Aberrant Mitochondrial Dna, Associated Fusion Transcripts And Hybridization Probes Therefor
Parr; Ryan ; et al.
U.S. patent application number 16/386381 was filed with the patent office on 2019-12-19 for aberrant mitochondrial dna, associated fusion transcripts and hybridization probes therefor. The applicant listed for this patent is MDNA LIFE SCIENCES INC.. Invention is credited to Jennifer Creed, Gabriel Dakubo, Ryan Parr, Brian Reguly, Kerry Robinson.
| Application Number | 20190382846 16/386381 |
| Document ID | / |
| Family ID | 41112880 |
| Filed Date | 2019-12-19 |



View All Diagrams
| United States Patent Application | 20190382846 |
| Kind Code | A1 |
| Parr; Ryan ; et al. | December 19, 2019 |
ABERRANT MITOCHONDRIAL DNA, ASSOCIATED FUSION TRANSCRIPTS AND HYBRIDIZATION PROBES THEREFOR
Abstract
The present invention provides novel mitochondrial fusion transcripts and the parent mutated mtDNA molecules that are useful for predicting, diagnosing and/or monitoring cancer. Hybridization probes complementary thereto for use in the methods of the invention are also provided.
| Inventors: | Parr; Ryan; (Thunder Bay, CA) ; Reguly; Brian; (Thunder Bay, CA) ; Dakubo; Gabriel; (Thunder Bay, CA) ; Creed; Jennifer; (Broomsfield, CO) ; Robinson; Kerry; (Thunder Bay, CA) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 41112880 | ||||||||||
| Appl. No.: | 16/386381 | ||||||||||
| Filed: | April 17, 2019 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 14627755 | Feb 20, 2015 | 10266899 | ||
| 16386381 | ||||
| 12935181 | Jan 17, 2011 | |||
| PCT/CA2009/000351 | Mar 27, 2009 | |||
| 14627755 | ||||
| 61040616 | Mar 28, 2008 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C07K 14/4748 20130101; C12Q 1/6886 20130101; C12Q 2600/158 20130101; A61P 35/00 20180101 |
| International Class: | C12Q 1/6886 20060101 C12Q001/6886; C07K 14/47 20060101 C07K014/47 |
Claims
1. An isolated mitochondrial fusion transcript associated with cancer, wherein the transcript has the nucleic acid sequence as set forth in any one of SEQ ID NOs:18 to 33 or 50.
2. An isolated mitochondrial DNA (mtDNA) molecule having a nucleic acid sequence encoding the fusion transcript of claim 1.
3. The isolated mtDNA of claim 2, wherein the mtDNA has the nucleic acid sequence as set forth in any one of SEQ ID NOs: 2-17 or 51.
4. A mitochondrial fusion protein, wherein the fusion protein is a translation product of the fusion transcript of claim 1.
5. The mitochondrial fusion protein of claim 4, wherein the fusion protein has the amino acid sequence as set forth in any one of SEQ ID NOs: 34 to 49 and 52.
6. A hybridization probe having a nucleic acid sequence complementary to a portion of the mitochondrial fusion transcript of claim 1.
7. The hybridization probe of claim 6, wherein the portion of the mitochondrial fusion transcript comprises an expressed sequence of mtDNA corresponding to a junction point resulting from a deletion of nucleotides of the human mtDNA genome.
8. The hybridization probe of claim 7, wherein the probe has a length of at least about 15 nucleotides, at least about 20 nucleotides, at least about 30 nucleotides, at least about 40 nucleotides, at least about 50 nucleotides, at least about 75 nucleotides, or at least about 150 nucleotides.
9. A kit for diagnosing cancer or for detecting the fusion transcript of claim 1, wherein the kit comprises the hybridization probe of claim 6.
10. The kit of claim 9, wherein the cancer is prostate cancer, testicular cancer, ovarian cancer, breast cancer, colorectal cancer, lung cancer, melanoma skin cancer or combinations thereof.
11. A method of detecting a cancer in a mammal, the method comprising assaying a tissue sample from the mammal for the presence of: (i) a mitochondrial fusion transcript having the nucleic acid sequence as set forth in any one of SEQ ID NOs:18 to 33 or 50; (ii) an mtDNA molecule encoding the fusion transcript of (i); (iii) an mtDNA molecule having the nucleic acid sequence as set forth in any one of SEQ ID NOs: 2-17 or 51; (iv) a fusion protein comprising a translation product of the fusion transcript of (i); or (v) a fusion protein having the amino acid sequence as set forth in any one of SEQ ID NOs: 34 to 49 and 52.
12. The method of claim 11, wherein the method comprises hybridizing the tissue sample with at least one hybridization probe having a nucleic acid sequence complementary to at least a portion of the mitochondrial fusion transcript of (i), the mtDNA of (ii) or the mtDNA of (iii).
13. The method of claim 12, wherein the portion of the mitochondrial fusion transcript comprises an expressed sequence of mtDNA corresponding to a junction point resulting from a deletion of nucleotides of the human mtDNA genome.
14. The method of claim 12, wherein the assay comprises: a) conducting a hybridization reaction using at least one of said probes to allow said at least one probe to hybridize to a complementary sequence of the mitochondrial fusion transcript or the mtDNA; and, b) quantifying the amount of the mitochondrial fusion transcript or the mtDNA in said sample by quantifying the amount of said transcript or mtDNA hybridized to the at least one probe.
15. The method of claim 14, wherein the step of quantifying is carried out using diagnostic imaging technology, branched DNA technology, or PCR.
16. The method of claim 11, wherein the cancer is prostate cancer, testicular cancer, ovarian cancer, breast cancer, colorectal cancer, lung cancer, melanoma skin cancer or combinations thereof.
Description
CROSS REFERENCE TO RELATED APPLICATIONS
[0001] This application is a Division of U.S. application Ser. No. 14/627,755, filed Feb. 20, 2015, now U.S. Pat. No. ______, issued ______, which is a Continuation of U.S. application Ser. No. 12/935,181, filed Jan. 17, 2011, now abandoned, which is a national entry of PCT Application No. PCT/CA2009/000351, filed Mar. 27, 2009, which claims priority under 35 U.S.C. .sctn. 119(e) from U.S. Provisional Application No. 61/040,616, filed Mar. 28, 2008. Each of the aforementioned applications is incorporated by reference herein as if set forth in its entirety.
FIELD OF THE INVENTION
[0002] The present invention relates to the field of mitochondrial genomics. In one aspect, the invention relates to the identification and use of mitochondrial genome fusion transcripts and probes that hybridize thereto.
BACKGROUND OF THE INVENTION
[0003] Mitochondrial Genome
[0004] The mitochondrial genome is a compact yet critical sequence of nucleic acids. Mitochondrial DNA, or "mtDNA", comprises a small genome of 16,569 nucleic acid base pairs (bp) (Anderson et al., 1981; Andrews et al., 1999) in contrast to the immense nuclear genome of 3.3 billion bp (haploid). Its genetic complement is substantially smaller than that of its nuclear cell mate (0.0005%). However, individual cells carry anywhere from 10.sup.3 to 10.sup.4 mitochondria depending on specific cellular functions (Singh and Modica-Napolitano 2002). Communication or chemical signalling routinely occurs between the nuclear and mitochondrial genomes (Sherratt et al., 1997). Moreover, specific nuclear components are responsible for the maintenance and integrity of mitochondrial sequences (Croteau et al., 1999). All mtDNA genomes in a given individual are identical due to the clonal expansion of mitochondria within the ovum, once fertilization has occurred. However mutagenic events can induce sequence diversity reflected as somatic mutations. These mutations may accumulate in different tissues throughout the body in a condition known as heteroplasmy.
[0005] Mitochondrial Proteome
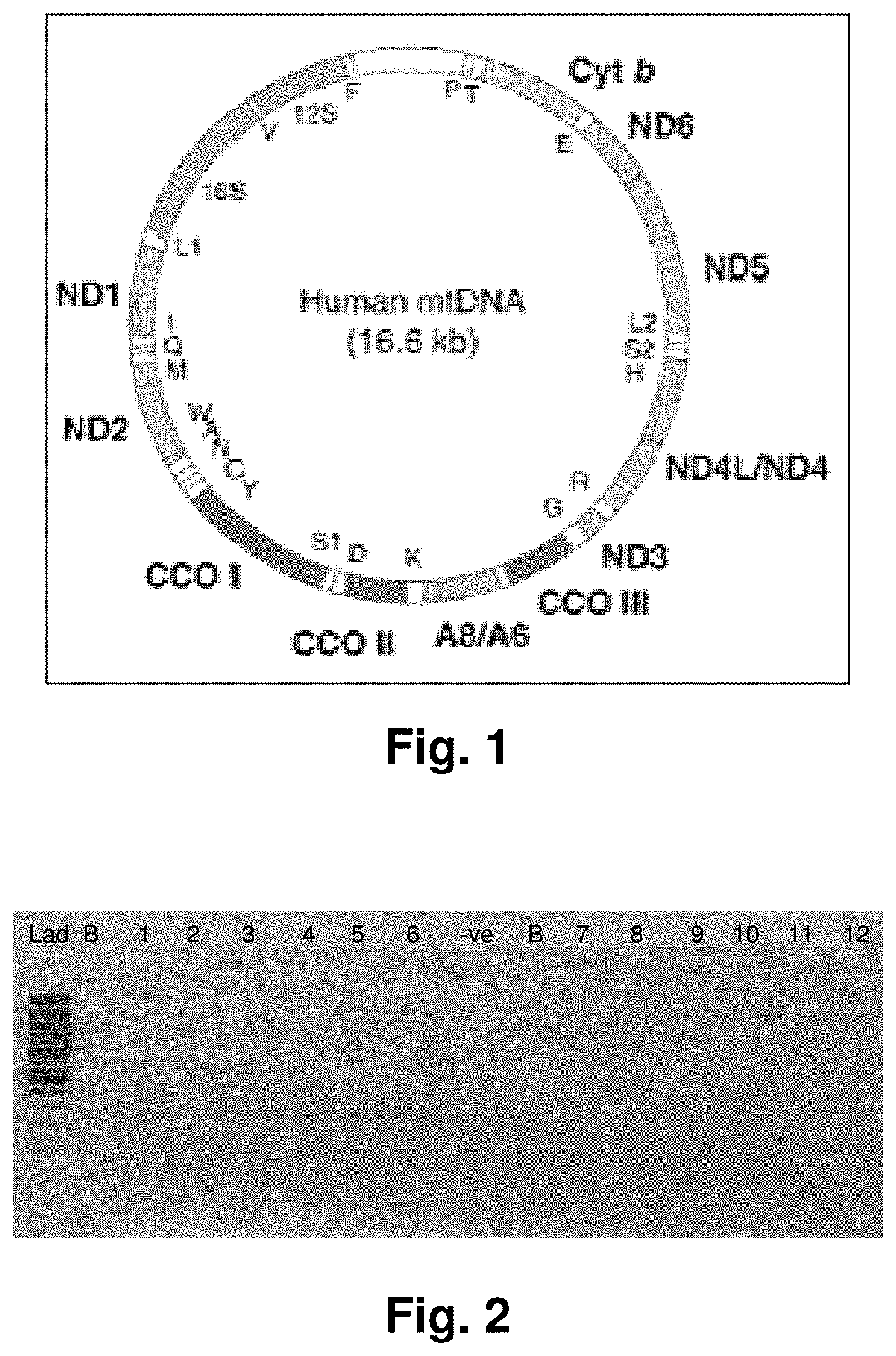
[0006] About 3,000 nuclear genes are required to construct, operate and maintain mitochondria, with only thirty-seven of these coded by the mitochondrial genome, indicating heavy mitochondrial dependence on nuclear loci. The mitochondrial genome codes for a complement of 24 genes, including 2 rRNAs and 22 tRNAs that ensure correct translation of the remaining 13 genes which are vital to electron transport (see FIG. 1). The mitochondrial genome is dependent on seventy nuclear encoded proteins to accomplish the oxidation and reduction reactions necessary for this vital function, in addition to the thirteen polypeptides supplied by the mitochondrial genome. Both nuclear and mitochondrial proteins form complexes spanning the inner mitochondrial membrane and collectively generate 80-90% of the chemical fuel adenosine triphosphate, or ATP, required for cellular metabolism. In addition to energy production, mitochondria play a central role in other metabolic pathways as well. A critical function of the mitochondria is mediation of cell death, or apoptosis (see Green and Kroemer, 2005). Essentially, there are signal pathways which permeabilize the outer mitochondrial membrane, or in addition, the inner mitochondrial membrane as well. When particular mitochondrial proteins are released into the cytosol, non-reversible cell death is set in motion. This process highlights the multi-functional role that some mitochondrial proteins have. These multi-tasking proteins suggest that there are other mitochondrial proteins as well which may have alternate functions.
[0007] Mitochondrial Fusion Transcriptome
[0008] The mitochondrial genome is unusual in that it is a circular, intron-less DNA molecule. The genome is interspersed with repeat motifs which flank specific lengths of sequences. Sequences between these repeats are prone to deletion under circumstances which are not well understood. Given the number of repeats in the mitochondrial genome, there are many possible deletions. The best known example is the 4977 "common deletion." This deletion has been associated with several purported conditions and diseases and is thought to increase in frequency with aging (Dai et al., 2004; Ro et al., 2003; Barron et al., 2001; Lewis et al., 2000; Muller-Hocker, 1998; Porteous et al., 1998) (FIG. 4). The current thinking in the field of mitochondrial genomics is that mitochondrial deletions are merely deleterious by-products of damage to the mitochondrial genome by such agents as reactive oxygen species and UVR. (Krishnan et al 2008, Nature Genetics). Further, though it is recognized that high levels of mtDNA deletions can have severe consequences on the cell's ability to produce energy in the form of ATP as a result of missing gene sequences necessary for cellular respiration, it is not anticipated that these deleted mitochondrial molecules may be a component of downstream pathways, have an intended functional role, and possibly may be more aptly viewed as alternate natural forms of the recognized genes of the mitochondria as has been anticipated by the Applicant.
[0009] The sequence dynamics of mtDNA are important diagnostic tools. Mutations in mtDNA are often preliminary indicators of developing disease. For example, it has been demonstrated that point mutations in the mitochondrial genome are characteristic of tumour foci in the prostate. This trend also extends to normal appearing tissue both adjacent to and distant from tumour tissue (Parr et al. 2006). This suggests that mitochondrial mutations occur early in the malignant transformation pathway.
[0010] For example, the frequency of a 3.4 kb mitochondrial deletion has excellent utility in discriminating between benign and malignant prostate tissues (Maki et al. 2008).
[0011] Mitochondrial fusion transcripts have been reported previously in the literature, first in soybeans (Morgens et al. 1984) and then later in two patients with Kearns-Sayre Syndrome, a rare neuromuscular disorder (Nakase et al 1990). Importantly, these transcripts were not found to have (or investigated regarding) association with any human cancers.
SUMMARY OF THE INVENTION
[0012] An object of the present invention to provide aberrant mitochondrial DNA, associated fusion transcripts and hybridization probes therefor.
[0013] In accordance with an aspect of the invention, there is provided an isolated mitochondrial fusion transcript associated with cancer.
[0014] In accordance with an aspect of the invention, there is provided a mitochondrial fusion protein corresponding to the above fusion transcript, having a sequence as set forth in any one of SEQ ID NOs: 34 to 49 and 52.
[0015] In accordance with another aspect of the invention, there is provided an isolated mtDNA encoding a fusion transcript of the invention.
[0016] In accordance with another aspect of the invention, there is provided a hybridization probe having a nucleic acid sequence complementary to at least a portion of a mitochondrial fusion transcript or an mtDNA of the invention.
[0017] In accordance with another aspect of the invention, there is provided a method of detecting a cancer in a mammal, the method comprising assaying a tissue sample from the mammal for the presence of at least one mitochondrial fusion transcript associated with cancer by hybridizing the sample with at least one hybridization probe having a nucleic acid sequence complementary to at least a portion of a mitochondrial fusion transcript according to the invention.
[0018] In accordance with another aspect of the invention, there is provided a method of detecting a cancer in a mammal, the method comprising assaying a tissue sample from the mammal for the presence of at least one aberrant mtDNA associated with cancer by hybridizing the sample with at least one hybridization probe having a nucleic acid sequence complementary to at least a portion of an mtDNA according to the invention.
[0019] In accordance with another aspect of the invention, there is provided a kit for conducting an assay for detecting the presence of a cancer in a mammal, said kit comprising at least one hybridization probe complementary to at least a portion of a fusion transcript or an mtDNA of the invention.
[0020] In accordance with another aspect of the invention, there is provided a screening tool comprised of a microarray having 10's, 100's, or 1000's of mitochondrial fusion transcripts for identification of those associated with cancer.
[0021] In accordance with another aspect of the invention, there is provided a screening tool comprised of a microarray having 10's, 100's, or 1000's of mitochondrial DNAs corresponding to mitochondrial fusion transcripts for identification of those associated with cancer.
[0022] In accordance with another aspect of the invention, there is provided a screening tool comprised of a multiplexed branched DNA assay having 10's, 100's, or 1000's of mitochondrial fusion transcripts for identification of those associated with cancer.
[0023] In accordance with another aspect of the invention, there is provided a screening tool comprised of a multiplexed branched DNA assay having 10's, 100's, or 1000's of mitochondrial DNAs corresponding to mitochondrial fusion transcripts for identification of those associated with cancer.
BRIEF DESCRIPTION OF THE DRAWINGS
[0024] The embodiments of the invention will now be described by way of example only with reference to the appended drawings wherein:
[0025] FIG. 1 is an illustration showing mitochondrial coding genes.
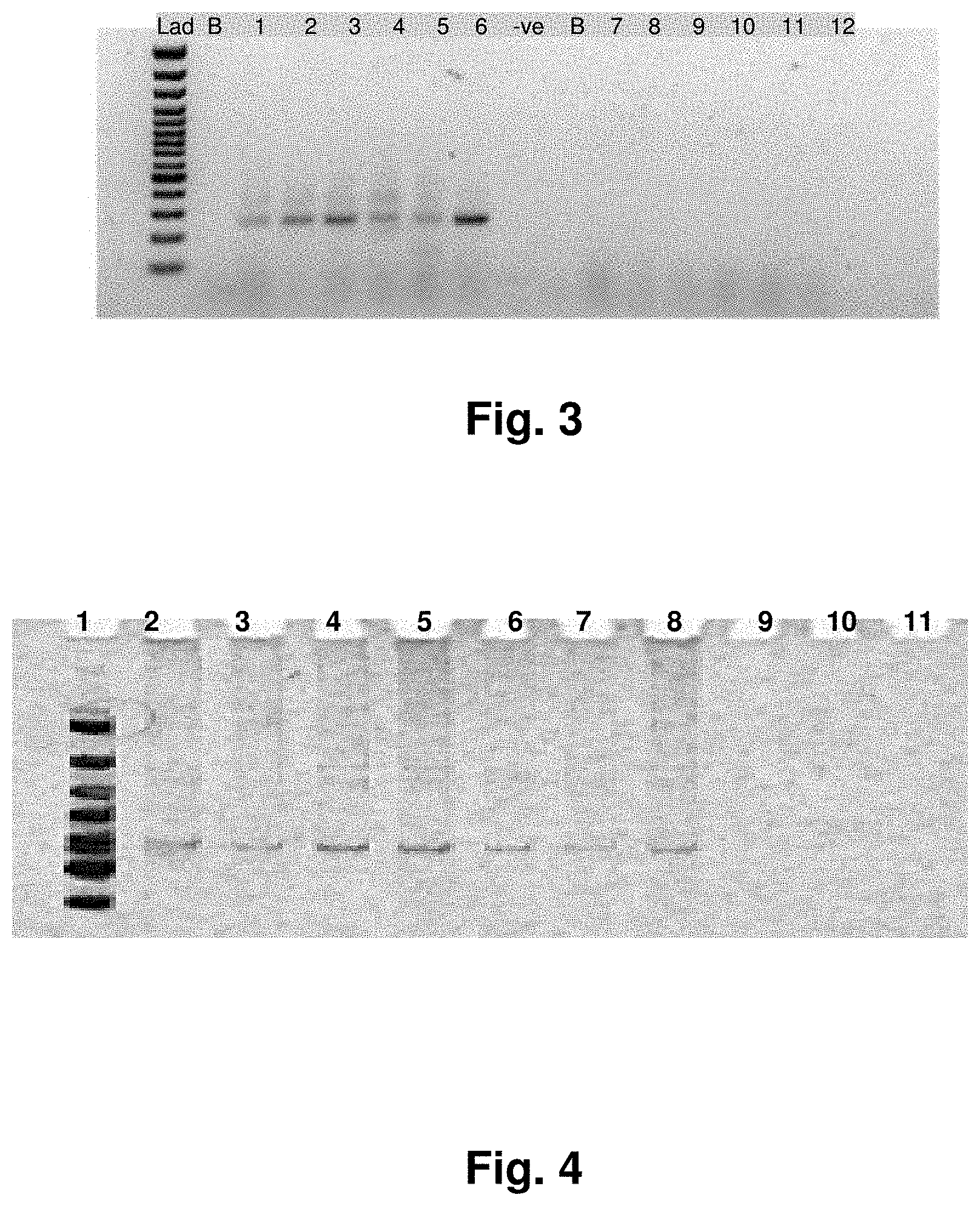
[0026] FIG. 2 shows polyadenalated fusion transcripts in prostate samples invoked by the loss of the 3.4 kb deletion.
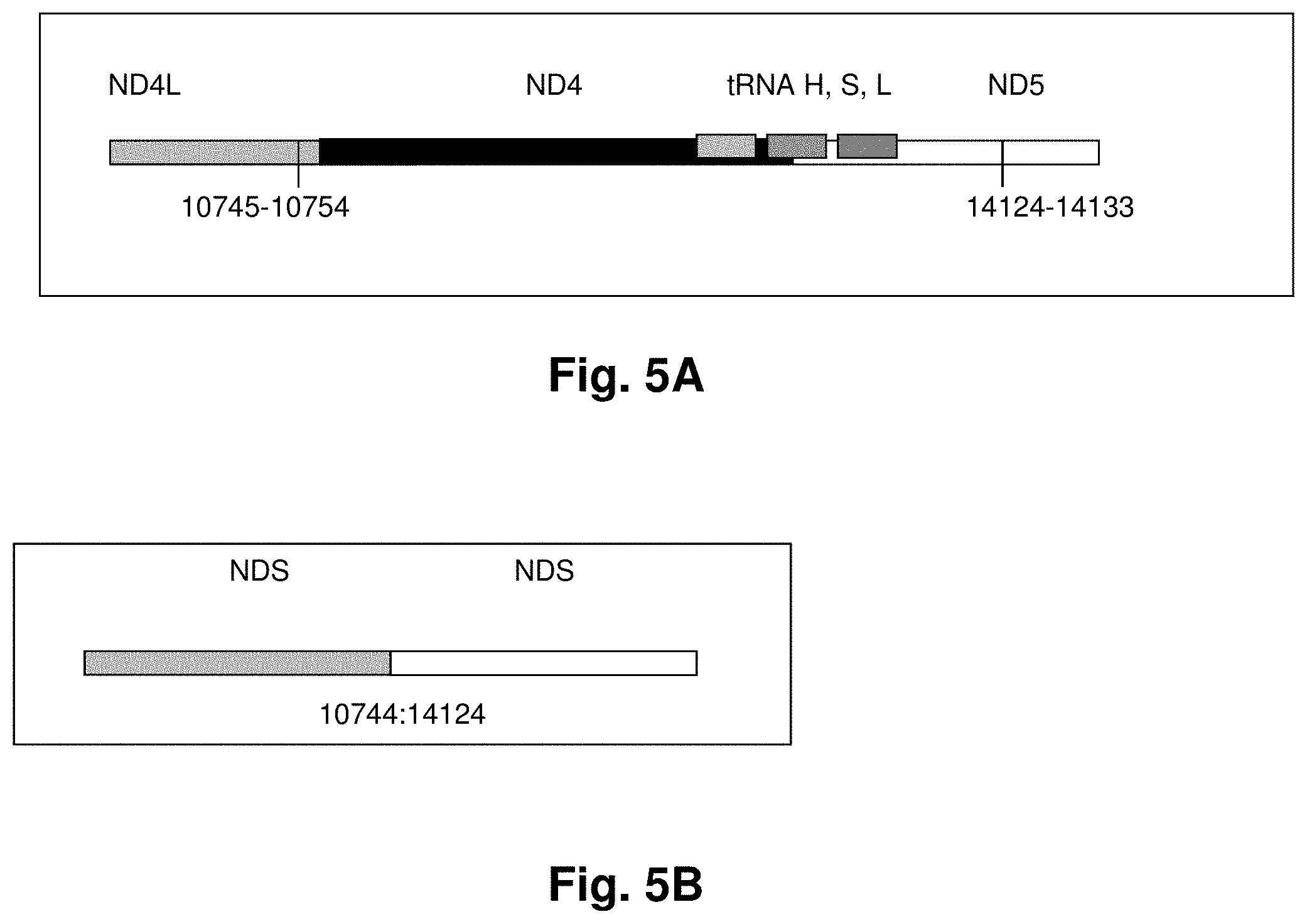
[0027] FIG. 3 shows polyadenalated fusion transcripts in prostate samples invoked by the loss of the 4977 kb common deletion.
[0028] FIG. 4 shows polyadenalated fusion transcripts in breast samples invoked by the loss of the 3.4 kb segment from the mtgenome.
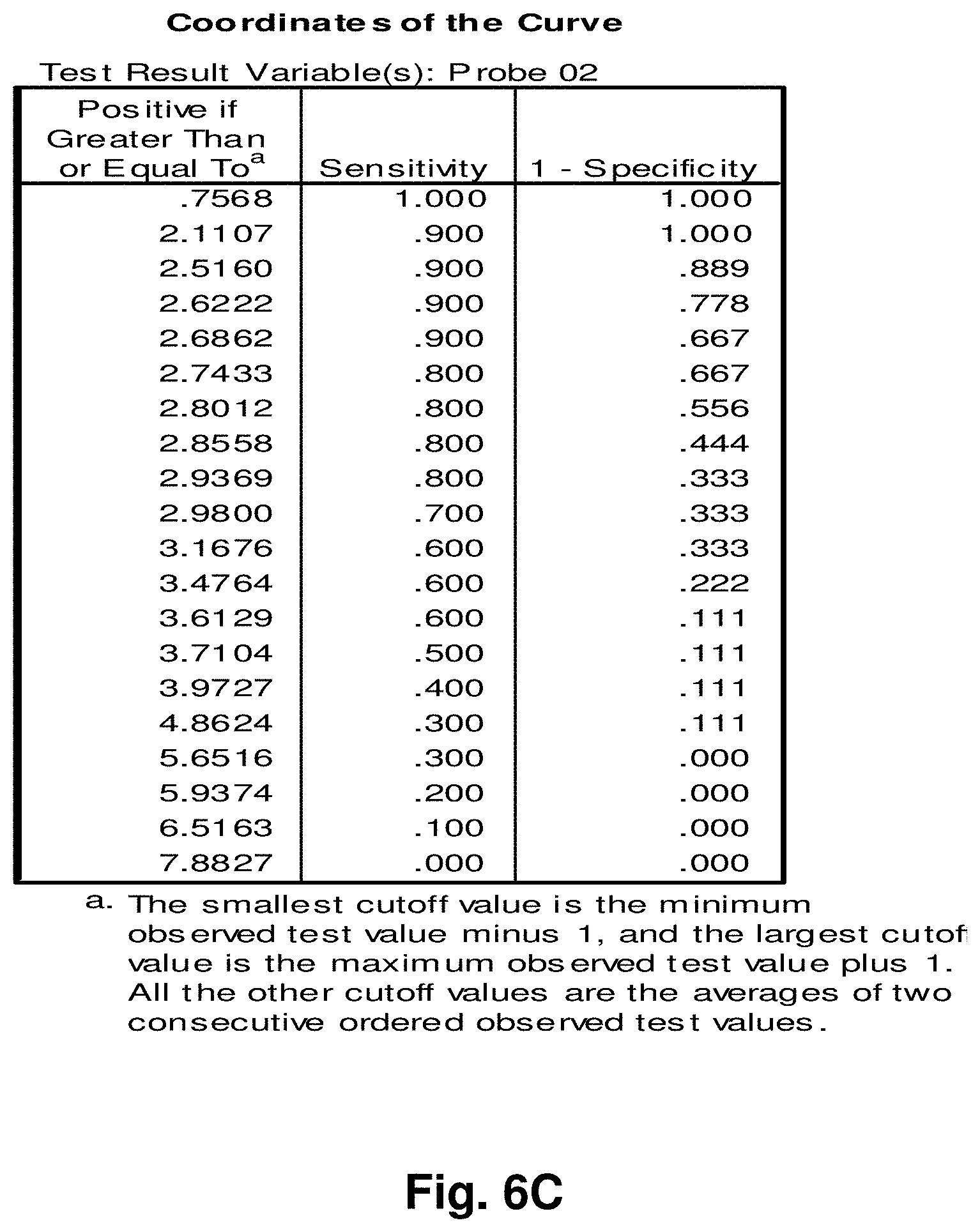
[0029] FIGS. 5A and 5B show an example of a mitochondrial DNA region before and after splicing of genes.
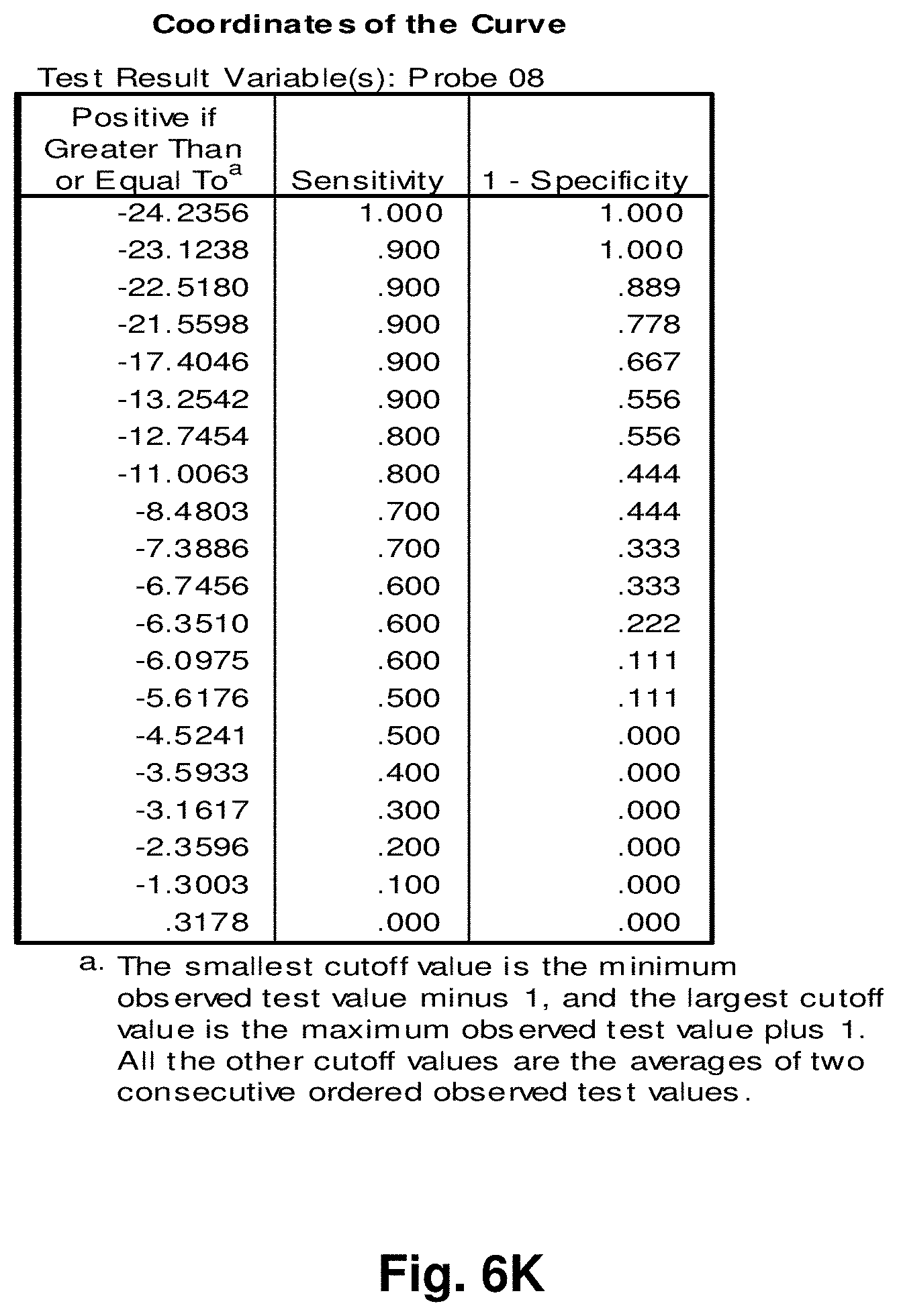
[0030] FIGS. 6A to 6BB illustrate the results for transcripts 2, 3, 8, 9, 10, 11 and 12 of the invention in the identification of colorectal cancer tumours.
[0031] FIGS. 7A to 7P illustrate the results for transcripts 6, 8, 10 and 20 of the invention in the identification of lung cancer tumours.
[0032] FIGS. 8A to 8BB illustrate the results for transcripts 6, 10, 11, 14, 15, 16 and 20 of the invention in the identification of melanomas.
[0033] FIGS. 9A to 9NN illustrate the results for transcripts 1, 2, 3, 6, 11, 12, 15 and 20 of the invention in the identification of ovarian cancer.
[0034] FIGS. 10A to 10K illustrate the results for transcripts 2, 3, 4, 11, 12, 13, 15, 16 and 20 of the invention in the identification of testicular cancer.
[0035] FIGS. 11A to 11I illustrate the results for transcripts 2, 3, 4, 11, 12, 13, 15, 16 and 20 of the invention in the identification of testicular cancer.
[0036] FIGS. 12A to 12E illustrate the results for transcripts 2, 3, 4, 11, 12, 13, 15, 16 and 20 of the invention in the identification of testicular cancer.
[0037] FIGS. 13A to 13I illustrate the results for transcripts 2, 3, 4, 11, 12, 13, 15, 16 and 20 of the invention in the identification of testicular cancer.
[0038] FIGS. 14A to 14I illustrate the results for transcripts 2, 3, 4, 11, 12, 13, 15, 16 and 20 of the invention in the identification of testicular cancer.
[0039] FIGS. 15A to 15N illustrate the results for transcripts 2, 3, 4, 11, 12, 13, 15, 16 and 20 of the invention in the identification of testicular cancer.
[0040] FIGS. 16A to 16E illustrate the results for transcripts 2, 3, 4, 11, 12, 13, 15, 16 and 20 of the invention in the identification of testicular cancer.
[0041] FIGS. 17A to 17J illustrate the results for transcripts 2, 3, 4, 11, 12, 13, 15, 16 and 20 of the invention in the identification of testicular cancer.
[0042] FIGS. 18A to 18I illustrate the results for transcripts 2, 3, 4, 11, 12, 13, 15, 16 and 20 of the invention in the identification of testicular cancer.
DETAILED DESCRIPTION OF THE INVENTION
[0043] The present invention provides novel mitochondrial fusion transcripts and the parent mutated mtDNA molecules that are useful for predicting, diagnosing and/or monitoring cancer. The invention further provides hybridization probes for the detection of fusion transcripts and associated mtDNA molecules and the use of such probes.
Definitions
[0044] Unless defined otherwise, all technical and scientific terms used herein have the same meaning as commonly understood by one of ordinary skill in the art to which this invention belongs.
[0045] As used herein, "aberration" or "mutation" encompasses any modification in the wild type mitochondrial DNA sequence that results in a fusion transcript and includes, without limitation, insertions, translocations, deletions, duplications, recombinations, rearrangements or combinations thereof.
[0046] As defined herein, "biological sample" refers to a tissue or bodily fluid containing cells from which a molecule of interest can be obtained. For example, the biological sample can be derived from tissue such as prostate, breast, colorectal, lung and skin, or from blood, saliva, cerebral spinal fluid, sputa, urine, mucous, synovial fluid, peritoneal fluid, amniotic fluid and the like. The biological sample may be a surgical specimen or a biopsy specimen. The biological sample can be used either directly as obtained from the source or following a pre-treatment to modify the character of the sample. Thus, the biological sample can be pre-treated prior to use by, for example, preparing plasma or serum from blood, disrupting cells, preparing liquids from solid materials, diluting viscous fluids, filtering liquids, distilling liquids, concentrating liquids, inactivating interfering components, adding reagents, and the like.
[0047] A "continuous" transcript is a fusion transcript that keeps the reading frame from the beginning to the end of both spliced genes. An "end" transcript is a fusion transcript that results in a premature termination codon before the original termination codon of a second spliced gene.
[0048] As used herein, "mitochondrial DNA" or "mtDNA" is DNA present in mitochondria.
[0049] As used herein, the expression "mitochondrial fusion transcript" or "fusion transcript" refers to an RNA transcription product produced as a result of the transcription of a mutated mitochondrial DNA sequence wherein such mutations may comprise mitochondrial deletions and other large-scale mitochondrial DNA rearrangements.
[0050] Computer Analysis and Sequence Targeting
[0051] As discussed above, mitochondrial fusion transcripts have been reported in soybeans (Morgens et al. 1984) and in humans suffering from a rare neuromuscular disorder (Nakase et al 1990). Fusion transcripts associated with human cancer have not, however, been described.
[0052] Using the knowledge gained from mapping the large-scale deletions of the human mitochondrial genome associated with cancer, the observation of high frequencies of these deletions, and the evidence in another organism and another disease type of trancriptionally active mutated mtDNA molecules, Applicant hypothesized that such deletions may have importance beyond the DNA molecule and the damage and repair processes as it relates to cancer. To test this hypothesis computer analysis of the mitochondrial genome was conducted, specific for repeat elements, which suggested many potential deletion sites. Following this initial step identifying unique repeats in the mitochondrial sequence having non-adjacent or non-tandem locations, a filter was then applied to identify those repeats that upon initiating a deletion event in the DNA molecule would then likely reclose or religate to produce a fused DNA sequence having an open reading frame (ORF). A subset of 18 molecules were then selected for targetting to investigate whether: 1) they existed in the natural biological state of humans and 2) they had relevance to malignancy. Results from these investigations are described hereinafter.
[0053] Genomic Mutations
[0054] Mitochondrial DNA (mtDNA) dynamics are an important diagnostic tool. Mutations in mtDNA are often preliminary indicators of developing disease and behave as biomarkers indicative of risk factors associated with disease onset. According to the present invention, large-scale rearrangement mutations in the mitochondrial genome result in the generation of fusion transcripts associated with cancer. Thus, the use of mtDNA encoding such transcripts and probes directed thereto for the detection, diagnosis and monitoring of cancer is provided.
[0055] One of skill in the art will appreciate that the mtDNA molecules for use in the methods of the present invention may be derived through the isolation of naturally-occurring mutants or may be based on the complementary sequence of any of the fusion transcripts described herein. Exemplary mtDNA sequences and fusion transcripts are disclosed in Applicant's U.S. priority application No. 61/040,616, herein incorporated in its entirety by reference.
[0056] Detection of Mutant Genomic Sequences
[0057] Mutant mtDNA sequences according to the present invention may comprise any modification that results in the generation of a fusion transcript. Non-limiting examples of such modifications include insertions, translocations, deletions, duplications, recombinations, rearrangements or combinations thereof. While the modification or change can vary greatly in size from only a few bases to several kilobases, preferably the modification results in a substantive deletion or other large-scale genomic aberration.
[0058] Extraction of DNA to detect the presence of such mutations may take place using art-recognized methods, followed by amplification of all or a region of the mitochondrial genome, and may include sequencing of the mitochondrial genome, as described in Current Protocols in Molecular Biology. Alternatively, crude tissue homogenates may be used as well as techniques not requiring amplification of specific fragments of interest.
[0059] The step of detecting the mutations can be selected from any technique as is known to those skilled in the art. For example, analyzing mtDNA can comprise selection of targets by branching DNA, sequencing the mtDNA, amplifying mtDNA by PCR, Southern, Northern, Western South-Western blot hybridizations, denaturing HPLC, hybridization to microarrays, biochips or gene chips, molecular marker analysis, biosensors, melting temperature profiling or a combination of any of the above.
[0060] Any suitable means to sequence mitochondrial DNA may be used. Preferably, mtDNA is amplified by PCR prior to sequencing. The method of PCR is well known in the art and may be performed as described in Mullis and Faloona, 1987, Methods Enzymol., 155: 335. PCR products can be sequenced directly or cloned into a vector which is then placed into a bacterial host. Examples of DNA sequencing methods are found in Brumley, R. L. Jr. and Smith, L. M., 1991, Rapid DNA sequencing by horizontal ultrathin gel electrophoresis, Nucleic Acids Res. 19:4121-4126 and Luckey, J. A., et al, 1993, High speed DNA sequencing by capillary gel electrophoresis, Methods Enzymol. 218: 154-172. The combined use of PCR and sequencing of mtDNA is described in Hopgood, R., et al, 1992, Strategies for automated sequencing of human mtDNA directly from PCR products, Biotechniques 13:82-92 and Tanaka, M. et al, 1996, Automated sequencing of mtDNA, Methods Enzymol. 264: 407-421.
[0061] Methods of selecting appropriate sequences for preparing various primers are also known in the art. For example, the primer can be prepared using conventional solid-phase synthesis using commercially available equipment, such as that available from Applied Biosystems USA Inc. (Foster City, Calif.), DuPont, (Wilmington, Del.), or Milligen (Bedford, Mass.).
[0062] According to an aspect of the invention, to determine candidate genomic sequences, a junction point of a sequence deletion is first identified. Sequence deletions are primarily identified by direct and indirect repetitive elements which flank the sequence to be deleted at the 5' and 3' end. The removal of a section of the nucleotides from the genome followed by the ligation of the genome results in the creation of a novel junction point.
[0063] Upon identification of the junction point, the nucleotides of the genes flanking the junction point are determined in order to identify a spliced gene. Typically the spliced gene comprises the initiation codon from the first gene and the termination codon of the second gene, and may be expressed as a continuous transcript, i.e. one that keeps the reading frame from the beginning to the end of both spliced genes. It is also possible that alternate initiation or termination codons contained within the gene sequences may be used as is evidenced by SEQ ID No:2 and SEQ ID No: 17 disclosed herein. Some known mitochondrial deletions discovered to have an open reading frame (ORF) when the rearranged sequences are rejoined at the splice site are provided in Table 1.
[0064] Exemplary mtDNA molecules for use in the methods of the present invention, which have been verified to exist in the lab, are provided below. These mtDNAs are based on modifications of the known mitochondrial genome (SEQ ID NO: 1) and have been assigned a fusion or "FUS" designation, wherein A:B represents the junction point between the last mitochondrial nucleotide of the first spliced gene and the first mitochondrial nucleotide of the second spliced gene. The identification of the spliced genes is provided in parentheses followed by the corresponding sequence identifier. Where provided below, (AltMet) and (OrigMet) refer to alternate and original translation start sites, respectively. [0065] FUS 8469:13447 (AltMet) (ATP synthase FO subunit 8 to NADH dehydrogenase subunit) (SEQ ID No: 2) [0066] FUS 10744:14124 (NADH dehydrogenase subunit 4L (ND4L) to NADH dehydrogenase subunit 5 (ND5)) (SEQ ID No: 3) [0067] FUS 7974:15496 (Cytochrome c oxidase subunit II (COII) to Cytochrome b (Cytb)) (SEQ ID No: 4) [0068] FUS 7992:15730 (Cytochrome c oxidase subunit II (COII) to Cytochrome b (Cytb)) (SEQ ID No: 5) [0069] FUS 8210:15339 (Cytochrome c oxidase subunit II (COII) to Cytochrome b (Cytb)) (SEQ ID No: 6) [0070] FUS 8828:14896 (ATP synthase FO subunit 6 (ATPase6) to Cytochrome b (Cytb)) (SEQ ID No: 7) [0071] FUS 10665:14856 (NADH dehydrogenase subunit 4L (ND4L) to Cytochrome b (Cytb)) (SEQ ID No: 8) [0072] FUS 6075:13799 (Cytochrome c oxidase subunit I (COI) to NADH dehydrogenase subunit 5 (ND5)) (SEQ ID No: 9) [0073] FUS 6325:13989 (Cytochrome c oxidase subunit I (COI) to NADH dehydrogenase subunit 5 (ND5)) (SEQ ID No: 10) [0074] FUS 7438:13476 (Cytochrome c oxidase subunit I (COI) to NADH dehydrogenase subunit 5 (ND5)) (SEQ ID No: 11) [0075] FUS 7775:13532 (Cytochrome c oxidase subunit II (COII) to NADH dehydrogenase subunit 5 (ND5)) (SEQ ID No: 12) [0076] FUS 8213:13991 (Cytochrome c oxidase subunit II (COII) to NADH dehydrogenase subunit 5 (ND5)) (SEQ ID No: 13) [0077] FUS 9191:12909 (ATP synthase FO subunit 6 (ATPase6) to NADH dehydrogenase subunit 5 (ND5)) (SEQ ID No: 14) [0078] FUS 9574:12972 (Cytochrome c oxidase subunit III (COIII) to NADH dehydrogenase subunit 5 (ND5)) (SEQ ID No: 15) [0079] FUS 10367:12829 (NADH dehydrogenase subunit 3 (ND3) to NADH dehydrogenase subunit 5 (ND5)) (SEQ ID No: 16) [0080] FUS 8469:13447 (OrigMet) (ATP synthase FO subunit 8 to NADH dehydrogenase subunit) (SEQ ID No: 17) [0081] FUS 9144:13816 ((ATP synthase FO subunit 6 (ATPase6) to NADH dehydrogenase subunit 5 (ND5)) (SEQ ID No: 51)
[0082] The present invention also provides the use of variants or fragments of these sequences for predicting, diagnosing and/or monitoring cancer.
[0083] "Variant", as used herein, refers to a nucleic acid differing from a mtDNA sequence of the present invention, but retaining essential properties thereof. Generally, variants are overall closely similar, and, in many regions, identical to a select mtDNA sequence. Specifically, the variants of the present invention comprise at least one of the nucleotides of the junction point of the spliced genes, and may further comprise one or more nucleotides adjacent thereto. In one embodiment of the invention, the variant sequence is at least 80%, 85%, 90%, 95%, 96%, 97%, 98% or 99% identical to any one of the mtDNA sequences of the invention, or the complementary strand thereto.
[0084] In the present invention, "fragment" refers to a short nucleic acid sequence which is a portion of that contained in the disclosed genomic sequences, or the complementary strand thereto. This portion includes at least one of the nucleotides comprising the junction point of the spliced genes, and may further comprise one or more nucleotides adjacent thereto. The fragments of the invention are preferably at least about 15 nt, and more preferably at least about 20 nt, still more preferably at least about 30 nt, and even more preferably, at least about 40 nt, at least about 50 nt, at least about 75 nt, or at least about 150 nt in length. A fragment "at least 20 nt in length," for example, is intended to include 20 or more contiguous bases of any one of the mtDNA sequences listed above. In this context "about" includes the particularly recited value, a value larger or smaller by several (5, 4, 3, 2, or 1) nucleotides, at either terminus or at both termini. These fragments have uses that include, but are not limited to, as diagnostic probes and primers as discussed herein. Of course, larger fragments (e.g., 50, 150, 500, 600, 2000 nucleotides) are also contemplated.
[0085] Thus, in specific embodiments of the invention, the mtDNA sequences are selected from the group consisting of:
[0086] SEQ ID NO: 2 (FUS 8469:13447; AltMet)
[0087] SEQ ID NO: 3 (FUS 10744:14124)
[0088] SEQ ID NO: 4 (FUS 7974:15496)
[0089] SEQ ID NO: 5 (FUS 7992:15730)
[0090] SEQ ID NO: 6 (FUS 8210:15339)
[0091] SEQ ID NO: 7 (FUS 8828:14896)
[0092] SEQ ID NO: 8 (FUS 10665:14856)
[0093] SEQ ID NO: 9 (FUS 6075:13799)
[0094] SEQ ID NO: 10 (FUS 6325:13989)
[0095] SEQ ID NO: 11 (FUS 7438:13476)
[0096] SEQ ID NO: 12 (FUS 7775:13532)
[0097] SEQ ID NO: 13 (FUS 8213:13991)
[0098] SEQ ID NO: 14 (FUS 9191:12909)
[0099] SEQ ID NO: 15 (FUS 9574:12972)
[0100] SEQ ID NO: 16 (FUS 10367:12829)
[0101] SEQ ID NO: 17 (FUS 8469:13447; OrigMet)
[0102] SEQ ID NO: 51 (FUS 9144:13816), and
fragments or variants thereof.
[0103] Probes
[0104] Another aspect of the invention is to provide a hybridization probe capable of recognizing an aberrant mtDNA sequence of the invention. As used herein, the term "probe" refers to an oligonucleotide which forms a duplex structure with a sequence in the target nucleic acid, due to complementarity of at least one sequence in the probe with a sequence in the target region. The probe may be labeled, according to methods known in the art.
[0105] Once aberrant mtDNA associated with a particular disease is identified, hybridization of mtDNA to, for example, an array of oligonucleotides can be used to identify particular mutations, however, any known method of hybridization may be used.
[0106] As with the primers of the present invention, probes may be generated directly against exemplary mtDNA fusion molecules of the invention, or to a fragment or variant thereof. For instance, the sequences set forth in SEQ ID NOs: 2-17 and 51 and those disclosed in Table 1 can be used to design primers or probes that will detect a nucleic acid sequence comprising a fusion sequence of interest. As would be understood by those of skill in the art, primers or probes which hybridize to these nucleic acid molecules may do so under highly stringent hybridization conditions or lower stringency conditions, such conditions known to those skilled in the art and found, for example, in Current Protocols in Molecular Biology (John Wiley & Sons, New York (1989)), 6.3.1-6.3.6.
[0107] In specific embodiments of the invention, the probes of the invention contain a sequence complementary to at least a portion of the aberrant mtDNA comprising the junction point of the spliced genes. This portion includes at least one of the nucleotides involved in the junction point A:B, and may further comprise one or more nucleotides adjacent thereto. In this regard, the present invention encompasses any suitable targeting mechanism that will select an mtDNA molecule using the nucleotides involved and/or adjacent to the junction point A:B.
[0108] Various types of probes known in the art are contemplated by the present invention. For example, the probe may be a hybridization probe, the binding of which to a target nucleotide sequence can be detected using a general DNA binding dye such as ethidium bromide, SYBR.RTM. Green, SYBR.RTM. Gold and the like. Alternatively, the probe can incorporate one or more detectable labels. Detectable labels are molecules or moieties a property or characteristic of which can be detected directly or indirectly and are chosen such that the ability of the probe to hybridize with its target sequence is not affected. Methods of labelling nucleic acid sequences are well-known in the art (see, for example, Ausubel et al., (1997 & updates) Current Protocols in Molecular Biology, Wiley & Sons, New York).
[0109] Labels suitable for use with the probes of the present invention include those that can be directly detected, such as radioisotopes, fluorophores, chemiluminophores, enzymes, colloidal particles, fluorescent microparticles, and the like. One skilled in the art will understand that directly detectable labels may require additional components, such as substrates, triggering reagents, light, and the like to enable detection of the label. The present invention also contemplates the use of labels that are detected indirectly.
[0110] The probes of the invention are preferably at least about 15 nt, and more preferably at least about 20 nt, still more preferably at least about 30 nt, and even more preferably, at least about 40 nt, at least about 50 nt, at least about 75 nt, or at least about 150 nt in length. A probe of "at least 20 nt in length," for example, is intended to include 20 or more contiguous bases that are complementary to an mtDNA sequence of the invention. Of course, larger probes (e.g., 50, 150, 500, 600, 2000 nucleotides) may be preferable.
[0111] The probes of the invention will also hybridize to nucleic acid molecules in biological samples, thereby enabling the methods of the invention. Accordingly, in one aspect of the invention, there is provided a hybridization probe for use in the detection of cancer, wherein the probe is complementary to at least a portion of an aberrant mtDNA molecule. In another aspect the present invention provides probes and a use of (or a method of using) such probes for the detection of colorectal cancer, lung cancer, breast cancer, ovarian cancer, testicular, cancer, prostate cancer and/or melanoma skin cancer.
[0112] Assays
[0113] Measuring the level of aberrant mtDNA in a biological sample can determine the presence of one or more cancers in a subject. The present invention, therefore, encompasses methods for predicting, diagnosing or monitoring cancer, comprising obtaining one or more biological samples, extracting mtDNA from the samples, and assaying the samples for aberrant mtDNA by: quantifying the amount of one or more aberrant mtDNA sequences in the sample and comparing the quantity detected with a reference value. As would be understood by those of skill in the art, the reference value is based on whether the method seeks to predict, diagnose or monitor cancer. Accordingly, the reference value may relate to mtDNA data collected from one or more known non-cancerous biological samples, from one or more known cancerous biological samples, and/or from one or more biological samples taken over time.
[0114] In one aspect, the invention provides a method of detecting cancer in a mammal, the method comprising assaying a tissue sample from the mammal for the presence of an aberrant mitochondrial DNA described above. The present invention also provides for methods comprising assaying a tissue sample from the mammal by hybridizing the sample with at least one hybridization probe. The probe may be generated against a mutant mitochondrial DNA sequence of the invention as described herein.
[0115] In another aspect, the invention provides a method as above, wherein the assay comprises:
[0116] a) conducting a hybridization reaction using at least one of the probes to allow the at least one probe to hybridize to a complementary aberrant mitochondrial DNA sequence;
[0117] b) quantifying the amount of the at least one aberrant mitochondrial DNA sequence in the sample by quantifying the amount of the mitochondrial DNA hybridized to the at least one probe; and,
[0118] c) comparing the amount of the mitochondrial DNA in the sample to at least one known reference value.
[0119] Also included in the present invention are methods for predicting, diagnosing or monitoring cancer comprising diagnostic imaging assays as described below. The diagnostic assays of the invention can be readily adapted for high-throughput. High-throughput assays provide the advantage of processing many samples simultaneously and significantly decrease the time required to screen a large number of samples. The present invention, therefore, contemplates the use of the nucleotides of the present invention in high-throughput screening or assays to detect and/or quantitate target nucleotide sequences in a plurality of test samples.
[0120] Fusion Transcripts
[0121] The present invention further provides the identification of fusion transcripts and associated hybridization probes useful in methods for predicting, diagnosing and/or monitoring cancer. One of skill in the art will appreciate that such molecules may be derived through the isolation of naturally-occurring transcripts or, alternatively, by the recombinant expression of mtDNAs isolated according to the methods of the invention. As discussed, such mtDNAs typically comprise a spliced gene having the initiation codon from the first gene and the termination codon of the second gene. Accordingly, fusion transcripts derived therefrom comprise a junction point associated with the spliced genes.
[0122] Detection of Fusion Transcripts
[0123] Naturally occurring fusion transcripts can be extracted from a biological sample and identified according to any suitable method known in the art, or may be conducted according to the methods described in the examples. In one embodiment of the invention, stable polyadenylated fusion transcripts are identified using Oligo(dT) primers that target transcripts with poly-A tails, followed by RT-PCR using primer pairs designed against the target transcript.
[0124] The following exemplary fusion transcripts were detected using such methods and found useful in predicting, diagnosing and/or monitoring cancer as indicated in the examples. Likewise, fusion transcripts derived from the ORF sequences identified in Table 1 may be useful in predicting, diagnosing and/or monitoring cancer according to the assays and methods of the present invention.
[0125] SEQ ID NO: 18 (Transcripts 1;8469:13447; AltMet)
[0126] SEQ ID NO: 19 (Transcript 2;10744:14124)
[0127] SEQ ID NO: 20 (Transcript 3;7974:15496)
[0128] SEQ ID NO: 21 (Transcript 4;7992:15730)
[0129] SEQ ID NO: 22 (Transcript 5; 8210:15339)
[0130] SEQ ID NO: 23 (Transcript 6;8828:14896)
[0131] SEQ ID NO: 24 (Transcript 7;10665:14856)
[0132] SEQ ID NO: 25 (Transcript 8;6075:13799)
[0133] SEQ ID NO: 26 (Transcript 9;6325:13989)
[0134] SEQ ID NO: 27 (Transcript 10;7438:13476)
[0135] SEQ ID NO: 28 (Transcript 11;7775:13532)
[0136] SEQ ID NO: 29 (Transcript 12;8213:13991)
[0137] SEQ ID NO: 30 (Transcript 14;9191:12909)
[0138] SEQ ID NO: 31 (Transcript 15;9574:12972)
[0139] SEQ ID NO: 32 (Transcript 16;10367:12829)
[0140] SEQ ID NO: 33 (Transcript 20; 8469:13447; OrigMet)
[0141] SEQ ID NO: 50 (Transcript 13; 9144:13816)
[0142] Further, fusion transcripts of like character to those described herein are contemplated for use in the field of clinical oncology.
[0143] Fusion transcripts can also be produced by recombinant techniques known in the art. Typically this involves transformation (including transfection, transduction, or infection) of a suitable host cell with an expression vector comprising an mtDNA sequence of interest.
[0144] Variants or fragments of the fusion transcripts identified herein are also provided. Such sequences may adhere to the size limitations and percent identities described above with respect to genomic variants and fragments, or as determined suitable by a skilled technician.
[0145] In addition, putative protein sequences corresponding to transcripts 1-16 and 20 are listed below. These sequences, which encode hypothetical fusion proteins, are provided as a further embodiment of the present invention.
[0146] SEQ ID NO: 34 (Transcripts 1)
[0147] SEQ ID NO: 35 (Transcript 2)
[0148] SEQ ID NO: 36 (Transcript 3)
[0149] SEQ ID NO: 37 (Transcript 4)
[0150] SEQ ID NO: 38 (Transcript 5)
[0151] SEQ ID NO: 39 (Transcript 6)
[0152] SEQ ID NO: 40 (Transcript 7)
[0153] SEQ ID NO: 41 (Transcript 8)
[0154] SEQ ID NO: 42 (Transcript 9)
[0155] SEQ ID NO: 43 (Transcript 10)
[0156] SEQ ID NO: 44 (Transcript 11)
[0157] SEQ ID NO: 45 (Transcript 12)
[0158] SEQ ID NO: 46 (Transcript 14)
[0159] SEQ ID NO: 47 (Transcript 15)
[0160] SEQ ID NO: 48 (Transcript 16)
[0161] SEQ ID NO: 49 (Transcripts 20)
[0162] SEQ ID NO: 52 (Transcript 13)
[0163] Probes
[0164] Once a fusion transcript has been characterized, primers or probes can be developed to target the transcript in a biological sample. Such primers and probes may be prepared using any known method (as described above) or as set out in the examples provided below. A probe may, for example, be generated for the fusion transcript, and detection technologies, such as QuantiGene 2.0.TM. by Panomics.TM., used to detect the presence of the transcript in a sample. Primers and probes may be generated directly against exemplary fusion transcripts of the invention, or to a fragment or variant thereof. For instance, the sequences set forth in SEQ ID NOs: 18-33 and 50 as well as those disclosed in Table 1 can be used to design probes that will detect a nucleic acid sequence comprising a fusion sequence of interest.
[0165] As would be understood by those skilled in the art, probes designed to hybridize to the fusion transcripts of the invention contain a sequence complementary to at least a portion of the transcript expressing the junction point of the spliced genes. This portion includes at least one of the nucleotides complementary to the expressed junction point, and may further comprise one or more complementary nucleotides adjacent thereto. In this regard, the present invention encompasses any suitable targeting mechanism that will select a fusion transcript that uses the nucleotides involved and adjacent to the junction point of the spliced genes.
[0166] Various types of probes and methods of labelling known in the art are contemplated for the preparation of transcript probes. Such types and methods have been described above with respect to the detection of genomic sequences. The transcript probes of the invention are preferably at least about 15 nt, and more preferably at least about 20 nt, still more preferably at least about 30 nt, and even more preferably, at least about 40 nt, at least about 50 nt, at least about 75 nt, or at least about 150 nt in length. A probe of "at least 20 nt in length," for example, is intended to include 20 or more contiguous bases that are complementary to an mtDNA sequence of the invention. Of course, larger probes (e.g., 50, 150, 500, 600, 2000 nucleotides) may be preferable.
[0167] In one aspect, the invention provides a hybridization probe for use in the detection of cancer, wherein the probe is complementary to at least a portion of a mitochondrial fusion transcript provided above.
[0168] In another aspect, the present invention provides probes and a use of (or a method of using) such probes for the detection of colorectal cancer, lung cancer, breast cancer, ovarian cancer, testicular cancer, prostate cancer or melanoma skin cancer.
[0169] Assays
[0170] Measuring the level of mitochondrial fusion transcripts in a biological sample can determine the presence of one or more cancers in a subject. The present invention, therefore, provides methods for predicting, diagnosing or monitoring cancer, comprising obtaining one or more biological samples, extracting mitochondrial RNA from the samples, and assaying the samples for fusion transcripts by: quantifying the amount of one or more fusion transcripts in the sample and comparing the quantity detected with a reference value. As would be understood by those of skill in the art, the reference value is based on whether the method seeks to predict, diagnose or monitor cancer. Accordingly, the reference value may relate to transcript data collected from one or more known non-cancerous biological samples, from one or more known cancerous biological samples, and/or from one or more biological samples taken over time.
[0171] In one aspect, the invention provides a method of detecting a cancer in a mammal, the method comprising assaying a tissue sample from said mammal for the presence of at least one fusion transcript of the invention by hybridizing said sample with at least one hybridization probe having a nucleic acid sequence complementary to at least a portion of the mitochondrial fusion transcript.
[0172] In another aspect, the invention provides a method as above, wherein the assay comprises:
[0173] a) conducting a hybridization reaction using at least one of the above-noted probes to allow the at least one probe to hybridize to a complementary mitochondrial fusion transcript;
[0174] b) quantifying the amount of the at least one mitochondrial fusion transcript in the sample by quantifying the amount of the transcript hybridized to the at least one probe; and,
[0175] c) comparing the amount of the mitochondrial fusion transcript in the sample to at least one known reference value.
[0176] As discussed above, the diagnostic assays of the invention may also comprise diagnostic methods and screening tools as described herein and can be readily adapted for high-throughput. The present invention, therefore, contemplates the use of the fusion transcripts and associated probes of the present invention in high-throughput screening or assays to detect and/or quantitate target nucleotide sequences in a plurality of test samples.
[0177] Diagnostic Methods and Screening Tools
[0178] Methods and screening tools for diagnosing specific diseases or identifying specific mitochondrial mutations are also herein contemplated. Any known method of hybridization may be used to carry out such methods including, without limitation, probe/primer based technologies such as branched DNA and qPCR, both single-plex and multi-plex. Array technology, which has oligonucleotide probes matching the wild type or mutated region, and a control probe, may also be used. Commercially available arrays such as microarrays or gene chips are suitable. These arrays contain thousands of matched and control pairs of probes on a slide or microchip, and are capable of sequencing the entire genome very quickly. Review articles describing the use of microarrays in genome and DNA sequence analysis are available on-line.
[0179] Screening tools designed to identify targets which are relevant to a given biological condition may include specific arrangements of nucleic acids associated with a particular disease or disorder. Thus, in accordance with one embodiment of the invention, there is provided a screening tool comprised of a microarray having 10's, 100's, or 1000's of mitochondrial fusion transcripts for identification of those associated with one or more cancers. In accordance with another embodiment, there is provided a screening tool comprised of a microarray having 10's, 100's, or 1000's of mitochondrial DNAs corresponding to mitochondrial fusion transcripts for identification of those associated with one or more cancers. In a further embodiment, there is provided a screening tool comprised of a multiplexed branched DNA assay having 10's, 100's, or 1000's of mitochondrial fusion transcripts for identification of those associated with one or more cancers. In yet another embodiment of the invention, there is provided a screening tool comprised of a multiplexed branched DNA assay having 10's, 100's, or 1000's of mitochondrial DNAs corresponding to mitochondrial fusion transcripts for identification of those associated with one or more cancers.
[0180] Approaches useful in the field of clinical oncology are also herein contemplated and may include such diagnostic imaging techniques as Positron Emission Tomography (PET), contrast Magnetic Resonance Imaging (MRI) or the like. These diagnostic methods are well known to those of skill in the art and are useful in the diagnosis and prognosis of cancer.
