Antigen-binding Proteins That Activate The Leptin Receptor
GROMADA; Jesper ; et al.
U.S. patent application number 16/593761 was filed with the patent office on 2020-01-30 for antigen-binding proteins that activate the leptin receptor. The applicant listed for this patent is Regeneron Pharmaceuticals, Inc.. Invention is credited to Judith ALTAREJOS, Jesper GROMADA, Panayiotis STEVIS.
| Application Number | 20200031946 16/593761 |
| Document ID | / |
| Family ID | 57184864 |
| Filed Date | 2020-01-30 |






| United States Patent Application | 20200031946 |
| Kind Code | A1 |
| GROMADA; Jesper ; et al. | January 30, 2020 |
ANTIGEN-BINDING PROTEINS THAT ACTIVATE THE LEPTIN RECEPTOR
Abstract
The present invention provides antibodies and antigen-binding fragments of antibodies that bind to leptin receptor (LEPR), and methods of using the same. According to certain embodiments, the invention includes antibodies and antigen-binding fragments of antibodies that bind LEPR and activate LEPR signaling. In other embodiments, the invention includes antibodies and antigen-binding fragments of antibodies that bind to LEPR and enhance sensitization of LEPR to an antigen. In certain embodiments, the invention includes antibodies and antigen-binding fragments of antibodies that bind LEPR in the presence and absence of leptin. In certain embodiments, the invention includes antibodies and antigen-binding fragments of antibodies that induce signaling in cells expressing LEPR mutants that otherwise exhibit defective or impaired signaling in the presence of leptin. The antibodies and antigen-binding fragments of the present invention are useful for the treatment of lipodystrophies and other diseases and disorders associated with or caused by leptin deficiency or leptin resistance.
| Inventors: | GROMADA; Jesper; (Scarsdale, NY) ; STEVIS; Panayiotis; (West Orange, NJ) ; ALTAREJOS; Judith; (Chappaqua, NY) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 57184864 | ||||||||||
| Appl. No.: | 16/593761 | ||||||||||
| Filed: | October 4, 2019 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 16007812 | Jun 13, 2018 | |||
| 16593761 | ||||
| 15290967 | Oct 11, 2016 | 10023644 | ||
| 16007812 | ||||
| 62240021 | Oct 12, 2015 | |||
| 62359757 | Jul 8, 2016 | |||
| 62375495 | Aug 16, 2016 | |||
| 62393143 | Sep 12, 2016 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | A61P 3/06 20180101; A61P 5/48 20180101; A61P 3/04 20180101; A61P 25/28 20180101; C07K 2317/34 20130101; A61P 5/00 20180101; A61P 3/00 20180101; A61P 15/00 20180101; A61P 43/00 20180101; A61P 5/50 20180101; C07K 2317/33 20130101; A61P 5/02 20180101; C07K 2317/565 20130101; A61K 2039/505 20130101; C07K 2317/21 20130101; A61K 39/395 20130101; C07K 16/2869 20130101; A61P 3/10 20180101; C07K 2317/75 20130101; C07K 2317/92 20130101 |
| International Class: | C07K 16/28 20060101 C07K016/28; A61K 39/395 20060101 A61K039/395 |
Claims
1-14.(canceled)
15. A method for treating a disease or condition associated with or caused by a signaling-defective or signaling-impaired leptin receptor (LEPR) mutation in a subject in need thereof, the method comprising administering, to the subject, a pharmaceutical composition comprising a pharmaceutically acceptable carrier or diluent and an antibody or antigen-binding fragment thereof that binds LEPR and activates LEPR signaling which comprises a light chain variable region that comprises the LCDRs of a light chain variable region comprising the amino acid sequence set forth in SEQ ID NO: 10; and a heavy chain variable region that comprises the HCDRs of a heavy chain variable region comprising the amino acid sequence set forth in SEQ ID NO: 2, 26 or 34.
16. The method of claim 15, wherein the signaling-defective or signaling impaired LEPR mutation is LEPR-A409E or LEPR-P316T.
17. The method of claim 15, wherein the disease or condition associated with or caused by a signaling-defective or signaling-impaired LEPR mutation is early-onset obesity.
18-24. (canceled)
25. The method of claim 15 wherein the antibody or antigen-binding fragment comprises a heavy chain variable region that comprises a HCDR1 that comprises the amino acid sequence set forth in SEQ ID NO: 28, a HCDR2 that comprises the amino acid sequence set forth in SEQ ID NO: 30, and a HCDR3 that comprises the amino acid sequence set forth in SEQ ID NO: 32; and a light chain variable region that comprises a LCDR1 that comprises the amino acid sequence set forth in SEQ ID NO: 12, a LCDR2 that comprises the amino acid sequence set forth in SEQ ID NO: 14, and a LCDR3 that comprises the amino acid sequence set forth in SEQ ID NO: 16.
26. The method of claim 15 wherein the antibody or antigen-binding fragment thereof comprises: a light chain variable region that comprises the amino acid sequence set forth in SEQ ID NO: 10; and a heavy chain variable region that comprises the amino acid sequence set forth in SEQ ID NO: 26.
27. The method of claim 26 wherein the antibody or antigen-binding fragment thereof is an antibody.
28. The method of claim 27 wherein the heavy chain variable region is linked to a human heavy chain constant domain; and the light chain variable region is linked to a human light chain constant domain.
29. The method of claim 28 wherein the disease or condition is lipodystrophy.
30. The method of claim 29 wherein the antibody is administered subcutaneously.
31. The method of claim 28 wherein the disease or condition is obesity.
32. The method of claim 31 wherein the antibody is administered subcutaneously.
33. The method of claim 28 wherein the disease or condition is congenital generalized lipodystrophy.
34. The method of claim 28 wherein the disease or condition is acquired generalized lipodystrophy.
35. The method of claim 28 wherein the disease or condition is familial partial lipodystrophy.
36. The method of claim 28 wherein the disease or condition is acquired partial lipodystrophy.
37. The method of claim 28 wherein the disease or condition is abdominal lipodystrophy.
38. The method of claim 28 wherein the disease or condition is lipoatrophia annularis.
39. The method of claim 28 wherein the disease or condition is localized lipodystrophy.
40. The method of claim 28 wherein the disease or condition is HIV-associated lipodystrophy.
41. A method for treating a disease or condition associated with or caused by a signaling-defective or signaling-impaired leptin receptor (LEPR) mutation in a subject in need thereof, the method comprising administering, to the subject, a pharmaceutical composition comprising a pharmaceutically acceptable carrier or diluent and an antibody or antigen-binding fragment thereof that binds LEPR and activates LEPR signaling and competes for binding to LEPR with an antibody that comprises a light chain variable region that comprises the amino acid sequence set forth in SEQ ID NO: 10; and a heavy chain variable region that comprises the amino acid sequence set forth in SEQ ID NO: 26.
42. A method for treating a disease or condition associated with or caused by a signaling-defective or signaling-impaired leptin receptor (LEPR) mutation in a subject in need thereof, the method comprising administering, to the subject, a pharmaceutical composition comprising a pharmaceutically acceptable carrier or diluent and an antibody or antigen-binding fragment thereof that binds LEPR at the same epitope as an antibody that comprises a light chain variable region that comprises the amino acid sequence set forth in SEQ ID NO: 10; and a heavy chain variable region that comprises the amino acid sequence set forth in SEQ ID NO: 26 and activates LEPR signaling.
Description
[0001] This application is a divisional of U.S. patent application Ser. No. 15/290,967, filed Oct. 11, 2016, which claims the benefit of priority under 35 U.S.C. .sctn. 119(e) of U.S. Provisional Application Nos. 62/240,021, filed Oct. 12, 2015; 62/359,757, filed Jul. 8, 2016; 62/375,495, filed Aug. 16, 2016, and 62/393,143, filed Sep. 12, 2016, the disclosures of which are herein incorporated by reference in their entireties.
SEQUENCE LISTING
[0002] An official copy of the sequence listing is submitted concurrently with the specification electronically via EFS-Web as an ASCII formatted sequence listing with a file name of 10178US02_SEQ_LIST_ST25.TXT, a creation date of Jun. 13, 2018, and a size of about 105 kilobytes. The sequence listing contained in this ASCII formatted document is part of the specification and is herein incorporated by reference in its entirety.
FIELD OF INVENTION
[0003] The present invention is related to antibodies and antigen-binding fragments of antibodies that bind human leptin receptor (LEPR), and therapeutic and diagnostic methods of using those antibodies.
BACKGROUND
[0004] Leptin is a polypeptide hormone predominantly expressed by adipose tissue and is involved in the regulation of metabolism, energy balance and food intake. Leptin activity is mediated by interaction with, and signaling through, the leptin receptor. Leptin receptor, (also known as "LEPR," "WSX," "OB receptor," "OB-R," and "CD295") is a single-pass transmembrane receptor of the class I cytokine receptor family with a large (818 amino acid) extracellular domain. Leptin deficiency, leptin resistance, and certain LEPR signaling-defective/signaling impaired mutations, are associated with obesity, type 2 diabetes, dyslipidemia, lipodystrophies, hepatic steatosis, non-alcoholic and alcoholic fatty liver diseases, severe insulin resistance, Leprechaunism/Donohue syndrome, Rabson-Mendenhall syndrome, and related complications. Therapeutic approaches to address leptin resistance, leptin deficiency, and hypoleptinemia (e.g., lipodystrophy) have mostly focused on the delivery of supplemental leptin or leptin analogues to affected individuals. Such approaches, however, have generally shown limited efficacy, particularly in leptin-resistant individuals, and are frequently associated with adverse side effects. Thus, a need exists in the art for alternative approaches to treating leptin resistance and other conditions associated with leptin deficiency or hypoleptinemia.
BRIEF SUMMARY OF THE INVENTION
[0005] The present invention provides antibodies and antigen-binding fragments thereof that bind human leptin receptor (LEPR). The antibodies of the present invention are agonist antibodies; i.e., binding of the anti-LEPR antibodies of the invention to LEPR causes, inter alia, activation of leptin receptor signaling in cells. In certain embodiments, the antibodies of the present invention do not compete with leptin for binding to LEPR. The antibodies of the present invention are useful, e.g., for mimicking, substituting for, or supplementing the normal biological activity of leptin in a subject. The antibodies and antigen-binding fragments of the present invention are therefore useful in the therapeutic treatment of diseases and disorders associated with leptin resistance and leptin deficiency.
[0006] The antibodies of the invention can be full-length (for example, an IgG1 or IgG4 antibody) or may comprise only an antigen-binding portion (for example, a Fab, F(ab').sub.2 or scFv fragment), and may be modified to affect functionality, e.g., to eliminate residual effector functions (Reddy et al., 2000, J. Immunol. 164:1925-1933).
[0007] Exemplary anti-LEPR antibodies of the present invention are listed in Tables 1 and 2 herein. Table 1 sets forth the amino acid sequence identifiers of the heavy chain variable regions (HCVRs), light chain variable regions (LCVRs), heavy chain complementarity determining regions (HCDR1, HCDR2 and HCDR3), and light chain complementarity determining regions (LCDR1, LCDR2 and LCDR3) of the exemplary anti-LEPR antibodies. Table 2 sets forth the nucleic acid sequence identifiers of the HCVRs, LCVRs, HCDR1, HCDR2 HCDR3, LCDR1, LCDR2 and LCDR3 of the exemplary anti-LEPR antibodies.
[0008] The present invention provides antibodies or antigen-binding fragments thereof that specifically bind LEPR, comprising an HCVR comprising an amino acid sequence selected from any of the HCVR amino acid sequences listed in Table 1, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity thereto.
[0009] The present invention also provides antibodies or antigen-binding fragments thereof that specifically bind LEPR, comprising an LCVR comprising an amino acid sequence selected from any of the LCVR amino acid sequences listed in Table 1, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity thereto.
[0010] The present invention also provides antibodies or antigen-binding fragments thereof that specifically bind LEPR, comprising an HCVR and an LCVR amino acid sequence pair (HCVR/LCVR) comprising any of the HCVR amino acid sequences listed in Table 1 paired with any of the LCVR amino acid sequences listed in Table 1. According to certain embodiments, the present invention provides antibodies, or antigen-binding fragments thereof, comprising an HCVR/LCVR amino acid sequence pair contained within any of the exemplary anti-LEPR antibodies listed in Table 1. In certain embodiments, the HCVR/LCVR amino acid sequence pair is selected from the group consisting of SEQ ID NOs: 2/10, 18/10, 26/10, 34/10, 42/10, 50/10, 58/66, 74/66 and 82/66.
[0011] The present invention also provides antibodies or antigen-binding fragments thereof that specifically bind LEPR, comprising a heavy chain CDR1 (HCDR1) comprising an amino acid sequence selected from any of the HCDR1 amino acid sequences listed in Table 1 or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity.
[0012] The present invention also provides antibodies or antigen-binding fragments thereof that specifically bind LEPR, comprising a heavy chain CDR2 (HCDR2) comprising an amino acid sequence selected from any of the HCDR2 amino acid sequences listed in Table 1 or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity.
[0013] The present invention also provides antibodies or antigen-binding fragments thereof that specifically bind LEPR, comprising a heavy chain CDR3 (HCDR3) comprising an amino acid sequence selected from any of the HCDR3 amino acid sequences listed in Table 1 or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity.
[0014] The present invention also provides antibodies or antigen-binding fragments thereof that specifically bind LEPR, comprising a light chain CDR1 (LCDR1) comprising an amino acid sequence selected from any of the LCDR1 amino acid sequences listed in Table 1 or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity.
[0015] The present invention also provides antibodies or antigen-binding fragments thereof that specifically bind LEPR, comprising a light chain CDR2 (LCDR2) comprising an amino acid sequence selected from any of the LCDR2 amino acid sequences listed in Table 1 or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity.
[0016] The present invention also provides antibodies or antigen-binding fragments thereof that specifically bind LEPR, comprising a light chain CDR3 (LCDR3) comprising an amino acid sequence selected from any of the LCDR3 amino acid sequences listed in Table 1 or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity.
[0017] The present invention also provides antibodies or antigen-binding fragments thereof that specifically bind LEPR, comprising an HCDR3 and an LCDR3 amino acid sequence pair (HCDR3/LCDR3) comprising any of the HCDR3 amino acid sequences listed in Table 1 paired with any of the LCDR3 amino acid sequences listed in Table 1. According to certain embodiments, the present invention provides antibodies, or antigen-binding fragments thereof, comprising an HCDR3/LCDR3 amino acid sequence pair contained within any of the exemplary anti-LEPR antibodies listed in Table 1. In certain embodiments, the HCDR3/LCDR3 amino acid sequence pair is selected from the group consisting of SEQ ID NOs: 8/16, 24/16, 32/16, 40/16, 48/16, 56/16, 64/72, 80/72 and 88/72.
[0018] The present invention also provides antibodies or antigen-binding fragments thereof that specifically bind LEPR, comprising a set of six CDRs (i.e., HCDR1, HCDR2, HCDR3, LCDR1, LCDR2, and LCDR3) contained within any of the exemplary anti-LEPR antibodies listed in Table 1. In certain embodiments, the HCDR1, HCDR2, HCDR3, LCDR1, LCDR2, and LCDR3 amino acid sequences set is selected from the group consisting of SEQ ID NOs: 4, 6, 8, 12, 14, 16; 20, 22, 24, 12, 14, 16; 28, 30, 32, 12, 14, 16; 36, 38, 40, 12, 14, 16; 44, 46, 48, 12, 14, 16; 52, 54, 56, 12, 14, 16; 60, 62, 64, 68, 70, 72; 76, 78, 80, 68, 70, 72; and 84, 86, 88, 68, 70, 72.
[0019] In a related embodiment, the present invention provides antibodies, or antigen-binding fragments thereof that specifically bind LEPR, comprising a set of six CDRs (i.e., HCDR1, HCDR2, HCDR3, LCDR1, LCDR2, and LCDR3) contained within an HCVR/LCVR amino acid sequence pair as defined by any of the exemplary anti-LEPR antibodies listed in Table 1. For example, the present invention includes antibodies or antigen-binding fragments thereof that specifically bind LEPR, comprising the HCDR1, HCDR2, HCDR3, LCDR1, LCDR2, and LCDR3 amino acid sequences set contained within an HCVR/LCVR amino acid sequence pair selected from the group consisting of SEQ ID NOs: 2/10, 18/10, 26/10, 34/10, 42/10, 50/10, 58/66, 74/66 and 82/66. Methods and techniques for identifying CDRs within HCVR and LCVR amino acid sequences are well known in the art and can be used to identify CDRs within the specified HCVR and/or LCVR amino acid sequences disclosed herein. Exemplary conventions that can be used to identify the boundaries of CDRs include, e.g., the Kabat definition, the Chothia definition, and the AbM definition. In general terms, the Kabat definition is based on sequence variability, the Chothia definition is based on the location of the structural loop regions, and the AbM definition is a compromise between the Kabat and Chothia approaches. See, e.g., Kabat, "Sequences of Proteins of Immunological Interest," National Institutes of Health, Bethesda, Md. (1991); Al-Lazikani et al., J. Mol. Biol. 273:927-948 (1997); and Martin et al., Proc. Natl. Acad. Sci. USA 86:9268-9272 (1989). Public databases are also available for identifying CDR sequences within an antibody.
[0020] The present invention also provides nucleic acid molecules encoding anti-LEPR antibodies or portions thereof. For example, the present invention provides nucleic acid molecules encoding any of the HCVR amino acid sequences listed in Table 1; in certain embodiments the nucleic acid molecule comprises a polynucleotide sequence selected from any of the HCVR nucleic acid sequences listed in Table 2, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity thereto.
[0021] The present invention also provides nucleic acid molecules encoding any of the LCVR amino acid sequences listed in Table 1; in certain embodiments the nucleic acid molecule comprises a polynucleotide sequence selected from any of the LCVR nucleic acid sequences listed in Table 2, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity thereto.
[0022] The present invention also provides nucleic acid molecules encoding any of the HCDR1 amino acid sequences listed in Table 1; in certain embodiments the nucleic acid molecule comprises a polynucleotide sequence selected from any of the HCDR1 nucleic acid sequences listed in Table 2, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity thereto.
[0023] The present invention also provides nucleic acid molecules encoding any of the HCDR2 amino acid sequences listed in Table 1; in certain embodiments the nucleic acid molecule comprises a polynucleotide sequence selected from any of the HCDR2 nucleic acid sequences listed in Table 2, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity thereto.
[0024] The present invention also provides nucleic acid molecules encoding any of the HCDR3 amino acid sequences listed in Table 1; in certain embodiments the nucleic acid molecule comprises a polynucleotide sequence selected from any of the HCDR3 nucleic acid sequences listed in Table 2, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity thereto.
[0025] The present invention also provides nucleic acid molecules encoding any of the LCDR1 amino acid sequences listed in Table 1; in certain embodiments the nucleic acid molecule comprises a polynucleotide sequence selected from any of the LCDR1 nucleic acid sequences listed in Table 2, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity thereto.
[0026] The present invention also provides nucleic acid molecules encoding any of the LCDR2 amino acid sequences listed in Table 1; in certain embodiments the nucleic acid molecule comprises a polynucleotide sequence selected from any of the LCDR2 nucleic acid sequences listed in Table 2, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity thereto.
[0027] The present invention also provides nucleic acid molecules encoding any of the LCDR3 amino acid sequences listed in Table 1; in certain embodiments the nucleic acid molecule comprises a polynucleotide sequence selected from any of the LCDR3 nucleic acid sequences listed in Table 2, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity thereto.
[0028] The present invention also provides nucleic acid molecules encoding an HCVR, wherein the HCVR comprises a set of three CDRs (i.e., HCDR1, HCDR2, HCDR3), wherein the HCDR1, HCDR2, HCDR3 amino acid sequence set is as defined by any of the exemplary anti-LEPR antibodies listed in Table 1.
[0029] The present invention also provides nucleic acid molecules encoding an LCVR, wherein the LCVR comprises a set of three CDRs (i.e., LCDR1, LCDR2, LCDR3), wherein the LCDR1, LCDR2, LCDR3 amino acid sequence set is as defined by any of the exemplary anti-LEPR antibodies listed in Table 1.
[0030] The present invention also provides nucleic acid molecules encoding both an HCVR and an LCVR, wherein the HCVR comprises an amino acid sequence of any of the HCVR amino acid sequences listed in Table 1, and wherein the LCVR comprises an amino acid sequence of any of the LCVR amino acid sequences listed in Table 1. In certain embodiments, the nucleic acid molecule comprises a polynucleotide sequence selected from any of the HCVR nucleic acid sequences listed in Table 2, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity thereto, and a polynucleotide sequence selected from any of the LCVR nucleic acid sequences listed in Table 2, or a substantially similar sequence thereof having at least 90%, at least 95%, at least 98% or at least 99% sequence identity thereto. In certain embodiments according to this aspect of the invention, the nucleic acid molecule encodes an HCVR and LCVR, wherein the HCVR and LCVR are both derived from the same anti-LEPR antibody listed in Table 1.
[0031] The present invention also provides recombinant expression vectors capable of expressing a polypeptide comprising a heavy or light chain variable region of an anti-LEPR antibody. For example, the present invention includes recombinant expression vectors comprising any of the nucleic acid molecules mentioned above, i.e., nucleic acid molecules encoding any of the HCVR, LCVR, and/or CDR sequences as set forth in Table 1. Also included within the scope of the present invention are host cells into which such vectors have been introduced, as well as methods of producing the antibodies or portions thereof by culturing the host cells under conditions permitting production of the antibodies or antibody fragments, and recovering the antibodies and antibody fragments so produced.
[0032] In another aspect, the invention provides a pharmaceutical composition comprising a recombinant human antibody or fragment thereof which specifically binds LEPR and a pharmaceutically acceptable carrier. In a related aspect, the invention features a composition which is a combination of an anti-LEPR antibody and a second therapeutic agent. In one embodiment, the second therapeutic agent is any agent that is advantageously combined with an anti-LEPR antibody.
[0033] In yet another aspect, the invention provides therapeutic methods for enhancing or stimulating LEPR signaling using an anti-LEPR antibody or antigen-binding portion of an antibody of the invention. The therapeutic methods according to this aspect of the invention comprise administering a therapeutically effective amount of a pharmaceutical composition comprising an antibody or antigen-binding fragment of an antibody of the invention to a subject in need thereof. The disorder treated is any disease or condition which is improved, ameliorated, inhibited or prevented by stimulating or activating LEPR signaling, or otherwise mimicking the natural activity of leptin in vitro or in vivo.
[0034] Other embodiments will become apparent from a review of the ensuing detailed description.
BRIEF DESCRIPTION OF THE FIGURES
[0035] FIG. 1 depicts the binding of dimeric human LEPR to human Leptin in the presence of increasing concentrations of test anti-LEPR antibodies or control molecules, as measured by ELISA (absorbance at 450 nm).
[0036] FIGS. 2A-2C illustrates the extent of LEPR signaling in HEK293 cells expressing either wild-type LEPR (circles), a signaling-defective LEPR mutant (A409E, squares), or a signaling-impaired LEPR mutant (P316T, triangles). LEPR signaling is expressed as ratio of pSTAT3-Y705/STAT3, measured by densitometry from Western blots prepared from cells treated with increasing concentrations of leptin (FIG. 2A), H4H16650 (FIG. 2B), or H4H16679 (FIG. 2C).
[0037] FIG. 3 shows the average daily food intake of leptin-deficient mice dosed with either an isotype control antibody at 3 mg/kg, or a LEPR antibody selected from H4H16650P2, H4H16679P2, H4H17319P2 or H4H17321P2 at 3 mg/kg.
[0038] FIG. 4 shows the average percent change in body weight of mice dosed with either an isotype control antibody at 3 mg/kg, or a LEPR antibody selected from H4H16650P2, H4H16679P2, H4H17319P2 or H4H17321P2 at 3 mg/kg.
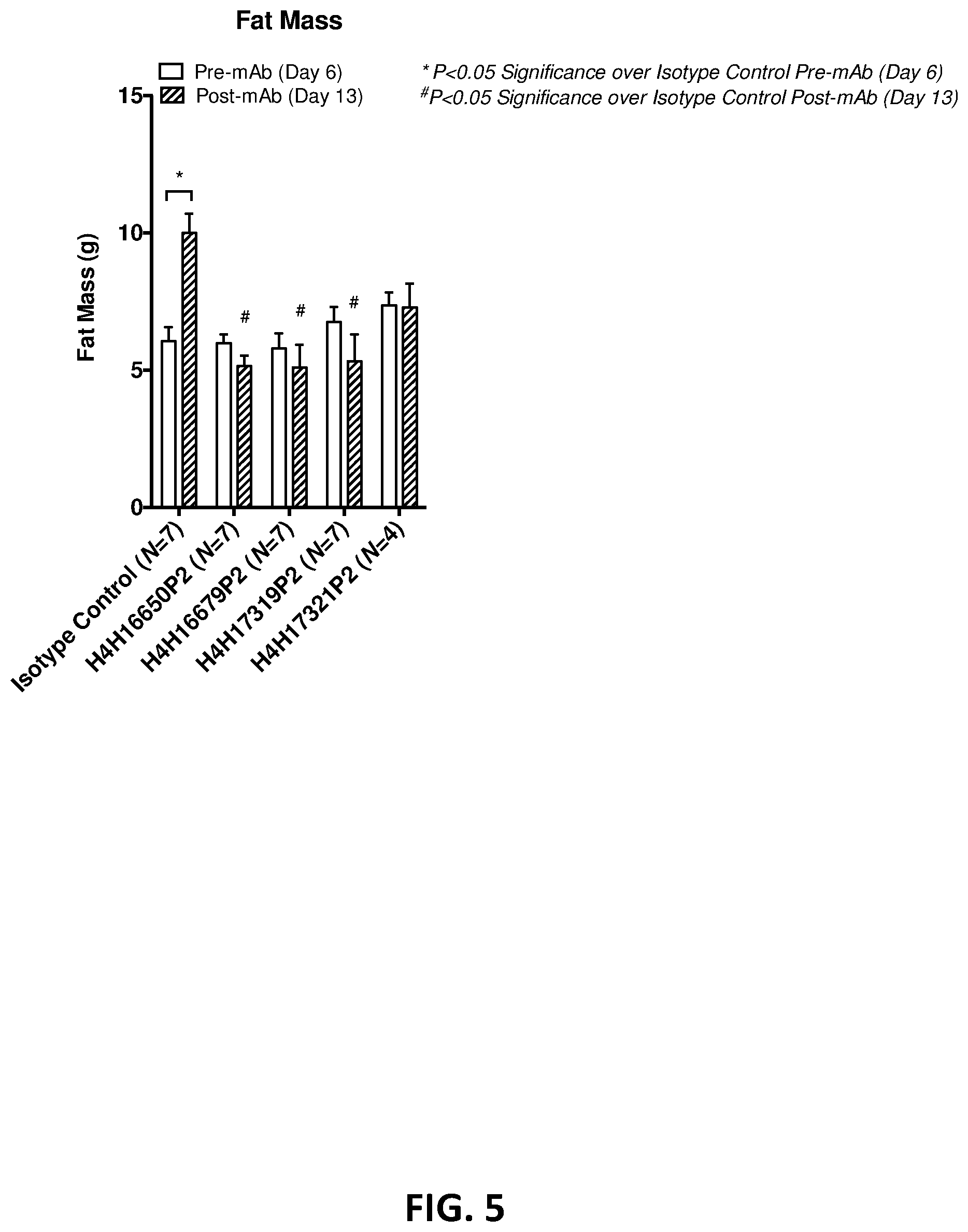
[0039] FIG. 5 shows the average fat mass for animals in each antibody treatment group quantified by .mu.CT 1 day prior to (bars not shaded) and 6 days following antibody treatment (shaded bars) expressed as mean .+-.SEM.
