACCGGATCGATTCACGGGAT CTGGGTCAGGTACTGATTTCACTTTGACTATCAGCTCTGTTCAGCCT GAAGATTTGGCAATTTACTACTGTCACCAATACTTGAGTAGCCGAAC TTTCGGCCAGGGCACGAAGCTCGAAATCAAGGGCGGAGGGGGAGG TTCTGGTGGGGGCGGTTCTGGCGGTGGAGGAAGCCAAGTACAGTTG CAACAGCCAGGGGCGGAGGTCGTAAAACCTGGGGCGTCTGTCAAGA TGAGCTGTAAAGCAAGTGGATACACCTTCACCTCCTACTATATACAT TGGATTAAGCAAACTCCGGGTCAGGGGCTGGAATGGGTTGGCGTTA TATACCCCGGGAACGATGATATATCATACAACCAAAAATTTCAAGG CAAGGCGACTCTGACTGCCGATAAGAGTAGCACAACAGCTTACATG CAGCTTTCTTCCCTGACCAGCGAAGATTCAGCAGTTTACTACTGCGC TCGGGAAGTGCGCCTGCGATACTTTGATGTCTGGGGTCAAGGAACT ACAGTTACTGTATCAAGCAGTGCTGCTGCCTTTGTCCCGGTATTTCT CCCAGCCAAACCGACCACGACTCCCGCCCCGCGCCCTCCGACACCC GCTCCCACCATCGCCTCTCAACCTCTTAGTCTTCGCCCCGAGGCATG CCGACCCGCCGCCGGGGGTGCTGTTCATACGAGGGGCTTGGACTTC GCTTGTGATATTTACATTTGGGCTCCGTTGGCGGGTACGTGCGGCGT CCTTTTGTTGTCACTCGTTATTACTTTGTATTGTAATCACAGGAATCG CAAACGGGGCAGAAAGAAACTCCTGTATATATTCAAACAACCATTT ATGAGACCAGTACAAACTACTCAAGAGGAAGATGGCTGTAGCTGCC GATTTCCAGAAGAAGAAGAAGGAGGATGTGAACTGCGAGTGAAGTT TTCCCGAAGCGCAGACGCTCCGGCATATCAGCAAGGACAGAATCAG CTGTATAACGAACTGAATTTGGGACGCCGCGAGGAGTATGACGTGC TTGATAAACGCCGGGGGAGAGACCCGGAAATGGGGGGTAAACCCC GAAGAAAGAATCCCCAAGAAGGACTCTACAATGAACTCCAGAAGG ATAAGATGGCGGAGGCCTACTCAGAAATAGGTATGAAGGGCGAAC GACGACGGGGAAAAGGTCACGATGGCCTCTACCAAGGGTTGAGTAC GGCAACCAAAGATACGTACGATGCACTGCATATGCAGGCCCTGCCT CCCAGATAATAATAAAATCGCTATCCATCGAAGATGGATGTGTGTT GGTTTTTTGTGTG 137 Anti-CD33 EIVLTQSPGSLAVSPGERVTMSCKSSQSVFFSSSQKNYLAWYQQIPGQS scFv PRLLIYWASTRESGVPDRFTGSGSGTDFTLTISSVQPEDLAIYYCHQYLS Linker SRTFGQGTKLEIKGGGGGSGGGGSGGGGSQVQLQQPGAEVVKPGASV underlined KMSCKASGYTFTSYYIHWIKQTPGQGLEWVGVIYPGNDDISYNQKFQG KATLTADKSSTTAYMQLSSLTSEDSAVYYCAREVRLRYFDVWGQGTT VTVSS 138 Anti-CD33 GAAATCGTCCTCACACAATCCCCGGGGAGCCTCGCAGTCAGTCCTG scFv GGGAACGAGTCACTATGAGCTGCAAATCCAGTCAGAGTGTTTTTTTC TCAAGTAGCCAGAAGAACTACCTCGCATGGTACCAACAAATACCGG GGCAATCTCCCCGCTTGCTTATATACTGGGCAAGTACCCGCGAATCC GGCGTACCGGATCGATTCACGGGATCTGGGTCAGGTACTGATTTCAC TTTGACTATCAGCTCTGTTCAGCCTGAAGATTTGGCAATTTACTACT GTCACCAATACTTGAGTAGCCGAACTTTCGGCCAGGGCACGAAGCT CGAAATCAAGGGCGGAGGGGGAGGTTCTGGTGGGGGCGGTTCTGGC GGTGGAGGAAGCCAAGTACAGTTGCAACAGCCAGGGGCGGAGGTC GTAAAACCTGGGGCGTCTGTCAAGATGAGCTGTAAAGCAAGTGGAT ACACCTTCACCTCCTACTATATACATTGGATTAAGCAAACTCCGGGT CAGGGGCTGGAATGGGTTGGCGTTATATACCCCGGGAACGATGATA TATCATACAACCAAAAATTTCAAGGCAAGGCGACTCTGACTGCCGA TAAGAGTAGCACAACAGCTTACATGCAGCTTTCTTCCCTGACCAGCG AAGATTCAGCAGTTTACTACTGCGCTCGGGAAGTGCGCCTGCGATA CTTTGATGTCTGGGGTCAAGGAACTACAGTTACTGTATCAAGC 139 Anti-CD33 MALPVTALLLPLALLLHAARPEIVLTQSPGSLAVSPGERVTMSCKSSQS CAR VFFSSSQKNYLAWYQQIPGQSPRLLIYWASTRESGVPDRFTGSGSGTDF 41BB TLTISSVQPEDLAIYYCHQYLSSRTFGQGTKLEIKGGGGGSGGGGSGGG costim. GSQVQLQQPGAEVVKPGASVKMSCKASGYTFTSYYIHWIKQTPGQGLE WVGVIYPGNDDISYNQKFQGKATLTADKSSTTAYMQLSSLTSEDSAVY YCAREVRLRYFDVWGQGTTVTVSSSAAAFVPVFLPAKPTTTPAPRPPTP APTIASQPLSLRPEACRPAAGGAVHTRGLDFACDIYIWAPLAGTCGVLL LSLVITLYCNHRNRKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEE EEGGCELRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRG RDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGH DGLYQGLSTATKDTYDALHMQALPPR 140 anti-CD33 QVQLQQPGAEVVKPGASVKMSCKASGYTFTSYYIHWIKQTPGQGLEW antibody VGVIYPGNDDISYNQKFQGKATLTADKSSTTAYMQLSSLTSEDSAVYY VH CAREVRLRYFDVWGQGTTVTVSS CDRs underlined and in bold 141 anti-CD33 EIVLTQSPGSLAVSPGERVTMSCKSSQSVFFSSSQKNYLAWYQQIPGQS antibody PRLLIYWASTRESGVPDRFTGSGSGTDFTLTISSVQPEDLAIYYCHQYLS VL SRTFGQGTKLEIK CDRs underlined and in bold 142 anti-CD33 SYYIH antibody VH CDR1 (Kabat) 143 anti-CD33 VIYPGNDDISYNQKFQG antibody ' VH CDR2 (Kabat) 144 anti-CD33 EVRLRYFDV antibody VH CDR3 (Kabat) 145 anti-CD33 KSSQSVFFSSSQKNYLA antibody VL CDR1 (Kabat & Chothia) 146 anti-CD33 WASTRES antibody VL CDR2 (Kabat & Chothia 147 anti-CD33 HQYLSSRT antibody VL CDR3 (Kabat & Chothia) 148 Anti-CD19 ATGCTTCTTTTGGTTACGTCTCTGTTGCTTTGCGAACTTCCTCATCCA CAR GCGTTCTTGCTGATCCCCGATATTCAGATGACTCAGACCACCAGTAG CD8[tm]- CTTGTCTGCCTCACTGGGAGACCGAGTAACAATCTCCTGCAGGGCAA CD28[co- GTCAAGACATTAGCAAATACCTCAATTGGTACCAGCAGAAGCCCGA stimulatory CGGAACGGTAAAACTCCTCATCTATCATACGTCAAGGTTGCATTCCG domain]- GAGTACCGTCACGATTTTCAGGTTCTGGGAGCGGAACTGACTATTCC CD3z) TTGACTATTTCAAACCTCGAGCAGGAGGACATTGCGACATATTTTTG TCAACAAGGTAATACCCTCCCTTACACTTTCGGAGGAGGAACCAAA CTCGAAATTACCGGGTCCACCAGTGGCTCTGGGAAGCCTGGCAGTG GAGAAGGTTCCACTAAAGGCGAGGTGAAGCTCCAGGAGAGCGGCCC CGGTCTCGTTGCCCCCAGTCAAAGCCTCTCTGTAACGTGCACAGTGA GTGGTGTATCATTGCCTGATTATGGCGTCTCCTGGATAAGGCAGCCC CCGCGAAAGGGTCTTGAATGGCTTGGGGTAATATGGGGCTCAGAGA CAACGTATTATAACTCCGCTCTCAAAAGTCGCTTGACGATAATAAAA GATAACTCCAAGAGTCAAGTTTTCCTTAAAATGAACAGTTTGCAGAC TGACGATACCGCTATATATTATTGTGCTAAACATTATTACTACGGCG GTAGTTACGCGATGGATTATTGGGGGCAGGGGACTTCTGTCACAGTC AGTAGTGCTGCTGCCTTTGTCCCGGTATTTCTCCCAGCCAAACCGAC CACGACTCCCGCCCCGCGCCCTCCGACACCCGCTCCCACCATCGCCT CTCAACCTCTTAGTCTTCGCCCCGAGGCATGCCGACCCGCCGCCGGG GGTGCTGTTCATACGAGGGGCTTGGACTTCGCTTGTGATATTTACAT TTGGGCTCCGTTGGCGGGTACGTGCGGCGTCCTTTTGTTGTCACTCGT TATTACTTTGTATTGTAATCACAGGAATCGCTCAAAGCGGAGTAGGT TGTTGCATTCCGATTACATGAATATGACTCCTCGCCGGCCTGGGCCG ACAAGAAAACATTACCAACCCTATGCCCCCCCACGAGACTTCGCTGC GTACAGGTCCCGAGTGAAGTTTTCCCGAAGCGCAGACGCTCCGGCA TATCAGCAAGGACAGAATCAGCTGTATAACGAACTGAATTTGGGAC GCCGCGAGGAGTATGACGTGCTTGATAAACGCCGGGGGAGAGACCC GGAAATGGGGGGTAAACCCCGAAGAAAGAATCCCCAAGAAGGACT CTACAATGAACTCCAGAAGGATAAGATGGCGGAGGCCTACTCAGAA ATAGGTATGAAGGGCGAACGACGACGGGGAAAAGGTCACGATGGC CTCTACCAAGGGTTGAGTACGGCAACCAAAGATACGTACGATGCAC TGCATATGCAGGCCCTGCCTCCCAGA 149 Anti-CD19 MLLLVTSLLLCELPHPAFLLIPDIQMTQTTSSLSASLGDRVTISCRASQDI CAR SKYLNWYQQKPDGTVKLLIYHTSRLHSGVPSRFSGSGSGTDYSLTISNL CD8[tm]- EQEDIATYFCQQGNTLPYTFGGGTKLEITGSTSGSGKPGSGEGSTKGEV CD28[co- KLQESGPGLVAPSQSLSVTCTVSGVSLPDYGVSWIRQPPRKGLEWLGVI stimulatory WGSETTYYNSALKSRLTIIKDNSKSQVFLKMNSLQTDDTAIYYCAKHY domain]- YYGGSYAMDYWGQGTSVTVSSAAAFVPVFLPAKPTTTPAPRPPTPAPTI CD3z) ASQPLSLRPEACRPAAGGAVHTRGLDFACDIYIWAPLAGTCGVLLLSLV Amino Acid ITLYCNHRNRSKRSRLLHSDYMNMTPRRPGPTRKHYQPYAPPRDFAAY RSRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEM GGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQ GLSTATKDTYDALHMQALPPR 150 Anti-CD19 GATATTCAGATGACTCAGACCACCAGTAGCTTGTCTGCCTCACTGGG scFv AGACCGAGTAACAATCTCCTGCAGGGCAAGTCAAGACATTAGCAAA TACCTCAATTGGTACCAGCAGAAGCCCGACGGAACGGTAAAACTCC TCATCTATCATACGTCAAGGTTGCATTCCGGAGTACCGTCACGATTT TCAGGTTCTGGGAGCGGAACTGACTATTCCTTGACTATTTCAAACCT CGAGCAGGAGGACATTGCGACATATTTTTGTCAACAAGGTAATACC CTCCCTTACACTTTCGGAGGAGGAACCAAACTCGAAATTACCGGGTC CACCAGTGGCTCTGGGAAGCCTGGCAGTGGAGAAGGTTCCACTAAA GGCGAGGTGAAGCTCCAGGAGAGCGGCCCCGGTCTCGTTGCCCCCA GTCAAAGCCTCTCTGTAACGTGCACAGTGAGTGGTGTATCATTGCCT GATTATGGCGTCTCCTGGATAAGGCAGCCCCCGCGAAAGGGTCTTG AATGGCTTGGGGTAATATGGGGCTCAGAGACAACGTATTATAACTC CGCTCTCAAAAGTCGCTTGACGATAATAAAAGATAACTCCAAGAGT CAAGTTTTCCTTAAAATGAACAGTTTGCAGACTGACGATACCGCTAT ATATTATTGTGCTAAACATTATTACTACGGCGGTAGTTACGCGATGG ATTATTGGGGGCAGGGGACTTCTGTCACAGTCAGTAGT 151 CD19 scFv DIQMTQTTSSLSASLGDRVTISCRASQDISKYLNWYQQKPDGTVKLLIY amino acid HTSRLHSGVPSRFSGSGSGTDYSLTISNLEQEDIATYFCQQGNTLPYTFG sequence GGTKLEITGSTSGSGKPGSGEGSTKGEVKLQESGPGLVAPSQSLSVTCTV Linker SGVSLPDYGVSWIRQPPRKGLEWLGVIWGSETTYYNSALKSRLTIIKDN underlined SKSQVFLKMNSLQTDDTAIYYCAKHYYYGGSYAMDYWGQGTSVTVSS 152 Anti-CD19 EVKLQESGPGLVAPSQSLSVTCTVSGVSLPDYGVSWIRQPPRKGLEWLG VH VIWGSETTYYNSALKSRLTIIKDNSKSQVFLKMNSLQTDDTAIYYCAKH YYYGGSYAMDYWGQGTSVTVSS 153 Anti-CD19 DIQMTQTTSSLSASLGDRVTISCRASQDISKYLNWYQQKPDGTVKLLIY VL HTSRLHSGVPSRFSGSGSGTDYSLTISNLEQEDIATYFCQQGNTLPYTFG GGTKLEIT 154 Anti-CD19 GSTSGSGKPGSGEGSTKG scFv linker 155 anti-CD19 CCTGCAGGCAGCTGCGCGCTCGCTCGCTCACTGAGGCCGCCCGGGC CAR rAAV GTCGGGCGACCTTTGGTCGCCCGGCCTCAGTGAGCGAGCGAGCGCG CAGAGAGGGAGTGGCCAACTCCATCACTAGGGGTTCCTGCGGCCGC ACGCGTGAGATGTAAGGAGCTGCTGTGACTTGCTCAAGGCCTTATAT CGAGTAAACGGTAGTGCTGGGGCTTAGACGCAGGTGTTCTGATTTAT AGTTCAAAACCTCTATCAATGAGAGAGCAATCTCCTGGTAATGTGAT AGATTTCCCAACTTAATGCCAACATACCATAAACCTCCCATTCTGCT AATGCCCAGCCTAAGTTGGGGAGACCACTCCAGATTCCAAGATGTA CAGTTTGCTTTGCTGGGCCTTTTTCCCATGCCTGCCTTTACTCTGCCA GAGTTATATTGCTGGGGTTTTGAAGAAGATCCTATTAAATAAAAGAA TAAGCAGTATTATTAAGTAGCCCTGCATTTCAGGTTTCCTTGAGTGG CAGGCCAGGCCTGGCCGTGAACGTTCACTGAAATCATGGCCTCTTGG CCAAGATTGATAGCTTGTGCCTGTCCCTGAGTCCCAGTCCATCACGA GCAGCTGGTTTCTAAGATGCTATTTCCCGTATAAAGCATGAGACCGT GACTTGCCAGCCCCACAGAGCCCCGCCCTTGTCCATCACTGGCATCT GGACTCCAGCCTGGGTTGGGGCAAAGAGGGAAATGAGATCATGTCC TAACCCTGATCCTCTTGTCCCACAGATATCCAGAACCCTGACCCTGC
CGTGTACCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCC TATTCACCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGAT TCTGATGTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTAT GGACTTCAGGCTCCGGTGCCCGTCAGTGGGCAGAGCGCACATCGCC CACAGTCCCCGAGAAGTTGGGGGGAGGGGTCGGCAATTGAACCGGT GCCTAGAGAAGGTGGCGCGGGGTAAACTGGGAAAGTGATGTCGTGT ACTGGCTCCGCCTTTTTCCCGAGGGTGGGGGAGAACCGTATATAAGT GCAGTAGTCGCCGTGAACGTTCTTTTTCGCAACGGGTTTGCCGCCAG AACACAGGTAAGTGCCGTGTGTGGTTCCCGCGGGCCTGGCCTCTTTA CGGGTTATGGCCCTTGCGTGCCTTGAATTACTTCCACTGGCTGCAGT ACGTGATTCTTGATCCCGAGCTTCGGGTTGGAAGTGGGTGGGAGAGT TCGAGGCCTTGCGCTTAAGGAGCCCCTTCGCCTCGTGCTTGAGTTGA GGCCTGGCCTGGGCGCTGGGGCCGCCGCGTGCGAATCTGGTGGCAC CTTCGCGCCTGTCTCGCTGCTTTCGATAAGTCTCTAGCCATTTAAAAT TTTTGATGACCTGCTGCGACGCTTTTTTTCTGGCAAGATAGTCTTGTA AATGCGGGCCAAGATCTGCACACTGGTATTTCGGTTTTTGGGGCCGC GGGCGGCGACGGGGCCCGTGCGTCCCAGCGCACATGTTCGGCGAGG CGGGGCCTGCGAGCGCGGCCACCGAGAATCGGACGGGGGTAGTCTC AAGCTGGCCGGCCTGCTCTGGTGCCTGGCCTCGCGCCGCCGTGTATC GCCCCGCCCTGGGCGGCAAGGCTGGCCCGGTCGGCACCAGTTGCGT GAGCGGAAAGATGGCCGCTTCCCGGCCCTGCTGCAGGGAGCTCAAA ATGGAGGACGCGGCGCTCGGGAGAGCGGGCGGGTGAGTCACCCACA CAAAGGAAAAGGGCCTTTCCGTCCTCAGCCGTCGCTTCATGTGACTC CACGGAGTACCGGGCGCCGTCCAGGCACCTCGATTAGTTCTCGAGCT TTTGGAGTACGTCGTCTTTAGGTTGGGGGGAGGGGTTTTATGCGATG GAGTTTCCCCACACTGAGTGGGTGGAGACTGAAGTTAGGCCAGCTT GGCACTTGATGTAATTCTCCTTGGAATTTGCCCTTTTTGAGTTTGGAT CTTGGTTCATTCTCAAGCCTCAGACAGTGGTTCAAAGTTTTTTTCTTC CATTTCAGGTGTCGTGACCACCATGCTTCTTTTGGTTACGTCTCTGTT GCTTTGCGAACTTCCTCATCCAGCGTTCTTGCTGATCCCCGATATTCA GATGACTCAGACCACCAGTAGCTTGTCTGCCTCACTGGGAGACCGA GTAACAATCTCCTGCAGGGCAAGTCAAGACATTAGCAAATACCTCA ATTGGTACCAGCAGAAGCCCGACGGAACGGTAAAACTCCTCATCTA TCATACGTCAAGGTTGCATTCCGGAGTACCGTCACGATTTTCAGGTT CTGGGAGCGGAACTGACTATTCCTTGACTATTTCAAACCTCGAGCAG GAGGACATTGCGACATATTTTTGTCAACAAGGTAATACCCTCCCTTA CACTTTCGGAGGAGGAACCAAACTCGAAATTACCGGGTCCACCAGT GGCTCTGGGAAGCCTGGCAGTGGAGAAGGTTCCACTAAAGGCGAGG TGAAGCTCCAGGAGAGCGGCCCCGGTCTCGTTGCCCCCAGTCAAAG CCTCTCTGTAACGTGCACAGTGAGTGGTGTATCATTGCCTGATTATG GCGTCTCCTGGATAAGGCAGCCCCCGCGAAAGGGTCTTGAATGGCTT GGGGTAATATGGGGCTCAGAGACAACGTATTATAACTCCGCTCTCA AAAGTCGCTTGACGATAATAAAAGATAACTCCAAGAGTCAAGTTTT CCTTAAAATGAACAGTTTGCAGACTGACGATACCGCTATATATTATT GTGCTAAACATTATTACTACGGCGGTAGTTACGCGATGGATTATTGG GGGCAGGGGACTTCTGTCACAGTCAGTAGTGCTGCTGCCTTTGTCCC GGTATTTCTCCCAGCCAAACCGACCACGACTCCCGCCCCGCGCCCTC CGACACCCGCTCCCACCATCGCCTCTCAACCTCTTAGTCTTCGCCCC GAGGCATGCCGACCCGCCGCCGGGGGTGCTGTTCATACGAGGGGCT TGGACTTCGCTTGTGATATTTACATTTGGGCTCCGTTGGCGGGTACG TGCGGCGTCCTTTTGTTGTCACTCGTTATTACTTTGTATTGTAATCAC AGGAATCGCTCAAAGCGGAGTAGGTTGTTGCATTCCGATTACATGA ATATGACTCCTCGCCGGCCTGGGCCGACAAGAAAACATTACCAACC CTATGCCCCCCCACGAGACTTCGCTGCGTACAGGTCCCGAGTGAAGT TTTCCCGAAGCGCAGACGCTCCGGCATATCAGCAAGGACAGAATCA GCTGTATAACGAACTGAATTTGGGACGCCGCGAGGAGTATGACGTG CTTGATAAACGCCGGGGGAGAGACCCGGAAATGGGGGGTAAACCCC GAAGAAAGAATCCCCAAGAAGGACTCTACAATGAACTCCAGAAGGA TAAGATGGCGGAGGCCTACTCAGAAATAGGTATGAAGGGCGAACGA CGACGGGGAAAAGGTCACGATGGCCTCTACCAAGGGTTGAGTACGG CAACCAAAGATACGTACGATGCACTGCATATGCAGGCCCTGCCTCCC AGATAATAATAAAATCGCTATCCATCGAAGATGGATGTGTGTTGGTT TTTTGTGTGTGGAGCAACAAATCTGACTTTGCATGTGCAAACGCCTT CAACAACAGCATTATTCCAGAAGACACCTTCTTCCCCAGCCCAGGTA AGGGCAGCTTTGGTGCCTTCGCAGGCTGTTTCCTTGCTTCAGGAATG GCCAGGTTCTGCCCAGAGCTCTGGTCAATGATGTCTAAAACTCCTCT GATTGGTGGTCTCGGCCTTATCCATTGCCACCAAAACCCTCTTTTTAC TAAGAAACAGTGAGCCTTGTTCTGGCAGTCCAGAGAATGACACGGG AAAAAAGCAGATGAAGAGAAGGTGGCAGGAGAGGGCACGTGGCCC AGCCTCAGTCTCTCCAACTGAGTTCCTGCCTGCCTGCCTTTGCTCAGA CTGTTTGCCCCTTACTGCTCTTCTAGGCCTCATTCTAAGCCCCTTCTC CAAGTTGCCTCTCCTTATTTCTCCCTGTCTGCCAAAAAATCTTTCCCA GCTCACTAAGTCAGTCTCACGCAGTCACTCATTAACCCACCAATCAC TGATTGTGCCGGCACATGAATGCACCAGGTGTTGAAGTGGAGGAAT TAAAAAGTCAGATGAGGGGTGTGCCCAGAGGAAGCACCATTCTAGT TGGGGGAGCCCATCTGTCAGCTGGGAAAAGTCCAAATAACTTCAGA TTGGAATGTGTTTTAACTCAGGGTTGAGAAAACAGCTACCTTCAGGA CAAAAGTCAGGGAAGGGCTCTCTGAAGAAATGCTACTTGAAGATAC CAGCCCTACCAAGGGCAGGGAGAGGACCCTATAGAGGCCTGGGACA GGAGCTCAATGAGAAAGGTAACCACGTGCGGACCGAGGCTGCAGCG TCGTCCTCCCTAGGAACCCCTAGTGATGGAGTTGGCCACTCCCTCTC TGCGCGCTCGCTCGCTCACTGAGGCCGGGCGACCAAAGGTCGCCCG ACGCCCGGGCTTTGCCCGGGCGGCCTCAGTGAGCGAGCGAGCGCGC AGCTGCCTGCAGG 156 Anti-CD19 GAGATGTAAGGAGCTGCTGTGACTTGCTCAAGGCCTTATATCGAGTA CAR LHA AACGGTAGTGCTGGGGCTTAGACGCAGGTGTTCTGATTTATAGTTCA to RHA AAACCTCTATCAATGAGAGAGCAATCTCCTGGTAATGTGATAGATTT CCCAACTTAATGCCAACATACCATAAACCTCCCATTCTGCTAATGCC CAGCCTAAGTTGGGGAGACCACTCCAGATTCCAAGATGTACAGTTTG CTTTGCTGGGCCTTTTTCCCATGCCTGCCTTTACTCTGCCAGAGTTAT ATTGCTGGGGTTTTGAAGAAGATCCTATTAAATAAAAGAATAAGCA GTATTATTAAGTAGCCCTGCATTTCAGGTTTCCTTGAGTGGCAGGCC AGGCCTGGCCGTGAACGTTCACTGAAATCATGGCCTCTTGGCCAAGA TTGATAGCTTGTGCCTGTCCCTGAGTCCCAGTCCATCACGAGCAGCT GGTTTCTAAGATGCTATTTCCCGTATAAAGCATGAGACCGTGACTTG CCAGCCCCACAGAGCCCCGCCCTTGTCCATCACTGGCATCTGGACTC CAGCCTGGGTTGGGGCAAAGAGGGAAATGAGATCATGTCCTAACCC TGATCCTCTTGTCCCACAGATATCCAGAACCCTGACCCTGCCGTGTA CCAGCTGAGAGACTCTAAATCCAGTGACAAGTCTGTCTGCCTATTCA CCGATTTTGATTCTCAAACAAATGTGTCACAAAGTAAGGATTCTGAT GTGTATATCACAGACAAAACTGTGCTAGACATGAGGTCTATGGACTT CAGGCTCCGGTGCCCGTCAGTGGGCAGAGCGCACATCGCCCACAGT CCCCGAGAAGTTGGGGGGAGGGGTCGGCAATTGAACCGGTGCCTAG AGAAGGTGGCGCGGGGTAAACTGGGAAAGTGATGTCGTGTACTGGC TCCGCCTTTTTCCCGAGGGTGGGGGAGAACCGTATATAAGTGCAGTA GTCGCCGTGAACGTTCTTTTTCGCAACGGGTTTGCCGCCAGAACACA GGTAAGTGCCGTGTGTGGTTCCCGCGGGCCTGGCCTCTTTACGGGTT ATGGCCCTTGCGTGCCTTGAATTACTTCCACTGGCTGCAGTACGTGA TTCTTGATCCCGAGCTTCGGGTTGGAAGTGGGTGGGAGAGTTCGAGG CCTTGCGCTTAAGGAGCCCCTTCGCCTCGTGCTTGAGTTGAGGCCTG GCCTGGGCGCTGGGGCCGCCGCGTGCGAATCTGGTGGCACCTTCGC GCCTGTCTCGCTGCTTTCGATAAGTCTCTAGCCATTTAAAATTTTTGA TGACCTGCTGCGACGCTTTTTTTCTGGCAAGATAGTCTTGTAAATGC GGGCCAAGATCTGCACACTGGTATTTCGGTTTTTGGGGCCGCGGGCG GCGACGGGGCCCGTGCGTCCCAGCGCACATGTTCGGCGAGGCGGGG CCTGCGAGCGCGGCCACCGAGAATCGGACGGGGGTAGTCTCAAGCT GGCCGGCCTGCTCTGGTGCCTGGCCTCGCGCCGCCGTGTATCGCCCC GCCCTGGGCGGCAAGGCTGGCCCGGTCGGCACCAGTTGCGTGAGCG GAAAGATGGCCGCTTCCCGGCCCTGCTGCAGGGAGCTCAAAATGGA GGACGCGGCGCTCGGGAGAGCGGGCGGGTGAGTCACCCACACAAA GGAAAAGGGCCTTTCCGTCCTCAGCCGTCGCTTCATGTGACTCCACG GAGTACCGGGCGCCGTCCAGGCACCTCGATTAGTTCTCGAGCTTTTG GAGTACGTCGTCTTTAGGTTGGGGGGAGGGGTTTTATGCGATGGAGT TTCCCCACACTGAGTGGGTGGAGACTGAAGTTAGGCCAGCTTGGCA CTTGATGTAATTCTCCTTGGAATTTGCCCTTTTTGAGTTTGGATCTTG GTTCATTCTCAAGCCTCAGACAGTGGTTCAAAGTTTTTTTCTTCCATT TCAGGTGTCGTGACCACCATGCTTCTTTTGGTTACGTCTCTGTTGCTT TGCGAACTTCCTCATCCAGCGTTCTTGCTGATCCCCGATATTCAGAT GACTCAGACCACCAGTAGCTTGTCTGCCTCACTGGGAGACCGAGTA ACAATCTCCTGCAGGGCAAGTCAAGACATTAGCAAATACCTCAATT GGTACCAGCAGAAGCCCGACGGAACGGTAAAACTCCTCATCTATCA TACGTCAAGGTTGCATTCCGGAGTACCGTCACGATTTTCAGGTTCTG GGAGCGGAACTGACTATTCCTTGACTATTTCAAACCTCGAGCAGGAG GACATTGCGACATATTTTTGTCAACAAGGTAATACCCTCCCTTACAC TTTCGGAGGAGGAACCAAACTCGAAATTACCGGGTCCACCAGTGGC TCTGGGAAGCCTGGCAGTGGAGAAGGTTCCACTAAAGGCGAGGTGA AGCTCCAGGAGAGCGGCCCCGGTCTCGTTGCCCCCAGTCAAAGCCTC TCTGTAACGTGCACAGTGAGTGGTGTATCATTGCCTGATTATGGCGT CTCCTGGATAAGGCAGCCCCCGCGAAAGGGTCTTGAATGGCTTGGG GTAATATGGGGCTCAGAGACAACGTATTATAACTCCGCTCTCAAAA GTCGCTTGACGATAATAAAAGATAACTCCAAGAGTCAAGTTTTCCTT AAAATGAACAGTTTGCAGACTGACGATACCGCTATATATTATTGTGC TAAACATTATTACTACGGCGGTAGTTACGCGATGGATTATTGGGGGC AGGGGACTTCTGTCACAGTCAGTAGTGCTGCTGCCTTTGTCCCGGTA TTTCTCCCAGCCAAACCGACCACGACTCCCGCCCCGCGCCCTCCGAC ACCCGCTCCCACCATCGCCTCTCAACCTCTTAGTCTTCGCCCCGAGG CATGCCGACCCGCCGCCGGGGGTGCTGTTCATACGAGGGGCTTGGA CTTCGCTTGTGATATTTACATTTGGGCTCCGTTGGCGGGTACGTGCG GCGTCCTTTTGTTGTCACTCGTTATTACTTTGTATTGTAATCACAGGA ATCGCTCAAAGCGGAGTAGGTTGTTGCATTCCGATTACATGAATATG ACTCCTCGCCGGCCTGGGCCGACAAGAAAACATTACCAACCCTATG CCCCCCCACGAGACTTCGCTGCGTACAGGTCCCGAGTGAAGTTTTCC CGAAGCGCAGACGCTCCGGCATATCAGCAAGGACAGAATCAGCTGT ATAACGAACTGAATTTGGGACGCCGCGAGGAGTATGACGTGCTTGA TAAACGCCGGGGGAGAGACCCGGAAATGGGGGGTAAACCCCGAAG AAAGAATCCCCAAGAAGGACTCTACAATGAACTCCAGAAGGATAAG ATGGCGGAGGCCTACTCAGAAATAGGTATGAAGGGCGAACGACGAC GGGGAAAAGGTCACGATGGCCTCTACCAAGGGTTGAGTACGGCAAC CAAAGATACGTACGATGCACTGCATATGCAGGCCCTGCCTCCCAGAT AATAATAAAATCGCTATCCATCGAAGATGGATGTGTGTTGGTTTTTT GTGTGTGGAGCAACAAATCTGACTTTGCATGTGCAAACGCCTTCAAC AACAGCATTATTCCAGAAGACACCTTCTTCCCCAGCCCAGGTAAGGG CAGCTTTGGTGCCTTCGCAGGCTGTTTCCTTGCTTCAGGAATGGCCA GGTTCTGCCCAGAGCTCTGGTCAATGATGTCTAAAACTCCTCTGATT GGTGGTCTCGGCCTTATCCATTGCCACCAAAACCCTCTTTTTACTAA GAAACAGTGAGCCTTGTTCTGGCAGTCCAGAGAATGACACGGGAAA AAAGCAGATGAAGAGAAGGTGGCAGGAGAGGGCACGTGGCCCAGC CTCAGTCTCTCCAACTGAGTTCCTGCCTGCCTGCCTTTGCTCAGACTG TTTGCCCCTTACTGCTCTTCTAGGCCTCATTCTAAGCCCCTTCTCCAA GTTGCCTCTCCTTATTTCTCCCTGTCTGCCAAAAAATCTTTCCCAGCT CACTAAGTCAGTCTCACGCAGTCACTCATTAACCCACCAATCACTGA TTGTGCCGGCACATGAATGCACCAGGTGTTGAAGTGGAGGAATTAA AAAGTCAGATGAGGGGTGTGCCCAGAGGAAGCACCATTCTAGTTGG GGGAGCCCATCTGTCAGCTGGGAAAAGTCCAAATAACTTCAGATTG GAATGTGTTTTAACTCAGGGTTGAGAAAACAGCTACCTTCAGGACA AAAGTCAGGGAAGGGCTCTCTGAAGAAATGCTACTTGAAGATACCA GCCCTACCAAGGGCAGGGAGAGGACCCTATAGAGGCCTGGGACAGG AGCTCAATGAGAAAGG 157 CD70 TCACCAAGCCCGCGACCAAT sgRNA (E1_T1) 158 CD70 ATCACCAAGCCCGCGACCAA sgRNA (E1_T3) 159 CD70 CGGTGCGGCGCAGGCCCTAT sgRNA (E1_T4) 160 CD70 GCTTTGGTCCCATTGGTCGC sgRNA (E1_T7) 161 CD70 GCCCGCAGGACGCACCCATA sgRNA (E1_T8) 162 CD70 GTGCATCCAGCGCTTCGCAC sgRNA (E1_T10) 163 CD70 CAGCTACGTATCCATCGTGA sgRNA (E3_T1) 164 .beta.2M sgRNA GCTACTCTCTCTTTCTGGCC 165 PD-1 sgRNA CTGCAGCTTCTCCAACACAT 166 anti-CD19 RASQDISKYLN VL CDR1 (Kabat) 167 anti-CD19 HTSRLHS VL CDR2 (Kabat) 168 anti-CD19 QQGNTLPYT VL CDR3 (Kabat) 169 anti-CD19 DYGVS VH CDR1 (Kabat) 170 anti-CD19 VIWGSETTYYNSALKS VH CDR2 (Kabat) 171 anti-CD19 HYYYGGSYAMDY VH CDR3 (Kabat) 172 anti-CD19 RASQDISKYLN VL CDR1 (Chothia) 173 anti-CD19 HTSRLHS VL CDR2 (Chothia) 174 anti-CD19 QQGNTLPYT VL CDR3 (Chothia) 175 anti-CD19 GVSLPDY VH CDR1 (Chothia)