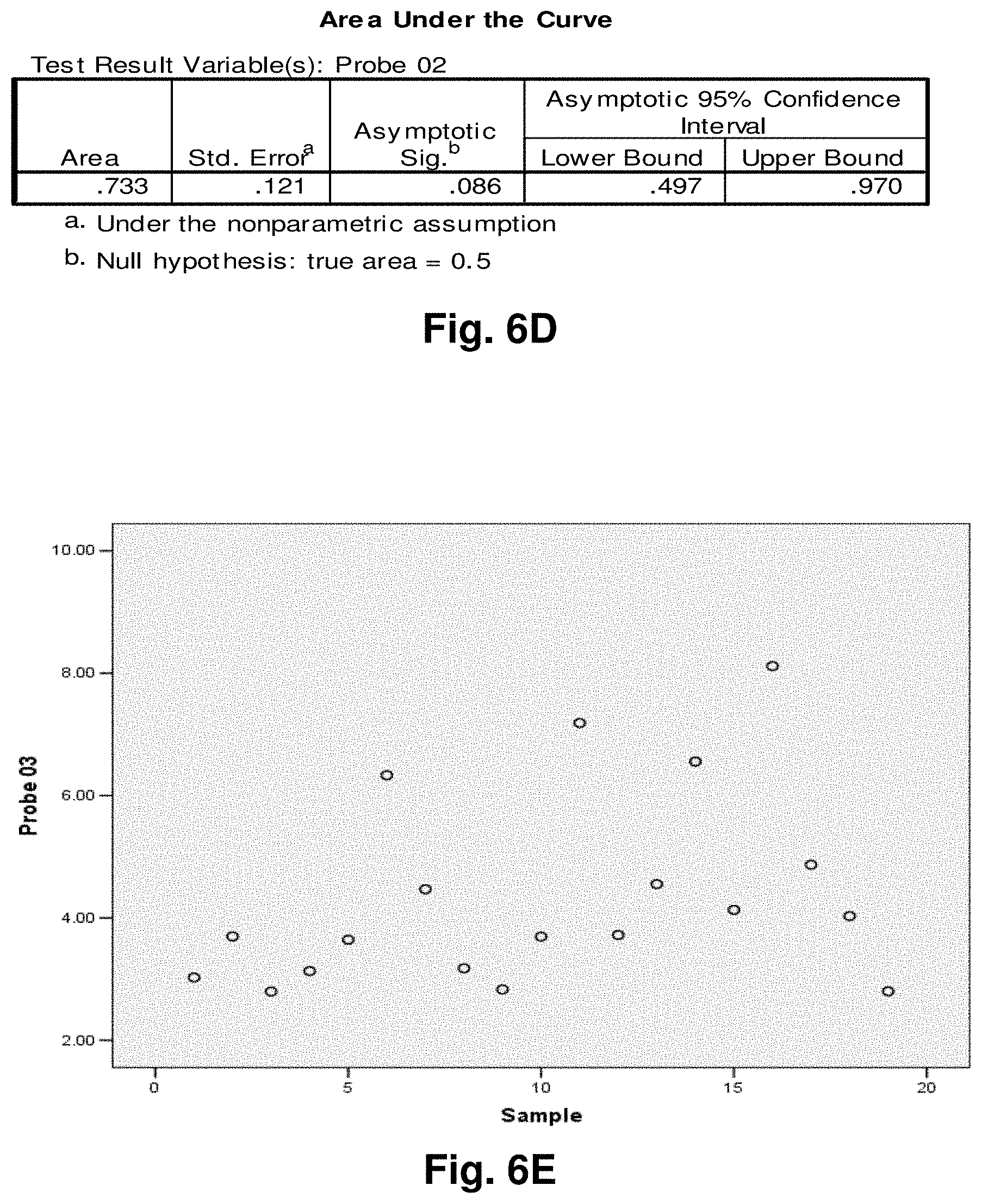
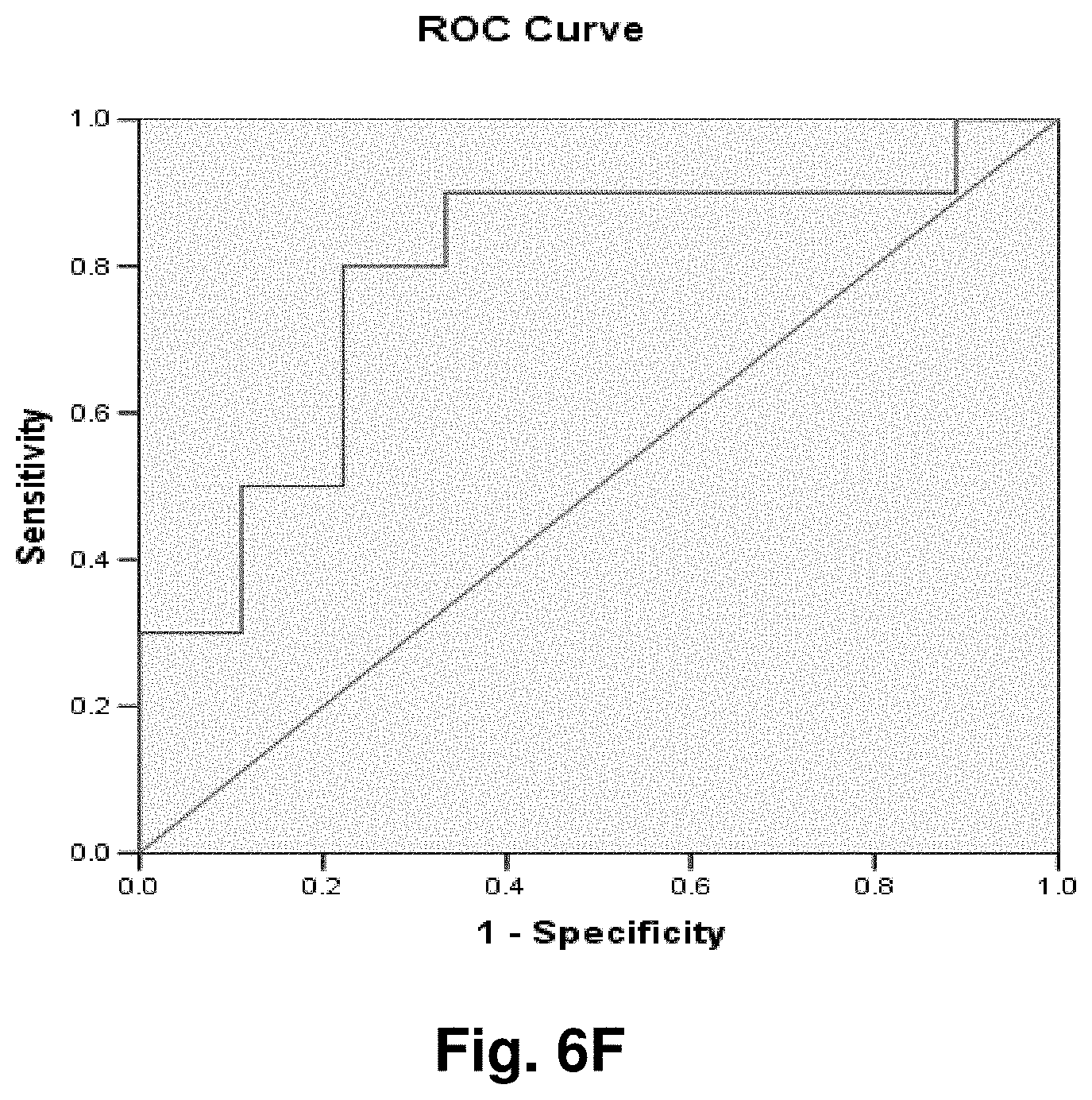
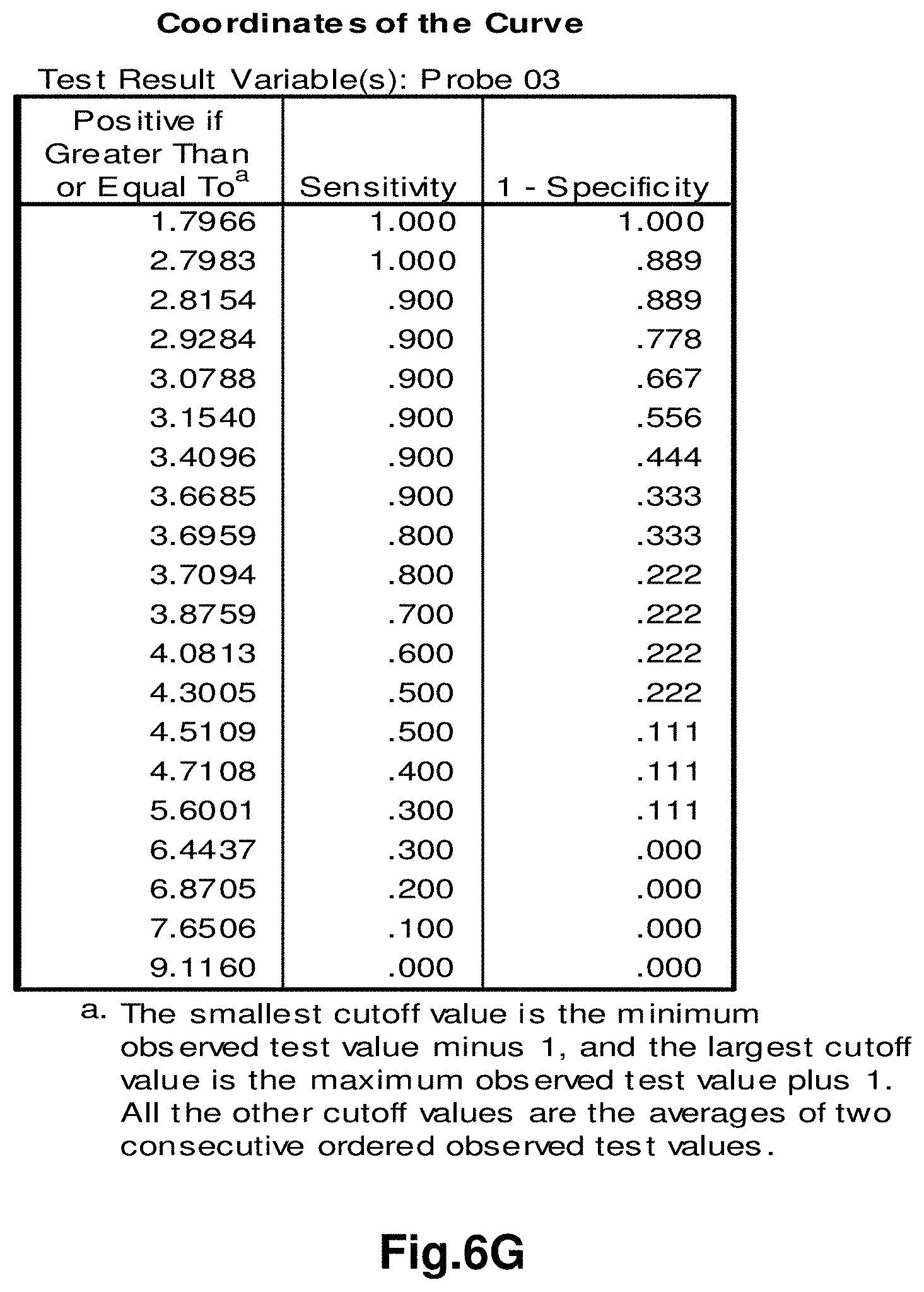
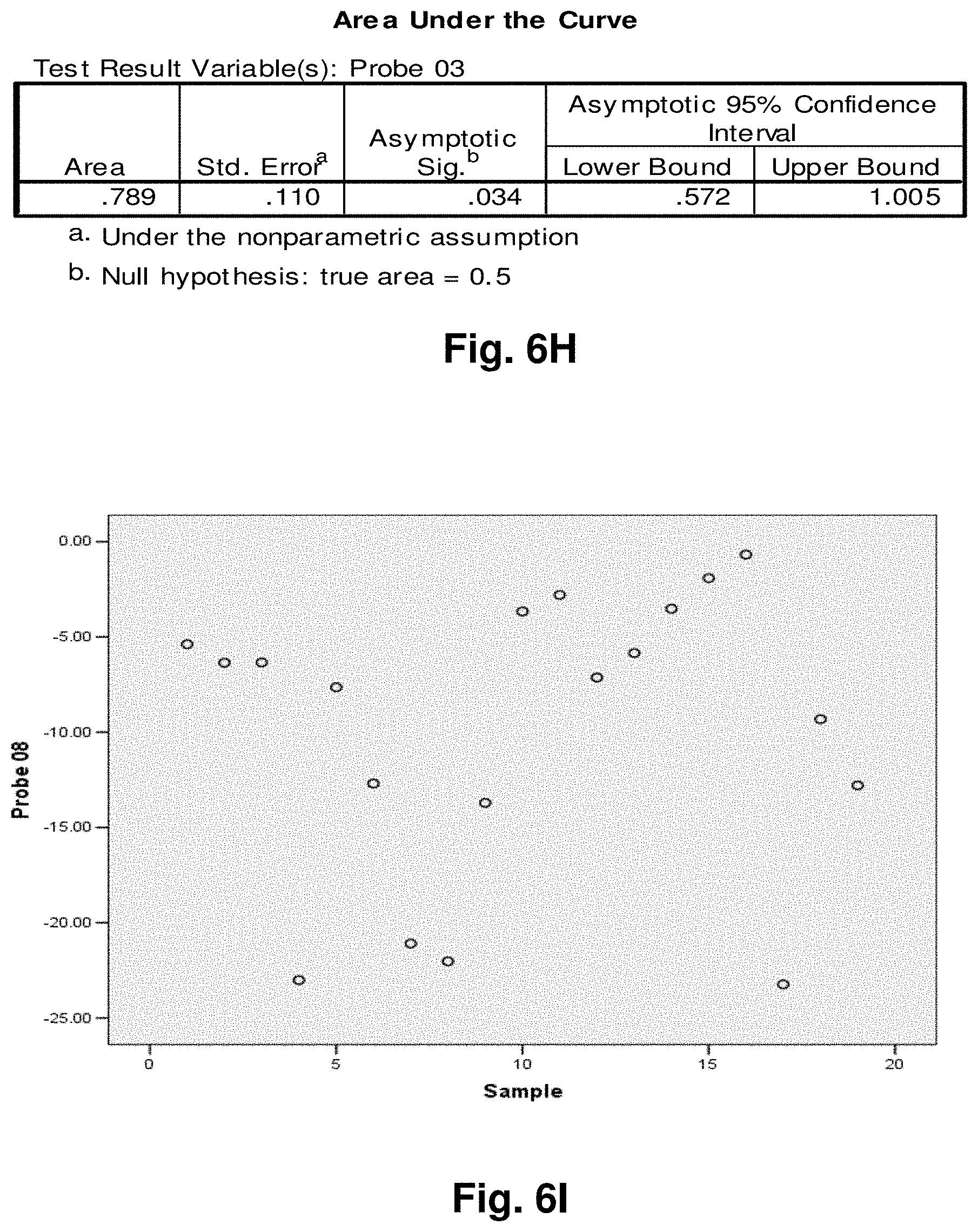
[0181] Diagnostic Monitoring
[0182] The methods of the present invention may further comprise the step of recommending a monitoring regime or course of therapy based on the outcome of one or more assays. This allows clinicians to practice personalized medicine; e.g. cancer therapy, by monitoring the progression of the patient's cancer (such as by recognizing when an initial or subsequent mutation occurs) or treatment (such as by recognizing when a mutation is stabilized).
[0183] With knowledge of the boundaries of the sequence variation in hand, the information can be used to diagnose a pre-cancerous condition or existing cancer condition. Further, by quantitating the amount of aberrant mtDNA in successive samples over time, the progression of a cancer condition can be monitored. For example, data provided by assaying the patient's tissues at one point in time to detect a first set of mutations from wild-type could be compared against data provided from a subsequent assay, to determine if changes in the aberration have occurred.
[0184] Where a mutation is found in an individual who has not yet developed symptoms of cancer, the mutation may be indicative of a genetic susceptibility to develop a cancer condition. A determination of susceptibility to disease or diagnosis of its presence can further be evaluated on a qualitative basis based on information concerning the prevalence, if any, of the cancer condition in the patient's family history and the presence of other risk factors, such as exposure to environmental factors and whether the patient's cells also carry a mutation of another sort.
[0185] Biological Sample
[0186] The present invention provides for diagnostic tests which involve obtaining or collecting one or more biological samples. In the context of the present invention, "biological sample" refers to a tissue or bodily fluid containing cells from which mtDNA and mtRNA can be obtained. For example, the biological sample can be derived from tissue including, but not limited to, skin, lung, breast, prostate, nervous, muscle, heart, stomach, colon, rectal tissue and the like; or from blood, saliva, cerebral spinal fluid, sputa, urine, mucous, synovial fluid, peritoneal fluid, amniotic fluid and the like. The biological sample may be obtained from a cancerous or non-cancerous tissue and may be, but is not limited to, a surgical specimen or a biopsy specimen.
[0187] The biological sample can be used either directly as obtained from the source or following a pre-treatment to modify the character of the sample. Thus, the biological sample can be pre-treated prior to use by, for example, preparing plasma or serum from blood, disrupting cells, preparing liquids from solid materials, diluting viscous fluids, filtering liquids, distilling liquids, concentrating liquids, inactivating interfering components, adding reagents, and the like.
[0188] One skilled in the art will understand that more than one sample type may be assayed at a single time (i.e. for the detection of more than one cancer). Furthermore, where a course of collections are required, for example, for the monitoring of cancer over time, a given sample may be diagnosed alone or together with other samples taken throughout a test period. In this regard, biological samples may be taken once only, or at regular intervals such as biweekly, monthly, semi-annually or annually.
[0189] Kits
[0190] The present invention provides diagnostic/screening kits for detecting cancer in a clinical environment. Such kits may include one or more sampling means, in combination with one or more probes according to the present invention.
[0191] The kits can optionally include reagents required to conduct a diagnostic assay, such as buffers, salts, detection reagents, and the like. Other components, such as buffers and solutions for the isolation and/or treatment of a biological sample, may also be included in the kit. One or more of the components of the kit may be lyophilised and the kit may further comprise reagents suitable for the reconstitution of the lyophilised components.
[0192] Where appropriate, the kit may also contain reaction vessels, mixing vessels and other components that facilitate the preparation of the test sample. The kit may also optionally include instructions for use, which may be provided in paper form or in computer-readable form, such as a disc, CD, DVD or the like.
[0193] In one embodiment of the invention there is provided a kit for diagnosing cancer comprising sampling means and a hybridization probe of the invention.
[0194] Various aspects of the invention will be described by illustration using the following examples. The examples provided herein serve only to illustrate certain specific embodiments of the invention and are not intended to limit the scope of the invention in any way.
EXAMPLES
Example 1: Detection of Mitochondrial Fusion Transcripts
[0195] The mitochondrial 4977 "common deletion" and a 3.4 kb deletion previously identified by the present Applicant in PCT application no. PCT/CA2007/001711 (the entire contents of which are incorporated by reference) result in unique open reading frames having active transcripts as identified by oligo-dT selection in prostate tissue (FIGS. 2 and 3). Examination of breast tissue samples also reveals the presence of a stable polyadenylated fusion transcript resulting from the 3.4 kb deletion (FIG. 4).
[0196] Reverse Transcriptase-PCR Protocol for Deletion Transcript Detection
[0197] RNA Isolation cDNA Synthesis
[0198] Total RNA was isolated from snap frozen prostate and breast tissue samples (both malignant and normal samples adjacent to tumours) using the Aurum.TM. Total RNA Fatty and Fibrous Tissue kit (Bio-Rad, Hercules, Calif.) following the manufacturer's instructions. Since in this experiment, genomic DNA contamination was to be avoided, a DNase I treatment step was included, using methods as commonly known in the art. RNA quantity and quality were determined with an ND-1000 spectrophotometer (NanoDrop.RTM. technologies). From a starting material of about 100 g, total RNA concentrations varied from 100-1000 ng/ul with a 260/280 ratio between 1.89-2.10. RNA concentrations were adjusted to 100 ng/ul and 2 ul of each template were used for first strand DNA synthesis with SuperScript.TM. First-Strand Synthesis System for RT-PCR (Invitrogen) following the manufacturer's instructions. In order to identify stable polyadenylated fusion transcripts, Oligo(dT) primers that target transcripts with poly-A tails were used.
[0199] PCR
[0200] Real time PCR was performed using 5 ul of each cDNA template with the iQ.TM. SYBR.RTM. Green Supermix (Bio-Rad, Hercules, Calif.) on DNA Engine Opticon.RTM. 2 Continuous Fluorescence Detection System (Bio-Rad, Hercules, Calif.). The primer pairs targeting the 4977 bp deletion are; 8416F 5'-CCTTACACTATTCCTCATCAC-3' (SEQ ID NO: 53), 13637R 5'-TGACCTGTTAGGGTGAGAAG-3' (SEQ ID NO: 54), and those for the 3.4 kb deletion are; ND4LF 5'-TCGCTCACACCTCATATCCTC-3' (SEQ ID NO: 55), ND5R 5'-TGTGATTAGGAGTAGGGTTAGG-3' (SEQ ID NO: 56). The reaction cocktail included: 2.times. SYBR.RTM. Green Supermix (100 mM KCL, 40 mM Tris-HCl, pH 8.4, 0.4 mM of each dNTP [dATP, dCTP, dGTP, and dTTP], iTaq.TM. DNA polymerase, 50 units/ml, 6 mM MgCl.sub.2, SYBR.RTM. Green 1, 20 nM flourescein, and stabilizers), 250 nM each of primers, and ddH.sub.2O. PCR cycling parameters were as follows; (1) 95.degree. C. for 2 min, (2) 95.degree. C. for 30 sec, (3) 55.degree. C. (for the 4977 bp deletion) and 63.degree. C. (for the 3.4 kb deletion) for 30 sec, (4) 72.degree. C. for 45 sec, (5) plate read, followed by 39 cycles of steps 3 to 5, and final incubation at 4.degree. C. Apart from cycling threshold and melting curve analysis, samples were run on agarose gels for specific visualization of amplification products (see FIGS. 2 to 4).
[0201] FIG. 2 is an agarose gel showing polyadenalated fusion transcripts in prostate samples invoked by the loss of 3.4 kb from the mitochondrial genome. Legend for FIG. 2: B-blank, Lanes 1-6 transcripts detected in cDNA; lanes 7-12 no reverse transcriptase (RT) controls for samples in lanes 1-6.
[0202] FIG. 3 shows polyadenalated fusion transcripts in prostate samples invoked by the loss of the 4977 kb common deletion. Legend for FIG. 3: B-blank, Lanes 1-6 transcripts detected in cDNA; lanes 7-12 no RT controls for samples in lanes 1-6.
[0203] FIG. 4 shows polyadenalated fusion transcripts in breast samples invoked by the loss of 3.4 kb from the mtgenome. Legend for FIG. 4: Lanes 2-8 transcripts from breast cDNAs; lane 9 negative (water) control; lanes 10 and 11, negative, no RT, controls for samples in lanes 2 and 3.
[0204] These results demonstrate the existence of stable mitochondrial fusion transcripts.
Example 2: Identification and Targeting of Fusion Products
[0205] Various hybridization probes were designed to detect, and further demonstrate the presence of novel transcripts resulting from mutated mitochondrial genomes, such as the 3.4 kb deletion. For this purpose, a single-plex branched DNA platform for quantitative gene expression analysis (QuantiGene 2.0.TM., Panomics.TM.) was utilized. The specific deletions and sequences listed in this example are based on their relative positions with the entire mtDNA genome, which is recited in SEQ ID NO: 1. The nucleic acid sequences of the four transcript to which the probes were designed in this example are identified herein as follows: Transcript 1 (SEQ ID NO: 18), Transcript 2 (SEQ ID NO: 19), Transcript 3 (SEQ ID NO: 20) and Transcript 4 (SEQ ID NO: 21).
[0206] An example of a continuous transcript from the 3.4 kb mitochondrial genome deletion occurs with the genes ND4L (NADH dehydrogenase subunit 4L) and ND5 (NADH dehydrogenase subunit 5). A probe having a complementary sequence to SEQ ID NO: 19, was used to detect transcript 2. The repetitive elements occur at positions 10745-10754 in ND4L and 14124-14133 in ND5.
[0207] The 3.4 kb deletion results in the removal of the 3' end of ND4L, the full ND4 gene, tRNA histidine, tRNA serine2, tRNA leucine2, and the majority of the 5' end of ND5 (see FIG. 5a), resulting in a gene splice of ND4L and ND5 with a junction point of 10744 (ND4L):14124 (ND5) (FIG. 5b). SEQ ID NO: 3 is the complementary DNA sequence to the RNA transcript (SEQ ID NO: 19) detected in the manner described above.
[0208] Similarly, transcript 1 is a fusion transcript between ATPase 8 and ND5 associated with positions 8469:13447 (SEQ ID NO: 18). Transcripts 3 and 4 (SEQ ID NO: 20 and SEQ ID NO: 21, respectively) are fusion transcripts between COII and Cytb associated with nucleotide positions 7974:15496 and 7992:15730 respectively. Table 3 provides a summary of the relationships between the various sequences used in this example. Table 3 includes the detected fusion transcript and the DNA sequence complementary to the fusion transcript detected.
Example 3: Application to Prostate Cancer
[0209] Using the four fusion transcripts, i.e. transcripts 1 to 4, discussed above, two prostate tissue samples from one patient were analyzed to assess the quantitative difference of the novel predicted fusion transcripts. The results of the experiment are provided in Table 2 below, wherein "Homog 1" refers to the homogenate of frozen prostate tumour tissue from a patient and "Homog 2" refers to the homogenate of frozen normal prostate tissue adjacent to the tumour of the patient. These samples were processed according to the manufacturer's protocol (QuantiGene.RTM. Sample Processing Kit for Fresh or Frozen Animal Tissues; and QuantiGene.RTM. 2.0 Reagent System User Manual) starting with 25.8 mg of Homog 1 and 28.9 mg of Homog 2 (the assay setup is shown in Tables 5a and 5b).
[0210] Clearly demonstrated is an increased presence of mitochondrial fusion transcripts in prostate cancer tissue compared to normal adjacent prostate tissue. The fusion transcript is present in the normal tissue, although at much lower levels. The relative luminescence units (RLU) generated by hybridization of a probe to a target transcript are directly proportional to the abundance of each transcript. Table 2 also indicates the coefficients of variation, CV, expressed as a percentage, of the readings taken for the samples. The CV comprises the Standard deviation divided by the average of the values. The significance of such stably transcribed mitochondrial gene products in cancer tissue has implications in disease evolution and progression.
Example 4: Application to Breast Cancer
[0211] Using the same protocol from Example 3 but focusing only on Transcript 2, the novel fusion transcript associated with the 3.4 kb mtgenome deletion, analyses were conducted on two samples of breast tumour tissue and two samples of tumour-free tissues adjacent to those tumours, as well as three samples of prostate tumour tissue, one sample comprising adjacent tumour-free tissue. Results for this example are provided in Table 4. The prostate tumour tissue sample having a corresponding normal tissue section demonstrated a similar pattern to the prostate sample analyzed in Example 3 in that the tumour tissue had approximately 2 times the amount of the fusion transcript than did the normal adjacent tissue. The breast tumour samples demonstrated a marked increase in the fusion transcript levels when compared to the adjacent non-tumour tissues. A 1:100 dilution of the homogenate was used for this analysis as it performed most reproducibly in the experiment cited in Example 3.
[0212] Thus, the above discussed results illustrate the application of the transcripts of the invention in the detection of tumours of both prostate and breast tissue.
Example 5: Application to Colorectal Cancer
[0213] This study sought to determine the effectiveness of several transcripts of the invention in detecting colorectal cancer. A total of 19 samples were prepared comprising nine control (benign) tissue samples (samples 1 to 9) and ten tumour (malignant) tissue samples (samples 10 to 19). The samples were homogenized according to the manufacturer's recommendations (Quantigene.RTM. Sample Processing Kit for Fresh or Frozen Animal Tissues; and Quantigene 2.0 Reagent System User Manual). Seven target transcripts and one housekeeper transcript were prepared in the manner as outlined above in previous examples. The characteristics of the transcripts are summarized as follows:
TABLE-US-00001 TABLE 7 Characteristics of Breast Cancer Transcripts Transcript ID Junction Site Gene Junction 2 10744:14124 ND4L:ND5 3 7974:15496 COII:Cytb 10 7438:13476 COI:ND5 11 7775:13532 COII:ND5 12 8213:13991 COII:ND5 Peptidylpropyl isomerase B (PPIB) N/A N/A ("housekeeper")
[0214] It is noted that transcripts 2 and 3 are the same as those discussed above with respect to Examples 3 and 4.
[0215] Homogenates were prepared using approximately 25 mg of tissue from OCT blocks and diluted 1:1 for transcripts 2 and 4, and 1:8 for transcripts 10 and 11. The quantity of the transcripts was measured in Relative Luminenscence Units RLU on a Glomax.TM. Multi Detection System (Promega). All samples were assayed in triplicate for each transcript. Background measurements (no template) were done in triplicate as well. The analysis accounted for background by subtracting the lower limit from the RLU values for the samples. Input RNA was accounted for by using the formula log.sub.2 a RLU-log.sub.2 h RLU where a is the target fusion transcript and h is the housekeeper transcript.
[0216] The analysis of the data comprised the following steps:
[0217] a) Establish CV's (coefficients of variation) for triplicate assays; acceptable if 15%.
[0218] b) Establish average RLU value for triplicate assays of target fusion transcript (a) and housekeeper transcript (h).
[0219] c) Establish lower limit from triplicate value of background RLU (I).
[0220] d) Subtract lower limit (I) from (a).
[0221] e) Calculate log.sub.2 a RLU-log 2 h RLU.
[0222] Summary of Results:
[0223] The results of the above analysis are illustrated in FIGS. 6a to 6g, which comprise plots of the log.sub.2 a RLU-log 2 h RLU against sample number. Also illustrated are the respective ROC (Receiver Operating Characteristic) curves determined from the results for each transcript.
[0224] Transcript 2:
[0225] There exists a statistically significant difference between the means (p<0.10) of the normal and malignant groups (p>0.09), using a cutoff value of 3.6129 as demonstrated by the ROC curve results in a sensitivity of 60% and specificity of 89% and the area under the curve is 0.73 indicating fair test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0226] Transcript 3:
[0227] There exists a statistically significant difference between the means (p<0.05) of the normal and malignant groups (p=0.03), using a cutoff value of 4.0813 as demonstrated by the ROC curve results in a sensitivity of 60% and specificity of 78% and the area under the curve is 0.79 indicating fair to good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0228] Transcript 8:
[0229] There exists a statistically significant difference between the means (p<0.1) of the normal and malignant groups (p=0.06). Using a cutoff value of -6.0975 as demonstrated by the ROC curve results in a sensitivity of 60% and specificity of 89% and the area under the curve is 0.76 indicating fair test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0230] Transcript 9:
[0231] There exists a statistically significant difference between the means (p<0.1) of the normal and malignant groups (p=0.06). Using a cutoff value of -7.5555 as demonstrated by the ROC curve results in a sensitivity of 60% and specificity of 89% and the area under the curve is 0.76 indicating fair to good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0232] Transcript 10:
[0233] There is a statistically significant difference between the means (100.01) of the normal and malignant groups (p=0.01). Using a cutoff value of -3.8272 as demonstrated by the ROC curve results in a sensitivity of 90% and specificity of 67% and the area under the curve is 0.84, indicating good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0234] Transcript 11:
[0235] There exists a statistically significant difference between the means (p<0.1) of the normal and malignant groups (p=0.06), using a cutoff value of 3.1753 as demonstrated by the ROC curve results in a sensitivity of 70% and specificity of 78% and the area under the curve is 0.76 indicating fair to good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0236] Transcript 12:
[0237] There exists a statistically significant difference between the means (p<0.1) of the normal and malignant groups (p=0.06), using a cut-off value of 3.2626 as demonstrated by the ROC curve results in a sensitivity of 70% and specificity of 78% and the area under the curve is 0.76 indicating fair to good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0238] Conclusions:
[0239] The above results illustrate the utility of transcripts 2, 3, 8, 9, 10, 11, and 12 in the detection of colorectal cancer and in distinguishing malignant from normal colorectal tissue. As indicated above, transcripts 2 and 3 were also found to have utility in the detection of prostate cancer. Transcript 2 was also found to have utility in the detection of breast cancer. Transcript 11 was also found to have utility in the detection of melanoma skin cancer. Transcript 10 was also found to have utility in the detection of lung cancer and melanoma. Transcript 8 was also found to have utility in the detection of lung cancer. Any of the 7 transcripts listed may be used individually or in combination as a tool for the detection of characterization of colorectal cancer in a clinical setting.
Example 6: Application to Lung Cancer
[0240] This study sought to determine the effectiveness of several transcripts of the invention in the detection of lung cancer. As in Example 5, nine control (benign) tissue samples (samples 1 to 9) and ten tumour (malignant) tissue samples (samples 10 to 19) were homogenized according to the manufacturer's recommendations (Quantigene.RTM. Sample Processing Kit for Fresh or Frozen Animal Tissues; and Quantigene 2.0 Reagent System User Manual). Homogenates were diluted 1:8 and the quantity of 4 target transcripts and 1 housekeeper transcript was measured in Relative Luminenscence Units RLU on a Glomax.TM. Multi Detection System (Promega). All samples were assayed in triplicate for each transcript. Background measurements (no template) were done in triplicate as well.
[0241] The following transcripts were prepared for this example:
TABLE-US-00002 TABLE 8 Characteristics of Lung Cancer Transcripts Transcript ID Junction Site Gene Junction 6 8828:14896 ATPase6:Cytb 8 6075:13799 COI:ND5 10 7438:13476 COI:ND5 20 8469:13447 ATPase8:ND5 Peptidylpropyl isomerase B (PPIB) N/A N/A ("housekeeper")
[0242] The tissue samples used in this example had the following characteristics:
TABLE-US-00003 TABLE 9 Characteristics of Lung Cancer Samples Sample Malignant Comments (source of tissue) 1 NO interstitial lung disease 2 NO emphysema 3 NO aneurysm 4 NO bronchopneumonia, COPD 5 NO malignant neoplasm in liver, origin unknown, calcified granulomas in lung 6 NO 12 hours post mortem, mild emphysema 7 NO 12 hours post mortem, large B cell lymphoma, pulmonary edema, pneumonia 8 NO pneumonia, edema, alveolar damage 9 NO congestion and edema 10 YES adenocarcinoma, non-small cell 11 YES small cell 12 YES squamous cell carcinoma, NSC, emphysema 13 YES adenocarcinoma, lung cancer, nsc, metastatic 14 YES squamous cell carcinoma, non-small cell 15 YES mixed squamous and adenocarcinoma 16 YES non-small cell carcinoma, squamous 17 YES small cell carcinoma 18 YES adenocarcinoma, lung cancer, nsc 19 YES adenocarcinoma, lung cancer, nsc, metastatic
[0243] The analysis of data was performed according to the method described in Example 5. The results are illustrated in FIGS. 7a, 7b, 7c and 7d.
[0244] Summary of Results:
[0245] Transcript 6:
[0246] There exists a statistically significant difference between the means (p<0.1) of the normal (benign) and malignant groups (p=0.06), using a cutoff value of -6.5691 as demonstrated by the ROC curve results in a sensitivity of 80% and specificity of 71% and the area under the curve is 0.77, indicating fair test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0247] Transcript 8:
[0248] The difference between the means of the normal and malignant groups is statistically significant, p<0.05 (p=0.02). Using a cutoff value of -9.6166 as demonstrated by the ROC curve results in a sensitivity of 90% and specificity of 86% and the area under the curve is 0.86 indicating good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0249] Transcript 10:
[0250] The difference between the means of the normal and malignant groups is statistically significant, p.ltoreq.0.01 (p=0.01). Using a cutoff value of -10.6717 as demonstrated by the ROC curve results in a sensitivity of 90% and specificity of 86% and the area under the curve is 0.89 indicating good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0251] Transcript 20:
[0252] The difference between the means of the normal and malignant groups is statistically significant, p.ltoreq.0.1 (p=0.1). Using a cutoff value of 2.5071 as demonstrated by the ROC curve results in a sensitivity of 70% and specificity of 71% and the area under the curve is 0.74 indicating fair test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0253] Conclusions:
[0254] The results from example 6 illustrate the utility of transcripts 6, 8, 10, and 20 of the invention in the detection of lung cancer tumours and the distinction between malignant and normal lung tissues. Any of these three transcripts may be used for the detection or characterization of lung cancer in a clinical setting.
Example 7: Application to Melanoma
[0255] This study sought to determine the effectiveness of several transcripts of the invention in the detection of melanomas. In this study a total of 14 samples were used, comprising five control (benign) tissue samples and nine malignant tissue samples. All samples were formalin fixed, paraffin embedded (FFPE). The FFPE tissue samples were sectioned into tubes and homogenized according to the manufacturer's recommendations (Quantigene.RTM. 2.0 Sample Processing Kit for FFPE Samples; and Quantigene 2.0 Reagent System User Manual) such that each sample approximated 20 microns prior to homogenization. Homogenates were diluted 1:4 and the quantity of 7 target transcripts and 1 housekeeper transcript was measured in Relative Luminenscence Units RLU on a Glomax.TM. Multi Detection System (Promega). All samples were assayed in triplicate for each transcript. Background measurements (no template) were done in triplicate as well.
[0256] The 14 tissue samples used in this example had the following characteristics:
TABLE-US-00004 TABLE 10 Characteristics of Melanoma Cancer Samples Sample Malignant Comments (source of tissue) 1 NO breast reduction tissue (skin) 2 NO breast reduction tissue (skin) 3 NO breast reduction tissue (skin) 4 NO breast reduction tissue (skin) 5 NO breast reduction tissue (skin) 6 YES lentigo maligna, (melanoma in situ) invasive melanoma not present 7 YES invasive malignant melanoma 8 YES nodular melanoma, pT3b, associated features of lentigo maligna 9 YES residual superficial spreading invasive malignant melanoma, Clark's level II 10 YES superficial spreading malignant melanoma, Clark's Level II 11 YES nodular malignant melanoma, Clark's level IV 12 YES superficial spreading malignant melanoma in situ, no evidence of invasion 13 YES superficial spreading malignant melanoma, Clark's level II, focally present vertical phase 14 YES superficial spreading malignant melanoma in situ, Clark's level I
[0257] The following transcripts were prepared for this example:
TABLE-US-00005 TABLE 11 Characteristics of Melanoma Cancer Transcripts Transcript ID Junction Site GeneJunction 6 8828:4896 ATPase6:Cytb 10 7438:13476 COI:ND5 11 7775:13532 COII:ND5 14 9191:12909 ATPase6:ND5 15 9574:12972 COIII:ND5 16 10367:12829 ND3:ND5 20 8469:13447 ATPase8:ND5 Peptidylpropyl isomerase B (PPIB) N/A N/A ("housekeeper")
[0258] As indicated, transcripts 10 and 11 were also used in Example 5. The analysis of data was performed according to the method described in Example 5. The results are illustrated in FIGS. 8a-8g.
[0259] Summary of Results:
[0260] Transcript 6:
[0261] There exists a statistically significant difference between the means (p.ltoreq.0.01) of the normal and malignant groups (p=0.01). Further, using a cutoff value of -5.9531 as demonstrated by the ROC curve results in a sensitivity of 89% and specificity of 80% and the area under the curve is 0.96, indicating very good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0262] Transcript 10:
[0263] There exists a statistically significant difference between the means (p.ltoreq.0.05) of the normal and malignant groups (p=0.05), using a cutoff value of -4.7572 as demonstrated by the ROC curve results in a sensitivity of 89% and specificity of 40% and the area under the curve is 0.82, indicating good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0264] Transcript 11:
[0265] There exists a statistically significant difference between the means (p<0.05) of the normal and malignant groups (p=0.02). Further, using a cutoff value of 1.6762 as demonstrated by the ROC curve results in a sensitivity of 78% and specificity of 100% and the area under the curve is 0.89, indicating good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0266] Transcript 14:
[0267] There exists a statistically significant difference between the means (p.ltoreq.0.05) of the normal and malignant groups (p=0.05). Further, using a cutoff value of -4.9118 as demonstrated by the ROC curve results in a sensitivity of 89% and specificity of 60% and the area under the curve is 0.82, indicating good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0268] Transcript 15:
[0269] There exists a statistically significant difference between the means (p<0.1) of the normal and malignant groups (p=0.07), using a cutoff value of -7.3107 as demonstrated by the ROC curve results in a sensitivity of 100% and specificity of 67% and the area under the curve is 0.80, indicating good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0270] Transcript 16:
[0271] There exists a statistically significant difference between the means (p<0.05) of the normal and malignant groups (p=0.03). Further, using a cutoff value of -10.5963 as demonstrated by the ROC curve results in a sensitivity of 89% and specificity of 80% and the area under the curve is 0.878, indicating good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0272] Transcript 20:
[0273] There exists a statistically significant difference between the means (p<0.05) of the normal and malignant groups (p=0.04). Further, using a cutoff value of -8.3543 as demonstrated by the ROC curve results in a sensitivity of 100% and specificity of 80% and the area under the curve is 0.89, indicating good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0274] Conclusions:
[0275] The results from example 7 illustrate the utility of transcripts 6, 10, 11, 14, 15, 16 and 20 of the invention in the detection of malignant melanomas. As indicated above, transcripts 10 and 11 were also found have utility in detecting colorectal cancer while transcript 6 has utility in the detection of lung cancer. A transcript summary by disease is provided at Table 6.
Example 8: Application to Ovarian Cancer
[0276] This study sought to determine the effectiveness of several transcripts of the invention in detecting ovarian cancer. A total of 20 samples were prepared comprising ten control (benign) tissue samples (samples 1 to 10) and ten tumour (malignant) tissue samples (samples 11 to 20). The samples were homogenized according to the manufacturer's recommendations (Quantigene.RTM. Sample Processing Kit for Fresh or Frozen Animal Tissues; and Quantigene 2.0 Reagent System User Manual). Eight target transcripts and one housekeeper transcript were prepared in the manner as outlined above in previous examples.
[0277] The 20 tissue samples used in this example had the following characteristics:
TABLE-US-00006 TABLE 12 Characteristics of Ovarian Cancer Samples Sample Diagnosis Comments 1 Normal follicular cyst 2 Normal fibroma 3 Normal No pathological change in ovaries 4 Normal follicular cysts 5 Normal cellular fibroma 6 Normal benign follicular and simple cysts 7 Normal leiomyomata, corpora albicantia 8 Normal copora albicantia and an epithelial inclusions cysts 9 Normal corpora albicantia 10 Normal corpora albicantia, surface inclusion cysts, follicullar cysts 11 Malignant high grade poorly differentiated papillary serous carcinoma involving omentum 12 Malignant endometrioid adenocarcinoma, well to moderately differentiated with focal serous differentiation 13 Malignant papillary serous carcinoma 14 Malignant mixed epithelial carcinoma predominantly papillary serous carcinoma 15 Malignant High grade: serous carcinoma, papillary and solid growth patterns 16 Malignant High Grade (3/3) Papillary serous carcinoma 17 Malignant papillary serous carcinoma, high nuclear grade 18 Malignant Papillary serous cystadenocarcinomas Grade: III 19 Malignant poorly differentiated papillary serous carcinoma 20 Malignant Well-differentiated adnocarcinoma, Endometrioid type, Grade 1
[0278] The characteristics of the transcripts are summarized as follows:
TABLE-US-00007 TABLE 13 Characteristics of Ovarian Cancer Transcripts Transcript ID Junction Site Gene Junction 1 8469:13447 ATPase8:ND5 2 10744:14124 ND4L:ND5 3 7974:15496 COII:Cytb 6 8828:14896 ATPase6:Cytb 11 7775:13532 COII:ND5 12 8213:13991 COII:ND5 15 9574:12972 COIII:ND5 20 8469:13447 ATPase8:ND5 Ribosomal Protein Large PO (LRP) N/A N/A Housekeeper
[0279] It is noted that transcripts 1, 2, 3, 6, 11, 12, 15 and 20 are the same as those discussed above with respect to Examples 3-7.
[0280] Homogenates were prepared using approximately 25 mg of frozen tissue and diluted 1:4. The quantity of the transcripts was measured in Relative Luminenscence Units RLU on a Glomax.TM. Multi Detection System (Promega). All samples were assayed in triplicate for each transcript. Background measurements (no template) were done in triplicate as well. The analysis accounted for background by subtracting the lower limit from the RLU values for the samples. Input RNA was accounted for by using the formula log.sub.2 a RLU-log.sub.2 h RLU where a is the target fusion transcript and h is the housekeeper transcript.
[0281] The analysis of the data comprised the following steps:
[0282] a) Establish CV's (coefficients of variation) for triplicate assays; acceptable if 15%.
[0283] b) Establish average RLU value for triplicate assays of target fusion transcript (a) and housekeeper transcript (h).
[0284] c) Establish lower limit from triplicate value of background RLU (I).
[0285] d) Subtract lower limit (I) from (a).
[0286] e) Calculate log.sub.2 a RLU-log 2 h RLU.
[0287] Summary of Results:
[0288] The results of the above analysis are illustrated in FIGS. 9a to 9h, which comprise plots of the log.sub.2 a RLU-log 2 h RLU against sample number. Also illustrated are the respective ROC (Receiver Operating Characteristic) curves determined from the results for each transcript.
[0289] Transcript 1:
[0290] There exists a statistically significant difference between the means (p<0.05) of the normal and malignant groups (p=0.002). Using a cutoff value of -11.1503 as demonstrated by the ROC curve results in a sensitivity of 90% and specificity of 80% and the area under the curve is 0.91 indicating very good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0291] Transcript 2:
[0292] There exists a statistically significant difference between the means (p<0.01) of the normal and malignant groups (p=0.001). Using a cutoff value of 0.6962 as demonstrated by the ROC curve results in a sensitivity of 90% and specificity of 100% and the area under the curve is 0.96 indicating very good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0293] Transcript 3:
[0294] There exists a statistically significant difference between the means (p<0.01) of the normal and malignant groups (p=0.000). Using a cutoff value of 0.6754 as demonstrated by the ROC curve results in a sensitivity of 100% and specificity of 100% and the area under the curve is 1.00 indicating excellent test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0295] Transcript 6:
[0296] There exists a statistically significant difference between the means (p<0.01) of the normal and malignant groups (p=0.007). Using a cutoff value of -9.6479 as demonstrated by the ROC curve results in a sensitivity of 90% and specificity of 70% and the area under the curve is 0.86 indicating good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0297] Transcript 11:
[0298] There is a statistically significant difference between the means (p<0.01) of the normal and malignant groups (p=0.000). Using a cutoff value of -1.3794 demonstrated by the ROC curve results in a sensitivity of 100% and specificity of 90% and the area under the curve is 0.99, indicating excellent test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0299] Transcript 12:
[0300] There exists a statistically significant difference between the means (p<0.01) of the normal and malignant groups (p=0.001). Using a cutoff value of -1.2379 as demonstrated by the ROC curve results in a sensitivity of 90% and specificity of 100% and the area under the curve is 0.96 indicating excellent test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0301] Transcript 15:
[0302] There exists a statistically significant difference between the means (p<0.05) of the normal and malignant groups (p=0.023). Using a cut-off value of -8.6926 as demonstrated by the ROC curve results in a sensitivity of 70% and specificity of 80% and the area under the curve is 0.80 indicating good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0303] Transcript 20:
[0304] There exists a statistically significant difference between the means (p<0.01) of the normal and malignant groups (p=0.000). Using a cut-off value of 0.6521 as demonstrated by the ROC curve results in a sensitivity of 100% and specificity of 100% and the area under the curve is 0.76 indicating fair to good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0305] Conclusions:
[0306] The above results illustrate the utility of transcripts 1, 2, 3, 6, 11, 12, 15, and 20 in the detection of ovarian cancer and in distinguishing malignant from normal ovarian tissue. Transcripts 1, 2 and 3 were also found to have utility in the detection of prostate cancer. Transcript 6 was also found to have utility in the detection of melanoma and lung cancer. Transcript 11 was also found to have utility in the detection of melanoma skin cancer, colorectal cancer and testicular cancer. Transcript 12 was also found to have utility in the detection of colorectal cancer and testicular cancer. Transcript 15 was also found to have utility in the detection of melanoma and testicular cancer. Transcript 20 was also found to have utility in the detection of colorectal cancer, melanoma, and testicular cancer. Any of the 8 transcripts listed may be used individually or in combination as a tool for the detection or characterization of ovarian cancer in a clinical setting.
Example 9: Application to Testicular Cancer
[0307] This study sought to determine the effectiveness of several transcripts of the invention in detecting testicular cancer. A total of 17 samples were prepared comprising eight control (benign) tissue samples (samples 1 to 8) and 9 tumour (malignant) tissue samples (samples 9 to 17), 5 of the malignant samples were non-seminomas (samples 9-13) and 4 were seminomas (samples 14-17). The samples were homogenized according to the manufacturer's recommendations (Quantigene.RTM. Sample Processing Kit for Fresh or Frozen Animal Tissues; and Quantigene 2.0 Reagent System User Manual). 10 target transcripts and one housekeeper transcript were prepared in the manner as outlined above in previous examples.
[0308] The 17 tissue samples used in this example had the following characteristics:
TABLE-US-00008 TABLE 14 Characteristics of Testicular Cancer Samples General Stratified Sample Diagnosis Malignant Diagnosis 1 Benign Benign 2 Benign Benign 3 Benign Benign 4 Benign Benign 5 Benign Benign 6 Benign Benign 7 Benign Benign 8 Benign Benign 9 Malignant Non-Seminoma 10 Malignant Non-Seminoma 11 Malignant Non-Seminoma 12 Malignant Non-Seminoma 13 Malignant Non-Seminoma 14 Malignant Seminoma 15 Malignant Seminoma 16 Malignant Seminoma 17 Malignant Seminoma
[0309] The characteristics of the transcripts are summarized as follows:
TABLE-US-00009 TABLE 15 Characteristics of Testicular Cancer Transcripts Transcript ID Junction Site Gene Junction 2 10744:14124 ND4L:ND5 3 7974:15496 COII:Cytb 4 7992:15730 COII:Cytb 11 7775:13532 COII:ND5 12 8213:13991 COII:ND5 13 9144:13816 ATPase6:ND5 15 9574:12972 COIII:ND5 16 10367:12829 ND3:ND5 20 8469:13447 ATPase8:ND5 Peptidylpropyl isomerase B (PPIB) N/A N/A
[0310] It is noted that transcripts 2, 3, 4, 7, 11, 12, 15, 16 and 20 are the same as those discussed above with respect to Examples 3-8.
[0311] Homogenates were prepared using approximately 25 mg of frozen tissue and diluted 1:4. The quantity of the transcripts was measured in Relative Luminenscence Units RLU on a Glomax.TM. Multi Detection System (Promega). All samples were assayed in triplicate for each transcript. Background measurements (no template) were done in triplicate as well. The analysis accounted for background by subtracting the lower limit from the RLU values for the samples. Input RNA was accounted for by using the formula log.sub.2 a RLU-log.sub.2 h RLU where a is the target fusion transcript and h is the housekeeper transcript.
[0312] The analysis of the data comprised the following steps:
[0313] a) Establish CV's (coefficients of variation) for triplicate assays; acceptable if 15%.
[0314] b) Establish average RLU value for triplicate assays of target fusion transcript (a) and housekeeper transcript (h).
[0315] c) Establish lower limit from triplicate value of background RLU (I).
[0316] d) Subtract lower limit (I) from (a).
[0317] e) Calculate log.sub.2 a RLU-log 2 h RLU.
[0318] Summary of Results:
[0319] The results of the above analysis are illustrated in FIGS. 10 to 18, which comprise plots of the log.sub.2 a RLU-log 2 h RLU against sample number. Also illustrated are the respective ROC (Receiver Operating Characteristic) curves determined from the results for each transcript.
[0320] While some transcripts distinguish between benign and malignant testicular tissue, others demonstrate distinction between the tumour subtypes of seminoma and non-seminoma and/or benign testicular tissue. It is therefore anticipated that combining transcripts from each class will facilitate not only detection of testicular cancer but also classification into subtype of seminoma or non-seminomas.
[0321] Transcript 2:
[0322] There exists a statistically significant difference between the means (p<0.05) of the normal group and the malignant seminomas (p=0.02). Using a cutoff value of 1.5621 as demonstrated by the ROC curve results in a sensitivity of 100% and specificity of 100% and the area under the curve is 1.00 indicating excellent test accuracy. There also exists a statistically significant difference between the means (p<0.05) of the malignant seminomas and the malignant non-seminomas (p=0.024). Using a cutoff value of 2.1006 as demonstrated by the ROC curve results in a sensitivity of 100% and specificity of 80% and the area under the curve is 0.90 indicating excellent test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0323] Transcript 3:
[0324] There exists a statistically significant difference between the means (p<0.05) of the normal group and the malignant seminomas (p=0.018). Using a cutoff value of 0.969 as demonstrated by the ROC curve results in a sensitivity of 100% and specificity of 87.5% and the area under the curve is 0.969 indicating excellent accuracy. There also exists a statistically significant difference between the means (p<0.05) of the malignant seminomas and the malignant non-seminomas (p=0.017). Using a cutoff value of 1.8181 as demonstrated by the ROC curve results in a sensitivity of 100% and specificity of 80% and the area under the curve is 0.9 indicating excellent test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0325] Transcript 4:
[0326] There exists a statistically significant difference between the means (p<0.05) of the normal and malignant groups (p=0.034). Using a cutoff value of -9.7628 as demonstrated by the ROC curve results in a sensitivity of 67% and specificity of 100% and the area under the curve is 0.833 indicating good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0327] Transcript 11:
[0328] There exists a statistically significant difference between the means (p<0.05) of the normal group and the malignant seminomas (p=0.016). Using a cutoff value of 0.732 as demonstrated by the ROC curve results in a sensitivity of 100% and specificity of 100% and the area under the curve is 1.00 indicating excellent test accuracy. There also exists a statistically significant difference between the means (p<0.05) of the malignant seminomas and the malignant non-seminomas (p=0.016). Using a cutoff value of 0.9884 as demonstrated by the ROC curve results in a sensitivity of 100% and specificity of 80% and the area under the curve is 0.90 indicating excellent test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0329] Transcript 12:
[0330] There exists a statistically significant difference between the means (p<0.1) of the normal group and the malignant seminomas (p=0.056). Using a cutoff value of 1.5361 as demonstrated by the ROC curve results in a sensitivity of 100% and specificity of 87.5% and the area under the curve is 0.969 indicating excellent test accuracy. There also exists a statistically significant difference between the means (p<0.05) of the malignant seminomas and the malignant non-seminomas (p=0.044). Using a cutoff value of 1.6039 as demonstrated by the ROC curve results in a sensitivity of 100% and specificity of 80% and the area under the curve is 0.9 indicating excellent test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0331] Transcript 13:
[0332] There exists a statistically significant difference between the means (p<0.05) of the normal group and the malignant group (p=0.019). Using a cutoff value of -9.8751 as demonstrated by the ROC curve results in a sensitivity of 87.5% and specificity of 78% and the area under the curve is 0.875 indicating very good test accuracy. There also exists a statistically significant difference between the means (p<0.01) of the malignant non-seminomas and the benign group (p=0.000). Using a cutoff value of -13.9519 as demonstrated by the ROC curve results in a sensitivity of 100% and specificity of 87.5% and the area under the curve is 0.975 indicating excellent test accuracy. There also exists a statistically significant difference between the means (p<0.01) of the malignant seminomas and the malignant non-seminomas (p=0.001). Using a cutoff value of -15.8501 as demonstrated by the ROC curve results in a sensitivity of 100% and specificity of 100% and the area under the curve is 1.00 indicating excellent test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0333] Transcript 15:
[0334] There exists a statistically significant difference between the means (p<0.1) of the normal and malignant groups (p=0.065). Using a cut-off value of -5.4916 as demonstrated by the ROC curve results in a sensitivity of 75% and specificity of 89% and the area under the curve is 0.833 indicating good test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0335] Transcript 16:
[0336] There exists a statistically significant difference between the means (p<0.05) of the normal and malignant groups including both seminomas and non-seminomas (p=0.037). Using a cut-off value of -6.448 as demonstrated by the ROC curve results in a sensitivity of 89% and specificity of 75% and the area under the curve is 0.806 indicating good test accuracy. There also exists a statistically significant difference between the means (p<0.05) of the normal and malignant seminomas (p=0.037). Using a cut-off value of -7.4575 as demonstrated by the ROC curve results in a sensitivity of 100% and specificity of 87.5% and the area under the curve is 0.938 indicating excellent test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0337] Transcript 20:
[0338] There exists a statistically significant difference between the means (p<0.01) of the normal group and the malignant seminomas (p=0.006). Using a cutoff value of 1.8364 as demonstrated by the ROC curve results in a sensitivity of 100% and specificity of 100% and the area under the curve is 1.00 indicating excellent test accuracy. There also exists a statistically significant difference between the means (p<0.01) of the malignant seminomas and the malignant non-seminomas (p=0.004). Using a cutoff value of 1.6065 as demonstrated by the ROC curve results in a sensitivity of 100% and specificity of 100% and the area under the curve is 1.00 indicating excellent test accuracy. The threshold value chosen may be adjusted to increase either the specificity or sensitivity of the test for a particular application.
[0339] Conclusions:
[0340] The above results illustrate the utility of transcripts 2, 3, 4, 11, 12, 13, 15, 16, and 20 in the detection of testicular cancer, and testicular cancer subtypes, and in distinguishing malignant from normal testicular tissue. Transcript 2 was also found to have utility in the detection of prostate, breast, colorectal and ovarian cancer. Transcript 3 was also found to have utility in the detection of prostate, breast, melanoma, colorectal, and ovarian cancers. Transcript 4 was also found to have utility in the detection of prostate and colorectal cancers. Transcript 11 was also found to have utility in the detection of colorectal, melanoma, and ovarian cancers. Transcript 12 was also found to have utility in the detection of colorectal and ovarian cancers. Transcript 15 was also found to have utility in the detection of melanoma and ovarian cancers. Transcript 16 was also found to have utility in the detection of melanoma skin cancer. Transcript 20 was also found to have utility in the detection of colorectal cancer, melanoma, and ovarian cancer. Any of the 9 transcripts listed may be used individually or in combination as a tool for the detection or characterization of testicular cancer in a clinical setting.
[0341] In one aspect, the invention provides a kit for conducting an assay for determining the presence of cancer in a tissue sample. The kit includes the required reagents for conducting the assay as described above. In particular, the kit includes one or more containers containing one or more hybridization probes corresponding to transcripts 1 to 17, and 20 described above. As will be understood, the reagents for conducting the assay may include any necessary buffers, salts, detection reagents etc. Further, the kit may include any necessary sample collection devices, containers etc. for obtaining the needed tissue samples, reagents or materials to prepare the tissue samples for example by homogenization or nucleic acid extraction, and for conducting the subject assay or assays. The kit may also include control tissues or samples to establish or validate acceptable values for diseased or non-diseased tissues.
[0342] Although the invention has been described with reference to certain specific embodiments, various modifications thereof will be apparent to those skilled in the art without departing from the spirit and scope of the invention as outlined in the claims appended hereto. All documents (articles, manuals, patent applications etc) referred to in the present application are incorporated herein in their entirety by reference.
BIBLIOGRAPHY
[0343] The following references, amongst others, were cited in the foregoing description. The entire contents of these references are incorporated herein by way of reference thereto.