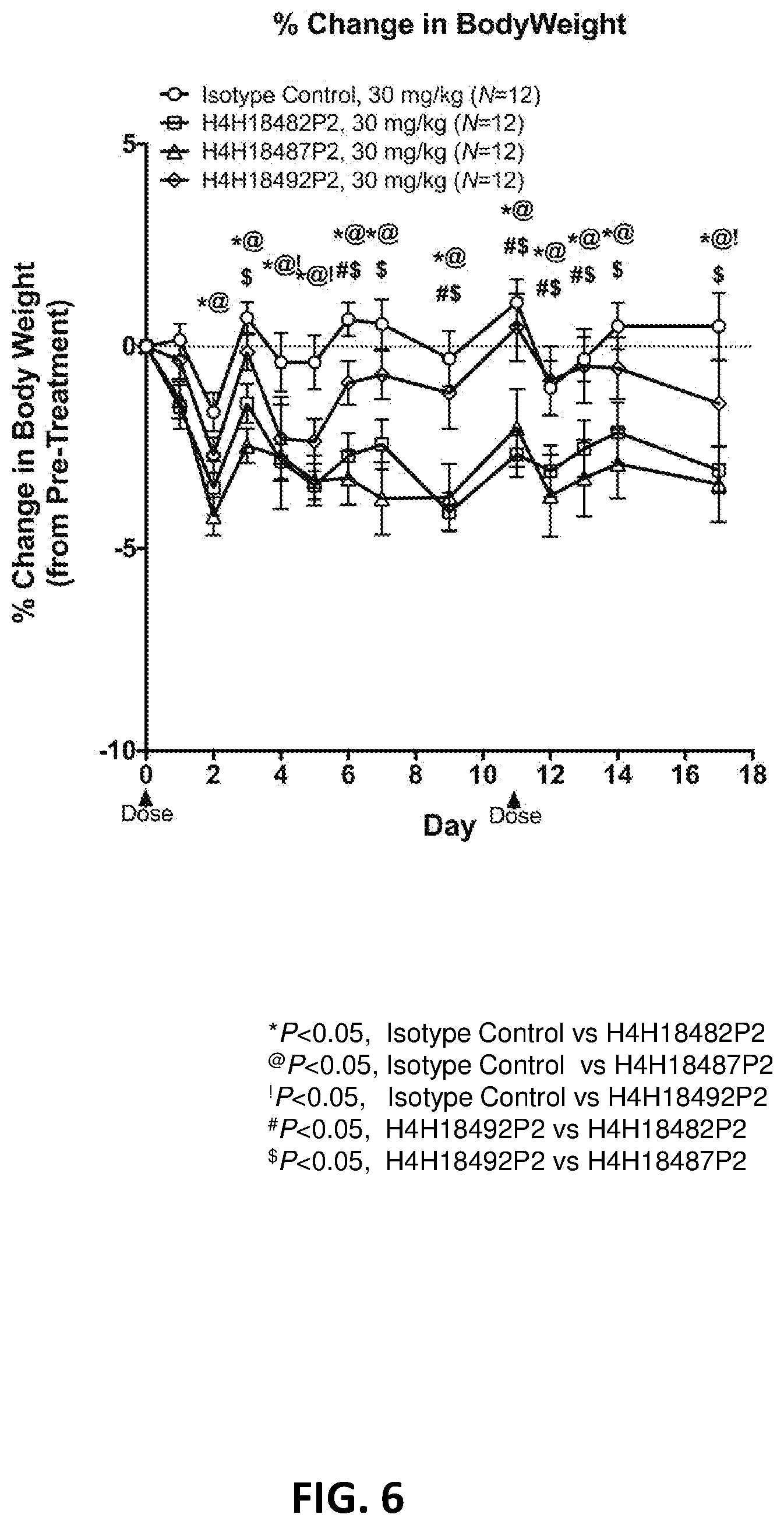
[0040] FIG. 6 shows the percent change in body weight of mice fed 30 mg/kg of an antibody selected from H4H18482P2, H4H18487P2, H4H18492P2 or an isotype control.
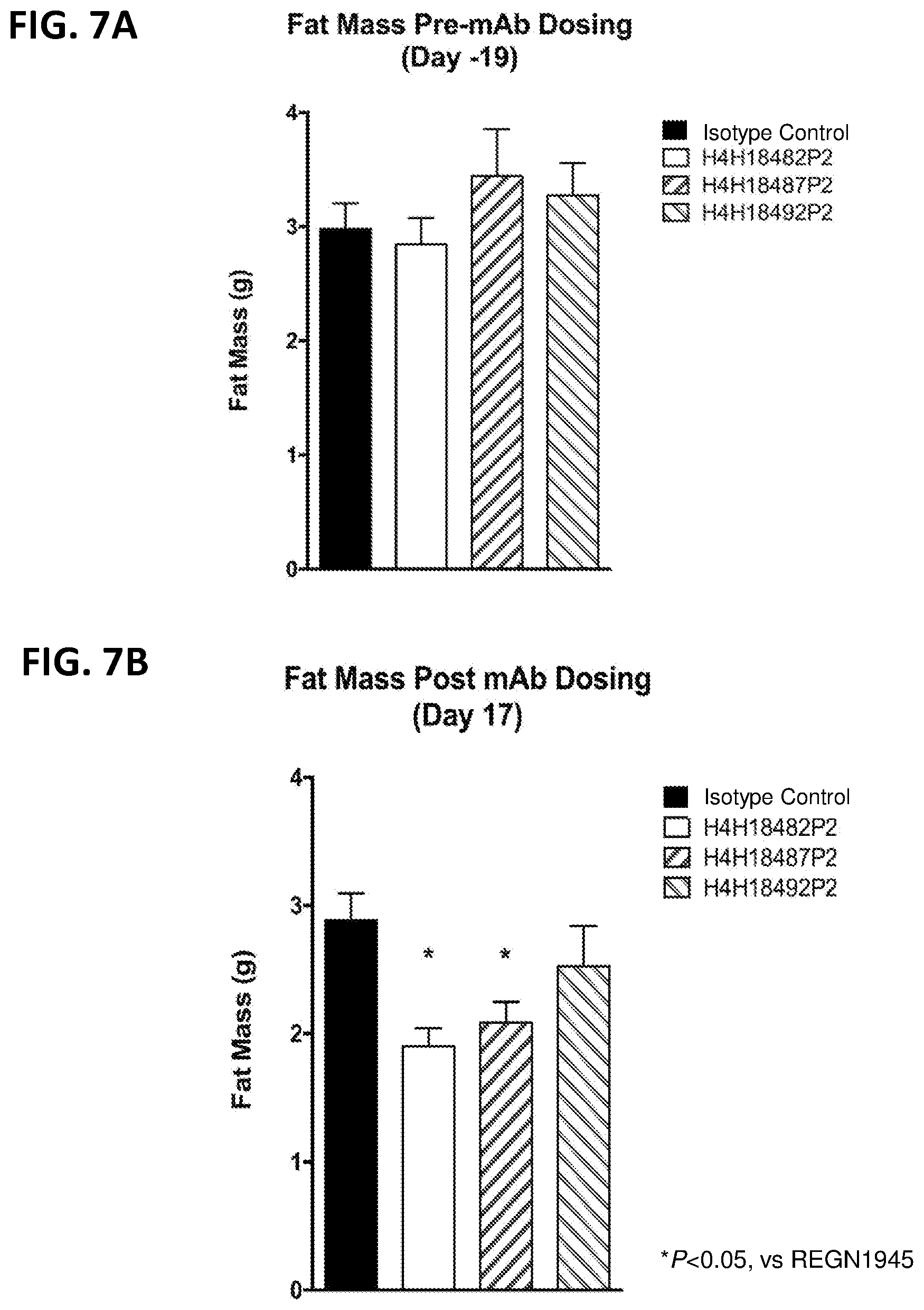
[0041] FIGS. 7A-7B. FIG. 7A shows the fat mass of mice before dosing with anti-LEPR antibodies H4H18482P2, H4H18487P2 or H4H18492P2. FIG. 7B shows the fat mass of mice treated with 30 mg/kg of H4H18482P2, H4H18487P2 or H4H18492P2.
[0042] FIG. 8. FIG. 8 shows that anti-LEPR antibodies tested activated monkey (Mf) LEPR in an IMR-32/STAT3-luc/Mf LEPR cell line.
DETAILED DESCRIPTION OF THE INVENTION
[0043] Before the present invention is described, it is to be understood that this invention is not limited to particular methods and experimental conditions described, as such methods and conditions may vary. It is also to be understood that the terminology used herein is for the purpose of describing particular embodiments only, and is not intended to be limiting, since the scope of the present invention will be limited only by the appended claims.
[0044] Unless defined otherwise, all technical and scientific terms used herein have the same meaning as commonly understood by one of ordinary skill in the art to which this invention belongs. As used herein, the term "about," when used in reference to a particular recited numerical value, means that the value may vary from the recited value by no more than 1%. For example, as used herein, the expression "about 100" includes 99 and 101 and all values in between (e.g., 99.1, 99.2, 99.3, 99.4, etc.).
[0045] Although any methods and materials similar or equivalent to those described herein can be used in the practice or testing of the present invention, the preferred methods and materials are now described. All patents, applications and non-patent publications mentioned in this specification are incorporated herein by reference in their entireties.
DEFINITIONS
[0046] The expression "leptin receptor," "LEPR," and the like, as used herein, refers to the human leptin receptor, comprising the amino acid sequence as set forth in SEQ ID NO:113 (see also UniProtKB/Swiss-Prot Accession No. P48357). Alternative names for LEPR used in the scientific literature include "OB receptor," "OB-R," and "CD295." LEPR is also referred to as "WSX" (see, e.g., U.S. Pat. No. 7,524,937). The expression "LEPR" includes both monomeric and multimeric (e.g., dimeric) LEPR molecules. As used herein, the expression "monomeric human LEPR" means a LEPR protein or portion thereof that does not contain or possess any multimerizing domains and that exists under normal conditions as a single LEPR molecule without a direct physical connection to another LEPR molecule. An exemplary monomeric LEPR molecule is the molecule referred to herein as "hLEPR.mmh" comprising the amino acid sequence of SEQ ID NO:114 (see, e.g., Example 3, herein). As used herein, the expression "dimeric human LEPR" means a construct comprising two LEPR molecules connected to one another through a linker, covalent bond, non-covalent bond, or through a multimerizing domain such as an antibody Fc domain. An exemplary dimeric LEPR molecule is the molecule referred to herein as "hLEPR.mFc" comprising the amino acid sequence of SEQ ID NO:115 (see, e.g., Example 3, herein), or the molecule referred to herein as "hLEPR.hFc" comprising the amino acid sequence of SEQ ID NO:116. As used herein, expressions such "anti-LEPR antibody," "antibody that specifically binds LEPR," "LEPR-specific binding protein," and the like, unless specifically indicated otherwise, refer to molecules that bind full length human LEPR, monomeric human LEPR, dimeric human LEPR, or other constructs that comprise or consist of the LEPR extracellular domain.
[0047] All references to proteins, polypeptides and protein fragments herein are intended to refer to the human version of the respective protein, polypeptide or protein fragment unless explicitly specified as being from a non-human species. Thus, the expression "LEPR" means human LEPR unless specified as being from a non-human species, e.g., "mouse LEPR," "monkey LEPR," etc.
[0048] As used herein, the expression "cell surface-expressed LEPR" means one or more LEPR protein(s), or the extracellular domain thereof, that is/are expressed on the surface of a cell in vitro or in vivo, such that at least a portion of a LEPR protein is exposed to the extracellular side of the cell membrane and is accessible to an antigen-binding portion of an antibody. A "cell surface-expressed LEPR" can comprise or consist of a LEPR protein expressed on the surface of a cell which normally (e.g., in the native or wild-type state) expresses LEPR protein. Alternatively, "cell surface-expressed LEPR" can comprise or consist of LEPR protein expressed on the surface of a cell that normally does not express human LEPR on its surface but has been artificially engineered to express LEPR on its surface.
[0049] As used herein, the expressions such as "anti-LEPR antibody," or "antibody that binds human leptin receptor," include both monovalent antibodies with a single specificity, as well as bispecific antibodies comprising a first arm that binds LEPR and a second arm that binds a second (target) antigen, wherein the anti-LEPR arm comprises any of the HCVR/LCVR or CDR sequences as set forth in Table 1 herein.
[0050] The term "antibody", as used herein, means any antigen-binding molecule or molecular complex comprising at least one complementarity determining region (CDR) that specifically binds to or interacts with a particular antigen (e.g., LEPR). The term "antibody" includes immunoglobulin molecules comprising four polypeptide chains, two heavy (H) chains and two light (L) chains inter-connected by disulfide bonds, as well as multimers thereof (e.g., IgM). Each heavy chain comprises a heavy chain variable region (abbreviated herein as HCVR or V.sub.H) and a heavy chain constant region. The heavy chain constant region comprises three domains, C.sub.H1, C.sub.H2 and C.sub.H3. Each light chain comprises a light chain variable region (abbreviated herein as LCVR or V.sub.L) and a light chain constant region. The light chain constant region comprises one domain (C.sub.L1). The V.sub.H and V.sub.L regions can be further subdivided into regions of hypervariability, termed complementarity determining regions (CDRs), interspersed with regions that are more conserved, termed framework regions (FR). Each V.sub.H and V.sub.L is composed of three CDRs and four FRs, arranged from amino-terminus to carboxy-terminus in the following order: FR1, CDR1, FR2, CDR2, FR3, CDR3, FR4. In different embodiments of the invention, the FRs of the anti-LEPR antibody (or antigen-binding portion thereof) may be identical to the human germline sequences, or may be naturally or artificially modified. An amino acid consensus sequence may be defined based on a side-by-side analysis of two or more CDRs.
[0051] The term "antibody", as used herein, also includes antigen-binding fragments of full antibody molecules. The terms "antigen-binding portion" of an antibody, "antigen-binding fragment" of an antibody, and the like, as used herein, include any naturally occurring, enzymatically obtainable, synthetic, or genetically engineered polypeptide or glycoprotein that specifically binds an antigen to form a complex. Antigen-binding fragments of an antibody may be derived, e.g., from full antibody molecules using any suitable standard techniques such as proteolytic digestion or recombinant genetic engineering techniques involving the manipulation and expression of DNA encoding antibody variable and optionally constant domains. Such DNA is known and/or is readily available from, e.g., commercial sources, DNA libraries (including, e.g., phage-antibody libraries), or can be synthesized. The DNA may be sequenced and manipulated chemically or by using molecular biology techniques, for example, to arrange one or more variable and/or constant domains into a suitable configuration, or to introduce codons, create cysteine residues, modify, add or delete amino acids, etc.
[0052] Non-limiting examples of antigen-binding fragments include: (i) Fab fragments; (ii) F(ab')2 fragments; (iii) Fd fragments; (iv) Fv fragments; (v) single-chain Fv (scFv) molecules; (vi) dAb fragments; and (vii) minimal recognition units consisting of the amino acid residues that mimic the hypervariable region of an antibody (e.g., an isolated complementarity determining region (CDR) such as a CDR3 peptide), or a constrained FR3-CDR3-FR4 peptide. Other engineered molecules, such as domain-specific antibodies, single domain antibodies, domain-deleted antibodies, chimeric antibodies, CDR-grafted antibodies, diabodies, triabodies, tetrabodies, minibodies, nanobodies (e.g. monovalent nanobodies, bivalent nanobodies, etc.), small modular immunopharmaceuticals (SMIPs), and shark variable IgNAR domains, are also encompassed within the expression "antigen-binding fragment," as used herein.
[0053] An antigen-binding fragment of an antibody will typically comprise at least one variable domain. The variable domain may be of any size or amino acid composition and will generally comprise at least one CDR which is adjacent to or in frame with one or more framework sequences. In antigen-binding fragments having a V.sub.H domain associated with a V.sub.L domain, the V.sub.H and V.sub.L domains may be situated relative to one another in any suitable arrangement. For example, the variable region may be dimeric and contain V.sub.H-V.sub.H, V.sub.H-V.sub.L or V.sub.L-V.sub.L dimers. Alternatively, the antigen-binding fragment of an antibody may contain a monomeric V.sub.H or V.sub.L domain.
[0054] In certain embodiments, an antigen-binding fragment of an antibody may contain at least one variable domain covalently linked to at least one constant domain. Non-limiting, exemplary configurations of variable and constant domains that may be found within an antigen-binding fragment of an antibody of the present invention include: (i) V.sub.H-C.sub.H1; (ii) V.sub.H-C.sub.H2; (iii) V.sub.H-C.sub.H3; (iv) V.sub.H-C.sub.H1-C.sub.H2; (v) V.sub.H-C.sub.H1-C.sub.H2-C.sub.H3; (vi) V.sub.H-C.sub.H2-C.sub.H3; (vii) V.sub.H-C.sub.L; (viii) V.sub.L-C.sub.H1; (ix) V.sub.L-C.sub.H2; (x) V.sub.L-C.sub.H3; (xi) V.sub.L-C.sub.H1-C.sub.H2; (xii) V.sub.L-C.sub.H1-C.sub.H2-C.sub.H3; (xiii) V.sub.L-C.sub.H2-C.sub.H3; and (xiv) V.sub.L-C.sub.L. In any configuration of variable and constant domains, including any of the exemplary configurations listed above, the variable and constant domains may be either directly linked to one another or may be linked by a full or partial hinge or linker region. A hinge region may consist of at least 2 (e.g., 5, 10, 15, 20, 40, 60 or more) amino acids which result in a flexible or semi-flexible linkage between adjacent variable and/or constant domains in a single polypeptide molecule. Moreover, an antigen-binding fragment of an antibody of the present invention may comprise a homo-dimer or hetero-dimer (or other multimer) of any of the variable and constant domain configurations listed above in non-covalent association with one another and/or with one or more monomeric V.sub.H or V.sub.L domain (e.g., by disulfide bond(s)).
[0055] As with full antibody molecules, antigen-binding fragments may be monospecific or multispecific (e.g., bispecific). A multispecific antigen-binding fragment of an antibody will typically comprise at least two different variable domains, wherein each variable domain is capable of specifically binding to a separate antigen or to a different epitope on the same antigen. Any multispecific antibody format, including the exemplary bispecific antibody formats disclosed herein, may be adapted for use in the context of an antigen-binding fragment of an antibody of the present invention using routine techniques available in the art.
[0056] In certain embodiments of the invention, the anti-LEPR antibodies of the invention are human antibodies. The term "human antibody", as used herein, is intended to include antibodies having variable and constant regions derived from human germline immunoglobulin sequences. The human antibodies of the invention may include amino acid residues not encoded by human germline immunoglobulin sequences (e.g., mutations introduced by random or site-specific mutagenesis in vitro or by somatic mutation in vivo), for example in the CDRs and in particular CDR3. However, the term "human antibody", as used herein, is not intended to include antibodies in which CDR sequences derived from the germline of another mammalian species, such as a mouse, have been grafted onto human framework sequences.
[0057] The antibodies of the invention may, in some embodiments, be recombinant human antibodies. The term "recombinant human antibody", as used herein, is intended to include all human antibodies that are prepared, expressed, created or isolated by recombinant means, such as antibodies expressed using a recombinant expression vector transfected into a host cell (described further below), antibodies isolated from a recombinant, combinatorial human antibody library (described further below), antibodies isolated from an animal (e.g., a mouse) that is transgenic for human immunoglobulin genes (see e.g., Taylor et al. (1992) Nucl. Acids Res. 20:6287-6295) or antibodies prepared, expressed, created or isolated by any other means that involves splicing of human immunoglobulin gene sequences to other DNA sequences. Such recombinant human antibodies have variable and constant regions derived from human germline immunoglobulin sequences. In certain embodiments, however, such recombinant human antibodies are subjected to in vitro mutagenesis (or, when an animal transgenic for human Ig sequences is used, in vivo somatic mutagenesis) and thus the amino acid sequences of the V.sub.H and V.sub.L regions of the recombinant antibodies are sequences that, while derived from and related to human germline V.sub.H and V.sub.L sequences, may not naturally exist within the human antibody germline repertoire in vivo.
[0058] The present invention encompasses antibodies having one or more mutations in the hinge, C.sub.H2 or C.sub.H3 region which may be desirable, for example, in production, to improve the yield of the desired antibody form.
[0059] The antibodies of the invention may be isolated antibodies. An "isolated antibody," as used herein, means an antibody that has been identified and separated and/or recovered from at least one component of its natural environment. For example, an antibody that has been separated or removed from at least one component of an organism, or from a tissue or cell in which the antibody naturally exists or is naturally produced, is an "isolated antibody" for purposes of the present invention. An isolated antibody also includes an antibody in situ within a recombinant cell. Isolated antibodies are antibodies that have been subjected to at least one purification or isolation step. According to certain embodiments, an isolated antibody may be substantially free of other cellular material and/or chemicals.
[0060] The present invention includes variants of the anti-LEPR antibodies disclosed herein comprising one or more amino acid substitutions, insertions and/or deletions in the framework and/or CDR regions of the heavy and light chain variable domains as compared to the corresponding germline sequences from which the antibodies were derived. Such mutations can be readily ascertained by comparing the amino acid sequences disclosed herein to germline sequences available from, for example, public antibody sequence databases. The present invention includes antibodies, and antigen-binding fragments thereof, which are derived from any of the amino acid sequences disclosed herein, wherein one or more amino acids within one or more framework and/or CDR regions are mutated to the corresponding residue(s) of the germline sequence from which the antibody was derived, or to the corresponding residue(s) of another human germline sequence, or to a conservative amino acid substitution of the corresponding germline residue(s) (such sequence changes are referred to herein collectively as "germline mutations"). A person of ordinary skill in the art, starting with the heavy and light chain variable region sequences disclosed herein, can easily produce numerous antibodies and antigen-binding fragments which comprise one or more individual germline mutations or combinations thereof. In certain embodiments, all of the framework and/or CDR residues within the V.sub.H and/or V.sub.L domains are mutated back to the residues found in the original germline sequence from which the antibody was derived. In other embodiments, only certain residues are mutated back to the original germline sequence, e.g., only the mutated residues found within the first 8 amino acids of FR1 or within the last 8 amino acids of FR4, or only the mutated residues found within CDR1, CDR2 or CDR3. In other embodiments, one or more of the framework and/or CDR residue(s) are mutated to the corresponding residue(s) of a different germline sequence (i.e., a germline sequence that is different from the germline sequence from which the antibody was originally derived). Furthermore, the antibodies of the present invention may contain any combination of two or more germline mutations within the framework and/or CDR regions, e.g., wherein certain individual residues are mutated to the corresponding residue of a particular germline sequence while certain other residues that differ from the original germline sequence are maintained or are mutated to the corresponding residue of a different germline sequence. Once obtained, antibodies and antigen-binding fragments that contain one or more germline mutations can be easily tested for one or more desired property such as, improved binding specificity, increased binding affinity, improved or enhanced antagonistic or agonistic biological properties (as the case may be), reduced immunogenicity, etc. Antibodies and antigen-binding fragments obtained in this general manner are encompassed within the present invention.
[0061] The present invention includes anti-LEPR antibodies and antigen-binding fragments thereof that comprise amino acid sequences that are substantially similar or substantially identical to one or more variable domain or CDR amino acid sequences as found in any of the exemplary anti-LEPR antibodies disclosed herein.
[0062] As applied to polypeptides, the term "substantial similarity" or "substantially similar" means that two peptide sequences, when optimally aligned, such as by the programs GAP or BESTFIT using default gap weights, share at least 95% sequence identity, even more preferably at least 98% or 99% sequence identity. Preferably, residue positions which are not identical differ by conservative amino acid substitutions. A "conservative amino acid substitution" is one in which an amino acid residue is substituted by another amino acid residue having a side chain (R group) with similar chemical properties (e.g., charge or hydrophobicity). In general, a conservative amino acid substitution will not substantially change the functional properties of a protein. In cases where two or more amino acid sequences differ from each other by conservative substitutions, the percent sequence identity or degree of similarity may be adjusted upwards to correct for the conservative nature of the substitution. Means for making this adjustment are well-known to those of skill in the art. See, e.g., Pearson (1994) Methods Mol. Biol. 24: 307-331, herein incorporated by reference. Examples of groups of amino acids that have side chains with similar chemical properties include (1) aliphatic side chains: glycine, alanine, valine, leucine and isoleucine; (2) aliphatic-hydroxyl side chains: serine and threonine; (3) amide-containing side chains: asparagine and glutamine; (4) aromatic side chains: phenylalanine, tyrosine, and tryptophan; (5) basic side chains: lysine, arginine, and histidine; (6) acidic side chains: aspartate and glutamate, and (7) sulfur-containing side chains are cysteine and methionine. Preferred conservative amino acids substitution groups are: valine-leucine-isoleucine, phenylalanine-tyrosine, lysine-arginine, alanine-valine, glutamate-aspartate, and asparagine-glutamine. Alternatively, a conservative replacement is any change having a positive value in the PAM250 log-likelihood matrix disclosed in Gonnet et al. (1992) Science 256: 1443-1445, herein incorporated by reference. A "moderately conservative" replacement is any change having a nonnegative value in the PAM250 log-likelihood matrix.
[0063] Sequence similarity for polypeptides, which is also referred to as sequence identity, is typically measured using sequence analysis software. Protein analysis software matches similar sequences using measures of similarity assigned to various substitutions, deletions and other modifications, including conservative amino acid substitutions. For instance, GCG software contains programs such as Gap and Bestfit which can be used with default parameters to determine sequence homology or sequence identity between closely related polypeptides, such as homologous polypeptides from different species of organisms or between a wild type protein and a mutein thereof. See, e.g., GCG Version 6.1. Polypeptide sequences also can be compared using FASTA using default or recommended parameters, a program in GCG Version 6.1. FASTA (e.g., FASTA2 and FASTA3) provides alignments and percent sequence identity of the regions of the best overlap between the query and search sequences (Pearson (2000) supra). Another preferred algorithm when comparing a sequence of the invention to a database containing a large number of sequences from different organisms is the computer program BLAST, especially BLASTP or TBLASTN, using default parameters. See, e.g., Altschul et al. (1990) J. Mol. Biol. 215:403-410 and Altschul et al. (1997) Nucleic Acids Res. 25:3389-402, each herein incorporated by reference.
Anti-LEPR Antibodies Comprising Fc Variants
[0064] According to certain embodiments of the present invention, anti-LEPR antibodies are provided comprising an Fc domain comprising one or more mutations which enhance or diminish antibody binding to the FcRn receptor, e.g., at acidic pH as compared to neutral pH. For example, the present invention includes anti-LEPR antibodies comprising a mutation in the C.sub.H2 or a C.sub.H3 region of the Fc domain, wherein the mutation(s) increases the affinity of the Fc domain to FcRn in an acidic environment (e.g., in an endosome where pH ranges from about 5.5 to about 6.0). Such mutations may result in an increase in serum half-life of the antibody when administered to an animal. Non-limiting examples of such Fc modifications include, e.g., a modification at position 250 (e.g., E or Q); 250 and 428 (e.g., L or F); 252 (e.g., L/Y/F/W or T), 254 (e.g., S or T), and 256 (e.g., S/R/Q/E/D or T); or a modification at position 428 and/or 433 (e.g., H/L/R/S/P/Q or K) and/or 434 (e.g., H/F or Y); or a modification at position 250 and/or 428; or a modification at position 307 or 308 (e.g., 308F, V308F), and 434. In one embodiment, the modification comprises a 428L (e.g., M428L) and 434S (e.g., N434S) modification; a 428L, 259I (e.g., V259I), and 308F (e.g., V308F) modification; a 433K (e.g., H433K) and a 434 (e.g., 434Y) modification; a 252, 254, and 256 (e.g., 252Y, 254T, and 256E) modification; a 250Q and 428L modification (e.g., T250Q and M428L); and a 307 and/or 308 modification (e.g., 308F or 308P).
[0065] For example, the present invention includes anti-LEPR antibodies comprising an Fc domain comprising one or more pairs or groups of mutations selected from the group consisting of: 250Q and 248L (e.g., T250Q and M248L); 252Y, 254T and 256E (e.g., M252Y, S254T and T256E); 428L and 434S (e.g., M428L and N434S); and 433K and 434F (e.g., H433K and N434F). All possible combinations of the foregoing Fc domain mutations, and other mutations within the antibody variable domains disclosed herein, are contemplated within the scope of the present invention.
[0066] The anti-LEPR antibodies of the present invention may comprise a modified Fc domain having reduced effector function. As used herein, a "modified Fc domain having reduced effector function" means any Fc portion of an immunoglobulin that has been modified, mutated, truncated, etc., relative to a wild-type, naturally occurring Fc domain such that a molecule comprising the modified Fc exhibits a reduction in the severity or extent of at least one effect selected from the group consisting of cell killing (e.g., ADCC and/or CDC), complement activation, phagocytosis and opsonization, relative to a comparator molecule comprising the wild-type, naturally occurring version of the Fc portion. In certain embodiments, a "modified Fc domain having reduced effector function" is an Fc domain with reduced or attenuated binding to an Fc receptor (e.g., Fc.gamma.R).
[0067] In certain embodiments of the present invention, the modified Fc domain is a variant IgG1 Fc or a variant IgG4 Fc comprising a substitution in the hinge region. For example, a modified Fc for use in the context of the present invention may comprise a variant IgG1 Fc wherein at least one amino acid of the IgG1 Fc hinge region is replaced with the corresponding amino acid from the IgG2 Fc hinge region. Alternatively, a modified Fc for use in the context of the present invention may comprise a variant IgG4 Fc wherein at least one amino acid of the IgG4 Fc hinge region is replaced with the corresponding amino acid from the IgG2 Fc hinge region. Non-limiting, exemplary modified Fc regions that can be used in the context of the present invention are set forth in U.S. Patent Application Publication No. 2014/0243504, the disclosure of which is hereby incorporated by reference in its entirety, as well as any functionally equivalent variants of the modified Fc regions set forth therein.
[0068] Other modified Fc domains and Fc modifications that can be used in the context of the present invention include any of the modifications as set forth in U.S. 2014/0171623; U.S. Pat. No. 8,697,396; U.S. 2014/0134162; WO 2014/043361, the disclosures of which are hereby incorporated by reference in their entireties. Methods of constructing antibodies or other antigen-binding fusion proteins comprising a modified Fc domain as described herein are known in the art.
Biological Characteristics of the Antibodies
[0069] The present invention includes antibodies and antigen-binding fragments thereof that bind human LEPR and activate LEPR signaling. Such antibodies may be referred to herein as "agonist antibodies." In the context of the present invention, "activation of LEPR signaling" means the stimulation of an intracellular effect that normally results from the interaction of leptin with LEPR in cells that express LEPR. In certain embodiments, "activation of LEPR signaling" means the transcriptional activation of STAT3, which can be detected using any method that can measure or identify, directly or indirectly, STAT3 activity, e.g., using a labeled version of STAT3 expressed in a reporter cell line. For example, the present invention includes antibodies and antigen-binding fragments thereof that activate LEPR signaling in a cell-based reporter assay, e.g., using a cell based assay format as defined in Example 7 herein, or a substantially similar assay. Cell-based reporter assays that detect LEPR activation, such as the assay set forth in Example 7 herein, can produce a detectable signal that may be expressed in terms of an EC.sub.50 value (i.e., the antibody concentration required to produce half-maximal signaling) and/or a percentage of the maximal signaling observed in the presence of leptin. In certain exemplary embodiments of the present invention, anti-LEPR antibodies are provided that activate LEPR signaling with an EC.sub.50 value of less than about 12.0 nM in a cell-based reporter assay, e.g., using an assay format as defined in Example 7 herein, or a substantially similar assay. In certain exemplary embodiments of the present invention, anti-LEPR antibodies are provided that activate LEPR signaling with maximum percent activation relative to leptin signaling of greater than about 65% in a cell-based reporter assay, e.g., using an assay format as defined in Example 7 herein, or a substantially similar assay.
