Anti-nr10 Antibody And Use Thereof
Kuramochi; Taichi ; et al.
U.S. patent application number 16/560143 was filed with the patent office on 2020-01-02 for anti-nr10 antibody and use thereof. This patent application is currently assigned to Chugai Seiyaku Kabushiki Kaisha. The applicant listed for this patent is Chugai Seiyaku Kabushiki Kaisha. Invention is credited to Keiko Esaki, Tomoyuki Igawa, Keiko Kasutani, Taichi Kuramochi, Souhei Ohyama, Hirotake Shiraiwa, Tatsuhiko Tachibana, Hiroyuki Tsunoda.
| Application Number | 20200002429 16/560143 |
| Document ID | / |
| Family ID | 40717782 |
| Filed Date | 2020-01-02 |






View All Diagrams
| United States Patent Application | 20200002429 |
| Kind Code | A1 |
| Kuramochi; Taichi ; et al. | January 2, 2020 |
ANTI-NR10 ANTIBODY AND USE THEREOF
Abstract
The present inventors successfully obtained anti-NR10 antibodies having an effective neutralizing activity against NR10. The anti-NR10 antibodies provided by the present invention are useful as, for example, pharmaceuticals for treating or preventing inflammatory diseases.
| Inventors: | Kuramochi; Taichi; (Shizuoka, JP) ; Kasutani; Keiko; (Shizuoka, JP) ; Ohyama; Souhei; (Shizuoka, JP) ; Tsunoda; Hiroyuki; (Shizuoka, JP) ; Igawa; Tomoyuki; (Shizuoka, JP) ; Tachibana; Tatsuhiko; (Shizuoka, JP) ; Shiraiwa; Hirotake; (Shizuoka, JP) ; Esaki; Keiko; (Shizuoka, JP) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Assignee: | Chugai Seiyaku Kabushiki
Kaisha Tokyo JP |
||||||||||
| Family ID: | 40717782 | ||||||||||
| Appl. No.: | 16/560143 | ||||||||||
| Filed: | September 4, 2019 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 14340883 | Jul 25, 2014 | |||
| 16560143 | ||||
| 12745781 | Sep 13, 2010 | |||
| PCT/JP2008/072152 | Dec 5, 2008 | |||
| 14340883 | ||||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C07K 2317/92 20130101; C07K 2317/76 20130101; G01N 2333/7155 20130101; A61P 37/00 20180101; G01N 33/6869 20130101; A61K 2039/505 20130101; C07K 2317/56 20130101; C07K 16/2866 20130101; C07K 2317/565 20130101; C07K 2317/567 20130101; C07K 2317/24 20130101; A61P 29/00 20180101 |
| International Class: | C07K 16/28 20060101 C07K016/28; G01N 33/68 20060101 G01N033/68 |
Foreign Application Data
| Date | Code | Application Number |
|---|---|---|
| Dec 5, 2007 | JP | 2007-315143 |
| Sep 26, 2008 | JP | 2008-247425 |
Claims
1. An antibody that recognizes domain 1 of NR10.
2. The antibody of claim 1, which has a neutralizing activity.
3. The antibody of claim 1 or 2, which is a humanized antibody.
4. An anti-NR10 antibody which is any one of: (1) an antibody comprising a heavy chain variable region which comprises CDR1 comprising the amino acid sequence of SEQ ID NO: 1, CDR2 comprising the amino acid sequence of SEQ ID NO: 2, and CDR3 comprising the amino acid sequence of SEQ ID NO: 3; (2) an antibody comprising the heavy chain variable region of SEQ ID NO: 4; (3) an antibody comprising a light chain variable region which comprises CDR1 comprising the amino acid sequence of SEQ ID NO: 5, CDR2 comprising the amino acid sequence of SEQ ID NO: 6, and CDR3 comprising the amino acid sequence of SEQ ID NO: 7; (4) an antibody comprising the light chain variable region of SEQ ID NO: 8; (5) an antibody comprising the heavy chain variable region of (1) and the light chain variable region of (3); (6) an antibody comprising the heavy chain variable region of (2) and the light chain variable region of (4); (7) an antibody in which one or more amino acids are substituted, deleted, added, and/or inserted in the antibody of any one of (1) to (6), which has an activity equivalent to that of the antibody of any one of (1) to (6); and (8) an antibody which binds to the same epitope as an epitope bound by the antibody of any one of (1) to (7).
5. An anti-NR10 antibody which is any one of: (1) an antibody comprising a heavy chain variable region which comprises CDR1 comprising the amino acid sequence of SEQ ID NO: 9, CDR2 comprising the amino acid sequence of SEQ ID NO: 10, and CDR3 comprising the amino acid sequence of SEQ ID NO: 11; (2) an antibody comprising the heavy chain variable region of SEQ ID NO: 12; (3) an antibody comprising a light chain variable region which comprises CDR1 comprising the amino acid sequence of SEQ ID NO: 13, CDR2 comprising the amino acid sequence of SEQ ID NO: 14, and CDR3 comprising the amino acid sequence of SEQ ID NO: 15; (4) an antibody comprising the light chain variable region of SEQ ID NO: 16; (5) an antibody comprising the heavy chain variable region of (1) and the light chain variable region of (3); (6) an antibody comprising the heavy chain variable region of (2) and the light chain variable region of (4); (7) an antibody in which one or more amino acids are substituted, deleted, added, and/or inserted in the antibody of any one of (1) to (6), which has an activity equivalent to that of the antibody of any one of (1) to (6); and (8) an antibody which binds to the same epitope as an epitope bound by the antibody of any one of (1) to (7).
6. An anti-NR10 antibody which is any one of: (1) an antibody comprising a heavy chain variable region which comprises CDR1 comprising the amino acid sequence of SEQ ID NO: 17, CDR2 comprising the amino acid sequence of SEQ ID NO: 18, and CDR3 comprising the amino acid sequence of SEQ ID NO: 19; (2) an antibody comprising the heavy chain variable region of SEQ ID NO: 20; (3) an antibody comprising a light chain variable region which comprises CDR1 comprising the amino acid sequence of SEQ ID NO: 21, CDR2 comprising the amino acid sequence of SEQ ID NO: 22, and CDR3 comprising the amino acid sequence of SEQ ID NO: 23; (4) an antibody comprising the light chain variable region of SEQ ID NO: 24; (5) an antibody comprising the heavy chain variable region of (1) and the light chain variable region of (3); (6) an antibody comprising the heavy chain variable region of (2) and the light chain variable region of (4); (7) an antibody in which one or more amino acids are substituted, deleted, added, and/or inserted in the antibody of any one of (1) to (6), which has an activity equivalent to that of the antibody of any one of (1) to (6); and (8) an antibody which binds to the same epitope as an epitope bound by the antibody of any one of (1) to (7).
7. An anti-NR10 antibody which is any one of: (1) an antibody comprising a heavy chain variable region which comprises CDR1 comprising the amino acid sequence of SEQ ID NO: 25, CDR2 comprising the amino acid sequence of SEQ ID NO: 26, and CDR3 comprising the amino acid sequence of SEQ ID NO: 27; (2) an antibody comprising the heavy chain variable region of SEQ ID NO: 28; (3) an antibody comprising a light chain variable region which comprises CDR1 comprising the amino acid sequence of SEQ ID NO: 29, CDR2 comprising the amino acid sequence of SEQ ID NO: 30, and CDR3 comprising the amino acid sequence of SEQ ID NO: 31; (4) an antibody comprising the light chain variable region of SEQ ID NO: 32; (5) an antibody comprising the heavy chain variable region of (1) and the light chain variable region of (3); (6) an antibody comprising the heavy chain variable region of (2) and the light chain variable region of (4); (7) an antibody in which one or more amino acids are substituted, deleted, added, and/or inserted in the antibody of any one of (1) to (6), which has an activity equivalent to that of the antibody of any one of (1) to (6); and (8) an antibody which binds to the same epitope as an epitope bound by the antibody of any one of (1) to (7).
8. An antibody or antibody variable region which is any one of: (1) a heavy chain variable region comprising CDR1 of SEQ ID NO: 196, CDR2 of SEQ ID NO: 197, and CDR3 of SEQ ID NO: 11 (H17); (2) a heavy chain variable region comprising CDR1 of SEQ ID NO: 176, CDR2 of SEQ ID NO: 197, and CDR3 of SEQ ID NO: 11 (H19); (3) a heavy chain variable region comprising CDR1 of SEQ ID NO: 196, CDR2 of SEQ ID NO: 197, and CDR3 of SEQ ID NO: 184 (H28, H42); (4) a heavy chain variable region comprising CDR1 of SEQ ID NO: 9, CDR2 of SEQ ID NO: 197, and CDR3 of SEQ ID NO: 184 (H30, H44); (5) a heavy chain variable region comprising CDR1 of SEQ ID NO: 176, CDR2 of SEQ ID NO: 197, CDR3 of SEQ ID NO: 184 (H34, H46); (6) a heavy chain variable region comprising CDR1 of SEQ ID NO: 9, CDR2 of SEQ ID NO: 198, and CDR3 of SEQ ID NO: 184 (H57, H78); (7) a heavy chain variable region comprising CDR1 of SEQ ID NO: 176, CDR2 of SEQ ID NO: 198, and CDR3 of SEQ ID NO: 184 (H71, H92); (8) a heavy chain variable region comprising CDR1 of SEQ ID NO: 9, CDR2 of SEQ ID NO: 199, and CDR3 of SEQ ID NO: 184 (H97, H98); (9) a light chain variable region comprising CDR1 of SEQ ID NO: 200, CDR2 of SEQ ID NO: 170, and CDR3 of SEQ ID NO: 193 (L11); (10) a light chain variable region comprising CDR1 of SEQ ID NO: 201, CDR2 of SEQ ID NO: 170, and CDR3 of SEQ ID NO: 193 (L12); (11) a light chain variable region comprising CDR1 of SEQ ID NO: 202, CDR2 of SEQ ID NO: 170, and CDR3 of SEQ ID NO: 193 (L17); (12) a light chain variable region comprising CDR1 of SEQ ID NO: 203, CDR2 of SEQ ID NO: 170, and CDR3 of SEQ ID NO: 193 (L50); (13) an antibody comprising the heavy chain variable region of (3) and the light chain variable region of (11); (14) an antibody comprising the heavy chain variable region of (4) and the light chain variable region of (11); (15) an antibody comprising the heavy chain variable region of (5) and the light chain variable region of (11); (16) an antibody comprising the heavy chain variable region of (6) and the light chain variable region of (11); (17) an antibody comprising the heavy chain variable region of (7) and the light chain variable region of (11); (18) an antibody comprising the heavy chain variable region of (8) and the light chain variable region of (12); (19) an antibody in which one or more amino acids are substituted, deleted, added, and/or inserted in the antibody of any one of (13) to (18), which has an activity equivalent to that of the antibody of any one of (13) to (18); and (20) an antibody which binds to the same epitope as an epitope bound by the antibody of any one of (13) to (18).
9. An antibody or antibody variable region which is any one of: (1) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 204 (H17); (2) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 205 (H19); (3) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 206 (H28); (4) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 207 (H30); (5) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 208 (H34), (6) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 209 (H42); (7) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 210 (H44); (8) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 211 (H46); (9) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 212 (H57); (10) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 213 (H71); (11) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 214 (H78); (12) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 215 (H92); (13) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 216 (H97); (14) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 217 (H98); (15) a light chain variable region comprising the amino acid sequence of SEQ ID NO: 218 (L11); (16) a light chain variable region comprising the amino acid sequence of SEQ ID NO: 219 (L12); (17) a light chain variable region comprising the amino acid sequence of SEQ ID NO: 220 (L17); (18) a light chain variable region comprising the amino acid sequence of SEQ ID NO: 221 (L50); (19) an antibody comprising the heavy chain variable region of (3) and the light chain variable region of (17) (H28L17); (20) an antibody comprising the heavy chain variable region of (4) and the light chain variable region of (17) (H30L17); (21) an antibody comprising the heavy chain variable region of (5) and the light chain variable region of (17) (H34L17); (22) an antibody comprising the heavy chain variable region of (6) and the light chain variable region of (17) (H42L17); (23) an antibody comprising the heavy chain variable region of (7) and the light chain variable region of (17) (H44L17); (24) an antibody comprising the heavy chain variable region of (8) and the light chain variable region of (17) (H46L17); (25) an antibody comprising the heavy chain variable region of (9) and the light chain variable region of (17) (H57L17); (26) an antibody comprising the heavy chain variable region of (10) and the light chain variable region of (17) (H71L17); (27) an antibody comprising the heavy chain variable region of (11) and the light chain variable region of (17) (H78L17); (28) an antibody comprising the heavy chain variable region of (12) and the light chain variable region of (17) (H92L17); (29) an antibody comprising the heavy chain variable region of (13) and the light chain variable region of (18) (H97L50); (30) an antibody comprising the heavy chain variable region of (14) and the light chain variable region of (18) (H98L50), (31) an antibody in which one or more amino acids are substituted, deleted, added, and/or inserted in the antibody of any one of (19) to (30), which has an activity equivalent to that of the antibody of any one of (19) to (30); and (32) an antibody which binds to the same epitope as an epitope bound by the antibody of any one of (19) to (30).
10. The anti-NR10 antibody of any one of claims 4 to 9, which is a humanized antibody.
11. An antibody, antibody heavy chain, or antibody light chain, which is any one of: (1) a heavy chain comprising the amino acid sequence of SEQ ID NO: 222 (H17); (2) a heavy chain comprising the amino acid sequence of SEQ ID NO: 223 (H19); (3) a heavy chain comprising the amino acid sequence of SEQ ID NO: 224 (H28); (4) a heavy chain comprising the amino acid sequence of SEQ ID NO: 225 (H30); (5) a heavy chain comprising the amino acid sequence of SEQ ID NO: 226 (H34); (6) a heavy chain comprising the amino acid sequence of SEQ ID NO: 227 (H42); (7) a heavy chain comprising the amino acid sequence of SEQ ID NO: 228 (H44); (8) a heavy chain comprising the amino acid sequence of SEQ ID NO: 229 (H46); (9) a heavy chain comprising the amino acid sequence of SEQ ID NO: 230 (H57); (10) a heavy chain comprising the amino acid sequence of SEQ ID NO: 231 (H71); (11) a heavy chain comprising the amino acid sequence of SEQ ID NO: 232 (H78); (12) a heavy chain comprising the amino acid sequence of SEQ ID NO: 233 (H92); (13) a heavy chain comprising the amino acid sequence of SEQ ID NO: 234 (H97); (14) a heavy chain comprising the amino acid sequence of SEQ ID NO: 235 (H98); (15) a light chain comprising the amino acid sequence of SEQ ID NO: 236 (L11); (16) a light chain comprising the amino acid sequence of SEQ ID NO: 237 (L12); (17) a light chain comprising the amino acid sequence of SEQ ID NO: 238 (L17); (18) a light chain comprising the amino acid sequence of SEQ ID NO: 239 (L50); (19) an antibody comprising the heavy chain of (3) and the light chain of (17) (H28L17); (20) an antibody comprising the heavy chain of (4) and the light chain of (17) (H30L17); (21) an antibody comprising the heavy chain of (5) and the light chain of (17) (H34L17); (22) an antibody comprising the heavy chain of (6) and the light chain of (17) (H42L17); (23) an antibody comprising the heavy chain of (7) and the light chain of (17) (H44L17); (24) an antibody comprising the heavy chain of (8) and the light chain of (17) (H46L17); (25) an antibody comprising the heavy chain of (9) and the light chain of (17) (H57L17); (26) an antibody comprising the heavy chain of (10) and the light chain of (17) (H71L17); (27) an antibody comprising the heavy chain of (11) and the light chain of (17) (H78L17); (28) an antibody comprising the heavy chain of (12) and the light chain of (17) (H92L17); (29) an antibody comprising the heavy chain of (13) and the light chain of (18) (H97L50); (30) an antibody comprising the heavy chain of (14) and the light chain of (18) (H98L50); (31) an antibody in which one or more amino acids are substituted, deleted, added, and/or inserted in the antibody of any one of (19) to (30), which has an activity equivalent to that of the antibody of any one of (19) to (30); and (32) an antibody which binds to the same epitope as an epitope bound by the antibody of any one of (19) to (30).
12. A pharmaceutical composition comprising the antibody of any one of claims 1 to 11.
13. The pharmaceutical composition of claim 12, which is an agent for treating an inflammatory disease.
Description
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] This application is a continuation of U.S. application Ser. No. 14/340,883, filed Jul. 25, 2014, which is a continuation of U.S. application Ser. No. 12/745,781, filed Sep. 13, 2010 (now abandoned), which is a 371 of PCT/JP2008/072152, filed Dec. 5, 2008, which in turn claims the benefit of Japanese Patent Application Nos. 2007-315143, filed Dec. 5, 2007, and 2008-247425, filed Sep. 26, 2008.
TECHNICAL FIELD
[0002] The present invention relates to anti-NR10 antibodies, and pharmaceutical compositions comprising an anti-NR10 antibody.
BACKGROUND ART
[0003] Many cytokines are known as humoral factors involved in the growth and differentiation of various types of cells, or in the activation of differentiated mature cell functions. Cytokine-stimulated cells produce different types of cytokines, thereby forming networks of multiple cytokines in the body. Biological homeostasis is maintained by a delicate balance of the mutual regulation between cytokines in these networks. Many inflammatory diseases are thought to result from a failure of such cytokine networks. Thus, monoclonal antibody-based anti-cytokine therapy is drawing much attention. For example, anti-TNF antibodies and anti-IL-6 receptor antibodies have been demonstrated to be highly effective clinically. On the other hand, there are many examples of failure where no therapeutic effects were produced when a single cytokine, such as IL-4, was blocked alone, due to the activation of compensatory pathways in actual pathological conditions.
[0004] The present inventors succeeded in isolating a novel cytokine receptor NR10 that was highly homologous to gp130, a receptor for IL-6 signal transduction (Patent Document 1). NR10 forms a heterodimer with oncostatin M receptor (OSMR) and functions as an IL-31 receptor (Non-patent Document 1). Regarding IL-31, it has been reported that transgenic mice overexpressing IL-31 spontaneously develop pruritic dermatitis (Patent Document 2).
[0005] Antibodies that bind to NR10 and inhibit the binding between NR10 and IL-31 may be effective in treating inflammatory diseases. For clinical use, anti-NR10 antibodies are required to have low immunogenicity. Furthermore, in order to achieve high therapeutic effects, antibodies with strong NR10-binding or neutralizing activity are desired.
[0006] Prior art documents of the present invention are described below. [0007] Patent Document 1: WO00/75314 [0008] Patent Document 2: WO03/060090 [0009] Non-patent Document 1: IL-31 is associated with cutaneous lymphocyte antigen-positive skin homing T cells in patients with atopic dermatitis, J Allergy Clin Immunol. 2006 February; 117(2): 418-25.
DISCLOSURE OF THE INVENTION
Problems to be Solved by the Invention
[0010] The present invention was achieved in view of the circumstances described above. An objective of the present invention is to provide anti-NR10 antibodies, and pharmaceutical compositions comprising an anti-NR10 antibody.
Means for Solving the Problems
[0011] The present inventors conducted dedicated studies to achieve the objective described above. The present inventors succeeded in obtaining anti-NR10 antibodies having an effective neutralizing activity against NR10. Furthermore, the present inventors succeeded in humanizing the antibodies while maintaining their activity. The present inventors also successfully produced antibodies with improved pharmacokinetics, enhanced antigen-binding activity, improved stability, and/or reduced risk of immunogenicity. These antibodies are useful as therapeutic agents for inflammatory diseases.
[0012] The present invention relates to anti-NR10 antibodies, and pharmaceutical compositions comprising an anti-NR10 antibody. More specifically, the present invention includes:
[1] an antibody that recognizes domain 1 of NR10; [2] the antibody of [1], which has a neutralizing activity; [3] the antibody of [1] or [2], which is a humanized antibody; [4] an anti-NR10 antibody which is any one of: (1) an antibody comprising a heavy chain variable region which comprises CDR1 comprising the amino acid sequence of SEQ ID NO: 1, CDR2 comprising the amino acid sequence of SEQ ID NO: 2, and CDR3 comprising the amino acid sequence of SEQ ID NO: 3; (2) an antibody comprising the heavy chain variable region of SEQ ID NO: 4; (3) an antibody comprising a light chain variable region which comprises CDR1 comprising the amino acid sequence of SEQ ID NO: 5, CDR2 comprising the amino acid sequence of SEQ ID NO: 6, and CDR3 comprising the amino acid sequence of SEQ ID NO: 7; (4) an antibody comprising the light chain variable region of SEQ ID NO: 8; (5) an antibody comprising the heavy chain variable region of (1) and the light chain variable region of (3); (6) an antibody comprising the heavy chain variable region of (2) and the light chain variable region of (4); (7) an antibody in which one or more amino acids are substituted, deleted, added, and/or inserted in the antibody of any one of (1) to (6), which has an activity equivalent to that of the antibody of any one of (1) to (6); and (8) an antibody which binds to the same epitope as an epitope bound by the antibody of any one of (1) to (7); [5] an anti-NR10 antibody which is any one of: (1) an antibody comprising a heavy chain variable region which comprises CDR1 comprising the amino acid sequence of SEQ ID NO: 9, CDR2 comprising the amino acid sequence of SEQ ID NO: 10, and CDR3 comprising the amino acid sequence of SEQ ID NO: 11; (2) an antibody comprising the heavy chain variable region of SEQ ID NO: 12; (3) an antibody comprising a light chain variable region which comprises CDR1 comprising the amino acid sequence of SEQ ID NO: 13, CDR2 comprising the amino acid sequence of SEQ ID NO: 14, and CDR3 comprising the amino acid sequence of SEQ ID NO: 15; (4) an antibody comprising the light chain variable region of SEQ ID NO: 16; (5) an antibody comprising the heavy chain variable region of (1) and the light chain variable region of (3); (6) an antibody comprising the heavy chain variable region of (2) and the light chain variable region of (4); (7) an antibody in which one or more amino acids are substituted, deleted, added, and/or inserted in the antibody of any one of (1) to (6), which has an activity equivalent to that of the antibody of any one of (1) to (6); and (8) an antibody which binds to the same epitope as an epitope bound by the antibody of any one of (1) to (7); [6] an anti-NR10 antibody which is any one of: (1) an antibody comprising a heavy chain variable region which comprises CDR1 comprising the amino acid sequence of SEQ ID NO: 17, CDR2 comprising the amino acid sequence of SEQ ID NO: 18, and CDR3 comprising the amino acid sequence of SEQ ID NO: 19; (2) an antibody comprising the heavy chain variable region of SEQ ID NO: 20; (3) an antibody comprising a light chain variable region which comprises CDR1 comprising the amino acid sequence of SEQ ID NO: 21, CDR2 comprising the amino acid sequence of SEQ ID NO: 22, and CDR3 comprising the amino acid sequence of SEQ ID NO: 23; (4) an antibody comprising the light chain variable region of SEQ ID NO: 24; (5) an antibody comprising the heavy chain variable region of (1) and the light chain variable region of (3); (6) an antibody comprising the heavy chain variable region of (2) and the light chain variable region of (4); (7) an antibody in which one or more amino acids are substituted, deleted, added, and/or inserted in the antibody of any one of (1) to (6), which has an activity equivalent to that of the antibody of any one of (1) to (6); and (8) an antibody which binds to the same epitope as an epitope bound by the antibody of any one of (1) to (7); [7] an anti-NR10 antibody which is any one of: (1) an antibody comprising a heavy chain variable region which comprises CDR1 comprising the amino acid sequence of SEQ ID NO: 25, CDR2 comprising the amino acid sequence of SEQ ID NO: 26, and CDR3 comprising the amino acid sequence of SEQ ID NO: 27; (2) an antibody comprising the heavy chain variable region of SEQ ID NO: 28; (3) an antibody comprising a light chain variable region which comprises CDR1 comprising the amino acid sequence of SEQ ID NO: 29, CDR2 comprising the amino acid sequence of SEQ ID NO: 30, and CDR3 comprising the amino acid sequence of SEQ ID NO: 31; (4) an antibody comprising the light chain variable region of SEQ ID NO: 32; (5) an antibody comprising the heavy chain variable region of (1) and the light chain variable region of (3); (6) an antibody comprising the heavy chain variable region of (2) and the light chain variable region of (4); (7) an antibody in which one or more amino acids are substituted, deleted, added, and/or inserted in the antibody of any one of (1) to (6), which has an activity equivalent to that of the antibody of any one of (1) to (6); and (8) an antibody which binds to the same epitope as an epitope bound by the antibody of any one of (1) to (7); [8] an antibody or antibody variable region which is any one of: (1) a heavy chain variable region comprising CDR1 of SEQ ID NO: 196, CDR2 of SEQ ID NO: 197, and CDR3 of SEQ ID NO: 11 (H17); (2) a heavy chain variable region comprising CDR1 of SEQ ID NO: 176, CDR2 of SEQ ID NO: 197, and CDR3 of SEQ ID NO: 11 (H19); (3) a heavy chain variable region comprising CDR1 of SEQ ID NO: 196, CDR2 of SEQ ID NO: 197, and CDR3 of SEQ ID NO: 184 (H28, H42); (4) a heavy chain variable region comprising CDR1 of SEQ ID NO: 9, CDR2 of SEQ ID NO: 197, and CDR3 of SEQ ID NO: 184 (H30, H44); (5) a heavy chain variable region comprising CDR1 of SEQ ID NO: 176, CDR2 of SEQ ID NO: 197, CDR3 of SEQ ID NO: 184 (H34, H46); (6) a heavy chain variable region comprising CDR1 of SEQ ID NO: 9, CDR2 of SEQ ID NO: 198, and CDR3 of SEQ ID NO: 184 (H57, H78); (7) a heavy chain variable region comprising CDR1 of SEQ ID NO: 176, CDR2 of SEQ ID NO: 198, and CDR3 of SEQ ID NO: 184 (H71, H92); (8) a heavy chain variable region comprising CDR1 of SEQ ID NO: 9, CDR2 of SEQ ID NO: 199, and CDR3 of SEQ ID NO: 184 (H97, H98); (9) a light chain variable region comprising CDR1 of SEQ ID NO: 200, CDR2 of SEQ ID NO: 170, and CDR3 of SEQ ID NO: 193 (L11); (10) a light chain variable region comprising CDR1 of SEQ ID NO: 201, CDR2 of SEQ ID NO: 170, and CDR3 of SEQ ID NO: 193 (L12); (11) a light chain variable region comprising CDR1 of SEQ ID NO: 202, CDR2 of SEQ ID NO: 170, and CDR3 of SEQ ID NO: 193 (L17); (12) a light chain variable region comprising CDR1 of SEQ ID NO: 203, CDR2 of SEQ ID NO: 170, and CDR3 of SEQ ID NO: 193 (L50); (13) an antibody comprising the heavy chain variable region of (3) and the light chain variable region of (11); (14) an antibody comprising the heavy chain variable region of (4) and the light chain variable region of (11); (15) an antibody comprising the heavy chain variable region of (5) and the light chain variable region of (11); (16) an antibody comprising the heavy chain variable region of (6) and the light chain variable region of (11); (17) an antibody comprising the heavy chain variable region of (7) and the light chain variable region of (11); (18) an antibody comprising the heavy chain variable region of (8) and the light chain variable region of (12); (19) an antibody in which one or more amino acids are substituted, deleted, added, and/or inserted in the antibody of any one of (13) to (18), which has an activity equivalent to that of the antibody of any one of (13) to (18); and (20) an antibody which binds to the same epitope as an epitope bound by the antibody of any one of (13) to (18); [9] an antibody or antibody variable region which is any one of: (1) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 204 (H17); (2) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 205 (H19); (3) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 206 (H28); (4) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 207 (H30); (5) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 208 (H34), (6) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 209 (H42); (7) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 210 (H44); (8) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 211 (H46); (9) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 212 (H57); (10) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 213 (H71); (11) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 214 (H78); (12) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 215 (H92); (13) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 216 (H97); (14) a heavy chain variable region comprising the amino acid sequence of SEQ ID NO: 217 (H98); (15) a light chain variable region comprising the amino acid sequence of SEQ ID NO: 218 (L11); (16) a light chain variable region comprising the amino acid sequence of SEQ ID NO: 219 (L12); (17) a light chain variable region comprising the amino acid sequence of SEQ ID NO: 220 (L17); (18) a light chain variable region comprising the amino acid sequence of SEQ ID NO: 221 (L50); (19) an antibody comprising the heavy chain variable region of (3) and the light chain variable region of (17) (H28L17); (20) an antibody comprising the heavy chain variable region of (4) and the light chain variable region of (17) (H30L17); (21) an antibody comprising the heavy chain variable region of (5) and the light chain variable region of (17) (H34L17); (22) an antibody comprising the heavy chain variable region of (6) and the light chain variable region of (17) (H42L17); (23) an antibody comprising the heavy chain variable region of (7) and the light chain variable region of (17) (H44L17); (24) an antibody comprising the heavy chain variable region of (8) and the light chain variable region of (17) (H46L17); (25) an antibody comprising the heavy chain variable region of (9) and the light chain variable region of (17) (H57L17); (26) an antibody comprising the heavy chain variable region of (10) and the light chain variable region of (17) (H71L17); (27) an antibody comprising the heavy chain variable region of (11) and the light chain variable region of (17) (H78L17); (28) an antibody comprising the heavy chain variable region of (12) and the light chain variable region of (17) (H92L17); (29) an antibody comprising the heavy chain variable region of (13) and the light chain variable region of (18) (H97L50); (30) an antibody comprising the heavy chain variable region of (14) and the light chain variable region of (18) (H98L50), (31) an antibody in which one or more amino acids are substituted, deleted, added, and/or inserted in the antibody of any one of (19) to (30), which has an activity equivalent to that of the antibody of any one of (19) to (30); and (32) an antibody which binds to the same epitope as an epitope bound by the antibody of any one of (19) to (30); [10] the anti-NR10 antibody of any one of [4] to [9], which is a humanized antibody; [11] an antibody, antibody heavy chain, or antibody light chain, which is any one of: (1) a heavy chain comprising the amino acid sequence of SEQ ID NO: 222 (H17); (2) a heavy chain comprising the amino acid sequence of SEQ ID NO: 223 (H19); (3) a heavy chain comprising the amino acid sequence of SEQ ID NO: 224 (H28); (4) a heavy chain comprising the amino acid sequence of SEQ ID NO: 225 (H30); (5) a heavy chain comprising the amino acid sequence of SEQ ID NO: 226 (H34); (6) a heavy chain comprising the amino acid sequence of SEQ ID NO: 227 (H42); (7) a heavy chain comprising the amino acid sequence of SEQ ID NO: 228 (H44); (8) a heavy chain comprising the amino acid sequence of SEQ ID NO: 229 (H46); (9) a heavy chain comprising the amino acid sequence of SEQ ID NO: 230 (H57); (10) a heavy chain comprising the amino acid sequence of SEQ ID NO: 231 (H71); (11) a heavy chain comprising the amino acid sequence of SEQ ID NO: 232 (H78); (12) a heavy chain comprising the amino acid sequence of SEQ ID NO: 233 (H92); (13) a heavy chain comprising the amino acid sequence of SEQ ID NO: 234 (H97); (14) a heavy chain comprising the amino acid sequence of SEQ ID NO: 235 (H98); (15) a light chain comprising the amino acid sequence of SEQ ID NO: 236 (L11); (16) a light chain comprising the amino acid sequence of SEQ ID NO: 237 (L12); (17) a light chain comprising the amino acid sequence of SEQ ID NO: 238 (L17); (18) a light chain comprising the amino acid sequence of SEQ ID NO: 239 (L50); (19) an antibody comprising the heavy chain of (3) and the light chain of (17) (H28L17); (20) an antibody comprising the heavy chain of (4) and the light chain of (17) (H30L17); (21) an antibody comprising the heavy chain of (5) and the light chain of (17) (H34L17); (22) an antibody comprising the heavy chain of (6) and the light chain of (17) (H42L17); (23) an antibody comprising the heavy chain of (7) and the light chain of (17) (H44L17); (24) an antibody comprising the heavy chain of (8) and the light chain of (17) (H46L17); (25) an antibody comprising the heavy chain of (9) and the light chain of (17) (H57L17); (26) an antibody comprising the heavy chain of (10) and the light chain of (17) (H71L17); (27) an antibody comprising the heavy chain of (11) and the light chain of (17) (H78L17); (28) an antibody comprising the heavy chain of (12) and the light chain of (17) (H92L17); (29) an antibody comprising the heavy chain of (13) and the light chain of (18) (H97L50); (30) an antibody comprising the heavy chain of (14) and the light chain of (18) (H98L50); (31) an antibody in which one or more amino acids are substituted, deleted, added, and/or inserted in the antibody of any one of (19) to (30), which has an activity equivalent to that of the antibody of any one of (19) to (30); and (32) an antibody which binds to the same epitope as an epitope bound by the antibody of any one of (19) to (30); [12] a pharmaceutical composition comprising the antibody of any one of [1] to [11]; [13] the pharmaceutical composition of [12], which is an agent for treating an inflammatory disease; [14] a method for treating or preventing an inflammatory disease, which comprises the step of administering the antibody of any one of [1] to [11]; and [15] use of the antibody of any one of [1] to [11] in the preparation of a therapeutic agent for an inflammatory disease.
BRIEF DESCRIPTION OF THE DRAWINGS
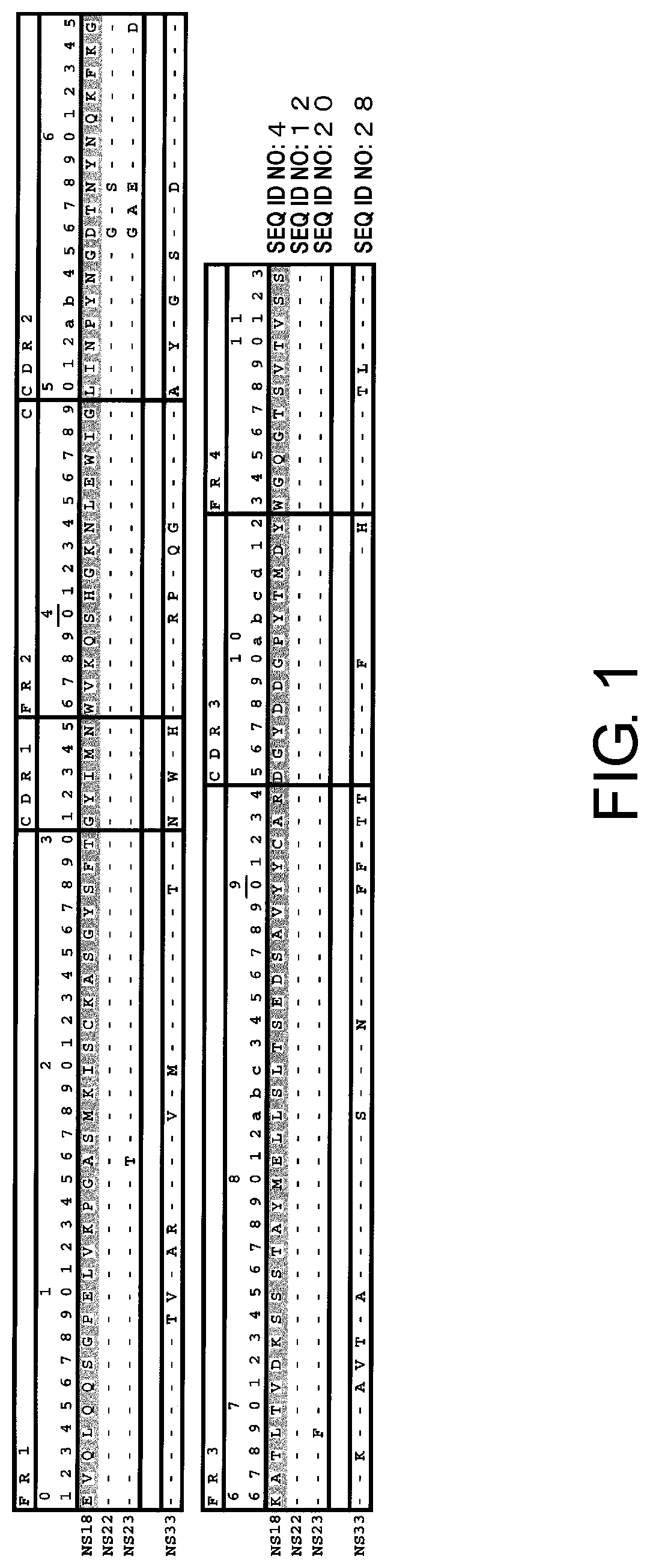
[0013] FIG. 1 shows the amino acid sequences of the heavy chain variable regions of mouse antibodies NS18, NS22, NS23, and NS33.
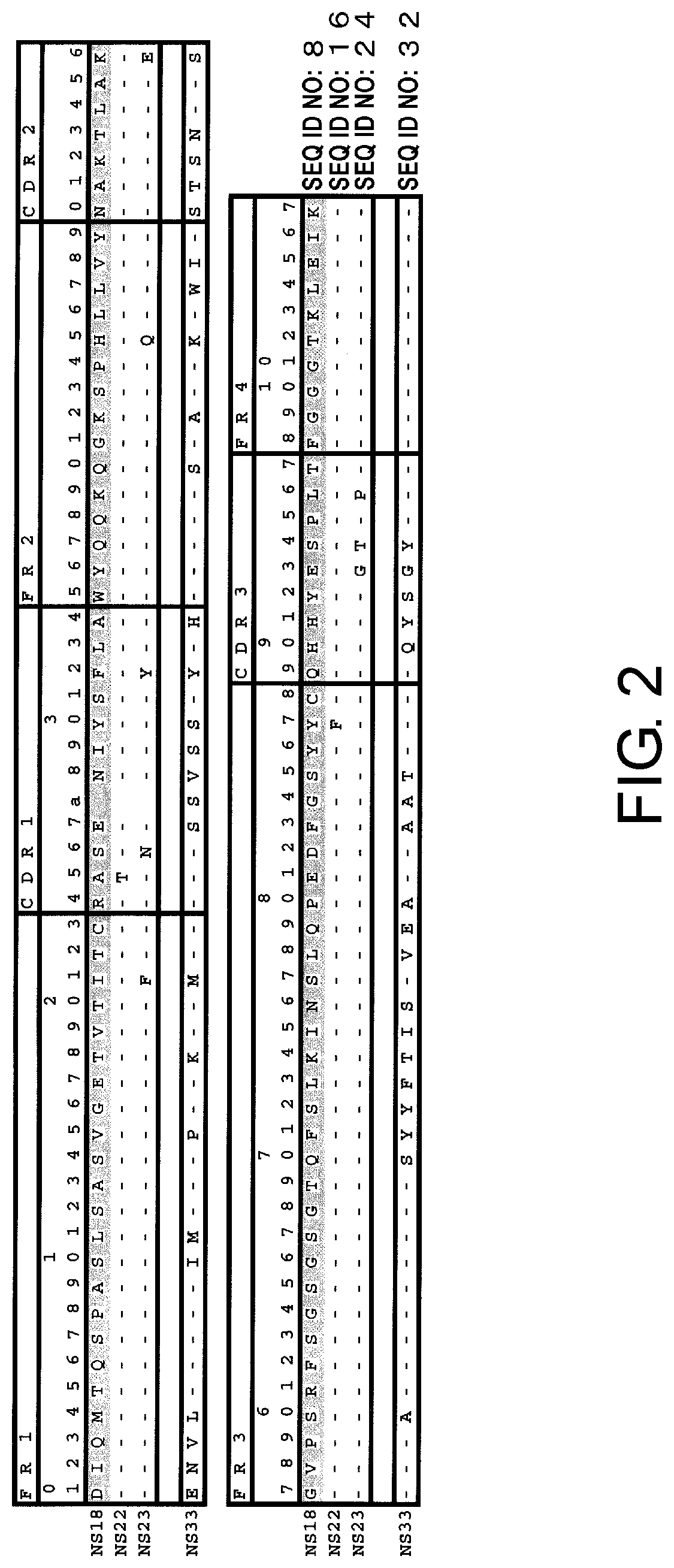
[0014] FIG. 2 shows the amino acid sequences of the light chain variable regions of mouse antibodies NS18, NS22, NS23, and NS33.
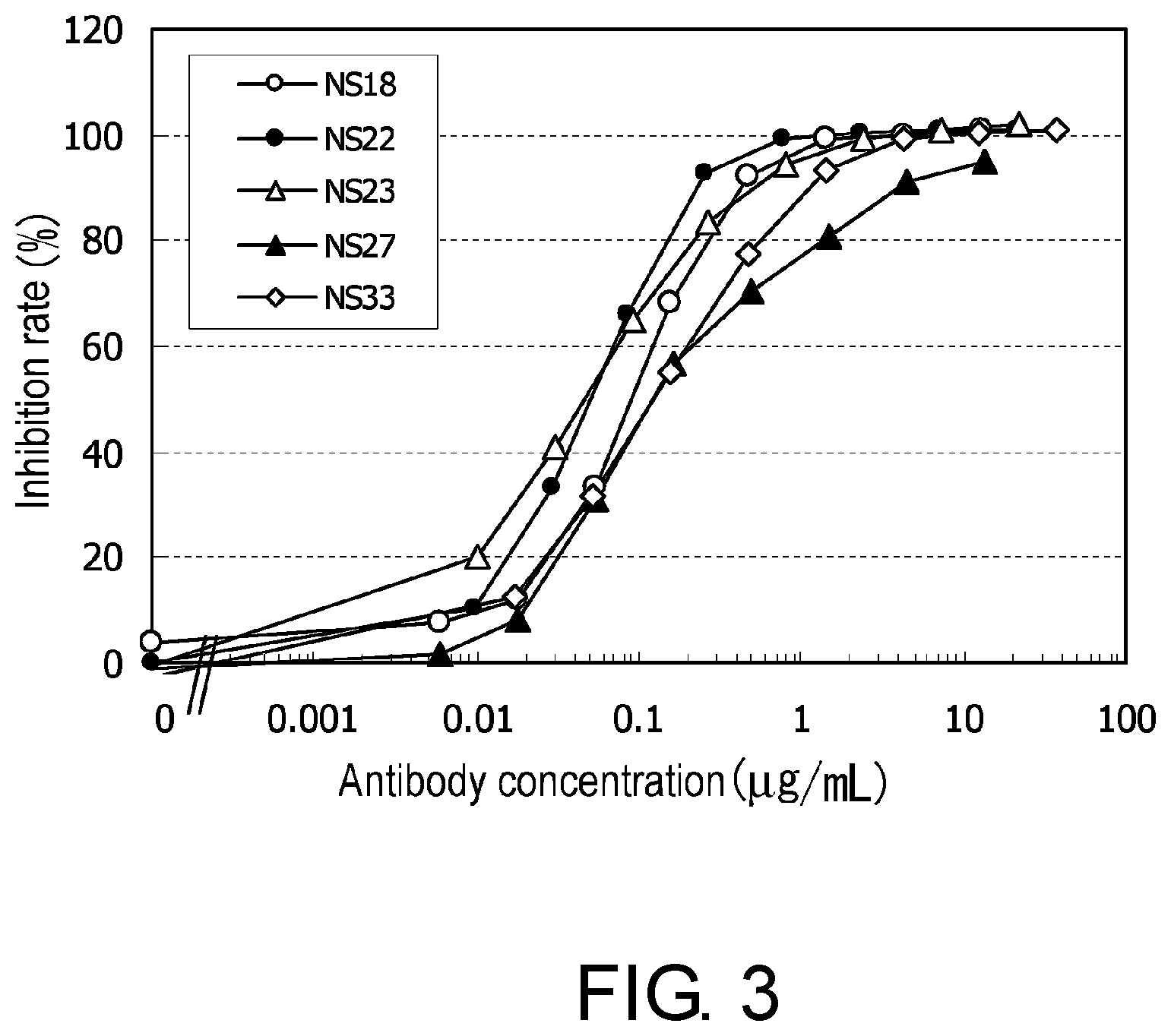
[0015] FIG. 3 is a graph showing the inhibition of hNR10/hOSMR/BaF3 cell growth by hybridoma culture supernatants.
[0016] FIG. 4 is a graph showing the inhibition of cynNR10/cynOSMR/BaF3 cell growth by hybridoma culture supernatants.
[0017] FIG. 5 is a graph showing the assessment of the activity of chimeric NS22 (BaF).
[0018] FIG. 6 is a graph showing the assessment of the activity of chimeric NS22 (DU-145).
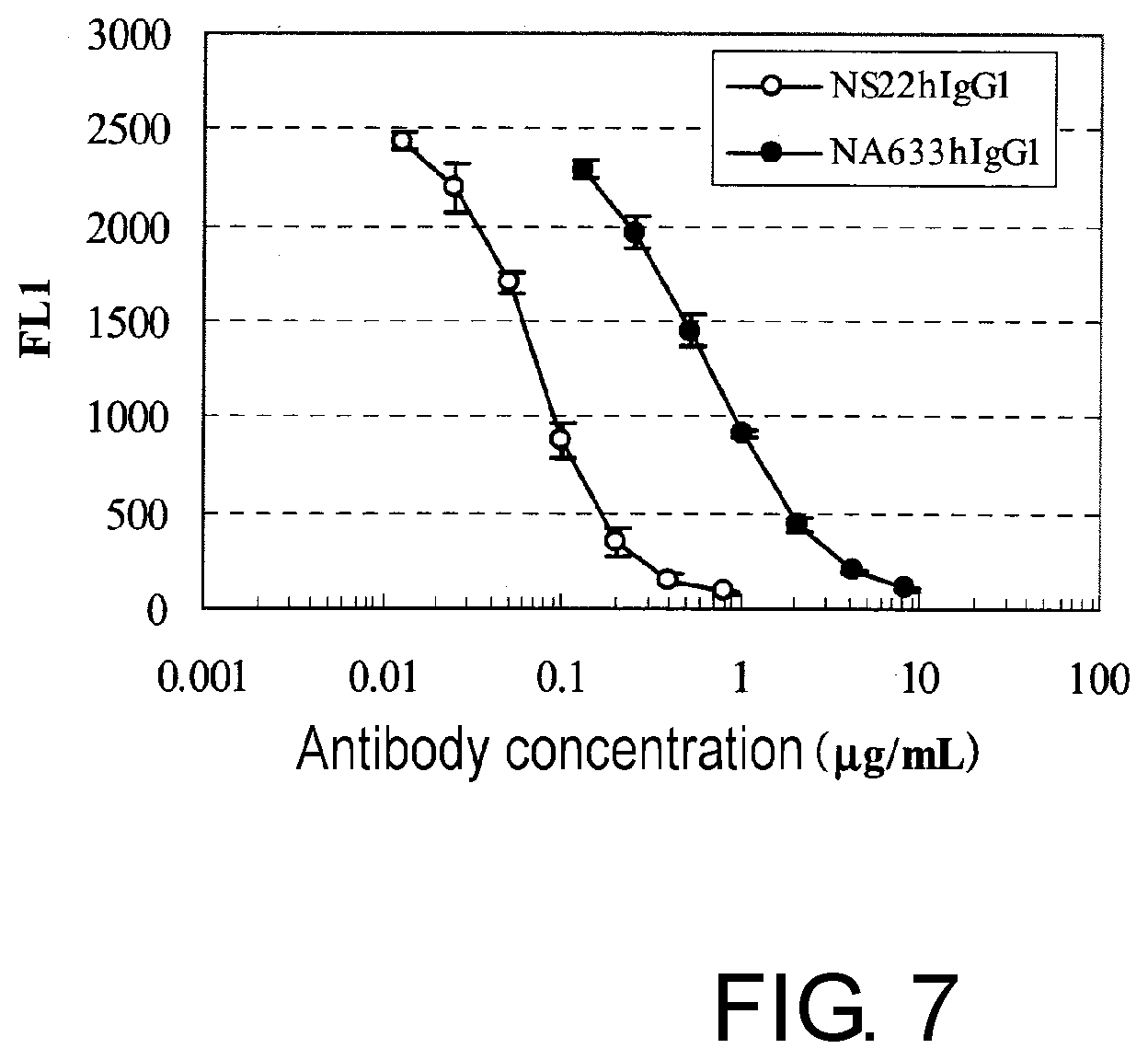
[0019] FIG. 7 is a graph showing the assessment of the competition of chimeric NS22 with IL-31.
[0020] FIG. 8 is a graph showing the NR10 competitive binding activity of anti-NR10 antibodies.
[0021] FIG. 9 is a set of graphs showing the assessment of the competition of humanized NS22 (H0L0) with IL-31.
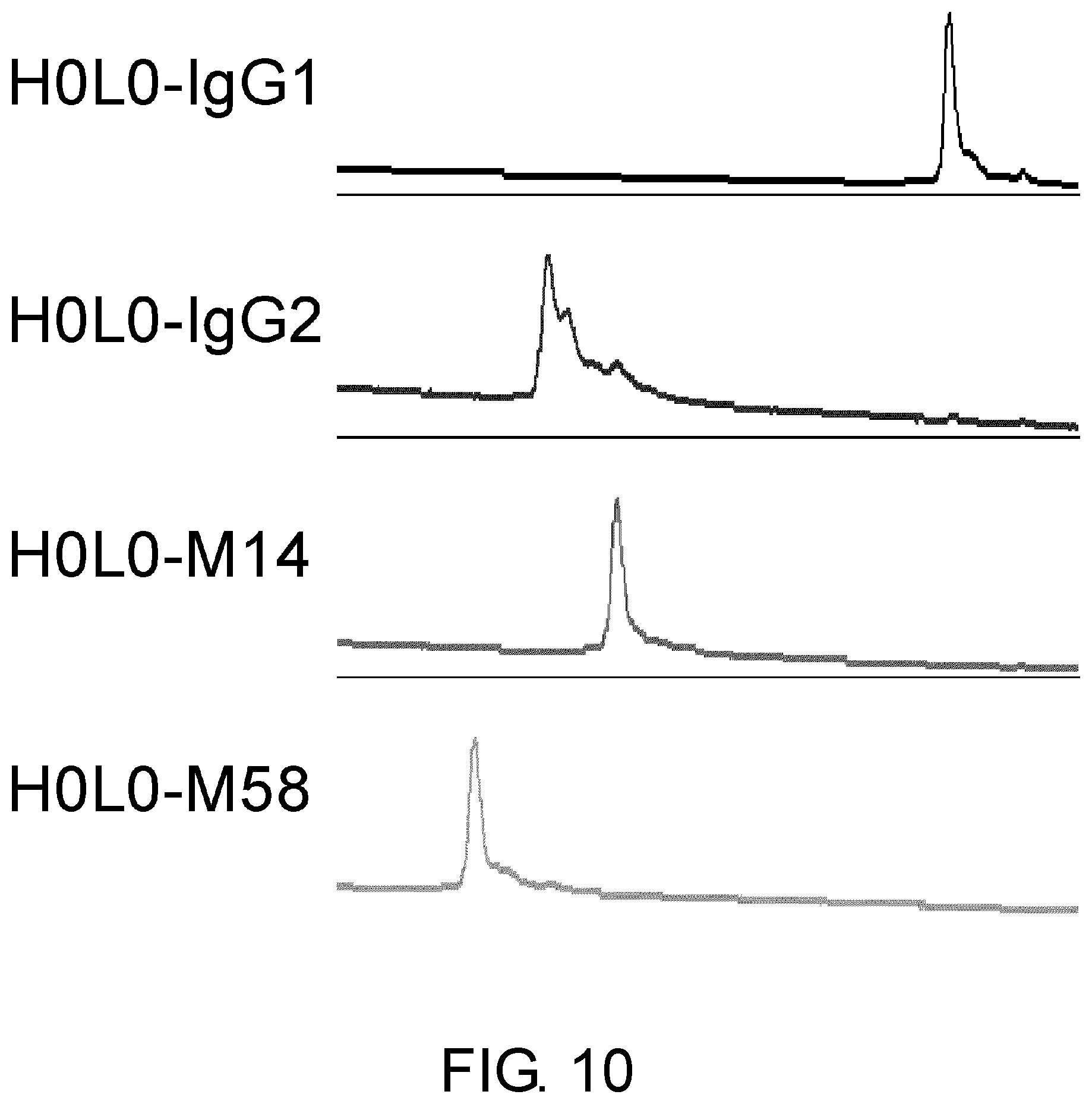
[0022] FIG. 10 shows the effect of the constant region of humanized anti-NR10 antibody H0L0 on the heterogeneity assessed by cation exchange chromatography.
[0023] FIG. 11 is a set of graphs showing the assessment of the competition of mutants of the humanized anti-NR10 antibody of which the isoelectric point of the variable regions is lowered without significant loss of the binding to NR10, with IL-31.
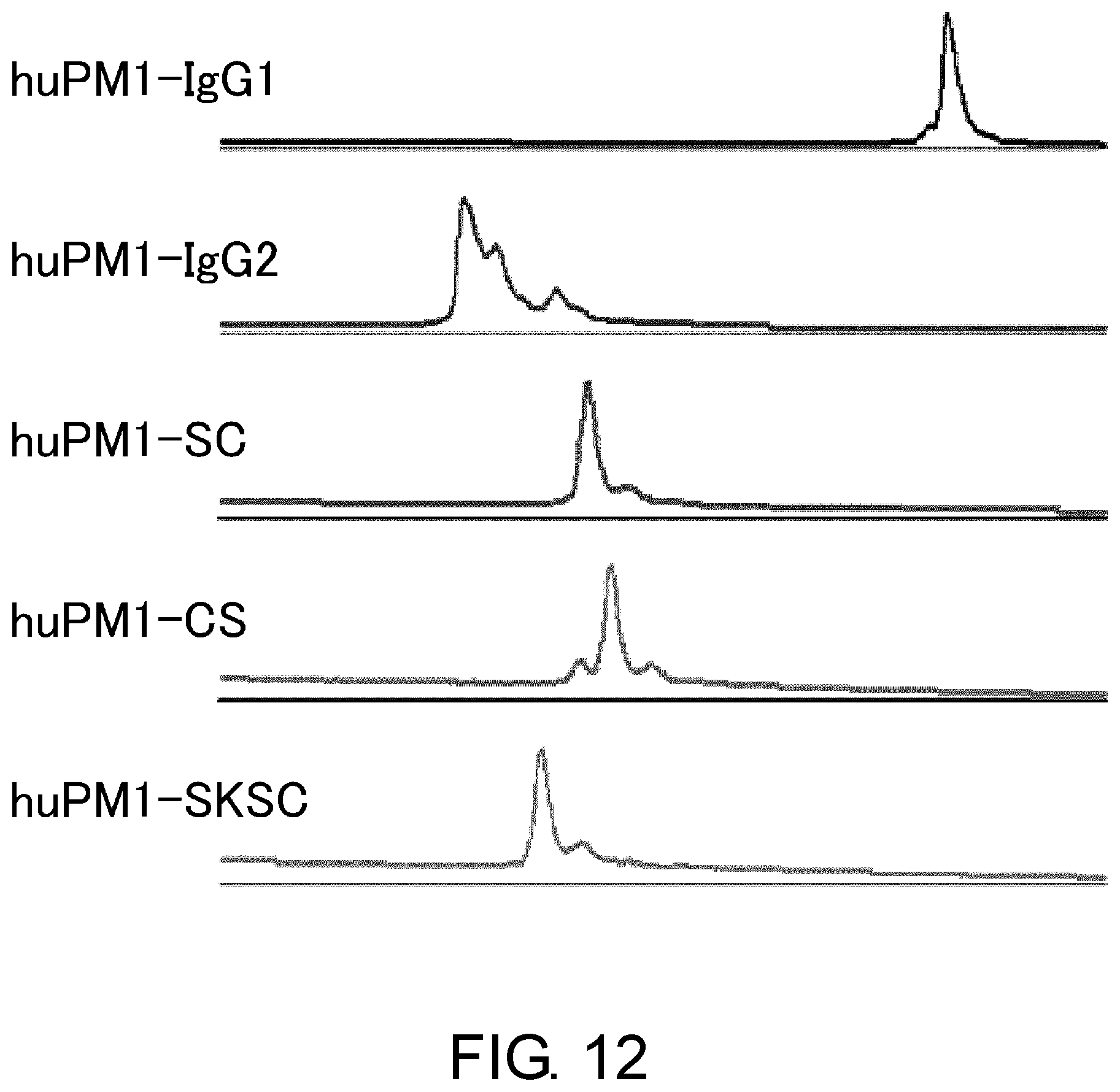
[0024] FIG. 12 shows the effect of the constant region of anti-IL-6 receptor antibody on the heterogeneity assessed by cation exchange chromatography.
[0025] FIG. 13 shows the effect of the constant region of anti-IL-6 receptor antibody on the denaturation peak assessed by DSC.
[0026] FIG. 14 shows the effect of the novel constant region M14 on the heterogeneity in an anti-IL-6 receptor antibody, assessed by cation exchange chromatography.
[0027] FIG. 15 shows the effect of the novel constant region M58 on the heterogeneity in an anti-IL-6 receptor antibody, assessed by cation exchange chromatography.
[0028] FIG. 16 shows the effect of the novel constant region M58 on the denaturation peak in an anti-IL-6 receptor antibody, assessed by DSC.
[0029] FIG. 17 shows the result of assaying the retention of huPM1-IgG1 and huPM1-M58 in the plasma of human FcRn transgenic mice.
[0030] FIG. 18 shows the biological activity of each antibody assessed using BaF/NR10.
[0031] FIG. 19 shows the analysis of thermally-accelerated (dotted line) and non-accelerated (solid line) samples of each modified antibody by cation exchange chromatography to compare the generation of degradation products between before and after thermal acceleration. Arrow indicates the peak position of basic component which was altered.
[0032] FIG. 20 is a set of graphs showing the assessment (BaF) of the activity of each variant.
[0033] FIG. 21 is a graph showing the assessment (BaF) of the activity of Ha401La402 and H0L0.
[0034] FIG. 22 is a graph showing the assessment (BaF) of the activity of H17L11 and H0L0.
[0035] FIG. 23 is a graph showing the assessment (BaF) of the activity of H19L12 and H0L0.
[0036] FIG. 24 is a graph showing the biological activity of H0L12 and H0L17 assessed using BaF/NR10.
[0037] FIG. 25-1 is a set of graphs showing the assessment (BaF) of the activity of each variant.
[0038] FIG. 25-2 is a continuation of FIG. 25-1.
[0039] FIG. 26 is a schematic diagram for human/mouse wild-type and chimeric NR10-ECD.
[0040] FIG. 27 is a set of photographs showing the detection of the binding domain by Western blotting. A is a photograph showing the result of detection using a humanized anti-human NR10 antibody; B is a photograph showing the result of detection using a mouse anti-human NR10 antibody; and C is a photograph showing the result of detection using an anti-Myc antibody. With the anti-human NR10 antibody a binding antigen was detected only in hhh, hhm, and hmm, but not in mmm, mmh, and mhm.
[0041] FIG. 28-1 shows the amino acid sequence of each variant of H0 (SEQ ID NO: 50).
[0042] FIG. 28-2 is a continuation of FIG. 28-1.
[0043] FIG. 28-3 is a continuation of FIG. 28-2.
[0044] FIG. 29-1 shows the amino acid sequence of each variant of L0 (SEQ ID NO: 52).
[0045] FIG. 29-2 is a continuation of FIG. 29-1.
MODE FOR CARRYING OUT THE INVENTION
NR10
[0046] NR10 is a protein that forms a heterodimer with oncostatin M receptor (OSMR) and functions as an IL-31 receptor. NR10 is also known as glm-r (J Biol Chem 277, 16831-6, 2002), GPL (J Biol Chem 278, 49850-9, 2003), IL31RA (Nat Immunol 5, 752-60, 2004), and such. Thus, NR10 in the present invention also includes proteins called by such names.
[0047] In the present invention, NR10 (also referred to as IL31RA, GPL, or glm-r) is not particularly limited in terms of its origin, and includes those derived from humans, mice, monkeys, and other mammals. NR10 derived from humans, mice, and monkeys is preferred, and human-derived NR10 is particularly preferred.
[0048] There are multiple known splicing variants of human-derived NR10 (WO 00/075314). Of the above-described splicing variants, NR10.1 consists of 662 amino acids and contains a transmembrane domain. NR10.2 is a soluble receptor-like protein consisting of 252 amino acids without the transmembrane domain. Meanwhile, known NR10 splicing variants that function as transmembrane receptor proteins include NR10.3 and IL-31RAv3. The human NR10 of the present invention is not particularly limited, as long as it forms a heterodimer with oncostatin M receptor (OSMR) and functions as an IL-31 receptor. Preferred NR10 includes NR10.3 (also referred to as ILRAv4 (Nat Immunol 5, 752-60, 2004)) and IL-31RAv3. NR 10.3 (IL31RAv4) consists of 662 amino acids (WO 00/075314; Nat Immunol 5, 752-60, 2004) and IL31RAv3 consists of 732 amino acids (GenBank Accession No: NM_139017). The amino acid sequence of IL31RAv4 is shown in SEQ ID NO: 79, and the amino acid sequence of IL31RAv3 is shown in SEQ ID NO: 80. Meanwhile, mouse-derived NR10 includes proteins comprising the amino acid sequence of SEQ ID NO: 81. In addition, cynomolgus monkey-derived NR10 includes proteins comprising the amino acid sequence of SEQ ID NO: 66.
Antibodies (Sequences)
[0049] Preferred embodiments of the anti-NR10 antibody of the present invention include the anti-NR10 antibodies of any one of (1) to (8) in (A) to (D) below.
(A)NS18
[0050] (1) antibodies having a heavy chain variable region that comprises CDR1 having the amino acid sequence of SEQ ID NO: 1 (HCDR1), CDR2 having the amino acid sequence of SEQ ID NO: 2 (HCDR2), and CDR3 having the amino acid sequence of SEQ ID NO: 3 (HCDR3); (2) antibodies having the heavy chain variable region of SEQ ID NO: 4 (VH); (3) antibodies having a light chain variable region that comprises CDR1 having the amino acid sequence of SEQ ID NO: 5 (LCDR1), CDR2 having the amino acid sequence of SEQ ID NO: 6 (LCDR2), and CDR3 having the amino acid sequence of SEQ ID NO: 7 (LCDR3); (4) antibodies having the light chain variable region of SEQ ID NO: 8 (VL); (5) antibodies having the heavy chain variable region of (1) and the light chain variable region of (3); (6) antibodies having the heavy chain variable region of (2) and the light chain variable region of (4); (7) antibodies in which one or more amino acids are substituted, deleted, added, and/or inserted in the antibodies of any one of (1) to (6), which have an activity equivalent to that of the antibodies of any one of (1) to (6); and (8) antibodies that bind to the same epitope as an epitope bound by the antibodies of any one of (1) to (7).
(B) NS22
[0051] (1) antibodies having a heavy chain variable region that comprises CDR1 having the amino acid sequence of SEQ ID NO: 9 (HCDR1), CDR2 having the amino acid sequence of SEQ ID NO: 10 (HCDR2), and CDR3 having the amino acid sequence of SEQ ID NO: 11 (HCDR3); (2) antibodies having the heavy chain variable region of SEQ ID NO: 12 (VH); (3) antibodies having a light chain variable region that comprises CDR1 having the amino acid sequence of SEQ ID NO: 13 (LCDR1), CDR2 having the amino acid sequence of SEQ ID NO: 14 (LCDR2), and CDR3 having the amino acid sequence of SEQ ID NO: 15 (LCDR3); (4) antibodies having the light chain variable region of SEQ ID NO: 16 (VL); (5) antibodies having the heavy chain variable region of (1) and the light chain variable region of (3); (6) antibodies having the heavy chain variable region of (2) and the light chain variable region of (4); (7) antibodies in which one or more amino acids are substituted, deleted, added, and/or inserted in the antibodies of any one of (1) to (6), which have an activity equivalent to that of the antibodies of any one of (1) to (6); and (8) antibodies that bind to the same epitope as an epitope bound by the antibodies of any one of (1) to (7).
[0052] Specific examples of the above-described substitution, deletion, addition, and/or insertion of one or more amino acids are not particularly limited and include, for example, the following modifications.
[0053] Substitution of Ile at position 3 in the heavy chain CDR1 of SEQ ID NO: 9 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Val.
[0054] Substitution of Met at position 4 in the heavy chain CDR1 of SEQ ID NO: 9 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Ile.
[0055] Substitution of Met at position 4 in the heavy chain CDR1 of SEQ ID NO: 9 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Leu.
[0056] Substitution of Ile at position 3 in the heavy chain CDR1 of SEQ ID NO: 9 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Ala.
[0057] Substitution of Leu at position 1 in the heavy chain CDR2 of SEQ ID NO: 10 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Glu.
[0058] Substitution of Asn at position 3 in the heavy chain CDR2 of SEQ ID NO: 10 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Asp.
[0059] Substitution of Gln at position 13 in the heavy chain CDR2 of SEQ ID NO: 10 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Asp.
[0060] Substitution of Lys at position 14 in the heavy chain CDR2 of SEQ ID NO: 10 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Gln.
[0061] Substitution of Lys at position 16 in the heavy chain CDR2 of SEQ ID NO: 10 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Gln.
[0062] Substitution of Gly at position 17 in the heavy chain CDR2 of SEQ ID NO: 10 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Asp.
[0063] Substitution of Lys and Gly at positions 16 and 17, respectively, in the heavy chain CDR2 of SEQ ID NO: 10 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples include substitution of Lys at position 16 with Gln, and Gly at position 17 with Asp.
[0064] Substitution of Lys, Lys, and Gly at positions 14, 16, and 17, respectively, in the heavy chain CDR2 of SEQ ID NO: 10 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples include substitution of Lys at position 14 with Gln, Lys at position 16 with Gln, and Gly at position 17 with Asp.
[0065] Substitution of Gln, Lys, Lys, and Gly at positions 13, 14, 16, and 17, respectively, in the heavy chain CDR2 of SEQ ID NO: 10 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples include substitution of Gln at position 13 with Asp, Lys at position 14 with Gln, Lys at position 16 with Gln, and Gly at position 17 with Asp.
[0066] Substitution of Ser at position 10 in the heavy chain CDR2 of SEQ ID NO: 10 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Asp.
[0067] Substitution of Gln at position 13 in the heavy chain CDR2 of SEQ ID NO: 10 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Pro.
[0068] Substitution of Tyr at position 3 in the heavy chain CDR3 of SEQ ID NO: 11 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Leu.
[0069] Substitution of Met at position 10 in the heavy chain CDR3 of SEQ ID NO: 11 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Leu.
[0070] Substitution of Asp at position 11 in the heavy chain CDR3 of SEQ ID NO: 11 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Glu.
[0071] Substitution of Tyr at position 12 in the heavy chain CDR3 of SEQ ID NO: 11 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Thr and Ser.
[0072] Substitution of Met, Asp, and Tyr at positions 10, 11, and 12, respectively, in the heavy chain CDR3 of SEQ ID NO: 11 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples include substitution of Met at position 10 with Leu, Asp at position 11 with Glu, and Tyr at position 12 with Thr.
[0073] Substitution of Asp and Tyr at positions 11 and 12, respectively, in the heavy chain CDR3 of SEQ ID NO: 11 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples include substitution of Asp at position 11 with Glu, and Tyr at position 12 with Thr.
[0074] Substitution of Tyr, Asp, and Tyr at positions 3, 11, and 12, respectively, in the heavy chain CDR3 of SEQ ID NO: 11 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples include substitution of Tyr at position 3 with Leu, Asp at position 11 with Glu, and Tyr at position 12 with Thr or Ser.
[0075] Substitution of Arg at position 1 in the light chain CDR1 of SEQ ID NO: 13 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Gln.
[0076] Substitution of Asn at position 5 in the light chain CDR1 of SEQ ID NO: 13 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Asp.
[0077] Substitution of Arg and Asn at positions 1 and 5, respectively, in the light chain CDR1 of SEQ ID NO: 13 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples include substitution of Arg at position 1 with Gln, and Asn at position 5 with Asp.
[0078] Substitution of Ser at position 8 in the light chain CDR1 of SEQ ID NO: 13 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Arg.
[0079] Substitution of Leu at position 10 in the light chain CDR1 of SEQ ID NO: 13 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Val.
[0080] Substitution of Ser and Leu at positions 8 and 10, respectively, in the light chain CDR1 of SEQ ID NO: 13 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples include substitution of Ser at position 8 with Arg, and Leu at position 10 with Val.
[0081] Substitution of Thr at position 2 in the light chain CDR1 of SEQ ID NO: 13 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Ala and Ser.
[0082] Substitution of Asn at position 1 in the light chain CDR2 of SEQ ID NO: 14 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Asp.
[0083] Substitution of Lys at position 3 in the light chain CDR2 of SEQ ID NO: 14 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Gln.
[0084] Substitution of Leu at position 5 in the light chain CDR2 of SEQ ID NO: 14 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Glu.
[0085] Substitution of Lys at position 7 in the light chain CDR2 of SEQ ID NO: 14 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Gln and Asp.
[0086] Substitution of Lys, Leu, and Lys at positions 3, 5, and 7, respectively, in the light chain CDR2 of SEQ ID NO: 14 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples include substitution of Lys at position 3 with Gln, Leu at position 5 with Glu, and Lys at position 7 with Gln.
[0087] Substitution of Glu at position 5 in the light chain CDR3 of SEQ ID NO: 15 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Asp.
[0088] Substitution of Ser at position 6 in the light chain CDR3 of SEQ ID NO: 15 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Asp.
[0089] Substitution of Thr at position 9 in the light chain CDR3 of SEQ ID NO: 15 with another amino acid. The amino acid after substitution is not particularly limited but preferred examples thereof include Phe.
[0090] Each of the above-described substitutions may be made alone, or multiple substitutions may be made in combination. Furthermore, the above substitutions may be combined with other substitutions. These substitutions can improve the antibody pharmacokinetics (retention in plasma), enhance the antigen-binding activity, improve the stability, and/or reduce the risk of immunogenicity.
[0091] In the present invention, specific examples of the variable regions having a combination of the above-described substitutions include, for example, heavy chain variable regions having the amino acid sequence of SEQ ID NO: 167 and light chain variable regions having the amino acid sequence of SEQ ID NO: 168. Moreover, examples of the antibodies having a combination of the above-described substitutions include, for example, antibodies that comprise a heavy chain variable region having the amino acid sequence of SEQ ID NO: 167 and a light chain variable region having the amino acid sequence of SEQ ID NO: 168.