TABLE-US-00010 Author Journal Title Volume Date Anderson et al Nature Sequence and Organization of the Human 290(5806): 457- 1981 Mitochondrial Genome 65 Andrews et al Nat Genet Reanalysis and revision of the Cambridge 23(2): 147 1999 reference sequence for human mitochondrial DNA. Modica- Expert Rev Mitochondria as targets for detection and 4: 1-19 2002 Napolitano et al Mol Med treatment of cancer Sherratt et al Clin Sci (Lond) Mitochondrial DNA defects: a widening 92(3): 225-35 1997 clinical spectrum of disorders. Croteau et al Mutat Res Mitochondrial DNA repair pathways. 434(3): 137-48 1999 Green and J Clin Invest Pharmacological manipulation of cell death: 115(10): 2610- 2005 Kroemer clinical applications in sight? 2617 Dai et al Acta Correlation of cochlear blood supply with 24(2): 130-6 2004 Otolaryngol mitochondrial DNA common deletion in presbyacusis. Ro et al Muscle Nerve Deleted 4977-bp mitochondrial DNA 28(6): 737-43 2003 mutation is associated with sporadic amyotrophic lateral sclerosis: a hospital- based case-control study. Barron et al Invest Mitochondrial abnormalities in ageing 42(12): 3016-22 2001 Ophthalmol macular photoreceptors. Vis Sci Lewis et al J Pathol Detection of damage to the mitochondrial 191(3): 274-81 2000 genome in the oncocytic cells of Warthin's tumour. Muller-Hocker Mod Pathol The common 4977 base pair deletion of 11(3): 295-301. 1998 et al mitochondrial DNA preferentially accumulates in the cardiac conduction system of patients with Kearns-Sayre syndrome. Porteous et al Eur J Biochem Bioenergetic consequences of accumulating 257(1): 192-201 1998 the common 4977-bp mitochondrial DNA deletion. Parr et al J Mol Diagn Somatic mitochondrial DNA mutations in 8(3): 312-9. 2006 prostate cancer and normal appearing adjacent glands in comparison to age- matched prostate samples without malignant histology. Maki et al Am J Clin Mitochondrial genome deletion aids in the 129(1): 57-66 2008 Pathol identification of false- and true-negative prostate needle core biopsy specimens. Nakase et al Am J Hum Transcription and translation of deleted 46(3): 418-27. 1990 Genet mitochondrial genomes in Kearns-Sayre syndrome: implications for pathogenesis. Libura et al Blood Therapy-related acute myeloid leukemia- 105(5): 2124-31 2005 like MLL rearrangements are induced by etoposide in primary human CD34+ cells and remain stable after clonal expansion. Meyer et al Proc Natl Diagnostic tool for the identification of MLL 102(2): 449-54 2005 Acad Sci rearrangements including unknown partner USA genes. Eguchi et al Genes MLL chimeric protein activation renders 45(8): 754-60 2006 Chromosomes cells vulnerable to chromosomal damage: Cancer an explanation for the very short latency of infant leukemia. Hayashi et al Proc Natl Introduction of disease-related 88: 10614- 1991 Acad Sci mitochondrial DNA deletions into HeLa cells 10618 USA lacking mitochondrial DNA results in mitochondrial dysfunction
TABLE-US-00011 TABLE 1 Known mitochondrial deletions having an ORF Deletion Deletion Repeat Number of Junction (nt:nt) Size (bp) Location (nt/nt) Repeats References COX I - ND5 6075:13799 -7723 6076-6084/13799- D, 9/9 Mita, S., Rizzuto, R., Moraes, C. T., Shanske, S., Arnaudo, E., Fabrizi, 13807 G. M., Koga, Y., DiMauro, S., Schon, E. A. (1990) "Recombination via flanking direct repeats is a major cause of large-scale deletions of human mitochondrial DNA" Nucleic Acids Research 18(3): 561-567 6238:14103 -7864 6235-6238/14099- D, 4/4 Blok, R. B., Thorburn, D.R., Thompson, G. N., Dahl, H. H. (1995) "A 14102 topoisomerase II cleavage site is associated with a novel mitochondrial DNA deletion" Human Genetics 95 (1): 75-81 6325:13989 -7663 6326-6341/13889- D, 16/17 Larsson, N. G., Holme, E., Kristiansson, B., Oldfors, A., Tulinius, M. 14004 (1990) "Progressive increase of the mutated mitochondrial DNA fraction in Kearns-Sayre syndrome" Pediatric Research 28 (2): 131- 136 Larsson, N. G., Holme, E. (1992) "Multiple short direct repeats associated with single mtDNA deletions " Biochimica et Biophysica Acta 1139(4): 311-314 6330:13994 -7663 6331-6341/13994- D, 11/11 Mita, S., Rizzuto, R., Moraes, C. T., Shanske, S., Arnaudo, E., Fabrizi, 14004 G. M., Koga, Y., DiMauro, S., Schon, E.A. (1990) "Recombination via flanking direct repeats is a major cause of large-scale deletions of human mitochondrial DNA" Nucleic Acids Research 18(3): 561-567 COX II - ND5 7829:14135 -6305 7824-7829/14129- D, 6/6 Bet, L., Moggio, M., Comi, G. P., Mariani, C., Prelle, A., Checcarelli, N., 14134 Bordoni, A., Bresolin, N., Scarpini, E., Scarlato, G. (1994) "Multiple sclerosis and mitochondrial myopathy: an unusual combination of diseases" Journal of Neurology 241 (8): 511-516 8213:13991 -5777 8214-8220/13991- D, 7/7 Hinokio, Y., Suzuki, S., Komatu, K., Ohtomo, M., Onoda, M., 13997 Matsumoto, M., Hirai, S., Sato, Y., Akai, H., Abe, K., Toyota, T. (1995) "A new mitochondrial DNA deletion associated with diabetic amyotrophy, diabetic myoatrophy and diabetic fatty liver" Muscle and Nerve 3 (9): S142-149 ATPase - ND5 8631:13513 -4881 8625-8631/13506- D, 7/7 Zhang, C., Baumer, A., Mackay, I. R., Linnane, A. W., Nagley, P. (1995) 13512 "Unusual pattern of mitochondrial DNA deletions in skeletal muscle of an adult human with chronic fatigue syndrome" Human Molecular Genetics 4 (4): 751 -754 9144:13816 -4671 9137-9144/13808- D, 8/8 Ota, Y., Tanaka, M., Sato, W., Ohno, K., Yamamoto, T., Maehara, M., 13815 Negoro, T., Watanabe, K., Awaya, S., Ozawa, T. (1991) "Detection of platelet mitochondrial DNA deletions in Kearns-Sayre syndrome" Investigative Ophthalmology and Visual Science 32 (10): 2667-2675 9191:12909 -3717 9189-9191/12906- D, 3/3 Tanaka, M., Sato, W., Ohno, K., Yamamoto, T., Ozawa, T. (1989) 12908 "Direct sequencing of mitochondrial DNA in myopathic patients" Biochemical and Biophysical Research Communications 164 ( ): 156- 163 COX III - ND5 10190:13753 -3562 10191-10190/13753- D, 8/8 Rotig, A., Bourgeron, T., Chretien. D., Rustin, P., Munnich, A. (1995) 13760 "Spectrum of mitochondrial DNA rearrangements in the Pearson marrow-pancreas syndrome" Human Molecular Genetics 4 (8): 1327- 1330 Rotig, A., Cormier, Y., Kol, F., Mize, C. E., Souslubray, J. M., Veerman, A., Pearson, H. A., Munnich, A. (1991) "Site-specific deletions of the mitochondrial genome in Pearson marrow-pancreas syndrome" Genomics 10 (2): 502-504 10067:12029 -2461 10365-10367/12825- D, 3/3 Kapsa, R., Thompson, G. N., Thorburn, D. R., Dahl, H. H., Marzuki, G., 12828 Byrna, E., Blair, R. B. (1984) "A novel mtDNA deletion in an infant with Pearson syndrome" Journal of Inherited Metabolic Disease 17 (5): 521- 526 ND4L - ND5 10744:14124 -3378 10745-10754/14124- D, 9/10 Cormier-Daire, V., Bonnefont, J. P., Rustin, P., Maurage, C., Ogler, H., 14133 Schmitz, J., Ricour, C., Saudubray, J. M., Munnich, A., Rotig, A. (1984) "Mitochondrial DNA, rearrangements with onset as chronic diarrhea with villous atrophy" Journal of Pediatrics 124 (1): 53-70 ND4 - ND5 11232:13980 -2747 1324-11242/13981- D, 9/9 Rotig, A., Cormier, V., Roll, F., Mize, C. E., Saudubray, J. M., Veerman, 13989 A., Pearson, H. A., Munnich, A. (1991) "Site-specific deletions of the mitochondrial genome in Pearson marrow-pancreas syndrome" Genomics 10 (2): 502-504 Rotig, A., Cormier, Y., Blanche, S., Bonnefont, J. P., Ledeist, F., Romero, N., Schmitz, J., Rustin, P., Fischer, A., Saudubray, J. M. (1990) "Pearson's marrow-pancreas syndrome. A multi-system mitochondrial disorder in infancy" Journal of Clinical Investigation 86 ( ): 1601-1608 Cormier, V., Rotig, A., Quartino, A. R., Forni, G. L., Cerane, R., Maier, M., Saudubray, J. M., Munnich, A. (1990) "Widespread multitissue deletions of the mitochondrial genome in Pearson marrow-pancreas syndrome" Journal of Pediatrics 117 (4): 599-602 Awata, T., Matsumata, T., Iwamoto, Y., Matsuda, A., Kuzuya, T., Saito, T. (1993) "Japanese case of diabetes mellitus and deafness with mutations in mitochondrial tRNALeu(UUR) gene [letter]" Lancet 341 (8855): 1281-1282
TABLE-US-00012 TABLE 2 Prostate Cancer Detection with Novel Mitochondrial Fusion Transcripts Homog Homog Homog Homog Homog Homog Homog Homog RNA 1 2 RNA 1 2 RNA 1 2 RNA 1 2 Transcript Tran- Tran- Tran- Tran- Tran- Tran- Tran- Tran- Tran- Tran- Tran- Tran- script script script script script script script script script script script script 1 1 1 2 2 2 3 3 3 4 4 4 1 2 3 4 5 6 7 8 9 10 11 12 No dilution A 2957 353 233 144838 75374 17192 348424 333189 213844 509 565 207 Replicate A B 3174 475 298 202793 100062 31750 320877 278137 210265 401 676 250 1:10 dilution C 1041 262 114 106195 98403 36191 238467 248677 123497 181 486 168 Replicate C D 1040 272 176 120308 116930 50323 239231 262520 129778 153 467 149 1:100 dilution E 318 170 110 25155 64823 27725 100345 164606 85287 72 265 119 Replicate E F 287 150 109 23500 50524 24629 100856 178527 84731 83 251 120 1:1000 dilution G 100 76 123 3002 12960 252 29203 102309 137 31 143 66 Replicate G H 94 83 91 1263 5796 285 29092 97257 96 45 110 94 % CV A 5.0 20.9 17.3 23.6 19.9 42.1 5.8 12.7 1.2 16.9 12.7 13.3 % CV C 0.1 2.5 30.1 8.8 12.2 23.1 0.2 3.8 3.5 12.0 2.8 8.3 % CV E 7.1 9.0 0.6 4.8 17.5 8.4 0.4 5.7 0.5 9.8 3.8 0.6 % CV G 4.7 6.0 20.8 57.7 54.0 8.8 0.3 3.6 25.0 27.0 18.2 24.9 * unit results in table are RLU (relative luminescence units); Data read on Glorunner .TM.. % CV = Coefficient of variation (as %). Legend: Homog = homogenate. Homog 1: Prostate tumour tissue sample from patient; Homog 2: Histologically normal tissue adjacent to tumour from patient. RNA: Control: Total RNA from prostate tissue (Ambion p/n 7988).
TABLE-US-00013 TABLE 3 Deletion/Transcript/DNA Complement DNA sequence with deletion complementary Deletion RNA transcript to RNA transcript Transcript No. ATP synthase F0 subunit 8 to NADH SEQ ID NO: 18 SEQ ID NO: 2 1 dehydrogenase subunit mitochondrial positions 8366-14148 (with reference to SEQ ID NO: 1). NADH dehydrogenase subunit 4L SEQ ID NO: 19 SEQ ID NO: 3 2 (ND4L) to NADH dehydrogenase subunit 5 (ND5); Mitochondrial positions 10470- 14148 (with reference to SEQ ID NO: 1) Cytochrome c oxidase subunit II (COII) to SEQ ID NO: 20 SEQ ID NO: 4 3 Cytochrome b (Cytb); Mitochondrial positions 7586-15887 (with reference to SEQ ID NO: 1) Cytochrome c oxidase subunit II (COII) to SEQ ID NO: 21 SEQ ID NO: 5 4 Cytochrome b (Cytb); Mitochondrial positions 7586-15887 (with reference to SEQ ID NO: 1)
TABLE-US-00014 TABLE 4 Breast and Prostate Cancer Detection Normal Normal Normal Adjacent Breast adjacent Breast Adjacent Prostate Prostate Prostate to Prostate Tumour Breast Tumour to Breast Tumour Tumour Tumour Tumour 1 Tumour 1 2 Tumour 2 3 4 5 5 1 2 3 4 5 6 7 8 1:100 dilution E 68920 2971 49108 1245 46723 56679 99836 35504 1:100 dilution F 92409 3017 60637 1512 53940 56155 100582 44221 replicate G 420 3 31 6 26 25 44 23 H 518 3 4 5 5 3 4 2 % CV 20.6 1.1 14.9 13.7 10.1 0.7 0.5 15.5
[0344] unit results in table are RLU (relative luminescence units) [0345] background G1, H1 [0346] empty well G2-G8, H2-H8
TABLE-US-00015 [0346] TABLE 5a Assay Conditions Template for the assay RNA Homogen 1 Homogen 2 RNA Homogen 1 Homogen 2 Transcript 1 Transcript 1 Transcript 1 Transcript 2 Transcript 2 Transcript 2 1 2 3 4 5 6 A RNA Homog 1 Homog 2 RNA Homog 1 Homog 2 B Dil 1 Dil 1 Dil 1 Dil 1 Dil 1 Dil 1 C RNA Homog 1 Homo 2 RNA Homog 1 Homog 2 D Dil 2 Dil 2 Dil 2 Dil 2 Dil 2 Dil 2 E RNA Homog 1 Homog 2 RNA Homog 1 Homog 2 F Dil 3 Dil 3 Dil 3 Dil 3 Dil 3 Dil 3 G RNA Homog 1 Transcript 1 RNA Homog 1 Transcript 1 H Dil 4 Dil 4 Background Dil 4 Dil 4 Background RNA Homogen 1 Homogen 2 RNA Homogen 1 Homogen 2 Transcript 3 Transcript 3 Transcript 3 Transcript 4 Transcript 4 Transcript 4 7 8 9 10 11 12 A RNA Homog 1 Homog 2 RNA Homog 1 Homog 2 B Dil 1 Dil 1 Dil 1 Dil 1 Dil 1 Dil 1 C RNA Homog 1 Homog 2 RNA Homog 1 Homog 2 D Dil 2 Dil 2 Dil 2 Dil 2 Dil 2 Dil 2 E RNA Homog 1 Homog 2 RNA Homog 1 Homog 2 F Dil 3 Dil 3 Dil 3 Dil 3 Dil 3 Dil 3 G RNA Homog 1 Transcript 1 RNA Homog 1 Transcript 1 H Dil 4 Dil 4 Background Dil 4 Dil 4 Background Homogenate1- Used 26 mg of tissue to homogenize in 700 ul H soln with Proteinase K (PK). Used Qiagen TissueRuptor. Used 40 ul homogenate supernatant, 20, 10 and 5 ul for dilution Homogenate1 = Tumour tissue from the tumorous Prostate Homogenate2- Used 29 mg of tissue to homogenize in 700 ul H soln with PK. Used Qiagen TissueRuptor. Used 40 ul homogenate supernatant, 20, 10 and 5 ul for dilution Homogenate2 = Normal tissue from the tumorous Prostate RNA dilution was made as below. RNA was from Prostate Normal from Ambion. Assay was done in duplicates.
TABLE-US-00016 TABLE 5b RNA dilution RNA Dilution ng/ul Dil 1 3000 1:3 dil Dil 2 1000 Serial dil Dil 3 333 Dil 4 111
TABLE-US-00017 TABLE 6 Transcript Summary by Disease Mela- Pros- Colo- noma Testi- tate Breast rectal Skin Lung Ovarian cular Probe Cancer Cancer Cancer Cancer Cancer Cancer Cancer 1 .cndot. .cndot. 2 .cndot. .cndot. .cndot. .cndot. .cndot. 3 .cndot. .cndot. .cndot. .cndot. 4 .cndot. .cndot. 5 6 .cndot. .cndot. .cndot. 7 8 .cndot. .cndot. 9 .cndot. 10 .cndot. .cndot. .cndot. 11 .cndot. .cndot. .cndot. .cndot. 12 .cndot. .cndot. .cndot. 13 .cndot. 14 .cndot. 15 .cndot. .cndot. .cndot. 16 .cndot. .cndot. 17 20 .cndot. .cndot. .cndot. .cndot.
Sequence CWU 1
1
56116568DNAHuman 1gatcacaggt ctatcaccct attaaccact cacgggagct
ctccatgcat ttggtatttt 60cgtctggggg gtatgcacgc gatagcattg cgagacgctg
gagccggagc accctatgtc 120gcagtatctg tctttgattc ctgcctcatc
ctattattta tcgcacctac gttcaatatt 180acaggcgaac atacttacta
aagtgtgtta attaattaat gcttgtagga cataataata 240acaattgaat
gtctgcacag ccactttcca cacagacatc ataacaaaaa atttccacca
300aaccccccct cccccgcttc tggccacagc acttaaacac atctctgcca
aaccccaaaa 360acaaagaacc ctaacaccag cctaaccaga tttcaaattt
tatcttttgg cggtatgcac 420ttttaacagt caccccccaa ctaacacatt
attttcccct cccactccca tactactaat 480ctcatcaata caacccccgc
ccatcctacc cagcacacac acaccgctgc taaccccata 540ccccgaacca
accaaacccc aaagacaccc cccacagttt atgtagctta cctcctcaaa
600gcaatacact gaaaatgttt agacgggctc acatcacccc ataaacaaat
aggtttggtc 660ctagcctttc tattagctct tagtaagatt acacatgcaa
gcatccccgt tccagtgagt 720tcaccctcta aatcaccacg atcaaaagga
acaagcatca agcacgcagc aatgcagctc 780aaaacgctta gcctagccac
acccccacgg gaaacagcag tgattaacct ttagcaataa 840acgaaagttt
aactaagcta tactaacccc agggttggtc aatttcgtgc cagccaccgc
900ggtcacacga ttaacccaag tcaatagaag ccggcgtaaa gagtgtttta
gatcaccccc 960tccccaataa agctaaaact cacctgagtt gtaaaaaact
ccagttgaca caaaatagac 1020tacgaaagtg gctttaacat atctgaacac
acaatagcta agacccaaac tgggattaga 1080taccccacta tgcttagccc
taaacctcaa cagttaaatc aacaaaactg ctcgccagaa 1140cactacgagc
cacagcttaa aactcaaagg acctggcggt gcttcatatc cctctagagg
1200agcctgttct gtaatcgata aaccccgatc aacctcacca cctcttgctc
agcctatata 1260ccgccatctt cagcaaaccc tgatgaaggc tacaaagtaa
gcgcaagtac ccacgtaaag 1320acgttaggtc aaggtgtagc ccatgaggtg
gcaagaaatg ggctacattt tctaccccag 1380aaaactacga tagcccttat
gaaacttaag ggtcgaaggt ggatttagca gtaaactaag 1440agtagagtgc
ttagttgaac agggccctga agcgcgtaca caccgcccgt caccctcctc
1500aagtatactt caaaggacat ttaactaaaa cccctacgca tttatataga
ggagacaagt 1560cgtaacatgg taagtgtact ggaaagtgca cttggacgaa
ccagagtgta gcttaacaca 1620aagcacccaa cttacactta ggagatttca
acttaacttg accgctctga gctaaaccta 1680gccccaaacc cactccacct
tactaccaga caaccttagc caaaccattt acccaaataa 1740agtataggcg
atagaaattg aaacctggcg caatagatat agtaccgcaa gggaaagatg
1800aaaaattata accaagcata atatagcaag gactaacccc tataccttct
gcataatgaa 1860ttaactagaa ataactttgc aaggagagcc aaagctaaga
cccccgaaac cagacgagct 1920acctaagaac agctaaaaga gcacacccgt
ctatgtagca aaatagtggg aagatttata 1980ggtagaggcg acaaacctac
cgagcctggt gatagctggt tgtccaagat agaatcttag 2040ttcaacttta
aatttgccca cagaaccctc taaatcccct tgtaaattta actgttagtc
2100caaagaggaa cagctctttg gacactagga aaaaaccttg tagagagagt
aaaaaattta 2160acacccatag taggcctaaa agcagccacc aattaagaaa
gcgttcaagc tcaacaccca 2220ctacctaaaa aatcccaaac atataactga
actcctcaca cccaattgga ccaatctatc 2280accctataga agaactaatg
ttagtataag taacatgaaa acattctcct ccgcataagc 2340ctgcgtcaga
ttaaaacact gaactgacaa ttaacagccc aatatctaca atcaaccaac
2400aagtcattat taccctcact gtcaacccaa cacaggcatg ctcataagga
aaggttaaaa 2460aaagtaaaag gaactcggca aatcttaccc cgcctgttta
ccaaaaacat cacctctagc 2520atcaccagta ttagaggcac cgcctgccca
gtgacacatg tttaacggcc gcggtaccct 2580aaccgtgcaa aggtagcata
atcacttgtt ccttaaatag ggacctgtat gaatggctcc 2640acgagggttc
agctgtctct tacttttaac cagtgaaatt gacctgcccg tgaagaggcg
2700ggcataacac agcaagacga gaagacccta tggagcttta atttattaat
gcaaacagta 2760cctaacaaac ccacaggtcc taaactacca aacctgcatt
aaaaatttcg gttggggcga 2820cctcggagca gaacccaacc tccgagcagt
acatgctaag acttcaccag tcaaagcgaa 2880ctactatact caattgatcc
aataacttga ccaacggaac aagttaccct agggataaca 2940gcgcaatcct
attctagagt ccatatcaac aatagggttt acgacctcga tgttggatca
3000ggacatcccg atggtgcagc cgctattaaa ggttcgtttg ttcaacgatt
aaagtcctac 3060gtgatctgag ttcagaccgg agtaatccag gtcggtttct
atctacttca aattcctccc 3120tgtacgaaag gacaagagaa ataaggccta
cttcacaaag cgccttcccc cgtaaatgat 3180atcatctcaa cttagtatta
tacccacacc cacccaagaa cagggtttgt taagatggca 3240gagcccggta
atcgcataaa acttaaaact ttacagtcag aggttcaatt cctcttctta
3300acaacatacc catggccaac ctcctactcc tcattgtacc cattctaatc
gcaatggcat 3360tcctaatgct taccgaacga aaaattctag gctatataca
actacgcaaa ggccccaacg 3420ttgtaggccc ctacgggcta ctacaaccct
tcgctgacgc cataaaactc ttcaccaaag 3480agcccctaaa acccgccaca
tctaccatca ccctctacat caccgccccg accttagctc 3540tcaccatcgc
tcttctacta tgaacccccc tccccatacc caaccccctg gtcaacctca
3600acctaggcct cctatttatt ctagccacct ctagcctagc cgtttactca
atcctctgat 3660cagggtgagc atcaaactca aactacgccc tgatcggcgc
actgcgagca gtagcccaaa 3720caatctcata tgaagtcacc ctagccatca
ttctactatc aacattacta ataagtggct 3780cctttaacct ctccaccctt
atcacaacac aagaacacct ctgattactc ctgccatcat 3840gacccttggc
cataatatga tttatctcca cactagcaga gaccaaccga acccccttcg
3900accttgccga aggggagtcc gaactagtct caggcttcaa catcgaatac
gccgcaggcc 3960ccttcgccct attcttcata gccgaataca caaacattat
tataataaac accctcacca 4020ctacaatctt cctaggaaca acatatgacg
cactctcccc tgaactctac acaacatatt 4080ttgtcaccaa gaccctactt
ctaacctccc tgttcttatg aattcgaaca gcataccccc 4140gattccgcta
cgaccaactc atacacctcc tatgaaaaaa cttcctacca ctcaccctag
4200cattacttat atgatatgtc tccataccca ttacaatctc cagcattccc
cctcaaacct 4260aagaaatatg tctgataaaa gagttacttt gatagagtaa
ataataggag cttaaacccc 4320cttatttcta ggactatgag aatcgaaccc
atccctgaga atccaaaatt ctccgtgcca 4380cctatcacac cccatcctaa
agtaaggtca gctaaataag ctatcgggcc cataccccga 4440aaatgttggt
tatacccttc ccgtactaat taatcccctg gcccaacccg tcatctactc
4500taccatcttt gcaggcacac tcatcacagc gctaagctcg cactgatttt
ttacctgagt 4560aggcctagaa ataaacatgc tagcttttat tccagttcta
accaaaaaaa taaaccctcg 4620ttccacagaa gctgccatca agtatttcct
cacgcaagca accgcatcca taatccttct 4680aatagctatc ctcttcaaca
atatactctc cggacaatga accataacca atactaccaa 4740tcaatactca
tcattaataa tcataatagc tatagcaata aaactaggaa tagccccctt
4800tcacttctga gtcccagagg ttacccaagg cacccctctg acatccggcc
tgcttcttct 4860cacatgacaa aaactagccc ccatctcaat catataccaa
atctctccct cactaaacgt 4920aagccttctc ctcactctct caatcttatc
catcatagca ggcagttgag gtggattaaa 4980ccaaacccag ctacgcaaaa
tcttagcata ctcctcaatt acccacatag gatgaataat 5040agcagttcta
ccgtacaacc ctaacataac cattcttaat ttaactattt atattatcct
5100aactactacc gcattcctac tactcaactt aaactccagc accacgaccc
tactactatc 5160tcgcacctga aacaagctaa catgactaac acccttaatt
ccatccaccc tcctctccct 5220aggaggcctg cccccgctaa ccggcttttt
gcccaaatgg gccattatcg aagaattcac 5280aaaaaacaat agcctcatca
tccccaccat catagccacc atcaccctcc ttaacctcta 5340cttctaccta
cgcctaatct actccacctc aatcacacta ctccccatat ctaacaacgt
5400aaaaataaaa tgacagtttg aacatacaaa acccacccca ttcctcccca
cactcatcgc 5460ccttaccacg ctactcctac ctatctcccc ttttatacta
ataatcttat agaaatttag 5520gttaaataca gaccaagagc cttcaaagcc
ctcagtaagt tgcaatactt aatttctgta 5580acagctaagg actgcaaaac
cccactctgc atcaactgaa cgcaaatcag ccactttaat 5640taagctaagc
ccttactaga ccaatgggac ttaaacccac aaacacttag ttaacagcta
5700agcaccctaa tcaactggct tcaatctact tctcccgccg ccgggaaaaa
aggcgggaga 5760agccccggca ggtttgaagc tgcttcttcg aatttgcaat
tcaatatgaa aatcacctcg 5820gagctggtaa aaagaggcct aacccctgtc
tttagattta cagtccaatg cttcactcag 5880ccattttacc tcacccccac
tgatgttcgc cgaccgttga ctattctcta caaaccacaa 5940agacattgga
acactatacc tattattcgg cgcatgagct ggagtcctag gcacagctct
6000aagcctcctt attcgagccg agctgggcca gccaggcaac cttctaggta
acgaccacat 6060ctacaacgtt atcgtcacag cccatgcatt tgtaataatc
ttcttcatag taatacccat 6120cataatcgga ggctttggca actgactagt
tcccctaata atcggtgccc ccgatatggc 6180gtttccccgc ataaacaaca
taagcttctg actcttacct ccctctctcc tactcctgct 6240cgcatctgct
atagtggagg ccggagcagg aacaggttga acagtctacc ctcccttagc
6300agggaactac tcccaccctg gagcctccgt agacctaacc atcttctcct
tacacctagc 6360aggtgtctcc tctatcttag gggccatcaa tttcatcaca
acaattatca atataaaacc 6420ccctgccata acccaatacc aaacgcccct
cttcgtctga tccgtcctaa tcacagcagt 6480cctacttctc ctatctctcc
cagtcctagc tgctggcatc actatactac taacagaccg 6540caacctcaac
accaccttct tcgaccccgc cggaggagga gaccccattc tataccaaca
6600cctattctga tttttcggtc accctgaagt ttatattctt atcctaccag
gcttcggaat 6660aatctcccat attgtaactt actactccgg aaaaaaagaa
ccatttggat acataggtat 6720ggtctgagct atgatatcaa ttggcttcct
agggtttatc gtgtgagcac accatatatt 6780tacagtagga atagacgtag
acacacgagc atatttcacc tccgctacca taatcatcgc 6840tatccccacc
ggcgtcaaag tatttagctg actcgccaca ctccacggaa gcaatatgaa
6900atgatctgct gcagtgctct gagccctagg attcatcttt cttttcaccg
taggtggcct 6960gactggcatt gtattagcaa actcatcact agacatcgta
ctacacgaca cgtactacgt 7020tgtagcccac ttccactatg tcctatcaat
aggagctgta tttgccatca taggaggctt 7080cattcactga tttcccctat
tctcaggcta caccctagac caaacctacg ccaaaatcca 7140tttcactatc
atattcatcg gcgtaaatct aactttcttc ccacaacact ttctcggcct
7200atccggaatg ccccgacgtt actcggacta ccccgatgca tacaccacat
gaaacatcct 7260atcatctgta ggctcattca tttctctaac agcagtaata
ttaataattt tcatgatttg 7320agaagccttc gcttcgaagc gaaaagtcct
aatagtagaa gaaccctcca taaacctgga 7380gtgactatat ggatgccccc
caccctacca cacattcgaa gaacccgtat acataaaatc 7440tagacaaaaa
aggaaggaat cgaacccccc aaagctggtt tcaagccaac cccatggcct
7500ccatgacttt ttcaaaaagg tattagaaaa accatttcat aactttgtca
aagttaaatt 7560ataggctaaa tcctatatat cttaatggca catgcagcgc
aagtaggtct acaagacgct 7620acttccccta tcatagaaga gcttatcacc
tttcatgatc acgccctcat aatcattttc 7680cttatctgct tcctagtcct
gtatgccctt ttcctaacac tcacaacaaa actaactaat 7740actaacatct
cagacgctca ggaaatagaa accgtctgaa ctatcctgcc cgccatcatc
7800ctagtcctca tcgccctccc atccctacgc atcctttaca taacagacga
ggtcaacgat 7860ccctccctta ccatcaaatc aattggccac caatggtact
gaacctacga gtacaccgac 7920tacggcggac taatcttcaa ctcctacata
cttcccccat tattcctaga accaggcgac 7980ctgcgactcc ttgacgttga
caatcgagta gtactcccga ttgaagcccc cattcgtata 8040ataattacat
cacaagacgt cttgcactca tgagctgtcc ccacattagg cttaaaaaca
8100gatgcaattc ccggacgtct aaaccaaacc actttcaccg ctacacgacc
gggggtatac 8160tacggtcaat gctctgaaat ctgtggagca aaccacagtt
tcatgcccat cgtcctagaa 8220ttaattcccc taaaaatctt tgaaataggg
cccgtattta ccctatagca ccccctctac 8280cccctctaga gcccactgta
aagctaactt agcattaacc ttttaagtta aagattaaga 8340gaaccaacac
ctctttacag tgaaatgccc caactaaata ctaccgtatg gcccaccata
8400attaccccca tactccttac actattcctc atcacccaac taaaaatatt
aaacacaaac 8460taccacctac ctccctcacc aaagcccata aaaataaaaa
attataacaa accctgagaa 8520ccaaaatgaa cgaaaatctg ttcgcttcat
tcattgcccc cacaatccta ggcctacccg 8580ccgcagtact gatcattcta
tttccccctc tattgatccc cacctccaaa tatctcatca 8640acaaccgact
aatcaccacc caacaatgac taatcaaact aacctcaaaa caaatgataa
8700ccatacacaa cactaaagga cgaacctgat ctcttatact agtatcctta
atcattttta 8760ttgccacaac taacctcctc ggactcctgc ctcactcatt
tacaccaacc acccaactat 8820ctataaacct agccatggcc atccccttat
gagcgggcac agtgattata ggctttcgct 8880ctaagattaa aaatgcccta
gcccacttct taccacaagg cacacctaca ccccttatcc 8940ccatactagt
tattatcgaa accatcagcc tactcattca accaatagcc ctggccgtac
9000gcctaaccgc taacattact gcaggccacc tactcatgca cctaattgga
agcgccaccc 9060tagcaatatc aaccattaac cttccctcta cacttatcat
cttcacaatt ctaattctac 9120tgactatcct agaaatcgct gtcgccttaa
tccaagccta cgttttcaca cttctagtaa 9180gcctctacct gcacgacaac
acataatgac ccaccaatca catgcctatc atatagtaaa 9240acccagccca
tgacccctaa caggggccct ctcagccctc ctaatgacct ccggcctagc
9300catgtgattt cacttccact ccataacgct cctcatacta ggcctactaa
ccaacacact 9360aaccatatac caatgatggc gcgatgtaac acgagaaagc
acataccaag gccaccacac 9420accacctgtc caaaaaggcc ttcgatacgg
gataatccta tttattacct cagaagtttt 9480tttcttcgca ggatttttct
gagcctttta ccactccagc ctagccccta ccccccaatt 9540aggagggcac
tggcccccaa caggcatcac cccgctaaat cccctagaag tcccactcct
9600aaacacatcc gtattactcg catcaggagt atcaatcacc tgagctcacc
atagtctaat 9660agaaaacaac cgaaaccaaa taattcaagc actgcttatt
acaattttac tgggtctcta 9720ttttaccctc ctacaagcct cagagtactt
cgagtctccc ttcaccattt ccgacggcat 9780ctacggctca acattttttg
tagccacagg cttccacgga cttcacgtca ttattggctc 9840aactttcctc
actatctgct tcatccgcca actaatattt cactttacat ccaaacatca
9900ctttggcttc gaagccgccg cctgatactg gcattttgta gatgtggttt
gactatttct 9960gtatgtctcc atctattgat gagggtctta ctcttttagt
ataaatagta ccgttaactt 10020ccaattaact agttttgaca acattcaaaa
aagagtaata aacttcgcct taattttaat 10080aatcaacacc ctcctagcct
tactactaat aattattaca ttttgactac cacaactcaa 10140cggctacata
gaaaaatcca ccccttacga gtgcggcttc gaccctatat cccccgcccg
10200cgtccctttc tccataaaat tcttcttagt agctattacc ttcttattat
ttgatctaga 10260aattgccctc cttttacccc taccatgagc cctacaaaca
actaacctgc cactaatagt 10320tatgtcatcc ctcttattaa tcatcatcct
agccctaagt ctggcctatg agtgactaca 10380aaaaggatta gactgaaccg
aattggtata tagtttaaac aaaacgaatg atttcgactc 10440attaaattat
gataatcata tttaccaaat gcccctcatt tacataaata ttatactagc
10500atttaccatc tcacttctag gaatactagt atatcgctca cacctcatat
cctccctact 10560atgcctagaa ggaataatac tatcgctgtt cattatagct
actctcataa ccctcaacac 10620ccactccctc ttagccaata ttgtgcctat
tgccatacta gtctttgccg cctgcgaagc 10680agcggtgggc ctagccctac
tagtctcaat ctccaacaca tatggcctag actacgtaca 10740taacctaaac
ctactccaat gctaaaacta atcgtcccaa caattatatt actaccactg
10800acatgacttt ccaaaaaaca cataatttga atcaacacaa ccacccacag
cctaattatt 10860agcatcatcc ctctactatt ttttaaccaa atcaacaaca
acctatttag ctgttcccca 10920accttttcct ccgaccccct aacaaccccc
ctcctaatac taactacctg actcctaccc 10980ctcacaatca tggcaagcca
acgccactta tccagtgaac cactatcacg aaaaaaactc 11040tacctctcta
tactaatctc cctacaaatc tccttaatta taacattcac agccacagaa
11100ctaatcatat tttatatctt cttcgaaacc acacttatcc ccaccttggc
tatcatcacc 11160cgatgaggca accagccaga acgcctgaac gcaggcacat
acttcctatt ctacacccta 11220gtaggctccc ttcccctact catcgcacta
atttacactc acaacaccct aggctcacta 11280aacattctac tactcactct
cactgcccaa gaactatcaa actcctgagc caacaactta 11340atatgactag
cttacacaat agcttttata gtaaagatac ctctttacgg actccactta
11400tgactcccta aagcccatgt cgaagccccc atcgctgggt caatagtact
tgccgcagta 11460ctcttaaaac taggcggcta tggtataata cgcctcacac
tcattctcaa ccccctgaca 11520aaacacatag cctacccctt ccttgtacta
tccctatgag gcataattat aacaagctcc 11580atctgcctac gacaaacaga
cctaaaatcg ctcattgcat actcttcaat cagccacata 11640gccctcgtag
taacagccat tctcatccaa accccctgaa gcttcaccgg cgcagtcatt
11700ctcataatcg cccacgggct tacatcctca ttactattct gcctagcaaa
ctcaaactac 11760gaacgcactc acagtcgcat cataatcctc tctcaaggac
ttcaaactct actcccacta 11820atagcttttt gatgacttct agcaagcctc
gctaacctcg ccttaccccc cactattaac 11880ctactgggag aactctctgt
gctagtaacc acgttctcct gatcaaatat cactctccta 11940cttacaggac
tcaacatact agtcacagcc ctatactccc tctacatatt taccacaaca
12000caatggggct cactcaccca ccacattaac aacataaaac cctcattcac
acgagaaaac 12060accctcatgt tcatacacct atcccccatt ctcctcctat
ccctcaaccc cgacatcatt 12120accgggtttt cctcttgtaa atatagttta
accaaaacat cagattgtga atctgacaac 12180agaggcttac gaccccttat
ttaccgagaa agctcacaag aactgctaac tcatgccccc 12240atgtctaaca
acatggcttt ctcaactttt aaaggataac agctatccat tggtcttagg
12300ccccaaaaat tttggtgcaa ctccaaataa aagtaataac catgcacact
actataacca 12360ccctaaccct gacttcccta attcccccca tccttaccac
cctcgttaac cctaacaaaa 12420aaaactcata cccccattat gtaaaatcca
ttgtcgcatc cacctttatt atcagtctct 12480tccccacaac aatattcatg
tgcctagacc aagaagttat tatctcgaac tgacactgag 12540ccacaaccca
aacaacccag ctctccctaa gcttcaaact agactacttc tccataatat
12600tcatccctgt agcattgttc gttacatggt ccatcataga attctcactg
tgatatataa 12660actcagaccc aaacattaat cagttcttca aatatctact
catcttccta attaccatac 12720taatcttagt taccgctaac aacctattcc
aactgttcat cggctgagag ggcgtaggaa 12780ttatatcctt cttgctcatc
agttgatgat acgcccgagc agatgccaac acagcagcca 12840ttcaagcaat
cctatacaac cgtatcggcg atatcggttt catcctcgcc ttagcatgat
12900ttatcctaca ctccaactca tgagacccac aacaaatagc ccttctaaac
gctaatccaa 12960gcctcacccc actactaggc ctcctcctag cagcagcagg
caaatcagcc caattaggtc 13020tccacccctg actcccctca gccatagaag
gccccacccc agtctcagcc ctactccact 13080caagcactat agttgtagca
ggaatcttct tactcatccg cttccacccc ctagcagaaa 13140atagcccact
aatccaaact ctaacactat gcttaggcgc tatcaccact ctgttcgcag
13200cagtctgcgc ccttacacaa aatgacatca aaaaaatcgt agccttctcc
acttcaagtc 13260aactaggact cataatagtt acaatcggca tcaaccaacc
acacctagca ttcctgcaca 13320tctgtaccca cgccttcttc aaagccatac
tatttatgtg ctccgggtcc atcatccaca 13380accttaacaa tgaacaagat
attcgaaaaa taggaggact actcaaaacc atacctctca 13440cttcaacctc
cctcaccatt ggcagcctag cattagcagg aatacctttc ctcacaggtt
13500tctactccaa agaccacatc atcgaaaccg caaacatatc atacacaaac
gcctgagccc 13560tatctattac tctcatcgct acctccctga caagcgccta
tagcactcga ataattcttc 13620tcaccctaac aggtcaacct cgcttcccca
cccttactaa cattaacgaa aataacccca 13680ccctactaaa ccccattaaa
cgcctggcag ccggaagcct attcgcagga tttctcatta 13740ctaacaacat
ttcccccgca tcccccttcc aaacaacaat ccccctctac ctaaaactca
13800cagccctcgc tgtcactttc ctaggacttc taacagccct agacctcaac
tacctaacca 13860acaaacttaa aataaaatcc ccactatgca cattttattt
ctccaacata ctcggattct 13920accctagcat cacacaccgc acaatcccct
atctaggcct tcttacgagc caaaacctgc 13980ccctactcct cctagaccta
acctgactag aaaagctatt acctaaaaca atttcacagc 14040accaaatctc
cacctccatc atcacctcaa cccaaaaagg cataattaaa ctttacttcc
14100tctctttctt cttcccactc atcctaaccc tactcctaat cacataacct
attcccccga 14160gcaatctcaa ttacaatata tacaccaaca aacaatgttc
aaccagtaac tactactaat 14220caacgcccat aatcatacaa agcccccgca
ccaataggat cctcccgaat caaccctgac 14280ccctctcctt cataaattat
tcagcttcct acactattaa agtttaccac aaccaccacc 14340ccatcatact
ctttcaccca cagcaccaat cctacctcca tcgctaaccc cactaaaaca
14400ctcaccaaga cctcaacccc tgacccccat gcctcaggat actcctcaat
agccatcgct 14460gtagtatatc caaagacaac catcattccc cctaaataaa
ttaaaaaaac tattaaaccc 14520atataacctc ccccaaaatt cagaataata
acacacccga ccacaccgct aacaatcaat 14580actaaacccc cataaatagg
agaaggctta gaagaaaacc ccacaaaccc cattactaaa 14640cccacactca
acagaaacaa agcatacatc attattctcg cacggactac aaccacgacc
14700aatgatatga aaaaccatcg ttgtatttca actacaagaa caccaatgac
cccaatacgc 14760aaaactaacc ccctaataaa attaattaac cactcattca
tcgacctccc caccccatcc 14820aacatctccg catgatgaaa cttcggctca
ctccttggcg cctgcctgat cctccaaatc 14880accacaggac tattcctagc
catgcactac tcaccagacg cctcaaccgc cttttcatca 14940atcgcccaca
tcactcgaga cgtaaattat ggctgaatca tccgctacct tcacgccaat
15000ggcgcctcaa tattctttat ctgcctcttc ctacacatcg ggcgaggcct
atattacgga 15060tcatttctct actcagaaac ctgaaacatc ggcattatcc
tcctgcttgc aactatagca 15120acagccttca taggctatgt cctcccgtga
ggccaaatat cattctgagg ggccacagta 15180attacaaact tactatccgc
catcccatac attgggacag acctagttca atgaatctga 15240ggaggctact
cagtagacag tcccaccctc acacgattct ttacctttca cttcatcttg
15300cccttcatta ttgcagccct agcaacactc cacctcctat tcttgcacga
aacgggatca 15360aacaaccccc taggaatcac ctcccattcc gataaaatca
ccttccaccc ttactacaca 15420atcaaagacg ccctcggctt acttctcttc
cttctctcct taatgacatt aacactattc 15480tcaccagacc tcctaggcga
cccagacaat tataccctag ccaacccctt aaacacccct 15540ccccacatca
agcccgaatg atatttccta ttcgcctaca caattctccg atccgtccct
15600aacaaactag gaggcgtcct tgccctatta ctatccatcc tcatcctagc
aataatcccc 15660atcctccata tatccaaaca acaaagcata atatttcgcc
cactaagcca atcactttat 15720tgactcctag ccgcagacct cctcattcta
acctgaatcg gaggacaacc agtaagctac 15780ccttttacca tcattggaca
agtagcatcc gtactatact tcacaacaat cctaatccta 15840ataccaacta
tctccctaat tgaaaacaaa atactcaaat gggcctgtcc ttgtagtata
15900aactaataca ccagtcttgt aaaccggaga tgaaaacctt tttccaagga
caaatcagag 15960aaaaagtctt taactccacc attagcaccc aaagctaaga
ttctaattta aactattctc 16020tgttctttca tggggaagca gatttgggta
ccacccaagt attgactcac ccatcaacaa 16080ccgctatgta tttcgtacat
tactgccagc caccatgaat attgtacggt accataaata 16140cttgaccacc
tgtagtacat aaaaacccaa tccacatcaa aaccccctcc ccatgcttac
16200aagcaagtac agcaatcaac cctcaactat cacacatcaa ctgcaactcc
aaagccaccc 16260ctcacccact aggataccaa caaacctacc cacccttaac
agtacatagt acataaagcc 16320atttaccgta catagcacat tacagtcaaa
tcccttctcg tccccatgga tgacccccct 16380cagatagggg tcccttgacc
accatcctcc gtgaaatcaa tatcccgcac aagagtgcta 16440ctctcctcgc
tccgggccca taacacttgg gggtagctaa agtgaactgt atccgacatc
16500tggttcctac ttcagggtca taaagcctaa atagcccaca cgttcccctt
aaataagaca 16560tcacgatg 165682783DNAArtificialcDNA 2atggcccacc
ataattaccc ccatactcct tacactattc ctcatcaccc aactaaaaat 60attaaacaca
aactaccacc tacctccctc accattggca gcctagcatt agcaggaata
120cctttcctca caggtttcta ctccaaagac cacatcatcg aaaccgcaaa
catatcatac 180acaaacgcct gagccctatc tattactctc atcgctacct
ccctgacaag cgcctatagc 240actcgaataa ttcttctcac cctaacaggt
caacctcgct tccccaccct tactaacatt 300aacgaaaata accccaccct
actaaacccc attaaacgcc tggcagccgg aagcctattc 360gcaggatttc
tcattactaa caacatttcc cccgcatccc ccttccaaac aacaatcccc
420ctctacctaa aactcacagc cctcgctgtc actttcctag gacttctaac
agccctagac 480ctcaactacc taaccaacaa acttaaaata aaatccccac
tatgcacatt ttatttctcc 540aacatactcg gattctaccc tagcatcaca
caccgcacaa tcccctatct aggccttctt 600acgagccaaa acctgcccct
actcctccta gacctaacct gactagaaaa gctattacct 660aaaacaattt
cacagcacca aatctccacc tccatcatca cctcaaccca aaaaggcata
720attaaacttt acttcctctc tttcttcttc ccactcatcc taaccctact
cctaatcaca 780taa 7833300DNAArtificialcDNA 3atgcccctca tttacataaa
tattatacta gcatttacca tctcacttct aggaatacta 60gtatatcgct cacacctcat
atcctcccta ctatgcctag aaggaataat actatcgctg 120ttcattatag
ctactctcat aaccctcaac acccactccc tcttagccaa tattgtgcct
180attgccatac tagtctttgc cgcctgcgaa gcagcggtgg gcctagccct
actagtctca 240atctccaaca catatggcct agactacgta cataacctaa
ccctactcct aatcacataa 3004781DNAArtificialcDNA 4atggcacatg
cagcgcaagt aggtctacaa gacgctactt cccctatcat agaagagctt 60atcacctttc
atgatcacgc cctcataatc attttcctta tctgcttcct agtcctgtat
120gcccttttcc taacactcac aacaaaacta actaatacta acatctcaga
cgctcaggaa 180atagaaaccg tctgaactat cctgcccgcc atcatcctag
tcctcatcgc cctcccatcc 240ctacgcatcc tttacataac agacgaggtc
aacgatccct cccttaccat caaatcaatt 300ggccaccaat ggtactgaac
ctacgagtac accgactacg gcggactaat cttcaactcc 360tacatacttc
ccccattatt cctagaacca ggcgacccag acaattatac cctagccaac
420cccttaaaca cccctcccca catcaagccc gaatgatatt tcctattcgc
ctacacaatt 480ctccgatccg tccctaacaa actaggaggc gtccttgccc
tattactatc catcctcatc 540ctagcaataa tccccatcct ccatatatcc
aaacaacaaa gcataatatt tcgcccacta 600agccaatcac tttattgact
cctagccgca gacctcctca ttctaacctg aatcggagga 660caaccagtaa
gctacccttt taccatcatt ggacaagtag catccgtact atacttcaca
720acaatcctaa tcctaatacc aactatctcc ctaattgaaa acaaaatact
caaatgggcc 780t 7815565DNAArtificialcDNA 5atggcacatg cagcgcaagt
aggtctacaa gacgctactt cccctatcat agaagagctt 60atcacctttc atgatcacgc
cctcataatc attttcctta tctgcttcct agtcctgtat 120gcccttttcc
taacactcac aacaaaacta actaatacta acatctcaga cgctcaggaa
180atagaaaccg tctgaactat cctgcccgcc atcatcctag tcctcatcgc
cctcccatcc 240ctacgcatcc tttacataac agacgaggtc aacgatccct
cccttaccat caaatcaatt 300ggccaccaat ggtactgaac ctacgagtac
accgactacg gcggactaat cttcaactcc 360tacatacttc ccccattatt
cctagaacca ggcgacctgc gactcctagc cgcagacctc 420ctcattctaa
cctgaatcgg aggacaacca gtaagctacc cttttaccat cattggacaa
480gtagcatccg tactatactt cacaacaatc ctaatcctaa taccaactat
ctccctaatt 540gaaaacaaaa tactcaaatg ggcct 56561174DNAArtificialcDNA
6atggcacatg cagcgcaagt aggtctacaa gacgctactt cccctatcat agaagagctt
60atcacctttc atgatcacgc cctcataatc attttcctta tctgcttcct agtcctgtat
120gcccttttcc taacactcac aacaaaacta actaatacta acatctcaga
cgctcaggaa 180atagaaaccg tctgaactat cctgcccgcc atcatcctag
tcctcatcgc cctcccatcc 240ctacgcatcc tttacataac agacgaggtc
aacgatccct cccttaccat caaatcaatt 300ggccaccaat ggtactgaac
ctacgagtac accgactacg gcggactaat cttcaactcc 360tacatacttc
ccccattatt cctagaacca ggcgacctgc gactccttga cgttgacaat
420cgagtagtac tcccgattga agcccccatt cgtataataa ttacatcaca
agacgtcttg 480cactcatgag ctgtccccac attaggctta aaaacagatg
caattcccgg acgtctaaac 540caaaccactt tcaccgctac acgaccgggg
gtatactacg gtcaatgctc tgaaatctgt 600ggagcaaacc acagtttcat
gcccatattc ttgcacgaaa cgggatcaaa caacccccta 660ggaatcacct
cccattccga taaaatcacc ttccaccctt actacacaat caaagacgcc
720ctcggcttac ttctcttcct tctctcctta atgacattaa cactattctc
accagacctc 780ctaggcgacc cagacaatta taccctagcc aaccccttaa
acacccctcc ccacatcaag 840cccgaatgat atttcctatt cgcctacaca
attctccgat ccgtccctaa caaactagga 900ggcgtccttg ccctattact
atccatcctc atcctagcaa taatccccat cctccatata 960tccaaacaac
aaagcataat atttcgccca ctaagccaat cactttattg actcctagcc
1020gcagacctcc tcattctaac ctgaatcgga ggacaaccag taagctaccc
ttttaccatc 1080attggacaag tagcatccgt actatacttc acaacaatcc
taatcctaat accaactatc 1140tccctaattg aaaacaaaat actcaaatgg gcct
117471294DNAArtificialcDNA 7atgaacgaaa atctgttcgc ttcattcatt
gcccccacaa tcctaggcct acccgccgca 60gtactgatca ttctatttcc ccctctattg
atccccacct ccaaatatct catcaacaac 120cgactaatca ccacccaaca
atgactaatc aaactaacct caaaacaaat gataaccata 180cacaacacta
aaggacgaac ctgatctctt atactagtat ccttaatcat ttttattgcc
240acaactaacc tcctcggact cctgcctcac tcatttacac caaccaccca
actatctata 300aacctagcca tgcactactc accagacgcc tcaaccgcct
tttcatcaat cgcccacatc 360actcgagacg taaattatgg ctgaatcatc
cgctaccttc acgccaatgg cgcctcaata 420ttctttatct gcctcttcct
acacatcggg cgaggcctat attacggatc atttctctac 480tcagaaacct
gaaacatcgg cattatcctc ctgcttgcaa ctatagcaac agccttcata
540ggctatgtcc tcccgtgagg ccaaatatca ttctgagggg ccacagtaat
tacaaactta 600ctatccgcca tcccatacat tgggacagac ctagttcaat
gaatctgagg aggctactca 660gtagacagtc ccaccctcac acgattcttt
acctttcact tcatcttgcc cttcattatt 720gcagccctag caacactcca
cctcctattc ttgcacgaaa cgggatcaaa caacccccta 780ggaatcacct
cccattccga taaaatcacc ttccaccctt actacacaat caaagacgcc
840ctcggcttac ttctcttcct tctctcctta atgacattaa cactattctc
accagacctc 900ctaggcgacc cagacaatta taccctagcc aaccccttaa
acacccctcc ccacatcaag 960cccgaatgat atttcctatt cgcctacaca
attctccgat ccgtccctaa caaactagga 1020ggcgtccttg ccctattact
atccatcctc atcctagcaa taatccccat cctccatata 1080tccaaacaac
aaagcataat atttcgccca ctaagccaat cactttattg actcctagcc
1140gcagacctcc tcattctaac ctgaatcgga ggacaaccag taagctaccc
ttttaccatc 1200attggacaag tagcatccgt actatacttc acaacaatcc
taatcctaat accaactatc 1260tccctaattg aaaacaaaat actcaaatgg gcct
129481228DNAArtificialcDNA 8atgcccctca tttacataaa tattatacta
gcatttacca tctcacttct aggaatacta 60gtatatcgct cacacctcat atcctcccta
ctatgcctag aaggaataat actatcgctg 120ttcattatag ctactctcat
aaccctcaac acccactccc tcttagccaa tattgtgcct 180attgccatac
tagtctttgg cgcctgcctg atcctccaaa tcaccacagg actattccta
240gccatgcact actcaccaga cgcctcaacc gccttttcat caatcgccca
catcactcga 300gacgtaaatt atggctgaat catccgctac cttcacgcca
atggcgcctc aatattcttt 360atctgcctct tcctacacat cgggcgaggc
ctatattacg gatcatttct ctactcagaa 420acctgaaaca tcggcattat
cctcctgctt gcaactatag caacagcctt cataggctat 480gtcctcccgt
gaggccaaat atcattctga ggggccacag taattacaaa cttactatcc
540gccatcccat acattgggac agacctagtt caatgaatct gaggaggcta
ctcagtagac 600agtcccaccc tcacacgatt ctttaccttt cacttcatct
tgcccttcat tattgcagcc 660ctagcaacac tccacctcct attcttgcac
gaaacgggat caaacaaccc cctaggaatc 720acctcccatt ccgataaaat
caccttccac ccttactaca caatcaaaga cgccctcggc 780ttacttctct
tccttctctc cttaatgaca ttaacactat tctcaccaga cctcctaggc
840gacccagaca attataccct agccaacccc ttaaacaccc ctccccacat
caagcccgaa 900tgatatttcc tattcgccta cacaattctc cgatccgtcc
ctaacaaact aggaggcgtc 960cttgccctat tactatccat cctcatccta
gcaataatcc ccatcctcca tatatccaaa 1020caacaaagca taatatttcg
cccactaagc caatcacttt attgactcct agccgcagac 1080ctcctcattc
taacctgaat cggaggacaa ccagtaagct acccttttac catcattgga
1140caagtagcat ccgtactata cttcacaaca atcctaatcc taataccaac
tatctcccta 1200attgaaaaca aaatactcaa atgggcct
12289522DNAArtificialcDNA 9atgttcgccg accgttgact attctctaca
aaccacaaag acattggaac actataccta 60ttattcggcg catgagctgg agtcctaggc
acagctctaa gcctccttat tcgagccgag 120ctgggccagc caggcaacct
tctaggtaac gaccacatct acaacgttat cgtcacagcc 180ctcgctgtca
ctttcctagg acttctaaca gccctagacc tcaactacct aaccaacaaa
240cttaaaataa aatccccact atgcacattt tatttctcca acatactcgg
attctaccct 300agcatcacac accgcacaat cccctatcta ggccttctta
cgagccaaaa cctgccccta 360ctcctcctag acctaacctg actagaaaag
ctattaccta aaacaatttc acagcaccaa 420atctccacct ccatcatcac
ctcaacccaa aaaggcataa ttaaacttta cttcctctct 480ttcttcttcc
cactcatcct aaccctactc ctaatcacat aa 52210582DNAArtificialcDNA
10atgttcgccg accgttgact attctctaca aaccacaaag acattggaac actataccta
60ttattcggcg catgagctgg agtcctaggc acagctctaa gcctccttat tcgagccgag
120ctgggccagc caggcaacct tctaggtaac gaccacatct acaacgttat
cgtcacagcc 180catgcatttg taataatctt cttcatagta atacccatca
taatcggagg ctttggcaac 240tgactagttc ccctaataat cggtgccccc
gatatggcgt ttccccgcat aaacaacata 300agcttctgac tcttacctcc
ctctctccta ctcctgctcg catctgctat agtggaggcc 360ggagcaggaa
caggttgaac agtctaccct cccttagcag ggaactactc ccaccctgga
420gccctcctag acctaacctg actagaaaag ctattaccta aaacaatttc
acagcaccaa 480atctccacct ccatcatcac ctcaacccaa aaaggcataa
ttaaacttta cttcctctct 540ttcttcttcc cactcatcct aaccctactc
ctaatcacat aa 582112208DNAArtificialcDNA 11atgttcgccg accgttgact
attctctaca aaccacaaag acattggaac actataccta 60ttattcggcg catgagctgg
agtcctaggc acagctctaa gcctccttat tcgagccgag 120ctgggccagc
caggcaacct tctaggtaac gaccacatct acaacgttat cgtcacagcc
180catgcatttg taataatctt cttcatagta atacccatca taatcggagg
ctttggcaac 240tgactagttc ccctaataat cggtgccccc gatatggcgt
ttccccgcat aaacaacata 300agcttctgac tcttacctcc ctctctccta
ctcctgctcg catctgctat agtggaggcc 360ggagcaggaa caggttgaac
agtctaccct cccttagcag ggaactactc ccaccctgga 420gcctccgtag
acctaaccat cttctcctta cacctagcag gtgtctcctc tatcttaggg
480gccatcaatt tcatcacaac aattatcaat ataaaacccc ctgccataac
ccaataccaa 540acgcccctct tcgtctgatc cgtcctaatc acagcagtcc
tacttctcct atctctccca 600gtcctagctg ctggcatcac tatactacta
acagaccgca acctcaacac caccttcttc 660gaccccgccg gaggaggaga
ccccattcta taccaacacc tattctgatt tttcggtcac 720cctgaagttt
atattcttat cctaccaggc ttcggaataa tctcccatat tgtaacttac
780tactccggaa aaaaagaacc atttggatac ataggtatgg tctgagctat
gatatcaatt 840ggcttcctag ggtttatcgt gtgagcacac catatattta
cagtaggaat agacgtagac 900acacgagcat atttcacctc cgctaccata
atcatcgcta tccccaccgg cgtcaaagta 960tttagctgac tcgccacact
ccacggaagc aatatgaaat gatctgctgc agtgctctga 1020gccctaggat
tcatctttct tttcaccgta ggtggcctga ctggcattgt attagcaaac
1080tcatcactag acatcgtact acacgacacg tactacgttg tagcccactt
ccactatgtc 1140ctatcaatag gagctgtatt tgccatcata ggaggcttca
ttcactgatt tcccctattc 1200tcaggctaca ccctagacca aacctacgcc
aaaatccatt tcactatcat attcatcggc 1260gtaaatctaa ctttcttccc
acaacacttt ctcggcctat ccggaatgcc ccgacgttac 1320tcggactacc
ccgatgcata caccacatga aacatcctat catctgtagg ctcattcatt
1380tctctaacag cagtaatatt aataattttc atgatttgag aagccttcgc
ttcgaagcga 1440aaagtcctaa tagtagaaga accctccata aacctggagt
gactatatgg atgcccccca 1500ccctaccaca cattcgaaga acccgtatac
ataaaagcag gaataccttt cctcacaggt 1560ttctactcca aagaccacat
catcgaaacc gcaaacatat catacacaaa cgcctgagcc 1620ctatctatta
ctctcatcgc tacctccctg acaagcgcct atagcactcg aataattctt
1680ctcaccctaa caggtcaacc tcgcttcccc acccttacta acattaacga
aaataacccc 1740accctactaa accccattaa acgcctggca gccggaagcc
tattcgcagg atttctcatt 1800actaacaaca tttcccccgc atcccccttc
caaacaacaa tccccctcta cctaaaactc 1860acagccctcg ctgtcacttt
cctaggactt ctaacagccc tagacctcaa ctacctaacc 1920aacaaactta
aaataaaatc cccactatgc acattttatt tctccaacat actcggattc
1980taccctagca tcacacaccg cacaatcccc tatctaggcc ttcttacgag
ccaaaacctg 2040cccctactcc tcctagacct aacctgacta gaaaagctat
tacctaaaac aatttcacag 2100caccaaatct ccacctccat catcacctca
acccaaaaag gcataattaa actttacttc 2160ctctctttct tcttcccact
catcctaacc ctactcctaa tcacataa 220812807DNAArtificialcDNA
12atggcacatg cagcgcaagt aggtctacaa gacgctactt cccctatcat agaagagctt
60atcacctttc atgatcacgc cctcataatc attttcctta tctgcttcct agtcctgtat
120gcccttttcc taacactcac aacaaaacta actaatacta acatctcaga
cgctcaggaa 180atagaaaccg caaacatatc atacacaaac gcctgagccc
tatctattac tctcatcgct 240acctccctga caagcgccta tagcactcga
ataattcttc tcaccctaac aggtcaacct 300cgcttcccca cccttactaa
cattaacgaa aataacccca ccctactaaa ccccattaaa 360cgcctggcag
ccggaagcct attcgcagga tttctcatta ctaacaacat ttcccccgca
420tcccccttcc aaacaacaat ccccctctac ctaaaactca cagccctcgc
tgtcactttc 480ctaggacttc taacagccct agacctcaac tacctaacca
acaaacttaa aataaaatcc 540ccactatgca cattttattt ctccaacata
ctcggattct accctagcat cacacaccgc 600acaatcccct atctaggcct
tcttacgagc caaaacctgc ccctactcct cctagaccta 660acctgactag
aaaagctatt acctaaaaca atttcacagc accaaatctc cacctccatc
720atcacctcaa cccaaaaagg cataattaaa ctttacttcc tctctttctt
cttcccactc 780atcctaaccc tactcctaat cacataa
80713786DNAArtificialcDNA 13atggcacatg cagcgcaagt aggtctacaa
gacgctactt cccctatcat agaagagctt 60atcacctttc atgatcacgc cctcataatc
attttcctta tctgcttcct agtcctgtat 120gcccttttcc taacactcac
aacaaaacta actaatacta acatctcaga cgctcaggaa 180atagaaaccg
tctgaactat cctgcccgcc atcatcctag tcctcatcgc cctcccatcc
240ctacgcatcc tttacataac agacgaggtc aacgatccct cccttaccat
caaatcaatt 300ggccaccaat ggtactgaac ctacgagtac accgactacg
gcggactaat cttcaactcc 360tacatacttc ccccattatt cctagaacca
ggcgacctgc gactccttga cgttgacaat 420cgagtagtac tcccgattga
agcccccatt cgtataataa ttacatcaca agacgtcttg 480cactcatgag
ctgtccccac attaggctta aaaacagatg caattcccgg acgtctaaac
540caaaccactt tcaccgctac acgaccgggg gtatactacg gtcaatgctc
tgaaatctgt 600ggagcaaacc acagtttcat gcccatcgtc ctagacctaa
cctgactaga aaagctatta 660cctaaaacaa tttcacagca ccaaatctcc
acctccatca tcacctcaac ccaaaaaggc 720ataattaaac tttacttcct
ctctttcttc ttcccactca tcctaaccct actcctaatc 780acataa
786141905DNAArtificialcDNA 14atgaacgaaa atctgttcgc ttcattcatt
gcccccacaa tcctaggcct acccgccgca 60gtactgatca ttctatttcc ccctctattg
atccccacct ccaaatatct catcaacaac 120cgactaatca ccacccaaca
atgactaatc aaactaacct caaaacaaat gataaccata 180cacaacacta
aaggacgaac ctgatctctt atactagtat ccttaatcat ttttattgcc
240acaactaacc tcctcggact cctgcctcac tcatttacac caaccaccca
actatctata 300aacctagcca tggccatccc cttatgagcg ggcacagtga
ttataggctt tcgctctaag 360attaaaaatg ccctagccca cttcttacca
caaggcacac ctacacccct tatccccata 420ctagttatta tcgaaaccat
cagcctactc attcaaccaa tagccctggc cgtacgccta 480accgctaaca
ttactgcagg ccacctactc atgcacctaa ttggaagcgc caccctagca
540atatcaacca ttaaccttcc ctctacactt atcatcttca caattctaat
tctactgact 600atcctagaaa tcgctgtcgc cttaatccaa gcctacgttt
tcacacttct agtaagcctc 660tacctacact ccaactcatg agacccacaa
caaatagccc ttctaaacgc taatccaagc 720ctcaccccac tactaggcct
cctcctagca gcagcaggca aatcagccca attaggtctc 780cacccctgac
tcccctcagc catagaaggc cccaccccag tctcagccct actccactca
840agcactatag ttgtagcagg aatcttctta ctcatccgct tccaccccct
agcagaaaat 900agcccactaa tccaaactct aacactatgc ttaggcgcta
tcaccactct gttcgcagca 960gtctgcgccc ttacacaaaa tgacatcaaa
aaaatcgtag ccttctccac ttcaagtcaa 1020ctaggactca taatagttac
aatcggcatc aaccaaccac acctagcatt cctgcacatc 1080tgtacccacg
ccttcttcaa agccatacta tttatgtgct ccgggtccat catccacaac
1140cttaacaatg aacaagatat tcgaaaaata ggaggactac tcaaaaccat
acctctcact 1200tcaacctccc tcaccattgg cagcctagca ttagcaggaa
tacctttcct cacaggtttc 1260tactccaaag accacatcat cgaaaccgca
aacatatcat acacaaacgc ctgagcccta 1320tctattactc tcatcgctac
ctccctgaca agcgcctata gcactcgaat aattcttctc 1380accctaacag
gtcaacctcg cttccccacc cttactaaca ttaacgaaaa taaccccacc
1440ctactaaacc ccattaaacg cctggcagcc ggaagcctat tcgcaggatt
tctcattact 1500aacaacattt cccccgcatc ccccttccaa acaacaatcc
ccctctacct aaaactcaca 1560gccctcgctg tcactttcct aggacttcta
acagccctag acctcaacta cctaaccaac 1620aaacttaaaa taaaatcccc
actatgcaca ttttatttct ccaacatact cggattctac 1680cctagcatca
cacaccgcac aatcccctat ctaggccttc ttacgagcca aaacctgccc
1740ctactcctcc tagacctaac ctgactagaa aagctattac ctaaaacaat
ttcacagcac
1800caaatctcca cctccatcat cacctcaacc caaaaaggca taattaaact
ttacttcctc 1860tctttcttct tcccactcat cctaacccta ctcctaatca cataa
1905151545DNAArtificialcDNA 15atgacccacc aatcacatgc ctatcatata
gtaaaaccca gcccatgacc cctaacaggg 60gccctctcag ccctcctaat gacctccggc
ctagccatgt gatttcactt ccactccata 120acgctcctca tactaggcct
actaaccaac acactaacca tataccaatg atggcgcgat 180gtaacacgag
aaagcacata ccaaggccac cacacaccac ctgtccaaaa aggccttcga
240tacgggataa tcctatttat tacctcagaa gtttttttct tcgcaggatt
tttctgagcc 300ttttaccact ccagcctagc ccctaccccc caattaggag
ggcactggcc cccaacaggc 360atcaccccac tactaggcct cctcctagca
gcagcaggca aatcagccca attaggtctc 420cacccctgac tcccctcagc
catagaaggc cccaccccag tctcagccct actccactca 480agcactatag
ttgtagcagg aatcttctta ctcatccgct tccaccccct agcagaaaat
540agcccactaa tccaaactct aacactatgc ttaggcgcta tcaccactct
gttcgcagca 600gtctgcgccc ttacacaaaa tgacatcaaa aaaatcgtag
ccttctccac ttcaagtcaa 660ctaggactca taatagttac aatcggcatc
aaccaaccac acctagcatt cctgcacatc 720tgtacccacg ccttcttcaa
agccatacta tttatgtgct ccgggtccat catccacaac 780cttaacaatg
aacaagatat tcgaaaaata ggaggactac tcaaaaccat acctctcact
840tcaacctccc tcaccattgg cagcctagca ttagcaggaa tacctttcct
cacaggtttc 900tactccaaag accacatcat cgaaaccgca aacatatcat
acacaaacgc ctgagcccta 960tctattactc tcatcgctac ctccctgaca
agcgcctata gcactcgaat aattcttctc 1020accctaacag gtcaacctcg
cttccccacc cttactaaca ttaacgaaaa taaccccacc 1080ctactaaacc
ccattaaacg cctggcagcc ggaagcctat tcgcaggatt tctcattact
1140aacaacattt cccccgcatc ccccttccaa acaacaatcc ccctctacct
aaaactcaca 1200gccctcgctg tcactttcct aggacttcta acagccctag
acctcaacta cctaaccaac 1260aaacttaaaa taaaatcccc actatgcaca
ttttatttct ccaacatact cggattctac 1320cctagcatca cacaccgcac
aatcccctat ctaggccttc ttacgagcca aaacctgccc 1380ctactcctcc
tagacctaac ctgactagaa aagctattac ctaaaacaat ttcacagcac
1440caaatctcca cctccatcat cacctcaacc caaaaaggca taattaaact
ttacttcctc 1500tctttcttct tcccactcat cctaacccta ctcctaatca cataa
1545161629DNAArtificialcDNA 16ataaacttcg ccttaatttt aataatcaac
accctcctag ccttactact aataattatt 60acattttgac taccacaact caacggctac
atagaaaaat ccacccctta cgagtgcggc 120ttcgacccta tatcccccgc
ccgcgtccct ttctccataa aattcttctt agtagctatt 180accttcttat
tatttgatct agaaattgcc ctccttttac ccctaccatg agccctacaa
240acaactaacc tgccactaat agttatgtca tccctcttat taatcatcat
cctagcccta 300agtctggcca acacagcagc cattcaagca atcctataca
accgtatcgg cgatatcggt 360ttcatcctcg ccttagcatg atttatccta
cactccaact catgagaccc acaacaaata 420gcccttctaa acgctaatcc
aagcctcacc ccactactag gcctcctcct agcagcagca 480ggcaaatcag
cccaattagg tctccacccc tgactcccct cagccataga aggccccacc
540ccagtctcag ccctactcca ctcaagcact atagttgtag caggaatctt
cttactcatc 600cgcttccacc ccctagcaga aaatagccca ctaatccaaa
ctctaacact atgcttaggc 660gctatcacca ctctgttcgc agcagtctgc
gcccttacac aaaatgacat caaaaaaatc 720gtagccttct ccacttcaag
tcaactagga ctcataatag ttacaatcgg catcaaccaa 780ccacacctag
cattcctgca catctgtacc cacgccttct tcaaagccat actatttatg
840tgctccgggt ccatcatcca caaccttaac aatgaacaag atattcgaaa
aataggagga 900ctactcaaaa ccatacctct cacttcaacc tccctcacca
ttggcagcct agcattagca 960ggaatacctt tcctcacagg tttctactcc
aaagaccaca tcatcgaaac cgcaaacata 1020tcatacacaa acgcctgagc
cctatctatt actctcatcg ctacctccct gacaagcgcc 1080tatagcactc
gaataattct tctcacccta acaggtcaac ctcgcttccc cacccttact
1140aacattaacg aaaataaccc caccctacta aaccccatta aacgcctggc
agccggaagc 1200ctattcgcag gatttctcat tactaacaac atttcccccg