[0070] The present invention includes antibodies and antigen-binding fragments thereof that bind monomeric human LEPR with high affinity. For example, the present invention includes anti-LEPR antibodies that bind monomeric human LEPR (e.g., hLEPR.mmh, SEQ ID NO:114) with a K.sub.D of less than about 150 nM as measured by surface plasmon resonance at 25.degree. C. or 37.degree. C., e.g., using an assay format as defined in Example 3 herein, or a substantially similar assay. According to certain embodiments, anti-LEPR antibodies are provided that bind monomeric human LEPR at 25.degree. C. with a K.sub.D of less than about 150 nM, less than about 140 nM, less than about 130 nM, less than about 120 nM, less than about 110 nM, less than about 100 nM, less than about 90 nM, less than about 80 nM, less than about 70 nM, less than about 60 nM, less than about 50 nM, less than about 40 nM, less than about 30 nM, less than about 20 nM, less than about 10 nM, less than about 9 nM, less than about 8 nM, less than about 7 nM, less than about 6 nM, less than about 5 nM, less than about 4 nM, less than about 3 nM, less than about 2 nM, less than about 1 nM, less than about 900 pM, less than about 800 pM, less than about 700 pM, less than about 600 pM, less than about 500 pM, less than about 400 pM, or less than about 300 pM, as measured by surface plasmon resonance, e.g., using an assay format as defined in Example 3 herein, or a substantially similar assay.
[0071] The present invention also includes antibodies and antigen-binding fragments thereof that bind monomeric human LEPR (e.g., hLEPR.mmh, SEQ ID NO:114) with a dissociative half-life (t1/2) of greater than about 50 minutes as measured by surface plasmon resonance at 25.degree. C. or 37.degree. C., e.g., using an assay format as defined in Example 3 herein, or a substantially similar assay. According to certain embodiments, anti-LEPR antibodies are provided that bind monomeric human LEPR at 25.degree. C. with a t1/2 of greater than about 50 minutes, greater than about 55 minutes, greater than about 60 minutes, greater than about 65 minutes, or longer, as measured by surface plasmon resonance, e.g., using an assay format as defined in Example 3 herein, or a substantially similar assay.
[0072] The present invention also includes antibodies and antigen-binding fragments thereof that bind dimeric human LEPR (e.g., hLEPR.mFc, SEQ ID NO:115) with high affinity. For example, the present invention includes anti-LEPR antibodies that bind dimeric human LEPR with a K.sub.D of less than about 1.5 nM as measured by surface plasmon resonance at 25.degree. C. or 37.degree. C., e.g., using an assay format as defined in Example 3 herein, or a substantially similar assay. According to certain embodiments, anti-LEPR antibodies are provided that bind dimeric human LEPR at 25.degree. C. with a K.sub.D of less than about 150 nM, less than about 130 nM, less than about 110 nM, less than about 80 nM, less than about 70 nM, less than about 60 nM, less than about 50 nM, less than about 40 nM, less than about 30 nM, less than about 20 nM, or less than about 10 nM, as measured by surface plasmon resonance, e.g., using an assay format as defined in Example 3 herein, or a substantially similar assay.
[0073] The present invention also includes antibodies and antigen-binding fragments thereof that bind dimeric human LEPR (e.g., hLEPR.mFc, SEQ ID NO:115) with a dissociative half-life (t1/2) of greater than about 10 minutes as measured by surface plasmon resonance at 25.degree. C. or 37.degree. C., e.g., using an assay format as defined in Example 3 herein, or a substantially similar assay. According to certain embodiments, anti-LEPR antibodies are provided that bind dimeric human LEPR at 25.degree. C. with a t1/2 of greater than about 10, greater than about 15 minutes, greater than about 20 minutes, greater than about 25 minutes, greater than about 30 minutes, greater than about 40 minutes, greater than about 50 minutes, greater than about 60 minutes, greater than about 70 minutes, or longer, as measured by surface plasmon resonance, e.g., using an assay format as defined in Example 3 herein, or a substantially similar assay.
[0074] The present invention also includes antibodies and antigen-binding fragments thereof that bind LEPR in complex with human leptin ("LEPR in complex with human leptin" may also be represented by the expression "leptin:LEPR"). For example, the present invention includes antibodies and antigen-binding fragments thereof that are capable of binding to a pre-formed complex comprising hLEPR and human leptin. That is, according to certain embodiments, the interaction between anti-LEPR antibodies and LEPR is not inhibited by the presence of leptin in complex with LEPR; likewise, the interaction between leptin and LEPR, according to this aspect of the invention, is not inhibited by the presence of an anti-LEPR antibody. An exemplary assay format for determining whether an antibody or antigen-binding fragment thereof binds to LEPR in complex with human leptin is set forth in Example 4 herein.
[0075] Similarly, the present invention also includes antibodies and antigen-binding fragments thereof that bind LEPR and do not block the LEPR:leptin interaction. For example, the present invention includes antibodies and antigen-binding fragments thereof that are capable of binding LEPR, thereby producing an antibody:LEPR complex, wherein the resulting antibody:LEPR complex is capable of interacting with leptin to produce a three-member complex comprising the antibody, LEPR and leptin. An exemplary assay format for determining whether an antibody or antigen-binding fragment thereof is capable of binding LEPR in a manner that does not block or interfere with the interaction between LEPR and leptin is set forth in Example 5 herein.
[0076] The present invention also includes antibodies and antigen-binding fragments thereof that bind cell surface-expressed LEPR in the presence and/or absence of human leptin. Cell surface-expressed LEPR means LEPR or a portion thereof (e.g., an extracellular portion of LEPR) expressed on the surface of a cell, either naturally or in an engineered cell line, such that an antibody or antigen-binding fragment thereof is capable of binding to the LEPR molecule. In certain embodiments, cell surface-expressed LEPR includes recombinant complexes comprising an extracellular domain of LEPR connected to a cell via a tag or anchor (e.g., a GPI anchor as illustrated in Example 6 herein). According to this aspect of the invention, antibodies are provided which are capable of binding cell surface-expressed LEPR in the absence of leptin, and are also capable of binding cell surface-expressed LEPR in the presence of leptin (i.e., under circumstances wherein leptin is capable of binding to cell surface-expressed leptin). That is, according to certain embodiments, the interaction between anti-LEPR antibodies and cell surface-expressed LEPR is not inhibited by the presence of leptin in complex with cell surface-expressed LEPR. Antibodies according to this aspect of the invention are capable of forming a three-member complex on the surface of a cell comprising the antibody, cell surface-expressed LEPR and leptin. An exemplary assay format for determining whether an antibody or antigen-binding fragment thereof is capable of binding cell surface-expressed LEPR in the presence and absence of human leptin is set forth in Example 6 herein.
[0077] The antibodies of the present invention may possess one or more of the aforementioned biological characteristics, or any combination thereof. The foregoing list of biological characteristics of the antibodies of the invention is not intended to be exhaustive. Other biological characteristics of the antibodies of the present invention will be evident to a person of ordinary skill in the art from a review of the present disclosure including the working Examples herein.
Epitope Mapping and Related Technologies
[0078] The present invention also includes anti-LEPR antibodies comprising variants of any of the HCVR, LCVR, and/or CDR amino acid sequences disclosed herein having one or more conservative substitutions. For example, the present invention includes anti-LEPR antibodies having HCVR, LCVR, and/or CDR amino acid sequences with, e.g., 10 or fewer, 8 or fewer, 6 or fewer, 4 or fewer, etc. conservative amino acid substitutions relative to any of the HCVR, LCVR, and/or CDR amino acid sequences set forth in Table 1 herein. In certain embodiments, the present invention provides anti-LEPR antibodies comprising variant HCVR, LCVR and/or CDR amino acid sequences relative to the sequences set forth in Table 1 herein (e.g., comprising conservative amino acid substitutions), wherein such variant antibodies nonetheless exhibit one or more functions and/or properties of the exemplary anti-LEPR antibodies disclosed herein.
[0079] The extracellular domain of human LEPR contains an N-terminal cytokine receptor homology domain (CRH-1), an immunoglobulin-like (Ig) domain, and a second CRH domain (CRH-2) that is referred to as the leptin-binding domain (LBD). (Carpenter et al. (2012) Structure 20:487-97). Furthermore, LEPR shares the greatest homology and similar extracellular domain size and organization with granulocyte colony stimulating factor (GCSF) and glycoprotein 130 (gp13). (Haniu et al. (1998) J Biol Chem 273(44): 28691-699).
[0080] The term "epitope" refers to an antigenic determinant that interacts with a specific antigen binding site in the variable region of an antibody molecule known as a paratope. A single antigen may have more than one epitope. Thus, different antibodies may bind to different areas on an antigen and may have different biological effects. Epitopes may be either conformational or linear. A conformational epitope is produced by spatially juxtaposed amino acids from different segments of the linear polypeptide chain. A linear epitope is one produced by adjacent amino acid residues in a polypeptide chain. In certain circumstance, an epitope may include moieties of saccharides, phosphoryl groups, or sulfonyl groups on the antigen.
[0081] The present invention includes anti-LEPR antibodies that interact with one or more epitopes found within amino acids M1-D839 of human LEPR (SEQ ID NO: 113). As set forth in Example 11, 201 peptides from human LEPR had significantly reduced deuteration uptake when bound to antibody H4H16650P2. The peptides corresponding to amino acids 162-169 (amino acids LYVLPEVL of human LEPR, SEQ ID NO: 113) and 170-191 (amino acids EDSPLVPQKGSF of human LEPR, SEQ ID NO: 113) had slower deuteration rates when bound to H4H16650P2, indicating that this antibody binds at least two human LEPR epitopes having the sequences LYVLPEVL or EDSPLVPQKGSF (amino acids 162-169 or 170-191, respectively of SEQ ID NO: 113).
[0082] The epitope to which the antibodies of the present invention bind may consist of a single contiguous sequence of 3 or more (e.g., 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20 or more) amino acids of a LEPR protein. Alternatively, the epitope may consist of a plurality of non-contiguous amino acids (or amino acid sequences) of LEPR. In some embodiments, the epitope is located on or near the leptin-binding domain of LEPR. In other embodiments, the epitope is located at a region distinct from the leptin-binding domain of LEPR, e.g., at a location on the surface of LEPR at which an antibody, when bound to such an epitope, does not interfere with leptin binding to LEPR.
[0083] Various techniques known to persons of ordinary skill in the art can be used to identify the amino acids within an epitope recognized by a particular antibody. Exemplary techniques include, e.g., alanine scanning mutational analysis, peptide blot analysis, and peptide cleavage analysis. In addition, methods such as epitope excision, epitope extraction and chemical modification of antigens can be employed (Tomer, 2000, Protein Science 9:487-496). Another method that can be used to identify the amino acids within a polypeptide with which an antibody interacts is hydrogen/deuterium exchange detected by mass spectrometry. In general terms, the hydrogen/deuterium exchange method involves deuterium-labeling the protein of interest, followed by binding the antibody to the deuterium-labeled protein. Next, the protein/antibody complex is transferred to water to allow hydrogen-deuterium exchange to occur at all residues except for the residues protected by the antibody (which remain deuterium-labeled). After dissociation of the antibody, the target protein is subjected to protease cleavage and mass spectrometry analysis, thereby revealing the deuterium-labeled residues which correspond to the specific amino acids with which the antibody interacts. See, e.g., Ehring (1999) Analytical Biochemistry 267(2):252-259; Engen and Smith (2001) Anal. Chem. 73:256A-265A. X-ray crystallography analysis of an antibody in complex with its antigen may also be used to identify the amino acids within a polypeptide with which an antibody interacts.
[0084] The present invention further includes anti-LEPR antibodies that bind to the same epitope as any of the specific exemplary antibodies described herein (e.g. antibodies comprising any of the amino acid sequences as set forth in Table 1 herein). Likewise, the present invention also includes anti-LEPR antibodies that compete for binding to LEPR with any of the specific exemplary antibodies described herein (e.g. antibodies comprising any of the amino acid sequences as set forth in Table 1 herein).
[0085] One can determine whether an antibody binds to the same epitope as, or competes for binding with, a reference anti-LEPR antibody by using routine methods known in the art and exemplified herein. For example, to determine if a test antibody binds to the same epitope as a reference anti-LEPR antibody of the invention, the reference antibody is allowed to bind to a LEPR protein. Next, the ability of a test antibody to bind to the LEPR molecule is assessed. If the test antibody is able to bind to LEPR following saturation binding with the reference anti-LEPR antibody, it can be concluded that the test antibody binds to a different epitope than the reference anti-LEPR antibody. On the other hand, if the test antibody is not able to bind to the LEPR molecule following saturation binding with the reference anti-LEPR antibody, then the test antibody may bind to the same epitope as the epitope bound by the reference anti-LEPR antibody of the invention. Additional routine experimentation (e.g., peptide mutation and binding analyses) can then be carried out to confirm whether the observed lack of binding of the test antibody is in fact due to binding to the same epitope as the reference antibody or if steric blocking (or another phenomenon) is responsible for the lack of observed binding. Experiments of this sort can be performed using ELISA, RIA, Biacore, flow cytometry or any other quantitative or qualitative antibody-binding assay available in the art. In accordance with certain embodiments of the present invention, two antibodies bind to the same (or overlapping) epitope if, e.g., a 1-, 5-, 10-, 20- or 100-fold excess of one antibody inhibits binding of the other by at least 50% but preferably 75%, 90% or even 99% as measured in a competitive binding assay (see, e.g., Junghans et al., Cancer Res. 1990:50:1495-1502). Alternatively, two antibodies are deemed to bind to the same epitope if essentially all amino acid mutations in the antigen that reduce or eliminate binding of one antibody reduce or eliminate binding of the other. Two antibodies are deemed to have "overlapping epitopes" if only a subset of the amino acid mutations that reduce or eliminate binding of one antibody reduce or eliminate binding of the other.
[0086] To determine if an antibody competes for binding (or cross-competes for binding) with a reference anti-LEPR antibody, the above-described binding methodology is performed in two orientations: In a first orientation, the reference antibody is allowed to bind to a LEPR protein under saturating conditions followed by assessment of binding of the test antibody to the LEPR molecule. In a second orientation, the test antibody is allowed to bind to a LEPR molecule under saturating conditions followed by assessment of binding of the reference antibody to the LEPR molecule. If, in both orientations, only the first (saturating) antibody is capable of binding to the LEPR molecule, then it is concluded that the test antibody and the reference antibody compete for binding to LEPR. As will be appreciated by a person of ordinary skill in the art, an antibody that competes for binding with a reference antibody may not necessarily bind to the same epitope as the reference antibody, but may sterically block binding of the reference antibody by binding an overlapping or adjacent epitope.
Preparation of Human Antibodies
[0087] The anti-LEPR antibodies of the present invention can be fully human antibodies. Methods for generating monoclonal antibodies, including fully human monoclonal antibodies are known in the art. Any such known methods can be used in the context of the present invention to make human antibodies that specifically bind to human LEPR.
[0088] Using VELOCIMMUNE.TM. technology, for example, or any other similar known method for generating fully human monoclonal antibodies, high affinity chimeric antibodies to LEPR are initially isolated having a human variable region and a mouse constant region. As in the experimental section below, the antibodies are characterized and selected for desirable characteristics, including affinity, ligand blocking activity, selectivity, epitope, etc. If necessary, mouse constant regions are replaced with a desired human constant region, for example wild-type or modified IgG1 or IgG4, to generate a fully human anti-LEPR antibody. While the constant region selected may vary according to specific use, high affinity antigen-binding and target specificity characteristics reside in the variable region. In certain instances, fully human anti-LEPR antibodies are isolated directly from antigen-positive B cells.
Bioequivalents
[0089] The anti-LEPR antibodies and antibody fragments of the present invention encompass proteins having amino acid sequences that vary from those of the described antibodies but that retain the ability to bind human LEPR. Such variant antibodies and antibody fragments comprise one or more additions, deletions, or substitutions of amino acids when compared to parent sequence, but exhibit biological activity that is essentially equivalent to that of the described antibodies. Likewise, the anti-LEPR antibody-encoding DNA sequences of the present invention encompass sequences that comprise one or more additions, deletions, or substitutions of nucleotides when compared to the disclosed sequence, but that encode an anti-LEPR antibody or antibody fragment that is essentially bioequivalent to an anti-LEPR antibody or antibody fragment of the invention. Examples of such variant amino acid and DNA sequences are discussed above.
[0090] Two antigen-binding proteins, or antibodies, are considered bioequivalent if, for example, they are pharmaceutical equivalents or pharmaceutical alternatives whose rate and extent of absorption do not show a significant difference when administered at the same molar dose under similar experimental conditions, either single does or multiple dose. Some antibodies will be considered equivalents or pharmaceutical alternatives if they are equivalent in the extent of their absorption but not in their rate of absorption and yet may be considered bioequivalent because such differences in the rate of absorption are intentional and are reflected in the labeling, are not essential to the attainment of effective body drug concentrations on, e.g., chronic use, and are considered medically insignificant for the particular drug product studied.
[0091] In one embodiment, two antigen-binding proteins are bioequivalent if there are no clinically meaningful differences in their safety, purity, and potency.
[0092] In one embodiment, two antigen-binding proteins are bioequivalent if a patient can be switched one or more times between the reference product and the biological product without an expected increase in the risk of adverse effects, including a clinically significant change in immunogenicity, or diminished effectiveness, as compared to continued therapy without such switching.
[0093] In one embodiment, two antigen-binding proteins are bioequivalent if they both act by a common mechanism or mechanisms of action for the condition or conditions of use, to the extent that such mechanisms are known.
[0094] Bioequivalence may be demonstrated by in vivo and in vitro methods. Bioequivalence measures include, e.g., (a) an in vivo test in humans or other mammals, in which the concentration of the antibody or its metabolites is measured in blood, plasma, serum, or other biological fluid as a function of time; (b) an in vitro test that has been correlated with and is reasonably predictive of human in vivo bioavailability data; (c) an in vivo test in humans or other mammals in which the appropriate acute pharmacological effect of the antibody (or its target) is measured as a function of time; and (d) in a well-controlled clinical trial that establishes safety, efficacy, or bioavailability or bioequivalence of an antibody.
[0095] Bioequivalent variants of anti-LEPR antibodies of the invention may be constructed by, for example, making various substitutions of residues or sequences or deleting terminal or internal residues or sequences not needed for biological activity. For example, cysteine residues not essential for biological activity can be deleted or replaced with other amino acids to prevent formation of unnecessary or incorrect intramolecular disulfide bridges upon renaturation. In other contexts, bioequivalent antibodies may include anti-LEPR antibody variants comprising amino acid changes which modify the glycosylation characteristics of the antibodies, e.g., mutations which eliminate or remove glycosylation.
Species Selectivity and Species Cross-Reactivity
[0096] The present invention, according to certain embodiments, provides anti-LEPR antibodies that bind to human LEPR but not to LEPR from other species. The present invention also includes anti-LEPR antibodies that bind to human LEPR and to LEPR from one or more non-human species. For example, the anti-LEPR antibodies of the invention may bind to human LEPR and may bind or not bind, as the case may be, to one or more of mouse, rat, guinea pig, hamster, gerbil, pig, cat, dog, rabbit, goat, sheep, cow, horse, camel, cynomologous, marmoset, rhesus or chimpanzee LEPR. According to certain exemplary embodiments of the present invention, anti-LEPR antibodies are provided which specifically bind human LEPR and cynomolgus monkey (e.g., Macaca fascicularis) LEPR. Other anti-LEPR antibodies of the invention bind human LEPR but do not bind, or bind only weakly, to cynomolgus monkey LEPR.
Multispecific Antibodies
[0097] The antibodies of the present invention may be monospecific or multispecific (e.g., bispecific). Multispecific antibodies may be specific for different epitopes of one target polypeptide or may contain antigen-binding domains specific for more than one target polypeptide. See, e.g., Tutt et al., 1991, J. Immunol. 147:60-69; Kufer et al., 2004, Trends Biotechnol. 22:238-244. The anti-LEPR antibodies of the present invention can be linked to or co-expressed with another functional molecule, e.g., another peptide or protein. For example, an antibody or fragment thereof can be functionally linked (e.g., by chemical coupling, genetic fusion, noncovalent association or otherwise) to one or more other molecular entities, such as another antibody or antibody fragment to produce a bi-specific or a multispecific antibody with a second binding specificity.
[0098] The present invention includes bispecific antibodies wherein one arm of an immunoglobulin binds human LEPR, and the other arm of the immunoglobulin is specific for a second antigen. The LEPR-binding arm can comprise any of the HCVR/LCVR or CDR amino acid sequences as set forth in Table 1 herein.
[0099] An exemplary bispecific antibody format that can be used in the context of the present invention involves the use of a first immunoglobulin (Ig) C.sub.H3 domain and a second Ig C.sub.H3 domain, wherein the first and second Ig C.sub.H3 domains differ from one another by at least one amino acid, and wherein at least one amino acid difference reduces binding of the bispecific antibody to Protein A as compared to a bi-specific antibody lacking the amino acid difference. In one embodiment, the first Ig C.sub.H3 domain binds Protein A and the second Ig C.sub.H3 domain contains a mutation that reduces or abolishes Protein A binding such as an H95R modification (by IMGT exon numbering; H435R by EU numbering). The second C.sub.H3 may further comprise a Y96F modification (by IMGT; Y436F by EU). Further modifications that may be found within the second C.sub.H3 include: D16E, L18M, N44S, K52N, V57M, and V82I (by IMGT; D356E, L358M, N384S, K392N, V397M, and V422I by EU) in the case of IgG1 antibodies; N44S, K52N, and V82I (IMGT; N384S, K392N, and V422I by EU) in the case of IgG2 antibodies; and Q15R, N44S, K52N, V57M, R69K, E79Q, and V82I (by IMGT; Q355R, N384S, K392N, V397M, R409K, E419Q, and V422I by EU) in the case of IgG4 antibodies. Variations on the bispecific antibody format described above are contemplated within the scope of the present invention.
[0100] Other exemplary bispecific formats that can be used in the context of the present invention include, without limitation, e.g., scFv-based or diabody bispecific formats, IgG-scFv fusions, dual variable domain (DVD)-Ig, Quadroma, knobs-into-holes, common light chain (e.g., common light chain with knobs-into-holes, etc.), CrossMab, CrossFab, (SEED)body, leucine zipper, Duobody, IgG1/IgG2, dual acting Fab (DAF)-IgG, and Mab.sup.2 bispecific formats (see, e.g., Klein et al. 2012, mAbs 4:6, 1-11, and references cited therein, for a review of the foregoing formats). Bispecific antibodies can also be constructed using peptide/nucleic acid conjugation, e.g., wherein unnatural amino acids with orthogonal chemical reactivity are used to generate site-specific antibody-oligonucleotide conjugates which then self-assemble into multimeric complexes with defined composition, valency and geometry. (See, e.g., Kazane et al., J. Am. Chem. Soc. [Epub: Dec. 4, 2012]).
Therapeutic Formulation and Administration
[0101] The invention provides pharmaceutical compositions comprising the anti-LEPR antibodies or antigen-binding fragments thereof of the present invention. The pharmaceutical compositions of the invention are formulated with suitable carriers, excipients, and other agents that provide improved transfer, delivery, tolerance, and the like. A multitude of appropriate formulations can be found in the formulary known to all pharmaceutical chemists: Remington's Pharmaceutical Sciences, Mack Publishing Company, Easton, Pa. These formulations include, for example, powders, pastes, ointments, jellies, waxes, oils, lipids, lipid (cationic or anionic) containing vesicles (such as LIPOFECTIN.TM., Life Technologies, Carlsbad, Calif.), DNA conjugates, anhydrous absorption pastes, oil-in-water and water-in-oil emulsions, emulsions carbowax (polyethylene glycols of various molecular weights), semi-solid gels, and semi-solid mixtures containing carbowax. See also Powell et al. "Compendium of excipients for parenteral formulations" PDA (1998) J Pharm Sci Technol 52:238-311.
[0102] The dose of antibody administered to a patient may vary depending upon the age and the size of the patient, target disease, conditions, route of administration, and the like. The preferred dose is typically calculated according to body weight or body surface area. In an adult patient, it may be advantageous to intravenously administer the antibody of the present invention normally at a single dose of about 0.01 to about 20 mg/kg body weight, more preferably about 0.02 to about 7, about 0.03 to about 5, or about 0.05 to about 3 mg/kg body weight. Depending on the severity of the condition, the frequency and the duration of the treatment can be adjusted. Effective dosages and schedules for administering anti-LEPR antibodies may be determined empirically; for example, patient progress can be monitored by periodic assessment, and the dose adjusted accordingly. Moreover, interspecies scaling of dosages can be performed using well-known methods in the art (e.g., Mordenti et al., 1991, Pharmaceut. Res. 8:1351).
[0103] Various delivery systems are known and can be used to administer the pharmaceutical composition of the invention, e.g., encapsulation in liposomes, microparticles, microcapsules, recombinant cells capable of expressing the mutant viruses, receptor mediated endocytosis (see, e.g., Wu et al., 1987, J. Biol. Chem. 262:4429-4432). Methods of introduction include, but are not limited to, intradermal, intramuscular, intraperitoneal, intravenous, subcutaneous, intranasal, epidural, and oral routes. The composition may be administered by any convenient route, for example by infusion or bolus injection, by absorption through epithelial or mucocutaneous linings (e.g., oral mucosa, rectal and intestinal mucosa, etc.) and may be administered together with other biologically active agents. Administration can be systemic or local.
[0104] A pharmaceutical composition of the present invention can be delivered subcutaneously or intravenously with a standard needle and syringe. In addition, with respect to subcutaneous delivery, a pen delivery device readily has applications in delivering a pharmaceutical composition of the present invention. Such a pen delivery device can be reusable or disposable. A reusable pen delivery device generally utilizes a replaceable cartridge that contains a pharmaceutical composition. Once all of the pharmaceutical composition within the cartridge has been administered and the cartridge is empty, the empty cartridge can readily be discarded and replaced with a new cartridge that contains the pharmaceutical composition. The pen delivery device can then be reused. In a disposable pen delivery device, there is no replaceable cartridge. Rather, the disposable pen delivery device comes prefilled with the pharmaceutical composition held in a reservoir within the device. Once the reservoir is emptied of the pharmaceutical composition, the entire device is discarded.
[0105] Numerous reusable pen and autoinjector delivery devices have applications in the subcutaneous delivery of a pharmaceutical composition of the present invention. Examples include, but are not limited to AUTOPEN.TM. (Owen Mumford, Inc., Woodstock, UK), DISETRONIC.TM. pen (Disetronic Medical Systems, Bergdorf, Switzerland), HUMALOG MIX 75/25.TM. pen, HUMALOG.TM. pen, HUMALIN 70/30.TM. pen (Eli Lilly and Co., Indianapolis, Ind.), NOVOPEN.TM. I, II and III (Novo Nordisk, Copenhagen, Denmark), NOVOPEN JUNIOR.TM. (Novo Nordisk, Copenhagen, Denmark), BD.TM. pen (Becton Dickinson, Franklin Lakes, N.J.), OPTIPEN.TM., OPTIPEN PRO.TM., OPTIPEN STARLET.TM., and OPTICLIK.TM. (Sanofi-Aventis, Frankfurt, Germany), to name only a few. Examples of disposable pen delivery devices having applications in subcutaneous delivery of a pharmaceutical composition of the present invention include, but are not limited to the SOLOSTAR.TM. pen (Sanofi-Aventis), the FLEXPEN.TM. (Novo Nordisk), and the KWIKPEN.TM. (Eli Lilly), the SURECLICK.TM. Autoinjector (Amgen, Thousand Oaks, Calif.), the PENLET.TM. (Haselmeier, Stuttgart, Germany), the EPIPEN (Dey, L. P.), and the HUMIRA.TM. Pen (Abbott Labs, Abbott Park Ill.), to name only a few.
[0106] In certain situations, the pharmaceutical composition can be delivered in a controlled release system. In one embodiment, a pump may be used (see Langer, supra; Sefton, 1987, CRC Crit. Ref. Biomed. Eng. 14:201). In another embodiment, polymeric materials can be used; see, Medical Applications of Controlled Release, Langer and Wise (eds.), 1974, CRC Pres., Boca Raton, Fla. In yet another embodiment, a controlled release system can be placed in proximity of the composition's target, thus requiring only a fraction of the systemic dose (see, e.g., Goodson, 1984, in Medical Applications of Controlled Release, supra, vol. 2, pp. 115-138). Other controlled release systems are discussed in the review by Langer, 1990, Science 249:1527-1533.