176 anti-CD19 WGSET VH CDR2 (Chothia) 177 anti-CD19 HYYYGGSYAMDY VH CDR3 (Chothia) 178 anti-CD33 GYTFTSY VH CDR1 (Chothia) 179 anti-CD33 YPGNDD VH CDR2 (Chothia) 180 anti-CD33 EVRLRYFDV VH CDR3 (Chothia)
[0782] All references, patents and patent applications disclosed herein are incorporated by reference with respect to the subject matter for which each is cited, which in some cases may encompass the entirety of the document.
[0783] The indefinite articles "a" and "an," as used herein in the specification and in the claims, unless clearly indicated to the contrary, should be understood to mean "at least one."
[0784] It should also be understood that, unless clearly indicated to the contrary, in any methods claimed herein that include more than one step or act, the order of the steps or acts of the method is not necessarily limited to the order in which the steps or acts of the method are recited.
[0785] In the claims, as well as in the specification above, all transitional phrases such as "comprising," "including," "carrying," "having," "containing," "involving," "holding," "composed of," and the like are to be understood to be open-ended, i.e., to mean including but not limited to. Only the transitional phrases "consisting of" and "consisting essentially of" shall be closed or semi-closed transitional phrases, respectively, as set forth in the United States Patent Office Manual of Patent Examining Procedures, Section 2111.03.
[0786] The terms "about" and "substantially" preceding a numerical value mean .+-.10% of the recited numerical value.
[0787] Where a range of values is provided, each value between the upper and lower ends of the range are specifically contemplated and described herein.
Sequence CWU 1
1
180119DNAArtificial SequenceSynthetic TRAC Indel 1aagagcaaca
aatctgact 19258DNAArtificial SequenceSynthetic TRAC Indel
2aagagcaaca gtgctgtgcc tggagcaaca aatctgacta agagcaacaa atctgact
58352DNAArtificial SequenceSynthetic TRAC Indel 3aagagcaaca
gtgctggagc aacaaatctg actaagagca acaaatctga ct 52453DNAArtificial
SequenceSynthetic TRAC Indel 4aagagcaaca gtgcctggag caacaaatct
gactaagagc aacaaatctg act 53538DNAArtificial SequenceSynthetic TRAC
Indel 5aagagcaaca gtgctgacta agagcaacaa atctgact 38660DNAArtificial
SequenceSynthetic TRAC Indel 6aagagcaaca gtgctgtggg cctggagcaa
caaatctgac taagagcaac aaatctgact 60757DNAArtificial
SequenceSynthetic TRAC Indel 7aagagcaaca gtgctggcct ggagcaacaa
atctgactaa gagcaacaaa tctgact 57860DNAArtificial SequenceSynthetic
TRAC Indel 8aagagcaaca gtgctgtgtg cctggagcaa caaatctgac taagagcaac
aaatctgact 60979DNAArtificial SequenceSynthetic B2M Indel
9cgtggcctta gctgtgctcg cgctactctc tctttctgcc tggaggctat ccagcgtgag
60tctctcctac cctcccgct 791078DNAArtificial SequenceSynthetic B2M
Indel 10cgtggcctta gctgtgctcg cgctactctc tctttcgcct ggaggctatc
cagcgtgagt 60ctctcctacc ctcccgct 781175DNAArtificial
SequenceSynthetic B2M Indel 11cgtggcctta gctgtgctcg cgctactctc
tctttctgga ggctatccag cgtgagtctc 60tcctaccctc ccgct
751284DNAArtificial SequenceSynthetic B2M Indel 12cgtggcctta
gctgtgctcg cgctactctc tctttctgga tagcctggag gctatccagc 60gtgagtctct
cctaccctcc cgct 841355DNAArtificial SequenceSynthetic B2M Indel
13cgtggcctta gctgtgctcg cgctatccag cgtgagtctc tcctaccctc ccgct
551482DNAArtificial SequenceSynthetic B2M Indel 14cgtggcctta
gctgtgctcg cgctactctc tctttctgtg gcctggaggc tatccagcgt 60gagtctctcc
taccctcccg ct 8215100RNAArtificial SequenceSynthetic
sgRNAmisc_feature(1)..(20)n is a, c, g, or u 15nnnnnnnnnn
nnnnnnnnnn guuuuagagc uagaaauagc aaguuaaaau aaggcuaguc 60cguuaucaac
uugaaaaagu ggcaccgagu cggugcuuuu 1001696RNAArtificial
SequenceSynthetic sgRNAmisc_feature(1)..(20)n is a, c, g, or u
16nnnnnnnnnn nnnnnnnnnn guuuuagagc uagaaauagc aaguuaaaau aaggcuaguc
60cguuaucaac uugaaaaagu ggcaccgagu cggugc 9617114RNAArtificial
SequenceSynthetic sgRNAmisc_feature(1)..(30)n is a, c, g, or
umisc_feature(18)..(30)n may be present or
absentmisc_feature(108)..(114)u may be present or absent
17nnnnnnnnnn nnnnnnnnnn nnnnnnnnnn guuuuagagc uagaaauagc aaguuaaaau
60aaggcuaguc cguuaucaac uugaaaaagu ggcaccgagu cggugcuuuu uuuu
11418126DNAArtificial SequenceSynthetic 4-1BB nucleotide sequence
18aaacggggca gaaagaaact cctgtatata ttcaaacaac catttatgag accagtacaa
60actactcaag aggaagatgg ctgtagctgc cgatttccag aagaagaaga aggaggatgt
120gaactg 1261942PRTArtificial SequenceSynthetic 4-1BB amino acid
sequence 19Lys Arg Gly Arg Lys Lys Leu Leu Tyr Ile Phe Lys Gln Pro
Phe Met1 5 10 15Arg Pro Val Gln Thr Thr Gln Glu Glu Asp Gly Cys Ser
Cys Arg Phe 20 25 30Pro Glu Glu Glu Glu Gly Gly Cys Glu Leu 35
402040PRTArtificial SequenceSynthetic CD28 amino acid sequence
20Ser Lys Arg Ser Arg Leu Leu His Ser Asp Tyr Met Asn Met Thr Pro1
5 10 15Arg Arg Pro Gly Pro Thr Arg Lys His Tyr Gln Pro Tyr Ala Pro
Pro 20 25 30Arg Asp Phe Ala Ala Tyr Arg Ser 35 4021336DNAArtificial
SequenceSynthetic CD3-z nucleotide sequence 21cgagtgaagt tttcccgaag
cgcagacgct ccggcatatc agcaaggaca gaatcagctg 60tataacgaac tgaatttggg
acgccgcgag gagtatgacg tgcttgataa acgccggggg 120agagacccgg
aaatgggggg taaaccccga agaaagaatc cccaagaagg actctacaat
180gaactccaga aggataagat ggcggaggcc tactcagaaa taggtatgaa
gggcgaacga 240cgacggggaa aaggtcacga tggcctctac caagggttga
gtacggcaac caaagatacg 300tacgatgcac tgcatatgca ggccctgcct cccaga
33622112PRTArtificial SequenceSynthetic CD3-z amino acid sequence
22Arg Val Lys Phe Ser Arg Ser Ala Asp Ala Pro Ala Tyr Gln Gln Gly1
5 10 15Gln Asn Gln Leu Tyr Asn Glu Leu Asn Leu Gly Arg Arg Glu Glu
Tyr 20 25 30Asp Val Leu Asp Lys Arg Arg Gly Arg Asp Pro Glu Met Gly
Gly Lys 35 40 45Pro Arg Arg Lys Asn Pro Gln Glu Gly Leu Tyr Asn Glu
Leu Gln Lys 50 55 60Asp Lys Met Ala Glu Ala Tyr Ser Glu Ile Gly Met
Lys Gly Glu Arg65 70 75 80Arg Arg Gly Lys Gly His Asp Gly Leu Tyr
Gln Gly Leu Ser Thr Ala 85 90 95Thr Lys Asp Thr Tyr Asp Ala Leu His
Met Gln Ala Leu Pro Pro Arg 100 105 11023100RNAArtificial
SequenceSynthetic CD70 sgRNA (E1_T1) 23ucaccaagcc cgcgaccaau
guuuuagagc uagaaauagc aaguuaaaau aaggcuaguc 60cguuaucaac uugaaaaagu
ggcaccgagu cggugcuuuu 10024100RNAArtificial SequenceSynthetic CD70
sgRNA (E1_T3) 24aucaccaagc ccgcgaccaa guuuuagagc uagaaauagc
aaguuaaaau aaggcuaguc 60cguuaucaac uugaaaaagu ggcaccgagu cggugcuuuu
10025100RNAArtificial SequenceSynthetic CD70 sgRNA (E1_T4)
25cggugcggcg caggcccuau guuuuagagc uagaaauagc aaguuaaaau aaggcuaguc
60cguuaucaac uugaaaaagu ggcaccgagu cggugcuuuu 10026100RNAArtificial
SequenceSynthetic CD70 sgRNA (E1_T7) 26gcuuuggucc cauuggucgc
guuuuagagc uagaaauagc aaguuaaaau aaggcuaguc 60cguuaucaac uugaaaaagu
ggcaccgagu cggugcuuuu 10027100RNAArtificial SequenceSynthetic CD70
sgRNA (E1_T8) 27gcccgcagga cgcacccaua guuuuagagc uagaaauagc
aaguuaaaau aaggcuaguc 60cguuaucaac uugaaaaagu ggcaccgagu cggugcuuuu
10028100RNAArtificial SequenceSynthetic CD70 sgRNA (E1_T10)
28gugcauccag cgcuucgcac guuuuagagc uagaaauagc aaguuaaaau aaggcuaguc
60cguuaucaac uugaaaaagu ggcaccgagu cggugcuuuu 10029100RNAArtificial
SequenceSynthetic CD70 sgRNA (E3_T1) 29cagcuacgua uccaucguga
guuuuagagc uagaaauagc aaguuaaaau aaggcuaguc 60cguuaucaac uugaaaaagu
ggcaccgagu cggugcuuuu 10030100RNAArtificial SequenceSynthetic TRAC
sgRNA 30agagcaacag ugcuguggcc guuuuagagc uagaaauagc aaguuaaaau
aaggcuaguc 60cguuaucaac uugaaaaagu ggcaccgagu cggugcuuuu
10031100RNAArtificial SequenceSynthetic b2M sgRNA 31gcuacucucu
cuuucuggcc guuuuagagc uagaaauagc aaguuaaaau aaggcuaguc 60cguuaucaac
uugaaaaagu ggcaccgagu cggugcuuuu 10032100RNAArtificial
SequenceSynthetic PD-1 sgRNA 32cugcagcuuc uccaacacau guuuuagagc
uagaaauagc aaguuaaaau aaggcuaguc 60cguuaucaac uugaaaaagu ggcaccgagu
cggugcuuuu 10033100RNAArtificial SequenceSynthetic CD70 sgRNA
(E1_T1)modified_base(1)..(3)2'-O-methyl
phosphorothioatemodified_base(97)..(99)2'-O-methyl phosphorothioate
33ucaccaagcc cgcgaccaau guuuuagagc uagaaauagc aaguuaaaau aaggcuaguc
60cguuaucaac uugaaaaagu ggcaccgagu cggugcuuuu 10034100RNAArtificial
SequenceSynthetic CD70 sgRNA
(E1_T3)modified_base(1)..(3)2'-O-methyl
phosphorothioatemodified_base(97)..(99)2'-O-methyl phosphorothioate
34aucaccaagc ccgcgaccaa guuuuagagc uagaaauagc aaguuaaaau aaggcuaguc
60cguuaucaac uugaaaaagu ggcaccgagu cggugcuuuu 10035100RNAArtificial
SequenceSynthetic CD70 sgRNA
(E1_T4)modified_base(1)..(3)2'-O-methyl
phosphorothioatemodified_base(97)..(99)2'-O-methyl phosphorothioate
35cggugcggcg caggcccuau guuuuagagc uagaaauagc aaguuaaaau aaggcuaguc
60cguuaucaac uugaaaaagu ggcaccgagu cggugcuuuu 10036100RNAArtificial
SequenceSynthetic CD70 sgRNA
(E1_T7)modified_base(1)..(3)2'-O-methyl
phosphorothioatemodified_base(97)..(99)2'-O-methyl phosphorothioate
36gcuuuggucc cauuggucgc guuuuagagc uagaaauagc aaguuaaaau aaggcuaguc
60cguuaucaac uugaaaaagu ggcaccgagu cggugcuuuu 10037100RNAArtificial
SequenceSynthetic CD70 sgRNA
(E1_T8)modified_base(1)..(3)2'-O-methyl
phosphorothioatemodified_base(97)..(99)2'-O-methyl phosphorothioate
37gcccgcagga cgcacccaua guuuuagagc uagaaauagc aaguuaaaau aaggcuaguc
60cguuaucaac uugaaaaagu ggcaccgagu cggugcuuuu 10038100RNAArtificial
SequenceSynthetic CD70 sgRNA
(E1_T10)modified_base(1)..(3)2'-O-methyl
phosphorothioatemodified_base(97)..(99)2'-O-methyl phosphorothioate
38gugcauccag cgcuucgcac guuuuagagc uagaaauagc aaguuaaaau aaggcuaguc
60cguuaucaac uugaaaaagu ggcaccgagu cggugcuuuu 10039100RNAArtificial
SequenceSynthetic CD70 sgRNA
(E3_T1)modified_base(1)..(3)2'-O-methyl
phosphorothioatemodified_base(97)..(99)2'-O-methyl phosphorothioate
39cagcuacgua uccaucguga guuuuagagc uagaaauagc aaguuaaaau aaggcuaguc
60cguuaucaac uugaaaaagu ggcaccgagu cggugcuuuu 10040100RNAArtificial
SequenceSynthetic TRAC sgRNAmodified_base(1)..(3)2'-O-methyl
phosphorothioatemodified_base(97)..(99)2'-O-methyl phosphorothioate
40agagcaacag ugcuguggcc guuuuagagc uagaaauagc aaguuaaaau aaggcuaguc
60cguuaucaac uugaaaaagu ggcaccgagu cggugcuuuu 10041100RNAArtificial
SequenceSynthetic b2M sgRNAmodified_base(1)..(3)2'-O-methyl
phosphorothioatemodified_base(97)..(99)2'-O-methyl phosphorothioate
41gcuacucucu cuuucuggcc guuuuagagc uagaaauagc aaguuaaaau aaggcuaguc
60cguuaucaac uugaaaaagu ggcaccgagu cggugcuuuu 10042100RNAArtificial
SequenceSynthetic PD-1 sgRNAmodified_base(1)..(3)2'-O-methyl
phosphorothioatemodified_base(97)..(99)2'-O-methyl phosphorothioate
42cugcagcuuc uccaacacau guuuuagagc uagaaauagc aaguuaaaau aaggcuaguc
60cguuaucaac uugaaaaagu ggcaccgagu cggugcuuuu
100434688DNAArtificial SequenceSynthetic CD70 rAAV (CD70B scFV with
41BB) 43cctgcaggca gctgcgcgct cgctcgctca ctgaggccgc ccgggcgtcg
ggcgaccttt 60ggtcgcccgg cctcagtgag cgagcgagcg cgcagagagg gagtggccaa
ctccatcact 120aggggttcct gcggccgcac gcgtgagatg taaggagctg
ctgtgacttg ctcaaggcct 180tatatcgagt aaacggtagt gctggggctt
agacgcaggt gttctgattt atagttcaaa 240acctctatca atgagagagc
aatctcctgg taatgtgata gatttcccaa cttaatgcca 300acataccata
aacctcccat tctgctaatg cccagcctaa gttggggaga ccactccaga
360ttccaagatg tacagtttgc tttgctgggc ctttttccca tgcctgcctt
tactctgcca 420gagttatatt gctggggttt tgaagaagat cctattaaat
aaaagaataa gcagtattat 480taagtagccc tgcatttcag gtttccttga
gtggcaggcc aggcctggcc gtgaacgttc 540actgaaatca tggcctcttg
gccaagattg atagcttgtg cctgtccctg agtcccagtc 600catcacgagc
agctggtttc taagatgcta tttcccgtat aaagcatgag accgtgactt
660gccagcccca cagagccccg cccttgtcca tcactggcat ctggactcca
gcctgggttg 720gggcaaagag ggaaatgaga tcatgtccta accctgatcc
tcttgtccca cagatatcca 780gaaccctgac cctgccgtgt accagctgag
agactctaaa tccagtgaca agtctgtctg 840cctattcacc gattttgatt
ctcaaacaaa tgtgtcacaa agtaaggatt ctgatgtgta 900tatcacagac
aaaactgtgc tagacatgag gtctatggac ttcaggctcc ggtgcccgtc
960agtgggcaga gcgcacatcg cccacagtcc ccgagaagtt ggggggaggg
gtcggcaatt 1020gaaccggtgc ctagagaagg tggcgcgggg taaactggga
aagtgatgtc gtgtactggc 1080tccgcctttt tcccgagggt gggggagaac
cgtatataag tgcagtagtc gccgtgaacg 1140ttctttttcg caacgggttt
gccgccagaa cacaggtaag tgccgtgtgt ggttcccgcg 1200ggcctggcct
ctttacgggt tatggccctt gcgtgccttg aattacttcc actggctgca
1260gtacgtgatt cttgatcccg agcttcgggt tggaagtggg tgggagagtt
cgaggccttg 1320cgcttaagga gccccttcgc ctcgtgcttg agttgaggcc
tggcctgggc gctggggccg 1380ccgcgtgcga atctggtggc accttcgcgc
ctgtctcgct gctttcgata agtctctagc 1440catttaaaat ttttgatgac
ctgctgcgac gctttttttc tggcaagata gtcttgtaaa 1500tgcgggccaa
gatctgcaca ctggtatttc ggtttttggg gccgcgggcg gcgacggggc
1560ccgtgcgtcc cagcgcacat gttcggcgag gcggggcctg cgagcgcggc
caccgagaat 1620cggacggggg tagtctcaag ctggccggcc tgctctggtg
cctggcctcg cgccgccgtg 1680tatcgccccg ccctgggcgg caaggctggc
ccggtcggca ccagttgcgt gagcggaaag 1740atggccgctt cccggccctg
ctgcagggag ctcaaaatgg aggacgcggc gctcgggaga 1800gcgggcgggt
gagtcaccca cacaaaggaa aagggccttt ccgtcctcag ccgtcgcttc
1860atgtgactcc acggagtacc gggcgccgtc caggcacctc gattagttct
cgagcttttg 1920gagtacgtcg tctttaggtt ggggggaggg gttttatgcg
atggagtttc cccacactga 1980gtgggtggag actgaagtta ggccagcttg
gcacttgatg taattctcct tggaatttgc 2040cctttttgag tttggatctt
ggttcattct caagcctcag acagtggttc aaagtttttt 2100tcttccattt
caggtgtcgt gaccaccatg gcgcttccgg tgacagcact gctcctcccc
2160ttggcgctgt tgctccacgc agcaaggccg caggtccagt tggtgcaaag
cggggcggag 2220gtgaaaaaac ccggcgcttc cgtgaaggtg tcctgtaagg
cgtccggtta tacgttcacg 2280aactacggga tgaattgggt tcgccaagcg
ccggggcagg gactgaaatg gatggggtgg 2340ataaatacct acaccggcga
acctacatac gccgacgctt ttaaagggcg agtcactatg 2400acgcgcgata
ccagcatatc caccgcatac atggagctgt cccgactccg gtcagacgac
2460acggctgtct actattgtgc tcgggactat ggcgattatg gcatggacta
ctggggtcag 2520ggtacgactg taacagttag tagtggtgga ggcggcagtg
gcgggggggg aagcggagga 2580gggggttctg gtgacatagt tatgacccaa
tccccagata gtttggcggt ttctctgggc 2640gagagggcaa cgattaattg
tcgcgcatca aagagcgttt caacgagcgg atattctttt 2700atgcattggt
accagcaaaa acccggacaa ccgccgaagc tgctgatcta cttggcttca
2760aatcttgagt ctggggtgcc ggaccgattt tctggtagtg gaagcggaac
tgactttacg 2820ctcacgatca gttcactgca ggctgaggat gtagcggtct
attattgcca gcacagtaga 2880gaagtcccct ggaccttcgg tcaaggcacg
aaagtagaaa ttaaaagtgc tgctgccttt 2940gtcccggtat ttctcccagc
caaaccgacc acgactcccg ccccgcgccc tccgacaccc 3000gctcccacca
tcgcctctca acctcttagt cttcgccccg aggcatgccg acccgccgcc
3060gggggtgctg ttcatacgag gggcttggac ttcgcttgtg atatttacat
ttgggctccg 3120ttggcgggta cgtgcggcgt ccttttgttg tcactcgtta
ttactttgta ttgtaatcac 3180aggaatcgca aacggggcag aaagaaactc
ctgtatatat tcaaacaacc atttatgaga 3240ccagtacaaa ctactcaaga
ggaagatggc tgtagctgcc gatttccaga agaagaagaa 3300ggaggatgtg
aactgcgagt gaagttttcc cgaagcgcag acgctccggc atatcagcaa
3360ggacagaatc agctgtataa cgaactgaat ttgggacgcc gcgaggagta
tgacgtgctt 3420gataaacgcc gggggagaga cccggaaatg gggggtaaac
cccgaagaaa gaatccccaa 3480gaaggactct acaatgaact ccagaaggat
aagatggcgg aggcctactc agaaataggt 3540atgaagggcg aacgacgacg
gggaaaaggt cacgatggcc tctaccaagg gttgagtacg 3600gcaaccaaag
atacgtacga tgcactgcat atgcaggccc tgcctcccag ataataataa
3660aatcgctatc catcgaagat ggatgtgtgt tggttttttg tgtgtggagc
aacaaatctg 3720actttgcatg tgcaaacgcc ttcaacaaca gcattattcc
agaagacacc ttcttcccca 3780gcccaggtaa gggcagcttt ggtgccttcg
caggctgttt ccttgcttca ggaatggcca 3840ggttctgccc agagctctgg
tcaatgatgt ctaaaactcc tctgattggt ggtctcggcc 3900ttatccattg
ccaccaaaac cctcttttta ctaagaaaca gtgagccttg ttctggcagt
3960ccagagaatg acacgggaaa aaagcagatg aagagaaggt ggcaggagag
ggcacgtggc 4020ccagcctcag tctctccaac tgagttcctg cctgcctgcc
tttgctcaga ctgtttgccc 4080cttactgctc ttctaggcct cattctaagc
cccttctcca agttgcctct ccttatttct 4140ccctgtctgc caaaaaatct
ttcccagctc actaagtcag tctcacgcag tcactcatta 4200acccaccaat
cactgattgt gccggcacat gaatgcacca ggtgttgaag tggaggaatt
4260aaaaagtcag atgaggggtg tgcccagagg aagcaccatt ctagttgggg
gagcccatct 4320gtcagctggg aaaagtccaa ataacttcag attggaatgt
gttttaactc agggttgaga 4380aaacagctac cttcaggaca aaagtcaggg
aagggctctc tgaagaaatg ctacttgaag 4440ataccagccc taccaagggc
agggagagga ccctatagag gcctgggaca ggagctcaat 4500gagaaaggta
accacgtgcg gaccgaggct gcagcgtcgt cctccctagg aacccctagt
4560gatggagttg gccactccct ctctgcgcgc tcgctcgctc actgaggccg
ggcgaccaaa 4620ggtcgcccga cgcccgggct ttgcccgggc ggcctcagtg
agcgagcgag cgcgcagctg 4680cctgcagg 4688444364DNAArtificial
SequenceSynthetic CD70 LHA to RHA (CD70B scFV with 41BB)
44gagatgtaag gagctgctgt gacttgctca aggccttata tcgagtaaac ggtagtgctg
60gggcttagac gcaggtgttc tgatttatag ttcaaaacct ctatcaatga gagagcaatc
120tcctggtaat gtgatagatt tcccaactta atgccaacat accataaacc
tcccattctg 180ctaatgccca gcctaagttg gggagaccac tccagattcc