catccccctt ccaaacaaca 1260atccccctct acctaaaact cacagccctc
gctgtcactt tcctaggact tctaacagcc 1320ctagacctca actacctaac
caacaaactt aaaataaaat ccccactatg cacattttat 1380ttctccaaca
tactcggatt ctaccctagc atcacacacc gcacaatccc ctatctaggc
1440cttcttacga gccaaaacct gcccctactc ctcctagacc taacctgact
agaaaagcta 1500ttacctaaaa caatttcaca gcaccaaatc tccacctcca
tcatcacctc aacccaaaaa 1560ggcataatta aactttactt cctctctttc
ttcttcccac tcatcctaac cctactccta 1620atcacataa
162917129DNAArtificialcDNA 17atgccccaac taaatactac cgtatggccc
accataatta cccccatact ccttacacta 60ttcctcatca cccaactaaa aatattaaac
acaaactacc acctacctcc ctcaccattg 120gcagcctag 12918783RNAHuman
18auggcccacc auaauuaccc ccauacuccu uacacuauuc cucaucaccc aacuaaaaau
60auuaaacaca aacuaccacc uaccucccuc accauuggca gccuagcauu agcaggaaua
120ccuuuccuca cagguuucua cuccaaagac cacaucaucg aaaccgcaaa
cauaucauac 180acaaacgccu gagcccuauc uauuacucuc aucgcuaccu
cccugacaag cgccuauagc 240acucgaauaa uucuucucac ccuaacaggu
caaccucgcu uccccacccu uacuaacauu 300aacgaaaaua accccacccu
acuaaacccc auuaaacgcc uggcagccgg aagccuauuc 360gcaggauuuc
ucauuacuaa caacauuucc cccgcauccc ccuuccaaac aacaaucccc
420cucuaccuaa aacucacagc ccucgcuguc acuuuccuag gacuucuaac
agcccuagac 480cucaacuacc uaaccaacaa acuuaaaaua aaauccccac
uaugcacauu uuauuucucc 540aacauacucg gauucuaccc uagcaucaca
caccgcacaa uccccuaucu aggccuucuu 600acgagccaaa accugccccu
acuccuccua gaccuaaccu gacuagaaaa gcuauuaccu 660aaaacaauuu
cacagcacca aaucuccacc uccaucauca ccucaaccca aaaaggcaua
720auuaaacuuu acuuccucuc uuucuucuuc ccacucaucc uaacccuacu
ccuaaucaca 780uaa 78319300RNAHuman 19augccccuca uuuacauaaa
uauuauacua gcauuuacca ucucacuucu aggaauacua 60guauaucgcu cacaccucau
auccucccua cuaugccuag aaggaauaau acuaucgcug 120uucauuauag
cuacucucau aacccucaac acccacuccc ucuuagccaa uauugugccu
180auugccauac uagucuuugc cgccugcgaa gcagcggugg gccuagcccu
acuagucuca 240aucuccaaca cauauggccu agacuacgua cauaaccuaa
cccuacuccu aaucacauaa 30020781RNAHuman 20auggcacaug cagcgcaagu
aggucuacaa gacgcuacuu ccccuaucau agaagagcuu 60aucaccuuuc augaucacgc
ccucauaauc auuuuccuua ucugcuuccu aguccuguau 120gcccuuuucc
uaacacucac aacaaaacua acuaauacua acaucucaga cgcucaggaa
180auagaaaccg ucugaacuau ccugcccgcc aucauccuag uccucaucgc
ccucccaucc 240cuacgcaucc uuuacauaac agacgagguc aacgaucccu
cccuuaccau caaaucaauu 300ggccaccaau gguacugaac cuacgaguac
accgacuacg gcggacuaau cuucaacucc 360uacauacuuc ccccauuauu
ccuagaacca ggcgacccag acaauuauac ccuagccaac 420cccuuaaaca
ccccucccca caucaagccc gaaugauauu uccuauucgc cuacacaauu
480cuccgauccg ucccuaacaa acuaggaggc guccuugccc uauuacuauc
cauccucauc 540cuagcaauaa uccccauccu ccauauaucc aaacaacaaa
gcauaauauu ucgcccacua 600agccaaucac uuuauugacu ccuagccgca
gaccuccuca uucuaaccug aaucggagga 660caaccaguaa gcuacccuuu
uaccaucauu ggacaaguag cauccguacu auacuucaca 720acaauccuaa
uccuaauacc aacuaucucc cuaauugaaa acaaaauacu caaaugggcc 780u
78121565RNAHuman 21auggcacaug cagcgcaagu aggucuacaa gacgcuacuu
ccccuaucau agaagagcuu 60aucaccuuuc augaucacgc ccucauaauc auuuuccuua
ucugcuuccu aguccuguau 120gcccuuuucc uaacacucac aacaaaacua
acuaauacua acaucucaga cgcucaggaa 180auagaaaccg ucugaacuau
ccugcccgcc aucauccuag uccucaucgc ccucccaucc 240cuacgcaucc
uuuacauaac agacgagguc aacgaucccu cccuuaccau caaaucaauu
300ggccaccaau gguacugaac cuacgaguac accgacuacg gcggacuaau
cuucaacucc 360uacauacuuc ccccauuauu ccuagaacca ggcgaccugc
gacuccuagc cgcagaccuc 420cucauucuaa ccugaaucgg aggacaacca
guaagcuacc cuuuuaccau cauuggacaa 480guagcauccg uacuauacuu
cacaacaauc cuaauccuaa uaccaacuau cucccuaauu 540gaaaacaaaa
uacucaaaug ggccu 565221174RNAHuman 22auggcacaug cagcgcaagu
aggucuacaa gacgcuacuu ccccuaucau agaagagcuu 60aucaccuuuc augaucacgc
ccucauaauc auuuuccuua ucugcuuccu aguccuguau 120gcccuuuucc
uaacacucac aacaaaacua acuaauacua acaucucaga cgcucaggaa
180auagaaaccg ucugaacuau ccugcccgcc aucauccuag uccucaucgc
ccucccaucc 240cuacgcaucc uuuacauaac agacgagguc aacgaucccu
cccuuaccau caaaucaauu 300ggccaccaau gguacugaac cuacgaguac
accgacuacg gcggacuaau cuucaacucc 360uacauacuuc ccccauuauu
ccuagaacca ggcgaccugc gacuccuuga cguugacaau 420cgaguaguac
ucccgauuga agcccccauu cguauaauaa uuacaucaca agacgucuug
480cacucaugag cuguccccac auuaggcuua aaaacagaug caauucccgg
acgucuaaac 540caaaccacuu ucaccgcuac acgaccgggg guauacuacg
gucaaugcuc ugaaaucugu 600ggagcaaacc acaguuucau gcccauauuc
uugcacgaaa cgggaucaaa caacccccua 660ggaaucaccu cccauuccga
uaaaaucacc uuccacccuu acuacacaau caaagacgcc 720cucggcuuac
uucucuuccu ucucuccuua augacauuaa cacuauucuc accagaccuc
780cuaggcgacc cagacaauua uacccuagcc aaccccuuaa acaccccucc
ccacaucaag 840cccgaaugau auuuccuauu cgccuacaca auucuccgau
ccgucccuaa caaacuagga 900ggcguccuug cccuauuacu auccauccuc
auccuagcaa uaauccccau ccuccauaua 960uccaaacaac aaagcauaau
auuucgccca cuaagccaau cacuuuauug acuccuagcc 1020gcagaccucc
ucauucuaac cugaaucgga ggacaaccag uaagcuaccc uuuuaccauc
1080auuggacaag uagcauccgu acuauacuuc acaacaaucc uaauccuaau
accaacuauc 1140ucccuaauug aaaacaaaau acucaaaugg gccu
1174231294RNAHuman 23augaacgaaa aucuguucgc uucauucauu gcccccacaa
uccuaggccu acccgccgca 60guacugauca uucuauuucc cccucuauug auccccaccu
ccaaauaucu caucaacaac 120cgacuaauca ccacccaaca augacuaauc
aaacuaaccu caaaacaaau gauaaccaua 180cacaacacua aaggacgaac
cugaucucuu auacuaguau ccuuaaucau uuuuauugcc 240acaacuaacc
uccucggacu ccugccucac ucauuuacac caaccaccca acuaucuaua
300aaccuagcca ugcacuacuc accagacgcc ucaaccgccu uuucaucaau
cgcccacauc 360acucgagacg uaaauuaugg cugaaucauc cgcuaccuuc
acgccaaugg cgccucaaua 420uucuuuaucu gccucuuccu acacaucggg
cgaggccuau auuacggauc auuucucuac 480ucagaaaccu gaaacaucgg
cauuauccuc cugcuugcaa cuauagcaac agccuucaua 540ggcuaugucc
ucccgugagg ccaaauauca uucugagggg ccacaguaau uacaaacuua
600cuauccgcca ucccauacau ugggacagac cuaguucaau gaaucugagg
aggcuacuca 660guagacaguc ccacccucac acgauucuuu accuuucacu
ucaucuugcc cuucauuauu 720gcagcccuag caacacucca ccuccuauuc
uugcacgaaa cgggaucaaa caacccccua 780ggaaucaccu cccauuccga
uaaaaucacc uuccacccuu acuacacaau caaagacgcc 840cucggcuuac
uucucuuccu ucucuccuua augacauuaa cacuauucuc accagaccuc
900cuaggcgacc cagacaauua uacccuagcc aaccccuuaa acaccccucc
ccacaucaag 960cccgaaugau auuuccuauu cgccuacaca auucuccgau
ccgucccuaa caaacuagga 1020ggcguccuug cccuauuacu auccauccuc
auccuagcaa uaauccccau ccuccauaua 1080uccaaacaac aaagcauaau
auuucgccca cuaagccaau cacuuuauug acuccuagcc 1140gcagaccucc
ucauucuaac cugaaucgga ggacaaccag uaagcuaccc uuuuaccauc
1200auuggacaag uagcauccgu acuauacuuc acaacaaucc uaauccuaau
accaacuauc 1260ucccuaauug aaaacaaaau acucaaaugg gccu
1294241228RNAHuman 24augccccuca uuuacauaaa uauuauacua gcauuuacca
ucucacuucu aggaauacua 60guauaucgcu cacaccucau auccucccua cuaugccuag
aaggaauaau acuaucgcug 120uucauuauag cuacucucau aacccucaac
acccacuccc ucuuagccaa uauugugccu 180auugccauac uagucuuugg
cgccugccug auccuccaaa ucaccacagg acuauuccua 240gccaugcacu
acucaccaga cgccucaacc gccuuuucau caaucgccca caucacucga
300gacguaaauu auggcugaau cauccgcuac cuucacgcca auggcgccuc
aauauucuuu 360aucugccucu uccuacacau cgggcgaggc cuauauuacg
gaucauuucu cuacucagaa 420accugaaaca ucggcauuau ccuccugcuu
gcaacuauag caacagccuu cauaggcuau 480guccucccgu gaggccaaau
aucauucuga ggggccacag uaauuacaaa cuuacuaucc 540gccaucccau
acauugggac agaccuaguu caaugaaucu gaggaggcua cucaguagac
600agucccaccc ucacacgauu cuuuaccuuu cacuucaucu ugcccuucau
uauugcagcc 660cuagcaacac uccaccuccu auucuugcac gaaacgggau
caaacaaccc ccuaggaauc 720accucccauu ccgauaaaau caccuuccac
ccuuacuaca caaucaaaga cgcccucggc 780uuacuucucu uccuucucuc
cuuaaugaca uuaacacuau ucucaccaga ccuccuaggc 840gacccagaca
auuauacccu agccaacccc uuaaacaccc cuccccacau caagcccgaa
900ugauauuucc uauucgccua cacaauucuc cgauccgucc cuaacaaacu
aggaggcguc 960cuugcccuau uacuauccau ccucauccua gcaauaaucc
ccauccucca uauauccaaa 1020caacaaagca uaauauuucg cccacuaagc
caaucacuuu auugacuccu agccgcagac 1080cuccucauuc uaaccugaau
cggaggacaa ccaguaagcu acccuuuuac caucauugga 1140caaguagcau
ccguacuaua cuucacaaca auccuaaucc uaauaccaac uaucucccua
1200auugaaaaca aaauacucaa augggccu 122825522RNAHuman 25auguucgccg
accguugacu auucucuaca aaccacaaag acauuggaac acuauaccua 60uuauucggcg
caugagcugg aguccuaggc acagcucuaa gccuccuuau ucgagccgag
120cugggccagc caggcaaccu ucuagguaac gaccacaucu acaacguuau
cgucacagcc 180cucgcuguca cuuuccuagg acuucuaaca gcccuagacc
ucaacuaccu aaccaacaaa 240cuuaaaauaa aauccccacu augcacauuu
uauuucucca acauacucgg auucuacccu 300agcaucacac accgcacaau
ccccuaucua ggccuucuua cgagccaaaa ccugccccua 360cuccuccuag
accuaaccug acuagaaaag cuauuaccua aaacaauuuc acagcaccaa
420aucuccaccu ccaucaucac cucaacccaa aaaggcauaa uuaaacuuua
cuuccucucu 480uucuucuucc cacucauccu aacccuacuc cuaaucacau aa
52226582RNAHuman 26auguucgccg accguugacu auucucuaca aaccacaaag
acauuggaac acuauaccua 60uuauucggcg caugagcugg aguccuaggc acagcucuaa
gccuccuuau ucgagccgag 120cugggccagc caggcaaccu ucuagguaac
gaccacaucu acaacguuau cgucacagcc 180caugcauuug uaauaaucuu
cuucauagua auacccauca uaaucggagg cuuuggcaac 240ugacuaguuc
cccuaauaau cggugccccc gauauggcgu uuccccgcau aaacaacaua
300agcuucugac ucuuaccucc cucucuccua cuccugcucg caucugcuau
aguggaggcc 360ggagcaggaa cagguugaac agucuacccu cccuuagcag
ggaacuacuc ccacccugga 420gcccuccuag accuaaccug acuagaaaag
cuauuaccua aaacaauuuc acagcaccaa 480aucuccaccu ccaucaucac
cucaacccaa aaaggcauaa uuaaacuuua cuuccucucu 540uucuucuucc
cacucauccu aacccuacuc cuaaucacau aa 582272208RNAHuman 27auguucgccg
accguugacu auucucuaca aaccacaaag acauuggaac acuauaccua 60uuauucggcg
caugagcugg aguccuaggc acagcucuaa gccuccuuau ucgagccgag
120cugggccagc caggcaaccu ucuagguaac gaccacaucu acaacguuau
cgucacagcc 180caugcauuug uaauaaucuu cuucauagua auacccauca
uaaucggagg cuuuggcaac 240ugacuaguuc cccuaauaau cggugccccc
gauauggcgu uuccccgcau aaacaacaua 300agcuucugac ucuuaccucc
cucucuccua cuccugcucg caucugcuau aguggaggcc 360ggagcaggaa
cagguugaac agucuacccu cccuuagcag ggaacuacuc ccacccugga
420gccuccguag accuaaccau cuucuccuua caccuagcag gugucuccuc
uaucuuaggg 480gccaucaauu ucaucacaac aauuaucaau auaaaacccc
cugccauaac ccaauaccaa 540acgccccucu ucgucugauc cguccuaauc
acagcagucc uacuucuccu aucucuccca 600guccuagcug cuggcaucac
uauacuacua acagaccgca accucaacac caccuucuuc 660gaccccgccg
gaggaggaga ccccauucua uaccaacacc uauucugauu uuucggucac
720ccugaaguuu auauucuuau ccuaccaggc uucggaauaa ucucccauau
uguaacuuac 780uacuccggaa aaaaagaacc auuuggauac auagguaugg
ucugagcuau gauaucaauu 840ggcuuccuag gguuuaucgu gugagcacac
cauauauuua caguaggaau agacguagac 900acacgagcau auuucaccuc
cgcuaccaua aucaucgcua uccccaccgg cgucaaagua 960uuuagcugac
ucgccacacu ccacggaagc aauaugaaau gaucugcugc agugcucuga
1020gcccuaggau ucaucuuucu uuucaccgua gguggccuga cuggcauugu
auuagcaaac 1080ucaucacuag acaucguacu acacgacacg uacuacguug
uagcccacuu ccacuauguc 1140cuaucaauag gagcuguauu ugccaucaua
ggaggcuuca uucacugauu uccccuauuc 1200ucaggcuaca cccuagacca
aaccuacgcc aaaauccauu ucacuaucau auucaucggc 1260guaaaucuaa
cuuucuuccc acaacacuuu cucggccuau ccggaaugcc ccgacguuac
1320ucggacuacc ccgaugcaua caccacauga aacauccuau caucuguagg
cucauucauu 1380ucucuaacag caguaauauu aauaauuuuc augauuugag
aagccuucgc uucgaagcga 1440aaaguccuaa uaguagaaga acccuccaua
aaccuggagu gacuauaugg augcccccca 1500cccuaccaca cauucgaaga
acccguauac auaaaagcag gaauaccuuu ccucacaggu 1560uucuacucca
aagaccacau caucgaaacc gcaaacauau cauacacaaa cgccugagcc
1620cuaucuauua cucucaucgc uaccucccug acaagcgccu auagcacucg
aauaauucuu 1680cucacccuaa caggucaacc ucgcuucccc acccuuacua
acauuaacga aaauaacccc 1740acccuacuaa accccauuaa acgccuggca
gccggaagcc uauucgcagg auuucucauu 1800acuaacaaca uuucccccgc
aucccccuuc caaacaacaa ucccccucua ccuaaaacuc 1860acagcccucg
cugucacuuu ccuaggacuu cuaacagccc uagaccucaa cuaccuaacc
1920aacaaacuua aaauaaaauc cccacuaugc acauuuuauu ucuccaacau
acucggauuc 1980uacccuagca ucacacaccg cacaaucccc uaucuaggcc
uucuuacgag ccaaaaccug 2040ccccuacucc uccuagaccu aaccugacua
gaaaagcuau uaccuaaaac aauuucacag 2100caccaaaucu ccaccuccau
caucaccuca acccaaaaag gcauaauuaa acuuuacuuc 2160cucucuuucu
ucuucccacu cauccuaacc cuacuccuaa ucacauaa 220828807RNAHuman
28auggcacaug cagcgcaagu aggucuacaa gacgcuacuu ccccuaucau agaagagcuu
60aucaccuuuc augaucacgc ccucauaauc auuuuccuua ucugcuuccu aguccuguau
120gcccuuuucc uaacacucac aacaaaacua acuaauacua acaucucaga
cgcucaggaa 180auagaaaccg caaacauauc auacacaaac gccugagccc
uaucuauuac ucucaucgcu 240accucccuga caagcgccua uagcacucga
auaauucuuc ucacccuaac aggucaaccu 300cgcuucccca cccuuacuaa
cauuaacgaa aauaacccca cccuacuaaa ccccauuaaa 360cgccuggcag
ccggaagccu auucgcagga uuucucauua cuaacaacau uucccccgca
420ucccccuucc aaacaacaau cccccucuac cuaaaacuca cagcccucgc
ugucacuuuc 480cuaggacuuc uaacagcccu agaccucaac uaccuaacca
acaaacuuaa aauaaaaucc 540ccacuaugca cauuuuauuu cuccaacaua
cucggauucu acccuagcau cacacaccgc 600acaauccccu aucuaggccu
ucuuacgagc caaaaccugc cccuacuccu ccuagaccua 660accugacuag
aaaagcuauu accuaaaaca auuucacagc accaaaucuc caccuccauc
720aucaccucaa cccaaaaagg cauaauuaaa cuuuacuucc ucucuuucuu
cuucccacuc 780auccuaaccc uacuccuaau cacauaa 80729786RNAHuman
29auggcacaug cagcgcaagu aggucuacaa gacgcuacuu ccccuaucau agaagagcuu
60aucaccuuuc augaucacgc ccucauaauc auuuuccuua ucugcuuccu aguccuguau
120gcccuuuucc uaacacucac aacaaaacua acuaauacua acaucucaga
cgcucaggaa 180auagaaaccg ucugaacuau ccugcccgcc aucauccuag
uccucaucgc ccucccaucc 240cuacgcaucc uuuacauaac agacgagguc
aacgaucccu cccuuaccau caaaucaauu 300ggccaccaau gguacugaac
cuacgaguac accgacuacg gcggacuaau cuucaacucc 360uacauacuuc
ccccauuauu ccuagaacca ggcgaccugc gacuccuuga cguugacaau
420cgaguaguac ucccgauuga agcccccauu cguauaauaa uuacaucaca
agacgucuug 480cacucaugag cuguccccac auuaggcuua aaaacagaug
caauucccgg acgucuaaac 540caaaccacuu ucaccgcuac acgaccgggg
guauacuacg gucaaugcuc ugaaaucugu 600ggagcaaacc acaguuucau
gcccaucguc cuagaccuaa ccugacuaga
aaagcuauua 660ccuaaaacaa uuucacagca ccaaaucucc accuccauca
ucaccucaac ccaaaaaggc 720auaauuaaac uuuacuuccu cucuuucuuc
uucccacuca uccuaacccu acuccuaauc 780acauaa 786301905RNAHuman
30augaacgaaa aucuguucgc uucauucauu gcccccacaa uccuaggccu acccgccgca
60guacugauca uucuauuucc cccucuauug auccccaccu ccaaauaucu caucaacaac
120cgacuaauca ccacccaaca augacuaauc aaacuaaccu caaaacaaau
gauaaccaua 180cacaacacua aaggacgaac cugaucucuu auacuaguau
ccuuaaucau uuuuauugcc 240acaacuaacc uccucggacu ccugccucac
ucauuuacac caaccaccca acuaucuaua 300aaccuagcca uggccauccc
cuuaugagcg ggcacaguga uuauaggcuu ucgcucuaag 360auuaaaaaug
cccuagccca cuucuuacca caaggcacac cuacaccccu uauccccaua
420cuaguuauua ucgaaaccau cagccuacuc auucaaccaa uagcccuggc
cguacgccua 480accgcuaaca uuacugcagg ccaccuacuc augcaccuaa
uuggaagcgc cacccuagca 540auaucaacca uuaaccuucc cucuacacuu
aucaucuuca caauucuaau ucuacugacu 600auccuagaaa ucgcugucgc
cuuaauccaa gccuacguuu ucacacuucu aguaagccuc 660uaccuacacu
ccaacucaug agacccacaa caaauagccc uucuaaacgc uaauccaagc
720cucaccccac uacuaggccu ccuccuagca gcagcaggca aaucagccca
auuaggucuc 780caccccugac uccccucagc cauagaaggc cccaccccag
ucucagcccu acuccacuca 840agcacuauag uuguagcagg aaucuucuua
cucauccgcu uccacccccu agcagaaaau 900agcccacuaa uccaaacucu
aacacuaugc uuaggcgcua ucaccacucu guucgcagca 960gucugcgccc
uuacacaaaa ugacaucaaa aaaaucguag ccuucuccac uucaagucaa
1020cuaggacuca uaauaguuac aaucggcauc aaccaaccac accuagcauu
ccugcacauc 1080uguacccacg ccuucuucaa agccauacua uuuaugugcu
ccggguccau cauccacaac 1140cuuaacaaug aacaagauau ucgaaaaaua
ggaggacuac ucaaaaccau accucucacu 1200ucaaccuccc ucaccauugg
cagccuagca uuagcaggaa uaccuuuccu cacagguuuc 1260uacuccaaag
accacaucau cgaaaccgca aacauaucau acacaaacgc cugagcccua
1320ucuauuacuc ucaucgcuac cucccugaca agcgccuaua gcacucgaau
aauucuucuc 1380acccuaacag gucaaccucg cuuccccacc cuuacuaaca
uuaacgaaaa uaaccccacc 1440cuacuaaacc ccauuaaacg ccuggcagcc
ggaagccuau ucgcaggauu ucucauuacu 1500aacaacauuu cccccgcauc
ccccuuccaa acaacaaucc cccucuaccu aaaacucaca 1560gcccucgcug
ucacuuuccu aggacuucua acagcccuag accucaacua ccuaaccaac
1620aaacuuaaaa uaaaaucccc acuaugcaca uuuuauuucu ccaacauacu
cggauucuac 1680ccuagcauca cacaccgcac aauccccuau cuaggccuuc
uuacgagcca aaaccugccc 1740cuacuccucc uagaccuaac cugacuagaa
aagcuauuac cuaaaacaau uucacagcac 1800caaaucucca ccuccaucau
caccucaacc caaaaaggca uaauuaaacu uuacuuccuc 1860ucuuucuucu
ucccacucau ccuaacccua cuccuaauca cauaa 1905311545RNAHuman
31augacccacc aaucacaugc cuaucauaua guaaaaccca gcccaugacc ccuaacaggg
60gcccucucag cccuccuaau gaccuccggc cuagccaugu gauuucacuu ccacuccaua
120acgcuccuca uacuaggccu acuaaccaac acacuaacca uauaccaaug
auggcgcgau 180guaacacgag aaagcacaua ccaaggccac cacacaccac
cuguccaaaa aggccuucga 240uacgggauaa uccuauuuau uaccucagaa
guuuuuuucu ucgcaggauu uuucugagcc 300uuuuaccacu ccagccuagc
cccuaccccc caauuaggag ggcacuggcc cccaacaggc 360aucaccccac
uacuaggccu ccuccuagca gcagcaggca aaucagccca auuaggucuc
420caccccugac uccccucagc cauagaaggc cccaccccag ucucagcccu
acuccacuca 480agcacuauag uuguagcagg aaucuucuua cucauccgcu
uccacccccu agcagaaaau 540agcccacuaa uccaaacucu aacacuaugc
uuaggcgcua ucaccacucu guucgcagca 600gucugcgccc uuacacaaaa
ugacaucaaa aaaaucguag ccuucuccac uucaagucaa 660cuaggacuca
uaauaguuac aaucggcauc aaccaaccac accuagcauu ccugcacauc
720uguacccacg ccuucuucaa agccauacua uuuaugugcu ccggguccau
cauccacaac 780cuuaacaaug aacaagauau ucgaaaaaua ggaggacuac
ucaaaaccau accucucacu 840ucaaccuccc ucaccauugg cagccuagca
uuagcaggaa uaccuuuccu cacagguuuc 900uacuccaaag accacaucau
cgaaaccgca aacauaucau acacaaacgc cugagcccua 960ucuauuacuc
ucaucgcuac cucccugaca agcgccuaua gcacucgaau aauucuucuc
1020acccuaacag gucaaccucg cuuccccacc cuuacuaaca uuaacgaaaa
uaaccccacc 1080cuacuaaacc ccauuaaacg ccuggcagcc ggaagccuau
ucgcaggauu ucucauuacu 1140aacaacauuu cccccgcauc ccccuuccaa
acaacaaucc cccucuaccu aaaacucaca 1200gcccucgcug ucacuuuccu
aggacuucua acagcccuag accucaacua ccuaaccaac 1260aaacuuaaaa
uaaaaucccc acuaugcaca uuuuauuucu ccaacauacu cggauucuac
1320ccuagcauca cacaccgcac aauccccuau cuaggccuuc uuacgagcca
aaaccugccc 1380cuacuccucc uagaccuaac cugacuagaa aagcuauuac
cuaaaacaau uucacagcac 1440caaaucucca ccuccaucau caccucaacc
caaaaaggca uaauuaaacu uuacuuccuc 1500ucuuucuucu ucccacucau
ccuaacccua cuccuaauca cauaa 1545321629RNAHuman 32auaaacuucg
ccuuaauuuu aauaaucaac acccuccuag ccuuacuacu aauaauuauu 60acauuuugac
uaccacaacu caacggcuac auagaaaaau ccaccccuua cgagugcggc
120uucgacccua uaucccccgc ccgcgucccu uucuccauaa aauucuucuu
aguagcuauu 180accuucuuau uauuugaucu agaaauugcc cuccuuuuac
cccuaccaug agcccuacaa 240acaacuaacc ugccacuaau aguuauguca
ucccucuuau uaaucaucau ccuagcccua 300agucuggcca acacagcagc
cauucaagca auccuauaca accguaucgg cgauaucggu 360uucauccucg
ccuuagcaug auuuauccua cacuccaacu caugagaccc acaacaaaua
420gcccuucuaa acgcuaaucc aagccucacc ccacuacuag gccuccuccu
agcagcagca 480ggcaaaucag cccaauuagg ucuccacccc ugacuccccu
cagccauaga aggccccacc 540ccagucucag cccuacucca cucaagcacu
auaguuguag caggaaucuu cuuacucauc 600cgcuuccacc cccuagcaga
aaauagccca cuaauccaaa cucuaacacu augcuuaggc 660gcuaucacca
cucuguucgc agcagucugc gcccuuacac aaaaugacau caaaaaaauc
720guagccuucu ccacuucaag ucaacuagga cucauaauag uuacaaucgg
caucaaccaa 780ccacaccuag cauuccugca caucuguacc cacgccuucu
ucaaagccau acuauuuaug 840ugcuccgggu ccaucaucca caaccuuaac
aaugaacaag auauucgaaa aauaggagga 900cuacucaaaa ccauaccucu
cacuucaacc ucccucacca uuggcagccu agcauuagca 960ggaauaccuu
uccucacagg uuucuacucc aaagaccaca ucaucgaaac cgcaaacaua
1020ucauacacaa acgccugagc ccuaucuauu acucucaucg cuaccucccu
gacaagcgcc 1080uauagcacuc gaauaauucu ucucacccua acaggucaac
cucgcuuccc cacccuuacu 1140aacauuaacg aaaauaaccc cacccuacua
aaccccauua aacgccuggc agccggaagc 1200cuauucgcag gauuucucau
uacuaacaac auuucccccg caucccccuu ccaaacaaca 1260aucccccucu
accuaaaacu cacagcccuc gcugucacuu uccuaggacu ucuaacagcc
1320cuagaccuca acuaccuaac caacaaacuu aaaauaaaau ccccacuaug
cacauuuuau 1380uucuccaaca uacucggauu cuacccuagc aucacacacc
gcacaauccc cuaucuaggc 1440cuucuuacga gccaaaaccu gccccuacuc
cuccuagacc uaaccugacu agaaaagcua 1500uuaccuaaaa caauuucaca
gcaccaaauc uccaccucca ucaucaccuc aacccaaaaa 1560ggcauaauua
aacuuuacuu ccucucuuuc uucuucccac ucauccuaac ccuacuccua
1620aucacauaa 162933129RNAHuman 33augccccaac uaaauacuac cguauggccc
accauaauua cccccauacu ccuuacacua 60uuccucauca cccaacuaaa aauauuaaac
acaaacuacc accuaccucc cucaccauug 120gcagccuag
12934261PRTArtificialputative protein
sequencemisc_feature(261)..(261)Xaa can be any naturally occurring
amino acid 34Met Ala His His Asn Tyr Pro His Thr Pro Tyr Thr Ile
Pro His His1 5 10 15Pro Thr Lys Asn Ile Lys His Lys Leu Pro Pro Thr
Ser Leu Thr Ile 20 25 30Gly Ser Leu Ala Leu Ala Gly Met Pro Phe Leu
Thr Gly Phe Tyr Ser 35 40 45Lys Asp His Ile Ile Glu Thr Ala Asn Met
Ser Tyr Thr Asn Ala Trp 50 55 60Ala Leu Ser Ile Thr Leu Ile Ala Thr
Ser Leu Thr Ser Ala Tyr Ser65 70 75 80Thr Arg Met Ile Leu Leu Thr
Leu Thr Gly Gln Pro Arg Phe Pro Thr 85 90 95Leu Thr Asn Ile Asn Glu
Asn Asn Pro Thr Leu Leu Asn Pro Ile Lys 100 105 110Arg Leu Ala Ala
Gly Ser Leu Phe Ala Gly Phe Leu Ile Thr Asn Asn 115 120 125Ile Ser
Pro Ala Ser Pro Phe Gln Thr Thr Ile Pro Leu Tyr Leu Lys 130 135
140Leu Thr Ala Leu Ala Val Thr Phe Leu Gly Leu Leu Thr Ala Leu
Asp145 150 155 160Leu Asn Tyr Leu Thr Asn Lys Leu Lys Met Lys Ser
Pro Leu Cys Thr 165 170 175Phe Tyr Phe Ser Asn Met Leu Gly Phe Tyr
Pro Ser Ile Thr His Arg 180 185 190Thr Ile Pro Tyr Leu Gly Leu Leu
Thr Ser Gln Asn Leu Pro Leu Leu 195 200 205Leu Leu Asp Leu Thr Trp
Leu Glu Lys Leu Leu Pro Lys Thr Ile Ser 210 215 220Gln His Gln Ile
Ser Thr Ser Ile Ile Thr Ser Thr Gln Lys Gly Met225 230 235 240Ile
Lys Leu Tyr Phe Leu Ser Phe Phe Phe Pro Leu Ile Leu Thr Leu 245 250
255Leu Leu Ile Thr Xaa 26035100PRTArtificialputative protein
sequencemisc_feature(100)..(100)Xaa can be any naturally occurring
amino acid 35Met Pro Leu Ile Tyr Met Asn Ile Met Leu Ala Phe Thr
Ile Ser Leu1 5 10 15Leu Gly Met Leu Val Tyr Arg Ser His Leu Met Ser
Ser Leu Leu Cys 20 25 30Leu Glu Gly Met Met Leu Ser Leu Phe Ile Met
Ala Thr Leu Met Thr 35 40 45Leu Asn Thr His Ser Leu Leu Ala Asn Ile
Val Pro Ile Ala Met Leu 50 55 60Val Phe Ala Ala Cys Glu Ala Ala Val
Gly Leu Ala Leu Leu Val Ser65 70 75 80Ile Ser Asn Thr Tyr Gly Leu
Asp Tyr Val His Asn Leu Thr Leu Leu 85 90 95Leu Ile Thr Xaa
10036261PRTArtificialputative protein
sequencemisc_feature(261)..(261)Xaa can be any naturally occurring
amino acid 36Met Ala His Ala Ala Gln Val Gly Leu Gln Asp Ala Thr
Ser Pro Ile1 5 10 15Met Glu Glu Leu Ile Thr Phe His Asp His Ala Leu
Met Ile Ile Phe 20 25 30Leu Ile Cys Phe Leu Val Leu Tyr Ala Leu Phe
Leu Thr Leu Thr Thr 35 40 45Lys Leu Thr Asn Thr Asn Ile Ser Asp Ala
Gln Glu Met Glu Thr Val 50 55 60Trp Thr Ile Leu Pro Ala Ile Ile Leu
Val Leu Ile Ala Leu Pro Ser65 70 75 80Leu Arg Ile Leu Tyr Met Thr
Asp Glu Val Asn Asp Pro Ser Leu Thr 85 90 95Ile Lys Ser Ile Gly His
Gln Trp Tyr Trp Thr Tyr Glu Tyr Thr Asp 100 105 110Tyr Gly Gly Leu
Ile Phe Asn Ser Tyr Met Leu Pro Pro Leu Phe Leu 115 120 125Glu Pro
Gly Asp Pro Asp Asn Tyr Thr Leu Ala Asn Pro Leu Asn Thr 130 135
140Pro Pro His Ile Lys Pro Glu Trp Tyr Phe Leu Phe Ala Tyr Thr
Ile145 150 155 160Leu Arg Ser Val Pro Asn Lys Leu Gly Gly Val Leu
Ala Leu Leu Leu 165 170 175Ser Ile Leu Ile Leu Ala Met Ile Pro Ile
Leu His Met Ser Lys Gln 180 185 190Gln Ser Met Met Phe Arg Pro Leu
Ser Gln Ser Leu Tyr Trp Leu Leu 195 200 205Ala Ala Asp Leu Leu Ile
Leu Thr Trp Ile Gly Gly Gln Pro Val Ser 210 215 220Tyr Pro Phe Thr
Ile Ile Gly Gln Val Ala Ser Val Leu Tyr Phe Thr225 230 235 240Thr
Ile Leu Ile Leu Met Pro Thr Ile Ser Leu Ile Glu Asn Lys Met 245 250
255Leu Lys Trp Ala Xaa 26037189PRTArtificialputative protein
sequencemisc_feature(189)..(189)Xaa can be any naturally occurring
amino acid 37Met Ala His Ala Ala Gln Val Gly Leu Gln Asp Ala Thr
Ser Pro Ile1 5 10 15Met Glu Glu Leu Ile Thr Phe His Asp His Ala Leu
Met Ile Ile Phe 20 25 30Leu Ile Cys Phe Leu Val Leu Tyr Ala Leu Phe
Leu Thr Leu Thr Thr 35 40 45Lys Leu Thr Asn Thr Asn Ile Ser Asp Ala
Gln Glu Met Glu Thr Val 50 55 60Trp Thr Ile Leu Pro Ala Ile Ile Leu
Val Leu Ile Ala Leu Pro Ser65 70 75 80Leu Arg Ile Leu Tyr Met Thr
Asp Glu Val Asn Asp Pro Ser Leu Thr 85 90 95Ile Lys Ser Ile Gly His
Gln Trp Tyr Trp Thr Tyr Glu Tyr Thr Asp 100 105 110Tyr Gly Gly Leu
Ile Phe Asn Ser Tyr Met Leu Pro Pro Leu Phe Leu 115 120 125Glu Pro
Gly Asp Leu Arg Leu Leu Ala Ala Asp Leu Leu Ile Leu Thr 130 135
140Trp Ile Gly Gly Gln Pro Val Ser Tyr Pro Phe Thr Ile Ile Gly
Gln145 150 155 160Val Ala Ser Val Leu Tyr Phe Thr Thr Ile Leu Ile
Leu Met Pro Thr 165 170 175Ile Ser Leu Ile Glu Asn Lys Met Leu Lys
Trp Ala Xaa 180 18538392PRTArtificialputative protein
sequencemisc_feature(392)..(392)Xaa can be any naturally occurring
amino acid 38Met Ala His Ala Ala Gln Val Gly Leu Gln Asp Ala Thr
Ser Pro Ile1 5 10 15Met Glu Glu Leu Ile Thr Phe His Asp His Ala Leu
Met Ile Ile Phe 20 25 30Leu Ile Cys Phe Leu Val Leu Tyr Ala Leu Phe
Leu Thr Leu Thr Thr 35 40 45Lys Leu Thr Asn Thr Asn Ile Ser Asp Ala
Gln Glu Met Glu Thr Val 50 55 60Trp Thr Ile Leu Pro Ala Ile Ile Leu
Val Leu Ile Ala Leu Pro Ser65 70 75 80Leu Arg Ile Leu Tyr Met Thr
Asp Glu Val Asn Asp Pro Ser Leu Thr 85 90 95Ile Lys Ser Ile Gly His
Gln Trp Tyr Trp Thr Tyr Glu Tyr Thr Asp 100 105 110Tyr Gly Gly Leu
Ile Phe Asn Ser Tyr Met Leu Pro Pro Leu Phe Leu 115 120 125Glu Pro
Gly Asp Leu Arg Leu Leu Asp Val Asp Asn Arg Val Val Leu 130 135
140Pro Ile Glu Ala Pro Ile Arg Met Met Ile Thr Ser Gln Asp Val
Leu145 150 155 160His Ser Trp Ala Val Pro Thr Leu Gly Leu Lys Thr
Asp Ala Ile Pro 165 170 175Gly Arg Leu Asn Gln Thr Thr Phe Thr Ala
Thr Arg Pro Gly Val Tyr 180 185 190Tyr Gly Gln Cys Ser Glu Ile Cys
Gly Ala Asn His Ser Phe Met Pro 195 200 205Met Phe Leu His Glu Thr
Gly Ser Asn Asn Pro Leu Gly Ile Thr Ser 210 215 220His Ser Asp Lys
Ile Thr Phe His Pro Tyr Tyr Thr Ile Lys Asp Ala225 230 235 240Leu
Gly Leu Leu Leu Phe Leu Leu Ser Leu Met Thr Leu Thr Leu Phe 245 250
255Ser Pro Asp Leu Leu Gly Asp Pro Asp Asn Tyr Thr Leu Ala Asn Pro
260 265 270Leu Asn Thr Pro Pro His Ile Lys Pro Glu Trp Tyr Phe Leu
Phe Ala 275 280 285Tyr Thr Ile Leu Arg Ser Val Pro Asn Lys Leu Gly
Gly Val Leu Ala 290 295 300Leu Leu Leu Ser Ile Leu Ile Leu Ala Met
Ile Pro Ile Leu His Met305 310 315 320Ser Lys Gln Gln Ser Met Met
Phe Arg Pro Leu Ser Gln Ser Leu Tyr 325 330 335Trp Leu Leu Ala Ala
Asp Leu Leu Ile Leu Thr Trp Ile Gly Gly Gln 340 345 350Pro Val Ser
Tyr Pro Phe Thr Ile Ile Gly Gln Val Ala Ser Val Leu 355 360 365Tyr
Phe Thr Thr Ile Leu Ile Leu Met Pro Thr Ile Ser Leu Ile Glu 370 375
380Asn Lys Met Leu Lys Trp Ala Xaa385 39039432PRTArtificialputative
protein sequencemisc_feature(432)..(432)Xaa can be any naturally
occurring amino acid 39Met Asn Glu Asn Leu Phe Ala Ser Phe Ile Ala
Pro Thr Ile Leu Gly1 5 10 15Leu Pro Ala Ala Val Leu Ile Ile Leu Phe
Pro Pro Leu Leu Ile Pro 20 25 30Thr Ser Lys Tyr Leu Ile Asn Asn Arg
Leu Ile Thr Thr Gln Gln Trp 35 40 45Leu Ile Lys Leu Thr Ser Lys Gln
Met Met Thr Met His Asn Thr Lys 50 55 60Gly Arg Thr Trp Ser Leu Met
Leu Val Ser Leu Ile Ile Phe Ile Ala65 70 75 80Thr Thr Asn Leu Leu
Gly Leu Leu Pro His Ser Phe Thr Pro Thr Thr 85 90 95Gln Leu Ser Met
Asn Leu Ala Met His Tyr Ser Pro Asp Ala Ser Thr 100 105 110Ala Phe
Ser Ser Ile Ala His Ile Thr Arg Asp Val Asn Tyr Gly Trp 115 120
125Ile Ile Arg Tyr Leu His Ala Asn Gly Ala Ser Met Phe Phe Ile Cys
130 135 140Leu Phe Leu His Ile Gly Arg Gly Leu Tyr Tyr Gly Ser Phe
Leu Tyr145 150 155 160Ser Glu Thr Trp Asn Ile Gly Ile Ile Leu Leu
Leu Ala Thr Met Ala 165 170 175Thr Ala Phe Met Gly Tyr Val Leu Pro
Trp Gly Gln Met Ser Phe Trp 180 185 190Gly Ala Thr Val Ile Thr Asn
Leu Leu Ser Ala Ile Pro Tyr Ile Gly 195 200 205Thr Asp Leu Val Gln
Trp Ile Trp Gly Gly Tyr Ser Val Asp Ser Pro 210 215 220Thr Leu Thr
Arg Phe Phe Thr Phe His Phe Ile Leu Pro Phe Ile Ile225 230 235
240Ala Ala Leu Ala Thr Leu His Leu Leu Phe Leu His Glu Thr Gly
Ser
245 250 255Asn Asn Pro Leu Gly Ile Thr Ser His Ser Asp Lys Ile Thr
Phe His 260 265 270Pro Tyr Tyr Thr Ile Lys Asp Ala Leu Gly Leu Leu
Leu Phe Leu Leu 275 280 285Ser Leu Met Thr Leu Thr Leu Phe Ser Pro
Asp Leu Leu Gly Asp Pro 290 295 300Asp Asn Tyr Thr Leu Ala Asn Pro
Leu Asn Thr Pro Pro His Ile Lys305 310 315 320Pro Glu Trp Tyr Phe
Leu Phe Ala Tyr Thr Ile Leu Arg Ser Val Pro 325 330 335Asn Lys Leu
Gly Gly Val Leu Ala Leu Leu Leu Ser Ile Leu Ile Leu 340 345 350Ala
Met Ile Pro Ile Leu His Met Ser Lys Gln Gln Ser Met Met Phe 355 360
365Arg Pro Leu Ser Gln Ser Leu Tyr Trp Leu Leu Ala Ala Asp Leu Leu
370 375 380Ile Leu Thr Trp Ile Gly Gly Gln Pro Val Ser Tyr Pro Phe
Thr Ile385 390 395 400Ile Gly Gln Val Ala Ser Val Leu Tyr Phe Thr
Thr Ile Leu Ile Leu 405 410 415Met Pro Thr Ile Ser Leu Ile Glu Asn
Lys Met Leu Lys Trp Ala Xaa 420 425 43040410PRTArtificialputative
protein sequencemisc_feature(410)..(410)Xaa can be any naturally
occurring amino acid 40Met Pro Leu Ile Tyr Met Asn Ile Met Leu Ala
Phe Thr Ile Ser Leu1 5 10 15Leu Gly Met Leu Val Tyr Arg Ser His Leu
Met Ser Ser Leu Leu Cys 20 25 30Leu Glu Gly Met Met Leu Ser Leu Phe
Ile Met Ala Thr Leu Met Thr 35 40 45Leu Asn Thr His Ser Leu Leu Ala
Asn Ile Val Pro Ile Ala Met Leu 50 55 60Val Phe Gly Ala Cys Leu Ile
Leu Gln Ile Thr Thr Gly Leu Phe Leu65 70 75 80Ala Met His Tyr Ser
Pro Asp Ala Ser Thr Ala Phe Ser Ser Ile Ala 85 90 95His Ile Thr Arg
Asp Val Asn Tyr Gly Trp Ile Ile Arg Tyr Leu His 100 105 110Ala Asn
Gly Ala Ser Met Phe Phe Ile Cys Leu Phe Leu His Ile Gly 115 120
125Arg Gly Leu Tyr Tyr Gly Ser Phe Leu Tyr Ser Glu Thr Trp Asn Ile
130 135 140Gly Ile Ile Leu Leu Leu Ala Thr Met Ala Thr Ala Phe Met
Gly Tyr145 150 155 160Val Leu Pro Trp Gly Gln Met Ser Phe Trp Gly
Ala Thr Val Ile Thr 165 170 175Asn Leu Leu Ser Ala Ile Pro Tyr Ile
Gly Thr Asp Leu Val Gln Trp 180 185 190Ile Trp Gly Gly Tyr Ser Val
Asp Ser Pro Thr Leu Thr Arg Phe Phe 195 200 205Thr Phe His Phe Ile
Leu Pro Phe Ile Ile Ala Ala Leu Ala Thr Leu 210 215 220His Leu Leu
Phe Leu His Glu Thr Gly Ser Asn Asn Pro Leu Gly Ile225 230 235
240Thr Ser His Ser Asp Lys Ile Thr Phe His Pro Tyr Tyr Thr Ile Lys
245 250 255Asp Ala Leu Gly Leu Leu Leu Phe Leu Leu Ser Leu Met Thr
Leu Thr 260 265 270Leu Phe Ser Pro Asp Leu Leu Gly Asp Pro Asp Asn
Tyr Thr Leu Ala 275 280 285Asn Pro Leu Asn Thr Pro Pro His Ile Lys
Pro Glu Trp Tyr Phe Leu 290 295 300Phe Ala Tyr Thr Ile Leu Arg Ser
Val Pro Asn Lys Leu Gly Gly Val305 310 315 320Leu Ala Leu Leu Leu
Ser Ile Leu Ile Leu Ala Met Ile Pro Ile Leu 325 330 335His Met Ser
Lys Gln Gln Ser Met Met Phe Arg Pro Leu Ser Gln Ser 340 345 350Leu
Tyr Trp Leu Leu Ala Ala Asp Leu Leu Ile Leu Thr Trp Ile Gly 355 360
365Gly Gln Pro Val Ser Tyr Pro Phe Thr Ile Ile Gly Gln Val Ala Ser
370 375 380Val Leu Tyr Phe Thr Thr Ile Leu Ile Leu Met Pro Thr Ile
Ser Leu385 390 395 400Ile Glu Asn Lys Met Leu Lys Trp Ala Xaa 405
41041174PRTArtificialputative protein
sequencemisc_feature(174)..(174)Xaa can be any naturally occurring
amino acid 41Met Phe Ala Asp Arg Trp Leu Phe Ser Thr Asn His Lys
Asp Ile Gly1 5 10 15Thr Leu Tyr Leu Leu Phe Gly Ala Trp Ala Gly Val
Leu Gly Thr Ala 20 25 30Leu Ser Leu Leu Ile Arg Ala Glu Leu Gly Gln
Pro Gly Asn Leu Leu 35 40 45Gly Asn Asp His Ile Tyr Asn Val Ile Val
Thr Ala Leu Ala Val Thr 50 55 60Phe Leu Gly Leu Leu Thr Ala Leu Asp
Leu Asn Tyr Leu Thr Asn Lys65 70 75 80Leu Lys Met Lys Ser Pro Leu
Cys Thr Phe Tyr Phe Ser Asn Met Leu 85 90 95Gly Phe Tyr Pro Ser Ile
Thr His Arg Thr Ile Pro Tyr Leu Gly Leu 100 105 110Leu Thr Ser Gln
Asn Leu Pro Leu Leu Leu Leu Asp Leu Thr Trp Leu 115 120 125Glu Lys
Leu Leu Pro Lys Thr Ile Ser Gln His Gln Ile Ser Thr Ser 130 135
140Ile Ile Thr Ser Thr Gln Lys Gly Met Ile Lys Leu Tyr Phe Leu
Ser145 150 155 160Phe Phe Phe Pro Leu Ile Leu Thr Leu Leu Leu Ile
Thr Xaa 165 17042194PRTArtificialputative protein
sequencemisc_feature(194)..(194)Xaa can be any naturally occurring
amino acid 42Met Phe Ala Asp Arg Trp Leu Phe Ser Thr Asn His Lys
Asp Ile Gly1 5 10 15Thr Leu Tyr Leu Leu Phe Gly Ala Trp Ala Gly Val
Leu Gly Thr Ala 20 25 30Leu Ser Leu Leu Ile Arg Ala Glu Leu Gly Gln
Pro Gly Asn Leu Leu 35 40 45Gly Asn Asp His Ile Tyr Asn Val Ile Val
Thr Ala His Ala Phe Val 50 55 60Met Ile Phe Phe Met Val Met Pro Ile
Met Ile Gly Gly Phe Gly Asn65 70 75 80Trp Leu Val Pro Leu Met Ile
Gly Ala Pro Asp Met Ala Phe Pro Arg 85 90 95Met Asn Asn Met Ser Phe
Trp Leu Leu Pro Pro Ser Leu Leu Leu Leu 100 105 110Leu Ala Ser Ala
Met Val Glu Ala Gly Ala Gly Thr Gly Trp Thr Val 115 120 125Tyr Pro
Pro Leu Ala Gly Asn Tyr Ser His Pro Gly Ala Leu Leu Asp 130 135
140Leu Thr Trp Leu Glu Lys Leu Leu Pro Lys Thr Ile Ser Gln His
Gln145 150 155 160Ile Ser Thr Ser Ile Ile Thr Ser Thr Gln Lys Gly
Met Ile Lys Leu 165 170 175Tyr Phe Leu Ser Phe Phe Phe Pro Leu Ile
Leu Thr Leu Leu Leu Ile 180 185 190Thr
Xaa43736PRTArtificialputative protein
sequencemisc_feature(736)..(736)Xaa can be any naturally occurring
amino acid 43Met Phe Ala Asp Arg Trp Leu Phe Ser Thr Asn His Lys
Asp Ile Gly1 5 10 15Thr Leu Tyr Leu Leu Phe Gly Ala Trp Ala Gly Val
Leu Gly Thr Ala 20 25 30Leu Ser Leu Leu Ile Arg Ala Glu Leu Gly Gln
Pro Gly Asn Leu Leu 35 40 45Gly Asn Asp His Ile Tyr Asn Val Ile Val
Thr Ala His Ala Phe Val 50 55 60Met Ile Phe Phe Met Val Met Pro Ile
Met Ile Gly Gly Phe Gly Asn65 70 75 80Trp Leu Val Pro Leu Met Ile
Gly Ala Pro Asp Met Ala Phe Pro Arg 85 90 95Met Asn Asn Met Ser Phe
Trp Leu Leu Pro Pro Ser Leu Leu Leu Leu 100 105 110Leu Ala Ser Ala
Met Val Glu Ala Gly Ala Gly Thr Gly Trp Thr Val 115 120 125Tyr Pro
Pro Leu Ala Gly Asn Tyr Ser His Pro Gly Ala Ser Val Asp 130 135
140Leu Thr Ile Phe Ser Leu His Leu Ala Gly Val Ser Ser Ile Leu
Gly145 150 155 160Ala Ile Asn Phe Ile Thr Thr Ile Ile Asn Met Lys
Pro Pro Ala Met 165 170 175Thr Gln Tyr Gln Thr Pro Leu Phe Val Trp
Ser Val Leu Ile Thr Ala 180 185 190Val Leu Leu Leu Leu Ser Leu Pro
Val Leu Ala Ala Gly Ile Thr Met 195 200 205Leu Leu Thr Asp Arg Asn
Leu Asn Thr Thr Phe Phe Asp Pro Ala Gly 210 215 220Gly Gly Asp Pro
Ile Leu Tyr Gln His Leu Phe Trp Phe Phe Gly His225 230 235 240Pro
Glu Val Tyr Ile Leu Ile Leu Pro Gly Phe Gly Met Ile Ser His 245 250
255Ile Val Thr Tyr Tyr Ser Gly Lys Lys Glu Pro Phe Gly Tyr Met Gly
260 265 270Met Val Trp Ala Met Met Ser Ile Gly Phe Leu Gly Phe Ile
Val Trp 275 280 285Ala His His Met Phe Thr Val Gly Met Asp Val Asp
Thr Arg Ala Tyr 290 295 300Phe Thr Ser Ala Thr Met Ile Ile Ala Ile
Pro Thr Gly Val Lys Val305 310 315 320Phe Ser Trp Leu Ala Thr Leu
His Gly Ser Asn Met Lys Trp Ser Ala 325 330 335Ala Val Leu Trp Ala
Leu Gly Phe Ile Phe Leu Phe Thr Val Gly Gly 340 345 350Leu Thr Gly
Ile Val Leu Ala Asn Ser Ser Leu Asp Ile Val Leu His 355 360 365Asp
Thr Tyr Tyr Val Val Ala His Phe His Tyr Val Leu Ser Met Gly 370 375
380Ala Val Phe Ala Ile Met Gly Gly Phe Ile His Trp Phe Pro Leu
Phe385 390 395 400Ser Gly Tyr Thr Leu Asp Gln Thr Tyr Ala Lys Ile
His Phe Thr Ile 405 410 415Met Phe Ile Gly Val Asn Leu Thr Phe Phe
Pro Gln His Phe Leu Gly 420 425 430Leu Ser Gly Met Pro Arg Arg Tyr
Ser Asp Tyr Pro Asp Ala Tyr Thr 435 440 445Thr Trp Asn Ile Leu Ser
Ser Val Gly Ser Phe Ile Ser Leu Thr Ala 450 455 460Val Met Leu Met
Ile Phe Met Ile Trp Glu Ala Phe Ala Ser Lys Arg465 470 475 480Lys
Val Leu Met Val Glu Glu Pro Ser Met Asn Leu Glu Trp Leu Tyr 485 490
495Gly Cys Pro Pro Pro Tyr His Thr Phe Glu Glu Pro Val Tyr Met Lys
500 505 510Ala Gly Met Pro Phe Leu Thr Gly Phe Tyr Ser Lys Asp His
Ile Ile 515 520 525Glu Thr Ala Asn Met Ser Tyr Thr Asn Ala Trp Ala
Leu Ser Ile Thr 530 535 540Leu Ile Ala Thr Ser Leu Thr Ser Ala Tyr
Ser Thr Arg Met Ile Leu545 550 555 560Leu Thr Leu Thr Gly Gln Pro
Arg Phe Pro Thr Leu Thr Asn Ile Asn 565 570 575Glu Asn Asn Pro Thr
Leu Leu Asn Pro Ile Lys Arg Leu Ala Ala Gly 580 585 590Ser Leu Phe
Ala Gly Phe Leu Ile Thr Asn Asn Ile Ser Pro Ala Ser 595 600 605Pro
Phe Gln Thr Thr Ile Pro Leu Tyr Leu Lys Leu Thr Ala Leu Ala 610 615
620Val Thr Phe Leu Gly Leu Leu Thr Ala Leu Asp Leu Asn Tyr Leu
Thr625 630 635 640Asn Lys Leu Lys Met Lys Ser Pro Leu Cys Thr Phe
Tyr Phe Ser Asn 645 650 655Met Leu Gly Phe Tyr Pro Ser Ile Thr His
Arg Thr Ile Pro Tyr Leu 660 665 670Gly Leu Leu Thr Ser Gln Asn Leu
Pro Leu Leu Leu Leu Asp Leu Thr 675 680 685Trp Leu Glu Lys Leu Leu
Pro Lys Thr Ile Ser Gln His Gln Ile Ser 690 695 700Thr Ser Ile Ile
Thr Ser Thr Gln Lys Gly Met Ile Lys Leu Tyr Phe705 710 715 720Leu
Ser Phe Phe Phe Pro Leu Ile Leu Thr Leu Leu Leu Ile Thr Xaa 725 730
73544269PRTArtificialputative protein
sequencemisc_feature(269)..(269)Xaa can be any naturally occurring
amino acid 44Met Ala His Ala Ala Gln Val Gly Leu Gln Asp Ala Thr
Ser Pro Ile1 5 10 15Met Glu Glu Leu Ile Thr Phe His Asp His Ala Leu
Met Ile Ile Phe 20 25 30Leu Ile Cys Phe Leu Val Leu Tyr Ala Leu Phe
Leu Thr Leu Thr Thr 35 40 45Lys Leu Thr Asn Thr Asn Ile Ser Asp Ala
Gln Glu Met Glu Thr Ala 50 55 60Asn Met Ser Tyr Thr Asn Ala Trp Ala
Leu Ser Ile Thr Leu Ile Ala65 70 75 80Thr Ser Leu Thr Ser Ala Tyr
Ser Thr Arg Met Ile Leu Leu Thr Leu 85 90 95Thr Gly Gln Pro Arg Phe
Pro Thr Leu Thr Asn Ile Asn Glu Asn Asn 100 105 110Pro Thr Leu Leu
Asn Pro Ile Lys Arg Leu Ala Ala Gly Ser Leu Phe 115 120 125Ala Gly
Phe Leu Ile Thr Asn Asn Ile Ser Pro Ala Ser Pro Phe Gln 130 135
140Thr Thr Ile Pro Leu Tyr Leu Lys Leu Thr Ala Leu Ala Val Thr
Phe145 150 155 160Leu Gly Leu Leu Thr Ala Leu Asp Leu Asn Tyr Leu
Thr Asn Lys Leu 165 170 175Lys Met Lys Ser Pro Leu Cys Thr Phe Tyr
Phe Ser Asn Met Leu Gly 180 185 190Phe Tyr Pro Ser Ile Thr His Arg
Thr Ile Pro Tyr Leu Gly Leu Leu 195 200 205Thr Ser Gln Asn Leu Pro
Leu Leu Leu Leu Asp Leu Thr Trp Leu Glu 210 215 220Lys Leu Leu Pro
Lys Thr Ile Ser Gln His Gln Ile Ser Thr Ser Ile225 230 235 240Ile
Thr Ser Thr Gln Lys Gly Met Ile Lys Leu Tyr Phe Leu Ser Phe 245 250
255Phe Phe Pro Leu Ile Leu Thr Leu Leu Leu Ile Thr Xaa 260
26545262PRTArtificialputative protein
sequencemisc_feature(262)..(262)Xaa can be any naturally occurring
amino acid 45Met Ala His Ala Ala Gln Val Gly Leu Gln Asp Ala Thr
Ser Pro Ile1 5 10 15Met Glu Glu Leu Ile Thr Phe His Asp His Ala Leu
Met Ile Ile Phe 20 25 30Leu Ile Cys Phe Leu Val Leu Tyr Ala Leu Phe
Leu Thr Leu Thr Thr 35 40 45Lys Leu Thr Asn Thr Asn Ile Ser Asp Ala
Gln Glu Met Glu Thr Val 50 55 60Trp Thr Ile Leu Pro Ala Ile Ile Leu
Val Leu Ile Ala Leu Pro Ser65 70 75 80Leu Arg Ile Leu Tyr Met Thr
Asp Glu Val Asn Asp Pro Ser Leu Thr 85 90 95Ile Lys Ser Ile Gly His
Gln Trp Tyr Trp Thr Tyr Glu Tyr Thr Asp 100 105 110Tyr Gly Gly Leu
Ile Phe Asn Ser Tyr Met Leu Pro Pro Leu Phe Leu 115 120 125Glu Pro
Gly Asp Leu Arg Leu Leu Asp Val Asp Asn Arg Val Val Leu 130 135
140Pro Ile Glu Ala Pro Ile Arg Met Met Ile Thr Ser Gln Asp Val
Leu145 150 155 160His Ser Trp Ala Val Pro Thr Leu Gly Leu Lys Thr
Asp Ala Ile Pro 165 170 175Gly Arg Leu Asn Gln Thr Thr Phe Thr Ala
Thr Arg Pro Gly Val Tyr 180 185 190Tyr Gly Gln Cys Ser Glu Ile Cys
Gly Ala Asn His Ser Phe Met Pro 195 200 205Ile Val Leu Asp Leu Thr
Trp Leu Glu Lys Leu Leu Pro Lys Thr Ile 210 215 220Ser Gln His Gln
Ile Ser Thr Ser Ile Ile Thr Ser Thr Gln Lys Gly225 230 235 240Met
Ile Lys Leu Tyr Phe Leu Ser Phe Phe Phe Pro Leu Ile Leu Thr 245 250
255Leu Leu Leu Ile Thr Xaa 26046635PRTArtificialputative protein
sequencemisc_feature(635)..(635)Xaa can be any naturally occurring
amino acid 46Met Asn Glu Asn Leu Phe Ala Ser Phe Ile Ala Pro Thr
Ile Leu Gly1 5 10 15Leu Pro Ala Ala Val Leu Ile Ile Leu Phe Pro Pro
Leu Leu Ile Pro 20 25 30Thr Ser Lys Tyr Leu Ile Asn Asn Arg Leu Ile
Thr Thr Gln Gln Trp 35 40 45Leu Ile Lys Leu Thr Ser Lys Gln Met Met
Thr Met His Asn Thr Lys 50 55 60Gly Arg Thr Trp Ser Leu Met Leu Val
Ser Leu Ile Ile Phe Ile Ala65 70 75 80Thr Thr Asn Leu Leu Gly Leu
Leu Pro His Ser Phe Thr Pro Thr Thr 85 90 95Gln Leu Ser Met Asn Leu
Ala Met Ala Ile Pro Leu Trp Ala Gly Thr 100 105 110Val Ile Met Gly
Phe Arg Ser Lys Ile Lys Asn Ala Leu Ala His Phe 115 120 125Leu Pro
Gln Gly Thr Pro Thr Pro Leu Ile Pro Met Leu Val Ile Ile 130
135 140Glu Thr Ile Ser Leu Leu Ile Gln Pro Met Ala Leu Ala Val Arg
Leu145 150 155 160Thr Ala Asn Ile Thr Ala Gly His Leu Leu Met His
Leu Ile Gly Ser 165 170 175Ala Thr Leu Ala Met Ser Thr Ile Asn Leu
Pro Ser Thr Leu Ile Ile 180 185 190Phe Thr Ile Leu Ile Leu Leu Thr
Ile Leu Glu Ile Ala Val Ala Leu 195 200 205Ile Gln Ala Tyr Val Phe
Thr Leu Leu Val Ser Leu Tyr Leu His Ser 210 215 220Asn Ser Trp Asp
Pro Gln Gln Met Ala Leu Leu Asn Ala Asn Pro Ser225 230 235 240Leu
Thr Pro Leu Leu Gly Leu Leu Leu Ala Ala Ala Gly Lys Ser Ala 245 250
255Gln Leu Gly Leu His Pro Trp Leu Pro Ser Ala Met Glu Gly Pro Thr
260 265 270Pro Val Ser Ala Leu Leu His Ser Ser Thr Met Val Val Ala
Gly Ile 275 280 285Phe Leu Leu Ile Arg Phe His Pro Leu Ala Glu Asn
Ser Pro Leu Ile 290 295 300Gln Thr Leu Thr Leu Cys Leu Gly Ala Ile
Thr Thr Leu Phe Ala Ala305 310 315 320Val Cys Ala Leu Thr Gln Asn
Asp Ile Lys Lys Ile Val Ala Phe Ser 325 330 335Thr Ser Ser Gln Leu
Gly Leu Met Met Val Thr Ile Gly Ile Asn Gln 340 345 350Pro His Leu
Ala Phe Leu His Ile Cys Thr His Ala Phe Phe Lys Ala 355 360 365Met
Leu Phe Met Cys Ser Gly Ser Ile Ile His Asn Leu Asn Asn Glu 370 375
380Gln Asp Ile Arg Lys Met Gly Gly Leu Leu Lys Thr Met Pro Leu
Thr385 390 395 400Ser Thr Ser Leu Thr Ile Gly Ser Leu Ala Leu Ala
Gly Met Pro Phe 405 410 415Leu Thr Gly Phe Tyr Ser Lys Asp His Ile
Ile Glu Thr Ala Asn Met 420 425 430Ser Tyr Thr Asn Ala Trp Ala Leu
Ser Ile Thr Leu Ile Ala Thr Ser 435 440 445Leu Thr Ser Ala Tyr Ser
Thr Arg Met Ile Leu Leu Thr Leu Thr Gly 450 455 460Gln Pro Arg Phe
Pro Thr Leu Thr Asn Ile Asn Glu Asn Asn Pro Thr465 470 475 480Leu
Leu Asn Pro Ile Lys Arg Leu Ala Ala Gly Ser Leu Phe Ala Gly 485 490
495Phe Leu Ile Thr Asn Asn Ile Ser Pro Ala Ser Pro Phe Gln Thr Thr
500 505 510Ile Pro Leu Tyr Leu Lys Leu Thr Ala Leu Ala Val Thr Phe
Leu Gly 515 520 525Leu Leu Thr Ala Leu Asp Leu Asn Tyr Leu Thr Asn
Lys Leu Lys Met 530 535 540Lys Ser Pro Leu Cys Thr Phe Tyr Phe Ser
Asn Met Leu Gly Phe Tyr545 550 555 560Pro Ser Ile Thr His Arg Thr
Ile Pro Tyr Leu Gly Leu Leu Thr Ser 565 570 575Gln Asn Leu Pro Leu
Leu Leu Leu Asp Leu Thr Trp Leu Glu Lys Leu 580 585 590Leu Pro Lys
Thr Ile Ser Gln His Gln Ile Ser Thr Ser Ile Ile Thr 595 600 605Ser
Thr Gln Lys Gly Met Ile Lys Leu Tyr Phe Leu Ser Phe Phe Phe 610 615
620Pro Leu Ile Leu Thr Leu Leu Leu Ile Thr Xaa625 630
63547515PRTArtificialputative protein
sequencemisc_feature(515)..(515)Xaa can be any naturally occurring
amino acid 47Met Thr His Gln Ser His Ala Tyr His Met Val Lys Pro
Ser Pro Trp1 5 10 15Pro Leu Thr Gly Ala Leu Ser Ala Leu Leu Met Thr
Ser Gly Leu Ala 20 25 30Met Trp Phe His Phe His Ser Met Thr Leu Leu
Met Leu Gly Leu Leu 35 40 45Thr Asn Thr Leu Thr Met Tyr Gln Trp Trp
Arg Asp Val Thr Arg Glu 50 55 60Ser Thr Tyr Gln Gly His His Thr Pro
Pro Val Gln Lys Gly Leu Arg65 70 75 80Tyr Gly Met Ile Leu Phe Ile
Thr Ser Glu Val Phe Phe Phe Ala Gly 85 90 95Phe Phe Trp Ala Phe Tyr
His Ser Ser Leu Ala Pro Thr Pro Gln Leu 100 105 110Gly Gly His Trp
Pro Pro Thr Gly Ile Thr Pro Leu Leu Gly Leu Leu 115 120 125Leu Ala
Ala Ala Gly Lys Ser Ala Gln Leu Gly Leu His Pro Trp Leu 130 135
140Pro Ser Ala Met Glu Gly Pro Thr Pro Val Ser Ala Leu Leu His
Ser145 150 155 160Ser Thr Met Val Val Ala Gly Ile Phe Leu Leu Ile
Arg Phe His Pro 165 170 175Leu Ala Glu Asn Ser Pro Leu Ile Gln Thr
Leu Thr Leu Cys Leu Gly 180 185 190Ala Ile Thr Thr Leu Phe Ala Ala
Val Cys Ala Leu Thr Gln Asn Asp 195 200 205Ile Lys Lys Ile Val Ala
Phe Ser Thr Ser Ser Gln Leu Gly Leu Met 210 215 220Met Val Thr Ile
Gly Ile Asn Gln Pro His Leu Ala Phe Leu His Ile225 230 235 240Cys
Thr His Ala Phe Phe Lys Ala Met Leu Phe Met Cys Ser Gly Ser 245 250
255Ile Ile His Asn Leu Asn Asn Glu Gln Asp Ile Arg Lys Met Gly Gly
260 265 270Leu Leu Lys Thr Met Pro Leu Thr Ser Thr Ser Leu Thr Ile
Gly Ser 275 280 285Leu Ala Leu Ala Gly Met Pro Phe Leu Thr Gly Phe
Tyr Ser Lys Asp 290 295 300His Ile Ile Glu Thr Ala Asn Met Ser Tyr
Thr Asn Ala Trp Ala Leu305 310 315 320Ser Ile Thr Leu Ile Ala Thr
Ser Leu Thr Ser Ala Tyr Ser Thr Arg 325 330 335Met Ile Leu Leu Thr
Leu Thr Gly Gln Pro Arg Phe Pro Thr Leu Thr 340 345 350Asn Ile Asn
Glu Asn Asn Pro Thr Leu Leu Asn Pro Ile Lys Arg Leu 355 360 365Ala
Ala Gly Ser Leu Phe Ala Gly Phe Leu Ile Thr Asn Asn Ile Ser 370 375
380Pro Ala Ser Pro Phe Gln Thr Thr Ile Pro Leu Tyr Leu Lys Leu
Thr385 390 395 400Ala Leu Ala Val Thr Phe Leu Gly Leu Leu Thr Ala
Leu Asp Leu Asn 405 410 415Tyr Leu Thr Asn Lys Leu Lys Met Lys Ser
Pro Leu Cys Thr Phe Tyr 420 425 430Phe Ser Asn Met Leu Gly Phe Tyr
Pro Ser Ile Thr His Arg Thr Ile 435 440 445Pro Tyr Leu Gly Leu Leu
Thr Ser Gln Asn Leu Pro Leu Leu Leu Leu 450 455 460Asp Leu Thr Trp
Leu Glu Lys Leu Leu Pro Lys Thr Ile Ser Gln His465 470 475 480Gln
Ile Ser Thr Ser Ile Ile Thr Ser Thr Gln Lys Gly Met Ile Lys 485 490
495Leu Tyr Phe Leu Ser Phe Phe Phe Pro Leu Ile Leu Thr Leu Leu Leu
500 505 510Ile Thr Xaa 51548543PRTArtificialputative protein
sequencemisc_feature(543)..(543)Xaa can be any naturally occurring
amino acid 48Met Asn Phe Ala Leu Ile Leu Met Ile Asn Thr Leu Leu
Ala Leu Leu1 5 10 15Leu Met Ile Ile Thr Phe Trp Leu Pro Gln Leu Asn
Gly Tyr Met Glu 20 25 30Lys Ser Thr Pro Tyr Glu Cys Gly Phe Asp Pro
Met Ser Pro Ala Arg 35 40 45Val Pro Phe Ser Met Lys Phe Phe Leu Val
Ala Ile Thr Phe Leu Leu 50 55 60Phe Asp Leu Glu Ile Ala Leu Leu Leu
Pro Leu Pro Trp Ala Leu Gln65 70 75 80Thr Thr Asn Leu Pro Leu Met
Val Met Ser Ser Leu Leu Leu Ile Ile 85 90 95Ile Leu Ala Leu Ser Leu
Ala Asn Thr Ala Ala Ile Gln Ala Ile Leu 100 105 110Tyr Asn Arg Ile
Gly Asp Ile Gly Phe Ile Leu Ala Leu Ala Trp Phe 115 120 125Ile Leu
His Ser Asn Ser Trp Asp Pro Gln Gln Met Ala Leu Leu Asn 130 135
140Ala Asn Pro Ser Leu Thr Pro Leu Leu Gly Leu Leu Leu Ala Ala
Ala145 150 155 160Gly Lys Ser Ala Gln Leu Gly Leu His Pro Trp Leu
Pro Ser Ala Met 165 170 175Glu Gly Pro Thr Pro Val Ser Ala Leu Leu
His Ser Ser Thr Met Val 180 185 190Val Ala Gly Ile Phe Leu Leu Ile
Arg Phe His Pro Leu Ala Glu Asn 195 200 205Ser Pro Leu Ile Gln Thr
Leu Thr Leu Cys Leu Gly Ala Ile Thr Thr 210 215 220Leu Phe Ala Ala
Val Cys Ala Leu Thr Gln Asn Asp Ile Lys Lys Ile225 230 235 240Val
Ala Phe Ser Thr Ser Ser Gln Leu Gly Leu Met Met Val Thr Ile 245 250
255Gly Ile Asn Gln Pro His Leu Ala Phe Leu His Ile Cys Thr His Ala
260 265 270Phe Phe Lys Ala Met Leu Phe Met Cys Ser Gly Ser Ile Ile
His Asn 275 280 285Leu Asn Asn Glu Gln Asp Ile Arg Lys Met Gly Gly
Leu Leu Lys Thr 290 295 300Met Pro Leu Thr Ser Thr Ser Leu Thr Ile
Gly Ser Leu Ala Leu Ala305 310 315 320Gly Met Pro Phe Leu Thr Gly
Phe Tyr Ser Lys Asp His Ile Ile Glu 325 330 335Thr Ala Asn Met Ser
Tyr Thr Asn Ala Trp Ala Leu Ser Ile Thr Leu 340 345 350Ile Ala Thr
Ser Leu Thr Ser Ala Tyr Ser Thr Arg Met Ile Leu Leu 355 360 365Thr
Leu Thr Gly Gln Pro Arg Phe Pro Thr Leu Thr Asn Ile Asn Glu 370 375
380Asn Asn Pro Thr Leu Leu Asn Pro Ile Lys Arg Leu Ala Ala Gly
Ser385 390 395 400Leu Phe Ala Gly Phe Leu Ile Thr Asn Asn Ile Ser
Pro Ala Ser Pro 405 410 415Phe Gln Thr Thr Ile Pro Leu Tyr Leu Lys
Leu Thr Ala Leu Ala Val 420 425 430Thr Phe Leu Gly Leu Leu Thr Ala
Leu Asp Leu Asn Tyr Leu Thr Asn 435 440 445Lys Leu Lys Met Lys Ser
Pro Leu Cys Thr Phe Tyr Phe Ser Asn Met 450 455 460Leu Gly Phe Tyr
Pro Ser Ile Thr His Arg Thr Ile Pro Tyr Leu Gly465 470 475 480Leu
Leu Thr Ser Gln Asn Leu Pro Leu Leu Leu Leu Asp Leu Thr Trp 485 490
495Leu Glu Lys Leu Leu Pro Lys Thr Ile Ser Gln His Gln Ile Ser Thr
500 505 510Ser Ile Ile Thr Ser Thr Gln Lys Gly Met Ile Lys Leu Tyr
Phe Leu 515 520 525Ser Phe Phe Phe Pro Leu Ile Leu Thr Leu Leu Leu
Ile Thr Xaa 530 535 5404943PRTArtificialputative protein
sequencemisc_feature(43)..(43)Xaa can be any naturally occurring
amino acid 49Met Pro Gln Leu Asn Thr Thr Val Trp Pro Thr Met Ile
Thr Pro Met1 5 10 15Leu Leu Thr Leu Phe Leu Ile Thr Gln Leu Lys Met
Leu Asn Thr Asn 20 25 30Tyr His Leu Pro Pro Ser Pro Leu Ala Ala Xaa
35 4050951RNAHuman 50augaacgaaa aucuguucgc uucauucauu gcccccacaa
uccuaggccu acccgccgca 60guacugauca uucuauuucc cccucuauug auccccaccu
ccaaauaucu caucaacaac 120cgacuaauca ccacccaaca augacuaauc
aaacuaaccu caaaacaaau gauaaccaua 180cacaacacua aaggacgaac
cugaucucuu auacuaguau ccuuaaucau uuuuauugcc 240acaacuaacc
uccucggacu ccugccucac ucauuuacac caaccaccca acuaucuaua
300aaccuagcca uggccauccc cuuaugagcg ggcacaguga uuauaggcuu
ucgcucuaag 360auuaaaaaug cccuagccca cuucuuacca caaggcacac
cuacaccccu uauccccaua 420cuaguuauua ucgaaaccau cagccuacuc
auucaaccaa uagcccuggc cguacgccua 480accgcuaaca uuacugcagg
ccaccuacuc augcaccuaa uuggaagcgc cacccuagca 540auaucaacca
uuaaccuucc cucuacacuu aucaucuuca caauucuaau ucuacugacu
600auccuagaaa ucgcugucac uuuccuagga cuucuaacag cccuagaccu
caacuaccua 660accaacaaac uuaaaauaaa auccccacua ugcacauuuu
auuucuccaa cauacucgga 720uucuacccua gcaucacaca ccgcacaauc
cccuaucuag gccuucuuac gagccaaaac 780cugccccuac uccuccuaga
ccuaaccuga cuagaaaagc uauuaccuaa aacaauuuca 840cagcaccaaa
ucuccaccuc caucaucacc ucaacccaaa aaggcauaau uaaacuuuac
900uuccucucuu ucuucuuccc acucauccua acccuacucc uaaucacaua a
95151951DNAArtificialcDNA 51atgaacgaaa atctgttcgc ttcattcatt
gcccccacaa tcctaggcct acccgccgca 60gtactgatca ttctatttcc ccctctattg
atccccacct ccaaatatct catcaacaac 120cgactaatca ccacccaaca
atgactaatc aaactaacct caaaacaaat gataaccata 180cacaacacta
aaggacgaac ctgatctctt atactagtat ccttaatcat ttttattgcc
240acaactaacc tcctcggact cctgcctcac tcatttacac caaccaccca
actatctata 300aacctagcca tggccatccc cttatgagcg ggcacagtga
ttataggctt tcgctctaag 360attaaaaatg ccctagccca cttcttacca
caaggcacac ctacacccct tatccccata 420ctagttatta tcgaaaccat
cagcctactc attcaaccaa tagccctggc cgtacgccta 480accgctaaca
ttactgcagg ccacctactc atgcacctaa ttggaagcgc caccctagca
540atatcaacca ttaaccttcc ctctacactt atcatcttca caattctaat
tctactgact 600atcctagaaa tcgctgtcac tttcctagga cttctaacag
ccctagacct caactaccta 660accaacaaac ttaaaataaa atccccacta
tgcacatttt atttctccaa catactcgga 720ttctacccta gcatcacaca
ccgcacaatc ccctatctag gccttcttac gagccaaaac 780ctgcccctac
tcctcctaga cctaacctga ctagaaaagc tattacctaa aacaatttca
840cagcaccaaa tctccacctc catcatcacc tcaacccaaa aaggcataat
taaactttac 900ttcctctctt tcttcttccc actcatccta accctactcc
taatcacata a 95152317PRTArtificialputative protein
sequencemisc_feature(317)..(317)Xaa can be any naturally occurring
amino acid 52Met Asn Glu Asn Leu Phe Ala Ser Phe Ile Ala Pro Thr
Ile Leu Gly1 5 10 15Leu Pro Ala Ala Val Leu Ile Ile Leu Phe Pro Pro
Leu Leu Ile Pro 20 25 30Thr Ser Lys Tyr Leu Ile Asn Asn Arg Leu Ile
Thr Thr Gln Gln Trp 35 40 45Leu Ile Lys Leu Thr Ser Lys Gln Met Met
Thr Met His Asn Thr Lys 50 55 60Gly Arg Thr Trp Ser Leu Met Leu Val
Ser Leu Ile Ile Phe Ile Ala65 70 75 80Thr Thr Asn Leu Leu Gly Leu
Leu Pro His Ser Phe Thr Pro Thr Thr 85 90 95Gln Leu Ser Met Asn Leu
Ala Met Ala Ile Pro Leu Trp Ala Gly Thr 100 105 110Val Ile Met Gly
Phe Arg Ser Lys Ile Lys Asn Ala Leu Ala His Phe 115 120 125Leu Pro
Gln Gly Thr Pro Thr Pro Leu Ile Pro Met Leu Val Ile Ile 130 135
140Glu Thr Ile Ser Leu Leu Ile Gln Pro Met Ala Leu Ala Val Arg
Leu145 150 155 160Thr Ala Asn Ile Thr Ala Gly His Leu Leu Met His
Leu Ile Gly Ser 165 170 175Ala Thr Leu Ala Met Ser Thr Ile Asn Leu
Pro Ser Thr Leu Ile Ile 180 185 190Phe Thr Ile Leu Ile Leu Leu Thr
Ile Leu Glu Ile Ala Val Thr Phe 195 200 205Leu Gly Leu Leu Thr Ala
Leu Asp Leu Asn Tyr Leu Thr Asn Lys Leu 210 215 220Lys Met Lys Ser
Pro Leu Cys Thr Phe Tyr Phe Ser Asn Met Leu Gly225 230 235 240Phe
Tyr Pro Ser Ile Thr His Arg Thr Ile Pro Tyr Leu Gly Leu Leu 245 250
255Thr Ser Gln Asn Leu Pro Leu Leu Leu Leu Asp Leu Thr Trp Leu Glu
260 265 270Lys Leu Leu Pro Lys Thr Ile Ser Gln His Gln Ile Ser Thr
Ser Ile 275 280 285Ile Thr Ser Thr Gln Lys Gly Met Ile Lys Leu Tyr
Phe Leu Ser Phe 290 295 300Phe Phe Pro Leu Ile Leu Thr Leu Leu Leu
Ile Thr Xaa305 310 3155321DNAArtificial Sequenceprimer 53ccttacacta
ttcctcatca c 215420DNAArtificial Sequenceprimer 54tgacctgtta
gggtgagaag 205521DNAArtificial Sequenceprimer 55tcgctcacac
ctcatatcct c 215622DNAArtificial Sequenceprimer 56tgtgattagg
agtagggtta gg 22
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013
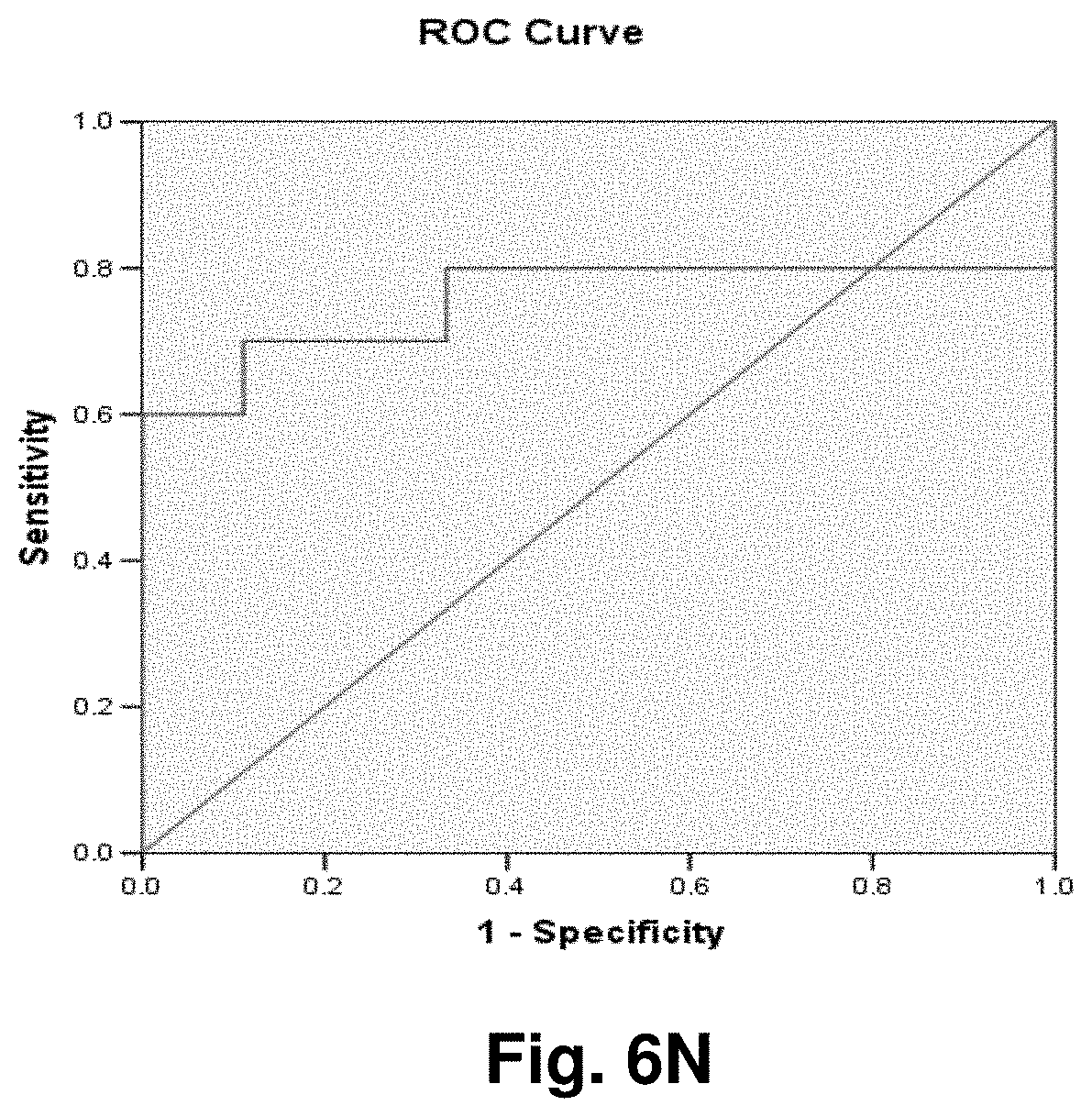
D00014
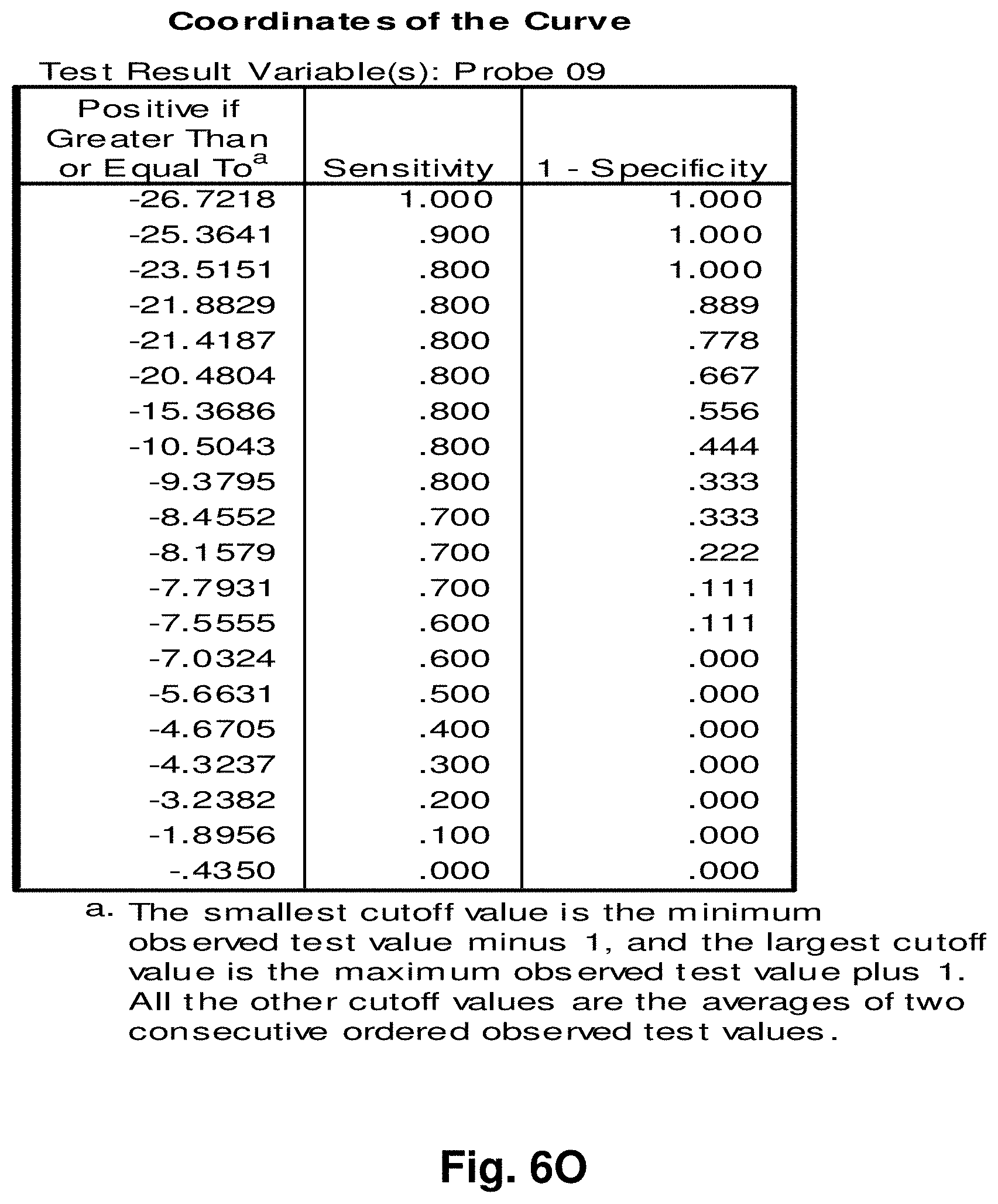
D00015
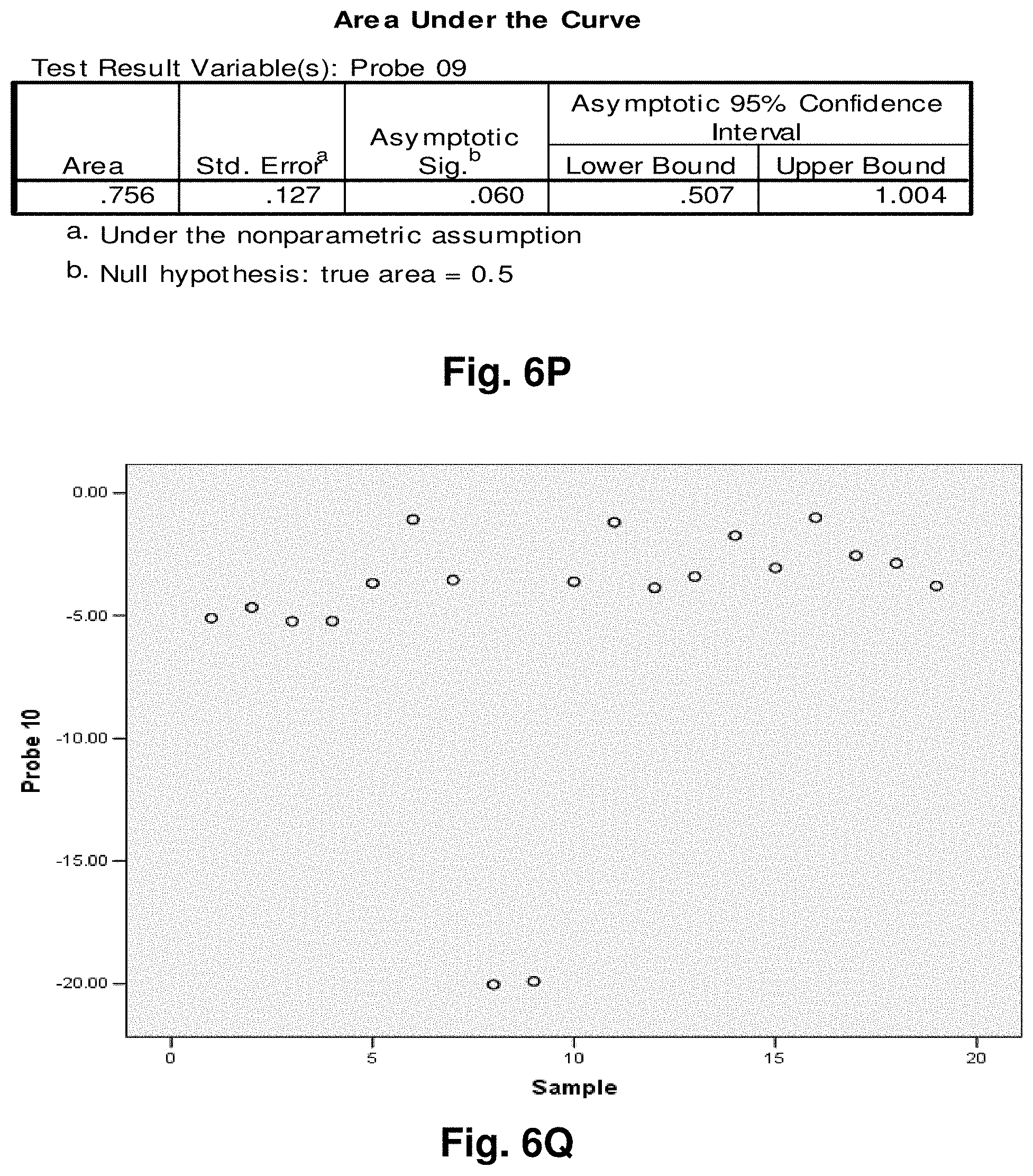
D00016