[0107] The injectable preparations may include dosage forms for intravenous, subcutaneous, intracutaneous and intramuscular injections, drip infusions, etc. These injectable preparations may be prepared by methods publicly known. For example, the injectable preparations may be prepared, e.g., by dissolving, suspending or emulsifying the antibody or its salt described above in a sterile aqueous medium or an oily medium conventionally used for injections. As the aqueous medium for injections, there are, for example, physiological saline, an isotonic solution containing glucose and other auxiliary agents, etc., which may be used in combination with an appropriate solubilizing agent such as an alcohol (e.g., ethanol), a polyalcohol (e.g., propylene glycol, polyethylene glycol), a nonionic surfactant [e.g., polysorbate 80, HCO-50 (polyoxyethylene (50 mol) adduct of hydrogenated castor oil)], etc. As the oily medium, there are employed, e.g., sesame oil, soybean oil, etc., which may be used in combination with a solubilizing agent such as benzyl benzoate, benzyl alcohol, etc. The injection thus prepared is preferably filled in an appropriate ampoule.
[0108] Advantageously, the pharmaceutical compositions for oral or parenteral use described above are prepared into dosage forms in a unit dose suited to fit a dose of the active ingredients. Such dosage forms in a unit dose include, for example, tablets, pills, capsules, injections (ampoules), suppositories, etc. The amount of the aforesaid antibody contained is generally about 5 to about 500 mg per dosage form in a unit dose; especially in the form of injection, it is preferred that the aforesaid antibody is contained in about 5 to about 100 mg and in about 10 to about 250 mg for the other dosage forms.
Therapeutic Uses of the Antibodies
[0109] The present invention includes methods comprising administering to a subject in need thereof a therapeutic composition comprising an anti-LEPR antibody (e.g., an anti-LEPR antibody comprising any of the HCVR/LCVR or CDR sequences as set forth in Table 1 herein). The therapeutic composition can comprise any of the anti-LEPR antibodies disclosed herein, or antigen-binding fragments thereof, and a pharmaceutically acceptable carrier or diluent.
[0110] The antibodies of the invention are useful, inter alia, for the treatment, prevention and/or amelioration of any disease or disorder associated with or mediated by leptin deficiency, leptin resistance, hypoleptinemia, or otherwise treatable by stimulating or activating LEPR signaling or mimicking the natural activity of leptin in vitro or in vivo. For example, the antibodies and antigen-binding fragments thereof of the present invention are useful for treating lipodystrophy conditions. Exemplary lipodystrophy conditions that are treatable by the antibodies and antigen-binding fragments of the present invention include, e.g., congenital generalized lipodystrophy, acquired generalized lipodystrophy, familial partial lipodystrophy, acquired partial lipodystrophy, centrifugal abdominal lipodystrophy, lipoatrophia annularis, localized lipodystrophy, and HIV-associated lipodystrophy.
[0111] The present invention also includes anti-LEPR antibodies and antigen-binding fragments thereof that are useful for restoring leptin signaling to cells, tissues and organs expressing one or more LEPR mutations associated with obesity. For example, certain LEPR mutants have been identified that exhibit no, or reduced signaling in the presence of leptin and are associated with obesity and related disorders. As used herein, a LEPR mutant that exhibits no signaling in the presence of leptin is referred to as a "signaling-defective LEPR mutant." An exemplary signaling-defective LEPR mutation is LEPR-A409E (Farooqi et al., 2007, N Engl J Med 356(3): 237-247). As used herein, a LEPR mutant that exhibits reduced signaling in the presence of leptin (as compared to wild-type LEPR) is referred to as a "signaling-impaired LEPR mutant." An exemplary signaling-impaired LEPR mutation is LEPR-P316T (Mazen et al., 2011, Mol Genet Metab 102:461-464). Thus, the present invention includes anti-LEPR antibodies and antigen-binding fragments thereof that are useful for the treatment, prevention and/or amelioration of diseases and disorders caused by or associated with one or more signaling-defective (e.g., A409E) and/or signaling-impaired (e.g., P316T) LEPR mutants.
[0112] The anti-LEPR antibodies and antigen-binding fragments thereof of the present invention are also useful for the treatment or prevention of one or more diseases or disorders selected from the group consisting of obesity, metabolic syndrome, diet-induced food craving, functional hypothalamic amenorrhea, type 1 diabetes, type 2 diabetes, insulin resistance, severe insulin resistance including severe insulin resistance due to mutation in insulin receptor, severe insulin resistance not caused by mutation in the insulin receptor, severe insulin resistance caused by a mutation in downstream signaling pathways or induced by other causes, non-alcoholic and alcoholic fatty liver diseases, Alzheimer's disease, leptin deficiency, leptin resistance, lipodystrophies, Leprechaunism/Donohue syndrome, Rabson-Mendenhall syndrome.
[0113] In the context of the methods of treatment described herein, the anti-LEPR antibody may be administered as a monotherapy (i.e., as the only therapeutic agent) or in combination with one or more additional therapeutic agents (examples of which are described elsewhere herein).
Combination Therapies and Formulations
[0114] The present invention includes compositions and therapeutic formulations comprising any of the anti-LEPR antibodies described herein in combination with one or more additional therapeutically active components, and methods of treatment comprising administering such combinations to subjects in need thereof.
[0115] The anti-LEPR antibodies of the present invention may be co-formulated with and/or administered in combination with one or more additional therapeutically active component(s), such as. e.g., pharmaceutical products prescribed for the treatment of obesity, hypercholesterolemia, hyperlipidemia, type 2 diabetes, type 1 diabetes, appetite control, infertility, etc. Examples of such additional therapeutically active components include, e.g., recombinant human leptin (e.g., metreleptin [MYALEPT]), PCSK9 inhibitors (e.g., anti-PCSK9 antibodies [alirocumab, evolocumab, bococizumab, lodelcizumab, ralpancizumab, etc.]), statins (atorvastatin, rosuvastatin, cerivastatin, pitavastatin, fluvastatin, simvastatin, lovastatin, pravastatin, etc.), ezetimibe, insulin, insulin variants, insulin secretagogues, metformin, sulfonylureas, sodium glucose cotransporter 2 (SGLT2) Inhibitors (e.g., dapaglifozin, canaglifozin, empagliflozin, etc.), GLP-1 agonists/analogues (e.g., extendin-4, exenatide, liraglutide, lixisenatide, albiglutide, dulaglutide, etc.), glucagon (GCG) inhibitors (e.g., anti-GCG antibodies), glucagon receptor (GCGR) inhibitors (e.g., anti-GCGR antibodies, small molecule GCGR antagonists, GCGR-specific antisense oligonucleotides, anti-GCGR aptamers [e.g., Spiegelmers], etc.), angiopoietin-like protein (ANGPTL) inhibitors (e.g., anti-ANGPTL3 antibodies, anti-ANGPTL4 antibodies, anti-ANGPTL8 antibodies, etc.), Phentermine, Orlistat, Topiramate, Bupropion, Topiramate/Phentermine, Bupropion/Naltrexone, Bupropion/Zonisamide, Pramlintide/Metrelepin, Lorcaserin, Cetilistat, Tesofensine, Velneperit, etc.
[0116] The additional therapeutically active component(s), e.g., any of the agents listed above or derivatives thereof, may be administered just prior to, concurrent with, or shortly after the administration of an anti-LEPR antibody of the present invention; (for purposes of the present disclosure, such administration regimens are considered the administration of an anti-LEPR antibody "in combination with" an additional therapeutically active component). The present invention includes pharmaceutical compositions in which an anti-LEPR antibody of the present invention is co-formulated with one or more of the additional therapeutically active component(s) as described elsewhere herein.
Administration Regimens
[0117] According to certain embodiments of the present invention, multiple doses of an anti-LEPR antibody (or a pharmaceutical composition comprising a combination of an anti-LEPR antibody and any of the additional therapeutically active agents mentioned herein) may be administered to a subject over a defined time course. The methods according to this aspect of the invention comprise sequentially administering to a subject multiple doses of an anti-LEPR antibody of the invention. As used herein, "sequentially administering" means that each dose of anti-LEPR antibody is administered to the subject at a different point in time, e.g., on different days separated by a predetermined interval (e.g., hours, days, weeks or months). The present invention includes methods which comprise sequentially administering to the patient a single initial dose of an anti-LEPR antibody, followed by one or more secondary doses of the anti-LEPR antibody, and optionally followed by one or more tertiary doses of the anti-LEPR antibody.
[0118] The terms "initial dose," "secondary doses," and "tertiary doses," refer to the temporal sequence of administration of the anti-LEPR antibody of the invention. Thus, the "initial dose" is the dose which is administered at the beginning of the treatment regimen (also referred to as the "baseline dose," "loading dose," "starting dose," and the like); the "secondary doses" are the doses which are administered after the initial dose; and the "tertiary doses" are the doses which are administered after the secondary doses. The initial, secondary, and tertiary doses may all contain the same amount of anti-LEPR antibody, but generally may differ from one another in terms of frequency of administration. In certain embodiments, however, the amount of anti-LEPR antibody contained in the initial, secondary and/or tertiary doses varies from one another (e.g., adjusted up or down as appropriate) during the course of treatment. In certain embodiments, two or more (e.g., 2, 3, 4, or 5) doses are administered at the beginning of the treatment regimen as "loading doses" followed by subsequent doses that are administered on a less frequent basis (e.g., "maintenance doses").
Diagnostic and Analytic Uses of the Antibodies
[0119] The anti-LEPR antibodies of the present invention may also be used to detect and/or measure LEPR, or LEPR-expressing cells in a sample, e.g., for diagnostic purposes. For example, an anti-LEPR antibody, or fragment thereof, may be used to diagnose a condition or disease characterized by aberrant expression (e.g., over-expression, under-expression, lack of expression, etc.) of LEPR. Exemplary diagnostic assays for LEPR may comprise, e.g., contacting a sample, obtained from a patient, with an anti-LEPR antibody of the invention, wherein the anti-LEPR antibody is labeled with a detectable label or reporter molecule. Alternatively, an unlabeled anti-LEPR antibody can be used in diagnostic applications in combination with a secondary antibody which is itself detectably labeled. The detectable label or reporter molecule can be a radioisotope, such as .sup.3H, .sup.14C, .sup.32P, .sup.35S, or .sup.125I; a fluorescent or chemiluminescent moiety such as fluorescein isothiocyanate, or rhodamine; or an enzyme such as alkaline phosphatase, beta-galactosidase, horseradish peroxidase, or luciferase. Specific exemplary assays that can be used to detect or measure LEPR in a sample include enzyme-linked immunosorbent assay (ELISA), radioimmunoassay (RIA), fluorescence-activated cell sorting (FACS), and positron emission tomography (PET) scanning.
[0120] Samples that can be used in LEPR diagnostic assays according to the present invention include any tissue or fluid sample obtainable from a patient which contains detectable quantities of LEPR protein, or fragments thereof, under normal or pathological conditions. Generally, levels of LEPR in a particular sample obtained from a healthy patient (e.g., a patient not afflicted with a disease or condition associated with abnormal LEPR levels or activity) will be measured to initially establish a baseline, or standard, level of LEPR. This baseline level of LEPR can then be compared against the levels of LEPR measured in samples obtained from individuals suspected of having a LEPR related disease or condition.
EXAMPLES
[0121] The following examples are put forth so as to provide those of ordinary skill in the art with a complete disclosure and description of how to make and use the methods and compositions of the invention, and are not intended to limit the scope of what the inventors regard as their invention. Efforts have been made to ensure accuracy with respect to numbers used (e.g., amounts, temperature, etc.) but some experimental errors and deviations should be accounted for. Unless indicated otherwise, parts are parts by weight, molecular weight is average molecular weight, temperature is in degrees Centigrade, and pressure is at or near atmospheric.
Example 1
Generation of Antigen-Binding Proteins that Specifically Bind the Leptin Receptor (LEPR)
[0122] Anti-LEPR antibodies were obtained by immunizing a VELOCIMMUNE.RTM. mouse (i.e., an engineered mouse comprising DNA encoding human immunoglobulin heavy and kappa light chain variable regions) with an immunogen comprising the extracellular domain of LEPR. The antibody immune response was monitored by a LEPR-specific immunoassay. Using previously described techniques, fully human anti-LEPR antibodies were isolated and purified.
[0123] Certain biological properties of the exemplary anti-LEPR antibodies generated in accordance with the methods of this Example are described in detail in the Examples set forth below.
Example 2
Heavy and Light Chain Variable Region Amino Acid and Nucleic Acid Sequences
[0124] Table 1 sets forth the amino acid sequence identifiers of the heavy and light chain variable regions and CDRs of selected anti-LEPR antibodies of the invention. The corresponding nucleic acid sequence identifiers are set forth in Table 2.
TABLE-US-00001 TABLE 1 Amino Acid Sequence Identifiers Antibody SEQ ID NOs: Designation HCVR HCDR1 HCDR2 HCDR3 LCVR LCDR1 LCDR2 LCDR3 H4H16650P2 2 4 6 8 10 12 14 16 H4H16679P2 18 20 22 24 10 12 14 16 H4H17319P2 26 28 30 32 10 12 14 16 H4H17321P2 34 36 38 40 10 12 14 16 H4H18417P2 42 44 46 48 10 12 14 16 H4H18438P2 50 52 54 56 10 12 14 16 H4H18445P2 58 60 62 64 10 12 14 16 H4H18446P2 66 68 70 72 10 12 14 16 H4H18449P2 74 76 78 80 10 12 14 16 H4H18482P2 82 84 86 88 90 92 94 96 H4H18487P2 98 100 102 104 90 92 94 96 H4H18492P2 106 108 110 112 90 92 94 96
TABLE-US-00002 TABLE 2 Nucleic Acid Sequence Identifiers Antibody SEQ ID NOs: Designation HCVR HCDR1 HCDR2 HCDR3 LCVR LCDR1 LCDR2 LCDR3 H4H16650P2 1 3 5 7 9 11 13 15 H4H16679P2 17 19 21 23 9 11 13 15 H4H17319P2 25 27 29 31 9 11 13 15 H4H17321P2 33 35 37 39 9 11 13 15 H4H18417P2 41 43 45 47 9 11 13 15 H4H18438P2 49 51 53 55 9 11 13 15 H4H18445P2 57 59 61 63 9 11 13 15 H4H18446P2 65 67 69 71 9 11 13 15 H4H18449P2 73 75 77 79 9 11 13 15 H4H18482P2 81 83 85 87 89 91 93 95 H4H18487P2 97 99 101 103 89 91 93 95 H4H18492P2 105 107 109 111 89 91 93 95
[0125] Antibodies are typically referred to herein according to the following nomenclature: Fc prefix (e.g. "H4H," "H1M," "H2M," etc.), followed by a numerical identifier (e.g. "16650," "16679," etc.), followed by a "P" or "N" suffix. Thus, according to this nomenclature, an antibody may be referred to herein as, e.g., "H4H16650P2," "H4H16679P2," etc. The Fc prefixes on the antibody designations used herein (H4H, H1M and H2M) indicate the particular Fc region isotype of the antibody. For example, an "H4H" antibody has a human IgG4 Fc, whereas an "H1M" antibody has a mouse IgG1 Fc, (all variable regions are fully human as denoted by the first `H` in the antibody designation). As will be appreciated by a person of ordinary skill in the art, an antibody having a particular Fc isotype can be converted to an antibody with a different Fc isotype (e.g., an antibody with a mouse IgG1 Fc can be converted to an antibody with a human IgG4, etc.), but in any event, the variable domains (including the CDRs)--which are indicated by the numerical identifiers shown in Tables 1 and 2--will remain the same, and the binding properties are expected to be identical or substantially similar regardless of the nature of the Fc domain.
[0126] "Comparator mAb" as used in Examples herein refers to Fab9F8 described in Fazeli et al. (2006) J Immunol Methods 312:190-200 and Carpenter et al. (2012) Structure 20(3):487-97.
Example 3
Surface Plasmon Resonance Derived Binding Affinities and Kinetic Constants of Human Monoclonal Anti-LEPR Antibodies
[0127] Equilibrium dissociation constants (KD values) for LEPR binding to purified anti-LEPR monoclonal antibodies were determined using a real-time surface plasmon resonance biosensor using a Biacore 4000 instrument. All binding studies were performed in 10 mM HEPES, 150 mM NaCl, 3 mM EDTA, and 0.05% v/v Surfactant Tween-20, pH 7.4 (HBS-ET) running buffer at 25.degree. C. and 37.degree. C. The Biacore sensor surface was first derivatized by amine coupling with a monoclonal mouse anti-human Fc antibody (GE, #BR-1008-39) to capture anti-LEPR monoclonal antibodies. Binding studies were performed on following LEPR reagents: human LEPR extracellular domain expressed with a C-terminal myc-myc-hexahistidine tag (hLEPR.mmh; SEQ ID NO: 114), macaca fascicularis LEPR extracellular domain expressed with a C-terminal myc-myc-hexahistidine tag (mfLEPR.mmh; SEQ ID NO: 117), human LEPR extracellular domain expressed with a C-terminal mouse IgG2a Fc tag (hLEPR.mFc; SEQ ID NO: 115), mouse LEPR extracellular domain expressed with a C-terminal myc-myc-hexahistidine tag (mLEPR.mmh; SEQ ID NO: 118) and rat LEPR extracellular domain expressed with a C-terminal myc-myc-hexahistidine tag (rLEPR.mmh; SEQ ID NO: 119). Different concentrations of LEPR reagents were first prepared in HBS-ET running buffer (100 nM-3.7 nM; 3-fold serial dilution) and were injected over anti-human Fc captured anti-LEPR monoclonal antibody surface for 4 minutes at a flow rate of 30 .mu.L/minute, while the dissociation of monoclonal antibody bound LEPR reagent was monitored for 10 minutes in HBS-ET running buffer. Kinetic association (k.sub.a) and dissociation (k.sub.d) rate constants were determined by fitting the real-time binding sensorgrams to a 1:1 binding model with mass transport limitation using Scrubber 2.0 c curve-fitting software. Binding dissociation equilibrium constants (K.sub.D) and dissociative half-lives (t1/2) were calculated from the kinetic rate constants as:
K D ( M ) = kd ka , and t 1 2 ( min ) = ln ( 2 ) 60 * kd ##EQU00001##
[0128] Binding kinetics parameters for hLEPR.mmh, mfLEPR.MMH or hLEPR.mFc, binding to different anti-LEPR monoclonal antibodies of the invention at 25.degree. C. and 37.degree. C. are shown in Tables 3 through 8.
TABLE-US-00003 TABLE 3 Binding kinetics parameters of hLEPR-MMH binding to LEPR monoclonal antibodies at 25.degree. C. 100 nM mAb Capture hLEPR-MMH k.sub.a k.sub.d K.sub.D t1/2 mAb Captured Level (RU) Bound (RU) (1/Ms) (1/s) (M) (min) H4H16650P2 167 .+-. 0.3 51 2.81E+04 2.23E-04 7.93E-09 52 H4H16679P2 192 .+-. 0.7 39 2.34E+04 2.46E-04 1.05E-08 47 H4H18417P2 163 .+-. 0.4 28 6.14E+04 7.90E-03 1.29E-07 1.5 H4H18438P2 166 .+-. 0.4 22 3.00E+04 2.26E-03 7.54E-08 5.1 H4H18445P2 194 .+-. 1.1 45 4.42E+04 4.78E-03 1.08E-07 2.4 H4H18446P2 163 .+-. 2.4 16 1.81E+04 9.51E-04 5.25E-08 12 H4H18449P2 176 .+-. 1.3 54 2.91E+04 2.35E-04 8.08E-09 49 H4H18482P2 163 .+-. 0.4 47 6.31E+04 6.77E-03 1.07E-07 1.7 H4H18487P2 190 .+-. 1.2 42 4.73E+04 7.03E-03 1.48E-07 1.6 H4H18492P2 167 .+-. 3.1 87 8.10E+04 8.98E-04 1.11E-08 13 H4H17319P2 200 .+-. 0.4 36 2.61E+04 5.29E-04 2.03E-08 22 H4H17321P2 221 .+-. 0.5 32 2.36E+04 1.96E-04 8.31E-09 59 Isotype 171 .+-. 0.4 4 NB* NB* NB* NB* Control mAb *NB indicates that no binding was observed under the current experimental conditions.
TABLE-US-00004 TABLE 4 Binding kinetics parameters of hLEPR-MMH binding to LEPR monoclonal antibodies at 37.degree. C. 100 nM mAb Capture hLEPR-MMH k.sub.a k.sub.d K.sub.D t1/2 mAb Captured Level (RU) Bound (RU) (1/Ms) (1/s) (M) (min) H4H16650P2 210 .+-. 2.5 77 4.85E+04 9.58E-04 1.98E-08 12 H4H16679P2 239 .+-. 2 61 3.84E+04 8.42E-04 2.19E-08 14 H4H18417P2 206 .+-. 3.2 22 7.70E+04 1.80E-02 2.33E-07 0.6 H4H18438P2 206 .+-. 2.4 32 3.38E+04 5.76E-03 1.70E-07 2.0 H4H18445P2 234 .+-. 2 38 5.13E+04 1.68E-02 3.26E-07 0.7 H4H18446P2 188 .+-. 3.4 21 2.12E+04 2.56E-03 1.21E-07 4.5 H4H18449P2 206 .+-. 2.1 73 3.94E+04 8.15E-04 2.07E-08 14 H4H18482P2 188 .+-. 0.8 38 9.53E+04 1.93E-02 2.03E-07 0.6 H4H18487P2 219 .+-. 1.7 30 6.51E+04 1.86E-02 2.86E-07 0.6 H4H18492P2 192 .+-. 2.2 93 1.17E+05 4.18E-03 3.59E-08 2.8 H4H17319P2 264 .+-. 0.3 44 3.54E+04 3.41E-03 9.63E-08 3.4 H4H17321P2 290 .+-. 0.4 61 2.95E+04 4.38E-04 1.48E-08 26 Isotype 193 .+-. 1.5 6 NB* NB* NB* NB* Control mAb *NB indicates that no binding was observed under the current experimental conditions.
TABLE-US-00005 TABLE 5 Binding kinetics parameters of mfLEPR.MMH binding to LEPR monoclonal antibodies at 25.degree. C. 100 nM mAb Capture mfLEP.MMH k.sub.a k.sub.d K.sub.D t1/2 mAb Captured Level (RU) Bound (RU) (1/Ms) (1/s) (M) (min) H4H16650P2 166 .+-. 0.6 93 6.02E+04 1.37E-04 2.27E-09 84 H4H16679P2 191 .+-. 0.7 66 4.37E+04 1.41E-04 3.22E-09 82 H4H18417P2 162 .+-. 0.3 33 8.83E+04 1.23E-02 1.39E-07 0.9 H4H18438P2 166 .+-. 0.6 5 IC* IC* IC* IC* H4H18445P2 193 .+-. 0.6 58 5.90E+04 4.86E-03 8.24E-08 2.4 H4H18446P2 163 .+-. 2.8 23 1.93E+04 1.12E-03 5.83E-08 10 H4H18449P2 175 .+-. 0.5 6 IC* IC* IC* IC* H4H18482P2 163 .+-. 0.8 63 1.01E+05 6.74E-03 6.66E-08 1.7 H4H18487P2 189 .+-. 0.5 59 7.37E+04 6.79E-03 9.21E-08 1.7 H4H18492P2 165 .+-. 2.4 52 1.10E+05 1.20E-02 1.10E-07 1.0 H4H17319P2 213 .+-. 0.5 83 4.00E+04 4.63E-04 1.16E-08 25 H4H17321P2 236 .+-. 0.4 75 3.26E+04 1.33E-04 4.07E-09 87 Isotype 171 .+-. 0.4 0 NB* NB* NB* NB* Control mAb *NB indicates that no binding was observed under the current experimental conditions. *IC indicates that observed binding was inclusive and was unable to fit the real time binding data under the current experimental conditions.
TABLE-US-00006 TABLE 6 Binding kinetics parameters of mfLEPR.MMH binding to LEPR monoclonal antibodies at 37.degree. C. 100 nM mAb Capture mfLEPR.MMH k.sub.a k.sub.d K.sub.D t1/2 mAb Captured Level (RU) Bound (RU) (1/Ms) (1/s) (M) (min) H4H16650P2 204 .+-. 1.7 134 1.22E+05 7.00E-04 5.76E-09 16 H4H16679P2 232 .+-. 1.1 104 6.49E+04 6.77E-04 1.04E-08 17 H4H18417P2 202 .+-. 1.3 28 1.22E+05 2.63E-02 2.17E-07 0.4 H4H18438P2 203 .+-. 1.3 7 IC* IC* IC* IC* H4H18445P2 232 .+-. 0.9 48 7.17E+04 1.90E-02 2.64E-07 0.6 H4H18446P2 188 .+-. 2.9 30 2.53E+04 3.54E-03 1.40E-07 3.3 H4H18449P2 202 .+-. 1 6 IC* IC* IC* IC* H4H18482P2 187 .+-. 1.2 52 1.52E+05 2.04E-02 1.34E-07 0.6 H4H18487P2 216 .+-. 0.7 44 1.10E+05 1.95E-02 1.78E-07 0.6 H4H18492P2 191 .+-. 1.4 34 2.34E+05 3.94E-02 1.69E-07 0.3 H4H17319P2 274 .+-. 0.5 113 5.39E+04 3.24E-03 6.01E-08 3.6 H4H17321P2 304 .+-. 0.7 143 4.97E+04 2.57E-04 5.18E-09 45 Isotype 190 .+-. 1 1 NB* NB* NB* NB* Control mAb *NB indicates that no binding was observed under the current experimental conditions. *IC indicates that observed binding was inclusive and was unable to fit the real time binding data under the current experimental conditions.
TABLE-US-00007 TABLE 7 Binding kinetics parameters of hLEPR.mFc binding to LEPR monoclonal antibodies at 25.degree. C. 100 nM mAb Capture hLEPR-mFc k.sub.a k.sub.d K.sub.D t1/2 mAb Captured Level (RU) Bound (RU) (1/Ms) (1/s) (M) (min) H4H16650P2 165 .+-. 0.2 102 1.06E+05 8.32E-05 7.85E-10 139 H4H16679P2 190 .+-. 1.2 78 5.84E+04 9.68E-05 1.66E-09 119 H4H18417P2 162 .+-. 0.6 90 1.40E+05 5.63E-04 4.04E-09 21 H4H18438P2 165 .+-. 1.2 51 5.19E+04 2.44E-04 4.70E-09 47 H4H18445P2 192 .+-. 0.4 76 1.22E+05 4.92E-04 4.03E-09 23 H4H18446P2 162 .+-. 2.8 20 3.20E+04 2.08E-04 6.48E-09 56 H4H18449P2 174 .+-. 0.6 116 7.05E+04 6.82E-05 9.64E-10 169 H4H18482P2 162 .+-. 0.5 88 1.44E+05 4.91E-04 3.42E-09 24 H4H18487P2 188 .+-. 0.6 85 1.06E+05 6.03E-04 5.70E-09 19 H4H18492P2 166 .+-. 3.2 129 2.27E+05 1.39E-04 6.13E-10 83 H4H17319P2 200 .+-. 0.5 69 4.77E+04 1.64E-04 3.45E-09 70 H4H17321P2 221 .+-. 0.4 65 4.10E+04 8.93E-05 2.18E-09 129 Isotype 170 .+-. 0.7 -2 NB* NB* NB* NB* Control mAb *NB indicates that no binding was observed under the current experimental conditions.
TABLE-US-00008 TABLE 8 Binding kinetics parameters of hLEPR.mFc binding to LEPR monoclonal antibodies at 37.degree. C. 100 nM mAb Capture hLEPR-mFc k.sub.a k.sub.d K.sub.D t1/2 mAb Captured Level (RU) Bound (RU) (1/Ms) (1/s) (M) (min) H4H16650P2 199 .+-. 1.9 145 1.57E+05 2.80E-04 1.79E-09 41 H4H16679P2 229 .+-. 2.3 116 1.21E+05 3.10E-04 2.56E-09 37 H4H18417P2 199 .+-. 1.1 111 1.85E+05 1.05E-03 5.64E-09 11 H4H18438P2 199 .+-. 0.6 82 7.02E+04 5.98E-04 8.53E-09 19 H4H18445P2 229 .+-. 2 104 1.56E+05 6.08E-04 3.89E-09 19 H4H18446P2 186 .+-. 2.5 34 4.27E+04 5.48E-04 1.28E-08 21 H4H18449P2 198 .+-. 1.6 148 1.33E+05 1.68E-04 1.26E-09 69 H4H18482P2 185 .+-. 1.3 109 1.89E+05 7.26E-04 3.84E-09 16 H4H18487P2 215 .+-. 1.5 99 1.23E+05 6.06E-04 4.93E-09 19 H4H18492P2 189 .+-. 1.8 160 4.33E+05 5.00E-04 1.16E-09 23 H4H17319P2 262 .+-. 0.5 100 8.51E+04 6.52E-04 7.66E-09 18 H4H17321P2 289 .+-. 0.4 110 5.53E+04 1.74E-04 3.15E-09 66 Isotype 188 .+-. 0.8 1 NB* NB* NB* NB* Control mAb *NB indicates that no binding was observed under the current experimental conditions.
[0129] At 25.degree. C., anti-LEPR monoclonal antibodies bound to hLEPR-MMH with K.sub.D values ranging from 7.93 nM to 148 nM, as shown in Table 5. At 37.degree. C., anti-LEPR monoclonal antibodies bound to hLEPR-MMH with K.sub.D values ranging from 14.8 nM to 326 nM, as shown in Table 4.
[0130] Ten out of 12 anti-LEPR monoclonal antibodies of the invention bound to mfLEPR.MMH. At 25.degree. C., anti-LEPR monoclonal antibodies bound to mfLEPR.MMH with K.sub.D values ranging from 2.27 nM to 139 nM, as shown in Table 7. At 37.degree. C., anti-LEPR monoclonal antibodies bound to mfLEPR.MMH with K.sub.D values ranging from 5.18 nM to 264 nM, as shown in Table 8.