[0092] Moreover, specific examples of the heavy chain or light chain variable regions having a combination of the above-described substitutions include, for example, the following variable regions:
(a) heavy chain variable regions that comprise CDR1 of SEQ ID NO: 196, CDR2 of SEQ ID NO: 197, and CDR3 of SEQ ID NO: 11 (H17); (b) heavy chain variable regions that comprise CDR1 of SEQ ID NO: 176, CDR2 of SEQ ID NO: 197, and CDR3 of SEQ ID NO: 11 (H19); (c) heavy chain variable regions that comprise CDR1 of SEQ ID NO: 196, CDR2 of SEQ ID NO: 197, and CDR3 of SEQ ID NO: 184 (H28, H42); (d) heavy chain variable regions that comprises CDR1 of SEQ ID NO: 9, CDR2 of SEQ ID NO: 197, and CDR3 of SEQ ID NO: 184 (H30, H44); (e) heavy chain variable regions that comprise CDR1 of SEQ ID NO: 176, CDR2 of SEQ ID NO: 197, and CDR3 of SEQ ID NO: 184 (H34, H46); (f) heavy chain variable regions that comprise CDR1 of SEQ ID NO: 9, CDR2 of SEQ ID NO: 198, and CDR3 of SEQ ID NO: 184 (H57, H78); (g) heavy chain variable regions that comprise CDR1 of SEQ ID NO: 176, CDR2 of SEQ ID NO: 198, and CDR3 of SEQ ID NO: 184 (H71, H92); (h) heavy chain variable regions that comprise CDR1 of SEQ ID NO: 9, CDR2 of SEQ ID NO: 199, and CDR3 of SEQ ID NO: 184 (H97, H98); (i) light chain variable regions that comprise CDR1 of SEQ ID NO: 200, CDR2 of SEQ ID NO: 170, and CDR3 of SEQ ID NO: 193 (L11); (j) light chain variable regions that comprise CDR1 of SEQ ID NO: 201, CDR2 of SEQ ID NO: 170, and CDR3 of SEQ ID NO: 193 (L12); (k) light chain variable regions that comprise CDR1 of SEQ ID NO: 202, CDR2 of SEQ ID NO: 170, and CDR3 of SEQ ID NO: 193 (L17); and (l) light chain variable regions that comprise CDR1 of SEQ ID NO: 203, CDR2 of SEQ ID NO: 170, and CDR3 of SEQ ID NO: 193 (L50).
[0093] Furthermore, specific examples of the antibodies having a combination of the above-described substitutions include, for example:
(i) antibodies that comprise the heavy chain variable region of (c) and the light chain variable region of (k); (ii) antibodies that comprise the heavy chain variable region of (d) and the light chain variable region of (k); (iii) antibodies that comprise the heavy chain variable region of (e) and the light chain variable region of (k); (iv) antibodies that comprise the heavy chain variable region of (f) and the light chain variable region of (k); (v) antibodies that comprise the heavy chain variable region of (g) and the light chain variable region of (k); and (vi) antibodies that comprise the heavy chain variable region of (h) and the light chain variable region of (1).
(C) NS23
[0094] (1) antibodies having a heavy chain variable region that comprises CDR1 having the amino acid sequence of SEQ ID NO: 17 (HCDR1), CDR2 having the amino acid sequence of SEQ ID NO: 18 (HCDR2), and CDR3 having the amino acid sequence of SEQ ID NO: 19 (HCDR3); (2) antibodies having the heavy chain variable region of SEQ ID NO: 20 (VH); (3) antibodies having a light chain variable region that comprises CDR1 having the amino acid sequence of SEQ ID NO: 21 (LCDR1), CDR2 having the amino acid sequence of SEQ ID NO: 22 (LCDR2), and CDR3 having the amino acid sequence of SEQ ID NO: 23 (LCDR3); (4) antibodies having the light chain variable region of SEQ ID NO: 24 (VL); (5) antibodies having the heavy chain variable region of (1) and the light chain variable region of (3); (6) antibodies having the heavy chain variable region of (2) and the light chain variable region of (4); (7) antibodies in which one or more amino acids are substituted, deleted, added, and/or inserted in the antibodies of any one of (1) to (6), which have an activity equivalent to that of the antibodies of any one of (1) to (6); and (8) antibodies that bind to the same epitope as an epitope bound by the antibodies of any one of (1) to (7).
(D) NS33
[0095] (1) antibodies having a heavy chain variable region that comprise CDR1 having the amino acid sequence of SEQ ID NO: 25 (HCDR1), CDR2 having the amino acid sequence of SEQ ID NO: 26 (HCDR2), and CDR3 having the amino acid sequence of SEQ ID NO: 27 (HCDR3); (2) antibodies having the heavy chain variable region of SEQ ID NO: 28 (VH); (3) antibodies having a light chain variable region that comprise CDR1 having the amino acid sequence of SEQ ID NO: 29 (LCDR1), CDR2 having the amino acid sequence of SEQ ID NO: 30 (LCDR2), and CDR3 having the amino acid sequence of SEQ ID NO: 31 (LCDR3); (4) antibodies having the light chain variable region of SEQ ID NO: 32 (VL); (5) antibodies having the heavy chain variable region of (1) and the light chain variable region of (3); (6) antibodies having the heavy chain variable region of (2) and the light chain variable region of (4); (7) antibodies in which one or more amino acids are substituted, deleted, added, and/or inserted in the antibodies of any one of (1) to (6), which have an activity equivalent to that of the antibodies of any one of (1) to (6); and (8) antibodies that bind to the same epitope as an epitope bound by the antibodies of any one of (1) to (7).
[0096] Any framework regions (FR) may be used for the above-described antibodies of (1) or (3); however, FRs derived from human are preferably used. Furthermore, any constant regions may be used for the above-described antibodies of (1) to (8); however, constant regions derived from human are preferably used. For the antibodies of the present invention, the amino acid sequence of the original FR or constant region may be used without modification, or after being modified to a different amino acid sequence by substitution, deletion, addition, and/or insertion of one or more amino acids.
[0097] The amino acid sequence of the heavy chain of the above-described NS18 is shown in SEQ ID NO: 34 and the nucleotide sequence encoding this amino acid sequence is shown in SEQ ID NO: 33. Meanwhile, the amino acid sequence of the light chain is shown in SEQ ID NO: 36 and the nucleotide sequence encoding this amino acid sequence is shown in SEQ ID NO: 35.
[0098] The amino acid sequence of the heavy chain of NS22 is shown in SEQ ID NO: 38 and the nucleotide sequence encoding this amino acid sequence is shown in SEQ ID NO: 37. Meanwhile, the amino acid sequence of the light chain is shown in SEQ ID NO: 40 and the nucleotide sequence encoding this amino acid sequence is shown in SEQ ID NO: 39.
[0099] The amino acid sequence of the heavy chain of NS23 is shown in SEQ ID NO: 42 and the nucleotide sequence encoding this amino acid sequence is shown in SEQ ID NO: 41. Meanwhile, the amino acid sequence of the light chain is shown in SEQ ID NO: 44 and the nucleotide sequence encoding this amino acid sequence is shown in SEQ ID NO: 43.
[0100] The amino acid sequence of the heavy chain of NS33 is shown in SEQ ID NO: 46 and the nucleotide sequence encoding this amino acid sequence is shown in SEQ ID NO: 45. Meanwhile, the amino acid sequence of the light chain is shown in SEQ ID NO: 48 and the nucleotide sequence encoding this amino acid sequence is shown in SEQ ID NO: 47.
[0101] In the present invention, the "activity equivalent to that of the antibody of any one of (1) to (6)" means that the activity of binding and/or neutralizing NR10 (for example, human NR10) is equivalent. In the present invention, the term "equivalent" means that the activity is not necessarily the same but may be enhanced or reduced as long as the activity is retained. Antibodies with a reduced activity include, for example, antibodies having an activity that is 30% or more, preferably 50% or more, and more preferably 80% or more of that of the original antibody.
[0102] The antibodies of any one of (1) to (6) mentioned above may have a substitution, deletion, addition, and/or insertion of one or more amino acids in the amino acid sequence of the variable regions (CDR sequences and/or FR sequences), as long as the NR10-binding and/or neutralizing activity is retained. Methods well known to those skilled in the art to prepare the amino acid sequence of an antibody that has a substitution, deletion, addition, and/or insertion of one or more amino acids in the amino acid sequence and retains NR10-binding and/or neutralizing activity, include methods for introducing mutations into proteins. For example, those skilled in the art can prepare mutants functionally equivalent to the antibody having NR10-binding and/or neutralizing activity by introducing appropriate mutations into the amino acid sequence of the antibody having NR10-binding and/or neutralizing activity using site-directed mutagenesis (Hashimoto-Gotoh, T, Mizuno, T, Ogasahara, Y, and Nakagawa, M. (1995) An oligodeoxyribonucleotide-directed dual amber method for site-directed mutagenesis. Gene 152, 271-275, Zoller, M J, and Smith, M. (1983) Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 100, 468-500, Kramer, W, Drutsa, V, Jansen, H W, Kramer, B, Pflugfelder, M, and Fritz, H J (1984) The gapped duplex DNA approach to oligonucleotide-directed mutation construction. Nucleic Acids Res. 12, 9441-9456, Kramer W, and Fritz H J (1987) Oligonucleotide-directed construction of mutations via gapped duplex DNA Methods. Enzymol. 154, 350-367, Kunkel, TA (1985) Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 82, 488-492) or the like. Thus, antibodies that contain one or more amino acid mutations in the variable regions and have NR10-binding and/or neutralizing activity are also included in the antibody of the present invention.
[0103] When an amino acid residue is altered, the amino acid is preferably mutated for a different amino acid(s) that conserves the properties of the amino acid side-chain. Examples of amino acid side chain properties are: hydrophobic amino acids (A, I, L, M, F, P, W, Y, and V), hydrophilic amino acids (R, D, N, C, E, Q, G, H, K, S, and T), amino acids containing aliphatic side chains (G, A, V, L, I, and P), amino acids containing hydroxyl group-containing side chains (S, T, and Y), amino acids containing sulfur-containing side chains (C and M), amino acids containing carboxylic acid- and amide-containing side chains (D, N, E, and Q), amino acids containing basic side chains (R, K, and H), and amino acids containing aromatic side chains (H, F, Y, and W) (amino acids are represented by one-letter codes in parentheses). Amino acid substitutions within each group are called conservative substitutions. It is already known that a polypeptide containing a modified amino acid sequence in which one or more amino acid residues in a given amino acid sequence are deleted, added, and/or substituted with other amino acids can retain the original biological activity (Mark, D. F. et al., Proc. Natl. Acad. Sci. USA; (1984) 81:5662-6; Zoller, M. J. and Smith, M., Nucleic Acids Res. (1982) 10:6487-500; Wang, A. et al., Science (1984) 224:1431-3; Dalbadie-McFarland, G. et al., Proc. Natl. Acad. Sci. USA (1982) 79:6409-13). Such mutants have an amino acid identity of at least 70%, more preferably at least 75%, even more preferably at least 80%, still more preferably at least 85%, yet more preferably at least 90%, and most preferably at least 95%, with the variable regions (for example, CDR sequences, FR sequences, or whole variable regions) of the present invention. Herein, sequence identity is defined as the percentage of residues identical to those in the original amino acid sequence of the heavy chain variable region or light chain variable region, determined after the sequences are aligned and gaps are appropriately introduced to maximize the sequence identity as necessary. The identity of amino acid sequences can be determined by the method described below.
[0104] Alternatively, the amino acid sequences of variable regions that have a substitution, deletion, addition, and/or insertion of one or more amino acids in the amino acid sequence of the variable regions (CDR sequences and/or FR sequences) and retain NR10-binding and/or neutralizing activity can be obtained from nucleic acids that hybridize under stringent conditions to nucleic acid composed of the nucleotide sequence encoding the amino acid sequence of the variable regions. Stringent hybridization conditions to isolate a nucleic acid that hybridizes under stringent conditions to a nucleic acid that includes the nucleotide sequence encoding the amino acid sequence of the variable regions include, for example, the conditions of 6M urea, 0.4% SDS, 0.5.times.SSC, and 37.degree. C., or hybridization conditions with stringencies equivalent thereto. With more stringent conditions, for example, the conditions of 6M urea, 0.4% SDS, 0.1.times.SSC, and 42.degree. C., isolation of nucleic acids with a much higher homology can be expected. The sequences of the isolated nucleic acids can be determined by the known methods described below. The overall nucleotide sequence homology of the isolated nucleic acid is at least 50% or higher sequence identity, preferably 70% or higher, more preferably 90% or higher (for example, 95%, 96%, 97%, 98%, 99%, or higher).
[0105] Nucleic acids that hybridize under stringent conditions to a nucleic acid composed of the nucleotide sequence encoding the amino acid sequence of the variable regions can also be isolated using, instead of the above-described methods using hybridization techniques, gene amplification methods such as polymerase chain reaction (PCR) using primers synthesized based on the information of nucleotide sequence encoding the amino acid sequence of the variable regions.
[0106] Specifically, the identity of one nucleotide sequence or amino acid sequence to another can be determined using the algorithm BLAST, by Karlin and Altschul (Proc. Natl. Acad. Sci. USA (1993) 90, 5873-7). Programs such as BLASTN and BLASTX were developed based on this algorithm (Altschul et al., J. Mol. Biol. (1990) 215, 403-10). To analyze nucleotide sequences according to BLASTN based on BLAST, the parameters are set, for example, as score=100 and wordlength=12. On the other hand, parameters used for the analysis of amino acid sequences by BLASTX based on BLAST include, for example, score=50 and wordlength=3. Default parameters for each program are used when using the BLAST and Gapped BLAST programs. Specific techniques for such analyses are known in the art (see the website of the National Center for Biotechnology Information (NCBI), Basic Local Alignment Search Tool (BLAST); http://www.ncbi.nlm.nih.gov).
[0107] The present invention also provides antibodies that bind to the same epitope as an epitope bound by the antibodies of any one of (1) to (7).
[0108] Whether an antibody recognizes the same epitope as that recognized by another antibody can be confirmed by the competition between the two antibodies against the epitope. Competition between the antibodies can be evaluated by competitive binding assays using means such as ELISA, fluorescence energy transfer method (FRET), and fluorometric microvolume assay technology (FMAT.RTM.). The amount of antibodies bound to an antigen indirectly correlate with the binding ability of candidate competitor antibodies (test antibodies) that competitively bind to the same epitope. In other words, as the amount of or the affinity of test antibodies against the same epitope increases, the amount of antibodies bound to the antigen decreases, and the amount of test antibodies bound to the antigen increases. Specifically, appropriately labeled antibodies and antibodies to be evaluated are simultaneously added to the antigens, and the thus bound antibodies are detected using the label. The amount of antibodies bound to the antigen can be easily determined by labeling the antibodies beforehand. This label is not particularly limited, and the labeling method is selected according to the assay technique used. The labeling method includes fluorescent labeling, radiolabeling, enzymatic labeling, and such.
[0109] For example, fluorescently labeled antibodies and unlabeled antibodies or test antibodies are simultaneously added to animal cells expressing NR10, and the labeled antibodies are detected by fluorometric microvolume assay technology.
[0110] Herein, the "antibody that recognizes the same epitope" refers to an antibody that can reduce the binding of the labeled antibody by at least 50% at a concentration that is usually 100 times higher, preferably 80 times higher, more preferably 50 times higher, even more preferably 30 times higher, and still more preferably 10 times higher than a concentration at which the non-labeled antibody reduces the binding of the labeled antibody by 50% (IC.sub.50).
[0111] Antibodies that bind to the epitope to which the antibodies set forth in any one of (1) to (7) above bind are useful because they have a particularly high neutralizing activity.
[0112] The antibodies set forth in any one of (1) to (8) above are preferably humanized antibodies, but are not particularly limited thereto.
[0113] Furthermore, the present invention provides genes encoding the anti-NR10 antibodies of any one of (1) to (8) of (A) to (D) above. The genes of the present invention may be any form of genes, for example, DNAs or RNAs.
Antibodies (Humanized)
[0114] Preferred embodiments of the antibodies of the present invention include humanized antibodies that bind to NR10. The humanized antibodies can be prepared by methods known to those skilled in the art.
[0115] The variable region of antibody is typically composed of three complementarity-determining regions (CDRs) sandwiched by four frames (FRs). The CDRs substantially determine the binding specificity of antibody. The amino acid sequences of CDRs are highly diverse. In contrast, the amino acid sequences of FRs often exhibit high homology between antibodies having different binding specificities. It is therefore said in general that the binding specificity of an antibody can be transplanted to a different antibody by grafting the CDRs.
[0116] Humanized antibodies are also referred to as reshaped human antibodies, and they are prepared by transferring the CDRs of an antibody derived from a non-human mammal such as a mouse, to the CDRs of a human antibody. General genetic recombination techniques for their preparation are also known (see European Patent Application Publication No. 125023 and WO 96/02576).
[0117] Specifically, for example, when the CDRs are derived from a mouse antibody, a DNA sequence designed such that the CDRs of the mouse antibody are linked with framework regions (FRs) of human antibody is synthesized by PCR using, as primers, several oligonucleotides that have portions overlapping the ends of both CDRs and FRs (see the method described in WO 98/13388). The resulting DNA is then ligated to a DNA encoding a human antibody constant region, inserted into an expression vector, and introduced into a host to produce the antibody (see European Patent Application Publication No. EP 239400 and International Patent Application Publication No. WO 96/02576).
[0118] Human antibody framework regions to be linked with CDRs are selected so that the CDRs form a favorable antigen-binding site. If needed, amino acid substitution, deletion, addition, and/or insertion may be introduced into the framework regions of antibody variable region so that the CDRs of the reshaped human antibody form a proper antigen-binding site. For example, mutations can be introduced into the amino acid sequence of FR by applying the PCR method which is used to graft mouse CDRs to human FRs. Specifically, mutations can be introduced into a portion of the nucleotide sequences of primers that anneal to the FRs. The mutations are introduced into FRs synthesized by such primers. The antigen-binding activity of mutant antibodies having amino acid substitutions can be determined and assessed by the method described above, and thereby mutant FR sequences having desired properties can be selected (Sato, K. et al., Cancer Res. (1993) 53, 851-856).
[0119] Constant (C) regions from human antibodies are used for those of humanized antibodies. For example, C.gamma.1, C.gamma.2, C.gamma.3, C.gamma.4, C.mu., C.delta., C.alpha.1, C.alpha.2, and C.epsilon. are used for H chains; and C.kappa. and C.lamda. are used for L chains. The amino acid sequence of C.kappa. is shown in SEQ ID NO: 58, and the nucleotide sequence encoding this amino acid sequence is shown in SEQ ID NO: 57. The amino acid sequence of C.gamma.1 is shown in SEQ ID NO: 60, and the nucleotide sequence encoding this amino acid sequence is shown in SEQ ID NO: 59. The amino acid sequence of C.gamma.2 is shown in SEQ ID NO: 62, and the nucleotide sequence encoding this amino acid sequence is shown in SEQ ID NO: 61. The amino acid sequence of C.gamma.4 is shown in SEQ ID NO: 64, and the nucleotide sequence encoding this amino acid sequence is shown in SEQ ID NO: 63. Furthermore, human antibody C regions may be modified to improve the stability of antibody or antibody production. Modified human antibody C regions include, for example, the C regions described herein below. Human antibodies used for humanization may be of any isotype such as IgG, IgM, IgA, IgE, or IgD; however, IgG is preferably used in the present invention. IgG that can be used includes IgG1, IgG2, IgG3, IgG4, and the like.
[0120] Moreover, after a humanized antibody is prepared, amino acids in the variable region (for example, CDR and FR) and constant region of the humanized antibody may be deleted, added, inserted, and/or substituted with other amino acids. The antibodies of the present invention also include such humanized antibodies with amino acid substitutions and the like.
[0121] The origin of CDRs of a humanized antibody is not particularly limited, and may be any animal. For example, it is possible to use the sequences of mouse antibodies, rat antibodies, rabbit antibodies, camel antibodies, and the like. CDR sequences of mouse antibodies are preferred.
[0122] In general, it is difficult to humanize antibodies while retaining the binding and neutralizing activities of the original antibodies. The present invention, however, succeeded in obtaining humanized antibodies having the binding and/or neutralizing activities equivalent to those of the original mouse antibodies. Humanized antibodies are useful when administered to humans for the therapeutic purposes, because they exhibit reduced immunogenicity in the human body.
[0123] Preferred examples of the humanized anti-NR10 antibodies of the present invention include, for example:
(a) humanized antibodies that comprise a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 (H0-VH); (b) humanized antibodies that comprise a heavy chain variable region having the amino acid sequence of SEQ ID NO: 112 (H1-VH); (c) humanized antibodies that comprise a light chain variable region having the amino acid sequence of SEQ ID NO: 52 (L0-VL); (d) humanized antibodies that comprise a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 (H0-VH) and a light chain variable region having the amino acid sequence of SEQ ID NO: 52 (L0-VL); and (e) humanized antibodies that comprise a heavy chain variable region having the amino acid sequence of SEQ ID NO: 112 and a light chain variable region having the amino acid sequence of SEQ ID NO: 52.
[0124] The heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 (H0-VH), heavy chain variable region having the amino acid sequence of SEQ ID NO: 112, and light chain variable region having the amino acid sequence of SEQ ID NO: 52 (L0-VL) may have a substitution, deletion, addition, and/or insertion of one or more amino acids. The substitution, deletion, addition, and/or insertion of amino acids may be made in either or both of the CDRs and FRs.
[0125] Thus, other preferred embodiments of the humanized anti-NR10 antibody of the present invention include, for example:
(f) antibodies that comprise a heavy chain variable region having an amino acid sequence in which one or more amino acids are substituted, deleted, added, and/or inserted in the amino acid sequence of SEQ ID NO: 50 (H0-VH); (g) antibodies that comprise a heavy chain variable region having an amino acid sequence in which one or more amino acids are substituted, deleted, added, and/or inserted in the amino acid sequence of SEQ ID NO: 112 (H1-VH); (h) antibodies that comprise a light chain variable region having an amino acid sequence in which one or more amino acids are substituted, deleted, added, and/or inserted in the amino acid sequence of SEQ ID NO: 52 (L0-VL); (i) antibodies that comprise a heavy chain variable region having an amino acid sequence in which one or more amino acids are substituted, deleted, added, and/or inserted in the amino acid sequence of SEQ ID NO: 50 (H0-VH), and a light chain variable region having an amino acid sequence in which one or more amino acids are substituted, deleted, added, and/or inserted in the amino acid sequence of SEQ ID NO: 52 (L0-VL); (j) antibodies that comprise a heavy chain variable region having an amino acid sequence in which one or more amino acids are substituted, deleted, added, and/or inserted in the amino acid sequence of SEQ ID NO: 112 (H1-VH), and a light chain variable region having an amino acid sequence in which one or more amino acids are substituted, deleted, added, and/or inserted in the amino acid sequence of SEQ ID NO: 52 (L0-VL);
[0126] Without particular limitation, the antibodies of any one of (f) to (j) preferably have an activity similar to that of the antibodies of any one of (a) to (e).
[0127] The substitution, deletion, addition, and/or insertion of amino acids are not particularly limited, but specific examples include, for example, the above-described amino acid substitutions.
[0128] More specifically, for example, the following amino acid substitutions may be included:
[0129] Substitution of Ile at position 3 of CDR1 (SEQ ID NO: 9) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Val (SEQ ID NO: 173). Thus, the present invention provides heavy chain variable regions in which CDR1 having the amino acid sequence of SEQ ID NO: 9 is substituted with CDR1 having the amino acid sequence of SEQ ID NO: 173 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0130] Substitution of Met at position 4 of CDR1 (SEQ ID NO: 9) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Ile (SEQ ID NO: 174). Thus, the present invention provides heavy chain variable regions in which CDR1 having the amino acid sequence of SEQ ID NO: 9 is substituted with CDR1 having the amino acid sequence of SEQ ID NO: 174 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0131] Substitution of Met at position 4 of CDR1 (SEQ ID NO: 9) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Leu (SEQ ID NO: 175). Thus, the present invention provides heavy chain variable regions in which CDR1 having the amino acid sequence of SEQ ID NO: 9 is substituted with CDR1 having the amino acid sequence of SEQ ID NO: 175 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0132] Substitution of Ile at position 3 of CDR1 (SEQ ID NO: 9) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Ala (SEQ ID NO: 176). Thus, the present invention provides heavy chain variable regions in which CDR1 having the amino acid sequence of SEQ ID NO: 9 is substituted with CDR1 having the amino acid sequence of SEQ ID NO: 176 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0133] Substitution of Leu at position 1 of CDR2 (SEQ ID NO: 10) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Glu (SEQ ID NO: 113). Thus, the present invention provides heavy chain variable regions in which CDR2 having the amino acid sequence of SEQ ID NO: 10 is substituted with CDR2 having the amino acid sequence of SEQ ID NO: 113 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0134] Substitution of Asn at position 3 of CDR2 (SEQ ID NO: 10) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Asp (SEQ ID NO: 114). Thus, the present invention provides heavy chain variable regions in which CDR2 having the amino acid sequence of SEQ ID NO: 10 is substituted with CDR2 having the amino acid sequence of SEQ ID NO: 114 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0135] Substitution of Gln at position 13 of CDR2 (SEQ ID NO: 10) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Asp (SEQ ID NO: 115). Thus, the present invention provides heavy chain variable regions in which CDR2 having the amino acid sequence of SEQ ID NO: 10 is substituted with CDR2 having the amino acid sequence of SEQ ID NO: 115 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0136] Substitution of Lys at position 14 of CDR2 (SEQ ID NO: 10) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Gln (SEQ ID NO: 116). Thus, the present invention provides heavy chain variable regions in which CDR2 having the amino acid sequence of SEQ ID NO: 10 is substituted with CDR2 having the amino acid sequence of SEQ ID NO: 116 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0137] Substitution of Lys at position 16 of CDR2 (SEQ ID NO: 10) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Gln (SEQ ID NO: 117). Thus, the present invention provides heavy chain variable regions in which CDR2 having the amino acid sequence of SEQ ID NO: 10 is substituted with CDR2 having the amino acid sequence of SEQ ID NO: 117 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0138] Substitution of Gly at position 17 of CDR2 (SEQ ID NO: 10) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Asp (SEQ ID NO: 118). Thus, the present invention provides heavy chain variable regions in which CDR2 having the amino acid sequence of SEQ ID NO: 10 is substituted with CDR2 having the amino acid sequence of SEQ ID NO: 118 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0139] Substitution of Lys at position 16 and Gly at position 17 of CDR2 (SEQ ID NO: 10) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Gln and Asp, respectively (SEQ ID NO: 119). Thus, the present invention provides heavy chain variable regions in which CDR2 having the amino acid sequence of SEQ ID NO: 10 is substituted with CDR2 having the amino acid sequence of SEQ ID NO: 119 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0140] Substitution of Lys at position 14, Lys at position 16, and Gly at position 17 of CDR2 (SEQ ID NO: 10) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Gln, Gln, and Asp, respectively (SEQ ID NO: 167). Thus, the present invention provides heavy chain variable regions in which CDR2 having the amino acid sequence of SEQ ID NO: 10 is substituted with CDR2 having the amino acid sequence of SEQ ID NO: 171 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0141] Substitution of Gln at position 13, Lys at position 14, Lys at position 16, and Gly at position 17 of CDR2 (SEQ ID NO: 10) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Asp, Gln, Gln, and Asp, respectively (SEQ ID NO: 172). Thus, the present invention provides heavy chain variable regions in which CDR2 having the amino acid sequence of SEQ ID NO: 10 is substituted with CDR2 having the amino acid sequence of SEQ ID NO: 172 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0142] Substitution of Ser at position 10 of CDR2 (SEQ ID NO: 10) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Asp (SEQ ID NO: 177). Thus, the present invention provides heavy chain variable regions in which CDR2 having the amino acid sequence of SEQ ID NO: 10 is substituted with CDR2 having the amino acid sequence of SEQ ID NO: 177 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0143] Substitution of Gln at position 13 of CDR2 (SEQ ID NO: 10) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Pro (SEQ ID NO: 178). Thus, the present invention provides heavy chain variable regions in which CDR2 having the amino acid sequence of SEQ ID NO: 10 is substituted with CDR2 having the amino acid sequence of SEQ ID NO: 178 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0144] Substitution of Tyr at position 3 of CDR3 (SEQ ID NO: 11) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Leu (SEQ ID NO: 179). Thus, the present invention provides heavy chain variable regions in which CDR3 having the amino acid sequence of SEQ ID NO: 11 is substituted with CDR3 having the amino acid sequence of SEQ ID NO: 179 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0145] Substitution of Met at position 10 of CDR3 (SEQ ID NO: 11) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Leu (SEQ ID NO: 180). Thus, the present invention provides heavy chain variable regions in which CDR3 having the amino acid sequence of SEQ ID NO: 11 is substituted with CDR3 having the amino acid sequence of SEQ ID NO: 180 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0146] Substitution of Asp at position 11 of CDR3 (SEQ ID NO: 11) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Glu (SEQ ID NO: 181). Thus, the present invention provides heavy chain variable regions in which CDR3 having the amino acid sequence of SEQ ID NO: 11 is substituted with CDR3 having the amino acid sequence of SEQ ID NO: 181 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0147] Substitution of Tyr at position 12 of CDR3 (SEQ ID NO: 11) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Thr (SEQ ID NO: 182). Thus, the present invention provides heavy chain variable regions in which CDR3 having the amino acid sequence of SEQ ID NO: 11 is substituted with CDR3 having the amino acid sequence of SEQ ID NO: 182 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0148] Substitution of Tyr at position 12 of CDR3 (SEQ ID NO: 11) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Ser (SEQ ID NO: 183). Thus, the present invention provides heavy chain variable regions in which CDR3 having the amino acid sequence of SEQ ID NO: 11 is substituted with CDR3 having the amino acid sequence of SEQ ID NO: 183 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0149] Substitution of Met at position 10, Asp at position 11, and Tyr at position 12 of CDR3 (SEQ ID NO: 11) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Leu, Glu, Thr, respectively (SEQ ID NO: 184). Thus, the present invention provides heavy chain variable regions in which CDR3 having the amino acid sequence of SEQ ID NO: 11 is substituted with CDR3 having the amino acid sequence of SEQ ID NO: 184 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0150] Substitution of Asp at position 11 and Tyr at position 12 of CDR3 (SEQ ID NO: 11) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Glu and Thr, respectively (SEQ ID NO: 185). Thus, the present invention provides heavy chain variable regions in which CDR3 having the amino acid sequence of SEQ ID NO: 11 is substituted with CDR3 having the amino acid sequence of SEQ ID NO: 185 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0151] Substitution of Tyr at position 3, Asp at position 11, and Tyr at position 12 of CDR3 (SEQ ID NO: 11) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Leu, Glu, and Thr, respectively (SEQ ID NO: 186). Thus, the present invention provides heavy chain variable regions in which CDR3 having the amino acid sequence of SEQ ID NO: 11 is substituted with CDR3 having the amino acid sequence of SEQ ID NO: 186 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0152] Substitution of Tyr at position 3, Asp at position 11, and Tyr at position 12 of CDR3 (SEQ ID NO: 11) in the heavy chain variable region of SEQ ID NO: 50 or 112 with Leu, Glu, and Ser, respectively (SEQ ID NO: 187). Thus, the present invention provides heavy chain variable regions in which CDR3 having the amino acid sequence of SEQ ID NO: 11 is substituted with CDR3 having the amino acid sequence of SEQ ID NO: 187 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0153] Substitution of Arg at position 1 of CDR1 (SEQ ID NO: 13) in the light chain variable region of SEQ ID NO: 52 with Gln (SEQ ID NO: 121). Thus, the present invention provides light chain variable regions in which CDR1 having the amino acid sequence of SEQ ID NO: 13 is substituted with CDR1 having the amino acid sequence of SEQ ID NO: 121 in a light chain variable region having the amino acid sequence of SEQ ID NO: 52.
[0154] Substitution of Asn at position 5 of CDR1 (SEQ ID NO: 13) in the light chain variable region of SEQ ID NO: 52 with Asp (SEQ ID NO: 122). Thus, the present invention provides light chain variable regions in which CDR1 having the amino acid sequence of SEQ ID NO: 13 is substituted with CDR1 having the amino acid sequence of SEQ ID NO: 122 in a light chain variable region having the amino acid sequence of SEQ ID NO: 52.
[0155] Substitution of Ser at position 8 of CDR1 (SEQ ID NO: 13) in the light chain variable region of SEQ ID NO: 52 with Arg (SEQ ID NO: 188). Thus, the present invention provides light chain variable regions in which CDR1 having the amino acid sequence of SEQ ID NO: 13 is substituted with CDR1 having the amino acid sequence of SEQ ID NO: 188 in a light chain variable region having the amino acid sequence of SEQ ID NO: 52.
[0156] Substitution of Leu at position 10 of CDR1 (SEQ ID NO: 13) of the light chain variable region of SEQ ID NO: 52 with Val (SEQ ID NO: 189). Thus, the present invention provides light chain variable regions in which CDR1 having the amino acid sequence of SEQ ID NO: 13 is substituted with CDR1 having the amino acid sequence of SEQ ID NO: 189 in a light chain variable region having the amino acid sequence of SEQ ID NO: 52.
[0157] Substitution of Ser at position 8 and Leu at position 10 of CDR1 (SEQ ID NO: 13) of the light chain variable region of SEQ ID NO: 52 with Arg and Val, respectively (SEQ ID NO: 190). Thus, the present invention provides light chain variable regions in which CDR1 having the amino acid sequence of SEQ ID NO: 13 is substituted with CDR1 having the amino acid sequence of SEQ ID NO: 190 in a light chain variable region having the amino acid sequence of SEQ ID NO: 52.
[0158] Substitution of Thr at position 2 of CDR1 (SEQ ID NO: 13) in the light chain variable region of SEQ ID NO: 52 with Ala (SEQ ID NO: 191). Thus, the present invention provides light chain variable regions in which CDR1 having the amino acid sequence of SEQ ID NO: 13 is substituted with CDR1 having the amino acid sequence of SEQ ID NO: 191 in a light chain variable region having the amino acid sequence of SEQ ID NO: 52.
[0159] Substitution of Thr at position 2 of CDR1 (SEQ ID NO: 13) in the light chain variable region of SEQ ID NO: 52 with Ser (SEQ ID NO: 192). Thus, the present invention provides light chain variable regions in which CDR1 having the amino acid sequence of SEQ ID NO: 13 is substituted with CDR1 having the amino acid sequence of SEQ ID NO: 192 in a light chain variable region having the amino acid sequence of SEQ ID NO: 52.
[0160] Substitution of Asn at position 1 of CDR2 (SEQ ID NO: 14) in the light chain variable region of SEQ ID NO: 52 with Asp (SEQ ID NO: 123). Thus, the present invention provides light chain variable regions in which CDR2 having the amino acid sequence of SEQ ID NO: 14 is substituted with CDR2 having the amino acid sequence of SEQ ID NO: 123 in a light chain variable region having the amino acid sequence of SEQ ID NO: 52.
[0161] Substitution of Lys at position 3 of CDR2 (SEQ ID NO: 14) in the light chain variable region of SEQ ID NO: 52 with Gln (SEQ ID NO: 124). Thus, the present invention provides light chain variable regions in which CDR2 having the amino acid sequence of SEQ ID NO: 14 is substituted with CDR2 having the amino acid sequence of SEQ ID NO: 124 in a light chain variable region having the amino acid sequence of SEQ ID NO: 52.
[0162] Substitution of Leu at position 5 of CDR2 (SEQ ID NO: 14) in the light chain variable region of SEQ ID NO: 52 with Glu (SEQ ID NO: 125). Thus, the present invention provides light chain variable regions in which CDR2 having the amino acid sequence of SEQ ID NO: 14 is substituted with CDR2 having the amino acid sequence of SEQ ID NO: 125 in a light chain variable region having the amino acid sequence of SEQ ID NO: 52.
[0163] Substitution of Lys at position 7 of CDR2 (SEQ ID NO: 14) in the light chain variable region of SEQ ID NO: 52 with Gln (SEQ ID NO: 126). Thus, the present invention provides light chain variable regions in which CDR2 having the amino acid sequence of SEQ ID NO: 14 is substituted with CDR2 having the amino acid sequence of SEQ ID NO: 126 in a light chain variable region having the amino acid sequence of SEQ ID NO: 52.
[0164] Substitution of Lys at position 7 of CDR2 (SEQ ID NO: 14) in the light chain variable region of SEQ ID NO: 52 with Asp (SEQ ID NO: 127). Thus, the present invention provides light chain variable regions in which CDR2 having the amino acid sequence of SEQ ID NO: 14 is substituted with CDR2 having the amino acid sequence of SEQ ID NO: 127 in a light chain variable region having the amino acid sequence of SEQ ID NO: 52.
[0165] Substitution of Arg at position 1 and Asn at position 5 of CDR1 (SEQ ID NO: 13) in the light chain variable region of SEQ ID NO: 52 with Gln and Asp, respectively (SEQ ID NO: 169). Thus, the present invention provides light chain variable regions in which CDR1 having the amino acid sequence of SEQ ID NO: 13 is substituted with CDR1 having the amino acid sequence of SEQ ID NO: 169 in a light chain variable region having the amino acid sequence of SEQ ID NO: 52.
[0166] Substitution of Lys at position 3, Leu at position 5, and Lys at position 7 of CDR2 (SEQ ID NO: 14) in the light chain variable region of SEQ ID NO: 52 with Gln, Glu, and Gln, respectively (SEQ ID NO: 170). Thus, the present invention provides light chain variable regions in which CDR2 having the amino acid sequence of SEQ ID NO: 14 is substituted with CDR2 having the amino acid sequence of SEQ ID NO: 170 in a light chain variable region having the amino acid sequence of SEQ ID NO: 52.
[0167] Substitution of Glu at position 5 of CDR3 (SEQ ID NO: 15) in the light chain variable region of SEQ ID NO: 52 with Asp (SEQ ID NO: 193). Thus, the present invention provides light chain variable regions in which CDR3 having the amino acid sequence of SEQ ID NO: 15 is substituted with CDR3 having the amino acid sequence of SEQ ID NO: 193 in a light chain variable region having the amino acid sequence of SEQ ID NO: 52.
[0168] Substitution of Ser at position 6 of CDR3 (SEQ ID NO: 15) in the light chain variable region of SEQ ID NO: 52 with Asp (SEQ ID NO: 194). Thus, the present invention provides light chain variable regions in which CDR3 having the amino acid sequence of SEQ ID NO: 15 is substituted with CDR3 having the amino acid sequence of SEQ ID NO: 194 in a light chain variable region having the amino acid sequence of SEQ ID NO: 52.
[0169] Substitution of Thr at position 9 of CDR3 (SEQ ID NO: 15) in the light chain variable region of SEQ ID NO: 52 with Phe (SEQ ID NO: 195). Thus, the present invention provides light chain variable regions in which CDR3 having the amino acid sequence of SEQ ID NO: 15 is substituted with CDR3 having the amino acid sequence of SEQ ID NO: 195 in a light chain variable region having the amino acid sequence of SEQ ID NO: 52.
[0170] In addition, the substitutions other than those described above include a substitution of Arg at position 3 of heavy chain FR2 having the amino acid sequence of SEQ ID NO: 97 with another amino acid. The amino acid after substitution is not particularly limited; but preferred examples thereof include Gln. When Arg at position 3 in SEQ ID NO: 97 has been replaced with Gln, Ala at position 5 may be substituted with Ser to produce a human FR2 sequence. The amino acid sequence in which Arg and Ala at positions 3 and 5 in the amino acid sequence of SEQ ID NO: 97 have been replaced with Gln and Ser, respectively, is shown in SEQ ID NO: 120. Thus, the present invention provides heavy chain variable regions in which FR2 having the amino acid sequence of SEQ ID NO: 97 is substituted with FR2 having the amino acid sequence of SEQ ID NO: 120 in a heavy chain variable region having the amino acid sequence of SEQ ID NO: 50 or 112.
[0171] Each of the above-described amino acid substitutions may be used alone or in combination with other amino acid substitutions described above. They also may be combined with amino acid substitutions other than those described above.
[0172] Specific examples of the antibodies in which the above-described substitutions have been carried out include, for example, antibodies that comprise a heavy chain variable region having the amino acid sequence of SEQ ID NO: 167, antibodies that comprise a light chain variable region having the amino acid sequence of SEQ ID NO: 168, and antibodies that comprise a heavy chain variable region having the amino acid sequence of SEQ ID NO: 167 and a light chain variable region having the amino acid sequence of SEQ ID NO: 168. Furthermore, specific examples of the heavy chain variable regions in which the above-described substitutions have been carried out include, for example, the following heavy chain variable regions:
(1) heavy chain variable regions having the amino acid sequence of SEQ ID NO: 204 (H17); (2) heavy chain variable regions having the amino acid sequence of SEQ ID NO: 205 (H19); (3) heavy chain variable regions having the amino acid sequence of SEQ ID NO: 206 (H28); (4) heavy chain variable regions having the amino acid sequence of SEQ ID NO: 207 (H30); (5) heavy chain variable regions having the amino acid sequence of SEQ ID NO: 208 (H34); (6) heavy chain variable regions having the amino acid sequence of SEQ ID NO: 209 (H42); (7) heavy chain variable regions having the amino acid sequence of SEQ ID NO: 210 (H44); (8) heavy chain variable regions having the amino acid sequence of SEQ ID NO: 211 (H46); (9) heavy chain variable regions having the amino acid sequence of SEQ ID NO: 212 (H57); (10) heavy chain variable regions having the amino acid sequence of SEQ ID NO: 213 (H71); (11) heavy chain variable regions having the amino acid sequence of SEQ ID NO: 214 (H78); (12) heavy chain variable regions having the amino acid sequence of SEQ ID NO: 215 (H92); (13) heavy chain variable regions having the amino acid sequence of SEQ ID NO: 216 (H97); and (14) heavy chain variable regions having the amino acid sequence of SEQ ID NO: 217 (H98).
[0173] Meanwhile, specific examples of the light chain variable regions in which the above-described substitutions carried out include, for example, the following light chain variable regions:
(15) light chain variable regions having the amino acid sequence of SEQ ID NO: 218 (L11); (16) light chain variable regions having the amino acid sequence of SEQ ID NO: 219 (L12); (17) light chain variable regions having the amino acid sequence of SEQ ID NO: 220 (L17); and (18) light chain variable regions having the amino acid sequence of SEQ ID NO: 221 (L50).
[0174] Furthermore, specific examples of the antibodies comprising the above-described heavy chain and light chain variable regions include, for example, the following antibodies:
(19) antibodies that comprise the heavy chain variable region of (3) and the light chain variable region of (17) (H28L17); (20) antibodies that comprise the heavy chain variable region of (4) and the light chain variable region of (17) (H30L17); (21) antibodies that comprise the heavy chain variable region of (5) and the light chain variable region of (17) (H34L17); (22) antibodies that comprise the heavy chain variable region of (6) and the light chain variable region of (17) (H42L17); (23) antibodies that comprise the heavy chain variable region of (7) and the light chain variable region of (17) (H44L17); (24) antibodies that comprise the heavy chain variable region of (8) and the light chain variable region of (17) (H46L17); (25) antibodies that comprise the heavy chain variable region of (9) and the light chain variable region of (17) (H57L17); (26) antibodies that comprise the heavy chain variable region of (10) and the light chain variable region of (17) (H71L17); (27) antibodies that comprise the heavy chain variable region of (11) and the light chain variable region of (17) (H78L17); (28) antibodies that comprise the heavy chain variable region of (12) and the light chain variable region of (17) (H92L17); (29) antibodies that comprise the heavy chain variable region of (13) and the light chain variable region of (18) (H97L50); and (30) antibodies that comprise the heavy chain variable region of (14) and the light chain variable region of (18) (H98L50).
[0175] The constant region used for the humanized antibodies of the present invention may be any constant region derived from a human antibody. Preferred examples of such constant regions derived from human antibodies include, for example, constant regions derived from IgG1 or IgG2. Moreover, constant regions in which one or more amino acids are substituted, deleted, added, and/or inserted in the constant region derived from a human antibody may also be used.
[0176] The constant regions in which one or more amino acids are substituted, deleted, added, and/or inserted in the constant region derived from a human antibody are not particularly limited, and include, for example, the following constant regions:
[0177] constant regions having the amino acid sequence of SEQ ID NO: 128 (M58);
[0178] constant regions having the amino acid sequence of SEQ ID NO: 129 (M14); and
[0179] constant regions having the amino acid sequence of SEQ ID NO: 62 (SKSC).
[0180] Specific examples of the heavy chains or antibodies having the above-described constant regions include, for example:
(1) heavy chains that comprise a variable region having the amino acid sequence of SEQ ID NO: 167 and a constant region having the amino acid sequence of SEQ ID NO: 128; (2) heavy chains in which CDR2 having the amino acid sequence of SEQ ID NO: 171 in the heavy chains of (1) is substituted with CDR2 having the amino acid sequence of SEQ ID NO: 172; (3) antibodies that comprise the heavy chain of (1) and a light chain having the amino acid sequence of SEQ ID NO: 152; and (4) antibodies that comprise the heavy chain of (2) and a light chain having the amino acid sequence of SEQ ID NO: 152.
[0181] More specific examples of the humanized anti-NR10 antibodies of the present invention include, for example, the following antibodies:
(k) antibodies that comprise a heavy chain having the amino acid sequence of SEQ ID NO: 54 (H0-VH+constant region); (l) antibodies that comprise a heavy chain having the amino acid sequence of SEQ ID NO: 130 (H1-VH+constant region); (m) antibodies that comprise a light chain having the amino acid sequence of SEQ ID NO: 56 (L0-VL+constant region); (n) antibodies that comprise a heavy chain having the amino acid sequence of SEQ ID NO: 54 (H0-VH+constant region) and a light chain having the amino acid sequence of SEQ ID NO: 56 (L0-VL+constant region); and (o) antibodies that comprise a heavy chain having the amino acid sequence of SEQ ID NO: 130 (H1-VH+constant region) and a light chain having the amino acid sequence of SEQ ID NO: 56 (L0-VL+constant region).
[0182] The heavy chain having the amino acid sequence of SEQ ID NO: 54 (H0-VH+constant region) and the light chain having the amino acid sequence of SEQ ID NO: 56 (L0-VL+constant region) may have a substitution, deletion, addition, and/or insertion of one or more amino acids. The substitution, deletion, addition, and/or insertion of amino acids may be carried out in either or both of the variable and constant regions.
[0183] Thus, the present invention provides:
(p) antibodies that comprise a heavy chain having an amino acid sequence in which one or more amino acids are substituted, deleted, added, and/or inserted in the amino acid sequence of SEQ ID NO: 54 (H0-VH+constant region); (q) antibodies that comprise a heavy chain having an amino acid sequence in which one or more amino acids are substituted, deleted, added, and/or inserted in the amino acid sequence of SEQ ID NO: 130 (H1-VH+constant region); (r) antibodies that comprise a light chain having an amino acid sequence in which one or more amino acids are substituted, deleted, added, and/or inserted in the amino acid sequence of SEQ ID NO: 56 (L0-VL+constant region); (s) antibodies that comprise a heavy chain having an amino acid sequence in which one or more amino acids are substituted, deleted, added, and/or inserted in the amino acid sequence of SEQ ID NO: 54 (H0-VH+constant region) and a light chain having an amino acid sequence in which one or more amino acids are substituted, deleted, added, and/or inserted in the amino acid sequence of SEQ ID NO: 56 (L0-VL+constant region); and (t) antibodies that comprise a heavy chain having an amino acid sequence in which one or more amino acids are substituted, deleted, added, and/or inserted in the amino acid sequence of SEQ ID NO: 130 (H1-VH+constant region) and a light chain having an amino acid sequence in which one or more amino acids are substituted, deleted, added, and/or inserted in the amino acid sequence of SEQ ID NO: 56 (L0-VL+constant region).
[0184] Without particular limitation, the antibodies of any one of (p) to (t) preferably have an activity similar to that of the antibodies of any one of (k) to (o).
[0185] The substitution, deletion, addition, and/or insertion of amino acids are not particularly limited, but specific examples thereof include, for example, the above-described amino acid substitutions.
[0186] Furthermore, the nucleotide sequence encoding the amino acid sequence of the above-described humanized heavy chain variable region (SEQ ID NO: 50) is shown in SEQ ID NO: 49. The nucleotide sequence encoding the amino acid sequence of the humanized light chain variable region (SEQ ID NO: 52) is shown in SEQ ID NO: 51. The nucleotide sequence encoding the amino acid sequence of the humanized heavy chain (SEQ ID NO: 54) is shown in SEQ ID NO: 53. The nucleotide sequence encoding the amino acid sequence of the humanized light chain (SEQ ID NO: 56) is shown in SEQ ID NO: 55.
[0187] Moreover, the present invention provides antibodies that recognize the same epitope as recognized by the antibodies of any one of (a) to (t) above. The binding to the same epitope is as already described above.
[0188] Furthermore, the present invention provides the following antibodies:
(u) antibodies that comprise a heavy chain having the amino acid sequence of SEQ ID NO: 151; (v) antibodies that comprise a light chain comprising the amino acid sequence of SEQ ID NO: 152; and (w) antibodies that comprise the heavy chain of (u) and the light chain of (v). Moreover, the present invention provides the following heavy and light chains and antibodies: (1) heavy chains having the amino acid sequence of SEQ ID NO: 222 (H17); (2) heavy chains having the amino acid sequence of SEQ ID NO: 223 (H19); (3) heavy chains having the amino acid sequence of SEQ ID NO: 224 (H28); (4) heavy chains having the amino acid sequence of SEQ ID NO: 225 (H30); (5) heavy chains having the amino acid sequence of SEQ ID NO: 226 (H34); (6) heavy chains having the amino acid sequence of SEQ ID NO: 227 (H42); (7) heavy chains having the amino acid sequence of SEQ ID NO: 228 (H44); (8) heavy chains having the amino acid sequence of SEQ ID NO: 229 (H46); (9) heavy chains having the amino acid sequence of SEQ ID NO: 230 (H57); (10) heavy chains having the amino acid sequence of SEQ ID NO: 231 (H71); (11) heavy chains having the amino acid sequence of SEQ ID NO: 232 (H78); (12) heavy chains having the amino acid sequence of SEQ ID NO: 233 (H92); (13) heavy chains having the amino acid sequence of SEQ ID NO: 234 (H97); (14) heavy chains having the amino acid sequence of SEQ ID NO: 235 (H98); (15) light chains having the amino acid sequence of SEQ ID NO: 236 (L11) (16) light chains having the amino acid sequence of SEQ ID NO: 237 (L12); (17) light chains having the amino acid sequence of SEQ ID NO: 238 (L17); (18) light chains having the amino acid sequence of SEQ ID NO: 239 (L50); (19) antibodies that comprise the heavy chain of (3) and the light chain of (17) (H28L17); (20) antibodies that comprise the heavy chain of (4) and the light chain of (17) (H30L17); (21) antibodies that comprise the heavy chain of (5) and the light chain of (17) (H34L17); (22) antibodies that comprise the heavy chain of (6) and the light chain of (17) (H42L17); (23) antibodies that comprise the heavy chain of (7) and the light chain of (17) (H44L17); (24) antibodies that comprise the heavy chain of (8) and the light chain of (17) (H46L17); (25) antibodies that comprise the heavy chain of (9) and the light chain of (17) (H57L17); (26) antibodies that comprise the heavy chain of (10) and the light chain of (17) (H71L17); (27) antibodies that comprise the heavy chain of (11) and the light chain of (17) (H78L17); (28) antibodies that comprise the heavy chain of (12) and the light chain of (17) (H92L17); (29) antibodies that comprise the heavy chain of (13) and the light chain of (18) (H97L50); (30) antibodies that comprise the heavy chain of (14) and the light chain of (18) (H98L50); (31) heavy chains having an amino acid sequence in which one or more amino acids are substituted, deleted, added and/or inserted in the heavy chains of any one of (1) to (14); (32) light chains having an amino acid sequence in which one or more amino acids are substituted, deleted, added and/or inserted in the light chains of any one of (15) to (18); (33) antibodies having an amino acid sequence in which one or more amino acids are substituted, deleted, added and/or inserted in the antibodies of any one of (19) to (30); and (34) antibodies that recognize the same epitope as recognized by the antibodies of any one of (19) to (33).
[0189] The substitution, deletion, addition, and/or insertion of amino acids are as described above. Antibodies that recognize the same epitope as recognized by an antibody are also described above.
[0190] The present invention also provides genes encoding the variable regions, heavy chains, light chains, or antibodies of the present invention.
[0191] The present invention also provides vectors carrying the above-described genes.
[0192] The present invention also provides host cells transformed with the above-described vectors.
[0193] The present invention also relates to methods for producing variable regions, heavy chains, light chains, or antibodies of the present invention, which comprise the step of culturing the above-described host cells.
[0194] The vectors, host cells, and culture of host cells are described herein below.
Antibodies that Recognize Domains
[0195] Preferred embodiments of the anti-NR10 antibody of the present invention include antibodies that recognize domain 1 or domain 2. In the present invention, domain 1 refers to the region of amino acids at positions 21 to 120 (LPAKP to LENIA) in the amino acid sequence of human NR10 of SEQ ID NO: 76, where the amino acid numbering is based on the sequence including the signal peptide. In addition, in the present invention, domain 2 refers to the region of amino acids at positions 121 to 227 (KTEPP to EEEAP) in the amino acid sequence of human NR10 of SEQ ID NO: 76, where the amino acid numbering is based on the sequence including the signal peptide.
[0196] Such antibodies are not particularly limited; however, in general, they have a neutralizing activity, and preferably are humanized antibodies.
[0197] Examples of the preferred antibodies in the present invention include antibodies that recognize domain 1. The antibodies that recognize domain 1 have a strong neutralizing activity, and thus are particularly useful as pharmaceuticals.
Antibodies (Neutralizing Activity)
[0198] The present invention also provides anti-NR10 antibodies having a neutralizing activity.
[0199] In the present invention, the neutralizing activity against NR10 refers to an activity of inhibiting the binding between NR10 and its ligand IL-31, and preferably an activity of suppressing a biological activity based on NR10.
[0200] Antibodies having a NR10-neutralizing activity can be selected, for example, by adding candidate antibodies to an IL-31-dependent cell line and observing their growth-suppressing effect on the cell line. In this method, antibodies that suppress the growth of the IL-31-dependent cell line are determined as antibodies having a neutralizing activity against NR10.
Antibodies (General)
[0201] The antibodies of the present invention are not limited in terms of their origin, and may be derived from any animals such as humans, mice, and rats. Moreover, the antibodies may be recombinant antibodies such as chimeric antibodies and humanized antibodies. As described above, the preferred antibodies of the present invention include humanized antibodies.
[0202] The chimeric antibodies contain, for example, the heavy and light chain constant regions of a human antibody, and the heavy and light chain variable regions of an antibody of a non-human mammal, such as mouse. The chimeric antibodies can be produced by known methods. For example, the antibodies can be produced by cloning an antibody gene from hybridomas, inserting it into an appropriate vector, and introducing the construct into hosts (see, for example, Carl, A. K. Borrebaeck, James, W. Larrick, THERAPEUTIC MONOCLONAL ANTIBODIES, Published in the United Kingdom by MACMILLAN PUBLISHERS LTD, 1990). Specifically, cDNAs of the antibody variable regions (V regions) are synthesized from mRNA of hybridomas using reverse transcriptase. Once DNAs encoding the V regions of an antibody of interest are obtained, these are linked with DNAs encoding the constant regions (C regions) of a desired human antibody. The resulting constructs are inserted into expression vectors. Alternatively, the DNAs encoding the antibody V regions may be inserted into expression vectors comprising DNAs encoding the C regions of a human antibody. The DNAs are inserted into expression vectors so that they are expressed under the regulation of the expression regulatory regions, for example, enhancers and promoters. In the next step, host cells can be transformed with the expression vectors to allow expression of chimeric antibodies.
[0203] Methods for obtaining human antibodies are also known. For example, desired human antibodies with antigen-binding activity can be obtained by (1) sensitizing human lymphocytes with antigens of interest or cells expressing antigens of interest in vitro; and (2) fusing the sensitized lymphocytes with human myeloma cells such as U266 (see Japanese Patent Application Kokoku Publication No. (JP-B) H01-59878 (examined, approved Japanese patent application published for opposition)). Alternatively, the desired human antibody can also be obtained by immunizing a transgenic animal having an entire repertoire of human antibody genes with a desired antigen (see International Patent Application Publication Nos. WO 93/12227, WO 92/03918, WO 94/02602, WO 94/25585, WO 96/34096, and WO 96/33735).
[0204] Furthermore, techniques to obtain human antibodies by panning with a human antibody phage library are known. For example, the variable region of a human antibody is expressed as a single chain antibody (scFv) on the surface of a phage, using a phage display method, and phages that bind to the antigen can be selected. By analyzing the genes of selected phages, the DNA sequences encoding the variable regions of human antibodies that bind to the antigen can be determined. If the DNA sequences of scFvs that bind to the antigen are identified, appropriate expression vectors comprising these sequences can be constructed to obtain human antibodies. Such methods are well known. Reference can be made to WO 92/01047, WO 92/20791, WO 93/06213, WO 93/11236, WO 93/19172, WO 95/01438, WO 95/15388, and such.
[0205] The antibodies of the present invention include not only divalent antibodies as represented by IgG, but also monovalent antibodies, multivalent antibodies as represented by IgM, and bispecific antibodies capable of binding to different antigens, as long as they have a NR10-binding activity and/or neutralizing activity. The multivalent antibodies of the present invention include multivalent antibodies in which the antigen-binding sites are all identical, and multivalent antibodies in which all or some of the antigen-binding sites are different. The antibodies of the present invention are not limited to full-length antibody molecules, but may also be low-molecular-weight antibodies or modified products thereof, as long as they bind to NR10 protein.
[0206] Alternatively, the antibodies of the present invention may be low-molecular-weight antibodies. Such low-molecular-weight antibodies are antibodies including antibody fragments lacking some portions of a whole antibody (for example, whole IgG), and are not particularly limited as long as they retain NR10-binding and/or neutralizing activity. In the present invention, the low-molecular-weight antibodies are not particularly limited, as long as they contain a portion of whole antibodies. The low-molecular-weight antibodies preferably contain a heavy chain variable region (VH) or light chain variable region (VL). Particularly preferred low-molecular-weight antibodies contain both VH and VL. In addition, preferred examples of the low-molecular-weight antibodies of the present invention include low-molecular-weight antibodies containing CDRs of an antibody. The CDRs contained in the low-molecular-weight antibodies may include some or all of the six CDRs of an antibody.
[0207] The low-molecular-weight antibodies of the present invention preferably have a smaller molecular weight than whole antibodies. However, the low-molecular-weight antibodies may form multimers, for example, dimers, trimers, or tetramers, and thus their molecular weights can be greater than those of whole antibodies.
[0208] Specific examples of the antibody fragments include, for example, Fab, Fab', F(ab')2, and Fv. Meanwhile, specific examples of the low-molecular-weight antibodies include, for example, Fab, Fab', F(ab')2, Fv, scFv (single chain Fv), diabodies, and sc(Fv)2 (single chain (Fv)2). Multimers (for example, dimers, trimers, tetramers, and polymers) of these antibodies are also included in the low-molecular-weight antibodies of the present invention.
[0209] Antibody fragments can be obtained, for example, by treating antibodies with enzymes to produce antibody fragments. Enzymes known to generate antibody fragments include, for example, papain, pepsin, and plasmin. Alternatively, a gene encoding such an antibody fragment can be constructed, introduced into an expression vector, and expressed in appropriate host cells (see, for example, Co, M. S. et al., J. Immunol. (1994)152, 2968-2976; Better, M. & Horwitz, A. H. Methods in Enzymology (1989)178, 476-496; Plueckthun, A. & Skerra, A. Methods in Enzymology (1989)178, 476-496; Lamoyi, E., Methods in Enzymology (1989)121, 652-663; Rousseaux, J. et al., Methods in Enzymology (1989)121, 663-669; Bird, R. E. et al., TIBTECH (1991)9, 132-137).
[0210] Digestive enzymes cleave a specific site of an antibody fragment, yielding antibody fragments of specific structures shown below. Genetic engineering techniques can be applied to such enzymatically-obtained antibody fragments to delete an arbitrary portion of the antibody.
[0211] Antibody fragments obtained by using the above-described digestive enzymes are as follows:
Papain digestion: F(ab)2 or Fab Pepsin digestion: F(ab')2 or Fab' Plasmin digestion: Facb
[0212] The low-molecular-weight antibodies of the present invention include antibody fragments lacking an arbitrary region, as long as they have a NR10-binding activity and/or neutralizing activity.
[0213] "Diabody" refers to a bivalent antibody fragment constructed by gene fusion (Holliger P et al., Proc. Natl. Acad. Sci. USA 90: 6444-6448 (1993); EP 404,097; WO 93/11161, etc). Diabodies are dimers composed of two polypeptide chains. In each of the polypeptide chains forming a dimer, a VL and a VH are usually linked by a linker in the same chain. In general, the linker in a diabody is short enough such that the VL and VH cannot bind to each other. Specifically, the number of amino acid residues constituting the linker is, for example, about five residues. Thus, the VL and VH encoded on the same polypeptide cannot form a single-chain variable region fragment, and will form a dimer with another single-chain variable region fragment. As a result, the diabody has two antigen binding sites.
[0214] ScFv antibodies are single-chain polypeptides produced by linking a heavy chain variable region ([VH]) and a light chain variable region ([VL]) via a linker or such (Huston, J. S. et al., Proc. Natl. Acad. Sci. U.S.A. (1988) 85, 5879-5883; Pluckthun "The Pharmacology of Monoclonal Antibodies" Vol. 113, eds., Resenburg and Moore, Springer Verlag, New York, pp. 269-315, (1994)). The H-chain V region and L-chain V region of scFv may be derived from any antibody described herein. The peptide linker for linking the V regions is not particularly limited. For example, an arbitrary single-chain peptide containing about three to 25 residues can be used as the linker. Specifically, it is possible to use the peptide linkers or such described below.
[0215] The V regions of both chains can be linked, for example, by PCR as described above. First, among the following DNAs, a DNA encoding a complete or desired partial amino acid sequence is used as a template to link the V regions by PCR:
[0216] DNA sequence encoding an H chain or H-chain V region of an antibody, and
[0217] DNA sequence encoding an L chain or L-chain V region of an antibody.
[0218] DNAs encoding the V regions of H chain and L chain are amplified by PCR using a pair of primers having sequences corresponding to those at both ends of the DNA to be amplified. Then, a DNA encoding the peptide linker portion is prepared. The peptide linker-encoding DNA can also be synthesized by PCR. Here, nucleotide sequences that can be ligated to the amplification products of V regions synthesized separately are added to the 5' end of the primers to be used. Then, PCR is carried out using each DNA of the [H chain V region DNA]-[peptide linker DNA]-[L chain V region DNA], and assembly PCR primers.
[0219] The assembly PCR primers are composed of a combination of a primer that anneals to the 5' end of the [H chain V region DNA] and a primer that anneals to the 3' end of the [L chain V region DNA]. In other words, the assembly PCR primers are a set of primers that can be used to amplify DNA encoding the full-length sequence of scFv to be synthesized. Meanwhile, nucleotide sequences that can be ligated to the V-region DNAs have been added to the [peptide linker DNA]. Thus, these DNAs are linked together, and then the whole scFv is ultimately generated as an amplification product by the assembly PCR primers. Once the scFv-encoding DNAs are generated, expression vectors carrying these DNAs and recombinant cells transformed with these expression vectors can be obtained by conventional methods. Furthermore, the scFv can be obtained by culturing the resulting recombinant cells to express the scFv-encoding DNAs.
[0220] The order of the heavy chain and light chain variable regions to be linked together is not particularly limited, and they may be arranged in any order. Examples of the arrangement are listed below.
[0221] [VH] linker [VL]
[0222] [VL] linker [VH]
[0223] sc(Fv)2 is a single-chain low-molecular-weight antibody produced by linking two VHs and two VLs using linkers and such (Hudson et al., J Immunol. Methods 1999; 231: 177-189). For example, sc(Fv)2 can be produced by linking scFvs via a linker.
[0224] Antibodies in which two VHs and two VLs are arranged in the order of VH-VL-VH-VL ([VH] linker [VL] linker [VH] linker [VL]) from the N terminus of the single-chain polypeptide are preferred. However, the order of the two VHs and two VLs is not limited to the above arrangement, and they may be arranged in any order. Examples of the arrangement are listed below:
[0225] [VL] linker [VH] linker [VH] linker [VL]
[0226] [VH] linker [VL] linker [VL] linker [VH]
[0227] [VH] linker [VH] linker [VL] linker [VL]
[0228] [VL] linker [VL] linker [VH] linker [VH]
[0229] [VL] linker [VH] linker [VL] linker [VH]
[0230] The amino acid sequence of the heavy chain variable region or light chain variable region in a low-molecular-weight antibody may contain a substitution, deletion, addition, and/or insertion. Furthermore, the heavy chain variable region and light chain variable region may also lack some portions or be added with other polypeptides, as long as they have antigen binding ability when linked together. Alternatively, the variable regions may be chimerized or humanized.
[0231] In the present invention, linkers which bind the variable regions of the antibody include arbitrary peptide linkers that can be introduced using genetic engineering, or synthetic linkers such as those disclosed in Protein Engineering, 9(3), 299-305, 1996.
[0232] The preferred linkers in the present invention are peptide linkers. The lengths of the peptide linkers are not particularly limited and those skilled in the art can appropriately select the lengths depending on the purpose. Typical lengths are one to 100 amino acids, preferably 3 to 50 amino acids, more preferably 5 to 30 amino acids, and particularly preferably 12 to 18 amino acids (for example, 15 amino acids).
[0233] Amino acid sequences of such peptide linkers include, for example:
TABLE-US-00001 Ser; Gly-Ser; Gly-Gly-Ser; Ser-Gly-Gly; (SEQ ID NO: 82) Gly-Gly-Gly-Ser; (SEQ ID NO: 83) Ser-Gly-Gly-Gly; (SEQ ID NO: 84) Gly-Gly-Gly-Gly-Ser; (SEQ ID NO: 85) Ser-Gly-Gly-Gly-Gly; (SEQ ID NO: 86) Gly-Gly-Gly-Gly-Gly-Ser; (SEQ ID NO: 87) Ser-Gly-Gly-Gly-Gly-Gly; (SEQ ID NO: 88) Gly-Gly-Gly-Gly-Gly-Gly-Ser; (SEQ ID NO: 89) Ser-Gly-Gly-Gly-Gly-Gly-Gly; (SEQ ID NO: 84) (Gly-Gly-Gly-Gly-Ser )n; and (SEQ ID NO: 85) (Ser-Gly-Gly-Gly-Gly )n,
where n is an integer of 1 or larger.
[0234] The amino acid sequence of peptide linker can be appropriately selected by those skilled in the art depending on the purpose. For example, the above-mentioned "n", which determines the length of the peptide linker, is usually 1 to 5, preferably 1 to 3, and more preferably 1 or 2.
[0235] Synthetic linkers (chemical crosslinking agents) include crosslinking agents that are routinely used to crosslink peptides, for example, N-hydroxy succinimide (NHS), disuccinimidyl suberate (DSS), bis(sulfosuccinimidyl) suberate (BS.sup.3), dithiobis(succinimidyl propionate) (DSP), dithiobis(sulfosuccinimidyl propionate) (DTSSP), ethylene glycol bis(succinimidyl succinate) (EGS), ethylene glycol bis(sulfosuccinimidyl succinate) (sulfo-EGS), disuccinimidyl tartrate (DST), disulfosuccinimidyl tartrate (sulfo-DST), bis[2-(succinimidoxycarbonyloxy)ethyl] sulfone (BSOCOES), and bis[2-(sulfosuccinimidoxycarbonyloxy)ethyl] sulfone (sulfo-BSOCOES). These crosslinking agents are commercially available.
[0236] When four antibody variable regions are linked, three linkers are usually required. Such multiple linkers may be the same or different.
[0237] The antibodies of the present invention include antibodies in which one or more amino acid residues have been added to the amino acid sequence of an antibody of the present invention. Further, fusion proteins which result from a fusion between one of the above antibodies and a second peptide or protein is included in the present invention. The fusion proteins can be prepared by ligating a polynucleotide encoding an antibody of the present invention and a polynucleotide encoding a second peptide or polypeptide in frame, inserting this into an expression vector, and expressing the fusion construct in a host. Some techniques known to those skilled in the art are available for this purpose. The partner peptide or polypeptide to be fused with an antibody of the present invention may be a known peptide, for example, FLAG (Hopp, T. P. et al., BioTechnology 6, 1204-1210 (1988)), 6.times. His consisting of six His (histidine) residues, 10.times. His, influenza hemagglutinin (HA), human c-myc fragment, VSV-GP fragment, p18HIV fragment, T7-tag, HSV-tag, E-tag, SV40 T antigen fragment, lck tag, .alpha.-tubulin fragment, B-tag, Protein C fragment. Other partner polypeptides to be fused with the antibodies of the present invention include, for example, GST (glutathione-S-transferase), HA (influenza hemagglutinin), immunoglobulin constant region, .beta.-galactosidase, and MBP (maltose-binding protein). A polynucleotide encoding one of these commercially available peptides or polypeptides can be fused with a polynucleotide encoding an antibody of the present invention. The fusion polypeptide can be prepared by expressing the fusion construct.
[0238] Furthermore, the antibodies of the present invention may be conjugated antibodies which are linked to any of various molecules including polymeric substances such as polyethylene glycol (PEG) and hyaluronic acid, radioactive substances, fluorescent substances, luminescent substances, enzymes, and toxins. Such conjugated antibodies can be obtained by chemically modifying the obtained antibodies. Methods for modifying antibodies have been established in this field (for example, U.S. Pat. Nos. 5,057,313 and 5,156,840). The "antibodies" of the present invention also include such conjugated antibodies.
[0239] Furthermore, the antibodies used in the present invention may be bispecific antibodies. The bispecific antibody refers to an antibody that has variable regions recognizing different epitopes in the same antibody molecule. In the present invention, the bispecific antibodies may recognize different epitopes on an NR10 molecule, or recognize NR10 with one antigen-binding site and a different substance with the other antigen-binding site.
[0240] Methods for producing bispecific antibodies are known. Bispecific antibodies can be prepared, for example, by linking two antibodies that recognize different antigens. Antibodies to be linked together may be half molecules each of which contains an H chain and an L chain, or quarter molecules that consist of only one H chain. Alternatively, hybridomas producing different monoclonal antibodies can be fused to produce a bispecific antibody-producing fused cell. Furthermore, bispecific antibodies can be produced by genetic engineering techniques.
[0241] The antibodies of the present invention may differ in amino acid sequence, molecular weight, isoelectric point, presence/absence of sugar chains, and conformation depending on the cell or host producing the antibody or the purification method as described below. However, a resulting antibody is included in the present invention, as long as it is functionally equivalent to an antibody of the present invention. For example, when an antibody of the present invention is expressed in prokaryotic cells, for example E. coli, a methionine residue is added to the N terminus of the original antibody amino acid sequence. Such antibodies are included in the present invention.
Antibody Production
[0242] The antibodies of the present invention may be polyclonal or monoclonal antibodies. Such monoclonal antibodies having NR10-binding and/or neutralizing activity can be obtained, for example, by the following procedure: anti-NR10 monoclonal antibodies are prepared by using as an antigen NR10 or a fragment thereof that is derived from a mammal such as human or mouse by known methods, and then antibodies having NR10-binding and/or neutralizing activity are selected from the thus obtained anti-NR10 monoclonal antibodies. Specifically, a desired antigen or cells expressing the desired antigen are used as a sensitizing antigen for immunization according to conventional immunization methods. Anti-NR10 monoclonal antibodies can be prepared by fusing the obtained immune cells with known parental cells using conventional cell fusion methods, and screening them for monoclonal antibody-producing cells (hybridomas) by conventional screening methods. Animals to be immunized include, for example, mammals such as mice, rats, rabbits, sheep, monkeys, goats, donkeys, cows, horses, and pigs. The antigen can be prepared using the known NR10 gene sequence according to known methods, for example, by methods using baculovirus (for example, WO 98/46777).