D00017
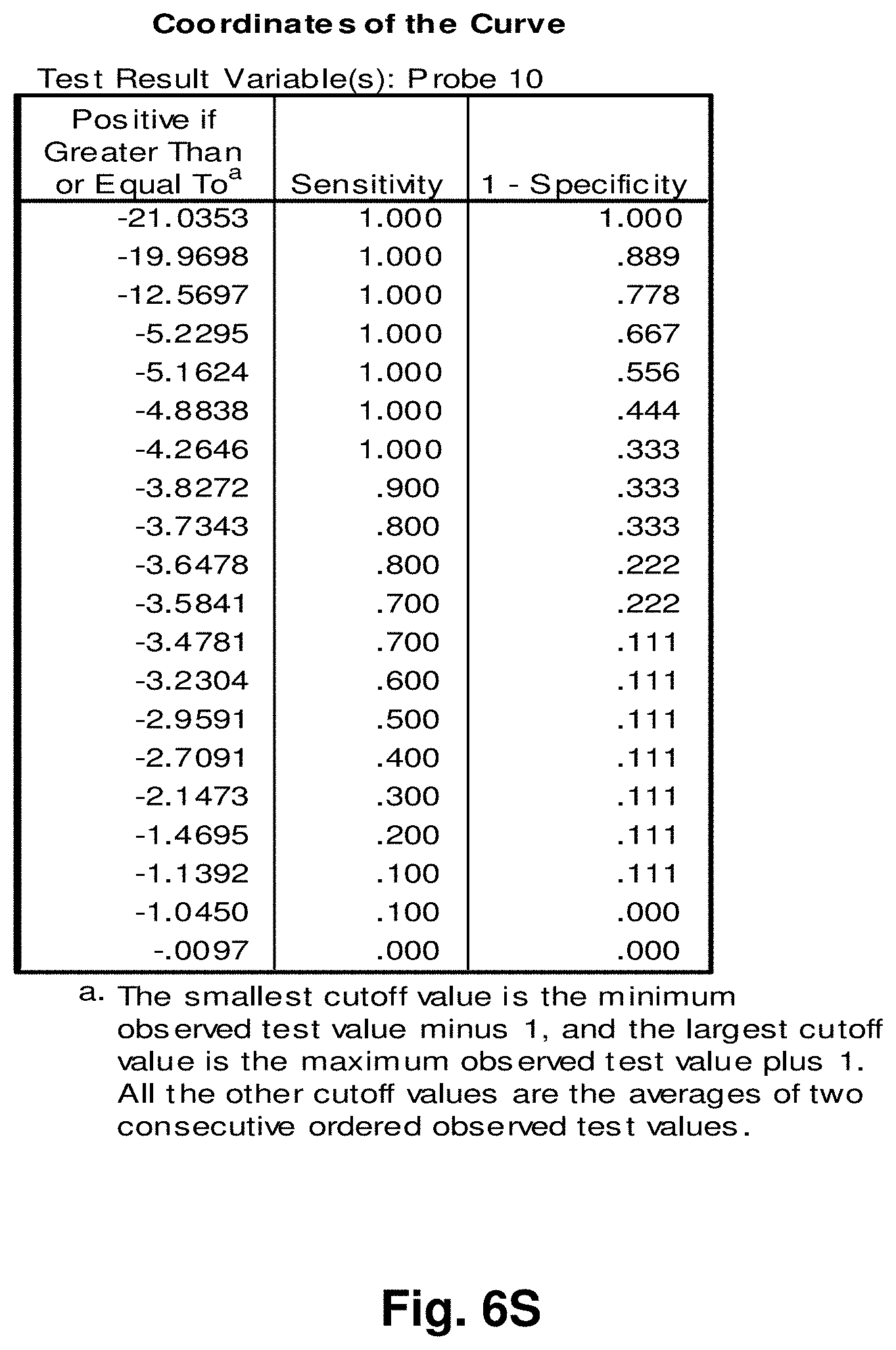
D00018

D00019
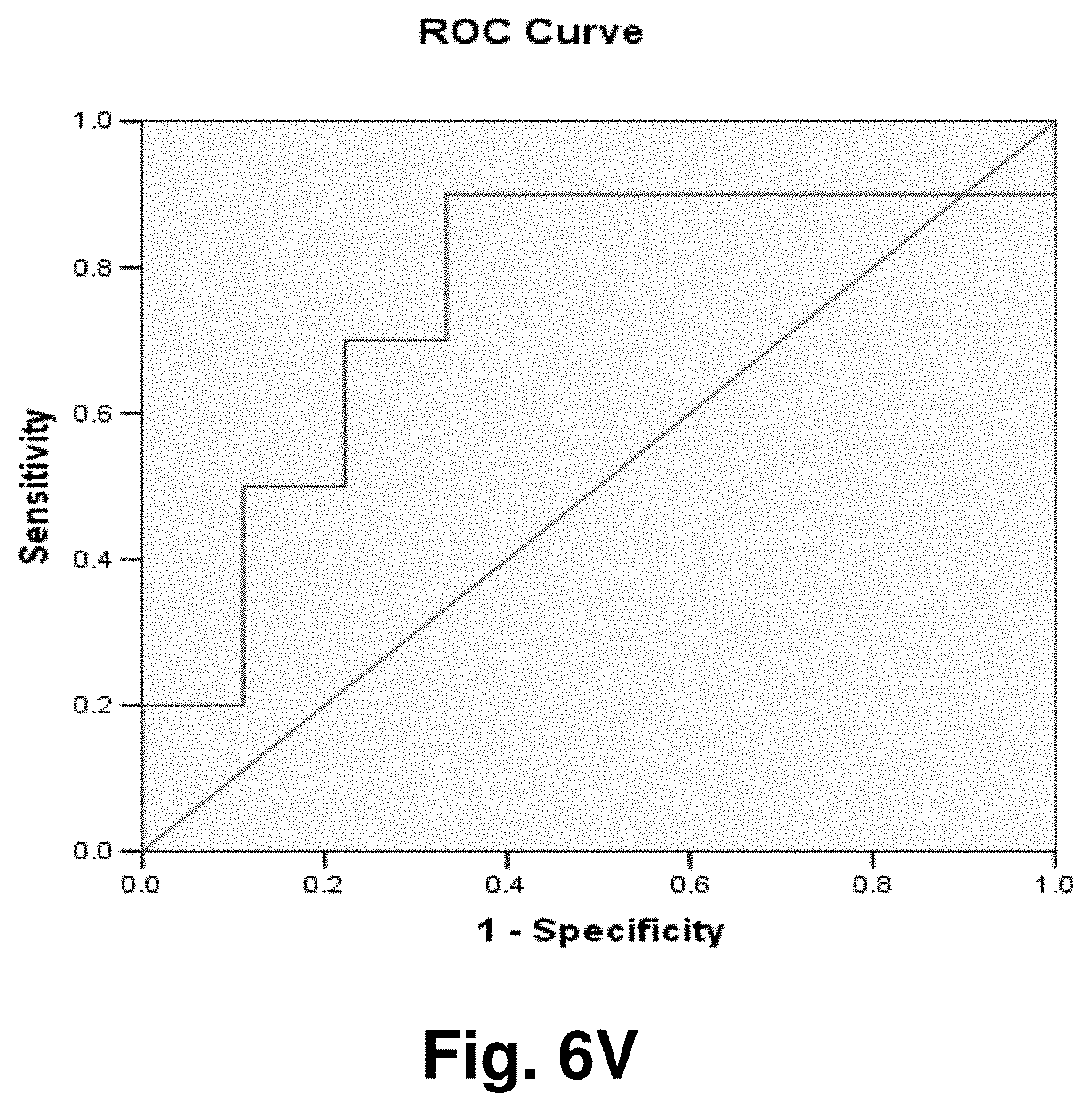
D00020
D00021
D00022
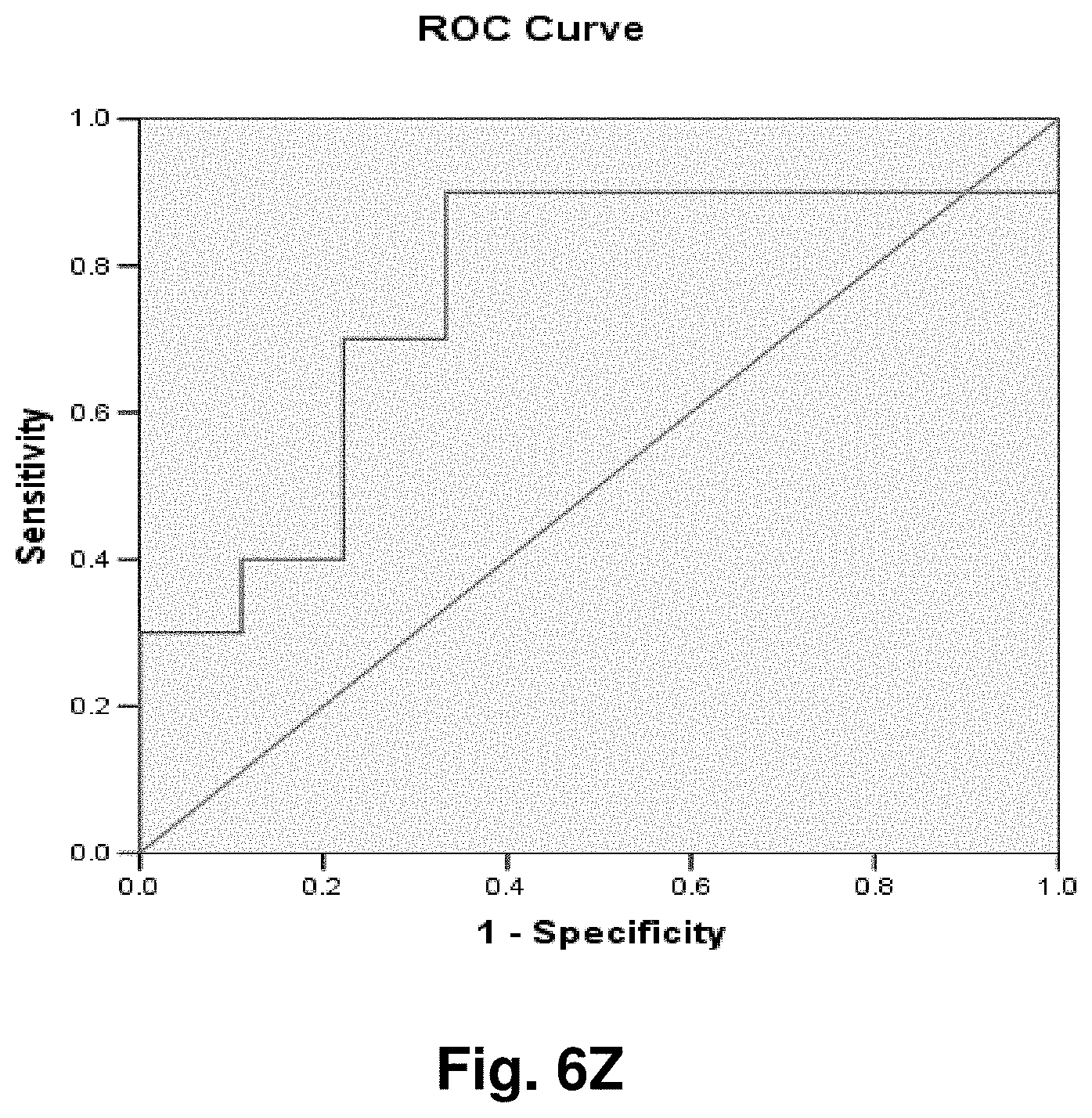
D00023

D00024

D00025
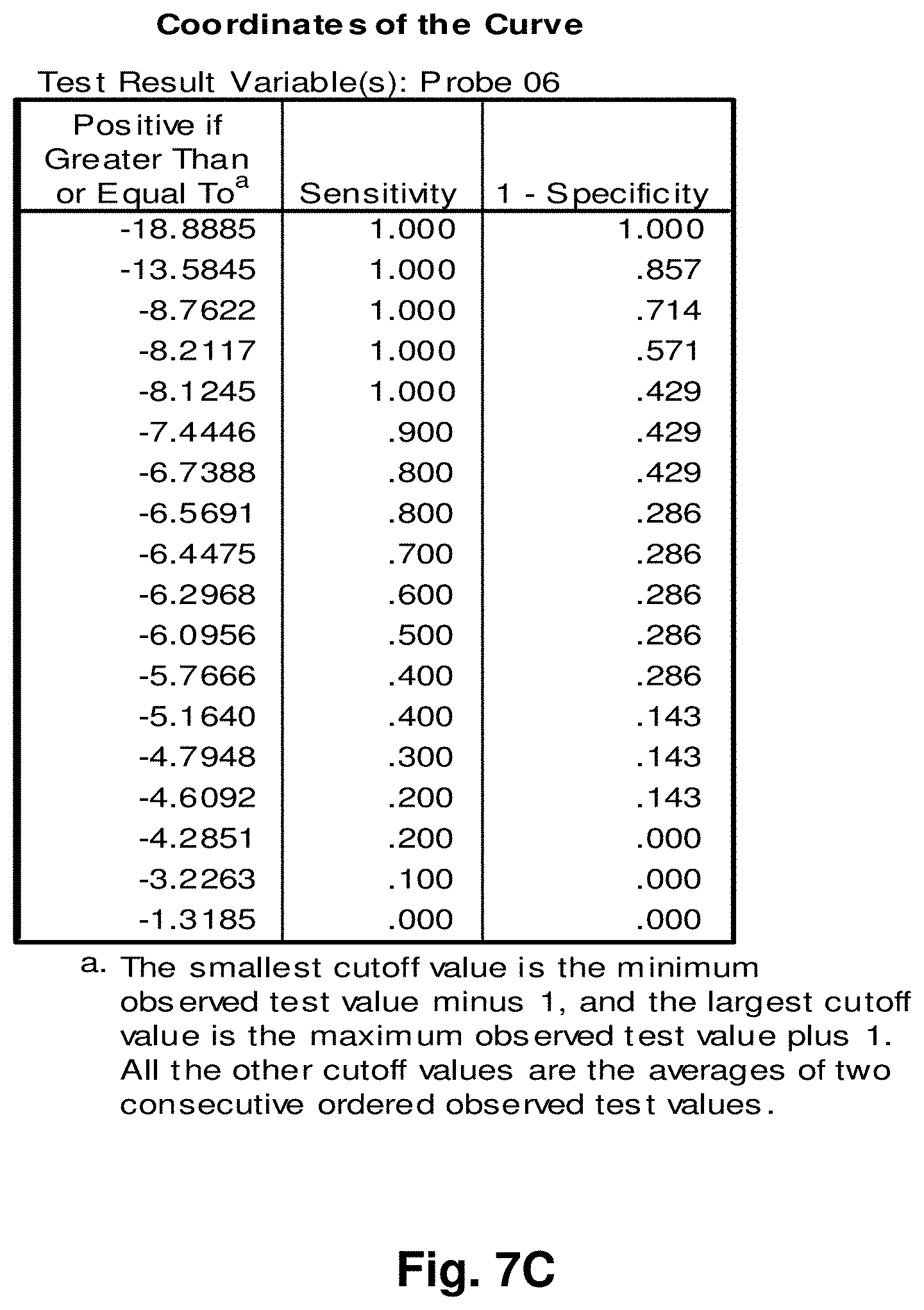
D00026
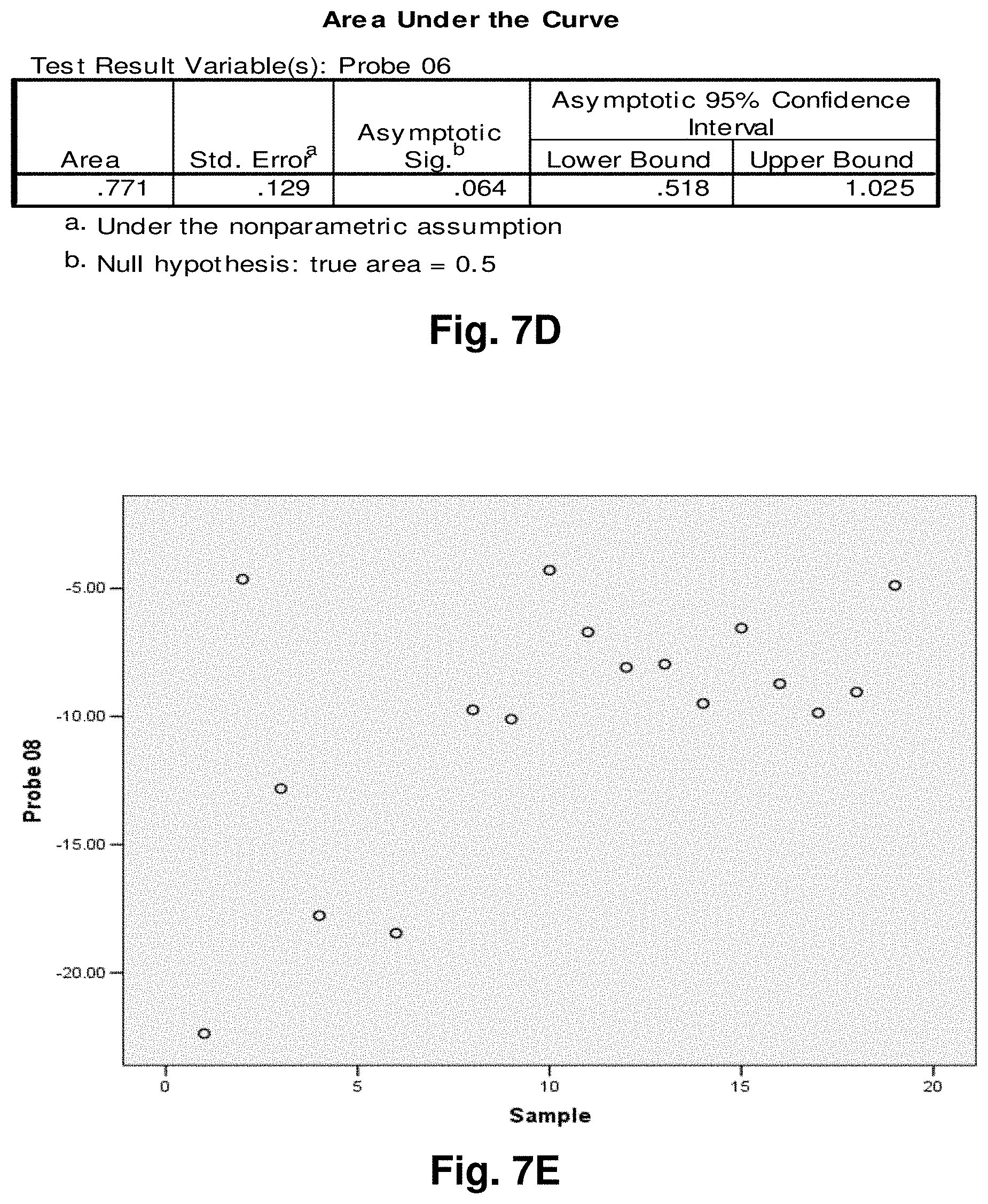
D00027
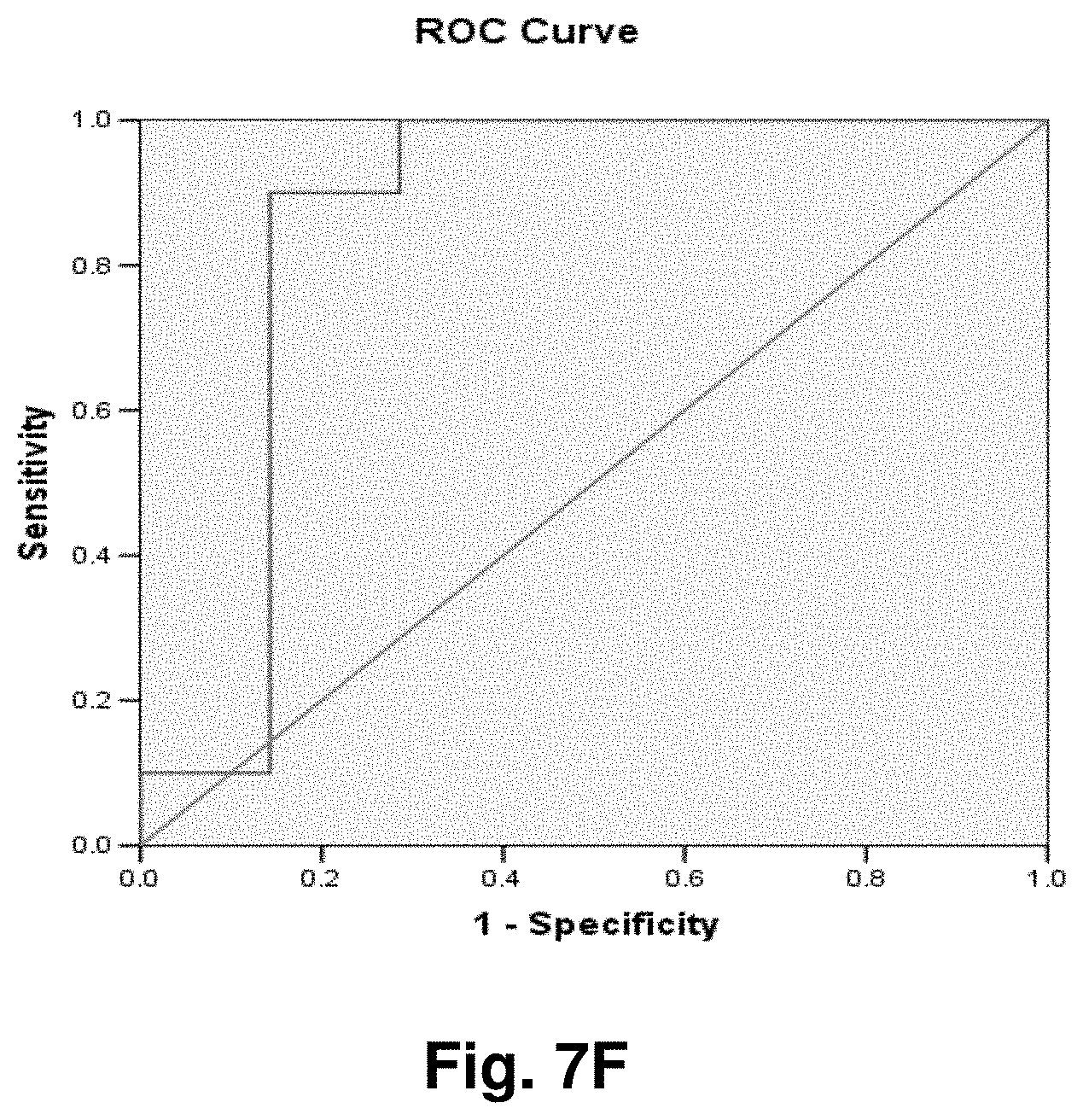
D00028