[0131] At 25.degree. C., anti-LEPR monoclonal antibodies bound to hLEPR-mFc with K.sub.D values ranging from 613 pM to 5.7 nM, as shown in Table 7. At 37.degree. C., anti-LEPR monoclonal antibodies bound to hLEPR-mFc with K.sub.D values ranging from 1.16 nM to 12.8 nM, as shown in Table 8.
[0132] None of the anti-LEPR monoclonal antibodies of the invention bound to mLEPR.MMH or rLEPR.MMH at 25.degree. C. or at 37.degree. C. (data not shown).
Example 4
Anti-LEPR Antibodies of the Invention Bind LEPR in the Presence of Leptin:LEPR Binding
[0133] Blocking of anti-LEPR antibodies from binding to LEPR by human Leptin was evaluated using a real-time surface plasmon resonance biosensor on a Biacore T200 instrument. The entire study was performed in 10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.05% v/v Surfactant Tween-20 (HBS-ET running buffer) at 25.degree. C. The Biacore CM5 sensor surface was first derivatized by amine coupling human Leptin (R&D Systems, #398-LP) using standard EDC/NHS surface chemistry. A complex of human LEPR and human Leptin, was formed by injecting 20 nM of human LEPR extracellular domain expressed with a C-terminal myc-myc-hexahistidine tag (hLEPR-MMH; SEQ ID NO: 114), over the human Leptin immobilized Biacore sensor surface at a flow rate of 10 .mu.L/minute or 25 .mu.L/minute for 4 minutes, to achieve a binding response of approximately 200 RU. To evaluate whether antibody binding to hLEPR-MMH is blocked by human Leptin, 200 nM of anti-LEPR monoclonal antibodies were injected over the preformed hLEPR-MMH:human Leptin complex, at a flow rate of 50 .mu.L/minute or 25 .mu.L/minute for 4-5 minutes. All of the anti-LEPR antibodies of this invention that were tested bound to the complex of hLEPR-MMH and human Leptin ("Leptn;LEPR") with nearly similar signal strength and the observed binding, expressed in RUs, are reported in Table 9. This result indicates that human Leptin does not block the binding of hLEPR-MMH to anti-LEPR antibodies tested.
TABLE-US-00009 TABLE 9 Binding of anti-LEPR monoclonal antibodies to the pre-complex of hLEPR-MMH and human Leptin. hLEPR-MMH 200 nM mAb Antibody Bound (RU) Bound (RU) H4H16650P2 196 81 H4H16679P2 195 90 H4H17319P2 196 92
Example 5
Human Leptin receptor blocking ELISA
[0134] For the ELISA, human Leptin (hLeptin; R&D Systems, #398-LP-01M) was coated at a concentration of 5 .mu.g/mL in PBS on a 96-well microtiter plate overnight at 4.degree. C. Nonspecific binding sites were subsequently blocked using a 0.5% (w/v) solution of BSA in PBS. A constant amount of 10 nM of extracellular domain portion of LEPR protein that was expressed with a C-terminal human Fc tag (hLEPR.hFc; SEQ ID NO: 116) was titrated with anti-LEPR antibodies, hLeptin protein, or an isotype control antibody ranging from 8.5 pM to 500 nM in serial dilution. These antibody-protein or protein-protein complexes were then incubated for 1.5 hour at room temperature (RT). Complexes were subsequently transferred to microtiter plates coated with hLeptin and incubated for 2 hours at RT, the wells were washed, and plate-bound hLEPR.hFc was detected with an anti-human IgG polyclonal antibody conjugated with horseradish peroxidase (Jackson ImmunoResearch Inc, #109-035-098). Samples were developed with a TMB solution (BD Biosciences, #555214; substrate A and B mixed at 1:1 ratio as per manufacturer's instructions) to produce a colorimetric reaction and then neutralized with 1M sulfuric acid before measuring absorbance at 450 nm on a Victor X5 plate reader.
[0135] Data analysis was performed using a sigmoidal dose-response model within Prism.TM. software (Graph Pad). Percent blockade at maximum concentration of the antibody tested was calculated as an indicator of the ability of the antibodies to block the binding of 10 nM of hLEPR.hFc to human Leptin on the plate. In the calculation, binding signal of 10 nM of hLEPR.hFc without the presence of the antibody was referenced as 100% binding or 0% blocking; and the baseline signal of buffer alone without the presence of hLEPR.hFc was referenced as 0% binding or 100% blocking. The blocking data at 500 nM antibody concentration is summarized in Table 10.
[0136] As shown in Table 10, none of the anti-LEPR antibodies of the invention demonstrated >28% blocking of the binding of hLEPR.hFc to the hLeptin coated surface. However, the Comparator Antibody and the hLeptin, as the positive control, were able to block 99% of the hLEPR.hFc binding to the hLeptin coated surface. The isotype control antibody demonstrated no measurable blocking at concentrations up to 500 nM.
TABLE-US-00010 TABLE 10 ELISA blocking of hLEPR.hFc binding to hLeptin by anti-LEPR antibodies 500 nM Ab Blocking of 10 nM hLEPR.hFc Binding to hLeptin Antibody (% blockade) H4H18487P2 5 H4H18417P2 16 H4H18482P2 25 H4H18492P2 -3 H4H18445P2 28 H4H18446P2 -5 H4H18449P2 8 H4H18438P2 15 H4H16650P2 -7 H4H16679P2 7 H4H17319P2 9 H4H17321P2 6 Controls Isotype control antibody -3 Human Leptin 99 Comparator Antibody 99 Mouse IgG2a Isotype 32 control
Example 6
Cell binding by FACS analysis with HEK293/Mycx2-hLepR(ecto)-GPI Anchored Cells
[0137] Leptin receptor, LEPR, is a single-pass transmembrane receptor of the class I cytokine receptor family (Tartaglia et al. (1997) J Biol Chem 7:272(10):6093-6). LEPR can bind to Leptin, a protein predominantly expressed by adipose tissue that is involved in regulation of food intake and metabolism (Friedman et al. (2014) J Endocrinol 223(1):T1-8).
[0138] In order to assess cell binding by anti-LEPR antibodies HEK293 stable cell lines were generated. One cell line, known hereafter as HEK293/hLEPR-GPI, stably expressed the extracellular domain of human LEPR (amino acids 22-839 of accession #P48357 (SEQ ID NO:113), Isoform B) with an N-terminal myc-myc tag and C-terminal peptide sequence from human carboxypeptidase M that guides the addition of GPI (Glycosylphosphatidylinositol) (Deddish et al. (1990) J. Biological Chemistry 265:25:15083-89) such that the protein can be GPI-anchored to the membrane. Another HEK293 cell line was generated to stably express the full length human LEPR (amino acids 1-1165 of accession #P48357(SEQ ID NO:113), Isoform B) along with a luciferase reporter (Stat3-luciferase, Stat3-luc, SA Bioscience, #CLS-6028L), and is known hereafter as HEK293/Stat3-luc/hLEPR-FL. HEK293 cells with the Stat3-luciferase reporter only (HEK293/Stat3-luc) were also generated as a control cell line.
[0139] For the FACS analysis, HEK293 parental cells and HEK293/hLEPR-GPI cells were dissociated and plated onto 96-well v-bottom plates at 5.times.10.sup.5 cells/well in PBS containing 2% FBS (FACS buffer). In order to test whether the ability of anti-hLEPR antibodies to bind to cells is affected by the presence of Leptin, FACS buffer with or without 1 .mu.M human Leptin (R&D Systems, #398-LP) was incubated with the cells for 30 minutes at 4.degree. C., followed by the addition of anti-LEPR antibodies or control antibodies at 10 nM in FACS buffer. The cells were subsequently incubated for 30 minutes at 4.degree. C., followed by washing and then incubation with 16 .mu.g/mL of Alexa Fluor.RTM.-647 conjugated secondary antibody (Jackson ImmunoResearch Laboratories Inc., #109-547-003) for 30 minutes at 4.degree. C. Cells were subsequently fixed using BD CytoFix.TM. (Becton Dickinson, #554655), filtered, and analyzed on a HyperCyt Flow Cytometer (Beckman Coulter). Unstained and secondary antibody alone controls were also tested for all cell lines. The results were analyzed using ForeCyt (IntelliCyt) and FlowJo version 10 software to determine the geometric means of fluorescence for viable cells. The geometric mean of fluorescence for each sample was then normalized to the geometric mean of unstained cells to obtain relative binding per condition referred to as "binding ratios", and these binding ratios were recorded for each antibody tested.
[0140] As shown in Table 11, 9 anti-LEPR antibodies of the invention tested at 10 nM demonstrated binding to HEK293/hLEPR-GPI cells with binding ratios ranging from 824 to 3374 fold without Leptin. The anti-LEPR antibodies also bound in the presence of 1 .mu.M Leptin with binding ratios of 398 and 4184 fold. As shown in Table 11, he Comparator Antibody tested at 10 nM demonstrated binding to HEK293/hLEPR-GPI cells with a binding ratio of 2349-fold without Leptin but showed significantly less binding to cells in the presence of 1 .mu.M Leptin with binding ratio of 112. The anti-LEPR antibodies did not demonstrate any significant binding to the HEK293 parental cells with binding ratios with and without 1 .mu.M Leptin ranging from 1 to 9 fold. The isotype control antibodies and secondary antibodies alone samples also did not demonstrate significant binding to either cell line with or without Leptin, with binding ratios ranging from 1 to 6 fold.
[0141] As shown in Table 12, four antibodies of the invention tested at 70 nM without Leptin, demonstrated binding to HEK293/hLEPR-GPI cells with binding ratios ranging from 707 to 1131 fold and to HEK293/Stat3-luc/hLEPR-FL cells with binding ratios ranging from 42 to 51. The anti-LEPR antibodies did not demonstrate any significant binding to the HEK293/Stat3-luc cells with binding ratios ranging from 1 to 8 fold. The isotype control antibodies and secondary antibodies alone samples also did not demonstrate significant binding to any of the cell lines tested, with binding ratios ranging from 1 to 2 fold.
TABLE-US-00011 TABLE 11 Binding of 10 nM anti-LEPR antibodies to HEK293/hLEPR-GPI and HEK293 parental cells +/- 1 .mu.M Human Leptin Binding Ratio: Normalized to Unstained Sample of Each Cell Line No added Leptin 1 .mu.M Leptin HEK293 HEK293/ HEK293 HEK293/ Antibody Antibody parental hLEPR-GPI parental hLEPR-GPI Type H4H16650P2 5 2420 4 3124 Agonist H4H16679P2 5 2058 8 2223 Agonist H4H18417P2 1 1835 2 2604 Potentiator H4H18438P2 2 1486 3 2414 Potentiator H4H18445P2 2 2016 3 2488 Potentiator H4H18449P2 5 3374 9 3113 Potentiator H4H18482P2 1 1966 3 2704 Potentiator H4H18487P2 1 2422 3 2670 Potentiator H4H18492P2 3 2603 7 4184 Potentiator Comparitor 6 2349 3 112 N/A Isotype control 1 6 2 4 N/A antibody Secondary 1 3 2 3 N/A antibody alone Unstained 1 1 1 1 N/A *Classification of antibodies as "Agonist" or "Potentiator" is based in part on the results observed in Examples 7 and 8 herein.
TABLE-US-00012 TABLE 12 Binding of 70 nM anti-LEPR antibodies to HEK293/hLEPR-GPI, HEK293/Stat3-hLEPR-FL, and HEK293/Stat3-luc parental cells Binding Ratio: Normalized to Unstained Sample of Each Cell Line HEK293/ HEK293/ HEK293/ Stat3-luc Antibody Antibody Stat3-luc hLEPR-GPI hLEPR-FL Type H4H16650P2 6 707 42 Agonist H4H16679P2 8 1078 51 Agonist H4H17319P2 7 1131 47 Agonist H4H17321P2 7 1126 46 Agonist Isotype control 2 2 2 antibody Secondary 1 1 1 antibody alone Unstained 1 1 1
Example 7
Anti-LEPR Antibodies of the Invention Activate LEPR Signaling in the Presence or Absence of Leptin
[0142] A bioassay was developed to detect the transcriptional activation of STAT3 via LEPR activation using a reporter cell line that stably expresses full-length human LEPR (hLEPR; amino acids 1 through 1165 of accession number NP_002294.2) along with a luciferase reporter (STAT3-Luc; Qiagen, #CLS-6028L) in an IMR-32 cell line, a human neuroblastoma cell line. The resulting stable cell line, referred to as IMR-32/STAT3-Luc/hLEPR, was isolated and maintained in MEM-Earl medium supplemented with 10% FBS, NEAA, 1 ug/mL Puromycin, 100 ug/mL of Hygromycin B and Penicillin/Streptomycin/L-Glutamine (Complete Medium).
[0143] The resulting bioassay was used to measure the effect of anti-LEPR antibodies of the invention on LEPR signaling in the presence or absence of Leptin. For the bioassay, IMR-32/STAT3-Luc/hLEPR cells were plated at the density of 20,000 cells/100 ul/well for 96 well format in the complete medium, and the following day replaced with the appropriate volume of Opti-MEM medium supplemented with 1% BSA and 0.1% FBS (Assay Buffer) for 30 minutes. To measure the effect of the antibodies of the invention in the absence of Leptin, the anti-LEPR antibodies or an isotype control antibody and human Leptin (hLeptin; R&D Systems, #398-LP) were half-log serially diluted to final concentrations ranging from 100 nM to 300 fM in Assay Buffer, which were added to the cells and subsequently incubated overnight at 37.degree. C. in 5% CO2.
[0144] To measure the effect of the antibodies of the invention in the presence of Leptin, a fixed concentration of human Leptin at 200 pM in Assay Buffer was added to the cells, immediately followed by the addition of anti-LEPR antibodies or isotype control antibody that were half-log serially diluted to final concentrations ranging from 100 nM to 300 fM. The samples were then incubated overnight at 37.degree. C. in 5% CO2. OneGlo reagent (Promega, #E6051) was then added to the samples and luciferase activity was measured on an Envision Multilable Plate Reader (Perkin Elmer) in Luminescent mode. The relative light unit (RLU) values were obtained and the results were analyzed using nonlinear regression with GraphPad Prism software (GraphPad). The maximum RLU value obtained from the hLeptin dose response was defined as 100% activation in the IMR-32/STAT3-LudhLEPR assay.
[0145] As shown in Table 13, in Study 1, in the absence of hLeptin, all of the anti-LEPR antibodies tested demonstrated weak stimulation of the IMR-32/STAT3-Luc/hLEPR cells with EC.sub.50 values ranging from 134 pM to 11.9 nM and maximal activation ranging from 5% to 13% respectively that of maximum activation obtained from the hLeptin dose response. In Study 2, in the absence of hLeptin, the 4 anti-LEPR antibodies tested demonstrated stimulation of the IMR-32/STAT3-Luc/hLEPR cells with EC.sub.50 values ranging from 61.9 pM to 206.9 pM and maximal activation ranging from 65% to 68% respective to the maximum activation obtained from the hLeptin dose response. In Study 1, in the presence of 200 pM of hLeptin, all of the anti-LEPR antibodies tested demonstrated stimulation of the IMR-32/STAT3-Luc/hLEPR cells with EC.sub.50 values ranging from 20.2 pM to 523 pM and maximal activation ranging from 66% to 107% respectively that of maximum activation obtained from the hLeptin dose response. Because these antibodies enhanced leptin-induced LEPR signaling, these antibodies were classified as "potentiators", as defined herein. In Study 2, in the presence of 200 pM of hLeptin, the 4 anti-LEPR antibodies tested demonstrated stimulation of the IMR-32/STAT3-Luc/hLEPR cells with EC.sub.50 values ranging from 51.9 pM to 257.3 pM with maximal activation ranging from 76% to 88% that of maximum activation obtained from the hLeptin dose response. LEPR signaling was not appreciably enhanced by these antibodies in the presence of leptin. The isotype control antibody did not demonstrate any measurable stimulation of the IMR-32/STAT3-Luc/hLEPR cells in any of the assays.
TABLE-US-00013 TABLE 13 Activation of hLEPR by anti-LEPR Antibodies IMR-32/LEPR without IMR-32/LEPR with human Leptin 200 pM human Leptin % % Antibody EC.sub.50 (M) activation EC.sub.50 (M) activation Study 1 H4H18445P2 1.19E-08 5 4.10E-10 97 H4H18446P2 3.73E-10 6 3.42E-11 68 H4H18449P2 2.12E-10 13 5.23E-11 66 H4H18438P2 1.49E-09 5 2.02E-11 76 H4H18482P2 2.69E-10 7 1.69E-10 94 H4H18487P2 8.01E-10 6 4.10E-10 107 H4H18492P2 1.34E-10 5 2.74E-11 94 H4H18417P2 1.53E-10 5 5.23E-10 87 Study 2 H4H16650P2 6.19E-11 68 5.19E-11 88 H4H16679P2 8.62E-11 65 7.37E-11 88 H4H17319P2 1.867E-10 68 1.914E-10 76 H4H17321P2 2.069E-10 66 2.573E-10 76
Example 8
Anti-LEPR Antibodies of the Invention Activate Signaling in Cells Expressing Signaling-Defective or Signaling-Impaired LEPR Mutants
[0146] LEPR mutants have been identified that exhibit defective or impaired leptin-mediated signaling and are associated with early-onset obesity. For example, LEPR-A409E is a signaling-defective mutant LEPR protein that does not transduce leptin signals to STAT3; the A409E mutant was originally identified as a monogenic cause of early onset obesity. (Farooqi et al., 2007, N Engl J Med 356(3): 237-247). LEPR-P316T is a signaling-impaired mutant LEPR protein that has also been shown to be associated with early-onset obesity. (Mazen et al., 2011, Mol Genet Metab 102:461-464).
[0147] In this Example, the ability of anti-LEPR antibodies of the invention to stimulate LEPR signaling in cell lines expressing signaling-defective or signaling-impaired LEPR mutants was assessed. In particular, reporter cell lines (HEK293) were constructed expressing either wild-type LEPR, LEPR-A409E (signaling-defective) or LEPR-P316T (signaling-impaired). Cells were treated with either vehicle only, recombinant human leptin, control IgG, or agonist anti-LEPR antibodies of the present invention (H4H16650 or H4H16679), and the extent of LEPR signaling (as measured by Western blot detection of pSTAT3-Y705 expression relative to STAT3 expression) was determined.
[0148] The agonist anti-LEPR antibodies of the present invention (H4H16650 and H4H16679) were shown in these experiments to stimulate LEPR signaling in cells expressing the LEPR-A409E mutant or the LEPR-P316T mutant (as measured by STAT3 expression) in a dose-dependent manner (FIG. 2, panels B and C). By contrast, leptin treatment induced only modest signaling in cells expressing the LEPR-P316T mutant, and no signaling in cells expressing the LEPR-A409E mutant. (FIG. 2, panel A). Moreover, no LEPR signaling was detected in any of the cell lines treated with vehicle or IgG control antibody (data not shown). Other signaling-defective or signaling-impaired LEPR mutants were tested in this assay but were not activated by anti-LEPR mutants (data not shown), suggesting that this rescue effect may be mutant-dependent.
[0149] The results of this Example indicate that the agonist anti-LEPR antibodies of the present invention may be useful in the treatment of diseases and disorders (e.g., early-onset obesity) that are caused by or associated with certain signaling-defective or signaling-impaired LEPR mutants (e.g., LEPR-P316T or LEPR-A409E).
Example 9
Octet Cross-Competition Between Different Anti-LEPR Monoclonal Antibodies
[0150] Binding competition between a panel of different anti-LEPR monoclonal antibodies was determined using a real time, label-free bio-layer interferometry assay on the Octet HTX biosensor platform (Pall ForteBio Corp.). The entire experiment was performed at 25.degree. C. in buffer containing 10 mM HEPES, 150 mM NaCl, 3 mM EDTA, and 0.05% v/v Surfactant Tween-20, 1 mg/mL BSA, pH7.4 (HBS-EBT) with the plate shaking at the speed of 1000 rpm. To assess whether two antibodies were able to compete with one another for binding to their respective epitopes on recombinant human LEPR expressed with a C-terminal myc-myc-hexahistidine tag (hLEPR.mmh; SEQ ID NO: 114), around 0.25 nm or 0.34 nm of hLEPR-MMH was first captured onto anti-penta-His antibody coated Octet biosensor tips (Fortebio Inc, #18-5122) by submerging the biosensor tips for 5 minutes in wells containing 20.mu.g/mL of hLEPR-MMH. The antigen captured biosensor tips were then saturated with a first anti-LEPR monoclonal antibody (subsequently referred to as mAb-1) by dipping into wells containing 50 .mu.g/mL solution of mAb-1 for 210 seconds. The biosensor tips were then subsequently dipped into wells containing a 50 .mu.g/mL solution of a second anti-LEPR monoclonal antibody (subsequently referred to as mAb-2) for 150 seconds. The biosensor tips were washed in HBS-EBT buffer in between every step of the experiment. The real-time binding response was monitored during the entire course of the experiment and the binding response at the end of every step was recorded. The response of mAb-2 binding to hLEPR-MMH pre-complexed with mAb-1 was compared and competitive/non-competitive behavior of different anti-LEPR monoclonal antibodies was determined as shown in Table 14 and Table 15.
TABLE-US-00014 TABLE 14 Cross-competition between anti-LEPR monoclonal antibodies First antibody Second antibody (mAb-1) binding to (mAb-2) shown to captured hLEPR-MMH compete with mAb-1 H4H18492P2 H4H18417P2 H4H18438P2 H4H18417P2 H4H18492P2 H4H18438P2 H4H18438P2 H4H18492P2 H4H18417P2 H4H16650P2 H4H16679P2 H4H16679P2 H4H16650P2 H4H18445P2 H4H18482P2 H4H18487P2 H4H18446P2 H4H18446P2 H4H18482P2 H4H18487P2 H4H18445P2 H4H18482P2 H4H18445P2 H4H18487P2 H4H18487P2 H4H18445P2 H4H18482P2 H4H18449P2 None Comparator Antibody None
TABLE-US-00015 TABLE 15 Cross-competition between anti-LEPR monoclonal antibodies mAb-2 that competes mAb-1 with mAb-1 H4H17319P2 H4H17321P2 H4H16650P2 H4H16679P2 H4H17321P2 H4H17319P2 H4H16650P2 H4H16679P2 H4H16650P2 H4H17319P2 H4H17321P2 H4H16679P2 H4H16679P2 H4H17319P2 H4H17321P2 H4H16650P2
Example 10
In vivo Efficacy of LEPR Agonist Antibodies H4H16650P2, H4H16679P2, H4H17319P2 and H4H17321P2 in an Inducible Mouse Model of Leptin Deficiency
[0151] The effects of four specific agonist anti-LEPR antibodies of the invention, H4H16650P2, H4H16679P2, H4H17319P2, and H4H17321P2 on food intake, body weight and adiposity were determined in an inducible model of leptin deficiency in genetically engineered LEPR.sup.Hu/Hu mice, that express a leptin receptor which is composed of the human LEPR ectodomain sequence in place of the murine LEPR ectodomain sequence. The model of leptin deficiency was induced by hydrodynamic DNA delivery (HDD) of a plasmid encoding an hFc-tagged mouse LEPR ectodomain (referred to herein as mLEPR.hFc or "Leptin trap"; SEQ ID NO: 120). The Leptin trap when expressed is secreted and binds circulating Leptin. After HDD of 50 .mu.g of the DNA construct encoding the Leptin trap, mice exhibited increased food consumption and increased adiposity and body weight.
[0152] Baseline daily food intake was measured between 7 and 4 days prior to administration of the Leptin trap (days -7 and -4). On day 0, thirty-five 13- to 17- week old male LEPR.sup.Hu/Hu mice were successfully subjected to HDD with the Leptin trap. On days 6 and 13 post HDD, retro-orbital bleeds were collected and body composition including adiposity was quantified by .mu.CT. On day 7 post HDD, mice were randomized into five groups of 7 mice based on percent body weight change from day 0. Each group received via subcutaneous injection either a single dose of isotype control antibody at 3 mg/kg, H4H16650P2 at 3 mg/kg, H4H16679P2 at 3 mg/kg, H4H17319P2 at 3 mg/kg, or H4H17321 at 3 mg/kg. The isotype control antibody did not bind any known mouse protein. Food intake and body weight were measured for each animal for the duration of the study. FIG. 3 summarizes the average daily food intake for each treatment group. In FIG. 3, the dotted line represents the average baseline food intake prior to HDD injection. The percent change in body weight from day 0 was calculated for each animal at each time point. FIG. 4 summarizes the average percent change in body weight for animals in each antibody treatment group. FIG. 5 summarizes the average fat mass for animals in each antibody treatment group quantified by .mu.CT 1 day prior to and 6 days following antibody treatment. All results are expressed as mean .+-.SEM.
[0153] As shown in FIGS. 3 and 4, following HDD with the Leptin trap, similar increases in food intake and percent change in body weight were observed among the groups of mice before antibody treatment. As shown in FIG. 3, mice treated with antibodies H4H16650P2 or H4H16679P2 at 3 mg/kg exhibited significant reductions in food intake starting at one day after antibody treatment (day 8 post HDD) and at subsequent time points measured as compared to mice injected with the isotype control antibody. Mice treated with antibodies H4H17319P2 or H4H17321P2 at 3 mg/kg exhibited a significant reduction in food intake at two days post antibody treatment (day 9 post HDD) and at the other subsequent time points measured as compared to mice injected with isotype control antibody. As shown in FIG. 4, mice treated with antibody H4H16650P2 at 3 mg/kg exhibited a significant reduction in percent body weight change one day after antibody treatment (day 8 post HDD) and at other subsequent time points measured as compared to mice injected with isotype control antibody. One day after antibody treatment, on day 8, mice treated with the isotype control showed a 21.16.+-.1.27% increase in body weight from day 0, whereas mice treated with H4H16650P2 had a 15.57.+-.0.9% increase in body weight from day 0. Mice treated with antibodies H4H16679P2, H4H17319P2 or H4H17321P2 at 3 mg/kg exhibited a significant reduction in percent body weight change two days after antibody treatment (day 9 post HDD) and at other subsequent time points measured as compared to mice injected with isotype control antibody. On day 9, the % body weight changes from day 0 were 23.18.+-.1.22, 13.17.+-.1.05, 12.95.+-.1.26, 15.98.+-.1.78 and 15.83.+-.2.01 for mice treated with isotype control, H4H16650P2, H4H16679P2, H4H17319P2, or H4H17321 P2, respectively. As shown in FIG. 5, mice treated with isotype control antibody at 3 mg/kg demonstrated a significant increase in fat mass 6 days after antibody treatment (day 13 post HDD) as compared to 1 day prior to antibody treatment (day 6 post HDD). Mice treated with antibodies H4H16650P2, H4H16679P2, H4H17319P2, or H4H17321 P2 at 3 mg/kg did not gain adipose mass after antibody treatment as compared to pre-antibody treatment. After 6 days of treatment (day 13 post HDD), mice treated with antibodies H4H16650P2, H4H16679P2 or H4H17319P2 at 3 mg/kg demonstrated significant decreases in fat mass as compared to mice treated with isotype control antibody at 3 mg/kg.
Example 11
Epitope Mapping of H4H16650P2 Binding to Human Leptin Receptor (hLEPR.mmh) by hydrogen deuterium exchange.
[0154] Experiments were conducted to determine the amino acid residues of hLEPR.mmh (amino acids M1-D839 of SEQ ID NO: 114) with which H4H16650P2 interacts. For this purpose H/D exchange epitope mapping with mass spectrometry was carried out. A general description of the H/D exchange method is set forth in, e.g., Ehring (1999) Analytical Biochemistry 267(2):252-259; and Engen and Smith (2001) Anal. Chem. 73:256A-265A.
[0155] Experimental procedure. HDX-MS experiments were performed on an integrated Waters HDX/MS platform, consisting of a Leaptec HDX PAL system for the deuterium labeling, a Waters Acquity M-Class (Auxiliary solvent manager) for the sample digestion and loading, a Waters Acquity M-Class (.mu.Binary solvent manager) for the analytical column gradient, and Synapt G2-Si mass spectrometer for peptic peptide mass measurement.
[0156] The labeling solution was prepared in 10 mM PBS buffer in D2O at pD 7.0 (equivalent to pH 6.6). For deterium labeling, 3.8 .mu.L of hLEPR.mmh (8 pmol/.mu.L) or hLEPR.mmh premixed with the antibody in 2:1 molar ratio was incubated with 56.2 .mu.L D2O labeling solution for various time-points (e.g., undeuterated control=0 sec, labeled for 1 min and 20 min). The deuteration was quenched by transferring 50 .mu.L sample to 50 .mu.L pre-chilled quench buffer (0.2 M TCEP, 6 M guanidine chloride in 100 mM phosphate buffer, pH 2.5) and the mixed sample was incubated at 1.0.degree. C. for two minutes. The quenched sample was then injected into a Waters HDX Manager for online pepsin/protease XIII digestion. The digested peptides were trapped onto an ACQUITY UPLC BEH C18 1.7-.mu.m, 2.1.times.5 mm VanGuard pre-column at 0.degree. C. and eluted to an analytical column ACQUITY UPLC BEH C18 1.7-.mu.m, 1.0.times.50 mm for a 9-minute gradient separation of 5%-40% B (mobile phase A: 0.1% formic acid in water, mobile phase B: 0.1% formic acid in acetonitrile). The mass spectrometer was set at cone voltage of 37 V, scan time of 0.5 s, and mass/charge range of 50-1700 Th.