[0243] Hybridomas can be prepared, for example, according to the method of Milstein et al. (Kohler, G. and Milstein, C., Methods Enzymol. (1981) 73: 3-46) or such. When the immunogenicity of an antigen is low, immunization may be performed after linking the antigen with a macromolecule having immunogenicity, such as albumin.
[0244] Embodiments of the antibodies of the present invention that have a binding and/or neutralizing activity against NR10 include monoclonal antibodies that have a binding and/or neutralizing activity against human NR10. Antigens used to prepare monoclonal antibodies that have a binding and/or neutralizing activity against human NR10 are not particularly limited, as long as they enable preparation of antibodies that have a binding and/or neutralizing activity against human NR10. For example, it is known that there are a number of variants of human NR10, and any variant may be used as an immunogen as long as it enables preparation of antibodies that have a binding and/or neutralizing activity against human NR10. Alternatively, under the same condition, a peptide fragment of NR10 or a protein in which artificial mutations have been introduced into the natural NR10 sequence may be used as an immunogen. Human NR10.3 is one of preferred immunogens in preparing antibodies that have an activity of binding and/or neutralizing NR10 in the present invention.
[0245] Furthermore, the binding and/or neutralizing activity of antibody against NR10 can be measured, for example, by observing the effect of suppressing the growth of the IL-31-dependent cell line as described in the Examples.
[0246] Meanwhile, monoclonal antibodies can also be obtained by DNA immunization. DNA immunization is a method in which a vector DNA constructed such that the gene encoding an antigen protein can be expressed in an animal to be immunized is administered to the animal, and the immunogen is expressed within the body of the animal to provide immunostimulation. As compared to common immunization methods based on the administration of protein antigens, the DNA immunization is expected to be advantageous in that:
[0247] it enables immunostimulation while retaining the structure of a membrane protein; and
[0248] the immunogen does not need to be purified.
[0249] On the other hand, it is difficult to combine DNA immunization with an immunostimulating means such as an adjuvant.
[0250] In order to obtain a monoclonal antibody by DNA immunization, first, DNA encoding NR10 is administered to an animal to be immunized. The DNA encoding NR10 can be synthesized by known methods such as PCR. The resulting DNA is inserted into an appropriate expression vector, and administered to the animal to be immunized. Expression vectors that can be used include commercially available expression vectors such as pcDNA3.1. The vector can be administered to the living body by conventional methods. For example, DNA immunization can be carried out by introducing gold particles coated with the expression vector into cells by gene gun. Booster using NR10-expressing cells after DNA immunization is a preferred method to yield a monoclonal antibody.
[0251] Once the mammal is immunized as described above and the serum level of a desired antibody is confirmed to be increased, immune cells are collected from the mammal and subjected to cell fusion. Preferred immune cells are spleen cells in particular.
[0252] Mammalian myeloma cells are used for fusion with the above immune cells. It is preferred that myeloma cells have appropriate selection markers for screening. The selection marker refers to a phenotype that allows (or does not allow) survival under particular culture conditions. Known selection markers include hypoxanthine-guanine phosphoribosyltransferase deficiency (hereinafter abbreviated as "HGPRT deficiency") and thymidine kinase deficiency (hereinafter abbreviated as "TK deficiency"). HGPRT- or TK-deficient cells exhibit hypoxanthine-aminopterin-thymidine sensitivity (hereinafter abbreviated as "HAT sensitivity"). In HAT selection medium, HAT-sensitive cells cannot synthesize DNA and thus will die. However, when fused with normal cells, they can continue to synthesize DNA via the salvage pathway of the normal cells and thus can grow even in HAT selection medium.
[0253] HGPRT- or TK-deficient cells can be selected using a medium containing 6-thioguanine, 8-azaguanine (hereinafter abbreviated as "8AG"), or 5'-bromodeoxyuridine. While normal cells are killed due to incorporation of these pyrimidine analogs into DNA, cells lacking these enzymes can survive in the selection medium because they cannot incorporate these pyrimidine analogs. Another selection marker called G418 resistance confers resistance to 2-deoxystreptamine antibiotics (gentamicin analogs) due to the neomycin resistance gene. Various myeloma cells suitable for cell fusion are known.
[0254] Cell fusion between immune cells and myeloma cells can be essentially carried out according to known methods, for example, the method by Kohler and Milstein (Kohler. G. and Milstein, C., Methods Enzymol. (1981) 73, 3-46).
[0255] More specifically, cell fusion can be carried out, for example, in a common culture medium in the presence of a cell fusion-promoting agent. The fusion-promoting agent includes, for example, polyethylene glycol (PEG) and Sendai virus (HVJ). If required, an auxiliary agent such as dimethyl sulfoxide may also be added to improve fusion efficiency.
[0256] The immune cells and myeloma cells may be used at an arbitrarily determined ratio. For example, the ratio of immune cells to myeloma cells is preferably from 1 to 10. Culture media to be used for cell fusion include, for example, media that are suitable for the cell growth of myeloma cell line, such as RPMI 1640 and MEM, and other common culture media used for this type of cell culture. In addition, the culture media may also be supplemented with serum supplement such as fetal calf serum (FCS).
[0257] Predetermined amounts of immune cells and myeloma cells are mixed well in the culture medium, and then mixed with a PEG solution pre-heated to 37.degree. C. to produce fused cells (hybridomas). In the cell fusion method, for example, PEG with mean molecular weight of about 1,000-6,000 can be added to the cells typically at a concentration of 30% to 60% (w/v). Then, successive addition of the appropriate culture medium listed above and removal of supernatant by centrifugation are repeated to eliminate the cell fusion agent and such, which are unfavorable to the growth of hybridomas.
[0258] The resulting hybridomas can be screened using a selection medium according to the selection marker possessed by myeloma cells used in the cell fusion. For example, HGPRT- or TK-deficient cells can be screened by culturing them in a HAT medium (a medium containing hypoxanthine, aminopterin, and thymidine). Specifically, when HAT-sensitive myeloma cells are used in cell fusion, cells successfully fused with normal cells can be selectively grown in the HAT medium. The cell culture using the above HAT medium is continued for a sufficient period of time to allow all cells except the desired hybridomas (non-fused cells) to die. Specifically, in general, the desired hybridomas can be selected by culturing the cells for several days to several weeks. Then, screening and single cloning of hybridomas that produce an antibody of interest can be carried out by performing ordinary limiting dilution methods. Alternatively, antibodies that recognize NR10 can be prepared by the method described in WO 03/104453.
[0259] Screening and single cloning of an antibody of interest can be suitably carried out by known screening methods based on antigen-antibody reaction. For example, an antigen is bound to a carrier such as beads made of polystyrene or such and commercially available 96-well microtiter plates, and then reacted with the culture supernatant of hybridoma. Next, the carrier is washed and then reacted with an enzyme-labeled secondary antibody or such. When the culture supernatant contains an antibody of interest reactive to the sensitizing antigen, the secondary antibody binds to the carrier via this antibody. Finally, the secondary antibody bound to the carrier is detected to determine whether the culture supernatant contains the antibody of interest. Hybridomas producing a desired antibody capable of binding to the antigen can be cloned by the limiting dilution method or such. Not only the antigen used for immunization but also an NR10 protein substantially equivalent thereto can be preferably used as an antigen for this purpose. For example, a cell line expressing NR10, the extracellular domain of NR10, or an oligopeptide composed of a partial amino acid sequence constituting the domain may be used as the antigen.
[0260] In addition to the above-described method for preparing hybridomas through immunization of a nonhuman animal with an antigen, antibodies of interest can also be obtained by sensitizing human lymphocytes with an antigen. Specifically, first, human lymphocytes are sensitized with an NR10 protein in vitro. Then, the sensitized lymphocytes are fused with an appropriate fusion partner. For example, human-derived myeloma cells with the ability to divide permanently can be used as the fusion partner (see Japanese Patent Application Kokoku Publication No. (JP-B) H1-59878 (examined, approved Japanese patent application published for opposition). Antibodies obtained by this method are human antibodies having an activity of binding to the NR10 protein.
[0261] The nucleotide sequence encoding an anti-NR10 antibody obtained by the above-described method or such, and its amino acid sequence can be obtained by methods known to those skilled in the art.
[0262] Based on the obtained sequence of the anti-NR10 antibody, the anti-NR10 antibody can be prepared, for example, by genetic recombination techniques known to those skilled in the art. Specifically, a polynucleotide encoding an antibody can be constructed based on the sequence of the NR10-recognizing antibody, inserted into an expression vector, and then expressed in appropriate host cells (see for example, Co, M. S. et al., J. Immunol. (1994) 152, 2968-2976; Better, M. and Horwitz, A. H., Methods Enzymol. (1989) 178, 476-496; Pluckthun, A. and Skerra, A., Methods Enzymol. (1989) 178, 497-515; Lamoyi, E., Methods Enzymol. (1986) 121, 652-663; Rousseaux, J. et al., Methods Enzymol. (1986) 121, 663-669; Bird, R. E. and Walker, B. W., Trends Biotechnol. (1991) 9, 132-137).
[0263] The vectors include M13 vectors, pUC vectors, pBR322, pBluescript, and pCR-Script. Alternatively, when aiming to subclone and excise cDNA, the vectors include, for example, pGEM-T, pDIRECT, and pT7, in addition to the vectors described above. Expression vectors are particularly useful when using vectors for producing the antibodies of the present invention. For example, when aiming for expression in E. coli such as JM109, DH5.alpha., HB101, and XL1-Blue, the expression vectors not only have the above-described characteristics that allow vector amplification in E. coli, but must also carry a promoter that allows efficient expression in E. coli, for example, lacZ promoter (Ward et al., Nature (1989) 341, 544-546; FASEB J. (1992) 6, 2422-2427), araB promoter (Better et al., Science (1988) 240, 1041-1043), T7 promoter or such. Such vectors include pGEX-5X-1 (Pharmacia), "QIAexpress system" (Qiagen), pEGFP, or pET (in this case, the host is preferably BL21 that expresses T7 RNA polymerase) in addition to the vectors described above.
[0264] The vectors may contain signal sequences for antibody secretion. As a signal sequence for antibody secretion, a pelB signal sequence (Lei, S. P. et al J. Bacteriol. (1987) 169, 4379) may be used when a protein is secreted into the E. coli periplasm. The vector can be introduced into host cells by calcium chloride or electroporation methods, for example.
[0265] In addition to vectors for E. coli, the vectors for producing the antibodies of the present invention include mammalian expression vectors (for example, pcDNA3 (Invitrogen), pEF-BOS (Nucleic Acids. Res. 1990, 18(17), p5322), pEF, and pCDM8), insect cell-derived expression vectors (for example, the "Bac-to-BAC baculovirus expression system" (Gibco-BRL) and pBacPAK8), plant-derived expression vectors (for example, pMH1 and pMH2), animal virus-derived expression vectors (for example, pHSV, pMV, and pAdexLcw), retroviral expression vectors (for example, pZIPneo), yeast expression vectors (for example, "Pichia Expression Kit" (Invitrogen), pNV11, and SP-Q01), and Bacillus subtilis expression vectors (for example, pPL608 and pKTH50), for example.
[0266] When aiming for expression in animal cells such as CHO, COS, and NIH3T3 cells, the vectors must have a promoter essential for expression in cells, for example, SV40 promoter (Mulligan et al., Nature (1979) 277, 108), MMLV-LTR promoter, EF1.alpha. promoter (Mizushima et al., Nucleic Acids Res. (1990) 18, 5322), and CMV promoter, and more preferably they have a gene for selecting transformed cells (for example, a drug resistance gene that allows evaluation using an agent (neomycin, G418, or such). Vectors with such characteristics include pMAM, pDR2, pBK-RSV, pBK-CMV, pOPRSV, and pOP13, for example.
[0267] In addition, the following method can be used for stable gene expression and gene amplification in cells: CHO cells deficient in a nucleic acid synthesis pathway are introduced with a vector (for example, pSV2-dhfr (Molecular Cloning 2.sup.nd edition, Cold Spring Harbor Laboratory Press, 1989)) that carries a DHFR gene which compensates for the deficiency, and the vector is amplified using methotrexate (MTX). Alternatively, the following method can be used for transient gene expression: COS cells with a gene expressing SV40 T antigen on their chromosome are transformed with a vector (pcD and such) with an SV40 replication origin. Replication origins derived from polyoma virus, adenovirus, bovine papilloma virus (BPV), and such can also be used. To amplify gene copy number in host cells, the expression vectors may further carry selection markers such as aminoglycoside transferase (APH) gene, thymidine kinase (TK) gene, E. coli xanthine-guanine phosphoribosyltransferase (Ecogpt) gene, and dihydrofolate reductase (dhfr) gene.
[0268] The antibodies of the present invention obtained by the methods described above can be isolated from inside host cells or from outside the cells (the medium, or such), and purified to homogeneity. The antibodies can be isolated and purified by methods routinely used for isolating and purifying antibodies, and the type of method is not limited. For example, the antibodies can be isolated and purified by appropriately selecting and combining column chromatography, filtration, ultrafiltration, salting out, solvent precipitation, solvent extraction, distillation, immunoprecipitation, SDS-polyacrylamide gel electrophoresis, isoelectrofocusing, dialysis, recrystallization, and such.
[0269] The chromatographies include, for example, affinity chromatography, ion exchange chromatography, hydrophobic chromatography, gel filtration, reverse phase chromatography, and adsorption chromatography (Strategies for Protein Purification and Characterization: A Laboratory Course Manual. Ed Daniel R. Marshak et al., Cold Spring Harbor Laboratory Press, 1996). The chromatographic methods described above can be conducted using liquid chromatography, for example, HPLC and FPLC. Columns that can be used for affinity chromatography include protein A columns and protein G columns. Columns using protein A include, for example, Hyper D, POROS, and Sepharose FF (GE Amersham Biosciences). The present invention includes antibodies that are highly purified using these purification methods.
[0270] The NR10-binding activity of the obtained antibodies can be determined by methods known to those skilled in the art. Methods for determining the antigen-binding activity of an antibody include, for example, ELISA (enzyme-linked immunosorbent assay), EIA (enzyme immunoassay), RIA (radioimmunoassay), and fluorescent antibody method. For example, when enzyme immunoassay is used, antibody-containing samples, such as purified antibodies and culture supernatants of antibody-producing cells, are added to antigen-coated plates. A secondary antibody labeled with an enzyme, such as alkaline phosphatase, is added and the plates are incubated. After washing, an enzyme substrate, such as p-nitrophenyl phosphate, is added, and the absorbance is measured to evaluate the antigen-binding activity.
Pharmaceutical Compositions
[0271] The present invention also provides pharmaceutical compositions comprising the antibody mentioned above as an active ingredient. Moreover, the present invention provides therapeutic agents for inflammatory diseases which comprise the antibody mentioned above as an active ingredient.
[0272] In the present invention, inflammatory disease refers to diseases with pathological features involved in cytological and histological reactions that occur in affected blood vessels and adjacent tissues in response to an injury or abnormal stimulation caused by physical, chemical, or biological agents (Stedman's Medical Dictionary, 5th Ed., MEDICAL VIEW CO., 2005). Generally, inflammatory diseases include, dermatitis (atopic dermatitis, chronic dermatitis, and such), inflammatory bowel diseases (colitis and such), asthma, arthritis (rheumatoid arthritis, osteoarthritis, and such), bronchitis, Th2 autoimmune diseases, systemic lupus erythematosus, myasthenia gravis, chronic GVHD, Crohn's disease, spondylitis deformans, lumbar pain, gout, inflammation after surgery or injury, swelling, neuralgia, laryngopharyngitis, cystitis, hepatitis (non-alcoholic steatohepatitis, alcoholic hepatitis, and such), hepatitis B, hepatitis C, arteriosclerosis, and pruritus.
[0273] Preferred examples of inflammatory diseases that are subjects of the present invention include atopic dermatitis, chronic dermatitis, rheumatism, osteoarthritis, chronic asthma, and pruritus.
[0274] The phrase "comprise(s) an anti-NR10 antibody as an active ingredient" means comprising an anti-NR10 antibody as at least one of the active ingredients, and does not limit the proportion of the antibody. In addition, the therapeutic agents for inflammatory diseases in the present invention may also comprise, in combination with the anti-NR10 antibody mentioned above, other ingredients that enhance the treatment of inflammatory diseases.
[0275] The therapeutic agents of the present invention may also be used for preventive purposes.
[0276] The anti-NR10 antibody of the present invention may be prepared as formulations according to standard methods (see, for example, Remington's Pharmaceutical Science, latest edition, Mark Publishing Company, Easton, USA). Further, they may contain pharmaceutically acceptable carriers and/or additives if necessary. For example, they may contain surfactants (for example, PEG and Tween), excipients, antioxidants (for example, ascorbic acid), coloring agents, flavoring agents, preservatives, stabilizers, buffering agents (for example, phosphoric acid, citric acid, and other organic acids), chelating agents (for example, EDTA), suspending agents, isotonizing agents, binders, disintegrators, lubricants, fluidity promoters, and corrigents. However, without limitation to these, the agents for preventing or treating inflammatory diseases of the present invention may contain other commonly used carriers. Such carriers specifically include light anhydrous silicic acid, lactose, crystalline cellulose, mannitol, starch, carmelose calcium, carmelose sodium, hydroxypropylcellulose, hydroxypropylmethylcellulose, polyvinylacetaldiethylaminoacetate, polyvinylpyrrolidone, gelatin, medium chain fatty acid triglyceride, polyoxyethylene hydrogenated castor oil 60, sucrose, carboxymethylcellulose, corn starch, and inorganic salt. The agents may also contain other low-molecular-weight polypeptides, proteins such as serum albumin, gelatin, and immunoglobulin, and amino acids such as glycine, glutamine, asparagine, arginine, and lysine. When the anti-NR10 antibody is prepared as an aqueous solution for injection, the anti-NR10 antibody may be dissolved in an isotonic solution containing, for example, physiological saline, dextrose, or other adjuvants. The adjuvants may include, for example, D-sorbitol, D-mannose, D-mannitol, and sodium chloride. In addition, appropriate solubilizing agents, for example, alcohols (for example, ethanol), polyalcohols (for example, propylene glycols and PEGs), and non-ionic detergents (polysorbate 80 and HCO-50) may be used concomitantly.
[0277] If necessary, anti-NR10 antibodies may be encapsulated in microcapsules (microcapsules made of hydroxymethylcellulose, gelatin, polymethylmethacrylate, and the like), and made into components of colloidal drug delivery systems (liposomes, albumin microspheres, microemulsions, nano-particles, and nano-capsules) (for example, see "Remington's Pharmaceutical Science 16th edition" &, Oslo Ed. (1980)). Moreover, methods for making sustained-release drugs are known, and these can be applied for anti-NR10 antibodies (Langer et al., J. Biomed. Mater. Res. (1981) 15, 167-277; Langer, Chem. Tech. (1982) 12, 98-105; U.S. Pat. No. 3,773,919; European Patent Application (EP) No. 58,481; Sidman et al., Biopolymers (1983) 22, 547-56; EP 133,988).
[0278] The pharmaceutical compositions of the present invention can be administered either orally or parenterally, but are preferably administered parenterally. Specifically, the agents are administered to patients by injection or percutaneous administration. Injections include, for example, intravenous injections, intramuscular injections, and subcutaneous injections, for systemic or local administration. The agents may be given to sites where inflammation is to be suppressed, or areas surrounding the sites by local infusion, intramuscular injection in particular. The administration methods can be properly selected according to the patient's age and condition. The single-administration dose can be selected, for example, from within the range of 0.0001 to 100 mg of the active ingredient per kg body weight. Alternatively, for example, when the agents are administered to human patients, the dose of the active ingredient can be selected from within the range of 0.001 to 1,000 mg/kg body weight. The single-administration dose preferably contains, for example, about 0.01 to 50 mg/kg body weight of the antibody of the present invention. However, the dose of an agent for preventing or treating inflammatory diseases of the present invention is not limited to these examples.
[0279] All prior-art documents cited in the present specification are herein incorporated by reference.
EXAMPLES
[0280] Herein below, the present invention will be specifically described with reference to Examples, but it is not to be construed as being limited thereto.
[Example 1] Preparation of Hybridomas
1.1. Preparation of Human and Cynomolgus Monkey NR10 Plasmids for DNA Immunization
[0281] 1.1.1. Preparation of Expression Vectors for hNR10 and cynNR10
[0282] Human NR10 (nucleotide sequence, SEQ ID NO: 75; amino acid sequence, SEQ ID NO: 76) was inserted into the expression vector pMacII, which expresses a protein under the control of mouse .beta.-actin promoter (WO2005/054467), to prepare an expression vector for hNR10. In the same manner, an expression vector for cynNR10 was constructed from cynomolgus monkey NR10 (nucleotide sequence, SEQ ID NO: 65; amino acid sequence, SEQ ID NO: 66).
1.1.2. Preparation of DNA Cartridge
[0283] In order to use the hNR10 or cynNR10 expression vector prepared in 1.1.1 for DNA immunization of mice, the Helios Gene Gun Cartridge Kit (BIO-RAD) was used to produce a DNA cartridge for each DNA that allows immunization with 1 .mu.g of DNA at one time.
1.2. Preparation of Hybridomas Producing Anti-Human NR10 Antibody
[0284] 1.2.1. Preparation of Hybridomas Using Mice Immunized with Human or Cynomolgus Monkey
NR10
[0285] Ten Balb/c mice (female; six weeks old at the beginning of immunization; Charles River Laboratories Japan) were immunized with human or cynomolgus monkey NR10 by the following procedure. For primary immunization, the mice were immunized with the DNA cartridge prepared with the hNR10 expression vector using the Helios Gene Gun System (BIO-RAD). One week later, secondary immunization was performed by the Helios Gene Gun System (BIO-RAD) using the DNA cartridge prepared with the cynNR10 expression vector. The third and subsequent immunizations were carried out at one-week intervals using the hNR10 and cynNR10 expression vectors alternately. After the titer of serum antibody against human NR10 was confirmed to be elevated, a human NR10 protein (extracellular domain) (Referential Example 4) diluted with PBS(-) was intravenously administered at 10 ng/head as the final immunization. Four days after the final immunization, mouse spleen cells were fused with mouse myeloma P3X63Ag8U.1 cells (abbreviated as P3U1; ATCC CRL-1597) by a conventional method using PEG1500 (Roche Diagnostics). The resulting fused cells, i.e., hybridomas, were cultured in RPMI1640 supplemented with 10% FBS (hereinafter abbreviated as 10% FBS/RPMI1640).
1.2.2. Selection of Hybridomas
[0286] On the next day of fusion, the fused cells were suspended in a semisolid medium (StemCells), and cultured for selection as well as colonization of hybridomas.
[0287] After nine or ten days of fusion, hybridoma colonies were picked up and each colony was seeded into each well of 96-well plates containing the HAT selection medium (10% FBS/RPMI1640, 2 vol % of HAT 50.times. concentrate (Dainippon Pharmaceutical), and 5 vol % of BM-Condimed H1 (Roche Diagnostics)). After three to four days of culture, the culture supernatant was collected from each well to determine the concentration of mouse IgG in the supernatant. The culture supernatants in which mouse IgG was detected were assessed for a neutralizing activity using a human IL-31-dependent cell line (hNR10/hOSMR/BaF3 cells; Referential Example 2), and several clones having a strong NR10-neutralizing activity were obtained (FIG. 3). Clones that suppress the human IL-31-induced growth of cells in a concentration-dependent manner and suppress the cynomolgus monkey IL-31-induced growth of cells (cynNR10/cynOSMR/BaF3 cells; Referential Example 2) in a concentration-dependent manner were obtained (FIG. 4).
[Example 2] Preparation of Chimeric Antibodies
Preparation of Expression Vectors for Chimeric Antibodies
[0288] Total RNAs were extracted from the hybridomas using RNeasy Mini Kits (QIAGEN), and cDNAs were synthesized from them using SMART RACE cDNA Amplification Kit (BD Biosciences). Antibody variable region genes were isolated by PCR using PrimeSTAR HS DNA polymerase (TaKaRa), 10.times. Universal Primer A Mix attached to SMART RACE cDNA Amplification Kit (BD Biosciences), and primers designed for each antibody constant region (H chain, mIgG1-rnot; L chain, mIgK-rnot). The nucleotide sequence of each isolated DNA fragment was determined with ABI PRISM 3730xL DNA Sequencer or ABI PRISM 3700 DNA Sequencer (Applied Biosystems), using BigDye Terminator Cycle Sequencing Kit (Applied Biosystems) according to the method described in the appended instruction manual. The determined amino acid sequences of H chain and L chain variable regions in the mouse antibodies NS18, NS22, NS23, and NS33 were shown in FIGS. 1 and 2, respectively.
[0289] Each of the resulting H and L chain fragments was subjected to PCR using PrimeSTAR HS DNA Polymerase (TaKaRa) and the primer sets shown in Table 1. The resulting amplified fragments were ligated with the constant region (human .gamma.1 or .gamma.2, and human .kappa., respectively), and then inserted into an animal cell expression vector. The nucleotide sequence of each DNA fragment was determined with ABI PRISM 3730xL DNA Sequencer or ABI PRISM 3700 DNA Sequencer (Applied Biosystems), using BigDye Terminator Cycle Sequencing Kit (Applied Biosystems) according to the method described in the appended instruction manual.
TABLE-US-00002 TABLE 1 Sequence (5' .fwdarw.3') SEQ ID NO: mIgG1-rnot TAATAGCGGCCGCTCATTATTTACCAGGAGAGTGGGAGAG 90 mIgK-rnot TAATAGCGGCCGCTCATTAACACTCATTCCTGTTGAAGCT 91 mNS18H-feco GACGAATTCCACCATGGGATGGAGCTGGATCTT 92 mNS18L-feco GACGAATTCCACCATGAGTGTGCCCACTCAGGT 93 mNS33H-feco GACGAATTCCACCATGGAATGTAACTGGATACT 94 mNS33L-feco GACGAATTCCACCATGGATTTTCTGGTGCAGAT 95 Forward primer Reverse primer NS18 H chain mNS18H-feco mIG1-rnot NS18 L chain mNS18L-feco mIGK-rnot NS22 H chain Mns18H-feco mIG1-rnot NS22 L chain mNS18L-feco mIGK-rnot NS23 H chain mNS18H-feco mIG1-rnot NS23 L chain mNS18L-feco mIGK-rnot NS33 H chain mNS33H-feco mIG1-rnot NS33 L chain mNS33L-feco mIGK-rnot
Preparation of Chimeric Antibodies
[0290] Human embryonic kidney cancer cell line HEK293H (Invitrogen) was suspended in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), and 10 ml of cells were seeded into dishes for adherent cells (10 cm in diameter; CORNING) at a cell density of 6.times.10.sup.5 cells/ml. The cells were incubated in a CO.sub.2 incubator (37.degree. C., 5% CO.sub.2) for one whole day and night. Then, the medium was removed by aspiration, and 6.9 ml of CHO-S-SFMII medium (Invitrogen) was added. CHO-S-SFMII medium was added to the prepared plasmid DNA mixture (13.8 .mu.g in total) to a volume of 700 .mu.l. This was mixed with 20.7 .mu.l of 1 .mu.g/ml polyethyleneimine (Polysciences Inc.), and allowed to stand at room temperature for 10 minutes. The solution was added to the cells in each dish. The cells were incubated in a CO.sub.2 incubator (37.degree. C., 5% CO.sub.2) for four to five hours. Then, 6.9 ml of CHO-S-SFMII medium (Invitrogen) was added, and the cells were incubated in a CO.sub.2 incubator for three to four days. The culture supernatants were collected and then centrifuged (approx. 2000 g, five minutes, room temperature) to remove the cells. The supernatants were filtered through 0.22-.mu.m filter MILLEX.RTM.-GV (Millipore). Each sample was stored at 4.degree. C. until use. Antibodies were purified from the supernatants using Protein G Sepharose (Amersham Biosciences). The purified antibodies were concentrated with Amicon Ultra 15 (Millipore), and then the solvent was replaced with PBS(-) containing 0.05% NaN.sub.3 using PD-10 Desalting columns (Amersham Biosciences. The absorbance at 280 nm was measured with ND-1000 Spectrophotometer (NanoDrop), and the concentrations were determined by the method of Pace et al. (Protein Science (1995) 4: 2411-2423).
Assessment of the Activity of Chimeric NS22
[0291] The activity of neutralizing hIL-31 was assessed using the hNR10/hOSMR/BaF3 cell line, which grows in an hIL-31 dose-dependent manner, as described below.
[0292] hNR10/hOSMR/BaF3 cells were prepared at 1.5.times.10.sup.5 cells/ml using RPMI1640 medium (GIBCO) containing 10% FBS (MOREGATE) and 1% Penicillin-Streptomycin (Invitrogen). hIL-31 (R&D Systems) was added to an aliquot of the cells to a final concentration of 4 ng/ml (IL-31(+); final conc.: 2 ng/ml). The remaining cell suspension was used as IL-31(-). The purified NS22 was adjusted to 2 .mu.g/ml using the medium, and eight serial dilutions were prepared at a common dilution ratio of 3 (final conc.: 1 .mu.g/ml or less). 50 .mu.l each of the cell suspension and the dilution of chimeric NS22 (human .gamma.1, .kappa.) was added to each well of 96-well flat-bottom plates (CORNING), and the cells were cultured in a 5% CO.sub.2 incubator at 37.degree. C. for two days. After culture, 20 .mu.l of a mixture of equal amounts of Cell Counting Kit-8 (Dojindo) and PBS was added to each well, and the absorbance (450 nm/620 nm) was measured (TECAN, SUNRISE CLASSIC). After the reaction was allowed to continue for two hours in a 5% CO.sub.2 incubator at 37.degree. C., the absorbance was measured again. The neutralizing activity of NS22 was presented as an inhibition rate using a value obtained by subtracting the 0-hour value from the 2-hour value. The result showed that NS22 suppressed the IL-31-induced growth of the hNR10/hOSMR/BaF3 cell line in a concentration-dependent manner. This demonstrates that NS22 has a neutralizing activity against the human IL-31 signaling (FIG. 5).
[0293] The IL-31-neutralizing activity was assessed as described below using the DU145 cell line (human prostate cancer cell line), in which IL-6 production is induced upon IL-31 stimulation.
[0294] DU145 cells were prepared at 2.5.times.10.sup.5 cells/ml in MEM (Invitrogen) containing 10% FBS (MOREGATE), 2 mmol/1 L-glutamine (Invitrogen), and 1 mmol/1 sodium pyruvate (SIGMA), and 200-.mu.1 aliquots were dispensed into each well of 48-well plates (CORNING). The cells were incubated at 37.degree. C. under 5% CO.sub.2 overnight. The purified chimeric NS22 (human .gamma.1, .kappa.) was diluted to 100 .mu.g/ml with MEM containing 10% FBS, 2 mmol/1 L-glutamine, and sodium pyruvate. Using this solution, six serial dilutions were prepared at a common dilution ratio of 5. Each dilution was combined with 100 ng/ml human interleukin-31 (R&D systems) at a ratio of 1:1, and a 50-.mu.l aliquot was added to each well. After two days of culture at 37.degree. C. under 5% CO.sub.2, the concentration of IL-6 in the culture supernatant was determined using DuoSet ELISA Development kit (R&D systems). The neutralizing activity of NS22 was assessed by determining the inhibition rate (%). Specifically, assuming the IL-6 concentration in the absence of IL-31 (A) as the maximal inhibitory activity (100% inhibition) and the IL-6 concentration in the presence of IL-31 without NS22 (B) as no inhibitory activity (0% inhibition), the IL-6 concentration in the presence of IL-31 and NS22 (C) was determined according to the following formula:
Inhibition rate (%)=(B-C)/(B-A).times.100
[0295] The result showed that NS22 suppressed the IL-31-induced IL-6 production in the DU145 cell line in a concentration-dependent manner and thus demonstrated that NS22 had a neutralizing activity against the human IL-31 signaling (FIG. 6).
Assessment of Competition of Chimeric Anti-NR10 Antibody with IL-31
[0296] Human IL-31 (R&D Systems) was labeled with FMAT Blue Monofunctional Reactive Dye (Applied Biosystems). 100 .mu.l of hIL-31 prepared at 0.5 mg/ml using 50 mM sodium phosphate buffer (pH 8.0) was mixed with 5.25 .mu.l of 25 nmoles FMAT Blue dissolved in DMSO (Junsei). After vortexing, the mixture was allowed to stand at room temperature for 15 minutes. The FMAT Blue-conjugating reaction with hIL-31 was terminated by adding 5 .mu.l of 1 M Tris-HCl (pH 7.4) and 1.1 .mu.l of 10% Tween20, and then FMAT Blue-labeled hIL-31 and unreacted FMAT Blue were separated by gel filtration using Superdex 75 (GE Healthcare, 17-0771-01) column with 0.1% Tween20/PBS developing solution.
[0297] Antibodies were assessed for the activity of inhibiting the IL-31/NR10 binding by using hNR10-expressing CHO cells as described below.
[0298] NS22 and NA633 (the constant region of each is .gamma.1, .kappa.) were diluted at an appropriate concentration using Assay buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl.sub.2, 3 mM MgCl.sub.2, 2% FBS, 0.01% NaN.sub.3), and then seven serial dilutions were prepared at a common dilution ratio of 2. The dilutions were added at 40 .mu.l/well to plates (96-Well FMAT Plates; Applied Biosystems). Then, FMAT Blue-labeled hIL-31 was diluted 400 times with Assay buffer and added at 20 .mu.l/well. Finally, cell suspensions adjusted to 2.5.times.10.sup.5 cells/ml using Assay buffer were added at 40 .mu.l/well (final 1.times.10.sup.4 cells/well). Two hours after addition of cells, the fluorescence (FL1) was determined using the 8200 Cellular Detection System (Applied Biosystems). The result showed that NS22 inhibited the binding of hIL-31/hNR10 in a dose-dependent manner, and demonstrated that its activity was superior to that of NA633 (FIG. 7).
[Example 3] Competition of Anti-NR10 Antibody Against NR10
[0299] The antibody NS22 purified from a hybridoma culture supernatant was labeled with FMAT Blue (Applied Biosystems, 4328853). 170 .mu.l of NS22 prepared at 1 mg/ml in PBS was mixed with 17 .mu.l of 1 M NaHCO.sub.3 solution and 3.4 .mu.l of FMAT Blue (17 nmoles) dissolved in DMSO. After vortexing, the mixture was allowed to stand at room temperature for 30 minutes. The FMAT Blue conjugating reaction with NS22 was terminated by adding 8 .mu.l of 1 M Tris-HCl (pH 7.4) and 1.9 .mu.l of 1% Tween 20, and then FMAT Blue-labeled NS22 (FMAT Blue-NS22) and unreacted FMAT Blue were separated by gel filtration using Superdex 75 (GE Healthcare, 17-0771-01) column with 0.01% Tween20/PBS developing solution.
[0300] Each antibody was examined for inhibition of the binding of the prepared FMAT Blue-NS22 to hNR10-expressing CHO cells (Referential Example 3) using the 8200 Cellular Detection System (Applied Biosystems, 4342920). The chimeric anti-NR10 antibodies (the constant region of each is .gamma.1, .kappa.) were added at various concentrations to each well containing 7500 cells and 8.8.times.10.sup.-2 .mu.g/ml FMAT Blue-NS22. The cells were allowed to stand in the dark for four hours, and then the fluorescent signal from FMAT Blue bound to the cells was measured. The reaction was carried out in 10 mM Hepes-KOH containing 2.5 mM CaCl.sub.2, 3 mM MgCl.sub.2, 140 mM NaCl, 2% FBS, and 0.01% NaNO.sub.3. The result is shown in FIG. 8. The fluorescence value FL1, which represents the binding of FMAT Blue-NS22 to NR10-expressing cells, was reduced with the increase in the concentration of antibody NS22 or NS23. On the other hand, FL1 was hardly reduced with the increase in the concentration of antibody NA633 (Referential Example 6) (FIG. 8).
[Example 4] Humanization of NS22 Antibody
[0301] Selection of Each Framework Sequence
[0302] The variable regions of mouse NS22 antibody were compared with human germline sequences. FR sequences used for humanization are summarized in Table 2. CDRs and FRs were determined based on the Kabat numbering. The humanized variable region sequences of H chain composed of FR1, FR2, FR3_1, and FR4, and composed of FR1, FR2, FR3_2, and FR4, which are listed in Table 2, are designated as H0-VH (SEQ ID NO: 50) and H1-VH (SEQ ID NO: 112), respectively. Meanwhile, the sequence of L chain composed of FR1, FR2, FR3, and FR4 is designated as L0 (SEQ ID NO: 52).
Preparation of Variable Region for Humanized NS22 H0L0
[0303] Synthetic oligo DNAs were designed for each of the H and L chains to construct the variable regions of humanized NS22 in which the CDRs of NS22 are grafted onto the FRs used for humanization. The respective synthetic oligo DNAs were mixed, and then subjected to assembly PCR to construct a gene encoding the variable region of humanized NS22. The assembly PCR was carried out using KOD-Plus (TOYOBO) according to the following conditions. A reaction mixture containing 10 pmol synthetic oligo DNAs and the appended PCR Buffer, dNTPs, MgSO.sub.4, and KOD-Plus was heated at 94.degree. C. for five minutes. The mixture was then subjected to two PCR cycles of 94.degree. C. for two minutes, 55.degree. C. for two minutes, and 68.degree. C. for two minutes. Then, 10 pmol each of a primer in which a restriction site and Kozak sequence has been added to the 5' end of the variable region, and a primer in which a restriction site has been added to the 3' end of the variable region, was added and subjected to 35 PCR cycles of 94.degree. C. for 30 seconds, 55.degree. C. for 30 seconds, and 68.degree. C. for one minute to yield a amplified fragment. The resulting amplified fragment was cloned into TOPO TA Cloning vector (TOYOBO), and its nucleotide sequence was determined by sequencing. The constructed variable regions were combined with the constant regions to prepare H0-SKSC (SEQ ID NO: 54) and L0 (SEQ ID NO: 56). The resulting construct was inserted into an expression vector capable of expressing the inserted gene in animal cells. The nucleotide sequence of each DNA fragment was determined using BigDye Terminator Cycle Sequencing Kit (Applied Biosystems) with ABI PRISM 3730xL DNA Sequencer or ABI PRISM 3700 DNA Sequencer (Applied Biosystems) according to the method described in the appended instruction manual.
Preparation of Variable Region for Humanized NS22 H1
[0304] H1-SKSC (SEQ ID NO: 130) was generated by substituting the glutamine (E) at Kabat-numbering position 73 in FR3 of H0-SKSC (SEQ ID NO: 54) with lysine (K). The mutant was prepared using commercially available QuikChange Site-Directed Mutagenesis Kit (Stratagene) according to the appended instruction manual.
Expression of IgG-Converted Antibody
[0305] Antibody expression was performed by the method described below. Human fetal renal cancer cell-derived cell line HEK293H (Invitrogen) was suspended in DMEM (Invitrogen) containing 10% fetal bovine serum (Invitrogen), and 10 ml of cells at a density of 5-6.times.10.sup.5 cells/ml was seeded onto dishes for adherent cells (10 cm in diameter; CORNING). The cells were incubated in a CO.sub.2 incubator (37.degree. C., 5% CO.sub.2) for one whole day and night. Then, the medium was removed by aspiration, and 6.9 ml of CHO-S-SFMII medium (Invitrogen) was added to the cells. The prepared plasmid DNA mixture (13.8 .mu.g in total) was mixed with 20.7 .mu.l of 1 .mu.g/ml polyethyleneimine (Polysciences Inc.) and 690 .mu.l of CHO-S-SFMII medium, and allowed to stand at room temperature for 10 minutes. The mixture was added to the cells in each dish, and the cells were incubated in a CO.sub.2 incubator (5% CO.sub.2, 37.degree. C.) for four to five hours. Then, 6.9 ml of CHO-S-SFMII medium (Invitrogen) was added, and the cells were incubated in a CO.sub.2 incubator for three days. The culture supernatant was collected and centrifuged (approx. 2000 g, five minutes, room temperature) to remove the cells. The supernatant was then sterilized by filtration through 0.22-.mu.m filter MILLEX.RTM.-GV (Millipore). Each sample was stored at 4.degree. C. until use.
Purification of IgG-Converted Antibody
[0306] 50 .mu.l of rProtein A Sepharose.TM. Fast Flow (Amersham Biosciences) suspended in TBS was added to the obtained culture supernatant, and mixed by inversion at 4.degree. C. for four hours or more. The solution was transferred to 0.22-.mu.m filter cup of Ultrafree.RTM.-MC (Millipore). After three washes with 500 .mu.l of TBS, rProtein A Sepharose.TM. resin was suspended in 100 .mu.l of aqueous solution of 50 mM sodium acetate (pH 3.3), and allowed to stand for three minutes to elute the antibody. The solution was immediately neutralized by adding 6.7 .mu.l of 1.5 M Tris-HCl (pH 7.8). The elution was performed twice and 200 .mu.l of purified antibody was obtained. 2 .mu.l of the antibody-containing solution was subjected to ND-1000 Spectrophotometer (NanoDrop)(Thermo Scientific NanoDrop.TM. 1000 Spectrophotometer (Thermo Scientific)) or 50 .mu.l was subjected to Spectrophotometer DU-600 (BECKMAN) to measure absorbance at 280 nm, and the antibody concentration was calculated by the method of Pace et al. (Protein Science (1995) 4: 2411-2423).
Measurement of Competition with IL-31 Using FMAT
[0307] Antibodies were assessed for the activity of inhibiting the IL-31/NR10 binding by using hNR10-expressing CHO cells as described below. The chimeric NS22 antibody and NS22_H0L0 (H chain, H0-SKSC/SEQ ID NO: 54; L chain, L0/SEQ ID NO: 56) were diluted at an appropriate concentration using Assay buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl.sub.2, 3 mM MgCl.sub.2, 2% FBS, 0.01% NaN.sub.3, pH7.4), and further eight serial dilutions were prepared at a common dilution ration of 2. The dilutions were added at 40 .mu.l/well to plates (96-Well FMAT Plates, Applied Biosystems). Then, FMAT Blue-labeled hIL-31 was diluted 400 times with Assay buffer, and added at 20 .mu.l/well. Finally, a cell suspension adjusted to 2.5.times.10.sup.5 cells/ml using Assay buffer was added at 40 .mu.l/well (final 1.times.10.sup.4 cells/well). Two hours after addition of cells, the fluorescence (FL1) was measured using the 8200 Cellular Detection System (Applied Biosystems).
[0308] The result showed that, as shown in FIG. 9, humanized NS22 antibodies H0L0 (H chain, H0-SKSC/SEQ ID NO: 54; L chain, L0/SEQ ID NO: 56), and H1L0 (H chain, H1-SKSC/SEQ ID NO: 130; L chain, L0/SEQ ID NO: 56) exhibited a competition activity comparable to that of the chimeric antibody, suggesting that both H0L0 and H1L0 are humanized anti-IL-31 receptor antibodies. In addition, it is considered that the FRs used for H0L0 and H1L0 can be used for humanization.
[0309] Accordingly, all of the mutations in CDRs described in the Examples hereinafter can be introduced into both H0 and H1.
TABLE-US-00003 TABLE 2 HO Germline Human FR sequence FR1 Germline: hVH_1_46(Accession No. X92343) QVQLVQSGAEVKKPGASVKVSCKASGYTFT (SEQ ID NO: 96) FR2 Germline: hVH_1_46 (Accession No. X92343) WVRQAPGQGLEWMG (SEQ ID NO: 97) FR3_1 Germline: hVH_1_69 (Accession No. L22582) RVTITADESTSTAYMELSSLRSEDTAVYYCAR (SEQ ID NO: 98) FR3_2 Germline: hVH_1_69 (Accession No. Z27506) RVTITADKSTSTAYMELSSLRSEDTAVYYCAR (SEQ ID NO: 131) FR4 Germline: JH1 WGQGTLVTVSS (SEQ ID NO: 99) LO Germline Human FR sequence FR1 Germline: hVK_1_39 (Accession No. X59315) DIQMTQSPSSLSASVGDRVTITC (SEQ ID NO: 100) FR2 Germline : hVK_1_39 (Accession No. X59315) WYQQKPGKAPKLLIY (SEQ ID NO: 101) FR3 Germline: hVK_1_39 (Accession No. X59315) GVPSRFSGSGSGTDFTLTISSLQPEDFATYYC (SEQ ID NO: 102) FR4 Germline: JK4 FGGGTKVEIK (SEQ ID NO: 103)
[Example 5] Heterogeneity-Reducing Effect of Novel Constant Regions M14 and M58 in Humanized Anti-IL31 Receptor Antibody
[0310] As shown in Referential Examples 7 to 9, it was demonstrated that the conversion of the constant region from IgG2 to M14 or M58 in the huPM1 antibody, a humanized anti-IL-6 receptor antibody, could reduce the heterogeneity derived from the IgG2 hinge region without loss of stability. Thus, humanized anti-IL-31 receptor antibodies were also tested to assess whether the heterogeneity can be reduced by converting their constant regions from the wild-type IgG2 to M14 or M58.
[0311] H0-M14, H0-M58, H0-IgG1, and H0-IgG2, which were generated by combining IgG1 (SEQ ID NO: 60), IgG2 (SEQ ID NO: 132), M14 (SEQ ID NO: 129) and M58 (SEQ ID NO: 128) generated in Referential Examples 8 and 9, with H chain variable region H0 (H0-VH/SEQ ID NO: 50) of humanized anti-IL-31 receptor antibody generated in Example 4, were used as H chains, and L0 (L0/SEQ ID NO: 56) produced in Example 4 was used as an L chain, to generate H0L0-IgG1 (H chain, H0-IgG1/SEQ ID NO: 133; L chain, L0/SEQ ID NO: 56), H0L0-IgG2 (H chain, H0-IgG2/SEQ ID NO: 134; L chain, L0/SEQ ID NO: 56), H0L0-M14 (H chain, H0-M14/SEQ ID NO: 135; L chain, L0/SEQ ID NO: 56), and H0L0-M58 (H chain, H0-M58/SEQ ID NO: 136; L chain, L0/SEQ ID NO: 56). Each antibody was expressed and purified by the method described in Example 4.
[0312] The heterogeneity was assessed by cation exchange chromatography. The prepared antibodies were assessed for heterogeneity using ProPac WCX-10 (Dionex) column, 20 mM sodium acetate (pH 5.0) as mobile phase A, and 20 mM sodium acetate/1M NaCl (pH 5.0) as mobile phase B, with an appropriate flow rate and gradient. The result of assessment by cation exchange chromatography (IEC) is shown in FIG. 10.
[0313] As shown in FIG. 10, the heterogeneity was increased by conversion of the constant region from IgG1 to IgG2 in the anti-IL-31 receptor antibody, and the heterogeneity can be reduced by conversion of the constant region to M14 or M58 in any antibody.
[Example 6] Pharmacokinetics Improving Effect of Novel Constant Region M58 in Anti-IL-31 Receptor Antibodies
[0314] As shown in Referential Example 9, conversion of the constant region from IgG1 to M58 in anti-IL-6 receptor antibody huPM1 was found to improve its human FcRn-binding activity and the pharmacokinetics in human FcRn transgenic mice. Thus, anti-IL-31 receptor antibodies were also tested to assess whether conversion of the constant region to M58 improves their pharmacokinetics.
[0315] H0L0-IgG1 (H chain: H0-IgG1/SEQ ID NO: 133; L chain: L0/SEQ ID NO: 56) and H0L0-M58 (H chain: H0-M58/SEQ ID NO: 136; L chain L0/SEQ ID NO: 56) prepared as described in Examples 4 and 5 were assessed for the human FcRn-binding activity by the method described in Referential Example 9. The result is shown in Table 3.
TABLE-US-00004 TABLE 3 KD (.mu.M) H0L0-IgG1 1.07 H0L0-M58 0.91
[0316] As shown in Table 3, conversion of the constant region from IgG1 to M58 also improved the human FcRn-binding activity of the anti-IL-31 receptor antibody H0L0 as in the anti-IL-6 receptor antibody hPM1. This suggests that conversion of the constant region from IgG1 to M58 may improve the pharmacokinetics of anti-IL-31 receptor antibody in human.
[Example 7] Identification of Mutation Sites Reducing the Isoelectric Point Production of Mutants
[0317] Each mutant was produced by the method described in Example 4 or by assembly PCR. In the method using assembly PCR, oligo DNAs are synthesized based on forward and reverse sequences including an altered site. Forward oligo DNA including an altered site and reverse oligo DNA binding to the vector in which the gene to be altered was inserted were combined, and reverse oligo DNA including an altered site and forward oligo DNA binding to the vector in which the gene to be altered was inserted were combined. PCR was carried out using PrimeSTAR (Takara) to produce 5'-end and 3'-end fragments including the altered site. The two fragments were assembled by assembly PCR to produce each mutant. The produced mutant was inserted into an expression vector capable of expressing the insert gene in animal cells. The nucleotide sequence of the resulting expression vector was determined by a method known to those skilled in the art. Antibodies were produced and purified by the method described in Example 4.
Identification of Mutation Sites
[0318] To improve the pharmacokinetics of H0L0 (H chain, H0-SKSC/SEQ ID NO: 54; L chain, L0/SEQ ID NO: 56), altered sites capable of reducing the isoelectric point of the variable region were examined. Screening of mutation sites in the variable regions predicted from the three-dimensional structure model revealed mutation sites that would decrease the isoelectric point of the variable regions without significantly reducing its binding to NR10. These are summarized in Table 4 (Hp5-VH/SEQ ID NO: 137, Hp7-VH/SEQ ID NO: 138, Hp8-VH/SEQ ID NO: 139, Hp6-VH/SEQ ID NO: 140, Hp9-VH/SEQ ID NO: 141, Hp1-VH/SEQ ID NO: 142, Hp13-VH/SEQ ID NO: 143, Lp1-VL/SEQ ID NO: 144, Lp2-VL/SEQ ID NO: 145, Lp3-VL/SEQ ID NO: 146, Lp4-VL/SEQ ID NO: 147, Lp7-VL/SEQ ID NO: 148, Lp5-VL/SEQ ID NO: 149, Lp6-VL/SEQ ID NO: 150). Each variant was produced and purified by the method described in Example 4.
[0319] Each variant was tested for the activity of inhibiting the hIL-31/hNR10 binding by using FMAT. The test was carried out according to the method as described in Example 4. As shown in FIG. 11, the competition activity of each variant was not greatly reduced as compared to that of H0L0.
TABLE-US-00005 TABLE 4 Mutation site H0 Amino acid Sequence after Name Type H0 sequence (kabat No) sequence after mutation mutation Hp5 FR2 WVRQAPGQGLEWMG 38 R Q WVQQSPGQGLEWMG (SEQ ID NO: 97) 40 *A S (SEQ ID NO: 120) Hp7 CDR2 LINPYNGGTSYNQKFKG 50 L E EINPYNGGTSYNQKFKG (SEQ ID NO: 10) (SEQ ID NO: 113) Hp8 CDR2 LINPYNGGTSYNQKFKG 52 N D LIDPYNGGTSYNQKFKG (SEQ ID NO: 10) (SEQ ID NO: 114) Hp6 CDR2 LINPYNGGTSYNQKFKG 61 Q D LINPYNGGTSYNDKFKG (SEQ ID NO: 10) (SEQ ID NO: 115) Hp9 CDR2 LINPYNGGTSYNQKFKG 62 K Q LINPYNGGTSYNQQFKG (SEQ ID NO: 10) (SEQ ID NO: 116) Hp1 CDR2 LINPYNGGTSYNQKFKG 64 K Q LINPYNGGTSYNQKFQG (SEQ ID NO: 10) (SEQ ID NO: 117) Hp13 CDR2 LINPYNGGTSYNQKFKG 64 K Q LINPYNGGTSYNQKFQD (SEQ ID NO: 10) 65 G D (SEQ ID NO: 119) Mutation site L0 Amino acid Sequence after Name Type L0 sequence (kabat No) sequence after mutation mutation Lp1 CDR1 RTSENIYSFLA 24 R Q QTSENIYSFLA (SEQ ID NO: 13) (SEQ ID NO: 121) Lp2 CDR1 RTSENIYSFLA 28 N D RTSEDIYSFLA (SEQ ID NO: 13) (SEQ ID NO: 122) Lp3 CDR2 NAKTLAK 50 N D DAKTLAK (SEQ ID NO: 14) (SEQ ID NO: 123) Lp4 CDR2 NAKTLAK 52 K Q NAQTLAK (SEQ ID NO: 14) (SEQ ID NO: 124) Lp7 CDR2 NAKTLAK 54 L E NAKTEAK (SEQ ID NO: 14) (SEQ ID NO: 125) Lp5 CDR2 NAKTLAK 56 K Q NAKTLAQ (SEQ ID NO: 14) (SEQ ID NO: 126) Lp6 CDR2 NAKTLAK 56 K D NAKTLAD (SEQ ID NO: 14) (SEQ ID NO: 127)
[0320] Asterisk (*) in Table 4 above indicates a site that was not relevant to the isoelectric point but altered for conversion into a human sequence.
[0321] Examples of the humanized NS22 antibodies whose isoelectric point has been reduced by combining these alterations include Hp3Lp15 (H chain: Hp3-SKSC/SEQ ID NO: 151; L chain: Lp15/SEQ ID NO: 152). Affinity for NR10, isoelectric point, and plasma retention in mice were compared between Hp3Lp15 and H0L0.
Measurement of Affinity
[0322] The affinity of each antibody for NR10 was determined by the method described in Referential Example 10.
[0323] The result of affinity measurement is shown in Table 5. The affinity of Hp3Lp15 was shown to be almost the same as that of H0L0.
TABLE-US-00006 TABLE 5 ka (1/Ms) kd (1/s) KD (M) H0L0 3.7E+05 1.2E-03 3.3E-09 Hp3Lp15 4.2E+05 1.6E-03 3.9E-09
Measurement of Isoelectric Point
[0324] Each antibody was analyzed by isoelectric focusing to assess changes in the isoelectric point of the whole antibody due to the amino acid alterations in its variable region. Isoelectric focusing was performed by the following method.
[0325] Phast-Gel Dry IEF gel (Amersham Biosciences) was swollen in Phastsystem Cassette (Amersham Biosciences) for about 30 minutes using the swelling solution shown below.
[0326] MilliQ water 1.5 ml
[0327] Pharmalyte 5-8 for IEF (Amersham Biosciences) 100 .mu.l
[0328] Electrophoresis was carried out in PhastSystem (Amersham Biosciences) using the swollen gel according to the program indicated below. The samples were loaded onto the gel in Step 2. Calibration Kit for pI (Amersham Biosciences) was used as a pI marker.
[0329] Step 1: 2000 V 2.5 mA 3.5 W 15.degree. C. 75 Vh
[0330] Step 2: 200 V 2.5 mA 3.5 W 15.degree. C. 15 Vh
[0331] Step 3: 2000 V 2.5 mA 3.5 W 15.degree. C. 410 Vh
[0332] After electrophoresis, the gel was fixed with 20% TCA, and then silver-stained using the Silver Staining Kit, Protein (Amersham Biosciences), according to the protocol attached to the kit. After staining, the isoelectric point of the sample (the whole antibody) was calculated from the known isoelectric points of the pI markers.
[0333] The result of isoelectric point measurement by isoelectric focusing showed that the isoelectric point of H0L0 was about 7.8, and the isoelectric point of Hp3Lp15 was about 5.5, showing that the isoelectric point of Hp3Lp15 was decreased by about 2.3 as compared to H0L0. When the theoretical isoelectric point of the variable region VH/VL was calculated by GENETYX (GENETYX CORPORATION), the theoretical isoelectric points of the variable regions of H0L0 and Hp3Lp15 were 7.76 and 4.63, respectively. Thus, the theoretical isoelectric point of Hp3Lp15 was decreased by 3.13 as compared to H0L0.
Assessment of Pharmacokinetics of Antibody with Reduced Isoelectric Point Using Mice
[0334] In order to assess the plasma retention of Hp3Lp15, a modified antibody with reduced isoelectric point, the plasma retention of H0L0 and Hp3Lp15 was compared in normal mice. A single dose of H0L0 or Hp3Lp15 was intravenously administered at 1 mg/kg to mice (C57BL/6J, Charles River Japan, Inc.) to compare the time course of the plasma concentration. The plasma concentrations were determined by ELISA. Appropriate concentrations of a calibration sample and test plasma samples were dispensed into immunoplates (Nunc-Immuno Plate, MaxiSorp (Nalge Nunc International)) coated with anti-human IgG (Fc-specific) antibody (Sigma). The samples were allowed to stand at room temperature for one hour. After reaction with Goat Anti-Human IgG-ALP (Sigma) at room temperature for one hour, color developing reaction was carried out using BluePhos Microwell Phosphatase Substrates System (Kirkegaard & Perry Laboratories) as a substrate. The absorbance at 650 nm was measured with a microplate reader. The plasma concentrations were determined based on the absorbance of the calibration curve using the analytical software SOFTmax PRO (Molecular Devices).
[0335] Pharmacokinetic parameters (AUC and systemic clearance (CL)) were calculated from the obtained time-course data of the plasma concentration using the pharmacokinetics analysis software WinNonlin (Pharsight). The parameters are shown in Table 6. AUC and the clearance of Hp3Lp15 after the intravenous administration were reduced by about 14% and about 12%, respectively, as compared to H0L0. Thus, it was demonstrated that Hp3Lp15, in which the isoelectric point of H0L0 has been reduced, had improved pharmacokinetics.
TABLE-US-00007 TABLE 6 AUC CL (.mu.g d/kg) (ml/d/kg) Mean SD Mean SD H0L0 281.8 13.1 3.6 0.2 Hp3Lp15 321.1 26.1 3.1 0.3
[Example 8] Effect of Combinations of Variable Region and Constant Region on the Biological Activity
[0336] In order to assess the effects of different constant regions on the biological activity, the following variants were produced.
[0337] SKSC (SEQ ID NO: 62) and M58 (SEQ ID NO: 128), constant regions prepared in Referential Examples 7 and 9, were combined with Hp3 (Hp3-VH/SEQ ID NO: 167), a variable region prepared in Example 7, to produce Hp3-M58 (SEQ ID NO: 240) and Hp3-SKSC (SEQ ID NO: 151) as H chains. The prepared H chains were combined with Lp15 (Lp15/SEQ ID NO: 152), an L chain prepared in Example 7, to produce Hp3Lp15-SKSC (H chain, Hp3-SKSC/SEQ ID NO: 151; L chain, Lp15/SEQ ID NO: 152) and Hp3Lp15-M58 (H chain, Hp3-M58/SEQ ID NO: 240; L chain, Lp15/SEQ ID NO: 152). Each antibody was expressed and purified by the method described in Example 4.
[0338] The antibodies produced as described above, H0L0-SKSC (H chain, H0-SKSC/SEQ ID NO: 54; L chain, L0/SEQ ID NO: 56) prepared using the constant region SKSC (SEQ ID NO: 62) described in Referential Example 7, and H0L0-M58 (H chain, H0-M58/SEQ ID NO: 136; L chain, L0/SEQ ID NO: 56) and H0L0-IgG2 (H chain, H0-IgG2/SEQ ID NO: 134; L chain, L0/SEQ ID NO: 56) prepared in Example 5, were assessed for the biological activity by the method described in Example 2 using BaF/NR10. The result is summarized in FIG. 18.
[0339] As shown in FIG. 18, no significant difference in the biological activity was detected between the constant regions. Since the biological activity was not affected when combining the two variable regions H0 and Hp3 with each constant region, combining variable regions created in future with any constant region would not result in alteration in the biological activity.
[Example 9] Identification of Mutation Sites Suppressing Degradation by Thermal Acceleration Study
[0340] Antibodies used for pharmaceuticals have heterogeneity even though they are monoclonal antibodies obtained from clones derived from single antibody-producing cells. Such antibody heterogeneity is known to result from modification such as oxidation or deamidation, and to be increased during long-term storage or upon exposure to stress conditions, such as heat stress or light stress (see "Heterogeneity of Monoclonal Antibodies", Journal of pharmaceutical sciences, vol. 97, No. 7, 2426-2447). However, when an antibody is developed as a pharmaceutical, physical properties of the protein, particularly homogeneity and stability, are highly important. Thus, it is desired that the heterogeneity of desired/related substances be reduced and the substance be composed of a single substance as much as possible. In this context, the experiment described below was conducted to assess the antibody heterogeneity under stress conditions and to reduce the heterogeneity.
[0341] To assess degradation products, an accelerated sample of H0L0 (H chain, H0-SKSC/SEQ ID NO: 54; L chain, L0/SEQ ID NO: 56) was prepared by the method described below. The prepared accelerated sample and non-accelerated sample (initial) were analyzed by cation exchange chromatography using the method described below.
[0342] Method for preparing accelerated samples
Buffer: PBS
[0343] Antibody concentration: 0.2 to 1.0 mg/ml
[0344] Acceleration temperature: 60.degree. C.
[0345] Acceleration period: one day
[0346] Method for analysis by cation exchange chromatography
[0347] Column: ProPac WCX-10, 4.times.250 mm (Dionex)
[0348] Mobile phase: (A) 25 mmol/1 MES/NaOH, pH 6.1 [0349] (B) 25 mmol/1 MES/NaOH, 250 mmol/l NaCl, pH 6.1
[0350] Flow rate: 0.5 ml/min
[0351] Column temperature: 40.degree. C.
[0352] Gradient: % B 0 to 0 (0-5 min).fwdarw.0 to 30 (5-80 min)
[0353] Detection: 280 nm
[0354] The resulting chromatograms for H0L0 samples before and after acceleration are shown in FIG. 19. The H0L0 sample after acceleration had a tendency to show an increased basic peak.
[0355] Then, screening was carried out to reduce this peak. As a result, Ha355, Ha356, Ha360, and Ha362 were found. These H chain variants were combined with L0 to produce Ha355L0 (H chain, Ha355-SKSC/SEQ ID NO: 242; L chain, L0/SEQ ID NO: 56), Ha356L0 (H chain, Ha356-SKSC/SEQ ID NO: 243; L chain, L0/SEQ ID NO: 56), Ha360L0 (H chain, Ha360-SKSC/SEQ ID NO: 244; L chain, L0/SEQ ID NO: 56), and Ha362L0 (H chain, Ha362-SKSC/SEQ ID NO: 245; L chain, L0/SEQ ID NO: 56). The sequence of each variant is shown in Table 7.
TABLE-US-00008 TABLE 7 Mutation site H0 Amino acid Sequence after Name Type H0 sequence (kabat No) sequence after mutation mutation Ha355 CDR3 DGYDDGPYTMDY 100d M L DGYDDGPYTLET (SEQ ID NO: 265) 101 D E (SEQ ID NO: 266) 102 Y T Ha356 CDR3 DGYDDGPYTMDY 101 D E DGYDDGPYTMET (SEQ ID NO: 265) 102 Y T (SEQ ID NO: 267) Ha360 CDR3 DGYDDGPYTMDY 97 Y L DGLDDGPYTMET (SEQ ID NO: 265) 101 D E (SEQ ID NO: 268) 102 Y T Ha362 CDR3 DGYDDGPYTMDY 97 Y L DGLDDGPYTMES (SEQ ID NO: 265) 101 D E (SEQ ID NO: 269) 102 Y S
[0356] Each of the identified antibodies was expressed and purified by the method described in Example 4. As with H0L0, a accelerated sample of each prepared antibody was prepared, and analyzed by cation exchange chromatography. The result is shown in FIG. 19.
[0357] The result showed that the generation of the basic peak increased after acceleration was reduced in the modified antibody containing a substitution of aspartic acid with glutamic acid at position 101 in the H chain, as compared to H0L0. The modified antibodies were assessed for the biological activity by the method described in Example 2 using BaF/NR10. The result is shown in FIG. 20. As shown in FIG. 20, the biological activities of the modified antibodies were comparable to or stronger than that of H0L0. These findings demonstrated that the modifications of Ha355, Ha356, Ha360, and Ha362 suppressed the generation of degradation products by acceleration, and therefore are effective in improving the stability of antibody.
[Example 10] Identification of Mutation Sites Increasing the Affinity
[0358] A library in which mutations were introduced into CDR sequences was constructed and examined to improve the affinity of H0L0 for NR10. As a result of screening of the library in which mutations were introduced into CDRs, mutations that improve the affinity for NR10 were found. The mutations are shown in Table 8. Each of the H chain variants Ha101-SKSC (SEQ ID NO: 246), Ha103-SKSC (SEQ ID NO: 247), Ha111-SKSC (SEQ ID NO: 248), Ha204-SKSC (SEQ ID NO: 249), and Ha219-SKSC (SEQ ID NO: 250) was combined with L0 (L0/SEQ ID NO: 56); and each of the modified L chains La134 (SEQ ID NO: 251), La130 (SEQ ID NO: 252), La303 (SEQ ID NO: 253), and La328 (SEQ ID NO: 254) was combined with H0 (H0-SKSC/SEQ ID NO: 54), to construct an antibody. Each variant was produced and purified by the method described in Example 4.
[0359] The affinity of each antibody for NR10 was assessed using Biacore. The result is shown in Table 9. The assay was carried out using the method described in Referential Example 10. As shown in Table 9, the KD value for each variant was found to be improved as compared to that of H0L0 (H chain, H0-SKSC/SEQ ID NO: 54; L chain, L0/SEQ ID NO: 56).
TABLE-US-00009 TABLE 8 Mutation site H0 Amino acid Sequence Name Type H0 sequence (kabat No) sequence after mutation after mutation Ha101 CDR1 GYIMN 33 I V GYVMN (SEQ ID NO: 270) (SEQ ID NO: 272) Ha103 CDR1 GYIMN 34 M I GYIIN (SEQ ID NO: 270) (SEQ ID NO: 273) Ha111 CDR1 GYIMN 34 M L GYILN (SEQ ID NO: 270) (SEQ ID NO: 274) Ha204 CDR2 LINPYNGGTSYNQKFKG 58 S D LINPYNGGTDYNQKFKG (SEQ ID NO: 271) (SEQ ID NO: 275) Ha219 CDR2 LINPYNGGTSYNQKFKG 61 Q P LINPYNGGTSYNPKFKG (SEQ ID NO: 271) (SEQ ID NO: 276) Mutation site L0 Amino acid Sequence Name Type L0 sequence (kabat No) sequence after mutation after mutation La134 CDR1 RTSENIYSFLA 31 S R RTSENIYRFLA (SEQ ID NO: 277) (SEQ ID NO: 279) La130 CDR1 RTSENIYSFLA 31 S R RTSENIYRFVA (SEQ ID NO: 277) 33 L V (SEQ ID NO: 280) Ls303 CDR3 QHHYESPLT 93 E D QHHYDSPLT (SEQ ID NO: 278) (SEQ ID NO: 281) La328 CDR3 QHHYESPLT 94 S D QHHYEDPLT (SEQ ID NO: 278) (SEQ ID NO: 282) La326 CDR3 QHHYESPLT 97 T F QHHYESPLF (SEQ ID NO: 278) (SEQ ID NO: 283)
TABLE-US-00010 TABLE 9 Name ka (1/Ms) kd (1/s) KD (M) H0L0 1.9E+05 6.2E-04 3.2E-09 Ha101L0 2.0E+05 3.1E-04 1.5E-09 Ha103L0 2.2E+05 5.3E-04 2.4E-09 Ha111L0 2.6E+05 5.6E-04 2.1E-09 Ha204L0 3.7E+05 4.8E-04 1.3E-09 Ha219L0 3.2E+05 9.6E-04 3.0E-09 H0L0 1.5E+05 7.4E-04 5.1E-09 H0La134 2.5E+05 4.4E-04 1.8E-09 H0La130 2.6E+05 4.0E-04 1.5E-09 H0La303 2.2E+05 4.6E-04 2.1E-09 H0La328 1.8E+05 5.2E-04 2.9E-09 H0La326 1.4E+05 5.2E-04 3.7E-09
[0360] Examples of combinations of these affinity-improving mutations with the isoelectric point-lowering mutations generated in Example 7 include, for example, Ha401La402 (H chain, Ha401-SKSC/SEQ ID NO: 255; L chain, La402/SEQ ID NO: 256) and H17L11 (H chain, H17-M58/SEQ ID NO: 222; L chain, L11/SEQ ID NO: 236). Each variant was produced and purified by the method described in Example 4.
[0361] Ha401La402 (H chain, Ha401-SKSC/SEQ ID NO: 255; L chain, La402/SEQ ID NO: 256) was assessed for its affinity for NR10 and its biological activity by the method described in Referential Example 10 and the method using BaF/NR10 as described in Example 2, respectively, and they were compared to those of H0L0 (H chain, H0-SKSC/SEQ ID NO: 54; L chain, L0/SEQ ID NO: 56). The result of affinity measurement is shown in Table 10, and the biological activity determined using BaF/NR10 is shown in FIG. 21. Both affinity and biological activity were found to be improved as compared to those of H0L0 (H chain, H0-SKSC/SEQ ID NO: 54; L chain, L0/SEQ ID NO: 56).
TABLE-US-00011 TABLE 10 ka (1/Ms) kd (1/s) KD (M) H0L0 2.9E+05 9.1E-04 3.2E-09 Ha401La402 5.8E+05 2.9E-04 5.0E-10
[0362] Furthermore, H17L11 (H chain, H17-M58/SEQ ID NO: 222; L chain, L11/SEQ ID NO: 236) was assessed for its affinity for NR10 and its biological activity by the method described in Example 7 and the method using BaF/NR10 as described in Example 2, respectively, and they were compared to those of H0L0 (H chain, H0-M58/SEQ ID NO: 136; L chain, L0/SEQ ID NO: 56). The result of affinity measurement is shown in Table 11, and the biological activity determined using BaF/NR10 is shown in FIG. 22. Both affinity and biological activity were found to be improved as compared to those of H0L0 (H chain, H0-M58/SEQ ID NO: 136; L chain, L0/SEQ ID NO: 56).
TABLE-US-00012 TABLE 11 ka (1/Ms) kd (1/s) KD (M) H0L0 1.4E+05 6.9E-04 4.8E-09 H17L11 4.3E+05 2.6E-04 6.2E-10
[Example 11] Identification of Mutation Sites Reducing Immunogenicity Risk
Reduction of Immunogenicity Risk in H Chain CDR1
[0363] T-cell epitopes present in the variable region sequence of H0L0 were analyzed using TEPITOPE (Methods 2004 December; 34(4): 468-75). As a result, CDR1 of the H chain was predicted to have many T-cell epitopes that bind to HLA (i.e. have sequences with a high immunogenicity risk). Then, TEPITOPE analysis was carried out to examine substitutions that would reduce the immunogenicity risk of the H chain CDR1. As a result, the immunogenicity risk was found to be greatly reduced by substituting isoleucine at position 33 in kabat numbering with alanine (A) (Table 12). Then, this alteration was added to H17 generated in Example 10 to produce H19 (H19-M58/SEQ ID NO: 223). The generated H19 was combined with L12 to produce H19L12 (H chain, H19-M58/SEQ ID NO: 223; L chain, L12/SEQ ID NO: 237). Each variant was produced and purified by the method described in Example 4.