D00029
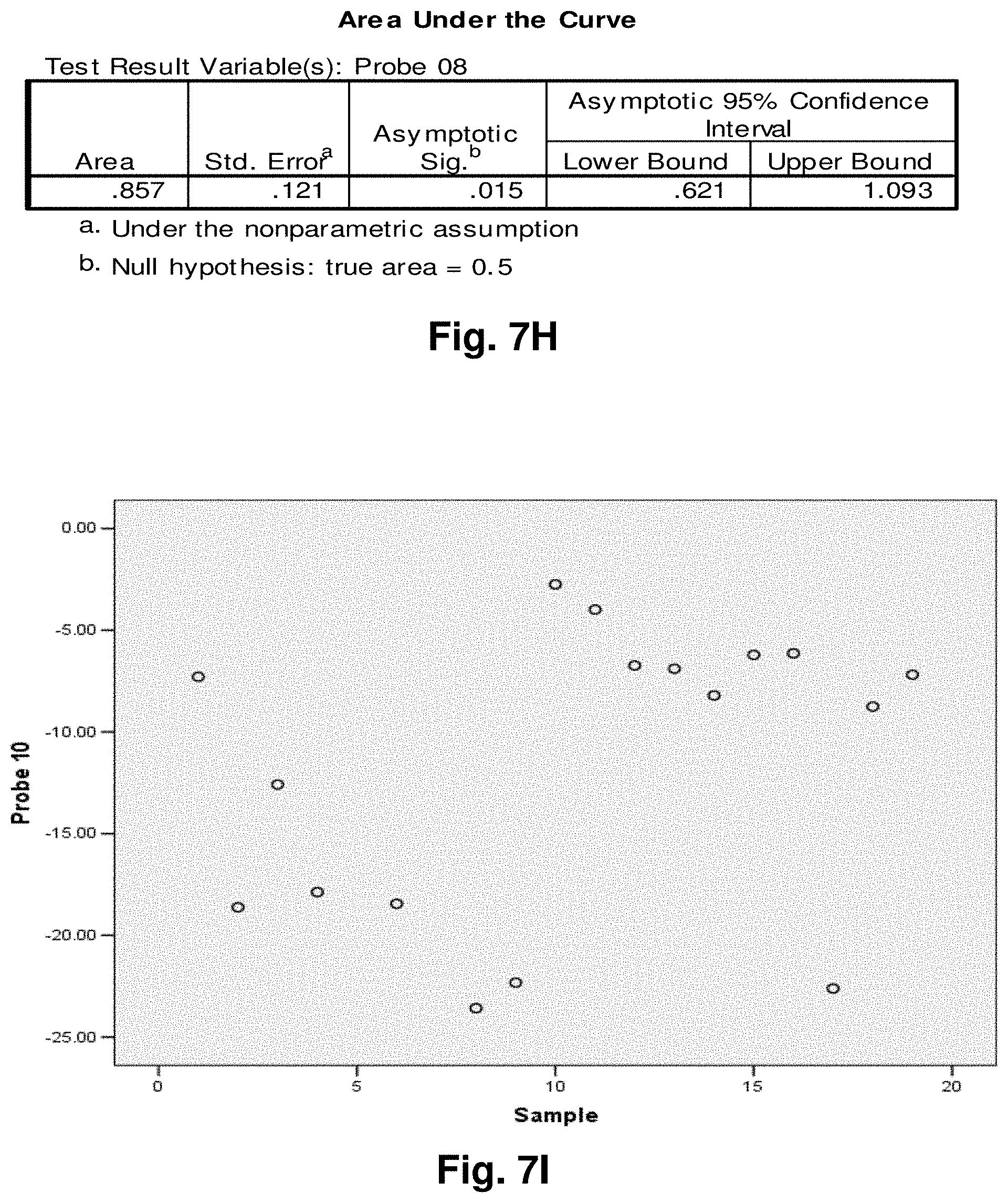
D00030

D00031
D00032
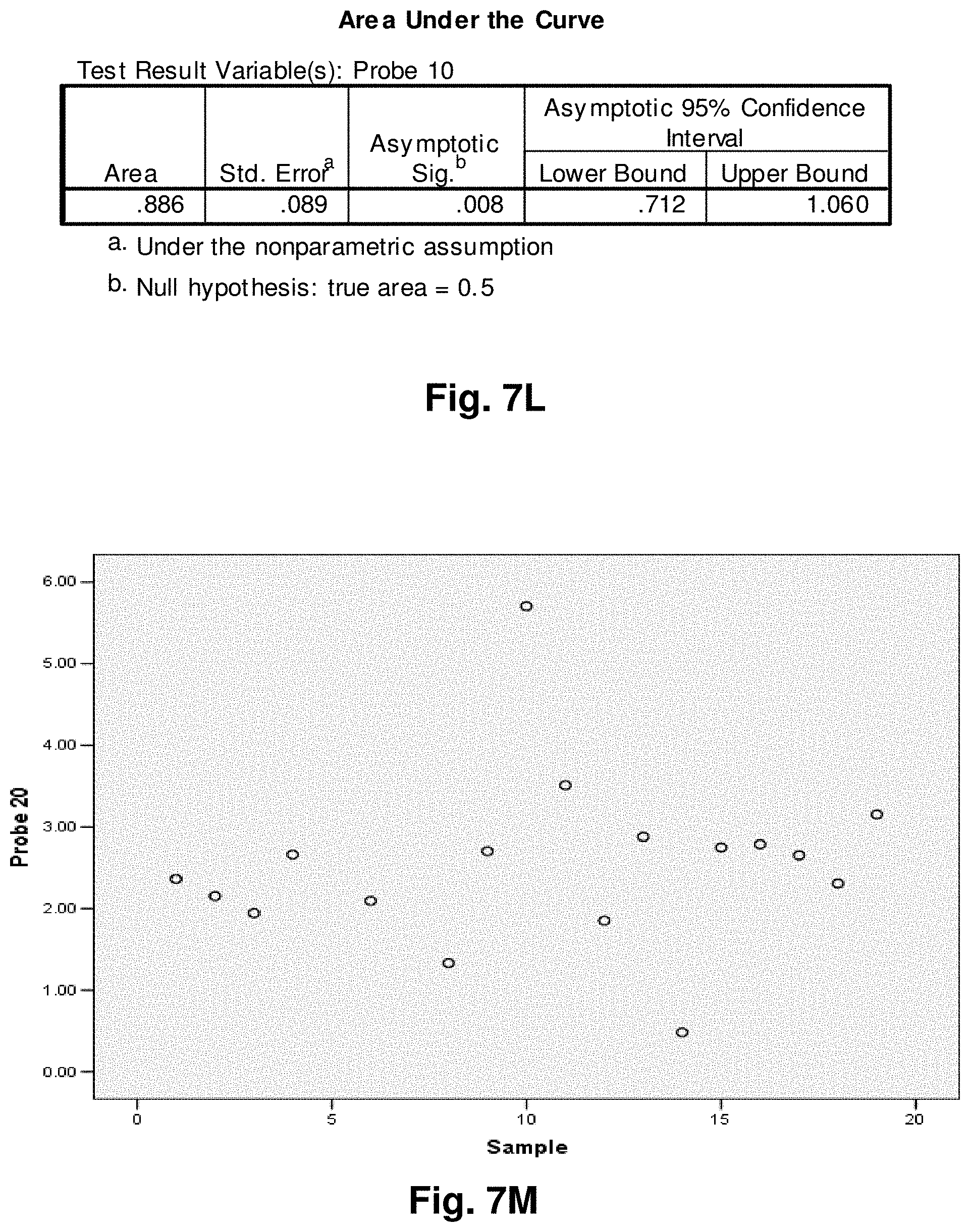
D00033

D00034

D00035
D00036
D00037

D00038

D00039
D00040
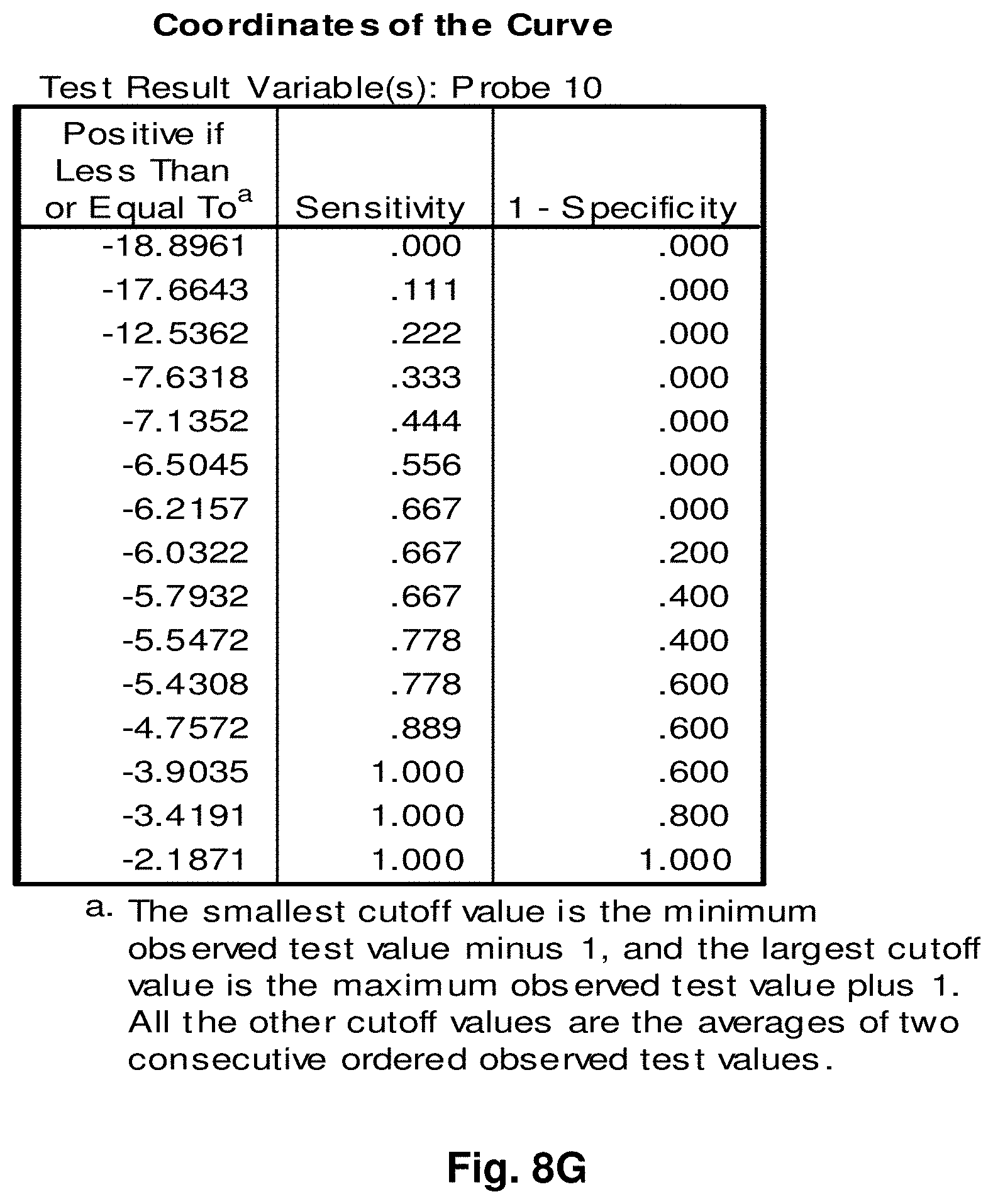
D00041
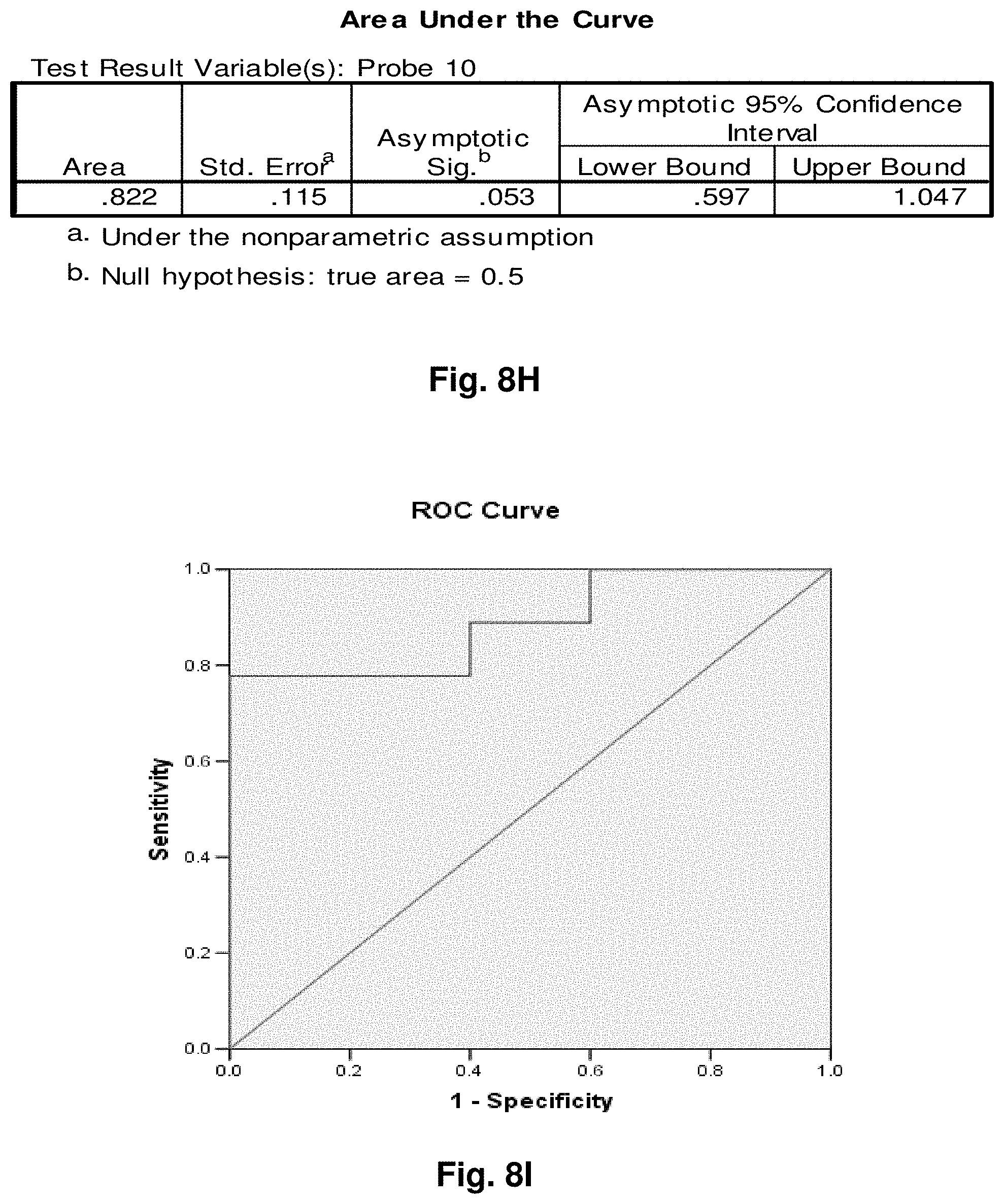
D00042

D00043
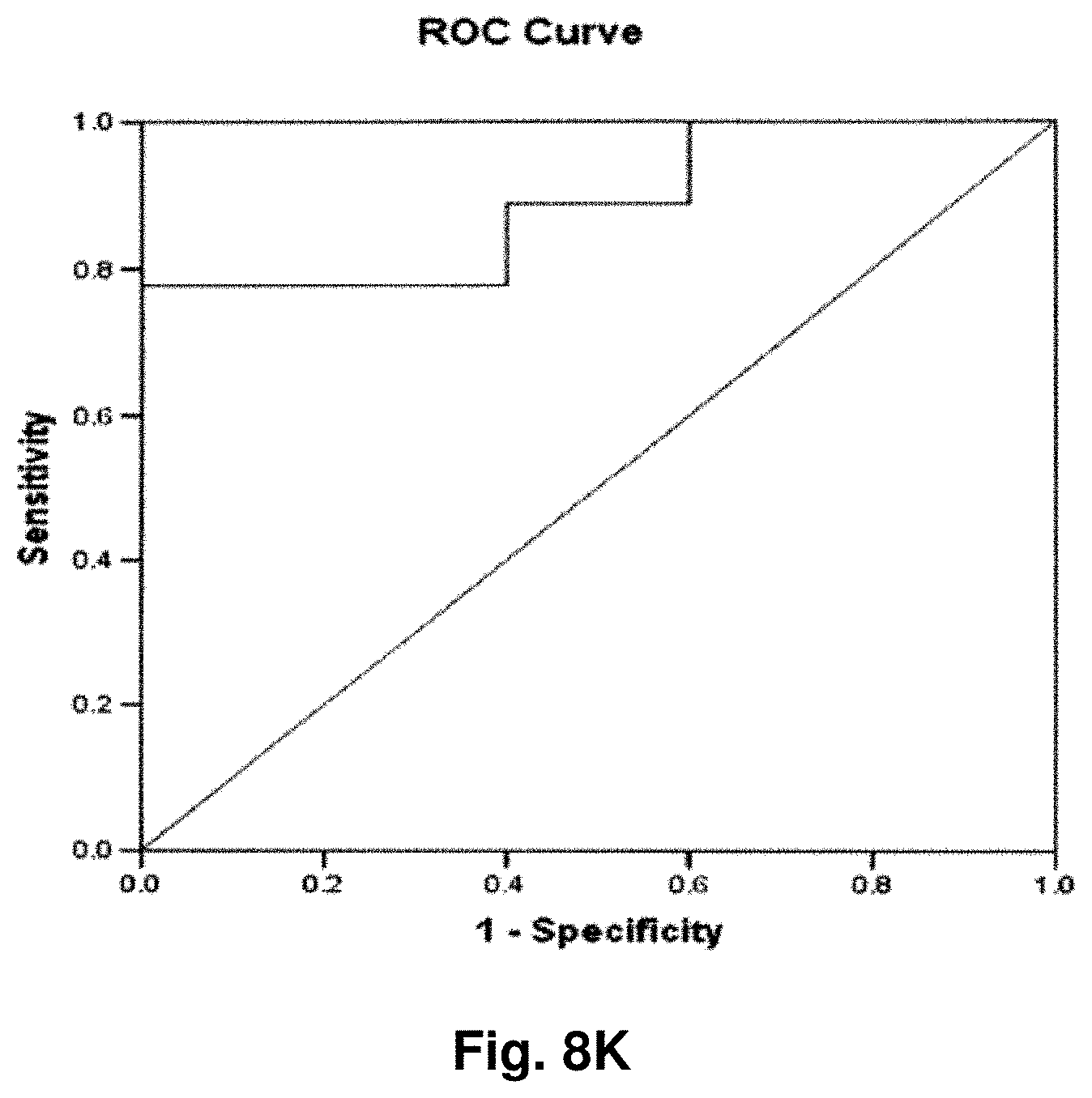
D00044

D00045
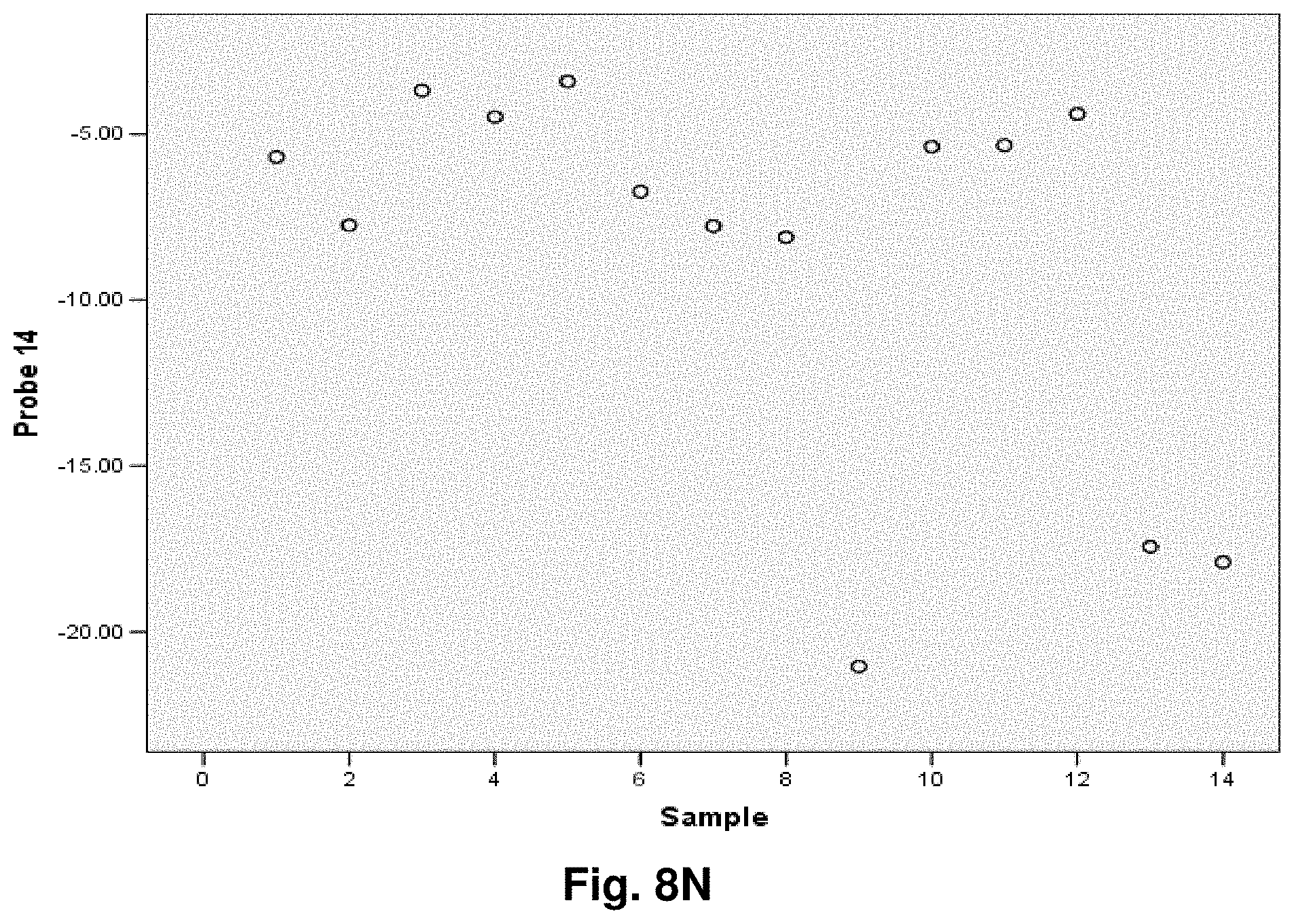
D00046

D00047
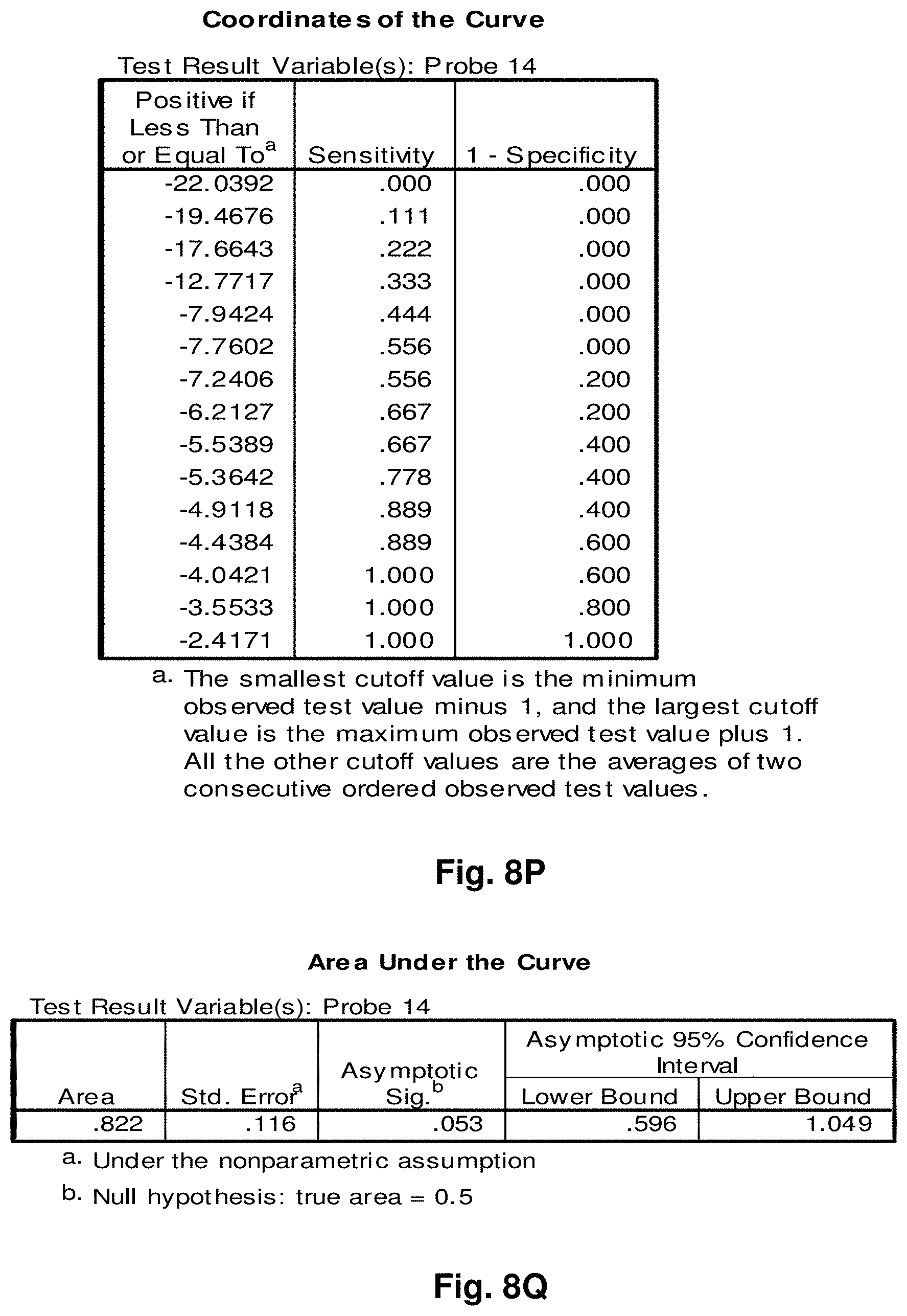
D00048
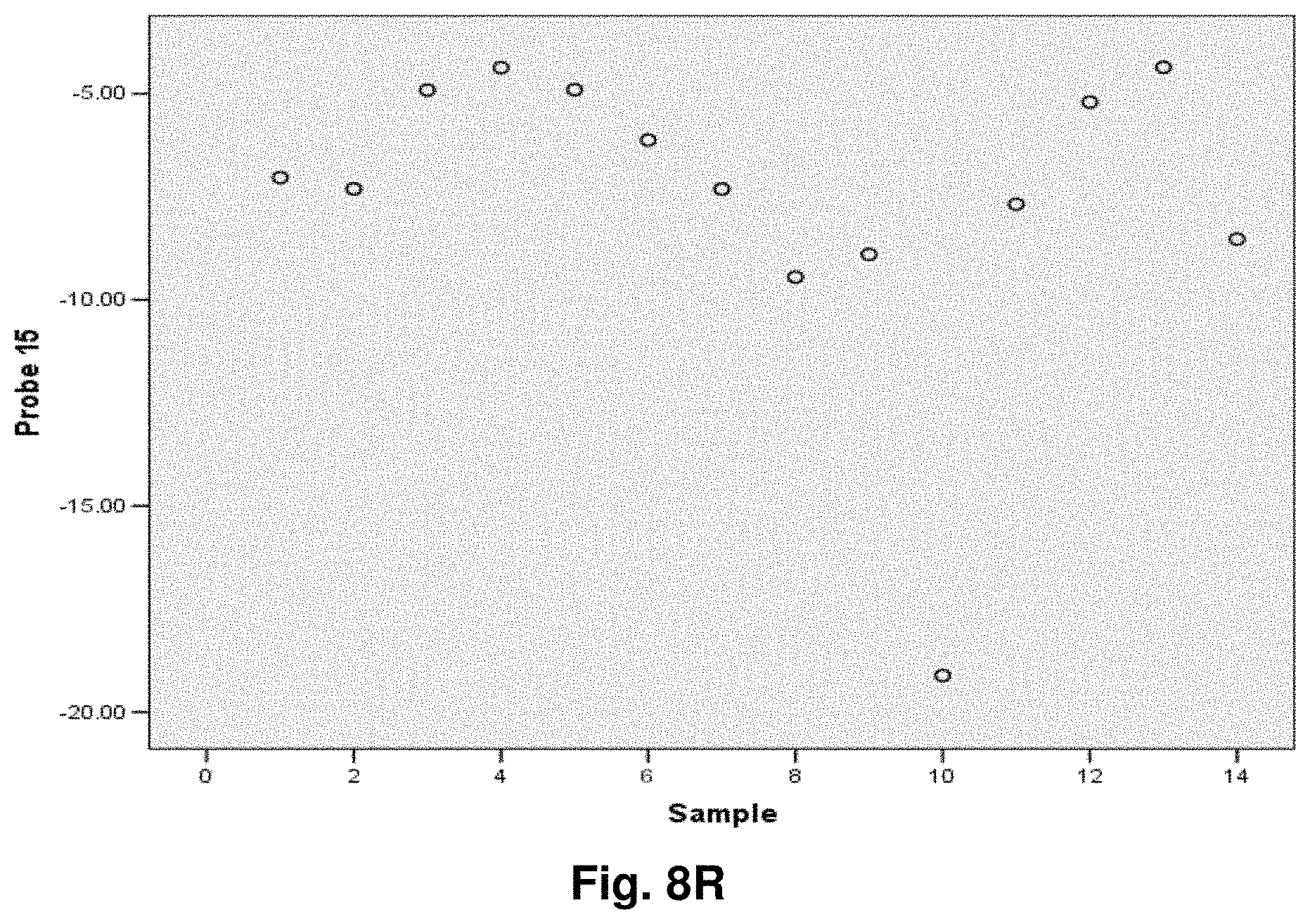
D00049
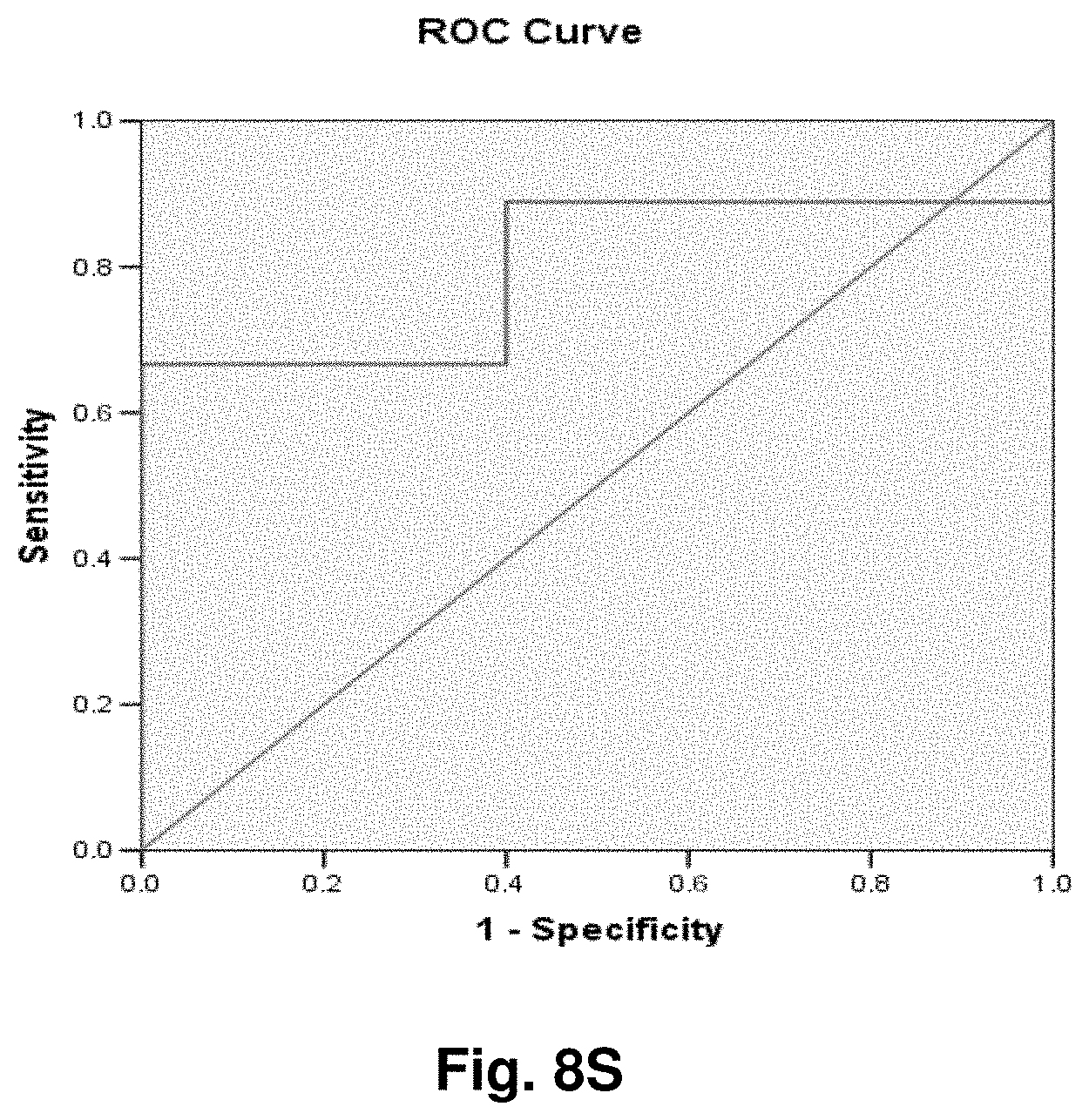
D00050

D00051
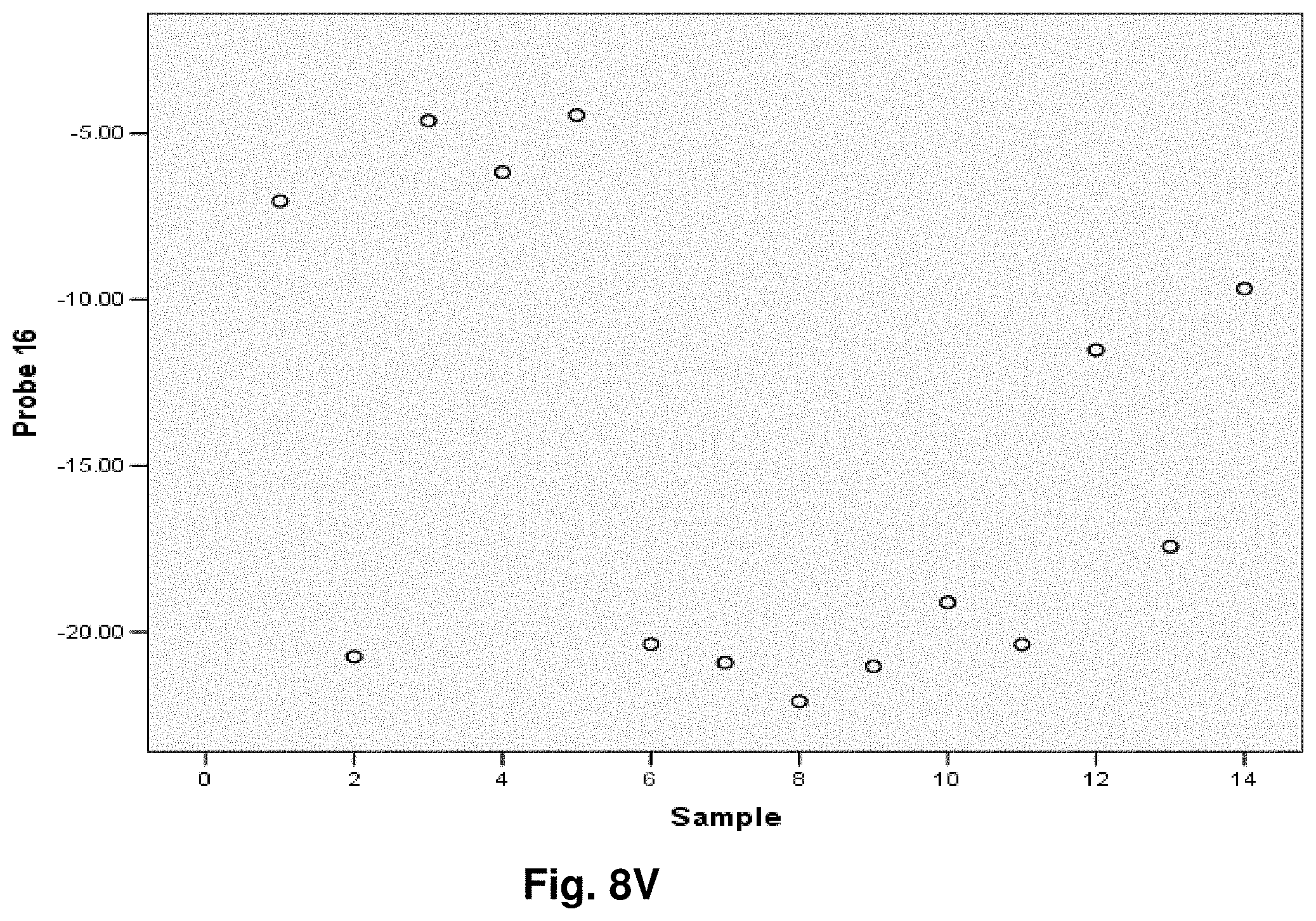
D00052
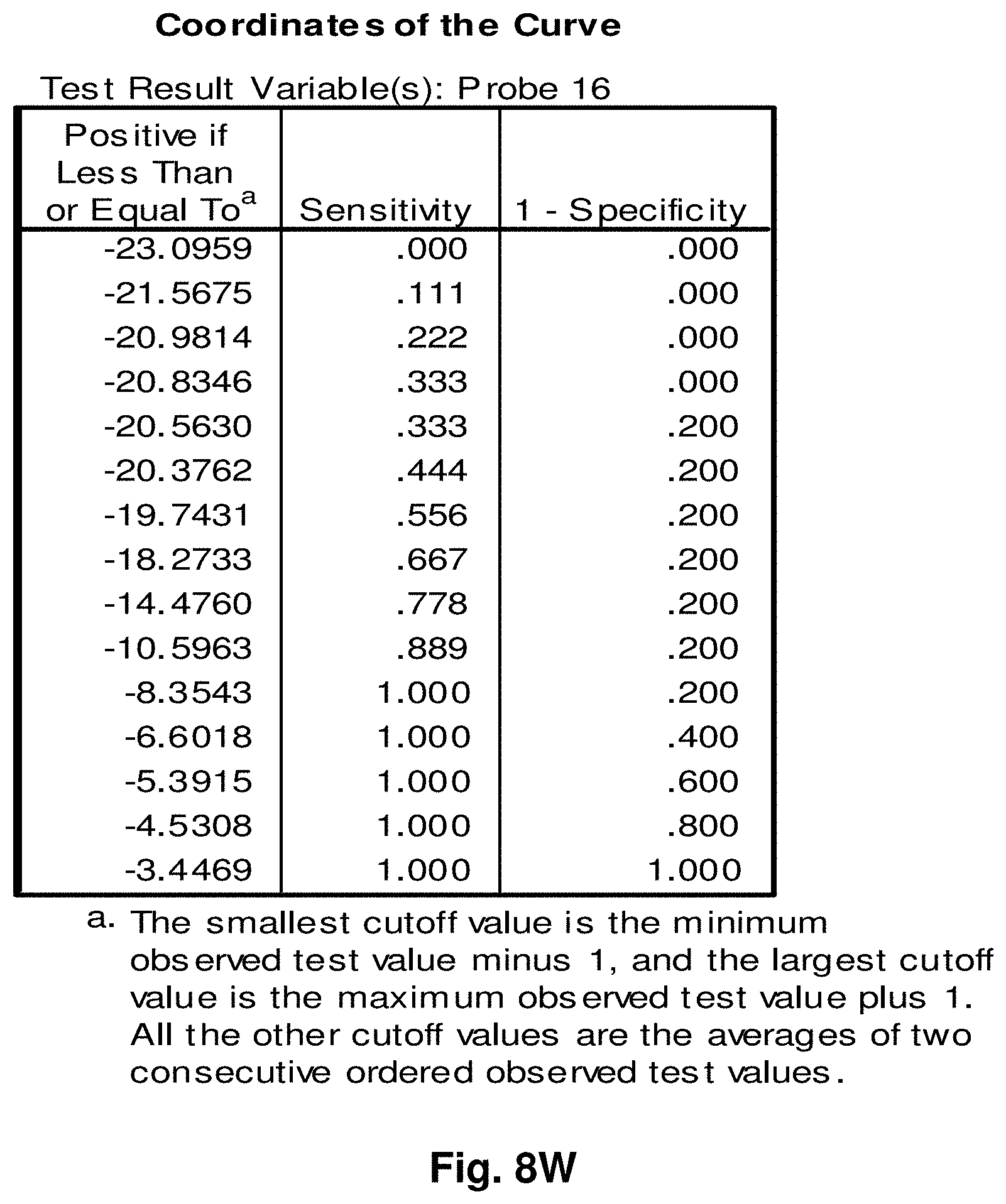
D00053

D00054

D00055
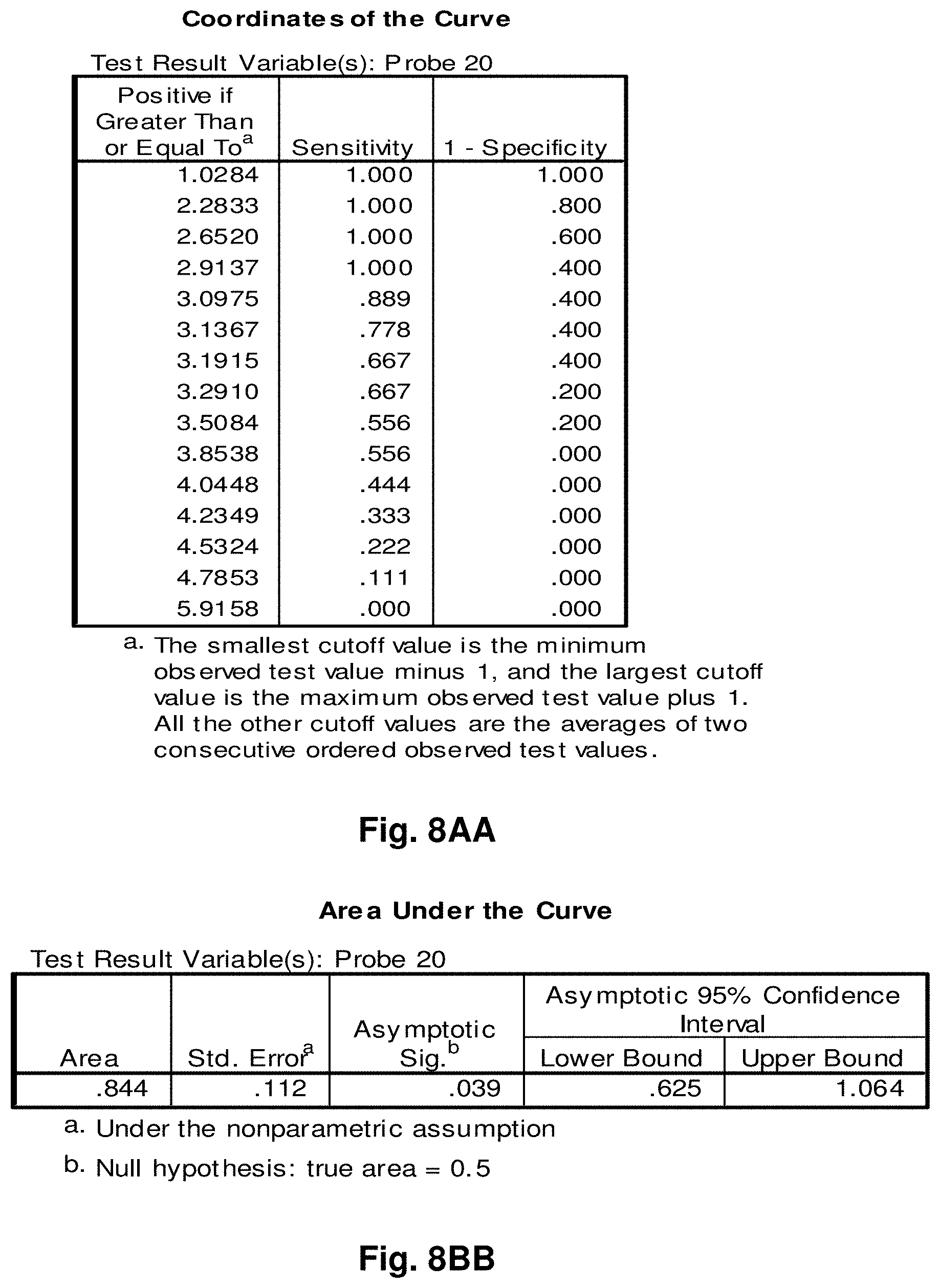
D00056

D00057

D00058
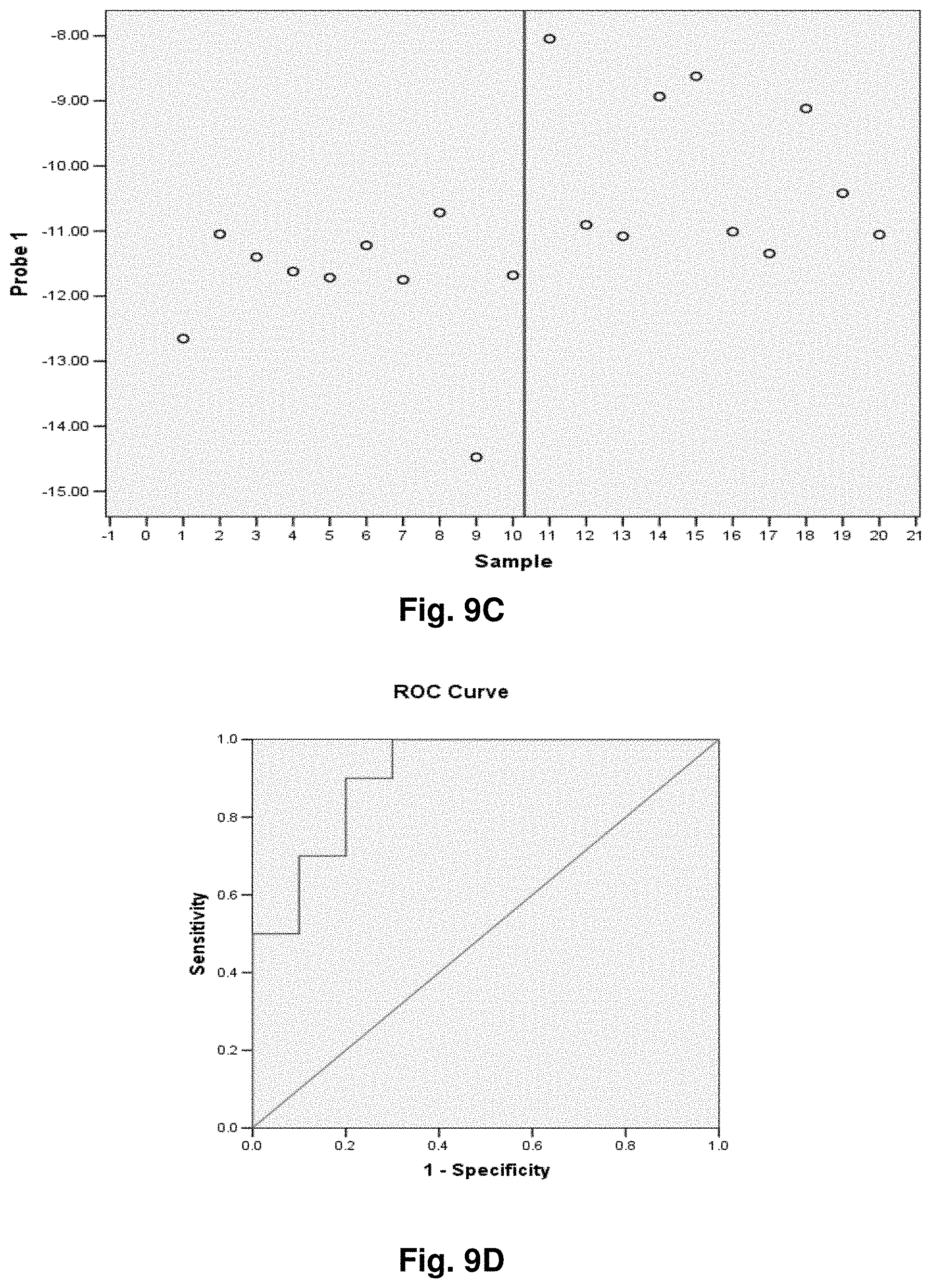
D00059

D00060
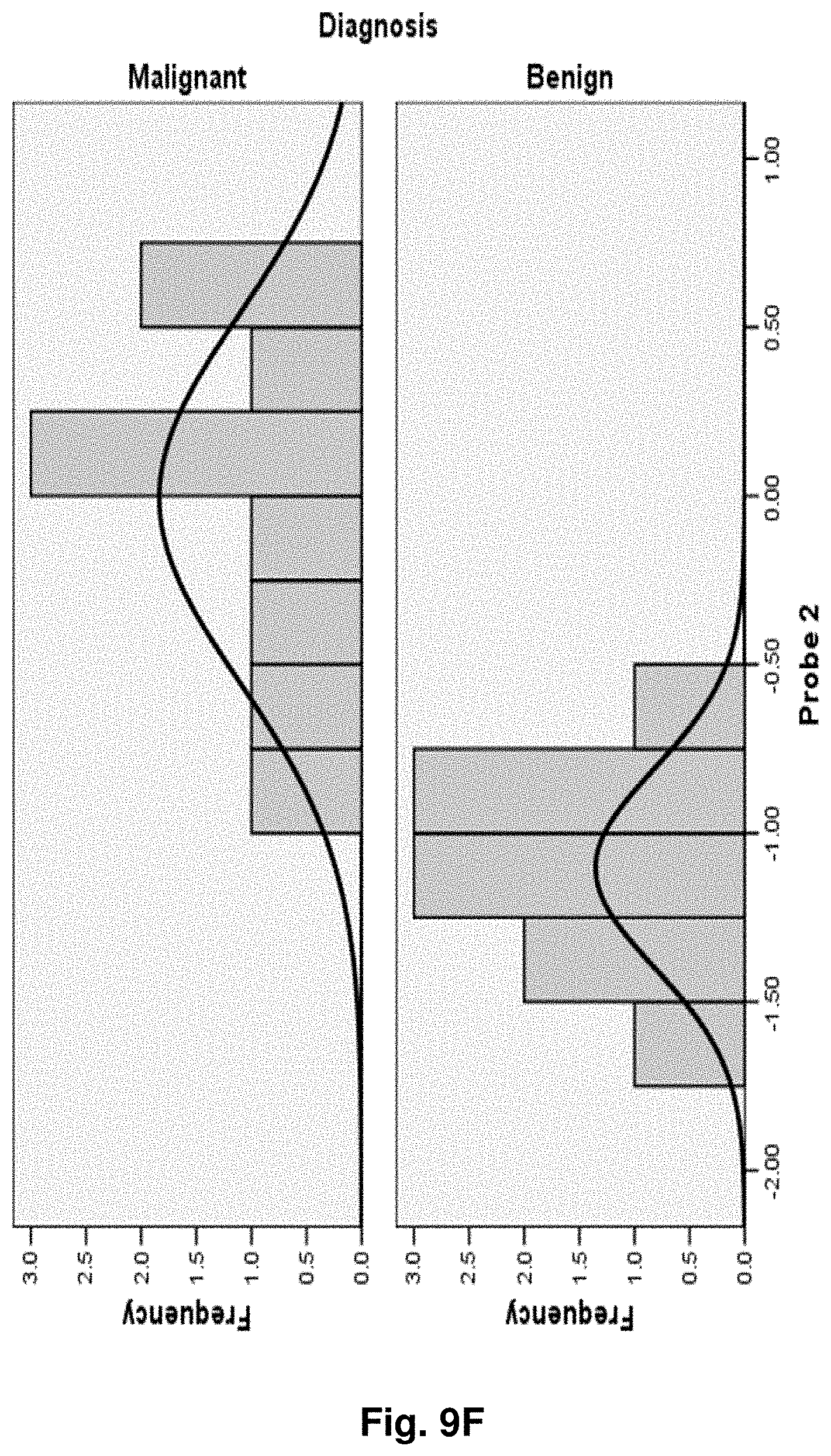
D00061

D00062

D00063
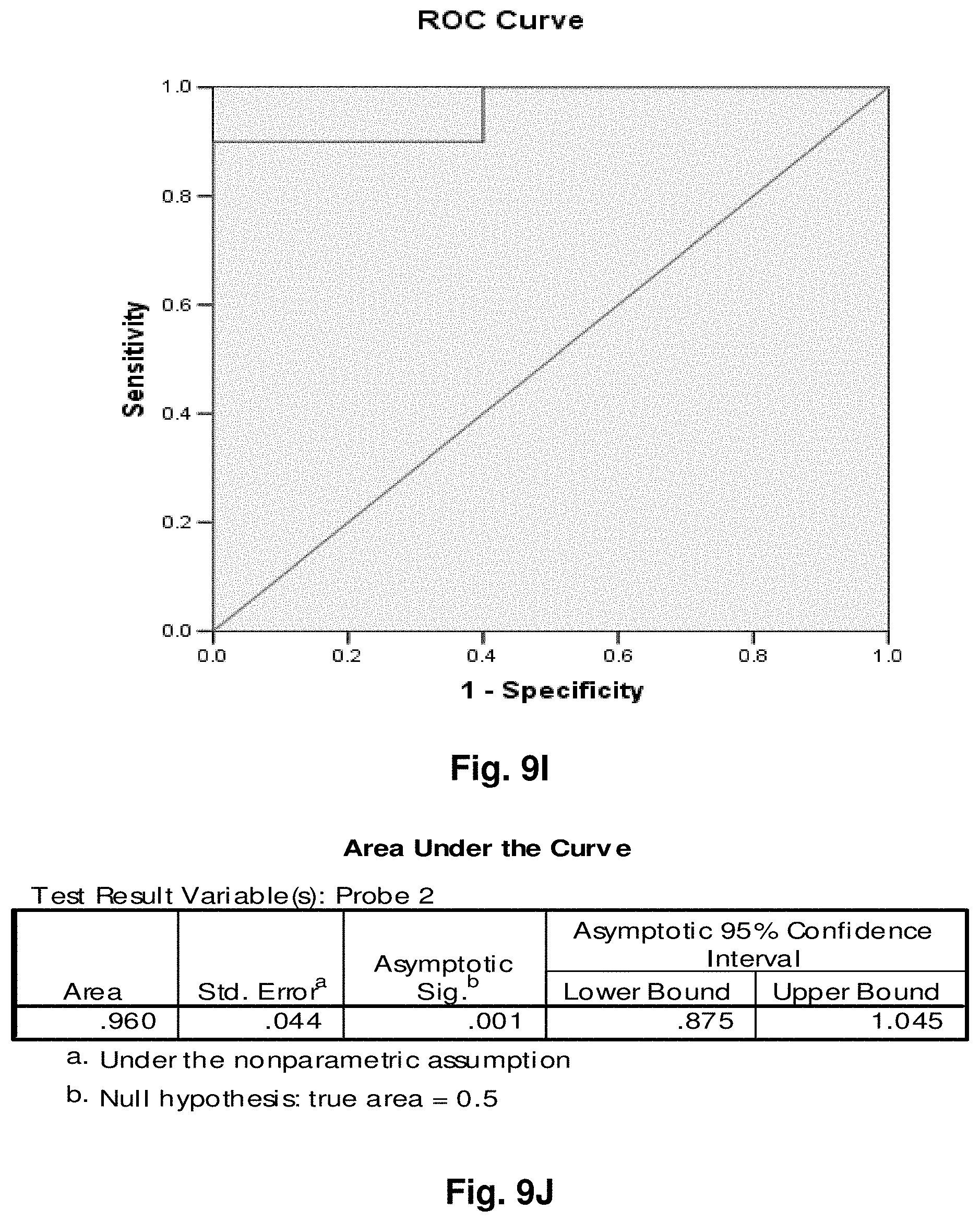
D00064

D00065
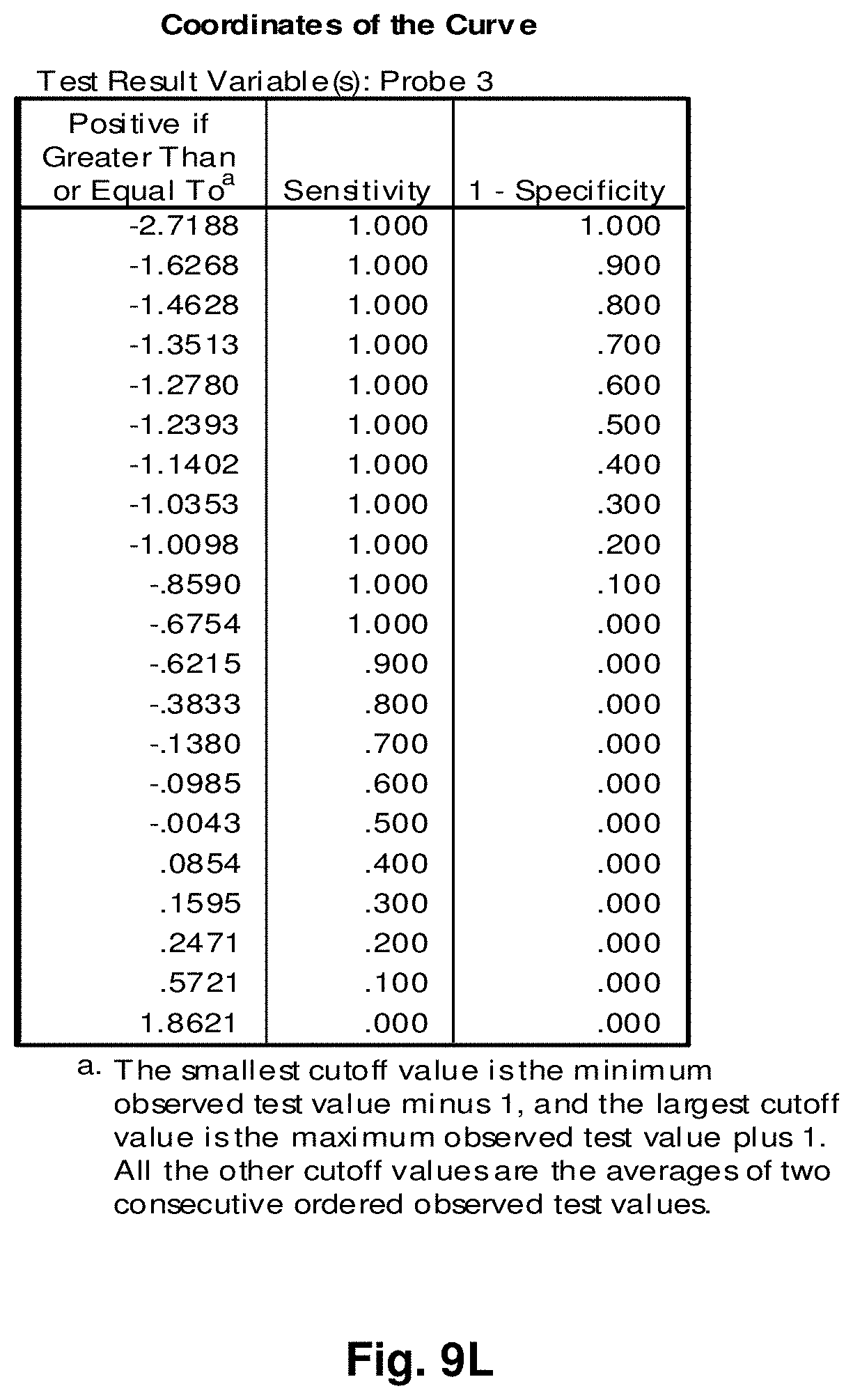
D00066
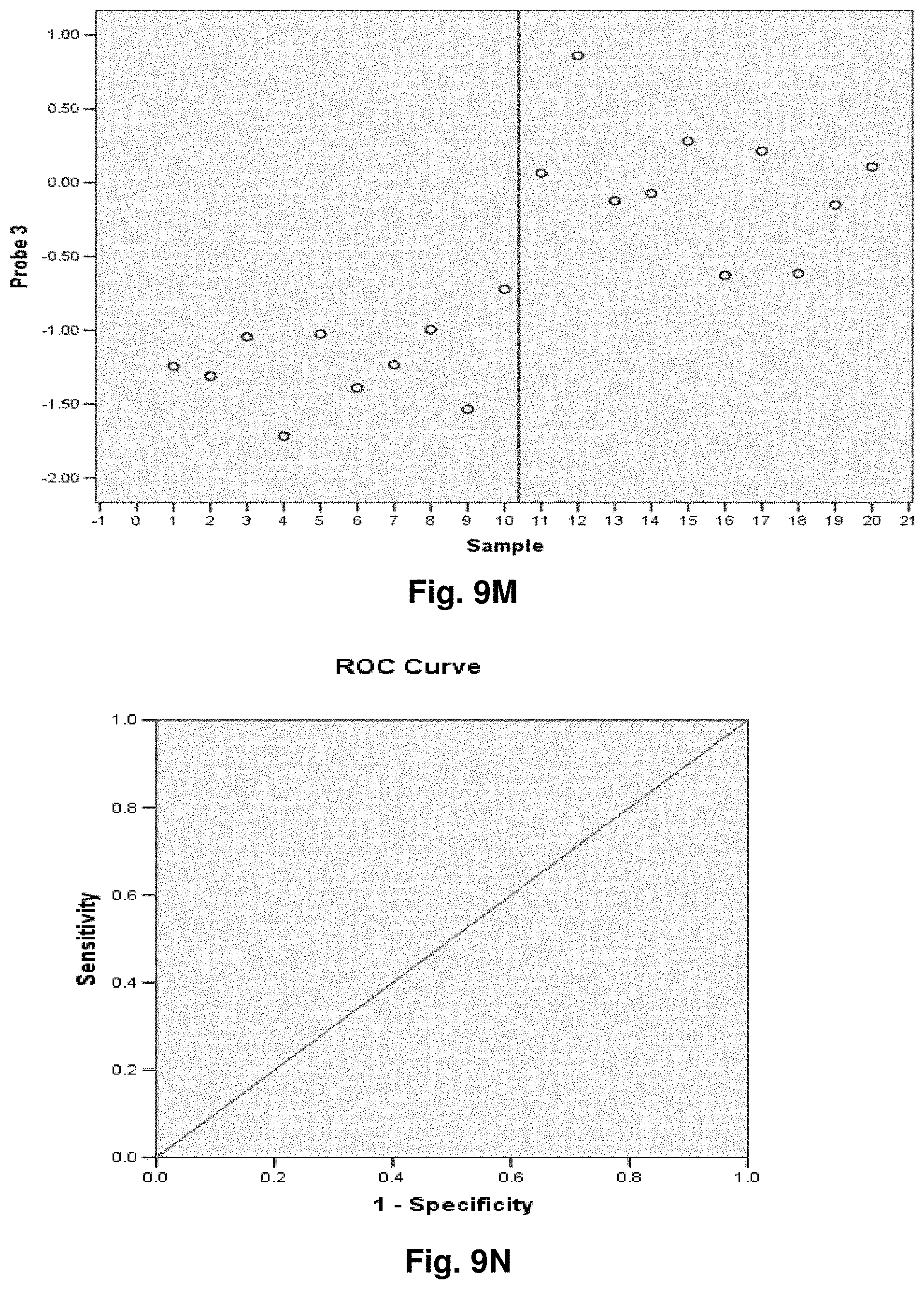
D00067

D00068

D00069

D00070
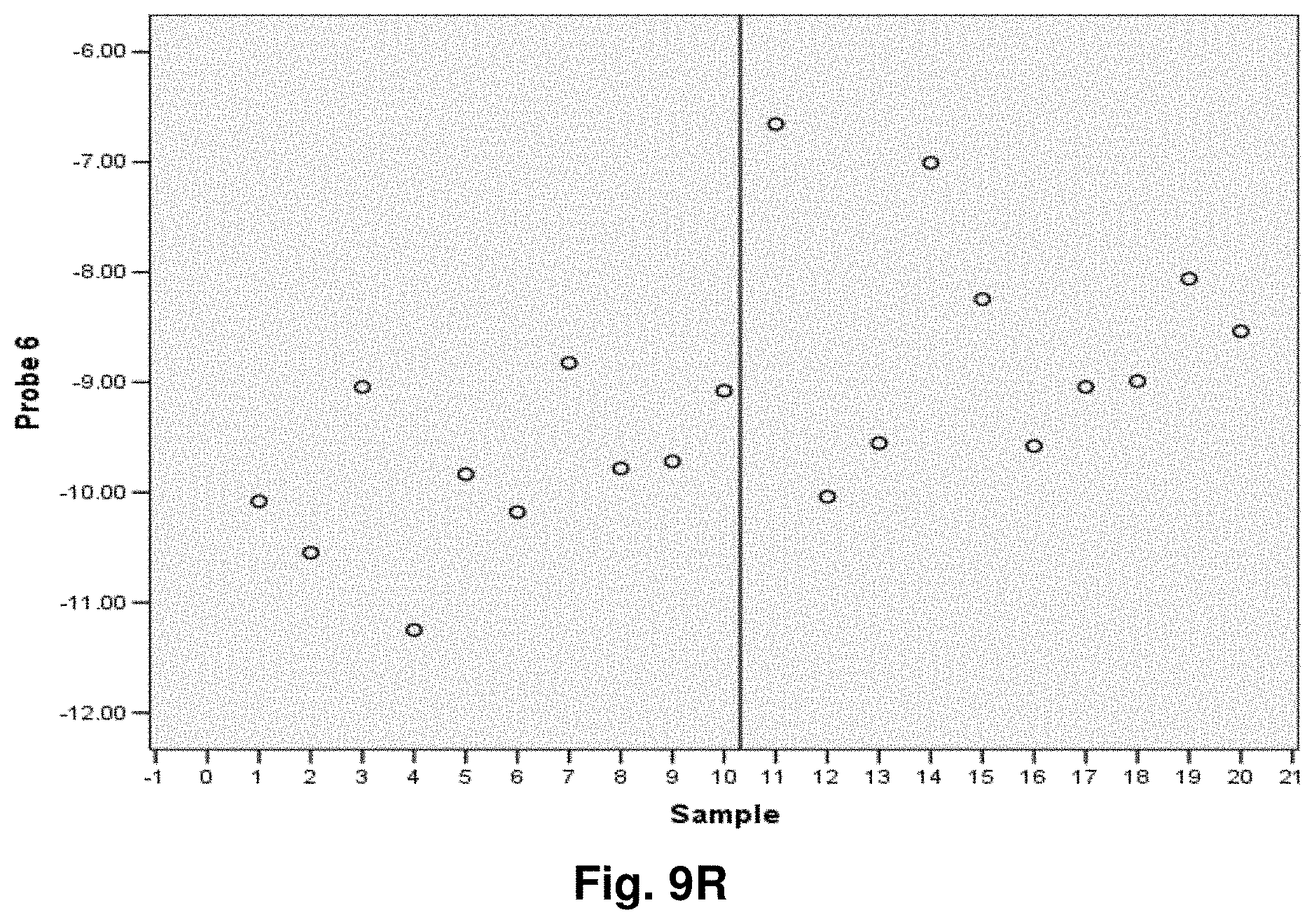
D00071
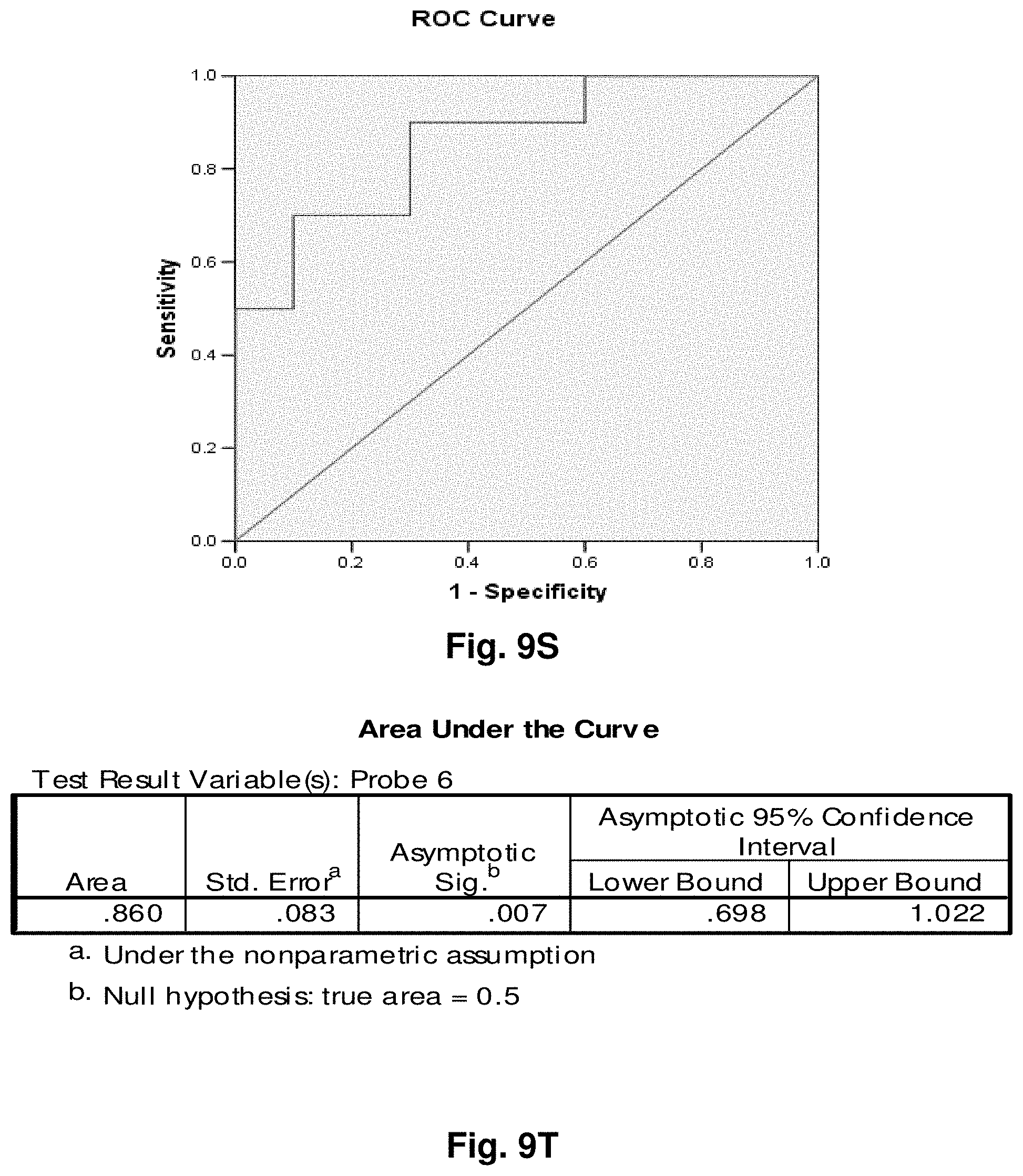
D00072