[0157] For the identification of the peptides from human LEPR, LC-MSE data from undeuterated sample were processed and searched against the database including human LEPR, pepsin, and their randomized sequences via Waters ProteinLynx Global Server (PLGS) software. The identified peptides were imported to DynamX software and filtered by two criteria: 1) minimum products per amino acid: 0.2, and 2) replication file threshold: 3. DynamX software then automatically determined deuterium uptake of each peptide based on retention time and high mass accuracy (<10 ppm) across multiple time points with 3 replicates at each time.
[0158] Results. Using the online pepsin/protease XIII column coupled with MS.sup.E data acquisition, total 201 peptides from human LEPR were reproducibly identified in the absence or presence of the antibody, representing 70% sequence coverage. Five peptides had significantly reduced deuteration uptake (centroid delta values >0.4 daltons with p-values <0.05) when bound to H4H16650P2 as shown in the Table 16. The recorded peptide mass corresponds to the average value of the centroid MH+ mass from three replicates. These peptides, corresponding to amino acids 162-169 (amino acids LYVLPEVL of human LEPR; SEQ ID NO: 113), and to amino acids 170-181 (amino acids EDSPLVPQKGSF of human LEPR; SEQ ID NO: 113), had a slower deuteration rate when bound to H4H16650P2. These identified residues also correspond to residues acids 162-169 and 170-181 of human LEPR as defined by Uniprot entry P48357 (SEQ ID NO. 113; Human leptin receptor)
TABLE-US-00016 TABLE 16 Human Leptin receptor peptides with significant protection upon binding to antibody H4H16650P2 1 min Deuteration 20 min Deuteration hLEPR.mmh + hLEPR.mmh + Residues hLEPR.mmh H4H16650P2 .DELTA. hLEPR.mmh H4H16650P2 .DELTA. 162-169 949.03 .+-. 0.03 947.99 .+-. 0.02 -1.04 949.23 .+-. 0.02 948.16 .+-. 0.02 -1.03 163-169 835.82 .+-. 0.03 834.79 .+-. 0.02 -1.03 836.03 .+-. 0.02 834.94 .+-. 0.02 -1.08 170-181 1310.02 .+-. 0.05 1309.12 .+-. 0.03 -0.89 1309.77 .+-. 0.02 1309.38 .+-. 0.02 -0.39
Example 12
In vivo Efficacy Testing of LEPR Potentiator Antibodies in Humanized LEPR Mice
[0159] The effects of three specific potentiator anti-LEPR antibodies of the invention, H4H18482P2, H4H18487P2 and H4H18492P2, on body weight and adiposity were determined in singly-housed genetically engineered LEPR.sup.Hu/Hu mice, that express a leptin receptor which is composed of the human LEPR ectodomain sequence in place of the murine LEPR ectodomain sequence (mLEPR.hFc, SEQ ID NO: 120).
[0160] On days -19 body composition including adiposity was quantified by .mu.CT. On days 0, forty-eight 14 to 16-week old female LEPR.sup.Hu/Hu mice were randomized to four groups of 12 mice based on body weight. On days 0 and 11, mice from each group received via subcutaneous injection a single dose of isotype control antibody at 30 mg/kg, H4H18482P2 at 30 mg/kg, H4H18487P2 at 30 mg/kg or H4H18492P2 at 30 mg/kg. The isotype control antibody does not bind any known mouse protein. Body weight was measured for the duration of the study for each animal. The percent change in body weight from day 0 was calculated for each animal at each time point. FIG. 6 summarizes the average percent change in body weight for animals in each treatment group. FIG. 6 summarizes the average fat mass for animals in each antibody treatment group quantified by .mu.CT 19 days prior to and 11 days following antibody treatment. All results are expressed as mean .+-.SEM.
[0161] As shown in FIG. 6, decreases in percent change in body weight were observed following dosing with the LEPR potentiator antibodies, but not the isotype control antibody. As shown in FIG. 6, mice treated with H4H18482P2 at 30 mg/kg exhibited significant decreases in percent body weight change starting two days after treatment (day 2), and at the other time points compared to mice injected with an isotype control antibody. Mice treated with H4H18487P2 at 30 mg/kg exhibited significant decreases in percent body weight change starting at day 2 and at the other time points compared to mice injected with isotype control antibody. Mice treated with H4H18492P2 at 30 mg/kg exhibited a significant reduction in percent body weight change on days 4, 5 and 17 but not at other time points compared to mice injected with isotype control antibody. Mice treated with H4H18482P2 at 30 mg/kg exhibited a significant decrease in percent body weight change starting at day 6 and on subsequent days but not days 7, 14 and 17, compared to mice injected with H4H18492P2. Mice treated with H4H18487P2 at 30 mg/kg exhibited a significant decrease in percent body weight change starting at day 3 and at the other time points, but not days 4 and 5, compared to mice injected with H4H18492P2.
[0162] As shown in FIG. 7A, there were no differences in fat mass between the groups prior to treatment (day-19). As shown in FIG. 7B, mice treated with antibodies H4H18482 and H4H18487, but not H4H18492, at 30 mg/kg showed a statistically significant decrease in fat mass 17 days after treatment (day 12) as compared to the isotype control antibody.
[0163] The present invention is not to be limited in scope by the specific embodiments described herein. Indeed, various modifications of the invention in addition to those described herein will become apparent to those skilled in the art from the foregoing description and the accompanying figures. Such modifications are intended to fall within the scope of the appended claims.
Example 13
Effect of Anti-LEPR Antibodies of the Invention on Monkey LEPR Signaling
[0164] In order to assess transcriptional activation of monkey Leptin receptor, a stable cell line was developed. IMR-32 cells (human Neuroblastoma ATCC) were generated to stably express the extracellular domain of Macaca fascicularis LEPR (MfLEPR; amino acids 22 through 837 of accession number XP_005543194.1 with threonine at 827 changed to alanine) fused with the transmembrane and cytosolic domains of human LEPR (hLEPR; amino acids 840 through 1165 of accession number NP_002294.2) along with a luciferase reporter (STAT3-Luc; SABiosciences, #CLS-6028L). The resulting cell line, referred to hereafter as IMR-32/STAT3-Luc/MfLEPR was isolated and maintained in MEM-Earl medium supplemented with 10% FBS, NEAA, 1 ug/mL Puromycin, 100 ug/mL of Hygromycin B and Penicillin/Streptomycin/L-Glutamine.
[0165] The bioassay was performed to measure the effect of anti-LEPR antibodies of the invention on monkey LEPR signaling in the absence of Leptin. For the bioassay, IMR-32/STAT3-Luc/MfLEPR cells were plated at 10,000 cells/well in a 96-well plate in 0.1% FBS in Optimem with penicillin/streptomycin (assay buffer) and incubated overnight at 37.degree. C. in 5% CO.sub.2. The following day human leptin (hLeptin), anti-LEPR antibodies or an isotype control antibody were serially diluted from 50 nM to 0.8 pM in the assay buffer (plus a sample containing buffer alone without test molecule) and added to the cells. After 5.5 hours at 37.degree. C. in 5% CO.sub.2, luciferase activity was measured with OneGlo.TM. reagent (Promega, #E6031) and Victor.TM. X multilabel plate reader (Perkin Elmer). The results were analyzed using nonlinear regression (4-parameter logistics) with Prism.TM.6 software (Graph Pad) to obtain EC.sub.50 values. Percentage of activation of antibodies was calculated as the maximum range of RLU achieved by the antibody relative to that of maximum range of RLU achieved by hLeptin.
[0166] As shown in Table 17, in the absence of hLeptin, all of the anti-LEPR antibodies tested showed activation of monkey LEPR signaling in IMR-32/STAT3-Luc/mfLEPR cells with EC.sub.50 values ranging from 266 pM to 368 pM and maximal activation ranging from 76% to 82% where 100% activation was obtained with hLeptin. hLeptin activated with an EC.sub.50 value of 333 pM. The isotype control antibody did not demonstrate any measurable stimulation of the IMR-32/STAT3-Luc/mfLEPR cells.
TABLE-US-00017 TABLE 17 Activation of Macaca fascicularis LEPR by anti-LEPR antibodies % Leptin or Antibody EC.sub.50 (M) Activation Human Leptin 3.33E-11 100 H4H16650P2 2.66E-10 82 H4H16679P2 2.49E-10 80 H4H17319P2 3.65E-10 76 H4H17321P2 3.68E-10 78 Isotype control antibody No Activation No Activation
Example 14
Epitope Binding to the Full-Length Extracellular Domain of Human LEPR Using Luminex MFI Signal
[0167] To determine the epitope of human LEPR on which anti-LEPR antibodies of the invention bind, a Luminex FLEXMAP (FM3DD, LuminexCorp) flow cytometry based analysis was utilized to characterize the interaction of anti-LEPR antibodies with recombinant human LEPR protein domains. For the assay, approximately 3 million carboxylated Microplex.sup.R microspheres (Luminex, Cat #LC1000A), were washed, vortexed and sonicated in 0.1 M NaPO.sub.4, pH 6.2 (activation buffer) and then centrifuged to remove the supernatant. The microspheres were resuspended in 120 .mu.L of activation buffer and the carboxylate groups (--COOH) were activated by addition of 15 .mu.L of 50 mg/mL of N-hydroxysuccinimide (NHS, Thermo Scientific, Cat #24500) followed by addition of 15 .mu.L of 50 mg/mL of 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide (EDC, ThermoScientific, Cat #22980) at 25.degree. C. After 10 minutes, the pH of the reaction was reduced to 5.0 with the addition of 600 .mu.L of 50 mM MES, pH 5 (coupling buffer), and the microspheres were vortexed, and centrifuged to remove supernatant. The activated beads were immediately mixed with 500 .mu.L of 20 .mu.g/mL monoclonal anti-myc antibodies with either a mouse IgG or a human IgG, in coupling buffer and incubated for two hours at 25.degree. C. The coupling reaction was quenched by addition of 50 .mu.L of 1M Tris-HCl, pH 8.0 and the microspheres were rapidly vortexed, centrifuged, and washed four times with 1 mL of DPBS, to remove uncoupled proteins and other reaction components.
[0168] The transiently expressed LEPR proteins, including human LEPR extracellular domain expressed with a C-terminal myc-myc hexahistidine tag (human LEPR-MMH, SEQ ID NO: 113), human LEPR CRH1 (D1) expressed with a C-terminal myc-myc hexahistidine tag (human LEPR CRH1 (D1)-MMH, amino acids 1-208 of SEQ ID NO: 113 with a myc-myc hexahistidine tag, amino acids 209-236), human LEPR CRH1 (D1,D2) domain expressed with a C-terminal myc-myc hexahistidine tag (human LEPR CRH1 (D1,D2)-MMH, amino acids 1-318 of SEQ ID NO: 113 with a myc-myc hexahistidine tag, amino acids 319-346), human LEPR CRH1-Ig (D1,D2,D3) domain expressed with a C-terminal myc-myc hexahistidine tag (human LEPR CRH1 (D1,D2,D3)-MMH, amino acids 1-278 of SEQ ID NO: 113 with a myc-myc hexahistidine tag, amino acids 279-306), human LEPR CRH1-Ig (D2,D3) domain expressed with a C-terminal myc-myc hexahistidine tag (human LEPR CRH1-Ig (D2,D3)-MMH, amino acids 1-198 of SEQ ID NO: 113 with a myc-myc hexahistidine tag, amino acids 199-226), human LEPR Ig (D3) domain expressed with a C-terminal myc-myc hexahistidine tag (human LEPR Ig (D3)-MMH, amino acids 1-88 of SEQ ID NO: 113 with a myc-myc hexahistidine tag, amino acids 89-116), human LEPR CRH2 domain expressed with a C-terminal myc-myc hexahistidine tag (human LEPR CRH2-MMH, amino acids 1-207 of SEQ ID NO: 113 with a myc-myc-hexahistidine tag, amino acids 208-235), human LEPR FNIII domain expressed with a C-terminal myc-myc hexahistidine tag (human LEPR FNIII-MMH, amino acids 1-204 of SEQ ID NO: 113 with a myc-myc hexahistidine tage, amino acids 205-232), and human LEPR Ig-CRH2-FNIII domain expressed with a C-terminal myc-myc hexahistidine tag (human LEPR Ig-CRH2-FNIII-MMH, amino acids 1-510 of SEQ ID NO: 113 with a myc-myc-hexahistidine tag, amino acids 511-538), were suspended in serum free CHO-S-SFM II Medium (Thermo Fisher, Cat #31033020) and were then clarified by centrifugation. Aliquots of microspheres with immobilized anti-myc monoclonal antibodies, prepared as described above, were added individually to 1 mL of the each of these protein supernatants. The microspheres were gently mixed, incubated for two hours at 25.degree. C., washed twice with 1 mL of DBPS, centrifuged to remove the supernatant and finally resuspended in 1 mL of DPBS buffer. Forty-eight .mu.L of anti-myc IgG coupled microspheres from individual reactions with full length human LEPR and with each of the human LEPR domain proteins were withdrawn and mixed together in 3.6 mL of PBS+20 mg/mL BSA+0.05% sodium azide (blocking buffer).
[0169] From this mixed pool, 75 .mu.L of microspheres were plated per well on a 96 well filter plate (Millipore, Cat. No: MSBVN1250) and mixed with 25 .mu.L of individual anti- human LEPR monoclonal antibodies (0.5 or 5 .mu.g/mL), incubated for two hours at 25.degree. C. and then washed twice with 200 .mu.L of DPBS with 0.05% Tween 20 (washing buffer). To detect and quantify the amounts of bound anti-LEPR antibody levels to individual microspheres, either 100 .mu.L of 2.5 .mu.g/mL R-Phycoerythrin conjugated goat F(ab')2 anti-human kappa (Southern Biotech, Cat #2063-09) in blocking buffer or 100 .mu.L of 1.25 .mu.g/mL R-Phycoerythrin AffiniPure F(ab')2 Fragment Goat Anti-Mouse IgG, F(ab')2 Fragment Specific (Jackson Immunoresearch, Cat. No: 115-116-072) in blocking buffer, was added and incubated for 30 minutes at 25.degree. C. After 30 minutes, the samples were washed twice with 200 .mu.L of washing buffer and resuspended in 150 .mu.L of wash buffer. The Median Fluorescence intensity (MFI) of the microspheres was measured in a Luminex Analyzer.
TABLE-US-00018 TABLE 18 Luminex MFI signal of anti-LEPR antibodies binding to myc tag captured full- length extracellular domain of human LEPR and isonalted human LEPR domains Full Length CRH1 CRH1 CRH1-Ig CRH1-Ig Ig Ig-CRH2- extracellular Probable Antibody (D1) (D1, D2) (D1, D2, D3) (D2, D3) (D3) CRH2 FNIII FNIII domain Binding site H4H18445P2 12 30 22 40 19 17 230 14544 6573 FNIII H4H18446P2 17 682 205 645 25 65 32 16852 10536 Ig-CRH2- FNIII H4H18482P2 13 40 21 52 27 23 167 15316 7311 Ig-CRH2- FNIII H4H18487P2 12 51 29 62 22 27 174 16320 7329 Ig-CRH2- FNIII H4H18417P2 10 16048 3334 5502 17 39 14 37 4887 CRH1 D2 H4H18438P2 13 18931 6572 8884 30 165 25 468 6251 CRH1 D2 H4H18492P2 11 19371 6354 8685 19 18 16 186 6382 CRH1 D2 H4H18449P2 20 2934 2056 42 24 15 13 43 7976 CRH1(D1-2) H4H16650P2 8 4722 2562 74 10 16 6 110 7603 CRH1(D1-2) H4H16679P2 12 4388 2797 34 14 33 10 42 7507 CRH1(D1-2) H4H17319P2 8 1246 938 14 8 91 20 8 3305 CRH1(D1-2) H4H17321P2 9 2649 1752 15 7 116 40 14 4696 CRH1(D1-2) Comparator -14 19 -57 27 10 9404 73 7112 3908 CRH2 mAb
[0170] The results of the Luminex based analysis are tabulated in Table 18. Luminex MFI signal intensities indicate that the twelve anti-LEPR antibodies of the invention bound to the complete human LEPR extracellular domain. Anti-LEPR antibodies H4H18417P2, H4H18438P2, and H4H18492P2, bound to epitopes within the CRH1 D2 domain of human LEPR. Anti-LEPR antibodies H4H18449P2, H4H16650P and H4H16679P, bound to epitopes within the CRH1(D1-2) domain of human LEPR. Anti-LEPR antibody Comparator mAB, bound to epitopes within the CRH2 domain of human LEPR. Anti-LEPR antibody H4H18445P2 bound to epitopes within the FNIII domain of human LEPR. Anti-LEPR antibodies H4H18446P2, H4H18482P2 and H4H18487P2, bound to epitopes within the Ig-CRH2-FNIII domain of human LEPR.
Sequence CWU 1
1
1201357DNAArtificial Sequencesynthetic 1caggtgcagc tggtggagtc
tgggggaggc gtggtccagc ctgggaagtc cctgagactc 60tcctgtgtag cgtctggatt
caccttcagt tccgatgcca tgtactgggt ccgccaggct 120ccaggcaagg
ggctggaatg ggtggcagtt atttattatg atggaaatta tcaatactat
180gaagactccg ttaagggtcg attcaccatc tccagagaca attcccagaa
cacgctggat 240ctgcaaatga acagcctgag agtcgacgac acggctgtat
atttctgtgc gcgtctcaac 300tgggattact ggtatctcga tctctggggc
cgtggcaccc tggtcactgt ctcctca 3572119PRTArtificial
Sequencesynthetic 2Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val Val
Gln Pro Gly Lys1 5 10 15Ser Leu Arg Leu Ser Cys Val Ala Ser Gly Phe
Thr Phe Ser Ser Asp 20 25 30Ala Met Tyr Trp Val Arg Gln Ala Pro Gly
Lys Gly Leu Glu Trp Val 35 40 45Ala Val Ile Tyr Tyr Asp Gly Asn Tyr
Gln Tyr Tyr Glu Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg
Asp Asn Ser Gln Asn Thr Leu Asp65 70 75 80Leu Gln Met Asn Ser Leu
Arg Val Asp Asp Thr Ala Val Tyr Phe Cys 85 90 95Ala Arg Leu Asn Trp
Asp Tyr Trp Tyr Leu Asp Leu Trp Gly Arg Gly 100 105 110Thr Leu Val
Thr Val Ser Ser 115324DNAArtificial Sequencesynthetic 3ggattcacct
tcagttccga tgcc 2448PRTArtificial Sequencesynthetic 4Gly Phe Thr
Phe Ser Ser Asp Ala1 5524DNAArtificial Sequencesynthetic
5atttattatg atggaaatta tcaa 2468PRTArtificial Sequencesynthetic
6Ile Tyr Tyr Asp Gly Asn Tyr Gln1 5736DNAArtificial
Sequencesynthetic 7gcgcgtctca actgggatta ctggtatctc gatctc
36812PRTArtificial Sequencesynthetic 8Ala Arg Leu Asn Trp Asp Tyr
Trp Tyr Leu Asp Leu1 5 109324DNAArtificial Sequencesynthetic
9gacatccaga tgacccagtc tccatcctcc ctgtctgcat ctgtaggaga cagagtcacc
60atcacttgcc gggcaagtca gagcattagc agctatttaa attggtatca gcagaaacca
120gggaaagccc ctaagctcct gatctatgct gcatccagtt tgcaaagtgg
ggtcccgtca 180aggttcagtg gcagtggatc tgggacagat ttcactctca
ccatcagcag tctgcaacct 240gaagattttg caacttacta ctgtcaacag
agttacagta cccctccgat caccttcggc 300caagggacac gactggagat taaa
32410108PRTArtificial Sequencesynthetic 10Asp Ile Gln Met Thr Gln
Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile
Thr Cys Arg Ala Ser Gln Ser Ile Ser Ser Tyr 20 25 30Leu Asn Trp Tyr
Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Ala Ala
Ser Ser Leu Gln Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly
Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75
80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln Ser Tyr Ser Thr Pro Pro
85 90 95Ile Thr Phe Gly Gln Gly Thr Arg Leu Glu Ile Lys 100
1051118DNAArtificial Sequencesynthetic 11cagagcatta gcagctat
18126PRTArtificial Sequencesynthetic 12Gln Ser Ile Ser Ser Tyr1
5139DNAArtificial Sequencesynthetic 13gctgcatcc 9143PRTArtificial
Sequencesynthetic 14Ala Ala Ser11530DNAArtificial Sequencesynthetic
15caacagagtt acagtacccc tccgatcacc 301610PRTArtificial
Sequencesynthetic 16Gln Gln Ser Tyr Ser Thr Pro Pro Ile Thr1 5
1017357DNAArtificial Sequencesynthetic 17caggtgcagc tggtggagtc
cgggggaggc gtggtccagc ctgggaggtc cctgagactc 60tcctgtacag cgtctggatt
caccttcagt agttatgcca tgtactgggt ccgccaggct 120ccaggcaagg
ggctggagtg ggtgtcagtt atatactatg atggaagtta taaatactat
180gcagactccg tgaagggccg attcaccatc tccagagaca attccaagaa
cacgctgtat 240ctgcaaatgg acagcctgag agccgaggac acggctgtct
attactgtgc gagttataac 300tggaactact ggtacttcga tttctggggc
cgtggcaccc tggtcactgt ctcctca 35718119PRTArtificial
Sequencesynthetic 18Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val Val
Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Thr Ala Ser Gly Phe
Thr Phe Ser Ser Tyr 20 25 30Ala Met Tyr Trp Val Arg Gln Ala Pro Gly
Lys Gly Leu Glu Trp Val 35 40 45Ser Val Ile Tyr Tyr Asp Gly Ser Tyr
Lys Tyr Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg
Asp Asn Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asp Ser Leu
Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Ser Tyr Asn Trp
Asn Tyr Trp Tyr Phe Asp Phe Trp Gly Arg Gly 100 105 110Thr Leu Val
Thr Val Ser Ser 1151924DNAArtificial Sequencesynthetic 19ggattcacct
tcagtagtta tgcc 24208PRTArtificial Sequencesynthetic 20Gly Phe Thr
Phe Ser Ser Tyr Ala1 52124DNAArtificial Sequencesynthetic
21atatactatg atggaagtta taaa 24228PRTArtificial Sequencesynthetic
22Ile Tyr Tyr Asp Gly Ser Tyr Lys1 52336DNAArtificial
Sequencesynthetic 23gcgagttata actggaacta ctggtacttc gatttc
362412PRTArtificial Sequencesynthetic 24Ala Ser Tyr Asn Trp Asn Tyr
Trp Tyr Phe Asp Phe1 5 1025357DNAArtificial Sequencesynthetic
25caggtgcagc tggtggagtc tgggggaagc gtggtccagc ctgggaggtc cctgagactc
60tcctgtgcag cgtctggatt caccttcagt acatatgcca tgtactgggt ccgccagact
120ccaggcaagg ggctggagtg ggtggctgtt ttatactctg atggaagtaa
taaatactat 180atagactccg tgaagggccg attcaccatc tccagagaca
cttccacgaa cactctgtat 240ctgcaaatga gcagcctgcg agccgacgac
tcggctctat attactgtgc gcgtctcaac 300tgggattact ggtacttcga
tctctggggc cgtggcaccc tggtcactgt ctcctca 35726119PRTArtificial
Sequencesynthetic 26Gln Val Gln Leu Val Glu Ser Gly Gly Ser Val Val
Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe
Thr Phe Ser Thr Tyr 20 25 30Ala Met Tyr Trp Val Arg Gln Thr Pro Gly