aagatgtaca gtttgctttg 240ctgggccttt ttcccatgcc tgcctttact
ctgccagagt tatattgctg gggttttgaa 300gaagatccta ttaaataaaa
gaataagcag tattattaag tagccctgca tttcaggttt 360ccttgagtgg
caggccaggc ctggccgtga acgttcactg aaatcatggc ctcttggcca
420agattgatag cttgtgcctg tccctgagtc ccagtccatc acgagcagct
ggtttctaag 480atgctatttc ccgtataaag catgagaccg tgacttgcca
gccccacaga gccccgccct 540tgtccatcac tggcatctgg actccagcct
gggttggggc aaagagggaa atgagatcat 600gtcctaaccc tgatcctctt
gtcccacaga tatccagaac cctgaccctg ccgtgtacca 660gctgagagac
tctaaatcca gtgacaagtc tgtctgccta ttcaccgatt ttgattctca
720aacaaatgtg tcacaaagta aggattctga tgtgtatatc acagacaaaa
ctgtgctaga 780catgaggtct atggacttca ggctccggtg cccgtcagtg
ggcagagcgc acatcgccca 840cagtccccga gaagttgggg ggaggggtcg
gcaattgaac cggtgcctag agaaggtggc 900gcggggtaaa ctgggaaagt
gatgtcgtgt actggctccg cctttttccc gagggtgggg 960gagaaccgta
tataagtgca gtagtcgccg tgaacgttct ttttcgcaac gggtttgccg
1020ccagaacaca ggtaagtgcc gtgtgtggtt cccgcgggcc tggcctcttt
acgggttatg 1080gcccttgcgt gccttgaatt acttccactg gctgcagtac
gtgattcttg atcccgagct 1140tcgggttgga agtgggtggg agagttcgag
gccttgcgct taaggagccc cttcgcctcg 1200tgcttgagtt gaggcctggc
ctgggcgctg gggccgccgc gtgcgaatct ggtggcacct 1260tcgcgcctgt
ctcgctgctt tcgataagtc tctagccatt taaaattttt gatgacctgc
1320tgcgacgctt tttttctggc aagatagtct tgtaaatgcg ggccaagatc
tgcacactgg 1380tatttcggtt tttggggccg cgggcggcga cggggcccgt
gcgtcccagc gcacatgttc 1440ggcgaggcgg ggcctgcgag cgcggccacc
gagaatcgga cgggggtagt ctcaagctgg 1500ccggcctgct ctggtgcctg
gcctcgcgcc gccgtgtatc gccccgccct gggcggcaag 1560gctggcccgg
tcggcaccag ttgcgtgagc ggaaagatgg ccgcttcccg gccctgctgc
1620agggagctca aaatggagga cgcggcgctc gggagagcgg gcgggtgagt
cacccacaca 1680aaggaaaagg gcctttccgt cctcagccgt cgcttcatgt
gactccacgg agtaccgggc 1740gccgtccagg cacctcgatt agttctcgag
cttttggagt acgtcgtctt taggttgggg 1800ggaggggttt tatgcgatgg
agtttcccca cactgagtgg gtggagactg aagttaggcc 1860agcttggcac
ttgatgtaat tctccttgga atttgccctt tttgagtttg gatcttggtt
1920cattctcaag cctcagacag tggttcaaag tttttttctt ccatttcagg
tgtcgtgacc 1980accatggcgc ttccggtgac agcactgctc ctccccttgg
cgctgttgct ccacgcagca 2040aggccgcagg tccagttggt gcaaagcggg
gcggaggtga aaaaacccgg cgcttccgtg 2100aaggtgtcct gtaaggcgtc
cggttatacg ttcacgaact acgggatgaa ttgggttcgc 2160caagcgccgg
ggcagggact gaaatggatg gggtggataa atacctacac cggcgaacct
2220acatacgccg acgcttttaa agggcgagtc actatgacgc gcgataccag
catatccacc 2280gcatacatgg agctgtcccg actccggtca gacgacacgg
ctgtctacta ttgtgctcgg 2340gactatggcg attatggcat ggactactgg
ggtcagggta cgactgtaac agttagtagt 2400ggtggaggcg gcagtggcgg
ggggggaagc ggaggagggg gttctggtga catagttatg 2460acccaatccc
cagatagttt ggcggtttct ctgggcgaga gggcaacgat taattgtcgc
2520gcatcaaaga gcgtttcaac gagcggatat tcttttatgc attggtacca
gcaaaaaccc 2580ggacaaccgc cgaagctgct gatctacttg gcttcaaatc
ttgagtctgg ggtgccggac 2640cgattttctg gtagtggaag cggaactgac
tttacgctca cgatcagttc actgcaggct 2700gaggatgtag cggtctatta
ttgccagcac agtagagaag tcccctggac cttcggtcaa 2760ggcacgaaag
tagaaattaa aagtgctgct gcctttgtcc cggtatttct cccagccaaa
2820ccgaccacga ctcccgcccc gcgccctccg acacccgctc ccaccatcgc
ctctcaacct 2880cttagtcttc gccccgaggc atgccgaccc gccgccgggg
gtgctgttca tacgaggggc 2940ttggacttcg cttgtgatat ttacatttgg
gctccgttgg cgggtacgtg cggcgtcctt 3000ttgttgtcac tcgttattac
tttgtattgt aatcacagga atcgcaaacg gggcagaaag 3060aaactcctgt
atatattcaa acaaccattt atgagaccag tacaaactac tcaagaggaa
3120gatggctgta gctgccgatt tccagaagaa gaagaaggag gatgtgaact
gcgagtgaag 3180ttttcccgaa gcgcagacgc tccggcatat cagcaaggac
agaatcagct gtataacgaa 3240ctgaatttgg gacgccgcga ggagtatgac
gtgcttgata aacgccgggg gagagacccg 3300gaaatggggg gtaaaccccg
aagaaagaat ccccaagaag gactctacaa tgaactccag 3360aaggataaga
tggcggaggc ctactcagaa ataggtatga agggcgaacg acgacgggga
3420aaaggtcacg atggcctcta ccaagggttg agtacggcaa ccaaagatac
gtacgatgca 3480ctgcatatgc aggccctgcc tcccagataa taataaaatc
gctatccatc gaagatggat 3540gtgtgttggt tttttgtgtg tggagcaaca
aatctgactt tgcatgtgca aacgccttca 3600acaacagcat tattccagaa
gacaccttct tccccagccc aggtaagggc agctttggtg 3660ccttcgcagg
ctgtttcctt gcttcaggaa tggccaggtt ctgcccagag ctctggtcaa
3720tgatgtctaa aactcctctg attggtggtc tcggccttat ccattgccac
caaaaccctc 3780tttttactaa gaaacagtga gccttgttct ggcagtccag
agaatgacac gggaaaaaag 3840cagatgaaga gaaggtggca ggagagggca
cgtggcccag cctcagtctc tccaactgag 3900ttcctgcctg cctgcctttg
ctcagactgt ttgcccctta ctgctcttct aggcctcatt 3960ctaagcccct
tctccaagtt gcctctcctt atttctccct gtctgccaaa aaatctttcc
4020cagctcacta agtcagtctc acgcagtcac tcattaaccc accaatcact
gattgtgccg 4080gcacatgaat gcaccaggtg ttgaagtgga ggaattaaaa
agtcagatga ggggtgtgcc 4140cagaggaagc accattctag ttgggggagc
ccatctgtca gctgggaaaa gtccaaataa 4200cttcagattg gaatgtgttt
taactcaggg ttgagaaaac agctaccttc aggacaaaag 4260tcagggaagg
gctctctgaa gaaatgctac ttgaagatac cagccctacc aagggcaggg
4320agaggaccct atagaggcct gggacaggag ctcaatgaga aagg
4364451527DNAArtificial SequenceSynthetic CD70 CAR nucleotide
sequence (CD70B scFV with 41BB) 45atggcgcttc cggtgacagc actgctcctc
cccttggcgc tgttgctcca cgcagcaagg 60ccgcaggtcc agttggtgca aagcggggcg
gaggtgaaaa aacccggcgc ttccgtgaag 120gtgtcctgta aggcgtccgg
ttatacgttc acgaactacg ggatgaattg ggttcgccaa 180gcgccggggc
agggactgaa atggatgggg tggataaata cctacaccgg cgaacctaca
240tacgccgacg cttttaaagg gcgagtcact atgacgcgcg ataccagcat
atccaccgca 300tacatggagc tgtcccgact ccggtcagac gacacggctg
tctactattg tgctcgggac 360tatggcgatt atggcatgga ctactggggt
cagggtacga ctgtaacagt tagtagtggt 420ggaggcggca gtggcggggg
gggaagcgga ggagggggtt ctggtgacat agttatgacc 480caatccccag
atagtttggc ggtttctctg ggcgagaggg caacgattaa ttgtcgcgca
540tcaaagagcg tttcaacgag cggatattct tttatgcatt ggtaccagca
aaaacccgga 600caaccgccga agctgctgat ctacttggct tcaaatcttg
agtctggggt gccggaccga 660ttttctggta gtggaagcgg aactgacttt
acgctcacga tcagttcact gcaggctgag 720gatgtagcgg tctattattg
ccagcacagt agagaagtcc cctggacctt cggtcaaggc 780acgaaagtag
aaattaaaag tgctgctgcc tttgtcccgg tatttctccc agccaaaccg
840accacgactc ccgccccgcg ccctccgaca cccgctccca ccatcgcctc
tcaacctctt 900agtcttcgcc ccgaggcatg ccgacccgcc gccgggggtg
ctgttcatac gaggggcttg 960gacttcgctt gtgatattta catttgggct
ccgttggcgg gtacgtgcgg cgtccttttg 1020ttgtcactcg ttattacttt
gtattgtaat cacaggaatc gcaaacgggg cagaaagaaa 1080ctcctgtata
tattcaaaca accatttatg agaccagtac aaactactca agaggaagat
1140ggctgtagct gccgatttcc agaagaagaa gaaggaggat gtgaactgcg
agtgaagttt 1200tcccgaagcg cagacgctcc ggcatatcag caaggacaga
atcagctgta taacgaactg 1260aatttgggac gccgcgagga gtatgacgtg
cttgataaac gccgggggag agacccggaa 1320atggggggta aaccccgaag
aaagaatccc caagaaggac tctacaatga actccagaag 1380gataagatgg
cggaggccta ctcagaaata ggtatgaagg gcgaacgacg acggggaaaa
1440ggtcacgatg gcctctacca agggttgagt acggcaacca aagatacgta
cgatgcactg 1500catatgcagg ccctgcctcc cagataa 152746508PRTArtificial
SequenceSynthetic CD70 CAR amino acid sequence (CD70B scFV with
41BB) 46Met Ala Leu Pro Val Thr Ala Leu Leu Leu Pro Leu Ala Leu Leu
Leu1 5 10 15His Ala Ala Arg Pro Gln Val Gln Leu Val Gln Ser Gly Ala
Glu Val 20 25 30Lys Lys Pro Gly Ala Ser Val Lys Val Ser Cys Lys Ala
Ser Gly Tyr 35 40 45Thr Phe Thr Asn Tyr Gly Met Asn Trp Val Arg Gln
Ala Pro Gly Gln 50 55 60Gly Leu Lys Trp Met Gly Trp Ile Asn Thr Tyr
Thr Gly Glu Pro Thr65 70 75 80Tyr Ala Asp Ala Phe Lys Gly Arg Val
Thr Met Thr Arg Asp Thr Ser 85 90 95Ile Ser Thr Ala Tyr Met Glu Leu
Ser Arg Leu Arg Ser Asp Asp Thr 100 105 110Ala Val Tyr Tyr Cys Ala
Arg Asp Tyr Gly Asp Tyr Gly Met Asp Tyr 115 120 125Trp Gly Gln Gly
Thr Thr Val Thr Val Ser Ser Gly Gly Gly Gly Ser 130 135 140Gly Gly
Gly Gly Ser Gly Gly Gly Gly Ser Gly Asp Ile Val Met Thr145 150 155
160Gln Ser Pro Asp Ser Leu Ala Val Ser Leu Gly Glu Arg Ala Thr Ile
165 170 175Asn Cys Arg Ala Ser Lys Ser Val Ser Thr Ser Gly Tyr Ser
Phe Met 180 185 190His Trp Tyr Gln Gln Lys Pro Gly Gln Pro Pro Lys
Leu Leu Ile Tyr 195 200 205Leu Ala Ser Asn Leu Glu Ser Gly Val Pro
Asp Arg Phe Ser Gly Ser 210 215 220Gly Ser Gly Thr Asp Phe Thr Leu
Thr Ile Ser Ser Leu Gln Ala Glu225 230 235 240Asp Val Ala Val Tyr
Tyr Cys Gln His Ser Arg Glu Val Pro Trp Thr 245 250 255Phe Gly Gln
Gly Thr Lys Val Glu Ile Lys Ser Ala Ala Ala Phe Val 260 265 270Pro
Val Phe Leu Pro Ala Lys Pro Thr Thr Thr Pro Ala Pro Arg Pro 275 280
285Pro Thr Pro Ala Pro Thr Ile Ala Ser Gln Pro Leu Ser Leu Arg Pro
290 295 300Glu Ala Cys Arg Pro Ala Ala Gly Gly Ala Val His Thr Arg
Gly Leu305 310 315 320Asp Phe Ala Cys Asp Ile Tyr Ile Trp Ala Pro
Leu Ala Gly Thr Cys 325 330 335Gly Val Leu Leu Leu Ser Leu Val Ile
Thr Leu Tyr Cys Asn His Arg 340 345 350Asn Arg Lys Arg Gly Arg Lys
Lys Leu Leu Tyr Ile Phe Lys Gln Pro 355 360 365Phe Met Arg Pro Val
Gln Thr Thr Gln Glu Glu Asp Gly Cys Ser Cys 370 375 380Arg Phe Pro
Glu Glu Glu Glu Gly Gly Cys Glu Leu Arg Val Lys Phe385 390 395
400Ser Arg Ser Ala Asp Ala Pro Ala Tyr Gln Gln Gly Gln Asn Gln Leu
405 410 415Tyr Asn Glu Leu Asn Leu Gly Arg Arg Glu Glu Tyr Asp Val
Leu Asp 420 425 430Lys Arg Arg Gly Arg Asp Pro Glu Met Gly Gly Lys
Pro Arg Arg Lys 435 440 445Asn Pro Gln Glu Gly Leu Tyr Asn Glu Leu
Gln Lys Asp Lys Met Ala 450 455 460Glu Ala Tyr Ser Glu Ile Gly Met
Lys Gly Glu Arg Arg Arg Gly Lys465 470 475 480Gly His Asp Gly Leu
Tyr Gln Gly Leu Ser Thr Ala Thr Lys Asp Thr 485 490 495Tyr Asp Ala
Leu His Met Gln Ala Leu Pro Pro Arg 500 50547735DNAArtificial
SequenceSynthetic CD70A scFv nucleotide sequence 47gatatagtta
tgacccaatc acccgatagt cttgcggtaa gcctggggga gcgagcaaca 60ataaactgtc
gggcatcaaa atccgtcagt acaagcgggt attcattcat gcactggtat
120caacagaaac ccggtcagcc acccaagctc ctgatttatc ttgcgtctaa
tcttgagtcc 180ggcgtcccag accggttttc cggctccggg agcggcacgg
attttactct tactatttct 240agccttcagg ccgaagatgt ggcggtatac
tactgccagc attcaaggga agttccttgg 300acgttcggtc agggcacgaa
agtggaaatt aaaggcgggg ggggatccgg cgggggaggg 360tctggaggag
gtggcagtgg tcaggtccaa ctggtgcagt ccggggcaga ggtaaaaaaa
420cccggcgcgt ctgttaaggt ttcatgcaag gccagtggat atactttcac
caattacgga 480atgaactggg tgaggcaggc ccctggtcaa ggcctgaaat
ggatgggatg gataaacacg 540tacaccggtg aacctaccta tgccgatgcc
tttaagggtc gggttacgat gacgagagac 600acctccatat caacagccta
catggagctc agcagattga ggagtgacga tacggcagtc 660tattactgtg
caagagacta cggcgattat ggcatggatt actggggcca gggcactaca
720gtaaccgttt ccagc 73548245PRTArtificial SequenceSynthetic CD70A
scFv amino acid sequence 48Asp Ile Val Met Thr Gln Ser Pro Asp Ser
Leu Ala Val Ser Leu Gly1 5 10 15Glu Arg Ala Thr Ile Asn Cys Arg Ala
Ser Lys Ser Val Ser Thr Ser 20 25 30Gly Tyr Ser Phe Met His Trp Tyr
Gln Gln Lys Pro Gly Gln Pro Pro 35 40 45Lys Leu Leu Ile Tyr Leu Ala
Ser Asn Leu Glu Ser Gly Val Pro Asp 50 55 60Arg Phe Ser Gly Ser Gly
Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser65 70 75 80Ser Leu Gln Ala
Glu Asp Val Ala Val Tyr Tyr Cys Gln His Ser Arg 85 90 95Glu Val Pro
Trp Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Gly 100 105 110Gly
Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly Gln 115 120
125Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala Ser
130 135 140Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Asn
Tyr Gly145 150 155 160Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly
Leu Lys Trp Met Gly 165 170 175Trp Ile Asn Thr Tyr Thr Gly Glu Pro
Thr Tyr Ala Asp Ala Phe Lys 180 185 190Gly Arg Val Thr Met Thr Arg
Asp Thr Ser Ile Ser Thr Ala Tyr Met 195 200 205Glu Leu Ser Arg Leu
Arg Ser Asp Asp Thr Ala Val Tyr Tyr Cys Ala 210 215 220Arg Asp Tyr
Gly Asp Tyr Gly Met Asp Tyr Trp Gly Gln Gly Thr Thr225 230 235
240Val Thr Val Ser Ser 24549735DNAArtificial SequenceSynthetic
CD70B scFv nucleotide sequence 49caggtccagt tggtgcaaag cggggcggag
gtgaaaaaac ccggcgcttc cgtgaaggtg 60tcctgtaagg cgtccggtta tacgttcacg
aactacggga tgaattgggt tcgccaagcg 120ccggggcagg gactgaaatg
gatggggtgg ataaatacct acaccggcga acctacatac 180gccgacgctt
ttaaagggcg agtcactatg acgcgcgata ccagcatatc caccgcatac
240atggagctgt cccgactccg gtcagacgac acggctgtct actattgtgc
tcgggactat 300ggcgattatg gcatggacta ctggggtcag ggtacgactg
taacagttag tagtggtgga 360ggcggcagtg gcgggggggg aagcggagga
gggggttctg gtgacatagt tatgacccaa 420tccccagata gtttggcggt
ttctctgggc gagagggcaa cgattaattg tcgcgcatca 480aagagcgttt
caacgagcgg atattctttt atgcattggt accagcaaaa acccggacaa
540ccgccgaagc tgctgatcta cttggcttca aatcttgagt ctggggtgcc
ggaccgattt 600tctggtagtg gaagcggaac tgactttacg ctcacgatca
gttcactgca ggctgaggat 660gtagcggtct attattgcca gcacagtaga
gaagtcccct ggaccttcgg tcaaggcacg 720aaagtagaaa ttaaa
73550245PRTArtificial SequenceSynthetic CD70B scFv amino acid
sequence 50Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro
Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe
Thr Asn Tyr 20 25 30Gly Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly
Leu Lys Trp Met 35 40 45Gly Trp Ile Asn Thr Tyr Thr Gly Glu Pro Thr
Tyr Ala Asp Ala Phe 50 55 60Lys Gly Arg Val Thr Met Thr Arg Asp Thr
Ser Ile Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Arg Leu Arg Ser
Asp Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Tyr Gly Asp Tyr
Gly Met Asp Tyr Trp Gly Gln Gly Thr 100 105 110Thr Val Thr Val Ser
Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser 115 120 125Gly Gly Gly
Gly Ser Gly Asp Ile Val Met Thr Gln Ser Pro Asp Ser 130 135 140Leu
Ala Val Ser Leu Gly Glu Arg Ala Thr Ile Asn Cys Arg Ala Ser145 150
155 160Lys Ser Val Ser Thr Ser Gly Tyr Ser Phe Met His Trp Tyr Gln
Gln 165 170 175Lys Pro Gly Gln Pro Pro Lys Leu Leu Ile Tyr Leu Ala
Ser Asn Leu 180 185 190Glu Ser Gly Val Pro Asp Arg Phe Ser Gly Ser
Gly Ser Gly Thr Asp 195 200 205Phe Thr Leu Thr Ile Ser Ser Leu Gln
Ala Glu Asp Val Ala Val Tyr 210 215 220Tyr Cys Gln His Ser Arg Glu
Val Pro Trp Thr Phe Gly Gln Gly Thr225 230 235 240Lys Val Glu Ile
Lys 24551118PRTArtificial SequenceSynthetic CD70 VH 51Gln Val Gln
Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val
Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Asn Tyr 20 25 30Gly
Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Lys Trp Met 35 40
45Gly Trp Ile Asn Thr Tyr Thr Gly Glu Pro Thr Tyr Ala Asp Ala Phe
50 55 60Lys Gly Arg Val Thr Met Thr Arg Asp Thr Ser Ile Ser Thr Ala
Tyr65 70 75 80Met Glu Leu Ser Arg Leu Arg Ser Asp Asp Thr Ala Val
Tyr Tyr Cys 85 90 95Ala Arg Asp Tyr Gly Asp Tyr Gly Met Asp Tyr Trp
Gly Gln Gly Thr 100 105 110Thr Val Thr Val Ser Ser
11552111PRTArtificial SequenceSynthetic CD70 VL 52Asp Ile Val Met
Thr Gln Ser Pro Asp Ser Leu Ala Val Ser Leu Gly1 5 10 15Glu Arg Ala
Thr Ile Asn Cys Arg Ala Ser Lys Ser Val Ser Thr Ser 20 25 30Gly Tyr
Ser Phe Met His Trp Tyr Gln Gln Lys Pro Gly Gln Pro Pro 35 40 45Lys
Leu Leu Ile Tyr Leu Ala Ser Asn Leu Glu Ser Gly Val Pro Asp 50 55
60Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser65
70 75 80Ser Leu Gln Ala Glu Asp Val Ala Val Tyr Tyr Cys Gln His Ser
Arg 85 90 95Glu Val Pro Trp Thr Phe Gly Gln Gly Thr Lys Val Glu Ile
Lys 100 105 1105316PRTArtificial SequenceSynthetic Linker 53Gly Gly
Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly1 5 10
15544688DNAArtificial SequenceSynthetic BCMA rAAV 54cctgcaggca
gctgcgcgct cgctcgctca ctgaggccgc ccgggcgtcg ggcgaccttt 60ggtcgcccgg
cctcagtgag cgagcgagcg cgcagagagg gagtggccaa ctccatcact
120aggggttcct gcggccgcac gcgtgagatg taaggagctg ctgtgacttg
ctcaaggcct 180tatatcgagt aaacggtagt gctggggctt agacgcaggt
gttctgattt atagttcaaa 240acctctatca atgagagagc aatctcctgg
taatgtgata gatttcccaa cttaatgcca 300acataccata aacctcccat
tctgctaatg cccagcctaa gttggggaga ccactccaga 360ttccaagatg
tacagtttgc tttgctgggc ctttttccca tgcctgcctt tactctgcca
420gagttatatt gctggggttt tgaagaagat cctattaaat aaaagaataa
gcagtattat 480taagtagccc tgcatttcag gtttccttga gtggcaggcc
aggcctggcc gtgaacgttc
540actgaaatca tggcctcttg gccaagattg atagcttgtg cctgtccctg
agtcccagtc 600catcacgagc agctggtttc taagatgcta tttcccgtat
aaagcatgag accgtgactt 660gccagcccca cagagccccg cccttgtcca
tcactggcat ctggactcca gcctgggttg 720gggcaaagag ggaaatgaga
tcatgtccta accctgatcc tcttgtccca cagatatcca 780gaaccctgac
cctgccgtgt accagctgag agactctaaa tccagtgaca agtctgtctg
840cctattcacc gattttgatt ctcaaacaaa tgtgtcacaa agtaaggatt
ctgatgtgta 900tatcacagac aaaactgtgc tagacatgag gtctatggac
ttcaggctcc ggtgcccgtc 960agtgggcaga gcgcacatcg cccacagtcc
ccgagaagtt ggggggaggg gtcggcaatt 1020gaaccggtgc ctagagaagg
tggcgcgggg taaactggga aagtgatgtc gtgtactggc 1080tccgcctttt
tcccgagggt gggggagaac cgtatataag tgcagtagtc gccgtgaacg
1140ttctttttcg caacgggttt gccgccagaa cacaggtaag tgccgtgtgt
ggttcccgcg 1200ggcctggcct ctttacgggt tatggccctt gcgtgccttg
aattacttcc actggctgca 1260gtacgtgatt cttgatcccg agcttcgggt
tggaagtggg tgggagagtt cgaggccttg 1320cgcttaagga gccccttcgc
ctcgtgcttg agttgaggcc tggcctgggc gctggggccg 1380ccgcgtgcga
atctggtggc accttcgcgc ctgtctcgct gctttcgata agtctctagc
1440catttaaaat ttttgatgac ctgctgcgac gctttttttc tggcaagata
gtcttgtaaa 1500tgcgggccaa gatctgcaca ctggtatttc ggtttttggg
gccgcgggcg gcgacggggc 1560ccgtgcgtcc cagcgcacat gttcggcgag
gcggggcctg cgagcgcggc caccgagaat 1620cggacggggg tagtctcaag
ctggccggcc tgctctggtg cctggcctcg cgccgccgtg 1680tatcgccccg
ccctgggcgg caaggctggc ccggtcggca ccagttgcgt gagcggaaag
1740atggccgctt cccggccctg ctgcagggag ctcaaaatgg aggacgcggc