[0364] The antibody was assessed for the affinity for NR10 and the biological activity by the method described in Referential Example 10 and the method using BaF/NR10 as described in Example 2, respectively, and they were compared to those of H0L0 (H chain, H0-M58/SEQ ID NO: 136; L chain, L0/SEQ ID NO: 56). The result of affinity measurement is shown in Table 13, and the biological activity determined using BaF/NR10 is shown in FIG. 23. Both affinity and biological activity were shown to be almost equal to those of H0L0.
TABLE-US-00013 TABLE 12 Mutation site H0 Amino acid Sequence Name Type H0 sequence (kabat No) sequence after mutation after mutation H19 CDR1 GYIMN 33 I A GYAMN (SEQ ID NO: 270) (SEQ ID NO: 284)
TABLE-US-00014 TABLE 13 ka (1/Ms) kd (1/s) KD (M) H0L0 1.8E+05 8.7E-04 4.8E-09 H19L12 2.3E+05 1.2E-03 5.1E-09
Reduction of Immunogenicity Risk in L Chain CDR1
[0365] Threonine (T) present at kabat-numbering position 25 in CDR1 of the L chain corresponds to alanine (A) or serine (S) in the germline sequence. Thus, it is predicted that the immunogenicity risk is reduced by substituting threonine (T) at position 25 with alanine (A) or serine (S) (Table 14). Therefore, the above substitution was added to L12 to produce L17 (SEQ ID NO: 238). The produced L17 was combined with H0 to produce H0L17 (H chain, H0-M58/SEQ ID NO: 136; L chain, L17/SEQ ID NO: 238). Each variant was produced and purified by the method described in Example 4.
[0366] Each variant was assessed for the affinity for NR10 and the biological activity by the method described in Referential Example 10 and the method using BaF/NR10 as described in Example 2, respectively, and they were compared to those of H0L0 (H chain, H0-M58/SEQ ID NO: 136; L chain, L0/SEQ ID NO: 56) and H0L12 (H chain, H0-M58/SEQ ID NO: 136; L chain, L12/SEQ ID NO: 237). Since L12 contains a sequence that improves the affinity, it exhibits about two times higher affinity than H0L0. The result of affinity measurement is shown in Table 15, and the biological activity determined using BaF/NR10 is shown in FIG. 24. Both affinity and biological activity were shown to be almost equal to those of H0L12.
TABLE-US-00015 TABLE 14 Mutation site L0 Amino acid Sequence Name Type L0 sequence (kabat No) sequence after mutation after mutation Ld-1 CDR1 RTSENIYSFLA 25 T A RASENIYSFLA (SEQ ID NO: 277) (SEQ ID NO: 285) Ld-2 CDR1 RTSENIYSFLA 25 T S RSSENIYSFLA (SEQ ID NO: 277) (SEQ ID NO: 286)
TABLE-US-00016 TABLE 15 ka (1/Ms) kd (1/s) KD (M) H0L0 1.6E+05 7.8E-04 4.8E-09 H0L12 3.8E+05 7.4E-04 2.0E-09 H0L17 3.9E+05 8.1E-04 2.1E-09
[Example 12] Preparation of Completely Humanized NS22 Antibody
[0367] Variable regions of NS22 variants were prepared by combining the multiple mutations that reduce the pI, increase the affinity, suppress the degradation of H chain, and reduce the immunogenicity risk, all of which were found in the above Examples, in H0 (H0-M58/SEQ ID NO: 136), H1 (H1-M58/SEQ ID NO: 257), or L0 (L0/SEQ ID NO: 56), and subjected to various screening procedures. As a result, H28L17 (H chain, H28-M58/SEQ ID NO: 224; L chain, L17/SEQ ID NO: 238), H30L17 (H chain, H30-M58/SEQ ID NO: 225; L chain, L17/SEQ ID NO: 238), H34L17 (H chain, H34-M58/SEQ ID NO: 226, L chain, L17/SEQ ID NO: 238), H42L17 (H chain, H42-M58/SEQ ID NO: 227; L chain, L17/SEQ ID NO: 238), H44L17 (H chain, H44-M58/SEQ ID NO: 228; L chain, L17/SEQ ID NO: 238), H46L17 (H chain, H46-M58/SEQ ID NO: 229; L chain, L17/SEQ ID NO: 238), H57L17 (H chain, H57-M58/SEQ ID NO: 230; L chain, L17/SEQ ID NO: 238), H71L17 (H chain, H71-M58/SEQ ID NO: 231; L chain, L17/SEQ ID NO: 238), H78L17 (H chain, H78-M58/SEQ ID NO: 232; L chain, L17/SEQ ID NO: 238), H92L17 (H chain, H92-M58/SEQ ID NO: 233; L chain, L17/SEQ ID NO: 238), H97L50 (H chain, H97-M58/SEQ ID NO: 234; L chain, L50/SEQ ID NO: 239), and H98L50 (H chain, H98-M58/SEQ ID NO: 235; L chain, L50/SEQ ID NO: 239) were found. Each variant was produced and purified by the method described in Example 4.
[0368] Each variant was assessed for the affinity for NR10 and the biological activity by the method described in Referential Example 10 and the method using BaF/NR10 as described in Example 2, respectively, and they were compared to those of H0L0 (H chain, H0-M58/SEQ ID NO: 136; L chain, L0/SEQ ID NO: 56). The result of affinity measurement is shown in Table 16, and the biological activity determined using BaF/NR10 is shown in FIGS. 25-1 and 25-2. Both affinity and biological activity of each antibody were shown to be almost equal to or greater than those of H0L0.
TABLE-US-00017 TABLE 16 Sample ka (1/Ms) kd (1/s) KD (M) H0L0 2.1E+05 8.8E-04 4.2E-09 H28L17 6.4E+05 3.3E-04 5.2E-10 H30L17 6.8E+05 5.7E-04 8.3E-10 H34L17 3.4E+05 1.2E-03 3.6E-09 H42L17 5.7E+05 3.7E-04 6.5E-10 H44L17 6.1E+05 7.2E-04 1.2E-09 H46L17 2.9E+05 1.3E-03 4.6E-09 H57L17 7.1E+05 5.5E-04 7.7E-10 H71L17 3.7E+05 1.2E-03 3.3E-09 H78L17 6.1E+05 7.0E-04 1.1E-09 H92L17 3.1E+05 1.3E-03 4.1E-09 H97L50 3.6E+05 1.3E-03 3.5E-09 H98L50 2.9E+05 1.3E-03 4.6E-09
[Example 13] Analysis of the Binding Domain of Anti-NR10 Neutralizing Antibody
(1) Preparation of Human/Mouse Wild-Type and Chimeric Antigens
[0369] The genes encoding human and mouse wild-type extracellular domains and chimeric extracellular domains of NR10 (hhh (SEQ ID NO: 258), mmm (SEQ ID NO: 259), hhm (SEQ ID NO: 260), mmh (SEQ ID NO: 261), hmm (SEQ ID NO: 262), mhm (SEQ ID NO: 263), and mhh (SEQ ID NO: 264)), were fused to His tag and Myc tag (HHHHHHEQKLISEEDUSEQ ID NO: 287) at their C termini, inserted into an animal expression vector, and transiently expressed using FreeStyle 293 Expression System (Invitrogen.TM.). Schematic diagrams for the human/mouse wild-type and chimeric NR10-ECDs are shown in FIG. 26.
[0370] The human/mouse wild-type and chimeric antigens (hhh, mmm, hhm, mmh, hmm, mhm, and mhh) were purified from culture supernatants by Ni-NTA Superflow column chromatography. Specifically, 1 ml of Ni-NTA Superflow (QIAGEN) was loaded onto Poly-Prep Empty Column (BioRad), and 30 ml of each culture supernatant was added thereto. After washing with D-PBS (Dulbecco's phosphate-buffered saline) containing 150 mM sodium chloride and 20 mM imidazole, the column was eluted with D-PBS containing 150 mM sodium chloride and 250 mM imidazole. The eluted fractions were buffer-exchanged with D-PBS and concentrated using Amicon-Ultra (Millipore) with a molecular weight cut-off of 10K.
(2) Detection of Binding Antigen by Western Blotting
[0371] Each of the prepared human/mouse wild-type and chimeric antigens was electrophoresed at 0.5 .mu.g/lane on three 4-20% polyacrylamide gels (Daiichi Pure Chemicals Co.). The proteins were electro-transferred onto PVDF membranes (Millipore) in a semi-dry blotting apparatus, and the membranes were blocked with TBS containing 5% skim milk. One membrane was incubated with 5 .mu.g/ml of H44M58L17 (detection system for humanized anti-human NR10 antibody); another with 5 .mu.g/ml of ND41 (detection system for mouse anti-human NR10 antibody); and the other one with anti-Myc antibody (SantaCruz, Cat.#sc-789) 500-times diluted with TBS containing 5% skim milk (detection system for Myc tag) at room temperature for one hour.
[0372] After washing three times with TBS containing 0.05% Tween.TM. 20, the secondary antibodies were incubated with the membranes. Alkaline phosphatase-labeled goat anti-human IgG.gamma. (BIOSOURCE, Cat. #AHI0305) was used to detect humanized anti-human NR10 antibody; alkaline phosphatase-labeled goat anti-mouse IgG (SantaCruz, Cat. #sc-2008) was used to detect mouse anti-human NR10 antibody; and alkaline phosphatase-labeled goat anti-rabbit IgG (SantaCruz, Cat. #sc-2057) was used to detect Myc tag. The reaction was carried out at room temperature for one hour. After washing four times with TBS containing 0.05% Tween.TM. 20 for three minutes, color development was carried out using BCIP/NBT Phosphatase substrate, 1-Component System (KPL). TBS (Tris-buffered saline) used here was prepared by dissolving a pack of TBS (Tris buffered saline) powder (TaKaRa) in 1 L of distilled water. The result is shown in FIG. 27.
[0373] When the humanized antibody or mouse antibody was used, the binding was detected only for hhh, hhm, and hmm, which are NR10 extracellular domains.
[Referential Example 1] Isolation of Cynomolgus Monkey NR10, OSMR, and IL-31 Genes
[0374] Since the cross-reactivity and neutralizing activity in cynomolgus monkeys were considered important for safety assessment at a pre-clinical stage, the cynomolgus monkey NR10 and OSMR genes were isolated. Primers were designed based on published information of Rhesus monkey genome and others, and the NR10 and OSMR genes were successfully amplified by PCR from cynomolgus monkey pancreatic cDNA. The sequences of the isolated cynomolgus monkey NR10, OSMR, and IL-31 genes are shown in SEQ ID NOs: 65, 69, and 67, respectively, and the amino acid sequences of cynomolgus monkey NR10, OSMR, and IL-31 are shown in SEQ ID NOs: 66, 70, and 68, respectively.
[Referential Example 2] Establishment of NR10- and OSMR-Expressing Ba/F3 Cell Lines
[0375] The full-length human NR10 cDNA (SEQ ID NO: 75) was inserted into the expression vector pCOS1 (Biochem. Biophys. Res. Commun. 228, p838-45, 1996), and the resulting vector was named pCosNR10.3. An oncostatin M receptor cDNA (OSMR, GenBank accession No. NM003999) was isolated by PCR from a human placental library, and the expression vector pCos1-hOSMR was constructed in the same manner. 10 .mu.g each of the vectors were simultaneously introduced into mouse IL-3-dependent pro-B cell-derived cell line Ba/F3 by electroporation (BioRad Gene Pulser, 960 .mu.F, 0.33 kV). After introduction, human IL-31 (R&D Systems) was added, and the cells were cultured to obtain a cell line (hNR10/hOSMR/BaF3 cell) that proliferates in an IL-31-dependent manner. Furthermore, the cynomolgus monkey IL-31 gene (SEQ ID NO: 67) was inserted into a mammalian cell expression vector and introduced into CHO cell line DG44. The resulting culture supernatant was obtained as cynomolgus monkey IL-31. As with hNR10/hOSMR/BaF3, the full-length cynomolgus monkey NR10 and OSMR genes were inserted into the expression vector pCOS1 and expressed in Ba/F3 cells, and a cynomolgus monkey IL-31-dependent cell line (cynNR10/cynOSMR/BaF3 cell) was established using the culture supernatant described above.
[Referential Example 3] Establishment of NR10-Expressing CHO Cell Lines
[0376] The genes for cytoplasmic domain-lacking human NR10 (SEQ ID NO: 73) and cytoplasmic domain-lacking cynomolgus monkey NR10 (SEQ ID NO: 71) were each inserted to a mammalian cell expression vector. The resulting vectors were linearized with a restriction enzyme, and then introduced into CHO cell line DG44 by electroporation (BioRad Gene Pulser, 25 .mu.F, 1.5 kV). After drug selection, NR10-expressing cells were selected and established by FCM analysis using anti-human NR10 antibody. The amino acid sequence encoded by the nucleotide sequence of cytoplasmic domain-lacking human NR10 gene (SEQ ID NO: 73) is shown in SEQ ID NO: 74, and the amino acid sequence encoded by the nucleotide sequence of cytoplasmic domain-lacking cynomolgus monkey NR10 gene (SEQ ID NO: 71) is shown in SEQ ID NO: 72.
[Referential Example 4] Preparation of NR10 Protein (Extracellular Domain)
[0377] The human NR10 cDNA was used as a template to amplify only the extracellular domain by PCR. The amplified region was then fused to a FLAG tag sequence at the C terminus and inserted to a mammalian cell expression vector. Ten .mu.g of the linearized vector was introduced into Chinese hamster ovary cell line DG44 by electroporation (BioRad Gene PulserII, 25 .mu.F, 1.5 kV). A cell line showing high level expression was obtained. The supernatant of the cell line cultured on a large scale was purified using anti-FLAG antibody column (Sigma) and gel filtration to obtain soluble NR10. The nucleotide sequence of soluble NR10 is shown in SEQ ID NO: 77, and the amino acid sequence is shown in SEQ ID NO: 78.
[Referential Example 5] Preparation of Anti-Human NR10 Antibodies
[0378] Mice were immunized with human NR10 protein (extracellular domain) (described in Referential Example 4), and hybridomas were prepared by a conventional method. The culture supernatants of these hybridomas were assessed for the neutralizing activity using the human IL-31-dependent cell line (hNR10/hOSMR/BaF3 cell) described in Referential Example 2, and thereby NA633 which has an NR10-neuralizing activity was obtained.
[0379] Furthermore, DNA immunization was carried out by He gas-driven gene gun using a mammalian expression vector carrying the full-length human NR10 gene (SEQ ID NO: 75), and hybridomas were prepared by a conventional method. The culture supernatants of these hybridomas were assessed for the neutralizing activity using the human IL-31-dependent cell line (hNR10/hOSMR/BaF3 cell) described in Referential Example 2, and thereby ND41 which has an NR10-neuralizing activity was obtained.
[Referential Example 6] Preparation of Human Chimeric Antibody
[0380] The amino acid sequences of heavy chain and light chain variable regions of NA633 are shown in SEQ ID NOs: 104 and 108, respectively. The amino acid sequences of CDR1, CDR2, and CDR3 of the heavy chain variable region of NA633 are shown in SEQ ID NOs: 105, 106, and 107, respectively, while those of CDR1, CDR2, and CDR3 of the light chain variable region are shown in SEQ ID NOs: 109, 110, and 111, respectively. Furthermore, a chimeric antibody between these mouse variable regions and human constant region (H chain, .gamma.1; L chain, .kappa.) was produced by a conventional method.
[Referential Example 7] Preparation of huPM1-SKSC in which the Heterogeneity of Wild Type IgG2 is Reduced without Loss of Stability
[0381] Since the NS22 antibody is an NR10-neutralizing antibody, its binding to Fc.gamma. receptor may be unfavorable in consideration of the immunogenicity and adverse effects. A possible method for reducing the binding to Fc.gamma. receptor is to select IgG2 or IgG4 instead of IgG1 as the isotype of the constant region (Ann Hematol. 1998 June; 76(6): 231-48). From the viewpoint of Fc.gamma. receptor I and retention in plasma, IgG2 has been considered more desirable than IgG4 (Nat Biotechnol. 2007 December; 25(12): 1369-72). Meanwhile, when an antibody is developed as a pharmaceutical, properties of the protein, particularly homogeneity and stability, are highly important. The IgG2 isotype has been reported to have very high heterogeneity resulting from the disulfide bonds in the hinge region (J Biol Chem. 2008 Jun. 6; 283(23): 16206-15). It is not easy and would be more costly to manufacture it as pharmaceutical in a large scale while maintaining difference in the heterogeneity of desired/related substances among products resulting from the above. Accordingly, it is desired that the substance be composed of a single substance as much as possible. Thus, when antibodies of IgG2 isotype are developed as pharmaceuticals, it is preferred to reduce the heterogeneity resulting from disulfide bonds, without lowering the stability.
[0382] In order to reduce the heterogeneity of the wild type IgG2, cysteines in the hinge region and CH1 domain of IgG2 were substituted. As a result of examination of various variants, SKSC (SEQ ID NO: 62), which is a constant region obtained by altering cysteine at position 131 and arginine at position 133 in the EU numbering (Sequences of proteins of immunological interest, NIH Publication No. 91-3242) within the H-chain CH1 domain of the wild type IgG2 constant region sequence to serine and lysine, respectively, and altering cysteine at EU-numbering position 219 in the H-chain upper hinge to serine could reduce the heterogeneity without decreasing the stability. Meanwhile, other possible methods for decreasing heterogeneity are to alter only cysteine at EU-numbering position 219 in the H-chain upper hinge to serine, and to alter only cysteine at EU-numbering position 220 to serine. Thus, constant region SC (SEQ ID NO: 153) in which cysteine at EU-numbering position 219 in IgG2 has been altered to serine, and constant region CS (SEQ ID NO: 154) in which cysteine at EU-numbering position 220 in IgG2 has been altered to serine, were produced.
[0383] huPM1-SC (SEQ ID NO: 157), huPM1-CS (SEQ ID NO: 158), huPM1-IgG1 (SEQ ID NO: 159), huPM1-IgG2 (SEQ ID NO: 160), and huPM1-SKSC (SEQ ID NO: 161), which were prepared by combining the constant regions produced as above, IgG1 (SEQ ID NO: 60), and IgG2 (SEQ ID NO: 132) with the variable region of the humanized anti-IL-6 receptor antibody (H chain variable region, huPM1-VH/SEQ ID NO: 155; L chain variable region huPM1-VL/SEQ ID NO: 156)(Cancer Res. 1993 Feb. 15; 53(4): 851-6), were used as an H chain, and huPM1-L (SEQ ID NO: 162) was used as an L chain, to produce each antibody. Each antibody was expressed and purified by the method described in Example 4.
[0384] The antibodies were compared to each other in terms of the heterogeneity. The heterogeneity of huPM1-IgG1, huPM1-IgG2, huPM1-SC, huPM1-CS, and huPM1-SKSC was assessed by cation exchange chromatography. The chromatography was carried out using a ProPac WCX-10 (Dionex) column, 20 mM sodium acetate (pH 5.0) as mobile phase A, and 20 mM sodium acetate/1M NaCl (pH 5.0) as mobile phase B, with an appropriate flow rate and gradient. The result of assessment by cation exchange chromatography is shown in FIG. 12.
[0385] As shown in FIG. 12, conversion of the constant region from IgG1 into IgG2 increased the heterogeneity. In contrast, the heterogeneity was markedly reduced by converting the constant region into SKSC. While constant region SC resulted in considerable reduction of the heterogeneity as in SKSC, constant region CS did not sufficiently improve the heterogeneity.
[0386] When an antibody is developed as a pharmaceutical, it is generally desired that the antibody have high stability in addition to low heterogeneity for the production of stable preparations. Thus, to assess the stability, the thermal denaturation midpoint temperature (Tm value) was determined by differential scanning calorimetry (DSC) (VP-DSC; Microcal). The thermal denaturation midpoint temperature (Tm value) serves as an indicator of stability. In order to prepare stable preparations as pharmaceuticals, a higher thermal denaturation midpoint temperature (Tm value) is preferred (J Pharm Sci. 2008 April; 97(4): 1414-26). Thus, huPM1-IgG1, huPM1-IgG2, huPM1-SC, huPM1-CS, and huPM1-SKSC were dialyzed against a solution of 20 mM sodium acetate/150 mM NaCl (pH 6.0) (EasySEP; TOMY), and DSC measurement was carried out using about 0.1 mg/ml of protein at a heating rate of 1.degree. C./min between 40 and 100.degree. C. The denaturation curves obtained by DSC are shown in FIG. 13. The Tm values of the Fab domains are listed in Table 17 below.
TABLE-US-00018 TABLE 17 Name Tm/.degree. C. huPM1-IgG1 94.8 huPM1-IgG2 93.9 huPM1-SC 86.7 huPM1-CS 86.4 huPM1-SKSC 93.7
[0387] The Tm values of huPM1-IgG1 and huPM1-IgG2 were almost the same, namely, about 94.degree. C. (IgG2 was lower by about 1.degree. C.). Meanwhile, the Tm values of huPM1-SC and huPM1-CS were about 86.degree. C., which was significantly lower than those of huPM1-IgG1 and huPM1-IgG2. On the other hand, the Tm value of huPM1-SKSC was about 94.degree. C., and almost the same as huPM1-IgG1 and huPM1-IgG2. Since the stability of huPM1-SC and huPM1-CS was markedly lower than that of IgG2, huPM1-SKSC in which cysteine in the CH1 domain have also been altered to serine may be more preferred in the development of pharmaceuticals. The significant decrease in Tm value of huPM1-SC and huPM1-CS as compared to IgG2 may be due to the disulfide-bonding pattern of huPM1-SC and huPM1-CS that is different from that of IgG2.
[0388] Furthermore, comparison of the DSC denaturation curves showed that the denaturation peak for the Fab domain was sharp in huPM1-IgG1 and huPM1-SKSC, while it was broader in huPM1-SC and huPM1-CS than the above two, and huPM1-IgG2 gave a shoulder peak on the lower temperature side of the Fab domain denaturation peak. The denaturation peak in DSC generally becomes sharp in the case of a single component, but may become broad when two or more components with different Tm values (namely, heterogeneity) are present. Thus, it was suggested that huPM1-IgG2, huPM1-SC, and huPM1-CS contained two or more components, and the heterogeneity of natural IgG2 was not reduced in huPM1-SC and huPM1-CS. This finding suggests that cysteines present in both the hinge region and the CH1 domain are involved in the heterogeneity of natural IgG2, and it is necessary to alter not only cysteine in the hinge region but also that in the CH1 domain to decrease the heterogeneity on DSC. Furthermore, as described above, it is only possible to attain stability equivalent to that of natural IgG2 by altering not only cysteine in the hinge region but also that in the CH1 domain.
[0389] As described above, as to the constant regions in which the heterogeneity resulting from the hinge region of IgG2 has been reduced, it was discovered that SC and CS, which are constant regions in which only cysteine in the hinge region has been substituted with serine, may be insufficient from the viewpoint of heterogeneity and stability, and that it is only possible to significantly reduce the heterogeneity while maintaining the stability comparable to IgG2 by additionally substituting cysteine at EU-numbering position 131 in the CH1 domain with serine. Such constant regions include SKSC.
[Referential Example 8] Production and Assessment of Optimized, Non-Fc.gamma. Receptor-Binding Constant Region M14
[0390] In the Fc.gamma. receptor-binding domain of IgG2 constant region, the residues at EU-numbering positions 233, 234, 235, and 236 are of non-binding type, while the residues at EU-numbering positions 327, 330, and 331 are different from those of IgG4, which are of non-binding type. Thus, it is necessary to alter the amino acids at EU-numbering positions 327, 330, and 331 to the sequence of IgG4 (G2.DELTA.a in Eur J Immunol. 1999 August; 29(8):2613-24). However, since the amino acid at EU-numbering position 339 is alanine in IgG4 while it is threonine in IgG2, mere alteration of the amino acids at EU-numbering positions 327, 330, and 331 to the sequence of IgG4 will generate a novel non-naturally occurring 9-amino acid peptide sequence that could be a T-cell epitope peptide, thereby causing a risk of immunogenicity. Thus, it was found that the occurrence of the novel peptide sequence could be prevented by altering threonine at EU-numbering position 339 in IgG2 to alanine, in addition to the alterations described above. In addition to the mutations described above, methionine at EU-numbering position 397 was mutated into valine to improve the stability of IgG2 under acidic condition. Furthermore, in SKSC (SEQ ID NO: 62) produced in Referential Example 7, in which the heterogeneity resulting from the disulfide bonds in the hinge region has been improved, introduction of mutations at positions 131 and 133 will generate a novel non-naturally occurring 9-amino acid peptide sequence that could be a T-cell epitope peptide, thereby causing a risk of immunogenicity. Thus, the peptide sequence around positions 131 to 139 was converted into the same as IgG1 by mutating glutamic acid at EU-numbering position 137 into glycine and mutating serine at EU-numbering position 138 into glycine. The constant region sequence M14 (SEQ ID NO: 129) was produced by introducing all the above mutations.
[0391] The expression and purification of huPM1-M14, prepared by using huPM1-M14 as an H chain and huPM1-L (SEQ ID NO: 162) as an L chain, was carried out by the method described in Referential Example 7. The prepared huPM1-M14 (SEQ ID NO: 163), huPM1-IgG1, and huPM1-IgG2 were assessed for the heterogeneity using cation exchange chromatography by the method described in Referential Example 7.
[0392] As shown in FIG. 14, the heterogeneity was also reduced in huPM1-M14 as in huPM1-SKSC.
[Referential Example 9] Preparation of huPM1-M58 with Reduced H-Chain C-Terminal Heterogeneity and Improved Pharmacokinetics
[0393] Preparation of huPM1-M58 Molecule
[0394] huPM1 is an IgG1 antibody. For the heterogeneity in the C-terminal sequence of the H chain of IgG antibody, the deletion of the C-terminal lysine residue and the amidation of the C-terminal amino group due to deletion of the two C-terminal amino acids, glycine and lysine, have been reported (Anal Biochem. 2007 Jan. 1; 360(1): 75-83). Also in huPM1, while the major component is a sequence in which the C-terminal lysine encoded by the nucleotide sequence has been deleted by post-translational modification, there are also a minor component in which the lysine remains and a minor component in which the C-terminal amino group is amidated due to deletion of both glycine and lysine, which contribute to heterogeneity. Producing a pharmaceutical in a large scale while maintaining the difference in the heterogeneity of desired/related substances between products is not easy but rather results in increase of cost, and it is thus desired that the substance be composed of a single substance as much as possible. When an antibody is developed as a pharmaceutical, reduction of the heterogeneity is desired. Thus, it is desired that the C-terminal of the H chain has no heterogeneity when developed as pharmaceuticals. It is also desirable to prolong the plasma half-life of the antibody in order to reduce the antibody dose.
[0395] Thus, the alterations described below were introduced to prepare a novel constant region in which the heterogeneity at C-terminal of the H chain has been reduced, the pharmacokinetics has been improved as compared to huPM1-IgG1, and the heterogeneity derived from wild-type IgG2 has also been reduced without loss of stability.
[0396] Specifically, in huPM1-SKSC, which has high stability and in which the above-mentioned heterogeneity related to antibodies with IgG2-isotype constant regions is reduced, glutamic acid at EU-numbering position 137 was substituted with glycine; serine at position 138 with glycine; histidine at position 268 with glutamine; arginine at position 355 with glutamine; and glutamine at position 419 with glutamic acid. In addition to the above substitutions, glycine and lysine at positions 446 and 447 were deleted to reduce the heterogeneity of the H-chain C terminus, thereby obtaining huPM1-M58 (SEQ ID NO: 164). huPM1-M58 prepared by using huPM1-M58 as an H chain and huPM1-L (SEQ ID NO: 162) as an L chain was expressed and purified by the method described in Example 4.
[0397] The huPM1-M58, huPM1-IgG1, and huPM1-IgG2 were assessed for the heterogeneity and stability by the methods described in Example 5 using cation exchange chromatography and DSC, respectively.
[0398] The result of DSC is shown in Table 18. As shown in FIGS. 13 and 16, huPM1-M58 was found to show reduced heterogeneity without loss of stability as in huPM1-SKSC.
TABLE-US-00019 TABLE 18 Name Tm/.degree. C. huPM1-IgG1 94.8 huPM1-IgG2 93.9 huPM1-SKSC 93.7 huPM1-M58 93.7
Assessment of huPM1-M58 for Plasma Retention
[0399] The prolonged retention (slow elimination) of IgG molecule in plasma is due to the function of FcRn, which is known as a salvage receptor of IgG molecule (Nat Rev Immunol. 2007 September; 7(9): 715-25). When incorporated into endosomes via pinocytosis, IgG molecules bind to FcRn expressed in endosomes under the acidic conditions within the endosome (approx. pH 6.0). While IgG molecules that are not bound to FcRn are transferred to and degraded in lysosomes, those bound to FcRn are translocated to the cell surface and then released from FcRn into plasma again under the neutral conditions in plasma (approx. pH 7.4).
[0400] IgG-type antibodies are known to include IgG1, IgG2, IgG3, and IgG4 isotypes. The plasma half-lives of these isotypes in human are reported to be about 36 days for IgG1 and IgG2; about 29 days for IgG3; and 16 days for IgG4 (Nat. Biotechnol. 2007 December; 25(12): 1369-72). Thus, the retention of IgG1 and IgG2 in plasma is believed to be the longest. In general, the isotypes of antibodies used as pharmaceutical agents are IgG1, IgG2, and IgG4. Reported methods for further improving the pharmacokinetics of these IgG antibodies include methods for improving the above-described binding activity to human FcRn by altering the sequence of IgG constant region (J. Biol. Chem. 2007 Jan. 19; 282(3): 1709-17; J. Immunol. 2006 Jan. 1; 176(1): 346-56).
[0401] There are species differences between mouse FcRn and human FcRn (Proc. Natl. Acad. Sci. USA. 2006 Dec. 5; 103(49): 18709-14). Therefore, to predict the retention of IgG antibodies having an altered constant region sequence in human plasma, it may be desirable to assess the binding to human FcRn and the plasma retention in human FcRn transgenic mice (Int. Immunol. 2006 December; 18(12): 1759-69).
Assessment of the Binding to Human FcRn
[0402] FcRn is a complex of FcRn and .beta.2-microglobulin. Oligo-DNA primers were prepared based on the published human FcRn gene sequence (J. Exp. Med. (1994) 180 (6), 2377-2381). A DNA fragment encoding the whole gene was prepared by PCR using human cDNA (Human Placenta Marathon-Ready cDNA, Clontech) as a template and the prepared primers. Using the obtained DNA fragment as a template, a DNA fragment encoding the extracellular domain containing the signal region (Met1-Leu290) was amplified by PCR, and inserted into an animal cell expression vector (the amino acid sequence of human FcRn/SEQ ID NO: 165). Likewise, oligo-DNA primers were prepared based on the published human .beta.2-microglobulin gene sequence (Proc. Natl. Acad. Sci. U.S.A. 99 (26), 16899-16903 (2002)). A DNA fragment encoding the whole gene was prepared by PCR using human cDNA (Hu-Placenta Marathon-Ready cDNA, CLONTECH) as a template and the prepared primers. Using the obtained DNA fragment as a template, a DNA fragment encoding the whole .beta.2-microglobulin containing the signal region (Met1-Met119) was amplified by PCR and inserted into an animal cell expression vector (the amino acid sequence of human .beta.2-microglobulin/SEQ ID NO: 166).
[0403] Soluble human FcRn was expressed by the following procedure. The prepared plasmids for human FcRn and .beta.2-microglobulin were introduced into the human embryonic kidney cancer-derived cell line HEK293H (Invitrogen) using 10% fetal bovine serum (Invitrogen) by lipofection. The resulting culture supernatant was collected and purified using IgG Sepharose 6 Fast Flow (Amersham Biosciences) by the method described in J. Immunol. 2002 Nov. 1; 169(9):5171-80. Then further purification was carried out using HiTrap Q HP (GE Healthcare).
[0404] The binding to human FcRn was assessed using Biacore 3000. An antibody was bound to Protein L or rabbit anti-human IgG Kappa chain antibody immobilized onto a sensor chip, human FcRn was added as an analyte for interaction with the antibody, and the affinity (KD) was calculated from the amount of bound human FcRn. Specifically, Protein L was immobilized onto sensor chip CMS (BIACORE) by the amine coupling method using 50 mM Na-phosphate buffer (pH 6.0) containing 150 mM NaCl as the running buffer. Then, an antibody was diluted with the running buffer containing 0.02% Tween20, and injected and allowed to bind to the chip. Human FcRn was then injected to assess the binding activity of the antibody to the human FcRn.
[0405] The affinity was calculated using BIAevaluation software. The obtained sensorgram was used to calculate the amount of hFcRn bound to the antibody immediately before the end of human FcRn injection. This was fitted by the steady state affinity method to calculate the affinity of human FcRn for the antibody.
Predictive Assessment of Plasma Retention of huPM1-IgG1 and huPM1-M58 in Human Using Human FcRn
[0406] The binding activities of huPM1-IgG1 and huPM1-M58 to human FcRn were assessed using BIAcore. As shown in Table 19, the binding activity of huPM1-M58 was greater than that of huPM1-IgG1 by about 1.4 times.
TABLE-US-00020 TABLE 19 KD (.mu.M) huPM1-IgG1 1.62 huPM1-M58 1.17
Assessment of the Plasma Retention in Human FcRn Transgenic Mice
[0407] The pharmacokinetics in human FcRn transgenic mice (B6.mFcRn-/-.hFcRn Tg line 276+/+ mice; Jackson Laboratories) was assessed by the following procedure. An antibody was intravenously administered once at a dose of 1 mg/kg to mice, and blood was collected at appropriate time points. The collected blood was immediately centrifuged at 15,000 rpm for 15 minutes at 4.degree. C. to obtain plasma. The separated plasma was stored in a freezer at -20.degree. C. or below until use. The plasma concentration was determined by ELISA.
Predictive Assessment of the Plasma Retention of huPM1-IgG1 and huPM1-M58 in Human Using Human FcRn Transgenic Mice
[0408] The plasma retention of huPM1-IgG1 and huPM1-M58 in human FcRn transgenic mice was assessed. As shown in FIG. 17, the result demonstrated that the pharmacokinetics of huPM1-M58 was improved as compared to huPM1-IgG1. It was suggested that the human FcRn-binding activity was correlated to the plasma retention in human FcRn transgenic mice.
[Referential Example 10] Measurement of the Affinity in Antigen-Antibody Reaction Using Biacore
[0409] Kinetic analysis of the antigen-antibody reaction was carried out using Biacore T100 (GE Healthcare Biosciences). The antigen-antibody interaction was measured by immobilizing rec-Protein A (hereinafter Protein A) (ZYMED) onto a sensor chip, capturing an antibody on the immobilized Protein A, and then reacting the antigen as an analyte. Various concentrations of rhNR10 were used as the antigen. The kinetic parameters, association rate constant k.sub.a (1/Ms) and dissociation rate constant k.sub.d (1/s), were calculated from the sensorgrams obtained by the measurement. Then, K.sub.D (M) was determined based on the rate constants. Each parameter was determined using Biacore T100 Evaluation Software version 1.1 (GE Healthcare Biosciences).
Immobilization of Protein A onto Sensor Chip
[0410] Protein A was immobilized onto all flow cells of sensor chip CMS (GE Healthcare Bioscinences) by the amine coupling method. The experiment was carried out using HBS-EP+ (10 mM HEPES, 0.15 M NaCl, 3 mM EDTA, 0.05% v/v Surfactant P20) as a running buffer at a flow rate of 10 .mu.L/min. The carboxyl groups of carboxymethyl dextran on the sensor chip were activated with 1004 of a 1:1 mixture of 75 mg/ml EDC (N-ethyl-N'-(3-dimethylaminopropyl)carbodiimide hydrochloride) and 11.5 mg/ml NHS (N-hydroxysuccinimide), and Protein A prepared at 50 .mu.g/ml using 10 mM acetate buffer (pH 4.5) was allowed to flow for reaction. Then, 1004 of 1 M ethanolamine hydrochloride (pH 8.5) was allowed to flow to inactivate the unreacted active groups. Ultimately, about 4000 to 5000 RU were immobilized. The experiment was carried out at 25.degree. C. at all times.
Measurement of Affinity in Antigen-Antibody Reaction Between rhNR10 and Antibody Captured on Protein A
[0411] The running buffer used was HBS-EP+. Each antibody was prepared at 0.25 .mu.g/ml, or prepared so that about 100 RU would bind to Protein A. rhNR10 used as an analyte was prepared at 0, 38.5, 77.0, and 154 nM, or at 0, 19.25, and 77.01 nM using HBS-EP+. In the measurement, first, the antibody solution was captured on Protein A, and an analyte solution was reacted at a flow rate of 20 .mu.L/min for three minutes. Then, the solution was switched to HBS-EP+, and the dissociation phase was measured for five minutes. After measurement of the dissociation phase, the sensor chip was regenerated by washing with 10 mM glycine-HCl (pH 1.5). The obtained sensorgrams were kinetically analyzed using the Biacore-specific data analysis software, Biacore T100 Evaluation Software Version 1.1.
INDUSTRIAL APPLICABILITY
[0412] The anti-NR10 antibodies obtained by the present inventors exhibit an effective neutralizing activity against NR10, and are useful as, for example, therapeutic agents for inflammatory diseases.
Sequence CWU 1
1
28715PRTMus musculus 1Gly Tyr Ile Met Asn1 5217PRTMus musculus 2Leu
Ile Asn Pro Tyr Asn Gly Asp Thr Asn Tyr Asn Gln Lys Phe Lys1 5 10
15Gly312PRTMus musculus 3Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met
Asp Tyr1 5 104121PRTMus musculus 4Glu Val Gln Leu Gln Gln Ser Gly
Pro Glu Leu Val Lys Pro Gly Ala1 5 10 15Ser Met Lys Ile Ser Cys Lys
Ala Ser Gly Tyr Ser Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Lys
Gln Ser His Gly Lys Asn Leu Glu Trp Ile 35 40 45Gly Leu Ile Asn Pro
Tyr Asn Gly Asp Thr Asn Tyr Asn Gln Lys Phe 50 55 60Lys Gly Lys Ala
Thr Leu Thr Val Asp Lys Ser Ser Ser Thr Ala Tyr65 70 75 80Met Glu
Leu Leu Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Tyr Cys 85 90 95Ala
Arg Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Asp Tyr Trp Gly 100 105
110Gln Gly Thr Ser Val Thr Val Ser Ser 115 120511PRTMus musculus
5Arg Ala Ser Glu Asn Ile Tyr Ser Phe Leu Ala1 5 1067PRTMus musculus
6Asn Ala Lys Thr Leu Ala Lys1 579PRTMus musculus 7Gln His His Tyr
Glu Ser Pro Leu Thr1 58107PRTMus musculus 8Asp Ile Gln Met Thr Gln
Ser Pro Ala Ser Leu Ser Ala Ser Val Gly1 5 10 15Glu Thr Val Thr Ile
Thr Cys Arg Ala Ser Glu Asn Ile Tyr Ser Phe 20 25 30Leu Ala Trp Tyr
Gln Gln Lys Gln Gly Lys Ser Pro His Leu Leu Val 35 40 45Tyr Asn Ala
Lys Thr Leu Ala Lys Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly
Ser Gly Thr Gln Phe Ser Leu Lys Ile Asn Ser Leu Gln Pro65 70 75
80Glu Asp Phe Gly Ser Tyr Tyr Cys Gln His His Tyr Glu Ser Pro Leu
85 90 95Thr Phe Gly Gly Gly Thr Lys Leu Glu Ile Lys 100 10595PRTMus
musculus 9Gly Tyr Ile Met Asn1 51017PRTMus musculus 10Leu Ile Asn
Pro Tyr Asn Gly Gly Thr Ser Tyr Asn Gln Lys Phe Lys1 5 10
15Gly1112PRTMus musculus 11Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met
Asp Tyr1 5 1012121PRTMus musculus 12Glu Val Gln Leu Gln Gln Ser Gly
Pro Glu Leu Val Lys Pro Gly Ala1 5 10 15Ser Met Lys Ile Ser Cys Lys
Ala Ser Gly Tyr Ser Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Lys
Gln Ser His Gly Lys Asn Leu Glu Trp Ile 35 40 45Gly Leu Ile Asn Pro
Tyr Asn Gly Gly Thr Ser Tyr Asn Gln Lys Phe 50 55 60Lys Gly Lys Ala
Thr Leu Thr Val Asp Lys Ser Ser Ser Thr Ala Tyr65 70 75 80Met Glu
Leu Leu Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Tyr Cys 85 90 95Ala
Arg Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Asp Tyr Trp Gly 100 105
110Gln Gly Thr Ser Val Thr Val Ser Ser 115 1201311PRTMus musculus
13Arg Thr Ser Glu Asn Ile Tyr Ser Phe Leu Ala1 5 10147PRTMus
musculus 14Asn Ala Lys Thr Leu Ala Lys1 5159PRTMus musculus 15Gln
His His Tyr Glu Ser Pro Leu Thr1 516107PRTMus musculus 16Asp Ile
Gln Met Thr Gln Ser Pro Ala Ser Leu Ser Ala Ser Val Gly1 5 10 15Glu
Thr Val Thr Ile Thr Cys Arg Thr Ser Glu Asn Ile Tyr Ser Phe 20 25
30Leu Ala Trp Tyr Gln Gln Lys Gln Gly Lys Ser Pro His Leu Leu Val
35 40 45Tyr Asn Ala Lys Thr Leu Ala Lys Gly Val Pro Ser Arg Phe Ser
Gly 50 55 60Ser Gly Ser Gly Thr Gln Phe Ser Leu Lys Ile Asn Ser Leu
Gln Pro65 70 75 80Glu Asp Phe Gly Ser Tyr Phe Cys Gln His His Tyr
Glu Ser Pro Leu 85 90 95Thr Phe Gly Gly Gly Thr Lys Leu Glu Ile Lys
100 105175PRTMus musculus 17Gly Tyr Ile Met Asn1 51817PRTMus
musculus 18Leu Ile Asn Pro Tyr Asn Gly Gly Ala Glu Tyr Asn Gln Lys
Phe Lys1 5 10 15Asp1912PRTMus musculus 19Asp Gly Tyr Asp Asp Gly
Pro Tyr Thr Met Asp Tyr1 5 1020121PRTMus musculus 20Glu Val Gln Leu
Gln Gln Ser Gly Pro Glu Leu Val Lys Pro Gly Thr1 5 10 15Ser Met Lys
Ile Ser Cys Lys Ala Ser Gly Tyr Ser Phe Thr Gly Tyr 20 25 30Ile Met
Asn Trp Val Lys Gln Ser His Gly Lys Asn Leu Glu Trp Ile 35 40 45Gly
Leu Ile Asn Pro Tyr Asn Gly Gly Ala Glu Tyr Asn Gln Lys Phe 50 55
60Lys Asp Lys Ala Thr Phe Thr Val Asp Lys Ser Ser Ser Thr Ala Tyr65
70 75 80Met Glu Leu Leu Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Tyr
Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Asp Tyr
Trp Gly 100 105 110Gln Gly Thr Ser Val Thr Val Ser Ser 115
1202111PRTMus musculus 21Arg Ala Asn Glu Asn Ile Tyr Ser Tyr Leu
Ala1 5 10227PRTMus musculus 22Asn Ala Lys Thr Leu Ala Glu1
5239PRTMus musculus 23Gln His His Tyr Gly Thr Pro Pro Thr1
524107PRTMus musculus 24Asp Ile Gln Met Thr Gln Ser Pro Ala Ser Leu
Ser Ala Ser Val Gly1 5 10 15Glu Thr Val Thr Phe Thr Cys Arg Ala Asn
Glu Asn Ile Tyr Ser Tyr 20 25 30Leu Ala Trp Tyr Gln Gln Lys Gln Gly
Lys Ser Pro Gln Leu Leu Val 35 40 45Tyr Asn Ala Lys Thr Leu Ala Glu
Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Gln Phe
Ser Leu Lys Ile Asn Ser Leu Gln Pro65 70 75 80Glu Asp Phe Gly Ser
Tyr Tyr Cys Gln His His Tyr Gly Thr Pro Pro 85 90 95Thr Phe Gly Gly
Gly Thr Lys Leu Glu Ile Lys 100 105255PRTMus musculus 25Asn Tyr Trp
Met His1 52617PRTMus musculus 26Ala Ile Tyr Pro Gly Asn Ser Asp Thr
Asp Tyr Asn Gln Lys Phe Lys1 5 10 15Gly278PRTMus musculus 27Asp Gly
Tyr Asp Asp Phe Asp His1 528116PRTMus musculus 28Glu Val Gln Leu
Gln Gln Ser Gly Thr Val Leu Ala Arg Pro Gly Ala1 5 10 15Ser Val Lys
Met Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Asn Tyr 20 25 30Trp Met
His Trp Val Lys Gln Arg Pro Gly Gln Gly Leu Glu Trp Ile 35 40 45Gly
Ala Ile Tyr Pro Gly Asn Ser Asp Thr Asp Tyr Asn Gln Lys Phe 50 55
60Lys Gly Lys Ala Lys Leu Thr Ala Val Thr Ser Ala Ser Thr Ala Tyr65
70 75 80Met Glu Leu Ser Ser Leu Thr Asn Glu Asp Ser Ala Val Phe Phe
Cys 85 90 95Thr Thr Gly Tyr Asp Asp Phe Asp His Trp Gly Gln Gly Thr
Thr Leu 100 105 110Thr Val Ser Ser 1152912PRTMus musculus 29Arg Ala
Ser Ser Ser Val Ser Ser Ser Tyr Leu His1 5 10307PRTMus musculus
30Ser Thr Ser Asn Leu Ala Ser1 5319PRTMus musculus 31Gln Gln Tyr
Ser Gly Tyr Pro Leu Thr1 532108PRTMus musculus 32Glu Asn Val Leu
Thr Gln Ser Pro Ala Ile Met Ser Ala Ser Pro Gly1 5 10 15Glu Lys Val
Thr Met Thr Cys Arg Ala Ser Ser Ser Val Ser Ser Ser 20 25 30Tyr Leu
His Trp Tyr Gln Gln Lys Ser Gly Ala Ser Pro Lys Leu Trp 35 40 45Ile
Tyr Ser Thr Ser Asn Leu Ala Ser Gly Val Pro Ala Arg Phe Ser 50 55
60Gly Ser Gly Ser Gly Thr Ser Tyr Tyr Phe Thr Ile Ser Ser Val Glu65
70 75 80Ala Glu Asp Ala Ala Thr Tyr Tyr Cys Gln Gln Tyr Ser Gly Tyr
Pro 85 90 95Leu Thr Phe Gly Gly Gly Thr Lys Leu Glu Ile Lys 100
105331406DNAMus musculus 33atgggatgga gctggatctt tctcttcctc
ctgtcaggaa ctgcaggtgt ccactctgag 60gtccagctgc aacagtctgg acctgagctg
gtgaagcctg gagcttcaat gaagatctcc 120tgcaaggctt ctggttactc
attcactggc tacatcatga actgggtgaa gcagagccat 180ggaaagaacc
ttgagtggat tggacttatt aatccttaca atggtgatac taactacaac
240cagaagttca agggcaaggc cacattaact gtagacaagt catccagcac
agcctacatg 300gaactcctca gtctgacatc agaggactct gcagtctatt
actgtgcaag ggatggttac 360gacgacggac cctatactat ggactactgg
ggtcaaggaa cctcagtcac cgtctcctca 420gccaaaacga cacccccatc
tgtctatcca ctggcccctg gatctgctgc ccaaactaac 480tccatggtga
ccctgggatg cctggtcaag ggctatttcc ctgagccagt gacagtgacc
540tggaactctg gatccctgtc cagcggtgtg cacaccttcc cagctgtcct
gcagtctgac 600ctctacactc tgagcagctc agtgactgtc ccctccagca
cctggcccag cgagaccgtc 660acctgcaacg ttgcccaccc ggccagcagc
accaaggtgg acaagaaaat tgtgcccagg 720gattgtggtt gtaagccttg
catatgtaca gtcccagaag tatcatctgt cttcatcttc 780cccccaaagc
ccaaggatgt gctcaccatt actctgactc ctaaggtcac gtgtgttgtg
840gtagacatca gcaaggatga tcccgaggtc cagttcagct ggtttgtaga
tgatgtggag 900gtgcacacag ctcagacgca accccgggag gagcagttca
acagcacttt ccgctcagtc 960agtgaacttc ccatcatgca ccaggactgg
ctcaatggca aggagttcaa atgcagggtc 1020aacagtgcag ctttccctgc
ccccatcgag aaaaccatct ccaaaaccaa aggcagaccg 1080aaggctccac
aggtgtacac cattccacct cccaaggagc agatggccaa ggataaagtc
1140agtctgacct gcatgataac agacttcttc cctgaagaca ttactgtgga
gtggcagtgg 1200aatgggcagc cagcggagaa ctacaagaac actcagccca
tcatggacac agatggctct 1260tacttcgtct acagcaagct caatgtgcag
aagagcaact gggaggcagg aaatactttc 1320acctgctctg tgttacatga
gggcctgcac aaccaccata ctgagaagag cctctcccac 1380tctcctggta
aataatgagc ggccgc 140634445PRTMus musculus 34Glu Val Gln Leu Gln
Gln Ser Gly Pro Glu Leu Val Lys Pro Gly Ala1 5 10 15Ser Met Lys Ile
Ser Cys Lys Ala Ser Gly Tyr Ser Phe Thr Gly Tyr 20 25 30Ile Met Asn
Trp Val Lys Gln Ser His Gly Lys Asn Leu Glu Trp Ile 35 40 45Gly Leu
Ile Asn Pro Tyr Asn Gly Asp Thr Asn Tyr Asn Gln Lys Phe 50 55 60Lys
Gly Lys Ala Thr Leu Thr Val Asp Lys Ser Ser Ser Thr Ala Tyr65 70 75
80Met Glu Leu Leu Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Tyr Cys
85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Asp Tyr Trp
Gly 100 105 110Gln Gly Thr Ser Val Thr Val Ser Ser Ala Lys Thr Thr
Pro Pro Ser 115 120 125Val Tyr Pro Leu Ala Pro Gly Ser Ala Ala Gln
Thr Asn Ser Met Val 130 135 140Thr Leu Gly Cys Leu Val Lys Gly Tyr
Phe Pro Glu Pro Val Thr Val145 150 155 160Thr Trp Asn Ser Gly Ser
Leu Ser Ser Gly Val His Thr Phe Pro Ala 165 170 175Val Leu Gln Ser
Asp Leu Tyr Thr Leu Ser Ser Ser Val Thr Val Pro 180 185 190Ser Ser
Thr Trp Pro Ser Glu Thr Val Thr Cys Asn Val Ala His Pro 195 200
205Ala Ser Ser Thr Lys Val Asp Lys Lys Ile Val Pro Arg Asp Cys Gly
210 215 220Cys Lys Pro Cys Ile Cys Thr Val Pro Glu Val Ser Ser Val
Phe Ile225 230 235 240Phe Pro Pro Lys Pro Lys Asp Val Leu Thr Ile
Thr Leu Thr Pro Lys 245 250 255Val Thr Cys Val Val Val Asp Ile Ser
Lys Asp Asp Pro Glu Val Gln 260 265 270Phe Ser Trp Phe Val Asp Asp
Val Glu Val His Thr Ala Gln Thr Gln 275 280 285Pro Arg Glu Glu Gln
Phe Asn Ser Thr Phe Arg Ser Val Ser Glu Leu 290 295 300Pro Ile Met
His Gln Asp Trp Leu Asn Gly Lys Glu Phe Lys Cys Arg305 310 315
320Val Asn Ser Ala Ala Phe Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys
325 330 335Thr Lys Gly Arg Pro Lys Ala Pro Gln Val Tyr Thr Ile Pro
Pro Pro 340 345 350Lys Glu Gln Met Ala Lys Asp Lys Val Ser Leu Thr
Cys Met Ile Thr 355 360 365Asp Phe Phe Pro Glu Asp Ile Thr Val Glu
Trp Gln Trp Asn Gly Gln 370 375 380Pro Ala Glu Asn Tyr Lys Asn Thr
Gln Pro Ile Met Asp Thr Asp Gly385 390 395 400Ser Tyr Phe Val Tyr
Ser Lys Leu Asn Val Gln Lys Ser Asn Trp Glu 405 410 415Ala Gly Asn
Thr Phe Thr Cys Ser Val Leu His Glu Gly Leu His Asn 420 425 430His
His Thr Glu Lys Ser Leu Ser His Ser Pro Gly Lys 435 440
44535716DNAMus musculus 35atgagtgtgc ccactcaggt cctggggttg
ctgctgctgt ggcttacagg tgccagatgt 60gacatccaga tgactcagtc tccagcctcc
ctatctgcat ctgtgggaga aactgtcacc 120atcacatgtc gagcaagtga
gaatatttac agttttttag catggtatca gcagaaacag 180ggaaaatctc
ctcacctcct ggtctataat gcaaaaacct tagcaaaagg tgtgccatca
240aggttcagtg gcagtggatc tggcacacag ttttctctga agatcaacag
cctgcagcct 300gaagattttg ggagttatta ctgtcaacat cattatgaga
gtcctctgac gttcggtgga 360ggcaccaagc tggaaatcaa acgggctgat
gctgcaccaa ctgtatccat cttcccacca 420tccagtgagc agttaacatc
tggaggtgcc tcagtcgtgt gcttcttgaa caacttctac 480cccaaagaca
tcaatgtcaa gtggaagatt gatggcagtg aacgacaaaa tggcgtcctg
540aacagttgga ctgatcagga cagcaaagac agcacctaca gcatgagcag
caccctcacg 600ttgaccaagg acgagtatga acgacataac agctatacct
gtgaggccac tcacaagaca 660tcaacttcac ccattgtcaa gagcttcaac
aggaatgagt gttaatgagc ggccgc 71636214PRTMus musculus 36Asp Ile Gln
Met Thr Gln Ser Pro Ala Ser Leu Ser Ala Ser Val Gly1 5 10 15Glu Thr
Val Thr Ile Thr Cys Arg Ala Ser Glu Asn Ile Tyr Ser Phe 20 25 30Leu
Ala Trp Tyr Gln Gln Lys Gln Gly Lys Ser Pro His Leu Leu Val 35 40
45Tyr Asn Ala Lys Thr Leu Ala Lys Gly Val Pro Ser Arg Phe Ser Gly
50 55 60Ser Gly Ser Gly Thr Gln Phe Ser Leu Lys Ile Asn Ser Leu Gln
Pro65 70 75 80Glu Asp Phe Gly Ser Tyr Tyr Cys Gln His His Tyr Glu
Ser Pro Leu 85 90 95Thr Phe Gly Gly Gly Thr Lys Leu Glu Ile Lys Arg
Ala Asp Ala Ala 100 105 110Pro Thr Val Ser Ile Phe Pro Pro Ser Ser
Glu Gln Leu Thr Ser Gly 115 120 125Gly Ala Ser Val Val Cys Phe Leu
Asn Asn Phe Tyr Pro Lys Asp Ile 130 135 140Asn Val Lys Trp Lys Ile
Asp Gly Ser Glu Arg Gln Asn Gly Val Leu145 150 155 160Asn Ser Trp
Thr Asp Gln Asp Ser Lys Asp Ser Thr Tyr Ser Met Ser 165 170 175Ser
Thr Leu Thr Leu Thr Lys Asp Glu Tyr Glu Arg His Asn Ser Tyr 180 185
190Thr Cys Glu Ala Thr His Lys Thr Ser Thr Ser Pro Ile Val Lys Ser
195 200 205Phe Asn Arg Asn Glu Cys 210371406DNAMus musculus
37atgggatgga gctggatctt tctcttcctc ctgtcaggaa ctgcaggtgt ccactctgag
60gtccagctgc aacagtctgg acctgagctg gtgaagcctg gagcttcaat gaagatctcc
120tgcaaggctt ctggttactc attcactggc tacatcatga actgggtgaa
gcagagccat 180ggaaagaacc ttgagtggat tggacttatt aatccttaca
atggtggtac tagctacaac 240cagaagttca agggcaaggc cacattaact
gtagacaagt catccagtac agcctacatg 300gaactcctca gtctgacatc
agaggactct gcagtctatt actgtgcaag ggatggttac 360gacgacggac
cctatactat ggactactgg ggtcaaggaa cctcagtcac cgtctcctca
420gccaaaacga cacccccatc tgtctatcca ctggcccctg gatctgctgc
ccaaactaac 480tccatggtga ccctgggatg cctggtcaag ggctatttcc
ctgagccagt gacagtgacc 540tggaactctg gatccctgtc cagcggtgtg
cacaccttcc cagctgtcct gcagtctgac 600ctctacactc tgagcagctc
agtgactgtc ccctccagca cctggcccag cgagaccgtc 660acctgcaacg
ttgcccaccc ggccagcagc accaaggtgg acaagaaaat tgtgcccagg
720gattgtggtt gtaagccttg catatgtaca gtcccagaag tatcatctgt
cttcatcttc 780cccccaaagc ccaaggatgt gctcaccatt actctgactc
ctaaggtcac gtgtgttgtg 840gtagacatca gcaaggatga tcccgaggtc
cagttcagct ggtttgtaga tgatgtggag 900gtgcacacag ctcagacgca
accccgggag gagcagttca acagcacttt ccgctcagtc 960agtgaacttc
ccatcatgca ccaggactgg ctcaatggca aggagttcaa atgcagggtc
1020aacagtgcag ctttccctgc ccccatcgag aaaaccatct ccaaaaccaa
aggcagaccg 1080aaggctccac aggtgtacac cattccacct cccaaggagc
agatggccaa ggataaagtc 1140agtctgacct gcatgataac agacttcttc
cctgaagaca ttactgtgga gtggcagtgg 1200aatgggcagc cagcggagaa
ctacaagaac actcagccca tcatggacac agatggctct 1260tacttcgtct
acagcaagct caatgtgcag aagagcaact gggaggcagg aaatactttc
1320acctgctctg tgttacatga gggcctgcac aaccaccata
ctgagaagag cctctcccac 1380tctcctggta aataatgagc ggccgc
140638445PRTMus musculus 38Glu Val Gln Leu Gln Gln Ser Gly Pro Glu
Leu Val Lys Pro Gly Ala1 5 10 15Ser Met Lys Ile Ser Cys Lys Ala Ser
Gly Tyr Ser Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Lys Gln Ser
His Gly Lys Asn Leu Glu Trp Ile 35 40 45Gly Leu Ile Asn Pro Tyr Asn
Gly Gly Thr Ser Tyr Asn Gln Lys Phe 50 55 60Lys Gly Lys Ala Thr Leu
Thr Val Asp Lys Ser Ser Ser Thr Ala Tyr65 70 75 80Met Glu Leu Leu
Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp
Gly Tyr Asp Asp Gly Pro Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln
Gly Thr Ser Val Thr Val Ser Ser Ala Lys Thr Thr Pro Pro Ser 115 120
125Val Tyr Pro Leu Ala Pro Gly Ser Ala Ala Gln Thr Asn Ser Met Val
130 135 140Thr Leu Gly Cys Leu Val Lys Gly Tyr Phe Pro Glu Pro Val
Thr Val145 150 155 160Thr Trp Asn Ser Gly Ser Leu Ser Ser Gly Val
His Thr Phe Pro Ala 165 170 175Val Leu Gln Ser Asp Leu Tyr Thr Leu
Ser Ser Ser Val Thr Val Pro 180 185 190Ser Ser Thr Trp Pro Ser Glu
Thr Val Thr Cys Asn Val Ala His Pro 195 200 205Ala Ser Ser Thr Lys
Val Asp Lys Lys Ile Val Pro Arg Asp Cys Gly 210 215 220Cys Lys Pro
Cys Ile Cys Thr Val Pro Glu Val Ser Ser Val Phe Ile225 230 235
240Phe Pro Pro Lys Pro Lys Asp Val Leu Thr Ile Thr Leu Thr Pro Lys
245 250 255Val Thr Cys Val Val Val Asp Ile Ser Lys Asp Asp Pro Glu
Val Gln 260 265 270Phe Ser Trp Phe Val Asp Asp Val Glu Val His Thr
Ala Gln Thr Gln 275 280 285Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe
Arg Ser Val Ser Glu Leu 290 295 300Pro Ile Met His Gln Asp Trp Leu
Asn Gly Lys Glu Phe Lys Cys Arg305 310 315 320Val Asn Ser Ala Ala
Phe Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys 325 330 335Thr Lys Gly
Arg Pro Lys Ala Pro Gln Val Tyr Thr Ile Pro Pro Pro 340 345 350Lys
Glu Gln Met Ala Lys Asp Lys Val Ser Leu Thr Cys Met Ile Thr 355 360
365Asp Phe Phe Pro Glu Asp Ile Thr Val Glu Trp Gln Trp Asn Gly Gln
370 375 380Pro Ala Glu Asn Tyr Lys Asn Thr Gln Pro Ile Met Asp Thr
Asp Gly385 390 395 400Ser Tyr Phe Val Tyr Ser Lys Leu Asn Val Gln
Lys Ser Asn Trp Glu 405 410 415Ala Gly Asn Thr Phe Thr Cys Ser Val
Leu His Glu Gly Leu His Asn 420 425 430His His Thr Glu Lys Ser Leu
Ser His Ser Pro Gly Lys 435 440 44539716DNAMus musculus
39atgagtgtgc ccactcaggt cctggggttg ctgctgctgt ggcttacagg tgccagatgt
60gacatccaga tgactcagtc tccagcctcc ctatctgcat ctgtgggaga aactgtcacc
120atcacatgtc gaacaagtga gaatatttac agttttttag catggtatca
gcagaaacag 180ggaaaatctc ctcacctcct ggtctataat gcaaaaacct
tagcaaaagg tgtgccatca 240aggttcagtg gcagtggatc tggcacacag
ttttctctga agatcaacag cctgcagcct 300gaagattttg ggagttattt
ctgtcaacat cattatgaga gtcctctgac gttcggtgga 360ggcaccaagc
tggaaatcaa acgggctgat gctgcaccaa ctgtatccat cttcccacca
420tccagtgagc agttaacatc tggaggtgcc tcagtcgtgt gcttcttgaa
caacttctac 480cccaaagaca tcaatgtcaa gtggaagatt gatggcagtg
aacgacaaaa tggcgtcctg 540aacagttgga ctgatcagga cagcaaagac
agcacctaca gcatgagcag caccctcacg 600ttgaccaagg acgagtatga
acgacataac agctatacct gtgaggccac tcacaagaca 660tcaacttcac
ccattgtcaa gagcttcaac aggaatgagt gttaatgagc ggccgc 71640214PRTMus
musculus 40Asp Ile Gln Met Thr Gln Ser Pro Ala Ser Leu Ser Ala Ser
Val Gly1 5 10 15Glu Thr Val Thr Ile Thr Cys Arg Thr Ser Glu Asn Ile
Tyr Ser Phe 20 25 30Leu Ala Trp Tyr Gln Gln Lys Gln Gly Lys Ser Pro
His Leu Leu Val 35 40 45Tyr Asn Ala Lys Thr Leu Ala Lys Gly Val Pro
Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Gln Phe Ser Leu Lys
Ile Asn Ser Leu Gln Pro65 70 75 80Glu Asp Phe Gly Ser Tyr Phe Cys
Gln His His Tyr Glu Ser Pro Leu 85 90 95Thr Phe Gly Gly Gly Thr Lys
Leu Glu Ile Lys Arg Ala Asp Ala Ala 100 105 110Pro Thr Val Ser Ile
Phe Pro Pro Ser Ser Glu Gln Leu Thr Ser Gly 115 120 125Gly Ala Ser
Val Val Cys Phe Leu Asn Asn Phe Tyr Pro Lys Asp Ile 130 135 140Asn
Val Lys Trp Lys Ile Asp Gly Ser Glu Arg Gln Asn Gly Val Leu145 150
155 160Asn Ser Trp Thr Asp Gln Asp Ser Lys Asp Ser Thr Tyr Ser Met
Ser 165 170 175Ser Thr Leu Thr Leu Thr Lys Asp Glu Tyr Glu Arg His
Asn Ser Tyr 180 185 190Thr Cys Glu Ala Thr His Lys Thr Ser Thr Ser
Pro Ile Val Lys Ser 195 200 205Phe Asn Arg Asn Glu Cys
210411406DNAMus musculus 41atgggatgga gctggatctt tctcttcctc
ctgtcaggaa ctgcaggtgt ccactctgag 60gtccagctgc aacagtctgg acctgagctg
gtgaagcctg gaacttcaat gaagatatcc 120tgcaaggctt ctggttactc
attcactggc tacatcatga actgggtgaa gcagagccat 180ggaaagaacc
ttgagtggat tggacttatt aatccttaca atggtggtgc tgagtacaac
240cagaagttca aggacaaggc cacattcact gtagacaagt catccagcac
agcctacatg 300gagctcctca gtctgacatc tgaagactct gcagtctatt
actgtgcaag ggatggttac 360gacgacggac cctatactat ggactactgg
ggtcaaggaa cctcagtcac cgtctcctca 420gccaaaacga cacccccatc
tgtctatcca ctggcccctg gatctgctgc ccaaactaac 480tccatggtga