Lys Gly Leu Glu Trp Val 35 40 45Ala Val Leu Tyr Ser Asp Gly Ser Asn
Lys Tyr Tyr Ile Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg
Asp Thr Ser Thr Asn Thr Leu Tyr65 70 75 80Leu Gln Met Ser Ser Leu
Arg Ala Asp Asp Ser Ala Leu Tyr Tyr Cys 85 90 95Ala Arg Leu Asn Trp
Asp Tyr Trp Tyr Phe Asp Leu Trp Gly Arg Gly 100 105 110Thr Leu Val
Thr Val Ser Ser 1152724DNAArtificial Sequencesynthetic 27ggattcacct
tcagtacata tgcc 24288PRTArtificial Sequencesynthetic 28Gly Phe Thr
Phe Ser Thr Tyr Ala1 52924DNAArtificial Sequencesynthetic
29ttatactctg atggaagtaa taaa 24308PRTArtificial Sequencesynthetic
30Leu Tyr Ser Asp Gly Ser Asn Lys1 53136DNAArtificial
Sequencesynthetic 31gcgcgtctca actgggatta ctggtacttc gatctc
363212PRTArtificial Sequencesynthetic 32Ala Arg Leu Asn Trp Asp Tyr
Trp Tyr Phe Asp Leu1 5 1033357DNAArtificial Sequencesynthetic
33caggtgcagc tggtggagtc tgggggaggc gtggtccagc ctgggaggtc cctgagactc
60tcctgtgaag cgtctggatt cagcagcagt gacaatgcca tgtactgggt ccgccaggct
120ccaggcaagg ggctggagtg ggtgtcagtt atatatcatg atggaagtta
taaatactat 180gaagactccg tgaagggccg attcaccatc gccagagaca
attccaagaa cacgctttat 240ttgcaaatga acagcctgag agccgaggac
acggctgtat attactgtgc gaggtataac 300tggaaccact ggtacttcga
tgtctggggc cgtggcaccc tggtcactgt ctcctca 35734119PRTArtificial
Sequencesynthetic 34Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val Val
Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Glu Ala Ser Gly Phe
Ser Ser Ser Asp Asn 20 25 30Ala Met Tyr Trp Val Arg Gln Ala Pro Gly
Lys Gly Leu Glu Trp Val 35 40 45Ser Val Ile Tyr His Asp Gly Ser Tyr
Lys Tyr Tyr Glu Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ala Arg
Asp Asn Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu
Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Tyr Asn Trp
Asn His Trp Tyr Phe Asp Val Trp Gly Arg Gly 100 105 110Thr Leu Val
Thr Val Ser Ser 1153524DNAArtificial Sequencesynthetic 35ggattcagca
gcagtgacaa tgcc 24368PRTArtificial Sequencesynthetic 36Gly Phe Ser
Ser Ser Asp Asn Ala1 53724DNAArtificial Sequencesynthetic
37atatatcatg atggaagtta taaa 24388PRTArtificial Sequencesynthetic
38Ile Tyr His Asp Gly Ser Tyr Lys1 53936DNAArtificial
Sequencesynthetic 39gcgaggtata actggaacca ctggtacttc gatgtc
364012PRTArtificial Sequencesynthetic 40Ala Arg Tyr Asn Trp Asn His
Trp Tyr Phe Asp Val1 5 1041366DNAArtificial Sequencesynthetic
41caggtgcagc tggtggagtc tgggggaggc gtggtccagc ctgggaggtc cctgagactc
60tcctgtgcag cctctggatt caccttcagt acctatggca tgcactgggt ccgccaggct
120ccaggcaagg ggctggagtg ggtgtcagtt atatcatatg acgaaagtaa
taagtactat 180gcagactccg tgaagggccg attcaccatt tctagagaca
attccaagaa cgcgctgtat 240ttacaaatga acagcctgag aaatgaggac
acggctgtgt attactgtgc gagagatcgg 300ccttttggat tggttaccgg
atggttcgac ccctggggcc agggaaccct ggtcaccgtc 360tcctca
36642122PRTArtificial Sequencesynthetic 42Gln Val Gln Leu Val Glu
Ser Gly Gly Gly Val Val Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser
Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr Tyr 20 25 30Gly Met His Trp
Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ser Val Ile
Ser Tyr Asp Glu Ser Asn Lys Tyr Tyr Ala Asp Ser Val 50 55 60Lys Gly
Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Ala Leu Tyr65 70 75
80Leu Gln Met Asn Ser Leu Arg Asn Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Arg Asp Arg Pro Phe Gly Leu Val Thr Gly Trp Phe Asp Pro
Trp 100 105 110Gly Gln Gly Thr Leu Val Thr Val Ser Ser 115
1204324DNAArtificial Sequencesynthetic 43ggattcacct tcagtaccta tggc
24448PRTArtificial Sequencesynthetic 44Gly Phe Thr Phe Ser Thr Tyr
Gly1 54524DNAArtificial Sequencesynthetic 45atatcatatg acgaaagtaa
taag 24468PRTArtificial Sequencesynthetic 46Ile Ser Tyr Asp Glu Ser
Asn Lys1 54745DNAArtificial Sequencesynthetic 47gcgagagatc
ggccttttgg attggttacc ggatggttcg acccc 454815PRTArtificial
Sequencesynthetic 48Ala Arg Asp Arg Pro Phe Gly Leu Val Thr Gly Trp
Phe Asp Pro1 5 10 1549360DNAArtificial Sequencesynthetic
49caggtgcagc tggtggagtc tgggggaggc gtggtccagc ctgggaggtc cctgagactc
60tcctgtgcag cgtctggatt cagtttcaat acctatggca tgcactgggt ccgccaggct
120ccaggcaagg ggctggagtg ggtgacaatt atatggtatg atggaagtat
taaatactat 180gcagactccg tgaagggccg attcaccatc tccagagaca
attccaagaa cacgctgtat 240ctgcaaatga acagcctgag agccgaggac
acggctgtgt attattgtgc gagaggtgga 300tatagtggct acctctactt
tgactactgg ggccagggaa ccctggtcac cgtctcctca 36050120PRTArtificial
Sequencesynthetic 50Gln Val Gln Leu Val Glu Ser Gly Gly Gly Val Val
Gln Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe
Ser Phe Asn Thr Tyr 20 25 30Gly Met His Trp Val Arg Gln Ala Pro Gly
Lys Gly Leu Glu Trp Val 35 40 45Thr Ile Ile Trp Tyr Asp Gly Ser Ile
Lys Tyr Tyr Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg
Asp Asn Ser Lys Asn Thr Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu
Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Gly Gly Tyr
Ser Gly Tyr Leu Tyr Phe Asp Tyr Trp Gly Gln 100 105 110Gly Thr Leu
Val Thr Val Ser Ser 115 1205124DNAArtificial Sequencesynthetic
51ggattcagtt tcaataccta tggc 24528PRTArtificial Sequencesynthetic
52Gly Phe Ser Phe Asn Thr Tyr Gly1 55324DNAArtificial
Sequencesynthetic 53atatggtatg atggaagtat taaa 24548PRTArtificial
Sequencesynthetic 54Ile Trp Tyr Asp Gly Ser Ile Lys1
55539DNAArtificial Sequencesynthetic 55gcgagaggtg gatatagtgg
ctacctctac tttgactac 395613PRTArtificial Sequencesynthetic 56Ala
Arg Gly Gly Tyr Ser Gly Tyr Leu Tyr Phe Asp Tyr1 5
1057372DNAArtificial Sequencesynthetic 57caggtgcagc tgcaggagtc
gggcccagga ctggtgaagc cttcacagac cctgtccctc 60acctgcactg tctctggtgg
ctccatcagc agcggtggtg actactggag ctggatccgc 120cagctcccag
ggaagggcct ggagtggatt gggtacatct attacagtgg gagcgcctac
180tataatccgt ccctcaagag tcgaggtacc atatcaatag acacgtctaa
gaaccagttc 240tccctgaagc tgacctctgt gactgccgcg gacacggccg
tatatttctg tgtgaaatta 300cgatttttgg agtggttctt ggggggctgg
ttcggcccct ggggccaggg aaccctggtc 360accgtctcct ca
37258124PRTArtificial Sequencesynthetic 58Gln Val Gln Leu Gln Glu
Ser Gly Pro Gly Leu Val Lys Pro Ser Gln1 5 10 15Thr Leu Ser Leu Thr
Cys Thr Val Ser Gly Gly Ser Ile Ser Ser Gly 20 25 30Gly Asp Tyr Trp
Ser Trp Ile Arg Gln Leu Pro Gly Lys Gly Leu Glu 35 40 45Trp Ile Gly
Tyr Ile Tyr Tyr Ser Gly Ser Ala Tyr Tyr Asn Pro Ser 50 55 60Leu Lys
Ser Arg Gly Thr Ile Ser Ile Asp Thr Ser Lys Asn Gln Phe65 70 75
80Ser Leu Lys Leu Thr Ser Val Thr Ala Ala Asp Thr Ala Val Tyr Phe
85 90 95Cys Val Lys Leu Arg Phe Leu Glu Trp Phe Leu Gly Gly Trp Phe
Gly 100 105 110Pro Trp Gly Gln Gly Thr Leu Val Thr Val Ser Ser 115
1205930DNAArtificial Sequencesynthetic 59ggtggctcca tcagcagcgg
tggtgactac 306010PRTArtificial Sequencesynthetic 60Gly Gly Ser Ile
Ser Ser Gly Gly Asp Tyr1 5 106121DNAArtificial Sequencesynthetic
61atctattaca gtgggagcgc c 21627PRTArtificial Sequencesynthetic
62Ile Tyr Tyr Ser Gly Ser Ala1 56348DNAArtificial Sequencesynthetic
63gtgaaattac gatttttgga gtggttcttg gggggctggt tcggcccc
486416PRTArtificial Sequencesynthetic 64Val Lys Leu Arg Phe Leu Glu
Trp Phe Leu Gly Gly Trp Phe Gly Pro1 5 10 1565351DNAArtificial
Sequencesynthetic 65gaggtgcagc tggtggagtc tgggggaggc ttggtacagc
cgggggggtc cctgagactc 60tcctgtgcag cctctggatt cacctttagc aactatggca
tgacctgggt ccgccaggct 120ccagggaagg gcctggaatg ggtctcagct
attactggtg gtggtggtag cacatactac 180tcaaactccg tgaagggccg
gttcaccatc tccagagaca attccaagaa cacggtgtat 240ctgcgaatga
acagtgtgag agccgaggac acggccgtat attactgtgc gaaatataag
300tggaacttcg tggacgactg gggccaggga accacggtca ccgtctcctc a
35166117PRTArtificial Sequencesynthetic 66Glu Val Gln Leu Val Glu
Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser
Cys Ala Ala Ser Gly Phe Thr Phe Ser Asn Tyr 20 25 30Gly Met Thr Trp
Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ser Ala Ile
Thr Gly Gly Gly Gly Ser Thr Tyr Tyr Ser Asn Ser Val 50 55 60Lys Gly
Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Val Tyr65 70 75
80Leu Arg Met Asn Ser Val Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Lys Tyr Lys Trp Asn Phe Val Asp Asp Trp Gly Gln Gly Thr
Thr 100 105 110Val Thr Val Ser Ser 1156724DNAArtificial
Sequencesynthetic 67ggattcacct ttagcaacta tggc 24688PRTArtificial
Sequencesynthetic 68Gly Phe Thr Phe Ser Asn Tyr Gly1
56924DNAArtificial Sequencesynthetic 69attactggtg gtggtggtag caca
24708PRTArtificial Sequencesynthetic 70Ile Thr Gly Gly
Gly Gly Ser Thr1 57130DNAArtificial Sequencesynthetic 71gcgaaatata
agtggaactt cgtggacgac 307210PRTArtificial Sequencesynthetic 72Ala
Lys Tyr Lys Trp Asn Phe Val Asp Asp1 5 1073360DNAArtificial
Sequencesynthetic 73gaggtgcagc tggtggagtc tgggggaggc ttggtacagc
ctggggggtc cctgagactc 60tcctgtgttg cctctggatt caccttcaat aaatacgaca
tgcactgggt ccgccaaact 120actggaaaag gtctagagtg ggtctcaggt
attgatactg atggtgacac atactatcca 180ggctccgtga agggccgatt
caccatctcc agagaaaatg ccgagaactc cctgtatctt 240caaatgaacg
gcctgagagt cggggacacg gctgtgtatt actgtgcaag atggccttgg
300agtggtttct atggtgcttt tgatatctgg ggccaaggga caatggtcac
cgtctcttca 36074120PRTArtificial Sequencesynthetic 74Glu Val Gln
Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu
Arg Leu Ser Cys Val Ala Ser Gly Phe Thr Phe Asn Lys Tyr 20 25 30Asp
Met His Trp Val Arg Gln Thr Thr Gly Lys Gly Leu Glu Trp Val 35 40
45Ser Gly Ile Asp Thr Asp Gly Asp Thr Tyr Tyr Pro Gly Ser Val Lys
50 55 60Gly Arg Phe Thr Ile Ser Arg Glu Asn Ala Glu Asn Ser Leu Tyr
Leu65 70 75 80Gln Met Asn Gly Leu Arg Val Gly Asp Thr Ala Val Tyr
Tyr Cys Ala 85 90 95Arg Trp Pro Trp Ser Gly Phe Tyr Gly Ala Phe Asp
Ile Trp Gly Gln 100 105 110Gly Thr Met Val Thr Val Ser Ser 115
1207524DNAArtificial Sequencesynthetic 75ggattcacct tcaataaata cgac
24768PRTArtificial Sequencesynthetic 76Gly Phe Thr Phe Asn Lys Tyr
Asp1 57721DNAArtificial Sequencesynthetic 77attgatactg atggtgacac a
21787PRTArtificial Sequencesynthetic 78Ile Asp Thr Asp Gly Asp Thr1
57942DNAArtificial Sequencesynthetic 79gcaagatggc cttggagtgg
tttctatggt gcttttgata tc 428014PRTArtificial Sequencesynthetic
80Ala Arg Trp Pro Trp Ser Gly Phe Tyr Gly Ala Phe Asp Ile1 5
1081366DNAArtificial Sequencesynthetic 81caggtgcagc tgcaggagtc
gggcccagga ctggtgaagc cttcacagac cctgtccctc 60acctgcactg tctctggtgg
ctccatcagc agtggtaatt actactggaa ctggatccgc 120caacagccag
gagagggcct ggagtggatt gcttacatct atcacaatgg ggtcaccaac
180ttcaatccgt ccctcaagag tcgacttact atatcagtag acacgtctaa
gactcagttc 240tccctgaagt tgaggtctgt gactgccgcg gacacggccg
tttattactg tgcgagatca 300ggcagctggt tcgagaactg gtacttcgat
ctctggggcc gtggcaccct ggtcactgtc 360tcctca 36682122PRTArtificial
Sequencesynthetic 82Gln Val Gln Leu Gln Glu Ser Gly Pro Gly Leu Val
Lys Pro Ser Gln1 5 10 15Thr Leu Ser Leu Thr Cys Thr Val Ser Gly Gly
Ser Ile Ser Ser Gly 20 25 30Asn Tyr Tyr Trp Asn Trp Ile Arg Gln Gln
Pro Gly Glu Gly Leu Glu 35 40 45Trp Ile Ala Tyr Ile Tyr His Asn Gly
Val Thr Asn Phe Asn Pro Ser 50 55 60Leu Lys Ser Arg Leu Thr Ile Ser
Val Asp Thr Ser Lys Thr Gln Phe65 70 75 80Ser Leu Lys Leu Arg Ser
Val Thr Ala Ala Asp Thr Ala Val Tyr Tyr 85 90 95Cys Ala Arg Ser Gly
Ser Trp Phe Glu Asn Trp Tyr Phe Asp Leu Trp 100 105 110Gly Arg Gly
Thr Leu Val Thr Val Ser Ser 115 1208330DNAArtificial
Sequencesynthetic 83ggtggctcca tcagcagtgg taattactac
308410PRTArtificial Sequencesynthetic 84Gly Gly Ser Ile Ser Ser Gly
Asn Tyr Tyr1 5 108521DNAArtificial Sequencesynthetic 85atctatcaca
atggggtcac c 21867PRTArtificial Sequencesynthetic 86Ile Tyr His Asn
Gly Val Thr1 58742DNAArtificial Sequencesynthetic 87gcgagatcag
gcagctggtt cgagaactgg tacttcgatc tc 428814PRTArtificial
Sequencesynthetic 88Ala Arg Ser Gly Ser Trp Phe Glu Asn Trp Tyr Phe
Asp Leu1 5 1089324DNAArtificial Sequencesynthetic 89gaaattgtgt
tgacgcagtc tccaggcacc ctgtctttgt ctccagggga aagagccacc 60ctctcctgca
gggccagtca gagtgttagc agcagctact tagcctggta ccagcagaaa
120cctggccagg ctcccaggct cctcatctat ggtgcatcca gcagggccac
tggcatccca 180gacaggttca gtggcagtgg gtctgggaca gacttcactc
tcaccatcag cagactggag 240cctgaagatt ttgcagtgta ttactgtcag
cagtatggta gctcaccttg gacgttcggc 300caagggacca aggtggaaat caaa
32490108PRTArtificial Sequencesynthetic 90Glu Ile Val Leu Thr Gln
Ser Pro Gly Thr Leu Ser Leu Ser Pro Gly1 5 10 15Glu Arg Ala Thr Leu
Ser Cys Arg Ala Ser Gln Ser Val Ser Ser Ser 20 25 30Tyr Leu Ala Trp
Tyr Gln Gln Lys Pro Gly Gln Ala Pro Arg Leu Leu 35 40 45Ile Tyr Gly
Ala Ser Ser Arg Ala Thr Gly Ile Pro Asp Arg Phe Ser 50 55 60Gly Ser
Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Arg Leu Glu65 70 75
80Pro Glu Asp Phe Ala Val Tyr Tyr Cys Gln Gln Tyr Gly Ser Ser Pro
85 90 95Trp Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys 100
1059121DNAArtificial Sequencesynthetic 91cagagtgtta gcagcagcta c
21927PRTArtificial Sequencesynthetic 92Gln Ser Val Ser Ser Ser Tyr1
5939DNAArtificial Sequencesynthetic 93ggtgcatcc 9943PRTArtificial
Sequencesynthetic 94Gly Ala Ser19527DNAArtificial Sequencesynthetic
95cagcagtatg gtagctcacc ttggacg 27969PRTArtificial
Sequencesynthetic 96Gln Gln Tyr Gly Ser Ser Pro Trp Thr1
597360DNAArtificial Sequencesynthetic 97caggtgcagc tgcaggagtc
gggcccagga ctggtgaagc cttcggagac cctgtccctc 60acctgcactg tctctggtgg
ctccatcagt aattcctact ggagctggat ccggcagccc 120ccagggaagg
gactggagtg gattggatat gtctattccc gtgggaacac caagtacaac
180ccctccctca cgagtcgagt caccatgtca tttgacacgt ccaagaacca
gttctccctg 240aaactgaggt ctgtgaccgc cgcagacacg gccgtgtatt
actgtgcgag aagcagcagc 300tggtacgagg actggtactt cgatctctgg
ggccgtggca ccctggtcac tgtctcctca 36098120PRTArtificial
Sequencesynthetic 98Gln Val Gln Leu Gln Glu Ser Gly Pro Gly Leu Val
Lys Pro Ser Glu1 5 10 15Thr Leu Ser Leu Thr Cys Thr Val Ser Gly Gly
Ser Ile Ser Asn Ser 20 25 30Tyr Trp Ser Trp Ile Arg Gln Pro Pro Gly
Lys Gly Leu Glu Trp Ile 35 40 45Gly Tyr Val Tyr Ser Arg Gly Asn Thr
Lys Tyr Asn Pro Ser Leu Thr 50 55 60Ser Arg Val Thr Met Ser Phe Asp
Thr Ser Lys Asn Gln Phe Ser Leu65 70 75 80Lys Leu Arg Ser Val Thr
Ala Ala Asp Thr Ala Val Tyr Tyr Cys Ala 85 90 95Arg Ser Ser Ser Trp
Tyr Glu Asp Trp Tyr Phe Asp Leu Trp Gly Arg 100 105 110Gly Thr Leu
Val Thr Val Ser Ser 115 1209924DNAArtificial Sequencesynthetic
99ggtggctcca tcagtaattc ctac 241008PRTArtificial Sequencesynthetic
100Gly Gly Ser Ile Ser Asn Ser Tyr1 510121DNAArtificial
Sequencesynthetic 101gtctattccc gtgggaacac c 211027PRTArtificial
Sequencesynthetic 102Val Tyr Ser Arg Gly Asn Thr1
510342DNAArtificial Sequencesynthetic 103gcgagaagca gcagctggta
cgaggactgg tacttcgatc tc 4210414PRTArtificial Sequencesynthetic
104Ala Arg Ser Ser Ser Trp Tyr Glu Asp Trp Tyr Phe Asp Leu1 5
10105354DNAArtificial Sequencesynthetic 105caggtgcagc tacagcagtg
gggcgcaggg ctgtttaagc cttcggagac cctgtccctc 60acctgcgatg tctatggtgg
gtccttcaga ggttattatt ggagttggat ccgccagccc 120ccagggaagg
ggctggagtg gattggggaa atcagttata gtggtttcac caattacaac
180ccgtccctca agagtcgagt catcatatca atagatacgt ccaagaacca
gttctccctg 240aagatgagct ctgtgaccgc cgcggacacg gctgtttatt
actgtgcgag agttacctat 300ggttatggga cctttgatta ttggggccag
ggaaccctgg tcaccgtctc ctca 354106118PRTArtificial Sequencesynthetic
106Gln Val Gln Leu Gln Gln Trp Gly Ala Gly Leu Phe Lys Pro Ser Glu1
5 10 15Thr Leu Ser Leu Thr Cys Asp Val Tyr Gly Gly Ser Phe Arg Gly
Tyr 20 25 30Tyr Trp Ser Trp Ile Arg Gln Pro Pro Gly Lys Gly Leu Glu
Trp Ile 35 40 45Gly Glu Ile Ser Tyr Ser Gly Phe Thr Asn Tyr Asn Pro
Ser Leu Lys 50 55 60Ser Arg Val Ile Ile Ser Ile Asp Thr Ser Lys Asn
Gln Phe Ser Leu65 70 75 80Lys Met Ser Ser Val Thr Ala Ala Asp Thr
Ala Val Tyr Tyr Cys Ala 85 90 95Arg Val Thr Tyr Gly Tyr Gly Thr Phe
Asp Tyr Trp Gly Gln Gly Thr 100 105 110Leu Val Thr Val Ser Ser
11510724DNAArtificial Sequencesynthetic 107ggtgggtcct tcagaggtta
ttat 241088PRTArtificial Sequencesynthetic 108Gly Gly Ser Phe Arg
Gly Tyr Tyr1 510921DNAArtificial Sequencesynthetic 109atcagttata
gtggtttcac c 211107PRTArtificial Sequencesynthetic 110Ile Ser Tyr
Ser Gly Phe Thr1 511136DNAArtificial Sequencesynthetic
111gcgagagtta cctatggtta tgggaccttt gattat 3611212PRTArtificial
Sequencesynthetic 112Ala Arg Val Thr Tyr Gly Tyr Gly Thr Phe Asp
Tyr1 5 101131165PRTArtificial SequencehLEPR Accession No P48357
113Met Ile Cys Gln Lys Phe Cys Val Val Leu Leu His Trp Glu Phe Ile1
5 10 15Tyr Val Ile Thr Ala Phe Asn Leu Ser Tyr Pro Ile Thr Pro Trp
Arg 20 25 30Phe Lys Leu Ser Cys Met Pro Pro Asn Ser Thr Tyr Asp Tyr
Phe Leu 35 40 45Leu Pro Ala Gly Leu Ser Lys Asn Thr Ser Asn Ser Asn
Gly His Tyr 50 55 60Glu Thr Ala Val Glu Pro Lys Phe Asn Ser Ser Gly
Thr His Phe Ser65 70 75 80Asn Leu Ser Lys Thr Thr Phe His Cys Cys
Phe Arg Ser Glu Gln Asp 85 90 95Arg Asn Cys Ser Leu Cys Ala Asp Asn
Ile Glu Gly Lys Thr Phe Val 100 105 110Ser Thr Val Asn Ser Leu Val
Phe Gln Gln Ile Asp Ala Asn Trp Asn 115 120 125Ile Gln Cys Trp Leu
Lys Gly Asp Leu Lys Leu Phe Ile Cys Tyr Val 130 135 140Glu Ser Leu
Phe Lys Asn Leu Phe Arg Asn Tyr Asn Tyr Lys Val His145 150 155
160Leu Leu Tyr Val Leu Pro Glu Val Leu Glu Asp Ser Pro Leu Val Pro
165 170 175Gln Lys Gly Ser Phe Gln Met Val His Cys Asn Cys Ser Val
His Glu 180 185 190Cys Cys Glu Cys Leu Val Pro Val Pro Thr Ala Lys
Leu Asn Asp Thr 195 200 205Leu Leu Met Cys Leu Lys Ile Thr Ser Gly
Gly Val Ile Phe Gln Ser 210 215 220Pro Leu Met Ser Val Gln Pro Ile
Asn Met Val Lys Pro Asp Pro Pro225 230 235 240Leu Gly Leu His Met
Glu Ile Thr Asp Asp Gly Asn Leu Lys Ile Ser 245 250 255Trp Ser Ser
Pro Pro Leu Val Pro Phe Pro Leu Gln Tyr Gln Val Lys 260 265 270Tyr
Ser Glu Asn Ser Thr Thr Val Ile Arg Glu Ala Asp Lys Ile Val 275 280
285Ser Ala Thr Ser Leu Leu Val Asp Ser Ile Leu Pro Gly Ser Ser Tyr
290 295 300Glu Val Gln Val Arg Gly Lys Arg Leu Asp Gly Pro Gly Ile
Trp Ser305 310 315 320Asp Trp Ser Thr Pro Arg Val Phe Thr Thr Gln
Asp Val Ile Tyr Phe 325 330 335Pro Pro Lys Ile Leu Thr Ser Val Gly
Ser Asn Val Ser Phe His Cys 340 345 350Ile Tyr Lys Lys Glu Asn Lys
Ile Val Pro Ser Lys Glu Ile Val Trp 355 360 365Trp Met Asn Leu Ala
Glu Lys Ile Pro Gln Ser Gln Tyr Asp Val Val 370 375 380Ser Asp His
Val Ser Lys Val Thr Phe Phe Asn Leu Asn Glu Thr Lys385 390 395
400Pro Arg Gly Lys Phe Thr Tyr Asp Ala Val Tyr Cys Cys Asn Glu His
405 410 415Glu Cys His His Arg Tyr Ala Glu Leu Tyr Val Ile Asp Val
Asn Ile 420 425 430Asn Ile Ser Cys Glu Thr Asp Gly Tyr Leu Thr Lys
Met Thr Cys Arg 435 440 445Trp Ser Thr Ser Thr Ile Gln Ser Leu Ala
Glu Ser Thr Leu Gln Leu 450 455 460Arg Tyr His Arg Ser Ser Leu Tyr
Cys Ser Asp Ile Pro Ser Ile His465 470 475 480Pro Ile Ser Glu Pro
Lys Asp Cys Tyr Leu Gln Ser Asp Gly Phe Tyr 485 490 495Glu Cys Ile
Phe Gln Pro Ile Phe Leu Leu Ser Gly Tyr Thr Met Trp 500 505 510Ile
Arg Ile Asn His Ser Leu Gly Ser Leu Asp Ser Pro Pro Thr Cys 515 520
525Val Leu Pro Asp Ser Val Val Lys Pro Leu Pro Pro Ser Ser Val Lys
530 535 540Ala Glu Ile Thr Ile Asn Ile Gly Leu Leu Lys Ile Ser Trp
Glu Lys545 550 555 560Pro Val Phe Pro Glu Asn Asn Leu Gln Phe Gln
Ile Arg Tyr Gly Leu 565 570 575Ser Gly Lys Glu Val Gln Trp Lys Met
Tyr Glu Val Tyr Asp Ala Lys 580 585 590Ser Lys Ser Val Ser Leu Pro
Val Pro Asp Leu Cys Ala Val Tyr Ala 595 600 605Val Gln Val Arg Cys
Lys Arg Leu Asp Gly Leu Gly Tyr Trp Ser Asn 610 615 620Trp Ser Asn
Pro Ala Tyr Thr Val Val Met Asp Ile Lys Val Pro Met625 630 635
640Arg Gly Pro Glu Phe Trp Arg Ile Ile Asn Gly Asp Thr Met Lys Lys
645 650 655Glu Lys Asn Val Thr Leu Leu Trp Lys Pro Leu Met Lys Asn
Asp Ser 660 665 670Leu Cys Ser Val Gln Arg Tyr Val Ile Asn His His
Thr Ser Cys Asn 675 680 685Gly Thr Trp Ser Glu Asp Val Gly Asn His
Thr Lys Phe Thr Phe Leu 690 695 700Trp Thr Glu Gln Ala His Thr Val
Thr Val Leu Ala Ile Asn Ser Ile705 710 715 720Gly Ala Ser Val Ala
Asn Phe Asn Leu Thr Phe Ser Trp Pro Met Ser 725 730 735Lys Val Asn
Ile Val Gln Ser Leu Ser Ala Tyr Pro Leu Asn Ser Ser 740 745 750Cys
Val Ile Val Ser Trp Ile Leu Ser Pro Ser Asp Tyr Lys Leu Met 755 760
765Tyr Phe Ile Ile Glu Trp Lys Asn Leu Asn Glu Asp Gly Glu Ile Lys
770 775 780Trp Leu Arg Ile Ser Ser Ser Val Lys Lys Tyr Tyr Ile His
Asp His785 790 795 800Phe Ile Pro Ile Glu Lys Tyr Gln Phe Ser Leu
Tyr Pro Ile Phe Met 805 810 815Glu Gly Val Gly Lys Pro Lys Ile Ile
Asn Ser Phe Thr Gln Asp Asp 820 825 830Ile Glu Lys His Gln Ser Asp
Ala Gly Leu Tyr Val Ile Val Pro Val 835 840 845Ile Ile Ser Ser Ser
Ile Leu Leu Leu Gly Thr Leu Leu Ile Ser His 850 855 860Gln Arg Met
Lys Lys Leu Phe Trp Glu Asp Val Pro Asn Pro Lys Asn865 870 875
880Cys Ser Trp Ala Gln Gly Leu Asn Phe Gln Lys Pro Glu Thr Phe Glu
885 890 895His Leu Phe Ile Lys His Thr Ala Ser Val Thr Cys Gly Pro
Leu Leu 900 905 910Leu Glu Pro Glu Thr Ile Ser Glu Asp Ile Ser Val
Asp Thr Ser Trp 915 920 925Lys Asn Lys Asp Glu Met Met Pro Thr Thr
Val Val Ser Leu Leu Ser 930 935 940Thr Thr Asp Leu Glu Lys Gly Ser
Val Cys Ile Ser Asp Gln Phe Asn945 950 955 960Ser Val Asn Phe Ser
Glu Ala Glu Gly Thr Glu Val Thr Tyr Glu Asp 965 970 975Glu Ser Gln