gctcgggaga 1800gcgggcgggt gagtcaccca cacaaaggaa aagggccttt
ccgtcctcag ccgtcgcttc 1860atgtgactcc acggagtacc gggcgccgtc
caggcacctc gattagttct cgagcttttg 1920gagtacgtcg tctttaggtt
ggggggaggg gttttatgcg atggagtttc cccacactga 1980gtgggtggag
actgaagtta ggccagcttg gcacttgatg taattctcct tggaatttgc
2040cctttttgag tttggatctt ggttcattct caagcctcag acagtggttc
aaagtttttt 2100tcttccattt caggtgtcgt gaccaccatg gcgcttccgg
tgacagcact gctcctcccc 2160ttggcgctgt tgctccacgc agcaaggccg
caggtgcagc tggtgcagag cggagccgag 2220ctcaagaagc ccggagcctc
cgtgaaggtg agctgcaagg ccagcggcaa caccctgacc 2280aactacgtga
tccactgggt gagacaagcc cccggccaaa ggctggagtg gatgggctac
2340atcctgccct acaacgacct gaccaagtac agccagaagt tccagggcag
ggtgaccatc 2400accagggata agagcgcctc caccgcctat atggagctga
gcagcctgag gagcgaggac 2460accgctgtgt actactgtac aaggtgggac
tgggacggct tctttgaccc ctggggccag 2520ggcacaacag tgaccgtcag
cagcggcggc ggaggcagcg gcggcggcgg cagcggcgga 2580ggcggaagcg
aaatcgtgat gacccagagc cccgccacac tgagcgtgag ccctggcgag
2640agggccagca tctcctgcag ggctagccaa agcctggtgc acagcaacgg
caacacccac 2700ctgcactggt accagcagag acccggacag gctcccaggc
tgctgatcta cagcgtgagc 2760aacaggttct ccgaggtgcc tgccaggttt
agcggcagcg gaagcggcac cgactttacc 2820ctgaccatca gcagcgtgga
gtccgaggac ttcgccgtgt attactgcag ccagaccagc 2880cacatccctt
acaccttcgg cggcggcacc aagctggaga tcaaaagtgc tgctgccttt
2940gtcccggtat ttctcccagc caaaccgacc acgactcccg ccccgcgccc
tccgacaccc 3000gctcccacca tcgcctctca acctcttagt cttcgccccg
aggcatgccg acccgccgcc 3060gggggtgctg ttcatacgag gggcttggac
ttcgcttgtg atatttacat ttgggctccg 3120ttggcgggta cgtgcggcgt
ccttttgttg tcactcgtta ttactttgta ttgtaatcac 3180aggaatcgca
aacggggcag aaagaaactc ctgtatatat tcaaacaacc atttatgaga
3240ccagtacaaa ctactcaaga ggaagatggc tgtagctgcc gatttccaga
agaagaagaa 3300ggaggatgtg aactgcgagt gaagttttcc cgaagcgcag
acgctccggc atatcagcaa 3360ggacagaatc agctgtataa cgaactgaat
ttgggacgcc gcgaggagta tgacgtgctt 3420gataaacgcc gggggagaga
cccggaaatg gggggtaaac cccgaagaaa gaatccccaa 3480gaaggactct
acaatgaact ccagaaggat aagatggcgg aggcctactc agaaataggt
3540atgaagggcg aacgacgacg gggaaaaggt cacgatggcc tctaccaagg
gttgagtacg 3600gcaaccaaag atacgtacga tgcactgcat atgcaggccc
tgcctcccag ataataataa 3660aatcgctatc catcgaagat ggatgtgtgt
tggttttttg tgtgtggagc aacaaatctg 3720actttgcatg tgcaaacgcc
ttcaacaaca gcattattcc agaagacacc ttcttcccca 3780gcccaggtaa
gggcagcttt ggtgccttcg caggctgttt ccttgcttca ggaatggcca
3840ggttctgccc agagctctgg tcaatgatgt ctaaaactcc tctgattggt
ggtctcggcc 3900ttatccattg ccaccaaaac cctcttttta ctaagaaaca
gtgagccttg ttctggcagt 3960ccagagaatg acacgggaaa aaagcagatg
aagagaaggt ggcaggagag ggcacgtggc 4020ccagcctcag tctctccaac
tgagttcctg cctgcctgcc tttgctcaga ctgtttgccc 4080cttactgctc
ttctaggcct cattctaagc cccttctcca agttgcctct ccttatttct
4140ccctgtctgc caaaaaatct ttcccagctc actaagtcag tctcacgcag
tcactcatta 4200acccaccaat cactgattgt gccggcacat gaatgcacca
ggtgttgaag tggaggaatt 4260aaaaagtcag atgaggggtg tgcccagagg
aagcaccatt ctagttgggg gagcccatct 4320gtcagctggg aaaagtccaa
ataacttcag attggaatgt gttttaactc agggttgaga 4380aaacagctac
cttcaggaca aaagtcaggg aagggctctc tgaagaaatg ctacttgaag
4440ataccagccc taccaagggc agggagagga ccctatagag gcctgggaca
ggagctcaat 4500gagaaaggta accacgtgcg gaccgaggct gcagcgtcgt
cctccctagg aacccctagt 4560gatggagttg gccactccct ctctgcgcgc
tcgctcgctc actgaggccg ggcgaccaaa 4620ggtcgcccga cgcccgggct
ttgcccgggc ggcctcagtg agcgagcgag cgcgcagctg 4680cctgcagg
4688554364DNAArtificial SequenceSynthetic BCMA RHA to LHA
55gagatgtaag gagctgctgt gacttgctca aggccttata tcgagtaaac ggtagtgctg
60gggcttagac gcaggtgttc tgatttatag ttcaaaacct ctatcaatga gagagcaatc
120tcctggtaat gtgatagatt tcccaactta atgccaacat accataaacc
tcccattctg 180ctaatgccca gcctaagttg gggagaccac tccagattcc
aagatgtaca gtttgctttg 240ctgggccttt ttcccatgcc tgcctttact
ctgccagagt tatattgctg gggttttgaa 300gaagatccta ttaaataaaa
gaataagcag tattattaag tagccctgca tttcaggttt 360ccttgagtgg
caggccaggc ctggccgtga acgttcactg aaatcatggc ctcttggcca
420agattgatag cttgtgcctg tccctgagtc ccagtccatc acgagcagct
ggtttctaag 480atgctatttc ccgtataaag catgagaccg tgacttgcca
gccccacaga gccccgccct 540tgtccatcac tggcatctgg actccagcct
gggttggggc aaagagggaa atgagatcat 600gtcctaaccc tgatcctctt
gtcccacaga tatccagaac cctgaccctg ccgtgtacca 660gctgagagac
tctaaatcca gtgacaagtc tgtctgccta ttcaccgatt ttgattctca
720aacaaatgtg tcacaaagta aggattctga tgtgtatatc acagacaaaa
ctgtgctaga 780catgaggtct atggacttca ggctccggtg cccgtcagtg
ggcagagcgc acatcgccca 840cagtccccga gaagttgggg ggaggggtcg
gcaattgaac cggtgcctag agaaggtggc 900gcggggtaaa ctgggaaagt
gatgtcgtgt actggctccg cctttttccc gagggtgggg 960gagaaccgta
tataagtgca gtagtcgccg tgaacgttct ttttcgcaac gggtttgccg
1020ccagaacaca ggtaagtgcc gtgtgtggtt cccgcgggcc tggcctcttt
acgggttatg 1080gcccttgcgt gccttgaatt acttccactg gctgcagtac
gtgattcttg atcccgagct 1140tcgggttgga agtgggtggg agagttcgag
gccttgcgct taaggagccc cttcgcctcg 1200tgcttgagtt gaggcctggc
ctgggcgctg gggccgccgc gtgcgaatct ggtggcacct 1260tcgcgcctgt
ctcgctgctt tcgataagtc tctagccatt taaaattttt gatgacctgc
1320tgcgacgctt tttttctggc aagatagtct tgtaaatgcg ggccaagatc
tgcacactgg 1380tatttcggtt tttggggccg cgggcggcga cggggcccgt
gcgtcccagc gcacatgttc 1440ggcgaggcgg ggcctgcgag cgcggccacc
gagaatcgga cgggggtagt ctcaagctgg 1500ccggcctgct ctggtgcctg
gcctcgcgcc gccgtgtatc gccccgccct gggcggcaag 1560gctggcccgg
tcggcaccag ttgcgtgagc ggaaagatgg ccgcttcccg gccctgctgc
1620agggagctca aaatggagga cgcggcgctc gggagagcgg gcgggtgagt
cacccacaca 1680aaggaaaagg gcctttccgt cctcagccgt cgcttcatgt
gactccacgg agtaccgggc 1740gccgtccagg cacctcgatt agttctcgag
cttttggagt acgtcgtctt taggttgggg 1800ggaggggttt tatgcgatgg
agtttcccca cactgagtgg gtggagactg aagttaggcc 1860agcttggcac
ttgatgtaat tctccttgga atttgccctt tttgagtttg gatcttggtt
1920cattctcaag cctcagacag tggttcaaag tttttttctt ccatttcagg
tgtcgtgacc 1980accatggcgc ttccggtgac agcactgctc ctccccttgg
cgctgttgct ccacgcagca 2040aggccgcagg tgcagctggt gcagagcgga
gccgagctca agaagcccgg agcctccgtg 2100aaggtgagct gcaaggccag
cggcaacacc ctgaccaact acgtgatcca ctgggtgaga 2160caagcccccg
gccaaaggct ggagtggatg ggctacatcc tgccctacaa cgacctgacc
2220aagtacagcc agaagttcca gggcagggtg accatcacca gggataagag
cgcctccacc 2280gcctatatgg agctgagcag cctgaggagc gaggacaccg
ctgtgtacta ctgtacaagg 2340tgggactggg acggcttctt tgacccctgg
ggccagggca caacagtgac cgtcagcagc 2400ggcggcggag gcagcggcgg
cggcggcagc ggcggaggcg gaagcgaaat cgtgatgacc 2460cagagccccg
ccacactgag cgtgagccct ggcgagaggg ccagcatctc ctgcagggct
2520agccaaagcc tggtgcacag caacggcaac acccacctgc actggtacca
gcagagaccc 2580ggacaggctc ccaggctgct gatctacagc gtgagcaaca
ggttctccga ggtgcctgcc 2640aggtttagcg gcagcggaag cggcaccgac
tttaccctga ccatcagcag cgtggagtcc 2700gaggacttcg ccgtgtatta
ctgcagccag accagccaca tcccttacac cttcggcggc 2760ggcaccaagc
tggagatcaa aagtgctgct gcctttgtcc cggtatttct cccagccaaa
2820ccgaccacga ctcccgcccc gcgccctccg acacccgctc ccaccatcgc
ctctcaacct 2880cttagtcttc gccccgaggc atgccgaccc gccgccgggg
gtgctgttca tacgaggggc 2940ttggacttcg cttgtgatat ttacatttgg
gctccgttgg cgggtacgtg cggcgtcctt 3000ttgttgtcac tcgttattac
tttgtattgt aatcacagga atcgcaaacg gggcagaaag 3060aaactcctgt
atatattcaa acaaccattt atgagaccag tacaaactac tcaagaggaa
3120gatggctgta gctgccgatt tccagaagaa gaagaaggag gatgtgaact
gcgagtgaag 3180ttttcccgaa gcgcagacgc tccggcatat cagcaaggac
agaatcagct gtataacgaa 3240ctgaatttgg gacgccgcga ggagtatgac
gtgcttgata aacgccgggg gagagacccg 3300gaaatggggg gtaaaccccg
aagaaagaat ccccaagaag gactctacaa tgaactccag 3360aaggataaga
tggcggaggc ctactcagaa ataggtatga agggcgaacg acgacgggga
3420aaaggtcacg atggcctcta ccaagggttg agtacggcaa ccaaagatac
gtacgatgca 3480ctgcatatgc aggccctgcc tcccagataa taataaaatc
gctatccatc gaagatggat 3540gtgtgttggt tttttgtgtg tggagcaaca
aatctgactt tgcatgtgca aacgccttca 3600acaacagcat tattccagaa
gacaccttct tccccagccc aggtaagggc agctttggtg 3660ccttcgcagg
ctgtttcctt gcttcaggaa tggccaggtt ctgcccagag ctctggtcaa
3720tgatgtctaa aactcctctg attggtggtc tcggccttat ccattgccac
caaaaccctc 3780tttttactaa gaaacagtga gccttgttct ggcagtccag
agaatgacac gggaaaaaag 3840cagatgaaga gaaggtggca ggagagggca
cgtggcccag cctcagtctc tccaactgag 3900ttcctgcctg cctgcctttg
ctcagactgt ttgcccctta ctgctcttct aggcctcatt 3960ctaagcccct
tctccaagtt gcctctcctt atttctccct gtctgccaaa aaatctttcc
4020cagctcacta agtcagtctc acgcagtcac tcattaaccc accaatcact
gattgtgccg 4080gcacatgaat gcaccaggtg ttgaagtgga ggaattaaaa
agtcagatga ggggtgtgcc 4140cagaggaagc accattctag ttgggggagc
ccatctgtca gctgggaaaa gtccaaataa 4200cttcagattg gaatgtgttt
taactcaggg ttgagaaaac agctaccttc aggacaaaag 4260tcagggaagg
gctctctgaa gaaatgctac ttgaagatac cagccctacc aagggcaggg
4320agaggaccct atagaggcct gggacaggag ctcaatgaga aagg
4364561524DNAArtificial SequenceSynthetic BCMA CAR nucleotide
sequence 56atggcgcttc cggtgacagc actgctcctc cccttggcgc tgttgctcca
cgcagcaagg 60ccgcaggtgc agctggtgca gagcggagcc gagctcaaga agcccggagc
ctccgtgaag 120gtgagctgca aggccagcgg caacaccctg accaactacg
tgatccactg ggtgagacaa 180gcccccggcc aaaggctgga gtggatgggc
tacatcctgc cctacaacga cctgaccaag 240tacagccaga agttccaggg
cagggtgacc atcaccaggg ataagagcgc ctccaccgcc 300tatatggagc
tgagcagcct gaggagcgag gacaccgctg tgtactactg tacaaggtgg
360gactgggacg gcttctttga cccctggggc cagggcacaa cagtgaccgt
cagcagcggc 420ggcggaggca gcggcggcgg cggcagcggc ggaggcggaa
gcgaaatcgt gatgacccag 480agccccgcca cactgagcgt gagccctggc
gagagggcca gcatctcctg cagggctagc 540caaagcctgg tgcacagcaa
cggcaacacc cacctgcact ggtaccagca gagacccgga 600caggctccca
ggctgctgat ctacagcgtg agcaacaggt tctccgaggt gcctgccagg
660tttagcggca gcggaagcgg caccgacttt accctgacca tcagcagcgt
ggagtccgag 720gacttcgccg tgtattactg cagccagacc agccacatcc
cttacacctt cggcggcggc 780accaagctgg agatcaaaag tgctgctgcc
tttgtcccgg tatttctccc agccaaaccg 840accacgactc ccgccccgcg
ccctccgaca cccgctccca ccatcgcctc tcaacctctt 900agtcttcgcc
ccgaggcatg ccgacccgcc gccgggggtg ctgttcatac gaggggcttg
960gacttcgctt gtgatattta catttgggct ccgttggcgg gtacgtgcgg
cgtccttttg 1020ttgtcactcg ttattacttt gtattgtaat cacaggaatc
gcaaacgggg cagaaagaaa 1080ctcctgtata tattcaaaca accatttatg
agaccagtac aaactactca agaggaagat 1140ggctgtagct gccgatttcc
agaagaagaa gaaggaggat gtgaactgcg agtgaagttt 1200tcccgaagcg
cagacgctcc ggcatatcag caaggacaga atcagctgta taacgaactg
1260aatttgggac gccgcgagga gtatgacgtg cttgataaac gccgggggag
agacccggaa 1320atggggggta aaccccgaag aaagaatccc caagaaggac
tctacaatga actccagaag 1380gataagatgg cggaggccta ctcagaaata
ggtatgaagg gcgaacgacg acggggaaaa 1440ggtcacgatg gcctctacca
agggttgagt acggcaacca aagatacgta cgatgcactg 1500catatgcagg
ccctgcctcc caga 152457508PRTArtificial SequenceSynthetic BCMA CAR
amino acid sequence 57Met Ala Leu Pro Val Thr Ala Leu Leu Leu Pro
Leu Ala Leu Leu Leu1 5 10 15His Ala Ala Arg Pro Gln Val Gln Leu Val
Gln Ser Gly Ala Glu Leu 20 25 30Lys Lys Pro Gly Ala Ser Val Lys Val
Ser Cys Lys Ala Ser Gly Asn 35 40 45Thr Leu Thr Asn Tyr Val Ile His
Trp Val Arg Gln Ala Pro Gly Gln 50 55 60Arg Leu Glu Trp Met Gly Tyr
Ile Leu Pro Tyr Asn Asp Leu Thr Lys65 70 75 80Tyr Ser Gln Lys Phe
Gln Gly Arg Val Thr Ile Thr Arg Asp Lys Ser 85 90 95Ala Ser Thr Ala
Tyr Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr 100 105 110Ala Val
Tyr Tyr Cys Thr Arg Trp Asp Trp Asp Gly Phe Phe Asp Pro 115 120
125Trp Gly Gln Gly Thr Thr Val Thr Val Ser Ser Gly Gly Gly Gly Ser
130 135 140Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Glu Ile Val Met
Thr Gln145 150 155 160Ser Pro Ala Thr Leu Ser Val Ser Pro Gly Glu
Arg Ala Ser Ile Ser 165 170 175Cys Arg Ala Ser Gln Ser Leu Val His
Ser Asn Gly Asn Thr His Leu 180 185 190His Trp Tyr Gln Gln Arg Pro
Gly Gln Ala Pro Arg Leu Leu Ile Tyr 195 200 205Ser Val Ser Asn Arg
Phe Ser Glu Val Pro Ala Arg Phe Ser Gly Ser 210 215 220Gly Ser Gly
Thr Asp Phe Thr Leu Thr Ile Ser Ser Val Glu Ser Glu225 230 235
240Asp Phe Ala Val Tyr Tyr Cys Ser Gln Thr Ser His Ile Pro Tyr Thr
245 250 255Phe Gly Gly Gly Thr Lys Leu Glu Ile Lys Ser Ala Ala Ala
Phe Val 260 265 270Pro Val Phe Leu Pro Ala Lys Pro Thr Thr Thr Pro
Ala Pro Arg Pro 275 280 285Pro Thr Pro Ala Pro Thr Ile Ala Ser Gln
Pro Leu Ser Leu Arg Pro 290 295 300Glu Ala Cys Arg Pro Ala Ala Gly
Gly Ala Val His Thr Arg Gly Leu305 310 315 320Asp Phe Ala Cys Asp
Ile Tyr Ile Trp Ala Pro Leu Ala Gly Thr Cys 325 330 335Gly Val Leu
Leu Leu Ser Leu Val Ile Thr Leu Tyr Cys Asn His Arg 340 345 350Asn
Arg Lys Arg Gly Arg Lys Lys Leu Leu Tyr Ile Phe Lys Gln Pro 355 360
365Phe Met Arg Pro Val Gln Thr Thr Gln Glu Glu Asp Gly Cys Ser Cys
370 375 380Arg Phe Pro Glu Glu Glu Glu Gly Gly Cys Glu Leu Arg Val
Lys Phe385 390 395 400Ser Arg Ser Ala Asp Ala Pro Ala Tyr Gln Gln
Gly Gln Asn Gln Leu 405 410 415Tyr Asn Glu Leu Asn Leu Gly Arg Arg
Glu Glu Tyr Asp Val Leu Asp 420 425 430Lys Arg Arg Gly Arg Asp Pro
Glu Met Gly Gly Lys Pro Arg Arg Lys 435 440 445Asn Pro Gln Glu Gly
Leu Tyr Asn Glu Leu Gln Lys Asp Lys Met Ala 450 455 460Glu Ala Tyr
Ser Glu Ile Gly Met Lys Gly Glu Arg Arg Arg Gly Lys465 470 475
480Gly His Asp Gly Leu Tyr Gln Gly Leu Ser Thr Ala Thr Lys Asp Thr
485 490 495Tyr Asp Ala Leu His Met Gln Ala Leu Pro Pro Arg 500
50558735DNAArtificial SequenceSynthetic BCMA scFv nucleotide
sequence 58caggtgcagc tggtgcagag cggagccgag ctcaagaagc ccggagcctc
cgtgaaggtg 60agctgcaagg ccagcggcaa caccctgacc aactacgtga tccactgggt
gagacaagcc 120cccggccaaa ggctggagtg gatgggctac atcctgccct
acaacgacct gaccaagtac 180agccagaagt tccagggcag ggtgaccatc
accagggata agagcgcctc caccgcctat 240atggagctga gcagcctgag
gagcgaggac accgctgtgt actactgtac aaggtgggac 300tgggacggct
tctttgaccc ctggggccag ggcacaacag tgaccgtcag cagcggcggc
360ggaggcagcg gcggcggcgg cagcggcgga ggcggaagcg aaatcgtgat
gacccagagc 420cccgccacac tgagcgtgag ccctggcgag agggccagca
tctcctgcag ggctagccaa 480agcctggtgc acagcaacgg caacacccac
ctgcactggt accagcagag acccggacag 540gctcccaggc tgctgatcta
cagcgtgagc aacaggttct ccgaggtgcc tgccaggttt 600agcggcagcg
gaagcggcac cgactttacc ctgaccatca gcagcgtgga gtccgaggac
660ttcgccgtgt attactgcag ccagaccagc cacatccctt acaccttcgg
cggcggcacc 720aagctggaga tcaaa 73559245PRTArtificial
SequenceSynthetic BCMA scFv amino acid sequence 59Gln Val Gln Leu
Val Gln Ser Gly Ala Glu Leu Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys
Val Ser Cys Lys Ala Ser Gly Asn Thr Leu Thr Asn Tyr 20 25 30Val Ile
His Trp Val Arg Gln Ala Pro Gly Gln Arg Leu Glu Trp Met 35 40 45Gly
Tyr Ile Leu Pro Tyr Asn Asp Leu Thr Lys Tyr Ser Gln Lys Phe 50 55
60Gln Gly Arg Val Thr Ile Thr Arg Asp Lys Ser Ala Ser Thr Ala Tyr65
70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr
Cys 85 90 95Thr Arg Trp Asp Trp Asp Gly Phe Phe Asp Pro Trp Gly Gln
Gly Thr 100 105 110Thr Val Thr Val Ser Ser Gly Gly Gly Gly Ser Gly
Gly Gly Gly Ser 115 120
125Gly Gly Gly Gly Ser Glu Ile Val Met Thr Gln Ser Pro Ala Thr Leu
130 135 140Ser Val Ser Pro Gly Glu Arg Ala Ser Ile Ser Cys Arg Ala
Ser Gln145 150 155 160Ser Leu Val His Ser Asn Gly Asn Thr His Leu
His Trp Tyr Gln Gln 165 170 175Arg Pro Gly Gln Ala Pro Arg Leu Leu
Ile Tyr Ser Val Ser Asn Arg 180 185 190Phe Ser Glu Val Pro Ala Arg
Phe Ser Gly Ser Gly Ser Gly Thr Asp 195 200 205Phe Thr Leu Thr Ile
Ser Ser Val Glu Ser Glu Asp Phe Ala Val Tyr 210 215 220Tyr Cys Ser
Gln Thr Ser His Ile Pro Tyr Thr Phe Gly Gly Gly Thr225 230 235
240Lys Leu Glu Ile Lys 24560118PRTArtificial SequenceSynthetic BCMA
VH 60Gln Val Gln Leu Val Gln Ser Gly Ala Glu Leu Lys Lys Pro Gly
Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Asn Thr Leu Thr
Asn Tyr 20 25 30Val Ile His Trp Val Arg Gln Ala Pro Gly Gln Arg Leu
Glu Trp Met 35 40 45Gly Tyr Ile Leu Pro Tyr Asn Asp Leu Thr Lys Tyr
Ser Gln Lys Phe 50 55 60Gln Gly Arg Val Thr Ile Thr Arg Asp Lys Ser
Ala Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu
Asp Thr Ala Val Tyr Tyr Cys 85 90 95Thr Arg Trp Asp Trp Asp Gly Phe
Phe Asp Pro Trp Gly Gln Gly Thr 100 105 110Thr Val Thr Val Ser Ser
11561112PRTArtificial SequenceSynthetic BCMA VL 61Glu Ile Val Met
Thr Gln Ser Pro Ala Thr Leu Ser Val Ser Pro Gly1 5 10 15Glu Arg Ala
Ser Ile Ser Cys Arg Ala Ser Gln Ser Leu Val His Ser 20 25 30Asn Gly
Asn Thr His Leu His Trp Tyr Gln Gln Arg Pro Gly Gln Ala 35 40 45Pro
Arg Leu Leu Ile Tyr Ser Val Ser Asn Arg Phe Ser Glu Val Pro 50 55
60Ala Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile65
70 75 80Ser Ser Val Glu Ser Glu Asp Phe Ala Val Tyr Tyr Cys Ser Gln
Thr 85 90 95Ser His Ile Pro Tyr Thr Phe Gly Gly Gly Thr Lys Leu Glu
Ile Lys 100 105 1106215PRTArtificial SequenceSynthetic CD70 VL CDR1
(Kabat) 62Arg Ala Ser Lys Ser Val Ser Thr Ser Gly Tyr Ser Phe Met
His1 5 10 156311PRTArtificial SequenceSynthetic CD70 VL CDR1
(Chothia) 63Ser Lys Ser Val Ser Thr Ser Gly Tyr Ser Phe1 5
10647PRTArtificial SequenceSynthetic CD70 VL CDR2 (Kabat) 64Leu Ala
Ser Asn Leu Glu Ser1 5653PRTArtificial SequenceSynthetic CD70 VL
CDR2 (Chothia) 65Leu Ala Ser1669PRTArtificial SequenceSynthetic
CD70 VL CDR3 (Kabat) 66Gln His Ser Arg Glu Val Pro Trp Thr1
5676PRTArtificial SequenceSynthetic CD70 VL CDR3 (Chothia) 67Ser
Arg Glu Val Pro Trp1 5685PRTArtificial SequenceSynthetic CD70 VH
CDR1 (Kabat) 68Asn Tyr Gly Met Asn1 56910PRTArtificial