ccctgggatg cctggtcaag ggctatttcc ctgagccagt gacagtgacc
540tggaactctg gatccctgtc cagcggtgtg cacaccttcc cagctgtcct
gcagtctgac 600ctctacactc tgagcagctc agtgactgtc ccctccagca
cctggcccag cgagaccgtc 660acctgcaacg ttgcccaccc ggccagcagc
accaaggtgg acaagaaaat tgtgcccagg 720gattgtggtt gtaagccttg
catatgtaca gtcccagaag tatcatctgt cttcatcttc 780cccccaaagc
ccaaggatgt gctcaccatt actctgactc ctaaggtcac gtgtgttgtg
840gtagacatca gcaaggatga tcccgaggtc cagttcagct ggtttgtaga
tgatgtggag 900gtgcacacag ctcagacgca accccgggag gagcagttca
acagcacttt ccgctcagtc 960agtgaacttc ccatcatgca ccaggactgg
ctcaatggca aggagttcaa atgcagggtc 1020aacagtgcag ctttccctgc
ccccatcgag aaaaccatct ccaaaaccaa aggcagaccg 1080aaggctccac
aggtgtacac cattccacct cccaaggagc agatggccaa ggataaagtc
1140agtctgacct gcatgataac agacttcttc cctgaagaca ttactgtgga
gtggcagtgg 1200aatgggcagc cagcggagaa ctacaagaac actcagccca
tcatggacac agatggctct 1260tacttcgtct acagcaagct caatgtgcag
aagagcaact gggaggcagg aaatactttc 1320acctgctctg tgttacatga
gggcctgcac aaccaccata ctgagaagag cctctcccac 1380tctcctggta
aataatgagc ggccgc 140642445PRTMus musculus 42Glu Val Gln Leu Gln
Gln Ser Gly Pro Glu Leu Val Lys Pro Gly Thr1 5 10 15Ser Met Lys Ile
Ser Cys Lys Ala Ser Gly Tyr Ser Phe Thr Gly Tyr 20 25 30Ile Met Asn
Trp Val Lys Gln Ser His Gly Lys Asn Leu Glu Trp Ile 35 40 45Gly Leu
Ile Asn Pro Tyr Asn Gly Gly Ala Glu Tyr Asn Gln Lys Phe 50 55 60Lys
Asp Lys Ala Thr Phe Thr Val Asp Lys Ser Ser Ser Thr Ala Tyr65 70 75
80Met Glu Leu Leu Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Tyr Cys
85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Asp Tyr Trp
Gly 100 105 110Gln Gly Thr Ser Val Thr Val Ser Ser Ala Lys Thr Thr
Pro Pro Ser 115 120 125Val Tyr Pro Leu Ala Pro Gly Ser Ala Ala Gln
Thr Asn Ser Met Val 130 135 140Thr Leu Gly Cys Leu Val Lys Gly Tyr
Phe Pro Glu Pro Val Thr Val145 150 155 160Thr Trp Asn Ser Gly Ser
Leu Ser Ser Gly Val His Thr Phe Pro Ala 165 170 175Val Leu Gln Ser
Asp Leu Tyr Thr Leu Ser Ser Ser Val Thr Val Pro 180 185 190Ser Ser
Thr Trp Pro Ser Glu Thr Val Thr Cys Asn Val Ala His Pro 195 200
205Ala Ser Ser Thr Lys Val Asp Lys Lys Ile Val Pro Arg Asp Cys Gly
210 215 220Cys Lys Pro Cys Ile Cys Thr Val Pro Glu Val Ser Ser Val
Phe Ile225 230 235 240Phe Pro Pro Lys Pro Lys Asp Val Leu Thr Ile
Thr Leu Thr Pro Lys 245 250 255Val Thr Cys Val Val Val Asp Ile Ser
Lys Asp Asp Pro Glu Val Gln 260 265 270Phe Ser Trp Phe Val Asp Asp
Val Glu Val His Thr Ala Gln Thr Gln 275 280 285Pro Arg Glu Glu Gln
Phe Asn Ser Thr Phe Arg Ser Val Ser Glu Leu 290 295 300Pro Ile Met
His Gln Asp Trp Leu Asn Gly Lys Glu Phe Lys Cys Arg305 310 315
320Val Asn Ser Ala Ala Phe Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys
325 330 335Thr Lys Gly Arg Pro Lys Ala Pro Gln Val Tyr Thr Ile Pro
Pro Pro 340 345 350Lys Glu Gln Met Ala Lys Asp Lys Val Ser Leu Thr
Cys Met Ile Thr 355 360 365Asp Phe Phe Pro Glu Asp Ile Thr Val Glu
Trp Gln Trp Asn Gly Gln 370 375 380Pro Ala Glu Asn Tyr Lys Asn Thr
Gln Pro Ile Met Asp Thr Asp Gly385 390 395 400Ser Tyr Phe Val Tyr
Ser Lys Leu Asn Val Gln Lys Ser Asn Trp Glu 405 410 415Ala Gly Asn
Thr Phe Thr Cys Ser Val Leu His Glu Gly Leu His Asn 420 425 430His
His Thr Glu Lys Ser Leu Ser His Ser Pro Gly Lys 435 440
44543716DNAMus musculus 43atgagtgtgc ccactcaggt cctggggttg
ctgctgctgt ggcttacagg tgccagatgt 60gacatccaga tgactcagtc tccagcctcc
ctatctgcat ctgtgggaga aactgtcacc 120ttcacatgtc gagcaaatga
gaatatttac agttatttag catggtatca gcagaaacag 180ggaaaatctc
ctcagctcct ggtctataat gcaaaaacct tagcagaagg tgtgccatca
240aggttcagtg gcagtggatc aggcacacag ttttctctga agatcaacag
cctgcagcct 300gaagattttg ggagttatta ctgtcaacat cattatggaa
ctcctccgac gttcggtgga 360ggcaccaagc tggaaatcaa acgggctgat
gctgcaccaa ctgtatccat cttcccacca 420tccagtgagc agttaacatc
tggaggtgcc tcagtcgtgt gcttcttgaa caacttctac 480cccaaagaca
tcaatgtcaa gtggaagatt gatggcagtg aacgacaaaa tggcgtcctg
540aacagttgga ctgatcagga cagcaaagac agcacctaca gcatgagcag
caccctcacg 600ttgaccaagg acgagtatga acgacataac agctatacct
gtgaggccac tcacaagaca 660tcaacttcac ccattgtcaa gagcttcaac
aggaatgagt gttaatgagc ggccgc 71644214PRTMus musculus 44Asp Ile Gln
Met Thr Gln Ser Pro Ala Ser Leu Ser Ala Ser Val Gly1 5 10 15Glu Thr
Val Thr Phe Thr Cys Arg Ala Asn Glu Asn Ile Tyr Ser Tyr 20 25 30Leu
Ala Trp Tyr Gln Gln Lys Gln Gly Lys Ser Pro Gln Leu Leu Val 35 40
45Tyr Asn Ala Lys Thr Leu Ala Glu Gly Val Pro Ser Arg Phe Ser Gly
50 55 60Ser Gly Ser Gly Thr Gln Phe Ser Leu Lys Ile Asn Ser Leu Gln
Pro65 70 75 80Glu Asp Phe Gly Ser Tyr Tyr Cys Gln His His Tyr Gly
Thr Pro Pro 85 90 95Thr Phe Gly Gly Gly Thr Lys Leu Glu Ile Lys Arg
Ala Asp Ala Ala 100 105 110Pro Thr Val Ser Ile Phe Pro Pro Ser Ser
Glu Gln Leu Thr Ser Gly 115 120 125Gly Ala Ser Val Val Cys Phe Leu
Asn Asn Phe Tyr Pro Lys Asp Ile 130 135 140Asn Val Lys Trp Lys Ile
Asp Gly Ser Glu Arg Gln Asn Gly Val Leu145 150 155 160Asn Ser Trp
Thr Asp Gln Asp Ser Lys Asp Ser Thr Tyr Ser Met Ser 165 170 175Ser
Thr Leu Thr Leu Thr Lys Asp Glu Tyr Glu Arg His Asn Ser Tyr 180 185
190Thr Cys Glu Ala Thr His Lys Thr Ser Thr Ser Pro Ile Val Lys Ser
195 200 205Phe Asn Arg Asn Glu Cys 210451391DNAMus musculus
45atggaatgta actggatact tccttttatt ctgtcggtaa tttcaggggt ctactcagag
60gttcagctcc agcagtctgg gactgtgctg gcaaggcctg gggcttccgt gaagatgtcc
120tgcaaggctt ctggctacac ctttaccaac tactggatgc actgggtaaa
acagaggcct 180ggacagggtc tagaatggat tggtgctatt tatcctggaa
atagtgatac tgactacaac 240cagaagttca agggcaaggc caaactgact
gcagtcacat ccgccagcac tgcctacatg 300gaactcagca gcctgacaaa
tgaggactct gcggtctttt tctgtaccac tggttacgac 360gacttcgacc
actggggcca aggcaccact ctcacagtct cctcagccaa aacgacaccc
420ccatctgtct atccactggc ccctggatct gctgcccaaa ctaactccat
ggtgaccctg 480ggatgcctgg tcaagggcta tttccctgag ccagtgacag
tgacctggaa ctctggatcc 540ctgtccagcg gtgtgcacac cttcccagct
gtcctgcagt ctgacctcta cactctgagc 600agctcagtga ctgtcccctc
cagcacctgg cccagcgaga ccgtcacctg caacgttgcc 660cacccggcca
gcagcaccaa ggtggacaag aaaattgtgc ccagggattg tggttgtaag
720ccttgcatat gtacagtccc agaagtatca tctgtcttca tcttcccccc
aaagcccaag 780gatgtgctca ccattactct gactcctaag gtcacgtgtg
ttgtggtaga catcagcaag 840gatgatcccg aggtccagtt cagctggttt
gtagatgatg tggaggtgca cacagctcag 900acgcaacccc gggaggagca
gttcaacagc actttccgct cagtcagtga acttcccatc 960atgcaccagg
actggctcaa tggcaaggag ttcaaatgca gggtcaacag tgcagctttc
1020cctgccccca tcgagaaaac catctccaaa accaaaggca gaccgaaggc
tccacaggtg 1080tacaccattc cacctcccaa ggagcagatg gccaaggata
aagtcagtct gacctgcatg 1140ataacagact tcttccctga agacattact
gtggagtggc agtggaatgg gcagccagcg 1200gagaactaca agaacactca
gcccatcatg gacacagatg gctcttactt cgtctacagc 1260aagctcaatg
tgcagaagag caactgggag gcaggaaata ctttcacctg ctctgtgtta
1320catgagggcc tgcacaacca ccatactgag aagagcctct cccactctcc
tggtaaataa 1380tgagcggccg c 139146440PRTMus musculus 46Glu Val Gln
Leu Gln Gln Ser Gly Thr Val Leu Ala Arg Pro Gly Ala1 5 10 15Ser Val
Lys Met Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Asn Tyr 20 25 30Trp
Met His Trp Val Lys Gln Arg Pro Gly Gln Gly Leu Glu Trp Ile 35 40
45Gly Ala Ile Tyr Pro Gly Asn Ser Asp Thr Asp Tyr Asn Gln Lys Phe
50 55 60Lys Gly Lys Ala Lys Leu Thr Ala Val Thr Ser Ala Ser Thr Ala
Tyr65 70 75 80Met Glu Leu Ser Ser Leu Thr Asn Glu Asp Ser Ala Val
Phe Phe Cys 85 90 95Thr Thr Gly Tyr Asp Asp Phe Asp His Trp Gly Gln
Gly Thr Thr Leu 100 105 110Thr Val Ser Ser Ala Lys Thr Thr Pro Pro
Ser Val Tyr Pro Leu Ala 115 120 125Pro Gly Ser Ala Ala Gln Thr Asn
Ser Met Val Thr Leu Gly Cys Leu 130 135 140Val Lys Gly Tyr Phe Pro
Glu Pro Val Thr Val Thr Trp Asn Ser Gly145 150 155 160Ser Leu Ser
Ser Gly Val His Thr Phe Pro Ala Val Leu Gln Ser Asp 165 170 175Leu
Tyr Thr Leu Ser Ser Ser Val Thr Val Pro Ser Ser Thr Trp Pro 180 185
190Ser Glu Thr Val Thr Cys Asn Val Ala His Pro Ala Ser Ser Thr Lys
195 200 205Val Asp Lys Lys Ile Val Pro Arg Asp Cys Gly Cys Lys Pro
Cys Ile 210 215 220Cys Thr Val Pro Glu Val Ser Ser Val Phe Ile Phe
Pro Pro Lys Pro225 230 235 240Lys Asp Val Leu Thr Ile Thr Leu Thr
Pro Lys Val Thr Cys Val Val 245 250 255Val Asp Ile Ser Lys Asp Asp
Pro Glu Val Gln Phe Ser Trp Phe Val 260 265 270Asp Asp Val Glu Val
His Thr Ala Gln Thr Gln Pro Arg Glu Glu Gln 275 280 285Phe Asn Ser
Thr Phe Arg Ser Val Ser Glu Leu Pro Ile Met His Gln 290 295 300Asp
Trp Leu Asn Gly Lys Glu Phe Lys Cys Arg Val Asn Ser Ala Ala305 310
315 320Phe Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Thr Lys Gly Arg
Pro 325 330 335Lys Ala Pro Gln Val Tyr Thr Ile Pro Pro Pro Lys Glu
Gln Met Ala 340 345 350Lys Asp Lys Val Ser Leu Thr Cys Met Ile Thr
Asp Phe Phe Pro Glu 355 360 365Asp Ile Thr Val Glu Trp Gln Trp Asn
Gly Gln Pro Ala Glu Asn Tyr 370 375 380Lys Asn Thr Gln Pro Ile Met
Asp Thr Asp Gly Ser Tyr Phe Val Tyr385 390 395 400Ser Lys Leu Asn
Val Gln Lys Ser Asn Trp Glu Ala Gly Asn Thr
Phe 405 410 415Thr Cys Ser Val Leu His Glu Gly Leu His Asn His His
Thr Glu Lys 420 425 430Ser Leu Ser His Ser Pro Gly Lys 435
44047725DNAMus musculus 47atggattttc tggtgcagat tttcagcttc
ttgctaatca gtgcctcagt tgcaatgtcc 60agaggagaaa atgtgctcac ccagtctcca
gcaatcatgt ctgcatctcc aggggaaaag 120gtcaccatga cctgcagggc
cagctcaagt gtaagttcca gttacttgca ctggtaccag 180cagaagtcag
gtgcctcccc caaactctgg atttatagca cttccaactt ggcttctgga
240gtccctgctc gcttcagtgg cagtgggtct gggacctctt actatttcac
aatcagcagt 300gtggaggctg aagatgctgc cacttattac tgccagcaat
acagtggtta cccactcacg 360ttcggagggg ggaccaagct ggaaataaaa
cgggctgatg ctgcaccaac tgtatccatc 420ttcccaccat ccagtgagca
gttaacatct ggaggtgcct cagtcgtgtg cttcttgaac 480aacttctacc
ccaaagacat caatgtcaag tggaagattg atggcagtga acgacaaaat
540ggcgtcctga acagttggac tgatcaggac agcaaagaca gcacctacag
catgagcagc 600accctcacgt tgaccaagga cgagtatgaa cgacataaca
gctatacctg tgaggccact 660cacaagacat caacttcacc cattgtcaag
agcttcaaca ggaatgagtg ttaatgagcg 720gccgc 72548215PRTMus musculus
48Glu Asn Val Leu Thr Gln Ser Pro Ala Ile Met Ser Ala Ser Pro Gly1
5 10 15Glu Lys Val Thr Met Thr Cys Arg Ala Ser Ser Ser Val Ser Ser
Ser 20 25 30Tyr Leu His Trp Tyr Gln Gln Lys Ser Gly Ala Ser Pro Lys
Leu Trp 35 40 45Ile Tyr Ser Thr Ser Asn Leu Ala Ser Gly Val Pro Ala
Arg Phe Ser 50 55 60Gly Ser Gly Ser Gly Thr Ser Tyr Tyr Phe Thr Ile
Ser Ser Val Glu65 70 75 80Ala Glu Asp Ala Ala Thr Tyr Tyr Cys Gln
Gln Tyr Ser Gly Tyr Pro 85 90 95Leu Thr Phe Gly Gly Gly Thr Lys Leu
Glu Ile Lys Arg Ala Asp Ala 100 105 110Ala Pro Thr Val Ser Ile Phe
Pro Pro Ser Ser Glu Gln Leu Thr Ser 115 120 125Gly Gly Ala Ser Val
Val Cys Phe Leu Asn Asn Phe Tyr Pro Lys Asp 130 135 140Ile Asn Val
Lys Trp Lys Ile Asp Gly Ser Glu Arg Gln Asn Gly Val145 150 155
160Leu Asn Ser Trp Thr Asp Gln Asp Ser Lys Asp Ser Thr Tyr Ser Met
165 170 175Ser Ser Thr Leu Thr Leu Thr Lys Asp Glu Tyr Glu Arg His
Asn Ser 180 185 190Tyr Thr Cys Glu Ala Thr His Lys Thr Ser Thr Ser
Pro Ile Val Lys 195 200 205Ser Phe Asn Arg Asn Glu Cys 210
21549425DNAArtificialan artificially synthesized sequence
49ccaccatgga ctggacctgg agggtcttct gcttgctggc tgtagctcca ggtgctcact
60cccaggtgca gctggtgcag tctggggctg aggtgaagaa gcctggggcc tcagtgaagg
120tttcctgcaa ggcatctgga tacaccttca ccggctacat catgaactgg
gtgcgacagg 180cccctggaca agggcttgag tggatgggac ttattaatcc
ttacaatggt ggtactagct 240acaaccagaa gttcaagggc agagtcacga
ttaccgcgga cgaatccacg agcacagcct 300acatggagct gagcagcctg
agatctgagg acacggccgt gtattactgt gcgagagatg 360gttacgacga
cggaccctat actatggact actggggcca gggcaccctc gtcacagtct 420cctca
42550121PRTArtificialan artificially synthesized sequence 50Gln Val
Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser
Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25
30Ile Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Met
35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn Gln Lys
Phe 50 55 60Lys Gly Arg Val Thr Ile Thr Ala Asp Glu Ser Thr Ser Thr
Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr Ala
Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro Tyr Thr
Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val Ser Ser
115 12051398DNAArtificialan artificially synthesized sequence
51ccaccatgga catgagggtc cccgctcagc tcctggggct cctgctactc tggctccgag
60gtgccagatg tgacatccag atgacccagt ctccatcctc cctgtctgca tctgtaggag
120acagagtcac catcacttgc cgaacaagtg agaatattta cagtttttta
gcatggtatc 180agcagaaacc agggaaagcc cctaagctcc tgatctataa
tgcaaaaacc ttagcaaaag 240gggtcccatc aaggttcagt ggcagtggat
ctgggacaga tttcactctc accatcagca 300gtctgcaacc tgaagatttt
gcaacttact actgtcaaca tcattatgag agtcctctga 360cgttcggcgg
agggaccaag gtggagatca aacgtacg 39852107PRTArtificialan artificially
synthesized sequence 52Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu
Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Thr Ser
Glu Asn Ile Tyr Ser Phe 20 25 30Leu Ala Trp Tyr Gln Gln Lys Pro Gly
Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Asn Ala Lys Thr Leu Ala Lys
Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe
Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr
Tyr Tyr Cys Gln His His Tyr Glu Ser Pro Leu 85 90 95Thr Phe Gly Gly
Gly Thr Lys Val Glu Ile Lys 100 105531422DNAArtificialan
artificially synthesized sequence 53gaattccacc atggactgga
cctggagggt cttctgcttg ctggctgtag ctccaggtgc 60tcactcccag gtgcagctgg
tgcagtctgg ggctgaggtg aagaagcctg gggcctcagt 120gaaggtttcc
tgcaaggcat ctggatacac cttcaccggc tacatcatga actgggtgcg
180acaggcccct ggacaagggc ttgagtggat gggacttatt aatccttaca
atggtggtac 240tagctacaac cagaagttca agggcagagt cacgattacc
gcggacgaat ccacgagcac 300agcctacatg gagctgagca gcctgagatc
tgaggacacg gccgtgtatt actgtgcgag 360agatggttac gacgacggac
cctatactat ggactactgg ggccagggca ccctcgtcac 420agtctcctca
gctagcacca agggcccatc ggtcttcccc ctggcgccct cctccaagag
480cacctccgag agcacagcgg ccctgggctg cctggtcaag gactacttcc
ccgaaccggt 540gacggtgtcg tggaactcag gcgctctgac cagcggcgtg
cacaccttcc cggctgtcct 600acagtcctca ggactctact ccctcagcag
cgtggtgacc gtgccctcca gcaacttcgg 660cacccagacc tacacctgca
acgtagatca caagcccagc aacaccaagg tggacaagac 720agttgagcgc
aaatcttgtg tcgagtgccc accgtgccca gcaccacctg tggcaggacc
780gtcagtcttc ctcttccccc caaaacccaa ggacaccctc atgatctccc
ggacccctga 840ggtcacgtgc gtggtggtgg acgtgagcca cgaagacccc
gaggtccagt tcaactggta 900cgtggacggc gtggaggtgc ataatgccaa
gacaaagcca cgggaggagc agttcaacag 960cacgttccgt gtggtcagcg
tcctcaccgt cgtgcaccag gactggctga acggcaagga 1020gtacaagtgc
aaggtctcca acaaaggcct cccagccccc atcgagaaaa ccatctccaa
1080aaccaaaggg cagccccgag aaccacaggt gtacaccctg cccccatccc
gggaggagat 1140gaccaagaac caggtcagcc tgacctgcct ggtcaaaggc
ttctacccca gcgacatcgc 1200cgtggagtgg gagagcaatg ggcagccgga
gaacaactac aagaccacac ctcccatgct 1260ggactccgac ggctccttct
tcctctacag caagctcacc gtggacaaga gcaggtggca 1320gcaggggaac
gtcttctcat gctccgtgat gcatgaggct ctgcacaacc actacacaca
1380gaagagcctc tccctgtctc cgggtaaatg ataagcggcc gc
142254447PRTArtificialan artificially synthesized sequence 54Gln
Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10
15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly Tyr
20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp
Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn Gln
Lys Phe 50 55 60Lys Gly Arg Val Thr Ile Thr Ala Asp Glu Ser Thr Ser
Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr
Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro Tyr
Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val Ser
Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro Leu Ala Pro
Ser Ser Lys Ser Thr Ser Glu Ser Thr Ala 130 135 140Ala Leu Gly Cys
Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150 155 160Ser
Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala 165 170
175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val
180 185 190Pro Ser Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys Asn Val
Asp His 195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys Thr Val Glu
Arg Lys Ser Cys 210 215 220Val Glu Cys Pro Pro Cys Pro Ala Pro Pro
Val Ala Gly Pro Ser Val225 230 235 240Phe Leu Phe Pro Pro Lys Pro
Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250 255Pro Glu Val Thr Cys
Val Val Val Asp Val Ser His Glu Asp Pro Glu 260 265 270Val Gln Phe
Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 275 280 285Thr
Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser 290 295
300Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu Tyr
Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro Ile
Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly Gln Pro Arg Glu Pro
Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Arg Glu Glu Met Thr Lys
Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr Pro
Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro Glu
Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser385 390 395 400Asp
Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg 405 410
415Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu
420 425 430His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly
Lys 435 440 44555719DNAArtificialan artificially synthesized
sequence 55ccaccatgga catgagggtc cccgctcagc tcctggggct cctgctactc
tggctccgag 60gtgccagatg tgacatccag atgacccagt ctccatcctc cctgtctgca
tctgtaggag 120acagagtcac catcacttgc cgaacaagtg agaatattta
cagtttttta gcatggtatc 180agcagaaacc agggaaagcc cctaagctcc
tgatctataa tgcaaaaacc ttagcaaaag 240gggtcccatc aaggttcagt
ggcagtggat ctgggacaga tttcactctc accatcagca 300gtctgcaacc
tgaagatttt gcaacttact actgtcaaca tcattatgag agtcctctga
360cgttcggcgg agggaccaag gtggagatca aacgtacggt ggctgcacca
tctgtcttca 420tcttcccgcc atctgatgag cagttgaaat ctggaactgc
ctctgttgtg tgcctgctga 480ataacttcta tcccagagag gccaaagtac
agtggaaggt ggataacgcc ctccaatcgg 540gtaactccca ggagagtgtc
acagagcagg acagcaagga cagcacctac agcctcagca 600gcaccctgac
gctgagcaaa gcagactacg agaaacacaa agtctacgcc tgcgaagtca
660cccatcaggg cctgagctcg cccgtcacaa agagcttcaa caggggagag tgttgataa
71956214PRTArtificialan artificially synthesized sequence 56Asp Ile
Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp
Arg Val Thr Ile Thr Cys Arg Thr Ser Glu Asn Ile Tyr Ser Phe 20 25
30Leu Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile
35 40 45Tyr Asn Ala Lys Thr Leu Ala Lys Gly Val Pro Ser Arg Phe Ser
Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu
Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln His His Tyr
Glu Ser Pro Leu 85 90 95Thr Phe Gly Gly Gly Thr Lys Val Glu Ile Lys
Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser
Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu
Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys
Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser
Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170
175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr
180 185 190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr
Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys 21057327DNAHomo sapiens
57cgtacggtgg ctgcaccatc tgtcttcatc ttcccgccat ctgatgagca gttgaaatct
60ggaactgcct ctgttgtgtg cctgctgaat aacttctatc ccagagaggc caaagtacag
120tggaaggtgg ataacgccct ccaatcgggt aactcccagg agagtgtcac
agagcaggac 180agcaaggaca gcacctacag cctcagcagc accctgacgc
tgagcaaagc agactacgag 240aaacacaaag tctacgcctg cgaagtcacc
catcagggcc tgagctcgcc cgtcacaaag 300agcttcaaca ggggagagtg ttgataa
32758107PRTHomo sapiens 58Arg Thr Val Ala Ala Pro Ser Val Phe Ile
Phe Pro Pro Ser Asp Glu1 5 10 15Gln Leu Lys Ser Gly Thr Ala Ser Val
Val Cys Leu Leu Asn Asn Phe 20 25 30Tyr Pro Arg Glu Ala Lys Val Gln
Trp Lys Val Asp Asn Ala Leu Gln 35 40 45Ser Gly Asn Ser Gln Glu Ser
Val Thr Glu Gln Asp Ser Lys Asp Ser 50 55 60Thr Tyr Ser Leu Ser Ser
Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu65 70 75 80Lys His Lys Val
Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser 85 90 95Pro Val Thr
Lys Ser Phe Asn Arg Gly Glu Cys 100 10559990DNAHomo sapiens
59gctagcacca agggcccatc ggtcttcccc ctggcaccct cctccaagag cacctctggg
60ggcacagcgg ccctgggctg cctggtcaag gactacttcc ccgaaccggt gacggtgtcg
120tggaactcag gcgccctgac cagcggcgtg cacaccttcc cggctgtcct
acagtcctca 180ggactctact ccctcagcag cgtggtgacc gtgccctcca
gcagcttggg cacccagacc 240tacatctgca acgtgaatca caagcccagc
aacaccaagg tggacaagaa agttgagccc 300aaatcttgtg acaaaactca
cacatgccca ccgtgcccag cacctgaact cctgggggga 360ccgtcagtct
tcctcttccc cccaaaaccc aaggacaccc tcatgatctc ccggacccct
420gaggtcacat gcgtggtggt ggacgtgagc cacgaagacc ctgaggtcaa
gttcaactgg 480tacgtggacg gcgtggaggt gcataatgcc aagacaaagc
cgcgggagga gcagtacaac 540agcacgtacc gtgtggtcag cgtcctcacc
gtcctgcacc aggactggct gaatggcaag 600gagtacaagt gcaaggtctc
caacaaagcc ctcccagccc ccatcgagaa aaccatctcc 660aaagccaaag
ggcagccccg agaaccacag gtgtacaccc tgcccccatc ccgggatgag
720ctgaccaaga accaggtcag cctgacctgc ctggtcaaag gcttctatcc
cagcgacatc 780gccgtggagt gggagagcaa tgggcagccg gagaacaact
acaagaccac gcctcccgtg 840ctggactccg acggctcctt cttcctctac
agcaagctca ccgtggacaa gagcaggtgg 900cagcagggga acgtcttctc
atgctccgtg atgcatgagg ctctgcacaa ccactacacg 960cagaagagcc
tctccctgtc tccgggtaaa 99060330PRTHomo sapiens 60Ala Ser Thr Lys Gly
Pro Ser Val Phe Pro Leu Ala Pro Ser Ser Lys1 5 10 15Ser Thr Ser Gly
Gly Thr Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr 20 25 30Phe Pro Glu
Pro Val Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser 35 40 45Gly Val
His Thr Phe Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser 50 55 60Leu
Ser Ser Val Val Thr Val Pro Ser Ser Ser Leu Gly Thr Gln Thr65 70 75
80Tyr Ile Cys Asn Val Asn His Lys Pro Ser Asn Thr Lys Val Asp Lys
85 90 95Lys Val Glu Pro Lys Ser Cys Asp Lys Thr His Thr Cys Pro Pro
Cys 100 105 110Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe Leu
Phe Pro Pro 115 120 125Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr
Pro Glu Val Thr Cys 130 135 140Val Val Val Asp Val Ser His Glu Asp
Pro Glu Val Lys Phe Asn Trp145 150 155 160Tyr Val Asp Gly Val Glu
Val His Asn Ala Lys Thr Lys Pro Arg Glu 165 170 175Glu Gln Tyr Asn
Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu 180 185 190His Gln
Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn 195 200
205Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly
210 215 220Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg
Asp Glu225 230 235 240Leu Thr Lys Asn Gln Val Ser Leu Thr Cys Leu
Val Lys Gly Phe Tyr 245 250 255Pro Ser Asp Ile Ala Val Glu Trp Glu
Ser Asn Gly Gln Pro Glu Asn 260 265 270Asn Tyr Lys Thr Thr Pro Pro
Val Leu Asp Ser Asp Gly Ser Phe Phe 275 280 285Leu Tyr Ser Lys Leu
Thr Val Asp Lys Ser Arg Trp
Gln Gln Gly Asn 290 295 300Val Phe Ser Cys Ser Val Met His Glu Ala
Leu His Asn His Tyr Thr305 310 315 320Gln Lys Ser Leu Ser Leu Ser
Pro Gly Lys 325 33061984DNAHomo sapiens 61gctagcacca agggcccatc
ggtcttcccc ctggcgccct cctccaagag cacctccgag 60agcacagcgg ccctgggctg
cctggtcaag gactacttcc ccgaaccggt gacggtgtcg 120tggaactcag
gcgctctgac cagcggcgtg cacaccttcc cggctgtcct acagtcctca
180ggactctact ccctcagcag cgtggtgacc gtgccctcca gcaacttcgg
cacccagacc 240tacacctgca acgtagatca caagcccagc aacaccaagg
tggacaagac agttgagcgc 300aaatcttgtg tcgagtgccc accgtgccca
gcaccacctg tggcaggacc gtcagtcttc 360ctcttccccc caaaacccaa
ggacaccctc atgatctccc ggacccctga ggtcacgtgc 420gtggtggtgg
acgtgagcca cgaagacccc gaggtccagt tcaactggta cgtggacggc
480gtggaggtgc ataatgccaa gacaaagcca cgggaggagc agttcaacag
cacgttccgt 540gtggtcagcg tcctcaccgt cgtgcaccag gactggctga
acggcaagga gtacaagtgc 600aaggtctcca acaaaggcct cccagccccc
atcgagaaaa ccatctccaa aaccaaaggg 660cagccccgag aaccacaggt
gtacaccctg cccccatccc gggaggagat gaccaagaac 720caggtcagcc
tgacctgcct ggtcaaaggc ttctacccca gcgacatcgc cgtggagtgg
780gagagcaatg ggcagccgga gaacaactac aagaccacac ctcccatgct
ggactccgac 840ggctccttct tcctctacag caagctcacc gtggacaaga
gcaggtggca gcaggggaac 900gtcttctcat gctccgtgat gcatgaggct
ctgcacaacc actacacaca gaagagcctc 960tccctgtctc cgggtaaatg ataa
98462326PRTHomo sapiens 62Ala Ser Thr Lys Gly Pro Ser Val Phe Pro
Leu Ala Pro Ser Ser Lys1 5 10 15Ser Thr Ser Glu Ser Thr Ala Ala Leu
Gly Cys Leu Val Lys Asp Tyr 20 25 30Phe Pro Glu Pro Val Thr Val Ser
Trp Asn Ser Gly Ala Leu Thr Ser 35 40 45Gly Val His Thr Phe Pro Ala
Val Leu Gln Ser Ser Gly Leu Tyr Ser 50 55 60Leu Ser Ser Val Val Thr
Val Pro Ser Ser Asn Phe Gly Thr Gln Thr65 70 75 80Tyr Thr Cys Asn
Val Asp His Lys Pro Ser Asn Thr Lys Val Asp Lys 85 90 95Thr Val Glu
Arg Lys Ser Cys Val Glu Cys Pro Pro Cys Pro Ala Pro 100 105 110Pro
Val Ala Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp 115 120
125Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp
130 135 140Val Ser His Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr Val
Asp Gly145 150 155 160Val Glu Val His Asn Ala Lys Thr Lys Pro Arg
Glu Glu Gln Phe Asn 165 170 175Ser Thr Phe Arg Val Val Ser Val Leu
Thr Val Val His Gln Asp Trp 180 185 190Leu Asn Gly Lys Glu Tyr Lys
Cys Lys Val Ser Asn Lys Gly Leu Pro 195 200 205Ala Pro Ile Glu Lys
Thr Ile Ser Lys Thr Lys Gly Gln Pro Arg Glu 210 215 220Pro Gln Val
Tyr Thr Leu Pro Pro Ser Arg Glu Glu Met Thr Lys Asn225 230 235
240Gln Val Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile
245 250 255Ala Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr
Lys Thr 260 265 270Thr Pro Pro Met Leu Asp Ser Asp Gly Ser Phe Phe
Leu Tyr Ser Lys 275 280 285Leu Thr Val Asp Lys Ser Arg Trp Gln Gln
Gly Asn Val Phe Ser Cys 290 295 300Ser Val Met His Glu Ala Leu His
Asn His Tyr Thr Gln Lys Ser Leu305 310 315 320Ser Leu Ser Pro Gly
Lys 32563995DNAHomo sapiens 63gctagcacca agggcccatc cgtcttcccc
ctggcgccct gctccaggag cacctccgag 60agcacagccg ccctgggctg cctggtcaag
gactacttcc ccgaaccggt gacggtgtcg 120tggaactcag gcgccctgac
cagcggcgtg cacaccttcc cggctgtcct acagtcctca 180ggactctact
ccctcagcag cgtggtgacc gtgccctcca gcagcttggg cacgaagacc
240tacacctgca acgtagatca caagcccagc aacaccaagg tggacaagag
agttgagtcc 300aaatatggtc ccccatgccc accatgccca gcacctgagt
tcctgggggg accatcagtc 360ttcctgttcc ccccaaaacc caaggacact
ctcatgatct cccggacccc tgaggtcacg 420tgcgtggtgg tggacgtgag
ccaggaagac cccgaggtcc agttcaactg gtacgtggat 480ggcgtggagg
tgcataatgc caagacaaag ccgcgggagg agcagttcaa cagcacgtac
540cgtgtggtca gcgtcctcac cgtcctgcac caggactggc tgaacggcaa
ggagtacaag 600tgcaaggtct ccaacaaagg cctcccgtcc tccatcgaga
aaaccatctc caaagccaaa 660gggcagcccc gagagccaca ggtgtacacc
ctgcccccat cccaggagga gatgaccaag 720aaccaggtca gcctgacctg
cctggtcaaa ggcttctacc ccagcgacat cgccgtggag 780tgggagagca
atgggcagcc ggagaacaac tacaagacca cgcctcccgt gctggactcc
840gacggctcct tcttcctcta cagcaggcta accgtggaca agagcaggtg
gcaggagggg 900aatgtcttct catgctccgt gatgcatgag gctctgcaca
accactacac acagaagagc 960ctctccctgt ctctgggtta atgataagcg gccgc
99564326PRTHomo sapiens 64Ala Ser Thr Lys Gly Pro Ser Val Phe Pro
Leu Ala Pro Cys Ser Arg1 5 10 15Ser Thr Ser Glu Ser Thr Ala Ala Leu
Gly Cys Leu Val Lys Asp Tyr 20 25 30Phe Pro Glu Pro Val Thr Val Ser
Trp Asn Ser Gly Ala Leu Thr Ser 35 40 45Gly Val His Thr Phe Pro Ala
Val Leu Gln Ser Ser Gly Leu Tyr Ser 50 55 60Leu Ser Ser Val Val Thr
Val Pro Ser Ser Ser Leu Gly Thr Lys Thr65 70 75 80Tyr Thr Cys Asn
Val Asp His Lys Pro Ser Asn Thr Lys Val Asp Lys 85 90 95Arg Val Glu
Ser Lys Tyr Gly Pro Pro Cys Pro Pro Cys Pro Ala Pro 100 105 110Glu
Phe Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys 115 120
125Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val
130 135 140Asp Val Ser Gln Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr
Val Asp145 150 155 160Gly Val Glu Val His Asn Ala Lys Thr Lys Pro
Arg Glu Glu Gln Phe 165 170 175Asn Ser Thr Tyr Arg Val Val Ser Val
Leu Thr Val Leu His Gln Asp 180 185 190Trp Leu Asn Gly Lys Glu Tyr
Lys Cys Lys Val Ser Asn Lys Gly Leu 195 200 205Pro Ser Ser Ile Glu
Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg 210 215 220Glu Pro Gln
Val Tyr Thr Leu Pro Pro Ser Gln Glu Glu Met Thr Lys225 230 235
240Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp
245 250 255Ile Ala Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn
Tyr Lys 260 265 270Thr Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe
Phe Leu Tyr Ser 275 280 285Arg Leu Thr Val Asp Lys Ser Arg Trp Gln
Glu Gly Asn Val Phe Ser 290 295 300Cys Ser Val Met His Glu Ala Leu
His Asn His Tyr Thr Gln Lys Ser305 310 315 320Leu Ser Leu Ser Leu
Gly 325652208DNAMacaca fascicularis 65atgatgtgga cctgggcact
gtggatgttc cctttactct gcaaattcgg cctggcagct 60ctgccagcta agcctgagaa
catttcctgt gtctactact ataggaaaaa tttaacctgc 120acttggagtc
caggaaagga aactagttat acccagtaca cagctaagag aacttacgct
180tttggaaaaa aacatgataa ttgtacaacc agtagttcta caagtgaaaa
tcgtgcttcg 240tgctcttttt tccttccaag aataacgatc ccagataatt
ataccattga ggtggaagct 300gaaaatggag atggtgtaat taaatctgat
atgacatgtt ggagattaga ggacatagcg 360aaaactgaac cacctgagat
tttcagtgtg aaaccagttt tgggcatcaa acgaatgatt 420cggattgaat
ggataaagcc tgagttggca cctgtttcat ctgatttaaa atatgcactt
480cgattcagga cagtcaatag taccagctgg atggaagtca acttcgctaa
gaaccgtaaa 540gatacaaacc aaacctacaa ccttatgggg ctgcaggctt
ttacagagta tgtcgtagct 600ctgcgatgtg cggtcaagga gtcaaagttc
tggagtgact ggagccaaga aaaaatggga 660atgactgagg aagaagctcc
atgtggcctg gaactgtgga gagtcctgaa accaactgag 720gtggatggaa
gaaggccagt gcggttgtta tggaagaagg caagaggagc cccagtccta
780gagaaaacac ttggctacaa catatggtac tttccagaaa acaacactaa
cctcacagag 840acagtgaaca ccactaacca gcagcttgaa ctgcatctgg
gaggcgagag ctattgggtg 900tctatgattt cttataattc tcttgggaag
tctccagtga ccaccctgag gattccagcc 960attcaggaaa agtcatttcg
gtgcattgag gtcatgcagg cctgccttgc tgaggaccag 1020ctagtggtga
agtggcaaag ctctgctcta gacgtgaaca cttggatgat tgaatggttt
1080ccggacatgg actcagagca ccccactctt tcctgggaat ctgtgtctca
ggccacgaac 1140tggacaatcc agcaagataa attaaaacct ttctggtgct
ataacatctc tgtgtatcca 1200atgttgcacg acaaagttgg cgagccatat
tccatccagg cttatgccaa agaaggcatt 1260ccatcaaaag gtcctgagac
caaggtggag aacattggcg tgaagacggt cacgatcaca 1320tggaaagaga
ttcccaagag tgagagaaag ggtatcatct gcaactacac catcttttac
1380caagctgaag gtggaaaagg attctccaag acagtcaact ccagcatctt
gcagtatggc 1440ctggagtccc tgaaacgaaa gacctcttac actgttcggg
tcatggccag caccagtgct 1500gggggaatca acgggaccag cataaatttc
aagacattgt cattcagtgt ttttgagatt 1560atccttataa cttctctgat
tggtggaggc cttcttattc tcattatcct gacggtggca 1620tatggtctca
aaaaacccaa caaattgact cacctgtgtt ggcccagtgt tcccaaccct
1680gctgaaagta gtatagccac atggcgtgga gatgatttca aggataagct
aaacctgaag 1740gagtctgatg actctgtgaa cacagaagac aggatcttaa
aaccatgttc cacccccagt 1800gacaagttgg ttattgacaa gtcggtggtg
aactttggga atgttctgca agaaatgttc 1860acagatgaag ccagaacggg
tcaggaaaac aatttaggag gggaaaagaa tgagtatgtg 1920acccacccct
tcagggctga ctgtcccctg gggaaaagtt ttgaggagct cccagtttca
1980cctgagattc ctcccagaaa atcccaatac ctacgttcga ggatgccaga
agggacctgc 2040ctagaagccg aagagcagct tctcgtttct ggtcaaagtc
tagaaagtct agcaccagac 2100catgtgcggg aggcagcggc cccaaatccg
tatttgaaaa attcagtgac aaccagggaa 2160tttcttgtgt ctcaaaaact
tccagagcac accaaaggag aagtctaa 220866735PRTMacaca fascicularis
66Met Met Trp Thr Trp Ala Leu Trp Met Phe Pro Leu Leu Cys Lys Phe1
5 10 15Gly Leu Ala Ala Leu Pro Ala Lys Pro Glu Asn Ile Ser Cys Val
Tyr 20 25 30Tyr Tyr Arg Lys Asn Leu Thr Cys Thr Trp Ser Pro Gly Lys
Glu Thr 35 40 45Ser Tyr Thr Gln Tyr Thr Ala Lys Arg Thr Tyr Ala Phe
Gly Lys Lys 50 55 60His Asp Asn Cys Thr Thr Ser Ser Ser Thr Ser Glu
Asn Arg Ala Ser65 70 75 80Cys Ser Phe Phe Leu Pro Arg Ile Thr Ile
Pro Asp Asn Tyr Thr Ile 85 90 95Glu Val Glu Ala Glu Asn Gly Asp Gly
Val Ile Lys Ser Asp Met Thr 100 105 110Cys Trp Arg Leu Glu Asp Ile
Ala Lys Thr Glu Pro Pro Glu Ile Phe 115 120 125Ser Val Lys Pro Val
Leu Gly Ile Lys Arg Met Ile Arg Ile Glu Trp 130 135 140Ile Lys Pro
Glu Leu Ala Pro Val Ser Ser Asp Leu Lys Tyr Ala Leu145 150 155
160Arg Phe Arg Thr Val Asn Ser Thr Ser Trp Met Glu Val Asn Phe Ala
165 170 175Lys Asn Arg Lys Asp Thr Asn Gln Thr Tyr Asn Leu Met Gly
Leu Gln 180 185 190Ala Phe Thr Glu Tyr Val Val Ala Leu Arg Cys Ala
Val Lys Glu Ser 195 200 205Lys Phe Trp Ser Asp Trp Ser Gln Glu Lys
Met Gly Met Thr Glu Glu 210 215 220Glu Ala Pro Cys Gly Leu Glu Leu
Trp Arg Val Leu Lys Pro Thr Glu225 230 235 240Val Asp Gly Arg Arg
Pro Val Arg Leu Leu Trp Lys Lys Ala Arg Gly 245 250 255Ala Pro Val
Leu Glu Lys Thr Leu Gly Tyr Asn Ile Trp Tyr Phe Pro 260 265 270Glu
Asn Asn Thr Asn Leu Thr Glu Thr Val Asn Thr Thr Asn Gln Gln 275 280
285Leu Glu Leu His Leu Gly Gly Glu Ser Tyr Trp Val Ser Met Ile Ser
290 295 300Tyr Asn Ser Leu Gly Lys Ser Pro Val Thr Thr Leu Arg Ile
Pro Ala305 310 315 320Ile Gln Glu Lys Ser Phe Arg Cys Ile Glu Val
Met Gln Ala Cys Leu 325 330 335Ala Glu Asp Gln Leu Val Val Lys Trp
Gln Ser Ser Ala Leu Asp Val 340 345 350Asn Thr Trp Met Ile Glu Trp
Phe Pro Asp Met Asp Ser Glu His Pro 355 360 365Thr Leu Ser Trp Glu
Ser Val Ser Gln Ala Thr Asn Trp Thr Ile Gln 370 375 380Gln Asp Lys
Leu Lys Pro Phe Trp Cys Tyr Asn Ile Ser Val Tyr Pro385 390 395
400Met Leu His Asp Lys Val Gly Glu Pro Tyr Ser Ile Gln Ala Tyr Ala
405 410 415Lys Glu Gly Ile Pro Ser Lys Gly Pro Glu Thr Lys Val Glu
Asn Ile 420 425 430Gly Val Lys Thr Val Thr Ile Thr Trp Lys Glu Ile
Pro Lys Ser Glu 435 440 445Arg Lys Gly Ile Ile Cys Asn Tyr Thr Ile
Phe Tyr Gln Ala Glu Gly 450 455 460Gly Lys Gly Phe Ser Lys Thr Val
Asn Ser Ser Ile Leu Gln Tyr Gly465 470 475 480Leu Glu Ser Leu Lys
Arg Lys Thr Ser Tyr Thr Val Arg Val Met Ala 485 490 495Ser Thr Ser
Ala Gly Gly Ile Asn Gly Thr Ser Ile Asn Phe Lys Thr 500 505 510Leu
Ser Phe Ser Val Phe Glu Ile Ile Leu Ile Thr Ser Leu Ile Gly 515 520
525Gly Gly Leu Leu Ile Leu Ile Ile Leu Thr Val Ala Tyr Gly Leu Lys
530 535 540Lys Pro Asn Lys Leu Thr His Leu Cys Trp Pro Ser Val Pro
Asn Pro545 550 555 560Ala Glu Ser Ser Ile Ala Thr Trp Arg Gly Asp
Asp Phe Lys Asp Lys 565 570 575Leu Asn Leu Lys Glu Ser Asp Asp Ser
Val Asn Thr Glu Asp Arg Ile 580 585 590Leu Lys Pro Cys Ser Thr Pro
Ser Asp Lys Leu Val Ile Asp Lys Ser 595 600 605Val Val Asn Phe Gly
Asn Val Leu Gln Glu Met Phe Thr Asp Glu Ala 610 615 620Arg Thr Gly
Gln Glu Asn Asn Leu Gly Gly Glu Lys Asn Glu Tyr Val625 630 635
640Thr His Pro Phe Arg Ala Asp Cys Pro Leu Gly Lys Ser Phe Glu Glu
645 650 655Leu Pro Val Ser Pro Glu Ile Pro Pro Arg Lys Ser Gln Tyr
Leu Arg 660 665 670Ser Arg Met Pro Glu Gly Thr Cys Leu Glu Ala Glu
Glu Gln Leu Leu 675 680 685Val Ser Gly Gln Ser Leu Glu Ser Leu Ala
Pro Asp His Val Arg Glu 690 695 700Ala Ala Ala Pro Asn Pro Tyr Leu
Lys Asn Ser Val Thr Thr Arg Glu705 710 715 720Phe Leu Val Ser Gln
Lys Leu Pro Glu His Thr Lys Gly Glu Val 725 730 73567495DNAMacaca
fascicularis 67atggcctctc actcagcagg ccccgcgacg tccgtgctgt
ttctgctctg ctgcctggga 60ggctggctga cctcccacac gttgcccgtc catttcctac
aaccaagtga tatacagaaa 120atagtcgagg aattacagtc cctctcgaag
atgcttttga aagatgtgaa ggaagacaag 180ggggtgctcg tgtcccagaa
ttacacgctg ccgtgtctca cccctgacgc ccagccgcca 240aacatcatcc
acagcccagc catccgggca tatctcaaga caatcagaca gttagacaac
300aaatctgtta ttgatgagat catagagcac ctcgacaaac tcatatttca
agatgcacca 360gaaacaaaca tttctgtgcc aacagacacc catgaatgta
aacgcttcat cctgactatt 420tctcaacagt tttcagagtg catggacctt
gcattaaaat cgttgacttc tggagcccag 480caggccacca cttaa
49568164PRTMacaca fascicularis 68Met Ala Ser His Ser Ala Gly Pro
Ala Thr Ser Val Leu Phe Leu Leu1 5 10 15Cys Cys Leu Gly Gly Trp Leu
Thr Ser His Thr Leu Pro Val His Phe 20 25 30Leu Gln Pro Ser Asp Ile
Gln Lys Ile Val Glu Glu Leu Gln Ser Leu 35 40 45Ser Lys Met Leu Leu
Lys Asp Val Lys Glu Asp Lys Gly Val Leu Val 50 55 60Ser Gln Asn Tyr
Thr Leu Pro Cys Leu Thr Pro Asp Ala Gln Pro Pro65 70 75 80Asn Ile
Ile His Ser Pro Ala Ile Arg Ala Tyr Leu Lys Thr Ile Arg 85 90 95Gln
Leu Asp Asn Lys Ser Val Ile Asp Glu Ile Ile Glu His Leu Asp 100 105
110Lys Leu Ile Phe Gln Asp Ala Pro Glu Thr Asn Ile Ser Val Pro Thr
115 120 125Asp Thr His Glu Cys Lys Arg Phe Ile Leu Thr Ile Ser Gln
Gln Phe 130 135 140Ser Glu Cys Met Asp Leu Ala Leu Lys Ser Leu Thr
Ser Gly Ala Gln145 150 155 160Gln Ala Thr Thr692934DNAMacaca
fascicularis 69atggctctat ttgtagtctt tcagacaaca ttcttcttaa
cattgctgtc cttgaggact 60taccagagtg aagtcttggc tgaacgttta ccattgactc
ctgtgtcact taaagtttcc 120accaattcta tacatcagag tttgcattta
caatggactg tccacaacct tccttatcat 180caggaattga aaatggtatt
tcagatccag atcagtagga ttgaaacatc caatgtcgtc 240tgggtgggga
attacagcac cactgtgaag tggaaccagg ttctgcattg gagctgggaa
300tcggaactcc ctttggaatg tgccacacac tttgtaagaa tcaagagtgt
gatagacgat 360gccagtttcc ctgagccaaa tttctggagc
aactggagtt cctgggagga agtcagtgta 420caagattatc ttggacgggg
cactttgttc gttttcccta aagataagct ggtggaagaa 480ggctccaatg
ttaccatttg ttatgtttct aggaacattc aaaataatgt atcctgttat
540ttggaaggga aacagattca cggagaacaa cttgatccac atgtaactgc
attcaacttg 600aatagtgtgc ctttcattag gaatagaggg acaaatatct
attgtgaggc gagtcaagga 660aatgtcagta aaggcataga aggcatcgtt
ctctttgtct caaaagtact tgaggagccc 720aaggactttt cttgtgaatc
ccaggacttc aacactttgc actgtacttg ggatcctggg 780acggacactg
ccttggggtg gtctaaacaa ccttcccaaa gctacacttt atttgaatca
840ttttctgggg aaaagaaact ttgtacgcac aaaaactggt gtaattggca
aataactcaa 900gactcacaag aaatgtataa cttcacactc atagctgaaa
attacttaag gaagagaagt 960gtcaatatcc tttttaacct gactcatcga
gtttatttaa tgaatccttt tagtgtcaac 1020tttgaaaatg taaatgccac
aaatgccatc atgacctgga aggtgcactc catgaggaat 1080aatttcacat
atttgtgtca gattgaactc catggtgaag gaaaaatgat gcaatacgat
1140gtttctatca acgtgaacgg tgagtacttc ttaagtgaac tggaacctgc
cacagaatat 1200atggcccgag tacgctgtgc tgatgccagc cacttctgga
aatggactga atggagtggt 1260cagaacttca ccacacttga agctgctccg
tcagaggccc ctgatgtctg gagaagtgtg 1320aactcagagc caggaaatca
tactgtgacc ttattctgga agccattatc aaaactgcat 1380gccaatggaa
agatcctgtt ctataatgta gttgtagaaa acctagacaa accgtccagg
1440tcagagctcc gttccattcc ggcaccagcc aacagcacaa aactaatcct
cgacaggtgt 1500tcctaccaaa tctgcgtcac agctaacaac agtgtgggcg
cttctcctgc ttctataata 1560gtcatctctg cggaccctga aaacaaagag
gttgaggaag aaagaattgc aggcacagag 1620ggtggattct ctctgtcttg
gaaaccccag cctggagatg ttataggcta tgttgtggac 1680tggtgtgacc
atccccagga tgtgctccag tggaagaatg taggtcccaa taccacaagc
1740acagtcatta gcacagatgc ttttaggcca ggagttcgat acgacttcag
aatctatggg 1800ttatctacaa aaaggattgc ttgtttatta gagaaaaaaa
caggatactc tcaggaactg 1860gctccttcag acaaccctca cgtgctggta
gatatgttga catcccactc cttcactctg 1920agttggaaag attactctac
tgaatctcaa cctggtttta tacaagggta ccatgtctat 1980ctgaaatcca
aggcgaggca gtgccaccca cgatttcaaa aggcagttct ttcagatggt
2040tcagaatgtt gcaaatacaa aattgacaac ccagaagaaa aggcattgat
tgtggacaac 2100ctaaagccag aatccttcta tgagtttttc gttactccat
tcactagtgc tggcgagggc 2160cccaatgcta cgttcacgaa ggtcacgact
ccggatgaac actcctccat gttgattcgt 2220atcctactgc ccatggtttt
ctgcgtcttg ctcatcatga tcgtgtgcta cttgaaaagt 2280cagtggatca
aggagacgtg ttatcctgac atccctgacc cttacaagag cagcatcctg
2340tcgttaataa aattcaagga gaaccctcac ctaacaataa tgaatgtcag
tgactgtatc 2400ccagatgcta ttgaagttgt cagcaagcca gaagggacaa
agatacagct cctaggcact 2460aggaagtcac tcacagaaac tgagttaact
aagcctaact acctttatct ccttccaaca 2520gaaaagaatc actctggccc
tggcccctgc atctgttttg agaactttac ctacaaccag 2580gcagcttctg
acgctggctc ttgtggccat gttccagtac cccccaaagc cccaccaagt
2640atgctaggac taatgacctc acctgaaaat gtactaaagg cgctagaaaa
aaactacatg 2700aactccctgg gagaagtccc agctggagaa acaagtttga
attatgtgtc ccagttggct 2760tcacccatgt ctggagacaa ggacagtctc
ccaacaaacc cagtggagcc accacactgt 2820tcagagtata aaatgcaaat
ggcagtcccc ctgcgtcttg ccctgcctcc cccgaccgag 2880aatagcagcc
tttcctcaat taccctttta gatccaggtg aacactaccg ctaa 293470977PRTMacaca
fascicularis 70Met Ala Leu Phe Val Val Phe Gln Thr Thr Phe Phe Leu
Thr Leu Leu1 5 10 15Ser Leu Arg Thr Tyr Gln Ser Glu Val Leu Ala Glu
Arg Leu Pro Leu 20 25 30Thr Pro Val Ser Leu Lys Val Ser Thr Asn Ser
Ile His Gln Ser Leu 35 40 45His Leu Gln Trp Thr Val His Asn Leu Pro
Tyr His Gln Glu Leu Lys 50 55 60Met Val Phe Gln Ile Gln Ile Ser Arg
Ile Glu Thr Ser Asn Val Val65 70 75 80Trp Val Gly Asn Tyr Ser Thr
Thr Val Lys Trp Asn Gln Val Leu His 85 90 95Trp Ser Trp Glu Ser Glu
Leu Pro Leu Glu Cys Ala Thr His Phe Val 100 105 110Arg Ile Lys Ser
Val Ile Asp Asp Ala Ser Phe Pro Glu Pro Asn Phe 115 120 125Trp Ser
Asn Trp Ser Ser Trp Glu Glu Val Ser Val Gln Asp Tyr Leu 130 135
140Gly Arg Gly Thr Leu Phe Val Phe Pro Lys Asp Lys Leu Val Glu
Glu145 150 155 160Gly Ser Asn Val Thr Ile Cys Tyr Val Ser Arg Asn
Ile Gln Asn Asn 165 170 175Val Ser Cys Tyr Leu Glu Gly Lys Gln Ile
His Gly Glu Gln Leu Asp 180 185 190Pro His Val Thr Ala Phe Asn Leu
Asn Ser Val Pro Phe Ile Arg Asn 195 200 205Arg Gly Thr Asn Ile Tyr
Cys Glu Ala Ser Gln Gly Asn Val Ser Lys 210 215 220Gly Ile Glu Gly
Ile Val Leu Phe Val Ser Lys Val Leu Glu Glu Pro225 230 235 240Lys
Asp Phe Ser Cys Glu Ser Gln Asp Phe Asn Thr Leu His Cys Thr 245 250
255Trp Asp Pro Gly Thr Asp Thr Ala Leu Gly Trp Ser Lys Gln Pro Ser
260 265 270Gln Ser Tyr Thr Leu Phe Glu Ser Phe Ser Gly Glu Lys Lys
Leu Cys 275 280 285Thr His Lys Asn Trp Cys Asn Trp Gln Ile Thr Gln
Asp Ser Gln Glu 290 295 300Met Tyr Asn Phe Thr Leu Ile Ala Glu Asn
Tyr Leu Arg Lys Arg Ser305 310 315 320Val Asn Ile Leu Phe Asn Leu
Thr His Arg Val Tyr Leu Met Asn Pro 325 330 335Phe Ser Val Asn Phe
Glu Asn Val Asn Ala Thr Asn Ala Ile Met Thr 340 345 350Trp Lys Val
His Ser Met Arg Asn Asn Phe Thr Tyr Leu Cys Gln Ile 355 360 365Glu
Leu His Gly Glu Gly Lys Met Met Gln Tyr Asp Val Ser Ile Asn 370 375
380Val Asn Gly Glu Tyr Phe Leu Ser Glu Leu Glu Pro Ala Thr Glu
Tyr385 390 395 400Met Ala Arg Val Arg Cys Ala Asp Ala Ser His Phe
Trp Lys Trp Thr 405 410 415Glu Trp Ser Gly Gln Asn Phe Thr Thr Leu
Glu Ala Ala Pro Ser Glu 420 425 430Ala Pro Asp Val Trp Arg Ser Val
Asn Ser Glu Pro Gly Asn His Thr 435 440 445Val Thr Leu Phe Trp Lys
Pro Leu Ser Lys Leu His Ala Asn Gly Lys 450 455 460Ile Leu Phe Tyr
Asn Val Val Val Glu Asn Leu Asp Lys Pro Ser Arg465 470 475 480Ser
Glu Leu Arg Ser Ile Pro Ala Pro Ala Asn Ser Thr Lys Leu Ile 485 490
495Leu Asp Arg Cys Ser Tyr Gln Ile Cys Val Thr Ala Asn Asn Ser Val
500 505 510Gly Ala Ser Pro Ala Ser Ile Ile Val Ile Ser Ala Asp Pro
Glu Asn 515 520 525Lys Glu Val Glu Glu Glu Arg Ile Ala Gly Thr Glu
Gly Gly Phe Ser 530 535 540Leu Ser Trp Lys Pro Gln Pro Gly Asp Val
Ile Gly Tyr Val Val Asp545 550 555 560Trp Cys Asp His Pro Gln Asp
Val Leu Gln Trp Lys Asn Val Gly Pro 565 570 575Asn Thr Thr Ser Thr
Val Ile Ser Thr Asp Ala Phe Arg Pro Gly Val 580 585 590Arg Tyr Asp
Phe Arg Ile Tyr Gly Leu Ser Thr Lys Arg Ile Ala Cys 595 600 605Leu
Leu Glu Lys Lys Thr Gly Tyr Ser Gln Glu Leu Ala Pro Ser Asp 610 615
620Asn Pro His Val Leu Val Asp Met Leu Thr Ser His Ser Phe Thr
Leu625 630 635 640Ser Trp Lys Asp Tyr Ser Thr Glu Ser Gln Pro Gly
Phe Ile Gln Gly 645 650 655Tyr His Val Tyr Leu Lys Ser Lys Ala Arg
Gln Cys His Pro Arg Phe 660 665 670Gln Lys Ala Val Leu Ser Asp Gly
Ser Glu Cys Cys Lys Tyr Lys Ile 675 680 685Asp Asn Pro Glu Glu Lys
Ala Leu Ile Val Asp Asn Leu Lys Pro Glu 690 695 700Ser Phe Tyr Glu
Phe Phe Val Thr Pro Phe Thr Ser Ala Gly Glu Gly705 710 715 720Pro
Asn Ala Thr Phe Thr Lys Val Thr Thr Pro Asp Glu His Ser Ser 725 730
735Met Leu Ile Arg Ile Leu Leu Pro Met Val Phe Cys Val Leu Leu Ile
740 745 750Met Ile Val Cys Tyr Leu Lys Ser Gln Trp Ile Lys Glu Thr
Cys Tyr 755 760 765Pro Asp Ile Pro Asp Pro Tyr Lys Ser Ser Ile Leu
Ser Leu Ile Lys 770 775 780Phe Lys Glu Asn Pro His Leu Thr Ile Met
Asn Val Ser Asp Cys Ile785 790 795 800Pro Asp Ala Ile Glu Val Val
Ser Lys Pro Glu Gly Thr Lys Ile Gln 805 810 815Leu Leu Gly Thr Arg
Lys Ser Leu Thr Glu Thr Glu Leu Thr Lys Pro 820 825 830Asn Tyr Leu
Tyr Leu Leu Pro Thr Glu Lys Asn His Ser Gly Pro Gly 835 840 845Pro
Cys Ile Cys Phe Glu Asn Phe Thr Tyr Asn Gln Ala Ala Ser Asp 850 855
860Ala Gly Ser Cys Gly His Val Pro Val Pro Pro Lys Ala Pro Pro
Ser865 870 875 880Met Leu Gly Leu Met Thr Ser Pro Glu Asn Val Leu
Lys Ala Leu Glu 885 890 895Lys Asn Tyr Met Asn Ser Leu Gly Glu Val
Pro Ala Gly Glu Thr Ser 900 905 910Leu Asn Tyr Val Ser Gln Leu Ala
Ser Pro Met Ser Gly Asp Lys Asp 915 920 925Ser Leu Pro Thr Asn Pro
Val Glu Pro Pro His Cys Ser Glu Tyr Lys 930 935 940Met Gln Met Ala
Val Pro Leu Arg Leu Ala Leu Pro Pro Pro Thr Glu945 950 955 960Asn
Ser Ser Leu Ser Ser Ile Thr Leu Leu Asp Pro Gly Glu His Tyr 965 970
975Arg711665DNAMacaca fascicularis 71atgatgtgga cctgggcact
gtggatgttc cctttactct gcaaattcgg cctggcagct 60ctgccagcta agcctgagaa
catttcctgt gtctactact ataggaaaaa tttaacctgc 120acttggagtc
caggaaagga aactagttat acccagtaca cagctaagag aacttacgct
180tttggaaaaa aacatgataa ttgtacaacc agtagttcta caagtgaaaa
tcgtgcttcg 240tgctcttttt tccttccaag aataacgatc ccagataatt
ataccattga ggtggaagct 300gaaaatggag atggtgtaat taaatctgat
atgacatgtt ggagattaga ggacatagcg 360aaaactgaac cacctgagat
tttcagtgtg aaaccagttt tgggcatcaa acgaatgatt 420cggattgaat
ggataaagcc tgagttggca cctgtttcat ctgatttaaa atatgcactt
480cgattcagga cagtcaatag taccagctgg atggaagtca acttcgctaa
gaaccgtaaa 540gatacaaacc aaacctacaa ccttatgggg ctgcaggctt
ttacagagta tgtcgtagct 600ctgcgatgtg cggtcaagga gtcaaagttc
tggagtgact ggagccaaga aaaaatggga 660atgactgagg aagaagctcc
atgtggcctg gaactgtgga gagtcctgaa accaactgag 720gtggatggaa
gaaggccagt gcggttgtta tggaagaagg caagaggagc cccagtccta
780gagaaaacac ttggctacaa catatggtac tttccagaaa acaacactaa
cctcacagag 840acagtgaaca ccactaacca gcagcttgaa ctgcatctgg
gaggcgagag ctattgggtg 900tctatgattt cttataattc tcttgggaag
tctccagtga ccaccctgag gattccagcc 960attcaggaaa agtcatttcg
gtgcattgag gtcatgcagg cctgccttgc tgaggaccag 1020ctagtggtga
agtggcaaag ctctgctcta gacgtgaaca cttggatgat tgaatggttt
1080ccggacatgg actcagagca ccccactctt tcctgggaat ctgtgtctca
ggccacgaac 1140tggacaatcc agcaagataa attaaaacct ttctggtgct
ataacatctc tgtgtatcca 1200atgttgcacg acaaagttgg cgagccatat
tccatccagg cttatgccaa agaaggcatt 1260ccatcaaaag gtcctgagac
caaggtggag aacattggcg tgaagacggt cacgatcaca 1320tggaaagaga
ttcccaagag tgagagaaag ggtatcatct gcaactacac catcttttac
1380caagctgaag gtggaaaagg attctccaag acagtcaact ccagcatctt
gcagtatggc 1440ctggagtccc tgaaacgaaa gacctcttac actgttcggg
tcatggccag caccagtgct 1500gggggaatca acgggaccag cataaatttc
aagacattgt cattcagtgt ttttgagatt 1560atccttataa cttctctgat
tggtggaggc cttcttattc tcattatcct gacggtggca 1620tatggtctca
aaaaacccaa caaattgact cacctgtgtt aatga 166572553PRTMacaca
fascicularis 72Met Met Trp Thr Trp Ala Leu Trp Met Phe Pro Leu Leu
Cys Lys Phe1 5 10 15Gly Leu Ala Ala Leu Pro Ala Lys Pro Glu Asn Ile
Ser Cys Val Tyr 20 25 30Tyr Tyr Arg Lys Asn Leu Thr Cys Thr Trp Ser
Pro Gly Lys Glu Thr 35 40 45Ser Tyr Thr Gln Tyr Thr Ala Lys Arg Thr
Tyr Ala Phe Gly Lys Lys 50 55 60His Asp Asn Cys Thr Thr Ser Ser Ser
Thr Ser Glu Asn Arg Ala Ser65 70 75 80Cys Ser Phe Phe Leu Pro Arg
Ile Thr Ile Pro Asp Asn Tyr Thr Ile 85 90 95Glu Val Glu Ala Glu Asn
Gly Asp Gly Val Ile Lys Ser Asp Met Thr 100 105 110Cys Trp Arg Leu
Glu Asp Ile Ala Lys Thr Glu Pro Pro Glu Ile Phe 115 120 125Ser Val
Lys Pro Val Leu Gly Ile Lys Arg Met Ile Arg Ile Glu Trp 130 135
140Ile Lys Pro Glu Leu Ala Pro Val Ser Ser Asp Leu Lys Tyr Ala
Leu145 150 155 160Arg Phe Arg Thr Val Asn Ser Thr Ser Trp Met Glu
Val Asn Phe Ala 165 170 175Lys Asn Arg Lys Asp Thr Asn Gln Thr Tyr
Asn Leu Met Gly Leu Gln 180 185 190Ala Phe Thr Glu Tyr Val Val Ala
Leu Arg Cys Ala Val Lys Glu Ser 195 200 205Lys Phe Trp Ser Asp Trp
Ser Gln Glu Lys Met Gly Met Thr Glu Glu 210 215 220Glu Ala Pro Cys
Gly Leu Glu Leu Trp Arg Val Leu Lys Pro Thr Glu225 230 235 240Val
Asp Gly Arg Arg Pro Val Arg Leu Leu Trp Lys Lys Ala Arg Gly 245 250
255Ala Pro Val Leu Glu Lys Thr Leu Gly Tyr Asn Ile Trp Tyr Phe Pro
260 265 270Glu Asn Asn Thr Asn Leu Thr Glu Thr Val Asn Thr Thr Asn
Gln Gln 275 280 285Leu Glu Leu His Leu Gly Gly Glu Ser Tyr Trp Val
Ser Met Ile Ser 290 295 300Tyr Asn Ser Leu Gly Lys Ser Pro Val Thr
Thr Leu Arg Ile Pro Ala305 310 315 320Ile Gln Glu Lys Ser Phe Arg
Cys Ile Glu Val Met Gln Ala Cys Leu 325 330 335Ala Glu Asp Gln Leu
Val Val Lys Trp Gln Ser Ser Ala Leu Asp Val 340 345 350Asn Thr Trp
Met Ile Glu Trp Phe Pro Asp Met Asp Ser Glu His Pro 355 360 365Thr
Leu Ser Trp Glu Ser Val Ser Gln Ala Thr Asn Trp Thr Ile Gln 370 375
380Gln Asp Lys Leu Lys Pro Phe Trp Cys Tyr Asn Ile Ser Val Tyr
Pro385 390 395 400Met Leu His Asp Lys Val Gly Glu Pro Tyr Ser Ile
Gln Ala Tyr Ala 405 410 415Lys Glu Gly Ile Pro Ser Lys Gly Pro Glu
Thr Lys Val Glu Asn Ile 420 425 430Gly Val Lys Thr Val Thr Ile Thr
Trp Lys Glu Ile Pro Lys Ser Glu 435 440 445Arg Lys Gly Ile Ile Cys
Asn Tyr Thr Ile Phe Tyr Gln Ala Glu Gly 450 455 460Gly Lys Gly Phe
Ser Lys Thr Val Asn Ser Ser Ile Leu Gln Tyr Gly465 470 475 480Leu
Glu Ser Leu Lys Arg Lys Thr Ser Tyr Thr Val Arg Val Met Ala 485 490
495Ser Thr Ser Ala Gly Gly Ile Asn Gly Thr Ser Ile Asn Phe Lys Thr
500 505 510Leu Ser Phe Ser Val Phe Glu Ile Ile Leu Ile Thr Ser Leu
Ile Gly 515 520 525Gly Gly Leu Leu Ile Leu Ile Ile Leu Thr Val Ala
Tyr Gly Leu Lys 530 535 540Lys Pro Asn Lys Leu Thr His Leu Cys545
550731665DNAHomo sapiens 73atgatgtgga cctgggcact gtggatgctc
ccctcactct gcaaattcag cctggcagct 60ctgccagcta agcctgagaa catttcctgt
gtctactact ataggaaaaa tttaacctgc 120acttggagtc caggaaagga
aaccagttat acccagtaca cagttaagag aacttacgct 180tttggagaaa
aacatgataa ttgtacaacc aatagttcta caagtgaaaa tcgtgcttcg
240tgctcttttt tccttccaag aataacgatc ccagataatt ataccattga
ggtggaagct 300gaaaatggag atggtgtaat taaatctcat atgacatact
ggagattaga gaacatagcg 360aaaactgaac cacctaagat tttccgtgtg
aaaccagttt tgggcatcaa acgaatgatt 420caaattgaat ggataaagcc
tgagttggcg cctgtttcat ctgatttaaa atacacactt 480cgattcagga
cagtcaacag taccagctgg atggaagtca acttcgctaa gaaccgtaag
540gataaaaacc aaacgtacaa cctcacgggg ctgcagcctt ttacagaata
tgtcatagct 600ctgcgatgtg cggtcaagga gtcaaagttc tggagtgact
ggagccaaga aaaaatggga 660atgactgagg aagaagctcc atgtggcctg
gaactgtgga gagtcctgaa accagctgag 720gcggatggaa gaaggccagt
gcggttgtta tggaagaagg caagaggagc cccagtccta 780gagaaaacac
ttggctacaa catatggtac tatccagaaa gcaacactaa cctcacagaa
840acaatgaaca ctactaacca gcagcttgaa ctgcatctgg gaggcgagag
cttttgggtg 900tctatgattt cttataattc tcttgggaag tctccagtgg
ccaccctgag gattccagct 960attcaagaaa aatcgtttca gtgcattgag
gtcatgcagg cctgcgttgc tgaggaccag 1020ctagtggtga agtggcaaag
ctctgctcta gacgtgaaca cttggatgat tgaatggttt 1080ccggatgtgg
actcagagcc caccaccctt tcctgggaat ctgtgtctca ggccacgaac
1140tggacgatcc agcaagataa attaaaacct ttctggtgct ataacatctc
tgtgtatcca 1200atgttgcatg acaaagttgg cgagccatat tccatccagg
cttatgccaa agaaggcgtt 1260ccatcagaag gtcctgagac caaggtggag
aacattggcg tgaagacggt cacgatcaca 1320tggaaagaga ttcccaagag
tgagagaaag ggtatcatct gcaactacac catcttttac 1380caagctgaag
gtggaaaagg attctccaag acagtcaatt
ccagcatctt gcagtacggc 1440ctggagtccc tgaaacgaaa gacctcttac
attgttcagg tcatggccag caccagtgct 1500gggggaacca acgggaccag
cataaatttc aagacattgt cattcagtgt ctttgagatt 1560atcctcataa
cttctctgat tggtggaggc cttcttattc tcattatcct gacagtggca
1620tatggtctca aaaaacccaa caaattgact catctgtgtt aatga
166574553PRTHomo sapiens 74Met Met Trp Thr Trp Ala Leu Trp Met Leu
Pro Ser Leu Cys Lys Phe1 5 10 15Ser Leu Ala Ala Leu Pro Ala Lys Pro
Glu Asn Ile Ser Cys Val Tyr 20 25 30Tyr Tyr Arg Lys Asn Leu Thr Cys
Thr Trp Ser Pro Gly Lys Glu Thr 35 40 45Ser Tyr Thr Gln Tyr Thr Val
Lys Arg Thr Tyr Ala Phe Gly Glu Lys 50 55 60His Asp Asn Cys Thr Thr
Asn Ser Ser Thr Ser Glu Asn Arg Ala Ser65 70 75 80Cys Ser Phe Phe
Leu Pro Arg Ile Thr Ile Pro Asp Asn Tyr Thr Ile 85 90 95Glu Val Glu
Ala Glu Asn Gly Asp Gly Val Ile Lys Ser His Met Thr 100 105 110Tyr
Trp Arg Leu Glu Asn Ile Ala Lys Thr Glu Pro Pro Lys Ile Phe 115 120
125Arg Val Lys Pro Val Leu Gly Ile Lys Arg Met Ile Gln Ile Glu Trp
130 135 140Ile Lys Pro Glu Leu Ala Pro Val Ser Ser Asp Leu Lys Tyr
Thr Leu145 150 155 160Arg Phe Arg Thr Val Asn Ser Thr Ser Trp Met
Glu Val Asn Phe Ala 165 170 175Lys Asn Arg Lys Asp Lys Asn Gln Thr
Tyr Asn Leu Thr Gly Leu Gln 180 185 190Pro Phe Thr Glu Tyr Val Ile
Ala Leu Arg Cys Ala Val Lys Glu Ser 195 200 205Lys Phe Trp Ser Asp
Trp Ser Gln Glu Lys Met Gly Met Thr Glu Glu 210 215 220Glu Ala Pro
Cys Gly Leu Glu Leu Trp Arg Val Leu Lys Pro Ala Glu225 230 235
240Ala Asp Gly Arg Arg Pro Val Arg Leu Leu Trp Lys Lys Ala Arg Gly
245 250 255Ala Pro Val Leu Glu Lys Thr Leu Gly Tyr Asn Ile Trp Tyr
Tyr Pro 260 265 270Glu Ser Asn Thr Asn Leu Thr Glu Thr Met Asn Thr
Thr Asn Gln Gln 275 280 285Leu Glu Leu His Leu Gly Gly Glu Ser Phe
Trp Val Ser Met Ile Ser 290 295 300Tyr Asn Ser Leu Gly Lys Ser Pro
Val Ala Thr Leu Arg Ile Pro Ala305 310 315 320Ile Gln Glu Lys Ser
Phe Gln Cys Ile Glu Val Met Gln Ala Cys Val 325 330 335Ala Glu Asp
Gln Leu Val Val Lys Trp Gln Ser Ser Ala Leu Asp Val 340 345 350Asn
Thr Trp Met Ile Glu Trp Phe Pro Asp Val Asp Ser Glu Pro Thr 355 360
365Thr Leu Ser Trp Glu Ser Val Ser Gln Ala Thr Asn Trp Thr Ile Gln
370 375 380Gln Asp Lys Leu Lys Pro Phe Trp Cys Tyr Asn Ile Ser Val
Tyr Pro385 390 395 400Met Leu His Asp Lys Val Gly Glu Pro Tyr Ser
Ile Gln Ala Tyr Ala 405 410 415Lys Glu Gly Val Pro Ser Glu Gly Pro
Glu Thr Lys Val Glu Asn Ile 420 425 430Gly Val Lys Thr Val Thr Ile
Thr Trp Lys Glu Ile Pro Lys Ser Glu 435 440 445Arg Lys Gly Ile Ile
Cys Asn Tyr Thr Ile Phe Tyr Gln Ala Glu Gly 450 455 460Gly Lys Gly
Phe Ser Lys Thr Val Asn Ser Ser Ile Leu Gln Tyr Gly465 470 475
480Leu Glu Ser Leu Lys Arg Lys Thr Ser Tyr Ile Val Gln Val Met Ala
485 490 495Ser Thr Ser Ala Gly Gly Thr Asn Gly Thr Ser Ile Asn Phe
Lys Thr 500 505 510Leu Ser Phe Ser Val Phe Glu Ile Ile Leu Ile Thr
Ser Leu Ile Gly 515 520 525Gly Gly Leu Leu Ile Leu Ile Ile Leu Thr
Val Ala Tyr Gly Leu Lys 530 535 540Lys Pro Asn Lys Leu Thr His Leu
Cys545 550752199DNAHomo sapiens 75atgatgtgga cctgggcact gtggatgctc
ccctcactct gcaaattcag cctggcagct 60ctgccagcta agcctgagaa catttcctgt
gtctactact ataggaaaaa tttaacctgc 120acttggagtc caggaaagga
aaccagttat acccagtaca cagttaagag aacttacgct 180tttggagaaa
aacatgataa ttgtacaacc aatagttcta caagtgaaaa tcgtgcttcg
240tgctcttttt tccttccaag aataacgatc ccagataatt ataccattga
ggtggaagct 300gaaaatggag atggtgtaat taaatctcat atgacatact
ggagattaga gaacatagcg 360aaaactgaac cacctaagat tttccgtgtg
aaaccagttt tgggcatcaa acgaatgatt 420caaattgaat ggataaagcc
tgagttggcg cctgtttcat ctgatttaaa atacacactt 480cgattcagga
cagtcaacag taccagctgg atggaagtca acttcgctaa gaaccgtaag
540gataaaaacc aaacgtacaa cctcacgggg ctgcagcctt ttacagaata
tgtcatagct 600ctgcgatgtg cggtcaagga gtcaaagttc tggagtgact
ggagccaaga aaaaatggga 660atgactgagg aagaagctcc atgtggcctg
gaactgtgga gagtcctgaa accagctgag 720gcggatggaa gaaggccagt
gcggttgtta tggaagaagg caagaggagc cccagtccta 780gagaaaacac
ttggctacaa catatggtac tatccagaaa gcaacactaa cctcacagaa
840acaatgaaca ctactaacca gcagcttgaa ctgcatctgg gaggcgagag
cttttgggtg 900tctatgattt cttataattc tcttgggaag tctccagtgg
ccaccctgag gattccagct 960attcaagaaa aatcgtttca gtgcattgag
gtcatgcagg cctgcgttgc tgaggaccag 1020ctagtggtga agtggcaaag
ctctgctcta gacgtgaaca cttggatgat tgaatggttt 1080ccggatgtgg
actcagagcc caccaccctt tcctgggaat ctgtgtctca ggccacgaac
1140tggacgatcc agcaagataa attaaaacct ttctggtgct ataacatctc
tgtgtatcca 1200atgttgcatg acaaagttgg cgagccatat tccatccagg
cttatgccaa agaaggcgtt 1260ccatcagaag gtcctgagac caaggtggag
aacattggcg tgaagacggt cacgatcaca 1320tggaaagaga ttcccaagag
tgagagaaag ggtatcatct gcaactacac catcttttac 1380caagctgaag
gtggaaaagg attctccaag acagtcaatt ccagcatctt gcagtacggc
1440ctggagtccc tgaaacgaaa gacctcttac attgttcagg tcatggccag
caccagtgct 1500gggggaacca acgggaccag cataaatttc aagacattgt
cattcagtgt ctttgagatt 1560atcctcataa cttctctgat tggtggaggc
cttcttattc tcattatcct gacagtggca 1620tatggtctca aaaaacccaa