D00073

D00074
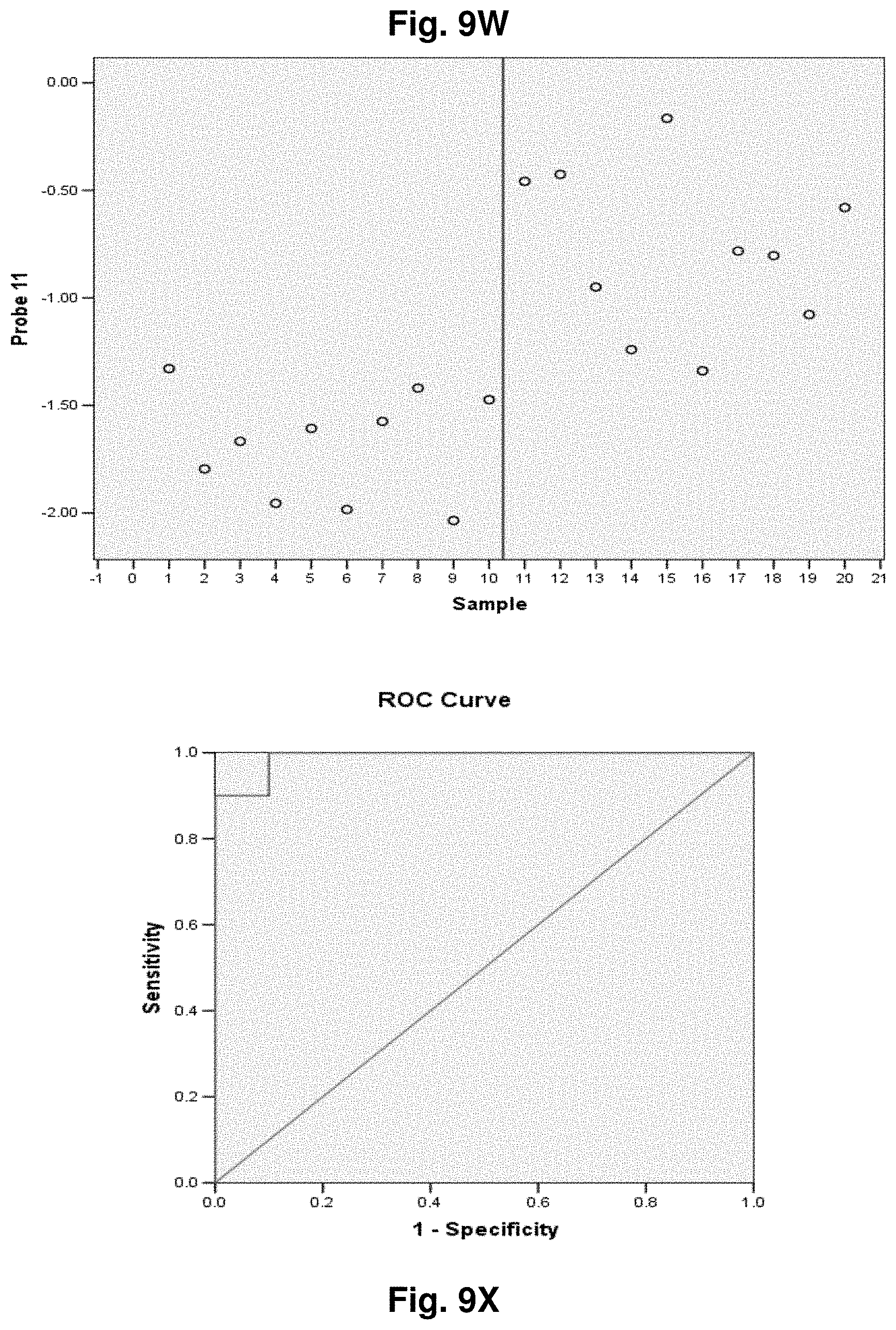
D00075

D00076
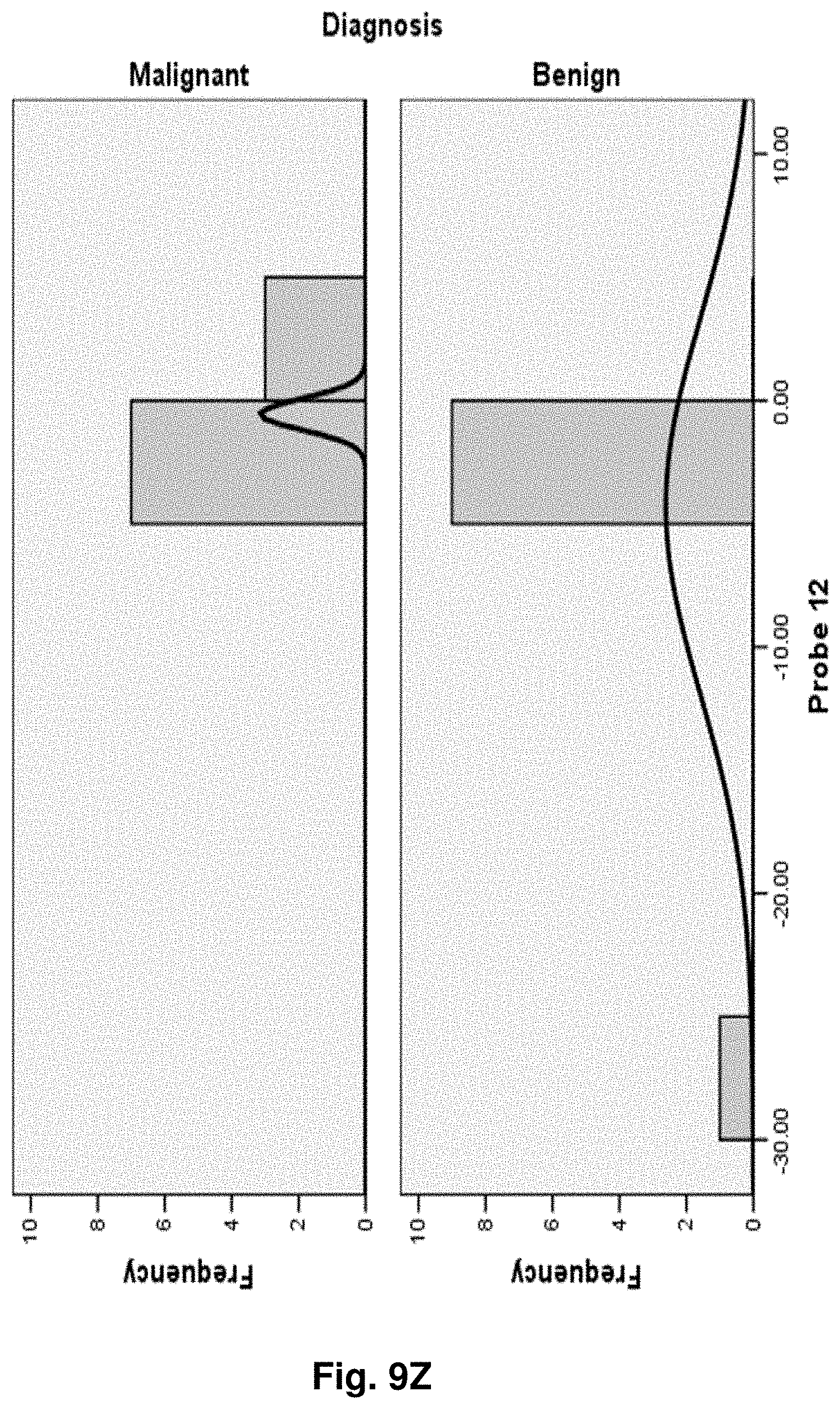
D00077
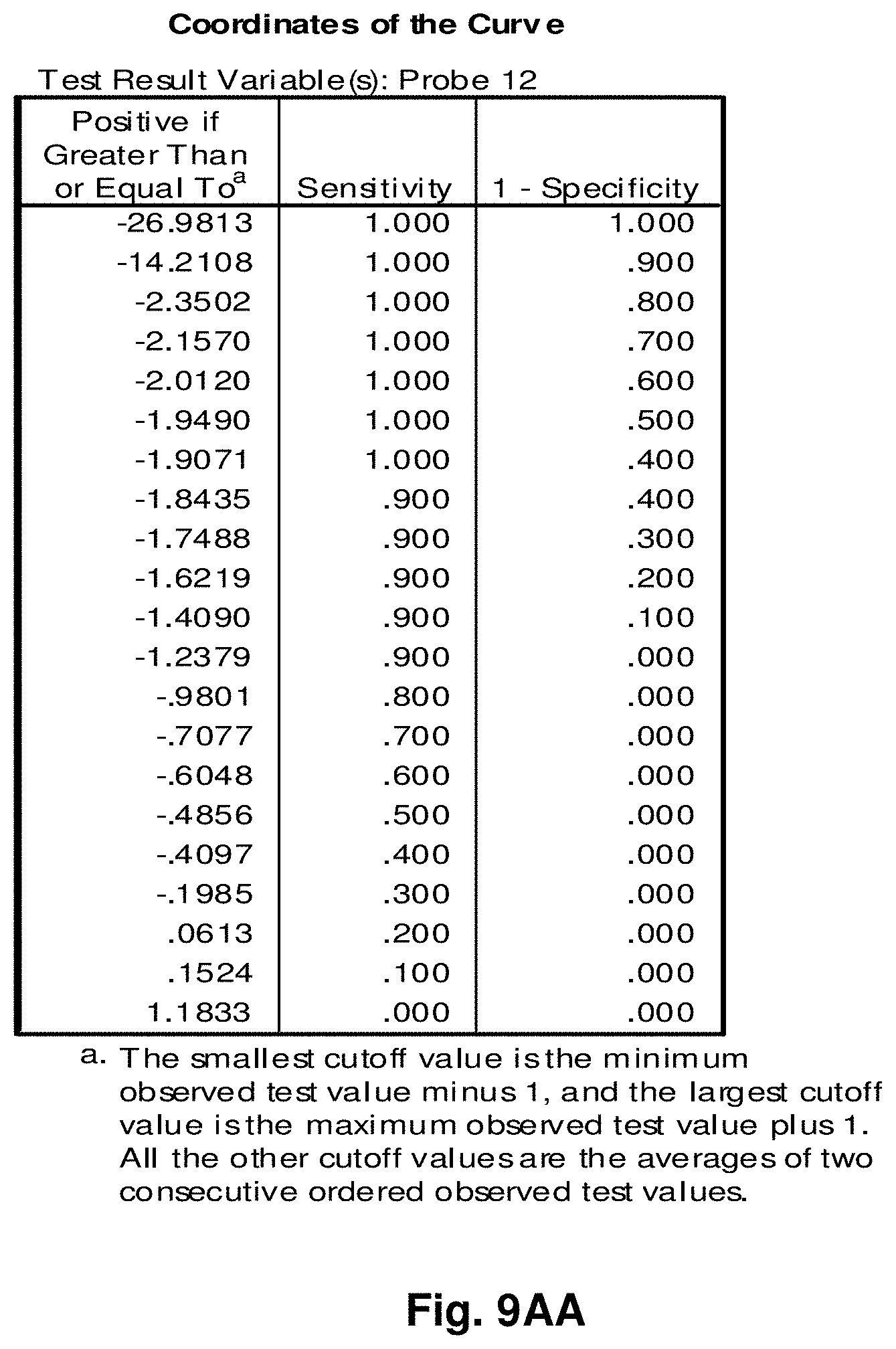
D00078

D00079

D00080
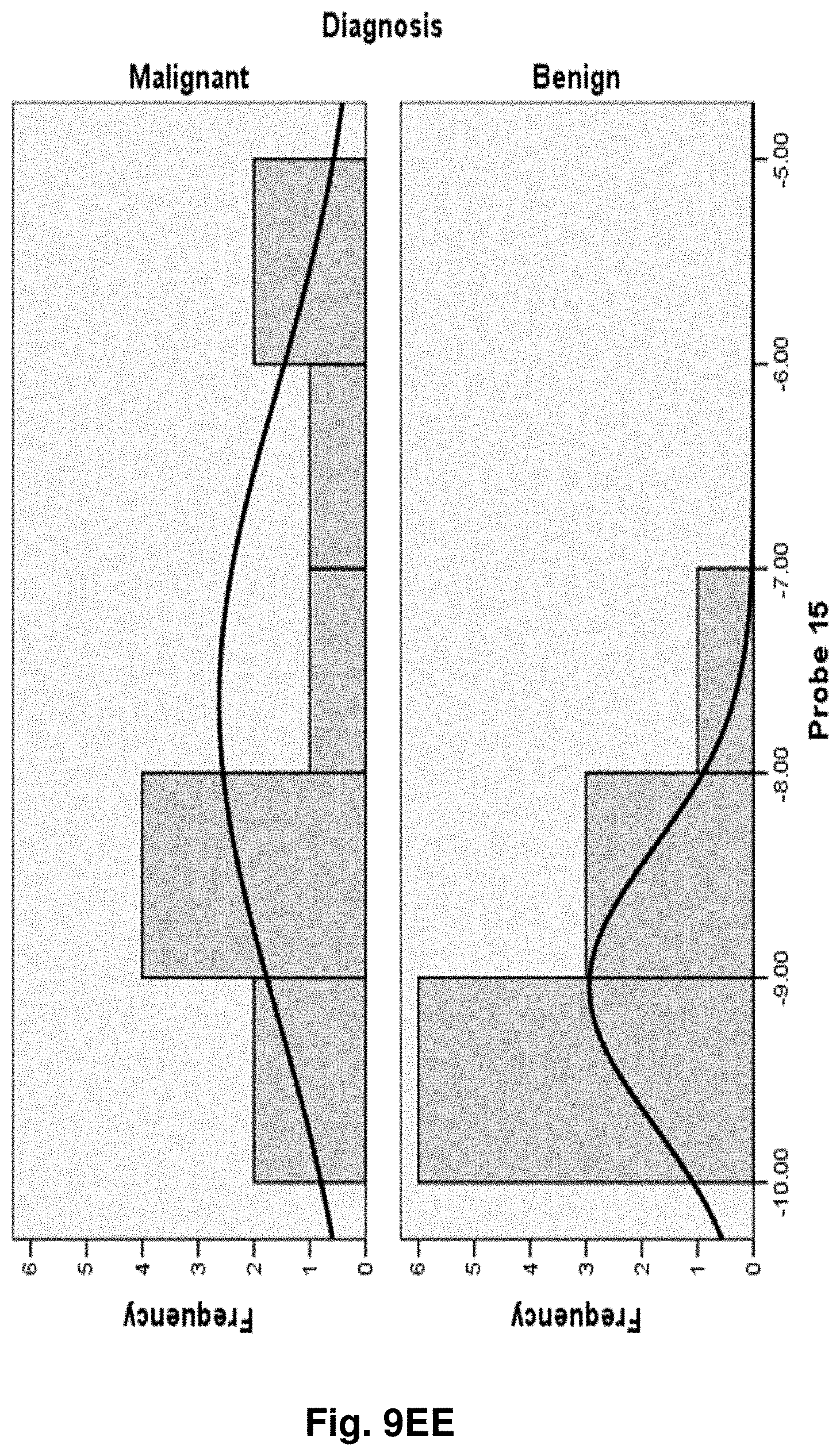
D00081
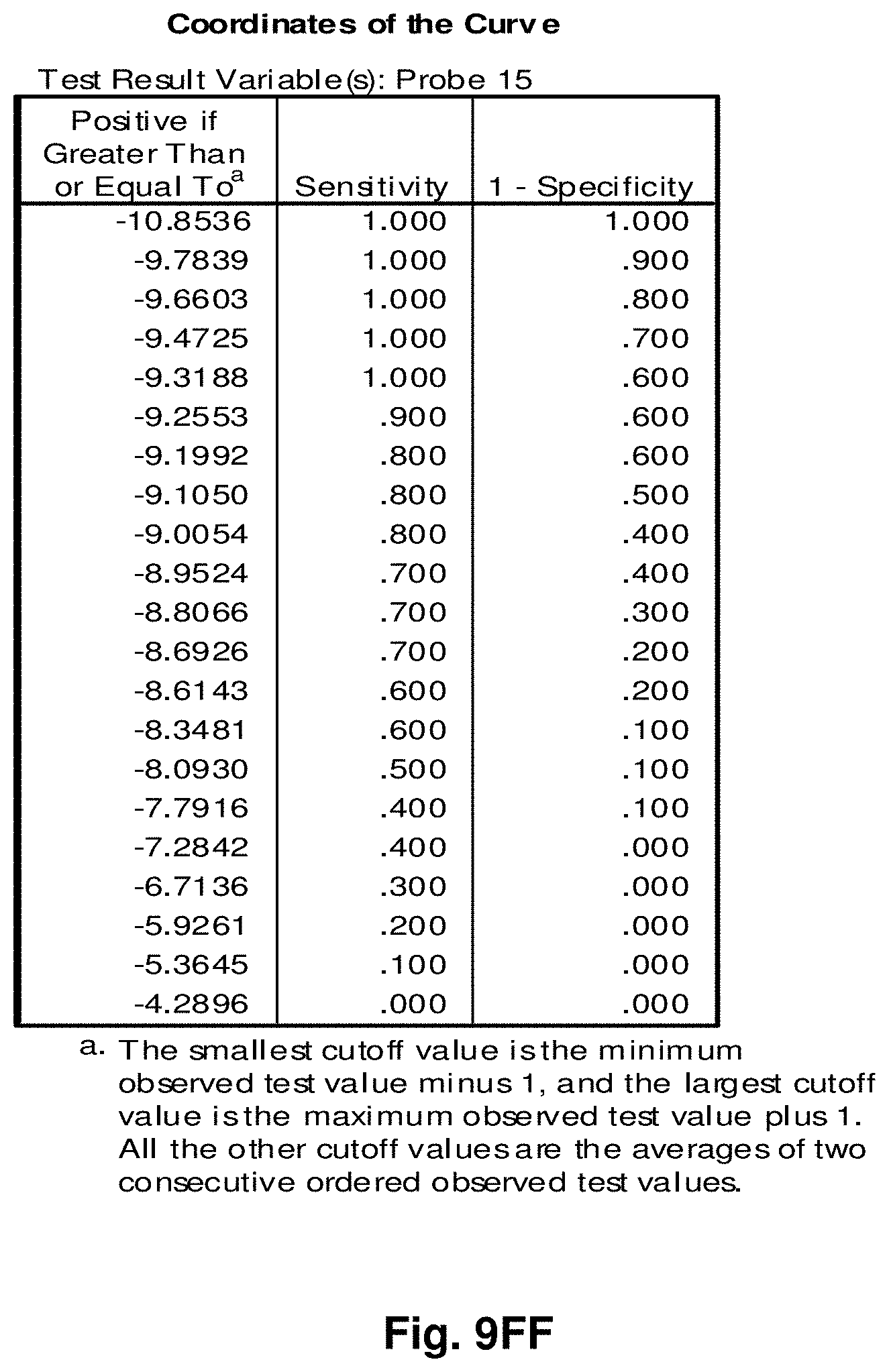
D00082

D00083
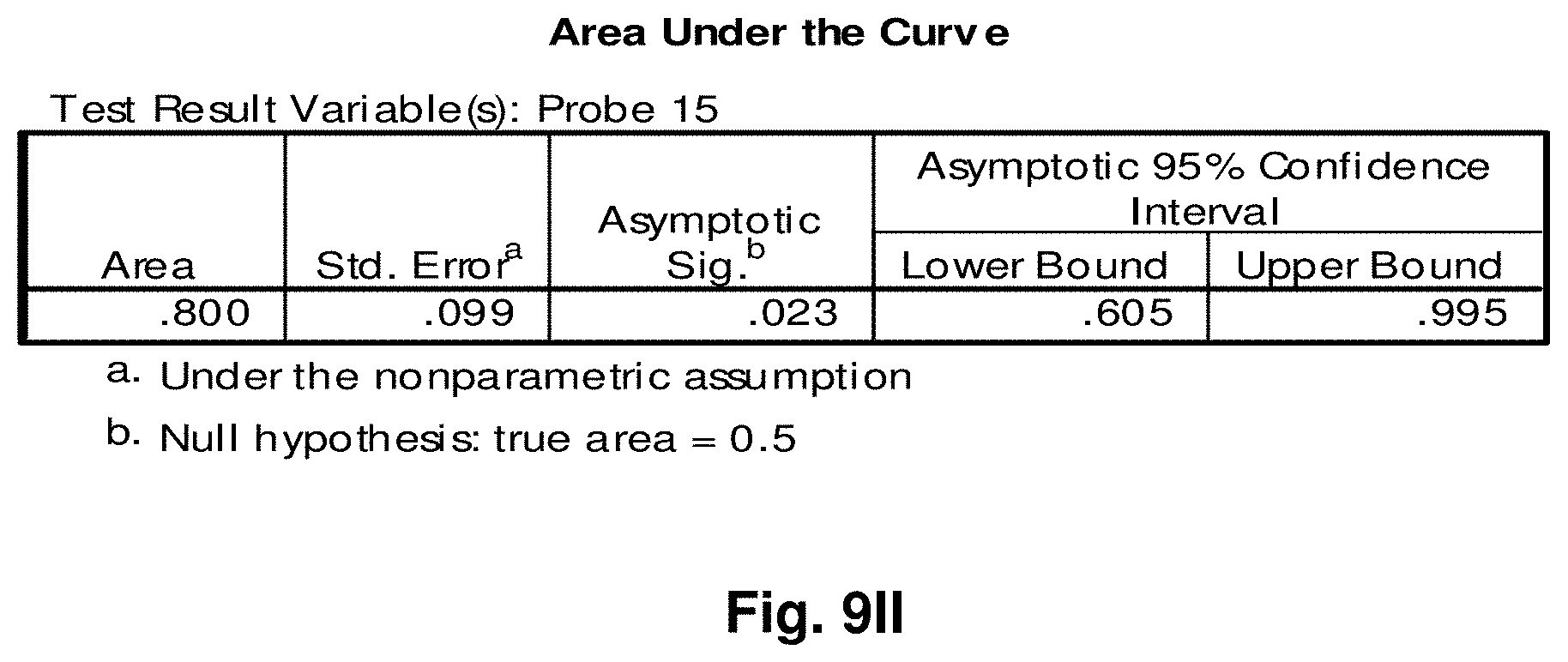
D00084

D00085
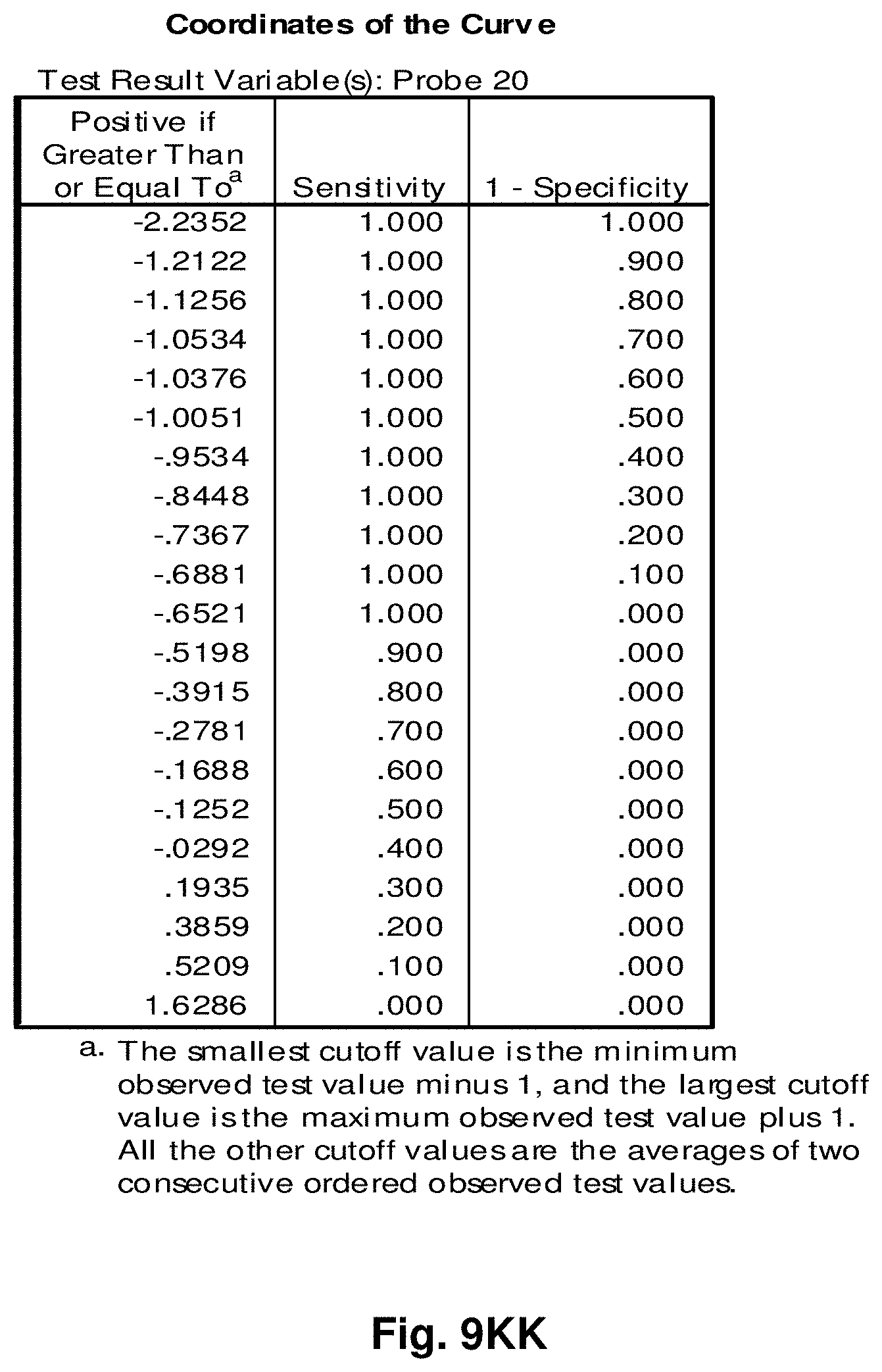
D00086
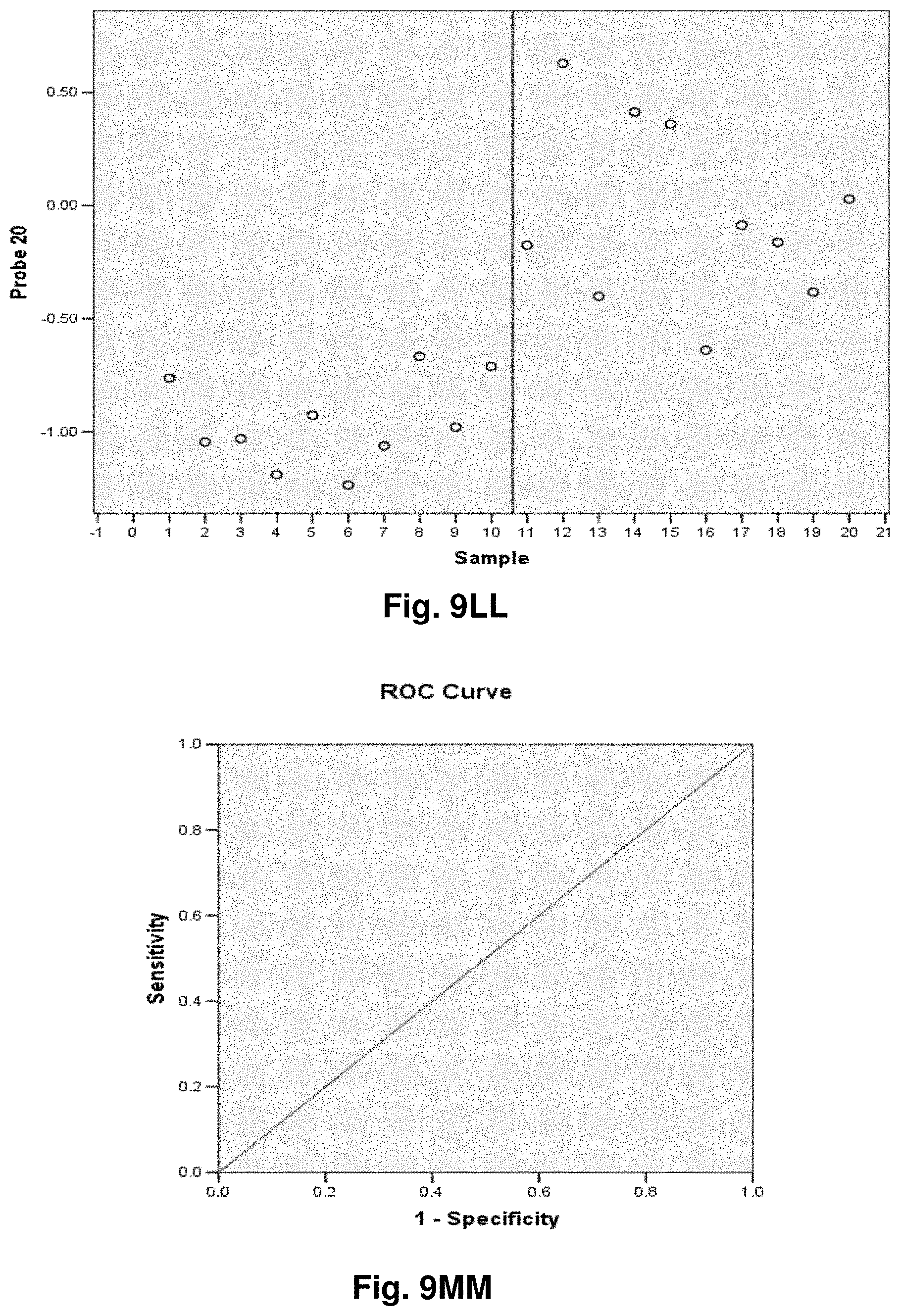
D00087

D00088
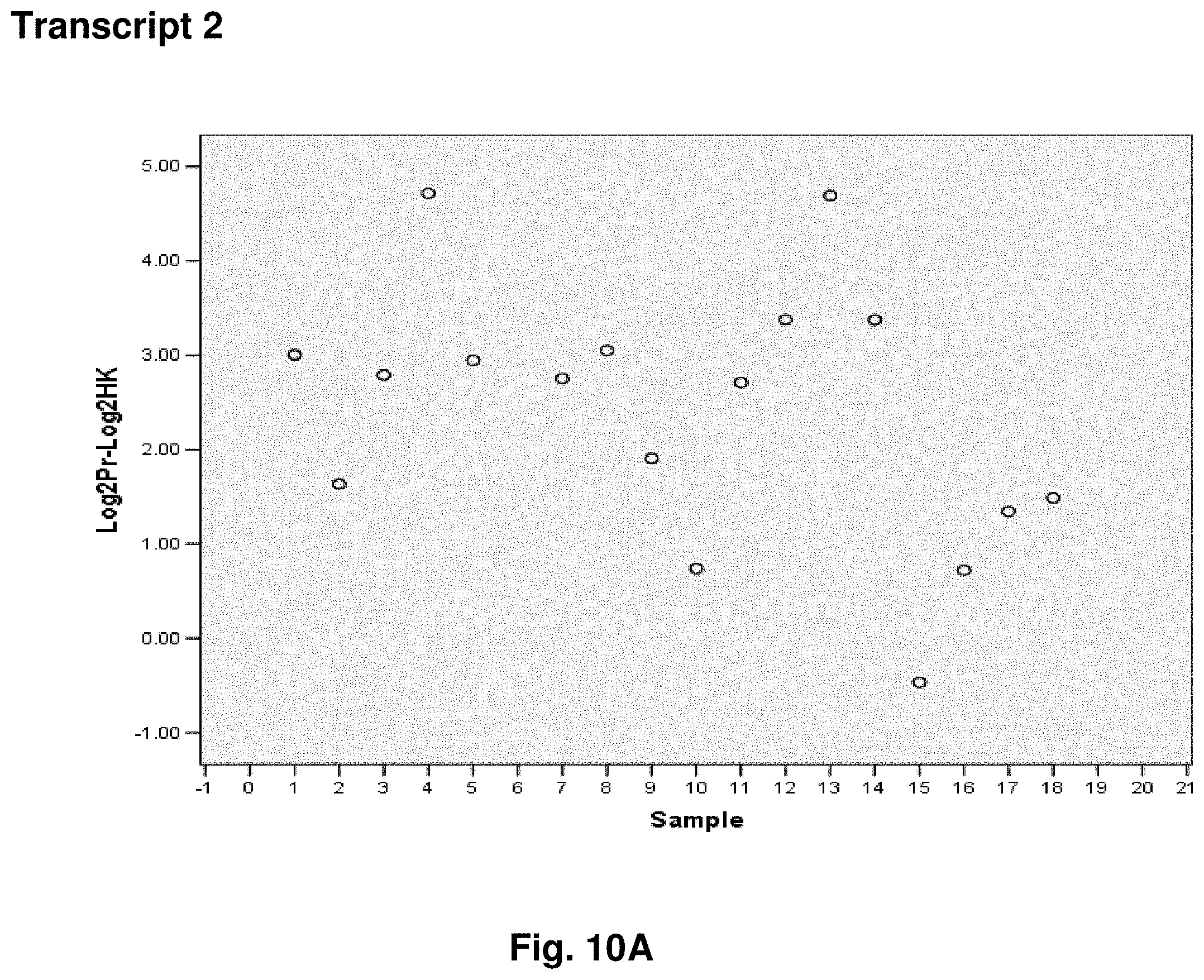
D00089
D00090
D00091

D00092

D00093
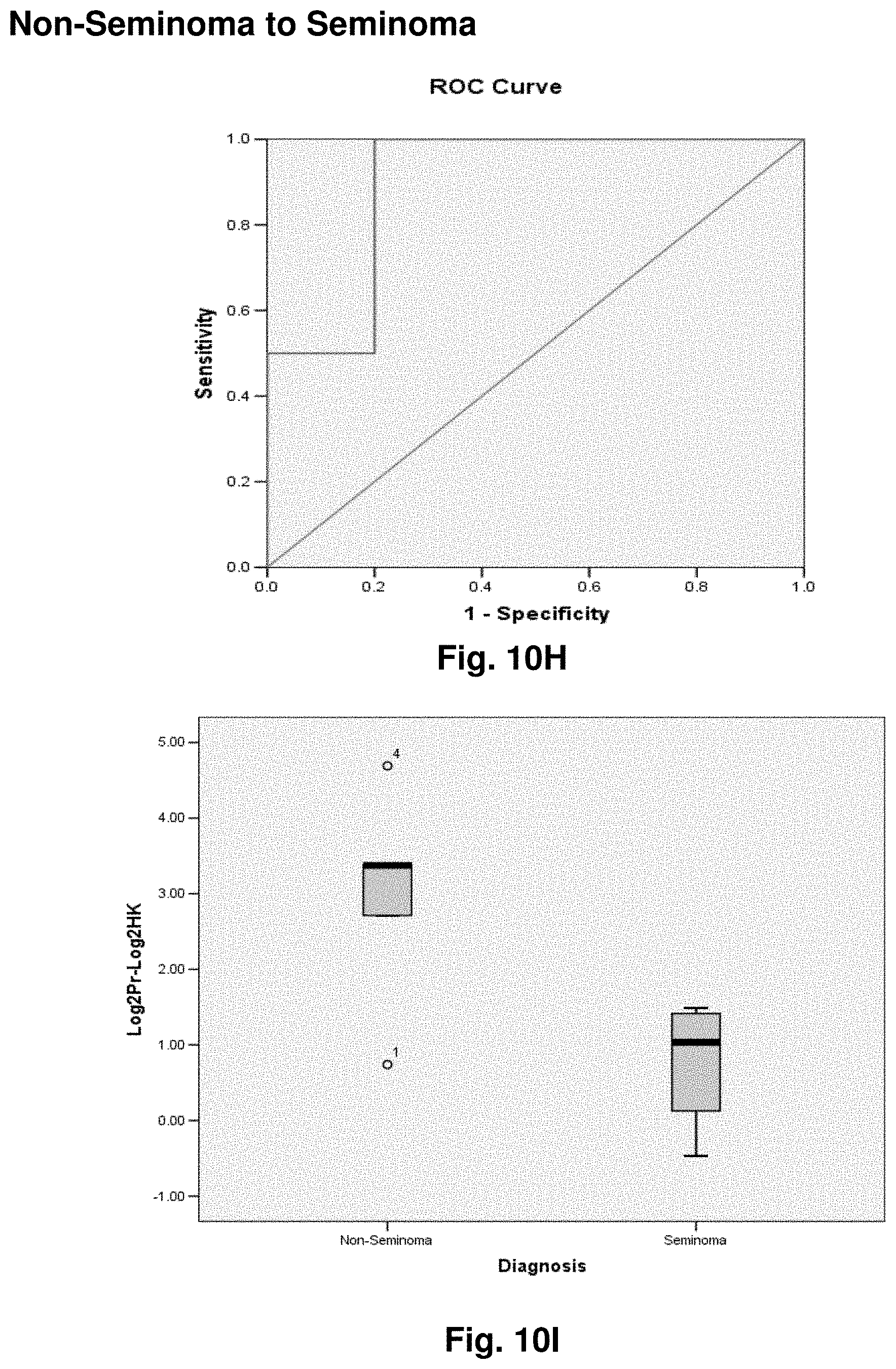
D00094
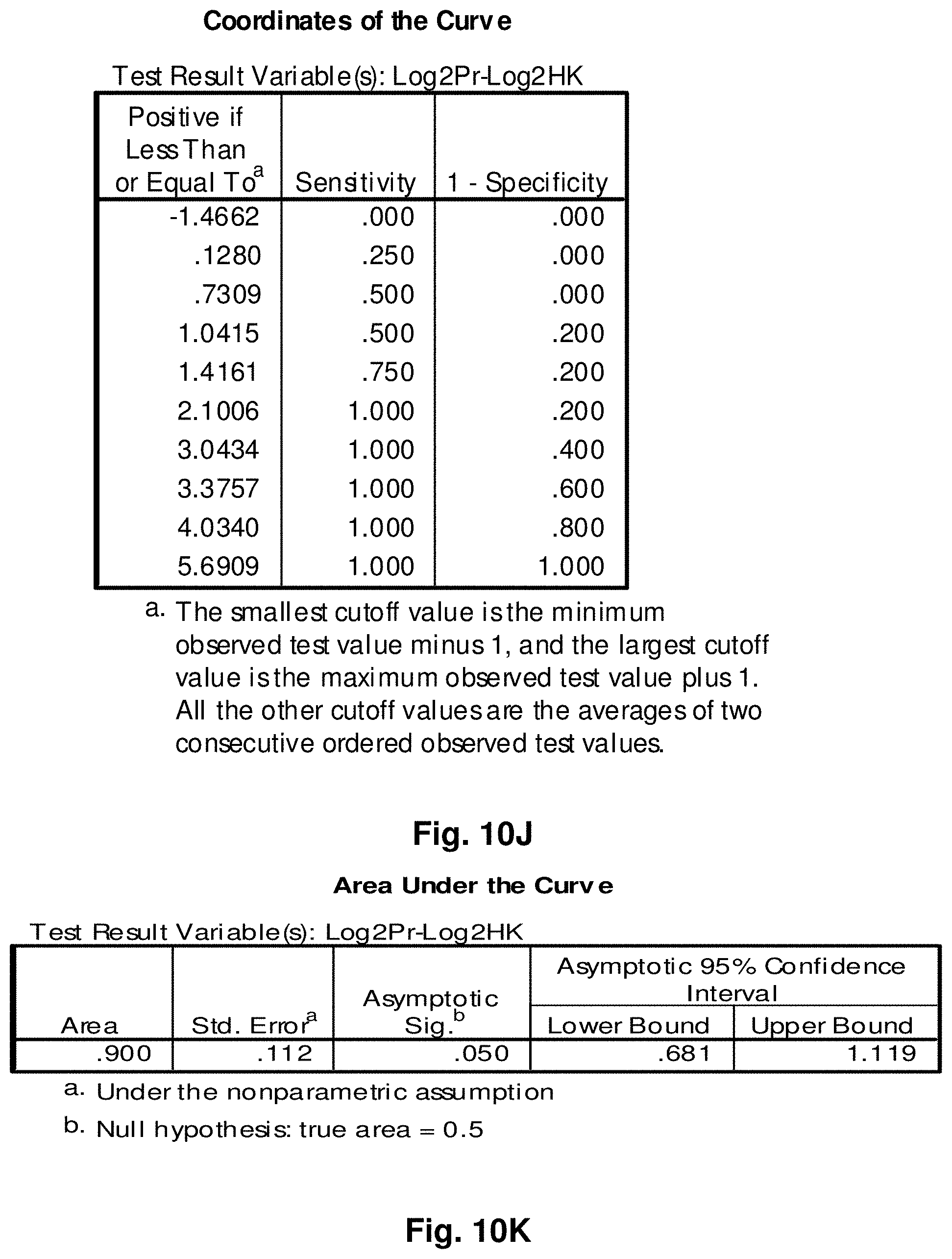
D00095

D00096
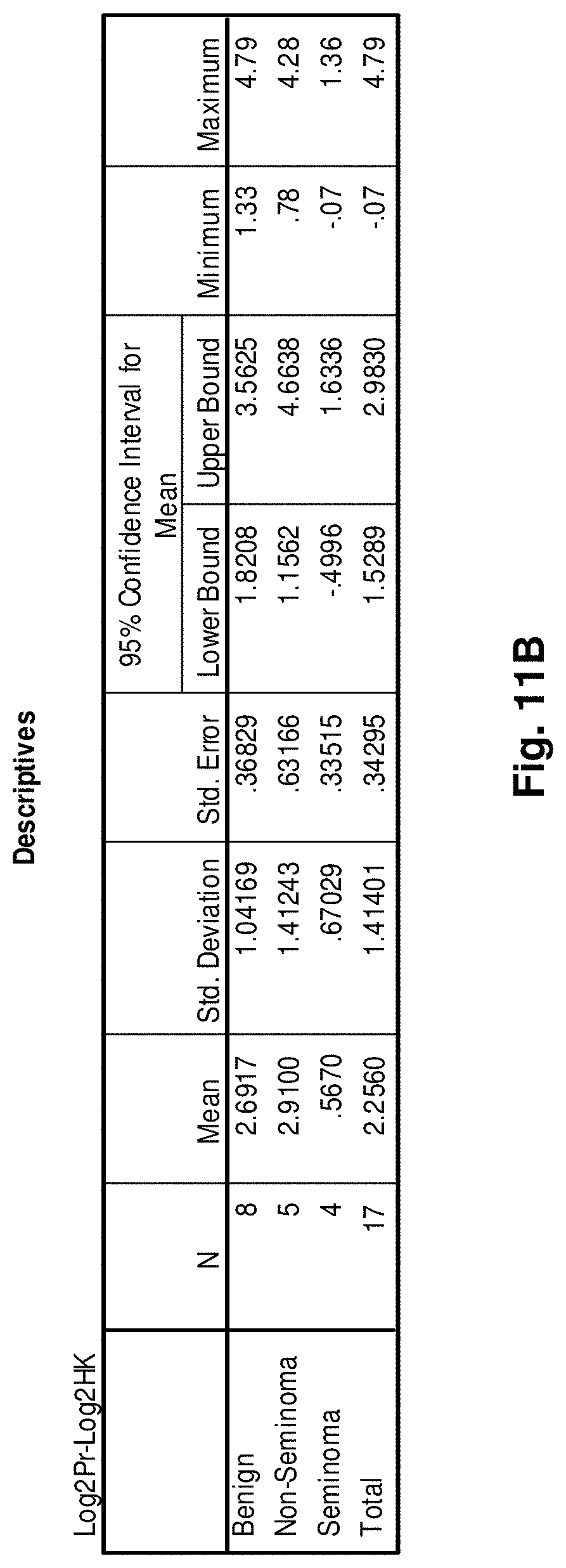
D00097
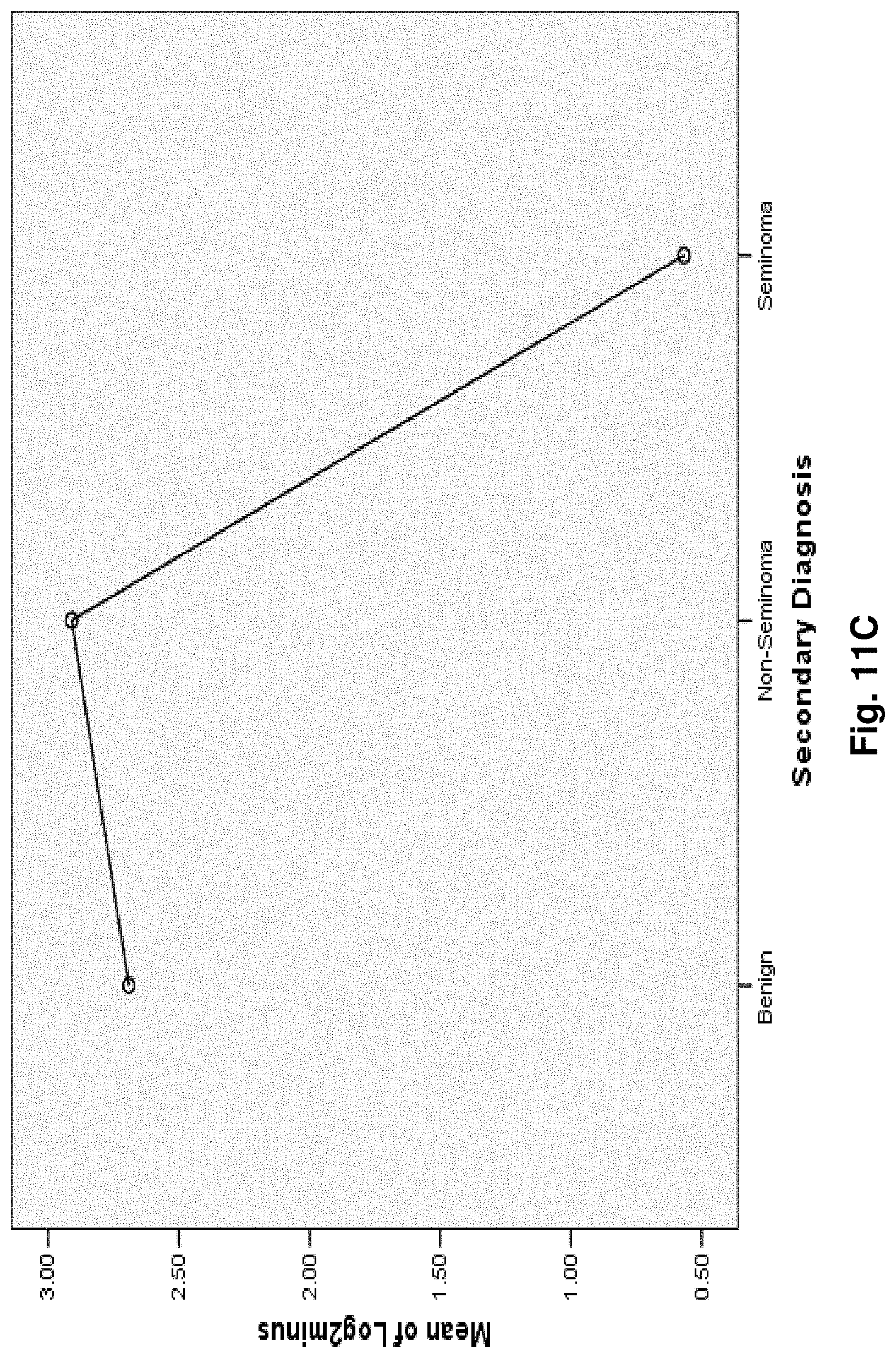
D00098
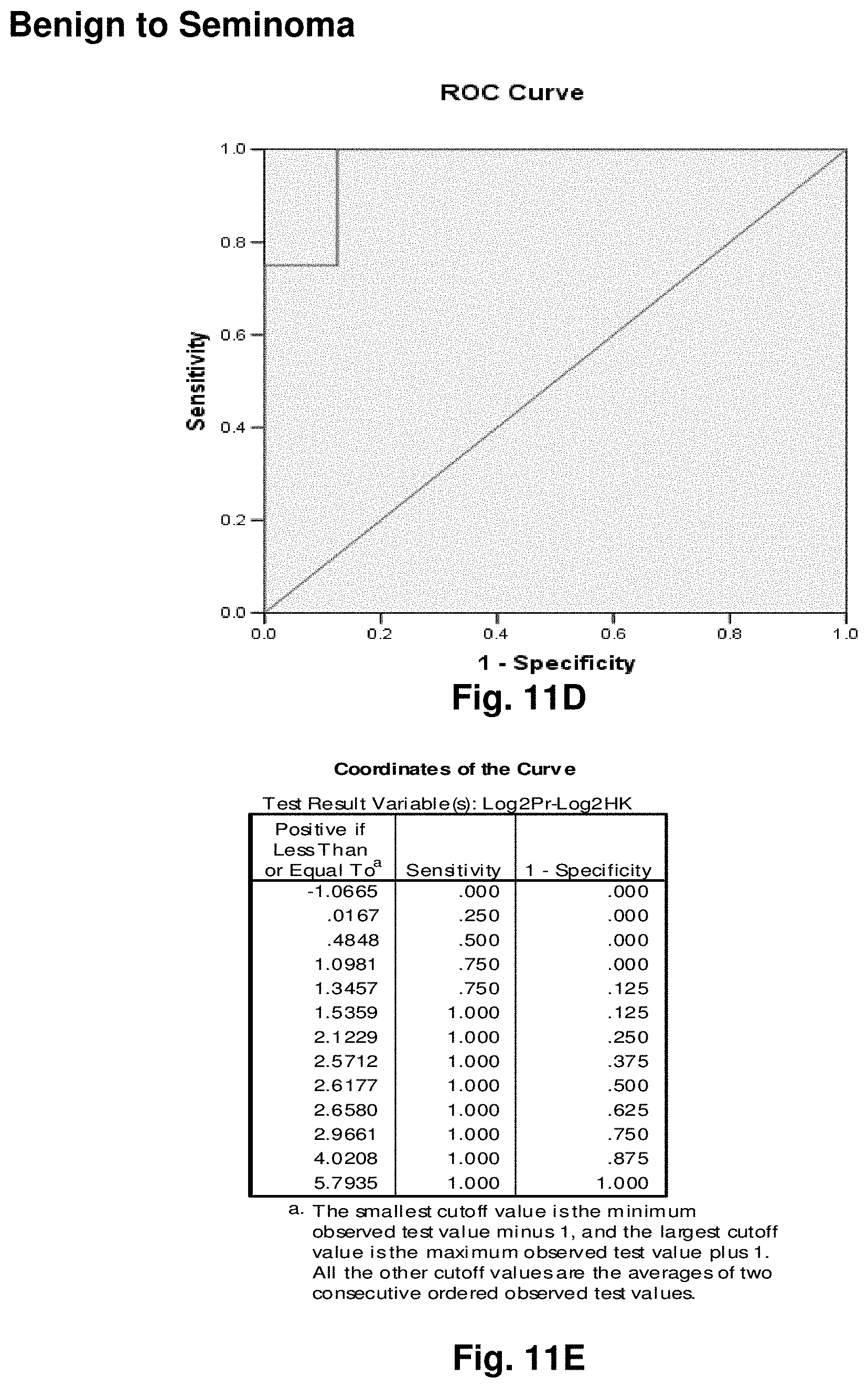
D00099
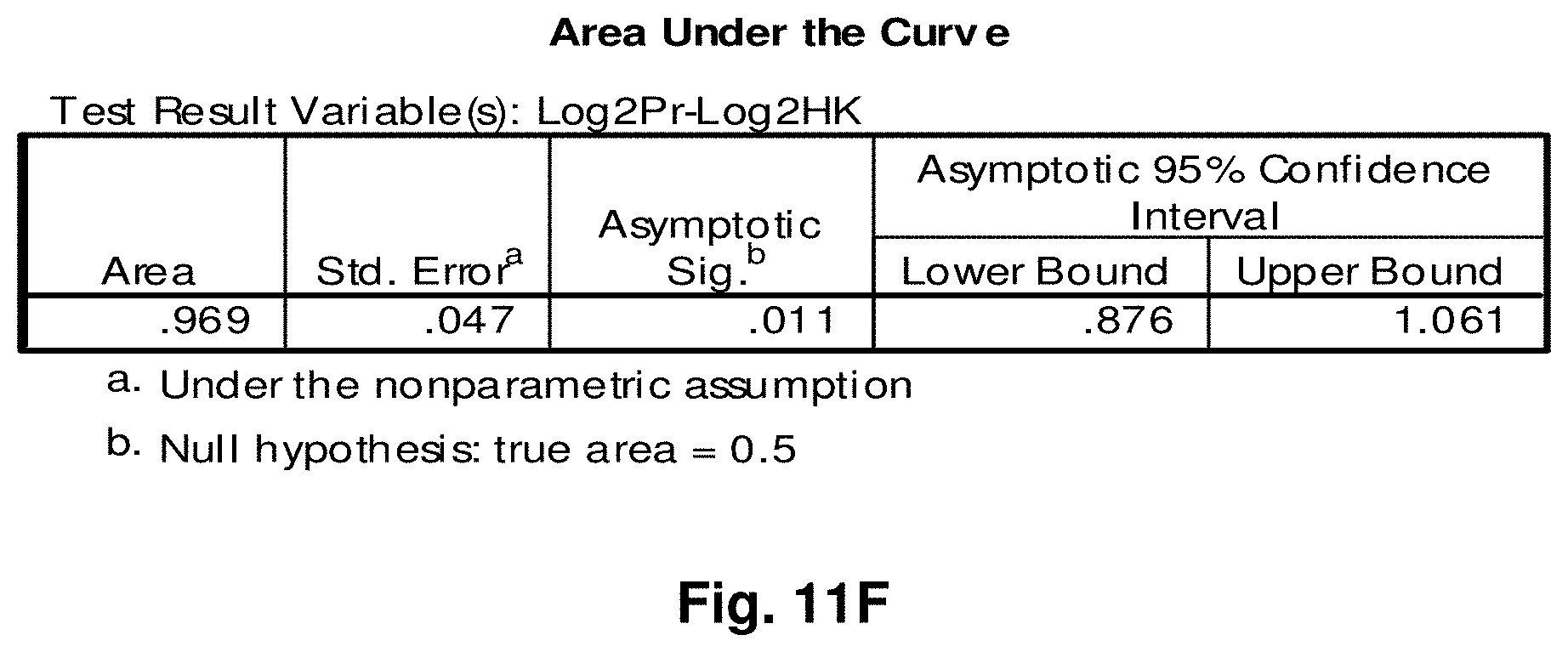
D00100

D00101
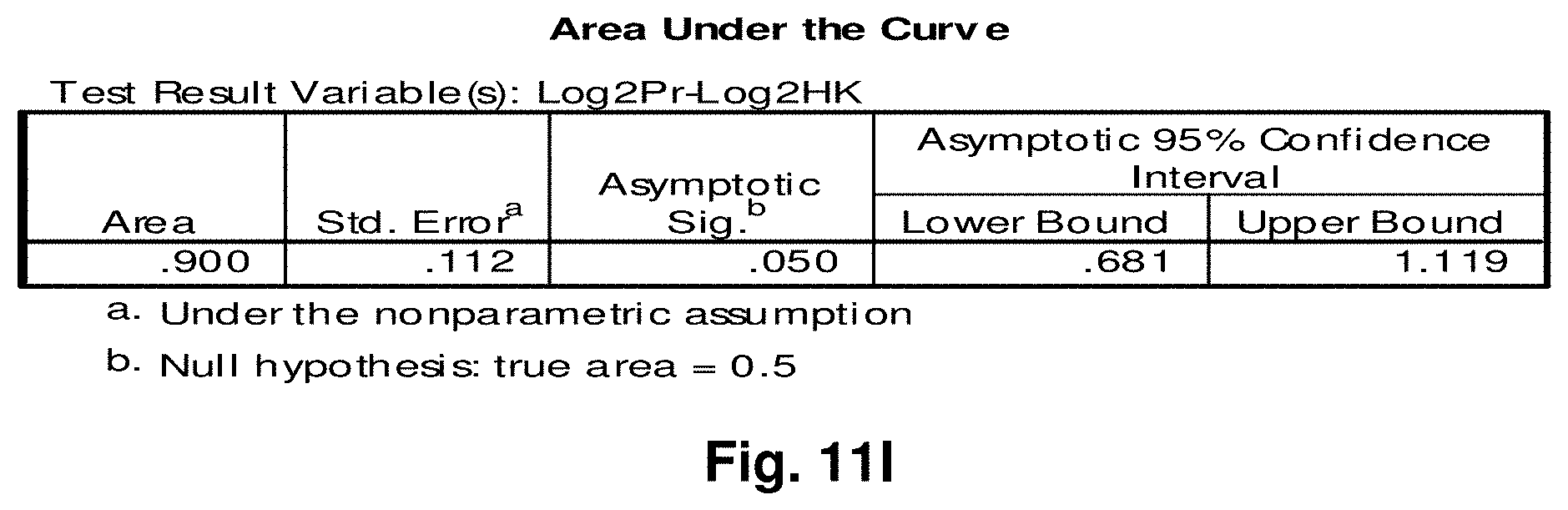
D00102

D00103

D00104
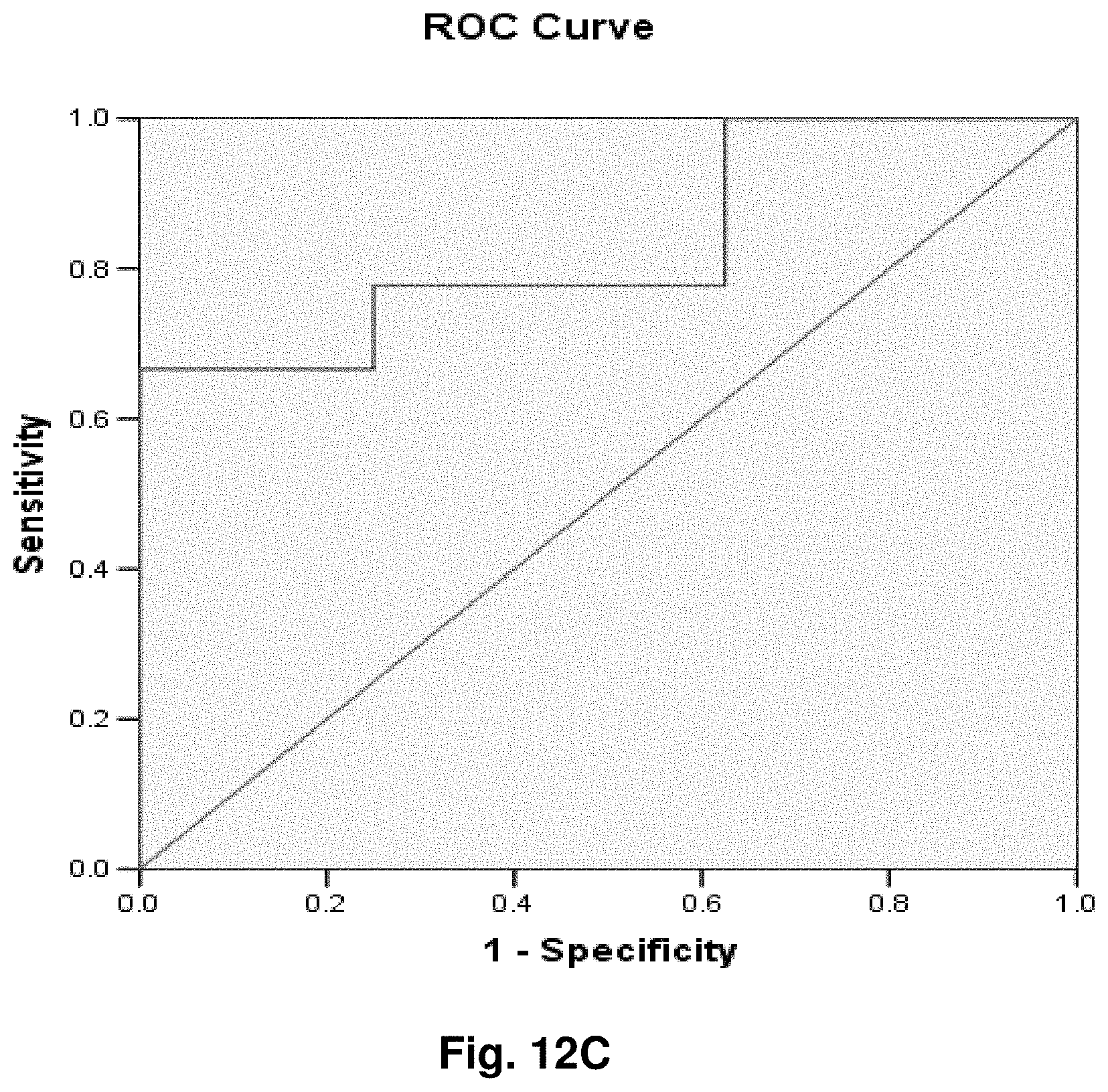
D00105

D00106
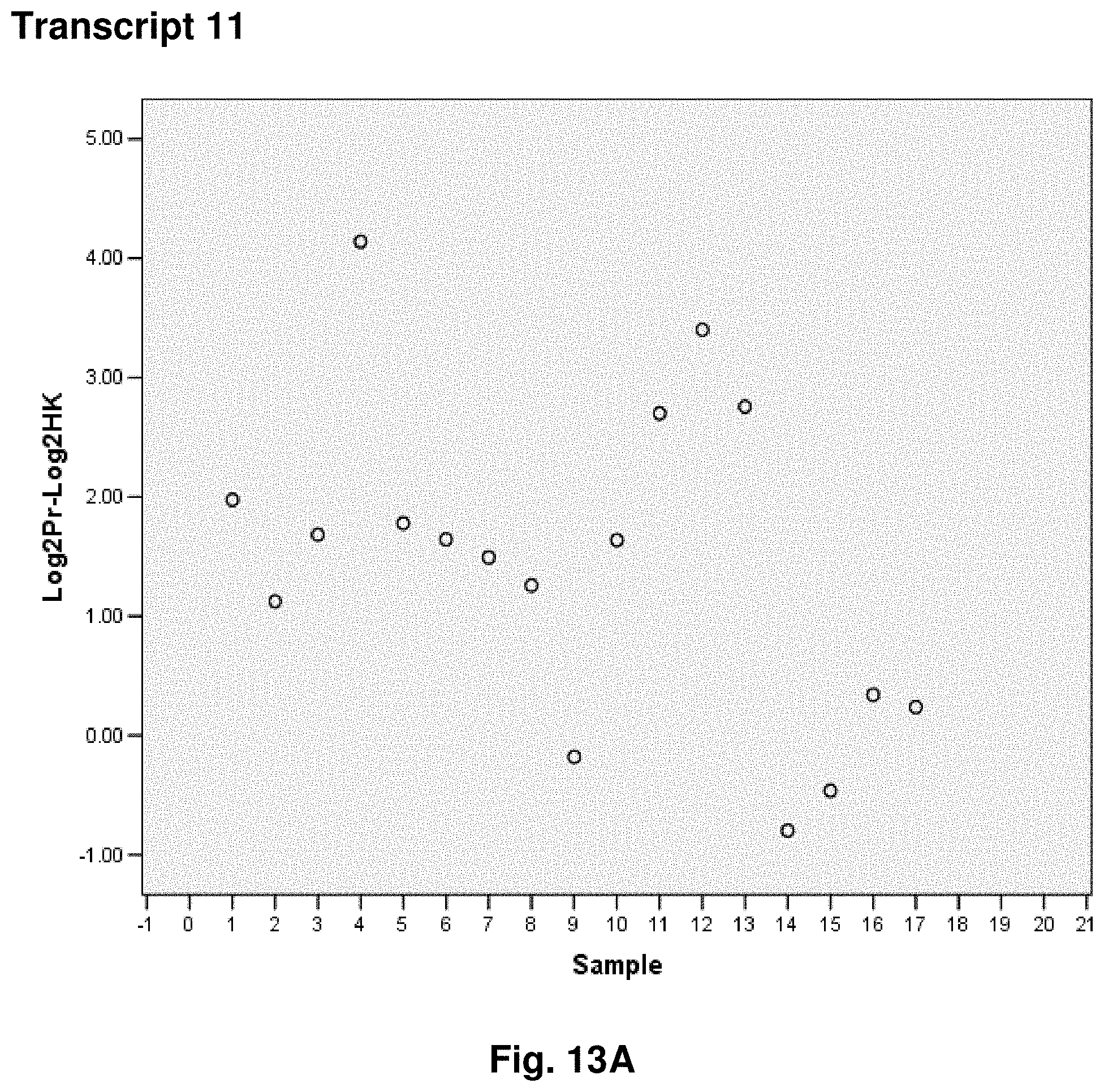
D00107
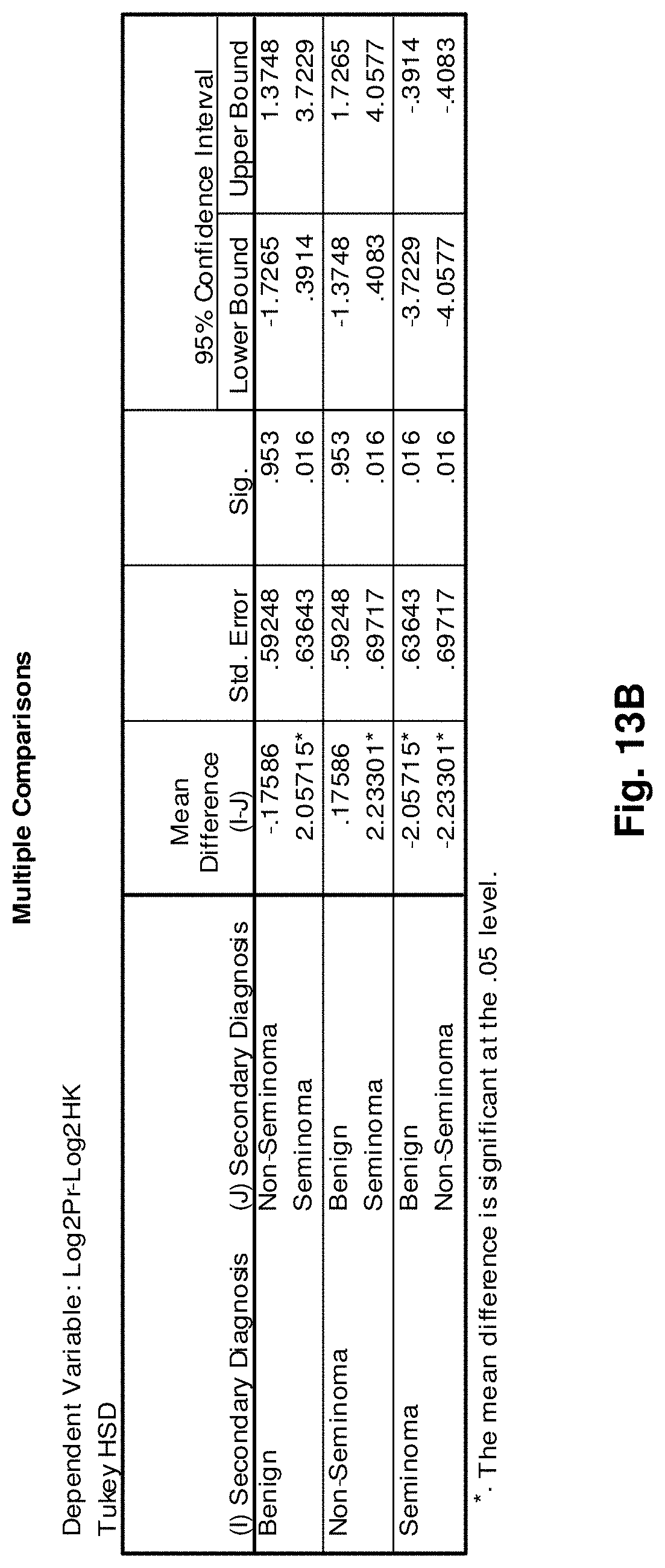
D00108