SequenceSynthetic CD70 VH CDR1 (Chothia) 69Gly Tyr Thr Phe Thr Asn
Tyr Gly Met Asn1 5 107017PRTArtificial SequenceSynthetic CD70 VH
CDR2 (Kabat) 70Trp Ile Asn Thr Tyr Thr Gly Glu Pro Thr Tyr Ala Asp
Ala Phe Lys1 5 10 15Gly716PRTArtificial SequenceSynthetic CD70 VH
CDR2 (Chothia) 71Asn Thr Tyr Thr Gly Glu1 5729PRTArtificial
SequenceSynthetic CD70 VH CDR3 (Kabat) 72Asp Tyr Gly Asp Tyr Gly
Met Asp Tyr1 57314PRTArtificial SequenceSynthetic CD70 VH CDR3
(Chothia) 73Cys Ala Arg Asp Tyr Gly Asp Tyr Gly Met Asp Tyr Trp
Gly1 5 107416PRTArtificial SequenceSynthetic BCMA VL CDR1 (Kabat)
74Arg Ala Ser Gln Ser Leu Val His Ser Asn Gly Asn Thr His Leu His1
5 10 157516PRTArtificial SequenceSynthetic BCMA VL CDR1 (Chothia)
75Arg Ala Ser Gln Ser Leu Val His Ser Asn Gly Asn Thr His Leu His1
5 10 15765PRTArtificial SequenceSynthetic BCMA VL CDR2 (Kabat)
76Ser Val Ser Asn Arg1 5775PRTArtificial SequenceSynthetic BCMA VL
CDR2 (Chothia) 77Ser Val Ser Asn Arg1 5789PRTArtificial
SequenceSynthetic BCMA VL CDR3 (Kabat) 78Ser Gln Thr Ser His Ile
Pro Tyr Thr1 5799PRTArtificial SequenceSynthetic BCMA VL CDR3
(Chothia) 79Ser Gln Thr Ser His Ile Pro Tyr Thr1 5805PRTArtificial
SequenceSynthetic BCMA VH CDR1 (Kabat) 80Asn Tyr Val Ile His1
5817PRTArtificial SequenceSynthetic BCMA VH CDR1 (Chothia) 81Gly
Asn Thr Leu Thr Asn Tyr1 58217PRTArtificial SequenceSynthetic BCMA
VH CDR2 (Kabat) 82Tyr Ile Leu Pro Tyr Asn Asp Leu Thr Lys Tyr Ser
Gln Lys Phe Gln1 5 10 15Gly836PRTArtificial SequenceSynthetic BCMA
VH CDR2 (Chothia) 83Leu Pro Tyr Asn Asp Leu1 5849PRTArtificial
SequenceSynthetic BCMA VH CDR3 (Kabat) 84Trp Asp Trp Asp Gly Phe
Phe Asp Pro1 5859PRTArtificial SequenceSynthetic BCMA VH CDR3
(Chothia) 85Trp Asp Trp Asp Gly Phe Phe Asp Pro1 58620DNAArtificial
SequenceSynthetic TRAC target sequence 86agagcaacag tgctgtggcc
20874691DNAArtificial SequenceSynthetic anti-CD33 CAR rAAV
87cctgcaggca gctgcgcgct cgctcgctca ctgaggccgc ccgggcgtcg ggcgaccttt
60ggtcgcccgg cctcagtgag cgagcgagcg cgcagagagg gagtggccaa ctccatcact
120aggggttcct gcggccgcac gcgtgagatg taaggagctg ctgtgacttg
ctcaaggcct 180tatatcgagt aaacggtagt gctggggctt agacgcaggt
gttctgattt atagttcaaa 240acctctatca atgagagagc aatctcctgg
taatgtgata gatttcccaa cttaatgcca 300acataccata aacctcccat
tctgctaatg cccagcctaa gttggggaga ccactccaga 360ttccaagatg
tacagtttgc tttgctgggc ctttttccca tgcctgcctt tactctgcca
420gagttatatt gctggggttt tgaagaagat cctattaaat aaaagaataa
gcagtattat 480taagtagccc tgcatttcag gtttccttga gtggcaggcc
aggcctggcc gtgaacgttc 540actgaaatca tggcctcttg gccaagattg
atagcttgtg cctgtccctg agtcccagtc 600catcacgagc agctggtttc
taagatgcta tttcccgtat aaagcatgag accgtgactt 660gccagcccca
cagagccccg cccttgtcca tcactggcat ctggactcca gcctgggttg
720gggcaaagag ggaaatgaga tcatgtccta accctgatcc tcttgtccca
cagatatcca 780gaaccctgac cctgccgtgt accagctgag agactctaaa
tccagtgaca agtctgtctg 840cctattcacc gattttgatt ctcaaacaaa
tgtgtcacaa agtaaggatt ctgatgtgta 900tatcacagac aaaactgtgc
tagacatgag gtctatggac ttcaggctcc ggtgcccgtc 960agtgggcaga
gcgcacatcg cccacagtcc ccgagaagtt ggggggaggg gtcggcaatt
1020gaaccggtgc ctagagaagg tggcgcgggg taaactggga aagtgatgtc
gtgtactggc 1080tccgcctttt tcccgagggt gggggagaac cgtatataag
tgcagtagtc gccgtgaacg 1140ttctttttcg caacgggttt gccgccagaa
cacaggtaag tgccgtgtgt ggttcccgcg 1200ggcctggcct ctttacgggt
tatggccctt gcgtgccttg aattacttcc actggctgca 1260gtacgtgatt
cttgatcccg agcttcgggt tggaagtggg tgggagagtt cgaggccttg
1320cgcttaagga gccccttcgc ctcgtgcttg agttgaggcc tggcctgggc
gctggggccg 1380ccgcgtgcga atctggtggc accttcgcgc ctgtctcgct
gctttcgata agtctctagc 1440catttaaaat ttttgatgac ctgctgcgac
gctttttttc tggcaagata gtcttgtaaa 1500tgcgggccaa gatctgcaca
ctggtatttc ggtttttggg gccgcgggcg gcgacggggc 1560ccgtgcgtcc
cagcgcacat gttcggcgag gcggggcctg cgagcgcggc caccgagaat
1620cggacggggg tagtctcaag ctggccggcc tgctctggtg cctggcctcg
cgccgccgtg 1680tatcgccccg ccctgggcgg caaggctggc ccggtcggca
ccagttgcgt gagcggaaag 1740atggccgctt cccggccctg ctgcagggag
ctcaaaatgg aggacgcggc gctcgggaga 1800gcgggcgggt gagtcaccca
cacaaaggaa aagggccttt ccgtcctcag ccgtcgcttc 1860atgtgactcc
acggagtacc gggcgccgtc caggcacctc gattagttct cgagcttttg
1920gagtacgtcg tctttaggtt ggggggaggg gttttatgcg atggagtttc
cccacactga 1980gtgggtggag actgaagtta ggccagcttg gcacttgatg
taattctcct tggaatttgc 2040cctttttgag tttggatctt ggttcattct
caagcctcag acagtggttc aaagtttttt 2100tcttccattt caggtgtcgt
gaccaccatg gcgcttccgg tgacagcact gctcctcccc 2160ttggcgctgt
tgctccacgc agcaaggccg gaaatcgtcc tcacacaatc cccggggagc
2220ctcgcagtca gtcctgggga acgagtcact atgagctgca aatccagtca
gagtgttttt 2280ttctcaagta gccagaagaa ctacctcgca tggtaccaac
aaataccggg gcaatctccc 2340cgcttgctta tatactgggc aagtacccgc
gaatccggcg taccggatcg attcacggga 2400tctgggtcag gtactgattt
cactttgact atcagctctg ttcagcctga agatttggca 2460atttactact
gtcaccaata cttgagtagc cgaactttcg gccagggcac gaagctcgaa
2520atcaagggcg gagggggagg ttctggtggg ggcggttctg gcggtggagg
aagccaagta 2580cagttgcaac agccaggggc ggaggtcgta aaacctgggg
cgtctgtcaa gatgagctgt 2640aaagcaagtg gatacacctt cacctcctac
tatatacatt ggattaagca aactccgggt 2700caggggctgg aatgggttgg
cgttatatac cccgggaacg atgatatatc atacaaccaa 2760aaatttcaag
gcaaggcgac tctgactgcc gataagagta gcacaacagc ttacatgcag
2820ctttcttccc tgaccagcga agattcagca gtttactact gcgctcggga
agtgcgcctg 2880cgatactttg atgtctgggg tcaaggaact acagttactg
tatcaagcag tgctgctgcc 2940tttgtcccgg tatttctccc agccaaaccg
accacgactc ccgccccgcg ccctccgaca 3000cccgctccca ccatcgcctc
tcaacctctt agtcttcgcc ccgaggcatg ccgacccgcc 3060gccgggggtg
ctgttcatac gaggggcttg gacttcgctt gtgatattta catttgggct
3120ccgttggcgg gtacgtgcgg cgtccttttg ttgtcactcg ttattacttt
gtattgtaat 3180cacaggaatc gcaaacgggg cagaaagaaa ctcctgtata
tattcaaaca accatttatg 3240agaccagtac aaactactca agaggaagat
ggctgtagct gccgatttcc agaagaagaa 3300gaaggaggat gtgaactgcg
agtgaagttt tcccgaagcg cagacgctcc ggcatatcag 3360caaggacaga
atcagctgta taacgaactg aatttgggac gccgcgagga gtatgacgtg
3420cttgataaac gccgggggag agacccggaa atggggggta aaccccgaag
aaagaatccc 3480caagaaggac tctacaatga actccagaag gataagatgg
cggaggccta ctcagaaata 3540ggtatgaagg gcgaacgacg acggggaaaa
ggtcacgatg gcctctacca agggttgagt 3600acggcaacca aagatacgta
cgatgcactg catatgcagg ccctgcctcc cagataataa 3660taaaatcgct
atccatcgaa gatggatgtg tgttggtttt ttgtgtgtgg agcaacaaat
3720ctgactttgc atgtgcaaac gccttcaaca acagcattat tccagaagac
accttcttcc 3780ccagcccagg taagggcagc tttggtgcct tcgcaggctg
tttccttgct tcaggaatgg 3840ccaggttctg cccagagctc tggtcaatga
tgtctaaaac tcctctgatt ggtggtctcg 3900gccttatcca ttgccaccaa
aaccctcttt ttactaagaa acagtgagcc ttgttctggc 3960agtccagaga
atgacacggg aaaaaagcag atgaagagaa ggtggcagga gagggcacgt
4020ggcccagcct cagtctctcc aactgagttc ctgcctgcct gcctttgctc
agactgtttg 4080ccccttactg ctcttctagg cctcattcta agccccttct
ccaagttgcc tctccttatt 4140tctccctgtc tgccaaaaaa tctttcccag
ctcactaagt cagtctcacg cagtcactca 4200ttaacccacc aatcactgat
tgtgccggca catgaatgca ccaggtgttg aagtggagga 4260attaaaaagt
cagatgaggg gtgtgcccag aggaagcacc attctagttg ggggagccca
4320tctgtcagct gggaaaagtc caaataactt cagattggaa tgtgttttaa
ctcagggttg 4380agaaaacagc taccttcagg acaaaagtca gggaagggct
ctctgaagaa atgctacttg 4440aagataccag ccctaccaag ggcagggaga
ggaccctata gaggcctggg acaggagctc 4500aatgagaaag gtaaccacgt
gcggaccgag gctgcagcgt cgtcctccct aggaacccct 4560agtgatggag
ttggccactc cctctctgcg cgctcgctcg ctcactgagg ccgggcgacc
4620aaaggtcgcc cgacgcccgg gctttgcccg ggcggcctca gtgagcgagc
gagcgcgcag 4680ctgcctgcag g 46918822PRTArtificial SequenceSynthetic
signal peptide 88Met Leu Leu Leu Val Thr Ser Leu Leu Leu Cys Glu
Leu Pro His Pro1 5 10 15Ala Phe Leu Leu Ile Pro 208921PRTArtificial
SequenceSynthetic signal peptide 89Met Ala Leu Pro Val Thr Ala Leu
Leu Leu Pro Leu Ala Leu Leu Leu1 5 10 15His Ala Ala Arg Pro
209084PRTArtificial SequenceSynthetic CD8a transmembrane domain
90Phe Val Pro Val Phe Leu Pro Ala Lys Pro Thr Thr Thr Pro Ala Pro1
5 10 15Arg Pro Pro Thr Pro Ala Pro Thr Ile Ala Ser Gln Pro Leu Ser
Leu 20 25 30Arg Pro Glu Ala Cys Arg Pro Ala Ala Gly Gly Ala Val His
Thr Arg 35 40 45Gly Leu Asp Phe Ala Cys Asp Ile Tyr Ile Trp Ala Pro
Leu Ala Gly 50 55 60Thr Cys Gly Val Leu Leu Leu Ser Leu Val Ile Thr
Leu Tyr Cys Asn65 70 75 80His Arg Asn Arg9120RNAArtificial
SequenceSynthetic CD70 sgRNA (E1_T1) spacer 91ucaccaagcc cgcgaccaau
209220RNAArtificial SequenceSynthetic CD70 sgRNA (E1_T3) spacer
92aucaccaagc ccgcgaccaa 209320RNAArtificial SequenceSynthetic CD70
sgRNA (E1_T4) spacer 93cggugcggcg caggcccuau 209420RNAArtificial
SequenceSynthetic CD70 sgRNA (E1_T7) spacer 94gcuuuggucc cauuggucgc
209520RNAArtificial SequenceSynthetic CD70 sgRNA (E1_T8) spacer
95gcccgcagga cgcacccaua 209620RNAArtificial SequenceSynthetic CD70
sgRNA (E1_T10) spacer 96gugcauccag cgcuucgcac 209720RNAArtificial
SequenceSynthetic CD70 sgRNA (E3_T1) spacer 97cagcuacgua uccaucguga
209820RNAArtificial SequenceSynthetic TRAC spacer 98agagcaacag
ugcuguggcc 209920RNAArtificial SequenceSynthetic b2M sgRNA spacer
99gcuacucucu cuuucuggcc 2010020RNAArtificial SequenceSynthetic PD-1
sgRNA spacer 100cugcagcuuc uccaacacau 2010120RNAArtificial
SequenceSynthetic CD70 sgRNA (E1_T3)
spacermodified_base(1)..(3)2'-O-methyl phosphorothioate
101ucaccaagcc cgcgaccaau 2010220RNAArtificial SequenceSynthetic
CD70 sgRNA (E1_T4) spacermodified_base(1)..(3)2'-O-methyl
phosphorothioate 102aucaccaagc ccgcgaccaa 2010320RNAArtificial
SequenceSynthetic CD70 sgRNA (E1_T7)
spacermodified_base(1)..(3)2'-O-methyl phosphorothioate
103cggugcggcg caggcccuau 2010420RNAArtificial SequenceSynthetic
CD70 sgRNA (E1_T8) spacermodified_base(1)..(3)2'-O-methyl
phosphorothioate 104gcuuuggucc cauuggucgc 2010520RNAArtificial
SequenceSynthetic CD70 sgRNA (E1_T10)
spacermodified_base(1)..(3)2'-O-methyl phosphorothioate
105gcccgcagga cgcacccaua 2010620RNAArtificial SequenceSynthetic
CD70 sgRNA (E3_T10) spacermodified_base(1)..(3)2'-O-methyl
phosphorothioate 106gugcauccag cgcuucgcac 2010720RNAArtificial
SequenceSynthetic CD70 sgRNA (E1_T3)
spacermodified_base(1)..(3)2'-O-methyl phosphorothioate
107cagcuacgua uccaucguga 2010820RNAArtificial SequenceSynthetic
TRAC spacermodified_base(1)..(3)2'-O-methyl phosphorothioate
108agagcaacag ugcuguggcc 2010920RNAArtificial SequenceSynthetic b2M
sgRNA spacermodified_base(1)..(3)2'-O-methyl phosphorothioate
109gcuacucucu cuuucuggcc 2011020RNAArtificial SequenceSynthetic
PD-1 sgRNA spacermodified_base(1)..(3)2'-O-methyl phosphorothioate
110cugcagcuuc uccaacacau 2011123DNAArtificial SequenceSynthetic
CD70 sgRNA (E1_T1) with PAM 111tcaccaagcc cgcgaccaat ggg
2311223DNAArtificial SequenceSynthetic CD70 sgRNA (E1_T3) with PAM
112atcaccaagc ccgcgaccaa tgg 2311323DNAArtificial SequenceSynthetic
CD70 sgRNA (E1_T4) with PAM 113cggtgcggcg caggccctat ggg
2311423DNAArtificial SequenceSynthetic CD70 sgRNA (E1_T7) with PAM
114gctttggtcc cattggtcgc ggg 2311523DNAArtificial SequenceSynthetic
CD70 sgRNA (E1_T8) with PAM 115gcccgcagga cgcacccata ggg
2311623DNAArtificial SequenceSynthetic CD70 sgRNA (E1_T10) with PAM
116gtgcatccag cgcttcgcac agg 2311723DNAArtificial SequenceSynthetic
CD70 sgRNA (E3_T1) with PAM 117cagctacgta tccatcgtga tgg
2311823DNAArtificial SequenceSynthetic TRAC sgRNA with PAM
118agagcaacag tgctgtggcc tgg 2311923DNAArtificial SequenceSynthetic
b2M sgRNA with PAM 119gctactctct ctttctggcc tgg
2312023DNAArtificial SequenceSynthetic PD-1 sgRNA with PAM
120ctgcagcttc tccaacacat cgg 23121120DNAArtificial
SequenceSynthetic CD28 nucleotide sequence 121tcaaagcgga gtaggttgtt
gcattccgat tacatgaata tgactcctcg ccggcctggg 60ccgacaagaa aacattacca
accctatgcc cccccacgag acttcgctgc gtacaggtcc 120122800DNAArtificial
SequenceSynthetic TRAC-LHA 122gagatgtaag gagctgctgt gacttgctca
aggccttata tcgagtaaac ggtagtgctg 60gggcttagac gcaggtgttc tgatttatag
ttcaaaacct ctatcaatga gagagcaatc 120tcctggtaat
gtgatagatt tcccaactta atgccaacat accataaacc tcccattctg
180ctaatgccca gcctaagttg gggagaccac tccagattcc aagatgtaca
gtttgctttg 240ctgggccttt ttcccatgcc tgcctttact ctgccagagt
tatattgctg gggttttgaa 300gaagatccta ttaaataaaa gaataagcag
tattattaag tagccctgca tttcaggttt 360ccttgagtgg caggccaggc
ctggccgtga acgttcactg aaatcatggc ctcttggcca 420agattgatag
cttgtgcctg tccctgagtc ccagtccatc acgagcagct ggtttctaag
480atgctatttc ccgtataaag catgagaccg tgacttgcca gccccacaga
gccccgccct 540tgtccatcac tggcatctgg actccagcct gggttggggc
aaagagggaa atgagatcat 600gtcctaaccc tgatcctctt gtcccacaga
tatccagaac cctgaccctg ccgtgtacca 660gctgagagac tctaaatcca
gtgacaagtc tgtctgccta ttcaccgatt ttgattctca 720aacaaatgtg
tcacaaagta aggattctga tgtgtatatc acagacaaaa ctgtgctaga
780catgaggtct atggacttca 8001231178DNAArtificial SequenceSynthetic
EF1-alpha promoter 123ggctccggtg cccgtcagtg ggcagagcgc acatcgccca
cagtccccga gaagttgggg 60ggaggggtcg gcaattgaac cggtgcctag agaaggtggc
gcggggtaaa ctgggaaagt 120gatgtcgtgt actggctccg cctttttccc
gagggtgggg gagaaccgta tataagtgca 180gtagtcgccg tgaacgttct
ttttcgcaac gggtttgccg ccagaacaca ggtaagtgcc 240gtgtgtggtt
cccgcgggcc tggcctcttt acgggttatg gcccttgcgt gccttgaatt
300acttccactg gctgcagtac gtgattcttg atcccgagct tcgggttgga
agtgggtggg 360agagttcgag gccttgcgct taaggagccc cttcgcctcg
tgcttgagtt gaggcctggc 420ctgggcgctg gggccgccgc gtgcgaatct
ggtggcacct tcgcgcctgt ctcgctgctt 480tcgataagtc tctagccatt
taaaattttt gatgacctgc tgcgacgctt tttttctggc 540aagatagtct
tgtaaatgcg ggccaagatc tgcacactgg tatttcggtt tttggggccg
600cgggcggcga cggggcccgt gcgtcccagc gcacatgttc ggcgaggcgg
ggcctgcgag 660cgcggccacc gagaatcgga cgggggtagt ctcaagctgg
ccggcctgct ctggtgcctg 720gcctcgcgcc gccgtgtatc gccccgccct
gggcggcaag gctggcccgg tcggcaccag 780ttgcgtgagc ggaaagatgg
ccgcttcccg gccctgctgc agggagctca aaatggagga 840cgcggcgctc
gggagagcgg gcgggtgagt cacccacaca aaggaaaagg gcctttccgt
900cctcagccgt cgcttcatgt gactccacgg agtaccgggc gccgtccagg
cacctcgatt 960agttctcgag cttttggagt acgtcgtctt taggttgggg
ggaggggttt tatgcgatgg 1020agtttcccca cactgagtgg gtggagactg
aagttaggcc agcttggcac ttgatgtaat 1080tctccttgga atttgccctt
tttgagtttg gatcttggtt cattctcaag cctcagacag 1140tggttcaaag
tttttttctt ccatttcagg tgtcgtga 117812449DNAArtificial
SequenceSynthetic Synthetic poly(A) signal 124aataaaatcg ctatccatcg
aagatggatg tgtgttggtt ttttgtgtg 49125804DNAArtificial
SequenceSynthetic TRACRHA 125tggagcaaca aatctgactt tgcatgtgca
aacgccttca acaacagcat tattccagaa 60gacaccttct tccccagccc aggtaagggc
agctttggtg ccttcgcagg ctgtttcctt 120gcttcaggaa tggccaggtt
ctgcccagag ctctggtcaa tgatgtctaa aactcctctg 180attggtggtc
tcggccttat ccattgccac caaaaccctc tttttactaa gaaacagtga
240gccttgttct ggcagtccag agaatgacac gggaaaaaag cagatgaaga
gaaggtggca 300ggagagggca cgtggcccag cctcagtctc tccaactgag
ttcctgcctg cctgcctttg 360ctcagactgt ttgcccctta ctgctcttct
aggcctcatt ctaagcccct tctccaagtt 420gcctctcctt atttctccct
gtctgccaaa aaatctttcc cagctcacta agtcagtctc 480acgcagtcac
tcattaaccc accaatcact gattgtgccg gcacatgaat gcaccaggtg
540ttgaagtgga ggaattaaaa agtcagatga ggggtgtgcc cagaggaagc
accattctag 600ttgggggagc ccatctgtca gctgggaaaa gtccaaataa
cttcagattg gaatgtgttt 660taactcaggg ttgagaaaac agctaccttc
aggacaaaag tcagggaagg gctctctgaa 720gaaatgctac ttgaagatac
cagccctacc aagggcaggg agaggaccct atagaggcct 780gggacaggag
ctcaatgaga aagg 80412623PRTArtificial SequenceSynthetic CD8a
transmembrane 126Ile Tyr Ile Trp Ala Pro Leu Ala Gly Thr Cys Gly
Val Leu Leu Leu1 5 10 15Ser Leu Val Ile Thr Leu Tyr
2012765DNAArtificial SequenceSynthetic CD70 forward primer
127tcgtcggcag cgtcagatgt gtataagaga cagcccaact tttccatctc
aactcacccc 60aagtg 6512853DNAArtificial SequenceSynthetic CD70
reverse primer 128gtctcgtggg ctcggagatg tgtataagag acagcccctc
ctgcgctagc gga 5312958DNAArtificial SequenceSynthetic CD70 Indel
129cacaccacga ggcagatcac caagcccgcg caatgggacc aaagcagccc gcaggacg
5813061DNAArtificial SequenceSynthetic CD70 Indel 130cacaccacga
ggcagatcac caagcccgcg aaccaatggg accaaagcag cccgcaggac 60g
6113148DNAArtificial SequenceSynthetic CD70 Indel 131cacaccacga
ggcagatcac caatgggacc aaagcagccc gcaggacg 4813259DNAArtificial
SequenceSynthetic CD70 Indel 132cacaccacga ggcagatcac caagcccgcg
ccaatgggac caaagcagcc cgcaggacg 5913359DNAArtificial
SequenceSynthetic CD70 Indel 133cacaccacga ggcagatcac caagcccgca
ccaatgggac caaagcagcc cgcaggacg 5913435DNAArtificial
SequenceSynthetic CD70 Indel 134cacaccacga ggcagatcac caagcccgca
ggacg 351354367DNAArtificial SequenceSynthetic Anti-CD33 CAR Donor
LHA to RHA 41BB costim. 135gagatgtaag gagctgctgt gacttgctca
aggccttata tcgagtaaac ggtagtgctg 60gggcttagac gcaggtgttc tgatttatag
ttcaaaacct ctatcaatga gagagcaatc 120tcctggtaat gtgatagatt
tcccaactta atgccaacat accataaacc tcccattctg 180ctaatgccca
gcctaagttg gggagaccac tccagattcc aagatgtaca gtttgctttg
240ctgggccttt ttcccatgcc tgcctttact ctgccagagt tatattgctg
gggttttgaa 300gaagatccta ttaaataaaa gaataagcag tattattaag
tagccctgca tttcaggttt 360ccttgagtgg caggccaggc ctggccgtga
acgttcactg aaatcatggc ctcttggcca 420agattgatag cttgtgcctg
tccctgagtc ccagtccatc acgagcagct ggtttctaag 480atgctatttc
ccgtataaag catgagaccg tgacttgcca gccccacaga gccccgccct
540tgtccatcac tggcatctgg actccagcct gggttggggc aaagagggaa