caaattgact catctgtgtt ggcccaccgt tcccaaccct 1680gctgaaagta
gtatagccac atggcatgga gatgatttca aggataagct aaacctgaag
1740gagtctgatg actctgtgaa cacagaagac aggatcttaa aaccatgttc
cacccccagt 1800gacaagttgg tgattgacaa gttggtggtg aactttggga
atgttctgca agaaattttc 1860acagatgaag ccagaacggg tcaggaaaac
aatttaggag gggaaaagaa tgggtatgtg 1920acctgcccct tcaggcctga
ttgtcccctg gggaaaagtt ttgaggagct cccagtttca 1980cctgagattc
cgcccagaaa atcccaatac ctacgttcga ggatgccaga ggggacccgc
2040ccagaagcca aagagcagct tctcttttct ggtcaaagtt tagtaccaga
tcatctgtgt 2100gaggaaggag ccccaaatcc atatttgaaa aattcagtga
cagccaggga atttcttgtg 2160tctgaaaaac ttccagagca caccaaggga
gaagtctaa 219976732PRTHomo sapiens 76Met Met Trp Thr Trp Ala Leu
Trp Met Leu Pro Ser Leu Cys Lys Phe1 5 10 15Ser Leu Ala Ala Leu Pro
Ala Lys Pro Glu Asn Ile Ser Cys Val Tyr 20 25 30Tyr Tyr Arg Lys Asn
Leu Thr Cys Thr Trp Ser Pro Gly Lys Glu Thr 35 40 45Ser Tyr Thr Gln
Tyr Thr Val Lys Arg Thr Tyr Ala Phe Gly Glu Lys 50 55 60His Asp Asn
Cys Thr Thr Asn Ser Ser Thr Ser Glu Asn Arg Ala Ser65 70 75 80Cys
Ser Phe Phe Leu Pro Arg Ile Thr Ile Pro Asp Asn Tyr Thr Ile 85 90
95Glu Val Glu Ala Glu Asn Gly Asp Gly Val Ile Lys Ser His Met Thr
100 105 110Tyr Trp Arg Leu Glu Asn Ile Ala Lys Thr Glu Pro Pro Lys
Ile Phe 115 120 125Arg Val Lys Pro Val Leu Gly Ile Lys Arg Met Ile
Gln Ile Glu Trp 130 135 140Ile Lys Pro Glu Leu Ala Pro Val Ser Ser
Asp Leu Lys Tyr Thr Leu145 150 155 160Arg Phe Arg Thr Val Asn Ser
Thr Ser Trp Met Glu Val Asn Phe Ala 165 170 175Lys Asn Arg Lys Asp
Lys Asn Gln Thr Tyr Asn Leu Thr Gly Leu Gln 180 185 190Pro Phe Thr
Glu Tyr Val Ile Ala Leu Arg Cys Ala Val Lys Glu Ser 195 200 205Lys
Phe Trp Ser Asp Trp Ser Gln Glu Lys Met Gly Met Thr Glu Glu 210 215
220Glu Ala Pro Cys Gly Leu Glu Leu Trp Arg Val Leu Lys Pro Ala
Glu225 230 235 240Ala Asp Gly Arg Arg Pro Val Arg Leu Leu Trp Lys
Lys Ala Arg Gly 245 250 255Ala Pro Val Leu Glu Lys Thr Leu Gly Tyr
Asn Ile Trp Tyr Tyr Pro 260 265 270Glu Ser Asn Thr Asn Leu Thr Glu
Thr Met Asn Thr Thr Asn Gln Gln 275 280 285Leu Glu Leu His Leu Gly
Gly Glu Ser Phe Trp Val Ser Met Ile Ser 290 295 300Tyr Asn Ser Leu
Gly Lys Ser Pro Val Ala Thr Leu Arg Ile Pro Ala305 310 315 320Ile
Gln Glu Lys Ser Phe Gln Cys Ile Glu Val Met Gln Ala Cys Val 325 330
335Ala Glu Asp Gln Leu Val Val Lys Trp Gln Ser Ser Ala Leu Asp Val
340 345 350Asn Thr Trp Met Ile Glu Trp Phe Pro Asp Val Asp Ser Glu
Pro Thr 355 360 365Thr Leu Ser Trp Glu Ser Val Ser Gln Ala Thr Asn
Trp Thr Ile Gln 370 375 380Gln Asp Lys Leu Lys Pro Phe Trp Cys Tyr
Asn Ile Ser Val Tyr Pro385 390 395 400Met Leu His Asp Lys Val Gly
Glu Pro Tyr Ser Ile Gln Ala Tyr Ala 405 410 415Lys Glu Gly Val Pro
Ser Glu Gly Pro Glu Thr Lys Val Glu Asn Ile 420 425 430Gly Val Lys
Thr Val Thr Ile Thr Trp Lys Glu Ile Pro Lys Ser Glu 435 440 445Arg
Lys Gly Ile Ile Cys Asn Tyr Thr Ile Phe Tyr Gln Ala Glu Gly 450 455
460Gly Lys Gly Phe Ser Lys Thr Val Asn Ser Ser Ile Leu Gln Tyr
Gly465 470 475 480Leu Glu Ser Leu Lys Arg Lys Thr Ser Tyr Ile Val
Gln Val Met Ala 485 490 495Ser Thr Ser Ala Gly Gly Thr Asn Gly Thr
Ser Ile Asn Phe Lys Thr 500 505 510Leu Ser Phe Ser Val Phe Glu Ile
Ile Leu Ile Thr Ser Leu Ile Gly 515 520 525Gly Gly Leu Leu Ile Leu
Ile Ile Leu Thr Val Ala Tyr Gly Leu Lys 530 535 540Lys Pro Asn Lys
Leu Thr His Leu Cys Trp Pro Thr Val Pro Asn Pro545 550 555 560Ala
Glu Ser Ser Ile Ala Thr Trp His Gly Asp Asp Phe Lys Asp Lys 565 570
575Leu Asn Leu Lys Glu Ser Asp Asp Ser Val Asn Thr Glu Asp Arg Ile
580 585 590Leu Lys Pro Cys Ser Thr Pro Ser Asp Lys Leu Val Ile Asp
Lys Leu 595 600 605Val Val Asn Phe Gly Asn Val Leu Gln Glu Ile Phe
Thr Asp Glu Ala 610 615 620Arg Thr Gly Gln Glu Asn Asn Leu Gly Gly
Glu Lys Asn Gly Tyr Val625 630 635 640Thr Cys Pro Phe Arg Pro Asp
Cys Pro Leu Gly Lys Ser Phe Glu Glu 645 650 655Leu Pro Val Ser Pro
Glu Ile Pro Pro Arg Lys Ser Gln Tyr Leu Arg 660 665 670Ser Arg Met
Pro Glu Gly Thr Arg Pro Glu Ala Lys Glu Gln Leu Leu 675 680 685Phe
Ser Gly Gln Ser Leu Val Pro Asp His Leu Cys Glu Glu Gly Ala 690 695
700Pro Asn Pro Tyr Leu Lys Asn Ser Val Thr Ala Arg Glu Phe Leu
Val705 710 715 720Ser Glu Lys Leu Pro Glu His Thr Lys Gly Glu Val
725 730771542DNAHomo sapiens 77atgatgtgga cctgggcact gtggatgctc
ccttcactct gcaaattcag cctggcagct 60ctgccagcta agcctgagaa catttcctgt
gtctactact ataggaaaaa tttaacctgc 120acttggagtc caggaaagga
aaccagttat acccagtaca cagttaagag aacttacgct 180tttggagaaa
aacatgataa ttgtacaacc aatagttcta caagtgaaaa tcgtgcttcg
240tgctcttttt tccttccaag aataacgatc ccagataatt ataccattga
ggtggaagct 300gaaaatggag atggtgtaat taaatctcat atgacatact
ggagattaga gaacatagcg 360aaaactgaac cacctaagat tttccgtgtg
aaaccagttt tgggcatcaa acgaatgatt 420caaattgaat ggataaagcc
tgagttggcg cctgtttcat ctgatttaaa atacacactt 480cgattcagga
cagtcaacag taccagctgg atggaagtca acttcgctaa gaaccgtaag
540gataaaaacc aaacgtacaa cctcacgggg ctgcagcctt ttacagaata
tgtcatagct 600ctgcgatgtg cggtcaagga gtcaaagttc tggagtgact
ggagccaaga aaaaatggga 660atgactgagg aagaagctcc atgtggcctg
gaactgtgga gagtcctgaa accagctgag 720gcggatggaa gaaggccagt
gcggttgtta tggaagaagg caagaggagc cccagtccta 780gagaaaacac
ttggctacaa catatggtac tatccagaaa gcaacactaa cctcacagaa
840acaatgaaca ctactaacca gcagcttgaa ctgcatctgg gaggcgagag
cttttgggtg 900tctatgattt cttataattc tcttgggaag tctccagtgg
ccaccctgag gattccagct 960attcaagaaa aatcgtttca gtgcattgag
gtcatgcagg cctgcgttgc tgaggaccag 1020ctagtggtga agtggcaaag
ctctgctcta gacgtgaaca cttggatgat tgaatggttt 1080ccggatgtgg
actcagagcc caccaccctt tcctgggaat ctgtgtctca ggccacgaac
1140tggacgatcc agcaagataa attaaaacct ttctggtgct ataacatctc
tgtgtatcca 1200atgttgcatg acaaagttgg cgagccatat tccatccagg
cttatgccaa agaaggcgtt 1260ccatcagaag gtcctgagac caaggtggag
aacattggcg tgaagacggt cacgatcaca 1320tggaaagaga ttcccaagag
tgagagaaag ggtatcatct gcaactacac catcttttac 1380caagctgaag
gtggaaaagg attctccaag acagtcaatt ccagcatctt gcagtacggc
1440ctggagtccc tgaaacgaaa gacctcttac attgttcagg tcatggccag
caccagtgct 1500gggggaacca acgggaccag cataaatttc aagacattgt ca
154278514PRTHomo sapiens 78Met Met Trp Thr Trp Ala Leu Trp Met Leu
Pro Ser Leu Cys Lys Phe1 5 10 15Ser Leu Ala Ala Leu Pro Ala Lys Pro
Glu Asn Ile Ser Cys Val Tyr 20 25 30Tyr Tyr Arg Lys Asn Leu Thr Cys
Thr Trp Ser Pro Gly Lys Glu Thr 35 40 45Ser Tyr Thr Gln Tyr Thr Val
Lys Arg Thr Tyr Ala Phe Gly Glu Lys 50 55 60His Asp Asn Cys Thr Thr
Asn Ser Ser Thr Ser Glu Asn Arg Ala Ser65 70 75 80Cys Ser Phe Phe
Leu Pro Arg Ile Thr Ile Pro Asp Asn Tyr Thr Ile 85 90 95Glu Val Glu
Ala Glu Asn Gly Asp Gly Val Ile Lys Ser His Met Thr 100 105 110Tyr
Trp Arg Leu Glu Asn Ile Ala Lys Thr Glu Pro Pro Lys Ile Phe 115 120
125Arg Val Lys Pro Val Leu Gly Ile Lys Arg Met Ile Gln Ile Glu Trp
130 135 140Ile Lys Pro Glu Leu Ala Pro Val Ser Ser Asp Leu Lys Tyr
Thr Leu145 150 155 160Arg Phe Arg Thr Val Asn Ser Thr Ser Trp Met
Glu Val Asn Phe Ala 165 170 175Lys Asn Arg Lys Asp Lys Asn Gln Thr
Tyr Asn Leu Thr Gly Leu Gln 180 185 190Pro Phe Thr Glu Tyr Val Ile
Ala Leu Arg Cys Ala Val Lys Glu Ser 195 200 205Lys Phe Trp Ser Asp
Trp Ser Gln Glu Lys Met Gly Met Thr Glu Glu 210 215 220Glu Ala Pro
Cys Gly Leu Glu Leu Trp Arg Val Leu Lys Pro Ala Glu225 230 235
240Ala Asp Gly Arg Arg Pro Val Arg Leu Leu Trp Lys Lys Ala Arg Gly
245 250 255Ala Pro Val Leu Glu Lys Thr Leu Gly Tyr Asn Ile Trp Tyr
Tyr Pro 260 265 270Glu Ser Asn Thr Asn Leu Thr Glu Thr Met Asn Thr
Thr Asn Gln Gln 275 280 285Leu Glu Leu His Leu Gly Gly Glu Ser Phe
Trp Val Ser Met Ile Ser 290 295 300Tyr Asn Ser Leu Gly Lys Ser Pro
Val Ala Thr Leu Arg Ile Pro Ala305 310 315 320Ile Gln Glu Lys Ser
Phe Gln Cys Ile Glu Val Met Gln Ala Cys Val 325 330 335Ala Glu Asp
Gln Leu Val Val Lys Trp Gln Ser Ser Ala Leu Asp Val 340 345 350Asn
Thr Trp Met Ile Glu Trp Phe Pro Asp Val Asp Ser Glu Pro Thr 355 360
365Thr Leu Ser Trp Glu Ser Val Ser Gln Ala Thr Asn Trp Thr Ile Gln
370 375 380Gln Asp Lys Leu Lys Pro Phe Trp Cys Tyr Asn Ile Ser Val
Tyr Pro385 390 395 400Met Leu His Asp Lys Val Gly Glu Pro Tyr Ser
Ile Gln Ala Tyr Ala 405 410 415Lys Glu Gly Val Pro Ser Glu Gly Pro
Glu Thr Lys Val Glu Asn Ile 420 425 430Gly Val Lys Thr Val Thr Ile
Thr Trp Lys Glu Ile Pro Lys Ser Glu 435 440 445Arg Lys Gly Ile Ile
Cys Asn Tyr Thr Ile Phe Tyr Gln Ala Glu Gly 450 455 460Gly Lys Gly
Phe Ser Lys Thr Val Asn Ser Ser Ile Leu Gln Tyr Gly465 470 475
480Leu Glu Ser Leu Lys Arg Lys Thr Ser Tyr Ile Val Gln Val Met Ala
485 490 495Ser Thr Ser Ala Gly Gly Thr Asn Gly Thr Ser Ile Asn Phe
Lys Thr 500 505 510Leu
Ser79662PRTHomo sapiens 79Met Lys Leu Ser Pro Gln Pro Ser Cys Val
Asn Leu Gly Met Met Trp1 5 10 15Thr Trp Ala Leu Trp Met Leu Pro Ser
Leu Cys Lys Phe Ser Leu Ala 20 25 30Ala Leu Pro Ala Lys Pro Glu Asn
Ile Ser Cys Val Tyr Tyr Tyr Arg 35 40 45Lys Asn Leu Thr Cys Thr Trp
Ser Pro Gly Lys Glu Thr Ser Tyr Thr 50 55 60Gln Tyr Thr Val Lys Arg
Thr Tyr Ala Phe Gly Glu Lys His Asp Asn65 70 75 80Cys Thr Thr Asn
Ser Ser Thr Ser Glu Asn Arg Ala Ser Cys Ser Phe 85 90 95Phe Leu Pro
Arg Ile Thr Ile Pro Asp Asn Tyr Thr Ile Glu Val Glu 100 105 110Ala
Glu Asn Gly Asp Gly Val Ile Lys Ser His Met Thr Tyr Trp Arg 115 120
125Leu Glu Asn Ile Ala Lys Thr Glu Pro Pro Lys Ile Phe Arg Val Lys
130 135 140Pro Val Leu Gly Ile Lys Arg Met Ile Gln Ile Glu Trp Ile
Lys Pro145 150 155 160Glu Leu Ala Pro Val Ser Ser Asp Leu Lys Tyr
Thr Leu Arg Phe Arg 165 170 175Thr Val Asn Ser Thr Ser Trp Met Glu
Val Asn Phe Ala Lys Asn Arg 180 185 190Lys Asp Lys Asn Gln Thr Tyr
Asn Leu Thr Gly Leu Gln Pro Phe Thr 195 200 205Glu Tyr Val Ile Ala
Leu Arg Cys Ala Val Lys Glu Ser Lys Phe Trp 210 215 220Ser Asp Trp
Ser Gln Glu Lys Met Gly Met Thr Glu Glu Glu Ala Pro225 230 235
240Cys Gly Leu Glu Leu Trp Arg Val Leu Lys Pro Ala Glu Ala Asp Gly
245 250 255Arg Arg Pro Val Arg Leu Leu Trp Lys Lys Ala Arg Gly Ala
Pro Val 260 265 270Leu Glu Lys Thr Leu Gly Tyr Asn Ile Trp Tyr Tyr
Pro Glu Ser Asn 275 280 285Thr Asn Leu Thr Glu Thr Met Asn Thr Thr
Asn Gln Gln Leu Glu Leu 290 295 300His Leu Gly Gly Glu Ser Phe Trp
Val Ser Met Ile Ser Tyr Asn Ser305 310 315 320Leu Gly Lys Ser Pro
Val Ala Thr Leu Arg Ile Pro Ala Ile Gln Glu 325 330 335Lys Ser Phe
Gln Cys Ile Glu Val Met Gln Ala Cys Val Ala Glu Asp 340 345 350Gln
Leu Val Val Lys Trp Gln Ser Ser Ala Leu Asp Val Asn Thr Trp 355 360
365Met Ile Glu Trp Phe Pro Asp Val Asp Ser Glu Pro Thr Thr Leu Ser
370 375 380Trp Glu Ser Val Ser Gln Ala Thr Asn Trp Thr Ile Gln Gln
Asp Lys385 390 395 400Leu Lys Pro Phe Trp Cys Tyr Asn Ile Ser Val
Tyr Pro Met Leu His 405 410 415Asp Lys Val Gly Glu Pro Tyr Ser Ile
Gln Ala Tyr Ala Lys Glu Gly 420 425 430Val Pro Ser Glu Gly Pro Glu
Thr Lys Val Glu Asn Ile Gly Val Lys 435 440 445Thr Val Thr Ile Thr
Trp Lys Glu Ile Pro Lys Ser Glu Arg Lys Gly 450 455 460Ile Ile Cys
Asn Tyr Thr Ile Phe Tyr Gln Ala Glu Gly Gly Lys Gly465 470 475
480Phe Ser Lys Thr Val Asn Ser Ser Ile Leu Gln Tyr Gly Leu Glu Ser
485 490 495Leu Lys Arg Lys Thr Ser Tyr Ile Val Gln Val Met Ala Ser
Thr Ser 500 505 510Ala Gly Gly Thr Asn Gly Thr Ser Ile Asn Phe Lys
Thr Leu Ser Phe 515 520 525Ser Val Phe Glu Ile Ile Leu Ile Thr Ser
Leu Ile Gly Gly Gly Leu 530 535 540Leu Ile Leu Ile Ile Leu Thr Val
Ala Tyr Gly Leu Lys Lys Pro Asn545 550 555 560Lys Leu Thr His Leu
Cys Trp Pro Thr Val Pro Asn Pro Ala Glu Ser 565 570 575Ser Ile Ala
Thr Trp His Gly Asp Asp Phe Lys Asp Lys Leu Asn Leu 580 585 590Lys
Glu Ser Asp Asp Ser Val Asn Thr Glu Asp Arg Ile Leu Lys Pro 595 600
605Cys Ser Thr Pro Ser Asp Lys Leu Val Ile Asp Lys Leu Val Val Asn
610 615 620Phe Gly Asn Val Leu Gln Glu Ile Phe Thr Asp Glu Ala Arg
Thr Gly625 630 635 640Gln Glu Asn Asn Leu Gly Gly Glu Lys Asn Gly
Thr Arg Ile Leu Ser 645 650 655Ser Cys Pro Thr Ser Ile
66080732PRTHomo sapiensSIGNAL(1)..(20)TRANSMEM(520)..(543) 80Met
Met Trp Thr Trp Ala Leu Trp Met Leu Pro Ser Leu Cys Lys Phe1 5 10
15Ser Leu Ala Ala Leu Pro Ala Lys Pro Glu Asn Ile Ser Cys Val Tyr
20 25 30Tyr Tyr Arg Lys Asn Leu Thr Cys Thr Trp Ser Pro Gly Lys Glu
Thr 35 40 45Ser Tyr Thr Gln Tyr Thr Val Lys Arg Thr Tyr Ala Phe Gly
Glu Lys 50 55 60His Asp Asn Cys Thr Thr Asn Ser Ser Thr Ser Glu Asn
Arg Ala Ser65 70 75 80Cys Ser Phe Phe Leu Pro Arg Ile Thr Ile Pro
Asp Asn Tyr Thr Ile 85 90 95Glu Val Glu Ala Glu Asn Gly Asp Gly Val
Ile Lys Ser His Met Thr 100 105 110Tyr Trp Arg Leu Glu Asn Ile Ala
Lys Thr Glu Pro Pro Lys Ile Phe 115 120 125Arg Val Lys Pro Val Leu
Gly Ile Lys Arg Met Ile Gln Ile Glu Trp 130 135 140Ile Lys Pro Glu
Leu Ala Pro Val Ser Ser Asp Leu Lys Tyr Thr Leu145 150 155 160Arg
Phe Arg Thr Val Asn Ser Thr Ser Trp Met Glu Val Asn Phe Ala 165 170
175Lys Asn Arg Lys Asp Lys Asn Gln Thr Tyr Asn Leu Thr Gly Leu Gln
180 185 190Pro Phe Thr Glu Tyr Val Ile Ala Leu Arg Cys Ala Val Lys
Glu Ser 195 200 205Lys Phe Trp Ser Asp Trp Ser Gln Glu Lys Met Gly
Met Thr Glu Glu 210 215 220Glu Ala Pro Cys Gly Leu Glu Leu Trp Arg
Val Leu Lys Pro Ala Glu225 230 235 240Ala Asp Gly Arg Arg Pro Val
Arg Leu Leu Trp Lys Lys Ala Arg Gly 245 250 255Ala Pro Val Leu Glu
Lys Thr Leu Gly Tyr Asn Ile Trp Tyr Tyr Pro 260 265 270Glu Ser Asn
Thr Asn Leu Thr Glu Thr Met Asn Thr Thr Asn Gln Gln 275 280 285Leu
Glu Leu His Leu Gly Gly Glu Ser Phe Trp Val Ser Met Ile Ser 290 295
300Tyr Asn Ser Leu Gly Lys Ser Pro Val Ala Thr Leu Arg Ile Pro
Ala305 310 315 320Ile Gln Glu Lys Ser Phe Gln Cys Ile Glu Val Met
Gln Ala Cys Val 325 330 335Ala Glu Asp Gln Leu Val Val Lys Trp Gln
Ser Ser Ala Leu Asp Val 340 345 350Asn Thr Trp Met Ile Glu Trp Phe
Pro Asp Val Asp Ser Glu Pro Thr 355 360 365Thr Leu Ser Trp Glu Ser
Val Ser Gln Ala Thr Asn Trp Thr Ile Gln 370 375 380Gln Asp Lys Leu
Lys Pro Phe Trp Cys Tyr Asn Ile Ser Val Tyr Pro385 390 395 400Met
Leu His Asp Lys Val Gly Glu Pro Tyr Ser Ile Gln Ala Tyr Ala 405 410
415Lys Glu Gly Val Pro Ser Glu Gly Pro Glu Thr Lys Val Glu Asn Ile
420 425 430Gly Val Lys Thr Val Thr Ile Thr Trp Lys Glu Ile Pro Lys
Ser Glu 435 440 445Arg Lys Gly Ile Ile Cys Asn Tyr Thr Ile Phe Tyr
Gln Ala Glu Gly 450 455 460Gly Lys Gly Phe Ser Lys Thr Val Asn Ser
Ser Ile Leu Gln Tyr Gly465 470 475 480Leu Glu Ser Leu Lys Arg Lys
Thr Ser Tyr Ile Val Gln Val Met Ala 485 490 495Ser Thr Ser Ala Gly
Gly Thr Asn Gly Thr Ser Ile Asn Phe Lys Thr 500 505 510Leu Ser Phe
Ser Val Phe Glu Ile Ile Leu Ile Thr Ser Leu Ile Gly 515 520 525Gly
Gly Leu Leu Ile Leu Ile Ile Leu Thr Val Ala Tyr Gly Leu Lys 530 535
540Lys Pro Asn Lys Leu Thr His Leu Cys Trp Pro Thr Val Pro Asn
Pro545 550 555 560Ala Glu Ser Ser Ile Ala Thr Trp His Gly Asp Asp
Phe Lys Asp Lys 565 570 575Leu Asn Leu Lys Glu Ser Asp Asp Ser Val
Asn Thr Glu Asp Arg Ile 580 585 590Leu Lys Pro Cys Ser Thr Pro Ser
Asp Lys Leu Val Ile Asp Lys Leu 595 600 605Val Val Asn Phe Gly Asn
Val Leu Gln Glu Ile Phe Thr Asp Glu Ala 610 615 620Arg Thr Gly Gln
Glu Asn Asn Leu Gly Gly Glu Lys Asn Gly Tyr Val625 630 635 640Thr
Cys Pro Phe Arg Pro Asp Cys Pro Leu Gly Lys Ser Phe Glu Glu 645 650
655Leu Pro Val Ser Pro Glu Ile Pro Pro Arg Lys Ser Gln Tyr Leu Arg
660 665 670Ser Arg Met Pro Glu Gly Thr Arg Pro Glu Ala Lys Glu Gln
Leu Leu 675 680 685Phe Ser Gly Gln Ser Leu Val Pro Asp His Leu Cys
Glu Glu Gly Ala 690 695 700Pro Asn Pro Tyr Leu Lys Asn Ser Val Thr
Ala Arg Glu Phe Leu Val705 710 715 720Ser Glu Lys Leu Pro Glu His
Thr Lys Gly Glu Val 725 73081716PRTMus musculus 81Met Trp Thr Leu
Ala Leu Trp Ala Phe Ser Phe Leu Cys Lys Phe Ser1 5 10 15Leu Ala Val
Leu Pro Thr Lys Pro Glu Asn Ile Ser Cys Val Phe Tyr 20 25 30Phe Asp
Arg Asn Leu Thr Cys Thr Trp Arg Pro Glu Lys Glu Thr Asn 35 40 45Asp
Thr Ser Tyr Ile Val Thr Leu Thr Tyr Ser Tyr Gly Lys Ser Asn 50 55
60Tyr Ser Asp Asn Ala Thr Glu Ala Ser Tyr Ser Phe Pro Arg Ser Cys65
70 75 80Ala Met Pro Pro Asp Ile Cys Ser Val Glu Val Gln Ala Gln Asn
Gly 85 90 95Asp Gly Lys Val Lys Ser Asp Ile Thr Tyr Trp His Leu Ile
Ser Ile 100 105 110Ala Lys Thr Glu Pro Pro Ile Ile Leu Ser Val Asn
Pro Ile Cys Asn 115 120 125Arg Met Phe Gln Ile Gln Trp Lys Pro Arg
Glu Lys Thr Arg Gly Phe 130 135 140Pro Leu Val Cys Met Leu Arg Phe
Arg Thr Val Asn Ser Ser Arg Trp145 150 155 160Thr Glu Val Asn Phe
Glu Asn Cys Lys Gln Val Cys Asn Leu Thr Gly 165 170 175Leu Gln Ala
Phe Thr Glu Tyr Val Leu Ala Leu Arg Phe Arg Phe Asn 180 185 190Asp
Ser Arg Tyr Trp Ser Lys Trp Ser Lys Glu Glu Thr Arg Val Thr 195 200
205Met Glu Glu Val Pro His Val Leu Asp Leu Trp Arg Ile Leu Glu Pro
210 215 220Ala Asp Met Asn Gly Asp Arg Lys Val Arg Leu Leu Trp Lys
Lys Ala225 230 235 240Arg Gly Ala Pro Val Leu Glu Lys Thr Phe Gly
Tyr His Ile Gln Tyr 245 250 255Phe Ala Glu Asn Ser Thr Asn Leu Thr
Glu Ile Asn Asn Ile Thr Thr 260 265 270Gln Gln Tyr Glu Leu Leu Leu
Met Ser Gln Ala His Ser Val Ser Val 275 280 285Thr Ser Phe Asn Ser
Leu Gly Lys Ser Gln Glu Ala Ile Leu Arg Ile 290 295 300Pro Asp Val
His Glu Lys Thr Phe Gln Tyr Ile Lys Ser Met Lys Ala305 310 315
320Tyr Ile Ala Glu Pro Leu Leu Val Val Asn Trp Gln Ser Ser Ile Pro
325 330 335Ala Val Asp Thr Trp Ile Val Glu Trp Leu Pro Glu Ala Ala
Met Ser 340 345 350Lys Phe Pro Ala Leu Ser Trp Glu Ser Val Ser Gln
Val Thr Asn Trp 355 360 365Thr Ile Glu Gln Asp Lys Leu Lys Pro Phe
Thr Cys Tyr Asn Ile Ser 370 375 380Val Tyr Pro Val Leu Gly His Arg
Val Gly Glu Pro Tyr Ser Ile Gln385 390 395 400Ala Tyr Ala Lys Glu
Gly Thr Pro Leu Lys Gly Pro Glu Thr Arg Val 405 410 415Glu Asn Ile
Gly Leu Arg Thr Ala Thr Ile Thr Trp Lys Glu Ile Pro 420 425 430Lys
Ser Ala Arg Asn Gly Phe Ile Asn Asn Tyr Thr Val Phe Tyr Gln 435 440
445Ala Glu Gly Gly Lys Glu Leu Ser Lys Thr Val Asn Ser His Ala Leu
450 455 460Gln Cys Asp Leu Glu Ser Leu Thr Arg Arg Thr Ser Tyr Thr
Val Trp465 470 475 480Val Met Ala Ser Thr Arg Ala Gly Gly Thr Asn
Gly Val Arg Ile Asn 485 490 495Phe Lys Thr Leu Ser Ile Ser Val Phe
Glu Ile Val Leu Leu Thr Ser 500 505 510Leu Val Gly Gly Gly Leu Leu
Leu Leu Ser Ile Lys Thr Val Thr Phe 515 520 525Gly Leu Arg Lys Pro
Asn Arg Leu Thr Pro Leu Cys Cys Pro Asp Val 530 535 540Pro Asn Pro
Ala Glu Ser Ser Leu Ala Thr Trp Leu Gly Asp Gly Phe545 550 555
560Lys Lys Ser Asn Met Lys Glu Thr Gly Asn Ser Gly Asp Thr Glu Asp
565 570 575Val Val Leu Lys Pro Cys Pro Val Pro Ala Asp Leu Ile Asp
Lys Leu 580 585 590Val Val Asn Phe Glu Asn Phe Leu Glu Val Val Leu
Thr Glu Glu Ala 595 600 605Gly Lys Gly Gln Ala Ser Ile Leu Gly Gly
Glu Ala Asn Glu Tyr Val 610 615 620Thr Ser Pro Ser Arg Pro Asp Gly
Pro Pro Gly Lys Ser Phe Lys Glu625 630 635 640Pro Ser Val Leu Thr
Glu Val Ala Ser Glu Asp Ser His Ser Thr Cys 645 650 655Ser Arg Met
Ala Asp Glu Ala Tyr Ser Glu Leu Ala Arg Gln Pro Ser 660 665 670Ser
Ser Cys Gln Ser Pro Gly Leu Ser Pro Pro Arg Glu Asp Gln Ala 675 680
685Gln Asn Pro Tyr Leu Lys Asn Ser Val Thr Thr Arg Glu Phe Leu Val
690 695 700His Glu Asn Ile Pro Glu His Ser Lys Gly Glu Val705 710
715824PRTArtificialan artificially synthesized peptide sequence
82Gly Gly Gly Ser1834PRTArtificialan artificially synthesized
peptide sequence 83Ser Gly Gly Gly1845PRTArtificialan artificially
synthesized peptide sequence 84Gly Gly Gly Gly Ser1
5855PRTArtificialan artificially synthesized peptide sequence 85Ser
Gly Gly Gly Gly1 5866PRTArtificialan artificially synthesized
peptide sequence 86Gly Gly Gly Gly Gly Ser1 5876PRTArtificialan
artificially synthesized peptide sequence 87Ser Gly Gly Gly Gly
Gly1 5887PRTArtificialan artificially synthesized peptide sequence
88Gly Gly Gly Gly Gly Gly Ser1 5897PRTArtificialan artificially
synthesized peptide sequence 89Ser Gly Gly Gly Gly Gly Gly1
59040DNAArtificialan artificially synthesized primer sequence
90taatagcggc cgctcattat ttaccaggag agtgggagag 409140DNAArtificialan
artificially synthesized primer sequence 91taatagcggc cgctcattaa
cactcattcc tgttgaagct 409233DNAArtificialan artificially
synthesized primer sequence 92gacgaattcc accatgggat ggagctggat ctt
339333DNAArtificialan artificially synthesized primer sequence
93gacgaattcc accatgagtg tgcccactca ggt 339433DNAArtificialan
artificially synthesized primer sequence 94gacgaattcc accatggaat
gtaactggat act 339533DNAArtificialan artificially synthesized
primer sequence 95gacgaattcc accatggatt ttctggtgca gat
339630PRTHomo sapiens 96Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val
Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly
Tyr Thr Phe Thr 20 25 309714PRTHomo sapiens 97Trp Val Arg Gln Ala
Pro Gly Gln Gly Leu Glu Trp Met Gly1 5 109832PRTHomo sapiens 98Arg
Val Thr Ile Thr Ala Asp Glu Ser Thr Ser Thr Ala Tyr Met Glu1 5 10
15Leu Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys Ala Arg
20 25 309911PRTHomo sapiens 99Trp Gly Gln Gly Thr Leu Val Thr Val
Ser Ser1 5 1010023PRTHomo sapiens 100Asp Ile Gln Met Thr Gln Ser
Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10
15Asp Arg Val Thr Ile Thr Cys 2010115PRTHomo sapiens 101Trp Tyr Gln
Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile Tyr1 5 10
1510232PRTHomo sapiens 102Gly Val Pro Ser Arg Phe Ser Gly Ser Gly
Ser Gly Thr Asp Phe Thr1 5 10 15Leu Thr Ile Ser Ser Leu Gln Pro Glu
Asp Phe Ala Thr Tyr Tyr Cys 20 25 3010310PRTHomo sapiens 103Phe Gly
Gly Gly Thr Lys Val Glu Ile Lys1 5 10104119PRTMus musculus 104Asp
Val Gln Leu Arg Glu Ser Gly Pro Gly Leu Val Lys Pro Ser Gln1 5 10
15Ser Leu Ser Leu Thr Cys Thr Val Thr Gly Tyr Ser Ile Thr Ser Asp
20 25 30Tyr Ala Trp Asn Trp Ile Arg Gln Phe Pro Gly Asn Lys Leu Glu
Trp 35 40 45Met Gly Tyr Ile Ser Tyr Ser Gly Ser Thr Ser Tyr Asn Pro
Ser Leu 50 55 60Lys Ser Arg Val Ser Ile Thr Arg Asp Thr Ser Lys Asn
Gln Phe Phe65 70 75 80Leu Gln Leu Asn Ser Val Thr Thr Glu Asp Thr
Ala Thr Tyr Tyr Cys 85 90 95Ala Pro Met Ile Thr Thr Asp Trp Phe Phe
Asp Val Trp Gly Ala Gly 100 105 110Thr Thr Val Thr Val Ser Ser
1151056PRTMus musculus 105Ser Asp Tyr Ala Trp Asn1 510615PRTMus
musculus 106Tyr Ile Ser Tyr Ser Gly Ser Thr Ser Tyr Asn Pro Ser Leu
Lys1 5 10 1510710PRTMus musculus 107Met Ile Thr Thr Asp Trp Phe Phe
Asp Val1 5 10108107PRTMus musculus 108Asp Ile Val Met Thr Gln Ser
Pro Ala Thr Leu Ser Val Thr Pro Gly1 5 10 15Asp Arg Val Ser Leu Ser
Cys Arg Ala Ser His Asp Ile Ser Asp Phe 20 25 30Leu His Trp Tyr Gln
Gln Lys Ser His Glu Ser Pro Arg Leu Leu Ile 35 40 45Lys Tyr Ala Ser
Gln Ser Ile Ser Gly Ile Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser
Gly Ser Asp Phe Thr Leu Ser Ile Asn Ser Val Glu Pro65 70 75 80Glu
Asp Val Gly Val Tyr Tyr Cys Gln Asn Gly His Ser Phe Pro Trp 85 90
95Thr Phe Gly Gly Gly Thr Lys Leu Glu Ile Lys 100 10510911PRTMus
musculus 109Arg Ala Ser His Asp Ile Ser Asp Phe Leu His1 5
101107PRTMus musculus 110Tyr Ala Ser Gln Ser Ile Ser1 51119PRTMus
musculus 111Gln Asn Gly His Ser Phe Pro Trp Thr1
5112121PRTArtificialAn artificially synthesized peptide sequence
112Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn
Gln Lys Phe 50 55 60Lys Gly Arg Val Thr Ile Thr Ala Asp Lys Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser 115 12011317PRTArtificialAn artificially synthesized
peptide sequence 113Glu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn
Gln Lys Phe Lys1 5 10 15Gly11417PRTArtificialAn artificially
synthesized peptide sequence 114Leu Ile Asp Pro Tyr Asn Gly Gly Thr
Ser Tyr Asn Gln Lys Phe Lys1 5 10 15Gly11517PRTArtificialAn
artificially synthesized peptide sequence 115Leu Ile Asn Pro Tyr
Asn Gly Gly Thr Ser Tyr Asn Asp Lys Phe Lys1 5 10
15Gly11617PRTArtificialAn artificially synthesized peptide sequence
116Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn Gln Gln Phe Lys1
5 10 15Gly11717PRTArtificialAn artificially synthesized peptide
sequence 117Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn Gln Lys
Phe Gln1 5 10 15Gly11817PRTArtificialAn artificially synthesized
peptide sequence 118Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn
Gln Lys Phe Lys1 5 10 15Asp11917PRTArtificialAn artificially
synthesized peptide sequence 119Leu Ile Asn Pro Tyr Asn Gly Gly Thr
Ser Tyr Asn Gln Lys Phe Gln1 5 10 15Asp12014PRTArtificialAn
artificially synthesized peptide sequence 120Trp Val Gln Gln Ser
Pro Gly Gln Gly Leu Glu Trp Met Gly1 5 1012111PRTArtificialAn
artificially synthesized peptide sequence 121Gln Thr Ser Glu Asn
Ile Tyr Ser Phe Leu Ala1 5 1012211PRTArtificialAn artificially
synthesized peptide sequence 122Arg Thr Ser Glu Asp Ile Tyr Ser Phe
Leu Ala1 5 101237PRTArtificialAn artificially synthesized peptide
sequence 123Asp Ala Lys Thr Leu Ala Lys1 51247PRTArtificialAn
artificially synthesized peptide sequence 124Asn Ala Gln Thr Leu
Ala Lys1 51257PRTArtificialAn artificially synthesized peptide
sequence 125Asn Ala Lys Thr Glu Ala Lys1 51267PRTArtificialAn
artificially synthesized peptide sequence 126Asn Ala Lys Thr Leu
Ala Gln1 51277PRTArtificialAn artificially synthesized peptide
sequence 127Asn Ala Lys Thr Leu Ala Asp1 5128324PRTArtificialAn
artificially synthesized peptide sequence 128Ala Ser Thr Lys Gly
Pro Ser Val Phe Pro Leu Ala Pro Ser Ser Lys1 5 10 15Ser Thr Ser Gly
Gly Thr Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr 20 25 30Phe Pro Glu
Pro Val Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser 35 40 45Gly Val
His Thr Phe Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser 50 55 60Leu
Ser Ser Val Val Thr Val Pro Ser Ser Asn Phe Gly Thr Gln Thr65 70 75
80Tyr Thr Cys Asn Val Asp His Lys Pro Ser Asn Thr Lys Val Asp Lys
85 90 95Thr Val Glu Arg Lys Ser Cys Val Glu Cys Pro Pro Cys Pro Ala
Pro 100 105 110Pro Val Ala Gly Pro Ser Val Phe Leu Phe Pro Pro Lys
Pro Lys Asp 115 120 125Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr
Cys Val Val Val Asp 130 135 140Val Ser Gln Glu Asp Pro Glu Val Gln
Phe Asn Trp Tyr Val Asp Gly145 150 155 160Val Glu Val His Asn Ala
Lys Thr Lys Pro Arg Glu Glu Gln Phe Asn 165 170 175Ser Thr Phe Arg
Val Val Ser Val Leu Thr Val Val His Gln Asp Trp 180 185 190Leu Asn
Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Gly Leu Pro 195 200
205Ala Pro Ile Glu Lys Thr Ile Ser Lys Thr Lys Gly Gln Pro Arg Glu
210 215 220Pro Gln Val Tyr Thr Leu Pro Pro Ser Gln Glu Glu Met Thr
Lys Asn225 230 235 240Gln Val Ser Leu Thr Cys Leu Val Lys Gly Phe
Tyr Pro Ser Asp Ile 245 250 255Ala Val Glu Trp Glu Ser Asn Gly Gln
Pro Glu Asn Asn Tyr Lys Thr 260 265 270Thr Pro Pro Met Leu Asp Ser
Asp Gly Ser Phe Phe Leu Tyr Ser Lys 275 280 285Leu Thr Val Asp Lys
Ser Arg Trp Gln Glu Gly Asn Val Phe Ser Cys 290 295 300Ser Val Met
His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu305 310 315
320Ser Leu Ser Pro129326PRTArtificialAn artificially synthesized
peptide sequence 129Ala Ser Thr Lys Gly Pro Ser Val Phe Pro Leu Ala
Pro Ser Ser Lys1 5 10 15Ser Thr Ser Gly Gly Thr Ala Ala Leu Gly Cys
Leu Val Lys Asp Tyr 20 25 30Phe Pro Glu Pro Val Thr Val Ser Trp Asn
Ser Gly Ala Leu Thr Ser 35 40 45Gly Val His Thr Phe Pro Ala Val Leu
Gln Ser Ser Gly Leu Tyr Ser 50 55 60Leu Ser Ser Val Val Thr Val Pro
Ser Ser Asn Phe Gly Thr Gln Thr65 70 75 80Tyr Thr Cys Asn Val Asp
His Lys Pro Ser Asn Thr Lys Val Asp Lys 85 90 95Thr Val Glu Arg Lys
Ser Cys Val Glu Cys Pro Pro Cys Pro Ala Pro 100 105 110Pro Val Ala
Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp 115 120 125Thr
Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp 130 135
140Val Ser His Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr Val Asp
Gly145 150 155 160Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu
Glu Gln Phe Asn 165 170 175Ser Thr Phe Arg Val Val Ser Val Leu Thr
Val Val His Gln Asp Trp 180 185 190Leu Asn Gly Lys Glu Tyr Lys Cys
Lys Val Ser Asn Lys Gly Leu Pro 195 200 205Ser Ser Ile Glu Lys Thr
Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu 210 215 220Pro Gln Val Tyr
Thr Leu Pro Pro Ser Arg Glu Glu Met Thr Lys Asn225 230 235 240Gln
Val Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile 245 250
255Ala Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr
260 265 270Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr
Ser Lys 275 280 285Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn
Val Phe Ser Cys 290 295 300Ser Val Met His Glu Ala Leu His Asn His
Tyr Thr Gln Lys Ser Leu305 310 315 320Ser Leu Ser Pro Gly Lys
325130447PRTArtificialAn artificially synthesized peptide sequence
130Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn
Gln Lys Phe 50 55 60Lys Gly Arg Val Thr Ile Thr Ala Asp Lys Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Glu Ser Thr Ala 130 135 140Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150 155
160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala
165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys
Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys Thr
Val Glu Arg Lys Ser Cys 210 215 220Val Glu Cys Pro Pro Cys Pro Ala
Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250 255Pro Glu Val
Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu 260 265 270Val
Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 275 280
285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser
290 295 300Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu
Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro
Ile Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Arg Glu Glu Met Thr
Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser385 390 395
400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg
405 410 415Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu
Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
Pro Gly Lys 435 440 44513132PRTArtificialAn artificially
synthesized peptide sequence 131Arg Val Thr Ile Thr Ala Asp Lys Ser
Thr Ser Thr Ala Tyr Met Glu1 5 10 15Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys Ala Arg 20 25 30132326PRTArtificialAn
artificially synthesized peptide sequence 132Ala Ser Thr Lys Gly
Pro Ser Val Phe Pro Leu Ala Pro Cys Ser Arg1 5 10 15Ser Thr Ser Glu
Ser Thr Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr 20 25 30Phe Pro Glu
Pro Val Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser 35 40 45Gly Val
His Thr Phe Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser 50 55 60Leu
Ser Ser Val Val Thr Val Pro Ser Ser Asn Phe Gly Thr Gln Thr65 70 75
80Tyr Thr Cys Asn Val Asp His Lys Pro Ser Asn Thr Lys Val Asp Lys
85 90 95Thr Val Glu Arg Lys Cys Cys Val Glu Cys Pro Pro Cys Pro Ala
Pro 100 105 110Pro Val Ala Gly Pro Ser Val Phe Leu Phe Pro Pro Lys
Pro Lys Asp 115 120 125Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr
Cys Val Val Val Asp 130 135 140Val Ser His Glu Asp Pro Glu Val Gln
Phe Asn Trp Tyr Val Asp Gly145 150 155 160Val Glu Val His Asn Ala
Lys Thr Lys Pro Arg Glu Glu Gln Phe Asn 165 170 175Ser Thr Phe Arg
Val Val Ser Val Leu Thr Val Val His Gln Asp Trp 180 185 190Leu Asn
Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Gly Leu Pro 195 200
205Ala Pro Ile Glu Lys Thr Ile Ser Lys Thr Lys Gly Gln Pro Arg Glu
210 215 220Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu Met Thr
Lys Asn225 230 235 240Gln Val Ser Leu Thr Cys Leu Val Lys Gly Phe
Tyr Pro Ser Asp Ile 245 250 255Ala Val Glu Trp Glu Ser Asn Gly Gln
Pro Glu Asn Asn Tyr Lys Thr 260 265 270Thr Pro Pro Met Leu Asp Ser
Asp Gly Ser Phe Phe Leu Tyr Ser Lys 275 280 285Leu Thr Val Asp Lys
Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys 290 295 300Ser Val Met
His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu305 310 315
320Ser Leu Ser Pro Gly Lys 325133451PRTArtificialAn artificially
synthesized peptide sequence 133Gln Val Gln Leu Val Gln Ser Gly Ala
Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala
Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Arg Gln
Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr
Asn Gly Gly Thr Ser Tyr Asn Gln Lys Phe 50 55 60Lys Gly Arg Val Thr
Ile Thr Ala Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu
Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg
Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Asp Tyr Trp Gly 100 105
110Gln Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser
115 120 125Val Phe
Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135
140Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr
Val145 150 155 160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His
Thr Phe Pro Ala 165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu
Ser Ser Val Val Thr Val 180 185 190Pro Ser Ser Ser Leu Gly Thr Gln
Thr Tyr Ile Cys Asn Val Asn His 195 200 205Lys Pro Ser Asn Thr Lys
Val Asp Lys Lys Val Glu Pro Lys Ser Cys 210 215 220Asp Lys Thr His
Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly225 230 235 240Gly
Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 245 250
255Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His
260 265 270Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val
Glu Val 275 280 285His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr
Asn Ser Thr Tyr 290 295 300Arg Val Val Ser Val Leu Thr Val Leu His
Gln Asp Trp Leu Asn Gly305 310 315 320Lys Glu Tyr Lys Cys Lys Val
Ser Asn Lys Ala Leu Pro Ala Pro Ile 325 330 335Glu Lys Thr Ile Ser
Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val 340 345 350Tyr Thr Leu
Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser 355 360 365Leu
Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu 370 375
380Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro
Pro385 390 395 400Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser
Lys Leu Thr Val 405 410 415Asp Lys Ser Arg Trp Gln Gln Gly Asn Val
Phe Ser Cys Ser Val Met 420 425 430His Glu Ala Leu His Asn His Tyr
Thr Gln Lys Ser Leu Ser Leu Ser 435 440 445Pro Gly Lys
450134447PRTArtificialAn artificially synthesized peptide sequence
134Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn
Gln Lys Phe 50 55 60Lys Gly Arg Val Thr Ile Thr Ala Asp Glu Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro Leu Ala
Pro Cys Ser Arg Ser Thr Ser Glu Ser Thr Ala 130 135 140Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150 155
160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala
165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys
Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys Thr
Val Glu Arg Lys Cys Cys 210 215 220Val Glu Cys Pro Pro Cys Pro Ala
Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250 255Pro Glu Val
Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu 260 265 270Val
Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 275 280
285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser
290 295 300Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu
Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro
Ile Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Arg Glu Glu Met Thr
Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser385 390 395
400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg
405 410 415Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu
Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
Pro Gly Lys 435 440 445135447PRTArtificialAn artificially
synthesized peptide sequence 135Gln Val Gln Leu Val Gln Ser Gly Ala
Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala
Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Arg Gln
Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr
Asn Gly Gly Thr Ser Tyr Asn Gln Lys Phe 50 55 60Lys Gly Arg Val Thr
Ile Thr Ala Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu
Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg
Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Asp Tyr Trp Gly 100 105
110Gln Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser
115 120 125Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly
Thr Ala 130 135 140Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu
Pro Val Thr Val145 150 155 160Ser Trp Asn Ser Gly Ala Leu Thr Ser
Gly Val His Thr Phe Pro Ala 165 170 175Val Leu Gln Ser Ser Gly Leu
Tyr Ser Leu Ser Ser Val Val Thr Val 180 185 190Pro Ser Ser Asn Phe
Gly Thr Gln Thr Tyr Thr Cys Asn Val Asp His 195 200 205Lys Pro Ser
Asn Thr Lys Val Asp Lys Thr Val Glu Arg Lys Ser Cys 210 215 220Val
Glu Cys Pro Pro Cys Pro Ala Pro Pro Val Ala Gly Pro Ser Val225 230
235 240Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg
Thr 245 250 255Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu
Asp Pro Glu 260 265 270Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu
Val His Asn Ala Lys 275 280 285Thr Lys Pro Arg Glu Glu Gln Phe Asn
Ser Thr Phe Arg Val Val Ser 290 295 300Val Leu Thr Val Val His Gln
Asp Trp Leu Asn Gly Lys Glu Tyr Lys305 310 315 320Cys Lys Val Ser
Asn Lys Gly Leu Pro Ser Ser Ile Glu Lys Thr Ile 325 330 335Ser Lys
Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro 340 345
350Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu
355 360 365Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu
Ser Asn 370 375 380Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro
Val Leu Asp Ser385 390 395 400Asp Gly Ser Phe Phe Leu Tyr Ser Lys
Leu Thr Val Asp Lys Ser Arg 405 410 415Trp Gln Gln Gly Asn Val Phe
Ser Cys Ser Val Met His Glu Ala Leu 420 425 430His Asn His Tyr Thr
Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 435 440
445136445PRTArtificialAn artificially synthesized peptide sequence
136Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn
Gln Lys Phe 50 55 60Lys Gly Arg Val Thr Ile Thr Ala Asp Glu Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135 140Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150 155
160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala
165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys
Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys Thr
Val Glu Arg Lys Ser Cys 210 215 220Val Glu Cys Pro Pro Cys Pro Ala
Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250 255Pro Glu Val
Thr Cys Val Val Val Asp Val Ser Gln Glu Asp Pro Glu 260 265 270Val
Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 275 280
285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser
290 295 300Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu
Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro
Ile Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Gln Glu Glu Met Thr
Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser385 390 395
400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg
405 410 415Trp Gln Glu Gly Asn Val Phe Ser Cys Ser Val Met His Glu
Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
Pro 435 440 445137121PRTArtificialAn artificially synthesized
peptide sequence 137Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Gln Gln Ser Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly
Thr Ser Tyr Asn Gln Lys Phe 50 55 60Lys Gly Arg Val Thr Ile Thr Ala
Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu
Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr
Asp Asp Gly Pro Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr
Leu Val Thr Val Ser Ser 115 120138121PRTArtificialAn artificially
synthesized peptide sequence 138Gln Val Gln Leu Val Gln Ser Gly Ala
Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala
Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Arg Gln
Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Glu Ile Asn Pro Tyr
Asn Gly Gly Thr Ser Tyr Asn Gln Lys Phe 50 55 60Lys Gly Arg Val Thr
Ile Thr Ala Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu
Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg
Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Asp Tyr Trp Gly 100 105
110Gln Gly Thr Leu Val Thr Val Ser Ser 115 120139121PRTArtificialAn
artificially synthesized peptide sequence 139Gln Val Gln Leu Val
Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val
Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn
Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu
Ile Asp Pro Tyr Asn Gly Gly Thr Ser Tyr Asn Gln Lys Phe 50 55 60Lys
Gly Arg Val Thr Ile Thr Ala Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75
80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Asp Tyr Trp
Gly 100 105 110Gln Gly Thr Leu Val Thr Val Ser Ser 115
120140121PRTArtificialAn artificially synthesized peptide sequence
140Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn
Asp Lys Phe 50 55 60Lys Gly Arg Val Thr Ile Thr Ala Asp Glu Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser 115 120141121PRTArtificialAn artificially synthesized
peptide sequence 141Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly
Thr Ser Tyr Asn Gln Gln Phe 50 55 60Lys Gly Arg Val Thr Ile Thr Ala
Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu
Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr
Asp Asp Gly Pro Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr
Leu Val Thr Val Ser Ser 115 120142121PRTArtificialAn artificially
synthesized peptide sequence 142Gln Val Gln Leu Val Gln Ser Gly Ala
Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala
Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Arg Gln
Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr
Asn Gly Gly Thr Ser Tyr Asn Gln Lys Phe 50 55 60Gln Gly Arg Val Thr
Ile Thr Ala Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu
Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg
Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Asp Tyr Trp Gly 100 105
110Gln Gly Thr Leu Val Thr Val Ser Ser 115
120143121PRTArtificialAn artificially synthesized peptide sequence
143Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn
Gln Lys Phe 50 55 60Gln Asp Arg Val Thr Ile Thr Ala Asp Glu Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser 115 120144107PRTArtificialAn artificially synthesized
peptide sequence 144Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser
Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Gln Thr Ser Glu
Asn Ile Tyr Ser Phe 20 25 30Leu Ala Trp Tyr Gln Gln Lys Pro Gly Lys
Ala Pro Lys Leu Leu Ile 35 40 45Tyr Asn Ala Lys Thr Leu Ala Lys Gly
Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr
Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr
Tyr Cys Gln His His Tyr Glu Ser Pro Leu 85 90 95Thr Phe Gly Gly Gly
Thr Lys Val Glu Ile Lys 100 105145107PRTArtificialAn artificially
synthesized peptide sequence 145Asp Ile Gln Met Thr Gln Ser Pro Ser
Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg
Thr Ser Glu Asp Ile Tyr Ser Phe 20 25 30Leu Ala Trp Tyr Gln Gln Lys
Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Asn Ala Lys Thr Leu
Ala Lys Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr
Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe
Ala Thr Tyr Tyr Cys Gln His His Tyr Glu Ser Pro Leu 85 90 95Thr Phe
Gly Gly Gly Thr Lys Val Glu Ile Lys 100 105146107PRTArtificialAn
artificially synthesized peptide sequence 146Asp Ile Gln Met Thr
Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr
Ile Thr Cys Arg Thr Ser Glu Asn Ile Tyr Ser Phe 20 25 30Leu Ala Trp
Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Asp
Ala Lys Thr Leu Ala Lys Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser
Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75
80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln His His Tyr Glu Ser Pro Leu
85 90 95Thr Phe Gly Gly Gly Thr Lys Val Glu Ile Lys 100
105147107PRTArtificialAn artificially synthesized peptide sequence
147Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1
5 10 15Asp Arg Val Thr Ile Thr Cys Arg Thr Ser Glu Asn Ile Tyr Ser
Phe 20 25 30Leu Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu
Leu Ile 35 40 45Tyr Asn Ala Gln Thr Leu Ala Lys Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln His
His Tyr Glu Ser Pro Leu 85 90 95Thr Phe Gly Gly Gly Thr Lys Val Glu
Ile Lys 100 105148107PRTArtificialAn artificially synthesized
peptide sequence 148Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser
Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Thr Ser Glu
Asn Ile Tyr Ser Phe 20 25 30Leu Ala Trp Tyr Gln Gln Lys Pro Gly Lys
Ala Pro Lys Leu Leu Ile 35 40 45Tyr Asn Ala Lys Thr Glu Ala Lys Gly
Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr
Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr
Tyr Cys Gln His His Tyr Glu Ser Pro Leu 85 90 95Thr Phe Gly Gly Gly
Thr Lys Val Glu Ile Lys 100 105149107PRTArtificialAn artificially
synthesized peptide sequence 149Asp Ile Gln Met Thr Gln Ser Pro Ser
Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg
Thr Ser Glu Asn Ile Tyr Ser Phe 20 25 30Leu Ala Trp Tyr Gln Gln Lys
Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Asn Ala Lys Thr Leu
Ala Gln Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr
Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe
Ala Thr Tyr Tyr Cys Gln His His Tyr Glu Ser Pro Leu 85 90 95Thr Phe
Gly Gly Gly Thr Lys Val Glu Ile Lys 100 105150107PRTArtificialAn
artificially synthesized peptide sequence 150Asp Ile Gln Met Thr
Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr
Ile Thr Cys Arg Thr Ser Glu Asn Ile Tyr Ser Phe 20 25 30Leu Ala Trp
Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Asn
Ala Lys Thr Leu Ala Asp Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser
Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75
80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln His His Tyr Glu Ser Pro Leu
85 90 95Thr Phe Gly Gly Gly Thr Lys Val Glu Ile Lys 100
105151447PRTArtificialAn artificially synthesized peptide sequence
151Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn
Gln Gln Phe 50 55 60Gln Asp Arg Val Thr Ile Thr Ala Asp Glu Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Glu Ser Thr Ala 130 135 140Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150 155
160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala
165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys
Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys Thr
Val Glu Arg Lys Ser Cys 210 215 220Val Glu Cys Pro Pro Cys Pro Ala
Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250 255Pro Glu Val
Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu 260 265 270Val
Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 275 280
285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser
290 295 300Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu
Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro
Ile Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Arg Glu Glu Met Thr
Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser385 390 395
400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg
405 410 415Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu
Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
Pro Gly Lys 435 440 445152214PRTArtificialAn artificially
synthesized peptide sequence 152Asp Ile Gln Met Thr Gln Ser Pro Ser
Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Gln
Thr Ser Glu Asp Ile Tyr Ser Phe 20 25 30Leu Ala Trp Tyr Gln Gln Lys
Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Asn Ala Gln Thr Glu
Ala Gln Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr
Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe
Ala Thr Tyr Tyr Cys Gln His His Tyr Glu Ser Pro Leu 85 90 95Thr Phe
Gly Gly Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala Ala 100 105
110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly
115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg
Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser
Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu Gln Asp Ser Lys
Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys
Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala Cys Glu Val Thr
His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200 205Phe Asn Arg
Gly Glu Cys 210153326PRTArtificialAn artificially synthesized
peptide sequence 153Ala Ser Thr Lys Gly Pro Ser Val Phe Pro Leu Ala
Pro Cys Ser Arg1 5 10 15Ser Thr Ser Glu Ser Thr Ala Ala Leu Gly Cys
Leu Val Lys Asp Tyr 20 25 30Phe Pro Glu Pro Val Thr Val Ser Trp Asn
Ser Gly Ala Leu Thr Ser 35 40 45Gly Val His Thr Phe Pro Ala Val Leu
Gln Ser Ser Gly Leu Tyr Ser 50 55 60Leu Ser Ser Val Val Thr Val Pro
Ser Ser Asn Phe Gly Thr Gln Thr65 70 75 80Tyr Thr Cys Asn Val Asp
His Lys Pro Ser Asn Thr Lys Val Asp Lys 85 90 95Thr Val Glu Arg Lys
Ser Cys Val Glu Cys Pro Pro Cys Pro Ala Pro 100 105 110Pro Val Ala
Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp 115 120 125Thr
Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp 130 135
140Val Ser His Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr Val Asp
Gly145 150 155 160Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu
Glu Gln Phe Asn 165 170 175Ser Thr Phe Arg Val Val Ser Val Leu Thr
Val Val His Gln Asp Trp 180 185 190Leu Asn Gly Lys Glu Tyr Lys Cys
Lys Val Ser Asn Lys Gly Leu Pro 195 200 205Ala Pro Ile Glu Lys Thr
Ile Ser Lys Thr Lys Gly Gln Pro Arg Glu 210 215 220Pro Gln Val Tyr
Thr Leu Pro Pro Ser Arg Glu Glu Met Thr Lys Asn225 230 235 240Gln
Val Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile 245 250
255Ala Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr
260 265 270Thr Pro Pro Met Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr
Ser Lys 275 280 285Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn
Val Phe Ser Cys 290 295 300Ser Val Met His Glu Ala Leu His Asn His
Tyr Thr Gln Lys Ser Leu305 310 315 320Ser Leu Ser Pro Gly Lys
325154326PRTArtificialAn artificially synthesized peptide sequence
154Ala Ser Thr Lys Gly Pro Ser Val Phe Pro Leu Ala Pro Cys Ser Arg1
5 10 15Ser Thr Ser Glu Ser Thr Ala Ala Leu Gly Cys Leu Val Lys Asp
Tyr 20 25 30Phe Pro Glu Pro Val Thr Val Ser Trp Asn Ser Gly Ala Leu
Thr Ser 35 40 45Gly Val His Thr Phe Pro Ala Val Leu Gln Ser Ser Gly
Leu Tyr Ser 50 55 60Leu Ser Ser Val Val Thr Val Pro Ser Ser Asn Phe
Gly Thr Gln Thr65 70 75 80Tyr Thr Cys Asn Val Asp His Lys Pro Ser
Asn Thr Lys Val Asp Lys 85 90 95Thr Val Glu Arg Lys Cys Ser Val Glu
Cys Pro Pro Cys Pro Ala Pro 100 105 110Pro Val Ala Gly Pro Ser Val
Phe Leu Phe Pro Pro Lys Pro Lys Asp 115 120 125Thr Leu Met Ile Ser
Arg Thr Pro Glu Val Thr Cys Val Val Val Asp 130 135 140Val Ser His
Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr Val Asp Gly145 150 155
160Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Phe Asn
165 170 175Ser Thr Phe Arg Val Val Ser Val Leu Thr Val Val His Gln
Asp Trp 180 185 190Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn
Lys Gly Leu Pro 195 200 205Ala Pro Ile Glu Lys Thr Ile Ser Lys Thr
Lys Gly Gln Pro Arg Glu 210 215 220Pro Gln Val Tyr Thr Leu Pro Pro
Ser Arg Glu Glu Met Thr Lys Asn225 230 235 240Gln Val Ser Leu Thr
Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile 245 250 255Ala Val Glu
Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr 260 265 270Thr
Pro Pro Met Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys 275 280
285Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys
290 295 300Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys
Ser Leu305 310 315 320Ser Leu Ser Pro Gly Lys
325155119PRTArtificialAn artificially synthesized peptide sequence
155Gln Val Gln Leu Gln Glu Ser Gly Pro Gly Leu Val Arg Pro Ser Gln1
5 10 15Thr Leu Ser Leu Thr Cys Thr Val Ser Gly Tyr Ser Ile Thr Ser
Asp 20 25 30His Ala Trp Ser Trp Val Arg Gln Pro Pro Gly Arg Gly Leu
Glu Trp 35 40 45Ile Gly Tyr Ile Ser Tyr Ser Gly Ile Thr Thr Tyr Asn
Pro Ser Leu 50 55 60Lys Ser Arg Val Thr Met Leu Arg Asp Thr Ser Lys
Asn Gln Phe Ser65 70 75 80Leu Arg Leu Ser Ser Val Thr Ala Ala Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Ser Leu Ala Arg Thr Thr Ala
Met Asp Tyr Trp Gly Gln Gly 100 105 110Ser Leu Val Thr Val Ser Ser
115156107PRTArtificialAn artificially synthesized peptide sequence
156Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1
5 10 15Asp Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Asp Ile Ser Ser
Tyr 20 25 30Leu Asn Trp Tyr Gln Gln
Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Tyr Thr Ser Arg
Leu His Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly
Thr Asp Phe Thr Phe Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp
Ile Ala Thr Tyr Tyr Cys Gln Gln Gly Asn Thr Leu Pro Tyr 85 90 95Thr
Phe Gly Gln Gly Thr Lys Val Glu Ile Lys 100
105157445PRTArtificialAn artificially synthesized peptide sequence
157Gln Val Gln Leu Gln Glu Ser Gly Pro Gly Leu Val Arg Pro Ser Gln1
5 10 15Thr Leu Ser Leu Thr Cys Thr Val Ser Gly Tyr Ser Ile Thr Ser
Asp 20 25 30His Ala Trp Ser Trp Val Arg Gln Pro Pro Gly Arg Gly Leu
Glu Trp 35 40 45Ile Gly Tyr Ile Ser Tyr Ser Gly Ile Thr Thr Tyr Asn
Pro Ser Leu 50 55 60Lys Ser Arg Val Thr Met Leu Arg Asp Thr Ser Lys
Asn Gln Phe Ser65 70 75 80Leu Arg Leu Ser Ser Val Thr Ala Ala Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Ser Leu Ala Arg Thr Thr Ala
Met Asp Tyr Trp Gly Gln Gly 100 105 110Ser Leu Val Thr Val Ser Ser
Ala Ser Thr Lys Gly Pro Ser Val Phe 115 120 125Pro Leu Ala Pro Cys
Ser Arg Ser Thr Ser Glu Ser Thr Ala Ala Leu 130 135 140Gly Cys Leu
Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp145 150 155
160Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val Leu
165 170 175Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val
Pro Ser 180 185 190Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys Asn Val
Asp His Lys Pro 195 200 205Ser Asn Thr Lys Val Asp Lys Thr Val Glu
Arg Lys Ser Cys Val Glu 210 215 220Cys Pro Pro Cys Pro Ala Pro Pro
Val Ala Gly Pro Ser Val Phe Leu225 230 235 240Phe Pro Pro Lys Pro
Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu 245 250 255Val Thr Cys
Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Gln 260 265 270Phe
Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys 275 280
285Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser Val Leu
290 295 300Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys
Cys Lys305 310 315 320Val Ser Asn Lys Gly Leu Pro Ala Pro Ile Glu
Lys Thr Ile Ser Lys 325 330 335Thr Lys Gly Gln Pro Arg Glu Pro Gln
Val Tyr Thr Leu Pro Pro Ser 340 345 350Arg Glu Glu Met Thr Lys Asn
Gln Val Ser Leu Thr Cys Leu Val Lys 355 360 365Gly Phe Tyr Pro Ser
Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln 370 375 380Pro Glu Asn
Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser Asp Gly385 390 395
400Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln
405 410 415Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu
His Asn 420 425 430His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly
Lys 435 440 445158445PRTArtificialAn artificially synthesized
peptide sequence 158Gln Val Gln Leu Gln Glu Ser Gly Pro Gly Leu Val
Arg Pro Ser Gln1 5 10 15Thr Leu Ser Leu Thr Cys Thr Val Ser Gly Tyr
Ser Ile Thr Ser Asp 20 25 30His Ala Trp Ser Trp Val Arg Gln Pro Pro
Gly Arg Gly Leu Glu Trp 35 40 45Ile Gly Tyr Ile Ser Tyr Ser Gly Ile
Thr Thr Tyr Asn Pro Ser Leu 50 55 60Lys Ser Arg Val Thr Met Leu Arg
Asp Thr Ser Lys Asn Gln Phe Ser65 70 75 80Leu Arg Leu Ser Ser Val
Thr Ala Ala Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Ser Leu Ala
Arg Thr Thr Ala Met Asp Tyr Trp Gly Gln Gly 100 105 110Ser Leu Val
Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser Val Phe 115 120 125Pro
Leu Ala Pro Cys Ser Arg Ser Thr Ser Glu Ser Thr Ala Ala Leu 130 135
140Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser
Trp145 150 155 160Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe
Pro Ala Val Leu 165 170 175Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser
Val Val Thr Val Pro Ser 180 185 190Ser Asn Phe Gly Thr Gln Thr Tyr
Thr Cys Asn Val Asp His Lys Pro 195 200 205Ser Asn Thr Lys Val Asp
Lys Thr Val Glu Arg Lys Cys Ser Val Glu 210 215 220Cys Pro Pro Cys
Pro Ala Pro Pro Val Ala Gly Pro Ser Val Phe Leu225 230 235 240Phe
Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu 245 250
255Val Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Gln
260 265 270Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys
Thr Lys 275 280 285Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val
Val Ser Val Leu 290 295 300Thr Val Val His Gln Asp Trp Leu Asn Gly
Lys Glu Tyr Lys Cys Lys305 310 315 320Val Ser Asn Lys Gly Leu Pro
Ala Pro Ile Glu Lys Thr Ile Ser Lys 325 330 335Thr Lys Gly Gln Pro
Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser 340 345 350Arg Glu Glu
Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys 355 360 365Gly
Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln 370 375
380Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser Asp
Gly385 390 395 400Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys
Ser Arg Trp Gln 405 410 415Gln Gly Asn Val Phe Ser Cys Ser Val Met
His Glu Ala Leu His Asn 420 425 430His Tyr Thr Gln Lys Ser Leu Ser
Leu Ser Pro Gly Lys 435 440 445159449PRTArtificialAn artificially
synthesized peptide sequence 159Gln Val Gln Leu Gln Glu Ser Gly Pro
Gly Leu Val Arg Pro Ser Gln1 5 10 15Thr Leu Ser Leu Thr Cys Thr Val
Ser Gly Tyr Ser Ile Thr Ser Asp 20 25 30His Ala Trp Ser Trp Val Arg
Gln Pro Pro Gly Arg Gly Leu Glu Trp 35 40 45Ile Gly Tyr Ile Ser Tyr
Ser Gly Ile Thr Thr Tyr Asn Pro Ser Leu 50 55 60Lys Ser Arg Val Thr
Met Leu Arg Asp Thr Ser Lys Asn Gln Phe Ser65 70 75 80Leu Arg Leu
Ser Ser Val Thr Ala Ala Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg
Ser Leu Ala Arg Thr Thr Ala Met Asp Tyr Trp Gly Gln Gly 100 105
110Ser Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser Val Phe
115 120 125Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala
Ala Leu 130 135 140Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val
Thr Val Ser Trp145 150 155 160Asn Ser Gly Ala Leu Thr Ser Gly Val
His Thr Phe Pro Ala Val Leu 165 170 175Gln Ser Ser Gly Leu Tyr Ser
Leu Ser Ser Val Val Thr Val Pro Ser 180 185 190Ser Ser Leu Gly Thr
Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro 195 200 205Ser Asn Thr
Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp Lys 210 215 220Thr
His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro225 230
235 240Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile
Ser 245 250 255Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser
His Glu Asp 260 265 270Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly
Val Glu Val His Asn 275 280 285Ala Lys Thr Lys Pro Arg Glu Glu Gln
Tyr Asn Ser Thr Tyr Arg Val 290 295 300Val Ser Val Leu Thr Val Leu
His Gln Asp Trp Leu Asn Gly Lys Glu305 310 315 320Tyr Lys Cys Lys
Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys 325 330 335Thr Ile
Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr 340 345
350Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr
355 360 365Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu
Trp Glu 370 375 380Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr
Pro Pro Val Leu385 390 395 400Asp Ser Asp Gly Ser Phe Phe Leu Tyr
Ser Lys Leu Thr Val Asp Lys 405 410 415Ser Arg Trp Gln Gln Gly Asn
Val Phe Ser Cys Ser Val Met His Glu 420 425 430Ala Leu His Asn His
Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly 435 440
445Lys160445PRTArtificialAn artificially synthesized peptide
sequence 160Gln Val Gln Leu Gln Glu Ser Gly Pro Gly Leu Val Arg Pro
Ser Gln1 5 10 15Thr Leu Ser Leu Thr Cys Thr Val Ser Gly Tyr Ser Ile
Thr Ser Asp 20 25 30His Ala Trp Ser Trp Val Arg Gln Pro Pro Gly Arg
Gly Leu Glu Trp 35 40 45Ile Gly Tyr Ile Ser Tyr Ser Gly Ile Thr Thr
Tyr Asn Pro Ser Leu 50 55 60Lys Ser Arg Val Thr Met Leu Arg Asp Thr
Ser Lys Asn Gln Phe Ser65 70 75 80Leu Arg Leu Ser Ser Val Thr Ala
Ala Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Ser Leu Ala Arg Thr
Thr Ala Met Asp Tyr Trp Gly Gln Gly 100 105 110Ser Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser Val Phe 115 120 125Pro Leu Ala
Pro Cys Ser Arg Ser Thr Ser Glu Ser Thr Ala Ala Leu 130 135 140Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp145 150
155 160Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val
Leu 165 170 175Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr
Val Pro Ser 180 185 190Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys Asn
Val Asp His Lys Pro 195 200 205Ser Asn Thr Lys Val Asp Lys Thr Val
Glu Arg Lys Cys Cys Val Glu 210 215 220Cys Pro Pro Cys Pro Ala Pro
Pro Val Ala Gly Pro Ser Val Phe Leu225 230 235 240Phe Pro Pro Lys
Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu 245 250 255Val Thr
Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Gln 260 265
270Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys
275 280 285Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser
Val Leu 290 295 300Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu
Tyr Lys Cys Lys305 310 315 320Val Ser Asn Lys Gly Leu Pro Ala Pro
Ile Glu Lys Thr Ile Ser Lys 325 330 335Thr Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr Leu Pro Pro Ser 340 345 350Arg Glu Glu Met Thr
Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys 355 360 365Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln 370 375 380Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser Asp Gly385 390
395 400Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp
Gln 405 410 415Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala
Leu His Asn 420 425 430His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro
Gly Lys 435 440 445161445PRTArtificialAn artificially synthesized
peptide sequence 161Gln Val Gln Leu Gln Glu Ser Gly Pro Gly Leu Val
Arg Pro Ser Gln1 5 10 15Thr Leu Ser Leu Thr Cys Thr Val Ser Gly Tyr
Ser Ile Thr Ser Asp 20 25 30His Ala Trp Ser Trp Val Arg Gln Pro Pro
Gly Arg Gly Leu Glu Trp 35 40 45Ile Gly Tyr Ile Ser Tyr Ser Gly Ile
Thr Thr Tyr Asn Pro Ser Leu 50 55 60Lys Ser Arg Val Thr Met Leu Arg
Asp Thr Ser Lys Asn Gln Phe Ser65 70 75 80Leu Arg Leu Ser Ser Val
Thr Ala Ala Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Ser Leu Ala
Arg Thr Thr Ala Met Asp Tyr Trp Gly Gln Gly 100 105 110Ser Leu Val
Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser Val Phe 115 120 125Pro
Leu Ala Pro Ser Ser Lys Ser Thr Ser Glu Ser Thr Ala Ala Leu 130 135
140Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser
Trp145 150 155 160Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe
Pro Ala Val Leu 165 170 175Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser
Val Val Thr Val Pro Ser 180 185 190Ser Asn Phe Gly Thr Gln Thr Tyr
Thr Cys Asn Val Asp His Lys Pro 195 200 205Ser Asn Thr Lys Val Asp
Lys Thr Val Glu Arg Lys Ser Cys Val Glu 210 215 220Cys Pro Pro Cys
Pro Ala Pro Pro Val Ala Gly Pro Ser Val Phe Leu225 230 235 240Phe
Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu 245 250
255Val Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Gln
260 265 270Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys
Thr Lys 275 280 285Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val
Val Ser Val Leu 290 295 300Thr Val Val His Gln Asp Trp Leu Asn Gly
Lys Glu Tyr Lys Cys Lys305 310 315 320Val Ser Asn Lys Gly Leu Pro
Ala Pro Ile Glu Lys Thr Ile Ser Lys 325 330 335Thr Lys Gly Gln Pro
Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser 340 345 350Arg Glu Glu
Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys 355 360 365Gly
Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln 370 375
380Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser Asp
Gly385 390 395 400Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys
Ser Arg Trp Gln 405 410 415Gln Gly Asn Val Phe Ser Cys Ser Val Met
His Glu Ala Leu His Asn 420 425 430His Tyr Thr Gln Lys Ser Leu Ser
Leu Ser Pro Gly Lys 435 440 445162214PRTArtificialAn artificially
synthesized peptide sequence 162Asp Ile Gln Met Thr Gln Ser Pro Ser
Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg
Ala Ser Gln Asp Ile Ser Ser Tyr 20 25 30Leu Asn Trp Tyr Gln Gln Lys
Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Tyr Thr Ser Arg Leu
His Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr
Asp Phe Thr Phe Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Ile
Ala Thr Tyr Tyr Cys Gln Gln Gly Asn Thr Leu Pro Tyr 85 90 95Thr Phe
Gly Gln Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala Ala 100
105 110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser
Gly 115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro
Arg Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln
Ser Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu Gln Asp Ser
Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr Leu Ser
Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala Cys Glu Val
Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200 205Phe Asn
Arg Gly Glu Cys 210163445PRTArtificialAn artificially synthesized
peptide sequence 163Gln Val Gln Leu Gln Glu Ser Gly Pro Gly Leu Val
Arg Pro Ser Gln1 5 10 15Thr Leu Ser Leu Thr Cys Thr Val Ser Gly Tyr
Ser Ile Thr Ser Asp 20 25 30His Ala Trp Ser Trp Val Arg Gln Pro Pro
Gly Arg Gly Leu Glu Trp 35 40 45Ile Gly Tyr Ile Ser Tyr Ser Gly Ile
Thr Thr Tyr Asn Pro Ser Leu 50 55 60Lys Ser Arg Val Thr Met Leu Arg
Asp Thr Ser Lys Asn Gln Phe Ser65 70 75 80Leu Arg Leu Ser Ser Val
Thr Ala Ala Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Ser Leu Ala
Arg Thr Thr Ala Met Asp Tyr Trp Gly Gln Gly 100 105 110Ser Leu Val
Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser Val Phe 115 120 125Pro
Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala Leu 130 135
140Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser
Trp145 150 155 160Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe
Pro Ala Val Leu 165 170 175Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser
Val Val Thr Val Pro Ser 180 185 190Ser Asn Phe Gly Thr Gln Thr Tyr
Thr Cys Asn Val Asp His Lys Pro 195 200 205Ser Asn Thr Lys Val Asp
Lys Thr Val Glu Arg Lys Ser Cys Val Glu 210 215 220Cys Pro Pro Cys
Pro Ala Pro Pro Val Ala Gly Pro Ser Val Phe Leu225 230 235 240Phe
Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu 245 250
255Val Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Gln
260 265 270Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys
Thr Lys 275 280 285Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val
Val Ser Val Leu 290 295 300Thr Val Val His Gln Asp Trp Leu Asn Gly
Lys Glu Tyr Lys Cys Lys305 310 315 320Val Ser Asn Lys Gly Leu Pro
Ser Ser Ile Glu Lys Thr Ile Ser Lys 325 330 335Ala Lys Gly Gln Pro
Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser 340 345 350Arg Glu Glu
Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys 355 360 365Gly
Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln 370 375
380Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser Asp
Gly385 390 395 400Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys
Ser Arg Trp Gln 405 410 415Gln Gly Asn Val Phe Ser Cys Ser Val Met
His Glu Ala Leu His Asn 420 425 430His Tyr Thr Gln Lys Ser Leu Ser
Leu Ser Pro Gly Lys 435 440 445164443PRTArtificialAn artificially
synthesized peptide sequence 164Gln Val Gln Leu Gln Glu Ser Gly Pro
Gly Leu Val Arg Pro Ser Gln1 5 10 15Thr Leu Ser Leu Thr Cys Thr Val
Ser Gly Tyr Ser Ile Thr Ser Asp 20 25 30His Ala Trp Ser Trp Val Arg
Gln Pro Pro Gly Arg Gly Leu Glu Trp 35 40 45Ile Gly Tyr Ile Ser Tyr
Ser Gly Ile Thr Thr Tyr Asn Pro Ser Leu 50 55 60Lys Ser Arg Val Thr
Met Leu Arg Asp Thr Ser Lys Asn Gln Phe Ser65 70 75 80Leu Arg Leu
Ser Ser Val Thr Ala Ala Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg
Ser Leu Ala Arg Thr Thr Ala Met Asp Tyr Trp Gly Gln Gly 100 105
110Ser Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser Val Phe
115 120 125Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala
Ala Leu 130 135 140Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val
Thr Val Ser Trp145 150 155 160Asn Ser Gly Ala Leu Thr Ser Gly Val
His Thr Phe Pro Ala Val Leu 165 170 175Gln Ser Ser Gly Leu Tyr Ser
Leu Ser Ser Val Val Thr Val Pro Ser 180 185 190Ser Asn Phe Gly Thr
Gln Thr Tyr Thr Cys Asn Val Asp His Lys Pro 195 200 205Ser Asn Thr
Lys Val Asp Lys Thr Val Glu Arg Lys Ser Cys Val Glu 210 215 220Cys
Pro Pro Cys Pro Ala Pro Pro Val Ala Gly Pro Ser Val Phe Leu225 230
235 240Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro
Glu 245 250 255Val Thr Cys Val Val Val Asp Val Ser Gln Glu Asp Pro
Glu Val Gln 260 265 270Phe Asn Trp Tyr Val Asp Gly Val Glu Val His
Asn Ala Lys Thr Lys 275 280 285Pro Arg Glu Glu Gln Phe Asn Ser Thr
Phe Arg Val Val Ser Val Leu 290 295 300Thr Val Val His Gln Asp Trp
Leu Asn Gly Lys Glu Tyr Lys Cys Lys305 310 315 320Val Ser Asn Lys
Gly Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys 325 330 335Thr Lys
Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser 340 345
350Gln Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys
355 360 365Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn
Gly Gln 370 375 380Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu
Asp Ser Asp Gly385 390 395 400Ser Phe Phe Leu Tyr Ser Lys Leu Thr
Val Asp Lys Ser Arg Trp Gln 405 410 415Glu Gly Asn Val Phe Ser Cys
Ser Val Met His Glu Ala Leu His Asn 420 425 430His Tyr Thr Gln Lys
Ser Leu Ser Leu Ser Pro 435 440165267PRTArtificialAn artificially
synthesized peptide sequence 165Ala Glu Ser His Leu Ser Leu Leu Tyr
His Leu Thr Ala Val Ser Ser1 5 10 15Pro Ala Pro Gly Thr Pro Ala Phe
Trp Val Ser Gly Trp Leu Gly Pro 20 25 30Gln Gln Tyr Leu Ser Tyr Asn
Ser Leu Arg Gly Glu Ala Glu Pro Cys 35 40 45Gly Ala Trp Val Trp Glu
Asn Gln Val Ser Trp Tyr Trp Glu Lys Glu 50 55 60Thr Thr Asp Leu Arg
Ile Lys Glu Lys Leu Phe Leu Glu Ala Phe Lys65 70 75 80Ala Leu Gly
Gly Lys Gly Pro Tyr Thr Leu Gln Gly Leu Leu Gly Cys 85 90 95Glu Leu
Gly Pro Asp Asn Thr Ser Val Pro Thr Ala Lys Phe Ala Leu 100 105
110Asn Gly Glu Glu Phe Met Asn Phe Asp Leu Lys Gln Gly Thr Trp Gly
115 120 125Gly Asp Trp Pro Glu Ala Leu Ala Ile Ser Gln Arg Trp Gln
Gln Gln 130 135 140Asp Lys Ala Ala Asn Lys Glu Leu Thr Phe Leu Leu
Phe Ser Cys Pro145 150 155 160His Arg Leu Arg Glu His Leu Glu Arg
Gly Arg Gly Asn Leu Glu Trp 165 170 175Lys Glu Pro Pro Ser Met Arg
Leu Lys Ala Arg Pro Ser Ser Pro Gly 180 185 190Phe Ser Val Leu Thr
Cys Ser Ala Phe Ser Phe Tyr Pro Pro Glu Leu 195 200 205Gln Leu Arg
Phe Leu Arg Asn Gly Leu Ala Ala Gly Thr Gly Gln Gly 210 215 220Asp
Phe Gly Pro Asn Ser Asp Gly Ser Phe His Ala Ser Ser Ser Leu225 230
235 240Thr Val Lys Ser Gly Asp Glu His His Tyr Cys Cys Ile Val Gln
His 245 250 255Ala Gly Leu Ala Gln Pro Leu Arg Val Glu Leu 260
26516699PRTArtificialAn artificially synthesized peptide sequence
166Ile Gln Arg Thr Pro Lys Ile Gln Val Tyr Ser Arg His Pro Ala Glu1
5 10 15Asn Gly Lys Ser Asn Phe Leu Asn Cys Tyr Val Ser Gly Phe His
Pro 20 25 30Ser Asp Ile Glu Val Asp Leu Leu Lys Asn Gly Glu Arg Ile
Glu Lys 35 40 45Val Glu His Ser Asp Leu Ser Phe Ser Lys Asp Trp Ser
Phe Tyr Leu 50 55 60Leu Tyr Tyr Thr Glu Phe Thr Pro Thr Glu Lys Asp
Glu Tyr Ala Cys65 70 75 80Arg Val Asn His Val Thr Leu Ser Gln Pro
Lys Ile Val Lys Trp Asp 85 90 95Arg Asp Met167121PRTArtificialAn
artificially synthesized peptide sequence 167Gln Val Gln Leu Val
Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val
Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn
Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu
Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn Gln Gln Phe 50 55 60Gln
Asp Arg Val Thr Ile Thr Ala Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75
80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Asp Tyr Trp
Gly 100 105 110Gln Gly Thr Leu Val Thr Val Ser Ser 115
120168107PRTArtificialAn artificially synthesized peptide sequence
168Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1
5 10 15Asp Arg Val Thr Ile Thr Cys Gln Thr Ser Glu Asp Ile Tyr Ser
Phe 20 25 30Leu Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu
Leu Ile 35 40 45Tyr Asn Ala Gln Thr Glu Ala Gln Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln His
His Tyr Glu Ser Pro Leu 85 90 95Thr Phe Gly Gly Gly Thr Lys Val Glu
Ile Lys 100 10516911PRTArtificialAn artificially synthesized
peptide sequence 169Gln Thr Ser Glu Asp Ile Tyr Ser Phe Leu Ala1 5
101707PRTArtificialAn artificially synthesized peptide sequence
170Asn Ala Gln Thr Glu Ala Gln1 517117PRTArtificialAn artificially
synthesized peptide sequence 171Leu Ile Asn Pro Tyr Asn Gly Gly Thr
Ser Tyr Asn Gln Gln Phe Gln1 5 10 15Asp17217PRTArtificialAn
artificially synthesized peptide sequence 172Leu Ile Asn Pro Tyr
Asn Gly Gly Thr Ser Tyr Asn Asp Gln Phe Gln1 5 10
15Asp1735PRTArtificialAn artificially synthesized peptide sequence
173Gly Tyr Val Met Asn1 51745PRTArtificialAn artificially
synthesized peptide sequence 174Gly Tyr Ile Ile Asn1
51755PRTArtificialAn artificially synthesized peptide sequence
175Gly Tyr Ile Leu Asn1 51765PRTArtificialAn artificially
synthesized peptide sequence 176Gly Tyr Ala Met Asn1
517717PRTArtificialAn artificially synthesized peptide sequence
177Leu Ile Asn Pro Tyr Asn Gly Gly Thr Asp Tyr Asn Gln Lys Phe Lys1
5 10 15Gly17817PRTArtificialAn artificially synthesized peptide
sequence 178Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn Pro Lys
Phe Lys1 5 10 15Gly17912PRTArtificialAn artificially synthesized
peptide sequence 179Asp Gly Leu Asp Asp Gly Pro Tyr Thr Met Asp
Tyr1 5 1018012PRTArtificialAn artificially synthesized peptide
sequence 180Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Leu Asp Tyr1 5
1018112PRTArtificialAn artificially synthesized peptide sequence
181Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Glu Tyr1 5
1018212PRTArtificialAn artificially synthesized peptide sequence
182Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Asp Thr1 5
1018312PRTArtificialAn artificially synthesized peptide sequence
183Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Asp Ser1 5
1018412PRTArtificialAn artificially synthesized peptide sequence
184Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Leu Glu Thr1 5
1018512PRTArtificialAn artificially synthesized peptide sequence
185Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Glu Thr1 5
1018612PRTArtificialAn artificially synthesized peptide sequence
186Asp Gly Leu Asp Asp Gly Pro Tyr Thr Met Glu Thr1 5
1018712PRTArtificialAn artificially synthesized peptide sequence
187Asp Gly Leu Asp Asp Gly Pro Tyr Thr Met Glu Ser1 5
1018811PRTArtificialAn artificially synthesized peptide sequence
188Arg Thr Ser Glu Asn Ile Tyr Arg Phe Leu Ala1 5
1018911PRTArtificialAn artificially synthesized peptide sequence
189Arg Thr Ser Glu Asn Ile Tyr Ser Phe Val Ala1 5
1019011PRTArtificialAn artificially synthesized peptide sequence
190Arg Thr Ser Glu Asn Ile Tyr Arg Phe Val Ala1 5
1019111PRTArtificialAn artificially synthesized peptide sequence
191Arg Ala Ser Glu Asn Ile Tyr Ser Phe Leu Ala1 5
1019211PRTArtificialAn artificially synthesized peptide sequence
192Arg Ser Ser Glu Asn Ile Tyr Ser Phe Leu Ala1 5
101939PRTArtificialAn artificially synthesized peptide sequence
193Gln His His Tyr Asp Ser Pro Leu Thr1 51949PRTArtificialAn
artificially synthesized peptide sequence 194Gln His His Tyr Glu
Asp Pro Leu Thr1 51959PRTArtificialAn artificially synthesized
peptide sequence 195Gln His His Tyr Glu Ser Pro Leu Phe1
51965PRTArtificialAn artificially synthesized peptide sequence
196Gly Tyr Val Leu Asn1 519717PRTArtificialAn artificially
synthesized peptide sequence 197Leu Ile Asn Pro Tyr Asn Gly Gly Thr
Asp Tyr Asn Pro Gln Phe Gln1 5 10 15Asp19817PRTArtificialAn
artificially synthesized peptide sequence 198Leu Ile Asn Pro Tyr
Asn Gly Gly Thr Asp Tyr Asn Pro Gln Phe Gln1 5 10
15Gly19917PRTArtificialAn artificially synthesized peptide sequence
199Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn Gln Gln Phe Gln1
5 10 15Asp20011PRTArtificialAn artificially synthesized peptide
sequence 200Gln Thr Ser Glu Asp Ile Tyr Arg Phe Val Ala1 5
1020111PRTArtificialAn artificially synthesized peptide sequence
201Gln Thr Ser Glu Asp Ile Tyr Ser Phe Val Ala1 5
1020211PRTArtificialAn artificially synthesized peptide sequence
202Gln Ala Ser Glu Asp Ile Tyr Ser Phe Val Ala1 5
1020311PRTArtificialAn artificially synthesized peptide sequence
203Gln Ala Ser Glu Asp Ile Tyr Ser Phe Leu Ala1 5
10204121PRTArtificialAn artificially synthesized peptide sequence
204Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Val Leu Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Asp Tyr Asn
Pro Gln Phe 50 55 60Gln Asp Arg Val Thr Ile Thr Ala Asp Glu Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser 115
120205121PRTArtificialAn artificially synthesized peptide sequence
205Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Asp Tyr Asn
Pro Gln Phe 50 55 60Gln Asp Arg Val Thr Ile Thr Ala Asp Glu Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser 115 120206121PRTArtificialAn artificially synthesized
peptide sequence 206Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Thr Phe Thr Gly Tyr 20 25 30Val Leu Asn Trp Val Arg Gln Ala Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly
Thr Asp Tyr Asn Pro Gln Phe 50 55 60Gln Asp Arg Val Thr Ile Thr Ala
Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu
Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr
Asp Asp Gly Pro Tyr Thr Leu Glu Thr Trp Gly 100 105 110Gln Gly Thr
Leu Val Thr Val Ser Ser 115 120207121PRTArtificialAn artificially
synthesized peptide sequence 207Gln Val Gln Leu Val Gln Ser Gly Ala
Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala
Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Arg Gln
Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr
Asn Gly Gly Thr Asp Tyr Asn Pro Gln Phe 50 55 60Gln Asp Arg Val Thr
Ile Thr Ala Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu
Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg
Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Leu Glu Thr Trp Gly 100 105
110Gln Gly Thr Leu Val Thr Val Ser Ser 115 120208121PRTArtificialAn
artificially synthesized peptide sequence 208Gln Val Gln Leu Val
Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val
Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ala Met Asn
Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu
Ile Asn Pro Tyr Asn Gly Gly Thr Asp Tyr Asn Pro Gln Phe 50 55 60Gln
Asp Arg Val Thr Ile Thr Ala Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75
80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Leu Glu Thr Trp
Gly 100 105 110Gln Gly Thr Leu Val Thr Val Ser Ser 115
120209121PRTArtificialAn artificially synthesized peptide sequence
209Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Val Leu Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Asp Tyr Asn
Pro Gln Phe 50 55 60Gln Asp Arg Val Thr Ile Thr Ala Asp Lys Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Leu Glu Thr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser 115 120210121PRTArtificialAn artificially synthesized
peptide sequence 210Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly
Thr Asp Tyr Asn Pro Gln Phe 50 55 60Gln Asp Arg Val Thr Ile Thr Ala
Asp Lys Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu
Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr
Asp Asp Gly Pro Tyr Thr Leu Glu Thr Trp Gly 100 105 110Gln Gly Thr
Leu Val Thr Val Ser Ser 115 120211121PRTArtificialAn artificially
synthesized peptide sequence 211Gln Val Gln Leu Val Gln Ser Gly Ala
Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala
Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ala Met Asn Trp Val Arg Gln
Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr
Asn Gly Gly Thr Asp Tyr Asn Pro Gln Phe 50 55 60Gln Asp Arg Val Thr
Ile Thr Ala Asp Lys Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu
Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg
Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Leu Glu Thr Trp Gly 100 105
110Gln Gly Thr Leu Val Thr Val Ser Ser 115 120212121PRTArtificialAn
artificially synthesized peptide sequence 212Gln Val Gln Leu Val
Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val
Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn
Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu
Ile Asn Pro Tyr Asn Gly Gly Thr Asp Tyr Asn Pro Gln Phe 50 55 60Gln
Gly Arg Val Thr Ile Thr Ala Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75
80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Leu Glu Thr Trp
Gly 100 105 110Gln Gly Thr Leu Val Thr Val Ser Ser 115
120213121PRTArtificialAn artificially synthesized peptide sequence
213Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Asp Tyr Asn
Pro Gln Phe 50 55 60Gln Gly Arg Val Thr Ile Thr Ala Asp Glu Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Leu Glu Thr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser 115 120214121PRTArtificialAn artificially synthesized
peptide sequence 214Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly
Thr Asp Tyr Asn Pro Gln Phe 50 55 60Gln Gly Arg Val Thr Ile Thr Ala
Asp Lys Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu
Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr
Asp Asp Gly Pro Tyr Thr Leu Glu Thr Trp Gly 100 105 110Gln Gly Thr
Leu Val Thr Val Ser Ser 115 120215121PRTArtificialAn artificially
synthesized peptide sequence 215Gln Val Gln Leu Val Gln Ser Gly Ala
Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala
Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ala Met Asn Trp Val Arg Gln
Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr
Asn Gly Gly Thr Asp Tyr Asn Pro Gln Phe 50 55 60Gln Gly Arg Val Thr
Ile Thr Ala Asp Lys Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu
Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg
Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Leu Glu Thr Trp Gly 100 105
110Gln Gly Thr Leu Val Thr Val Ser Ser 115 120216121PRTArtificialAn
artificially synthesized peptide sequence 216Gln Val Gln Leu Val
Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val
Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn
Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu
Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn Gln Gln Phe 50 55 60Gln
Asp Arg Val Thr Ile Thr Ala Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75
80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Leu Glu Thr Trp
Gly 100 105 110Gln Gly Thr Leu Val Thr Val Ser Ser 115
120217121PRTArtificialAn artificially synthesized peptide sequence
217Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn
Gln Gln Phe 50 55 60Gln Asp Arg Val Thr Ile Thr Ala Asp Lys Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Leu Glu Thr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser 115 120218107PRTArtificialAn artificially synthesized
peptide sequence 218Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser
Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Gln Thr Ser Glu
Asp Ile Tyr Arg Phe 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys
Ala Pro Lys Leu Leu Ile 35 40 45Tyr Asn Ala Gln Thr Glu Ala Gln Gly
Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr
Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr
Tyr Cys Gln His His Tyr Asp Ser Pro Leu 85 90 95Thr Phe Gly Gly Gly
Thr Lys Val Glu Ile Lys 100 105219107PRTArtificialAn artificially
synthesized peptide sequence 219Asp Ile Gln Met Thr Gln Ser Pro Ser
Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Gln
Thr Ser Glu Asp Ile Tyr Ser Phe 20 25 30Val Ala Trp Tyr Gln Gln Lys
Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Asn Ala Gln Thr Glu
Ala Gln Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr
Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe
Ala Thr Tyr Tyr Cys Gln His His Tyr Asp Ser Pro Leu 85 90 95Thr Phe
Gly Gly Gly Thr Lys Val Glu Ile Lys 100 105220107PRTArtificialAn
artificially synthesized peptide sequence 220Asp Ile Gln Met Thr
Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr
Ile Thr Cys Gln Ala Ser Glu Asp Ile Tyr Ser Phe 20 25 30Val Ala Trp
Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Asn
Ala Gln Thr Glu Ala Gln Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser
Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75
80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln His His Tyr Asp Ser Pro Leu
85 90 95Thr Phe Gly Gly Gly Thr Lys Val Glu Ile Lys 100
105221107PRTArtificialAn artificially synthesized peptide sequence
221Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1
5 10 15Asp Arg Val Thr Ile Thr Cys Gln Ala Ser Glu Asp Ile Tyr Ser
Phe 20 25 30Leu Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu
Leu Ile 35 40 45Tyr Asn Ala Gln Thr Glu Ala Gln Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln His
His Tyr Asp Ser Pro Leu 85 90 95Thr Phe Gly Gly Gly Thr Lys Val Glu
Ile Lys 100 105222445PRTArtificialAn artificially synthesized
peptide sequence 222Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Thr Phe Thr Gly Tyr 20 25 30Val Leu Asn Trp Val Arg Gln Ala Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly
Thr Asp Tyr Asn Pro Gln Phe 50 55 60Gln Asp Arg Val Thr Ile Thr Ala
Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu
Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr
Asp Asp Gly Pro Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr
Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val
Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135
140Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr
Val145 150 155 160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His
Thr Phe Pro Ala 165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu
Ser Ser Val Val Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln
Thr Tyr Thr Cys Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys
Val Asp Lys Thr Val Glu Arg Lys Ser Cys 210 215 220Val Glu Cys Pro
Pro Cys Pro Ala Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250
255Pro Glu Val Thr Cys Val Val Val Asp Val Ser Gln Glu Asp Pro Glu
260 265 270Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn
Ala Lys 275 280 285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe
Arg Val Val Ser 290
295 300Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu Tyr
Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro Ile
Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly Gln Pro Arg Glu Pro
Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Gln Glu Glu Met Thr Lys
Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr Pro
Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro Glu
Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser385 390 395 400Asp
Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg 405 410
415Trp Gln Glu Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu
420 425 430His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro 435
440 445223445PRTArtificialAn artificially synthesized peptide
sequence 223Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro
Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe
Thr Gly Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly
Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Asp
Tyr Asn Pro Gln Phe 50 55 60Gln Asp Arg Val Thr Ile Thr Ala Asp Glu
Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser
Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp
Gly Pro Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val
Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro
Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135 140Ala
Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150
155 160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro
Ala 165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val
Val Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln Thr Tyr Thr
Cys Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys
Thr Val Glu Arg Lys Ser Cys 210 215 220Val Glu Cys Pro Pro Cys Pro
Ala Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe Leu Phe Pro
Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250 255Pro Glu
Val Thr Cys Val Val Val Asp Val Ser Gln Glu Asp Pro Glu 260 265
270Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys
275 280 285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val
Val Ser 290 295 300Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly
Lys Glu Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly Leu Pro
Ala Pro Ile Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly Gln Pro
Arg Glu Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Gln Glu Glu
Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375 380Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser385 390
395 400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser
Arg 405 410 415Trp Gln Glu Gly Asn Val Phe Ser Cys Ser Val Met His
Glu Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu
Ser Pro 435 440 445224445PRTArtificialAn artificially synthesized
peptide sequence 224Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Thr Phe Thr Gly Tyr 20 25 30Val Leu Asn Trp Val Arg Gln Ala Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly
Thr Asp Tyr Asn Pro Gln Phe 50 55 60Gln Asp Arg Val Thr Ile Thr Ala
Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu
Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr
Asp Asp Gly Pro Tyr Thr Leu Glu Thr Trp Gly 100 105 110Gln Gly Thr
Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val
Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135
140Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr
Val145 150 155 160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His
Thr Phe Pro Ala 165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu
Ser Ser Val Val Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln
Thr Tyr Thr Cys Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys
Val Asp Lys Thr Val Glu Arg Lys Ser Cys 210 215 220Val Glu Cys Pro
Pro Cys Pro Ala Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250
255Pro Glu Val Thr Cys Val Val Val Asp Val Ser Gln Glu Asp Pro Glu
260 265 270Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn
Ala Lys 275 280 285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe
Arg Val Val Ser 290 295 300Val Leu Thr Val Val His Gln Asp Trp Leu
Asn Gly Lys Glu Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly
Leu Pro Ala Pro Ile Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly
Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Gln
Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val
Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375
380Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp
Ser385 390 395 400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val
Asp Lys Ser Arg 405 410 415Trp Gln Glu Gly Asn Val Phe Ser Cys Ser
Val Met His Glu Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser
Leu Ser Leu Ser Pro 435 440 445225445PRTArtificialAn artificially
synthesized peptide sequence 225Gln Val Gln Leu Val Gln Ser Gly Ala
Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala
Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Arg Gln
Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr
Asn Gly Gly Thr Asp Tyr Asn Pro Gln Phe 50 55 60Gln Asp Arg Val Thr
Ile Thr Ala Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu
Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg
Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Leu Glu Thr Trp Gly 100 105
110Gln Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser
115 120 125Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly
Thr Ala 130 135 140Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu
Pro Val Thr Val145 150 155 160Ser Trp Asn Ser Gly Ala Leu Thr Ser
Gly Val His Thr Phe Pro Ala 165 170 175Val Leu Gln Ser Ser Gly Leu
Tyr Ser Leu Ser Ser Val Val Thr Val 180 185 190Pro Ser Ser Asn Phe
Gly Thr Gln Thr Tyr Thr Cys Asn Val Asp His 195 200 205Lys Pro Ser
Asn Thr Lys Val Asp Lys Thr Val Glu Arg Lys Ser Cys 210 215 220Val
Glu Cys Pro Pro Cys Pro Ala Pro Pro Val Ala Gly Pro Ser Val225 230
235 240Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg
Thr 245 250 255Pro Glu Val Thr Cys Val Val Val Asp Val Ser Gln Glu
Asp Pro Glu 260 265 270Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu
Val His Asn Ala Lys 275 280 285Thr Lys Pro Arg Glu Glu Gln Phe Asn
Ser Thr Phe Arg Val Val Ser 290 295 300Val Leu Thr Val Val His Gln
Asp Trp Leu Asn Gly Lys Glu Tyr Lys305 310 315 320Cys Lys Val Ser
Asn Lys Gly Leu Pro Ala Pro Ile Glu Lys Thr Ile 325 330 335Ser Lys
Thr Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro 340 345
350Pro Ser Gln Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu
355 360 365Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu
Ser Asn 370 375 380Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro
Met Leu Asp Ser385 390 395 400Asp Gly Ser Phe Phe Leu Tyr Ser Lys
Leu Thr Val Asp Lys Ser Arg 405 410 415Trp Gln Glu Gly Asn Val Phe
Ser Cys Ser Val Met His Glu Ala Leu 420 425 430His Asn His Tyr Thr
Gln Lys Ser Leu Ser Leu Ser Pro 435 440 445226445PRTArtificialAn
artificially synthesized peptide sequence 226Gln Val Gln Leu Val
Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val
Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ala Met Asn
Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu
Ile Asn Pro Tyr Asn Gly Gly Thr Asp Tyr Asn Pro Gln Phe 50 55 60Gln
Asp Arg Val Thr Ile Thr Ala Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75
80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Leu Glu Thr Trp
Gly 100 105 110Gln Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys
Gly Pro Ser 115 120 125Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr
Ser Gly Gly Thr Ala 130 135 140Ala Leu Gly Cys Leu Val Lys Asp Tyr
Phe Pro Glu Pro Val Thr Val145 150 155 160Ser Trp Asn Ser Gly Ala
Leu Thr Ser Gly Val His Thr Phe Pro Ala 165 170 175Val Leu Gln Ser
Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val 180 185 190Pro Ser
Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys Asn Val Asp His 195 200
205Lys Pro Ser Asn Thr Lys Val Asp Lys Thr Val Glu Arg Lys Ser Cys
210 215 220Val Glu Cys Pro Pro Cys Pro Ala Pro Pro Val Ala Gly Pro
Ser Val225 230 235 240Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu
Met Ile Ser Arg Thr 245 250 255Pro Glu Val Thr Cys Val Val Val Asp
Val Ser Gln Glu Asp Pro Glu 260 265 270Val Gln Phe Asn Trp Tyr Val
Asp Gly Val Glu Val His Asn Ala Lys 275 280 285Thr Lys Pro Arg Glu
Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser 290 295 300Val Leu Thr
Val Val His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys305 310 315
320Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro Ile Glu Lys Thr Ile
325 330 335Ser Lys Thr Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr
Leu Pro 340 345 350Pro Ser Gln Glu Glu Met Thr Lys Asn Gln Val Ser
Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala
Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro Glu Asn Asn Tyr Lys
Thr Thr Pro Pro Met Leu Asp Ser385 390 395 400Asp Gly Ser Phe Phe
Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg 405 410 415Trp Gln Glu
Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu 420 425 430His
Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro 435 440
445227445PRTArtificialAn artificially synthesized peptide sequence
227Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Val Leu Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Asp Tyr Asn
Pro Gln Phe 50 55 60Gln Asp Arg Val Thr Ile Thr Ala Asp Lys Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Leu Glu Thr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135 140Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150 155
160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala
165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys
Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys Thr
Val Glu Arg Lys Ser Cys 210 215 220Val Glu Cys Pro Pro Cys Pro Ala
Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250 255Pro Glu Val
Thr Cys Val Val Val Asp Val Ser Gln Glu Asp Pro Glu 260 265 270Val
Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 275 280
285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser
290 295 300Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu
Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro
Ile Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Gln Glu Glu Met Thr
Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser385 390 395
400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg
405 410 415Trp Gln Glu Gly Asn Val Phe Ser Cys Ser Val Met His Glu
Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
Pro 435 440 445228445PRTArtificialAn artificially synthesized
peptide sequence 228Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile
Asn Pro Tyr Asn Gly Gly Thr Asp Tyr Asn Pro Gln Phe 50 55 60Gln Asp
Arg Val Thr Ile Thr Ala Asp Lys Ser Thr Ser Thr Ala Tyr65 70 75
80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Leu Glu Thr Trp
Gly 100 105 110Gln Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys
Gly Pro Ser 115 120 125Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr
Ser Gly Gly Thr Ala 130 135 140Ala Leu Gly Cys Leu Val Lys Asp Tyr
Phe Pro Glu Pro Val Thr Val145 150 155 160Ser Trp Asn Ser Gly Ala
Leu Thr Ser Gly Val His Thr Phe Pro Ala 165 170 175Val Leu Gln Ser
Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val 180 185 190Pro Ser
Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys Asn Val Asp His 195 200
205Lys Pro Ser Asn Thr Lys Val Asp Lys Thr Val Glu Arg Lys Ser Cys
210 215 220Val Glu Cys Pro Pro Cys Pro Ala Pro Pro Val Ala Gly Pro
Ser Val225 230 235 240Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu
Met Ile Ser Arg Thr 245 250 255Pro Glu Val Thr Cys Val Val Val Asp
Val Ser Gln Glu Asp Pro Glu 260 265 270Val Gln Phe Asn Trp Tyr Val
Asp Gly Val Glu Val His Asn Ala Lys 275 280 285Thr Lys Pro Arg Glu
Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser 290 295 300Val Leu Thr
Val Val His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys305 310 315
320Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro Ile Glu Lys Thr Ile
325 330 335Ser Lys Thr Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr
Leu Pro 340 345 350Pro Ser Gln Glu Glu Met Thr Lys Asn Gln Val Ser
Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala
Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro Glu Asn Asn Tyr Lys
Thr Thr Pro Pro Met Leu Asp Ser385 390 395 400Asp Gly Ser Phe Phe
Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg 405 410 415Trp Gln Glu
Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu 420 425 430His
Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro 435 440
445229445PRTArtificialAn artificially synthesized peptide sequence
229Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Asp Tyr Asn
Pro Gln Phe 50 55 60Gln Asp Arg Val Thr Ile Thr Ala Asp Lys Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Leu Glu Thr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135 140Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150 155
160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala
165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys
Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys Thr
Val Glu Arg Lys Ser Cys 210 215 220Val Glu Cys Pro Pro Cys Pro Ala
Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250 255Pro Glu Val
Thr Cys Val Val Val Asp Val Ser Gln Glu Asp Pro Glu 260 265 270Val
Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 275 280
285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser
290 295 300Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu
Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro
Ile Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Gln Glu Glu Met Thr
Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser385 390 395
400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg
405 410 415Trp Gln Glu Gly Asn Val Phe Ser Cys Ser Val Met His Glu
Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
Pro 435 440 445230445PRTArtificialAn artificially synthesized
peptide sequence 230Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly
Thr Asp Tyr Asn Pro Gln Phe 50 55 60Gln Gly Arg Val Thr Ile Thr Ala
Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu
Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr
Asp Asp Gly Pro Tyr Thr Leu Glu Thr Trp Gly 100 105 110Gln Gly Thr
Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val
Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135
140Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr
Val145 150 155 160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His
Thr Phe Pro Ala 165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu
Ser Ser Val Val Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln
Thr Tyr Thr Cys Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys
Val Asp Lys Thr Val Glu Arg Lys Ser Cys 210 215 220Val Glu Cys Pro
Pro Cys Pro Ala Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250
255Pro Glu Val Thr Cys Val Val Val Asp Val Ser Gln Glu Asp Pro Glu
260 265 270Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn
Ala Lys 275 280 285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe
Arg Val Val Ser 290 295 300Val Leu Thr Val Val His Gln Asp Trp Leu
Asn Gly Lys Glu Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly
Leu Pro Ala Pro Ile Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly
Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Gln
Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val
Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375
380Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp
Ser385 390 395 400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val
Asp Lys Ser Arg 405 410 415Trp Gln Glu Gly Asn Val Phe Ser Cys Ser
Val Met His Glu Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser
Leu Ser Leu Ser Pro 435 440 445231445PRTArtificialAn artificially
synthesized peptide sequence 231Gln Val Gln Leu Val Gln Ser Gly Ala
Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala
Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ala Met Asn Trp Val Arg Gln
Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr
Asn Gly Gly Thr Asp Tyr Asn Pro Gln Phe 50 55 60Gln Gly Arg Val Thr
Ile Thr Ala Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu
Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg
Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Leu Glu Thr Trp Gly 100 105
110Gln Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser
115 120 125Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly
Thr Ala 130 135 140Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu
Pro Val Thr Val145 150 155 160Ser Trp Asn Ser Gly Ala Leu Thr Ser
Gly Val His Thr Phe Pro Ala 165 170 175Val Leu Gln Ser Ser Gly Leu
Tyr Ser Leu Ser Ser Val Val Thr Val 180 185 190Pro Ser Ser Asn Phe
Gly Thr Gln Thr Tyr Thr Cys Asn Val Asp His 195 200 205Lys Pro Ser
Asn Thr Lys Val Asp Lys Thr Val Glu Arg Lys Ser Cys 210 215 220Val
Glu Cys Pro Pro Cys Pro Ala Pro Pro Val Ala Gly Pro Ser Val225 230
235 240Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg
Thr 245 250 255Pro Glu Val Thr Cys Val Val Val Asp Val Ser Gln Glu
Asp Pro Glu 260 265 270Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu
Val His Asn Ala Lys 275 280 285Thr Lys Pro Arg Glu Glu Gln Phe Asn
Ser Thr Phe Arg Val Val Ser 290 295 300Val Leu Thr Val Val His Gln
Asp Trp Leu Asn Gly Lys Glu Tyr Lys305 310 315 320Cys Lys Val Ser
Asn Lys Gly Leu Pro Ala Pro Ile Glu Lys Thr Ile 325 330 335Ser Lys
Thr Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro 340 345
350Pro Ser Gln Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu
355 360 365Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu
Ser Asn 370 375 380Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro
Met Leu Asp Ser385 390 395 400Asp Gly Ser Phe Phe Leu Tyr Ser Lys
Leu Thr Val Asp Lys Ser Arg 405 410 415Trp Gln Glu Gly Asn Val Phe
Ser Cys Ser Val Met His Glu Ala Leu 420 425 430His Asn His Tyr Thr
Gln Lys Ser Leu Ser Leu Ser Pro 435 440 445232445PRTArtificialAn
artificially synthesized peptide sequence 232Gln Val Gln Leu Val
Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val
Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn
Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu
Ile Asn Pro Tyr Asn Gly Gly Thr Asp Tyr Asn Pro Gln Phe 50 55 60Gln
Gly Arg Val Thr Ile Thr Ala Asp Lys Ser Thr Ser Thr Ala Tyr65 70 75
80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Leu Glu Thr Trp
Gly 100 105 110Gln Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys
Gly Pro Ser 115 120 125Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr
Ser Gly Gly Thr Ala 130 135 140Ala Leu Gly Cys Leu Val Lys Asp Tyr
Phe Pro Glu Pro Val Thr Val145 150 155 160Ser Trp Asn Ser Gly Ala
Leu Thr Ser Gly Val His Thr Phe Pro Ala 165 170 175Val Leu Gln Ser
Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val 180 185 190Pro Ser
Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys Asn Val Asp His 195 200
205Lys Pro Ser Asn Thr Lys Val Asp Lys Thr Val Glu Arg Lys Ser Cys
210 215 220Val Glu Cys Pro Pro Cys Pro Ala Pro Pro Val Ala Gly Pro
Ser Val225 230 235 240Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu
Met Ile Ser Arg Thr 245 250 255Pro Glu Val Thr Cys Val Val Val Asp
Val Ser Gln Glu Asp Pro Glu 260 265 270Val Gln Phe Asn Trp Tyr Val
Asp Gly Val Glu Val His Asn Ala Lys 275 280 285Thr Lys Pro Arg Glu
Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser 290 295 300Val Leu Thr
Val Val His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys305 310 315
320Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro Ile Glu Lys Thr Ile
325 330 335Ser Lys Thr Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr
Leu Pro 340 345 350Pro Ser Gln Glu Glu Met Thr Lys Asn Gln Val Ser
Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala
Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro Glu Asn Asn Tyr Lys
Thr Thr Pro Pro Met Leu Asp Ser385 390 395 400Asp Gly Ser Phe Phe
Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg 405 410 415Trp Gln Glu
Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu 420 425 430His
Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro 435 440
445233445PRTArtificialAn artificially synthesized peptide sequence
233Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Asp Tyr Asn
Pro Gln Phe 50 55 60Gln Gly Arg Val Thr Ile Thr Ala Asp Lys Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Leu Glu Thr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135 140Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150 155
160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala
165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys
Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys Thr
Val Glu Arg Lys Ser Cys 210 215 220Val Glu Cys Pro Pro Cys Pro Ala
Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250 255Pro Glu
Val
Thr Cys Val Val Val Asp Val Ser Gln Glu Asp Pro Glu 260 265 270Val
Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 275 280
285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser
290 295 300Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu
Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro
Ile Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Gln Glu Glu Met Thr
Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser385 390 395
400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg
405 410 415Trp Gln Glu Gly Asn Val Phe Ser Cys Ser Val Met His Glu
Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
Pro 435 440 445234445PRTArtificialAn artificially synthesized
peptide sequence 234Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly
Thr Ser Tyr Asn Gln Gln Phe 50 55 60Gln Asp Arg Val Thr Ile Thr Ala
Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu
Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr
Asp Asp Gly Pro Tyr Thr Leu Glu Thr Trp Gly 100 105 110Gln Gly Thr
Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val
Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135
140Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr
Val145 150 155 160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His
Thr Phe Pro Ala 165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu
Ser Ser Val Val Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln
Thr Tyr Thr Cys Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys
Val Asp Lys Thr Val Glu Arg Lys Ser Cys 210 215 220Val Glu Cys Pro
Pro Cys Pro Ala Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250
255Pro Glu Val Thr Cys Val Val Val Asp Val Ser Gln Glu Asp Pro Glu
260 265 270Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn
Ala Lys 275 280 285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe
Arg Val Val Ser 290 295 300Val Leu Thr Val Val His Gln Asp Trp Leu
Asn Gly Lys Glu Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly
Leu Pro Ala Pro Ile Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly
Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Gln
Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val
Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375
380Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp
Ser385 390 395 400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val
Asp Lys Ser Arg 405 410 415Trp Gln Glu Gly Asn Val Phe Ser Cys Ser
Val Met His Glu Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser
Leu Ser Leu Ser Pro 435 440 445235445PRTArtificialAn artificially
synthesized peptide sequence 235Gln Val Gln Leu Val Gln Ser Gly Ala
Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala
Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Arg Gln
Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr
Asn Gly Gly Thr Ser Tyr Asn Gln Gln Phe 50 55 60Gln Asp Arg Val Thr
Ile Thr Ala Asp Lys Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu
Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg
Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Leu Glu Thr Trp Gly 100 105
110Gln Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser
115 120 125Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly
Thr Ala 130 135 140Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu
Pro Val Thr Val145 150 155 160Ser Trp Asn Ser Gly Ala Leu Thr Ser
Gly Val His Thr Phe Pro Ala 165 170 175Val Leu Gln Ser Ser Gly Leu