D00109

D00110

D00111
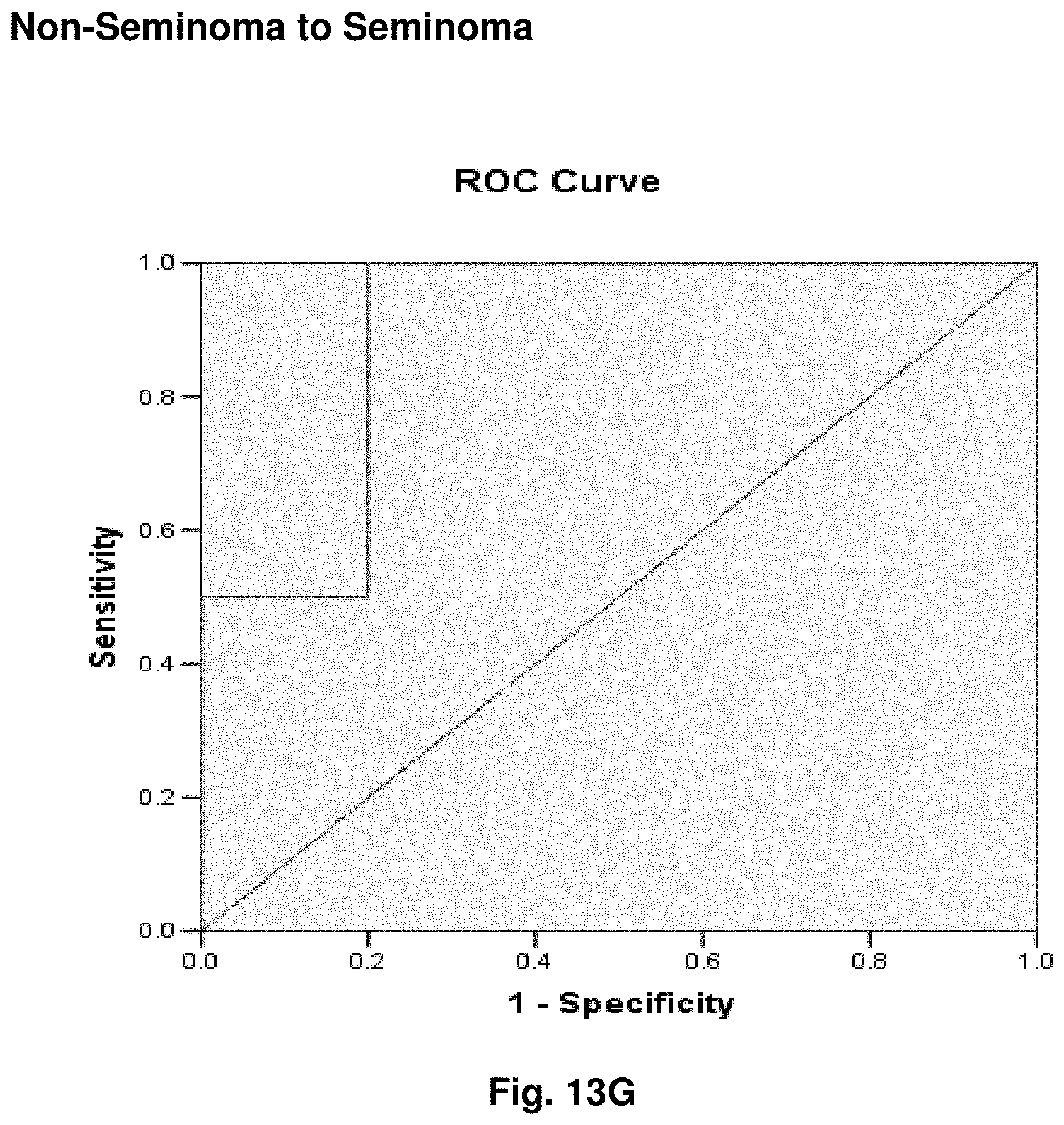
D00112

D00113
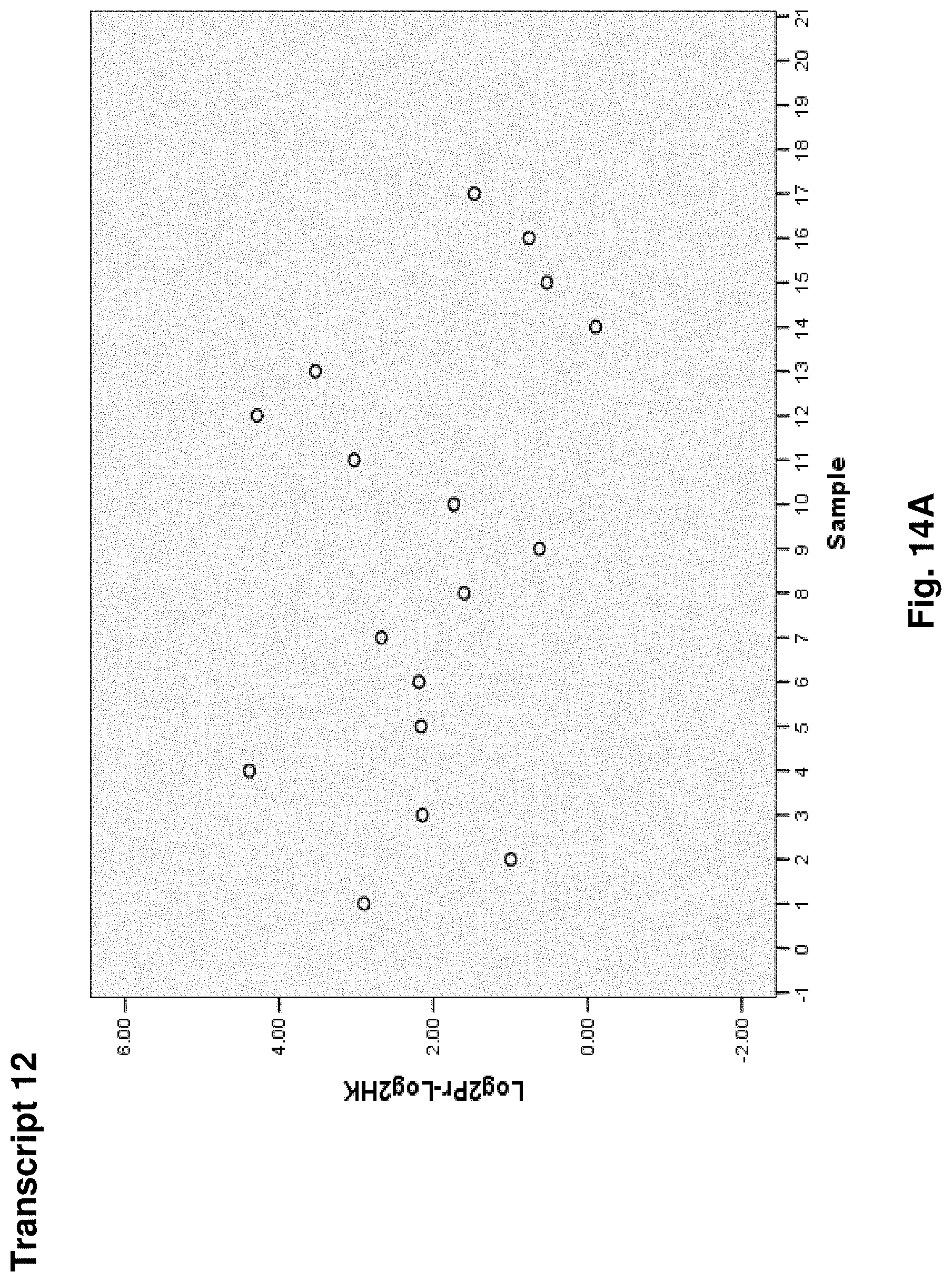
D00114

D00115
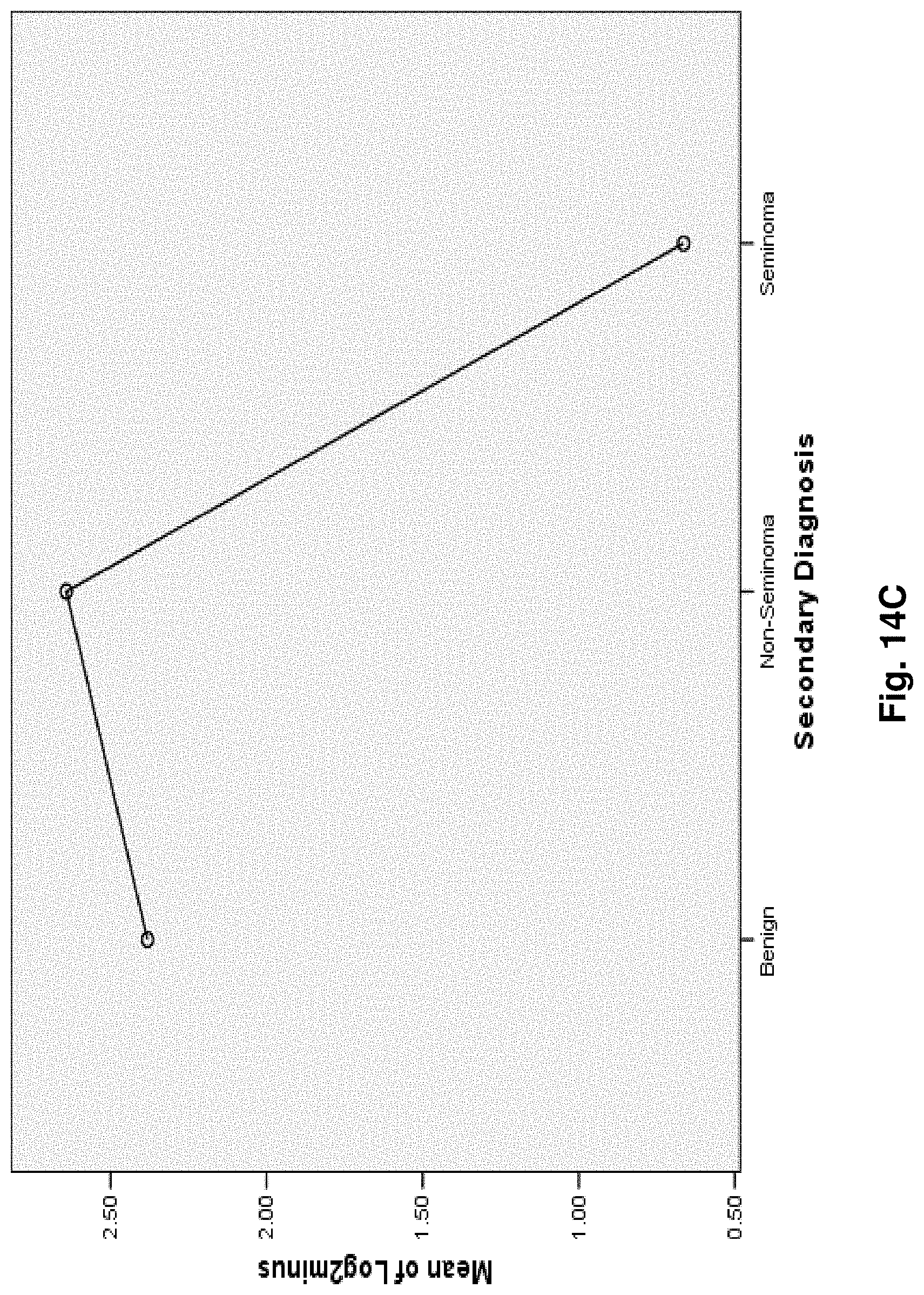
D00116

D00117
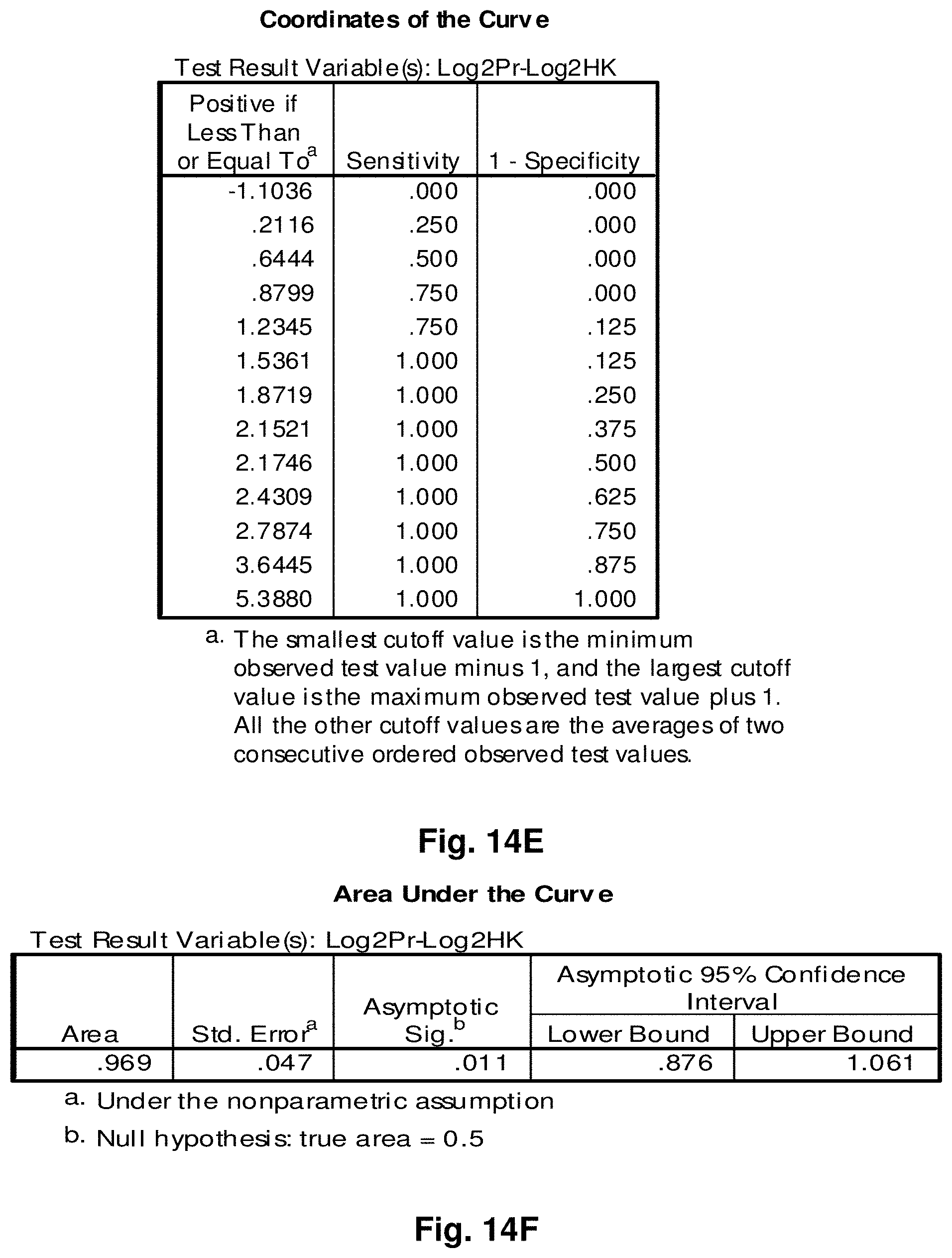
D00118

D00119
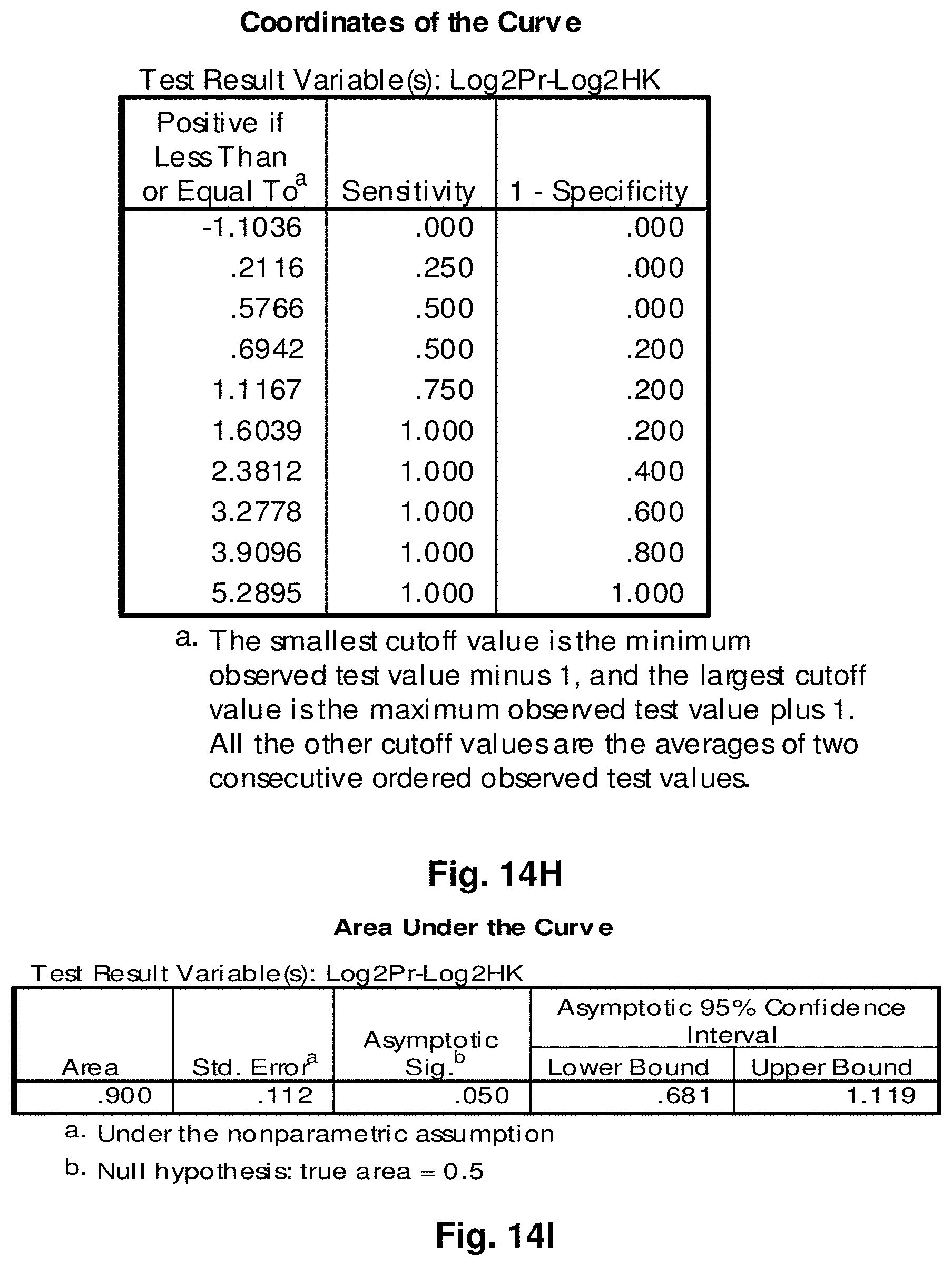
D00120
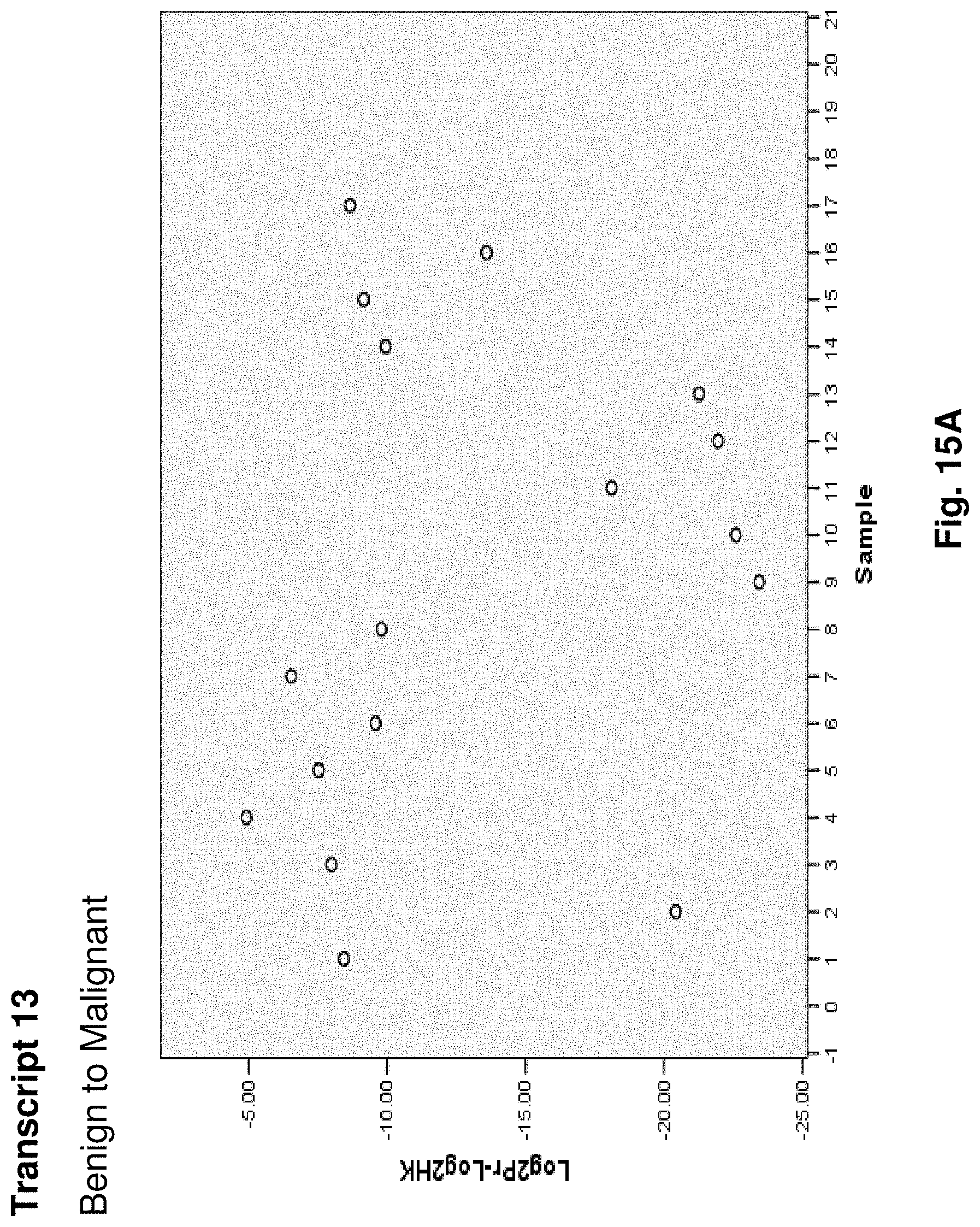
D00121

D00122
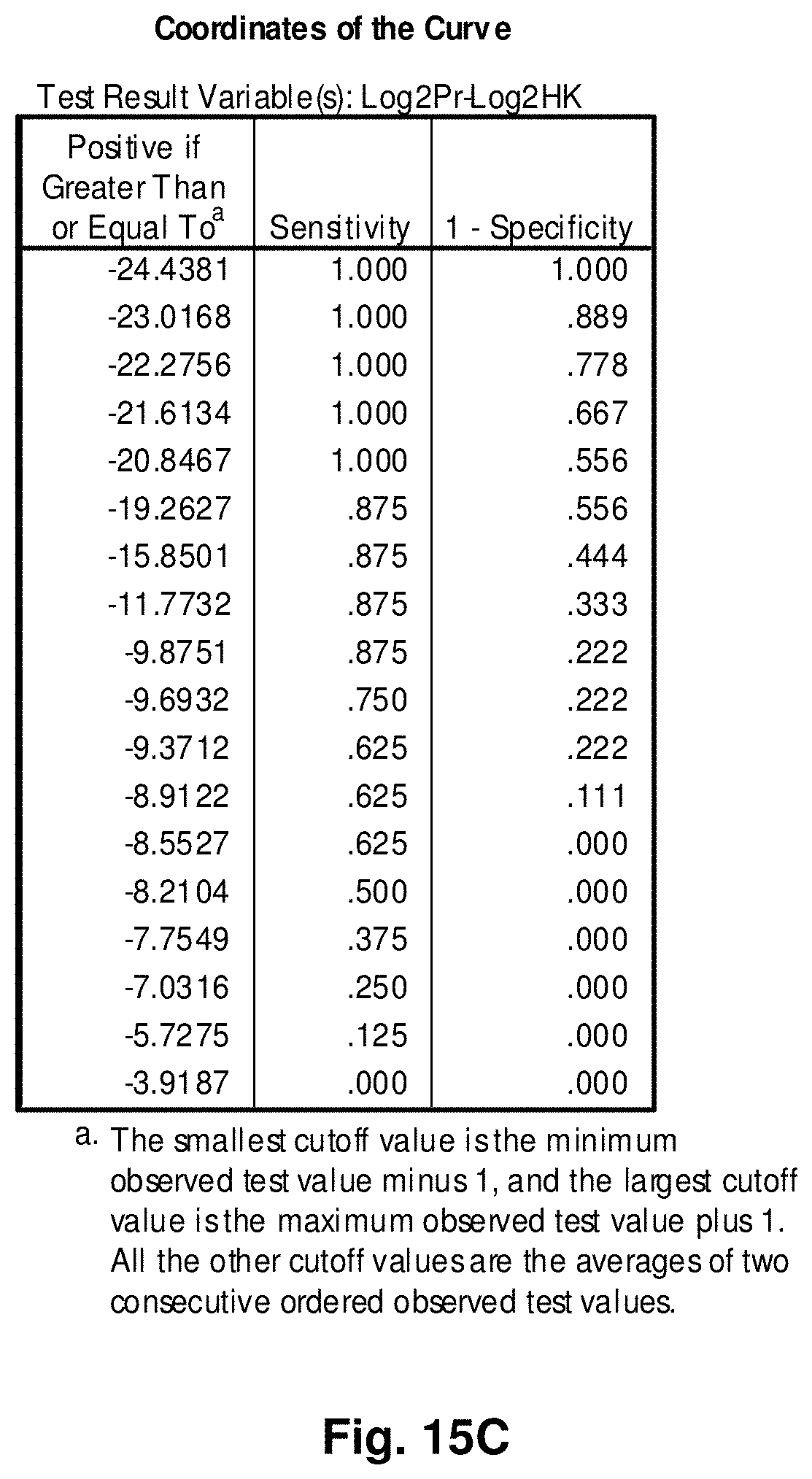
D00123
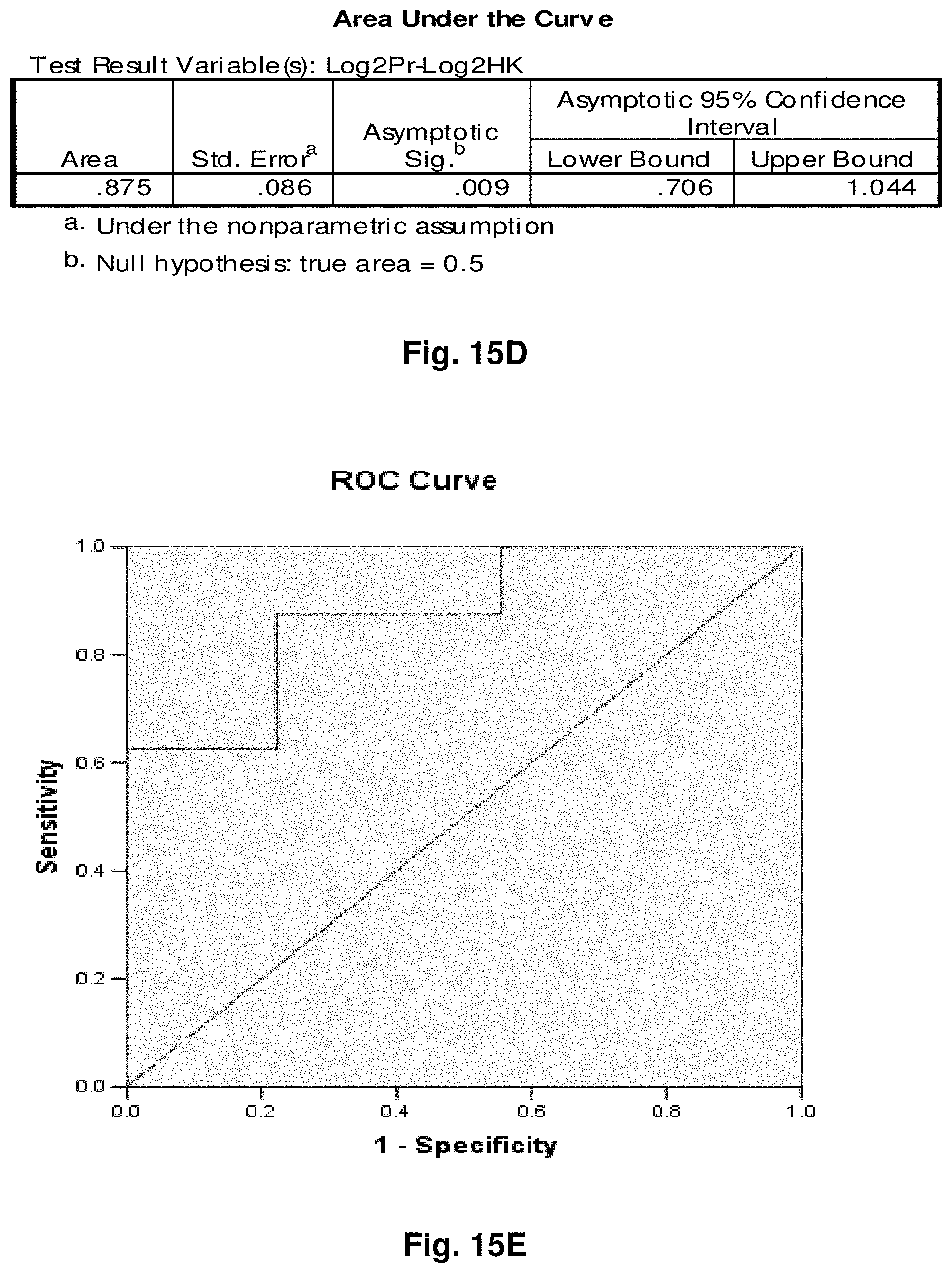
D00124

D00125

D00126
D00127
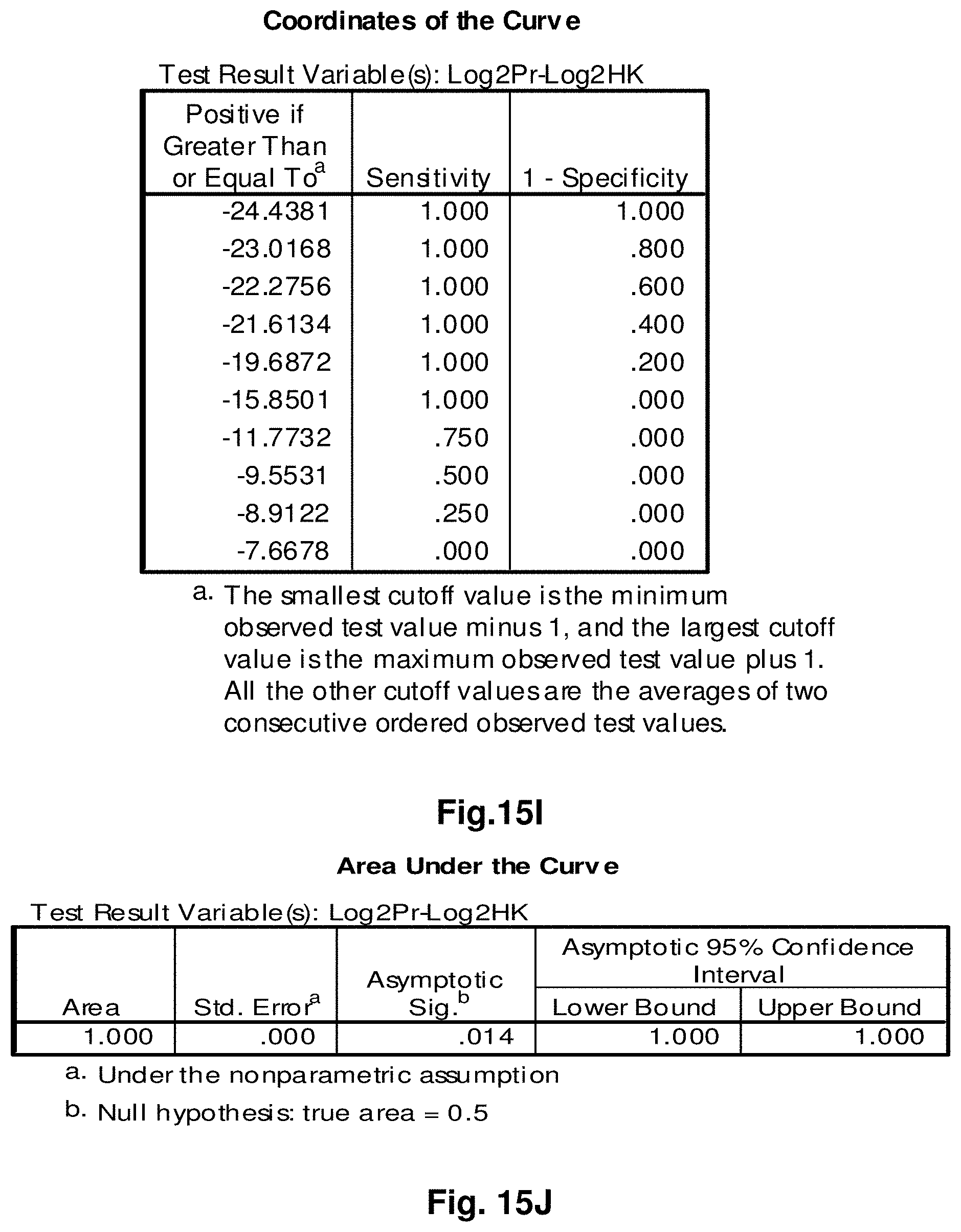
D00128
D00129
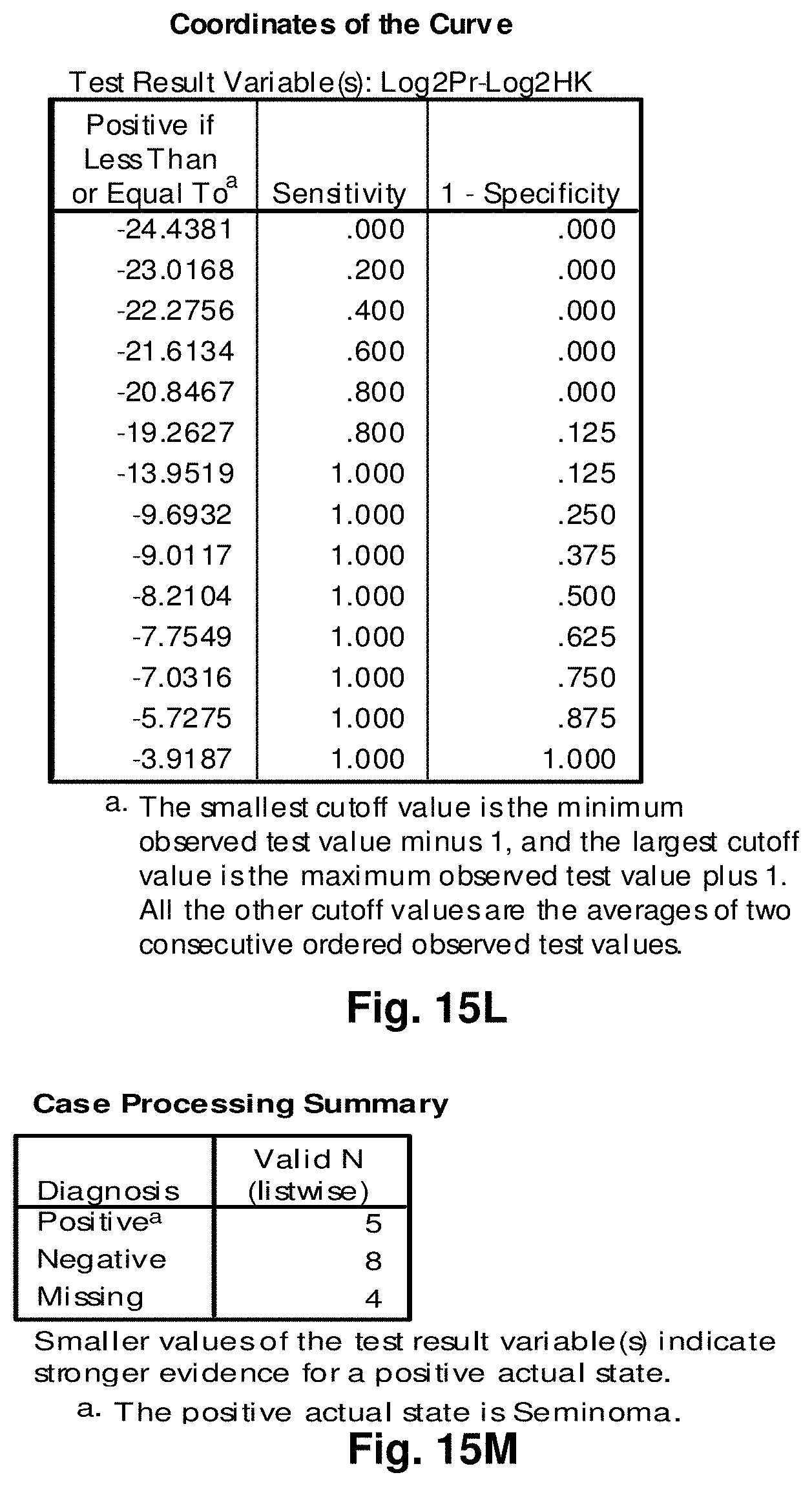
D00130

D00131
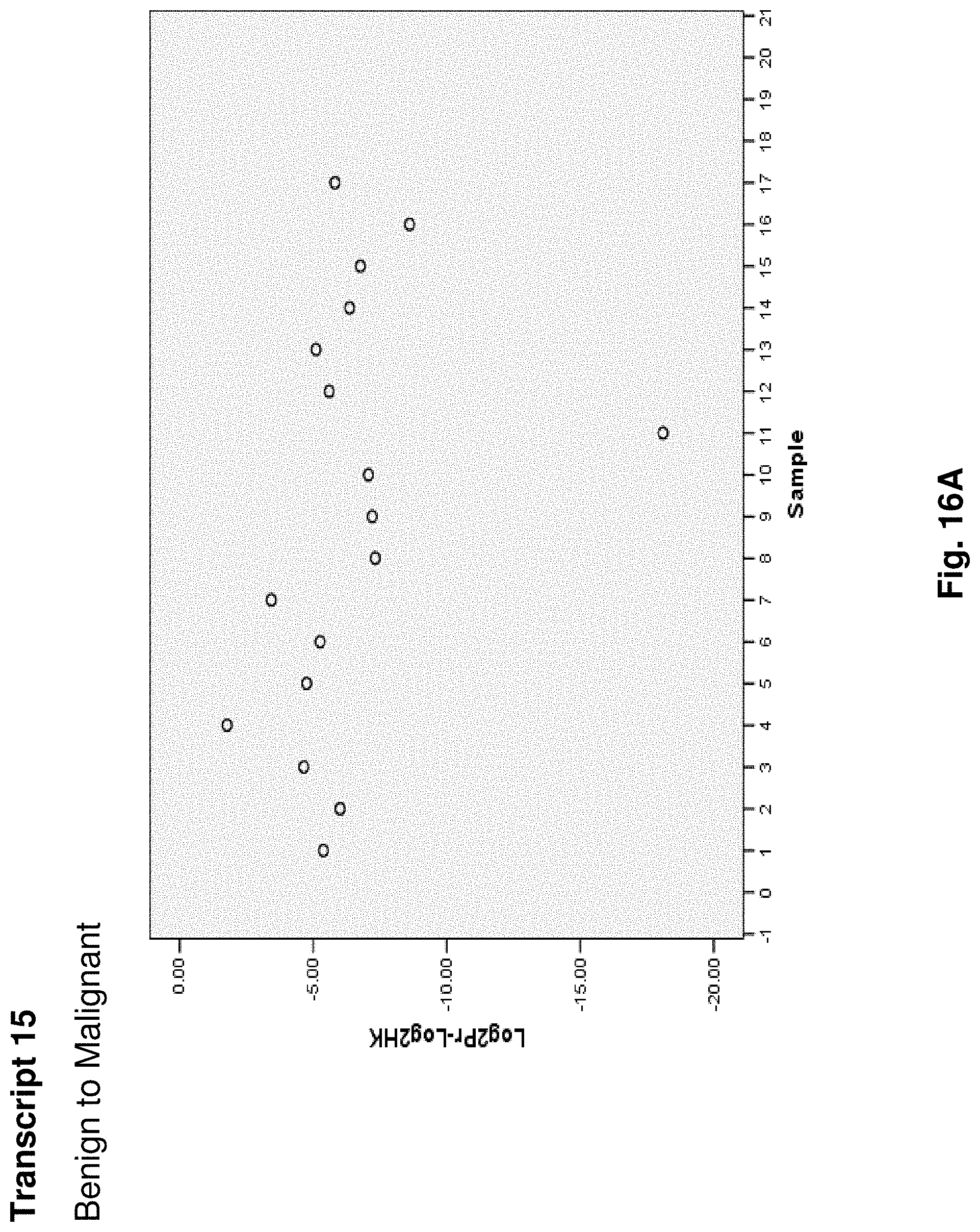
D00132
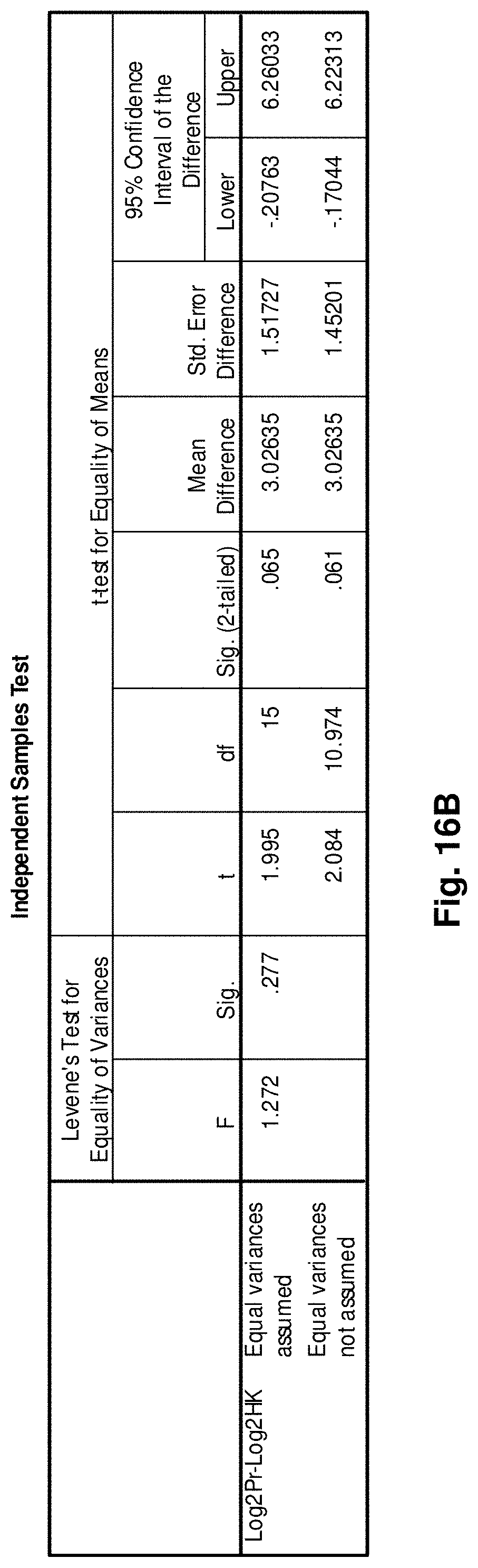
D00133
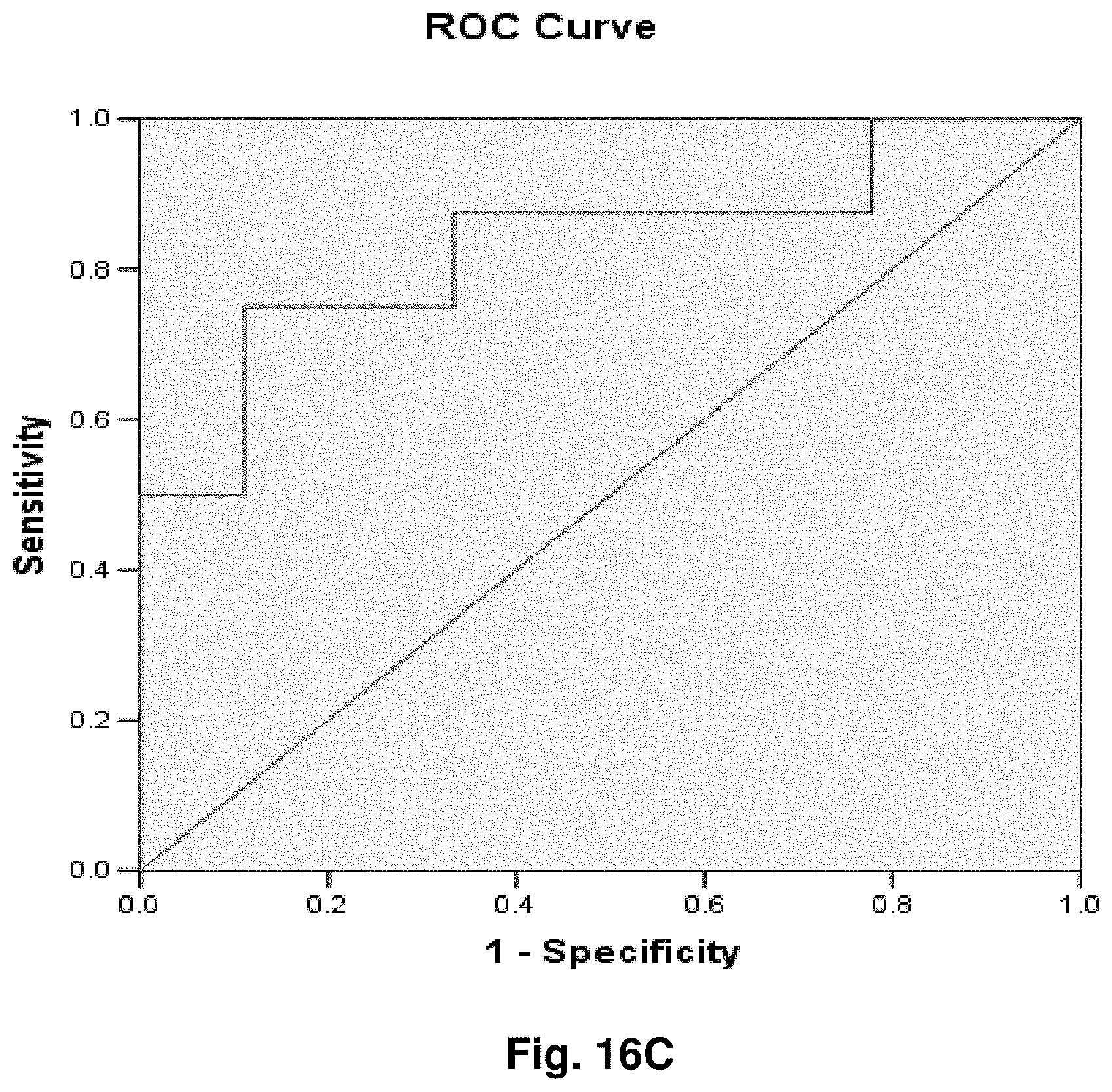
D00134

D00135

D00136
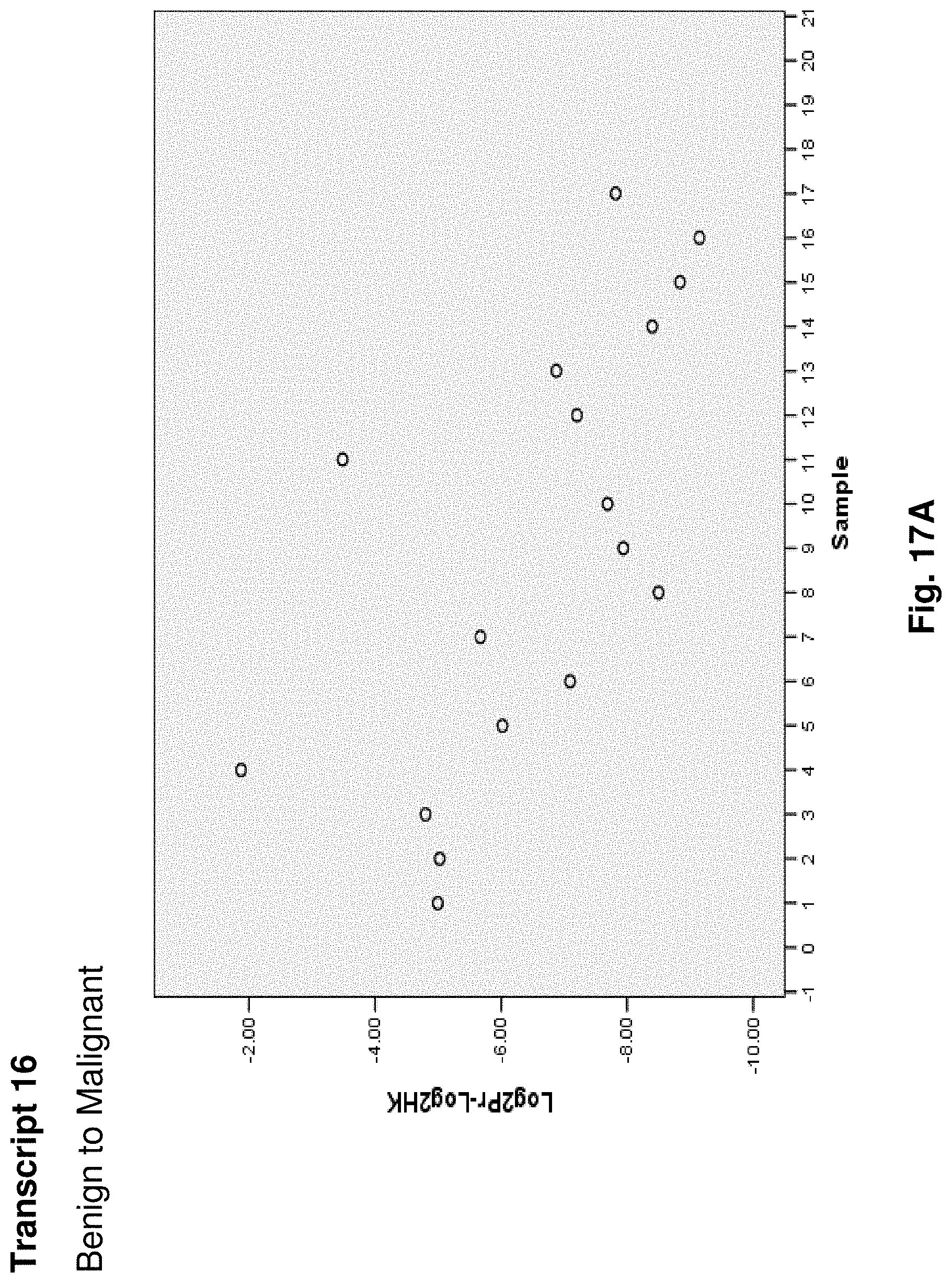
D00137

D00138

D00139

D00140

D00141

D00142

D00143

D00144

D00145
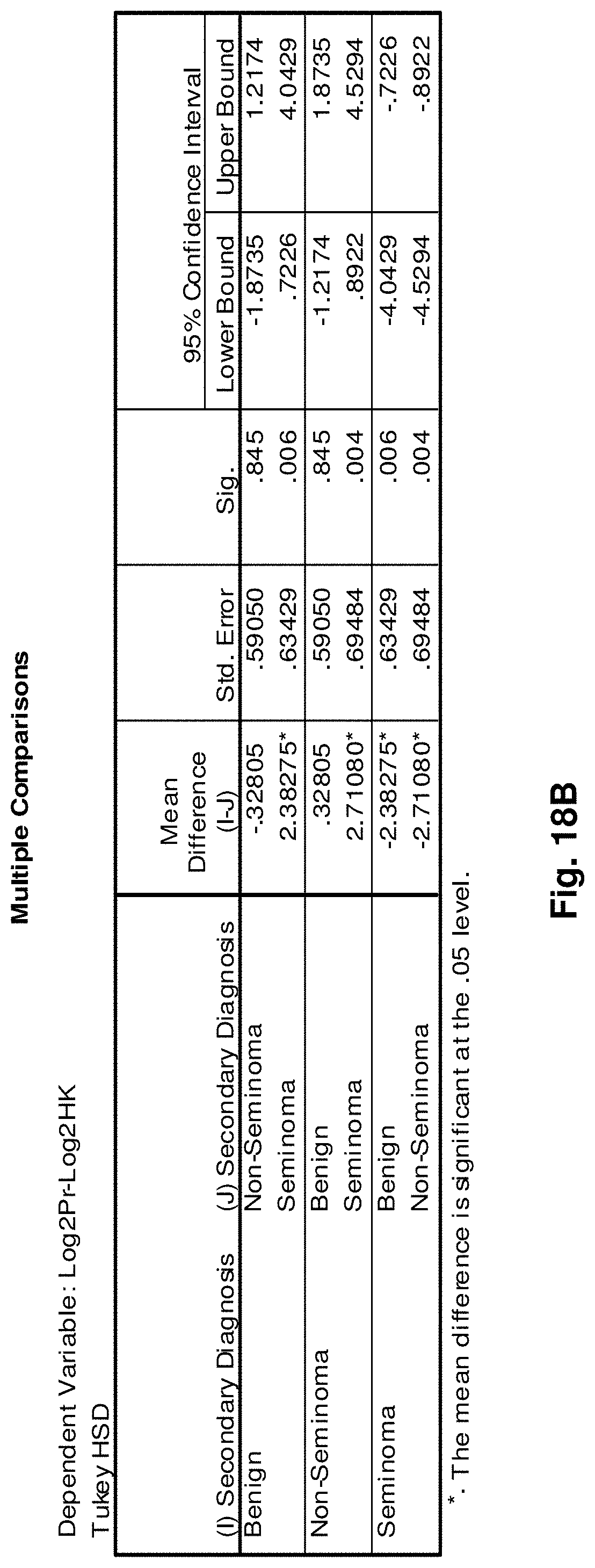
D00146

D00147

D00148

D00149
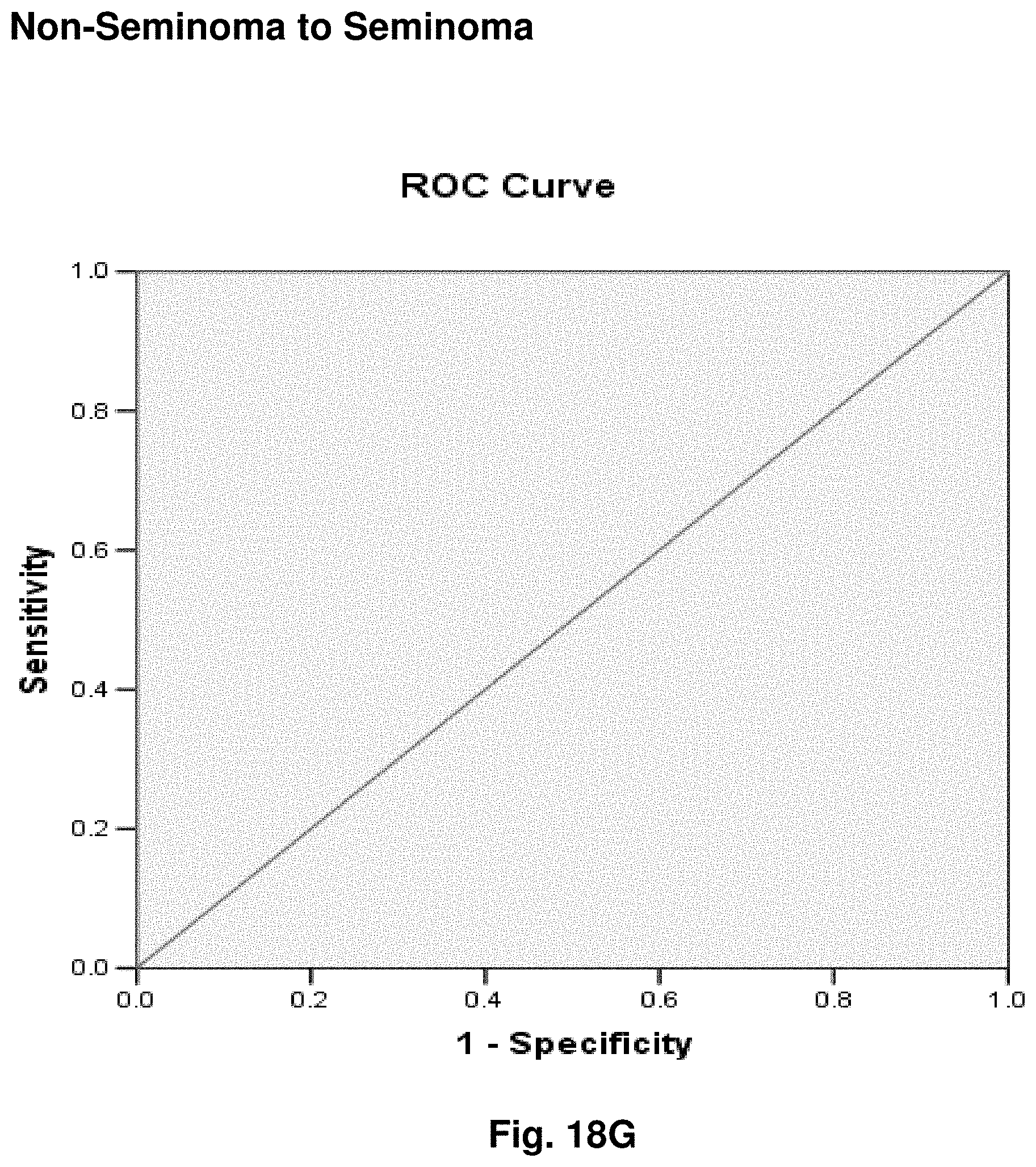
D00150

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.