atgagatcat 600gtcctaaccc tgatcctctt gtcccacaga tatccagaac
cctgaccctg ccgtgtacca 660gctgagagac tctaaatcca gtgacaagtc
tgtctgccta ttcaccgatt ttgattctca 720aacaaatgtg tcacaaagta
aggattctga tgtgtatatc acagacaaaa ctgtgctaga 780catgaggtct
atggacttca ggctccggtg cccgtcagtg ggcagagcgc acatcgccca
840cagtccccga gaagttgggg ggaggggtcg gcaattgaac cggtgcctag
agaaggtggc 900gcggggtaaa ctgggaaagt gatgtcgtgt actggctccg
cctttttccc gagggtgggg 960gagaaccgta tataagtgca gtagtcgccg
tgaacgttct ttttcgcaac gggtttgccg 1020ccagaacaca ggtaagtgcc
gtgtgtggtt cccgcgggcc tggcctcttt acgggttatg 1080gcccttgcgt
gccttgaatt acttccactg gctgcagtac gtgattcttg atcccgagct
1140tcgggttgga agtgggtggg agagttcgag gccttgcgct taaggagccc
cttcgcctcg 1200tgcttgagtt gaggcctggc ctgggcgctg gggccgccgc
gtgcgaatct ggtggcacct 1260tcgcgcctgt ctcgctgctt tcgataagtc
tctagccatt taaaattttt gatgacctgc 1320tgcgacgctt tttttctggc
aagatagtct tgtaaatgcg ggccaagatc tgcacactgg 1380tatttcggtt
tttggggccg cgggcggcga cggggcccgt gcgtcccagc gcacatgttc
1440ggcgaggcgg ggcctgcgag cgcggccacc gagaatcgga cgggggtagt
ctcaagctgg 1500ccggcctgct ctggtgcctg gcctcgcgcc gccgtgtatc
gccccgccct gggcggcaag 1560gctggcccgg tcggcaccag ttgcgtgagc
ggaaagatgg ccgcttcccg gccctgctgc 1620agggagctca aaatggagga
cgcggcgctc gggagagcgg gcgggtgagt cacccacaca 1680aaggaaaagg
gcctttccgt cctcagccgt cgcttcatgt gactccacgg agtaccgggc
1740gccgtccagg cacctcgatt agttctcgag cttttggagt acgtcgtctt
taggttgggg 1800ggaggggttt tatgcgatgg agtttcccca cactgagtgg
gtggagactg aagttaggcc 1860agcttggcac ttgatgtaat tctccttgga
atttgccctt tttgagtttg gatcttggtt 1920cattctcaag cctcagacag
tggttcaaag tttttttctt ccatttcagg tgtcgtgacc 1980accatggcgc
ttccggtgac agcactgctc ctccccttgg cgctgttgct ccacgcagca
2040aggccggaaa tcgtcctcac acaatccccg gggagcctcg cagtcagtcc
tggggaacga 2100gtcactatga gctgcaaatc cagtcagagt gtttttttct
caagtagcca gaagaactac 2160ctcgcatggt accaacaaat accggggcaa
tctccccgct tgcttatata ctgggcaagt 2220acccgcgaat ccggcgtacc
ggatcgattc acgggatctg ggtcaggtac tgatttcact 2280ttgactatca
gctctgttca gcctgaagat ttggcaattt actactgtca ccaatacttg
2340agtagccgaa ctttcggcca gggcacgaag ctcgaaatca agggcggagg
gggaggttct 2400ggtgggggcg gttctggcgg tggaggaagc caagtacagt
tgcaacagcc aggggcggag 2460gtcgtaaaac ctggggcgtc tgtcaagatg
agctgtaaag caagtggata caccttcacc 2520tcctactata tacattggat
taagcaaact ccgggtcagg ggctggaatg ggttggcgtt 2580atataccccg
ggaacgatga tatatcatac aaccaaaaat ttcaaggcaa ggcgactctg
2640actgccgata agagtagcac aacagcttac atgcagcttt cttccctgac
cagcgaagat 2700tcagcagttt actactgcgc tcgggaagtg cgcctgcgat
actttgatgt ctggggtcaa 2760ggaactacag ttactgtatc aagcagtgct
gctgcctttg tcccggtatt tctcccagcc 2820aaaccgacca cgactcccgc
cccgcgccct ccgacacccg ctcccaccat cgcctctcaa 2880cctcttagtc
ttcgccccga ggcatgccga cccgccgccg ggggtgctgt tcatacgagg
2940ggcttggact tcgcttgtga tatttacatt tgggctccgt tggcgggtac
gtgcggcgtc 3000cttttgttgt cactcgttat tactttgtat tgtaatcaca
ggaatcgcaa acggggcaga 3060aagaaactcc tgtatatatt caaacaacca
tttatgagac cagtacaaac tactcaagag 3120gaagatggct gtagctgccg
atttccagaa gaagaagaag gaggatgtga actgcgagtg 3180aagttttccc
gaagcgcaga cgctccggca tatcagcaag gacagaatca gctgtataac
3240gaactgaatt tgggacgccg cgaggagtat gacgtgcttg ataaacgccg
ggggagagac 3300ccggaaatgg ggggtaaacc ccgaagaaag aatccccaag
aaggactcta caatgaactc 3360cagaaggata agatggcgga ggcctactca
gaaataggta tgaagggcga acgacgacgg 3420ggaaaaggtc acgatggcct
ctaccaaggg ttgagtacgg caaccaaaga tacgtacgat 3480gcactgcata
tgcaggccct gcctcccaga taataataaa atcgctatcc atcgaagatg
3540gatgtgtgtt ggttttttgt gtgtggagca acaaatctga ctttgcatgt
gcaaacgcct 3600tcaacaacag cattattcca gaagacacct tcttccccag
cccaggtaag ggcagctttg 3660gtgccttcgc aggctgtttc cttgcttcag
gaatggccag gttctgccca gagctctggt 3720caatgatgtc taaaactcct
ctgattggtg gtctcggcct tatccattgc caccaaaacc 3780ctctttttac
taagaaacag tgagccttgt tctggcagtc cagagaatga cacgggaaaa
3840aagcagatga agagaaggtg gcaggagagg gcacgtggcc cagcctcagt
ctctccaact 3900gagttcctgc ctgcctgcct ttgctcagac tgtttgcccc
ttactgctct tctaggcctc 3960attctaagcc ccttctccaa gttgcctctc
cttatttctc cctgtctgcc aaaaaatctt 4020tcccagctca ctaagtcagt
ctcacgcagt cactcattaa cccaccaatc actgattgtg 4080ccggcacatg
aatgcaccag gtgttgaagt ggaggaatta aaaagtcaga tgaggggtgt
4140gcccagagga agcaccattc tagttggggg agcccatctg tcagctggga
aaagtccaaa 4200taacttcaga ttggaatgtg ttttaactca gggttgagaa
aacagctacc ttcaggacaa 4260aagtcaggga agggctctct gaagaaatgc
tacttgaaga taccagccct accaagggca 4320gggagaggac cctatagagg
cctgggacag gagctcaatg agaaagg 43671361585DNAArtificial
SequenceSynthetic Anti-CD33 CAR 41BB costim 136ccaccatggc
gcttccggtg acagcactgc tcctcccctt ggcgctgttg ctccacgcag 60caaggccgga
aatcgtcctc acacaatccc cggggagcct cgcagtcagt cctggggaac
120gagtcactat gagctgcaaa tccagtcaga gtgttttttt ctcaagtagc
cagaagaact 180acctcgcatg gtaccaacaa ataccggggc aatctccccg
cttgcttata tactgggcaa 240gtacccgcga atccggcgta ccggatcgat
tcacgggatc tgggtcaggt actgatttca 300ctttgactat cagctctgtt
cagcctgaag atttggcaat ttactactgt caccaatact 360tgagtagccg
aactttcggc cagggcacga agctcgaaat caagggcgga gggggaggtt
420ctggtggggg cggttctggc ggtggaggaa gccaagtaca gttgcaacag
ccaggggcgg 480aggtcgtaaa acctggggcg tctgtcaaga tgagctgtaa
agcaagtgga tacaccttca 540cctcctacta tatacattgg attaagcaaa
ctccgggtca ggggctggaa tgggttggcg 600ttatataccc cgggaacgat
gatatatcat acaaccaaaa atttcaaggc aaggcgactc 660tgactgccga
taagagtagc acaacagctt acatgcagct ttcttccctg accagcgaag
720attcagcagt ttactactgc gctcgggaag tgcgcctgcg atactttgat
gtctggggtc 780aaggaactac agttactgta tcaagcagtg ctgctgcctt
tgtcccggta tttctcccag 840ccaaaccgac cacgactccc gccccgcgcc
ctccgacacc cgctcccacc atcgcctctc 900aacctcttag tcttcgcccc
gaggcatgcc gacccgccgc cgggggtgct gttcatacga 960ggggcttgga
cttcgcttgt gatatttaca tttgggctcc gttggcgggt acgtgcggcg
1020tccttttgtt gtcactcgtt attactttgt attgtaatca caggaatcgc
aaacggggca 1080gaaagaaact cctgtatata ttcaaacaac catttatgag
accagtacaa actactcaag 1140aggaagatgg ctgtagctgc cgatttccag
aagaagaaga aggaggatgt gaactgcgag 1200tgaagttttc ccgaagcgca
gacgctccgg catatcagca aggacagaat cagctgtata 1260acgaactgaa
tttgggacgc cgcgaggagt atgacgtgct tgataaacgc cgggggagag
1320acccggaaat ggggggtaaa ccccgaagaa agaatcccca agaaggactc
tacaatgaac 1380tccagaagga taagatggcg gaggcctact cagaaatagg
tatgaagggc gaacgacgac 1440ggggaaaagg tcacgatggc ctctaccaag
ggttgagtac ggcaaccaaa gatacgtacg 1500atgcactgca tatgcaggcc
ctgcctccca gataataata aaatcgctat ccatcgaaga 1560tggatgtgtg
ttggtttttt gtgtg 1585137246PRTArtificial SequenceSynthetic
Anti-CD33 scFv Linker 137Glu Ile Val Leu Thr Gln Ser Pro Gly Ser
Leu Ala Val Ser Pro Gly1 5 10 15Glu Arg Val Thr Met Ser Cys Lys Ser
Ser Gln Ser Val Phe Phe Ser 20 25 30Ser Ser Gln Lys Asn Tyr Leu Ala
Trp Tyr Gln Gln Ile Pro Gly Gln 35 40 45Ser Pro Arg Leu Leu Ile Tyr
Trp Ala Ser Thr Arg Glu Ser Gly Val 50 55 60Pro Asp Arg Phe Thr Gly
Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr65 70 75 80Ile Ser Ser Val
Gln Pro Glu Asp Leu Ala Ile Tyr Tyr Cys His Gln 85 90 95Tyr Leu Ser
Ser Arg Thr Phe Gly Gln Gly Thr Lys Leu Glu Ile Lys 100 105 110Gly
Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser 115 120
125Gln Val Gln Leu Gln Gln Pro Gly Ala Glu Val Val Lys Pro Gly Ala
130 135 140Ser Val Lys Met Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr
Ser Tyr145 150 155 160Tyr Ile His Trp Ile Lys Gln Thr Pro Gly Gln
Gly Leu Glu Trp Val 165 170 175Gly Val Ile Tyr Pro Gly Asn Asp Asp
Ile Ser Tyr Asn Gln Lys Phe 180 185 190Gln Gly Lys Ala Thr Leu Thr
Ala Asp Lys Ser Ser Thr Thr Ala Tyr 195 200 205Met Gln Leu Ser Ser
Leu Thr Ser Glu Asp Ser Ala Val Tyr Tyr Cys 210 215 220Ala Arg Glu
Val Arg Leu Arg Tyr Phe Asp Val Trp Gly Gln Gly Thr225 230 235
240Thr Val Thr Val Ser Ser 245138738DNAArtificial SequenceSynthetic
Anti-CD33 scFv 138gaaatcgtcc tcacacaatc cccggggagc ctcgcagtca
gtcctgggga acgagtcact 60atgagctgca aatccagtca gagtgttttt ttctcaagta
gccagaagaa ctacctcgca 120tggtaccaac aaataccggg gcaatctccc
cgcttgctta tatactgggc aagtacccgc 180gaatccggcg taccggatcg
attcacggga tctgggtcag gtactgattt cactttgact 240atcagctctg
ttcagcctga agatttggca atttactact gtcaccaata cttgagtagc
300cgaactttcg gccagggcac gaagctcgaa atcaagggcg gagggggagg
ttctggtggg 360ggcggttctg gcggtggagg aagccaagta cagttgcaac
agccaggggc ggaggtcgta 420aaacctgggg cgtctgtcaa gatgagctgt
aaagcaagtg gatacacctt cacctcctac 480tatatacatt ggattaagca
aactccgggt caggggctgg aatgggttgg cgttatatac 540cccgggaacg
atgatatatc atacaaccaa aaatttcaag gcaaggcgac tctgactgcc
600gataagagta gcacaacagc ttacatgcag ctttcttccc tgaccagcga
agattcagca 660gtttactact gcgctcggga agtgcgcctg cgatactttg
atgtctgggg tcaaggaact 720acagttactg tatcaagc 738139509PRTArtificial
SequenceSynthetic Anti-CD33 CAR 41BB costim. 139Met Ala Leu Pro Val
Thr Ala Leu Leu Leu Pro Leu Ala Leu Leu Leu1 5 10 15His Ala Ala Arg
Pro Glu Ile Val Leu Thr Gln Ser Pro Gly Ser Leu 20 25 30Ala Val Ser
Pro Gly Glu Arg Val Thr Met Ser Cys Lys Ser Ser Gln 35 40 45Ser Val
Phe Phe Ser Ser Ser Gln Lys Asn Tyr Leu Ala Trp Tyr Gln 50 55 60Gln
Ile Pro Gly Gln Ser Pro Arg Leu Leu Ile Tyr Trp Ala Ser Thr65 70 75
80Arg Glu Ser Gly Val Pro Asp Arg Phe Thr Gly Ser Gly Ser Gly Thr
85 90 95Asp Phe Thr Leu Thr Ile Ser Ser Val Gln Pro Glu Asp Leu Ala
Ile 100 105 110Tyr Tyr Cys His Gln Tyr Leu Ser Ser Arg Thr Phe Gly
Gln Gly Thr 115 120 125Lys Leu Glu Ile Lys Gly Gly Gly Gly Gly Ser
Gly Gly Gly Gly Ser 130 135 140Gly Gly Gly Gly Ser Gln Val Gln Leu
Gln Gln Pro Gly Ala Glu Val145 150 155 160Val Lys Pro Gly Ala Ser
Val Lys Met Ser Cys Lys Ala Ser Gly Tyr 165 170 175Thr Phe Thr Ser
Tyr Tyr Ile His Trp Ile Lys Gln Thr Pro Gly Gln 180 185 190Gly Leu
Glu Trp Val Gly Val Ile Tyr Pro Gly Asn Asp Asp Ile Ser 195 200
205Tyr Asn Gln Lys Phe Gln Gly Lys Ala Thr Leu Thr Ala Asp Lys Ser
210 215 220Ser Thr Thr Ala Tyr Met Gln Leu Ser Ser Leu Thr Ser Glu
Asp Ser225 230 235 240Ala Val Tyr Tyr Cys Ala Arg Glu Val Arg Leu
Arg Tyr Phe Asp Val 245 250 255Trp Gly Gln Gly Thr Thr Val Thr Val
Ser Ser Ser Ala Ala Ala Phe 260 265 270Val Pro Val Phe Leu Pro Ala
Lys Pro Thr Thr Thr Pro Ala Pro Arg 275 280 285Pro Pro Thr Pro Ala
Pro Thr Ile Ala Ser Gln Pro Leu Ser Leu Arg 290 295 300Pro Glu Ala
Cys Arg Pro Ala Ala Gly Gly Ala Val His Thr Arg Gly305 310 315
320Leu Asp Phe Ala Cys Asp Ile Tyr Ile Trp Ala Pro Leu Ala Gly Thr
325 330 335Cys Gly Val Leu Leu Leu Ser Leu Val Ile Thr Leu Tyr Cys
Asn His 340 345 350Arg Asn Arg Lys Arg Gly Arg Lys Lys Leu Leu Tyr
Ile Phe Lys Gln 355 360
365Pro Phe Met Arg Pro Val Gln Thr Thr Gln Glu Glu Asp Gly Cys Ser
370 375 380Cys Arg Phe Pro Glu Glu Glu Glu Gly Gly Cys Glu Leu Arg
Val Lys385 390 395 400Phe Ser Arg Ser Ala Asp Ala Pro Ala Tyr Gln
Gln Gly Gln Asn Gln 405 410 415Leu Tyr Asn Glu Leu Asn Leu Gly Arg
Arg Glu Glu Tyr Asp Val Leu 420 425 430Asp Lys Arg Arg Gly Arg Asp
Pro Glu Met Gly Gly Lys Pro Arg Arg 435 440 445Lys Asn Pro Gln Glu
Gly Leu Tyr Asn Glu Leu Gln Lys Asp Lys Met 450 455 460Ala Glu Ala
Tyr Ser Glu Ile Gly Met Lys Gly Glu Arg Arg Arg Gly465 470 475
480Lys Gly His Asp Gly Leu Tyr Gln Gly Leu Ser Thr Ala Thr Lys Asp
485 490 495Thr Tyr Asp Ala Leu His Met Gln Ala Leu Pro Pro Arg 500
505140118PRTArtificial SequenceSynthetic anti-CD33 antibody VH CDRs
140Gln Val Gln Leu Gln Gln Pro Gly Ala Glu Val Val Lys Pro Gly Ala1
5 10 15Ser Val Lys Met Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Ser
Tyr 20 25 30Tyr Ile His Trp Ile Lys Gln Thr Pro Gly Gln Gly Leu Glu
Trp Val 35 40 45Gly Val Ile Tyr Pro Gly Asn Asp Asp Ile Ser Tyr Asn
Gln Lys Phe 50 55 60Gln Gly Lys Ala Thr Leu Thr Ala Asp Lys Ser Ser
Thr Thr Ala Tyr65 70 75 80Met Gln Leu Ser Ser Leu Thr Ser Glu Asp
Ser Ala Val Tyr Tyr Cys 85 90 95Ala Arg Glu Val Arg Leu Arg Tyr Phe
Asp Val Trp Gly Gln Gly Thr 100 105 110Thr Val Thr Val Ser Ser
115141112PRTArtificial SequenceSynthetic antiCD33 antibody VL CDRs
141Glu Ile Val Leu Thr Gln Ser Pro Gly Ser Leu Ala Val Ser Pro Gly1
5 10 15Glu Arg Val Thr Met Ser Cys Lys Ser Ser Gln Ser Val Phe Phe
Ser 20 25 30Ser Ser Gln Lys Asn Tyr Leu Ala Trp Tyr Gln Gln Ile Pro
Gly Gln 35 40 45Ser Pro Arg Leu Leu Ile Tyr Trp Ala Ser Thr Arg Glu
Ser Gly Val 50 55 60Pro Asp Arg Phe Thr Gly Ser Gly Ser Gly Thr Asp
Phe Thr Leu Thr65 70 75 80Ile Ser Ser Val Gln Pro Glu Asp Leu Ala
Ile Tyr Tyr Cys His Gln 85 90 95Tyr Leu Ser Ser Arg Thr Phe Gly Gln
Gly Thr Lys Leu Glu Ile Lys 100 105 1101425PRTArtificial
SequenceSynthetic anti-CD33 antibody VH CDR1 (Kabat) 142Ser Tyr Tyr
Ile His1 514317PRTArtificial SequenceSynthetic anti-CD33 antibody
VH CDR2 (Kabat) 143Val Ile Tyr Pro Gly Asn Asp Asp Ile Ser Tyr Asn
Gln Lys Phe Gln1 5 10 15Gly1449PRTArtificial SequenceSynthetic
anti-CD33 antibody VH CDR3 (Kabat) 144Glu Val Arg Leu Arg Tyr Phe
Asp Val1 514517PRTArtificial SequenceSynthetic anti-CD33 antibody
VL CDR1 (Kabat & Chothia) 145Lys Ser Ser Gln Ser Val Phe Phe
Ser Ser Ser Gln Lys Asn Tyr Leu1 5 10 15Ala1467PRTArtificial
SequenceSynthetic anti-CD33 antibody VL CDR2 (Kabat & Chothia
146Trp Ala Ser Thr Arg Glu Ser1 51478PRTArtificial
SequenceSynthetic anti-CD33 antibody VL CDR3 (Kabat & Chothia)
147His Gln Tyr Leu Ser Ser Arg Thr1 51481518DNAArtificial
SequenceSynthetic Anti-CD19 CAR CD8[tm]-CD28[co- stimulatory
domain]-CD3z) 148atgcttcttt tggttacgtc tctgttgctt tgcgaacttc
ctcatccagc gttcttgctg 60atccccgata ttcagatgac tcagaccacc agtagcttgt
ctgcctcact gggagaccga 120gtaacaatct cctgcagggc aagtcaagac
attagcaaat acctcaattg gtaccagcag 180aagcccgacg gaacggtaaa
actcctcatc tatcatacgt caaggttgca ttccggagta 240ccgtcacgat
tttcaggttc tgggagcgga actgactatt ccttgactat ttcaaacctc
300gagcaggagg acattgcgac atatttttgt caacaaggta ataccctccc
ttacactttc 360ggaggaggaa ccaaactcga aattaccggg tccaccagtg
gctctgggaa gcctggcagt 420ggagaaggtt ccactaaagg cgaggtgaag
ctccaggaga gcggccccgg tctcgttgcc 480cccagtcaaa gcctctctgt
aacgtgcaca gtgagtggtg tatcattgcc tgattatggc 540gtctcctgga
taaggcagcc cccgcgaaag ggtcttgaat ggcttggggt aatatggggc
600tcagagacaa cgtattataa ctccgctctc aaaagtcgct tgacgataat
aaaagataac 660tccaagagtc aagttttcct taaaatgaac agtttgcaga
ctgacgatac cgctatatat 720tattgtgcta aacattatta ctacggcggt
agttacgcga tggattattg ggggcagggg 780acttctgtca cagtcagtag
tgctgctgcc tttgtcccgg tatttctccc agccaaaccg 840accacgactc
ccgccccgcg ccctccgaca cccgctccca ccatcgcctc tcaacctctt
900agtcttcgcc ccgaggcatg ccgacccgcc gccgggggtg ctgttcatac
gaggggcttg 960gacttcgctt gtgatattta catttgggct ccgttggcgg
gtacgtgcgg cgtccttttg 1020ttgtcactcg ttattacttt gtattgtaat
cacaggaatc gctcaaagcg gagtaggttg 1080ttgcattccg attacatgaa
tatgactcct cgccggcctg ggccgacaag aaaacattac 1140caaccctatg
cccccccacg agacttcgct gcgtacaggt cccgagtgaa gttttcccga
1200agcgcagacg ctccggcata tcagcaagga cagaatcagc tgtataacga
actgaatttg 1260ggacgccgcg aggagtatga cgtgcttgat aaacgccggg
ggagagaccc ggaaatgggg 1320ggtaaacccc gaagaaagaa tccccaagaa
ggactctaca atgaactcca gaaggataag 1380atggcggagg cctactcaga
aataggtatg aagggcgaac gacgacgggg aaaaggtcac 1440gatggcctct
accaagggtt gagtacggca accaaagata cgtacgatgc actgcatatg
1500caggccctgc ctcccaga 1518149506PRTArtificial SequenceSynthetic
Anti-CD19 CAR CD8[tm]-CD28[co- stimulatory domain]-CD3z) Amino Acid
149Met Leu Leu Leu Val Thr Ser Leu Leu Leu Cys Glu Leu Pro His Pro1
5 10 15Ala Phe Leu Leu Ile Pro Asp Ile Gln Met Thr Gln Thr Thr Ser
Ser 20 25 30Leu Ser Ala Ser Leu Gly Asp Arg Val Thr Ile Ser Cys Arg
Ala Ser 35 40 45Gln Asp Ile Ser Lys Tyr Leu Asn Trp Tyr Gln Gln Lys
Pro Asp Gly 50 55 60Thr Val Lys Leu Leu Ile Tyr His Thr Ser Arg Leu
His Ser Gly Val65 70 75 80Pro Ser Arg Phe Ser Gly Ser Gly Ser Gly
Thr Asp Tyr Ser Leu Thr 85 90 95Ile Ser Asn Leu Glu Gln Glu Asp Ile
Ala Thr Tyr Phe Cys Gln Gln 100 105 110Gly Asn Thr Leu Pro Tyr Thr
Phe Gly Gly Gly Thr Lys Leu Glu Ile 115 120 125Thr Gly Ser Thr Ser
Gly Ser Gly Lys Pro Gly Ser Gly Glu Gly Ser 130 135 140Thr Lys Gly
Glu Val Lys Leu Gln Glu Ser Gly Pro Gly Leu Val Ala145 150 155
160Pro Ser Gln Ser Leu Ser Val Thr Cys Thr Val Ser Gly Val Ser Leu
165 170 175Pro Asp Tyr Gly Val Ser Trp Ile Arg Gln Pro Pro Arg Lys
Gly Leu 180 185 190Glu Trp Leu Gly Val Ile Trp Gly Ser Glu Thr Thr
Tyr Tyr Asn Ser 195 200 205Ala Leu Lys Ser Arg Leu Thr Ile Ile Lys
Asp Asn Ser Lys Ser Gln 210 215 220Val Phe Leu Lys Met Asn Ser Leu
Gln Thr Asp Asp Thr Ala Ile Tyr225 230 235 240Tyr Cys Ala Lys His
Tyr Tyr Tyr Gly Gly Ser Tyr Ala Met Asp Tyr 245 250 255Trp Gly Gln
Gly Thr Ser Val Thr Val Ser Ser Ala Ala Ala Phe Val 260 265 270Pro
Val Phe Leu Pro Ala Lys Pro Thr Thr Thr Pro Ala Pro Arg Pro 275 280
285Pro Thr Pro Ala Pro Thr Ile Ala Ser Gln Pro Leu Ser Leu Arg Pro
290 295 300Glu Ala Cys Arg Pro Ala Ala Gly Gly Ala Val His Thr Arg
Gly Leu305 310 315 320Asp Phe Ala Cys Asp Ile Tyr Ile Trp Ala Pro
Leu Ala Gly Thr Cys 325 330 335Gly Val Leu Leu Leu Ser Leu Val Ile
Thr Leu Tyr Cys Asn His Arg 340 345 350Asn Arg Ser Lys Arg Ser Arg
Leu Leu His Ser Asp Tyr Met Asn Met 355 360 365Thr Pro Arg Arg Pro
Gly Pro Thr Arg Lys His Tyr Gln Pro Tyr Ala 370 375 380Pro Pro Arg
Asp Phe Ala Ala Tyr Arg Ser Arg Val Lys Phe Ser Arg385 390 395
400Ser Ala Asp Ala Pro Ala Tyr Gln Gln Gly Gln Asn Gln Leu Tyr Asn
405 410 415Glu Leu Asn Leu Gly Arg Arg Glu Glu Tyr Asp Val Leu Asp