Tyr Ser Leu Ser Ser Val Val Thr Val 180 185 190Pro Ser Ser Asn Phe
Gly Thr Gln Thr Tyr Thr Cys Asn Val Asp His 195 200 205Lys Pro Ser
Asn Thr Lys Val Asp Lys Thr Val Glu Arg Lys Ser Cys 210 215 220Val
Glu Cys Pro Pro Cys Pro Ala Pro Pro Val Ala Gly Pro Ser Val225 230
235 240Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg
Thr 245 250 255Pro Glu Val Thr Cys Val Val Val Asp Val Ser Gln Glu
Asp Pro Glu 260 265 270Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu
Val His Asn Ala Lys 275 280 285Thr Lys Pro Arg Glu Glu Gln Phe Asn
Ser Thr Phe Arg Val Val Ser 290 295 300Val Leu Thr Val Val His Gln
Asp Trp Leu Asn Gly Lys Glu Tyr Lys305 310 315 320Cys Lys Val Ser
Asn Lys Gly Leu Pro Ala Pro Ile Glu Lys Thr Ile 325 330 335Ser Lys
Thr Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro 340 345
350Pro Ser Gln Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu
355 360 365Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu
Ser Asn 370 375 380Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro
Met Leu Asp Ser385 390 395 400Asp Gly Ser Phe Phe Leu Tyr Ser Lys
Leu Thr Val Asp Lys Ser Arg 405 410 415Trp Gln Glu Gly Asn Val Phe
Ser Cys Ser Val Met His Glu Ala Leu 420 425 430His Asn His Tyr Thr
Gln Lys Ser Leu Ser Leu Ser Pro 435 440 445236214PRTArtificialAn
artificially synthesized peptide sequence 236Asp Ile Gln Met Thr
Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr
Ile Thr Cys Gln Thr Ser Glu Asp Ile Tyr Arg Phe 20 25 30Val Ala Trp
Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Asn
Ala Gln Thr Glu Ala Gln Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser
Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75
80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln His His Tyr Asp Ser Pro Leu
85 90 95Thr Phe Gly Gly Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala
Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu
Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe
Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn Ala
Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu Gln
Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr
Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala Cys
Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200
205Phe Asn Arg Gly Glu Cys 210237214PRTArtificialAn artificially
synthesized peptide sequence 237Asp Ile Gln Met Thr Gln Ser Pro Ser
Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Gln
Thr Ser Glu Asp Ile Tyr Ser Phe 20 25 30Val Ala Trp Tyr Gln Gln Lys
Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Asn Ala Gln Thr Glu
Ala Gln Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr
Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe
Ala Thr Tyr Tyr Cys Gln His His Tyr Asp Ser Pro Leu 85 90 95Thr Phe
Gly Gly Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala Ala 100 105
110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly
115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg
Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser
Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu Gln Asp Ser Lys
Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys
Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala Cys Glu Val Thr
His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200 205Phe Asn Arg
Gly Glu Cys 210238214PRTArtificialAn artificially synthesized
peptide sequence 238Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser
Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Gln Ala Ser Glu
Asp Ile Tyr Ser Phe 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys
Ala Pro Lys Leu Leu Ile 35 40 45Tyr Asn Ala Gln Thr Glu Ala Gln Gly
Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr
Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr
Tyr Cys Gln His His Tyr Asp Ser Pro Leu 85 90 95Thr Phe Gly Gly Gly
Thr Lys Val Glu Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val
Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr
Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135
140Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser
Gln145 150 155 160Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr
Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr
Glu Lys His Lys Val Tyr 180 185 190Ala Cys Glu Val Thr His Gln Gly
Leu Ser Ser Pro Val Thr Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys
210239214PRTArtificialAn artificially synthesized peptide sequence
239Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1
5 10 15Asp Arg Val Thr Ile Thr Cys Gln Ala Ser Glu Asp Ile Tyr Ser
Phe 20 25 30Leu Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu
Leu Ile 35 40 45Tyr Asn Ala Gln Thr Glu Ala Gln Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln His
His Tyr Asp Ser Pro Leu 85 90 95Thr Phe Gly Gly Gly Thr Lys Val Glu
Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro
Pro Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val
Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln
Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150 155
160Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser
165 170 175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys
Val Tyr 180 185 190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro
Val Thr Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys
210240445PRTArtificialAn artificially synthesized peptide sequence
240Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn
Gln Gln Phe 50 55 60Gln Asp Arg Val Thr Ile Thr Ala Asp Glu Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135 140Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150 155
160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala
165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys
Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys Thr
Val Glu Arg Lys Ser Cys 210 215 220Val Glu Cys Pro Pro Cys Pro Ala
Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250 255Pro Glu Val
Thr Cys Val Val Val Asp Val Ser Gln Glu Asp Pro Glu 260 265 270Val
Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 275 280
285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser
290 295 300Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu
Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro
Ile Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Gln Glu Glu Met Thr
Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser385 390 395
400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg
405 410 415Trp Gln Glu Gly Asn Val Phe Ser Cys Ser Val Met His Glu
Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
Pro 435 440 445241447PRTArtificialAn artificially synthesized
peptide sequence 241Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Arg
Gln Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro
Tyr Asn Gly Gly Thr Ser Tyr Asn Gln Gln Phe 50 55 60Gln Asp Arg Val
Thr Ile Thr Ala Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu
Leu Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala
Arg Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Asp Tyr Trp Gly 100 105
110Gln Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser
115 120 125Val Phe Pro Leu Ala Pro Cys Ser Arg Ser Thr Ser Glu Ser
Thr Ala 130 135 140Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu
Pro Val Thr Val145 150 155 160Ser Trp Asn Ser Gly Ala Leu Thr Ser
Gly Val His Thr Phe Pro Ala 165 170 175Val Leu Gln Ser Ser Gly Leu
Tyr Ser Leu Ser Ser Val Val Thr Val 180 185 190Pro Ser Ser Asn Phe
Gly Thr Gln Thr Tyr Thr Cys Asn Val Asp His 195 200 205Lys Pro Ser
Asn Thr Lys Val Asp Lys Thr Val Glu Arg Lys Cys Cys 210 215 220Val
Glu Cys Pro Pro Cys Pro Ala Pro Pro Val Ala Gly Pro Ser Val225 230
235 240Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg
Thr 245 250 255Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu
Asp Pro Glu 260 265 270Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu
Val His Asn Ala Lys 275 280 285Thr Lys Pro Arg Glu Glu Gln Phe Asn
Ser Thr Phe Arg Val Val Ser 290 295 300Val Leu Thr Val Val His Gln
Asp Trp Leu Asn Gly Lys Glu Tyr Lys305 310 315 320Cys Lys Val Ser
Asn Lys Gly Leu Pro Ala Pro Ile Glu Lys Thr Ile 325 330 335Ser Lys
Thr Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro 340 345
350Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu
355 360 365Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu
Ser Asn 370 375 380Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro
Met Leu Asp Ser385 390 395 400Asp Gly Ser Phe Phe Leu Tyr Ser Lys
Leu Thr Val Asp Lys Ser Arg 405 410 415Trp Gln Gln Gly Asn Val Phe
Ser Cys Ser Val Met His Glu Ala Leu 420 425 430His Asn His Tyr Thr
Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 435 440
445242447PRTArtificialAn artificially synthesized peptide sequence
242Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn
Gln Lys Phe 50 55 60Lys Gly Arg Val Thr Ile Thr Ala Asp Glu Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Leu Glu Thr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Glu Ser Thr Ala 130 135 140Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150 155
160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala
165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys
Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys Thr
Val Glu Arg Lys Ser Cys 210 215 220Val Glu Cys Pro Pro Cys Pro Ala
Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250 255Pro Glu Val
Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu 260 265 270Val
Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 275 280
285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser
290 295 300Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu
Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro
Ile Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Arg Glu Glu Met Thr
Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser385 390 395
400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg
405 410 415Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu
Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
Pro Gly Lys 435 440 445243447PRTArtificialAn artificially
synthesized peptide sequence 243Gln Val Gln Leu Val Gln Ser Gly Ala
Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala
Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Arg Gln
Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr
Asn Gly Gly Thr Ser Tyr Asn Gln Lys Phe 50 55 60Lys Gly Arg Val Thr
Ile Thr Ala Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu
Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg
Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Glu Thr Trp Gly 100 105
110Gln Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser
115 120 125Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Glu Ser
Thr Ala 130 135 140Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu
Pro Val Thr Val145 150 155 160Ser Trp Asn Ser Gly Ala Leu Thr Ser
Gly Val His Thr Phe Pro Ala 165 170 175Val Leu Gln Ser Ser Gly Leu
Tyr Ser Leu Ser Ser Val Val Thr Val 180 185 190Pro Ser Ser Asn Phe
Gly Thr Gln Thr Tyr Thr Cys Asn Val Asp His 195 200 205Lys Pro Ser
Asn Thr Lys Val Asp Lys Thr Val Glu Arg Lys Ser Cys 210 215 220Val
Glu Cys Pro Pro Cys Pro Ala Pro Pro Val Ala Gly Pro Ser Val225 230
235 240Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg
Thr 245 250 255Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu
Asp Pro Glu 260 265 270Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu
Val His Asn Ala Lys 275 280 285Thr Lys Pro Arg Glu Glu Gln Phe Asn
Ser Thr Phe Arg Val Val Ser 290 295 300Val Leu Thr Val Val His Gln
Asp Trp Leu Asn Gly Lys Glu Tyr Lys305 310 315 320Cys Lys Val Ser
Asn Lys Gly Leu Pro Ala Pro Ile Glu Lys Thr Ile 325 330 335Ser Lys
Thr Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro 340 345
350Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu
355 360 365Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu
Ser Asn 370 375 380Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro
Met Leu Asp Ser385 390 395 400Asp Gly Ser Phe Phe Leu Tyr Ser Lys
Leu Thr Val Asp Lys Ser Arg 405 410 415Trp Gln Gln Gly Asn Val Phe
Ser Cys Ser Val Met His Glu Ala Leu 420 425 430His Asn His Tyr Thr
Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 435 440
445244447PRTArtificialAn artificially synthesized peptide sequence
244Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn
Gln Lys Phe 50 55 60Lys Gly Arg Val Thr Ile Thr Ala Asp Glu Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Leu Asp Asp Gly Pro
Tyr Thr Met Glu Thr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Glu Ser Thr Ala 130 135 140Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150 155
160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala
165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys
Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys Thr
Val Glu Arg Lys Ser Cys 210 215 220Val Glu Cys Pro Pro Cys Pro Ala
Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250 255Pro Glu Val
Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu 260 265 270Val
Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 275 280
285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser
290 295 300Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu
Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro
Ile Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Arg Glu Glu Met Thr
Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser385 390 395
400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg
405 410 415Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu
Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
Pro Gly Lys 435 440 445245447PRTArtificialAn artificially
synthesized peptide sequence 245Gln Val Gln Leu Val Gln Ser Gly Ala
Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala
Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Arg Gln
Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr
Asn Gly Gly Thr Ser Tyr Asn Gln Lys Phe 50 55 60Lys Gly Arg Val Thr
Ile Thr Ala Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu
Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg
Asp Gly Leu Asp Asp Gly Pro Tyr Thr Met Glu Ser Trp Gly 100 105
110Gln Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser
115 120 125Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Glu Ser
Thr Ala 130 135 140Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu
Pro Val Thr Val145 150 155 160Ser Trp Asn Ser Gly Ala Leu Thr Ser
Gly Val His Thr Phe Pro Ala 165 170 175Val Leu Gln Ser Ser Gly Leu
Tyr Ser Leu Ser Ser Val Val Thr Val 180 185 190Pro Ser Ser Asn Phe
Gly Thr Gln Thr Tyr Thr Cys Asn Val Asp His 195 200 205Lys Pro Ser
Asn Thr Lys Val Asp Lys Thr Val Glu Arg Lys Ser Cys 210 215 220Val
Glu Cys Pro Pro Cys Pro Ala Pro Pro Val Ala Gly Pro Ser Val225 230
235 240Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg
Thr 245 250 255Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu
Asp Pro Glu 260 265 270Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu
Val His Asn Ala Lys 275 280 285Thr Lys Pro Arg Glu Glu Gln Phe Asn
Ser Thr Phe Arg Val Val Ser 290 295 300Val Leu Thr Val Val His Gln
Asp Trp Leu Asn Gly Lys Glu Tyr Lys305 310 315 320Cys Lys Val Ser
Asn Lys Gly Leu Pro Ala Pro Ile Glu Lys Thr Ile 325 330 335Ser Lys
Thr Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro 340 345
350Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu
355 360 365Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu
Ser Asn 370 375 380Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro
Met Leu Asp Ser385 390 395 400Asp Gly Ser Phe Phe Leu Tyr Ser Lys
Leu Thr Val Asp Lys Ser Arg 405 410 415Trp Gln Gln Gly Asn Val Phe
Ser Cys Ser Val Met His Glu Ala Leu 420 425 430His Asn His Tyr Thr
Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 435 440
445246447PRTArtificialAn artificially synthesized peptide sequence
246Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Val Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn
Gln Lys Phe 50 55 60Lys Gly Arg Val Thr Ile Thr Ala Asp Glu Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Glu Ser Thr Ala 130 135 140Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150 155
160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala
165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys
Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys Thr
Val Glu Arg Lys Ser Cys 210 215 220Val Glu Cys Pro Pro
Cys Pro Ala Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe Leu
Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250
255Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu
260 265 270Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn
Ala Lys 275 280 285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe
Arg Val Val Ser 290 295 300Val Leu Thr Val Val His Gln Asp Trp Leu
Asn Gly Lys Glu Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly
Leu Pro Ala Pro Ile Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly
Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Arg
Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val
Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375
380Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp
Ser385 390 395 400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val
Asp Lys Ser Arg 405 410 415Trp Gln Gln Gly Asn Val Phe Ser Cys Ser
Val Met His Glu Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser
Leu Ser Leu Ser Pro Gly Lys 435 440 445247447PRTArtificialAn
artificially synthesized peptide sequence 247Gln Val Gln Leu Val
Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val
Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ile Ile Asn
Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu
Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn Gln Lys Phe 50 55 60Lys
Gly Arg Val Thr Ile Thr Ala Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75
80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Asp Tyr Trp
Gly 100 105 110Gln Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys
Gly Pro Ser 115 120 125Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr
Ser Glu Ser Thr Ala 130 135 140Ala Leu Gly Cys Leu Val Lys Asp Tyr
Phe Pro Glu Pro Val Thr Val145 150 155 160Ser Trp Asn Ser Gly Ala
Leu Thr Ser Gly Val His Thr Phe Pro Ala 165 170 175Val Leu Gln Ser
Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val 180 185 190Pro Ser
Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys Asn Val Asp His 195 200
205Lys Pro Ser Asn Thr Lys Val Asp Lys Thr Val Glu Arg Lys Ser Cys
210 215 220Val Glu Cys Pro Pro Cys Pro Ala Pro Pro Val Ala Gly Pro
Ser Val225 230 235 240Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu
Met Ile Ser Arg Thr 245 250 255Pro Glu Val Thr Cys Val Val Val Asp
Val Ser His Glu Asp Pro Glu 260 265 270Val Gln Phe Asn Trp Tyr Val
Asp Gly Val Glu Val His Asn Ala Lys 275 280 285Thr Lys Pro Arg Glu
Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser 290 295 300Val Leu Thr
Val Val His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys305 310 315
320Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro Ile Glu Lys Thr Ile
325 330 335Ser Lys Thr Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr
Leu Pro 340 345 350Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser
Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala
Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro Glu Asn Asn Tyr Lys
Thr Thr Pro Pro Met Leu Asp Ser385 390 395 400Asp Gly Ser Phe Phe
Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg 405 410 415Trp Gln Gln
Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu 420 425 430His
Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 435 440
445248447PRTArtificialAn artificially synthesized peptide sequence
248Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Ile Leu Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn
Gln Lys Phe 50 55 60Lys Gly Arg Val Thr Ile Thr Ala Asp Glu Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Glu Ser Thr Ala 130 135 140Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150 155
160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala
165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys
Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys Thr
Val Glu Arg Lys Ser Cys 210 215 220Val Glu Cys Pro Pro Cys Pro Ala
Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250 255Pro Glu Val
Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu 260 265 270Val
Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 275 280
285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser
290 295 300Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu
Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro
Ile Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Arg Glu Glu Met Thr
Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser385 390 395
400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg
405 410 415Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu
Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
Pro Gly Lys 435 440 445249447PRTArtificialAn artificially
synthesized peptide sequence 249Gln Val Gln Leu Val Gln Ser Gly Ala
Glu Val Lys Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala
Ser Gly Tyr Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Arg Gln
Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr
Asn Gly Gly Thr Asp Tyr Asn Gln Lys Phe 50 55 60Lys Gly Arg Val Thr
Ile Thr Ala Asp Glu Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu
Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg
Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Asp Tyr Trp Gly 100 105
110Gln Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser
115 120 125Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Glu Ser
Thr Ala 130 135 140Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu
Pro Val Thr Val145 150 155 160Ser Trp Asn Ser Gly Ala Leu Thr Ser
Gly Val His Thr Phe Pro Ala 165 170 175Val Leu Gln Ser Ser Gly Leu
Tyr Ser Leu Ser Ser Val Val Thr Val 180 185 190Pro Ser Ser Asn Phe
Gly Thr Gln Thr Tyr Thr Cys Asn Val Asp His 195 200 205Lys Pro Ser
Asn Thr Lys Val Asp Lys Thr Val Glu Arg Lys Ser Cys 210 215 220Val
Glu Cys Pro Pro Cys Pro Ala Pro Pro Val Ala Gly Pro Ser Val225 230
235 240Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg
Thr 245 250 255Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu
Asp Pro Glu 260 265 270Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu
Val His Asn Ala Lys 275 280 285Thr Lys Pro Arg Glu Glu Gln Phe Asn
Ser Thr Phe Arg Val Val Ser 290 295 300Val Leu Thr Val Val His Gln
Asp Trp Leu Asn Gly Lys Glu Tyr Lys305 310 315 320Cys Lys Val Ser
Asn Lys Gly Leu Pro Ala Pro Ile Glu Lys Thr Ile 325 330 335Ser Lys
Thr Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro 340 345
350Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu
355 360 365Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu
Ser Asn 370 375 380Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro
Met Leu Asp Ser385 390 395 400Asp Gly Ser Phe Phe Leu Tyr Ser Lys
Leu Thr Val Asp Lys Ser Arg 405 410 415Trp Gln Gln Gly Asn Val Phe
Ser Cys Ser Val Met His Glu Ala Leu 420 425 430His Asn His Tyr Thr
Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 435 440
445250447PRTArtificialAn artificially synthesized peptide sequence
250Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn
Pro Lys Phe 50 55 60Lys Gly Arg Val Thr Ile Thr Ala Asp Glu Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Glu Ser Thr Ala 130 135 140Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150 155
160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala
165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys
Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys Thr
Val Glu Arg Lys Ser Cys 210 215 220Val Glu Cys Pro Pro Cys Pro Ala
Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250 255Pro Glu Val
Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu 260 265 270Val
Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 275 280
285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser
290 295 300Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu
Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro
Ile Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Arg Glu Glu Met Thr
Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser385 390 395
400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg
405 410 415Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu
Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
Pro Gly Lys 435 440 445251214PRTArtificialAn artificially
synthesized peptide sequence 251Asp Ile Gln Met Thr Gln Ser Pro Ser
Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg
Thr Ser Glu Asn Ile Tyr Arg Phe 20 25 30Leu Ala Trp Tyr Gln Gln Lys
Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Asn Ala Lys Thr Leu
Ala Lys Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr
Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe
Ala Thr Tyr Tyr Cys Gln His His Tyr Glu Ser Pro Leu 85 90 95Thr Phe
Gly Gly Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala Ala 100 105
110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly
115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg
Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser
Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu Gln Asp Ser Lys
Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys
Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala Cys Glu Val Thr
His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200 205Phe Asn Arg
Gly Glu Cys 210252214PRTArtificialAn artificially synthesized
peptide sequence 252Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser
Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Thr Ser Glu
Asn Ile Tyr Arg Phe 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys
Ala Pro Lys Leu Leu Ile 35 40 45Tyr Asn Ala Lys Thr Leu Ala Lys Gly
Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr
Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr
Tyr Cys Gln His His Tyr Glu Ser Pro Leu 85 90 95Thr Phe Gly Gly Gly
Thr Lys Val Glu Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val
Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr
Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135
140Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser
Gln145 150 155 160Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr
Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr
Glu Lys His Lys Val Tyr 180 185 190Ala Cys Glu Val Thr His
Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200 205Phe Asn Arg Gly
Glu Cys 210253214PRTArtificialAn artificially synthesized peptide
sequence 253Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser
Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Arg Thr Ser Glu Asn Ile
Tyr Ser Phe 20 25 30Leu Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro
Lys Leu Leu Ile 35 40 45Tyr Asn Ala Lys Thr Leu Ala Lys Gly Val Pro
Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr
Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys
Gln His His Tyr Asp Ser Pro Leu 85 90 95Thr Phe Gly Gly Gly Thr Lys
Val Glu Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile
Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser
Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys
Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150
155 160Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu
Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His
Lys Val Tyr 180 185 190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser
Pro Val Thr Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys
210254214PRTArtificialAn artificially synthesized peptide sequence
254Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly1
5 10 15Asp Arg Val Thr Ile Thr Cys Arg Thr Ser Glu Asn Ile Tyr Ser
Phe 20 25 30Leu Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu
Leu Ile 35 40 45Tyr Asn Ala Lys Thr Leu Ala Lys Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln His
His Tyr Glu Asp Pro Leu 85 90 95Thr Phe Gly Gly Gly Thr Lys Val Glu
Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile Phe Pro
Pro Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser Val Val
Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln
Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150 155
160Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser
165 170 175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys
Val Tyr 180 185 190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro
Val Thr Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys
210255447PRTArtificialAn artificially synthesized peptide sequence
255Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ala1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Gly
Tyr 20 25 30Val Ile Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly Thr Asp Tyr Asn
Gln Lys Phe 50 55 60Lys Gly Arg Val Thr Ile Thr Ala Asp Glu Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr Asp Asp Gly Pro
Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Glu Ser Thr Ala 130 135 140Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val145 150 155
160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala
165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val
Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln Thr Tyr Thr Cys
Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys Val Asp Lys Thr
Val Glu Arg Lys Ser Cys 210 215 220Val Glu Cys Pro Pro Cys Pro Ala
Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe Leu Phe Pro Pro
Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250 255Pro Glu Val
Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu 260 265 270Val
Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys 275 280
285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe Arg Val Val Ser
290 295 300Val Leu Thr Val Val His Gln Asp Trp Leu Asn Gly Lys Glu
Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly Leu Pro Ala Pro
Ile Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Arg Glu Glu Met Thr
Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375 380Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp Ser385 390 395
400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg
405 410 415Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu
Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
Pro Gly Lys 435 440 445256214PRTArtificialAn artificially
synthesized peptide sequence 256Asp Ile Gln Met Thr Gln Ser Pro Ser
Ser Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Gln
Thr Ser Glu Asn Ile Tyr Arg Phe 20 25 30Val Ala Trp Tyr Gln Gln Lys
Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45Tyr Asn Ala Lys Thr Leu
Ala Lys Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr
Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe
Ala Thr Tyr Tyr Cys Gln His His Tyr Asp Ser Pro Leu 85 90 95Thr Phe
Gly Gly Gly Thr Lys Val Glu Ile Lys Arg Thr Val Ala Ala 100 105
110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly
115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg
Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser
Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu Gln Asp Ser Lys
Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr Leu Ser Lys
Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala Cys Glu Val Thr
His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200 205Phe Asn Arg
Gly Glu Cys 210257445PRTArtificialAn artificially synthesized
peptide sequence 257Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ala1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Thr Phe Thr Gly Tyr 20 25 30Ile Met Asn Trp Val Arg Gln Ala Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Leu Ile Asn Pro Tyr Asn Gly Gly
Thr Ser Tyr Asn Gln Lys Phe 50 55 60Lys Gly Arg Val Thr Ile Thr Ala
Asp Lys Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu
Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asp Gly Tyr
Asp Asp Gly Pro Tyr Thr Met Asp Tyr Trp Gly 100 105 110Gln Gly Thr
Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125Val
Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135
140Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr
Val145 150 155 160Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His
Thr Phe Pro Ala 165 170 175Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu
Ser Ser Val Val Thr Val 180 185 190Pro Ser Ser Asn Phe Gly Thr Gln
Thr Tyr Thr Cys Asn Val Asp His 195 200 205Lys Pro Ser Asn Thr Lys
Val Asp Lys Thr Val Glu Arg Lys Ser Cys 210 215 220Val Glu Cys Pro
Pro Cys Pro Ala Pro Pro Val Ala Gly Pro Ser Val225 230 235 240Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr 245 250
255Pro Glu Val Thr Cys Val Val Val Asp Val Ser Gln Glu Asp Pro Glu
260 265 270Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn
Ala Lys 275 280 285Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Phe
Arg Val Val Ser 290 295 300Val Leu Thr Val Val His Gln Asp Trp Leu
Asn Gly Lys Glu Tyr Lys305 310 315 320Cys Lys Val Ser Asn Lys Gly
Leu Pro Ala Pro Ile Glu Lys Thr Ile 325 330 335Ser Lys Thr Lys Gly
Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro 340 345 350Pro Ser Gln
Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu 355 360 365Val
Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn 370 375
380Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Met Leu Asp
Ser385 390 395 400Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val
Asp Lys Ser Arg 405 410 415Trp Gln Glu Gly Asn Val Phe Ser Cys Ser
Val Met His Glu Ala Leu 420 425 430His Asn His Tyr Thr Gln Lys Ser
Leu Ser Leu Ser Pro 435 440 445258530PRTHomo sapiens 258Met Met Trp
Thr Trp Ala Leu Trp Met Leu Pro Ser Leu Cys Lys Phe1 5 10 15Ser Leu
Ala Ala Leu Pro Ala Lys Pro Glu Asn Ile Ser Cys Val Tyr 20 25 30Tyr
Tyr Arg Lys Asn Leu Thr Cys Thr Trp Ser Pro Gly Lys Glu Thr 35 40
45Ser Tyr Thr Gln Tyr Thr Val Lys Arg Thr Tyr Ala Phe Gly Glu Lys
50 55 60His Asp Asn Cys Thr Thr Asn Ser Ser Thr Ser Glu Asn Arg Ala
Ser65 70 75 80Cys Ser Phe Phe Leu Pro Arg Ile Thr Ile Pro Asp Asn
Tyr Thr Ile 85 90 95Glu Val Glu Ala Glu Asn Gly Asp Gly Val Ile Lys
Ser His Met Thr 100 105 110Tyr Trp Arg Leu Glu Asn Ile Ala Lys Thr
Glu Pro Pro Lys Ile Phe 115 120 125Arg Val Lys Pro Val Leu Gly Ile
Lys Arg Met Ile Gln Ile Glu Trp 130 135 140Ile Lys Pro Glu Leu Ala
Pro Val Ser Ser Asp Leu Lys Tyr Thr Leu145 150 155 160Arg Phe Arg
Thr Val Asn Ser Thr Ser Trp Met Glu Val Asn Phe Ala 165 170 175Lys
Asn Arg Lys Asp Lys Asn Gln Thr Tyr Asn Leu Thr Gly Leu Gln 180 185
190Pro Phe Thr Glu Tyr Val Ile Ala Leu Arg Cys Ala Val Lys Glu Ser
195 200 205Lys Phe Trp Ser Asp Trp Ser Gln Glu Lys Met Gly Met Thr
Glu Glu 210 215 220Glu Ala Pro Cys Gly Leu Glu Leu Trp Arg Val Leu
Lys Pro Ala Glu225 230 235 240Ala Asp Gly Arg Arg Pro Val Arg Leu
Leu Trp Lys Lys Ala Arg Gly 245 250 255Ala Pro Val Leu Glu Lys Thr
Leu Gly Tyr Asn Ile Trp Tyr Tyr Pro 260 265 270Glu Ser Asn Thr Asn
Leu Thr Glu Thr Met Asn Thr Thr Asn Gln Gln 275 280 285Leu Glu Leu
His Leu Gly Gly Glu Ser Phe Trp Val Ser Met Ile Ser 290 295 300Tyr
Asn Ser Leu Gly Lys Ser Pro Val Ala Thr Leu Arg Ile Pro Ala305 310
315 320Ile Gln Glu Lys Ser Phe Gln Cys Ile Glu Val Met Gln Ala Cys
Val 325 330 335Ala Glu Asp Gln Leu Val Val Lys Trp Gln Ser Ser Ala
Leu Asp Val 340 345 350Asn Thr Trp Met Ile Glu Trp Phe Pro Asp Val
Asp Ser Glu Pro Thr 355 360 365Thr Leu Ser Trp Glu Ser Val Ser Gln
Ala Thr Asn Trp Thr Ile Gln 370 375 380Gln Asp Lys Leu Lys Pro Phe
Trp Cys Tyr Asn Ile Ser Val Tyr Pro385 390 395 400Met Leu His Asp
Lys Val Gly Glu Pro Tyr Ser Ile Gln Ala Tyr Ala 405 410 415Lys Glu
Gly Val Pro Ser Glu Gly Pro Glu Thr Lys Val Glu Asn Ile 420 425
430Gly Val Lys Thr Val Thr Ile Thr Trp Lys Glu Ile Pro Lys Ser Glu
435 440 445Arg Lys Gly Ile Ile Cys Asn Tyr Thr Ile Phe Tyr Gln Ala
Glu Gly 450 455 460Gly Lys Gly Phe Ser Lys Thr Val Asn Ser Ser Ile
Leu Gln Tyr Gly465 470 475 480Leu Glu Ser Leu Lys Arg Lys Thr Ser
Tyr Ile Val Gln Val Met Ala 485 490 495Ser Thr Ser Ala Gly Gly Thr
Asn Gly Thr Ser Ile Asn Phe Lys Thr 500 505 510Leu Ser His His His
His His His Glu Gln Lys Leu Ile Ser Glu Glu 515 520 525Asp Leu
530259517PRTMus musculus 259Met Trp Thr Leu Ala Leu Trp Ala Phe Ser
Phe Leu Cys Lys Phe Ser1 5 10 15Leu Ala Val Leu Pro Thr Lys Pro Glu
Asn Ile Ser Cys Val Phe Tyr 20 25 30Phe Asp Arg Asn Leu Thr Cys Thr
Trp Arg Pro Glu Lys Glu Thr Asn 35 40 45Asp Thr Ser Tyr Ile Val Thr
Leu Thr Tyr Ser Tyr Gly Lys Ser Asn 50 55 60Tyr Ser Asp Asn Ala Thr
Glu Ala Ser Tyr Ser Phe Pro Arg Ser Cys65 70 75 80Ala Met Pro Pro
Asp Ile Cys Ser Val Glu Val Gln Ala Gln Asn Gly 85 90 95Asp Gly Lys
Val Lys Ser Asp Ile Thr Tyr Trp His Leu Ile Ser Ile 100 105 110Ala
Lys Thr Glu Pro Pro Ile Ile Leu Ser Val Asn Pro Ile Cys Asn 115 120
125Arg Met Phe Gln Ile Gln Trp Lys Pro Arg Glu Lys Thr Arg Gly Phe
130 135 140Pro Leu Val Cys Met Leu Arg Phe Arg Thr Val Asn Ser Ser
Arg Trp145 150 155 160Thr Glu Val Asn Phe Glu Asn Cys Lys Gln Val
Cys Asn Leu Thr Gly 165 170 175Leu Gln Ala Phe Thr Glu Tyr Val Leu
Ala Leu Arg Phe Arg Phe Asn 180 185 190Asp Ser Arg Tyr Trp Ser Lys
Trp Ser Lys Glu Glu Thr Arg Val Thr 195 200 205Met Glu Glu Val Pro
His Val Leu Asp Leu Trp Arg Ile Leu Glu Pro 210 215 220Ala Asp Met
Asn Gly Asp Arg Lys Val Arg Leu Leu Trp Lys Lys Ala225 230 235
240Arg Gly Ala Pro Val Leu Glu Lys Thr Phe Gly Tyr His Ile Gln Tyr
245 250 255Phe Ala Glu Asn Ser Thr Asn Leu Thr Glu Ile Asn Asn Ile
Thr Thr 260 265 270Gln Gln Tyr Glu Leu Leu Leu Met Ser Gln Ala His
Ser Val Ser Val 275 280 285Thr Ser Phe Asn Ser Leu Gly Lys Ser Gln
Glu Thr Ile Leu Arg Ile 290 295 300Pro Asp Val His Glu Lys Thr Phe
Gln Tyr Ile Lys Ser Met Gln Ala305 310 315 320Tyr Ile Ala Glu Pro
Leu Leu Val Val Asn Trp Gln Ser Ser Ile Pro 325 330 335Ala Val Asp
Thr Trp Ile Val Glu Trp Leu Pro Glu Ala Ala Met Ser 340 345
350Lys Phe Pro Ala Leu Ser Trp Glu Ser Val Ser Gln Val Thr Asn Trp
355 360 365Thr Ile Glu Gln Asp Lys Leu Lys Pro Phe Thr Cys Tyr Asn
Ile Ser 370 375 380Val Tyr Pro Val Leu Gly His Arg Val Gly Glu Pro
Tyr Ser Ile Gln385 390 395 400Ala Tyr Ala Lys Glu Gly Thr Pro Leu
Lys Gly Pro Glu Thr Arg Val 405 410 415Glu Asn Ile Gly Leu Arg Thr
Ala Thr Ile Thr Trp Lys Glu Ile Pro 420 425 430Lys Ser Ala Arg Asn
Gly Phe Ile Asn Asn Tyr Thr Val Phe Tyr Gln 435 440 445Ala Glu Gly
Gly Lys Glu Leu Ser Lys Thr Val Asn Ser His Ala Leu 450 455 460Gln
Cys Asp Leu Glu Ser Leu Thr Arg Arg Thr Ser Tyr Thr Val Trp465 470
475 480Val Met Ala Ser Thr Arg Ala Gly Gly Thr Asn Gly Val Arg Ile
Asn 485 490 495Phe Lys Thr Leu Ser His His His His His His Glu Gln
Lys Leu Ile 500 505 510Ser Glu Glu Asp Leu 515260531PRTArtificialAn
artificially synthesized peptide sequence 260Met Met Trp Thr Trp
Ala Leu Trp Met Leu Pro Ser Leu Cys Lys Phe1 5 10 15Ser Leu Ala Ala
Leu Pro Ala Lys Pro Glu Asn Ile Ser Cys Val Tyr 20 25 30Tyr Tyr Arg
Lys Asn Leu Thr Cys Thr Trp Ser Pro Gly Lys Glu Thr 35 40 45Ser Tyr
Thr Gln Tyr Thr Val Lys Arg Thr Tyr Ala Phe Gly Glu Lys 50 55 60His
Asp Asn Cys Thr Thr Asn Ser Ser Thr Ser Glu Asn Arg Ala Ser65 70 75
80Cys Ser Phe Phe Leu Pro Arg Ile Thr Ile Pro Asp Asn Tyr Thr Ile
85 90 95Glu Val Glu Ala Glu Asn Gly Asp Gly Val Ile Lys Ser His Met
Thr 100 105 110Tyr Trp Arg Leu Glu Asn Ile Ala Lys Thr Glu Pro Pro
Lys Ile Phe 115 120 125Arg Val Lys Pro Val Leu Gly Ile Lys Arg Met
Ile Gln Ile Glu Trp 130 135 140Ile Lys Pro Glu Leu Ala Pro Val Ser
Ser Asp Leu Lys Tyr Thr Leu145 150 155 160Arg Phe Arg Thr Val Asn
Ser Thr Ser Trp Met Glu Val Asn Phe Ala 165 170 175Lys Asn Arg Lys
Asp Lys Asn Gln Thr Tyr Asn Leu Thr Gly Leu Gln 180 185 190Pro Phe
Thr Glu Tyr Val Ile Ala Leu Arg Cys Ala Val Lys Glu Ser 195 200
205Lys Phe Trp Ser Asp Trp Ser Gln Glu Lys Met Gly Met Thr Glu Glu
210 215 220Glu Ala Pro His Val Leu Asp Leu Trp Arg Ile Leu Glu Pro
Ala Asp225 230 235 240Met Asn Gly Asp Arg Lys Val Arg Leu Leu Trp
Lys Lys Ala Arg Gly 245 250 255Ala Pro Val Leu Glu Lys Thr Phe Gly
Tyr His Ile Gln Tyr Phe Ala 260 265 270Glu Asn Ser Thr Asn Leu Thr
Glu Ile Asn Asn Ile Thr Thr Gln Gln 275 280 285Tyr Glu Leu Leu Leu
Met Ser Gln Ala His Ser Val Ser Val Thr Ser 290 295 300Phe Asn Ser
Leu Gly Lys Ser Gln Glu Thr Ile Leu Arg Ile Pro Asp305 310 315
320Val His Glu Lys Thr Phe Gln Tyr Ile Lys Ser Met Gln Ala Tyr Ile
325 330 335Ala Glu Pro Leu Leu Val Val Asn Trp Gln Ser Ser Ile Pro
Ala Val 340 345 350Asp Thr Trp Ile Val Glu Trp Leu Pro Glu Ala Ala
Met Ser Lys Phe 355 360 365Pro Ala Leu Ser Trp Glu Ser Val Ser Gln
Val Thr Asn Trp Thr Ile 370 375 380Glu Gln Asp Lys Leu Lys Pro Phe
Thr Cys Tyr Asn Ile Ser Val Tyr385 390 395 400Pro Val Leu Gly His
Arg Val Gly Glu Pro Tyr Ser Ile Gln Ala Tyr 405 410 415Ala Lys Glu
Gly Thr Pro Leu Lys Gly Pro Glu Thr Arg Val Glu Asn 420 425 430Ile
Gly Leu Arg Thr Ala Thr Ile Thr Trp Lys Glu Ile Pro Lys Ser 435 440
445Ala Arg Asn Gly Phe Ile Asn Asn Tyr Thr Val Phe Tyr Gln Ala Glu
450 455 460Gly Gly Lys Glu Leu Ser Lys Thr Val Asn Ser His Ala Leu
Gln Cys465 470 475 480Asp Leu Glu Ser Leu Thr Arg Arg Thr Ser Tyr
Thr Val Trp Val Met 485 490 495Ala Ser Thr Arg Ala Gly Gly Thr Asn
Gly Val Arg Ile Asn Phe Lys 500 505 510Thr Leu Ser His His His His
His His Glu Gln Lys Leu Ile Ser Glu 515 520 525Glu Asp Leu
530261516PRTArtificialAn artificially synthesized peptide sequence
261Met Trp Thr Leu Ala Leu Trp Ala Phe Ser Phe Leu Cys Lys Phe Ser1
5 10 15Leu Ala Val Leu Pro Thr Lys Pro Glu Asn Ile Ser Cys Val Phe
Tyr 20 25 30Phe Asp Arg Asn Leu Thr Cys Thr Trp Arg Pro Glu Lys Glu
Thr Asn 35 40 45Asp Thr Ser Tyr Ile Val Thr Leu Thr Tyr Ser Tyr Gly
Lys Ser Asn 50 55 60Tyr Ser Asp Asn Ala Thr Glu Ala Ser Tyr Ser Phe
Pro Arg Ser Cys65 70 75 80Ala Met Pro Pro Asp Ile Cys Ser Val Glu
Val Gln Ala Gln Asn Gly 85 90 95Asp Gly Lys Val Lys Ser Asp Ile Thr
Tyr Trp His Leu Ile Ser Ile 100 105 110Ala Lys Thr Glu Pro Pro Ile
Ile Leu Ser Val Asn Pro Ile Cys Asn 115 120 125Arg Met Phe Gln Ile
Gln Trp Lys Pro Arg Glu Lys Thr Arg Gly Phe 130 135 140Pro Leu Val
Cys Met Leu Arg Phe Arg Thr Val Asn Ser Ser Arg Trp145 150 155
160Thr Glu Val Asn Phe Glu Asn Cys Lys Gln Val Cys Asn Leu Thr Gly
165 170 175Leu Gln Ala Phe Thr Glu Tyr Val Leu Ala Leu Arg Phe Arg
Phe Asn 180 185 190Asp Ser Arg Tyr Trp Ser Lys Trp Ser Lys Glu Glu
Thr Arg Val Thr 195 200 205Met Glu Glu Val Pro Cys Gly Leu Glu Leu
Trp Arg Val Leu Lys Pro 210 215 220Ala Glu Ala Asp Gly Arg Arg Pro
Val Arg Leu Leu Trp Lys Lys Ala225 230 235 240Arg Gly Ala Pro Val
Leu Glu Lys Thr Leu Gly Tyr Asn Ile Trp Tyr 245 250 255Tyr Pro Glu
Ser Asn Thr Asn Leu Thr Glu Thr Met Asn Thr Thr Asn 260 265 270Gln
Gln Leu Glu Leu His Leu Gly Gly Glu Ser Phe Trp Val Ser Met 275 280
285Ile Ser Tyr Asn Ser Leu Gly Lys Ser Pro Val Ala Thr Leu Arg Ile
290 295 300Pro Ala Ile Gln Glu Lys Ser Phe Gln Cys Ile Glu Val Met
Gln Ala305 310 315 320Cys Val Ala Glu Asp Gln Leu Val Val Lys Trp
Gln Ser Ser Ala Leu 325 330 335Asp Val Asn Thr Trp Met Ile Glu Trp
Phe Pro Asp Val Asp Ser Glu 340 345 350Pro Thr Thr Leu Ser Trp Glu
Ser Val Ser Gln Ala Thr Asn Trp Thr 355 360 365Ile Gln Gln Asp Lys
Leu Lys Pro Phe Trp Cys Tyr Asn Ile Ser Val 370 375 380Tyr Pro Met
Leu His Asp Lys Val Gly Glu Pro Tyr Ser Ile Gln Ala385 390 395
400Tyr Ala Lys Glu Gly Val Pro Ser Glu Gly Pro Glu Thr Lys Val Glu
405 410 415Asn Ile Gly Val Lys Thr Val Thr Ile Thr Trp Lys Glu Ile
Pro Lys 420 425 430Ser Glu Arg Lys Gly Ile Ile Cys Asn Tyr Thr Ile
Phe Tyr Gln Ala 435 440 445Glu Gly Gly Lys Gly Phe Ser Lys Thr Val
Asn Ser Ser Ile Leu Gln 450 455 460Tyr Gly Leu Glu Ser Leu Lys Arg
Lys Thr Ser Tyr Ile Val Gln Val465 470 475 480Met Ala Ser Thr Ser
Ala Gly Gly Thr Asn Gly Thr Ser Ile Asn Phe 485 490 495Lys Thr Leu
Ser His His His His His His Glu Gln Lys Leu Ile Ser 500 505 510Glu
Glu Asp Leu 515262524PRTArtificialAn artificially synthesized
peptide sequence 262Met Met Trp Thr Trp Ala Leu Trp Met Leu Pro Ser
Leu Cys Lys Phe1 5 10 15Ser Leu Ala Ala Leu Pro Ala Lys Pro Glu Asn
Ile Ser Cys Val Tyr 20 25 30Tyr Tyr Arg Lys Asn Leu Thr Cys Thr Trp
Ser Pro Gly Lys Glu Thr 35 40 45Ser Tyr Thr Gln Tyr Thr Val Lys Arg
Thr Tyr Ala Phe Gly Glu Lys 50 55 60His Asp Asn Cys Thr Thr Asn Ser
Ser Thr Ser Glu Asn Arg Ala Ser65 70 75 80Cys Ser Phe Phe Leu Pro
Arg Ile Thr Ile Pro Asp Asn Tyr Thr Ile 85 90 95Glu Val Glu Ala Glu
Asn Gly Asp Gly Val Ile Lys Ser His Met Thr 100 105 110Tyr Trp Arg
Leu Glu Asn Ile Ala Lys Thr Glu Pro Pro Ile Ile Leu 115 120 125Ser
Val Asn Pro Ile Cys Asn Arg Met Phe Gln Ile Gln Trp Lys Pro 130 135
140Arg Glu Lys Thr Arg Gly Phe Pro Leu Val Cys Met Leu Arg Phe
Arg145 150 155 160Thr Val Asn Ser Ser Arg Trp Thr Glu Val Asn Phe
Glu Asn Cys Lys 165 170 175Gln Val Cys Asn Leu Thr Gly Leu Gln Ala
Phe Thr Glu Tyr Val Leu 180 185 190Ala Leu Arg Phe Arg Phe Asn Asp
Ser Arg Tyr Trp Ser Lys Trp Ser 195 200 205Lys Glu Glu Thr Arg Val
Thr Met Glu Glu Val Pro His Val Leu Asp 210 215 220Leu Trp Arg Ile
Leu Glu Pro Ala Asp Met Asn Gly Asp Arg Lys Val225 230 235 240Arg
Leu Leu Trp Lys Lys Ala Arg Gly Ala Pro Val Leu Glu Lys Thr 245 250
255Phe Gly Tyr His Ile Gln Tyr Phe Ala Glu Asn Ser Thr Asn Leu Thr
260 265 270Glu Ile Asn Asn Ile Thr Thr Gln Gln Tyr Glu Leu Leu Leu
Met Ser 275 280 285Gln Ala His Ser Val Ser Val Thr Ser Phe Asn Ser
Leu Gly Lys Ser 290 295 300Gln Glu Thr Ile Leu Arg Ile Pro Asp Val
His Glu Lys Thr Phe Gln305 310 315 320Tyr Ile Lys Ser Met Gln Ala
Tyr Ile Ala Glu Pro Leu Leu Val Val 325 330 335Asn Trp Gln Ser Ser
Ile Pro Ala Val Asp Thr Trp Ile Val Glu Trp 340 345 350Leu Pro Glu
Ala Ala Met Ser Lys Phe Pro Ala Leu Ser Trp Glu Ser 355 360 365Val
Ser Gln Val Thr Asn Trp Thr Ile Glu Gln Asp Lys Leu Lys Pro 370 375
380Phe Thr Cys Tyr Asn Ile Ser Val Tyr Pro Val Leu Gly His Arg
Val385 390 395 400Gly Glu Pro Tyr Ser Ile Gln Ala Tyr Ala Lys Glu
Gly Thr Pro Leu 405 410 415Lys Gly Pro Glu Thr Arg Val Glu Asn Ile
Gly Leu Arg Thr Ala Thr 420 425 430Ile Thr Trp Lys Glu Ile Pro Lys
Ser Ala Arg Asn Gly Phe Ile Asn 435 440 445Asn Tyr Thr Val Phe Tyr
Gln Ala Glu Gly Gly Lys Glu Leu Ser Lys 450 455 460Thr Val Asn Ser
His Ala Leu Gln Cys Asp Leu Glu Ser Leu Thr Arg465 470 475 480Arg
Thr Ser Tyr Thr Val Trp Val Met Ala Ser Thr Arg Ala Gly Gly 485 490
495Thr Asn Gly Val Arg Ile Asn Phe Lys Thr Leu Ser His His His His
500 505 510His His Glu Gln Lys Leu Ile Ser Glu Glu Asp Leu 515
520263524PRTArtificialAn artificially synthesized peptide sequence
263Met Trp Thr Leu Ala Leu Trp Ala Phe Ser Phe Leu Cys Lys Phe Ser1
5 10 15Leu Ala Val Leu Pro Thr Lys Pro Glu Asn Ile Ser Cys Val Phe
Tyr 20 25 30Phe Asp Arg Asn Leu Thr Cys Thr Trp Arg Pro Glu Lys Glu
Thr Asn 35 40 45Asp Thr Ser Tyr Ile Val Thr Leu Thr Tyr Ser Tyr Gly
Lys Ser Asn 50 55 60Tyr Ser Asp Asn Ala Thr Glu Ala Ser Tyr Ser Phe
Pro Arg Ser Cys65 70 75 80Ala Met Pro Pro Asp Ile Cys Ser Val Glu
Val Gln Ala Gln Asn Gly 85 90 95Asp Gly Lys Val Lys Ser Asp Ile Thr
Tyr Trp His Leu Ile Ser Ile 100 105 110Ala Lys Thr Glu Pro Pro Lys
Ile Phe Arg Val Lys Pro Val Leu Gly 115 120 125Ile Lys Arg Met Ile
Gln Ile Glu Trp Ile Lys Pro Glu Leu Ala Pro 130 135 140Val Ser Ser
Asp Leu Lys Tyr Thr Leu Arg Phe Arg Thr Val Asn Ser145 150 155
160Thr Ser Trp Met Glu Val Asn Phe Ala Lys Asn Arg Lys Asp Lys Asn
165 170 175Gln Thr Tyr Asn Leu Thr Gly Leu Gln Pro Phe Thr Glu Tyr
Val Ile 180 185 190Ala Leu Arg Cys Ala Val Lys Glu Ser Lys Phe Trp
Ser Asp Trp Ser 195 200 205Gln Glu Lys Met Gly Met Thr Glu Glu Glu
Ala Pro His Val Leu Asp 210 215 220Leu Trp Arg Ile Leu Glu Pro Ala
Asp Met Asn Gly Asp Arg Lys Val225 230 235 240Arg Leu Leu Trp Lys
Lys Ala Arg Gly Ala Pro Val Leu Glu Lys Thr 245 250 255Phe Gly Tyr
His Ile Gln Tyr Phe Ala Glu Asn Ser Thr Asn Leu Thr 260 265 270Glu
Ile Asn Asn Ile Thr Thr Gln Gln Tyr Glu Leu Leu Leu Met Ser 275 280
285Gln Ala His Ser Val Ser Val Thr Ser Phe Asn Ser Leu Gly Lys Ser
290 295 300Gln Glu Thr Ile Leu Arg Ile Pro Asp Val His Glu Lys Thr
Phe Gln305 310 315 320Tyr Ile Lys Ser Met Gln Ala Tyr Ile Ala Glu
Pro Leu Leu Val Val 325 330 335Asn Trp Gln Ser Ser Ile Pro Ala Val
Asp Thr Trp Ile Val Glu Trp 340 345 350Leu Pro Glu Ala Ala Met Ser
Lys Phe Pro Ala Leu Ser Trp Glu Ser 355 360 365Val Ser Gln Val Thr
Asn Trp Thr Ile Glu Gln Asp Lys Leu Lys Pro 370 375 380Phe Thr Cys
Tyr Asn Ile Ser Val Tyr Pro Val Leu Gly His Arg Val385 390 395
400Gly Glu Pro Tyr Ser Ile Gln Ala Tyr Ala Lys Glu Gly Thr Pro Leu
405 410 415Lys Gly Pro Glu Thr Arg Val Glu Asn Ile Gly Leu Arg Thr
Ala Thr 420 425 430Ile Thr Trp Lys Glu Ile Pro Lys Ser Ala Arg Asn
Gly Phe Ile Asn 435 440 445Asn Tyr Thr Val Phe Tyr Gln Ala Glu Gly
Gly Lys Glu Leu Ser Lys 450 455 460Thr Val Asn Ser His Ala Leu Gln
Cys Asp Leu Glu Ser Leu Thr Arg465 470 475 480Arg Thr Ser Tyr Thr
Val Trp Val Met Ala Ser Thr Arg Ala Gly Gly 485 490 495Thr Asn Gly
Val Arg Ile Asn Phe Lys Thr Leu Ser His His His His 500 505 510His
His Glu Gln Lys Leu Ile Ser Glu Glu Asp Leu 515
520264523PRTArtificialAn artificially synthesized peptide sequence
264Met Trp Thr Leu Ala Leu Trp Ala Phe Ser Phe Leu Cys Lys Phe Ser1
5 10 15Leu Ala Val Leu Pro Thr Lys Pro Glu Asn Ile Ser Cys Val Phe
Tyr 20 25 30Phe Asp Arg Asn Leu Thr Cys Thr Trp Arg Pro Glu Lys Glu
Thr Asn 35 40 45Asp Thr Ser Tyr Ile Val Thr Leu Thr Tyr Ser Tyr Gly
Lys Ser Asn 50 55 60Tyr Ser Asp Asn Ala Thr Glu Ala Ser Tyr Ser Phe
Pro Arg Ser Cys65 70 75 80Ala Met Pro Pro Asp Ile Cys Ser Val Glu
Val Gln Ala Gln Asn Gly 85 90 95Asp Gly Lys Val Lys Ser Asp Ile Thr
Tyr Trp His Leu Ile Ser Ile 100 105 110Ala Lys Thr Glu Pro Pro Lys
Ile Phe Arg Val Lys Pro Val Leu Gly 115 120 125Ile Lys Arg Met Ile
Gln Ile Glu Trp Ile Lys Pro Glu Leu Ala Pro 130 135 140Val Ser Ser
Asp Leu Lys Tyr Thr Leu Arg Phe Arg Thr Val Asn Ser145 150 155
160Thr Ser Trp Met Glu Val Asn Phe Ala Lys Asn
Arg Lys Asp Lys Asn 165 170 175Gln Thr Tyr Asn Leu Thr Gly Leu Gln
Pro Phe Thr Glu Tyr Val Ile 180 185 190Ala Leu Arg Cys Ala Val Lys
Glu Ser Lys Phe Trp Ser Asp Trp Ser 195 200 205Gln Glu Lys Met Gly
Met Thr Glu Glu Glu Ala Pro Cys Gly Leu Glu 210 215 220Leu Trp Arg
Val Leu Lys Pro Ala Glu Ala Asp Gly Arg Arg Pro Val225 230 235
240Arg Leu Leu Trp Lys Lys Ala Arg Gly Ala Pro Val Leu Glu Lys Thr
245 250 255Leu Gly Tyr Asn Ile Trp Tyr Tyr Pro Glu Ser Asn Thr Asn
Leu Thr 260 265 270Glu Thr Met Asn Thr Thr Asn Gln Gln Leu Glu Leu
His Leu Gly Gly 275 280 285Glu Ser Phe Trp Val Ser Met Ile Ser Tyr
Asn Ser Leu Gly Lys Ser 290 295 300Pro Val Ala Thr Leu Arg Ile Pro
Ala Ile Gln Glu Lys Ser Phe Gln305 310 315 320Cys Ile Glu Val Met
Gln Ala Cys Val Ala Glu Asp Gln Leu Val Val 325 330 335Lys Trp Gln
Ser Ser Ala Leu Asp Val Asn Thr Trp Met Ile Glu Trp 340 345 350Phe
Pro Asp Val Asp Ser Glu Pro Thr Thr Leu Ser Trp Glu Ser Val 355 360
365Ser Gln Ala Thr Asn Trp Thr Ile Gln Gln Asp Lys Leu Lys Pro Phe
370 375 380Trp Cys Tyr Asn Ile Ser Val Tyr Pro Met Leu His Asp Lys
Val Gly385 390 395 400Glu Pro Tyr Ser Ile Gln Ala Tyr Ala Lys Glu
Gly Val Pro Ser Glu 405 410 415Gly Pro Glu Thr Lys Val Glu Asn Ile
Gly Val Lys Thr Val Thr Ile 420 425 430Thr Trp Lys Glu Ile Pro Lys
Ser Glu Arg Lys Gly Ile Ile Cys Asn 435 440 445Tyr Thr Ile Phe Tyr
Gln Ala Glu Gly Gly Lys Gly Phe Ser Lys Thr 450 455 460Val Asn Ser
Ser Ile Leu Gln Tyr Gly Leu Glu Ser Leu Lys Arg Lys465 470 475
480Thr Ser Tyr Ile Val Gln Val Met Ala Ser Thr Ser Ala Gly Gly Thr
485 490 495Asn Gly Thr Ser Ile Asn Phe Lys Thr Leu Ser His His His
His His 500 505 510His Glu Gln Lys Leu Ile Ser Glu Glu Asp Leu 515
52026512PRTArtificialAn artificially synthesized peptide sequence
265Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Asp Tyr1 5
1026612PRTArtificialAn artificially synthesized peptide sequence
266Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Leu Glu Thr1 5
1026712PRTArtificialAn artificially synthesized peptide sequence
267Asp Gly Tyr Asp Asp Gly Pro Tyr Thr Met Glu Thr1 5
1026812PRTArtificialAn artificially synthesized peptide sequence
268Asp Gly Leu Asp Asp Gly Pro Tyr Thr Met Glu Thr1 5
1026912PRTArtificialAn artificially synthesized peptide sequence
269Asp Gly Leu Asp Asp Gly Pro Tyr Thr Met Glu Ser1 5
102705PRTArtificialAn artificially synthesized peptide sequence
270Gly Tyr Ile Met Asn1 527117PRTArtificialAn artificially
synthesized peptide sequence 271Leu Ile Asn Pro Tyr Asn Gly Gly Thr
Ser Tyr Asn Gln Lys Phe Lys1 5 10 15Gly2725PRTArtificialAn
artificially synthesized peptide sequence 272Gly Tyr Val Met Asn1
52735PRTArtificialAn artificially synthesized peptide sequence
273Gly Tyr Ile Ile Asn1 52745PRTArtificialAn artificially
synthesized peptide sequence 274Gly Tyr Ile Leu Asn1
527517PRTArtificialAn artificially synthesized peptide sequence
275Leu Ile Asn Pro Tyr Asn Gly Gly Thr Asp Tyr Asn Gln Lys Phe Lys1
5 10 15Gly27617PRTArtificialAn artificially synthesized peptide
sequence 276Leu Ile Asn Pro Tyr Asn Gly Gly Thr Ser Tyr Asn Pro Lys
Phe Lys1 5 10 15Gly27711PRTArtificialAn artificially synthesized
peptide sequence 277Arg Thr Ser Glu Asn Ile Tyr Ser Phe Leu Ala1 5
102789PRTArtificialAn artificially synthesized peptide sequence
278Gln His His Tyr Glu Ser Pro Leu Thr1 527911PRTArtificialAn
artificially synthesized peptide sequence 279Arg Thr Ser Glu Asn
Ile Tyr Arg Phe Leu Ala1 5 1028011PRTArtificialAn artificially
synthesized peptide sequence 280Arg Thr Ser Glu Asn Ile Tyr Arg Phe
Val Ala1 5 102819PRTArtificialAn artificially synthesized peptide
sequence 281Gln His His Tyr Asp Ser Pro Leu Thr1
52829PRTArtificialAn artificially synthesized peptide sequence
282Gln His His Tyr Glu Asp Pro Leu Thr1 52839PRTArtificialAn
artificially synthesized peptide sequence 283Gln His His Thr Glu
Ser Pro Leu Phe1 52845PRTArtificialAn artificially synthesized
peptide sequence 284Gly Tyr Ala Met Asn1 528511PRTArtificialAn
artificially synthesized peptide sequence 285Arg Ala Ser Glu Asn
Ile Tyr Ser Phe Leu Ala1 5 1028611PRTArtificialAn artificially
synthesized peptide sequence 286Arg Ser Ser Glu Asn Ile Tyr Ser Phe
Leu Ala1 5 1028716PRTArtificialAn artificially synthesized sequence
287His His His His His His Glu Gln Lys Leu Ile Ser Glu Glu Asp Leu1
5 10 15
References
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013
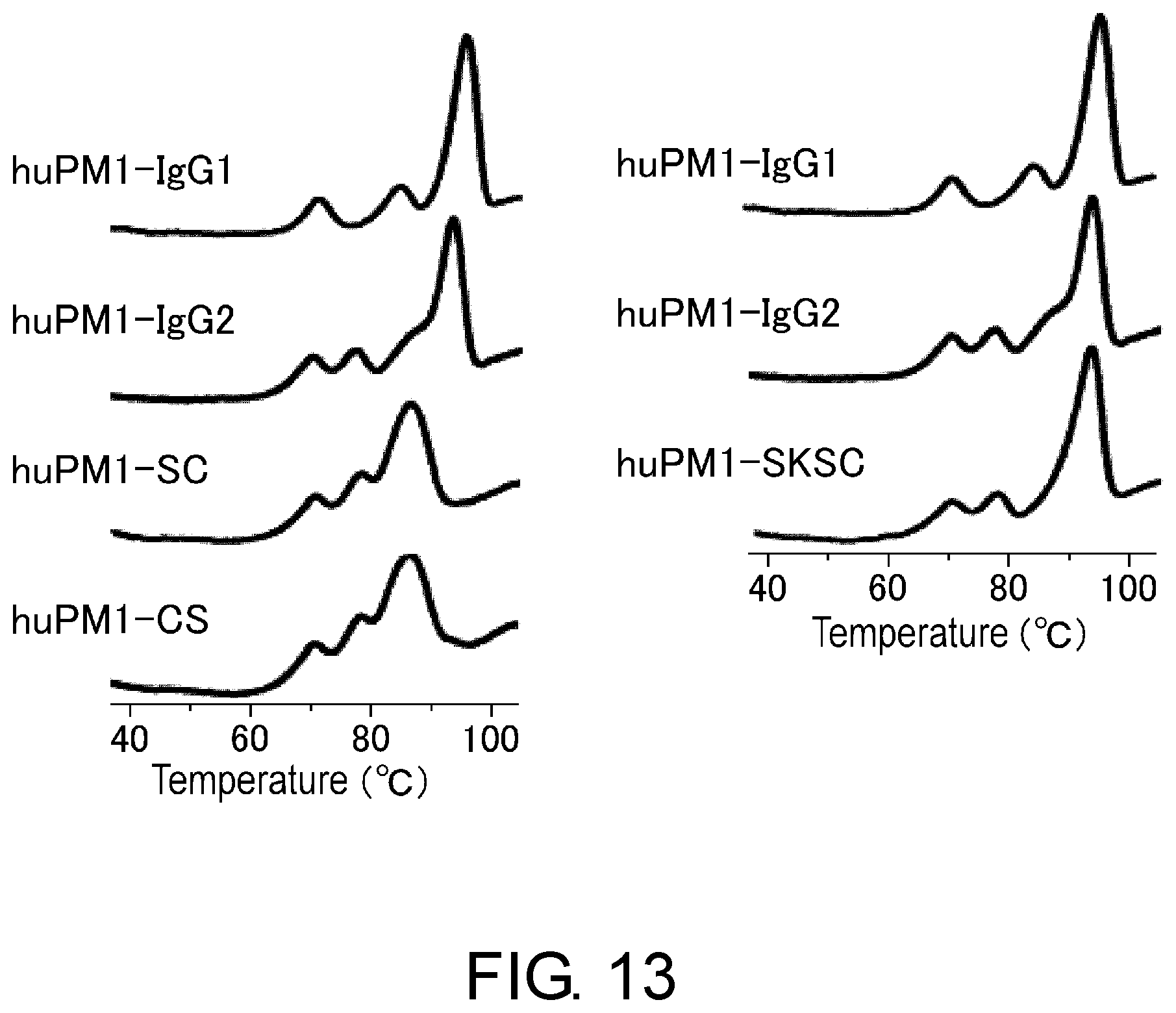
D00014

D00015
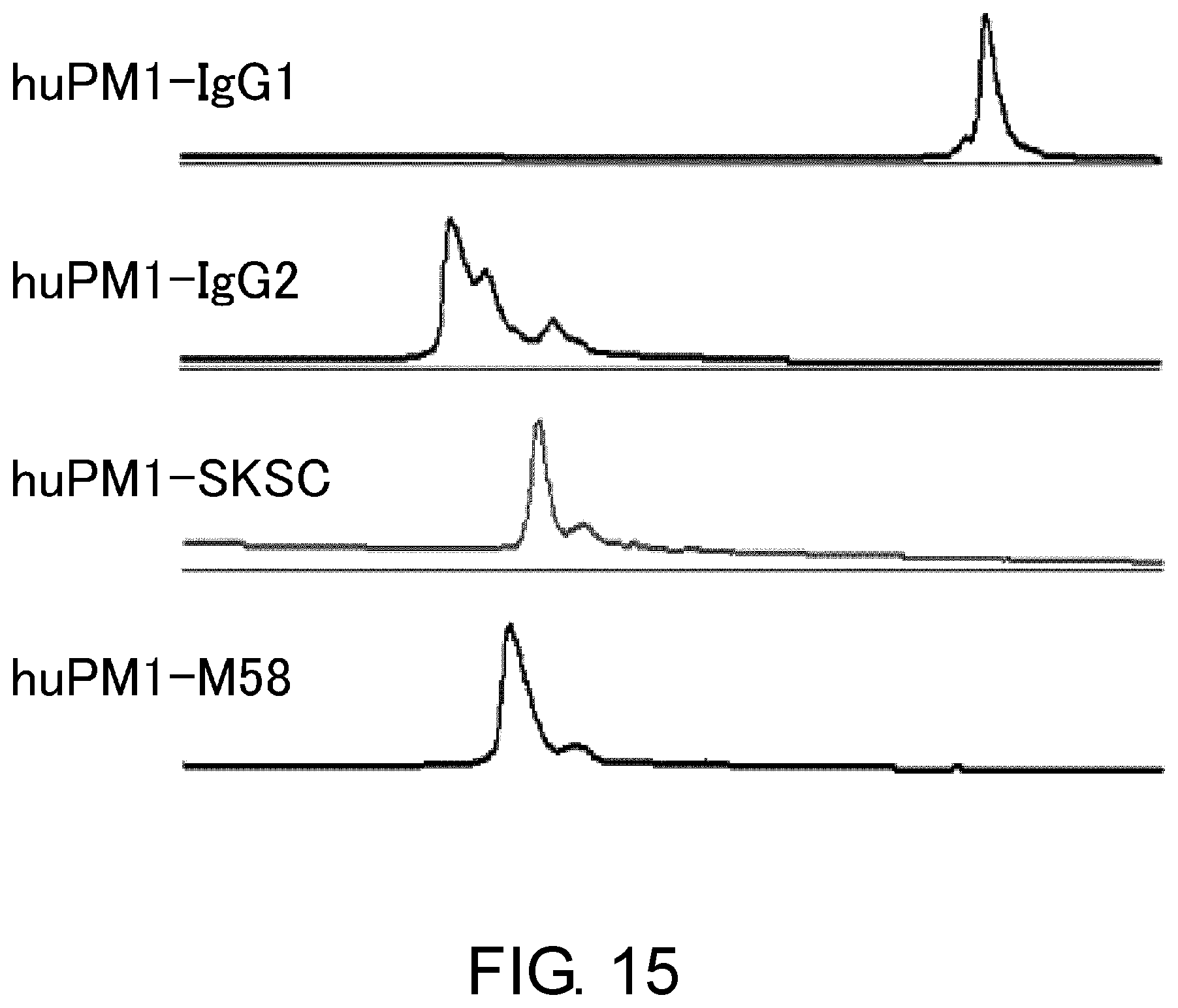
D00016

D00017

D00018
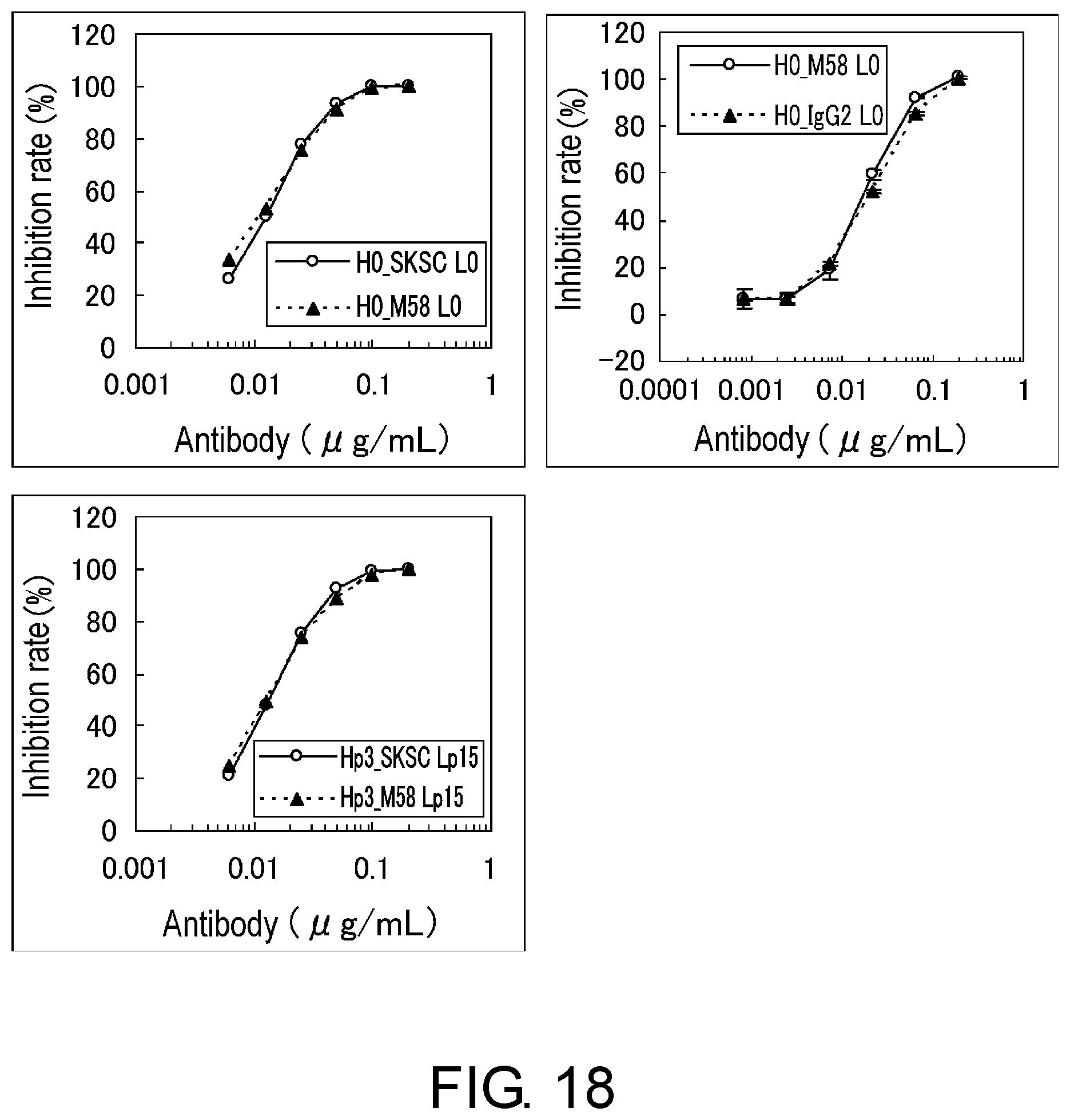
D00019

D00020

D00021

D00022
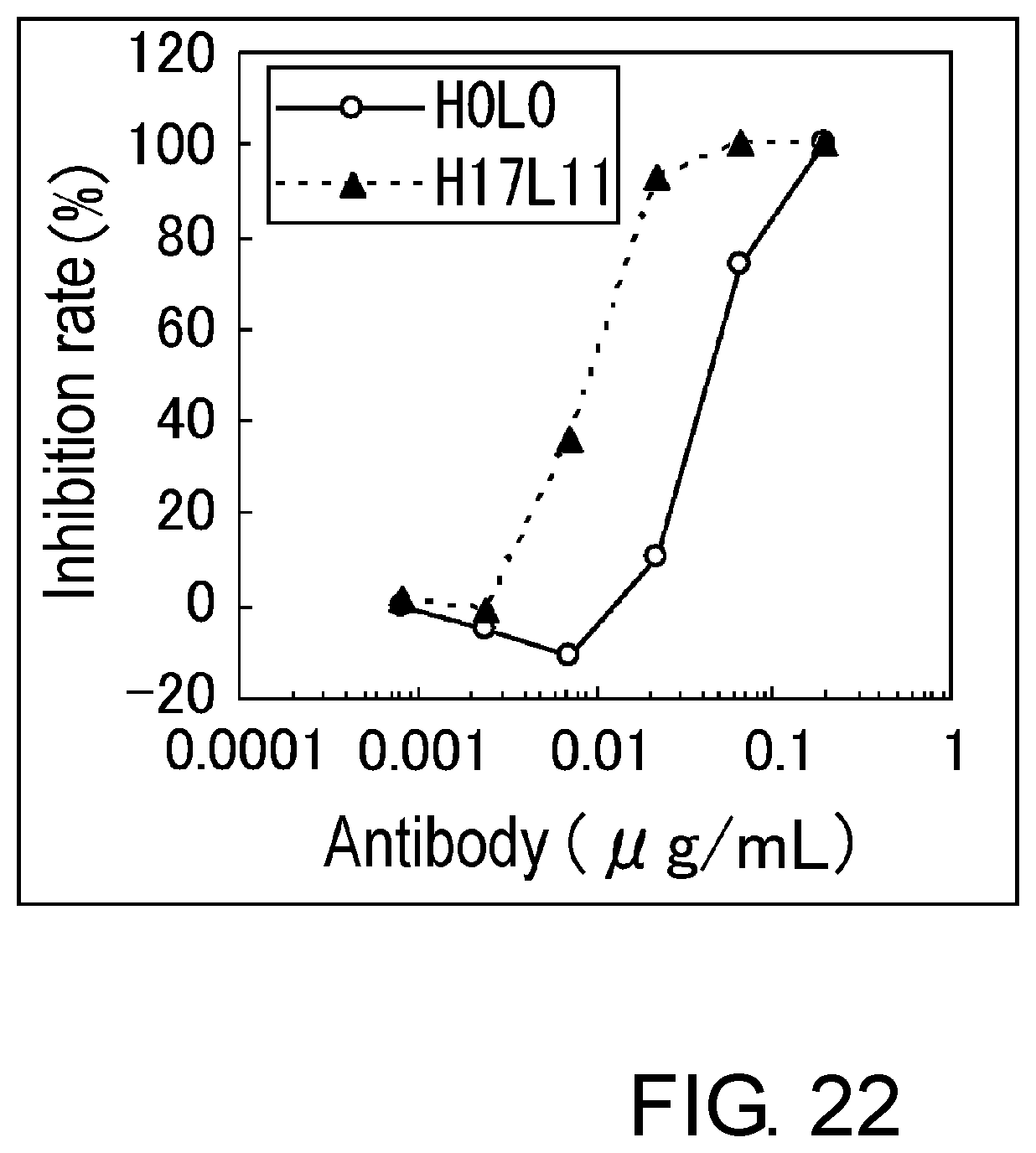
D00023
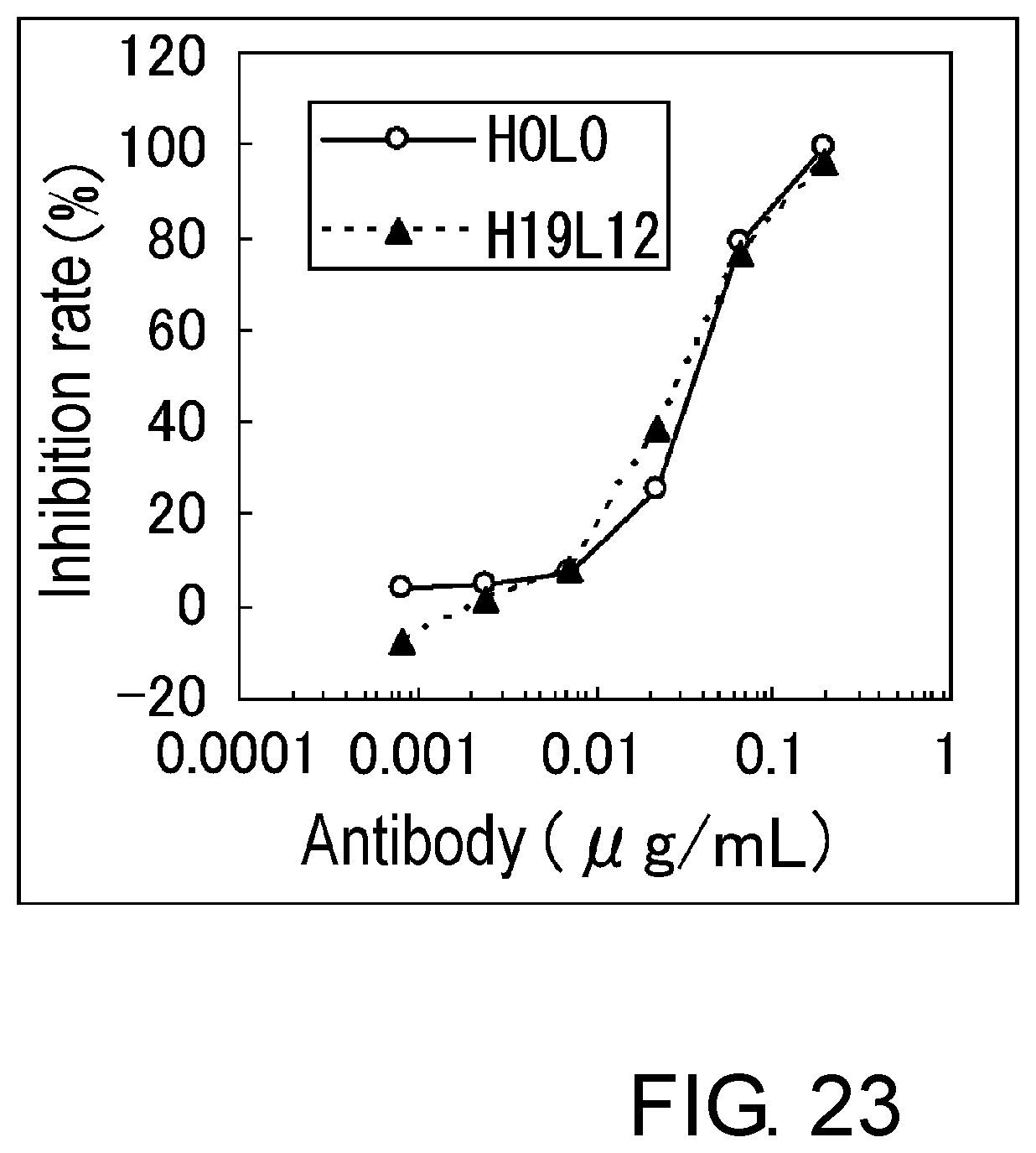
D00024

D00025

D00026
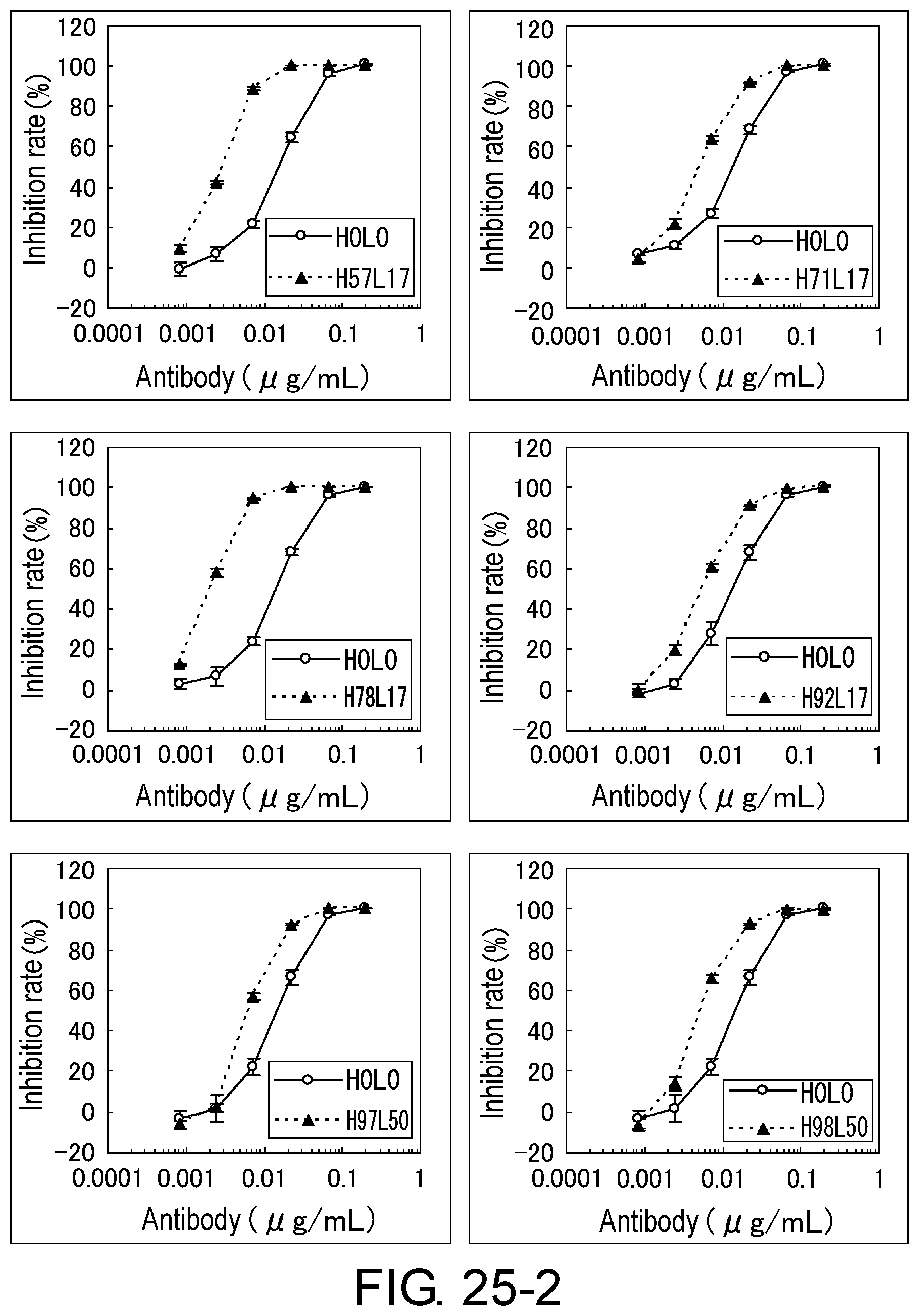
D00027
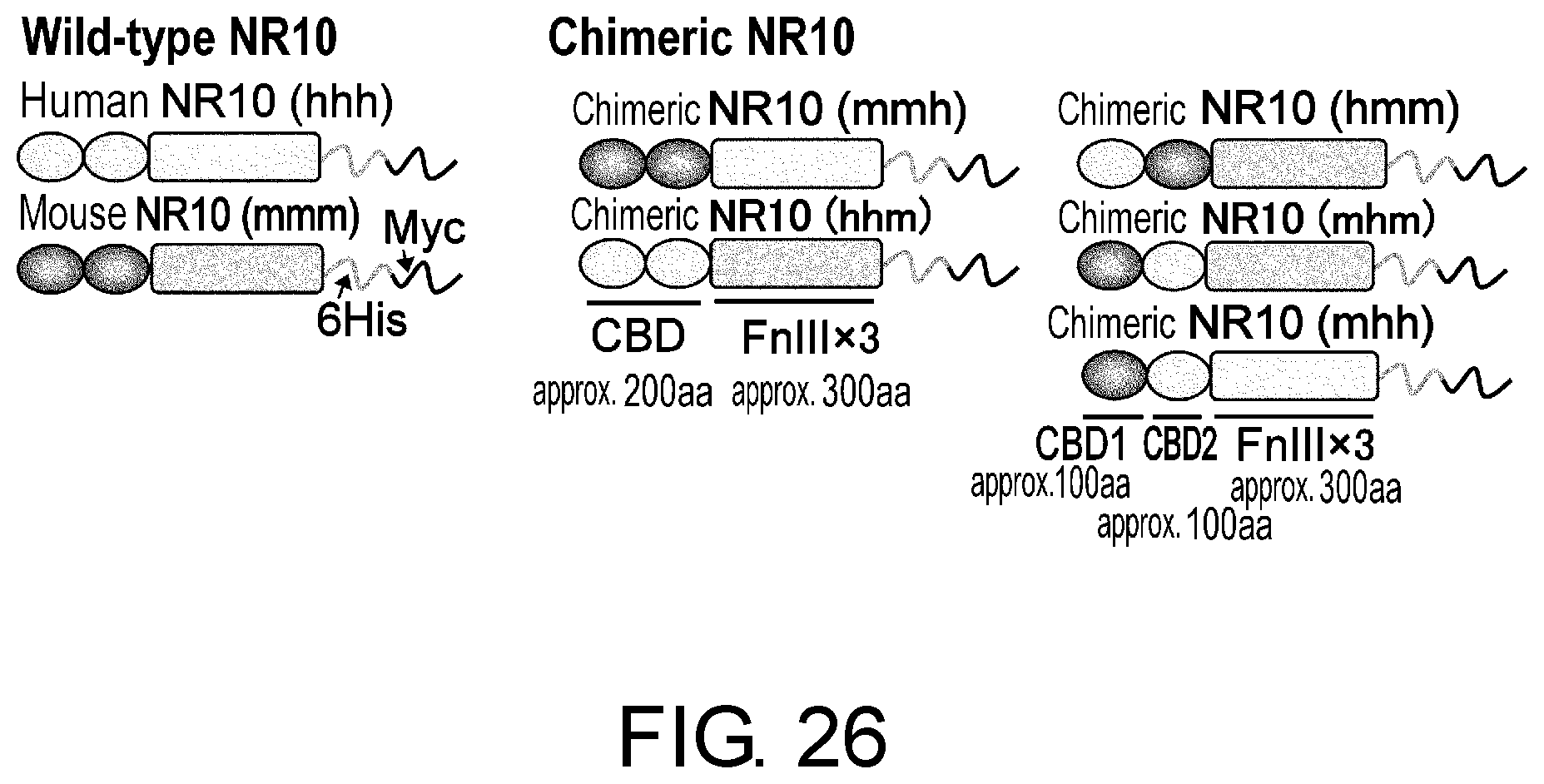
D00028

D00029

D00030
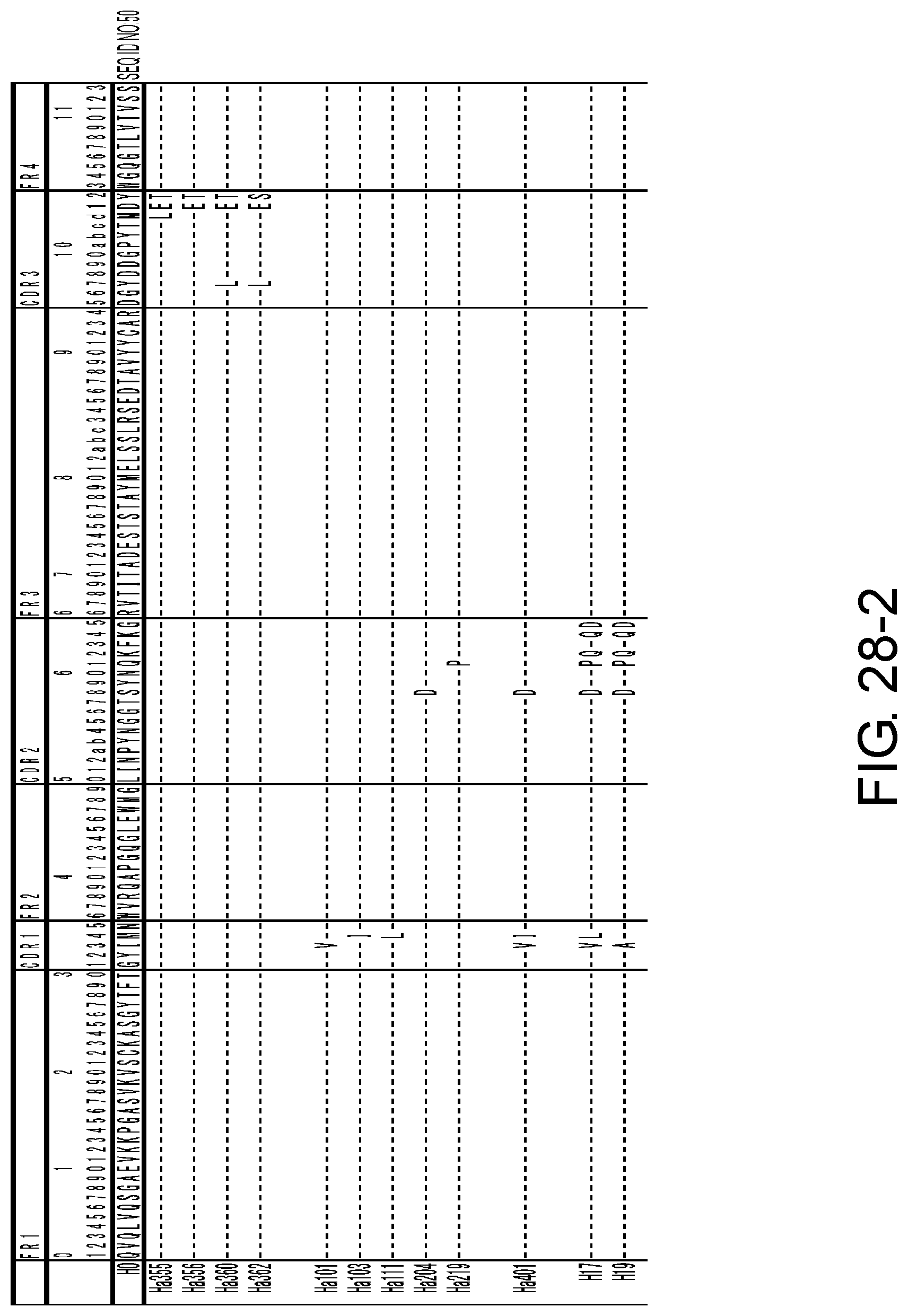
D00031

D00032
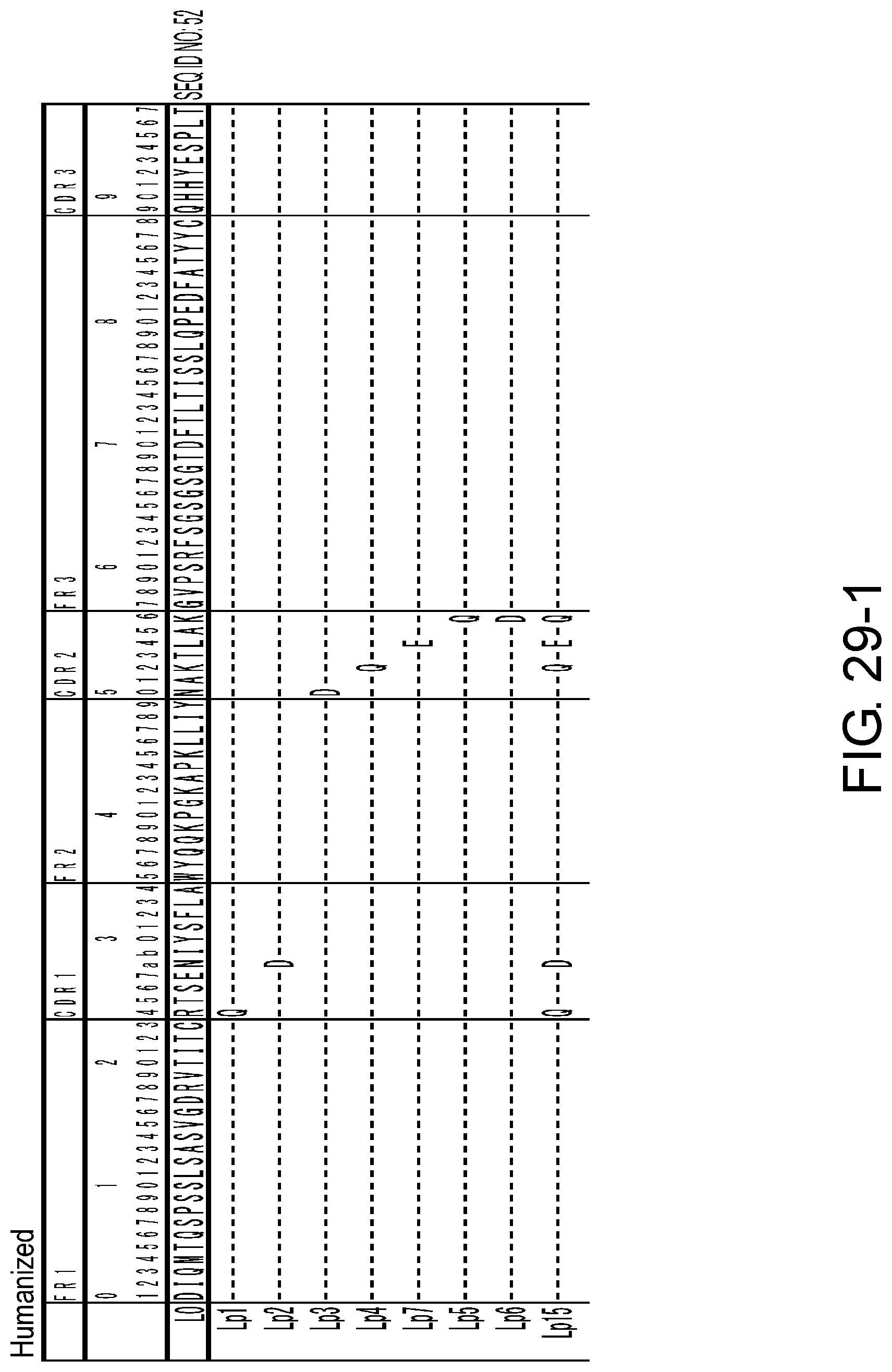
D00033

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.