Arg Gln Pro Phe Val Lys Tyr Ala Thr Leu Ile Ser Asn 980 985 990Ser
Lys Pro Ser Glu Thr Gly Glu Glu Gln Gly Leu Ile Asn Ser Ser 995
1000 1005Val Thr Lys Cys Phe Ser Ser Lys Asn Ser Pro Leu Lys Asp
Ser 1010 1015 1020Phe Ser Asn Ser Ser Trp Glu Ile Glu Ala Gln Ala
Phe Phe
Ile 1025 1030 1035Leu Ser Asp Gln His Pro Asn Ile Ile Ser Pro His
Leu Thr Phe 1040 1045 1050Ser Glu Gly Leu Asp Glu Leu Leu Lys Leu
Glu Gly Asn Phe Pro 1055 1060 1065Glu Glu Asn Asn Asp Lys Lys Ser
Ile Tyr Tyr Leu Gly Val Thr 1070 1075 1080Ser Ile Lys Lys Arg Glu
Ser Gly Val Leu Leu Thr Asp Lys Ser 1085 1090 1095Arg Val Ser Cys
Pro Phe Pro Ala Pro Cys Leu Phe Thr Asp Ile 1100 1105 1110Arg Val
Leu Gln Asp Ser Cys Ser His Phe Val Glu Asn Asn Ile 1115 1120
1125Asn Leu Gly Thr Ser Ser Lys Lys Thr Phe Ala Ser Tyr Met Pro
1130 1135 1140Gln Phe Gln Thr Cys Ser Thr Gln Thr His Lys Ile Met
Glu Asn 1145 1150 1155Lys Met Cys Asp Leu Thr Val 1160
1165114846PRTArtificial SequencehLEPR.mmh aa 1-818 F22-D839 of
P48357 aa 819-846 myc-myc-hexahistidine tag 114Phe Asn Leu Ser Tyr
Pro Ile Thr Pro Trp Arg Phe Lys Leu Ser Cys1 5 10 15Met Pro Pro Asn
Ser Thr Tyr Asp Tyr Phe Leu Leu Pro Ala Gly Leu 20 25 30Ser Lys Asn
Thr Ser Asn Ser Asn Gly His Tyr Glu Thr Ala Val Glu 35 40 45Pro Lys
Phe Asn Ser Ser Gly Thr His Phe Ser Asn Leu Ser Lys Thr 50 55 60Thr
Phe His Cys Cys Phe Arg Ser Glu Gln Asp Arg Asn Cys Ser Leu65 70 75
80Cys Ala Asp Asn Ile Glu Gly Lys Thr Phe Val Ser Thr Val Asn Ser
85 90 95Leu Val Phe Gln Gln Ile Asp Ala Asn Trp Asn Ile Gln Cys Trp
Leu 100 105 110Lys Gly Asp Leu Lys Leu Phe Ile Cys Tyr Val Glu Ser
Leu Phe Lys 115 120 125Asn Leu Phe Arg Asn Tyr Asn Tyr Lys Val His
Leu Leu Tyr Val Leu 130 135 140Pro Glu Val Leu Glu Asp Ser Pro Leu
Val Pro Gln Lys Gly Ser Phe145 150 155 160Gln Met Val His Cys Asn
Cys Ser Val His Glu Cys Cys Glu Cys Leu 165 170 175Val Pro Val Pro
Thr Ala Lys Leu Asn Asp Thr Leu Leu Met Cys Leu 180 185 190Lys Ile
Thr Ser Gly Gly Val Ile Phe Gln Ser Pro Leu Met Ser Val 195 200
205Gln Pro Ile Asn Met Val Lys Pro Asp Pro Pro Leu Gly Leu His Met
210 215 220Glu Ile Thr Asp Asp Gly Asn Leu Lys Ile Ser Trp Ser Ser
Pro Pro225 230 235 240Leu Val Pro Phe Pro Leu Gln Tyr Gln Val Lys
Tyr Ser Glu Asn Ser 245 250 255Thr Thr Val Ile Arg Glu Ala Asp Lys
Ile Val Ser Ala Thr Ser Leu 260 265 270Leu Val Asp Ser Ile Leu Pro
Gly Ser Ser Tyr Glu Val Gln Val Arg 275 280 285Gly Lys Arg Leu Asp
Gly Pro Gly Ile Trp Ser Asp Trp Ser Thr Pro 290 295 300Arg Val Phe
Thr Thr Gln Asp Val Ile Tyr Phe Pro Pro Lys Ile Leu305 310 315
320Thr Ser Val Gly Ser Asn Val Ser Phe His Cys Ile Tyr Lys Lys Glu
325 330 335Asn Lys Ile Val Pro Ser Lys Glu Ile Val Trp Trp Met Asn
Leu Ala 340 345 350Glu Lys Ile Pro Gln Ser Gln Tyr Asp Val Val Ser
Asp His Val Ser 355 360 365Lys Val Thr Phe Phe Asn Leu Asn Glu Thr
Lys Pro Arg Gly Lys Phe 370 375 380Thr Tyr Asp Ala Val Tyr Cys Cys
Asn Glu His Glu Cys His His Arg385 390 395 400Tyr Ala Glu Leu Tyr
Val Ile Asp Val Asn Ile Asn Ile Ser Cys Glu 405 410 415Thr Asp Gly
Tyr Leu Thr Lys Met Thr Cys Arg Trp Ser Thr Ser Thr 420 425 430Ile
Gln Ser Leu Ala Glu Ser Thr Leu Gln Leu Arg Tyr His Arg Ser 435 440
445Ser Leu Tyr Cys Ser Asp Ile Pro Ser Ile His Pro Ile Ser Glu Pro
450 455 460Lys Asp Cys Tyr Leu Gln Ser Asp Gly Phe Tyr Glu Cys Ile
Phe Gln465 470 475 480Pro Ile Phe Leu Leu Ser Gly Tyr Thr Met Trp
Ile Arg Ile Asn His 485 490 495Ser Leu Gly Ser Leu Asp Ser Pro Pro
Thr Cys Val Leu Pro Asp Ser 500 505 510Val Val Lys Pro Leu Pro Pro
Ser Ser Val Lys Ala Glu Ile Thr Ile 515 520 525Asn Ile Gly Leu Leu
Lys Ile Ser Trp Glu Lys Pro Val Phe Pro Glu 530 535 540Asn Asn Leu
Gln Phe Gln Ile Arg Tyr Gly Leu Ser Gly Lys Glu Val545 550 555
560Gln Trp Lys Met Tyr Glu Val Tyr Asp Ala Lys Ser Lys Ser Val Ser
565 570 575Leu Pro Val Pro Asp Leu Cys Ala Val Tyr Ala Val Gln Val
Arg Cys 580 585 590Lys Arg Leu Asp Gly Leu Gly Tyr Trp Ser Asn Trp
Ser Asn Pro Ala 595 600 605Tyr Thr Val Val Met Asp Ile Lys Val Pro
Met Arg Gly Pro Glu Phe 610 615 620Trp Arg Ile Ile Asn Gly Asp Thr
Met Lys Lys Glu Lys Asn Val Thr625 630 635 640Leu Leu Trp Lys Pro
Leu Met Lys Asn Asp Ser Leu Cys Ser Val Gln 645 650 655Arg Tyr Val
Ile Asn His His Thr Ser Cys Asn Gly Thr Trp Ser Glu 660 665 670Asp
Val Gly Asn His Thr Lys Phe Thr Phe Leu Trp Thr Glu Gln Ala 675 680
685His Thr Val Thr Val Leu Ala Ile Asn Ser Ile Gly Ala Ser Val Ala
690 695 700Asn Phe Asn Leu Thr Phe Ser Trp Pro Met Ser Lys Val Asn
Ile Val705 710 715 720Gln Ser Leu Ser Ala Tyr Pro Leu Asn Ser Ser
Cys Val Ile Val Ser 725 730 735Trp Ile Leu Ser Pro Ser Asp Tyr Lys
Leu Met Tyr Phe Ile Ile Glu 740 745 750Trp Lys Asn Leu Asn Glu Asp
Gly Glu Ile Lys Trp Leu Arg Ile Ser 755 760 765Ser Ser Val Lys Lys
Tyr Tyr Ile His Asp His Phe Ile Pro Ile Glu 770 775 780Lys Tyr Gln
Phe Ser Leu Tyr Pro Ile Phe Met Glu Gly Val Gly Lys785 790 795
800Pro Lys Ile Ile Asn Ser Phe Thr Gln Asp Asp Ile Glu Lys His Gln
805 810 815Ser Asp Glu Gln Lys Leu Ile Ser Glu Glu Asp Leu Gly Gly
Glu Gln 820 825 830Lys Leu Ile Ser Glu Glu Asp Leu His His His His
His His 835 840 8451151051PRTArtificial SequencehLEPR.mFc aa 1-818
F22-D839 of P48357 aa 819-1051 mouse IgG2a (E98-K330 of P01863)
115Phe Asn Leu Ser Tyr Pro Ile Thr Pro Trp Arg Phe Lys Leu Ser Cys1
5 10 15Met Pro Pro Asn Ser Thr Tyr Asp Tyr Phe Leu Leu Pro Ala Gly
Leu 20 25 30Ser Lys Asn Thr Ser Asn Ser Asn Gly His Tyr Glu Thr Ala
Val Glu 35 40 45Pro Lys Phe Asn Ser Ser Gly Thr His Phe Ser Asn Leu
Ser Lys Thr 50 55 60Thr Phe His Cys Cys Phe Arg Ser Glu Gln Asp Arg
Asn Cys Ser Leu65 70 75 80Cys Ala Asp Asn Ile Glu Gly Lys Thr Phe
Val Ser Thr Val Asn Ser 85 90 95Leu Val Phe Gln Gln Ile Asp Ala Asn
Trp Asn Ile Gln Cys Trp Leu 100 105 110Lys Gly Asp Leu Lys Leu Phe
Ile Cys Tyr Val Glu Ser Leu Phe Lys 115 120 125Asn Leu Phe Arg Asn
Tyr Asn Tyr Lys Val His Leu Leu Tyr Val Leu 130 135 140Pro Glu Val
Leu Glu Asp Ser Pro Leu Val Pro Gln Lys Gly Ser Phe145 150 155
160Gln Met Val His Cys Asn Cys Ser Val His Glu Cys Cys Glu Cys Leu
165 170 175Val Pro Val Pro Thr Ala Lys Leu Asn Asp Thr Leu Leu Met
Cys Leu 180 185 190Lys Ile Thr Ser Gly Gly Val Ile Phe Gln Ser Pro
Leu Met Ser Val 195 200 205Gln Pro Ile Asn Met Val Lys Pro Asp Pro
Pro Leu Gly Leu His Met 210 215 220Glu Ile Thr Asp Asp Gly Asn Leu
Lys Ile Ser Trp Ser Ser Pro Pro225 230 235 240Leu Val Pro Phe Pro
Leu Gln Tyr Gln Val Lys Tyr Ser Glu Asn Ser 245 250 255Thr Thr Val
Ile Arg Glu Ala Asp Lys Ile Val Ser Ala Thr Ser Leu 260 265 270Leu
Val Asp Ser Ile Leu Pro Gly Ser Ser Tyr Glu Val Gln Val Arg 275 280
285Gly Lys Arg Leu Asp Gly Pro Gly Ile Trp Ser Asp Trp Ser Thr Pro
290 295 300Arg Val Phe Thr Thr Gln Asp Val Ile Tyr Phe Pro Pro Lys
Ile Leu305 310 315 320Thr Ser Val Gly Ser Asn Val Ser Phe His Cys
Ile Tyr Lys Lys Glu 325 330 335Asn Lys Ile Val Pro Ser Lys Glu Ile
Val Trp Trp Met Asn Leu Ala 340 345 350Glu Lys Ile Pro Gln Ser Gln
Tyr Asp Val Val Ser Asp His Val Ser 355 360 365Lys Val Thr Phe Phe
Asn Leu Asn Glu Thr Lys Pro Arg Gly Lys Phe 370 375 380Thr Tyr Asp
Ala Val Tyr Cys Cys Asn Glu His Glu Cys His His Arg385 390 395
400Tyr Ala Glu Leu Tyr Val Ile Asp Val Asn Ile Asn Ile Ser Cys Glu
405 410 415Thr Asp Gly Tyr Leu Thr Lys Met Thr Cys Arg Trp Ser Thr
Ser Thr 420 425 430Ile Gln Ser Leu Ala Glu Ser Thr Leu Gln Leu Arg
Tyr His Arg Ser 435 440 445Ser Leu Tyr Cys Ser Asp Ile Pro Ser Ile
His Pro Ile Ser Glu Pro 450 455 460Lys Asp Cys Tyr Leu Gln Ser Asp
Gly Phe Tyr Glu Cys Ile Phe Gln465 470 475 480Pro Ile Phe Leu Leu
Ser Gly Tyr Thr Met Trp Ile Arg Ile Asn His 485 490 495Ser Leu Gly
Ser Leu Asp Ser Pro Pro Thr Cys Val Leu Pro Asp Ser 500 505 510Val
Val Lys Pro Leu Pro Pro Ser Ser Val Lys Ala Glu Ile Thr Ile 515 520
525Asn Ile Gly Leu Leu Lys Ile Ser Trp Glu Lys Pro Val Phe Pro Glu
530 535 540Asn Asn Leu Gln Phe Gln Ile Arg Tyr Gly Leu Ser Gly Lys
Glu Val545 550 555 560Gln Trp Lys Met Tyr Glu Val Tyr Asp Ala Lys
Ser Lys Ser Val Ser 565 570 575Leu Pro Val Pro Asp Leu Cys Ala Val
Tyr Ala Val Gln Val Arg Cys 580 585 590Lys Arg Leu Asp Gly Leu Gly
Tyr Trp Ser Asn Trp Ser Asn Pro Ala 595 600 605Tyr Thr Val Val Met
Asp Ile Lys Val Pro Met Arg Gly Pro Glu Phe 610 615 620Trp Arg Ile
Ile Asn Gly Asp Thr Met Lys Lys Glu Lys Asn Val Thr625 630 635
640Leu Leu Trp Lys Pro Leu Met Lys Asn Asp Ser Leu Cys Ser Val Gln
645 650 655Arg Tyr Val Ile Asn His His Thr Ser Cys Asn Gly Thr Trp
Ser Glu 660 665 670Asp Val Gly Asn His Thr Lys Phe Thr Phe Leu Trp
Thr Glu Gln Ala 675 680 685His Thr Val Thr Val Leu Ala Ile Asn Ser
Ile Gly Ala Ser Val Ala 690 695 700Asn Phe Asn Leu Thr Phe Ser Trp
Pro Met Ser Lys Val Asn Ile Val705 710 715 720Gln Ser Leu Ser Ala
Tyr Pro Leu Asn Ser Ser Cys Val Ile Val Ser 725 730 735Trp Ile Leu
Ser Pro Ser Asp Tyr Lys Leu Met Tyr Phe Ile Ile Glu 740 745 750Trp
Lys Asn Leu Asn Glu Asp Gly Glu Ile Lys Trp Leu Arg Ile Ser 755 760
765Ser Ser Val Lys Lys Tyr Tyr Ile His Asp His Phe Ile Pro Ile Glu
770 775 780Lys Tyr Gln Phe Ser Leu Tyr Pro Ile Phe Met Glu Gly Val
Gly Lys785 790 795 800Pro Lys Ile Ile Asn Ser Phe Thr Gln Asp Asp
Ile Glu Lys His Gln 805 810 815Ser Asp Glu Pro Arg Gly Pro Thr Ile
Lys Pro Cys Pro Pro Cys Lys 820 825 830Cys Pro Ala Pro Asn Leu Leu
Gly Gly Pro Ser Val Phe Ile Phe Pro 835 840 845Pro Lys Ile Lys Asp
Val Leu Met Ile Ser Leu Ser Pro Ile Val Thr 850 855 860Cys Val Val
Val Asp Val Ser Glu Asp Asp Pro Asp Val Gln Ile Ser865 870 875
880Trp Phe Val Asn Asn Val Glu Val His Thr Ala Gln Thr Gln Thr His
885 890 895Arg Glu Asp Tyr Asn Ser Thr Leu Arg Val Val Ser Ala Leu
Pro Ile 900 905 910Gln His Gln Asp Trp Met Ser Gly Lys Glu Phe Lys
Cys Lys Val Asn 915 920 925Asn Lys Asp Leu Pro Ala Pro Ile Glu Arg
Thr Ile Ser Lys Pro Lys 930 935 940Gly Ser Val Arg Ala Pro Gln Val
Tyr Val Leu Pro Pro Pro Glu Glu945 950 955 960Glu Met Thr Lys Lys
Gln Val Thr Leu Thr Cys Met Val Thr Asp Phe 965 970 975Met Pro Glu
Asp Ile Tyr Val Glu Trp Thr Asn Asn Gly Lys Thr Glu 980 985 990Leu
Asn Tyr Lys Asn Thr Glu Pro Val Leu Asp Ser Asp Gly Ser Tyr 995
1000 1005Phe Met Tyr Ser Lys Leu Arg Val Glu Lys Lys Asn Trp Val
Glu 1010 1015 1020Arg Asn Ser Tyr Ser Cys Ser Val Val His Glu Gly
Leu His Asn 1025 1030 1035His His Thr Thr Lys Ser Phe Ser Arg Thr
Pro Gly Lys 1040 1045 10501161045PRTArtificial SequencehLEPR.hFc aa
1-818 F22-D839 of P48357 aa 819-1045 human IgG1 tag (D104-K330 of
P01857) 116Phe Asn Leu Ser Tyr Pro Ile Thr Pro Trp Arg Phe Lys Leu
Ser Cys1 5 10 15Met Pro Pro Asn Ser Thr Tyr Asp Tyr Phe Leu Leu Pro
Ala Gly Leu 20 25 30Ser Lys Asn Thr Ser Asn Ser Asn Gly His Tyr Glu
Thr Ala Val Glu 35 40 45Pro Lys Phe Asn Ser Ser Gly Thr His Phe Ser
Asn Leu Ser Lys Thr 50 55 60Thr Phe His Cys Cys Phe Arg Ser Glu Gln
Asp Arg Asn Cys Ser Leu65 70 75 80Cys Ala Asp Asn Ile Glu Gly Lys
Thr Phe Val Ser Thr Val Asn Ser 85 90 95Leu Val Phe Gln Gln Ile Asp
Ala Asn Trp Asn Ile Gln Cys Trp Leu 100 105 110Lys Gly Asp Leu Lys
Leu Phe Ile Cys Tyr Val Glu Ser Leu Phe Lys 115 120 125Asn Leu Phe
Arg Asn Tyr Asn Tyr Lys Val His Leu Leu Tyr Val Leu 130 135 140Pro
Glu Val Leu Glu Asp Ser Pro Leu Val Pro Gln Lys Gly Ser Phe145 150
155 160Gln Met Val His Cys Asn Cys Ser Val His Glu Cys Cys Glu Cys
Leu 165 170 175Val Pro Val Pro Thr Ala Lys Leu Asn Asp Thr Leu Leu
Met Cys Leu 180 185 190Lys Ile Thr Ser Gly Gly Val Ile Phe Gln Ser
Pro Leu Met Ser Val 195 200 205Gln Pro Ile Asn Met Val Lys Pro Asp
Pro Pro Leu Gly Leu His Met 210 215 220Glu Ile Thr Asp Asp Gly Asn
Leu Lys Ile Ser Trp Ser Ser Pro Pro225 230 235 240Leu Val Pro Phe
Pro Leu Gln Tyr Gln Val Lys Tyr Ser Glu Asn Ser 245 250 255Thr Thr
Val Ile Arg Glu Ala Asp Lys Ile Val Ser Ala Thr Ser Leu 260 265
270Leu Val Asp Ser Ile Leu Pro Gly Ser Ser Tyr Glu Val Gln Val Arg
275 280 285Gly Lys Arg Leu Asp Gly Pro Gly Ile Trp Ser Asp Trp Ser
Thr Pro 290 295 300Arg Val Phe Thr Thr Gln Asp Val Ile Tyr Phe Pro
Pro Lys Ile Leu305 310 315 320Thr Ser Val Gly Ser Asn Val Ser Phe
His Cys Ile Tyr Lys Lys Glu 325 330 335Asn Lys Ile Val Pro Ser Lys
Glu Ile Val Trp Trp Met Asn Leu Ala 340 345 350Glu Lys Ile Pro Gln
Ser Gln Tyr Asp Val Val Ser Asp His Val Ser 355 360 365Lys Val Thr
Phe Phe Asn Leu Asn Glu Thr Lys Pro Arg Gly Lys Phe 370 375 380Thr
Tyr Asp Ala Val Tyr Cys Cys Asn Glu
His Glu Cys His His Arg385 390 395 400Tyr Ala Glu Leu Tyr Val Ile
Asp Val Asn Ile Asn Ile Ser Cys Glu 405 410 415Thr Asp Gly Tyr Leu
Thr Lys Met Thr Cys Arg Trp Ser Thr Ser Thr 420 425 430Ile Gln Ser
Leu Ala Glu Ser Thr Leu Gln Leu Arg Tyr His Arg Ser 435 440 445Ser
Leu Tyr Cys Ser Asp Ile Pro Ser Ile His Pro Ile Ser Glu Pro 450 455
460Lys Asp Cys Tyr Leu Gln Ser Asp Gly Phe Tyr Glu Cys Ile Phe
Gln465 470 475 480Pro Ile Phe Leu Leu Ser Gly Tyr Thr Met Trp Ile
Arg Ile Asn His 485 490 495Ser Leu Gly Ser Leu Asp Ser Pro Pro Thr
Cys Val Leu Pro Asp Ser 500 505 510Val Val Lys Pro Leu Pro Pro Ser
Ser Val Lys Ala Glu Ile Thr Ile 515 520 525Asn Ile Gly Leu Leu Lys
Ile Ser Trp Glu Lys Pro Val Phe Pro Glu 530 535 540Asn Asn Leu Gln
Phe Gln Ile Arg Tyr Gly Leu Ser Gly Lys Glu Val545 550 555 560Gln
Trp Lys Met Tyr Glu Val Tyr Asp Ala Lys Ser Lys Ser Val Ser 565 570
575Leu Pro Val Pro Asp Leu Cys Ala Val Tyr Ala Val Gln Val Arg Cys
580 585 590Lys Arg Leu Asp Gly Leu Gly Tyr Trp Ser Asn Trp Ser Asn
Pro Ala 595 600 605Tyr Thr Val Val Met Asp Ile Lys Val Pro Met Arg
Gly Pro Glu Phe 610 615 620Trp Arg Ile Ile Asn Gly Asp Thr Met Lys
Lys Glu Lys Asn Val Thr625 630 635 640Leu Leu Trp Lys Pro Leu Met
Lys Asn Asp Ser Leu Cys Ser Val Gln 645 650 655Arg Tyr Val Ile Asn
His His Thr Ser Cys Asn Gly Thr Trp Ser Glu 660 665 670Asp Val Gly
Asn His Thr Lys Phe Thr Phe Leu Trp Thr Glu Gln Ala 675 680 685His
Thr Val Thr Val Leu Ala Ile Asn Ser Ile Gly Ala Ser Val Ala 690 695
700Asn Phe Asn Leu Thr Phe Ser Trp Pro Met Ser Lys Val Asn Ile
Val705 710 715 720Gln Ser Leu Ser Ala Tyr Pro Leu Asn Ser Ser Cys
Val Ile Val Ser 725 730 735Trp Ile Leu Ser Pro Ser Asp Tyr Lys Leu
Met Tyr Phe Ile Ile Glu 740 745 750Trp Lys Asn Leu Asn Glu Asp Gly
Glu Ile Lys Trp Leu Arg Ile Ser 755 760 765Ser Ser Val Lys Lys Tyr
Tyr Ile His Asp His Phe Ile Pro Ile Glu 770 775 780Lys Tyr Gln Phe
Ser Leu Tyr Pro Ile Phe Met Glu Gly Val Gly Lys785 790 795 800Pro
Lys Ile Ile Asn Ser Phe Thr Gln Asp Asp Ile Glu Lys His Gln 805 810
815Ser Asp Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu
820 825 830Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys
Asp Thr 835 840 845Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val
Val Val Asp Val 850 855 860Ser His Glu Asp Pro Glu Val Lys Phe Asn
Trp Tyr Val Asp Gly Val865 870 875 880Glu Val His Asn Ala Lys Thr
Lys Pro Arg Glu Glu Gln Tyr Asn Ser 885 890 895Thr Tyr Arg Val Val
Ser Val Leu Thr Val Leu His Gln Asp Trp Leu 900 905 910Asn Gly Lys
Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala 915 920 925Pro
Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro 930 935
940Gln Val Tyr Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn
Gln945 950 955 960Val Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro
Ser Asp Ile Ala 965 970 975Val Glu Trp Glu Ser Asn Gly Gln Pro Glu
Asn Asn Tyr Lys Thr Thr 980 985 990Pro Pro Val Leu Asp Ser Asp Gly
Ser Phe Phe Leu Tyr Ser Lys Leu 995 1000 1005Thr Val Asp Lys Ser
Arg Trp Gln Gln Gly Asn Val Phe Ser Cys 1010 1015 1020Ser Val Met
His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser 1025 1030 1035Leu
Ser Leu Ser Pro Gly Lys 1040 1045117844PRTArtificial
SequenceMfLEPR.mmh aa 1-816 Macaca fascicularis F22-D837 with a
T827A substitution from XP_005543194.1 aa 817-844
myc-myc-hexahistidine tag 117Phe Asn Leu Ser Tyr Pro Ile Thr Pro
Trp Arg Phe Lys Leu Ser Cys1 5 10 15Met Pro Pro Asn Ser Thr Tyr Asp
Tyr Phe Leu Leu Pro Ala Gly Leu 20 25 30Ser Lys Asn Thr Ser Asn Leu
Asn Gly His Tyr Glu Thr Ala Val Glu 35 40 45Phe Asn Ser Ser Asp Thr
His Phe Ser Asn Leu Ser Lys Thr Thr Phe 50 55 60His Cys Cys Phe Arg
Ser Glu Gln Asp Arg Asn Cys Ser Leu Cys Ala65 70 75 80Asp Asn Ile
Glu Gly Lys Thr Phe Val Ser Thr Val Asn Ser Ser Val 85 90 95Phe Gln
Gln Met Gly Ala Asn Trp Asn Ile Gln Cys Trp Leu Lys Gly 100 105
110Asp Leu Lys Leu Phe Ile Cys Tyr Val Glu Ser Leu Phe Lys Asn Pro
115 120 125Phe Lys Asn Tyr Lys His Lys Val His Leu Leu Tyr Val Leu
Pro Glu 130 135 140Val Leu Glu Asp Ser Pro Leu Val Pro Gln Lys Gly
Ser Phe Gln Met145 150 155 160Val His Cys Asn Cys Ser Val His Glu
Arg Cys Glu Cys Leu Val Pro 165 170 175Val Pro Thr Ala Lys Leu Asn
Asp Thr Leu Leu Met Cys Leu Lys Ile 180 185 190Thr Ser Gly Gly Val
Ile Phe Gln Ser Pro Leu Met Ser Val Gln Pro 195 200 205Ile Asn Met
Val Lys Pro Asp Pro Pro Leu Gly Leu Arg Met Glu Ile 210 215 220Thr
Asp Asp Gly Asn Leu Lys Ile Ser Trp Ser Ser Pro Pro Leu Val225 230
235 240Pro Phe Pro Leu Gln Tyr Glu Val Lys Tyr Ser Glu Asn Ser Thr
Thr 245 250 255Val Ile Arg Glu Ala Asp Lys Ile Val Ser Ala Thr Ser
Leu Leu Val 260 265 270Asp Gly Ile Leu Pro Gly Ser Ser Tyr Glu Val
Gln Val Arg Gly Lys 275 280 285Arg Leu Asp Gly Pro Gly Ile Trp Ser
Asp Trp Ser Thr Pro His Val 290 295 300Phe Thr Thr Gln Asp Val Ile
Tyr Phe Pro Pro Lys Ile Leu Thr Ser305 310 315 320Val Gly Ser Asn
Val Ser Phe His Cys Ile Tyr Lys Asn Glu Asn Lys 325 330 335Ile Val
Ser Ser Lys Lys Ile Val Trp Trp Met Asn Leu Ala Glu Lys 340 345
350Ile Pro Gln Ser Gln Tyr Asp Val Val Ser Asp His Val Ser Lys Val
355 360 365Thr Phe Phe Asn Leu Asn Glu Thr Lys Pro Arg Gly Lys Phe
Thr Tyr 370 375 380Asp Ala Val Tyr Cys Cys Asn Glu His Glu Cys His
His Arg Tyr Ala385 390 395 400Glu Leu Tyr Val Ile Asp Val Asn Ile
Asn Ile Ser Cys Glu Thr Asp 405 410 415Gly His Leu Thr Lys Met Thr
Cys Arg Trp Ser Thr Asn Thr Ile Gln 420 425 430Ser Leu Ala Gly Ser
Thr Leu Gln Leu Arg Tyr Arg Arg Ser Ser Leu 435 440 445Tyr Cys Phe
Asp Ile Pro Ser Ile His Pro Ile Ser Lys Pro Lys Asp 450 455 460Cys
Tyr Leu Gln Ser Asp Gly Phe Tyr Glu Cys Val Phe Gln Pro Ile465 470
475 480Phe Leu Leu Ser Gly Tyr Thr Met Trp Ile Arg Ile Asn His Pro
Leu 485 490 495Gly Ser Leu Asp Ser Pro Pro Thr Cys Val Leu Pro Asp
Ser Val Val 500 505 510Lys Pro Leu Pro Pro Ser Ser Val Lys Ala Glu
Ile Ile Lys Asn Ile 515 520 525Gly Leu Leu Lys Ile Ser Trp Glu Lys
Pro Val Phe Pro Glu Asn Asn 530 535 540Leu Gln Phe Gln Ile Arg Tyr
Gly Leu Ser Gly Lys Glu Ile Gln Trp545 550 555 560Lys Met Tyr Asp
Val Tyr Asp Ala Lys Ser Lys Ser Val Ser Leu Pro 565 570 575Val Pro
Asp Phe Cys Ala Val Tyr Ala Val Gln Val Arg Cys Lys Arg 580 585
590Ser Asp Gly Leu Gly Leu Trp Ser Asn Trp Ser Asn Pro Ala Tyr Thr
595 600 605Val Val Met Asp Ile Lys Val Pro Met Arg Gly Pro Glu Phe
Trp Arg 610 615 620Ile Ile Asn Gly Asp Thr Met Lys Lys Glu Lys Asn
Val Thr Leu Leu625 630 635 640Trp Lys Pro Leu Met Lys Asn Asp Ser
Leu Cys Ser Val Gln Arg Tyr 645 650 655Val Ile Asn His His Thr Ser
Cys Asn Gly Thr Trp Ser Glu Asp Val 660 665 670Gly Asn His Thr Lys
Phe Thr Phe Leu Trp Thr Glu Gln Ala His Thr 675 680 685Val Thr Val
Leu Ala Ile Asn Ser Ile Gly Ala Ser Val Ala Asn Phe 690 695 700Asn
Leu Thr Phe Ser Trp Pro Met Ser Lys Val Asn Ile Val Gln Ser705 710
715 720Leu Ser Ala Tyr Pro Leu Asn Ser Ser Cys Val Ile Leu Ser Trp
Ile 725 730 735Leu Ser Pro Ser Asp Tyr Lys Leu Met Tyr Phe Ile Ile
Glu Trp Lys 740 745 750Asn Leu Asn Glu Asp Gly Glu Ile Lys Trp Leu
Arg Ile Ser Ser Ser 755 760 765Val Lys Lys Tyr Tyr Ile His Asp His
Phe Ile Pro Ile Glu Lys Tyr 770 775 780Gln Phe Ser Leu Tyr Pro Ile
Phe Met Glu Gly Val Gly Lys Pro Lys785 790 795 800Ile Ile Asn Ser
Phe Ala Gln Asp Asn Thr Glu Lys His Gln Asn Asp 805 810 815Glu Gln
Lys Leu Ile Ser Glu Glu Asp Leu Gly Gly Glu Gln Lys Leu 820 825
830Ile Ser Glu Glu Asp Leu His His His His His His 835
840118846PRTArtificial SequencemLEPR.mmh aa 1-818 Mouse LEPR
(L22-G839 of NP_666258.2) aa 817-846 myc-myc-hexahistidine tag
118Leu Asn Leu Ala Tyr Pro Ile Ser Pro Trp Lys Phe Lys Leu Phe Cys1
5 10 15Gly Pro Pro Asn Thr Thr Asp Asp Ser Phe Leu Ser Pro Ala Gly
Ala 20 25 30Pro Asn Asn Ala Ser Ala Leu Lys Gly Ala Ser Glu Ala Ile
Val Glu 35 40 45Ala Lys Phe Asn Ser Ser Gly Ile Tyr Val Pro Glu Leu
Ser Lys Thr 50 55 60Val Phe His Cys Cys Phe Gly Asn Glu Gln Gly Gln
Asn Cys Ser Ala65 70 75 80Leu Thr Asp Asn Thr Glu Gly Lys Thr Leu
Ala Ser Val Val Lys Ala 85 90 95Ser Val Phe Arg Gln Leu Gly Val Asn
Trp Asp Ile Glu Cys Trp Met 100 105 110Lys Gly Asp Leu Thr Leu Phe
Ile Cys His Met Glu Pro Leu Pro Lys 115 120 125Asn Pro Phe Lys Asn
Tyr Asp Ser Lys Val His Leu Leu Tyr Asp Leu 130 135 140Pro Glu Val
Ile Asp Asp Ser Pro Leu Pro Pro Leu Lys Asp Ser Phe145 150 155
160Gln Thr Val Gln Cys Asn Cys Ser Leu Arg Gly Cys Glu Cys His Val
165 170 175Pro Val Pro Arg Ala Lys Leu Asn Tyr Ala Leu Leu Met Tyr
Leu Glu
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008


S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.