Lys Arg 420 425 430Arg Gly Arg Asp Pro Glu Met Gly Gly Lys Pro Arg
Arg Lys Asn Pro 435 440 445Gln Glu Gly Leu Tyr Asn Glu Leu Gln Lys
Asp Lys Met Ala Glu Ala 450 455 460Tyr Ser Glu Ile Gly Met Lys Gly
Glu Arg Arg Arg Gly Lys Gly His465 470 475 480Asp Gly Leu Tyr Gln
Gly Leu Ser Thr Ala Thr Lys Asp Thr Tyr Asp 485 490 495Ala Leu His
Met Gln Ala Leu Pro Pro Arg 500 505150735DNAArtificial
SequenceSynthetic Anti-CD19 scFv 150gatattcaga tgactcagac
caccagtagc ttgtctgcct cactgggaga ccgagtaaca 60atctcctgca gggcaagtca
agacattagc aaatacctca attggtacca gcagaagccc 120gacggaacgg
taaaactcct catctatcat acgtcaaggt tgcattccgg agtaccgtca
180cgattttcag gttctgggag cggaactgac tattccttga ctatttcaaa
cctcgagcag 240gaggacattg cgacatattt ttgtcaacaa ggtaataccc
tcccttacac tttcggagga 300ggaaccaaac tcgaaattac cgggtccacc
agtggctctg ggaagcctgg cagtggagaa 360ggttccacta aaggcgaggt
gaagctccag gagagcggcc ccggtctcgt tgcccccagt 420caaagcctct
ctgtaacgtg cacagtgagt ggtgtatcat tgcctgatta tggcgtctcc
480tggataaggc agcccccgcg aaagggtctt gaatggcttg gggtaatatg
gggctcagag 540acaacgtatt ataactccgc tctcaaaagt cgcttgacga
taataaaaga taactccaag 600agtcaagttt tccttaaaat gaacagtttg
cagactgacg ataccgctat atattattgt 660gctaaacatt attactacgg
cggtagttac gcgatggatt attgggggca ggggacttct 720gtcacagtca gtagt
735151245PRTArtificial SequenceSynthetic CD19 scFv amino acid
sequence Linker 151Asp Ile Gln Met Thr Gln Thr Thr Ser Ser Leu Ser
Ala Ser Leu Gly1 5 10 15Asp Arg Val Thr Ile Ser Cys Arg Ala Ser Gln
Asp Ile Ser Lys Tyr 20 25 30Leu Asn Trp Tyr Gln Gln Lys Pro Asp Gly
Thr Val Lys Leu Leu Ile 35 40 45Tyr His Thr Ser Arg Leu His Ser Gly
Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Tyr Ser
Leu Thr Ile Ser Asn Leu Glu Gln65 70 75 80Glu Asp Ile Ala Thr Tyr
Phe Cys Gln Gln Gly Asn Thr Leu Pro Tyr 85 90 95Thr Phe Gly Gly Gly
Thr Lys Leu Glu Ile Thr Gly Ser Thr Ser Gly 100 105 110Ser Gly Lys
Pro Gly Ser Gly Glu Gly Ser Thr Lys Gly Glu Val Lys 115 120 125Leu
Gln Glu Ser Gly Pro Gly Leu Val Ala Pro Ser Gln Ser Leu Ser 130 135
140Val Thr Cys Thr Val Ser Gly Val Ser Leu Pro Asp Tyr Gly Val
Ser145 150 155 160Trp Ile Arg Gln Pro Pro Arg Lys Gly Leu Glu Trp
Leu Gly Val Ile 165 170 175Trp Gly Ser Glu Thr Thr Tyr Tyr Asn Ser
Ala Leu Lys Ser Arg Leu 180 185 190Thr Ile Ile Lys Asp Asn Ser Lys
Ser Gln Val Phe Leu Lys Met Asn 195 200 205Ser Leu Gln Thr Asp Asp
Thr Ala Ile Tyr Tyr Cys Ala Lys His Tyr 210 215 220Tyr Tyr Gly Gly
Ser Tyr Ala Met Asp Tyr Trp Gly Gln Gly Thr Ser225 230 235 240Val
Thr Val Ser Ser 245152120PRTArtificial SequenceSynthetic Anti-CD19
VH 152Glu Val Lys Leu Gln Glu Ser Gly Pro Gly Leu Val Ala Pro Ser
Gln1 5 10 15Ser Leu Ser Val Thr Cys Thr Val Ser Gly Val Ser Leu Pro
Asp Tyr 20 25 30Gly Val Ser Trp Ile Arg Gln Pro Pro Arg Lys Gly Leu
Glu Trp Leu 35 40 45Gly Val Ile Trp Gly Ser Glu Thr Thr Tyr Tyr Asn
Ser Ala Leu Lys 50 55 60Ser Arg Leu Thr Ile Ile Lys Asp Asn Ser Lys
Ser Gln Val Phe Leu65 70 75 80Lys Met Asn Ser Leu Gln Thr Asp Asp
Thr Ala Ile Tyr Tyr Cys Ala 85 90 95Lys His Tyr Tyr Tyr Gly Gly Ser
Tyr Ala Met Asp Tyr Trp Gly Gln 100 105 110Gly Thr Ser Val Thr Val
Ser Ser 115 120153107PRTArtificial SequenceSynthetic Anti-CD19 VL
153Asp Ile Gln Met Thr Gln Thr Thr Ser Ser Leu Ser Ala Ser Leu Gly1
5 10 15Asp Arg Val Thr Ile Ser Cys Arg Ala Ser Gln Asp Ile Ser Lys
Tyr 20 25 30Leu Asn Trp Tyr Gln Gln Lys Pro Asp Gly Thr Val Lys Leu
Leu Ile 35 40 45Tyr His Thr Ser Arg Leu His Ser Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Tyr Ser Leu Thr Ile Ser
Asn Leu Glu Gln65 70 75 80Glu Asp Ile Ala Thr Tyr Phe Cys Gln Gln
Gly Asn Thr Leu Pro Tyr 85 90 95Thr Phe Gly Gly Gly Thr Lys Leu Glu
Ile Thr 100 10515418PRTArtificial SequenceSynthetic Anti-CD19 scFv
linker 154Gly Ser Thr Ser Gly Ser Gly Lys Pro Gly Ser Gly Glu Gly
Ser Thr1 5 10 15Lys Gly1554682DNAArtificial SequenceSynthetic
anti-CD19 CAR rAAV 155cctgcaggca gctgcgcgct cgctcgctca ctgaggccgc
ccgggcgtcg ggcgaccttt 60ggtcgcccgg cctcagtgag cgagcgagcg cgcagagagg
gagtggccaa ctccatcact 120aggggttcct gcggccgcac gcgtgagatg
taaggagctg ctgtgacttg ctcaaggcct 180tatatcgagt aaacggtagt
gctggggctt agacgcaggt gttctgattt atagttcaaa 240acctctatca
atgagagagc aatctcctgg taatgtgata gatttcccaa cttaatgcca
300acataccata aacctcccat tctgctaatg cccagcctaa gttggggaga
ccactccaga 360ttccaagatg tacagtttgc tttgctgggc ctttttccca
tgcctgcctt tactctgcca 420gagttatatt gctggggttt tgaagaagat
cctattaaat aaaagaataa gcagtattat 480taagtagccc tgcatttcag
gtttccttga gtggcaggcc aggcctggcc gtgaacgttc 540actgaaatca
tggcctcttg gccaagattg atagcttgtg cctgtccctg agtcccagtc
600catcacgagc agctggtttc taagatgcta tttcccgtat aaagcatgag
accgtgactt 660gccagcccca cagagccccg cccttgtcca tcactggcat
ctggactcca gcctgggttg 720gggcaaagag ggaaatgaga tcatgtccta
accctgatcc tcttgtccca cagatatcca 780gaaccctgac cctgccgtgt
accagctgag agactctaaa tccagtgaca agtctgtctg 840cctattcacc
gattttgatt ctcaaacaaa tgtgtcacaa agtaaggatt ctgatgtgta
900tatcacagac aaaactgtgc tagacatgag gtctatggac ttcaggctcc
ggtgcccgtc 960agtgggcaga gcgcacatcg cccacagtcc ccgagaagtt
ggggggaggg gtcggcaatt 1020gaaccggtgc ctagagaagg tggcgcgggg
taaactggga aagtgatgtc gtgtactggc 1080tccgcctttt tcccgagggt
gggggagaac cgtatataag tgcagtagtc gccgtgaacg 1140ttctttttcg
caacgggttt gccgccagaa cacaggtaag tgccgtgtgt ggttcccgcg
1200ggcctggcct ctttacgggt tatggccctt gcgtgccttg aattacttcc
actggctgca 1260gtacgtgatt cttgatcccg agcttcgggt tggaagtggg
tgggagagtt cgaggccttg 1320cgcttaagga gccccttcgc ctcgtgcttg
agttgaggcc tggcctgggc gctggggccg 1380ccgcgtgcga atctggtggc
accttcgcgc ctgtctcgct gctttcgata agtctctagc 1440catttaaaat
ttttgatgac ctgctgcgac gctttttttc tggcaagata gtcttgtaaa
1500tgcgggccaa gatctgcaca ctggtatttc ggtttttggg gccgcgggcg
gcgacggggc 1560ccgtgcgtcc cagcgcacat gttcggcgag gcggggcctg
cgagcgcggc caccgagaat 1620cggacggggg tagtctcaag ctggccggcc
tgctctggtg cctggcctcg cgccgccgtg 1680tatcgccccg ccctgggcgg
caaggctggc ccggtcggca ccagttgcgt gagcggaaag 1740atggccgctt
cccggccctg ctgcagggag ctcaaaatgg aggacgcggc gctcgggaga
1800gcgggcgggt gagtcaccca cacaaaggaa aagggccttt ccgtcctcag
ccgtcgcttc 1860atgtgactcc acggagtacc gggcgccgtc caggcacctc
gattagttct cgagcttttg 1920gagtacgtcg tctttaggtt ggggggaggg
gttttatgcg atggagtttc cccacactga 1980gtgggtggag actgaagtta
ggccagcttg gcacttgatg taattctcct tggaatttgc 2040cctttttgag
tttggatctt ggttcattct caagcctcag acagtggttc aaagtttttt
2100tcttccattt caggtgtcgt gaccaccatg cttcttttgg ttacgtctct
gttgctttgc 2160gaacttcctc atccagcgtt cttgctgatc cccgatattc
agatgactca gaccaccagt 2220agcttgtctg cctcactggg agaccgagta
acaatctcct gcagggcaag tcaagacatt 2280agcaaatacc tcaattggta
ccagcagaag cccgacggaa cggtaaaact cctcatctat 2340catacgtcaa
ggttgcattc cggagtaccg tcacgatttt caggttctgg gagcggaact
2400gactattcct tgactatttc aaacctcgag caggaggaca ttgcgacata
tttttgtcaa 2460caaggtaata ccctccctta cactttcgga ggaggaacca
aactcgaaat taccgggtcc 2520accagtggct ctgggaagcc tggcagtgga
gaaggttcca ctaaaggcga ggtgaagctc 2580caggagagcg gccccggtct
cgttgccccc agtcaaagcc tctctgtaac gtgcacagtg 2640agtggtgtat
cattgcctga ttatggcgtc tcctggataa ggcagccccc gcgaaagggt
2700cttgaatggc ttggggtaat atggggctca gagacaacgt attataactc
cgctctcaaa 2760agtcgcttga cgataataaa agataactcc aagagtcaag
ttttccttaa aatgaacagt 2820ttgcagactg acgataccgc tatatattat
tgtgctaaac attattacta cggcggtagt 2880tacgcgatgg attattgggg
gcaggggact tctgtcacag tcagtagtgc tgctgccttt
2940gtcccggtat ttctcccagc caaaccgacc acgactcccg ccccgcgccc
tccgacaccc 3000gctcccacca tcgcctctca acctcttagt cttcgccccg
aggcatgccg acccgccgcc 3060gggggtgctg ttcatacgag gggcttggac
ttcgcttgtg atatttacat ttgggctccg 3120ttggcgggta cgtgcggcgt
ccttttgttg tcactcgtta ttactttgta ttgtaatcac 3180aggaatcgct
caaagcggag taggttgttg cattccgatt acatgaatat gactcctcgc
3240cggcctgggc cgacaagaaa acattaccaa ccctatgccc ccccacgaga
cttcgctgcg 3300tacaggtccc gagtgaagtt ttcccgaagc gcagacgctc
cggcatatca gcaaggacag 3360aatcagctgt ataacgaact gaatttggga
cgccgcgagg agtatgacgt gcttgataaa 3420cgccggggga gagacccgga
aatggggggt aaaccccgaa gaaagaatcc ccaagaagga 3480ctctacaatg
aactccagaa ggataagatg gcggaggcct actcagaaat aggtatgaag
3540ggcgaacgac gacggggaaa aggtcacgat ggcctctacc aagggttgag
tacggcaacc 3600aaagatacgt acgatgcact gcatatgcag gccctgcctc
ccagataata ataaaatcgc 3660tatccatcga agatggatgt gtgttggttt
tttgtgtgtg gagcaacaaa tctgactttg 3720catgtgcaaa cgccttcaac
aacagcatta ttccagaaga caccttcttc cccagcccag 3780gtaagggcag
ctttggtgcc ttcgcaggct gtttccttgc ttcaggaatg gccaggttct
3840gcccagagct ctggtcaatg atgtctaaaa ctcctctgat tggtggtctc
ggccttatcc 3900attgccacca aaaccctctt tttactaaga aacagtgagc
cttgttctgg cagtccagag 3960aatgacacgg gaaaaaagca gatgaagaga
aggtggcagg agagggcacg tggcccagcc 4020tcagtctctc caactgagtt
cctgcctgcc tgcctttgct cagactgttt gccccttact 4080gctcttctag
gcctcattct aagccccttc tccaagttgc ctctccttat ttctccctgt
4140ctgccaaaaa atctttccca gctcactaag tcagtctcac gcagtcactc
attaacccac 4200caatcactga ttgtgccggc acatgaatgc accaggtgtt
gaagtggagg aattaaaaag 4260tcagatgagg ggtgtgccca gaggaagcac
cattctagtt gggggagccc atctgtcagc 4320tgggaaaagt ccaaataact
tcagattgga atgtgtttta actcagggtt gagaaaacag 4380ctaccttcag
gacaaaagtc agggaagggc tctctgaaga aatgctactt gaagatacca
4440gccctaccaa gggcagggag aggaccctat agaggcctgg gacaggagct
caatgagaaa 4500ggtaaccacg tgcggaccga ggctgcagcg tcgtcctccc
taggaacccc tagtgatgga 4560gttggccact ccctctctgc gcgctcgctc
gctcactgag gccgggcgac caaaggtcgc 4620ccgacgcccg ggctttgccc
gggcggcctc agtgagcgag cgagcgcgca gctgcctgca 4680gg
46821564358DNAArtificial SequenceSynthetic Anti-CD19 CAR LHA to RHA
156gagatgtaag gagctgctgt gacttgctca aggccttata tcgagtaaac
ggtagtgctg 60gggcttagac gcaggtgttc tgatttatag ttcaaaacct ctatcaatga
gagagcaatc 120tcctggtaat gtgatagatt tcccaactta atgccaacat
accataaacc tcccattctg 180ctaatgccca gcctaagttg gggagaccac
tccagattcc aagatgtaca gtttgctttg 240ctgggccttt ttcccatgcc
tgcctttact ctgccagagt tatattgctg gggttttgaa 300gaagatccta
ttaaataaaa gaataagcag tattattaag tagccctgca tttcaggttt
360ccttgagtgg caggccaggc ctggccgtga acgttcactg aaatcatggc
ctcttggcca 420agattgatag cttgtgcctg tccctgagtc ccagtccatc
acgagcagct ggtttctaag 480atgctatttc ccgtataaag catgagaccg
tgacttgcca gccccacaga gccccgccct 540tgtccatcac tggcatctgg
actccagcct gggttggggc aaagagggaa atgagatcat 600gtcctaaccc
tgatcctctt gtcccacaga tatccagaac cctgaccctg ccgtgtacca
660gctgagagac tctaaatcca gtgacaagtc tgtctgccta ttcaccgatt
ttgattctca 720aacaaatgtg tcacaaagta aggattctga tgtgtatatc
acagacaaaa ctgtgctaga 780catgaggtct atggacttca ggctccggtg
cccgtcagtg ggcagagcgc acatcgccca 840cagtccccga gaagttgggg
ggaggggtcg gcaattgaac cggtgcctag agaaggtggc 900gcggggtaaa
ctgggaaagt gatgtcgtgt actggctccg cctttttccc gagggtgggg
960gagaaccgta tataagtgca gtagtcgccg tgaacgttct ttttcgcaac
gggtttgccg 1020ccagaacaca ggtaagtgcc gtgtgtggtt cccgcgggcc
tggcctcttt acgggttatg 1080gcccttgcgt gccttgaatt acttccactg
gctgcagtac gtgattcttg atcccgagct 1140tcgggttgga agtgggtggg
agagttcgag gccttgcgct taaggagccc cttcgcctcg 1200tgcttgagtt
gaggcctggc ctgggcgctg gggccgccgc gtgcgaatct ggtggcacct
1260tcgcgcctgt ctcgctgctt tcgataagtc tctagccatt taaaattttt
gatgacctgc 1320tgcgacgctt tttttctggc aagatagtct tgtaaatgcg
ggccaagatc tgcacactgg 1380tatttcggtt tttggggccg cgggcggcga
cggggcccgt gcgtcccagc gcacatgttc 1440ggcgaggcgg ggcctgcgag
cgcggccacc gagaatcgga cgggggtagt ctcaagctgg 1500ccggcctgct
ctggtgcctg gcctcgcgcc gccgtgtatc gccccgccct gggcggcaag
1560gctggcccgg tcggcaccag ttgcgtgagc ggaaagatgg ccgcttcccg
gccctgctgc 1620agggagctca aaatggagga cgcggcgctc gggagagcgg
gcgggtgagt cacccacaca 1680aaggaaaagg gcctttccgt cctcagccgt
cgcttcatgt gactccacgg agtaccgggc 1740gccgtccagg cacctcgatt
agttctcgag cttttggagt acgtcgtctt taggttgggg 1800ggaggggttt
tatgcgatgg agtttcccca cactgagtgg gtggagactg aagttaggcc
1860agcttggcac ttgatgtaat tctccttgga atttgccctt tttgagtttg
gatcttggtt 1920cattctcaag cctcagacag tggttcaaag tttttttctt
ccatttcagg tgtcgtgacc 1980accatgcttc ttttggttac gtctctgttg
ctttgcgaac ttcctcatcc agcgttcttg 2040ctgatccccg atattcagat
gactcagacc accagtagct tgtctgcctc actgggagac 2100cgagtaacaa
tctcctgcag ggcaagtcaa gacattagca aatacctcaa ttggtaccag
2160cagaagcccg acggaacggt aaaactcctc atctatcata cgtcaaggtt
gcattccgga 2220gtaccgtcac gattttcagg ttctgggagc ggaactgact
attccttgac tatttcaaac 2280ctcgagcagg aggacattgc gacatatttt
tgtcaacaag gtaataccct cccttacact 2340ttcggaggag gaaccaaact
cgaaattacc gggtccacca gtggctctgg gaagcctggc 2400agtggagaag
gttccactaa aggcgaggtg aagctccagg agagcggccc cggtctcgtt
2460gcccccagtc aaagcctctc tgtaacgtgc acagtgagtg gtgtatcatt
gcctgattat 2520ggcgtctcct ggataaggca gcccccgcga aagggtcttg
aatggcttgg ggtaatatgg 2580ggctcagaga caacgtatta taactccgct
ctcaaaagtc gcttgacgat aataaaagat 2640aactccaaga gtcaagtttt
ccttaaaatg aacagtttgc agactgacga taccgctata 2700tattattgtg
ctaaacatta ttactacggc ggtagttacg cgatggatta ttgggggcag
2760gggacttctg tcacagtcag tagtgctgct gcctttgtcc cggtatttct
cccagccaaa 2820ccgaccacga ctcccgcccc gcgccctccg acacccgctc
ccaccatcgc ctctcaacct 2880cttagtcttc gccccgaggc atgccgaccc
gccgccgggg gtgctgttca tacgaggggc 2940ttggacttcg cttgtgatat
ttacatttgg gctccgttgg cgggtacgtg cggcgtcctt 3000ttgttgtcac
tcgttattac tttgtattgt aatcacagga atcgctcaaa gcggagtagg
3060ttgttgcatt ccgattacat gaatatgact cctcgccggc ctgggccgac
aagaaaacat 3120taccaaccct atgccccccc acgagacttc gctgcgtaca
ggtcccgagt gaagttttcc 3180cgaagcgcag acgctccggc atatcagcaa
ggacagaatc agctgtataa cgaactgaat 3240ttgggacgcc gcgaggagta
tgacgtgctt gataaacgcc gggggagaga cccggaaatg 3300gggggtaaac
cccgaagaaa gaatccccaa gaaggactct acaatgaact ccagaaggat
3360aagatggcgg aggcctactc agaaataggt atgaagggcg aacgacgacg
gggaaaaggt 3420cacgatggcc tctaccaagg gttgagtacg gcaaccaaag
atacgtacga tgcactgcat 3480atgcaggccc tgcctcccag ataataataa
aatcgctatc catcgaagat ggatgtgtgt 3540tggttttttg tgtgtggagc
aacaaatctg actttgcatg tgcaaacgcc ttcaacaaca 3600gcattattcc
agaagacacc ttcttcccca gcccaggtaa gggcagcttt ggtgccttcg
3660caggctgttt ccttgcttca ggaatggcca ggttctgccc agagctctgg
tcaatgatgt 3720ctaaaactcc tctgattggt ggtctcggcc ttatccattg
ccaccaaaac cctcttttta 3780ctaagaaaca gtgagccttg ttctggcagt
ccagagaatg acacgggaaa aaagcagatg 3840aagagaaggt ggcaggagag
ggcacgtggc ccagcctcag tctctccaac tgagttcctg 3900cctgcctgcc
tttgctcaga ctgtttgccc cttactgctc ttctaggcct cattctaagc
3960cccttctcca agttgcctct ccttatttct ccctgtctgc caaaaaatct
ttcccagctc 4020actaagtcag tctcacgcag tcactcatta acccaccaat
cactgattgt gccggcacat 4080gaatgcacca ggtgttgaag tggaggaatt
aaaaagtcag atgaggggtg tgcccagagg 4140aagcaccatt ctagttgggg
gagcccatct gtcagctggg aaaagtccaa ataacttcag 4200attggaatgt
gttttaactc agggttgaga aaacagctac cttcaggaca aaagtcaggg
4260aagggctctc tgaagaaatg ctacttgaag ataccagccc taccaagggc
agggagagga 4320ccctatagag gcctgggaca ggagctcaat gagaaagg
435815720DNAArtificial SequenceSynthetic CD70 sgRNA (E1_T1)
157tcaccaagcc cgcgaccaat 2015820DNAArtificial SequenceSynthetic
CD70 sgRNA (E1_T3) 158atcaccaagc ccgcgaccaa 2015920DNAArtificial
SequenceSynthetic CD70 sgRNA (E1_T4) 159cggtgcggcg caggccctat
2016020DNAArtificial SequenceSynthetic CD70 sgRNA (E1_T7)
160gctttggtcc cattggtcgc 2016120DNAArtificial SequenceSynthetic
CD70 sgRNA (E1_T8) 161gcccgcagga cgcacccata 2016220DNAArtificial
SequenceSynthetic CD70 sgRNA (E1_T10) 162gtgcatccag cgcttcgcac
2016320DNAArtificial SequenceSynthetic CD70 sgRNA (E3_T1)
163cagctacgta tccatcgtga 2016420DNAArtificial SequenceSynthetic b2M
sgRNA 164gctactctct ctttctggcc 2016520DNAArtificial
SequenceSynthetic PD-1 sgRNA 165ctgcagcttc tccaacacat
2016611PRTArtificial SequenceSynthetic anti-CD19 VL CDR1 (Kabat)
166Arg Ala Ser Gln Asp Ile Ser Lys Tyr Leu Asn1 5
101677PRTArtificial SequenceSynthetic anti-CD19 VL CDR2 (Kabat)
167His Thr Ser Arg Leu His Ser1 51689PRTArtificial
SequenceSynthetic anti-CD19 VL CDR3 (Kabat) 168Gln Gln Gly Asn Thr
Leu Pro Tyr Thr1 51695PRTArtificial SequenceSynthetic anti-CD19 VH
CDR1 (Kabat) 169Asp Tyr Gly Val Ser1 517016PRTArtificial
SequenceSynthetic anti-CD19 VH CDR2 (Kabat) 170Val Ile Trp Gly Ser
Glu Thr Thr Tyr Tyr Asn Ser Ala Leu Lys Ser1 5 10
1517112PRTArtificial SequenceSynthetic anti-CD19 VH CDR3 (Kabat)
171His Tyr Tyr Tyr Gly Gly Ser Tyr Ala Met Asp Tyr1 5
1017211PRTArtificial SequenceSynthetic anti-CD19 VL CDR1 (Chothia)
172Arg Ala Ser Gln Asp Ile Ser Lys Tyr Leu Asn1 5
101737PRTArtificial SequenceSynthetic anti-CD19 VL CDR2 (Chothia)
173His Thr Ser Arg Leu His Ser1 51749PRTArtificial
SequenceSynthetic anti-CD19 VL CDR3 (Chothia) 174Gln Gln Gly Asn
Thr Leu Pro Tyr Thr1 51757PRTArtificial SequenceSynthetic anti-CD19
VH CDR1 (Chothia) 175Gly Val Ser Leu Pro Asp Tyr1
51765PRTArtificial SequenceSynthetic anti-CD19 VH CDR2 (Chothia)
176Trp Gly Ser Glu Thr1 517712PRTArtificial SequenceSynthetic
anti-CD19 VH CDR3 (Chothia) 177His Tyr Tyr Tyr Gly Gly Ser Tyr Ala
Met Asp Tyr1 5 101787PRTArtificial SequenceSynthetic anti-CD33 VH
CDR1 (Chothia) 178Gly Tyr Thr Phe Thr Ser Tyr1 51796PRTArtificial
SequenceSynthetic anti-CD33 VH CDR2 (Chothia) 179Tyr Pro Gly Asn
Asp Asp1 51809PRTArtificial SequenceSynthetic anti-CD33 VH CDR3
(Chothia) 180Glu Val Arg Leu Arg Tyr Phe Asp Val1 5
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014

D00015

D00016

D00017

D00018

D00019

D00020

D00021

D00022

D00023

D00024

D00025

D00026

D00027

D00028

D00029

D00030

D00031

D00032

D00033

D00034

D00035

D00036

D00037

D00038

D00039

D00040

D00041

D00042

D00043

D00044

D00045

D00046

D00047

D00048
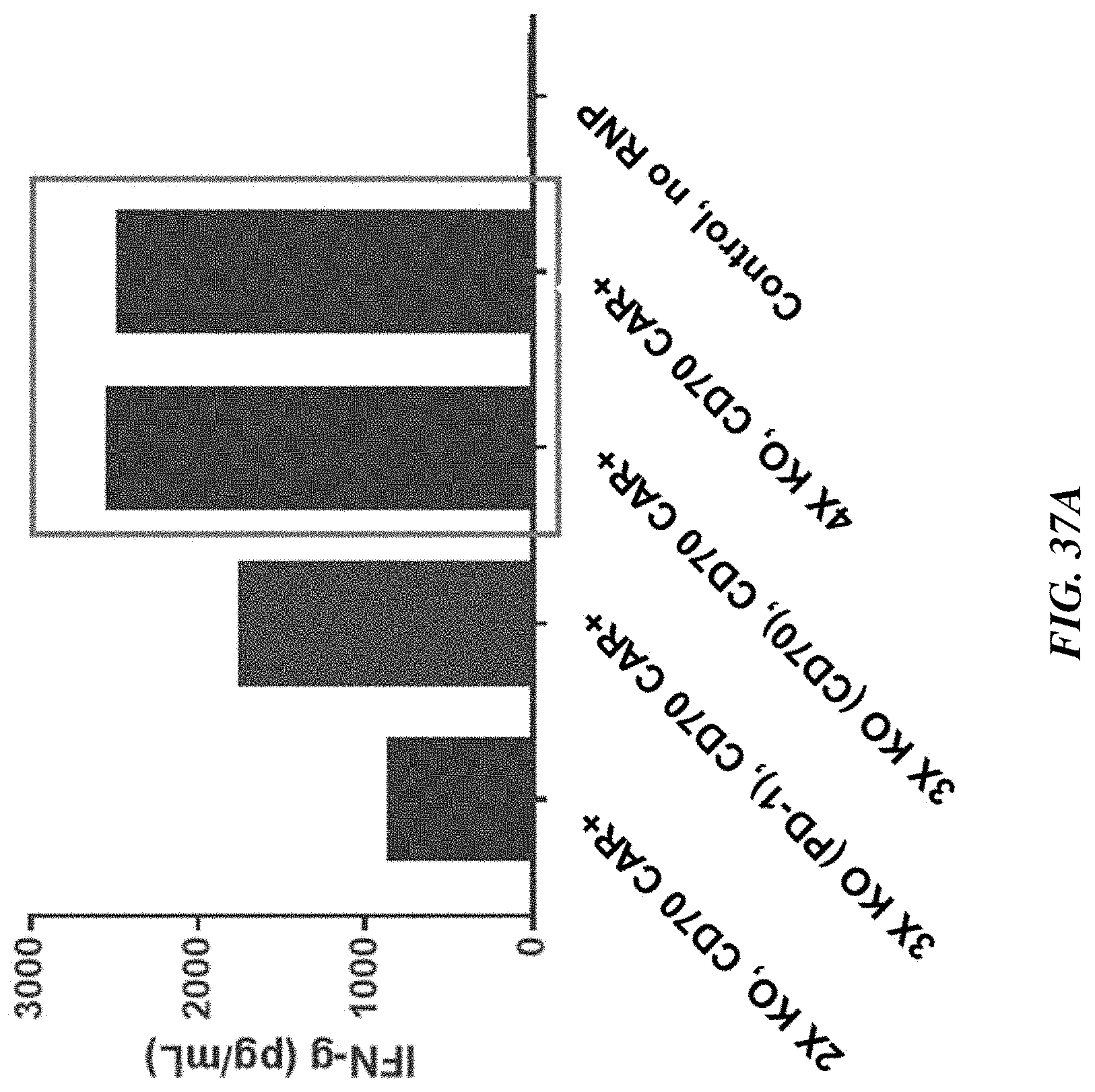
D00049

D00050

D00051

D00052

D00053

D00054

D00055

D00056

D00057

D00058

D00059

D00060

D00061

D00062

D00063
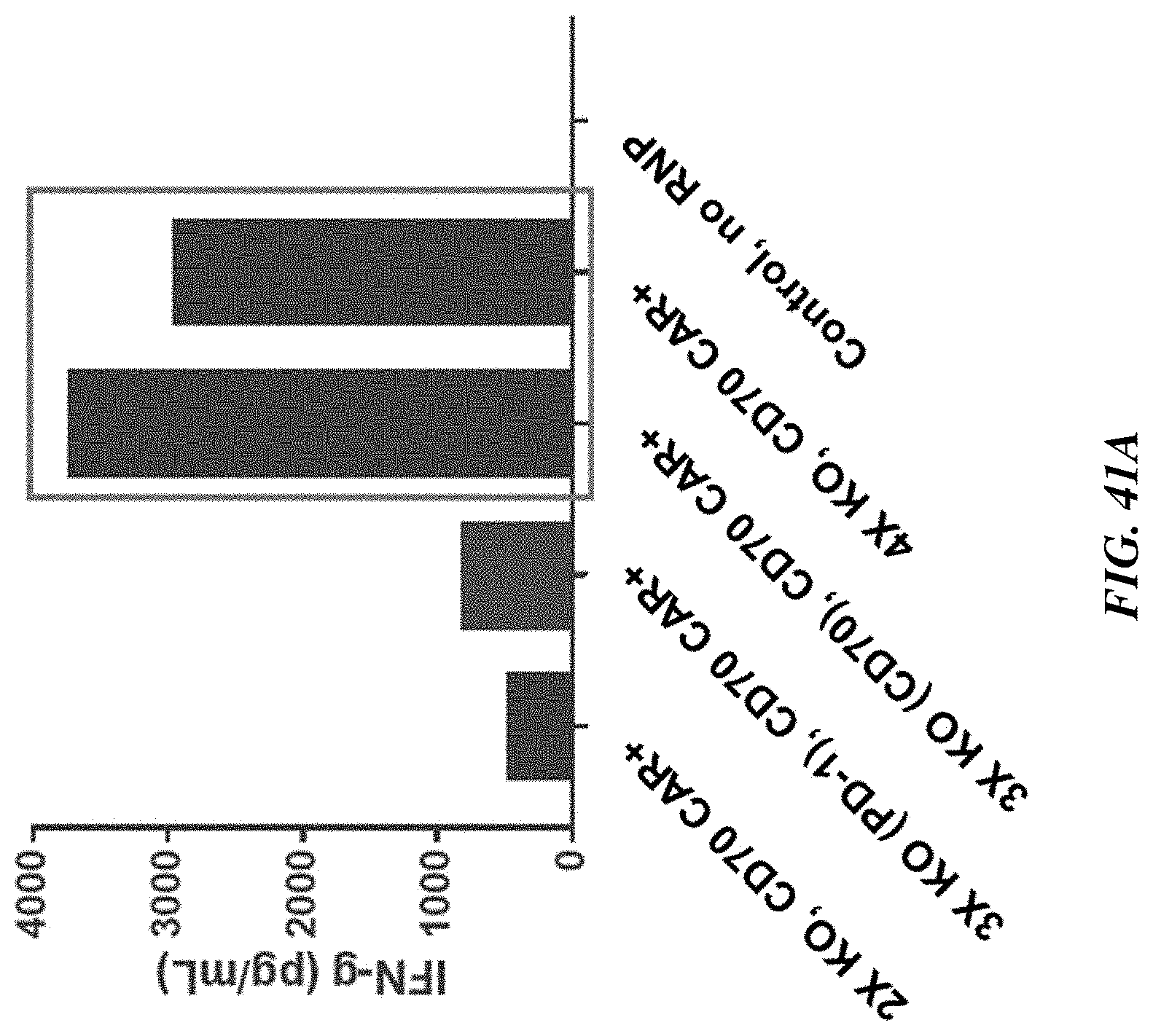
D00064

D00065

D00066

D00067
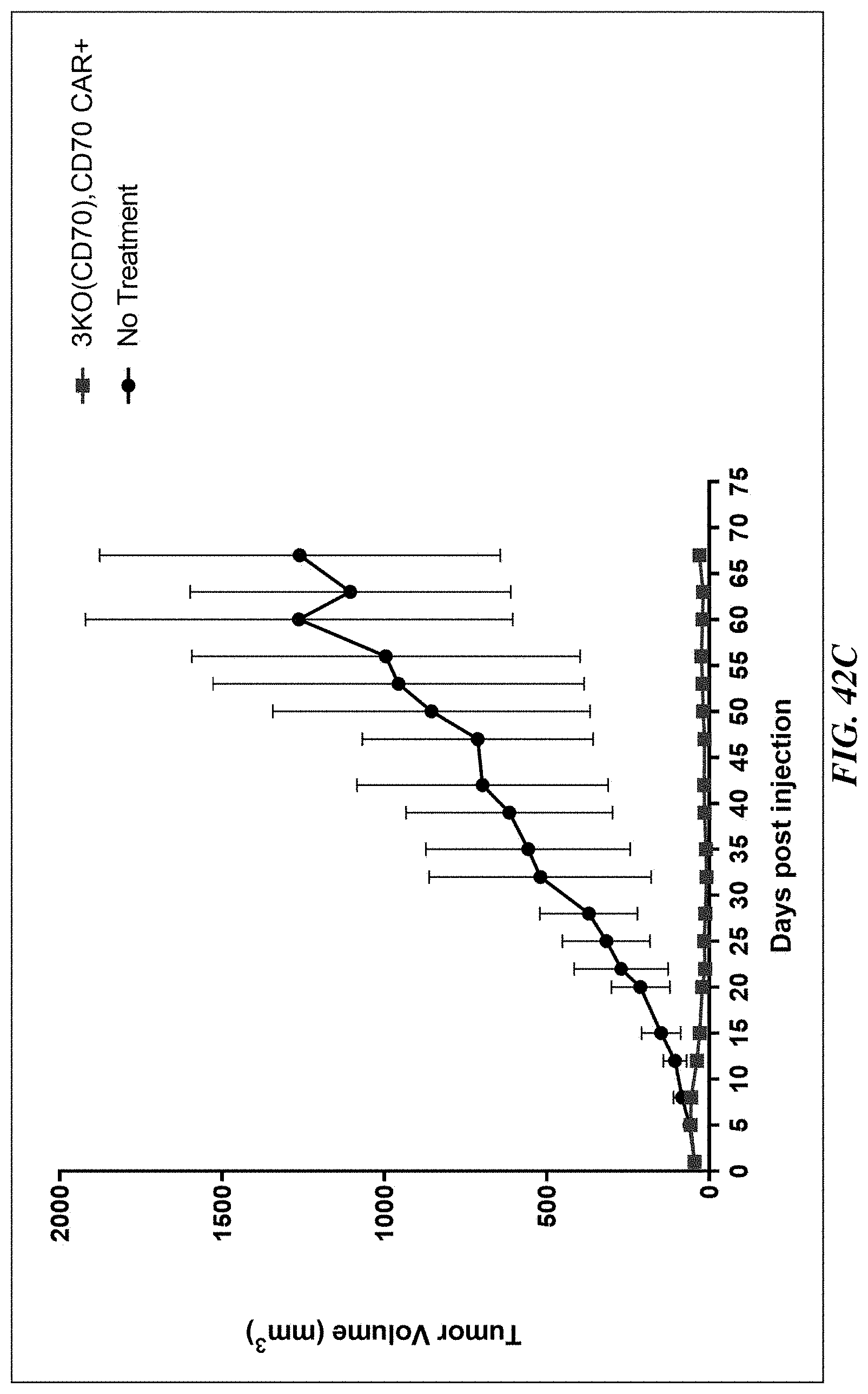
D00068

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.